code
stringlengths 2.5k
6.36M
| kind
stringclasses 2
values | parsed_code
stringlengths 0
404k
| quality_prob
float64 0
0.98
| learning_prob
float64 0.03
1
|
|---|---|---|---|---|
```
import pandas as pd
```
### Generate a brief statistic summary for corresponding data
* Sort daily gain/loss for January of 2018 and store the result back to a .csv file
```
FILE = r"C:\Users\pavan\Desktop\SP500 (1).csv"
data = pd.read_csv(FILE)
data.shape
data.columns
data.head()
data['Gain'] = data.Close - data.Open
data.head()
data['Date'] = pd.to_datetime(data.Date)
data.loc[data.Date.dt.year==2018,].sort_values(['Gain'],ascending=False).to_csv("./2018_Gain_Loss.csv",index=False)
```
* Find all of the daily gains reach 20% and above and display them
```
data['Gain%'] = (data['Gain'] /data['Open'])*100
data[data['Gain%']>=20]
```
* the highest daily gain and its date, the highest daily loss and its date,
```
data.sort_values(['Gain%'],ascending=False).head(1)[['Date','Gain%']]
data.sort_values(['Gain%'],ascending=True).head(1)[['Date','Gain%']]
```
* the most daily transaction volume and its date,
```
data.sort_values(['Volume'],ascending=False).head(1)[['Date','Volume']]
```
* a monthly report for year 2017-2018, which has monthly average open price, close price, transaction volume and gain/loss, and a query to find all of the months which have certain range of open prices
```
dataA = data.loc[(data.Date.dt.year==2018) | (data.Date.dt.year==2017),]
dataA['Month'] = dataA.Date.dt.month
dataA.groupby(['Month']).agg({'Open': "mean", 'Close': "mean","Volume":'mean',"Gain":'mean'}).reset_index()
```
* a yearly report which has annual average open price, close price, transaction volume and gain/loss from 1950 to 2018, and the most profitable year
```
data['Year'] = data.Date.dt.year
data.groupby(['Year']).agg({'Open': "mean", 'Close': "mean","Volume":'mean',"Gain":'mean'}).reset_index()
```
* a every other five year report which has every five year average open price, close price, transaction volume and gain/loss from 1950 to 2018, and the most profitable five year,
```
cnt=0
start=end=1950
while end<2018:
end=start+5
print(start,end)
data.loc[(data.Year>=start) & (data.Year<=end),'Bin']=str(start)+'-'+str(end)
start=end
cnt=cnt+1
data.groupby(['Bin']).agg({'Open': "mean", 'Close': "mean","Volume":'mean',"Gain":'mean'}).reset_index()
FILE=r"C:\Users\pavan\Desktop\Sacramentorealestatetransactions (2).csv"
dataB = pd.read_csv(FILE)
```
* Regroup the data first by city name, then by type
```
dataB.head()
dataB_agg = dataB.groupby(['city','type']).agg({'price': "mean"}).reset_index()
dataB_agg
# higest
dataB_agg[dataB_agg.price == max(dataB_agg.price)]
#lowest
dataB_agg[dataB_agg.price == min(dataB_agg.price)]
#median
dataB_agg.median()
dataC_agg = dataB.groupby(['zip','type']).agg({'price': "mean"}).reset_index()
dataC_agg
```
|
github_jupyter
|
import pandas as pd
FILE = r"C:\Users\pavan\Desktop\SP500 (1).csv"
data = pd.read_csv(FILE)
data.shape
data.columns
data.head()
data['Gain'] = data.Close - data.Open
data.head()
data['Date'] = pd.to_datetime(data.Date)
data.loc[data.Date.dt.year==2018,].sort_values(['Gain'],ascending=False).to_csv("./2018_Gain_Loss.csv",index=False)
data['Gain%'] = (data['Gain'] /data['Open'])*100
data[data['Gain%']>=20]
data.sort_values(['Gain%'],ascending=False).head(1)[['Date','Gain%']]
data.sort_values(['Gain%'],ascending=True).head(1)[['Date','Gain%']]
data.sort_values(['Volume'],ascending=False).head(1)[['Date','Volume']]
dataA = data.loc[(data.Date.dt.year==2018) | (data.Date.dt.year==2017),]
dataA['Month'] = dataA.Date.dt.month
dataA.groupby(['Month']).agg({'Open': "mean", 'Close': "mean","Volume":'mean',"Gain":'mean'}).reset_index()
data['Year'] = data.Date.dt.year
data.groupby(['Year']).agg({'Open': "mean", 'Close': "mean","Volume":'mean',"Gain":'mean'}).reset_index()
cnt=0
start=end=1950
while end<2018:
end=start+5
print(start,end)
data.loc[(data.Year>=start) & (data.Year<=end),'Bin']=str(start)+'-'+str(end)
start=end
cnt=cnt+1
data.groupby(['Bin']).agg({'Open': "mean", 'Close': "mean","Volume":'mean',"Gain":'mean'}).reset_index()
FILE=r"C:\Users\pavan\Desktop\Sacramentorealestatetransactions (2).csv"
dataB = pd.read_csv(FILE)
dataB.head()
dataB_agg = dataB.groupby(['city','type']).agg({'price': "mean"}).reset_index()
dataB_agg
# higest
dataB_agg[dataB_agg.price == max(dataB_agg.price)]
#lowest
dataB_agg[dataB_agg.price == min(dataB_agg.price)]
#median
dataB_agg.median()
dataC_agg = dataB.groupby(['zip','type']).agg({'price': "mean"}).reset_index()
dataC_agg
| 0.113801 | 0.90291 |
```
import keras
keras.__version__
```
# 5.2 - Using convnets with small datasets
This notebook contains the code sample found in Chapter 5, Section 2 of [Deep Learning with Python](https://www.manning.com/books/deep-learning-with-python?a_aid=keras&a_bid=76564dff). Note that the original text features far more content, in particular further explanations and figures: in this notebook, you will only find source code and related comments.
## Training a convnet from scratch on a small dataset
Having to train an image classification model using only very little data is a common situation, which you likely encounter yourself in
practice if you ever do computer vision in a professional context.
Having "few" samples can mean anywhere from a few hundreds to a few tens of thousands of images. As a practical example, we will focus on
classifying images as "dogs" or "cats", in a dataset containing 4000 pictures of cats and dogs (2000 cats, 2000 dogs). We will use 2000
pictures for training, 1000 for validation, and finally 1000 for testing.
In this section, we will review one basic strategy to tackle this problem: training a new model from scratch on what little data we have. We
will start by naively training a small convnet on our 2000 training samples, without any regularization, to set a baseline for what can be
achieved. This will get us to a classification accuracy of 71%. At that point, our main issue will be overfitting. Then we will introduce
*data augmentation*, a powerful technique for mitigating overfitting in computer vision. By leveraging data augmentation, we will improve
our network to reach an accuracy of 82%.
In the next section, we will review two more essential techniques for applying deep learning to small datasets: *doing feature extraction
with a pre-trained network* (this will get us to an accuracy of 90% to 93%), and *fine-tuning a pre-trained network* (this will get us to
our final accuracy of 95%). Together, these three strategies -- training a small model from scratch, doing feature extracting using a
pre-trained model, and fine-tuning a pre-trained model -- will constitute your future toolbox for tackling the problem of doing computer
vision with small datasets.
## The relevance of deep learning for small-data problems
You will sometimes hear that deep learning only works when lots of data is available. This is in part a valid point: one fundamental
characteristic of deep learning is that it is able to find interesting features in the training data on its own, without any need for manual
feature engineering, and this can only be achieved when lots of training examples are available. This is especially true for problems where
the input samples are very high-dimensional, like images.
However, what constitutes "lots" of samples is relative -- relative to the size and depth of the network you are trying to train, for
starters. It isn't possible to train a convnet to solve a complex problem with just a few tens of samples, but a few hundreds can
potentially suffice if the model is small and well-regularized and if the task is simple.
Because convnets learn local, translation-invariant features, they are very
data-efficient on perceptual problems. Training a convnet from scratch on a very small image dataset will still yield reasonable results
despite a relative lack of data, without the need for any custom feature engineering. You will see this in action in this section.
But what's more, deep learning models are by nature highly repurposable: you can take, say, an image classification or speech-to-text model
trained on a large-scale dataset then reuse it on a significantly different problem with only minor changes. Specifically, in the case of
computer vision, many pre-trained models (usually trained on the ImageNet dataset) are now publicly available for download and can be used
to bootstrap powerful vision models out of very little data. That's what we will do in the next section.
For now, let's get started by getting our hands on the data.
## Downloading the data
The cats vs. dogs dataset that we will use isn't packaged with Keras. It was made available by Kaggle.com as part of a computer vision
competition in late 2013, back when convnets weren't quite mainstream. You can download the original dataset at:
`https://www.kaggle.com/c/dogs-vs-cats/data` (you will need to create a Kaggle account if you don't already have one -- don't worry, the
process is painless).
The pictures are medium-resolution color JPEGs. They look like this:

Unsurprisingly, the cats vs. dogs Kaggle competition in 2013 was won by entrants who used convnets. The best entries could achieve up to
95% accuracy. In our own example, we will get fairly close to this accuracy (in the next section), even though we will be training our
models on less than 10% of the data that was available to the competitors.
This original dataset contains 25,000 images of dogs and cats (12,500 from each class) and is 543MB large (compressed). After downloading
and uncompressing it, we will create a new dataset containing three subsets: a training set with 1000 samples of each class, a validation
set with 500 samples of each class, and finally a test set with 500 samples of each class.
Here are a few lines of code to do this:
```
import os, shutil
# The path to the directory where the original
# dataset was uncompressed
original_dataset_dir = '/Users/fchollet/Downloads/kaggle_original_data'
# The directory where we will
# store our smaller dataset
base_dir = '/Users/fchollet/Downloads/cats_and_dogs_small'
os.mkdir(base_dir)
# Directories for our training,
# validation and test splits
train_dir = os.path.join(base_dir, 'train')
os.mkdir(train_dir)
validation_dir = os.path.join(base_dir, 'validation')
os.mkdir(validation_dir)
test_dir = os.path.join(base_dir, 'test')
os.mkdir(test_dir)
# Directory with our training cat pictures
train_cats_dir = os.path.join(train_dir, 'cats')
os.mkdir(train_cats_dir)
# Directory with our training dog pictures
train_dogs_dir = os.path.join(train_dir, 'dogs')
os.mkdir(train_dogs_dir)
# Directory with our validation cat pictures
validation_cats_dir = os.path.join(validation_dir, 'cats')
os.mkdir(validation_cats_dir)
# Directory with our validation dog pictures
validation_dogs_dir = os.path.join(validation_dir, 'dogs')
os.mkdir(validation_dogs_dir)
# Directory with our validation cat pictures
test_cats_dir = os.path.join(test_dir, 'cats')
os.mkdir(test_cats_dir)
# Directory with our validation dog pictures
test_dogs_dir = os.path.join(test_dir, 'dogs')
os.mkdir(test_dogs_dir)
# Copy first 1000 cat images to train_cats_dir
fnames = ['cat.{}.jpg'.format(i) for i in range(1000)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(train_cats_dir, fname)
shutil.copyfile(src, dst)
# Copy next 500 cat images to validation_cats_dir
fnames = ['cat.{}.jpg'.format(i) for i in range(1000, 1500)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(validation_cats_dir, fname)
shutil.copyfile(src, dst)
# Copy next 500 cat images to test_cats_dir
fnames = ['cat.{}.jpg'.format(i) for i in range(1500, 2000)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(test_cats_dir, fname)
shutil.copyfile(src, dst)
# Copy first 1000 dog images to train_dogs_dir
fnames = ['dog.{}.jpg'.format(i) for i in range(1000)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(train_dogs_dir, fname)
shutil.copyfile(src, dst)
# Copy next 500 dog images to validation_dogs_dir
fnames = ['dog.{}.jpg'.format(i) for i in range(1000, 1500)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(validation_dogs_dir, fname)
shutil.copyfile(src, dst)
# Copy next 500 dog images to test_dogs_dir
fnames = ['dog.{}.jpg'.format(i) for i in range(1500, 2000)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(test_dogs_dir, fname)
shutil.copyfile(src, dst)
```
As a sanity check, let's count how many pictures we have in each training split (train/validation/test):
```
print('total training cat images:', len(os.listdir(train_cats_dir)))
print('total training dog images:', len(os.listdir(train_dogs_dir)))
print('total validation cat images:', len(os.listdir(validation_cats_dir)))
print('total validation dog images:', len(os.listdir(validation_dogs_dir)))
print('total test cat images:', len(os.listdir(test_cats_dir)))
print('total test dog images:', len(os.listdir(test_dogs_dir)))
```
So we have indeed 2000 training images, and then 1000 validation images and 1000 test images. In each split, there is the same number of
samples from each class: this is a balanced binary classification problem, which means that classification accuracy will be an appropriate
measure of success.
## Building our network
We've already built a small convnet for MNIST in the previous example, so you should be familiar with them. We will reuse the same
general structure: our convnet will be a stack of alternated `Conv2D` (with `relu` activation) and `MaxPooling2D` layers.
However, since we are dealing with bigger images and a more complex problem, we will make our network accordingly larger: it will have one
more `Conv2D` + `MaxPooling2D` stage. This serves both to augment the capacity of the network, and to further reduce the size of the
feature maps, so that they aren't overly large when we reach the `Flatten` layer. Here, since we start from inputs of size 150x150 (a
somewhat arbitrary choice), we end up with feature maps of size 7x7 right before the `Flatten` layer.
Note that the depth of the feature maps is progressively increasing in the network (from 32 to 128), while the size of the feature maps is
decreasing (from 148x148 to 7x7). This is a pattern that you will see in almost all convnets.
Since we are attacking a binary classification problem, we are ending the network with a single unit (a `Dense` layer of size 1) and a
`sigmoid` activation. This unit will encode the probability that the network is looking at one class or the other.
```
from keras import layers
from keras import models
model = models.Sequential()
model.add(layers.Conv2D(32, (3, 3), activation='relu',
input_shape=(150, 150, 3)))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(128, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(128, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Flatten())
model.add(layers.Dense(512, activation='relu'))
model.add(layers.Dense(1, activation='sigmoid'))
```
Let's take a look at how the dimensions of the feature maps change with every successive layer:
```
model.summary()
```
For our compilation step, we'll go with the `RMSprop` optimizer as usual. Since we ended our network with a single sigmoid unit, we will
use binary crossentropy as our loss (as a reminder, check out the table in Chapter 4, section 5 for a cheatsheet on what loss function to
use in various situations).
```
from keras import optimizers
model.compile(loss='binary_crossentropy',
optimizer=optimizers.RMSprop(lr=1e-4),
metrics=['acc'])
```
## Data preprocessing
As you already know by now, data should be formatted into appropriately pre-processed floating point tensors before being fed into our
network. Currently, our data sits on a drive as JPEG files, so the steps for getting it into our network are roughly:
* Read the picture files.
* Decode the JPEG content to RBG grids of pixels.
* Convert these into floating point tensors.
* Rescale the pixel values (between 0 and 255) to the [0, 1] interval (as you know, neural networks prefer to deal with small input values).
It may seem a bit daunting, but thankfully Keras has utilities to take care of these steps automatically. Keras has a module with image
processing helper tools, located at `keras.preprocessing.image`. In particular, it contains the class `ImageDataGenerator` which allows to
quickly set up Python generators that can automatically turn image files on disk into batches of pre-processed tensors. This is what we
will use here.
```
from keras.preprocessing.image import ImageDataGenerator
# All images will be rescaled by 1./255
train_datagen = ImageDataGenerator(rescale=1./255)
test_datagen = ImageDataGenerator(rescale=1./255)
train_generator = train_datagen.flow_from_directory(
# This is the target directory
train_dir,
# All images will be resized to 150x150
target_size=(150, 150),
batch_size=20,
# Since we use binary_crossentropy loss, we need binary labels
class_mode='binary')
validation_generator = test_datagen.flow_from_directory(
validation_dir,
target_size=(150, 150),
batch_size=20,
class_mode='binary')
```
Let's take a look at the output of one of these generators: it yields batches of 150x150 RGB images (shape `(20, 150, 150, 3)`) and binary
labels (shape `(20,)`). 20 is the number of samples in each batch (the batch size). Note that the generator yields these batches
indefinitely: it just loops endlessly over the images present in the target folder. For this reason, we need to `break` the iteration loop
at some point.
```
for data_batch, labels_batch in train_generator:
print('data batch shape:', data_batch.shape)
print('labels batch shape:', labels_batch.shape)
break
```
Let's fit our model to the data using the generator. We do it using the `fit_generator` method, the equivalent of `fit` for data generators
like ours. It expects as first argument a Python generator that will yield batches of inputs and targets indefinitely, like ours does.
Because the data is being generated endlessly, the generator needs to know example how many samples to draw from the generator before
declaring an epoch over. This is the role of the `steps_per_epoch` argument: after having drawn `steps_per_epoch` batches from the
generator, i.e. after having run for `steps_per_epoch` gradient descent steps, the fitting process will go to the next epoch. In our case,
batches are 20-sample large, so it will take 100 batches until we see our target of 2000 samples.
When using `fit_generator`, one may pass a `validation_data` argument, much like with the `fit` method. Importantly, this argument is
allowed to be a data generator itself, but it could be a tuple of Numpy arrays as well. If you pass a generator as `validation_data`, then
this generator is expected to yield batches of validation data endlessly, and thus you should also specify the `validation_steps` argument,
which tells the process how many batches to draw from the validation generator for evaluation.
```
history = model.fit_generator(
train_generator,
steps_per_epoch=100,
epochs=30,
validation_data=validation_generator,
validation_steps=50)
```
It is good practice to always save your models after training:
```
model.save('cats_and_dogs_small_1.h5')
```
Let's plot the loss and accuracy of the model over the training and validation data during training:
```
import matplotlib.pyplot as plt
acc = history.history['acc']
val_acc = history.history['val_acc']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(len(acc))
plt.plot(epochs, acc, 'bo', label='Training acc')
plt.plot(epochs, val_acc, 'b', label='Validation acc')
plt.title('Training and validation accuracy')
plt.legend()
plt.figure()
plt.plot(epochs, loss, 'bo', label='Training loss')
plt.plot(epochs, val_loss, 'b', label='Validation loss')
plt.title('Training and validation loss')
plt.legend()
plt.show()
```
These plots are characteristic of overfitting. Our training accuracy increases linearly over time, until it reaches nearly 100%, while our
validation accuracy stalls at 70-72%. Our validation loss reaches its minimum after only five epochs then stalls, while the training loss
keeps decreasing linearly until it reaches nearly 0.
Because we only have relatively few training samples (2000), overfitting is going to be our number one concern. You already know about a
number of techniques that can help mitigate overfitting, such as dropout and weight decay (L2 regularization). We are now going to
introduce a new one, specific to computer vision, and used almost universally when processing images with deep learning models: *data
augmentation*.
## Using data augmentation
Overfitting is caused by having too few samples to learn from, rendering us unable to train a model able to generalize to new data.
Given infinite data, our model would be exposed to every possible aspect of the data distribution at hand: we would never overfit. Data
augmentation takes the approach of generating more training data from existing training samples, by "augmenting" the samples via a number
of random transformations that yield believable-looking images. The goal is that at training time, our model would never see the exact same
picture twice. This helps the model get exposed to more aspects of the data and generalize better.
In Keras, this can be done by configuring a number of random transformations to be performed on the images read by our `ImageDataGenerator`
instance. Let's get started with an example:
```
datagen = ImageDataGenerator(
rotation_range=40,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
fill_mode='nearest')
```
These are just a few of the options available (for more, see the Keras documentation). Let's quickly go over what we just wrote:
* `rotation_range` is a value in degrees (0-180), a range within which to randomly rotate pictures.
* `width_shift` and `height_shift` are ranges (as a fraction of total width or height) within which to randomly translate pictures
vertically or horizontally.
* `shear_range` is for randomly applying shearing transformations.
* `zoom_range` is for randomly zooming inside pictures.
* `horizontal_flip` is for randomly flipping half of the images horizontally -- relevant when there are no assumptions of horizontal
asymmetry (e.g. real-world pictures).
* `fill_mode` is the strategy used for filling in newly created pixels, which can appear after a rotation or a width/height shift.
Let's take a look at our augmented images:
```
# This is module with image preprocessing utilities
from keras.preprocessing import image
fnames = [os.path.join(train_cats_dir, fname) for fname in os.listdir(train_cats_dir)]
# We pick one image to "augment"
img_path = fnames[3]
# Read the image and resize it
img = image.load_img(img_path, target_size=(150, 150))
# Convert it to a Numpy array with shape (150, 150, 3)
x = image.img_to_array(img)
# Reshape it to (1, 150, 150, 3)
x = x.reshape((1,) + x.shape)
# The .flow() command below generates batches of randomly transformed images.
# It will loop indefinitely, so we need to `break` the loop at some point!
i = 0
for batch in datagen.flow(x, batch_size=1):
plt.figure(i)
imgplot = plt.imshow(image.array_to_img(batch[0]))
i += 1
if i % 4 == 0:
break
plt.show()
```
If we train a new network using this data augmentation configuration, our network will never see twice the same input. However, the inputs
that it sees are still heavily intercorrelated, since they come from a small number of original images -- we cannot produce new information,
we can only remix existing information. As such, this might not be quite enough to completely get rid of overfitting. To further fight
overfitting, we will also add a Dropout layer to our model, right before the densely-connected classifier:
```
model = models.Sequential()
model.add(layers.Conv2D(32, (3, 3), activation='relu',
input_shape=(150, 150, 3)))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(128, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(128, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Flatten())
model.add(layers.Dropout(0.5))
model.add(layers.Dense(512, activation='relu'))
model.add(layers.Dense(1, activation='sigmoid'))
model.compile(loss='binary_crossentropy',
optimizer=optimizers.RMSprop(lr=1e-4),
metrics=['acc'])
```
Let's train our network using data augmentation and dropout:
```
train_datagen = ImageDataGenerator(
rescale=1./255,
rotation_range=40,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,)
# Note that the validation data should not be augmented!
test_datagen = ImageDataGenerator(rescale=1./255)
train_generator = train_datagen.flow_from_directory(
# This is the target directory
train_dir,
# All images will be resized to 150x150
target_size=(150, 150),
batch_size=32,
# Since we use binary_crossentropy loss, we need binary labels
class_mode='binary')
validation_generator = test_datagen.flow_from_directory(
validation_dir,
target_size=(150, 150),
batch_size=32,
class_mode='binary')
history = model.fit_generator(
train_generator,
steps_per_epoch=100,
epochs=100,
validation_data=validation_generator,
validation_steps=50)
```
Let's save our model -- we will be using it in the section on convnet visualization.
```
model.save('cats_and_dogs_small_2.h5')
```
Let's plot our results again:
```
acc = history.history['acc']
val_acc = history.history['val_acc']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(len(acc))
plt.plot(epochs, acc, 'bo', label='Training acc')
plt.plot(epochs, val_acc, 'b', label='Validation acc')
plt.title('Training and validation accuracy')
plt.legend()
plt.figure()
plt.plot(epochs, loss, 'bo', label='Training loss')
plt.plot(epochs, val_loss, 'b', label='Validation loss')
plt.title('Training and validation loss')
plt.legend()
plt.show()
```
Thanks to data augmentation and dropout, we are no longer overfitting: the training curves are rather closely tracking the validation
curves. We are now able to reach an accuracy of 82%, a 15% relative improvement over the non-regularized model.
By leveraging regularization techniques even further and by tuning the network's parameters (such as the number of filters per convolution
layer, or the number of layers in the network), we may be able to get an even better accuracy, likely up to 86-87%. However, it would prove
very difficult to go any higher just by training our own convnet from scratch, simply because we have so little data to work with. As a
next step to improve our accuracy on this problem, we will have to leverage a pre-trained model, which will be the focus of the next two
sections.
|
github_jupyter
|
import keras
keras.__version__
import os, shutil
# The path to the directory where the original
# dataset was uncompressed
original_dataset_dir = '/Users/fchollet/Downloads/kaggle_original_data'
# The directory where we will
# store our smaller dataset
base_dir = '/Users/fchollet/Downloads/cats_and_dogs_small'
os.mkdir(base_dir)
# Directories for our training,
# validation and test splits
train_dir = os.path.join(base_dir, 'train')
os.mkdir(train_dir)
validation_dir = os.path.join(base_dir, 'validation')
os.mkdir(validation_dir)
test_dir = os.path.join(base_dir, 'test')
os.mkdir(test_dir)
# Directory with our training cat pictures
train_cats_dir = os.path.join(train_dir, 'cats')
os.mkdir(train_cats_dir)
# Directory with our training dog pictures
train_dogs_dir = os.path.join(train_dir, 'dogs')
os.mkdir(train_dogs_dir)
# Directory with our validation cat pictures
validation_cats_dir = os.path.join(validation_dir, 'cats')
os.mkdir(validation_cats_dir)
# Directory with our validation dog pictures
validation_dogs_dir = os.path.join(validation_dir, 'dogs')
os.mkdir(validation_dogs_dir)
# Directory with our validation cat pictures
test_cats_dir = os.path.join(test_dir, 'cats')
os.mkdir(test_cats_dir)
# Directory with our validation dog pictures
test_dogs_dir = os.path.join(test_dir, 'dogs')
os.mkdir(test_dogs_dir)
# Copy first 1000 cat images to train_cats_dir
fnames = ['cat.{}.jpg'.format(i) for i in range(1000)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(train_cats_dir, fname)
shutil.copyfile(src, dst)
# Copy next 500 cat images to validation_cats_dir
fnames = ['cat.{}.jpg'.format(i) for i in range(1000, 1500)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(validation_cats_dir, fname)
shutil.copyfile(src, dst)
# Copy next 500 cat images to test_cats_dir
fnames = ['cat.{}.jpg'.format(i) for i in range(1500, 2000)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(test_cats_dir, fname)
shutil.copyfile(src, dst)
# Copy first 1000 dog images to train_dogs_dir
fnames = ['dog.{}.jpg'.format(i) for i in range(1000)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(train_dogs_dir, fname)
shutil.copyfile(src, dst)
# Copy next 500 dog images to validation_dogs_dir
fnames = ['dog.{}.jpg'.format(i) for i in range(1000, 1500)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(validation_dogs_dir, fname)
shutil.copyfile(src, dst)
# Copy next 500 dog images to test_dogs_dir
fnames = ['dog.{}.jpg'.format(i) for i in range(1500, 2000)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(test_dogs_dir, fname)
shutil.copyfile(src, dst)
print('total training cat images:', len(os.listdir(train_cats_dir)))
print('total training dog images:', len(os.listdir(train_dogs_dir)))
print('total validation cat images:', len(os.listdir(validation_cats_dir)))
print('total validation dog images:', len(os.listdir(validation_dogs_dir)))
print('total test cat images:', len(os.listdir(test_cats_dir)))
print('total test dog images:', len(os.listdir(test_dogs_dir)))
from keras import layers
from keras import models
model = models.Sequential()
model.add(layers.Conv2D(32, (3, 3), activation='relu',
input_shape=(150, 150, 3)))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(128, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(128, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Flatten())
model.add(layers.Dense(512, activation='relu'))
model.add(layers.Dense(1, activation='sigmoid'))
model.summary()
from keras import optimizers
model.compile(loss='binary_crossentropy',
optimizer=optimizers.RMSprop(lr=1e-4),
metrics=['acc'])
from keras.preprocessing.image import ImageDataGenerator
# All images will be rescaled by 1./255
train_datagen = ImageDataGenerator(rescale=1./255)
test_datagen = ImageDataGenerator(rescale=1./255)
train_generator = train_datagen.flow_from_directory(
# This is the target directory
train_dir,
# All images will be resized to 150x150
target_size=(150, 150),
batch_size=20,
# Since we use binary_crossentropy loss, we need binary labels
class_mode='binary')
validation_generator = test_datagen.flow_from_directory(
validation_dir,
target_size=(150, 150),
batch_size=20,
class_mode='binary')
for data_batch, labels_batch in train_generator:
print('data batch shape:', data_batch.shape)
print('labels batch shape:', labels_batch.shape)
break
history = model.fit_generator(
train_generator,
steps_per_epoch=100,
epochs=30,
validation_data=validation_generator,
validation_steps=50)
model.save('cats_and_dogs_small_1.h5')
import matplotlib.pyplot as plt
acc = history.history['acc']
val_acc = history.history['val_acc']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(len(acc))
plt.plot(epochs, acc, 'bo', label='Training acc')
plt.plot(epochs, val_acc, 'b', label='Validation acc')
plt.title('Training and validation accuracy')
plt.legend()
plt.figure()
plt.plot(epochs, loss, 'bo', label='Training loss')
plt.plot(epochs, val_loss, 'b', label='Validation loss')
plt.title('Training and validation loss')
plt.legend()
plt.show()
datagen = ImageDataGenerator(
rotation_range=40,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
fill_mode='nearest')
# This is module with image preprocessing utilities
from keras.preprocessing import image
fnames = [os.path.join(train_cats_dir, fname) for fname in os.listdir(train_cats_dir)]
# We pick one image to "augment"
img_path = fnames[3]
# Read the image and resize it
img = image.load_img(img_path, target_size=(150, 150))
# Convert it to a Numpy array with shape (150, 150, 3)
x = image.img_to_array(img)
# Reshape it to (1, 150, 150, 3)
x = x.reshape((1,) + x.shape)
# The .flow() command below generates batches of randomly transformed images.
# It will loop indefinitely, so we need to `break` the loop at some point!
i = 0
for batch in datagen.flow(x, batch_size=1):
plt.figure(i)
imgplot = plt.imshow(image.array_to_img(batch[0]))
i += 1
if i % 4 == 0:
break
plt.show()
model = models.Sequential()
model.add(layers.Conv2D(32, (3, 3), activation='relu',
input_shape=(150, 150, 3)))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(128, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(128, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Flatten())
model.add(layers.Dropout(0.5))
model.add(layers.Dense(512, activation='relu'))
model.add(layers.Dense(1, activation='sigmoid'))
model.compile(loss='binary_crossentropy',
optimizer=optimizers.RMSprop(lr=1e-4),
metrics=['acc'])
train_datagen = ImageDataGenerator(
rescale=1./255,
rotation_range=40,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,)
# Note that the validation data should not be augmented!
test_datagen = ImageDataGenerator(rescale=1./255)
train_generator = train_datagen.flow_from_directory(
# This is the target directory
train_dir,
# All images will be resized to 150x150
target_size=(150, 150),
batch_size=32,
# Since we use binary_crossentropy loss, we need binary labels
class_mode='binary')
validation_generator = test_datagen.flow_from_directory(
validation_dir,
target_size=(150, 150),
batch_size=32,
class_mode='binary')
history = model.fit_generator(
train_generator,
steps_per_epoch=100,
epochs=100,
validation_data=validation_generator,
validation_steps=50)
model.save('cats_and_dogs_small_2.h5')
acc = history.history['acc']
val_acc = history.history['val_acc']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(len(acc))
plt.plot(epochs, acc, 'bo', label='Training acc')
plt.plot(epochs, val_acc, 'b', label='Validation acc')
plt.title('Training and validation accuracy')
plt.legend()
plt.figure()
plt.plot(epochs, loss, 'bo', label='Training loss')
plt.plot(epochs, val_loss, 'b', label='Validation loss')
plt.title('Training and validation loss')
plt.legend()
plt.show()
| 0.528533 | 0.980205 |
# Qiskit Pulseで高エネルギー状態へアクセスする
ほとんどの量子アルゴリズム/アプリケーションでは、$|0\rangle$と$|1\rangle$によって張られた2次元空間で計算が実行されます。ただし、IBMのハードウェアでは、通常は使用されない、より高いエネルギー状態も存在します。このセクションでは、Qiskit Pulseを使ってこれらの状態を探索することにフォーカスを当てます。特に、$|2\rangle$ 状態を励起し、$|0\rangle$、$|1\rangle$、$|2\rangle$の状態を分類するための識別器を作成する方法を示します。
このノートブックを読む前に、[前の章](./calibrating-qubits-openpulse.html)を読むことをお勧めします。また、Qiskit Pulseのスペック(Ref [1](#refs))も読むことをお勧めします。
### 物理学的背景
ここで、IBMの量子ハードウェアの多くの基礎となっている、トランズモンキュービットの物理学的な背景を説明します。このシステムには、ジョセフソン接合とコンデンサーで構成される超伝導回路が含まれています。超伝導回路に不慣れな方は、[こちらのレビュー](https://arxiv.org/pdf/1904.06560.pdf) (Ref. [2](#refs))を参照してください。このシステムのハミルトニアンは以下で与えられます。
$$
H = 4 E_C n^2 - E_J \cos(\phi),
$$
ここで、$E_C, E_J$はコンデンサーのエネルギーとジョセフソンエネルギーを示し、$n$は減衰した電荷数演算子で、$\phi$はジャンクションのために減衰した磁束です。$\hbar=1$として扱います。
トランズモンキュービットは$\phi$が小さい領域で定義されるため、$E_J \cos(\phi)$をテイラー級数で展開できます(定数項を無視します)。
$$
E_J \cos(\phi) \approx \frac{1}{2} E_J \phi^2 - \frac{1}{24} E_J \phi^4 + \mathcal{O}(\phi^6).
$$
$\phi$の二次の項$\phi^2$は、標準の調和振動子を定義します。その他の追加の項はそれぞれ非調和性をもたらします。
$n \sim (a-a^\dagger), \phi \sim (a+a^\dagger)$の関係を使うと($a^\dagger,a$は生成消滅演算子)、システムは以下のハミルトニアンを持つダフィング(Duffing)振動子に似ていることを示せます。
$$
H = \omega a^\dagger a + \frac{\alpha}{2} a^\dagger a^\dagger a a,
$$
$\omega$は、$0\rightarrow1$の励起周波数($\omega \equiv \omega^{0\rightarrow1}$)を与え、$\alpha$は$0\rightarrow1$の周波数と$1\rightarrow2$の周波数の間の非調和です。必要に応じて駆動の条件を追加できます。
標準の2次元部分空間へ特化したい場合は、$|\alpha|$ を十分に大きくとるか、高エネルギー状態を抑制する特別な制御テクニックを使います。
# 目次
0. [はじめに](#importing)
1. [0と1の状態の識別](#discrim01)
1. [0->1の周波数スイープ](#freqsweep01)
2. [0->1のラビ実験](#rabi01)
3. [0,1の識別器を構築する](#builddiscrim01)
2. [0,1,2の状態の識別](#discrim012)
1. [1->2の周波数の計算](#freq12)
1. [サイドバンド法を使った1->2の周波数スイープ](#sideband12)
2. [1->2のラビ実験](#rabi12)
3. [0,1,2の識別器を構築する](#builddiscrim012)
4. [参考文献](#refs)
## 0. はじめに <a id="importing"></a>
まず、依存関係をインポートし、いくつかのデフォルトの変数を定義します。量子ビット0を実験に使います。公開されている単一量子ビットデバイスである`ibmq_armonk`で実験を行います。
```
import numpy as np
import matplotlib.pyplot as plt
from scipy.optimize import curve_fit
from scipy.signal import find_peaks
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.model_selection import train_test_split
import qiskit.pulse as pulse
import qiskit.pulse.library as pulse_lib
from qiskit.compiler import assemble
from qiskit.pulse.library import SamplePulse
from qiskit.tools.monitor import job_monitor
import warnings
warnings.filterwarnings('ignore')
from qiskit.tools.jupyter import *
%matplotlib inline
from qiskit import IBMQ
IBMQ.load_account()
provider = IBMQ.get_provider(hub='ibm-q', group='open', project='main')
backend = provider.get_backend('ibmq_armonk')
backend_config = backend.configuration()
assert backend_config.open_pulse, "Backend doesn't support Pulse"
dt = backend_config.dt
backend_defaults = backend.defaults()
# 単位変換係数 -> すべてのバックエンドのプロパティーがSI単位系(Hz, sec, etc)で返される
GHz = 1.0e9 # Gigahertz
MHz = 1.0e6 # Megahertz
us = 1.0e-6 # Microseconds
ns = 1.0e-9 # Nanoseconds
qubit = 0 # 分析に使う量子ビット
default_qubit_freq = backend_defaults.qubit_freq_est[qubit] # デフォルトの量子ビット周波数単位はHz
print(f"Qubit {qubit} has an estimated frequency of {default_qubit_freq/ GHz} GHz.")
#(各デバイスに固有の)データをスケーリング
scale_factor = 1e-14
# 実験のショット回数
NUM_SHOTS = 1024
### 必要なチャネルを収集する
drive_chan = pulse.DriveChannel(qubit)
meas_chan = pulse.MeasureChannel(qubit)
acq_chan = pulse.AcquireChannel(qubit)
```
いくつか便利な関数を追加で定義します。
```
def get_job_data(job, average):
"""すでに実行されているジョブからデータを取得します。
引数:
job (Job): データが必要なジョブ
average (bool): Trueの場合、データが平均であると想定してデータを取得。
Falseの場合、シングルショット用と想定してデータを取得。
返し値:
list: ジョブの結果データを含むリスト
"""
job_results = job.result(timeout=120) # タイムアウトパラメーターは120秒にセット
result_data = []
for i in range(len(job_results.results)):
if average: # 平均データを得る
result_data.append(job_results.get_memory(i)[qubit]*scale_factor)
else: # シングルデータを得る
result_data.append(job_results.get_memory(i)[:, qubit]*scale_factor)
return result_data
def get_closest_multiple_of_16(num):
"""16の倍数に最も近いものを計算します。
パルスが使えるデバイスが16サンプルの倍数の期間が必要なためです。
"""
return (int(num) - (int(num)%16))
```
次に、駆動パルスと測定のためのいくつかのデフォルトパラメーターを含めます。命令スケジュールマップから(バックエンドデフォルトから)`measure`コマンドをプルして、新しいキャリブレーションでアップデートされるようにします。
```
# 駆動パルスのパラメーター (us = マイクロ秒)
drive_sigma_us = 0.075 # ガウシアンの実際の幅を決めます
drive_samples_us = drive_sigma_us*8 # 切り捨てパラメーター
# ガウシアンには自然な有限長がないためです。
drive_sigma = get_closest_multiple_of_16(drive_sigma_us * us /dt) # ガウシアンの幅の単位はdt
drive_samples = get_closest_multiple_of_16(drive_samples_us * us /dt) # 切り捨てパラメーターの単位はdt
# この量子ビットに必要な測定マップインデックスを見つける
meas_map_idx = None
for i, measure_group in enumerate(backend_config.meas_map):
if qubit in measure_group:
meas_map_idx = i
break
assert meas_map_idx is not None, f"Couldn't find qubit {qubit} in the meas_map!"
# 命令スケジュールマップからデフォルトの測定パルスを取得
inst_sched_map = backend_defaults.instruction_schedule_map
measure = inst_sched_map.get('measure', qubits=backend_config.meas_map[meas_map_idx])
```
## 1. $|0\rangle$ と $|1\rangle$の状態の識別 <a id="discrim01"></a>
このセクションでは、標準の$|0\rangle$と$|1\rangle$の状態の識別器を構築します。識別器のジョブは、`meas_level=1`の複素数データを取得し、標準の$|0\rangle$の$|1\rangle$の状態(`meas_level=2`)に分類することです。これは、前の[章](./calibrating-qubits-openpulse.html)の多くと同じ作業です。この結果は、このNotebookがフォーカスしている高エネルギー状態に励起するために必要です。
### 1A. 0->1 周波数のスイープ <a id="freqsweep01"></a>
識別器の構築の最初のステップは、前の章でやったのと同じように、我々の量子ビット周波数をキャリブレーションすることです。
```
def create_ground_freq_sweep_program(freqs, drive_power):
"""基底状態を励起して周波数掃引を行うプログラムを作成します。
ドライブパワーに応じて、これは0->1の周波数なのか、または0->2の周波数なのかを明らかにすることができます。
引数:
freqs (np.ndarray(dtype=float)):スイープする周波数のNumpy配列。
drive_power (float):ドライブ振幅の値。
レイズ:
ValueError:75を超える頻度を使用すると発生します。
現在、これを実行しようとすると、バックエンドでエラーが投げられます。
戻り値:
Qobj:基底状態の周波数掃引実験のプログラム。
"""
if len(freqs) > 75:
raise ValueError("You can only run 75 schedules at a time.")
# スイープ情報を表示
print(f"The frequency sweep will go from {freqs[0] / GHz} GHz to {freqs[-1]/ GHz} GHz \
using {len(freqs)} frequencies. The drive power is {drive_power}.")
# 駆動パルスを定義
ground_sweep_drive_pulse = pulse_lib.gaussian(duration=drive_samples,
sigma=drive_sigma,
amp=drive_power,
name='ground_sweep_drive_pulse')
# スイープのための周波数を定義
schedule = pulse.Schedule(name='Frequency sweep starting from ground state.')
schedule |= pulse.Play(ground_sweep_drive_pulse, drive_chan)
schedule |= measure << schedule.duration
# define frequencies for the sweep
schedule_freqs = [{drive_chan: freq} for freq in freqs]
# プログラムを組み立てる
# 注:それぞれが同じことを行うため、必要なスケジュールは1つだけです;
# スケジュールごとに、ドライブをミックスダウンするLO周波数が変化します
# これにより周波数掃引が可能になります
ground_freq_sweep_program = assemble(schedule,
backend=backend,
meas_level=1,
meas_return='avg',
shots=NUM_SHOTS,
schedule_los=schedule_freqs)
return ground_freq_sweep_program
# 75個の周波数で推定周波数の周りに40MHzを掃引します
num_freqs = 75
ground_sweep_freqs = default_qubit_freq + np.linspace(-20*MHz, 20*MHz, num_freqs)
ground_freq_sweep_program = create_ground_freq_sweep_program(ground_sweep_freqs, drive_power=0.3)
ground_freq_sweep_job = backend.run(ground_freq_sweep_program)
print(ground_freq_sweep_job.job_id())
job_monitor(ground_freq_sweep_job)
# ジョブのデータ(平均)を取得する
ground_freq_sweep_data = get_job_data(ground_freq_sweep_job, average=True)
```
データをローレンツ曲線に適合させ、キャリブレーションされた周波数を抽出します。
```
def fit_function(x_values, y_values, function, init_params):
"""Fit a function using scipy curve_fit."""
fitparams, conv = curve_fit(function, x_values, y_values, init_params)
y_fit = function(x_values, *fitparams)
return fitparams, y_fit
# Hz単位でのフィッティングをします
(ground_sweep_fit_params,
ground_sweep_y_fit) = fit_function(ground_sweep_freqs,
ground_freq_sweep_data,
lambda x, A, q_freq, B, C: (A / np.pi) * (B / ((x - q_freq)**2 + B**2)) + C,
[7, 4.975*GHz, 1*GHz, 3*GHz] # フィッティングのための初期パラメーター
)
# 注:シグナルの実数部のみをプロットしています
plt.scatter(ground_sweep_freqs/GHz, ground_freq_sweep_data, color='black')
plt.plot(ground_sweep_freqs/GHz, ground_sweep_y_fit, color='red')
plt.xlim([min(ground_sweep_freqs/GHz), max(ground_sweep_freqs/GHz)])
plt.xlabel("Frequency [GHz]", fontsize=15)
plt.ylabel("Measured Signal [a.u.]", fontsize=15)
plt.title("0->1 Frequency Sweep", fontsize=15)
plt.show()
_, cal_qubit_freq, _, _ = ground_sweep_fit_params
print(f"We've updated our qubit frequency estimate from "
f"{round(default_qubit_freq/GHz, 7)} GHz to {round(cal_qubit_freq/GHz, 7)} GHz.")
```
### 1B. 0->1 のラビ実験 <a id="rabi01"></a>
次に、$0\rightarrow1 ~ \pi$パルスの振幅を計算するラビ実験を実行します。$\pi$パルスは、$|0\rangle$から$|1\rangle$の状態へ移動させるパルス(ブロッホ球上での$\pi$回転)だということを思い出してください。
```
# 実験の構成
num_rabi_points = 50 # 実験の数(つまり、掃引の振幅)
# 反復する駆動パルスの振幅値:0から0.75まで等間隔に配置された50の振幅
drive_amp_min = 0
drive_amp_max = 0.75
drive_amps = np.linspace(drive_amp_min, drive_amp_max, num_rabi_points)
# スケジュールを作成
rabi_01_schedules = []
# 駆動振幅すべてにわたってループ
for ii, drive_amp in enumerate(drive_amps):
# 駆動パルス
rabi_01_pulse = pulse_lib.gaussian(duration=drive_samples,
amp=drive_amp,
sigma=drive_sigma,
name='rabi_01_pulse_%d' % ii)
# スケジュールにコマンドを追加
schedule = pulse.Schedule(name='Rabi Experiment at drive amp = %s' % drive_amp)
schedule |= pulse.Play(rabi_01_pulse, drive_chan)
schedule |= measure << schedule.duration # 測定をドライブパルスの後にシフト
rabi_01_schedules.append(schedule)
# プログラムにスケジュールを組み込む
# 注:較正された周波数で駆動します。
rabi_01_expt_program = assemble(rabi_01_schedules,
backend=backend,
meas_level=1,
meas_return='avg',
shots=NUM_SHOTS,
schedule_los=[{drive_chan: cal_qubit_freq}]
* num_rabi_points)
rabi_01_job = backend.run(rabi_01_expt_program)
print(rabi_01_job.job_id())
job_monitor(rabi_01_job)
# ジョブのデータ(平均)を取得する
rabi_01_data = get_job_data(rabi_01_job, average=True)
def baseline_remove(values):
"""Center data around 0."""
return np.array(values) - np.mean(values)
# 注:データの実数部のみがプロットされます
rabi_01_data = np.real(baseline_remove(rabi_01_data))
(rabi_01_fit_params,
rabi_01_y_fit) = fit_function(drive_amps,
rabi_01_data,
lambda x, A, B, drive_01_period, phi: (A*np.cos(2*np.pi*x/drive_01_period - phi) + B),
[4, -4, 0.5, 0])
plt.scatter(drive_amps, rabi_01_data, color='black')
plt.plot(drive_amps, rabi_01_y_fit, color='red')
drive_01_period = rabi_01_fit_params[2]
# piの振幅計算でphiを計算
pi_amp_01 = (drive_01_period/2/np.pi) *(np.pi+rabi_01_fit_params[3])
plt.axvline(pi_amp_01, color='red', linestyle='--')
plt.axvline(pi_amp_01+drive_01_period/2, color='red', linestyle='--')
plt.annotate("", xy=(pi_amp_01+drive_01_period/2, 0), xytext=(pi_amp_01,0), arrowprops=dict(arrowstyle="<->", color='red'))
plt.annotate("$\pi$", xy=(pi_amp_01-0.03, 0.1), color='red')
plt.xlabel("Drive amp [a.u.]", fontsize=15)
plt.ylabel("Measured signal [a.u.]", fontsize=15)
plt.title('0->1 Rabi Experiment', fontsize=15)
plt.show()
print(f"Pi Amplitude (0->1) = {pi_amp_01}")
```
この結果を使って、$0\rightarrow1$ $\pi$パルスを定義します。
```
pi_pulse_01 = pulse_lib.gaussian(duration=drive_samples,
amp=pi_amp_01,
sigma=drive_sigma,
name='pi_pulse_01')
```
### 1C. 0,1 の識別器を構築する <a id="builddiscrim01"></a>
これで、キャリブレーションされた周波数と$\pi$パルスを得たので、$|0\rangle$と$1\rangle$の状態の識別器を構築できます。識別器は、IQ平面において`meas_level=1`のデータを取って、それを$|0\rangle$または$1\rangle$を判別することで機能します。
$|0\rangle$と$|1\rangle$の状態は、IQ平面上で重心として知られているコヒーレントな円形の"ブロブ"を形成します。重心の中心は、各状態の正確なノイズのないIQポイントを定義します。周囲の雲は、様々なノイズ源から生成されたデータの分散を示します。
$|0\rangle$と$|1\rangle$間を識別(判別)するために、機械学習のテクニック、線形判別分析を適用します。この方法は量子ビットの状態を判別する一般的なテクニックです。
最初のステップは、重心データを得ることです。そのために、2つのスケジュールを定義します(システムが$|0\rangle$の状態から始まることを思い出しましょう。):
1. $|0\rangle$の状態を直接測定します($|0\rangle$の重心を得ます)。
2. $\pi$パルスを適用して、測定します($|1\rangle$の重心を得ます)。
```
# 2つのスケジュールを作る
# 基底状態のスケジュール
zero_schedule = pulse.Schedule(name="zero schedule")
zero_schedule |= measure
# 励起状態のスケジュール
one_schedule = pulse.Schedule(name="one schedule")
one_schedule |= pulse.Play(pi_pulse_01, drive_chan)
one_schedule |= measure << one_schedule.duration
# スケジュールをプログラムにアセンブルする
IQ_01_program = assemble([zero_schedule, one_schedule],
backend=backend,
meas_level=1,
meas_return='single',
shots=NUM_SHOTS,
schedule_los=[{drive_chan: cal_qubit_freq}] * 2)
IQ_01_job = backend.run(IQ_01_program)
print(IQ_01_job.job_id())
job_monitor(IQ_01_job)
# (単一の)ジョブデータを取得します;0と1に分割します
IQ_01_data = get_job_data(IQ_01_job, average=False)
zero_data = IQ_01_data[0]
one_data = IQ_01_data[1]
def IQ_01_plot(x_min, x_max, y_min, y_max):
"""Helper function for plotting IQ plane for |0>, |1>. Limits of plot given
as arguments."""
# 0のデータは青でプロット
plt.scatter(np.real(zero_data), np.imag(zero_data),
s=5, cmap='viridis', c='blue', alpha=0.5, label=r'$|0\rangle$')
# 1のデータは赤でプロット
plt.scatter(np.real(one_data), np.imag(one_data),
s=5, cmap='viridis', c='red', alpha=0.5, label=r'$|1\rangle$')
# 0状態と1状態の平均に大きなドットをプロットします。
mean_zero = np.mean(zero_data) # 実部と虚部両方の平均を取ります。
mean_one = np.mean(one_data)
plt.scatter(np.real(mean_zero), np.imag(mean_zero),
s=200, cmap='viridis', c='black',alpha=1.0)
plt.scatter(np.real(mean_one), np.imag(mean_one),
s=200, cmap='viridis', c='black',alpha=1.0)
plt.xlim(x_min, x_max)
plt.ylim(y_min,y_max)
plt.legend()
plt.ylabel('I [a.u.]', fontsize=15)
plt.xlabel('Q [a.u.]', fontsize=15)
plt.title("0-1 discrimination", fontsize=15)
```
以下のように、IQプロットを表示します。青の重心は$|0\rangle$状態で、赤の重心は$|1\rangle$状態です。(注:プロットが見えないときは、Notebookを再実行してください。)
```
x_min = -5
x_max = 15
y_min = -5
y_max = 10
IQ_01_plot(x_min, x_max, y_min, y_max)
```
さて、実際に識別器を構築する時が来ました。先に述べたように、線形判別分析(Linear Discriminant Analysis, LDA)と呼ばれる機械学習のテクニックを使います。LDAは、任意のデータセットをカテゴリーのセット(ここでは$|0\rangle$と$|1\rangle$)に分類するために、各カテゴリーの平均の間の距離を最大化し、各カテゴリーの分散を最小化します。より詳しくは、[こちら](https://scikit-learn.org/stable/modules/lda_qda.html#id4) (Ref. [3](#refs))をご覧ください。
LDAは、セパラトリックス(separatrix)と呼ばれるラインを生成します。与えられたデータポイントがどちら側のセパラトリックスにあるかに応じて、それがどのカテゴリーに属しているかを判別できます。我々の場合、セパラトリックスの片側が$|0\rangle$状態で、もう一方の側が$|1\rangle$の状態です。
我々は、最初の半分のデータを学習用に使い、残りの半分をテスト用に使います。LDAの実装のために`scikit.learn`を使います:将来のリリースでは、この機能は、Qiskit-Ignisに直接実装されてリリースされる予定です([ここ](https://github.com/Qiskit/qiskit-ignis/tree/master/qiskit/ignis/measurement/discriminator)を参照)。
結果データを判別に適したフォーマットになるように再形成します。
```
def reshape_complex_vec(vec):
"""
複素数ベクトルvecを取り込んで、実際のimagエントリーを含む2d配列を返します。
これは学習に必要なデータです。
Args:
vec (list):データの複素数ベクトル
戻り値:
list:(real(vec], imag(vec))で指定されたエントリー付きのベクトル
"""
length = len(vec)
vec_reshaped = np.zeros((length, 2))
for i in range(len(vec)):
vec_reshaped[i]=[np.real(vec[i]), np.imag(vec[i])]
return vec_reshaped
# IQベクトルを作成します(実部と虚部で構成されています)
zero_data_reshaped = reshape_complex_vec(zero_data)
one_data_reshaped = reshape_complex_vec(one_data)
IQ_01_data = np.concatenate((zero_data_reshaped, one_data_reshaped))
print(IQ_01_data.shape) # IQデータの形を確認します
```
次に、学習用データとテスト用データを分割します。期待される結果(基底状態のスケジュールは`0`の配列、励起状態のスケジュールは`1`の配列)を含む状態ベクトルを使ってテストします。
```
#(テスト用に)0と1でベクトルを構築する
state_01 = np.zeros(NUM_SHOTS) # shotsは実験の回数
state_01 = np.concatenate((state_01, np.ones(NUM_SHOTS)))
print(len(state_01))
# データをシャッフルしてトレーニングセットとテストセットに分割します
IQ_01_train, IQ_01_test, state_01_train, state_01_test = train_test_split(IQ_01_data, state_01, test_size=0.5)
```
最後に、モデルを設定して、学習します。学習精度が表示されます。
```
# LDAをセットアップします
LDA_01 = LinearDiscriminantAnalysis()
LDA_01.fit(IQ_01_train, state_01_train)
# シンプルなデータでテストします
print(LDA_01.predict([[0,0], [10, 0]]))
# 精度を計算します
score_01 = LDA_01.score(IQ_01_test, state_01_test)
print(score_01)
```
最後のステップは、セパラトリックスをプロットすることです。
```
# セパラトリックスを表示データの上にプロットします
def separatrixPlot(lda, x_min, x_max, y_min, y_max, shots):
nx, ny = shots, shots
xx, yy = np.meshgrid(np.linspace(x_min, x_max, nx),
np.linspace(y_min, y_max, ny))
Z = lda.predict_proba(np.c_[xx.ravel(), yy.ravel()])
Z = Z[:, 1].reshape(xx.shape)
plt.contour(xx, yy, Z, [0.5], linewidths=2., colors='black')
IQ_01_plot(x_min, x_max, y_min, y_max)
separatrixPlot(LDA_01, x_min, x_max, y_min, y_max, NUM_SHOTS)
```
セラパトリックスのどちらのサイドがどの重心(つまり状態)に対応しているか確認します。IQ平面上の点が与えられると、このモデルはセラパトリックスのどちらの側にそれが置かれているかチェックし、対応する状態を返します。
## 2. $|0\rangle$, $|1\rangle$, $|2\rangle$ の状態の識別 <a id="discrim012"></a>
$0, 1$の識別器をキャリブレーションしたので、高エネルギー状態の励起に移ります。特に、$|2\rangle$の状態の励起にフォーカスし、$|0\rangle$と$|1\rangle$と$|2\rangle$の状態をそれぞれのIQデータポイントから判別する識別器を構築することに焦点を当てます。さらに高い状態($|3\rangle$、$|4\rangle$など)の手順も同様ですが、明示的にテストはしていません。
高い状態の識別器を構築する手順は以下の通りです:
1. $1\rightarrow2$周波数を計算します。
2. $1\rightarrow2$のための$\pi$パルスの振幅を得るためにラビ実験を行います。そのためには、まず、$0\rightarrow1$ $\pi$パルスを適用して、$|0\rangle$から$|1\rangle$の状態にします。次に、上記で得た$1\rightarrow2$周波数において、駆動振幅のスイープを行います。
3. 3つのスケジュールを構成します:\
a. 0スケジュール:基底状態を測定するだけです。\
b. 1スケジュール:$0\rightarrow1$ $\pi$パルスを適用し、測定します。\
c. 2スケジュール:$0\rightarrow1$ $\pi$パルスを適用し、次に$1\rightarrow2$ $\pi$パルスを適用し測定します。
4. 各スケジュールのデータを学習用データとテスト用データのセットに分け、判別用のLDAモデルを構築します。
### 2A. 1->2 周波数の計算 <a id="freq12"></a>
キャリブレーションの最初のステップは、$1\rightarrow2$ の状態に移行するために必要な周波数を計算することです。これを行うには2つの方法があります:
1. 基底状態から周波数をスイープし、非常に高い電力をかけます。印加電力が十分に高い場合には、2つのピークが観測されます。1つはセクション [1](#discrim01)で見つかった $0\rightarrow1$周波数で、もう一つは、$0\rightarrow2$周波数です。$1\rightarrow2$周波数は2つの差を取ることで得られます。残念ながら、`ibmq_armonk`では、最大駆動電力$1.0$はこの遷移を起こすのに十分ではありません。代わりに、2番目の方法を使います。
2. $0\rightarrow1$ $\pi$パルスを適用して、$|1\rangle$状態を励起します。その後、$|1\rangle$状態のさらに上の励起に対して、周波数スイープを実行します。$0\rightarrow1$周波数より低いところで、$1\rightarrow2$周波数に対応した単一ピークが観測されるはずです。
#### サイドバンド法を使用した1->2 の周波数スイープ <a id="sideband12"></a>
上記の2番目の方法に従いましょう。$0\rightarrow 1$ $\pi$パルスを駆動するために、ローカル共振(local oscilattor, LO)周波数が必要です。これは、キャリブレーションされた$0\rightarrow1$周波数`cal_qubit_freq`(セクション[1](#discrim01)のラビ$\pi$パルスの構築を参照)によって与えられます。ただし、$1\rightarrow2$周波数の範囲をスイープするために、LO周波数を変化させる必要があります。残念ながら、Pulseのスペックでは、各スケジュールごとに、一つのLO周波数が必要です。
これを解決するには、LO周波数を`cal_qubit_freq`にセットし、を`freq-cal_qubit_freq`で$1\rightarrow2$パルスの上にサイン関数を乗算します。ここで`freq`は目的のスキャン周波数です。知られているように、正弦波サイドバンドを適用すると、プログラムのアセンブル時に手動で設定せずにLO周波数を変更可能です。
```
def apply_sideband(pulse, freq):
"""freq周波数でこのパルスに正弦波サイドバンドを適用します。
引数:
pulse (SamplePulse):対象のパルス。
freq (float):スイープを適用するLO周波数。
戻り値:
SamplePulse:サイドバンドが適用されたパルス(freqとcal_qubit_freqの差で振動します)。
"""
# 時間は0からdt*drive_samplesで、2*pi*f*tの形の正弦波引数になります
t_samples = np.linspace(0, dt*drive_samples, drive_samples)
sine_pulse = np.sin(2*np.pi*(freq-cal_qubit_freq)*t_samples) # no amp for the sine
# サイドバンドが適用されたサンプルパルスを作成
# 注:sq_pulse.samplesを実数にし、要素ごとに乗算する必要があります
sideband_pulse = SamplePulse(np.multiply(np.real(pulse.samples), sine_pulse), name='sideband_pulse')
return sideband_pulse
```
プログラムをアセンブルするためのロジックをメソッドにラップして、プログラムを実行します。
```
def create_excited_freq_sweep_program(freqs, drive_power):
"""|1>状態を励起することにより、周波数掃引を行うプログラムを作成します。
これにより、1-> 2の周波数を取得できます。
較正された量子ビット周波数を使用して、piパルスを介して|0>から|1>の状態になります。
|1>から|2>への周波数掃引を行うには、正弦係数を掃引駆動パルスに追加することにより、サイドバンド法を使用します。
引数:
freqs (np.ndarray(dtype=float)):掃引周波数のNumpy配列。
drive_power (float):駆動振幅の値。
レイズ:
ValueError:75を超える頻度を使用するとスローされます; 現在、75個を超える周波数を試行すると、
バックエンドでエラーがスローされます。
戻り値:
Qobj:周波数掃引実験用のプログラム。
"""
if len(freqs) > 75:
raise ValueError("You can only run 75 schedules at a time.")
print(f"The frequency sweep will go from {freqs[0] / GHz} GHz to {freqs[-1]/ GHz} GHz \
using {len(freqs)} frequencies. The drive power is {drive_power}.")
base_12_pulse = pulse_lib.gaussian(duration=drive_samples,
sigma=drive_sigma,
amp=drive_power,
name='base_12_pulse')
schedules = []
for jj, freq in enumerate(freqs):
# ガウシアンパルスにサイドバンドを追加
freq_sweep_12_pulse = apply_sideband(base_12_pulse, freq)
# スケジュールのコマンドを追加
schedule = pulse.Schedule(name="Frequency = {}".format(freq))
# 0->1のパルス、掃引パルスの周波数、測定を追加
schedule |= pulse.Play(pi_pulse_01, drive_chan)
schedule |= pulse.Play(freq_sweep_12_pulse, drive_chan) << schedule.duration
schedule |= measure << schedule.duration # 駆動パルスの後に測定をシフト
schedules.append(schedule)
num_freqs = len(freqs)
# スケジュールを表示します
display(schedules[-1].draw(channels=[drive_chan, meas_chan], label=True, scale=1.0))
# 周波数掃引プログラムを組み込みます
# 注:LOは各スケジュールでのcal_qubit_freqです;サイドバンドによって組み込みます
excited_freq_sweep_program = assemble(schedules,
backend=backend,
meas_level=1,
meas_return='avg',
shots=NUM_SHOTS,
schedule_los=[{drive_chan: cal_qubit_freq}]
* num_freqs)
return excited_freq_sweep_program
# 0->1周波数より下で1->2の周波数を見つけるために400 MHzを掃引します
num_freqs = 75
excited_sweep_freqs = cal_qubit_freq + np.linspace(-400*MHz, 30*MHz, num_freqs)
excited_freq_sweep_program = create_excited_freq_sweep_program(excited_sweep_freqs, drive_power=0.3)
# 確認のためにスケジュールの一例をプロットします
excited_freq_sweep_job = backend.run(excited_freq_sweep_program)
print(excited_freq_sweep_job.job_id())
job_monitor(excited_freq_sweep_job)
# (平均の)ジョブデータを取得します
excited_freq_sweep_data = get_job_data(excited_freq_sweep_job, average=True)
# 注:シグナルの実部だけをプロットします
plt.scatter(excited_sweep_freqs/GHz, excited_freq_sweep_data, color='black')
plt.xlim([min(excited_sweep_freqs/GHz)+0.01, max(excited_sweep_freqs/GHz)]) # ignore min point (is off)
plt.xlabel("Frequency [GHz]", fontsize=15)
plt.ylabel("Measured Signal [a.u.]", fontsize=15)
plt.title("1->2 Frequency Sweep (first pass)", fontsize=15)
plt.show()
```
最小値が$4.64$ GHz近辺に見られます。いくつかの偽の最大値がありますが、それらは、$1\rightarrow2$周波数には大きすぎます。最小値が$1\rightarrow2$周波数に対応します。
相対最小関数を使って、この点の値を正確に計算します。これで、$1\rightarrow2$周波数の推定値が得られます。
```
# output_dataに相対的最小周波数を表示します;高さは下限(絶対値)を示します
def rel_maxima(freqs, output_data, height):
"""output_dataに相対的な最小周波数を出力します(ピークを確認できます);
高さは上限(絶対値)を示します。
高さを正しく設定しないと、ピークが無視されます。
引数:
freqs (list):周波数リスト
output_data (list):結果のシグナルのリスト
height (float):ピークの上限(絶対値)
戻り値:
list:相対的な最小周波数を含むリスト
"""
peaks, _ = find_peaks(output_data, height)
print("Freq. dips: ", freqs[peaks])
return freqs[peaks]
maxima = rel_maxima(excited_sweep_freqs, np.real(excited_freq_sweep_data), 10)
approx_12_freq = maxima
```
上記で得られた推定値を使って、より正確な掃引を行います(つまり、大幅に狭い範囲で掃引を行います)。これによって、$1\rightarrow2$周波数のより正確な値を得ることができます。上下$20$ MHzをスイープします。
```
# 狭い範囲での掃引
num_freqs = 75
refined_excited_sweep_freqs = approx_12_freq + np.linspace(-20*MHz, 20*MHz, num_freqs)
refined_excited_freq_sweep_program = create_excited_freq_sweep_program(refined_excited_sweep_freqs, drive_power=0.3)
refined_excited_freq_sweep_job = backend.run(refined_excited_freq_sweep_program)
print(refined_excited_freq_sweep_job.job_id())
job_monitor(refined_excited_freq_sweep_job)
# より正確な(平均)データを取得する
refined_excited_freq_sweep_data = get_job_data(refined_excited_freq_sweep_job, average=True)
```
標準ローレンツ曲線を用いて、このより正確な信号をプロットしてフィットします。
```
# Hzの単位でフィッティングする
(refined_excited_sweep_fit_params,
refined_excited_sweep_y_fit) = fit_function(refined_excited_sweep_freqs,
refined_excited_freq_sweep_data,
lambda x, A, q_freq, B, C: (A / np.pi) * (B / ((x - q_freq)**2 + B**2)) + C,
[-12, 4.625*GHz, 0.05*GHz, 3*GHz] # フィッティングのための初期パラメーター
)
# 注:シグナルの実数部のみをプロットしています
plt.scatter(refined_excited_sweep_freqs/GHz, refined_excited_freq_sweep_data, color='black')
plt.plot(refined_excited_sweep_freqs/GHz, refined_excited_sweep_y_fit, color='red')
plt.xlim([min(refined_excited_sweep_freqs/GHz), max(refined_excited_sweep_freqs/GHz)])
plt.xlabel("Frequency [GHz]", fontsize=15)
plt.ylabel("Measured Signal [a.u.]", fontsize=15)
plt.title("1->2 Frequency Sweep (refined pass)", fontsize=15)
plt.show()
_, qubit_12_freq, _, _ = refined_excited_sweep_fit_params
print(f"Our updated estimate for the 1->2 transition frequency is "
f"{round(qubit_12_freq/GHz, 7)} GHz.")
```
### 2B. 1->2 ラビ実験 <a id="rabi12"></a>
これで、$1\rightarrow2$周波数の良い推定が得られたので、$1\rightarrow2$遷移のための$\pi$パルス振幅を得るためのラビ実験を行います。そのために、$0\rightarrow1$ $\pi$ パルスを適用してから、$1\rightarrow2$周波数において駆動振幅をスイープします(サイドバンド法を使います)。
```
# 実験の構成
num_rabi_points = 75 # 実験数(つまり掃引する振幅)
# 駆動振幅の繰り返し値:0から1.0の間で均等に配置された75個の振幅
drive_amp_min = 0
drive_amp_max = 1.0
drive_amps = np.linspace(drive_amp_min, drive_amp_max, num_rabi_points)
# スケジュールの作成
rabi_12_schedules = []
# すべての駆動振幅をループします
for ii, drive_amp in enumerate(drive_amps):
base_12_pulse = pulse_lib.gaussian(duration=drive_samples,
sigma=drive_sigma,
amp=drive_amp,
name='base_12_pulse')
# 1->2の周波数においてサイドバンドを適用
rabi_12_pulse = apply_sideband(base_12_pulse, qubit_12_freq)
# スケジュールにコマンドを追加
schedule = pulse.Schedule(name='Rabi Experiment at drive amp = %s' % drive_amp)
schedule |= pulse.Play(pi_pulse_01, drive_chan) # 0->1
schedule |= pulse.Play(rabi_12_pulse, drive_chan) << schedule.duration # 1->2のラビパルス
schedule |= measure << schedule.duration # 駆動パルスの後に測定をシフト
rabi_12_schedules.append(schedule)
# プログラムにスケジュールを組み込みます
# 注:LO周波数はcal_qubit_freqであり、0->1のpiパルスを作ります;
# サイドバンドを使って、1->2のパルス用に変更されます
rabi_12_expt_program = assemble(rabi_12_schedules,
backend=backend,
meas_level=1,
meas_return='avg',
shots=NUM_SHOTS,
schedule_los=[{drive_chan: cal_qubit_freq}]
* num_rabi_points)
rabi_12_job = backend.run(rabi_12_expt_program)
print(rabi_12_job.job_id())
job_monitor(rabi_12_job)
# ジョブデータ(平均)を取得します
rabi_12_data = get_job_data(rabi_12_job, average=True)
```
We plot and fit our data as before.
```
# 注:信号の実部のみプロットします。
rabi_12_data = np.real(baseline_remove(rabi_12_data))
(rabi_12_fit_params,
rabi_12_y_fit) = fit_function(drive_amps,
rabi_12_data,
lambda x, A, B, drive_12_period, phi: (A*np.cos(2*np.pi*x/drive_12_period - phi) + B),
[3, 0.5, 0.9, 0])
plt.scatter(drive_amps, rabi_12_data, color='black')
plt.plot(drive_amps, rabi_12_y_fit, color='red')
drive_12_period = rabi_12_fit_params[2]
# piパルス用の振幅のためにphiを考慮します
pi_amp_12 = (drive_12_period/2/np.pi) *(np.pi+rabi_12_fit_params[3])
plt.axvline(pi_amp_12, color='red', linestyle='--')
plt.axvline(pi_amp_12+drive_12_period/2, color='red', linestyle='--')
plt.annotate("", xy=(pi_amp_12+drive_12_period/2, 0), xytext=(pi_amp_12,0), arrowprops=dict(arrowstyle="<->", color='red'))
plt.annotate("$\pi$", xy=(pi_amp_12-0.03, 0.1), color='red')
plt.xlabel("Drive amp [a.u.]", fontsize=15)
plt.ylabel("Measured signal [a.u.]", fontsize=15)
plt.title('Rabi Experiment (1->2)', fontsize=20)
plt.show()
print(f"Our updated estimate for the 1->2 transition frequency is "
f"{round(qubit_12_freq/GHz, 7)} GHz.")
print(f"Pi Amplitude (1->2) = {pi_amp_12}")
```
この情報を使って、$1\rightarrow2$ $\pi$パルスを定義できます。(必ず、$1\rightarrow2$周波数でサイドバンドを追加してください。)
```
pi_pulse_12 = pulse_lib.gaussian(duration=drive_samples,
amp=pi_amp_12,
sigma=drive_sigma,
name='pi_pulse_12')
# このパルスがサイドバンドであることを再確認してください
pi_pulse_12 = apply_sideband(pi_pulse_12, qubit_12_freq)
```
### 2C. 0,1,2の識別器を構築する <a id="builddiscrim012"></a>
とうとう、 $|0\rangle$と$|1\rangle$と$|2\rangle$状態の識別器を構築できます。手順はセクション[1](#discrim01)と同様ですが、$|2\rangle$状態のためにスケジュールを追加します。
3つのスケジュールがあります。(再度、私たちのシステムが$|0\rangle$から開始することを思い出してください):
1. $|0\rangle$状態を直接測定します。($|0\rangle$の重心を得ます。)
2. $0\rightarrow1$ $\pi$パルスを適用し、測定します。($|1\rangle$の重心を得ます。)
3. $0\rightarrow1$ $\pi$パルスを適用した後、$1\rightarrow2$ $\pi$パルスを適用しそして測定します。($|2\rangle$の重心を得ます。)
```
# 3つのスケジュールを作ります
# 基底状態のスケジュール
zero_schedule = pulse.Schedule(name="zero schedule")
zero_schedule |= measure
# 励起状態のスケジュール
one_schedule = pulse.Schedule(name="one schedule")
one_schedule |= pulse.Play(pi_pulse_01, drive_chan)
one_schedule |= measure << one_schedule.duration
# 励起状態のスケジュール
two_schedule = pulse.Schedule(name="two schedule")
two_schedule |= pulse.Play(pi_pulse_01, drive_chan)
two_schedule |= pulse.Play(pi_pulse_12, drive_chan) << two_schedule.duration
two_schedule |= measure << two_schedule.duration
```
プログラムを構築し、IQ平面上に重心をプロットします。
```
# プログラムにスケジュールを組み込みます
IQ_012_program = assemble([zero_schedule, one_schedule, two_schedule],
backend=backend,
meas_level=1,
meas_return='single',
shots=NUM_SHOTS,
schedule_los=[{drive_chan: cal_qubit_freq}] * 3)
IQ_012_job = backend.run(IQ_012_program)
print(IQ_012_job.job_id())
job_monitor(IQ_012_job)
# (単一の)ジョブデータを取得します;0,1,2に分割します
IQ_012_data = get_job_data(IQ_012_job, average=False)
zero_data = IQ_012_data[0]
one_data = IQ_012_data[1]
two_data = IQ_012_data[2]
def IQ_012_plot(x_min, x_max, y_min, y_max):
"""0、1、2のIQ平面をプロットするための補助関数。引数としてプロットの制限を与えます。
"""
# 0のデータは青でプロット
plt.scatter(np.real(zero_data), np.imag(zero_data),
s=5, cmap='viridis', c='blue', alpha=0.5, label=r'$|0\rangle$')
# 1のデータは赤でプロット
plt.scatter(np.real(one_data), np.imag(one_data),
s=5, cmap='viridis', c='red', alpha=0.5, label=r'$|1\rangle$')
# 2のデータは緑でプロット
plt.scatter(np.real(two_data), np.imag(two_data),
s=5, cmap='viridis', c='green', alpha=0.5, label=r'$|2\rangle$')
# 0、1、2の状態の結果の平均を大きなドットでプロット
mean_zero = np.mean(zero_data) # 実部と虚部それぞれの平均をとる
mean_one = np.mean(one_data)
mean_two = np.mean(two_data)
plt.scatter(np.real(mean_zero), np.imag(mean_zero),
s=200, cmap='viridis', c='black',alpha=1.0)
plt.scatter(np.real(mean_one), np.imag(mean_one),
s=200, cmap='viridis', c='black',alpha=1.0)
plt.scatter(np.real(mean_two), np.imag(mean_two),
s=200, cmap='viridis', c='black',alpha=1.0)
plt.xlim(x_min, x_max)
plt.ylim(y_min,y_max)
plt.legend()
plt.ylabel('I [a.u.]', fontsize=15)
plt.xlabel('Q [a.u.]', fontsize=15)
plt.title("0-1-2 discrimination", fontsize=15)
x_min = -10
x_max = 20
y_min = -25
y_max = 10
IQ_012_plot(x_min, x_max, y_min, y_max)
```
今回は、$|2\rangle$状態に対応した3個目の重心が観測されます。(注:プロットが見えない場合は、Notebookを再実行してください。)
このデータで、識別器を構築します。再び`scikit.learn` を使って線形判別分析(LDA)を使います。
LDAのためにデータを形成することから始めます。
```
# IQベクトルを作成します(実部と虚部で構成されています)
zero_data_reshaped = reshape_complex_vec(zero_data)
one_data_reshaped = reshape_complex_vec(one_data)
two_data_reshaped = reshape_complex_vec(two_data)
IQ_012_data = np.concatenate((zero_data_reshaped, one_data_reshaped, two_data_reshaped))
print(IQ_012_data.shape) # IQデータの形を確認します
```
次に、学習用データとテスト用データを分割します(前回と同じように半分ずつです)。テスト用データは、0スケジュールの場合、`0`の配列が含まれたベクトルで、1スケジュールの場合、`1`の配列が含まれたベクトルで、2スケジュールの場合`2`の配列が含まれたベクトルです。
```
# (テスト用に)0と1と2の値が含まれたベクトルを構築します
state_012 = np.zeros(NUM_SHOTS) # 実験のショット数
state_012 = np.concatenate((state_012, np.ones(NUM_SHOTS)))
state_012 = np.concatenate((state_012, 2*np.ones(NUM_SHOTS)))
print(len(state_012))
# データをシャッフルして学習用セットとテスト用セットに分割します
IQ_012_train, IQ_012_test, state_012_train, state_012_test = train_test_split(IQ_012_data, state_012, test_size=0.5)
```
最後に、モデルを設定して学習します。学習の精度が出力されます。
```
# LDAを設定します
LDA_012 = LinearDiscriminantAnalysis()
LDA_012.fit(IQ_012_train, state_012_train)
# シンプルなデータでテストします
print(LDA_012.predict([[0, 0], [-10, 0], [-15, -5]]))
# 精度を計算します
score_012 = LDA_012.score(IQ_012_test, state_012_test)
print(score_012)
```
最後のステップは、セパラトリックスのプロットです。
```
IQ_012_plot(x_min, x_max, y_min, y_max)
separatrixPlot(LDA_012, x_min, x_max, y_min, y_max, NUM_SHOTS)
```
3つの重心を得たので、セラパトリックスは線ではなく、2つの線の組み合わせを含む曲線になります。$|0\rangle$、$|1\rangle$と$|2\rangle$の状態を区別するために、私たちのモデルは、IQ上の点がセラパトリックスのどの側にあるかどこにあるかチェックし、それに応じて分類します。
## 3. 参考文献 <a id="refs"></a>
1. D. C. McKay, T. Alexander, L. Bello, M. J. Biercuk, L. Bishop, J. Chen, J. M. Chow, A. D. C ́orcoles, D. Egger, S. Filipp, J. Gomez, M. Hush, A. Javadi-Abhari, D. Moreda, P. Nation, B. Paulovicks, E. Winston, C. J. Wood, J. Wootton, and J. M. Gambetta, “Qiskit backend specifications for OpenQASM and OpenPulse experiments,” 2018, https://arxiv.org/abs/1809.03452.
2. Krantz, P. et al. “A Quantum Engineer’s Guide to Superconducting Qubits.” Applied Physics Reviews 6.2 (2019): 021318, https://arxiv.org/abs/1904.06560.
3. Scikit-learn: Machine Learning in Python, Pedregosa et al., JMLR 12, pp. 2825-2830, 2011, https://scikit-learn.org/stable/modules/lda_qda.html#id4.
```
import qiskit.tools.jupyter
%qiskit_version_table
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
from scipy.optimize import curve_fit
from scipy.signal import find_peaks
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.model_selection import train_test_split
import qiskit.pulse as pulse
import qiskit.pulse.library as pulse_lib
from qiskit.compiler import assemble
from qiskit.pulse.library import SamplePulse
from qiskit.tools.monitor import job_monitor
import warnings
warnings.filterwarnings('ignore')
from qiskit.tools.jupyter import *
%matplotlib inline
from qiskit import IBMQ
IBMQ.load_account()
provider = IBMQ.get_provider(hub='ibm-q', group='open', project='main')
backend = provider.get_backend('ibmq_armonk')
backend_config = backend.configuration()
assert backend_config.open_pulse, "Backend doesn't support Pulse"
dt = backend_config.dt
backend_defaults = backend.defaults()
# 単位変換係数 -> すべてのバックエンドのプロパティーがSI単位系(Hz, sec, etc)で返される
GHz = 1.0e9 # Gigahertz
MHz = 1.0e6 # Megahertz
us = 1.0e-6 # Microseconds
ns = 1.0e-9 # Nanoseconds
qubit = 0 # 分析に使う量子ビット
default_qubit_freq = backend_defaults.qubit_freq_est[qubit] # デフォルトの量子ビット周波数単位はHz
print(f"Qubit {qubit} has an estimated frequency of {default_qubit_freq/ GHz} GHz.")
#(各デバイスに固有の)データをスケーリング
scale_factor = 1e-14
# 実験のショット回数
NUM_SHOTS = 1024
### 必要なチャネルを収集する
drive_chan = pulse.DriveChannel(qubit)
meas_chan = pulse.MeasureChannel(qubit)
acq_chan = pulse.AcquireChannel(qubit)
def get_job_data(job, average):
"""すでに実行されているジョブからデータを取得します。
引数:
job (Job): データが必要なジョブ
average (bool): Trueの場合、データが平均であると想定してデータを取得。
Falseの場合、シングルショット用と想定してデータを取得。
返し値:
list: ジョブの結果データを含むリスト
"""
job_results = job.result(timeout=120) # タイムアウトパラメーターは120秒にセット
result_data = []
for i in range(len(job_results.results)):
if average: # 平均データを得る
result_data.append(job_results.get_memory(i)[qubit]*scale_factor)
else: # シングルデータを得る
result_data.append(job_results.get_memory(i)[:, qubit]*scale_factor)
return result_data
def get_closest_multiple_of_16(num):
"""16の倍数に最も近いものを計算します。
パルスが使えるデバイスが16サンプルの倍数の期間が必要なためです。
"""
return (int(num) - (int(num)%16))
# 駆動パルスのパラメーター (us = マイクロ秒)
drive_sigma_us = 0.075 # ガウシアンの実際の幅を決めます
drive_samples_us = drive_sigma_us*8 # 切り捨てパラメーター
# ガウシアンには自然な有限長がないためです。
drive_sigma = get_closest_multiple_of_16(drive_sigma_us * us /dt) # ガウシアンの幅の単位はdt
drive_samples = get_closest_multiple_of_16(drive_samples_us * us /dt) # 切り捨てパラメーターの単位はdt
# この量子ビットに必要な測定マップインデックスを見つける
meas_map_idx = None
for i, measure_group in enumerate(backend_config.meas_map):
if qubit in measure_group:
meas_map_idx = i
break
assert meas_map_idx is not None, f"Couldn't find qubit {qubit} in the meas_map!"
# 命令スケジュールマップからデフォルトの測定パルスを取得
inst_sched_map = backend_defaults.instruction_schedule_map
measure = inst_sched_map.get('measure', qubits=backend_config.meas_map[meas_map_idx])
def create_ground_freq_sweep_program(freqs, drive_power):
"""基底状態を励起して周波数掃引を行うプログラムを作成します。
ドライブパワーに応じて、これは0->1の周波数なのか、または0->2の周波数なのかを明らかにすることができます。
引数:
freqs (np.ndarray(dtype=float)):スイープする周波数のNumpy配列。
drive_power (float):ドライブ振幅の値。
レイズ:
ValueError:75を超える頻度を使用すると発生します。
現在、これを実行しようとすると、バックエンドでエラーが投げられます。
戻り値:
Qobj:基底状態の周波数掃引実験のプログラム。
"""
if len(freqs) > 75:
raise ValueError("You can only run 75 schedules at a time.")
# スイープ情報を表示
print(f"The frequency sweep will go from {freqs[0] / GHz} GHz to {freqs[-1]/ GHz} GHz \
using {len(freqs)} frequencies. The drive power is {drive_power}.")
# 駆動パルスを定義
ground_sweep_drive_pulse = pulse_lib.gaussian(duration=drive_samples,
sigma=drive_sigma,
amp=drive_power,
name='ground_sweep_drive_pulse')
# スイープのための周波数を定義
schedule = pulse.Schedule(name='Frequency sweep starting from ground state.')
schedule |= pulse.Play(ground_sweep_drive_pulse, drive_chan)
schedule |= measure << schedule.duration
# define frequencies for the sweep
schedule_freqs = [{drive_chan: freq} for freq in freqs]
# プログラムを組み立てる
# 注:それぞれが同じことを行うため、必要なスケジュールは1つだけです;
# スケジュールごとに、ドライブをミックスダウンするLO周波数が変化します
# これにより周波数掃引が可能になります
ground_freq_sweep_program = assemble(schedule,
backend=backend,
meas_level=1,
meas_return='avg',
shots=NUM_SHOTS,
schedule_los=schedule_freqs)
return ground_freq_sweep_program
# 75個の周波数で推定周波数の周りに40MHzを掃引します
num_freqs = 75
ground_sweep_freqs = default_qubit_freq + np.linspace(-20*MHz, 20*MHz, num_freqs)
ground_freq_sweep_program = create_ground_freq_sweep_program(ground_sweep_freqs, drive_power=0.3)
ground_freq_sweep_job = backend.run(ground_freq_sweep_program)
print(ground_freq_sweep_job.job_id())
job_monitor(ground_freq_sweep_job)
# ジョブのデータ(平均)を取得する
ground_freq_sweep_data = get_job_data(ground_freq_sweep_job, average=True)
def fit_function(x_values, y_values, function, init_params):
"""Fit a function using scipy curve_fit."""
fitparams, conv = curve_fit(function, x_values, y_values, init_params)
y_fit = function(x_values, *fitparams)
return fitparams, y_fit
# Hz単位でのフィッティングをします
(ground_sweep_fit_params,
ground_sweep_y_fit) = fit_function(ground_sweep_freqs,
ground_freq_sweep_data,
lambda x, A, q_freq, B, C: (A / np.pi) * (B / ((x - q_freq)**2 + B**2)) + C,
[7, 4.975*GHz, 1*GHz, 3*GHz] # フィッティングのための初期パラメーター
)
# 注:シグナルの実数部のみをプロットしています
plt.scatter(ground_sweep_freqs/GHz, ground_freq_sweep_data, color='black')
plt.plot(ground_sweep_freqs/GHz, ground_sweep_y_fit, color='red')
plt.xlim([min(ground_sweep_freqs/GHz), max(ground_sweep_freqs/GHz)])
plt.xlabel("Frequency [GHz]", fontsize=15)
plt.ylabel("Measured Signal [a.u.]", fontsize=15)
plt.title("0->1 Frequency Sweep", fontsize=15)
plt.show()
_, cal_qubit_freq, _, _ = ground_sweep_fit_params
print(f"We've updated our qubit frequency estimate from "
f"{round(default_qubit_freq/GHz, 7)} GHz to {round(cal_qubit_freq/GHz, 7)} GHz.")
# 実験の構成
num_rabi_points = 50 # 実験の数(つまり、掃引の振幅)
# 反復する駆動パルスの振幅値:0から0.75まで等間隔に配置された50の振幅
drive_amp_min = 0
drive_amp_max = 0.75
drive_amps = np.linspace(drive_amp_min, drive_amp_max, num_rabi_points)
# スケジュールを作成
rabi_01_schedules = []
# 駆動振幅すべてにわたってループ
for ii, drive_amp in enumerate(drive_amps):
# 駆動パルス
rabi_01_pulse = pulse_lib.gaussian(duration=drive_samples,
amp=drive_amp,
sigma=drive_sigma,
name='rabi_01_pulse_%d' % ii)
# スケジュールにコマンドを追加
schedule = pulse.Schedule(name='Rabi Experiment at drive amp = %s' % drive_amp)
schedule |= pulse.Play(rabi_01_pulse, drive_chan)
schedule |= measure << schedule.duration # 測定をドライブパルスの後にシフト
rabi_01_schedules.append(schedule)
# プログラムにスケジュールを組み込む
# 注:較正された周波数で駆動します。
rabi_01_expt_program = assemble(rabi_01_schedules,
backend=backend,
meas_level=1,
meas_return='avg',
shots=NUM_SHOTS,
schedule_los=[{drive_chan: cal_qubit_freq}]
* num_rabi_points)
rabi_01_job = backend.run(rabi_01_expt_program)
print(rabi_01_job.job_id())
job_monitor(rabi_01_job)
# ジョブのデータ(平均)を取得する
rabi_01_data = get_job_data(rabi_01_job, average=True)
def baseline_remove(values):
"""Center data around 0."""
return np.array(values) - np.mean(values)
# 注:データの実数部のみがプロットされます
rabi_01_data = np.real(baseline_remove(rabi_01_data))
(rabi_01_fit_params,
rabi_01_y_fit) = fit_function(drive_amps,
rabi_01_data,
lambda x, A, B, drive_01_period, phi: (A*np.cos(2*np.pi*x/drive_01_period - phi) + B),
[4, -4, 0.5, 0])
plt.scatter(drive_amps, rabi_01_data, color='black')
plt.plot(drive_amps, rabi_01_y_fit, color='red')
drive_01_period = rabi_01_fit_params[2]
# piの振幅計算でphiを計算
pi_amp_01 = (drive_01_period/2/np.pi) *(np.pi+rabi_01_fit_params[3])
plt.axvline(pi_amp_01, color='red', linestyle='--')
plt.axvline(pi_amp_01+drive_01_period/2, color='red', linestyle='--')
plt.annotate("", xy=(pi_amp_01+drive_01_period/2, 0), xytext=(pi_amp_01,0), arrowprops=dict(arrowstyle="<->", color='red'))
plt.annotate("$\pi$", xy=(pi_amp_01-0.03, 0.1), color='red')
plt.xlabel("Drive amp [a.u.]", fontsize=15)
plt.ylabel("Measured signal [a.u.]", fontsize=15)
plt.title('0->1 Rabi Experiment', fontsize=15)
plt.show()
print(f"Pi Amplitude (0->1) = {pi_amp_01}")
pi_pulse_01 = pulse_lib.gaussian(duration=drive_samples,
amp=pi_amp_01,
sigma=drive_sigma,
name='pi_pulse_01')
# 2つのスケジュールを作る
# 基底状態のスケジュール
zero_schedule = pulse.Schedule(name="zero schedule")
zero_schedule |= measure
# 励起状態のスケジュール
one_schedule = pulse.Schedule(name="one schedule")
one_schedule |= pulse.Play(pi_pulse_01, drive_chan)
one_schedule |= measure << one_schedule.duration
# スケジュールをプログラムにアセンブルする
IQ_01_program = assemble([zero_schedule, one_schedule],
backend=backend,
meas_level=1,
meas_return='single',
shots=NUM_SHOTS,
schedule_los=[{drive_chan: cal_qubit_freq}] * 2)
IQ_01_job = backend.run(IQ_01_program)
print(IQ_01_job.job_id())
job_monitor(IQ_01_job)
# (単一の)ジョブデータを取得します;0と1に分割します
IQ_01_data = get_job_data(IQ_01_job, average=False)
zero_data = IQ_01_data[0]
one_data = IQ_01_data[1]
def IQ_01_plot(x_min, x_max, y_min, y_max):
"""Helper function for plotting IQ plane for |0>, |1>. Limits of plot given
as arguments."""
# 0のデータは青でプロット
plt.scatter(np.real(zero_data), np.imag(zero_data),
s=5, cmap='viridis', c='blue', alpha=0.5, label=r'$|0\rangle$')
# 1のデータは赤でプロット
plt.scatter(np.real(one_data), np.imag(one_data),
s=5, cmap='viridis', c='red', alpha=0.5, label=r'$|1\rangle$')
# 0状態と1状態の平均に大きなドットをプロットします。
mean_zero = np.mean(zero_data) # 実部と虚部両方の平均を取ります。
mean_one = np.mean(one_data)
plt.scatter(np.real(mean_zero), np.imag(mean_zero),
s=200, cmap='viridis', c='black',alpha=1.0)
plt.scatter(np.real(mean_one), np.imag(mean_one),
s=200, cmap='viridis', c='black',alpha=1.0)
plt.xlim(x_min, x_max)
plt.ylim(y_min,y_max)
plt.legend()
plt.ylabel('I [a.u.]', fontsize=15)
plt.xlabel('Q [a.u.]', fontsize=15)
plt.title("0-1 discrimination", fontsize=15)
x_min = -5
x_max = 15
y_min = -5
y_max = 10
IQ_01_plot(x_min, x_max, y_min, y_max)
def reshape_complex_vec(vec):
"""
複素数ベクトルvecを取り込んで、実際のimagエントリーを含む2d配列を返します。
これは学習に必要なデータです。
Args:
vec (list):データの複素数ベクトル
戻り値:
list:(real(vec], imag(vec))で指定されたエントリー付きのベクトル
"""
length = len(vec)
vec_reshaped = np.zeros((length, 2))
for i in range(len(vec)):
vec_reshaped[i]=[np.real(vec[i]), np.imag(vec[i])]
return vec_reshaped
# IQベクトルを作成します(実部と虚部で構成されています)
zero_data_reshaped = reshape_complex_vec(zero_data)
one_data_reshaped = reshape_complex_vec(one_data)
IQ_01_data = np.concatenate((zero_data_reshaped, one_data_reshaped))
print(IQ_01_data.shape) # IQデータの形を確認します
#(テスト用に)0と1でベクトルを構築する
state_01 = np.zeros(NUM_SHOTS) # shotsは実験の回数
state_01 = np.concatenate((state_01, np.ones(NUM_SHOTS)))
print(len(state_01))
# データをシャッフルしてトレーニングセットとテストセットに分割します
IQ_01_train, IQ_01_test, state_01_train, state_01_test = train_test_split(IQ_01_data, state_01, test_size=0.5)
# LDAをセットアップします
LDA_01 = LinearDiscriminantAnalysis()
LDA_01.fit(IQ_01_train, state_01_train)
# シンプルなデータでテストします
print(LDA_01.predict([[0,0], [10, 0]]))
# 精度を計算します
score_01 = LDA_01.score(IQ_01_test, state_01_test)
print(score_01)
# セパラトリックスを表示データの上にプロットします
def separatrixPlot(lda, x_min, x_max, y_min, y_max, shots):
nx, ny = shots, shots
xx, yy = np.meshgrid(np.linspace(x_min, x_max, nx),
np.linspace(y_min, y_max, ny))
Z = lda.predict_proba(np.c_[xx.ravel(), yy.ravel()])
Z = Z[:, 1].reshape(xx.shape)
plt.contour(xx, yy, Z, [0.5], linewidths=2., colors='black')
IQ_01_plot(x_min, x_max, y_min, y_max)
separatrixPlot(LDA_01, x_min, x_max, y_min, y_max, NUM_SHOTS)
def apply_sideband(pulse, freq):
"""freq周波数でこのパルスに正弦波サイドバンドを適用します。
引数:
pulse (SamplePulse):対象のパルス。
freq (float):スイープを適用するLO周波数。
戻り値:
SamplePulse:サイドバンドが適用されたパルス(freqとcal_qubit_freqの差で振動します)。
"""
# 時間は0からdt*drive_samplesで、2*pi*f*tの形の正弦波引数になります
t_samples = np.linspace(0, dt*drive_samples, drive_samples)
sine_pulse = np.sin(2*np.pi*(freq-cal_qubit_freq)*t_samples) # no amp for the sine
# サイドバンドが適用されたサンプルパルスを作成
# 注:sq_pulse.samplesを実数にし、要素ごとに乗算する必要があります
sideband_pulse = SamplePulse(np.multiply(np.real(pulse.samples), sine_pulse), name='sideband_pulse')
return sideband_pulse
def create_excited_freq_sweep_program(freqs, drive_power):
"""|1>状態を励起することにより、周波数掃引を行うプログラムを作成します。
これにより、1-> 2の周波数を取得できます。
較正された量子ビット周波数を使用して、piパルスを介して|0>から|1>の状態になります。
|1>から|2>への周波数掃引を行うには、正弦係数を掃引駆動パルスに追加することにより、サイドバンド法を使用します。
引数:
freqs (np.ndarray(dtype=float)):掃引周波数のNumpy配列。
drive_power (float):駆動振幅の値。
レイズ:
ValueError:75を超える頻度を使用するとスローされます; 現在、75個を超える周波数を試行すると、
バックエンドでエラーがスローされます。
戻り値:
Qobj:周波数掃引実験用のプログラム。
"""
if len(freqs) > 75:
raise ValueError("You can only run 75 schedules at a time.")
print(f"The frequency sweep will go from {freqs[0] / GHz} GHz to {freqs[-1]/ GHz} GHz \
using {len(freqs)} frequencies. The drive power is {drive_power}.")
base_12_pulse = pulse_lib.gaussian(duration=drive_samples,
sigma=drive_sigma,
amp=drive_power,
name='base_12_pulse')
schedules = []
for jj, freq in enumerate(freqs):
# ガウシアンパルスにサイドバンドを追加
freq_sweep_12_pulse = apply_sideband(base_12_pulse, freq)
# スケジュールのコマンドを追加
schedule = pulse.Schedule(name="Frequency = {}".format(freq))
# 0->1のパルス、掃引パルスの周波数、測定を追加
schedule |= pulse.Play(pi_pulse_01, drive_chan)
schedule |= pulse.Play(freq_sweep_12_pulse, drive_chan) << schedule.duration
schedule |= measure << schedule.duration # 駆動パルスの後に測定をシフト
schedules.append(schedule)
num_freqs = len(freqs)
# スケジュールを表示します
display(schedules[-1].draw(channels=[drive_chan, meas_chan], label=True, scale=1.0))
# 周波数掃引プログラムを組み込みます
# 注:LOは各スケジュールでのcal_qubit_freqです;サイドバンドによって組み込みます
excited_freq_sweep_program = assemble(schedules,
backend=backend,
meas_level=1,
meas_return='avg',
shots=NUM_SHOTS,
schedule_los=[{drive_chan: cal_qubit_freq}]
* num_freqs)
return excited_freq_sweep_program
# 0->1周波数より下で1->2の周波数を見つけるために400 MHzを掃引します
num_freqs = 75
excited_sweep_freqs = cal_qubit_freq + np.linspace(-400*MHz, 30*MHz, num_freqs)
excited_freq_sweep_program = create_excited_freq_sweep_program(excited_sweep_freqs, drive_power=0.3)
# 確認のためにスケジュールの一例をプロットします
excited_freq_sweep_job = backend.run(excited_freq_sweep_program)
print(excited_freq_sweep_job.job_id())
job_monitor(excited_freq_sweep_job)
# (平均の)ジョブデータを取得します
excited_freq_sweep_data = get_job_data(excited_freq_sweep_job, average=True)
# 注:シグナルの実部だけをプロットします
plt.scatter(excited_sweep_freqs/GHz, excited_freq_sweep_data, color='black')
plt.xlim([min(excited_sweep_freqs/GHz)+0.01, max(excited_sweep_freqs/GHz)]) # ignore min point (is off)
plt.xlabel("Frequency [GHz]", fontsize=15)
plt.ylabel("Measured Signal [a.u.]", fontsize=15)
plt.title("1->2 Frequency Sweep (first pass)", fontsize=15)
plt.show()
# output_dataに相対的最小周波数を表示します;高さは下限(絶対値)を示します
def rel_maxima(freqs, output_data, height):
"""output_dataに相対的な最小周波数を出力します(ピークを確認できます);
高さは上限(絶対値)を示します。
高さを正しく設定しないと、ピークが無視されます。
引数:
freqs (list):周波数リスト
output_data (list):結果のシグナルのリスト
height (float):ピークの上限(絶対値)
戻り値:
list:相対的な最小周波数を含むリスト
"""
peaks, _ = find_peaks(output_data, height)
print("Freq. dips: ", freqs[peaks])
return freqs[peaks]
maxima = rel_maxima(excited_sweep_freqs, np.real(excited_freq_sweep_data), 10)
approx_12_freq = maxima
# 狭い範囲での掃引
num_freqs = 75
refined_excited_sweep_freqs = approx_12_freq + np.linspace(-20*MHz, 20*MHz, num_freqs)
refined_excited_freq_sweep_program = create_excited_freq_sweep_program(refined_excited_sweep_freqs, drive_power=0.3)
refined_excited_freq_sweep_job = backend.run(refined_excited_freq_sweep_program)
print(refined_excited_freq_sweep_job.job_id())
job_monitor(refined_excited_freq_sweep_job)
# より正確な(平均)データを取得する
refined_excited_freq_sweep_data = get_job_data(refined_excited_freq_sweep_job, average=True)
# Hzの単位でフィッティングする
(refined_excited_sweep_fit_params,
refined_excited_sweep_y_fit) = fit_function(refined_excited_sweep_freqs,
refined_excited_freq_sweep_data,
lambda x, A, q_freq, B, C: (A / np.pi) * (B / ((x - q_freq)**2 + B**2)) + C,
[-12, 4.625*GHz, 0.05*GHz, 3*GHz] # フィッティングのための初期パラメーター
)
# 注:シグナルの実数部のみをプロットしています
plt.scatter(refined_excited_sweep_freqs/GHz, refined_excited_freq_sweep_data, color='black')
plt.plot(refined_excited_sweep_freqs/GHz, refined_excited_sweep_y_fit, color='red')
plt.xlim([min(refined_excited_sweep_freqs/GHz), max(refined_excited_sweep_freqs/GHz)])
plt.xlabel("Frequency [GHz]", fontsize=15)
plt.ylabel("Measured Signal [a.u.]", fontsize=15)
plt.title("1->2 Frequency Sweep (refined pass)", fontsize=15)
plt.show()
_, qubit_12_freq, _, _ = refined_excited_sweep_fit_params
print(f"Our updated estimate for the 1->2 transition frequency is "
f"{round(qubit_12_freq/GHz, 7)} GHz.")
# 実験の構成
num_rabi_points = 75 # 実験数(つまり掃引する振幅)
# 駆動振幅の繰り返し値:0から1.0の間で均等に配置された75個の振幅
drive_amp_min = 0
drive_amp_max = 1.0
drive_amps = np.linspace(drive_amp_min, drive_amp_max, num_rabi_points)
# スケジュールの作成
rabi_12_schedules = []
# すべての駆動振幅をループします
for ii, drive_amp in enumerate(drive_amps):
base_12_pulse = pulse_lib.gaussian(duration=drive_samples,
sigma=drive_sigma,
amp=drive_amp,
name='base_12_pulse')
# 1->2の周波数においてサイドバンドを適用
rabi_12_pulse = apply_sideband(base_12_pulse, qubit_12_freq)
# スケジュールにコマンドを追加
schedule = pulse.Schedule(name='Rabi Experiment at drive amp = %s' % drive_amp)
schedule |= pulse.Play(pi_pulse_01, drive_chan) # 0->1
schedule |= pulse.Play(rabi_12_pulse, drive_chan) << schedule.duration # 1->2のラビパルス
schedule |= measure << schedule.duration # 駆動パルスの後に測定をシフト
rabi_12_schedules.append(schedule)
# プログラムにスケジュールを組み込みます
# 注:LO周波数はcal_qubit_freqであり、0->1のpiパルスを作ります;
# サイドバンドを使って、1->2のパルス用に変更されます
rabi_12_expt_program = assemble(rabi_12_schedules,
backend=backend,
meas_level=1,
meas_return='avg',
shots=NUM_SHOTS,
schedule_los=[{drive_chan: cal_qubit_freq}]
* num_rabi_points)
rabi_12_job = backend.run(rabi_12_expt_program)
print(rabi_12_job.job_id())
job_monitor(rabi_12_job)
# ジョブデータ(平均)を取得します
rabi_12_data = get_job_data(rabi_12_job, average=True)
# 注:信号の実部のみプロットします。
rabi_12_data = np.real(baseline_remove(rabi_12_data))
(rabi_12_fit_params,
rabi_12_y_fit) = fit_function(drive_amps,
rabi_12_data,
lambda x, A, B, drive_12_period, phi: (A*np.cos(2*np.pi*x/drive_12_period - phi) + B),
[3, 0.5, 0.9, 0])
plt.scatter(drive_amps, rabi_12_data, color='black')
plt.plot(drive_amps, rabi_12_y_fit, color='red')
drive_12_period = rabi_12_fit_params[2]
# piパルス用の振幅のためにphiを考慮します
pi_amp_12 = (drive_12_period/2/np.pi) *(np.pi+rabi_12_fit_params[3])
plt.axvline(pi_amp_12, color='red', linestyle='--')
plt.axvline(pi_amp_12+drive_12_period/2, color='red', linestyle='--')
plt.annotate("", xy=(pi_amp_12+drive_12_period/2, 0), xytext=(pi_amp_12,0), arrowprops=dict(arrowstyle="<->", color='red'))
plt.annotate("$\pi$", xy=(pi_amp_12-0.03, 0.1), color='red')
plt.xlabel("Drive amp [a.u.]", fontsize=15)
plt.ylabel("Measured signal [a.u.]", fontsize=15)
plt.title('Rabi Experiment (1->2)', fontsize=20)
plt.show()
print(f"Our updated estimate for the 1->2 transition frequency is "
f"{round(qubit_12_freq/GHz, 7)} GHz.")
print(f"Pi Amplitude (1->2) = {pi_amp_12}")
pi_pulse_12 = pulse_lib.gaussian(duration=drive_samples,
amp=pi_amp_12,
sigma=drive_sigma,
name='pi_pulse_12')
# このパルスがサイドバンドであることを再確認してください
pi_pulse_12 = apply_sideband(pi_pulse_12, qubit_12_freq)
# 3つのスケジュールを作ります
# 基底状態のスケジュール
zero_schedule = pulse.Schedule(name="zero schedule")
zero_schedule |= measure
# 励起状態のスケジュール
one_schedule = pulse.Schedule(name="one schedule")
one_schedule |= pulse.Play(pi_pulse_01, drive_chan)
one_schedule |= measure << one_schedule.duration
# 励起状態のスケジュール
two_schedule = pulse.Schedule(name="two schedule")
two_schedule |= pulse.Play(pi_pulse_01, drive_chan)
two_schedule |= pulse.Play(pi_pulse_12, drive_chan) << two_schedule.duration
two_schedule |= measure << two_schedule.duration
# プログラムにスケジュールを組み込みます
IQ_012_program = assemble([zero_schedule, one_schedule, two_schedule],
backend=backend,
meas_level=1,
meas_return='single',
shots=NUM_SHOTS,
schedule_los=[{drive_chan: cal_qubit_freq}] * 3)
IQ_012_job = backend.run(IQ_012_program)
print(IQ_012_job.job_id())
job_monitor(IQ_012_job)
# (単一の)ジョブデータを取得します;0,1,2に分割します
IQ_012_data = get_job_data(IQ_012_job, average=False)
zero_data = IQ_012_data[0]
one_data = IQ_012_data[1]
two_data = IQ_012_data[2]
def IQ_012_plot(x_min, x_max, y_min, y_max):
"""0、1、2のIQ平面をプロットするための補助関数。引数としてプロットの制限を与えます。
"""
# 0のデータは青でプロット
plt.scatter(np.real(zero_data), np.imag(zero_data),
s=5, cmap='viridis', c='blue', alpha=0.5, label=r'$|0\rangle$')
# 1のデータは赤でプロット
plt.scatter(np.real(one_data), np.imag(one_data),
s=5, cmap='viridis', c='red', alpha=0.5, label=r'$|1\rangle$')
# 2のデータは緑でプロット
plt.scatter(np.real(two_data), np.imag(two_data),
s=5, cmap='viridis', c='green', alpha=0.5, label=r'$|2\rangle$')
# 0、1、2の状態の結果の平均を大きなドットでプロット
mean_zero = np.mean(zero_data) # 実部と虚部それぞれの平均をとる
mean_one = np.mean(one_data)
mean_two = np.mean(two_data)
plt.scatter(np.real(mean_zero), np.imag(mean_zero),
s=200, cmap='viridis', c='black',alpha=1.0)
plt.scatter(np.real(mean_one), np.imag(mean_one),
s=200, cmap='viridis', c='black',alpha=1.0)
plt.scatter(np.real(mean_two), np.imag(mean_two),
s=200, cmap='viridis', c='black',alpha=1.0)
plt.xlim(x_min, x_max)
plt.ylim(y_min,y_max)
plt.legend()
plt.ylabel('I [a.u.]', fontsize=15)
plt.xlabel('Q [a.u.]', fontsize=15)
plt.title("0-1-2 discrimination", fontsize=15)
x_min = -10
x_max = 20
y_min = -25
y_max = 10
IQ_012_plot(x_min, x_max, y_min, y_max)
# IQベクトルを作成します(実部と虚部で構成されています)
zero_data_reshaped = reshape_complex_vec(zero_data)
one_data_reshaped = reshape_complex_vec(one_data)
two_data_reshaped = reshape_complex_vec(two_data)
IQ_012_data = np.concatenate((zero_data_reshaped, one_data_reshaped, two_data_reshaped))
print(IQ_012_data.shape) # IQデータの形を確認します
# (テスト用に)0と1と2の値が含まれたベクトルを構築します
state_012 = np.zeros(NUM_SHOTS) # 実験のショット数
state_012 = np.concatenate((state_012, np.ones(NUM_SHOTS)))
state_012 = np.concatenate((state_012, 2*np.ones(NUM_SHOTS)))
print(len(state_012))
# データをシャッフルして学習用セットとテスト用セットに分割します
IQ_012_train, IQ_012_test, state_012_train, state_012_test = train_test_split(IQ_012_data, state_012, test_size=0.5)
# LDAを設定します
LDA_012 = LinearDiscriminantAnalysis()
LDA_012.fit(IQ_012_train, state_012_train)
# シンプルなデータでテストします
print(LDA_012.predict([[0, 0], [-10, 0], [-15, -5]]))
# 精度を計算します
score_012 = LDA_012.score(IQ_012_test, state_012_test)
print(score_012)
IQ_012_plot(x_min, x_max, y_min, y_max)
separatrixPlot(LDA_012, x_min, x_max, y_min, y_max, NUM_SHOTS)
import qiskit.tools.jupyter
%qiskit_version_table
| 0.473414 | 0.957873 |
# Inaugural Project - Housing demand and taxation
### - *Mathilde Pilgaard, Klara Krogh Hammerum, Louise Albæk Jensen og Oluf Kelkjær*
A given household can spend cash $m$ on either housing or consumption $c$. Quality of housing, $h$, grants household utility and has the cost $p_{h}$ which is subject to progressive taxation and morgage cost as by equation (\ref{eq4}). The household aims to maximize utility such that optimal amounts of housing, $h^{*}$, and consumption, $c^{*}$, are met according to equation (\ref{eq1}).
\begin{align}
c^{*},h^{*} & = \underset{c, h}{\operatorname{argmax}} c^{1-\phi}h^{\phi} \label{eq1}\tag{1} \\
&s.t.\\
\tilde{p}_{h} &= p_{h}\varepsilon \label{eq2}\tag{2} \\
m &= \tau(p_{h},\tilde{p}_{h})+c \label{eq3}\tag{3} \\
\tau(p_{h},\tilde{p}_{h})&=r p_{h}+\tau^{g}\tilde{p}_{h}+\tau^{p}max\{\tilde{p}_{h}-\bar{p},0\} \label{eq4}\tag{4}
\end{align}
The homes are taxed according to the public assesment being $\tilde{p}_{h}$. The assesment is politically decided according to equation (\ref{eq2}) by $\varepsilon$. Available cash for a given household is split between housing costs and consumption. $r$ denotes the carrying interest for purchasing a home, and $\tau^{g}$ denotes the base housing tax while the last element in equation (\ref{eq4}) homes valued above a certain cutoff, $\bar{p}$ are taxed at an additional rate $\tau^{p}$
# Q1
Construct a function that solves household's problem above. We let the market price of a home be equal to its quality:
$$ p_{h}=h $$
and assume that the household in question has cash-on-hand $m=0.5$. Notive that the monetary units is in millions DKK. In addition use the given parameter-values.
Knowing that the marketprice of a home is equal to its quality, we can use a scalar solver using the problems monotonicity.
$$ c = m - \tau(p_{h},\tilde{p}_{h})\Leftrightarrow $$
$$ c = m - rh-\tau^{g}h\varepsilon+\tau^{p}max\{h\varepsilon-\bar{p},0 \} $$
meaning $c$ is implicit for a given level of $h$ through the budget constraint
```
# Importing relevant packages
from scipy import optimize
import numpy as np
par1 = {'m':0.5,
'phi':0.3,
'epsilon': 0.5,
'r': 0.03,
'tau_g': 0.012,
'tau_p': 0.004,
'p_bar': 3
}
# Creating utility function
def u_func(c, h, phi):
return c**(1-phi)*h**phi
# Creating objective function to minimize
def value_of_choice(h,m,phi,epsilon,r,tau_g, tau_p, p_bar):
c = m - r*h-tau_g*h*epsilon-tau_p*max(h*epsilon-p_bar,0)
return -u_func(c,h,phi)
from scipy import optimize
par1 = {'m':0.5,
'phi':0.3,
'epsilon': 0.5,
'r': 0.03,
'tau_g': 0.012,
'tau_p': 0.004,
'p_bar': 3
}
def u_optimizer(phi, m, epsilon, r, tau_g, tau_p, p_bar, print_res = False, **kwargs):
sol = optimize.minimize_scalar(lambda h: value_of_choice(h,phi, m, epsilon, r, tau_g, tau_p, p_bar),
method = 'bounded', bounds = (0,m*10))
h = sol.x
h_cost = r*h+tau_g*h*epsilon+tau_p*max(h*epsilon-p_bar,0)
c = m - h_cost
u = u_func(c,h,phi)
if print_res == True:
excess_dkk = m - h_cost - c
text = f'---'*35
text += '\nFor parameters:\n'
text += f'\u03C6={phi:.3f}'
text += f' m={m:.3f}'
text += f' \u03B5={epsilon:.3f}'
text += f' r={r:.3f}'
text += f' \u03C4ᵍ={tau_g:.3f}'
text += f' \u03C4ᵖ={tau_p:.3f}'
text += f'\nThe optimal allocation of ressources is consuming c={c:.3f} while buying a house of quality h={h:.3f} \n'
text += f'As the household pays no downpayment and only has to service interest-rate and housing taxes, the total spending amounts to:\n'
text += f'c={c:.3f} and buying a house of quality h={h:.3f} at a cost of {h_cost:.3f} leaving excess mDKK of {excess_dkk:.3f}\n\n'
text += f'Above allocation of ressources results in a utility of {u:.3f}\n'
text += f'---'*35
print(text)
return u, h, c
u_star, h_star, c_star = u_optimizer(**par1, print_res = True)
import numpy as np
N = 10000
# Initializing storage
m_array = np.linspace(0.4,4,N)
h_stars = np.empty(N)
c_stars = np.empty(N)
u_stars = np.empty(N)
# Excluding m in a new dictionary, such that it can be varied.
par1_exc = par1.copy()
del par1_exc['m']
par1_exc
for i, v in enumerate(m_array):
result_vector = u_optimizer(**par1_exc, m=v, print_res = False)
# loading results
u_stars[i] = result_vector[0]
h_stars[i] = result_vector[1]
c_stars[i] = result_vector[2]
# print(u_stars)
# Plot housing costs
# I will probably sky-rocket or at least kink at some point.
# Problem: c can be negative, shouldn't happen
# Create bounds on h, such that cost of h can't exceed some limit.
# Perhaps make the unknown h, such that c is selected and put boundary on such that c can't exceed m.
c_stars
import matplotlib.pyplot as plt
fig = plt.figure(figsize=(20,5))# figsize is in inches
ax = fig.add_subplot(1,2,1)
ax.plot(c_stars,h_stars)
ax.set_xlabel(f"$c^\star$")
ax.set_ylabel(f"$h^\star$")
ax.set_title(f'Value function')
ax_right = fig.add_subplot(1,2,2)
ax_right.plot(m_array,u_stars)
ax_right.set_xlabel(f"$m$")
ax_right.set_ylabel(f"$u^\star$")
ax_right.set_title(f'Consumption function')
plt.show()
par1_exc = par1.copy()
del par1_exc['m']
par1_exc
u_star, h_star, c_star = u_optimizer(**par1_exc, m=2)
u_star
```
|
github_jupyter
|
# Importing relevant packages
from scipy import optimize
import numpy as np
par1 = {'m':0.5,
'phi':0.3,
'epsilon': 0.5,
'r': 0.03,
'tau_g': 0.012,
'tau_p': 0.004,
'p_bar': 3
}
# Creating utility function
def u_func(c, h, phi):
return c**(1-phi)*h**phi
# Creating objective function to minimize
def value_of_choice(h,m,phi,epsilon,r,tau_g, tau_p, p_bar):
c = m - r*h-tau_g*h*epsilon-tau_p*max(h*epsilon-p_bar,0)
return -u_func(c,h,phi)
from scipy import optimize
par1 = {'m':0.5,
'phi':0.3,
'epsilon': 0.5,
'r': 0.03,
'tau_g': 0.012,
'tau_p': 0.004,
'p_bar': 3
}
def u_optimizer(phi, m, epsilon, r, tau_g, tau_p, p_bar, print_res = False, **kwargs):
sol = optimize.minimize_scalar(lambda h: value_of_choice(h,phi, m, epsilon, r, tau_g, tau_p, p_bar),
method = 'bounded', bounds = (0,m*10))
h = sol.x
h_cost = r*h+tau_g*h*epsilon+tau_p*max(h*epsilon-p_bar,0)
c = m - h_cost
u = u_func(c,h,phi)
if print_res == True:
excess_dkk = m - h_cost - c
text = f'---'*35
text += '\nFor parameters:\n'
text += f'\u03C6={phi:.3f}'
text += f' m={m:.3f}'
text += f' \u03B5={epsilon:.3f}'
text += f' r={r:.3f}'
text += f' \u03C4ᵍ={tau_g:.3f}'
text += f' \u03C4ᵖ={tau_p:.3f}'
text += f'\nThe optimal allocation of ressources is consuming c={c:.3f} while buying a house of quality h={h:.3f} \n'
text += f'As the household pays no downpayment and only has to service interest-rate and housing taxes, the total spending amounts to:\n'
text += f'c={c:.3f} and buying a house of quality h={h:.3f} at a cost of {h_cost:.3f} leaving excess mDKK of {excess_dkk:.3f}\n\n'
text += f'Above allocation of ressources results in a utility of {u:.3f}\n'
text += f'---'*35
print(text)
return u, h, c
u_star, h_star, c_star = u_optimizer(**par1, print_res = True)
import numpy as np
N = 10000
# Initializing storage
m_array = np.linspace(0.4,4,N)
h_stars = np.empty(N)
c_stars = np.empty(N)
u_stars = np.empty(N)
# Excluding m in a new dictionary, such that it can be varied.
par1_exc = par1.copy()
del par1_exc['m']
par1_exc
for i, v in enumerate(m_array):
result_vector = u_optimizer(**par1_exc, m=v, print_res = False)
# loading results
u_stars[i] = result_vector[0]
h_stars[i] = result_vector[1]
c_stars[i] = result_vector[2]
# print(u_stars)
# Plot housing costs
# I will probably sky-rocket or at least kink at some point.
# Problem: c can be negative, shouldn't happen
# Create bounds on h, such that cost of h can't exceed some limit.
# Perhaps make the unknown h, such that c is selected and put boundary on such that c can't exceed m.
c_stars
import matplotlib.pyplot as plt
fig = plt.figure(figsize=(20,5))# figsize is in inches
ax = fig.add_subplot(1,2,1)
ax.plot(c_stars,h_stars)
ax.set_xlabel(f"$c^\star$")
ax.set_ylabel(f"$h^\star$")
ax.set_title(f'Value function')
ax_right = fig.add_subplot(1,2,2)
ax_right.plot(m_array,u_stars)
ax_right.set_xlabel(f"$m$")
ax_right.set_ylabel(f"$u^\star$")
ax_right.set_title(f'Consumption function')
plt.show()
par1_exc = par1.copy()
del par1_exc['m']
par1_exc
u_star, h_star, c_star = u_optimizer(**par1_exc, m=2)
u_star
| 0.591959 | 0.938857 |
# Introduction
This notebook shows how to run a strategy against feeds from the Binance crypto exchange using the **roboquant** algo-trading framework.
<img src="https://upload.wikimedia.org/wikipedia/commons/1/12/Binance_logo.svg" alt="Binance" width="400"/>
Roboquant includes a dedicated module for crypto trading and this module also includes the support for Binance.
There are two types of feed supported, both based on the spot market prices:
- Historic feed with price bars (candlesticks)
- Live feed with price bars
We start with the following three steps to load everything required:
1. As always, we load roboquant using the ```%use``` cell magic. This will retrieve all the required roboquant packages the first time it is invoked, and imports the most commonly used functionality. Depending on your internet speed, this might take a few seconds.
2. The Welcome() statement is optional, but it provides some insights into the used environment and available memory.
```
%use @http://roboquant.org/roboquant-crypto.json
Welcome()
```
# Binance Historic Feed
We start with retrieving historic data from Binance. We have to pass two types of parameters:
1. One or more currency pairs we are interested in. In this example we are interested in Bitcoin (BTC) and ether (ETH), both trading against the Binance USD (BUSD).
> Please note how we defined a currency-pair: currency1-currency2
2. The timeframe that we want to retrieve, in this case we select the past calendar 500 days.
> Please note that the Binance API has limits imposed, so too large timeframes are not allowed.
### Optional parameters
There are also several optional parameters available:
- The interval, default being DAILY. But you can define many other values. You can use code completion to see the available values.
- Limit the number of candelsticks to be returned per currency pair, default is 1000
So a more eloborate call could look something like this:
```Kotlin
feed.retrieve("BTCBUSD", "ETHBUSD", interval = Interval.FIVE_MINUTES, timeframe = tf, limit = 250)
```
```
val feed = BinanceHistoricFeed()
val tf = Timeframe.past(500.days)
feed.retrieve("BTCBUSD", "ETHBUSD", timeframe = tf)
feed.timeframe
```
## Charts
Lets plot the data we just received. Since the feed can contain multiple assets, we also need to tell the chart which asset to draw.
Chart in roboquant are fully interactive, you can zoom and move the mouse to see details. In the top right part of the chart there is a toolbox with more options available.
> There is a dedicated notebook called ```visualization.ipynb``` that shows the different available charts
```
for (asset in feed.assets) PriceBarChart(feed, asset).render()
```
# Run Back-test
We now create the other components we want to use.
- We start with defining a simple strategy that uses moving average crossover to generate BUY and SELL signals.
- Because we won't be using USD as a base currency, we create a new SimBroker instance with an initial deposit of 1.000.000,- Binance USD (BUSD). We could also have used the default ```SimBroker``` with 1 million USD initial deposit. But in that case we would have needed to define the exchange rate from USD to BUSD, since we going to trade Bitcoin and Ether denominated in BUSD.
- We also define a metric that will help afterwards to validate the performance: ```AccountSummary()```
```
val strategy = EMACrossover.EMA_12_26
val broker = SimBroker(1_000_000, "BUSD")
val roboquant = Roboquant(strategy, AccountSummary(), broker = broker)
roboquant.run(feed)
roboquant.broker.account.summary()
val logger = roboquant.logger
logger.metricNames.summary()
```
Lets see how the overal value of our account (= cash + positions) did during the back test.
```
MetricChart(logger.getMetric("account.equity"))
```
We will now plot again the historic prices but this time also plot the actual trades that happened as part of our run. So you can easily see when a trade occured in time and the amount traded.
```
PriceBarChart(feed, feed.assets.last(), broker.account.trades)
```
# Binance Live Feed
Now we create an instance of the `BinanceLiveFeed` and subscribe to pricebar events (candlesticks) of the the BTC/USDT currency pair. It is also possible to subscribe to multiple currency pairs by providing additional parameters to the same ```subscribePriceBar``` method. Optional you can also specify an interval, the default being 1 minute price-bars.
```
val feed = BinanceLiveFeed()
feed.subscribePriceBar("BTCUSDT")
```
# Run Live Test
We use almost the same setup as in the previous section using the historic data, but this time we also add the EventRecorder. This metric will capture the received events from the live feed, so we can inspect them afterwards.
```
val strategy = EMACrossover(3, 5, minEvents = 3)
val broker = SimBroker(1_000_000.00, "USDT")
val eventRecorder = EventRecorder()
val roboquant = Roboquant(strategy, AccountSummary(), eventRecorder, broker = broker)
```
Because it is a live feed we have to specify for how long we want to run it otherwise it would run forever until interrupt the execution. In this case we run it for 60 minutes, but you can run it of course for any duration. If you run it for a short time, it is less likely that the used strategy will generate signals and as a result you won't see any trades or changes to the portfolio.
All that remains is to start the run and get some coffee.
```
val timeframe = Timeframe.next(45.minutes)
roboquant.run(feed, timeframe)
val asset = feed.assets.first()
val account = roboquant.broker.account
PriceBarChart(eventRecorder, asset)
PriceChart(eventRecorder, asset, account.trades)
val accountValue = roboquant.logger.getMetric("account.equity")
MetricChart(accountValue)
```
# Binance Paper Trading
This is still very much work in progress and is currently not available. However the goal is that once released, you can also place and manage orders on Binance using roboquant. Instead of the ```SimBroker``` used in the above examples, you only need to specify ```BinanceBroker``` and you will be able to do paper- and live trading.
There is already some ground work done, so have a look at the roboquant source code if you want to see what is already there and perhaps contribute some code.
|
github_jupyter
|
%use @http://roboquant.org/roboquant-crypto.json
Welcome()
feed.retrieve("BTCBUSD", "ETHBUSD", interval = Interval.FIVE_MINUTES, timeframe = tf, limit = 250)
val feed = BinanceHistoricFeed()
val tf = Timeframe.past(500.days)
feed.retrieve("BTCBUSD", "ETHBUSD", timeframe = tf)
feed.timeframe
for (asset in feed.assets) PriceBarChart(feed, asset).render()
Lets see how the overal value of our account (= cash + positions) did during the back test.
We will now plot again the historic prices but this time also plot the actual trades that happened as part of our run. So you can easily see when a trade occured in time and the amount traded.
# Binance Live Feed
Now we create an instance of the `BinanceLiveFeed` and subscribe to pricebar events (candlesticks) of the the BTC/USDT currency pair. It is also possible to subscribe to multiple currency pairs by providing additional parameters to the same ```subscribePriceBar``` method. Optional you can also specify an interval, the default being 1 minute price-bars.
# Run Live Test
We use almost the same setup as in the previous section using the historic data, but this time we also add the EventRecorder. This metric will capture the received events from the live feed, so we can inspect them afterwards.
Because it is a live feed we have to specify for how long we want to run it otherwise it would run forever until interrupt the execution. In this case we run it for 60 minutes, but you can run it of course for any duration. If you run it for a short time, it is less likely that the used strategy will generate signals and as a result you won't see any trades or changes to the portfolio.
All that remains is to start the run and get some coffee.
| 0.709019 | 0.985608 |
```
import os
os.environ['GOOGLE_APPLICATION_CREDENTIALS'] = '/home/ubuntu/mesolitica-tpu.json'
import string
char_vocabs = [''] + list(string.ascii_lowercase + string.digits) + [' ']
sr = 16000
maxlen = 18
maxlen_subwords = 100
minlen_text = 1
global_count = 0
from google.cloud import storage
import numpy as np
import six
def to_example(dictionary):
"""Helper: build tf.Example from (string -> int/float/str list) dictionary."""
features = {}
for (k, v) in six.iteritems(dictionary):
if not v:
raise ValueError('Empty generated field: %s' % str((k, v)))
# Subtly in PY2 vs PY3, map is not scriptable in py3. As a result,
# map objects will fail with TypeError, unless converted to a list.
if six.PY3 and isinstance(v, map):
v = list(v)
if isinstance(v[0], six.integer_types) or np.issubdtype(
type(v[0]), np.integer
):
features[k] = tf.train.Feature(
int64_list=tf.train.Int64List(value=v)
)
elif isinstance(v[0], float):
features[k] = tf.train.Feature(
float_list=tf.train.FloatList(value=v)
)
elif isinstance(v[0], six.string_types):
if not six.PY2: # Convert in python 3.
v = [bytes(x, 'utf-8') for x in v]
features[k] = tf.train.Feature(
bytes_list=tf.train.BytesList(value=v)
)
elif isinstance(v[0], bytes):
features[k] = tf.train.Feature(
bytes_list=tf.train.BytesList(value=v)
)
else:
raise ValueError(
'Value for %s is not a recognized type; v: %s type: %s'
% (k, str(v[0]), str(type(v[0])))
)
return tf.train.Example(features=tf.train.Features(feature=features))
from glob import glob
from tqdm import tqdm
files = glob('ST-CMDS-20170001_1-OS/*.txt')
len(files)
texts = []
for f in tqdm(files):
with open(f) as fopen:
data = fopen.read()
texts.append(data)
files = list(zip(files, texts))
files[0]
import tensorflow as tf
import malaya_speech
pinyin.get(files[0][1], format="strip", delimiter=" ")
os.path.exists(files[0][0].replace('.txt', '.wav'))
import re
def loop(files):
import pinyin
client = storage.Client()
bucket = client.bucket('mesolitica-tpu-general')
files, index = files
output_file = f'{index}-{global_count}.tfrecord'
writer = tf.io.TFRecordWriter(output_file)
for s in tqdm(files):
try:
t = pinyin.get(s[1], format="strip", delimiter=" ")
f = s[0].replace('.txt', '.wav')
if len(s[1]) < minlen_text:
continue
y, _ = malaya_speech.load(f)
if (len(y) / sr) > maxlen:
continue
t = ''.join([c if c in char_vocabs else ' ' for c in t])
t = re.sub(r'[ ]+', ' ', t).strip()
new_t = [char_vocabs.index(c) for c in t]
example = to_example({'waveforms': y.tolist(),
'targets': new_t,
'targets_length': [len(new_t)],
'lang': [2]})
writer.write(example.SerializeToString())
except Exception as e:
print(e)
pass
writer.close()
blob = bucket.blob(f'mandarin-v2/{output_file}')
blob.upload_from_filename(output_file)
os.system(f'rm {output_file}')
# loop((files[:100], 0))
import mp
batch_size = 5000
for i in range(0, len(files), batch_size):
batch = files[i: i + batch_size]
mp.multiprocessing(batch, loop, cores = 8, returned = False)
global_count += 1
```
|
github_jupyter
|
import os
os.environ['GOOGLE_APPLICATION_CREDENTIALS'] = '/home/ubuntu/mesolitica-tpu.json'
import string
char_vocabs = [''] + list(string.ascii_lowercase + string.digits) + [' ']
sr = 16000
maxlen = 18
maxlen_subwords = 100
minlen_text = 1
global_count = 0
from google.cloud import storage
import numpy as np
import six
def to_example(dictionary):
"""Helper: build tf.Example from (string -> int/float/str list) dictionary."""
features = {}
for (k, v) in six.iteritems(dictionary):
if not v:
raise ValueError('Empty generated field: %s' % str((k, v)))
# Subtly in PY2 vs PY3, map is not scriptable in py3. As a result,
# map objects will fail with TypeError, unless converted to a list.
if six.PY3 and isinstance(v, map):
v = list(v)
if isinstance(v[0], six.integer_types) or np.issubdtype(
type(v[0]), np.integer
):
features[k] = tf.train.Feature(
int64_list=tf.train.Int64List(value=v)
)
elif isinstance(v[0], float):
features[k] = tf.train.Feature(
float_list=tf.train.FloatList(value=v)
)
elif isinstance(v[0], six.string_types):
if not six.PY2: # Convert in python 3.
v = [bytes(x, 'utf-8') for x in v]
features[k] = tf.train.Feature(
bytes_list=tf.train.BytesList(value=v)
)
elif isinstance(v[0], bytes):
features[k] = tf.train.Feature(
bytes_list=tf.train.BytesList(value=v)
)
else:
raise ValueError(
'Value for %s is not a recognized type; v: %s type: %s'
% (k, str(v[0]), str(type(v[0])))
)
return tf.train.Example(features=tf.train.Features(feature=features))
from glob import glob
from tqdm import tqdm
files = glob('ST-CMDS-20170001_1-OS/*.txt')
len(files)
texts = []
for f in tqdm(files):
with open(f) as fopen:
data = fopen.read()
texts.append(data)
files = list(zip(files, texts))
files[0]
import tensorflow as tf
import malaya_speech
pinyin.get(files[0][1], format="strip", delimiter=" ")
os.path.exists(files[0][0].replace('.txt', '.wav'))
import re
def loop(files):
import pinyin
client = storage.Client()
bucket = client.bucket('mesolitica-tpu-general')
files, index = files
output_file = f'{index}-{global_count}.tfrecord'
writer = tf.io.TFRecordWriter(output_file)
for s in tqdm(files):
try:
t = pinyin.get(s[1], format="strip", delimiter=" ")
f = s[0].replace('.txt', '.wav')
if len(s[1]) < minlen_text:
continue
y, _ = malaya_speech.load(f)
if (len(y) / sr) > maxlen:
continue
t = ''.join([c if c in char_vocabs else ' ' for c in t])
t = re.sub(r'[ ]+', ' ', t).strip()
new_t = [char_vocabs.index(c) for c in t]
example = to_example({'waveforms': y.tolist(),
'targets': new_t,
'targets_length': [len(new_t)],
'lang': [2]})
writer.write(example.SerializeToString())
except Exception as e:
print(e)
pass
writer.close()
blob = bucket.blob(f'mandarin-v2/{output_file}')
blob.upload_from_filename(output_file)
os.system(f'rm {output_file}')
# loop((files[:100], 0))
import mp
batch_size = 5000
for i in range(0, len(files), batch_size):
batch = files[i: i + batch_size]
mp.multiprocessing(batch, loop, cores = 8, returned = False)
global_count += 1
| 0.311846 | 0.178025 |
<script async src="https://www.googletagmanager.com/gtag/js?id=UA-59152712-8"></script>
<script>
window.dataLayer = window.dataLayer || [];
function gtag(){dataLayer.push(arguments);}
gtag('js', new Date());
gtag('config', 'UA-59152712-8');
</script>
# `GiRaFFE_NRPy` C code library: Conservative-to-Primitive and Primitive-to-Conservative Solvers
## Author: Patrick Nelson
<a id='intro'></a>
**Notebook Status:** <font color=Green><b> Validated </b></font>
**Validation Notes:** These functions have been validated to round-off precision against the coresponding functions in the original `GiRaFFE`.
### NRPy+ Source Code for this module:
* [GiRaFFE_NRPy/GiRaFFE_NRPy_C2P_P2C.py](../../edit/in_progress/GiRaFFE_NRPy/GiRaFFE_NRPy_C2P_P2C.py)
## Introduction:
This writes and documents the C code that `GiRaFFE_NRPy` uses in order to update the Valencia 3-velocity at each timestep. It also computes corrections to the densitized Poynting flux in order to keep the physical quantities from violating the GRFFE constraints.
These algorithms are adapted from the original `GiRaFFE` code (see [arxiv:1704.00599v2](https://arxiv.org/abs/1704.00599v2)), based on the description in [arXiv:1310.3274v2](https://arxiv.org/abs/1310.3274v2). They have been fully NRPyfied and modified to use the Valencia 3-velocity instead of the drift velocity.
The algorithm to do this is as follows:
1. Apply fixes to ${\tilde S}_i$
1. Enforce the orthogonality of ${\tilde S}_i$ and $B^i$
* ${\tilde S}_i \rightarrow {\tilde S}_i - ({\tilde S}_j {\tilde B}^j) {\tilde B}_i/{\tilde B}^2$
1. Rescale ${\tilde S}_i$ to limit the Lorentz factor and enforce the velocity cap
* $f = \sqrt{(1-\gamma_{\max}^{-2}){\tilde B}^4/(16 \pi^2 \gamma {\tilde S}^2)}$
* ${\tilde S}_i \rightarrow {\tilde S}_i \min(1,f)$
1. Recompute the velocities at the new timestep
* $v^i = 4 \pi \gamma^{ij} {\tilde S}_j \gamma^{-1/2} B^{-2}$
1. Enforce the Current Sheet prescription
1. Zero the velocity normal to the sheet
* ${\tilde n}_i v^i = 0$
1. Recompute the Poynting flux to be consistent.
Each of these steps can be toggled on/off by changing the following NRPy+ parameters, specified in the python module:
```python
par.initialize_param(par.glb_param(type="bool", module=thismodule, parname="enforce_orthogonality_StildeD_BtildeU", defaultval=True))
par.initialize_param(par.glb_param(type="bool", module=thismodule, parname="enforce_speed_limit_StildeD", defaultval=True))
par.initialize_param(par.glb_param(type="bool", module=thismodule, parname="enforce_current_sheet_prescription", defaultval=True))
```
<a id='toc'></a>
# Table of Contents
$$\label{toc}$$
This notebook is organized as follows
1. [Step 1](#c2p): The conservative-to-primitive solver
1. [Step 1.a](#ortho_s_b): Enforce the orthogonality of $\tilde{S}_i$ and $B^i$
1. [Step 1.b](#vel_cap): Rescale ${\tilde S}_i$ to limit the Lorentz factor and enforce the velocity cap
1. [Step 1.c](#update_vel): Recompute the velocities at the new timestep
1. [Step 1.d](#current_sheet): Enforce the Current Sheet prescription
1. [Step 2](#p2c): The primitive-to-conservative solver
1. [Step 3](#code_validation): Code Validation against
1. [Step 4](#latex_pdf_output): Output this notebook to $\LaTeX$-formatted PDF file
```
# Step 0: Add NRPy's directory to the path
# https://stackoverflow.com/questions/16780014/import-file-from-parent-directory
import os,sys
nrpy_dir_path = os.path.join("..")
if nrpy_dir_path not in sys.path:
sys.path.append(nrpy_dir_path)
import os
import cmdline_helper as cmd
outdir = "GiRaFFE_NRPy/GiRaFFE_Ccode_validation/"
cmd.mkdir(outdir)
```
<a id='c2p'></a>
# Step 1: The conservative-to-primitive solver \[Back to [top](#toc)\]
$$\label{c2p}$$
We start with the Conservative-to-Primitive solver. This function is called after the vector potential and Poynting vector have been evolved at a timestep and updates the velocities. The algorithm will be as follows:
1. Apply fixes to ${\tilde S}_i$
1. Enforce the orthogonality of ${\tilde S}_i$ and $B^i$
* ${\tilde S}_i \rightarrow {\tilde S}_i - ({\tilde S}_j {\tilde B}^j) {\tilde B}_i/{\tilde B}^2$
1. Rescale ${\tilde S}_i$ to limit the Lorentz factor and enforce the velocity cap
* $f = \sqrt{(1-\gamma_{\max}^{-2}){\tilde B}^4/(16 \pi^2 \gamma {\tilde S}^2)}$
* ${\tilde S}_i \rightarrow {\tilde S}_i \min(1,f)$
1. Recompute the velocities at the new timestep
* $v^i = 4 \pi \gamma^{ij} {\tilde S}_j \gamma^{-1/2} B^{-2}$
1. Enforce the Current Sheet prescription
* ${\tilde n}_i v^i = 0$
We will begin simply by creating the file. We will also `#include` the header file `<sys/time.h>` and define $\pi$.
```
from outputC import nrpyAbs # NRPy+: Core C code output module
import sympy as sp # SymPy: The Python computer algebra package upon which NRPy+ depends
import NRPy_param_funcs as par # NRPy+: Parameter interface
import grid as gri # NRPy+: Functions having to do with numerical grids
import indexedexp as ixp # NRPy+: Symbolic indexed expression (e.g., tensors, vectors, etc.) support
import reference_metric as rfm # NRPy+: Reference metric support
import GRHD.equations as GRHD # NRPy+: Generate general relativistic hydrodynamics equations
import GRFFE.equations as GRFFE # NRPy+: Generate general relativisitic force-free electrodynamics equations
thismodule = "GiRaFFE_NRPy-C2P_P2C"
# There are several C parameters that we will need in this module:
M_PI = par.Cparameters("#define",thismodule,["M_PI"], "")
GAMMA_SPEED_LIMIT = par.Cparameters("REAL",thismodule,"GAMMA_SPEED_LIMIT",10.0) # Default value based on
# IllinoisGRMHD.
# GiRaFFE default = 2000.0
gammaDD = ixp.declarerank2("gammaDD","sym01")
betaU = ixp.declarerank1("betaU")
StildeD = ixp.declarerank1("StildeD")
BU = ixp.declarerank1("BU")
alpha = sp.symbols('alpha',real=True)
sqrt4pi = sp.symbols('sqrt4pi', real=True)
ValenciavU = ixp.declarerank1("ValenciavU")
GRHD.compute_sqrtgammaDET(gammaDD)
gammaUU,unusedgammadet = ixp.symm_matrix_inverter3x3(gammaDD)
import GiRaFFE_NRPy.GiRaFFE_NRPy_C2P_P2C as C2P_P2C
```
<a id='ortho_s_b'></a>
## Step 1.a: Enforce the orthogonality of $\tilde{S}_i$ and $B^i$ \[Back to [top](#toc)\]
$$\label{ortho_s_b}$$
Now, we will enforce the orthogonality of the magnetic field and densitized poynting flux using Eq. 22 of [arxiv:1704.00599v2](https://arxiv.org/abs/1704.00599v2):
$${\tilde S}_i \rightarrow {\tilde S}_i - ({\tilde S}_j {\tilde B}^j) {\tilde B}_i/{\tilde B}^2$$
First, we compute the inner products ${\tilde S}_j {\tilde B}^j$ and ${\tilde B}^2 = \gamma_{ij} {\tilde B}^i {\tilde B}^j,$ where $\tilde{B}^i = B^i \sqrt{\gamma}$. Then, we subtract $({\tilde S}_j {\tilde B}^j) {\tilde B}_i/{\tilde B}^2$ from ${\tilde S}_i$. We thus guarantee that ${\tilde S}_i B^i=0$.
Having fixed ${\tilde S}_i$, we will also compute the related quantities ${\tilde S}^i = \gamma^{ij} {\tilde S}_j$ and ${\tilde S}^2 = {\tilde S}_i {\tilde S}^i$.
```
# First, we need to obtain the index-lowered form of Btilde^i and (Btilde^i)^2
# Recall that Btilde^i = sqrtgamma*B^i
BtildeU = ixp.zerorank1()
for i in range(3):
# \tilde{B}^i = B^i \sqrt{\gamma}
BtildeU[i] = GRHD.sqrtgammaDET*BU[i]
BtildeD = ixp.zerorank1()
for i in range(3):
for j in range(3):
BtildeD[j] += gammaDD[i][j]*BtildeU[i]
Btilde2 = sp.sympify(0)
for i in range(3):
Btilde2 += BtildeU[i]*BtildeD[i]
# Then, enforce the orthogonality:
if par.parval_from_str("enforce_orthogonality_StildeD_BtildeU"):
StimesB = sp.sympify(0)
for i in range(3):
StimesB += StildeD[i]*BtildeU[i]
for i in range(3):
# {\tilde S}_i = {\tilde S}_i - ({\tilde S}_j {\tilde B}^j) {\tilde B}_i/{\tilde B}^2
StildeD[i] -= StimesB*BtildeD[i]/Btilde2
```
<a id='vel_cap'></a>
## Step 1.b: Rescale ${\tilde S}_i$ to limit the Lorentz factor and enforce the velocity cap \[Back to [top](#toc)\]
$$\label{vel_cap}$$
The next fix that we will apply limits the Lorentz factor using Eqs. 92 and 93 of [arXiv:1310.3274v2](https://arxiv.org/abs/1310.3274v2). That is, we define the factor $f$ as
$$f = \sqrt{(1-\Gamma_{\max}^{-2}){\tilde B}^4/(16 \pi^2 \gamma {\tilde S}^2)}.$$
If $f<1$, we rescale the components of ${\tilde S}_i$ by $f$. That is, we must set
$${\tilde S}_i \rightarrow {\tilde S}_i \min(1,f).$$
Here, we will use a formulation of the `min()` function that does not use `if`:
\begin{equation}
\min(a,b) = \frac{1}{2} \left( a+b - \lvert a-b \rvert \right),
\end{equation}
or, in code,
```
min_noif(a,b) = sp.Rational(1,2)*(a+b-nrpyAbs(a-b))
```
```
# Calculate \tilde{S}^2:
Stilde2 = sp.sympify(0)
for i in range(3):
for j in range(3):
Stilde2 += gammaUU[i][j]*StildeD[i]*StildeD[j]
# First we need to compute the factor f:
# f = \sqrt{(1-\Gamma_{\max}^{-2}){\tilde B}^4/(16 \pi^2 \gamma {\tilde S}^2)}
speed_limit_factor = sp.sqrt((sp.sympify(1)-GAMMA_SPEED_LIMIT**(-2.0))*Btilde2*Btilde2*sp.Rational(1,16)/\
(M_PI*M_PI*GRHD.sqrtgammaDET*GRHD.sqrtgammaDET*Stilde2))
import Min_Max_and_Piecewise_Expressions as noif
# Calculate B^2
B2 = sp.sympify(0)
for i in range(3):
for j in range(3):
B2 += gammaDD[i][j]*BU[i]*BU[j]
# Enforce the speed limit on StildeD:
if par.parval_from_str("enforce_speed_limit_StildeD"):
for i in range(3):
StildeD[i] *= noif.min_noif(sp.sympify(1),speed_limit_factor)
```
<a id='update_vel'></a>
## Step 1.c: Recompute the velocities at the new timestep \[Back to [top](#toc)\]
$$\label{update_vel}$$
Finally, we can calculate the velocities. In [arxiv:1704.00599v2](https://arxiv.org/abs/1704.00599v2), Eq. 16 gives the drift velocity as
$$v^i = 4 \pi \alpha \gamma^{ij} {\tilde S}_j \gamma^{-1/2} B^{-2} - \beta^i.$$
However, we wish to use the Valencia velocity instead. Since the Valencia velocity $\bar{v}^i = \frac{1}{\alpha} \left( v^i + \beta^i \right)$, we will code
$$\bar{v}^i = 4 \pi \frac{\gamma^{ij} {\tilde S}_j}{\sqrt{\gamma} B^2}.$$
```
ValenciavU = ixp.zerorank1()
# Recompute 3-velocity:
for i in range(3):
for j in range(3):
# \bar{v}^i = 4 \pi \gamma^{ij} {\tilde S}_j / (\sqrt{\gamma} B^2)
ValenciavU[i] += sp.sympify(4)*M_PI*gammaUU[i][j]*StildeD[j]/(GRHD.sqrtgammaDET*B2)
```
<a id='current_sheet'></a>
## Step 1.d: Enforce the Current Sheet prescription \[Back to [top](#toc)\]
$$\label{current_sheet}$$
Now, we seek to handle any current sheets (a physically important phenomenon) that might form. This algorithm, given as Eq. 23 in [arxiv:1704.00599v2](https://arxiv.org/abs/1704.00599v2), will preserve current sheets that form in the xy-plane by preventing our numerical scheme from dissipating them. After fixing the z-component of the velocity, we recompute the conservative variables $\tilde{S}_i$ to be consistent with the new velocities.
Thus, if we are within four gridpoints of $z=0$, we set the component of the velocity perpendicular to the current sheet to zero by $n_i v^i = 0$, where $n_i = \gamma_{ij} n^j$ is a unit normal to the current sheet and $n^j = \delta^{jz} = (0\ 0\ 1)$. For drift velocity, this means we just set
$$
v^z = -\frac{\gamma_{xz} v^x + \gamma_{yz} v^y}{\gamma_{zz}}.
$$
This reduces to $v^z = 0$ in flat space, as one would expect. We then express the Valencia velocity in terms of the drift velocity as $\bar{v}^i = \frac{1}{\alpha} \left( v^i + \beta^i \right)$.
```
# This number determines how far away (in grid points) we will apply the fix.
grid_points_from_z_plane = par.Cparameters("REAL",thismodule,"grid_points_from_z_plane",4.0)
if par.parval_from_str("enforce_current_sheet_prescription"):
# Calculate the drift velocity
driftvU = ixp.zerorank1()
for i in range(3):
driftvU[i] = alpha*ValenciavU[i] - betaU[i]
# The direct approach, used by the original GiRaFFE:
# v^z = -(\gamma_{xz} v^x + \gamma_{yz} v^y) / \gamma_{zz}
newdriftvU2 = -(gammaDD[0][2]*driftvU[0] + gammaDD[1][2]*driftvU[1])/gammaDD[2][2]
# Now that we have the z component, it's time to substitute its Valencia form in.
# Remember, we only do this if abs(z) < (k+0.01)*dz. Note that we add 0.01; this helps
# avoid floating point errors and division by zero. This is the same as abs(z) - (k+0.01)*dz<0
coord = nrpyAbs(rfm.xx[2])
bound =(grid_points_from_z_plane+sp.Rational(1,100))*gri.dxx[2]
ValenciavU[2] = noif.coord_leq_bound(coord,bound)*(newdriftvU2+betaU[2])/alpha \
+ noif.coord_greater_bound(coord,bound)*ValenciavU[2]
```
Below is an experiment in coding this more abstractly. While it works, it's a bit harder to follow than the direct approach, which is what is coded above
```python
# Set the Cartesian normal vector. This can be expanded later to arbitrary sheets and coordinate systems.
nU = ixp.zerorank1()
nU[2] = 1
# Lower the index, as usual:
nD = ixp.zerorank1()
for i in range(3):
for j in range(3):
nD[i] = gammaDD[i][j]*nU[j]
if par.parval_from_str("enforce_current_sheet_prescription"):
# Calculate the drift velocity
driftvU = ixp.declarerank1("driftvU")
inner_product = sp.sympify(0)
for i in range(3):
inner_product += driftvU[i]*nD[i] # This is the portion of the drift velocity normal to the z plane
# In flat space, this is just v^z
# We'll use a sympy utility to solve for v^z. This should make it easier to generalize later
newdriftvU2 = sp.solve(inner_product,driftvU[2]) # This outputs a list with a single element.
# Take the 0th element so .subs() works right.
newdriftvU2 = newdriftvU2[0] # In flat space this reduces to v^z=0
for i in range(3):
# Now, we substitute drift velocity in terms of our preferred Valencia velocity
newdriftvU2 = newdriftvU2.subs(driftvU[i],alpha*ValenciavU[i]-betaU[i])
# Now that we have the z component, it's time to substitute its Valencia form in.
# Remember, we only do this if abs(z) < (k+0.01)*dz. Note that we add 0.01; this helps
# avoid floating point errors and division by zero. This is the same as abs(z) - (k+0.01)*dz<0
boundary = nrpyAbs(rfm.xx[2]) - (grid_points_from_z_plane+sp.Rational(1,100))*gri.dxx[2]
ValenciavU[2] = min_normal0(boundary)*(newdriftvU2+betaU[2])/alpha \
+ max_normal0(boundary)*ValenciavU[2]
```
<a id='p2c'></a>
# Step 2: The primitive-to-conservative solver \[Back to [top](#toc)\]
$$\label{p2c}$$
This function is used to recompute the conservatives $\tilde{S}_i$ after the 3-velocity is changed as part of the current sheet prescription using Eq. 21 of [arxiv:1704.00599v2](https://arxiv.org/abs/1704.00599v2). It implements the same equation used to compute the initial Poynting flux from the initial velocity: $$\tilde{S}_i = \gamma_{ij} \frac{\bar{v}^j \sqrt{\gamma}B^2}{4 \pi}$$ in terms of the Valencia 3-velocity. In the implementation here, we first calculate $B^2 = \gamma_{ij} B^i B^j$, then $v_i = \gamma_{ij} v^j$ before we calculate the equivalent expression $$\tilde{S}_i = \frac{\bar{v}_i \sqrt{\gamma}B^2}{4 \pi}.$$
Here, we will simply let the NRPy+ `GRFFE` module handle this part, since that is already validated.
```
def GiRaFFE_NRPy_P2C(gammaDD,betaU,alpha, ValenciavU,BU, sqrt4pi):
# After recalculating the 3-velocity, we need to update the poynting flux:
# We'll reset the Valencia velocity, since this will be part of a second call to outCfunction.
# First compute stress-energy tensor T4UU and T4UD:
GRHD.compute_sqrtgammaDET(gammaDD)
GRHD.u4U_in_terms_of_ValenciavU__rescale_ValenciavU_by_applying_speed_limit(alpha, betaU, gammaDD, ValenciavU)
GRFFE.compute_smallb4U_with_driftvU_for_FFE(gammaDD, betaU, alpha, GRHD.u4U_ito_ValenciavU, BU, sqrt4pi)
GRFFE.compute_smallbsquared(gammaDD, betaU, alpha, GRFFE.smallb4_with_driftv_for_FFE_U)
GRFFE.compute_TEM4UU(gammaDD, betaU, alpha, GRFFE.smallb4_with_driftv_for_FFE_U, GRFFE.smallbsquared, GRHD.u4U_ito_ValenciavU)
GRFFE.compute_TEM4UD(gammaDD, betaU, alpha, GRFFE.TEM4UU)
# Compute conservative variables in terms of primitive variables
GRHD.compute_S_tildeD(alpha, GRHD.sqrtgammaDET, GRFFE.TEM4UD)
global StildeD
StildeD = GRHD.S_tildeD
```
<a id='code_validation'></a>
# Step 3: Code Validation \[Back to [top](#toc)\]
$$\label{code_validation}$$
As a code validation check, we will verify agreement in the SymPy expressions between
1. this tutorial and
2. the NRPy+ [GiRaFFE_NRPy/GiRaFFE_NRPy_C2P_P2C.py](../../edit/in_progress/GiRaFFE_NRPy/GiRaFFE_NRPy_C2P_P2C.py) module.
```
all_passed=True
def comp_func(expr1,expr2,basename,prefixname2="C2P_P2C."):
if str(expr1-expr2)!="0":
print(basename+" - "+prefixname2+basename+" = "+ str(expr1-expr2))
all_passed=False
def gfnm(basename,idx1,idx2=None,idx3=None):
if idx2 is None:
return basename+"["+str(idx1)+"]"
if idx3 is None:
return basename+"["+str(idx1)+"]["+str(idx2)+"]"
return basename+"["+str(idx1)+"]["+str(idx2)+"]["+str(idx3)+"]"
notebook_StildeD = StildeD
StildeD = ixp.declarerank1("StildeD")
C2P_P2C.GiRaFFE_NRPy_C2P(StildeD,BU,gammaDD,betaU,alpha)
expr_list = []
exprcheck_list = []
namecheck_list = []
for i in range(3):
namecheck_list.extend([gfnm("StildeD",i),gfnm("ValenciavU",i)])
exprcheck_list.extend([C2P_P2C.outStildeD[i],C2P_P2C.ValenciavU[i]])
expr_list.extend([notebook_StildeD[i],ValenciavU[i]])
for i in range(len(expr_list)):
comp_func(expr_list[i],exprcheck_list[i],namecheck_list[i])
import sys
if all_passed:
print("ALL TESTS PASSED!")
else:
print("ERROR: AT LEAST ONE TEST DID NOT PASS")
sys.exit(1)
all_passed=True
gammaDD = ixp.declarerank2("gammaDD","sym01")
betaU = ixp.declarerank1("betaU")
ValenciavU = ixp.declarerank1("ValenciavU")
BU = ixp.declarerank1("BU")
alpha = sp.symbols('alpha',real=True)
sqrt4pi = sp.symbols('sqrt4pi', real=True)
GiRaFFE_NRPy_P2C(gammaDD,betaU,alpha, ValenciavU,BU, sqrt4pi)
C2P_P2C.GiRaFFE_NRPy_P2C(gammaDD,betaU,alpha, ValenciavU,BU, sqrt4pi)
expr_list = []
exprcheck_list = []
namecheck_list = []
for i in range(3):
namecheck_list.extend([gfnm("StildeD",i)])
exprcheck_list.extend([C2P_P2C.StildeD[i]])
expr_list.extend([StildeD[i]])
for i in range(len(expr_list)):
comp_func(expr_list[i],exprcheck_list[i],namecheck_list[i])
import sys
if all_passed:
print("ALL TESTS PASSED!")
else:
print("ERROR: AT LEAST ONE TEST DID NOT PASS")
sys.exit(1)
```
<a id='latex_pdf_output'></a>
# Step 4: Output this notebook to $\LaTeX$-formatted PDF file \[Back to [top](#toc)\]
$$\label{latex_pdf_output}$$
The following code cell converts this Jupyter notebook into a proper, clickable $\LaTeX$-formatted PDF file. After the cell is successfully run, the generated PDF may be found in the root NRPy+ tutorial directory, with filename
[Tutorial-GiRaFFE_NRPy-C2P_P2C.pdf](Tutorial-GiRaFFE_NRPy-C2P_P2C.pdf) (Note that clicking on this link may not work; you may need to open the PDF file through another means.)
```
import cmdline_helper as cmd # NRPy+: Multi-platform Python command-line interface
cmd.output_Jupyter_notebook_to_LaTeXed_PDF("Tutorial-GiRaFFE_NRPy-C2P_P2C")
```
|
github_jupyter
|
par.initialize_param(par.glb_param(type="bool", module=thismodule, parname="enforce_orthogonality_StildeD_BtildeU", defaultval=True))
par.initialize_param(par.glb_param(type="bool", module=thismodule, parname="enforce_speed_limit_StildeD", defaultval=True))
par.initialize_param(par.glb_param(type="bool", module=thismodule, parname="enforce_current_sheet_prescription", defaultval=True))
# Step 0: Add NRPy's directory to the path
# https://stackoverflow.com/questions/16780014/import-file-from-parent-directory
import os,sys
nrpy_dir_path = os.path.join("..")
if nrpy_dir_path not in sys.path:
sys.path.append(nrpy_dir_path)
import os
import cmdline_helper as cmd
outdir = "GiRaFFE_NRPy/GiRaFFE_Ccode_validation/"
cmd.mkdir(outdir)
from outputC import nrpyAbs # NRPy+: Core C code output module
import sympy as sp # SymPy: The Python computer algebra package upon which NRPy+ depends
import NRPy_param_funcs as par # NRPy+: Parameter interface
import grid as gri # NRPy+: Functions having to do with numerical grids
import indexedexp as ixp # NRPy+: Symbolic indexed expression (e.g., tensors, vectors, etc.) support
import reference_metric as rfm # NRPy+: Reference metric support
import GRHD.equations as GRHD # NRPy+: Generate general relativistic hydrodynamics equations
import GRFFE.equations as GRFFE # NRPy+: Generate general relativisitic force-free electrodynamics equations
thismodule = "GiRaFFE_NRPy-C2P_P2C"
# There are several C parameters that we will need in this module:
M_PI = par.Cparameters("#define",thismodule,["M_PI"], "")
GAMMA_SPEED_LIMIT = par.Cparameters("REAL",thismodule,"GAMMA_SPEED_LIMIT",10.0) # Default value based on
# IllinoisGRMHD.
# GiRaFFE default = 2000.0
gammaDD = ixp.declarerank2("gammaDD","sym01")
betaU = ixp.declarerank1("betaU")
StildeD = ixp.declarerank1("StildeD")
BU = ixp.declarerank1("BU")
alpha = sp.symbols('alpha',real=True)
sqrt4pi = sp.symbols('sqrt4pi', real=True)
ValenciavU = ixp.declarerank1("ValenciavU")
GRHD.compute_sqrtgammaDET(gammaDD)
gammaUU,unusedgammadet = ixp.symm_matrix_inverter3x3(gammaDD)
import GiRaFFE_NRPy.GiRaFFE_NRPy_C2P_P2C as C2P_P2C
# First, we need to obtain the index-lowered form of Btilde^i and (Btilde^i)^2
# Recall that Btilde^i = sqrtgamma*B^i
BtildeU = ixp.zerorank1()
for i in range(3):
# \tilde{B}^i = B^i \sqrt{\gamma}
BtildeU[i] = GRHD.sqrtgammaDET*BU[i]
BtildeD = ixp.zerorank1()
for i in range(3):
for j in range(3):
BtildeD[j] += gammaDD[i][j]*BtildeU[i]
Btilde2 = sp.sympify(0)
for i in range(3):
Btilde2 += BtildeU[i]*BtildeD[i]
# Then, enforce the orthogonality:
if par.parval_from_str("enforce_orthogonality_StildeD_BtildeU"):
StimesB = sp.sympify(0)
for i in range(3):
StimesB += StildeD[i]*BtildeU[i]
for i in range(3):
# {\tilde S}_i = {\tilde S}_i - ({\tilde S}_j {\tilde B}^j) {\tilde B}_i/{\tilde B}^2
StildeD[i] -= StimesB*BtildeD[i]/Btilde2
min_noif(a,b) = sp.Rational(1,2)*(a+b-nrpyAbs(a-b))
# Calculate \tilde{S}^2:
Stilde2 = sp.sympify(0)
for i in range(3):
for j in range(3):
Stilde2 += gammaUU[i][j]*StildeD[i]*StildeD[j]
# First we need to compute the factor f:
# f = \sqrt{(1-\Gamma_{\max}^{-2}){\tilde B}^4/(16 \pi^2 \gamma {\tilde S}^2)}
speed_limit_factor = sp.sqrt((sp.sympify(1)-GAMMA_SPEED_LIMIT**(-2.0))*Btilde2*Btilde2*sp.Rational(1,16)/\
(M_PI*M_PI*GRHD.sqrtgammaDET*GRHD.sqrtgammaDET*Stilde2))
import Min_Max_and_Piecewise_Expressions as noif
# Calculate B^2
B2 = sp.sympify(0)
for i in range(3):
for j in range(3):
B2 += gammaDD[i][j]*BU[i]*BU[j]
# Enforce the speed limit on StildeD:
if par.parval_from_str("enforce_speed_limit_StildeD"):
for i in range(3):
StildeD[i] *= noif.min_noif(sp.sympify(1),speed_limit_factor)
ValenciavU = ixp.zerorank1()
# Recompute 3-velocity:
for i in range(3):
for j in range(3):
# \bar{v}^i = 4 \pi \gamma^{ij} {\tilde S}_j / (\sqrt{\gamma} B^2)
ValenciavU[i] += sp.sympify(4)*M_PI*gammaUU[i][j]*StildeD[j]/(GRHD.sqrtgammaDET*B2)
# This number determines how far away (in grid points) we will apply the fix.
grid_points_from_z_plane = par.Cparameters("REAL",thismodule,"grid_points_from_z_plane",4.0)
if par.parval_from_str("enforce_current_sheet_prescription"):
# Calculate the drift velocity
driftvU = ixp.zerorank1()
for i in range(3):
driftvU[i] = alpha*ValenciavU[i] - betaU[i]
# The direct approach, used by the original GiRaFFE:
# v^z = -(\gamma_{xz} v^x + \gamma_{yz} v^y) / \gamma_{zz}
newdriftvU2 = -(gammaDD[0][2]*driftvU[0] + gammaDD[1][2]*driftvU[1])/gammaDD[2][2]
# Now that we have the z component, it's time to substitute its Valencia form in.
# Remember, we only do this if abs(z) < (k+0.01)*dz. Note that we add 0.01; this helps
# avoid floating point errors and division by zero. This is the same as abs(z) - (k+0.01)*dz<0
coord = nrpyAbs(rfm.xx[2])
bound =(grid_points_from_z_plane+sp.Rational(1,100))*gri.dxx[2]
ValenciavU[2] = noif.coord_leq_bound(coord,bound)*(newdriftvU2+betaU[2])/alpha \
+ noif.coord_greater_bound(coord,bound)*ValenciavU[2]
# Set the Cartesian normal vector. This can be expanded later to arbitrary sheets and coordinate systems.
nU = ixp.zerorank1()
nU[2] = 1
# Lower the index, as usual:
nD = ixp.zerorank1()
for i in range(3):
for j in range(3):
nD[i] = gammaDD[i][j]*nU[j]
if par.parval_from_str("enforce_current_sheet_prescription"):
# Calculate the drift velocity
driftvU = ixp.declarerank1("driftvU")
inner_product = sp.sympify(0)
for i in range(3):
inner_product += driftvU[i]*nD[i] # This is the portion of the drift velocity normal to the z plane
# In flat space, this is just v^z
# We'll use a sympy utility to solve for v^z. This should make it easier to generalize later
newdriftvU2 = sp.solve(inner_product,driftvU[2]) # This outputs a list with a single element.
# Take the 0th element so .subs() works right.
newdriftvU2 = newdriftvU2[0] # In flat space this reduces to v^z=0
for i in range(3):
# Now, we substitute drift velocity in terms of our preferred Valencia velocity
newdriftvU2 = newdriftvU2.subs(driftvU[i],alpha*ValenciavU[i]-betaU[i])
# Now that we have the z component, it's time to substitute its Valencia form in.
# Remember, we only do this if abs(z) < (k+0.01)*dz. Note that we add 0.01; this helps
# avoid floating point errors and division by zero. This is the same as abs(z) - (k+0.01)*dz<0
boundary = nrpyAbs(rfm.xx[2]) - (grid_points_from_z_plane+sp.Rational(1,100))*gri.dxx[2]
ValenciavU[2] = min_normal0(boundary)*(newdriftvU2+betaU[2])/alpha \
+ max_normal0(boundary)*ValenciavU[2]
def GiRaFFE_NRPy_P2C(gammaDD,betaU,alpha, ValenciavU,BU, sqrt4pi):
# After recalculating the 3-velocity, we need to update the poynting flux:
# We'll reset the Valencia velocity, since this will be part of a second call to outCfunction.
# First compute stress-energy tensor T4UU and T4UD:
GRHD.compute_sqrtgammaDET(gammaDD)
GRHD.u4U_in_terms_of_ValenciavU__rescale_ValenciavU_by_applying_speed_limit(alpha, betaU, gammaDD, ValenciavU)
GRFFE.compute_smallb4U_with_driftvU_for_FFE(gammaDD, betaU, alpha, GRHD.u4U_ito_ValenciavU, BU, sqrt4pi)
GRFFE.compute_smallbsquared(gammaDD, betaU, alpha, GRFFE.smallb4_with_driftv_for_FFE_U)
GRFFE.compute_TEM4UU(gammaDD, betaU, alpha, GRFFE.smallb4_with_driftv_for_FFE_U, GRFFE.smallbsquared, GRHD.u4U_ito_ValenciavU)
GRFFE.compute_TEM4UD(gammaDD, betaU, alpha, GRFFE.TEM4UU)
# Compute conservative variables in terms of primitive variables
GRHD.compute_S_tildeD(alpha, GRHD.sqrtgammaDET, GRFFE.TEM4UD)
global StildeD
StildeD = GRHD.S_tildeD
all_passed=True
def comp_func(expr1,expr2,basename,prefixname2="C2P_P2C."):
if str(expr1-expr2)!="0":
print(basename+" - "+prefixname2+basename+" = "+ str(expr1-expr2))
all_passed=False
def gfnm(basename,idx1,idx2=None,idx3=None):
if idx2 is None:
return basename+"["+str(idx1)+"]"
if idx3 is None:
return basename+"["+str(idx1)+"]["+str(idx2)+"]"
return basename+"["+str(idx1)+"]["+str(idx2)+"]["+str(idx3)+"]"
notebook_StildeD = StildeD
StildeD = ixp.declarerank1("StildeD")
C2P_P2C.GiRaFFE_NRPy_C2P(StildeD,BU,gammaDD,betaU,alpha)
expr_list = []
exprcheck_list = []
namecheck_list = []
for i in range(3):
namecheck_list.extend([gfnm("StildeD",i),gfnm("ValenciavU",i)])
exprcheck_list.extend([C2P_P2C.outStildeD[i],C2P_P2C.ValenciavU[i]])
expr_list.extend([notebook_StildeD[i],ValenciavU[i]])
for i in range(len(expr_list)):
comp_func(expr_list[i],exprcheck_list[i],namecheck_list[i])
import sys
if all_passed:
print("ALL TESTS PASSED!")
else:
print("ERROR: AT LEAST ONE TEST DID NOT PASS")
sys.exit(1)
all_passed=True
gammaDD = ixp.declarerank2("gammaDD","sym01")
betaU = ixp.declarerank1("betaU")
ValenciavU = ixp.declarerank1("ValenciavU")
BU = ixp.declarerank1("BU")
alpha = sp.symbols('alpha',real=True)
sqrt4pi = sp.symbols('sqrt4pi', real=True)
GiRaFFE_NRPy_P2C(gammaDD,betaU,alpha, ValenciavU,BU, sqrt4pi)
C2P_P2C.GiRaFFE_NRPy_P2C(gammaDD,betaU,alpha, ValenciavU,BU, sqrt4pi)
expr_list = []
exprcheck_list = []
namecheck_list = []
for i in range(3):
namecheck_list.extend([gfnm("StildeD",i)])
exprcheck_list.extend([C2P_P2C.StildeD[i]])
expr_list.extend([StildeD[i]])
for i in range(len(expr_list)):
comp_func(expr_list[i],exprcheck_list[i],namecheck_list[i])
import sys
if all_passed:
print("ALL TESTS PASSED!")
else:
print("ERROR: AT LEAST ONE TEST DID NOT PASS")
sys.exit(1)
import cmdline_helper as cmd # NRPy+: Multi-platform Python command-line interface
cmd.output_Jupyter_notebook_to_LaTeXed_PDF("Tutorial-GiRaFFE_NRPy-C2P_P2C")
| 0.540681 | 0.932944 |
# Computing Galactic Orbits of Stars with Gala
## Authors
Adrian Price-Whelan, Stephanie T. Douglas
## Learning Goals
* Query the Gaia data release 2 catalog to retrieve data for a sample of well-measured, nearby stars
* Define high-mass and low-mass stellar samples using color-magnitude selections
* Calculate orbits of the high-mass and low-mass stars within the Galaxy to show that the typically younger stars (high-mass) have smaller vertical excursions
## Keywords
coordinates, astroquery, gala, galactic dynamics, astrometry, matplotlib, scatter plot, histogram
## Companion Content
Astropy Docs: [Description of the Galactocentric frame in astropy coordinates](
http://docs.astropy.org/en/latest/generated/examples/coordinates/plot_galactocentric-frame.html#sphx-glr-generated-examples-coordinates-plot-galactocentric-frame-py)
## Summary
We will use data from the [Gaia mission](https://www.cosmos.esa.int/web/gaia) to get sky positions, distances (parallaxes), proper motions, and radial velocities for a set of stars that are close to the Sun. We will then transform these observed, heliocentric kinematic measurements to Galactocentric Cartesian coordinates and use the positions and velocities as initial conditions to compute the orbits of these stars in the galaxy using the [gala](http://gala.adrian.pw) Python package. We will compare the orbits of high-mass main sequence (i.e. young) stars to the orbits of lower-mass main sequence stars to show that young stars have smaller vertical amplitudes.
## Installing Dependencies
This tutorial depends on the Astropy affiliated packages `gala` and `astroquery`. Both of these packages can be pip-installed with:
pip install gala astroquery
## Imports
```
# astropy imports
import astropy.coordinates as coord
from astropy.table import QTable
import astropy.units as u
from astroquery.gaia import Gaia
# Third-party imports
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
%matplotlib inline
# gala imports
import gala.coordinates as gc
import gala.dynamics as gd
import gala.potential as gp
from gala.units import galactic
```
## Scientific Background
[The Gaia mission](https://www.cosmos.esa.int/web/gaia/) is an ESA mission that aims to measure the 3D positions and velocities of a large number of stars throughout the Milky Way. The primary mission objective is to enable studying the formation, structure, and evolutionary history of our Galaxy by measuring astrometry (sky position, parallax, and proper motion) for about 2 billion stars brighter than the Gaia $G$-band photometric magnitude $G \lesssim 21$. By end of mission (~2022), Gaia will also provide multi-band photometry and low-resolution spectrophotometry for these sources, along with radial or line-of-sight velocities for a subsample of about 100 million stars.
In April 2018, Gaia publicly released its first major catalog of data — data release 2 or DR2 — which provides a subset of these data to anyone with an internet connection. In this tutorial, we will use astrometry, radial velocities, and photometry for a small subset of DR2 to study the kinematics of different types of stars in the Milky Way.
## Using `astroquery` to retrieve Gaia data
We'll start by querying the [Gaia science archive](http://gea.esac.esa.int/archive/) to download astrometric and kinematic data (parallax, proper motion, radial velocity) for a sample of stars near the Sun. We'll use data exclusively from [data release 2 (DR2)](https://www.cosmos.esa.int/web/gaia/data-release-2) from the *Gaia* mission. For the demonstration here, let's grab data for a random subset of 4096 stars within a distance of 100 pc from the Sun that have high signal-to-noise astrometric measurements.
To perform the query and to retrieve the data, we'll use the *Gaia* module in the [astroquery](https://astroquery.readthedocs.io/en/latest/gaia/gaia.html) package, `astroquery.gaia`. This module expects us to provide an SQL query to select the data we want (technically it should be an [ADQL](https://gea.esac.esa.int/archive-help/adql/index.html) query, which is similar to SQL but provides some additional functionality for astronomy; to learn more about ADQL syntax and options, [this guide](https://www.gaia.ac.uk/data/gaia-data-release-1/adql-cookbook) provides an introduction). We don't need all of the columns that are available in DR2, so we'll limit our query to request the sky position (`ra`, `dec`), parallax, proper motion components (`pmra`, `pmdec`), radial velocity, and magnitudes (`phot_*_mean_mag`). More information about the available columns is in the [Gaia DR2 data model](https://gea.esac.esa.int/archive/documentation/GDR2/Gaia_archive/chap_datamodel/sec_dm_main_tables/ssec_dm_gaia_source.html).
To select stars that have high signal-to-noise parallaxes, we'll use the filter ``parallax_over_error > 10`` to select stars that have small fractional uncertainties. We'll also use the filter ``radial_velocity IS NOT null`` to only select stars that have measured radial velocities.
```
query_text = '''SELECT TOP 4096 ra, dec, parallax, pmra, pmdec, radial_velocity,
phot_g_mean_mag, phot_bp_mean_mag, phot_rp_mean_mag
FROM gaiadr2.gaia_source
WHERE parallax_over_error > 10 AND
parallax > 10 AND
radial_velocity IS NOT null
ORDER BY random_index
'''
```
We now pass this query to the `Gaia.launch_job()` class method to create an anonymous job in the `Gaia` science archive to run our query. To retrieve the results of this query as an Astropy `Table` object, we then use the `job.get_results()` method. Note that you may receive a number of warnings (output lines that begin with ``WARNING:``) from the ``astropy.io.votable`` package — these are expected, and it's OK to ignore these warnings (the `Gaia` archive returns a slightly invalid VOTable).
```
# Note: the following lines require an internet connection, so we have
# provided the results of this query as a FITS file included with the
# tutorials repository. If you have an internet connection, feel free
# to uncomment these lines to retrieve the data with `astroquery`:
# job = Gaia.launch_job(query_text)
# gaia_data = job.get_results()
# gaia_data.write('gaia_data.fits')
gaia_data = QTable.read('gaia_data.fits')
```
The `data` object is now an Astropy `Table` called `gaia_data` that contains `Gaia` data for 4096 random stars within 100 pc (or with a parallax > 10 mas) of the Sun, as we requested. Let's look at the first four rows of the table:
```
gaia_data[:4]
```
Note that the table columns already contain units! They are indicated in the second row of the header.
## Using `astropy.coordinates` to represent and transform stellar positions and velocities
Let's double check that the farthest star is still within 100 pc, as we expect from the parallax selection we did in the query above. To do this, we'll create an Astropy `Distance` object using the parallax (*Note: this inverts the parallax to compute the distance! This is only a good approximation when the parallax signal to noise is large, as we ensured in the query above with `parallax_over_error > 10`*):
```
dist = coord.Distance(parallax=u.Quantity(gaia_data['parallax']))
dist.min(), dist.max()
```
It looks like the closest star in our sample is about 9 pc away, and the farthest is almost 100 pc, as we expected.
We next want to convert the coordinate position and velocity data from heliocentric, spherical values to Galactocentric, Cartesian values. We'll do this using the [Astropy coordinates](http://docs.astropy.org/en/latest/coordinates/index.html) transformation machinery. To make use of this functionality, we first have to create a `SkyCoord` object from the `Gaia` data we downloaded. The `Gaia` DR2 data are in the ICRS (equatorial) reference frame, which is also the default frame when creating new `SkyCoord` objects, so we don't need to specify the frame below:
```
c = coord.SkyCoord(ra=gaia_data['ra'],
dec=gaia_data['dec'],
distance=dist,
pm_ra_cosdec=gaia_data['pmra'],
pm_dec=gaia_data['pmdec'],
radial_velocity=gaia_data['radial_velocity'])
```
Note: as described in the [Gaia DR2 data model](https://gea.esac.esa.int/archive/documentation/GDR2/Gaia_archive/chap_datamodel/sec_dm_main_tables/ssec_dm_gaia_source.html), the Gaia column `pmra` contains the cos(dec) term. In Astropy coordinates, the name of this component is `pm_ra_cosdec`.
Let's again look at the first four coordinates in the `SkyCoord` object:
```
c[:4]
```
Now that we have a `SkyCoord` object with the Gaia data, we can transform to other coordinate systems. For example, we can transform to the `Galactic` coordinate system (centered on the Sun but with the zero latitude approximately aligned with the Galactic plane) using the `.galactic` attribute (this works for any of the [built-in Astropy coordinate frames](http://docs.astropy.org/en/latest/coordinates/index.html#reference-api), e.g., `.fk5` should also work):
```
c.galactic[:4]
```
The `Galactic` frame is still centered on the solar system barycenter, whereas we want to compute the positions and velocities of our sample of stars in a Galactocentric frame, centered on the center of the Milky Way. To do this transformation, Astropy provides the `Galactocentric` frame class, which allows us to use our own conventions for, e.g., the distance from the sun to the Galactic center (`galcen_distance`) or the height of the Sun over the Galactic midplane (`z_sun`). Let's look at the default values for the solar position and velocity:
```
coord.Galactocentric()
```
We'll instead use a distance of 8.1 kpc — more consistent with the [recent results from the GRAVITY collaboration](https://arxiv.org/abs/1807.09409) — and a solar height of 0 pc. We'll use the default solar velocity (see output above). We can transform our data to this frame using the `transform_to()` method by specifying the `Galactocentric` frame with our adopted values:
```
galcen = c.transform_to(coord.Galactocentric(z_sun=0*u.pc,
galcen_distance=8.1*u.kpc))
```
The `galcen` object now contains the data for our sample, but in the Galactocentric frame:
```
galcen[:4]
```
We can access the positions of the stars using the `.x`, `.y`, and `.z` attributes, for example:
```
plt.hist(galcen.z.value, bins=np.linspace(-110, 110, 32))
plt.xlabel('$z$ [{0:latex_inline}]'.format(galcen.z.unit));
```
Similarly, for the velocity components, we can use `.v_x`, `.v_y`, and `.v_z`. For example, to create a classic "UV" plane velocity plot:
```
fig, ax = plt.subplots(1, 1, figsize=(6, 6))
ax.plot(galcen.v_x.value, galcen.v_y.value,
marker='.', linestyle='none', alpha=0.5)
ax.set_xlim(-125, 125)
ax.set_ylim(200-125, 200+125)
ax.set_xlabel('$v_x$ [{0:latex_inline}]'.format(u.km/u.s))
ax.set_ylabel('$v_y$ [{0:latex_inline}]'.format(u.km/u.s))
```
Along with astrometric and radial velocity data, `Gaia` also provides photometric data for three photometric bandpasses: the broad-band `G`, the blue `BP`, and the red `RP` magnitudes. Let's make a Gaia color-magnitude diagram using the $G_{\rm BP}-G_{\rm RP}$ color and the absolute $G$-band magnitude $M_G$. We'll compute the absolute magnitude using the distances we computed earlier — Astropy `Distance` objects have a convenient `.distmod` attribute that provides the distance modulus:
```
M_G = gaia_data['phot_g_mean_mag'] - dist.distmod
BP_RP = gaia_data['phot_bp_mean_mag'] - gaia_data['phot_rp_mean_mag']
fig, ax = plt.subplots(1, 1, figsize=(6, 6))
ax.plot(BP_RP, M_G,
marker='.', linestyle='none', alpha=0.3)
ax.set_xlim(0, 3)
ax.set_ylim(11, 1)
ax.set_xlabel('$G_{BP}-G_{RP}$')
ax.set_ylabel('$M_G$')
```
In the above, there is a wide range of main sequence star masses which have a range of lifetimes. The most massive stars were likely born in the thin disk and their orbits therefore likely have smaller vertical amplitudes than the typical old main sequence star. To compare, we'll create two sub-selections of the Gaia CMD to select massive and low-mass main sequence stars from the CMD for comparison. You may see two ``RuntimeWarning``(s) from running the next cell — these are expected and it's safe to ignore them.
```
np.seterr(invalid="ignore")
hi_mass_mask = ((BP_RP > 0.5*u.mag) & (BP_RP < 0.7*u.mag) &
(M_G > 2*u.mag) & (M_G < 3.75*u.mag) &
(np.abs(galcen.v_y - 220*u.km/u.s) < 50*u.km/u.s))
lo_mass_mask = ((BP_RP > 2*u.mag) & (BP_RP < 2.4*u.mag) &
(M_G > 8.2*u.mag) & (M_G < 9.7*u.mag) &
(np.abs(galcen.v_y - 220*u.km/u.s) < 50*u.km/u.s))
```
Let's also define default colors to use when visualizing the high- and low-mass stars:
```
hi_mass_color = 'tab:red'
lo_mass_color = 'tab:purple'
```
Let's now visualize these two CMD selections:
```
fig, ax = plt.subplots(1, 1, figsize=(6, 6))
ax.plot(BP_RP, M_G,
marker='.', linestyle='none', alpha=0.1)
for mask, color in zip([lo_mass_mask, hi_mass_mask],
[lo_mass_color, hi_mass_color]):
ax.plot(BP_RP[mask], M_G[mask],
marker='.', linestyle='none',
alpha=0.5, color=color)
ax.set_xlim(0, 3)
ax.set_ylim(11, 1)
ax.set_xlabel('$G_{BP}-G_{RP}$')
ax.set_ylabel('$M_G$')
```
Thus far, we've used the color-magnitude diagram (using parallaxes and photometry from Gaia to compute absolute magnitudes) to select samples of high- and low-mass stars based on their colors.
In what follows, we'll compute Galactic orbits for stars in the high- and low-mass star selections above and compare.
## Using `gala` to numerically integrate Galactic stellar orbits
`gala` is an Astropy affiliated package for Galactic dynamics. `gala` provides functionality for representing analytic mass models that are commonly used in Galactic dynamics contexts for numerically integrating stellar orbits. For examples, see Chapter 3 of Binney and Tremaine (2008). The gravitational potential models are defined by specifying parameters like mass, scale radii, or shape parameters and can be combined. Once defined, they can be used in combination with numerical integrators provided in `gala` to compute orbits. `gala` comes with a pre-defined, multi-component, but [simple model for the Milky Way](http://gala.adrian.pw/en/latest/potential/define-milky-way-model.html) that can be used for orbit integrations. Let's create an instance of the `MilkyWayPotential` model and integrate orbits for the high- and low-mass main sequence stars selected above:
```
milky_way = gp.MilkyWayPotential()
milky_way
```
This model has mass components for the Galactic disk, bulge, nucleus, and halo, and the parameters were defined by fitting measurements of the Milky Way enclosed mass at various radii. See [this document](http://gala.adrian.pw/en/latest/potential/define-milky-way-model.html) for more details. The parameters of the `MilkyWayPotential` can be changed by passing in a dictionary of parameter values to argument names set by the component names. For example, to change the disk mass to make it slightly more massive (the choice `8e10` is arbitrary!):
```
different_disk_potential = gp.MilkyWayPotential(disk=dict(m=8e10*u.Msun))
different_disk_potential
```
To integrate orbits, we have to combine the mass model with a reference frame into a `Hamiltonian` object. If no reference frame is passed in, it's assumed that we are in a static inertial frame moving with the center of the mass model:
```
H = gp.Hamiltonian(milky_way)
```
Now that we have the mass model, we can integrate orbits. Let's now define initial conditions for subsets of the high- and low-mass star selections we did above. Initial conditions in `gala` are specified by creating `PhaseSpacePosition` objects. We can create these objects directly from a `Galactocentric` object, like we have defined above from transforming the Gaia data — we first have to extract the data with a Cartesian representation. We can do this by calling `galcen.cartesian`:
```
w0_hi = gd.PhaseSpacePosition(galcen[hi_mass_mask].cartesian)
w0_lo = gd.PhaseSpacePosition(galcen[lo_mass_mask].cartesian)
w0_hi.shape, w0_lo.shape
```
From the above, we can see that we have 185 high-mass star and 577 low-mass stars in our selections. To integrate orbits, we call the `.integrate_orbit()` method on the Hamiltonian object we defined above, and pass in initial conditions. We also have to specify the timestep for integration, and how long we want to integrate for. We can do this by either specifying the amount of time to integrate for, or by specifying the number of timesteps. Let's specify a timestep of 1 Myr and a time of 500 Myr (approximately two revolutions around the Galaxy for a Sun-like orbit):
```
orbits_hi = H.integrate_orbit(w0_hi, dt=1*u.Myr,
t1=0*u.Myr, t2=500*u.Myr)
orbits_lo = H.integrate_orbit(w0_lo, dt=1*u.Myr,
t1=0*u.Myr, t2=500*u.Myr)
```
By default this uses a [Leapfrog](https://en.wikipedia.org/wiki/Leapfrog_integration) numerical integration scheme, but the integrator can be customized — see the `gala` [examples](http://gala.adrian.pw/en/latest/examples/integrate-potential-example.html) for more details.
With the orbit objects in hand, we can continue our comparison of the orbits of high-mass and low-mass main sequence stars in the solar neighborhood. Let's start by plotting a few orbits. The `.plot()` convenience function provides a quick way to visualize orbits in three Cartesian projections. For example, let's plot the first orbit in each subsample on the same figure:
```
fig = orbits_hi[:, 0].plot(color=hi_mass_color)
_ = orbits_lo[:, 0].plot(axes=fig.axes, color=lo_mass_color)
```
Note in the above figure that the orbits are almost constrained to the x-y plane: the excursions are much larger in the x and y directions as compared to the z direction.
The default plots show all Cartesian projections. This can be customized to, for example, only show specified components (including velocity components):
```
fig = orbits_hi[:, 0].plot(['x', 'v_x'],
auto_aspect=False,
color=hi_mass_color)
```
The representation can also be changed, for example, to a cylindrical representation:
```
fig = orbits_hi[:, 0].cylindrical.plot(['rho', 'z'],
color=hi_mass_color,
label='high mass')
_ = orbits_lo[:, 0].cylindrical.plot(['rho', 'z'], color=lo_mass_color,
axes=fig.axes,
label='low mass')
fig.axes[0].legend(loc='upper left')
fig.axes[0].set_ylim(-0.3, 0.3)
```
Already in the above plot we can see that the high-mass star has an orbit with smaller eccentricity (smaller radial variations) and smaller vertical oscillations as compared to the low-mass star. Below, we'll quantify this and look at the vertical excursions of all of the high- and low-mass stars, respectively.
Let's now compare the vertical amplitudes of the orbits in each of our sub-selections! We can compute the (approximate) maximum vertical height of each orbit using the convenience method `.zmax()` (you can see a list of all convenience methods on the `Orbit` object [in the Gala documentation here](http://gala.adrian.pw/en/latest/api/gala.dynamics.Orbit.html#gala.dynamics.Orbit)):
```
zmax_hi = orbits_hi.zmax(approximate=True)
zmax_lo = orbits_lo.zmax(approximate=True)
```
Let's make histograms of the maximum $z$ heights for these two samples:
```
bins = np.linspace(0, 2, 50)
plt.hist(zmax_hi.value, bins=bins,
alpha=0.4, density=True, label='high-mass',
color=hi_mass_color)
plt.hist(zmax_lo.value, bins=bins,
alpha=0.4, density=True, label='low-mass',
color=lo_mass_color);
plt.legend(loc='best', fontsize=14)
plt.yscale('log')
plt.xlabel(r"$z_{\rm max}$" + " [{0:latex}]".format(zmax_hi.unit))
```
The distribution of $z$-heights for the low-mass (i.e. typically older) stars is more extended, as we predicted!
In this tutorial, we've used `astroquery` to query the Gaia science archive to retrieve kinematic and photometric data for a small sample of stars with well-measured parallaxes from Gaia DR2. We used the colors and absolute magnitudes of these stars to select subsamples of high- and low-mass stars, which, on average, will provide us with subsamples of stars that are younger and older, respectively. We then constructed a model for the gravitational field of the Milky Way and numerically integrated the orbits of all stars in each of the two subsamples. Finally, we used the orbits to compute the maximum height that each star reaches above the Galactic midplane and showed that the younger (higher-mass) stars tend to have smaller excursions from the Galactic plane, consistent with the idea that stars are either born in a "thinner" disk and dynamically "heated," or that older stars formed with a larger vertical scale-height.
## Exercises
1. Some of the low-mass star orbits have large vertical excursions from the Galactic disk (up to and above 1.5 kpc) and could therefore be stellar halo stars rather than part of the Galactic disk. Use the zmax values to select a few of these stars and plot their full orbits. Do these stars look like they are part of the disk? Why / why not?
2. [Orbit](http://gala.adrian.pw/en/latest/dynamics/orbits-in-detail.html) objects also provide methods for computing apocenter and pericenter distances and eccentricities. Which types of stars (high-mass or low-mass) tend to have high eccentricity orbits within the Galaxy? Similar to the plot above, make a plot showing the two distributions of eccentricity values.
|
github_jupyter
|
# astropy imports
import astropy.coordinates as coord
from astropy.table import QTable
import astropy.units as u
from astroquery.gaia import Gaia
# Third-party imports
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
%matplotlib inline
# gala imports
import gala.coordinates as gc
import gala.dynamics as gd
import gala.potential as gp
from gala.units import galactic
query_text = '''SELECT TOP 4096 ra, dec, parallax, pmra, pmdec, radial_velocity,
phot_g_mean_mag, phot_bp_mean_mag, phot_rp_mean_mag
FROM gaiadr2.gaia_source
WHERE parallax_over_error > 10 AND
parallax > 10 AND
radial_velocity IS NOT null
ORDER BY random_index
'''
# Note: the following lines require an internet connection, so we have
# provided the results of this query as a FITS file included with the
# tutorials repository. If you have an internet connection, feel free
# to uncomment these lines to retrieve the data with `astroquery`:
# job = Gaia.launch_job(query_text)
# gaia_data = job.get_results()
# gaia_data.write('gaia_data.fits')
gaia_data = QTable.read('gaia_data.fits')
gaia_data[:4]
dist = coord.Distance(parallax=u.Quantity(gaia_data['parallax']))
dist.min(), dist.max()
c = coord.SkyCoord(ra=gaia_data['ra'],
dec=gaia_data['dec'],
distance=dist,
pm_ra_cosdec=gaia_data['pmra'],
pm_dec=gaia_data['pmdec'],
radial_velocity=gaia_data['radial_velocity'])
c[:4]
c.galactic[:4]
coord.Galactocentric()
galcen = c.transform_to(coord.Galactocentric(z_sun=0*u.pc,
galcen_distance=8.1*u.kpc))
galcen[:4]
plt.hist(galcen.z.value, bins=np.linspace(-110, 110, 32))
plt.xlabel('$z$ [{0:latex_inline}]'.format(galcen.z.unit));
fig, ax = plt.subplots(1, 1, figsize=(6, 6))
ax.plot(galcen.v_x.value, galcen.v_y.value,
marker='.', linestyle='none', alpha=0.5)
ax.set_xlim(-125, 125)
ax.set_ylim(200-125, 200+125)
ax.set_xlabel('$v_x$ [{0:latex_inline}]'.format(u.km/u.s))
ax.set_ylabel('$v_y$ [{0:latex_inline}]'.format(u.km/u.s))
M_G = gaia_data['phot_g_mean_mag'] - dist.distmod
BP_RP = gaia_data['phot_bp_mean_mag'] - gaia_data['phot_rp_mean_mag']
fig, ax = plt.subplots(1, 1, figsize=(6, 6))
ax.plot(BP_RP, M_G,
marker='.', linestyle='none', alpha=0.3)
ax.set_xlim(0, 3)
ax.set_ylim(11, 1)
ax.set_xlabel('$G_{BP}-G_{RP}$')
ax.set_ylabel('$M_G$')
np.seterr(invalid="ignore")
hi_mass_mask = ((BP_RP > 0.5*u.mag) & (BP_RP < 0.7*u.mag) &
(M_G > 2*u.mag) & (M_G < 3.75*u.mag) &
(np.abs(galcen.v_y - 220*u.km/u.s) < 50*u.km/u.s))
lo_mass_mask = ((BP_RP > 2*u.mag) & (BP_RP < 2.4*u.mag) &
(M_G > 8.2*u.mag) & (M_G < 9.7*u.mag) &
(np.abs(galcen.v_y - 220*u.km/u.s) < 50*u.km/u.s))
hi_mass_color = 'tab:red'
lo_mass_color = 'tab:purple'
fig, ax = plt.subplots(1, 1, figsize=(6, 6))
ax.plot(BP_RP, M_G,
marker='.', linestyle='none', alpha=0.1)
for mask, color in zip([lo_mass_mask, hi_mass_mask],
[lo_mass_color, hi_mass_color]):
ax.plot(BP_RP[mask], M_G[mask],
marker='.', linestyle='none',
alpha=0.5, color=color)
ax.set_xlim(0, 3)
ax.set_ylim(11, 1)
ax.set_xlabel('$G_{BP}-G_{RP}$')
ax.set_ylabel('$M_G$')
milky_way = gp.MilkyWayPotential()
milky_way
different_disk_potential = gp.MilkyWayPotential(disk=dict(m=8e10*u.Msun))
different_disk_potential
H = gp.Hamiltonian(milky_way)
w0_hi = gd.PhaseSpacePosition(galcen[hi_mass_mask].cartesian)
w0_lo = gd.PhaseSpacePosition(galcen[lo_mass_mask].cartesian)
w0_hi.shape, w0_lo.shape
orbits_hi = H.integrate_orbit(w0_hi, dt=1*u.Myr,
t1=0*u.Myr, t2=500*u.Myr)
orbits_lo = H.integrate_orbit(w0_lo, dt=1*u.Myr,
t1=0*u.Myr, t2=500*u.Myr)
fig = orbits_hi[:, 0].plot(color=hi_mass_color)
_ = orbits_lo[:, 0].plot(axes=fig.axes, color=lo_mass_color)
fig = orbits_hi[:, 0].plot(['x', 'v_x'],
auto_aspect=False,
color=hi_mass_color)
fig = orbits_hi[:, 0].cylindrical.plot(['rho', 'z'],
color=hi_mass_color,
label='high mass')
_ = orbits_lo[:, 0].cylindrical.plot(['rho', 'z'], color=lo_mass_color,
axes=fig.axes,
label='low mass')
fig.axes[0].legend(loc='upper left')
fig.axes[0].set_ylim(-0.3, 0.3)
zmax_hi = orbits_hi.zmax(approximate=True)
zmax_lo = orbits_lo.zmax(approximate=True)
bins = np.linspace(0, 2, 50)
plt.hist(zmax_hi.value, bins=bins,
alpha=0.4, density=True, label='high-mass',
color=hi_mass_color)
plt.hist(zmax_lo.value, bins=bins,
alpha=0.4, density=True, label='low-mass',
color=lo_mass_color);
plt.legend(loc='best', fontsize=14)
plt.yscale('log')
plt.xlabel(r"$z_{\rm max}$" + " [{0:latex}]".format(zmax_hi.unit))
| 0.673621 | 0.991836 |
# Running PARC for clustering analysis of Covid-19 scRNA cells
### Introduction
Parc is a fast clustering algorithm designed to effectively cluster heterogeneity in large single cell data. We show how PARC enables downstream analysis on the recent dataset published by [Liao. et al (2020)](https://www.nature.com/articles/s41591-020-0901-9)
### Load Libraries
```
import matplotlib.pyplot as plt
import warnings
from numba.errors import NumbaPerformanceWarning
import numpy as np
import pandas as pd
import scanpy as sc
import parc
import harmonypy as hm
```
### Load Data
The data is available on [GEO GSE145926](https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145926) with each of the 12 patients in a separate .h5 file.
The result should be a matrix of shape (n_cells x n_genes) (108230, 33538).
```
datadir = "/home/shobi/Thesis/Data/Covid/GSE145926_RAW/"
file_batches = ['GSM4475051_C148_filtered_feature_bc_matrix.h5','GSM4475052_C149_filtered_feature_bc_matrix.h5','GSM4475053_C152_filtered_feature_bc_matrix.h5','GSM4475048_C51_filtered_feature_bc_matrix.h5','GSM4475049_C52_filtered_feature_bc_matrix.h5','GSM4339769_C141_filtered_feature_bc_matrix.h5','GSM4339770_C142_filtered_feature_bc_matrix.h5','GSM4339771_C143_filtered_feature_bc_matrix.h5','GSM4339772_C144_filtered_feature_bc_matrix.h5','GSM4475050_C100_filtered_feature_bc_matrix.h5','GSM4339773_C145_filtered_feature_bc_matrix.h5','GSM4339774_C146_filtered_feature_bc_matrix.h5']
patient_type =['S4','S5','S6','HC1','HC2','M1','M2','S2','M3','HC3','S1','S3']
patient_health = ['S','S','S','H','H','M','M','S','M','H','S','S']
for i in range(0,len(patient_type)):
if i ==0:
adata = sc.read_10x_h5('/home/shobi/Thesis/Data/Covid/GSE145926_RAW/'+file_batches[i])
adata.obs['patient_type'] = [patient_type[i] for i_range in range(adata.shape[0])]
adata.obs['patient_health'] = [patient_health[i] for i_range in range(adata.shape[0])]
adata.var_names_make_unique()
else:
temp = sc.read_10x_h5('/home/shobi/Thesis/Data/Covid/GSE145926_RAW/'+file_batches[i] )
temp.var_names_make_unique()
temp.obs['patient_type'] = [patient_type[i] for i_range in range(temp.shape[0])]
temp.obs['patient_health'] = [patient_health[i] for i_range in range(temp.shape[0])]
adata = adata.concatenate(temp, join='inner') #we want the genes in common
```
### Filtering and pre-processing
Following the filters used by Liao et.al (2020) and removing cells with mitochondrial gene proportion > 0.1.
After filtering the n_cells = 63753 and n_genes = 25668.
```
min_cells=3
min_genes=200
max_genes = 6000
min_counts=1000
n_top_genes=2000
n_comps_pca= 50
sc.pp.filter_genes(adata, min_cells=min_cells) # only consider genes expressed in more than min_cells
sc.pp.filter_cells(adata, min_genes=min_genes) #only consider cells with more than min_genes
sc.pp.filter_cells(adata,max_genes=max_genes) #only consider cells with less than max_cells
sc.pp.filter_cells(adata, min_counts=min_counts) #only consider cells with more than min_counts
mito_genes = adata.var_names.str.startswith('MT-')
adata.obs['percent_mito'] = np.sum(adata[:, mito_genes].X, axis=1).A1/ np.sum(adata.X, axis=1).A1
adata = adata[adata.obs.percent_mito < 0.1, :] #filter cells with high mito
adata.obs['n_counts'] = adata.X.sum(axis=1).A1 #add the total counts per cell as observations-annotation to adata
print('shape after filtering', adata.shape)
sc.pp.normalize_per_cell(adata, key_n_counts='n_counts_all' )# normalize with total UMI count per cell
sc.pp.log1p(adata)
adata.raw = adata
#select HVG
filter_result = sc.pp.filter_genes_dispersion(adata.X, flavor='cell_ranger', n_top_genes=n_top_genes, log=False ) # select highly-variable genes
adata = adata[:, filter_result.gene_subset] # subset the genes
sc.pp.scale(adata, max_value=5) # scale to unit variance and shift to zero mean. Clip values exceeding standard deviation 10.
sc.tl.pca(adata, svd_solver='arpack', n_comps=n_comps_pca)
```
### Harmony PCA to integrate the batches
```
df_meta = pd.DataFrame()
df_meta['patient_type'] = adata.obs['patient_type']
harmony_out = hm.run_harmony(adata.obsm['X_pca'], df_meta, 'patient_type')
res = harmony_out.Z_corr.T
print('size of harmony corrected output', res.shape, type(res))
```
### Run PARC clustering
```
p = parc.PARC(res, random_seed=42)
p.run_PARC()
adata.obs['parc'] = [str(i) for i in p.labels]
marker_genes = {"macrophages": ['CD68','FCN1','SPP1','FABP4'], "neutrophils": ['FCGR3B'],
"mDC": ['CD1C', 'CLEC9A'], "pDC": ['LILRA4'],
"NK": ['KLRD1'], 'T-cell': ['CD3D'], 'B-cell': ['MS4A1','IGHD', 'CD22'], 'plasma': ['IGHG4'], #CD19 doesnt show up for B
'epithel': ['TPPP3', 'KRT18']}
print('Plot cluster average expression of marker genes')
ax_mat = sc.pl.matrixplot(adata, marker_genes, groupby='parc')
graph = p.knngraph_full()
embedding= p.run_umap_hnsw(res, graph, random_state = 1)
print('completed embedding')
color_views = ['FCN1','SPP1','FABP4']
fig, axs = plt.subplots(2,3 ,figsize=(24,24))
for i in range(3):
axs[1,i].scatter(embedding[:,0], embedding[:,1], c = adata[:,color_views[i]].X.flatten(), alpha=0.4, s =3)
axs[1,i].set_title(color_views[i])
color_views_categ = ['patient_type', 'patient_health', 'parc']
import matplotlib.colors as colors
import matplotlib.cm as cmx
for i_ax in range(3):
uniq = list(set(adata.obs[color_views_categ[i_ax]]))
# Set the color map to match the number of species
z = range(1,len(uniq))
hot = plt.get_cmap('hsv')
cNorm = colors.Normalize(vmin=0, vmax=len(uniq))
scalarMap = cmx.ScalarMappable(norm=cNorm, cmap='hsv')
# Plot each species
for i in range(len(uniq)):
indx = adata.obs[color_views_categ[i_ax]] == uniq[i]
axs[0,i_ax].scatter(embedding[indx,0], embedding[indx,1], color =scalarMap.to_rgba(i), label=uniq[i], alpha=0.4, s =3)
axs[0,i_ax].set_title(color_views_categ[i_ax])
axs[0,i_ax].legend()
plt.show()
```
|
github_jupyter
|
import matplotlib.pyplot as plt
import warnings
from numba.errors import NumbaPerformanceWarning
import numpy as np
import pandas as pd
import scanpy as sc
import parc
import harmonypy as hm
datadir = "/home/shobi/Thesis/Data/Covid/GSE145926_RAW/"
file_batches = ['GSM4475051_C148_filtered_feature_bc_matrix.h5','GSM4475052_C149_filtered_feature_bc_matrix.h5','GSM4475053_C152_filtered_feature_bc_matrix.h5','GSM4475048_C51_filtered_feature_bc_matrix.h5','GSM4475049_C52_filtered_feature_bc_matrix.h5','GSM4339769_C141_filtered_feature_bc_matrix.h5','GSM4339770_C142_filtered_feature_bc_matrix.h5','GSM4339771_C143_filtered_feature_bc_matrix.h5','GSM4339772_C144_filtered_feature_bc_matrix.h5','GSM4475050_C100_filtered_feature_bc_matrix.h5','GSM4339773_C145_filtered_feature_bc_matrix.h5','GSM4339774_C146_filtered_feature_bc_matrix.h5']
patient_type =['S4','S5','S6','HC1','HC2','M1','M2','S2','M3','HC3','S1','S3']
patient_health = ['S','S','S','H','H','M','M','S','M','H','S','S']
for i in range(0,len(patient_type)):
if i ==0:
adata = sc.read_10x_h5('/home/shobi/Thesis/Data/Covid/GSE145926_RAW/'+file_batches[i])
adata.obs['patient_type'] = [patient_type[i] for i_range in range(adata.shape[0])]
adata.obs['patient_health'] = [patient_health[i] for i_range in range(adata.shape[0])]
adata.var_names_make_unique()
else:
temp = sc.read_10x_h5('/home/shobi/Thesis/Data/Covid/GSE145926_RAW/'+file_batches[i] )
temp.var_names_make_unique()
temp.obs['patient_type'] = [patient_type[i] for i_range in range(temp.shape[0])]
temp.obs['patient_health'] = [patient_health[i] for i_range in range(temp.shape[0])]
adata = adata.concatenate(temp, join='inner') #we want the genes in common
min_cells=3
min_genes=200
max_genes = 6000
min_counts=1000
n_top_genes=2000
n_comps_pca= 50
sc.pp.filter_genes(adata, min_cells=min_cells) # only consider genes expressed in more than min_cells
sc.pp.filter_cells(adata, min_genes=min_genes) #only consider cells with more than min_genes
sc.pp.filter_cells(adata,max_genes=max_genes) #only consider cells with less than max_cells
sc.pp.filter_cells(adata, min_counts=min_counts) #only consider cells with more than min_counts
mito_genes = adata.var_names.str.startswith('MT-')
adata.obs['percent_mito'] = np.sum(adata[:, mito_genes].X, axis=1).A1/ np.sum(adata.X, axis=1).A1
adata = adata[adata.obs.percent_mito < 0.1, :] #filter cells with high mito
adata.obs['n_counts'] = adata.X.sum(axis=1).A1 #add the total counts per cell as observations-annotation to adata
print('shape after filtering', adata.shape)
sc.pp.normalize_per_cell(adata, key_n_counts='n_counts_all' )# normalize with total UMI count per cell
sc.pp.log1p(adata)
adata.raw = adata
#select HVG
filter_result = sc.pp.filter_genes_dispersion(adata.X, flavor='cell_ranger', n_top_genes=n_top_genes, log=False ) # select highly-variable genes
adata = adata[:, filter_result.gene_subset] # subset the genes
sc.pp.scale(adata, max_value=5) # scale to unit variance and shift to zero mean. Clip values exceeding standard deviation 10.
sc.tl.pca(adata, svd_solver='arpack', n_comps=n_comps_pca)
df_meta = pd.DataFrame()
df_meta['patient_type'] = adata.obs['patient_type']
harmony_out = hm.run_harmony(adata.obsm['X_pca'], df_meta, 'patient_type')
res = harmony_out.Z_corr.T
print('size of harmony corrected output', res.shape, type(res))
p = parc.PARC(res, random_seed=42)
p.run_PARC()
adata.obs['parc'] = [str(i) for i in p.labels]
marker_genes = {"macrophages": ['CD68','FCN1','SPP1','FABP4'], "neutrophils": ['FCGR3B'],
"mDC": ['CD1C', 'CLEC9A'], "pDC": ['LILRA4'],
"NK": ['KLRD1'], 'T-cell': ['CD3D'], 'B-cell': ['MS4A1','IGHD', 'CD22'], 'plasma': ['IGHG4'], #CD19 doesnt show up for B
'epithel': ['TPPP3', 'KRT18']}
print('Plot cluster average expression of marker genes')
ax_mat = sc.pl.matrixplot(adata, marker_genes, groupby='parc')
graph = p.knngraph_full()
embedding= p.run_umap_hnsw(res, graph, random_state = 1)
print('completed embedding')
color_views = ['FCN1','SPP1','FABP4']
fig, axs = plt.subplots(2,3 ,figsize=(24,24))
for i in range(3):
axs[1,i].scatter(embedding[:,0], embedding[:,1], c = adata[:,color_views[i]].X.flatten(), alpha=0.4, s =3)
axs[1,i].set_title(color_views[i])
color_views_categ = ['patient_type', 'patient_health', 'parc']
import matplotlib.colors as colors
import matplotlib.cm as cmx
for i_ax in range(3):
uniq = list(set(adata.obs[color_views_categ[i_ax]]))
# Set the color map to match the number of species
z = range(1,len(uniq))
hot = plt.get_cmap('hsv')
cNorm = colors.Normalize(vmin=0, vmax=len(uniq))
scalarMap = cmx.ScalarMappable(norm=cNorm, cmap='hsv')
# Plot each species
for i in range(len(uniq)):
indx = adata.obs[color_views_categ[i_ax]] == uniq[i]
axs[0,i_ax].scatter(embedding[indx,0], embedding[indx,1], color =scalarMap.to_rgba(i), label=uniq[i], alpha=0.4, s =3)
axs[0,i_ax].set_title(color_views_categ[i_ax])
axs[0,i_ax].legend()
plt.show()
| 0.31363 | 0.88642 |
# Diversificación y fuentes de riesgo en un portafolio II - Una ilustración con mercados internacionales.
<img style="float: right; margin: 0px 0px 15px 15px;" src="https://upload.wikimedia.org/wikipedia/commons/5/5f/Map_International_Markets.jpg" width="500px" height="300px" />
> Entonces, la clase pasada vimos cómo afecta la correlación entre pares de activos en un portafolio. Dijimos que como un par de activos nunca tienen correlación perfecta, al combinarlos en un portafolio siempre conseguimos diversificación del riesgo.
> Vimos también que no todo el riesgo se puede diversificar. Dos fuentes de riesgo:
> - Sistemático: afecta de igual manera a todos los activos. No se puede diversificar.
> - Idiosincrático: afecta a cada activo en particular por razones específicas. Se puede diversificar.
En esta clase veremos un ejemplo de diversificación en un portafolio, usando datos de mercados de activos internacionales.
En el camino, definiremos términos como *frontera de mínima varianza*, *portafolio de mínima varianza* y *portafolios eficientes*, los cuales son básicos para la construcción de la **teoría moderna de portafolios**.
Estos portafolios los aprenderemos a obtener formalmente en el siguiente módulo. Por ahora nos bastará con agarrar intuición.
**Objetivo:**
- Ver los beneficios de la diversificación ilustrativamente.
- ¿Qué es la frontera de mínima varianza?
- ¿Qué son el portafolio de varianza mínima y portafolios eficientes?
*Referencia:*
- Notas del curso "Portfolio Selection and Risk Management", Rice University, disponible en Coursera.
___
## 1. Ejemplo
**Los datos:** tenemos el siguiente reporte de rendimientos esperados y volatilidad (anuales) para los mercados de acciones en los países integrantes del $G5$: EU, RU, Francia, Alemania y Japón.
```
# Importamos pandas y numpy
import pandas as pd
import numpy as np
# Resumen en base anual de rendimientos esperados y volatilidades
annual_ret_summ = pd.DataFrame(columns=['EU', 'RU', 'Francia', 'Alemania', 'Japon'], index=['Media', 'Volatilidad'])
annual_ret_summ.loc['Media'] = np.array([0.1355, 0.1589, 0.1519, 0.1435, 0.1497])
annual_ret_summ.loc['Volatilidad'] = np.array([0.1535, 0.2430, 0.2324, 0.2038, 0.2298])
annual_ret_summ.round(4)
```
¿Qué podemos notar?
- En cuanto al rendimiento esperado: son similares. El mínimo es de 13.5% y el máximo es de 15.9%.
- En cuanto al riesgo: hay mucha dispersión. Varía desde 15.3% hasta 24.3%.
Además, tenemos el siguiente reporte de la matriz de correlación:
```
# Matriz de correlación
corr = pd.DataFrame(data= np.array([[1.0000, 0.5003, 0.4398, 0.3681, 0.2663],
[0.5003, 1.0000, 0.5420, 0.4265, 0.3581],
[0.4398, 0.5420, 1.0000, 0.6032, 0.3923],
[0.3681, 0.4265, 0.6032, 1.0000, 0.3663],
[0.2663, 0.3581, 0.3923, 0.3663, 1.0000]]),
columns=annual_ret_summ.columns, index=annual_ret_summ.columns)
corr.round(4)
```
¿Qué se puede observar acerca de la matriz de correlación?
- Los índices con mayor correlación son: Francia y Alemania.
- Los índices con menor correlación son: Japón y Estados Unidos.
Recordar: correlaciones bajas significan una gran oportunidad para diversificación.
### Nos enfocaremos entonces únicamente en dos mercados: EU y Japón
- ¿Cómo construiríamos un portafolio que consiste de los mercados de acciones de EU y Japón?
- ¿Cuáles serían las posibles combinaciones?
#### 1. Supongamos que $w$ es la participación del mercado de EU en nuestro portafolio.
- ¿Cuál es la participación del mercado de Japón entonces?: $1-w$
- Luego, nuestras fórmulas de rendimiento esperado y varianza de portafolios son:
$$E[r_p]=wE[r_{EU}]+(1-w)E[r_J]$$
$$\sigma_p^2=w^2\sigma_{EU}^2+(1-w)^2\sigma_J^2+2w(1-w)\sigma_{EU,J}$$
#### 2. Con lo anterior...
- podemos variar $w$ con pasos pequeños entre $0$ y $1$, y
- calcular el rendimiento esperado y volatilidad para cada valor de $w$.
```
# Vector de w variando entre 0 y 1 con N pasos
# Rendimientos esperados individuales
# Activo1: EU, Activo2:Japon
# Volatilidades individuales
# Correlacion
# Covarianza
# Crear un DataFrame cuyas columnas sean rendimiento esperado
# y volatilidad del portafolio para cada una de las w
# generadas
```
#### 3. Finalmente,
- cada una de las combinaciones las podemos graficar en el espacio de rendimiento esperado (eje $y$) contra volatilidad (eje $x$).
```
# Importar matplotlib.pyplot
# Graficar el lugar geométrico de los portafolios en el
# espacio rendimiento esperado vs. volatilidad.
# Especificar también los puntos relativos a los casos
# extremos.
```
#### De la gráfica,
1. Ver casos extremos.
2. ¿Conviene invertir 100% en el mercado de EU? ¿Porqué?
3. ¿Porqué ocurre esto?
4. Definición: frontera de mínima varianza. Caso particular: dos activos.
5. Definición: portafolio de varianza mínima.
6. Definición: portafolios eficientes.
#### 1. Definición (frontera de mínima varianza): es el lugar geométrico de los portafolios en el espacio de rendimiento esperado contra volatilidad, que para cada nivel de rendimiento esperado provee la menor varianza (volatilidad). Para dos activos, la frontera de mínima varianza son, simplemente, todos los posibles portafolios que se pueden formar con esos dos activos.
#### 2. Definición (portafolio de mínima varianza): es el portafolio que posee la menor varianza. No podemos encontrar ningún portafolio más a la izquierda de éste, en el espacio de rendimiento esperado contra volatilidad.
#### 3. Definición (portafolios eficientes): son los portafolios que están en la parte superior de la frontera de mínima varianza, partiendo desde el portafolio de mínima varianza.
___
## 2. ¿Cómo hallar el portafolio de varianza mínima?
Bien, esta será nuestra primera selección de portafolio. Si bien se hace de manera básica e intuitiva, nos servirá como introducción al siguiente módulo.
**Comentario:** estrictamente, el portafolio que está más a la izquierda en la curva de arriba es el de *volatilidad mínima*. Sin embargo, como tanto la volatilidad es una medida siempre positiva, minimizar la volatilidad equivale a minimizar la varianza. Por lo anterior, llamamos a dicho portafolio, el portafolio de *varianza mínima*.
De modo que la búsqueda del portafolio de varianza mínima corresponde a la solución del siguiente problema de optimización:
- Para un portafolio con $n$ activos ($\boldsymbol{w}=[w_1,\dots,w_n]^T\in\mathbb{R}^n$):
\begin{align*}
&\min_{\boldsymbol{w}} & \sigma_p^2=\boldsymbol{w}^T\Sigma\boldsymbol{w}\\
&\text{s.t.} \qquad & \boldsymbol{w}\geq0,\\
& & w_1+\dots+w_n=1
\end{align*}
donde $\Sigma$ es la matriz de varianza-covarianza de los rendimientos de los $n$ activos.
- En particular, para un portafolio con dos activos el problema anterior se reduce a:
\begin{align*}
&\min_{w_1,w_2}\sigma_p^2=w_1^2\sigma_1^2+w_2^2\sigma_2^2+2w_1w_2\sigma_{12}\\
&\text{s.t.} \qquad w_1,w_2\geq0, \qquad w_1 + w_2 = 1
\end{align*}
donde $\sigma_1,\sigma_2$ son las volatilidades de los activos individuales y $\sigma_{12}$ es la covarianza entre los activos. Equivalentemente, haciendo $w_1=w$ y $w_2=1-w$, el problema anterior se puede reescribir de la siguiente manera:
\begin{align*}
&\min_{w}\sigma_p^2=w^2\sigma_1^2+(1-w)^2\sigma_2^2+2w(1-w)\rho_{12}\sigma_1\sigma_2\\
&\text{s.t.} \qquad 0\leq w\leq1,
\end{align*}
La solución al anterior problema de optimización es
$$w^{\ast}=\frac{\sigma_2^2 - \sigma_{12}}{\sigma_1^2+\sigma_2^2-2\sigma_{12}}$$
1. Los anteriores son problemas de **programación cuadrática** (función convexa sobre dominio convexo: mínimo absoluto asegurado).
2. Existen diversos algoritmos para problemas de programación cuadrática. Por ejemplo, en la librería cvxopt. Más adelante la instalaremos y la usaremos.
3. En scipy.optimize no hay un algoritmo dedicado a la solución de este tipo de problemas de optimización. Sin embargo, la función mínimize nos permite resolver problemas de optimización en general (es un poco limitada, pero nos sirve por ahora).
### 2.1. Antes de resolver el problema con la función minimize: resolverlo a mano en el tablero.
```
# Calcular w_minvar y mostrar...
```
**Conclusiones:**
- Para obtener el portafolio de mínima varianza, deberíamos invertir aproximadamente el 75.40% en el índice de EU, y el 24.60% restante en el índice de Japón.
### 2.2. Ahora sí, con la función scipy.optimize.minimize
```
# Importar la función minimize
# Función minimize
# Función objetivo
# Dato inicial
# Volatilidades individuales
# s1, s2
# Covarianza
# s12
# Cota de w
# Restricciones
# --
# Solución
# Peso del portafolio de minima varianza
# Graficar el portafolio de varianza mínima
# sobre el mismo gráfico realizado anteriormente
```
___
## 3. Ahora, para tres activos, obtengamos la frontera de mínima varianza
```
import scipy.optimize as opt
## Construcción de parámetros
## Activo 1: EU, Activo 2: Japon, Activo 3: RU
# 1. Sigma: matriz de varianza-covarianza
s1 = annual_ret_summ['EU']['Volatilidad']
s2 = annual_ret_summ['Japon']['Volatilidad']
s3 = annual_ret_summ['RU']['Volatilidad']
s12 = corr['EU']['Japon'] * s1 * s2
s13 = corr['EU']['RU'] * s1 * s3
s23 = corr['Japon']['RU'] * s2 * s3
Sigma = np.array([[s1**2, s12, s13],
[s12, s2**2, s23],
[s13, s23, s3**2]])
# 2. Eind: rendimientos esperados activos individuales
E1 = annual_ret_summ['EU']['Media']
E2 = annual_ret_summ['Japon']['Media']
E3 = annual_ret_summ['RU']['Media']
Eind = np.array([E1, E2, E3])
# 3. Ereq: rendimientos requeridos para el portafolio
# Número de portafolios
N = 100
Ereq = np.linspace(Eind.min(), Eind.max(), N)
def varianza(w, Sigma):
return w.T.dot(Sigma).dot(w)
def rendimiento_req(w, Eind, Ereq):
return Eind.T.dot(w) - Ereq
# Dato inicial
w0 = np.ones(3,) / 3
# Cotas de las variables
bnds = ((0, None), (0, None), (0, None))
# DataFrame de portafolios de la frontera
portfolios3 = pd.DataFrame(index=range(N), columns=['w1', 'w2', 'w3', 'Ret', 'Vol'])
# Construcción de los N portafolios de la frontera
for i in range(N):
# Restricciones
cons = ({'type': 'eq', 'fun': rendimiento_req, 'args': (Eind, Ereq[i])},
{'type': 'eq', 'fun': lambda w: w.sum() - 1})
# Portafolio de mínima varianza para nivel de rendimiento esperado Ereq[i]
min_var = opt.minimize(fun=varianza,
x0=w0,
args=(Sigma,),
bounds=bnds,
constraints=cons)
# Pesos, rendimientos y volatilidades de los portafolio
portfolios3.loc[i, ['w1','w2','w3']] = min_var.x
portfolios3['Ret'][i] = Eind.dot(min_var.x)
portfolios3['Vol'][i] = np.sqrt(varianza(min_var.x, Sigma))
# Portafolios de la frontera
portfolios3
# Portafolio de mínima varianza
cons = ({'type': 'eq', 'fun': lambda w: np.sum(w)-1},)
min_var3 = opt.minimize(varianza, w0, args=(Sigma,), bounds=bnds, constraints=cons)
min_var3
w_minvar3 = min_var3.x
E_minvar3 = Eind.dot(w_minvar3)
s_minvar3 = np.sqrt(varianza(w_minvar3, Sigma))
# Graficamos junto a los portafolios de solo EU y Japón
plt.figure(figsize=(12,8))
plt.plot(portafolios2['Vol'], portafolios2['Media'], label='Portafolios 2 act')
plt.plot(portfolios3.Vol, portfolios3.Ret, 'k-', lw=2, label='Portafolios 3 act')
plt.plot(s1, E1, 'b*', ms=10, label='EU')
plt.plot(s2, E2, 'r*', ms=10, label='Japon')
plt.plot(s3, E3, 'c*', ms=10, label='RU')
plt.plot(s_minvar, E_minvar, 'oy', ms=10, label='Port. Min. Var. 2')
plt.plot(s_minvar3, E_minvar3, 'om', ms=10, label='Port. Min. Var. 3')
plt.grid()
plt.legend(loc='best')
plt.xlabel('Volatilidad ($\sigma$)')
plt.ylabel('Rendimiento esperado ($E[r]$)')
#plt.axis([0.14, 0.16, 0.135, 0.14])
E_minvar, s_minvar
E_minvar3, s_minvar3
```
**Conclusión.** Mayor diversificación.
___
## 4. Comentarios acerca de la Teoría Moderna de Portafolios.
- Todo lo anterior es un abrebocas de lo que llamamos análisis de media-varianza, y que es la base de la teoría moderna de portafolios.
- El análisis de media-varianza transformó el mundo de las inversiones cuando fué presentada por primera vez.
- Claro, tiene ciertas limitaciones, pero se mantiene como una de las ideas principales en la selección óptima de portafolios.
### Historia.
1. Fue presentada por primera vez por Harry Markowitz en 1950. Acá su [artículo](https://www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=1&ved=2ahUKEwjd0cOTx8XdAhUVo4MKHcLoBhcQFjAAegQICBAC&url=https%3A%2F%2Fwww.math.ust.hk%2F~maykwok%2Fcourses%2Fma362%2F07F%2Fmarkowitz_JF.pdf&usg=AOvVaw3d29hQoNJVqXvC8zPuixYG).
2. Era un joven estudiante de Doctorado en la Universidad de Chicago.
3. Publicó su tesis doctoral en selección de portafolios en "Journal of Finance" en 1952.
4. Su contribución transformó por completo la forma en la que entendemos el riesgo.
5. Básicamente obtuvo una teoría que analiza como los inversionistas deberían escoger de manera óptima sus portafolios, en otras palabras, cómo distribuir la riqueza de manera óptima en diferentes activos.
6. Casi 40 años después, Markowitz ganó el Premio Nobel en economía por esta idea.
- La suposición detrás del análisis media-varianza es que los rendimientos de los activos pueden ser caracterizados por completo por sus rendimientos esperados y volatilidad.
- Por eso es que graficamos activos y sus combinaciones (portafolios) en el espacio de rendimiento esperado contra volatilidad.
- El análisis media-varianza es básicamente acerca de la diversificación: la interacción de activos permite que las ganancias de unos compensen las pérdidas de otros.
- La diversificación reduce el riesgo total mientras combinemos activos imperfectamente correlacionados.
- En el siguiente módulo revisaremos cómo elegir portafolios óptimos como si los inversionistas sólo se preocuparan por medias y varianzas.
- ¿Qué pasa si un inversionista también se preocupa por otros momentos (asimetría, curtosis...)?
- La belleza del análisis media-varianza es que cuando combinamos activos correlacionados imperfectamente, las varianzas siempre decrecen (no sabemos que pasa con otras medidas de riesgo).
- Si a un inversionista le preocupan otras medidas de riesgo, el análisis media-varianza no es el camino.
- Además, si eres una persona que le gusta el riesgo: quieres encontrar la próxima compañía top que apenas va arrancando (como Google en los 2000) e invertir todo en ella para generar ganancias extraordinarias; entonces la diversificación no es tampoco el camino.
- La diversificación, por definición, elimina el riesgo idiosincrático (de cada compañía), y por tanto elimina estos rendimientos altísimos que brindaría un portafolio altamente concentrado.
# Anuncios parroquiales
## 1. Recordar quiz la próxima clase.
## 2. Revisar archivo de la Tarea 5.
## 3. La próxima clase es de repaso, sin embargo, el repaso no lo hago yo, lo hacen ustedes. Estaremos resolviendo todo tipo de dudas que ustedes planteen acerca de lo visto hasta ahora. Si no hay dudas, dedicarán el tiempo de la clase a tareas del curso.
## 4. Fin Módulo 2: revisar Clase0 para ver objetivos.
<script>
$(document).ready(function(){
$('div.prompt').hide();
$('div.back-to-top').hide();
$('nav#menubar').hide();
$('.breadcrumb').hide();
$('.hidden-print').hide();
});
</script>
<footer id="attribution" style="float:right; color:#808080; background:#fff;">
Created with Jupyter by Esteban Jiménez Rodríguez.
</footer>
|
github_jupyter
|
# Importamos pandas y numpy
import pandas as pd
import numpy as np
# Resumen en base anual de rendimientos esperados y volatilidades
annual_ret_summ = pd.DataFrame(columns=['EU', 'RU', 'Francia', 'Alemania', 'Japon'], index=['Media', 'Volatilidad'])
annual_ret_summ.loc['Media'] = np.array([0.1355, 0.1589, 0.1519, 0.1435, 0.1497])
annual_ret_summ.loc['Volatilidad'] = np.array([0.1535, 0.2430, 0.2324, 0.2038, 0.2298])
annual_ret_summ.round(4)
# Matriz de correlación
corr = pd.DataFrame(data= np.array([[1.0000, 0.5003, 0.4398, 0.3681, 0.2663],
[0.5003, 1.0000, 0.5420, 0.4265, 0.3581],
[0.4398, 0.5420, 1.0000, 0.6032, 0.3923],
[0.3681, 0.4265, 0.6032, 1.0000, 0.3663],
[0.2663, 0.3581, 0.3923, 0.3663, 1.0000]]),
columns=annual_ret_summ.columns, index=annual_ret_summ.columns)
corr.round(4)
# Vector de w variando entre 0 y 1 con N pasos
# Rendimientos esperados individuales
# Activo1: EU, Activo2:Japon
# Volatilidades individuales
# Correlacion
# Covarianza
# Crear un DataFrame cuyas columnas sean rendimiento esperado
# y volatilidad del portafolio para cada una de las w
# generadas
# Importar matplotlib.pyplot
# Graficar el lugar geométrico de los portafolios en el
# espacio rendimiento esperado vs. volatilidad.
# Especificar también los puntos relativos a los casos
# extremos.
# Calcular w_minvar y mostrar...
# Importar la función minimize
# Función minimize
# Función objetivo
# Dato inicial
# Volatilidades individuales
# s1, s2
# Covarianza
# s12
# Cota de w
# Restricciones
# --
# Solución
# Peso del portafolio de minima varianza
# Graficar el portafolio de varianza mínima
# sobre el mismo gráfico realizado anteriormente
import scipy.optimize as opt
## Construcción de parámetros
## Activo 1: EU, Activo 2: Japon, Activo 3: RU
# 1. Sigma: matriz de varianza-covarianza
s1 = annual_ret_summ['EU']['Volatilidad']
s2 = annual_ret_summ['Japon']['Volatilidad']
s3 = annual_ret_summ['RU']['Volatilidad']
s12 = corr['EU']['Japon'] * s1 * s2
s13 = corr['EU']['RU'] * s1 * s3
s23 = corr['Japon']['RU'] * s2 * s3
Sigma = np.array([[s1**2, s12, s13],
[s12, s2**2, s23],
[s13, s23, s3**2]])
# 2. Eind: rendimientos esperados activos individuales
E1 = annual_ret_summ['EU']['Media']
E2 = annual_ret_summ['Japon']['Media']
E3 = annual_ret_summ['RU']['Media']
Eind = np.array([E1, E2, E3])
# 3. Ereq: rendimientos requeridos para el portafolio
# Número de portafolios
N = 100
Ereq = np.linspace(Eind.min(), Eind.max(), N)
def varianza(w, Sigma):
return w.T.dot(Sigma).dot(w)
def rendimiento_req(w, Eind, Ereq):
return Eind.T.dot(w) - Ereq
# Dato inicial
w0 = np.ones(3,) / 3
# Cotas de las variables
bnds = ((0, None), (0, None), (0, None))
# DataFrame de portafolios de la frontera
portfolios3 = pd.DataFrame(index=range(N), columns=['w1', 'w2', 'w3', 'Ret', 'Vol'])
# Construcción de los N portafolios de la frontera
for i in range(N):
# Restricciones
cons = ({'type': 'eq', 'fun': rendimiento_req, 'args': (Eind, Ereq[i])},
{'type': 'eq', 'fun': lambda w: w.sum() - 1})
# Portafolio de mínima varianza para nivel de rendimiento esperado Ereq[i]
min_var = opt.minimize(fun=varianza,
x0=w0,
args=(Sigma,),
bounds=bnds,
constraints=cons)
# Pesos, rendimientos y volatilidades de los portafolio
portfolios3.loc[i, ['w1','w2','w3']] = min_var.x
portfolios3['Ret'][i] = Eind.dot(min_var.x)
portfolios3['Vol'][i] = np.sqrt(varianza(min_var.x, Sigma))
# Portafolios de la frontera
portfolios3
# Portafolio de mínima varianza
cons = ({'type': 'eq', 'fun': lambda w: np.sum(w)-1},)
min_var3 = opt.minimize(varianza, w0, args=(Sigma,), bounds=bnds, constraints=cons)
min_var3
w_minvar3 = min_var3.x
E_minvar3 = Eind.dot(w_minvar3)
s_minvar3 = np.sqrt(varianza(w_minvar3, Sigma))
# Graficamos junto a los portafolios de solo EU y Japón
plt.figure(figsize=(12,8))
plt.plot(portafolios2['Vol'], portafolios2['Media'], label='Portafolios 2 act')
plt.plot(portfolios3.Vol, portfolios3.Ret, 'k-', lw=2, label='Portafolios 3 act')
plt.plot(s1, E1, 'b*', ms=10, label='EU')
plt.plot(s2, E2, 'r*', ms=10, label='Japon')
plt.plot(s3, E3, 'c*', ms=10, label='RU')
plt.plot(s_minvar, E_minvar, 'oy', ms=10, label='Port. Min. Var. 2')
plt.plot(s_minvar3, E_minvar3, 'om', ms=10, label='Port. Min. Var. 3')
plt.grid()
plt.legend(loc='best')
plt.xlabel('Volatilidad ($\sigma$)')
plt.ylabel('Rendimiento esperado ($E[r]$)')
#plt.axis([0.14, 0.16, 0.135, 0.14])
E_minvar, s_minvar
E_minvar3, s_minvar3
| 0.303113 | 0.987129 |
<div style="width: 100%; overflow: hidden;">
<div style="width: 150px; float: left;"> <img src="https://raw.githubusercontent.com/DataForScience/Networks/master/data/D4Sci_logo_ball.png" alt="Data For Science, Inc" align="left" border="0" width=150px> </div>
<div style="float: left; margin-left: 10px;"> <h1>Transforming Excel Analysis into pandas Data Models</h1>
<h1>Basic Excel Spreadsheets</h1>
<p>Bruno Gonçalves<br/>
<a href="http://www.data4sci.com/">www.data4sci.com</a><br/>
@bgoncalves, @data4sci</p></div>
</div>
```
from collections import Counter
from pprint import pprint
import pandas as pd
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
import openpyxl
import watermark
%load_ext watermark
%matplotlib inline
```
We start by print out the versions of the libraries we're using for future reference
```
%watermark -n -v -m -g -iv
```
Load default figure style
```
plt.style.use('./d4sci.mplstyle')
```
## Create a new empty workbook
```
book = openpyxl.Workbook()
```
Workbooks are always created with at least one sheet
```
book.sheetnames
```
We can index it directly by name:
```
sheet = book['Sheet']
print(sheet)
```
Or simply get the currently active one
```
sheet = book.active
print(sheet)
```
And to rename it we can doo
```
sheet.title = 'My Data'
```
And now we see that the name has changed, as expected
```
book.sheetnames
```
The sheet is currently empty
```
sheet.dimensions
```
Let's add some data
```
sheet['A3'] = "Hello"
sheet['B3'] = "World"
```
Now we see that we have a few more rows/columns
```
sheet.dimensions
rows = np.arange(20).reshape(10, 2)
rows
for row in rows:
sheet.append(list(row)) # Rows have to be lists or tuples
sheet.dimensions
```
And finally we can save our new workbooks
```
book.save('data/Simple.xlsx')
!open data/Simple.xlsx
```
## Load an existing Workbook
```
book = openpyxl.load_workbook('data/Simple.xlsx')
```
List all the available worksheets
```
book.sheetnames
```
Get the worksheet by name
```
sheet = book['My Data']
```
Print some statistics
```
print(sheet.dimensions)
print("Min row: %u Max row: %u" % (sheet.min_row, sheet.max_row))
print("Min col: %u Max col: %u" % (sheet.min_column, sheet.max_column))
```
Print all the values:
```
for c1, c2 in sheet[sheet.dimensions]:
print(c1.value, c2.value)
```
## Add a new sheet
```
sheet = book.create_sheet(title="My analysis", index=0)
book.sheetnames
sheet
sheet["B4"] = "The best results"
sheet["B5"] = "guaranteed!"
```
and the file again:
```
book.save("data/Simple2.xlsx")
!open data/Simple2.xlsx
```
## Remove sheets from an existing file
```
book = openpyxl.load_workbook("data/movies.xlsx")
book.sheetnames
book.remove(book["3000s"])
book.sheetnames
book.save('data/movies2.xlsx')
!open data/movies2.xlsx
```
## Extract formulas
```
book = openpyxl.load_workbook('data/excel-mortgage-calculator.xlsx')
book.sheetnames
sheet = book.active
sheet.dimensions
```
The computed values start at B17, so we print that row plus the previous one for the headers.
```
for row in sheet["B16:K17"]:
for cell in row:
print("%s%s: '%s'" % (cell.column_letter, cell.row, cell.value))
print("")
```
If instead of opened the file with data_only=True, we would get the numerical values instead
```
book = openpyxl.load_workbook('data/excel-mortgage-calculator.xlsx', data_only=True)
sheet = book.active
for row in sheet["B16:K17"]:
for cell in row:
print("%s%s: '%s'" % (cell.column_letter, cell.row, cell.value))
print("")
```
<div style="width: 100%; overflow: hidden;">
<img src="data/D4Sci_logo_full.png" alt="Data For Science, Inc" align="center" border="0" width=300px>
</div>
|
github_jupyter
|
from collections import Counter
from pprint import pprint
import pandas as pd
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
import openpyxl
import watermark
%load_ext watermark
%matplotlib inline
%watermark -n -v -m -g -iv
plt.style.use('./d4sci.mplstyle')
book = openpyxl.Workbook()
book.sheetnames
sheet = book['Sheet']
print(sheet)
sheet = book.active
print(sheet)
sheet.title = 'My Data'
book.sheetnames
sheet.dimensions
sheet['A3'] = "Hello"
sheet['B3'] = "World"
sheet.dimensions
rows = np.arange(20).reshape(10, 2)
rows
for row in rows:
sheet.append(list(row)) # Rows have to be lists or tuples
sheet.dimensions
book.save('data/Simple.xlsx')
!open data/Simple.xlsx
book = openpyxl.load_workbook('data/Simple.xlsx')
book.sheetnames
sheet = book['My Data']
print(sheet.dimensions)
print("Min row: %u Max row: %u" % (sheet.min_row, sheet.max_row))
print("Min col: %u Max col: %u" % (sheet.min_column, sheet.max_column))
for c1, c2 in sheet[sheet.dimensions]:
print(c1.value, c2.value)
sheet = book.create_sheet(title="My analysis", index=0)
book.sheetnames
sheet
sheet["B4"] = "The best results"
sheet["B5"] = "guaranteed!"
book.save("data/Simple2.xlsx")
!open data/Simple2.xlsx
book = openpyxl.load_workbook("data/movies.xlsx")
book.sheetnames
book.remove(book["3000s"])
book.sheetnames
book.save('data/movies2.xlsx')
!open data/movies2.xlsx
book = openpyxl.load_workbook('data/excel-mortgage-calculator.xlsx')
book.sheetnames
sheet = book.active
sheet.dimensions
for row in sheet["B16:K17"]:
for cell in row:
print("%s%s: '%s'" % (cell.column_letter, cell.row, cell.value))
print("")
book = openpyxl.load_workbook('data/excel-mortgage-calculator.xlsx', data_only=True)
sheet = book.active
for row in sheet["B16:K17"]:
for cell in row:
print("%s%s: '%s'" % (cell.column_letter, cell.row, cell.value))
print("")
| 0.274643 | 0.969498 |
```
#Importing the required libraries
import pandas as pd
```
## Dynamic Asset Allocation or Balanced Advantage Fund
These mutual funds invest in both Stocks and Debt/Bonds. Allocation between debt and socks can vary as per market conditions.
## Exctracting Dynamic Asset Allocation or Balanced Advantage Mutual Fund's Historical Investment Returns Data
Data in this table: Get Absolute historical returns for ₹1000 investment. If 1Y column value is 1234.5 that means, your ₹1000 investment 1 year back would have grown to ₹1234.5.
```
daa_lump_sum_rtn = pd.read_html(
"https://www.moneycontrol.com/mutual-funds/performance-tracker/returns/dynamic-asset-allocation-or-balanced-advantage.html")
df1 = pd.DataFrame(daa_lump_sum_rtn[0])
#Renaming historical returns column names
df1.rename({'1W': '1W_RTN(%)', '1M': '1M_RTN(%)', '3M': '3M_RTN(%)', '6M': '6M_RTN(%)',
'YTD': 'YTD_RTN(%)', '1Y': '1Y_RTN(%)', '2Y': '2Y_RTN(%)', '3Y': '3Y_RTN(%)',
'5Y': '5Y_RTN(%)', '10Y': '10Y_RTN(%)'
}, axis=1, inplace=True)
print("Shape of the dataframe:", df1.shape)
df1.head()
```
## Exctracting Dynamic Asset Allocation or Balanced Advantage Mutual Fund's Monthly Returns Data
Data in this table: Get monthly returns. If Jan month column value is 5.4% that means, fund has given 5.4% returns in Jan month.
```
daa_monthly_rtn = pd.read_html(
"https://www.moneycontrol.com/mutual-funds/performance-tracker/monthly-returns/dynamic-asset-allocation-or-balanced-advantage.html")
df2 = pd.DataFrame(daa_monthly_rtn[0])
#Renaming df1 column names
df1.rename({"Apr'21": "Apr'21(%)", "Apr'21": "Apr'21(%)", "Apr'21": "Apr'21(%)", "Apr'21": "Apr'21(%)",
'MTD': 'MTD_RTN(%)', "Apr'21": "Apr'21(%)", "Apr'21": "Apr'21(%)", "Apr'21": "Apr'21(%)",
"Apr'21": "Apr'21(%)", "Apr'21": "Apr'21(%)"
}, axis=1, inplace=True)
print("Shape of the dataframe:", df2.shape)
df2.head()
```
## Exctracting Dynamic Asset Allocation or Balanced Advantage Mutual Fund's Quarterly Returns Data
Data in this table: Get quarterly returns. If Q1 column value is 5.4% that means, fund has given 5.4% returns from 1st Jan to 31st Mar.
```
daa_quarterly_rtn = pd.read_html(
"https://www.moneycontrol.com/mutual-funds/performance-tracker/quarterly-returns/dynamic-asset-allocation-or-balanced-advantage.html")
df3 = pd.DataFrame(daa_quarterly_rtn[0])
print("Shape of the dataframe:", df3.shape)
df3.head()
```
## Exctracting Dynamic Asset Allocation or Balanced Advantage Mutual Fund's Annual Investment Returns Data
Data in this table: Get annual returns. If 2018 year column value is 5.4% that means, fund has given 5.4% returns from 1st Jan to 31st Dec/Last date.
```
daa_annual_rtn = pd.read_html(
"https://www.moneycontrol.com/mutual-funds/performance-tracker/annual-returns/dynamic-asset-allocation-or-balanced-advantage.html")
df4 = pd.DataFrame(daa_annual_rtn[0])
#Renaming yearly returns column names
df4.rename({'2020': '2020_RTN(%)', '2019': '2019_RTN(%)', '2018': '2018_RTN(%)', '2017': '2017_RTN(%)',
'2016': '2016_RTN(%)', '2015': '2015_RTN(%)', '2014': '2014_RTN(%)', '2013': '2013_RTN(%)',
'2012': '2012_RTN(%)', '2011': '2011_RTN(%)', '2010': '2010_RTN(%)'
}, axis=1, inplace=True)
print("Shape of the dataframe:", df4.shape)
df4.head()
```
## Exctracting Dynamic Asset Allocation or Balanced Advantage Mutual Fund's Rank Within Category Data
Data in this table: Get performance rank within category. If 1Y column value is 3/45 that means, Fund ranked 3rd in terms of performance out of 45 funds in that category.
```
daa_rank_in_category = pd.read_html(
"https://www.moneycontrol.com/mutual-funds/performance-tracker/ranks/dynamic-asset-allocation-or-balanced-advantage.html")
df5 = pd.DataFrame(daa_rank_in_category[0])
#Renaming df5 column names
df5.rename({'1W': '1W_Rank', '1M': '1M_Rank', '3M': '3M_Rank', '6M': '6M_Rank', 'YTD': 'YTD_Rank',
'1Y': '1Y_Rank', '2Y': '2Y_Rank', '3Y': '3Y_Rank', '5Y': '5Y_Rank', '10Y': '10Y_Rank'
}, axis=1, inplace=True)
print("Shape of the dataframe:", df5.shape)
df5.head()
```
## Exctracting Dynamic Asset Allocation or Balanced Advantage Mutual Fund's Risk Ratios Data
Data in this table: Get values of risk ratios calculated on daily returns for last 3 years.
```
daa_risk_ratio = pd.read_html(
"https://www.moneycontrol.com/mutual-funds/performance-tracker/risk-ratios/dynamic-asset-allocation-or-balanced-advantage.html")
df6 = pd.DataFrame(daa_risk_ratio[0])
#Droping the 'Category' column
df6.drop('Category', inplace=True, axis=1)
print("Shape of the dataframe:", df6.shape)
df6.head()
```
## Exctracting Dynamic Asset Allocation or Balanced Advantage Mutual Fund's Portfolio Data
Data in this table: Compare how schemes have invested money across various asset class and number of instruments.
```
daa_portfolio = pd.read_html(
"https://www.moneycontrol.com/mutual-funds/performance-tracker/portfolioassets/dynamic-asset-allocation-or-balanced-advantage.html")
df7 = pd.DataFrame(daa_portfolio[0])
#Renaming SIP returns column names
df7.rename({'Turnover ratio': 'Turnover ratio(%)'}, axis=1, inplace=True)
print("Shape of the dataframe:", df7.shape)
df7.head()
```
## Exctracting Dynamic Asset Allocation or Balanced Advantage Mutual Fund's Latest NAV Data
Data in this table: Get the latest values of NAV for the mutual funds.
```
daa_nav = pd.read_html(
"https://www.moneycontrol.com/mutual-funds/performance-tracker/navs/dynamic-asset-allocation-or-balanced-advantage.html")
df8 = pd.DataFrame(daa_nav[0])
df8.rename({'1D Change' : '1D Change(%)'}, axis=1, inplace=True)
print("Shape of the dataframe:", df8.shape)
df8.head()
```
## Exctracting Dynamic Asset Allocation or Balanced Advantage Mutual Fund's SIP Returns Data
Data in this table: Get absolute SIP returns. If 1Y column value is 10%, that means fund has given 10% returns on your SIP investments started 1 year back from latest NAV date.
```
daa_sip_rtns = pd.read_html(
"https://www.moneycontrol.com/mutual-funds/performance-tracker/sip-returns/dynamic-asset-allocation-or-balanced-advantage.html")
df9 = pd.DataFrame(daa_sip_rtns[0])
#Renaming SIP returns column names
df9.rename({'1Y': '1Y_SIP_RTN(%)', '2Y': '2Y_SIP_RTN(%)', '3Y': '3Y_SIP_RTN(%)',
'5Y': '5Y_SIP_RTN(%)', '10Y': '10Y_SIP_RTN(%)', 'YTD' : 'YTD_SIP_RTN(%)'
}, axis=1, inplace=True)
print("Shape of the dataframe:", df9.shape)
df9.head()
df_final = pd.concat([df1,df2,df3,df4,df5,df6,df7,df8,df9],axis=1,sort=False)
print("Shape of the dataframe:", df_final.shape)
# Remove duplicate columns by name in Pandas
df_final = df_final.loc[:,~df_final.columns.duplicated()]
# Removing spaces in the column names
#df_final.columns = df_final.columns.str.replace(' ','_')
print("Shape of the dataframe:", df_final.shape)
df_final.head()
#Exporting the consolidated Dynamic Asset Allocation or Balanced Advantage mf data as a csv file
#print("Shape of the dataframe:", df_final.shape)
#df_final.to_csv('daa_mf_data('+ str(pd.to_datetime('today').strftime('%d-%b-%Y %H:%M:%S')) + ').csv',
# index=False)
#Exporting the Dynamic Asset Allocation or Balanced Advantage mf data columns with its datatype as a csv file
#df_dtypes.to_csv('daa_mf_col_data_types('+ str(pd.to_datetime('today').strftime('%d-%b-%Y %H:%M:%S')) + '.csv)')
```
|
github_jupyter
|
#Importing the required libraries
import pandas as pd
daa_lump_sum_rtn = pd.read_html(
"https://www.moneycontrol.com/mutual-funds/performance-tracker/returns/dynamic-asset-allocation-or-balanced-advantage.html")
df1 = pd.DataFrame(daa_lump_sum_rtn[0])
#Renaming historical returns column names
df1.rename({'1W': '1W_RTN(%)', '1M': '1M_RTN(%)', '3M': '3M_RTN(%)', '6M': '6M_RTN(%)',
'YTD': 'YTD_RTN(%)', '1Y': '1Y_RTN(%)', '2Y': '2Y_RTN(%)', '3Y': '3Y_RTN(%)',
'5Y': '5Y_RTN(%)', '10Y': '10Y_RTN(%)'
}, axis=1, inplace=True)
print("Shape of the dataframe:", df1.shape)
df1.head()
daa_monthly_rtn = pd.read_html(
"https://www.moneycontrol.com/mutual-funds/performance-tracker/monthly-returns/dynamic-asset-allocation-or-balanced-advantage.html")
df2 = pd.DataFrame(daa_monthly_rtn[0])
#Renaming df1 column names
df1.rename({"Apr'21": "Apr'21(%)", "Apr'21": "Apr'21(%)", "Apr'21": "Apr'21(%)", "Apr'21": "Apr'21(%)",
'MTD': 'MTD_RTN(%)', "Apr'21": "Apr'21(%)", "Apr'21": "Apr'21(%)", "Apr'21": "Apr'21(%)",
"Apr'21": "Apr'21(%)", "Apr'21": "Apr'21(%)"
}, axis=1, inplace=True)
print("Shape of the dataframe:", df2.shape)
df2.head()
daa_quarterly_rtn = pd.read_html(
"https://www.moneycontrol.com/mutual-funds/performance-tracker/quarterly-returns/dynamic-asset-allocation-or-balanced-advantage.html")
df3 = pd.DataFrame(daa_quarterly_rtn[0])
print("Shape of the dataframe:", df3.shape)
df3.head()
daa_annual_rtn = pd.read_html(
"https://www.moneycontrol.com/mutual-funds/performance-tracker/annual-returns/dynamic-asset-allocation-or-balanced-advantage.html")
df4 = pd.DataFrame(daa_annual_rtn[0])
#Renaming yearly returns column names
df4.rename({'2020': '2020_RTN(%)', '2019': '2019_RTN(%)', '2018': '2018_RTN(%)', '2017': '2017_RTN(%)',
'2016': '2016_RTN(%)', '2015': '2015_RTN(%)', '2014': '2014_RTN(%)', '2013': '2013_RTN(%)',
'2012': '2012_RTN(%)', '2011': '2011_RTN(%)', '2010': '2010_RTN(%)'
}, axis=1, inplace=True)
print("Shape of the dataframe:", df4.shape)
df4.head()
daa_rank_in_category = pd.read_html(
"https://www.moneycontrol.com/mutual-funds/performance-tracker/ranks/dynamic-asset-allocation-or-balanced-advantage.html")
df5 = pd.DataFrame(daa_rank_in_category[0])
#Renaming df5 column names
df5.rename({'1W': '1W_Rank', '1M': '1M_Rank', '3M': '3M_Rank', '6M': '6M_Rank', 'YTD': 'YTD_Rank',
'1Y': '1Y_Rank', '2Y': '2Y_Rank', '3Y': '3Y_Rank', '5Y': '5Y_Rank', '10Y': '10Y_Rank'
}, axis=1, inplace=True)
print("Shape of the dataframe:", df5.shape)
df5.head()
daa_risk_ratio = pd.read_html(
"https://www.moneycontrol.com/mutual-funds/performance-tracker/risk-ratios/dynamic-asset-allocation-or-balanced-advantage.html")
df6 = pd.DataFrame(daa_risk_ratio[0])
#Droping the 'Category' column
df6.drop('Category', inplace=True, axis=1)
print("Shape of the dataframe:", df6.shape)
df6.head()
daa_portfolio = pd.read_html(
"https://www.moneycontrol.com/mutual-funds/performance-tracker/portfolioassets/dynamic-asset-allocation-or-balanced-advantage.html")
df7 = pd.DataFrame(daa_portfolio[0])
#Renaming SIP returns column names
df7.rename({'Turnover ratio': 'Turnover ratio(%)'}, axis=1, inplace=True)
print("Shape of the dataframe:", df7.shape)
df7.head()
daa_nav = pd.read_html(
"https://www.moneycontrol.com/mutual-funds/performance-tracker/navs/dynamic-asset-allocation-or-balanced-advantage.html")
df8 = pd.DataFrame(daa_nav[0])
df8.rename({'1D Change' : '1D Change(%)'}, axis=1, inplace=True)
print("Shape of the dataframe:", df8.shape)
df8.head()
daa_sip_rtns = pd.read_html(
"https://www.moneycontrol.com/mutual-funds/performance-tracker/sip-returns/dynamic-asset-allocation-or-balanced-advantage.html")
df9 = pd.DataFrame(daa_sip_rtns[0])
#Renaming SIP returns column names
df9.rename({'1Y': '1Y_SIP_RTN(%)', '2Y': '2Y_SIP_RTN(%)', '3Y': '3Y_SIP_RTN(%)',
'5Y': '5Y_SIP_RTN(%)', '10Y': '10Y_SIP_RTN(%)', 'YTD' : 'YTD_SIP_RTN(%)'
}, axis=1, inplace=True)
print("Shape of the dataframe:", df9.shape)
df9.head()
df_final = pd.concat([df1,df2,df3,df4,df5,df6,df7,df8,df9],axis=1,sort=False)
print("Shape of the dataframe:", df_final.shape)
# Remove duplicate columns by name in Pandas
df_final = df_final.loc[:,~df_final.columns.duplicated()]
# Removing spaces in the column names
#df_final.columns = df_final.columns.str.replace(' ','_')
print("Shape of the dataframe:", df_final.shape)
df_final.head()
#Exporting the consolidated Dynamic Asset Allocation or Balanced Advantage mf data as a csv file
#print("Shape of the dataframe:", df_final.shape)
#df_final.to_csv('daa_mf_data('+ str(pd.to_datetime('today').strftime('%d-%b-%Y %H:%M:%S')) + ').csv',
# index=False)
#Exporting the Dynamic Asset Allocation or Balanced Advantage mf data columns with its datatype as a csv file
#df_dtypes.to_csv('daa_mf_col_data_types('+ str(pd.to_datetime('today').strftime('%d-%b-%Y %H:%M:%S')) + '.csv)')
| 0.466846 | 0.866698 |
# Produce Eastern Hydro Profile Using Multiple Data Sources
Following data sources are used to generate eastern_hydro_v3.csv
* EIA monthly net generation for conventional hydro plants from Form 923
* Hourly total hydro generation profiles of 4 Independent System Operators (ISO): ISONE, NYISO, PJM and SWPP in 2016
* Hourly net demand profile of base grid
Features of this methodology
* Pumped storage hydro (HPS) and conventional hydro (HYC) are handled separately
* Historical hourly hydro profiles of ISONE, NYISO, PJM and SWPP, are used directly
* For the rest of areas, which are not covered by the 4 ISOs, the hydro profile is generated by scaling the net demand of the corresponding state from the results of our Eastern 2016 basecase scenario based on the monthly total net generation from EIA 923. This is similar to western hydro profile v2.
*Note that in order to run this demo properly, **timezonefinder** package is required to be installed as a dependency in order to find out the local timezone of a given geographic coordination in (lat, lon) pair.*
```
import json
import pytz
import pandas as pd
from tqdm import tqdm
from collections import defaultdict
from timezonefinder import TimezoneFinder
from powersimdata.input.grid import Grid
from powersimdata.network.usa_tamu.constants.zones import (interconnect2loadzone,
loadzone2state,
state2abv)
from prereise.gather.helpers import (trim_eia_form_923,
get_monthly_net_generation)
from prereise.gather.hydrodata.eia.helpers import scale_profile
from prereise.gather.demanddata.eia.map_ba import map_buses_to_county
from prereise.gather.hydrodata.eia.decompose_profile import get_profile_by_plant, get_normalized_profile
from prereise.gather.hydrodata.eia.net_demand import get_net_demand_profile
# Note that using current version of the grid won't be able to reproduce the eastern_hydro_v3 profile.
# The purpose of this notebook is to illustrate the methodology we used to generate this profile.
eastern = Grid(['Eastern'])
```
## 1. Generate eastern pumped storage hydro profiles
Generate hourly profile for HPS based on a deterministic model described in 'hps_plants_eastern.xlsx'
```
# This step takes 30 sec to finish
eastern_hps = pd.read_excel(io='./hps_plants_eastern.xlsx',sheet_name = 'all_plantIDs',header = 0)
time_index = pd.date_range(start='2016-01-01 00:00:00', end='2016-12-31 23:00:00', freq='H')
eastern_hydro_v3_hps = pd.DataFrame(index = time_index, columns = sorted(eastern_hps['PlantIDs']))
utc = pytz.utc
tf = TimezoneFinder()
for plantid in tqdm(eastern_hydro_v3_hps.columns):
lat = eastern.plant.loc[plantid,'lat']
lon = eastern.plant.loc[plantid,'lon']
capacity = eastern.plant.loc[plantid,'Pmax']
tz_target = pytz.timezone(tf.certain_timezone_at(lat=lat, lng=lon))
for time_ind in time_index:
time_utc = utc.localize(time_ind)
time_local = time_utc.astimezone(tz_target)
# weekday, 0:Monday, 1:Tuesday, 2:Wednesday, 3:Thursday, 4:Friday
if time_local.weekday() <= 4:
if time_local.hour in {11,18}:
eastern_hydro_v3_hps.loc[time_ind,plantid] = capacity*0.5
if 11 < time_local.hour < 18:
eastern_hydro_v3_hps.loc[time_ind,plantid] = capacity
eastern_hydro_v3_hps.fillna(0,inplace = True)
```
Total HPS generation during the year turns out to be 30301.6 GWh based on current approach, which is 50% higher than the reported number in EIA 923, i.e. 19884GWh. Hence, we decided to scale the current HPS profile down by 35%
```
eastern_hydro_v3_hps = eastern_hydro_v3_hps.apply(lambda x: x*0.65)
eastern_hydro_v3_hps.sum().sum()
```
## 2. Generate eastern conventional hydro profiles
### a) Generate a mapping between each conventional hydro generator and BAs via counties
This is a similar procedure as generating the mapping between bus to BA via county in eastern demand v5.
```
eastern_hyc_id_list = set(eastern.plant[eastern.plant['type'] == 'hydro'].index) - set(eastern_hps['PlantIDs'])
eastern_hyc = eastern.plant.loc[sorted(eastern_hyc_id_list)][['Pmax','lat','lon','zone_name']].copy()
eastern_hyc, eastern_hyc_no_county_match = map_buses_to_county(eastern_hyc)
eastern_hyc_no_county_match
data = json.load(open('../../../../data/ba_to_county.txt'))
ba_county_list = {}
for val in data['groups'].values():
ba_county_list[val['label']] = set(val['paths'])
eastern_hyc['BA'] = None
for index, row in eastern_hyc.iterrows():
for BA, clist in ba_county_list.items():
try:
county = row['County'].replace(' ','_')
county = county.replace('.','')
county = county.replace('-','')
county = county.replace('\'','_')
if row['County'] == 'LaSalle__IL':
county = 'La_Salle__IL'
if row['County'] == 'Lac Qui Parle__MN':
county = 'Lac_qui_Parle__MN'
if row['County'] == 'Baltimore__MD':
county = 'Baltimore_County__MD'
if row['County'] == 'District of Columbia__DC':
county = 'Washington__DC'
if row['County'] == 'St. Louis City__MO':
county = 'St_Louis_Co__MO'
if county in clist:
eastern_hyc.loc[index,'BA'] = BA
break
except:
continue
eastern_hyc_no_BA_match = list(eastern_hyc[eastern_hyc['BA'].isna()].index)
# Fix mismatch county names in Virginia Mountains
for ind in eastern_hyc_no_BA_match:
if eastern_hyc.loc[ind,'zone_name'] == 'Virginia Mountains':
eastern_hyc.loc[ind,'BA'] = 'PJM'
eastern_hyc_no_BA_match = list(eastern_hyc[eastern_hyc['BA'].isna()].index)
# Assign the rest no-ba-match buses to SWPP
for ind in eastern_hyc_no_BA_match:
eastern_hyc.loc[ind,'BA'] = 'SWPP'
eastern_hyc_no_BA_match = list(eastern_hyc[eastern_hyc['BA'].isna()].index)
eastern_hyc_no_BA_match
eastern_hyc.BA.unique()
eastern_hyc.to_csv('eastern_hyc_to_BA.csv')
```
### b) Decompose 2016 total hydro profiles of ISONE, NYISO, PJM, SWPP into plant level profiles in the corresponding region
Load total profiles of ISONE, NYISO, PJM and SWPP
```
isone_hydro = pd.read_csv('../../../data/neiso_hydro_2016.csv', index_col = 0)
nyiso_hydro = pd.read_csv('../../../data/nyiso_hydro_2016.csv', index_col = 0)
pjm_hydro = pd.read_csv('../../../data/pjm_hydro_2016.csv', index_col = 0)
swpp_hydro = pd.read_csv('../../../data/spp_hydro_2016.csv', index_col = 0)
hydro_v3_isone = get_profile_by_plant(eastern_hyc[eastern_hyc['BA'] == 'ISONE'], isone_hydro['hydro'])
hydro_v3_nyiso = get_profile_by_plant(eastern_hyc[eastern_hyc['BA'] == 'NYISO'], nyiso_hydro['GenMWh'])
hydro_v3_pjm = get_profile_by_plant(eastern_hyc[eastern_hyc['BA'] == 'PJM'], pjm_hydro['hydro'])
hydro_v3_swpp = get_profile_by_plant(eastern_hyc[eastern_hyc['BA'] == 'SWPP'], swpp_hydro['hydro'])
hydro_v3_isone.index = eastern_hydro_v3_hps.index
hydro_v3_nyiso.index = eastern_hydro_v3_hps.index
hydro_v3_pjm.index = eastern_hydro_v3_hps.index
hydro_v3_swpp.index = eastern_hydro_v3_hps.index
```
### c) For the hydro plants in the rest of the area, using the same methodology as in western hydro profile v2, i.e. scale the hourly net demand profile based on the monthly total net generation of conventional hydro reported in EIA 923 in each state, then decompose into plant level profile based on the corresponding plant capacities.
```
eastern_loadzone_to_state_abbrev = {}
for lz in interconnect2loadzone['Eastern']:
eastern_loadzone_to_state_abbrev[lz] = state2abv[loadzone2state[lz]]
state_ba_fraction = defaultdict(lambda: defaultdict(float))
ba_name = {'ISONE','NYISO','SWPP','PJM'}
for index,row in eastern_hyc.iterrows():
if row['BA'] in ba_name:
state_ba_fraction[eastern_loadzone_to_state_abbrev[row['zone_name']]][row['BA']] += row['Pmax']
state_ba_fraction[eastern_loadzone_to_state_abbrev[row['zone_name']]]['total'] += row['Pmax']
state_ba_fraction
```
* Observing from state_ba_fraction, there is no such state that overlaps with PJM and SWPP simultaneously
* Observing from state_ba_fraction, there is no such state that partially overlaps with ISONE or NYISO and partially doesn't overlap with any other BAs
* We only need to consider states not overlapping with any of the 4 ISOs or partailly overlap with either PJM or SWPP to generate the hydro profile of the rest states.
```
eastern_monthly_hyc_rest = {}
eia_923_filename = 'EIA923_Schedules_2_3_4_5_M_12_2016_Final_Revision.xlsx'
eia_923_form = trim_eia_form_923(eia_923_filename)
for state, ba in tqdm(state_ba_fraction.items()):
if len(ba) == 1:
eastern_monthly_hyc_rest[state] = get_monthly_net_generation(state, eia_923_form, 'hydro', hps=False)
elif 'PJM' in ba:
total_state_profile = get_monthly_net_generation(state, eia_923_form, 'hydro', hps=False)
frac = 1-(ba['PJM']/ba['total'])
if frac > 0:
eastern_monthly_hyc_rest[state] = [val*frac for val in total_state_profile]
elif 'SWPP' in ba:
total_state_profile = get_monthly_net_generation(state, eia_923_form, 'hydro', hps=False)
frac = 1-(ba['SWPP']/ba['total'])
if frac > 0:
eastern_monthly_hyc_rest[state] = [val*frac for val in total_state_profile]
```
For Montana, we only have 5 HYC generators (1 plant) in eastern, which we found the corresponding real plant in EIA 923. Comparing with EIA 923, all hydro plants in MT are connected to WECC, so we zero out the hydro generation for eastern in MT here.
```
eastern_monthly_hyc_rest['MT'] = [0]*12
eastern_hyc[eastern_hyc['zone_name'] == 'Montana Eastern']
```
For Eastern Texas, we only have 5 hyc generators (2 plants) in Eastern, which we found the corresponding real plants in EIA 923
```
eastern_monthly_hyc_rest['TX'] = [8430, 8091, 9172, 16705, 23493, 26282, 5259, 7536, 7229, 2376, 5503, 2665]
eastern_hyc[eastern_hyc['zone_name'] == 'East Texas']
```
### d) Get net demand for each state to define the hourly shape of HYC in the rest of states not covered by the 4 ISOs.
```
eastern_net_demand_state_rest = {}
for state in eastern_monthly_hyc_rest:
eastern_net_demand_state_rest[state] = get_net_demand_profile(state, interconnect="Eastern")
```
Scale hourly net demand profile based on monthly net generation to get the HYC hourly total profile of the corresponding state
```
eastern_hyc_hourly_total_state_rest = {}
for state in eastern_net_demand_state_rest:
eastern_hyc_hourly_total_state_rest[state] = scale_profile(pd.Series(eastern_net_demand_state_rest[state], index=eastern_hydro_v3_hps.index), eastern_monthly_hyc_rest[state])
```
Decompose HYC hourly total profile into plant level profiles proportional to plant capacities
* Two HYC generators [9209,9210] in Louisiana are placed in East Texas loadzone. We put them back to LA when generating plant level profiles.
```
hydro_v3_rest_state = {}
for state in eastern_hyc_hourly_total_state_rest:
plantlist = list(eastern_hyc[(eastern_hyc['zone_name'].apply(lambda x: eastern_loadzone_to_state_abbrev[x]) == state) & (~eastern_hyc['BA'].isin({'ISONE','NYISO','SWPP','PJM'}))].index)
if state == 'TX':
plantlist.remove(9209)
plantlist.remove(9210)
if state == 'LA':
plantlist.append(9209)
plantlist.append(9210)
plant_df = eastern_hyc.loc[plantlist].copy()
hydro_v3_rest_state[state] = get_profile_by_plant(plant_df,eastern_hyc_hourly_total_state_rest[state])
# Generate final eastern hydro v3
eastern_hydro_v3 = pd.concat(list(hydro_v3_rest_state.values()),axis = 1)
eastern_hydro_v3.index = eastern_hydro_v3_hps.index
eastern_hydro_v3 = pd.concat([eastern_hydro_v3, eastern_hydro_v3_hps],axis = 1)
eastern_hydro_v3 = pd.concat([eastern_hydro_v3, hydro_v3_isone, hydro_v3_nyiso, hydro_v3_pjm, hydro_v3_swpp],axis = 1)
eastern_hydro_v3 = eastern_hydro_v3[sorted(eastern_hydro_v3.columns)]
eastern_hydro_v3_normalize = get_normalized_profile(eastern.plant[eastern.plant.type == "hydro"], eastern_hydro_v3)
eastern_hydro_v3_normalize.to_csv('eastern_hydro_v3_normalize.csv')
```
|
github_jupyter
|
import json
import pytz
import pandas as pd
from tqdm import tqdm
from collections import defaultdict
from timezonefinder import TimezoneFinder
from powersimdata.input.grid import Grid
from powersimdata.network.usa_tamu.constants.zones import (interconnect2loadzone,
loadzone2state,
state2abv)
from prereise.gather.helpers import (trim_eia_form_923,
get_monthly_net_generation)
from prereise.gather.hydrodata.eia.helpers import scale_profile
from prereise.gather.demanddata.eia.map_ba import map_buses_to_county
from prereise.gather.hydrodata.eia.decompose_profile import get_profile_by_plant, get_normalized_profile
from prereise.gather.hydrodata.eia.net_demand import get_net_demand_profile
# Note that using current version of the grid won't be able to reproduce the eastern_hydro_v3 profile.
# The purpose of this notebook is to illustrate the methodology we used to generate this profile.
eastern = Grid(['Eastern'])
# This step takes 30 sec to finish
eastern_hps = pd.read_excel(io='./hps_plants_eastern.xlsx',sheet_name = 'all_plantIDs',header = 0)
time_index = pd.date_range(start='2016-01-01 00:00:00', end='2016-12-31 23:00:00', freq='H')
eastern_hydro_v3_hps = pd.DataFrame(index = time_index, columns = sorted(eastern_hps['PlantIDs']))
utc = pytz.utc
tf = TimezoneFinder()
for plantid in tqdm(eastern_hydro_v3_hps.columns):
lat = eastern.plant.loc[plantid,'lat']
lon = eastern.plant.loc[plantid,'lon']
capacity = eastern.plant.loc[plantid,'Pmax']
tz_target = pytz.timezone(tf.certain_timezone_at(lat=lat, lng=lon))
for time_ind in time_index:
time_utc = utc.localize(time_ind)
time_local = time_utc.astimezone(tz_target)
# weekday, 0:Monday, 1:Tuesday, 2:Wednesday, 3:Thursday, 4:Friday
if time_local.weekday() <= 4:
if time_local.hour in {11,18}:
eastern_hydro_v3_hps.loc[time_ind,plantid] = capacity*0.5
if 11 < time_local.hour < 18:
eastern_hydro_v3_hps.loc[time_ind,plantid] = capacity
eastern_hydro_v3_hps.fillna(0,inplace = True)
eastern_hydro_v3_hps = eastern_hydro_v3_hps.apply(lambda x: x*0.65)
eastern_hydro_v3_hps.sum().sum()
eastern_hyc_id_list = set(eastern.plant[eastern.plant['type'] == 'hydro'].index) - set(eastern_hps['PlantIDs'])
eastern_hyc = eastern.plant.loc[sorted(eastern_hyc_id_list)][['Pmax','lat','lon','zone_name']].copy()
eastern_hyc, eastern_hyc_no_county_match = map_buses_to_county(eastern_hyc)
eastern_hyc_no_county_match
data = json.load(open('../../../../data/ba_to_county.txt'))
ba_county_list = {}
for val in data['groups'].values():
ba_county_list[val['label']] = set(val['paths'])
eastern_hyc['BA'] = None
for index, row in eastern_hyc.iterrows():
for BA, clist in ba_county_list.items():
try:
county = row['County'].replace(' ','_')
county = county.replace('.','')
county = county.replace('-','')
county = county.replace('\'','_')
if row['County'] == 'LaSalle__IL':
county = 'La_Salle__IL'
if row['County'] == 'Lac Qui Parle__MN':
county = 'Lac_qui_Parle__MN'
if row['County'] == 'Baltimore__MD':
county = 'Baltimore_County__MD'
if row['County'] == 'District of Columbia__DC':
county = 'Washington__DC'
if row['County'] == 'St. Louis City__MO':
county = 'St_Louis_Co__MO'
if county in clist:
eastern_hyc.loc[index,'BA'] = BA
break
except:
continue
eastern_hyc_no_BA_match = list(eastern_hyc[eastern_hyc['BA'].isna()].index)
# Fix mismatch county names in Virginia Mountains
for ind in eastern_hyc_no_BA_match:
if eastern_hyc.loc[ind,'zone_name'] == 'Virginia Mountains':
eastern_hyc.loc[ind,'BA'] = 'PJM'
eastern_hyc_no_BA_match = list(eastern_hyc[eastern_hyc['BA'].isna()].index)
# Assign the rest no-ba-match buses to SWPP
for ind in eastern_hyc_no_BA_match:
eastern_hyc.loc[ind,'BA'] = 'SWPP'
eastern_hyc_no_BA_match = list(eastern_hyc[eastern_hyc['BA'].isna()].index)
eastern_hyc_no_BA_match
eastern_hyc.BA.unique()
eastern_hyc.to_csv('eastern_hyc_to_BA.csv')
isone_hydro = pd.read_csv('../../../data/neiso_hydro_2016.csv', index_col = 0)
nyiso_hydro = pd.read_csv('../../../data/nyiso_hydro_2016.csv', index_col = 0)
pjm_hydro = pd.read_csv('../../../data/pjm_hydro_2016.csv', index_col = 0)
swpp_hydro = pd.read_csv('../../../data/spp_hydro_2016.csv', index_col = 0)
hydro_v3_isone = get_profile_by_plant(eastern_hyc[eastern_hyc['BA'] == 'ISONE'], isone_hydro['hydro'])
hydro_v3_nyiso = get_profile_by_plant(eastern_hyc[eastern_hyc['BA'] == 'NYISO'], nyiso_hydro['GenMWh'])
hydro_v3_pjm = get_profile_by_plant(eastern_hyc[eastern_hyc['BA'] == 'PJM'], pjm_hydro['hydro'])
hydro_v3_swpp = get_profile_by_plant(eastern_hyc[eastern_hyc['BA'] == 'SWPP'], swpp_hydro['hydro'])
hydro_v3_isone.index = eastern_hydro_v3_hps.index
hydro_v3_nyiso.index = eastern_hydro_v3_hps.index
hydro_v3_pjm.index = eastern_hydro_v3_hps.index
hydro_v3_swpp.index = eastern_hydro_v3_hps.index
eastern_loadzone_to_state_abbrev = {}
for lz in interconnect2loadzone['Eastern']:
eastern_loadzone_to_state_abbrev[lz] = state2abv[loadzone2state[lz]]
state_ba_fraction = defaultdict(lambda: defaultdict(float))
ba_name = {'ISONE','NYISO','SWPP','PJM'}
for index,row in eastern_hyc.iterrows():
if row['BA'] in ba_name:
state_ba_fraction[eastern_loadzone_to_state_abbrev[row['zone_name']]][row['BA']] += row['Pmax']
state_ba_fraction[eastern_loadzone_to_state_abbrev[row['zone_name']]]['total'] += row['Pmax']
state_ba_fraction
eastern_monthly_hyc_rest = {}
eia_923_filename = 'EIA923_Schedules_2_3_4_5_M_12_2016_Final_Revision.xlsx'
eia_923_form = trim_eia_form_923(eia_923_filename)
for state, ba in tqdm(state_ba_fraction.items()):
if len(ba) == 1:
eastern_monthly_hyc_rest[state] = get_monthly_net_generation(state, eia_923_form, 'hydro', hps=False)
elif 'PJM' in ba:
total_state_profile = get_monthly_net_generation(state, eia_923_form, 'hydro', hps=False)
frac = 1-(ba['PJM']/ba['total'])
if frac > 0:
eastern_monthly_hyc_rest[state] = [val*frac for val in total_state_profile]
elif 'SWPP' in ba:
total_state_profile = get_monthly_net_generation(state, eia_923_form, 'hydro', hps=False)
frac = 1-(ba['SWPP']/ba['total'])
if frac > 0:
eastern_monthly_hyc_rest[state] = [val*frac for val in total_state_profile]
eastern_monthly_hyc_rest['MT'] = [0]*12
eastern_hyc[eastern_hyc['zone_name'] == 'Montana Eastern']
eastern_monthly_hyc_rest['TX'] = [8430, 8091, 9172, 16705, 23493, 26282, 5259, 7536, 7229, 2376, 5503, 2665]
eastern_hyc[eastern_hyc['zone_name'] == 'East Texas']
eastern_net_demand_state_rest = {}
for state in eastern_monthly_hyc_rest:
eastern_net_demand_state_rest[state] = get_net_demand_profile(state, interconnect="Eastern")
eastern_hyc_hourly_total_state_rest = {}
for state in eastern_net_demand_state_rest:
eastern_hyc_hourly_total_state_rest[state] = scale_profile(pd.Series(eastern_net_demand_state_rest[state], index=eastern_hydro_v3_hps.index), eastern_monthly_hyc_rest[state])
hydro_v3_rest_state = {}
for state in eastern_hyc_hourly_total_state_rest:
plantlist = list(eastern_hyc[(eastern_hyc['zone_name'].apply(lambda x: eastern_loadzone_to_state_abbrev[x]) == state) & (~eastern_hyc['BA'].isin({'ISONE','NYISO','SWPP','PJM'}))].index)
if state == 'TX':
plantlist.remove(9209)
plantlist.remove(9210)
if state == 'LA':
plantlist.append(9209)
plantlist.append(9210)
plant_df = eastern_hyc.loc[plantlist].copy()
hydro_v3_rest_state[state] = get_profile_by_plant(plant_df,eastern_hyc_hourly_total_state_rest[state])
# Generate final eastern hydro v3
eastern_hydro_v3 = pd.concat(list(hydro_v3_rest_state.values()),axis = 1)
eastern_hydro_v3.index = eastern_hydro_v3_hps.index
eastern_hydro_v3 = pd.concat([eastern_hydro_v3, eastern_hydro_v3_hps],axis = 1)
eastern_hydro_v3 = pd.concat([eastern_hydro_v3, hydro_v3_isone, hydro_v3_nyiso, hydro_v3_pjm, hydro_v3_swpp],axis = 1)
eastern_hydro_v3 = eastern_hydro_v3[sorted(eastern_hydro_v3.columns)]
eastern_hydro_v3_normalize = get_normalized_profile(eastern.plant[eastern.plant.type == "hydro"], eastern_hydro_v3)
eastern_hydro_v3_normalize.to_csv('eastern_hydro_v3_normalize.csv')
| 0.451568 | 0.872728 |
# Functions
One of the core principles of any programming language is, **"Don't Repeat Yourself"**.
If you have an action that should occur many times, you can define that action once and then call that code whenever you need to carry out that action.
We are already repeating ourselves in our code, so this is a good time to introduce simple functions. Functions mean less work for us as programmers, and effective use of functions results in code that is less error-prone.
So far, we have only been using the functions that come with Python, but it is also possible to **add new functions**.
A **function definition** specifies the name of a new function and the sequence of statements that execute when the function is called.
<a name='general_syntax'></a>General Syntax
---
A general function looks something like this:
```python
# Let's define a function.
def function_name(argument_1, argument_2):
# Do whatever we want this function to do,
# using argument_1 and argument_2
# Use function_name to call the function.
function_name(value_1, value_2)
```
This code will not run, but it shows how functions are used in general.
- **Defining a function**
- Give the keyword `def`, which tells Python that you are about to *define* a function.
- Give your function a name. A variable name tells you what kind of value the variable contains; a function name should tell you what the function does.
- Give names for each value the function needs in order to do its work.
- These are called the function's *arguments*.
- Make sure the function definition line ends with a colon.
- Inside the function, write whatever code you need to make the function do its work.
- **Calling (i.e. _Using_ ) your function**
- To *call* your function, write its name followed by parentheses.
- Inside the parentheses, give the values you want the function to work with.
<a name='examples'></a>Basic Examples
===
For a simple first example, we will look at a program that executes the sum of two numbers `[1]`
Let's look at the example, and then try to understand the code.
First we will look at a version of this program as we would have written without using functions.
<span class="fn"><i>[1]</i> *I know*, I will think of better examples later. However even simple examples may lead to interesting discussions ;) [2]</span>
<span class="fn"><i>[2]</i> We won't end up being re-implementing the calculator, I promise.
```
print("2+2 is equal to: ", 2+2)
print("3+2 is equal to: ", 3+2)
print("3+3 is equal to: ", 3+3)
```
Functions take repeated code, put it in one place, and then you call that code when you want to use it. Here's what the same program looks like with a function.
```
# Here is the definition of function
def sum(n: int, m: int):
print(f"{n}+{m} is equal to: ", n+m)
# Note: here we are using f-strings
# Alternatively, we could've written
# print("{}+{} is equal to: ".format(n, m), n+m)
sum(2, 2)
sum(3, 2)
sum(3, 3)
```
In our original code, each pair of print statements was run three times, and the only difference was the two numbers we were summing up.
When you see repetition like this, you can usually make your program more efficient by defining a **function**.
The keyword `def` tells Python that we are about to define a function.
We give our function a name, `sum` in this case.
A variable's name should tell us what kind of information it holds; a function's name should tell us what the variable does.
We then put parentheses.
Inside these parenthese we assign a name for any **parameter** the function will need in order to do its job.
In this case the function will need two numbers sum. The variables `n` and `m` will hold the value that is passed into the function `sum`.
**Note:**
The function signature
```python
def sum(n: int, m: int):
```
contains **type-hints annotations** ([doc](https://www.python.org/dev/peps/pep-0484/#rationale-and-goals)).
This is for **documentation purposes** only, not to confuse with **static type** assignments.
#### Warm up Exercise
###### Ex. 1
Try to re-implement the previous function replacing the f-string with the format alternative.
**Note**: it is vital that you **do not** copy&paste the code, but you re-write the whole code again, instead. This will help you in bearing in mind what you are doing.
###### Side note
From now on, we will try to also levarage on VSCode [Live Share Extension](https://code.visualstudio.com/blogs/2017/11/15/live-share).
This will be mostly for the hands-on parts, and exercises.
So, in these cases you are supposed to follow the five steps reported below:
1. Open your VS Code Editor on your local machine
2. Join the live session (link will be provided live)
3. Create a folder with your name
2. Create a new Python file called the way you fancy the most (e.g. `functions.py` in this case)
3. Write the instructions in the file (Python module)
4. Save the file
5. **Execute** your code on your local machine.
- To execute, prompt in a terminal: `$ python functions.py` (for example)
###### Ex. 2
Try to call the function with two arguments of type `string`, and see what happens.
### A common error
A function must be defined before you use it in your program. For example, putting the function at the end of the program would **not** simply work.
```python
sum(2, 2)
sum(3, 2)
sum(3, 3)
def sum(n: int, m: int):
print(f"{n}+{m} is equal to: ", n+m)
```
On the first line we ask Python to run the function `sum`, but Python does not yet know how to do this function. We define our functions at the beginning of our programs, and then we can use them when we need to.
### Advantages of using functions
You might be able to see some advantages of using functions, through this example:
- We write a set of instructions once. We save some work in this simple example, and we save even more work in larger programs.
- When our function works, we don't have to worry about that code anymore. Every time you repeat code in your program, you introduce an opportunity to make a mistake. Writing a function means there is one place to fix mistakes, and when those bugs are fixed, we can be confident that this function will continue to work correctly.
- We can modify our function's behavior, and that change takes effect every time the function is called. This is much better than deciding we need some new behavior, and then having to change code in many different places in our program.
<a name='return_value'></a>Returning a Value
---
Each function you create can **return** a value.
This can be in addition to the primary work the function does, or it can be the function's main job.
The following function takes in a number, and returns the corresponding **english word** of that number:
```python
def get_number_word(number):
# Takes in a numerical value, and returns
# the word corresponding to that number.
if number == 1:
return 'one'
elif number == 2:
return 'two'
elif number == 3:
return 'three'
# ...
```
This function be like:
<blockquote class="twitter-tweet"><p lang="en" dir="ltr">God I wish there was an easier way to do this <a href="https://t.co/8UrBNKdTRW">pic.twitter.com/8UrBNKdTRW</a></p>— Kat Maddox (@ctrlshifti) <a href="https://twitter.com/ctrlshifti/status/1288745146759000064?ref_src=twsrc%5Etfw">July 30, 2020</a></blockquote> <script async src="https://platform.twitter.com/widgets.js" charset="utf-8"></script>
##### Better `encapsulation`
Let's try to re-work our previous example by using the `return` statement.
This will lead also to better **encapsulation**, namely **you had one job!**
```
def sum(n: int, m: int) -> int:
return n+m
print("10+2 is equal to: ", sum(10, 2))
```
###### Few Notes:
1. The function signature now also includes a _type-hint_ for the return type, i.e. `int`:
```python
def sum(n: int, m: int) -> int:
```
The `->` sign refers to the `type` of the **returned value**.
2. This implementation looks quite similar to the previsou one, with the only (BIG) difference that the definition of function's responsibility is better defined in this case:
- The function indeed implements the **sum of two numbers**, rather than **printing** the sum of two numbers! Can you see the difference?
###### Ex 3. Not that kind of calculator
Try to implement a new function called `multiply` which implements the multiplication of two numbers.
Try to run the function with the following arguments:
```python
multiply(3, 3)
multiply("2", 3)
multiply(2, "3")
```
###### Ex 3.1 (Optional)
Implement also the `substract` function and test it using the same input.
#### Getting along with functions
There is much more to learn about functions, but we will get to those details later. For now, feel free to use functions whenever you find yourself writing the same code several times in a program. Some of the things you will learn when we focus on functions:
- How to give the arguments in your function default values.
- How to let your functions accept different numbers of arguments.
---
In previous sections, we learnt the most bare-boned versions of functions.
Now we will learn more general concepts about functions, like how to handle parameters in functions.
### Default argument values
When we first introduced functions, we started with this example:
```python
def sum(n: int, m: int):
print(f"{n}+{m} is equal to: ", n+m)
# Note: here we are using f-strings
# Alternatively, we could've written
# print("{}+{} is equal to: ".format(n, m), n+m)
sum(2, 2)
sum(3, 2)
sum(3, 3)
```
This function works fine, but it fails if you don't pass in **all** the paramenters:
```
sum(3)
```
That makes sense; the function needs to have **both** `n` and `m` in order to do its work, so without either the two, it stucks.
If you want your function to do something by **default**, to cover the case when some information is not (intentionally) provided, you can do so by giving your arguments `default values`.
The default values of parameters are specified in function signature:
```
def sum(n: int, m: int = 0):
"""Implements the sum of two numbers, n and m.
If not provided, m=0 by default so implementing the
identify function.
"""
return n+m
print("This won't fail, assuming 'm=0' -> sum(5) =", sum(5))
```
This is particularly useful when you have a number of arguments in your function, and some of those arguments almost always have the same value. This allows people who use the function to only specify the values that are unique to their use of the function.
##### Ex 4. Identity for Binary Operators
Even if it shouldn't be allowed to define a binary operator like `sum` and `multiply` with one argument, let's assume that our requirement is to implement the identity for those operators when conditions apply.
Therefore, please implement the `identity` function for both `sum` and `multiply` in case either `n` or `m` are not provided in function call.
### Positional Arguments
Much of what you will have to learn about using functions involves how to pass values from your calling statement to the function itself.
The example we just looked at is pretty simple, and only makes sense with numbers.
Let's try to exit a bit the confort zone and let's try to implement a more "sophisticated" function.
#### String Padding
We want to implement a `pad` function which pads two input strings with a given pad character.
Example:
```python
>>> pad("Python", "language", "-")
'Python-language'
```
```
def pad(w1: str, w2: str, p: str) -> str:
"""String padding leveraging on f-strings
Paramenters
-----------
w1: str
First word
w2: str
Second word
p: str
Padding character or string
Returns
-------
The two strings w1 and w2 padded with p
"""
return f"{w1}{p}{w2}"
pad("Python", "language", "-")
```
The arguments in this function are `w1`, `w1`, and `p` for the padding.
This function implements also the bare bones of a [docstring](https://www.python.org/dev/peps/pep-0257/).
Whenever we are calling a function like
```python
pad("Python", "language", "-")
```
we are leveraging on **Positional Arguments**: Python matches the first value `"Valerio"` with the first argument, `"Maggio"` as the second `w2` argument, and so on.
This is pretty straightforward, but it means we have to make sure to get the arguments in the right order.
If we mess up the order, we get nonsense results or (sometimes) errors:
```
pad("Python", "-", "language")
```
### Keyword arguments
Python also allows for another syntax called *keyword arguments*.
In this case, we can give the arguments in any order when we **call the function**, as long as we use the name of the arguments in our calling statement.
Here is how the previous code can be made to work using keyword arguments:
```
pad(w1="Python", p="-", w2="language")
```
This works, because Python does not have to match values to arguments by position, but it matches values to corresponding paramenters by name.
**This also** makes the code more readable when we have to deal with functions with lots of parameters.
#### Mixing positional and keyword arguments
It can make good sense sometimes to mix positional and keyword arguments.
In our previous example, we can expect this function to always take in a first name and a last name.
Before we start mixing positional and keyword arguments, let's add another piece of information (i.e. **requirements**).
We want to modify the `pad` function by also specifying an exact **number of times** we want the padding character to be inserted; `1` time by default.
The function stub would look like this:
```python
def pad(w1: str, w2: str, p: str, times: int = 1):
```
###### I need your help:
Could you please suggest a possible implementation for this function?
###### When you're done
Try to call the new function with the following input:
```python
>>> pad("Python", "language", times=2, p="-")
>>> pad("Python", "language", p="//")
>>> pad(w1="Python", w2="language", p=" ")
>>> pad(w2="Python", "language", times=2, p="-")
```
---
**Before we conclude...**
# The "main" function
The last function in a program like this is **usually** called `main` and it
runs the program using other functions.
**Please note** that this is just a code convention, it is **not** a rule.
So, imagine that we defined a function called `main`:
```python
def main():
...
```
In order to instruct the interpreter to properly define a **main** section
in our Python module, we need to add the following two lines at the end of the
file:
```python
if __name__ == '__main__':
main()
```
The `__name__` variable is set differently depending on how we run the
file, and it's `'__main__'` when we run the file directly instead of
importing.
So if we run the file, the code will enter the main section and will execute; otherwise (in the case in which we import the module) we can still run the functions one by one
but the **main section** won't be executed.
# More than one
Differently from other programming languages, in Python a function may return **more** than one value. For example:
```python
def return_two_values():
return 1, 2
```
When called, this function will return **two** values (i.e. `1` and `2`, respectively). So, if we want to get those values, we can do:
```python
>>> values = return_two_values()
>>> print(values)
(1, 2)
```
In this case, the `type` of the variable `values` will be automagically mapped to a **tuple** (more on this in the next section).
**Moreover**, another possibility is to take the two return values separately in **two** different variables like this:
```python
>>> first, second = return_two_values()
>>> print(first)
1
>>> print(second)
2
```
This particular operation of assigning "at the same time" values to more-than-one variables is called **tuple unpacking** (more on this in the section about tuples)
There are **many** built-in functions in Python returning more than one value. Perhaps, the most common one is `enumerate` which is
used to `enumerate` a sequence (e.g. `list`, `tuple`...)
|
github_jupyter
|
# Let's define a function.
def function_name(argument_1, argument_2):
# Do whatever we want this function to do,
# using argument_1 and argument_2
# Use function_name to call the function.
function_name(value_1, value_2)
print("2+2 is equal to: ", 2+2)
print("3+2 is equal to: ", 3+2)
print("3+3 is equal to: ", 3+3)
# Here is the definition of function
def sum(n: int, m: int):
print(f"{n}+{m} is equal to: ", n+m)
# Note: here we are using f-strings
# Alternatively, we could've written
# print("{}+{} is equal to: ".format(n, m), n+m)
sum(2, 2)
sum(3, 2)
sum(3, 3)
def sum(n: int, m: int):
sum(2, 2)
sum(3, 2)
sum(3, 3)
def sum(n: int, m: int):
print(f"{n}+{m} is equal to: ", n+m)
def get_number_word(number):
# Takes in a numerical value, and returns
# the word corresponding to that number.
if number == 1:
return 'one'
elif number == 2:
return 'two'
elif number == 3:
return 'three'
# ...
def sum(n: int, m: int) -> int:
return n+m
print("10+2 is equal to: ", sum(10, 2))
def sum(n: int, m: int) -> int:
multiply(3, 3)
multiply("2", 3)
multiply(2, "3")
def sum(n: int, m: int):
print(f"{n}+{m} is equal to: ", n+m)
# Note: here we are using f-strings
# Alternatively, we could've written
# print("{}+{} is equal to: ".format(n, m), n+m)
sum(2, 2)
sum(3, 2)
sum(3, 3)
sum(3)
def sum(n: int, m: int = 0):
"""Implements the sum of two numbers, n and m.
If not provided, m=0 by default so implementing the
identify function.
"""
return n+m
print("This won't fail, assuming 'm=0' -> sum(5) =", sum(5))
>>> pad("Python", "language", "-")
'Python-language'
def pad(w1: str, w2: str, p: str) -> str:
"""String padding leveraging on f-strings
Paramenters
-----------
w1: str
First word
w2: str
Second word
p: str
Padding character or string
Returns
-------
The two strings w1 and w2 padded with p
"""
return f"{w1}{p}{w2}"
pad("Python", "language", "-")
pad("Python", "language", "-")
pad("Python", "-", "language")
pad(w1="Python", p="-", w2="language")
###### I need your help:
Could you please suggest a possible implementation for this function?
###### When you're done
Try to call the new function with the following input:
---
**Before we conclude...**
# The "main" function
The last function in a program like this is **usually** called `main` and it
runs the program using other functions.
**Please note** that this is just a code convention, it is **not** a rule.
So, imagine that we defined a function called `main`:
In order to instruct the interpreter to properly define a **main** section
in our Python module, we need to add the following two lines at the end of the
file:
The `__name__` variable is set differently depending on how we run the
file, and it's `'__main__'` when we run the file directly instead of
importing.
So if we run the file, the code will enter the main section and will execute; otherwise (in the case in which we import the module) we can still run the functions one by one
but the **main section** won't be executed.
# More than one
Differently from other programming languages, in Python a function may return **more** than one value. For example:
When called, this function will return **two** values (i.e. `1` and `2`, respectively). So, if we want to get those values, we can do:
In this case, the `type` of the variable `values` will be automagically mapped to a **tuple** (more on this in the next section).
**Moreover**, another possibility is to take the two return values separately in **two** different variables like this:
| 0.444565 | 0.974629 |
<a href="https://colab.research.google.com/github/BaiganKing/DS-Unit-2-Kaggle-Challenge/blob/master/module1/assignment_kaggle_challenge_1.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
Lambda School Data Science, Unit 2: Predictive Modeling
# Kaggle Challenge, Module 1
## Assignment
- [ ] Do train/validate/test split with the Tanzania Waterpumps data.
- [ ] Define a function to wrangle train, validate, and test sets in the same way. Clean outliers and engineer features. (For example, [what other columns have zeros and shouldn't?](https://github.com/Quartz/bad-data-guide#zeros-replace-missing-values) What other columns are duplicates, or nearly duplicates? Can you extract the year from date_recorded? Can you engineer new features, such as the number of years from waterpump construction to waterpump inspection?)
- [ ] Select features. Use a scikit-learn pipeline to encode categoricals, impute missing values, and fit a decision tree classifier.
- [ ] Get your validation accuracy score.
- [ ] Get and plot your feature importances.
- [ ] Submit your predictions to our Kaggle competition. (Go to our Kaggle InClass competition webpage. Use the blue **Submit Predictions** button to upload your CSV file. Or you can use the Kaggle API to submit your predictions.)
- [ ] Commit your notebook to your fork of the GitHub repo.
## Stretch Goals
### Reading
- A Visual Introduction to Machine Learning
- [Part 1: A Decision Tree](http://www.r2d3.us/visual-intro-to-machine-learning-part-1/)
- [Part 2: Bias and Variance](http://www.r2d3.us/visual-intro-to-machine-learning-part-2/)
- [Decision Trees: Advantages & Disadvantages](https://christophm.github.io/interpretable-ml-book/tree.html#advantages-2)
- [How a Russian mathematician constructed a decision tree — by hand — to solve a medical problem](http://fastml.com/how-a-russian-mathematician-constructed-a-decision-tree-by-hand-to-solve-a-medical-problem/)
- [How decision trees work](https://brohrer.github.io/how_decision_trees_work.html)
- [Let’s Write a Decision Tree Classifier from Scratch](https://www.youtube.com/watch?v=LDRbO9a6XPU) — _Don’t worry about understanding the code, just get introduced to the concepts. This 10 minute video has excellent diagrams and explanations._
- [Random Forests for Complete Beginners: The definitive guide to Random Forests and Decision Trees](https://victorzhou.com/blog/intro-to-random-forests/)
### Doing
- [ ] Add your own stretch goal(s) !
- [ ] Try other [scikit-learn imputers](https://scikit-learn.org/stable/modules/impute.html).
- [ ] Try other [scikit-learn scalers](https://scikit-learn.org/stable/modules/preprocessing.html).
- [ ] Make exploratory visualizations and share on Slack.
#### Exploratory visualizations
Visualize the relationships between feature(s) and target. I recommend you do this with your training set, after splitting your data.
For this problem, you may want to create a new column to represent the target as a number, 0 or 1. For example:
```python
train['functional'] = (train['status_group']=='functional').astype(int)
```
You can try [Seaborn "Categorical estimate" plots](https://seaborn.pydata.org/tutorial/categorical.html) for features with reasonably few unique values. (With too many unique values, the plot is unreadable.)
- Categorical features. (If there are too many unique values, you can replace less frequent values with "OTHER.")
- Numeric features. (If there are too many unique values, you can [bin with pandas cut / qcut functions](https://pandas.pydata.org/pandas-docs/stable/getting_started/basics.html?highlight=qcut#discretization-and-quantiling).)
You can try [Seaborn linear model plots](https://seaborn.pydata.org/tutorial/regression.html) with numeric features. For this problem, you may want to use the parameter `logistic=True`
You do _not_ need to use Seaborn, but it's nice because it includes confidence intervals to visualize uncertainty.
#### High-cardinality categoricals
This code from a previous assignment demonstrates how to replace less frequent values with 'OTHER'
```python
# Reduce cardinality for NEIGHBORHOOD feature ...
# Get a list of the top 10 neighborhoods
top10 = train['NEIGHBORHOOD'].value_counts()[:10].index
# At locations where the neighborhood is NOT in the top 10,
# replace the neighborhood with 'OTHER'
train.loc[~train['NEIGHBORHOOD'].isin(top10), 'NEIGHBORHOOD'] = 'OTHER'
test.loc[~test['NEIGHBORHOOD'].isin(top10), 'NEIGHBORHOOD'] = 'OTHER'
```
```
# If you're in Colab...
import os, sys
in_colab = 'google.colab' in sys.modules
if in_colab:
# Install required python packages:
# category_encoders, version >= 2.0
# pandas-profiling, version >= 2.0
# plotly, version >= 4.0
!pip install --upgrade category_encoders pandas-profiling plotly
# Pull files from Github repo
os.chdir('/content')
!git init .
!git remote add origin https://github.com/LambdaSchool/DS-Unit-2-Kaggle-Challenge.git
!git pull origin master
# Change into directory for module
os.chdir('module1')
import pandas as pd
from sklearn.model_selection import train_test_split
train = pd.merge(pd.read_csv('../data/tanzania/train_features.csv'),
pd.read_csv('../data/tanzania/train_labels.csv'))
test = pd.read_csv('../data/tanzania/test_features.csv')
sample_submission = pd.read_csv('../data/tanzania/sample_submission.csv')
print(train.shape, test.shape)
train, val = train_test_split(train, train_size=0.8, test_size=0.2,
stratify=train['status_group'],random_state=42)
print(train.shape, val.shape)
import numpy as np
def wrangle(df):
df = df.copy()
df['latitude'] = df['latitude'].replace(-2e-08,0)
col_zero = ['longitude','latitude','date_recorded','subvillage',
'installer','region','basin']
for col in col_zero:
df[col] = df[col].replace(0,np.nan)
df[col] = df[col].replace('0',np.nan)
df = df.drop(columns=['quantity_group','scheme_management',
'extraction_type_group','payment_type'])
df['date_recorded'] = pd.to_datetime(df['date_recorded'],infer_datetime_format=True)
return df
import category_encoders as ce
from sklearn.impute import SimpleImputer
from sklearn.experimental import enable_iterative_imputer
from sklearn.impute import IterativeImputer
from sklearn.linear_model import LogisticRegression
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import StandardScaler
train = wrangle(train)
val = wrangle(val)
test = wrangle(test)
target = 'status_group'
train_features = train.drop(columns=[target, 'id'])
numeric_features = train_features.select_dtypes(include='number').columns.tolist()
cardinality = train_features.select_dtypes(exclude='number').nunique()
categorical_features = cardinality[cardinality <= 50].index.tolist()
features = numeric_features + categorical_features
X_train = train[features]
y_train = train[target]
X_val = val[features]
y_val = val[target]
X_test = test[features]
pipeline = make_pipeline(
ce.OneHotEncoder(use_cat_names=True),
SimpleImputer(),
StandardScaler(),
LogisticRegression(solver='lbfgs', multi_class='auto', max_iter=1000)
)
pipeline.fit(X_train, y_train)
print('Validation Accuracy', pipeline.score(X_val, y_val))
y_pred = pipeline.predict(X_test)
submission = sample_submission.copy()
submission['status_group'] = y_pred
submission.to_csv('submission-02.csv', index=False)
from google.colab import files
files.download('submission-02.csv')
```
|
github_jupyter
|
train['functional'] = (train['status_group']=='functional').astype(int)
# Reduce cardinality for NEIGHBORHOOD feature ...
# Get a list of the top 10 neighborhoods
top10 = train['NEIGHBORHOOD'].value_counts()[:10].index
# At locations where the neighborhood is NOT in the top 10,
# replace the neighborhood with 'OTHER'
train.loc[~train['NEIGHBORHOOD'].isin(top10), 'NEIGHBORHOOD'] = 'OTHER'
test.loc[~test['NEIGHBORHOOD'].isin(top10), 'NEIGHBORHOOD'] = 'OTHER'
# If you're in Colab...
import os, sys
in_colab = 'google.colab' in sys.modules
if in_colab:
# Install required python packages:
# category_encoders, version >= 2.0
# pandas-profiling, version >= 2.0
# plotly, version >= 4.0
!pip install --upgrade category_encoders pandas-profiling plotly
# Pull files from Github repo
os.chdir('/content')
!git init .
!git remote add origin https://github.com/LambdaSchool/DS-Unit-2-Kaggle-Challenge.git
!git pull origin master
# Change into directory for module
os.chdir('module1')
import pandas as pd
from sklearn.model_selection import train_test_split
train = pd.merge(pd.read_csv('../data/tanzania/train_features.csv'),
pd.read_csv('../data/tanzania/train_labels.csv'))
test = pd.read_csv('../data/tanzania/test_features.csv')
sample_submission = pd.read_csv('../data/tanzania/sample_submission.csv')
print(train.shape, test.shape)
train, val = train_test_split(train, train_size=0.8, test_size=0.2,
stratify=train['status_group'],random_state=42)
print(train.shape, val.shape)
import numpy as np
def wrangle(df):
df = df.copy()
df['latitude'] = df['latitude'].replace(-2e-08,0)
col_zero = ['longitude','latitude','date_recorded','subvillage',
'installer','region','basin']
for col in col_zero:
df[col] = df[col].replace(0,np.nan)
df[col] = df[col].replace('0',np.nan)
df = df.drop(columns=['quantity_group','scheme_management',
'extraction_type_group','payment_type'])
df['date_recorded'] = pd.to_datetime(df['date_recorded'],infer_datetime_format=True)
return df
import category_encoders as ce
from sklearn.impute import SimpleImputer
from sklearn.experimental import enable_iterative_imputer
from sklearn.impute import IterativeImputer
from sklearn.linear_model import LogisticRegression
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import StandardScaler
train = wrangle(train)
val = wrangle(val)
test = wrangle(test)
target = 'status_group'
train_features = train.drop(columns=[target, 'id'])
numeric_features = train_features.select_dtypes(include='number').columns.tolist()
cardinality = train_features.select_dtypes(exclude='number').nunique()
categorical_features = cardinality[cardinality <= 50].index.tolist()
features = numeric_features + categorical_features
X_train = train[features]
y_train = train[target]
X_val = val[features]
y_val = val[target]
X_test = test[features]
pipeline = make_pipeline(
ce.OneHotEncoder(use_cat_names=True),
SimpleImputer(),
StandardScaler(),
LogisticRegression(solver='lbfgs', multi_class='auto', max_iter=1000)
)
pipeline.fit(X_train, y_train)
print('Validation Accuracy', pipeline.score(X_val, y_val))
y_pred = pipeline.predict(X_test)
submission = sample_submission.copy()
submission['status_group'] = y_pred
submission.to_csv('submission-02.csv', index=False)
from google.colab import files
files.download('submission-02.csv')
| 0.385375 | 0.974797 |
# Imports
```
import math
import pandas as pd
import pennylane as qml
import time
from keras.datasets import mnist
from matplotlib import pyplot as plt
from pennylane import numpy as np
from pennylane.templates import AmplitudeEmbedding, AngleEmbedding
from pennylane.templates.subroutines import ArbitraryUnitary
from sklearn.decomposition import PCA
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
```
# Model Params
```
np.random.seed(131)
initial_params = np.random.random([15])
INITIALIZATION_METHOD = 'Amplitude'
BATCH_SIZE = 20
EPOCHS = 400
STEP_SIZE = 0.01
BETA_1 = 0.9
BETA_2 = 0.99
EPSILON = 0.00000001
TRAINING_SIZE = 0.78
VALIDATION_SIZE = 0.07
TEST_SIZE = 1-TRAINING_SIZE-VALIDATION_SIZE
initial_time = time.time()
```
# Import dataset
```
(train_X, train_y), (test_X, test_y) = mnist.load_data()
examples = np.append(train_X, test_X, axis=0)
examples = examples.reshape(70000, 28*28)
classes = np.append(train_y, test_y)
x = []
y = []
for (example, label) in zip(examples, classes):
if label in [0, 2, 4, 6, 8]:
x.append(example)
y.append(-1)
else:
x.append(example)
y.append(1)
x = np.array(x)
y = np.array(y)
# Normalize pixels values
x = x / 255
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size=TEST_SIZE, shuffle=True)
validation_indexes = np.random.random_integers(len(X_train), size=(math.floor(len(X_train)*VALIDATION_SIZE),))
X_validation = [X_train[n-1] for n in validation_indexes]
y_validation = [y_train[n-1] for n in validation_indexes]
pca = PCA(n_components=8)
pca.fit(X_train)
X_train = pca.transform(X_train)
X_validation = pca.transform(X_validation)
X_test = pca.transform(X_test)
preprocessing_time = time.time()
```
# Circuit creation
```
device = qml.device("default.qubit", wires=3)
@qml.qnode(device)
def circuit(features, params):
# Load state
if INITIALIZATION_METHOD == 'Amplitude':
AmplitudeEmbedding(features=features, wires=range(3), normalize=True, pad_with=0.)
else:
AngleEmbedding(features=features, wires=range(3), rotation='Y')
# First layer
qml.U3(params[0], params[1], params[2], wires=0)
qml.U3(params[3], params[4], params[5], wires=1)
qml.CNOT(wires=[0, 1])
# Second layer
qml.U3(params[6], params[7], params[8], wires=1)
qml.U3(params[9], params[10], params[11], wires=2)
qml.CNOT(wires=[1, 2])
# Third layer
qml.U3(params[12], params[13], params[14], wires=2)
# Measurement
return qml.expval(qml.PauliZ(2))
```
## Circuit example
```
features = X_train[0]
print(f"Inital parameters: {initial_params}\n")
print(f"Example features: {features}\n")
print(f"Expectation value: {circuit(features, initial_params)}\n")
print(circuit.draw())
```
# Accuracy test definition
```
def measure_accuracy(x, y, circuit_params):
class_errors = 0
for example, example_class in zip(x, y):
predicted_value = circuit(example, circuit_params)
if (example_class > 0 and predicted_value <= 0) or (example_class <= 0 and predicted_value > 0):
class_errors += 1
return 1 - (class_errors/len(y))
```
# Training
```
params = initial_params
opt = qml.AdamOptimizer(stepsize=STEP_SIZE, beta1=BETA_1, beta2=BETA_2, eps=EPSILON)
test_accuracies = []
best_validation_accuracy = 0.0
best_params = []
for i in range(len(X_train)):
features = X_train[i]
expected_value = y_train[i]
def cost(circuit_params):
value = circuit(features, circuit_params)
return ((expected_value - value) ** 2)/len(X_train)
params = opt.step(cost, params)
if i % BATCH_SIZE == 0:
print(f"epoch {i//BATCH_SIZE}")
if i % (10*BATCH_SIZE) == 0:
current_accuracy = measure_accuracy(X_validation, y_validation, params)
test_accuracies.append(current_accuracy)
print(f"accuracy: {current_accuracy}")
if current_accuracy > best_validation_accuracy:
print("best accuracy so far!")
best_validation_accuracy = current_accuracy
best_params = params
if len(test_accuracies) == 30:
print(f"test_accuracies: {test_accuracies}")
if np.allclose(best_validation_accuracy, test_accuracies[0]):
params = best_params
break
del test_accuracies[0]
print("Optimized rotation angles: {}".format(params))
training_time = time.time()
```
# Testing
```
accuracy = measure_accuracy(X_test, y_test, params)
print(accuracy)
test_time = time.time()
print(f"pre-processing time: {preprocessing_time-initial_time}")
print(f"training time: {training_time - preprocessing_time}")
print(f"test time: {test_time - training_time}")
print(f"total time: {test_time - initial_time}")
```
|
github_jupyter
|
import math
import pandas as pd
import pennylane as qml
import time
from keras.datasets import mnist
from matplotlib import pyplot as plt
from pennylane import numpy as np
from pennylane.templates import AmplitudeEmbedding, AngleEmbedding
from pennylane.templates.subroutines import ArbitraryUnitary
from sklearn.decomposition import PCA
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
np.random.seed(131)
initial_params = np.random.random([15])
INITIALIZATION_METHOD = 'Amplitude'
BATCH_SIZE = 20
EPOCHS = 400
STEP_SIZE = 0.01
BETA_1 = 0.9
BETA_2 = 0.99
EPSILON = 0.00000001
TRAINING_SIZE = 0.78
VALIDATION_SIZE = 0.07
TEST_SIZE = 1-TRAINING_SIZE-VALIDATION_SIZE
initial_time = time.time()
(train_X, train_y), (test_X, test_y) = mnist.load_data()
examples = np.append(train_X, test_X, axis=0)
examples = examples.reshape(70000, 28*28)
classes = np.append(train_y, test_y)
x = []
y = []
for (example, label) in zip(examples, classes):
if label in [0, 2, 4, 6, 8]:
x.append(example)
y.append(-1)
else:
x.append(example)
y.append(1)
x = np.array(x)
y = np.array(y)
# Normalize pixels values
x = x / 255
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size=TEST_SIZE, shuffle=True)
validation_indexes = np.random.random_integers(len(X_train), size=(math.floor(len(X_train)*VALIDATION_SIZE),))
X_validation = [X_train[n-1] for n in validation_indexes]
y_validation = [y_train[n-1] for n in validation_indexes]
pca = PCA(n_components=8)
pca.fit(X_train)
X_train = pca.transform(X_train)
X_validation = pca.transform(X_validation)
X_test = pca.transform(X_test)
preprocessing_time = time.time()
device = qml.device("default.qubit", wires=3)
@qml.qnode(device)
def circuit(features, params):
# Load state
if INITIALIZATION_METHOD == 'Amplitude':
AmplitudeEmbedding(features=features, wires=range(3), normalize=True, pad_with=0.)
else:
AngleEmbedding(features=features, wires=range(3), rotation='Y')
# First layer
qml.U3(params[0], params[1], params[2], wires=0)
qml.U3(params[3], params[4], params[5], wires=1)
qml.CNOT(wires=[0, 1])
# Second layer
qml.U3(params[6], params[7], params[8], wires=1)
qml.U3(params[9], params[10], params[11], wires=2)
qml.CNOT(wires=[1, 2])
# Third layer
qml.U3(params[12], params[13], params[14], wires=2)
# Measurement
return qml.expval(qml.PauliZ(2))
features = X_train[0]
print(f"Inital parameters: {initial_params}\n")
print(f"Example features: {features}\n")
print(f"Expectation value: {circuit(features, initial_params)}\n")
print(circuit.draw())
def measure_accuracy(x, y, circuit_params):
class_errors = 0
for example, example_class in zip(x, y):
predicted_value = circuit(example, circuit_params)
if (example_class > 0 and predicted_value <= 0) or (example_class <= 0 and predicted_value > 0):
class_errors += 1
return 1 - (class_errors/len(y))
params = initial_params
opt = qml.AdamOptimizer(stepsize=STEP_SIZE, beta1=BETA_1, beta2=BETA_2, eps=EPSILON)
test_accuracies = []
best_validation_accuracy = 0.0
best_params = []
for i in range(len(X_train)):
features = X_train[i]
expected_value = y_train[i]
def cost(circuit_params):
value = circuit(features, circuit_params)
return ((expected_value - value) ** 2)/len(X_train)
params = opt.step(cost, params)
if i % BATCH_SIZE == 0:
print(f"epoch {i//BATCH_SIZE}")
if i % (10*BATCH_SIZE) == 0:
current_accuracy = measure_accuracy(X_validation, y_validation, params)
test_accuracies.append(current_accuracy)
print(f"accuracy: {current_accuracy}")
if current_accuracy > best_validation_accuracy:
print("best accuracy so far!")
best_validation_accuracy = current_accuracy
best_params = params
if len(test_accuracies) == 30:
print(f"test_accuracies: {test_accuracies}")
if np.allclose(best_validation_accuracy, test_accuracies[0]):
params = best_params
break
del test_accuracies[0]
print("Optimized rotation angles: {}".format(params))
training_time = time.time()
accuracy = measure_accuracy(X_test, y_test, params)
print(accuracy)
test_time = time.time()
print(f"pre-processing time: {preprocessing_time-initial_time}")
print(f"training time: {training_time - preprocessing_time}")
print(f"test time: {test_time - training_time}")
print(f"total time: {test_time - initial_time}")
| 0.549157 | 0.879665 |
# cMLP Lagged VAR Demo
- In this notebook, we train a cMLP model on linear VAR data with lagged interactions.
- After examining the Granger causality discovery, we train a debiased model using only the discovered interactions.
```
import torch
import numpy as np
import matplotlib.pyplot as plt
from synthetic import simulate_lorenz_96
from models.cmlp import cMLP, cMLPSparse, train_model_ista, train_unregularized
# For GPU acceleration
device = torch.device('cuda')
# Simulate data
p = 10
X_np, GC = simulate_lorenz_96(p=p, F=5, T=1000, delta_t=1)
X = torch.tensor(X_np[np.newaxis], dtype=torch.float32, device=device)
for i in range(p):
GC[i][i] = 0
# Plot data
fig, axarr = plt.subplots(1, 2, figsize=(16, 5))
axarr[0].plot(X_np)
axarr[0].set_xlabel('T')
axarr[0].set_title('Entire time series')
axarr[1].plot(X_np[:50])
axarr[1].set_xlabel('T')
axarr[1].set_title('First 50 time points')
plt.tight_layout()
plt.show()
```
# Still need to tune $\lambda$ and perhaps lr
```
# Set up model
cmlp = cMLP(X.shape[-1], lag=1, hidden=[100]).cuda(device=device)
# Train with ISTA
train_loss_list = train_model_ista(
cmlp, X, lam=2.5, lam_ridge=0.008, lr=5e-4, penalty='H', max_iter=100000,
check_every=100)
# Loss function plot
plt.figure(figsize=(8, 5))
plt.plot(50 * np.arange(len(train_loss_list)), train_loss_list)
plt.title('cMLP training')
plt.ylabel('Loss')
plt.xlabel('Training steps')
plt.tight_layout()
plt.show()
# Verify learned Granger causality
GC_est = cmlp.GC().cpu().data.numpy()
print('True variable usage = %.2f%%' % (100 * np.mean(GC)))
print('Estimated variable usage = %.2f%%' % (100 * np.mean(GC_est)))
print('Accuracy = %.2f%%' % (100 * np.mean(GC == GC_est)))
# Make figures
fig, axarr = plt.subplots(1, 2, figsize=(16, 5))
axarr[0].imshow(GC, cmap='Blues')
axarr[0].set_title('GC actual')
axarr[0].set_ylabel('Affected series')
axarr[0].set_xlabel('Causal series')
axarr[0].set_xticks([])
axarr[0].set_yticks([])
axarr[1].imshow(GC_est, cmap='Blues', vmin=0, vmax=1, extent=(0, len(GC_est), len(GC_est), 0))
axarr[1].set_title('GC estimated')
axarr[1].set_ylabel('Affected series')
axarr[1].set_xlabel('Causal series')
axarr[1].set_xticks([])
axarr[1].set_yticks([])
# Mark disagreements
for i in range(len(GC_est)):
for j in range(len(GC_est)):
if GC[i, j] != GC_est[i, j]:
rect = plt.Rectangle((j, i-0.05), 1, 1, facecolor='none', edgecolor='red', linewidth=1)
axarr[1].add_patch(rect)
plt.show()
# Verify lag selection
for i in range(len(GC_est)):
# Get true GC
GC_lag = np.zeros((1, len(GC_est)))
GC_lag[:3, GC[i].astype(bool)] = 1.0
# Get estimated GC
GC_est_lag = cmlp.GC(ignore_lag=False, threshold=False)[i].cpu().data.numpy().T[::-1]
# Make figures
fig, axarr = plt.subplots(1, 2, figsize=(16, 5))
axarr[0].imshow(GC_lag, cmap='Blues', extent=(0, len(GC_est), 1, 0))
axarr[0].set_title('Series %d true GC' % (i + 1))
axarr[0].set_ylabel('Lag')
axarr[0].set_xlabel('Series')
axarr[0].set_xticks(np.arange(len(GC_est)) + 0.5)
axarr[0].set_xticklabels(range(len(GC_est)))
axarr[0].set_yticks(np.arange(1) + 0.5)
axarr[0].set_yticklabels(range(1, 1 + 1))
axarr[0].tick_params(axis='both', length=0)
axarr[1].imshow(GC_est_lag, cmap='Blues', extent=(0, len(GC_est), 1, 0))
axarr[1].set_title('Series %d estimated GC' % (i + 1))
axarr[1].set_ylabel('Lag')
axarr[1].set_xlabel('Series')
axarr[1].set_xticks(np.arange(len(GC_est)) + 0.5)
axarr[1].set_xticklabels(range(len(GC_est)))
axarr[1].set_yticks(np.arange(1) + 0.5)
axarr[1].set_yticklabels(range(1, 1 + 1))
axarr[1].tick_params(axis='both', length=0)
# Mark nonzeros
for i in range(len(GC_est)):
for j in range(1):
if GC_est_lag[j, i] > 0.0:
rect = plt.Rectangle((i, j), 1, 1, facecolor='none', edgecolor='green', linewidth=1.0)
axarr[1].add_patch(rect)
plt.show()
```
# Train sparsified model
```
# Create a debiased model
sparsity = cmlp.GC().bool()
cmlp_sparse = cMLPSparse(X.shape[-1], sparsity, lag=1, hidden=[100]).cuda(device=device)
# Train
train_loss_list = train_unregularized(cmlp_sparse, X, lr=1e-3, max_iter=10000,
check_every=100, verbose=1)
# Plot loss function
plt.figure(figsize=(10, 5))
plt.title('Debiased model training')
plt.ylabel('Loss')
plt.xlabel('Training steps')
plt.plot(100 * np.arange(len(train_loss_list)), train_loss_list)
plt.show()
```
|
github_jupyter
|
import torch
import numpy as np
import matplotlib.pyplot as plt
from synthetic import simulate_lorenz_96
from models.cmlp import cMLP, cMLPSparse, train_model_ista, train_unregularized
# For GPU acceleration
device = torch.device('cuda')
# Simulate data
p = 10
X_np, GC = simulate_lorenz_96(p=p, F=5, T=1000, delta_t=1)
X = torch.tensor(X_np[np.newaxis], dtype=torch.float32, device=device)
for i in range(p):
GC[i][i] = 0
# Plot data
fig, axarr = plt.subplots(1, 2, figsize=(16, 5))
axarr[0].plot(X_np)
axarr[0].set_xlabel('T')
axarr[0].set_title('Entire time series')
axarr[1].plot(X_np[:50])
axarr[1].set_xlabel('T')
axarr[1].set_title('First 50 time points')
plt.tight_layout()
plt.show()
# Set up model
cmlp = cMLP(X.shape[-1], lag=1, hidden=[100]).cuda(device=device)
# Train with ISTA
train_loss_list = train_model_ista(
cmlp, X, lam=2.5, lam_ridge=0.008, lr=5e-4, penalty='H', max_iter=100000,
check_every=100)
# Loss function plot
plt.figure(figsize=(8, 5))
plt.plot(50 * np.arange(len(train_loss_list)), train_loss_list)
plt.title('cMLP training')
plt.ylabel('Loss')
plt.xlabel('Training steps')
plt.tight_layout()
plt.show()
# Verify learned Granger causality
GC_est = cmlp.GC().cpu().data.numpy()
print('True variable usage = %.2f%%' % (100 * np.mean(GC)))
print('Estimated variable usage = %.2f%%' % (100 * np.mean(GC_est)))
print('Accuracy = %.2f%%' % (100 * np.mean(GC == GC_est)))
# Make figures
fig, axarr = plt.subplots(1, 2, figsize=(16, 5))
axarr[0].imshow(GC, cmap='Blues')
axarr[0].set_title('GC actual')
axarr[0].set_ylabel('Affected series')
axarr[0].set_xlabel('Causal series')
axarr[0].set_xticks([])
axarr[0].set_yticks([])
axarr[1].imshow(GC_est, cmap='Blues', vmin=0, vmax=1, extent=(0, len(GC_est), len(GC_est), 0))
axarr[1].set_title('GC estimated')
axarr[1].set_ylabel('Affected series')
axarr[1].set_xlabel('Causal series')
axarr[1].set_xticks([])
axarr[1].set_yticks([])
# Mark disagreements
for i in range(len(GC_est)):
for j in range(len(GC_est)):
if GC[i, j] != GC_est[i, j]:
rect = plt.Rectangle((j, i-0.05), 1, 1, facecolor='none', edgecolor='red', linewidth=1)
axarr[1].add_patch(rect)
plt.show()
# Verify lag selection
for i in range(len(GC_est)):
# Get true GC
GC_lag = np.zeros((1, len(GC_est)))
GC_lag[:3, GC[i].astype(bool)] = 1.0
# Get estimated GC
GC_est_lag = cmlp.GC(ignore_lag=False, threshold=False)[i].cpu().data.numpy().T[::-1]
# Make figures
fig, axarr = plt.subplots(1, 2, figsize=(16, 5))
axarr[0].imshow(GC_lag, cmap='Blues', extent=(0, len(GC_est), 1, 0))
axarr[0].set_title('Series %d true GC' % (i + 1))
axarr[0].set_ylabel('Lag')
axarr[0].set_xlabel('Series')
axarr[0].set_xticks(np.arange(len(GC_est)) + 0.5)
axarr[0].set_xticklabels(range(len(GC_est)))
axarr[0].set_yticks(np.arange(1) + 0.5)
axarr[0].set_yticklabels(range(1, 1 + 1))
axarr[0].tick_params(axis='both', length=0)
axarr[1].imshow(GC_est_lag, cmap='Blues', extent=(0, len(GC_est), 1, 0))
axarr[1].set_title('Series %d estimated GC' % (i + 1))
axarr[1].set_ylabel('Lag')
axarr[1].set_xlabel('Series')
axarr[1].set_xticks(np.arange(len(GC_est)) + 0.5)
axarr[1].set_xticklabels(range(len(GC_est)))
axarr[1].set_yticks(np.arange(1) + 0.5)
axarr[1].set_yticklabels(range(1, 1 + 1))
axarr[1].tick_params(axis='both', length=0)
# Mark nonzeros
for i in range(len(GC_est)):
for j in range(1):
if GC_est_lag[j, i] > 0.0:
rect = plt.Rectangle((i, j), 1, 1, facecolor='none', edgecolor='green', linewidth=1.0)
axarr[1].add_patch(rect)
plt.show()
# Create a debiased model
sparsity = cmlp.GC().bool()
cmlp_sparse = cMLPSparse(X.shape[-1], sparsity, lag=1, hidden=[100]).cuda(device=device)
# Train
train_loss_list = train_unregularized(cmlp_sparse, X, lr=1e-3, max_iter=10000,
check_every=100, verbose=1)
# Plot loss function
plt.figure(figsize=(10, 5))
plt.title('Debiased model training')
plt.ylabel('Loss')
plt.xlabel('Training steps')
plt.plot(100 * np.arange(len(train_loss_list)), train_loss_list)
plt.show()
| 0.715821 | 0.895294 |
# Predict phonon DoS for new materials and evaluate their specific heat capacities
- `ComprehensiveEvaluation`: the function that evaluate phonon DoS and heat capacities with the input `*.cif` file
- `AtomEmbeddingAndSumLastLayer`: the model function
```
import glob
import torch
import torch_geometric
import torch_scatter
import e3nn
from e3nn import rs, o3
from e3nn.point.data_helpers import DataPeriodicNeighbors
from e3nn.networks import GatedConvParityNetwork
from e3nn.kernel_mod import Kernel
from e3nn.point.message_passing import Convolution
import pymatgen
from pymatgen.core.structure import Structure
import time, os
import datetime
import pickle
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
import numpy as np
import h5py
from class_evaluate_MPdata import ComprehensiveEvaluation, AtomEmbeddingAndSumLastLayer
torch.set_default_dtype(torch.float64)
if torch.cuda.is_available():
device = "cuda"
else:
device = "cpu"
print('torch device:' , device)
```
### Import model and trained model parameters
```
model_data = torch.load('models/200803-1018_len51max1000_fwin101ord3_trial_run_full_data.torch', map_location=device)
model = AtomEmbeddingAndSumLastLayer(model_data['state']['linear.weight'].shape[1],
model_data['state']['linear.weight'].shape[0],
GatedConvParityNetwork(**model_data['model_kwargs']))
model.load_state_dict(model_data['state'])
model.to(device)
model.eval()
```
### Import frequency points to evaluate heat capacities
Please make sure to unzip the file `models/phdos_e3nn_len51max1000_fwin101ord3.zip`
```
with open('models/phdos_e3nn_len51max1000_fwin101ord3.pkl', 'rb') as f:
data_dict = pickle.load(f)
phfre = data_dict['phfre']
```
Load cif file names and filter out those materials with more than 13 atoms inside the unit cells.
```
with open('models/cif_unique_files.pkl', 'rb') as f:
ciflist_dict = pickle.load(f)
cif_name = ciflist_dict.get('cif_name')
cif_id = ciflist_dict.get('cif_id')
num_sites = ciflist_dict.get('num_sites')
cif_name_suc = [cif_name[i] for i in range(len(cif_name)) if num_sites[i] <= 13]
cif_id_suc = [cif_id[i] for i in range(len(cif_id)) if num_sites[i] <= 13]
```
- `T_lst`: temperature list that you want to evaluate $C_{V}$ at
- `cif_path`: where you stored all downloaded `*.cif` files
- `h5_file`: the `*.h5` file that you want to store calculated phonon DoS and heat capacities
```
T_lst = [273.15, 293.15]
cif_path = 'data/mp_data/'
h5_file = 'data/phdos_maxsites13_2020Aug27.h5'
for i in range(0,len(cif_id_suc)):
material_id = cif_id_suc[i]
chunk_evaluation = ComprehensiveEvaluation([cif_name_suc[i]], model_data['model_kwargs'], cif_path=cif_path, chunk_id=material_id)
chunk_evaluation.predict_phdos(chunk_evaluation.data,model,device=device)
chunk_evaluation.cal_heatcap(chunk_evaluation.phdos,phfre.tolist(),T_lst,chunk_evaluation.structures)
if os.path.exists(h5_file):
with h5py.File(h5_file, 'a') as hf:
hf["material_id"].resize((hf["material_id"].shape[0]+np.array([material_id])[None,:].shape[0]),axis=0)
hf["material_id"][-np.array([material_id])[None,:].shape[0]:] = np.array([material_id])[None,:]
hf["phdos_max1"].resize((hf["phdos_max1"].shape[0]+np.array(chunk_evaluation.phdos).shape[0]),axis=0)
hf["phdos_max1"][-np.array(chunk_evaluation.phdos).shape[0]:] = np.array(chunk_evaluation.phdos)
hf["phdos_norm"].resize((hf["phdos_norm"].shape[0]+np.array(chunk_evaluation.phdos_norm).shape[0]),axis=0)
hf["phdos_norm"][-np.array(chunk_evaluation.phdos_norm).shape[0]:] = np.array(chunk_evaluation.phdos_norm)
hf["heat_cap_mol"].resize((hf["heat_cap_mol"].shape[0]+np.array(chunk_evaluation.C_v_mol).shape[0]),axis=0)
hf["heat_cap_mol"][-np.array(chunk_evaluation.C_v_mol).shape[0]:] = np.array(chunk_evaluation.C_v_mol)
hf["heat_cap_kg"].resize((hf["heat_cap_kg"].shape[0]+np.array(chunk_evaluation.C_v_kg).shape[0]),axis=0)
hf["heat_cap_kg"][-np.array(chunk_evaluation.C_v_kg).shape[0]:] = np.array(chunk_evaluation.C_v_kg)
print("{} Calculating mp-{} ".format(i, cif_id_suc[i]), end="\r", flush=True)
else:
with h5py.File(h5_file, 'w') as hf:
hf.create_dataset("material_id", data=np.array([material_id])[None,:],
compression="gzip", compression_opts=9, chunks=True, maxshape=(None,None))
hf.create_dataset("phdos_max1", data=np.array(chunk_evaluation.phdos),
compression="gzip", compression_opts=9, chunks=True, maxshape=(None,None))
hf.create_dataset("phdos_norm", data=np.array(chunk_evaluation.phdos_norm),
compression="gzip", compression_opts=9, chunks=True, maxshape=(None,None))
hf.create_dataset("heat_cap_mol", data=np.array(chunk_evaluation.C_v_mol),
compression="gzip", compression_opts=9, chunks=True, maxshape=(None,None))
hf.create_dataset("heat_cap_kg", data=np.array(chunk_evaluation.C_v_kg),
compression="gzip", compression_opts=9, chunks=True, maxshape=(None,None))
hf.create_dataset("phfre", data=phfre,
compression="gzip", compression_opts=9, chunks=True, maxshape=(None))
print("Created new h5py data\n")
```
|
github_jupyter
|
import glob
import torch
import torch_geometric
import torch_scatter
import e3nn
from e3nn import rs, o3
from e3nn.point.data_helpers import DataPeriodicNeighbors
from e3nn.networks import GatedConvParityNetwork
from e3nn.kernel_mod import Kernel
from e3nn.point.message_passing import Convolution
import pymatgen
from pymatgen.core.structure import Structure
import time, os
import datetime
import pickle
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
import numpy as np
import h5py
from class_evaluate_MPdata import ComprehensiveEvaluation, AtomEmbeddingAndSumLastLayer
torch.set_default_dtype(torch.float64)
if torch.cuda.is_available():
device = "cuda"
else:
device = "cpu"
print('torch device:' , device)
model_data = torch.load('models/200803-1018_len51max1000_fwin101ord3_trial_run_full_data.torch', map_location=device)
model = AtomEmbeddingAndSumLastLayer(model_data['state']['linear.weight'].shape[1],
model_data['state']['linear.weight'].shape[0],
GatedConvParityNetwork(**model_data['model_kwargs']))
model.load_state_dict(model_data['state'])
model.to(device)
model.eval()
with open('models/phdos_e3nn_len51max1000_fwin101ord3.pkl', 'rb') as f:
data_dict = pickle.load(f)
phfre = data_dict['phfre']
with open('models/cif_unique_files.pkl', 'rb') as f:
ciflist_dict = pickle.load(f)
cif_name = ciflist_dict.get('cif_name')
cif_id = ciflist_dict.get('cif_id')
num_sites = ciflist_dict.get('num_sites')
cif_name_suc = [cif_name[i] for i in range(len(cif_name)) if num_sites[i] <= 13]
cif_id_suc = [cif_id[i] for i in range(len(cif_id)) if num_sites[i] <= 13]
T_lst = [273.15, 293.15]
cif_path = 'data/mp_data/'
h5_file = 'data/phdos_maxsites13_2020Aug27.h5'
for i in range(0,len(cif_id_suc)):
material_id = cif_id_suc[i]
chunk_evaluation = ComprehensiveEvaluation([cif_name_suc[i]], model_data['model_kwargs'], cif_path=cif_path, chunk_id=material_id)
chunk_evaluation.predict_phdos(chunk_evaluation.data,model,device=device)
chunk_evaluation.cal_heatcap(chunk_evaluation.phdos,phfre.tolist(),T_lst,chunk_evaluation.structures)
if os.path.exists(h5_file):
with h5py.File(h5_file, 'a') as hf:
hf["material_id"].resize((hf["material_id"].shape[0]+np.array([material_id])[None,:].shape[0]),axis=0)
hf["material_id"][-np.array([material_id])[None,:].shape[0]:] = np.array([material_id])[None,:]
hf["phdos_max1"].resize((hf["phdos_max1"].shape[0]+np.array(chunk_evaluation.phdos).shape[0]),axis=0)
hf["phdos_max1"][-np.array(chunk_evaluation.phdos).shape[0]:] = np.array(chunk_evaluation.phdos)
hf["phdos_norm"].resize((hf["phdos_norm"].shape[0]+np.array(chunk_evaluation.phdos_norm).shape[0]),axis=0)
hf["phdos_norm"][-np.array(chunk_evaluation.phdos_norm).shape[0]:] = np.array(chunk_evaluation.phdos_norm)
hf["heat_cap_mol"].resize((hf["heat_cap_mol"].shape[0]+np.array(chunk_evaluation.C_v_mol).shape[0]),axis=0)
hf["heat_cap_mol"][-np.array(chunk_evaluation.C_v_mol).shape[0]:] = np.array(chunk_evaluation.C_v_mol)
hf["heat_cap_kg"].resize((hf["heat_cap_kg"].shape[0]+np.array(chunk_evaluation.C_v_kg).shape[0]),axis=0)
hf["heat_cap_kg"][-np.array(chunk_evaluation.C_v_kg).shape[0]:] = np.array(chunk_evaluation.C_v_kg)
print("{} Calculating mp-{} ".format(i, cif_id_suc[i]), end="\r", flush=True)
else:
with h5py.File(h5_file, 'w') as hf:
hf.create_dataset("material_id", data=np.array([material_id])[None,:],
compression="gzip", compression_opts=9, chunks=True, maxshape=(None,None))
hf.create_dataset("phdos_max1", data=np.array(chunk_evaluation.phdos),
compression="gzip", compression_opts=9, chunks=True, maxshape=(None,None))
hf.create_dataset("phdos_norm", data=np.array(chunk_evaluation.phdos_norm),
compression="gzip", compression_opts=9, chunks=True, maxshape=(None,None))
hf.create_dataset("heat_cap_mol", data=np.array(chunk_evaluation.C_v_mol),
compression="gzip", compression_opts=9, chunks=True, maxshape=(None,None))
hf.create_dataset("heat_cap_kg", data=np.array(chunk_evaluation.C_v_kg),
compression="gzip", compression_opts=9, chunks=True, maxshape=(None,None))
hf.create_dataset("phfre", data=phfre,
compression="gzip", compression_opts=9, chunks=True, maxshape=(None))
print("Created new h5py data\n")
| 0.413004 | 0.808823 |
# Deep Deterministic Policy Gradients (DDPG)
---
In this notebook, we train DDPG with OpenAI Gym's Pendulum-v0 environment.
### 1. Import the Necessary Packages
```
import gym
import random
import torch
import numpy as np
from collections import deque
import matplotlib.pyplot as plt
%matplotlib inline
from ddpg_agent import Agent
```
### 2. Instantiate the Environment and Agent
```
env = gym.make('Pendulum-v1')
env.seed(2)
agent = Agent(state_size=3, action_size=1, random_seed=2)
```
### 3. Train the Agent with DDPG
```
def ddpg(n_episodes=1000, max_t=300, print_every=100):
scores_deque = deque(maxlen=print_every)
scores = []
for i_episode in range(1, n_episodes+1): # loop through episodes
state = env.reset() # reset environment
agent.reset() # reset agent
score = 0
for t in range(max_t): # loop through the tip step
action = agent.act(state) # get action from actor network
next_state, reward, done, _ = env.step(action) # get next state, reward from env
agent.step(state, action, reward, next_state, done) # update agent
state = next_state
score += reward
if done:
break
scores_deque.append(score)
scores.append(score)
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_deque)), end="")
torch.save(agent.actor_local.state_dict(), 'checkpoint_actor.pth')
torch.save(agent.critic_local.state_dict(), 'checkpoint_critic.pth')
if i_episode % print_every == 0:
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_deque)))
return scores
scores = ddpg()
fig = plt.figure()
ax = fig.add_subplot(111)
plt.plot(np.arange(1, len(scores)+1), scores)
plt.ylabel('Score')
plt.xlabel('Episode #')
plt.show()
```
### 4. Watch a Smart Agent!
```
agent.actor_local.load_state_dict(torch.load('checkpoint_actor.pth'))
agent.critic_local.load_state_dict(torch.load('checkpoint_critic.pth'))
state = env.reset()
for t in range(200):
action = agent.act(state, add_noise=False)
env.render()
state, reward, done, _ = env.step(action)
if done:
break
env.close()
```
### 6. Explore
In this exercise, we have provided a sample DDPG agent and demonstrated how to use it to solve an OpenAI Gym environment. To continue your learning, you are encouraged to complete any (or all!) of the following tasks:
- Amend the various hyperparameters and network architecture to see if you can get your agent to solve the environment faster than this benchmark implementation. Once you build intuition for the hyperparameters that work well with this environment, try solving a different OpenAI Gym task!
- Write your own DDPG implementation. Use this code as reference only when needed -- try as much as you can to write your own algorithm from scratch.
- You may also like to implement prioritized experience replay, to see if it speeds learning.
- The current implementation adds Ornsetein-Uhlenbeck noise to the action space. However, it has [been shown](https://blog.openai.com/better-exploration-with-parameter-noise/) that adding noise to the parameters of the neural network policy can improve performance. Make this change to the code, to verify it for yourself!
- Write a blog post explaining the intuition behind the DDPG algorithm and demonstrating how to use it to solve an RL environment of your choosing.
|
github_jupyter
|
import gym
import random
import torch
import numpy as np
from collections import deque
import matplotlib.pyplot as plt
%matplotlib inline
from ddpg_agent import Agent
env = gym.make('Pendulum-v1')
env.seed(2)
agent = Agent(state_size=3, action_size=1, random_seed=2)
def ddpg(n_episodes=1000, max_t=300, print_every=100):
scores_deque = deque(maxlen=print_every)
scores = []
for i_episode in range(1, n_episodes+1): # loop through episodes
state = env.reset() # reset environment
agent.reset() # reset agent
score = 0
for t in range(max_t): # loop through the tip step
action = agent.act(state) # get action from actor network
next_state, reward, done, _ = env.step(action) # get next state, reward from env
agent.step(state, action, reward, next_state, done) # update agent
state = next_state
score += reward
if done:
break
scores_deque.append(score)
scores.append(score)
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_deque)), end="")
torch.save(agent.actor_local.state_dict(), 'checkpoint_actor.pth')
torch.save(agent.critic_local.state_dict(), 'checkpoint_critic.pth')
if i_episode % print_every == 0:
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_deque)))
return scores
scores = ddpg()
fig = plt.figure()
ax = fig.add_subplot(111)
plt.plot(np.arange(1, len(scores)+1), scores)
plt.ylabel('Score')
plt.xlabel('Episode #')
plt.show()
agent.actor_local.load_state_dict(torch.load('checkpoint_actor.pth'))
agent.critic_local.load_state_dict(torch.load('checkpoint_critic.pth'))
state = env.reset()
for t in range(200):
action = agent.act(state, add_noise=False)
env.render()
state, reward, done, _ = env.step(action)
if done:
break
env.close()
| 0.454472 | 0.936401 |
```
import pandas as pd
import numpy as np
df = pd.read_csv('./data/in_micro_PERSONS_PerthOnly_2011.csv')
df.rename(columns={
'AREAENUM':'sample_geog',
"ABSHID":'serialno'
},inplace=True)
zonecol = 'sample_geog'
df.columns
df
def produce_one_marginal(col, df):
gdf = pd.DataFrame(df[[zonecol,col]])
gdf = gdf.groupby([zonecol,col])[col].count()
return list(gdf.iteritems())
def merge_one_marginal(resdf, newd, col):
for ad in newd:
zoneid = ad[0][0]
catname = col + ';' + str(ad[0][1])
val = ad[1]
resdf.at[zoneid, catname] = val
def get_twolevel_cols(resdf):
cols1 = []
cols2 = []
for acol in resdf.columns:
col1 = acol
col2 = ''
if ';' in acol:
acolsplit = acol.split(';')
col1 = acolsplit[0]
col2 = acolsplit[1]
cols1.append(col1)
cols2.append(col2)
resdf.columns = cols1
resdf.loc[-1] = cols2
resdf.index = resdf.index + 1
return resdf.sort_index()
def produce_list_marginals(listofmar, df,times = 1, insertgeog = False):
resdf = pd.DataFrame()
for col in listofmar:
newd = produce_one_marginal(col, df)
merge_one_marginal(resdf,newd,col)
resdf = resdf.reset_index().rename({"index":"zone"},axis = 1).fillna(0.0)
resdf = resdf * times
resdf['zone'] = resdf['zone']/times
if insertgeog:
resdf['sample_geog'] = resdf['zone']
resdf = get_twolevel_cols(resdf)
return resdf
resdf = produce_list_marginals(['INCP', 'INDP', 'LFSP', 'OCCP','RLHP', 'SEXP'], df, 2)
resdf
df.to_csv('./data/person_sample.csv',index=False)
resdf.to_csv('./data/person_marginals.csv',index=False)
# start households data things
hdf = pd.read_csv('./data/in_micro_DWELLINGS_PerthOnly_2011.csv')
geomapdf = pd.read_csv('./data/census2011/geocodemap.csv')
geomap = {
'Bendigo and Shepparton':20,
'Cairns and Queensland - Outback':35,
'North West, Warrnambool and South West':31,
'Sydney - Baulkham Hills and Hawkesbury, Sydney - Ryde':8,
'Sydney - Inner West':13,
'Bunbury, Mandurah, Western Australia - Wheat Belt':48,
}
for idx,row in geomapdf.iterrows():
geomap[row['Name']] = int(row['code'])
geomap
# hdf = hdf.replace({'AREAENUM':geomap})
hdf.rename(columns={
'AREAENUM':'sample_geog',
"ABSHID":'serialno'
},inplace=True)
hdf.columns
hresdf = produce_list_marginals(['BEDRD', 'MRERD', 'RNTRD', 'STRD', 'VEHRD'], hdf, 2, True)
hresdf
hdf.to_csv('./data/household_sample.csv',index=False)
hresdf.to_csv('./data/hh_marginals.csv',index=False)
```
|
github_jupyter
|
import pandas as pd
import numpy as np
df = pd.read_csv('./data/in_micro_PERSONS_PerthOnly_2011.csv')
df.rename(columns={
'AREAENUM':'sample_geog',
"ABSHID":'serialno'
},inplace=True)
zonecol = 'sample_geog'
df.columns
df
def produce_one_marginal(col, df):
gdf = pd.DataFrame(df[[zonecol,col]])
gdf = gdf.groupby([zonecol,col])[col].count()
return list(gdf.iteritems())
def merge_one_marginal(resdf, newd, col):
for ad in newd:
zoneid = ad[0][0]
catname = col + ';' + str(ad[0][1])
val = ad[1]
resdf.at[zoneid, catname] = val
def get_twolevel_cols(resdf):
cols1 = []
cols2 = []
for acol in resdf.columns:
col1 = acol
col2 = ''
if ';' in acol:
acolsplit = acol.split(';')
col1 = acolsplit[0]
col2 = acolsplit[1]
cols1.append(col1)
cols2.append(col2)
resdf.columns = cols1
resdf.loc[-1] = cols2
resdf.index = resdf.index + 1
return resdf.sort_index()
def produce_list_marginals(listofmar, df,times = 1, insertgeog = False):
resdf = pd.DataFrame()
for col in listofmar:
newd = produce_one_marginal(col, df)
merge_one_marginal(resdf,newd,col)
resdf = resdf.reset_index().rename({"index":"zone"},axis = 1).fillna(0.0)
resdf = resdf * times
resdf['zone'] = resdf['zone']/times
if insertgeog:
resdf['sample_geog'] = resdf['zone']
resdf = get_twolevel_cols(resdf)
return resdf
resdf = produce_list_marginals(['INCP', 'INDP', 'LFSP', 'OCCP','RLHP', 'SEXP'], df, 2)
resdf
df.to_csv('./data/person_sample.csv',index=False)
resdf.to_csv('./data/person_marginals.csv',index=False)
# start households data things
hdf = pd.read_csv('./data/in_micro_DWELLINGS_PerthOnly_2011.csv')
geomapdf = pd.read_csv('./data/census2011/geocodemap.csv')
geomap = {
'Bendigo and Shepparton':20,
'Cairns and Queensland - Outback':35,
'North West, Warrnambool and South West':31,
'Sydney - Baulkham Hills and Hawkesbury, Sydney - Ryde':8,
'Sydney - Inner West':13,
'Bunbury, Mandurah, Western Australia - Wheat Belt':48,
}
for idx,row in geomapdf.iterrows():
geomap[row['Name']] = int(row['code'])
geomap
# hdf = hdf.replace({'AREAENUM':geomap})
hdf.rename(columns={
'AREAENUM':'sample_geog',
"ABSHID":'serialno'
},inplace=True)
hdf.columns
hresdf = produce_list_marginals(['BEDRD', 'MRERD', 'RNTRD', 'STRD', 'VEHRD'], hdf, 2, True)
hresdf
hdf.to_csv('./data/household_sample.csv',index=False)
hresdf.to_csv('./data/hh_marginals.csv',index=False)
| 0.132739 | 0.210604 |
# Titanic's data analysis and machine learning
## By Jérémy P. Schneider
As someone new in this field, I decided to take my first challenge with the Titanic dataset from Kaggle (https://www.kaggle.com/c/titanic)
## My OS
For this work I used a computer with :
* Windows 7
* Intel(R) Core(TM) i5-2500K CPU @ 3.30GHz
* 16 Go RAM (4 x 4 Go)
* NVIDIA GeForce GTX 1050 Ti
This notebook will be split in different part :
## [Setting our environnement :](#ENV)
### [Libraries Import](#lib)
### [Personnalized functions and tools](#func)
### [Data import](#import)
## [Data cleaning and analysis :](#CLEAN)
### [Data analysis](#analysis)
### [Handeling missing data](#MISSING)
### [Converting all data in number](#convert)
## [Test and train a model](#test_train)
## [Conclusion and trial over the test sample](#conclusion)
<a id="ENV"></a>
# Setting our environnement
<a id="lib"></a>
## Libraries Import
```
import time
import pandas as pd
import matplotlib.pyplot as plt
import math
import seaborn as sns
import numpy as np
from sklearn import svm
from sklearn.preprocessing import MinMaxScaler
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split, GridSearchCV
from sklearn.metrics import confusion_matrix
```
<a id="func"></a>
## Personalized functions and tools
Since the Titanic dataset contains a name with a title, I used the idea of Ertuğrul Demir to create a dictionary to replace them (https://www.kaggle.com/datafan07/titanic-eda-and-several-modelling-approaches)
```
dict_title = {
'Capt': 'Dr/Clerc/Mil',
'Col': 'Dr/Clerc/Mil',
'Major': 'Dr/Clerc/Mil',
'Jonkheer': 'Honor',
'Don': 'Honor',
'Dona': 'Honor',
'Sir': 'Honor',
'Dr': 'Dr/Clerc/Mil',
'Rev': 'Dr/Clerc/Mil',
'the Countess': 'Honor',
'Mme': 'Mrs',
'Mlle': 'Miss',
'Ms': 'Mrs',
'Mr': 'Mr',
'Mrs': 'Mrs',
'Miss': 'Miss',
'Master': 'Master',
'Lady': 'Honor'
}
def extractTitle(df, nameCol, dictTitle):
'''
extractTitle(df, nameCol, dictTitle)
Input : df : dataframe, will be copied.
nameCol : name of the columns where to extract titles.
dictTitle : dictionary of title and their conversion.
This fonction extract title from a specific column with a custom dict and remove nameCol.
'''
df_new = df.copy()
df_new["Title"] = ""
for row in range(df_new.shape[0]):
name = df_new.loc[row][nameCol]
for title in dictTitle:
if title in name:
df_new["Title"][row] = dictTitle[title]
return df_new.drop([nameCol], axis=1)
def getDummiesTitanic(df, dummies):
'''
getDummiesTitanic(df, dummies)
Input : df : dataframe, will be copied.
dummies : list of dummies to transform.
dictTitle : dictionary of title and their conversion
This fonction get dummies for a given list and drop the original column.
'''
df_new = df.copy()
for dummy in dummies:
try :
df_new = df_new.join(pd.get_dummies(df_new[dummy], prefix = dummy))
df_new = df_new.drop([dummy], axis=1)
except KeyError:
print("Warning : column {} is missing".format(dummy))
return df_new
def drawConfusionMatrix(y_test, y_pred):
'''
drawConfusionMatrix(y_test, y_pred)
Input : y_test : list of real target.
y_pred : list of predicted target.
This fonction draw a confusion matrix from y_test and y_pred.
'''
cf_matrix = confusion_matrix(y_test, y_pred)
cm_sum = np.sum(cf_matrix, axis=1, keepdims=True)
cm_perc = cf_matrix / cm_sum.astype(float) * 100
annot = np.empty_like(cf_matrix).astype(str)
nrows, ncols = cf_matrix.shape
labels = ["Died", "Survived"]
sns.heatmap(cf_matrix/np.sum(cf_matrix),
xticklabels=labels,
yticklabels=labels,
annot=True)
plt.yticks(rotation=0)
plt.ylabel('Predicted values', rotation=0)
plt.xlabel('Actual values')
plt.show()
```
<a id="import"></a>
## Data import
```
df_train_org = pd.read_csv("data/train.csv")
df_test_org = pd.read_csv("data/test.csv")
```
<a id="CLEAN"></a>
# Data cleaning and analysis
<a id="analysis"></a>
## Data analysis
From Kaggle we know how the data is ordered and it define our goal :
The data has been split into two groups:
training set (train.csv)
test set (test.csv)
The training set should be used to build your machine learning models. For the training set, we provide the outcome (also known as the “ground truth”) for each passenger. Your model will be based on “features” like passengers’ gender and class. You can also use feature engineering to create new features.
The test set should be used to see how well your model performs on unseen data. For the test set, we do not provide the ground truth for each passenger. It is your job to predict these outcomes. For each passenger in the test set, use the model you trained to predict whether or not they survived the sinking of the Titanic.
We also include gender_submission.csv, a set of predictions that assume all and only female passengers survive, as an example of what a submission file should look like.
### Data Dictionary
|Variable |Definition |Key
| ------------- |:-----------------:| -----:|
|survival |Survival |0 = No, 1 = Yes
|pclass |Ticket class |1 = 1st, 2 = 2nd, 3 = 3rd
|sex |Sex
|Age |Age in years
|sibsp |# of siblings / spouses aboard the Titanic
|parch |# of parents / children aboard the Titanic
|ticket |Ticket number
|fare |Passenger fare
|cabin |Cabin number
|embarked |Port of Embarkation|C = Cherbourg, Q = Queenstown, S = Southampton
### Variable Notes
pclass: A proxy for socio-economic status (SES)
1st = Upper
2nd = Middle
3rd = Lower
age: Age is fractional if less than 1. If the age is estimated, is it in the form of xx.5
sibsp: The dataset defines family relations in this way...
Sibling = brother, sister, stepbrother, stepsister
Spouse = husband, wife (mistresses and fiancés were ignored)
parch: The dataset defines family relations in this way...
Parent = mother, father
Child = daughter, son, stepdaughter, stepson
Some children travelled only with a nanny, therefore parch=0 for them.
If we watch how the data is in our ile, wa saw that the information is correct.
* First the train data
```
df_train_org.dtypes
print("In the train data we have {} rows and {} columns".format(df_train_org.shape[0], df_train_org.shape[1]))
```
* Then the test data
```
df_test_org.dtypes
print("In the test data we have {} rows and {} columns".format(df_test_org.shape[0], df_test_org.shape[1]))
```
As we see the two set have the same topology :
* int and float for numerical values.
* object for text data.
We'll need to transform the text data in numerical values in order to feed our machine learning model.
### Checking correlation
We first remplace __Sex__ and __Embarked__ with numerical values based on the occurencies on the train data.
Since we don't want to have biais we won't look too deep in the test data.
```
df_train_org["Sex"].value_counts()
df_train_org["Embarked"].value_counts()
df_train = df_train_org.copy()
df_train = df_train.drop(["PassengerId", "Ticket"],axis=1) # Remove unique ID
df_train["SexNum"] = df_train["Sex"]
df_train["SexNum"].loc[df_train["SexNum"] == "male"] = 1
df_train["SexNum"].loc[df_train["SexNum"] == "female"] = 0
df_train["EmbarkedNum"] = df_train["Embarked"]
df_train["EmbarkedNum"] = df_train["EmbarkedNum"].fillna(0)
df_train["EmbarkedNum"].loc[df_train["EmbarkedNum"] == "S"] = 2
df_train["EmbarkedNum"].loc[df_train["EmbarkedNum"] == "C"] = 1
df_train["EmbarkedNum"].loc[df_train["EmbarkedNum"] == "Q"] = 0
df_train["EmbarkedNum"] = df_train["EmbarkedNum"].astype(int)
df_test= df_test_org.copy()
df_test= df_test.drop(["PassengerId", "Ticket"],axis=1) # Remove unique ID
df_test["SexNum"] = df_test["Sex"]
df_test["SexNum"].loc[df_test["SexNum"] == "male"] = 1
df_test["SexNum"].loc[df_test["SexNum"] == "female"] = 0
df_test["EmbarkedNum"] = df_test["Embarked"]
df_test["EmbarkedNum"] = df_test["EmbarkedNum"].fillna(0)
df_test["EmbarkedNum"].loc[df_test["EmbarkedNum"] == "S"] = 2
df_test["EmbarkedNum"].loc[df_test["EmbarkedNum"] == "C"] = 1
df_test["EmbarkedNum"].loc[df_test["EmbarkedNum"] == "Q"] = 0
df_test["EmbarkedNum"] = df_test["EmbarkedNum"].astype(int)
```
Then we compute the correlation
```
start_time = time.time()
plt.figure(figsize=(8,8))
sns.heatmap(df_train.corr(), annot=True, linewidths=.5, annot_kws={"size":10})
plt.show()
elapsed_time = time.time() - start_time
print("This graphic took me : {}".format(time.strftime("%H:%M:%S", time.gmtime(elapsed_time))))
```
We can se that we have :
* a weak negative correclation between __PClass__ and __Survived__ : 1st class survived more.
* a weak positive correlation between __Survived__ and __Fare__ : if you have more money you wuold have survived.
* a weak negative correlation between __Survived__ and __SexNum__ : more women survived.
* a weak negative correlation between __Pclass__ and __Fare__ : 1st class have more money than 3rd.
* a weak positive correlation between __SibSp__ and __Parch__ : since it's the number of family onboard it's logical.
<a id="MISSING"></a>
### Handeling missing data
After checking the type of the data we use, we need to see where missing values are, to do so we can simply calculated them this way :
```
df_train.isna().mean()
df_test.isna().mean()
```
From this step we know :
* __Cabin__ information is missing in more than 75% of our data, so we'll not use it.
* __Age__ information is missing at 20% of the time, we'll try to remplace the missing values
* __Embarked__ information is missing several values in the trainset
* __Fare__ information is missing serveral values in the testset
We can use three method to take care of the missing data points
* Remove columns
```
df_train_remove = df_train.drop(["Cabin", "Age", "Embarked", "Fare"], axis=1)
df_test_remove = df_test.drop(["Cabin", "Age", "Embarked", "Fare"], axis=1)
```
* Compute MEAN for numerical, and most occurent for categorical
For Embarked we take at look at the data
```
df_train["Embarked"].value_counts()
start_time = time.time()
df_train_mean = df_train.drop(["Cabin"], axis=1).copy()
df_train_mean["Age"] = df_train_mean["Age"].fillna(df_train_mean["Age"].mean())
df_train_mean["Embarked"] = df_train_mean["Embarked"].fillna("S")
df_test_mean = df_test.drop(["Cabin"], axis=1).copy()
df_test_mean["Age"] = df_test_mean["Age"].fillna(df_test_mean["Age"].mean())
df_test_mean["Fare"] = df_test_mean["Fare"].fillna(df_test_mean["Fare"].mean())
elapsed_time = time.time() - start_time
print("This calculations took me : {}".format(time.strftime("%H:%M:%S", time.gmtime(elapsed_time))))
df_train_mean.isna().sum()
```
* Compute MEDIAN for numerical and most occurent for categorical
```
df_train_median = df_train.drop(["Cabin"], axis=1).copy()
df_train_median["Age"] = df_train_median["Age"].fillna(df_train_median["Age"].median())
df_train_median["Embarked"] = df_train_median["Embarked"].fillna("S")
df_test_median = df_test.drop(["Cabin"], axis=1).copy()
df_test_median["Age"] = df_test_median["Age"].fillna(df_test_median["Age"].median())
df_test_median["Fare"] = df_test_median["Fare"].fillna(df_test_median["Fare"].median())
```
<a id="convert"></a>
### Converting all data in number
We'll first take care of the name column. We'll extract the title from the names. (REF)
The steps are :
* Create a new column
* Read each names
* Check if any title exist in the name
* Add the title in the new column
We'll need to do it on all our datasets, so we can use the fonction defined before.
```
start_time = time.time()
df_train_remove = extractTitle(df_train_remove, "Name", dict_title)
df_test_remove = extractTitle(df_test_remove, "Name", dict_title)
df_train_mean = extractTitle(df_train_mean, "Name", dict_title)
df_test_mean = extractTitle(df_test_mean, "Name", dict_title)
df_train_median = extractTitle(df_train_median, "Name", dict_title)
df_test_median = extractTitle(df_test_median, "Name", dict_title)
elapsed_time = time.time() - start_time
print("This calculations took me : {}".format(time.strftime("%H:%M:%S", time.gmtime(elapsed_time))))
```
One of the most current way to deal with categorical data is to [get dummies](https://towardsdatascience.com/the-dummys-guide-to-creating-dummy-variables-f21faddb1d40)
```
df_train_remove.head()
start_time = time.time()
list_dummies = ["Sex", "Embarked", "Title"]
df_train_remove = getDummiesTitanic(df_train_remove, list_dummies)
df_test_remove = getDummiesTitanic(df_test_remove, list_dummies)
df_train_mean = getDummiesTitanic(df_train_mean, list_dummies)
df_test_mean = getDummiesTitanic(df_test_mean, list_dummies)
df_train_median = getDummiesTitanic(df_train_median, list_dummies)
df_test_median = getDummiesTitanic(df_test_median, list_dummies)
elapsed_time = time.time() - start_time
print("This calculations took me : {}".format(time.strftime("%H:%M:%S", time.gmtime(elapsed_time))))
df_train_remove.head()
```
## MinMaxScaler
```
df_train_remove_minmax = df_train_remove.copy()
df_test_remove_minmax = df_test_remove.copy()
df_train_mean_minmax = df_train_mean.copy()
df_test_mean_minmax = df_test_mean.copy()
df_train_median_minmax = df_train_median.copy()
df_test_median_minmax = df_test_median.copy()
```
### Searching features to standardize
```
list_remove = []
list_mean = []
list_median = []
scaler = MinMaxScaler()
for column in df_train_remove_minmax.columns:
if df_train_remove_minmax[column].max() > 1:
list_remove.append(column)
df_train_remove_minmax[list_remove] = scaler.fit_transform(df_train_remove_minmax[list_remove])
df_test_remove_minmax[list_remove] = scaler.fit_transform(df_test_remove_minmax[list_remove])
for column in df_train_mean_minmax.columns:
if df_train_mean_minmax[column].max() > 1:
list_mean.append(column)
df_train_mean_minmax[list_mean] = scaler.fit_transform(df_train_mean_minmax[list_mean])
df_test_mean_minmax[list_mean] = scaler.fit_transform(df_test_mean_minmax[list_mean])
for column in df_train_median_minmax.columns:
if df_train_median_minmax[column].max() > 1:
list_median.append(column)
df_train_median_minmax[list_median] = scaler.fit_transform(df_train_median_minmax[list_median])
df_test_median_minmax[list_median] = scaler.fit_transform(df_test_median_minmax[list_median])
```
<a id="test_train"></a>
## Test and train a model
First we need to split the datasets in train and test. The mesure the preformance of our models.
We also create a dict to record all the scores.
```
X_train_remove, X_test_remove, y_train_remove, y_test_remove = train_test_split(df_train_remove.drop(["Survived"], axis=1),
df_train_remove["Survived"],
test_size=0.2,
random_state=0)
X_train_mean, X_test_mean, y_train_mean, y_test_mean = train_test_split(df_train_mean.drop(["Survived"], axis=1),
df_train_mean["Survived"],
test_size=0.2,
random_state=0)
X_train_median, X_test_median, y_train_median, y_test_median = train_test_split(df_train_median.drop(["Survived"], axis=1),
df_train_median["Survived"],
test_size=0.2,
random_state=0)
X_train_remove_minmax, X_test_remove_minmax, y_train_remove_minmax, y_test_remove_minmax = train_test_split(df_train_remove_minmax.drop(["Survived"], axis=1),
df_train_remove_minmax["Survived"],
test_size=0.2,
random_state=0)
X_train_mean_minmax, X_test_mean_minmax, y_train_mean_minmax, y_test_mean_minmax = train_test_split(df_train_mean_minmax.drop(["Survived"], axis=1),
df_train_mean_minmax["Survived"],
test_size=0.2,
random_state=0)
X_train_median_minmax, X_test_median_minmax, y_train_median_minmax, y_test_median_minmax = train_test_split(df_train_median_minmax.drop(["Survived"], axis=1),
df_train_median_minmax["Survived"],
test_size=0.2,
random_state=0)
SCORES = {"Remove":{},"Mean":{},"Median":{},"Remove_minmax":{},"Mean_minmax":{},"Median_minmax":{}}
```
### With the SVM (Support Vector Machines [see documentation](https://scikit-learn.org/stable/modules/svm.html)) :
* If we use the df_train_remove :
```
start_time = time.time()
clf = svm.SVC(kernel='linear', C = 1.0) #Check other models
clf.fit(X_train_remove, y_train_remove)
y_pred_remove = clf.predict(X_test_remove)
drawConfusionMatrix(y_test_remove, y_pred_remove)
score = (((y_pred_remove == y_test_remove).sum())/y_test_remove.shape[0])
score = round(score*100,2)
SCORES["Remove"]["SVM"] = score
print("Perfomace is : {}% for SVM_Remove".format(score))
elapsed_time = time.time() - start_time
print("This calculations took me : {}".format(time.strftime("%H:%M:%S", time.gmtime(elapsed_time))))
```
* If we use the df_train_mean
```
start_time = time.time()
clf.fit(X_train_mean, y_train_mean)
y_pred_mean = clf.predict(X_test_mean)
drawConfusionMatrix(y_test_mean, y_pred_mean)
score = (((y_pred_mean == y_test_mean).sum())/y_test_mean.shape[0])
score = round(score*100,2)
SCORES["Mean"]["SVM"] = score
print("Perfomace is : {}% for SVM_Mean".format(score))
elapsed_time = time.time() - start_time
print("This calculations took me : {}".format(time.strftime("%H:%M:%S", time.gmtime(elapsed_time))))
```
* If we use the df_train_median
```
start_time = time.time()
clf.fit(X_train_median, y_train_median)
y_pred_median = clf.predict(X_test_median)
drawConfusionMatrix(y_test_median, y_pred_median)
score = (((y_pred_median == y_test_median).sum())/y_test_median.shape[0])
score = round(score*100,2)
SCORES["Median"]["SVM"] = score
print("Perfomace is : {}% for SVM_Median".format(score))
elapsed_time = time.time() - start_time
print("This calculations took me : {}".format(time.strftime("%H:%M:%S", time.gmtime(elapsed_time))))
SCORES
```
## GridSearch to optimized Hyperparameters [see documentation](https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.GridSearchCV.html)
The results with basic parameters for SVM are not bad, but we can find better hyperparameters with GridSearch.
```
# Dict to save best parameters
BEST_PARAMS = {"Remove":{},"Mean":{},"Median":{},"Remove_minmax":{},"Mean_minmax":{},"Median_minmax":{}}
# defining parameter range
param_grid = [{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["rbf", "sigmoid"]},
{"C": [0.1, 1, 10, 100, 1000],
"kernel": ["linear"]},
{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["poly"],
"degree" : [1,2,3,4,5,6,7,8,9,10]}]
# tol and max_iter because It's taking too long to train
grid = GridSearchCV(svm.SVC(max_iter=1000000), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_remove, y_train_remove)
print("The best parameters are : {} with removed features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Remove"]["SVM_BestParam"] = score
BEST_PARAMS["Remove"]["SVM"] = grid.best_params_
# defining parameter range
param_grid = [{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["rbf", "sigmoid"]},
{"C": [0.1, 1, 10, 100, 1000],
"kernel": ["linear"]},
{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["poly"],
"degree" : [1,2,3,4,5,6,7,8,9,10]}]
# tol and max_iter because It's taking too long to train
grid = GridSearchCV(svm.SVC(max_iter=1000000), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_mean, y_train_mean)
print("The best parameters are : {} with mean features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Mean"]["SVM_BestParam"] = score
BEST_PARAMS["Mean"]["SVM"] = grid.best_params_
# defining parameter range
param_grid = [{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["rbf", "sigmoid"]},
{"C": [0.1, 1, 10, 100, 1000],
"kernel": ["linear"]},
{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["poly"],
"degree" : [1,2,3,4,5,6,7,8,9,10]}]
# tol and max_iter because It's taking too long to train
grid = GridSearchCV(svm.SVC(max_iter=1000000), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_median, y_train_median)
print("The best parameters are : {} with median featuresand the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Median"]["SVM_BestParam"] = score
BEST_PARAMS["Median"]["SVM"] = grid.best_params_
SCORES
BEST_PARAMS
```
As we can see with the best scores we almost gain 2% with optimized hyperparameters.
If we plot them :
```
pd.DataFrame(SCORES).plot(kind='bar')
plt.ylim(0, 120)
plt.ylabel('Precision')
plt.show()
```
## With Standardized values
```
# defining parameter range
param_grid = [{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["rbf", "sigmoid"]},
{"C": [0.1, 1, 10, 100, 1000],
"kernel": ["linear"]},
{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["poly"],
"degree" : [1,2,3,4,5,6,7,8,9,10]}]
# tol and max_iter because It's taking too long to train
grid = GridSearchCV(svm.SVC(max_iter=1000000), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_remove_minmax, y_train_remove_minmax)
print("The best parameters are : {} with removed features and MinMaxScaler also the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Remove_minmax"]["SVM_BestParam"] = score
BEST_PARAMS["Remove_minmax"]["SVM"] = grid.best_params_
# defining parameter range
param_grid = [{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["rbf", "sigmoid"]},
{"C": [0.1, 1, 10, 100, 1000],
"kernel": ["linear"]},
{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["poly"],
"degree" : [1,2,3,4,5,6,7,8,9,10]}]
# tol and max_iter because It's taking too long to train
grid = GridSearchCV(svm.SVC(max_iter=1000000), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_mean_minmax, y_train_mean_minmax)
print("The best parameters are : {} with mean features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Mean_minmax"]["SVM_BestParam"] = score
BEST_PARAMS["Mean_minmax"]["SVM"] = grid.best_params_
# defining parameter range
param_grid = [{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["rbf", "sigmoid"]},
{"C": [0.1, 1, 10, 100, 1000],
"kernel": ["linear"]},
{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["poly"],
"degree" : [1,2,3,4,5,6,7,8,9,10]}]
# tol and max_iter because It's taking too long to train
grid = GridSearchCV(svm.SVC(max_iter=1000000), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_median_minmax, y_train_median_minmax)
print("The best parameters are : {} with median featuresand the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Median_minmax"]["SVM_BestParam"] = score
BEST_PARAMS["Median_minmax"]["SVM"] = grid.best_params_
```
## With the KNeighborsClassifier [see documentation](https://scikit-learn.org/stable/modules/generated/sklearn.neighbors.KNeighborsClassifier.html)
```
knn = KNeighborsClassifier(n_neighbors=3)
print("Train/Test/Record for df_train_remove")
knn.fit(X_train_remove, y_train_remove)
y_pred_remove = knn.predict(X_test_remove)
print(confusion_matrix(y_test_remove, y_pred_remove))
score = (((y_pred_remove == y_test_remove).sum())/y_test_remove.shape[0])
score = round(score*100,2)
SCORES["Remove"]["KNN_3"] = score
print("Performace is : {}% for KNN_3_Remove".format(score))
print("Train/Test/Record for df_train_mean")
knn.fit(X_train_mean, y_train_mean)
y_pred_mean = knn.predict(X_test_mean)
print(confusion_matrix(y_test_mean, y_pred_mean))
score = (((y_pred_mean == y_test_mean).sum())/y_test_mean.shape[0])
score = round(score*100,2)
SCORES["Mean"]["KNN_3"] = score
print("Performace is : {}% for KNN_3_Mean".format(score))
print("Train/Test/Record for df_train_median")
knn.fit(X_train_median, y_train_median)
y_pred_median = knn.predict(X_test_median)
print(confusion_matrix(y_test_median, y_pred_median))
score = (((y_pred_median == y_test_median).sum())/y_test_median.shape[0])
score = round(score*100,2)
SCORES["Median"]["KNN_3"] = score
print("Performace is : {}% for KNN_3_Median".format(score))
SCORES
```
## Trying differents hyperparameters (again)
```
# defining parameter range
param_grid = [{"n_neighbors": range(1,101),
"weights": ["uniform", "distance"],
"algorithm": ["auto", "brute"],
"p" : [1,2]},
{"n_neighbors": range(1,101),
"weights": ["uniform", "distance"],
"algorithm": ["ball_tree", "kd_tree"],
"p" : [1,2],
"leaf_size": [1,2,3,4,5,10,15,20,25,30]}]
# With Remove features
grid = GridSearchCV(KNeighborsClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_remove, y_train_remove)
print("The best parameters are : {} with remove features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Remove"]["KNN_BestParam"] = score
BEST_PARAMS["Remove"]["KNN"] = grid.best_params_
# With Mean features
grid = GridSearchCV(KNeighborsClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_mean, y_train_mean)
print("The best parameters are : {} with mean features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Mean"]["KNN_BestParam"] = score
BEST_PARAMS["Mean"]["KNN"] = grid.best_params_
# With Median features
grid = GridSearchCV(KNeighborsClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_median, y_train_median)
print("The best parameters are : {} with mean features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Median"]["KNN_BestParam"] = score
BEST_PARAMS["Median"]["KNN"] = grid.best_params_
# With Remove_minmax features
grid = GridSearchCV(KNeighborsClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_remove_minmax, y_train_remove_minmax)
print("The best parameters are : {} with remove features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Remove_minmax"]["KNN_BestParam"] = score
BEST_PARAMS["Remove_minmax"]["KNN"] = grid.best_params_
# With Mean_minmax features
grid = GridSearchCV(KNeighborsClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_mean_minmax, y_train_mean_minmax)
print("The best parameters are : {} with mean features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Mean_minmax"]["KNN_BestParam"] = score
BEST_PARAMS["Mean_minmax"]["KNN"] = grid.best_params_
# With Median_minmax features
grid = GridSearchCV(KNeighborsClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_median_minmax, y_train_median_minmax)
print("The best parameters are : {} with mean features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Median_minmax"]["KNN_BestParam"] = score
BEST_PARAMS["Median_minmax"]["KNN"] = grid.best_params_
```
# Random Forest Classifier
```
RF = RandomForestClassifier(n_estimators=100)
print("Train/Test/Record for df_train_remove")
RF.fit(X_train_remove, y_train_remove)
y_pred_remove = RF.predict(X_test_remove)
print(confusion_matrix(y_test_remove, y_pred_remove))
score = (((y_pred_remove == y_test_remove).sum())/y_test_remove.shape[0])
score = round(score*100,2)
SCORES["Remove"]["RF_100"] = score
print("Performace is : {}% for RF_100_Remove".format(score))
print("Train/Test/Record for df_train_mean")
RF.fit(X_train_mean, y_train_mean)
y_pred_mean = RF.predict(X_test_mean)
print(confusion_matrix(y_test_mean, y_pred_mean))
score = (((y_pred_mean == y_test_mean).sum())/y_test_mean.shape[0])
score = round(score*100,2)
SCORES["Mean"]["RF_100"] = score
print("Performace is : {}% for RF_100_Mean".format(score))
print("Train/Test/Record for df_train_median")
RF.fit(X_train_median, y_train_median)
y_pred_median = RF.predict(X_test_median)
print(confusion_matrix(y_test_median, y_pred_median))
score = (((y_pred_median == y_test_median).sum())/y_test_median.shape[0])
score = round(score*100,2)
SCORES["Median"]["RF_100"] = score
print("Performace is : {}% for RF_100_Median".format(score))
```
## Trying differents hyperparameters (and again)
```
# defining parameter range
param_grid = [{"n_estimators": range(1,251),
"criterion": ["gini", "entropy"],
"max_depth": [1,2,3,4,5, None]
}]
# With Remove features
grid = GridSearchCV(RandomForestClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_remove, y_train_remove)
print("The best parameters are : {} with remove features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Remove"]["RF_BestParam"] = score
BEST_PARAMS["Remove"]["RF"] = grid.best_params_
# With Mean features
grid = GridSearchCV(RandomForestClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_mean, y_train_mean)
print("The best parameters are : {} with mean features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Mean"]["RF_BestParam"] = score
BEST_PARAMS["Mean"]["RF"] = grid.best_params_
# With Median features
grid = GridSearchCV(RandomForestClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_median, y_train_median)
print("The best parameters are : {} with mean features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Median"]["RF_BestParam"] = score
BEST_PARAMS["Median"]["RF"] = grid.best_params_
# With Remove_minmax features
grid = GridSearchCV(RandomForestClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_remove_minmax, y_train_remove_minmax)
print("The best parameters are : {} with remove features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Remove_minmax"]["RF_BestParam"] = score
BEST_PARAMS["Remove_minmax"]["RF"] = grid.best_params_
# With Mean_minmax features
grid = GridSearchCV(RandomForestClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_mean_minmax, y_train_mean_minmax)
print("The best parameters are : {} with mean features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Mean_minmax"]["RF_BestParam"] = score
BEST_PARAMS["Mean_minmax"]["RF"] = grid.best_params_
# With Median_minmax features
grid = GridSearchCV(RandomForestClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_median_minmax, y_train_median_minmax)
print("The best parameters are : {} with mean features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Median_minmax"]["RF_BestParam"] = score
BEST_PARAMS["Median_minmax"]["RF"] = grid.best_params_
```
If we plot the results
```
pd.DataFrame(SCORES).plot(kind='bar')
plt.ylim(0, 120)
plt.ylabel('Precision')
plt.show()
SCORES
BEST_PARAMS
```
<a id="conclusion"></a>
## Conclusion and trial over the test sample
From all these tests we can conclude that the best model is the SVM with best_param with 83.15% and remove features.
So if we do it with the real test data :
```
X_train = df_train_remove.drop(["Survived"], axis=1)
Y_train = df_train_remove["Survived"]
X_test = df_test_remove
BEST_PARAMS["Remove"]["SVM"]
start_time = time.time()
clf = svm.SVC(C = 1, gamma = 0.1, kernel = 'rbf') #Use best params
clf.fit(X_train, Y_train)
y_pred = clf.predict(X_test)
# Creation of the submission file :
DF_Fin = pd.DataFrame(columns=["PassengerId","Survived"])
DF_Fin["PassengerId"] = df_test_org["PassengerId"]
DF_Fin["Survived"] = y_pred
DF_Fin.head()
```
|
github_jupyter
|
import time
import pandas as pd
import matplotlib.pyplot as plt
import math
import seaborn as sns
import numpy as np
from sklearn import svm
from sklearn.preprocessing import MinMaxScaler
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split, GridSearchCV
from sklearn.metrics import confusion_matrix
dict_title = {
'Capt': 'Dr/Clerc/Mil',
'Col': 'Dr/Clerc/Mil',
'Major': 'Dr/Clerc/Mil',
'Jonkheer': 'Honor',
'Don': 'Honor',
'Dona': 'Honor',
'Sir': 'Honor',
'Dr': 'Dr/Clerc/Mil',
'Rev': 'Dr/Clerc/Mil',
'the Countess': 'Honor',
'Mme': 'Mrs',
'Mlle': 'Miss',
'Ms': 'Mrs',
'Mr': 'Mr',
'Mrs': 'Mrs',
'Miss': 'Miss',
'Master': 'Master',
'Lady': 'Honor'
}
def extractTitle(df, nameCol, dictTitle):
'''
extractTitle(df, nameCol, dictTitle)
Input : df : dataframe, will be copied.
nameCol : name of the columns where to extract titles.
dictTitle : dictionary of title and their conversion.
This fonction extract title from a specific column with a custom dict and remove nameCol.
'''
df_new = df.copy()
df_new["Title"] = ""
for row in range(df_new.shape[0]):
name = df_new.loc[row][nameCol]
for title in dictTitle:
if title in name:
df_new["Title"][row] = dictTitle[title]
return df_new.drop([nameCol], axis=1)
def getDummiesTitanic(df, dummies):
'''
getDummiesTitanic(df, dummies)
Input : df : dataframe, will be copied.
dummies : list of dummies to transform.
dictTitle : dictionary of title and their conversion
This fonction get dummies for a given list and drop the original column.
'''
df_new = df.copy()
for dummy in dummies:
try :
df_new = df_new.join(pd.get_dummies(df_new[dummy], prefix = dummy))
df_new = df_new.drop([dummy], axis=1)
except KeyError:
print("Warning : column {} is missing".format(dummy))
return df_new
def drawConfusionMatrix(y_test, y_pred):
'''
drawConfusionMatrix(y_test, y_pred)
Input : y_test : list of real target.
y_pred : list of predicted target.
This fonction draw a confusion matrix from y_test and y_pred.
'''
cf_matrix = confusion_matrix(y_test, y_pred)
cm_sum = np.sum(cf_matrix, axis=1, keepdims=True)
cm_perc = cf_matrix / cm_sum.astype(float) * 100
annot = np.empty_like(cf_matrix).astype(str)
nrows, ncols = cf_matrix.shape
labels = ["Died", "Survived"]
sns.heatmap(cf_matrix/np.sum(cf_matrix),
xticklabels=labels,
yticklabels=labels,
annot=True)
plt.yticks(rotation=0)
plt.ylabel('Predicted values', rotation=0)
plt.xlabel('Actual values')
plt.show()
df_train_org = pd.read_csv("data/train.csv")
df_test_org = pd.read_csv("data/test.csv")
df_train_org.dtypes
print("In the train data we have {} rows and {} columns".format(df_train_org.shape[0], df_train_org.shape[1]))
df_test_org.dtypes
print("In the test data we have {} rows and {} columns".format(df_test_org.shape[0], df_test_org.shape[1]))
df_train_org["Sex"].value_counts()
df_train_org["Embarked"].value_counts()
df_train = df_train_org.copy()
df_train = df_train.drop(["PassengerId", "Ticket"],axis=1) # Remove unique ID
df_train["SexNum"] = df_train["Sex"]
df_train["SexNum"].loc[df_train["SexNum"] == "male"] = 1
df_train["SexNum"].loc[df_train["SexNum"] == "female"] = 0
df_train["EmbarkedNum"] = df_train["Embarked"]
df_train["EmbarkedNum"] = df_train["EmbarkedNum"].fillna(0)
df_train["EmbarkedNum"].loc[df_train["EmbarkedNum"] == "S"] = 2
df_train["EmbarkedNum"].loc[df_train["EmbarkedNum"] == "C"] = 1
df_train["EmbarkedNum"].loc[df_train["EmbarkedNum"] == "Q"] = 0
df_train["EmbarkedNum"] = df_train["EmbarkedNum"].astype(int)
df_test= df_test_org.copy()
df_test= df_test.drop(["PassengerId", "Ticket"],axis=1) # Remove unique ID
df_test["SexNum"] = df_test["Sex"]
df_test["SexNum"].loc[df_test["SexNum"] == "male"] = 1
df_test["SexNum"].loc[df_test["SexNum"] == "female"] = 0
df_test["EmbarkedNum"] = df_test["Embarked"]
df_test["EmbarkedNum"] = df_test["EmbarkedNum"].fillna(0)
df_test["EmbarkedNum"].loc[df_test["EmbarkedNum"] == "S"] = 2
df_test["EmbarkedNum"].loc[df_test["EmbarkedNum"] == "C"] = 1
df_test["EmbarkedNum"].loc[df_test["EmbarkedNum"] == "Q"] = 0
df_test["EmbarkedNum"] = df_test["EmbarkedNum"].astype(int)
start_time = time.time()
plt.figure(figsize=(8,8))
sns.heatmap(df_train.corr(), annot=True, linewidths=.5, annot_kws={"size":10})
plt.show()
elapsed_time = time.time() - start_time
print("This graphic took me : {}".format(time.strftime("%H:%M:%S", time.gmtime(elapsed_time))))
df_train.isna().mean()
df_test.isna().mean()
df_train_remove = df_train.drop(["Cabin", "Age", "Embarked", "Fare"], axis=1)
df_test_remove = df_test.drop(["Cabin", "Age", "Embarked", "Fare"], axis=1)
df_train["Embarked"].value_counts()
start_time = time.time()
df_train_mean = df_train.drop(["Cabin"], axis=1).copy()
df_train_mean["Age"] = df_train_mean["Age"].fillna(df_train_mean["Age"].mean())
df_train_mean["Embarked"] = df_train_mean["Embarked"].fillna("S")
df_test_mean = df_test.drop(["Cabin"], axis=1).copy()
df_test_mean["Age"] = df_test_mean["Age"].fillna(df_test_mean["Age"].mean())
df_test_mean["Fare"] = df_test_mean["Fare"].fillna(df_test_mean["Fare"].mean())
elapsed_time = time.time() - start_time
print("This calculations took me : {}".format(time.strftime("%H:%M:%S", time.gmtime(elapsed_time))))
df_train_mean.isna().sum()
df_train_median = df_train.drop(["Cabin"], axis=1).copy()
df_train_median["Age"] = df_train_median["Age"].fillna(df_train_median["Age"].median())
df_train_median["Embarked"] = df_train_median["Embarked"].fillna("S")
df_test_median = df_test.drop(["Cabin"], axis=1).copy()
df_test_median["Age"] = df_test_median["Age"].fillna(df_test_median["Age"].median())
df_test_median["Fare"] = df_test_median["Fare"].fillna(df_test_median["Fare"].median())
start_time = time.time()
df_train_remove = extractTitle(df_train_remove, "Name", dict_title)
df_test_remove = extractTitle(df_test_remove, "Name", dict_title)
df_train_mean = extractTitle(df_train_mean, "Name", dict_title)
df_test_mean = extractTitle(df_test_mean, "Name", dict_title)
df_train_median = extractTitle(df_train_median, "Name", dict_title)
df_test_median = extractTitle(df_test_median, "Name", dict_title)
elapsed_time = time.time() - start_time
print("This calculations took me : {}".format(time.strftime("%H:%M:%S", time.gmtime(elapsed_time))))
df_train_remove.head()
start_time = time.time()
list_dummies = ["Sex", "Embarked", "Title"]
df_train_remove = getDummiesTitanic(df_train_remove, list_dummies)
df_test_remove = getDummiesTitanic(df_test_remove, list_dummies)
df_train_mean = getDummiesTitanic(df_train_mean, list_dummies)
df_test_mean = getDummiesTitanic(df_test_mean, list_dummies)
df_train_median = getDummiesTitanic(df_train_median, list_dummies)
df_test_median = getDummiesTitanic(df_test_median, list_dummies)
elapsed_time = time.time() - start_time
print("This calculations took me : {}".format(time.strftime("%H:%M:%S", time.gmtime(elapsed_time))))
df_train_remove.head()
df_train_remove_minmax = df_train_remove.copy()
df_test_remove_minmax = df_test_remove.copy()
df_train_mean_minmax = df_train_mean.copy()
df_test_mean_minmax = df_test_mean.copy()
df_train_median_minmax = df_train_median.copy()
df_test_median_minmax = df_test_median.copy()
list_remove = []
list_mean = []
list_median = []
scaler = MinMaxScaler()
for column in df_train_remove_minmax.columns:
if df_train_remove_minmax[column].max() > 1:
list_remove.append(column)
df_train_remove_minmax[list_remove] = scaler.fit_transform(df_train_remove_minmax[list_remove])
df_test_remove_minmax[list_remove] = scaler.fit_transform(df_test_remove_minmax[list_remove])
for column in df_train_mean_minmax.columns:
if df_train_mean_minmax[column].max() > 1:
list_mean.append(column)
df_train_mean_minmax[list_mean] = scaler.fit_transform(df_train_mean_minmax[list_mean])
df_test_mean_minmax[list_mean] = scaler.fit_transform(df_test_mean_minmax[list_mean])
for column in df_train_median_minmax.columns:
if df_train_median_minmax[column].max() > 1:
list_median.append(column)
df_train_median_minmax[list_median] = scaler.fit_transform(df_train_median_minmax[list_median])
df_test_median_minmax[list_median] = scaler.fit_transform(df_test_median_minmax[list_median])
X_train_remove, X_test_remove, y_train_remove, y_test_remove = train_test_split(df_train_remove.drop(["Survived"], axis=1),
df_train_remove["Survived"],
test_size=0.2,
random_state=0)
X_train_mean, X_test_mean, y_train_mean, y_test_mean = train_test_split(df_train_mean.drop(["Survived"], axis=1),
df_train_mean["Survived"],
test_size=0.2,
random_state=0)
X_train_median, X_test_median, y_train_median, y_test_median = train_test_split(df_train_median.drop(["Survived"], axis=1),
df_train_median["Survived"],
test_size=0.2,
random_state=0)
X_train_remove_minmax, X_test_remove_minmax, y_train_remove_minmax, y_test_remove_minmax = train_test_split(df_train_remove_minmax.drop(["Survived"], axis=1),
df_train_remove_minmax["Survived"],
test_size=0.2,
random_state=0)
X_train_mean_minmax, X_test_mean_minmax, y_train_mean_minmax, y_test_mean_minmax = train_test_split(df_train_mean_minmax.drop(["Survived"], axis=1),
df_train_mean_minmax["Survived"],
test_size=0.2,
random_state=0)
X_train_median_minmax, X_test_median_minmax, y_train_median_minmax, y_test_median_minmax = train_test_split(df_train_median_minmax.drop(["Survived"], axis=1),
df_train_median_minmax["Survived"],
test_size=0.2,
random_state=0)
SCORES = {"Remove":{},"Mean":{},"Median":{},"Remove_minmax":{},"Mean_minmax":{},"Median_minmax":{}}
start_time = time.time()
clf = svm.SVC(kernel='linear', C = 1.0) #Check other models
clf.fit(X_train_remove, y_train_remove)
y_pred_remove = clf.predict(X_test_remove)
drawConfusionMatrix(y_test_remove, y_pred_remove)
score = (((y_pred_remove == y_test_remove).sum())/y_test_remove.shape[0])
score = round(score*100,2)
SCORES["Remove"]["SVM"] = score
print("Perfomace is : {}% for SVM_Remove".format(score))
elapsed_time = time.time() - start_time
print("This calculations took me : {}".format(time.strftime("%H:%M:%S", time.gmtime(elapsed_time))))
start_time = time.time()
clf.fit(X_train_mean, y_train_mean)
y_pred_mean = clf.predict(X_test_mean)
drawConfusionMatrix(y_test_mean, y_pred_mean)
score = (((y_pred_mean == y_test_mean).sum())/y_test_mean.shape[0])
score = round(score*100,2)
SCORES["Mean"]["SVM"] = score
print("Perfomace is : {}% for SVM_Mean".format(score))
elapsed_time = time.time() - start_time
print("This calculations took me : {}".format(time.strftime("%H:%M:%S", time.gmtime(elapsed_time))))
start_time = time.time()
clf.fit(X_train_median, y_train_median)
y_pred_median = clf.predict(X_test_median)
drawConfusionMatrix(y_test_median, y_pred_median)
score = (((y_pred_median == y_test_median).sum())/y_test_median.shape[0])
score = round(score*100,2)
SCORES["Median"]["SVM"] = score
print("Perfomace is : {}% for SVM_Median".format(score))
elapsed_time = time.time() - start_time
print("This calculations took me : {}".format(time.strftime("%H:%M:%S", time.gmtime(elapsed_time))))
SCORES
# Dict to save best parameters
BEST_PARAMS = {"Remove":{},"Mean":{},"Median":{},"Remove_minmax":{},"Mean_minmax":{},"Median_minmax":{}}
# defining parameter range
param_grid = [{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["rbf", "sigmoid"]},
{"C": [0.1, 1, 10, 100, 1000],
"kernel": ["linear"]},
{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["poly"],
"degree" : [1,2,3,4,5,6,7,8,9,10]}]
# tol and max_iter because It's taking too long to train
grid = GridSearchCV(svm.SVC(max_iter=1000000), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_remove, y_train_remove)
print("The best parameters are : {} with removed features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Remove"]["SVM_BestParam"] = score
BEST_PARAMS["Remove"]["SVM"] = grid.best_params_
# defining parameter range
param_grid = [{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["rbf", "sigmoid"]},
{"C": [0.1, 1, 10, 100, 1000],
"kernel": ["linear"]},
{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["poly"],
"degree" : [1,2,3,4,5,6,7,8,9,10]}]
# tol and max_iter because It's taking too long to train
grid = GridSearchCV(svm.SVC(max_iter=1000000), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_mean, y_train_mean)
print("The best parameters are : {} with mean features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Mean"]["SVM_BestParam"] = score
BEST_PARAMS["Mean"]["SVM"] = grid.best_params_
# defining parameter range
param_grid = [{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["rbf", "sigmoid"]},
{"C": [0.1, 1, 10, 100, 1000],
"kernel": ["linear"]},
{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["poly"],
"degree" : [1,2,3,4,5,6,7,8,9,10]}]
# tol and max_iter because It's taking too long to train
grid = GridSearchCV(svm.SVC(max_iter=1000000), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_median, y_train_median)
print("The best parameters are : {} with median featuresand the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Median"]["SVM_BestParam"] = score
BEST_PARAMS["Median"]["SVM"] = grid.best_params_
SCORES
BEST_PARAMS
pd.DataFrame(SCORES).plot(kind='bar')
plt.ylim(0, 120)
plt.ylabel('Precision')
plt.show()
# defining parameter range
param_grid = [{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["rbf", "sigmoid"]},
{"C": [0.1, 1, 10, 100, 1000],
"kernel": ["linear"]},
{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["poly"],
"degree" : [1,2,3,4,5,6,7,8,9,10]}]
# tol and max_iter because It's taking too long to train
grid = GridSearchCV(svm.SVC(max_iter=1000000), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_remove_minmax, y_train_remove_minmax)
print("The best parameters are : {} with removed features and MinMaxScaler also the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Remove_minmax"]["SVM_BestParam"] = score
BEST_PARAMS["Remove_minmax"]["SVM"] = grid.best_params_
# defining parameter range
param_grid = [{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["rbf", "sigmoid"]},
{"C": [0.1, 1, 10, 100, 1000],
"kernel": ["linear"]},
{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["poly"],
"degree" : [1,2,3,4,5,6,7,8,9,10]}]
# tol and max_iter because It's taking too long to train
grid = GridSearchCV(svm.SVC(max_iter=1000000), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_mean_minmax, y_train_mean_minmax)
print("The best parameters are : {} with mean features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Mean_minmax"]["SVM_BestParam"] = score
BEST_PARAMS["Mean_minmax"]["SVM"] = grid.best_params_
# defining parameter range
param_grid = [{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["rbf", "sigmoid"]},
{"C": [0.1, 1, 10, 100, 1000],
"kernel": ["linear"]},
{"C": [0.1, 1, 10, 100, 1000],
"gamma": [1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["poly"],
"degree" : [1,2,3,4,5,6,7,8,9,10]}]
# tol and max_iter because It's taking too long to train
grid = GridSearchCV(svm.SVC(max_iter=1000000), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_median_minmax, y_train_median_minmax)
print("The best parameters are : {} with median featuresand the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Median_minmax"]["SVM_BestParam"] = score
BEST_PARAMS["Median_minmax"]["SVM"] = grid.best_params_
knn = KNeighborsClassifier(n_neighbors=3)
print("Train/Test/Record for df_train_remove")
knn.fit(X_train_remove, y_train_remove)
y_pred_remove = knn.predict(X_test_remove)
print(confusion_matrix(y_test_remove, y_pred_remove))
score = (((y_pred_remove == y_test_remove).sum())/y_test_remove.shape[0])
score = round(score*100,2)
SCORES["Remove"]["KNN_3"] = score
print("Performace is : {}% for KNN_3_Remove".format(score))
print("Train/Test/Record for df_train_mean")
knn.fit(X_train_mean, y_train_mean)
y_pred_mean = knn.predict(X_test_mean)
print(confusion_matrix(y_test_mean, y_pred_mean))
score = (((y_pred_mean == y_test_mean).sum())/y_test_mean.shape[0])
score = round(score*100,2)
SCORES["Mean"]["KNN_3"] = score
print("Performace is : {}% for KNN_3_Mean".format(score))
print("Train/Test/Record for df_train_median")
knn.fit(X_train_median, y_train_median)
y_pred_median = knn.predict(X_test_median)
print(confusion_matrix(y_test_median, y_pred_median))
score = (((y_pred_median == y_test_median).sum())/y_test_median.shape[0])
score = round(score*100,2)
SCORES["Median"]["KNN_3"] = score
print("Performace is : {}% for KNN_3_Median".format(score))
SCORES
# defining parameter range
param_grid = [{"n_neighbors": range(1,101),
"weights": ["uniform", "distance"],
"algorithm": ["auto", "brute"],
"p" : [1,2]},
{"n_neighbors": range(1,101),
"weights": ["uniform", "distance"],
"algorithm": ["ball_tree", "kd_tree"],
"p" : [1,2],
"leaf_size": [1,2,3,4,5,10,15,20,25,30]}]
# With Remove features
grid = GridSearchCV(KNeighborsClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_remove, y_train_remove)
print("The best parameters are : {} with remove features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Remove"]["KNN_BestParam"] = score
BEST_PARAMS["Remove"]["KNN"] = grid.best_params_
# With Mean features
grid = GridSearchCV(KNeighborsClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_mean, y_train_mean)
print("The best parameters are : {} with mean features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Mean"]["KNN_BestParam"] = score
BEST_PARAMS["Mean"]["KNN"] = grid.best_params_
# With Median features
grid = GridSearchCV(KNeighborsClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_median, y_train_median)
print("The best parameters are : {} with mean features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Median"]["KNN_BestParam"] = score
BEST_PARAMS["Median"]["KNN"] = grid.best_params_
# With Remove_minmax features
grid = GridSearchCV(KNeighborsClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_remove_minmax, y_train_remove_minmax)
print("The best parameters are : {} with remove features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Remove_minmax"]["KNN_BestParam"] = score
BEST_PARAMS["Remove_minmax"]["KNN"] = grid.best_params_
# With Mean_minmax features
grid = GridSearchCV(KNeighborsClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_mean_minmax, y_train_mean_minmax)
print("The best parameters are : {} with mean features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Mean_minmax"]["KNN_BestParam"] = score
BEST_PARAMS["Mean_minmax"]["KNN"] = grid.best_params_
# With Median_minmax features
grid = GridSearchCV(KNeighborsClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_median_minmax, y_train_median_minmax)
print("The best parameters are : {} with mean features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Median_minmax"]["KNN_BestParam"] = score
BEST_PARAMS["Median_minmax"]["KNN"] = grid.best_params_
RF = RandomForestClassifier(n_estimators=100)
print("Train/Test/Record for df_train_remove")
RF.fit(X_train_remove, y_train_remove)
y_pred_remove = RF.predict(X_test_remove)
print(confusion_matrix(y_test_remove, y_pred_remove))
score = (((y_pred_remove == y_test_remove).sum())/y_test_remove.shape[0])
score = round(score*100,2)
SCORES["Remove"]["RF_100"] = score
print("Performace is : {}% for RF_100_Remove".format(score))
print("Train/Test/Record for df_train_mean")
RF.fit(X_train_mean, y_train_mean)
y_pred_mean = RF.predict(X_test_mean)
print(confusion_matrix(y_test_mean, y_pred_mean))
score = (((y_pred_mean == y_test_mean).sum())/y_test_mean.shape[0])
score = round(score*100,2)
SCORES["Mean"]["RF_100"] = score
print("Performace is : {}% for RF_100_Mean".format(score))
print("Train/Test/Record for df_train_median")
RF.fit(X_train_median, y_train_median)
y_pred_median = RF.predict(X_test_median)
print(confusion_matrix(y_test_median, y_pred_median))
score = (((y_pred_median == y_test_median).sum())/y_test_median.shape[0])
score = round(score*100,2)
SCORES["Median"]["RF_100"] = score
print("Performace is : {}% for RF_100_Median".format(score))
# defining parameter range
param_grid = [{"n_estimators": range(1,251),
"criterion": ["gini", "entropy"],
"max_depth": [1,2,3,4,5, None]
}]
# With Remove features
grid = GridSearchCV(RandomForestClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_remove, y_train_remove)
print("The best parameters are : {} with remove features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Remove"]["RF_BestParam"] = score
BEST_PARAMS["Remove"]["RF"] = grid.best_params_
# With Mean features
grid = GridSearchCV(RandomForestClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_mean, y_train_mean)
print("The best parameters are : {} with mean features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Mean"]["RF_BestParam"] = score
BEST_PARAMS["Mean"]["RF"] = grid.best_params_
# With Median features
grid = GridSearchCV(RandomForestClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_median, y_train_median)
print("The best parameters are : {} with mean features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Median"]["RF_BestParam"] = score
BEST_PARAMS["Median"]["RF"] = grid.best_params_
# With Remove_minmax features
grid = GridSearchCV(RandomForestClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_remove_minmax, y_train_remove_minmax)
print("The best parameters are : {} with remove features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Remove_minmax"]["RF_BestParam"] = score
BEST_PARAMS["Remove_minmax"]["RF"] = grid.best_params_
# With Mean_minmax features
grid = GridSearchCV(RandomForestClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_mean_minmax, y_train_mean_minmax)
print("The best parameters are : {} with mean features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Mean_minmax"]["RF_BestParam"] = score
BEST_PARAMS["Mean_minmax"]["RF"] = grid.best_params_
# With Median_minmax features
grid = GridSearchCV(RandomForestClassifier(), param_grid, refit = True, verbose=3, n_jobs=-1, cv=5)
grid.fit(X_train_median_minmax, y_train_median_minmax)
print("The best parameters are : {} with mean features and the score is {}".format(grid.best_params_, grid.best_score_))
score = round((grid.best_score_*100),2)
SCORES["Median_minmax"]["RF_BestParam"] = score
BEST_PARAMS["Median_minmax"]["RF"] = grid.best_params_
pd.DataFrame(SCORES).plot(kind='bar')
plt.ylim(0, 120)
plt.ylabel('Precision')
plt.show()
SCORES
BEST_PARAMS
X_train = df_train_remove.drop(["Survived"], axis=1)
Y_train = df_train_remove["Survived"]
X_test = df_test_remove
BEST_PARAMS["Remove"]["SVM"]
start_time = time.time()
clf = svm.SVC(C = 1, gamma = 0.1, kernel = 'rbf') #Use best params
clf.fit(X_train, Y_train)
y_pred = clf.predict(X_test)
# Creation of the submission file :
DF_Fin = pd.DataFrame(columns=["PassengerId","Survived"])
DF_Fin["PassengerId"] = df_test_org["PassengerId"]
DF_Fin["Survived"] = y_pred
DF_Fin.head()
| 0.242475 | 0.945801 |
```
import matplotlib.pyplot as plt
import matplotlib.ticker as mtick
import numpy as np
import os
import pandas as pd
import math
%matplotlib inline
# Load data from filesystem
df = pd.read_csv('/kaggle/input/survey_results_public.csv', delimiter=',', nrows = None)
df.dataframeName = 'survey_results_public.csv'
pandasVersion = pd.__version__
print(f'Pandas version is {pandasVersion}')
# Some classes we will be using for convenience and to keep the code shorter.
class DataFrameHelper:
''' Keeps a copy of the origin (unless filter is applied) so we can always use as many rows as possible. Has often used helper methods. '''
def __init__(self, dataframe):
''' Constructor.
Args:
dataframe: The dataframe to operate on
'''
self.dataframe = dataframe
def printShape(self):
''' Print the shape of the dataframe '''
nRow, nCol = self.dataframe.shape
print(f'Shape: there are {nRow} rows and {nCol} columns in the dataframe.')
def printMissingValuesSum(self, columnNames):
''' Prints the missing value of the passed columns
Args:
columnNames: Names of the columgs to print the sum of missing values of
'''
for columnName in columnNames:
print("{} has {} missing values.".format(columnName, self.dataframe[columnName].isnull().sum()))
def applyFilter(self, columnNames):
''' Alters the contained dataframe by keeping only the passed columns
Args:
columnNames: Names of the columns to apply the filter to
'''
self.dataframe = self.dataframe.filter(columnNames)
return self
def dropNa(self, columnNames):
''' Remove NaN values from the passed columns and return a fresh dataframe
Args:
columnNames: Names of the columns to drop NaN values of
'''
return self.dataframe.dropna(subset = columnNames)
def dropZero(self, columnName):
''' Remove zero (0) values from the passed columns and return a fresh dataframe
Args:
columnName: Name of the column to drop zero values of
'''
return self.dataframe[self.dataframe[columnName] != 0]
def stackAggregatedValues(self, columnName, separator = ";"):
'''Count the values of the given column.
Used when column values are aggregated (contain multiple values which are delimited)
Args:
columnName (str): Name of the column to split and count the values of.
delimiter (str, optional): The delimiter to splie by. Defaults to ';'.
Returns:
A dataframe with the value counts.
'''
# Split each row's values by the delimiter into new columns, then stack these into rows and count the values.
# strip() is used because f.i. data can have the values "Take online courses" and " Take online courses".
return self.dataframe[columnName].str.split(';', expand = True).stack().str.strip()
class PlotHelper:
''' Plot helper has some predefined values for quicker and easier plotting '''
def __init__(self, dataframe):
''' Constructor.
Args:
dataframe: Used to calculate the total number of items
xAxisLabelValue: Label of the x axis
yAxisLabelValue: Label of the y axis
yAxisFactorValue: Scale the y axis, useful if there are percentages
yAxisFormatterValue: The y axis formatter to use
showGridValue: Whether to show the grid
numberOfItemsValue: Limit the number of items to plot
kindValue: What kind of plot to display
'''
self.dataframe = dataframe
self.xAxisLabelValue = ""
self.yAxisLabelValue = ""
self.yAxisFactorValue = 1
self.yAxisFormatterValue = mtick.PercentFormatter()
self.showGridValue = True
self.numberOfItemsValue = 0
self.kindValue = "bar"
def xAxisLabel(self, xAxisLabel):
''' Set the label of the x axis
Args:
xAxisLabelValue: Label of the x axis
'''
self.xAxisLabelValue = xAxisLabel
return self
def yAxisLabel(self, yAxisLabel):
''' Set the label of the y axis
Args:
yAxisLabelValue: Label of the y axis
'''
self.yAxisLabelValue = yAxisLabel
return self
def yAxisFactor(self, yAxisFactor):
''' Set the scaling of the y axis
Args:
yAxisFactorValue: Scale the y axis, useful if there are percentages
'''
self.yAxisFactorValue = yAxisFactor
return self
def yAxisFormatter(self, yAxisFormatter):
''' Set the y axis formatter
Args:
yAxisFormatterValue: The y axis formatter to use
'''
self.yAxisFormatterValue = yAxisFormatter
return self
def showGridValue(self, showGridValue):
''' Whether to show the grid in the plot
Args:
showGridValue: Whether to show the grid
'''
self.showGridValue = showGridValue
return self
def top(self, numberOfItems):
''' Limit plot items
Args:
numberOfItemsValue: Limit the number of items to plot
'''
self.numberOfItemsValue = numberOfItems
return self
def kind(self, kind):
''' Set the kind of plot we want to show
Args:
kindValue: What kind of plot to display
'''
self.kindValue = kind
return self
def showPlot(self, data, title = "Title"):
''' Plot with the set instance values
Args:
title: The plot's main title
'''
if (self.numberOfItemsValue > 0):
# Limit the number of items shown
data = data.head(self.numberOfItemsValue)
# Calcluate the plot values
resultingPlot = (data * self.yAxisFactorValue / self.dataframe.shape[0]).plot(kind = self.kindValue);
# Set formatter, labels, grid, and title
resultingPlot.yaxis.set_major_formatter(self.yAxisFormatterValue)
plt.xlabel(self.xAxisLabelValue)
plt.ylabel(self.yAxisLabelValue)
plt.grid(self.showGridValue)
plt.title(title);
# Show the plot
plt.show()
# Some preliminary settings, like what columns are most relevant and how many of the top languages to look at, create initial dataset
relevantColumns = ["HaveWorkedLanguage", "WantWorkLanguage", "CareerSatisfaction", "JobSatisfaction", "JobSeekingStatus", "DeveloperType", "Salary"]
topLanguages = 15
origin = DataFrameHelper(df)
# Print some information on the original dataset
origin.printShape()
origin.printMissingValuesSum(relevantColumns)
# Only use the columns we're interested in
origin.applyFilter(relevantColumns)
origin.printShape()
# Question 1: What languages are most represented?
# Drop the null values and use the resulting dataset.
hwl = DataFrameHelper(origin.dropNa(["HaveWorkedLanguage"]))
# Plot the results
PlotHelper(hwl.dataframe).top(topLanguages).yAxisFactor(100).xAxisLabel("Language").yAxisLabel("Usage in percent (multiple nominations allowed)") \
.showPlot(hwl.stackAggregatedValues("HaveWorkedLanguage").value_counts(), f"Top {topLanguages} Languages")
# Question 2: Refine the languages by developer type.
# This is achieved in several passes:
# 1. For simplicity's sake, assume the first given role is the developer's primary role. Store this in the DeveloperType column.
# 2. Build a list of all possible developer types.
# 3. Build a list of all possible language types.
# 4. In a dictionary that has all developer types, keep a counted dictionary of each language per that developer type.
# As always clean the data. Also apply filter to make working with the dataset a bit easier.
lbt = DataFrameHelper(origin.dropNa(["HaveWorkedLanguage", "DeveloperType"])).applyFilter(["HaveWorkedLanguage", "DeveloperType"]).dataframe
# Only use the primary role.
for (index, roles) in lbt["DeveloperType"].iteritems():
lbt.at[index, "DeveloperType"] = roles.split(";")[0]
# Skip roles "Other" and "Graphic designer" (there are only three results for graphic designer, this is not representative)
lbt = lbt[lbt.DeveloperType != "Other"]
lbt = lbt[lbt.DeveloperType != "Graphic designer"]
# Find all unique developer types and languages.
exsitingDeveloperTypes = pd.unique(lbt["DeveloperType"])
existingLanguages = pd.unique(DataFrameHelper(hwl.dataframe.head(topLanguages)).stackAggregatedValues("HaveWorkedLanguage"))
def createLanguageCounter(languages):
# Build a dictionary of all possible languages and counts.
languagesInUse = {}
for language in languages:
languagesInUse[language] = 0
return languagesInUse
# Build a dictionary of all possible developer types and their respective language counts.
developerTypes = {}
for developerType in exsitingDeveloperTypes:
developerTypes[developerType] = createLanguageCounter(existingLanguages)
# Perform the count (using list comprehension with lambda instead of iterrows)
def countByDeveloperType(d, l):
for language in l.split(";"):
if language in developerTypes[d]:
developerTypes[d][language.strip()] += 1
[countByDeveloperType(d, l) for d, l in zip(lbt['DeveloperType'], lbt['HaveWorkedLanguage'])]
# Plot and do a bit of scaling.
# Unfortunately I found no better way to remove titles.
pd.DataFrame(developerTypes).plot(subplots = True, kind = 'bar', title = [''] * len(exsitingDeveloperTypes))
fig_size = plt.gcf().get_size_inches()
sizefactor = 3
plt.gcf().set_size_inches(sizefactor * fig_size);
plt.show()
# Question 3: Now we are looking at the languages with the highest job satisfaction and salaries at the same time.
# For highest job satisfaction and salaries we will be looking at all values above the mean.
# Prepare the data for selection.
salsat = DataFrameHelper(origin.dropNa(["HaveWorkedLanguage", "JobSatisfaction", "Salary"])).applyFilter(["HaveWorkedLanguage", "JobSatisfaction", "Salary"])
# Save language counts to later proportionalize the filtered value counts
salaryAndSatisfactionCountsBefore = salsat.stackAggregatedValues("HaveWorkedLanguage").value_counts()
# Determine the means.
salsat = salsat.dataframe
salaryMean = salsat["Salary"].mean()
jobSatisfactionMean = salsat["JobSatisfaction"].mean()
salsat["Salary"] = salsat["Salary"].astype("int64")
# Drop any records below the mean.
salsat.drop(salsat[salsat["Salary"] < salaryMean].index, inplace = True)
salsat.drop(salsat[salsat["JobSatisfaction"] < jobSatisfactionMean].index, inplace = True)
# Count the langueage values present in the dataset.
salaryAndSatisfactionCountsAfter = DataFrameHelper(salsat).stackAggregatedValues("HaveWorkedLanguage").value_counts()
# Create a new dataframe for easier working.
salaryAndSatisfactionChart = DataFrameHelper(pd.DataFrame.from_dict(salaryAndSatisfactionCountsAfter))
# Proportionalize the language counts, then for plotting, simplify again and sort.
salaryAndSatisfactionChart.dataframe["HaveWorkedLanguage"] = (salaryAndSatisfactionChart.dataframe.iloc[:,0] / salaryAndSatisfactionCountsBefore[salaryAndSatisfactionChart.dataframe.index]) * 100
salaryAndSatisfactionChart.applyFilter(["HaveWorkedLanguage"])
salaryAndSatisfactionChart = salaryAndSatisfactionChart.dataframe.head(topLanguages + 5)
salaryAndSatisfactionChart = salaryAndSatisfactionChart.sort_values(by = ['HaveWorkedLanguage'], ascending = False)
# Plot.
PlotHelper(salaryAndSatisfactionChart).yAxisFactor(10).xAxisLabel("Language").yAxisLabel("High job satisfaction and salary in percent") \
.showPlot(salaryAndSatisfactionChart, f"Languages with the highest job satisfaction and salary")
# Question 4 examines what language developers that consider getting a new job are currently using.
jss = DataFrameHelper(origin.dropNa(["HaveWorkedLanguage", "JobSeekingStatus"])).applyFilter(["HaveWorkedLanguage", "JobSeekingStatus"]).dataframe
isNotSeekingJob = "no"
isSeekingJob = "yes"
# Filter by developers actively looking for a new job or who may be open to new opportunities.
jss["LookingForJob"] = np.where(jss["JobSeekingStatus"] != "I am not interested in new job opportunities", isSeekingJob, isNotSeekingJob)
# Get total number of jobs for those who answered the "JobSeekingStatus" question
seekingLanguageCounts = DataFrameHelper(jss).stackAggregatedValues("HaveWorkedLanguage").value_counts()
# Build a dictionary of whether developers are looking for a new job and their respective language counts.
jobSeekers = { isSeekingJob: createLanguageCounter(existingLanguages) }
def countByJobSeekers(s, l):
if s == isSeekingJob:
for language in l.split(";"):
if language in jobSeekers[s]:
jobSeekers[s][language.strip()] += 1
[countByJobSeekers(s, l) for s, l in zip(jss['LookingForJob'], jss['HaveWorkedLanguage'])]
# Create a new dataframe for easier working. Proportionalize and prepare for plotting as we did in Question 3.
jobSeekersChart = DataFrameHelper(pd.DataFrame.from_dict(jobSeekers))
jobSeekersChart.dataframe["MaySeekNewJob"] = (jobSeekersChart.dataframe.iloc[:,0] / seekingLanguageCounts[jobSeekersChart.dataframe.index]) * 100
jobSeekersChart.applyFilter(["MaySeekNewJob"])
jobSeekersChart = jobSeekersChart.dataframe.sort_values(by = ['MaySeekNewJob'], ascending = False)
# Plot
PlotHelper(jobSeekersChart).yAxisFactor(10).xAxisLabel("Language").yAxisLabel("Seeking new job in percent") \
.showPlot(jobSeekersChart, f"Developers of these languages may be seeking new jobs")
#Some data that was used for selection, pre-validation, lookup, reference and result verification
#print(pd.unique(lbt["DeveloperType"]))
#print(pd.unique(hwl.stackAggregatedValues("HaveWorkedLanguage")))
#print(pd.unique(jss["JobSeekingStatus"]))
#print(hwl.stackAggregatedValues("HaveWorkedLanguage").value_counts())
#print(DataFrameHelper(jss).stackAggregatedValues("HaveWorkedLanguage").value_counts())
#print(print(len(jss)))
```
|
github_jupyter
|
import matplotlib.pyplot as plt
import matplotlib.ticker as mtick
import numpy as np
import os
import pandas as pd
import math
%matplotlib inline
# Load data from filesystem
df = pd.read_csv('/kaggle/input/survey_results_public.csv', delimiter=',', nrows = None)
df.dataframeName = 'survey_results_public.csv'
pandasVersion = pd.__version__
print(f'Pandas version is {pandasVersion}')
# Some classes we will be using for convenience and to keep the code shorter.
class DataFrameHelper:
''' Keeps a copy of the origin (unless filter is applied) so we can always use as many rows as possible. Has often used helper methods. '''
def __init__(self, dataframe):
''' Constructor.
Args:
dataframe: The dataframe to operate on
'''
self.dataframe = dataframe
def printShape(self):
''' Print the shape of the dataframe '''
nRow, nCol = self.dataframe.shape
print(f'Shape: there are {nRow} rows and {nCol} columns in the dataframe.')
def printMissingValuesSum(self, columnNames):
''' Prints the missing value of the passed columns
Args:
columnNames: Names of the columgs to print the sum of missing values of
'''
for columnName in columnNames:
print("{} has {} missing values.".format(columnName, self.dataframe[columnName].isnull().sum()))
def applyFilter(self, columnNames):
''' Alters the contained dataframe by keeping only the passed columns
Args:
columnNames: Names of the columns to apply the filter to
'''
self.dataframe = self.dataframe.filter(columnNames)
return self
def dropNa(self, columnNames):
''' Remove NaN values from the passed columns and return a fresh dataframe
Args:
columnNames: Names of the columns to drop NaN values of
'''
return self.dataframe.dropna(subset = columnNames)
def dropZero(self, columnName):
''' Remove zero (0) values from the passed columns and return a fresh dataframe
Args:
columnName: Name of the column to drop zero values of
'''
return self.dataframe[self.dataframe[columnName] != 0]
def stackAggregatedValues(self, columnName, separator = ";"):
'''Count the values of the given column.
Used when column values are aggregated (contain multiple values which are delimited)
Args:
columnName (str): Name of the column to split and count the values of.
delimiter (str, optional): The delimiter to splie by. Defaults to ';'.
Returns:
A dataframe with the value counts.
'''
# Split each row's values by the delimiter into new columns, then stack these into rows and count the values.
# strip() is used because f.i. data can have the values "Take online courses" and " Take online courses".
return self.dataframe[columnName].str.split(';', expand = True).stack().str.strip()
class PlotHelper:
''' Plot helper has some predefined values for quicker and easier plotting '''
def __init__(self, dataframe):
''' Constructor.
Args:
dataframe: Used to calculate the total number of items
xAxisLabelValue: Label of the x axis
yAxisLabelValue: Label of the y axis
yAxisFactorValue: Scale the y axis, useful if there are percentages
yAxisFormatterValue: The y axis formatter to use
showGridValue: Whether to show the grid
numberOfItemsValue: Limit the number of items to plot
kindValue: What kind of plot to display
'''
self.dataframe = dataframe
self.xAxisLabelValue = ""
self.yAxisLabelValue = ""
self.yAxisFactorValue = 1
self.yAxisFormatterValue = mtick.PercentFormatter()
self.showGridValue = True
self.numberOfItemsValue = 0
self.kindValue = "bar"
def xAxisLabel(self, xAxisLabel):
''' Set the label of the x axis
Args:
xAxisLabelValue: Label of the x axis
'''
self.xAxisLabelValue = xAxisLabel
return self
def yAxisLabel(self, yAxisLabel):
''' Set the label of the y axis
Args:
yAxisLabelValue: Label of the y axis
'''
self.yAxisLabelValue = yAxisLabel
return self
def yAxisFactor(self, yAxisFactor):
''' Set the scaling of the y axis
Args:
yAxisFactorValue: Scale the y axis, useful if there are percentages
'''
self.yAxisFactorValue = yAxisFactor
return self
def yAxisFormatter(self, yAxisFormatter):
''' Set the y axis formatter
Args:
yAxisFormatterValue: The y axis formatter to use
'''
self.yAxisFormatterValue = yAxisFormatter
return self
def showGridValue(self, showGridValue):
''' Whether to show the grid in the plot
Args:
showGridValue: Whether to show the grid
'''
self.showGridValue = showGridValue
return self
def top(self, numberOfItems):
''' Limit plot items
Args:
numberOfItemsValue: Limit the number of items to plot
'''
self.numberOfItemsValue = numberOfItems
return self
def kind(self, kind):
''' Set the kind of plot we want to show
Args:
kindValue: What kind of plot to display
'''
self.kindValue = kind
return self
def showPlot(self, data, title = "Title"):
''' Plot with the set instance values
Args:
title: The plot's main title
'''
if (self.numberOfItemsValue > 0):
# Limit the number of items shown
data = data.head(self.numberOfItemsValue)
# Calcluate the plot values
resultingPlot = (data * self.yAxisFactorValue / self.dataframe.shape[0]).plot(kind = self.kindValue);
# Set formatter, labels, grid, and title
resultingPlot.yaxis.set_major_formatter(self.yAxisFormatterValue)
plt.xlabel(self.xAxisLabelValue)
plt.ylabel(self.yAxisLabelValue)
plt.grid(self.showGridValue)
plt.title(title);
# Show the plot
plt.show()
# Some preliminary settings, like what columns are most relevant and how many of the top languages to look at, create initial dataset
relevantColumns = ["HaveWorkedLanguage", "WantWorkLanguage", "CareerSatisfaction", "JobSatisfaction", "JobSeekingStatus", "DeveloperType", "Salary"]
topLanguages = 15
origin = DataFrameHelper(df)
# Print some information on the original dataset
origin.printShape()
origin.printMissingValuesSum(relevantColumns)
# Only use the columns we're interested in
origin.applyFilter(relevantColumns)
origin.printShape()
# Question 1: What languages are most represented?
# Drop the null values and use the resulting dataset.
hwl = DataFrameHelper(origin.dropNa(["HaveWorkedLanguage"]))
# Plot the results
PlotHelper(hwl.dataframe).top(topLanguages).yAxisFactor(100).xAxisLabel("Language").yAxisLabel("Usage in percent (multiple nominations allowed)") \
.showPlot(hwl.stackAggregatedValues("HaveWorkedLanguage").value_counts(), f"Top {topLanguages} Languages")
# Question 2: Refine the languages by developer type.
# This is achieved in several passes:
# 1. For simplicity's sake, assume the first given role is the developer's primary role. Store this in the DeveloperType column.
# 2. Build a list of all possible developer types.
# 3. Build a list of all possible language types.
# 4. In a dictionary that has all developer types, keep a counted dictionary of each language per that developer type.
# As always clean the data. Also apply filter to make working with the dataset a bit easier.
lbt = DataFrameHelper(origin.dropNa(["HaveWorkedLanguage", "DeveloperType"])).applyFilter(["HaveWorkedLanguage", "DeveloperType"]).dataframe
# Only use the primary role.
for (index, roles) in lbt["DeveloperType"].iteritems():
lbt.at[index, "DeveloperType"] = roles.split(";")[0]
# Skip roles "Other" and "Graphic designer" (there are only three results for graphic designer, this is not representative)
lbt = lbt[lbt.DeveloperType != "Other"]
lbt = lbt[lbt.DeveloperType != "Graphic designer"]
# Find all unique developer types and languages.
exsitingDeveloperTypes = pd.unique(lbt["DeveloperType"])
existingLanguages = pd.unique(DataFrameHelper(hwl.dataframe.head(topLanguages)).stackAggregatedValues("HaveWorkedLanguage"))
def createLanguageCounter(languages):
# Build a dictionary of all possible languages and counts.
languagesInUse = {}
for language in languages:
languagesInUse[language] = 0
return languagesInUse
# Build a dictionary of all possible developer types and their respective language counts.
developerTypes = {}
for developerType in exsitingDeveloperTypes:
developerTypes[developerType] = createLanguageCounter(existingLanguages)
# Perform the count (using list comprehension with lambda instead of iterrows)
def countByDeveloperType(d, l):
for language in l.split(";"):
if language in developerTypes[d]:
developerTypes[d][language.strip()] += 1
[countByDeveloperType(d, l) for d, l in zip(lbt['DeveloperType'], lbt['HaveWorkedLanguage'])]
# Plot and do a bit of scaling.
# Unfortunately I found no better way to remove titles.
pd.DataFrame(developerTypes).plot(subplots = True, kind = 'bar', title = [''] * len(exsitingDeveloperTypes))
fig_size = plt.gcf().get_size_inches()
sizefactor = 3
plt.gcf().set_size_inches(sizefactor * fig_size);
plt.show()
# Question 3: Now we are looking at the languages with the highest job satisfaction and salaries at the same time.
# For highest job satisfaction and salaries we will be looking at all values above the mean.
# Prepare the data for selection.
salsat = DataFrameHelper(origin.dropNa(["HaveWorkedLanguage", "JobSatisfaction", "Salary"])).applyFilter(["HaveWorkedLanguage", "JobSatisfaction", "Salary"])
# Save language counts to later proportionalize the filtered value counts
salaryAndSatisfactionCountsBefore = salsat.stackAggregatedValues("HaveWorkedLanguage").value_counts()
# Determine the means.
salsat = salsat.dataframe
salaryMean = salsat["Salary"].mean()
jobSatisfactionMean = salsat["JobSatisfaction"].mean()
salsat["Salary"] = salsat["Salary"].astype("int64")
# Drop any records below the mean.
salsat.drop(salsat[salsat["Salary"] < salaryMean].index, inplace = True)
salsat.drop(salsat[salsat["JobSatisfaction"] < jobSatisfactionMean].index, inplace = True)
# Count the langueage values present in the dataset.
salaryAndSatisfactionCountsAfter = DataFrameHelper(salsat).stackAggregatedValues("HaveWorkedLanguage").value_counts()
# Create a new dataframe for easier working.
salaryAndSatisfactionChart = DataFrameHelper(pd.DataFrame.from_dict(salaryAndSatisfactionCountsAfter))
# Proportionalize the language counts, then for plotting, simplify again and sort.
salaryAndSatisfactionChart.dataframe["HaveWorkedLanguage"] = (salaryAndSatisfactionChart.dataframe.iloc[:,0] / salaryAndSatisfactionCountsBefore[salaryAndSatisfactionChart.dataframe.index]) * 100
salaryAndSatisfactionChart.applyFilter(["HaveWorkedLanguage"])
salaryAndSatisfactionChart = salaryAndSatisfactionChart.dataframe.head(topLanguages + 5)
salaryAndSatisfactionChart = salaryAndSatisfactionChart.sort_values(by = ['HaveWorkedLanguage'], ascending = False)
# Plot.
PlotHelper(salaryAndSatisfactionChart).yAxisFactor(10).xAxisLabel("Language").yAxisLabel("High job satisfaction and salary in percent") \
.showPlot(salaryAndSatisfactionChart, f"Languages with the highest job satisfaction and salary")
# Question 4 examines what language developers that consider getting a new job are currently using.
jss = DataFrameHelper(origin.dropNa(["HaveWorkedLanguage", "JobSeekingStatus"])).applyFilter(["HaveWorkedLanguage", "JobSeekingStatus"]).dataframe
isNotSeekingJob = "no"
isSeekingJob = "yes"
# Filter by developers actively looking for a new job or who may be open to new opportunities.
jss["LookingForJob"] = np.where(jss["JobSeekingStatus"] != "I am not interested in new job opportunities", isSeekingJob, isNotSeekingJob)
# Get total number of jobs for those who answered the "JobSeekingStatus" question
seekingLanguageCounts = DataFrameHelper(jss).stackAggregatedValues("HaveWorkedLanguage").value_counts()
# Build a dictionary of whether developers are looking for a new job and their respective language counts.
jobSeekers = { isSeekingJob: createLanguageCounter(existingLanguages) }
def countByJobSeekers(s, l):
if s == isSeekingJob:
for language in l.split(";"):
if language in jobSeekers[s]:
jobSeekers[s][language.strip()] += 1
[countByJobSeekers(s, l) for s, l in zip(jss['LookingForJob'], jss['HaveWorkedLanguage'])]
# Create a new dataframe for easier working. Proportionalize and prepare for plotting as we did in Question 3.
jobSeekersChart = DataFrameHelper(pd.DataFrame.from_dict(jobSeekers))
jobSeekersChart.dataframe["MaySeekNewJob"] = (jobSeekersChart.dataframe.iloc[:,0] / seekingLanguageCounts[jobSeekersChart.dataframe.index]) * 100
jobSeekersChart.applyFilter(["MaySeekNewJob"])
jobSeekersChart = jobSeekersChart.dataframe.sort_values(by = ['MaySeekNewJob'], ascending = False)
# Plot
PlotHelper(jobSeekersChart).yAxisFactor(10).xAxisLabel("Language").yAxisLabel("Seeking new job in percent") \
.showPlot(jobSeekersChart, f"Developers of these languages may be seeking new jobs")
#Some data that was used for selection, pre-validation, lookup, reference and result verification
#print(pd.unique(lbt["DeveloperType"]))
#print(pd.unique(hwl.stackAggregatedValues("HaveWorkedLanguage")))
#print(pd.unique(jss["JobSeekingStatus"]))
#print(hwl.stackAggregatedValues("HaveWorkedLanguage").value_counts())
#print(DataFrameHelper(jss).stackAggregatedValues("HaveWorkedLanguage").value_counts())
#print(print(len(jss)))
| 0.788217 | 0.689423 |
<a href="https://colab.research.google.com/github/aly202012/Teaching/blob/master/Copy_of_dataexploration.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
data = [50,50,47,97,49,3,53,42,26,74,82,62,37,15,70,27,36,35,48,52,63,64]
print(data)
import numpy as np
grades = np.array(data)
print(grades)
# حدثت مضاعفه للبيانات الاصليه من حيث العدد
print (type(data),'x 2:', data * 2)
print('---')
# تم تطبيق عمليه حسابيه علي القيم الموجوده وبالتالي تضاعفت الارقام من حيث القيمه
print (type(grades),'x 2:', grades * 2)
grades.shape
# تلك البيانات مكونه من بعد واحد
grades[0]
grades.mean()
# تم اجراء عمليه احصائيه علي تلك البيانات
# Define an array of study hours
study_hours = [10.0,11.5,9.0,16.0,9.25,1.0,11.5,9.0,8.5,14.5,15.5,
13.75,9.0,8.0,15.5,8.0,9.0,6.0,10.0,12.0,12.5,12.0]
# Create a 2D array (an array of arrays)
student_data = np.array([study_hours, grades])
# display the array
print(student_data)
#في الناتج نلاح انه تم دمج المصفوفتان معا علي شكل مصفوفه ثنائيه الابعاد
# Show shape of 2D array
student_data.shape
# يمكننا اضافه العديد / المزيد من الابعاد
student_names=["ali","b","d","e","f","g","h","i","j","k","l","m","n","o","p","q","r","s","t","u","v","w"]
print(student_names)
newst_name=np.array(student_names)
print(newst_name)
mostudent_data=np.array([newst_name,study_hours, grades])
print(mostudent_data)
print(mostudent_data.shape)
# Show the first element of the first element
mostudent_data[0][0][0]
# نعود لي موضوعنا الاساسي
# Show the first element of the first element
student_data[0][0]
# Get the mean value of each sub-array
avg_study = student_data[0].mean()
avg_grade = student_data[1].mean()
# هذا الكود يشبه كود اخر بالاسفل والغرض منه ايضا ايجاد فورم معين لمجموعه من العمليات الاحصائيه
#print('Minimum:{:.2f}\nMean:{:.2f}\nMedian:{:.2f}\nMode:{:.2f}\nMaximum:{:.2f}\n'.format(min_val,
# mean_val,
# med_val,
# mod_val,
# max_val))
print('Average study hours: {:.2f}\nAverage grade: {:.2f}'.format(avg_study, avg_grade))
```
starting with data frame
```
import pandas as pd
df_students = pd.DataFrame({'Name': ['Dan', 'Joann', 'Pedro', 'Rosie', 'Ethan', 'Vicky', 'Frederic', 'Jimmie',
'Rhonda', 'Giovanni', 'Francesca', 'Rajab', 'Naiyana', 'Kian', 'Jenny',
'Jakeem','Helena','Ismat','Anila','Skye','Daniel','Aisha'],
'StudyHours':student_data[0],
'Grade':student_data[1]})
df_students
# تم الامر يا صديقي
import pandas as pd
modf_students = pd.DataFrame({'Name': mostudent_data[0],
'StudyHours':mostudent_data[1],
'Grade':mostudent_data[2]})
modf_students
# Get the data for index value 5
df_students.loc[5]
# Get the rows with index values from 0 to 5
# The loc method returned rows with index label in the list of values from 0 to 5 - which includes 0, 1, 2, 3, 4, and 5 (six rows).
df_students.loc[0:5]
# Get data in the first five rows
# the iloc method returns the rows in the positions included in the range 0 to 5
df_students.iloc[0:5]
df_students.iloc[0,[1,2]]
df_students.iloc[0,[0,1]]
df_students.loc[df_students['Name']=='Aisha']
df_students[df_students['Name']=='Aisha']
df_students.query('Name=="Aisha"')
df_students = pd.read_csv('flipkart_com-ecommerce_sample.csv',delimiter=',',header='infer')
df_students.head()
df_students.isnull()
# للحصول علي القيم المفقوده لكل عمود
df_students.isnull().sum()
# القيم المفقوده تظهر علي شكل نان NaN وليس شيء اخر
df_students[df_students.isnull().any(axis=1)]
# تم جمع القيم جميعها للعمود retail_price وتم احتساب المتوسط الخاص بهم واستبدال القيم المفقوده بالقيم المتوسطه
df_students.retail_price = df_students.retail_price.fillna(df_students.retail_price.mean())
df_students
#print(df_students.shape)
df_students = df_students.dropna(axis=0, how='any')
df_students
#print(df_students.shape)
print(df_students.shape)
df_students = df_students.dropna(axis=0, how='any')
print(df_students)
print(df_students.shape)
# Get the mean study hours using to column name as an index
mean_study = df_students['StudyHours'].mean()
# Get the mean grade using the column name as a property (just to make the point!)
mean_grade = df_students.Grade.mean()
# Print the mean study hours and mean grade
# هنا اواجه مشكله وهي عدم التعرف علي ذلك الكود ماذا يفعل
print('Average weekly study hours: {:.2f}\nAverage grade: {:.2f}'.format(mean_study, mean_grade))
# Get students who studied for the mean or more hours
# العثور علي الطلاب الذين درسوا اكثر من نصف الوقت
df_students[df_students.StudyHours > mean_study]
#print("----------------------------------------------")
#df_students[df_students.StudyHours < mean_study]
# على سبيل المثال ، دعنا نعثر على متوسط الدرجات للطلاب الذين قضوا أكثر من متوسط وقت الدراسة.
df_students[df_students.StudyHours > mean_study].Grade.mean()
# تمييز الطلاب بالنجاح او الفشل حسب الدرجه المحدده للنجاح وهي ال 60
passes = pd.Series(df_students['Grade'] >= 60)
df_students = pd.concat([df_students, passes.rename("Pass")], axis=1)
df_students
print(df_students.groupby(df_students.Pass).Name.count())
# يمكنك تجميع عدة حقول في مجموعة باستخدام أي وظيفة تجميع متاحة. على سبيل المثال ، يمكنك العثور على متوسط وقت الدراسة والتقدير لمجموعات الطلاب الذين اجتازوا الدورة التدريبية وفشلوا فيها.
print(df_students.groupby(df_students.Pass)['StudyHours', 'Grade'].mean())
# يقوم الكود التالي بفرز بيانات الطالب بترتيب تنازلي من الدرجة ، ويعين DataFrame الناتج إلى متغير df_students الأصلي.
# Create a DataFrame with the data sorted by Grade (descending)
df_students = df_students.sort_values('Grade', ascending=False)
# Show the DataFrame
df_students
```
Visualizing data with Matplotlib¶
```
# Ensure plots are displayed inline in the notebook
#%matplotlib inline
from matplotlib import pyplot as plt
# Create a bar plot of name vs grade
plt.bar(x=df_students.Name, height=df_students.Grade)
# Display the plot
plt.show()
# Ensure plots are displayed inline in the notebook
#%matplotlib inline
from matplotlib import pyplot as plt
# Create a bar plot of name vs grade
plt.bar(x=modf_students.Name, height=modf_students.Grade,color="orange")
# Display the plot
plt.show()
# Create a bar plot of name vs grade
fig = plt.figure(figsize=(10,3))
plt.bar(x=df_students.Name, height=df_students.Grade, color='orange')
# Customize the chart
plt.title('Student Grades')
plt.xlabel('Student')
plt.ylabel('Grade')
plt.grid(color='#95a5a6', linestyle='--', linewidth=2, axis='y', alpha=1.0)
plt.xticks(rotation=90)
# Display the plot
plt.show()
# Create a Figure
# ال10 لمحور اكس وال ال3 لمحور واي
fig = plt.figure(figsize=(10,3))
# Create a bar plot of name vs grade
plt.bar(x=df_students.Name, height=df_students.Grade, color='orange')
# Customize the chart
plt.title('Student Grades')
plt.xlabel('Student')
plt.ylabel('Grade')
plt.grid(color='#95a5a6', linestyle='--', linewidth=2, axis='y', alpha=0.7)
plt.xticks(rotation=90)
# Show the figure
plt.show()
# Create a figure for 2 subplots (1 row, 2 columns)
fig, ax = plt.subplots(1,2, figsize = (10,4))
# Create a bar plot of name vs grade on the first axis
# ال 0 تدل علي الشكل البياني الاول وال ال1 تدل علي الشكل البياني الثاني
ax[0].bar(x=df_students.Name, height=df_students.Grade, color='orange')
ax[0].set_title('Grades')
ax[0].set_xticklabels(df_students.Name, rotation=90)
# Create a pie chart of pass counts on the second axis
pass_counts = df_students['Pass'].value_counts()
ax[1].pie(pass_counts, labels=pass_counts)
ax[1].set_title('Passing Grades')
ax[1].legend(pass_counts.keys().tolist())
# Add a title to the Figure
fig.suptitle('Student Data')
# Show the figure
fig.show()
df_students.plot.bar(x='Name', y='StudyHours', color='red', figsize=(6,4))
```
Getting started with statistical analysis
Descriptive statistics and data distribution
> Indented block
الاحصاء الوصفيه وتوزيع البيانات
عند فحص متغير (على سبيل المثال ، عينة من درجات الطلاب) ، يهتم علماء البيانات بشكل خاص بتوزيعه (بمعنى آخر ، كيف تنتشر جميع قيم التقدير المختلفة عبر العينة). غالبًا ما تكون نقطة البداية لهذا الاستكشاف هي تصور البيانات كرسم بياني ، ومعرفة مدى تكرار حدوث كل قيمة للمتغير.
```
# Get the variable to examine
var_data = df_students['Grade']
# Create a Figure
fig = plt.figure(figsize=(10,4))
# Plot a histogram
plt.hist(var_data)
# Add titles and labels
plt.title('Data Distribution')
plt.xlabel('Value')
plt.ylabel('Frequency')
# Show the figure
fig.show()
```
Measures of central tendency¶
مقياس النزعه المركزيه والميل
```
# Get the variable to examine
var = df_students['Grade']
# Get statistics
min_val = var.min()
max_val = var.max()
mean_val = var.mean()
med_val = var.median()
mod_val = var.mode()[0]
print('Minimum:{:.2f}\nMean:{:.2f}\nMedian:{:.2f}\nMode:{:.2f}\nMaximum:{:.2f}\n'.format(min_val,
mean_val,
med_val,
mod_val,
max_val))
# Create a Figure
fig = plt.figure(figsize=(10,4))
# Plot a histogram
#وذلك من دون اي بيانات علي الشكل
plt.hist(var)
# Add lines for the statistics
# رقم 2 هو من اجل اظهار الخط بشكل اوضح
plt.axvline(x=min_val, color = 'gray', linestyle='dashed', linewidth = 2)
plt.axvline(x=mean_val, color = 'cyan', linestyle='dashed', linewidth = 2)
plt.axvline(x=med_val, color = 'red', linestyle='dashed', linewidth = 2)
plt.axvline(x=mod_val, color = 'yellow', linestyle='dashed', linewidth = 2)
plt.axvline(x=max_val, color = 'gray', linestyle='dashed', linewidth = 2)
# Add titles and labels
plt.title('Data Distribution')
plt.xlabel('Value')
plt.ylabel('Frequency')
# Show the figure
fig.show()
# تفصيل من الكود السابق
print('Minimum:{:.2f}\nMean:{:.2f}\nMedian:{:.2f}\nMode:{:.2f}\nMaximum:{:.2f}\n'.format(min_val,
mean_val,
med_val,
mod_val,
max_val))
plt.hist(var)
# طريقه اخري لاظهار البيانات كالكود السابق
# Get the variable to examine
var = df_students['Grade']
# Create a Figure
fig = plt.figure(figsize=(10,4))
# Plot a histogram
plt.boxplot(var)
# Add titles and labels
plt.title('Data Distribution')
# Show the figure
fig.show()
# نلاحظ ان اكبر توزيع للبيانات عند 50 لمحور واي وهو مطابق تماما للشكل السابق من حيث المعلومات التي يعطيها لنا الشكل
# هي داله للدمج بين المخططين السابقين
# Create a function that we can re-use
def show_distribution(var_data):
from matplotlib import pyplot as plt
# Get statistics
min_val = var_data.min()
max_val = var_data.max()
mean_val = var_data.mean()
med_val = var_data.median()
mod_val = var_data.mode()[0]
print('Minimum:{:.2f}\nMean:{:.2f}\nMedian:{:.2f}\nMode:{:.2f}\nMaximum:{:.2f}\n'.format(min_val,
mean_val,
med_val,
mod_val,
max_val))
# Create a figure for 2 subplots (2 rows, 1 column)
fig, ax = plt.subplots(2, 1, figsize = (10,4))
# Plot the histogram
ax[0].hist(var_data)
ax[0].set_ylabel('Frequency')
# Add lines for the mean, median, and mode
ax[0].axvline(x=min_val, color = 'gray', linestyle='dashed', linewidth = 2)
ax[0].axvline(x=mean_val, color = 'cyan', linestyle='dashed', linewidth = 2)
ax[0].axvline(x=med_val, color = 'red', linestyle='dashed', linewidth = 2)
ax[0].axvline(x=mod_val, color = 'yellow', linestyle='dashed', linewidth = 2)
ax[0].axvline(x=max_val, color = 'gray', linestyle='dashed', linewidth = 2)
# Plot the boxplot
ax[1].boxplot(var_data, vert=False)
ax[1].set_xlabel('Value')
# Add a title to the Figure
fig.suptitle('Data Distribution')
# Show the figure
fig.show()
# Get the variable to examine
col = df_students['Grade']
# Call the function
show_distribution(col)
```
التالي هو اظهار داله كثافه الاحتمال باستخدام عدد كافٍ من هذه المتغيرات العشوائية ، يمكنك حساب ما يسمى دالة كثافة الاحتمال ، والتي تقدر توزيع الدرجات لكافة السكان.
probability density function, which estimates the distribution of grades for the full population.
```
def show_density(var_data):
from matplotlib import pyplot as plt
fig = plt.figure(figsize=(10,4))
# Plot density
var_data.plot.density()
# Add titles and labels
plt.title('Data Density')
# Show the mean, median, and mode
plt.axvline(x=var_data.mean(), color = 'cyan', linestyle='dashed', linewidth = 2)
plt.axvline(x=var_data.median(), color = 'red', linestyle='dashed', linewidth = 2)
plt.axvline(x=var_data.mode()[0], color = 'yellow', linestyle='dashed', linewidth = 2)
# Show the figure
plt.show()
# Get the density of Grade
col = df_students['Grade']
show_density(col)
# Get the variable to examine
# القاء نظره علي توزيع ساعات الدراسه
col = df_students['StudyHours']
# Call the function
show_distribution(col)
# نلاحظ وجود شذوذ في التوزيع
# Get the variable to examine
col = df_students[df_students.StudyHours>1]['StudyHours']
# Call the function
show_distribution(col)
# التوزيع الطبيعي بدون شذوذ في التوزيع
```
يستخدم الكود التالي وظيفة Pandas quantile لاستبعاد الملاحظات التي تقل عن النسبة المئوية 0.01 (القيمة التي يوجد فوقها 99٪ من البيانات).
```
q01 = df_students.StudyHours.quantile(0.01)
# Get the variable to examine
col = df_students[df_students.StudyHours>q01]['StudyHours']
# Call the function
show_distribution(col)
```
نصيحة: يمكنك أيضًا التخلص من القيم المتطرفة في الطرف العلوي للتوزيع عن طريق تحديد عتبة بقيمة مئوية عالية - على سبيل المثال ، يمكنك استخدام الدالة الكمية للعثور على النسبة المئوية 0.99 التي يوجد تحتها 99٪ من البيانات.
مع إزالة القيم المتطرفة ، يعرض مخطط الصندوق جميع البيانات داخل الأرباع الأربعة. لاحظ أن التوزيع ليس متماثلًا كما هو الحال بالنسبة لبيانات الصف رغم ذلك - هناك بعض الطلاب لديهم أوقات دراسة عالية جدًا تبلغ حوالي 16 ساعة ، ولكن الجزء الأكبر من البيانات يتراوح بين 7 و 13 ساعة ؛ تسحب القيم القليلة العالية للغاية المتوسط نحو النهاية الأعلى للمقياس.
```
# Get the density of StudyHours
show_density(col)
#يسمى هذا النوع من التوزيع بالانحراف الصحيح.
#توجد كتلة البيانات على الجانب الأيسر من التوزيع
# مما يؤدي إلى إنشاء ذيل طويل إلى اليمين بسبب القيم الموجودة في الطرف الأعلى الأقصى ؛ التي تسحب الوسيلة إلى اليمين.
```
مقياس التباين
Measures of variance¶
بعد معرفه مكان توزيع البيانات
Typical statistics that measure variability in the data include:
Range: The difference between the maximum and minimum. There's no built-in function for this, but it's easy to calculate using the min and max functions.
Variance: The average of the squared difference from the mean. You can use the built-in var function to find this.
Standard Deviation: The square root of the variance. You can use the built-in std function to find this.
```
for col_name in ['Grade','StudyHours']:
col = df_students[col_name]
rng = col.max() - col.min()
var = col.var()
std = col.std()
print('\n{}:\n - Range: {:.2f}\n - Variance: {:.2f}\n - Std.Dev: {:.2f}'.format(col_name, rng, var, std))
```
من بين هذه الإحصائيات ، يكون الانحراف المعياري هو الأكثر فائدة بشكل عام. يوفر مقياسًا للتباين في البيانات على نفس المقياس مثل البيانات نفسها (أي نقاط الدرجات لتوزيع الدرجة وساعات توزيع ساعات الدراسة). كلما زاد الانحراف المعياري ، زاد التباين عند مقارنة القيم في التوزيع مع يعني التوزيع - وبعبارة أخرى ، يتم توزيع البيانات بشكل أكبر.
عند العمل مع التوزيع الطبيعي ، يعمل الانحراف المعياري مع الخصائص المعينة للتوزيع الطبيعي لتوفير رؤية أكبر
```
import scipy.stats as stats
# Get the Grade column
col = df_students['Grade']
# get the density
density = stats.gaussian_kde(col)
# Plot the density
# بدون اي تفاصيل
col.plot.density()
# Get the mean and standard deviation
s = col.std()
m = col.mean()
# Annotate 1 stdev
x1 = [m-s, m+s]
y1 = density(x1)
plt.plot(x1,y1, color='magenta')
plt.annotate('1 std (68.26%)', (x1[1],y1[1]))
# Annotate 2 stdevs
x2 = [m-(s*2), m+(s*2)]
y2 = density(x2)
plt.plot(x2,y2, color='green')
plt.annotate('2 std (95.45%)', (x2[1],y2[1]))
# Annotate 3 stdevs
x3 = [m-(s*3), m+(s*3)]
y3 = density(x3)
plt.plot(x3,y3, color='orange')
plt.annotate('3 std (99.73%)', (x3[1],y3[1]))
# Show the location of the mean
plt.axvline(col.mean(), color='cyan', linestyle='dashed', linewidth=1)
plt.axis('off')
plt.show()
```
توضح الخطوط الأفقية النسبة المئوية للبيانات ضمن الانحرافات المعيارية 1 و 2 و 3 للمتوسط (زائد أو ناقص).
في أي توزيع طبيعي:
ما يقرب من 68.26٪ من القيم تقع ضمن انحراف معياري واحد عن المتوسط.
يقع 95.45٪ تقريبًا من القيم ضمن انحرافين معياريين عن المتوسط.
ما يقرب من 99.73٪ من القيم تقع ضمن ثلاثة انحرافات معيارية عن المتوسط.
لذلك ، بما أننا نعلم أن متوسط الدرجة هو 49.18 ، فإن الانحراف المعياري هو 21.74 ، وتوزيع الدرجات طبيعي تقريبًا ؛ يمكننا حساب أن 68.26٪ من الطلاب يجب أن يحصلوا على درجة بين 27.44 و 70.92.
الإحصائيات الوصفية التي استخدمناها لفهم توزيع متغيرات بيانات الطلاب هي أساس التحليل الإحصائي ؛ ولأنها جزء مهم من استكشاف بياناتك ، فهناك طريقة وصف مضمنة لكائن DataFrame تُرجع الإحصائيات الوصفية الرئيسية لجميع الأعمدة الرقمية.
```
df_students.describe()
```
Comparing data¶
مقارنه البيانات
الآن بعد أن عرفت شيئًا عن التوزيع الإحصائي للبيانات في مجموعة البيانات الخاصة بك ، فأنت على استعداد لفحص بياناتك لتحديد أي علاقات ظاهرة بين المتغيرات.
```
# هذا العمود يحتوي علي قيم متطرفه ذات قيم منخفضه للغايه لذا وجب التخلص منه في البدايه
# حتي نحصل علي عينه من البيانات نموذجيه
df_sample = df_students[df_students['StudyHours']>1]
df_sample
```
Comparing numeric and categorical variables¶
مقارنه الفئات العدديه والفرديه
تتضمن البيانات متغيرين رقميين (StudyHours و Grade) ومتغيرين فئويين (Name and Pass). لنبدأ بمقارنة عمود StudyHours الرقمي بعمود Pass الفئوي لمعرفة ما إذا كانت هناك علاقة واضحة بين عدد الساعات التي تمت دراستها ودرجة النجاح.
```
# لإجراء هذه المقارنة ، دعنا ننشئ مخططات مربعة توضح توزيع ساعات الدراسة لكل قيمة مرور ممكنة (صواب وخطأ).
df_sample.boxplot(column='StudyHours', by='Pass', figsize=(8,5))
# بمقارنة توزيعات StudyHours ، من الواضح على الفور (إن لم يكن مفاجئًا بشكل خاص) أن الطلاب الذين اجتازوا الدورة كانوا يميلون إلى الدراسة لساعات أكثر من الطلاب الذين لم ينجحوا.
# لذلك ، إذا كنت تريد التنبؤ بما إذا كان من المحتمل أن يجتاز الطالب الدورة التدريبية أم لا ، فقد يكون مقدار الوقت الذي يقضونه في الدراسة ميزة تنبؤية جيدة.
# سوف نبدا الان بالمقارنه بين متغيرين رقميين
#لنقارن الآن بين متغيرين رقميين. سنبدأ بإنشاء مخطط شريطي يعرض كلاً من الصف وساعات الدراسة.
# Create a bar plot of name vs grade and study hours
df_sample.plot(x='Name', y=['Grade','StudyHours'], kind='bar', figsize=(8,5))
```
يعرض الرسم البياني أشرطة لكل من الصف الدراسي وساعات الدراسة لكل طالب ؛ لكن ليس من السهل مقارنتها لأن القيم بمقاييس مختلفة. يتم قياس الدرجات بنقاط التقدير وتتراوح من 3 إلى 97 ؛ بينما يقاس وقت الدراسة بالساعات ويتراوح من 1 إلى 16.
من الأساليب الشائعة عند التعامل مع البيانات الرقمية بمقاييس مختلفة تسوية البيانات بحيث تحتفظ القيم بتوزيعها النسبي ، ولكن يتم قياسها على نفس المقياس. لتحقيق ذلك ، سنستخدم تقنية تسمى MinMax scaling التي توزع القيم بشكل متناسب على مقياس من 0 إلى 1. يمكنك كتابة الكود لتطبيق هذا التحويل ؛ لكن مكتبة Scikit-Learn توفر أداة قياس للقيام بذلك نيابة عنك.
```
from sklearn.preprocessing import MinMaxScaler
# Get a scaler object
scaler = MinMaxScaler()
# Create a new dataframe for the scaled values
df_normalized = df_sample[['Name', 'Grade', 'StudyHours']].copy()
# Normalize the numeric columns
df_normalized[['Grade','StudyHours']] = scaler.fit_transform(df_normalized[['Grade','StudyHours']])
# Plot the normalized values
df_normalized.plot(x='Name', y=['Grade','StudyHours'], kind='bar', figsize=(8,5))
```
مع تسوية البيانات ، يصبح من السهل رؤية علاقة واضحة بين الصف الدراسي ووقت الدراسة. إنها ليست مطابقة تامة ، ولكن يبدو بالتأكيد أن الطلاب الحاصلين على درجات أعلى يميلون إلى الدراسة أكثر.
```
df_normalized.Grade.corr(df_normalized.StudyHours)
#لذلك يبدو أن هناك علاقة بين وقت الدراسة والصف ؛ وفي الواقع ، هناك قياس ارتباط إحصائي يمكننا استخدامه لتحديد العلاقة بين هذه الأعمدة.
```
إحصائية الارتباط هي قيمة بين -1 و 1 تشير إلى قوة العلاقة. تشير القيم الأعلى من 0 إلى ارتباط إيجابي (تميل القيم العالية لمتغير واحد إلى التطابق مع القيم العالية لمتغير آخر) ، بينما تشير القيم الأقل من 0 إلى ارتباط سلبي (القيم العالية لمتغير واحد تميل إلى التطابق مع القيم المنخفضة للمتغير الآخر). في هذه الحالة ، تكون قيمة الارتباط قريبة من 1 ؛ إظهار ارتباط إيجابي قوي بين وقت الدراسة والدرجة.
ملاحظة: غالبًا ما يقتبس علماء البيانات مبدأ "الارتباط ليس علاقة سببية". بعبارة أخرى ، مهما كان الأمر مغريًا ، يجب ألا تفسر الارتباط الإحصائي على أنه يوضح سبب ارتفاع إحدى القيم. في حالة بيانات الطلاب ، توضح الإحصائيات أن الطلاب الحاصلين على درجات عالية يميلون أيضًا إلى قضاء وقت دراسي كبير ؛ لكن هذا ليس هو نفسه إثبات حصولهم على درجات عالية لأنهم درسوا كثيرًا. يمكن استخدام الإحصاء على حد سواء كدليل لدعم الاستنتاج غير المنطقي بأن الطلاب درسوا كثيرًا لأن درجاتهم كانت ستكون عالية.
هناك طريقة أخرى لتصور الارتباط الظاهر بين عمودين رقميين وهي استخدام مخطط مبعثر.
```
# Create a scatter plot
df_sample.plot.scatter(title='Study Time vs Grade', x='StudyHours', y='Grade')
```
مرة أخرى ، يبدو أن هناك نمطًا واضحًا يكون فيه الطلاب الذين درسوا معظم الساعات هم أيضًا الطلاب الذين حصلوا على أعلى الدرجات.
يمكننا رؤية ذلك بشكل أكثر وضوحًا عن طريق إضافة خط انحدار (أو خط أفضل ملاءمة) إلى الرسم البياني الذي يُظهر الاتجاه العام في البيانات. للقيام بذلك ، سنستخدم تقنية إحصائية تسمى انحدار المربعات الصغرى
تحذير - الرياضيات إلى الأمام!
عد بعقلك إلى الوقت الذي كنت تتعلم فيه كيفية حل المعادلات الخطية في المدرسة ، وتذكر أن صيغة الميل والمقطع للمعادلة الخطية تبدو كما يلي:
$$ y = mx + b $$
في هذه المعادلة ، y و x هما متغيرات الإحداثيات ، و m هو ميل الخط ، و b هو تقاطع y (حيث يمر الخط عبر المحور Y).
في حالة مخطط التبعثر الخاص بنا لبيانات الطلاب ، لدينا بالفعل قيمنا لـ x (ساعات الدراسة) و y (الدرجة) ، لذلك نحتاج فقط إلى حساب تقاطع وانحدار الخط المستقيم الأقرب إلى تلك النقاط. ثم يمكننا تكوين معادلة خطية تحسب قيمة y جديدة على هذا السطر لكل من قيم x (StudyHours) الخاصة بنا - لتجنب الالتباس ، سنسمي هذه القيمة y الجديدة f (x) (لأنها ناتج خطي دالة المعادلة على أساس x). الفرق بين قيمة y (التقدير) الأصلية وقيمة f (x) هو الخطأ بين خط الانحدار الخاص بنا والدرجة الفعلية التي حققها الطالب. هدفنا هو حساب الميل والاعتراض لخط به أدنى خطأ إجمالي.
على وجه التحديد ، نحدد الخطأ الإجمالي عن طريق أخذ الخطأ لكل نقطة ، وتربيعه ، وإضافة جميع الأخطاء التربيعية معًا. السطر الأفضل ملاءمة هو السطر الذي يعطينا أدنى قيمة لمجموع الأخطاء التربيعية - ومن هنا جاء اسم انحدار المربعات الصغرى.
لحسن الحظ ، لا تحتاج إلى ترميز حساب الانحدار بنفسك - تتضمن حزمة SciPy فئة الإحصائيات التي توفر طريقة linregress للقيام بالعمل الشاق نيابة عنك. هذا يعيد (من بين أشياء أخرى) المعامِلات التي تحتاجها لمعادلة الميل - الميل (م) والتقاطع (ب) بناءً على زوج معين من العينات المتغيرة التي تريد مقارنتها.
```
from scipy import stats
#
df_regression = df_sample[['Grade', 'StudyHours']].copy()
# Get the regression slope and intercept
m, b, r, p, se = stats.linregress(df_regression['StudyHours'], df_regression['Grade'])
print('slope: {:.4f}\ny-intercept: {:.4f}'.format(m,b))
print('so...\n f(x) = {:.4f}x + {:.4f}'.format(m,b))
# Use the function (mx + b) to calculate f(x) for each x (StudyHours) value
df_regression['fx'] = (m * df_regression['StudyHours']) + b
# Calculate the error between f(x) and the actual y (Grade) value
df_regression['error'] = df_regression['fx'] - df_regression['Grade']
# Create a scatter plot of Grade vs Salary
df_regression.plot.scatter(x='StudyHours', y='Grade')
# Plot the regression line
plt.plot(df_regression['StudyHours'],df_regression['fx'], color='cyan')
# Display the plot
plt.show()
# Show the original x,y values, the f(x) value, and the error
df_regression[['StudyHours', 'Grade', 'fx', 'error']]
```
Using the regression coefficients for prediction Now that you have the regression coefficients for the study time and grade relationship, you can use them in a function to estimate the expected grade for a given amount of study.
ARABIC
استخدام معاملات الانحدار للتنبؤ الآن بعد أن أصبح لديك معاملات الانحدار لوقت الدراسة وعلاقة الدرجة ، يمكنك استخدامها في دالة لتقدير الدرجة المتوقعة لمقدار معين من الدراسة.
```
# Define a function based on our regression coefficients
def f(x):
m = 6.3134
b = -17.9164
return m*x + b
study_time = 14
# Get f(x) for study time
prediction = f(study_time)
# Grade can't be less than 0 or more than 100
expected_grade = max(0,min(100,prediction))
#Print the estimated grade
print ('Studying for {} hours per week may result in a grade of {:.0f}'.format(study_time, expected_grade))
```
|
github_jupyter
|
data = [50,50,47,97,49,3,53,42,26,74,82,62,37,15,70,27,36,35,48,52,63,64]
print(data)
import numpy as np
grades = np.array(data)
print(grades)
# حدثت مضاعفه للبيانات الاصليه من حيث العدد
print (type(data),'x 2:', data * 2)
print('---')
# تم تطبيق عمليه حسابيه علي القيم الموجوده وبالتالي تضاعفت الارقام من حيث القيمه
print (type(grades),'x 2:', grades * 2)
grades.shape
# تلك البيانات مكونه من بعد واحد
grades[0]
grades.mean()
# تم اجراء عمليه احصائيه علي تلك البيانات
# Define an array of study hours
study_hours = [10.0,11.5,9.0,16.0,9.25,1.0,11.5,9.0,8.5,14.5,15.5,
13.75,9.0,8.0,15.5,8.0,9.0,6.0,10.0,12.0,12.5,12.0]
# Create a 2D array (an array of arrays)
student_data = np.array([study_hours, grades])
# display the array
print(student_data)
#في الناتج نلاح انه تم دمج المصفوفتان معا علي شكل مصفوفه ثنائيه الابعاد
# Show shape of 2D array
student_data.shape
# يمكننا اضافه العديد / المزيد من الابعاد
student_names=["ali","b","d","e","f","g","h","i","j","k","l","m","n","o","p","q","r","s","t","u","v","w"]
print(student_names)
newst_name=np.array(student_names)
print(newst_name)
mostudent_data=np.array([newst_name,study_hours, grades])
print(mostudent_data)
print(mostudent_data.shape)
# Show the first element of the first element
mostudent_data[0][0][0]
# نعود لي موضوعنا الاساسي
# Show the first element of the first element
student_data[0][0]
# Get the mean value of each sub-array
avg_study = student_data[0].mean()
avg_grade = student_data[1].mean()
# هذا الكود يشبه كود اخر بالاسفل والغرض منه ايضا ايجاد فورم معين لمجموعه من العمليات الاحصائيه
#print('Minimum:{:.2f}\nMean:{:.2f}\nMedian:{:.2f}\nMode:{:.2f}\nMaximum:{:.2f}\n'.format(min_val,
# mean_val,
# med_val,
# mod_val,
# max_val))
print('Average study hours: {:.2f}\nAverage grade: {:.2f}'.format(avg_study, avg_grade))
import pandas as pd
df_students = pd.DataFrame({'Name': ['Dan', 'Joann', 'Pedro', 'Rosie', 'Ethan', 'Vicky', 'Frederic', 'Jimmie',
'Rhonda', 'Giovanni', 'Francesca', 'Rajab', 'Naiyana', 'Kian', 'Jenny',
'Jakeem','Helena','Ismat','Anila','Skye','Daniel','Aisha'],
'StudyHours':student_data[0],
'Grade':student_data[1]})
df_students
# تم الامر يا صديقي
import pandas as pd
modf_students = pd.DataFrame({'Name': mostudent_data[0],
'StudyHours':mostudent_data[1],
'Grade':mostudent_data[2]})
modf_students
# Get the data for index value 5
df_students.loc[5]
# Get the rows with index values from 0 to 5
# The loc method returned rows with index label in the list of values from 0 to 5 - which includes 0, 1, 2, 3, 4, and 5 (six rows).
df_students.loc[0:5]
# Get data in the first five rows
# the iloc method returns the rows in the positions included in the range 0 to 5
df_students.iloc[0:5]
df_students.iloc[0,[1,2]]
df_students.iloc[0,[0,1]]
df_students.loc[df_students['Name']=='Aisha']
df_students[df_students['Name']=='Aisha']
df_students.query('Name=="Aisha"')
df_students = pd.read_csv('flipkart_com-ecommerce_sample.csv',delimiter=',',header='infer')
df_students.head()
df_students.isnull()
# للحصول علي القيم المفقوده لكل عمود
df_students.isnull().sum()
# القيم المفقوده تظهر علي شكل نان NaN وليس شيء اخر
df_students[df_students.isnull().any(axis=1)]
# تم جمع القيم جميعها للعمود retail_price وتم احتساب المتوسط الخاص بهم واستبدال القيم المفقوده بالقيم المتوسطه
df_students.retail_price = df_students.retail_price.fillna(df_students.retail_price.mean())
df_students
#print(df_students.shape)
df_students = df_students.dropna(axis=0, how='any')
df_students
#print(df_students.shape)
print(df_students.shape)
df_students = df_students.dropna(axis=0, how='any')
print(df_students)
print(df_students.shape)
# Get the mean study hours using to column name as an index
mean_study = df_students['StudyHours'].mean()
# Get the mean grade using the column name as a property (just to make the point!)
mean_grade = df_students.Grade.mean()
# Print the mean study hours and mean grade
# هنا اواجه مشكله وهي عدم التعرف علي ذلك الكود ماذا يفعل
print('Average weekly study hours: {:.2f}\nAverage grade: {:.2f}'.format(mean_study, mean_grade))
# Get students who studied for the mean or more hours
# العثور علي الطلاب الذين درسوا اكثر من نصف الوقت
df_students[df_students.StudyHours > mean_study]
#print("----------------------------------------------")
#df_students[df_students.StudyHours < mean_study]
# على سبيل المثال ، دعنا نعثر على متوسط الدرجات للطلاب الذين قضوا أكثر من متوسط وقت الدراسة.
df_students[df_students.StudyHours > mean_study].Grade.mean()
# تمييز الطلاب بالنجاح او الفشل حسب الدرجه المحدده للنجاح وهي ال 60
passes = pd.Series(df_students['Grade'] >= 60)
df_students = pd.concat([df_students, passes.rename("Pass")], axis=1)
df_students
print(df_students.groupby(df_students.Pass).Name.count())
# يمكنك تجميع عدة حقول في مجموعة باستخدام أي وظيفة تجميع متاحة. على سبيل المثال ، يمكنك العثور على متوسط وقت الدراسة والتقدير لمجموعات الطلاب الذين اجتازوا الدورة التدريبية وفشلوا فيها.
print(df_students.groupby(df_students.Pass)['StudyHours', 'Grade'].mean())
# يقوم الكود التالي بفرز بيانات الطالب بترتيب تنازلي من الدرجة ، ويعين DataFrame الناتج إلى متغير df_students الأصلي.
# Create a DataFrame with the data sorted by Grade (descending)
df_students = df_students.sort_values('Grade', ascending=False)
# Show the DataFrame
df_students
# Ensure plots are displayed inline in the notebook
#%matplotlib inline
from matplotlib import pyplot as plt
# Create a bar plot of name vs grade
plt.bar(x=df_students.Name, height=df_students.Grade)
# Display the plot
plt.show()
# Ensure plots are displayed inline in the notebook
#%matplotlib inline
from matplotlib import pyplot as plt
# Create a bar plot of name vs grade
plt.bar(x=modf_students.Name, height=modf_students.Grade,color="orange")
# Display the plot
plt.show()
# Create a bar plot of name vs grade
fig = plt.figure(figsize=(10,3))
plt.bar(x=df_students.Name, height=df_students.Grade, color='orange')
# Customize the chart
plt.title('Student Grades')
plt.xlabel('Student')
plt.ylabel('Grade')
plt.grid(color='#95a5a6', linestyle='--', linewidth=2, axis='y', alpha=1.0)
plt.xticks(rotation=90)
# Display the plot
plt.show()
# Create a Figure
# ال10 لمحور اكس وال ال3 لمحور واي
fig = plt.figure(figsize=(10,3))
# Create a bar plot of name vs grade
plt.bar(x=df_students.Name, height=df_students.Grade, color='orange')
# Customize the chart
plt.title('Student Grades')
plt.xlabel('Student')
plt.ylabel('Grade')
plt.grid(color='#95a5a6', linestyle='--', linewidth=2, axis='y', alpha=0.7)
plt.xticks(rotation=90)
# Show the figure
plt.show()
# Create a figure for 2 subplots (1 row, 2 columns)
fig, ax = plt.subplots(1,2, figsize = (10,4))
# Create a bar plot of name vs grade on the first axis
# ال 0 تدل علي الشكل البياني الاول وال ال1 تدل علي الشكل البياني الثاني
ax[0].bar(x=df_students.Name, height=df_students.Grade, color='orange')
ax[0].set_title('Grades')
ax[0].set_xticklabels(df_students.Name, rotation=90)
# Create a pie chart of pass counts on the second axis
pass_counts = df_students['Pass'].value_counts()
ax[1].pie(pass_counts, labels=pass_counts)
ax[1].set_title('Passing Grades')
ax[1].legend(pass_counts.keys().tolist())
# Add a title to the Figure
fig.suptitle('Student Data')
# Show the figure
fig.show()
df_students.plot.bar(x='Name', y='StudyHours', color='red', figsize=(6,4))
# Get the variable to examine
var_data = df_students['Grade']
# Create a Figure
fig = plt.figure(figsize=(10,4))
# Plot a histogram
plt.hist(var_data)
# Add titles and labels
plt.title('Data Distribution')
plt.xlabel('Value')
plt.ylabel('Frequency')
# Show the figure
fig.show()
# Get the variable to examine
var = df_students['Grade']
# Get statistics
min_val = var.min()
max_val = var.max()
mean_val = var.mean()
med_val = var.median()
mod_val = var.mode()[0]
print('Minimum:{:.2f}\nMean:{:.2f}\nMedian:{:.2f}\nMode:{:.2f}\nMaximum:{:.2f}\n'.format(min_val,
mean_val,
med_val,
mod_val,
max_val))
# Create a Figure
fig = plt.figure(figsize=(10,4))
# Plot a histogram
#وذلك من دون اي بيانات علي الشكل
plt.hist(var)
# Add lines for the statistics
# رقم 2 هو من اجل اظهار الخط بشكل اوضح
plt.axvline(x=min_val, color = 'gray', linestyle='dashed', linewidth = 2)
plt.axvline(x=mean_val, color = 'cyan', linestyle='dashed', linewidth = 2)
plt.axvline(x=med_val, color = 'red', linestyle='dashed', linewidth = 2)
plt.axvline(x=mod_val, color = 'yellow', linestyle='dashed', linewidth = 2)
plt.axvline(x=max_val, color = 'gray', linestyle='dashed', linewidth = 2)
# Add titles and labels
plt.title('Data Distribution')
plt.xlabel('Value')
plt.ylabel('Frequency')
# Show the figure
fig.show()
# تفصيل من الكود السابق
print('Minimum:{:.2f}\nMean:{:.2f}\nMedian:{:.2f}\nMode:{:.2f}\nMaximum:{:.2f}\n'.format(min_val,
mean_val,
med_val,
mod_val,
max_val))
plt.hist(var)
# طريقه اخري لاظهار البيانات كالكود السابق
# Get the variable to examine
var = df_students['Grade']
# Create a Figure
fig = plt.figure(figsize=(10,4))
# Plot a histogram
plt.boxplot(var)
# Add titles and labels
plt.title('Data Distribution')
# Show the figure
fig.show()
# نلاحظ ان اكبر توزيع للبيانات عند 50 لمحور واي وهو مطابق تماما للشكل السابق من حيث المعلومات التي يعطيها لنا الشكل
# هي داله للدمج بين المخططين السابقين
# Create a function that we can re-use
def show_distribution(var_data):
from matplotlib import pyplot as plt
# Get statistics
min_val = var_data.min()
max_val = var_data.max()
mean_val = var_data.mean()
med_val = var_data.median()
mod_val = var_data.mode()[0]
print('Minimum:{:.2f}\nMean:{:.2f}\nMedian:{:.2f}\nMode:{:.2f}\nMaximum:{:.2f}\n'.format(min_val,
mean_val,
med_val,
mod_val,
max_val))
# Create a figure for 2 subplots (2 rows, 1 column)
fig, ax = plt.subplots(2, 1, figsize = (10,4))
# Plot the histogram
ax[0].hist(var_data)
ax[0].set_ylabel('Frequency')
# Add lines for the mean, median, and mode
ax[0].axvline(x=min_val, color = 'gray', linestyle='dashed', linewidth = 2)
ax[0].axvline(x=mean_val, color = 'cyan', linestyle='dashed', linewidth = 2)
ax[0].axvline(x=med_val, color = 'red', linestyle='dashed', linewidth = 2)
ax[0].axvline(x=mod_val, color = 'yellow', linestyle='dashed', linewidth = 2)
ax[0].axvline(x=max_val, color = 'gray', linestyle='dashed', linewidth = 2)
# Plot the boxplot
ax[1].boxplot(var_data, vert=False)
ax[1].set_xlabel('Value')
# Add a title to the Figure
fig.suptitle('Data Distribution')
# Show the figure
fig.show()
# Get the variable to examine
col = df_students['Grade']
# Call the function
show_distribution(col)
def show_density(var_data):
from matplotlib import pyplot as plt
fig = plt.figure(figsize=(10,4))
# Plot density
var_data.plot.density()
# Add titles and labels
plt.title('Data Density')
# Show the mean, median, and mode
plt.axvline(x=var_data.mean(), color = 'cyan', linestyle='dashed', linewidth = 2)
plt.axvline(x=var_data.median(), color = 'red', linestyle='dashed', linewidth = 2)
plt.axvline(x=var_data.mode()[0], color = 'yellow', linestyle='dashed', linewidth = 2)
# Show the figure
plt.show()
# Get the density of Grade
col = df_students['Grade']
show_density(col)
# Get the variable to examine
# القاء نظره علي توزيع ساعات الدراسه
col = df_students['StudyHours']
# Call the function
show_distribution(col)
# نلاحظ وجود شذوذ في التوزيع
# Get the variable to examine
col = df_students[df_students.StudyHours>1]['StudyHours']
# Call the function
show_distribution(col)
# التوزيع الطبيعي بدون شذوذ في التوزيع
q01 = df_students.StudyHours.quantile(0.01)
# Get the variable to examine
col = df_students[df_students.StudyHours>q01]['StudyHours']
# Call the function
show_distribution(col)
# Get the density of StudyHours
show_density(col)
#يسمى هذا النوع من التوزيع بالانحراف الصحيح.
#توجد كتلة البيانات على الجانب الأيسر من التوزيع
# مما يؤدي إلى إنشاء ذيل طويل إلى اليمين بسبب القيم الموجودة في الطرف الأعلى الأقصى ؛ التي تسحب الوسيلة إلى اليمين.
for col_name in ['Grade','StudyHours']:
col = df_students[col_name]
rng = col.max() - col.min()
var = col.var()
std = col.std()
print('\n{}:\n - Range: {:.2f}\n - Variance: {:.2f}\n - Std.Dev: {:.2f}'.format(col_name, rng, var, std))
import scipy.stats as stats
# Get the Grade column
col = df_students['Grade']
# get the density
density = stats.gaussian_kde(col)
# Plot the density
# بدون اي تفاصيل
col.plot.density()
# Get the mean and standard deviation
s = col.std()
m = col.mean()
# Annotate 1 stdev
x1 = [m-s, m+s]
y1 = density(x1)
plt.plot(x1,y1, color='magenta')
plt.annotate('1 std (68.26%)', (x1[1],y1[1]))
# Annotate 2 stdevs
x2 = [m-(s*2), m+(s*2)]
y2 = density(x2)
plt.plot(x2,y2, color='green')
plt.annotate('2 std (95.45%)', (x2[1],y2[1]))
# Annotate 3 stdevs
x3 = [m-(s*3), m+(s*3)]
y3 = density(x3)
plt.plot(x3,y3, color='orange')
plt.annotate('3 std (99.73%)', (x3[1],y3[1]))
# Show the location of the mean
plt.axvline(col.mean(), color='cyan', linestyle='dashed', linewidth=1)
plt.axis('off')
plt.show()
df_students.describe()
# هذا العمود يحتوي علي قيم متطرفه ذات قيم منخفضه للغايه لذا وجب التخلص منه في البدايه
# حتي نحصل علي عينه من البيانات نموذجيه
df_sample = df_students[df_students['StudyHours']>1]
df_sample
# لإجراء هذه المقارنة ، دعنا ننشئ مخططات مربعة توضح توزيع ساعات الدراسة لكل قيمة مرور ممكنة (صواب وخطأ).
df_sample.boxplot(column='StudyHours', by='Pass', figsize=(8,5))
# بمقارنة توزيعات StudyHours ، من الواضح على الفور (إن لم يكن مفاجئًا بشكل خاص) أن الطلاب الذين اجتازوا الدورة كانوا يميلون إلى الدراسة لساعات أكثر من الطلاب الذين لم ينجحوا.
# لذلك ، إذا كنت تريد التنبؤ بما إذا كان من المحتمل أن يجتاز الطالب الدورة التدريبية أم لا ، فقد يكون مقدار الوقت الذي يقضونه في الدراسة ميزة تنبؤية جيدة.
# سوف نبدا الان بالمقارنه بين متغيرين رقميين
#لنقارن الآن بين متغيرين رقميين. سنبدأ بإنشاء مخطط شريطي يعرض كلاً من الصف وساعات الدراسة.
# Create a bar plot of name vs grade and study hours
df_sample.plot(x='Name', y=['Grade','StudyHours'], kind='bar', figsize=(8,5))
from sklearn.preprocessing import MinMaxScaler
# Get a scaler object
scaler = MinMaxScaler()
# Create a new dataframe for the scaled values
df_normalized = df_sample[['Name', 'Grade', 'StudyHours']].copy()
# Normalize the numeric columns
df_normalized[['Grade','StudyHours']] = scaler.fit_transform(df_normalized[['Grade','StudyHours']])
# Plot the normalized values
df_normalized.plot(x='Name', y=['Grade','StudyHours'], kind='bar', figsize=(8,5))
df_normalized.Grade.corr(df_normalized.StudyHours)
#لذلك يبدو أن هناك علاقة بين وقت الدراسة والصف ؛ وفي الواقع ، هناك قياس ارتباط إحصائي يمكننا استخدامه لتحديد العلاقة بين هذه الأعمدة.
# Create a scatter plot
df_sample.plot.scatter(title='Study Time vs Grade', x='StudyHours', y='Grade')
from scipy import stats
#
df_regression = df_sample[['Grade', 'StudyHours']].copy()
# Get the regression slope and intercept
m, b, r, p, se = stats.linregress(df_regression['StudyHours'], df_regression['Grade'])
print('slope: {:.4f}\ny-intercept: {:.4f}'.format(m,b))
print('so...\n f(x) = {:.4f}x + {:.4f}'.format(m,b))
# Use the function (mx + b) to calculate f(x) for each x (StudyHours) value
df_regression['fx'] = (m * df_regression['StudyHours']) + b
# Calculate the error between f(x) and the actual y (Grade) value
df_regression['error'] = df_regression['fx'] - df_regression['Grade']
# Create a scatter plot of Grade vs Salary
df_regression.plot.scatter(x='StudyHours', y='Grade')
# Plot the regression line
plt.plot(df_regression['StudyHours'],df_regression['fx'], color='cyan')
# Display the plot
plt.show()
# Show the original x,y values, the f(x) value, and the error
df_regression[['StudyHours', 'Grade', 'fx', 'error']]
# Define a function based on our regression coefficients
def f(x):
m = 6.3134
b = -17.9164
return m*x + b
study_time = 14
# Get f(x) for study time
prediction = f(study_time)
# Grade can't be less than 0 or more than 100
expected_grade = max(0,min(100,prediction))
#Print the estimated grade
print ('Studying for {} hours per week may result in a grade of {:.0f}'.format(study_time, expected_grade))
| 0.187207 | 0.810028 |
<div align="right" style="text-align:right"><i>Peter Norvig<br>May 2015</i></div>
# When Cheryl Met Eve: A Birthday Story
The *Cheryl's Birthday* logic puzzle [made the rounds](https://www.google.com/webhp?#q=cheryl%27s+birthday),
and I wrote [code](Cheryl.ipynb) that solves it. In that notebook I said that one reason for solving the problem with code rather than pencil and paper is that you can do more with code.
**[Gabe Gaster](http://www.gabegaster.com/)** proved me right when he [tweeted](https://twitter.com/gabegaster/status/593976413314777089/photo/1) that he had extended my code to generate a new list of dates that satisfies the constraints of the puzzle:
January 15, January 4,
July 13, July 24, July 30,
March 13, March 24,
May 11, May 17, May 30
In this notebook, I verify Gabe's result, and find some other variations on the puzzle.
First, let's recap [the puzzle](https://en.wikipedia.org/wiki/Cheryl%27s_Birthday):
> 1. Albert and Bernard became friends with Cheryl, and want to know when her birthday is. Cheryl gave them a list of 10 possible dates:
May 15 May 16 May 19
June 17 June 18
July 14 July 16
August 14 August 15 August 17
> 2. **Cheryl** then privately tells Albert the month and Bernard the day of her birthday.
> 3. **Albert**: "I don't know when Cheryl's birthday is, and I know that Bernard does not know."
> 4. **Bernard**: "At first I don't know when Cheryl's birthday is, but I know now."
> 5. **Albert**: "Then I also know when Cheryl's birthday is."
> 6. So when is Cheryl's birthday?
# Code for Original Cheryl's Birthday Puzzle
This is a slight modification of my [previous code](Cheryl.ipynb), and I'll give a slight modification of the explanation. The puzzle concerns these concepts:
- **Possible dates** that might be Cheryl's birthday.
- **Knowing** which dates are still possible; knowing for sure when only one is possible.
- **Telling** Albert and Bernard specific facts about the birthday.
- **Statements** about knowledge.
- **Hearing** the statements about knowledge.
I implement them as follows:
- `dates` is a set of all possible dates (each date is a string); we also consider subsets of `dates`.
- `know(possible_dates)` is a function that returns `True` when there is only one possible date.
- `told(part)` is a function that returns the set of possible dates after Cheryl tells a part (month or day).
- *`statement`*`(date)` returns true if the statement is true given that `date` is Cheryl's birthday.
- `satisfy(possible_dates, statement,...)` returns a subset of possible_dates that are still possible after hearing the statements.
In the [previous code](Cheryl.ipynb) I treated `dates` as a constant, but in this version the whole point is exploring different possible sets of dates, so now `dates` is a global variable, and the function `set_dates` is used to set the value of the global variable.
```
# Albert and Bernard just became friends with Cheryl, and they want to know when her birthday is.
# Cheryl gave them a list of 10 possible dates:
dates = ['May 15', 'May 16', 'May 19',
'June 17', 'June 18',
'July 14', 'July 16',
'August 14', 'August 15', 'August 17']
def month(date): return date.split()[0]
def day(date): return date.split()[1]
# Cheryl then tells Albert and Bernard separately
# the month and the day of the birthday respectively.
BeliefState = set
def told(part: str) -> BeliefState:
"""Cheryl told a part of her birthdate to someone; return a belief state of possible dates."""
return {date for date in dates if part in date}
def know(beliefs: BeliefState) -> bool:
"""A person `knows` the answer if their belief state has only one possibility."""
return len(beliefs) == 1
def satisfy(some_dates, *statements) -> BeliefState:
"""Return the subset of dates that satisfy all the statements."""
return {date for date in some_dates
if all(statement(date) for statement in statements)}
# Albert and Bernard make three statements:
def albert1(date) -> bool:
"""Albert: I don't know when Cheryl's birthday is, but I know that Bernard does not know too."""
albert_beliefs = told(month(date))
return not know(albert_beliefs) and not satisfy(albert_beliefs, bernard_knows)
def bernard_knows(date) -> bool: return know(told(day(date)))
def bernard1(date) -> bool:
"""Bernard: At first I don't know when Cheryl's birthday is, but I know now."""
at_first_beliefs = told(day(date))
after_beliefs = satisfy(at_first_beliefs, albert1)
return not know(at_first_beliefs) and know(after_beliefs)
def albert2(date) -> bool:
"""Albert: Then I also know when Cheryl's birthday is."""
then = satisfy(told(month(date)), bernard1)
return know(then)
# So when is Cheryl's birthday?
def cheryls_birthday(dates) -> BeliefState:
"""Return a subset of the global `dates` for which all three statements are true."""
return satisfy(set_dates(dates), albert1, bernard1, albert2)
def set_dates(new_dates):
"""Set the value of the global `dates` to `new_dates`"""
global dates
dates = new_dates
return dates
# Some tests
assert month('May 19') == 'May'
assert day('May 19') == '19'
assert albert1('May 19') == False
assert albert1('July 14') == True
assert know(told('17')) == False
assert know(told('19')) == True
cheryls_birthday(dates)
satisfy(dates, albert1)
satisfy(dates, albert1, bernard1)
satisfy(dates, albert1, bernard1, albert2)
```
# Verifying Gabe's Version
Gabe tweeted these ten dates:
```
gabe_dates = [
'January 15', 'January 4',
'July 13', 'July 24', 'July 30',
'March 13', 'March 24',
'May 11', 'May 17', 'May 30']
```
We can verify that they do indeed make the puzzle work, giving a single known birthdate:
```
cheryls_birthday(gabe_dates)
```
# Creating Our Own Versions
If Gabe can do it, we can do it! Our strategy will be to repeatedly pick a random sample of dates, and check if they solve the puzzle. We'll limit ourselves to a subset of dates (not all 366) to make it more likely that a random selection will have multiple dates with the same month and day (otherwise Albert and Bernard would know right away):
```
many_dates = {mo + ' ' + d1 + d2
for mo in ('March', 'April', 'May', 'June', 'July')
for d1 in '12'
for d2 in '3456789'}
```
Now we need to cycle through random samples of these possible dates until we hit one that works. I anticipate wanting to solve other puzzles besides the original `cheryls_birthday`, so I'll make the `puzzle` be a parameter of the function `pick_dates`. Note that `pick_dates` returns two things: the one date that is the solution (the birthday), and the `k` (default 10) dates that form the puzzle.
```
import random
def pick_dates(puzzle=cheryls_birthday, k=10):
"Pick a set of `k` dates for which the `puzzle` has a unique solution."
while True:
random_dates = random.sample(many_dates, k)
solutions = puzzle(random_dates)
if know(solutions):
return solutions.pop(), random_dates
pick_dates()
pick_dates(k=6)
pick_dates(k=12)
```
Great! We can make a new puzzle, just like Gabe. But how often do we get a unique solution to the puzzle (that is, the puzzle returns a set of size 1)? How often do we get a solution where Albert and Bernard know, but we the puzzle solver doesn't (that is, a set of size greater than 1)? How often is there no solution (size 0)? Let's make a Counter of the number of times each length-of-solution occurs:
```
from collections import Counter
def solution_lengths(puzzle=cheryls_birthday, N=10000, k=10, many_dates=many_dates):
"Try N random samples and count how often each possible length-of-puzzle-solution appears."
return Counter(len(puzzle(random.sample(many_dates, k)))
for _ in range(N))
solution_lengths(cheryls_birthday)
```
This says that about 2% of the time we get a unique solution (a set of `len` 1). With similar frequency we get an ambiguous solution (with 2 or more possible birth dates). And about 95% of the time, the sample of dates leads to no solution dates.
What happens if Cheryl changes the number of possible dates?
```
solution_lengths(cheryls_birthday, k=6)
solution_lengths(cheryls_birthday, k=12)
```
It is really hard (but not impossible) to find a set of 6 dates that work for the puzzle, and much easier to find a solution with 12 dates.
# A New Puzzle: All About Eve
Now let's see if we can create a more complicated puzzle. We'll introduce a new character, Eve, give her a statement, and alter the rest of the puzzle slightly:
> 1. Albert and Bernard just became friends with Cheryl, and they want to know when her birthday is. Cheryl wrote down a list of 10 possible dates for all to see.
> 2. **Cheryl** then writes down the month and shows it just to Albert, and also writes down the day and shows it just to Bernard.
> 3. **Albert**: I don't know when Cheryl's birthday is, but I know that Bernard does not know either.
> 4. **Bernard**: At first I didn't know when Cheryl's birthday is, but I know now.
> 5. **Albert**: Then I also know when Cheryl's birthday is.
> 6. **Eve**: Hi, Everybody. My name is Eve and I'm an evesdropper. It's what I do! I peeked and saw the first letter of the month and the first digit of the day. When I peeked, I didn't know Cheryl's birthday, but after listening to Albert and Bernard I do. And it's a good thing I peeked, because otherwise I couldn't have
figured it out.
> 7. So when is Cheryl's birthday?
We can easily code this up:
```
def cheryls_birthday_with_eve(dates):
"Return a set of the dates for which Albert, Bernard, and Eve's statements are true."
return satisfy(set_dates(dates), albert1, bernard1, albert2, eve1)
def eve1(date):
"""Eve: I peeked and saw the first letter of the month and the first digit of the day.
When I peeked, I didn't know Cheryl's birthday, but after listening to Albert and Bernard
I do. And it's a good thing I peeked, because otherwise I couldn't have figured it out."""
at_first = told(first(day(date))) & told(first(month(date)))
otherwise = told('')
return (not know(at_first) and
know(satisfy(at_first, albert1, bernard1, albert2)) and
not know(satisfy(otherwise, albert1, bernard1, albert2)))
def first(seq): return seq[0]
```
*Note*: I admit I "cheated" a bit here. Remember that the function `told` tests for `(part in date)`. For that to work for Eve, we have to make sure that the first letter is distinct from any other character in the date (it is—because only the first letter is uppercase) and that the first digit is distinct from any other character (it is—because in `many_dates` I carefully made sure that the first digit is always 1 or 2, and the second digit is never 1 or 2). Also note that `told('')` denotes the hypothetical situation where Cheryl "told" Eve nothing.
I have no idea if it is possible to find a set of dates that works for this puzzle. But I can try:
```
pick_dates(puzzle=cheryls_birthday_with_eve)
```
That was easy. How often is a random sample of dates a solution to this puzzle?
```
solution_lengths(cheryls_birthday_with_eve)
```
About half as often as for the original puzzle.
# An Even More Complex Puzzle
Let's make the puzzle even more complicated by making Albert wait one more time before he finally knows:
> 1. Albert and Bernard just became friends with Cheryl, and they want to know when her birtxhday is. Cheryl wrote down a list of 10 possible dates for all to see.
> 2. **Cheryl** then writes down the month and shows it just to Albert, and also writes down the day and shows it just to Bernard.
> 3. **Albert**: I don't know when Cheryl's birthday is, but I know that Bernard does not know either.
> 4. **Bernard**: At first I didn't know when Cheryl's birthday is, but I know now.
> 5. **Albert**: I still don't know.
> 6. **Eve**: Hi, Everybody. My name is Eve and I'm an evesdropper. It's what I do! I peeked and saw the first letter of the month and the first digit of the day. When I peeked, I didn't know Cheryl's birthday, but after listening to Albert and Bernard I do. And it's a good thing I peeked, because otherwise I couldn't have
figured it out.
> 7. **Albert**: OK, now I know.
> 8. So when is Cheryl's birthday?
Let's be careful in coding this up; Albert's second statement is different; he has a new third statement; and Eve's statement uses the same words, but it now implicitly refers to a different statement by Albert. We'll use the names `albert2c`, `eve1c`, and `albert3c` (`c` for "complex") to represent the new statements:
```
def cheryls_birthday_complex(dates):
"Return a set of the dates for which Albert, Bernard, and Eve's statements are true."
return satisfy(set_dates(dates), albert1, bernard1, albert2c, eve1c, albert3c)
def albert2c(date):
"Albert: I still don't know."
return not know(satisfy(told(month(date)), bernard1))
def eve1c(date):
"""Eve: I peeked and saw the first letter of the month and the first digit of the day.
When I peeked, I didn't know Cheryl's birthday, but after listening to Albert and Bernard
I do. And it's a good thing I peeked, because otherwise I couldn't have figured it out."""
at_first = told(first(day(date))) & told(first(month(date)))
otherwise = told('')
return (not know(at_first)
and know(satisfy(at_first, albert1, bernard1, albert2c)) and
not know(satisfy(otherwise, albert1, bernard1, albert2c)))
def albert3c(date):
"Albert: OK, now I know."
return know(satisfy(told(month(date)), bernard1, eve1c))
```
Again, I don't know if it is possible to find dates that works with this story, but I can try:
```
pick_dates(puzzle=cheryls_birthday_complex)
```
It worked! Were we just lucky, or are there many sets of dates that work?
```
solution_lengths(cheryls_birthday_complex)
```
Interesting. It was actually easier to find dates that work for this story than for either of the other stories.
## Analyzing a Solution to the Complex Puzzle
Now we will go through a solution step-by-step. We'll use a set of dates selected in a previous run:
```
previous_run_dates = {
'April 28',
'July 27',
'June 19',
'June 16',
'July 15',
'April 15',
'June 29',
'July 16',
'May 24',
'May 27'}
```
Let's find the solution:
```
cheryls_birthday_complex(previous_run_dates)
```
Now the first step is that Albert was told "July":
```
told('July')
```
And no matter which of these three dates is the actual birthday, Albert knows that Bernard would not know the birthday, because each of the days (15, 16, 27) appears twice in the list of possible dates.
```
not know(told('15')) and not know(told('16')) and not know(told('27'))
```
Next, Bernard is told the day:
```
told('27')
```
There are two dates with a 27, so Bernard did not know then. But only one of these dates is still consistent after hearing Albert's statement:
```
satisfy(told('27'), albert1)
```
So after Albert's statement, Bernard knows. Poor Albert still doesn't know (after being told `'July'` and hearing Bernard's statement):
```
satisfy(told('July'), bernard1)
```
Then along comes Eve. She evesdrops the "J" and the "2":
```
told('J') & told('2')
```
Two dates, so Eve doesn't know yet. But only one of the dates works after hearing the three statements made by Albert and Bernard:
```
satisfy(told('J') & told('2'), albert1, bernard1, albert2c)
```
But Eve wouldn't have known if she had been told nothing:
```
satisfy(told(''), albert1, bernard1, albert2c)
```
What about Albert? After hearing Eve's statement he finally knows:
```
satisfy(told('July'), eve1c)
```
# Three Children
Here's another puzzle:
> 1. A parent has the following conversation with a friend:
> 2. **Parent:** the product of my three childrens' ages is 36.
> 3. **Friend**: I don't know their ages.
> 4. **Parent**: The sum of their ages is the same as the number of people in this room.
> 5. **Friend**: I still don't know their ages.
> 6. **Parent**: The oldest one likes bananas.
> 7. **Friend**: Now I know their ages.
Let's follow the same methodology to solve this puzzle. Except this time, we're not dealing with sets of possible dates, we're dealing with set of possible *states* of the world. We'll define a state as a tuple of 4 numbers: the ages of the three children (in increasing order), and the number of people in the room.
Note: We'll limit the children's ages to be below 30 and the number of people in the room to be below 90. Also, in `friend2` and `friend3` we'll compute the `possible_states` and cache them, since the computation does not depend on the `date`.
```
N = 30
states = {(a, b, c, n)
for a in range(1, N)
for b in range(a, N)
for c in range(b, N) if a * b * c == 36
for n in range(2, 90)}
def ages(state): return state[:-1]
def room(state): return state[-1]
def parent1(state):
"""The product of my three childrens' ages is 36."""
a, b, c = ages(state)
return a * b * c == 36
def friend1(state):
"""I don't know their ages."""
possible_ages = {ages(s) for s in satisfy(states, parent1)}
return not know(possible_ages)
def parent2(state):
"""The sum of their ages is the same as the number of people in this room."""
return sum(ages(state)) == room(state)
def friend2(state, possible_states=satisfy(states, parent1, friend1, parent2)):
"""I still don't know their ages."""
# Given there are room(state) people in the room, I still don't know the ages.
possible_ages = {ages(s) for s in possible_states if room(s) == room(state)}
return not know(possible_ages)
def parent3(state):
"""The oldest one likes bananas."""
# I.e., there is an oldest one (and not twins of the same age)
a, b, c = ages(state)
return c > b
def friend3(state, possible_states=satisfy(states, parent1, friend1, parent2, friend2, parent3)):
"Now I know their ages."
possible_ages = {ages(s) for s in possible_states}
return know(possible_ages)
def child_age_puzzle(states):
return satisfy(states, parent1, friend1, parent2, friend2, parent3, friend3)
child_age_puzzle(states)
```
The tricky part of this puzzle comes after the `parent2` statement:
```
satisfy(states, parent1, friend1, parent2)
```
We see that out of these 7 possibilities, if the number of people in the room (the last number in each tuple)
were anything other than 13, then the friend (who can observe the number of people in the room) would know the ages. Since the `friend2` statement professes continued ignorance, it must be that the number of people in the room is 13. Then the `parent3` statement makes it clear that there can't be 6-year-old twins as the oldest children; it must be 2-year-old twins with an oldest age 9.
# What Next?
If you like, there are many other directions you could take this:
- Could you create a puzzle that goes one or two rounds more before everyone knows?
- Could you add new characters: Faith, and then George, and maybe even a new Hope?
- Would it be more interesting with a different number of possible dates (not 10)?
- Should we include the year or the day of the week, as well as the month and day?
- Perhaps a puzzle that starts with [Richard Smullyan](http://en.wikipedia.org/wiki/Raymond_Smullyan) announcing that one of the characters is a liar.
- Or you could make a puzzle harder than [the hardest logic puzzle ever](https://en.wikipedia.org/wiki/The_Hardest_Logic_Puzzle_Ever).
- Try the "black and white hats" [Riddler Express](https://fivethirtyeight.com/features/can-you-solve-these-colorful-puzzles/) stumper.
- It's up to you ...
|
github_jupyter
|
# Albert and Bernard just became friends with Cheryl, and they want to know when her birthday is.
# Cheryl gave them a list of 10 possible dates:
dates = ['May 15', 'May 16', 'May 19',
'June 17', 'June 18',
'July 14', 'July 16',
'August 14', 'August 15', 'August 17']
def month(date): return date.split()[0]
def day(date): return date.split()[1]
# Cheryl then tells Albert and Bernard separately
# the month and the day of the birthday respectively.
BeliefState = set
def told(part: str) -> BeliefState:
"""Cheryl told a part of her birthdate to someone; return a belief state of possible dates."""
return {date for date in dates if part in date}
def know(beliefs: BeliefState) -> bool:
"""A person `knows` the answer if their belief state has only one possibility."""
return len(beliefs) == 1
def satisfy(some_dates, *statements) -> BeliefState:
"""Return the subset of dates that satisfy all the statements."""
return {date for date in some_dates
if all(statement(date) for statement in statements)}
# Albert and Bernard make three statements:
def albert1(date) -> bool:
"""Albert: I don't know when Cheryl's birthday is, but I know that Bernard does not know too."""
albert_beliefs = told(month(date))
return not know(albert_beliefs) and not satisfy(albert_beliefs, bernard_knows)
def bernard_knows(date) -> bool: return know(told(day(date)))
def bernard1(date) -> bool:
"""Bernard: At first I don't know when Cheryl's birthday is, but I know now."""
at_first_beliefs = told(day(date))
after_beliefs = satisfy(at_first_beliefs, albert1)
return not know(at_first_beliefs) and know(after_beliefs)
def albert2(date) -> bool:
"""Albert: Then I also know when Cheryl's birthday is."""
then = satisfy(told(month(date)), bernard1)
return know(then)
# So when is Cheryl's birthday?
def cheryls_birthday(dates) -> BeliefState:
"""Return a subset of the global `dates` for which all three statements are true."""
return satisfy(set_dates(dates), albert1, bernard1, albert2)
def set_dates(new_dates):
"""Set the value of the global `dates` to `new_dates`"""
global dates
dates = new_dates
return dates
# Some tests
assert month('May 19') == 'May'
assert day('May 19') == '19'
assert albert1('May 19') == False
assert albert1('July 14') == True
assert know(told('17')) == False
assert know(told('19')) == True
cheryls_birthday(dates)
satisfy(dates, albert1)
satisfy(dates, albert1, bernard1)
satisfy(dates, albert1, bernard1, albert2)
gabe_dates = [
'January 15', 'January 4',
'July 13', 'July 24', 'July 30',
'March 13', 'March 24',
'May 11', 'May 17', 'May 30']
cheryls_birthday(gabe_dates)
many_dates = {mo + ' ' + d1 + d2
for mo in ('March', 'April', 'May', 'June', 'July')
for d1 in '12'
for d2 in '3456789'}
import random
def pick_dates(puzzle=cheryls_birthday, k=10):
"Pick a set of `k` dates for which the `puzzle` has a unique solution."
while True:
random_dates = random.sample(many_dates, k)
solutions = puzzle(random_dates)
if know(solutions):
return solutions.pop(), random_dates
pick_dates()
pick_dates(k=6)
pick_dates(k=12)
from collections import Counter
def solution_lengths(puzzle=cheryls_birthday, N=10000, k=10, many_dates=many_dates):
"Try N random samples and count how often each possible length-of-puzzle-solution appears."
return Counter(len(puzzle(random.sample(many_dates, k)))
for _ in range(N))
solution_lengths(cheryls_birthday)
solution_lengths(cheryls_birthday, k=6)
solution_lengths(cheryls_birthday, k=12)
def cheryls_birthday_with_eve(dates):
"Return a set of the dates for which Albert, Bernard, and Eve's statements are true."
return satisfy(set_dates(dates), albert1, bernard1, albert2, eve1)
def eve1(date):
"""Eve: I peeked and saw the first letter of the month and the first digit of the day.
When I peeked, I didn't know Cheryl's birthday, but after listening to Albert and Bernard
I do. And it's a good thing I peeked, because otherwise I couldn't have figured it out."""
at_first = told(first(day(date))) & told(first(month(date)))
otherwise = told('')
return (not know(at_first) and
know(satisfy(at_first, albert1, bernard1, albert2)) and
not know(satisfy(otherwise, albert1, bernard1, albert2)))
def first(seq): return seq[0]
pick_dates(puzzle=cheryls_birthday_with_eve)
solution_lengths(cheryls_birthday_with_eve)
def cheryls_birthday_complex(dates):
"Return a set of the dates for which Albert, Bernard, and Eve's statements are true."
return satisfy(set_dates(dates), albert1, bernard1, albert2c, eve1c, albert3c)
def albert2c(date):
"Albert: I still don't know."
return not know(satisfy(told(month(date)), bernard1))
def eve1c(date):
"""Eve: I peeked and saw the first letter of the month and the first digit of the day.
When I peeked, I didn't know Cheryl's birthday, but after listening to Albert and Bernard
I do. And it's a good thing I peeked, because otherwise I couldn't have figured it out."""
at_first = told(first(day(date))) & told(first(month(date)))
otherwise = told('')
return (not know(at_first)
and know(satisfy(at_first, albert1, bernard1, albert2c)) and
not know(satisfy(otherwise, albert1, bernard1, albert2c)))
def albert3c(date):
"Albert: OK, now I know."
return know(satisfy(told(month(date)), bernard1, eve1c))
pick_dates(puzzle=cheryls_birthday_complex)
solution_lengths(cheryls_birthday_complex)
previous_run_dates = {
'April 28',
'July 27',
'June 19',
'June 16',
'July 15',
'April 15',
'June 29',
'July 16',
'May 24',
'May 27'}
cheryls_birthday_complex(previous_run_dates)
told('July')
not know(told('15')) and not know(told('16')) and not know(told('27'))
told('27')
satisfy(told('27'), albert1)
satisfy(told('July'), bernard1)
told('J') & told('2')
satisfy(told('J') & told('2'), albert1, bernard1, albert2c)
satisfy(told(''), albert1, bernard1, albert2c)
satisfy(told('July'), eve1c)
N = 30
states = {(a, b, c, n)
for a in range(1, N)
for b in range(a, N)
for c in range(b, N) if a * b * c == 36
for n in range(2, 90)}
def ages(state): return state[:-1]
def room(state): return state[-1]
def parent1(state):
"""The product of my three childrens' ages is 36."""
a, b, c = ages(state)
return a * b * c == 36
def friend1(state):
"""I don't know their ages."""
possible_ages = {ages(s) for s in satisfy(states, parent1)}
return not know(possible_ages)
def parent2(state):
"""The sum of their ages is the same as the number of people in this room."""
return sum(ages(state)) == room(state)
def friend2(state, possible_states=satisfy(states, parent1, friend1, parent2)):
"""I still don't know their ages."""
# Given there are room(state) people in the room, I still don't know the ages.
possible_ages = {ages(s) for s in possible_states if room(s) == room(state)}
return not know(possible_ages)
def parent3(state):
"""The oldest one likes bananas."""
# I.e., there is an oldest one (and not twins of the same age)
a, b, c = ages(state)
return c > b
def friend3(state, possible_states=satisfy(states, parent1, friend1, parent2, friend2, parent3)):
"Now I know their ages."
possible_ages = {ages(s) for s in possible_states}
return know(possible_ages)
def child_age_puzzle(states):
return satisfy(states, parent1, friend1, parent2, friend2, parent3, friend3)
child_age_puzzle(states)
satisfy(states, parent1, friend1, parent2)
| 0.621196 | 0.921145 |
<title>Learn Quantum Computation using Qiskit</title>
<div class="preface-top">
<div class="preface-checker-pattern"></div>
<div class="preface-summary">
<aside class="preface-summary-image"><img src="images/preface_illustration_2.svg"></aside>
<div class="preface-summary-text">
<p>
Greetings from the Qiskit Community team! This textbook is a university quantum algorithms/computation course supplement based on Qiskit to help learn:</p>
<ol>
<li>The mathematics behind quantum algorithms</li>
<li>Details about today's non-fault-tolerant quantum devices</li>
<li>Writing code in Qiskit to implement quantum algorithms on IBM's cloud quantum systems</li>
</ol>
</div>
<a href="https://qiskit.org/textbook/ch-states/introduction.html"><button class="preface-button read-textbook">Read the textbook <span class="rangle"><img src="/textbook/assets/images/rightarrow.svg"></span></button></a>
</div>
# About the Textbook
<p>This is a free digital textbook that will teach you the concepts of quantum computing while you learn to use the Qiskit SDK.</p>
## Run the Code Inline
<p>This textbook is built on a jupyter notebook framework that allows for easy reading, but it also allows readers to edit and run the code right in the textbook. The chapters can also be opened as Jupyter notebooks in the <a href="https://quantum-computing.ibm.com/jupyter">IBM Quantum Experience</a>, no installs required!</p>
```
# Click 'try', then 'run' to see the output,
# you can change the code and run it again.
print("This code works!")
from qiskit import QuantumCircuit
qc = QuantumCircuit(2) # Create circuit with 2 qubits
qc.h(0) # Do H-gate on q0
qc.cx(0,1) # Do CNOT on q1 controlled by q0
qc.measure_all()
qc.draw()
```
<a href="https://qiskit.org/textbook/widgets-index.html"><button class="preface-button">Interactivity Tour<span class="rangle"><img src="/textbook/assets/images/rightarrow.svg"></span></button></a>
## Learn with Real Quantum Systems
<p>The best way to learn is by doing. Qiskit allows users to run experiments on state-of-the-art quantum devices from the comfort of their homes. The textbook teaches not only theoretical quantum computing but the experimental quantum physics that realises it.</p>
<img src="images/preface-hw-example.png" class="preface-image">
<a href="https://qiskit.org/textbook/ch-quantum-hardware/accessing_higher_energy_states.html"><button class="preface-button">See Example: Accessing Higher Level States<span class="rangle"><img src="/textbook/assets/images/rightarrow.svg"></span></button></a>
# Using the Textbook
<p>If you're reading the textbook independently, you don't have to read it all in order, but we recommend you read chapters 1-3 first.</p>
## Curriculum Integration
<p>The textbook can be followed as an independent course, however, it has been designed to accompany a traditional university course. The textbook shows students how to use Qiskit to experiment with quantum algorithms and hardware, and uses this to reinforce their understanding.
</p>
<img src="images/curriculum.svg" class="preface-image">
## Use the Textbook in Your Course
If you are using the Qiskit Textbook in your course, you can join the IBM Quantum Educators Program. The Program provides:
<ul class="preface-list">
<li> The ability to reserve time for priority access to our open systems for in-class demonstrations </li>
<li> Access to additional premium systems beyond our open systems</li>
<li> Access to a 5-qubit system with full microwave control using Qiskit Pulse</li>
</ul>
<a href="https://quantum-computing.ibm.com/programs/educators"><button class="preface-button">Sign Up for the IBM Quantum Educators Program<span class="rangle"><img src="/textbook/assets/images/rightarrow.svg"></span></button></a>
# Contact
<p> If you have any questions or suggestions about the textbook or would like to incorporate it into your curriculum, please contact Frank Harkins <a href="mailto:[email protected]">([email protected])</a>. In the true spirit of open-source, any chapter contributions are welcome in this GitHub repository.</p>
# Contributors
<p> Learn Quantum Computation using Qiskit is the work of several individuals. If you use it in your work, cite it using <a href="https://github.com/qiskit-community/qiskit-textbook/blob/master/content/qiskit-textbook.bib">this bib file</a> or directly as:</p>
<p><i>
Abraham Asfaw, Luciano Bello, Yael Ben-Haim, Sergey Bravyi, Nicholas Bronn, Lauren Capelluto, Almudena Carrera Vazquez, Jack Ceroni, Richard Chen, Albert Frisch, Jay Gambetta, Shelly Garion, Leron Gil, Salvador De La Puente Gonzalez, Francis Harkins, Takashi Imamichi, David McKay, Antonio Mezzacapo, Zlatko Minev, Ramis Movassagh, Giacomo Nannicni, Paul Nation, Anna Phan, Marco Pistoia, Arthur Rattew, Joachim Schaefer, Javad Shabani, John Smolin, John Stenger, Kristan Temme, Madeleine Tod, Stephen Wood, James Wootton.</i></p>
|
github_jupyter
|
# Click 'try', then 'run' to see the output,
# you can change the code and run it again.
print("This code works!")
from qiskit import QuantumCircuit
qc = QuantumCircuit(2) # Create circuit with 2 qubits
qc.h(0) # Do H-gate on q0
qc.cx(0,1) # Do CNOT on q1 controlled by q0
qc.measure_all()
qc.draw()
| 0.302288 | 0.92912 |
```
# install keras, tensorflow and tflearn
# (1) Importing dependency
import keras
from keras.models import Sequential
from keras.layers import Dense, Activation, Dropout, Flatten,\
Conv2D, MaxPooling2D
from keras.layers.normalization import BatchNormalization
import numpy as np
np.random.seed(1000)
# (2) Get Data
import tflearn.datasets.oxflower17 as oxflower17
x, y = oxflower17.load_data(one_hot=True)
# (3) Create a sequential model
model = Sequential()
# 1st Convolutional Layer
model.add(Conv2D(filters=96, input_shape=(224,224,3), kernel_size=(11,11),\
strides=(4,4), padding='valid'))
model.add(Activation('relu'))
# Pooling
model.add(MaxPooling2D(pool_size=(2,2), strides=(2,2), padding='valid'))
# Batch Normalisation before passing it to the next layer
model.add(BatchNormalization())
# 2nd Convolutional Layer
model.add(Conv2D(filters=256, kernel_size=(11,11), strides=(1,1), padding='valid'))
model.add(Activation('relu'))
# Pooling
model.add(MaxPooling2D(pool_size=(2,2), strides=(2,2), padding='valid'))
# Batch Normalisation
model.add(BatchNormalization())
# 3rd Convolutional Layer
model.add(Conv2D(filters=384, kernel_size=(3,3), strides=(1,1), padding='valid'))
model.add(Activation('relu'))
# Batch Normalisation
model.add(BatchNormalization())
# 4th Convolutional Layer
model.add(Conv2D(filters=384, kernel_size=(3,3), strides=(1,1), padding='valid'))
model.add(Activation('relu'))
# Batch Normalisation
model.add(BatchNormalization())
# 5th Convolutional Layer
model.add(Conv2D(filters=256, kernel_size=(3,3), strides=(1,1), padding='valid'))
model.add(Activation('relu'))
# Pooling
model.add(MaxPooling2D(pool_size=(2,2), strides=(2,2), padding='valid'))
# Batch Normalisation
model.add(BatchNormalization())
# Passing it to a dense layer
model.add(Flatten())
# 1st Dense Layer
model.add(Dense(4096, input_shape=(224*224*3,)))
model.add(Activation('relu'))
# Add Dropout to prevent overfitting
model.add(Dropout(0.4))
# Batch Normalisation
model.add(BatchNormalization())
# 2nd Dense Layer
model.add(Dense(4096))
model.add(Activation('relu'))
# Add Dropout
model.add(Dropout(0.4))
# Batch Normalisation
model.add(BatchNormalization())
# 3rd Dense Layer
model.add(Dense(1000))
model.add(Activation('relu'))
# Add Dropout
model.add(Dropout(0.4))
# Batch Normalisation
model.add(BatchNormalization())
# Output Layer
model.add(Dense(17))
model.add(Activation('softmax'))
model.summary()
# (4) Compile
model.compile(loss='categorical_crossentropy', optimizer='adam',\
metrics=['accuracy'])
# (5) Train
model.fit(x, y, batch_size=64, epochs=10, verbose=1, \
validation_split=0.2, shuffle=True)
```
|
github_jupyter
|
# install keras, tensorflow and tflearn
# (1) Importing dependency
import keras
from keras.models import Sequential
from keras.layers import Dense, Activation, Dropout, Flatten,\
Conv2D, MaxPooling2D
from keras.layers.normalization import BatchNormalization
import numpy as np
np.random.seed(1000)
# (2) Get Data
import tflearn.datasets.oxflower17 as oxflower17
x, y = oxflower17.load_data(one_hot=True)
# (3) Create a sequential model
model = Sequential()
# 1st Convolutional Layer
model.add(Conv2D(filters=96, input_shape=(224,224,3), kernel_size=(11,11),\
strides=(4,4), padding='valid'))
model.add(Activation('relu'))
# Pooling
model.add(MaxPooling2D(pool_size=(2,2), strides=(2,2), padding='valid'))
# Batch Normalisation before passing it to the next layer
model.add(BatchNormalization())
# 2nd Convolutional Layer
model.add(Conv2D(filters=256, kernel_size=(11,11), strides=(1,1), padding='valid'))
model.add(Activation('relu'))
# Pooling
model.add(MaxPooling2D(pool_size=(2,2), strides=(2,2), padding='valid'))
# Batch Normalisation
model.add(BatchNormalization())
# 3rd Convolutional Layer
model.add(Conv2D(filters=384, kernel_size=(3,3), strides=(1,1), padding='valid'))
model.add(Activation('relu'))
# Batch Normalisation
model.add(BatchNormalization())
# 4th Convolutional Layer
model.add(Conv2D(filters=384, kernel_size=(3,3), strides=(1,1), padding='valid'))
model.add(Activation('relu'))
# Batch Normalisation
model.add(BatchNormalization())
# 5th Convolutional Layer
model.add(Conv2D(filters=256, kernel_size=(3,3), strides=(1,1), padding='valid'))
model.add(Activation('relu'))
# Pooling
model.add(MaxPooling2D(pool_size=(2,2), strides=(2,2), padding='valid'))
# Batch Normalisation
model.add(BatchNormalization())
# Passing it to a dense layer
model.add(Flatten())
# 1st Dense Layer
model.add(Dense(4096, input_shape=(224*224*3,)))
model.add(Activation('relu'))
# Add Dropout to prevent overfitting
model.add(Dropout(0.4))
# Batch Normalisation
model.add(BatchNormalization())
# 2nd Dense Layer
model.add(Dense(4096))
model.add(Activation('relu'))
# Add Dropout
model.add(Dropout(0.4))
# Batch Normalisation
model.add(BatchNormalization())
# 3rd Dense Layer
model.add(Dense(1000))
model.add(Activation('relu'))
# Add Dropout
model.add(Dropout(0.4))
# Batch Normalisation
model.add(BatchNormalization())
# Output Layer
model.add(Dense(17))
model.add(Activation('softmax'))
model.summary()
# (4) Compile
model.compile(loss='categorical_crossentropy', optimizer='adam',\
metrics=['accuracy'])
# (5) Train
model.fit(x, y, batch_size=64, epochs=10, verbose=1, \
validation_split=0.2, shuffle=True)
| 0.91282 | 0.697879 |
```
#使用seaborn
%matplotlib inline
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_context("paper",font_scale=1.5,rc={'figure.dpi':300})
sns.set_style("ticks") # 风格选择包括:"white", "dark", "whitegrid", "darkgrid", "ticks"
sns.set_style({'font.sans-serif': ['SimHei', 'Calibri']}) #设置中文设定
from pandas import Series,DataFrame
import numpy as np
np.random.seed(10000)
import imp
import input_data_class
import os
import configparser
import argparse
import logging
import logging.config
from scipy.stats import entropy
import math
from sklearn.manifold import TSNE
from sklearn import metrics
#paper imgs dir
paper_imgs_dir="paper_imgs/"
def draw_tsner(data,labels,paper_img_name=""):
color={0:'r',1:'b'}
labels=[ color[i] for i in labels ]
x_embedded = TSNE(n_components=2,random_state=0).fit_transform(data)
plt.figure()
plt.scatter(x_embedded[:,0],x_embedded[:,1],c=labels, s=1.0, alpha = 0.5)
plt.savefig(paper_img_name,format="pdf",bbox_inches="tight",dpi=600)
plt.show()
print(metrics.silhouette_score(x_embedded, labels, sample_size=len(data), metric='euclidean'))
def Interpretability(dataset="location",TOP=2):
defense="MemGuard"
bins=30
xlim=(0, 1.0)
ylim=(0, 15.0)
input_data=input_data_class.InputData(dataset=dataset)
config = configparser.ConfigParser()
config.read('config.ini')
user_label_dim=int(config[dataset]["num_classes"])
result_folder=config[dataset]["result_folder"]
(x_evaluate,y_evaluate,l_evaluate)=input_data.input_data_attacker_evaluate()
evaluation_noise_filepath=result_folder+"/attack/MemGuard_noise_data_evaluation.npz"
print(evaluation_noise_filepath)
if not os.path.isfile(evaluation_noise_filepath):
raise FileNotFoundError
npz_defense=np.load(evaluation_noise_filepath)
f_evaluate_noise=npz_defense['defense_output']
f_evaluate_origin=npz_defense['tc_output']
f_evaluate_noise=np.sort(f_evaluate_noise,axis=1)
f_evaluate_origin=np.sort(f_evaluate_origin,axis=1)
print("*"*100)
print("nn attack+no defense")
data=f_evaluate_origin.copy()
labels=l_evaluate
draw_tsner(data,labels,paper_imgs_dir+"v_nn_no.pdf")
print("*"*100)
print("nn attack+MemGuard defense")
data=f_evaluate_noise.copy()
labels=l_evaluate
draw_tsner(data,labels,paper_imgs_dir+"v_nn_m.pdf")
print("*"*100)
print("our attack+no defense")
data=f_evaluate_origin.copy()
data[:,:-TOP]=0
labels=l_evaluate
draw_tsner(data,labels,paper_imgs_dir+"v_our_no.pdf")
print("*"*100)
print("our attack+MemGuard defense")
data=f_evaluate_noise.copy()
data[:,:-TOP]=0
labels=l_evaluate
draw_tsner(data,labels,paper_imgs_dir+"v_our_m.pdf")
Interpretability(dataset="location",TOP=2)
"""
(x_evaluate,y_evaluate,l_evaluate)=input_data.input_data_attacker_evaluate()
evaluation_noise_filepath=result_folder+"/attack/MemGuard_noise_data_evaluation.npz"
print(evaluation_noise_filepath)
if not os.path.isfile(evaluation_noise_filepath):
raise FileNotFoundError
npz_defense=np.load(evaluation_noise_filepath)
f_evaluate_noise=npz_defense['defense_output']
f_evaluate_origin=npz_defense['tc_output']
f_evaluate_noise=np.sort(f_evaluate_noise,axis=1)
f_evaluate_origin=np.sort(f_evaluate_origin,axis=1)
for TOP in [1,2,3,5,10,20,30]:
data=f_evaluate_noise.copy()
data[:,:-TOP]=0
x_embedded = TSNE(n_components=2,random_state=0).fit_transform(data)
labels=l_evaluate
print("TOP={} silhouette_score={}".format( TOP, metrics.silhouette_score(x_embedded, labels, sample_size=len(data), metric='euclidean')))
"""
```
The gap between the two curves in a graph corresponds to the information leakage of the target classifier’s training dataset. Our defense substantially reduces such gaps.
|
github_jupyter
|
#使用seaborn
%matplotlib inline
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_context("paper",font_scale=1.5,rc={'figure.dpi':300})
sns.set_style("ticks") # 风格选择包括:"white", "dark", "whitegrid", "darkgrid", "ticks"
sns.set_style({'font.sans-serif': ['SimHei', 'Calibri']}) #设置中文设定
from pandas import Series,DataFrame
import numpy as np
np.random.seed(10000)
import imp
import input_data_class
import os
import configparser
import argparse
import logging
import logging.config
from scipy.stats import entropy
import math
from sklearn.manifold import TSNE
from sklearn import metrics
#paper imgs dir
paper_imgs_dir="paper_imgs/"
def draw_tsner(data,labels,paper_img_name=""):
color={0:'r',1:'b'}
labels=[ color[i] for i in labels ]
x_embedded = TSNE(n_components=2,random_state=0).fit_transform(data)
plt.figure()
plt.scatter(x_embedded[:,0],x_embedded[:,1],c=labels, s=1.0, alpha = 0.5)
plt.savefig(paper_img_name,format="pdf",bbox_inches="tight",dpi=600)
plt.show()
print(metrics.silhouette_score(x_embedded, labels, sample_size=len(data), metric='euclidean'))
def Interpretability(dataset="location",TOP=2):
defense="MemGuard"
bins=30
xlim=(0, 1.0)
ylim=(0, 15.0)
input_data=input_data_class.InputData(dataset=dataset)
config = configparser.ConfigParser()
config.read('config.ini')
user_label_dim=int(config[dataset]["num_classes"])
result_folder=config[dataset]["result_folder"]
(x_evaluate,y_evaluate,l_evaluate)=input_data.input_data_attacker_evaluate()
evaluation_noise_filepath=result_folder+"/attack/MemGuard_noise_data_evaluation.npz"
print(evaluation_noise_filepath)
if not os.path.isfile(evaluation_noise_filepath):
raise FileNotFoundError
npz_defense=np.load(evaluation_noise_filepath)
f_evaluate_noise=npz_defense['defense_output']
f_evaluate_origin=npz_defense['tc_output']
f_evaluate_noise=np.sort(f_evaluate_noise,axis=1)
f_evaluate_origin=np.sort(f_evaluate_origin,axis=1)
print("*"*100)
print("nn attack+no defense")
data=f_evaluate_origin.copy()
labels=l_evaluate
draw_tsner(data,labels,paper_imgs_dir+"v_nn_no.pdf")
print("*"*100)
print("nn attack+MemGuard defense")
data=f_evaluate_noise.copy()
labels=l_evaluate
draw_tsner(data,labels,paper_imgs_dir+"v_nn_m.pdf")
print("*"*100)
print("our attack+no defense")
data=f_evaluate_origin.copy()
data[:,:-TOP]=0
labels=l_evaluate
draw_tsner(data,labels,paper_imgs_dir+"v_our_no.pdf")
print("*"*100)
print("our attack+MemGuard defense")
data=f_evaluate_noise.copy()
data[:,:-TOP]=0
labels=l_evaluate
draw_tsner(data,labels,paper_imgs_dir+"v_our_m.pdf")
Interpretability(dataset="location",TOP=2)
"""
(x_evaluate,y_evaluate,l_evaluate)=input_data.input_data_attacker_evaluate()
evaluation_noise_filepath=result_folder+"/attack/MemGuard_noise_data_evaluation.npz"
print(evaluation_noise_filepath)
if not os.path.isfile(evaluation_noise_filepath):
raise FileNotFoundError
npz_defense=np.load(evaluation_noise_filepath)
f_evaluate_noise=npz_defense['defense_output']
f_evaluate_origin=npz_defense['tc_output']
f_evaluate_noise=np.sort(f_evaluate_noise,axis=1)
f_evaluate_origin=np.sort(f_evaluate_origin,axis=1)
for TOP in [1,2,3,5,10,20,30]:
data=f_evaluate_noise.copy()
data[:,:-TOP]=0
x_embedded = TSNE(n_components=2,random_state=0).fit_transform(data)
labels=l_evaluate
print("TOP={} silhouette_score={}".format( TOP, metrics.silhouette_score(x_embedded, labels, sample_size=len(data), metric='euclidean')))
"""
| 0.219505 | 0.237267 |
# Lesson 3 Exercise 1: Three Queries Three Tables
<img src="images/cassandralogo.png" width="250" height="250">
### Walk through the basics of creating a table in Apache Cassandra, inserting rows of data, and doing a simple CQL query to validate the information. You will practice Denormalization, and the concept of 1 table per query, which is an encouraged practice with Apache Cassandra.
### Remember, replace ##### with your answer.
#### We will use a python wrapper/ python driver called cassandra to run the Apache Cassandra queries. This library should be preinstalled but in the future to install this library you can run this command in a notebook to install locally:
! pip install cassandra-driver
#### More documentation can be found here: https://datastax.github.io/python-driver/
#### Import Apache Cassandra python package
```
import cassandra
```
### Create a connection to the database
```
from cassandra.cluster import Cluster
try:
cluster = Cluster(['127.0.0.1']) #If you have a locally installed Apache Cassandra instance
session = cluster.connect()
except Exception as e:
print(e)
```
### Create a keyspace to work in
```
try:
session.execute("""
CREATE KEYSPACE IF NOT EXISTS udacity
WITH REPLICATION =
{ 'class' : 'SimpleStrategy', 'replication_factor' : 1 }"""
)
except Exception as e:
print(e)
```
#### Connect to our Keyspace. Compare this to how we had to create a new session in PostgreSQL.
```
try:
session.set_keyspace('udacity')
except Exception as e:
print(e)
```
### Let's imagine we would like to start creating a Music Library of albums.
### We want to ask 3 questions of the data
#### 1. Give every album in the music library that was released in a given year
`select * from music_library WHERE YEAR=1970`
#### 2. Give every album in the music library that was created by a given artist
`select * from artist_library WHERE artist_name="The Beatles"`
#### 3. Give all the information from the music library about a given album
`select * from album_library WHERE album_name="Close To You"`
### Because we want to do three different queries, we will need different tables that partition the data differently.
<img src="images/table1.png" width="350" height="350">
<img src="images/table2.png" width="350" height="350">
<img src="images/table0.png" width="550" height="550">
### TO-DO: Create the tables.
```
query = "CREATE TABLE IF NOT EXISTS music_library"
query = query + "(year int, artist_name text, album_name text, PRIMARY KEY (year, artist_name))"
try:
session.execute(query)
except Exception as e:
print(e)
query1 = "CREATE TABLE IF NOT EXISTS artist_library"
query1 = query1 + "(artist_name text, year int, album_name text, PRIMARY KEY (year, artist_name))"
try:
session.execute(query1)
except Exception as e:
print(e)
query2 = "CREATE TABLE IF NOT EXISTS album_library"
query2 = query2 + "(album_name text, artist_name text, year int, PRIMARY KEY (year, artist_name))"
try:
session.execute(query2)
except Exception as e:
print(e)
```
### TO-DO: Insert data into the tables
```
query = "INSERT INTO music_library (year, artist_name, album_name)"
query = query + " VALUES (%s, %s, %s)"
query1 = "INSERT INTO artist_library (artist_name, year, album_name)"
query1 = query1 + " VALUES (%s, %s, %s)"
query2 = "INSERT INTO album_library (album_name, artist_name, year)"
query2 = query2 + " VALUES (%s, %s, %s)"
try:
session.execute(query, (1970, "The Beatles", "Let it Be"))
except Exception as e:
print(e)
try:
session.execute(query, (1965, "The Beatles", "Rubber Soul"))
except Exception as e:
print(e)
try:
session.execute(query, (1965, "The Who", "My Generation"))
except Exception as e:
print(e)
try:
session.execute(query, (1966, "The Monkees", "The Monkees"))
except Exception as e:
print(e)
try:
session.execute(query, (1970, "The Carpenters", "Close To You"))
except Exception as e:
print(e)
try:
session.execute(query1, ("The Beatles", 1970, "Let it Be"))
except Exception as e:
print(e)
try:
session.execute(query1, ("The Beatles", 1965, "Rubber Soul"))
except Exception as e:
print(e)
try:
session.execute(query1, ("The Who", 1965, "My Generation"))
except Exception as e:
print(e)
try:
session.execute(query1, ("The Monkees", 1966, "The Monkees"))
except Exception as e:
print(e)
try:
session.execute(query1, ("The Carpenters", 1970, "Close To You"))
except Exception as e:
print(e)
try:
session.execute(query2, ("Let it Be", "The Beatles", 1970))
except Exception as e:
print(e)
try:
session.execute(query2, ("Rubber Soul", "The Beatles", 1965))
except Exception as e:
print(e)
try:
session.execute(query2, ("My Generation", "The Who", 1965))
except Exception as e:
print(e)
try:
session.execute(query2, ("The Monkees", "The Monkees", 1966))
except Exception as e:
print(e)
try:
session.execute(query2, ("Close To You", "The Carpenters", 1970))
except Exception as e:
print(e)
```
This might have felt unnatural to insert duplicate data into the tables. If I just normalized these tables, I wouldn't have to have extra copies! While this is true, remember there are no `JOINS` in Apache Cassandra. For the benefit of high availibity and scalabity, denormalization must be how this is done.
### TO-DO: Validate the Data Model
```
query = "select * from music_library WHERE year=1970"
try:
rows = session.execute(query)
except Exception as e:
print(e)
for row in rows:
print (row.year, row.artist_name, row.album_name)
```
### Your output should be:
1970 The Beatles Let it Be<br>
1970 The Carpenters Close To You
### TO-DO: Validate the Data Model
```
query = "select * from album_library WHERE artist_name='The Beatles' ALLOW FILTERING"
try:
rows = session.execute(query)
except Exception as e:
print(e)
for row in rows:
print (row.artist_name, row.album_name, row.year)
```
### Your output should be:
The Beatles Rubber Soul 1965 <br>
The Beatles Let it Be 1970
### TO-DO: Validate the Data Model
```
query = "select * from artist_library WHERE artist_name='The Carpenters' ALLOW FILTERING"
try:
rows = session.execute(query)
except Exception as e:
print(e)
for row in rows:
print (row.artist_name, row.year, row.album_name)
```
### Your output should be:
The Carpenters 1970 Close To You
### And finally close the session and cluster connection
```
session.shutdown()
cluster.shutdown()
```
|
github_jupyter
|
import cassandra
from cassandra.cluster import Cluster
try:
cluster = Cluster(['127.0.0.1']) #If you have a locally installed Apache Cassandra instance
session = cluster.connect()
except Exception as e:
print(e)
try:
session.execute("""
CREATE KEYSPACE IF NOT EXISTS udacity
WITH REPLICATION =
{ 'class' : 'SimpleStrategy', 'replication_factor' : 1 }"""
)
except Exception as e:
print(e)
try:
session.set_keyspace('udacity')
except Exception as e:
print(e)
query = "CREATE TABLE IF NOT EXISTS music_library"
query = query + "(year int, artist_name text, album_name text, PRIMARY KEY (year, artist_name))"
try:
session.execute(query)
except Exception as e:
print(e)
query1 = "CREATE TABLE IF NOT EXISTS artist_library"
query1 = query1 + "(artist_name text, year int, album_name text, PRIMARY KEY (year, artist_name))"
try:
session.execute(query1)
except Exception as e:
print(e)
query2 = "CREATE TABLE IF NOT EXISTS album_library"
query2 = query2 + "(album_name text, artist_name text, year int, PRIMARY KEY (year, artist_name))"
try:
session.execute(query2)
except Exception as e:
print(e)
query = "INSERT INTO music_library (year, artist_name, album_name)"
query = query + " VALUES (%s, %s, %s)"
query1 = "INSERT INTO artist_library (artist_name, year, album_name)"
query1 = query1 + " VALUES (%s, %s, %s)"
query2 = "INSERT INTO album_library (album_name, artist_name, year)"
query2 = query2 + " VALUES (%s, %s, %s)"
try:
session.execute(query, (1970, "The Beatles", "Let it Be"))
except Exception as e:
print(e)
try:
session.execute(query, (1965, "The Beatles", "Rubber Soul"))
except Exception as e:
print(e)
try:
session.execute(query, (1965, "The Who", "My Generation"))
except Exception as e:
print(e)
try:
session.execute(query, (1966, "The Monkees", "The Monkees"))
except Exception as e:
print(e)
try:
session.execute(query, (1970, "The Carpenters", "Close To You"))
except Exception as e:
print(e)
try:
session.execute(query1, ("The Beatles", 1970, "Let it Be"))
except Exception as e:
print(e)
try:
session.execute(query1, ("The Beatles", 1965, "Rubber Soul"))
except Exception as e:
print(e)
try:
session.execute(query1, ("The Who", 1965, "My Generation"))
except Exception as e:
print(e)
try:
session.execute(query1, ("The Monkees", 1966, "The Monkees"))
except Exception as e:
print(e)
try:
session.execute(query1, ("The Carpenters", 1970, "Close To You"))
except Exception as e:
print(e)
try:
session.execute(query2, ("Let it Be", "The Beatles", 1970))
except Exception as e:
print(e)
try:
session.execute(query2, ("Rubber Soul", "The Beatles", 1965))
except Exception as e:
print(e)
try:
session.execute(query2, ("My Generation", "The Who", 1965))
except Exception as e:
print(e)
try:
session.execute(query2, ("The Monkees", "The Monkees", 1966))
except Exception as e:
print(e)
try:
session.execute(query2, ("Close To You", "The Carpenters", 1970))
except Exception as e:
print(e)
query = "select * from music_library WHERE year=1970"
try:
rows = session.execute(query)
except Exception as e:
print(e)
for row in rows:
print (row.year, row.artist_name, row.album_name)
query = "select * from album_library WHERE artist_name='The Beatles' ALLOW FILTERING"
try:
rows = session.execute(query)
except Exception as e:
print(e)
for row in rows:
print (row.artist_name, row.album_name, row.year)
query = "select * from artist_library WHERE artist_name='The Carpenters' ALLOW FILTERING"
try:
rows = session.execute(query)
except Exception as e:
print(e)
for row in rows:
print (row.artist_name, row.year, row.album_name)
session.shutdown()
cluster.shutdown()
| 0.187579 | 0.902695 |
```
import pandas as pd
import numpy as np
import requests
import bs4 as bs
import urllib.request
```
## Extracting features of 2020 movies from Wikipedia
```
link = "https://en.wikipedia.org/wiki/List_of_American_films_of_2020"
source = urllib.request.urlopen(link).read()
soup = bs.BeautifulSoup(source,'lxml')
tables = soup.find_all('table',class_='wikitable sortable')
len(tables)
type(tables[0])
df1 = pd.read_html(str(tables[0]))[0]
df2 = pd.read_html(str(tables[1]))[0]
df3 = pd.read_html(str(tables[2]))[0]
df4 = pd.read_html(str(tables[3]).replace("'1\"\'",'"1"'))[0] # avoided "ValueError: invalid literal for int() with base 10: '1"'
df = df1.append(df2.append(df3.append(df4,ignore_index=True),ignore_index=True),ignore_index=True)
df
df_2020 = df[['Title','Cast and crew']]
df_2020
!pip install tmdbv3api
from tmdbv3api import TMDb
import json
import requests
tmdb = TMDb()
tmdb.api_key = ''
from tmdbv3api import Movie
tmdb_movie = Movie()
def get_genre(x):
genres = []
result = tmdb_movie.search(x)
if not result:
return np.NaN
else:
movie_id = result[0].id
response = requests.get('https://api.themoviedb.org/3/movie/{}?api_key={}'.format(movie_id,tmdb.api_key))
data_json = response.json()
if data_json['genres']:
genre_str = " "
for i in range(0,len(data_json['genres'])):
genres.append(data_json['genres'][i]['name'])
return genre_str.join(genres)
else:
return np.NaN
df_2020['genres'] = df_2020['Title'].map(lambda x: get_genre(str(x)))
df_2020
def get_director(x):
if " (director)" in x:
return x.split(" (director)")[0]
elif " (directors)" in x:
return x.split(" (directors)")[0]
else:
return x.split(" (director/screenplay)")[0]
df_2020['director_name'] = df_2020['Cast and crew'].map(lambda x: get_director(str(x)))
def get_actor1(x):
return ((x.split("screenplay); ")[-1]).split(", ")[0])
df_2020['actor_1_name'] = df_2020['Cast and crew'].map(lambda x: get_actor1(str(x)))
def get_actor2(x):
if len((x.split("screenplay); ")[-1]).split(", ")) < 2:
return np.NaN
else:
return ((x.split("screenplay); ")[-1]).split(", ")[1])
df_2020['actor_2_name'] = df_2020['Cast and crew'].map(lambda x: get_actor2(str(x)))
def get_actor3(x):
if len((x.split("screenplay); ")[-1]).split(", ")) < 3:
return np.NaN
else:
return ((x.split("screenplay); ")[-1]).split(", ")[2])
df_2020['actor_3_name'] = df_2020['Cast and crew'].map(lambda x: get_actor3(str(x)))
df_2020
df_2020 = df_2020.rename(columns={'Title':'movie_title'})
new_df20 = df_2020.loc[:,['director_name','actor_1_name','actor_2_name','actor_3_name','genres','movie_title']]
new_df20
new_df20['comb'] = new_df20['actor_1_name'] + ' ' + new_df20['actor_2_name'] + ' '+ new_df20['actor_3_name'] + ' '+ new_df20['director_name'] +' ' + new_df20['genres']
new_df20.isna().sum()
new_df20 = new_df20.dropna(how='any')
new_df20.isna().sum()
new_df20['movie_title'] = new_df20['movie_title'].str.lower()
new_df20
old_df = pd.read_csv('final_data.csv')
old_df
final_df = old_df.append(new_df20,ignore_index=True)
final_df
final_df.to_csv('main_data.csv',index=False)
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import requests
import bs4 as bs
import urllib.request
link = "https://en.wikipedia.org/wiki/List_of_American_films_of_2020"
source = urllib.request.urlopen(link).read()
soup = bs.BeautifulSoup(source,'lxml')
tables = soup.find_all('table',class_='wikitable sortable')
len(tables)
type(tables[0])
df1 = pd.read_html(str(tables[0]))[0]
df2 = pd.read_html(str(tables[1]))[0]
df3 = pd.read_html(str(tables[2]))[0]
df4 = pd.read_html(str(tables[3]).replace("'1\"\'",'"1"'))[0] # avoided "ValueError: invalid literal for int() with base 10: '1"'
df = df1.append(df2.append(df3.append(df4,ignore_index=True),ignore_index=True),ignore_index=True)
df
df_2020 = df[['Title','Cast and crew']]
df_2020
!pip install tmdbv3api
from tmdbv3api import TMDb
import json
import requests
tmdb = TMDb()
tmdb.api_key = ''
from tmdbv3api import Movie
tmdb_movie = Movie()
def get_genre(x):
genres = []
result = tmdb_movie.search(x)
if not result:
return np.NaN
else:
movie_id = result[0].id
response = requests.get('https://api.themoviedb.org/3/movie/{}?api_key={}'.format(movie_id,tmdb.api_key))
data_json = response.json()
if data_json['genres']:
genre_str = " "
for i in range(0,len(data_json['genres'])):
genres.append(data_json['genres'][i]['name'])
return genre_str.join(genres)
else:
return np.NaN
df_2020['genres'] = df_2020['Title'].map(lambda x: get_genre(str(x)))
df_2020
def get_director(x):
if " (director)" in x:
return x.split(" (director)")[0]
elif " (directors)" in x:
return x.split(" (directors)")[0]
else:
return x.split(" (director/screenplay)")[0]
df_2020['director_name'] = df_2020['Cast and crew'].map(lambda x: get_director(str(x)))
def get_actor1(x):
return ((x.split("screenplay); ")[-1]).split(", ")[0])
df_2020['actor_1_name'] = df_2020['Cast and crew'].map(lambda x: get_actor1(str(x)))
def get_actor2(x):
if len((x.split("screenplay); ")[-1]).split(", ")) < 2:
return np.NaN
else:
return ((x.split("screenplay); ")[-1]).split(", ")[1])
df_2020['actor_2_name'] = df_2020['Cast and crew'].map(lambda x: get_actor2(str(x)))
def get_actor3(x):
if len((x.split("screenplay); ")[-1]).split(", ")) < 3:
return np.NaN
else:
return ((x.split("screenplay); ")[-1]).split(", ")[2])
df_2020['actor_3_name'] = df_2020['Cast and crew'].map(lambda x: get_actor3(str(x)))
df_2020
df_2020 = df_2020.rename(columns={'Title':'movie_title'})
new_df20 = df_2020.loc[:,['director_name','actor_1_name','actor_2_name','actor_3_name','genres','movie_title']]
new_df20
new_df20['comb'] = new_df20['actor_1_name'] + ' ' + new_df20['actor_2_name'] + ' '+ new_df20['actor_3_name'] + ' '+ new_df20['director_name'] +' ' + new_df20['genres']
new_df20.isna().sum()
new_df20 = new_df20.dropna(how='any')
new_df20.isna().sum()
new_df20['movie_title'] = new_df20['movie_title'].str.lower()
new_df20
old_df = pd.read_csv('final_data.csv')
old_df
final_df = old_df.append(new_df20,ignore_index=True)
final_df
final_df.to_csv('main_data.csv',index=False)
| 0.208098 | 0.413773 |
# Everything is object in Python
# 1 Pass through arguments in constructor
```
class Passthrough:
def __init__(self, **kwargs):
for k, v in kwargs.items():
setattr(self, k, v)
pt = Passthrough(name="Zhaokang", age=35)
pt.name
```
# 2 Inheritance
> The order of base class defined method resolution order in case of multiple inheritance
```
class Base:
# pass
def __init__(self, name, **kwargs):
print(name)
print(kwargs)
class Derived(Base):
def __init__(self, name, **kwargs):
super().__init__(name, **kwargs)
d = Derived(name="Zhaokang", age=35)
```
# 3 Class related methods
```
isinstance(3, float)
isinstance(3, (float, int))
isinstance(True, int)
issubclass(Derived, Base)
issubclass(bool, int)
type(d)
d.__class__
d.__class__.__name__
```
# 4 Magic methods
> define \__init\__(self) for mutables and \__new\__(self) for immutables
> attr package for full automation
```
d.__str__
d.__repr__
d.__int__
d.__float__
d.__init__
d.__new__
d.__eq__
d.__ne__
d.__gt__
d.__lt__
d.__ge__
d.__le__
```
# 5 Math operation methods
> \__add\__
> \__radd\__ (Reflextive)
> \__iadd\__ (In place)
> Similar for \__mul\__
# 6 Emulate built-in methods
> \__len\__
> \__contains\__(self, item)
> \__iter\__(self)
> \__getattribute\__(self, item) (for dot operator)
```
class Iterable:
def __init__(self):
self.items = [1, 2, 3]
def __iter__(self):
yield from self.items
i = Iterable()
for e in i:
print(e)
class Dottable(object):
def __init__(self):
self.name = "Zhaokang"
def __getattribute__(self, name):
return object.__getattribute__(self, name)
d = Dottable()
d.name
```
# 7 Class method and static method
> [Difference between class method and static method](https://www.geeksforgeeks.org/class-method-vs-static-method-python/)
```
class Foo():
def __init__(self, name):
self.name = name
@classmethod
def foo(cls, arg):
return cls(arg)
f = Foo.foo("Zhaokang")
print(f.name)
```
# 8 Encapsulation
> __ prefix variable and method name
> They are not truly private accessibility, they are prefixed with _Private
```
class Private():
__name = "Zhaokang"
def __age():
return 35
dir(Private)
```
# 9 Getter and Setter
```
class Getter():
@property
def name(self):
return "Zhaokang"
Getter().name
class Setter():
@property
def age(self):
return self.__age
@age.setter
def age(self, value):
self.__age = value
s = Setter()
s.age = 34
s.age
```
|
github_jupyter
|
class Passthrough:
def __init__(self, **kwargs):
for k, v in kwargs.items():
setattr(self, k, v)
pt = Passthrough(name="Zhaokang", age=35)
pt.name
class Base:
# pass
def __init__(self, name, **kwargs):
print(name)
print(kwargs)
class Derived(Base):
def __init__(self, name, **kwargs):
super().__init__(name, **kwargs)
d = Derived(name="Zhaokang", age=35)
isinstance(3, float)
isinstance(3, (float, int))
isinstance(True, int)
issubclass(Derived, Base)
issubclass(bool, int)
type(d)
d.__class__
d.__class__.__name__
d.__str__
d.__repr__
d.__int__
d.__float__
d.__init__
d.__new__
d.__eq__
d.__ne__
d.__gt__
d.__lt__
d.__ge__
d.__le__
class Iterable:
def __init__(self):
self.items = [1, 2, 3]
def __iter__(self):
yield from self.items
i = Iterable()
for e in i:
print(e)
class Dottable(object):
def __init__(self):
self.name = "Zhaokang"
def __getattribute__(self, name):
return object.__getattribute__(self, name)
d = Dottable()
d.name
class Foo():
def __init__(self, name):
self.name = name
@classmethod
def foo(cls, arg):
return cls(arg)
f = Foo.foo("Zhaokang")
print(f.name)
class Private():
__name = "Zhaokang"
def __age():
return 35
dir(Private)
class Getter():
@property
def name(self):
return "Zhaokang"
Getter().name
class Setter():
@property
def age(self):
return self.__age
@age.setter
def age(self, value):
self.__age = value
s = Setter()
s.age = 34
s.age
| 0.682574 | 0.647032 |
```
%load_ext autoreload
%autoreload 2
# default_exp pod.client
```
# Pod Client
```
# export
from pyintegrators.data.itembase import Edge, ItemBase
from pyintegrators.indexers.facerecognition.photo import resize
from pyintegrators.data.schema import *
from pyintegrators.imports import *
from hashlib import sha256
# export
DEFAULT_POD_ADDRESS = "http://localhost:3030"
POD_VERSION = "v2"
# export
class PodClient:
def __init__(self, url=DEFAULT_POD_ADDRESS, version=POD_VERSION, database_key=None, owner_key=None):
self.url = url
self.version = POD_VERSION
self.test_connection(verbose=False)
self.database_key=database_key if database_key is not None else self.generate_random_key()
self.owner_key=owner_key if owner_key is not None else self.generate_random_key()
self.base_url = f"{url}/{version}/{self.owner_key}"
@staticmethod
def generate_random_key():
return "".join([str(random.randint(0, 9)) for i in range(64)])
def test_connection(self, verbose=True):
try:
res = requests.get(self.url)
if verbose: print("Succesfully connected to pod")
return True
except requests.exceptions.RequestException as e:
print("Could no connect to backend")
return False
def create(self, node):
if isinstance(node, Photo) and not self.create_photo_file(node): return False
try:
body = {"databaseKey": self.database_key, "payload":self.get_properties_json(node) }
result = requests.post(f"{self.base_url}/create_item", json=body)
if result.status_code != 200:
print(result, result.content)
return False
else:
uid = int(result.json())
node.uid = uid
ItemBase.add_to_db(node)
return True
except requests.exceptions.RequestException as e:
print(e)
return False
def create_photo_file(self, photo):
file = photo.file[0]
self.create(file)
return self.upload_photo(photo.data)
def upload_photo(self, arr):
return self.upload_file(arr.tobytes())
def upload_file(self, file):
# TODO: currently this only works for numpy images
try:
sha = sha256(file).hexdigest()
result = requests.post(f"{self.base_url}/upload_file/{self.database_key}/{sha}", data=file)
if result.status_code != 200:
print(result, result.content)
return False
else:
return True
except requests.exceptions.RequestException as e:
print(e)
return False
def get_file(self, sha):
# TODO: currently this only works for numpy images
try:
body= {"databaseKey": self.database_key, "payload": {"sha256": sha}}
result = requests.post(f"{self.base_url}/get_file", json=body)
if result.status_code != 200:
print(result, result.content)
return None
else:
return result.content
except requests.exceptions.RequestException as e:
print(e)
return None
def get_photo(self, uid, size=640):
photo = self.get(uid)
self._load_photo_data(photo, size=size)
return photo
def _load_photo_data(self, photo, size=None):
if len(photo.file) > 0 and photo.data is None:
file = self.get_file(photo.file[0].sha256)
if file is None:
print(f"Could not load data of {photo} attached file item does not have data in pod")
return
data = np.frombuffer(file, dtype=np.uint8)
c = photo.channels
shape = (photo.height,photo.width, c) if c is not None and c > 1 else (photo.height, photo.width)
data = data.reshape(shape)
if size is not None: data = resize(data, size)
photo.data = data
return
print(f"could not load data of {photo}, no file attached")
def create_if_external_id_not_exists(self, node):
if not self.external_id_exists(node):
self.create(node)
def external_id_exists(self, node):
if node.externalId is None: return False
existing = self.search_by_fields({"externalId": node.externalId})
return len(existing) > 0
def create_edges(self, edges):
"""Create edges between nodes, edges should be of format [{"_type": "friend", "_source": 1, "_target": 2}]"""
create_edges = []
for e in edges:
src, target = e.source.uid, e.target.uid
if src is None or target is None:
print(f"Could not create edge {e} missing source or target uid")
return False
data = {"_source": src, "_target": target, "_type": e._type}
if e.label is not None: data[LABEL] = e.label
if e.sequence is not None: data[SEQUENCE] = e.sequence
if e.reverse:
data2 = copy(data)
data2["_source"] = target
data2["_target"] = src
data2["_type"] = "~" + data2["_type"]
create_edges.append(data2)
create_edges.append(data)
return self.bulk_action(create_items=[], update_items=[],create_edges=create_edges)
def delete_items(self, items):
uids = [i.uid for i in items]
return self.bulk_action(delete_items=uids)
def delete_all(self):
items = self.get_all_items()
self.delete_items(items)
def bulk_action(self, create_items=None, update_items=None, create_edges=None, delete_items=None):
create_items = create_items if create_items is not None else []
update_items = update_items if update_items is not None else []
create_edges = create_edges if create_edges is not None else []
delete_items = delete_items if delete_items is not None else []
edges_data = {"databaseKey": self.database_key, "payload": {
"createItems": create_items, "updateItems": update_items,
"createEdges": create_edges, "deleteItems": delete_items}}
try:
result = requests.post(f"{self.base_url}/bulk_action",
json=edges_data)
if result.status_code != 200:
if "UNIQUE constraint failed" in str(result.content):
print(result.status_code, "Edge already exists")
else:
print(result, result.content)
return False
else:
return True
except requests.exceptions.RequestException as e:
print(e)
return False
def create_edge(self, edge):
return self.create_edges([edge])
def get(self, uid, expanded=True):
if not expanded:
res = self._get_item_with_properties(uid)
else:
res = self._get_item_expanded(uid)
if res is None:
return None
elif res.deleted == True:
print(f"Item with uid {uid} does not exist anymore")
return None
else:
return res
def get_all_items(self):
try:
body = { "databaseKey": self.database_key, "payload":None}
result = requests.post(f"{self.base_url}/get_all_items", json=body)
if result.status_code != 200:
print(result, result.content)
return None
else:
json = result.json()
res = [self.item_from_json(x) for x in json]
return self.filter_deleted(res)
except requests.exceptions.RequestException as e:
print(e)
return None
def filter_deleted(self, items):
return [i for i in items if not i.deleted == True]
def _get_item_expanded(self, uid):
body = {"payload": [uid],
"databaseKey": self.database_key}
try:
result = requests.post(f"{self.base_url}/get_items_with_edges",
json=body)
if result.status_code != 200:
print(result, result.content)
return None
else:
json = result.json()[0]
res = self.item_from_json(json)
return res
except requests.exceptions.RequestException as e:
print(e)
return None
def _get_item_with_properties(uid):
try:
result = requests.get(f"{self.base_url}/items/{uid}")
if result.status_code != 200:
print(result, result.content)
return None
else:
json = result.json()
if json == []:
return None
else:
return json
except requests.exceptions.RequestException as e:
print(e)
return None
def get_properties_json(self, node):
res = dict()
private = getattr(node, "private", [])
for k,v in node.__dict__.items():
if k[:1] != '_' and k != "private" and k not in private and not (isinstance(v, list)\
and len(v)>0 and isinstance(v[0], Edge)) and v is not None:
res[k] = v
res["_type"] = self._get_schema_type(node)
return res
@staticmethod
def _get_schema_type(node):
for cls in node.__class__.mro():
if cls.__module__ == "pyintegrators.data.schema" and cls.__name__ != "ItemBase":
return cls.__name__
raise ValueError
def update_item(self, node):
data = self.get_properties_json(node)
uid = data["uid"]
body = {"payload": data,
"databaseKey": self.database_key}
try:
result = requests.post(f"{self.base_url}/update_item",
json=body)
if result.status_code != 200:
print(result, result.content)
except requests.exceptions.RequestException as e:
print(e)
def search_by_fields(self, fields_data):
body = {"payload": fields_data,
"databaseKey": self.database_key}
try:
result = requests.post(f"{self.base_url}/search_by_fields", json=body)
json = result.json()
res = [self.item_from_json(item) for item in json]
return self.filter_deleted(res)
except requests.exceptions.RequestException as e:
return None
def item_from_json(self, json):
indexer_class = json.get("indexerClass", None)
constructor = get_constructor(json["_type"], indexer_class)
new_item = constructor.from_json(json)
existing = ItemBase.global_db.get(new_item.uid)
# TODO: cleanup
if existing is not None:
if not existing.is_expanded() and new_item.is_expanded():
for edge_name in new_item.get_all_edge_names():
edges = new_item.get_edges(edge_name)
for e in edges:
e.source = existing
existing.__setattr__(edge_name, edges)
for prop_name in new_item.get_property_names():
existing.__setattr__(prop_name, new_item.__getattribute__(prop_name))
return existing
else:
return new_item
def get_properties(self, expanded):
properties = copy(expanded)
if ALL_EDGES in properties: del properties[ALL_EDGES]
return properties
def run_importer(self, uid, servicePayload):
body = dict()
body["databaseKey"] = servicePayload["databaseKey"]
body["payload"] = {"uid": uid, "servicePayload": servicePayload}
print(body)
try:
res = requests.post(f"{self.base_url}/run_importer", json=body)
# res = requests.post(self.url)
if res.status_code != 200:
print(f"Failed to start importer on {url}:\n{res.status_code}: {res.text}")
else:
print("Starting importer")
except requests.exceptions.RequestException as e:
print("Error with calling importer {e}")
```
Pyintegrators communicate with the pod via the PodClient. The PodClient requires you to provide a [database key](https://gitlab.memri.io/memri/pod/-/blob/dev/docs/HTTP_API.md#user-content-api-authentication-credentials) and an [owner key](https://gitlab.memri.io/memri/pod/-/blob/dev/docs/HTTP_API.md#user-content-api-authentication-credentials). During development, you don't have to worry about these keys, you can just omit the keys when initializing the PodClient, which creates a new user by defining random keys. When you are using the app, setting the keys in the pod, and passing them when calling an integrator is handled for you by the app itself.
```
client = PodClient()
success = client.test_connection()
assert success
```
## Creating Items and Edges
Now that we have access to the pod, we can create items here and upload them to the pod. All items are defined in the memri [schema](https://gitlab.memri.io/memri/schema). When the schema is changed it automatically generates all the class definitions for the different languages used in memri, the python schema file lives in [schema.py](https://gitlab.memri.io/memri/pyintegrators/-/blob/master/integrators/schema.py) in the integrators package. When Initializing an Item, always make sure to use the from_data classmethod to initialize.
```
email_item = EmailMessage.from_data(content="example content field")
email_item
success = client.create(email_item)
assert success
email_item
```
We can connect items using edges. Let's create another item, a person, and connect the email and the person.
```
person_item = Person.from_data(firstName="Alice")
item_succes = client.create(person_item)
edge = Edge(person_item, email_item, "author")
edge_succes = client.create_edge(edge)
assert item_succes and edge_succes
edge
```
# Fetching and updating Items
We can use the client to fetch data from the database. This is in particular usefull for indexers, which often use data in the database as input for their models. The simplest form of querying the database is by querying items in the pod by their uid (unique identifier).
```
person_item = Person.from_data(firstName="Alice")
client.create(person_item)
person_from_db = client.get(person_item.uid)
assert person_from_db is not None
assert person_from_db == person_item
person_from_db
```
Appart from creating, we might want to update existing items:
```
person_item.lastName = "Awesome"
client.update_item(person_item)
person_from_db = client.get(person_item.uid)
assert person_from_db.lastName == "Awesome"
person_from_db
```
Sometimes, we might not know the uids of the items we want to fetch. We can also search by a certain property. We can use this for instance when we want to query all items from a particular type to perform some indexing on.
```
person_item2 = Person.from_data(firstName="Bob")
client.create(person_item2);
all_people = client.search_by_fields({"_type": "Person"})
assert all([isinstance(p, Person) for p in all_people]) and len(all_people) > 0
all_people[:3]
```
## Uploading & downloading files
The file API is currently only tested for images.
```
from pyintegrators.indexers.facerecognition.photo import *
x = np.random.randint(0, 255+1, size=(640, 640), dtype=np.uint8)
photo = IPhoto.from_np(x)
assert client.create(photo)
res = client.get_photo(photo.uid, size=640)
assert (res.data == x).all()
```
# Check if an item exists
```
person_item = Person.from_data(firstName="Eve", externalId="gmail_1")
person_item2 = Person.from_data(firstName="Eve2", externalId="gmail_1")
client.create_if_external_id_not_exists(person_item)
client.create_if_external_id_not_exists(person_item2)
existing = client.search_by_fields({"externalId": "gmail_1"})
assert len(existing) == 1
client.delete_all()
```
# Resetting the db
```
client.delete_all()
```
# Export -
```
# hide
from nbdev.export import *
notebook2script()
```
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
# default_exp pod.client
# export
from pyintegrators.data.itembase import Edge, ItemBase
from pyintegrators.indexers.facerecognition.photo import resize
from pyintegrators.data.schema import *
from pyintegrators.imports import *
from hashlib import sha256
# export
DEFAULT_POD_ADDRESS = "http://localhost:3030"
POD_VERSION = "v2"
# export
class PodClient:
def __init__(self, url=DEFAULT_POD_ADDRESS, version=POD_VERSION, database_key=None, owner_key=None):
self.url = url
self.version = POD_VERSION
self.test_connection(verbose=False)
self.database_key=database_key if database_key is not None else self.generate_random_key()
self.owner_key=owner_key if owner_key is not None else self.generate_random_key()
self.base_url = f"{url}/{version}/{self.owner_key}"
@staticmethod
def generate_random_key():
return "".join([str(random.randint(0, 9)) for i in range(64)])
def test_connection(self, verbose=True):
try:
res = requests.get(self.url)
if verbose: print("Succesfully connected to pod")
return True
except requests.exceptions.RequestException as e:
print("Could no connect to backend")
return False
def create(self, node):
if isinstance(node, Photo) and not self.create_photo_file(node): return False
try:
body = {"databaseKey": self.database_key, "payload":self.get_properties_json(node) }
result = requests.post(f"{self.base_url}/create_item", json=body)
if result.status_code != 200:
print(result, result.content)
return False
else:
uid = int(result.json())
node.uid = uid
ItemBase.add_to_db(node)
return True
except requests.exceptions.RequestException as e:
print(e)
return False
def create_photo_file(self, photo):
file = photo.file[0]
self.create(file)
return self.upload_photo(photo.data)
def upload_photo(self, arr):
return self.upload_file(arr.tobytes())
def upload_file(self, file):
# TODO: currently this only works for numpy images
try:
sha = sha256(file).hexdigest()
result = requests.post(f"{self.base_url}/upload_file/{self.database_key}/{sha}", data=file)
if result.status_code != 200:
print(result, result.content)
return False
else:
return True
except requests.exceptions.RequestException as e:
print(e)
return False
def get_file(self, sha):
# TODO: currently this only works for numpy images
try:
body= {"databaseKey": self.database_key, "payload": {"sha256": sha}}
result = requests.post(f"{self.base_url}/get_file", json=body)
if result.status_code != 200:
print(result, result.content)
return None
else:
return result.content
except requests.exceptions.RequestException as e:
print(e)
return None
def get_photo(self, uid, size=640):
photo = self.get(uid)
self._load_photo_data(photo, size=size)
return photo
def _load_photo_data(self, photo, size=None):
if len(photo.file) > 0 and photo.data is None:
file = self.get_file(photo.file[0].sha256)
if file is None:
print(f"Could not load data of {photo} attached file item does not have data in pod")
return
data = np.frombuffer(file, dtype=np.uint8)
c = photo.channels
shape = (photo.height,photo.width, c) if c is not None and c > 1 else (photo.height, photo.width)
data = data.reshape(shape)
if size is not None: data = resize(data, size)
photo.data = data
return
print(f"could not load data of {photo}, no file attached")
def create_if_external_id_not_exists(self, node):
if not self.external_id_exists(node):
self.create(node)
def external_id_exists(self, node):
if node.externalId is None: return False
existing = self.search_by_fields({"externalId": node.externalId})
return len(existing) > 0
def create_edges(self, edges):
"""Create edges between nodes, edges should be of format [{"_type": "friend", "_source": 1, "_target": 2}]"""
create_edges = []
for e in edges:
src, target = e.source.uid, e.target.uid
if src is None or target is None:
print(f"Could not create edge {e} missing source or target uid")
return False
data = {"_source": src, "_target": target, "_type": e._type}
if e.label is not None: data[LABEL] = e.label
if e.sequence is not None: data[SEQUENCE] = e.sequence
if e.reverse:
data2 = copy(data)
data2["_source"] = target
data2["_target"] = src
data2["_type"] = "~" + data2["_type"]
create_edges.append(data2)
create_edges.append(data)
return self.bulk_action(create_items=[], update_items=[],create_edges=create_edges)
def delete_items(self, items):
uids = [i.uid for i in items]
return self.bulk_action(delete_items=uids)
def delete_all(self):
items = self.get_all_items()
self.delete_items(items)
def bulk_action(self, create_items=None, update_items=None, create_edges=None, delete_items=None):
create_items = create_items if create_items is not None else []
update_items = update_items if update_items is not None else []
create_edges = create_edges if create_edges is not None else []
delete_items = delete_items if delete_items is not None else []
edges_data = {"databaseKey": self.database_key, "payload": {
"createItems": create_items, "updateItems": update_items,
"createEdges": create_edges, "deleteItems": delete_items}}
try:
result = requests.post(f"{self.base_url}/bulk_action",
json=edges_data)
if result.status_code != 200:
if "UNIQUE constraint failed" in str(result.content):
print(result.status_code, "Edge already exists")
else:
print(result, result.content)
return False
else:
return True
except requests.exceptions.RequestException as e:
print(e)
return False
def create_edge(self, edge):
return self.create_edges([edge])
def get(self, uid, expanded=True):
if not expanded:
res = self._get_item_with_properties(uid)
else:
res = self._get_item_expanded(uid)
if res is None:
return None
elif res.deleted == True:
print(f"Item with uid {uid} does not exist anymore")
return None
else:
return res
def get_all_items(self):
try:
body = { "databaseKey": self.database_key, "payload":None}
result = requests.post(f"{self.base_url}/get_all_items", json=body)
if result.status_code != 200:
print(result, result.content)
return None
else:
json = result.json()
res = [self.item_from_json(x) for x in json]
return self.filter_deleted(res)
except requests.exceptions.RequestException as e:
print(e)
return None
def filter_deleted(self, items):
return [i for i in items if not i.deleted == True]
def _get_item_expanded(self, uid):
body = {"payload": [uid],
"databaseKey": self.database_key}
try:
result = requests.post(f"{self.base_url}/get_items_with_edges",
json=body)
if result.status_code != 200:
print(result, result.content)
return None
else:
json = result.json()[0]
res = self.item_from_json(json)
return res
except requests.exceptions.RequestException as e:
print(e)
return None
def _get_item_with_properties(uid):
try:
result = requests.get(f"{self.base_url}/items/{uid}")
if result.status_code != 200:
print(result, result.content)
return None
else:
json = result.json()
if json == []:
return None
else:
return json
except requests.exceptions.RequestException as e:
print(e)
return None
def get_properties_json(self, node):
res = dict()
private = getattr(node, "private", [])
for k,v in node.__dict__.items():
if k[:1] != '_' and k != "private" and k not in private and not (isinstance(v, list)\
and len(v)>0 and isinstance(v[0], Edge)) and v is not None:
res[k] = v
res["_type"] = self._get_schema_type(node)
return res
@staticmethod
def _get_schema_type(node):
for cls in node.__class__.mro():
if cls.__module__ == "pyintegrators.data.schema" and cls.__name__ != "ItemBase":
return cls.__name__
raise ValueError
def update_item(self, node):
data = self.get_properties_json(node)
uid = data["uid"]
body = {"payload": data,
"databaseKey": self.database_key}
try:
result = requests.post(f"{self.base_url}/update_item",
json=body)
if result.status_code != 200:
print(result, result.content)
except requests.exceptions.RequestException as e:
print(e)
def search_by_fields(self, fields_data):
body = {"payload": fields_data,
"databaseKey": self.database_key}
try:
result = requests.post(f"{self.base_url}/search_by_fields", json=body)
json = result.json()
res = [self.item_from_json(item) for item in json]
return self.filter_deleted(res)
except requests.exceptions.RequestException as e:
return None
def item_from_json(self, json):
indexer_class = json.get("indexerClass", None)
constructor = get_constructor(json["_type"], indexer_class)
new_item = constructor.from_json(json)
existing = ItemBase.global_db.get(new_item.uid)
# TODO: cleanup
if existing is not None:
if not existing.is_expanded() and new_item.is_expanded():
for edge_name in new_item.get_all_edge_names():
edges = new_item.get_edges(edge_name)
for e in edges:
e.source = existing
existing.__setattr__(edge_name, edges)
for prop_name in new_item.get_property_names():
existing.__setattr__(prop_name, new_item.__getattribute__(prop_name))
return existing
else:
return new_item
def get_properties(self, expanded):
properties = copy(expanded)
if ALL_EDGES in properties: del properties[ALL_EDGES]
return properties
def run_importer(self, uid, servicePayload):
body = dict()
body["databaseKey"] = servicePayload["databaseKey"]
body["payload"] = {"uid": uid, "servicePayload": servicePayload}
print(body)
try:
res = requests.post(f"{self.base_url}/run_importer", json=body)
# res = requests.post(self.url)
if res.status_code != 200:
print(f"Failed to start importer on {url}:\n{res.status_code}: {res.text}")
else:
print("Starting importer")
except requests.exceptions.RequestException as e:
print("Error with calling importer {e}")
client = PodClient()
success = client.test_connection()
assert success
email_item = EmailMessage.from_data(content="example content field")
email_item
success = client.create(email_item)
assert success
email_item
person_item = Person.from_data(firstName="Alice")
item_succes = client.create(person_item)
edge = Edge(person_item, email_item, "author")
edge_succes = client.create_edge(edge)
assert item_succes and edge_succes
edge
person_item = Person.from_data(firstName="Alice")
client.create(person_item)
person_from_db = client.get(person_item.uid)
assert person_from_db is not None
assert person_from_db == person_item
person_from_db
person_item.lastName = "Awesome"
client.update_item(person_item)
person_from_db = client.get(person_item.uid)
assert person_from_db.lastName == "Awesome"
person_from_db
person_item2 = Person.from_data(firstName="Bob")
client.create(person_item2);
all_people = client.search_by_fields({"_type": "Person"})
assert all([isinstance(p, Person) for p in all_people]) and len(all_people) > 0
all_people[:3]
from pyintegrators.indexers.facerecognition.photo import *
x = np.random.randint(0, 255+1, size=(640, 640), dtype=np.uint8)
photo = IPhoto.from_np(x)
assert client.create(photo)
res = client.get_photo(photo.uid, size=640)
assert (res.data == x).all()
person_item = Person.from_data(firstName="Eve", externalId="gmail_1")
person_item2 = Person.from_data(firstName="Eve2", externalId="gmail_1")
client.create_if_external_id_not_exists(person_item)
client.create_if_external_id_not_exists(person_item2)
existing = client.search_by_fields({"externalId": "gmail_1"})
assert len(existing) == 1
client.delete_all()
client.delete_all()
# hide
from nbdev.export import *
notebook2script()
| 0.240329 | 0.400544 |
We left off with the disturbing realization that even though we are satisfied the requirements of the sampling theorem, we still have errors in our approximating formula. We can resolve this by examining the Whittaker interpolating functions which are used to reconstruct the signal from its samples.
```
%pylab inline
from __future__ import division
t = linspace(-5,5,300) # redefine this here for convenience
fig,ax = subplots()
fs=5.0
ax.plot(t,sinc(fs * t))
ax.grid()
ax.annotate('This keeps going...',
xy=(-4,0),
xytext=(-5+.1,0.5),
arrowprops={'facecolor':'green','shrink':0.05},fontsize=14)
ax.annotate('... and going...',
xy=(4,0),
xytext=(3+.1,0.5),
arrowprops={'facecolor':'green','shrink':0.05},fontsize=14)
# fig.savefig('[email protected]', bbox_inches='tight', dpi=300)
```
Notice in the above plot that the function extends to infinity in either direction. This basically means that the signals we can represent must also extend to infinity in either direction which then means that we have to sample forever to exactly reconstruct the signal! So, on the one hand the sampling theorem says we only need a sparse density of samples, this result says we need to sample forever. No free lunch here!
This is a deep consequence of dealing with band-limited functions which, as we have just demonstrated, are **not** time-limited. Now, the new question is how to get these signals into a computer with finite memory. How can we use what we have learned about the sampling theorem with these finite-duration signals?
## Approximately Time-Limited Functions
Let's back off a bit and settle for functions that are *approximately* time-limited in the sense that almost all of their energy is concentrated in a finite time-window:
$$ \int_{-\tau}^\tau |f(t)|^2 dt = E-\epsilon$$
where $E$ is the total energy of the signal:
$$ \int_{-\infty}^\infty |f(t)|^2 dt = E$$
Now, with this new definition, we can seek out functions that are band-limited but come very, very (i.e. within $\epsilon$) close to being time-limited as well. In other words, we want functions $\phi(t)$ so that they are band-limited:
$$ \phi(t) = \int_{-W}^W \Phi(\nu) e^{2 \pi j \nu t} dt $$
and coincidentally maximize the following:
$$ \int_{-\tau}^\tau |\phi(t) |^2 dt$$
After a complicated derivation, this boils down to solving the following eigenvalue equation:
$$ \int_{-\tau}^\tau \phi(x)\frac{\sin(2\pi W(t-x))}{\pi(t-x)} dx = \lambda \phi(t)$$
The set of $\phi_k(t)$ eigenfunctions form the basis for arbitrary
approximately time-limited functions. In other words, we can express
$$ f(t) = \sum_k a_k \phi_k(t) $$
Note that
the $\phi_k(t)$ functions are not time-limited, but only time-concentrated in the $[-\tau,\tau]$ interval. With a change of variables, we can write this in normalized form as
$$ \int_{-1}^1 \psi(x)\frac{\sin(2\pi\sigma(t-x)/4)}{\pi(t-x)} dx = \lambda \psi(t)$$
where we define$\sigma = (2\tau)(2W)$ as the time-bandwidth product.The advantage of this change of variables is that $\tau$ and $W$ are expressed as a single term. Furthermore, this is the form of a classic problem where the $\psi$ functions turn out to be the angular prolate spheroidal wave functions. Let's see what these $\psi$ functions look like but solving this form of
the eigenvalue problem
```
def kernel(x,sigma=1):
'convenient function to compute kernel of eigenvalue problem'
x = np.asanyarray(x)
y = pi*where(x == 0,1.0e-20, x)
return sin(sigma/2*y)/y
```
Now, we are ready to setup the eigenvalues and see how they change with the time-bandwidth product.
```
nstep=100 # quick and dirty integral quantization
t = linspace(-1,1,nstep) # quantization of time
dt = diff(t)[0] # differential step size
def eigv(sigma):
return eigvalsh(kernel(t-t[:,None],sigma)).max() # compute max eigenvalue
sigma = linspace(0.01,4,15) # range of time-bandwidth products to consider
fig,ax = subplots()
ax.plot(sigma, dt*array([eigv(i) for i in sigma]),'-o')
ax.set_xlabel('time-bandwidth product $\sigma$',fontsize=14)
ax.set_ylabel('max eigenvalue',fontsize=14)
ax.axis(ymax=1.01)
ax.grid()
# fig.savefig('[email protected]', bbox_inches='tight', dpi=300)
```
The largest eigenvalue is the fraction of the energy of the contained in the interval $[-1,1]$. Thus, this means
that for $\sigma \gt 3$, $\psi_0(t)$ is the eigenfunction that is most concentrated in that interval. Now, let's look at the this eigenfunction under those conditions.
```
sigma=3
w,v=eigh(kernel(t-t[:,None],sigma))
maxv=v[:, w.argmax()]
fig,ax=subplots()
ax.plot(t,maxv)
ax.set_xlabel('time',fontsize=18)
ax.set_ylabel('$\psi_0(t)$',fontsize=22)
ax.set_title('Eigenvector corresponding to e-value=%3.4f;$\sigma$=%3.2f'%(w.max()*dt,sigma))
# fig.savefig('[email protected]', bbox_inches='tight', dpi=300)
```
Note that we'll see this shape again when we take up window functions.
What does this all mean? By framing our problem this way, we made a connection between the quality of our reconstruction via the Whittaker interpolant and the time-bandwidth product. Up until now, we did not have a concrete way of relating limitations in time to limitations in frequency. Now that we know how to use the time-bandwidth product, let's go back to the original formulation with the separate $\tau$ and $W$ terms as in the following:
$$ \int_{-\tau}^\tau \phi(x)\frac{\sin(2\pi W (t-x))}{\pi(t-x)} dx = \lambda \phi(t)$$
and then re-solve the eigenvalue problem.
```
def kernel_tau(x,W=1):
'convenient function to compute kernel of eigenvalue problem'
x = np.asanyarray(x)
y = pi*where(x == 0,1.0e-20, x)
return sin(2*W*y)/y
nstep=300 # quick and dirty integral quantization
t = linspace(-1,1,nstep) # quantization of time
tt = linspace(-2,2,nstep)# extend interval
sigma = 5
W = sigma/2./2./t.max()
w,v=eig(kernel_tau(t-tt[:,None],W))
ii = argsort(w.real)
maxv=v[:, w.real.argmax()].real
fig,ax = subplots()
ax.plot(tt,maxv/sign(maxv[nstep/2])) # normalize to keep orientation upwards
ax.set_xlabel('time',fontsize=14)
ax.set_ylabel(r'$\phi_{max}(t)$',fontsize=18)
ax.set_title('$\sigma=%d$'%(2*W*2*t.max()),fontsize=16)
# fig.savefig('[email protected]', bbox_inches='tight', dpi=300)
```
$\DeclareMathOperator{\sinc}{sinc}$
This looks suspicously like the $\sinc$ function. In fact, in the limit as $\sigma \rightarrow \infty$, the eigenfunctions devolve into time-shifted versions of the $\sinc$ function. These are the same functions used in the Whittaker interpolant. Now we have a way to justify the interpolant by appealing to large $\sigma$ values.
## Summary
In this section, at first blush, it may look like we accomplished nothing. We started out investigating why is it that we have some residual error in the reconstruction formula using the Whittaker approximation functions. Then, we recognized that we cannot have signals that are simultaneously time-limited and band-limited. This realization drove us to investigate "approximately" time-limited functions. Through carefully examining the resulting eigenvalue problem, we determined the time-bandwidth conditions under which the
Whittaker interopolant is asymptotically valid. As you can imagine, there is much more to this story and many powerful theorems place bounds on the quality and dimensionality of this reconstruction, but for us, the qualifying concept of time-bandwidth product is enough for now.
## References
* This is in the [IPython Notebook format](http://ipython.org/) and was converted to HTML using [nbconvert](https://github.com/ipython/nbconvert).
* See [Signal analysis](http://books.google.com/books?id=Re5SAAAAMAAJ) for more detailed mathematical development.
* The IPython notebook corresponding to this post can be found [here](https://github.com/unpingco/Python-for-Signal-Processing/blob/master/Sampling_Theorem_Part_2.ipynb).
```
%qtconsole
```
|
github_jupyter
|
%pylab inline
from __future__ import division
t = linspace(-5,5,300) # redefine this here for convenience
fig,ax = subplots()
fs=5.0
ax.plot(t,sinc(fs * t))
ax.grid()
ax.annotate('This keeps going...',
xy=(-4,0),
xytext=(-5+.1,0.5),
arrowprops={'facecolor':'green','shrink':0.05},fontsize=14)
ax.annotate('... and going...',
xy=(4,0),
xytext=(3+.1,0.5),
arrowprops={'facecolor':'green','shrink':0.05},fontsize=14)
# fig.savefig('[email protected]', bbox_inches='tight', dpi=300)
def kernel(x,sigma=1):
'convenient function to compute kernel of eigenvalue problem'
x = np.asanyarray(x)
y = pi*where(x == 0,1.0e-20, x)
return sin(sigma/2*y)/y
nstep=100 # quick and dirty integral quantization
t = linspace(-1,1,nstep) # quantization of time
dt = diff(t)[0] # differential step size
def eigv(sigma):
return eigvalsh(kernel(t-t[:,None],sigma)).max() # compute max eigenvalue
sigma = linspace(0.01,4,15) # range of time-bandwidth products to consider
fig,ax = subplots()
ax.plot(sigma, dt*array([eigv(i) for i in sigma]),'-o')
ax.set_xlabel('time-bandwidth product $\sigma$',fontsize=14)
ax.set_ylabel('max eigenvalue',fontsize=14)
ax.axis(ymax=1.01)
ax.grid()
# fig.savefig('[email protected]', bbox_inches='tight', dpi=300)
sigma=3
w,v=eigh(kernel(t-t[:,None],sigma))
maxv=v[:, w.argmax()]
fig,ax=subplots()
ax.plot(t,maxv)
ax.set_xlabel('time',fontsize=18)
ax.set_ylabel('$\psi_0(t)$',fontsize=22)
ax.set_title('Eigenvector corresponding to e-value=%3.4f;$\sigma$=%3.2f'%(w.max()*dt,sigma))
# fig.savefig('[email protected]', bbox_inches='tight', dpi=300)
def kernel_tau(x,W=1):
'convenient function to compute kernel of eigenvalue problem'
x = np.asanyarray(x)
y = pi*where(x == 0,1.0e-20, x)
return sin(2*W*y)/y
nstep=300 # quick and dirty integral quantization
t = linspace(-1,1,nstep) # quantization of time
tt = linspace(-2,2,nstep)# extend interval
sigma = 5
W = sigma/2./2./t.max()
w,v=eig(kernel_tau(t-tt[:,None],W))
ii = argsort(w.real)
maxv=v[:, w.real.argmax()].real
fig,ax = subplots()
ax.plot(tt,maxv/sign(maxv[nstep/2])) # normalize to keep orientation upwards
ax.set_xlabel('time',fontsize=14)
ax.set_ylabel(r'$\phi_{max}(t)$',fontsize=18)
ax.set_title('$\sigma=%d$'%(2*W*2*t.max()),fontsize=16)
# fig.savefig('[email protected]', bbox_inches='tight', dpi=300)
%qtconsole
| 0.622574 | 0.986218 |
<h1 align='center'>WebScraping TripAdvisor</h1>
---
El código a continuación tiene por objetivo extraer la **[información solicitada](https://github.com/mozilla/geckodriver/releases/download/v0.28.0/geckodriver-v0.28.0-win64.zip "Word en Google Drive")**, desde la página de **[TripAdvisor](https://www.tripadvisor.cl/Restaurants-g294305-Santiago_Santiago_Metropolitan_Region.html "Web TripAdvisor")** para Ximena. La siguiente celda sólo cumple con el propósito de **silenciar las posibles advertencias** que pudieran levantarse al correr el código, pero no aportan mayormente a la comprensión del proceso por parte del usuario.
```
%%capture --no-display
import warnings
warnings.filterwarnings('ignore')
```
La celda anterior asegurará que no se desplieguen advertencias innecesarias para la correcta comprensión y lectura de este informe. A continuación se darán las **instrucciones para instalar las librerías** necesarias para correr el código, cuestión que requiere de un comando para ello, por lo que las instrucciones se despliegan como impresión de una celda.
```
import os
print(f'Si es la primera vez que corre este programa, por favor abra la terminal PowerShell de Anaconda' +
f' e ingrese el siguiente comando: "\033[4mpip install -r {os.getcwd()}\\requirements.txt\033[4m"')
```
La primera parte fundamental de todo programa, corresponde a la **importación de librerías de Python**. Si acaso hubiera errores en esta primera celda, se aconseja contactar a Nicolás Ganter a su correo: [email protected]
```
import time
import pickle
import pandas as pd
from tqdm import tqdm
from datetime import datetime
from dask.distributed import Client, progress
import utils
```
Dentro del código, hay ciertas **variables que es preferible tener en especial consideración**. Entre ellas, encontramos la ubicación del *driver* para *Selenium* que permitirá lanzar una instancia de *Firefox* para navegar la página y extraer los enlaces requeridos en la primera etapa de *webcrawling*. Si aún no ha instalado el driver, acceda a este **[link de descarga](https://github.com/mozilla/geckodriver/releases/download/v0.28.0/geckodriver-v0.28.0-win64.zip "geckodriver download link")**, extraiga el paquete y mueva los documentos a la carpeta de binarios de las librerías de Python.
```
geckodriver_path = r'C:\Users\nicol\anaconda3\Library\bin\geckodriver'
time_id = datetime.today().strftime('%Y%m%d')
basic_url = 'https://www.tripadvisor.cl'
client = Client()
client
```
<h2>Webcrawling de los restaurantes</h2>
La siguiente celda se encargará de la **extracción de los enlaces** asociados a cada restaurante en las páginas especificadas mediante el enlace de la segunda línea. En este caso, se extraerán los restaurantes de Santiago de Chile. Al final de la celda se imprime la cantidad de restaurantes extraídos, la cantidad de restaurantes disponibles según la página, y el porcentaje capturado por el programa. Nótese que el proceso toma algo así como 10 minutos, por lo que se utilizará un atajo mediante *pickles* (estructura de datos propia de este lenguaje de programación) y se especificará la fecha de captura de la información asociada a éste.
```
start = time.time()
url = basic_url + '/Restaurants-g294305-Santiago_Santiago_Metropolitan_Region.html'
info = utils.info_restaurants(url, geckodriver_path)
cwd = os.getcwd()
dict_pickles = utils.check_files(dir_files=cwd, keyword='urls')
if len(dict_pickles) == 0:
urls = utils.gen_pickle(url, geckodriver_path, info['pages'], basic_url, time_id)
else:
last_pickle = utils.last_pickle(dict_pickles)
with open(last_pickle, 'rb') as file:
urls = pickle.load(file)
print('Se obtuvieron {} restaurantes de {} lo que corresponde a una extracción del {}%'
.format(len(urls), info['max_restaurants'], round(len(urls) / info['max_restaurants'] * 100, 2)))
stop = time.time()
print(f'Este proceso tomó {round(stop-start, 2)} segundos en correr.\n')
```
<h2>Webscraping de los restaurantes</h2>
Con esto concluye la parte más compleja y crítica de la recopilación de enlaces para los restaurantes. No obstante esta tarea continúa luego a nivel de comentarios, **a continuación se procederá a extraer la información solicitada** para cada uno de los restaurantes en la lista. Dado que se utilizan estrategias de computación paralela, no es posible observar el avance, sino abriendo el *Dashboard* cuyo link se encuentra bajo la cuarta celda del código.
```
start = time.time()
dict_dataframes = utils.check_files(dir_files=cwd, keyword='dataframe')
if len(dict_dataframes) == 0:
futures = [client.submit(utils.get_restaurant, url_restaurant) for url_restaurant in list(set(urls))]
results = client.gather(futures)
dict_structure = {'id':[], 'Nombre restaurante':[], 'Promedio de calificaciones':[],
'N° de opiniones':[], 'Calificación de viajeros por categoría':[],
'Toman medidas de seguridad':[], 'Rankings':[],
'Tipo de comida y servicios':[], 'url':[]}
df_restaurants = utils.build_dataframe(dict_structure, results, time_id)
df_restaurants.to_pickle(f'{time_id}_dataframe_of_{df_restaurants.shape[0]}_restaurants.pickle')
print(f'Se guardó "{time_id}_dataframe_of_{df_restaurants.shape[0]}_restaurants.pickle" en "{os.getcwd()}".')
else:
last_pickle = utils.last_pickle(dict_dataframes)
with open(last_pickle, 'rb') as file:
df_restaurants = pickle.load(file)
stop = time.time()
print(f'Este proceso tomó {round(stop-start, 2)} segundos en correr.\n')
```
<h2>Webcrawling de los comentarios</h2>
```
start = time.time()
dict_files = utils.check_files(dir_files=cwd, keyword='review_urls')
if len(dict_files) == 0:
futures = [client.submit(utils.review_urls, url_restaurant) for url_restaurant in list(set(urls))]
results = client.gather(futures)
dict_reviews = {key:value for key, value in results if isinstance(value, list)}
n_reviews = len(dict_reviews.values())
with open(f'{time_id}_{n_reviews}_review_urls.pickle', 'wb') as file:
pickle.dump(dict_reviews, file)
print(f'Se guardó "{time_id}_{n_reviews}_review_urls.pickle" en "{os.getcwd()}".')
else:
last_pickle = utils.last_pickle(dict_files)
with open(last_pickle, 'rb') as file:
dict_reviews = pickle.load(file)
stop = time.time()
print(f'Este proceso tomó {round(stop-start, 2)} segundos en correr.',
'Se dispone aproximadamente de {} comentarios para extraer.\n'.format(len(dict_reviews.values())*10))
```
<h2>Webscraping de los comentarios</h2>
```
start = time.time()
dict_files = utils.check_files(dir_files=cwd, keyword='scraped_reviews')
url_reviews = utils.prepare_urls(dict_reviews)
if len(dict_files) == 0:
futures = [client.submit(utils.get_reviews, url) for url in url_reviews]
results = client.gather(futures)
dict_structure = {'id':[], 'date_review':[], 'comments':[], 'date_stayed':[], 'response_body':[],
'user_name':[], 'user_reviews':[], 'useful_votes':[]}
df_reviews = utils.build_dataframe(dict_structure, results, time_id)
df_pathname = f'{time_id}_dataframe_of_{df_reviews.shape[0]}_scraped_reviews.pickle'
df_reviews.to_pickle(df_pathname)
print(f'Se guardó "{df_pathname}" en "{os.getcwd()}"')
else:
last_pickle = utils.last_pickle(dict_files)
df_reviews = pd.read_pickle(last_pickle)
stop = time.time()
print(f'Este proceso tomó {round(stop-start, 2)} segundos en correr.',
'Se extrajeron {} comentarios.\n'.format(df_reviews.shape[0]))
```
<h2>Generación de tablas</h2>
```
start = time.time()
df_restaurants.to_excel(f'{time_id}_excel_with_{df_restaurants.shape[0]}_restaurants.xlsx')
df_reviews.to_excel(f'{time_id}_excel_with_{df_reviews.shape[0]}_reviews.xlsx')
stop = time.time()
print(f'Este proceso tomó {round(stop-start, 2)} segundos en correr.',
'Se extrajeron {} comentarios.\n'.format(df_reviews.shape[0]))
```
|
github_jupyter
|
%%capture --no-display
import warnings
warnings.filterwarnings('ignore')
import os
print(f'Si es la primera vez que corre este programa, por favor abra la terminal PowerShell de Anaconda' +
f' e ingrese el siguiente comando: "\033[4mpip install -r {os.getcwd()}\\requirements.txt\033[4m"')
import time
import pickle
import pandas as pd
from tqdm import tqdm
from datetime import datetime
from dask.distributed import Client, progress
import utils
geckodriver_path = r'C:\Users\nicol\anaconda3\Library\bin\geckodriver'
time_id = datetime.today().strftime('%Y%m%d')
basic_url = 'https://www.tripadvisor.cl'
client = Client()
client
start = time.time()
url = basic_url + '/Restaurants-g294305-Santiago_Santiago_Metropolitan_Region.html'
info = utils.info_restaurants(url, geckodriver_path)
cwd = os.getcwd()
dict_pickles = utils.check_files(dir_files=cwd, keyword='urls')
if len(dict_pickles) == 0:
urls = utils.gen_pickle(url, geckodriver_path, info['pages'], basic_url, time_id)
else:
last_pickle = utils.last_pickle(dict_pickles)
with open(last_pickle, 'rb') as file:
urls = pickle.load(file)
print('Se obtuvieron {} restaurantes de {} lo que corresponde a una extracción del {}%'
.format(len(urls), info['max_restaurants'], round(len(urls) / info['max_restaurants'] * 100, 2)))
stop = time.time()
print(f'Este proceso tomó {round(stop-start, 2)} segundos en correr.\n')
start = time.time()
dict_dataframes = utils.check_files(dir_files=cwd, keyword='dataframe')
if len(dict_dataframes) == 0:
futures = [client.submit(utils.get_restaurant, url_restaurant) for url_restaurant in list(set(urls))]
results = client.gather(futures)
dict_structure = {'id':[], 'Nombre restaurante':[], 'Promedio de calificaciones':[],
'N° de opiniones':[], 'Calificación de viajeros por categoría':[],
'Toman medidas de seguridad':[], 'Rankings':[],
'Tipo de comida y servicios':[], 'url':[]}
df_restaurants = utils.build_dataframe(dict_structure, results, time_id)
df_restaurants.to_pickle(f'{time_id}_dataframe_of_{df_restaurants.shape[0]}_restaurants.pickle')
print(f'Se guardó "{time_id}_dataframe_of_{df_restaurants.shape[0]}_restaurants.pickle" en "{os.getcwd()}".')
else:
last_pickle = utils.last_pickle(dict_dataframes)
with open(last_pickle, 'rb') as file:
df_restaurants = pickle.load(file)
stop = time.time()
print(f'Este proceso tomó {round(stop-start, 2)} segundos en correr.\n')
start = time.time()
dict_files = utils.check_files(dir_files=cwd, keyword='review_urls')
if len(dict_files) == 0:
futures = [client.submit(utils.review_urls, url_restaurant) for url_restaurant in list(set(urls))]
results = client.gather(futures)
dict_reviews = {key:value for key, value in results if isinstance(value, list)}
n_reviews = len(dict_reviews.values())
with open(f'{time_id}_{n_reviews}_review_urls.pickle', 'wb') as file:
pickle.dump(dict_reviews, file)
print(f'Se guardó "{time_id}_{n_reviews}_review_urls.pickle" en "{os.getcwd()}".')
else:
last_pickle = utils.last_pickle(dict_files)
with open(last_pickle, 'rb') as file:
dict_reviews = pickle.load(file)
stop = time.time()
print(f'Este proceso tomó {round(stop-start, 2)} segundos en correr.',
'Se dispone aproximadamente de {} comentarios para extraer.\n'.format(len(dict_reviews.values())*10))
start = time.time()
dict_files = utils.check_files(dir_files=cwd, keyword='scraped_reviews')
url_reviews = utils.prepare_urls(dict_reviews)
if len(dict_files) == 0:
futures = [client.submit(utils.get_reviews, url) for url in url_reviews]
results = client.gather(futures)
dict_structure = {'id':[], 'date_review':[], 'comments':[], 'date_stayed':[], 'response_body':[],
'user_name':[], 'user_reviews':[], 'useful_votes':[]}
df_reviews = utils.build_dataframe(dict_structure, results, time_id)
df_pathname = f'{time_id}_dataframe_of_{df_reviews.shape[0]}_scraped_reviews.pickle'
df_reviews.to_pickle(df_pathname)
print(f'Se guardó "{df_pathname}" en "{os.getcwd()}"')
else:
last_pickle = utils.last_pickle(dict_files)
df_reviews = pd.read_pickle(last_pickle)
stop = time.time()
print(f'Este proceso tomó {round(stop-start, 2)} segundos en correr.',
'Se extrajeron {} comentarios.\n'.format(df_reviews.shape[0]))
start = time.time()
df_restaurants.to_excel(f'{time_id}_excel_with_{df_restaurants.shape[0]}_restaurants.xlsx')
df_reviews.to_excel(f'{time_id}_excel_with_{df_reviews.shape[0]}_reviews.xlsx')
stop = time.time()
print(f'Este proceso tomó {round(stop-start, 2)} segundos en correr.',
'Se extrajeron {} comentarios.\n'.format(df_reviews.shape[0]))
| 0.220091 | 0.844601 |
# Rates calculation
This notebooks demonstrates basic rates calculation method. We say "basic"
because we have also implemented a PyTorch module for rates calculation that can work on GPU.
```
import sys
import numpy as np
import pandas as pd
from datetime import timedelta
sys.path.append('..')
from deepfield import Field
```
The provided `norne_simplified` model (in the `/open_data` directory) contains an archive `RESULTS.zip` with model states precomputed. Unzip it before loading the model and load the model as usual:
```
model = Field('../open_data/norne_simplified/norne_simplified.data').load()
```
Required well's preprocessing:
```
(model.wells
.drop_incomplete()
.get_wellblocks(model.grid)
.drop_outside()
.apply_perforations()
.calculate_cf(rock=model.rock, grid=model.grid)
.compute_events(grid=model.grid))
```
Get rates (by default CPU multiprocessing is used):
```
model.calculate_rates()
```
Now we can check daily rates data:
```
model.wells.total_rates.head()
```
and cumulative rates:
```
model.wells.cum_rates.head()
```
Note: to get detailed rates for each block see the `BLOCKS_DYNAMICS` attribute.
Rates can be quickly visualized using `show_rates` method:
```python
model.wells.show_rates()
```
It activates an interactive widget where one can specify wellname, phase, time interval and cumulative mode as shown below:
<p><img src="data:image/png;base64,iVBORw0KGgoAAAANSUhEUgAABK4AAAKzCAYAAAA3AkUAAAAAAXNSR0IArs4c6QAAAARnQU1BAACxjwv8YQUAAAAJcEhZcwAAEnQAABJ0Ad5mH3gAAOMqSURBVHhe7N0JeBVVurd9FG3FdmqPim1rOyvOKCrOEyIqKo4MgjjhKygKelScRwRFnHB8nZj0QxRFReaZBgQabPAgDRxAoAGBAzRyhBe7lfXxX6kKOzuVkEBSVtVz/65rCdTeiUlIdlI3q55dxQEAAAAAAAAJRLgCAAAAAABAIhGuAAAAAAAAkEiEKwAAAAAAACQS4QoAAAAAAACJRLhKgffff5/FYrFKXAAAAACQVYSrFODEFEBJeHwAAAAAkGWEqxTgxBRASXh8AAAAAJBlhKsU4MQUQEl4fAAAAACQZYSrFODEFEBJeHwAAAAAkGWEqxTgxBRASXh8AAAAAJBlhKsU4MQUQEl4fAAAAACQZYSrFODEFEBJeHwAAAAAkGWEqxTgxBRASXh8AAAAAJBlhKsU4MQUQEl4fAAAAACQZYSrFODEFEBJeHwAAAAAkGWEqxTgxBRASXh8AAAAAJBlKQ9XX7uOVaq4jl+Hf+zoqlTpuPFotpT3xPTvXWu72rXbuZH/DA7k+udI1652bdf178Gf8/29q6vdbqQr/qJ/d103vlztwhX1+v/pRrbLvU/XjS8FoDIRrgAAAABkGeEqBbYsXG1cUQGqtHAV3Fb85YJolfNC/xzZrli88v/fnJcteDuIV0C+77//3rVt2zb4U+natGnj/va3vwV/Ko5wBQAAACDLCFcpsMXhauNql78tqoRwVRCigpfLD1fahVUsQAW7q8JXFPl6C4JXZCQDjPrnP//patasufGxqoq78cYbg6PRbrjhBn+/3XffvcR4RbgCAAAAkGUxhasfXK8GVVyVwsIkEcd+6OUaVGngev0Q/HmjrztuvM/GE7eCVfS20sNVwetv0Ovrgv9P+Doa9Np4S1FF/x/F7+Nv3/g/KXq/gv9PkWNF3r8Cpb/9ZbNF4ardSDcyasdTRGAKo5WO5e+aKo2/b2lVqrTdXYBhYZDSKile5d7n+OOP98ErCuEKAAAAQJbFtuPqh14NigYhH6k2npTlHsvbMVUQfXJ2UPnbc0JVGcJVkfsH/88GOfXI/z9y34bgdeZGqDA+Fb5c+LZHHCv2ukt9+8tmS8PVPyMu8dtcTCp7uCp43cV2dOUoTwQDrCktXpU1WgnhCgAAAECWxXepoI82m3YcFYSsBq5BfqgKq04QgvIjT9HQVIZwlfcKSn35QNH7BH/ODVDh6y4SvPL+f2V6+8tmy8NV0d1UXgWFq6gZV6Hcyw7ZbQWULCpelSdaCeEKAAAAQJbFOOOqaCRSwOn4de6xorf7sFUkFgWKxKnNh6vcHVBScjgqeF3hCWOxcJX3Mv5YkSpVNFyV7e0vm60JV27jfwue6S+4ZLAiwpWfeVX6bqsCm9+VBViXG6oOPPDAwt+XJVoJ4QoAAABAlsU6nH1T7FEkKog3Oubjkt+htCnoFISfghO44iu839aHK//n8PUGxyPvs0XhauOfI1ec4WqjIFb5Swa3NlyVOVoVKNh9xTMLAqXJjVdaZY1WQrgCAAAAkGWxhquCywN7uR8UqYIQFB77Oryt4K5B+Nlc4NnKcFXGy/m2PFyVL1CVZKvD1UaFl/b9fSvCVTmjlRCugLIJ41V5opUQrgAAAABkWazhqiAUNXAdOzbYFJR8bNr45/zIFBzP606Foavg8FaGqyL3DwWXDG5luCp43Zt7+8umIsKV2/ingksGC1a5w9XmopW/vfjMq83u4AJQ6PHHHy9XtBLCFQAAAIAsizdchXGnyC6nIBRFRB4fh3LDUrEdUhWz4yr3PgX/z6L/3y0KVxtt/u0vm4oJVxuFlwyWN1wFL1f6TqsgjOW+bBC7Svp/Adh6hCsAAAAAWRZzuIqIOWHwyQtDoU0hqWAVjT5bGa7Ev0zO/2PjKyu4zG9TSCv2Mhv5Y0XemOLhSkp/+8umwsLVRv62coar8GUiV5H7Ft3VVdKzDgKoOIQrAAAAAFkWe7hC+XFiCqAkPD4AAAAAyDLCVQpwYgqgJDw+AAAAAMgywlUKcGIKoCQ8PgAAAADIMsJVCnBiCqAkPD4AAAAAyDLCVQpwYgqgJDw+AAAAAMgywlUKcGIKoCQ8PgAAAADIMsJVCnBiCqAkPD4AAAAAyDLCVQpwYgqgJDw+AAAAAMgywlUKcGIKoCQ8PgAAAADIMsJVCnBiCqAkPD4AAAAAyDLCVQroxJTFYrFKWgAAAACQVYQrAAAAAAAAJBLhCgAAAAAAAIlEuAIAAAAAAEAiEa4AAAAAAACQSIQrAAAAAAAAJBLhKgWinkWMxWKxwgUAAAAAWUW4SgFOTAGUhMcHAAAAAFlGuEoBTkwBlITHBwAAAABZRrhKAU5MAZSExwcAAAAAWUa4SgFOTAGUhMcHAAAAAFlGuEoBTkwBlITHBwAAAABZRrhKAU5MAZSExwcAAAAAWUa4SgFOTAGUhMcHAAAAAFlGuEoBTkwBlITHBwAAAABZRrhKAU5MAZSExwcAAAAAWUa4SgFOTAGUhMcHAAAAAFlGuEoBTkwBlITHBwAAAABZRriqQF93bOB6/RD8oQJt+Ynp313X2rVd7dzV9e/BbTn+3nXjbV033rvAP0e2c7XbjXT/DP4MILkIVwAAAACyjHBVUb7u6KpUSVC48jGqtms3Mjc//dONbKeAtSlSRSFcAZXr+++/d23btg3+VLo2bdq4v/3tb8GfiiNcAQAAAMgywlVFSVK4+udI1652bRe1uUr+3rV2qWGKcAVUnn/+85+uZs2aGx8vqrgbb7wxOBrthhtu8PfbfffdS4xXhCsAAAAAWUa4KuJr11HxqZciVJWiIcqHKR0L16bbfujVoMhtDXLq1dcdo1+mPMp7Yrq5MFUsbHGpIBCrMEhplRSvcu9z/PHH++AVhXAFAAAAIMsIV0UoXOlEsePG3+WI2E1VEKRy7lfm+1RxHYu88s0r34lpweWARS8RzJd3H8IVELvS4lVZo5UQrgAAAABkGeGqiIJwlbtjSnyAyq9N+aEq/88/9HINNr6u4i+28XU16OWK/h9KV74T04KB7GUJV4WD2glXwG8iKl6VJ1oJ4QoAAABAlhGuiigIV6XtiCrx0r+8cFVw+WDezi3x94s4XgrCFZBduaHqwAMPLPx9WaKVEK4AAAAAZBnhqogSwlVwiV/BCuLUZnZc5c+9KroqM1xxqSCQNrnxSqus0UoIVwAAAACyjHBVRFS4+sH1arDxZLKclwqWuONqC5T3xJTh7ED6hPGqPNFKCFcAAAAAsoxwVURUuIrehVVwyWDJ4arYnwM+aFXqjKuN8sNUnmJhi3AFJMLjjz9ermglhCsAAAAAWUa4KqKUHVe5sclHqYJLegrvGxGqij2rYAkD2zdni05MfYzKv2QwmG2VE6k8whWQWoQrAAAAAFlGuCqihBlXwfEwVvkYFUSoTc9AGAQu3Z7zCooOcy9/tJItPzEtGNSugFW4orZhEa6A1CJcAQAAAMgywlUKcGIKoCQ8PgAAAADIMsJVCnBiCqAkPD4AAAAAyDLCVQpwYgqgJDw+AAAAAMgywlUKcGIKoCQ8PgAAAADIMsJVCnBiCqAkPD4AAAAAyDLCVQpwYgqgJDw+AAAAAMgywlUKcGIKoCQ8PgAAAADIMsJVCnBiCqAkPD4AAAAAyDLCVQpwYgqgJDw+AAAAAMgywlUKcGIKoCQ8PgAAAADIMsJVCujElMVisUpaAAAAAJBVhCsAAAAAAAAkEuEKAAAAAAAAiUS4AgAAAAAAQCIRrgAAAAAAAJBIhCsAAAAAAAAkEuEKAAAAAAAAiUS4AgAAAAAAQCIRrgAAAAAAAJBIhCsAAAAAAAAkEuEKAAAAAAAAiUS4AgAAAAAAQCIRrgAAAAAAAJBIhKuU+Mtf/uI6d+7s7r///jIt3VcvAwAAAAAAkFaEqxRQgOrRo4dbunRpcGTzdN/u3bsTrwAAAAAAQGoRrlJAu6fKE61Cehm9LAAAAAAAQBoRrlJAl/5tqa15WQAAAAAAgN8S4SoFfpNw9UMv16BKFVclZ3X8OrgtEX5wvRpUcQ16/RD8uYz0fuW8Iz/0auCqNOi18bUBAAAAAICkIVylQNzh6uuOClUNXJEmFIasxNSrLQlXBS+TnPcBAAAAAACUhnCVArGGq687Fo9WIX9bUnZeEa4AAAAAAMg6wlUKxBeuNhd2vnZf597kQ1bHjUdzBDuzwlcRXor3tX4NLjkMw5i/LTxW5HK96Cjld4IV3i/qPsHbX/j/2bgK35evXcfc48HbvelSwZLe9+L/nyJv98ZFBwMAAAAAoHIQrlIgvnBVEHfKHGLKGq4UeKICUv6xwj9vSbiKCE/B21LafTaFq+D3ke/Pph1oBffJ2ZFW7P8BAAAAAAAqCuEqBWILV3nRabPKHK6KXnpYMEOr6MttfjfVZu6TF5gK5Ieq0sNV1Ptf5PYgsOW/XZHBCwAAAAAAbDXCVQqkP1wVvU/RGFRgq8NVruBtKLycr6zhqtjteaHKv7/5cWyjyGgGAAAAAAC2FuEqBWILV0GoSWe4CqJTkViVH6Ly/7zxSN7bUuTtzQ9V/s85/48ii3AFAAAAAEBFI1ylQHzhqnjYKSrv9iSFq8jdUPnvT/H3r9jbkvP2F/3/bRT5/wAAAAAAAJWFcJUCnTt3dkuXLg3+VHZ6Gb1suZQWZ4IdR4XdJ+q+effZmnBVNKAFx0oIV1GvM4xQ5QpXhffpWHyeVV6UK+Tf57yABwAAAAAAthrhKgX+8pe/uB49epQrXum+3bt39y9bXj4i5QepYhFoo+DYpriz6RkDw7ttWbgK34ZNL1fwenLvUzRcFcSj3KgUBKiIlyk9XOX8vyICXsFtucejB7YDAAAAAICtR7hKCQUo7Z7SpX9lWbrvlkSrQkEIyl3FdhpJkfsp6BSdk1UQesofrjbea1N40tr4Cou+XF642mhTcCpYui0/gG16ewviU9TbsvFgQaTLPx6I+v8AAAAAAICKR7gCAAAAAABAIhGuAAAAAAAAkEiEKwAAAAAAACQS4QoAAAAAAACJRLgCAAAAAABAIhGuAAAAAAAAkEiEKwAAAAAAACQS4QoAAAAAAACJRLgCAAAAAABAIhGuAAAAAAAAkEiEKwAAAAAAACQS4QoAAAAAAACJRLgCAAAAAABAIhGuAAAAAAAAkEiEKwAAAAAAACQS4QoAAAAAAACJRLgCAAAAAABAIhGuAAAAAAAAkEiEKwAAAAAAACQS4QoAAAAAAACJRLgCAAAAAABAIhGuAAAAAAAAkEiEKwAAAAAAACQS4QoAAAAAAACJRLhKsXnz5rm//vWvbvDgwa5Xr15+6fc69v333wf3AgAAAAAASCfCVQr961//cgMHDnSTJk1y06ZNc//4xz/c2rVr/dLvdUy3DRo0yN8XAAAAAAAgjQhXKbN48WLXo0cP/+vmLFq0yN/3hx9+CI4AAAAAAACkB+EqRRSrhg4dGvyp7IYMGeKWLFkS/AkAAAAAACAdYglXP/Rq4Dp+HfzB/eB6NajiGvRKzy6grzs2cBX15m7p69Ilf9o9taW6d+/u/v3vfwd/AlDU165jlSquSrg2PWABAAAAAH5DlR+ufujlGmw8EUzteeDXHTeeyFZQuNqK16WZVmW5PLAkumxQM68A5CuIVptiev6fAQAAAAC/FcLV5iQgXOnZAzVsfWtNnDiRZxsE8mhHaJUqHV2Rhyj/tZp3DAAAAAAQu8oNV/7kL//ym6KXCvqTxga93Nf+5DG8b0HcKTihDI5tvE9+7yly+8a1ZXGs4O2Jej35r3/TDoziL1P00iLt2Nj4PvQK3/8G7s03S3pdm/fXv/7VP1Pg1po6daqbPHly8CcA8nXH/K9fKdh1tWWPKQAAAACAivIb7LiKCFeKOYV3yJk1k38s5yyy4OVydi8F/5/yXd4TBKjcs9P8XVHFdklFvEyx/3f4PkTt4sh9XWUzePBg949//CP405bT6+jVq5cbPnw4i1XulU1FH482KfgaLt/jCQAAAACgoiUkXBWNOX4HRF708ccKd11Fn1QWvK7yXN5ThpPT/Njk35/8+JQfs0p4vVsYrhSb1q5dG/xpy+l1aMB7VJRgsTa3solwBQAAAABJlpBwVTQ2+WN5lwYWCVclBaDIqFS6gkiWG8XylBabgvctvPwvP1zlbsryCFesFK9sIlwBAAAAQJKlOFzlBKMiq/xhqOBtyHkducWpWGwKdlcVuW9wrJLCVUVdKrhw4UL30UcfRUYJFmtzK5sIVwAAAACQZNnacVUBwohVeMKa//+K/H9XbriqqOHsGszep0+fyCjBYm1uZZV/bCn+xRr9NQwAAAAAiFU6w1Wx1xnwYag8M66iFTmRzYtNUW9b+PZUVriaN2+emzhxYvCnLafXMX/+/OBPACTqMaiiHksAAAAAAFsnneFqo4KXy41AW3JpT8TL5L+9+bHJ/7n4++MvGyx8+yo2XMmgQYPcokWLgj+Vny4THDJkSPAnAJsUfL3mh+fyPZYAAAAAACpD5YerjXx0Kgw7FROupOBlg9e9xSeawUlria8nJ0wFJ7ZR/9+C9zF8P0oIVxGvq6z+/e9/u+7duwd/Kr9u3bq5X375JfgTgKLyHgfK+fUJAAAAAKgcsYQrVIwffvhhi3ZN6WX0sgAAAAAAAGlCuEqZJUuW+J1XZblsUM9EqJ1WRCsAAAAAAJBG2QxX4bD0UteWzZpKAl02qJlXGrY+depUH6jWrl3rl2ZZ6Zhu004rLg8EAAAAAABpxY6rFPv+++/d5MmT3eDBg12vXr38UqzSMZ49EAAAAAAApB3hCgAAAAAAAIlEuAIAAAAAAEAiEa4AAAAAAACQSIQrAAAAAAAAJBLhCgAAAAAAAIlEuAIAAAAAAEAiEa4AAAAAAACQSIQrAAAAAAAAJBLhCgAAAAAAAIlEuAIAAAAAAEAiEa4AAAAAAACQSIQrAAAAAAAAJBLhCgAAAAAAAIlEuAIAAAAAAEAiEa4AAAAAAACQSIQrAAAAAAAAJBLhCgAAAAAAAIlEuAIAAAAAAEAiEa4AAAAAAACQSIQrAAAAAAAAJBLhCkCkefPmub/+9a9u8ODBrlevXn7p9zr2/fffB/cCAAAAAKDyEK4AFPGvf/3LDRw40E2aNMlNmzbN/eMf/3Br1671S7/XMd02aNAg9+9//zt4KQAAAAAAKh7hCkChxYsXux49evhfN2fRokWue/fu7ocffgiOAAAAAABQsQhXADzFqqFDhwZ/KrshQ4a4JUuWBH8CAAAAAKDi/Ebh6mvXsUEvV7hP4+uOrkqVjhuPZt0PrleDKq5Kx+y/p0gXXR6onVZbSjuvUnHZoH+saeB6lbhJrOBrNPpLdOPjVpWNX7/hyn0MAwAAAABUit8kXH3d0epJH+EKyaSZVmW5PLAkumxQM6+SLQxPJYcr/9i08T7Fv0SDly28IfhaJl4BAAAAQKUiXMWKcIXk0bMHatj61po4cWKin20wjFLR4arobqr8L9EfejUo/pj1Qy/XoNTdWwAAAACArRVzuArCTeEJYnDSV+RSwYITyI5f512W488kSz+5zL99y+PY5l5PaW9jrrz3t2MvwhUS569//at/psCtNXXqVDd58uTgTwkTPsZEXioYfB3r69zHqPzHloKv4wYUKgAAAACIXTJ2XEWEq9yTS7/bIfJY7glo8HI5Z5wFOyzKOzurLK+n+NtY8D7knvAWv5SocMdHBYar4cOHs1hbtXr16uX+8Y9/BJ9RW06vY/DgwcGfkqTg69WHp8hwlSMyXBW8vI5t2rVV9GsbAAAAAFA5khuucs8cg5PJIjse8k4wC0JWfqTKOWEto7K9nqjXm3cZYGk7O4qeFW+VqBDBYpVnaSj72rVrg8+oLafXoQiWNEUu89uScBUc21yYBgAAAABUvMSGq6JRqOBY1MlkwbG8aJTD/78ijkcr6+uJeHvyXrbIyXKhkl//looKESxWeVamw5V/nMjfGbll4apYAI+6LwAAAACgQmUrXG38c+Qq85llWV/P5sNVsfcxUL6QtnlRIYLFKs+qqEsFFy5c6IYMGRL8KQkKviaLPJZsRbgq/mUb9VgFAAAAAKhImd9xVT5lfT2bD1fsuGKlZfXp08f97W9/Cz6jtlzihrMHjxGREXrjigxOkZEq6utdCFcAAAAAUNkyEq4iXqcXseNiM8r2ejYfrgrep8qfcQVsrXnz5rmJEycGf9pyeh3z588P/pRQW7LjaqPInZIl3BcAAAAAUHEyE67C++SeXEYPWt+csryeiLcnP1yFf855P/37nfe6gSQYNGiQW7RoUfCn8kveZYIl2MJwVeLjTbHIDQAAAACoSL9JuApPAhVx/IlghYQrCU4mC1d5o1Voc68n4u0pFq4kOBa+no69Iu4D/Pb+/e9/u+7duwd/Kr9u3bq5X375JfhTgm1puPLyHhf4OgYAAACASvfbhCsAifPDDz9s0a4pvYxeFgAAAACAika4AlBoyZIlfudVWS4b1OWB2mlFtAIAAAAAVBYz4apwvlRpi0t/AH/ZoGZeadi6ninwH//4h1u7dq1filU6ptu00yoVlwcCAAAAAFKLHVcAIn3//fdu8uTJbvDgwa5Xr15+KVbpWOKfPRAAAAAAkAmEKwAAAAAAACQS4QoAAAAAAACJRLgCAAAAAABAIhGuAAAAAAAAkEiEKwAAAAAAACQS4QoAAAAAAACJRLgCAAAAAABAIhGuAAAAAAAAkEiEKwAAAAAAACQS4QoAAAAAAACJRLgCAAAAAABAIhGuAAAAAAAAkEiEKwAAAAAAACQS4QoAAAAAAACJRLgCAAAAAABAIhGuAAAAAAAAkEiEKwAAAAAAACQS4QoAAAAAAACJRLgCAAAAAABAIhGuAAAAAAAAkEiEKwAAAAAAACQS4QoAAAAAAACJRLgCAAAAAABAIhGuAAAAAAAAkEiEKwAAAAAAACQS4QoAAKAc1q5d62bOnOnmzZvnf79+/Xq3ePFiN2nSJP97FPj555/dkiVL3JQpU/zHSX744Qc3bdo0t3r1avfLL7/4YwAAAKUhXAEAgMzYsGGDj0cKJflh5KeffnL/+7//6/71r38FRwqsW7fOr3//+9/BkdL9/e9/dzfddJN74IEH3IwZM9w//vEP98Ybb7ijjjrK/74kCjkKNnob9HaGfv31V/f//t//829D+DbrmP68Zs0a/zJ62/PfPt1H76te348//uiX7qf/Ty69nI7rdr0u/aqPT1nf33x62/UxDP+/ep16O/X26m0KKVr16NHDnXfeeW7WrFn+2HvvvecuvPBCN2rUKP/yAAAAm0O4AgAAmaGdT126dHG33Xab++abb4KjBdGqcePGrmHDhu7zzz8PjhZ45JFH3IMPPujGjx8fHCndloar999/39/nqquu8juPQnqZTp06uXbt2rnp06f7Y9rNdd9997mLL77YHX300a558+Y+9uT67//+b/faa6+5Sy65xNWsWdOddNJJ7rrrrnMffPBBkWg3evRo//aefPLJ7thjj3WnnXaau+uuu9yIESOCe5TPsmXL/MdQH8sTTjjBHXPMMe6CCy5wTz75pJs7d25hENPboDilgBXGQsIVAAAoL8IVAADIjBUrVriPPvrI1apVyw0ePNjvDlJIWbhwoTvjjDPcYYcd5sOWjmtpx9DVV1/t2rRp46NLWWxpuHr55ZfdDjvs4N+GPn36+LdVFKkeeughd8stt/jL6mTBggWuQ4cO7oUXXnB169b1QWrgwIH+ttB//dd/uf/7f/+ve/PNN/3r6927t49wetu++OKLwsvz5syZ40NTv3793FdffeV69erl7r//fteyZUv3/fffl2vnlYLbW2+95ePbww8/7COZ/r8vvviiu/XWW30c/O677/wOsiiEKwAAUF6EKwAAkBkKJn/729/c4Ycf7nr27OnjiNZf/vIXd/nll7vjjz/ePfvss27lypV+R5DiT/369d0TTzzhQ49ili59GzNmjH/5t99+28cZ3U+7tmRrwtUee+zhd1EpGmnWk0SFK70N48aN8/+v22+/3Ueh/HClnUx//etffXzSJYN6+/r27etuuOEGv3tL76Po/VIkC3dhKT61b9/eX8Kn3VglRaYoAwYMKAxUevsU/nSJoC4FVBDUDix9LPQ+6eP+7bff+ssFw7eFcAUAAMqLcAUAADJD4UmRRJfNPffccz6gKPBoV5IuvVNweeaZZ/xlhJoFpd1H2nGlnUuiiKOdSf/5n//pj1922WX+18cee8xHGF3ytjXhqkaNGv6+tWvX9juVFKiiwlUuXcaoSx/zw1WUr7/+2t17773u5ptvdsuXLw+OFszx0kB5xabPPvvMtWrVqjAglTVcaX6VdnRdeeWVrmvXrsHRTfRxuf76630gHDZsWJGPi3ZhCeEKAACUF+EKAABkinYeNWvWzIcqhZrZs2e7O+64w++eUszSbiNdTqiYo8vd/s//+T/uyy+/9DuSFFs0A0rHp06d6ncU6XVohpQijHYrbU24UrDSTqsGDRr4l9dOsIoKV7rkT1GqSZMmrmPHju6f//xncEvB5YJ33nmnO/TQQ90f/vAHd8ABB/j3O9xlVhb6eClMXXvttf5jk2/p0qX+MkLFOX18CVcAAKAiEK4AAECmaAeRApV2HWn2k2LQueee60aOHOmGDh3qw9VTTz3lw4kCkqLR5MmT/eV0H3/8sbvooov8fCztyFLUUQBq27at320UXr63NeFKO5/0dmknlwKTwlpFhCu9bZrVpR1R+bOr9Hu9f/Pnz/dD2V9//XW/yyz/GRZLoo+DhrI3atTIxyu9/nza6aaP3/777+8vDyRcAQCAikC4AgAAmaIY079/f3fFFVf4oeEKVno2Pc1h0mWDCldNmzb1v9fxV155xT8b4aJFi9zzzz/v52OFA9G1FGuOO+44v5Np0KBBWx2utPtJO7e0C0zzqz788MOtClfaKaa5Xnp9mp2lweylBSkFI82q0rMA6nVqJ9XmKFxp95k+buEA9nwKWwpTGj6vSzAJVwAAoCIQrgAAQKZoFpPikOZTKeS8//77fseVQpWCjp6p75JLLvHRRpfOKbIo3ihevfTSS/6yQO0qUpgKl4ad61JDBZitDVe6NFA7oLp16+Zjk4KVnuVPO8TKG640lF1BTuFL91EYCgehl0YzqHTpYKdOnfycrbLQx047urRTTHPA8un90uWZZ599tn/2QsIVAACoCIQrAACQOYos2i11zjnn+F1NijS6VE40WFzPJKiopSHuw4cP98dXrVrlZ13pNv2qsKKdRgphmpulpddbUeFKv2r3l0KZ3lY9G2BuuNL/W0v/H0Up7ZLSn0OKVnpbdFmkQpzmSmnXUy7txtL7ofdN74deXu+H7qv/Z/4srM1RvNOOK4U2hUBdTqn/h16HdrkpWmmwvYbfE64AAEBFIFwBAIBM0i6kI4880p188snu3XffdWvWrPHHdbmfos2+++7rB42HsUgBRpfw1alTx+9y0swoxS7FID1bnwaS/8///E+FhSvRZX2KPXvvvbdr2LBh4dui2xWb9P++6667XPPmzf3OMP1ZM7wUoDRnSrvH9ttvPz9TSq9Xu6d0SV84dF3v86RJk3yIU2jSbiy9L9rpdeaZZ7ohQ4b43Wa6r0KYolZuHMunnWz6uCr4denSxe/2Wrhwod+BpY+JBrPr46b/L+EKAABUBMIVAADIJEWS008/3UcWxRuFGdGz+im+7Lzzzu7xxx93c+fO9cdFO6omTpzoLw1UwDrhhBPciSee6C6//HLXu3dv/8x5FRmuNCxdc7WqVavmrrnmmsJwpdCjS/I0b2u33XZzu+yyi6tevbr/fyhCKWopFp111lluhx128M8SqGik2/X/aNWqlY9sCkiaoXXBBRe4Y445xh199NH+du3u+uCDDwqjlXZMvfrqq36Ol+KdIl4Uvd16/1977TU/xF4zwhQHzzjjDB/DNE8s3KlGuAIAABWBcAUAADJJO4H+8pe/+EiiXUjhTiLFHMUXhR/tGNLupFz6s0KLnn1Pl+dpjRkzxocYxS8FF10Kp/vo99oBpd1PuuRQvy+JAtno0aMLd0OJXp9eVjFKcS2cN6Vf9f/U8c8//7xwaXaUhsjrEj3N5FIo0q4tvS/h0o6yMNQpNC1YsMB/DPSyWtplpUCmCBdSsNPHYuzYsYU7ukqi2/Wx1dunywP1OjUz69tvvy2MVuH9wo9LuNtNb8v48eP9TraS4hgAAEAuwhUAAAAAAAASiXAFAAAAAACARCJcAQAAAAAAIJEIVwAAAAAAAEgkwhUAAAAAAAASiXAFAAAAAACARDIZrpYsWeKfinny5MmsFK+//vWvfkXdxsrW0tO6Rx1nZWvp65m/axtLf888fmd/8TVtZ/E1bWPxNW1n8TVtZ1XW17R6y8KFC4MCs/VMhqv/+3//rzvggAPcOeeck/p19tln+xV1W9bXKaec4mrXrh15Gys766yzznInnniiO/PMMyNvZ2VnnXrqqe7kk0+OvI2VnaXvWbVq1XJnnHFG5O2s7KzTTjuNr2kj66STTnKnn3565G2s7Cz9Hevv2uq5h6Wlx279XBZ1G6vyVtzn9jq/0nmWzreibt+adcghh7jnn38+KDBbz2S4+uCDD1yzZs3c4sWLWSleo0ePduPGjYu8jZWdNXv2bNe3b1/397//PfJ2VnbWhAkT3IgRIyJvY2VnLViwwPXr18/913/9V+TtrOws/YvrkCFDIm9jZWsNGDDA/e1vf4u8jZWdpb9j/V3rcTzqdlZ2lh679XNZ1G2s7CydX+k8a968eZG3b81q2bKle+utt4ICs/VMhqv/7//7/9z/+T//J/hTeq1Zs8Zvv9P6+eefg6N26Avtv//7v4M/Iav+93//1/3lL39x//znP4MjyKrvv//exwxk27///W/39ddfu//5n/8JjiCr/vGPf/gTXWSfLin64Ycfgj8hq/R3rL9rPY4j2/TYrZ/LEJ9ff/3VBx+d269evTo4Wrl0fqXzrP/3//5fcKTi/Od//qe/0q2iEK5STP/aMXToUL8UsawhXNlAuLKDcGUD4coOwpUdhCsbCFd2EK7i969//cuf8+jcfu7cucHRykW4SrishCs9mGi7rtaPP/4YHLWDcGUD4coOwpUNhCs7CFd2EK5sIFzZQbiKn8LVyJEj/bm9RqXEgXCVcFkJV9pO+Msvv/i1YcOG4KgdhCsbCFd2EK5sIFzZQbiyg3BlA+HKDsJV/HQ+H57b6zw/DoSrhMtKuNInd+6yhnBlA+HKDsKVDYQrOwhXdhCubCBc2UG4it9vcW5PuEq4rIQrXR6oOVdaDGdHVhGu7CBc2UC4soNwZQfhygbClR2Eq/hpl9WiRYv8uT3D2YsjXKXY/Pnz/VOVajGcHVlFuLKDcGUD4coOwpUdhCsbCFd2EK7ipxlXY8aM8ef2c+bMCY5WLsJVwmVpOPvAgQP9Yjg7sopwZQfhygbClR2EKzsIVzYQruwgXMVP4WrUqFH+3D6uc1zCVcJlJVxZR7iygXBlB+HKBsKVHYQrOwhXNhCu7CBc2UC4SjjCVTYQrmwgXNlBuLKBcGUH4coOwpUNhCs7CFc2EK4SLkvD2fWAosVwdmQV4coOwpUNhCs7CFd2EK5sIFzZQbiKn4az6/umPu5xnfcQrhIuS8PZBw8e7BczrpBVhCs7CFc2EK7sIFzZQbiygXBlB+EqfppxNXr0aH9uz3D24ghXKaZwNWjQIL+iwtWGDRvcsmXL3IwZM9y0adPc3Llz/SeljmcB4coGwpUdhCsbCFd2EK7sIFzZQLiyg3AVvzBc6dw+rnNcwlXCWZlxtXbtWtejRw/XsGFDV6dOHXfPPfe42bNn+y+KLCBc2UC4soNwZQPhyg7ClR2EKxsIV3YQrmwgXCWclXDVtWtXd95557nddtvN7bTTTq569eru3nvvzcyJIeHKBsKVHYQrGwhXdhCu7CBc2UC4soNwZQPhKuGyEq5Wr17tL//TWr9+fXB0k7Zt27oDDzzQbbPNNq5KlSquatWqPmQNHz48uEe6Ea5sIFzZQbiygXBlB+HKDsKVDYQrOwhX8fvll1/cwoUL/bn9qlWrgqOVi3CVcFaGs+uT5YADDvDRKgxXdevWdSNGjAjukW6EKxsIV3YQrmwgXNlBuLKDcGUD4coOwlX8NM5nzJgxDGcvAeEqxRYsWOCGDBni15o1a4Kjm/Tp08eddtppPlr9/ve/d4cddphr3769mzVrVnCPdCNc2UC4soNwZQPhyg7ClR2EKxsIV3YQruKncKVzHp3bE66KI1xlmL6pNG/e3G233XauRo0a7qmnnnKLFi1yv/76a3CPdCNc2UC4soNwZQPhyg7ClR2EKxsIV3YQrmwgXCWclXC1YcMGd8MNN7jdd9/dNWrUyF8rq280Op4FhCsbCFd2EK5sIFzZQbiyg3BlA+HKDsKVDYSrhMtKuNInmrYRakUNZ5cwXDVr1iw4kh2EKxsIV3YQrmwgXNlBuLKDcGUD4coOwlX8NJxdM6x1br9y5crgaOUiXCVcloazDxo0yK+o4eyicLXbbru5Jk2aBEeyg3BlA+HKDsKVDYQrOwhXdhCubCBc2UG4ip9mXI0ePdqf28d1jku4SrgsDWcfNmyYX1HD2UXhapdddvGXCmYN4coGwpUdhCsbCFd2EK7sIFzZQLiyg3AVP4WrsWPH+nP7uXPnBkcrF+Eq4azMuBKFq5133tldc801wZHsIFzZQLiyg3BlA+HKDsKVHYQrGwhXdhCubCBcJZy1cLXTTju5q666KjiSHYQrGwhXdhCubCBc2UG4soNwZQPhyg7ClQ2Eq4TL0nD22bNn+1XacPZq1aq5Bg0aBEeyg3BlA+HKDsKVDYQrOwhXdhCubCBc2UG4ip+Gs+tjrnP7FStWBEcrF+Eq4awNZ99hhx3cZZddFhzJDsKVDYQrOwhXNhCu7CBc2UG4soFwZQfhKn4MZy8d4SrFFi5c6EaMGOFXacPZf/e737lLLrkkOJIdhCsbCFd2EK5sIFzZQbiyg3BlA+HKDsJV/BSuxo8f78/t582bFxytXISrhLM242q77bZz9erVC45kB+HKBsKVHYQrGwhXdhCu7CBc2UC4soNwZQPhKuGshauqVau6unXrBkeyg3BlA+HKDsKVDYQrOwhXdhCubCBc2UG4soFwlXBZCVerVq1ys2bN8qukTzaFq2222cbVqVMnOJIdhCsbCFd2EK5sIFzZQbiyg3BlA+HKDsJV/DScXZcI6tye4ezFEa5SbHPD2Tds2ODDVZUqVdx5550XHM0OwpUNhCs7CFc2EK7sIFzZQbiygXBlB+EqfgxnLx3hKsX0A+GoUaP80sl9vl9//bUwXJ1zzjnB0ewgXNlAuLKDcGUD4coOwpUdhCsbCFd2EK7iF/58pHP7uD72hKuEszLjSuGqefPmhCukGuHKDsKVDYQrOwhXdhCubCBc2UG4soFwlXBWwpWukyVcIe0IV3YQrmwgXNlBuLKDcGUD4coOwpUNhKuEy0q4WrlypY83WlGfbPqmcv311xOukGqEKzsIVzYQruwgXNlBuLKBcGUH4Sp+2nQyZ84cf44b189IhKuEy0q40oPJwIED/Yoazq4Bb4QrpB3hyg7ClQ2EKzsIV3YQrmwgXNlBuIqfzt0130rn9nGd4xKuEi4r4WrRokX+mQe0dHKfb/369a5Zs2aEK6Qa4coOwpUNhCs7CFd2EK5sIFzZQbiKn76uJk6c6M/t58+fHxytXISrhLMy40qfgIQrpB3hyg7ClQ2EKzsIV3YQrmwgXNlBuLKBcJVwVsLVunXrXNOmTQlXSDXClR2EKxsIV3YQruwgXNlAuLKDcGUD4SrhsjScfcaMGX5FfbKtXbuWcIXUI1zZQbiygXBlB+HKDsKVDYQrOwhX8dNwdp3b6tx++fLlwdHKRbhKOCvD2X/66Sd33XXXEa6QaoQrOwhXNhCu7CBc2UG4soFwZQfhKn4MZy8d4SrFFi9e7D/RtHRyn0/HCFdIO8KVHYQrGwhXdhCu7CBc2UC4soNwFT99XenrS+c9CxYsCI5WLsJVwlmZcaVdWE2aNCFcIdUIV3YQrmwgXNlBuLKDcGUD4coOwpUNhKuEsxKuVq9eTbhC6hGu7CBc2UC4soNwZQfhygbClR2EKxsIVwmXlXC1YsUKN336dL+iPtn0idi4cWPCFVKNcGUH4coGwpUdhCs7CFc2EK7sIFzFT8PZZ82a5c/tly1bFhytXISrhLMynH3VqlWuUaNGhCukGuHKDsKVDYQrOwhXdhCubCBc2UG4ih/D2UtHuEoxDWcfN26cX3oGwXzakZUbrjZs2OBXVhCubCBc2UG4soFwZQfhyg7ClQ2EKzsIV/HT19XkyZP9uf3ChQuDo5WLcJVwVmZcKVw1bNiwMFxp+yHhCmlDuLKDcGUD4coOwpUdhCsbCFd2EK5sIFwlnJVwtXz58sJwdfbZZ7v169e7X3/9Nbg1/QhXNhCu7CBc2UC4soNwZQfhygbClR2EKxsIVwmXlXClH/p1kqe1bt264OgmGup27bXX+nB11lln+csJtesqKwhXNhCu7CBc2UC4soNwZQfhygbClR2Eq/jpPH3mzJn+Z2GGsxdHuEoxPZgMGDDAr6jh7EuXLi0MV2eccYb/xCRcIW0IV3YQrmwgXNlBuLKDcGUD4coOwlX8NJx95MiR/tx+9uzZwdHKRbhKuKyEqyVLlrjx48f7FTWcXd9crrnmGh+uTj/9dH/pYJa+0RCubCBc2UG4soFwZQfhyg7ClQ2EKzsIV/HT19WUKVP8uT3D2YsjXGWYnnXw6quv9uHqtNNO839Wyc0KwpUNhCs7CFc2EK7sIFzZQbiygXBlB+HKBsJVwlkMV6eeeqpbsGCB+/nnn4Nb049wZQPhyg7ClQ2EKzsIV3YQrmwgXNlBuLKBcJVwWRrO/u233/oVNZxdPzCG4ap27dpuzpw5/pkFs4JwZQPhyg7ClQ2EKzsIV3YQrmwgXNlBuIqfZlHr/Fbn9ppVHQfCVQkUTRRPPvroI9etWzf35Zdf+sn5+XSiqgfFXr16uZ49e/r7jxgxwh/fsGGDv8/KlSvdhAkT/OvRGjNmTJn/grM0nH3gwIF+RQ1n17WxV111lQ9XJ598sv9CqIxPyt8K4coGwpUdhCsbCFd2EK7sIFzZQLiyg3AVP430GTVqlD+3j+scl3BVgkWLFrkXX3zRNWvWzDVu3Ni1aNHCdenSxe8WCoOUaOj4G2+84W+/8cYb3fXXX+/uvvtuH6f0Qf3111/90LL777/fNWrUyC/drgn8ZXnWvKyEK33zmDhxol9Rw9l1aeCVV17pw9VJJ51U4s6stCJc2UC4soNwZQPhyg7ClR2EKxsIV3YQruKnryt93HVur24SB8JVBMWmsWPHupo1a/qdUmvWrPG7qZo2bepPVHKHhq9evdqHKf2qEKXbn3vuOXfDDTf4Z8bT7qLXXnvNxy99sLUUrh555JEyndxamXE1f/78wnBVq1Yt/ywFa9euDW5NP8KVDYQrOwhXNhCu7CBc2UG4soFwZQfhygbCVQT9gNqnTx9Xv359fwmbQpY+SI899ph79913fcgKKVZpB1G4e0pDxhW5Lr30Un854NSpU13nzp3dU0895e+j9eabb7onn3zS/zC8OVbC1bx589wVV1zhw9WJJ55Y4s6stCJc2UC4soNwZQPhyg7ClR2EKxsIV3YQrmwgXEXQJ/7777/vbrnllsIfVnXp2ksvveTj1apVq/yxKPrC6dChg7vnnnv8bKvhw4f7HVhvv/12cA/nPvnkE/fMM8+4zz//PDiyiXZuDRs2zD377LPuiSeecA0bNnSXX365Dx9pXpMnT3YjR470Sx/L/NuHDh3q6tSp48NV9erVXfPmzd0LL7zg54V99913xe6ftqUvsnHjxkXexsrOmjZtmhs8eLB/HIi6nZWdpZgxevToyNtY2Vn6/qPvT9oFHHU7KztL/2Cmn1GibmNla+lncwWNqNtY2Vn6O9bfdRbOI1ilLz126+eyqNtYlbP0daXRSPrYx/V4qvMrnWfpH46jbt+addttt6UzXGlnjN7wO++8061YscIf0zv0+uuvu/vuu88HqSiad6XgpZfTD7qa0aSBZYpQPXr0CO7l3BdffOE6duzoevfuHRzZRJcWKnLo8kKFGwUcXUKntynNS5deaoC9lk7u82/XN5YLLrjAh6vtttvO/eEPf3DHH3+8a9eunf8Ezb9/2pailR5Qo25jZWdNnz7dDRkyxMfZqNtZ2Vk6ydVjddRtrOwsPSmL/jFJu6ejbmdlZ+kHbw2ajbqNla2lfxRVjI66jZWdpb9j/V3rcTzqdlZ2lh679XNZ1G2syllqI59++qk/t9c/5Ebdp6KXzq90njVjxozI27dmtWzZMp3hau7cuf6SQF2iF+640gnpK6+84mdT5e+40rB2BSddXqgh7J06dSrclqqApXD13nvv+T+L/pK1K+uzzz4LjpQsK5cK6rLJSZMm+RU1u0qfMA0aNPDhKlzbb7+9j1kff/xxcK/00he33kdkG5cK2sGlgjZwqaAdXCpohyIllwpmH5cK2sGlgvHT15U2o+jcnuHsxcUWrvRAp2B01VVX+eAieuBTbNIzDepyvlz6i9OOIj2r4KOPPlrkpFXD3RWytHsq1K1bNz/zSruMNsfKjKvZs2f7SyJzw5XWwQcf7D/maUe4soFwZQfhygbClR2EKzsIVzYQruwgXNlAuIrw888/++s1TzrpJL/NVEPCdVnfTTfd5Lchrl+/Prin85cDfvPNNz5yPf/8837rWjioXXSpYZcuXfyzDGqou5Yuf9POrTCKlcZKuJo1a1axcKUdV/Xq1fM71NKOcGUD4coOwpUNhCs7CFd2EK5sIFzZQbiygXBVAj3LnXZPaV7V3Xff7X/VXCr98DpgwAA/s0jb4nQ/DWI/4ogj/LPi6VJB7abSMwcqUunBUvMx2rRp49q2beuXfq8B7f/617+C/1vJshKuli1b5gOfVtSlggo7l112WWGw2muvvfxlgrpkc8GCBcG90otwZQPhyg7ClQ2EKzsIV3YQrmwgXNlBuIqfvq40Sknn9przHQfCVQkUV3Tdpp5JsH379u6dd97xz4ynvyQNgNP1nHpAXLx4sR+krmcA1DMF6rJAvYyGseskVnQ/DWnX69Hq27dvmWNMVsKVHkz69+/vl+aB5dNOtTBcKVpdc801/mOoj9Ovv/4a3Cu9CFc2EK7sIFzZQLiyg3BlB+HKBsKVHYSr+GkDjq5Q07m9Rv7EgXCVcFkJV7osUuFPK2rHlZ5S89JLL3XbbLON37326quvuuXLlwe3ph/hygbClR2EKxsIV3YQruwgXNlAuLKDcBU/fV3pWf50bq+NPHEgXCWclRlX2mqocLXddtu5o48+2g+z1+WFWUG4soFwZQfhygbClR2EKzsIVzYQruwgXNlAuEo4K+FKJ4D169d3O+64o6tZs6a/pLIsw+vTgnBlA+HKDsKVDYQrOwhXdhCubCBc2UG4soFwlXBZGs6uZ2jUirpUUFsNL7nkErfzzju7k08+2Q/Gj2vQWxwIVzYQruwgXNlAuLKDcGUH4coGwpUdhKv46etKPwfr3J7h7MURrlJMDyZ6NkatqOHsU6dO9eFq9913d6effrq77777YrteNg6EKxsIV3YQrmwgXNlBuLKDcGUD4coOwlX8wuHsOrdnOHtxhKsU044rPV2mVtSOKz3gXHzxxW7PPfd055xzjrvzzjv9D5FZQbiygXBlB+HKBsKVHYQrOwhXNhCu7CBcxU9fV5pRrXN7dlwVR7jKsDBc7bPPPq5OnTr+fV64cGFwa/oRrmwgXNlBuLKBcGUH4coOwpUNhCs7CFc2EK4Szkq40vWxF110kdtvv/38r82bN3fz588Pbk0/wpUNhCs7CFc2EK7sIFzZQbiygXBlB+HKBsJVwmUlXOkZAvXNQyvqUsHJkyf7YHXggQe6yy67zDVs2DBTD0CEKxsIV3YQrmwgXNlBuLKDcGUD4coOwlX89HWlJ1fT11hcc6kJVwmXlXA1b948179/f7+ihrPrk75evXru0EMPdVdddZVr0KCBf5msIFzZQLiyg3BlA+HKDsKVHYQrGwhXdhCu4qfh7CNGjPDn9gxnL45wlWLLly/3Dypa69atC45uMnHiRB+uatSo4Ro1auR/P2fOnODW9CNc2UC4soNwZQPhyg7ClR2EKxsIV3YQruL3yy+/uO+++85/7ON6PCVcJZyVGVcTJkxwF154oTvmmGNc06ZN3bnnnpup0EO4soFwZQfhygbClR2EKzsIVzYQruwgXNlAuEo4K+FKJwYKVzVr1nQ33nijO/XUU92sWbOCW9OPcGUD4coOwpUNhCs7CFd2EK5sIFzZQbiygXCVcFkazj5p0iS/ooazjx8/3tWtW9eddNJJ7tZbb/UBa+bMmcGt6Ue4soFwZQfhygbClR2EKzsIVzYQruwgXMVPX1fTpk3z5/aLFi0KjlYuwlXCWRnOPm7cOB+utNPq9ttvd0ceeaSbMWNGcGv6Ea5sIFzZQbiygXBlB+HKDsKVDYQrOwhX8WM4e+kIVymmH/r1lJlaUcPZx44d6y644AJ35plnujZt2riDDjrID3zLCsKVDYQrOwhXNhCu7CBc2UG4soFwZQfhKn4azq7zW53b68qqOBCuEs7KjKsxY8a4OnXq+KHs9957r9t3333d9OnTg1vTj3BlA+HKDsKVDYQrOwhXdhCubCBc2UG4soFwlXBWwtXo0aN9uNKuqwcffNDtsccevuBmBeHKBsKVHYQrGwhXdhCu7CBc2UC4soNwZQPhKuGyEq70zWPixIl+/fTTT8HRTUaNGuXOP/98V69ePffoo4+6nXbayQ98ywrClQ2EKzsIVzYQruwgXNlBuLKBcGUH4Sp++rrSx13n9gxnL45wlWKbG84+cuRIH67q16/vnnrqKVe1alX/xbBhw4bgHulGuLKBcGUH4coGwpUdhCs7CFc2EK7sIFzFj+HspSNcpZh+6NdJnlbUcHZ94itcNWjQwHXs2NFVqVLFf7P59ddfg3ukG+HKBsKVHYQrGwhXdhCu7CBc2UC4soNwFT8NZ585c6b/WXjZsmXB0cpFuEo4KzOuhg8f7mdcXX311e6FF17w4Wr8+PG+5mYB4coGwpUdhCsbCFd2EK7sIFzZQLiyg3BlA+Eq4ayEq6FDh/pw1bhxY9elSxcfrjT3av369cE90o1wZQPhyg7ClQ2EKzsIV3YQrmwgXNlBuLKBcJVwWQlXS5Ys8T/8a0UNZx8yZIgPV82aNXNvvfWWD1eDBw92a9euDe6RboQrGwhXdhCubCBc2UG4soNwZQPhyg7CVfz0dfXNN9/4n5H0/TMOhKuEy9Jw9q+++sqvqOHsilQKVzfeeKN7//33fbjq16+fW7NmTXCPdCNc2UC4soNwZQPhyg7ClR2EKxsIV3YQruKncT4a9aNz+1mzZgVHKxfhKuGyNJx92rRpfkUNZx80aJC74IILXIsWLVzPnj19uPr0008zEwAIVzYQruwgXNlAuLKDcGUH4coGwpUdhKv4aTi7zm91br906dLgaOUiXCWclRlXAwcO9OGqZcuWrnfv3j5c9erVy61YsSK4R7oRrmwgXNlBuLKBcGUH4coOwpUNhCs7CFc2EK4Szkq4GjBggKtbt65r3bq1++yzz3y46t69e2xPr1nZCFc2EK7sIFzZQLiyg3BlB+HKBsKVHYQrGwhXCZel4ezjx4/3K2o4u66PVbhq27at/73C1dtvv+1fLgsIVzYQruwgXNlAuLKDcGUH4coGwpUdhKv46etqypQp/tx+4cKFwdHKRbhKOCvD2TWIXeHq3nvv9c8wqHD12muvuUWLFgX3SDfClQ2EKzsIVzYQruwgXNlBuLKBcGUH4Sp+DGcvHeEqxTSrSid5WlHD2b/88ksfrtq1a+e/CBSuHn74YTdhwgS3evVqt2HDhuCe6US4soFwZQfhygbClR2EKzsIVzYQruwgXMVPw9kVrPSzcFyjfQhXCWdlxtUXX3zhLrzwwsJwVbVqVbf//vu7Jk2a+Nv0xZFmhCsbCFd2EK5sIFzZQbiyg3BlA+HKDsKVDYSrhLMSrj7//HNXr149d8kll7g777zT77jafvvt3V577eWuuOIK9+mnnwb3TCfClQ2EKzsIVzYQruwgXNlBuLKBcGUH4coGwlXCZSVcLV682I0dO9Yvndzn69u3rw9Xxx13nDvjjDN8uNLadttt3aGHHuovG0wzwpUNhCs7CFc2EK7sIFzZQbiygXBlB+Eqfvq60teXzu0XLFgQHK1chKuEy9Jwdg1g19LMqnzaUaVwVbNmTXfOOecUCVdHHHGEe+yxx4J7phPhygbClR2EKxsIV3YQruwgXNlAuLKDcBW/cDi7zu0Zzl4c4SrFNJz9u+++8yvqk61Pnz4+XF166aXurrvucr/73e/cdttt5/bcc08uFURqEK7sIFzZQLiyg3BlB+HKBsKVHYSr+Gn+9OzZs/25/fLly4OjlYtwlXBWZlx98skn7qKLLvKXBOoTcr/99nN//OMf3bXXXuufcZDh7EgDwpUdhCsbCFd2EK7sIFzZQLiyg3BlA+Eq4ayEq48//tiHq0cffdQ/+NSpU8ddeeWVfvaVLi3csGFDcM90IlzZQLiyg3BlA+HKDsKVHYQrGwhXdhCubCBcJVxWwtWiRYv8J5qWTu7z9e7d21188cV+ltX06dNdw4YNXePGjd2oUaOCe6Qb4coGwpUdhCsbCFd2EK7sIFzZQLiyg3AVP31dTZo0yZ/3MJy9OMJVimk4+1dffeVX1HD2jz76yIerxx9/3M2cOdO1aNHCz7YaOnRocI90I1zZQLiyg3BlA+HKDsKVHYQrGwhXdhCu4hcOZ9e5PcPZiyNcpdjKlSt9kNKK+mTT+6lw9cQTT/jA07ZtW//ngQMHBvdIN8KVDYQrOwhXNhCu7CBc2UG4soFwZQfhKn6aPz1nzhx/bh/Xz0iEq4SzMuPqww8/9KHqySef9LuzNOvq3HPP9YPZs4BwZQPhyg7ClQ2EKzsIV3YQrmwgXNlBuLKBcJVwWQlXv/76qy+zWlGD1j/44AN3ySWX+HCl62Q7derkTj31VPfpp58G90g3wpUNhCs7CFc2EK7sIFzZQbiygXBlB+EqfjqfD8/tdZ4fB8JVwmUlXGk4++jRo/3SyX2+nj17+nD11FNPucWLF7s333zT1axZ08++ygLClQ2EKzsIVzYQruwgXNlBuLKBcGUH4Sp++rqaOHGiP7efP39+cLRyEa4SLivhKnc4+48//hgc3aRHjx6ufv367umnn3ZLly71O7Bq1Kjhg1bUDq20IVzZQLiyg3BlA+HKDsKVHYQrGwhXdhCu4qfh7CNGjGA4ewkIVym2atUq/0mtFfXJ1r17dx+u2rdv708QPv/8c3fwwQe7999/P7bth5WJcGUD4coOwpUNhCs7CFd2EK5sIFzZQbiKny4R1MYUnduvWLEiOFq5CFcJZ2U4e9euXd2ll17qnnnmGR+59PSahxxyiL9kcP369cG90otwZQPhyg7ClQ2EKzsIV3YQrmwgXNlBuLKBcJVwWRrOri2FWlGX/mlnVRiudCnhpEmT3GGHHeZeeuklt3r16uBe6UW4soFwZQfhygbClR2EKzsIVzYQruwgXMVP5/P62tK5vXZfxYFwlXBZCVf6gbC04ezvvfeeD1cdOnRwa9eu9aFHM646duzoli1bFtwrvQhXNhCu7CBc2UC4soNwZQfhygbClR2Eq/jp62rChAkMZy8B4SrFdA1s//79/Yoazv7uu+/6cKVQpUsD9SyERx11lHv88cfdwoULg3ulF+HKBsKVHYQrGwhXdhCu7CBc2UC4soNwFb9wOLvO7WfPnh0crVyEq4TLSrjSJ5rCjVbUzKp33nnHXXbZZe7ZZ5/132B0/2OOOcY98MADmQg+hCsbCFd2EK5sIFzZQbiyg3BlA+HKDsJV/HR5oD7mOr9duXJlcLRyEa4Szspw9rffftuHq+eee67wmtnjjz/etWnTxk2fPj24V3oRrmwgXNlBuLKBcGUH4coOwpUNhCs7CFc2EK4SLivhSlVWO620NKg931tvveUuv/xy16lTJx+utE499VR3xx13uMmTJwf3Si/ClQ2EKzsIVzYQruwgXNlBuLKBcGUH4Sp+Olf/+eef/bl9XF9jhKuEy9Jw9lGjRvmlk/t8b775ZmG4CtWpU8e1bNnSjRs3LjiSXoQrGwhXdhCubCBc2UG4soNwZQPhyg7CVfzCn490bh/Xx55wlXBZGs7er18/v1avXh0c3eSNN97w4er5558Pjjh/6eCtt97qB7+lHeHKBsKVHYQrGwhXdhCu7CBc2UC4soNwFT8NZx8+fLg/t581a1ZwtHIRrhIuK+FKsWru3Ll+RQ1nf/3113246ty5c3DEuUaNGrmbb77ZDRw4MDiSXoQrGwhXdhCubCBc2UG4soNwZQPhyg7CVfw0BmjBggX+3H7VqlXB0cpFuEo4K8PZX3vtNdegQQP3wgsvBEecj1bNmzd3ffv2DY6kF+HKBsKVHYQrGwhXdhCu7CBc2UC4soNwZQPhKuGyEq70TWPdunV+RQ1nf/XVV90VV1zhXnzxxeCIc61bt3ZNmzZ1H330UXAkvQhXNhCu7CBc2UC4soNwZQfhygbClR2Eq/hpOLsCks7tddlgHAhXCZel4ewjR470Syf3+V555RUfrl566aXgiHPt2rVzjRs3dt27dw+OpBfhygbClR2EKxsIV3YQruwgXNlAuLKDcBW/8OcjndvH9bEnXCVcloazf/XVV379+OOPwdFNXn755WLh6sknn3TXXnute/vtt4Mj6UW4soFwZQfhygbClR2EKzsIVzYQruwgXMUvHM6uc3uGsxdHuEoxxSo9oGj9/PPPwdFNFKwUrhSwQnqGwSuvvNJfRph2hCsbCFd2EK5sIFzZQbiyg3BlA+HKDsJV/DT6R9839XGP67yHcJVwVoaza7aVIpUuGQy98cYb/pkGFbDSjnBlA+HKDsKVDYQrOwhXdhCubCBc2UG4soFwlXBZCVfaTrh27Vq/ooaz69kEFa66dOkSHHGuW7durn79+q59+/bBkfQiXNlAuLKDcGUD4coOwpUdhCsbCFd2EK7ip+HsGsyuc3uGsxdHuEqxhQsXuhEjRvi1Zs2a4OgmnTt3dldddVWRywI/+eQTd9FFF7lHH300OJJehCsbCFd2EK5sIFzZQbiyg3BlA+HKDsJV/BSrxo8f78/tNcs6DoSrhMvScPb+/fv7FTWcXZcD5ocr3bdu3bru/vvv91U3zQhXNhCu7CBc2UC4soNwZQfhygbClR2Eq/gpXCla6Xx99uzZwdHKRbhKuKyEK+2yWrBggV9Rw9mfe+45H65ee+214IjzT69Zp04d16ZNG/fLL78ER9OJcGUD4coOwpUNhCs7CFd2EK5sIFzZQbiKn0b/LF682J/br169OjhauQhXCWdlOPuzzz7rw9Xrr78eHHFu0qRJrl69eq5Vq1b++tk0I1zZQLiyg3BlA+HKDsKVHYQrGwhXdhCubCBcJVxWwpV2WWnXlVbU7qmOHTu6q6++2j+TYGj69Onu0ksvdS1atEj9SQPhygbClR2EKxsIV3YQruwgXNlAuLKDcBU/jfH56aef/Ln9+vXrg6OVi3CVcFkazj58+HC/ooazd+jQwYerN998MzhScGKoZxps3ry534aYZoQrGwhXdhCubCBc2UG4soNwZQPhyg7CVfw042rcuHH+3J7h7MURrlJMDyYDBgzwK2o4+zPPPOOuueaaIuFq2bJl7tprr3VNmjRxM2fODI6mE+HKBsKVHYQrGwhXdhCu7CBc2UC4soNwFT+FK82j1rk9w9mLizVc6XI2DRrTX8SMGTN8SYw6GdVgMu0gmjNnjj+J0f20bS6XfuD99ttvfXzR0n1XrlwZ3Fq6LA1n1w+FWlHD2du3b+/D1VtvvRUccf7jeN111/mdWGn/YZJwZQPhyg7ClQ2EKzsIV3YQrmwgXNlBuIqfGsiSJUv8986oTSmVgXBVAoWlzz77zJ122mnu0EMPdQ0aNHAffvhhcOsmiluDBg3yz3631157ucsuu8yNGjUquLXAK6+84nbddVd3/PHH+3X++ee7d955J7i1dFaGsz/11FN+d1XuJ4y+IG644QZ3+eWX+62IaUa4soFwZQfhygbClR2EKzsIVzYQruwgXNlAuIqgYWNTpkxxF154oevTp49/djvFp9tuu83Nnz+/yAPgunXrfJRQvNI7fP/99xcLV6+99po766yzTO+40tA21VitqOHsTz75pA9Xb7/9dnCkYNfbTTfd5Pbff3//jINdunTx34QUtNKGcGUD4coOwpUNhCs7CFd2EK5sIFzZQbiKn3qJrqjSuT3D2YuLLVzpg/Lll1+6unXr+sikaziHDh3q7rvvPvfxxx8XuRRQcUUnq3qZ119/3T3xxBPFwpWOX3TRRcGfyicr4UrD1YcNG+ZX1HB2fdwUrnJ3og0ePNidd955bocddnB77723q1Wrlnv11VdT+cBEuLKBcGUH4coGwpUdhCs7CFc2EK7sIFzFT31k7Nix/tx+7ty5wdHKRbiKoGfA69Gjh2vWrJlbvny5P6YviM6dO/sh4iWdlCq6aOdQfrhSbFF06devn/vqq6/ctGnTSnwduTu4dH/t4GrcuLF/8E3z0q41XWqppblh+bfrk0WXWb7wwguFxxQKDz74YFelShW3zTbbuO23397Vr1/fR8Xcl03D0jdO7eKLuo2VnaUZd3p2DUXKqNtZ2VlTp051EyZMiLyNlZ21aNEi/z191qxZkbezsrMUojWWIOo2VrbWmDFj/PzaqNtY2Vn6O9bftR7Ho25nZWfpsVs/l0XdxqqcpV7y6aef+nP78ePHR96nopfOr3SepQ0xUbdvzWrdunU6w5U+KHrD77zzTrdixQp/TDFJO6cUU0q6zK+kcPXBBx/4WVkaNN6oUSP36KOP+vtE/QuA/lW3V69erkWLFv7+55xzjqtXr54PH2leekAZMWKEXzrZy7+9xcb394ILLnCPPPJI4TENZdfcMIWrMF4de+yxPgTmvmwalr7I9L5H3cbKztIDt55dQ5/vUbezsrP0GK5/ZYq6jZWdNXHiRDdw4ED/r4pRt7Oys0aPHu2GDBkSeRsrW0s7+hU0om5jZWfp71h/13ocj7qdlZ2lx279XBZ1G6tyljal6FkFdX4b189IOr/SeVZUS9jadcstt6QzXGlHUEnhSjugyhuudGmhXo9mM6lOalbWPffc4xYvXhzco2RWhrMr5jVs2NC99957wRHnZ1odd9xxPlhtu+22rlq1aj5waedS2nCpoA1cKmgHlwrawKWCdnCpoB06SdG/sCPb9Hesv2suFcw+LhW0gUsFI2hLqXZJaXfUsmXL/DHFkueee84vPZNglJLClYJVOJBc14MqyDz22GP+h+HNyUq40ifYqlWr/Ir6BvLwww/7j/f7778fHCn4hnPHHXe4fffd1x1yyCE+WukbUO6MsbQgXNlAuLKDcGUD4coOwpUdhCsbCFd2EK7ip76hJqJze406igPhKsLatWv9lsMzzjjDn5joL+Pzzz93rVq18peGlPSXExWufv75Zz9pPwxXet0aRN6uXTs3ffp0f6w0WRrOrgH3WlHD2R966CEfrrp27RocKfiCePPNN/2QfF02qHioaKVnMUgbwpUNhCs7CFc2EK7sIFzZQbiygXBlB+EqftqMo3MendsznL242MKVwoii0vXXX+9eeeUVv/tKQeree+/1P9houLqebVAfPD0YLl261A8Mb9mypb/cTfft37+/D1z6YVfXVmt4meKXQlTbtm39rqvwMsTSZCVc6cFEA+e19LSZ+R588MFi4Uo+++wzPyS/RYsWwZF0IlzZQLiyg3BlA+HKDsKVHYQrGwhXdhCu4qdwpc06OreP6xyXcFUCbX3T7irFKz3bXZs2bXyM0g6qnj17+mf8U7zSDqrJkyf7Z/47++yz/S4tDRnXy+kZCTXT6u2333bXXHONj1r6Vc+c9+233wb/p9JlJVxpp5S+gWjpEz3fAw884D+G3bp1C44U0M635s2bu2uvvTY4kk6EKxsIV3YQrmwgXNlBuLKDcGUD4coOwlX8dGWUWoe+znT+EwfCVcJlJVxtji6dVLjq3r17cKSAnqXtpptu8s+smGaEKxsIV3YQrmwgXNlBuLKDcGUD4coOwpUNhKuEy0q40ieYno1RK+obyH333eeaNGnievToERwpoJ1pugRTu9m0u011N40IVzYQruwgXNlAuLKDcGUH4coGwpUdhKv46Zxc5zs6t2c4e3GEqxTTcHZd9qcVNZxd88MUrnQZZq558+a5e+65x5122mn+2R51qWYaEa5sIFzZQbiygXBlB+HKDsKVDYQrOwhX8QuHs+vcXuOT4kC4SrishKv58+eXOpxdnywKVxqEn2vZsmXukUcecSeddJLffRVX0a1ohCsbCFd2EK5sIFzZQbiyg3BlA+HKDsJV/BSuRo8ezXD2EhCuUkyX+SlCaUUNZ9euqqhwpZfr0KGDq1mzphs5cmRsw98qGuHKBsKVHYQrGwhXdhCu7CBc2UC4soNwFT9dKqifjXRurydhiwPhKuGsDGe/++673XXXXec+/PDD4EgBfVG8/PLL7rjjjnN9+vRJbRAgXNlAuLKDcGUD4coOwpUdhCsbCFd2EK5sIFwlXFbClS7x0w/+WlHfQNq2bevDld7fXBs2bPCfRLVr13ZvvfVWak8eCFc2EK7sIFzZQLiyg3BlB+HKBsKVHYSr+GlziQaz6+cjXSEVB8JVwmUlXC1cuNCNGDHCr6jh7G3atHFNmzZ1vXr1Co5s0q1bN3fOOee4jh07uqVLlwZH04VwZQPhyg7ClQ2EKzsIV3YQrmwgXNlBuIqfRv+MHz/en9vrydTiQLhKuKyEKw1nL+1ZBe+8804frj766KPgyCa9e/d2F198sX/mQT2zYBoRrmwgXNlBuLKBcGUH4coOwpUNhCs7CFfxU7gaM2YMzypYAsJVim3uUsHWrVv7cKVIle+LL75wV199tbvxxhvdggULgqPpQriygXBlB+HKBsKVHYQrOwhXNhCu7CBcxY9LBUtHuMqwO+64o8RwpZJ7/fXXu8svv9zNnTs3OJouhCsbCFd2EK5sIFzZQbiyg3BlA+HKDsKVDYSrhMtKuFKJ1dNlamlrYb7bb7/dNWvWzH388cfBkU20DVEfg9NPP93Nnj07OJouhCsbCFd2EK5sIFzZQbiyg3BlA+HKDsJV/LTjSj8b6dz+p59+Co5WLsJVwmVpOPvw4cP9ippx1apVKx+uPvnkk+BIAX1RDBgwwNWvX9/96U9/cm+++aa/XFDH04RwZQPhyg7ClQ2EKzsIV3YQrmwgXNlBuIqfNqKMGzfOn9sznL04wlWKKTaVNpy9ZcuW/nLAPn36BEcK6AvhmWeecQcffLCrWrWqO/fcc927776bullXhCsbCFd2EK5sIFzZQbiyg3BlA+HKDsJV/BSudM7DcPZohKsU0yeYBrhpRX0D0fuocPXpp58GRwr07NnTnXPOOT5aValSxW2//fauXr16xe6XdIQrGwhXdhCubCBc2UG4soNwZQPhyg7CVfx09ZPOd3RurydhiwPhKuGsDGe/9dZbXfPmzd1nn30WHCnw1FNPuT//+c8+WoVLu69efPHF4B7pQLiygXBlB+HKBsKVHYQrOwhXNhCu7CBc2UC4SrishCsNbdM3EK2o4ewtWrSIDFfhjqvtttuucMfVhRdeyI4rJBLhyg7ClQ2EKzsIV3YQrmwgXNlBuIqfdlwtX77cf53p/CcOhKuEy9KMq2HDhvkVNePqlltu8eGqb9++wZECM2fOdJ06dXInnniiD1fHHHOM3201d+7c4B7pQLiygXBlB+HKBsKVHYQrOwhXNhCu7CBcxU8bUcaOHevP7eM6LydcJVyWwtXQoUP9igpXN998s7vhhhvc559/HhwpoJo7Y8YM9/DDD/twddppp7m33nrLzZ8/P1XPLEi4soFwZQfhygbClR2EKzsIVzYQruwgXMUvHM6uc3vCVXGEqxTTJ9iqVav8ivoGctNNN/lw9cUXXwRHNlHo6t27t9tmm23cPvvs4y6++GL3zjvvuGXLlgX3SD7ClQ2EKzsIVzYQruwgXNlBuLKBcGUH4Sp+2kCyevVqf27PcPbiCFcZpmhVUrjSNsSmTZv6cBXOuTr99NN9vEoLwpUNhCs7CFc2EK7sIFzZQbiygXBlB+HKBsJVwmUlXOmEfvHixX5FDWfXfKsbb7zRffnll8GRTfr06VM440pLAUvPNHjPPfcE90g+wpUNhCs7CFc2EK7sIFzZQbiygXBlB+EqftpxtXTpUn9uHzUGqDIQrhLOyoyr66+/3oerfv36BUc2GThwoKtbt27hjquqVau6o48+2j3zzDPBPZKPcGUD4coOwpUNhCs7CFd2EK5sIFzZQbiKHzOuSke4SrHNPatgs2bNfLj66quvgiObzJo1y0eq6tWrux122MEddNBB7rbbbnMjR44M7pF8hCsbCFd2EK5sIFzZQbiyg3BlA+HKDsJV/HhWwdIRrlJs/fr17scff/Trl19+CY5uohlWGtAeFa60FVEPSJdeeqkfzq7ANWrUqMjXk1SEKxsIV3YQrmwgXNlBuLKDcGUD4coOwlX8NmzY4Dej6Nxe5/lxIFwlnJXh7Nddd50PV/379w+OFLVkyRI/jP2YY47xH5O0IVzZQLiyg3BlA+HKDsKVHYQrGwhXdhCubCBcJVxWwpWKrH4o1Pr555+Do5s0btzY3XzzzW7AgAHBkaJWrFjho9ahhx7qP6niKrsVhXBlA+HKDsKVDYQrOwhXdhCubCBc2UG4ip+uiNLGEn3v1K6rOBCuEi4r4Wr+/PluyJAhfkXNuGrUqJEPVxrEHkVBQA9Khx9+uHv++ed9yEoTwpUNhCs7CFc2EK7sIFzZQbiygXBlB+EqfppxNWbMGH9uP2fOnOBo5SJcJVxWwtXChQvd8OHD/YoKVw0bNvThatCgQcGRorRLS0+5qUsFH3roodiGwFUUwpUNhCs7CFc2EK7sIFzZQbiygXBlB+EqfgpX48aN8+f28+bNC45WLsJVwmUlXCk8KVhpRQ1Vv/baa0sNVxoAp288p5xyimvdurX75ptvglvSgXBlA+HKDsKVDYQrOwhXdhCubCBc2UG4ip/OzX/66Sd/bs9w9uIIVxl2zTXXuFtuucUNHjw4OBLt4osvdi1atHAjRowIjqQD4coGwpUdhCsbCFd2EK7sIFzZQLiyg3BlA+Eq4bI0nH3BggV+RQ1nv/rqq3240nWypbnxxhvd9ddf7/r27RscSQfClQ2EKzsIVzYQruwgXNlBuLKBcGUH4Sp+Gs6+ePFif26/evXq4GjlIlwlXJaGs2s3lVbUMw9ceeWVfifV0KFDgyPR7r33Xj/IvXv37sGRdCBc2UC4soNwZQPhyg7ClR2EKxsIV3YQruKnGVejR4/25/YMZy+OcJViGs6uy/u0ooazX3HFFT5cDRs2LDgSrVOnTv6+Xbp0CY6kA+HKBsKVHYQrGwhXdhCu7CBc2UC4soNwFT+Fq/Hjx/tze4azF0e4SjF9cq9du9YvbS3M16BBAx+u9MwEpenWrZufc/X000/7oXBpQbiygXBlB+HKBsKVHYQrOwhXNhCu7CBcxU/n4evWrfPn9jrPjwPhKuGsDGe/7LLLyhSuBgwY4OrUqeMvGSRcIWkIV3YQrmwgXNlBuLKDcGUD4coOwpUNhKuEszKcXeHq1ltv3eyzBU6aNMlddNFFrmXLlpGzspKKcGUD4coOwpUNhCs7CFd2EK5sIFzZQbiKH8PZS0e4SjE9mAwcONCvqOB06aWX+nA1cuTI4Eg0DXnXIPdmzZql6gGKcGUD4coOwpUNhCs7CFd2EK5sIFzZQbiKny4PHDVqlD+3j+scl3CVcFkJV/qBUFFKSyf3+S655BL/fuoLoDSKXtddd50788wz/SfX5MmT3U8//ZT4ywYJVzYQruwgXNlAuLKDcGUH4coGwpUdhKv4hT8f6dw+ro894SrhshKu9MmtAW5aUcPZNXBd76eeVrM0+ibUsGFDt+uuu7rjjjvOz8XSNyXFqyQjXNlAuLKDcGUD4coOwpUdhCsbCFd2EK7ip00jCkg6t2c4e3GEqwzT3KrbbrvNjRkzJjgSrUuXLj5YValSxW277bauWrVqPl5NmTIluEcyEa5sIFzZQbiygXBlB+HKDsKVDYQrOwhXNhCuEi4r4UqX+Gk+lVbUcPYLL7ywTOGqVatWbt999/XhKoxXp59+uhsyZEhwj2QiXNlAuLKDcGUD4coOwpUdhCsbCFd2EK7ipyuoFi1a5M/t4zrvyWy4UiiZNWuWn4GkUKJ3UMf07HZpYmU4exiu9MlYmoceesgdcsghbptttvFr++2394PdNxe8fmuEKxsIV3YQrmwgXNlBuLKDcGUD4coOwlX8GM5eujKHqzCSvPDCCz50rFq1yj9N48SJE93gwYODe6VDloaz65NbSyf3+erWretatmzpxo4dGxyJpr+/q6++2u2xxx5ul112cbVq1XKvvvpq4h+sCFc2EK7sIFzZQLiyg3BlB+HKBsKVHYSr+IU/H+ncPq6PfSbD1fvvv+/atm3rmjVr5k4++WS3ePFiv8vn3Xffdbfffntwr3TISrj65Zdf3Pr16/2KGs5ep04dH67GjRsXHImm16O/39NOO83vvHr++efdkiVLIl9nkhCubCBc2UG4soFwZQfhyg7ClQ2EKzsIV/HTcHZd1aZz+7i+xjIZrhStOnXq5B+szjrrLB825IMPPnA333yz/31aWBnOfv755/twNX78+OBIyRYsWODuvfdeV7t2bTd37lz/BZN0hCsbCFd2EK5sIFzZQbiyg3BlA+HKDsKVDZkMV4oaukxQ863OPvtsH650HWbXrl0JV78RXao5b948v6JC03nnnecHr+sEYXMUB5555hlXs2ZNN2zYsFTMLSNc2UC4soNwZQPhyg7ClR2EKxsIV3YQruKnq6AWLlzoz+01likOmQxXb7zxhnv66afdc88950488UTXr18/995777mHH37Yde7cObhXOmQlXOmTun///n5FDWc/55xzyhyu9IWiv+NTTjnFf4KtWLEiuCW5CFc2EK7sIFzZQLiyg3BlB+HKBsKVHYSr+GlT0IgRI/y5/ezZs4OjlSuT4eqbb75xPXr0cPfdd58f+v3UU08V2YWVJlkazj569Gi/dHKfTzvjFK4mTJgQHCmdPi76u73//vv9DLOkI1zZQLiyg3BlA+HKDsKVHYQrGwhXdhCu4qevK52369x+/vz5wdHKlclwpe1qs2bN8s9A99JLL7kXX3zR18DvvvsuFbtzcmUlXGl4usqsloa55VO40uB8PfNjWejvs1GjRv4ZBrWbK+kIVzYQruwgXNlAuLKDcGUH4coGwpUdhKv46XxeX1s6t9fVUHHIZLjq3r27+/LLL4M/bTJ27NgKfYPikJVwtTlnnnmmD1eTJk0KjpROzz6oYe4nnHCCmzlzZnA0uQhXNhCu7CBc2UC4soNwZQfhygbClR2EKxsyGa4ee+wx9/rrrwd/2qRPnz7+GQfTJCvhSp9oc+bM8StqOPsZZ5zh7rjjjjKHK50wPvjgg26PPfZwU6dOjdzFlSSEKxsIV3YQrmwgXNlBuLKDcGUD4coOwlX8tMtKlwjq3H7lypXB0cqVmXClgd+6xvLll1/2l51deumlfjh7uNq3b+936GjuVZpkJVzpcr6vvvrKr6jh7KeffroPV/oGUxYLFizwM8uqVq3q2rRp43r27OlPInVJYhIRrmwgXNlBuLKBcGUH4coOwpUNhCs7CFfxC4ez69xeI5rikJlwpblW+sBpF47C1UUXXeTuvvvuwnXXXXe5J554wj/DYJpkJVwtWrSo1OHsp556qg9XZR2er2cv0N9nlSpVXPXq1f2OLT2TpAJREhGubCBc2UG4soFwZQfhyg7ClQ2EKzsIV/HT15VmUzOcPVqp4Urb1dasWeMfpN5++2330Ucf+WebW7hwod+ds3TpUn972h68sjScXX9HWlGX9dWuXduHqylTpgRHSjdkyBB38cUX+3Clte2227rjjjvOde7cObhHshCubCBc2UG4soFwZQfhyg7ClQ2EKzsIV/HT+Xx4bh/XFU+ZCVdRtIVt2bJlhfFKS39OEyvD2RWuWrdu7b755pvgSOn0iaX5VmG40tppp53cVVddFdwjWQhXNhCu7CBc2UC4soNwZQfhygbClR2EKxsyGa70LHNvvvmmu/baa12dOnVcgwYN3Hnnnefq1avnnnrqqeBe6ZCl4ewKN1pRw9lPPvnkcoUrDd9XqMoNV7/73e9c/fr1g3skC+HKBsKVHYQrGwhXdhCu7CBc2UC4soNwFT/ttNLHXOe3DGcvrszh6q233nKPPPKIe+CBB1yNGjXciy++6B5++GHXvHlzPwMrTbI4nH316tXB0U1OOukkd+edd5b5h0aFyV133bVIuNpxxx3d5ZdfHtwjWQhXNhCu7CBc2UC4soNwZQfhygbClR2Eq/jpyrbhw4cznL0EZQ5XGsbeoUMH/46dddZZftaVfiB577333GOPPRbcKx2yNJxdfx9aOrnPV6tWLR+upk6dGhwpXe/evd2xxx7rttlmm8Jw9ac//cm/jiQiXNlAuLKDcGUD4coOwpUdhCsbCFd2EK7ip6+rSZMm+fMejWOKQybD1X333edeeOEF/wx1F1xwgf+g6gcS7cRq1apVcK90sDLj6oQTTvDRadq0acGR0umSwnvuucftu+++bvvtt3fVqlXzl4L26dMnuEeyEK5sIFzZQbiygXBlB+HKDsKVDYQrOwhXNmQyXGlX1csvv+y+/fZb/0x1TZs2dS1btnSNGjXyf04TK+GqZs2aPlzp76ws9OwFs2fPdg899JDbb7/93NFHH+26dOnir7dNIsKVDYQrOwhXNhCu7CBc2UG4soFwZQfhyoZMhisFDc1UWrNmjT+x0E6r559/3nXv3r3Ml6IlRVbC1apVq/zQfK2oTzaFq7vuuqvM4UpPwbl27Vo3d+5cP7tMQ9nfffddfzyJCFc2EK7sIFzZQLiyg3BlB+HKBsKVHYSr+GmziM7DdW4f189ImQxX+kBqKWLowUrBQMFq4sSJqTvRyNJw9n79+vmVO5xdf0daxx9/vGvTps0W/f106tTJD2XXLrukIlzZQLiyg3BlA+HKDsKVHYQrGwhXdhCu4hcOZ9e5PcPZiys1XCl+rF+/3gcCPUh99913bsWKFf6Ssjlz5rhBgwb5N6Znz57BS6RDVsKVBuSPHTvWL53ch/T3pr8jhau2bdu66dOnB7eUneZaab6VnjFSX0RJRLiygXBlB+HKBsKVHYQrOwhXNhCu7CBcxU9fV/r60rk9w9mLKzVc/fTTT35gt3bftGvXzv+qp2ecMWOGe+mll9y1117r12uvvRa8RDpkfcZVuCtO4UrPBqngWF5TpkxxF110kbv11lvdypUrg6PJQriygXBlB+HKBsKVHYQrOwhXNhCu7CBc2ZCZcKVA1aRJE3fTTTe5Dh06uEceecT/uUGDBq5u3bp+/pG2sVXGO1qZsh6utNtKu6SOO+64LQ5XOqFo2LChD5NlnZEVN8KVDYQrOwhXNhCu7CBc2UG4soFwZQfhyobMhCvtujnnnHP8O6PtahoW9uGHH7oaNWq4zz//3F+q9vPPPwf3To+shCvthFK80cr9ZFO40iWeClf33HOPD5DlpZe/+eab3aGHHuqfQfLtt992y5Yt8687KQhXNhCu7CBc2UC4soNwZQfhygbClR2Eq/hpnrjGMekcN66fkTITrvTAdOaZZ/pAFdIPm3q2On0i65K0NMrScHZduqn1448/BkcLwpU++RSu9AmjT/7yGjVqlJ9xVa1aNbfPPvu4008/3b3zzjtu4cKFwT1+e4QrGwhXdhCubCBc2UG4soNwZQPhyg7CVfzC4ew6t2c4e3GlhqsJEyb43VWaYaWdVlpPP/20O/LII/2xDz74wB8bOnRo8BLpkJVwtWTJEjd+/Hi/NI8spFq7bt06H67uvfde/5Sa5fXYY4+5ww47zG2zzTZ+bb/99v4SUX1iJwXhygbClR2EKxsIV3YQruwgXNlAuLKDcBU/fV3pijed28e1WSQz4Wry5MnuxBNP9LttzjjjDL9OPfVUV6tWLX8sXK1btw5eIh2yPuNK4Uoh69hjj3X33XffFhVbfXz23XdfV6VKFb8Ur2rXru2fSTIpCFc2EK7sIFzZQLiyg3BlB+HKBsKVHYQrGzITrhRAVq9evdmVu9snDSyEK53sK1zdf//9bvbs2cEtZffiiy/6ZyWsWrWqj1Y77LCDH9Kvb1ZJQbiygXBlB+HKBsKVHYQrOwhXNhCu7CBc2ZCZcFXR1qxZ4z8wt99+ux/4/fDDD7thw4YFt26iEDZp0iQ/WFyXpz300ENu6tSpwa0F5s+f77p37+5fj9brr79e5llOWQlXK1as8M8YqJX7yaZvJpp5pXDVrl27LQpXGsbfqVMnv7tut9128zvr9HeVO0vrt0a4soFwZQfhygbClR2EKzsIVzYQruwgXMVPm0903q5z++XLlwdHKxfhqgQKDQpGzz77rHvzzTfdo48+6qNU/rPVKVx98803fo5Ww4YNXatWrfyw8JCeyfCLL75wLVu2dG+88YZfbdu2de+//75/NrzNyfpwdn0z0SfhMccc4x544IEtijv6+9Alho8//riPV/p70P8vSQP5CVc2EK7sIFzZQLiyg3BlB+HKBsKVHYSr+DGcvXSxhau1a9e6IUOG+Gcp1ImJhod//vnnfveVdvLkfrD0l6YfaBctWuRefvll98QTTxQJVzr+yiuv+JdV5NLrfvLJJ/1AccWVzcnScHb98K+Ve7mmPn6rVq3y4erBBx/0T6u5pTTTqnHjxu6iiy7aqtdTGQhXNhCu7CBc2UC4soNwZQfhygbClR2Eq/jp60qbd/Qzkr5/xoFwFUGxSc9C2KhRI7/DSjQ1/7nnnvNLs7KivPPOOz5K5YarcePG+cvYFK9CPXv29M94OHjw4OBIybI+40rhauXKlT5caUfb3Llzg1vKT188+qTToHY9gGkLY1IQrmwgXNlBuLKBcGUH4coOwpUNhCs7CFc2EK4iKDC8/fbb/hkINZtJZsyY4S8H1ABxhZYoUeFKu4B0uaFmXIW0e6tDhw7u448/Do6UzEK40sdY4UpzxMqyC60k+qFTu9623XZbHwWTNIifcGUD4coOwpUNhCs7CFd2EK5sIFzZQbiygXAVQYPG9IbfeeedheFK4UFD1e+7775yhauBAwf6cNWjR4/giPMzrzp27Oh69+4dHNlE/7/PPvvMz8FSsKpXr5675JJL/Bdkmpd2nikmaWmYfXhcvx8xYoQ75JBDXIsWLVz//v2LvFx5lv4fnTt39s8sePbZZ7u77rrL727Tbrmo+8e5Ro4c6T8vom5jZWfp81lf8xMmTIi8nZWdNWbMGH9tf9RtrOwsff/Q9y3Fq6jbWdlZY8eO9eMgom5jZWtpHIh+Zoy6jZWdpb9j/V0n4TyAVblLj936uSzqNlblLH1dDR061P+MpO+fUfep6KXzK51nKUhH3b41Sx0ileFKz1LXrVs317x588Ip+XqmwBdeeMFf4lfSboqocKXf6/JCDWUPffTRR659+/Z+mFk+7RLS/6tPnz5+t5Xiy7XXXuv/JTDNS59oH374oV+KgOFx7bDSJ8vhhx/u2rRpU3id7JYsfaz1OrTjqlq1au7AAw/0z+KoQBh1/zjXxIkT/RdZ1G2s7CztqtM3z5kzZ0bezsrO0qXJ48ePj7yNlZ2lZwXWP65o13XU7azsLP3spR++o25jZWuNHj3a75iNuo2VnaW/Y/1d63E86nZWdpYeu/VzWdRtrMpZ2uH26aef+nP7uL536vxK51kaLRR1+9YszSNPZbhSmPryyy/dBRdc4Id863I2FUXttlIEKekStKhwpSn7L774omvXrp1/hkEtBTA9A960adOCe5UsK5cKaruu4o1W7sdPHw/ddvTRR/tnbtQ3ly2l4qvB7NpxVaVKFVe1alV37LHH+h1vvzUuFbSBSwXt4FJBG7hU0A794Kp/SEP2camgDVwqaIceu7lUMF76utLHXef2mg8eBy4VjLBhwwY3efJkV7duXb/zSQ96Gq6ugKTCp2fI0zPh6YP266+/+hCjQPXUU0+5O+64w8+z0uWG+gvVMxJq91azZs38riMt7aLSLiw9w+DmZH3G1fr1693ixYt9uNIzLWq325bS39WJJ55YGK60/vznP7t77rknuMdvh3BlA+HKDsKVDYQrOwhXdhCubCBc2UG4soFwVQJdIqitb+eee66rWbOmu/rqq13Xrl19bNKsq169evlLB3Siqu1xmqmkZ7Pba6+9/Lwm7dYKvylqZ9UjjzziX4+W5lfpZcrCSrg66qij/C60hQsXBreUn65tbtKkifvd737n49V2223nateu7d56663gHr8dwpUNhCs7CFc2EK7sIFzZQbiygXBlB+HKBsJVCfQgpx9UNXhMu6R0grJ06VK/w0o/3Oj3ili//PKLW716td+hpRilD6bmnug6W10GJ7qfvpjCHVe6/HDNmjX+ts3JSrjSx/Lbb7/1S7vQQgpX+ngqXD3xxBP+91tKn8y6pFO72/bcc0930EEH+WC4Na+zohCubCBc2UG4soFwZQfhyg7ClQ2EKzsIV/FTA9H5rc7t1UXiQLhKuKyEKw1h1zMGav3444/BUec/8XR5oMKV5oNtzTWyusRTn9AKiFdeeaW//FDPyvjQQw+5l156yV/iqfD4WyBc2UC4soNwZQPhyg7ClR2EKxsIV3YQruKnGeB68hqd22tEUhwIVwmXlXClEjtp0iS/cmd76RNPA9kVrjQjrKKGu+mZAfSsgjvssIPbdddd/eWbusRza2ZobQ3ClQ2EKzsIVzYQruwgXNlBuLKBcGUH4Sp++rrSOCSd2zOcvTjCVQbpskHtxjryyCN9uNK8q4qgZxf8/e9/XzikXevwww93X3zxRXCPeBGubCBc2UG4soFwZQfhyg7ClQ2EKzsIVzYQrhLOQrjSMzUqXD399NP+cr6KcNJJJ7ltt922SLjS0PYePXoE94gX4coGwpUdhCsbCFd2EK7sIFzZQLiyg3BlA+Eq4bISrvQsjXpQ0codzq7fa1i9wlX79u0r7AcJPZtg1apVi4SrHXfc0fXs2TO4R7wIVzYQruwgXNlAuLKDcGUH4coGwpUdhKv4aTj7d9995z/2cT2eEq4SLuvD2TXvSgPdFK6eeeaZCntWglatWrk//elPRcLV6aef7oYMGRLcI16EKxsIV3YQrmwgXNlBuLKDcGUD4coOwlX8GM5eOsJViilI6ZuHVu5wdv1+1qxZrkaNGq5Dhw4VFq4GDhzomjZt6uOVdlrtueeernPnzj6g/RYIVzYQruwgXNlAuLKDcGUH4coGwpUdhKv46evq22+/9V9jFTWjenMIVwmX9RlXCleKOgpXHTt2dMuWLQtu2TravqjdVTfccIM7+OCD3bHHHuv69u3rtzTq//Hrr78G94wH4coGwpUdhCsbCFd2EK7sIFzZQLiyg3BlA+Eq4bIern766Sc3Y8YMH66effZZPwurImzYsMH9/PPPbujQoa5hw4Zup512ctWrV3cnn3yye/XVV/1tcSJc2UC4soNwZQPhyg7ClR2EKxsIV3YQrmwgXCVcVsKVdjl98803fuVeKqhwNX36dHfEEUe45557rsJPDj7++GN33HHH+WcY1KpWrZqPVy+++GJwj3gQrmwgXNlBuLKBcGUH4coOwpUNhCs7CFfx09eVzuF1br9kyZLgaOUiXCVcVsKVHkwGDBjgV+5wdoUrnfwpXHXq1KnCTw7eeustt9tuuxUZ0r7zzju7a665JrhHPAhXNhCu7CBc2UC4soNwZQfhygbClR2Eq/hpOPvIkSP9uT3D2YsjXKWYdlxNmTLFr9wdVzrR12A3havnn3/erVixIrilYnTp0sVtv/32RcJV1apVXd26dYN7xINwZQPhyg7ClQ2EKzsIV3YQrmwgXNlBuIqfvq70c7DO7dlxVRzhKoN0oj916lQfrvSsfytXrgxuqRhvv/2222OPPYqEq9///vfuqquuCu4RD8KVDYQrOwhXNhCu7CBc2UG4soFwZQfhygbCVcJZCFd6sDn88MPdCy+84FatWhXcUjG0hfHqq68uEq5q1qzpB7THiXBlA+HKDsKVDYQrOwhXdhCubCBc2UG4soFwlXBZCVdLly51kydP9iv3UsE1a9b4LYZhuKroE36FsCFDhrimTZu6gw46yM+70sdTP6DGiXBlA+HKDsKVDYQrOwhXdhCubCBc2UG4ip++rjTuR+f2ixcvDo5WLsJVwmVpOHv//v39yh3OrnClbyoKV3qmv4o+4d+wYYN/nWPHjnXPPvusO/HEE90dd9zhVq9eHdwjHoQrGwhXdhCubCBc2UG4soNwZQPhyg7CVfzC4ew6t2c4e3GEqxTTcHY9XaZW7o4rRaxJkya5ww47zL300kuVGpTmz5/vrr32WtekSZPYTzgJVzYQruwgXNlAuLKDcGUH4coGwpUdhKv46etq+vTp/tye4ezFEa4ySOFq4sSJPly9/PLLRXZjVTR9gbVq1co/o2CPHj38ZYQ6pl1ZlY1wZQPhyg7ClQ2EKzsIV3YQrmwgXNlBuLKBcJVwFsLVhAkTfLh65ZVX/KWDlUXfuO699153wAEHuFq1arknn3zSLVq0yP3666/BPSoP4coGwpUdhCsbCFd2EK7sIFzZQLiyg3BlA+Eq4bI0nF2XBGrlXiqoSwPHjx/vw1WXLl38iX9l6dOnjzv//PNdtWrV3M477+z/n+3bt3ezZs0K7lF5CFc2EK7sIFzZQLiyg3BlB+HKBsKVHYSr+Onratq0af7cXhtB4kC4SrgsDWcfMGCAX7mXAypcaXD6oYce6l599VX3008/BbdUPH1C6pkFt9lmG1elShVXtWpVd8IJJ7gOHTr4E9DK3HlFuLKBcGUH4coGwpUdhCs7CFc2EK7sIFzFLxzOrnN7hrMXR7hKseXLl7upU6f6tW7duuBoQbjSJ2Ac4apt27buwAMPLAxXWttvv72/bPCZZ57xA+YqC+HKBsKVHYQrGwhXdhCu7CBc2UC4soNwFb9ffvnFfffdd/7cPq7HU8JVwmV9xpU+AUePHu3D1WuvvVbkMsKK1rVrV3feeee5XXbZpTBcaW233XbulFNOcZ07dy64YyUgXNlAuLKDcGUD4coOwpUdhCsbCFd2EK5sIFwlnKVw9frrrxfZjVXRFMX0bIJ16tTxO61y49Uf//hHd8MNNwT3rHiEKxsIV3YQrmwgXNlBuLKDcGUD4coOwpUNhKuEy0q40jePiRMn+pV7OaA+AXV97CGHHOLeeOONSvlEDG3YsMEtW7bMvfTSS37WVW642meffVzz5s2De1Y8wpUNhCs7CFc2EK7sIFzZQbiygXBlB+Eqfvq60sdd5/YMZy+OcJViejAZOHCgX7nD2VetWuWGDx/uw9Wbb77p1q9fH9xSeXr16uWOPPLIYuHq+uuvD+5R8QhXNhCu7CBc2UC4soNwZQfhygbClR2Eq/hpOPuoUaP8uX1c57iEq4TLSrjSD/3ffvutX7mXAypcDRs2zIert956y/3888/BLZXnyy+/dGeeeaZ/VsEwXGkH1l133RXco+IRrmwgXNlBuLKBcGUH4coOwpUNhCs7CFfx03B2nd/q3H7p0qXB0cpFuEq4rM+4WrlypRsyZIgPV/pkiSNc6ZtY69atXfXq1V21atXctttu64477jj39NNP+5Cmb3C6rLAiEa5sIFzZQbiygXBlB+HKDsKVDYQrOwhXNhCuEs5CuBo8eLA7+OCD3dtvv+23HVY2xTHV4XvuucedccYZ7g9/+INfp512mnvyySf9dbq//vprcO+KQbiygXBlB+HKBsKVHYQrOwhXNhCu7CBc2UC4SrishKslS5a48ePH+5U7nF3hatCgQT5cvfPOO7F8c9FuKj3D4Ny5c92jjz7qatSo4Z9lcNddd3WHHXaYa9++vZs1a1Zw74pBuLKBcGUH4coGwpUdhCs7CFc2EK7sIFzFT19XU6ZM8ef2CxcuDI5WLsJVwmVpOPuAAQP8yh3OvmLFCn9M4erdd9/118vGSZ+kBx54YOGsK829qlu3rhsxYkRwj4pBuLKBcGUH4coGwpUdhCs7CFc2EK7sIFzFT1dJjRw50p/Hz549OzhauQhXCZel4ew6ydPKHc6ucNW/f38/HP29996r8Ev0Nqdt27Y+XG2zzTaF8Urztl5++WVfjyvq7SFc2UC4soNwZQPhyg7ClR2EKxsIV3YQruKnzSYzZ870PwsvW7YsOFq5CFcJl/UZVwpX/fr18+Hq/fffjz1cde3a1Z133nlul112KYxXe+yxh7vkkkv826Nverq0cGuHtROubCBc2UG4soFwZQfhyg7ClQ2EKzsIVzYQrhIu6+FKJwNffvllYbiq6Gfz2xzNuurRo4erV6+e23nnnX24UsDSzCtdvqiPvb7hEa5QFoQrOwhXNhCu7CBc2UG4soFwZQfhygbCVcJlJVwtXrzYjRs3zq/c4ew6Gfjiiy98uNLup7jDlf5/2t7YoUMHv9MqvFxQ63e/+50f3K5nO9TOsK1BuLKBcGUH4coGwpUdhCs7CFc2EK7sIFzFT19XkydP9uf2DGcvjnCVYnowGThwoF+5w9l1MtC3b9/CcPVbUZzac889i4Qrrd12281dc801bt68ecE9twzhygbClR2EKxsIV3YQruwgXNlAuLKDcBU/DWcfNWqUP7eP6xyXcJVwWQlX2rE0ffp0v3I/2ZYvX+4+++wzf1let27dgqPx6927tzvhhBOKhauddtrJ1a5d2w+f25rdYIQrGwhXdhCubCBc2UG4soNwZQPhyg7CVfw0nH3WrFn+3J7h7MURrjJI4apPnz7+mfx+y3A1adIkd/fdd/sdVtttt11huKpWrZqrVauWmzBhwlZ9kRCubCBc2UG4soFwZQfhyg7ClQ2EKzsIVzYQrhLOQrj65JNP3KGHHuq6d+8eHI3fzz//7GbPnu3atm3r9t9/f1e1alUfrrbddls/tP2OO+5wU6dODe5dfoQrGwhXdhCubCBc2UG4soNwZQPhyg7ClQ2Eq4TL0nB2faJp6eQ+pK2FukzvsMMO+03DlS4D1BfB3Llz3WWXXeZ23333wl1Xilenn366GzJkSHDv8iNc2UC4soNwZQPhyg7ClR2EKxsIV3YQruKnryt9fem8Z8GCBcHRykW4SrisD2dXuProo4/c4Ycf7nr06BEc/W099NBDPqQpWIXx6pJLLnGjR48O7lF+hCsbCFd2EK5sIFzZQbiyg3BlA+HKDsJV/BjOXjrCVYqtXLnSzZgxw6/cTzaFq169erkjjjjC9ezZMzj62xo8eLC74YYb/OWLu+66qw9X5557ruvYsaMbNGiQ++6779yvv/5armHthCsbCFd2EK5sIFzZQbiyg3BlA+HKDsJV/DScXee2OrfX6J84EK4SLuszrpYuXeo+/PBDV6NGjcSEK30hfvXVV/7jftxxx/lwpSHtf/jDH/wlg08++aT/Ai3PN0LClQ2EKzsIVzYQruwgXNlBuLKBcGUH4coGwlXCWQhXH3zwgTvyyCP9r0mgnVQa1j58+HDXvHnzwssFt9lmG7f99tu7gw46yD311FPl+qGHcGUD4coOwpUNhCs7CFd2EK5sIFzZQbiygXCVcFkJV4sWLfIzorR0ch/SNxXNtjrqqKMSE65Cn332md9hFYarcO20007uhBNOKLxksCwIVzYQruwgXNlAuLKDcGUH4coGwpUdhKv46etq4sSJ/tx+/vz5wdHKRbhKuKwPZ9c3lW7duvlwpUsGk0Rva/369f1Oq9xwpaHtu+yyi3v11VfdwoULg3uXjnBlA+HKDsKVDYQrOwhXdhCubCBc2UG4ih/D2UtHuEqxFStW+JM8rXXr1gVHC76pdO3a1R199NGJC1caNtepUyc/OH7HHXcsFq+uuuoqN27cuODepSNc2UC4soNwZQPhyg7ClR2EKxsIV3YQruKnmdCzZs3yPwvrydbiQLhKuKzPuFqyZIl7//333THHHOPf1yTRZYB6+5599lk/gys/XumYopt+2NWwds3GKumZBglXNhCu7CBc2UC4soNwZQfhygbClR2EKxsIVwlnIVy999577thjj01cuFKEUrzSDjHtvDr++OPddtttVxiu9tprL7fvvvv6t11/R/oiKmnmFeHKBsKVHYQrGwhXdhCu7CBc2UC4soNwZQPhKuGyEq70A6Gug9XSyX1I4eqdd95xxx13nOvVq1dwNHkWLFjgOnfu7OrWresOOOCAwmcYrFq1qtthhx38sVatWpU484pwZQPhyg7ClQ2EKzsIV3YQrmwgXNlBuIpf+PORzu3j+tgTrhIuK+FKzzYwaNAgv3KHsy9evNh/kmg300cffRQcTR7tpNJ1vBpAp0sHFa1yh7brz4pXeqbBKIQrGwhXdhCubCBc2UG4soNwZQPhyg7CVfw0nF3PKKhz+7jOcQlXCZeVcKXh7Io6WrmfbApXb731lqtZs2aiw1Uunazus88+RS4b1LD23//+965v375u1apVxWZdEa5sIFzZQbiygXBlB+HKDsKVDYQrOwhX8dNw9tmzZ/tze816jgPhKuGyPuNq0aJF7o033khVuJo3b5674oor/IwrBSuFK+2+2mmnnVzjxo1dv379fJDTzrJwYDvhygbClR2EKxsIV3YQruwgXNlAuLKDcGUD4SrhrISrE044wfXu3Ts4mmz6BqiTGEUqxavcSwb1zIP77befa9Cggd9JpvsSruwgXNlBuLKBcGUH4coOwpUNhCs7CFc2EK4SLivhSkPLR4wY4deaNWuCowXh6rXXXnO1atVyH3/8cXA02cJnG5wxY4a77bbbXPXq1QvDlZYuIdxjjz1c7dq1fbzSCQ/hygbClR2EKxsIV3YQruwgXNlAuLKDcBU/zbgaP368P7fX1UhxIFwlXNaHs+sHxS5duriTTjopNeEqlz7BTznllCLzrsK16667ujp16vhgRbiygXBlB+HKBsKVHYQrOwhXNhCu7CBcxY/h7KUjXKXYypUrfbzRyv1k0w+Kr7zyig9Xn3zySXA0PcaOHevuuusud+ihhxa5ZFCratWq7j/+4z/c559/7ou0Btgh2whXdhCubCBc2UG4soNwZQPhyg7CVfw0nH3OnDn+3D6un5EIVwmX9RlX+kHxpZdecieffHIqw5W+aCdNmuTuvfdef3mgYlVuvNJOrEaNGvk5Xt9++23wUsgqwpUdhCsbCFd2EK7sIFzZQLiyg3BlA+Eq4bIerjT76sUXX/SX2/Xp0yc4mh6ad6VviHPnznVt2rRx++yzT7GdV7vssos7//zz/d8lso1wZQfhygbClR2EKzsIVzYQruwgXNlAuEq4rISrBQsWuGHDhvmVO5xd4eqFF17wg8zTGK5C69ev95cCaufVEUccUWTmlUKWdmNdffXV/n3ULi0FL2QP4coOwpUNhCs7CFd2EK5sIFzZQbiKn2ZcaWSOzu21gSMOhKuEy0q4Kmk4u8LV888/70499VT36aefBkfTJ3ymQT1wtmvXzh111FFFLhvcdttt3Z/+9Cd3xRVXuC+//NKtXr06eElkCeHKDsKVDYQrOwhXdhCubCBc2UG4ih/D2UtHuEqxVatWuZkzZ/qV+8mmnVidOnVyp512WqrDVa5wYPshhxzig1VuvNprr738zKvhw4f7jwk7r7KFcGUH4coGwpUdhCs7CFc2EK7sIFzFT1cQaaeVzu3j+hmJcJVwWZ9xpXD17LPPutNPP9199tlnwdF0yx3YXr16dbf99tsXxitdNlitWjXXuHFjN2TIEB86kB2EKzsIVzYQruwgXNlBuLKBcGUH4coGwlXCWQhXHTt2dGeccYbr27dvcDTdcge233PPPW7fffctsvNKSwPbtfNK2yuRHYQrOwhXNhCu7CBc2UG4soFwZQfhygbCVcJlaTi7dhhp5Q5n1+yrZ555JlPhKpQ7sP2www4rEq+08+qPf/yja9KkCQPbM4RwZQfhygbClR2EKzsIVzYQruwgXMVPM650zqNz+zlz5gRHKxfhKuGyNJx98ODBfuUOZ9fx9u3buzPPPDNz4Sp3YPsdd9zhDjjggCID2/X7/fbbj4HtGUK4soNwZQPhyg7ClR2EKxsIV3YQruKncDVmzBh/bk+4Ko5wlWL6RNPuIy3tRAopXD399NPurLPOcp9//nlwNHs++eQT17BhQ3fooYcWe7ZBBrZnB+HKDsKVDYQrOwhXdhCubCBc2UG4ip+uFtLHXOf2K1asCI5WLsJVwmV9xpU+4Z988kl39tlnuy+++CI4mj3fffed/7u87777NjuwPfdSSqQL4coOwpUNhCs7CFd2EK5sIFzZQbiygXCVcBbC1RNPPOHOOeecTIerGTNmuL///e+FA9t1iWDuziutcGB7lneeZR3hyg7ClQ2EKzsIV3YQrmwgXNlBuLKBcJVwWZpxpWfQ08qdcTVv3jz32GOPuXPPPdfPecoqRav//u//LhzYrp1XNWrUcNttt12RnVca2H7VVVcxsD2lCFd2EK5sIFzZQbiyg3BlA+HKDsJV/DTjavTo0f7cXue4cSBclWDt2rXu22+/dS+//LIfHv7uu++6KVOmBLcWtW7dOh8aXnrpJffss8/62KQPrAZz//TTT35o2UMPPeQ6derk1/vvv1/mH46yFK6ihrMrXD3yyCPuvPPOMxGucge2t2vXzh199NFF4pV2YelSwjp16viPS+/evf0zMuplkHyEKzsIVzYQruwgXNlBuLKBcGUH4Sp+YbhiOHu0WMOVYkGHDh3crbfe6lq0aOFatWrlXnjhBX9imrsLRtFKgat169b+fjfffLO7++67/V+ibtMPu3o9ChRt27b167nnnnPjxo0LXkPpshKu9ImmcBPuOgopXD388MM+XPXr1y84mj1huMo1duxYd9dddxUb2K6lGVh/+MMfXL169Xw8HTVqlH9QUMBiF1ZyEa7sIFzZQLiyg3BlB+HKBsKVHYSr+OnKIG1M0fntypUrg6OVi3AVQQ9wenrHWrVquUmTJvmTUQWk5s2b+y+Mn3/+ObhnwQ862kV1//33+wdHhRi9002aNHHLli3zP+y+8sor7sYbbwxeonyyEq5KoplP2o2mHUbWwpW+4PX5pcsG99lnnyID28OlZx3cbbfd3CmnnOKeeuopt3z5cr4BJxjhyg7ClQ2EKzsIV3YQrmwgXNlBuLKBcBVh6dKl7qOPPnJXXHGFW7JkiT82YcIEf8ngq6++WuRSN+22uvrqq/1lbno5PTiOHDnSHX744W7hwoWEq81QuHrwwQfdBRdc4L766qvgaPZEhSvtnNLnS2kD27U0+2qHHXZwhx12mI9X/LCVXIQrOwhXNhCu7CBc2UG4soFwZQfhygbCVQTtmnrvvfd8MAp/WJ0+fboPULqsbdWqVf6YTJ482Z111ln+B9s1a9b4Y/r9Mccc459JTuFLlwoeccQR/rLDO++803366aeFQWxzshKu9GAycOBAv3LDn6KNZj3VrVvXXLgK5Q5sP+6449zOO+/sd1rlB6xq1ar5IKqAqtfH3KvkIVzZQbiygXBlB+HKDsKVDYQrOwhX8dOMK42y0bl9See4FY1wFUEffL3hikwrVqzwxxQKXn/9dR8Xcq/j1KVetWvX9gFLA91Fu7OOP/54f1Kjlx86dKh7+umn3ZtvvumHtz/wwAPuk08+8X/h+XTiO3HiRNetWzf/Ntx2221+R5e+GNO8NM+pZ8+efunjEh7XUDe9j2eeeaYfWp/7MllaOvHR32vUbQqlCnja5ffoo4+6a665xj/joEJV7g4s7bzS5YS6hPWOO+7wn4/9+/f3L6vXEfW6WfEuPU7o6/27776LvJ2VnaXHfD2uRd3Gys7SbMHhw4cX+b7Fyub65ptv/JiIqNtY2Vo62Zo6dWrkbazsLP0dhzNio25nZWfpsVs/l0XdxqqcpV7y8ccf+3N7nc9H3aeil86vdJ6lDR9Rt2/N0jzzVO+40g6p8F9Z9UOrhmQrLOTvuDr77LPd+PHjC3dc6fcaxj5z5kxfBLU0qF07ZDT0XdFBO7fCKJZLr1sxQs8op/J3+eWXu0svvdT//9O89C8eelDRUhUPjw8YMMDdcMMN7vTTT3dvvPFGkZfJ0tIXtApx1G25a9q0ab5ca1dV06ZN3QknnOCHtOfuwFLA2nHHHd0BBxzgPzc07F8PGvrY6tLVqNfLimfpmUf1tLD6fI+6nZWdpWily8KjbmNlZ+kxeciQIf4fqaJuZ2Vn6We3MFKysr2GDRvm/5E56jZWdpb+jvV3rcfxqNtZ2Vl67NbPZVG3sSpn6etKH3Odf8b1M5LOr3SeldsSKmqp+6QyXGmougriZZdd5hYtWuRnEelZAJ988km/ayoMVKJLCBs1alR4+Z8Gt+tB8qijjvIzrvSyuc8Cp4Hc2n2lMFWWp47MyqWC4cch/+OhWqv5Tnr2PEWsrCrtUsEo+hhpa3PXrl394HoNaA/DVe763e9+5/bee28fuXRylfu5ifhxqaAd+tcZfaNDtnGpoB1cKmgHlwrawKWCdnCpYPxKOrevTFwqGEFxSR+UmjVr+su7FAO0o0VxQCcquZf4KWy98MILPr7ovooT2jnUrFkz/wxwOpHV0m4r/aXq/gpRmuu0evXq4LWULCvhSu+/Pq5auZ/cYbi66KKLCFc5wgcBzb/S594ll1ziI1VUvNIOrF122cUH1M8//zx4DfgtEK7sIFzZQLiyg3BlB+HKBsKVHYSr+Ok8NTy3j2vuMuGqBPoBRkFKsUpB4Oabb/aXCuoD1rt3bzd48GD/BaIPnIaw33777e66667z923durW/plq36VpMPROhnlVQr+P66693Dz30kN/SqL/ozcnScHaFKa3c4eyKOW3btnUXX3yxv0Quq8obrnLp2So1E61BgwZujz328HOuFKvy49U+++zjrrzyStenT59igRDxIFzZQbiygXBlB+HKDsKVDYQrOwhX8dNGHo3M0Lm9Zk7FgXBVAs2kUpB65513/LMJKiDpWs7wh1gN/NOOKsUB3VfPiKf7amB23759/W4q1UddPqhLuHRclxm+9dZbft6RXrYsshSudE2qVn64atOmjd9RpNuyamvClT6PtFOvX79+rlOnTv4SVs230pyr3IClQe777ruvq1+/vp979eWXX/qXi6uCg3BlCeHKBsKVHYQrOwhXNhCu7CBcxS98VkGdv2/pOW55Ea4SLivhSrFKDyhamgMWUqHVszcqthCuShduyVQkffDBB/3H7KCDDiryzINhwNp9993dhRde6F566aXCZ1RRwGIXVuUiXNlBuLKBcGUH4coOwpUNhCs7CFfx03mlvm/q4x7XeQ/hKuGyEq4UTHJXSOFKz7JIuCofBSztALzvvvv8JYK6fDA3XoVLs69OOukk9/jjj/tny9TJl+ZmEbAqB+HKDsKVDYQrOwhXdhCubCBc2UG4il9J5/aViXCVcFkJVwot2lKolfvJPWvWLD8f7NJLL/Vzw7KqosOVPob6Rjx37lw/3H6//fYrtvMqXNttt53bdddd3RFHHOHniU2ZMqVM89VQfoQrOwhXNhCu7CBc2UG4soFwZQfhKn46Fw3P7eM6ryRcJVxWwpV2+/Tv39+v3BlXCletWrXyc5s0CyyrKjpchbR7SrvWtPPquOOOczvvvLPbdttti8UrzcLSsxJqBtZZZ53ld7kxxL3iEa7sIFzZQLiyg3BlB+HKBsKVHYSr+ClYjRgxwp/bM5y9OMJVis2fP9/vqNLKD1ctW7Z0l19+OeFqCyg66RrjCRMmuHfffdfPCzv55JN9wCppB9YOO+zg9t9/fz8Qv2PHjv4JA/TEAr169fKvh1lYW45wZQfhygbClR2EKzsIVzYQruwgXMVP4UpPOKdze81SjgPhKuGyNJxd8Uordzj7zJkz/fvXoEEDN3To0OBo9lRWuMql4KTi/cYbb7gbbrjB1apVy+25555+/lXusw+GSzuzNAPr4IMPdocddpg75ZRT/N+Fhr/rG8CqVasIWOVEuLKDcGUD4coOwpUdhCsbCFd2EK7ip/NOPXu9zu3jOu8hXCVc1oezK1y1aNHCXXHFFYSrCqKPr75Jd+3a1V177bWuRo0abrfdditxB1bu0uWEe++9t7vuuutcv379/AOSomP+3xuiEa7sIFzZQLiyg3BlB+HKBsKVHYSr+IXnhnGeIxKuEi4r4UrfNPRJpqVCG1K4uuWWW9yVV17phg0bFhzNnrjDlZbmX61evdqNHDnS3XTTTa569er+MsGo3Ve5S7fvuOOOfuD7xRdf7C8l1N9fXA9KaUa4soNwZQPhyg7ClR2EKxsIV3YQruIXnmvq3D6urzHCVcJlfTi7go6iylVXXUW4qgR6UNHHW4FwzJgxrk2bNqUOcc9dejZC7dQ68sgjfVjs3LmzmzZtWpHwiKIIV3YQrmwgXNlBuLKDcGUD4coOwlX8GM5eOsJVii1YsMAPX9das2ZNcLQg6Nx4443u6quvdsOHDw+OZs9vFa5CClgKTrlD3E899VT3pz/9ye+s2mOPPfwuq6jdWLrEUKFLQ9/btm3rd2C9/fbb/vXo108//dRNnz6doLUR4coOwpUNhCs7CFd2EK5sIFzZQbiKn8KVznl0bs9w9uIIVymmWLVw4UK/coezK+hokPg111xDuIqJApPKuKLTvffe69q1a+ef2bF+/fp+ULsuJ4zajaVje+21lzv00EP9MPcjjjjC/3rOOee4Bx54wD9wfffdd27dunVmLyskXNlBuLKBcGUH4coOwpUNhCs7CFfx0/nkkiVL/Lm9RtPEgXCVcFkazq5PcK3cqDFjxgx3/fXX+yHi2m6YVUkKV1F++eUXfxngQw895HdW/fGPf/Q7sDZ3OaGWdmnttNNO7vDDD3e33367mzhxov9h4aeffjIXsAhXdhCubCBc2UG4soNwZQPhyg7CVfxKOrevTISrhMtKuNJ2wrVr1/qlT/CQwlWzZs1cw4YN/RDxrEp6uNIDjr6xKzatWLHCPffcc+60005zu+++e5mejVBL91PA0q4tPSth3759zf2wQLiyg3BlA+HKDsKVHYQrGwhXdhCu4qdzR11lo3N7nefHgXCVcFkazv7VV1/5lTucXZeWKXIQrpJBD0Ja+gF+8uTJfo6VBudrQPtRRx3l9t57bz+0PSpchWv77bd3e+65pzvxxBP9TjrtwtKliO3bt/d/x9rdlVWEKzsIVzYQruwgXNlBuLKBcGUH4Sp+ilUa86Nz+1mzZgVHKxfhKuGyNJx96NChfuUOZ1e4atKkiWvUqJEbNWpUcDR70hKucmln3KJFi9yAAQPcO++842diKUDVqlXLD3Xff//9/bMO/u53v4sc6q6Apds1/F3317MZKlK+8MILmR3sTriyg3BlA+HKDsKVHYQrGwhXdhCu4hcOZ9e5/dy5c4OjlYtwlXBZCVc6oVcE0codzq5wpWjVuHFjwlXCaafUpEmTXJcuXdx9993nh7q3aNHCnXvuuT5OlRSwwqXbNPhdA96zOtidcGUH4coGwpUdhCs7CFc2EK7sIFzFT5sO9DWmc/vcq6kqE+Eq4bI+nF27bXSZoHZdjR49OjiaPVkIV1EUs4YNG+YvB9SOqj322GOzASt35Q92nzBhgn92Cj1LhR4MV61a5X/gSEvMIlzZQbiygXBlB+HKDsKVDYQrOwhX8Svp3L4yEa4SLkvD2XVSr5U740jhSnOQdAkZ4Sp99EAV/t1qx9Q999zjatSo4XbeeecyPSNhuMLB7gceeKCrXbu2u+CCC9zll1/unnzySV/y9aCYBoQrOwhXNhCu7CBc2UG4soFwZQfhKn46B9Rgdp375F5NVZkIVwmXxeHsq1evDo4WhKurr77aNW3a1I0ZMyY4mj1ZDVchPXjpQUTXOOskT89KePHFF/uh7kcffbSrXr26q1at2mZ3YmkulgLWLrvs4v7whz/4kHXRRRe5G264wbVq1crdfffdfjaWTjCSGLMIV3YQrmwgXNlBuLKDcGUD4coOwlX8GM5eOsJViunyL31ya+UOZ9eJn561rlmzZoSrjFBQmjlzpuvbt6+PTBrEroHsik8nnHBCmQa7hyu8lFBzsfRyBx10kDv77LP9fK3XXnvND43v1q2bv1xxxYoVsW1VLQnhyg7ClQ2EKzsIV3YQrmwgXNlBuIqfwtW4ceP8ub02qMSBcJVwWQlXP/30k59bpKVP9JBO/K644gofrvSJmFWWwlWUrR3snrt0CaJ2ZCl+aTbWscce66655hofyMaPH/+bXlpIuLKDcGUD4coOwpUdhCsbCFd2EK7ip3OtZcuW+XP73E0plYlwlXBZH87+7bff+llG119/PeHKmKjB7rqcUM88qEsGyzMjS/fVpYV169Z1r7/+uv/mpQfSxYsXFwZT/QCjYe8Kp5W1M4twZQfhygbClR2EKzsIVzYQruwgXMWvpHP7ykS4SrishKv169f72VZaucPZw3DVvHlzN3bs2OBo9hCuitODXP5gd10GeNJJJ7mjjjrK/cd//Icf2h4VqqKWdmxp55ZeTi+viHX++ee7OnXqFBn2PmfOHP8gWxkIV3YQrmwgXNlBuLKDcGUD4coOwlX8dB6nnVY6t6+MkBSFcJVwWQlX8+fPd4MGDfLrxx9/DI4WhKtLL73Uzz/SdbJZRbgqmR741q1b5+OV4uWoUaP8TqwePXr4Z5zUbixdEnjAAQe4XXfddbM7sXT7jjvu6Hdg7b777v5XLc3JOuKII9yVV17pWrdu7e6//3736KOP+v+PdmZVRMwiXNlBuLKBcGUH4coOwpUNhCs7CFfx0+aD0aNH+3P7uM5xCVcJl6Xh7CNGjPAr9zpYhav69eu7G2+8kXCFQopZKvgKWBq+riHsugTw4Ycfduecc4479NBD/Wysvffe2w9vL+tlhdqVpcsRK2vYO+HKDsKVDYQrOwhXdhCubCBc2UG4ip/ClWYL69ye4ezFEa5STMPZ9Q1EK3c4+7Rp09zFF1/sbrrpJv/Jn1WEq62nHVE6eezZs6d74okn/I6pu+66y+/KqlGjht9dtd1220UGq9KWoldpw96nT5/upk6d6neC6Ruj3oaSghbhyg7ClQ2EKzsIV3YQrmwgXNlBuIqfzsuWL1/uv850/hMHwlXCZWk4u2ZbaeWe9Ctc1atXz918882EK5SbHjQ1eL1Tp07ukksu8TuodDmhZl1pyLt+1drSYe+ajdWyZUv/5AE1a9Z0TZs2dZ988olbsGBB4fB37czSA6g+rwlXdhCubCBc2UG4soNwZQPhyg7CVfx03qPzMJ3bV8S4lbIgXCVcloaz65NNK3c4u3ayXHjhhe6WW27xJwdZRbiqHOGDpmZk6XPro48+ctddd52rVauWO+WUU9yJJ564VcPetYNL87K09LL6dd999/WvV1FLw9/vvvtuf5mrPq8JV3YQrmwgXNlBuLKDcGUD4coOwlX8dA6mudU656mMkBRF/y/CVYJlJVzpwWTgwIF+5Q5nV7jSs7+1aNGCcIWtogdQ/ZAyZcoUf1mfBgbq1/xh7yeccEK5hr3nL8Ws3//+94XD3w8++GAfX2+77TbXtm1b17hxY38J43333ec6dOjg+vXr58Oa3j5kB+HKBsKVHYQrOwhXNhCu7CBcxU+jf3SOpXP7uM5xCVcJl5VwpR8IR44c6VfudbAKV9q5cuutt7oJEyYER7OHcPXbUjQKh71rRlZ5hr1r55VWbrzKXbpNlyXusccefjfWnnvu6X/V8HfN3tKzZj7zzDPuzTff9P/fXr16+c917UIkZqUX4coGwpUdhCs7CFc2EK7sIFzFL/z5SOf2cX3sCVcJl5VwtXbtWrds2TK/coez64FGl1vpfSRcIU66xFAnoyUNe69evbof2H7EEUf4XVWKU+XdnaWlHVramaX5W9rppUsY9fn+2WefueHDhxfuCguXvib0dhG1ko1wZQPhyg7ClR2EKxsIV3YQruIXnkfp3F5PwhYHwlXCZSVchcPb8oez64Hm3HPP9ZdZTZw4MTiaPYSrdNDnaTjsvXnz5q5Nmzbu2WefdQ8++KA7+eST/S6qnXfeuXDou+ZelbYbq6Sll9XMLc3eOv744/3SJYz6dXMD4JEMhCsbCFd2EK7sIFzZQLiyg3AVP52ThOf2On+KA+Eq4bISrjTjRyffWrnfQPRAo0u1CFdIAj0I68FXn6+6pFX/gqDf61d97r7yyivu8ssv9wPf9SyD2pWlge1bEq/0Mgpf+WtzA+CRDIQrGwhXdhCu7CBc2UC4soNwFT+dL61atcqfH+nKqjgQrhIuS8PZBwwY4FfucPZvvvnGnXXWWa5ly5Zu0qRJwdHsIVylWxi09HmswKpL+kaMGOG++uor17p1a3f66af7ywAPO+wwvytLM7P0q4a4b+nlhbkD4HWZoZ7EQI8FGvp+7733+ksbGQD/2yFc2UC4soNwZQfhygbClR2Eq/hp9I/mW+ncfvbs2cHRykW4SrgsDWcPZ/jkDmdXuDrzzDNdq1atCFdIlTBmTZ482fXu3dsPXn/ttdd8TOrSpYtfd955pzv11FP9rCwNf9flgVu6Q0uXFypkKYhpR5Zen36vGVz169d37du3d2+88YZ7++23/dLbo18VtebPnx/bNl4rCFc2EK7sIFzZQbiygXBlB+Eqfvq60nxqndvH9bEnXCVcli4V1A/+WrnfQBSuzjjjDHf77bf7by5ZRbiyQVFWD6h6YFUo0r9AKB61a9fO75DS57kik3Zk6VkIFaLCpZ1VilpbukNr1113dQceeKDf9aUV7gC76KKL3HPPPecGDRrkv7noX0fCiKxvOHob9TXJbq3yIVzZQLiyg3BlB+HKBsKVHYSr+Ok8Z+XKlf7nIy4VLI5wlWL65I4azj5lyhR32mmnuTvuuMPvXMkqwpUNueEqir4Opk2b5h5//HF31VVXuSuvvNJdccUVrkGDBu6yyy5ztWvXrrAB8OHSy+qywz//+c9+Ltdxxx3nB8Hr97r88IEHHvCfn3pWEH0DWr58uR8IHw6G16/h0u3aGkzkIlxZQbiyg3BlB+HKBsKVHYSr+OlcgOHsJSNcpZhKrE6ItXK/gShc6VIqzQkiXCHtNheu9CCvz399PaxZs6bI0uw3haH8AfB//OMft/jywtylnVz5g+C33357H8kOPvhg/wQJjz32mJ83p4Hwilr6tU6dOn5I/CWXXOIeffRRN2fOnNi+QSUZ4coGwpUdhCs7CFc2EK7sIFzFT+cCGsyuc3s9iVUcCFcJl/Xh7IpVp5xyip8FpIiVVYQrGzYXrkqjqKVvAvpayR0A379//yID4DXTSpca6tetGQAfLgUx7ezSHC69Xv2qyxbDyxfDSxk1n0v/f+0S09uTPyT+xRdfdGPHji22qzKrCFc2EK7sIFzZQbiygXBlB+EqfgxnLx3hKsUWLVrkRo8e7VfucHaFq5NPPplwhUzYmnAVJYxZuQPg33333cLh61ED4BWztLRTa5dddvHzr6KC1ZYsRa5q1aq5vfbay/8/cofE6xLE6667znXu3LnIgPhwSHz466effuqmTp3q3680By7ClQ2EKzsIV3YQrmwgXNlBuIqfvq70D+06t9eTQMWBcJVwWRrOru2EWrnfQHRCXqtWLXfXXXf5Qe1ZRbiyoaLD1eYo/uQPgNfuJy09AN90001+R6PCUu4QeP2qSwR1qWDuJYhbczmiXnaHHXbwO7PCAfHhkPhwh5j+fM4557h77rnH7yQLh8SHa8yYMT4G6eOY9KhFuLKBcGUH4coOwpUNhCs7CFfx0znIqlWr/Lk9w9mLI1ylmD65o4az6xvKiSee6Nq2bUu4QurFHa5Ko68z/bCmx5BWrVr5IfAaCK/5WbrcT/OratSo4SOWgpOelVDPdKhfdemgjoUD4hW4tuZyxNylwKWZXdoRphlexx9/vP9VA+MV2fR4p3+9WbhwoR8On9RB8YQrGwhXdhCu7CBc2UC4soNwFT/9/M1w9pIRrlJMJXbp0qV+6WQzpG8oOmG9++67M/0DI+HKhqSFK63169f7oYn5w+D1Nura9Ntvv90/m6F2ZykgN2nSxO+CVERSVNaQ+KOOOsrtueeeFX7ZYf6weL1+XYqoyw91CbHiWlIHxROubCBc2UG4soNwZQPhyg7CVfz087d+NtK5vc5/4kC4SrishKt58+b5S4O0coez6xuKdlzo0iHCFdIuSeFqcxS1tMX322+/9Tuc9PU3ffp0P2tOl+3pWHgJ37Bhw9xHH33kmjZt6kOzLv/LvQRw77339sFpay41zF3hsx3qssaSBsXrssMGDRr48JY7KF6/6s/6/UMPPeS6du3qd2tVdOAiXNlAuLKDcGUH4coGwpUdhKv4aSNK+CRSDGcvjnCVYhrOrk80rdwqO2nSJD/UWeFKA5uzinBlQ5rCVXkocmnXliJWz549iwxd17B4PaPgDTfc4KOW5mnlDonX78Mh7opPuvSwogKXLjnUTjD9f3IHxYf/3wMOOMCdccYZPmRpkL3e3txB8X369HFDhw51AwcOdO+9957r169fmXdxEa5sIFzZQbiyg3BlA+HKDsJV/PR1pfN4nfcsWLAgOFq5CFcJl5VwpU8w7e7Qyv0Gok/4Y445xn+yEK6QdlkNV5uj69v1taw4pEikQfHhDqhwF5SO3Xrrre7cc891Bx10kN85pZla4U6qcLZWRUWtcOn1/f73v/cxS7u0cgfFn3322X7u16WXXurnfV100UWuffv2btCgQT7S6VLKcNdZuMJj2oH24Ycf+n9l0mOa4h6yh3BlB+HKDsKVDYQrOwhX8dM/8q5evdqf2+tJ2OJAuEq4rIQrfXLrG0f+CZ6eRlPzc3RiO23atOBo9hCubLAarspKgUuXHT7wwAP+Mj9FI/2qwfEXX3yxH9KuHVS67FDD4cMB8ZU1KF5LYSuMZWHk+vOf/+x3j2k3qH4Nh8jnHtPj1mmnnebfF319a1dp7hD53GHyucf07Cv6Bk/oSgfClR2EKzsIVzYQruwgXMVPP8fq53p9fVX0OI6SEK4SLivhSif0OmnTyh3OHoYr7dIgXCHtCFel0zc5ff3ryRryh8XrX23mzp3r51JpELuGwmtpQHzuoHjt0KzoQfH5S2Esf3B81NLbsMsuu/hdXGeddZYfHq+3XQPkzzvvPP9reCwcMK8notBMAH2zR/IRruwgXNlBuLKBcGUH4Sp+ilUazK5ze/0cHwfCVcJlaTj7V1995VfucHaFK12io8uJNCQ6qwhXNhCutpyilp4BUV8r48aNK3J5Xu6g+OHDh5c6KF6XAOrPBx54oNttt918YIqKU1raYaVItTU7uPQ6tCNM/6/cIfK5w+Rzjx188ME+ZulxPbyMUr/mD5h/8803/Q9icf0rFqIRruwgXNlBuLKBcGUH4Sp++odo/Uyuc/tZs2YFRysX4SrhshKuVGPHjh3rl07uQwpXOtnU/JssDzomXNlAuKp8ClylDYoPh6+/8cYb7oknnvCRSFFLw9pzB8Vr5pUu+9NOqdq1a/vwpB1U4WWDlbXC0KWQlf/2hEPl9at2l918883uhRde8N9ItXLf19xf84998cUXZR4yj9IRruwgXNlBuLKBcGUH4Sp++rrS15fO7RnOXhzhKsW0k0KfbFq5l8hMmDDB75DQnBjCFdKOcJUcClza3dm7d28fsLSTKdzNpFCuHU56NsQePXr4b1S33HKLvyRR4Sh/p1TuCo9pFlZlhq4wcO21117ukEMO8fFNO8n0eBnuKos6pl91WeLTTz/tny0x3Kn2zTffuOnTp/tf9efcXWzhCo9p0L5+CFH40sfRMsKVHYQrOwhXNhCu7CBcxS/8OVvnPJURkqIQrhIu68PZFa50Uka4QhYQrtJJj0l6bNLjbevWrf2weA2Ov/zyy92VV17p/xwOkw+PnXnmmf5yREUsBabcAfLh73OPaemyxcre0aWl/4fC2v777184WL5hw4auZcuW/tdw0Lx+1e1auYPo9SyLinr6ITCcTRg1aD7q2PLly/3XgT6mWYhehCs7CFd2EK5sIFzZQbiKX/izs1ZcO/wJVwmXlXClExmd1GjlDmfXCYFmvjz44IN+N0BWEa5sIFylUxhZNFxS4SU/yOTHGf06fvx499prr/mdWrVq1SqMPopBGiSf/4yEWtWrV3c77rhjbPFKO8LCaLbzzjv7yyH1q/6cH9Vyj+k+2nl26qmn+mAXPvOjVhjuSjqm8Pfpp5/6H2QIV0gTwpUdhCsbCFd2EK7ip1jFcPaSEa5STMPZ+/Xr55eePSykE4KDDjrIP5PYd999FxzNHsKVDYQrO/SYpiHyelIJXWKXe+mdfj9y5Mgix/Tn/v37+7CjuVq6pE/z/cKh8rmX/O2xxx4+IMURuEpa2267rY9suZdH5v4+6piWgtdpp53mbrrpJv9DgC7N1GWZuYPoo47pz88//3zhMy4mJXoRruwgXNlBuLKBcGUH4Sp+4XB2ndsznL04wlWKqcbqJE9Lg5VDOiHQ5TYPP/ww4QqpR7iyQz8glefyZoUY/evU5MmT3SeffOIHqecOkw8Hq2sp4GgH07HHHls4sD1qiHv+Mf15l112cdtvv32x6KU/hyv3eGUs7fTadddd/dujlfv25b69+ceOOeYYd80117hOnTr5Z1XUDxC5Q+dzfy3LMX2cp06dulWzughXdhCu7CBc2UC4soNwFT99XelnWp3bL1y4MDhauQhXCZeVcKXh7BrgppU7nF2X2/z5z392jzzyiJsxY0ZwNHsIVzYQruwob7gqKwUWPUYOHjzYde7c2e9E0jD53J1JJR3Tn7XTSTueFINyd0TpHwi0syucy6VdXeHt4X10LBw6HxWk4liKbnp7tBNX8w/DofNlHU6fe0xzyO655x6/000/1OrvS4Pnw11x4W64cEUd0w6w119/3T9To/6s71l6PNf3tCxcColNCFd2EK5sIFzZQbiKn34G0iWCOrfXz0RxIFwlXFbCVUnD2XUSoBOsRx99lHCF1CNc2VFZ4Wpr6LFVj7GaMZU7ZF6zpxS4nn32Wf+r/hwe1320u0tzqnTs7LPP9vFH87By517l/r6kY5X5LItbsvS26HLHP/7xj65Jkyb+e2n9+vX93DHNH9McMv0+nEMWdUy/6uOh3WD6/Xnnnefuvvtu/0OyQod2E+fPP4uaiVaWY9rVtW7dOoLYb4RwZQfhygbClR2Eq/iFP3NqMZy9OMJViqnI6odCrZ9//jk4WhCudInIY4895uNOVhGubCBc2ZHUcKWlz8P8IfOKIqtWrfK/lhZQFixY4D788EPXuHHjwkHzYcAJw05Jx7R7VoPdt91228iQ9FssxSs9m6PeLl2+uNNOOxWLb+GKOqalXWDh7/Xy//Ef/+Hf/3r16vn4pxXGv3BgfXmP6VftohszZkyRXcmID+HKDsKVDYQrOwhX8VOs0s+N+t6pXVdxIFwlXFbC1dy5cyOHs+u6WP1rOOEKWUC4siOJ4aoiKHwtW7bMffPNN/7SuLJeUqdjgwYNcu3bt3cXXnihv1RPlyaGl/CFl/GVdGyfffZx1apVS9SOrZKW3kZFLIWw8DLL3F+39Jgu4zz//PP9M1Xqsk/9EJV7Gah2y+VeHpr7+1dffdV1797dvfHGG0Xuq9vyX04773QpapKG4CcB4coOwpUNhCs7CFfx03D2YcOGMZy9BISrFFOR1e4qrdzh7ApXOmF54okn3MyZM4Oj2UO4soFwZUdWw9XW0L++zZkzx8+D0nD0qOHzJR176aWXXIsWLVytWrX8LtzcIe76ff5g980dUwzSDqowhOnXcOWHqKSs3CCm90MrfN9y37/8Y/pVO9800+ukk04qcjz8fe7H6qijjvK7vZ577rlSh+BX9LHevXv7Qa5bMyy/MhGu7CBc2UC4soNwFT99XU2ZMsWf2zOcvTjCVYrp8kCd1GvlXgahcFW9enX35JNPEq6QeoQrOwhXFUsxQz94vvbaa35XkC6b06+5O4bKc+y2225zF1xwgR/yrqHz2tlVo0YNH28UtXQsd+dTSce0dJmhjuvXqGdsTMoqT5jT+6H3qSKG4Jf1mJ404I477vD/Oqun0Nbg+5EjR/ode+GvuWtzxyZOnOimT5/uvw51iWXu/cqyU1A/f+h7czhon3BlB+HKBsKVHYSr+On7pjaj6NwndwxQZSJcJVyWhrNrS6FW7r+0jh071u21117uqaeeim2b4W+BcGUD4coOwlWy6R9I9LX4wAMPuKuvvtr/44gukcsdWq9ZU5ovFc6aijqm35911lnukksucXXq1PG7lRS4NPQ9dyZW+PstOaalOVxJ3g1WESvcUaYZYYqIRx99tDv22GP9vDDNSdOuMf1eK5ydVtIxzVe76KKLXMuWLf1OvVNOOcUf08q9b2nH9EQE4aB9/WuxdoMNGTKkyMy33N9X5jGdYK9cudIv/Vlvj27X9xKddHNJZ8UiXNlAuLKDcBU/fV/S15bO7eOazUm4SrishCsNbdPQX63cKqtwteeee7qnn37azZ49OziaPYQrGwhXdhCukk0/UOkZ+jSMXiFAQUDD6fOH1uf/Pv+YAoJ2CGlnj36vsKHdXAolYUBRFNFw+tyoUp5j+lWzHrdmxpdeTvFr2wQNxi9p6W3MHXgfrvJGPw3K17NfaudY7v1y71PaMX28FdH096EoqR16ilmKllszaH9Ljl1//fWuY8eOrkOHDj60nnzyyT6W6hJafR7qH/9QcQhXNhCu7CBcxU/flxYtWuTP7XPnV1cmwlXCZWk4+5dffulX7ie3Pvn0r9eEK2QB4coOwpUNOuH5+uuvfQBTDNM/wihi6dK03EvPynJpWknHtAYMGODuuecev7tLl9VpYH14mV14qV1Jx/SroouCh3Yh5R7X/cKX06+aKangk4bAFccKd4GFzzq5yy67+MtDcy8ZDX9fmcf23ntv/3elvyf9TKTIpiinnWn6e9WTHmgWXJcuXfwP17o0tqTh+0k6pl2OCr9JeyIAwpUNhCs7CFfx006roUOH+nP7uMb9EK4SLkvD2fXDv1bucHZ98umHNv1QluWwQ7iygXBlB+HKhtxwVVl0Qq9/uZw6dar75JNP/DDz8gy2168aiK8fIBUJSruvdvC0atXK7xjb3BD3ijym7/OKMRV9OaReX0W/zqQt7QxT1Kpdu7bfnaePafgxLsvH/rc8pktr69ev7wPW66+/7p8M4K233vInB1r6fNUzXH744Yfu+eef90u36/M1/PzVr1999ZX//O7Tp0/h53J4e/7XQlmOPfTQQ65z585b9LLlOda3b18futkx99sgXNlBuIqfvq70DNT6GUkzIuNAuEq4rIQrVVkFK63cb+D65NO/Kj7zzDOEK6Qe4coOwpUNcYSrOOn773fffecDgnbGbO0Q/LIe0+WV9erVcwcffHCRnUbh2pJjCiTaRaaoo0v+cu+TP2g//5h2VmmnVdajVxLWdttt53ez/fnPf3YHHnhg4RMCaF144YX+0kk9G2b496TPEe0WDHcM6teLL77YX16pSznDHYW5g//DHYW5OxJLO7b//vv7/8/m7rc1x/R2nX/++f7zv3///v7JCLQmTJjgpk2b5meqaaflwIED/bNy6fuJ4nXUjswtefKCJB3TY2j4RAr6GOh91u5T/VnH9UQJuS+7NTtYw2MKVvoYT5o0yZ+DINsIV/HTP7qtXbvWn9sznL04wlWK6YdlfePQyt0ursst9AON5jroadSzinBlA+HKDsKVDVkLV78VXSqmE8iHH37YhwoNwtevmu8UznsqzzH9WYPZtZNH//DVsGFDf7uOh68/f65U7rG6deu6Y445ptigfc3eCn+fOxOrMo9paVdVGG7052233ZaoloGlv0N9fulSUO0+01LAbd68uf881IxXPUHReeed52699VbXrFmzwicS0K/h78O5eGk8pl2CupRZX686n9HHQO+zYqX+rOO6RFr3C+f+ha9ra47p4/vEE0/4y5g0g0ez4ipqRc1DDH/VHEXNU1yxYkWR2/NfLivHtJYuXerfZy3tctMxzT7SXEgtfUw0Z1LfR6NeviKOaf6knml2c/fbkmNa4ZzMZcuWlfnl9HHReBydAyfpUumKovdJPyPp3J7h7MURrlJMc0F0oqeVW2UVrvQvnxpKSrhC2hGu7CBc2UC4qhj6AVc/aOpkrqw/9Jd2TEsnEDqRCJ+JL/8+pb0uXdagnRm5g/b1LIfaMaMB+uEJcHgyXJnHtHTirks49faEu48UPDRwPwwgaRm+zyq6wr+7MFpqxpx+7v3973/v/z51m8JlOGMtN2zm/j7Nx/Q+K8rqfdSf9T7rNl1xoc91vf/hy+a+3NYc09w6zfTT17Vm/2nXnp7wIFyXXnqpv4xVv4ZLYVu/XnbZZf73+lXRW7/Xr+HvFcn1+zCM5x5r06aND+qKZorl4XGtML7rZbJyTOvGG2/077OWnmhCx/Qx15NMaGl+o87ztAs3fHn9Gr68XtfWHjvnnHP8P0hsyctu7pj+YUQjbfT+3XHHHf423Uf3De8Tdeymm27yu5vzrzbKCr1P+l6qn4fjOu8hXCVcloazawaHVu5wdoUrffPWg4Huk1WEKxsIV3YQrmwgXGWTQpoer8NB+7q0KJwvVhGXKZX3mC6V0tsSvj2a6aQfoHXiq50ruuQsavh+7mVzSTumZ8rc3BMBKOqUZWeZ7sMuNFZ5lz5nFLEUzLQUyqKWolr+r+EKd0Lm/j731/zfa76bvhZ0Kap2dIbHw5V73605Fv4+CceqV6/u32eFf+0u1DF9vBURtXRprm4/4IADyvw6y3tM0Vd/d5u735Yc06Xo4eOb/n7z75v7+9xjCqd6/G7SpIm7/fbbfdTUuvvuu/2vbdu2LVw6Fv4aLgU/HdOv4VJcCX8Nf69L8vV7/Rr+Xpfrh79q6Xh4+b5+Lc+xp556ynXv3t0v/V7Hwts0K1CzM/X9SpcBV/YTcRCuEi4r4UpbR7WFUyt3OLt+eNMPNs899xzhCqlHuLKDcGUD4coO/cux5qQkgX7w1+40Xf6ikwUN+S7L8P3f+lj4e/368ssv+11kp556auHg9twh7vpVA+fPPPNMf0KoHTJaCl5R99OuNJ0E6s86Ec4dAq/f5x7TKu2YLlXT/2dz99uaYzqJ1qWnUbGtpGNZDnMlvW/bsouQlZK1JV+f+vzWjkLFLwU+LQWt3F/zj4VLj1Hhr/m/1+NM/u/1a7jCx0X9Gq7w8am8xxTtzjjjDL/0+9z76fFdj+GaP6heoXi1bt264DtZxSNcJVxWwpV++NcnslbudkmFK22HV7iaN29ecDR7CFc2EK7sIFzZQLiyI0nhKgv0s56eCEAhS/96nzu4X7/XMZ0kKMxpTpkuq9ElR9pBEHW/Hj16uBdffLHIcf2rf7g7oDzHNEtKl/xsycuW9ViLFi18bNMuE+2+CHffKGppZ4oG1evnXy2dkOqYBtbn7tgIV9qP6aRacVInvQp6ep91Iq/3uUaNGoW7dMKX29yTK2zumP5BXE8KEBUS4lhZj5Alray/zxb/Tje3tKNOX8/6VY9fd911V6X+vES4KoG+4WpSvnYKacCcZink7hTKpfuGMxY0P0H/SpY7pEwznXS7Xo+WLpXTD8NlkZVwpY9H1HB2bZHXNt5OnToRrpB6hCs7CFc2EK7sIFzZoWec08/3lUk/9+qST4Uszb/RZZ/6NZyB9MADD/j5alq33HKLnwH02GOPFc7KyZ+bk+Zjinh6n/U+ar6U3udrrrnG/zmci5T78uHHSn/ekmPaAaJnr9TlY7lPuJA7A6uyjmnXYBjSdKy0+2blmEKh3l9Fw3BemT7ummWmFX5Mwhln5X39ZTmm/19Z7rclxxRmwiiq96W0+4a/6u3Z1sBuQgViLb2verIJzY2rzMdWwlUJFJcGDx7sH0jPPfdc/2wfms+UTxFGg8cfffRR/6Cpp/bVvwYpYIXxSk97qyF9GhynZw555ZVXynxZXFbClT6eGr6utX79+uDopnD1/PPP+xPBrCJc2UC4soNwZQPhyg7ClR1xhCudH+gfvPXYkfvkAOGzzul4+KxrevYx/QN37pMX5D6ZQNqPhc86p/dRx/U+67j+nPtMdLkvt7nXWdqx+fPnu/fee8+fl2mXl54AQU+GoCdHCH8fPkFCeY/pctWSjunPime6RPbmm2/2x3S7jmvpz7pvRRwL37YkHNO5rd5nrfAZIo888kh/Sa5202k+n549slGjRoUvr191P70Ova6tPabde3rGyop6fbnHFFrVAfT+aXi/jus+uq9u1591/9xjmvWn2KVwFxV80rYUpvS+5O460zHtIFUkVrTS7smHHnrIf11XFsJVCb799lv/LALa4qxgpX8V0IA07ZjK3U2lvxzNHmjdurW/b8+ePd3TTz/tL31TrNHt3bp185/w/fv3d7169fID1/SAWpbhZVkIV9pxph/89a8q2g6uIXUKgo0bN/aDRvVFoKinj5tmYOV+fLOCcGUD4coOwpUNhCs7CFd2xBGu8NvR+ZX+fjUXTieimruT+2QIWlv6RAqbO6bPLT3Jgs4jdSz3vvr9yJEjM3ds/PjxhU8uoSea0DF9zAcOHOjXlClT/G3ffPNN4cuHrytcW3tMM/Y++uijIsdy77Olx/R7PWGGft7T+zBp0qTC4/oYhPfLP6b5hG+88YYPmQo6ulRWcS0c8q6wpT+n4VitWrV8fNTS78Pjup/O5xUk1Uy0iUeXh+u8v7IQriKsWbPGDRgwwO+0UnDQDiH9Wdt7NRhTlxCGFCMUorp06eJmzpzpf7j9+OOP/TPAqPpPnjzZRyxFGf0ArN1ZqpHagaXLDzcnC+FKHzNtE9ZwTT0tqraRKlZpp5V+r2qr7aOq1ZoPoAf9rCFc2UC4soNwZQPhyg7ClR2EKxv0d6y/az2OI9v02J2kK3cUT7XTcNCgQf7JKkp6Qos0HPvggw98BNbS78PjWp999pnf4KNOMnXqVD8+qSwbc7YU4SqCfnjREMimTZv6Lb2iSqzL2Tp06FDkpFRfKPXq1XPDhw/32131lzV27FhfInU5YJ8+ffzL6NeQLhVUuNLrzKdIpllPeh0qtrre/brrrvM/NKd16dp9XWceXgNc0nA7bUE89thjfeSLej1pXvrXBn1BR93Gys7S9nf9K5O+eUbdzsrOUrTSv7xF3cbKztIPnvpXVF3mHnU7KztrxowZhZGSle2ln7FnzZoVeRsrO0t/x/q71uN41O2s7Cw9duvnsqjbWJWz1EjC+d2KxFH3qeil8yudZ+my4Kjbt2bdeeed6QxX2hmjN1zvgK43F+2Yef311/38qtxrN3Xiop1E2lkV7sSaMGGC3z2kbaKqke3bt/dbBkO6TPCpp57yPwzn0y4sPcOKLqO77LLL/OuuW7eu/4JM69L7ocFtGlSnFRWtwqWnAtX7HvV60rxUqYcNGxZ5Gys7S/8KoEuCta046nZWdpb+sUKP61G3sbKzdNmD/iVR36+jbmdlZ+mHYc02jbqNla2ly5f0j8NRt7Gys/R3rL9rPY5H3c7KztJjt34ui7qNVTlLl4jqfEc/HykQR92nopf+fzrPqoz/n+bSpTJczZ49u8RwpUvZ8sOVhrZFhSuV35LClXYVRYUrzXfSpYr61wFdaqjrY/UUwdoSl9Z1++23u/32288/TaaWdl1FRSvFLV1KqB1pUa8nzUufC/rX3KjbWNlZerzQ17W+fqNuZ2Vn6fuEdtxG3cbKztLlv/oBSf+6F3U7KztLu+R1WVHUbaxsLf2cvmDBgsjbWNlZ+jvW37Uex6NuZ2Vn6bFbP5dF3caqnKUnn1Af0TluXD8jhbvgdfVb1O1bszT6iUsFS7lUsCyzFLIw40qXyeljoAFuenpczbPSMxFovpWebULD3TQM7oILLnCdO3cu8zMupgkzrmxgxpUdzLiygRlXdjDjyg5mXNnAjCs79NidpBlXFvzrX//yG3M+//xzf54bB2ZcRcgdzq6B65sbzq5nG8wfzq6nBmU4ewG93/rYfPrpp+6TTz7xz9D45JNP+pj10ksvFQ6B07Mz6n4a7JY1hCsbCFd2EK5sIFzZQbiyg3BlA+HKDsJV/PR1NW3aNP81ph1XcSBcRdCuKf1FXHvttT6oaFq+YosClbadKq5oCLMCly4N0kT91q1b+3e2Z8+ePlI9++yzbvXq1f6ywq5du/pn1dM1mb169fKvR7GmLFP3sxCuRJdA6ukxtbIYpjaHcGUD4coOwpUNhCs7CFd2EK5sIFzZQbiKnzqGdl3p3F7n+XEgXJVAHxgN9Pv/27vTYLmqqo3jH/0oDh+wsJxKKZVRBTSAgRAMqExxwAGUAKIkYMAYhmgUwhwJMYEwiDKJOJOAqCQIyqCiSJREE5FBhcJAAgpoAGMR9lu/7T2p9nLVt8ri3E6f5191ir59TjcFa6+113r22rv322+/sssuu1ThSUeQ7iuik+4hv1bBaASqmTNnln333bdMmDCh/oc7Yb8xol+T8+uAY8eOrZ1Y8+bNq79Q9P8hwtVgEOGqG0S46g4RrrpBhKvuEOGqO0S46gYRrrpDhKv2iXD1n2lVuCKu2BJoux8RyrY+h5Axku1+XjNW86zOKs82P8/Ya0AGdb/5yUhCV/PZ/8agCFfO/9Kp5nouBlu/E+GqG0S46g4RrrpBhKvuEOGqO0S46gYRrrpDhKv2oXU4l1pt31aOFOGqzxkU4UqHmY41F+Gua0S46gYRrrpDhKtuEOGqO0S46g4RrrpBhKvuEOGqfTThLFmypNb26tw2iHDV5wyKcOXnKx1U7+o93L4rRLjqBhGuukOEq24Q4ao7RLjqDhGuukGEq+4Q4ap9+NXy5ctrbZ/D2Z9NhKuNGNspDXDX/+dQ+kEjwlU3iHDVHSJcdYMIV90hwlV3iHDVDSJcdYcIV+2jnm9q+7bOr45w1ecMknBlL6wrwlUYVCJcdYcIV90gwlV3iHDVHSJcdYMIV90hwlX7qOeb2j7C1bOJcLUR43D6lStX1uu5GGz9ToSrbhDhqjtEuOoGEa66Q4Sr7hDhqhtEuOoOEa7ah2CltlXbr1mzZujd55YIV33OoAhXBvbChQvr1cWiPsJVN4hw1R0iXHWDCFfdIcJVd4hw1Q0iXHWHCFfts27durJ48eJa2xOv2iDCVZ8zKMLV6tWry9KlS+uVw9nDoBLhqjtEuOoGEa66Q4Sr7hDhqhtEuOoOEa7ah1/Jg9X2q1atGnr3uSXCVZ8zKMJV7z7YnHEVBpUIV90hwlU3iHDVHSJcdYcIV90gwlV3iHDVPur55gzrnHH1bCJcbcTkcPYIV10gwlV3iHDVDSJcdYcIV90hwlU3iHDVHSJctU8OZ//PRLjaiHnkkUfKihUr6vVcDLZ+J8JVN4hw1R0iXHWDCFfdIcJVd4hw1Q0iXHWHCFftQ7C66667am2fw9mfTYSrjRiiTQ5nj3A16ES46g4RrrpBhKvuEOGqO0S46gYRrrpDhKv2yeHs/5lOCleXXHJJGTduXP0fuTFfp5xySpk8eXK95s2bN+Izg3ydcMIJZdasWSPeyzU41/z588u0adPK3LlzR7yfa3AuMW3mzJkj3ss1ONd5551Xpk+fXubMmTPi/VyDc51++ullxowZI97LNVjXcccdV2bPnj3ivVyDc7ExW4vjI93PNTiX2C0vG+lerufm4ldHHXVUre3bqnHVV+qsBQsWjHj/f7n23HPPcvbZZw8pMP87nRSulixZUg499NCqAubaeK/x48dXhxjpXq7BuQ477LCyxRZblEmTJo14P9fgXHvttVcZO3bsiPdyDc41derUss0225QDDjhgxPu5BueaOHFiGTNmzIj3cg3Wtd1225X9999/xHu5BudiY7YWx0e6n2twLrFbXjbSvVyDc6mv1FlTpkwZ8f7/cqnhFi1aNKTA/O90UrgKg8GJJ55YzjrrrKG/wqDyu9/9ruy6667l9ttvH3onDCpWZ0x0YbCx/XfvvfcuN95449A7YVAZlKMZwn/ngx/8YLnmmmuG/gqDChuztTgeBhuxW14WBhv1lTpr1apVQ+/0LxGuwkZLhKtuEOGqO0S46gYRrrpDhKvuEOGqG0S46g4RrrpBhKsQWiDCVTeIcNUdIlx1gwhX3SHCVXeIcNUNIlx1hwhX3SDCVQgtcOmll5ZvfOMbQ3+FQcWvUh199NFVwAqDjYTYwZRhsHnyySfLpz71qXLHHXcMvRMGlRtuuCELTB3h5JNPLj/96U+H/gqDChuztTgeBhuxO2L04KO+Umc98sgjQ+/0LxGuwkbLb37zm3LnnXcO/RUGlb/+9a/lhz/8YfnLX/4y9E4YVO655578dH4H8DPqN998c1mzZs3QO2FQue++++pP54fB59Zbby1/+tOfhv4Kgwobs7U4HgYbsVteFgYb9ZU666mnnhp6p3+JcBVCCCGEEEIIIYQQ+pIIVyGEEEIIIYQQQgihL4lwFUYVhzv+4Ac/KDNmzChHHnlk+fjHP14PAlyxYkVZv359vX/uueeWT3ziE/XQZq/Xrl1b79kmeNFFF5WjjjqqXscee2xZvHhx+fOf/zz07f/kmWeeKT//+c/LMcccU0455ZTy0EMPDd0JbfH000/XvdMXXHBBtSNbO+PG3nnvr1u3rvzsZz8rn/70p8vkyZPreLjpppvK3//+92rvn/zkJ9V27Dxt2rRy5pln1j3ZTVvr/fffX6644oo6TqZOnVpOOOGE8v3vfz+t7KMAe/E39uXPrnnz5tWWc77o/pe+9KUyffr0uqd+7ty51Wf59AMPPFCuvPLKamf3jBV2dc4ZW9p2NH/+/DoG2Nm/Q3vz448/PvRvD23BltrLL7nkkmpLPs3/vvnNb9YYy6dt+/zsZz9b7x1//PFlyZIl1WfFA+db8WN29jk+f9ttt2048NfnjQdnbLC375gzZ055+OGH61gJ7cFm7CU+8zs+zRbNeUZPPPFE+da3vlXnWPdPO+20up2oN/6ymTh96qmn1lhuq3/Db3/72zrvi/0+v3DhwvLggw8O3Q1t8uijj9YD9dmyycn83cTge++9t86v3j/uuOOq3YefdWR7v/n7Xe96V7nqqquetSXYvM7ePi/eh/ZhA37nrCoxmK1PP/30ajd2ZtPvfOc71UZsLY57/h//+MfQN/wTMdoc/pnPfKYsW7asxnaI5WK3z/ruz3/+8+XXv/51vRfaRX703e9+t9qy8ekvf/nLNYdmL7FazBZ7zeWXX355jen4/e9/X84///xy8MEH13Hi+uIXv1jjtznafWPIHO67vXauYXLv9mEPtvzc5z73L/ZQF/Nn87gcTK41ZcqUMnPmzJqj8Wl5uQPazcEudhbnzcXNWPDcXXfdVeOEHP2II46o48i83iYRrsKoIkm6/vrry+zZs8uCBQtqgSt4XnjhheWPf/xjufrqq2sgnTVrVnUir02mArFJ8itf+Ur9jAOdTayeue6664a+/Z/cfffdVfCaMGFCeec731kDbWgXAU+Cc/HFF9ck5+yzz64FzAEHHFDPSpDksBHbuyfoEigEVUkwmxK9zjnnnFrsEiwEZGNEAiYwH3LIITVRMo4Uuu4TQJtEKrTDY489Vm3Kp9mLfyqCzjjjjFr8fO9736sTp0lTAWuCNDkSMFevXl1+9KMfVX+WLPmM5y677LI6hiTORC9JsPHh84ceemhZvnx5EqWWIVwRqMRgfsdv+aYEl5j4y1/+ssZxSRCRQ+z2TwJms/DgxzXYmS1POumkmlg3goYCmZ29L2Z4hohpnES4ahdJ7dKlS6s/sjNbECLNtwQmyTA78WtJrYSWbfl7g8SZ777jHe8okyZN2vCLko2IIWaLGT5vzIgD8en2ITIRn/m0+M33DjvssBq3zaeKWr80pjiyWGieJXb0Ijfj75tvvnkdL+bpXtjWd/hlUfYO7aMYJSSxM392EabZVCHKP9nG32zNJy1S9ObP5gDxf7/99ivvf//76yK0edoYavI5Y8hlnlbkWngI7eL/uYVc8zNbmFcPP/zw6svEq69//es1ZrO3fEstpS4Tf+XgxsA+++xT52qX7+LThBCfN4bkecaQzxLGjCFCSmgP9vjDH/6wIe9u7PGRj3yk2umWW26pOZj6yhhgcyKke2pxdt1ll12qvdmZv/74xz+uczTEC8+b532/2O4zars2iXAVRhWTp8S3GfgcRODkOMSID33oQ1WwMFkqdLz2ns6L5nPNap/iVTA2yTb4PquFCiIFcoSr0YF4pDuD3Qgb/mavN77xjdU+kmOFkCAIk6YVPIUvG1L5deVIikzCVnnf/OY31yLY9wmme+yxR/0pV99NIFMIGUM+E9qDPxKg2JrAIHkhJkpcFSwmURMr+ytsFbNER6KURKnXXuwumTbBsiufZ2NdOZ4jcmy55Zbl2muvrcV1aA9FC9+ToPJttlawiLE6YRUuOub4Ktsobi1ASHjArxtR2fdIkN761rdW3zdmiNV8uumoMzZ8Jslw+/j/zraS4sZHickf+MAHatcV/zTvErf4vnuECYkyxAT+bjy8973vrbFeYWzM+E7vE634NTHUT+2LC74rtIuFInkVgZiPK4be97731Zit0LVST0BW6FigYHc5WwP78Xljg6DRLEKCvfm6OKAbgLAR4Wp0EEflUvJhNubTut/23Xff6reKU104bMzWbKrLgkjdfN7igvmZT/PhRrgyL8vh2bmJF3JzwncO+m4fcVXexDcbn5aPnXjiiVWkFqvFbPFWnCZqyJ/VZ4Srpkt2OOwqThhDnmVr40OTgDHE10N7sIc6yZzaa4+dd9655lPszSflWJ7VDWuBwcIxH1cvmXv5/HDkauL/xz72sZrHG0PiudzA6zaJcBX6CkGViEElFkhf85rX1ADIMTii4sZ7itzhcFgJNMdsUDz72yRqVSHCVf8ggWEPdpbUSGJNglAI66oiOA7HWDAmCBYCrKJWoazIlTApsCTZPquTS/AOo0ezKivplfxus802G4Qmk54tSG94wxvq9sLmeUmxZMt77CiJGo7niCY77bRT7cw0ZsLoouglQipWrep5zR8lOBKjppumQTJE1BCnbRseP358TYok0MbMnnvuWeO1sSD+R7TqD/iepFfctgXlLW95S+3GYH8JsdhOhJQQg32t9lt4agpawpVnLVbo3FJAwXhQKMkDbF0Iowvf1UFhTmUXHXMKYPOquZfddWPAuGBPK/0E6oMOOqjOzY1wJebL4YgcRAyr/xGu+gO2IzzpmDU/y83YvFk0EMf333//cumll9bnjQEdFzoz5GrEy0a4kp+bt4lfnrPg1CxYeR1GF7a2eNDEWbFazGY7PipP23XXXWs8J1wRrSwkm4fdNx58x3C8ZyGD+GEMpbtudGns8e53v7vO1wRm8Vau7B77yNH4cSNcWaSw1ZOtzcXiP9jd4pIY4HWTkxkzbRPhKvQVChYrcVZx/Fz6C1/4wlrYcjKXFvQXvehFI+6VF2wVS1ThBtsXdONwRIlxhKv+QMBTuLz61a+uKwLsZiW3CZL+KekxFoajrVXC++EPf7isXLmyjgvbknzHVlttVYURIpYuH8JIGF0kvBIfEyRffsUrXlF9m91gdehVr3pVLWggBvBVBRJbKppG+jl9CbVEebvttqtnoDXdO2F0ICjrirW69+1vf7suPtgK3OvTilUdsw2SYs/tuOOOVbwkYit0dNopcsRrBY/Ei0hCzGq+L4weEl/ChERY14W4qwun8Wn3t9122/K1r31tQ6xnQ/6vE68RrsRnNlVASZphHCmGJcmeCaOHokRXLAGZYEGQHjt27IZYy97srsiFhSexXnFkEWm4cGWsTJw4sc75jnyIcNU/6I7R5fj2t7+95shsbst+49PiLoGS+MxHLQryaYWxDvde4cplq/B73vOeusC4xRZb1O3BFiUyT48u/v87B4lt+J4YLVY3C3/sbTHCfCzHMkfLwV772tfW+Vjezc7DzxKG5zUIyAGMoSw0jS6NPdhXJx3Ric17c7IDDzywzrWEK9vBt95665qPGR9yM4ImodLik23hFhfVWjozLUJaqGi7zopwFfoCwdJkyDGaLWMC5vOf//zqcA0Eq0022eRfhCuf87fCmFghOaIUK3at7gqygmiEq/5AECRcmAy1rgqMVud6OzFAuCJm9aLd2QqvyVPrq2TL5Oi1wlZyvWjRoro6JJGyYp9EaXTg0yY0xYkzriRIfO9lL3vZhi1E0DX18pe/fINw5TMmSUmuApaIrWDqhc8raq0OWy2SiIXRg0+Lt/xVsuMsHGKElddeCFfErAbJssUIW1QkT7Zzi/cEaeKGgodPK3Sdo6GtvWlTD6MD/1So6poRi4kVjXDVIGGWANsG7kc32N2CkmctJPUKV8RKW5LEcHiPvxsPbB1GB/Omudm8bC61DZQNFaW9sLGODTirTDy2LVgM7xWuCGCKKN2z5gH2j3DVH8ihzM9sY87VTWHhiHDVy1577VXvE6DZjS/Lyfh+r3BF1PBZeZ1FDJdz0oyPzNWjB6HC/3/Cg7zKNnzd6uJ377a+RsAQxy0kiuGEZvMwu4oJw39Ugd119ZjDLWoYQ43oGdqn1x7mX3Ov7dvD4618zH3zrkVkoiU7E57lcLaR6sxSV/su3dXu2W7ovu/zfJtEuAqjjuDGMSQ4tvoRmKy6K34222yzDdu9OKLiyHvuwWquQodDKZg86zlOKsD6VRsJts4rSdTrX//6uuIrqQrtw9a29ylUiIoKVDa0VUR3ldcgPGo/N8E2KHIlSJIhyZMVAhOxhFgB5VnjyFgxyRoPxpSAHNpH0qPQsUVQgstOrte97nW1A1KyrDjSpeO95hfKehEHFLC+oxEr+DfhU/FL5FZcxcajB59W1DrrxvkHklxJK6GJwKyt3DPsJ8lxtsZw+LtYYJVf542tJoraN73pTfV9Y6U5986Ws5ydMTqwoaRVDLZIZB7l55JZB7nyQz6tmN1hhx3qXO55XRzEa2PEVgR2Fp915ei4Mj+L2c2/wxkr7D9STAjPPfzVYiARqjlIn8+xpxV3/m3uZW92d6YN5Fh8nm3N6dtvv309tFs3vK3A8jRdOrqyHA1g1d7KPsHDd4b2YUdCBSGDzxEVvccuTde6v8Vx3XK6siwsv+1tb9tQFOuiNQbEBaKWWO19ArfY7WLz5sys0D58mjCh/tHFTGyQL5tvxWqCltjNVgQPMV1sl295T17tImA4u85Y8T6MDz7M74kZcjLvhdFhuD3sUmFHzR1yMPkW1FtitlzN+Gh81WviM8HZrgc/cGaB2RzOz83vxoox5N/Re650G0S4CqOO4lRSI6Gxus5BwFn8woEOGgHWc157zz2OaIuYPdq2LBCtGockcjRFsyCtHVLCpduDINIIX6FdFKQCJ9GqdxuIbUEExqaTTrEqkfJsU/QqbHRcKFxtP/M+2FKhQ7iyJ9/7VuolSlZ+GsEjtAf/M3FazdE9QZyC8y10PVqldwCw5yROClkdlny6V4SSLBsDOquaYkkXncnYWEhH3ehjW58YzP8UNE0MJibrwGIjyZDn+CS/huca20mIiZpjxoypSbMkWgeAJIk4wu4KHsWQLk2idWgXsZVgLPll7yZW80tiVLMg5DmxXXHjnBNFrFVbHRcuW80Ikl4TOcVq3bXEbf5PIFEQK5qNmdA+xAsLf2ykI0MBC/a0au/sQUWPIpbdGzFax3xja92WflVQvmahSY5HuJCPuW/bmeJ43LhxVfwyF4R2YUOLDmK32GoObpA7EyfY2HPiON8nQhKU2bmx9W677Va3lhGw+L4uSsJls33YRQQ1b+vyCe0j32IPQgUbWtyH7nexmt3Ebs8RHMV0sV1MbuZp87D4rFYzXszh8jVjyCKiBQh1WBg92Ip4PJI95F5yMPVV85wcTa7GR9myqavUTXJ4W33VbewuHsjpm8VIOZmY79/TJhGuwqgiEEpoOINuK8qw4CkgKmS0H3MKCa72c6+tuiuAm24MiZHk2N8CbW/h2+D5bBUcXRSnik5JromRnV2KWu2pEh3nJ3hPIPWsw33Z0wqOyVWBY2sRO7sEX0Knw2B110myFLU+T9iwYtCsCoV24NP8VeFiUmQvNpX8EqBNnLqlbA2SKDc+bTXQff9UKPmMidUqj9Z03+s7dOYpfvhzM4YkV82EG9qDTxOSFTQK3cYefLZZ8bMaJxkmVvJp/2RLiw+2IXjefeOBcOVzbEnQUBBJvCTZOncUU4oivh/ag710NhMt+GJT5PBHtpDQsq04LimWzLJ9byHc0LtVkJ19j0ULsV0s93nztE6sRgQN7WFOJUbptCI09Pq0jnexmn3NtbYLsaXnhzN8q+BwFMzGjYWJ0D58T14ldhOgiJLsrCg1n+qs4tNszNbiuBxtpC7I4VsFCdx82vghSOreELv93Xv0R2gHPm3+1A3JThZ+2Zo4YQGi8Wmx1zzMp3XhuO/Z5nnzsCMdiM4Wm80Lum+MIQKmxoLeMZScrF38/5Y7y5F77WGOZg9zKp9WH8m11U18mpgsvqu51U8+oxa36KxrVn4GzSCOZJGX82u5mPEkxrdJhKswqihStRe/+MUvru2qVthtKxBIHbhswrT9ZPfdd6+tyV4TnogRzkvw7Etf+tJa4OjasAdfYjycCFejiwJE0ut8DAeyOxOD7VyCn0Cow8LfDnplS+KW4Kr7QnB03o1998aB8xf23nvvOumalCn/RJJmnFgRan4pI5Nnuyg+dcC94AUvqJ0V7MGutoYoViTLOiq8rwtSMqVrTpJkJd+qUBMH+KtYQAhTJBO5TaTGkHHSjCHdeJKr0B78jijJV1/5yldW327sIeHRDat7iq/qqvCcApdYJX7rrrVC74cUPGMbis4LiTAkSxIlgrTxwKcl1xImCXNoD4tC5lVnTuqsMN+ys/nYgpLOKAI0O/Fp24x0aIwkMPYKVxCfdV2K3+YF3y2Zto0wsbt9xGL2051OSG58WqFKRCZKiMvsxHfZfaRziyJc9TdECDnXpptuWnMrMZqd2YyIRcSwKMHGbC1GEzWa7rtehgtX5mp5ncOgxQN5GXHD1qIsJLaPjneNAS95yUtql6O5mK0tIsqt5OZ8nq3kZRYRiBjmWV3UOvKanMw40EHpO+XXcjJjyHEPTU5me3B+VbB92MOCn/y41x5yK7mYOVUnM/vzd7b0vs+pkTUOaBDg8z6nycA24mYBSe0sb/N5l++V6/lsm0S4CqMKZd4KjMKT6ttcCiITq0lQAcQZXV43Sr7ESPIjQDaf84zCeDjUZMkVgcOkGtpFkWvVjX0UKb22Fgwp/ApW9pT8SI4JID4naBoj3pc4NXZW+DRn3finZ9xzfpJVQaJoCp/24V8KWT7NFo2dFbISGb7IVuznfUkT0YqtjBG+33xOUaxNuemu48PuDR9DRA7fG9qDvSQ8w2OwSzeVAodIZYVWRyyf5pPsKCH2mp96nj97TgIkvkOB42/f7xnPGlfx6fbhn+K0bWNs2diZ0MzG7itk+DT/VAzx2ZEERiKGZ5utKvDa4oUY7/PsLDcI7WMulScpWBs7u6zA83f3+So7ic9sqfNyOOZtW4jkaSN1zon14kS2g44O5ku20SXZ5FUucVYuZh6XS7MxW7M5m43k00QOQlfvEQ7mZHFAvPB5Y6HX50N7mIvVTmzd69MWfi0EidVsxU5iOPG68Wljge3cM08bD3JzPm2ulpM1B3o332us+FxysnZhD/OxfKzXHmzGR82p8i65GL+UW3meT5vDG3/3GeNAzi7mNz7Pnvy6yd01l4ghcro2iXAVQgghhBBCCCGEEPqSCFchhBBCCCGEEEIIoS+JcBVCCCGEEEIIIYQQ+pIIVyGEEEIIIYQQQgihL4lwFUIIIYQQQgghhBD6kghXIYQQQgghhBBCCKEviXAVQgghhNASjz32WP3Z6dtuu6384he/qNeyZcvqT4s3P0P+3/DT1M1PmYcQQgghDDoRrkIIIYQQWuLGG28sz3ve88r2229fxowZU6999tmnzJs3rzz00EPl6aefHnpyZNavX1/uv//+cs0111TxK4QQQghh0IlwFUIIIYTQEjfffHPZdNNNq+iky+ree+8tV199dZk4cWI57bTTyj333DP05MgQtnRpzZgxo1x22WVD74YQQgghDC4RrkIIIYQQWoJwtdlmm5X77ruv/k2IWrVqVfnqV79axo8fXzuynnjiibJ06dIqTn3yk58s06ZNKxdffHHttHryySfLscceW3bYYYey++67l1mzZpVrr722rF27ttxwww3lrLPOqp+ZPn16WbRoURXHQgghhBA2ZiJchRBCCCG0xHDhCs628vdWW21VrrzyynoO1sqVK8uFF15YvvCFL5Q5c+aUk046qVx00UXlqaeeqn9PmDChHHjggeXyyy8vt956a+3gOvfcc8vs2bPLBRdcULceHnnkkeX666+vYlcIIYQQwsZKhKsQQgghhJYYSbjC448/XrbeeutyxRVX1IPXH3744XLHHXfUbYGLFy8uxxxzTDnooIOqyEWoOvroo6uwBYe1n3/++fWZ8847r4pYuq922223+r6zs0IIIYQQNlYiXIUQQgghtMS/E64effTR2nFly+CaNWuqOKWjauzYsWWnnXYqm2++edljjz1GFK5sLZw8eXLZcccd66HvO++8c7223XbbcsYZZ5S77767PhdCCCGEsDES4SqEEEIIoSVGEq50TK1YsaJsueWW9dcCb7/99npQ+/HHH1/fd7aVs6ycgfXvhKvDDz+8biH0/c61cj3wwAN126HPhBBCCCFsrES4CiGEEEJoieHC1bp168ry5cvrgeqTJk2qotUtt9xSD2YnRBGeli1bVqZMmVLGjRtXD3P/1a9+Vc+vcqYVCF/nnHNOPbTdGVnr168vzzzzTFm9enXdghjhKoQQQggbMxGuQgghhBBa4qabbiqbbLJJOfXUU+t5VPPnzy8nn3xymTp1au22crbVnXfeWYWogw8+uCxYsKAetD5x4sT6K4IEqQcffLAKXR/96Ec3HM7uLCzfSfDyGaLW3LlzqxBG2AohhBBC2FiJcBVCCCGE0BIOXCdAOWidMOWaPn16WbhwYfnb3/5Wn/FPXVeHHHJIPbvqzDPPrN1ULui68guDRxxxRD2Q/aqrripr164t1113Xf31weZ7iWG+R1dXCCGEEMLGSoSrEEIIIYQQQgghhNCXRLgKIYQQQgghhBBCCH1JhKsQQgghhBBCCCGE0JdEuAohhBBCCCGEEEIIfUmEqxBCCCGEEEIIIYTQl0S4CiGEEEIIIYQQQgh9SYSrEEIIIYQQQgghhNCXRLgKIYQQQgghhBBCCH1JhKsQQgghhBBCCCGE0IeU8n9dEB6odBOagwAAAABJRU5ErkJggg=="></p>
Note: to get detailed plots for each block use
```python
model.wells.show_blocks_dynamics()
```
Done!
|
github_jupyter
|
import sys
import numpy as np
import pandas as pd
from datetime import timedelta
sys.path.append('..')
from deepfield import Field
model = Field('../open_data/norne_simplified/norne_simplified.data').load()
(model.wells
.drop_incomplete()
.get_wellblocks(model.grid)
.drop_outside()
.apply_perforations()
.calculate_cf(rock=model.rock, grid=model.grid)
.compute_events(grid=model.grid))
model.calculate_rates()
model.wells.total_rates.head()
model.wells.cum_rates.head()
model.wells.show_rates()
model.wells.show_blocks_dynamics()
| 0.198841 | 0.839767 |
```
%matplotlib inline
```
# Pyplot tutorial
An introduction to the pyplot interface.
Intro to pyplot
===============
:mod:`matplotlib.pyplot` is a collection of command style functions
that make matplotlib work like MATLAB.
Each ``pyplot`` function makes
some change to a figure: e.g., creates a figure, creates a plotting area
in a figure, plots some lines in a plotting area, decorates the plot
with labels, etc.
In :mod:`matplotlib.pyplot` various states are preserved
across function calls, so that it keeps track of things like
the current figure and plotting area, and the plotting
functions are directed to the current axes (please note that "axes" here
and in most places in the documentation refers to the *axes*
`part of a figure <figure_parts>`
and not the strict mathematical term for more than one axis).
<div class="alert alert-info"><h4>Note</h4><p>the pyplot API is generally less-flexible than the object-oriented API.
Most of the function calls you see here can also be called as methods
from an ``Axes`` object. We recommend browsing the tutorials and
examples to see how this works.</p></div>
Generating visualizations with pyplot is very quick:
```
import matplotlib.pyplot as plt
plt.plot([1, 2, 3, 4])
plt.ylabel('some numbers')
plt.show()
```
You may be wondering why the x-axis ranges from 0-3 and the y-axis
from 1-4. If you provide a single list or array to the
:func:`~matplotlib.pyplot.plot` command, matplotlib assumes it is a
sequence of y values, and automatically generates the x values for
you. Since python ranges start with 0, the default x vector has the
same length as y but starts with 0. Hence the x data are
``[0,1,2,3]``.
:func:`~matplotlib.pyplot.plot` is a versatile command, and will take
an arbitrary number of arguments. For example, to plot x versus y,
you can issue the command:
```
plt.plot([1, 2, 3, 4], [1, 4, 9, 16])
```
Formatting the style of your plot
---------------------------------
For every x, y pair of arguments, there is an optional third argument
which is the format string that indicates the color and line type of
the plot. The letters and symbols of the format string are from
MATLAB, and you concatenate a color string with a line style string.
The default format string is 'b-', which is a solid blue line. For
example, to plot the above with red circles, you would issue
```
plt.plot([1, 2, 3, 4], [1, 4, 9, 16], 'ro')
plt.axis([0, 6, 0, 20])
plt.show()
```
See the :func:`~matplotlib.pyplot.plot` documentation for a complete
list of line styles and format strings. The
:func:`~matplotlib.pyplot.axis` command in the example above takes a
list of ``[xmin, xmax, ymin, ymax]`` and specifies the viewport of the
axes.
If matplotlib were limited to working with lists, it would be fairly
useless for numeric processing. Generally, you will use `numpy
<http://www.numpy.org>`_ arrays. In fact, all sequences are
converted to numpy arrays internally. The example below illustrates a
plotting several lines with different format styles in one command
using arrays.
```
import numpy as np
# evenly sampled time at 200ms intervals
t = np.arange(0., 5., 0.2)
# red dashes, blue squares and green triangles
plt.plot(t, t, 'r--', t, t**2, 'bs', t, t**3, 'g^')
plt.show()
```
Plotting with keyword strings
=============================
There are some instances where you have data in a format that lets you
access particular variables with strings. For example, with
:class:`numpy.recarray` or :class:`pandas.DataFrame`.
Matplotlib allows you provide such an object with
the ``data`` keyword argument. If provided, then you may generate plots with
the strings corresponding to these variables.
```
data = {'a': np.arange(50),
'c': np.random.randint(0, 50, 50),
'd': np.random.randn(50)}
data['b'] = data['a'] + 10 * np.random.randn(50)
data['d'] = np.abs(data['d']) * 100
plt.scatter('a', 'b', c='c', s='d', data=data)
plt.xlabel('entry a')
plt.ylabel('entry b')
plt.show()
```
Plotting with categorical variables
===================================
It is also possible to create a plot using categorical variables.
Matplotlib allows you to pass categorical variables directly to
many plotting functions. For example:
```
names = ['group_a', 'group_b', 'group_c']
values = [1, 10, 100]
plt.figure(1, figsize=(9, 3))
plt.subplot(131)
plt.bar(names, values)
plt.subplot(132)
plt.scatter(names, values)
plt.subplot(133)
plt.plot(names, values)
plt.suptitle('Categorical Plotting')
plt.show()
```
Controlling line properties
===========================
Lines have many attributes that you can set: linewidth, dash style,
antialiased, etc; see :class:`matplotlib.lines.Line2D`. There are
several ways to set line properties
* Use keyword args::
plt.plot(x, y, linewidth=2.0)
* Use the setter methods of a ``Line2D`` instance. ``plot`` returns a list
of ``Line2D`` objects; e.g., ``line1, line2 = plot(x1, y1, x2, y2)``. In the code
below we will suppose that we have only
one line so that the list returned is of length 1. We use tuple unpacking with
``line,`` to get the first element of that list::
line, = plt.plot(x, y, '-')
line.set_antialiased(False) # turn off antialising
* Use the :func:`~matplotlib.pyplot.setp` command. The example below
uses a MATLAB-style command to set multiple properties
on a list of lines. ``setp`` works transparently with a list of objects
or a single object. You can either use python keyword arguments or
MATLAB-style string/value pairs::
lines = plt.plot(x1, y1, x2, y2)
# use keyword args
plt.setp(lines, color='r', linewidth=2.0)
# or MATLAB style string value pairs
plt.setp(lines, 'color', 'r', 'linewidth', 2.0)
Here are the available :class:`~matplotlib.lines.Line2D` properties.
====================== ==================================================
Property Value Type
====================== ==================================================
alpha float
animated [True | False]
antialiased or aa [True | False]
clip_box a matplotlib.transform.Bbox instance
clip_on [True | False]
clip_path a Path instance and a Transform instance, a Patch
color or c any matplotlib color
contains the hit testing function
dash_capstyle [``'butt'`` | ``'round'`` | ``'projecting'``]
dash_joinstyle [``'miter'`` | ``'round'`` | ``'bevel'``]
dashes sequence of on/off ink in points
data (np.array xdata, np.array ydata)
figure a matplotlib.figure.Figure instance
label any string
linestyle or ls [ ``'-'`` | ``'--'`` | ``'-.'`` | ``':'`` | ``'steps'`` | ...]
linewidth or lw float value in points
lod [True | False]
marker [ ``'+'`` | ``','`` | ``'.'`` | ``'1'`` | ``'2'`` | ``'3'`` | ``'4'`` ]
markeredgecolor or mec any matplotlib color
markeredgewidth or mew float value in points
markerfacecolor or mfc any matplotlib color
markersize or ms float
markevery [ None | integer | (startind, stride) ]
picker used in interactive line selection
pickradius the line pick selection radius
solid_capstyle [``'butt'`` | ``'round'`` | ``'projecting'``]
solid_joinstyle [``'miter'`` | ``'round'`` | ``'bevel'``]
transform a matplotlib.transforms.Transform instance
visible [True | False]
xdata np.array
ydata np.array
zorder any number
====================== ==================================================
To get a list of settable line properties, call the
:func:`~matplotlib.pyplot.setp` function with a line or lines
as argument
.. sourcecode:: ipython
In [69]: lines = plt.plot([1, 2, 3])
In [70]: plt.setp(lines)
alpha: float
animated: [True | False]
antialiased or aa: [True | False]
...snip
Working with multiple figures and axes
======================================
MATLAB, and :mod:`~matplotlib.pyplot`, have the concept of the current
figure and the current axes. All plotting commands apply to the
current axes. The function :func:`~matplotlib.pyplot.gca` returns the
current axes (a :class:`matplotlib.axes.Axes` instance), and
:func:`~matplotlib.pyplot.gcf` returns the current figure
(:class:`matplotlib.figure.Figure` instance). Normally, you don't have
to worry about this, because it is all taken care of behind the
scenes. Below is a script to create two subplots.
```
def f(t):
return np.exp(-t) * np.cos(2*np.pi*t)
t1 = np.arange(0.0, 5.0, 0.1)
t2 = np.arange(0.0, 5.0, 0.02)
plt.figure(1)
plt.subplot(211)
plt.plot(t1, f(t1), 'bo', t2, f(t2), 'k')
plt.subplot(212)
plt.plot(t2, np.cos(2*np.pi*t2), 'r--')
plt.show()
```
The :func:`~matplotlib.pyplot.figure` command here is optional because
``figure(1)`` will be created by default, just as a ``subplot(111)``
will be created by default if you don't manually specify any axes. The
:func:`~matplotlib.pyplot.subplot` command specifies ``numrows,
numcols, plot_number`` where ``plot_number`` ranges from 1 to
``numrows*numcols``. The commas in the ``subplot`` command are
optional if ``numrows*numcols<10``. So ``subplot(211)`` is identical
to ``subplot(2, 1, 1)``.
You can create an arbitrary number of subplots
and axes. If you want to place an axes manually, i.e., not on a
rectangular grid, use the :func:`~matplotlib.pyplot.axes` command,
which allows you to specify the location as ``axes([left, bottom,
width, height])`` where all values are in fractional (0 to 1)
coordinates. See :doc:`/gallery/subplots_axes_and_figures/axes_demo` for an example of
placing axes manually and :doc:`/gallery/subplots_axes_and_figures/subplot_demo` for an
example with lots of subplots.
You can create multiple figures by using multiple
:func:`~matplotlib.pyplot.figure` calls with an increasing figure
number. Of course, each figure can contain as many axes and subplots
as your heart desires::
import matplotlib.pyplot as plt
plt.figure(1) # the first figure
plt.subplot(211) # the first subplot in the first figure
plt.plot([1, 2, 3])
plt.subplot(212) # the second subplot in the first figure
plt.plot([4, 5, 6])
plt.figure(2) # a second figure
plt.plot([4, 5, 6]) # creates a subplot(111) by default
plt.figure(1) # figure 1 current; subplot(212) still current
plt.subplot(211) # make subplot(211) in figure1 current
plt.title('Easy as 1, 2, 3') # subplot 211 title
You can clear the current figure with :func:`~matplotlib.pyplot.clf`
and the current axes with :func:`~matplotlib.pyplot.cla`. If you find
it annoying that states (specifically the current image, figure and axes)
are being maintained for you behind the scenes, don't despair: this is just a thin
stateful wrapper around an object oriented API, which you can use
instead (see :doc:`/tutorials/intermediate/artists`)
If you are making lots of figures, you need to be aware of one
more thing: the memory required for a figure is not completely
released until the figure is explicitly closed with
:func:`~matplotlib.pyplot.close`. Deleting all references to the
figure, and/or using the window manager to kill the window in which
the figure appears on the screen, is not enough, because pyplot
maintains internal references until :func:`~matplotlib.pyplot.close`
is called.
Working with text
=================
The :func:`~matplotlib.pyplot.text` command can be used to add text in
an arbitrary location, and the :func:`~matplotlib.pyplot.xlabel`,
:func:`~matplotlib.pyplot.ylabel` and :func:`~matplotlib.pyplot.title`
are used to add text in the indicated locations (see :doc:`/tutorials/text/text_intro`
for a more detailed example)
```
mu, sigma = 100, 15
x = mu + sigma * np.random.randn(10000)
# the histogram of the data
n, bins, patches = plt.hist(x, 50, density=1, facecolor='g', alpha=0.75)
plt.xlabel('Smarts')
plt.ylabel('Probability')
plt.title('Histogram of IQ')
plt.text(60, .025, r'$\mu=100,\ \sigma=15$')
plt.axis([40, 160, 0, 0.03])
plt.grid(True)
plt.show()
```
All of the :func:`~matplotlib.pyplot.text` commands return an
:class:`matplotlib.text.Text` instance. Just as with with lines
above, you can customize the properties by passing keyword arguments
into the text functions or using :func:`~matplotlib.pyplot.setp`::
t = plt.xlabel('my data', fontsize=14, color='red')
These properties are covered in more detail in :doc:`/tutorials/text/text_props`.
Using mathematical expressions in text
--------------------------------------
matplotlib accepts TeX equation expressions in any text expression.
For example to write the expression $\sigma_i=15$ in the title,
you can write a TeX expression surrounded by dollar signs::
plt.title(r'$\sigma_i=15$')
The ``r`` preceding the title string is important -- it signifies
that the string is a *raw* string and not to treat backslashes as
python escapes. matplotlib has a built-in TeX expression parser and
layout engine, and ships its own math fonts -- for details see
:doc:`/tutorials/text/mathtext`. Thus you can use mathematical text across platforms
without requiring a TeX installation. For those who have LaTeX and
dvipng installed, you can also use LaTeX to format your text and
incorporate the output directly into your display figures or saved
postscript -- see :doc:`/tutorials/text/usetex`.
Annotating text
---------------
The uses of the basic :func:`~matplotlib.pyplot.text` command above
place text at an arbitrary position on the Axes. A common use for
text is to annotate some feature of the plot, and the
:func:`~matplotlib.pyplot.annotate` method provides helper
functionality to make annotations easy. In an annotation, there are
two points to consider: the location being annotated represented by
the argument ``xy`` and the location of the text ``xytext``. Both of
these arguments are ``(x,y)`` tuples.
```
ax = plt.subplot(111)
t = np.arange(0.0, 5.0, 0.01)
s = np.cos(2*np.pi*t)
line, = plt.plot(t, s, lw=2)
plt.annotate('local max', xy=(2, 1), xytext=(3, 1.5),
arrowprops=dict(facecolor='black', shrink=0.05),
)
plt.ylim(-2, 2)
plt.show()
```
In this basic example, both the ``xy`` (arrow tip) and ``xytext``
locations (text location) are in data coordinates. There are a
variety of other coordinate systems one can choose -- see
`annotations-tutorial` and `plotting-guide-annotation` for
details. More examples can be found in
:doc:`/gallery/text_labels_and_annotations/annotation_demo`.
Logarithmic and other nonlinear axes
====================================
:mod:`matplotlib.pyplot` supports not only linear axis scales, but also
logarithmic and logit scales. This is commonly used if data spans many orders
of magnitude. Changing the scale of an axis is easy:
plt.xscale('log')
An example of four plots with the same data and different scales for the y axis
is shown below.
```
from matplotlib.ticker import NullFormatter # useful for `logit` scale
# Fixing random state for reproducibility
np.random.seed(19680801)
# make up some data in the interval ]0, 1[
y = np.random.normal(loc=0.5, scale=0.4, size=1000)
y = y[(y > 0) & (y < 1)]
y.sort()
x = np.arange(len(y))
# plot with various axes scales
plt.figure(1)
# linear
plt.subplot(221)
plt.plot(x, y)
plt.yscale('linear')
plt.title('linear')
plt.grid(True)
# log
plt.subplot(222)
plt.plot(x, y)
plt.yscale('log')
plt.title('log')
plt.grid(True)
# symmetric log
plt.subplot(223)
plt.plot(x, y - y.mean())
plt.yscale('symlog', linthreshy=0.01)
plt.title('symlog')
plt.grid(True)
# logit
plt.subplot(224)
plt.plot(x, y)
plt.yscale('logit')
plt.title('logit')
plt.grid(True)
# Format the minor tick labels of the y-axis into empty strings with
# `NullFormatter`, to avoid cumbering the axis with too many labels.
plt.gca().yaxis.set_minor_formatter(NullFormatter())
# Adjust the subplot layout, because the logit one may take more space
# than usual, due to y-tick labels like "1 - 10^{-3}"
plt.subplots_adjust(top=0.92, bottom=0.08, left=0.10, right=0.95, hspace=0.25,
wspace=0.35)
plt.show()
```
It is also possible to add your own scale, see `adding-new-scales` for
details.
|
github_jupyter
|
%matplotlib inline
import matplotlib.pyplot as plt
plt.plot([1, 2, 3, 4])
plt.ylabel('some numbers')
plt.show()
plt.plot([1, 2, 3, 4], [1, 4, 9, 16])
plt.plot([1, 2, 3, 4], [1, 4, 9, 16], 'ro')
plt.axis([0, 6, 0, 20])
plt.show()
import numpy as np
# evenly sampled time at 200ms intervals
t = np.arange(0., 5., 0.2)
# red dashes, blue squares and green triangles
plt.plot(t, t, 'r--', t, t**2, 'bs', t, t**3, 'g^')
plt.show()
data = {'a': np.arange(50),
'c': np.random.randint(0, 50, 50),
'd': np.random.randn(50)}
data['b'] = data['a'] + 10 * np.random.randn(50)
data['d'] = np.abs(data['d']) * 100
plt.scatter('a', 'b', c='c', s='d', data=data)
plt.xlabel('entry a')
plt.ylabel('entry b')
plt.show()
names = ['group_a', 'group_b', 'group_c']
values = [1, 10, 100]
plt.figure(1, figsize=(9, 3))
plt.subplot(131)
plt.bar(names, values)
plt.subplot(132)
plt.scatter(names, values)
plt.subplot(133)
plt.plot(names, values)
plt.suptitle('Categorical Plotting')
plt.show()
def f(t):
return np.exp(-t) * np.cos(2*np.pi*t)
t1 = np.arange(0.0, 5.0, 0.1)
t2 = np.arange(0.0, 5.0, 0.02)
plt.figure(1)
plt.subplot(211)
plt.plot(t1, f(t1), 'bo', t2, f(t2), 'k')
plt.subplot(212)
plt.plot(t2, np.cos(2*np.pi*t2), 'r--')
plt.show()
mu, sigma = 100, 15
x = mu + sigma * np.random.randn(10000)
# the histogram of the data
n, bins, patches = plt.hist(x, 50, density=1, facecolor='g', alpha=0.75)
plt.xlabel('Smarts')
plt.ylabel('Probability')
plt.title('Histogram of IQ')
plt.text(60, .025, r'$\mu=100,\ \sigma=15$')
plt.axis([40, 160, 0, 0.03])
plt.grid(True)
plt.show()
ax = plt.subplot(111)
t = np.arange(0.0, 5.0, 0.01)
s = np.cos(2*np.pi*t)
line, = plt.plot(t, s, lw=2)
plt.annotate('local max', xy=(2, 1), xytext=(3, 1.5),
arrowprops=dict(facecolor='black', shrink=0.05),
)
plt.ylim(-2, 2)
plt.show()
from matplotlib.ticker import NullFormatter # useful for `logit` scale
# Fixing random state for reproducibility
np.random.seed(19680801)
# make up some data in the interval ]0, 1[
y = np.random.normal(loc=0.5, scale=0.4, size=1000)
y = y[(y > 0) & (y < 1)]
y.sort()
x = np.arange(len(y))
# plot with various axes scales
plt.figure(1)
# linear
plt.subplot(221)
plt.plot(x, y)
plt.yscale('linear')
plt.title('linear')
plt.grid(True)
# log
plt.subplot(222)
plt.plot(x, y)
plt.yscale('log')
plt.title('log')
plt.grid(True)
# symmetric log
plt.subplot(223)
plt.plot(x, y - y.mean())
plt.yscale('symlog', linthreshy=0.01)
plt.title('symlog')
plt.grid(True)
# logit
plt.subplot(224)
plt.plot(x, y)
plt.yscale('logit')
plt.title('logit')
plt.grid(True)
# Format the minor tick labels of the y-axis into empty strings with
# `NullFormatter`, to avoid cumbering the axis with too many labels.
plt.gca().yaxis.set_minor_formatter(NullFormatter())
# Adjust the subplot layout, because the logit one may take more space
# than usual, due to y-tick labels like "1 - 10^{-3}"
plt.subplots_adjust(top=0.92, bottom=0.08, left=0.10, right=0.95, hspace=0.25,
wspace=0.35)
plt.show()
| 0.741487 | 0.987017 |
In this notebook, we will use a multi-layer perceptron to develop time series forecasting models.
The dataset used for the examples of this notebook is on air pollution measured by concentration of
particulate matter (PM) of diameter less than or equal to 2.5 micrometers. There are other variables
such as air pressure, air temparature, dewpoint and so on.
Two time series models are developed - one on air pressure and the other on pm2.5.
The dataset has been downloaded from UCI Machine Learning Repository.
https://archive.ics.uci.edu/ml/datasets/Beijing+PM2.5+Data
```
from __future__ import print_function
import os
import sys
import pandas as pd
import numpy as np
%matplotlib inline
from matplotlib import pyplot as plt
import seaborn as sns
import datetime
#set current working directory
os.chdir('D:/Practical Time Series')
#Read the dataset into a pandas.DataFrame
df = pd.read_csv('datasets/PRSA_data_2010.1.1-2014.12.31.csv')
print('Shape of the dataframe:', df.shape)
#Let's see the first five rows of the DataFrame
df.head()
```
To make sure that the rows are in the right order of date and time of observations,
a new column datetime is created from the date and time related columns of the DataFrame.
The new column consists of Python's datetime.datetime objects. The DataFrame is sorted in ascending order
over this column.
```
df['datetime'] = df[['year', 'month', 'day', 'hour']].apply(lambda row: datetime.datetime(year=row['year'], month=row['month'], day=row['day'],
hour=row['hour']), axis=1)
df.sort_values('datetime', ascending=True, inplace=True)
#Let us draw a box plot to visualize the central tendency and dispersion of PRES
plt.figure(figsize=(5.5, 5.5))
g = sns.boxplot(df['PRES'])
g.set_title('Box plot of PRES')
plt.figure(figsize=(5.5, 5.5))
g = sns.tsplot(df['PRES'])
g.set_title('Time series of PRES')
g.set_xlabel('Index')
g.set_ylabel('PRES readings')
```
Gradient descent algorithms perform better (for example converge faster) if the variables are wihtin range [-1, 1]. Many sources relax the boundary to even [-3, 3]. The PRES variable is mixmax scaled to bound the tranformed variable within [0,1].
```
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler(feature_range=(0, 1))
df['scaled_PRES'] = scaler.fit_transform(np.array(df['PRES']).reshape(-1, 1))
```
Before training the model, the dataset is split in two parts - train set and validation set.
The neural network is trained on the train set. This means computation of the loss function, back propagation
and weights updated by a gradient descent algorithm is done on the train set. The validation set is
used to evaluate the model and to determine the number of epochs in model training. Increasing the number of
epochs will further decrease the loss function on the train set but might not neccesarily have the same effect
for the validation set due to overfitting on the train set.Hence, the number of epochs is controlled by keeping
a tap on the loss function computed for the validation set. We use Keras with Tensorflow backend to define and train
the model. All the steps involved in model training and validation is done by calling appropriate functions
of the Keras API.
```
"""
Let's start by splitting the dataset into train and validation. The dataset's time period if from
Jan 1st, 2010 to Dec 31st, 2014. The first fours years - 2010 to 2013 is used as train and
2014 is kept for validation.
"""
split_date = datetime.datetime(year=2014, month=1, day=1, hour=0)
df_train = df.loc[df['datetime']<split_date]
df_val = df.loc[df['datetime']>=split_date]
print('Shape of train:', df_train.shape)
print('Shape of test:', df_val.shape)
#First five rows of train
df_train.head()
#First five rows of validation
df_val.head()
#Reset the indices of the validation set
df_val.reset_index(drop=True, inplace=True)
"""
The train and validation time series of scaled PRES is also plotted.
"""
plt.figure(figsize=(5.5, 5.5))
g = sns.tsplot(df_train['scaled_PRES'], color='b')
g.set_title('Time series of scaled PRES in train set')
g.set_xlabel('Index')
g.set_ylabel('Scaled PRES readings')
plt.figure(figsize=(5.5, 5.5))
g = sns.tsplot(df_val['scaled_PRES'], color='r')
g.set_title('Time series of scaled PRES in validation set')
g.set_xlabel('Index')
g.set_ylabel('Scaled PRES readings')
```
Now we need to generate regressors (X) and target variable (y) for train and validation. 2-D array of regressor and 1-D array of target is created from the original 1-D array of columm standardized_PRES in the DataFrames. For the time series forecasting model, Past seven days of observations are used to predict for the next day. This is equivalent to a AR(7) model. We define a function which takes the original time series and the number of timesteps in regressors as input to generate the arrays of X and y.
```
def makeXy(ts, nb_timesteps):
"""
Input:
ts: original time series
nb_timesteps: number of time steps in the regressors
Output:
X: 2-D array of regressors
y: 1-D array of target
"""
X = []
y = []
for i in range(nb_timesteps, ts.shape[0]):
X.append(list(ts.loc[i-nb_timesteps:i-1]))
y.append(ts.loc[i])
X, y = np.array(X), np.array(y)
return X, y
X_train, y_train = makeXy(df_train['scaled_PRES'], 7)
print('Shape of train arrays:', X_train.shape, y_train.shape)
X_val, y_val = makeXy(df_val['scaled_PRES'], 7)
print('Shape of validation arrays:', X_val.shape, y_val.shape)
```
The input to convolution layers must be of shape (number of samples, number of timesteps, number of features per timestep). In this case we are modeling only PRES hence number of features per timestep is one. Number of timesteps is seven and number of samples is same as the number of samples in X_train and X_val, which are reshaped to 3D arrays.
```
#X_train and X_val are reshaped to 3D arrays
X_train, X_val = X_train.reshape((X_train.shape[0], X_train.shape[1], 1)),\
X_val.reshape((X_val.shape[0], X_val.shape[1], 1))
print('Shape of arrays after reshaping:', X_train.shape, X_val.shape)
```
Now we define the MLP using the Keras Functional API. In this approach a layer can be declared as the input of the following layer at the time of defining the next layer.
```
from keras.layers import Dense
from keras.layers import Input
from keras.layers import Dropout
from keras.layers import Flatten
from keras.layers.convolutional import ZeroPadding1D
from keras.layers.convolutional import Conv1D
from keras.layers.pooling import AveragePooling1D
from keras.optimizers import SGD
from keras.models import Model
from keras.models import load_model
from keras.callbacks import ModelCheckpoint
#Define input layer which has shape (None, 7) and of type float32. None indicates the number of instances
input_layer = Input(shape=(7,1), dtype='float32')
```
ZeroPadding1D layer is added next to add zeros at the beginning and end of each series. Zeropadding ensure that the downstream convolution layer does not reduce the dimension of the output sequences. Pooling layer, added after the convolution layer is used to downsampling the input.
```
#Add zero padding
zeropadding_layer = ZeroPadding1D(padding=1)(input_layer)
```
The first argument of Conv1D is the number of filters, which determine the number of features in the output. Second argument indicates length of the 1D convolution window. The third argument is strides and represent the number of places to shift the convolution window. Lastly, setting use_bias as True, add a bias value during computation of an output feature. Here, the 1D convolution can be thought of as generating local AR models over rolling window of three time units.
```
#Add 1D convolution layer
conv1D_layer = Conv1D(64, 3, strides=1, use_bias=True)(zeropadding_layer)
```
AveragePooling1D is added next to downsample the input by taking average over pool size of three with stride of one timesteps. The average pooling in this case can be thought of as taking moving averages over a rolling window of three time units. We have used average pooling instead of max pooling to generate the moving averages.
```
#Add AveragePooling1D layer
avgpooling_layer = AveragePooling1D(pool_size=3, strides=1)(conv1D_layer)
```
The preceeding pooling layer returns 3D output. Hence before passing to the output layer, a Flatten layer is added. The Flatten layer reshapes the input to (number of samples, number of timesteps*number of features per timestep), which is then fed to the output layer
```
#Add Flatten layer
flatten_layer = Flatten()(avgpooling_layer)
dropout_layer = Dropout(0.2)(flatten_layer)
#Finally the output layer gives prediction for the next day's air pressure.
output_layer = Dense(1, activation='linear')(dropout_layer)
```
The input, dense and output layers will now be packed inside a Model, which is wrapper class for training and making
predictions. Mean squared error (MSE) is used as the loss function.
The network's weights are optimized by the Adam algorithm. Adam stands for adaptive moment estimation
and has been a popular choice for training deep neural networks. Unlike, stochastic gradient descent, adam uses
different learning rates for each weight and separately updates the same as the training progresses. The learning rate of a weight is updated based on exponentially weighted moving averages of the weight's gradients and the squared gradients.
```
ts_model = Model(inputs=input_layer, outputs=output_layer)
ts_model.compile(loss='mean_absolute_error', optimizer='adam')#SGD(lr=0.001, decay=1e-5))
ts_model.summary()
```
The model is trained by calling the fit function on the model object and passing the X_train and y_train. The training
is done for a predefined number of epochs. Additionally, batch_size defines the number of samples of train set to be
used for a instance of back propagation.The validation dataset is also passed to evaluate the model after every epoch
completes. A ModelCheckpoint object tracks the loss function on the validation set and saves the model for the epoch,
at which the loss function has been minimum.
```
save_weights_at = os.path.join('keras_models', 'PRSA_data_Air_Pressure_1DConv_weights.{epoch:02d}-{val_loss:.4f}.hdf5')
save_best = ModelCheckpoint(save_weights_at, monitor='val_loss', verbose=0,
save_best_only=True, save_weights_only=False, mode='min',
period=1)
ts_model.fit(x=X_train, y=y_train, batch_size=16, epochs=20,
verbose=1, callbacks=[save_best], validation_data=(X_val, y_val),
shuffle=True)
```
Prediction are made for the PRES from the best saved model. The model's predictions, which are on the standardized PRES, are inverse transformed to get predictions of original PRES.
```
best_model = load_model(os.path.join('keras_models', 'PRSA_data_Air_Pressure_1DConv_weights.16-0.0097.hdf5'))
preds = best_model.predict(X_val)
pred_PRES = np.squeeze(scaler.inverse_transform(preds))
from sklearn.metrics import r2_score
r2 = r2_score(df_val['PRES'].loc[7:], pred_PRES)
print('R-squared for the validation set:', round(r2, 4))
#Let's plot the first 50 actual and predicted values of PRES.
plt.figure(figsize=(5.5, 5.5))
plt.plot(range(50), df_val['PRES'].loc[7:56], linestyle='-', marker='*', color='r')
plt.plot(range(50), pred_PRES[:50], linestyle='-', marker='.', color='b')
plt.legend(['Actual','Predicted'], loc=2)
plt.title('Actual vs Predicted PRES')
plt.ylabel('PRES')
plt.xlabel('Index')
```
|
github_jupyter
|
from __future__ import print_function
import os
import sys
import pandas as pd
import numpy as np
%matplotlib inline
from matplotlib import pyplot as plt
import seaborn as sns
import datetime
#set current working directory
os.chdir('D:/Practical Time Series')
#Read the dataset into a pandas.DataFrame
df = pd.read_csv('datasets/PRSA_data_2010.1.1-2014.12.31.csv')
print('Shape of the dataframe:', df.shape)
#Let's see the first five rows of the DataFrame
df.head()
df['datetime'] = df[['year', 'month', 'day', 'hour']].apply(lambda row: datetime.datetime(year=row['year'], month=row['month'], day=row['day'],
hour=row['hour']), axis=1)
df.sort_values('datetime', ascending=True, inplace=True)
#Let us draw a box plot to visualize the central tendency and dispersion of PRES
plt.figure(figsize=(5.5, 5.5))
g = sns.boxplot(df['PRES'])
g.set_title('Box plot of PRES')
plt.figure(figsize=(5.5, 5.5))
g = sns.tsplot(df['PRES'])
g.set_title('Time series of PRES')
g.set_xlabel('Index')
g.set_ylabel('PRES readings')
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler(feature_range=(0, 1))
df['scaled_PRES'] = scaler.fit_transform(np.array(df['PRES']).reshape(-1, 1))
"""
Let's start by splitting the dataset into train and validation. The dataset's time period if from
Jan 1st, 2010 to Dec 31st, 2014. The first fours years - 2010 to 2013 is used as train and
2014 is kept for validation.
"""
split_date = datetime.datetime(year=2014, month=1, day=1, hour=0)
df_train = df.loc[df['datetime']<split_date]
df_val = df.loc[df['datetime']>=split_date]
print('Shape of train:', df_train.shape)
print('Shape of test:', df_val.shape)
#First five rows of train
df_train.head()
#First five rows of validation
df_val.head()
#Reset the indices of the validation set
df_val.reset_index(drop=True, inplace=True)
"""
The train and validation time series of scaled PRES is also plotted.
"""
plt.figure(figsize=(5.5, 5.5))
g = sns.tsplot(df_train['scaled_PRES'], color='b')
g.set_title('Time series of scaled PRES in train set')
g.set_xlabel('Index')
g.set_ylabel('Scaled PRES readings')
plt.figure(figsize=(5.5, 5.5))
g = sns.tsplot(df_val['scaled_PRES'], color='r')
g.set_title('Time series of scaled PRES in validation set')
g.set_xlabel('Index')
g.set_ylabel('Scaled PRES readings')
def makeXy(ts, nb_timesteps):
"""
Input:
ts: original time series
nb_timesteps: number of time steps in the regressors
Output:
X: 2-D array of regressors
y: 1-D array of target
"""
X = []
y = []
for i in range(nb_timesteps, ts.shape[0]):
X.append(list(ts.loc[i-nb_timesteps:i-1]))
y.append(ts.loc[i])
X, y = np.array(X), np.array(y)
return X, y
X_train, y_train = makeXy(df_train['scaled_PRES'], 7)
print('Shape of train arrays:', X_train.shape, y_train.shape)
X_val, y_val = makeXy(df_val['scaled_PRES'], 7)
print('Shape of validation arrays:', X_val.shape, y_val.shape)
#X_train and X_val are reshaped to 3D arrays
X_train, X_val = X_train.reshape((X_train.shape[0], X_train.shape[1], 1)),\
X_val.reshape((X_val.shape[0], X_val.shape[1], 1))
print('Shape of arrays after reshaping:', X_train.shape, X_val.shape)
from keras.layers import Dense
from keras.layers import Input
from keras.layers import Dropout
from keras.layers import Flatten
from keras.layers.convolutional import ZeroPadding1D
from keras.layers.convolutional import Conv1D
from keras.layers.pooling import AveragePooling1D
from keras.optimizers import SGD
from keras.models import Model
from keras.models import load_model
from keras.callbacks import ModelCheckpoint
#Define input layer which has shape (None, 7) and of type float32. None indicates the number of instances
input_layer = Input(shape=(7,1), dtype='float32')
#Add zero padding
zeropadding_layer = ZeroPadding1D(padding=1)(input_layer)
#Add 1D convolution layer
conv1D_layer = Conv1D(64, 3, strides=1, use_bias=True)(zeropadding_layer)
#Add AveragePooling1D layer
avgpooling_layer = AveragePooling1D(pool_size=3, strides=1)(conv1D_layer)
#Add Flatten layer
flatten_layer = Flatten()(avgpooling_layer)
dropout_layer = Dropout(0.2)(flatten_layer)
#Finally the output layer gives prediction for the next day's air pressure.
output_layer = Dense(1, activation='linear')(dropout_layer)
ts_model = Model(inputs=input_layer, outputs=output_layer)
ts_model.compile(loss='mean_absolute_error', optimizer='adam')#SGD(lr=0.001, decay=1e-5))
ts_model.summary()
save_weights_at = os.path.join('keras_models', 'PRSA_data_Air_Pressure_1DConv_weights.{epoch:02d}-{val_loss:.4f}.hdf5')
save_best = ModelCheckpoint(save_weights_at, monitor='val_loss', verbose=0,
save_best_only=True, save_weights_only=False, mode='min',
period=1)
ts_model.fit(x=X_train, y=y_train, batch_size=16, epochs=20,
verbose=1, callbacks=[save_best], validation_data=(X_val, y_val),
shuffle=True)
best_model = load_model(os.path.join('keras_models', 'PRSA_data_Air_Pressure_1DConv_weights.16-0.0097.hdf5'))
preds = best_model.predict(X_val)
pred_PRES = np.squeeze(scaler.inverse_transform(preds))
from sklearn.metrics import r2_score
r2 = r2_score(df_val['PRES'].loc[7:], pred_PRES)
print('R-squared for the validation set:', round(r2, 4))
#Let's plot the first 50 actual and predicted values of PRES.
plt.figure(figsize=(5.5, 5.5))
plt.plot(range(50), df_val['PRES'].loc[7:56], linestyle='-', marker='*', color='r')
plt.plot(range(50), pred_PRES[:50], linestyle='-', marker='.', color='b')
plt.legend(['Actual','Predicted'], loc=2)
plt.title('Actual vs Predicted PRES')
plt.ylabel('PRES')
plt.xlabel('Index')
| 0.628635 | 0.982757 |
```
# %load setup.py
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import matplotlib.ticker as mtick
from itertools import product
%run helpers.ipynb
params_suffstat = pd.read_csv('output/params_suffstat.csv')
params_sim = pd.read_csv('output/params_sim.csv')
params_full = pd.concat([params_suffstat, params_sim])
params_full = dict(params_full.values)
params_full
# Range of aggregate demand
ALPHA = np.arange(start=0.97, step=0.005, stop=1.03)
# Grid to search for equilibrium tightness x
x0 = np.arange(start=0.001, step=0.001, stop=2)
xad, Gad = np.empty(len(ALPHA)), np.empty(len(ALPHA))
G0 = params_full['GY_bar']*Y_func(x0, **params_full) # G such that G/Y=16.5%
for i, alpha in enumerate(ALPHA):
eva = find_eq(G0, x0, alpha, **params_full)
# Finding where AS = AD
ind = np.argmin(eva)
# Record equlibrium tightness and public expenditure
xad[i] = x0[ind]
Gad[i] = G0[ind]
ad = pd.DataFrame({'Y':Y_func(xad, **params_full),
'u':u_func(xad, **params_full),
'M':M_func(G=Gad, x=xad, **params_full),
'G/Y':Gad/Y_func(xad, **params_full)}, index=ALPHA)
ad_axes = ad.plot(subplots=True, layout=(2, 2), title=['Output', 'Unemployment', 'Output Multiplier', 'Public Expenditure'],
legend=False, figsize=(10, 10), grid=True, color='red')
ad_axes[0, 0].set(xlabel=r'$\alpha$', ylabel='Output = measured productivity')
ad_axes[0, 1].set(xlabel=r'$\alpha$', ylabel='Unemployment/idleness rate')
ad_axes[0, 1].yaxis.set_major_formatter(mtick.PercentFormatter(1.0, decimals=0))
ad_axes[1, 0].set(xlabel='Aggregate Demand', ylabel='Output multiplier (dY/dG)')
ad_axes[1, 1].set(xlabel='Aggregate Demand', ylabel='Public Expenditure, % of GDP')
ad_axes[1, 1].yaxis.set_major_formatter(mtick.PercentFormatter(1.0, decimals=0))
xoptimal, Goptimal = np.empty(len(ALPHA)), np.empty(len(ALPHA))
# We now use grid search over a 2-d grid to find eq G and x
GY0 = np.arange(start=0.07, step=0.0005, stop=0.25)
x1, GY1 = np.meshgrid(x0, GY0)
G1 = GY1*Y_func(x1, **params_full)
for i, alpha in enumerate(ALPHA):
eva = find_eq(G=G1, x=x1, alpha=alpha, **params_full)
# Finding all x's where AS = AD
ind = np.argmin(eva, axis=1)
x2 = x0[ind]
G2 = GY0*Y_func(x2, **params_full)
# Finding values of x and G for which the optimality equation is satisfied
eva = optimal_func(G=G2, x=x2, **params_full)
# Finding where AS = AD
ind = np.argmin(eva)
# Record equlibrium tightness and public expenditure
xoptimal[i] = x2[ind]
Goptimal[i] = G2[ind]
exact_opt = pd.DataFrame({'Y':Y_func(x=xoptimal, **params_full),
'u':u_func(x=xoptimal, **params_full),
'M':M_func(G=Goptimal, x=xoptimal, **params_full),
'G/Y':Goptimal/Y_func(x=xoptimal, **params_full)}, index=ALPHA)
ad_axes = ad.plot(subplots=True, layout=(2, 2), title=['Output', 'Unemployment', 'Output Multiplier', 'Public Expenditure'],
legend=False, figsize=(10, 10), grid=True, color='blue', label='G/Y = 15%')
exact_opt['Y'].plot(ax=ad_axes[0, 0], grid=True)
exact_opt['u'].plot(ax=ad_axes[0, 1], grid=True)
exact_opt['M'].plot(ax=ad_axes[1, 0], grid=True)
exact_opt['G/Y'].plot(ax=ad_axes[1, 1], grid=True)
ad_axes[0, 0].set(ylabel='Output = measured productivity')
ad_axes[0, 1].set(ylabel='Unemployment/idleness rate')
ad_axes[0, 1].yaxis.set_major_formatter(mtick.PercentFormatter(1.0, decimals=0))
ad_axes[1, 0].set(xlabel='Aggregate Demand', ylabel='Output multiplier (dY/dG)')
ad_axes[1, 1].set(xlabel='Aggregate Demand', ylabel='Public Expenditure, % of GDP')
ad_axes[0, 1].legend(['G/Y = 16.5%', 'Optimal G: Exact'])
ad_axes[1, 1].yaxis.set_major_formatter(mtick.PercentFormatter(1.0, decimals=0))
xsuffstat, Gsuffstat = np.empty(len(ALPHA)), np.empty(len(ALPHA))
for i, alpha in enumerate(ALPHA):
u = u_func(xad[i], **params_full)
# Calculate optimal stimulus with eq 23
suffstat0 = suffstat_func(u0=u, m=m_func(which='dlnxdlng', G=Gad[i], x=xad[i], **params_full), **params_full)
G0 = GY_func(GC=(1 + suffstat0)*params_full['GC_bar'])*Y_func(x0, **params_full)
eva = find_eq(G0, x0, alpha, **params_full)
# Finding where AS = AD
ind = np.argmin(eva)
# Record equlibrium tightness and public expenditure
xsuffstat[i] = x0[ind]
Gsuffstat[i] = G0[ind]
ss = pd.DataFrame({'Y':Y_func(x=xsuffstat, **params_full),
'u':u_func(x=xsuffstat, **params_full),
'M':M_func(G=Gsuffstat, x=xsuffstat, **params_full),
'G/Y':Gsuffstat/Y_func(x=xsuffstat, **params_full)}, index=ALPHA)
ad_axes = ad.plot(subplots=True, layout=(2, 2), title=['Output', 'Unemployment', 'Output Multiplier', 'Public Expenditure'],
legend=False, figsize=(10, 10), grid=True, color='blue', label='G/Y = 15%')
ss['Y'].plot(ax=ad_axes[0, 0], grid=True)
ss['u'].plot(ax=ad_axes[0, 1], grid=True)
ss['M'].plot(ax=ad_axes[1, 0], grid=True)
ss['G/Y'].plot(ax=ad_axes[1, 1], grid=True)
ad_axes[0, 0].set(ylabel='Output = measured productivity')
ad_axes[0, 1].set(ylabel='Unemployment/idleness rate')
ad_axes[0, 1].yaxis.set_major_formatter(mtick.PercentFormatter(1.0, decimals=0))
ad_axes[1, 0].set(xlabel='Aggregate Demand', ylabel='Output multiplier (dY/dG)')
ad_axes[1, 1].set(xlabel='Aggregate Demand', ylabel='Public Expenditure, % of GDP')
ad_axes[0, 1].legend(['G/Y = 16.5%', 'Optimal G: Sufficient Statistics'])
ad_axes[1, 1].yaxis.set_major_formatter(mtick.PercentFormatter(1.0, decimals=0))
ss_ax = ss['G/Y'].plot(label='Optimal G: Sufficient Statistics', grid=True)
ss_ax.set(xlabel='Aggregate Demand', ylabel='Public Expenditure, % of GDP')
ss_ax.yaxis.set_major_formatter(mtick.PercentFormatter(1.0, decimals=0))
exact_opt['G/Y'].plot(ax=ss_ax, grid=True, label='Optimal G: Exact Solution')
ss_ax.legend()
```
|
github_jupyter
|
# %load setup.py
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import matplotlib.ticker as mtick
from itertools import product
%run helpers.ipynb
params_suffstat = pd.read_csv('output/params_suffstat.csv')
params_sim = pd.read_csv('output/params_sim.csv')
params_full = pd.concat([params_suffstat, params_sim])
params_full = dict(params_full.values)
params_full
# Range of aggregate demand
ALPHA = np.arange(start=0.97, step=0.005, stop=1.03)
# Grid to search for equilibrium tightness x
x0 = np.arange(start=0.001, step=0.001, stop=2)
xad, Gad = np.empty(len(ALPHA)), np.empty(len(ALPHA))
G0 = params_full['GY_bar']*Y_func(x0, **params_full) # G such that G/Y=16.5%
for i, alpha in enumerate(ALPHA):
eva = find_eq(G0, x0, alpha, **params_full)
# Finding where AS = AD
ind = np.argmin(eva)
# Record equlibrium tightness and public expenditure
xad[i] = x0[ind]
Gad[i] = G0[ind]
ad = pd.DataFrame({'Y':Y_func(xad, **params_full),
'u':u_func(xad, **params_full),
'M':M_func(G=Gad, x=xad, **params_full),
'G/Y':Gad/Y_func(xad, **params_full)}, index=ALPHA)
ad_axes = ad.plot(subplots=True, layout=(2, 2), title=['Output', 'Unemployment', 'Output Multiplier', 'Public Expenditure'],
legend=False, figsize=(10, 10), grid=True, color='red')
ad_axes[0, 0].set(xlabel=r'$\alpha$', ylabel='Output = measured productivity')
ad_axes[0, 1].set(xlabel=r'$\alpha$', ylabel='Unemployment/idleness rate')
ad_axes[0, 1].yaxis.set_major_formatter(mtick.PercentFormatter(1.0, decimals=0))
ad_axes[1, 0].set(xlabel='Aggregate Demand', ylabel='Output multiplier (dY/dG)')
ad_axes[1, 1].set(xlabel='Aggregate Demand', ylabel='Public Expenditure, % of GDP')
ad_axes[1, 1].yaxis.set_major_formatter(mtick.PercentFormatter(1.0, decimals=0))
xoptimal, Goptimal = np.empty(len(ALPHA)), np.empty(len(ALPHA))
# We now use grid search over a 2-d grid to find eq G and x
GY0 = np.arange(start=0.07, step=0.0005, stop=0.25)
x1, GY1 = np.meshgrid(x0, GY0)
G1 = GY1*Y_func(x1, **params_full)
for i, alpha in enumerate(ALPHA):
eva = find_eq(G=G1, x=x1, alpha=alpha, **params_full)
# Finding all x's where AS = AD
ind = np.argmin(eva, axis=1)
x2 = x0[ind]
G2 = GY0*Y_func(x2, **params_full)
# Finding values of x and G for which the optimality equation is satisfied
eva = optimal_func(G=G2, x=x2, **params_full)
# Finding where AS = AD
ind = np.argmin(eva)
# Record equlibrium tightness and public expenditure
xoptimal[i] = x2[ind]
Goptimal[i] = G2[ind]
exact_opt = pd.DataFrame({'Y':Y_func(x=xoptimal, **params_full),
'u':u_func(x=xoptimal, **params_full),
'M':M_func(G=Goptimal, x=xoptimal, **params_full),
'G/Y':Goptimal/Y_func(x=xoptimal, **params_full)}, index=ALPHA)
ad_axes = ad.plot(subplots=True, layout=(2, 2), title=['Output', 'Unemployment', 'Output Multiplier', 'Public Expenditure'],
legend=False, figsize=(10, 10), grid=True, color='blue', label='G/Y = 15%')
exact_opt['Y'].plot(ax=ad_axes[0, 0], grid=True)
exact_opt['u'].plot(ax=ad_axes[0, 1], grid=True)
exact_opt['M'].plot(ax=ad_axes[1, 0], grid=True)
exact_opt['G/Y'].plot(ax=ad_axes[1, 1], grid=True)
ad_axes[0, 0].set(ylabel='Output = measured productivity')
ad_axes[0, 1].set(ylabel='Unemployment/idleness rate')
ad_axes[0, 1].yaxis.set_major_formatter(mtick.PercentFormatter(1.0, decimals=0))
ad_axes[1, 0].set(xlabel='Aggregate Demand', ylabel='Output multiplier (dY/dG)')
ad_axes[1, 1].set(xlabel='Aggregate Demand', ylabel='Public Expenditure, % of GDP')
ad_axes[0, 1].legend(['G/Y = 16.5%', 'Optimal G: Exact'])
ad_axes[1, 1].yaxis.set_major_formatter(mtick.PercentFormatter(1.0, decimals=0))
xsuffstat, Gsuffstat = np.empty(len(ALPHA)), np.empty(len(ALPHA))
for i, alpha in enumerate(ALPHA):
u = u_func(xad[i], **params_full)
# Calculate optimal stimulus with eq 23
suffstat0 = suffstat_func(u0=u, m=m_func(which='dlnxdlng', G=Gad[i], x=xad[i], **params_full), **params_full)
G0 = GY_func(GC=(1 + suffstat0)*params_full['GC_bar'])*Y_func(x0, **params_full)
eva = find_eq(G0, x0, alpha, **params_full)
# Finding where AS = AD
ind = np.argmin(eva)
# Record equlibrium tightness and public expenditure
xsuffstat[i] = x0[ind]
Gsuffstat[i] = G0[ind]
ss = pd.DataFrame({'Y':Y_func(x=xsuffstat, **params_full),
'u':u_func(x=xsuffstat, **params_full),
'M':M_func(G=Gsuffstat, x=xsuffstat, **params_full),
'G/Y':Gsuffstat/Y_func(x=xsuffstat, **params_full)}, index=ALPHA)
ad_axes = ad.plot(subplots=True, layout=(2, 2), title=['Output', 'Unemployment', 'Output Multiplier', 'Public Expenditure'],
legend=False, figsize=(10, 10), grid=True, color='blue', label='G/Y = 15%')
ss['Y'].plot(ax=ad_axes[0, 0], grid=True)
ss['u'].plot(ax=ad_axes[0, 1], grid=True)
ss['M'].plot(ax=ad_axes[1, 0], grid=True)
ss['G/Y'].plot(ax=ad_axes[1, 1], grid=True)
ad_axes[0, 0].set(ylabel='Output = measured productivity')
ad_axes[0, 1].set(ylabel='Unemployment/idleness rate')
ad_axes[0, 1].yaxis.set_major_formatter(mtick.PercentFormatter(1.0, decimals=0))
ad_axes[1, 0].set(xlabel='Aggregate Demand', ylabel='Output multiplier (dY/dG)')
ad_axes[1, 1].set(xlabel='Aggregate Demand', ylabel='Public Expenditure, % of GDP')
ad_axes[0, 1].legend(['G/Y = 16.5%', 'Optimal G: Sufficient Statistics'])
ad_axes[1, 1].yaxis.set_major_formatter(mtick.PercentFormatter(1.0, decimals=0))
ss_ax = ss['G/Y'].plot(label='Optimal G: Sufficient Statistics', grid=True)
ss_ax.set(xlabel='Aggregate Demand', ylabel='Public Expenditure, % of GDP')
ss_ax.yaxis.set_major_formatter(mtick.PercentFormatter(1.0, decimals=0))
exact_opt['G/Y'].plot(ax=ss_ax, grid=True, label='Optimal G: Exact Solution')
ss_ax.legend()
| 0.527803 | 0.551815 |
```
import clustertools as ctools
import numpy as np
from astropy.table import QTable
import matplotlib.pyplot as plt
```
# Loading and Advancing
**Loading**
To manually load a snapshot of a cluster, simply read in the file via your preferred method, declare a StarCluster with the appropriate units and origin, and add stars. For example, consider a snapshot of a star cluster in a file named 00000.dat with columns of stellar mass, position (x,y,z) in pc , and velocity (vx,vy,vz) in km/s. The origin of the system is the cluster's centre. A StarCluster can be initialized via:
```
m,x,y,z,vx,vy,vz=np.loadtxt('00000.dat',unpack=True)
cluster=ctools.StarCluster(units='pckms',origin='cluster')
cluster.add_stars(x,y,z,vx,vy,vz,m)
```
The snapshot can be quickly viewed using the ``starplot`` function
```
ctools.starplot(cluster)
```
When manually setting up a cluster, key cluster parameters will not be calculated unless the function ``key_params()`` is called.
```
cluster.analyze()
print('Total Number of Stars = ',cluster.ntot)
print('Total Mass = ',cluster.mtot)
print('Mean Mass = ',cluster.mmean)
print('Mean Radius = ',cluster.rmean)
print('Maximum Radius = ',cluster.rmax)
print('Half-mass radius = ',cluster.rm)
print('Projected Half-mass radius = ',cluster.rmpro)
print('10% Lagrange radius = ',cluster.r10)
print('Projected 10% Lagrange radius = ',cluster.r10pro)
```
Alternatively, several functions have been written to more easily load a snapshot of a cluster. For example, snapshot 00000.dat could more easily be loaded via:
```
cluster=ctools.load_cluster('snapshot',filename='00000.dat',units='pckms',origin='cluster')
```
When using load_cluster, key cluster parameters are instantly calculated.
```
print('Total Number of Stars = ',cluster.ntot)
print('Total Mass = ',cluster.mtot)
print('Mean Mass = ',cluster.mmean)
print('Mean Radius = ',cluster.rmean)
print('Maximum Radius = ',cluster.rmax)
print('Half-mass radius = ',cluster.rm)
print('Projected Half-mass radius = ',cluster.rmpro)
print('10% Lagrange radius = ',cluster.r10)
print('Projected 10% Lagrange radius = ',cluster.r10pro)
```
If the columns in your snapshot do not have the format mass,x,y,z,vx,vy,vz,i_d,kw, then use ``col_names`` and ``col_nums`` when using ``load_cluster``
```
cluster=ctools.load_cluster('snapshot',filename='00000.dat',units='pckms',origin='cluster',
col_names=["m", "x", "y", "z", "vx", "vy", "vz"],col_nums=[0, 1, 2, 3, 4, 5, 6],)
```
If the orbital information of your simulated cluster is known, and is contained in a file with columns of time, x, y, z, vx, vy, vz then either the filename can be given to the ``load_cluster`` command using the ``ofilename`` flag:
```
cluster=ctools.load_cluster('snapshot',filename='00000.dat',units='pckms',origin='cluster',
col_names=["m", "x", "y", "z", "vx", "vy", "vz"],
col_nums=[0, 1, 2, 3, 4, 5, 6], ofilename='orbit.dat')
```
Alternatively, orbital information can be read in and added separately. If the units in ``orbit.dat`` are not the same as ``cluster.units``, one can use the ``ounits`` flag in ``add_orbit``. They will be converted to cluster.units.
```
t,xgc,ygc,zgc,vxgc,vygc,vzgc=np.loadtxt('orbit.dat',unpack=True)
cluster.tphys=t
cluster.add_orbit(xgc,ygc,zgc,vxgc,vygc,vzgc,ounits='kpckms')
print(cluster.tphys,cluster.xgc,cluster.ygc,cluster.zgc,cluster.vxgc,cluster.vygc,cluster.vzgc)
```
Functions have also been written to automatically load snapshots from the commonly used codes NBODY6 and gyrfalcon. To load an NBODY6 snapshot from OUT33 and OUT9:
```
cluster=ctools.load_cluster('nbody6',wdir='./nbody6_sim/')
ctools.starplot(cluster)
```
``clustertools`` knows that when reading in NBODY6 snapshots, the units will be ``'nbody'`` units and the origin will be ``'cluster'``. For an NBODY6 simulation with stellar evolution, fort.82 and fort.83 will also be read.
Finally, for a gyrfalcon simulation. the first cluster snapshot can be read in using:
```
cluster=ctools.load_cluster('gyrfalcon',filename='cluster.nemo.dat')
ctools.starplot(cluster)
```
Note that since gyrfalcon does not have a default output filename like NBODY6, the filename must be specifified. Furthermore, gyrfalcon's output is a binary file that must be converted to ascii using NEMO's ``s2a`` command before it can be read in by ``clustertools``.
Finally, clusters can be read in from astropy tables aswell. The below example reads in an N-body simulation of Pal 5 in units of ``kpckms`` in Galactocentric coordinates. The snapshot corresponds to Pal 5's most recent pericentre pass.
```
data = QTable.read("pal5_rp.dat", format="ascii")
cluster = ctools.load_cluster('astropy_table',particles=data, units='kpckms',origin='galaxy')
ctools.starplot(cluster)
```
**Advancing**
In most cases, it is desirable to read more than just the first timestep of a simulation. ``clustertools`` has been set up to easily read in a snapshot, calculate key cluster parameters, and then advance to the next snapshot. An advance can easily be first reading in the cluster:
```
cluster=ctools.load_cluster(ctype='nbody6',wdir='./nbody6_sim/')
```
``advance_cluster`` will then move to the next timestep. One can then keep track of properties like time and mass to follow a cluster's evolution.
```
t=[]
m=[]
while cluster.ntot>0:
cluster.to_pckms()
t.append(cluster.tphys)
m.append(np.sum(cluster.m))
cluster=ctools.advance_cluster(cluster,wdir='./nbody6_sim/')
plt.plot(t,m)
plt.xlabel(r'$ \rm Time \ (Myr)$')
plt.ylabel(r'$ \rm Mass \ (M_{\odot})$')
```
For the previous case above where the first snapshot had a filename of 00000.dat, ``advance_cluster`` will look for a filename titled 00001.dat. If your filenames have a different naming convention, just specify some of the keyword arguments when loading the first snapshot. For example, if your snapshots are named snap_cluster_001.txt and snap_cluster_002.txt, they can be loaded via:
```
cluster=ctools.load_cluster('snapshot',snapbase='snap_cluster_',nsnap=1,nzfill=3,snapend='.txt',snapdir='',units='pckms',origin='cluster')
cluster=ctools.advance_cluster(cluster)
```
|
github_jupyter
|
import clustertools as ctools
import numpy as np
from astropy.table import QTable
import matplotlib.pyplot as plt
m,x,y,z,vx,vy,vz=np.loadtxt('00000.dat',unpack=True)
cluster=ctools.StarCluster(units='pckms',origin='cluster')
cluster.add_stars(x,y,z,vx,vy,vz,m)
ctools.starplot(cluster)
cluster.analyze()
print('Total Number of Stars = ',cluster.ntot)
print('Total Mass = ',cluster.mtot)
print('Mean Mass = ',cluster.mmean)
print('Mean Radius = ',cluster.rmean)
print('Maximum Radius = ',cluster.rmax)
print('Half-mass radius = ',cluster.rm)
print('Projected Half-mass radius = ',cluster.rmpro)
print('10% Lagrange radius = ',cluster.r10)
print('Projected 10% Lagrange radius = ',cluster.r10pro)
cluster=ctools.load_cluster('snapshot',filename='00000.dat',units='pckms',origin='cluster')
print('Total Number of Stars = ',cluster.ntot)
print('Total Mass = ',cluster.mtot)
print('Mean Mass = ',cluster.mmean)
print('Mean Radius = ',cluster.rmean)
print('Maximum Radius = ',cluster.rmax)
print('Half-mass radius = ',cluster.rm)
print('Projected Half-mass radius = ',cluster.rmpro)
print('10% Lagrange radius = ',cluster.r10)
print('Projected 10% Lagrange radius = ',cluster.r10pro)
cluster=ctools.load_cluster('snapshot',filename='00000.dat',units='pckms',origin='cluster',
col_names=["m", "x", "y", "z", "vx", "vy", "vz"],col_nums=[0, 1, 2, 3, 4, 5, 6],)
cluster=ctools.load_cluster('snapshot',filename='00000.dat',units='pckms',origin='cluster',
col_names=["m", "x", "y", "z", "vx", "vy", "vz"],
col_nums=[0, 1, 2, 3, 4, 5, 6], ofilename='orbit.dat')
t,xgc,ygc,zgc,vxgc,vygc,vzgc=np.loadtxt('orbit.dat',unpack=True)
cluster.tphys=t
cluster.add_orbit(xgc,ygc,zgc,vxgc,vygc,vzgc,ounits='kpckms')
print(cluster.tphys,cluster.xgc,cluster.ygc,cluster.zgc,cluster.vxgc,cluster.vygc,cluster.vzgc)
cluster=ctools.load_cluster('nbody6',wdir='./nbody6_sim/')
ctools.starplot(cluster)
cluster=ctools.load_cluster('gyrfalcon',filename='cluster.nemo.dat')
ctools.starplot(cluster)
data = QTable.read("pal5_rp.dat", format="ascii")
cluster = ctools.load_cluster('astropy_table',particles=data, units='kpckms',origin='galaxy')
ctools.starplot(cluster)
cluster=ctools.load_cluster(ctype='nbody6',wdir='./nbody6_sim/')
t=[]
m=[]
while cluster.ntot>0:
cluster.to_pckms()
t.append(cluster.tphys)
m.append(np.sum(cluster.m))
cluster=ctools.advance_cluster(cluster,wdir='./nbody6_sim/')
plt.plot(t,m)
plt.xlabel(r'$ \rm Time \ (Myr)$')
plt.ylabel(r'$ \rm Mass \ (M_{\odot})$')
cluster=ctools.load_cluster('snapshot',snapbase='snap_cluster_',nsnap=1,nzfill=3,snapend='.txt',snapdir='',units='pckms',origin='cluster')
cluster=ctools.advance_cluster(cluster)
| 0.556159 | 0.896523 |
```
import torch
import numpy as np
from torch import nn, optim
import pandas as pd
from sklearn.preprocessing import StandardScaler, RobustScaler
from sklearn.model_selection import StratifiedKFold
import torch.nn.functional as F
import torchvision
from sklearn.metrics import accuracy_score, confusion_matrix,f1_score, precision_recall_curve, average_precision_score, roc_auc_score, confusion_matrix
import matplotlib.pyplot as plt
%matplotlib inline
plt.style.use('ggplot')
cuda = torch.cuda.is_available()
if cuda:
device = "cuda"
print("cuda available")
torch.cuda.get_device_name(0)
%%time
df_train = pd.read_csv('train.csv.zip')
df_test = pd.read_csv('test.csv.zip')
train_cols = [col for col in df_train.columns if col not in ['ID_code', 'target']]
y_train = df_train['target']
df_train.shape
ss = StandardScaler()
rs = RobustScaler()
df_train[train_cols] = ss.fit_transform(df_train[train_cols])
df_test[train_cols] = ss.fit_transform(df_test[train_cols])
df_train.head()
class classifier(nn.Module):
def __init__(self, hidden_dim, dropout = 0.3):
super().__init__()
self.fc1 = nn.Linear(200, hidden_dim)
self.fc2 = nn.Linear(hidden_dim, hidden_dim)
self.fc3 = nn.Linear(hidden_dim, 1)
self.dropout = nn.Dropout(p = dropout)
def forward(self,x):
x = self.dropout(F.relu(self.fc1(x)))
x = self.dropout(F.relu(self.fc2(x)))
x = self.fc3(x)
return x
model = classifier(100)
print(model)
del model
batch_size = 1000
train_x = torch.from_numpy(df_train[train_cols].values).float().cuda()
train_y = torch.from_numpy(y_train.values).float().cuda()
test_x = torch.from_numpy(df_test[train_cols].values).float().cuda()
type(train_x)
train_x_dataset = torch.utils.data.TensorDataset(train_x, train_y)
# te_x_dataset = torch.utils.data.TensorDataset(te_x)
trainloader = torch.utils.data.DataLoader(train_x_dataset, batch_size = batch_size, shuffle = True)
model = classifier(100)
model.cuda()
optimizer = optim.SGD(model.parameters(), lr = 0.005)
# criterion = nn.CrossEntropyLoss() # also number of outputs should be 2
criterion = nn.BCEWithLogitsLoss()
print(model)
x,y = next(iter(trainloader))
x
x.shape, x.view(x.shape[0],1,-1).shape, y.shape, y.view(-1,1).shape
o = model(x)
l = criterion(o, y.view(-1,1))
l
epochs = 2
train_losses, test_losses = [],[]
for e in range(epochs):
running_loss = 0
for step, (x_batch,y_batch) in enumerate(trainloader):
optimizer.zero_grad()
# print(step)
# print(x_batch)
# print(y_batch)
model.train()
output = model(x_batch)
loss = criterion(output, y_batch.view(-1,1))
loss.backward()
optimizer.step()
running_loss += loss
if step % 100 ==0:
print(running_loss.item())
train_losses.append(running_loss.item())
print(train_losses)
with torch.no_grad():
model.eval()
pred_y = model(test_x)
print(pred_y.shape)
pred_y
pred_y = pred_y.cpu().numpy()
pred_y[:,0]
def sigmoid(x):
return 1 / (1 + np.exp(-x))
sigmoid(pred_y).squeeze().shape
pred_y = pred_y[:,1]
min(pred_y), max(pred_y)
(pred_y-min(pred_y))/(max(pred_y)-min(pred_y))
```
### NN with kfold
```
folds = StratifiedKFold(n_splits = 5, shuffle = True)
for n_fold, (train_idx, val_idx) in enumerate(folds.split(df_train, y_train)):
print(n_fold, train_idx.shape, val_idx.shape)
def sigmoid(x):
return 1/(1 + np.exp(-x))
%%time
oof = np.zeros(len(df_train))
predictions = np.zeros(len(df_test))
n_epochs = 30
test_x = torch.from_numpy(df_test[train_cols].values).float()#.cuda()
test_dataset = torch.utils.data.TensorDataset(test_x)
testloader = torch.utils.data.DataLoader(test_dataset, batch_size = batch_size, shuffle = False)
for n_fold, (train_idx, val_idx) in enumerate(folds.split(df_train, y_train)):
running_train_loss, running_val_loss = [],[]
val_loss_min = np.Inf
# print(i, train_idx.shape, val_idx.shape)
print("Fold number: ", n_fold+1)
train_x_fold = torch.from_numpy(df_train.iloc[train_idx][train_cols].values).float().cuda()
train_y_fold = torch.from_numpy(y_train[train_idx].values).float().cuda()
train_fold_dataset = torch.utils.data.TensorDataset(train_x_fold,train_y_fold)
trainloader = torch.utils.data.DataLoader(train_fold_dataset, batch_size = batch_size, shuffle = True)
val_x_fold = torch.from_numpy(df_train.iloc[val_idx][train_cols].values).float().cuda()
val_y_fold = torch.from_numpy(y_train[val_idx].values).float().cuda()
val_fold_dataset = torch.utils.data.TensorDataset(val_x_fold,val_y_fold )
valloader = torch.utils.data.DataLoader(val_fold_dataset, batch_size = batch_size, shuffle = False)
#Initiating model
model = classifier(100)
model.cuda()
optimizer = optim.Adam(model.parameters(), lr = 0.005)
# criterion = nn.CrossEntropyLoss() # also number of outputs should be 2
criterion = nn.BCEWithLogitsLoss()
for epoch in range(n_epochs):
train_loss = 0
for train_x_batch, train_y_batch in trainloader:
model.train()
optimizer.zero_grad()
output = model(train_x_batch)
loss = criterion(output, train_y_batch.view(-1,1))
loss.backward()
optimizer.step()
train_loss += loss.item()/len(trainloader)
with torch.no_grad():
val_loss = 0
model.eval()
val_preds = []
val_true = []
for i, (val_x_batch, val_y_batch) in enumerate(valloader):
val_output = model(val_x_batch)
val_loss += (criterion(val_output, val_y_batch.view(-1,1)).item())/len(valloader)
batch_output = sigmoid(val_output.cpu().numpy().squeeze())
try:
batch_output = list(batch_output)
except TypeError:
batch_output =[batch_output]
val_preds.extend(batch_output)
# batch_true = val_y_batch.cpu().numpy().squeeze()
# try:
# batch_true = list(batch_true)
# except TypeError:
# batch_true =[batch_true]
# val_true.extend(batch_true)
running_train_loss.append(train_loss)
running_val_loss.append(val_loss)
print("Epoch: {} Training loss: {:.6f} Validation Loss: {:.6f} Val_auc:{:.5f}".format(epoch+1,
train_loss,
val_loss,
roc_auc_score(y_train[val_idx].values,
val_preds))
)
if val_loss <= val_loss_min:
print("Validation loss decresed from {:.6f} ----> {:.6f} Saving Model".format(val_loss_min,val_loss))
torch.save(model.state_dict(), "san_cust_tran_torch.pt")
val_loss_min = val_loss
oof[val_idx] = val_preds
print("Fold {} metrics: Avg Training loss: {:.4f} Avg Validation Loss: {:.4f} Val_auc:{:.5f}".format(n_fold+1,
np.mean(running_train_loss),
np.mean(running_val_loss),
roc_auc_score(y_train[val_idx].values,
oof[val_idx])))
y_test_pred_fold = []
print("Saving test results for best model")
for (test_x_batch,) in testloader:
model.load_state_dict(torch.load("san_cust_tran_torch.pt"))
model.cpu()
test_output = model(test_x_batch)
test_batch_output = sigmoid(test_output.detach().numpy().squeeze())
try:
test_batch_output = list(test_batch_output)
except TypeError:
test_batch_output =[test_batch_output]
y_test_pred_fold.extend(test_batch_output)
predictions += np.array(y_test_pred_fold)/folds.n_splits
print("end of fold: ",n_fold+1,"\n")
plt.figure(figsize = (8,4))
plt.title("Train vs val loss on last epoch")
plt.plot(running_train_loss, label = "train")
plt.plot(running_val_loss, label = "val")
plt.legend()
plt.show()
sub = pd.DataFrame({'ID_code': df_test.ID_code.values,
'target': predictions})
sub.to_csv('sub_pytorch_simplenn.csv', index = False)
y_train[np.arange(0,10)].shape
model.state_dict()
for i, (val_x_batch, val_y_batch) in enumerate(valloader):
print(i, val_x_batch.shape, val_y_batch.shape)
sigmoid(val_output.cpu().numpy()[:,0])
a = 10
print(a)
b = np.arange(2)
print(b)
list(b)
[b]
```
|
github_jupyter
|
import torch
import numpy as np
from torch import nn, optim
import pandas as pd
from sklearn.preprocessing import StandardScaler, RobustScaler
from sklearn.model_selection import StratifiedKFold
import torch.nn.functional as F
import torchvision
from sklearn.metrics import accuracy_score, confusion_matrix,f1_score, precision_recall_curve, average_precision_score, roc_auc_score, confusion_matrix
import matplotlib.pyplot as plt
%matplotlib inline
plt.style.use('ggplot')
cuda = torch.cuda.is_available()
if cuda:
device = "cuda"
print("cuda available")
torch.cuda.get_device_name(0)
%%time
df_train = pd.read_csv('train.csv.zip')
df_test = pd.read_csv('test.csv.zip')
train_cols = [col for col in df_train.columns if col not in ['ID_code', 'target']]
y_train = df_train['target']
df_train.shape
ss = StandardScaler()
rs = RobustScaler()
df_train[train_cols] = ss.fit_transform(df_train[train_cols])
df_test[train_cols] = ss.fit_transform(df_test[train_cols])
df_train.head()
class classifier(nn.Module):
def __init__(self, hidden_dim, dropout = 0.3):
super().__init__()
self.fc1 = nn.Linear(200, hidden_dim)
self.fc2 = nn.Linear(hidden_dim, hidden_dim)
self.fc3 = nn.Linear(hidden_dim, 1)
self.dropout = nn.Dropout(p = dropout)
def forward(self,x):
x = self.dropout(F.relu(self.fc1(x)))
x = self.dropout(F.relu(self.fc2(x)))
x = self.fc3(x)
return x
model = classifier(100)
print(model)
del model
batch_size = 1000
train_x = torch.from_numpy(df_train[train_cols].values).float().cuda()
train_y = torch.from_numpy(y_train.values).float().cuda()
test_x = torch.from_numpy(df_test[train_cols].values).float().cuda()
type(train_x)
train_x_dataset = torch.utils.data.TensorDataset(train_x, train_y)
# te_x_dataset = torch.utils.data.TensorDataset(te_x)
trainloader = torch.utils.data.DataLoader(train_x_dataset, batch_size = batch_size, shuffle = True)
model = classifier(100)
model.cuda()
optimizer = optim.SGD(model.parameters(), lr = 0.005)
# criterion = nn.CrossEntropyLoss() # also number of outputs should be 2
criterion = nn.BCEWithLogitsLoss()
print(model)
x,y = next(iter(trainloader))
x
x.shape, x.view(x.shape[0],1,-1).shape, y.shape, y.view(-1,1).shape
o = model(x)
l = criterion(o, y.view(-1,1))
l
epochs = 2
train_losses, test_losses = [],[]
for e in range(epochs):
running_loss = 0
for step, (x_batch,y_batch) in enumerate(trainloader):
optimizer.zero_grad()
# print(step)
# print(x_batch)
# print(y_batch)
model.train()
output = model(x_batch)
loss = criterion(output, y_batch.view(-1,1))
loss.backward()
optimizer.step()
running_loss += loss
if step % 100 ==0:
print(running_loss.item())
train_losses.append(running_loss.item())
print(train_losses)
with torch.no_grad():
model.eval()
pred_y = model(test_x)
print(pred_y.shape)
pred_y
pred_y = pred_y.cpu().numpy()
pred_y[:,0]
def sigmoid(x):
return 1 / (1 + np.exp(-x))
sigmoid(pred_y).squeeze().shape
pred_y = pred_y[:,1]
min(pred_y), max(pred_y)
(pred_y-min(pred_y))/(max(pred_y)-min(pred_y))
folds = StratifiedKFold(n_splits = 5, shuffle = True)
for n_fold, (train_idx, val_idx) in enumerate(folds.split(df_train, y_train)):
print(n_fold, train_idx.shape, val_idx.shape)
def sigmoid(x):
return 1/(1 + np.exp(-x))
%%time
oof = np.zeros(len(df_train))
predictions = np.zeros(len(df_test))
n_epochs = 30
test_x = torch.from_numpy(df_test[train_cols].values).float()#.cuda()
test_dataset = torch.utils.data.TensorDataset(test_x)
testloader = torch.utils.data.DataLoader(test_dataset, batch_size = batch_size, shuffle = False)
for n_fold, (train_idx, val_idx) in enumerate(folds.split(df_train, y_train)):
running_train_loss, running_val_loss = [],[]
val_loss_min = np.Inf
# print(i, train_idx.shape, val_idx.shape)
print("Fold number: ", n_fold+1)
train_x_fold = torch.from_numpy(df_train.iloc[train_idx][train_cols].values).float().cuda()
train_y_fold = torch.from_numpy(y_train[train_idx].values).float().cuda()
train_fold_dataset = torch.utils.data.TensorDataset(train_x_fold,train_y_fold)
trainloader = torch.utils.data.DataLoader(train_fold_dataset, batch_size = batch_size, shuffle = True)
val_x_fold = torch.from_numpy(df_train.iloc[val_idx][train_cols].values).float().cuda()
val_y_fold = torch.from_numpy(y_train[val_idx].values).float().cuda()
val_fold_dataset = torch.utils.data.TensorDataset(val_x_fold,val_y_fold )
valloader = torch.utils.data.DataLoader(val_fold_dataset, batch_size = batch_size, shuffle = False)
#Initiating model
model = classifier(100)
model.cuda()
optimizer = optim.Adam(model.parameters(), lr = 0.005)
# criterion = nn.CrossEntropyLoss() # also number of outputs should be 2
criterion = nn.BCEWithLogitsLoss()
for epoch in range(n_epochs):
train_loss = 0
for train_x_batch, train_y_batch in trainloader:
model.train()
optimizer.zero_grad()
output = model(train_x_batch)
loss = criterion(output, train_y_batch.view(-1,1))
loss.backward()
optimizer.step()
train_loss += loss.item()/len(trainloader)
with torch.no_grad():
val_loss = 0
model.eval()
val_preds = []
val_true = []
for i, (val_x_batch, val_y_batch) in enumerate(valloader):
val_output = model(val_x_batch)
val_loss += (criterion(val_output, val_y_batch.view(-1,1)).item())/len(valloader)
batch_output = sigmoid(val_output.cpu().numpy().squeeze())
try:
batch_output = list(batch_output)
except TypeError:
batch_output =[batch_output]
val_preds.extend(batch_output)
# batch_true = val_y_batch.cpu().numpy().squeeze()
# try:
# batch_true = list(batch_true)
# except TypeError:
# batch_true =[batch_true]
# val_true.extend(batch_true)
running_train_loss.append(train_loss)
running_val_loss.append(val_loss)
print("Epoch: {} Training loss: {:.6f} Validation Loss: {:.6f} Val_auc:{:.5f}".format(epoch+1,
train_loss,
val_loss,
roc_auc_score(y_train[val_idx].values,
val_preds))
)
if val_loss <= val_loss_min:
print("Validation loss decresed from {:.6f} ----> {:.6f} Saving Model".format(val_loss_min,val_loss))
torch.save(model.state_dict(), "san_cust_tran_torch.pt")
val_loss_min = val_loss
oof[val_idx] = val_preds
print("Fold {} metrics: Avg Training loss: {:.4f} Avg Validation Loss: {:.4f} Val_auc:{:.5f}".format(n_fold+1,
np.mean(running_train_loss),
np.mean(running_val_loss),
roc_auc_score(y_train[val_idx].values,
oof[val_idx])))
y_test_pred_fold = []
print("Saving test results for best model")
for (test_x_batch,) in testloader:
model.load_state_dict(torch.load("san_cust_tran_torch.pt"))
model.cpu()
test_output = model(test_x_batch)
test_batch_output = sigmoid(test_output.detach().numpy().squeeze())
try:
test_batch_output = list(test_batch_output)
except TypeError:
test_batch_output =[test_batch_output]
y_test_pred_fold.extend(test_batch_output)
predictions += np.array(y_test_pred_fold)/folds.n_splits
print("end of fold: ",n_fold+1,"\n")
plt.figure(figsize = (8,4))
plt.title("Train vs val loss on last epoch")
plt.plot(running_train_loss, label = "train")
plt.plot(running_val_loss, label = "val")
plt.legend()
plt.show()
sub = pd.DataFrame({'ID_code': df_test.ID_code.values,
'target': predictions})
sub.to_csv('sub_pytorch_simplenn.csv', index = False)
y_train[np.arange(0,10)].shape
model.state_dict()
for i, (val_x_batch, val_y_batch) in enumerate(valloader):
print(i, val_x_batch.shape, val_y_batch.shape)
sigmoid(val_output.cpu().numpy()[:,0])
a = 10
print(a)
b = np.arange(2)
print(b)
list(b)
[b]
| 0.867148 | 0.692408 |
# Text Generation with LSTM
Recurrent neural networks are also known for their ability to generate text. As a result, the output of the neural network can be free-form text. In this section, we will see how to train an LSTM can on a textual document, such as classic literature, and learn to output new text that appears to be of the same form as the training material. If you train your LSTM on [Shakespeare](https://en.wikipedia.org/wiki/William_Shakespeare), it will learn to crank out new prose similar to what Shakespeare had written.
Don't get your hopes up. You are not going to teach your deep neural network to write the next [Pulitzer Prize for Fiction](https://en.wikipedia.org/wiki/Pulitzer_Prize_for_Fiction). The prose generated by your neural network will be nonsensical. However, it will usually be nearly grammatically and of a similar style as the source training documents.
A neural network generating nonsensical text based on literature may not seem useful at first glance. However, this technology gets so much interest because it forms the foundation for many more advanced technologies. The fact that the LSTM will typically learn human grammar from the source document opens a wide range of possibilities. You can use similar technology to complete sentences when a user is entering text. Simply the ability to output free-form text becomes the foundation of many other technologies. In the next part, we will use this technique to create a neural network that can write captions for images to describe what is going on in the picture.
### Additional Information
The following are some of the articles that I found useful in putting this section together.
* [The Unreasonable Effectiveness of Recurrent Neural Networks](http://karpathy.github.io/2015/05/21/rnn-effectiveness/)
* [Keras LSTM Generation Example](https://keras.io/examples/lstm_text_generation/)
### Character-Level Text Generation
There are several different approaches to teaching a neural network to output free-form text. The most basic question is if you wish the neural network to learn at the word or character level. In many ways, learning at the character level is the more interesting of the two. The LSTM is learning to construct its own words without even being shown what a word is. We will begin with character-level text generation. In the next module, we will see how we can use nearly the same technique to operate at the word level. We will implement word-level automatic captioning in the next module.
We begin by importing the needed Python packages and defining the sequence length, named **maxlen**. Time-series neural networks always accept their input as a fixed-length array. Because you might not use all of the sequence elements, it is common to fill extra elements with zeros. You will divide the text into sequences of this length, and the neural network will train to predict what comes after this sequence.
```
from tensorflow.keras.callbacks import LambdaCallback
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense
from tensorflow.keras.layers import LSTM
from tensorflow.keras.optimizers import RMSprop
from tensorflow.keras.utils import get_file
import numpy as np
import random
import sys
import io
import requests
import re
```
For this simple example, we will train the neural network on the classic children's book [Treasure Island](https://en.wikipedia.org/wiki/Treasure_Island). We begin by loading this text into a Python string and displaying the first 1,000 characters.
```
r = requests.get("https://data.heatonresearch.com/data/t81-558/text/"\
"treasure_island.txt")
raw_text = r.text
print(raw_text[0:1000])
```
We will extract all unique characters from the text and sort them. This technique allows us to assign a unique ID to each character. Because we sorted the characters, these IDs should remain the same. If we add new characters to the original text, then the IDs would change. We build two dictionaries. The first **char2idx** is used to convert a character into its ID. The second **idx2char** converts an ID back into its character.
```
processed_text = raw_text.lower()
processed_text = re.sub(r'[^\x00-\x7f]',r'', processed_text)
print('corpus length:', len(processed_text))
chars = sorted(list(set(processed_text)))
print('total chars:', len(chars))
char_indices = dict((c, i) for i, c in enumerate(chars))
indices_char = dict((i, c) for i, c in enumerate(chars))
```
We are now ready to build the actual sequences. Just like previous neural networks, there will be an $x$ and $y$. However, for the LSTM, $x$ and $y$ will both be sequences. The $x$ input will specify the sequences where $y$ are the expected output. The following code generates all possible sequences.
```
# cut the text in semi-redundant sequences of maxlen characters
maxlen = 40
step = 3
sentences = []
next_chars = []
for i in range(0, len(processed_text) - maxlen, step):
sentences.append(processed_text[i: i + maxlen])
next_chars.append(processed_text[i + maxlen])
print('nb sequences:', len(sentences))
sentences
print('Vectorization...')
x = np.zeros((len(sentences), maxlen, len(chars)), dtype=np.bool)
y = np.zeros((len(sentences), len(chars)), dtype=np.bool)
for i, sentence in enumerate(sentences):
for t, char in enumerate(sentence):
x[i, t, char_indices[char]] = 1
y[i, char_indices[next_chars[i]]] = 1
x.shape
y.shape
```
The dummy variables for $y$ are shown below.
```
y[0:10]
```
Next, we create the neural network. This neural network's primary feature is the LSTM layer, which allows the sequences to be processed.
```
# build the model: a single LSTM
print('Build model...')
model = Sequential()
model.add(LSTM(128, input_shape=(maxlen, len(chars))))
model.add(Dense(len(chars), activation='softmax'))
optimizer = RMSprop(lr=0.01)
model.compile(loss='categorical_crossentropy', optimizer=optimizer)
model.summary()
```
The LSTM will produce new text character by character. We will need to sample the correct letter from the LSTM predictions each time. The **sample** function accepts the following two parameters:
* **preds** - The output neurons.
* **temperature** - 1.0 is the most conservative, 0.0 is the most confident (willing to make spelling and other errors).
The sample function below is essentially performing a [softmax]() on the neural network predictions. This causes each output neuron to become a probability of its particular letter.
```
def sample(preds, temperature=1.0):
# helper function to sample an index from a probability array
preds = np.asarray(preds).astype('float64')
preds = np.log(preds) / temperature
exp_preds = np.exp(preds)
preds = exp_preds / np.sum(exp_preds)
probas = np.random.multinomial(1, preds, 1)
return np.argmax(probas)
```
Keras calls the following function at the end of each training Epoch. The code generates sample text generations that visually demonstrate the neural network better at text generation. As the neural network trains, the generations should look more realistic.
```
def on_epoch_end(epoch, _):
# Function invoked at end of each epoch. Prints generated text.
print("******************************************************")
print('----- Generating text after Epoch: %d' % epoch)
start_index = random.randint(0, len(processed_text) - maxlen - 1)
for temperature in [0.2, 0.5, 1.0, 1.2]:
print('----- temperature:', temperature)
generated = ''
sentence = processed_text[start_index: start_index + maxlen]
generated += sentence
print('----- Generating with seed: "' + sentence + '"')
sys.stdout.write(generated)
for i in range(400):
x_pred = np.zeros((1, maxlen, len(chars)))
for t, char in enumerate(sentence):
x_pred[0, t, char_indices[char]] = 1.
preds = model.predict(x_pred, verbose=0)[0]
next_index = sample(preds, temperature)
next_char = indices_char[next_index]
generated += next_char
sentence = sentence[1:] + next_char
sys.stdout.write(next_char)
sys.stdout.flush()
print()
```
We are now ready to train. It can take up to an hour to train this network, depending on how fast your computer is. If you have a GPU available, please make sure to use it.
```
# Ignore useless W0819 warnings generated by TensorFlow 2.0. Hopefully can remove this ignore in the future.
# See https://github.com/tensorflow/tensorflow/issues/31308
import logging, os
logging.disable(logging.WARNING)
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "3"
# Fit the model
print_callback = LambdaCallback(on_epoch_end=on_epoch_end)
model.fit(x, y,
batch_size=128,
epochs=60,
callbacks=[print_callback])
```
|
github_jupyter
|
from tensorflow.keras.callbacks import LambdaCallback
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense
from tensorflow.keras.layers import LSTM
from tensorflow.keras.optimizers import RMSprop
from tensorflow.keras.utils import get_file
import numpy as np
import random
import sys
import io
import requests
import re
r = requests.get("https://data.heatonresearch.com/data/t81-558/text/"\
"treasure_island.txt")
raw_text = r.text
print(raw_text[0:1000])
processed_text = raw_text.lower()
processed_text = re.sub(r'[^\x00-\x7f]',r'', processed_text)
print('corpus length:', len(processed_text))
chars = sorted(list(set(processed_text)))
print('total chars:', len(chars))
char_indices = dict((c, i) for i, c in enumerate(chars))
indices_char = dict((i, c) for i, c in enumerate(chars))
# cut the text in semi-redundant sequences of maxlen characters
maxlen = 40
step = 3
sentences = []
next_chars = []
for i in range(0, len(processed_text) - maxlen, step):
sentences.append(processed_text[i: i + maxlen])
next_chars.append(processed_text[i + maxlen])
print('nb sequences:', len(sentences))
sentences
print('Vectorization...')
x = np.zeros((len(sentences), maxlen, len(chars)), dtype=np.bool)
y = np.zeros((len(sentences), len(chars)), dtype=np.bool)
for i, sentence in enumerate(sentences):
for t, char in enumerate(sentence):
x[i, t, char_indices[char]] = 1
y[i, char_indices[next_chars[i]]] = 1
x.shape
y.shape
y[0:10]
# build the model: a single LSTM
print('Build model...')
model = Sequential()
model.add(LSTM(128, input_shape=(maxlen, len(chars))))
model.add(Dense(len(chars), activation='softmax'))
optimizer = RMSprop(lr=0.01)
model.compile(loss='categorical_crossentropy', optimizer=optimizer)
model.summary()
def sample(preds, temperature=1.0):
# helper function to sample an index from a probability array
preds = np.asarray(preds).astype('float64')
preds = np.log(preds) / temperature
exp_preds = np.exp(preds)
preds = exp_preds / np.sum(exp_preds)
probas = np.random.multinomial(1, preds, 1)
return np.argmax(probas)
def on_epoch_end(epoch, _):
# Function invoked at end of each epoch. Prints generated text.
print("******************************************************")
print('----- Generating text after Epoch: %d' % epoch)
start_index = random.randint(0, len(processed_text) - maxlen - 1)
for temperature in [0.2, 0.5, 1.0, 1.2]:
print('----- temperature:', temperature)
generated = ''
sentence = processed_text[start_index: start_index + maxlen]
generated += sentence
print('----- Generating with seed: "' + sentence + '"')
sys.stdout.write(generated)
for i in range(400):
x_pred = np.zeros((1, maxlen, len(chars)))
for t, char in enumerate(sentence):
x_pred[0, t, char_indices[char]] = 1.
preds = model.predict(x_pred, verbose=0)[0]
next_index = sample(preds, temperature)
next_char = indices_char[next_index]
generated += next_char
sentence = sentence[1:] + next_char
sys.stdout.write(next_char)
sys.stdout.flush()
print()
# Ignore useless W0819 warnings generated by TensorFlow 2.0. Hopefully can remove this ignore in the future.
# See https://github.com/tensorflow/tensorflow/issues/31308
import logging, os
logging.disable(logging.WARNING)
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "3"
# Fit the model
print_callback = LambdaCallback(on_epoch_end=on_epoch_end)
model.fit(x, y,
batch_size=128,
epochs=60,
callbacks=[print_callback])
| 0.625896 | 0.992349 |
# スキーム確認ツール for Navis(I1,I2,I3,I4,A26)
#### excelの全シートを読み込んでDFに格納
新しいフロートの生データ(テキスト版)からJAMSTECのデコード用DBに登録用のフィールドが存在するかを判定する
```
import os
import pandas as pd
import re
import termcolor
import Levenshtein # レーベンシュタイン距離ライブラリにある、ジャロ・ウインクラー距離を計算するのに使う
# jaro_dist = Levenshtein.jaro_winkler(srt1 , str2)
navis_excel = pd.read_excel('Navis.xlsx' , sheet_name=None) # sheet_name=Noneで全てのシート読み込む
```
## ジャロ・ウィンクラー距離法の関数
2つの文字列の類似度を返す。1で完全一致
#### Winkler, W. E. (1990). "String Comparator Metrics and Enhanced Decision Rules in the Fellegi-Sunter Model of Record Linkage". Proceedings of the Section on Survey Research Methods. American Statistical Association: 354–359.
```
def jaro_dist(str1,str2):
return Levenshtein.jaro_winkler(str1,str2)
```
#### DFに格納、検索しやすいようにカラムにも名前をつける
シート名が日本語でも指定すればちゃんと読める。
必要な列を抜き出してscheme,techという新しいDFを作る
```
#print(navis_excel['技術情報'].columns)
# df.loc[行名:列名]
# df.iloc[行番号:列番号]
msgcol = navis_excel['scheme'].iloc[:,1]
i12msg = navis_excel['scheme'].iloc[:,7] # 7列目がI1/I2のmsg , 8列目はlog。以降同様にカウントする
i3msg = navis_excel['scheme'].iloc[:,9]
i4msg = navis_excel['scheme'].iloc[:,12]
scheme = pd.concat([msgcol,i12msg,i3msg,i4msg], axis=1).rename(columns={'Unnamed: 1':'field_name' , 'Unnamed: 7':'i1i2a26' , 'Unnamed: 9':'i3X1A27' , 'Unnamed: 12':'i4'})
#print(len(scheme))
#print(scheme.iloc[7,:])
#print(scheme['i1i2a26'])
techcol = navis_excel['tech'].iloc[:,1]
i1msg = navis_excel['tech'].iloc[:,7]
i1log = navis_excel['tech'].iloc[:,8]
i2msg = navis_excel['tech'].iloc[:,9]
i2log = navis_excel['tech'].iloc[:,10]
i3msg = navis_excel['tech'].iloc[:,11]
i3log = navis_excel['tech'].iloc[:,12]
i3isus = navis_excel['tech'].iloc[:,13]
i4msg = navis_excel['tech'].iloc[:,14]
i4log = navis_excel['tech'].iloc[:,15]
tech = pd.concat([techcol,i1msg,i1log,i2msg,i2log,i3msg,i3log,i3isus,i4msg,i4log], axis=1).rename(columns={'Unnamed: 1':'field_name', 'Unnamed: 7':'i1msg' , 'Unnamed: 8':'i1log' , 'Unnamed: 9':'i2msg' , 'Unnamed: 10':'i2log' , 'Unnamed: 11':'i3msg' , 'Unnamed: 12':'i3log' , 'Unnamed: 13':'i3isus' , 'Unnamed: 14':'i4msg' , 'Unnamed: 15':'i4log'})
#print(tech.iloc[56,:])
#print(type(tech))
```
## 生データファイル読み込み
バイナリからアスキーへの変換はメーカー提供のツールによる
```
with open('i0394/0394.001.msg','r') as msg:
msgline = msg.readlines() # prof,techで使う
with open('i0394/0394.001.log','r') as log:
logline = log.readlines()
# msgline,loglineとも1行毎に読み込んでる。
# 安直にループさせて新しいDataFrameを作ることは出来ないので辞書リストに一度保存する(Pythonの仕様)
msgdata = {}
i = 0
for orig_msg in msgline:
regex = r'^\$' # 先頭が$ならその行をパラメータと判断する
regex2 = r'^[A-Z]' # 先頭が大文字アルファベットの場合もパラメータと判断
pattern = re.compile(regex)
pattern2 = re.compile(regex2)
# 行頭$スペースの2文字を除いて(の手前までをパラメータとして保存
if (pattern.match(orig_msg) ): # ’(’以降は削除
pos = orig_msg.find('(')
# print(orig_msg[2:pos])
msgdata[i] = (orig_msg[2:pos])
i += 1
elif (pattern2.match(orig_msg)): # 先頭大文字、=までもパラメータとする
pos = orig_msg.find('=')
#print(orig_msg[:pos])
msgdata[i] = orig_msg[:pos]
i += 1
# dict_valuesをPythonの listに変換する必要はなかった。
#print(data.values())
msgdf = pd.DataFrame(msgdata.values() , columns={'field_name'})
#print(len(msgdata))
```
## logfileも同様にDataFrameにする
### 上記の様に安直にループさせて新規のDFは仕様上作れないので一度辞書リストに保存する
```
logdata = {}
i = 0
for orig_log in logline:
# sec)が見えたらその次の文字列をパラメータとみなす
pos = orig_log.find('sec) ')
pos2 = orig_log.find('()')
#print(orig_log[pos+5:pos2].split(' ')[0])
logdata[i] = orig_log[pos+5:pos2].split(' ')[0]
i += 1
logdf = pd.DataFrame(logdata.values(),columns={'field_name'})
#print(logdf)
#(len(scheme.dropna()))
```
### 比較する
#### schee,techのDataframeと生データを読み込んだリストで文字列比較
- scheme エクセルのスキーム情報タブ
- カラム名 field_name , i1i2A26 , i3 , i4
- tech エクセルの技術情報タブ
- カラム名 field_name , i1msg , i1log , i2msg , i2log , i3msg , i3log , i3isus , i4msg , i4log
#### とりあえずDataframeも準備したけど辞書リストと比較しても良い
- msgdf テキスト生データのメッセージファイルに書いてあるパラメータ名のdataframe
- カラム名 field_name
- msgdata 生データのパラメータを抜き出した辞書リストdict
- logdf テキスト生データのログファイルに書いてあるパラメータ名のdataframe
- カラム名 field_name
- logdata 生データのパラメータを抜き出した辞書リストdict
#### dataframe は検索結果はQuery(Series)で返る
文字列が一致したIndexにはTrue、それ以外にはFalseが入ったqueryが返るので
Trueがある場合そのIndexを取得して有無を判定する(無いばあいはNoneが返る)
#### msgとスキーマ情報の比較
dataframe(scheme)の1列はSeriesになる。
queryが全部Falseだった場合はDBフィールドが無いということ。
もう一度ループを回して生データの1行と比較する(12行目あたり)
scoreを一旦保存して一番高いものを一つ表示する(result というDataFrameに保存)
```
result = pd.DataFrame(columns=['index','msg','xls','score'])
for line in msgdata.values():
#print(line)
query = scheme['i4'].dropna().str.startswith(line , na=False)
#print(query.values)
#print(scheme['i1i2a26'].dropna()) # NaNのところは削除
if (query[query == True].first_valid_index()):
exist = line + ' field exists.'
print(termcolor.colored(exist,'blue'))
else:
nf = line + ' is not found.'
print(termcolor.colored(nf,'red'))
for index,item in scheme.dropna().iterrows():
score = jaro_dist(str(item['i4']) , line) # 引数を入れ替えると結果が多少変わる。
#print(line + ' is probably ' + str(item['i4']) + ' ( ' + str(round(score,2)*100) + '%)' )
record = pd.Series([index , line , item['i4'] , score] , index=result.columns)
result = result.append(record , ignore_index=True)
#print(result)
items = len(scheme.dropna()) # Index数、この数分ジャロ・ウインクラー距離を計算したら次のマッチしなかった語句になる
kazu = int( len(result) / items ) # クエリーのリストになくnot foundで表示した数
for count in range(kazu):
res = result[items*count : items*(count+1)]
ranking = res.sort_values('score',ascending=False)[:3] # Score降順にソートして上から3つを表示
if((re.match('^.',ranking.iat[0,1])) is not None ):
# print(ranking.iat[0,3])
# score が0と1の時は抜く(アルゴリズム?で100%が結構出てる。
disp_res = str(ranking.iat[0,1]) + ' is probably ' + str(ranking.iat[0,2]) + ' ( ' + str(round(ranking.iat[0,3] , 2) * 100 ) + '% )' + '\n' \
+ ' or ' + str(ranking.iat[1,2]) + ' ( ' + str(round(ranking.iat[1,3] , 2) * 100 ) + '% )' + '\n' \
+ ' or ' + str(ranking.iat[2,2]) + ' ( ' + str(round(ranking.iat[2,3] , 2) * 100 ) + '% )' + '\n'
print( disp_res )
```
#### ジャロ・ウィンクラー距離法で文字列比較
##### 80%以上なら表示、以下のループだとフィールドがあるやつも表示してしまう。
ジャロウィンクラー距離法でNaNは文字列でないためそのまま使えない。面倒なのでdropna()でNaNは削除している。
scoreはround()で偶数丸め(四捨五入ではない)をして%表記
## debug用ブロック
for line in msgdata.values():
for index, item in scheme.dropna().iterrows():
score = jaro_dist(line,str(item['i4']))
if ( score > 0.1 and score < 1.0 ): # 80%以上なら候補を表示
print (line + ' is probably ' + str(item['i4']) + ' ( ' + str(round(score,2)*100) + '% ) ')
#print(line,item['i1i2a26'],index)
#print(jaro_dist('AirPressure', 4))
```
# debug用ブロック
str1 = 'AirPressure'
str2 = 'AirPres'
lev_dist = Levenshtein.distance(str1,str2)
# 標準化、長い方の文字列の長さで割る
devider = len(str1) if len(str1) > len(str2) else len(str2)
lev_dist = lev_dist / devider
# 指標を合わせる(0:完全不一致>1:完全一致)
lev_dist = 1 - lev_dist
#print(lev_dist)
```
#### msgと技術情報タブの比較
```
# ジャロウィンクラー距離のスコアを一度DFに保存してソートする
diff_result = pd.DataFrame(columns=['index','msg','xls','score'])
for line in logdata.values():
#print(line)
query = tech['i4msg'].dropna().str.startswith(line , na=False)
#print(query.values)
if (query[query == True].first_valid_index()):
exist = line + ' field exists.'
print(termcolor.colored(exist,'blue'))
else:
nf = line + ' is not found.'
print(termcolor.colored(nf,'red'))
for index,item in tech.iterrows():
score = jaro_dist(str(item['i4msg']) , line) # 引数を入れ替えると結果が多少変わる。
#print(score)
#print(line + ' is probably ' + str(item['i4msg']) + ' ( ' + str(round(score,2)*100) + '%)' )
diff_record = pd.Series([index , line , item['i4msg'] , score] , index=diff_result.columns)
diff_result = diff_result.append(diff_record , ignore_index=True)
# スコアの結果DF(上ではdiff_resultをソートして上から3つを表示
techlength = len(tech) # Index数、この数分ジャロ・ウインクラー距離を計算したら次のマッチしなかった語句になる
num = int( len(diff_result) / techlength ) # クエリーのリストになくnot foundで表示した数
for count in range(num):
diff_res = diff_result[techlength*count : techlength*(count+1)]
diff_ranking = diff_res.sort_values('score',ascending=False)[:3] # Score降順にソートして上から3つを表示
#print(diff_result)
if((re.match('^.',diff_ranking.iat[0,1])) is not None ):
#print(diff_ranking.iat[0,3])
# score が0と1の時は抜く(アルゴリズム?で100%が結構出てる。
disp_res2 = str(diff_ranking.iat[0,1]) + ' is probably ' + str(diff_ranking.iat[0,2]) + ' ( ' + str(round(diff_ranking.iat[0,3] , 2) * 100 ) + '% )' + '\n' \
+ ' or ' + str(diff_ranking.iat[1,2]) + ' ( ' + str(round(diff_ranking.iat[1,3] , 2) * 100 ) + '% )' + '\n' \
+ ' or ' + str(diff_ranking.iat[2,2]) + ' ( ' + str(round(diff_ranking.iat[2,3] , 2) * 100 ) + '% )' + '\n'
print( disp_res2 )
```
##### logと技術情報の比較
```
# 保存用のDFを準備
# ジャロウィンクラー距離のスコアを一度DFに保存してソートする
# Pythonは参照渡しなのでメモリ上にデータは全部残っているから変数名はキチン特別しておくこと
tech_result = pd.DataFrame(columns=['index','msg','xls','score'])
for line in logdata.values():
#print(line)
query = tech['i4log'].str.startswith(line , na=False)
#print(query.values)
if (query[query == True].first_valid_index()):
exist = line + ' field exists.'
print(termcolor.colored(exist,'blue'))
else:
nf = line + ' is not found.'
print(termcolor.colored(nf,'red'))
for index,item in tech.iterrows():
score = jaro_dist(str(item['i4log']) , line) # 引数を入れ替えると結果が多少変わる。
#print(score)
#print(line + ' is probably ' + str(item['i4msg']) + ' ( ' + str(round(score,2)*100) + '%)' )
tech_record = pd.Series([index , line , item['i4msg'] , score] , index=tech_result.columns)
tech_result = tech_result.append(tech_record , ignore_index=True)
```
### 類似度の高い方から3つを表示する
```
# スコアの結果DF(上ではtech_resultをソートして上から3つを表示
techloglen = len(tech) # Index数、この数分ジャロ・ウインクラー距離を計算したら次のマッチしなかった語句になる
num = int( len(tech_result) / techloglen ) # クエリーのリストになくnot foundで表示した数
for count in range(num):
tech_res = tech_result[techloglen*count : techloglen*(count+1)]
tech_ranking = tech_res.sort_values('score',ascending=False)[:3] # Score降順にソートして上から3つを表示
#print(diff_result)
if((re.match('^.',tech_ranking.iat[0,1])) is not None ):
#print(diff_ranking.iat[0,3])
# score が0と1の時は抜く(アルゴリズム?で100%が結構出てる。
disp_res3 = str(tech_ranking.iat[0,1]) + ' is probably ' + str(tech_ranking.iat[0,2]) + ' ( ' + str(round(tech_ranking.iat[0,3] , 2) * 100 ) + '% )' + '\n' \
+ ' or ' + str(tech_ranking.iat[1,2]) + ' ( ' + str(round(tech_ranking.iat[1,3] , 2) * 100 ) + '% )' + '\n' \
+ ' or ' + str(tech_ranking.iat[2,2]) + ' ( ' + str(round(tech_ranking.iat[2,3] , 2) * 100 ) + '% )' + '\n'
print( disp_res3 )
```
|
github_jupyter
|
import os
import pandas as pd
import re
import termcolor
import Levenshtein # レーベンシュタイン距離ライブラリにある、ジャロ・ウインクラー距離を計算するのに使う
# jaro_dist = Levenshtein.jaro_winkler(srt1 , str2)
navis_excel = pd.read_excel('Navis.xlsx' , sheet_name=None) # sheet_name=Noneで全てのシート読み込む
def jaro_dist(str1,str2):
return Levenshtein.jaro_winkler(str1,str2)
#print(navis_excel['技術情報'].columns)
# df.loc[行名:列名]
# df.iloc[行番号:列番号]
msgcol = navis_excel['scheme'].iloc[:,1]
i12msg = navis_excel['scheme'].iloc[:,7] # 7列目がI1/I2のmsg , 8列目はlog。以降同様にカウントする
i3msg = navis_excel['scheme'].iloc[:,9]
i4msg = navis_excel['scheme'].iloc[:,12]
scheme = pd.concat([msgcol,i12msg,i3msg,i4msg], axis=1).rename(columns={'Unnamed: 1':'field_name' , 'Unnamed: 7':'i1i2a26' , 'Unnamed: 9':'i3X1A27' , 'Unnamed: 12':'i4'})
#print(len(scheme))
#print(scheme.iloc[7,:])
#print(scheme['i1i2a26'])
techcol = navis_excel['tech'].iloc[:,1]
i1msg = navis_excel['tech'].iloc[:,7]
i1log = navis_excel['tech'].iloc[:,8]
i2msg = navis_excel['tech'].iloc[:,9]
i2log = navis_excel['tech'].iloc[:,10]
i3msg = navis_excel['tech'].iloc[:,11]
i3log = navis_excel['tech'].iloc[:,12]
i3isus = navis_excel['tech'].iloc[:,13]
i4msg = navis_excel['tech'].iloc[:,14]
i4log = navis_excel['tech'].iloc[:,15]
tech = pd.concat([techcol,i1msg,i1log,i2msg,i2log,i3msg,i3log,i3isus,i4msg,i4log], axis=1).rename(columns={'Unnamed: 1':'field_name', 'Unnamed: 7':'i1msg' , 'Unnamed: 8':'i1log' , 'Unnamed: 9':'i2msg' , 'Unnamed: 10':'i2log' , 'Unnamed: 11':'i3msg' , 'Unnamed: 12':'i3log' , 'Unnamed: 13':'i3isus' , 'Unnamed: 14':'i4msg' , 'Unnamed: 15':'i4log'})
#print(tech.iloc[56,:])
#print(type(tech))
with open('i0394/0394.001.msg','r') as msg:
msgline = msg.readlines() # prof,techで使う
with open('i0394/0394.001.log','r') as log:
logline = log.readlines()
# msgline,loglineとも1行毎に読み込んでる。
# 安直にループさせて新しいDataFrameを作ることは出来ないので辞書リストに一度保存する(Pythonの仕様)
msgdata = {}
i = 0
for orig_msg in msgline:
regex = r'^\$' # 先頭が$ならその行をパラメータと判断する
regex2 = r'^[A-Z]' # 先頭が大文字アルファベットの場合もパラメータと判断
pattern = re.compile(regex)
pattern2 = re.compile(regex2)
# 行頭$スペースの2文字を除いて(の手前までをパラメータとして保存
if (pattern.match(orig_msg) ): # ’(’以降は削除
pos = orig_msg.find('(')
# print(orig_msg[2:pos])
msgdata[i] = (orig_msg[2:pos])
i += 1
elif (pattern2.match(orig_msg)): # 先頭大文字、=までもパラメータとする
pos = orig_msg.find('=')
#print(orig_msg[:pos])
msgdata[i] = orig_msg[:pos]
i += 1
# dict_valuesをPythonの listに変換する必要はなかった。
#print(data.values())
msgdf = pd.DataFrame(msgdata.values() , columns={'field_name'})
#print(len(msgdata))
logdata = {}
i = 0
for orig_log in logline:
# sec)が見えたらその次の文字列をパラメータとみなす
pos = orig_log.find('sec) ')
pos2 = orig_log.find('()')
#print(orig_log[pos+5:pos2].split(' ')[0])
logdata[i] = orig_log[pos+5:pos2].split(' ')[0]
i += 1
logdf = pd.DataFrame(logdata.values(),columns={'field_name'})
#print(logdf)
#(len(scheme.dropna()))
result = pd.DataFrame(columns=['index','msg','xls','score'])
for line in msgdata.values():
#print(line)
query = scheme['i4'].dropna().str.startswith(line , na=False)
#print(query.values)
#print(scheme['i1i2a26'].dropna()) # NaNのところは削除
if (query[query == True].first_valid_index()):
exist = line + ' field exists.'
print(termcolor.colored(exist,'blue'))
else:
nf = line + ' is not found.'
print(termcolor.colored(nf,'red'))
for index,item in scheme.dropna().iterrows():
score = jaro_dist(str(item['i4']) , line) # 引数を入れ替えると結果が多少変わる。
#print(line + ' is probably ' + str(item['i4']) + ' ( ' + str(round(score,2)*100) + '%)' )
record = pd.Series([index , line , item['i4'] , score] , index=result.columns)
result = result.append(record , ignore_index=True)
#print(result)
items = len(scheme.dropna()) # Index数、この数分ジャロ・ウインクラー距離を計算したら次のマッチしなかった語句になる
kazu = int( len(result) / items ) # クエリーのリストになくnot foundで表示した数
for count in range(kazu):
res = result[items*count : items*(count+1)]
ranking = res.sort_values('score',ascending=False)[:3] # Score降順にソートして上から3つを表示
if((re.match('^.',ranking.iat[0,1])) is not None ):
# print(ranking.iat[0,3])
# score が0と1の時は抜く(アルゴリズム?で100%が結構出てる。
disp_res = str(ranking.iat[0,1]) + ' is probably ' + str(ranking.iat[0,2]) + ' ( ' + str(round(ranking.iat[0,3] , 2) * 100 ) + '% )' + '\n' \
+ ' or ' + str(ranking.iat[1,2]) + ' ( ' + str(round(ranking.iat[1,3] , 2) * 100 ) + '% )' + '\n' \
+ ' or ' + str(ranking.iat[2,2]) + ' ( ' + str(round(ranking.iat[2,3] , 2) * 100 ) + '% )' + '\n'
print( disp_res )
# debug用ブロック
str1 = 'AirPressure'
str2 = 'AirPres'
lev_dist = Levenshtein.distance(str1,str2)
# 標準化、長い方の文字列の長さで割る
devider = len(str1) if len(str1) > len(str2) else len(str2)
lev_dist = lev_dist / devider
# 指標を合わせる(0:完全不一致>1:完全一致)
lev_dist = 1 - lev_dist
#print(lev_dist)
# ジャロウィンクラー距離のスコアを一度DFに保存してソートする
diff_result = pd.DataFrame(columns=['index','msg','xls','score'])
for line in logdata.values():
#print(line)
query = tech['i4msg'].dropna().str.startswith(line , na=False)
#print(query.values)
if (query[query == True].first_valid_index()):
exist = line + ' field exists.'
print(termcolor.colored(exist,'blue'))
else:
nf = line + ' is not found.'
print(termcolor.colored(nf,'red'))
for index,item in tech.iterrows():
score = jaro_dist(str(item['i4msg']) , line) # 引数を入れ替えると結果が多少変わる。
#print(score)
#print(line + ' is probably ' + str(item['i4msg']) + ' ( ' + str(round(score,2)*100) + '%)' )
diff_record = pd.Series([index , line , item['i4msg'] , score] , index=diff_result.columns)
diff_result = diff_result.append(diff_record , ignore_index=True)
# スコアの結果DF(上ではdiff_resultをソートして上から3つを表示
techlength = len(tech) # Index数、この数分ジャロ・ウインクラー距離を計算したら次のマッチしなかった語句になる
num = int( len(diff_result) / techlength ) # クエリーのリストになくnot foundで表示した数
for count in range(num):
diff_res = diff_result[techlength*count : techlength*(count+1)]
diff_ranking = diff_res.sort_values('score',ascending=False)[:3] # Score降順にソートして上から3つを表示
#print(diff_result)
if((re.match('^.',diff_ranking.iat[0,1])) is not None ):
#print(diff_ranking.iat[0,3])
# score が0と1の時は抜く(アルゴリズム?で100%が結構出てる。
disp_res2 = str(diff_ranking.iat[0,1]) + ' is probably ' + str(diff_ranking.iat[0,2]) + ' ( ' + str(round(diff_ranking.iat[0,3] , 2) * 100 ) + '% )' + '\n' \
+ ' or ' + str(diff_ranking.iat[1,2]) + ' ( ' + str(round(diff_ranking.iat[1,3] , 2) * 100 ) + '% )' + '\n' \
+ ' or ' + str(diff_ranking.iat[2,2]) + ' ( ' + str(round(diff_ranking.iat[2,3] , 2) * 100 ) + '% )' + '\n'
print( disp_res2 )
# 保存用のDFを準備
# ジャロウィンクラー距離のスコアを一度DFに保存してソートする
# Pythonは参照渡しなのでメモリ上にデータは全部残っているから変数名はキチン特別しておくこと
tech_result = pd.DataFrame(columns=['index','msg','xls','score'])
for line in logdata.values():
#print(line)
query = tech['i4log'].str.startswith(line , na=False)
#print(query.values)
if (query[query == True].first_valid_index()):
exist = line + ' field exists.'
print(termcolor.colored(exist,'blue'))
else:
nf = line + ' is not found.'
print(termcolor.colored(nf,'red'))
for index,item in tech.iterrows():
score = jaro_dist(str(item['i4log']) , line) # 引数を入れ替えると結果が多少変わる。
#print(score)
#print(line + ' is probably ' + str(item['i4msg']) + ' ( ' + str(round(score,2)*100) + '%)' )
tech_record = pd.Series([index , line , item['i4msg'] , score] , index=tech_result.columns)
tech_result = tech_result.append(tech_record , ignore_index=True)
# スコアの結果DF(上ではtech_resultをソートして上から3つを表示
techloglen = len(tech) # Index数、この数分ジャロ・ウインクラー距離を計算したら次のマッチしなかった語句になる
num = int( len(tech_result) / techloglen ) # クエリーのリストになくnot foundで表示した数
for count in range(num):
tech_res = tech_result[techloglen*count : techloglen*(count+1)]
tech_ranking = tech_res.sort_values('score',ascending=False)[:3] # Score降順にソートして上から3つを表示
#print(diff_result)
if((re.match('^.',tech_ranking.iat[0,1])) is not None ):
#print(diff_ranking.iat[0,3])
# score が0と1の時は抜く(アルゴリズム?で100%が結構出てる。
disp_res3 = str(tech_ranking.iat[0,1]) + ' is probably ' + str(tech_ranking.iat[0,2]) + ' ( ' + str(round(tech_ranking.iat[0,3] , 2) * 100 ) + '% )' + '\n' \
+ ' or ' + str(tech_ranking.iat[1,2]) + ' ( ' + str(round(tech_ranking.iat[1,3] , 2) * 100 ) + '% )' + '\n' \
+ ' or ' + str(tech_ranking.iat[2,2]) + ' ( ' + str(round(tech_ranking.iat[2,3] , 2) * 100 ) + '% )' + '\n'
print( disp_res3 )
| 0.043437 | 0.80567 |
# Source contributions
```
import pickle
import numpy as np
import netCDF4 as nc
import pandas as pd
from calendar import monthrange
%matplotlib inline
```
###### Parameters:
```
# domain dimensions:
imin, imax = 1479, 2179
jmin, jmax = 159, 799
isize = imax-imin
jsize = jmax-jmin
# Mn model result folders:
folder_ref = '/data/brogalla/run_storage/Mn-reference-202110/'
folder_cleanice = '/data/brogalla/run_storage/Mn-clean-ice-202110/'
folder_spm = '/data/brogalla/run_storage/Mn-spm-202110/'
folder_bio = '/data/brogalla/run_storage/Mn-bio-202110/'
colors = ['#ccb598', '#448d90', '#739f78', '#CC8741', '#cee7fd', '#b9c1c7']
years = [2002, 2003, 2004, 2005, 2006, 2007, 2008, 2009, 2010, 2011, 2012, 2013, 2014, 2015, 2016, \
2017, 2018, 2019]
```
###### Load files:
```
# ANHA12 grid:
mask = nc.Dataset('/ocean/brogalla/GEOTRACES/data/ANHA12/ANHA12_mesh1.nc')
tmask = np.array(mask.variables['tmask'])[0,:,imin:imax,jmin:jmax]
land_mask = np.ma.masked_where((tmask[:,:,:] > 0.1), tmask[:,:,:])
e1t_base = np.array(mask.variables['e1t'])[0,imin:imax,jmin:jmax]
e2t_base = np.array(mask.variables['e2t'])[0,imin:imax,jmin:jmax]
e3t = np.array(mask.variables['e3t_0'])[0,:,imin:imax,jmin:jmax]
e3t_masked = np.ma.masked_where((tmask[:,:,:] < 0.1), e3t)
nav_lev = np.array(mask.variables['nav_lev'])
lons = np.array(mask.variables['nav_lon'])
e1t = np.tile(e1t_base, (50,1,1))
e2t = np.tile(e2t_base, (50,1,1))
volume = e1t*e2t*e3t
area_base = e1t_base*e2t_base
volume_masked = np.ma.masked_where((tmask[:,:,:] < 0.1), volume)
area_masked = np.ma.masked_where((tmask[0,:,:] < 0.1), area_base)
```
##### Functions:
```
def load_results(folder_ref, year, experiment):
months = ['01', '02', '03', '04', '05', '06', '07', '08', '09', '10', '11', '12']
dmn_riv = np.empty((12,isize,jsize)) ; dmn_sed = np.empty((12,isize,jsize));
dmn_atm = np.empty((12,isize,jsize)) ; dmn_ice = np.empty((12,isize,jsize));
dmn_sedice = np.empty((12,isize,jsize)); dmn_priv = np.empty((12,isize,jsize));
dmn_red = np.empty((12,50,isize,jsize)); dmn_oxi = np.empty((12,50,isize,jsize));
dmn_bio = np.empty((12,50,isize,jsize));
for i, month in enumerate(months):
file = f'ANHA12_EXH006_1m_{year}0101_{year}1231_comp_{year}{month}-{year}{month}.nc'
ref_monthly = nc.Dataset(folder_ref+file)
dmn_riv[i,:,:] = np.array(ref_monthly.variables['dmnriv'])[0,:,:]
if experiment=='spm':
dmn_priv[i,:,:] = np.array(ref_monthly.variables['pmnriv'])[0,:,:]
dmn_bio[i,:,:] = 0
elif experiment=='bio':
dmn_priv[i,:,:] = 0
dmn_bio[i,:,:] = np.array(ref_monthly.variables['dmnbio'])[0,:,:,:]
else:
dmn_priv[i,:,:] = 0
dmn_bio[i,:,:] = 0
dmn_sed[i,:,:] = np.array(ref_monthly.variables['dmnsed'])[0,:,:]
dmn_sedice[i,:,:] = np.array(ref_monthly.variables['dmnsedice'])[0,:,:]
dmn_atm[i,:,:] = np.array(ref_monthly.variables['dmnatm'])[0,:,:]
dmn_ice[i,:,:] = np.array(ref_monthly.variables['dmnice'])[0,:,:]
dmn_red[i,:,:,:] = np.array(ref_monthly.variables['dmnred'])[0,:,:,:]
dmn_oxi[i,:,:,:] = np.array(ref_monthly.variables['dmnoxi'])[0,:,:,:]
tmask_surf = np.empty(dmn_riv.shape)
tmask_surf[:] = tmask[0,:,:]
tmask_full = np.empty(dmn_red.shape)
tmask_full[:] = tmask
# Mask points on land:
dmn_riv = np.ma.masked_where((tmask_surf < 0.1), dmn_riv);
dmn_priv = np.ma.masked_where((tmask_surf < 0.1), dmn_priv);
dmn_sed = np.ma.masked_where((tmask_surf < 0.1), dmn_sed)
dmn_sedice = np.ma.masked_where((tmask_surf < 0.1), dmn_sedice)
dmn_atm = np.ma.masked_where((tmask_surf < 0.1), dmn_atm)
dmn_ice = np.ma.masked_where((tmask_surf < 0.1), dmn_ice)
dmn_red = np.ma.masked_where((tmask_full < 0.1), dmn_red)
dmn_oxi = np.ma.masked_where((tmask_full < 0.1), dmn_oxi)
dmn_bio = np.ma.masked_where((tmask_full < 0.1), dmn_bio)
return dmn_riv, dmn_priv, dmn_sed, dmn_sedice, dmn_atm, dmn_ice, dmn_red, dmn_oxi, dmn_bio
def calculate_contribution(year, mask, experiment='ref'):
# Calculate the contribution of the model components by region:
if experiment=='spm':
folder_year = folder_spm + f'ANHA12_spm-{year}_20211026/'
elif experiment=='bio':
folder_year = folder_bio + f'ANHA12_bio-{year}_20211022/'
else:
folder_year = folder_ref + f'ANHA12_ref-{year}_20211012/'
if experiment=='bio' and year>2015:
total_yearly_contribution = np.empty((9))
ave_yearly_contribution = np.empty((9))
total_yearly_contribution[:] = np.NaN
ave_yearly_contribution[:] = np.NaN
return total_yearly_contribution, ave_yearly_contribution
dmn_mriv, dmn_mpriv, dmn_msed, dmn_msedice, dmn_matm, dmn_mice, dmn_mred, dmn_moxi, dmn_mbio = \
load_results(folder_year, year, experiment)
# Calculate contributions to the upper water column, so mask locations where sediment resuspension is added in the deep.
indexes_bottom = np.tile(index_bottom, (12,1,1))
# Momentary contribution: moles / second (from moles / L / s --> moles / m3 / s --> moles / s)
# Mask is for the points associated with the specified region.
priv = np.ma.masked_where(mask==0, dmn_mpriv) * volume_masked[0,:,:] * 1e3
riv = np.ma.masked_where(mask==0, dmn_mriv) * volume_masked[0,:,:] * 1e3
sed = np.ma.masked_where(mask==0, dmn_msed) * volume_bottom * 1e3
sedice = np.ma.masked_where(mask==0, dmn_msedice) * volume_masked[0,:,:] * 1e3
atm = np.ma.masked_where(mask==0, dmn_matm) * volume_masked[0,:,:] * 1e3
ice = np.ma.masked_where(mask==0, dmn_mice) * volume_masked[0,:,:] * 1e3
# 3D fields only calculated over polar mixed layer:
mask_with_depths = np.tile(mask, (50,1,1,1)).reshape((12,50,700,640))
bio = np.ma.masked_where(mask_with_depths==0, dmn_mbio)[:,0:17,:,:] * volume_masked[0:17,:,:] * 1e3
red = np.ma.masked_where(mask_with_depths==0, dmn_mred)[:,0:17,:,:] * volume_masked[0:17,:,:] * 1e3
oxi = np.ma.masked_where(mask_with_depths==0, dmn_moxi)[:,0:17,:,:] * volume_masked[0:17,:,:] * 1e3
# Yearly contribution to domain: moles / year (from moles / s / month --> moles / month --> moles / year)
atm_year = 0; riv_year = 0; priv_year = 0; sed_year = 0; sedice_year = 0;
ice_year = 0; bio_year = 0; red_year = 0; oxi_year = 0;
for month in range(1,13):
index = month-1
days_in_month = monthrange(year, month)[1]
if days_in_month == 29: # ignore leap year
days_in_month = 28
atm_year = atm_year + np.ma.sum(atm[index,:,:],axis=(0,1)) *3600*24*days_in_month
riv_year = riv_year + np.ma.sum(riv[index,:,:],axis=(0,1)) *3600*24*days_in_month
priv_year = priv_year + np.ma.sum(priv[index,:,:],axis=(0,1)) *3600*24*days_in_month
sed_year = sed_year + np.ma.sum(sed[index,:,:],axis=(0,1)) *3600*24*days_in_month
sedice_year = sedice_year + np.ma.sum(sedice[index,:,:],axis=(0,1))*3600*24*days_in_month
ice_year = ice_year + np.ma.sum(ice[index,:,:],axis=(0,1)) *3600*24*days_in_month
bio_year = bio_year + np.ma.sum(bio[index,:,:,:],axis=(0,1,2)) *3600*24*days_in_month
red_year = red_year + np.ma.sum(red[index,:,:,:],axis=(0,1,2)) *3600*24*days_in_month
oxi_year = oxi_year + np.ma.sum(oxi[index,:,:,:],axis=(0,1,2)) *3600*24*days_in_month
# Average yearly contribution over domain: moles / m2 / year
total_area = np.ma.sum(np.ma.masked_where(mask[0,:,:]==0, area_masked[:,:]))
atmm2_year = atm_year / total_area
rivm2_year = riv_year / total_area
privm2_year = priv_year / total_area
sedm2_year = sed_year / total_area
sedicem2_year = sedice_year / total_area
icem2_year = ice_year / total_area
# Polar mixed layer average contribution:
PML_depth = nav_lev[17]
biom2_year = bio_year / (total_area*PML_depth)
redm2_year = red_year / (total_area*PML_depth)
oxim2_year = oxi_year / (total_area*PML_depth)
total_yearly_contribution = np.array([priv_year, riv_year, sed_year, sedice_year, ice_year, atm_year, \
bio_year, red_year, oxi_year]) # mol/yr
ave_yearly_contribution = np.array([privm2_year, rivm2_year, sedm2_year, sedicem2_year, icem2_year, \
atmm2_year, biom2_year, redm2_year, oxim2_year]) # mol/m2/yr
return total_yearly_contribution, ave_yearly_contribution
def calculate_regional_contributions(mask):
totals_ref = np.empty((len(years),9)); totals_spm = np.empty((len(years),9)); totals_bio = np.empty((len(years),9));
averages_ref = np.empty((len(years),9)); averages_spm = np.empty((len(years),9)); averages_bio = np.empty((len(years),9));
totals_ref[:] = np.NaN; totals_spm[:] = np.NaN; totals_bio[:] = np.NaN;
averages_ref[:] = np.NaN; averages_spm[:] = np.NaN; averages_spm[:] = np.NaN;
for i, year in enumerate(years):
total_ref_year, ave_ref_year = calculate_contribution(year, mask, experiment='ref')
total_spm_year, ave_spm_year = calculate_contribution(year, mask, experiment='spm')
total_bio_year, ave_bio_year = calculate_contribution(year, mask, experiment='bio')
totals_ref[i,:] = total_ref_year
totals_spm[i,:] = total_spm_year
totals_bio[i,:] = total_bio_year
averages_ref[i,:] = ave_ref_year
averages_spm[i,:] = ave_spm_year
averages_bio[i,:] = ave_bio_year
return totals_ref, totals_spm, totals_bio, averages_ref, averages_spm, averages_bio
def reshape_arrays(array_in):
fixed = np.zeros((len(years),8))
for i in range(0,8):
if i==0: # Sum the particulate and dissolved river contributions
fixed[:,i] = array_in[:,i]+array_in[:,i+1]
else:
fixed[:,i] = array_in[:,i+1]
return fixed
def pipeline(totals_input_ref, totals_input_spm, totals_input_bio,
averages_input_ref, averages_input_spm, averages_input_bio):
regions = ['full', 'CB', 'CAA', 'wCAA', 'eCAA']
totals_fixed_ref = totals_input_ref.copy(); averages_fixed_ref = averages_input_ref.copy();
totals_fixed_spm = totals_input_spm.copy(); averages_fixed_spm = averages_input_spm.copy();
totals_fixed_bio = totals_input_bio.copy(); averages_fixed_bio = averages_input_bio.copy();
# Reshape arrays so that particulate and dissolved river contributions are combined into one.
for region in regions:
totals_fixed_ref[region] = reshape_arrays(totals_input_ref[region])
totals_fixed_spm[region] = reshape_arrays(totals_input_spm[region])
totals_fixed_bio[region] = reshape_arrays(totals_input_bio[region])
averages_fixed_ref[region] = reshape_arrays(averages_input_ref[region])
averages_fixed_spm[region] = reshape_arrays(averages_input_spm[region])
averages_fixed_bio[region] = reshape_arrays(averages_input_bio[region])
return totals_fixed_ref, totals_fixed_spm, totals_fixed_bio, averages_fixed_ref, averages_fixed_spm, averages_fixed_bio
def print_interannual_averages(totals_ref, totals_spm, totals_bio,
averages_ref, averages_spm, averages_bio,
location='Full'):
print(f'Interannual average values in {location}')
print('----------------------------------------------------------------')
total_ref_annual = np.sum(totals_ref[location][:,0:5], axis=1)
total_spm_annual = np.sum(totals_spm[location][:,0:5], axis=1)
total_bio_annual = np.sum(totals_bio[location][:,0:5], axis=1)
# Total annual contribution is averaged over the 18-year time series and multiplied by 1e-6 to convert to Mmol
print(f"{'Total annual contribution of Mn [Mmol/yr]:':<55}" +
f"{np.average(total_ref_annual)*1e-6:<5.0f} ({np.average(total_spm_annual)*1e-6:<5.0f})")
print(f"{'-- River discharge ---':<40}" +
f" {np.average(totals_ref[location][:,0])*1e-6:<8.2f} ({np.average(totals_spm[location][:,0])*1e-6:<5.2f})")
print(f"{'-- Sediment resuspension ---':<40}" +
f" {np.average(totals_ref[location][:,1])*1e-6:<8.2f} ({np.average(totals_spm[location][:,1])*1e-6:<5.2f})")
print(f"{'-- Sediment released by sea ice ---':<40}" +
f" {np.average(totals_ref[location][:,2])*1e-6:<8.2f} ({np.average(totals_spm[location][:,2])*1e-6:<5.2f})")
print(f"{'-- Dust released by sea ice ---':<40}" +
f" {np.average(totals_ref[location][:,3])*1e-6:<8.2f} ({np.average(totals_spm[location][:,3])*1e-6:<5.2f})")
print(f"{'-- Dust deposition ---':<40}" +
f" {np.average(totals_ref[location][:,4])*1e-6:<8.2f} ({np.average(totals_spm[location][:,4])*1e-6:<5.2f})")
tref = np.sum(totals_ref[location][:,0:5], axis=(0,1))
tspm = np.sum(totals_spm[location][:,0:5], axis=(0,1))
tbio = np.sum(totals_bio[location][:,0:5], axis=(0,1))
# Percent is calculated from sum of component contribution over all months / total contributions over full time period
print('----------------------------------------------------------------')
print('Total annual contribution of Mn [%]:')
print(f"{'-- River discharge ---':<40}" +
f"{np.sum(totals_ref[location][:,0])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,0])*100/tspm:<5.2f})")
print(f"{'-- Sediment resuspension ---':<40}" +
f"{np.sum(totals_ref[location][:,1])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,1])*100/tspm:<5.2f})")
print(f"{'-- Sediment released by sea ice ---':<40}" +
f"{np.sum(totals_ref[location][:,2])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,2])*100/tspm:<5.2f})")
print(f"{'-- Dust released by sea ice ---':<40}" +
f"{np.sum(totals_ref[location][:,3])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,3])*100/tspm:<5.2f})")
print(f"{'-- Dust deposition ---':<40}" +
f"{np.sum(totals_ref[location][:,4])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,4])*100/tspm:<5.2f})")
# Average annual contribution is calculated as an average over the full time series and converted to millimol
print('----------------------------------------------------------------')
print(f"{'Average annual contribution of Mn [micromol/m2/yr]:':<55}" +
f"{np.average(np.sum(averages_ref[location][:,0:5], axis=1)*1e6):<5.0f}" +
f"({np.average(np.sum(averages_spm[location][:,0:5], axis=1)*1e6):<5.0f})")
print(f"{'-- River discharge ---':<40}" +
f"{np.average(averages_ref[location][:,0])*1e6:<8.2f} ({np.average(averages_spm[location][:,0])*1e6:<5.2f})")
print(f"{'-- Sediment resuspension ---':<40}" +
f"{np.average(averages_ref[location][:,1])*1e6:<8.2f} ({np.average(averages_spm[location][:,1])*1e6:<5.2f})")
print(f"{'-- Sediment released by sea ice ---':<40}" +
f"{np.average(averages_ref[location][:,2])*1e6:<8.2f} ({np.average(averages_spm[location][:,2])*1e6:<5.2f})")
print(f"{'-- Dust released by sea ice ---':<40}" +
f"{np.average(averages_ref[location][:,3])*1e6:<8.2f} ({np.average(averages_spm[location][:,3])*1e6:<5.2f})")
print(f"{'-- Dust deposition ---':<40}" +
f"{np.average(averages_ref[location][:,4])*1e6:<8.2f} ({np.average(averages_spm[location][:,4])*1e6:<5.2f})")
return
def print_interannual_averages_3D(totals_ref, totals_spm, totals_bio,
averages_ref, averages_spm, averages_bio,
location='Full'):
# include reduction, oxidation, and bio components calculated over the polar mixed layer
print(f'Interannual average values in {location}')
print('----------------------------------------------------------------')
total_ref_annual = np.sum(totals_ref[location], axis=1)
total_spm_annual = np.sum(totals_spm[location], axis=1)
total_bio_annual = np.nansum(totals_bio[location], axis=1)
print(f"{'Total annual contribution of Mn [Mmol/yr]:':<55}" +
f"{np.average(total_ref_annual)*1e-6:<5.0f} ({np.average(total_spm_annual)*1e-6:<5.0f})" + \
f" {np.nanmean(total_bio_annual)*1e-6:<8.2f}")
print(f"{'-- River discharge ---':<40}" +
f" {np.average(totals_ref[location][:,0])*1e-6:<8.2f} ({np.average(totals_spm[location][:,0])*1e-6:<5.2f})" + \
f" {np.nanmean(totals_bio[location][:,0])*1e-6:<8.2f}")
print(f"{'-- Sediment resuspension ---':<40}" +
f" {np.average(totals_ref[location][:,1])*1e-6:<8.2f} ({np.average(totals_spm[location][:,1])*1e-6:<5.2f})" + \
f" {np.nanmean(totals_bio[location][:,1])*1e-6:<8.2f}")
print(f"{'-- Sediment released by sea ice ---':<40}" +
f" {np.average(totals_ref[location][:,2])*1e-6:<8.2f} ({np.average(totals_spm[location][:,2])*1e-6:<5.2f})" + \
f" {np.nanmean(totals_bio[location][:,2])*1e-6:<8.2f}")
print(f"{'-- Dust released by sea ice ---':<40}" +
f" {np.average(totals_ref[location][:,3])*1e-6:<8.2f} ({np.average(totals_spm[location][:,3])*1e-6:<5.2f})" + \
f" {np.nanmean(totals_bio[location][:,3])*1e-6:<8.2f}")
print(f"{'-- Dust deposition ---':<40}" +
f" {np.average(totals_ref[location][:,4])*1e-6:<8.2f} ({np.average(totals_spm[location][:,4])*1e-6:<5.2f})" + \
f" {np.nanmean(totals_bio[location][:,4])*1e-6:<8.2f}")
print(f"{'-- Uptake/remineralization ---':<40}" +
f" {np.average(totals_ref[location][:,5])*1e-6:<8.2f} ({np.average(totals_spm[location][:,5])*1e-6:<5.2f})" + \
f" {np.nanmean(totals_bio[location][:,5])*1e-6:<8.2f}")
print(f"{'-- Scavenging ---':<40}" +
f" {(np.average(totals_ref[location][:,6])-np.average(totals_ref[location][:,7]))*1e-6:<8.2f}" + \
f"({(np.average(totals_spm[location][:,6])-np.average(totals_spm[location][:,7]))*1e-6:<5.2f})" + \
f" {(np.nanmean(totals_bio[location][:,6])-np.nanmean(totals_bio[location][:,7]))*1e-6:<8.2f}")
tref = np.sum(totals_ref[location], axis=(0,1))
tspm = np.sum(totals_spm[location], axis=(0,1))
tbio = np.nansum(totals_bio[location], axis=(0,1))
print('----------------------------------------------------------------')
print('Total annual contribution of Mn [%]:')
print(f"{'-- River discharge ---':<40}" +
f"{np.sum(totals_ref[location][:,0])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,0])*100/tspm:<5.2f})" + \
f"{np.nansum(totals_bio[location][:,0])*100/tref:<5.2f}")
print(f"{'-- Sediment resuspension ---':<40}" +
f"{np.sum(totals_ref[location][:,1])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,1])*100/tspm:<5.2f})" + \
f"{np.nansum(totals_bio[location][:,1])*100/tref:<5.2f}")
print(f"{'-- Sediment released by sea ice ---':<40}" +
f"{np.sum(totals_ref[location][:,2])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,2])*100/tspm:<5.2f})" + \
f"{np.nansum(totals_bio[location][:,2])*100/tref:<5.2f}")
print(f"{'-- Dust released by sea ice ---':<40}" +
f"{np.sum(totals_ref[location][:,3])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,3])*100/tspm:<5.2f})" + \
f"{np.nansum(totals_bio[location][:,3])*100/tref:<5.2f}")
print(f"{'-- Dust deposition ---':<40}" +
f"{np.sum(totals_ref[location][:,4])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,4])*100/tspm:<5.2f})" + \
f"{np.nansum(totals_bio[location][:,4])*100/tref:<5.2f}")
print(f"{'-- Uptake/remineralization ---':<40}" +
f"{np.sum(totals_ref[location][:,5])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,5])*100/tspm:<5.2f})" + \
f"{np.nansum(totals_bio[location][:,5])*100/tref:<5.2f}")
print(f"{'-- Scavenging ---':<40}" +
f"{np.sum(totals_ref[location][:,6] - totals_ref[location][:,7])*100/tref:<5.2f} "+ \
f"({np.sum(totals_spm[location][:,6] - totals_spm[location][:,7])*100/tspm:<5.2f})" + \
f"{np.nansum(totals_bio[location][:,6]- totals_bio[location][:,7])*100/tref:<5.2f}")
print('----------------------------------------------------------------')
print(f"{'Average annual contribution of Mn [micromol/m2/yr]:':<55}" +
f"{np.average(np.sum(averages_ref[location], axis=1)*1e6):<5.0f}" +
f"({np.average(np.sum(averages_spm[location], axis=1)*1e6):<5.0f})"+
f"{np.nanmean(np.nansum(averages_bio[location], axis=1)*1e6):<5.0f}")
print(f"{'-- River discharge ---':<40}" +
f"{np.average(averages_ref[location][:,0])*1e6:<8.2f} ({np.average(averages_spm[location][:,0])*1e6:<5.2f})" +\
f"{np.nanmean(averages_bio[location][:,0])*1e6:<8.2f}")
print(f"{'-- Sediment resuspension ---':<40}" +
f"{np.average(averages_ref[location][:,1])*1e6:<8.2f} ({np.average(averages_spm[location][:,1])*1e6:<5.2f})" +\
f"{np.nanmean(averages_bio[location][:,1])*1e6:<8.2f}")
print(f"{'-- Sediment released by sea ice ---':<40}" +
f"{np.average(averages_ref[location][:,2])*1e6:<8.2f} ({np.average(averages_spm[location][:,2])*1e6:<5.2f})" +\
f"{np.nanmean(averages_bio[location][:,2])*1e6:<8.2f}")
print(f"{'-- Dust released by sea ice ---':<40}" +
f"{np.average(averages_ref[location][:,3])*1e6:<8.2f} ({np.average(averages_spm[location][:,3])*1e6:<5.2f})" +\
f"{np.nanmean(averages_bio[location][:,3])*1e6:<8.2f}")
print(f"{'-- Dust deposition ---':<40}" +
f"{np.average(averages_ref[location][:,4])*1e6:<8.2f} ({np.average(averages_spm[location][:,4])*1e6:<5.2f})" +\
f"{np.nanmean(averages_bio[location][:,4])*1e6:<8.2f}")
print(f"{'-- Uptake/remineralization ---':<40}" +
f"{np.average(averages_ref[location][:,5])*1e6:<8.2f} ({np.average(averages_spm[location][:,5])*1e6:<5.2f})" +\
f"{np.nanmean(averages_bio[location][:,5])*1e6:<8.2f}")
print(f"{'-- Scavenging ---':<40}" +
f"{(np.average(averages_ref[location][:,6]) - np.average(averages_ref[location][:,7]))*1e6:<8.2f}"+ \
f"({(np.average(averages_spm[location][:,6])- np.average(averages_spm[location][:,7]))*1e6:<5.2f})" +\
f"{(np.nanmean(averages_bio[location][:,6]) - np.nanmean(averages_bio[location][:,7]))*1e6:<8.2f}")
return
```
### Calculations:
Find grid cell volume and thickness at ocean floor:
```
tmask_bot = np.copy(tmask)
tmask_bot[0,:,:] = 1
e3t_mask = np.ma.masked_where((tmask_bot[:,:,:] < 0.1), e3t[:,:,:])
# find bottom grid cell index:
ind = np.array(np.ma.notmasked_edges(e3t_mask[:,:,:], axis=0))
# array of shape (isize, jsize) containing bottom grid cell index
index_bottom = np.ma.masked_where((tmask[0,:,:] < 0.1), ind[1][0][:].reshape(isize,jsize))
e3t_bot = np.zeros((isize,jsize))
vol_bot = np.zeros((isize,jsize))
for i in range(0, isize):
for j in range(0,jsize):
k = index_bottom[i,j]
try:
e3t_bot[i,j] = e3t[k,i,j]
vol_bot[i,j] = volume[k,i,j]
except:
e3t_bot[i,j] = np.nan
vol_bot[i,j] = np.nan
e3t_bottom = np.ma.masked_where((tmask[0,:,:] < 0.1), np.ma.masked_where(np.isnan(e3t_bot), e3t_bot))
volume_bottom = np.ma.masked_where((tmask[0,:,:] < 0.1), np.ma.masked_where(np.isnan(vol_bot), vol_bot))
```
Create masks for specific subregions:
- Canada Basin
- CAA
- Full domain
- Western CAA (separated by Barrow Sill)
- Eastern CAA (separated by Barrow Sill)
```
# Find indices to separate out Canada Basin:
x_ind = np.arange(1621, 2100, 1)
y_ind = (-7/8)*x_ind + 1517 + 700
CB_indx = []
CB_indy = []
for index in range(0,len(x_ind)):
CB_x = np.arange(x_ind[index],2179,1)
CB_y = np.ones(CB_x.shape)*y_ind[index]
CB_indx = np.append(CB_x, CB_indx)
CB_indy = np.append(CB_y, CB_indy)
# Separate Canada Basin and the CAA:
mask_ini_CB = np.zeros((isize,jsize))
mask_ini_CAA = np.ones((isize,jsize))
for i, j in zip(CB_indx, CB_indy):
mask_ini_CB[int(i-imin),int(j-jmin)] = 1
mask_ini_CAA[int(i-imin),int(j-jmin)] = 0
mask_ini_CB[150:-1 ,-8:-1] = 1
mask_ini_CAA[150:-1,-8:-1] = 0
mask_ini_wCAA = np.zeros((isize,jsize))
mask_ini_eCAA = np.zeros((isize,jsize))
# Separate the western and eastern CAA:
mask_ini_wCAA = np.where(lons[imin:imax,jmin:jmax] < -100, 1, 0)
mask_ini_eCAA = np.where(lons[imin:imax,jmin:jmax] > -100, 1, 0)
for i, j in zip(CB_indx, CB_indy):
mask_ini_wCAA[int(i-imin),int(j-jmin)] = 0
mask_ini_eCAA[int(i-imin),int(j-jmin)] = 0
mask_ini_wCAA[150:-1,-8:-1] = 0
mask_ini_eCAA[150:-1,-8:-1] = 0
mask_CB_yr = np.tile(mask_ini_CB , (12,1,1))
mask_CAA_yr = np.tile(mask_ini_CAA , (12,1,1))
mask_full_yr = np.ones((12, isize, jsize))
mask_wCAA_yr = np.tile(mask_ini_wCAA, (12,1,1))
mask_eCAA_yr = np.tile(mask_ini_eCAA, (12,1,1))
mask_CB_depth = np.tile(mask_CB_yr , (50,1,1,1))
mask_CB_depth = mask_CB_depth.reshape(12,50,700,640)
```
### Calculate and pickle the results:
```
full_totals_ref, full_totals_spm, full_totals_bio, \
full_averages_ref, full_averages_spm, full_averages_bio = calculate_regional_contributions(mask_full_yr)
pickle.dump((full_totals_ref , full_totals_spm, full_totals_bio ), open('calculations/full-domain-totals-202110.pickle','wb'))
pickle.dump((full_averages_ref, full_averages_spm, full_averages_bio), open('calculations/full-domain-averages-202110.pickle','wb'))
CB_totals_ref, CB_totals_spm, CB_totals_bio, \
CB_averages_ref, CB_averages_spm, CB_averages_bio = calculate_regional_contributions(mask_CB_yr)
pickle.dump((CB_totals_ref , CB_totals_spm, CB_totals_bio ), open('calculations/Canada-Basin-totals-202110--fullcolumn.pickle','wb'))
pickle.dump((CB_averages_ref, CB_averages_spm, CB_averages_bio), open('calculations/Canada-Basin-averages-202110--fullcolumn.pickle','wb'))
CAA_totals_ref, CAA_totals_spm, CAA_totals_bio, \
CAA_averages_ref, CAA_averages_spm, CAA_averages_bio = calculate_regional_contributions(mask_CAA_yr)
pickle.dump((CAA_totals_ref , CAA_totals_spm, CAA_totals_bio ), open('calculations/CAA-totals-202110--fullcolumn.pickle','wb'))
pickle.dump((CAA_averages_ref, CAA_averages_spm, CAA_averages_bio), open('calculations/CAA-averages-202110--fullcolumn.pickle','wb'))
wCAA_totals_ref, wCAA_totals_spm, wCAA_totals_bio, \
wCAA_averages_ref, wCAA_averages_spm, wCAA_averages_bio = calculate_regional_contributions(mask_wCAA_yr)
pickle.dump((wCAA_totals_ref , wCAA_totals_spm, wCAA_totals_bio ), open('calculations/wCAA-totals-202110.pickle','wb'))
pickle.dump((wCAA_averages_ref, wCAA_averages_spm, wCAA_averages_bio), open('calculations/wCAA-averages-202110.pickle','wb'))
eCAA_totals_ref, eCAA_totals_spm, eCAA_totals_bio, \
eCAA_averages_ref, eCAA_averages_spm, eCAA_averages_bio = calculate_regional_contributions(mask_eCAA_yr)
pickle.dump((eCAA_totals_ref , eCAA_totals_spm, eCAA_totals_bio ), open('calculations/eCAA-totals-202110.pickle','wb'))
pickle.dump((eCAA_averages_ref, eCAA_averages_spm, eCAA_averages_bio), open('calculations/eCAA-averages-202110.pickle','wb'))
```
### Overview:
```
# Component contribution calculations partial water column:
CB_totals_ref, CB_totals_spm, CB_totals_bio = pickle.load(open('calculations/Canada-Basin-totals-202110.pickle','rb'))
CAA_totals_ref, CAA_totals_spm, CAA_totals_bio = pickle.load(open('calculations/CAA-totals-202110.pickle' ,'rb'))
wCAA_totals_ref, wCAA_totals_spm, wCAA_totals_bio = pickle.load(open('calculations/wCAA-totals-202110.pickle','rb'))
eCAA_totals_ref, eCAA_totals_spm, eCAA_totals_bio = pickle.load(open('calculations/eCAA-totals-202110.pickle','rb'))
totals_ref , totals_spm , totals_bio = pickle.load(open('calculations/full-domain-totals-202110.pickle','rb'))
ref_totals = {'full': totals_ref, 'CB': CB_totals_ref, 'CAA':CAA_totals_ref, 'wCAA':wCAA_totals_ref, 'eCAA':eCAA_totals_ref}
spm_totals = {'full': totals_spm, 'CB': CB_totals_spm, 'CAA':CAA_totals_spm, 'wCAA':wCAA_totals_spm, 'eCAA':eCAA_totals_spm}
bio_totals = {'full': totals_bio, 'CB': CB_totals_bio, 'CAA':CAA_totals_bio, 'wCAA':wCAA_totals_bio, 'eCAA':eCAA_totals_bio}
# Component contribution calculations partial water column:
CB_averages_ref, CB_averages_spm, CB_averages_bio = pickle.load(open('calculations/Canada-Basin-averages-202110.pickle','rb'))
CAA_averages_ref, CAA_averages_spm, CAA_averages_bio = pickle.load(open('calculations/CAA-averages-202110.pickle' ,'rb'))
wCAA_averages_ref, wCAA_averages_spm, wCAA_averages_bio = pickle.load(open('calculations/wCAA-averages-202110.pickle','rb'))
eCAA_averages_ref, eCAA_averages_spm, eCAA_averages_bio = pickle.load(open('calculations/eCAA-averages-202110.pickle','rb'))
averages_ref , averages_spm , averages_bio = pickle.load(open('calculations/full-domain-averages-202110.pickle','rb'))
ref_averages = {'full': averages_ref, 'CB': CB_averages_ref, 'CAA':CAA_averages_ref, 'wCAA':wCAA_averages_ref, 'eCAA':eCAA_averages_ref}
spm_averages = {'full': averages_spm, 'CB': CB_averages_spm, 'CAA':CAA_averages_spm, 'wCAA':wCAA_averages_spm, 'eCAA':eCAA_averages_spm}
bio_averages = {'full': averages_bio, 'CB': CB_averages_bio, 'CAA':CAA_averages_bio, 'wCAA':wCAA_averages_bio, 'eCAA':eCAA_averages_bio}
# Component contribution calculations full water column:
CB_totals_ref, CB_totals_spm, CB_totals_bio = pickle.load(open('calculations/Canada-Basin-totals-202110--fullcolumn.pickle','rb'))
CAA_totals_ref, CAA_totals_spm, CAA_totals_bio = pickle.load(open('calculations/CAA-totals-202110--fullcolumn.pickle' ,'rb'))
wCAA_totals_ref, wCAA_totals_spm, wCAA_totals_bio = pickle.load(open('calculations/wCAA-totals-202110.pickle','rb'))
eCAA_totals_ref, eCAA_totals_spm, eCAA_totals_bio = pickle.load(open('calculations/eCAA-totals-202110.pickle','rb'))
totals_ref , totals_spm , totals_bio = pickle.load(open('calculations/full-domain-totals-202110.pickle','rb'))
ref_totals = {'full': totals_ref, 'CB': CB_totals_ref, 'CAA':CAA_totals_ref, 'wCAA':wCAA_totals_ref, 'eCAA':eCAA_totals_ref}
spm_totals = {'full': totals_spm, 'CB': CB_totals_spm, 'CAA':CAA_totals_spm, 'wCAA':wCAA_totals_spm, 'eCAA':eCAA_totals_spm}
bio_totals = {'full': totals_bio, 'CB': CB_totals_bio, 'CAA':CAA_totals_bio, 'wCAA':wCAA_totals_bio, 'eCAA':eCAA_totals_bio}
# Component contribution calculations full water column:
CB_averages_ref, CB_averages_spm, CB_averages_bio = pickle.load(open('calculations/Canada-Basin-averages-202110--fullcolumn.pickle','rb'))
CAA_averages_ref, CAA_averages_spm, CAA_averages_bio = pickle.load(open('calculations/CAA-averages-202110--fullcolumn.pickle' ,'rb'))
wCAA_averages_ref, wCAA_averages_spm, wCAA_averages_bio = pickle.load(open('calculations/wCAA-averages-202110.pickle','rb'))
eCAA_averages_ref, eCAA_averages_spm, eCAA_averages_bio = pickle.load(open('calculations/eCAA-averages-202110.pickle','rb'))
averages_ref , averages_spm , averages_bio = pickle.load(open('calculations/full-domain-averages-202110.pickle','rb'))
ref_averages = {'full': averages_ref, 'CB': CB_averages_ref, 'CAA':CAA_averages_ref, 'wCAA':wCAA_averages_ref, 'eCAA':eCAA_averages_ref}
spm_averages = {'full': averages_spm, 'CB': CB_averages_spm, 'CAA':CAA_averages_spm, 'wCAA':wCAA_averages_spm, 'eCAA':eCAA_averages_spm}
bio_averages = {'full': averages_bio, 'CB': CB_averages_bio, 'CAA':CAA_averages_bio, 'wCAA':wCAA_averages_bio, 'eCAA':eCAA_averages_bio}
final_ref_totals, final_spm_totals, final_bio_totals, \
final_ref_averages, final_spm_averages, final_bio_averages = pipeline(ref_totals, spm_totals, bio_totals, \
ref_averages, spm_averages, bio_averages)
print_interannual_averages(final_ref_totals, final_spm_totals, final_bio_totals, \
final_ref_averages, final_spm_averages, final_bio_averages, \
location='full')
print_interannual_averages(final_ref_totals, final_spm_totals, final_bio_totals, \
final_ref_averages, final_spm_averages, final_bio_averages, \
location='CB')
# full water column:
print_interannual_averages(final_ref_totals, final_spm_totals, final_bio_totals, \
final_ref_averages, final_spm_averages, final_bio_averages, \
location='CB')
# full water column:
print_interannual_averages(final_ref_totals, final_spm_totals, final_bio_totals, \
final_ref_averages, final_spm_averages, final_bio_averages, \
location='CAA')
print_interannual_averages(final_ref_totals, final_spm_totals, final_bio_totals, \
final_ref_averages, final_spm_averages, final_bio_averages, \
location='CAA')
print_interannual_averages(final_ref_totals, final_spm_totals, final_bio_totals, \
final_ref_averages, final_spm_averages, final_bio_averages, \
location='wCAA')
print_interannual_averages(final_ref_totals, final_spm_totals, final_bio_totals, \
final_ref_averages, final_spm_averages, final_bio_averages, \
location='eCAA')
# biology runs only go to 2015, so later years have NaNs.
print_interannual_averages_3D(final_ref_totals, final_spm_totals, final_bio_totals, \
final_ref_averages, final_spm_averages, final_bio_averages, \
location='CB')
print_interannual_averages_3D(final_ref_totals, final_spm_totals, final_bio_totals, \
final_ref_averages, final_spm_averages, final_bio_averages, \
location='CAA')
```
|
github_jupyter
|
import pickle
import numpy as np
import netCDF4 as nc
import pandas as pd
from calendar import monthrange
%matplotlib inline
# domain dimensions:
imin, imax = 1479, 2179
jmin, jmax = 159, 799
isize = imax-imin
jsize = jmax-jmin
# Mn model result folders:
folder_ref = '/data/brogalla/run_storage/Mn-reference-202110/'
folder_cleanice = '/data/brogalla/run_storage/Mn-clean-ice-202110/'
folder_spm = '/data/brogalla/run_storage/Mn-spm-202110/'
folder_bio = '/data/brogalla/run_storage/Mn-bio-202110/'
colors = ['#ccb598', '#448d90', '#739f78', '#CC8741', '#cee7fd', '#b9c1c7']
years = [2002, 2003, 2004, 2005, 2006, 2007, 2008, 2009, 2010, 2011, 2012, 2013, 2014, 2015, 2016, \
2017, 2018, 2019]
# ANHA12 grid:
mask = nc.Dataset('/ocean/brogalla/GEOTRACES/data/ANHA12/ANHA12_mesh1.nc')
tmask = np.array(mask.variables['tmask'])[0,:,imin:imax,jmin:jmax]
land_mask = np.ma.masked_where((tmask[:,:,:] > 0.1), tmask[:,:,:])
e1t_base = np.array(mask.variables['e1t'])[0,imin:imax,jmin:jmax]
e2t_base = np.array(mask.variables['e2t'])[0,imin:imax,jmin:jmax]
e3t = np.array(mask.variables['e3t_0'])[0,:,imin:imax,jmin:jmax]
e3t_masked = np.ma.masked_where((tmask[:,:,:] < 0.1), e3t)
nav_lev = np.array(mask.variables['nav_lev'])
lons = np.array(mask.variables['nav_lon'])
e1t = np.tile(e1t_base, (50,1,1))
e2t = np.tile(e2t_base, (50,1,1))
volume = e1t*e2t*e3t
area_base = e1t_base*e2t_base
volume_masked = np.ma.masked_where((tmask[:,:,:] < 0.1), volume)
area_masked = np.ma.masked_where((tmask[0,:,:] < 0.1), area_base)
def load_results(folder_ref, year, experiment):
months = ['01', '02', '03', '04', '05', '06', '07', '08', '09', '10', '11', '12']
dmn_riv = np.empty((12,isize,jsize)) ; dmn_sed = np.empty((12,isize,jsize));
dmn_atm = np.empty((12,isize,jsize)) ; dmn_ice = np.empty((12,isize,jsize));
dmn_sedice = np.empty((12,isize,jsize)); dmn_priv = np.empty((12,isize,jsize));
dmn_red = np.empty((12,50,isize,jsize)); dmn_oxi = np.empty((12,50,isize,jsize));
dmn_bio = np.empty((12,50,isize,jsize));
for i, month in enumerate(months):
file = f'ANHA12_EXH006_1m_{year}0101_{year}1231_comp_{year}{month}-{year}{month}.nc'
ref_monthly = nc.Dataset(folder_ref+file)
dmn_riv[i,:,:] = np.array(ref_monthly.variables['dmnriv'])[0,:,:]
if experiment=='spm':
dmn_priv[i,:,:] = np.array(ref_monthly.variables['pmnriv'])[0,:,:]
dmn_bio[i,:,:] = 0
elif experiment=='bio':
dmn_priv[i,:,:] = 0
dmn_bio[i,:,:] = np.array(ref_monthly.variables['dmnbio'])[0,:,:,:]
else:
dmn_priv[i,:,:] = 0
dmn_bio[i,:,:] = 0
dmn_sed[i,:,:] = np.array(ref_monthly.variables['dmnsed'])[0,:,:]
dmn_sedice[i,:,:] = np.array(ref_monthly.variables['dmnsedice'])[0,:,:]
dmn_atm[i,:,:] = np.array(ref_monthly.variables['dmnatm'])[0,:,:]
dmn_ice[i,:,:] = np.array(ref_monthly.variables['dmnice'])[0,:,:]
dmn_red[i,:,:,:] = np.array(ref_monthly.variables['dmnred'])[0,:,:,:]
dmn_oxi[i,:,:,:] = np.array(ref_monthly.variables['dmnoxi'])[0,:,:,:]
tmask_surf = np.empty(dmn_riv.shape)
tmask_surf[:] = tmask[0,:,:]
tmask_full = np.empty(dmn_red.shape)
tmask_full[:] = tmask
# Mask points on land:
dmn_riv = np.ma.masked_where((tmask_surf < 0.1), dmn_riv);
dmn_priv = np.ma.masked_where((tmask_surf < 0.1), dmn_priv);
dmn_sed = np.ma.masked_where((tmask_surf < 0.1), dmn_sed)
dmn_sedice = np.ma.masked_where((tmask_surf < 0.1), dmn_sedice)
dmn_atm = np.ma.masked_where((tmask_surf < 0.1), dmn_atm)
dmn_ice = np.ma.masked_where((tmask_surf < 0.1), dmn_ice)
dmn_red = np.ma.masked_where((tmask_full < 0.1), dmn_red)
dmn_oxi = np.ma.masked_where((tmask_full < 0.1), dmn_oxi)
dmn_bio = np.ma.masked_where((tmask_full < 0.1), dmn_bio)
return dmn_riv, dmn_priv, dmn_sed, dmn_sedice, dmn_atm, dmn_ice, dmn_red, dmn_oxi, dmn_bio
def calculate_contribution(year, mask, experiment='ref'):
# Calculate the contribution of the model components by region:
if experiment=='spm':
folder_year = folder_spm + f'ANHA12_spm-{year}_20211026/'
elif experiment=='bio':
folder_year = folder_bio + f'ANHA12_bio-{year}_20211022/'
else:
folder_year = folder_ref + f'ANHA12_ref-{year}_20211012/'
if experiment=='bio' and year>2015:
total_yearly_contribution = np.empty((9))
ave_yearly_contribution = np.empty((9))
total_yearly_contribution[:] = np.NaN
ave_yearly_contribution[:] = np.NaN
return total_yearly_contribution, ave_yearly_contribution
dmn_mriv, dmn_mpriv, dmn_msed, dmn_msedice, dmn_matm, dmn_mice, dmn_mred, dmn_moxi, dmn_mbio = \
load_results(folder_year, year, experiment)
# Calculate contributions to the upper water column, so mask locations where sediment resuspension is added in the deep.
indexes_bottom = np.tile(index_bottom, (12,1,1))
# Momentary contribution: moles / second (from moles / L / s --> moles / m3 / s --> moles / s)
# Mask is for the points associated with the specified region.
priv = np.ma.masked_where(mask==0, dmn_mpriv) * volume_masked[0,:,:] * 1e3
riv = np.ma.masked_where(mask==0, dmn_mriv) * volume_masked[0,:,:] * 1e3
sed = np.ma.masked_where(mask==0, dmn_msed) * volume_bottom * 1e3
sedice = np.ma.masked_where(mask==0, dmn_msedice) * volume_masked[0,:,:] * 1e3
atm = np.ma.masked_where(mask==0, dmn_matm) * volume_masked[0,:,:] * 1e3
ice = np.ma.masked_where(mask==0, dmn_mice) * volume_masked[0,:,:] * 1e3
# 3D fields only calculated over polar mixed layer:
mask_with_depths = np.tile(mask, (50,1,1,1)).reshape((12,50,700,640))
bio = np.ma.masked_where(mask_with_depths==0, dmn_mbio)[:,0:17,:,:] * volume_masked[0:17,:,:] * 1e3
red = np.ma.masked_where(mask_with_depths==0, dmn_mred)[:,0:17,:,:] * volume_masked[0:17,:,:] * 1e3
oxi = np.ma.masked_where(mask_with_depths==0, dmn_moxi)[:,0:17,:,:] * volume_masked[0:17,:,:] * 1e3
# Yearly contribution to domain: moles / year (from moles / s / month --> moles / month --> moles / year)
atm_year = 0; riv_year = 0; priv_year = 0; sed_year = 0; sedice_year = 0;
ice_year = 0; bio_year = 0; red_year = 0; oxi_year = 0;
for month in range(1,13):
index = month-1
days_in_month = monthrange(year, month)[1]
if days_in_month == 29: # ignore leap year
days_in_month = 28
atm_year = atm_year + np.ma.sum(atm[index,:,:],axis=(0,1)) *3600*24*days_in_month
riv_year = riv_year + np.ma.sum(riv[index,:,:],axis=(0,1)) *3600*24*days_in_month
priv_year = priv_year + np.ma.sum(priv[index,:,:],axis=(0,1)) *3600*24*days_in_month
sed_year = sed_year + np.ma.sum(sed[index,:,:],axis=(0,1)) *3600*24*days_in_month
sedice_year = sedice_year + np.ma.sum(sedice[index,:,:],axis=(0,1))*3600*24*days_in_month
ice_year = ice_year + np.ma.sum(ice[index,:,:],axis=(0,1)) *3600*24*days_in_month
bio_year = bio_year + np.ma.sum(bio[index,:,:,:],axis=(0,1,2)) *3600*24*days_in_month
red_year = red_year + np.ma.sum(red[index,:,:,:],axis=(0,1,2)) *3600*24*days_in_month
oxi_year = oxi_year + np.ma.sum(oxi[index,:,:,:],axis=(0,1,2)) *3600*24*days_in_month
# Average yearly contribution over domain: moles / m2 / year
total_area = np.ma.sum(np.ma.masked_where(mask[0,:,:]==0, area_masked[:,:]))
atmm2_year = atm_year / total_area
rivm2_year = riv_year / total_area
privm2_year = priv_year / total_area
sedm2_year = sed_year / total_area
sedicem2_year = sedice_year / total_area
icem2_year = ice_year / total_area
# Polar mixed layer average contribution:
PML_depth = nav_lev[17]
biom2_year = bio_year / (total_area*PML_depth)
redm2_year = red_year / (total_area*PML_depth)
oxim2_year = oxi_year / (total_area*PML_depth)
total_yearly_contribution = np.array([priv_year, riv_year, sed_year, sedice_year, ice_year, atm_year, \
bio_year, red_year, oxi_year]) # mol/yr
ave_yearly_contribution = np.array([privm2_year, rivm2_year, sedm2_year, sedicem2_year, icem2_year, \
atmm2_year, biom2_year, redm2_year, oxim2_year]) # mol/m2/yr
return total_yearly_contribution, ave_yearly_contribution
def calculate_regional_contributions(mask):
totals_ref = np.empty((len(years),9)); totals_spm = np.empty((len(years),9)); totals_bio = np.empty((len(years),9));
averages_ref = np.empty((len(years),9)); averages_spm = np.empty((len(years),9)); averages_bio = np.empty((len(years),9));
totals_ref[:] = np.NaN; totals_spm[:] = np.NaN; totals_bio[:] = np.NaN;
averages_ref[:] = np.NaN; averages_spm[:] = np.NaN; averages_spm[:] = np.NaN;
for i, year in enumerate(years):
total_ref_year, ave_ref_year = calculate_contribution(year, mask, experiment='ref')
total_spm_year, ave_spm_year = calculate_contribution(year, mask, experiment='spm')
total_bio_year, ave_bio_year = calculate_contribution(year, mask, experiment='bio')
totals_ref[i,:] = total_ref_year
totals_spm[i,:] = total_spm_year
totals_bio[i,:] = total_bio_year
averages_ref[i,:] = ave_ref_year
averages_spm[i,:] = ave_spm_year
averages_bio[i,:] = ave_bio_year
return totals_ref, totals_spm, totals_bio, averages_ref, averages_spm, averages_bio
def reshape_arrays(array_in):
fixed = np.zeros((len(years),8))
for i in range(0,8):
if i==0: # Sum the particulate and dissolved river contributions
fixed[:,i] = array_in[:,i]+array_in[:,i+1]
else:
fixed[:,i] = array_in[:,i+1]
return fixed
def pipeline(totals_input_ref, totals_input_spm, totals_input_bio,
averages_input_ref, averages_input_spm, averages_input_bio):
regions = ['full', 'CB', 'CAA', 'wCAA', 'eCAA']
totals_fixed_ref = totals_input_ref.copy(); averages_fixed_ref = averages_input_ref.copy();
totals_fixed_spm = totals_input_spm.copy(); averages_fixed_spm = averages_input_spm.copy();
totals_fixed_bio = totals_input_bio.copy(); averages_fixed_bio = averages_input_bio.copy();
# Reshape arrays so that particulate and dissolved river contributions are combined into one.
for region in regions:
totals_fixed_ref[region] = reshape_arrays(totals_input_ref[region])
totals_fixed_spm[region] = reshape_arrays(totals_input_spm[region])
totals_fixed_bio[region] = reshape_arrays(totals_input_bio[region])
averages_fixed_ref[region] = reshape_arrays(averages_input_ref[region])
averages_fixed_spm[region] = reshape_arrays(averages_input_spm[region])
averages_fixed_bio[region] = reshape_arrays(averages_input_bio[region])
return totals_fixed_ref, totals_fixed_spm, totals_fixed_bio, averages_fixed_ref, averages_fixed_spm, averages_fixed_bio
def print_interannual_averages(totals_ref, totals_spm, totals_bio,
averages_ref, averages_spm, averages_bio,
location='Full'):
print(f'Interannual average values in {location}')
print('----------------------------------------------------------------')
total_ref_annual = np.sum(totals_ref[location][:,0:5], axis=1)
total_spm_annual = np.sum(totals_spm[location][:,0:5], axis=1)
total_bio_annual = np.sum(totals_bio[location][:,0:5], axis=1)
# Total annual contribution is averaged over the 18-year time series and multiplied by 1e-6 to convert to Mmol
print(f"{'Total annual contribution of Mn [Mmol/yr]:':<55}" +
f"{np.average(total_ref_annual)*1e-6:<5.0f} ({np.average(total_spm_annual)*1e-6:<5.0f})")
print(f"{'-- River discharge ---':<40}" +
f" {np.average(totals_ref[location][:,0])*1e-6:<8.2f} ({np.average(totals_spm[location][:,0])*1e-6:<5.2f})")
print(f"{'-- Sediment resuspension ---':<40}" +
f" {np.average(totals_ref[location][:,1])*1e-6:<8.2f} ({np.average(totals_spm[location][:,1])*1e-6:<5.2f})")
print(f"{'-- Sediment released by sea ice ---':<40}" +
f" {np.average(totals_ref[location][:,2])*1e-6:<8.2f} ({np.average(totals_spm[location][:,2])*1e-6:<5.2f})")
print(f"{'-- Dust released by sea ice ---':<40}" +
f" {np.average(totals_ref[location][:,3])*1e-6:<8.2f} ({np.average(totals_spm[location][:,3])*1e-6:<5.2f})")
print(f"{'-- Dust deposition ---':<40}" +
f" {np.average(totals_ref[location][:,4])*1e-6:<8.2f} ({np.average(totals_spm[location][:,4])*1e-6:<5.2f})")
tref = np.sum(totals_ref[location][:,0:5], axis=(0,1))
tspm = np.sum(totals_spm[location][:,0:5], axis=(0,1))
tbio = np.sum(totals_bio[location][:,0:5], axis=(0,1))
# Percent is calculated from sum of component contribution over all months / total contributions over full time period
print('----------------------------------------------------------------')
print('Total annual contribution of Mn [%]:')
print(f"{'-- River discharge ---':<40}" +
f"{np.sum(totals_ref[location][:,0])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,0])*100/tspm:<5.2f})")
print(f"{'-- Sediment resuspension ---':<40}" +
f"{np.sum(totals_ref[location][:,1])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,1])*100/tspm:<5.2f})")
print(f"{'-- Sediment released by sea ice ---':<40}" +
f"{np.sum(totals_ref[location][:,2])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,2])*100/tspm:<5.2f})")
print(f"{'-- Dust released by sea ice ---':<40}" +
f"{np.sum(totals_ref[location][:,3])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,3])*100/tspm:<5.2f})")
print(f"{'-- Dust deposition ---':<40}" +
f"{np.sum(totals_ref[location][:,4])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,4])*100/tspm:<5.2f})")
# Average annual contribution is calculated as an average over the full time series and converted to millimol
print('----------------------------------------------------------------')
print(f"{'Average annual contribution of Mn [micromol/m2/yr]:':<55}" +
f"{np.average(np.sum(averages_ref[location][:,0:5], axis=1)*1e6):<5.0f}" +
f"({np.average(np.sum(averages_spm[location][:,0:5], axis=1)*1e6):<5.0f})")
print(f"{'-- River discharge ---':<40}" +
f"{np.average(averages_ref[location][:,0])*1e6:<8.2f} ({np.average(averages_spm[location][:,0])*1e6:<5.2f})")
print(f"{'-- Sediment resuspension ---':<40}" +
f"{np.average(averages_ref[location][:,1])*1e6:<8.2f} ({np.average(averages_spm[location][:,1])*1e6:<5.2f})")
print(f"{'-- Sediment released by sea ice ---':<40}" +
f"{np.average(averages_ref[location][:,2])*1e6:<8.2f} ({np.average(averages_spm[location][:,2])*1e6:<5.2f})")
print(f"{'-- Dust released by sea ice ---':<40}" +
f"{np.average(averages_ref[location][:,3])*1e6:<8.2f} ({np.average(averages_spm[location][:,3])*1e6:<5.2f})")
print(f"{'-- Dust deposition ---':<40}" +
f"{np.average(averages_ref[location][:,4])*1e6:<8.2f} ({np.average(averages_spm[location][:,4])*1e6:<5.2f})")
return
def print_interannual_averages_3D(totals_ref, totals_spm, totals_bio,
averages_ref, averages_spm, averages_bio,
location='Full'):
# include reduction, oxidation, and bio components calculated over the polar mixed layer
print(f'Interannual average values in {location}')
print('----------------------------------------------------------------')
total_ref_annual = np.sum(totals_ref[location], axis=1)
total_spm_annual = np.sum(totals_spm[location], axis=1)
total_bio_annual = np.nansum(totals_bio[location], axis=1)
print(f"{'Total annual contribution of Mn [Mmol/yr]:':<55}" +
f"{np.average(total_ref_annual)*1e-6:<5.0f} ({np.average(total_spm_annual)*1e-6:<5.0f})" + \
f" {np.nanmean(total_bio_annual)*1e-6:<8.2f}")
print(f"{'-- River discharge ---':<40}" +
f" {np.average(totals_ref[location][:,0])*1e-6:<8.2f} ({np.average(totals_spm[location][:,0])*1e-6:<5.2f})" + \
f" {np.nanmean(totals_bio[location][:,0])*1e-6:<8.2f}")
print(f"{'-- Sediment resuspension ---':<40}" +
f" {np.average(totals_ref[location][:,1])*1e-6:<8.2f} ({np.average(totals_spm[location][:,1])*1e-6:<5.2f})" + \
f" {np.nanmean(totals_bio[location][:,1])*1e-6:<8.2f}")
print(f"{'-- Sediment released by sea ice ---':<40}" +
f" {np.average(totals_ref[location][:,2])*1e-6:<8.2f} ({np.average(totals_spm[location][:,2])*1e-6:<5.2f})" + \
f" {np.nanmean(totals_bio[location][:,2])*1e-6:<8.2f}")
print(f"{'-- Dust released by sea ice ---':<40}" +
f" {np.average(totals_ref[location][:,3])*1e-6:<8.2f} ({np.average(totals_spm[location][:,3])*1e-6:<5.2f})" + \
f" {np.nanmean(totals_bio[location][:,3])*1e-6:<8.2f}")
print(f"{'-- Dust deposition ---':<40}" +
f" {np.average(totals_ref[location][:,4])*1e-6:<8.2f} ({np.average(totals_spm[location][:,4])*1e-6:<5.2f})" + \
f" {np.nanmean(totals_bio[location][:,4])*1e-6:<8.2f}")
print(f"{'-- Uptake/remineralization ---':<40}" +
f" {np.average(totals_ref[location][:,5])*1e-6:<8.2f} ({np.average(totals_spm[location][:,5])*1e-6:<5.2f})" + \
f" {np.nanmean(totals_bio[location][:,5])*1e-6:<8.2f}")
print(f"{'-- Scavenging ---':<40}" +
f" {(np.average(totals_ref[location][:,6])-np.average(totals_ref[location][:,7]))*1e-6:<8.2f}" + \
f"({(np.average(totals_spm[location][:,6])-np.average(totals_spm[location][:,7]))*1e-6:<5.2f})" + \
f" {(np.nanmean(totals_bio[location][:,6])-np.nanmean(totals_bio[location][:,7]))*1e-6:<8.2f}")
tref = np.sum(totals_ref[location], axis=(0,1))
tspm = np.sum(totals_spm[location], axis=(0,1))
tbio = np.nansum(totals_bio[location], axis=(0,1))
print('----------------------------------------------------------------')
print('Total annual contribution of Mn [%]:')
print(f"{'-- River discharge ---':<40}" +
f"{np.sum(totals_ref[location][:,0])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,0])*100/tspm:<5.2f})" + \
f"{np.nansum(totals_bio[location][:,0])*100/tref:<5.2f}")
print(f"{'-- Sediment resuspension ---':<40}" +
f"{np.sum(totals_ref[location][:,1])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,1])*100/tspm:<5.2f})" + \
f"{np.nansum(totals_bio[location][:,1])*100/tref:<5.2f}")
print(f"{'-- Sediment released by sea ice ---':<40}" +
f"{np.sum(totals_ref[location][:,2])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,2])*100/tspm:<5.2f})" + \
f"{np.nansum(totals_bio[location][:,2])*100/tref:<5.2f}")
print(f"{'-- Dust released by sea ice ---':<40}" +
f"{np.sum(totals_ref[location][:,3])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,3])*100/tspm:<5.2f})" + \
f"{np.nansum(totals_bio[location][:,3])*100/tref:<5.2f}")
print(f"{'-- Dust deposition ---':<40}" +
f"{np.sum(totals_ref[location][:,4])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,4])*100/tspm:<5.2f})" + \
f"{np.nansum(totals_bio[location][:,4])*100/tref:<5.2f}")
print(f"{'-- Uptake/remineralization ---':<40}" +
f"{np.sum(totals_ref[location][:,5])*100/tref:<5.2f} ({np.sum(totals_spm[location][:,5])*100/tspm:<5.2f})" + \
f"{np.nansum(totals_bio[location][:,5])*100/tref:<5.2f}")
print(f"{'-- Scavenging ---':<40}" +
f"{np.sum(totals_ref[location][:,6] - totals_ref[location][:,7])*100/tref:<5.2f} "+ \
f"({np.sum(totals_spm[location][:,6] - totals_spm[location][:,7])*100/tspm:<5.2f})" + \
f"{np.nansum(totals_bio[location][:,6]- totals_bio[location][:,7])*100/tref:<5.2f}")
print('----------------------------------------------------------------')
print(f"{'Average annual contribution of Mn [micromol/m2/yr]:':<55}" +
f"{np.average(np.sum(averages_ref[location], axis=1)*1e6):<5.0f}" +
f"({np.average(np.sum(averages_spm[location], axis=1)*1e6):<5.0f})"+
f"{np.nanmean(np.nansum(averages_bio[location], axis=1)*1e6):<5.0f}")
print(f"{'-- River discharge ---':<40}" +
f"{np.average(averages_ref[location][:,0])*1e6:<8.2f} ({np.average(averages_spm[location][:,0])*1e6:<5.2f})" +\
f"{np.nanmean(averages_bio[location][:,0])*1e6:<8.2f}")
print(f"{'-- Sediment resuspension ---':<40}" +
f"{np.average(averages_ref[location][:,1])*1e6:<8.2f} ({np.average(averages_spm[location][:,1])*1e6:<5.2f})" +\
f"{np.nanmean(averages_bio[location][:,1])*1e6:<8.2f}")
print(f"{'-- Sediment released by sea ice ---':<40}" +
f"{np.average(averages_ref[location][:,2])*1e6:<8.2f} ({np.average(averages_spm[location][:,2])*1e6:<5.2f})" +\
f"{np.nanmean(averages_bio[location][:,2])*1e6:<8.2f}")
print(f"{'-- Dust released by sea ice ---':<40}" +
f"{np.average(averages_ref[location][:,3])*1e6:<8.2f} ({np.average(averages_spm[location][:,3])*1e6:<5.2f})" +\
f"{np.nanmean(averages_bio[location][:,3])*1e6:<8.2f}")
print(f"{'-- Dust deposition ---':<40}" +
f"{np.average(averages_ref[location][:,4])*1e6:<8.2f} ({np.average(averages_spm[location][:,4])*1e6:<5.2f})" +\
f"{np.nanmean(averages_bio[location][:,4])*1e6:<8.2f}")
print(f"{'-- Uptake/remineralization ---':<40}" +
f"{np.average(averages_ref[location][:,5])*1e6:<8.2f} ({np.average(averages_spm[location][:,5])*1e6:<5.2f})" +\
f"{np.nanmean(averages_bio[location][:,5])*1e6:<8.2f}")
print(f"{'-- Scavenging ---':<40}" +
f"{(np.average(averages_ref[location][:,6]) - np.average(averages_ref[location][:,7]))*1e6:<8.2f}"+ \
f"({(np.average(averages_spm[location][:,6])- np.average(averages_spm[location][:,7]))*1e6:<5.2f})" +\
f"{(np.nanmean(averages_bio[location][:,6]) - np.nanmean(averages_bio[location][:,7]))*1e6:<8.2f}")
return
tmask_bot = np.copy(tmask)
tmask_bot[0,:,:] = 1
e3t_mask = np.ma.masked_where((tmask_bot[:,:,:] < 0.1), e3t[:,:,:])
# find bottom grid cell index:
ind = np.array(np.ma.notmasked_edges(e3t_mask[:,:,:], axis=0))
# array of shape (isize, jsize) containing bottom grid cell index
index_bottom = np.ma.masked_where((tmask[0,:,:] < 0.1), ind[1][0][:].reshape(isize,jsize))
e3t_bot = np.zeros((isize,jsize))
vol_bot = np.zeros((isize,jsize))
for i in range(0, isize):
for j in range(0,jsize):
k = index_bottom[i,j]
try:
e3t_bot[i,j] = e3t[k,i,j]
vol_bot[i,j] = volume[k,i,j]
except:
e3t_bot[i,j] = np.nan
vol_bot[i,j] = np.nan
e3t_bottom = np.ma.masked_where((tmask[0,:,:] < 0.1), np.ma.masked_where(np.isnan(e3t_bot), e3t_bot))
volume_bottom = np.ma.masked_where((tmask[0,:,:] < 0.1), np.ma.masked_where(np.isnan(vol_bot), vol_bot))
# Find indices to separate out Canada Basin:
x_ind = np.arange(1621, 2100, 1)
y_ind = (-7/8)*x_ind + 1517 + 700
CB_indx = []
CB_indy = []
for index in range(0,len(x_ind)):
CB_x = np.arange(x_ind[index],2179,1)
CB_y = np.ones(CB_x.shape)*y_ind[index]
CB_indx = np.append(CB_x, CB_indx)
CB_indy = np.append(CB_y, CB_indy)
# Separate Canada Basin and the CAA:
mask_ini_CB = np.zeros((isize,jsize))
mask_ini_CAA = np.ones((isize,jsize))
for i, j in zip(CB_indx, CB_indy):
mask_ini_CB[int(i-imin),int(j-jmin)] = 1
mask_ini_CAA[int(i-imin),int(j-jmin)] = 0
mask_ini_CB[150:-1 ,-8:-1] = 1
mask_ini_CAA[150:-1,-8:-1] = 0
mask_ini_wCAA = np.zeros((isize,jsize))
mask_ini_eCAA = np.zeros((isize,jsize))
# Separate the western and eastern CAA:
mask_ini_wCAA = np.where(lons[imin:imax,jmin:jmax] < -100, 1, 0)
mask_ini_eCAA = np.where(lons[imin:imax,jmin:jmax] > -100, 1, 0)
for i, j in zip(CB_indx, CB_indy):
mask_ini_wCAA[int(i-imin),int(j-jmin)] = 0
mask_ini_eCAA[int(i-imin),int(j-jmin)] = 0
mask_ini_wCAA[150:-1,-8:-1] = 0
mask_ini_eCAA[150:-1,-8:-1] = 0
mask_CB_yr = np.tile(mask_ini_CB , (12,1,1))
mask_CAA_yr = np.tile(mask_ini_CAA , (12,1,1))
mask_full_yr = np.ones((12, isize, jsize))
mask_wCAA_yr = np.tile(mask_ini_wCAA, (12,1,1))
mask_eCAA_yr = np.tile(mask_ini_eCAA, (12,1,1))
mask_CB_depth = np.tile(mask_CB_yr , (50,1,1,1))
mask_CB_depth = mask_CB_depth.reshape(12,50,700,640)
full_totals_ref, full_totals_spm, full_totals_bio, \
full_averages_ref, full_averages_spm, full_averages_bio = calculate_regional_contributions(mask_full_yr)
pickle.dump((full_totals_ref , full_totals_spm, full_totals_bio ), open('calculations/full-domain-totals-202110.pickle','wb'))
pickle.dump((full_averages_ref, full_averages_spm, full_averages_bio), open('calculations/full-domain-averages-202110.pickle','wb'))
CB_totals_ref, CB_totals_spm, CB_totals_bio, \
CB_averages_ref, CB_averages_spm, CB_averages_bio = calculate_regional_contributions(mask_CB_yr)
pickle.dump((CB_totals_ref , CB_totals_spm, CB_totals_bio ), open('calculations/Canada-Basin-totals-202110--fullcolumn.pickle','wb'))
pickle.dump((CB_averages_ref, CB_averages_spm, CB_averages_bio), open('calculations/Canada-Basin-averages-202110--fullcolumn.pickle','wb'))
CAA_totals_ref, CAA_totals_spm, CAA_totals_bio, \
CAA_averages_ref, CAA_averages_spm, CAA_averages_bio = calculate_regional_contributions(mask_CAA_yr)
pickle.dump((CAA_totals_ref , CAA_totals_spm, CAA_totals_bio ), open('calculations/CAA-totals-202110--fullcolumn.pickle','wb'))
pickle.dump((CAA_averages_ref, CAA_averages_spm, CAA_averages_bio), open('calculations/CAA-averages-202110--fullcolumn.pickle','wb'))
wCAA_totals_ref, wCAA_totals_spm, wCAA_totals_bio, \
wCAA_averages_ref, wCAA_averages_spm, wCAA_averages_bio = calculate_regional_contributions(mask_wCAA_yr)
pickle.dump((wCAA_totals_ref , wCAA_totals_spm, wCAA_totals_bio ), open('calculations/wCAA-totals-202110.pickle','wb'))
pickle.dump((wCAA_averages_ref, wCAA_averages_spm, wCAA_averages_bio), open('calculations/wCAA-averages-202110.pickle','wb'))
eCAA_totals_ref, eCAA_totals_spm, eCAA_totals_bio, \
eCAA_averages_ref, eCAA_averages_spm, eCAA_averages_bio = calculate_regional_contributions(mask_eCAA_yr)
pickle.dump((eCAA_totals_ref , eCAA_totals_spm, eCAA_totals_bio ), open('calculations/eCAA-totals-202110.pickle','wb'))
pickle.dump((eCAA_averages_ref, eCAA_averages_spm, eCAA_averages_bio), open('calculations/eCAA-averages-202110.pickle','wb'))
# Component contribution calculations partial water column:
CB_totals_ref, CB_totals_spm, CB_totals_bio = pickle.load(open('calculations/Canada-Basin-totals-202110.pickle','rb'))
CAA_totals_ref, CAA_totals_spm, CAA_totals_bio = pickle.load(open('calculations/CAA-totals-202110.pickle' ,'rb'))
wCAA_totals_ref, wCAA_totals_spm, wCAA_totals_bio = pickle.load(open('calculations/wCAA-totals-202110.pickle','rb'))
eCAA_totals_ref, eCAA_totals_spm, eCAA_totals_bio = pickle.load(open('calculations/eCAA-totals-202110.pickle','rb'))
totals_ref , totals_spm , totals_bio = pickle.load(open('calculations/full-domain-totals-202110.pickle','rb'))
ref_totals = {'full': totals_ref, 'CB': CB_totals_ref, 'CAA':CAA_totals_ref, 'wCAA':wCAA_totals_ref, 'eCAA':eCAA_totals_ref}
spm_totals = {'full': totals_spm, 'CB': CB_totals_spm, 'CAA':CAA_totals_spm, 'wCAA':wCAA_totals_spm, 'eCAA':eCAA_totals_spm}
bio_totals = {'full': totals_bio, 'CB': CB_totals_bio, 'CAA':CAA_totals_bio, 'wCAA':wCAA_totals_bio, 'eCAA':eCAA_totals_bio}
# Component contribution calculations partial water column:
CB_averages_ref, CB_averages_spm, CB_averages_bio = pickle.load(open('calculations/Canada-Basin-averages-202110.pickle','rb'))
CAA_averages_ref, CAA_averages_spm, CAA_averages_bio = pickle.load(open('calculations/CAA-averages-202110.pickle' ,'rb'))
wCAA_averages_ref, wCAA_averages_spm, wCAA_averages_bio = pickle.load(open('calculations/wCAA-averages-202110.pickle','rb'))
eCAA_averages_ref, eCAA_averages_spm, eCAA_averages_bio = pickle.load(open('calculations/eCAA-averages-202110.pickle','rb'))
averages_ref , averages_spm , averages_bio = pickle.load(open('calculations/full-domain-averages-202110.pickle','rb'))
ref_averages = {'full': averages_ref, 'CB': CB_averages_ref, 'CAA':CAA_averages_ref, 'wCAA':wCAA_averages_ref, 'eCAA':eCAA_averages_ref}
spm_averages = {'full': averages_spm, 'CB': CB_averages_spm, 'CAA':CAA_averages_spm, 'wCAA':wCAA_averages_spm, 'eCAA':eCAA_averages_spm}
bio_averages = {'full': averages_bio, 'CB': CB_averages_bio, 'CAA':CAA_averages_bio, 'wCAA':wCAA_averages_bio, 'eCAA':eCAA_averages_bio}
# Component contribution calculations full water column:
CB_totals_ref, CB_totals_spm, CB_totals_bio = pickle.load(open('calculations/Canada-Basin-totals-202110--fullcolumn.pickle','rb'))
CAA_totals_ref, CAA_totals_spm, CAA_totals_bio = pickle.load(open('calculations/CAA-totals-202110--fullcolumn.pickle' ,'rb'))
wCAA_totals_ref, wCAA_totals_spm, wCAA_totals_bio = pickle.load(open('calculations/wCAA-totals-202110.pickle','rb'))
eCAA_totals_ref, eCAA_totals_spm, eCAA_totals_bio = pickle.load(open('calculations/eCAA-totals-202110.pickle','rb'))
totals_ref , totals_spm , totals_bio = pickle.load(open('calculations/full-domain-totals-202110.pickle','rb'))
ref_totals = {'full': totals_ref, 'CB': CB_totals_ref, 'CAA':CAA_totals_ref, 'wCAA':wCAA_totals_ref, 'eCAA':eCAA_totals_ref}
spm_totals = {'full': totals_spm, 'CB': CB_totals_spm, 'CAA':CAA_totals_spm, 'wCAA':wCAA_totals_spm, 'eCAA':eCAA_totals_spm}
bio_totals = {'full': totals_bio, 'CB': CB_totals_bio, 'CAA':CAA_totals_bio, 'wCAA':wCAA_totals_bio, 'eCAA':eCAA_totals_bio}
# Component contribution calculations full water column:
CB_averages_ref, CB_averages_spm, CB_averages_bio = pickle.load(open('calculations/Canada-Basin-averages-202110--fullcolumn.pickle','rb'))
CAA_averages_ref, CAA_averages_spm, CAA_averages_bio = pickle.load(open('calculations/CAA-averages-202110--fullcolumn.pickle' ,'rb'))
wCAA_averages_ref, wCAA_averages_spm, wCAA_averages_bio = pickle.load(open('calculations/wCAA-averages-202110.pickle','rb'))
eCAA_averages_ref, eCAA_averages_spm, eCAA_averages_bio = pickle.load(open('calculations/eCAA-averages-202110.pickle','rb'))
averages_ref , averages_spm , averages_bio = pickle.load(open('calculations/full-domain-averages-202110.pickle','rb'))
ref_averages = {'full': averages_ref, 'CB': CB_averages_ref, 'CAA':CAA_averages_ref, 'wCAA':wCAA_averages_ref, 'eCAA':eCAA_averages_ref}
spm_averages = {'full': averages_spm, 'CB': CB_averages_spm, 'CAA':CAA_averages_spm, 'wCAA':wCAA_averages_spm, 'eCAA':eCAA_averages_spm}
bio_averages = {'full': averages_bio, 'CB': CB_averages_bio, 'CAA':CAA_averages_bio, 'wCAA':wCAA_averages_bio, 'eCAA':eCAA_averages_bio}
final_ref_totals, final_spm_totals, final_bio_totals, \
final_ref_averages, final_spm_averages, final_bio_averages = pipeline(ref_totals, spm_totals, bio_totals, \
ref_averages, spm_averages, bio_averages)
print_interannual_averages(final_ref_totals, final_spm_totals, final_bio_totals, \
final_ref_averages, final_spm_averages, final_bio_averages, \
location='full')
print_interannual_averages(final_ref_totals, final_spm_totals, final_bio_totals, \
final_ref_averages, final_spm_averages, final_bio_averages, \
location='CB')
# full water column:
print_interannual_averages(final_ref_totals, final_spm_totals, final_bio_totals, \
final_ref_averages, final_spm_averages, final_bio_averages, \
location='CB')
# full water column:
print_interannual_averages(final_ref_totals, final_spm_totals, final_bio_totals, \
final_ref_averages, final_spm_averages, final_bio_averages, \
location='CAA')
print_interannual_averages(final_ref_totals, final_spm_totals, final_bio_totals, \
final_ref_averages, final_spm_averages, final_bio_averages, \
location='CAA')
print_interannual_averages(final_ref_totals, final_spm_totals, final_bio_totals, \
final_ref_averages, final_spm_averages, final_bio_averages, \
location='wCAA')
print_interannual_averages(final_ref_totals, final_spm_totals, final_bio_totals, \
final_ref_averages, final_spm_averages, final_bio_averages, \
location='eCAA')
# biology runs only go to 2015, so later years have NaNs.
print_interannual_averages_3D(final_ref_totals, final_spm_totals, final_bio_totals, \
final_ref_averages, final_spm_averages, final_bio_averages, \
location='CB')
print_interannual_averages_3D(final_ref_totals, final_spm_totals, final_bio_totals, \
final_ref_averages, final_spm_averages, final_bio_averages, \
location='CAA')
| 0.323594 | 0.668124 |
# Traveling Salesman Problem
## Objective and Prerequisites
In this notebook, you will learn how to:
1. Formulate the Traveling Salesman Problem (TSP) as a MIP model.
2. Use lazy constraints to identify solutions of the TSP problem that are infeasible.
This modeling example is at the advanced level, where we assume that you know Python and the Gurobi Python API and you have advanced knowledge of building mathematical optimization models. Typically, the objective function and/or constraints of these examples are complex or require advanced features of the Gurobi Python API.
**Note:** You can download the repository containing this and other examples by clicking [here](https://github.com/Gurobi/modeling-examples/archive/master.zip). In order to run this Jupyter Notebook properly, you must have a Gurobi license. If you do not have one, you can request an [evaluation license](https://www.gurobi.com/downloads/request-an-evaluation-license/?utm_source=Github&utm_medium=website_JupyterME&utm_campaign=CommercialDataScience) as a *commercial user*, or download a [free license](https://www.gurobi.com/academia/academic-program-and-licenses/?utm_source=Github&utm_medium=website_JupyterME&utm_campaign=AcademicDataScience) as an *academic user*.
## Motivation
The Traveling Salesman Problem (TSP) is one of the most famous combinatorial optimization problems. This problem is very easy to explain, but very complicated to solve – even for instances with a small number of cities. More detailed information on the TSP can be found in the book The Traveling Salesman Problem: A Computational Study [1], or at the TSP home page [2]. If you are interested in the history and mathematical background of the TSP, we recommend that you watch the video by William Cook [3].
The origin of the traveling salesman problem is not very clear; it is mentioned in an 1832 manual for traveling salesman, which included example tours of 45 German cities but was not formulated as a mathematical problem. However, in the 1800s, mathematicians William Rowan Hamilton and Thomas Kirkman devised mathematical formulations of the problem.
It seems that the general form of the Traveling Salesman Problem was first studied by Karl Menger in Vienna and Harvard in the 1930s.
The problem became more and more popular in the 1950s and 1960s. In particular, George Dantzig, D. Ray Fulkerson, and Selmer M. Johnson at the RAND Corporation solved the 48-state problem by formulating it as a linear programming problem. The methods they described in their paper on this topic set the foundation for future work in combinatorial optimization, especially highlighting the importance of cutting planes.
In the early 1970s, the concept of P vs. NP problems created excitement in the theoretical computer science community. In 1972, Richard Karp demonstrated that the Hamiltonian cycle problem was NP-complete, implying that the traveling salesman problem was NP-hard.
Increasingly sophisticated codes led to rapid increases in the sizes of the traveling salesman problems solved. Dantzig, Fulkerson, and Johnson had solved a 48-city instance of the problem in 1954. Martin Grötechel more than doubled this 23 years later, solving a 120-city instance in 1977. Harlan Crowder and Manfred W. Padberg again more than doubled this in just 3 years, with a 318-city solution.
In 1987, rapid improvements were made, culminating in a 2,392-city solution by Padberg and Giovanni Rinaldi. In the following two decades, great strides were made with David L. Applegate, Robert E. Bixby, Vasek Chvátal, & William J. Cook solving a 3,308-city instance in 1992, a 7,397-city instance in 1994, a 24,978-city instance in 2004, and an 85,900-city instance in 2006 – which is the largest 2-D Euclidean TSP instance ever solved. William Cook et. al. wrote a program called Concorde TSP Solver for solving the TSP [4]. Concorde is a computer code for the symmetric TSP and some related network optimization problems. The code is written in the ANSI C programming language and it has been used to obtain the optimal solutions to the full set of 110 TSPLIB instances, the largest instance is a 109,399 node 3-D “star” instance.
The continued interest in the TSP can be explained by its success as a general engine of discovery and a steady stream of new applications. Some of the general applications of the TSP are as follows:
* Scheduling and routing problems.
* Genome sequencing.
* Drilling problems.
* Aiming telescopes and x-rays.
* Data clustering.
* Machine scheduling.
We use this classic combinatorial optimization problem to demonstrate how Gurobi can be used to easily and effectively solve small-sized problem instances of the TSP. However, in order to be able to solve larger instances, one needs more sophisticated techniques – such as those implemented in the Concord TSP Solver.
## Problem Description
The TSP can be defined as follows: for a given list of cities and the distances between each pair of them, we want to find the shortest possible route that goes to each city once and returns to the origin city.
There is a class of Traveling Salesman Problems that assumes that the distance of going from city $i$ to city $j$ is the same as going form city $j$ to city $i$, this type of Traveling Salesman Problem is also known as the symmetric Traveling Salesman Problem. In this example, we use Euclidean distances, but the TSP model formulation is valid independent of the way in which the individual distances are determined.
## Solution Approach
Mathematical programming is a declarative approach where the modeler formulates a mathematical optimization model that captures the key aspects of a complex decision problem. The Gurobi Optimizer solves such models using state-of-the-art mathematics and computer science.
A mathematical optimization model has five components, namely:
* Sets and indices.
* Parameters.
* Decision variables.
* Objective function(s).
* Constraints.
We now present a MIP formulation of the TSP that identifies the shortest route that goes to all the cities once and returns to the origin city.
## TSP Model Formulation
### Sets and Indices
$i, j \in Capitals $: indices and set of US capital cities.
$\text{Pairings}= \{(i,j) \in Capitals \times Capitals \}$: Set of allowed pairings
$S \subset Capitals$: A subset of the set of US capital cities.
$G = (Capitals, Pairings)$: A graph where the set $Capitals$ defines the set of nodes and the set $Pairings$ defines the set of edges.
### Parameters
$d_{i, j} \in \mathbb{R}^+$: Distance from capital city $i$ to capital city $j$, for all $(i, j) \in Pairings$.
Notice that the distance from capital city $i$ to capital city $j$ is the same as the distance from capital city $j$ to capital city $i$, i.e. $d_{i, j} = d_{j, i}$. For this reason, this TSP is also called the symmetric Traveling Salesman Problem.
### Decision Variables
$x_{i, j} \in \{0, 1\}$: This variable is equal to 1, if we decide to connect city $i$ with city $j$. Otherwise, the decision variable is equal to zero.
### Objective Function
- **Shortest Route**. Minimize the total distance of a route. A route is a sequence of capital cities where the salesperson visits each city only once and returns to the starting capital city.
\begin{equation}
\text{Min} \quad Z = \sum_{(i,j) \in \text{Pairings}}d_{i,j} \cdot x_{i,j}
\tag{0}
\end{equation}
### Constraints
- **Symmetry Constraints**. For each edge $(i,j)$, ensure that the city capitals $i$ and $j$ are connected, if the former is visited immediately before or after visiting the latter.
\begin{equation}
x_{i, j} = x_{j, i} \quad \forall (i, j) \in Pairings
\tag{1}
\end{equation}
- **Entering and leaving a capital city**. For each capital city $i$, ensure that this city is connected to two other cities.
\begin{equation}
\sum_{(i,j) \in \text{Pairings}}x_{i,j} = 2 \quad \forall i \in Capitals
\tag{2}
\end{equation}
- **Subtour elimination**. These constraints ensure that for any subset of cities $S$ of the set of $Capitals$, there is no cycle. That is, there is no route that visits all the cities in the subset and returns to the origin city.
\begin{equation}
\sum_{(i \neq j) \in S}x_{i,j} \leq |S|-1 \quad \forall S \subset Capitals
\tag{3}
\end{equation}
- **Remark**. In general, if the number of cities of the TSP is $n$, then the possible number of routes is n\!.
Since there are an exponential number of constraints ($2^{n} - 2$) to eliminate cycles, we use lazy constraints to dynamically eliminate those cycles.
## Python Implementation
Consider a salesperson that needs to visit customers at each state capital of the continental US. The salesperson wants to identify the shortest route that goes to all the state capitals.
This modeling example requires to import the following libraries.
* **math** access to mathematical functions.
* **itertools** implements a number of iterator building blocks.
* **folium** creates maps.
* **gurobipy** calls Gurobi algorithms to solve MIP models.
### Reading Input Data
The capital names and coordinates are read from a json file.
```
import json
# Read capital names and coordinates from json file
capitals_json = json.load(open('capitals.json'))
capitals = []
coordinates = {}
for state in capitals_json:
if state not in ['AK', 'HI']:
capital = capitals_json[state]['capital']
capitals.append(capital)
coordinates[capital] = (float(capitals_json[state]['lat']), float(capitals_json[state]['long']))
```
### Preprocessing
The following function calculates the distance of each pair of state capitals combination.
```
import math
from itertools import combinations,product
# Compute pairwise distance matrix
def distance(city1, city2):
c1 = coordinates[city1]
c2 = coordinates[city2]
diff = (c1[0]-c2[0], c1[1]-c2[1])
return math.sqrt(diff[0]*diff[0]+diff[1]*diff[1])
dist = {(c1, c2): distance(c1, c2) for c1, c2 in product(capitals, capitals) if c1 != c2}
```
### Callback Definition
The following function determines lazy constraints to eliminate sub-tours.
```
# Callback - use lazy constraints to eliminate sub-tours
def subtourelim(model, where):
if where == GRB.Callback.MIPSOL:
# make a list of edges selected in the solution
vals = model.cbGetSolution(model._vars)
selected = gp.tuplelist((i, j) for i, j in model._vars.keys()
if vals[i, j] > 0.5)
# find the shortest cycle in the selected edge list
tour = subtour(selected)
if len(tour) < len(capitals):
# add subtour elimination constr. for every pair of cities in subtour
model.cbLazy(gp.quicksum(model._vars[i, j] for i, j in combinations(tour, 2))
<= len(tour)-1)
```
### Finding Shortest Route
The following function determines the shortest route from a given set of edges.
```
# Given a tuplelist of edges, find the shortest subtour
def subtour(edges):
unvisited = capitals[:]
cycle = capitals[:] # Dummy - guaranteed to be replaced
while unvisited: # true if list is non-empty
thiscycle = []
neighbors = unvisited
while neighbors:
current = neighbors[0]
thiscycle.append(current)
unvisited.remove(current)
neighbors = [j for i, j in edges.select(current, '*')
if j in unvisited]
if len(thiscycle) <= len(cycle):
cycle = thiscycle # New shortest subtour
return cycle
```
### Model Deployment
We now determine the model for the TSP, by defining decision variables, constraints, and objective function. Next, we start the optimization and Gurobi finds the optimal route for the TSP.
```
import gurobipy as gp
from gurobipy import GRB
# tested with Python 3.7 & Gurobi 9.0.0
m = gp.Model()
# Variables: is city 'i' adjacent to city 'j' on the tour?
vars = m.addVars(dist.keys(), obj=dist, vtype=GRB.BINARY, name='e')
for i, j in vars.keys():
m.addConstr(vars[j, i] == vars[i, j]) # edge in opposite direction
# Constraints: two edges incident to each city
m.addConstrs(vars.sum(c, '*') == 2 for c in capitals)
# Optimize the model
m._vars = vars
m.Params.lazyConstraints = 1
m.optimize(subtourelim)
```
## Analysis
We retrieve the optimal solution of the TSP and verify that the optimal route (or tour) goes to all the cities and returns to the origin city.
```
# Retrieve solution
vals = m.getAttr('x', vars)
selected = gp.tuplelist((i, j) for i, j in vals.keys() if vals[i, j] > 0.5)
tour = subtour(selected)
assert len(tour) == len(capitals)
```
The optimal route is displayed in the following map.
```
# Map the solution
import folium
map = folium.Map(location=[40,-95], zoom_start = 4)
points = []
for city in tour:
points.append(coordinates[city])
points.append(points[0])
folium.PolyLine(points).add_to(map)
map
```
## Conclusions
The Traveling Salesman Problem (TSP) is the most popular combinatorial optimization problem. This problem is very easy to explain, although it is very complicated to solve. The largest TSP problem solved has 85,900 cities. The TSP is a source of discovery for new approaches to solve complex combinatorial optimization problems and has led to many applications.
In this modeling example, we have shown how to formulate the symmetric Traveling Salesman Problem as a MIP problem. We also showed how to dynamically eliminate subtours by using lazy constraints.
## References
[1] D. L. Applegate, R. E. Bixby, V. Chvatal and W. J. Cook , The Traveling Salesman Problem: A Computational Study, Princeton University Press, Princeton, 2006.
[2] http://www.math.uwaterloo.ca/tsp/index.html
[3] https://www.youtube.com/watch?v=q8nQTNvCrjE&t=35s
[4] http://www.math.uwaterloo.ca/tsp/concorde.html
Copyright © 2020 Gurobi Optimization, LLC
|
github_jupyter
|
import json
# Read capital names and coordinates from json file
capitals_json = json.load(open('capitals.json'))
capitals = []
coordinates = {}
for state in capitals_json:
if state not in ['AK', 'HI']:
capital = capitals_json[state]['capital']
capitals.append(capital)
coordinates[capital] = (float(capitals_json[state]['lat']), float(capitals_json[state]['long']))
import math
from itertools import combinations,product
# Compute pairwise distance matrix
def distance(city1, city2):
c1 = coordinates[city1]
c2 = coordinates[city2]
diff = (c1[0]-c2[0], c1[1]-c2[1])
return math.sqrt(diff[0]*diff[0]+diff[1]*diff[1])
dist = {(c1, c2): distance(c1, c2) for c1, c2 in product(capitals, capitals) if c1 != c2}
# Callback - use lazy constraints to eliminate sub-tours
def subtourelim(model, where):
if where == GRB.Callback.MIPSOL:
# make a list of edges selected in the solution
vals = model.cbGetSolution(model._vars)
selected = gp.tuplelist((i, j) for i, j in model._vars.keys()
if vals[i, j] > 0.5)
# find the shortest cycle in the selected edge list
tour = subtour(selected)
if len(tour) < len(capitals):
# add subtour elimination constr. for every pair of cities in subtour
model.cbLazy(gp.quicksum(model._vars[i, j] for i, j in combinations(tour, 2))
<= len(tour)-1)
# Given a tuplelist of edges, find the shortest subtour
def subtour(edges):
unvisited = capitals[:]
cycle = capitals[:] # Dummy - guaranteed to be replaced
while unvisited: # true if list is non-empty
thiscycle = []
neighbors = unvisited
while neighbors:
current = neighbors[0]
thiscycle.append(current)
unvisited.remove(current)
neighbors = [j for i, j in edges.select(current, '*')
if j in unvisited]
if len(thiscycle) <= len(cycle):
cycle = thiscycle # New shortest subtour
return cycle
import gurobipy as gp
from gurobipy import GRB
# tested with Python 3.7 & Gurobi 9.0.0
m = gp.Model()
# Variables: is city 'i' adjacent to city 'j' on the tour?
vars = m.addVars(dist.keys(), obj=dist, vtype=GRB.BINARY, name='e')
for i, j in vars.keys():
m.addConstr(vars[j, i] == vars[i, j]) # edge in opposite direction
# Constraints: two edges incident to each city
m.addConstrs(vars.sum(c, '*') == 2 for c in capitals)
# Optimize the model
m._vars = vars
m.Params.lazyConstraints = 1
m.optimize(subtourelim)
# Retrieve solution
vals = m.getAttr('x', vars)
selected = gp.tuplelist((i, j) for i, j in vals.keys() if vals[i, j] > 0.5)
tour = subtour(selected)
assert len(tour) == len(capitals)
# Map the solution
import folium
map = folium.Map(location=[40,-95], zoom_start = 4)
points = []
for city in tour:
points.append(coordinates[city])
points.append(points[0])
folium.PolyLine(points).add_to(map)
map
| 0.504883 | 0.983391 |
## Deep Learning Challenge
### Loading the CIFAR10 data
The data can be loaded directly from keras (`keras.datasets.cifar10`).
```python
cifar10 = keras.datasets.cifar10
(train_images, train_labels), (test_images, test_labels) = cifar10.load_data()
```
```
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Convolution2D
from tensorflow.keras.layers import MaxPooling2D
from tensorflow.keras.layers import Flatten
from tensorflow.keras.layers import Dense
from sklearn.model_selection import KFold
from matplotlib import pyplot
import tensorflow as tf
from tensorflow import keras
from keras.datasets import cifar10
cifar10 = keras.datasets.cifar10
(train_images, train_labels), (test_images, test_labels) = cifar10.load_data()
```
#### Task: Build a convulution neural network (CNN) to predict the object in the images.
Try to do it on your own first before consulting with peers or tutorials on the internet. If you are stuck early, reach out to a mentor who will point you in the right direction.
```
classifier = Sequential()
classifier.add(Convolution2D(32, 3, 3, # 32 no. of filters, then filter dimension
input_shape=(32, 32, 3), # this is image dimensionality
activation='relu'))
# Convolution2D check documentation
classifier.add(MaxPooling2D(pool_size=(2, 2)))
classifier.add(Flatten())
classifier.add(Dense(128, activation='relu'))
classifier.add(Dense(10, activation='softmax'))
classifier.compile(optimizer='adam', loss='sparse_categorical_crossentropy', metrics=['accuracy'])
def load_dataset():
(train_images, train_labels), (test_images, test_labels) = cifar10.load_data()
# how to know what to reshape as?
trainX = train_images
testX = test_images
trainY = train_labels
testY = test_labels
return trainX, trainY, testX, testY
def prep_pixels(train,test):
train_norm = train.astype('float32')
test_norm = test.astype('float32')
# why / 255? RGB
train_norm = train_norm / 255.0
test_norm = test_norm / 255.0
return train_norm, test_norm
def define_model():
model = Sequential()
# explain shapes here
model.add(Convolution2D(32, 3, 3, input_shape=(32, 32, 3), activation='relu'))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Flatten())
model.add(Dense(128, activation='relu'))
# how do I tell how many categories
model.add(Dense(10, activation='softmax'))
model.compile(optimizer='adam', loss='sparse_categorical_crossentropy', metrics=['accuracy'])
return model
def evaluate_model(dataX, dataY, n_folds=5):
scores, histories = list(), list()
kfold = KFold(n_folds, shuffle=True, random_state=42)
for train_ix, test_ix in kfold.split(dataX):
model = define_model()
trainX, trainY, testX, testY = dataX[train_ix], dataY[train_ix], dataX[test_ix], dataY[test_ix]
history = model.fit(trainX, trainY, epochs=10, steps_per_epoch = 80, batch_size=32, validation_data=(testX, testY), verbose=0)
_, acc = model.evaluate(testX, testY, verbose=0)
print('> %.3f' % (acc * 100.0))
scores.append(acc)
histories.append(history)
return scores, histories
def summarize_diagnostics(histories):
for i in range(len(histories)):
pyplot.subplot(211)
pyplot.title('Cross Entropy Loss')
pyplot.plot(histories[i].history['loss'], color='blue', label='train')
pyplot.plot(histories[i].history['val_loss'], color='orange', label='test')
pyplot.subplot(212)
pyplot.title('Classification Accuracy')
pyplot.plot(histories[i].history['accuracy'], color='blue', label='train')
pyplot.plot(histories[i].history['val_accuracy'], color='orange', label='test')
pyplot.show()
def summarize_performance(scores):
print('Accuracy: mean=%.3f std=%.3f, n=%d' % (mean(scores)*100, std(scores)*100, len(scores)))
pyplot.boxplot(scores)
pyplot.show()
def run_test_harness():
trainX, trainY, testX, testY = load_dataset()
trainX, testX = prep_pixels(trainX, testX)
scores, histories = evaluate_model(trainX, trainY)
summarize_performance(scores)
summarize_diagnostics(histories)
run_test_harness()
```
|
github_jupyter
|
cifar10 = keras.datasets.cifar10
(train_images, train_labels), (test_images, test_labels) = cifar10.load_data()
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Convolution2D
from tensorflow.keras.layers import MaxPooling2D
from tensorflow.keras.layers import Flatten
from tensorflow.keras.layers import Dense
from sklearn.model_selection import KFold
from matplotlib import pyplot
import tensorflow as tf
from tensorflow import keras
from keras.datasets import cifar10
cifar10 = keras.datasets.cifar10
(train_images, train_labels), (test_images, test_labels) = cifar10.load_data()
classifier = Sequential()
classifier.add(Convolution2D(32, 3, 3, # 32 no. of filters, then filter dimension
input_shape=(32, 32, 3), # this is image dimensionality
activation='relu'))
# Convolution2D check documentation
classifier.add(MaxPooling2D(pool_size=(2, 2)))
classifier.add(Flatten())
classifier.add(Dense(128, activation='relu'))
classifier.add(Dense(10, activation='softmax'))
classifier.compile(optimizer='adam', loss='sparse_categorical_crossentropy', metrics=['accuracy'])
def load_dataset():
(train_images, train_labels), (test_images, test_labels) = cifar10.load_data()
# how to know what to reshape as?
trainX = train_images
testX = test_images
trainY = train_labels
testY = test_labels
return trainX, trainY, testX, testY
def prep_pixels(train,test):
train_norm = train.astype('float32')
test_norm = test.astype('float32')
# why / 255? RGB
train_norm = train_norm / 255.0
test_norm = test_norm / 255.0
return train_norm, test_norm
def define_model():
model = Sequential()
# explain shapes here
model.add(Convolution2D(32, 3, 3, input_shape=(32, 32, 3), activation='relu'))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Flatten())
model.add(Dense(128, activation='relu'))
# how do I tell how many categories
model.add(Dense(10, activation='softmax'))
model.compile(optimizer='adam', loss='sparse_categorical_crossentropy', metrics=['accuracy'])
return model
def evaluate_model(dataX, dataY, n_folds=5):
scores, histories = list(), list()
kfold = KFold(n_folds, shuffle=True, random_state=42)
for train_ix, test_ix in kfold.split(dataX):
model = define_model()
trainX, trainY, testX, testY = dataX[train_ix], dataY[train_ix], dataX[test_ix], dataY[test_ix]
history = model.fit(trainX, trainY, epochs=10, steps_per_epoch = 80, batch_size=32, validation_data=(testX, testY), verbose=0)
_, acc = model.evaluate(testX, testY, verbose=0)
print('> %.3f' % (acc * 100.0))
scores.append(acc)
histories.append(history)
return scores, histories
def summarize_diagnostics(histories):
for i in range(len(histories)):
pyplot.subplot(211)
pyplot.title('Cross Entropy Loss')
pyplot.plot(histories[i].history['loss'], color='blue', label='train')
pyplot.plot(histories[i].history['val_loss'], color='orange', label='test')
pyplot.subplot(212)
pyplot.title('Classification Accuracy')
pyplot.plot(histories[i].history['accuracy'], color='blue', label='train')
pyplot.plot(histories[i].history['val_accuracy'], color='orange', label='test')
pyplot.show()
def summarize_performance(scores):
print('Accuracy: mean=%.3f std=%.3f, n=%d' % (mean(scores)*100, std(scores)*100, len(scores)))
pyplot.boxplot(scores)
pyplot.show()
def run_test_harness():
trainX, trainY, testX, testY = load_dataset()
trainX, testX = prep_pixels(trainX, testX)
scores, histories = evaluate_model(trainX, trainY)
summarize_performance(scores)
summarize_diagnostics(histories)
run_test_harness()
| 0.912974 | 0.976602 |
## Exploring `Series` and `DataFrame` Objects
### Working with pandas
*Curtis Miller*
Let's create some `Series`.
```
import pandas as pd
from pandas import Series, DataFrame
import numpy as np
ser1 = Series([1, 2, 3, 4])
ser2 = Series(['a', 'b', 'c'])
print(ser1)
print(ser2)
# Create a pandas Index
idx = pd.Index(["New York", "Los Angeles", "Chicago",
"Houston", "Philadelphia", "Phoenix", "San Antonio",
"San Diego", "Dallas"])
print(idx)
pops = Series([8550, 3972, 2721, 2296, 1567, np.nan, 1470, 1395, 1300],
index=idx, name="Population")
print(pops)
state = Series({"New York": "New York", "Los Angeles": "California", "Phoenix": "Arizona", "San Antonio": "Texas",
"San Diego": "California", "Dallas": "Texas"}, name = "State")
print(state)
area = Series({"New York": 302.6, "Los Angeles": 468.7, "Philadelphia": 134.1, "Phoenix": 516.7, "Austin": 322.48},
name = "Area")
print(area)
```
Let's see some of the ways we can create `DataFrame`s, first without indices.
```
# From a NumPy array
mat = np.arange(0,9).reshape(3, 3)
print(mat)
print(DataFrame(mat))
# Adding labels
print(DataFrame(mat, index=['a', 'b', 'c'], columns = ['alpha', 'beta', 'gamma']))
# What amounts to a 2D array (each tuple a row)
arr = [(1, 'a'), (2, 'b'), (3, 'c')]
print(arr)
print(DataFrame(arr, columns = ["Numbers", "Letters"]))
# Creating from a dict
print(DataFrame({"Numbers": [1, 2, 3], "Letters": ['a', 'b', 'c']}))
# What if not all lists are the same length?
# We get an error
print(DataFrame({"Numbers": [1, 2, 3, 4], "Letters": ['a', 'b', 'c']}))
# Do we get an error?
DataFrame({"Numbers": ser1, "Letters": ser2}) # nan fills in "missing" information (Series not of same length)
```
Let's now create a DataFrame containing information about cities.
```
# When passed as a list, series are treated as rows
# Notice that these Series are not the same length nor all have the same entries; nan will be generated
print(DataFrame([pops, state, area]))
print(DataFrame({"Population": pops, "State": state, "Area": area}))
# Or, we could use DataFrame's T (transpose) method
print(DataFrame([pops, state, area]).T)
```
How can we add new data to `Series` or `DataFrame`s?
```
# Let's append new data to each Series
pops.append(Series({"Seattle": 684, "Denver": 683})) # Not done in place
df = DataFrame([pops, state, area]).T
df.append(DataFrame({"Population": Series({"Seattle": 684, "Denver": 683}),
"State": Series({"Seattle": "Washington", "Denver": "Colorado"}),
"Area": Series({"Seattle": np.nan, "Denver": np.nan})}))
pd.concat([df, DataFrame({"Numbers": Series(np.arange(9), index=pops.index),
"Letters": Series(['a', 'c', 'd', 'h', 'l', 'n', 'p', 'p', 's'], index=pops.index)})],
axis=1)
```
Finally we save the data to a CSV file for later use.
```
df = DataFrame([pops, state, area]).T
# Saving data to csv file
df.to_csv("cities.csv")
```
|
github_jupyter
|
import pandas as pd
from pandas import Series, DataFrame
import numpy as np
ser1 = Series([1, 2, 3, 4])
ser2 = Series(['a', 'b', 'c'])
print(ser1)
print(ser2)
# Create a pandas Index
idx = pd.Index(["New York", "Los Angeles", "Chicago",
"Houston", "Philadelphia", "Phoenix", "San Antonio",
"San Diego", "Dallas"])
print(idx)
pops = Series([8550, 3972, 2721, 2296, 1567, np.nan, 1470, 1395, 1300],
index=idx, name="Population")
print(pops)
state = Series({"New York": "New York", "Los Angeles": "California", "Phoenix": "Arizona", "San Antonio": "Texas",
"San Diego": "California", "Dallas": "Texas"}, name = "State")
print(state)
area = Series({"New York": 302.6, "Los Angeles": 468.7, "Philadelphia": 134.1, "Phoenix": 516.7, "Austin": 322.48},
name = "Area")
print(area)
# From a NumPy array
mat = np.arange(0,9).reshape(3, 3)
print(mat)
print(DataFrame(mat))
# Adding labels
print(DataFrame(mat, index=['a', 'b', 'c'], columns = ['alpha', 'beta', 'gamma']))
# What amounts to a 2D array (each tuple a row)
arr = [(1, 'a'), (2, 'b'), (3, 'c')]
print(arr)
print(DataFrame(arr, columns = ["Numbers", "Letters"]))
# Creating from a dict
print(DataFrame({"Numbers": [1, 2, 3], "Letters": ['a', 'b', 'c']}))
# What if not all lists are the same length?
# We get an error
print(DataFrame({"Numbers": [1, 2, 3, 4], "Letters": ['a', 'b', 'c']}))
# Do we get an error?
DataFrame({"Numbers": ser1, "Letters": ser2}) # nan fills in "missing" information (Series not of same length)
# When passed as a list, series are treated as rows
# Notice that these Series are not the same length nor all have the same entries; nan will be generated
print(DataFrame([pops, state, area]))
print(DataFrame({"Population": pops, "State": state, "Area": area}))
# Or, we could use DataFrame's T (transpose) method
print(DataFrame([pops, state, area]).T)
# Let's append new data to each Series
pops.append(Series({"Seattle": 684, "Denver": 683})) # Not done in place
df = DataFrame([pops, state, area]).T
df.append(DataFrame({"Population": Series({"Seattle": 684, "Denver": 683}),
"State": Series({"Seattle": "Washington", "Denver": "Colorado"}),
"Area": Series({"Seattle": np.nan, "Denver": np.nan})}))
pd.concat([df, DataFrame({"Numbers": Series(np.arange(9), index=pops.index),
"Letters": Series(['a', 'c', 'd', 'h', 'l', 'n', 'p', 'p', 's'], index=pops.index)})],
axis=1)
df = DataFrame([pops, state, area]).T
# Saving data to csv file
df.to_csv("cities.csv")
| 0.457621 | 0.938969 |
# R: Impact of 401(k) on Financial Wealth
In this real-data example, we illustrate how the [DoubleML](https://docs.doubleml.org/stable/index.html) package can be used to estimate the effect of 401(k) eligibility and participation on accumulated assets. The 401(k) data set has been analyzed in several studies, among others [Chernozhukov et al. (2018)](https://arxiv.org/abs/1608.00060).
401(k) plans are pension accounts sponsored by employers. The key problem in determining the effect of participation in 401(k) plans on accumulated assets is saver heterogeneity coupled with the fact that the decision to enroll in a 401(k) is non-random. It is generally recognized that some people have a higher preference for saving than others. It also seems likely that those individuals with high unobserved preference for saving would be most likely to choose to participate in tax-advantaged retirement savings plans and would tend to have otherwise high amounts of accumulated assets. The presence of unobserved savings preferences with these properties then implies that conventional estimates that do not account for saver heterogeneity and endogeneity of participation will be biased upward, tending to overstate the savings effects of 401(k) participation.
One can argue that eligibility for enrolling in a 401(k) plan in this data can be taken as exogenous after conditioning on a few observables of which the most important for their argument is income. The basic idea is that, at least around the time 401(k)’s initially became available, people were unlikely to be basing their employment decisions on whether an employer offered a 401(k) but would instead focus on income and other aspects of the job.
## Data
The preprocessed data can be fetched by calling [fetch_401k()](https://docs.doubleml.org/r/stable/reference/fetch_401k.html). The arguments `polynomial_features` and `instrument` can be used to replicate the models used in [Chernozhukov et al. (2018)](https://arxiv.org/abs/1608.00060). Note that an internet connection is required for loading the data. We start with a baseline specification of the regression model and reload the data later in case we want to use another specification.
```
# Load required packages for this tutorial
library(DoubleML)
library(mlr3)
library(mlr3learners)
library(data.table)
library(ggplot2)
# suppress messages during fitting
lgr::get_logger("mlr3")$set_threshold("warn")
# load data as a data.table
data = fetch_401k(return_type = "data.table", instrument = TRUE)
dim(data)
str(data)
```
See the "Details" section on the description of the data set, which can be accessed by typing [help(fetch_401k)](https://docs.doubleml.org/r/stable/reference/fetch_401k.html).
The data consist of 9,915 observations at the household level drawn from the 1991 Survey of Income and Program Participation (SIPP). All the variables are referred to 1990. We use net financial assets (*net\_tfa*) as the outcome variable, $Y$, in our analysis. The net financial assets are computed as the sum of IRA balances, 401(k) balances, checking accounts, saving bonds, other interest-earning accounts, other interest-earning assets, stocks, and mutual funds less non mortgage debts.
Among the $9915$ individuals, $3682$ are eligible to participate in the program. The variable *e401* indicates eligibility and *p401* indicates participation, respectively.
```
hist_e401 = ggplot(data, aes(x = e401, fill = factor(e401))) +
geom_bar() + theme_minimal() +
ggtitle("Eligibility, 401(k)") +
theme(legend.position = "bottom", plot.title = element_text(hjust = 0.5),
text = element_text(size = 20))
hist_e401
hist_p401 = ggplot(data, aes(x = p401, fill = factor(p401))) +
geom_bar() + theme_minimal() +
ggtitle("Participation, 401(k)") +
theme(legend.position = "bottom", plot.title = element_text(hjust = 0.5),
text = element_text(size = 20))
hist_p401
```
Eligibility is highly associated with financial wealth:
```
dens_net_tfa = ggplot(data, aes(x = net_tfa, color = factor(e401), fill = factor(e401)) ) +
geom_density() + xlim(c(-20000, 150000)) +
facet_wrap(.~e401) + theme_minimal() +
theme(legend.position = "bottom", text = element_text(size = 20))
dens_net_tfa
```
As a first estimate, we calculate the unconditional average predictive effect (APE) of 401(k) eligibility on accumulated assets. This effect corresponds to the average treatment effect if 401(k) eligibility would be assigned to individuals in an entirely randomized way. The unconditional APE of e401 is about $19559$:
```
APE_e401_uncond = data[e401==1, mean(net_tfa)] - data[e401==0, mean(net_tfa)]
round(APE_e401_uncond, 2)
```
Among the $3682$ individuals that are eligible, $2594$ decided to participate in the program. The unconditional APE of p401 is about $27372$:
```
APE_p401_uncond = data[p401==1, mean(net_tfa)] - data[p401==0, mean(net_tfa)]
round(APE_p401_uncond, 2)
```
As discussed, these estimates are biased since they do not account for saver heterogeneity and endogeneity of participation.
## The `DoubleML` package
Let's use the package [DoubleML](https://docs.doubleml.org/stable/index.html) to estimate the average treatment effect of 401(k) eligibility, i.e. `e401`, and participation, i.e. `p401`, on net financial assets `net_tfa`.
## Estimating the Average Treatment Effect of 401(k) Eligibility on Net Financial Assets
We first look at the treatment effect of `e401` on net total financial assets. We give estimates of the ATE in the linear model
\begin{equation*}
Y = D \alpha + f(X)'\beta+ \epsilon,
\end{equation*}
where $f(X)$ is a dictonary applied to the raw regressors. $X$ contains variables on marital status, two-earner status, defined benefit pension status, IRA participation, home ownership, family size, education, age, and income.
In the following, we will consider two different models,
* a basic model specification that includes the raw regressors, i.e., $f(X) = X$, and
* a flexible model specification, where $f(X)$ includes the raw regressors $X$ and the orthogonal polynomials of degree 2 for the variables family size education, age, and income.
We will use the basic model specification whenever we use nonlinear methods, for example regression trees or random forests, and use the flexible model for linear methods such as the lasso. There are, of course, multiple ways how the model can be specified even more flexibly, for example including interactions of variable and higher order interaction. However, for the sake of simplicity we stick to the specification above. Users who are interested in varying the model can manipulate the model formula below (`formula_flex`), for example implementing the orignal specification in [Chernozhukov et al. (2018)](https://arxiv.org/abs/1608.00060).
In the first step, we report estimates of the average treatment effect (ATE) of 401(k) eligibility on net financial assets both in the partially linear regression (PLR) model and in the interactive regression model (IRM) allowing for heterogeneous treatment effects.
### The Data Backend: `DoubleMLData`
To start our analysis, we initialize the data backend, i.e., a new instance of a [DoubleMLData](https://docs.doubleml.org/r/stable/reference/DoubleMLData.html) object. Here, we manually implement the regression model by using R's formula interface. A shortcut would be to directly specify the options `polynomial_features` and `instrument` when calling [fetch_401k()](https://docs.doubleml.org/r/stable/reference/fetch_401k.html).$^{**}$
To implement both models (basic and flexible), we generate two data backends: `data_dml_base` and `data_dml_flex`.
$^{**}$ Note that the model specification using `polynomial_features` differs from the one used in our example.
```
# Set up basic model: Specify variables for data-backend
features_base = c("age", "inc", "educ", "fsize",
"marr", "twoearn", "db", "pira", "hown")
# Initialize DoubleMLData (data-backend of DoubleML)
data_dml_base = DoubleMLData$new(data,
y_col = "net_tfa",
d_cols = "e401",
x_cols = features_base)
data_dml_base
# Set up a model according to regression formula with polynomials
formula_flex = formula(" ~ -1 + poly(age, 2, raw=TRUE) +
poly(inc, 2, raw=TRUE) + poly(educ, 2, raw=TRUE) +
poly(fsize, 2, raw=TRUE) + marr + twoearn +
db + pira + hown")
features_flex = data.frame(model.matrix(formula_flex, data))
model_data = data.table("net_tfa" = data[, net_tfa],
"e401" = data[, e401],
features_flex)
# Initialize DoubleMLData (data-backend of DoubleML)
data_dml_flex = DoubleMLData$new(model_data,
y_col = "net_tfa",
d_cols = "e401")
data_dml_flex
```
### Partially Linear Regression Model (PLR)
We start using lasso to estimate the function $g_0$ and $m_0$ in the following PLR model:
\begin{eqnarray}
& Y = D\theta_0 + g_0(X) + \zeta, &\quad E[\zeta \mid D,X]= 0,\\
& D = m_0(X) + V, &\quad E[V \mid X] = 0.
\end{eqnarray}
To estimate the causal parameter $\theta_0$ here, we use double machine learning with 3-fold cross-fitting.
Estimation of the nuisance components $g_0$ and $m_0$, is based on the lasso with cross-validated choice of the penalty term , $\lambda$, as provided by the [glmnet package](https://glmnet.stanford.edu/reference/cv.glmnet.html). We load the learner by using the [mlr3](https://mlr3.mlr-org.com/) function [lrn()](https://mlr3.mlr-org.com/reference/Learner.html). Hyperparameters and options can be set during instantiation of the learner. Here we specify that the lasso should use that value of $\lambda$ that minimizes the cross-validated mean squared error which is based on 5-fold cross validation.
In order to use a learner, the underlying R packages have to be installed. In this case, the package [glmnet package](https://glmnet.stanford.edu/reference/cv.glmnet.html) needs to be installed. Moreover, installation of the package [mlr3learners](https://mlr3learners.mlr-org.com/) is required.
We start by estimation the ATE in the basic model and then repeat the estimation in the flexible model.
```
# Initialize learners
set.seed(123)
lasso = lrn("regr.cv_glmnet", nfolds = 5, s = "lambda.min")
lasso_class = lrn("classif.cv_glmnet", nfolds = 5, s = "lambda.min")
# Initialize DoubleMLPLR model
dml_plr_lasso = DoubleMLPLR$new(data_dml_base,
ml_g = lasso,
ml_m = lasso_class,
n_folds = 3)
dml_plr_lasso$fit()
dml_plr_lasso$summary()
# Initialize learners
set.seed(123)
lasso = lrn("regr.cv_glmnet", nfolds = 5, s = "lambda.min")
lasso_class = lrn("classif.cv_glmnet", nfolds = 5, s = "lambda.min")
# Initialize DoubleMLPLR model
dml_plr_lasso = DoubleMLPLR$new(data_dml_flex,
ml_g = lasso,
ml_m = lasso_class,
n_folds = 3)
dml_plr_lasso$fit()
dml_plr_lasso$summary()
```
Alternatively, we can repeat this procedure with other machine learning methods, for example a random forest learner as provided by the [ranger](https://github.com/imbs-hl/ranger) package for R. The website of the [mlr3extralearners](https://mlr3extralearners.mlr-org.com/articles/learners/list_learners.html) package has a searchable list of all learners that are available in the [mlr3verse](https://mlr3verse.mlr-org.com/).
```
# Random Forest
randomForest = lrn("regr.ranger", max.depth = 7,
mtry = 3, min.node.size = 3)
randomForest_class = lrn("classif.ranger", max.depth = 5,
mtry = 4, min.node.size = 7)
set.seed(123)
dml_plr_forest = DoubleMLPLR$new(data_dml_base,
ml_g = randomForest,
ml_m = randomForest_class,
n_folds = 3)
dml_plr_forest$fit()
dml_plr_forest$summary()
```
Now, let's use a regression tree as provided by the R package [rpart](https://github.com/bethatkinson/rpart).
```
# Trees
trees = lrn("regr.rpart", cp = 0.0047, minsplit = 203)
trees_class = lrn("classif.rpart", cp = 0.0042, minsplit = 104)
set.seed(123)
dml_plr_tree = DoubleMLPLR$new(data_dml_base,
ml_g = trees,
ml_m = trees_class,
n_folds = 3)
dml_plr_tree$fit()
dml_plr_tree$summary()
```
We can also experiment with extreme gradient boosting as provided by [xgboost](https://xgboost.readthedocs.io/en/latest/).
```
# Boosted trees
boost = lrn("regr.xgboost",
objective = "reg:squarederror",
eta = 0.1, nrounds = 35)
boost_class = lrn("classif.xgboost",
objective = "binary:logistic", eval_metric = "logloss",
eta = 0.1, nrounds = 34)
set.seed(123)
dml_plr_boost = DoubleMLPLR$new(data_dml_base,
ml_g = boost,
ml_m = boost_class,
n_folds = 3)
dml_plr_boost$fit()
dml_plr_boost$summary()
```
Let's sum up the results:
```
confints = rbind(dml_plr_lasso$confint(), dml_plr_forest$confint(),
dml_plr_tree$confint(), dml_plr_boost$confint())
estimates = c(dml_plr_lasso$coef, dml_plr_forest$coef,
dml_plr_tree$coef, dml_plr_boost$coef)
result_plr = data.table("model" = "PLR",
"ML" = c("glmnet", "ranger", "rpart", "xgboost"),
"Estimate" = estimates,
"lower" = confints[,1],
"upper" = confints[,2])
result_plr
g_ci = ggplot(result_plr, aes(x = ML, y = Estimate, color = ML)) +
geom_point() +
geom_errorbar(aes(ymin = lower, ymax = upper, color = ML)) +
geom_hline(yintercept = 0, color = "grey") +
theme_minimal() + ylab("Coefficients and 0.95- confidence interval") +
xlab("") +
theme(axis.text.x = element_text(angle = 90), legend.position = "none",
text = element_text(size = 20))
g_ci
```
### Interactive Regression Model (IRM)
Next, we consider estimation of average treatment effects when treatment effects are fully heterogeneous:
\begin{eqnarray}
& Y = g_0(D,X) + U, &\quad E[U\mid X,D] = 0,\\
& D = m_0(X) + V, &\quad E[V\mid X] = 0.
\end{eqnarray}
To reduce the disproportionate impact of extreme propensity score weights in the interactive model
we trim the propensity scores which are close to the bounds.
```
set.seed(123)
# Initialize DoubleMLIRM model
dml_irm_lasso = DoubleMLIRM$new(data_dml_flex,
ml_g = lasso,
ml_m = lasso_class,
trimming_threshold = 0.01,
n_folds = 3)
dml_irm_lasso$fit()
dml_irm_lasso$summary()
# Initialize Learner
randomForest = lrn("regr.ranger")
randomForest_class = lrn("classif.ranger")
# Random Forest
set.seed(123)
dml_irm_forest = DoubleMLIRM$new(data_dml_base,
ml_g = randomForest,
ml_m = randomForest_class,
trimming_threshold = 0.01,
n_folds = 3)
# Set nuisance-part specific parameters
dml_irm_forest$set_ml_nuisance_params(
"ml_g0", "e401", list(max.depth = 6, mtry = 4, min.node.size = 7))
dml_irm_forest$set_ml_nuisance_params(
"ml_g1", "e401", list(max.depth = 6, mtry = 3, min.node.size = 5))
dml_irm_forest$set_ml_nuisance_params(
"ml_m", "e401", list(max.depth = 6, mtry = 3, min.node.size = 6))
dml_irm_forest$fit()
dml_irm_forest$summary()
# Initialize Learner
trees = lrn("regr.rpart")
trees_class = lrn("classif.rpart")
# Trees
set.seed(123)
dml_irm_tree = DoubleMLIRM$new(data_dml_base,
ml_g = trees,
ml_m = trees_class,
trimming_threshold = 0.01,
n_folds = 3)
# Set nuisance-part specific parameters
dml_irm_tree$set_ml_nuisance_params(
"ml_g0", "e401", list(cp = 0.0016, minsplit = 74))
dml_irm_tree$set_ml_nuisance_params(
"ml_g1", "e401", list(cp = 0.0018, minsplit = 70))
dml_irm_tree$set_ml_nuisance_params(
"ml_m", "e401", list(cp = 0.0028, minsplit = 167))
dml_irm_tree$fit()
dml_irm_tree$summary()
# Initialize Learners
boost = lrn("regr.xgboost", objective = "reg:squarederror")
boost_class = lrn("classif.xgboost", objective = "binary:logistic", eval_metric = "logloss")
# Boosted Trees
set.seed(123)
dml_irm_boost = DoubleMLIRM$new(data_dml_base,
ml_g = boost,
ml_m = boost_class,
trimming_threshold = 0.01,
n_folds = 3)
# Set nuisance-part specific parameters
if (compareVersion(as.character(packageVersion("DoubleML")), "0.2.1") > 0) {
dml_irm_boost$set_ml_nuisance_params(
"ml_g0", "e401", list(nrounds = 8, eta = 0.1))
dml_irm_boost$set_ml_nuisance_params(
"ml_g1", "e401", list(nrounds = 29, eta = 0.1))
dml_irm_boost$set_ml_nuisance_params(
"ml_m", "e401", list(nrounds = 23, eta = 0.1))
} else {
# behavior of set_ml_nuisance_params() changed in https://github.com/DoubleML/doubleml-for-r/pull/89
dml_irm_boost$set_ml_nuisance_params(
"ml_g0", "e401", list(nrounds = 8, eta = 0.1, objective = "reg:squarederror", verbose=0))
dml_irm_boost$set_ml_nuisance_params(
"ml_g1", "e401", list(nrounds = 29, eta = 0.1, objective = "reg:squarederror", verbose=0))
dml_irm_boost$set_ml_nuisance_params(
"ml_m", "e401", list(nrounds = 23, eta = 0.1, objective = "binary:logistic", eval_metric = "logloss", verbose=0))
}
dml_irm_boost$fit()
dml_irm_boost$summary()
confints = rbind(dml_irm_lasso$confint(), dml_irm_forest$confint(),
dml_irm_tree$confint(), dml_irm_boost$confint())
estimates = c(dml_irm_lasso$coef, dml_irm_forest$coef,
dml_irm_tree$coef, dml_irm_boost$coef)
result_irm = data.table("model" = "IRM",
"ML" = c("glmnet", "ranger", "rpart", "xgboost"),
"Estimate" = estimates,
"lower" = confints[,1],
"upper" = confints[,2])
result_irm
g_ci = ggplot(result_irm, aes(x = ML, y = Estimate, color = ML)) +
geom_point() +
geom_errorbar(aes(ymin = lower, ymax = upper, color = ML)) +
geom_hline(yintercept = 0, color = "grey") +
theme_minimal() + ylab("Coefficients and 0.95- confidence interval") +
xlab("") +
theme(axis.text.x = element_text(angle = 90), legend.position = "none",
text = element_text(size = 20))
g_ci
```
These estimates that flexibly account for confounding are
substantially attenuated relative to the baseline estimate (*19559*) that does not account for confounding. They suggest much smaller causal effects of 401(k) eligiblity on financial asset holdings. The best model with lowest RMSE in both equations is the PLR model estimated via lasso. It gives the following estimate:
## Local Average Treatment Effects of 401(k) Participation on Net Financial Assets
### Interactive IV Model (IIVM)
In the examples above, we estimated the average treatment effect of *eligibility* on financial asset holdings. Now, we consider estimation of local average treatment effects (LATE) of *participation* using eligibility as an instrument for the participation decision. Under appropriate assumptions, the LATE identifies the treatment effect for so-called compliers, i.e., individuals who would only participate if eligible and otherwise not participate in the program.
As before, $Y$ denotes the outcome `net_tfa`, and $X$ is the vector of covariates. We use `e401` as a binary instrument for the treatment variable `p401`. Here the structural equation model is:
\begin{eqnarray}
& Y = g_0(Z,X) + U, &\quad E[U\mid Z,X] = 0,\\
& D = r_0(Z,X) + V, &\quad E[V\mid Z, X] = 0,\\
& Z = m_0(X) + \zeta, &\quad E[\zeta \mid X] = 0.
\end{eqnarray}
```
# Initialize DoubleMLData with an instrument
# Basic model
data_dml_base_iv = DoubleMLData$new(data,
y_col = "net_tfa",
d_cols = "p401",
x_cols = features_base,
z_cols = "e401")
data_dml_base_iv
# Flexible model
model_data = data.table("net_tfa" = data[, net_tfa],
"e401" = data[, e401],
"p401" = data[, p401],
features_flex)
data_dml_flex_iv = DoubleMLData$new(model_data,
y_col = "net_tfa",
d_cols = "p401",
z_cols = "e401")
set.seed(123)
dml_iivm_lasso = DoubleMLIIVM$new(data_dml_flex_iv,
ml_g = lasso,
ml_m = lasso_class,
ml_r = lasso_class,
n_folds = 3,
trimming_threshold = 0.01,
subgroups = list(always_takers = FALSE,
never_takers = TRUE))
dml_iivm_lasso$fit()
dml_iivm_lasso$summary()
```
Again, we repeat the procedure for the other machine learning methods:
```
# Initialize Learner
randomForest = lrn("regr.ranger")
randomForest_class = lrn("classif.ranger")
# Random Forest
set.seed(123)
dml_iivm_forest = DoubleMLIIVM$new(data_dml_base_iv,
ml_g = randomForest,
ml_m = randomForest_class,
ml_r = randomForest_class,
n_folds = 3,
trimming_threshold = 0.01,
subgroups = list(always_takers = FALSE,
never_takers = TRUE))
# Set nuisance-part specific parameters
dml_iivm_forest$set_ml_nuisance_params(
"ml_g0", "p401",
list(max.depth = 6, mtry = 4, min.node.size = 7))
dml_iivm_forest$set_ml_nuisance_params(
"ml_g1", "p401",
list(max.depth = 6, mtry = 3, min.node.size = 5))
dml_iivm_forest$set_ml_nuisance_params(
"ml_m", "p401",
list(max.depth = 6, mtry = 3, min.node.size = 6))
dml_iivm_forest$set_ml_nuisance_params(
"ml_r1", "p401",
list(max.depth = 4, mtry = 7, min.node.size = 6))
dml_iivm_forest$fit()
dml_iivm_forest$summary()
# Initialize Learner
trees = lrn("regr.rpart")
trees_class = lrn("classif.rpart")
# Trees
set.seed(123)
dml_iivm_tree = DoubleMLIIVM$new(data_dml_base_iv,
ml_g = trees,
ml_m = trees_class,
ml_r = trees_class,
n_folds = 3,
trimming_threshold = 0.01,
subgroups = list(always_takers = FALSE,
never_takers = TRUE))
# Set nuisance-part specific parameters
dml_iivm_tree$set_ml_nuisance_params(
"ml_g0", "p401",
list(cp = 0.0016, minsplit = 74))
dml_iivm_tree$set_ml_nuisance_params(
"ml_g1", "p401",
list(cp = 0.0018, minsplit = 70))
dml_iivm_tree$set_ml_nuisance_params(
"ml_m", "p401",
list(cp = 0.0028, minsplit = 167))
dml_iivm_tree$set_ml_nuisance_params(
"ml_r1", "p401",
list(cp = 0.0576, minsplit = 55))
dml_iivm_tree$fit()
dml_iivm_tree$summary()
# Initialize Learner
boost = lrn("regr.xgboost", objective = "reg:squarederror")
boost_class = lrn("classif.xgboost", objective = "binary:logistic", eval_metric = "logloss")
# Boosted Trees
set.seed(123)
dml_iivm_boost = DoubleMLIIVM$new(data_dml_base_iv,
ml_g = boost,
ml_m = boost_class,
ml_r = boost_class,
n_folds = 3,
trimming_threshold = 0.01,
subgroups = list(always_takers = FALSE,
never_takers = TRUE))
# Set nuisance-part specific parameters
if (compareVersion(as.character(packageVersion("DoubleML")), "0.2.1") > 0) {
dml_iivm_boost$set_ml_nuisance_params(
"ml_g0", "p401",
list(nrounds = 9, eta = 0.1))
dml_iivm_boost$set_ml_nuisance_params(
"ml_g1", "p401",
list(nrounds = 33, eta = 0.1))
dml_iivm_boost$set_ml_nuisance_params(
"ml_m", "p401",
list(nrounds = 12, eta = 0.1))
dml_iivm_boost$set_ml_nuisance_params(
"ml_r1", "p401",
list(nrounds = 25, eta = 0.1))
} else {
# behavior of set_ml_nuisance_params() changed in https://github.com/DoubleML/doubleml-for-r/pull/89
dml_iivm_boost$set_ml_nuisance_params(
"ml_g0", "p401",
list(nrounds = 9, eta = 0.1, objective = "reg:squarederror", verbose=0))
dml_iivm_boost$set_ml_nuisance_params(
"ml_g1", "p401",
list(nrounds = 33, eta = 0.1, objective = "reg:squarederror", verbose=0))
dml_iivm_boost$set_ml_nuisance_params(
"ml_m", "p401",
list(nrounds = 12, eta = 0.1, objective = "binary:logistic", eval_metric = "logloss", verbose=0))
dml_iivm_boost$set_ml_nuisance_params(
"ml_r1", "p401",
list(nrounds = 25, eta = 0.1, objective = "binary:logistic", eval_metric = "logloss", verbose=0))
}
dml_iivm_boost$fit()
dml_iivm_boost$summary()
confints = rbind(dml_iivm_lasso$confint(), dml_iivm_forest$confint(),
dml_iivm_tree$confint(), dml_iivm_boost$confint())
estimates = c(dml_iivm_lasso$coef, dml_iivm_forest$coef,
dml_iivm_tree$coef, dml_iivm_boost$coef)
result_iivm = data.table("model" = "IIVM",
"ML" = c("glmnet", "ranger", "rpart", "xgboost"),
"Estimate" = estimates,
"lower" = confints[,1],
"upper" = confints[,2])
result_iivm
g_ci = ggplot(result_iivm, aes(x = ML, y = Estimate, color = ML)) +
geom_point() +
geom_errorbar(aes(ymin = lower, ymax = upper, color = ML)) +
geom_hline(yintercept = 0, color = "grey") +
theme_minimal() + ylab("Coefficients and 0.95- confidence interval") +
xlab("") +
theme(axis.text.x = element_text(angle = 90), legend.position = "none",
text = element_text(size = 20))
g_ci
```
## Summary of Results
To sum up, let's merge all our results so far and illustrate them in a plot.
```
summary_result = rbindlist(list(result_plr, result_irm, result_iivm))
summary_result[, model := factor(model, levels = c("PLR", "IRM", "IIVM"))]
g_all = ggplot(summary_result, aes(x = ML, y = Estimate, color = ML)) +
geom_point() +
geom_errorbar(aes(ymin = lower, ymax = upper, color = ML)) +
geom_hline(yintercept = 0, color = "grey") +
theme_minimal() + ylab("Coefficients and 0.95- confidence interval") +
xlab("") +
theme(axis.text.x = element_text(angle = 90), legend.position = "none",
text = element_text(size = 20)) +
facet_wrap(model ~., ncol = 1)
g_all
```
We report results based on four ML methods for estimating the nuisance functions used in
forming the orthogonal estimating equations. We find again that the estimates of the treatment effect are stable across ML methods. The estimates are highly significant, hence we would reject the hypothesis
that 401(k) participation has no effect on financial wealth.
______
**Acknowledgement**
We would like to thank [Jannis Kueck](https://www.bwl.uni-hamburg.de/en/statistik/team/kueck.html) for sharing [the kaggle notebook](https://www.kaggle.com/janniskueck/pm5-401k). The pension data set has been analyzed in several studies, among others [Chernozhukov et al. (2018)](https://arxiv.org/abs/1608.00060).
|
github_jupyter
|
# Load required packages for this tutorial
library(DoubleML)
library(mlr3)
library(mlr3learners)
library(data.table)
library(ggplot2)
# suppress messages during fitting
lgr::get_logger("mlr3")$set_threshold("warn")
# load data as a data.table
data = fetch_401k(return_type = "data.table", instrument = TRUE)
dim(data)
str(data)
hist_e401 = ggplot(data, aes(x = e401, fill = factor(e401))) +
geom_bar() + theme_minimal() +
ggtitle("Eligibility, 401(k)") +
theme(legend.position = "bottom", plot.title = element_text(hjust = 0.5),
text = element_text(size = 20))
hist_e401
hist_p401 = ggplot(data, aes(x = p401, fill = factor(p401))) +
geom_bar() + theme_minimal() +
ggtitle("Participation, 401(k)") +
theme(legend.position = "bottom", plot.title = element_text(hjust = 0.5),
text = element_text(size = 20))
hist_p401
dens_net_tfa = ggplot(data, aes(x = net_tfa, color = factor(e401), fill = factor(e401)) ) +
geom_density() + xlim(c(-20000, 150000)) +
facet_wrap(.~e401) + theme_minimal() +
theme(legend.position = "bottom", text = element_text(size = 20))
dens_net_tfa
APE_e401_uncond = data[e401==1, mean(net_tfa)] - data[e401==0, mean(net_tfa)]
round(APE_e401_uncond, 2)
APE_p401_uncond = data[p401==1, mean(net_tfa)] - data[p401==0, mean(net_tfa)]
round(APE_p401_uncond, 2)
# Set up basic model: Specify variables for data-backend
features_base = c("age", "inc", "educ", "fsize",
"marr", "twoearn", "db", "pira", "hown")
# Initialize DoubleMLData (data-backend of DoubleML)
data_dml_base = DoubleMLData$new(data,
y_col = "net_tfa",
d_cols = "e401",
x_cols = features_base)
data_dml_base
# Set up a model according to regression formula with polynomials
formula_flex = formula(" ~ -1 + poly(age, 2, raw=TRUE) +
poly(inc, 2, raw=TRUE) + poly(educ, 2, raw=TRUE) +
poly(fsize, 2, raw=TRUE) + marr + twoearn +
db + pira + hown")
features_flex = data.frame(model.matrix(formula_flex, data))
model_data = data.table("net_tfa" = data[, net_tfa],
"e401" = data[, e401],
features_flex)
# Initialize DoubleMLData (data-backend of DoubleML)
data_dml_flex = DoubleMLData$new(model_data,
y_col = "net_tfa",
d_cols = "e401")
data_dml_flex
# Initialize learners
set.seed(123)
lasso = lrn("regr.cv_glmnet", nfolds = 5, s = "lambda.min")
lasso_class = lrn("classif.cv_glmnet", nfolds = 5, s = "lambda.min")
# Initialize DoubleMLPLR model
dml_plr_lasso = DoubleMLPLR$new(data_dml_base,
ml_g = lasso,
ml_m = lasso_class,
n_folds = 3)
dml_plr_lasso$fit()
dml_plr_lasso$summary()
# Initialize learners
set.seed(123)
lasso = lrn("regr.cv_glmnet", nfolds = 5, s = "lambda.min")
lasso_class = lrn("classif.cv_glmnet", nfolds = 5, s = "lambda.min")
# Initialize DoubleMLPLR model
dml_plr_lasso = DoubleMLPLR$new(data_dml_flex,
ml_g = lasso,
ml_m = lasso_class,
n_folds = 3)
dml_plr_lasso$fit()
dml_plr_lasso$summary()
# Random Forest
randomForest = lrn("regr.ranger", max.depth = 7,
mtry = 3, min.node.size = 3)
randomForest_class = lrn("classif.ranger", max.depth = 5,
mtry = 4, min.node.size = 7)
set.seed(123)
dml_plr_forest = DoubleMLPLR$new(data_dml_base,
ml_g = randomForest,
ml_m = randomForest_class,
n_folds = 3)
dml_plr_forest$fit()
dml_plr_forest$summary()
# Trees
trees = lrn("regr.rpart", cp = 0.0047, minsplit = 203)
trees_class = lrn("classif.rpart", cp = 0.0042, minsplit = 104)
set.seed(123)
dml_plr_tree = DoubleMLPLR$new(data_dml_base,
ml_g = trees,
ml_m = trees_class,
n_folds = 3)
dml_plr_tree$fit()
dml_plr_tree$summary()
# Boosted trees
boost = lrn("regr.xgboost",
objective = "reg:squarederror",
eta = 0.1, nrounds = 35)
boost_class = lrn("classif.xgboost",
objective = "binary:logistic", eval_metric = "logloss",
eta = 0.1, nrounds = 34)
set.seed(123)
dml_plr_boost = DoubleMLPLR$new(data_dml_base,
ml_g = boost,
ml_m = boost_class,
n_folds = 3)
dml_plr_boost$fit()
dml_plr_boost$summary()
confints = rbind(dml_plr_lasso$confint(), dml_plr_forest$confint(),
dml_plr_tree$confint(), dml_plr_boost$confint())
estimates = c(dml_plr_lasso$coef, dml_plr_forest$coef,
dml_plr_tree$coef, dml_plr_boost$coef)
result_plr = data.table("model" = "PLR",
"ML" = c("glmnet", "ranger", "rpart", "xgboost"),
"Estimate" = estimates,
"lower" = confints[,1],
"upper" = confints[,2])
result_plr
g_ci = ggplot(result_plr, aes(x = ML, y = Estimate, color = ML)) +
geom_point() +
geom_errorbar(aes(ymin = lower, ymax = upper, color = ML)) +
geom_hline(yintercept = 0, color = "grey") +
theme_minimal() + ylab("Coefficients and 0.95- confidence interval") +
xlab("") +
theme(axis.text.x = element_text(angle = 90), legend.position = "none",
text = element_text(size = 20))
g_ci
set.seed(123)
# Initialize DoubleMLIRM model
dml_irm_lasso = DoubleMLIRM$new(data_dml_flex,
ml_g = lasso,
ml_m = lasso_class,
trimming_threshold = 0.01,
n_folds = 3)
dml_irm_lasso$fit()
dml_irm_lasso$summary()
# Initialize Learner
randomForest = lrn("regr.ranger")
randomForest_class = lrn("classif.ranger")
# Random Forest
set.seed(123)
dml_irm_forest = DoubleMLIRM$new(data_dml_base,
ml_g = randomForest,
ml_m = randomForest_class,
trimming_threshold = 0.01,
n_folds = 3)
# Set nuisance-part specific parameters
dml_irm_forest$set_ml_nuisance_params(
"ml_g0", "e401", list(max.depth = 6, mtry = 4, min.node.size = 7))
dml_irm_forest$set_ml_nuisance_params(
"ml_g1", "e401", list(max.depth = 6, mtry = 3, min.node.size = 5))
dml_irm_forest$set_ml_nuisance_params(
"ml_m", "e401", list(max.depth = 6, mtry = 3, min.node.size = 6))
dml_irm_forest$fit()
dml_irm_forest$summary()
# Initialize Learner
trees = lrn("regr.rpart")
trees_class = lrn("classif.rpart")
# Trees
set.seed(123)
dml_irm_tree = DoubleMLIRM$new(data_dml_base,
ml_g = trees,
ml_m = trees_class,
trimming_threshold = 0.01,
n_folds = 3)
# Set nuisance-part specific parameters
dml_irm_tree$set_ml_nuisance_params(
"ml_g0", "e401", list(cp = 0.0016, minsplit = 74))
dml_irm_tree$set_ml_nuisance_params(
"ml_g1", "e401", list(cp = 0.0018, minsplit = 70))
dml_irm_tree$set_ml_nuisance_params(
"ml_m", "e401", list(cp = 0.0028, minsplit = 167))
dml_irm_tree$fit()
dml_irm_tree$summary()
# Initialize Learners
boost = lrn("regr.xgboost", objective = "reg:squarederror")
boost_class = lrn("classif.xgboost", objective = "binary:logistic", eval_metric = "logloss")
# Boosted Trees
set.seed(123)
dml_irm_boost = DoubleMLIRM$new(data_dml_base,
ml_g = boost,
ml_m = boost_class,
trimming_threshold = 0.01,
n_folds = 3)
# Set nuisance-part specific parameters
if (compareVersion(as.character(packageVersion("DoubleML")), "0.2.1") > 0) {
dml_irm_boost$set_ml_nuisance_params(
"ml_g0", "e401", list(nrounds = 8, eta = 0.1))
dml_irm_boost$set_ml_nuisance_params(
"ml_g1", "e401", list(nrounds = 29, eta = 0.1))
dml_irm_boost$set_ml_nuisance_params(
"ml_m", "e401", list(nrounds = 23, eta = 0.1))
} else {
# behavior of set_ml_nuisance_params() changed in https://github.com/DoubleML/doubleml-for-r/pull/89
dml_irm_boost$set_ml_nuisance_params(
"ml_g0", "e401", list(nrounds = 8, eta = 0.1, objective = "reg:squarederror", verbose=0))
dml_irm_boost$set_ml_nuisance_params(
"ml_g1", "e401", list(nrounds = 29, eta = 0.1, objective = "reg:squarederror", verbose=0))
dml_irm_boost$set_ml_nuisance_params(
"ml_m", "e401", list(nrounds = 23, eta = 0.1, objective = "binary:logistic", eval_metric = "logloss", verbose=0))
}
dml_irm_boost$fit()
dml_irm_boost$summary()
confints = rbind(dml_irm_lasso$confint(), dml_irm_forest$confint(),
dml_irm_tree$confint(), dml_irm_boost$confint())
estimates = c(dml_irm_lasso$coef, dml_irm_forest$coef,
dml_irm_tree$coef, dml_irm_boost$coef)
result_irm = data.table("model" = "IRM",
"ML" = c("glmnet", "ranger", "rpart", "xgboost"),
"Estimate" = estimates,
"lower" = confints[,1],
"upper" = confints[,2])
result_irm
g_ci = ggplot(result_irm, aes(x = ML, y = Estimate, color = ML)) +
geom_point() +
geom_errorbar(aes(ymin = lower, ymax = upper, color = ML)) +
geom_hline(yintercept = 0, color = "grey") +
theme_minimal() + ylab("Coefficients and 0.95- confidence interval") +
xlab("") +
theme(axis.text.x = element_text(angle = 90), legend.position = "none",
text = element_text(size = 20))
g_ci
# Initialize DoubleMLData with an instrument
# Basic model
data_dml_base_iv = DoubleMLData$new(data,
y_col = "net_tfa",
d_cols = "p401",
x_cols = features_base,
z_cols = "e401")
data_dml_base_iv
# Flexible model
model_data = data.table("net_tfa" = data[, net_tfa],
"e401" = data[, e401],
"p401" = data[, p401],
features_flex)
data_dml_flex_iv = DoubleMLData$new(model_data,
y_col = "net_tfa",
d_cols = "p401",
z_cols = "e401")
set.seed(123)
dml_iivm_lasso = DoubleMLIIVM$new(data_dml_flex_iv,
ml_g = lasso,
ml_m = lasso_class,
ml_r = lasso_class,
n_folds = 3,
trimming_threshold = 0.01,
subgroups = list(always_takers = FALSE,
never_takers = TRUE))
dml_iivm_lasso$fit()
dml_iivm_lasso$summary()
# Initialize Learner
randomForest = lrn("regr.ranger")
randomForest_class = lrn("classif.ranger")
# Random Forest
set.seed(123)
dml_iivm_forest = DoubleMLIIVM$new(data_dml_base_iv,
ml_g = randomForest,
ml_m = randomForest_class,
ml_r = randomForest_class,
n_folds = 3,
trimming_threshold = 0.01,
subgroups = list(always_takers = FALSE,
never_takers = TRUE))
# Set nuisance-part specific parameters
dml_iivm_forest$set_ml_nuisance_params(
"ml_g0", "p401",
list(max.depth = 6, mtry = 4, min.node.size = 7))
dml_iivm_forest$set_ml_nuisance_params(
"ml_g1", "p401",
list(max.depth = 6, mtry = 3, min.node.size = 5))
dml_iivm_forest$set_ml_nuisance_params(
"ml_m", "p401",
list(max.depth = 6, mtry = 3, min.node.size = 6))
dml_iivm_forest$set_ml_nuisance_params(
"ml_r1", "p401",
list(max.depth = 4, mtry = 7, min.node.size = 6))
dml_iivm_forest$fit()
dml_iivm_forest$summary()
# Initialize Learner
trees = lrn("regr.rpart")
trees_class = lrn("classif.rpart")
# Trees
set.seed(123)
dml_iivm_tree = DoubleMLIIVM$new(data_dml_base_iv,
ml_g = trees,
ml_m = trees_class,
ml_r = trees_class,
n_folds = 3,
trimming_threshold = 0.01,
subgroups = list(always_takers = FALSE,
never_takers = TRUE))
# Set nuisance-part specific parameters
dml_iivm_tree$set_ml_nuisance_params(
"ml_g0", "p401",
list(cp = 0.0016, minsplit = 74))
dml_iivm_tree$set_ml_nuisance_params(
"ml_g1", "p401",
list(cp = 0.0018, minsplit = 70))
dml_iivm_tree$set_ml_nuisance_params(
"ml_m", "p401",
list(cp = 0.0028, minsplit = 167))
dml_iivm_tree$set_ml_nuisance_params(
"ml_r1", "p401",
list(cp = 0.0576, minsplit = 55))
dml_iivm_tree$fit()
dml_iivm_tree$summary()
# Initialize Learner
boost = lrn("regr.xgboost", objective = "reg:squarederror")
boost_class = lrn("classif.xgboost", objective = "binary:logistic", eval_metric = "logloss")
# Boosted Trees
set.seed(123)
dml_iivm_boost = DoubleMLIIVM$new(data_dml_base_iv,
ml_g = boost,
ml_m = boost_class,
ml_r = boost_class,
n_folds = 3,
trimming_threshold = 0.01,
subgroups = list(always_takers = FALSE,
never_takers = TRUE))
# Set nuisance-part specific parameters
if (compareVersion(as.character(packageVersion("DoubleML")), "0.2.1") > 0) {
dml_iivm_boost$set_ml_nuisance_params(
"ml_g0", "p401",
list(nrounds = 9, eta = 0.1))
dml_iivm_boost$set_ml_nuisance_params(
"ml_g1", "p401",
list(nrounds = 33, eta = 0.1))
dml_iivm_boost$set_ml_nuisance_params(
"ml_m", "p401",
list(nrounds = 12, eta = 0.1))
dml_iivm_boost$set_ml_nuisance_params(
"ml_r1", "p401",
list(nrounds = 25, eta = 0.1))
} else {
# behavior of set_ml_nuisance_params() changed in https://github.com/DoubleML/doubleml-for-r/pull/89
dml_iivm_boost$set_ml_nuisance_params(
"ml_g0", "p401",
list(nrounds = 9, eta = 0.1, objective = "reg:squarederror", verbose=0))
dml_iivm_boost$set_ml_nuisance_params(
"ml_g1", "p401",
list(nrounds = 33, eta = 0.1, objective = "reg:squarederror", verbose=0))
dml_iivm_boost$set_ml_nuisance_params(
"ml_m", "p401",
list(nrounds = 12, eta = 0.1, objective = "binary:logistic", eval_metric = "logloss", verbose=0))
dml_iivm_boost$set_ml_nuisance_params(
"ml_r1", "p401",
list(nrounds = 25, eta = 0.1, objective = "binary:logistic", eval_metric = "logloss", verbose=0))
}
dml_iivm_boost$fit()
dml_iivm_boost$summary()
confints = rbind(dml_iivm_lasso$confint(), dml_iivm_forest$confint(),
dml_iivm_tree$confint(), dml_iivm_boost$confint())
estimates = c(dml_iivm_lasso$coef, dml_iivm_forest$coef,
dml_iivm_tree$coef, dml_iivm_boost$coef)
result_iivm = data.table("model" = "IIVM",
"ML" = c("glmnet", "ranger", "rpart", "xgboost"),
"Estimate" = estimates,
"lower" = confints[,1],
"upper" = confints[,2])
result_iivm
g_ci = ggplot(result_iivm, aes(x = ML, y = Estimate, color = ML)) +
geom_point() +
geom_errorbar(aes(ymin = lower, ymax = upper, color = ML)) +
geom_hline(yintercept = 0, color = "grey") +
theme_minimal() + ylab("Coefficients and 0.95- confidence interval") +
xlab("") +
theme(axis.text.x = element_text(angle = 90), legend.position = "none",
text = element_text(size = 20))
g_ci
summary_result = rbindlist(list(result_plr, result_irm, result_iivm))
summary_result[, model := factor(model, levels = c("PLR", "IRM", "IIVM"))]
g_all = ggplot(summary_result, aes(x = ML, y = Estimate, color = ML)) +
geom_point() +
geom_errorbar(aes(ymin = lower, ymax = upper, color = ML)) +
geom_hline(yintercept = 0, color = "grey") +
theme_minimal() + ylab("Coefficients and 0.95- confidence interval") +
xlab("") +
theme(axis.text.x = element_text(angle = 90), legend.position = "none",
text = element_text(size = 20)) +
facet_wrap(model ~., ncol = 1)
g_all
| 0.792986 | 0.984351 |
# Create a Local Docker Image
In this section, we will create an IoT Edge module, a Docker container image with an HTTP web server that has a scoring REST endpoint.
## Get Global Variables
```
import sys
sys.path.append('../../../common')
from env_variables import *
```
## Create Web Application & Inference Server for Our ML Solution
```
%%writefile $lvaExtensionPath/app.py
import threading
import cv2
import numpy as np
import io
import onnxruntime
import json
import logging
import linecache
import sys
from score import MLModel, PrintGetExceptionDetails
from flask import Flask, request, jsonify, Response
logging.basicConfig(level=logging.DEBUG)
app = Flask(__name__)
inferenceEngine = MLModel()
@app.route("/score", methods = ['POST'])
def scoreRRS():
global inferenceEngine
try:
# get request as byte stream
reqBody = request.get_data(False)
# convert from byte stream
inMemFile = io.BytesIO(reqBody)
# load a sample image
inMemFile.seek(0)
fileBytes = np.asarray(bytearray(inMemFile.read()), dtype=np.uint8)
cvImage = cv2.imdecode(fileBytes, cv2.IMREAD_COLOR)
if cvImage.shape[:2] != (416, 416):
return Response(response='Image must be 416x416', status=400)
# Infer Image
detectedObjects = inferenceEngine.Score(cvImage)
if len(detectedObjects) > 0:
respBody = {
"inferences" : detectedObjects
}
respBody = json.dumps(respBody)
logging.info("[LVAX] Sending response.")
return Response(respBody, status= 200, mimetype ='application/json')
else:
logging.info("[LVAX] Sending empty response.")
return Response(status= 204)
except:
PrintGetExceptionDetails()
return Response(response='Exception occured while processing the image.', status=500)
@app.route("/")
def healthy():
return "Healthy"
if __name__ == "__main__":
app.run(host='127.0.0.1', port=8888)
```
8888 is the internal port of the webserver app that listens the requests. Next, we will map it to different ports to expose it externally.
```
%%writefile $lvaExtensionPath/wsgi.py
from app import app as application
def create():
application.run(host='127.0.0.1', port=8888)
import os
os.makedirs(os.path.join(lvaExtensionPath, "nginx"), exist_ok=True)
```
The exposed port of the web app is now 80, while the internal one is still 8888.
```
%%writefile $lvaExtensionPath/nginx/app
server {
listen 80;
server_name _;
location / {
include proxy_params;
proxy_pass http://127.0.0.1:8888;
proxy_connect_timeout 5000s;
proxy_read_timeout 5000s;
}
}
%%writefile $lvaExtensionPath/gunicorn_logging.conf
[loggers]
keys=root, gunicorn.error
[handlers]
keys=console
[formatters]
keys=json
[logger_root]
level=INFO
handlers=console
[logger_gunicorn.error]
level=ERROR
handlers=console
propagate=0
qualname=gunicorn.error
[handler_console]
class=StreamHandler
formatter=json
args=(sys.stdout, )
[formatter_json]
class=jsonlogging.JSONFormatter
%%writefile $lvaExtensionPath/kill_supervisor.py
import sys
import os
import signal
def write_stdout(s):
sys.stdout.write(s)
sys.stdout.flush()
# this function is modified from the code and knowledge found here: http://supervisord.org/events.html#example-event-listener-implementation
def main():
while 1:
write_stdout('[LVAX] READY\n')
# wait for the event on stdin that supervisord will send
line = sys.stdin.readline()
write_stdout('[LVAX] Terminating supervisor with this event: ' + line);
try:
# supervisord writes its pid to its file from which we read it here, see supervisord.conf
pidfile = open('/tmp/supervisord.pid','r')
pid = int(pidfile.readline());
os.kill(pid, signal.SIGQUIT)
except Exception as e:
write_stdout('[LVAX] Could not terminate supervisor: ' + e.strerror + '\n')
write_stdout('[LVAX] RESULT 2\nOK')
main()
import os
os.makedirs(os.path.join(lvaExtensionPath, "etc"), exist_ok=True)
%%writefile $lvaExtensionPath/etc/supervisord.conf
[supervisord]
logfile=/tmp/supervisord.log ; (main log file;default $CWD/supervisord.log)
logfile_maxbytes=50MB ; (max main logfile bytes b4 rotation;default 50MB)
logfile_backups=10 ; (num of main logfile rotation backups;default 10)
loglevel=info ; (log level;default info; others: debug,warn,trace)
pidfile=/tmp/supervisord.pid ; (supervisord pidfile;default supervisord.pid)
nodaemon=true ; (start in foreground if true;default false)
minfds=1024 ; (min. avail startup file descriptors;default 1024)
minprocs=200 ; (min. avail process descriptors;default 200)
[program:gunicorn]
command=bash -c "gunicorn --workers 1 -m 007 --timeout 100000 --capture-output --error-logfile - --log-level debug --log-config gunicorn_logging.conf \"wsgi:create()\""
directory=/lvaExtension
redirect_stderr=true
stdout_logfile =/dev/stdout
stdout_logfile_maxbytes=0
startretries=2
startsecs=20
[program:nginx]
command=/usr/sbin/nginx -g "daemon off;"
startretries=2
startsecs=5
priority=3
[eventlistener:program_exit]
command=python kill_supervisor.py
directory=/lvaExtension
events=PROCESS_STATE_FATAL
priority=2
```
## Create a Docker File to Containerize the ML Solution and Web App Server
```
%%writefile $lvaExtensionPath/Dockerfile
FROM ubuntu:18.04
ENV WORK_DIR=/lvaExtension
WORKDIR ${WORK_DIR}
RUN apt-get update && apt-get install -y --no-install-recommends wget ca-certificates
COPY etc /etc
RUN apt-get update && apt-get install -y --no-install-recommends \
python3-pip python3-dev libglib2.0-0 libsm6 libxext6 libxrender-dev nginx supervisor runit nginx python3-setuptools
RUN cd /usr/local/bin \
&& ln -s /usr/bin/python3 python \
&& pip3 install --upgrade pip \
&& pip install numpy onnxruntime flask pillow gunicorn opencv-python json-logging-py
RUN apt-get update && apt-get install -y --no-install-recommends \
libgl1-mesa-dev
COPY . ${WORK_DIR}/
RUN rm -rf /var/lib/apt/lists/* \
&& apt-get clean \
&& rm /etc/nginx/sites-enabled/default \
&& cp ${WORK_DIR}/nginx/app /etc/nginx/sites-available/ \
&& ln -s /etc/nginx/sites-available/app /etc/nginx/sites-enabled/
EXPOSE 80
CMD ["supervisord", "-c", "/lvaExtension/etc/supervisord.conf"]
```
## Create a Local Docker Image
Finally, we will create a Docker image locally. We will later host the image in a container registry like Docker Hub, Azure Container Registry, or a local registry.
To run the following code snippet, you must have the pre-requisities mentioned in [the requirements page](../../../common/requirements.md). Most notably, we are running the `docker` command without `sudo`.
> <span>[!WARNING]</span>
> Please ensure that Docker is running before executing the cell below. Execution of the cell below may take several minutes.
```
!docker build -t $containerImageName --file ./$lvaExtensionPath/Dockerfile ./$lvaExtensionPath
```
## Next Steps
If all the code cells above have successfully finished running, return to the Readme page to continue.
|
github_jupyter
|
import sys
sys.path.append('../../../common')
from env_variables import *
%%writefile $lvaExtensionPath/app.py
import threading
import cv2
import numpy as np
import io
import onnxruntime
import json
import logging
import linecache
import sys
from score import MLModel, PrintGetExceptionDetails
from flask import Flask, request, jsonify, Response
logging.basicConfig(level=logging.DEBUG)
app = Flask(__name__)
inferenceEngine = MLModel()
@app.route("/score", methods = ['POST'])
def scoreRRS():
global inferenceEngine
try:
# get request as byte stream
reqBody = request.get_data(False)
# convert from byte stream
inMemFile = io.BytesIO(reqBody)
# load a sample image
inMemFile.seek(0)
fileBytes = np.asarray(bytearray(inMemFile.read()), dtype=np.uint8)
cvImage = cv2.imdecode(fileBytes, cv2.IMREAD_COLOR)
if cvImage.shape[:2] != (416, 416):
return Response(response='Image must be 416x416', status=400)
# Infer Image
detectedObjects = inferenceEngine.Score(cvImage)
if len(detectedObjects) > 0:
respBody = {
"inferences" : detectedObjects
}
respBody = json.dumps(respBody)
logging.info("[LVAX] Sending response.")
return Response(respBody, status= 200, mimetype ='application/json')
else:
logging.info("[LVAX] Sending empty response.")
return Response(status= 204)
except:
PrintGetExceptionDetails()
return Response(response='Exception occured while processing the image.', status=500)
@app.route("/")
def healthy():
return "Healthy"
if __name__ == "__main__":
app.run(host='127.0.0.1', port=8888)
%%writefile $lvaExtensionPath/wsgi.py
from app import app as application
def create():
application.run(host='127.0.0.1', port=8888)
import os
os.makedirs(os.path.join(lvaExtensionPath, "nginx"), exist_ok=True)
%%writefile $lvaExtensionPath/nginx/app
server {
listen 80;
server_name _;
location / {
include proxy_params;
proxy_pass http://127.0.0.1:8888;
proxy_connect_timeout 5000s;
proxy_read_timeout 5000s;
}
}
%%writefile $lvaExtensionPath/gunicorn_logging.conf
[loggers]
keys=root, gunicorn.error
[handlers]
keys=console
[formatters]
keys=json
[logger_root]
level=INFO
handlers=console
[logger_gunicorn.error]
level=ERROR
handlers=console
propagate=0
qualname=gunicorn.error
[handler_console]
class=StreamHandler
formatter=json
args=(sys.stdout, )
[formatter_json]
class=jsonlogging.JSONFormatter
%%writefile $lvaExtensionPath/kill_supervisor.py
import sys
import os
import signal
def write_stdout(s):
sys.stdout.write(s)
sys.stdout.flush()
# this function is modified from the code and knowledge found here: http://supervisord.org/events.html#example-event-listener-implementation
def main():
while 1:
write_stdout('[LVAX] READY\n')
# wait for the event on stdin that supervisord will send
line = sys.stdin.readline()
write_stdout('[LVAX] Terminating supervisor with this event: ' + line);
try:
# supervisord writes its pid to its file from which we read it here, see supervisord.conf
pidfile = open('/tmp/supervisord.pid','r')
pid = int(pidfile.readline());
os.kill(pid, signal.SIGQUIT)
except Exception as e:
write_stdout('[LVAX] Could not terminate supervisor: ' + e.strerror + '\n')
write_stdout('[LVAX] RESULT 2\nOK')
main()
import os
os.makedirs(os.path.join(lvaExtensionPath, "etc"), exist_ok=True)
%%writefile $lvaExtensionPath/etc/supervisord.conf
[supervisord]
logfile=/tmp/supervisord.log ; (main log file;default $CWD/supervisord.log)
logfile_maxbytes=50MB ; (max main logfile bytes b4 rotation;default 50MB)
logfile_backups=10 ; (num of main logfile rotation backups;default 10)
loglevel=info ; (log level;default info; others: debug,warn,trace)
pidfile=/tmp/supervisord.pid ; (supervisord pidfile;default supervisord.pid)
nodaemon=true ; (start in foreground if true;default false)
minfds=1024 ; (min. avail startup file descriptors;default 1024)
minprocs=200 ; (min. avail process descriptors;default 200)
[program:gunicorn]
command=bash -c "gunicorn --workers 1 -m 007 --timeout 100000 --capture-output --error-logfile - --log-level debug --log-config gunicorn_logging.conf \"wsgi:create()\""
directory=/lvaExtension
redirect_stderr=true
stdout_logfile =/dev/stdout
stdout_logfile_maxbytes=0
startretries=2
startsecs=20
[program:nginx]
command=/usr/sbin/nginx -g "daemon off;"
startretries=2
startsecs=5
priority=3
[eventlistener:program_exit]
command=python kill_supervisor.py
directory=/lvaExtension
events=PROCESS_STATE_FATAL
priority=2
%%writefile $lvaExtensionPath/Dockerfile
FROM ubuntu:18.04
ENV WORK_DIR=/lvaExtension
WORKDIR ${WORK_DIR}
RUN apt-get update && apt-get install -y --no-install-recommends wget ca-certificates
COPY etc /etc
RUN apt-get update && apt-get install -y --no-install-recommends \
python3-pip python3-dev libglib2.0-0 libsm6 libxext6 libxrender-dev nginx supervisor runit nginx python3-setuptools
RUN cd /usr/local/bin \
&& ln -s /usr/bin/python3 python \
&& pip3 install --upgrade pip \
&& pip install numpy onnxruntime flask pillow gunicorn opencv-python json-logging-py
RUN apt-get update && apt-get install -y --no-install-recommends \
libgl1-mesa-dev
COPY . ${WORK_DIR}/
RUN rm -rf /var/lib/apt/lists/* \
&& apt-get clean \
&& rm /etc/nginx/sites-enabled/default \
&& cp ${WORK_DIR}/nginx/app /etc/nginx/sites-available/ \
&& ln -s /etc/nginx/sites-available/app /etc/nginx/sites-enabled/
EXPOSE 80
CMD ["supervisord", "-c", "/lvaExtension/etc/supervisord.conf"]
!docker build -t $containerImageName --file ./$lvaExtensionPath/Dockerfile ./$lvaExtensionPath
| 0.171616 | 0.461077 |
## Heat flow estimation from aerial radiometric measurements.
In this notebook we are concerned with the image processing of radiometric maps from Nova Scotia https://novascotia.ca/natr/meb/download/dp163.asp and the subsequent estimate of heat flow following the equations in Beamish and Busby (2016).
### About the data
This data set consists of 7 JPEG images of radiometric data for the province of Nova Scotia. They include images showing Potassium (K, %), equivalent Thorium (eTh, ppm), equivalent Uranium (eU, ppm), the ratio Thorium/Potassium (eTh/K, ppm/%), the ratio Uranium/Potassium (eU/K, ppm/%), the ratio Uranium/Thorium (eU/eTh) and the Total Count at a 50m resolution. The images were created by combining radiometrics data provided by the Geological Survey of Canada (GSC) from their surveys flown at 1 km line-spacing across the entire province, and 7 detailed surveys flown at 250 m line spacing by the GSC in the following areas: East Kemptville, Liscomb, Ship Harbor, Gibraltor Hill, Granite Lake, Big Indian Lake, and Tantallon Lake. The images were produced by contractor M. S. King using funds provided under the Natural Resources Canada and Nova Scotia Department of Natural Resources joint project 'Mapping, Mineral and Energy Resource Evaluation, Central Nova Scotia', part of Natural Resources Canada's Targeted Geoscience Initiative (TGI-2) 2003-2005.
### Image processing
Unfortunately there are two problems with the data as delivered. The first is the images have had a hillshading effect applied obscuring data values from a pure radiometric measurement. The second issue is the images don't have a colorscale, so it's impossible to convert the colours back into physical quantities without using some external heuristic or calibration.
We deal with the first issue by converting the R,G,B channels in the image to H, S, V, and then dripping the V channel – which is where the hillshade effect lies. This is following the code from Matt Hall in a Gist called, Ripping data from pseudocolour images:
https://gist.github.com/kwinkunks/485190adcf3239341d8bebac94de3a2b#file-rip-data-2-py
(notebook saved without cell outputs because rendered outputs are too large)
```
"""
If the colourmap matches all or part of the colour wheel or hue circle,
we can decompose the image to HSV and use H as a proxy for the data.
"""
from io import BytesIO
import matplotlib.pyplot as plt
import numpy as np
from PIL import Image
import requests
from skimage.color import rgb2hsv
import glob
def heat_equation(Cu, Cth, Ck, density=2700, c1=9.52, c2=2.56, c3=3.48):
"""
Heat production equation from Beamish and Busby (2016)
density is the density of the
density: rock density in kg/m3
Cu: weight of uranium in ppm
Cth: weight of thorium in ppm
Ck: weight of potassium in %
Returns: Radioactive heat production in W/m3
"""
return (10e-5)*density*(c1 * Cu + c2 * Cth + c3 * Ck)
def heat_equation_no_density(Cu, Cth, Ck, c1=0.26, c2=0.07, c3=0.10):
"""
Heat production equation from Beamish and Busby (2016)
density is the density of the
Cu: weight of uranium in ppm
Cth: weight of thorium in ppm
Ck: weight of potassium in %
Returns: Radioactive heat production in W/m3
"""
return c1 * Cu + c2 * Cth + c3 * Ck
# Get the 7 radiometric maps and their names
fnames = glob.glob('i163nsaa_NS_Radiometric_Images_50m/jpg/*.jpg')
names = [fname.split('/')[-1].split('.')[0] for fname in fnames]
names
fnames
print(fname, name)
img = Image.open(fnames[1])
# Read the image and transform to HSV and save fig.
for fname, name in zip(fnames, names):
print(fname, name)
img = Image.open(fname)
img_size = img.size
rgb_im = np.asarray(img)[..., :3] / 255.
hsv_im = rgb2hsv(rgb_im)
hue = hsv_im[..., 0]
# val = hsv_im[..., 2]
# Make a new figure.
my_dpi = 96
plt.figure(figsize=(img_size[0]/my_dpi, img_size[1]/my_dpi), dpi=my_dpi)
plt.imshow(hue, cmap='Greys_r')
plt.axis('off')
plt.tight_layout()
plt.savefig(f'{name}_fixed.jpg')
```
## Fixed images
7 new maps with hillshading removed
```
fixed_images = glob.glob('*fixed.jpg')
fixed_images
imgs = [Image.open(fixed_image) for fixed_image in fixed_images]
imgs[1]
```
# Composite map
```
for img in imgs:
print (img.size, img.filename)
```
The images aren't the same size. Dang it. Fortunately the "Potassium", "eThorium" and "eUranium" are the same size.
```
potassium = np.array(imgs[3])
eThorium = np.array(imgs[4])
eUranium = np.array(imgs[1])
potassium_gray = np.mean(potassium, axis=2)
eThorium_gray = np.mean(eThorium, axis=2)
eUranium_gray = np.mean(eUranium, axis=2)
print('Grayscale image shape:', potassium_gray.shape)
print('Grayscale image shape:', eThorium_gray.shape)
print('Grayscale image shape:', eUranium_gray.shape)
# Set the water to np.nan instead of 255
potassium_gray[potassium_gray == np.amax(potassium_gray)] = np.nan
eThorium_gray[eThorium_gray == np.amax(eThorium_gray)] = np.nan
eUranium_gray[eUranium_gray == np.amax(eUranium_gray)] = np.nan
plt.figure(figsize=(10,10))
plt.imshow(eUranium_gray[::10, ::10], cmap='Greys')
plt.colorbar(shrink=0.5)
```
# Corendering K (reds), Th (greens), U (blues)
```
c1, c2, c3 = 255, 255, 255
U_Th_K_stack = np.stack((potassium_gray/c1, eThorium_gray/c2, eUranium_gray/c3), axis=-1)
U_Th_K_stack.shape
step = 20 # change this for faster / slower rendering
plt.figure(figsize=(20,20))
plt.imshow(U_Th_K_stack[::step, ::step, :])
plt.savefig('Radiometric_corendering_NS.png')
def normU(u):
"""
A function to scale Uranium map. We don't know what this function should be
"""
return u
def normTh(th):
"""
A function to scale thorium. We don't know what this function should be
"""
return th
def normK(k):
"""
A function to scale potassium. We don't know what this function should be
"""
return k
heat_gen1 = heat_equation_no_density(eUranium_gray, eThorium_gray, potassium_gray)
heat_gen1[heat_gen1 == np.amax(heat_gen1)] = np.nan
plt.figure(figsize=(20,20))
plt.imshow(heat_gen1[::step, ::step], cmap='plasma')
plt.colorbar(shrink=0.5)
heat_gen2 = heat_equation(eUranium_gray, eThorium_gray, potassium_gray)
heat_gen2[heat_gen2 == np.amax(heat_gen2)] = np.nan
plt.figure(figsize=(20,20))
plt.imshow(heat_gen2[::step, ::step], cmap='plasma')
plt.colorbar(shrink=0.5)
plt.savefig('Heat_flow_estimate_NS.png')
```
The maps look similar, but are off by about an order of magnitude. Does that help in how they should be scaled?
|
github_jupyter
|
"""
If the colourmap matches all or part of the colour wheel or hue circle,
we can decompose the image to HSV and use H as a proxy for the data.
"""
from io import BytesIO
import matplotlib.pyplot as plt
import numpy as np
from PIL import Image
import requests
from skimage.color import rgb2hsv
import glob
def heat_equation(Cu, Cth, Ck, density=2700, c1=9.52, c2=2.56, c3=3.48):
"""
Heat production equation from Beamish and Busby (2016)
density is the density of the
density: rock density in kg/m3
Cu: weight of uranium in ppm
Cth: weight of thorium in ppm
Ck: weight of potassium in %
Returns: Radioactive heat production in W/m3
"""
return (10e-5)*density*(c1 * Cu + c2 * Cth + c3 * Ck)
def heat_equation_no_density(Cu, Cth, Ck, c1=0.26, c2=0.07, c3=0.10):
"""
Heat production equation from Beamish and Busby (2016)
density is the density of the
Cu: weight of uranium in ppm
Cth: weight of thorium in ppm
Ck: weight of potassium in %
Returns: Radioactive heat production in W/m3
"""
return c1 * Cu + c2 * Cth + c3 * Ck
# Get the 7 radiometric maps and their names
fnames = glob.glob('i163nsaa_NS_Radiometric_Images_50m/jpg/*.jpg')
names = [fname.split('/')[-1].split('.')[0] for fname in fnames]
names
fnames
print(fname, name)
img = Image.open(fnames[1])
# Read the image and transform to HSV and save fig.
for fname, name in zip(fnames, names):
print(fname, name)
img = Image.open(fname)
img_size = img.size
rgb_im = np.asarray(img)[..., :3] / 255.
hsv_im = rgb2hsv(rgb_im)
hue = hsv_im[..., 0]
# val = hsv_im[..., 2]
# Make a new figure.
my_dpi = 96
plt.figure(figsize=(img_size[0]/my_dpi, img_size[1]/my_dpi), dpi=my_dpi)
plt.imshow(hue, cmap='Greys_r')
plt.axis('off')
plt.tight_layout()
plt.savefig(f'{name}_fixed.jpg')
fixed_images = glob.glob('*fixed.jpg')
fixed_images
imgs = [Image.open(fixed_image) for fixed_image in fixed_images]
imgs[1]
for img in imgs:
print (img.size, img.filename)
potassium = np.array(imgs[3])
eThorium = np.array(imgs[4])
eUranium = np.array(imgs[1])
potassium_gray = np.mean(potassium, axis=2)
eThorium_gray = np.mean(eThorium, axis=2)
eUranium_gray = np.mean(eUranium, axis=2)
print('Grayscale image shape:', potassium_gray.shape)
print('Grayscale image shape:', eThorium_gray.shape)
print('Grayscale image shape:', eUranium_gray.shape)
# Set the water to np.nan instead of 255
potassium_gray[potassium_gray == np.amax(potassium_gray)] = np.nan
eThorium_gray[eThorium_gray == np.amax(eThorium_gray)] = np.nan
eUranium_gray[eUranium_gray == np.amax(eUranium_gray)] = np.nan
plt.figure(figsize=(10,10))
plt.imshow(eUranium_gray[::10, ::10], cmap='Greys')
plt.colorbar(shrink=0.5)
c1, c2, c3 = 255, 255, 255
U_Th_K_stack = np.stack((potassium_gray/c1, eThorium_gray/c2, eUranium_gray/c3), axis=-1)
U_Th_K_stack.shape
step = 20 # change this for faster / slower rendering
plt.figure(figsize=(20,20))
plt.imshow(U_Th_K_stack[::step, ::step, :])
plt.savefig('Radiometric_corendering_NS.png')
def normU(u):
"""
A function to scale Uranium map. We don't know what this function should be
"""
return u
def normTh(th):
"""
A function to scale thorium. We don't know what this function should be
"""
return th
def normK(k):
"""
A function to scale potassium. We don't know what this function should be
"""
return k
heat_gen1 = heat_equation_no_density(eUranium_gray, eThorium_gray, potassium_gray)
heat_gen1[heat_gen1 == np.amax(heat_gen1)] = np.nan
plt.figure(figsize=(20,20))
plt.imshow(heat_gen1[::step, ::step], cmap='plasma')
plt.colorbar(shrink=0.5)
heat_gen2 = heat_equation(eUranium_gray, eThorium_gray, potassium_gray)
heat_gen2[heat_gen2 == np.amax(heat_gen2)] = np.nan
plt.figure(figsize=(20,20))
plt.imshow(heat_gen2[::step, ::step], cmap='plasma')
plt.colorbar(shrink=0.5)
plt.savefig('Heat_flow_estimate_NS.png')
| 0.845209 | 0.972934 |
# Distirbuted Training of Mask-RCNN in Amazon SageMaker using EFS
This notebook is a step-by-step tutorial on distributed tranining of [Mask R-CNN](https://arxiv.org/abs/1703.06870) implemented in [TensorFlow](https://www.tensorflow.org/) framework. Mask R-CNN is also referred to as heavy weight object detection model and it is part of [MLPerf](https://www.mlperf.org/training-results-0-6/).
Concretely, we will describe the steps for training [TensorPack Faster-RCNN/Mask-RCNN](https://github.com/tensorpack/tensorpack/tree/master/examples/FasterRCNN) and [AWS Samples Mask R-CNN](https://github.com/aws-samples/mask-rcnn-tensorflow) in [Amazon SageMaker](https://aws.amazon.com/sagemaker/) using [Amazon EFS](https://aws.amazon.com/efs/) file-system as data source.
The outline of steps is as follows:
1. Stage COCO 2017 dataset in [Amazon S3](https://aws.amazon.com/s3/)
2. Copy COCO 2017 dataset from S3 to Amazon EFS file-system mounted on this notebook instance
3. Build Docker training image and push it to [Amazon ECR](https://aws.amazon.com/ecr/)
4. Configure data input channels
5. Configure hyper-prarameters
6. Define training metrics
7. Define training job and start training
Before we get started, let us initialize two python variables ```aws_region``` and ```s3_bucket``` that we will use throughout the notebook:
```
aws_region = # <aws-region>
s3_bucket = # <your-s3_bucket>
```
## Stage COCO 2017 dataset in Amazon S3
We use [COCO 2017 dataset](http://cocodataset.org/#home) for training. We download COCO 2017 training and validation dataset to this notebook instance, extract the files from the dataset archives, and upload the extracted files to your Amazon [S3 bucket](https://docs.aws.amazon.com/AmazonS3/latest/gsg/CreatingABucket.html). The ```prepare-s3-bucket.sh``` script executes this step.
```
!cat ./prepare-s3-bucket.sh
```
Using your *Amazon S3 bucket* as argument, run the cell below. If you have already uploaded COCO 2017 dataset to your Amazon S3 bucket, you may skip this step.
```
%%time
!./prepare-s3-bucket.sh {s3_bucket}
```
## Copy COCO 2017 dataset from S3 to Amazon EFS
Next, we copy COCO 2017 dataset from S3 to EFS file-system. The ```prepare-efs.sh``` script executes this step.
```
!cat ./prepare-efs.sh
```
If you have already copied COCO 2017 dataset from S3 to your EFS file-system, skip this step.
```
%%time
!./prepare-efs.sh {s3_bucket}
```
## Build and push SageMaker training images
For this step, the [IAM Role](https://docs.aws.amazon.com/IAM/latest/UserGuide/id_roles.html) attached to this notebook instance needs full access to Amazon ECR service. If you created this notebook instance using the ```./stack-sm.sh``` script in this repository, the IAM Role attached to this notebook instance is already setup with full access to ECR service.
Below, we have a choice of two different implementations:
1. [TensorPack Faster-RCNN/Mask-RCNN](https://github.com/tensorpack/tensorpack/tree/master/examples/FasterRCNN) implementation supports a maximum per-GPU batch size of 1, and does not support mixed precision. It can be used with mainstream TensorFlow releases.
2. [AWS Samples Mask R-CNN](https://github.com/aws-samples/mask-rcnn-tensorflow) is an optimized implementation that supports a maximum batch size of 4 and supports mixed precision. This implementation uses TensorFlow base version 1.13 augmented with custom TensorFlow ops.
It is recommended that you build and push both SageMaker training images and use either image for training later.
### TensorPack Faster-RCNN/Mask-RCNN
Use ```./container/build_tools/build_and_push.sh``` script to build and push the TensorPack Faster-RCNN/Mask-RCNN training image to Amazon ECR.
```
!cat ./container/build_tools/build_and_push.sh
```
Using your *AWS region* as argument, run the cell below.
```
%%time
! ./container/build_tools/build_and_push.sh {aws_region}
```
Set ```tensorpack_image``` below to Amazon ECR URI of the image you pushed above.
```
tensorpack_image = #<amazon-ecr-uri>
```
### AWS Samples Mask R-CNN
Use ```./container-optimized/build_tools/build_and_push.sh``` script to build and push the AWS Samples Mask R-CNN training image to Amazon ECR.
```
!cat ./container-optimized/build_tools/build_and_push.sh
```
Using your *AWS region* as argument, run the cell below.
```
%%time
! ./container-optimized/build_tools/build_and_push.sh {aws_region}
```
Set ```aws_samples_image``` below to Amazon ECR URI of the image you pushed above.
```
aws_samples_image = #<amazon-ecr-uri>
```
## SageMaker Initialization
We have staged the data and we have built and pushed the training docker image to Amazon ECR. Now we are ready to start using Amazon SageMaker.
```
%%time
import os
import time
import boto3
import sagemaker
from sagemaker import get_execution_role
from sagemaker.estimator import Estimator
role = get_execution_role() # provide a pre-existing role ARN as an alternative to creating a new role
print(f'SageMaker Execution Role:{role}')
client = boto3.client('sts')
account = client.get_caller_identity()['Account']
print(f'AWS account:{account}')
session = boto3.session.Session()
region = session.region_name
print(f'AWS region:{region}')
```
Next, we set the Amazon ECR image URI used for training. You saved this URI in a previous step.
```
training_image = # set to tensorpack_image or aws_samples_image
print(f'Training image: {training_image}')
```
## Define SageMaker Data Channels
Next, we define the *train* and *log* data channels using EFS file-system. To do so, we need to specify the EFS file-system id, which is shown in the output of the command below.
```
!df -kh | grep 'fs-' | sed 's/\(fs-[0-9a-z]*\).*/\1/'
```
Set the EFS ```file_system_id``` below to the ouput of the command shown above. In the cell below, we define the `train` data input channel.
```
from sagemaker.inputs import FileSystemInput
# Specify EFS ile system id.
file_system_id = # 'fs-xxxxxxxx'
print(f"EFS file-system-id: {file_system_id}")
# Specify directory path for input data on the file system.
# You need to provide normalized and absolute path below.
file_system_directory_path = '/mask-rcnn/sagemaker/input/train'
print(f'EFS file-system data input path: {file_system_directory_path}')
# Specify the access mode of the mount of the directory associated with the file system.
# Directory must be mounted 'ro'(read-only).
file_system_access_mode = 'ro'
# Specify your file system type
file_system_type = 'EFS'
train = FileSystemInput(file_system_id=file_system_id,
file_system_type=file_system_type,
directory_path=file_system_directory_path,
file_system_access_mode=file_system_access_mode)
```
Below we create the log output directory and define the `log` data output channel.
```
# Specify directory path for log output on the EFS file system.
# You need to provide normalized and absolute path below.
# For example, '/mask-rcnn/sagemaker/output/log'
# Log output directory must not exist
file_system_directory_path = f'/mask-rcnn/sagemaker/output/log-{int(time.time())}'
# Create the log output directory.
# EFS file-system is mounted on '$HOME/efs' mount point for this notebook.
home_dir=os.environ['HOME']
local_efs_path = os.path.join(home_dir,'efs', file_system_directory_path[1:])
print(f"Creating log directory on EFS: {local_efs_path}")
assert not os.path.isdir(local_efs_path)
! sudo mkdir -p -m a=rw {local_efs_path}
assert os.path.isdir(local_efs_path)
# Specify the access mode of the mount of the directory associated with the file system.
# Directory must be mounted 'rw'(read-write).
file_system_access_mode = 'rw'
log = FileSystemInput(file_system_id=file_system_id,
file_system_type=file_system_type,
directory_path=file_system_directory_path,
file_system_access_mode=file_system_access_mode)
data_channels = {'train': train, 'log': log}
```
Next, we define the model output location in S3. Set ```s3_bucket``` to your S3 bucket name prior to running the cell below.
The model checkpoints, logs and Tensorboard events will be written to the log output directory on the EFS file system you created above. At the end of the model training, they will be copied from the log output directory to the `s3_output_location` defined below.
```
prefix = "mask-rcnn/sagemaker" #prefix in your bucket
s3_output_location = f's3://{s3_bucket}/{prefix}/output'
print(f'S3 model output location: {s3_output_location}')
```
## Configure Hyper-parameters
Next we define the hyper-parameters.
Note, some hyper-parameters are different between the two implementations. The batch size per GPU in TensorPack Faster-RCNN/Mask-RCNN is fixed at 1, but is configurable in AWS Samples Mask-RCNN. The learning rate schedule is specified in units of steps in TensorPack Faster-RCNN/Mask-RCNN, but in epochs in AWS Samples Mask-RCNN.
The detault learning rate schedule values shown below correspond to training for a total of 24 epochs, at 120,000 images per epoch.
<table align='left'>
<caption>TensorPack Faster-RCNN/Mask-RCNN Hyper-parameters</caption>
<tr>
<th style="text-align:center">Hyper-parameter</th>
<th style="text-align:center">Description</th>
<th style="text-align:center">Default</th>
</tr>
<tr>
<td style="text-align:center">mode_fpn</td>
<td style="text-align:left">Flag to indicate use of Feature Pyramid Network (FPN) in the Mask R-CNN model backbone</td>
<td style="text-align:center">"True"</td>
</tr>
<tr>
<td style="text-align:center">mode_mask</td>
<td style="text-align:left">A value of "False" means Faster-RCNN model, "True" means Mask R-CNN moodel</td>
<td style="text-align:center">"True"</td>
</tr>
<tr>
<td style="text-align:center">eval_period</td>
<td style="text-align:left">Number of epochs period for evaluation during training</td>
<td style="text-align:center">1</td>
</tr>
<tr>
<td style="text-align:center">lr_schedule</td>
<td style="text-align:left">Learning rate schedule in training steps</td>
<td style="text-align:center">'[240000, 320000, 360000]'</td>
</tr>
<tr>
<td style="text-align:center">batch_norm</td>
<td style="text-align:left">Batch normalization option ('FreezeBN', 'SyncBN', 'GN', 'None') </td>
<td style="text-align:center">'FreezeBN'</td>
</tr>
<tr>
<td style="text-align:center">images_per_epoch</td>
<td style="text-align:left">Images per epoch </td>
<td style="text-align:center">120000</td>
</tr>
<tr>
<td style="text-align:center">data_train</td>
<td style="text-align:left">Training data under data directory</td>
<td style="text-align:center">'coco_train2017'</td>
</tr>
<tr>
<td style="text-align:center">data_val</td>
<td style="text-align:left">Validation data under data directory</td>
<td style="text-align:center">'coco_val2017'</td>
</tr>
<tr>
<td style="text-align:center">resnet_arch</td>
<td style="text-align:left">Must be 'resnet50' or 'resnet101'</td>
<td style="text-align:center">'resnet50'</td>
</tr>
<tr>
<td style="text-align:center">backbone_weights</td>
<td style="text-align:left">ResNet backbone weights</td>
<td style="text-align:center">'ImageNet-R50-AlignPadding.npz'</td>
</tr>
<tr>
<td style="text-align:center">load_model</td>
<td style="text-align:left">Pre-trained model to load</td>
<td style="text-align:center"></td>
</tr>
<tr>
<td style="text-align:center">config:</td>
<td style="text-align:left">Any hyperparamter prefixed with <b>config:</b> is set as a model config parameter</td>
<td style="text-align:center"></td>
</tr>
</table>
<table align='left'>
<caption>AWS Samples Mask-RCNN Hyper-parameters</caption>
<tr>
<th style="text-align:center">Hyper-parameter</th>
<th style="text-align:center">Description</th>
<th style="text-align:center">Default</th>
</tr>
<tr>
<td style="text-align:center">mode_fpn</td>
<td style="text-align:left">Flag to indicate use of Feature Pyramid Network (FPN) in the Mask R-CNN model backbone</td>
<td style="text-align:center">"True"</td>
</tr>
<tr>
<td style="text-align:center">mode_mask</td>
<td style="text-align:left">A value of "False" means Faster-RCNN model, "True" means Mask R-CNN moodel</td>
<td style="text-align:center">"True"</td>
</tr>
<tr>
<td style="text-align:center">eval_period</td>
<td style="text-align:left">Number of epochs period for evaluation during training</td>
<td style="text-align:center">1</td>
</tr>
<tr>
<td style="text-align:center">lr_epoch_schedule</td>
<td style="text-align:left">Learning rate schedule in epochs</td>
<td style="text-align:center">'[(16, 0.1), (20, 0.01), (24, None)]'</td>
</tr>
<tr>
<td style="text-align:center">batch_size_per_gpu</td>
<td style="text-align:left">Batch size per gpu ( Minimum 1, Maximum 4)</td>
<td style="text-align:center">4</td>
</tr>
<tr>
<td style="text-align:center">batch_norm</td>
<td style="text-align:left">Batch normalization option ('FreezeBN', 'SyncBN', 'GN', 'None') </td>
<td style="text-align:center">'FreezeBN'</td>
</tr>
<tr>
<td style="text-align:center">images_per_epoch</td>
<td style="text-align:left">Images per epoch </td>
<td style="text-align:center">120000</td>
</tr>
<tr>
<td style="text-align:center">data_train</td>
<td style="text-align:left">Training data under data directory</td>
<td style="text-align:center">'train2017'</td>
</tr>
<tr>
<td style="text-align:center">backbone_weights</td>
<td style="text-align:left">ResNet backbone weights</td>
<td style="text-align:center">'ImageNet-R50-AlignPadding.npz'</td>
</tr>
<tr>
<td style="text-align:center">load_model</td>
<td style="text-align:left">Pre-trained model to load</td>
<td style="text-align:center"></td>
</tr>
<tr>
<td style="text-align:center">config:</td>
<td style="text-align:left">Any hyperparamter prefixed with <b>config:</b> is set as a model config parameter</td>
<td style="text-align:center"></td>
</tr>
</table>
```
hyperparameters = {
"mode_fpn": "True",
"mode_mask": "True",
"eval_period": 1,
"batch_norm": "FreezeBN"
}
```
## Define Training Metrics
Next, we define the regular expressions that SageMaker uses to extract algorithm metrics from training logs and send them to [AWS CloudWatch metrics](https://docs.aws.amazon.com/en_pv/AmazonCloudWatch/latest/monitoring/working_with_metrics.html). These algorithm metrics are visualized in SageMaker console.
```
metric_definitions=[
{
"Name": "fastrcnn_losses/box_loss",
"Regex": ".*fastrcnn_losses/box_loss:\\s*(\\S+).*"
},
{
"Name": "fastrcnn_losses/label_loss",
"Regex": ".*fastrcnn_losses/label_loss:\\s*(\\S+).*"
},
{
"Name": "fastrcnn_losses/label_metrics/accuracy",
"Regex": ".*fastrcnn_losses/label_metrics/accuracy:\\s*(\\S+).*"
},
{
"Name": "fastrcnn_losses/label_metrics/false_negative",
"Regex": ".*fastrcnn_losses/label_metrics/false_negative:\\s*(\\S+).*"
},
{
"Name": "fastrcnn_losses/label_metrics/fg_accuracy",
"Regex": ".*fastrcnn_losses/label_metrics/fg_accuracy:\\s*(\\S+).*"
},
{
"Name": "fastrcnn_losses/num_fg_label",
"Regex": ".*fastrcnn_losses/num_fg_label:\\s*(\\S+).*"
},
{
"Name": "maskrcnn_loss/accuracy",
"Regex": ".*maskrcnn_loss/accuracy:\\s*(\\S+).*"
},
{
"Name": "maskrcnn_loss/fg_pixel_ratio",
"Regex": ".*maskrcnn_loss/fg_pixel_ratio:\\s*(\\S+).*"
},
{
"Name": "maskrcnn_loss/maskrcnn_loss",
"Regex": ".*maskrcnn_loss/maskrcnn_loss:\\s*(\\S+).*"
},
{
"Name": "maskrcnn_loss/pos_accuracy",
"Regex": ".*maskrcnn_loss/pos_accuracy:\\s*(\\S+).*"
},
{
"Name": "mAP(bbox)/IoU=0.5",
"Regex": ".*mAP\\(bbox\\)/IoU=0\\.5:\\s*(\\S+).*"
},
{
"Name": "mAP(bbox)/IoU=0.5:0.95",
"Regex": ".*mAP\\(bbox\\)/IoU=0\\.5:0\\.95:\\s*(\\S+).*"
},
{
"Name": "mAP(bbox)/IoU=0.75",
"Regex": ".*mAP\\(bbox\\)/IoU=0\\.75:\\s*(\\S+).*"
},
{
"Name": "mAP(bbox)/large",
"Regex": ".*mAP\\(bbox\\)/large:\\s*(\\S+).*"
},
{
"Name": "mAP(bbox)/medium",
"Regex": ".*mAP\\(bbox\\)/medium:\\s*(\\S+).*"
},
{
"Name": "mAP(bbox)/small",
"Regex": ".*mAP\\(bbox\\)/small:\\s*(\\S+).*"
},
{
"Name": "mAP(segm)/IoU=0.5",
"Regex": ".*mAP\\(segm\\)/IoU=0\\.5:\\s*(\\S+).*"
},
{
"Name": "mAP(segm)/IoU=0.5:0.95",
"Regex": ".*mAP\\(segm\\)/IoU=0\\.5:0\\.95:\\s*(\\S+).*"
},
{
"Name": "mAP(segm)/IoU=0.75",
"Regex": ".*mAP\\(segm\\)/IoU=0\\.75:\\s*(\\S+).*"
},
{
"Name": "mAP(segm)/large",
"Regex": ".*mAP\\(segm\\)/large:\\s*(\\S+).*"
},
{
"Name": "mAP(segm)/medium",
"Regex": ".*mAP\\(segm\\)/medium:\\s*(\\S+).*"
},
{
"Name": "mAP(segm)/small",
"Regex": ".*mAP\\(segm\\)/small:\\s*(\\S+).*"
}
]
```
## Define SageMaker Training Job
Next, we use SageMaker [Estimator](https://sagemaker.readthedocs.io/en/stable/estimators.html) API to define a SageMaker Training Job.
We recommned using 32 GPUs, so we set ```train_instance_count=4``` and ```train_instance_type='ml.p3.16xlarge'```, because there are 8 Tesla V100 GPUs per ```ml.p3.16xlarge``` instance. We recommend using 100 GB [Amazon EBS](https://aws.amazon.com/ebs/) storage volume with each training instance, so we set ```train_volume_size = 100```.
We run the training job in your private VPC, so we need to set the ```subnets``` and ```security_group_ids``` prior to running the cell below. You may specify multiple subnet ids in the ```subnets``` list. The subnets included in the ```sunbets``` list must be part of the output of ```./stack-sm.sh``` CloudFormation stack script used to create this notebook instance. Specify only one security group id in ```security_group_ids``` list. The security group id must be part of the output of ```./stack-sm.sh``` script.
For ```train_instance_type``` below, you have the option to use ```ml.p3.16xlarge``` with 16 GB per-GPU memory and 25 Gbs network interconnectivity, or ```ml.p3dn.24xlarge``` with 32 GB per-GPU memory and 100 Gbs network interconnectivity. The ```ml.p3dn.24xlarge``` instance type offers significantly better performance than ```ml.p3.16xlarge``` for Mask R-CNN distributed TensorFlow training.
```
# Give Amazon SageMaker Training Jobs Access to FileSystem Resources in Your Amazon VPC.
security_group_ids = # ['sg-xxxxxxxx']
subnets = # [ 'subnet-xxxxxxx']
sagemaker_session = sagemaker.session.Session(boto_session=session)
mask_rcnn_estimator = Estimator(training_image,
role,
train_instance_count=4,
train_instance_type='ml.p3.16xlarge',
train_volume_size = 100,
train_max_run = 400000,
output_path=s3_output_location,
sagemaker_session=sagemaker_session,
hyperparameters = hyperparameters,
metric_definitions = metric_definitions,
base_job_name="mask-rcnn-efs",
subnets=subnets,
security_group_ids=security_group_ids)
```
Finally, we launch the SageMaker training job.
The time to complete the training depends on type and number of training instances, and the training image used for training.
```
mask_rcnn_estimator.fit(inputs=data_channels, logs=True)
```
|
github_jupyter
|
aws_region = # <aws-region>
s3_bucket = # <your-s3_bucket>
!cat ./prepare-s3-bucket.sh
%%time
!./prepare-s3-bucket.sh {s3_bucket}
!cat ./prepare-efs.sh
%%time
!./prepare-efs.sh {s3_bucket}
!cat ./container/build_tools/build_and_push.sh
%%time
! ./container/build_tools/build_and_push.sh {aws_region}
tensorpack_image = #<amazon-ecr-uri>
!cat ./container-optimized/build_tools/build_and_push.sh
%%time
! ./container-optimized/build_tools/build_and_push.sh {aws_region}
aws_samples_image = #<amazon-ecr-uri>
%%time
import os
import time
import boto3
import sagemaker
from sagemaker import get_execution_role
from sagemaker.estimator import Estimator
role = get_execution_role() # provide a pre-existing role ARN as an alternative to creating a new role
print(f'SageMaker Execution Role:{role}')
client = boto3.client('sts')
account = client.get_caller_identity()['Account']
print(f'AWS account:{account}')
session = boto3.session.Session()
region = session.region_name
print(f'AWS region:{region}')
training_image = # set to tensorpack_image or aws_samples_image
print(f'Training image: {training_image}')
!df -kh | grep 'fs-' | sed 's/\(fs-[0-9a-z]*\).*/\1/'
from sagemaker.inputs import FileSystemInput
# Specify EFS ile system id.
file_system_id = # 'fs-xxxxxxxx'
print(f"EFS file-system-id: {file_system_id}")
# Specify directory path for input data on the file system.
# You need to provide normalized and absolute path below.
file_system_directory_path = '/mask-rcnn/sagemaker/input/train'
print(f'EFS file-system data input path: {file_system_directory_path}')
# Specify the access mode of the mount of the directory associated with the file system.
# Directory must be mounted 'ro'(read-only).
file_system_access_mode = 'ro'
# Specify your file system type
file_system_type = 'EFS'
train = FileSystemInput(file_system_id=file_system_id,
file_system_type=file_system_type,
directory_path=file_system_directory_path,
file_system_access_mode=file_system_access_mode)
# Specify directory path for log output on the EFS file system.
# You need to provide normalized and absolute path below.
# For example, '/mask-rcnn/sagemaker/output/log'
# Log output directory must not exist
file_system_directory_path = f'/mask-rcnn/sagemaker/output/log-{int(time.time())}'
# Create the log output directory.
# EFS file-system is mounted on '$HOME/efs' mount point for this notebook.
home_dir=os.environ['HOME']
local_efs_path = os.path.join(home_dir,'efs', file_system_directory_path[1:])
print(f"Creating log directory on EFS: {local_efs_path}")
assert not os.path.isdir(local_efs_path)
! sudo mkdir -p -m a=rw {local_efs_path}
assert os.path.isdir(local_efs_path)
# Specify the access mode of the mount of the directory associated with the file system.
# Directory must be mounted 'rw'(read-write).
file_system_access_mode = 'rw'
log = FileSystemInput(file_system_id=file_system_id,
file_system_type=file_system_type,
directory_path=file_system_directory_path,
file_system_access_mode=file_system_access_mode)
data_channels = {'train': train, 'log': log}
prefix = "mask-rcnn/sagemaker" #prefix in your bucket
s3_output_location = f's3://{s3_bucket}/{prefix}/output'
print(f'S3 model output location: {s3_output_location}')
hyperparameters = {
"mode_fpn": "True",
"mode_mask": "True",
"eval_period": 1,
"batch_norm": "FreezeBN"
}
metric_definitions=[
{
"Name": "fastrcnn_losses/box_loss",
"Regex": ".*fastrcnn_losses/box_loss:\\s*(\\S+).*"
},
{
"Name": "fastrcnn_losses/label_loss",
"Regex": ".*fastrcnn_losses/label_loss:\\s*(\\S+).*"
},
{
"Name": "fastrcnn_losses/label_metrics/accuracy",
"Regex": ".*fastrcnn_losses/label_metrics/accuracy:\\s*(\\S+).*"
},
{
"Name": "fastrcnn_losses/label_metrics/false_negative",
"Regex": ".*fastrcnn_losses/label_metrics/false_negative:\\s*(\\S+).*"
},
{
"Name": "fastrcnn_losses/label_metrics/fg_accuracy",
"Regex": ".*fastrcnn_losses/label_metrics/fg_accuracy:\\s*(\\S+).*"
},
{
"Name": "fastrcnn_losses/num_fg_label",
"Regex": ".*fastrcnn_losses/num_fg_label:\\s*(\\S+).*"
},
{
"Name": "maskrcnn_loss/accuracy",
"Regex": ".*maskrcnn_loss/accuracy:\\s*(\\S+).*"
},
{
"Name": "maskrcnn_loss/fg_pixel_ratio",
"Regex": ".*maskrcnn_loss/fg_pixel_ratio:\\s*(\\S+).*"
},
{
"Name": "maskrcnn_loss/maskrcnn_loss",
"Regex": ".*maskrcnn_loss/maskrcnn_loss:\\s*(\\S+).*"
},
{
"Name": "maskrcnn_loss/pos_accuracy",
"Regex": ".*maskrcnn_loss/pos_accuracy:\\s*(\\S+).*"
},
{
"Name": "mAP(bbox)/IoU=0.5",
"Regex": ".*mAP\\(bbox\\)/IoU=0\\.5:\\s*(\\S+).*"
},
{
"Name": "mAP(bbox)/IoU=0.5:0.95",
"Regex": ".*mAP\\(bbox\\)/IoU=0\\.5:0\\.95:\\s*(\\S+).*"
},
{
"Name": "mAP(bbox)/IoU=0.75",
"Regex": ".*mAP\\(bbox\\)/IoU=0\\.75:\\s*(\\S+).*"
},
{
"Name": "mAP(bbox)/large",
"Regex": ".*mAP\\(bbox\\)/large:\\s*(\\S+).*"
},
{
"Name": "mAP(bbox)/medium",
"Regex": ".*mAP\\(bbox\\)/medium:\\s*(\\S+).*"
},
{
"Name": "mAP(bbox)/small",
"Regex": ".*mAP\\(bbox\\)/small:\\s*(\\S+).*"
},
{
"Name": "mAP(segm)/IoU=0.5",
"Regex": ".*mAP\\(segm\\)/IoU=0\\.5:\\s*(\\S+).*"
},
{
"Name": "mAP(segm)/IoU=0.5:0.95",
"Regex": ".*mAP\\(segm\\)/IoU=0\\.5:0\\.95:\\s*(\\S+).*"
},
{
"Name": "mAP(segm)/IoU=0.75",
"Regex": ".*mAP\\(segm\\)/IoU=0\\.75:\\s*(\\S+).*"
},
{
"Name": "mAP(segm)/large",
"Regex": ".*mAP\\(segm\\)/large:\\s*(\\S+).*"
},
{
"Name": "mAP(segm)/medium",
"Regex": ".*mAP\\(segm\\)/medium:\\s*(\\S+).*"
},
{
"Name": "mAP(segm)/small",
"Regex": ".*mAP\\(segm\\)/small:\\s*(\\S+).*"
}
]
# Give Amazon SageMaker Training Jobs Access to FileSystem Resources in Your Amazon VPC.
security_group_ids = # ['sg-xxxxxxxx']
subnets = # [ 'subnet-xxxxxxx']
sagemaker_session = sagemaker.session.Session(boto_session=session)
mask_rcnn_estimator = Estimator(training_image,
role,
train_instance_count=4,
train_instance_type='ml.p3.16xlarge',
train_volume_size = 100,
train_max_run = 400000,
output_path=s3_output_location,
sagemaker_session=sagemaker_session,
hyperparameters = hyperparameters,
metric_definitions = metric_definitions,
base_job_name="mask-rcnn-efs",
subnets=subnets,
security_group_ids=security_group_ids)
mask_rcnn_estimator.fit(inputs=data_channels, logs=True)
| 0.452536 | 0.966663 |
# Analyzing a PAC
I'm trying to understand the spending habits of the Microsoft PAC since I work there. The following is primarily based on the code from [this workbook](https://github.com/boblannon/blogpost_fec-api-howto/blob/master/fec_api.ipynb) from [Bob Lannon](https://github.com/boblannon) that explains how to use the FEC API.
First I exported the [spending data for the Microsoft PAC from 2017-18](https://www.fec.gov/data/committee/C00227546/?tab=spending). This got me a csv file used later. This wasn't really enough to sort out what I was interested in as there is no party affiliation in the exported data. So that is what I wanted to use the FEC API to fill in.
```
%pylab inline
import matplotlib.pyplot as plt
matplotlib.style.use('ggplot')
import numpy as np
import pandas as pd
import requests
import os
import json
from copy import deepcopy
import logging
logging.basicConfig(format='%(asctime)s : %(levelname)s : %(message)s')
logging.getLogger("requests").setLevel(logging.ERROR) # silencing requests logging
# Logging for this notebook
logger = logging.getLogger()
logger.setLevel(logging.INFO) # set this to whatever you'd like
```
Globals for the base url of the web api and a file to store your API key.
```
BASE_URL = 'http://api.open.fec.gov/v1'
API_KEY = open(os.path.expanduser('~/.api-keys/data.gov'),'r').read().strip()
```
Helper functions from Bob's original notebook, no changes.
```
def all_results(endpoint, params):
_params = deepcopy(params)
_params.update({'api_key': API_KEY})
_url = BASE_URL+endpoint
logging.info('querying endpoint: {}'.format(_url))
initial_resp = requests.get(_url, params=_params)
logging.debug('full url eg: {}'.format(initial_resp.url))
initial_data = initial_resp.json()
num_pages = initial_data['pagination']['pages']
num_records = initial_data['pagination']['count']
logging.info('{p} pages to be retrieved, with {n} records'.format(
p=num_pages, n=num_records))
current_page = initial_data['pagination']['page']
logging.debug('page {} retrieved'.format(current_page))
for record in initial_data['results']:
yield record
while current_page < num_pages:
current_page += 1
_params.update({'page': current_page})
_data = requests.get(_url, params=_params).json()
logging.debug('page {} retrieved'.format(current_page))
for record in _data['results']:
yield record
logging.info('all pages retrieved')
def count_results(endpoint, params):
_params = deepcopy(params)
_params.update({'api_key': API_KEY})
_url = BASE_URL+endpoint
_data = requests.get(_url, params=_params).json()
return _data['pagination']['count']
```
First let'sfind the Microsoft PAC using the API
```
q_committee = {
'q': 'microsoft',
}
[r for r in all_results('/committees/', q_committee)]
```
Pretty clear one of those is not Microsoft... so let's get the record for the one that is.
```
q_committee = {
'committee_id': 'C00227546',
}
[r for r in all_results('/committee/C00227546/', q_committee)]
```
Let's try getting some history data for 2018
```
count_results('/committee/C00227546/history/',{'cycle':2018})
[r for r in all_results('/committee/C00227546/history/',{'cycle':2018})]
```
Not to interesting...
At this point I pivoted to start looking at data from within the exported csv, first to figure out how to query committee data.
```
[r for r in all_results('/committee/C00008664/', {})]
```
Lot of stuff in there, but I just want a few fields.
```
committee = [r for r in all_results('/committee/C00403592/', {})]
committee_df = pd.DataFrame(committee)
committee_df.head()
```
And to get just specific fields...
```
committee_df[['name','state', 'party_full', 'party_type_full', 'designation_full', 'committee_type_full', 'website']]
```
The trouble with that is I really just want the value to put into a new CSV to merge with the original one.
There is a reason for this if statement. In working through the export to csv I hit a problem where some of the values come back with the text "None". Python interprets that as the None type. I don't really know Python so it took some finagling (and begging for help on Twitter) to get the syntax right on this check.
```
if committee_df.iloc[0]['party_full'] is not None:
print(committee_df.iloc[0]['party_full'] )
else:
print('yes')
```
Now on to getting candidate data
```
candidate = [r for r in all_results('/candidate/H0CA27085/', {})]
candidate_df = pd.DataFrame(candidate)
candidate_df.head()
candidate_df[['name', 'office', 'state', 'party_full']]
party = candidate_df.iloc[0]['party_full']
print(party)
```
On to building a csv with the data I want. I first prepped a file of just unique committee and candidate ids using Excel.
The code is pretty straightforward, now that it works. I worked through this by figuring out how toloop over my existing csv and output to a new one first. Then I went back and worked the API calls in. There was a lot of fussing with this and I know it isn't optimal. I'm sure I'll learn better ways later.
```
import csv
csv_file = open("mspac_with_party.csv", 'w', newline='')
writer = csv.writer(csv_file, escapechar=' ', quoting=csv.QUOTE_NONE)
writer.writerow(['recipient_committee_id', 'recipient_name', 'recipient_state', 'recipient_city', 'recipient_zip', \
'entity_type', 'entity_type_desc', 'election_type', 'fec_election_type_desc', 'fec_election_year', \
'committee_party_full', 'party_type_full', 'designation_full', 'committee_type_full', \
'candidate_party_full', 'candidate_id', 'candidate_name', 'candidate_office_description', 'candidate_office_district'])
current_line = []
with open('mspac-full.csv') as csvfile:
reader = csv.DictReader(csvfile)
for row in reader:
current_line = row['recipient_committee_id'] + ',' + row['recipient_name'].replace(',', '') + ',' + row['recipient_state'] + ',' \
+ row['recipient_city'] + ',' + row['recipient_zip'] + ',' \
+ row['entity_type'] + ',' + row['entity_type_desc'] + ',' \
+ row['election_type'] + ',' + row['fec_election_type_desc'] + ',' + row['fec_election_year']
# committee info
endpoint = '/committee/{c}'.format(c=row['recipient_committee_id'])
committee = all_results(endpoint, {})
committee_df = pd.DataFrame(committee)
if committee_df.iloc[0]['party_full'] is not None:
current_line = current_line + ',' + committee_df.iloc[0]['party_full']
else:
current_line = current_line + ','
if committee_df.iloc[0]['party_type_full'] is not None:
current_line = current_line + ',' + committee_df.iloc[0]['party_type_full']
else:
current_line = current_line + ','
if committee_df.iloc[0]['designation_full'] is not None:
current_line = current_line + ',' + committee_df.iloc[0]['designation_full']
else:
current_line = current_line + ','
if committee_df.iloc[0]['committee_type_full'] is not None:
current_line = current_line + ',' + committee_df.iloc[0]['committee_type_full']
else:
current_line = current_line + ','
#when there is a candidate get info
if row['candidate_id'] is not '':
c_endpoint = '/candidate/{c}'.format(c=row['candidate_id'])
candidate = all_results(c_endpoint, {})
candidate_df = pd.DataFrame(candidate)
if candidate_df.iloc[0]['party_full'] is not None:
current_line = current_line + ',' + candidate_df.iloc[0]['party_full']
else:
current_line = current_line + ','
current_line = current_line + ',' + row['candidate_id'] + ',' + row['candidate_name'].replace(',', '') + ',' \
+ row['candidate_office_description'] + ',' + row['candidate_office_district']
writer.writerow([current_line])
csv_file.close()
print('Done')
```
## Outcome
This got me most of the way there. I pulled this data back into Excel in a new table then created a data model to use pivot tables to analyze the spending data with political parties included. Some of the PACs did not have a party affiliation in the FEC data. I came to my own conclusions based on their spending records. I've made that clear with a column next to the committee party in the worksheet.
I accept that my analysis here is rough, but it seems fair based on what I'm seeing.
So far in the 2018 cycle the Microsoft PAC is giving more money and more contributions to Republicans. This is pretty shocking given how much legal assistance our company is providing to defend our own employees from policies put in place by the Republican party which controls all branches of government in the United States. So, I don't know who this PAC is representing, but it does not seem to represent the interests of Microsoft or its employees very well.
## Next steps
I'd like to go at this again more methodically, probably with bulk data exports. I'd love to get it to the point with any committee id as a starting input it could spit out the full analysis on its own. I think that would be extremely interesting to look at other corporate PACs to get some transparency on them. I suspect many of them, such as the Microsoft PAC, don't represent what the public face of the company is.
I'd also like to algorithmically determine the political orientation of a PAC by analyzing its spending. It would also be interesting to look at effectiveness of a PAC. Some of the unaffiliated ones I looked up on [OpenSecrets](https://www.opensecrets.org/) were shocking in how little they spent on campaigns, it was almost all fundraising and administrative overhead. It would be interesting to know if your corporate PAC was giving to such groups wouldn't it?
I'd also like to look at contributions to the PAC and cross check the political orientation of the contributors. That would be interesting to see if the PAC is at least representing its contributors even if it may be doing a poor job of representing its company affiliation. Which would be a good reason for these things to drop the corporate name they bear since that isn't who they really represent.
|
github_jupyter
|
%pylab inline
import matplotlib.pyplot as plt
matplotlib.style.use('ggplot')
import numpy as np
import pandas as pd
import requests
import os
import json
from copy import deepcopy
import logging
logging.basicConfig(format='%(asctime)s : %(levelname)s : %(message)s')
logging.getLogger("requests").setLevel(logging.ERROR) # silencing requests logging
# Logging for this notebook
logger = logging.getLogger()
logger.setLevel(logging.INFO) # set this to whatever you'd like
BASE_URL = 'http://api.open.fec.gov/v1'
API_KEY = open(os.path.expanduser('~/.api-keys/data.gov'),'r').read().strip()
def all_results(endpoint, params):
_params = deepcopy(params)
_params.update({'api_key': API_KEY})
_url = BASE_URL+endpoint
logging.info('querying endpoint: {}'.format(_url))
initial_resp = requests.get(_url, params=_params)
logging.debug('full url eg: {}'.format(initial_resp.url))
initial_data = initial_resp.json()
num_pages = initial_data['pagination']['pages']
num_records = initial_data['pagination']['count']
logging.info('{p} pages to be retrieved, with {n} records'.format(
p=num_pages, n=num_records))
current_page = initial_data['pagination']['page']
logging.debug('page {} retrieved'.format(current_page))
for record in initial_data['results']:
yield record
while current_page < num_pages:
current_page += 1
_params.update({'page': current_page})
_data = requests.get(_url, params=_params).json()
logging.debug('page {} retrieved'.format(current_page))
for record in _data['results']:
yield record
logging.info('all pages retrieved')
def count_results(endpoint, params):
_params = deepcopy(params)
_params.update({'api_key': API_KEY})
_url = BASE_URL+endpoint
_data = requests.get(_url, params=_params).json()
return _data['pagination']['count']
q_committee = {
'q': 'microsoft',
}
[r for r in all_results('/committees/', q_committee)]
q_committee = {
'committee_id': 'C00227546',
}
[r for r in all_results('/committee/C00227546/', q_committee)]
count_results('/committee/C00227546/history/',{'cycle':2018})
[r for r in all_results('/committee/C00227546/history/',{'cycle':2018})]
[r for r in all_results('/committee/C00008664/', {})]
committee = [r for r in all_results('/committee/C00403592/', {})]
committee_df = pd.DataFrame(committee)
committee_df.head()
committee_df[['name','state', 'party_full', 'party_type_full', 'designation_full', 'committee_type_full', 'website']]
if committee_df.iloc[0]['party_full'] is not None:
print(committee_df.iloc[0]['party_full'] )
else:
print('yes')
candidate = [r for r in all_results('/candidate/H0CA27085/', {})]
candidate_df = pd.DataFrame(candidate)
candidate_df.head()
candidate_df[['name', 'office', 'state', 'party_full']]
party = candidate_df.iloc[0]['party_full']
print(party)
import csv
csv_file = open("mspac_with_party.csv", 'w', newline='')
writer = csv.writer(csv_file, escapechar=' ', quoting=csv.QUOTE_NONE)
writer.writerow(['recipient_committee_id', 'recipient_name', 'recipient_state', 'recipient_city', 'recipient_zip', \
'entity_type', 'entity_type_desc', 'election_type', 'fec_election_type_desc', 'fec_election_year', \
'committee_party_full', 'party_type_full', 'designation_full', 'committee_type_full', \
'candidate_party_full', 'candidate_id', 'candidate_name', 'candidate_office_description', 'candidate_office_district'])
current_line = []
with open('mspac-full.csv') as csvfile:
reader = csv.DictReader(csvfile)
for row in reader:
current_line = row['recipient_committee_id'] + ',' + row['recipient_name'].replace(',', '') + ',' + row['recipient_state'] + ',' \
+ row['recipient_city'] + ',' + row['recipient_zip'] + ',' \
+ row['entity_type'] + ',' + row['entity_type_desc'] + ',' \
+ row['election_type'] + ',' + row['fec_election_type_desc'] + ',' + row['fec_election_year']
# committee info
endpoint = '/committee/{c}'.format(c=row['recipient_committee_id'])
committee = all_results(endpoint, {})
committee_df = pd.DataFrame(committee)
if committee_df.iloc[0]['party_full'] is not None:
current_line = current_line + ',' + committee_df.iloc[0]['party_full']
else:
current_line = current_line + ','
if committee_df.iloc[0]['party_type_full'] is not None:
current_line = current_line + ',' + committee_df.iloc[0]['party_type_full']
else:
current_line = current_line + ','
if committee_df.iloc[0]['designation_full'] is not None:
current_line = current_line + ',' + committee_df.iloc[0]['designation_full']
else:
current_line = current_line + ','
if committee_df.iloc[0]['committee_type_full'] is not None:
current_line = current_line + ',' + committee_df.iloc[0]['committee_type_full']
else:
current_line = current_line + ','
#when there is a candidate get info
if row['candidate_id'] is not '':
c_endpoint = '/candidate/{c}'.format(c=row['candidate_id'])
candidate = all_results(c_endpoint, {})
candidate_df = pd.DataFrame(candidate)
if candidate_df.iloc[0]['party_full'] is not None:
current_line = current_line + ',' + candidate_df.iloc[0]['party_full']
else:
current_line = current_line + ','
current_line = current_line + ',' + row['candidate_id'] + ',' + row['candidate_name'].replace(',', '') + ',' \
+ row['candidate_office_description'] + ',' + row['candidate_office_district']
writer.writerow([current_line])
csv_file.close()
print('Done')
| 0.195786 | 0.832985 |
# VacationPy
----
#### Note
* Keep an eye on your API usage. Use https://developers.google.com/maps/reporting/gmp-reporting as reference for how to monitor your usage and billing.
* Instructions have been included for each segment. You do not have to follow them exactly, but they are included to help you think through the steps.
```
# Dependencies and Setup
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import requests
import gmaps
import os
# Import API key
from api_keys import g_key
```
### Store Part I results into DataFrame
* Load the csv exported in Part I to a DataFrame
```
csvpath = '../WeatherPy/output_data/cities.csv'
cities_df = pd.read_csv(csvpath)
cities_df['Max Temp'] = cities_df['Max Temp']*9/5 -459.67
cities_df.head(10)
```
### Humidity Heatmap
* Configure gmaps.
* Use the Lat and Lng as locations and Humidity as the weight.
* Add Heatmap layer to map.
```
# Access maps with unique API key
gmaps.configure(api_key=g_key)
# Create a list containing coordinates
locations = cities_df[["Lat", "Lon"]]
# Fill NaN values and convert to float
rating = cities_df["Humidity"].astype(float)
#maximum intensity
H_max = max(cities_df['Humidity'])
print(H_max)
# Plot Heatmap
fig = gmaps.figure()
# Create heat layer
heat_layer = gmaps.heatmap_layer(locations, weights=rating,
dissipating=False, max_intensity=H_max,
point_radius=2)
# Add layer
fig.add_layer(heat_layer)
# Display figure
fig
```
### Create new DataFrame fitting weather criteria
* Narrow down the cities to fit weather conditions.
* Drop any rows will null values.
```
#Narrow down the DataFrame to find your ideal weather condition. For example:
# A max temperature lower than 80 degrees but higher than 70.
# Wind speed less than 10 mph.
# Zero cloudiness.
# Drop any rows that don't contain all three conditions. You want to be sure the weather is ideal.
T = 'Max Temp'
WS = 'Wind Speed'
C = 'Cloudiness'
H = 'Humidity'
hotel_df = cities_df.loc[(cities_df[T] >= 70) & (cities_df[T] <= 80) & (cities_df[WS]< 10) & (cities_df[C] == 0) & (cities_df[H] < 70)].dropna().reset_index(drop=True)
hotel_df
```
### Hotel Map
* Store into variable named `hotel_df`.
* Add a "Hotel Name" column to the DataFrame.
* Set parameters to search for hotels with 5000 meters.
* Hit the Google Places API for each city's coordinates.
* Store the first Hotel result into the DataFrame.
* Plot markers on top of the heatmap.
```
target_type = "Lodging"
radius = 5000
base_url = "https://maps.googleapis.com/maps/api/place/nearbysearch/json"
coords = [str(hotel_df['Lat'][x])+','+str(hotel_df['Lon'][x]) for x in range(len(hotel_df['Lat']))]
params = {
"location": coords,
"types": target_type,
"radius": radius,
"key": g_key
}
responses = []
for x in range(len(coords)):
params["location"] = coords[x]
# rewrite params dict
# Run request
response = requests.get(base_url, params)
responses.append(response)
#converting the resposenses to json format
respo = [responses[x].json() for x in range(len(responses))]
#populating hotels list
hotels = []
for x in range(len(responses)):
try:
hotels += [respo[x]['results'][0]['name'] ]
except:
hotels += [pd.NaT]
#adding hotels as a new column to the dataframe and dropping NA
hotel_df['Hotel Name'] = hotels
hotel_df = hotel_df.dropna().reset_index(drop=True)
hotel_df
# NOTE: Do not change any of the code in this cell
# Using the template add the hotel marks to the heatmap
info_box_template = """
<dl>
<dt>Name</dt><dd>{Hotel Name}</dd>
<dt>City</dt><dd>{City}</dd>
<dt>Country</dt><dd>{Country}</dd>
</dl>
"""
# Store the DataFrame Row
# NOTE: be sure to update with your DataFrame name
hotel_info = [info_box_template.format(**row) for index, row in hotel_df.iterrows()]
locations = hotel_df[["Lat", "Lon"]]
# Add marker layer ontop of heat map
markers = gmaps.marker_layer(locations, info_box_content=hotel_info)
fig.add_layer(markers)
# Display figure
fig
```
|
github_jupyter
|
# Dependencies and Setup
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import requests
import gmaps
import os
# Import API key
from api_keys import g_key
csvpath = '../WeatherPy/output_data/cities.csv'
cities_df = pd.read_csv(csvpath)
cities_df['Max Temp'] = cities_df['Max Temp']*9/5 -459.67
cities_df.head(10)
# Access maps with unique API key
gmaps.configure(api_key=g_key)
# Create a list containing coordinates
locations = cities_df[["Lat", "Lon"]]
# Fill NaN values and convert to float
rating = cities_df["Humidity"].astype(float)
#maximum intensity
H_max = max(cities_df['Humidity'])
print(H_max)
# Plot Heatmap
fig = gmaps.figure()
# Create heat layer
heat_layer = gmaps.heatmap_layer(locations, weights=rating,
dissipating=False, max_intensity=H_max,
point_radius=2)
# Add layer
fig.add_layer(heat_layer)
# Display figure
fig
#Narrow down the DataFrame to find your ideal weather condition. For example:
# A max temperature lower than 80 degrees but higher than 70.
# Wind speed less than 10 mph.
# Zero cloudiness.
# Drop any rows that don't contain all three conditions. You want to be sure the weather is ideal.
T = 'Max Temp'
WS = 'Wind Speed'
C = 'Cloudiness'
H = 'Humidity'
hotel_df = cities_df.loc[(cities_df[T] >= 70) & (cities_df[T] <= 80) & (cities_df[WS]< 10) & (cities_df[C] == 0) & (cities_df[H] < 70)].dropna().reset_index(drop=True)
hotel_df
target_type = "Lodging"
radius = 5000
base_url = "https://maps.googleapis.com/maps/api/place/nearbysearch/json"
coords = [str(hotel_df['Lat'][x])+','+str(hotel_df['Lon'][x]) for x in range(len(hotel_df['Lat']))]
params = {
"location": coords,
"types": target_type,
"radius": radius,
"key": g_key
}
responses = []
for x in range(len(coords)):
params["location"] = coords[x]
# rewrite params dict
# Run request
response = requests.get(base_url, params)
responses.append(response)
#converting the resposenses to json format
respo = [responses[x].json() for x in range(len(responses))]
#populating hotels list
hotels = []
for x in range(len(responses)):
try:
hotels += [respo[x]['results'][0]['name'] ]
except:
hotels += [pd.NaT]
#adding hotels as a new column to the dataframe and dropping NA
hotel_df['Hotel Name'] = hotels
hotel_df = hotel_df.dropna().reset_index(drop=True)
hotel_df
# NOTE: Do not change any of the code in this cell
# Using the template add the hotel marks to the heatmap
info_box_template = """
<dl>
<dt>Name</dt><dd>{Hotel Name}</dd>
<dt>City</dt><dd>{City}</dd>
<dt>Country</dt><dd>{Country}</dd>
</dl>
"""
# Store the DataFrame Row
# NOTE: be sure to update with your DataFrame name
hotel_info = [info_box_template.format(**row) for index, row in hotel_df.iterrows()]
locations = hotel_df[["Lat", "Lon"]]
# Add marker layer ontop of heat map
markers = gmaps.marker_layer(locations, info_box_content=hotel_info)
fig.add_layer(markers)
# Display figure
fig
| 0.677581 | 0.844216 |
```
%matplotlib inline
import datetime as dt
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from sklearn.metrics import mean_squared_error
from alphamind.api import *
from PyFin.api import *
plt.style.use('ggplot')
engine = SqlEngine('postgres+psycopg2://postgres:[email protected]/alpha')
u_name = 'zz500'
benchmark = 905
universe = Universe(u_name, [u_name])
factor_coverage = engine.fetch_factor_coverage()
flitered_coverage = factor_coverage[((factor_coverage.source == 'uqer'))
& (factor_coverage.universe == u_name)
& (factor_coverage.trade_date >= '2012-01-01')]
coverage_report = flitered_coverage.groupby(['factor'])['coverage'].mean()
alpha_factors = {
'eps': LAST('eps_q'),
'roe': LAST('roe_q'),
'bdto': LAST('BDTO'),
'cfinc1': LAST('CFinc1'),
'chv': LAST('CHV'),
'rvol': LAST('RVOL'),
'val': LAST('VAL'),
'grev': LAST('GREV'),
#'droeafternonorecurring': LAST('DROEAfterNonRecurring')
}
alpha_factors.__len__()
frequency = '2w'
batch = 8
start_date = '2012-01-01'
end_date = '2017-11-02'
method = 'risk_neutral'
portfolio_risk_neutralize = ['SIZE']
neutralize_risk = industry_styles + portfolio_risk_neutralize
industry_lower = 1.
industry_upper = 1.
data_package = fetch_data_package(engine,
alpha_factors=alpha_factors,
start_date=start_date,
end_date=end_date,
frequency=frequency,
universe=universe,
benchmark=benchmark,
batch=batch,
neutralized_risk=neutralize_risk,
pre_process=[winsorize_normal],
post_process=[winsorize_normal],
warm_start=batch)
train_x = data_package['train']['x']
train_y = data_package['train']['y']
predict_x = data_package['predict']['x']
predict_y = data_package['predict']['y']
features = data_package['x_names']
```
## 0. Train Score on a specific date
------------------------------------
```
ref_date = list(train_x.keys())[-2]
sample_train_x = train_x[ref_date]
sample_train_y = train_y[ref_date].flatten()
sample_test_x = predict_x[ref_date]
sample_test_y = predict_y[ref_date].flatten()
model = LinearRegression(features, fit_intercept=False)
model.fit(sample_train_x, sample_train_y)
model.impl.score(sample_train_x, sample_train_y)
model.impl.score(sample_test_x, sample_test_y)
```
## 1. Train and test accuracy trend (Linear Regression)
----------
```
dates = sorted(train_x.keys())
accuray_table = pd.DataFrame(columns=['train', 'predict'])
model_df = pd.Series()
for ref_date in dates:
sample_train_x = train_x[ref_date]
sample_train_y = train_y[ref_date].flatten()
model = LinearRegression(features, fit_intercept=False)
model.fit(sample_train_x, sample_train_y)
accuray_table.loc[ref_date, 'train'] = mean_squared_error(model.predict(sample_train_x), sample_train_y)
model_df.loc[ref_date] = model
alpha_logger.info('trade_date: {0} training finished'.format(ref_date))
portfolio_industry_neutralize = True
settlement = data_package['settlement']
industry_dummies = pd.get_dummies(settlement['industry'].values)
risk_styles = settlement[portfolio_risk_neutralize].values
final_res = np.zeros(len(dates))
method = 'risk_neutral'
for i, ref_date in enumerate(dates):
model = model_df[ref_date]
sample_test_x = predict_x[ref_date]
sample_test_y = predict_y[ref_date].flatten()
cons = Constraints()
index = settlement.trade_date == ref_date
benchmark_w = settlement[index]['weight'].values
realized_r = settlement[index]['dx'].values
industry_names = settlement[index]['industry'].values
is_tradable = settlement[index]['isOpen'].values
cons.add_exposure(['total'], np.ones((len(is_tradable), 1)))
cons.set_constraints('total', benchmark_w.sum(), benchmark_w.sum())
if portfolio_industry_neutralize:
ind_exp = industry_dummies[index]
risk_tags = ind_exp.columns
cons.add_exposure(risk_tags, ind_exp.values)
benchmark_exp = benchmark_w @ ind_exp.values
for k, name in enumerate(risk_tags):
cons.set_constraints(name, benchmark_exp[k]*industry_lower, benchmark_exp[k]*industry_upper)
if portfolio_risk_neutralize:
risk_exp = risk_styles[index]
risk_tags = np.array(portfolio_risk_neutralize)
cons.add_exposure(risk_tags, risk_exp)
benchmark_exp = benchmark_w @ risk_exp
for k, name in enumerate(risk_tags):
cons.set_constraints(name, benchmark_exp[k], benchmark_exp[k])
y = model.predict(sample_test_x)
accuray_table.loc[ref_date, 'predict'] = mean_squared_error(y, sample_test_y)
is_tradable[:] = True
weights, analysis = er_portfolio_analysis(y,
industry_names,
realized_r,
constraints=cons,
detail_analysis=True,
benchmark=benchmark_w,
is_tradable=is_tradable,
method=method)
final_res[i] = analysis['er']['total'] / benchmark_w.sum()
alpha_logger.info('trade_date: {0} predicting finished'.format(ref_date))
last_date = advanceDateByCalendar('china.sse', dates[-1], frequency)
df = pd.Series(final_res, index=dates[1:] + [last_date])
df.sort_index(inplace=True)
df['2012-01-01':].cumsum().plot(figsize=(12, 6))
plt.title('Prod factors model {1} \n ({0}, eps_q & roe_q, {2} - {3})'.format(method, model.__class__.__name__, start_date, end_date))
accuray_table.plot(figsize=(12, 6))
accuray_table.aggregate([np.mean, np.median, np.std])
```
|
github_jupyter
|
%matplotlib inline
import datetime as dt
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from sklearn.metrics import mean_squared_error
from alphamind.api import *
from PyFin.api import *
plt.style.use('ggplot')
engine = SqlEngine('postgres+psycopg2://postgres:[email protected]/alpha')
u_name = 'zz500'
benchmark = 905
universe = Universe(u_name, [u_name])
factor_coverage = engine.fetch_factor_coverage()
flitered_coverage = factor_coverage[((factor_coverage.source == 'uqer'))
& (factor_coverage.universe == u_name)
& (factor_coverage.trade_date >= '2012-01-01')]
coverage_report = flitered_coverage.groupby(['factor'])['coverage'].mean()
alpha_factors = {
'eps': LAST('eps_q'),
'roe': LAST('roe_q'),
'bdto': LAST('BDTO'),
'cfinc1': LAST('CFinc1'),
'chv': LAST('CHV'),
'rvol': LAST('RVOL'),
'val': LAST('VAL'),
'grev': LAST('GREV'),
#'droeafternonorecurring': LAST('DROEAfterNonRecurring')
}
alpha_factors.__len__()
frequency = '2w'
batch = 8
start_date = '2012-01-01'
end_date = '2017-11-02'
method = 'risk_neutral'
portfolio_risk_neutralize = ['SIZE']
neutralize_risk = industry_styles + portfolio_risk_neutralize
industry_lower = 1.
industry_upper = 1.
data_package = fetch_data_package(engine,
alpha_factors=alpha_factors,
start_date=start_date,
end_date=end_date,
frequency=frequency,
universe=universe,
benchmark=benchmark,
batch=batch,
neutralized_risk=neutralize_risk,
pre_process=[winsorize_normal],
post_process=[winsorize_normal],
warm_start=batch)
train_x = data_package['train']['x']
train_y = data_package['train']['y']
predict_x = data_package['predict']['x']
predict_y = data_package['predict']['y']
features = data_package['x_names']
ref_date = list(train_x.keys())[-2]
sample_train_x = train_x[ref_date]
sample_train_y = train_y[ref_date].flatten()
sample_test_x = predict_x[ref_date]
sample_test_y = predict_y[ref_date].flatten()
model = LinearRegression(features, fit_intercept=False)
model.fit(sample_train_x, sample_train_y)
model.impl.score(sample_train_x, sample_train_y)
model.impl.score(sample_test_x, sample_test_y)
dates = sorted(train_x.keys())
accuray_table = pd.DataFrame(columns=['train', 'predict'])
model_df = pd.Series()
for ref_date in dates:
sample_train_x = train_x[ref_date]
sample_train_y = train_y[ref_date].flatten()
model = LinearRegression(features, fit_intercept=False)
model.fit(sample_train_x, sample_train_y)
accuray_table.loc[ref_date, 'train'] = mean_squared_error(model.predict(sample_train_x), sample_train_y)
model_df.loc[ref_date] = model
alpha_logger.info('trade_date: {0} training finished'.format(ref_date))
portfolio_industry_neutralize = True
settlement = data_package['settlement']
industry_dummies = pd.get_dummies(settlement['industry'].values)
risk_styles = settlement[portfolio_risk_neutralize].values
final_res = np.zeros(len(dates))
method = 'risk_neutral'
for i, ref_date in enumerate(dates):
model = model_df[ref_date]
sample_test_x = predict_x[ref_date]
sample_test_y = predict_y[ref_date].flatten()
cons = Constraints()
index = settlement.trade_date == ref_date
benchmark_w = settlement[index]['weight'].values
realized_r = settlement[index]['dx'].values
industry_names = settlement[index]['industry'].values
is_tradable = settlement[index]['isOpen'].values
cons.add_exposure(['total'], np.ones((len(is_tradable), 1)))
cons.set_constraints('total', benchmark_w.sum(), benchmark_w.sum())
if portfolio_industry_neutralize:
ind_exp = industry_dummies[index]
risk_tags = ind_exp.columns
cons.add_exposure(risk_tags, ind_exp.values)
benchmark_exp = benchmark_w @ ind_exp.values
for k, name in enumerate(risk_tags):
cons.set_constraints(name, benchmark_exp[k]*industry_lower, benchmark_exp[k]*industry_upper)
if portfolio_risk_neutralize:
risk_exp = risk_styles[index]
risk_tags = np.array(portfolio_risk_neutralize)
cons.add_exposure(risk_tags, risk_exp)
benchmark_exp = benchmark_w @ risk_exp
for k, name in enumerate(risk_tags):
cons.set_constraints(name, benchmark_exp[k], benchmark_exp[k])
y = model.predict(sample_test_x)
accuray_table.loc[ref_date, 'predict'] = mean_squared_error(y, sample_test_y)
is_tradable[:] = True
weights, analysis = er_portfolio_analysis(y,
industry_names,
realized_r,
constraints=cons,
detail_analysis=True,
benchmark=benchmark_w,
is_tradable=is_tradable,
method=method)
final_res[i] = analysis['er']['total'] / benchmark_w.sum()
alpha_logger.info('trade_date: {0} predicting finished'.format(ref_date))
last_date = advanceDateByCalendar('china.sse', dates[-1], frequency)
df = pd.Series(final_res, index=dates[1:] + [last_date])
df.sort_index(inplace=True)
df['2012-01-01':].cumsum().plot(figsize=(12, 6))
plt.title('Prod factors model {1} \n ({0}, eps_q & roe_q, {2} - {3})'.format(method, model.__class__.__name__, start_date, end_date))
accuray_table.plot(figsize=(12, 6))
accuray_table.aggregate([np.mean, np.median, np.std])
| 0.5144 | 0.600423 |
# Iris species classification.
**Problem statement** -
Given _Sepal Length_ ,_Sepal width_, _Petal length_ and _Petal width_ classify
each instance into one of **Iris-setosa, Iris-versicolor or Iris-virginica** species using Machine learning Classification algorithms.
## Importing the required libraries.
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn import metrics
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score, precision_score, f1_score, confusion_matrix, classification_report
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.linear_model import LogisticRegression
sns.set()
%matplotlib inline
data = pd.read_csv('Iris.csv')
data.shape
data.head()
data.tail()
# Finding all the columns
data.columns
# Checking range of index
data.index
# Checking for any null entries in the dataset.
data[data.isnull().any(axis = 1)].shape
data.info()
data.describe()
# since Id column is not very useful to us we will drop it.
data.drop('Id', axis = 1, inplace = True)
# Checking if the Id column is successfully dropped or not.
data.columns
data.head(3)
# Lets check all the unique species of the flower
data['Species'].unique()
# Now lets check their count in the dataset.
data['Species'].value_counts()
```
## Doing some Exploratory data analysis for understanding the data better.
```
sns.distplot(data['SepalLengthCm'])
sns.distplot(data['SepalWidthCm'])
sns.distplot(data['PetalLengthCm'])
sns.distplot(data['PetalWidthCm'])
plt.figure(figsize = (10,10))
plt.title("Distribution of Sepal Length of all 3 species.", fontsize = 15)
sns.boxplot(x = 'Species', y = 'SepalLengthCm', data = data)
plt.figure(figsize = (10,10))
plt.title("Distribution of Sepal Width of all 3 species.", fontsize = 15)
sns.boxplot(x = 'Species', y = 'SepalWidthCm', data = data)
plt.figure(figsize = (10,10))
plt.title("Distribution of Petal Length of all 3 species.", fontsize = 15)
sns.boxplot(x = 'Species', y = 'PetalLengthCm', data = data)
plt.figure(figsize = (10,10))
plt.title("Distribution of Petal Width of all 3 species.", fontsize = 15)
sns.boxplot(x = 'Species', y = 'PetalWidthCm', data = data)
```
## Finding correlations.
Correlation is basically association/relation between two quantitative variables(here our features). A correlation coefficient can vary from -1 to +1. Where :
* -1 = Two variables(or features) are purely negatively correlated.
* 0 = Two variables are independent of each other.
* +1 = Two variables are purely positively correlated.
```
data.corr()
sns.heatmap(data.corr(), annot = True, cmap = 'coolwarm')
plt.title("Correlation between all the columns ", fontsize = 12)
feature_set = ['SepalLengthCm', 'SepalWidthCm', 'PetalLengthCm', 'PetalWidthCm']
X = data[feature_set].values
# Since species contains categorical features we will map the species category to 0,1 and 2 respectively.
y = data['Species'].map({'Iris-setosa': 0 , 'Iris-versicolor' :1 ,'Iris-virginica' : 2 }).values
X.shape, y.shape
type(X), type(y)
```
## Building the model.
## 1. Logistic Regression
It's a Linear model which predicts the probabilty that a certain instance will belong to which category.
Based on the probablity we decide the target class to which an instance will belong to.
For ex- The probability of a certain instance to be a spam message is 0.85 then it will belong to the Spam category, else if the probability is somewhat 0.40 then it won't.
Note - The rounding off of probability to binary values will depend on use case and context.

```
# Splitting the dataset into the test and train part.
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.4, random_state = 12)
print(X_train.shape)
print(X_test.shape)
print(y_train.shape)
print(y_test.shape)
# Creating a classifier object and fitting the training dataset.
clf = LogisticRegression()
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
type(y_pred)
cm = confusion_matrix(y_test, y_pred)
# converting the confusion matrix into a dataframe
cm_df = pd.DataFrame(cm,
index = ['setosa','versicolor','virginica'],
columns = ['setosa','versicolor','virginica'])
sns.heatmap(cm_df, annot = True)
plt.xlabel('True Label')
plt.ylabel('Predicted Label')
# Finding accuracy of our model
acc = accuracy_score(y_test, y_pred)
acc*100
```
*So the accuracy of our model using Logistic regression is = 95%*
## 2. K Nearest Neighbor
In this algorithm a new data point will belong to an existing class depending on where it's *K nearest* data points belong to. For ex - If we take K=3 here, the new data point(a star here) will belong to the class Red Circle(RC) since all the 3 nearest data points belong to the same class.
> Note that the default value of K in KNN is always taken as 5.

Steps involved in K nearest neighbour algorithm :
* Finding Euclidean or Manhattan distance between the new data point and the rest of the data points and sorting them in ascennding order.
* Finding the class of K nearest data points.
* Get the most frequent class in the K neighbors.
* Predict the class of new data point.
**To choose an optimal value of K we use elbow method i.e we plot few values of K with their error rates and then choose the one with least error rate.**

```
score = []
for n in range(1,26):
clf = KNeighborsClassifier(n_neighbors=n)
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
score.append(accuracy_score(y_test, y_pred))
plt.figure(figsize = (12,10))
plt.plot(range(1,26),score)
plt.title("Accuracy score based on different values of n")
plt.xlabel("Values of n")
plt.ylabel("Accuracy score ")
```
**The accuracy with n = 8,9 and 10 comes out to be 100% !**
```
clf = KNeighborsClassifier(n_neighbors=9)
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
acc = accuracy_score(y_test, y_pred)
acc*100
cm = confusion_matrix(y_test, y_pred)
# converting the confusion matrix into a dataframe
cm_df = pd.DataFrame(cm,
index = ['setosa','versicolor','virginica'],
columns = ['setosa','versicolor','virginica'])
sns.heatmap(cm_df, annot = True)
plt.xlabel('True Label')
plt.ylabel('Predicted Label')
```
*So the accuracy of our model using KNN is = 100%*
## 3. Decision Tree Classifier.
A decision tree classifier is a supervised classification technique and can be used in both the cases where the target label is categorical as well as continuous. The main aim of this algorithm is to split the root node(total data points) into smaller homogenous sub-nodes where homogenous sub-nodes are the ones that have all the data points belonging to only one particular target class. So, on what basis the splitting is done? The splitting is done on the basis of various algorithms, some of them are **Gini, Chi-square, Information gain etc**
### Gini
* It does only binary splits.
* Higher the gini value, higher is the homogenity of the sub-node.
### Advantages of a Tree based classidfier.
* Easy to understand.
* Less data cleaning required.
* Data type is not a constraint.
### Disadvantages
* Overfitting
* Not very good for continuous variable.
To read in more detail about the same here is one such article. Click [here](https://www.analyticsvidhya.com/blog/2016/04/complete-tutorial-tree-based-modeling-scratch-in-python/)
```
clf = DecisionTreeClassifier()
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
confusion_matrix(y_test, y_pred)
acc = accuracy_score(y_test, y_pred)
acc*100
```
*So the accuracy obtained from our Decision Tree classifier is = 95%*
## 4. Random forest classifier.
The literary meaning of word ‘ensemble’ is group. Ensemble methods involve group of predictive models to achieve a better accuracy and model stability. Ensemble methods are known to impart supreme boost to tree based models.
```
clf = RandomForestClassifier()
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
confusion_matrix(y_test, y_pred)
acc = accuracy_score(y_test, y_pred)
acc*100
```
*So the accuracy obtained by our Random forest classifier is = 93.333*
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn import metrics
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score, precision_score, f1_score, confusion_matrix, classification_report
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.linear_model import LogisticRegression
sns.set()
%matplotlib inline
data = pd.read_csv('Iris.csv')
data.shape
data.head()
data.tail()
# Finding all the columns
data.columns
# Checking range of index
data.index
# Checking for any null entries in the dataset.
data[data.isnull().any(axis = 1)].shape
data.info()
data.describe()
# since Id column is not very useful to us we will drop it.
data.drop('Id', axis = 1, inplace = True)
# Checking if the Id column is successfully dropped or not.
data.columns
data.head(3)
# Lets check all the unique species of the flower
data['Species'].unique()
# Now lets check their count in the dataset.
data['Species'].value_counts()
sns.distplot(data['SepalLengthCm'])
sns.distplot(data['SepalWidthCm'])
sns.distplot(data['PetalLengthCm'])
sns.distplot(data['PetalWidthCm'])
plt.figure(figsize = (10,10))
plt.title("Distribution of Sepal Length of all 3 species.", fontsize = 15)
sns.boxplot(x = 'Species', y = 'SepalLengthCm', data = data)
plt.figure(figsize = (10,10))
plt.title("Distribution of Sepal Width of all 3 species.", fontsize = 15)
sns.boxplot(x = 'Species', y = 'SepalWidthCm', data = data)
plt.figure(figsize = (10,10))
plt.title("Distribution of Petal Length of all 3 species.", fontsize = 15)
sns.boxplot(x = 'Species', y = 'PetalLengthCm', data = data)
plt.figure(figsize = (10,10))
plt.title("Distribution of Petal Width of all 3 species.", fontsize = 15)
sns.boxplot(x = 'Species', y = 'PetalWidthCm', data = data)
data.corr()
sns.heatmap(data.corr(), annot = True, cmap = 'coolwarm')
plt.title("Correlation between all the columns ", fontsize = 12)
feature_set = ['SepalLengthCm', 'SepalWidthCm', 'PetalLengthCm', 'PetalWidthCm']
X = data[feature_set].values
# Since species contains categorical features we will map the species category to 0,1 and 2 respectively.
y = data['Species'].map({'Iris-setosa': 0 , 'Iris-versicolor' :1 ,'Iris-virginica' : 2 }).values
X.shape, y.shape
type(X), type(y)
# Splitting the dataset into the test and train part.
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.4, random_state = 12)
print(X_train.shape)
print(X_test.shape)
print(y_train.shape)
print(y_test.shape)
# Creating a classifier object and fitting the training dataset.
clf = LogisticRegression()
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
type(y_pred)
cm = confusion_matrix(y_test, y_pred)
# converting the confusion matrix into a dataframe
cm_df = pd.DataFrame(cm,
index = ['setosa','versicolor','virginica'],
columns = ['setosa','versicolor','virginica'])
sns.heatmap(cm_df, annot = True)
plt.xlabel('True Label')
plt.ylabel('Predicted Label')
# Finding accuracy of our model
acc = accuracy_score(y_test, y_pred)
acc*100
score = []
for n in range(1,26):
clf = KNeighborsClassifier(n_neighbors=n)
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
score.append(accuracy_score(y_test, y_pred))
plt.figure(figsize = (12,10))
plt.plot(range(1,26),score)
plt.title("Accuracy score based on different values of n")
plt.xlabel("Values of n")
plt.ylabel("Accuracy score ")
clf = KNeighborsClassifier(n_neighbors=9)
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
acc = accuracy_score(y_test, y_pred)
acc*100
cm = confusion_matrix(y_test, y_pred)
# converting the confusion matrix into a dataframe
cm_df = pd.DataFrame(cm,
index = ['setosa','versicolor','virginica'],
columns = ['setosa','versicolor','virginica'])
sns.heatmap(cm_df, annot = True)
plt.xlabel('True Label')
plt.ylabel('Predicted Label')
clf = DecisionTreeClassifier()
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
confusion_matrix(y_test, y_pred)
acc = accuracy_score(y_test, y_pred)
acc*100
clf = RandomForestClassifier()
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
confusion_matrix(y_test, y_pred)
acc = accuracy_score(y_test, y_pred)
acc*100
| 0.758153 | 0.939526 |
## 实体链指比赛方案分享
### 1. **任务与难点介绍**
面向中文短文本的实体链指,简称 EL(Entity Linking),是NLP、知识图谱领域的基础任务之一,即对于给定的一个中文短文本(如搜索 Query、微博、对话内容、文章/视频/图片的标题等),EL将其中的实体与给定知识库中对应的实体进行关联。
此次任务的输入输出定义如下:
输入:中文短文本以及该短文本中的实体集合。
输出:输出文本此中文短文本的实体链指结果。每个结果包含:实体 mention、在中文短文本中的位置偏移、其在给定知识库中的 id,如果为 NIL 情况,需要再给出实体的上位概念类型。
传统的实体链指任务主要是针对长文档,长文档拥有在写的上下文信息能辅助实体的歧义消解并完成链指。相比之下,针对中文短文本的实体链指存在很大的挑战,主要原因如下:
(1)口语化严重,导致实体歧义消解困难;
(2)短文本上下文语境不丰富,须对上下文语境进行精准理解;
(3)相比英文,中文由于语言自身的特点,在短文本的链指问题上更有挑战。
### 2. **思路介绍**
可以将面向中文短文本的实体链指任务拆分为实体消歧与实体分类两个子任务,然后使用两个模型分别解决。针对实体消歧任务,沿用[1],[2]中的做法,将其转化为句对的二分类任务。针对实体分类任务,采用文本分类算法解决。
### 3. **数据处理方式**
#### 3.1 **候选实体获取**
采用字典匹配方式构建候选实体集合。字典的构造逻辑如下:
a.构建空字典D
b.遍历知识库,对每个mention,将其subject字段和alias字段中出现的实体作为key,subject_id作为value
c.若key已出现在字典D中,将value添加到对应的list中;否则,新建一个空的list作为key在D中的值,然后将value添加进去
按照上述方式得到字典D后,对每个实体,若其出现在字典中,取其在字典D中对应的list作为候选实体;否则,候选实体集合为空。
#### 3.2 **实体链指数据处理**
对知识库中的每个实体,可以借用[1]的做法,将其实体属性拼接得到实体的描述文本。考虑到Type,摘要,义项描述等信息比较重要,本文按照Type,摘要,义项描述,其他信息的顺序进行拼接。其中Type字段在拼接前会先转成中文。
针对实体消歧任务,对短文本中出现的某个实体(本文称之为中心实体),按照[2]的做法,将短文本与中心实体对应的知识库中的实体描述文本作为正样本(标签为1),从候选集合中随机选取与正样本不同的实体描述文本作为负样本(标签为0)。由于各实体对应的候选实体集合大小不一,为了保证正负样本的均衡,可以采用随机负采样的方法来减少负样本的数量。为了充分利用负样本,本文使用了基于动态采样的方法,即每个epoch训练时重新采样负样本。
### 4. **模型设计**
#### 4.1 **实体消歧模型**
本文采用基于预训练语言模型的句对分类算法解决实体消歧任务。具体的,将短文本作为text_a,实体的描述文本作为text_b,将其按照[CLS]text_a[SEP]text_b[SEP]的格式输入到BERT[3]中,然后取[CLS]对应位置的向量作二分类。考虑到每个短文本中可能出现多个实体,在中心实体前后添加特殊符号###以标记中心实体的位置。模型示意图如下:
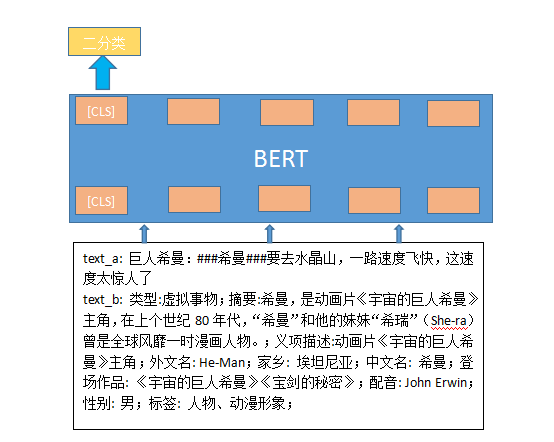
在模型推理阶段,从字典D中取出中心实体的候选集合S(若候选集合S为空时,将中心实体标记为NIL),得到每个候选实体的概率(预测标签为1的概率)。若其中的最大值大于某个阈值(可以作为一个超参数调节),则取概率最大的作为中心实体在知识库中的对应实体,否则将中心实体标记为NIL,然后使用实体分类模型对其进行分类。
#### 4.2 **实体分类模型**
类似地,本文采用基于预训练语言模型的文本分类算法解决实体分类任务。具体来说,将短文本作为text_a,将其按照[CLS]text_a[SEP]的格式输入到BERT中,然后取[CLS]对应位置的向量作多分类。在中心实体前后添加特殊符号###以标记中心实体的位置,这样模型能够利用实体的位置信息。模型示意图如下:
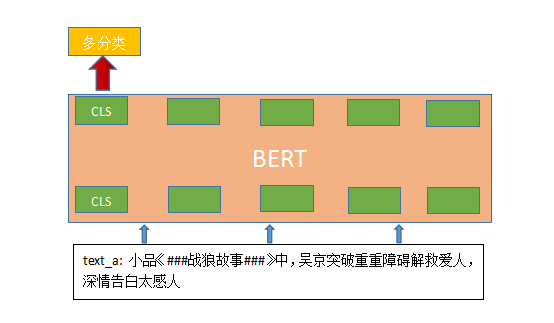
在训练阶段,本文使用中心实体不在知识库中的样本训练;在模型推理时,基于实体消歧模型的输出预测NIL的类别。考虑到训练集中包含大量非NIL实体(类别已知),为了充分利用这些信息,本文采用[1]中的两阶段训练的办法,即先在非NIL实体数据上微调预训练模型,然后在NIL实体数据上继续训练。为了提高模型的泛化能力,尝试了基于FGM[4],PGD[5]的对抗训练策略来训练实体分类模型。
#### 4.3 **模型融合**:
本文使用简单的概率平均办法进行模型融合。对实体消歧模型,本文基于RoBERT-wwm-base[6]和ERNIE-1.0[7]总共训练了7个模型(不同的训练数据,随机种子等)。对实体分类模型,本文基于ERNIE-1.0训练了5个模型(不同的训练数据)。
### 5. **实验与结果分析**
实验基于百度提供的数据,训练集包含7万条数据,验证集包含1万条数据。A榜测试集包含1万条数据,B榜测试数据包含2.5万条数据。本次竞赛的数据从自然网页标题、多模标题、搜索query中抽取得到,通过人工众包标注,知识库实体重复率约5%,实体上位概念准确率95.27%,数据集标注准确率95.32%。该任务的知识库来自百度百科知识库。知识库中的每个实体都包含一个subject_id(知识库id),一个subject名称,实体的别名,对应的概念类型,以及与此实体相关的一系列二元组< predicate,object>(<属性,属性值>)信息形式。知识库中每行代表知识库的一条记录(一个实体信息),每条记录为json数据格式。评估方式采用F1分值。特别注意,在计算评价指标时,对NIL实体的上位概念类型判断结果NIL_Type与实体的关联id等价处理。
实验参数设置如下:对ERNIE-1.0,学习率设置为5e-5,对RoBERT-wwm-base学习率取3e-5,epochs均为3,warmup比例设置为0.1。实体消歧模型采用动态负采样,每个中心实体采样的负样本数设置为2,batch size取64,最大序列长度为256。实体分类模型batch size为64,最大序列长度设置为72。
#### 5.1 **实体消歧模型结果**
下面从负采样的数目,是否使用动态负采样,阈值调整等方面进行实验,结果如下:
| 模型 | 验证集F1 | A榜F1 |
| -------- | -------- | -------- |
| ernie | 0.857 | - |
| ernie + 动态负采样 | 0.871 | 0.878 |
| ernie + 动态负采样 + 负采样2 | 0.872 | 0.880 |
| ernie + 动态负采样 + 负采样3 | 0.875 | 0.876 |
| ernie + 动态负采样 + 负采样2 + 阈值调整 | 0.878 | 0.882 |
从上表第一,第二行的结果可以看出,使用动态负采样优于静态负采样,因为动态负采样能更高效地利用负样本。从第二,第三,第四行的结果可以看出,当负样本个数取2时,模型效果最好。上面的实验结果与[2]中的结果基本一致。对比第三行与第五行,阈值调整能带来一定的性能提升。
#### 5.2 **实体分类模型结果**
| 模型 | 验证集F1 | A榜F1|
| -------- | -------- | -------- |
| 直接训练 | 0.870 | - |
| 两阶段训练 | 0.878 | 0.882 |
| 两阶段训练 + FGM | 0.880 | 0.883 |
| 两阶段训练 + PGD | 0.879 | 0.883 |
从上表可以看出,两阶段训练能够提升模型的性能。在此基础上使用对抗训练也能带来较小的提升。综合考虑下,本文在训练实体消歧模型时使用动态负采样,采样数为2,并且添加了阈值的调整;在训练实体分类时采用两阶段训练加FGM的策略。
### 6. **总结**
本文将面向中文短文本的实体链指任务拆分为实体消歧与实体分类两个子任务。对实体消歧任务,采用基于预训练语言模型的句子对分类算法;对实体分类任务,使用基于预训练语言模型的文本分类模型。此外,本文还使用了一些技巧提升模型的性能,比如:对抗学习,阈值调整,模型融合等。在A榜测试集上取得0.8889的成绩,在B榜测试集上取得0.90981的成绩。本文的方案仍有地方需要改进,当前知识库的使用只是简单的将各属性连接,由于输入长度的限制,在输入到BERT后可能会有信息的丢失,可以设计更好的方案提取最重要的属性。
最后说一下paddle的使用体验,个人感觉还是挺香的。paddle的动态图模式与torch的使用比较相似,熟悉torch的同学基本可以轻松入门。
### 7. **参考文献**
[1]吕荣荣, 王鹏程, 陈帅. 面向中文短文本的多因子融合实体链指研究
https://bj.bcebos.com/v1/conference/ccks2020/eval_paper/ccks2020_eval_paper_2_1.pdf
[2]潘春光, 王胜广, 罗志鹏. 知识增强的实体消歧与实体类别判断
https://bj.bcebos.com/v1/conference/ccks2020/eval_paper/ccks2020_eval_paper_2_2.pdf
[3]Devlin J, Chang M W, Lee K, et al. Bert: Pre-training of deep bidirectional
transformers for language understanding[J]. arXiv preprint arXiv:1810.04805, 2018.
[4]Goodfellow, Ian J, Shlens, Jonathon, and Szegedy, Christian. Explaining and harnessing adversarial examples.International Conference on Learning Representations(ICLR), 2015.
[5]Aleksander Madry, Aleksandar Makelov, Ludwig Schmidt,et al. Towards Deep Learning Models Resistant to Adversarial Attacks. https://arxiv.org/abs/1706.06083
[6]Cui Y, Che W, Liu T, et al. Pre-training with whole word masking for chinese bert[J].
arXiv preprint arXiv:1906.08101, 2019.
[7]Sun Y, Wang S, Li Y, et al. Ernie: Enhanced representation through knowledge
integration[J]. arXiv preprint arXiv:1904.09223, 2019.
### **代码部分**
```
## 环境配置:基于PaddlePaddle 1.8.4开发(python 3.7), 使用单块V100(32G)训练
## 各个文件的作用:
### eval.py 官方提供的评估脚本
### post_matching.py 实体消歧模型后处理,对每个实体选取概率最大的一个作为kb_id(若小于一个阈值,则取NIL)(单模)
### main_nil.py 实体分类模型的推理代码,对实体消歧模型预测为NIL的实体预测其类别
### post_nil.py 实体分类模型的后处理代码,生成提交文件(单模)
### utils.py 定义各种训练,推理过程中需要的函数等
### main_matching.py 实体消歧模型推理代码
### train_matching.py 实体消歧模型训练代码
### post_matching_ens.py 实体消歧模型后处理,对每个实体选取概率最大的一个作为kb_id(若小于一个阈值,则取NIL)(多个模型)
### train_nil.py 实体分类模型训练代码
### post_nil_ens.py 实体分类模型的后处理代码,生成提交文件(多个模型)
## 环境安装
!pip install paddle-ernie==0.0.4dev1
## 训练实体消歧模型(基于ernie),需要20G左右显存,若是分配的16G会出现OOM,可以传入--bsz 16
!python work/train_matching.py --from_pretrained ernie-1.0 --use_lr_decay --save_dir work/tmp/dynamic_neg1_all --max_seqlen 256
## 训练实体分类模型(基于ernie),两阶段训练,第一阶段利用非NIL实体,第二阶段使用第一阶段得到的模型在NIL上finetune
!python work/train_nil.py --from_pretrained ernie-1.0 --use_lr_decay --save_dir work/checkpoint/nil_preround --preround --epochs 2
!python work/train_nil.py --from_pretrained ernie-1.0 --use_lr_decay --save_dir work/checkpoint/nil_ft_preround --init_checkpoint work/checkpoint/nil_preround_debug.pdparams
## 单模型推理代码(1个matching, 1个nil)
### 实体消歧模型推理
!python work/main_matching.py --from_pretrained ernie-1.0 --init_checkpoint work/checkpoint/dynamic_neg2_all.pdparams --save_path work/result/test_matching.pkl --max_seqlen 256 --use_test_data
### 实体消歧模型后处理
!python work/post_matching.py --use_test_data --thres 0.2
### 实体分类模型推理
!python work/main_nil.py --from_pretrained ernie-1.0 --init_checkpoint work/checkpoint/nil_ft_ad.pdparams --save_path work/result/test_nil.pkl --use_test_data
### 实体分类模型后处理
!python work/post_nil.py --use_test_data
```
请点击[此处](https://ai.baidu.com/docs#/AIStudio_Project_Notebook/a38e5576)查看本环境基本用法. <br>
Please click [here ](https://ai.baidu.com/docs#/AIStudio_Project_Notebook/a38e5576) for more detailed instructions.
|
github_jupyter
|
## 环境配置:基于PaddlePaddle 1.8.4开发(python 3.7), 使用单块V100(32G)训练
## 各个文件的作用:
### eval.py 官方提供的评估脚本
### post_matching.py 实体消歧模型后处理,对每个实体选取概率最大的一个作为kb_id(若小于一个阈值,则取NIL)(单模)
### main_nil.py 实体分类模型的推理代码,对实体消歧模型预测为NIL的实体预测其类别
### post_nil.py 实体分类模型的后处理代码,生成提交文件(单模)
### utils.py 定义各种训练,推理过程中需要的函数等
### main_matching.py 实体消歧模型推理代码
### train_matching.py 实体消歧模型训练代码
### post_matching_ens.py 实体消歧模型后处理,对每个实体选取概率最大的一个作为kb_id(若小于一个阈值,则取NIL)(多个模型)
### train_nil.py 实体分类模型训练代码
### post_nil_ens.py 实体分类模型的后处理代码,生成提交文件(多个模型)
## 环境安装
!pip install paddle-ernie==0.0.4dev1
## 训练实体消歧模型(基于ernie),需要20G左右显存,若是分配的16G会出现OOM,可以传入--bsz 16
!python work/train_matching.py --from_pretrained ernie-1.0 --use_lr_decay --save_dir work/tmp/dynamic_neg1_all --max_seqlen 256
## 训练实体分类模型(基于ernie),两阶段训练,第一阶段利用非NIL实体,第二阶段使用第一阶段得到的模型在NIL上finetune
!python work/train_nil.py --from_pretrained ernie-1.0 --use_lr_decay --save_dir work/checkpoint/nil_preround --preround --epochs 2
!python work/train_nil.py --from_pretrained ernie-1.0 --use_lr_decay --save_dir work/checkpoint/nil_ft_preround --init_checkpoint work/checkpoint/nil_preround_debug.pdparams
## 单模型推理代码(1个matching, 1个nil)
### 实体消歧模型推理
!python work/main_matching.py --from_pretrained ernie-1.0 --init_checkpoint work/checkpoint/dynamic_neg2_all.pdparams --save_path work/result/test_matching.pkl --max_seqlen 256 --use_test_data
### 实体消歧模型后处理
!python work/post_matching.py --use_test_data --thres 0.2
### 实体分类模型推理
!python work/main_nil.py --from_pretrained ernie-1.0 --init_checkpoint work/checkpoint/nil_ft_ad.pdparams --save_path work/result/test_nil.pkl --use_test_data
### 实体分类模型后处理
!python work/post_nil.py --use_test_data
| 0.196094 | 0.63576 |
<a href="https://colab.research.google.com/github/szoha/test/blob/master/Copy_of_astho_ubaid.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Introduction
In this notebook, we implement [YOLOv4](https://arxiv.org/pdf/2004.10934.pdf) for training on your own dataset in PyTorch.
We also recommend reading our blog post on [Training YOLOv4 on custom data](https://blog.roboflow.ai/training-yolov4-on-a-custom-dataset/) side by side.
We will take the following steps to implement YOLOv4 on our custom data:
* Set up YOLOv4 environment
* Download Custom Dataset via Roboflow
* Train Custom YOLOv4 detector
* Reload Custom YOLOv4 detector weights for inference
When you are done you will have a custom detector that you can use. It will make inference like this:
#### 
### **Reach out for support**
If you run into any hurdles on your own data set or just want to share some cool results in your own domain, [reach out!](https://roboflow.ai/contact)
#### 
# Set up YOLOv4 Environment
```
!git clone https://github.com/roboflow-ai/pytorch-YOLOv4.git
%cd /content/pytorch-YOLOv4
!pip install -r requirements.txt
# download yolov4 weights that have already been converted to PyTorch
!gdown https://drive.google.com/uc?id=1fcbR0bWzYfIEdLJPzOsn4R5mlvR6IQyA
```
# Download Custom Dataset
## Export Your Dataset from Roboflow
Roboflow enables you to export your dataset in any format you need - including for this notebook.
Create a [free account](https://app.roboflow.ai). Upload your private dataset. Generate a version (applying any preprocessing and augmentations you desire). Create an export. Select **YOLOv4 PyTorch** as the export format. Click **"Show Download code"**, copy your link, and paste it in the next cell. Magic.
```
# REPLACE this link with your Roboflow dataset (export as YOLOv4 PyTorch format)
!curl -L "https://app.roboflow.com/ds/wdCtWCDJho?key=vIBLN5wWEO" > roboflow.zip; unzip roboflow.zip; rm roboflow.zip
%cp train/_annotations.txt train/train.txt
%cp train/_annotations.txt train.txt
%cp valid/_annotations.txt data/val.txt
%cp valid/*.jpg train/
def file_len(fname):
with open(fname) as f:
for i, l in enumerate(f):
pass
return i + 1
num_classes = file_len('train/_classes.txt')
print(num_classes)
%%writefile /content/pytorch-YOLOv4/dataset.py
# -*- coding: utf-8 -*-
'''
@Time : 2020/05/06 21:09
@Author : Tianxiaomo
@File : dataset.py
@Noice :
@Modificattion :
@Author :
@Time :
@Detail :
'''
from torch.utils.data.dataset import Dataset
import random
import cv2
import sys
import numpy as np
import os
import matplotlib.pyplot as plt
def rand_uniform_strong(min, max):
if min > max:
swap = min
min = max
max = swap
return random.random() * (max - min) + min
def rand_scale(s):
scale = rand_uniform_strong(1, s)
if random.randint(0, 1) % 2:
return scale
return 1. / scale
def rand_precalc_random(min, max, random_part):
if max < min:
swap = min
min = max
max = swap
return (random_part * (max - min)) + min
def fill_truth_detection(bboxes, num_boxes, classes, flip, dx, dy, sx, sy, net_w, net_h):
if bboxes.shape[0] == 0:
return bboxes, 10000
np.random.shuffle(bboxes)
bboxes[:, 0] -= dx
bboxes[:, 2] -= dx
bboxes[:, 1] -= dy
bboxes[:, 3] -= dy
bboxes[:, 0] = np.clip(bboxes[:, 0], 0, sx)
bboxes[:, 2] = np.clip(bboxes[:, 2], 0, sx)
bboxes[:, 1] = np.clip(bboxes[:, 1], 0, sy)
bboxes[:, 3] = np.clip(bboxes[:, 3], 0, sy)
out_box = list(np.where(((bboxes[:, 1] == sy) & (bboxes[:, 3] == sy)) |
((bboxes[:, 0] == sx) & (bboxes[:, 2] == sx)) |
((bboxes[:, 1] == 0) & (bboxes[:, 3] == 0)) |
((bboxes[:, 0] == 0) & (bboxes[:, 2] == 0)))[0])
list_box = list(range(bboxes.shape[0]))
for i in out_box:
list_box.remove(i)
bboxes = bboxes[list_box]
if bboxes.shape[0] == 0:
return bboxes, 10000
bboxes = bboxes[np.where((bboxes[:, 4] < classes) & (bboxes[:, 4] >= 0))[0]]
if bboxes.shape[0] > num_boxes:
bboxes = bboxes[:num_boxes]
min_w_h = np.array([bboxes[:, 2] - bboxes[:, 0], bboxes[:, 3] - bboxes[:, 1]]).min()
bboxes[:, 0] *= (net_w / sx)
bboxes[:, 2] *= (net_w / sx)
bboxes[:, 1] *= (net_h / sy)
bboxes[:, 3] *= (net_h / sy)
if flip:
temp = net_w - bboxes[:, 0]
bboxes[:, 0] = net_w - bboxes[:, 2]
bboxes[:, 2] = temp
return bboxes, min_w_h
def rect_intersection(a, b):
minx = max(a[0], b[0])
miny = max(a[1], b[1])
maxx = min(a[2], b[2])
maxy = min(a[3], b[3])
return [minx, miny, maxx, maxy]
def image_data_augmentation(mat, w, h, pleft, ptop, swidth, sheight, flip, dhue, dsat, dexp, gaussian_noise, blur,
truth):
try:
img = mat
oh, ow, _ = img.shape
pleft, ptop, swidth, sheight = int(pleft), int(ptop), int(swidth), int(sheight)
# crop
src_rect = [pleft, ptop, swidth + pleft, sheight + ptop] # x1,y1,x2,y2
img_rect = [0, 0, ow, oh]
new_src_rect = rect_intersection(src_rect, img_rect) # 交集
dst_rect = [max(0, -pleft), max(0, -ptop), max(0, -pleft) + new_src_rect[2] - new_src_rect[0],
max(0, -ptop) + new_src_rect[3] - new_src_rect[1]]
# cv2.Mat sized
if (src_rect[0] == 0 and src_rect[1] == 0 and src_rect[2] == img.shape[0] and src_rect[3] == img.shape[1]):
sized = cv2.resize(img, (w, h), cv2.INTER_LINEAR)
else:
cropped = np.zeros([sheight, swidth, 3])
cropped[:, :, ] = np.mean(img, axis=(0, 1))
cropped[dst_rect[1]:dst_rect[3], dst_rect[0]:dst_rect[2]] = \
img[new_src_rect[1]:new_src_rect[3], new_src_rect[0]:new_src_rect[2]]
# resize
sized = cv2.resize(cropped, (w, h), cv2.INTER_LINEAR)
# flip
if flip:
# cv2.Mat cropped
sized = cv2.flip(sized, 1) # 0 - x-axis, 1 - y-axis, -1 - both axes (x & y)
# HSV augmentation
# cv2.COLOR_BGR2HSV, cv2.COLOR_RGB2HSV, cv2.COLOR_HSV2BGR, cv2.COLOR_HSV2RGB
if dsat != 1 or dexp != 1 or dhue != 0:
if img.shape[2] >= 3:
hsv_src = cv2.cvtColor(sized.astype(np.float32), cv2.COLOR_RGB2HSV) # RGB to HSV
hsv = cv2.split(hsv_src)
hsv[1] *= dsat
hsv[2] *= dexp
hsv[0] += 179 * dhue
hsv_src = cv2.merge(hsv)
sized = np.clip(cv2.cvtColor(hsv_src, cv2.COLOR_HSV2RGB), 0, 255) # HSV to RGB (the same as previous)
else:
sized *= dexp
if blur:
if blur == 1:
dst = cv2.GaussianBlur(sized, (17, 17), 0)
# cv2.bilateralFilter(sized, dst, 17, 75, 75)
else:
ksize = (blur / 2) * 2 + 1
dst = cv2.GaussianBlur(sized, (ksize, ksize), 0)
if blur == 1:
img_rect = [0, 0, sized.cols, sized.rows]
for b in truth:
left = (b.x - b.w / 2.) * sized.shape[1]
width = b.w * sized.shape[1]
top = (b.y - b.h / 2.) * sized.shape[0]
height = b.h * sized.shape[0]
roi(left, top, width, height)
roi = roi & img_rect
dst[roi[0]:roi[0] + roi[2], roi[1]:roi[1] + roi[3]] = sized[roi[0]:roi[0] + roi[2],
roi[1]:roi[1] + roi[3]]
sized = dst
if gaussian_noise:
noise = np.array(sized.shape)
gaussian_noise = min(gaussian_noise, 127)
gaussian_noise = max(gaussian_noise, 0)
cv2.randn(noise, 0, gaussian_noise) # mean and variance
sized = sized + noise
except:
print("OpenCV can't augment image: " + str(w) + " x " + str(h))
sized = mat
return sized
def filter_truth(bboxes, dx, dy, sx, sy, xd, yd):
bboxes[:, 0] -= dx
bboxes[:, 2] -= dx
bboxes[:, 1] -= dy
bboxes[:, 3] -= dy
bboxes[:, 0] = np.clip(bboxes[:, 0], 0, sx)
bboxes[:, 2] = np.clip(bboxes[:, 2], 0, sx)
bboxes[:, 1] = np.clip(bboxes[:, 1], 0, sy)
bboxes[:, 3] = np.clip(bboxes[:, 3], 0, sy)
out_box = list(np.where(((bboxes[:, 1] == sy) & (bboxes[:, 3] == sy)) |
((bboxes[:, 0] == sx) & (bboxes[:, 2] == sx)) |
((bboxes[:, 1] == 0) & (bboxes[:, 3] == 0)) |
((bboxes[:, 0] == 0) & (bboxes[:, 2] == 0)))[0])
list_box = list(range(bboxes.shape[0]))
for i in out_box:
list_box.remove(i)
bboxes = bboxes[list_box]
bboxes[:, 0] += xd
bboxes[:, 2] += xd
bboxes[:, 1] += yd
bboxes[:, 3] += yd
return bboxes
def blend_truth_mosaic(out_img, img, bboxes, w, h, cut_x, cut_y, i_mixup,
left_shift, right_shift, top_shift, bot_shift):
left_shift = min(left_shift, w - cut_x)
top_shift = min(top_shift, h - cut_y)
right_shift = min(right_shift, cut_x)
bot_shift = min(bot_shift, cut_y)
if i_mixup == 0:
bboxes = filter_truth(bboxes, left_shift, top_shift, cut_x, cut_y, 0, 0)
out_img[:cut_y, :cut_x] = img[top_shift:top_shift + cut_y, left_shift:left_shift + cut_x]
if i_mixup == 1:
bboxes = filter_truth(bboxes, cut_x - right_shift, top_shift, w - cut_x, cut_y, cut_x, 0)
out_img[:cut_y, cut_x:] = img[top_shift:top_shift + cut_y, cut_x - right_shift:w - right_shift]
if i_mixup == 2:
bboxes = filter_truth(bboxes, left_shift, cut_y - bot_shift, cut_x, h - cut_y, 0, cut_y)
out_img[cut_y:, :cut_x] = img[cut_y - bot_shift:h - bot_shift, left_shift:left_shift + cut_x]
if i_mixup == 3:
bboxes = filter_truth(bboxes, cut_x - right_shift, cut_y - bot_shift, w - cut_x, h - cut_y, cut_x, cut_y)
out_img[cut_y:, cut_x:] = img[cut_y - bot_shift:h - bot_shift, cut_x - right_shift:w - right_shift]
return out_img, bboxes
def draw_box(img, bboxes):
for b in bboxes:
img = cv2.rectangle(img, (b[0], b[1]), (b[2], b[3]), (0, 255, 0), 2)
return img
class Yolo_dataset(Dataset):
def __init__(self, lable_path, cfg):
super(Yolo_dataset, self).__init__()
if cfg.mixup == 2:
print("cutmix=1 - isn't supported for Detector")
raise
elif cfg.mixup == 2 and cfg.letter_box:
print("Combination: letter_box=1 & mosaic=1 - isn't supported, use only 1 of these parameters")
raise
self.cfg = cfg
truth = {}
f = open(lable_path, 'r', encoding='utf-8')
for line in f.readlines():
data = line.split(" ")
truth[data[0]] = []
for i in data[1:]:
truth[data[0]].append([int(j) for j in i.split(',')])
self.truth = truth
def __len__(self):
return len(self.truth.keys())
def __getitem__(self, index):
img_path = list(self.truth.keys())[index]
bboxes = np.array(self.truth.get(img_path), dtype=np.float)
img_path = os.path.join(self.cfg.dataset_dir, img_path)
use_mixup = self.cfg.mixup
if random.randint(0, 1):
use_mixup = 0
if use_mixup == 3:
min_offset = 0.2
cut_x = random.randint(int(self.cfg.w * min_offset), int(self.cfg.w * (1 - min_offset)))
cut_y = random.randint(int(self.cfg.h * min_offset), int(self.cfg.h * (1 - min_offset)))
r1, r2, r3, r4, r_scale = 0, 0, 0, 0, 0
dhue, dsat, dexp, flip, blur = 0, 0, 0, 0, 0
gaussian_noise = 0
out_img = np.zeros([self.cfg.h, self.cfg.w, 3])
out_bboxes = []
for i in range(use_mixup + 1):
if i != 0:
img_path = random.choice(list(self.truth.keys()))
bboxes = np.array(self.truth.get(img_path), dtype=np.float)
img_path = os.path.join(self.cfg.dataset_dir, img_path)
#print(img_path)
img = cv2.imread(img_path)
#print(img)
if img is None:
continue
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
oh, ow, oc = img.shape
dh, dw, dc = np.array(np.array([oh, ow, oc]) * self.cfg.jitter, dtype=np.int)
dhue = rand_uniform_strong(-self.cfg.hue, self.cfg.hue)
dsat = rand_scale(self.cfg.saturation)
dexp = rand_scale(self.cfg.exposure)
pleft = random.randint(-dw, dw)
pright = random.randint(-dw, dw)
ptop = random.randint(-dh, dh)
pbot = random.randint(-dh, dh)
flip = random.randint(0, 1) if self.cfg.flip else 0
if (self.cfg.blur):
tmp_blur = random.randint(0, 2) # 0 - disable, 1 - blur background, 2 - blur the whole image
if tmp_blur == 0:
blur = 0
elif tmp_blur == 1:
blur = 1
else:
blur = self.cfg.blur
if self.cfg.gaussian and random.randint(0, 1):
gaussian_noise = self.cfg.gaussian
else:
gaussian_noise = 0
if self.cfg.letter_box:
img_ar = ow / oh
net_ar = self.cfg.w / self.cfg.h
result_ar = img_ar / net_ar
# print(" ow = %d, oh = %d, w = %d, h = %d, img_ar = %f, net_ar = %f, result_ar = %f \n", ow, oh, w, h, img_ar, net_ar, result_ar);
if result_ar > 1: # sheight - should be increased
oh_tmp = ow / net_ar
delta_h = (oh_tmp - oh) / 2
ptop = ptop - delta_h
pbot = pbot - delta_h
# print(" result_ar = %f, oh_tmp = %f, delta_h = %d, ptop = %f, pbot = %f \n", result_ar, oh_tmp, delta_h, ptop, pbot);
else: # swidth - should be increased
ow_tmp = oh * net_ar
delta_w = (ow_tmp - ow) / 2
pleft = pleft - delta_w
pright = pright - delta_w
# printf(" result_ar = %f, ow_tmp = %f, delta_w = %d, pleft = %f, pright = %f \n", result_ar, ow_tmp, delta_w, pleft, pright);
swidth = ow - pleft - pright
sheight = oh - ptop - pbot
truth, min_w_h = fill_truth_detection(bboxes, self.cfg.boxes, self.cfg.classes, flip, pleft, ptop, swidth,
sheight, self.cfg.w, self.cfg.h)
if (min_w_h / 8) < blur and blur > 1: # disable blur if one of the objects is too small
blur = min_w_h / 8
ai = image_data_augmentation(img, self.cfg.w, self.cfg.h, pleft, ptop, swidth, sheight, flip,
dhue, dsat, dexp, gaussian_noise, blur, truth)
if use_mixup == 0:
out_img = ai
out_bboxes = truth
if use_mixup == 1:
if i == 0:
old_img = ai.copy()
old_truth = truth.copy()
elif i == 1:
out_img = cv2.addWeighted(ai, 0.5, old_img, 0.5)
out_bboxes = np.concatenate([old_truth, truth], axis=0)
elif use_mixup == 3:
if flip:
tmp = pleft
pleft = pright
pright = tmp
left_shift = int(min(cut_x, max(0, (-int(pleft) * self.cfg.w / swidth))))
top_shift = int(min(cut_y, max(0, (-int(ptop) * self.cfg.h / sheight))))
right_shift = int(min((self.cfg.w - cut_x), max(0, (-int(pright) * self.cfg.w / swidth))))
bot_shift = int(min(self.cfg.h - cut_y, max(0, (-int(pbot) * self.cfg.h / sheight))))
out_img, out_bbox = blend_truth_mosaic(out_img, ai, truth.copy(), self.cfg.w, self.cfg.h, cut_x,
cut_y, i, left_shift, right_shift, top_shift, bot_shift)
out_bboxes.append(out_bbox)
# print(img_path)
if use_mixup == 3:
out_bboxes = np.concatenate(out_bboxes, axis=0)
out_bboxes1 = np.zeros([self.cfg.boxes, 5])
out_bboxes1[:min(out_bboxes.shape[0], self.cfg.boxes)] = out_bboxes[:min(out_bboxes.shape[0], self.cfg.boxes)]
return out_img, out_bboxes1
if __name__ == "__main__":
from cfg import Cfg
random.seed(2020)
np.random.seed(2020)
Cfg.dataset_dir = '/mnt/e/Dataset'
dataset = Yolo_dataset(Cfg.train_label, Cfg)
for i in range(100):
out_img, out_bboxes = dataset.__getitem__(i)
a = draw_box(out_img.copy(), out_bboxes.astype(np.int32))
plt.imshow(a.astype(np.int32))
plt.show()
```
# Train Custom Detector
```
#start training
#-b batch size (you should keep this low (2-4) for training to work properly)
#-s number of subdivisions in the batch, this was more relevant for the darknet framework
#-l learning rate
#-g direct training to the GPU device
#pretrained invoke the pretrained weights that we downloaded above
#classes - number of classes
#dir - where the training data is
#epoch - how long to train for
!python train.py -b 2 -s 1 -l 0.001 -g 0 -pretrained ./yolov4.conv.137.pth -classes {num_classes} -dir ./train -epochs 40
```
# Load Trained Weights for Custom Detection
You can also use this to load previously saved weights!
```
#looking at the weights that our model has saved during training
!ls checkpoints
#choose random test image
import os
test_images = [f for f in os.listdir('test') if f.endswith('.jpg')]
import random
img_path = "test/" + random.choice(test_images);
%%time
##change the epoch here to the one you would like to use for inference
!python models.py {num_classes} checkpoints/Yolov4_epoch35.pth {img_path} test/_classes.txt
#visualize inference
from IPython.display import Image
Image('predictions.jpg')
from google.colab import drive
drive.mount('/content/drive')
%cp /content/
```
|
github_jupyter
|
!git clone https://github.com/roboflow-ai/pytorch-YOLOv4.git
%cd /content/pytorch-YOLOv4
!pip install -r requirements.txt
# download yolov4 weights that have already been converted to PyTorch
!gdown https://drive.google.com/uc?id=1fcbR0bWzYfIEdLJPzOsn4R5mlvR6IQyA
# REPLACE this link with your Roboflow dataset (export as YOLOv4 PyTorch format)
!curl -L "https://app.roboflow.com/ds/wdCtWCDJho?key=vIBLN5wWEO" > roboflow.zip; unzip roboflow.zip; rm roboflow.zip
%cp train/_annotations.txt train/train.txt
%cp train/_annotations.txt train.txt
%cp valid/_annotations.txt data/val.txt
%cp valid/*.jpg train/
def file_len(fname):
with open(fname) as f:
for i, l in enumerate(f):
pass
return i + 1
num_classes = file_len('train/_classes.txt')
print(num_classes)
%%writefile /content/pytorch-YOLOv4/dataset.py
# -*- coding: utf-8 -*-
'''
@Time : 2020/05/06 21:09
@Author : Tianxiaomo
@File : dataset.py
@Noice :
@Modificattion :
@Author :
@Time :
@Detail :
'''
from torch.utils.data.dataset import Dataset
import random
import cv2
import sys
import numpy as np
import os
import matplotlib.pyplot as plt
def rand_uniform_strong(min, max):
if min > max:
swap = min
min = max
max = swap
return random.random() * (max - min) + min
def rand_scale(s):
scale = rand_uniform_strong(1, s)
if random.randint(0, 1) % 2:
return scale
return 1. / scale
def rand_precalc_random(min, max, random_part):
if max < min:
swap = min
min = max
max = swap
return (random_part * (max - min)) + min
def fill_truth_detection(bboxes, num_boxes, classes, flip, dx, dy, sx, sy, net_w, net_h):
if bboxes.shape[0] == 0:
return bboxes, 10000
np.random.shuffle(bboxes)
bboxes[:, 0] -= dx
bboxes[:, 2] -= dx
bboxes[:, 1] -= dy
bboxes[:, 3] -= dy
bboxes[:, 0] = np.clip(bboxes[:, 0], 0, sx)
bboxes[:, 2] = np.clip(bboxes[:, 2], 0, sx)
bboxes[:, 1] = np.clip(bboxes[:, 1], 0, sy)
bboxes[:, 3] = np.clip(bboxes[:, 3], 0, sy)
out_box = list(np.where(((bboxes[:, 1] == sy) & (bboxes[:, 3] == sy)) |
((bboxes[:, 0] == sx) & (bboxes[:, 2] == sx)) |
((bboxes[:, 1] == 0) & (bboxes[:, 3] == 0)) |
((bboxes[:, 0] == 0) & (bboxes[:, 2] == 0)))[0])
list_box = list(range(bboxes.shape[0]))
for i in out_box:
list_box.remove(i)
bboxes = bboxes[list_box]
if bboxes.shape[0] == 0:
return bboxes, 10000
bboxes = bboxes[np.where((bboxes[:, 4] < classes) & (bboxes[:, 4] >= 0))[0]]
if bboxes.shape[0] > num_boxes:
bboxes = bboxes[:num_boxes]
min_w_h = np.array([bboxes[:, 2] - bboxes[:, 0], bboxes[:, 3] - bboxes[:, 1]]).min()
bboxes[:, 0] *= (net_w / sx)
bboxes[:, 2] *= (net_w / sx)
bboxes[:, 1] *= (net_h / sy)
bboxes[:, 3] *= (net_h / sy)
if flip:
temp = net_w - bboxes[:, 0]
bboxes[:, 0] = net_w - bboxes[:, 2]
bboxes[:, 2] = temp
return bboxes, min_w_h
def rect_intersection(a, b):
minx = max(a[0], b[0])
miny = max(a[1], b[1])
maxx = min(a[2], b[2])
maxy = min(a[3], b[3])
return [minx, miny, maxx, maxy]
def image_data_augmentation(mat, w, h, pleft, ptop, swidth, sheight, flip, dhue, dsat, dexp, gaussian_noise, blur,
truth):
try:
img = mat
oh, ow, _ = img.shape
pleft, ptop, swidth, sheight = int(pleft), int(ptop), int(swidth), int(sheight)
# crop
src_rect = [pleft, ptop, swidth + pleft, sheight + ptop] # x1,y1,x2,y2
img_rect = [0, 0, ow, oh]
new_src_rect = rect_intersection(src_rect, img_rect) # 交集
dst_rect = [max(0, -pleft), max(0, -ptop), max(0, -pleft) + new_src_rect[2] - new_src_rect[0],
max(0, -ptop) + new_src_rect[3] - new_src_rect[1]]
# cv2.Mat sized
if (src_rect[0] == 0 and src_rect[1] == 0 and src_rect[2] == img.shape[0] and src_rect[3] == img.shape[1]):
sized = cv2.resize(img, (w, h), cv2.INTER_LINEAR)
else:
cropped = np.zeros([sheight, swidth, 3])
cropped[:, :, ] = np.mean(img, axis=(0, 1))
cropped[dst_rect[1]:dst_rect[3], dst_rect[0]:dst_rect[2]] = \
img[new_src_rect[1]:new_src_rect[3], new_src_rect[0]:new_src_rect[2]]
# resize
sized = cv2.resize(cropped, (w, h), cv2.INTER_LINEAR)
# flip
if flip:
# cv2.Mat cropped
sized = cv2.flip(sized, 1) # 0 - x-axis, 1 - y-axis, -1 - both axes (x & y)
# HSV augmentation
# cv2.COLOR_BGR2HSV, cv2.COLOR_RGB2HSV, cv2.COLOR_HSV2BGR, cv2.COLOR_HSV2RGB
if dsat != 1 or dexp != 1 or dhue != 0:
if img.shape[2] >= 3:
hsv_src = cv2.cvtColor(sized.astype(np.float32), cv2.COLOR_RGB2HSV) # RGB to HSV
hsv = cv2.split(hsv_src)
hsv[1] *= dsat
hsv[2] *= dexp
hsv[0] += 179 * dhue
hsv_src = cv2.merge(hsv)
sized = np.clip(cv2.cvtColor(hsv_src, cv2.COLOR_HSV2RGB), 0, 255) # HSV to RGB (the same as previous)
else:
sized *= dexp
if blur:
if blur == 1:
dst = cv2.GaussianBlur(sized, (17, 17), 0)
# cv2.bilateralFilter(sized, dst, 17, 75, 75)
else:
ksize = (blur / 2) * 2 + 1
dst = cv2.GaussianBlur(sized, (ksize, ksize), 0)
if blur == 1:
img_rect = [0, 0, sized.cols, sized.rows]
for b in truth:
left = (b.x - b.w / 2.) * sized.shape[1]
width = b.w * sized.shape[1]
top = (b.y - b.h / 2.) * sized.shape[0]
height = b.h * sized.shape[0]
roi(left, top, width, height)
roi = roi & img_rect
dst[roi[0]:roi[0] + roi[2], roi[1]:roi[1] + roi[3]] = sized[roi[0]:roi[0] + roi[2],
roi[1]:roi[1] + roi[3]]
sized = dst
if gaussian_noise:
noise = np.array(sized.shape)
gaussian_noise = min(gaussian_noise, 127)
gaussian_noise = max(gaussian_noise, 0)
cv2.randn(noise, 0, gaussian_noise) # mean and variance
sized = sized + noise
except:
print("OpenCV can't augment image: " + str(w) + " x " + str(h))
sized = mat
return sized
def filter_truth(bboxes, dx, dy, sx, sy, xd, yd):
bboxes[:, 0] -= dx
bboxes[:, 2] -= dx
bboxes[:, 1] -= dy
bboxes[:, 3] -= dy
bboxes[:, 0] = np.clip(bboxes[:, 0], 0, sx)
bboxes[:, 2] = np.clip(bboxes[:, 2], 0, sx)
bboxes[:, 1] = np.clip(bboxes[:, 1], 0, sy)
bboxes[:, 3] = np.clip(bboxes[:, 3], 0, sy)
out_box = list(np.where(((bboxes[:, 1] == sy) & (bboxes[:, 3] == sy)) |
((bboxes[:, 0] == sx) & (bboxes[:, 2] == sx)) |
((bboxes[:, 1] == 0) & (bboxes[:, 3] == 0)) |
((bboxes[:, 0] == 0) & (bboxes[:, 2] == 0)))[0])
list_box = list(range(bboxes.shape[0]))
for i in out_box:
list_box.remove(i)
bboxes = bboxes[list_box]
bboxes[:, 0] += xd
bboxes[:, 2] += xd
bboxes[:, 1] += yd
bboxes[:, 3] += yd
return bboxes
def blend_truth_mosaic(out_img, img, bboxes, w, h, cut_x, cut_y, i_mixup,
left_shift, right_shift, top_shift, bot_shift):
left_shift = min(left_shift, w - cut_x)
top_shift = min(top_shift, h - cut_y)
right_shift = min(right_shift, cut_x)
bot_shift = min(bot_shift, cut_y)
if i_mixup == 0:
bboxes = filter_truth(bboxes, left_shift, top_shift, cut_x, cut_y, 0, 0)
out_img[:cut_y, :cut_x] = img[top_shift:top_shift + cut_y, left_shift:left_shift + cut_x]
if i_mixup == 1:
bboxes = filter_truth(bboxes, cut_x - right_shift, top_shift, w - cut_x, cut_y, cut_x, 0)
out_img[:cut_y, cut_x:] = img[top_shift:top_shift + cut_y, cut_x - right_shift:w - right_shift]
if i_mixup == 2:
bboxes = filter_truth(bboxes, left_shift, cut_y - bot_shift, cut_x, h - cut_y, 0, cut_y)
out_img[cut_y:, :cut_x] = img[cut_y - bot_shift:h - bot_shift, left_shift:left_shift + cut_x]
if i_mixup == 3:
bboxes = filter_truth(bboxes, cut_x - right_shift, cut_y - bot_shift, w - cut_x, h - cut_y, cut_x, cut_y)
out_img[cut_y:, cut_x:] = img[cut_y - bot_shift:h - bot_shift, cut_x - right_shift:w - right_shift]
return out_img, bboxes
def draw_box(img, bboxes):
for b in bboxes:
img = cv2.rectangle(img, (b[0], b[1]), (b[2], b[3]), (0, 255, 0), 2)
return img
class Yolo_dataset(Dataset):
def __init__(self, lable_path, cfg):
super(Yolo_dataset, self).__init__()
if cfg.mixup == 2:
print("cutmix=1 - isn't supported for Detector")
raise
elif cfg.mixup == 2 and cfg.letter_box:
print("Combination: letter_box=1 & mosaic=1 - isn't supported, use only 1 of these parameters")
raise
self.cfg = cfg
truth = {}
f = open(lable_path, 'r', encoding='utf-8')
for line in f.readlines():
data = line.split(" ")
truth[data[0]] = []
for i in data[1:]:
truth[data[0]].append([int(j) for j in i.split(',')])
self.truth = truth
def __len__(self):
return len(self.truth.keys())
def __getitem__(self, index):
img_path = list(self.truth.keys())[index]
bboxes = np.array(self.truth.get(img_path), dtype=np.float)
img_path = os.path.join(self.cfg.dataset_dir, img_path)
use_mixup = self.cfg.mixup
if random.randint(0, 1):
use_mixup = 0
if use_mixup == 3:
min_offset = 0.2
cut_x = random.randint(int(self.cfg.w * min_offset), int(self.cfg.w * (1 - min_offset)))
cut_y = random.randint(int(self.cfg.h * min_offset), int(self.cfg.h * (1 - min_offset)))
r1, r2, r3, r4, r_scale = 0, 0, 0, 0, 0
dhue, dsat, dexp, flip, blur = 0, 0, 0, 0, 0
gaussian_noise = 0
out_img = np.zeros([self.cfg.h, self.cfg.w, 3])
out_bboxes = []
for i in range(use_mixup + 1):
if i != 0:
img_path = random.choice(list(self.truth.keys()))
bboxes = np.array(self.truth.get(img_path), dtype=np.float)
img_path = os.path.join(self.cfg.dataset_dir, img_path)
#print(img_path)
img = cv2.imread(img_path)
#print(img)
if img is None:
continue
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
oh, ow, oc = img.shape
dh, dw, dc = np.array(np.array([oh, ow, oc]) * self.cfg.jitter, dtype=np.int)
dhue = rand_uniform_strong(-self.cfg.hue, self.cfg.hue)
dsat = rand_scale(self.cfg.saturation)
dexp = rand_scale(self.cfg.exposure)
pleft = random.randint(-dw, dw)
pright = random.randint(-dw, dw)
ptop = random.randint(-dh, dh)
pbot = random.randint(-dh, dh)
flip = random.randint(0, 1) if self.cfg.flip else 0
if (self.cfg.blur):
tmp_blur = random.randint(0, 2) # 0 - disable, 1 - blur background, 2 - blur the whole image
if tmp_blur == 0:
blur = 0
elif tmp_blur == 1:
blur = 1
else:
blur = self.cfg.blur
if self.cfg.gaussian and random.randint(0, 1):
gaussian_noise = self.cfg.gaussian
else:
gaussian_noise = 0
if self.cfg.letter_box:
img_ar = ow / oh
net_ar = self.cfg.w / self.cfg.h
result_ar = img_ar / net_ar
# print(" ow = %d, oh = %d, w = %d, h = %d, img_ar = %f, net_ar = %f, result_ar = %f \n", ow, oh, w, h, img_ar, net_ar, result_ar);
if result_ar > 1: # sheight - should be increased
oh_tmp = ow / net_ar
delta_h = (oh_tmp - oh) / 2
ptop = ptop - delta_h
pbot = pbot - delta_h
# print(" result_ar = %f, oh_tmp = %f, delta_h = %d, ptop = %f, pbot = %f \n", result_ar, oh_tmp, delta_h, ptop, pbot);
else: # swidth - should be increased
ow_tmp = oh * net_ar
delta_w = (ow_tmp - ow) / 2
pleft = pleft - delta_w
pright = pright - delta_w
# printf(" result_ar = %f, ow_tmp = %f, delta_w = %d, pleft = %f, pright = %f \n", result_ar, ow_tmp, delta_w, pleft, pright);
swidth = ow - pleft - pright
sheight = oh - ptop - pbot
truth, min_w_h = fill_truth_detection(bboxes, self.cfg.boxes, self.cfg.classes, flip, pleft, ptop, swidth,
sheight, self.cfg.w, self.cfg.h)
if (min_w_h / 8) < blur and blur > 1: # disable blur if one of the objects is too small
blur = min_w_h / 8
ai = image_data_augmentation(img, self.cfg.w, self.cfg.h, pleft, ptop, swidth, sheight, flip,
dhue, dsat, dexp, gaussian_noise, blur, truth)
if use_mixup == 0:
out_img = ai
out_bboxes = truth
if use_mixup == 1:
if i == 0:
old_img = ai.copy()
old_truth = truth.copy()
elif i == 1:
out_img = cv2.addWeighted(ai, 0.5, old_img, 0.5)
out_bboxes = np.concatenate([old_truth, truth], axis=0)
elif use_mixup == 3:
if flip:
tmp = pleft
pleft = pright
pright = tmp
left_shift = int(min(cut_x, max(0, (-int(pleft) * self.cfg.w / swidth))))
top_shift = int(min(cut_y, max(0, (-int(ptop) * self.cfg.h / sheight))))
right_shift = int(min((self.cfg.w - cut_x), max(0, (-int(pright) * self.cfg.w / swidth))))
bot_shift = int(min(self.cfg.h - cut_y, max(0, (-int(pbot) * self.cfg.h / sheight))))
out_img, out_bbox = blend_truth_mosaic(out_img, ai, truth.copy(), self.cfg.w, self.cfg.h, cut_x,
cut_y, i, left_shift, right_shift, top_shift, bot_shift)
out_bboxes.append(out_bbox)
# print(img_path)
if use_mixup == 3:
out_bboxes = np.concatenate(out_bboxes, axis=0)
out_bboxes1 = np.zeros([self.cfg.boxes, 5])
out_bboxes1[:min(out_bboxes.shape[0], self.cfg.boxes)] = out_bboxes[:min(out_bboxes.shape[0], self.cfg.boxes)]
return out_img, out_bboxes1
if __name__ == "__main__":
from cfg import Cfg
random.seed(2020)
np.random.seed(2020)
Cfg.dataset_dir = '/mnt/e/Dataset'
dataset = Yolo_dataset(Cfg.train_label, Cfg)
for i in range(100):
out_img, out_bboxes = dataset.__getitem__(i)
a = draw_box(out_img.copy(), out_bboxes.astype(np.int32))
plt.imshow(a.astype(np.int32))
plt.show()
#start training
#-b batch size (you should keep this low (2-4) for training to work properly)
#-s number of subdivisions in the batch, this was more relevant for the darknet framework
#-l learning rate
#-g direct training to the GPU device
#pretrained invoke the pretrained weights that we downloaded above
#classes - number of classes
#dir - where the training data is
#epoch - how long to train for
!python train.py -b 2 -s 1 -l 0.001 -g 0 -pretrained ./yolov4.conv.137.pth -classes {num_classes} -dir ./train -epochs 40
#looking at the weights that our model has saved during training
!ls checkpoints
#choose random test image
import os
test_images = [f for f in os.listdir('test') if f.endswith('.jpg')]
import random
img_path = "test/" + random.choice(test_images);
%%time
##change the epoch here to the one you would like to use for inference
!python models.py {num_classes} checkpoints/Yolov4_epoch35.pth {img_path} test/_classes.txt
#visualize inference
from IPython.display import Image
Image('predictions.jpg')
from google.colab import drive
drive.mount('/content/drive')
%cp /content/
| 0.425605 | 0.973215 |
```
import requests
import simplejson as json
import pandas as pd
import numpy as np
import os
import json
import math
from openpyxl import load_workbook
notebook_path = os.path.abspath("OLS matching.ipynb")
# Path to config file
config_path = os.path.join(os.path.dirname(notebook_path), "Data/config.json")
# Path to asctb formatted azimuth data
az_path = os.path.join(os.path.dirname(notebook_path), "Data/asctb_formatted_azimuth_data/")
with open(config_path) as config_file:
config= json.load(config_file)
config
# Fetch ASCTB sheet ID from config file
asctb_sheet_id = config["asctb_sid"]
# Fetch Azimuth Data
# AS/2 - Represents Author label
# AS/2/Label - Represents Ontology label
def fetch_azimuth(az_url,name):
if name+'.csv' in os.listdir(az_path):
azimuth_df= pd.read_csv (az_path+'/'+name+'.csv',skiprows=10)
else:
azimuth_df= pd.read_csv (az_url,skiprows=10)
azimuth_all_cts=[]
azimuth_all_label=[]
azimuth_all_label_author=[]
# Filter CT/ID column
azimuth_ct = azimuth_df.filter(regex=("ID"))
# Filter CT Label column
azimuth_label = azimuth_df.filter(regex=("AS/[0-9]/LABEL$"))
# Filter Author Label column
azimuth_label_author = azimuth_df.filter(regex=("AS/[0-9]$"))
# Flatten dataframe to list. Append CT/IDs in all annotation level to a single list and convert it to dataframe.
for col in azimuth_ct:
azimuth_all_cts.extend(azimuth_ct[col].tolist())
azimuth_all_cts=pd.DataFrame(azimuth_all_cts)
azimuth_all_cts.rename(columns = {0:"CT/ID"},inplace = True)
# Flatten dataframe to list. Append CT Label in all annotation level to a single list and convert it to dataframe.
for col in azimuth_label:
azimuth_all_label.extend(azimuth_label[col].tolist())
azimuth_all_label=pd.DataFrame(azimuth_all_label)
azimuth_all_label.rename(columns = {0:"CT/LABEL"},inplace = True)
# Flatten dataframe to list. Append Author Label in all annotation level to a single list and convert it to dataframe.
for col in azimuth_label_author:
azimuth_all_label_author.extend(azimuth_label_author[col].tolist())
azimuth_all_label_author=pd.DataFrame(azimuth_all_label_author)
azimuth_all_label_author.rename(columns = {0:"CT/LABEL.Author"},inplace = True)
# Column bind CT/ID , CT/Label and Author Label column
azimuth_all_cts_label=pd.concat([azimuth_all_cts,azimuth_all_label,azimuth_all_label_author],axis=1)
# Remove duplicate rows
azimuth_all_cts_label_unique=azimuth_all_cts_label.drop_duplicates()
azimuth_all_cts_label_unique.reset_index(drop=True, inplace=True)
# Return flattend dataframe before and after removing duplicates.
return azimuth_all_cts_label,azimuth_all_cts_label_unique
# Fetch Asctb Data
# CT/2 - Represents Author label
# CT/2/Label - Represents Ontology label
def fetch_asctb(sheet_id,asctb_sheet_name):
# Read ASCT+B organ table from google sheet
asctb_df = pd.read_csv(f"https://docs.google.com/spreadsheets/d/{sheet_id}/gviz/tq?tqx=out:csv&sheet={asctb_sheet_name}",skiprows=3)
# Filter CT/ID column
asctb_ct = asctb_df.filter(regex=("^CT.*ID$"))
# Filter CT Label column
asctb_label = asctb_df.filter(regex=("CT/[0-9]/LABEL$"))
# Filter Author Label column
asctb_label_author = asctb_df.filter(regex=("CT/[0-9]$"))
asctb_all_cts=[]
asctb_all_label=[]
asctb_all_label_author=[]
# Flatten dataframe to list. Append CT/IDs in all annotation level to a single list and convert it to dataframe.
for col in asctb_ct:
asctb_all_cts.extend(asctb_ct[col].tolist())
asctb_all_cts=pd.DataFrame(asctb_all_cts)
asctb_all_cts.rename(columns = {0:"CT/ID"},inplace = True)
# Flatten dataframe to list. Append CT Labels in all annotation level to a single list and convert it to dataframe.
for col in asctb_label:
asctb_all_label.extend(asctb_label[col].tolist())
asctb_all_label=pd.DataFrame(asctb_all_label)
asctb_all_label.rename(columns = {0:"CT/LABEL"},inplace = True)
# Flatten dataframe to list. Append Author Label in all annotation level to a single list and convert it to dataframe.
for col in asctb_label_author:
asctb_all_label_author.extend(asctb_label_author[col].tolist())
asctb_all_label_author=pd.DataFrame(asctb_all_label_author)
asctb_all_label_author.rename(columns = {0:"CT/LABEL.Author"},inplace = True)
# Column bind CT/ID , CT/Label and Author Label column
asctb_all_cts_label=pd.concat([asctb_all_cts,asctb_all_label,asctb_all_label_author],axis=1)
# Remove duplicate rows
asctb_all_cts_label_unique=asctb_all_cts_label.drop_duplicates()
asctb_all_cts_label_unique.reset_index(drop=True, inplace=True)
# Return flattend dataframe before and after removing duplicates.
return asctb_all_cts_label,asctb_all_cts_label_unique
# Check whether the Azimuth CT ID (cl_az) is present in ASCT+B. asctb_all_cts_label_unique is a dataframe that contains
# unique CT/ID, Label and author label for a reference organ.
# i and j are the index pointing to corresponding row in Azimuth and ASCT+B dataframe respectively.
# i is the row number of cl_az in Azimuth dataframe.
# j points to the row number in ASCT+B where cl_az matches in ASCT+B.
# az_row_all,asctb_row_all are global lists used to store these the row number of Azimuth, ASCT+B
# not_matching_all is a list storing index of Azimuth CT(cl_az) that does not match to any CT in ASCT+B.
# And if there is a match then we append azimuth row to the list az_row_all and corresponding ASCT+B row number
# to asctb_row_all
def check_in_asctb(cl_az,i,asctb_all_cts_label_unique,az_row_all,asctb_row_all,not_matching_all):
flag=0
for j in range(len(asctb_all_cts_label_unique['CT/ID'])):
if cl_az == asctb_all_cts_label_unique['CT/ID'][j]:
az_row_all.append(i)
asctb_row_all.append(j)
flag=1
if flag==0:
not_matching_all.append(i)
# Check for perfect match between ASCT+B and Azimuth tables and return Azimuth CT mismatches
# For example- If the cell CL:000158 is present in Azimuth and the same cell is present in ASCT+B table for a
# respective organ then we call it a perfect match i.e CTs that are present in both the data sets.
# Matching is performed based on CT/ID
def perfect_match_for_azimuthct_in_asctb(azimuth_all_cts_label_unique,asctb_all_cts_label_unique):
# az_row_all ,asctb_row_all List to store index number of ASCTB, Azimuth row number where a match is occuring
# not_matching_all list stores Azimuth row number where CT/ID match is not found
az_row_all=[]
asctb_row_all=[]
not_matching_all=[]
for i in range(len(azimuth_all_cts_label_unique['CT/ID'])):
if type(azimuth_all_cts_label_unique['CT/ID'][i])!=np.float64 and type(azimuth_all_cts_label_unique['CT/ID'][i])!=float and azimuth_all_cts_label_unique['CT/ID'][i][:3]=="CL:":
check_in_asctb(azimuth_all_cts_label_unique['CT/ID'][i],i,asctb_all_cts_label_unique,az_row_all,asctb_row_all,not_matching_all)
else:
not_matching_all.append(i)
# Subset Azimuth and ASCTB dataframe by rows were a match is found.
az_matches_all=azimuth_all_cts_label_unique.loc[az_row_all]
asctb_matches_all=asctb_all_cts_label_unique.loc[asctb_row_all]
az_matches_all.reset_index(drop=True,inplace=True)
asctb_matches_all.reset_index(drop=True,inplace=True)
az_matches_all.rename(columns = {"CT/ID":"AZ.CT/ID","CT/LABEL":"AZ.CT/LABEL","CT/LABEL.Author":"AZ.CT/LABEL.Author"},inplace = True)
asctb_matches_all.rename(columns = {"CT/ID":"ASCTB.CT/ID","CT/LABEL":"ASCTB.CT/LABEL","CT/LABEL.Author":"ASCTB.CT/LABEL.Author"},inplace = True)
# Cbind both dataframes to show the perfect matches found in one dataframe
perfect_matches_all=pd.concat([az_matches_all,asctb_matches_all],axis=1)
perfect_matches_all=perfect_matches_all.drop_duplicates()
perfect_matches_all.reset_index(drop=True, inplace=True)
az_mismatches_all=azimuth_all_cts_label_unique.loc[not_matching_all]
az_mismatches_all=az_mismatches_all.drop_duplicates()
az_mismatches_all.reset_index(drop=True, inplace=True)
# retrun Perfect matches and azimuth mismatches
return perfect_matches_all,az_mismatches_all
# Check whether the ASCTB CT ID (cl_asctb) is present in Azimuth. az_all_cts_label_unique is a dataframe that contains
# unique CT/ID, Label and author label for a reference organ.
# i and j are the index(row number) pointing to corresponding row in ASCT+B and Azimuth dataframe respectively.
# i is the row number of cl_az in Azimuth dataframe.
# j points to the row number in azimuth where cl_asctb matches in Azimuth.
# az_row_all,asctb_row_all are global lists used to store these the row number of Azimuth, ASCT+B
# not_matching_all is a list storing index of Asctb CT(cl_asctb) that does not match to any CT in Azimuth.
# And if there is a match then we append Asctb row to the list asctb_row_all and corresponding Azimuth row number
# to az_row_all
def check_in_az(cl_asctb,i,az_kidney_all_cts_label_unique,az_row,asctb_row,not_matching):
flag=0
for j in range(len(az_kidney_all_cts_label_unique['CT/ID'])):
if cl_asctb == az_kidney_all_cts_label_unique['CT/ID'][j]:
az_row.append(j)
asctb_row.append(i)
flag=1
break
if flag==0:
not_matching.append(i)
# Check for mismatches between ASCT+B and Azimuth tables and return Asctb CT mismatches
def perfect_match_for_asctbct_in_azimuth(azimuth_all_cts_label_unique,asctb_kidney_all_cts_label_unique):
az_row=[]
asctb_row=[]
not_matching=[]
for i in range(len(asctb_kidney_all_cts_label_unique['CT/ID'])):
if type(asctb_kidney_all_cts_label_unique['CT/ID'][i])!=np.float64 and type(asctb_kidney_all_cts_label_unique['CT/ID'][i])!=float and asctb_kidney_all_cts_label_unique['CT/ID'][i][:3]=="CL:":
check_in_az(asctb_kidney_all_cts_label_unique['CT/ID'][i],i,azimuth_all_cts_label_unique,az_row,asctb_row,not_matching)
else:
not_matching.append(i)
az_matches=azimuth_all_cts_label_unique.loc[az_row]
asctb_matches=asctb_kidney_all_cts_label_unique.loc[asctb_row]
az_matches.reset_index(drop=True,inplace=True)
asctb_matches.reset_index(drop=True,inplace=True)
az_matches.rename(columns = {"CT/ID":"AZ.CT/ID","CT/LABEL":"AZ.CT/LABEL","CT/LABEL.Author":"AZ.CT/LABEL.Author"},inplace = True)
asctb_matches.rename(columns = {"CT/ID":"ASCTB.CT/ID","CT/LABEL":"ASCTB.CT/LABEL","CT/LABEL.Author":"ASCTB.CT/LABEL.Author"},inplace = True)
perfect_matches=pd.concat([asctb_matches,az_matches],axis=1)
asctb_mismatches=asctb_kidney_all_cts_label_unique.loc[not_matching]
asctb_mismatches.reset_index(drop=True,inplace=True)
return asctb_mismatches
# Filter out CTs that are not present in Ontology
def incorrect_cts_ebi(mismatches):
found_in_ols=[]
not_found_in_ols=[]
already_checked={}
for i in range(len(mismatches['CT/ID'])):
if type(mismatches['CT/ID'][i])!=np.float64 and type(mismatches['CT/ID'][i])!=float:
cl_az=mismatches['CT/ID'][i].replace(":","_")
if cl_az in already_checked:
if already_checked[cl_az]=='Y':
print("Already checked for mismatch",cl_az)
found_in_ols.append(i)
else:
print("Already checked for mismatch",cl_az)
not_found_in_ols.append(i)
continue
print("Checking for mismatch",cl_az)
url = "http://www.ebi.ac.uk/ols/api/ontologies/cl/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2F"
payload={}
headers = {
'Accept': 'application/json'
}
response = requests.request("GET", url+cl_az, headers=headers, data=payload)
if response.status_code!=200:
not_found_in_ols.append(i)
already_checked[cl_az] = 'N'
else:
found_in_ols.append(i)
already_checked[cl_az] = 'Y'
else:
not_found_in_ols.append(i)
az_not_found_in_ols=mismatches.loc[not_found_in_ols]
az_not_found_in_ols.reset_index(drop=True,inplace=True)
az_mismatch_asctb_all = mismatches.loc[found_in_ols]
az_mismatch_asctb_all.reset_index(drop=True,inplace=True)
return az_not_found_in_ols,az_mismatch_asctb_all
# Concat hierarchy of traversed CTs in a string and add it to the dataframe
# Example - CL:0000084 (T cell) >> CL:0000542 (lymphocyte) >> CL:0000842 (mononuclear cell) >> CL:0000738 (leukocyte)
def add_hier(azimuth_matches_tree,hierarchy_list_all):
found_match=[]
hier=[]
len_hier=[]
asctb_ct=[]
asctb_label=[]
for i in range(len(hierarchy_list_all)):
if len(hierarchy_list_all[i])==3:
found_match.append("Yes")
asctb_ct.append(list(hierarchy_list_all[i][0][0].items())[-1][0])
asctb_label.append(list(hierarchy_list_all[i][0][0].items())[-1][1])
else:
found_match.append("No")
asctb_ct.append("Not found")
asctb_label.append("Not found")
len_hier.append((len(hierarchy_list_all[i][0][0])))
x=[]
for k,v in hierarchy_list_all[i][0][0].items():
abc=str(k + " (" + v + ")")
x.append(abc)
hier.append(x)
hier_1=[]
for item in hier:
hier_1.append(str(" >> ".join(item)))
hier_1=pd.DataFrame(hier_1,columns=["Hierarchy"])
found_match=pd.DataFrame(found_match,columns=["Match Found"])
len_hier=pd.DataFrame(len_hier,columns=["Hierarchy Length"])
asctb_ct = pd.DataFrame(asctb_ct,columns=["ASCTB.CT/ID"])
asctb_label = pd.DataFrame(asctb_label,columns=["ASCTB.CT/LABEL"])
azimuth_matches_tree.rename(columns = {"CT/ID":"AZ.CT/ID","CT/LABEL":"AZ.CT/LABEL","CT/LABEL.Author":"AZ.CT/LABEL.Author"},inplace = True)
df_hier=pd.concat([azimuth_matches_tree,found_match,asctb_ct,asctb_label,len_hier,hier_1],axis=1)
return df_hier
# Concat hierarchy of traversed CTs in a string and add it to the dataframe
# Example - CL:0000084 (T cell) >> CL:0000542 (lymphocyte) >> CL:0000842 (mononuclear cell) >> CL:0000738 (leukocyte)
def add_hier_1(azimuth_matches_tree,hierarchy_list_all):
found_match=[]
hier=[]
len_hier=[]
asctb_ct=[]
asctb_label=[]
for i in range(len(hierarchy_list_all)):
if len(hierarchy_list_all[i])==3:
found_match.append("Yes")
asctb_ct.append(list(hierarchy_list_all[i][0][0].items())[-1][0])
asctb_label.append(list(hierarchy_list_all[i][0][0].items())[-1][1])
else:
found_match.append("No")
asctb_ct.append("Not found")
asctb_label.append("Not found")
len_hier.append((len(hierarchy_list_all[i][0][0])))
x=[]
for k,v in hierarchy_list_all[i][0][0].items():
abc=str(k + " (" + v + ")")
x.append(abc)
hier.append(x)
hier_1=[]
for item in hier:
hier_1.append(str(" >> ".join(item)))
hier_1=pd.DataFrame(hier_1,columns=["Hierarchy"])
found_match=pd.DataFrame(found_match,columns=["Match Found"])
len_hier=pd.DataFrame(len_hier,columns=["Hierarchy Length"])
asctb_ct = pd.DataFrame(asctb_ct,columns=["AZ.CT/ID"])
asctb_label = pd.DataFrame(asctb_label,columns=["AZ.CT/LABEL"])
azimuth_matches_tree.rename(columns = {"CT/ID":"ASCTB.CT/ID","CT/LABEL":"ASCTB.CT/LABEL","CT/LABEL.Author":"ASCTB.CT/LABEL.Author"},inplace = True)
df_hier=pd.concat([azimuth_matches_tree,found_match,asctb_ct,asctb_label,len_hier,hier_1],axis=1)
return df_hier
# Check for ASCTB CT in Azimuth
def check_in_az_1(az_kidney_all_cts_label_unique,cl_asctb,i,all_links_asctb,hierarchy):
flag=0
for j in range(len(az_kidney_all_cts_label_unique['CT/ID'])):
if cl_asctb == az_kidney_all_cts_label_unique['CT/ID'][j]:
tree_match_asctb_1.append(i)
tree_match_az_1.append(j)
flag=1
hierarchy_list_1.append([[hierarchy],[i],[j]])
print(cl_asctb,az_kidney_all_cts_label_unique['CT/ID'][j],"Match found")
break
if flag==0:
print(cl_asctb)
ols_call(az_kidney_all_cts_label_unique,cl_asctb,i,all_links_asctb,hierarchy)
# Check for Azimuth CT in ASCTB
def check_in_asctb_1(asctb_all_cts_label_unique,cl_az,i,all_links_az,hierarchy_all):
flag=0
for j in range(len(asctb_all_cts_label_unique['CT/ID'])):
if cl_az == asctb_all_cts_label_unique['CT/ID'][j]:
tree_match_asctb_all.append(j)
tree_match_az_all.append(i)
flag=1
hierarchy_list_all.append([[hierarchy_all],[i],[j]])
print(cl_az,asctb_all_cts_label_unique['CT/ID'][j],"Match found")
if flag==0:
print(cl_az)
ols_call_1(asctb_all_cts_label_unique,cl_az,i,all_links_az,hierarchy_all)
# Recursively look for parent CTs of Azimuth/ASCTB CT until a CT match in found ASCTB/ Azimuth.
def ols_call_1(asctb_all_cts_label_unique,cl_az,i,all_links_az,hierarchy_all):
url = "http://www.ebi.ac.uk/ols/api/ontologies/cl/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2F"
payload={}
headers = {
'Accept': 'application/json'
}
#Azimuth
try:
response = requests.request("GET", all_links_az['parents']['href'], headers=headers, data=payload)
except:
print("No parent")
tree_not_match_all.append(i)
hierarchy_list_all.append([[hierarchy_all],[i]])
return
if response.status_code!=200:
print("Status !=200")
tree_not_match_all.append(i)
hierarchy_list_all.append([[hierarchy_all],[i]])
else:
result_az= json.loads(response.text)
all_links_az=result_az['_embedded']['terms'][0]['_links']
ct_id_az=result_az['_embedded']['terms'][0]['obo_id']
label_az=result_az['_embedded']['terms'][0]['label']
hierarchy_all[ct_id_az]=label_az
if ct_id_az[:-8:-1]=='0000000':
hierarchy_all[ct_id_az]= label_az
tree_not_match_all.append(i)
hierarchy_list_all.append([[hierarchy_all],[i]])
print(ct_id_az, "No match")
else:
hierarchy_all[ct_id_az]= label_az
check_in_asctb_1(asctb_all_cts_label_unique,ct_id_az,i,all_links_az,hierarchy_all)
# Recursively look for parent CTs of Azimuth/ASCTB CT until a CT match in found ASCTB/ Azimuth.
def ols_call(azimuth_all_cts_label_unique,cl_asctb,i,all_links_asctb,hierarchy):
url = "http://www.ebi.ac.uk/ols/api/ontologies/cl/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2F"
payload={}
headers = {
'Accept': 'application/json'
}
#ASCTB
try:
response = requests.request("GET", all_links_asctb['parents']['href'], headers=headers, data=payload)
except:
print("No parent")
tree_not_match_1.append(i)
hierarchy_list_1.append([[hierarchy],[i]])
return
if response.status_code!=200:
print("Status !=200")
tree_not_match_1.append(i)
hierarchy_list_1.append([[hierarchy],[i]])
else:
result_asctb= json.loads(response.text)
all_links_asctb=result_asctb['_embedded']['terms'][0]['_links']
ct_id_asctb=result_asctb['_embedded']['terms'][0]['obo_id']
label_asctb=result_asctb['_embedded']['terms'][0]['label']
hierarchy[ct_id_asctb]=label_asctb
if ct_id_asctb[:-8:-1]=='0000000':
hierarchy[ct_id_asctb]= label_asctb
tree_not_match_1.append(i)
hierarchy_list_1.append([[hierarchy],[i]])
print(ct_id_asctb, "No match")
else:
hierarchy[ct_id_asctb]= label_asctb
check_in_az_1(azimuth_all_cts_label_unique,ct_id_asctb,i,all_links_asctb,hierarchy)
# Traverse up Azimuth CTs to get a match in ASCTB
def tree_traversal_azimuth(az_mismatch_asctb_all,asctb_all_cts_label_unique):
url = "http://www.ebi.ac.uk/ols/api/ontologies/cl/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2F"
payload={}
headers = {
'Accept': 'application/json'
}
for i in range(len(az_mismatch_asctb_all['CT/ID'])):
hierarchy_all={}
cl_az=az_mismatch_asctb_all['CT/ID'][i]
print(cl_az,"Original")
hierarchy_all[cl_az]=az_mismatch_asctb_all['CT/LABEL'][i]
cl_az=cl_az.replace(":","_")
response = requests.request("GET", url+cl_az, headers=headers, data=payload)
if response.status_code!=200:
tree_not_match_all.append(i)
hierarchy_list_all.append([hierarchy_all,i])
else:
result_az= json.loads(response.text)
all_links_az=result_az['_embedded']['terms'][0]['_links']
ols_call_1(asctb_all_cts_label_unique,cl_az,i,all_links_az,hierarchy_all)
az_matches_tree_all=az_mismatch_asctb_all.loc[tree_match_az_all]
az_matches_tree_all.reset_index(drop=True,inplace=True)
asctb_matches_tree_all=asctb_all_cts_label_unique.loc[tree_match_asctb_all]
asctb_matches_tree_all.reset_index(drop=True,inplace=True)
az_matches_tree_all.rename(columns = {"CT/ID":"AZ.CT/ID","CT/LABEL":"AZ.CT/LABEL","CT/LABEL.Author":"AZ.CT/LABEL.Author"},inplace = True)
asctb_matches_tree_all.rename(columns = {"CT/ID":"ASCTB.CT/ID","CT/LABEL":"ASCTB.CT/LABEL","CT/LABEL.Author":"ASCTB.CT/LABEL.Author"},inplace = True)
az_final_matches =pd.concat([az_matches_tree_all,asctb_matches_tree_all],axis=1)
az_mismatches_final_all=az_mismatch_asctb_all.loc[tree_not_match_all]
az_mismatches_final_all.reset_index(drop=True,inplace=True)
az_mismatches_final_all.rename(columns = {"CT/ID":"AZ.CT/ID","CT/LABEL":"AZ.CT/LABEL","CT/LABEL.Author":"AZ.CT/LABEL.Author"},inplace = True)
return az_final_matches,az_mismatches_final_all
# Traverse up ASCTB CT to get a match in Azimuth
def tree_traversal_asctb(asctb_mismatch_az,azimuth_all_cts_label_unique):
url = "http://www.ebi.ac.uk/ols/api/ontologies/cl/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2F"
payload={}
#hierarchy={}
headers = {
'Accept': 'application/json'
}
for i in range(len(asctb_mismatch_az['CT/ID'])):
hierarchy={}
cl_asctb=asctb_mismatch_az['CT/ID'][i]
print(cl_asctb,"Original")
hierarchy[cl_asctb]=asctb_mismatch_az['CT/LABEL'][i]
cl_asctb=cl_asctb.replace(":","_")
response = requests.request("GET", url+cl_asctb, headers=headers, data=payload)
if response.status_code!=200:
tree_not_match_1.append(i)
hierarchy_list_1.append([hierarchy,i])
else:
result_asctb= json.loads(response.text)
all_links_asctb=result_asctb['_embedded']['terms'][0]['_links']
ols_call(azimuth_all_cts_label_unique,cl_asctb,i,all_links_asctb,hierarchy)
asctb_matches_tree=asctb_mismatch_az.loc[tree_match_asctb_1]
asctb_matches_tree.reset_index(drop=True,inplace=True)
az_matches_tree=azimuth_all_cts_label_unique.loc[tree_match_az_1]
az_matches_tree.reset_index(drop=True,inplace=True)
az_matches_tree.rename(columns = {"CT/ID":"AZ.CT/ID","CT/LABEL":"AZ.CT/LABEL","CT/LABEL.Author":"AZ.CT/LABEL.Author"},inplace = True)
asctb_matches_tree.rename(columns = {"CT/ID":"ASCTB.CT/ID","CT/LABEL":"ASCTB.CT/LABEL","CT/LABEL.Author":"ASCTB.CT/LABEL.Author"},inplace = True)
az_final_matches =pd.concat([asctb_matches_tree,az_matches_tree],axis=1)
asctb_mismatches_final=asctb_mismatch_az.loc[tree_not_match_1]
asctb_mismatches_final.reset_index(drop=True,inplace=True)
asctb_mismatches_final.rename(columns = {"CT/ID":"ASCTB.CT/ID","CT/LABEL":"ASCTB.CT/LABEL","CT/LABEL.Author":"ASCTB.CT/LABEL.Author"},inplace = True)
return az_final_matches,asctb_mismatches_final
# Set differene between dataframes
def uq_ct_df(incorrect_ct_ebi,cts_uq):
set_diff_df = pd.concat([cts_uq, incorrect_ct_ebi, incorrect_ct_ebi]).drop_duplicates(keep=False)
return(set_diff_df)
# Generate summary for all organs
summary = pd.DataFrame(columns=["Organ","Az_missing_cts","Asctb_missing_cts","Az_Asctb_perfect_matches","Az_unique_CT","ASCTB_unique_CT","Az_cts_not_matched","Az_pecentage_not_matched","Asctb_cts_not_matched","Asctb_percentage_not_matched","Az_incorrect_cts","Asctb_incorrect_cts","Az_ct_match_found_crosswalk","Asctb_ct_match_found_crosswalk"])
ct=0
loc=0
# Loop over all organ in reference file
for ref in config['references']:
name= ref['name']
asctb_sheet_name = ref['asctb_sheet_name']
az_url= ref['url']
# Fetch Azimuth data
azimuth_all_cts_label,azimuth_all_cts_label_unique = fetch_azimuth(az_url,name)
azimuth_all_cts_label = azimuth_all_cts_label.dropna(axis = 0, how = 'all', inplace = False)
azimuth_all_cts_label_unique = azimuth_all_cts_label_unique.dropna(axis = 0, how = 'all', inplace = False)
azimuth_all_cts_label_unique.reset_index(drop=True, inplace=True)
# Fetch ASCTB data
asctb_all_cts_label,asctb_all_cts_label_unique = fetch_asctb(asctb_sheet_id,asctb_sheet_name)
asctb_all_cts_label = asctb_all_cts_label.dropna(axis = 0, how = 'all', inplace = False)
asctb_all_cts_label_unique = asctb_all_cts_label_unique.dropna(axis = 0, how = 'all', inplace = False)
asctb_all_cts_label_unique.reset_index(drop=True, inplace=True)
# Number of Azimuth cts without IDs
azimuth_missing_cts = azimuth_all_cts_label_unique[azimuth_all_cts_label_unique['CT/ID'].isna() & ~azimuth_all_cts_label_unique['CT/LABEL'].isna()].reset_index(drop=True)
# Number of ASCTB cts without IDs
asctb_missing_cts = asctb_all_cts_label_unique[asctb_all_cts_label_unique['CT/ID'].isna() & ~asctb_all_cts_label_unique['CT/LABEL'].isna()].reset_index(drop=True)
# Perfect Match and Mismatch for Azimuth CT in ASCTB (AZ - ASCTB)
azimuth_perfect_matches,azimuth_mismatches=perfect_match_for_azimuthct_in_asctb(azimuth_all_cts_label_unique,asctb_all_cts_label_unique)
azimuth_perfect_matches.sort_values(by=['AZ.CT/ID','AZ.CT/LABEL.Author'],inplace=True)
# Mismatch for ASCTB CT in Azimuth (ASCTB - Azimuth)
asctb_mismatches=perfect_match_for_asctbct_in_azimuth(azimuth_all_cts_label_unique,asctb_all_cts_label_unique)
# Remove rows having missing CT/IDs
azimuth_mismatches = azimuth_mismatches[~azimuth_mismatches['CT/ID'].isna()].reset_index(drop=True)
asctb_mismatches = asctb_mismatches[~asctb_mismatches['CT/ID'].isna()].reset_index(drop=True)
# Incorrect CT ID in Azimuth (EBI)
incorrect_ct_azimuth_ebi, azimuth_mismatches_filtered=incorrect_cts_ebi(azimuth_mismatches)
incorrect_ct_azimuth_ebi_ct = incorrect_ct_azimuth_ebi.iloc[:,0].drop_duplicates()
incorrect_ct_azimuth_ebi_ct.reset_index(drop=True, inplace=True)
incorrect_ct_azimuth_ebi_ct.dropna(axis = 0, how = 'all', inplace = True)
incorrect_ct_azimuth_ebi_ct.reset_index(drop=True, inplace=True)
# Incorrect CT ID in Asctb (EBI)
incorrect_ct_asctb_ebi, asctb_mismatches_filtered=incorrect_cts_ebi(asctb_mismatches)
incorrect_ct_asctb_ebi_ct = incorrect_ct_asctb_ebi.iloc[:,0].drop_duplicates()
incorrect_ct_asctb_ebi_ct.reset_index(drop=True, inplace=True)
incorrect_ct_asctb_ebi_ct.dropna(axis = 0, how = 'all', inplace = True)
incorrect_ct_asctb_ebi_ct.reset_index(drop=True, inplace=True)
asctb_cts_uq=asctb_all_cts_label_unique.iloc[:,0].drop_duplicates()
asctb_cts_uq.reset_index(drop=True, inplace=True)
azimuth_cts_uq=azimuth_all_cts_label_unique.iloc[:,0].drop_duplicates()
azimuth_cts_uq.reset_index(drop=True, inplace=True)
azimuth_cts_uq.dropna(axis = 0, how = 'all', inplace = True)
asctb_cts_uq.dropna(axis = 0, how = 'all', inplace = True)
ct_asctb = uq_ct_df(incorrect_ct_asctb_ebi_ct,asctb_cts_uq)
ct_azimuth = uq_ct_df(incorrect_ct_azimuth_ebi_ct,azimuth_cts_uq)
# Tree traversal for matching Az to Asctb. Traversing up Azmiuth
tree_match_asctb_all=[]
tree_match_az_all=[]
tree_not_match_all=[]
hierarchy_list_all=[]
azimuth_matches_tree,azimuth_mismatches_tree = tree_traversal_azimuth(azimuth_mismatches_filtered,asctb_all_cts_label_unique)
azimuth_matches_tree_hier = add_hier(azimuth_mismatches_filtered,hierarchy_list_all)
# Tree traversal for matching Asctb to Az. Traversing up Asctb
tree_match_asctb_1=[]
tree_match_az_1=[]
tree_not_match_1=[]
hierarchy_list_1=[]
asctb_matches_tree_all,asctb_mismatch_tree = tree_traversal_asctb(asctb_mismatches_filtered,azimuth_all_cts_label_unique)
asctb_matches_tree_hier = add_hier_1(asctb_mismatches_filtered,hierarchy_list_1)
set_diff_df = pd.concat([azimuth_mismatches_tree['AZ.CT/ID'], asctb_matches_tree_all['AZ.CT/ID'].drop_duplicates().reset_index(drop=True), asctb_matches_tree_all['AZ.CT/ID'].drop_duplicates().reset_index(drop=True)]).drop_duplicates(keep=False)
idx=set_diff_df.index.tolist()
azimuth_mismatches_tree= azimuth_mismatches_tree.filter(items=idx,axis=0)
set_diff_df = pd.concat([asctb_mismatch_tree['ASCTB.CT/ID'], azimuth_matches_tree['ASCTB.CT/ID'].drop_duplicates().reset_index(drop=True), azimuth_matches_tree['ASCTB.CT/ID'].drop_duplicates().reset_index(drop=True)]).drop_duplicates(keep=False)
idx=set_diff_df.index.tolist()
asctb_mismatch_tree= asctb_mismatch_tree.filter(items=idx,axis=0)
# Drop all na
azimuth_missing_cts.dropna(axis = 0, how = 'all', inplace = True)
asctb_missing_cts.dropna(axis = 0, how = 'all', inplace = True)
azimuth_perfect_matches.dropna(axis = 0, how = 'all', inplace = True)
incorrect_ct_azimuth_ebi.dropna(axis = 0, how = 'all', inplace = True)
incorrect_ct_asctb_ebi.dropna(axis = 0, how = 'all', inplace = True)
azimuth_matches_tree_hier.dropna(axis = 0, how = 'all', inplace = True)
asctb_matches_tree_hier.dropna(axis = 0, how = 'all', inplace = True)
asctb_mismatch_tree.dropna(axis = 0, how = 'all', inplace = True)
azimuth_mismatches_tree.dropna(axis = 0, how = 'all', inplace = True)
azimuth_matches_tree_hier_y= azimuth_matches_tree_hier[azimuth_matches_tree_hier["Match Found"]=="Yes"]
asctb_matches_tree_hier_y= asctb_matches_tree_hier[asctb_matches_tree_hier["Match Found"]=="Yes"]
azimuth_matches_tree_hier_n= azimuth_matches_tree_hier[azimuth_matches_tree_hier["Match Found"]=="No"]
asctb_matches_tree_hier_n= asctb_matches_tree_hier[asctb_matches_tree_hier["Match Found"]=="No"]
azimuth_matches_tree_hier_len = sum(azimuth_matches_tree_hier["Match Found"]=="Yes")
asctb_matches_tree_hier_len = sum(asctb_matches_tree_hier["Match Found"]=="Yes")
# Create a list of final matches i.e perfect match and crosswalk match
a = azimuth_perfect_matches[['AZ.CT/LABEL','ASCTB.CT/LABEL']].drop_duplicates().reset_index(drop=True)
b = azimuth_matches_tree_hier_y[['AZ.CT/LABEL','ASCTB.CT/LABEL']].drop_duplicates().reset_index(drop=True)
c = asctb_matches_tree_hier_y[['AZ.CT/LABEL','ASCTB.CT/LABEL']].drop_duplicates().reset_index(drop=True)
final_match = pd.concat([a,b,c])
final_match.drop_duplicates().reset_index(drop=True,inplace=True)
print("---------------------------------------------------------------------------------------------------")
print(name)
#print("ASCTB Mismatches",len(asctb_mismatches))
#print("Azimuth Mismatches",len(azimuth_mismatches))
print("Unique Azimuth Total CT&Label",len(azimuth_all_cts_label_unique))
print("Unique ASCTB Total CT&Label",len(asctb_all_cts_label_unique))
print("Perfect Matches",len(azimuth_perfect_matches))
print("Ct Matches (Traversing up Azmiuth)",len(azimuth_matches_tree_hier_y))
print("Ct Matches (Traversing up Asctb)",len(asctb_matches_tree_hier_y))
print("Final mismatch (Azimuth)",len(azimuth_mismatches_tree))
print("Final mismatch (ASCTB)",len(asctb_mismatch_tree))
# Calculate percentage missing
az_percent_not_matching = len(azimuth_mismatches_tree)*100/(len(azimuth_all_cts_label_unique) if len(ct_azimuth)!=0 else 1)
asctb_percent_not_matching = len(asctb_mismatch_tree)*100/(len(asctb_all_cts_label_unique) if len(ct_asctb)!=0 else 1)
print("Percentage not matching (Azimuth)",az_percent_not_matching)
print("Percentage not matching (Asctb)",asctb_percent_not_matching)
print("---------------------------------------------------------------------------------------------------")
with pd.ExcelWriter("./Data/Final/"+name+ ".xlsx") as writer:
# use to_excel function and specify the sheet_name and index
# to store the dataframe in specified sheet
azimuth_missing_cts.to_excel(writer, sheet_name="Az_missing_cts", index=False)
asctb_missing_cts.to_excel(writer, sheet_name="Asctb_missing_cts", index=False)
azimuth_perfect_matches.to_excel(writer, sheet_name="Az_Asctb_cts_perfect_matches", index=False)
incorrect_ct_azimuth_ebi.to_excel(writer, sheet_name="Az_incorrect_cts", index=False)
incorrect_ct_asctb_ebi.to_excel(writer, sheet_name="Asctb_incorrect_cts", index=False)
azimuth_matches_tree_hier_y = azimuth_matches_tree_hier_y.sort_values(by=['AZ.CT/ID','AZ.CT/LABEL.Author'],inplace=False)
asctb_matches_tree_hier_y = asctb_matches_tree_hier_y.sort_values(by=['ASCTB.CT/ID','ASCTB.CT/LABEL.Author'],inplace=False)
azimuth_matches_tree_hier_y.to_excel(writer, sheet_name="Az_match_tree_crosswalk", index=False)
asctb_matches_tree_hier_y.to_excel(writer, sheet_name="Asctb_match_tree_crosswalk", index=False)
azimuth_mismatches_tree = azimuth_mismatches_tree.sort_values(by=['AZ.CT/ID','AZ.CT/LABEL.Author'],inplace=False)
asctb_mismatch_tree = asctb_mismatch_tree.sort_values(by=['ASCTB.CT/ID','ASCTB.CT/LABEL.Author'],inplace=False)
azimuth_mismatches_tree.to_excel(writer, sheet_name="Az_cts_mismatch_final", index=False)
asctb_mismatch_tree.to_excel(writer, sheet_name="Asctb_cts_mismatch_final", index=False)
final_match.to_excel(writer, sheet_name="Final_Matches", index=False)
summary.loc[loc] = [name]+ [len(azimuth_missing_cts)]+[len(asctb_missing_cts)]+[len(azimuth_perfect_matches)] + [len(azimuth_all_cts_label_unique)] +[len(asctb_all_cts_label_unique)] + [len(azimuth_mismatches_tree)]+ [az_percent_not_matching]+[len(asctb_mismatch_tree)]+[asctb_percent_not_matching] + [len(incorrect_ct_azimuth_ebi)] + [len(incorrect_ct_asctb_ebi)]+ [azimuth_matches_tree_hier_len] + [asctb_matches_tree_hier_len]
loc+=1
with pd.ExcelWriter("./Data/Final/summary.xlsx") as writer:
summary.to_excel(writer, sheet_name="Summary", index=False)
```
|
github_jupyter
|
import requests
import simplejson as json
import pandas as pd
import numpy as np
import os
import json
import math
from openpyxl import load_workbook
notebook_path = os.path.abspath("OLS matching.ipynb")
# Path to config file
config_path = os.path.join(os.path.dirname(notebook_path), "Data/config.json")
# Path to asctb formatted azimuth data
az_path = os.path.join(os.path.dirname(notebook_path), "Data/asctb_formatted_azimuth_data/")
with open(config_path) as config_file:
config= json.load(config_file)
config
# Fetch ASCTB sheet ID from config file
asctb_sheet_id = config["asctb_sid"]
# Fetch Azimuth Data
# AS/2 - Represents Author label
# AS/2/Label - Represents Ontology label
def fetch_azimuth(az_url,name):
if name+'.csv' in os.listdir(az_path):
azimuth_df= pd.read_csv (az_path+'/'+name+'.csv',skiprows=10)
else:
azimuth_df= pd.read_csv (az_url,skiprows=10)
azimuth_all_cts=[]
azimuth_all_label=[]
azimuth_all_label_author=[]
# Filter CT/ID column
azimuth_ct = azimuth_df.filter(regex=("ID"))
# Filter CT Label column
azimuth_label = azimuth_df.filter(regex=("AS/[0-9]/LABEL$"))
# Filter Author Label column
azimuth_label_author = azimuth_df.filter(regex=("AS/[0-9]$"))
# Flatten dataframe to list. Append CT/IDs in all annotation level to a single list and convert it to dataframe.
for col in azimuth_ct:
azimuth_all_cts.extend(azimuth_ct[col].tolist())
azimuth_all_cts=pd.DataFrame(azimuth_all_cts)
azimuth_all_cts.rename(columns = {0:"CT/ID"},inplace = True)
# Flatten dataframe to list. Append CT Label in all annotation level to a single list and convert it to dataframe.
for col in azimuth_label:
azimuth_all_label.extend(azimuth_label[col].tolist())
azimuth_all_label=pd.DataFrame(azimuth_all_label)
azimuth_all_label.rename(columns = {0:"CT/LABEL"},inplace = True)
# Flatten dataframe to list. Append Author Label in all annotation level to a single list and convert it to dataframe.
for col in azimuth_label_author:
azimuth_all_label_author.extend(azimuth_label_author[col].tolist())
azimuth_all_label_author=pd.DataFrame(azimuth_all_label_author)
azimuth_all_label_author.rename(columns = {0:"CT/LABEL.Author"},inplace = True)
# Column bind CT/ID , CT/Label and Author Label column
azimuth_all_cts_label=pd.concat([azimuth_all_cts,azimuth_all_label,azimuth_all_label_author],axis=1)
# Remove duplicate rows
azimuth_all_cts_label_unique=azimuth_all_cts_label.drop_duplicates()
azimuth_all_cts_label_unique.reset_index(drop=True, inplace=True)
# Return flattend dataframe before and after removing duplicates.
return azimuth_all_cts_label,azimuth_all_cts_label_unique
# Fetch Asctb Data
# CT/2 - Represents Author label
# CT/2/Label - Represents Ontology label
def fetch_asctb(sheet_id,asctb_sheet_name):
# Read ASCT+B organ table from google sheet
asctb_df = pd.read_csv(f"https://docs.google.com/spreadsheets/d/{sheet_id}/gviz/tq?tqx=out:csv&sheet={asctb_sheet_name}",skiprows=3)
# Filter CT/ID column
asctb_ct = asctb_df.filter(regex=("^CT.*ID$"))
# Filter CT Label column
asctb_label = asctb_df.filter(regex=("CT/[0-9]/LABEL$"))
# Filter Author Label column
asctb_label_author = asctb_df.filter(regex=("CT/[0-9]$"))
asctb_all_cts=[]
asctb_all_label=[]
asctb_all_label_author=[]
# Flatten dataframe to list. Append CT/IDs in all annotation level to a single list and convert it to dataframe.
for col in asctb_ct:
asctb_all_cts.extend(asctb_ct[col].tolist())
asctb_all_cts=pd.DataFrame(asctb_all_cts)
asctb_all_cts.rename(columns = {0:"CT/ID"},inplace = True)
# Flatten dataframe to list. Append CT Labels in all annotation level to a single list and convert it to dataframe.
for col in asctb_label:
asctb_all_label.extend(asctb_label[col].tolist())
asctb_all_label=pd.DataFrame(asctb_all_label)
asctb_all_label.rename(columns = {0:"CT/LABEL"},inplace = True)
# Flatten dataframe to list. Append Author Label in all annotation level to a single list and convert it to dataframe.
for col in asctb_label_author:
asctb_all_label_author.extend(asctb_label_author[col].tolist())
asctb_all_label_author=pd.DataFrame(asctb_all_label_author)
asctb_all_label_author.rename(columns = {0:"CT/LABEL.Author"},inplace = True)
# Column bind CT/ID , CT/Label and Author Label column
asctb_all_cts_label=pd.concat([asctb_all_cts,asctb_all_label,asctb_all_label_author],axis=1)
# Remove duplicate rows
asctb_all_cts_label_unique=asctb_all_cts_label.drop_duplicates()
asctb_all_cts_label_unique.reset_index(drop=True, inplace=True)
# Return flattend dataframe before and after removing duplicates.
return asctb_all_cts_label,asctb_all_cts_label_unique
# Check whether the Azimuth CT ID (cl_az) is present in ASCT+B. asctb_all_cts_label_unique is a dataframe that contains
# unique CT/ID, Label and author label for a reference organ.
# i and j are the index pointing to corresponding row in Azimuth and ASCT+B dataframe respectively.
# i is the row number of cl_az in Azimuth dataframe.
# j points to the row number in ASCT+B where cl_az matches in ASCT+B.
# az_row_all,asctb_row_all are global lists used to store these the row number of Azimuth, ASCT+B
# not_matching_all is a list storing index of Azimuth CT(cl_az) that does not match to any CT in ASCT+B.
# And if there is a match then we append azimuth row to the list az_row_all and corresponding ASCT+B row number
# to asctb_row_all
def check_in_asctb(cl_az,i,asctb_all_cts_label_unique,az_row_all,asctb_row_all,not_matching_all):
flag=0
for j in range(len(asctb_all_cts_label_unique['CT/ID'])):
if cl_az == asctb_all_cts_label_unique['CT/ID'][j]:
az_row_all.append(i)
asctb_row_all.append(j)
flag=1
if flag==0:
not_matching_all.append(i)
# Check for perfect match between ASCT+B and Azimuth tables and return Azimuth CT mismatches
# For example- If the cell CL:000158 is present in Azimuth and the same cell is present in ASCT+B table for a
# respective organ then we call it a perfect match i.e CTs that are present in both the data sets.
# Matching is performed based on CT/ID
def perfect_match_for_azimuthct_in_asctb(azimuth_all_cts_label_unique,asctb_all_cts_label_unique):
# az_row_all ,asctb_row_all List to store index number of ASCTB, Azimuth row number where a match is occuring
# not_matching_all list stores Azimuth row number where CT/ID match is not found
az_row_all=[]
asctb_row_all=[]
not_matching_all=[]
for i in range(len(azimuth_all_cts_label_unique['CT/ID'])):
if type(azimuth_all_cts_label_unique['CT/ID'][i])!=np.float64 and type(azimuth_all_cts_label_unique['CT/ID'][i])!=float and azimuth_all_cts_label_unique['CT/ID'][i][:3]=="CL:":
check_in_asctb(azimuth_all_cts_label_unique['CT/ID'][i],i,asctb_all_cts_label_unique,az_row_all,asctb_row_all,not_matching_all)
else:
not_matching_all.append(i)
# Subset Azimuth and ASCTB dataframe by rows were a match is found.
az_matches_all=azimuth_all_cts_label_unique.loc[az_row_all]
asctb_matches_all=asctb_all_cts_label_unique.loc[asctb_row_all]
az_matches_all.reset_index(drop=True,inplace=True)
asctb_matches_all.reset_index(drop=True,inplace=True)
az_matches_all.rename(columns = {"CT/ID":"AZ.CT/ID","CT/LABEL":"AZ.CT/LABEL","CT/LABEL.Author":"AZ.CT/LABEL.Author"},inplace = True)
asctb_matches_all.rename(columns = {"CT/ID":"ASCTB.CT/ID","CT/LABEL":"ASCTB.CT/LABEL","CT/LABEL.Author":"ASCTB.CT/LABEL.Author"},inplace = True)
# Cbind both dataframes to show the perfect matches found in one dataframe
perfect_matches_all=pd.concat([az_matches_all,asctb_matches_all],axis=1)
perfect_matches_all=perfect_matches_all.drop_duplicates()
perfect_matches_all.reset_index(drop=True, inplace=True)
az_mismatches_all=azimuth_all_cts_label_unique.loc[not_matching_all]
az_mismatches_all=az_mismatches_all.drop_duplicates()
az_mismatches_all.reset_index(drop=True, inplace=True)
# retrun Perfect matches and azimuth mismatches
return perfect_matches_all,az_mismatches_all
# Check whether the ASCTB CT ID (cl_asctb) is present in Azimuth. az_all_cts_label_unique is a dataframe that contains
# unique CT/ID, Label and author label for a reference organ.
# i and j are the index(row number) pointing to corresponding row in ASCT+B and Azimuth dataframe respectively.
# i is the row number of cl_az in Azimuth dataframe.
# j points to the row number in azimuth where cl_asctb matches in Azimuth.
# az_row_all,asctb_row_all are global lists used to store these the row number of Azimuth, ASCT+B
# not_matching_all is a list storing index of Asctb CT(cl_asctb) that does not match to any CT in Azimuth.
# And if there is a match then we append Asctb row to the list asctb_row_all and corresponding Azimuth row number
# to az_row_all
def check_in_az(cl_asctb,i,az_kidney_all_cts_label_unique,az_row,asctb_row,not_matching):
flag=0
for j in range(len(az_kidney_all_cts_label_unique['CT/ID'])):
if cl_asctb == az_kidney_all_cts_label_unique['CT/ID'][j]:
az_row.append(j)
asctb_row.append(i)
flag=1
break
if flag==0:
not_matching.append(i)
# Check for mismatches between ASCT+B and Azimuth tables and return Asctb CT mismatches
def perfect_match_for_asctbct_in_azimuth(azimuth_all_cts_label_unique,asctb_kidney_all_cts_label_unique):
az_row=[]
asctb_row=[]
not_matching=[]
for i in range(len(asctb_kidney_all_cts_label_unique['CT/ID'])):
if type(asctb_kidney_all_cts_label_unique['CT/ID'][i])!=np.float64 and type(asctb_kidney_all_cts_label_unique['CT/ID'][i])!=float and asctb_kidney_all_cts_label_unique['CT/ID'][i][:3]=="CL:":
check_in_az(asctb_kidney_all_cts_label_unique['CT/ID'][i],i,azimuth_all_cts_label_unique,az_row,asctb_row,not_matching)
else:
not_matching.append(i)
az_matches=azimuth_all_cts_label_unique.loc[az_row]
asctb_matches=asctb_kidney_all_cts_label_unique.loc[asctb_row]
az_matches.reset_index(drop=True,inplace=True)
asctb_matches.reset_index(drop=True,inplace=True)
az_matches.rename(columns = {"CT/ID":"AZ.CT/ID","CT/LABEL":"AZ.CT/LABEL","CT/LABEL.Author":"AZ.CT/LABEL.Author"},inplace = True)
asctb_matches.rename(columns = {"CT/ID":"ASCTB.CT/ID","CT/LABEL":"ASCTB.CT/LABEL","CT/LABEL.Author":"ASCTB.CT/LABEL.Author"},inplace = True)
perfect_matches=pd.concat([asctb_matches,az_matches],axis=1)
asctb_mismatches=asctb_kidney_all_cts_label_unique.loc[not_matching]
asctb_mismatches.reset_index(drop=True,inplace=True)
return asctb_mismatches
# Filter out CTs that are not present in Ontology
def incorrect_cts_ebi(mismatches):
found_in_ols=[]
not_found_in_ols=[]
already_checked={}
for i in range(len(mismatches['CT/ID'])):
if type(mismatches['CT/ID'][i])!=np.float64 and type(mismatches['CT/ID'][i])!=float:
cl_az=mismatches['CT/ID'][i].replace(":","_")
if cl_az in already_checked:
if already_checked[cl_az]=='Y':
print("Already checked for mismatch",cl_az)
found_in_ols.append(i)
else:
print("Already checked for mismatch",cl_az)
not_found_in_ols.append(i)
continue
print("Checking for mismatch",cl_az)
url = "http://www.ebi.ac.uk/ols/api/ontologies/cl/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2F"
payload={}
headers = {
'Accept': 'application/json'
}
response = requests.request("GET", url+cl_az, headers=headers, data=payload)
if response.status_code!=200:
not_found_in_ols.append(i)
already_checked[cl_az] = 'N'
else:
found_in_ols.append(i)
already_checked[cl_az] = 'Y'
else:
not_found_in_ols.append(i)
az_not_found_in_ols=mismatches.loc[not_found_in_ols]
az_not_found_in_ols.reset_index(drop=True,inplace=True)
az_mismatch_asctb_all = mismatches.loc[found_in_ols]
az_mismatch_asctb_all.reset_index(drop=True,inplace=True)
return az_not_found_in_ols,az_mismatch_asctb_all
# Concat hierarchy of traversed CTs in a string and add it to the dataframe
# Example - CL:0000084 (T cell) >> CL:0000542 (lymphocyte) >> CL:0000842 (mononuclear cell) >> CL:0000738 (leukocyte)
def add_hier(azimuth_matches_tree,hierarchy_list_all):
found_match=[]
hier=[]
len_hier=[]
asctb_ct=[]
asctb_label=[]
for i in range(len(hierarchy_list_all)):
if len(hierarchy_list_all[i])==3:
found_match.append("Yes")
asctb_ct.append(list(hierarchy_list_all[i][0][0].items())[-1][0])
asctb_label.append(list(hierarchy_list_all[i][0][0].items())[-1][1])
else:
found_match.append("No")
asctb_ct.append("Not found")
asctb_label.append("Not found")
len_hier.append((len(hierarchy_list_all[i][0][0])))
x=[]
for k,v in hierarchy_list_all[i][0][0].items():
abc=str(k + " (" + v + ")")
x.append(abc)
hier.append(x)
hier_1=[]
for item in hier:
hier_1.append(str(" >> ".join(item)))
hier_1=pd.DataFrame(hier_1,columns=["Hierarchy"])
found_match=pd.DataFrame(found_match,columns=["Match Found"])
len_hier=pd.DataFrame(len_hier,columns=["Hierarchy Length"])
asctb_ct = pd.DataFrame(asctb_ct,columns=["ASCTB.CT/ID"])
asctb_label = pd.DataFrame(asctb_label,columns=["ASCTB.CT/LABEL"])
azimuth_matches_tree.rename(columns = {"CT/ID":"AZ.CT/ID","CT/LABEL":"AZ.CT/LABEL","CT/LABEL.Author":"AZ.CT/LABEL.Author"},inplace = True)
df_hier=pd.concat([azimuth_matches_tree,found_match,asctb_ct,asctb_label,len_hier,hier_1],axis=1)
return df_hier
# Concat hierarchy of traversed CTs in a string and add it to the dataframe
# Example - CL:0000084 (T cell) >> CL:0000542 (lymphocyte) >> CL:0000842 (mononuclear cell) >> CL:0000738 (leukocyte)
def add_hier_1(azimuth_matches_tree,hierarchy_list_all):
found_match=[]
hier=[]
len_hier=[]
asctb_ct=[]
asctb_label=[]
for i in range(len(hierarchy_list_all)):
if len(hierarchy_list_all[i])==3:
found_match.append("Yes")
asctb_ct.append(list(hierarchy_list_all[i][0][0].items())[-1][0])
asctb_label.append(list(hierarchy_list_all[i][0][0].items())[-1][1])
else:
found_match.append("No")
asctb_ct.append("Not found")
asctb_label.append("Not found")
len_hier.append((len(hierarchy_list_all[i][0][0])))
x=[]
for k,v in hierarchy_list_all[i][0][0].items():
abc=str(k + " (" + v + ")")
x.append(abc)
hier.append(x)
hier_1=[]
for item in hier:
hier_1.append(str(" >> ".join(item)))
hier_1=pd.DataFrame(hier_1,columns=["Hierarchy"])
found_match=pd.DataFrame(found_match,columns=["Match Found"])
len_hier=pd.DataFrame(len_hier,columns=["Hierarchy Length"])
asctb_ct = pd.DataFrame(asctb_ct,columns=["AZ.CT/ID"])
asctb_label = pd.DataFrame(asctb_label,columns=["AZ.CT/LABEL"])
azimuth_matches_tree.rename(columns = {"CT/ID":"ASCTB.CT/ID","CT/LABEL":"ASCTB.CT/LABEL","CT/LABEL.Author":"ASCTB.CT/LABEL.Author"},inplace = True)
df_hier=pd.concat([azimuth_matches_tree,found_match,asctb_ct,asctb_label,len_hier,hier_1],axis=1)
return df_hier
# Check for ASCTB CT in Azimuth
def check_in_az_1(az_kidney_all_cts_label_unique,cl_asctb,i,all_links_asctb,hierarchy):
flag=0
for j in range(len(az_kidney_all_cts_label_unique['CT/ID'])):
if cl_asctb == az_kidney_all_cts_label_unique['CT/ID'][j]:
tree_match_asctb_1.append(i)
tree_match_az_1.append(j)
flag=1
hierarchy_list_1.append([[hierarchy],[i],[j]])
print(cl_asctb,az_kidney_all_cts_label_unique['CT/ID'][j],"Match found")
break
if flag==0:
print(cl_asctb)
ols_call(az_kidney_all_cts_label_unique,cl_asctb,i,all_links_asctb,hierarchy)
# Check for Azimuth CT in ASCTB
def check_in_asctb_1(asctb_all_cts_label_unique,cl_az,i,all_links_az,hierarchy_all):
flag=0
for j in range(len(asctb_all_cts_label_unique['CT/ID'])):
if cl_az == asctb_all_cts_label_unique['CT/ID'][j]:
tree_match_asctb_all.append(j)
tree_match_az_all.append(i)
flag=1
hierarchy_list_all.append([[hierarchy_all],[i],[j]])
print(cl_az,asctb_all_cts_label_unique['CT/ID'][j],"Match found")
if flag==0:
print(cl_az)
ols_call_1(asctb_all_cts_label_unique,cl_az,i,all_links_az,hierarchy_all)
# Recursively look for parent CTs of Azimuth/ASCTB CT until a CT match in found ASCTB/ Azimuth.
def ols_call_1(asctb_all_cts_label_unique,cl_az,i,all_links_az,hierarchy_all):
url = "http://www.ebi.ac.uk/ols/api/ontologies/cl/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2F"
payload={}
headers = {
'Accept': 'application/json'
}
#Azimuth
try:
response = requests.request("GET", all_links_az['parents']['href'], headers=headers, data=payload)
except:
print("No parent")
tree_not_match_all.append(i)
hierarchy_list_all.append([[hierarchy_all],[i]])
return
if response.status_code!=200:
print("Status !=200")
tree_not_match_all.append(i)
hierarchy_list_all.append([[hierarchy_all],[i]])
else:
result_az= json.loads(response.text)
all_links_az=result_az['_embedded']['terms'][0]['_links']
ct_id_az=result_az['_embedded']['terms'][0]['obo_id']
label_az=result_az['_embedded']['terms'][0]['label']
hierarchy_all[ct_id_az]=label_az
if ct_id_az[:-8:-1]=='0000000':
hierarchy_all[ct_id_az]= label_az
tree_not_match_all.append(i)
hierarchy_list_all.append([[hierarchy_all],[i]])
print(ct_id_az, "No match")
else:
hierarchy_all[ct_id_az]= label_az
check_in_asctb_1(asctb_all_cts_label_unique,ct_id_az,i,all_links_az,hierarchy_all)
# Recursively look for parent CTs of Azimuth/ASCTB CT until a CT match in found ASCTB/ Azimuth.
def ols_call(azimuth_all_cts_label_unique,cl_asctb,i,all_links_asctb,hierarchy):
url = "http://www.ebi.ac.uk/ols/api/ontologies/cl/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2F"
payload={}
headers = {
'Accept': 'application/json'
}
#ASCTB
try:
response = requests.request("GET", all_links_asctb['parents']['href'], headers=headers, data=payload)
except:
print("No parent")
tree_not_match_1.append(i)
hierarchy_list_1.append([[hierarchy],[i]])
return
if response.status_code!=200:
print("Status !=200")
tree_not_match_1.append(i)
hierarchy_list_1.append([[hierarchy],[i]])
else:
result_asctb= json.loads(response.text)
all_links_asctb=result_asctb['_embedded']['terms'][0]['_links']
ct_id_asctb=result_asctb['_embedded']['terms'][0]['obo_id']
label_asctb=result_asctb['_embedded']['terms'][0]['label']
hierarchy[ct_id_asctb]=label_asctb
if ct_id_asctb[:-8:-1]=='0000000':
hierarchy[ct_id_asctb]= label_asctb
tree_not_match_1.append(i)
hierarchy_list_1.append([[hierarchy],[i]])
print(ct_id_asctb, "No match")
else:
hierarchy[ct_id_asctb]= label_asctb
check_in_az_1(azimuth_all_cts_label_unique,ct_id_asctb,i,all_links_asctb,hierarchy)
# Traverse up Azimuth CTs to get a match in ASCTB
def tree_traversal_azimuth(az_mismatch_asctb_all,asctb_all_cts_label_unique):
url = "http://www.ebi.ac.uk/ols/api/ontologies/cl/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2F"
payload={}
headers = {
'Accept': 'application/json'
}
for i in range(len(az_mismatch_asctb_all['CT/ID'])):
hierarchy_all={}
cl_az=az_mismatch_asctb_all['CT/ID'][i]
print(cl_az,"Original")
hierarchy_all[cl_az]=az_mismatch_asctb_all['CT/LABEL'][i]
cl_az=cl_az.replace(":","_")
response = requests.request("GET", url+cl_az, headers=headers, data=payload)
if response.status_code!=200:
tree_not_match_all.append(i)
hierarchy_list_all.append([hierarchy_all,i])
else:
result_az= json.loads(response.text)
all_links_az=result_az['_embedded']['terms'][0]['_links']
ols_call_1(asctb_all_cts_label_unique,cl_az,i,all_links_az,hierarchy_all)
az_matches_tree_all=az_mismatch_asctb_all.loc[tree_match_az_all]
az_matches_tree_all.reset_index(drop=True,inplace=True)
asctb_matches_tree_all=asctb_all_cts_label_unique.loc[tree_match_asctb_all]
asctb_matches_tree_all.reset_index(drop=True,inplace=True)
az_matches_tree_all.rename(columns = {"CT/ID":"AZ.CT/ID","CT/LABEL":"AZ.CT/LABEL","CT/LABEL.Author":"AZ.CT/LABEL.Author"},inplace = True)
asctb_matches_tree_all.rename(columns = {"CT/ID":"ASCTB.CT/ID","CT/LABEL":"ASCTB.CT/LABEL","CT/LABEL.Author":"ASCTB.CT/LABEL.Author"},inplace = True)
az_final_matches =pd.concat([az_matches_tree_all,asctb_matches_tree_all],axis=1)
az_mismatches_final_all=az_mismatch_asctb_all.loc[tree_not_match_all]
az_mismatches_final_all.reset_index(drop=True,inplace=True)
az_mismatches_final_all.rename(columns = {"CT/ID":"AZ.CT/ID","CT/LABEL":"AZ.CT/LABEL","CT/LABEL.Author":"AZ.CT/LABEL.Author"},inplace = True)
return az_final_matches,az_mismatches_final_all
# Traverse up ASCTB CT to get a match in Azimuth
def tree_traversal_asctb(asctb_mismatch_az,azimuth_all_cts_label_unique):
url = "http://www.ebi.ac.uk/ols/api/ontologies/cl/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2F"
payload={}
#hierarchy={}
headers = {
'Accept': 'application/json'
}
for i in range(len(asctb_mismatch_az['CT/ID'])):
hierarchy={}
cl_asctb=asctb_mismatch_az['CT/ID'][i]
print(cl_asctb,"Original")
hierarchy[cl_asctb]=asctb_mismatch_az['CT/LABEL'][i]
cl_asctb=cl_asctb.replace(":","_")
response = requests.request("GET", url+cl_asctb, headers=headers, data=payload)
if response.status_code!=200:
tree_not_match_1.append(i)
hierarchy_list_1.append([hierarchy,i])
else:
result_asctb= json.loads(response.text)
all_links_asctb=result_asctb['_embedded']['terms'][0]['_links']
ols_call(azimuth_all_cts_label_unique,cl_asctb,i,all_links_asctb,hierarchy)
asctb_matches_tree=asctb_mismatch_az.loc[tree_match_asctb_1]
asctb_matches_tree.reset_index(drop=True,inplace=True)
az_matches_tree=azimuth_all_cts_label_unique.loc[tree_match_az_1]
az_matches_tree.reset_index(drop=True,inplace=True)
az_matches_tree.rename(columns = {"CT/ID":"AZ.CT/ID","CT/LABEL":"AZ.CT/LABEL","CT/LABEL.Author":"AZ.CT/LABEL.Author"},inplace = True)
asctb_matches_tree.rename(columns = {"CT/ID":"ASCTB.CT/ID","CT/LABEL":"ASCTB.CT/LABEL","CT/LABEL.Author":"ASCTB.CT/LABEL.Author"},inplace = True)
az_final_matches =pd.concat([asctb_matches_tree,az_matches_tree],axis=1)
asctb_mismatches_final=asctb_mismatch_az.loc[tree_not_match_1]
asctb_mismatches_final.reset_index(drop=True,inplace=True)
asctb_mismatches_final.rename(columns = {"CT/ID":"ASCTB.CT/ID","CT/LABEL":"ASCTB.CT/LABEL","CT/LABEL.Author":"ASCTB.CT/LABEL.Author"},inplace = True)
return az_final_matches,asctb_mismatches_final
# Set differene between dataframes
def uq_ct_df(incorrect_ct_ebi,cts_uq):
set_diff_df = pd.concat([cts_uq, incorrect_ct_ebi, incorrect_ct_ebi]).drop_duplicates(keep=False)
return(set_diff_df)
# Generate summary for all organs
summary = pd.DataFrame(columns=["Organ","Az_missing_cts","Asctb_missing_cts","Az_Asctb_perfect_matches","Az_unique_CT","ASCTB_unique_CT","Az_cts_not_matched","Az_pecentage_not_matched","Asctb_cts_not_matched","Asctb_percentage_not_matched","Az_incorrect_cts","Asctb_incorrect_cts","Az_ct_match_found_crosswalk","Asctb_ct_match_found_crosswalk"])
ct=0
loc=0
# Loop over all organ in reference file
for ref in config['references']:
name= ref['name']
asctb_sheet_name = ref['asctb_sheet_name']
az_url= ref['url']
# Fetch Azimuth data
azimuth_all_cts_label,azimuth_all_cts_label_unique = fetch_azimuth(az_url,name)
azimuth_all_cts_label = azimuth_all_cts_label.dropna(axis = 0, how = 'all', inplace = False)
azimuth_all_cts_label_unique = azimuth_all_cts_label_unique.dropna(axis = 0, how = 'all', inplace = False)
azimuth_all_cts_label_unique.reset_index(drop=True, inplace=True)
# Fetch ASCTB data
asctb_all_cts_label,asctb_all_cts_label_unique = fetch_asctb(asctb_sheet_id,asctb_sheet_name)
asctb_all_cts_label = asctb_all_cts_label.dropna(axis = 0, how = 'all', inplace = False)
asctb_all_cts_label_unique = asctb_all_cts_label_unique.dropna(axis = 0, how = 'all', inplace = False)
asctb_all_cts_label_unique.reset_index(drop=True, inplace=True)
# Number of Azimuth cts without IDs
azimuth_missing_cts = azimuth_all_cts_label_unique[azimuth_all_cts_label_unique['CT/ID'].isna() & ~azimuth_all_cts_label_unique['CT/LABEL'].isna()].reset_index(drop=True)
# Number of ASCTB cts without IDs
asctb_missing_cts = asctb_all_cts_label_unique[asctb_all_cts_label_unique['CT/ID'].isna() & ~asctb_all_cts_label_unique['CT/LABEL'].isna()].reset_index(drop=True)
# Perfect Match and Mismatch for Azimuth CT in ASCTB (AZ - ASCTB)
azimuth_perfect_matches,azimuth_mismatches=perfect_match_for_azimuthct_in_asctb(azimuth_all_cts_label_unique,asctb_all_cts_label_unique)
azimuth_perfect_matches.sort_values(by=['AZ.CT/ID','AZ.CT/LABEL.Author'],inplace=True)
# Mismatch for ASCTB CT in Azimuth (ASCTB - Azimuth)
asctb_mismatches=perfect_match_for_asctbct_in_azimuth(azimuth_all_cts_label_unique,asctb_all_cts_label_unique)
# Remove rows having missing CT/IDs
azimuth_mismatches = azimuth_mismatches[~azimuth_mismatches['CT/ID'].isna()].reset_index(drop=True)
asctb_mismatches = asctb_mismatches[~asctb_mismatches['CT/ID'].isna()].reset_index(drop=True)
# Incorrect CT ID in Azimuth (EBI)
incorrect_ct_azimuth_ebi, azimuth_mismatches_filtered=incorrect_cts_ebi(azimuth_mismatches)
incorrect_ct_azimuth_ebi_ct = incorrect_ct_azimuth_ebi.iloc[:,0].drop_duplicates()
incorrect_ct_azimuth_ebi_ct.reset_index(drop=True, inplace=True)
incorrect_ct_azimuth_ebi_ct.dropna(axis = 0, how = 'all', inplace = True)
incorrect_ct_azimuth_ebi_ct.reset_index(drop=True, inplace=True)
# Incorrect CT ID in Asctb (EBI)
incorrect_ct_asctb_ebi, asctb_mismatches_filtered=incorrect_cts_ebi(asctb_mismatches)
incorrect_ct_asctb_ebi_ct = incorrect_ct_asctb_ebi.iloc[:,0].drop_duplicates()
incorrect_ct_asctb_ebi_ct.reset_index(drop=True, inplace=True)
incorrect_ct_asctb_ebi_ct.dropna(axis = 0, how = 'all', inplace = True)
incorrect_ct_asctb_ebi_ct.reset_index(drop=True, inplace=True)
asctb_cts_uq=asctb_all_cts_label_unique.iloc[:,0].drop_duplicates()
asctb_cts_uq.reset_index(drop=True, inplace=True)
azimuth_cts_uq=azimuth_all_cts_label_unique.iloc[:,0].drop_duplicates()
azimuth_cts_uq.reset_index(drop=True, inplace=True)
azimuth_cts_uq.dropna(axis = 0, how = 'all', inplace = True)
asctb_cts_uq.dropna(axis = 0, how = 'all', inplace = True)
ct_asctb = uq_ct_df(incorrect_ct_asctb_ebi_ct,asctb_cts_uq)
ct_azimuth = uq_ct_df(incorrect_ct_azimuth_ebi_ct,azimuth_cts_uq)
# Tree traversal for matching Az to Asctb. Traversing up Azmiuth
tree_match_asctb_all=[]
tree_match_az_all=[]
tree_not_match_all=[]
hierarchy_list_all=[]
azimuth_matches_tree,azimuth_mismatches_tree = tree_traversal_azimuth(azimuth_mismatches_filtered,asctb_all_cts_label_unique)
azimuth_matches_tree_hier = add_hier(azimuth_mismatches_filtered,hierarchy_list_all)
# Tree traversal for matching Asctb to Az. Traversing up Asctb
tree_match_asctb_1=[]
tree_match_az_1=[]
tree_not_match_1=[]
hierarchy_list_1=[]
asctb_matches_tree_all,asctb_mismatch_tree = tree_traversal_asctb(asctb_mismatches_filtered,azimuth_all_cts_label_unique)
asctb_matches_tree_hier = add_hier_1(asctb_mismatches_filtered,hierarchy_list_1)
set_diff_df = pd.concat([azimuth_mismatches_tree['AZ.CT/ID'], asctb_matches_tree_all['AZ.CT/ID'].drop_duplicates().reset_index(drop=True), asctb_matches_tree_all['AZ.CT/ID'].drop_duplicates().reset_index(drop=True)]).drop_duplicates(keep=False)
idx=set_diff_df.index.tolist()
azimuth_mismatches_tree= azimuth_mismatches_tree.filter(items=idx,axis=0)
set_diff_df = pd.concat([asctb_mismatch_tree['ASCTB.CT/ID'], azimuth_matches_tree['ASCTB.CT/ID'].drop_duplicates().reset_index(drop=True), azimuth_matches_tree['ASCTB.CT/ID'].drop_duplicates().reset_index(drop=True)]).drop_duplicates(keep=False)
idx=set_diff_df.index.tolist()
asctb_mismatch_tree= asctb_mismatch_tree.filter(items=idx,axis=0)
# Drop all na
azimuth_missing_cts.dropna(axis = 0, how = 'all', inplace = True)
asctb_missing_cts.dropna(axis = 0, how = 'all', inplace = True)
azimuth_perfect_matches.dropna(axis = 0, how = 'all', inplace = True)
incorrect_ct_azimuth_ebi.dropna(axis = 0, how = 'all', inplace = True)
incorrect_ct_asctb_ebi.dropna(axis = 0, how = 'all', inplace = True)
azimuth_matches_tree_hier.dropna(axis = 0, how = 'all', inplace = True)
asctb_matches_tree_hier.dropna(axis = 0, how = 'all', inplace = True)
asctb_mismatch_tree.dropna(axis = 0, how = 'all', inplace = True)
azimuth_mismatches_tree.dropna(axis = 0, how = 'all', inplace = True)
azimuth_matches_tree_hier_y= azimuth_matches_tree_hier[azimuth_matches_tree_hier["Match Found"]=="Yes"]
asctb_matches_tree_hier_y= asctb_matches_tree_hier[asctb_matches_tree_hier["Match Found"]=="Yes"]
azimuth_matches_tree_hier_n= azimuth_matches_tree_hier[azimuth_matches_tree_hier["Match Found"]=="No"]
asctb_matches_tree_hier_n= asctb_matches_tree_hier[asctb_matches_tree_hier["Match Found"]=="No"]
azimuth_matches_tree_hier_len = sum(azimuth_matches_tree_hier["Match Found"]=="Yes")
asctb_matches_tree_hier_len = sum(asctb_matches_tree_hier["Match Found"]=="Yes")
# Create a list of final matches i.e perfect match and crosswalk match
a = azimuth_perfect_matches[['AZ.CT/LABEL','ASCTB.CT/LABEL']].drop_duplicates().reset_index(drop=True)
b = azimuth_matches_tree_hier_y[['AZ.CT/LABEL','ASCTB.CT/LABEL']].drop_duplicates().reset_index(drop=True)
c = asctb_matches_tree_hier_y[['AZ.CT/LABEL','ASCTB.CT/LABEL']].drop_duplicates().reset_index(drop=True)
final_match = pd.concat([a,b,c])
final_match.drop_duplicates().reset_index(drop=True,inplace=True)
print("---------------------------------------------------------------------------------------------------")
print(name)
#print("ASCTB Mismatches",len(asctb_mismatches))
#print("Azimuth Mismatches",len(azimuth_mismatches))
print("Unique Azimuth Total CT&Label",len(azimuth_all_cts_label_unique))
print("Unique ASCTB Total CT&Label",len(asctb_all_cts_label_unique))
print("Perfect Matches",len(azimuth_perfect_matches))
print("Ct Matches (Traversing up Azmiuth)",len(azimuth_matches_tree_hier_y))
print("Ct Matches (Traversing up Asctb)",len(asctb_matches_tree_hier_y))
print("Final mismatch (Azimuth)",len(azimuth_mismatches_tree))
print("Final mismatch (ASCTB)",len(asctb_mismatch_tree))
# Calculate percentage missing
az_percent_not_matching = len(azimuth_mismatches_tree)*100/(len(azimuth_all_cts_label_unique) if len(ct_azimuth)!=0 else 1)
asctb_percent_not_matching = len(asctb_mismatch_tree)*100/(len(asctb_all_cts_label_unique) if len(ct_asctb)!=0 else 1)
print("Percentage not matching (Azimuth)",az_percent_not_matching)
print("Percentage not matching (Asctb)",asctb_percent_not_matching)
print("---------------------------------------------------------------------------------------------------")
with pd.ExcelWriter("./Data/Final/"+name+ ".xlsx") as writer:
# use to_excel function and specify the sheet_name and index
# to store the dataframe in specified sheet
azimuth_missing_cts.to_excel(writer, sheet_name="Az_missing_cts", index=False)
asctb_missing_cts.to_excel(writer, sheet_name="Asctb_missing_cts", index=False)
azimuth_perfect_matches.to_excel(writer, sheet_name="Az_Asctb_cts_perfect_matches", index=False)
incorrect_ct_azimuth_ebi.to_excel(writer, sheet_name="Az_incorrect_cts", index=False)
incorrect_ct_asctb_ebi.to_excel(writer, sheet_name="Asctb_incorrect_cts", index=False)
azimuth_matches_tree_hier_y = azimuth_matches_tree_hier_y.sort_values(by=['AZ.CT/ID','AZ.CT/LABEL.Author'],inplace=False)
asctb_matches_tree_hier_y = asctb_matches_tree_hier_y.sort_values(by=['ASCTB.CT/ID','ASCTB.CT/LABEL.Author'],inplace=False)
azimuth_matches_tree_hier_y.to_excel(writer, sheet_name="Az_match_tree_crosswalk", index=False)
asctb_matches_tree_hier_y.to_excel(writer, sheet_name="Asctb_match_tree_crosswalk", index=False)
azimuth_mismatches_tree = azimuth_mismatches_tree.sort_values(by=['AZ.CT/ID','AZ.CT/LABEL.Author'],inplace=False)
asctb_mismatch_tree = asctb_mismatch_tree.sort_values(by=['ASCTB.CT/ID','ASCTB.CT/LABEL.Author'],inplace=False)
azimuth_mismatches_tree.to_excel(writer, sheet_name="Az_cts_mismatch_final", index=False)
asctb_mismatch_tree.to_excel(writer, sheet_name="Asctb_cts_mismatch_final", index=False)
final_match.to_excel(writer, sheet_name="Final_Matches", index=False)
summary.loc[loc] = [name]+ [len(azimuth_missing_cts)]+[len(asctb_missing_cts)]+[len(azimuth_perfect_matches)] + [len(azimuth_all_cts_label_unique)] +[len(asctb_all_cts_label_unique)] + [len(azimuth_mismatches_tree)]+ [az_percent_not_matching]+[len(asctb_mismatch_tree)]+[asctb_percent_not_matching] + [len(incorrect_ct_azimuth_ebi)] + [len(incorrect_ct_asctb_ebi)]+ [azimuth_matches_tree_hier_len] + [asctb_matches_tree_hier_len]
loc+=1
with pd.ExcelWriter("./Data/Final/summary.xlsx") as writer:
summary.to_excel(writer, sheet_name="Summary", index=False)
| 0.469277 | 0.216239 |
# CSCE 5290 - Final Project
## Pre-trained model evaluation (BART, T5)
Dan Waters ([email protected])
```
!pip install transformers
from transformers import pipeline
summarizer = pipeline("summarization")
# Get the data (not the one with the start tokens)
!gdown --id 17u3TvSpRq17mFVJ1pEKf6D9fYgBFj4lI
import pandas as pd
cnn_df = pd.read_csv('cnn_cleaned_test_10k.csv')
cnn_df = cnn_df[['text', 'summary']]
cnn_df.head(5)
article = cnn_df.iloc[0]['text']
article
```
### Write the results to files to compare with ROUGE.
Limit of 100 documents to conserve time.
```
# Summarize a bunch of articles
score_max_count = 100
with open('cnn_pred_bart_test.txt', 'w') as f_pred:
for i, c in cnn_df.iterrows():
text = cnn_df.iloc[i]['text']
s = summarizer(text, max_length=250, min_length=25, do_sample=False)
f_pred.write(s[0]['summary_text'])
if i >= score_max_count:
break
with open('cnn_ref_test.txt', 'w') as f_ref:
for i, c in cnn_df.iterrows():
summary = cnn_df.iloc[i]['summary']
f_ref.write(summary)
if i >= score_max_count:
break
!pip install rouge
from rouge import FilesRouge
import sys
fr = FilesRouge()
#sys.setrecursionlimit(50 * 50 + 10)
scores = fr.get_scores('cnn_pred_bart_test.txt', 'cnn_ref_test.txt', avg=True)
scores
```
### Compare with Google's T5 transformer (pretrained on some data including CNN/Dailymail)
see: https://huggingface.co/transformers/task_summary.html
```
from transformers import AutoModelForSeq2SeqLM, AutoTokenizer
model = AutoModelForSeq2SeqLM.from_pretrained("t5-base")
tokenizer = AutoTokenizer.from_pretrained("t5-base")
# T5 uses a max_length of 512 so we cut the article to 512 tokens.
inputs = tokenizer("summarize: " + article, return_tensors="pt", max_length=512, truncation=True)
outputs = model.generate(inputs["input_ids"],
max_length=250, min_length=25, length_penalty=2.0,
num_beams=4, early_stopping=True)
print(tokenizer.decode(outputs[0]))
# Summarize a bunch of articles
from tqdm import tqdm
score_max_count = 100
with open('cnn_pred_t5_test.txt', 'w') as f_pred:
for i, c in tqdm(cnn_df.iterrows(), total = cnn_df.shape[0]):
text = cnn_df.iloc[i]['text']
inputs = tokenizer("summarize: " + text, return_tensors="pt",
max_length=512, truncation=True)
outputs = model.generate(inputs["input_ids"],
max_length=250, min_length=25, length_penalty=2.0,
num_beams=4, early_stopping=True)
s = tokenizer.decode(outputs[0]).replace('<pad>', '').replace('</s>', '')
f_pred.write(s)
if i >= score_max_count:
break
scores = fr.get_scores('cnn_pred_t5_test.txt', 'cnn_ref_test.txt', avg=True)
scores
```
|
github_jupyter
|
!pip install transformers
from transformers import pipeline
summarizer = pipeline("summarization")
# Get the data (not the one with the start tokens)
!gdown --id 17u3TvSpRq17mFVJ1pEKf6D9fYgBFj4lI
import pandas as pd
cnn_df = pd.read_csv('cnn_cleaned_test_10k.csv')
cnn_df = cnn_df[['text', 'summary']]
cnn_df.head(5)
article = cnn_df.iloc[0]['text']
article
# Summarize a bunch of articles
score_max_count = 100
with open('cnn_pred_bart_test.txt', 'w') as f_pred:
for i, c in cnn_df.iterrows():
text = cnn_df.iloc[i]['text']
s = summarizer(text, max_length=250, min_length=25, do_sample=False)
f_pred.write(s[0]['summary_text'])
if i >= score_max_count:
break
with open('cnn_ref_test.txt', 'w') as f_ref:
for i, c in cnn_df.iterrows():
summary = cnn_df.iloc[i]['summary']
f_ref.write(summary)
if i >= score_max_count:
break
!pip install rouge
from rouge import FilesRouge
import sys
fr = FilesRouge()
#sys.setrecursionlimit(50 * 50 + 10)
scores = fr.get_scores('cnn_pred_bart_test.txt', 'cnn_ref_test.txt', avg=True)
scores
from transformers import AutoModelForSeq2SeqLM, AutoTokenizer
model = AutoModelForSeq2SeqLM.from_pretrained("t5-base")
tokenizer = AutoTokenizer.from_pretrained("t5-base")
# T5 uses a max_length of 512 so we cut the article to 512 tokens.
inputs = tokenizer("summarize: " + article, return_tensors="pt", max_length=512, truncation=True)
outputs = model.generate(inputs["input_ids"],
max_length=250, min_length=25, length_penalty=2.0,
num_beams=4, early_stopping=True)
print(tokenizer.decode(outputs[0]))
# Summarize a bunch of articles
from tqdm import tqdm
score_max_count = 100
with open('cnn_pred_t5_test.txt', 'w') as f_pred:
for i, c in tqdm(cnn_df.iterrows(), total = cnn_df.shape[0]):
text = cnn_df.iloc[i]['text']
inputs = tokenizer("summarize: " + text, return_tensors="pt",
max_length=512, truncation=True)
outputs = model.generate(inputs["input_ids"],
max_length=250, min_length=25, length_penalty=2.0,
num_beams=4, early_stopping=True)
s = tokenizer.decode(outputs[0]).replace('<pad>', '').replace('</s>', '')
f_pred.write(s)
if i >= score_max_count:
break
scores = fr.get_scores('cnn_pred_t5_test.txt', 'cnn_ref_test.txt', avg=True)
scores
| 0.443841 | 0.752581 |
```
import numpy as np
import scipy.stats as stats
from matplotlib import pyplot as plt
x = np.arange(0, 1, 0.01)
uno = np.full(x.shape,1)
low = np.full(x.shape,0.01)
ph = 0.5
#plt.rcParams['figure.dpi'] = 72
plt.rcParams['figure.figsize'] = [10, 10]
plt.xticks([0,1 / 6, 0.25, 3/8, 0.5, 5/8, 0.75, 5/6,1])
plt.plot(x,uno)
plt.plot(x,low)
sig = 0.17
wave = stats.norm(scale = sig)
n100 = wave.pdf(x - ph) / wave.pdf(0)
plt.plot(x,n100)
sig = 0.125
wave = stats.norm(scale = sig)
n23 = wave.pdf(x - ph) / wave.pdf(0)
plt.plot(x,n23)
sig = 0.095
wave = stats.norm(scale = sig)
n12 = wave.pdf(x - ph) / wave.pdf(0)
plt.plot(x,n12)
sig = 0.05
wave = stats.norm(scale = sig)
n14 = wave.pdf(x - ph) / wave.pdf(0)
plt.plot(x,n14)
# http://elki.dbs.ifi.lmu.de/browser/elki/elki-core-math/src/main/java/de/lmu/ifi/dbs/elki/math/statistics/distribution/SkewGeneralizedNormalDistribution.java
def skewed(x, sigma, skew):
if (skew == 0):
return stats.norm(scale = sigma).pdf(x)
x = x / sigma
arg = -skew * x
if (arg <= -1):
return 0
y = -np.log1p(-skew * x) / skew
ONE_BY_SQRTTWOPI = 1. / np.sqrt(2. * np.pi)
return ONE_BY_SQRTTWOPI / sigma * np.exp(-0.5 * y * y) / (1 - skew * x)
plt.plot(x,uno)
plt.plot(x,low)
plt.plot(x,n100)
ph = 0.68
sigma = 0.4
skew = 1.2
wave = np.vectorize( lambda x: skewed(x, sigma, skew))
sk100 = wave(x - ph)
sk100 = sk100 -sk100.min()
sk100 = sk100 / sk100.max()
sk100 = np.roll(sk100,-40)
plt.plot(x,sk100)
ph = 0.76
sigma = 0.2
skew = 0.8
wave = np.vectorize( lambda x: skewed(x, sigma, skew))
sk100 = wave(x - ph)
sk100 = sk100 -sk100.min()
sk100 = sk100 / sk100.max()
sk100 = np.roll(sk100,-30)
plt.plot(x,sk100)
ph = 0.67
sigma = 0.18
skew = 0.4
wave = np.vectorize( lambda x: skewed(x, sigma, skew))
sk100 = wave(x - ph)
sk100 = sk100 -sk100.min()
sk100 = sk100 / sk100.max()
sk100 = np.roll(sk100,-20)
plt.plot(x,sk100)
#sk100
plt.plot(x,uno)
plt.plot(x,low)
plt.plot(x,n23)
ph = 0.72
sigma = 0.34
skew = 1.2
wave = np.vectorize( lambda x: skewed(x, sigma, skew))
sk100 = wave(x - ph)
sk100 = sk100 -sk100.min()
sk100 = sk100 / sk100.max()
sk100 = np.roll(sk100,-40)
plt.plot(x,sk100)
ph = 0.79
sigma = 0.17
skew = 0.8
wave = np.vectorize( lambda x: skewed(x, sigma, skew))
sk100 = wave(x - ph)
sk100 = sk100 -sk100.min()
sk100 = sk100 / sk100.max()
sk100 = np.roll(sk100,-35)
plt.plot(x,sk100)
ph = 0.76
sigma = 0.14
skew = 0.4
wave = np.vectorize( lambda x: skewed(x, sigma, skew))
sk100 = wave(x - ph)
sk100 = sk100 -sk100.min()
sk100 = sk100 / sk100.max()
sk100 = np.roll(sk100,-30)
plt.plot(x,sk100)
#sk100
plt.plot(x,uno)
plt.plot(x,low)
plt.plot(x,n12)
ph = 0.8
sigma = 0.25
skew = 1.2
wave = np.vectorize( lambda x: skewed(x, sigma, skew))
sk100 = wave(x - ph)
sk100 = sk100 -sk100.min()
sk100 = sk100 / sk100.max()
sk100 = np.roll(sk100,-45)
plt.plot(x,sk100)
ph = 0.84
sigma = 0.14
skew = 0.8
wave = np.vectorize( lambda x: skewed(x, sigma, skew))
sk100 = wave(x - ph)
sk100 = sk100 -sk100.min()
sk100 = sk100 / sk100.max()
sk100 = np.roll(sk100,-40)
plt.plot(x,sk100)
ph = 0.76
sigma = 0.1
skew = 0.4
wave = np.vectorize( lambda x: skewed(x, sigma, skew))
sk100 = wave(x - ph)
sk100 = sk100 -sk100.min()
sk100 = sk100 / sk100.max()
sk100 = np.roll(sk100,-30)
plt.plot(x,sk100)
#sk100
plt.plot(x,uno)
plt.plot(x,low)
plt.plot(x,n100)
ph = .04
shape = 10.1
scale = 0.05
wave = stats.gamma(a = shape, scale = scale, loc = 0 )
ga100 = wave.pdf(x - ph)
ga100 = ga100 / ga100.max()
plt.plot(x,ga100)
ph = 0.25
shape = 5.1
scale = 0.06
wave = stats.gamma(a = shape, scale = scale, loc = 0 )
ga200 = wave.pdf(x - ph)
ga200 = ga200 / ga200.max()
plt.plot(x,ga200)
#COS
xt=[0.0, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1, 0.11, 0.12, 0.13, 0.14, 0.15, 0.16, 0.17, 0.18, 0.19, 0.2, 0.21, 0.22, 0.23, 0.24, 0.25, 0.26, 0.27, 0.28, 0.29, 0.3, 0.31, 0.32, 0.33, 0.34, 0.35, 0.36, 0.37, 0.38, 0.39, 0.4, 0.41, 0.42, 0.43, 0.44, 0.45, 0.46, 0.47, 0.48, 0.49, 0.5, 0.51, 0.52, 0.53, 0.54, 0.55, 0.56, 0.57, 0.58, 0.59, 0.6, 0.61, 0.62, 0.63, 0.64, 0.65, 0.66, 0.67, 0.68, 0.69, 0.7, 0.71, 0.72, 0.73, 0.74, 0.75, 0.76, 0.77, 0.78, 0.79, 0.8, 0.81, 0.82, 0.83, 0.84, 0.85, 0.86, 0.87, 0.88, 0.89, 0.9, 0.91, 0.92, 0.93, 0.94, 0.95, 0.96, 0.97, 0.98, 0.99]
yt=[1.0, 0.9990133642141358, 0.996057350657239, 0.9911436253643444, 0.9842915805643155, 0.9755282581475768, 0.9648882429441257, 0.9524135262330098, 0.9381533400219317, 0.9221639627510075, 0.9045084971874737, 0.8852566213878946, 0.8644843137107058, 0.8422735529643444, 0.8187119948743449, 0.7938926261462366, 0.7679133974894983, 0.7408768370508576, 0.7128896457825363, 0.6840622763423391, 0.6545084971874737, 0.6243449435824275, 0.5936906572928624, 0.5626666167821521, 0.5313952597646567, 0.5, 0.4686047402353433, 0.4373333832178478, 0.4063093427071376, 0.3756550564175727, 0.34549150281252633, 0.31593772365766104, 0.28711035421746367, 0.2591231629491423, 0.23208660251050156, 0.2061073738537635, 0.18128800512565513, 0.15772644703565564, 0.13551568628929433, 0.11474337861210543, 0.09549150281252633, 0.07783603724899257, 0.06184665997806832, 0.04758647376699032, 0.035111757055874326, 0.024471741852423234, 0.015708419435684462, 0.008856374635655695, 0.0039426493427611176, 9.866357858642205E-4, 0.0, 9.866357858642205E-4, 0.0039426493427611176, 0.008856374635655695, 0.015708419435684517, 0.02447174185242329, 0.03511175705587444, 0.04758647376699021, 0.06184665997806821, 0.07783603724899246, 0.09549150281252633, 0.11474337861210543, 0.13551568628929433, 0.15772644703565564, 0.18128800512565513, 0.2061073738537635, 0.23208660251050178, 0.2591231629491425, 0.28711035421746384, 0.31593772365766093, 0.3454915028125262, 0.3756550564175726, 0.4063093427071376, 0.4373333832178478, 0.4686047402353433, 0.5, 0.5313952597646567, 0.5626666167821522, 0.5936906572928624, 0.6243449435824275, 0.6545084971874738, 0.6840622763423392, 0.7128896457825362, 0.7408768370508576, 0.7679133974894983, 0.7938926261462365, 0.8187119948743449, 0.8422735529643444, 0.8644843137107058, 0.8852566213878947, 0.9045084971874737, 0.9221639627510076, 0.9381533400219318, 0.9524135262330098, 0.9648882429441257, 0.9755282581475768, 0.9842915805643155, 0.9911436253643443, 0.9960573506572389, 0.9990133642141358]
plt.plot(xt, yt)
#WIDE_PEAK no fix min
# case NONE: return new GaussianWaveform(0.203125, phase, false);
# /*
# case LOW: return new SkewedGaussWaveform(0.21897347065651573, skew.skew, phase, false);
# case MID: return new SkewedGaussWaveform(0.2784780878770199, skew.skew, phase, false);
# case HIGH: return new SkewedGaussWaveform(0.415636603046377, skew.skew, phase, false);
xt=[0.0, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1, 0.11, 0.12, 0.13, 0.14, 0.15, 0.16, 0.17, 0.18, 0.19, 0.2, 0.21, 0.22, 0.23, 0.24, 0.25, 0.26, 0.27, 0.28, 0.29, 0.3, 0.31, 0.32, 0.33, 0.34, 0.35, 0.36, 0.37, 0.38, 0.39, 0.4, 0.41, 0.42, 0.43, 0.44, 0.45, 0.46, 0.47, 0.48, 0.49, 0.5, 0.51, 0.52, 0.53, 0.54, 0.55, 0.56, 0.57, 0.58, 0.59, 0.6, 0.61, 0.62, 0.63, 0.64, 0.65, 0.66, 0.67, 0.68, 0.69, 0.7, 0.71, 0.72, 0.73, 0.74, 0.75, 0.76, 0.77, 0.78, 0.79, 0.8, 0.81, 0.82, 0.83, 0.84, 0.85, 0.86, 0.87, 0.88, 0.89, 0.9, 0.91, 0.92, 0.93, 0.94, 0.95, 0.96, 0.97, 0.98, 0.99]
yt=[1.0, 0.9987888996551672, 0.9951643921014979, 0.9891527514554127, 0.9807974152891648, 0.9701584619536344, 0.9573118912015268, 0.9423487201601802, 0.9253739096952884, 0.9065051388774692, 0.8858714475589475, 0.8636117689435711, 0.8398733754565454, 0.8148102621687294, 0.7885814924939011, 0.7613495308577245, 0.7332785865473379, 0.7045329920144862, 0.6752756375567601, 0.6456664825833416, 0.6158611616335171, 0.5860097010137122, 0.5562553594115757, 0.5267336031957593, 0.49757122438015355, 0.46888560648317157, 0.44078414080532446, 0.4133637930369936, 0.3867108176429706, 0.36090061519481487, 0.3359977257732059, 0.31205594976959017, 0.2891185859010446, 0.26721877502808633, 0.2463799374380964, 0.22661629062572547, 0.20793343425796706, 0.19032898894126954, 0.17379327559160265, 0.1583100226219387, 0.14385708877784129, 0.1304071902410655, 0.11792862155203371, 0.10638596094299341, 0.09574075179308944, 0.0859521530840816, 0.076977552922306, 0.06877314037233147, 0.06129443199693493, 0.054496750595930094, 0.048335654665708876, 0.054496750595930094, 0.06129443199693493, 0.06877314037233147, 0.07697755292230607, 0.08595215308408165, 0.09574075179308947, 0.10638596094299331, 0.11792862155203365, 0.13040719024106545, 0.14385708877784129, 0.1583100226219387, 0.17379327559160265, 0.19032898894126954, 0.20793343425796706, 0.22661629062572547, 0.24637993743809652, 0.26721877502808644, 0.2891185859010447, 0.3120559497695901, 0.33599772577320586, 0.3609006151948148, 0.3867108176429706, 0.4133637930369936, 0.44078414080532446, 0.46888560648317157, 0.49757122438015355, 0.5267336031957595, 0.5562553594115758, 0.5860097010137123, 0.6158611616335173, 0.6456664825833417, 0.67527563755676, 0.704532992014486, 0.7332785865473377, 0.7613495308577244, 0.7885814924939011, 0.8148102621687294, 0.8398733754565454, 0.8636117689435712, 0.8858714475589475, 0.9065051388774692, 0.9253739096952885, 0.9423487201601803, 0.9573118912015268, 0.9701584619536343, 0.9807974152891648, 0.9891527514554127, 0.9951643921014979, 0.9987888996551672]
plt.plot(xt, yt)
yt=[1.0, 0.9985337870668837, 0.9940178136391471, 0.9862879138460792, 0.9751988723702859, 0.9606293631911291, 0.9424872367484814, 0.9207150779901252, 0.8952959269393653, 0.8662590179489144, 0.8336853540417393, 0.7977128894808132, 0.758541048362926, 0.7164342617787033, 0.6717241640662659, 0.6248100542016115, 0.5761572070119776, 0.5262926176620537, 0.47579779010002304, 0.4252982453321619, 0.37544953855460284, 0.3269197449059852, 0.28036860948936987, 0.2364238615847212, 0.19565556141136928, 0.1585497648119985, 0.12548322544227722, 0.09670125405191868, 0.0723011459542647, 0.06896702759489831, 0.07243961456383377, 0.0760841324091454, 0.07990856974616012, 0.08392122294723851, 0.08813070071714943, 0.09254592777989244, 0.09717614752876519, 0.10203092347562741, 0.10712013931816212, 0.11245399742539441, 0.11804301552174556, 0.12389802132839711, 0.13003014489767012, 0.13645080835143822, 0.143171712708246, 0.15020482145578343, 0.1575623404956575, 0.16525669405603718, 0.1733004961347555, 0.18170651700092888, 0.19048764424721906, 0.19965683784767513, 0.20922707863790907, 0.2192113095954468, 0.22962236925886825, 0.24047291658526712, 0.25177534650720934, 0.2635416954134839, 0.2757835357433708, 0.28851185885293595, 0.3017369452852588, 0.31546822155591203, 0.3297141025522071, 0.3444818186416254, 0.3597772265938246, 0.37560460344427626, 0.3919664224689946, 0.40886311050248597, 0.4262927859188711, 0.4442509767136279, 0.4627303182755495, 0.4817202306309141, 0.501206575180716, 0.5211712912439482, 0.5415920130727792, 0.5624416684271292, 0.5836880602952406, 0.6052934339324837, 0.6272140320723614, 0.6493996419512129, 0.671793138691142, 0.6943300306135921, 0.716938013217347, 0.7395365398568545, 0.7620364186049235, 0.7843394463805416, 0.8063380931662674, 0.8279152510237264, 0.8489440646266927, 0.8692878621469449, 0.8888002075155294, 0.907325097294831, 0.9246973275721614, 0.940743058340922, 0.955280604665043, 0.968121485393274, 0.9790717611376433, 0.9879336934552613, 0.9945077564356877, 0.9985950299162479]
plt.plot(xt, yt)
yt=[1.0, 0.9975510364251826, 0.9896552937276493, 0.9754250344672477, 0.9538932084329071, 0.9240306707199512, 0.8847799938978558, 0.835114983723763, 0.7741393327924341, 0.7012438153695731, 0.6163490256781425, 0.5202685262373395, 0.41522980271506027, 0.3055697381162451, 0.19852529695426535, 0.1047253536630324, 0.09952458775530065, 0.10217560350924919, 0.10490854206567754, 0.10772625862576309, 0.11063171672512416, 0.11362799245916687, 0.11671827885543067, 0.1199058903945872, 0.12319426768117116, 0.12658698226443943, 0.1300877416089389, 0.1337003942134063, 0.1374289348754912, 0.1412775100984766, 0.1452504236346249, 0.1493521421579854, 0.1535873010574145, 0.15796071033814574, 0.16247736061745424, 0.1671424291967366, 0.1719612861886087, 0.17693950067334172, 0.18208284685402804, 0.18739731017420438, 0.1928890933551486, 0.19856462230259295, 0.2044305518240226, 0.21049377108788095, 0.21676140874472888, 0.22324083761745803, 0.22993967885283223, 0.23686580540962618, 0.2440273447391467, 0.25143268049157985, 0.2590904530560047, 0.2670095587125516, 0.2751991471415242, 0.2836686169956862, 0.2924276091976282, 0.3014859975732905, 0.31085387637438305, 0.3205415441754416, 0.3305594835543096, 0.34091833587640685, 0.3516288704015034, 0.36270194681482765, 0.37414847014986563, 0.38597933691545794, 0.39820537106163145, 0.4108372482134034, 0.42388540636539196, 0.43735994095761155, 0.4512704819387737, 0.4656260500612941, 0.48043488923465555, 0.4957042712822553, 0.5114402688916571, 0.5276474919081199, 0.5443287813837295, 0.5614848549451181, 0.5791138960655363, 0.5972110787041198, 0.6157680174869952, 0.6347721321303812, 0.6542059131230359, 0.6740460737722095, 0.6942625715530278, 0.7148174792691809, 0.7356636838237534, 0.7567433874173666, 0.7779863827628906, 0.7993080704937563, 0.8206071834629612, 0.8417631792828963, 0.8626332595697469, 0.8830489724487088, 0.9028123547508695, 0.9216915732171409, 0.939416031778589, 0.9556709274046181, 0.9700912642965331, 0.9822553816292672, 0.9916781229819943, 0.9978038900420947]
plt.plot(xt, yt)
yt=[1.0, 0.994131529661843, 0.9732155761251555, 0.9305281794879899, 0.8561715724769962, 0.7356291648274507, 0.548881333664372, 0.27847354184296536, 0.11370490538739975, 0.11569997309245596, 0.11774244407738371, 0.11983376000538844, 0.12197541631448319, 0.1241689645950756, 0.12641601508854627, 0.12871823931370432, 0.13107737282843168, 0.133495218134278, 0.13597364773224727, 0.13851460733852752, 0.14112011926945378, 0.14379228600557217, 0.1465332939452796, 0.14934541735916013, 0.15223102255682408, 0.15519257227877842, 0.1582326303266232, 0.16135386644567384, 0.1645590614749623, 0.16785111278046055, 0.17123303998830966, 0.17470799103581677, 0.17827924855900681, 0.18195023663657303, 0.1857245279111725, 0.18960585111013656, 0.19359809898881764, 0.19770533672095816, 0.20193181076162856, 0.20628195820943157, 0.210760416695775, 0.21537203483006112, 0.22012188323058046, 0.2250152661716974, 0.23005773387851117, 0.2352550955005079, 0.24061343279569464, 0.24613911455622106, 0.2518388118054221, 0.2577195137943767, 0.26378854482328784, 0.27005358190898116, 0.27652267331428537, 0.2832042579476088, 0.2901071856311783, 0.29724073822356023, 0.30461465156548556, 0.3122391381967358, 0.3201249107647289, 0.32828320601105004, 0.3367258091786584, 0.34546507862760417, 0.3545139703779291, 0.36388606221139375, 0.3735955768541872, 0.3836574036250458, 0.3940871177598649, 0.4049009964055202, 0.4161160300002061, 0.4277499274096736, 0.4398211127484022, 0.4523487112561398, 0.46535252088987933, 0.47885296538536826, 0.4928710233831996, 0.5074281267265734, 0.5225460191204192, 0.5382465638606903, 0.5545514861184785, 0.5714820310535343, 0.5890585135060767, 0.607299727723445, 0.6262221758973758, 0.6458390613548075, 0.6661589748500951, 0.6871841788393103, 0.7089083624383448, 0.7313136954605031, 0.7543669483773343, 0.7780143587096144, 0.8021748019979849, 0.8267306501648432, 0.8515154458159513, 0.8762971475860714, 0.9007551456194267, 0.9244484063464173, 0.9467708173448146, 0.9668877952390316, 0.9836450403618952, 0.995435212698413]
plt.plot(xt, yt)
#WIDE_PEAK fix min
# case NONE: return new GaussianWaveform(0.203125, phase, false);
# case LOW: return new SkewedGaussWaveform(0.711034440272608, skew.skew, phase, true);
# case MID: return new SkewedGaussWaveform(0.6700816291266387, skew.skew, phase, true);
# case HIGH: return new SkewedGaussWaveform(.7442591767475941, skew.skew, phase, true);
xt=[0.0, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1, 0.11, 0.12, 0.13, 0.14, 0.15, 0.16, 0.17, 0.18, 0.19, 0.2, 0.21, 0.22, 0.23, 0.24, 0.25, 0.26, 0.27, 0.28, 0.29, 0.3, 0.31, 0.32, 0.33, 0.34, 0.35, 0.36, 0.37, 0.38, 0.39, 0.4, 0.41, 0.42, 0.43, 0.44, 0.45, 0.46, 0.47, 0.48, 0.49, 0.5, 0.51, 0.52, 0.53, 0.54, 0.55, 0.56, 0.57, 0.58, 0.59, 0.6, 0.61, 0.62, 0.63, 0.64, 0.65, 0.66, 0.67, 0.68, 0.69, 0.7, 0.71, 0.72, 0.73, 0.74, 0.75, 0.76, 0.77, 0.78, 0.79, 0.8, 0.81, 0.82, 0.83, 0.84, 0.85, 0.86, 0.87, 0.88, 0.89, 0.9, 0.91, 0.92, 0.93, 0.94, 0.95, 0.96, 0.97, 0.98, 0.99]
yt=[1.0, 0.9987888996551672, 0.9951643921014979, 0.9891527514554127, 0.9807974152891648, 0.9701584619536344, 0.9573118912015268, 0.9423487201601802, 0.9253739096952884, 0.9065051388774692, 0.8858714475589475, 0.8636117689435711, 0.8398733754565454, 0.8148102621687294, 0.7885814924939011, 0.7613495308577245, 0.7332785865473379, 0.7045329920144862, 0.6752756375567601, 0.6456664825833416, 0.6158611616335171, 0.5860097010137122, 0.5562553594115757, 0.5267336031957593, 0.49757122438015355, 0.46888560648317157, 0.44078414080532446, 0.4133637930369936, 0.3867108176429706, 0.36090061519481487, 0.3359977257732059, 0.31205594976959017, 0.2891185859010446, 0.26721877502808633, 0.2463799374380964, 0.22661629062572547, 0.20793343425796706, 0.19032898894126954, 0.17379327559160265, 0.1583100226219387, 0.14385708877784129, 0.1304071902410655, 0.11792862155203371, 0.10638596094299341, 0.09574075179308944, 0.0859521530840816, 0.076977552922306, 0.06877314037233147, 0.06129443199693493, 0.054496750595930094, 0.048335654665708876, 0.054496750595930094, 0.06129443199693493, 0.06877314037233147, 0.07697755292230607, 0.08595215308408165, 0.09574075179308947, 0.10638596094299331, 0.11792862155203365, 0.13040719024106545, 0.14385708877784129, 0.1583100226219387, 0.17379327559160265, 0.19032898894126954, 0.20793343425796706, 0.22661629062572547, 0.24637993743809652, 0.26721877502808644, 0.2891185859010447, 0.3120559497695901, 0.33599772577320586, 0.3609006151948148, 0.3867108176429706, 0.4133637930369936, 0.44078414080532446, 0.46888560648317157, 0.49757122438015355, 0.5267336031957595, 0.5562553594115758, 0.5860097010137123, 0.6158611616335173, 0.6456664825833417, 0.67527563755676, 0.704532992014486, 0.7332785865473377, 0.7613495308577244, 0.7885814924939011, 0.8148102621687294, 0.8398733754565454, 0.8636117689435712, 0.8858714475589475, 0.9065051388774692, 0.9253739096952885, 0.9423487201601803, 0.9573118912015268, 0.9701584619536343, 0.9807974152891648, 0.9891527514554127, 0.9951643921014979, 0.9987888996551672]
plt.plot(xt, yt)
yt=[1.0, 0.9995102843432049, 0.9980284607237517, 0.9955358001111833, 0.9920139820513978, 0.9874451365272697, 0.9818118871445073, 0.9750973956232678, 0.9672854075707601, 0.9583602995043832, 0.9483071270889154, 0.9371116745448073, 0.9247605051778345, 0.9112410129730932, 0.8965414751887001, 0.8806511058764787, 0.8635601102484152, 0.8452597397987818, 0.8257423480824496, 0.8050014470401782, 0.7830317637514589, 0.7598292974849418, 0.7353913769054206, 0.7097167172850146, 0.6828054775544402, 0.6546593170181222, 0.625281451544565, 0.594676709030621, 0.5628515839254216, 0.5298142905865504, 0.4955748152277632, 0.4601449662041548, 0.42353842236727235, 0.3857707792092918, 0.34685959250213566, 0.3068244191243798, 0.26568685475611564, 0.22347056810963686, 0.18020133135210678, 0.13590704636534567, 0.09061776647765024, 0.04436571329336314, 0.0, 0.02487450804866056, 0.049732928424932404, 0.07456586695834065, 0.09936364246441017, 0.1241162832847375, 0.14881352398102166, 0.17344480219628583, 0.19799925569713314, 0.22246571961140235, 0.24683272387620217, 0.27108849091189324, 0.29522093353817436, 0.31921765314905404, 0.3430659381641156, 0.3667527627740813, 0.3902647859993561, 0.4135883510808275, 0.4367094852228859, 0.4596138997092142, 0.4822869904126182, 0.5047138387207257, 0.5268792129001052, 0.5487675699219278, 0.5703630577729658, 0.5916495182763223, 0.6126104904468823, 0.6332292144071151, 0.6534886358893555, 0.6733714113513362, 0.6928599137321704, 0.7119362388765922, 0.7305822126556258, 0.7487793988123691, 0.7665091075619057, 0.7837524049747516, 0.8004901231734921, 0.8167028713725416, 0.832371047791083, 0.8474748524694163, 0.8619943010188972, 0.875909239335673, 0.8891993593082157, 0.9018442155484633, 0.9138232431760318, 0.9251157766844703, 0.9357010699180284, 0.9455583171866245, 0.9546666755459245, 0.963005288268412, 0.9705533095301959, 0.9772899303369702, 0.9831944057110563, 0.9882460831597464, 0.9924244324432898, 0.9957090766587328, 0.9980798246535053, 0.999516704780054]
plt.plot(xt, yt)
yt=[1.0, 0.9992157374601713, 0.9967910252496027, 0.9926142243124053, 0.9865687143537644, 0.9785329597780681, 0.9683806446621771, 0.9559808923664647, 0.9411985883372177, 0.9238948281292443, 0.9039275167632181, 0.881152150325474, 0.8554228163261821, 0.8265934558715815, 0.7945194382989864, 0.7590595076897714, 0.7200781707224045, 0.6774486067285324, 0.63105619357916, 0.5808027570491707, 0.5266116662874221, 0.46843391334248685, 0.4062553292449494, 0.3401051010220707, 0.2700657600888102, 0.19628480769377324, 0.11898811955698216, 0.0384952171087181, 0.007632393404751336, 0.019700894320261127, 0.0319227377906967, 0.044298724760434026, 0.05682957443558885, 0.06951591744942406, 0.08235828859330907, 0.09535711908684807, 0.1085127283592086, 0.1218253153119767, 0.1352949490320834, 0.1489215589214369, 0.16270492420789207, 0.17664466280004973, 0.19074021944612649, 0.2049908531547537, 0.21939562383303374, 0.23395337809452307, 0.24866273418698645, 0.2635220659867983, 0.2785294860037282, 0.29368282733653345, 0.30897962451630967, 0.3244170931708697, 0.3399921084395755, 0.3557011820640028, 0.3715404380755658, 0.3875055869968012, 0.40359189846836224, 0.41979417220893456, 0.43610670721025185, 0.45252326906416646, 0.46903705531331225, 0.48564065871132867, 0.502326028272891, 0.5190844279879271, 0.5359063930684612, 0.5527816835905127, 0.569699235387468, 0.5866471080453762, 0.6036124298447949, 0.6205813394881889, 0.6375389244466161, 0.6544691557546123, 0.6713548190780048, 0.6881774418760314, 0.704917216476835, 0.7215529188844547, 0.7380618231361371, 0.7544196110315892, 0.7706002770611112, 0.786576028367983, 0.8023171795926761, 0.8177920424632094, 0.8329668100182119, 0.847805435378083, 0.8622695050163403, 0.8763181065294406, 0.8899076909607181, 0.9029919298049421, 0.9155215669067377, 0.9274442655718558, 0.9387044513384791, 0.9492431510106848, 0.9589978287427351, 0.9679022201868954, 0.9758861659859396, 0.9828754462124326, 0.988791617740055, 0.9935518569888925, 0.997068811030219, 0.999250460682733]
plt.plot(xt, yt)
yt=[1.0, 0.9976422085879377, 0.9898718809541124, 0.9754599259642711, 0.9528888735734694, 0.9202782062213068, 0.8752911490583485, 0.8150225433499098, 0.7358758886066773, 0.6334633833725137, 0.502639436577133, 0.33801472869722626, 0.13608984970747226, 0.005513084648484356, 0.010483407259427995, 0.015544230096937344, 0.020697635431358056, 0.025945761644221275, 0.03129080481665839, 0.03673502035196645, 0.04228072463121288, 0.04793029670045044, 0.05368617998774652, 0.05955088404782143, 0.06552698633160717, 0.07161713397750648, 0.07782404562050979, 0.08415051321462684, 0.09059940386329679, 0.09717366165152644, 0.10387630947247785, 0.11071045084005456, 0.1176792716776992, 0.12478604207210241, 0.13203411797879788, 0.13942694286466323, 0.14696804927011622, 0.15466106027126558, 0.16250969081939204, 0.17051774893185784, 0.1786891367048084, 0.1870278511137712, 0.19553798456341284, 0.2042237251421776, 0.21308935653122146, 0.22213925750983857, 0.23137790099133565, 0.2408098525138736, 0.250439768099997, 0.26027239138618813, 0.2703125499095788, 0.28056515042263747, 0.2910351730878897, 0.30172766438313336, 0.3126477285227275, 0.32380051717180647, 0.3351912171970877, 0.3468250361595393, 0.3587071852096795, 0.37084285899463054, 0.3832372121260397, 0.3958953316880915, 0.40882220518334705, 0.42202268321894787, 0.43550143612429637, 0.44926290356067666, 0.46331123602978, 0.4776502270073615, 0.49228323421504544, 0.5072130882911485, 0.5224419868225776, 0.5379713713448412, 0.5538017844943979, 0.569932703992691, 0.5863623495367449, 0.603087457945416, 0.6201030210364852, 0.6374019796542482, 0.6549748659884129, 0.6728093847705899, 0.6908899220385872, 0.7091969678378942, 0.7277064363784859, 0.7463888636487767, 0.7652084581347872, 0.7841219748796451, 0.8030773763597929, 0.8220122351777226, 0.8408518228911681, 0.8595068157790919, 0.877870531147271, 0.8958155857917309, 0.9131898399932121, 0.929811453945427, 0.9454628361970487, 0.9598832019956413, 0.9727593786589126, 0.9837143890090183, 0.9922932042618913, 0.9979448739972219]
plt.plot(xt, yt)
#ONE_THIRD_PEAK no fix min
# case NONE: return new GaussianWaveform(0.140625, phase, false);
# /*case LOW: return new SkewedGaussWaveform(0.1516067989805035, skew.skew, phase, false);
# case MID: return new SkewedGaussWaveform(.19283827997555814, skew.skew, phase, false);
# case HIGH: return new SkewedGaussWaveform(.2878768391594375, skew.skew, phase, false);*/
xt=[0.0, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1, 0.11, 0.12, 0.13, 0.14, 0.15, 0.16, 0.17, 0.18, 0.19, 0.2, 0.21, 0.22, 0.23, 0.24, 0.25, 0.26, 0.27, 0.28, 0.29, 0.3, 0.31, 0.32, 0.33, 0.34, 0.35, 0.36, 0.37, 0.38, 0.39, 0.4, 0.41, 0.42, 0.43, 0.44, 0.45, 0.46, 0.47, 0.48, 0.49, 0.5, 0.51, 0.52, 0.53, 0.54, 0.55, 0.56, 0.57, 0.58, 0.59, 0.6, 0.61, 0.62, 0.63, 0.64, 0.65, 0.66, 0.67, 0.68, 0.69, 0.7, 0.71, 0.72, 0.73, 0.74, 0.75, 0.76, 0.77, 0.78, 0.79, 0.8, 0.81, 0.82, 0.83, 0.84, 0.85, 0.86, 0.87, 0.88, 0.89, 0.9, 0.91, 0.92, 0.93, 0.94, 0.95, 0.96, 0.97, 0.98, 0.99]
yt=[1.0, 0.9974747986368548, 0.9899373900304076, 0.9775013993578034, 0.9603530314874869, 0.938746432156527, 0.9129974218422362, 0.883475818227102, 0.8505966079942792, 0.8148102621687294, 0.776592510126528, 0.7364338951763066, 0.6948294293818791, 0.6522686477895945, 0.6092263337644984, 0.5661541495171976, 0.5234733612647516, 0.48156879918121415, 0.4407841408053246, 0.4014185552787085, 0.3637246969042015, 0.32790799197435705, 0.29412712419549963, 0.2624955924772389, 0.23308419108452028, 0.20592424643419877, 0.1810114370251389, 0.15831002262193872, 0.13775731507104333, 0.11926823499333838, 0.10273981490249434, 0.0880555288083265, 0.07508934984809916, 0.06370945978172436, 0.053781556242727124, 0.045171724564724666, 0.037748860091299, 0.031386643603503045, 0.02596508652980629, 0.021371673780233053, 0.01750214036840672, 0.014260923582095412, 0.011561335567520117, 0.009325502111768767, 0.0074841124933293325, 0.005976022895005943, 0.004747752405935435, 0.003752906429386003, 0.002951557676005975, 0.002309610130864761, 0.0017981666618475832, 0.002309610130864761, 0.002951557676005975, 0.003752906429386003, 0.004747752405935443, 0.005976022895005953, 0.0074841124933293385, 0.00932550211176875, 0.011561335567520096, 0.014260923582095386, 0.01750214036840672, 0.021371673780233053, 0.02596508652980629, 0.031386643603503045, 0.037748860091299, 0.045171724564724666, 0.05378155624272717, 0.06370945978172445, 0.07508934984809923, 0.08805552880832646, 0.1027398149024943, 0.11926823499333827, 0.13775731507104333, 0.15831002262193872, 0.1810114370251389, 0.20592424643419877, 0.23308419108452028, 0.262495592477239, 0.2941271241954997, 0.32790799197435705, 0.3637246969042018, 0.40141855527870873, 0.4407841408053243, 0.4815687991812141, 0.5234733612647515, 0.5661541495171974, 0.6092263337644984, 0.6522686477895945, 0.6948294293818791, 0.7364338951763068, 0.7765925101265282, 0.8148102621687295, 0.8505966079942793, 0.8834758182271021, 0.912997421842236, 0.938746432156527, 0.9603530314874867, 0.9775013993578034, 0.9899373900304076, 0.9974747986368548]
plt.plot(xt, yt)
yt=[1.0, 0.9969144020240112, 0.9873132505712133, 0.9707403135256472, 0.9468411303791583, 0.9153963475301403, 0.8763571387546044, 0.8298810204084439, 0.7763656397013128, 0.7164772943316775, 0.6511700977395593, 0.5816909502345677, 0.5095649899332954, 0.43655623072339234, 0.36459897280863635, 0.29569766904455874, 0.23179660367644392, 0.17462619710324762, 0.12553983583287942, 0.0853630244825286, 0.05428355812564234, 0.03181431065967187, 0.016855155175558616, 0.009616812969173738, 0.010311715794179629, 0.011058068317145602, 0.011859732512134544, 0.012720859468089997, 0.013645910353472105, 0.014639678761954062, 0.015707314505990277, 0.01685434892396994, 0.018086721764568214, 0.019410809708582905, 0.02083345658376502, 0.022362005321635094, 0.024004331696706765, 0.025768879877542856, 0.027664699805232052, 0.029701486397707606, 0.031889620557287064, 0.03424021193326941, 0.03676514336066708, 0.039477116859378394, 0.042389701034409956, 0.045517379666117345, 0.048875601218705406, 0.052480828924133985, 0.05635059101569209, 0.060503530589251356, 0.06495945445885726, 0.06973938024494675, 0.07486558078602443, 0.08036162479582923, 0.08625241249546979, 0.09256420473113523, 0.09932464384010198, 0.10656276424808873, 0.11430899046673222, 0.12259511980829721, 0.13145428674306497, 0.14092090539078958, 0.15103058615927817, 0.16182002201930662, 0.17332683933544366, 0.18558940755801656, 0.19864660142522886, 0.21253750863154763, 0.22730107519711795, 0.24297568003524794, 0.25959862947791, 0.27720556180579736, 0.295829751170397, 0.3155012997306196, 0.33624620640682856, 0.3580853004451419, 0.3810330280651537, 0.40509608093409183, 0.43027185619117697, 0.4565467393839419, 0.4838942041482799, 0.5122727259721258, 0.541623512168875, 0.5718680565261275, 0.6029055352982476, 0.6346100716197727, 0.666827908398138, 0.6993745456786437, 0.7320319177345702, 0.7645457080554008, 0.7966229272380713, 0.8279299096368556, 0.8580909193733472, 0.8866875944816707, 0.9132594986073665, 0.9373060911561782, 0.9582904665605484, 0.9756452476797082, 0.9887810420716499, 0.9970978759575706]
plt.plot(xt, yt)
yt=[1.0, 0.9947673420474036, 0.977359315191228, 0.9449447014890051, 0.8944232417815924, 0.8226660957255137, 0.7270368145751555, 0.6064162572813783, 0.4630852307714943, 0.30590394884325683, 0.15477662968437159, 0.04314905952461143, 0.03518796976901374, 0.036339310171593274, 0.03753457366329907, 0.03877567856899969, 0.04006463746770648, 0.04140356230937693, 0.04279466983227655, 0.04424028729945708, 0.045742858574052137, 0.047304950554298696, 0.048929259990460036, 0.05061862070715829, 0.05237601125602062, 0.05420456302499993, 0.056107568832251424, 0.05808849203402141, 0.06015097617763478, 0.06229885523234258, 0.06453616443250446, 0.06686715176931628, 0.06929629016904208, 0.07182829039744304, 0.07446811473179742, 0.07722099144353528, 0.0800924301360365, 0.08308823798350734, 0.08621453691800048, 0.08947778181250364, 0.09288477970849743, 0.09644271013637146, 0.10015914657644401, 0.10404207910690366, 0.10809993828257214, 0.11234162028474329, 0.11677651337719377, 0.12141452569642727, 0.12626611439489346, 0.13134231614379185, 0.1366547789865125, 0.14221579551403266, 0.1480383373087685, 0.1541360905723808, 0.16052349281452785, 0.16721577043193564, 0.17422897694847586, 0.1815800316148465, 0.1892867579780772, 0.19736792192296265, 0.20584326855545113, 0.21473355713683684, 0.22406059308110887, 0.2338472557883555, 0.24411752079546375, 0.2548964743701248, 0.26621031824151864, 0.2780863616340239, 0.2905529971280984, 0.30363965608957366, 0.31737673845367453, 0.3317955104845624, 0.3469279627076793, 0.36280661847219636, 0.3794642814724097, 0.39693370795112703, 0.4152471861153433, 0.43443600137960586, 0.4545297612487822, 0.47555554775428377, 0.4975368581141336, 0.520492285388978, 0.5444338799771884, 0.5693651193776602, 0.5952783972040833, 0.6221519223172526, 0.6499458944127683, 0.6785977926396467, 0.7080165779767833, 0.7380755673768572, 0.7686036876007216, 0.7993747593847523, 0.8300943996778805, 0.8603840654027469, 0.8897617057448423, 0.9176184595460687, 0.9431908642404713, 0.9655281957740405, 0.9834549486858607, 0.9955292956451746]
plt.plot(xt, yt)
yt=[1.0, 0.9869244025377764, 0.9363573387303759, 0.821525286116017, 0.5964929346110752, 0.20822109401974534, 0.057917930603761325, 0.0590891792730345, 0.06029208625855767, 0.0615277392963867, 0.06279727188753956, 0.06410186558920487, 0.06544275243937371, 0.06682121752376409, 0.06823860169456965, 0.06969630445128094, 0.07119578699460434, 0.07273857546534557, 0.07432626438103701, 0.07596052028407853, 0.07764308561623642, 0.07937578283551253, 0.08116051879266366, 0.08299928938603167, 0.08489418451484382, 0.08684739335277623, 0.08886120996535206, 0.09093803929668141, 0.09308040355316266, 0.09529094901406653, 0.09757245330143592, 0.09992783314447577, 0.1023601526766003, 0.10487263230657527, 0.1074686582087636, 0.11015179248138893, 0.11292578402600056, 0.11579458020599037, 0.11876233934711984, 0.12183344414859974, 0.1250125160793729, 0.12830443084093346, 0.1317143349853211, 0.1352476637849148, 0.13891016045937649, 0.14270789687462251, 0.14664729583908923, 0.1507351551338845, 0.15497867342573077, 0.15938547822497454, 0.1639636560654246, 0.16872178509841335, 0.17366897031029596, 0.17881488159060221, 0.18416979489720012, 0.18974463678502881, 0.1955510325860435, 0.20160135854971234, 0.20790879827531078, 0.21448740378874004, 0.22135216163680316, 0.22851906438953395, 0.23600518795458336, 0.24382877511440704, 0.25200932569378187, 0.26056769374747457, 0.26952619211951306, 0.2789087046580313, 0.2887408062615365, 0.29904989076794536, 0.30986530645531446, 0.3212184985733069, 0.33314315782652243, 0.34567537302884604, 0.35885378516442074, 0.3727197387178378, 0.38731742422284704, 0.4026940033132503, 0.4188997038416264, 0.4359878674337639, 0.4540149245558431, 0.47304026190030135, 0.49312593235018354, 0.5143361370532572, 0.5367363793873698, 0.560392147555079, 0.5853669197009236, 0.6117191927568713, 0.6394980979735889, 0.6687369573078026, 0.699443815138693, 0.7315874827346891, 0.7650768469304793, 0.7997299281925484, 0.8352270903280852, 0.8710392980589597, 0.9063162646835984, 0.9397085796321796, 0.9690782116027895, 0.9910145120396]
plt.plot(xt, yt)
#ONE_THIRD_PEAK fix min
# case NONE: return new GaussianWaveform(0.140625, phase, false);
# case LOW: return new SkewedGaussWaveform(0.16159421514938846, skew.skew, phase, true);
# case MID: return new SkewedGaussWaveform(0.2890010832949674, skew.skew, phase, true);
# case HIGH: return new SkewedGaussWaveform(0.4830298083311916, skew.skew, phase, true);
xt=[0.0, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1, 0.11, 0.12, 0.13, 0.14, 0.15, 0.16, 0.17, 0.18, 0.19, 0.2, 0.21, 0.22, 0.23, 0.24, 0.25, 0.26, 0.27, 0.28, 0.29, 0.3, 0.31, 0.32, 0.33, 0.34, 0.35, 0.36, 0.37, 0.38, 0.39, 0.4, 0.41, 0.42, 0.43, 0.44, 0.45, 0.46, 0.47, 0.48, 0.49, 0.5, 0.51, 0.52, 0.53, 0.54, 0.55, 0.56, 0.57, 0.58, 0.59, 0.6, 0.61, 0.62, 0.63, 0.64, 0.65, 0.66, 0.67, 0.68, 0.69, 0.7, 0.71, 0.72, 0.73, 0.74, 0.75, 0.76, 0.77, 0.78, 0.79, 0.8, 0.81, 0.82, 0.83, 0.84, 0.85, 0.86, 0.87, 0.88, 0.89, 0.9, 0.91, 0.92, 0.93, 0.94, 0.95, 0.96, 0.97, 0.98, 0.99]
yt=[1.0, 0.9974747986368548, 0.9899373900304076, 0.9775013993578034, 0.9603530314874869, 0.938746432156527, 0.9129974218422362, 0.883475818227102, 0.8505966079942792, 0.8148102621687294, 0.776592510126528, 0.7364338951763066, 0.6948294293818791, 0.6522686477895945, 0.6092263337644984, 0.5661541495171976, 0.5234733612647516, 0.48156879918121415, 0.4407841408053246, 0.4014185552787085, 0.3637246969042015, 0.32790799197435705, 0.29412712419549963, 0.2624955924772389, 0.23308419108452028, 0.20592424643419877, 0.1810114370251389, 0.15831002262193872, 0.13775731507104333, 0.11926823499333838, 0.10273981490249434, 0.0880555288083265, 0.07508934984809916, 0.06370945978172436, 0.053781556242727124, 0.045171724564724666, 0.037748860091299, 0.031386643603503045, 0.02596508652980629, 0.021371673780233053, 0.01750214036840672, 0.014260923582095412, 0.011561335567520117, 0.009325502111768767, 0.0074841124933293325, 0.005976022895005943, 0.004747752405935435, 0.003752906429386003, 0.002951557676005975, 0.002309610130864761, 0.0017981666618475832, 0.002309610130864761, 0.002951557676005975, 0.003752906429386003, 0.004747752405935443, 0.005976022895005953, 0.0074841124933293385, 0.00932550211176875, 0.011561335567520096, 0.014260923582095386, 0.01750214036840672, 0.021371673780233053, 0.02596508652980629, 0.031386643603503045, 0.037748860091299, 0.045171724564724666, 0.05378155624272717, 0.06370945978172445, 0.07508934984809923, 0.08805552880832646, 0.1027398149024943, 0.11926823499333827, 0.13775731507104333, 0.15831002262193872, 0.1810114370251389, 0.20592424643419877, 0.23308419108452028, 0.262495592477239, 0.2941271241954997, 0.32790799197435705, 0.3637246969042018, 0.40141855527870873, 0.4407841408053243, 0.4815687991812141, 0.5234733612647515, 0.5661541495171974, 0.6092263337644984, 0.6522686477895945, 0.6948294293818791, 0.7364338951763068, 0.7765925101265282, 0.8148102621687295, 0.8505966079942793, 0.8834758182271021, 0.912997421842236, 0.938746432156527, 0.9603530314874867, 0.9775013993578034, 0.9899373900304076, 0.9974747986368548]
plt.plot(xt, yt)
yt=[1.0, 0.9972501604946407, 0.9887106168440861, 0.9739923022112235, 0.9527829911885627, 0.9248715748263387, 0.8901739576959637, 0.8487595756970521, 0.8008771044420258, 0.7469774412155415, 0.6877315265240409, 0.6240400741229032, 0.5570318803212603, 0.48804719898906096, 0.4186028513432698, 0.3503364771591109, 0.2849288315054989, 0.22400547120818484, 0.16902264795974092, 0.12114661803043528, 0.08114043079164736, 0.04927660040510581, 0.02529631401746099, 0.00843386967274716, 3.1494052069963466E-4, 0.001306790253227645, 0.002367676566614052, 0.0035024358989461995, 0.004716238280933486, 0.006014609034375455, 0.007403451659743108, 0.008889071942220902, 0.010478203299712906, 0.012178033388995146, 0.013996231977155412, 0.01594098007443509, 0.01802100031127071, 0.02024558852637576, 0.022624646513731205, 0.025168715853915356, 0.027889012728828155, 0.03079746358799916, 0.033906741498707725, 0.03723030297042172, 0.04078242499582402, 0.04457824199513306, 0.04863378228662916, 0.05296600363328674, 0.05759282733212253, 0.06253317021813175, 0.06780697384726882, 0.07343523000150774, 0.07944000152219204, 0.08584443732421171, 0.0926727802715123, 0.0999503664025534, 0.10770361378109314, 0.1159599990116635, 0.12474801919903661, 0.13409713684577979, 0.14403770487091655, 0.1546008685954163, 0.16581844117703515, 0.1777227485890082, 0.19034643982639052, 0.20372225759393386, 0.2178827642854523, 0.2328600176139568, 0.24868518980437496, 0.2653881238295662, 0.28299681977271124, 0.30153684405683967, 0.3210306540228551, 0.341496830195189, 0.36294920859152974, 0.38539590566145276, 0.40883822894059707, 0.4332694673568074, 0.45867355641077007, 0.4850236152796651, 0.5122803553784219, 0.5403903631969167, 0.5692842644683849, 0.5988747820872101, 0.6290547068711685, 0.6596948084537295, 0.6906417235005612, 0.7217158702701209, 0.7527094524599653, 0.7833846314354725, 0.8134719643924972, 0.8426692267166211, 0.870640759564172, 0.8970175080719259, 0.9213979408612081, 0.9433500664845758, 0.9624147854843906, 0.9781108354371517, 0.9899415975681453, 0.9974040330828318]
plt.plot(xt, yt)
yt=[1.0, 0.9974610312914682, 0.9892963031082308, 0.9746217481337056, 0.9524759269821461, 0.9218348627378363, 0.8816409814770783, 0.8308536096459763, 0.7685318902080945, 0.6939656766893287, 0.6068760375248102, 0.5077137901110932, 0.3980889831016173, 0.2813568252508997, 0.16333857010256256, 0.05299428396959511, 0.0019035709936207914, 0.0050397401417521534, 0.008270323668280044, 0.011598518200481866, 0.015027637447712301, 0.01856111653795968, 0.0222025164888667, 0.025955528812906806, 0.029823980255625233, 0.033811837664932516, 0.03792321298836569, 0.04216236839399172, 0.04653372150918521, 0.05104185076984862, 0.05569150087072562, 0.06048758830524846, 0.06543520698082197, 0.07053963389253416, 0.0758063348349432, 0.0812409701277636, 0.08684940032689922, 0.09263769188726335, 0.09861212273810441, 0.10477918772502445, 0.11114560386541388, 0.11771831535551341, 0.12450449825759793, 0.1315115647846911, 0.13874716708758053, 0.14621920043447956, 0.15393580565723786, 0.16190537071924113, 0.17013653123874456, 0.17863816977697586, 0.18741941367248438, 0.19648963117143853, 0.20585842556728307, 0.21553562702175158, 0.22553128169191689, 0.23585563773391902, 0.2465191276922479, 0.25753234671285613, 0.2689060259376364, 0.2806510003454347, 0.29277817019907487, 0.3052984551368594, 0.3182227398084424, 0.3315618097962112, 0.34532627638138946, 0.35952648850552277, 0.3741724300388756, 0.3892736001930191, 0.40483887460029017, 0.42087634422191067, 0.4373931288325474, 0.45439516135418445, 0.4718869387675878, 0.48987123470534205, 0.5083487681152598, 0.527317821564331, 0.5467738018174919, 0.5667087342571612, 0.5871106814925352, 0.6079630751250396, 0.6292439480709782, 0.6509250530781165, 0.6729708510959478, 0.6953373509611247, 0.7179707794401072, 0.740806058045255, 0.7637650602458925, 0.7867546198051615, 0.8096642581128816, 0.8323635957600511, 0.8546994115362165, 0.8764923110254761, 0.8975329677857689, 0.9175779038513044, 0.9363447846835924, 0.9535072192056138, 0.968689081865807, 0.9814584162141771, 0.9913210462096186, 0.9977141241514784]
plt.plot(xt, yt)
yt=[1.0, 0.9950188274196835, 0.9777052505810897, 0.9434608948142958, 0.8858837876430168, 0.7960291725086978, 0.6615642634545562, 0.46669956100429777, 0.19833986481645313, 0.0014411207643317218, 0.004242415267491692, 0.007106383296551392, 0.010034821431083493, 0.013029589253297405, 0.016092611948024974, 0.019225883024369998, 0.022431467165198404, 0.025711503210962047, 0.029068207284678224, 0.03250387606523009, 0.03602089021650686, 0.03962171798027175, 0.043308918941022914, 0.047085147971502046, 0.05095315936790396, 0.054915811184243504, 0.058976069775746015, 0.06313701456153713, 0.06740184301731326, 0.07177387590907504, 0.07625656277938699, 0.08085348769799029, 0.0855683752889268, 0.09040509704661726, 0.09536767795356958, 0.1004603034125487, 0.10568732650610144, 0.11105327559627545, 0.116562862277164, 0.1222209896925213, 0.12803276123007432, 0.1340034896032634, 0.14013870632990263, 0.14644417161559892, 0.15292588464760323, 0.15959009430198948, 0.16644331026352663, 0.1734923145531766, 0.180744173452617, 0.1882062498083247, 0.19588621568928716, 0.20379206536199143, 0.21193212853356475, 0.22031508379831122, 0.22894997220380328, 0.23784621082939145, 0.24701360624161658, 0.25646236765641883, 0.26620311959590887, 0.27624691377612853, 0.2866052398996616, 0.29729003495062467, 0.3083136904963893, 0.31968905738649556, 0.33142944709982164, 0.3435486288202156, 0.35606082111098514, 0.3689806768005226, 0.382323259373095, 0.39610400876555346, 0.4103386939834872, 0.42504334934507665, 0.44023419040681894, 0.45592750468285953, 0.47213951108787716, 0.48888618054634964, 0.5061830083324554, 0.5240447263224248, 0.5424849403065423, 0.5615156736253574, 0.5811467934027421, 0.6013852891977256, 0.6222343655155531, 0.6436922986656907, 0.6657509940535937, 0.6883941609353365, 0.7115949962689279, 0.7353132352069496, 0.7594913796576906, 0.7840498534333176, 0.8088807459318786, 0.8338396860462229, 0.858735219291071, 0.883314821981491, 0.9072463435060798, 0.9300931707400997, 0.9512806795150966, 0.9700504556674066, 0.9853971431889703, 0.9959803139957485]
plt.plot(xt, yt)
#HALF_PEAK no fix min
# case NONE: return new GaussianWaveform(0.1015625, phase, false);
# /*case LOW: return new SkewedGaussWaveform(0.10950254478545297, skew.skew, phase, false);
# case MID: return new SkewedGaussWaveform(0.13931330292050073, skew.skew, phase, false);
# case HIGH: return new SkewedGaussWaveform(0.20802617814718816, skew.skew, phase, false);*/
xt=[0.0, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1, 0.11, 0.12, 0.13, 0.14, 0.15, 0.16, 0.17, 0.18, 0.19, 0.2, 0.21, 0.22, 0.23, 0.24, 0.25, 0.26, 0.27, 0.28, 0.29, 0.3, 0.31, 0.32, 0.33, 0.34, 0.35, 0.36, 0.37, 0.38, 0.39, 0.4, 0.41, 0.42, 0.43, 0.44, 0.45, 0.46, 0.47, 0.48, 0.49, 0.5, 0.51, 0.52, 0.53, 0.54, 0.55, 0.56, 0.57, 0.58, 0.59, 0.6, 0.61, 0.62, 0.63, 0.64, 0.65, 0.66, 0.67, 0.68, 0.69, 0.7, 0.71, 0.72, 0.73, 0.74, 0.75, 0.76, 0.77, 0.78, 0.79, 0.8, 0.81, 0.82, 0.83, 0.84, 0.85, 0.86, 0.87, 0.88, 0.89, 0.9, 0.91, 0.92, 0.93, 0.94, 0.95, 0.96, 0.97, 0.98, 0.99]
yt=[1.0, 0.9951643921014979, 0.9807974152891648, 0.9573118912015268, 0.9253739096952884, 0.8858714475589475, 0.8398733754565454, 0.7885814924939011, 0.7332785865473379, 0.6752756375567601, 0.6158611616335171, 0.5562553594115757, 0.49757122438015355, 0.44078414080532446, 0.3867108176429706, 0.3359977257732059, 0.2891185859010446, 0.2463799374380964, 0.20793343425796706, 0.17379327559160265, 0.14385708877784129, 0.11792862155203371, 0.09574075179308944, 0.076977552922306, 0.06129443199693493, 0.048335654665708876, 0.037748860091298964, 0.02919642888226213, 0.022363783736295544, 0.01696486818005936, 0.012745161746014099, 0.009482653239773929, 0.006987213991546008, 0.005098798802789249, 0.003684863141087681, 0.002637330188709126, 0.001869378768951731, 0.0013122596110907415, 9.122878163395304E-4, 6.281070149764964E-4, 4.2827731285284825E-4, 2.8920523548780385E-4, 1.9340909317879185E-4, 1.2809650571570262E-4, 8.402090085120773E-5, 5.457918428170992E-5, 3.511206759435065E-5, 2.23704862200293E-5, 1.411510455726985E-5, 8.820281214411466E-6, 5.458463624457845E-6, 8.820281214411466E-6, 1.411510455726985E-5, 2.23704862200293E-5, 3.5112067594350776E-5, 5.4579184281710016E-5, 8.402090085120788E-5, 1.2809650571570219E-4, 1.934090931787915E-4, 2.8920523548780336E-4, 4.2827731285284825E-4, 6.281070149764964E-4, 9.122878163395304E-4, 0.0013122596110907415, 0.001869378768951731, 0.002637330188709126, 0.0036848631410876876, 0.005098798802789258, 0.006987213991546021, 0.00948265323977392, 0.012745161746014089, 0.016964868180059347, 0.022363783736295544, 0.02919642888226213, 0.037748860091298964, 0.048335654665708876, 0.06129443199693493, 0.07697755292230607, 0.09574075179308947, 0.11792862155203376, 0.14385708877784148, 0.17379327559160276, 0.2079334342579669, 0.24637993743809636, 0.2891185859010444, 0.33599772577320586, 0.3867108176429706, 0.44078414080532446, 0.49757122438015355, 0.5562553594115758, 0.6158611616335173, 0.6752756375567603, 0.7332785865473381, 0.7885814924939014, 0.8398733754565452, 0.8858714475589473, 0.9253739096952883, 0.9573118912015268, 0.9807974152891648, 0.9951643921014979]
plt.plot(xt, yt)
yt=[1.0, 0.9940236973434217, 0.9752149343446068, 0.9425180099966111, 0.8953458032351831, 0.8337580701281446, 0.7586390557169136, 0.6718479358925181, 0.5763045847247216, 0.4759635273832647, 0.3756252495717562, 0.2805433413083743, 0.19581714790742324, 0.12562039511071912, 0.07240602825262826, 0.036325181498027354, 0.015148989175152127, 0.004898538788703484, 0.0010986052770816548, 9.014743498909495E-4, 9.86248778160585E-4, 0.0010793038039837056, 0.0011814762051475078, 0.0012936898094723333, 0.0014169647575591729, 0.0015524277646156274, 0.0017013234890879362, 0.0018650271271625387, 0.002045058364644113, 0.0022430968313535385, 0.002460999218107876, 0.00270081823263216, 0.0029648235884951798, 0.003255525240438412, 0.0035756991003493025, 0.003928415490674822, 0.00431707061631475, 0.004745421361986493, 0.005217623749685687, 0.005738275420102731, 0.006312462532552227, 0.006945811509906044, 0.00764454608786269, 0.008415550161171268, 0.009266436952530852, 0.010205625061967058, 0.011242421984453918, 0.012387115709989533, 0.013651075041448859, 0.015046859279050223, 0.01658833792333332, 0.018290821037599138, 0.020171200881412044, 0.02224810537359851, 0.02454206385956878, 0.027075685535667127, 0.029873850712839375, 0.03296391487138248, 0.036375925153740905, 0.04014284854629153, 0.044300810493684895, 0.048889342046778216, 0.05395163283953513, 0.05953478618883148, 0.06569007137606411, 0.07247316665736087, 0.07994438471063463, 0.08816887000720669, 0.09721675493184732, 0.10716325830116398, 0.11808870617575164, 0.13007845045427485, 0.1432226556077657, 0.1576159179978325, 0.17335667547725286, 0.19054635737747627, 0.20928821657087723, 0.22968577614844687, 0.2518408135641794, 0.27585079518903693, 0.30180566459941494, 0.32978387935780956, 0.3598475846198296, 0.392036809147292, 0.4263625722998535, 0.4627988020863099, 0.5012729879937974, 0.5416555326912573, 0.583747829575301, 0.6272691854734725, 0.671842837832683, 0.7169814925835262, 0.7620730422177054, 0.8063674224646807, 0.8489659368624597, 0.8888148226023018, 0.9247053394127536, 0.9552832104529706, 0.9790707789653794, 0.9945056778285049]
plt.plot(xt, yt)
yt=[1.0, 0.9896654119544297, 0.953944080080306, 0.8849154824271009, 0.7744151333052411, 0.6168199065852619, 0.4159073935639206, 0.19927615983210928, 0.037527206165594705, 0.01154519618277527, 0.011984921481107457, 0.012444121532649833, 0.012923784411774539, 0.013424954867305824, 0.013948737953718674, 0.01449630291930722, 0.015068887371186399, 0.01566780173864785, 0.01629443405819575, 0.016950255105554344, 0.01763682390207964, 0.018355793625343075, 0.019108917956197336, 0.01989805789740964, 0.020725189101971915, 0.021592409752496416, 0.022501949036705094, 0.023456176267946043, 0.02445761070395571, 0.02550893212175816, 0.02661299221169297, 0.027772826859125823, 0.028991669388463693, 0.030272964850713367, 0.031620385443034804, 0.03303784715659988, 0.03452952775762635, 0.03609988621577074, 0.03775368370419113, 0.03949600630659095, 0.04133228957848676, 0.043268345122867834, 0.04531038935438539, 0.047465074641281656, 0.04973952303047901, 0.05214136277863261, 0.054678767930512, 0.057360501205803105, 0.06019596047625976, 0.06319522913697563, 0.06636913069822721, 0.06972928794759443, 0.07328818705551995, 0.07705924702059579, 0.08105689487292615, 0.08529664707393012, 0.08979519756756756, 0.09457051294944059, 0.09964193522418455, 0.10503029261494173, 0.11075801886746896, 0.11684928145028428, 0.12333011898433505, 0.1302285881320081, 0.13757492002425245, 0.14540168609099216, 0.1537439728641784, 0.16263956491913625, 0.17212913457496062, 0.18225643624515664, 0.1930685023588448, 0.20461583648664233, 0.21695259760698166, 0.2301367672112333, 0.24423028800298455, 0.2592991590789613, 0.27541346739737665, 0.29264732866200793, 0.31107870197127385, 0.33078903102969753, 0.35186264950154794, 0.3743858680133733, 0.3984456337955216, 0.4241276188704604, 0.45151354621051176, 0.4806775016079255, 0.5116808970687529, 0.5445656426406905, 0.5793449388549723, 0.6159909098670495, 0.6544180432129085, 0.6944610677890142, 0.7358454665068452, 0.7781482637166659, 0.8207460381741739, 0.862746306715452, 0.9028975922935625, 0.9394728972114357, 0.9701216079867595, 0.9916876022440039]
plt.plot(xt, yt)
yt=[1.0, 0.9723893403017706, 0.8537948345986012, 0.5436276988565926, 0.02899169774820895, 0.029643458494955744, 0.03031455207009856, 0.031005701180265372, 0.03171766143779013, 0.03245122313860151, 0.03320721315179421, 0.03398649692890645, 0.03478998064157789, 0.035618613456971496, 0.03647338996111623, 0.037355352741173756, 0.0382655951385545, 0.03920526418581987, 0.0401755637414117, 0.041177757837461894, 0.04221317425726209, 0.043283208360429336, 0.044389327175403025, 0.045533073780664686, 0.046716071998005274, 0.04794003142329054, 0.04920675282251898, 0.0505181339235498, 0.05187617563673021, 0.05328298874080015, 0.054740801073935065, 0.05625196527364105, 0.057818967113484034, 0.059444434489368064, 0.06113114711332851, 0.06288204697864098, 0.06470024966653287, 0.06658905657200509, 0.06855196813431666, 0.07059269816665502, 0.07271518938953302, 0.0749236302836446, 0.07722247339043412, 0.07961645520265494, 0.08211061780290967, 0.08471033242580071, 0.08742132513912701, 0.09024970486183233, 0.09320199396147254, 0.09628516170219788, 0.09950666084607358, 0.10287446774647442, 0.10639712631284422, 0.1100837962719437, 0.1139443062025348, 0.11798921187908892, 0.12222986052647002, 0.12667846166267635, 0.1313481652917743, 0.13625314830541435, 0.14140871006018743, 0.1468313782210838, 0.1525390261000732, 0.15855100287498822, 0.16488827824907878, 0.1715736033072639, 0.17863168954227873, 0.18608940826294512, 0.19397601285677032, 0.202323386657095, 0.2111663194549718, 0.22054281598680028, 0.2304944400017904, 0.2410666977382077, 0.2523094647659167, 0.2642774601097554, 0.2770307712372228, 0.2906354326952396, 0.30516405963791915, 0.3206965347744528, 0.33732074272519075, 0.3551333383826564, 0.37424052403491137, 0.3947587912039422, 0.4168155533767092, 0.4405495486191905, 0.4661108159701426, 0.49365992917092355, 0.5233659765834353, 0.5554024573017874, 0.5899397336461619, 0.6271317845384727, 0.6670934593022424, 0.7098617038329841, 0.7553292844867537, 0.8031302818529311, 0.8524386864859369, 0.9016051748250998, 0.9474802824187716, 0.9841000184872646]
plt.plot(xt, yt)
#HALF_PEAK fix min
#case NONE: return new GaussianWaveform(0.1015625, phase, false);
#case LOW: return new SkewedGaussWaveform(0.11016841333997066, skew.skew, phase, true);
# case MID: return new SkewedGaussWaveform(0.1500836205989934, skew.skew, phase, true);
# case HIGH: return new SkewedGaussWaveform(0.28198739813204843, skew.skew, phase, true);
xt=[0.0, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1, 0.11, 0.12, 0.13, 0.14, 0.15, 0.16, 0.17, 0.18, 0.19, 0.2, 0.21, 0.22, 0.23, 0.24, 0.25, 0.26, 0.27, 0.28, 0.29, 0.3, 0.31, 0.32, 0.33, 0.34, 0.35, 0.36, 0.37, 0.38, 0.39, 0.4, 0.41, 0.42, 0.43, 0.44, 0.45, 0.46, 0.47, 0.48, 0.49, 0.5, 0.51, 0.52, 0.53, 0.54, 0.55, 0.56, 0.57, 0.58, 0.59, 0.6, 0.61, 0.62, 0.63, 0.64, 0.65, 0.66, 0.67, 0.68, 0.69, 0.7, 0.71, 0.72, 0.73, 0.74, 0.75, 0.76, 0.77, 0.78, 0.79, 0.8, 0.81, 0.82, 0.83, 0.84, 0.85, 0.86, 0.87, 0.88, 0.89, 0.9, 0.91, 0.92, 0.93, 0.94, 0.95, 0.96, 0.97, 0.98, 0.99]
yt=[1.0, 0.9951643921014979, 0.9807974152891648, 0.9573118912015268, 0.9253739096952884, 0.8858714475589475, 0.8398733754565454, 0.7885814924939011, 0.7332785865473379, 0.6752756375567601, 0.6158611616335171, 0.5562553594115757, 0.49757122438015355, 0.44078414080532446, 0.3867108176429706, 0.3359977257732059, 0.2891185859010446, 0.2463799374380964, 0.20793343425796706, 0.17379327559160265, 0.14385708877784129, 0.11792862155203371, 0.09574075179308944, 0.076977552922306, 0.06129443199693493, 0.048335654665708876, 0.037748860091298964, 0.02919642888226213, 0.022363783736295544, 0.01696486818005936, 0.012745161746014099, 0.009482653239773929, 0.006987213991546008, 0.005098798802789249, 0.003684863141087681, 0.002637330188709126, 0.001869378768951731, 0.0013122596110907415, 9.122878163395304E-4, 6.281070149764964E-4, 4.2827731285284825E-4, 2.8920523548780385E-4, 1.9340909317879185E-4, 1.2809650571570262E-4, 8.402090085120773E-5, 5.457918428170992E-5, 3.511206759435065E-5, 2.23704862200293E-5, 1.411510455726985E-5, 8.820281214411466E-6, 5.458463624457845E-6, 8.820281214411466E-6, 1.411510455726985E-5, 2.23704862200293E-5, 3.5112067594350776E-5, 5.4579184281710016E-5, 8.402090085120788E-5, 1.2809650571570219E-4, 1.934090931787915E-4, 2.8920523548780336E-4, 4.2827731285284825E-4, 6.281070149764964E-4, 9.122878163395304E-4, 0.0013122596110907415, 0.001869378768951731, 0.002637330188709126, 0.0036848631410876876, 0.005098798802789258, 0.006987213991546021, 0.00948265323977392, 0.012745161746014089, 0.016964868180059347, 0.022363783736295544, 0.02919642888226213, 0.037748860091298964, 0.048335654665708876, 0.06129443199693493, 0.07697755292230607, 0.09574075179308947, 0.11792862155203376, 0.14385708877784148, 0.17379327559160276, 0.2079334342579669, 0.24637993743809636, 0.2891185859010444, 0.33599772577320586, 0.3867108176429706, 0.44078414080532446, 0.49757122438015355, 0.5562553594115758, 0.6158611616335173, 0.6752756375567603, 0.7332785865473381, 0.7885814924939014, 0.8398733754565452, 0.8858714475589473, 0.9253739096952883, 0.9573118912015268, 0.9807974152891648, 0.9951643921014979]
plt.plot(xt, yt)
yt=[1.0, 0.9940917967813461, 0.9755016555693229, 0.9431883351404714, 0.896566285821943, 0.8356786804388897, 0.7613701823336441, 0.6754342658632283, 0.5806993921935479, 0.48100904688669854, 0.3810467925792234, 0.2859656400864687, 0.20080954660977055, 0.1297711773819257, 0.07541437152625977, 0.038083800510912084, 0.015778865588980326, 0.004702956388147249, 4.3996864488390366E-4, 5.944007716239042E-5, 1.4768547617681103E-4, 2.445087056690181E-4, 3.507722082273543E-4, 4.674276693575205E-4, 5.955254592598492E-4, 7.362250865350252E-4, 8.908067721244873E-4, 0.00106068426308129, 0.001247419018144052, 0.0014527359106323908, 0.0016785406089804503, 0.0019269388113572926, 0.0022002575283679195, 0.0025010686268579878, 0.0028322148684214584, 0.0031968386983790204, 0.0035984140647818356, 0.004040781572395402, 0.004528187303587714, 0.005065325666490441, 0.0056573866605590575, 0.0063101079804806365, 0.007029832410913148, 0.007823570996295536, 0.008699072501285158, 0.009664899707388062, 0.010730513118918376, 0.011906362675106379, 0.013203988083132584, 0.014636128396780922, 0.016216841464385307, 0.017961633854202966, 0.019887601830853873, 0.022013583897582678, 0.024360325329197425, 0.026950654991549566, 0.029809674565600332, 0.03296496005573468, 0.03644677514898309, 0.04028829558744655, 0.0445258432006309, 0.049199127594162645, 0.05435149267904196, 0.060030164219229555, 0.06628649333806495, 0.07317618941350595, 0.08075953396057153, 0.0891015648930861, 0.09827221791681492, 0.10834640866861765, 0.11940403551429829, 0.13152987858352633, 0.14481336558864252, 0.1593481691890764, 0.17523159408512792, 0.19256370464313624, 0.2114461357021882, 0.23198052039039344, 0.2542664594814495, 0.2783989473781218, 0.30446516072930496, 0.33254050772766175, 0.3626838303750709, 0.3949316499420343, 0.42929134951659836, 0.4657331996340432, 0.5041811570035668, 0.5445024067578943, 0.586495680981057, 0.6298784772084288, 0.6742734279551634, 0.7191942448299474, 0.7640318875088858, 0.8080418971852129, 0.8503341920261148, 0.889867049301426, 0.9254474862004681, 0.9557407740080416, 0.9792923287974417, 0.9945656308479225]
plt.plot(xt, yt)
yt=[1.0, 0.9910645969631764, 0.9604797208001989, 0.901911064936227, 0.8086545044845985, 0.6752231126489243, 0.5011402195503974, 0.2989587975824691, 0.10795079305353876, 0.0036073744711473043, 0.004156962002239292, 0.004730038933981712, 0.005327755709185523, 0.005951326451824011, 0.0066020329005100145, 0.007281228609788701, 0.007990343439093562, 0.008730888350791971, 0.00950446054045797, 0.010312748924363244, 0.011157540011188736, 0.012040724187137858, 0.012964302445995942, 0.01393039359824294, 0.014941241996103811, 0.015999225814433603, 0.017106865930599117, 0.01826683545005742, 0.01948196992816639, 0.02075527834291571, 0.02208995487776431, 0.023489391578636873, 0.024957191954395923, 0.026497185595794472, 0.028113443894055998, 0.029810296946851788, 0.03159235174658101, 0.03346451175352982, 0.03543199796472255, 0.0375003715980969, 0.03967555852105721, 0.04196387556249165, 0.04437205885798074, 0.046907294389160775, 0.04957725089000199, 0.05239011530506169, 0.055354630997483834, 0.05848013891751074, 0.06177662195535813, 0.0652547527152345, 0.06892594495970855, 0.072802408985091, 0.07689721119839968, 0.08122433817402626, 0.08579876547241092, 0.09063653150252553, 0.09575481670308693, 0.10117202830197608, 0.10690789088655468, 0.11298354297587045, 0.11942163972458697, 0.12624646180204052, 0.13348403037071427, 0.1411622279271637, 0.1493109245529578, 0.15796210883809844, 0.16715002236495355, 0.17691129615177292, 0.18728508681907982, 0.198313209418257, 0.2100402627963013, 0.22251374199564197, 0.23578413041498614, 0.24990496217265018, 0.2649328421706726, 0.2809274075656387, 0.29795120946318576, 0.31606948734588575, 0.3353498005994103, 0.35586147097245513, 0.3776747761767631, 0.40085981716899916, 0.4254849587310474, 0.45161471317527496, 0.47930689824810063, 0.5086088498505346, 0.5395524044806872, 0.5721472807430711, 0.6063723780187167, 0.6421643661505632, 0.6794027539561727, 0.7178903865116855, 0.7573280213925151, 0.7972812653398135, 0.8371377180519078, 0.8760516969766009, 0.912873491076894, 0.946059920854087, 0.9735635356347342, 0.9927000731645895]
plt.plot(xt, yt)
yt=[1.0, 0.9855598642699223, 0.9294001158280938, 0.8008933841728406, 0.5471612069668034, 0.11514789412643789, 0.004233260002191489, 0.005430813056036933, 0.006660972803801468, 0.007924867257817565, 0.009223672204021933, 0.010558613609415866, 0.011930970170655723, 0.013342076013221639, 0.014793323551322499, 0.016286166519465052, 0.017822123187450974, 0.019402779771468977, 0.021029794054933458, 0.022704899233785412, 0.024429908002132565, 0.026206716895362026, 0.02803731090923111, 0.02992376841493013, 0.0318682663917343, 0.03387308600063021, 0.03594061852422567, 0.03807337170035644, 0.04027397647909168, 0.04254519423534368, 0.04488992447201947, 0.04731121305163702, 0.049812260997593165, 0.05239643390983976, 0.05506727204362964, 0.057828501104269577, 0.06068404381549643, 0.06363803232421983, 0.06669482150999018, 0.06985900327370355, 0.07313542188679763, 0.07652919048958326, 0.08004570883545187, 0.08369068238657162, 0.08747014287640066, 0.09139047046498293, 0.09545841762462953, 0.09968113490630591, 0.10406619875093197, 0.10862164152493625, 0.11335598397587264, 0.1182782703217742, 0.1233981062072514, 0.12872569978016946, 0.1342719061650696, 0.14004827563328592, 0.14606710579483068, 0.15234149816336998, 0.15888541947261944, 0.16571376814972158, 0.17284244637779914, 0.1802884382047462, 0.18806989417674325, 0.19620622299063, 0.2047181906658708, 0.21362802772988526, 0.22295954488377262, 0.23273825756035485, 0.2429915196912378, 0.25374866684805963, 0.2650411686930146, 0.2769027903344478, 0.2893697616924809, 0.30248095327857927, 0.3162780558001528, 0.3308057596027666, 0.34611192799844, 0.36224775577319196, 0.3792679003000231, 0.39723056724690653, 0.4161975252034139, 0.43623401269673373, 0.4574084856255422, 0.47979213103489454, 0.503458041289441, 0.5284798963965457, 0.5549299343302808, 0.5828758886261959, 0.6123764218320279, 0.6434743547973376, 0.6761866401042755, 0.7104894784829502, 0.7462961037330157, 0.7834233468977864, 0.8215407494756561, 0.8600920297252698, 0.8981718099105672, 0.9343281677086588, 0.9662387642460433, 0.9901646958155236]
plt.plot(xt, yt)
#QUARTER_PEAK no fix min
#case NONE: return new GaussianWaveform(0.05078125, phase, false);
# *case LOW: return new SkewedGaussWaveform(0.05476715911828679, skew.skew, phase, false);
# case MID: return new SkewedGaussWaveform(0.06973107359499559, skew.skew, phase, false);
# case HIGH: return new SkewedGaussWaveform(0.10420928568117749, skew.skew, phase, false);
xt=[0.0, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1, 0.11, 0.12, 0.13, 0.14, 0.15, 0.16, 0.17, 0.18, 0.19, 0.2, 0.21, 0.22, 0.23, 0.24, 0.25, 0.26, 0.27, 0.28, 0.29, 0.3, 0.31, 0.32, 0.33, 0.34, 0.35, 0.36, 0.37, 0.38, 0.39, 0.4, 0.41, 0.42, 0.43, 0.44, 0.45, 0.46, 0.47, 0.48, 0.49, 0.5, 0.51, 0.52, 0.53, 0.54, 0.55, 0.56, 0.57, 0.58, 0.59, 0.6, 0.61, 0.62, 0.63, 0.64, 0.65, 0.66, 0.67, 0.68, 0.69, 0.7, 0.71, 0.72, 0.73, 0.74, 0.75, 0.76, 0.77, 0.78, 0.79, 0.8, 0.81, 0.82, 0.83, 0.84, 0.85, 0.86, 0.87, 0.88, 0.89, 0.9, 0.91, 0.92, 0.93, 0.94, 0.95, 0.96, 0.97, 0.98, 0.99]
yt=[1.0, 0.9807974152891648, 0.9253739096952884, 0.8398733754565454, 0.7332785865473379, 0.6158611616335171, 0.49757122438015355, 0.3867108176429706, 0.2891185859010446, 0.20793343425796706, 0.14385708877784129, 0.09574075179308944, 0.06129443199693493, 0.037748860091298964, 0.022363783736295544, 0.012745161746014099, 0.006987213991546008, 0.003684863141087681, 0.001869378768951731, 9.122878163395304E-4, 4.2827731285284825E-4, 1.9340909317879185E-4, 8.402090085120773E-5, 3.511206759435065E-5, 1.411510455726985E-5, 5.458463624457845E-6, 2.0305578494036798E-6, 7.266393931060717E-7, 2.501388422756234E-7, 8.283272670791912E-8, 2.6386476780894055E-8, 8.085734530356179E-9, 2.3835056016115253E-9, 6.758829650948635E-10, 1.84367959751066E-10, 4.837912665332134E-11, 1.2212068281778742E-11, 2.9653710887231095E-12, 6.926717879695354E-13, 1.55644785504638E-13, 3.364343077958482E-14, 6.995594102219332E-15, 1.3992894339418642E-15, 2.692459206189263E-16, 4.9836706569819453E-17, 8.873769884841599E-18, 1.5199370993560441E-18, 2.5043884613760726E-19, 3.969505409115469E-20, 6.0524293320881495E-21, 8.877316050951375E-22, 6.0524293320881495E-21, 3.969505409115469E-20, 2.5043884613760726E-19, 1.5199370993560657E-18, 8.873769884841662E-18, 4.983670656981981E-17, 2.692459206189225E-16, 1.3992894339418541E-15, 6.9955941022192825E-15, 3.364343077958482E-14, 1.55644785504638E-13, 6.926717879695354E-13, 2.9653710887231095E-12, 1.2212068281778742E-11, 4.837912665332134E-11, 1.843679597510673E-10, 6.758829650948683E-10, 2.3835056016115422E-9, 8.08573453035615E-9, 2.6386476780893963E-8, 8.283272670791883E-8, 2.501388422756234E-7, 7.266393931060717E-7, 2.0305578494036798E-6, 5.458463624457845E-6, 1.411510455726985E-5, 3.5112067594350776E-5, 8.402090085120788E-5, 1.934090931787922E-4, 4.282773128528506E-4, 9.122878163395336E-4, 0.0018693787689517244, 0.0036848631410876776, 0.006987213991545989, 0.012745161746014089, 0.022363783736295544, 0.037748860091298964, 0.06129443199693493, 0.09574075179308947, 0.14385708877784148, 0.20793343425796731, 0.289118585901045, 0.3867108176429712, 0.49757122438015294, 0.6158611616335167, 0.7332785865473375, 0.8398733754565452, 0.9253739096952883, 0.9807974152891648]
plt.plot(xt, yt)
yt=[1.0, 0.9753676230067523, 0.8957020385730722, 0.7592185188094147, 0.5770648858039882, 0.37644238586894246, 0.19650959653797195, 0.07282681734478977, 0.015299526824410523, 0.0011189385291109242, 7.447207054212331E-6, 4.97682116853634E-7, 5.691324475361825E-7, 6.513096798905048E-7, 7.458957922813491E-7, 8.548478704838678E-7, 9.804449865297402E-7, 1.1253433893178883E-6, 1.2926415383795032E-6, 1.485956804983433E-6, 1.709516015575801E-6, 1.968262433258445E-6, 2.2679822780347135E-6, 2.615454493365543E-6, 3.0186281964018206E-6, 3.4868331278744914E-6, 4.031029476392856E-6, 4.664104729003642E-6, 5.401226741826163E-6, 6.2602640881492355E-6, 7.262286995836017E-6, 8.432164915887675E-6, 9.799280073108679E-6, 1.1398380364736144E-5, 1.3270599848929666E-5, 1.546468099258224E-5, 1.8038440060686905E-5, 2.106052581449987E-5, 2.4612532395758238E-5, 2.879154034321649E-5, 3.3713175649910904E-5, 3.9515296282835956E-5, 4.636243946202219E-5, 5.44511922333508E-5, 6.401668370251589E-5, 7.534044124902891E-5, 8.875990698889159E-5, 1.0467997702382162E-4, 1.2358700747031975E-4, 1.4606583144318106E-4, 1.7282045444535728E-4, 2.046992473821641E-4, 2.427256433233694E-4, 2.8813557437606046E-4, 3.4242316868860487E-4, 4.0739657707138485E-4, 4.852462291852998E-4, 5.786283492932997E-4, 6.907672138410231E-4, 8.255804351232282E-4, 9.878325402073189E-4, 0.0011833233219626714, 0.0014191189168873942, 0.0017038353682276635, 0.0020479866273946062, 0.002464411605596174, 0.0029687980925593997, 0.0035803251971015158, 0.004322450521757658, 0.005223873637246493, 0.006319713622135463, 0.007652945475029957, 0.009276147991661182, 0.011253623967633483, 0.013663961821304344, 0.016603115008716727, 0.020188080385657284, 0.02456125649904, 0.029895553838586178, 0.03640030555679817, 0.0443279804705494, 0.0539816177713719, 0.06572276692370753, 0.0799795017611583, 0.09725375082022843, 0.11812670171038289, 0.14326033944604286, 0.1733922009937805, 0.2093191023073552, 0.2518638676220806, 0.30181696702429867, 0.3598425808538925, 0.42633635525125413, 0.5012208741480145, 0.5836663592540382, 0.6717313360217216, 0.7619358347676647, 0.8488151766126255, 0.9245640827654453, 0.9789753785315325]
plt.plot(xt, yt)
yt=[1.0, 0.9541581440522309, 0.7752607930958018, 0.41774022636943875, 0.03861388662357505, 6.163669107456331E-4, 6.46945829703247E-4, 6.7926732843687E-4, 7.134441574275566E-4, 7.495972680382372E-4, 7.87856477254498E-4, 8.283611920188841E-4, 8.712611990281531E-4, 9.167175264942839E-4, 9.649033850750898E-4, 0.0010160051959685586, 0.0010702237150468485, 0.0011277752628935272, 0.0011888930717145656, 0.0012538287613357182, 0.0013228539578937196, 0.0013962620703968306, 0.0014743702420947757, 0.0015575214955856193, 0.001646087092828338, 0.001740469133759401, 0.0018411034200702461, 0.00194846261393573, 0.002063059725145361, 0.0021854519642401, 0.0023162450039688662, 0.0024560976967325505, 0.0026057273017735475, 0.0027659152828057995, 0.0029375137446910216, 0.0031214525867992657, 0.0033187474610180865, 0.003530508634194921, 0.0037579508683457537, 0.004002404447512374, 0.004265327498020737, 0.0045483197694582474, 0.004853138067386734, 0.005181713556158119, 0.005536171181806774, 0.005918851501573101, 0.006332335249011181, 0.006779471012847139, 0.007263406464965887, 0.007787623639511858, 0.008355978842756821, 0.0089727478640889, 0.009642677264564381, 0.010371042643737288, 0.011163714931286487, 0.012027235921287177, 0.012968904468600403, 0.013996875004493776, 0.015120270309104072, 0.016349310809909638, 0.017695463067824355, 0.019171610577635483, 0.020792250561385464, 0.022573721088814846, 0.024534463638274998, 0.026695327138551526, 0.02907992063521242, 0.03171502303785641, 0.034631059966026376, 0.03786265956657653, 0.0414493013748587, 0.04543607489224432, 0.04987456761213559, 0.05482390580291565, 0.06035197549596939, 0.066536855850679, 0.0734685023439523, 0.08125072292854793, 0.09000349611618504, 0.0998656852591231, 0.11099820700314184, 0.1235877119725182, 0.13785082877308624, 0.15403900246927144, 0.17244391581042723, 0.19340339874545967, 0.21730758055859117, 0.24460477069580075, 0.27580608609829527, 0.31148703210251477, 0.3522828438690135, 0.3988719746829817, 0.4519379167298718, 0.5120922111999543, 0.5797286464613137, 0.6547559918084345, 0.7361167484218011, 0.8209301635352937, 0.9029827459629145, 0.9701233879685169]
plt.plot(xt, yt)
yt=[1.0, 0.8432949558297448, 0.005313093136040679, 0.005458863498210377, 0.005609774954350007, 0.005766051761061479, 0.005927929928314618, 0.0060956579432576, 0.006269497545491437, 0.0064497245579737935, 0.006636629778097021, 0.006830519933903186, 0.0070317187108593325, 0.007240567855124996, 0.007457428359805122, 0.00768268174130269, 0.007916731413572199, 0.008160004168836252, 0.008412951774171323, 0.00867605269430561, 0.008949813952012463, 0.009234773138641033, 0.009531500588614449, 0.00984060173316364, 0.010162719650168595, 0.010498537828771436, 0.010848783169430689, 0.011214229242332091, 0.011595699829590026, 0.01199407277950188, 0.012410284204297621, 0.012845333056407159, 0.013300286122304529, 0.01377628347754461, 0.014274544451760603, 0.014796374158223574, 0.01534317064918034, 0.0159164327656987, 0.01651776875929403, 0.017148905772347546, 0.017811700275437714, 0.01850814957240778, 0.01924040449853836, 0.020010783453881697, 0.020821787932992712, 0.021676119734378718, 0.022576700058470196, 0.023526690732374077, 0.02452951783379742, 0.025588898026145956, 0.026708867962895518, 0.02789381717307877, 0.029148524902544697, 0.03047820145923562, 0.03188853469714653, 0.033385742375377385, 0.03497663124878976, 0.0366686638889269, 0.03847003440256104, 0.040389754416027955, 0.04243775093319948, 0.04462497796193278, 0.04696354414852572, 0.04946685907503282, 0.05214980137639309, 0.055028912443387944, 0.0581226202188744, 0.06145149850054782, 0.06503856827423501, 0.06890964896894676, 0.07309376921439657, 0.07762364877764417, 0.08253626596618414, 0.08787352804974824, 0.09368306635280894, 0.1000191828382489, 0.10694398154368347, 0.11452872654006513, 0.12285547866918296, 0.13201907684330325, 0.1421295470023667, 0.15331504398723983, 0.16572545990436288, 0.1795368685339405, 0.19495702051343453, 0.21223215951141244, 0.23165549486551423, 0.2535777362558769, 0.2784201552102035, 0.3066906455459692, 0.33900311022710705, 0.3760999671089734, 0.4188760705618839, 0.46839849948186923, 0.525906956500606, 0.5927546788016804, 0.6701836635903726, 0.7586410785982922, 0.855767273462868, 0.9501830360203302]
plt.plot(xt, yt)
#QUARTER_PEAK fix min
#GaussianWaveform(0.05078125, phase, false);
#SkewedGaussWaveform(0.054767394902696165, skew.skew, phase, true);
#SkewedGaussWaveform(0.07037477639896425, skew.skew, phase, true);
#SkewedGaussWaveform(0.1147667366789808, skew.skew, phase, true);
xt=[0.0, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1, 0.11, 0.12, 0.13, 0.14, 0.15, 0.16, 0.17, 0.18, 0.19, 0.2, 0.21, 0.22, 0.23, 0.24, 0.25, 0.26, 0.27, 0.28, 0.29, 0.3, 0.31, 0.32, 0.33, 0.34, 0.35, 0.36, 0.37, 0.38, 0.39, 0.4, 0.41, 0.42, 0.43, 0.44, 0.45, 0.46, 0.47, 0.48, 0.49, 0.5, 0.51, 0.52, 0.53, 0.54, 0.55, 0.56, 0.57, 0.58, 0.59, 0.6, 0.61, 0.62, 0.63, 0.64, 0.65, 0.66, 0.67, 0.68, 0.69, 0.7, 0.71, 0.72, 0.73, 0.74, 0.75, 0.76, 0.77, 0.78, 0.79, 0.8, 0.81, 0.82, 0.83, 0.84, 0.85, 0.86, 0.87, 0.88, 0.89, 0.9, 0.91, 0.92, 0.93, 0.94, 0.95, 0.96, 0.97, 0.98, 0.99]
yt=[1.0, 0.9807974152891648, 0.9253739096952884, 0.8398733754565454, 0.7332785865473379, 0.6158611616335171, 0.49757122438015355, 0.3867108176429706, 0.2891185859010446, 0.20793343425796706, 0.14385708877784129, 0.09574075179308944, 0.06129443199693493, 0.037748860091298964, 0.022363783736295544, 0.012745161746014099, 0.006987213991546008, 0.003684863141087681, 0.001869378768951731, 9.122878163395304E-4, 4.2827731285284825E-4, 1.9340909317879185E-4, 8.402090085120773E-5, 3.511206759435065E-5, 1.411510455726985E-5, 5.458463624457845E-6, 2.0305578494036798E-6, 7.266393931060717E-7, 2.501388422756234E-7, 8.283272670791912E-8, 2.6386476780894055E-8, 8.085734530356179E-9, 2.3835056016115253E-9, 6.758829650948635E-10, 1.84367959751066E-10, 4.837912665332134E-11, 1.2212068281778742E-11, 2.9653710887231095E-12, 6.926717879695354E-13, 1.55644785504638E-13, 3.364343077958482E-14, 6.995594102219332E-15, 1.3992894339418642E-15, 2.692459206189263E-16, 4.9836706569819453E-17, 8.873769884841599E-18, 1.5199370993560441E-18, 2.5043884613760726E-19, 3.969505409115469E-20, 6.0524293320881495E-21, 8.877316050951375E-22, 6.0524293320881495E-21, 3.969505409115469E-20, 2.5043884613760726E-19, 1.5199370993560657E-18, 8.873769884841662E-18, 4.983670656981981E-17, 2.692459206189225E-16, 1.3992894339418541E-15, 6.9955941022192825E-15, 3.364343077958482E-14, 1.55644785504638E-13, 6.926717879695354E-13, 2.9653710887231095E-12, 1.2212068281778742E-11, 4.837912665332134E-11, 1.843679597510673E-10, 6.758829650948683E-10, 2.3835056016115422E-9, 8.08573453035615E-9, 2.6386476780893963E-8, 8.283272670791883E-8, 2.501388422756234E-7, 7.266393931060717E-7, 2.0305578494036798E-6, 5.458463624457845E-6, 1.411510455726985E-5, 3.5112067594350776E-5, 8.402090085120788E-5, 1.934090931787922E-4, 4.282773128528506E-4, 9.122878163395336E-4, 0.0018693787689517244, 0.0036848631410876776, 0.006987213991545989, 0.012745161746014089, 0.022363783736295544, 0.037748860091298964, 0.06129443199693493, 0.09574075179308947, 0.14385708877784148, 0.20793343425796731, 0.289118585901045, 0.3867108176429712, 0.49757122438015294, 0.6158611616335167, 0.7332785865473375, 0.8398733754565452, 0.9253739096952883, 0.9807974152891648]
plt.plot(xt, yt)
yt=[1.0, 0.9753678330400302, 0.8957029386322871, 0.7592205392683384, 0.5770681492873888, 0.3764464398577459, 0.19651339192307404, 0.07282923199147215, 0.015300276211152876, 0.0011187969225162503, 7.158147082828829E-6, 2.063159130771156E-7, 2.7776975468930587E-7, 3.5995099900633937E-7, 4.54541698853725E-7, 5.634990259717674E-7, 6.891021517061961E-7, 8.340074397696772E-7, 1.001313482516436E-6, 1.1946378049779337E-6, 1.4182074117262597E-6, 1.676965772355625E-6, 1.976699346411755E-6, 2.324187355431266E-6, 2.7273792395981214E-6, 3.195605115164805E-6, 3.7398256075855005E-6, 4.372928712446037E-6, 5.110082878281011E-6, 5.969157368989489E-6, 6.971223218080117E-6, 8.141150817053711E-6, 9.508323489406782E-6, 1.1107490416759891E-5, 1.2979787159767185E-5, 1.5173957944171167E-5, 1.774782109530682E-5, 2.0770027789566586E-5, 2.4322175001592924E-5, 2.8501346595251326E-5, 3.342317246891921E-5, 3.9225515179294784E-5, 4.607291734370979E-5, 5.416197235882599E-5, 6.372781680734144E-5, 7.505198687658212E-5, 8.847193506350956E-5, 1.0439256971101563E-4, 1.233002613770413E-4, 1.4577986022085496E-4, 1.7253539186985983E-4, 2.0441525099839145E-4, 2.424428987578237E-4, 2.87854300439322E-4, 3.421436234110205E-4, 4.0711906484016997E-4, 4.8497110917987966E-4, 5.783560454485291E-4, 6.904982266050508E-4, 8.253153553185698E-4, 9.87572065349832E-4, 0.00118306827544337, 0.0014188702704752431, 0.0017035942684059532, 0.0020477544260961524, 0.0024641898959489104, 0.0029685887502869245, 0.003580130428232627, 0.0043222729172705705, 0.005223716235186701, 0.006319579977573116, 0.007652839737925042, 0.00927607499223088, 0.011253589308098758, 0.013663971971842982, 0.01660317740395658, 0.020188203515966608, 0.02456144998804397, 0.02989582849457037, 0.036400673380745145, 0.04432845459730691, 0.05398221231274829, 0.0657234966731705, 0.07998038170962321, 0.09725479542207396, 0.11812792383015797, 0.1432617489126483, 0.17339380269576596, 0.20932089378911475, 0.2518658361991608, 0.3018190865448726, 0.3598448084158025, 0.4263386284651823, 0.5012231099364872, 0.5836684556869592, 0.671733179242945, 0.7619373134664291, 0.848816206314798, 0.9245646406982786, 0.9789755457990131]
plt.plot(xt, yt)
yt=[1.0, 0.9550457960911298, 0.779941360899498, 0.429172781321184, 0.045934162253628376, 1.70794435468167E-5, 4.888470210229897E-5, 8.249664178207782E-5, 1.1803180363918053E-4, 1.556151803587797E-4, 1.953808993657684E-4, 2.3747296700123467E-4, 2.8204607976606684E-4, 3.2926650927327865E-4, 3.793130682691003E-4, 4.3237816588570987E-4, 4.886689611864024E-4, 5.484086250693957E-4, 6.118377217229506E-4, 6.792157220880702E-4, 7.508226632036289E-4, 8.269609689030052E-4, 9.079574491246936E-4, 9.941654971186774E-4, 0.0010859675061066425, 0.001183777529522552, 0.001288044211861248, 0.0013992540204433776, 0.001517934812118571, 0.0016446597731373702, 0.0017800517751972499, 0.0019247881960915794, 0.0020796062595578764, 0.0022453089559436373, 0.0024227716133124, 0.0026129491977482364, 0.00281688443205618, 0.0030357168340008817, 0.003270692788910474, 0.0035231767871708093, 0.0037946639751684098, 0.004086794188982109, 0.0044013676640149236, 0.004740362641314088, 0.00510595512315683, 0.005500541067296695, 0.005926761351913287, 0.0063875298927837665, 0.0068860653516737745, 0.007425926941824745, 0.008011054914346444, 0.008645816400273926, 0.00933505738935011, 0.010084161751032153, 0.010899118349102751, 0.011786597472553666, 0.012754038006817497, 0.013809747006602596, 0.014963013611283504, 0.016224239574091835, 0.017605089066905884, 0.019118660884803945, 0.020779686722544356, 0.022604759845242975, 0.02461259924736341, 0.02682435531100368, 0.029263964063853315, 0.03195855843135789, 0.03493894641401778, 0.03824016794224998, 0.041902144315965874, 0.04597043667623084, 0.0504971329359878, 0.0555418860663515, 0.061173130632250335, 0.06746950900508128, 0.07452154370259433, 0.08243359766577656, 0.09132616964555915, 0.10133857659385306, 0.11263207786317408, 0.12539349506580083, 0.13983937310613292, 0.15622070620567113, 0.17482820761641776, 0.19599801621231813, 0.22011757869225, 0.24763117441462557, 0.2790440787852654, 0.31492355005636796, 0.3558934308312087, 0.4026167575483118, 0.4557566285478778, 0.5158983865737179, 0.5834036074026626, 0.6581443627580118, 0.739027677090902, 0.8231535765598103, 0.9043405223837573, 0.9705956911379466]
plt.plot(xt, yt)
yt=[1.0, 0.8944259017481836, 0.22291732467789546, 1.3116439246747355E-4, 3.2081635365400883E-4, 5.170494177516707E-4, 7.201468328908817E-4, 9.304065259206613E-4, 0.0011481419977043318, 0.001373683281542252, 0.0016073779697935088, 0.0018495923142275304, 0.0021007124061416184, 0.0023611454428373955, 0.0026313210876627554, 0.0029116929315040016, 0.003202740064361486, 0.0035049687664705253, 0.003818914329346159, 0.004145143018146697, 0.00448425418787802, 0.004836882567212742, 0.005203700725089625, 0.005585421736807596, 0.005982802068054013, 0.006396644697231115, 0.00682780249859407, 0.007277181911115927, 0.007745746920684598, 0.008234523386250267, 0.008744603743924316, 0.009277152126829906, 0.009833409942780424, 0.010414701956677644, 0.011022442929955599, 0.011658144875534555, 0.012323424993694758, 0.013020014362149217, 0.013749767462525039, 0.014514672635612723, 0.015316863569297701, 0.016158631936263695, 0.017042441313605987, 0.017970942533710944, 0.01894699063549286, 0.019973663607740312, 0.021054283142393077, 0.022192437645618998, 0.02339200778926139, 0.024657194925384818, 0.025992552733214384, 0.027403022521878987, 0.02889397267539415, 0.030471242799887863, 0.03214119321914541, 0.03391076056549235, 0.03578752033170804, 0.03777975738953147, 0.039896545645598795, 0.042147838201474344, 0.04454456961708473, 0.04709877215403859, 0.04982370820648982, 0.05273402152407098, 0.055845910308459965, 0.059177325840320164, 0.06274820098906234, 0.06658071380208978, 0.07069959239796955, 0.07513246864338863, 0.07991028963234763, 0.08506779787797136, 0.0906440934613917, 0.09668329427094906, 0.10323531405121864, 0.1103567824460085, 0.11811213679116198, 0.1265749223792385, 0.1358293466370175, 0.14597214356696028, 0.15711481842766487, 0.1693863595567397, 0.18293652507378633, 0.1979398374037531, 0.21460044811215745, 0.2331580681955405, 0.25389519065909033, 0.277145852702458, 0.30330617058381537, 0.33284677904046023, 0.3663270044186771, 0.40440984333021746, 0.44787503775111265, 0.4976234177059749, 0.5546560665274937, 0.6199887970341318, 0.694404626380419, 0.7777930012674013, 0.8673797517994262, 0.952719833837565]
plt.plot(xt, yt)
```
|
github_jupyter
|
import numpy as np
import scipy.stats as stats
from matplotlib import pyplot as plt
x = np.arange(0, 1, 0.01)
uno = np.full(x.shape,1)
low = np.full(x.shape,0.01)
ph = 0.5
#plt.rcParams['figure.dpi'] = 72
plt.rcParams['figure.figsize'] = [10, 10]
plt.xticks([0,1 / 6, 0.25, 3/8, 0.5, 5/8, 0.75, 5/6,1])
plt.plot(x,uno)
plt.plot(x,low)
sig = 0.17
wave = stats.norm(scale = sig)
n100 = wave.pdf(x - ph) / wave.pdf(0)
plt.plot(x,n100)
sig = 0.125
wave = stats.norm(scale = sig)
n23 = wave.pdf(x - ph) / wave.pdf(0)
plt.plot(x,n23)
sig = 0.095
wave = stats.norm(scale = sig)
n12 = wave.pdf(x - ph) / wave.pdf(0)
plt.plot(x,n12)
sig = 0.05
wave = stats.norm(scale = sig)
n14 = wave.pdf(x - ph) / wave.pdf(0)
plt.plot(x,n14)
# http://elki.dbs.ifi.lmu.de/browser/elki/elki-core-math/src/main/java/de/lmu/ifi/dbs/elki/math/statistics/distribution/SkewGeneralizedNormalDistribution.java
def skewed(x, sigma, skew):
if (skew == 0):
return stats.norm(scale = sigma).pdf(x)
x = x / sigma
arg = -skew * x
if (arg <= -1):
return 0
y = -np.log1p(-skew * x) / skew
ONE_BY_SQRTTWOPI = 1. / np.sqrt(2. * np.pi)
return ONE_BY_SQRTTWOPI / sigma * np.exp(-0.5 * y * y) / (1 - skew * x)
plt.plot(x,uno)
plt.plot(x,low)
plt.plot(x,n100)
ph = 0.68
sigma = 0.4
skew = 1.2
wave = np.vectorize( lambda x: skewed(x, sigma, skew))
sk100 = wave(x - ph)
sk100 = sk100 -sk100.min()
sk100 = sk100 / sk100.max()
sk100 = np.roll(sk100,-40)
plt.plot(x,sk100)
ph = 0.76
sigma = 0.2
skew = 0.8
wave = np.vectorize( lambda x: skewed(x, sigma, skew))
sk100 = wave(x - ph)
sk100 = sk100 -sk100.min()
sk100 = sk100 / sk100.max()
sk100 = np.roll(sk100,-30)
plt.plot(x,sk100)
ph = 0.67
sigma = 0.18
skew = 0.4
wave = np.vectorize( lambda x: skewed(x, sigma, skew))
sk100 = wave(x - ph)
sk100 = sk100 -sk100.min()
sk100 = sk100 / sk100.max()
sk100 = np.roll(sk100,-20)
plt.plot(x,sk100)
#sk100
plt.plot(x,uno)
plt.plot(x,low)
plt.plot(x,n23)
ph = 0.72
sigma = 0.34
skew = 1.2
wave = np.vectorize( lambda x: skewed(x, sigma, skew))
sk100 = wave(x - ph)
sk100 = sk100 -sk100.min()
sk100 = sk100 / sk100.max()
sk100 = np.roll(sk100,-40)
plt.plot(x,sk100)
ph = 0.79
sigma = 0.17
skew = 0.8
wave = np.vectorize( lambda x: skewed(x, sigma, skew))
sk100 = wave(x - ph)
sk100 = sk100 -sk100.min()
sk100 = sk100 / sk100.max()
sk100 = np.roll(sk100,-35)
plt.plot(x,sk100)
ph = 0.76
sigma = 0.14
skew = 0.4
wave = np.vectorize( lambda x: skewed(x, sigma, skew))
sk100 = wave(x - ph)
sk100 = sk100 -sk100.min()
sk100 = sk100 / sk100.max()
sk100 = np.roll(sk100,-30)
plt.plot(x,sk100)
#sk100
plt.plot(x,uno)
plt.plot(x,low)
plt.plot(x,n12)
ph = 0.8
sigma = 0.25
skew = 1.2
wave = np.vectorize( lambda x: skewed(x, sigma, skew))
sk100 = wave(x - ph)
sk100 = sk100 -sk100.min()
sk100 = sk100 / sk100.max()
sk100 = np.roll(sk100,-45)
plt.plot(x,sk100)
ph = 0.84
sigma = 0.14
skew = 0.8
wave = np.vectorize( lambda x: skewed(x, sigma, skew))
sk100 = wave(x - ph)
sk100 = sk100 -sk100.min()
sk100 = sk100 / sk100.max()
sk100 = np.roll(sk100,-40)
plt.plot(x,sk100)
ph = 0.76
sigma = 0.1
skew = 0.4
wave = np.vectorize( lambda x: skewed(x, sigma, skew))
sk100 = wave(x - ph)
sk100 = sk100 -sk100.min()
sk100 = sk100 / sk100.max()
sk100 = np.roll(sk100,-30)
plt.plot(x,sk100)
#sk100
plt.plot(x,uno)
plt.plot(x,low)
plt.plot(x,n100)
ph = .04
shape = 10.1
scale = 0.05
wave = stats.gamma(a = shape, scale = scale, loc = 0 )
ga100 = wave.pdf(x - ph)
ga100 = ga100 / ga100.max()
plt.plot(x,ga100)
ph = 0.25
shape = 5.1
scale = 0.06
wave = stats.gamma(a = shape, scale = scale, loc = 0 )
ga200 = wave.pdf(x - ph)
ga200 = ga200 / ga200.max()
plt.plot(x,ga200)
#COS
xt=[0.0, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1, 0.11, 0.12, 0.13, 0.14, 0.15, 0.16, 0.17, 0.18, 0.19, 0.2, 0.21, 0.22, 0.23, 0.24, 0.25, 0.26, 0.27, 0.28, 0.29, 0.3, 0.31, 0.32, 0.33, 0.34, 0.35, 0.36, 0.37, 0.38, 0.39, 0.4, 0.41, 0.42, 0.43, 0.44, 0.45, 0.46, 0.47, 0.48, 0.49, 0.5, 0.51, 0.52, 0.53, 0.54, 0.55, 0.56, 0.57, 0.58, 0.59, 0.6, 0.61, 0.62, 0.63, 0.64, 0.65, 0.66, 0.67, 0.68, 0.69, 0.7, 0.71, 0.72, 0.73, 0.74, 0.75, 0.76, 0.77, 0.78, 0.79, 0.8, 0.81, 0.82, 0.83, 0.84, 0.85, 0.86, 0.87, 0.88, 0.89, 0.9, 0.91, 0.92, 0.93, 0.94, 0.95, 0.96, 0.97, 0.98, 0.99]
yt=[1.0, 0.9990133642141358, 0.996057350657239, 0.9911436253643444, 0.9842915805643155, 0.9755282581475768, 0.9648882429441257, 0.9524135262330098, 0.9381533400219317, 0.9221639627510075, 0.9045084971874737, 0.8852566213878946, 0.8644843137107058, 0.8422735529643444, 0.8187119948743449, 0.7938926261462366, 0.7679133974894983, 0.7408768370508576, 0.7128896457825363, 0.6840622763423391, 0.6545084971874737, 0.6243449435824275, 0.5936906572928624, 0.5626666167821521, 0.5313952597646567, 0.5, 0.4686047402353433, 0.4373333832178478, 0.4063093427071376, 0.3756550564175727, 0.34549150281252633, 0.31593772365766104, 0.28711035421746367, 0.2591231629491423, 0.23208660251050156, 0.2061073738537635, 0.18128800512565513, 0.15772644703565564, 0.13551568628929433, 0.11474337861210543, 0.09549150281252633, 0.07783603724899257, 0.06184665997806832, 0.04758647376699032, 0.035111757055874326, 0.024471741852423234, 0.015708419435684462, 0.008856374635655695, 0.0039426493427611176, 9.866357858642205E-4, 0.0, 9.866357858642205E-4, 0.0039426493427611176, 0.008856374635655695, 0.015708419435684517, 0.02447174185242329, 0.03511175705587444, 0.04758647376699021, 0.06184665997806821, 0.07783603724899246, 0.09549150281252633, 0.11474337861210543, 0.13551568628929433, 0.15772644703565564, 0.18128800512565513, 0.2061073738537635, 0.23208660251050178, 0.2591231629491425, 0.28711035421746384, 0.31593772365766093, 0.3454915028125262, 0.3756550564175726, 0.4063093427071376, 0.4373333832178478, 0.4686047402353433, 0.5, 0.5313952597646567, 0.5626666167821522, 0.5936906572928624, 0.6243449435824275, 0.6545084971874738, 0.6840622763423392, 0.7128896457825362, 0.7408768370508576, 0.7679133974894983, 0.7938926261462365, 0.8187119948743449, 0.8422735529643444, 0.8644843137107058, 0.8852566213878947, 0.9045084971874737, 0.9221639627510076, 0.9381533400219318, 0.9524135262330098, 0.9648882429441257, 0.9755282581475768, 0.9842915805643155, 0.9911436253643443, 0.9960573506572389, 0.9990133642141358]
plt.plot(xt, yt)
#WIDE_PEAK no fix min
# case NONE: return new GaussianWaveform(0.203125, phase, false);
# /*
# case LOW: return new SkewedGaussWaveform(0.21897347065651573, skew.skew, phase, false);
# case MID: return new SkewedGaussWaveform(0.2784780878770199, skew.skew, phase, false);
# case HIGH: return new SkewedGaussWaveform(0.415636603046377, skew.skew, phase, false);
xt=[0.0, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1, 0.11, 0.12, 0.13, 0.14, 0.15, 0.16, 0.17, 0.18, 0.19, 0.2, 0.21, 0.22, 0.23, 0.24, 0.25, 0.26, 0.27, 0.28, 0.29, 0.3, 0.31, 0.32, 0.33, 0.34, 0.35, 0.36, 0.37, 0.38, 0.39, 0.4, 0.41, 0.42, 0.43, 0.44, 0.45, 0.46, 0.47, 0.48, 0.49, 0.5, 0.51, 0.52, 0.53, 0.54, 0.55, 0.56, 0.57, 0.58, 0.59, 0.6, 0.61, 0.62, 0.63, 0.64, 0.65, 0.66, 0.67, 0.68, 0.69, 0.7, 0.71, 0.72, 0.73, 0.74, 0.75, 0.76, 0.77, 0.78, 0.79, 0.8, 0.81, 0.82, 0.83, 0.84, 0.85, 0.86, 0.87, 0.88, 0.89, 0.9, 0.91, 0.92, 0.93, 0.94, 0.95, 0.96, 0.97, 0.98, 0.99]
yt=[1.0, 0.9987888996551672, 0.9951643921014979, 0.9891527514554127, 0.9807974152891648, 0.9701584619536344, 0.9573118912015268, 0.9423487201601802, 0.9253739096952884, 0.9065051388774692, 0.8858714475589475, 0.8636117689435711, 0.8398733754565454, 0.8148102621687294, 0.7885814924939011, 0.7613495308577245, 0.7332785865473379, 0.7045329920144862, 0.6752756375567601, 0.6456664825833416, 0.6158611616335171, 0.5860097010137122, 0.5562553594115757, 0.5267336031957593, 0.49757122438015355, 0.46888560648317157, 0.44078414080532446, 0.4133637930369936, 0.3867108176429706, 0.36090061519481487, 0.3359977257732059, 0.31205594976959017, 0.2891185859010446, 0.26721877502808633, 0.2463799374380964, 0.22661629062572547, 0.20793343425796706, 0.19032898894126954, 0.17379327559160265, 0.1583100226219387, 0.14385708877784129, 0.1304071902410655, 0.11792862155203371, 0.10638596094299341, 0.09574075179308944, 0.0859521530840816, 0.076977552922306, 0.06877314037233147, 0.06129443199693493, 0.054496750595930094, 0.048335654665708876, 0.054496750595930094, 0.06129443199693493, 0.06877314037233147, 0.07697755292230607, 0.08595215308408165, 0.09574075179308947, 0.10638596094299331, 0.11792862155203365, 0.13040719024106545, 0.14385708877784129, 0.1583100226219387, 0.17379327559160265, 0.19032898894126954, 0.20793343425796706, 0.22661629062572547, 0.24637993743809652, 0.26721877502808644, 0.2891185859010447, 0.3120559497695901, 0.33599772577320586, 0.3609006151948148, 0.3867108176429706, 0.4133637930369936, 0.44078414080532446, 0.46888560648317157, 0.49757122438015355, 0.5267336031957595, 0.5562553594115758, 0.5860097010137123, 0.6158611616335173, 0.6456664825833417, 0.67527563755676, 0.704532992014486, 0.7332785865473377, 0.7613495308577244, 0.7885814924939011, 0.8148102621687294, 0.8398733754565454, 0.8636117689435712, 0.8858714475589475, 0.9065051388774692, 0.9253739096952885, 0.9423487201601803, 0.9573118912015268, 0.9701584619536343, 0.9807974152891648, 0.9891527514554127, 0.9951643921014979, 0.9987888996551672]
plt.plot(xt, yt)
yt=[1.0, 0.9985337870668837, 0.9940178136391471, 0.9862879138460792, 0.9751988723702859, 0.9606293631911291, 0.9424872367484814, 0.9207150779901252, 0.8952959269393653, 0.8662590179489144, 0.8336853540417393, 0.7977128894808132, 0.758541048362926, 0.7164342617787033, 0.6717241640662659, 0.6248100542016115, 0.5761572070119776, 0.5262926176620537, 0.47579779010002304, 0.4252982453321619, 0.37544953855460284, 0.3269197449059852, 0.28036860948936987, 0.2364238615847212, 0.19565556141136928, 0.1585497648119985, 0.12548322544227722, 0.09670125405191868, 0.0723011459542647, 0.06896702759489831, 0.07243961456383377, 0.0760841324091454, 0.07990856974616012, 0.08392122294723851, 0.08813070071714943, 0.09254592777989244, 0.09717614752876519, 0.10203092347562741, 0.10712013931816212, 0.11245399742539441, 0.11804301552174556, 0.12389802132839711, 0.13003014489767012, 0.13645080835143822, 0.143171712708246, 0.15020482145578343, 0.1575623404956575, 0.16525669405603718, 0.1733004961347555, 0.18170651700092888, 0.19048764424721906, 0.19965683784767513, 0.20922707863790907, 0.2192113095954468, 0.22962236925886825, 0.24047291658526712, 0.25177534650720934, 0.2635416954134839, 0.2757835357433708, 0.28851185885293595, 0.3017369452852588, 0.31546822155591203, 0.3297141025522071, 0.3444818186416254, 0.3597772265938246, 0.37560460344427626, 0.3919664224689946, 0.40886311050248597, 0.4262927859188711, 0.4442509767136279, 0.4627303182755495, 0.4817202306309141, 0.501206575180716, 0.5211712912439482, 0.5415920130727792, 0.5624416684271292, 0.5836880602952406, 0.6052934339324837, 0.6272140320723614, 0.6493996419512129, 0.671793138691142, 0.6943300306135921, 0.716938013217347, 0.7395365398568545, 0.7620364186049235, 0.7843394463805416, 0.8063380931662674, 0.8279152510237264, 0.8489440646266927, 0.8692878621469449, 0.8888002075155294, 0.907325097294831, 0.9246973275721614, 0.940743058340922, 0.955280604665043, 0.968121485393274, 0.9790717611376433, 0.9879336934552613, 0.9945077564356877, 0.9985950299162479]
plt.plot(xt, yt)
yt=[1.0, 0.9975510364251826, 0.9896552937276493, 0.9754250344672477, 0.9538932084329071, 0.9240306707199512, 0.8847799938978558, 0.835114983723763, 0.7741393327924341, 0.7012438153695731, 0.6163490256781425, 0.5202685262373395, 0.41522980271506027, 0.3055697381162451, 0.19852529695426535, 0.1047253536630324, 0.09952458775530065, 0.10217560350924919, 0.10490854206567754, 0.10772625862576309, 0.11063171672512416, 0.11362799245916687, 0.11671827885543067, 0.1199058903945872, 0.12319426768117116, 0.12658698226443943, 0.1300877416089389, 0.1337003942134063, 0.1374289348754912, 0.1412775100984766, 0.1452504236346249, 0.1493521421579854, 0.1535873010574145, 0.15796071033814574, 0.16247736061745424, 0.1671424291967366, 0.1719612861886087, 0.17693950067334172, 0.18208284685402804, 0.18739731017420438, 0.1928890933551486, 0.19856462230259295, 0.2044305518240226, 0.21049377108788095, 0.21676140874472888, 0.22324083761745803, 0.22993967885283223, 0.23686580540962618, 0.2440273447391467, 0.25143268049157985, 0.2590904530560047, 0.2670095587125516, 0.2751991471415242, 0.2836686169956862, 0.2924276091976282, 0.3014859975732905, 0.31085387637438305, 0.3205415441754416, 0.3305594835543096, 0.34091833587640685, 0.3516288704015034, 0.36270194681482765, 0.37414847014986563, 0.38597933691545794, 0.39820537106163145, 0.4108372482134034, 0.42388540636539196, 0.43735994095761155, 0.4512704819387737, 0.4656260500612941, 0.48043488923465555, 0.4957042712822553, 0.5114402688916571, 0.5276474919081199, 0.5443287813837295, 0.5614848549451181, 0.5791138960655363, 0.5972110787041198, 0.6157680174869952, 0.6347721321303812, 0.6542059131230359, 0.6740460737722095, 0.6942625715530278, 0.7148174792691809, 0.7356636838237534, 0.7567433874173666, 0.7779863827628906, 0.7993080704937563, 0.8206071834629612, 0.8417631792828963, 0.8626332595697469, 0.8830489724487088, 0.9028123547508695, 0.9216915732171409, 0.939416031778589, 0.9556709274046181, 0.9700912642965331, 0.9822553816292672, 0.9916781229819943, 0.9978038900420947]
plt.plot(xt, yt)
yt=[1.0, 0.994131529661843, 0.9732155761251555, 0.9305281794879899, 0.8561715724769962, 0.7356291648274507, 0.548881333664372, 0.27847354184296536, 0.11370490538739975, 0.11569997309245596, 0.11774244407738371, 0.11983376000538844, 0.12197541631448319, 0.1241689645950756, 0.12641601508854627, 0.12871823931370432, 0.13107737282843168, 0.133495218134278, 0.13597364773224727, 0.13851460733852752, 0.14112011926945378, 0.14379228600557217, 0.1465332939452796, 0.14934541735916013, 0.15223102255682408, 0.15519257227877842, 0.1582326303266232, 0.16135386644567384, 0.1645590614749623, 0.16785111278046055, 0.17123303998830966, 0.17470799103581677, 0.17827924855900681, 0.18195023663657303, 0.1857245279111725, 0.18960585111013656, 0.19359809898881764, 0.19770533672095816, 0.20193181076162856, 0.20628195820943157, 0.210760416695775, 0.21537203483006112, 0.22012188323058046, 0.2250152661716974, 0.23005773387851117, 0.2352550955005079, 0.24061343279569464, 0.24613911455622106, 0.2518388118054221, 0.2577195137943767, 0.26378854482328784, 0.27005358190898116, 0.27652267331428537, 0.2832042579476088, 0.2901071856311783, 0.29724073822356023, 0.30461465156548556, 0.3122391381967358, 0.3201249107647289, 0.32828320601105004, 0.3367258091786584, 0.34546507862760417, 0.3545139703779291, 0.36388606221139375, 0.3735955768541872, 0.3836574036250458, 0.3940871177598649, 0.4049009964055202, 0.4161160300002061, 0.4277499274096736, 0.4398211127484022, 0.4523487112561398, 0.46535252088987933, 0.47885296538536826, 0.4928710233831996, 0.5074281267265734, 0.5225460191204192, 0.5382465638606903, 0.5545514861184785, 0.5714820310535343, 0.5890585135060767, 0.607299727723445, 0.6262221758973758, 0.6458390613548075, 0.6661589748500951, 0.6871841788393103, 0.7089083624383448, 0.7313136954605031, 0.7543669483773343, 0.7780143587096144, 0.8021748019979849, 0.8267306501648432, 0.8515154458159513, 0.8762971475860714, 0.9007551456194267, 0.9244484063464173, 0.9467708173448146, 0.9668877952390316, 0.9836450403618952, 0.995435212698413]
plt.plot(xt, yt)
#WIDE_PEAK fix min
# case NONE: return new GaussianWaveform(0.203125, phase, false);
# case LOW: return new SkewedGaussWaveform(0.711034440272608, skew.skew, phase, true);
# case MID: return new SkewedGaussWaveform(0.6700816291266387, skew.skew, phase, true);
# case HIGH: return new SkewedGaussWaveform(.7442591767475941, skew.skew, phase, true);
xt=[0.0, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1, 0.11, 0.12, 0.13, 0.14, 0.15, 0.16, 0.17, 0.18, 0.19, 0.2, 0.21, 0.22, 0.23, 0.24, 0.25, 0.26, 0.27, 0.28, 0.29, 0.3, 0.31, 0.32, 0.33, 0.34, 0.35, 0.36, 0.37, 0.38, 0.39, 0.4, 0.41, 0.42, 0.43, 0.44, 0.45, 0.46, 0.47, 0.48, 0.49, 0.5, 0.51, 0.52, 0.53, 0.54, 0.55, 0.56, 0.57, 0.58, 0.59, 0.6, 0.61, 0.62, 0.63, 0.64, 0.65, 0.66, 0.67, 0.68, 0.69, 0.7, 0.71, 0.72, 0.73, 0.74, 0.75, 0.76, 0.77, 0.78, 0.79, 0.8, 0.81, 0.82, 0.83, 0.84, 0.85, 0.86, 0.87, 0.88, 0.89, 0.9, 0.91, 0.92, 0.93, 0.94, 0.95, 0.96, 0.97, 0.98, 0.99]
yt=[1.0, 0.9987888996551672, 0.9951643921014979, 0.9891527514554127, 0.9807974152891648, 0.9701584619536344, 0.9573118912015268, 0.9423487201601802, 0.9253739096952884, 0.9065051388774692, 0.8858714475589475, 0.8636117689435711, 0.8398733754565454, 0.8148102621687294, 0.7885814924939011, 0.7613495308577245, 0.7332785865473379, 0.7045329920144862, 0.6752756375567601, 0.6456664825833416, 0.6158611616335171, 0.5860097010137122, 0.5562553594115757, 0.5267336031957593, 0.49757122438015355, 0.46888560648317157, 0.44078414080532446, 0.4133637930369936, 0.3867108176429706, 0.36090061519481487, 0.3359977257732059, 0.31205594976959017, 0.2891185859010446, 0.26721877502808633, 0.2463799374380964, 0.22661629062572547, 0.20793343425796706, 0.19032898894126954, 0.17379327559160265, 0.1583100226219387, 0.14385708877784129, 0.1304071902410655, 0.11792862155203371, 0.10638596094299341, 0.09574075179308944, 0.0859521530840816, 0.076977552922306, 0.06877314037233147, 0.06129443199693493, 0.054496750595930094, 0.048335654665708876, 0.054496750595930094, 0.06129443199693493, 0.06877314037233147, 0.07697755292230607, 0.08595215308408165, 0.09574075179308947, 0.10638596094299331, 0.11792862155203365, 0.13040719024106545, 0.14385708877784129, 0.1583100226219387, 0.17379327559160265, 0.19032898894126954, 0.20793343425796706, 0.22661629062572547, 0.24637993743809652, 0.26721877502808644, 0.2891185859010447, 0.3120559497695901, 0.33599772577320586, 0.3609006151948148, 0.3867108176429706, 0.4133637930369936, 0.44078414080532446, 0.46888560648317157, 0.49757122438015355, 0.5267336031957595, 0.5562553594115758, 0.5860097010137123, 0.6158611616335173, 0.6456664825833417, 0.67527563755676, 0.704532992014486, 0.7332785865473377, 0.7613495308577244, 0.7885814924939011, 0.8148102621687294, 0.8398733754565454, 0.8636117689435712, 0.8858714475589475, 0.9065051388774692, 0.9253739096952885, 0.9423487201601803, 0.9573118912015268, 0.9701584619536343, 0.9807974152891648, 0.9891527514554127, 0.9951643921014979, 0.9987888996551672]
plt.plot(xt, yt)
yt=[1.0, 0.9995102843432049, 0.9980284607237517, 0.9955358001111833, 0.9920139820513978, 0.9874451365272697, 0.9818118871445073, 0.9750973956232678, 0.9672854075707601, 0.9583602995043832, 0.9483071270889154, 0.9371116745448073, 0.9247605051778345, 0.9112410129730932, 0.8965414751887001, 0.8806511058764787, 0.8635601102484152, 0.8452597397987818, 0.8257423480824496, 0.8050014470401782, 0.7830317637514589, 0.7598292974849418, 0.7353913769054206, 0.7097167172850146, 0.6828054775544402, 0.6546593170181222, 0.625281451544565, 0.594676709030621, 0.5628515839254216, 0.5298142905865504, 0.4955748152277632, 0.4601449662041548, 0.42353842236727235, 0.3857707792092918, 0.34685959250213566, 0.3068244191243798, 0.26568685475611564, 0.22347056810963686, 0.18020133135210678, 0.13590704636534567, 0.09061776647765024, 0.04436571329336314, 0.0, 0.02487450804866056, 0.049732928424932404, 0.07456586695834065, 0.09936364246441017, 0.1241162832847375, 0.14881352398102166, 0.17344480219628583, 0.19799925569713314, 0.22246571961140235, 0.24683272387620217, 0.27108849091189324, 0.29522093353817436, 0.31921765314905404, 0.3430659381641156, 0.3667527627740813, 0.3902647859993561, 0.4135883510808275, 0.4367094852228859, 0.4596138997092142, 0.4822869904126182, 0.5047138387207257, 0.5268792129001052, 0.5487675699219278, 0.5703630577729658, 0.5916495182763223, 0.6126104904468823, 0.6332292144071151, 0.6534886358893555, 0.6733714113513362, 0.6928599137321704, 0.7119362388765922, 0.7305822126556258, 0.7487793988123691, 0.7665091075619057, 0.7837524049747516, 0.8004901231734921, 0.8167028713725416, 0.832371047791083, 0.8474748524694163, 0.8619943010188972, 0.875909239335673, 0.8891993593082157, 0.9018442155484633, 0.9138232431760318, 0.9251157766844703, 0.9357010699180284, 0.9455583171866245, 0.9546666755459245, 0.963005288268412, 0.9705533095301959, 0.9772899303369702, 0.9831944057110563, 0.9882460831597464, 0.9924244324432898, 0.9957090766587328, 0.9980798246535053, 0.999516704780054]
plt.plot(xt, yt)
yt=[1.0, 0.9992157374601713, 0.9967910252496027, 0.9926142243124053, 0.9865687143537644, 0.9785329597780681, 0.9683806446621771, 0.9559808923664647, 0.9411985883372177, 0.9238948281292443, 0.9039275167632181, 0.881152150325474, 0.8554228163261821, 0.8265934558715815, 0.7945194382989864, 0.7590595076897714, 0.7200781707224045, 0.6774486067285324, 0.63105619357916, 0.5808027570491707, 0.5266116662874221, 0.46843391334248685, 0.4062553292449494, 0.3401051010220707, 0.2700657600888102, 0.19628480769377324, 0.11898811955698216, 0.0384952171087181, 0.007632393404751336, 0.019700894320261127, 0.0319227377906967, 0.044298724760434026, 0.05682957443558885, 0.06951591744942406, 0.08235828859330907, 0.09535711908684807, 0.1085127283592086, 0.1218253153119767, 0.1352949490320834, 0.1489215589214369, 0.16270492420789207, 0.17664466280004973, 0.19074021944612649, 0.2049908531547537, 0.21939562383303374, 0.23395337809452307, 0.24866273418698645, 0.2635220659867983, 0.2785294860037282, 0.29368282733653345, 0.30897962451630967, 0.3244170931708697, 0.3399921084395755, 0.3557011820640028, 0.3715404380755658, 0.3875055869968012, 0.40359189846836224, 0.41979417220893456, 0.43610670721025185, 0.45252326906416646, 0.46903705531331225, 0.48564065871132867, 0.502326028272891, 0.5190844279879271, 0.5359063930684612, 0.5527816835905127, 0.569699235387468, 0.5866471080453762, 0.6036124298447949, 0.6205813394881889, 0.6375389244466161, 0.6544691557546123, 0.6713548190780048, 0.6881774418760314, 0.704917216476835, 0.7215529188844547, 0.7380618231361371, 0.7544196110315892, 0.7706002770611112, 0.786576028367983, 0.8023171795926761, 0.8177920424632094, 0.8329668100182119, 0.847805435378083, 0.8622695050163403, 0.8763181065294406, 0.8899076909607181, 0.9029919298049421, 0.9155215669067377, 0.9274442655718558, 0.9387044513384791, 0.9492431510106848, 0.9589978287427351, 0.9679022201868954, 0.9758861659859396, 0.9828754462124326, 0.988791617740055, 0.9935518569888925, 0.997068811030219, 0.999250460682733]
plt.plot(xt, yt)
yt=[1.0, 0.9976422085879377, 0.9898718809541124, 0.9754599259642711, 0.9528888735734694, 0.9202782062213068, 0.8752911490583485, 0.8150225433499098, 0.7358758886066773, 0.6334633833725137, 0.502639436577133, 0.33801472869722626, 0.13608984970747226, 0.005513084648484356, 0.010483407259427995, 0.015544230096937344, 0.020697635431358056, 0.025945761644221275, 0.03129080481665839, 0.03673502035196645, 0.04228072463121288, 0.04793029670045044, 0.05368617998774652, 0.05955088404782143, 0.06552698633160717, 0.07161713397750648, 0.07782404562050979, 0.08415051321462684, 0.09059940386329679, 0.09717366165152644, 0.10387630947247785, 0.11071045084005456, 0.1176792716776992, 0.12478604207210241, 0.13203411797879788, 0.13942694286466323, 0.14696804927011622, 0.15466106027126558, 0.16250969081939204, 0.17051774893185784, 0.1786891367048084, 0.1870278511137712, 0.19553798456341284, 0.2042237251421776, 0.21308935653122146, 0.22213925750983857, 0.23137790099133565, 0.2408098525138736, 0.250439768099997, 0.26027239138618813, 0.2703125499095788, 0.28056515042263747, 0.2910351730878897, 0.30172766438313336, 0.3126477285227275, 0.32380051717180647, 0.3351912171970877, 0.3468250361595393, 0.3587071852096795, 0.37084285899463054, 0.3832372121260397, 0.3958953316880915, 0.40882220518334705, 0.42202268321894787, 0.43550143612429637, 0.44926290356067666, 0.46331123602978, 0.4776502270073615, 0.49228323421504544, 0.5072130882911485, 0.5224419868225776, 0.5379713713448412, 0.5538017844943979, 0.569932703992691, 0.5863623495367449, 0.603087457945416, 0.6201030210364852, 0.6374019796542482, 0.6549748659884129, 0.6728093847705899, 0.6908899220385872, 0.7091969678378942, 0.7277064363784859, 0.7463888636487767, 0.7652084581347872, 0.7841219748796451, 0.8030773763597929, 0.8220122351777226, 0.8408518228911681, 0.8595068157790919, 0.877870531147271, 0.8958155857917309, 0.9131898399932121, 0.929811453945427, 0.9454628361970487, 0.9598832019956413, 0.9727593786589126, 0.9837143890090183, 0.9922932042618913, 0.9979448739972219]
plt.plot(xt, yt)
#ONE_THIRD_PEAK no fix min
# case NONE: return new GaussianWaveform(0.140625, phase, false);
# /*case LOW: return new SkewedGaussWaveform(0.1516067989805035, skew.skew, phase, false);
# case MID: return new SkewedGaussWaveform(.19283827997555814, skew.skew, phase, false);
# case HIGH: return new SkewedGaussWaveform(.2878768391594375, skew.skew, phase, false);*/
xt=[0.0, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1, 0.11, 0.12, 0.13, 0.14, 0.15, 0.16, 0.17, 0.18, 0.19, 0.2, 0.21, 0.22, 0.23, 0.24, 0.25, 0.26, 0.27, 0.28, 0.29, 0.3, 0.31, 0.32, 0.33, 0.34, 0.35, 0.36, 0.37, 0.38, 0.39, 0.4, 0.41, 0.42, 0.43, 0.44, 0.45, 0.46, 0.47, 0.48, 0.49, 0.5, 0.51, 0.52, 0.53, 0.54, 0.55, 0.56, 0.57, 0.58, 0.59, 0.6, 0.61, 0.62, 0.63, 0.64, 0.65, 0.66, 0.67, 0.68, 0.69, 0.7, 0.71, 0.72, 0.73, 0.74, 0.75, 0.76, 0.77, 0.78, 0.79, 0.8, 0.81, 0.82, 0.83, 0.84, 0.85, 0.86, 0.87, 0.88, 0.89, 0.9, 0.91, 0.92, 0.93, 0.94, 0.95, 0.96, 0.97, 0.98, 0.99]
yt=[1.0, 0.9974747986368548, 0.9899373900304076, 0.9775013993578034, 0.9603530314874869, 0.938746432156527, 0.9129974218422362, 0.883475818227102, 0.8505966079942792, 0.8148102621687294, 0.776592510126528, 0.7364338951763066, 0.6948294293818791, 0.6522686477895945, 0.6092263337644984, 0.5661541495171976, 0.5234733612647516, 0.48156879918121415, 0.4407841408053246, 0.4014185552787085, 0.3637246969042015, 0.32790799197435705, 0.29412712419549963, 0.2624955924772389, 0.23308419108452028, 0.20592424643419877, 0.1810114370251389, 0.15831002262193872, 0.13775731507104333, 0.11926823499333838, 0.10273981490249434, 0.0880555288083265, 0.07508934984809916, 0.06370945978172436, 0.053781556242727124, 0.045171724564724666, 0.037748860091299, 0.031386643603503045, 0.02596508652980629, 0.021371673780233053, 0.01750214036840672, 0.014260923582095412, 0.011561335567520117, 0.009325502111768767, 0.0074841124933293325, 0.005976022895005943, 0.004747752405935435, 0.003752906429386003, 0.002951557676005975, 0.002309610130864761, 0.0017981666618475832, 0.002309610130864761, 0.002951557676005975, 0.003752906429386003, 0.004747752405935443, 0.005976022895005953, 0.0074841124933293385, 0.00932550211176875, 0.011561335567520096, 0.014260923582095386, 0.01750214036840672, 0.021371673780233053, 0.02596508652980629, 0.031386643603503045, 0.037748860091299, 0.045171724564724666, 0.05378155624272717, 0.06370945978172445, 0.07508934984809923, 0.08805552880832646, 0.1027398149024943, 0.11926823499333827, 0.13775731507104333, 0.15831002262193872, 0.1810114370251389, 0.20592424643419877, 0.23308419108452028, 0.262495592477239, 0.2941271241954997, 0.32790799197435705, 0.3637246969042018, 0.40141855527870873, 0.4407841408053243, 0.4815687991812141, 0.5234733612647515, 0.5661541495171974, 0.6092263337644984, 0.6522686477895945, 0.6948294293818791, 0.7364338951763068, 0.7765925101265282, 0.8148102621687295, 0.8505966079942793, 0.8834758182271021, 0.912997421842236, 0.938746432156527, 0.9603530314874867, 0.9775013993578034, 0.9899373900304076, 0.9974747986368548]
plt.plot(xt, yt)
yt=[1.0, 0.9969144020240112, 0.9873132505712133, 0.9707403135256472, 0.9468411303791583, 0.9153963475301403, 0.8763571387546044, 0.8298810204084439, 0.7763656397013128, 0.7164772943316775, 0.6511700977395593, 0.5816909502345677, 0.5095649899332954, 0.43655623072339234, 0.36459897280863635, 0.29569766904455874, 0.23179660367644392, 0.17462619710324762, 0.12553983583287942, 0.0853630244825286, 0.05428355812564234, 0.03181431065967187, 0.016855155175558616, 0.009616812969173738, 0.010311715794179629, 0.011058068317145602, 0.011859732512134544, 0.012720859468089997, 0.013645910353472105, 0.014639678761954062, 0.015707314505990277, 0.01685434892396994, 0.018086721764568214, 0.019410809708582905, 0.02083345658376502, 0.022362005321635094, 0.024004331696706765, 0.025768879877542856, 0.027664699805232052, 0.029701486397707606, 0.031889620557287064, 0.03424021193326941, 0.03676514336066708, 0.039477116859378394, 0.042389701034409956, 0.045517379666117345, 0.048875601218705406, 0.052480828924133985, 0.05635059101569209, 0.060503530589251356, 0.06495945445885726, 0.06973938024494675, 0.07486558078602443, 0.08036162479582923, 0.08625241249546979, 0.09256420473113523, 0.09932464384010198, 0.10656276424808873, 0.11430899046673222, 0.12259511980829721, 0.13145428674306497, 0.14092090539078958, 0.15103058615927817, 0.16182002201930662, 0.17332683933544366, 0.18558940755801656, 0.19864660142522886, 0.21253750863154763, 0.22730107519711795, 0.24297568003524794, 0.25959862947791, 0.27720556180579736, 0.295829751170397, 0.3155012997306196, 0.33624620640682856, 0.3580853004451419, 0.3810330280651537, 0.40509608093409183, 0.43027185619117697, 0.4565467393839419, 0.4838942041482799, 0.5122727259721258, 0.541623512168875, 0.5718680565261275, 0.6029055352982476, 0.6346100716197727, 0.666827908398138, 0.6993745456786437, 0.7320319177345702, 0.7645457080554008, 0.7966229272380713, 0.8279299096368556, 0.8580909193733472, 0.8866875944816707, 0.9132594986073665, 0.9373060911561782, 0.9582904665605484, 0.9756452476797082, 0.9887810420716499, 0.9970978759575706]
plt.plot(xt, yt)
yt=[1.0, 0.9947673420474036, 0.977359315191228, 0.9449447014890051, 0.8944232417815924, 0.8226660957255137, 0.7270368145751555, 0.6064162572813783, 0.4630852307714943, 0.30590394884325683, 0.15477662968437159, 0.04314905952461143, 0.03518796976901374, 0.036339310171593274, 0.03753457366329907, 0.03877567856899969, 0.04006463746770648, 0.04140356230937693, 0.04279466983227655, 0.04424028729945708, 0.045742858574052137, 0.047304950554298696, 0.048929259990460036, 0.05061862070715829, 0.05237601125602062, 0.05420456302499993, 0.056107568832251424, 0.05808849203402141, 0.06015097617763478, 0.06229885523234258, 0.06453616443250446, 0.06686715176931628, 0.06929629016904208, 0.07182829039744304, 0.07446811473179742, 0.07722099144353528, 0.0800924301360365, 0.08308823798350734, 0.08621453691800048, 0.08947778181250364, 0.09288477970849743, 0.09644271013637146, 0.10015914657644401, 0.10404207910690366, 0.10809993828257214, 0.11234162028474329, 0.11677651337719377, 0.12141452569642727, 0.12626611439489346, 0.13134231614379185, 0.1366547789865125, 0.14221579551403266, 0.1480383373087685, 0.1541360905723808, 0.16052349281452785, 0.16721577043193564, 0.17422897694847586, 0.1815800316148465, 0.1892867579780772, 0.19736792192296265, 0.20584326855545113, 0.21473355713683684, 0.22406059308110887, 0.2338472557883555, 0.24411752079546375, 0.2548964743701248, 0.26621031824151864, 0.2780863616340239, 0.2905529971280984, 0.30363965608957366, 0.31737673845367453, 0.3317955104845624, 0.3469279627076793, 0.36280661847219636, 0.3794642814724097, 0.39693370795112703, 0.4152471861153433, 0.43443600137960586, 0.4545297612487822, 0.47555554775428377, 0.4975368581141336, 0.520492285388978, 0.5444338799771884, 0.5693651193776602, 0.5952783972040833, 0.6221519223172526, 0.6499458944127683, 0.6785977926396467, 0.7080165779767833, 0.7380755673768572, 0.7686036876007216, 0.7993747593847523, 0.8300943996778805, 0.8603840654027469, 0.8897617057448423, 0.9176184595460687, 0.9431908642404713, 0.9655281957740405, 0.9834549486858607, 0.9955292956451746]
plt.plot(xt, yt)
yt=[1.0, 0.9869244025377764, 0.9363573387303759, 0.821525286116017, 0.5964929346110752, 0.20822109401974534, 0.057917930603761325, 0.0590891792730345, 0.06029208625855767, 0.0615277392963867, 0.06279727188753956, 0.06410186558920487, 0.06544275243937371, 0.06682121752376409, 0.06823860169456965, 0.06969630445128094, 0.07119578699460434, 0.07273857546534557, 0.07432626438103701, 0.07596052028407853, 0.07764308561623642, 0.07937578283551253, 0.08116051879266366, 0.08299928938603167, 0.08489418451484382, 0.08684739335277623, 0.08886120996535206, 0.09093803929668141, 0.09308040355316266, 0.09529094901406653, 0.09757245330143592, 0.09992783314447577, 0.1023601526766003, 0.10487263230657527, 0.1074686582087636, 0.11015179248138893, 0.11292578402600056, 0.11579458020599037, 0.11876233934711984, 0.12183344414859974, 0.1250125160793729, 0.12830443084093346, 0.1317143349853211, 0.1352476637849148, 0.13891016045937649, 0.14270789687462251, 0.14664729583908923, 0.1507351551338845, 0.15497867342573077, 0.15938547822497454, 0.1639636560654246, 0.16872178509841335, 0.17366897031029596, 0.17881488159060221, 0.18416979489720012, 0.18974463678502881, 0.1955510325860435, 0.20160135854971234, 0.20790879827531078, 0.21448740378874004, 0.22135216163680316, 0.22851906438953395, 0.23600518795458336, 0.24382877511440704, 0.25200932569378187, 0.26056769374747457, 0.26952619211951306, 0.2789087046580313, 0.2887408062615365, 0.29904989076794536, 0.30986530645531446, 0.3212184985733069, 0.33314315782652243, 0.34567537302884604, 0.35885378516442074, 0.3727197387178378, 0.38731742422284704, 0.4026940033132503, 0.4188997038416264, 0.4359878674337639, 0.4540149245558431, 0.47304026190030135, 0.49312593235018354, 0.5143361370532572, 0.5367363793873698, 0.560392147555079, 0.5853669197009236, 0.6117191927568713, 0.6394980979735889, 0.6687369573078026, 0.699443815138693, 0.7315874827346891, 0.7650768469304793, 0.7997299281925484, 0.8352270903280852, 0.8710392980589597, 0.9063162646835984, 0.9397085796321796, 0.9690782116027895, 0.9910145120396]
plt.plot(xt, yt)
#ONE_THIRD_PEAK fix min
# case NONE: return new GaussianWaveform(0.140625, phase, false);
# case LOW: return new SkewedGaussWaveform(0.16159421514938846, skew.skew, phase, true);
# case MID: return new SkewedGaussWaveform(0.2890010832949674, skew.skew, phase, true);
# case HIGH: return new SkewedGaussWaveform(0.4830298083311916, skew.skew, phase, true);
xt=[0.0, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1, 0.11, 0.12, 0.13, 0.14, 0.15, 0.16, 0.17, 0.18, 0.19, 0.2, 0.21, 0.22, 0.23, 0.24, 0.25, 0.26, 0.27, 0.28, 0.29, 0.3, 0.31, 0.32, 0.33, 0.34, 0.35, 0.36, 0.37, 0.38, 0.39, 0.4, 0.41, 0.42, 0.43, 0.44, 0.45, 0.46, 0.47, 0.48, 0.49, 0.5, 0.51, 0.52, 0.53, 0.54, 0.55, 0.56, 0.57, 0.58, 0.59, 0.6, 0.61, 0.62, 0.63, 0.64, 0.65, 0.66, 0.67, 0.68, 0.69, 0.7, 0.71, 0.72, 0.73, 0.74, 0.75, 0.76, 0.77, 0.78, 0.79, 0.8, 0.81, 0.82, 0.83, 0.84, 0.85, 0.86, 0.87, 0.88, 0.89, 0.9, 0.91, 0.92, 0.93, 0.94, 0.95, 0.96, 0.97, 0.98, 0.99]
yt=[1.0, 0.9974747986368548, 0.9899373900304076, 0.9775013993578034, 0.9603530314874869, 0.938746432156527, 0.9129974218422362, 0.883475818227102, 0.8505966079942792, 0.8148102621687294, 0.776592510126528, 0.7364338951763066, 0.6948294293818791, 0.6522686477895945, 0.6092263337644984, 0.5661541495171976, 0.5234733612647516, 0.48156879918121415, 0.4407841408053246, 0.4014185552787085, 0.3637246969042015, 0.32790799197435705, 0.29412712419549963, 0.2624955924772389, 0.23308419108452028, 0.20592424643419877, 0.1810114370251389, 0.15831002262193872, 0.13775731507104333, 0.11926823499333838, 0.10273981490249434, 0.0880555288083265, 0.07508934984809916, 0.06370945978172436, 0.053781556242727124, 0.045171724564724666, 0.037748860091299, 0.031386643603503045, 0.02596508652980629, 0.021371673780233053, 0.01750214036840672, 0.014260923582095412, 0.011561335567520117, 0.009325502111768767, 0.0074841124933293325, 0.005976022895005943, 0.004747752405935435, 0.003752906429386003, 0.002951557676005975, 0.002309610130864761, 0.0017981666618475832, 0.002309610130864761, 0.002951557676005975, 0.003752906429386003, 0.004747752405935443, 0.005976022895005953, 0.0074841124933293385, 0.00932550211176875, 0.011561335567520096, 0.014260923582095386, 0.01750214036840672, 0.021371673780233053, 0.02596508652980629, 0.031386643603503045, 0.037748860091299, 0.045171724564724666, 0.05378155624272717, 0.06370945978172445, 0.07508934984809923, 0.08805552880832646, 0.1027398149024943, 0.11926823499333827, 0.13775731507104333, 0.15831002262193872, 0.1810114370251389, 0.20592424643419877, 0.23308419108452028, 0.262495592477239, 0.2941271241954997, 0.32790799197435705, 0.3637246969042018, 0.40141855527870873, 0.4407841408053243, 0.4815687991812141, 0.5234733612647515, 0.5661541495171974, 0.6092263337644984, 0.6522686477895945, 0.6948294293818791, 0.7364338951763068, 0.7765925101265282, 0.8148102621687295, 0.8505966079942793, 0.8834758182271021, 0.912997421842236, 0.938746432156527, 0.9603530314874867, 0.9775013993578034, 0.9899373900304076, 0.9974747986368548]
plt.plot(xt, yt)
yt=[1.0, 0.9972501604946407, 0.9887106168440861, 0.9739923022112235, 0.9527829911885627, 0.9248715748263387, 0.8901739576959637, 0.8487595756970521, 0.8008771044420258, 0.7469774412155415, 0.6877315265240409, 0.6240400741229032, 0.5570318803212603, 0.48804719898906096, 0.4186028513432698, 0.3503364771591109, 0.2849288315054989, 0.22400547120818484, 0.16902264795974092, 0.12114661803043528, 0.08114043079164736, 0.04927660040510581, 0.02529631401746099, 0.00843386967274716, 3.1494052069963466E-4, 0.001306790253227645, 0.002367676566614052, 0.0035024358989461995, 0.004716238280933486, 0.006014609034375455, 0.007403451659743108, 0.008889071942220902, 0.010478203299712906, 0.012178033388995146, 0.013996231977155412, 0.01594098007443509, 0.01802100031127071, 0.02024558852637576, 0.022624646513731205, 0.025168715853915356, 0.027889012728828155, 0.03079746358799916, 0.033906741498707725, 0.03723030297042172, 0.04078242499582402, 0.04457824199513306, 0.04863378228662916, 0.05296600363328674, 0.05759282733212253, 0.06253317021813175, 0.06780697384726882, 0.07343523000150774, 0.07944000152219204, 0.08584443732421171, 0.0926727802715123, 0.0999503664025534, 0.10770361378109314, 0.1159599990116635, 0.12474801919903661, 0.13409713684577979, 0.14403770487091655, 0.1546008685954163, 0.16581844117703515, 0.1777227485890082, 0.19034643982639052, 0.20372225759393386, 0.2178827642854523, 0.2328600176139568, 0.24868518980437496, 0.2653881238295662, 0.28299681977271124, 0.30153684405683967, 0.3210306540228551, 0.341496830195189, 0.36294920859152974, 0.38539590566145276, 0.40883822894059707, 0.4332694673568074, 0.45867355641077007, 0.4850236152796651, 0.5122803553784219, 0.5403903631969167, 0.5692842644683849, 0.5988747820872101, 0.6290547068711685, 0.6596948084537295, 0.6906417235005612, 0.7217158702701209, 0.7527094524599653, 0.7833846314354725, 0.8134719643924972, 0.8426692267166211, 0.870640759564172, 0.8970175080719259, 0.9213979408612081, 0.9433500664845758, 0.9624147854843906, 0.9781108354371517, 0.9899415975681453, 0.9974040330828318]
plt.plot(xt, yt)
yt=[1.0, 0.9974610312914682, 0.9892963031082308, 0.9746217481337056, 0.9524759269821461, 0.9218348627378363, 0.8816409814770783, 0.8308536096459763, 0.7685318902080945, 0.6939656766893287, 0.6068760375248102, 0.5077137901110932, 0.3980889831016173, 0.2813568252508997, 0.16333857010256256, 0.05299428396959511, 0.0019035709936207914, 0.0050397401417521534, 0.008270323668280044, 0.011598518200481866, 0.015027637447712301, 0.01856111653795968, 0.0222025164888667, 0.025955528812906806, 0.029823980255625233, 0.033811837664932516, 0.03792321298836569, 0.04216236839399172, 0.04653372150918521, 0.05104185076984862, 0.05569150087072562, 0.06048758830524846, 0.06543520698082197, 0.07053963389253416, 0.0758063348349432, 0.0812409701277636, 0.08684940032689922, 0.09263769188726335, 0.09861212273810441, 0.10477918772502445, 0.11114560386541388, 0.11771831535551341, 0.12450449825759793, 0.1315115647846911, 0.13874716708758053, 0.14621920043447956, 0.15393580565723786, 0.16190537071924113, 0.17013653123874456, 0.17863816977697586, 0.18741941367248438, 0.19648963117143853, 0.20585842556728307, 0.21553562702175158, 0.22553128169191689, 0.23585563773391902, 0.2465191276922479, 0.25753234671285613, 0.2689060259376364, 0.2806510003454347, 0.29277817019907487, 0.3052984551368594, 0.3182227398084424, 0.3315618097962112, 0.34532627638138946, 0.35952648850552277, 0.3741724300388756, 0.3892736001930191, 0.40483887460029017, 0.42087634422191067, 0.4373931288325474, 0.45439516135418445, 0.4718869387675878, 0.48987123470534205, 0.5083487681152598, 0.527317821564331, 0.5467738018174919, 0.5667087342571612, 0.5871106814925352, 0.6079630751250396, 0.6292439480709782, 0.6509250530781165, 0.6729708510959478, 0.6953373509611247, 0.7179707794401072, 0.740806058045255, 0.7637650602458925, 0.7867546198051615, 0.8096642581128816, 0.8323635957600511, 0.8546994115362165, 0.8764923110254761, 0.8975329677857689, 0.9175779038513044, 0.9363447846835924, 0.9535072192056138, 0.968689081865807, 0.9814584162141771, 0.9913210462096186, 0.9977141241514784]
plt.plot(xt, yt)
yt=[1.0, 0.9950188274196835, 0.9777052505810897, 0.9434608948142958, 0.8858837876430168, 0.7960291725086978, 0.6615642634545562, 0.46669956100429777, 0.19833986481645313, 0.0014411207643317218, 0.004242415267491692, 0.007106383296551392, 0.010034821431083493, 0.013029589253297405, 0.016092611948024974, 0.019225883024369998, 0.022431467165198404, 0.025711503210962047, 0.029068207284678224, 0.03250387606523009, 0.03602089021650686, 0.03962171798027175, 0.043308918941022914, 0.047085147971502046, 0.05095315936790396, 0.054915811184243504, 0.058976069775746015, 0.06313701456153713, 0.06740184301731326, 0.07177387590907504, 0.07625656277938699, 0.08085348769799029, 0.0855683752889268, 0.09040509704661726, 0.09536767795356958, 0.1004603034125487, 0.10568732650610144, 0.11105327559627545, 0.116562862277164, 0.1222209896925213, 0.12803276123007432, 0.1340034896032634, 0.14013870632990263, 0.14644417161559892, 0.15292588464760323, 0.15959009430198948, 0.16644331026352663, 0.1734923145531766, 0.180744173452617, 0.1882062498083247, 0.19588621568928716, 0.20379206536199143, 0.21193212853356475, 0.22031508379831122, 0.22894997220380328, 0.23784621082939145, 0.24701360624161658, 0.25646236765641883, 0.26620311959590887, 0.27624691377612853, 0.2866052398996616, 0.29729003495062467, 0.3083136904963893, 0.31968905738649556, 0.33142944709982164, 0.3435486288202156, 0.35606082111098514, 0.3689806768005226, 0.382323259373095, 0.39610400876555346, 0.4103386939834872, 0.42504334934507665, 0.44023419040681894, 0.45592750468285953, 0.47213951108787716, 0.48888618054634964, 0.5061830083324554, 0.5240447263224248, 0.5424849403065423, 0.5615156736253574, 0.5811467934027421, 0.6013852891977256, 0.6222343655155531, 0.6436922986656907, 0.6657509940535937, 0.6883941609353365, 0.7115949962689279, 0.7353132352069496, 0.7594913796576906, 0.7840498534333176, 0.8088807459318786, 0.8338396860462229, 0.858735219291071, 0.883314821981491, 0.9072463435060798, 0.9300931707400997, 0.9512806795150966, 0.9700504556674066, 0.9853971431889703, 0.9959803139957485]
plt.plot(xt, yt)
#HALF_PEAK no fix min
# case NONE: return new GaussianWaveform(0.1015625, phase, false);
# /*case LOW: return new SkewedGaussWaveform(0.10950254478545297, skew.skew, phase, false);
# case MID: return new SkewedGaussWaveform(0.13931330292050073, skew.skew, phase, false);
# case HIGH: return new SkewedGaussWaveform(0.20802617814718816, skew.skew, phase, false);*/
xt=[0.0, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1, 0.11, 0.12, 0.13, 0.14, 0.15, 0.16, 0.17, 0.18, 0.19, 0.2, 0.21, 0.22, 0.23, 0.24, 0.25, 0.26, 0.27, 0.28, 0.29, 0.3, 0.31, 0.32, 0.33, 0.34, 0.35, 0.36, 0.37, 0.38, 0.39, 0.4, 0.41, 0.42, 0.43, 0.44, 0.45, 0.46, 0.47, 0.48, 0.49, 0.5, 0.51, 0.52, 0.53, 0.54, 0.55, 0.56, 0.57, 0.58, 0.59, 0.6, 0.61, 0.62, 0.63, 0.64, 0.65, 0.66, 0.67, 0.68, 0.69, 0.7, 0.71, 0.72, 0.73, 0.74, 0.75, 0.76, 0.77, 0.78, 0.79, 0.8, 0.81, 0.82, 0.83, 0.84, 0.85, 0.86, 0.87, 0.88, 0.89, 0.9, 0.91, 0.92, 0.93, 0.94, 0.95, 0.96, 0.97, 0.98, 0.99]
yt=[1.0, 0.9951643921014979, 0.9807974152891648, 0.9573118912015268, 0.9253739096952884, 0.8858714475589475, 0.8398733754565454, 0.7885814924939011, 0.7332785865473379, 0.6752756375567601, 0.6158611616335171, 0.5562553594115757, 0.49757122438015355, 0.44078414080532446, 0.3867108176429706, 0.3359977257732059, 0.2891185859010446, 0.2463799374380964, 0.20793343425796706, 0.17379327559160265, 0.14385708877784129, 0.11792862155203371, 0.09574075179308944, 0.076977552922306, 0.06129443199693493, 0.048335654665708876, 0.037748860091298964, 0.02919642888226213, 0.022363783736295544, 0.01696486818005936, 0.012745161746014099, 0.009482653239773929, 0.006987213991546008, 0.005098798802789249, 0.003684863141087681, 0.002637330188709126, 0.001869378768951731, 0.0013122596110907415, 9.122878163395304E-4, 6.281070149764964E-4, 4.2827731285284825E-4, 2.8920523548780385E-4, 1.9340909317879185E-4, 1.2809650571570262E-4, 8.402090085120773E-5, 5.457918428170992E-5, 3.511206759435065E-5, 2.23704862200293E-5, 1.411510455726985E-5, 8.820281214411466E-6, 5.458463624457845E-6, 8.820281214411466E-6, 1.411510455726985E-5, 2.23704862200293E-5, 3.5112067594350776E-5, 5.4579184281710016E-5, 8.402090085120788E-5, 1.2809650571570219E-4, 1.934090931787915E-4, 2.8920523548780336E-4, 4.2827731285284825E-4, 6.281070149764964E-4, 9.122878163395304E-4, 0.0013122596110907415, 0.001869378768951731, 0.002637330188709126, 0.0036848631410876876, 0.005098798802789258, 0.006987213991546021, 0.00948265323977392, 0.012745161746014089, 0.016964868180059347, 0.022363783736295544, 0.02919642888226213, 0.037748860091298964, 0.048335654665708876, 0.06129443199693493, 0.07697755292230607, 0.09574075179308947, 0.11792862155203376, 0.14385708877784148, 0.17379327559160276, 0.2079334342579669, 0.24637993743809636, 0.2891185859010444, 0.33599772577320586, 0.3867108176429706, 0.44078414080532446, 0.49757122438015355, 0.5562553594115758, 0.6158611616335173, 0.6752756375567603, 0.7332785865473381, 0.7885814924939014, 0.8398733754565452, 0.8858714475589473, 0.9253739096952883, 0.9573118912015268, 0.9807974152891648, 0.9951643921014979]
plt.plot(xt, yt)
yt=[1.0, 0.9940236973434217, 0.9752149343446068, 0.9425180099966111, 0.8953458032351831, 0.8337580701281446, 0.7586390557169136, 0.6718479358925181, 0.5763045847247216, 0.4759635273832647, 0.3756252495717562, 0.2805433413083743, 0.19581714790742324, 0.12562039511071912, 0.07240602825262826, 0.036325181498027354, 0.015148989175152127, 0.004898538788703484, 0.0010986052770816548, 9.014743498909495E-4, 9.86248778160585E-4, 0.0010793038039837056, 0.0011814762051475078, 0.0012936898094723333, 0.0014169647575591729, 0.0015524277646156274, 0.0017013234890879362, 0.0018650271271625387, 0.002045058364644113, 0.0022430968313535385, 0.002460999218107876, 0.00270081823263216, 0.0029648235884951798, 0.003255525240438412, 0.0035756991003493025, 0.003928415490674822, 0.00431707061631475, 0.004745421361986493, 0.005217623749685687, 0.005738275420102731, 0.006312462532552227, 0.006945811509906044, 0.00764454608786269, 0.008415550161171268, 0.009266436952530852, 0.010205625061967058, 0.011242421984453918, 0.012387115709989533, 0.013651075041448859, 0.015046859279050223, 0.01658833792333332, 0.018290821037599138, 0.020171200881412044, 0.02224810537359851, 0.02454206385956878, 0.027075685535667127, 0.029873850712839375, 0.03296391487138248, 0.036375925153740905, 0.04014284854629153, 0.044300810493684895, 0.048889342046778216, 0.05395163283953513, 0.05953478618883148, 0.06569007137606411, 0.07247316665736087, 0.07994438471063463, 0.08816887000720669, 0.09721675493184732, 0.10716325830116398, 0.11808870617575164, 0.13007845045427485, 0.1432226556077657, 0.1576159179978325, 0.17335667547725286, 0.19054635737747627, 0.20928821657087723, 0.22968577614844687, 0.2518408135641794, 0.27585079518903693, 0.30180566459941494, 0.32978387935780956, 0.3598475846198296, 0.392036809147292, 0.4263625722998535, 0.4627988020863099, 0.5012729879937974, 0.5416555326912573, 0.583747829575301, 0.6272691854734725, 0.671842837832683, 0.7169814925835262, 0.7620730422177054, 0.8063674224646807, 0.8489659368624597, 0.8888148226023018, 0.9247053394127536, 0.9552832104529706, 0.9790707789653794, 0.9945056778285049]
plt.plot(xt, yt)
yt=[1.0, 0.9896654119544297, 0.953944080080306, 0.8849154824271009, 0.7744151333052411, 0.6168199065852619, 0.4159073935639206, 0.19927615983210928, 0.037527206165594705, 0.01154519618277527, 0.011984921481107457, 0.012444121532649833, 0.012923784411774539, 0.013424954867305824, 0.013948737953718674, 0.01449630291930722, 0.015068887371186399, 0.01566780173864785, 0.01629443405819575, 0.016950255105554344, 0.01763682390207964, 0.018355793625343075, 0.019108917956197336, 0.01989805789740964, 0.020725189101971915, 0.021592409752496416, 0.022501949036705094, 0.023456176267946043, 0.02445761070395571, 0.02550893212175816, 0.02661299221169297, 0.027772826859125823, 0.028991669388463693, 0.030272964850713367, 0.031620385443034804, 0.03303784715659988, 0.03452952775762635, 0.03609988621577074, 0.03775368370419113, 0.03949600630659095, 0.04133228957848676, 0.043268345122867834, 0.04531038935438539, 0.047465074641281656, 0.04973952303047901, 0.05214136277863261, 0.054678767930512, 0.057360501205803105, 0.06019596047625976, 0.06319522913697563, 0.06636913069822721, 0.06972928794759443, 0.07328818705551995, 0.07705924702059579, 0.08105689487292615, 0.08529664707393012, 0.08979519756756756, 0.09457051294944059, 0.09964193522418455, 0.10503029261494173, 0.11075801886746896, 0.11684928145028428, 0.12333011898433505, 0.1302285881320081, 0.13757492002425245, 0.14540168609099216, 0.1537439728641784, 0.16263956491913625, 0.17212913457496062, 0.18225643624515664, 0.1930685023588448, 0.20461583648664233, 0.21695259760698166, 0.2301367672112333, 0.24423028800298455, 0.2592991590789613, 0.27541346739737665, 0.29264732866200793, 0.31107870197127385, 0.33078903102969753, 0.35186264950154794, 0.3743858680133733, 0.3984456337955216, 0.4241276188704604, 0.45151354621051176, 0.4806775016079255, 0.5116808970687529, 0.5445656426406905, 0.5793449388549723, 0.6159909098670495, 0.6544180432129085, 0.6944610677890142, 0.7358454665068452, 0.7781482637166659, 0.8207460381741739, 0.862746306715452, 0.9028975922935625, 0.9394728972114357, 0.9701216079867595, 0.9916876022440039]
plt.plot(xt, yt)
yt=[1.0, 0.9723893403017706, 0.8537948345986012, 0.5436276988565926, 0.02899169774820895, 0.029643458494955744, 0.03031455207009856, 0.031005701180265372, 0.03171766143779013, 0.03245122313860151, 0.03320721315179421, 0.03398649692890645, 0.03478998064157789, 0.035618613456971496, 0.03647338996111623, 0.037355352741173756, 0.0382655951385545, 0.03920526418581987, 0.0401755637414117, 0.041177757837461894, 0.04221317425726209, 0.043283208360429336, 0.044389327175403025, 0.045533073780664686, 0.046716071998005274, 0.04794003142329054, 0.04920675282251898, 0.0505181339235498, 0.05187617563673021, 0.05328298874080015, 0.054740801073935065, 0.05625196527364105, 0.057818967113484034, 0.059444434489368064, 0.06113114711332851, 0.06288204697864098, 0.06470024966653287, 0.06658905657200509, 0.06855196813431666, 0.07059269816665502, 0.07271518938953302, 0.0749236302836446, 0.07722247339043412, 0.07961645520265494, 0.08211061780290967, 0.08471033242580071, 0.08742132513912701, 0.09024970486183233, 0.09320199396147254, 0.09628516170219788, 0.09950666084607358, 0.10287446774647442, 0.10639712631284422, 0.1100837962719437, 0.1139443062025348, 0.11798921187908892, 0.12222986052647002, 0.12667846166267635, 0.1313481652917743, 0.13625314830541435, 0.14140871006018743, 0.1468313782210838, 0.1525390261000732, 0.15855100287498822, 0.16488827824907878, 0.1715736033072639, 0.17863168954227873, 0.18608940826294512, 0.19397601285677032, 0.202323386657095, 0.2111663194549718, 0.22054281598680028, 0.2304944400017904, 0.2410666977382077, 0.2523094647659167, 0.2642774601097554, 0.2770307712372228, 0.2906354326952396, 0.30516405963791915, 0.3206965347744528, 0.33732074272519075, 0.3551333383826564, 0.37424052403491137, 0.3947587912039422, 0.4168155533767092, 0.4405495486191905, 0.4661108159701426, 0.49365992917092355, 0.5233659765834353, 0.5554024573017874, 0.5899397336461619, 0.6271317845384727, 0.6670934593022424, 0.7098617038329841, 0.7553292844867537, 0.8031302818529311, 0.8524386864859369, 0.9016051748250998, 0.9474802824187716, 0.9841000184872646]
plt.plot(xt, yt)
#HALF_PEAK fix min
#case NONE: return new GaussianWaveform(0.1015625, phase, false);
#case LOW: return new SkewedGaussWaveform(0.11016841333997066, skew.skew, phase, true);
# case MID: return new SkewedGaussWaveform(0.1500836205989934, skew.skew, phase, true);
# case HIGH: return new SkewedGaussWaveform(0.28198739813204843, skew.skew, phase, true);
xt=[0.0, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1, 0.11, 0.12, 0.13, 0.14, 0.15, 0.16, 0.17, 0.18, 0.19, 0.2, 0.21, 0.22, 0.23, 0.24, 0.25, 0.26, 0.27, 0.28, 0.29, 0.3, 0.31, 0.32, 0.33, 0.34, 0.35, 0.36, 0.37, 0.38, 0.39, 0.4, 0.41, 0.42, 0.43, 0.44, 0.45, 0.46, 0.47, 0.48, 0.49, 0.5, 0.51, 0.52, 0.53, 0.54, 0.55, 0.56, 0.57, 0.58, 0.59, 0.6, 0.61, 0.62, 0.63, 0.64, 0.65, 0.66, 0.67, 0.68, 0.69, 0.7, 0.71, 0.72, 0.73, 0.74, 0.75, 0.76, 0.77, 0.78, 0.79, 0.8, 0.81, 0.82, 0.83, 0.84, 0.85, 0.86, 0.87, 0.88, 0.89, 0.9, 0.91, 0.92, 0.93, 0.94, 0.95, 0.96, 0.97, 0.98, 0.99]
yt=[1.0, 0.9951643921014979, 0.9807974152891648, 0.9573118912015268, 0.9253739096952884, 0.8858714475589475, 0.8398733754565454, 0.7885814924939011, 0.7332785865473379, 0.6752756375567601, 0.6158611616335171, 0.5562553594115757, 0.49757122438015355, 0.44078414080532446, 0.3867108176429706, 0.3359977257732059, 0.2891185859010446, 0.2463799374380964, 0.20793343425796706, 0.17379327559160265, 0.14385708877784129, 0.11792862155203371, 0.09574075179308944, 0.076977552922306, 0.06129443199693493, 0.048335654665708876, 0.037748860091298964, 0.02919642888226213, 0.022363783736295544, 0.01696486818005936, 0.012745161746014099, 0.009482653239773929, 0.006987213991546008, 0.005098798802789249, 0.003684863141087681, 0.002637330188709126, 0.001869378768951731, 0.0013122596110907415, 9.122878163395304E-4, 6.281070149764964E-4, 4.2827731285284825E-4, 2.8920523548780385E-4, 1.9340909317879185E-4, 1.2809650571570262E-4, 8.402090085120773E-5, 5.457918428170992E-5, 3.511206759435065E-5, 2.23704862200293E-5, 1.411510455726985E-5, 8.820281214411466E-6, 5.458463624457845E-6, 8.820281214411466E-6, 1.411510455726985E-5, 2.23704862200293E-5, 3.5112067594350776E-5, 5.4579184281710016E-5, 8.402090085120788E-5, 1.2809650571570219E-4, 1.934090931787915E-4, 2.8920523548780336E-4, 4.2827731285284825E-4, 6.281070149764964E-4, 9.122878163395304E-4, 0.0013122596110907415, 0.001869378768951731, 0.002637330188709126, 0.0036848631410876876, 0.005098798802789258, 0.006987213991546021, 0.00948265323977392, 0.012745161746014089, 0.016964868180059347, 0.022363783736295544, 0.02919642888226213, 0.037748860091298964, 0.048335654665708876, 0.06129443199693493, 0.07697755292230607, 0.09574075179308947, 0.11792862155203376, 0.14385708877784148, 0.17379327559160276, 0.2079334342579669, 0.24637993743809636, 0.2891185859010444, 0.33599772577320586, 0.3867108176429706, 0.44078414080532446, 0.49757122438015355, 0.5562553594115758, 0.6158611616335173, 0.6752756375567603, 0.7332785865473381, 0.7885814924939014, 0.8398733754565452, 0.8858714475589473, 0.9253739096952883, 0.9573118912015268, 0.9807974152891648, 0.9951643921014979]
plt.plot(xt, yt)
yt=[1.0, 0.9940917967813461, 0.9755016555693229, 0.9431883351404714, 0.896566285821943, 0.8356786804388897, 0.7613701823336441, 0.6754342658632283, 0.5806993921935479, 0.48100904688669854, 0.3810467925792234, 0.2859656400864687, 0.20080954660977055, 0.1297711773819257, 0.07541437152625977, 0.038083800510912084, 0.015778865588980326, 0.004702956388147249, 4.3996864488390366E-4, 5.944007716239042E-5, 1.4768547617681103E-4, 2.445087056690181E-4, 3.507722082273543E-4, 4.674276693575205E-4, 5.955254592598492E-4, 7.362250865350252E-4, 8.908067721244873E-4, 0.00106068426308129, 0.001247419018144052, 0.0014527359106323908, 0.0016785406089804503, 0.0019269388113572926, 0.0022002575283679195, 0.0025010686268579878, 0.0028322148684214584, 0.0031968386983790204, 0.0035984140647818356, 0.004040781572395402, 0.004528187303587714, 0.005065325666490441, 0.0056573866605590575, 0.0063101079804806365, 0.007029832410913148, 0.007823570996295536, 0.008699072501285158, 0.009664899707388062, 0.010730513118918376, 0.011906362675106379, 0.013203988083132584, 0.014636128396780922, 0.016216841464385307, 0.017961633854202966, 0.019887601830853873, 0.022013583897582678, 0.024360325329197425, 0.026950654991549566, 0.029809674565600332, 0.03296496005573468, 0.03644677514898309, 0.04028829558744655, 0.0445258432006309, 0.049199127594162645, 0.05435149267904196, 0.060030164219229555, 0.06628649333806495, 0.07317618941350595, 0.08075953396057153, 0.0891015648930861, 0.09827221791681492, 0.10834640866861765, 0.11940403551429829, 0.13152987858352633, 0.14481336558864252, 0.1593481691890764, 0.17523159408512792, 0.19256370464313624, 0.2114461357021882, 0.23198052039039344, 0.2542664594814495, 0.2783989473781218, 0.30446516072930496, 0.33254050772766175, 0.3626838303750709, 0.3949316499420343, 0.42929134951659836, 0.4657331996340432, 0.5041811570035668, 0.5445024067578943, 0.586495680981057, 0.6298784772084288, 0.6742734279551634, 0.7191942448299474, 0.7640318875088858, 0.8080418971852129, 0.8503341920261148, 0.889867049301426, 0.9254474862004681, 0.9557407740080416, 0.9792923287974417, 0.9945656308479225]
plt.plot(xt, yt)
yt=[1.0, 0.9910645969631764, 0.9604797208001989, 0.901911064936227, 0.8086545044845985, 0.6752231126489243, 0.5011402195503974, 0.2989587975824691, 0.10795079305353876, 0.0036073744711473043, 0.004156962002239292, 0.004730038933981712, 0.005327755709185523, 0.005951326451824011, 0.0066020329005100145, 0.007281228609788701, 0.007990343439093562, 0.008730888350791971, 0.00950446054045797, 0.010312748924363244, 0.011157540011188736, 0.012040724187137858, 0.012964302445995942, 0.01393039359824294, 0.014941241996103811, 0.015999225814433603, 0.017106865930599117, 0.01826683545005742, 0.01948196992816639, 0.02075527834291571, 0.02208995487776431, 0.023489391578636873, 0.024957191954395923, 0.026497185595794472, 0.028113443894055998, 0.029810296946851788, 0.03159235174658101, 0.03346451175352982, 0.03543199796472255, 0.0375003715980969, 0.03967555852105721, 0.04196387556249165, 0.04437205885798074, 0.046907294389160775, 0.04957725089000199, 0.05239011530506169, 0.055354630997483834, 0.05848013891751074, 0.06177662195535813, 0.0652547527152345, 0.06892594495970855, 0.072802408985091, 0.07689721119839968, 0.08122433817402626, 0.08579876547241092, 0.09063653150252553, 0.09575481670308693, 0.10117202830197608, 0.10690789088655468, 0.11298354297587045, 0.11942163972458697, 0.12624646180204052, 0.13348403037071427, 0.1411622279271637, 0.1493109245529578, 0.15796210883809844, 0.16715002236495355, 0.17691129615177292, 0.18728508681907982, 0.198313209418257, 0.2100402627963013, 0.22251374199564197, 0.23578413041498614, 0.24990496217265018, 0.2649328421706726, 0.2809274075656387, 0.29795120946318576, 0.31606948734588575, 0.3353498005994103, 0.35586147097245513, 0.3776747761767631, 0.40085981716899916, 0.4254849587310474, 0.45161471317527496, 0.47930689824810063, 0.5086088498505346, 0.5395524044806872, 0.5721472807430711, 0.6063723780187167, 0.6421643661505632, 0.6794027539561727, 0.7178903865116855, 0.7573280213925151, 0.7972812653398135, 0.8371377180519078, 0.8760516969766009, 0.912873491076894, 0.946059920854087, 0.9735635356347342, 0.9927000731645895]
plt.plot(xt, yt)
yt=[1.0, 0.9855598642699223, 0.9294001158280938, 0.8008933841728406, 0.5471612069668034, 0.11514789412643789, 0.004233260002191489, 0.005430813056036933, 0.006660972803801468, 0.007924867257817565, 0.009223672204021933, 0.010558613609415866, 0.011930970170655723, 0.013342076013221639, 0.014793323551322499, 0.016286166519465052, 0.017822123187450974, 0.019402779771468977, 0.021029794054933458, 0.022704899233785412, 0.024429908002132565, 0.026206716895362026, 0.02803731090923111, 0.02992376841493013, 0.0318682663917343, 0.03387308600063021, 0.03594061852422567, 0.03807337170035644, 0.04027397647909168, 0.04254519423534368, 0.04488992447201947, 0.04731121305163702, 0.049812260997593165, 0.05239643390983976, 0.05506727204362964, 0.057828501104269577, 0.06068404381549643, 0.06363803232421983, 0.06669482150999018, 0.06985900327370355, 0.07313542188679763, 0.07652919048958326, 0.08004570883545187, 0.08369068238657162, 0.08747014287640066, 0.09139047046498293, 0.09545841762462953, 0.09968113490630591, 0.10406619875093197, 0.10862164152493625, 0.11335598397587264, 0.1182782703217742, 0.1233981062072514, 0.12872569978016946, 0.1342719061650696, 0.14004827563328592, 0.14606710579483068, 0.15234149816336998, 0.15888541947261944, 0.16571376814972158, 0.17284244637779914, 0.1802884382047462, 0.18806989417674325, 0.19620622299063, 0.2047181906658708, 0.21362802772988526, 0.22295954488377262, 0.23273825756035485, 0.2429915196912378, 0.25374866684805963, 0.2650411686930146, 0.2769027903344478, 0.2893697616924809, 0.30248095327857927, 0.3162780558001528, 0.3308057596027666, 0.34611192799844, 0.36224775577319196, 0.3792679003000231, 0.39723056724690653, 0.4161975252034139, 0.43623401269673373, 0.4574084856255422, 0.47979213103489454, 0.503458041289441, 0.5284798963965457, 0.5549299343302808, 0.5828758886261959, 0.6123764218320279, 0.6434743547973376, 0.6761866401042755, 0.7104894784829502, 0.7462961037330157, 0.7834233468977864, 0.8215407494756561, 0.8600920297252698, 0.8981718099105672, 0.9343281677086588, 0.9662387642460433, 0.9901646958155236]
plt.plot(xt, yt)
#QUARTER_PEAK no fix min
#case NONE: return new GaussianWaveform(0.05078125, phase, false);
# *case LOW: return new SkewedGaussWaveform(0.05476715911828679, skew.skew, phase, false);
# case MID: return new SkewedGaussWaveform(0.06973107359499559, skew.skew, phase, false);
# case HIGH: return new SkewedGaussWaveform(0.10420928568117749, skew.skew, phase, false);
xt=[0.0, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1, 0.11, 0.12, 0.13, 0.14, 0.15, 0.16, 0.17, 0.18, 0.19, 0.2, 0.21, 0.22, 0.23, 0.24, 0.25, 0.26, 0.27, 0.28, 0.29, 0.3, 0.31, 0.32, 0.33, 0.34, 0.35, 0.36, 0.37, 0.38, 0.39, 0.4, 0.41, 0.42, 0.43, 0.44, 0.45, 0.46, 0.47, 0.48, 0.49, 0.5, 0.51, 0.52, 0.53, 0.54, 0.55, 0.56, 0.57, 0.58, 0.59, 0.6, 0.61, 0.62, 0.63, 0.64, 0.65, 0.66, 0.67, 0.68, 0.69, 0.7, 0.71, 0.72, 0.73, 0.74, 0.75, 0.76, 0.77, 0.78, 0.79, 0.8, 0.81, 0.82, 0.83, 0.84, 0.85, 0.86, 0.87, 0.88, 0.89, 0.9, 0.91, 0.92, 0.93, 0.94, 0.95, 0.96, 0.97, 0.98, 0.99]
yt=[1.0, 0.9807974152891648, 0.9253739096952884, 0.8398733754565454, 0.7332785865473379, 0.6158611616335171, 0.49757122438015355, 0.3867108176429706, 0.2891185859010446, 0.20793343425796706, 0.14385708877784129, 0.09574075179308944, 0.06129443199693493, 0.037748860091298964, 0.022363783736295544, 0.012745161746014099, 0.006987213991546008, 0.003684863141087681, 0.001869378768951731, 9.122878163395304E-4, 4.2827731285284825E-4, 1.9340909317879185E-4, 8.402090085120773E-5, 3.511206759435065E-5, 1.411510455726985E-5, 5.458463624457845E-6, 2.0305578494036798E-6, 7.266393931060717E-7, 2.501388422756234E-7, 8.283272670791912E-8, 2.6386476780894055E-8, 8.085734530356179E-9, 2.3835056016115253E-9, 6.758829650948635E-10, 1.84367959751066E-10, 4.837912665332134E-11, 1.2212068281778742E-11, 2.9653710887231095E-12, 6.926717879695354E-13, 1.55644785504638E-13, 3.364343077958482E-14, 6.995594102219332E-15, 1.3992894339418642E-15, 2.692459206189263E-16, 4.9836706569819453E-17, 8.873769884841599E-18, 1.5199370993560441E-18, 2.5043884613760726E-19, 3.969505409115469E-20, 6.0524293320881495E-21, 8.877316050951375E-22, 6.0524293320881495E-21, 3.969505409115469E-20, 2.5043884613760726E-19, 1.5199370993560657E-18, 8.873769884841662E-18, 4.983670656981981E-17, 2.692459206189225E-16, 1.3992894339418541E-15, 6.9955941022192825E-15, 3.364343077958482E-14, 1.55644785504638E-13, 6.926717879695354E-13, 2.9653710887231095E-12, 1.2212068281778742E-11, 4.837912665332134E-11, 1.843679597510673E-10, 6.758829650948683E-10, 2.3835056016115422E-9, 8.08573453035615E-9, 2.6386476780893963E-8, 8.283272670791883E-8, 2.501388422756234E-7, 7.266393931060717E-7, 2.0305578494036798E-6, 5.458463624457845E-6, 1.411510455726985E-5, 3.5112067594350776E-5, 8.402090085120788E-5, 1.934090931787922E-4, 4.282773128528506E-4, 9.122878163395336E-4, 0.0018693787689517244, 0.0036848631410876776, 0.006987213991545989, 0.012745161746014089, 0.022363783736295544, 0.037748860091298964, 0.06129443199693493, 0.09574075179308947, 0.14385708877784148, 0.20793343425796731, 0.289118585901045, 0.3867108176429712, 0.49757122438015294, 0.6158611616335167, 0.7332785865473375, 0.8398733754565452, 0.9253739096952883, 0.9807974152891648]
plt.plot(xt, yt)
yt=[1.0, 0.9753676230067523, 0.8957020385730722, 0.7592185188094147, 0.5770648858039882, 0.37644238586894246, 0.19650959653797195, 0.07282681734478977, 0.015299526824410523, 0.0011189385291109242, 7.447207054212331E-6, 4.97682116853634E-7, 5.691324475361825E-7, 6.513096798905048E-7, 7.458957922813491E-7, 8.548478704838678E-7, 9.804449865297402E-7, 1.1253433893178883E-6, 1.2926415383795032E-6, 1.485956804983433E-6, 1.709516015575801E-6, 1.968262433258445E-6, 2.2679822780347135E-6, 2.615454493365543E-6, 3.0186281964018206E-6, 3.4868331278744914E-6, 4.031029476392856E-6, 4.664104729003642E-6, 5.401226741826163E-6, 6.2602640881492355E-6, 7.262286995836017E-6, 8.432164915887675E-6, 9.799280073108679E-6, 1.1398380364736144E-5, 1.3270599848929666E-5, 1.546468099258224E-5, 1.8038440060686905E-5, 2.106052581449987E-5, 2.4612532395758238E-5, 2.879154034321649E-5, 3.3713175649910904E-5, 3.9515296282835956E-5, 4.636243946202219E-5, 5.44511922333508E-5, 6.401668370251589E-5, 7.534044124902891E-5, 8.875990698889159E-5, 1.0467997702382162E-4, 1.2358700747031975E-4, 1.4606583144318106E-4, 1.7282045444535728E-4, 2.046992473821641E-4, 2.427256433233694E-4, 2.8813557437606046E-4, 3.4242316868860487E-4, 4.0739657707138485E-4, 4.852462291852998E-4, 5.786283492932997E-4, 6.907672138410231E-4, 8.255804351232282E-4, 9.878325402073189E-4, 0.0011833233219626714, 0.0014191189168873942, 0.0017038353682276635, 0.0020479866273946062, 0.002464411605596174, 0.0029687980925593997, 0.0035803251971015158, 0.004322450521757658, 0.005223873637246493, 0.006319713622135463, 0.007652945475029957, 0.009276147991661182, 0.011253623967633483, 0.013663961821304344, 0.016603115008716727, 0.020188080385657284, 0.02456125649904, 0.029895553838586178, 0.03640030555679817, 0.0443279804705494, 0.0539816177713719, 0.06572276692370753, 0.0799795017611583, 0.09725375082022843, 0.11812670171038289, 0.14326033944604286, 0.1733922009937805, 0.2093191023073552, 0.2518638676220806, 0.30181696702429867, 0.3598425808538925, 0.42633635525125413, 0.5012208741480145, 0.5836663592540382, 0.6717313360217216, 0.7619358347676647, 0.8488151766126255, 0.9245640827654453, 0.9789753785315325]
plt.plot(xt, yt)
yt=[1.0, 0.9541581440522309, 0.7752607930958018, 0.41774022636943875, 0.03861388662357505, 6.163669107456331E-4, 6.46945829703247E-4, 6.7926732843687E-4, 7.134441574275566E-4, 7.495972680382372E-4, 7.87856477254498E-4, 8.283611920188841E-4, 8.712611990281531E-4, 9.167175264942839E-4, 9.649033850750898E-4, 0.0010160051959685586, 0.0010702237150468485, 0.0011277752628935272, 0.0011888930717145656, 0.0012538287613357182, 0.0013228539578937196, 0.0013962620703968306, 0.0014743702420947757, 0.0015575214955856193, 0.001646087092828338, 0.001740469133759401, 0.0018411034200702461, 0.00194846261393573, 0.002063059725145361, 0.0021854519642401, 0.0023162450039688662, 0.0024560976967325505, 0.0026057273017735475, 0.0027659152828057995, 0.0029375137446910216, 0.0031214525867992657, 0.0033187474610180865, 0.003530508634194921, 0.0037579508683457537, 0.004002404447512374, 0.004265327498020737, 0.0045483197694582474, 0.004853138067386734, 0.005181713556158119, 0.005536171181806774, 0.005918851501573101, 0.006332335249011181, 0.006779471012847139, 0.007263406464965887, 0.007787623639511858, 0.008355978842756821, 0.0089727478640889, 0.009642677264564381, 0.010371042643737288, 0.011163714931286487, 0.012027235921287177, 0.012968904468600403, 0.013996875004493776, 0.015120270309104072, 0.016349310809909638, 0.017695463067824355, 0.019171610577635483, 0.020792250561385464, 0.022573721088814846, 0.024534463638274998, 0.026695327138551526, 0.02907992063521242, 0.03171502303785641, 0.034631059966026376, 0.03786265956657653, 0.0414493013748587, 0.04543607489224432, 0.04987456761213559, 0.05482390580291565, 0.06035197549596939, 0.066536855850679, 0.0734685023439523, 0.08125072292854793, 0.09000349611618504, 0.0998656852591231, 0.11099820700314184, 0.1235877119725182, 0.13785082877308624, 0.15403900246927144, 0.17244391581042723, 0.19340339874545967, 0.21730758055859117, 0.24460477069580075, 0.27580608609829527, 0.31148703210251477, 0.3522828438690135, 0.3988719746829817, 0.4519379167298718, 0.5120922111999543, 0.5797286464613137, 0.6547559918084345, 0.7361167484218011, 0.8209301635352937, 0.9029827459629145, 0.9701233879685169]
plt.plot(xt, yt)
yt=[1.0, 0.8432949558297448, 0.005313093136040679, 0.005458863498210377, 0.005609774954350007, 0.005766051761061479, 0.005927929928314618, 0.0060956579432576, 0.006269497545491437, 0.0064497245579737935, 0.006636629778097021, 0.006830519933903186, 0.0070317187108593325, 0.007240567855124996, 0.007457428359805122, 0.00768268174130269, 0.007916731413572199, 0.008160004168836252, 0.008412951774171323, 0.00867605269430561, 0.008949813952012463, 0.009234773138641033, 0.009531500588614449, 0.00984060173316364, 0.010162719650168595, 0.010498537828771436, 0.010848783169430689, 0.011214229242332091, 0.011595699829590026, 0.01199407277950188, 0.012410284204297621, 0.012845333056407159, 0.013300286122304529, 0.01377628347754461, 0.014274544451760603, 0.014796374158223574, 0.01534317064918034, 0.0159164327656987, 0.01651776875929403, 0.017148905772347546, 0.017811700275437714, 0.01850814957240778, 0.01924040449853836, 0.020010783453881697, 0.020821787932992712, 0.021676119734378718, 0.022576700058470196, 0.023526690732374077, 0.02452951783379742, 0.025588898026145956, 0.026708867962895518, 0.02789381717307877, 0.029148524902544697, 0.03047820145923562, 0.03188853469714653, 0.033385742375377385, 0.03497663124878976, 0.0366686638889269, 0.03847003440256104, 0.040389754416027955, 0.04243775093319948, 0.04462497796193278, 0.04696354414852572, 0.04946685907503282, 0.05214980137639309, 0.055028912443387944, 0.0581226202188744, 0.06145149850054782, 0.06503856827423501, 0.06890964896894676, 0.07309376921439657, 0.07762364877764417, 0.08253626596618414, 0.08787352804974824, 0.09368306635280894, 0.1000191828382489, 0.10694398154368347, 0.11452872654006513, 0.12285547866918296, 0.13201907684330325, 0.1421295470023667, 0.15331504398723983, 0.16572545990436288, 0.1795368685339405, 0.19495702051343453, 0.21223215951141244, 0.23165549486551423, 0.2535777362558769, 0.2784201552102035, 0.3066906455459692, 0.33900311022710705, 0.3760999671089734, 0.4188760705618839, 0.46839849948186923, 0.525906956500606, 0.5927546788016804, 0.6701836635903726, 0.7586410785982922, 0.855767273462868, 0.9501830360203302]
plt.plot(xt, yt)
#QUARTER_PEAK fix min
#GaussianWaveform(0.05078125, phase, false);
#SkewedGaussWaveform(0.054767394902696165, skew.skew, phase, true);
#SkewedGaussWaveform(0.07037477639896425, skew.skew, phase, true);
#SkewedGaussWaveform(0.1147667366789808, skew.skew, phase, true);
xt=[0.0, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1, 0.11, 0.12, 0.13, 0.14, 0.15, 0.16, 0.17, 0.18, 0.19, 0.2, 0.21, 0.22, 0.23, 0.24, 0.25, 0.26, 0.27, 0.28, 0.29, 0.3, 0.31, 0.32, 0.33, 0.34, 0.35, 0.36, 0.37, 0.38, 0.39, 0.4, 0.41, 0.42, 0.43, 0.44, 0.45, 0.46, 0.47, 0.48, 0.49, 0.5, 0.51, 0.52, 0.53, 0.54, 0.55, 0.56, 0.57, 0.58, 0.59, 0.6, 0.61, 0.62, 0.63, 0.64, 0.65, 0.66, 0.67, 0.68, 0.69, 0.7, 0.71, 0.72, 0.73, 0.74, 0.75, 0.76, 0.77, 0.78, 0.79, 0.8, 0.81, 0.82, 0.83, 0.84, 0.85, 0.86, 0.87, 0.88, 0.89, 0.9, 0.91, 0.92, 0.93, 0.94, 0.95, 0.96, 0.97, 0.98, 0.99]
yt=[1.0, 0.9807974152891648, 0.9253739096952884, 0.8398733754565454, 0.7332785865473379, 0.6158611616335171, 0.49757122438015355, 0.3867108176429706, 0.2891185859010446, 0.20793343425796706, 0.14385708877784129, 0.09574075179308944, 0.06129443199693493, 0.037748860091298964, 0.022363783736295544, 0.012745161746014099, 0.006987213991546008, 0.003684863141087681, 0.001869378768951731, 9.122878163395304E-4, 4.2827731285284825E-4, 1.9340909317879185E-4, 8.402090085120773E-5, 3.511206759435065E-5, 1.411510455726985E-5, 5.458463624457845E-6, 2.0305578494036798E-6, 7.266393931060717E-7, 2.501388422756234E-7, 8.283272670791912E-8, 2.6386476780894055E-8, 8.085734530356179E-9, 2.3835056016115253E-9, 6.758829650948635E-10, 1.84367959751066E-10, 4.837912665332134E-11, 1.2212068281778742E-11, 2.9653710887231095E-12, 6.926717879695354E-13, 1.55644785504638E-13, 3.364343077958482E-14, 6.995594102219332E-15, 1.3992894339418642E-15, 2.692459206189263E-16, 4.9836706569819453E-17, 8.873769884841599E-18, 1.5199370993560441E-18, 2.5043884613760726E-19, 3.969505409115469E-20, 6.0524293320881495E-21, 8.877316050951375E-22, 6.0524293320881495E-21, 3.969505409115469E-20, 2.5043884613760726E-19, 1.5199370993560657E-18, 8.873769884841662E-18, 4.983670656981981E-17, 2.692459206189225E-16, 1.3992894339418541E-15, 6.9955941022192825E-15, 3.364343077958482E-14, 1.55644785504638E-13, 6.926717879695354E-13, 2.9653710887231095E-12, 1.2212068281778742E-11, 4.837912665332134E-11, 1.843679597510673E-10, 6.758829650948683E-10, 2.3835056016115422E-9, 8.08573453035615E-9, 2.6386476780893963E-8, 8.283272670791883E-8, 2.501388422756234E-7, 7.266393931060717E-7, 2.0305578494036798E-6, 5.458463624457845E-6, 1.411510455726985E-5, 3.5112067594350776E-5, 8.402090085120788E-5, 1.934090931787922E-4, 4.282773128528506E-4, 9.122878163395336E-4, 0.0018693787689517244, 0.0036848631410876776, 0.006987213991545989, 0.012745161746014089, 0.022363783736295544, 0.037748860091298964, 0.06129443199693493, 0.09574075179308947, 0.14385708877784148, 0.20793343425796731, 0.289118585901045, 0.3867108176429712, 0.49757122438015294, 0.6158611616335167, 0.7332785865473375, 0.8398733754565452, 0.9253739096952883, 0.9807974152891648]
plt.plot(xt, yt)
yt=[1.0, 0.9753678330400302, 0.8957029386322871, 0.7592205392683384, 0.5770681492873888, 0.3764464398577459, 0.19651339192307404, 0.07282923199147215, 0.015300276211152876, 0.0011187969225162503, 7.158147082828829E-6, 2.063159130771156E-7, 2.7776975468930587E-7, 3.5995099900633937E-7, 4.54541698853725E-7, 5.634990259717674E-7, 6.891021517061961E-7, 8.340074397696772E-7, 1.001313482516436E-6, 1.1946378049779337E-6, 1.4182074117262597E-6, 1.676965772355625E-6, 1.976699346411755E-6, 2.324187355431266E-6, 2.7273792395981214E-6, 3.195605115164805E-6, 3.7398256075855005E-6, 4.372928712446037E-6, 5.110082878281011E-6, 5.969157368989489E-6, 6.971223218080117E-6, 8.141150817053711E-6, 9.508323489406782E-6, 1.1107490416759891E-5, 1.2979787159767185E-5, 1.5173957944171167E-5, 1.774782109530682E-5, 2.0770027789566586E-5, 2.4322175001592924E-5, 2.8501346595251326E-5, 3.342317246891921E-5, 3.9225515179294784E-5, 4.607291734370979E-5, 5.416197235882599E-5, 6.372781680734144E-5, 7.505198687658212E-5, 8.847193506350956E-5, 1.0439256971101563E-4, 1.233002613770413E-4, 1.4577986022085496E-4, 1.7253539186985983E-4, 2.0441525099839145E-4, 2.424428987578237E-4, 2.87854300439322E-4, 3.421436234110205E-4, 4.0711906484016997E-4, 4.8497110917987966E-4, 5.783560454485291E-4, 6.904982266050508E-4, 8.253153553185698E-4, 9.87572065349832E-4, 0.00118306827544337, 0.0014188702704752431, 0.0017035942684059532, 0.0020477544260961524, 0.0024641898959489104, 0.0029685887502869245, 0.003580130428232627, 0.0043222729172705705, 0.005223716235186701, 0.006319579977573116, 0.007652839737925042, 0.00927607499223088, 0.011253589308098758, 0.013663971971842982, 0.01660317740395658, 0.020188203515966608, 0.02456144998804397, 0.02989582849457037, 0.036400673380745145, 0.04432845459730691, 0.05398221231274829, 0.0657234966731705, 0.07998038170962321, 0.09725479542207396, 0.11812792383015797, 0.1432617489126483, 0.17339380269576596, 0.20932089378911475, 0.2518658361991608, 0.3018190865448726, 0.3598448084158025, 0.4263386284651823, 0.5012231099364872, 0.5836684556869592, 0.671733179242945, 0.7619373134664291, 0.848816206314798, 0.9245646406982786, 0.9789755457990131]
plt.plot(xt, yt)
yt=[1.0, 0.9550457960911298, 0.779941360899498, 0.429172781321184, 0.045934162253628376, 1.70794435468167E-5, 4.888470210229897E-5, 8.249664178207782E-5, 1.1803180363918053E-4, 1.556151803587797E-4, 1.953808993657684E-4, 2.3747296700123467E-4, 2.8204607976606684E-4, 3.2926650927327865E-4, 3.793130682691003E-4, 4.3237816588570987E-4, 4.886689611864024E-4, 5.484086250693957E-4, 6.118377217229506E-4, 6.792157220880702E-4, 7.508226632036289E-4, 8.269609689030052E-4, 9.079574491246936E-4, 9.941654971186774E-4, 0.0010859675061066425, 0.001183777529522552, 0.001288044211861248, 0.0013992540204433776, 0.001517934812118571, 0.0016446597731373702, 0.0017800517751972499, 0.0019247881960915794, 0.0020796062595578764, 0.0022453089559436373, 0.0024227716133124, 0.0026129491977482364, 0.00281688443205618, 0.0030357168340008817, 0.003270692788910474, 0.0035231767871708093, 0.0037946639751684098, 0.004086794188982109, 0.0044013676640149236, 0.004740362641314088, 0.00510595512315683, 0.005500541067296695, 0.005926761351913287, 0.0063875298927837665, 0.0068860653516737745, 0.007425926941824745, 0.008011054914346444, 0.008645816400273926, 0.00933505738935011, 0.010084161751032153, 0.010899118349102751, 0.011786597472553666, 0.012754038006817497, 0.013809747006602596, 0.014963013611283504, 0.016224239574091835, 0.017605089066905884, 0.019118660884803945, 0.020779686722544356, 0.022604759845242975, 0.02461259924736341, 0.02682435531100368, 0.029263964063853315, 0.03195855843135789, 0.03493894641401778, 0.03824016794224998, 0.041902144315965874, 0.04597043667623084, 0.0504971329359878, 0.0555418860663515, 0.061173130632250335, 0.06746950900508128, 0.07452154370259433, 0.08243359766577656, 0.09132616964555915, 0.10133857659385306, 0.11263207786317408, 0.12539349506580083, 0.13983937310613292, 0.15622070620567113, 0.17482820761641776, 0.19599801621231813, 0.22011757869225, 0.24763117441462557, 0.2790440787852654, 0.31492355005636796, 0.3558934308312087, 0.4026167575483118, 0.4557566285478778, 0.5158983865737179, 0.5834036074026626, 0.6581443627580118, 0.739027677090902, 0.8231535765598103, 0.9043405223837573, 0.9705956911379466]
plt.plot(xt, yt)
yt=[1.0, 0.8944259017481836, 0.22291732467789546, 1.3116439246747355E-4, 3.2081635365400883E-4, 5.170494177516707E-4, 7.201468328908817E-4, 9.304065259206613E-4, 0.0011481419977043318, 0.001373683281542252, 0.0016073779697935088, 0.0018495923142275304, 0.0021007124061416184, 0.0023611454428373955, 0.0026313210876627554, 0.0029116929315040016, 0.003202740064361486, 0.0035049687664705253, 0.003818914329346159, 0.004145143018146697, 0.00448425418787802, 0.004836882567212742, 0.005203700725089625, 0.005585421736807596, 0.005982802068054013, 0.006396644697231115, 0.00682780249859407, 0.007277181911115927, 0.007745746920684598, 0.008234523386250267, 0.008744603743924316, 0.009277152126829906, 0.009833409942780424, 0.010414701956677644, 0.011022442929955599, 0.011658144875534555, 0.012323424993694758, 0.013020014362149217, 0.013749767462525039, 0.014514672635612723, 0.015316863569297701, 0.016158631936263695, 0.017042441313605987, 0.017970942533710944, 0.01894699063549286, 0.019973663607740312, 0.021054283142393077, 0.022192437645618998, 0.02339200778926139, 0.024657194925384818, 0.025992552733214384, 0.027403022521878987, 0.02889397267539415, 0.030471242799887863, 0.03214119321914541, 0.03391076056549235, 0.03578752033170804, 0.03777975738953147, 0.039896545645598795, 0.042147838201474344, 0.04454456961708473, 0.04709877215403859, 0.04982370820648982, 0.05273402152407098, 0.055845910308459965, 0.059177325840320164, 0.06274820098906234, 0.06658071380208978, 0.07069959239796955, 0.07513246864338863, 0.07991028963234763, 0.08506779787797136, 0.0906440934613917, 0.09668329427094906, 0.10323531405121864, 0.1103567824460085, 0.11811213679116198, 0.1265749223792385, 0.1358293466370175, 0.14597214356696028, 0.15711481842766487, 0.1693863595567397, 0.18293652507378633, 0.1979398374037531, 0.21460044811215745, 0.2331580681955405, 0.25389519065909033, 0.277145852702458, 0.30330617058381537, 0.33284677904046023, 0.3663270044186771, 0.40440984333021746, 0.44787503775111265, 0.4976234177059749, 0.5546560665274937, 0.6199887970341318, 0.694404626380419, 0.7777930012674013, 0.8673797517994262, 0.952719833837565]
plt.plot(xt, yt)
| 0.411111 | 0.725041 |
<h1 style="text-align:center">Deep Learning </h1>
<h1 style="text-align:center"> Lab Session 2 - 3 Hours </h1>
<h1 style="text-align:center"> Convolutional Neural Network (CNN) for Handwritten Digits Recognition</h1>
<b> Student 1:</b> CANALE
<b> Student 2:</b> ELLENA
The aim of this session is to practice with Convolutional Neural Networks. Answers and experiments should be made by groups of one or two students. Each group should fill and run appropriate notebook cells.
Once you have completed all of the code implementations and successfully answered each question above, you may finalize your work by exporting the iPython Notebook as an pdf document using print as PDF (Ctrl+P). Do not forget to run all your cells before generating your final report and do not forget to include the names of all participants in the group. The lab session should be completed by May 29th 2017.
Send you pdf file to [email protected] and [email protected] using **[DeepLearning_lab2]** as Subject of your email.
# Introduction
In the last Lab Session, you built a Multilayer Perceptron for recognizing hand-written digits from the MNIST data-set. The best achieved accuracy on testing data was about 97%. Can you do better than these results using a deep CNN ?
In this Lab Session, you will build, train and optimize in TensorFlow one of the early Convolutional Neural Networks: **LeNet-5** to go to more than 99% of accuracy.
# Load MNIST Data in TensorFlow
Run the cell above to load the MNIST data that comes with TensorFlow. You will use this data in **Section 1** and **Section 2**.
```
import tensorflow as tf
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets("MNIST_data/", one_hot=True)
X_train, y_train = mnist.train.images, mnist.train.labels
X_validation, y_validation = mnist.validation.images, mnist.validation.labels
X_test, y_test = mnist.test.images, mnist.test.labels
print("Image Shape: {}".format(X_train[0].shape))
print("Training Set: {} samples".format(len(X_train)))
print("Validation Set: {} samples".format(len(X_validation)))
print("Test Set: {} samples".format(len(X_test)))
```
# Section 1 : My First Model in TensorFlow
Before starting with CNN, let's train and test in TensorFlow the example :
**y=softmax(Wx+b)** seen in the DeepLearing course last week.
This model reaches an accuracy of about 92 %.
You will also learn how to launch the tensorBoard https://www.tensorflow.org/get_started/summaries_and_tensorboard to visualize the computation graph, statistics and learning curves.
<b> Part 1 </b> : Read carefully the code in the cell below. Run it to perform training.
```
from __future__ import print_function
import tensorflow as tf
#STEP 1
# Parameters
learning_rate = 0.01
training_epochs = 100
batch_size = 128
display_step = 1
logs_path = 'log_files/' # useful for tensorboard
# tf Graph Input: mnist data image of shape 28*28=784
x = tf.placeholder(tf.float32, [None, 784], name='InputData')
# 0-9 digits recognition, 10 classes
y = tf.placeholder(tf.float32, [None, 10], name='LabelData')
# Set model weights
W = tf.Variable(tf.zeros([784, 10]), name='Weights')
b = tf.Variable(tf.zeros([10]), name='Bias')
# Construct model and encapsulating all ops into scopes, making Tensorboard's Graph visualization more convenient
with tf.name_scope('Model'):
# Model
pred = tf.nn.softmax(tf.matmul(x, W) + b) # Softmax
with tf.name_scope('Loss'):
# Minimize error using cross entropy
cost = tf.reduce_mean(-tf.reduce_sum(y*tf.log(pred), reduction_indices=1))
with tf.name_scope('SGD'):
# Gradient Descent
optimizer = tf.train.GradientDescentOptimizer(learning_rate).minimize(cost)
with tf.name_scope('Accuracy'):
# Accuracy
acc = tf.equal(tf.argmax(pred, 1), tf.argmax(y, 1))
acc = tf.reduce_mean(tf.cast(acc, tf.float32))
# Initializing the variables
init = tf.global_variables_initializer()
# Create a summary to monitor cost tensor
tf.summary.scalar("Loss", cost)
# Create a summary to monitor accuracy tensor
tf.summary.scalar("Accuracy", acc)
# Merge all summaries into a single op
merged_summary_op = tf.summary.merge_all()
#STEP 2
# Launch the graph for training
with tf.Session() as sess:
sess.run(init)
# op to write logs to Tensorboard
summary_writer = tf.summary.FileWriter(logs_path, graph=tf.get_default_graph())
# Training cycle
for epoch in range(training_epochs):
avg_cost = 0.
total_batch = int(mnist.train.num_examples/batch_size)
# Loop over all batches
for i in range(total_batch):
batch_xs, batch_ys = mnist.train.next_batch(batch_size)
# Run optimization op (backprop), cost op (to get loss value)
# and summary nodes
_, c, summary = sess.run([optimizer, cost, merged_summary_op],
feed_dict={x: batch_xs, y: batch_ys})
# Write logs at every iteration
summary_writer.add_summary(summary, epoch * total_batch + i)
# Compute average loss
avg_cost += c / total_batch
# Display logs per epoch step
if (epoch+1) % display_step == 0:
print("Epoch: ", '%02d' % (epoch+1), " =====> Loss=", "{:.9f}".format(avg_cost))
print("Optimization Finished!")
# Test model
# Calculate accuracy
print("Accuracy:", acc.eval({x: mnist.test.images, y: mnist.test.labels}))
```
<b> Part 2 </b>: Using Tensorboard, we can now visualize the created graph, giving you an overview of your architecture and how all of the major components are connected. You can also see and analyse the learning curves.
To launch tensorBoard:
- Go to the **TP2** folder,
- Open a Terminal and run the command line **"tensorboard --logdir= log_files/"**, it will generate an http link ,ex http://666.6.6.6:6006,
- Copy this link into your web browser
Enjoy It !!
# Section 2 : The 99% MNIST Challenge !
<b> Part 1 </b> : LeNet5 implementation
One you are now familar with **tensorFlow** and **tensorBoard**, you are in this section to build, train and test the baseline [LeNet-5](http://yann.lecun.com/exdb/lenet/) model for the MNIST digits recognition problem.
In more advanced step you will make some optimizations to get more than 99% of accuracy. The best model can get to over 99.7% accuracy!
For more information, have a look at this list of results : http://rodrigob.github.io/are_we_there_yet/build/classification_datasets_results.html
<img src="lenet.png",width="800" height="600" align="center">
<center><span>Figure 1: Lenet 5 </span></center>
The LeNet architecture accepts a 32x32xC image as input, where C is the number of color channels. Since MNIST images are grayscale, C is 1 in this case.
--------------------------
**Layer 1: Convolutional.** The output shape should be 28x28x6 **Activation.** sigmoid **Pooling.** The output shape should be 14x14x6.
**Layer 2: Convolutional.** The output shape should be 10x10x16. **Activation.** sigmoid **Pooling.** The output shape should be 5x5x16.
**Flatten.** Flatten the output shape of the final pooling layer such that it's 1D instead of 3D. You may need to use **flatten* from tensorflow.contrib.layers import flatten
**Layer 3: Fully Connected.** This should have 120 outputs. **Activation.** sigmoid
**Layer 4: Fully Connected.** This should have 84 outputs. **Activation.** sigmoid
**Layer 5: Fully Connected.** This should have 10 outputs. **Activation.** softmax
<b> Question 2.1.1 </b> Implement the Neural Network architecture described above.
For that, your will use classes and functions from https://www.tensorflow.org/api_docs/python/tf/nn.
We give you some helper functions for weigths and bias initilization. Also you can refer to section 1.
```
#Helper functions for weigths and bias initilization
def weight_variable(shape):
initial = tf.truncated_normal(shape, stddev=0.1)
return tf.Variable(initial)
def bias_variable(shape):
initial = tf.constant(0.1, shape=shape)
return tf.Variable(initial)
# https://adeshpande3.github.io/adeshpande3.github.io/A-Beginner's-Guide-To-Understanding-Convolutional-Neural-Networks/
# https://adeshpande3.github.io/A-Beginner%27s-Guide-To-Understanding-Convolutional-Neural-Networks-Part-2/
def LeNet5_Model(data,activation_function=tf.nn.sigmoid):
# layer 1 param
conv1_weights = weight_variable([5,5,1,6])
conv1_bias = bias_variable([6])
# layer 2 param
conv2_weights = weight_variable([5,5,6,16])
conv2_bias = bias_variable([16])
# layer 3 param
layer3_weights = weight_variable([400, 120])
layer3_bias = bias_variable([120])
# layer 4 param
layer4_weights = weight_variable([120, 84])
layer4_bias = bias_variable([84])
# layer 5 param
layer5_weights = weight_variable([84, 10])
layer5_bias = bias_variable([10])
with tf.name_scope('Model'):
with tf.name_scope('Layer1'):
conv1 = tf.nn.conv2d(input=data,filter=conv1_weights,strides=[1,1,1,1],padding='SAME')
print(conv1.shape)
sigmoid1 = activation_function(conv1 + conv1_bias)
pool1 = tf.nn.max_pool(sigmoid1,ksize=[1, 2, 2, 1],strides=[1, 2, 2, 1],padding='VALID')
print(pool1.shape)
with tf.name_scope('Layer2'):
conv2 = tf.nn.conv2d(input=pool1,filter=conv2_weights,strides=[1,1,1,1],padding='VALID')
print(conv2.shape)
sigmoid2 = activation_function(conv2 + conv2_bias)
pool2 = tf.nn.max_pool(sigmoid2,ksize=[1, 2, 2, 1],strides=[1, 2, 2, 1],padding='VALID')
print(pool2.shape)
with tf.name_scope('Flatten'):
flat_inputs = tf.contrib.layers.flatten(pool2)
print(flat_inputs.shape)
with tf.name_scope('Layer3'):
out3 = activation_function(tf.matmul(flat_inputs, layer3_weights) + layer3_bias)
with tf.name_scope('Layer4'):
out4 = activation_function(tf.matmul(out3, layer4_weights) + layer4_bias)
with tf.name_scope('Layer5'):
pred = tf.nn.softmax(tf.matmul(out4, layer5_weights) + layer5_bias) # Softmax
return pred
```
<b> Question 2.1.2. </b> Calculate the number of parameters of this model
```
total_parameters = 0
for variable in tf.trainable_variables():
# shape is an array of tf.Dimension
shape = variable.get_shape()
print(shape)
variable_parametes = 1
for dim in shape:
variable_parametes *= dim.value
print(variable_parametes)
total_parameters += variable_parametes
print(total_parameters)
layer1 = 5*5*1*6 + 6
layer2 = 5*5*6*16 + 16
layer3 = 400*120 + 120
layer4 = 120*84 + 84
layer5 = 84*10 + 10
tot = layer1 + layer2 + layer3 + layer4 + layer5
print('total number of parameters: %d' % tot)
```
Your answer goes here in details
<b> Question 2.1.3. </b> Start the training with the parameters cited below:
Learning rate =0.1
Loss Fucntion : Cross entropy
Optimisateur: SGD
Number of training iterations= 100
The batch size =128
```
from __future__ import print_function
import tensorflow as tf
from numpy import array
import numpy as np
#STEP 1
tf.reset_default_graph()
# Parameters
learning_rate = 0.1
training_epochs = 100
batch_size = 128
display_step = 1
logs_path = 'log_files/' # useful for tensorboard
# tf Graph Input: mnist data image of shape 28*28=784
x = tf.placeholder(tf.float32, [batch_size,28, 28,1], name='InputData')
# 0-9 digits recognition, 10 classes
y = tf.placeholder(tf.float32, [batch_size, 10], name='LabelData')
# Construct model and encapsulating all ops into scopes, making Tensorboard's Graph visualization more convenient
with tf.name_scope('Model'):
# Model
pred = LeNet5_Model(data=x)
with tf.name_scope('Loss'):
# Minimize error using cross entropy
cost = tf.reduce_mean(-tf.reduce_sum(y*tf.log(pred), reduction_indices=1))
with tf.name_scope('SGD'):
# Gradient Descent
optimizer = tf.train.GradientDescentOptimizer(learning_rate).minimize(cost)
with tf.name_scope('Accuracy'):
# Accuracy
acc = tf.equal(tf.argmax(pred, 1), tf.argmax(y, 1))
acc = tf.reduce_mean(tf.cast(acc, tf.float32))
# Initializing the variables
init = tf.global_variables_initializer()
# Create a summary to monitor cost tensor
tf.summary.scalar("Loss", cost)
# Create a summary to monitor accuracy tensor
tf.summary.scalar("Accuracy", acc)
# Merge all summaries into a single op
merged_summary_op = tf.summary.merge_all()
#STEP 2
# Launch the graph for training
with tf.Session() as sess:
sess.run(init)
# op to write logs to Tensorboard
summary_writer = tf.summary.FileWriter(logs_path, graph=tf.get_default_graph())
# Training cycle
for epoch in range(training_epochs):
avg_cost = 0.
total_batch = int(mnist.train.num_examples/batch_size)
# Loop over all batches
for i in range(total_batch):
batch_xs, batch_ys = mnist.train.next_batch(batch_size)
batch_xs = array(batch_xs).reshape(batch_size, 28,28,1)
#print(batch_xs.shape)
#print(batch_xs.dtype)
# Run optimization op (backprop), cost op (to get loss value)
# and summary nodes
_, c, summary = sess.run([optimizer, cost, merged_summary_op],
feed_dict={x: batch_xs, y: batch_ys})
# Write logs at every iteration
summary_writer.add_summary(summary, epoch * total_batch + i)
# Compute average loss
avg_cost += c / total_batch
# Display logs per epoch step
if (epoch+1) % display_step == 0:
print("Epoch: ", '%02d' % (epoch+1), " =====> Loss=", "{:.9f}".format(avg_cost))
print("Optimization Finished!")
```
<b> Question 2.1.4. </b> Implement the evaluation function for accuracy computation
```
def evaluate(model, y):
#your implementation goes here
correct_prediction = tf.equal(tf.argmax(model,1), tf.argmax(y,1))
accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))
#print(accuracy.eval(feed_dict={x: mnist.test.images, y_: mnist.test.labels}))
return accuracy
```
<b> Question 2.1.5. </b> Implement training pipeline and run the training data through it to train the model.
- Before each epoch, shuffle the training set.
- Print the loss per mini batch and the training/validation accuracy per epoch. (Display results every 100 epochs)
- Save the model after training
- Print after training the final testing accuracy
```
import numpy as np
# Initializing the variables
def train(learning_rate, training_epochs, batch_size, display_step, optimizer_method=tf.train.GradientDescentOptimizer,activation_function=tf.nn.sigmoid):
tf.reset_default_graph()
# Initializing the session
logs_path = 'log_files/' # useful for tensorboard
# tf Graph Input: mnist data image of shape 28*28=784
x = tf.placeholder(tf.float32, [None,28, 28,1], name='InputData')
# 0-9 digits recognition, 10 classes
y = tf.placeholder(tf.float32, [None, 10], name='LabelData')
# Construct model and encapsulating all ops into scopes, making Tensorboard's Graph visualization more convenient
with tf.name_scope('Model'):
# Model
pred = LeNet5_Model(data=x,activation_function=activation_function)
with tf.name_scope('Loss'):
# Minimize error using cross entropy
# Minimize error using cross entropy
if activation_function == tf.nn.sigmoid:
cost = tf.reduce_mean(-tf.reduce_sum(y*tf.log(pred), reduction_indices=1))
else:
cost = tf.reduce_mean(-tf.reduce_sum(y*tf.log(tf.clip_by_value(pred,-1.0,1.0)), reduction_indices=1))
#cost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits=pred, labels=y))
with tf.name_scope('SGD'):
# Gradient Descent
optimizer = optimizer_method(learning_rate).minimize(cost)
with tf.name_scope('Accuracy'):
# Accuracy
acc = evaluate(pred, y)
# Initializing the variables
init = tf.global_variables_initializer()
# Create a summary to monitor cost tensor
tf.summary.scalar("Loss", cost)
# Create a summary to monitor accuracy tensor
tf.summary.scalar("Accuracy", acc)
# Merge all summaries into a single op
merged_summary_op = tf.summary.merge_all()
saver = tf.train.Saver()
print ("Start Training!")
t0 = time()
X_train,Y_train = mnist.train.images.reshape((-1,28,28,1)), mnist.train.labels
X_val,Y_val = mnist.validation.images.reshape((-1,28,28,1)), mnist.validation.labels
# Launch the graph for training
with tf.Session() as sess:
sess.run(init)
# op to write logs to Tensorboard
summary_writer = tf.summary.FileWriter(logs_path, graph=tf.get_default_graph())
# Training cycle
for epoch in range(training_epochs):
avg_cost = 0.
total_batch = int(mnist.train.num_examples/batch_size)
# Loop over all batches
for i in range(total_batch):
# train_next_batch shuffle the images by default
batch_xs, batch_ys = mnist.train.next_batch(batch_size)
batch_xs = batch_xs.reshape((-1,28,28,1))
# Run optimization op (backprop), cost op (to get loss value)
# and summary nodes
_, c, summary = sess.run([optimizer, cost, merged_summary_op],
feed_dict={x: batch_xs,
y: batch_ys})
# Write logs at every iteration
summary_writer.add_summary(summary, epoch * total_batch + i)
# Compute average loss
avg_cost += c / total_batch
# Display logs per epoch step
if (epoch+1) % display_step == 0:
print("Epoch: ", '%02d' % (epoch+1), "=====> Loss=", "{:.9f}".format(avg_cost))
acc_train = acc.eval({x: X_train, y: Y_train})
print("Epoch: ", '%02d' % (epoch+1), "=====> Accuracy Train=", "{:.9f}".format(acc_train))
acc_val = acc.eval({x: X_val, y: Y_val})
print("Epoch: ", '%02d' % (epoch+1), "=====> Accuracy Validation=", "{:.9f}".format(acc_val))
print ("Training Finished!")
t1 = time()
# Save the variables to disk.
save_path = saver.save(sess, "model.ckpt")
print("Model saved in file: %s" % save_path)
#Your implementation for testing accuracy after training goes here
X_test,Y_test = mnist.test.images.reshape((-1,28,28,1)),mnist.test.labels
acc_test = acc.eval({x: X_test, y: Y_test})
print("Accuracy Test=", "{:.9f}".format(acc_test))
return acc_train,acc_val,acc_test,t1-t0
%time train (0.1,100,128,10,optimizer_method=tf.train.GradientDescentOptimizer)
```
<b> Question 2.1.6 </b> : Use tensorBoard to visualise and save the LeNet5 Graph and all learning curves.
Save all obtained figures in the folder **"TP2/MNIST_99_Challenge_Figures"**
<img src="graph_run.png">
<img src="loss1.png">
<img src="accuracy1.png">
## Comment:
Here we see how the accuracy rapidly increases after few epochs and then increases at an always slower rate. Regarding the accuracy, we see that it gets always nearer to zero epoch after epoch.
From the graph we can have a confirm of our network architecture.
<b> Part 2 </b> : LeNET 5 Optimization
<b> Question 2.2.1 </b> Change the sigmoid function with a Relu :
- Retrain your network with SGD and AdamOptimizer and then fill the table above :
| Optimizer | Gradient Descent |AdamOptimizer |
| ------------- |: -------------: | ---------:
| Validation Accuracy |0.99180001|0.097599998|
| Testing Accuracy |0.99089998|0.1032|
| Training Time |36min|36min| |
- Try with different learning rates for each Optimizer (0.0001 and 0.001 ) and different Batch sizes (50 and 128) for 20000 Epochs.
- For each optimizer, plot (on the same curve) the **testing accuracies** function to **(learning rate, batch size)**
- Did you reach the 99% accuracy ? What are the optimal parametres that gave you the best results?
## Comment:
- Relu: when we use the relu we need to change the cost function, in fact we need to do gradient clipping otherwise our network will crash.
- The Adam optimizer gives very bad results when used with high learning rate, in fact it works better with a low learning rate, such as 0.001
- When we use the stocastic gradient descent with the relu we obtain really good results: more than 99% test accuracy. So we could stop here.
```
from time import time
%time train (0.1,100,128,10,optimizer_method=tf.train.GradientDescentOptimizer,activation_function=tf.nn.relu)
%time train (0.1,100,128,10,optimizer_method=tf.train.AdamOptimizer,activation_function=tf.nn.relu)
# your answer goas here
columns = ['optimizer','learning_rate','activation_function','batch_size','training_accuracy','validation_accuracy','test_accuracy','elapsed_time']
optimizer_options = {'gradient_descent':tf.train.GradientDescentOptimizer,'adam':tf.train.AdamOptimizer}
learning_options = [0.001,0.0001]
activation_options = {'sigmoid':tf.nn.sigmoid,'relu':tf.nn.relu}
batch_options = [50,128]
final_results = []
for optimizer_label in optimizer_options:
optimizer = optimizer_options[optimizer_label]
for learning_rate in learning_options:
for activation_label in activation_options:
activation_function = activation_options[activation_label]
for batch_size in batch_options:
#TO DEFINE TrainAndTest
training_accuracy,validation_accuracy,test_accuracy,elapsed_time = train(
learning_rate = learning_rate,
training_epochs=100,
batch_size = batch_size,
display_step = 10,
optimizer_method = optimizer,
activation_function = activation_function
)
obj_test = {'optimizer':optimizer_label,
'learning_rate':learning_rate,
'activation_function':activation_label,
'batch_size':batch_size,
'training_accuracy':training_accuracy,
'validation_accuracy':training_accuracy,
'test_accuracy':test_accuracy,
'elapsed_time': elapsed_time
}
final_results.append(obj_test)
final_results
```
## Comment
Here we have seen that the relu performs the best. Then, we also seen that the the sigmoid, combined with a low learning rate, takes an infinite amount of time to reach the optimum.
{'activation_function': 'relu',
<br>
'batch_size': 128,
<br>
'elapsed_time': 2140.7165479660034,
<br>
'learning_rate': 0.001,
<br>
'optimizer': 'adam',
<br>
'test_accuracy': 0.99260002,
<br>
'training_accuracy': 1.0,
<br>
'validation_accuracy': 1.0},
Regarding the batch size, the best configuration (learning rate=0.001, adam optimizer) achieved a better result with a larger batch size. Generally, we have not seen great differences. The most important thing is that with a littler batch size we need more time to train.
Regarding the learning rate, we achieved the best result with a learning rate of 0.001, but also in this case the answer is not certain. The final accuracy depends from a combination of factors and we can't find something like a proportional behaviour.
Special cases: when we use the stocasthic gradient descent with a low learning rate and the sigmoid activation function, the accuracy is always really low.
As a general rule, the adam optimizer can achieve a better acuracy in a lower amount of time, but we need to take care of carefully choosing the learning rate.
```
with open('json.json','r') as input_fp:
results = json.load(input_fp)
import matplotlib.pyplot as plt
sigmoid = [x for x in results if x['activation_function']=='sigmoid']
relu =[x for x in results if x['activation_function'] !='sigmoid']
plt.figure(figsize=(15,5))
plt.plot(range(len(sigmoid)),[x['test_accuracy'] for x in sigmoid])
plt.plot(range(len(sigmoid)),[x['test_accuracy'] for x in relu])
plt.legend(['sigmoid','relu'])
plt.ylabel('test accuracy')
plt.xlabel('index run')
plt.show()
```
The relu is always equal or better than the sigmoid
```
import matplotlib.pyplot as plt
a = [x for x in results if x['batch_size']== 128]
b =[x for x in results if x['batch_size'] !=128]
plt.figure(figsize=(15,5))
plt.plot(range(len(a)),[x['test_accuracy'] for x in a])
plt.plot(range(len(a)),[x['test_accuracy'] for x in b])
plt.legend(['128','50'])
plt.ylabel('test accuracy')
plt.xlabel('index run')
plt.show()
```
Test accuracy does not change when we change batch size
```
import matplotlib.pyplot as plt
a = [x for x in results if x['learning_rate']== 0.0001]
b =[x for x in results if x['learning_rate'] !=0.0001]
plt.figure(figsize=(15,5))
plt.plot(range(len(a)),[x['test_accuracy'] for x in a])
plt.plot(range(len(a)),[x['test_accuracy'] for x in b])
plt.legend(['learning_rate = 0.0001','learning_rate = 0.001'])
plt.ylabel('test accuracy')
plt.xlabel('index run')
plt.show()
```
An higher learning rate is better, in this specifi case. Later we will see that this is not always the case.
```
import matplotlib.pyplot as plt
a = [x for x in results if x['optimizer']== 'adam']
b =[x for x in results if x['optimizer'] != 'adam']
plt.figure(figsize=(15,5))
plt.plot(range(len(a)),[x['test_accuracy'] for x in a])
plt.plot(range(len(a)),[x['test_accuracy'] for x in b])
plt.legend(['adam','sgd'])
plt.ylabel('test accuracy')
plt.xlabel('index run')
plt.show()
```
Adam is always better than Stocasthic gradient descent
```
import matplotlib.pyplot as plt
a = [x for x in results if x['optimizer']== 'adam']
b =[x for x in results if x['optimizer'] != 'adam']
plt.figure(figsize=(15,5))
plt.plot(range(len(a)),[x['elapsed_time'] for x in a])
plt.plot(range(len(a)),[x['elapsed_time'] for x in b])
plt.legend(['adam','sgd'])
plt.ylabel('elapsed_time')
plt.xlabel('index run')
plt.show()
```
Sometimes adam is vaster than SGD
```
import matplotlib.pyplot as plt
a = [x for x in results if x['batch_size']== 128]
b =[x for x in results if x['batch_size'] !=128]
plt.figure(figsize=(15,5))
plt.plot(range(len(a)),[x['elapsed_time'] for x in a])
plt.plot(range(len(a)),[x['elapsed_time'] for x in b])
plt.legend(['128','50'])
plt.ylabel('elapsed_time')
plt.xlabel('index run')
plt.show()
```
Bigger batches mean less training time, this is actually a good news if we consider that the batch dimension does not has a big influence on the final accuracy
<b> Question 2.2.2 </b> What about applying a dropout layer on the Fully conntected layer and then retraining the model with the best Optimizer and parameters(Learning rate and Batsh size) obtained in *Question 2.2.1* ? (probability to keep units=0.75). For this stage ensure that the keep prob is set to 1.0 to evaluate the
performance of the network including all nodes.
```
# https://adeshpande3.github.io/adeshpande3.github.io/A-Beginner's-Guide-To-Understanding-Convolutional-Neural-Networks/
# https://adeshpande3.github.io/A-Beginner%27s-Guide-To-Understanding-Convolutional-Neural-Networks-Part-2/
def LeNet5_Model(data,keep_prob,activation_function=tf.nn.sigmoid):
# layer 1 param
conv1_weights = weight_variable([5,5,1,6])
conv1_bias = bias_variable([6])
# layer 2 param
conv2_weights = weight_variable([5,5,6,16])
conv2_bias = bias_variable([16])
# layer 3 param
layer3_weights = weight_variable([400, 120])
layer3_bias = bias_variable([120])
# layer 4 param
layer4_weights = weight_variable([120, 84])
layer4_bias = bias_variable([84])
# layer 5 param
layer5_weights = weight_variable([84, 10])
layer5_bias = bias_variable([10])
with tf.name_scope('Model'):
with tf.name_scope('Layer1'):
conv1 = tf.nn.conv2d(input=data,filter=conv1_weights,strides=[1,1,1,1],padding='SAME')
print(conv1.shape)
sigmoid1 = activation_function(conv1 + conv1_bias)
pool1 = tf.nn.max_pool(sigmoid1,ksize=[1, 2, 2, 1],strides=[1, 2, 2, 1],padding='VALID')
print(pool1.shape)
with tf.name_scope('Layer2'):
conv2 = tf.nn.conv2d(input=pool1,filter=conv2_weights,strides=[1,1,1,1],padding='VALID')
print(conv2.shape)
sigmoid2 = activation_function(conv2 + conv2_bias)
pool2 = tf.nn.max_pool(sigmoid2,ksize=[1, 2, 2, 1],strides=[1, 2, 2, 1],padding='VALID')
print(pool2.shape)
with tf.name_scope('Flatten'):
flat_inputs = tf.contrib.layers.flatten(pool2)
print(flat_inputs.shape)
with tf.name_scope('Layer3'):
out3 = activation_function(tf.matmul(flat_inputs, layer3_weights) + layer3_bias)
with tf.name_scope('Layer4'):
out4 = activation_function(tf.matmul(out3, layer4_weights) + layer4_bias)
with tf.name_scope('Layer5'):
out_drop = tf.nn.dropout(out4, keep_prob)
pred = tf.nn.softmax(tf.matmul(out_drop, layer5_weights) + layer5_bias) # Softmax
return pred
import numpy as np
# Initializing the variables
def train(learning_rate, training_epochs, batch_size, display_step, optimizer_method=tf.train.GradientDescentOptimizer,activation_function=tf.nn.sigmoid):
tf.reset_default_graph()
# Initializing the session
logs_path = 'log_files/' # useful for tensorboard
# tf Graph Input: mnist data image of shape 28*28=784
x = tf.placeholder(tf.float32, [None,28, 28,1], name='InputData')
# 0-9 digits recognition, 10 classes
y = tf.placeholder(tf.float32, [None, 10], name='LabelData')
keep_prob = tf.placeholder(tf.float32)
# Construct model and encapsulating all ops into scopes, making Tensorboard's Graph visualization more convenient
with tf.name_scope('Model'):
# Model
pred = LeNet5_Model(x,keep_prob,activation_function=activation_function)
with tf.name_scope('Loss'):
# Minimize error using cross entropy
# Minimize error using cross entropy
if activation_function == tf.nn.sigmoid:
cost = tf.reduce_mean(-tf.reduce_sum(y*tf.log(pred), reduction_indices=1))
else:
cost = tf.reduce_mean(-tf.reduce_sum(y*tf.log(tf.clip_by_value(pred,-1.0,1.0)), reduction_indices=1))
#cost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits=pred, labels=y))
with tf.name_scope('SGD'):
# Gradient Descent
optimizer = optimizer_method(learning_rate).minimize(cost)
with tf.name_scope('Accuracy'):
# Accuracy
acc = evaluate(pred, y)
# Initializing the variables
init = tf.global_variables_initializer()
# Create a summary to monitor cost tensor
tf.summary.scalar("Loss", cost)
# Create a summary to monitor accuracy tensor
tf.summary.scalar("Accuracy", acc)
# Merge all summaries into a single op
merged_summary_op = tf.summary.merge_all()
saver = tf.train.Saver()
print ("Start Training!")
t0 = time()
X_train,Y_train = mnist.train.images.reshape((-1,28,28,1)), mnist.train.labels
X_val,Y_val = mnist.validation.images.reshape((-1,28,28,1)), mnist.validation.labels
# Launch the graph for training
with tf.Session() as sess:
sess.run(init)
# op to write logs to Tensorboard
summary_writer = tf.summary.FileWriter(logs_path, graph=tf.get_default_graph())
# Training cycle
for epoch in range(training_epochs):
avg_cost = 0.
total_batch = int(mnist.train.num_examples/batch_size)
# Loop over all batches
for i in range(total_batch):
# train_next_batch shuffle the images by default
batch_xs, batch_ys = mnist.train.next_batch(batch_size)
batch_xs = batch_xs.reshape((-1,28,28,1))
# Run optimization op (backprop), cost op (to get loss value)
# and summary nodes
_, c, summary = sess.run([optimizer, cost, merged_summary_op],
feed_dict={x: batch_xs,
y: batch_ys,keep_prob:0.75})
# Write logs at every iteration
summary_writer.add_summary(summary, epoch * total_batch + i)
# Compute average loss
avg_cost += c / total_batch
# Display logs per epoch step
if (epoch+1) % display_step == 0:
print("Epoch: ", '%02d' % (epoch+1), "=====> Loss=", "{:.9f}".format(avg_cost))
acc_train = acc.eval({x: X_train, y: Y_train,keep_prob:1.0})
print("Epoch: ", '%02d' % (epoch+1), "=====> Accuracy Train=", "{:.9f}".format(acc_train))
acc_val = acc.eval({x: X_val, y: Y_val,keep_prob:1.0})
print("Epoch: ", '%02d' % (epoch+1), "=====> Accuracy Validation=", "{:.9f}".format(acc_val))
print ("Training Finished!")
t1 = time()
# Save the variables to disk.
save_path = saver.save(sess, "model.ckpt")
print("Model saved in file: %s" % save_path)
#Your implementation for testing accuracy after training goes here
X_test,Y_test = mnist.test.images.reshape((-1,28,28,1)),mnist.test.labels
acc_test = acc.eval({x: X_test, y: Y_test,keep_prob:1.0})
print("Accuracy Test=", "{:.9f}".format(acc_test))
return acc_train,acc_val,acc_test,t1-t0
```
## Comment:
Here we managed the keep_prob using a placeholder, in this way we can change it dynamically during our run.
We are also quite sure that we can achieve good performances with a limited number of epochs.
## Note:
We have seen that using a learning rate = 0.001 is unstable, this behavior is even more visible when we add the dropout, thus we used a lower learning rate.
The adam optimizer uses an Adaptive Moment Estimation and with an high learning rate, combined with a big batch size can actually bring the network in a worse state. https://arxiv.org/pdf/1412.6980.pdf
```
train (0.0001,50,128,10,optimizer_method=tf.train.AdamOptimizer,activation_function=tf.nn.relu)
```
## Comment:
Here after 50 epochs the result is not what we expected, we think that using the dropout we might actually need to use different parameters.
```
train (0.0001,50,50,10,optimizer_method=tf.train.AdamOptimizer,activation_function=tf.nn.relu)
```
## Comment:
We managed to obtain 99% accuracy score over the test set in 50 epochs, this is a really good result. This actually explain the idea of the adam optimizer, that using the concept of moment can improve the model in a short amount of time and a low learning rate. This, combined with the relu function allows us to achieve high accuracies.
|
github_jupyter
|
import tensorflow as tf
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets("MNIST_data/", one_hot=True)
X_train, y_train = mnist.train.images, mnist.train.labels
X_validation, y_validation = mnist.validation.images, mnist.validation.labels
X_test, y_test = mnist.test.images, mnist.test.labels
print("Image Shape: {}".format(X_train[0].shape))
print("Training Set: {} samples".format(len(X_train)))
print("Validation Set: {} samples".format(len(X_validation)))
print("Test Set: {} samples".format(len(X_test)))
from __future__ import print_function
import tensorflow as tf
#STEP 1
# Parameters
learning_rate = 0.01
training_epochs = 100
batch_size = 128
display_step = 1
logs_path = 'log_files/' # useful for tensorboard
# tf Graph Input: mnist data image of shape 28*28=784
x = tf.placeholder(tf.float32, [None, 784], name='InputData')
# 0-9 digits recognition, 10 classes
y = tf.placeholder(tf.float32, [None, 10], name='LabelData')
# Set model weights
W = tf.Variable(tf.zeros([784, 10]), name='Weights')
b = tf.Variable(tf.zeros([10]), name='Bias')
# Construct model and encapsulating all ops into scopes, making Tensorboard's Graph visualization more convenient
with tf.name_scope('Model'):
# Model
pred = tf.nn.softmax(tf.matmul(x, W) + b) # Softmax
with tf.name_scope('Loss'):
# Minimize error using cross entropy
cost = tf.reduce_mean(-tf.reduce_sum(y*tf.log(pred), reduction_indices=1))
with tf.name_scope('SGD'):
# Gradient Descent
optimizer = tf.train.GradientDescentOptimizer(learning_rate).minimize(cost)
with tf.name_scope('Accuracy'):
# Accuracy
acc = tf.equal(tf.argmax(pred, 1), tf.argmax(y, 1))
acc = tf.reduce_mean(tf.cast(acc, tf.float32))
# Initializing the variables
init = tf.global_variables_initializer()
# Create a summary to monitor cost tensor
tf.summary.scalar("Loss", cost)
# Create a summary to monitor accuracy tensor
tf.summary.scalar("Accuracy", acc)
# Merge all summaries into a single op
merged_summary_op = tf.summary.merge_all()
#STEP 2
# Launch the graph for training
with tf.Session() as sess:
sess.run(init)
# op to write logs to Tensorboard
summary_writer = tf.summary.FileWriter(logs_path, graph=tf.get_default_graph())
# Training cycle
for epoch in range(training_epochs):
avg_cost = 0.
total_batch = int(mnist.train.num_examples/batch_size)
# Loop over all batches
for i in range(total_batch):
batch_xs, batch_ys = mnist.train.next_batch(batch_size)
# Run optimization op (backprop), cost op (to get loss value)
# and summary nodes
_, c, summary = sess.run([optimizer, cost, merged_summary_op],
feed_dict={x: batch_xs, y: batch_ys})
# Write logs at every iteration
summary_writer.add_summary(summary, epoch * total_batch + i)
# Compute average loss
avg_cost += c / total_batch
# Display logs per epoch step
if (epoch+1) % display_step == 0:
print("Epoch: ", '%02d' % (epoch+1), " =====> Loss=", "{:.9f}".format(avg_cost))
print("Optimization Finished!")
# Test model
# Calculate accuracy
print("Accuracy:", acc.eval({x: mnist.test.images, y: mnist.test.labels}))
#Helper functions for weigths and bias initilization
def weight_variable(shape):
initial = tf.truncated_normal(shape, stddev=0.1)
return tf.Variable(initial)
def bias_variable(shape):
initial = tf.constant(0.1, shape=shape)
return tf.Variable(initial)
# https://adeshpande3.github.io/adeshpande3.github.io/A-Beginner's-Guide-To-Understanding-Convolutional-Neural-Networks/
# https://adeshpande3.github.io/A-Beginner%27s-Guide-To-Understanding-Convolutional-Neural-Networks-Part-2/
def LeNet5_Model(data,activation_function=tf.nn.sigmoid):
# layer 1 param
conv1_weights = weight_variable([5,5,1,6])
conv1_bias = bias_variable([6])
# layer 2 param
conv2_weights = weight_variable([5,5,6,16])
conv2_bias = bias_variable([16])
# layer 3 param
layer3_weights = weight_variable([400, 120])
layer3_bias = bias_variable([120])
# layer 4 param
layer4_weights = weight_variable([120, 84])
layer4_bias = bias_variable([84])
# layer 5 param
layer5_weights = weight_variable([84, 10])
layer5_bias = bias_variable([10])
with tf.name_scope('Model'):
with tf.name_scope('Layer1'):
conv1 = tf.nn.conv2d(input=data,filter=conv1_weights,strides=[1,1,1,1],padding='SAME')
print(conv1.shape)
sigmoid1 = activation_function(conv1 + conv1_bias)
pool1 = tf.nn.max_pool(sigmoid1,ksize=[1, 2, 2, 1],strides=[1, 2, 2, 1],padding='VALID')
print(pool1.shape)
with tf.name_scope('Layer2'):
conv2 = tf.nn.conv2d(input=pool1,filter=conv2_weights,strides=[1,1,1,1],padding='VALID')
print(conv2.shape)
sigmoid2 = activation_function(conv2 + conv2_bias)
pool2 = tf.nn.max_pool(sigmoid2,ksize=[1, 2, 2, 1],strides=[1, 2, 2, 1],padding='VALID')
print(pool2.shape)
with tf.name_scope('Flatten'):
flat_inputs = tf.contrib.layers.flatten(pool2)
print(flat_inputs.shape)
with tf.name_scope('Layer3'):
out3 = activation_function(tf.matmul(flat_inputs, layer3_weights) + layer3_bias)
with tf.name_scope('Layer4'):
out4 = activation_function(tf.matmul(out3, layer4_weights) + layer4_bias)
with tf.name_scope('Layer5'):
pred = tf.nn.softmax(tf.matmul(out4, layer5_weights) + layer5_bias) # Softmax
return pred
total_parameters = 0
for variable in tf.trainable_variables():
# shape is an array of tf.Dimension
shape = variable.get_shape()
print(shape)
variable_parametes = 1
for dim in shape:
variable_parametes *= dim.value
print(variable_parametes)
total_parameters += variable_parametes
print(total_parameters)
layer1 = 5*5*1*6 + 6
layer2 = 5*5*6*16 + 16
layer3 = 400*120 + 120
layer4 = 120*84 + 84
layer5 = 84*10 + 10
tot = layer1 + layer2 + layer3 + layer4 + layer5
print('total number of parameters: %d' % tot)
from __future__ import print_function
import tensorflow as tf
from numpy import array
import numpy as np
#STEP 1
tf.reset_default_graph()
# Parameters
learning_rate = 0.1
training_epochs = 100
batch_size = 128
display_step = 1
logs_path = 'log_files/' # useful for tensorboard
# tf Graph Input: mnist data image of shape 28*28=784
x = tf.placeholder(tf.float32, [batch_size,28, 28,1], name='InputData')
# 0-9 digits recognition, 10 classes
y = tf.placeholder(tf.float32, [batch_size, 10], name='LabelData')
# Construct model and encapsulating all ops into scopes, making Tensorboard's Graph visualization more convenient
with tf.name_scope('Model'):
# Model
pred = LeNet5_Model(data=x)
with tf.name_scope('Loss'):
# Minimize error using cross entropy
cost = tf.reduce_mean(-tf.reduce_sum(y*tf.log(pred), reduction_indices=1))
with tf.name_scope('SGD'):
# Gradient Descent
optimizer = tf.train.GradientDescentOptimizer(learning_rate).minimize(cost)
with tf.name_scope('Accuracy'):
# Accuracy
acc = tf.equal(tf.argmax(pred, 1), tf.argmax(y, 1))
acc = tf.reduce_mean(tf.cast(acc, tf.float32))
# Initializing the variables
init = tf.global_variables_initializer()
# Create a summary to monitor cost tensor
tf.summary.scalar("Loss", cost)
# Create a summary to monitor accuracy tensor
tf.summary.scalar("Accuracy", acc)
# Merge all summaries into a single op
merged_summary_op = tf.summary.merge_all()
#STEP 2
# Launch the graph for training
with tf.Session() as sess:
sess.run(init)
# op to write logs to Tensorboard
summary_writer = tf.summary.FileWriter(logs_path, graph=tf.get_default_graph())
# Training cycle
for epoch in range(training_epochs):
avg_cost = 0.
total_batch = int(mnist.train.num_examples/batch_size)
# Loop over all batches
for i in range(total_batch):
batch_xs, batch_ys = mnist.train.next_batch(batch_size)
batch_xs = array(batch_xs).reshape(batch_size, 28,28,1)
#print(batch_xs.shape)
#print(batch_xs.dtype)
# Run optimization op (backprop), cost op (to get loss value)
# and summary nodes
_, c, summary = sess.run([optimizer, cost, merged_summary_op],
feed_dict={x: batch_xs, y: batch_ys})
# Write logs at every iteration
summary_writer.add_summary(summary, epoch * total_batch + i)
# Compute average loss
avg_cost += c / total_batch
# Display logs per epoch step
if (epoch+1) % display_step == 0:
print("Epoch: ", '%02d' % (epoch+1), " =====> Loss=", "{:.9f}".format(avg_cost))
print("Optimization Finished!")
def evaluate(model, y):
#your implementation goes here
correct_prediction = tf.equal(tf.argmax(model,1), tf.argmax(y,1))
accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))
#print(accuracy.eval(feed_dict={x: mnist.test.images, y_: mnist.test.labels}))
return accuracy
import numpy as np
# Initializing the variables
def train(learning_rate, training_epochs, batch_size, display_step, optimizer_method=tf.train.GradientDescentOptimizer,activation_function=tf.nn.sigmoid):
tf.reset_default_graph()
# Initializing the session
logs_path = 'log_files/' # useful for tensorboard
# tf Graph Input: mnist data image of shape 28*28=784
x = tf.placeholder(tf.float32, [None,28, 28,1], name='InputData')
# 0-9 digits recognition, 10 classes
y = tf.placeholder(tf.float32, [None, 10], name='LabelData')
# Construct model and encapsulating all ops into scopes, making Tensorboard's Graph visualization more convenient
with tf.name_scope('Model'):
# Model
pred = LeNet5_Model(data=x,activation_function=activation_function)
with tf.name_scope('Loss'):
# Minimize error using cross entropy
# Minimize error using cross entropy
if activation_function == tf.nn.sigmoid:
cost = tf.reduce_mean(-tf.reduce_sum(y*tf.log(pred), reduction_indices=1))
else:
cost = tf.reduce_mean(-tf.reduce_sum(y*tf.log(tf.clip_by_value(pred,-1.0,1.0)), reduction_indices=1))
#cost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits=pred, labels=y))
with tf.name_scope('SGD'):
# Gradient Descent
optimizer = optimizer_method(learning_rate).minimize(cost)
with tf.name_scope('Accuracy'):
# Accuracy
acc = evaluate(pred, y)
# Initializing the variables
init = tf.global_variables_initializer()
# Create a summary to monitor cost tensor
tf.summary.scalar("Loss", cost)
# Create a summary to monitor accuracy tensor
tf.summary.scalar("Accuracy", acc)
# Merge all summaries into a single op
merged_summary_op = tf.summary.merge_all()
saver = tf.train.Saver()
print ("Start Training!")
t0 = time()
X_train,Y_train = mnist.train.images.reshape((-1,28,28,1)), mnist.train.labels
X_val,Y_val = mnist.validation.images.reshape((-1,28,28,1)), mnist.validation.labels
# Launch the graph for training
with tf.Session() as sess:
sess.run(init)
# op to write logs to Tensorboard
summary_writer = tf.summary.FileWriter(logs_path, graph=tf.get_default_graph())
# Training cycle
for epoch in range(training_epochs):
avg_cost = 0.
total_batch = int(mnist.train.num_examples/batch_size)
# Loop over all batches
for i in range(total_batch):
# train_next_batch shuffle the images by default
batch_xs, batch_ys = mnist.train.next_batch(batch_size)
batch_xs = batch_xs.reshape((-1,28,28,1))
# Run optimization op (backprop), cost op (to get loss value)
# and summary nodes
_, c, summary = sess.run([optimizer, cost, merged_summary_op],
feed_dict={x: batch_xs,
y: batch_ys})
# Write logs at every iteration
summary_writer.add_summary(summary, epoch * total_batch + i)
# Compute average loss
avg_cost += c / total_batch
# Display logs per epoch step
if (epoch+1) % display_step == 0:
print("Epoch: ", '%02d' % (epoch+1), "=====> Loss=", "{:.9f}".format(avg_cost))
acc_train = acc.eval({x: X_train, y: Y_train})
print("Epoch: ", '%02d' % (epoch+1), "=====> Accuracy Train=", "{:.9f}".format(acc_train))
acc_val = acc.eval({x: X_val, y: Y_val})
print("Epoch: ", '%02d' % (epoch+1), "=====> Accuracy Validation=", "{:.9f}".format(acc_val))
print ("Training Finished!")
t1 = time()
# Save the variables to disk.
save_path = saver.save(sess, "model.ckpt")
print("Model saved in file: %s" % save_path)
#Your implementation for testing accuracy after training goes here
X_test,Y_test = mnist.test.images.reshape((-1,28,28,1)),mnist.test.labels
acc_test = acc.eval({x: X_test, y: Y_test})
print("Accuracy Test=", "{:.9f}".format(acc_test))
return acc_train,acc_val,acc_test,t1-t0
%time train (0.1,100,128,10,optimizer_method=tf.train.GradientDescentOptimizer)
from time import time
%time train (0.1,100,128,10,optimizer_method=tf.train.GradientDescentOptimizer,activation_function=tf.nn.relu)
%time train (0.1,100,128,10,optimizer_method=tf.train.AdamOptimizer,activation_function=tf.nn.relu)
# your answer goas here
columns = ['optimizer','learning_rate','activation_function','batch_size','training_accuracy','validation_accuracy','test_accuracy','elapsed_time']
optimizer_options = {'gradient_descent':tf.train.GradientDescentOptimizer,'adam':tf.train.AdamOptimizer}
learning_options = [0.001,0.0001]
activation_options = {'sigmoid':tf.nn.sigmoid,'relu':tf.nn.relu}
batch_options = [50,128]
final_results = []
for optimizer_label in optimizer_options:
optimizer = optimizer_options[optimizer_label]
for learning_rate in learning_options:
for activation_label in activation_options:
activation_function = activation_options[activation_label]
for batch_size in batch_options:
#TO DEFINE TrainAndTest
training_accuracy,validation_accuracy,test_accuracy,elapsed_time = train(
learning_rate = learning_rate,
training_epochs=100,
batch_size = batch_size,
display_step = 10,
optimizer_method = optimizer,
activation_function = activation_function
)
obj_test = {'optimizer':optimizer_label,
'learning_rate':learning_rate,
'activation_function':activation_label,
'batch_size':batch_size,
'training_accuracy':training_accuracy,
'validation_accuracy':training_accuracy,
'test_accuracy':test_accuracy,
'elapsed_time': elapsed_time
}
final_results.append(obj_test)
final_results
with open('json.json','r') as input_fp:
results = json.load(input_fp)
import matplotlib.pyplot as plt
sigmoid = [x for x in results if x['activation_function']=='sigmoid']
relu =[x for x in results if x['activation_function'] !='sigmoid']
plt.figure(figsize=(15,5))
plt.plot(range(len(sigmoid)),[x['test_accuracy'] for x in sigmoid])
plt.plot(range(len(sigmoid)),[x['test_accuracy'] for x in relu])
plt.legend(['sigmoid','relu'])
plt.ylabel('test accuracy')
plt.xlabel('index run')
plt.show()
import matplotlib.pyplot as plt
a = [x for x in results if x['batch_size']== 128]
b =[x for x in results if x['batch_size'] !=128]
plt.figure(figsize=(15,5))
plt.plot(range(len(a)),[x['test_accuracy'] for x in a])
plt.plot(range(len(a)),[x['test_accuracy'] for x in b])
plt.legend(['128','50'])
plt.ylabel('test accuracy')
plt.xlabel('index run')
plt.show()
import matplotlib.pyplot as plt
a = [x for x in results if x['learning_rate']== 0.0001]
b =[x for x in results if x['learning_rate'] !=0.0001]
plt.figure(figsize=(15,5))
plt.plot(range(len(a)),[x['test_accuracy'] for x in a])
plt.plot(range(len(a)),[x['test_accuracy'] for x in b])
plt.legend(['learning_rate = 0.0001','learning_rate = 0.001'])
plt.ylabel('test accuracy')
plt.xlabel('index run')
plt.show()
import matplotlib.pyplot as plt
a = [x for x in results if x['optimizer']== 'adam']
b =[x for x in results if x['optimizer'] != 'adam']
plt.figure(figsize=(15,5))
plt.plot(range(len(a)),[x['test_accuracy'] for x in a])
plt.plot(range(len(a)),[x['test_accuracy'] for x in b])
plt.legend(['adam','sgd'])
plt.ylabel('test accuracy')
plt.xlabel('index run')
plt.show()
import matplotlib.pyplot as plt
a = [x for x in results if x['optimizer']== 'adam']
b =[x for x in results if x['optimizer'] != 'adam']
plt.figure(figsize=(15,5))
plt.plot(range(len(a)),[x['elapsed_time'] for x in a])
plt.plot(range(len(a)),[x['elapsed_time'] for x in b])
plt.legend(['adam','sgd'])
plt.ylabel('elapsed_time')
plt.xlabel('index run')
plt.show()
import matplotlib.pyplot as plt
a = [x for x in results if x['batch_size']== 128]
b =[x for x in results if x['batch_size'] !=128]
plt.figure(figsize=(15,5))
plt.plot(range(len(a)),[x['elapsed_time'] for x in a])
plt.plot(range(len(a)),[x['elapsed_time'] for x in b])
plt.legend(['128','50'])
plt.ylabel('elapsed_time')
plt.xlabel('index run')
plt.show()
# https://adeshpande3.github.io/adeshpande3.github.io/A-Beginner's-Guide-To-Understanding-Convolutional-Neural-Networks/
# https://adeshpande3.github.io/A-Beginner%27s-Guide-To-Understanding-Convolutional-Neural-Networks-Part-2/
def LeNet5_Model(data,keep_prob,activation_function=tf.nn.sigmoid):
# layer 1 param
conv1_weights = weight_variable([5,5,1,6])
conv1_bias = bias_variable([6])
# layer 2 param
conv2_weights = weight_variable([5,5,6,16])
conv2_bias = bias_variable([16])
# layer 3 param
layer3_weights = weight_variable([400, 120])
layer3_bias = bias_variable([120])
# layer 4 param
layer4_weights = weight_variable([120, 84])
layer4_bias = bias_variable([84])
# layer 5 param
layer5_weights = weight_variable([84, 10])
layer5_bias = bias_variable([10])
with tf.name_scope('Model'):
with tf.name_scope('Layer1'):
conv1 = tf.nn.conv2d(input=data,filter=conv1_weights,strides=[1,1,1,1],padding='SAME')
print(conv1.shape)
sigmoid1 = activation_function(conv1 + conv1_bias)
pool1 = tf.nn.max_pool(sigmoid1,ksize=[1, 2, 2, 1],strides=[1, 2, 2, 1],padding='VALID')
print(pool1.shape)
with tf.name_scope('Layer2'):
conv2 = tf.nn.conv2d(input=pool1,filter=conv2_weights,strides=[1,1,1,1],padding='VALID')
print(conv2.shape)
sigmoid2 = activation_function(conv2 + conv2_bias)
pool2 = tf.nn.max_pool(sigmoid2,ksize=[1, 2, 2, 1],strides=[1, 2, 2, 1],padding='VALID')
print(pool2.shape)
with tf.name_scope('Flatten'):
flat_inputs = tf.contrib.layers.flatten(pool2)
print(flat_inputs.shape)
with tf.name_scope('Layer3'):
out3 = activation_function(tf.matmul(flat_inputs, layer3_weights) + layer3_bias)
with tf.name_scope('Layer4'):
out4 = activation_function(tf.matmul(out3, layer4_weights) + layer4_bias)
with tf.name_scope('Layer5'):
out_drop = tf.nn.dropout(out4, keep_prob)
pred = tf.nn.softmax(tf.matmul(out_drop, layer5_weights) + layer5_bias) # Softmax
return pred
import numpy as np
# Initializing the variables
def train(learning_rate, training_epochs, batch_size, display_step, optimizer_method=tf.train.GradientDescentOptimizer,activation_function=tf.nn.sigmoid):
tf.reset_default_graph()
# Initializing the session
logs_path = 'log_files/' # useful for tensorboard
# tf Graph Input: mnist data image of shape 28*28=784
x = tf.placeholder(tf.float32, [None,28, 28,1], name='InputData')
# 0-9 digits recognition, 10 classes
y = tf.placeholder(tf.float32, [None, 10], name='LabelData')
keep_prob = tf.placeholder(tf.float32)
# Construct model and encapsulating all ops into scopes, making Tensorboard's Graph visualization more convenient
with tf.name_scope('Model'):
# Model
pred = LeNet5_Model(x,keep_prob,activation_function=activation_function)
with tf.name_scope('Loss'):
# Minimize error using cross entropy
# Minimize error using cross entropy
if activation_function == tf.nn.sigmoid:
cost = tf.reduce_mean(-tf.reduce_sum(y*tf.log(pred), reduction_indices=1))
else:
cost = tf.reduce_mean(-tf.reduce_sum(y*tf.log(tf.clip_by_value(pred,-1.0,1.0)), reduction_indices=1))
#cost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits=pred, labels=y))
with tf.name_scope('SGD'):
# Gradient Descent
optimizer = optimizer_method(learning_rate).minimize(cost)
with tf.name_scope('Accuracy'):
# Accuracy
acc = evaluate(pred, y)
# Initializing the variables
init = tf.global_variables_initializer()
# Create a summary to monitor cost tensor
tf.summary.scalar("Loss", cost)
# Create a summary to monitor accuracy tensor
tf.summary.scalar("Accuracy", acc)
# Merge all summaries into a single op
merged_summary_op = tf.summary.merge_all()
saver = tf.train.Saver()
print ("Start Training!")
t0 = time()
X_train,Y_train = mnist.train.images.reshape((-1,28,28,1)), mnist.train.labels
X_val,Y_val = mnist.validation.images.reshape((-1,28,28,1)), mnist.validation.labels
# Launch the graph for training
with tf.Session() as sess:
sess.run(init)
# op to write logs to Tensorboard
summary_writer = tf.summary.FileWriter(logs_path, graph=tf.get_default_graph())
# Training cycle
for epoch in range(training_epochs):
avg_cost = 0.
total_batch = int(mnist.train.num_examples/batch_size)
# Loop over all batches
for i in range(total_batch):
# train_next_batch shuffle the images by default
batch_xs, batch_ys = mnist.train.next_batch(batch_size)
batch_xs = batch_xs.reshape((-1,28,28,1))
# Run optimization op (backprop), cost op (to get loss value)
# and summary nodes
_, c, summary = sess.run([optimizer, cost, merged_summary_op],
feed_dict={x: batch_xs,
y: batch_ys,keep_prob:0.75})
# Write logs at every iteration
summary_writer.add_summary(summary, epoch * total_batch + i)
# Compute average loss
avg_cost += c / total_batch
# Display logs per epoch step
if (epoch+1) % display_step == 0:
print("Epoch: ", '%02d' % (epoch+1), "=====> Loss=", "{:.9f}".format(avg_cost))
acc_train = acc.eval({x: X_train, y: Y_train,keep_prob:1.0})
print("Epoch: ", '%02d' % (epoch+1), "=====> Accuracy Train=", "{:.9f}".format(acc_train))
acc_val = acc.eval({x: X_val, y: Y_val,keep_prob:1.0})
print("Epoch: ", '%02d' % (epoch+1), "=====> Accuracy Validation=", "{:.9f}".format(acc_val))
print ("Training Finished!")
t1 = time()
# Save the variables to disk.
save_path = saver.save(sess, "model.ckpt")
print("Model saved in file: %s" % save_path)
#Your implementation for testing accuracy after training goes here
X_test,Y_test = mnist.test.images.reshape((-1,28,28,1)),mnist.test.labels
acc_test = acc.eval({x: X_test, y: Y_test,keep_prob:1.0})
print("Accuracy Test=", "{:.9f}".format(acc_test))
return acc_train,acc_val,acc_test,t1-t0
train (0.0001,50,128,10,optimizer_method=tf.train.AdamOptimizer,activation_function=tf.nn.relu)
train (0.0001,50,50,10,optimizer_method=tf.train.AdamOptimizer,activation_function=tf.nn.relu)
| 0.832509 | 0.985608 |
This notebook is part of the `nbsphinx` documentation: https://nbsphinx.readthedocs.io/.
# Code Cells
## Code, Output, Streams
An empty code cell:
Two empty lines:
```
```
Leading/trailing empty lines:
```
# 2 empty lines before, 1 after
```
A simple output:
```
6 * 7
```
The standard output stream:
```
print('Hello, world!')
```
Normal output + standard output
```
print('Hello, world!')
6 * 7
```
The standard error stream is highlighted and displayed just below the code cell.
The standard output stream comes afterwards (with no special highlighting).
Finally, the "normal" output is displayed.
```
import sys
print("I'll appear on the standard error stream", file=sys.stderr)
print("I'll appear on the standard output stream")
"I'm the 'normal' output"
```
<div class="alert alert-info">
Note
Using the IPython kernel, the order is actually mixed up,
see https://github.com/ipython/ipykernel/issues/280.
</div>
## Cell Magics
IPython can handle code in other languages by means of [cell magics](https://ipython.readthedocs.io/en/stable/interactive/magics.html#cell-magics):
```
%%bash
for i in 1 2 3
do
echo $i
done
```
## Special Display Formats
See [IPython example notebook](https://nbviewer.jupyter.org/github/ipython/ipython/blob/master/examples/IPython Kernel/Rich Output.ipynb).
### Local Image Files
```
from IPython.display import Image
i = Image(filename='images/notebook_icon.png')
i
display(i)
```
See also [SVG support for LaTeX](markdown-cells.ipynb#SVG-support-for-LaTeX).
```
from IPython.display import SVG
SVG(filename='images/python_logo.svg')
```
### Image URLs
```
Image(url='https://www.python.org/static/img/python-logo-large.png')
Image(url='https://www.python.org/static/img/python-logo-large.png', embed=True)
Image(url='https://jupyter.org/assets/nav_logo.svg')
```
### Math
```
from IPython.display import Math
eq = Math(r'\int\limits_{-\infty}^\infty f(x) \delta(x - x_0) dx = f(x_0)')
eq
display(eq)
from IPython.display import Latex
Latex(r'This is a \LaTeX{} equation: $a^2 + b^2 = c^2$')
%%latex
\begin{equation}
\int\limits_{-\infty}^\infty f(x) \delta(x - x_0) dx = f(x_0)
\end{equation}
```
### Plots
The output formats for Matplotlib plots can be customized.
You'll need separate settings for the Jupyter Notebook application and for `nbsphinx`.
If you want to use SVG images for Matplotlib plots,
add this line to your IPython configuration file:
```python
c.InlineBackend.figure_formats = {'svg'}
```
If you want SVG images, but also want nice plots when exporting to LaTeX/PDF, you can select:
```python
c.InlineBackend.figure_formats = {'svg', 'pdf'}
```
If you want to use the default PNG plots or HiDPI plots using `'png2x'` (a.k.a. `'retina'`),
make sure to set this:
```python
c.InlineBackend.rc = {'figure.dpi': 96}
```
This is needed because the default `'figure.dpi'` value of 72
is only valid for the [Qt Console](https://qtconsole.readthedocs.io/).
If you are planning to store your SVG plots as part of your notebooks,
you should also have a look at the `'svg.hashsalt'` setting.
For more details on these and other settings, have a look at
[Default Values for Matplotlib's "inline" Backend](https://nbviewer.jupyter.org/github/mgeier/python-audio/blob/master/plotting/matplotlib-inline-defaults.ipynb).
The configuration file `ipython_kernel_config.py` can be either
in the directory where your notebook is located
(see the [ipython_kernel_config.py](ipython_kernel_config.py) in this directory),
or in your profile directory
(typically `~/.ipython/profile_default/ipython_kernel_config.py`).
To find out your IPython profile directory, use this command:
python3 -m IPython profile locate
A local `ipython_kernel_config.py` in the notebook directory
also works on https://mybinder.org/.
Alternatively, you can create a file with those settings in a file named
`.ipython/profile_default/ipython_kernel_config.py` in your repository.
To get SVG and PDF plots for `nbsphinx`,
use something like this in your `conf.py` file:
```python
nbsphinx_execute_arguments = [
"--InlineBackend.figure_formats={'svg', 'pdf'}",
"--InlineBackend.rc=figure.dpi=96",
]
```
In the following example, `nbsphinx` should use an SVG image in the HTML output
and a PDF image for LaTeX/PDF output.
```
import matplotlib.pyplot as plt
fig, ax = plt.subplots(figsize=[6, 3])
ax.plot([4, 9, 7, 20, 6, 33, 13, 23, 16, 62, 8]);
```
Alternatively, the figure format(s) can also be chosen directly in the notebook
(which overrides the setting in `nbsphinx_execute_arguments` and in the IPython configuration):
```
%config InlineBackend.figure_formats = ['png']
fig
```
If you want to use PNG images, but with HiDPI resolution,
use the special `'png2x'` (a.k.a. `'retina'`) format
(which also looks nice in the LaTeX output):
```
%config InlineBackend.figure_formats = ['png2x']
fig
```
### Pandas Dataframes
[Pandas dataframes](https://pandas.pydata.org/pandas-docs/stable/user_guide/dsintro.html#dataframe)
should be displayed as nicely formatted HTML tables (if you are using HTML output).
```
import numpy as np
import pandas as pd
df = pd.DataFrame(np.random.randint(0, 100, size=[5, 4]),
columns=['a', 'b', 'c', 'd'])
df
```
For LaTeX output, however, the plain text output is used by default.
To get nice LaTeX tables, a few settings have to be changed:
```
pd.set_option('display.latex.repr', True)
```
This is not enabled by default because of
[Pandas issue #12182](https://github.com/pandas-dev/pandas/issues/12182).
The generated LaTeX tables utilize the `booktabs` package, so you have to make sure that package is [loaded in the preamble](https://www.sphinx-doc.org/en/master/latex.html) with:
\usepackage{booktabs}
In order to allow page breaks within tables, you should use:
```
pd.set_option('display.latex.longtable', True)
```
The `longtable` package is already used by Sphinx,
so you don't have to manually load it in the preamble.
Finally, if you want to use LaTeX math expressions in your dataframe, you'll have to disable escaping:
```
pd.set_option('display.latex.escape', False)
```
The above settings should have no influence on the HTML output, but the LaTeX output should now look nicer:
```
df = pd.DataFrame(np.random.randint(0, 100, size=[10, 4]),
columns=[r'$\alpha$', r'$\beta$', r'$\gamma$', r'$\delta$'])
df
```
### YouTube Videos
```
from IPython.display import YouTubeVideo
YouTubeVideo('WAikxUGbomY')
```
### Interactive Widgets (HTML only)
The basic widget infrastructure is provided by
the [ipywidgets](https://ipywidgets.readthedocs.io/) module.
More advanced widgets are available in separate packages,
see for example https://jupyter.org/widgets.
The JavaScript code which is needed to display Jupyter widgets
is loaded automatically (using RequireJS).
If you want to use non-default URLs or local files,
you can use the
[nbsphinx_widgets_path](usage.ipynb#nbsphinx_widgets_path) and
[nbsphinx_requirejs_path](usage.ipynb#nbsphinx_requirejs_path)
settings.
```
import ipywidgets as w
slider = w.IntSlider()
slider.value = 42
slider
```
A widget typically consists of a so-called "model" and a "view" into that model.
If you display a widget multiple times,
all instances act as a "view" into the same "model".
That means that their state is synchronized.
You can move either one of these sliders to try this out:
```
slider
```
You can also link different widgets.
Widgets can be linked via the kernel
(which of course only works while a kernel is running)
or directly in the client
(which even works in the rendered HTML pages).
Widgets can be linked uni- or bi-directionally.
Examples for all 4 combinations are shown here:
```
link = w.IntSlider(description='link')
w.link((slider, 'value'), (link, 'value'))
jslink = w.IntSlider(description='jslink')
w.jslink((slider, 'value'), (jslink, 'value'))
dlink = w.IntSlider(description='dlink')
w.dlink((slider, 'value'), (dlink, 'value'))
jsdlink = w.IntSlider(description='jsdlink')
w.jsdlink((slider, 'value'), (jsdlink, 'value'))
w.VBox([link, jslink, dlink, jsdlink])
```
<div class="alert alert-info">
Other Languages
The examples shown here are using Python,
but the widget technology can also be used with
different Jupyter kernels
(i.e. with different programming languages).
</div>
#### Troubleshooting
To obtain more information if widgets are not displayed as expected, you will need to look at the error message in the web browser console.
> To figure out how to open the web browser console, you may look at the web browser documentation:
> Chrome: https://developers.google.com/web/tools/chrome-devtools/open#console
> Firefox: https://developer.mozilla.org/en-US/docs/Tools/Web_Console#opening_the_web_console
The error is most probably linked to the JavaScript files not being loaded or loaded in the wrong order within the HTML file. To analyze the error, you can inspect the HTML file within the web browser (e.g.: right-click on the page and select *View Page Source*) and look at the `<head>` section of the page. That section should contain
some JavaScript libraries. Those relevant for widgets are:
```html
<!-- require.js is a mandatory dependency for jupyter-widgets -->
<script crossorigin="anonymous" integrity="sha256-Ae2Vz/4ePdIu6ZyI/5ZGsYnb+m0JlOmKPjt6XZ9JJkA=" src="https://cdnjs.cloudflare.com/ajax/libs/require.js/2.3.4/require.min.js"></script>
<!-- jupyter-widgets JavaScript -->
<script type="text/javascript" src="https://unpkg.com/@jupyter-widgets/html-manager@^0.18.0/dist/embed-amd.js"></script>
<!-- JavaScript containing custom Jupyter widgets -->
<script src="../_static/embed-widgets.js"></script>
```
The two first elements are mandatory. The third one is required only if you designed your own widgets but did not publish them on npm.js.
If those libraries appear in a different order, the widgets won't be displayed.
Here is a list of possible solutions:
- If the widgets are **not displayed**, see [#519](https://github.com/spatialaudio/nbsphinx/issues/519).
- If the widgets are **displayed multiple times**, see [#378](https://github.com/spatialaudio/nbsphinx/issues/378).
### Arbitrary JavaScript Output (HTML only)
```
%%javascript
var text = document.createTextNode("Hello, I was generated with JavaScript!");
// Content appended to "element" will be visible in the output area:
element.appendChild(text);
```
### Unsupported Output Types
If a code cell produces data with an unsupported MIME type, the Jupyter Notebook doesn't generate any output.
`nbsphinx`, however, shows a warning message.
```
display({
'text/x-python': 'print("Hello, world!")',
'text/x-haskell': 'main = putStrLn "Hello, world!"',
}, raw=True)
```
## ANSI Colors
The standard output and standard error streams may contain [ANSI escape sequences](https://en.wikipedia.org/wiki/ANSI_escape_code) to change the text and background colors.
```
print('BEWARE: \x1b[1;33;41mugly colors\x1b[m!', file=sys.stderr)
print('AB\x1b[43mCD\x1b[35mEF\x1b[1mGH\x1b[4mIJ\x1b[7m'
'KL\x1b[49mMN\x1b[39mOP\x1b[22mQR\x1b[24mST\x1b[27mUV')
```
The following code showing the 8 basic ANSI colors is based on https://tldp.org/HOWTO/Bash-Prompt-HOWTO/x329.html.
Each of the 8 colors has an "intense" variation, which is used for bold text.
```
text = ' XYZ '
formatstring = '\x1b[{}m' + text + '\x1b[m'
print(' ' * 6 + ' ' * len(text) +
''.join('{:^{}}'.format(bg, len(text)) for bg in range(40, 48)))
for fg in range(30, 38):
for bold in False, True:
fg_code = ('1;' if bold else '') + str(fg)
print(' {:>4} '.format(fg_code) + formatstring.format(fg_code) +
''.join(formatstring.format(fg_code + ';' + str(bg))
for bg in range(40, 48)))
```
ANSI also supports a set of 256 indexed colors.
The following code showing all of them is based on [http://bitmote.com/index.php?post/2012/11/19/Using-ANSI-Color-Codes-to-Colorize-Your-Bash-Prompt-on-Linux](https://web.archive.org/web/20190109005413/http://bitmote.com/index.php?post/2012/11/19/Using-ANSI-Color-Codes-to-Colorize-Your-Bash-Prompt-on-Linux).
```
formatstring = '\x1b[38;5;{0};48;5;{0}mX\x1b[1mX\x1b[m'
print(' + ' + ''.join('{:2}'.format(i) for i in range(36)))
print(' 0 ' + ''.join(formatstring.format(i) for i in range(16)))
for i in range(7):
i = i * 36 + 16
print('{:3} '.format(i) + ''.join(formatstring.format(i + j)
for j in range(36) if i + j < 256))
```
You can even use 24-bit RGB colors:
```
start = 255, 0, 0
end = 0, 0, 255
length = 79
out = []
for i in range(length):
rgb = [start[c] + int(i * (end[c] - start[c]) / length) for c in range(3)]
out.append('\x1b['
'38;2;{rgb[2]};{rgb[1]};{rgb[0]};'
'48;2;{rgb[0]};{rgb[1]};{rgb[2]}mX\x1b[m'.format(rgb=rgb))
print(''.join(out))
```
|
github_jupyter
|
```
Leading/trailing empty lines:
A simple output:
The standard output stream:
Normal output + standard output
The standard error stream is highlighted and displayed just below the code cell.
The standard output stream comes afterwards (with no special highlighting).
Finally, the "normal" output is displayed.
<div class="alert alert-info">
Note
Using the IPython kernel, the order is actually mixed up,
see https://github.com/ipython/ipykernel/issues/280.
</div>
## Cell Magics
IPython can handle code in other languages by means of [cell magics](https://ipython.readthedocs.io/en/stable/interactive/magics.html#cell-magics):
## Special Display Formats
See [IPython example notebook](https://nbviewer.jupyter.org/github/ipython/ipython/blob/master/examples/IPython Kernel/Rich Output.ipynb).
### Local Image Files
See also [SVG support for LaTeX](markdown-cells.ipynb#SVG-support-for-LaTeX).
### Image URLs
### Math
### Plots
The output formats for Matplotlib plots can be customized.
You'll need separate settings for the Jupyter Notebook application and for `nbsphinx`.
If you want to use SVG images for Matplotlib plots,
add this line to your IPython configuration file:
If you want SVG images, but also want nice plots when exporting to LaTeX/PDF, you can select:
If you want to use the default PNG plots or HiDPI plots using `'png2x'` (a.k.a. `'retina'`),
make sure to set this:
This is needed because the default `'figure.dpi'` value of 72
is only valid for the [Qt Console](https://qtconsole.readthedocs.io/).
If you are planning to store your SVG plots as part of your notebooks,
you should also have a look at the `'svg.hashsalt'` setting.
For more details on these and other settings, have a look at
[Default Values for Matplotlib's "inline" Backend](https://nbviewer.jupyter.org/github/mgeier/python-audio/blob/master/plotting/matplotlib-inline-defaults.ipynb).
The configuration file `ipython_kernel_config.py` can be either
in the directory where your notebook is located
(see the [ipython_kernel_config.py](ipython_kernel_config.py) in this directory),
or in your profile directory
(typically `~/.ipython/profile_default/ipython_kernel_config.py`).
To find out your IPython profile directory, use this command:
python3 -m IPython profile locate
A local `ipython_kernel_config.py` in the notebook directory
also works on https://mybinder.org/.
Alternatively, you can create a file with those settings in a file named
`.ipython/profile_default/ipython_kernel_config.py` in your repository.
To get SVG and PDF plots for `nbsphinx`,
use something like this in your `conf.py` file:
In the following example, `nbsphinx` should use an SVG image in the HTML output
and a PDF image for LaTeX/PDF output.
Alternatively, the figure format(s) can also be chosen directly in the notebook
(which overrides the setting in `nbsphinx_execute_arguments` and in the IPython configuration):
If you want to use PNG images, but with HiDPI resolution,
use the special `'png2x'` (a.k.a. `'retina'`) format
(which also looks nice in the LaTeX output):
### Pandas Dataframes
[Pandas dataframes](https://pandas.pydata.org/pandas-docs/stable/user_guide/dsintro.html#dataframe)
should be displayed as nicely formatted HTML tables (if you are using HTML output).
For LaTeX output, however, the plain text output is used by default.
To get nice LaTeX tables, a few settings have to be changed:
This is not enabled by default because of
[Pandas issue #12182](https://github.com/pandas-dev/pandas/issues/12182).
The generated LaTeX tables utilize the `booktabs` package, so you have to make sure that package is [loaded in the preamble](https://www.sphinx-doc.org/en/master/latex.html) with:
\usepackage{booktabs}
In order to allow page breaks within tables, you should use:
The `longtable` package is already used by Sphinx,
so you don't have to manually load it in the preamble.
Finally, if you want to use LaTeX math expressions in your dataframe, you'll have to disable escaping:
The above settings should have no influence on the HTML output, but the LaTeX output should now look nicer:
### YouTube Videos
### Interactive Widgets (HTML only)
The basic widget infrastructure is provided by
the [ipywidgets](https://ipywidgets.readthedocs.io/) module.
More advanced widgets are available in separate packages,
see for example https://jupyter.org/widgets.
The JavaScript code which is needed to display Jupyter widgets
is loaded automatically (using RequireJS).
If you want to use non-default URLs or local files,
you can use the
[nbsphinx_widgets_path](usage.ipynb#nbsphinx_widgets_path) and
[nbsphinx_requirejs_path](usage.ipynb#nbsphinx_requirejs_path)
settings.
A widget typically consists of a so-called "model" and a "view" into that model.
If you display a widget multiple times,
all instances act as a "view" into the same "model".
That means that their state is synchronized.
You can move either one of these sliders to try this out:
You can also link different widgets.
Widgets can be linked via the kernel
(which of course only works while a kernel is running)
or directly in the client
(which even works in the rendered HTML pages).
Widgets can be linked uni- or bi-directionally.
Examples for all 4 combinations are shown here:
<div class="alert alert-info">
Other Languages
The examples shown here are using Python,
but the widget technology can also be used with
different Jupyter kernels
(i.e. with different programming languages).
</div>
#### Troubleshooting
To obtain more information if widgets are not displayed as expected, you will need to look at the error message in the web browser console.
> To figure out how to open the web browser console, you may look at the web browser documentation:
> Chrome: https://developers.google.com/web/tools/chrome-devtools/open#console
> Firefox: https://developer.mozilla.org/en-US/docs/Tools/Web_Console#opening_the_web_console
The error is most probably linked to the JavaScript files not being loaded or loaded in the wrong order within the HTML file. To analyze the error, you can inspect the HTML file within the web browser (e.g.: right-click on the page and select *View Page Source*) and look at the `<head>` section of the page. That section should contain
some JavaScript libraries. Those relevant for widgets are:
The two first elements are mandatory. The third one is required only if you designed your own widgets but did not publish them on npm.js.
If those libraries appear in a different order, the widgets won't be displayed.
Here is a list of possible solutions:
- If the widgets are **not displayed**, see [#519](https://github.com/spatialaudio/nbsphinx/issues/519).
- If the widgets are **displayed multiple times**, see [#378](https://github.com/spatialaudio/nbsphinx/issues/378).
### Arbitrary JavaScript Output (HTML only)
### Unsupported Output Types
If a code cell produces data with an unsupported MIME type, the Jupyter Notebook doesn't generate any output.
`nbsphinx`, however, shows a warning message.
## ANSI Colors
The standard output and standard error streams may contain [ANSI escape sequences](https://en.wikipedia.org/wiki/ANSI_escape_code) to change the text and background colors.
The following code showing the 8 basic ANSI colors is based on https://tldp.org/HOWTO/Bash-Prompt-HOWTO/x329.html.
Each of the 8 colors has an "intense" variation, which is used for bold text.
ANSI also supports a set of 256 indexed colors.
The following code showing all of them is based on [http://bitmote.com/index.php?post/2012/11/19/Using-ANSI-Color-Codes-to-Colorize-Your-Bash-Prompt-on-Linux](https://web.archive.org/web/20190109005413/http://bitmote.com/index.php?post/2012/11/19/Using-ANSI-Color-Codes-to-Colorize-Your-Bash-Prompt-on-Linux).
You can even use 24-bit RGB colors:
| 0.779574 | 0.949389 |
**This notebook is an exercise in the [Introduction to Machine Learning](https://www.kaggle.com/learn/intro-to-machine-learning) course. You can reference the tutorial at [this link](https://www.kaggle.com/alexisbcook/machine-learning-competitions).**
---
# Introduction
In this exercise, you will create and submit predictions for a Kaggle competition. You can then improve your model (e.g. by adding features) to apply what you've learned and move up the leaderboard.
Begin by running the code cell below to set up code checking and the filepaths for the dataset.
```
# Set up code checking
from learntools.core import binder
binder.bind(globals())
from learntools.machine_learning.ex7 import *
# Set up filepaths
import os
if not os.path.exists("../input/train.csv"):
os.symlink("../input/home-data-for-ml-course/train.csv", "../input/train.csv")
os.symlink("../input/home-data-for-ml-course/test.csv", "../input/test.csv")
```
Here's some of the code you've written so far. Start by running it again.
```
# Import helpful libraries
import pandas as pd
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_absolute_error
from sklearn.model_selection import train_test_split
# Load the data, and separate the target
iowa_file_path = '../input/train.csv'
home_data = pd.read_csv(iowa_file_path)
y = home_data.SalePrice
# Create X (After completing the exercise, you can return to modify this line!)
features = ['LotArea', 'YearBuilt', '1stFlrSF', '2ndFlrSF', 'FullBath', 'BedroomAbvGr', 'TotRmsAbvGrd']
# Select columns corresponding to features, and preview the data
X = home_data[features]
X.head()
# Split into validation and training data
train_X, val_X, train_y, val_y = train_test_split(X, y, random_state=1)
# Define a random forest model
rf_model = RandomForestRegressor(random_state=1)
rf_model.fit(train_X, train_y)
rf_val_predictions = rf_model.predict(val_X)
rf_val_mae = mean_absolute_error(rf_val_predictions, val_y)
print("Validation MAE for Random Forest Model: {:,.0f}".format(rf_val_mae))
```
# Train a model for the competition
The code cell above trains a Random Forest model on **`train_X`** and **`train_y`**.
Use the code cell below to build a Random Forest model and train it on all of **`X`** and **`y`**.
```
# To improve accuracy, create a new Random Forest model which you will train on all training data
rf_model_on_full_data = RandomForestRegressor()
# fit rf_model_on_full_data on all data from the training data
rf_model_on_full_data.fit(X, y)
```
Now, read the file of "test" data, and apply your model to make predictions.
```
# path to file you will use for predictions
test_data_path = '../input/test.csv'
# read test data file using pandas
test_data = pd.read_csv(test_data_path)
# create test_X which comes from test_data but includes only the columns you used for prediction.
# The list of columns is stored in a variable called features
test_X = test_data[features]
# make predictions which we will submit.
test_preds = rf_model_on_full_data.predict(test_X)
```
Before submitting, run a check to make sure your `test_preds` have the right format.
```
# Check your answer (To get credit for completing the exercise, you must get a "Correct" result!)
step_1.check()
# step_1.solution()
```
# Generate a submission
Run the code cell below to generate a CSV file with your predictions that you can use to submit to the competition.
```
# Run the code to save predictions in the format used for competition scoring
output = pd.DataFrame({'Id': test_data.Id,
'SalePrice': test_preds})
output.to_csv('submission.csv', index=False)
```
# Submit to the competition
To test your results, you'll need to join the competition (if you haven't already). So open a new window by clicking on **[this link](https://www.kaggle.com/c/home-data-for-ml-course)**. Then click on the **Join Competition** button.

Next, follow the instructions below:
1. Begin by clicking on the **Save Version** button in the top right corner of the window. This will generate a pop-up window.
2. Ensure that the **Save and Run All** option is selected, and then click on the **Save** button.
3. This generates a window in the bottom left corner of the notebook. After it has finished running, click on the number to the right of the **Save Version** button. This pulls up a list of versions on the right of the screen. Click on the ellipsis **(...)** to the right of the most recent version, and select **Open in Viewer**. This brings you into view mode of the same page. You will need to scroll down to get back to these instructions.
4. Click on the **Output** tab on the right of the screen. Then, click on the file you would like to submit, and click on the blue **Submit** button to submit your results to the leaderboard.
You have now successfully submitted to the competition!
If you want to keep working to improve your performance, select the **Edit** button in the top right of the screen. Then you can change your code and repeat the process. There's a lot of room to improve, and you will climb up the leaderboard as you work.
# Continue Your Progress
There are many ways to improve your model, and **experimenting is a great way to learn at this point.**
The best way to improve your model is to add features. To add more features to the data, revisit the first code cell, and change this line of code to include more column names:
```python
features = ['LotArea', 'YearBuilt', '1stFlrSF', '2ndFlrSF', 'FullBath', 'BedroomAbvGr', 'TotRmsAbvGrd']
```
Some features will cause errors because of issues like missing values or non-numeric data types. Here is a complete list of potential columns that you might like to use, and that won't throw errors:
- 'MSSubClass'
- 'LotArea'
- 'OverallQual'
- 'OverallCond'
- 'YearBuilt'
- 'YearRemodAdd'
- 'BsmtFinSF1'
- 'BsmtFinSF2'
- 'BsmtUnfSF'
- 'TotalBsmtSF'
- '1stFlrSF'
- '2ndFlrSF'
- 'LowQualFinSF'
- 'GrLivArea'
- 'BsmtFullBath'
- 'BsmtHalfBath'
- 'FullBath'
- 'HalfBath'
- 'BedroomAbvGr'
- 'KitchenAbvGr'
- 'TotRmsAbvGrd'
- 'Fireplaces'
- 'GarageCars'
- 'GarageArea'
- 'WoodDeckSF'
- 'OpenPorchSF'
- 'EnclosedPorch'
- '3SsnPorch'
- 'ScreenPorch'
- 'PoolArea'
- 'MiscVal'
- 'MoSold'
- 'YrSold'
Look at the list of columns and think about what might affect home prices. To learn more about each of these features, take a look at the data description on the **[competition page](https://www.kaggle.com/c/home-data-for-ml-course/data)**.
After updating the code cell above that defines the features, re-run all of the code cells to evaluate the model and generate a new submission file.
# What's next?
As mentioned above, some of the features will throw an error if you try to use them to train your model. The **[Intermediate Machine Learning](https://www.kaggle.com/learn/intermediate-machine-learning)** course will teach you how to handle these types of features. You will also learn to use **xgboost**, a technique giving even better accuracy than Random Forest.
The **[Pandas](https://kaggle.com/Learn/Pandas)** course will give you the data manipulation skills to quickly go from conceptual idea to implementation in your data science projects.
You are also ready for the **[Deep Learning](https://kaggle.com/Learn/intro-to-Deep-Learning)** course, where you will build models with better-than-human level performance at computer vision tasks.
---
*Have questions or comments? Visit the [Learn Discussion forum](https://www.kaggle.com/learn-forum/161285) to chat with other Learners.*
|
github_jupyter
|
# Set up code checking
from learntools.core import binder
binder.bind(globals())
from learntools.machine_learning.ex7 import *
# Set up filepaths
import os
if not os.path.exists("../input/train.csv"):
os.symlink("../input/home-data-for-ml-course/train.csv", "../input/train.csv")
os.symlink("../input/home-data-for-ml-course/test.csv", "../input/test.csv")
# Import helpful libraries
import pandas as pd
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_absolute_error
from sklearn.model_selection import train_test_split
# Load the data, and separate the target
iowa_file_path = '../input/train.csv'
home_data = pd.read_csv(iowa_file_path)
y = home_data.SalePrice
# Create X (After completing the exercise, you can return to modify this line!)
features = ['LotArea', 'YearBuilt', '1stFlrSF', '2ndFlrSF', 'FullBath', 'BedroomAbvGr', 'TotRmsAbvGrd']
# Select columns corresponding to features, and preview the data
X = home_data[features]
X.head()
# Split into validation and training data
train_X, val_X, train_y, val_y = train_test_split(X, y, random_state=1)
# Define a random forest model
rf_model = RandomForestRegressor(random_state=1)
rf_model.fit(train_X, train_y)
rf_val_predictions = rf_model.predict(val_X)
rf_val_mae = mean_absolute_error(rf_val_predictions, val_y)
print("Validation MAE for Random Forest Model: {:,.0f}".format(rf_val_mae))
# To improve accuracy, create a new Random Forest model which you will train on all training data
rf_model_on_full_data = RandomForestRegressor()
# fit rf_model_on_full_data on all data from the training data
rf_model_on_full_data.fit(X, y)
# path to file you will use for predictions
test_data_path = '../input/test.csv'
# read test data file using pandas
test_data = pd.read_csv(test_data_path)
# create test_X which comes from test_data but includes only the columns you used for prediction.
# The list of columns is stored in a variable called features
test_X = test_data[features]
# make predictions which we will submit.
test_preds = rf_model_on_full_data.predict(test_X)
# Check your answer (To get credit for completing the exercise, you must get a "Correct" result!)
step_1.check()
# step_1.solution()
# Run the code to save predictions in the format used for competition scoring
output = pd.DataFrame({'Id': test_data.Id,
'SalePrice': test_preds})
output.to_csv('submission.csv', index=False)
features = ['LotArea', 'YearBuilt', '1stFlrSF', '2ndFlrSF', 'FullBath', 'BedroomAbvGr', 'TotRmsAbvGrd']
| 0.640299 | 0.955527 |
# Inverse Kinematics Optimization
The previous doc explained features and how they define objectives of a constrained optimization problem. Here we show how to use this to solve IK optimization problems.
At the bottom there is more general text explaining the basic concepts.
## Demo of features in Inverse Kinematics
Let's setup a standard configuration. (Lock the window with "Always on Top".)
```
import sys
sys.path.append('../../lib') #rai/lib')
import numpy as np
import libry as ry
C = ry.Config()
C.addFile('../../../rai-robotModels/pr2/pr2.g')
C.addFile('../../../rai-robotModels/objects/kitchen.g')
C.view()
```
For simplicity, let's add a frame that represents goals
```
goal = C.addFrame("goal")
goal.setShape(ry.ST.sphere, [.05])
goal.setColor([.5,1,1])
goal.setPosition([1,.5,1])
X0 = C.getFrameState() #store the initial configuration
```
We create an IK engine. The only objective is that the `positionDiff` (position difference in world coordinates) between `pr2L` (the yellow blob in the left hand) and `goal` is equal to zero:
```
IK = C.komo_IK(False)
IK.addObjective(type=ry.OT.eq, feature=ry.FS.positionDiff, frames=['pr2L', 'goal'])
```
We now call the optimizer (True means with random initialization/restart).
```
IK.optimize()
IK.getReport()
```
The best way to retrieve the result is to copy the optimized IK configuration back into your working configuration C, which is now also displayed
```
C.setFrameState( IK.getConfiguration(0) )
```
We can redo the optimization, but for a different configuration, namely a configuration where the goal is in another location.
For this we move goal in our working configuration C, then copy C back into the IK engine's configurations:
```
## (iterate executing this cell for different goal locations!)
# move goal
goal.setPosition([.8,.2,.5])
# copy C into the IK's internal configuration(s)
IK.setConfigurations(C)
# reoptimize
IK.optimize(0.) # 0: no adding of noise for a random restart
print(IK.getReport())
# grab result
C.setFrameState( IK.getConfiguration(1) )
```
Let's solve some other problems, always creating a novel IK engine:
The relative position of `goal` in `pr2R` coordinates equals [0,0,-.2] (which is 20cm straight in front of the yellow blob)
```
C.setFrameState(X0)
IK = C.komo_IK(False)
IK.addObjective(type=ry.OT.eq, feature=ry.FS.positionRel, frames=['goal','pr2R'], target=[0,0,-.2])
IK.optimize()
C.setFrameState( IK.getConfiguration(0) )
```
The distance between `pr2R` and `pr2L` is zero:
```
C.setFrameState(X0)
IK = C.komo_IK(False)
IK.addObjective(type=ry.OT.eq, feature=ry.FS.distance, frames=['pr2L','pr2R'])
IK.optimize()
C.setFrameState( IK.getConfiguration(0) )
```
The 3D difference between the z-vector of `pr2R` and the z-vector of `goal`:
```
C.setFrameState(X0)
IK = C.komo_IK(False)
IK.addObjective(type=ry.OT.eq, feature=ry.FS.vectorZDiff, frames=['pr2R', 'goal'])
IK.optimize()
C.setFrameState( IK.getConfiguration(0) )
```
The scalar product between the z-vector of `pr2R` and the z-vector of `goal` is zero:
```
C.setFrameState(X0)
IK = C.komo_IK(False)
IK.addObjective(type=ry.OT.eq, feature=ry.FS.scalarProductZZ, frames=['pr2R', 'goal'])
IK.optimize()
C.setFrameState( IK.getConfiguration(0) )
```
etc etc
## More explanations
All methods to compute paths or configurations solve constrained optimization problems. To use them, you need to learn to define constrained optimization problems. Some definitions:
* An objective defines either a sum-of-square cost term, or an equality constraint, or an inequality constraint in the optimization problem. An objective is defined by its type and its feature. The type can be `sos` (sum-of-squares), `eq`, or `ineq`, referring to the three cases.
* A feature is a (differentiable mapping) from the decision variable (the full path, or robot configuration) to a feature space. If the feature space is, e.g., 3-dimensional, this defines 3 sum-of-squares terms, or 3 inequality, or 3 equality constraints, one for each dimension. For instance, the feature can be the 3-dim robot hand position in the 15th time slice of a path optimization problem. If you put an equality constraint on this feature, then this adds 3 equality constraints to the overall path optimization problem.
* A feature is defined by the keyword for the feature map (e.g., `pos` for position), typically by a set of frame names that tell which objects we refer to (e.g., `pr2L` for the left hand of the pr2), optionally some modifiers (e.g., a scale or target, which linearly transform the feature map), and the set of configuration IDs or time slices the feature is to be computed from (e.g., `confs=[15]` if the feat is to be computed from the 15th time slice in a path optimization problem).
* In path optimization problems, we often want to add objectives for a whole time interval rather than for a single time slice or specific configuration. E.g., avoid collisions from start to end. When adding objectives to the optimization problem we can specify whole intervals, with `times=[1., 2.]`, so that the objective is added to each configuration in this time interval.
* Some features, especially velocity and acceleration, refer to a tuple of (consecutive) configurations. E.g., when you impose an acceleration feature, you need to specify `confs=[13, 14, 15]`. Or if you use `times=[1.,1.]`, the acceleration features is computed from the configuration that corresponds to time=1 and the two configurations *before*.
* All kinematic feature maps (that depend on only one configuration) can be modified to become a velocity or acceleration features. E.g., the position feature map can be modified to become a velocity or acceleration feature.
* The `sos`, `eq`, and `ineq` always refer to the feature map to be *zero*, e.g., constraining all features to be equal to zero with `eq`. This is natural for many features, esp. when they refer to differences (e.g. `posDiff` or `posRel`, which compute the relative position between two frames). However, when one aims to constrain the feature to a non-zero constant value, one can modify the objective with a `target` specification.
* Finally, all features can be rescaled with a `scale` specification. Rescaling changes the costs that arise from `sos` objectives. Rescaling also has significant impact on the convergence behavior for `eq` and `ineq` constraints. As a guide: scale constraints so that if they *would* be treated as squared penalties (squaredPenalty optim mode, to be made accessible) convergence to reasonable approximate solutions is efficient. Then the AugLag will also converge efficiently to precise constraints.
```
# Designing a cylinder grasp
D=0
C=0
import sys
sys.path.append('../../lib') #rai/lib')
import numpy as np
import libry as ry
C = ry.Config()
C.addFile('../../../rai-robotModels/pr2/pr2.g')
C.addFile('../../../rai-robotModels/objects/kitchen.g')
C.view()
C.setJointState([.7], ["l_gripper_l_finger_joint"])
C.setJointState( C.getJointState() )
goal = C.addFrame("goal")
goal.setShape(ry.ST.cylinder, [0,0,.2, .03])
goal.setColor([.5,1,1])
goal.setPosition([1,.5,1])
X0 = C.getFrameState()
C.setFrameState(X0)
goal.setPosition([1,.5,1.2])
IK = C.komo_IK(False)
IK.addObjective(type=ry.OT.eq, feature=ry.FS.positionDiff, frames=['pr2L', 'goal'], scaleTrans=[[1,0,0],[0,1,0]])
IK.addObjective(type=ry.OT.ineq, feature=ry.FS.positionDiff, frames=['pr2L', 'goal'], scaleTrans=[[0,0,1]], target=[0,0,.05])
IK.addObjective(type=ry.OT.ineq, feature=ry.FS.positionDiff, frames=['pr2L', 'goal'], scaleTrans=[[0,0,-1]], target=[0,0,-.05])
IK.addObjective(type=ry.OT.sos, feature=ry.FS.scalarProductZZ, frames=['pr2L', 'goal'], scale=[0.1])
IK.addObjective(type=ry.OT.eq, feature=ry.FS.scalarProductXZ, frames=['pr2L', 'goal'])
IK.optimize()
C.setFrameState( IK.getConfiguration(0) )
IK.getReport()
IK.view()
```
|
github_jupyter
|
import sys
sys.path.append('../../lib') #rai/lib')
import numpy as np
import libry as ry
C = ry.Config()
C.addFile('../../../rai-robotModels/pr2/pr2.g')
C.addFile('../../../rai-robotModels/objects/kitchen.g')
C.view()
goal = C.addFrame("goal")
goal.setShape(ry.ST.sphere, [.05])
goal.setColor([.5,1,1])
goal.setPosition([1,.5,1])
X0 = C.getFrameState() #store the initial configuration
IK = C.komo_IK(False)
IK.addObjective(type=ry.OT.eq, feature=ry.FS.positionDiff, frames=['pr2L', 'goal'])
IK.optimize()
IK.getReport()
C.setFrameState( IK.getConfiguration(0) )
## (iterate executing this cell for different goal locations!)
# move goal
goal.setPosition([.8,.2,.5])
# copy C into the IK's internal configuration(s)
IK.setConfigurations(C)
# reoptimize
IK.optimize(0.) # 0: no adding of noise for a random restart
print(IK.getReport())
# grab result
C.setFrameState( IK.getConfiguration(1) )
C.setFrameState(X0)
IK = C.komo_IK(False)
IK.addObjective(type=ry.OT.eq, feature=ry.FS.positionRel, frames=['goal','pr2R'], target=[0,0,-.2])
IK.optimize()
C.setFrameState( IK.getConfiguration(0) )
C.setFrameState(X0)
IK = C.komo_IK(False)
IK.addObjective(type=ry.OT.eq, feature=ry.FS.distance, frames=['pr2L','pr2R'])
IK.optimize()
C.setFrameState( IK.getConfiguration(0) )
C.setFrameState(X0)
IK = C.komo_IK(False)
IK.addObjective(type=ry.OT.eq, feature=ry.FS.vectorZDiff, frames=['pr2R', 'goal'])
IK.optimize()
C.setFrameState( IK.getConfiguration(0) )
C.setFrameState(X0)
IK = C.komo_IK(False)
IK.addObjective(type=ry.OT.eq, feature=ry.FS.scalarProductZZ, frames=['pr2R', 'goal'])
IK.optimize()
C.setFrameState( IK.getConfiguration(0) )
# Designing a cylinder grasp
D=0
C=0
import sys
sys.path.append('../../lib') #rai/lib')
import numpy as np
import libry as ry
C = ry.Config()
C.addFile('../../../rai-robotModels/pr2/pr2.g')
C.addFile('../../../rai-robotModels/objects/kitchen.g')
C.view()
C.setJointState([.7], ["l_gripper_l_finger_joint"])
C.setJointState( C.getJointState() )
goal = C.addFrame("goal")
goal.setShape(ry.ST.cylinder, [0,0,.2, .03])
goal.setColor([.5,1,1])
goal.setPosition([1,.5,1])
X0 = C.getFrameState()
C.setFrameState(X0)
goal.setPosition([1,.5,1.2])
IK = C.komo_IK(False)
IK.addObjective(type=ry.OT.eq, feature=ry.FS.positionDiff, frames=['pr2L', 'goal'], scaleTrans=[[1,0,0],[0,1,0]])
IK.addObjective(type=ry.OT.ineq, feature=ry.FS.positionDiff, frames=['pr2L', 'goal'], scaleTrans=[[0,0,1]], target=[0,0,.05])
IK.addObjective(type=ry.OT.ineq, feature=ry.FS.positionDiff, frames=['pr2L', 'goal'], scaleTrans=[[0,0,-1]], target=[0,0,-.05])
IK.addObjective(type=ry.OT.sos, feature=ry.FS.scalarProductZZ, frames=['pr2L', 'goal'], scale=[0.1])
IK.addObjective(type=ry.OT.eq, feature=ry.FS.scalarProductXZ, frames=['pr2L', 'goal'])
IK.optimize()
C.setFrameState( IK.getConfiguration(0) )
IK.getReport()
IK.view()
| 0.337749 | 0.95594 |
# ml lab6
```
import numpy as np
import matplotlib.pyplot as plt
import scipy.io
```
### 1. read data
```
data = scipy.io.loadmat('data/ex6data1.mat')
X = data['X']
X.shape
```
### 2. random init centroids
```
def rand_centroids(X, K):
rand_indices = np.arange(len(X))
np.random.shuffle(rand_indices)
centroids = X[rand_indices][:K]
return centroids
rand_centroids(X, 3)
```
### 3. closest centroids search
```
def find_closest_centroids(X, centroids):
distances = np.array([np.sqrt((X.T[0] - c[0])**2 + (X.T[1] - c[1])**2) for c in centroids])
return distances.argmin(axis=0)
```
### 4. centroids recompute
```
def compute_means(X, centroid_idx, K):
centroids = []
for k in range(K):
t = X[centroid_idx == k]
c = np.mean(t, axis=0) if t.size > 0 else np.zeros((X.shape[1],))
centroids.append(c)
return np.array(centroids)
```
### 5. k-means algorithm
```
def run_k_means(X, K, num_iter=10):
centroids = rand_centroids(X, K)
centroids_history = [centroids]
for i in range(num_iter):
centroid_idx = find_closest_centroids(X, centroids)
centroids = compute_means(X, centroid_idx, K)
centroids_history.append(centroids)
return centroids, centroid_idx, centroids_history
def k_means_distortion(X, centroids, idx):
K = centroids.shape[0]
distortion = 0
for i in range(K):
distortion += np.sum((X[idx == i] - centroids[i])**2)
distortion /= X.shape[0]
return distortion
def find_best_k_means(X, K, num_iter=100):
result = np.inf
r_centroids = None
r_idx = None
r_history = None
for i in range(num_iter):
centroids, idx, history = run_k_means(X, K)
d = k_means_distortion(X, centroids, idx)
if d < result:
print(f'> [{i}]: k-means improved with distortion: {d}')
r_centroids = centroids
r_idx = idx
r_history = history
result = d
return r_centroids, r_idx, r_history
```
### 6. plot data with `K=3`
```
import matplotlib.cm as cm
def plot_k_means(X, K, centroid_idx, centroids_history):
plt.figure(figsize=(15, 10))
colors = cm.rainbow(np.linspace(0, 1, K))
for k in range(K):
plt.scatter(X[centroid_idx == k][:, 0], X[centroid_idx == k][:, 1], c=[colors[k]])
for i in range(K):
vals = np.array([points[i] for points in centroids_history])
plt.plot(vals[:, 0], vals[:, 1], '-Xk', c=colors[i], markeredgecolor='black')
plt.title(f'K-Means with K={K}, {len(centroids_history)-1} iterations')
plt.show()
K = 3
centroids, idx, history = find_best_k_means(X, K)
plot_k_means(X, K, idx, history)
```
### 7. read `bird_small.mat` data
```
img = scipy.io.loadmat('data/bird_small.mat')
A = np.reshape(img['A'], newshape=(-1, 3))
A = A.astype('float') / 255.0
A.shape
```
### 8. compress image with k-means, 16 colors
```
def show_images(original, compressed):
fig, axs = plt.subplots(1, 2, figsize=(15, 10))
axs.flat[0].imshow(original)
axs.flat[1].imshow(compressed)
plt.show()
K = 16
centroids, idx, _ = find_best_k_means(A, K)
A_recon = centroids[idx]
A_recon = A_recon.reshape(-1, 128, 3)
show_images(img['A'], A_recon)
w, h = 128, 128
bits_per_pixel = 24
o_size = w * h * bits_per_pixel
print(f'Original:\t{o_size} bytes, {A.nbytes / 1024} kb')
colors = 16
comp_size = colors * bits_per_pixel + w * h * colors / 4
print(f'Compressed:\t{comp_size} bytes, {comp_size / 1024} kb, x{o_size / comp_size:.0f} times smaller')
```
### 9. test one more image
```
import matplotlib.image as mpimg
lena = mpimg.imread('data/lena.png')
lena = lena[:, :, :3]
A = np.reshape(lena, newshape=(-1, 3))
K = 16
centroids, idx, _ = find_best_k_means(A, K)
A_recon = centroids[idx]
A_recon = A_recon.reshape(-1, lena.shape[1], 3)
show_images(lena, A_recon)
```
### 10. hierarchical clustering algorithm
```
from sklearn.cluster import AgglomerativeClustering
clustering = AgglomerativeClustering(n_clusters=K).fit(A)
idx = clustering.labels_
centroids = compute_means(A, idx, K)
A_recon = centroids[idx]
A_recon = A_recon.reshape(-1, data.shape[1], 3)
show_images(lena, A_recon)
```
> На глаз результат сжатия методом `K-means` и иерахической кластеризации используя `scipy` реализацию `AgglomerativeClustering` получился одинаковым
### 12. conclusions
В данной работе рассмотрен метод кластеризации `K-means`, произведено сжатие изображения до 16 цветов
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import scipy.io
data = scipy.io.loadmat('data/ex6data1.mat')
X = data['X']
X.shape
def rand_centroids(X, K):
rand_indices = np.arange(len(X))
np.random.shuffle(rand_indices)
centroids = X[rand_indices][:K]
return centroids
rand_centroids(X, 3)
def find_closest_centroids(X, centroids):
distances = np.array([np.sqrt((X.T[0] - c[0])**2 + (X.T[1] - c[1])**2) for c in centroids])
return distances.argmin(axis=0)
def compute_means(X, centroid_idx, K):
centroids = []
for k in range(K):
t = X[centroid_idx == k]
c = np.mean(t, axis=0) if t.size > 0 else np.zeros((X.shape[1],))
centroids.append(c)
return np.array(centroids)
def run_k_means(X, K, num_iter=10):
centroids = rand_centroids(X, K)
centroids_history = [centroids]
for i in range(num_iter):
centroid_idx = find_closest_centroids(X, centroids)
centroids = compute_means(X, centroid_idx, K)
centroids_history.append(centroids)
return centroids, centroid_idx, centroids_history
def k_means_distortion(X, centroids, idx):
K = centroids.shape[0]
distortion = 0
for i in range(K):
distortion += np.sum((X[idx == i] - centroids[i])**2)
distortion /= X.shape[0]
return distortion
def find_best_k_means(X, K, num_iter=100):
result = np.inf
r_centroids = None
r_idx = None
r_history = None
for i in range(num_iter):
centroids, idx, history = run_k_means(X, K)
d = k_means_distortion(X, centroids, idx)
if d < result:
print(f'> [{i}]: k-means improved with distortion: {d}')
r_centroids = centroids
r_idx = idx
r_history = history
result = d
return r_centroids, r_idx, r_history
import matplotlib.cm as cm
def plot_k_means(X, K, centroid_idx, centroids_history):
plt.figure(figsize=(15, 10))
colors = cm.rainbow(np.linspace(0, 1, K))
for k in range(K):
plt.scatter(X[centroid_idx == k][:, 0], X[centroid_idx == k][:, 1], c=[colors[k]])
for i in range(K):
vals = np.array([points[i] for points in centroids_history])
plt.plot(vals[:, 0], vals[:, 1], '-Xk', c=colors[i], markeredgecolor='black')
plt.title(f'K-Means with K={K}, {len(centroids_history)-1} iterations')
plt.show()
K = 3
centroids, idx, history = find_best_k_means(X, K)
plot_k_means(X, K, idx, history)
img = scipy.io.loadmat('data/bird_small.mat')
A = np.reshape(img['A'], newshape=(-1, 3))
A = A.astype('float') / 255.0
A.shape
def show_images(original, compressed):
fig, axs = plt.subplots(1, 2, figsize=(15, 10))
axs.flat[0].imshow(original)
axs.flat[1].imshow(compressed)
plt.show()
K = 16
centroids, idx, _ = find_best_k_means(A, K)
A_recon = centroids[idx]
A_recon = A_recon.reshape(-1, 128, 3)
show_images(img['A'], A_recon)
w, h = 128, 128
bits_per_pixel = 24
o_size = w * h * bits_per_pixel
print(f'Original:\t{o_size} bytes, {A.nbytes / 1024} kb')
colors = 16
comp_size = colors * bits_per_pixel + w * h * colors / 4
print(f'Compressed:\t{comp_size} bytes, {comp_size / 1024} kb, x{o_size / comp_size:.0f} times smaller')
import matplotlib.image as mpimg
lena = mpimg.imread('data/lena.png')
lena = lena[:, :, :3]
A = np.reshape(lena, newshape=(-1, 3))
K = 16
centroids, idx, _ = find_best_k_means(A, K)
A_recon = centroids[idx]
A_recon = A_recon.reshape(-1, lena.shape[1], 3)
show_images(lena, A_recon)
from sklearn.cluster import AgglomerativeClustering
clustering = AgglomerativeClustering(n_clusters=K).fit(A)
idx = clustering.labels_
centroids = compute_means(A, idx, K)
A_recon = centroids[idx]
A_recon = A_recon.reshape(-1, data.shape[1], 3)
show_images(lena, A_recon)
| 0.430147 | 0.918663 |
# Pandoc Markdown Syntax
Pandoc supports a large number of input file formats for processing (including markdown, reStructuredText, textile, HTML, DocBook, LaTeX, MediaWiki markup, TWiki markup, OPML, Emacs Org-Mode, Txt2Tags, Microsoft Word docx, LibreOffice ODT, EPUB, or Haddock markup) into a vast number of output formats. For our purposes in this workshop we will focus on working with the extended markdown syntax supported by Pandoc as a language for creating content and generating multiple output representations.
For complete documentation of Pandoc's syntax refer to the [Pandoc User's Guide](http://pandoc.org/MANUAL.html).
```
3**4
```
## Title Block and Additional Metadata
Pandoc provides an option for providing document metadata in two forms: as a simple `title block` at the beginning of a document, and also as a `YAML` metadata block that you can provide anywhere in the document, or even as an externally referenced document.
The `title block` provides basic title, author, and date information about the document that is used by many of the default Pandoc templates to automatically fill in specific document elements. For example, the `title` provided in the title block is used to set the `title` meta element in a generated HTML file and for a `title` block in a LaTeX document. The authorship and date information is similarly used to automatically generate content for some output document types. The `title block` has a simple three part structure, each of which is required if a `title block` if a subsequent element is used. The format of a title block is:
```
% title
% author(s) (separated by semicolons)
% date (treated as a text string)
```
These elements (and any others that you would like to create and use) may also be placed in a `YAML` metadata block:
```
---
title: title
author: author(s) (separated by semicolons)
date: date (treated as a text string)
...
```
*Why would you use a `YAML` metadata block?* - additional metadata can be very useful for setting variable values that Pandoc uses when generating documents (such as specifying a bibliography file to use), or values that can be referred to within custom document templates.
## Outline Structure
The section of the Pandoc User's Guide relating to headers can be found [here](https://pandoc.org/MANUAL.html#headings).
A key element of any document that you create is the hierarchical structure representing the document outline. This hierarchy is used to deliniate content sections and the styles used to present the content of those sections. Examples of the use of the outline structure include:
* mapping of the hierarchy levels into corresponding HTML header elements - `<h1>`, `<h2>`,`<h3>`, etc.
* defining chapters and sections of generated PDF files
* defining title slides, slides and slide content for slide shows
When defining the outline structure of your documents focus on the structure and not intended representation of your content. Other elements of the markdown syntax can be used to create effects like emphasys or strong emphasys, and additional options are available through very granular control that you can exercise through customized templates (e.g. for HTML and LaTeX generated PDF files) and CSS (for HTML and EPubs) stylesheets.
There are a couple of options for specifying outline levels:
```
# Level One
## Level Two
### Level Three
...
###### Level Six
```
and
```
A Level One Header
==================
A Level Two Header
------------------
```
How youre selected header levels are rendered will depend upon the template and format used when translating the markdown into a specific representation.
Note: while standard markdown allows for having no empty line before a header, Pandoc required a blank line before a header.
### Optional Header Attributes
You can also specific optional header attributes as part of your document. These attributes can be used to assign `id` and `class` values to headers that can be used when creating custom HTML styles, provide labels used in automatically generated Table of Contents content, and other output-format-specific elements. The attributes for a header are specified using the following syntax:
```
{#identifier .class .class key=value key=value}
```
which is added following the end of the header text.
## Blocks of Content
Block of content fall into a number of categories including *paragraphs*, *block quotes*, a variety of *lists*, *verbatim* and *line blocks*. Each of these content block types has its own syntax for specifying the content.
### Paragraphs
The section of the Pandoc User's Guide relating to paragraphs can be found [here](https://pandoc.org/MANUAL.html#paragraphs).
The syntax for paragraphs is very simple - just blocks of text separated from each other by blank lines. By default line breaks will be ignored so you can type your paragraph as multiple (contiguous) lines and expect it to be rendered as a single paragraph in the output representation. Paragraph breaks are automatically created when a blank line is encountered at the end of a block of contiguous lines of text.
### Block Quotes
The section of the Pandoc User's Guide relating to block quotes can be found [here](https://pandoc.org/MANUAL.html#block-quotations).
Block quotes are defined through the use of the `>` character at the beginning of the text that is intended for inclusion in the block quote. The `>` may appear at the start of each line, or a *lazy* may be used in which only the first line of a block of contiguous text lines is preceeded by a `>`. For example:
```
> this is a
> multiline quote
```
> this is a
> multiline quote
which is treated similarly to
```
> this is another
multiline quote
```
> this is another
multiline quote
In either case, the quote will be styled according to the specifications of the output file format.
### Lists
The section of the Pandoc User's Guide relating to lists can be found [here](https://pandoc.org/MANUAL.html#lists).
A variety of list types are supported by Pandoc, including nesting of lists of different types. Nesting is defined through the use of four spaces or one tab to indent the list item. Individual list items may contain multiple paragraphs (each separated by a blank line) and preceeded by *four spaces*. Because of the indentation rule for multi-paragraph list items, `code blocks` included in a list item must be indented *eight spaces*. These lists include:
#### Unnumbered Lists
List items in unnumbered (bulleted) lists are preceeded by a `*` or `+` character followed by a space.
```
* List Item 1
* List Item 2
* List Item 2a
* List Item 2b
* List Item 3
```
Which will be rendered like:
* List Item 1
* List Item 2
* List Item 2a
* List Item 2b
* List Item 3
#### Numbered/lettered Lists
Numbered and lettered lists may be preceeded by numbers or lower- and upper-case letters, any of which may be enclosed in parentheses, or followed by a closing parenthesis or period followed by a space (or two-spaces if the preceeding character is an upper-case. The specific characters or numbers used aren't used (except for the first one in the list which defines the beginning of the sequence), but instead are used to define the desired auto-generated list item prefix values. To obtain the default list numbers you can preceed each item with a `#.` instead of a character or number.
```
1. List Item 1
1. List Item 2
a. List Item 2a
a. List Item 2b
1. List Item 3
```
Even when not indented, a new list will be created when the list type changes.
#### Definition Lists
Lists that are specifically designed for providing terms and their definitions are also supported by Pandoc's mardown syntax. An example (others are available) of a compact syntax for a defintion list is provided here:
```
Term 1
~ Definition 1
Term 2
~ Definition 2a
~ Definition 2b
```
#### Example Lists
Example lists may be created within a document. The elements of an example list may be created throughout the document with the examples being numbered sequentially throughout the document. Example list elements may be given labels that may be used to cross-reference examples elsewhere in the document.
```
(@) My first example will be numbered (1).
(@) My second example will be numbered (2).
Explanation of examples.
(@) My third example will be numbered (3).
(@good) This is a good example.
As (@good) illustrates, ...
```
### Code and Other Verbatim Text
The section of the Pandoc User's Guide relating to verbatim text can be found [here](https://pandoc.org/MANUAL.html#verbatim-code-blocks).
There are times when you want to display text exactly as entered instead of having line breaks or other modifications made to the text structure. While this is commonly used for displaying code, it is also helpful when you want to present a block of text where there is a specific line structure that needs to be maintained.
#### Code Blocks
Code blocks can be defined by either indenting each line by four spaces or a tab, or by creating a *fenced code block* by placing the text between lines with three or more tilde characters (`~`) and the closing set being at least as long as the opening set.
```
pandoc -s -S \
--normalize \
--filter pandoc-citeproc \
--csl ./science.csl \
--natbib \
--template=poster.tex \
-f markdown+raw_tex \
-o 2016-12_AGUPoster.tex \
2016-12_AGUPoster.md
```
This is an example of a *fenced code block*
```
~~~
pandoc -s -S \
--normalize \
--filter pandoc-citeproc \
--csl ./science.csl \
--natbib \
--template=poster.tex \
-f markdown+raw_tex \
-o 2016-12_AGUPoster.tex \
2016-12_AGUPoster.md
~~~
```
Some renderers support syntax coloring for different programming languages. Pandoc allows for the specification of attributes for a fenced code block to provide hints for renderers as to what language is being presented and whether line numbers should be provided in the output:
```
~~~~ {#mycode .bash .numberLines startFrom="15"}
pandoc -s -S \
--normalize \
--filter pandoc-citeproc \
--csl ./science.csl \
--natbib \
--template=poster.tex \
-f markdown+raw_tex \
-o 2016-12_AGUPoster.tex \
2016-12_AGUPoster.md
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
```
#### Line Blocks
An alternative method for controlling the flow of text without the semantic implication of `code` is to use *line blocks* to define blocks of text for which line breaks and included indentation is maintained.
```
| Karl Benedict
| MSC05 3020
| 1 University of New Mexico
| Albuquerque, NM
| 98131
```
## Inline Text Formatting
The section of the Pandoc User's Guide relating to inline text formatting can be found [here](https://pandoc.org/MANUAL.html#inline-formatting).
Pandoc markdown supports a number of methods for altering the appearance of text within heading, paragraph, block quote, and list blocks. These methods enable some of the standard text modifications you are accustomed to using in word processors, though with an emphasys on the *semantic* specification (for example *emphasys* or **strong emphasys**) of text modifications instead of directly specifying (*italic* or **bold**). While a bit of a nuanced point, it is important to think about this difference as these *semantic* directives get translated into different representations when output files are generated. You can specify the following inline text formats:
* *Emphasys*
```
Here is an emphasized *word*
This is another way to define an emphasized _word_
```
Here is an emphasized *word*
This is another way to define an emphasized _word_
* **Strong Emphasys**
```
Here is a strongly emphasized **word**
This is another way to define a strongly emphasized __word__
```
Here is a strongly emphasized **word**
This is another way to define a strongly emphasized __word__
* Superscript and subscript text
Superscript text is surrounded by `^` characters:
```
e=mc^2^
```
Subscript text is surrounded by `~` characters:
```
H~2~O
```
* Strikeout text
Strikeout text is surrounded by `~~` (double tilde) characters:
```
I meant this instead of ~~that~~
```
I meant this instead of ~~that~~
* Inline verbatim text
Verbatim text is surrounded by `\`` (back-tick) characters:
```
You should enter `pandoc` on the command line to start a document processing command.
```
You should enter `pandoc` on the command line to start a document processing command.
## Footnotes
The section of the Pandoc User's Guide relating to footnotes can be found [here](https://pandoc.org/MANUAL.html#footnotes).
Footnotes are supported by Pandoc through both *inline* footnotes and two-part footnote definitions. Inline footnotes are very convenient for defining a relatively short bit of text (or other content) within the flow of a text block while two-part footnotes may be used to define footnote content that is larger or more complex than you can place within the flow of a text block. Both inline and two-part footnotes may be mixed within a single document. The specific rendering of the footnotes would need to be modified through updates to the template used to generate the output.
### Inline Footnotes
The syntax for inline footnotes consists of a combination of a `^` followed by a pair of square brackets (`[]`) that contain the content of the footnote.
`The Google^[http://www.google.com] search engine may be used to ...`
where the contents of the inline footnote - `http://www.google.com` - would be added to the generated output using a representation appropriate for the specific output format.
### Two-part Footnotes
Two-part footnotes consist of an inline identifier that is placed within square brackets (`[]`) that starts with a `^` symbol followed by the remainder of the identifier text (which cannot include spaces). Elsewhere in the document you can place the actual footnote text on its own line. The footnotes will be automatically numbered based on the order of the references within the document.
```
This my text with a two-part footnote[^1]. The actual actual label text used
doesn't matter, except that it can't contain spaces, tabs or newlines[^ref].
[^1]: this is the text for the first footnote
[^ref]: this is the text for the secondd footnote.
```
## Tables
The section of the Pandoc User's Guide relating to tables can be found [here](https://pandoc.org/MANUAL.html#tables).
Pandoc supports a number of methods for defining the structure and content of tabular content within documents. A review of the different syntaxes for specifying tables ([here](https://pandoc.org/MANUAL.html#tables)) is recommended if you need to provide more than the most simple tables as part of your output. A basic table production example is provided here:
```
Right Left Center Default
------- ------ ---------- -------
12 12 12 12
123 123 123 123
1 1 1 1
```
This simple table format illustrates the use of column headings (which are optional), a row of dashed lines that define the top of the table and provide a reference for how the content of the table rows should be aligned within each column, and the content of each table row - note that the position of the header row columns determines the alignment of each column. If there are no headers the alignment of the first row of values determines the column alignment.
## An Example :
Following is an example of these various content elements within a markdown document.
```
% title
% author(s) (separated by semicolons)
% date (treated as a text string)
# Level One
Fatback labore cupidatat meatball quis. Consequat kevin commodo ipsum laborum ham hock aute jerky pastrami deserunt cillum non turducken. In irure bresaola cupidatat laborum alcatra. In flank do labore kevin, filet mignon in. Dolore chicken beef ribs ribeye cow. Commodo cupidatat shankle laboris, exercitation pastrami magna porchetta.
## Level Two
Chuck pork consequat, biltong pork loin meatball pancetta brisket commodo anim. Cupidatat capicola pancetta, excepteur ribeye ex hamburger prosciutto elit filet mignon. Aliquip voluptate est occaecat pancetta meatloaf sunt commodo laborum spare ribs. Cupidatat commodo shank, culpa fugiat veniam dolore dolor consequat. Beef shank landjaeger short loin. Nisi flank ad, alcatra prosciutto consequat ullamco.
### Level Three
Officia alcatra anim fugiat. Laborum bresaola shoulder beef doner pork belly et burgdoggen. Commodo swine culpa ad shank voluptate cow kevin elit strip steak minim. Swine ullamco burgdoggen chuck est occaecat dolore meatball reprehenderit deserunt jerky adipisicing. Sed venison aliquip officia short ribs. Irure drumstick tempor reprehenderit kielbasa brisket. Ea swine kielbasa pork chop picanha pancetta.
#### Level Four
Cillum turducken consectetur ut tri-tip short ribs t-bone meatloaf venison cupidatat labore. Aliqua tail esse, filet mignon cupim drumstick ut nostrud. Ribeye laborum aliquip, ad voluptate aliqua biltong commodo pig burgdoggen nulla. In boudin chuck aute labore ad alcatra pig deserunt strip steak picanha mollit shank nostrud burgdoggen. Commodo tongue deserunt, brisket ball tip voluptate magna turducken.
##### Level Five
Capicola hamburger duis minim. Andouille sed dolore sunt voluptate exercitation bacon anim eu capicola sausage burgdoggen brisket. T-bone pastrami lorem, in short loin dolore sed pork chop incididunt turducken exercitation. Quis ribeye boudin, dolore turkey shoulder do rump aliquip picanha adipisicing. Aute enim ex, ad corned beef aliqua in eiusmod culpa incididunt.
###### Level Six
Labore eiusmod magna chuck occaecat ribeye pig sed. Occaecat nisi id ut deserunt anim drumstick pastrami cow. Meatloaf culpa pork loin swine nisi, dolore tri-tip sirloin andouille nostrud salami tongue lorem porchetta. Pastrami kielbasa landjaeger tenderloin.
Level One
=========
Fatback labore cupidatat meatball quis. Consequat kevin commodo ipsum laborum ham hock aute jerky pastrami deserunt cillum non turducken. In irure bresaola cupidatat laborum alcatra. In flank do labore kevin, filet mignon in. Dolore chicken beef ribs ribeye cow. Commodo cupidatat shankle laboris, exercitation pastrami magna porchetta.
Level Two
---------
Chuck pork consequat, biltong pork loin meatball pancetta brisket commodo anim. Cupidatat capicola pancetta, excepteur ribeye ex hamburger prosciutto elit filet mignon. Aliquip voluptate est occaecat pancetta meatloaf sunt commodo laborum spare ribs. Cupidatat commodo shank, culpa fugiat veniam dolore dolor consequat. Beef shank landjaeger short loin. Nisi flank ad, alcatra prosciutto consequat ullamco.
------------------------------------------------
> this is a
> multiline quote
> this is another
multiline quote
* List Item 1
* List Item 2
* List Item 2a
* List Item 2b
* List Item 3
Term 1
~ Definition 1
Term 2
~ Definition 2a
~ Definition 2b
(@) My first example will be numbered (1).
(@) My second example will be numbered (2).
Explanation of examples.
(@) My third example will be numbered (3).
(@good) This is a good example.
As (@good) illustrates, ...
pandoc -s -S \
--normalize \
--filter pandoc-citeproc \
--csl ./science.csl \
--natbib \
--template=poster.tex \
-f markdown+raw_tex \
-o 2016-12_AGUPoster.tex \
2016-12_AGUPoster.md
~~~
pandoc -s -S \
--normalize \
--filter pandoc-citeproc \
--csl ./science.csl \
--natbib \
--template=poster.tex \
-f markdown+raw_tex \
-o 2016-12_AGUPoster.tex \
2016-12_AGUPoster.md
~~~
~~~~ {#mycode .bash .numberLines startFrom="15"}
pandoc -s -S \
--normalize \
--filter pandoc-citeproc \
--csl ./science.csl \
--natbib \
--template=poster.tex \
-f markdown+raw_tex \
-o 2016-12_AGUPoster.tex \
2016-12_AGUPoster.md
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
| Karl Benedict
| MSC05 3020
| 1 University of New Mexico
| Albuquerque, NM
| 98131
------------------------------------------------
Here is an emphasized *word*
This is another way to define an emphasized _word_
Here is a strongly emphasized **word**
This is another way to define a strongly emphasized __word__
e=mc^2^
H~2~O
I meant this instead of ~~that~~
You should enter `pandoc` on the command line to start a document processing command.
------------------------------------------------
The Google^[http://www.google.com] search engine may be used to ...
This my text with a two-part footnote[^1]. The actual actual label text used
doesn't matter, except that it can't contain spaces, tabs or newlines[^ref].
[^1]: this is the text for the first footnote
[^ref]: this is the text for the secondd footnote.
------------------------------------------------
Right Left Center Default
------- ------ ---------- -------
12 12 12 12
123 123 123 123
1 1 1 1
```
------------------------------------------------
% title
% author(s) (separated by semicolons)
% date (treated as a text string)
# Level One
Fatback labore cupidatat meatball quis. Consequat kevin commodo ipsum laborum ham hock aute jerky pastrami deserunt cillum non turducken. In irure bresaola cupidatat laborum alcatra. In flank do labore kevin, filet mignon in. Dolore chicken beef ribs ribeye cow. Commodo cupidatat shankle laboris, exercitation pastrami magna porchetta.
## Level Two
Chuck pork consequat, biltong pork loin meatball pancetta brisket commodo anim. Cupidatat capicola pancetta, excepteur ribeye ex hamburger prosciutto elit filet mignon. Aliquip voluptate est occaecat pancetta meatloaf sunt commodo laborum spare ribs. Cupidatat commodo shank, culpa fugiat veniam dolore dolor consequat. Beef shank landjaeger short loin. Nisi flank ad, alcatra prosciutto consequat ullamco.
### Level Three
Officia alcatra anim fugiat. Laborum bresaola shoulder beef doner pork belly et burgdoggen. Commodo swine culpa ad shank voluptate cow kevin elit strip steak minim. Swine ullamco burgdoggen chuck est occaecat dolore meatball reprehenderit deserunt jerky adipisicing. Sed venison aliquip officia short ribs. Irure drumstick tempor reprehenderit kielbasa brisket. Ea swine kielbasa pork chop picanha pancetta.
#### Level Four
Cillum turducken consectetur ut tri-tip short ribs t-bone meatloaf venison cupidatat labore. Aliqua tail esse, filet mignon cupim drumstick ut nostrud. Ribeye laborum aliquip, ad voluptate aliqua biltong commodo pig burgdoggen nulla. In boudin chuck aute labore ad alcatra pig deserunt strip steak picanha mollit shank nostrud burgdoggen. Commodo tongue deserunt, brisket ball tip voluptate magna turducken.
##### Level Five
Capicola hamburger duis minim. Andouille sed dolore sunt voluptate exercitation bacon anim eu capicola sausage burgdoggen brisket. T-bone pastrami lorem, in short loin dolore sed pork chop incididunt turducken exercitation. Quis ribeye boudin, dolore turkey shoulder do rump aliquip picanha adipisicing. Aute enim ex, ad corned beef aliqua in eiusmod culpa incididunt.
###### Level Six
Labore eiusmod magna chuck occaecat ribeye pig sed. Occaecat nisi id ut deserunt anim drumstick pastrami cow. Meatloaf culpa pork loin swine nisi, dolore tri-tip sirloin andouille nostrud salami tongue lorem porchetta. Pastrami kielbasa landjaeger tenderloin.
Level One
=========
Fatback labore cupidatat meatball quis. Consequat kevin commodo ipsum laborum ham hock aute jerky pastrami deserunt cillum non turducken. In irure bresaola cupidatat laborum alcatra. In flank do labore kevin, filet mignon in. Dolore chicken beef ribs ribeye cow. Commodo cupidatat shankle laboris, exercitation pastrami magna porchetta.
Level Two
---------
Chuck pork consequat, biltong pork loin meatball pancetta brisket commodo anim. Cupidatat capicola pancetta, excepteur ribeye ex hamburger prosciutto elit filet mignon. Aliquip voluptate est occaecat pancetta meatloaf sunt commodo laborum spare ribs. Cupidatat commodo shank, culpa fugiat veniam dolore dolor consequat. Beef shank landjaeger short loin. Nisi flank ad, alcatra prosciutto consequat ullamco.
------------------------------------------------
> this is a
> multiline quote
> this is another
multiline quote
* List Item 1
* List Item 2
* List Item 2a
* List Item 2b
* List Item 3
Term 1
~ Definition 1
Term 2
~ Definition 2a
~ Definition 2b
(@) My first example will be numbered (1).
(@) My second example will be numbered (2).
Explanation of examples.
(@) My third example will be numbered (3).
(@good) This is a good example.
As (@good) illustrates, ...
pandoc -s -S \
--normalize \
--filter pandoc-citeproc \
--csl ./science.csl \
--natbib \
--template=poster.tex \
-f markdown+raw_tex \
-o 2016-12_AGUPoster.tex \
2016-12_AGUPoster.md
~~~
pandoc -s -S \
--normalize \
--filter pandoc-citeproc \
--csl ./science.csl \
--natbib \
--template=poster.tex \
-f markdown+raw_tex \
-o 2016-12_AGUPoster.tex \
2016-12_AGUPoster.md
~~~
~~~~ {#mycode .bash .numberLines startFrom="15"}
pandoc -s -S \
--normalize \
--filter pandoc-citeproc \
--csl ./science.csl \
--natbib \
--template=poster.tex \
-f markdown+raw_tex \
-o 2016-12_AGUPoster.tex \
2016-12_AGUPoster.md
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
| Karl Benedict
| MSC05 3020
| 1 University of New Mexico
| Albuquerque, NM
| 98131
------------------------------------------------
Here is an emphasized *word*
This is another way to define an emphasized _word_
Here is a strongly emphasized **word**
This is another way to define a strongly emphasized __word__
e=mc^2^
H~2~O
I meant this instead of ~~that~~
You should enter `pandoc` on the command line to start a document processing command.
------------------------------------------------
The Google^[http://www.google.com] search engine may be used to ...
This my text with a two-part footnote[^1]. The actual actual label text used
doesn't matter, except that it can't contain spaces, tabs or newlines[^ref].
[^1]: this is the text for the first footnote
[^ref]: this is the text for the secondd footnote.
------------------------------------------------
Right Left Center Default
------- ------ ---------- -------
12 12 12 12
123 123 123 123
1 1 1 1
|
github_jupyter
|
3**4
% title
% author(s) (separated by semicolons)
% date (treated as a text string)
---
title: title
author: author(s) (separated by semicolons)
date: date (treated as a text string)
...
# Level One
## Level Two
### Level Three
...
###### Level Six
A Level One Header
==================
A Level Two Header
------------------
{#identifier .class .class key=value key=value}
> this is a
> multiline quote
> this is another
multiline quote
* List Item 1
* List Item 2
* List Item 2a
* List Item 2b
* List Item 3
1. List Item 1
1. List Item 2
a. List Item 2a
a. List Item 2b
1. List Item 3
Term 1
~ Definition 1
Term 2
~ Definition 2a
~ Definition 2b
(@) My first example will be numbered (1).
(@) My second example will be numbered (2).
Explanation of examples.
(@) My third example will be numbered (3).
(@good) This is a good example.
As (@good) illustrates, ...
pandoc -s -S \
--normalize \
--filter pandoc-citeproc \
--csl ./science.csl \
--natbib \
--template=poster.tex \
-f markdown+raw_tex \
-o 2016-12_AGUPoster.tex \
2016-12_AGUPoster.md
~~~
pandoc -s -S \
--normalize \
--filter pandoc-citeproc \
--csl ./science.csl \
--natbib \
--template=poster.tex \
-f markdown+raw_tex \
-o 2016-12_AGUPoster.tex \
2016-12_AGUPoster.md
~~~
~~~~ {#mycode .bash .numberLines startFrom="15"}
pandoc -s -S \
--normalize \
--filter pandoc-citeproc \
--csl ./science.csl \
--natbib \
--template=poster.tex \
-f markdown+raw_tex \
-o 2016-12_AGUPoster.tex \
2016-12_AGUPoster.md
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
| Karl Benedict
| MSC05 3020
| 1 University of New Mexico
| Albuquerque, NM
| 98131
Here is an emphasized *word*
This is another way to define an emphasized _word_
```
Here is an emphasized *word*
This is another way to define an emphasized _word_
* **Strong Emphasys**
```
Here is a strongly emphasized **word**
This is another way to define a strongly emphasized __word__
```
Here is a strongly emphasized **word**
This is another way to define a strongly emphasized __word__
* Superscript and subscript text
Superscript text is surrounded by `^` characters:
```
e=mc^2^
```
Subscript text is surrounded by `~` characters:
```
H~2~O
```
* Strikeout text
Strikeout text is surrounded by `~~` (double tilde) characters:
```
I meant this instead of ~~that~~
```
I meant this instead of ~~that~~
* Inline verbatim text
Verbatim text is surrounded by `\`` (back-tick) characters:
```
You should enter `pandoc` on the command line to start a document processing command.
```
You should enter `pandoc` on the command line to start a document processing command.
## Footnotes
The section of the Pandoc User's Guide relating to footnotes can be found [here](https://pandoc.org/MANUAL.html#footnotes).
Footnotes are supported by Pandoc through both *inline* footnotes and two-part footnote definitions. Inline footnotes are very convenient for defining a relatively short bit of text (or other content) within the flow of a text block while two-part footnotes may be used to define footnote content that is larger or more complex than you can place within the flow of a text block. Both inline and two-part footnotes may be mixed within a single document. The specific rendering of the footnotes would need to be modified through updates to the template used to generate the output.
### Inline Footnotes
The syntax for inline footnotes consists of a combination of a `^` followed by a pair of square brackets (`[]`) that contain the content of the footnote.
`The Google^[http://www.google.com] search engine may be used to ...`
where the contents of the inline footnote - `http://www.google.com` - would be added to the generated output using a representation appropriate for the specific output format.
### Two-part Footnotes
Two-part footnotes consist of an inline identifier that is placed within square brackets (`[]`) that starts with a `^` symbol followed by the remainder of the identifier text (which cannot include spaces). Elsewhere in the document you can place the actual footnote text on its own line. The footnotes will be automatically numbered based on the order of the references within the document.
## Tables
The section of the Pandoc User's Guide relating to tables can be found [here](https://pandoc.org/MANUAL.html#tables).
Pandoc supports a number of methods for defining the structure and content of tabular content within documents. A review of the different syntaxes for specifying tables ([here](https://pandoc.org/MANUAL.html#tables)) is recommended if you need to provide more than the most simple tables as part of your output. A basic table production example is provided here:
This simple table format illustrates the use of column headings (which are optional), a row of dashed lines that define the top of the table and provide a reference for how the content of the table rows should be aligned within each column, and the content of each table row - note that the position of the header row columns determines the alignment of each column. If there are no headers the alignment of the first row of values determines the column alignment.
## An Example :
Following is an example of these various content elements within a markdown document.
| 0.915427 | 0.982757 |
```
import torch
import torch.optim as optim
import torchvision.transforms as transforms
import torch.nn as nn
import torch.nn.functional as F
import numpy as np
import time
import dataset.dataset as dataset
import datasplit.datasplit as datasplit
import model.models as models
import trainer.trainer as trainer
import utils.utils as utils
torch.cuda.device_count()
cuda0 = torch.device('cuda:0')
cuda1 = torch.device('cuda:1')
cuda2 = torch.device('cuda:2')
cuda3 = torch.device('cuda:3')
device = torch.device(cuda0 if torch.cuda.is_available() else "cpu")
```
# INIT
```
# transforms
transform = transforms.Compose([
transforms.ToTensor(),
])
# dataset
root = '/Volumes/Macintosh HD/DATASETS/GUITAR-FX/Mono_Random_Bg'
excl_folders = ['MT2']
spectra_folder= 'mel_22050_1024_512'
proc_settings_csv = 'proc_settings.csv'
max_num_settings=3
dataset = dataset.FxDataset(root=root,
excl_folders=excl_folders,
spectra_folder=spectra_folder,
processed_settings_csv=proc_settings_csv,
max_num_settings=max_num_settings,
transform=transform)
dataset.init_dataset()
# dataset.generate_mel()
# split
# set test_train_split=0.0 and val_train_split=0.0 to test pre-trained model
split = datasplit.DataSplit(dataset, test_train_split=0.8, val_train_split=0.1, shuffle=True)
# loaders
train_loader, val_loader, test_loader = split.get_split(batch_size=100)
print('dataset size: ', len(dataset))
print('train set size: ', len(split.train_sampler))
print('val set size: ', len(split.val_sampler))
print('test set size: ', len(split.test_sampler))
dataset.fx_to_label
```
# TRAIN FxNET
```
# model
fxnet = models.FxNet(n_classes=dataset.num_fx).to(device)
# optimizer
optimizer_fxnet = optim.Adam(fxnet.parameters(), lr=0.001)
# loss function
loss_func_fxnet = nn.CrossEntropyLoss()
print(fxnet)
# SAVE
models_folder = '../../saved/models'
model_name = '20201024_fxnet_mono_cont_best'
results_folder = '../../saved/results'
results_subfolder = '20201024_fxnet_mono_cont'
# TRAIN and TEST FxNet OVER MULTIPLE EPOCHS
train_set_size = len(split.train_sampler)
val_set_size = len(split.val_sampler)
test_set_size = len(split.test_sampler)
all_train_losses, all_val_losses, all_test_losses = [],[],[]
all_train_correct, all_val_correct, all_test_correct = [],[],[]
all_train_results, all_val_results, all_test_results = [],[],[]
best_val_correct = 0
early_stop_counter = 0
start = time.time()
for epoch in range(100):
train_loss, train_correct, train_results = trainer.train_fx_net(
model=fxnet,
optimizer=optimizer_fxnet,
train_loader=train_loader,
train_sampler=split.train_sampler,
epoch=epoch,
device=device
)
val_loss, val_correct, val_results = trainer.val_fx_net(
model=fxnet,
val_loader=val_loader,
val_sampler=split.val_sampler,
device=device
)
test_loss, test_correct, test_results = trainer.test_fx_net(
model=fxnet,
test_loader=test_loader,
test_sampler=split.test_sampler,
device=device
)
# save model
if val_correct > best_val_correct:
best_val_correct = val_correct
torch.save(fxnet, '%s/%s' % (models_folder, model_name))
early_stop_counter = 0
print('\n=== saved best model ===\n')
else:
early_stop_counter += 1
# append results
all_train_losses.append(train_loss)
all_val_losses.append(val_loss)
all_test_losses.append(test_loss)
all_train_correct.append(train_correct)
all_val_correct.append(val_correct)
all_test_correct.append(test_correct)
all_train_results.append(train_results)
all_val_results.append(val_results)
all_test_results.append(test_results)
if early_stop_counter == 15:
print('\n--- early stop ---\n')
break
stop = time.time()
print(f"Training time: {stop - start}s")
# BEST RESULTS
print('Accuracy: ', 100 * max(all_train_correct) / train_set_size)
print('Epoch: ', np.argmax(all_train_correct))
print()
print('Accuracy: ', 100 * max(all_val_correct) / val_set_size)
print('Epoch: ', np.argmax(all_val_correct))
print()
print('Accuracy: ', 100 * max(all_test_correct) / test_set_size)
print('Epoch: ', np.argmax(all_test_correct))
print()
# SAVE RESULTS - all losses, all correct, best results
all_train_losses_npy = np.array(all_train_losses)
all_train_correct_npy = np.array(all_train_correct)
best_train_results_npy = np.array(all_train_results[67])
all_val_losses_npy = np.array(all_val_losses)
all_val_correct_npy = np.array(all_val_correct)
best_val_results_npy = np.array(all_val_results[67])
all_test_losses_npy = np.array(all_test_losses)
all_test_correct_npy = np.array(all_test_correct)
best_test_results_npy = np.array(all_test_results[67])
fx_labels_npy = np.array(list(dataset.fx_to_label.keys()))
np.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'all_train_losses')), arr=all_train_losses_npy)
np.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'all_train_correct')), arr=all_train_correct_npy)
np.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'best_train_results')), arr=best_train_results_npy)
np.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'all_val_losses')), arr=all_val_losses_npy)
np.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'all_val_correct')), arr=all_val_correct_npy)
np.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'best_val_results')), arr=best_val_results_npy)
np.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'all_test_losses')), arr=all_test_losses_npy)
np.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'all_test_correct')), arr=all_test_correct_npy)
np.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'best_test_results')), arr=best_test_results_npy)
np.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'fx_labels')), arr=fx_labels_npy)
```
|
github_jupyter
|
import torch
import torch.optim as optim
import torchvision.transforms as transforms
import torch.nn as nn
import torch.nn.functional as F
import numpy as np
import time
import dataset.dataset as dataset
import datasplit.datasplit as datasplit
import model.models as models
import trainer.trainer as trainer
import utils.utils as utils
torch.cuda.device_count()
cuda0 = torch.device('cuda:0')
cuda1 = torch.device('cuda:1')
cuda2 = torch.device('cuda:2')
cuda3 = torch.device('cuda:3')
device = torch.device(cuda0 if torch.cuda.is_available() else "cpu")
# transforms
transform = transforms.Compose([
transforms.ToTensor(),
])
# dataset
root = '/Volumes/Macintosh HD/DATASETS/GUITAR-FX/Mono_Random_Bg'
excl_folders = ['MT2']
spectra_folder= 'mel_22050_1024_512'
proc_settings_csv = 'proc_settings.csv'
max_num_settings=3
dataset = dataset.FxDataset(root=root,
excl_folders=excl_folders,
spectra_folder=spectra_folder,
processed_settings_csv=proc_settings_csv,
max_num_settings=max_num_settings,
transform=transform)
dataset.init_dataset()
# dataset.generate_mel()
# split
# set test_train_split=0.0 and val_train_split=0.0 to test pre-trained model
split = datasplit.DataSplit(dataset, test_train_split=0.8, val_train_split=0.1, shuffle=True)
# loaders
train_loader, val_loader, test_loader = split.get_split(batch_size=100)
print('dataset size: ', len(dataset))
print('train set size: ', len(split.train_sampler))
print('val set size: ', len(split.val_sampler))
print('test set size: ', len(split.test_sampler))
dataset.fx_to_label
# model
fxnet = models.FxNet(n_classes=dataset.num_fx).to(device)
# optimizer
optimizer_fxnet = optim.Adam(fxnet.parameters(), lr=0.001)
# loss function
loss_func_fxnet = nn.CrossEntropyLoss()
print(fxnet)
# SAVE
models_folder = '../../saved/models'
model_name = '20201024_fxnet_mono_cont_best'
results_folder = '../../saved/results'
results_subfolder = '20201024_fxnet_mono_cont'
# TRAIN and TEST FxNet OVER MULTIPLE EPOCHS
train_set_size = len(split.train_sampler)
val_set_size = len(split.val_sampler)
test_set_size = len(split.test_sampler)
all_train_losses, all_val_losses, all_test_losses = [],[],[]
all_train_correct, all_val_correct, all_test_correct = [],[],[]
all_train_results, all_val_results, all_test_results = [],[],[]
best_val_correct = 0
early_stop_counter = 0
start = time.time()
for epoch in range(100):
train_loss, train_correct, train_results = trainer.train_fx_net(
model=fxnet,
optimizer=optimizer_fxnet,
train_loader=train_loader,
train_sampler=split.train_sampler,
epoch=epoch,
device=device
)
val_loss, val_correct, val_results = trainer.val_fx_net(
model=fxnet,
val_loader=val_loader,
val_sampler=split.val_sampler,
device=device
)
test_loss, test_correct, test_results = trainer.test_fx_net(
model=fxnet,
test_loader=test_loader,
test_sampler=split.test_sampler,
device=device
)
# save model
if val_correct > best_val_correct:
best_val_correct = val_correct
torch.save(fxnet, '%s/%s' % (models_folder, model_name))
early_stop_counter = 0
print('\n=== saved best model ===\n')
else:
early_stop_counter += 1
# append results
all_train_losses.append(train_loss)
all_val_losses.append(val_loss)
all_test_losses.append(test_loss)
all_train_correct.append(train_correct)
all_val_correct.append(val_correct)
all_test_correct.append(test_correct)
all_train_results.append(train_results)
all_val_results.append(val_results)
all_test_results.append(test_results)
if early_stop_counter == 15:
print('\n--- early stop ---\n')
break
stop = time.time()
print(f"Training time: {stop - start}s")
# BEST RESULTS
print('Accuracy: ', 100 * max(all_train_correct) / train_set_size)
print('Epoch: ', np.argmax(all_train_correct))
print()
print('Accuracy: ', 100 * max(all_val_correct) / val_set_size)
print('Epoch: ', np.argmax(all_val_correct))
print()
print('Accuracy: ', 100 * max(all_test_correct) / test_set_size)
print('Epoch: ', np.argmax(all_test_correct))
print()
# SAVE RESULTS - all losses, all correct, best results
all_train_losses_npy = np.array(all_train_losses)
all_train_correct_npy = np.array(all_train_correct)
best_train_results_npy = np.array(all_train_results[67])
all_val_losses_npy = np.array(all_val_losses)
all_val_correct_npy = np.array(all_val_correct)
best_val_results_npy = np.array(all_val_results[67])
all_test_losses_npy = np.array(all_test_losses)
all_test_correct_npy = np.array(all_test_correct)
best_test_results_npy = np.array(all_test_results[67])
fx_labels_npy = np.array(list(dataset.fx_to_label.keys()))
np.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'all_train_losses')), arr=all_train_losses_npy)
np.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'all_train_correct')), arr=all_train_correct_npy)
np.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'best_train_results')), arr=best_train_results_npy)
np.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'all_val_losses')), arr=all_val_losses_npy)
np.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'all_val_correct')), arr=all_val_correct_npy)
np.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'best_val_results')), arr=best_val_results_npy)
np.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'all_test_losses')), arr=all_test_losses_npy)
np.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'all_test_correct')), arr=all_test_correct_npy)
np.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'best_test_results')), arr=best_test_results_npy)
np.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'fx_labels')), arr=fx_labels_npy)
| 0.494629 | 0.624752 |
```
# This Python 3 environment comes with many helpful analytics libraries installed
# It is defined by the kaggle/python docker image: https://github.com/kaggle/docker-python
# For example, here's several helpful packages to load in
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory
import os
print(os.listdir("../input"))
# Any results you write to the current directory are saved as output.
# importing packages
import warnings
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
warnings.filterwarnings('ignore')
train_df = pd.read_csv("../input/train.csv")
train_df.head()
test_df = pd.read_csv("../input/test.csv")
test_df.head()
# checking the summary of the data
train_df.info()
# checking the basic statistics on numeric column
train_df.describe()
# checking the null columns
train_df.isnull().sum()
# sum it up to check how many rows have all missing values
train_df.isnull().all(axis = 1).sum()
# summing up the missing values (column-wise) in percentage
round(100 * (train_df.isnull().sum() / len(train_df.index)), 2)
# Basic preprocessing, Feature extraction and EDA
# Some basic EDA
# looking at target variable
is_duplicate = train_df['is_duplicate'].value_counts()
is_duplicate.plot(kind='bar')
# is_duplicate ratio / checking the data is balance or not
data_ratio = is_duplicate / is_duplicate.sum()
data_ratio * 100
# importing nltk packages
import math
import re
import nltk
from collections import Counter
from nltk.corpus import stopwords
from nltk import (word_tokenize, ngrams)
WORD = re.compile(r'\w+')
english_stopwords = set(stopwords.words('english'))
# checking the total words in the question
def find_total_words(element):
"""method to find the number of words in the question"""
return len(str(element).split())
def find_length(element):
"method to find the length of question in the question"
return len(str(element))
def find_total_sentences(element):
"method to find the total number of sentences in the question"
return len(str(element).split("."))
def find_total_stopwords(element):
"method to find total number of stopwords in the question"
total_words = str(element).split()
return len([word for word in total_words if word in english_stopwords])
def get_unigrams(element):
"""method to find and return available unigrams in the question"""
lower_str = str(element).lower()
total_words = word_tokenize(lower_str)
total_unigrams = [word for word in total_words if word not in english_stopwords]
return total_unigrams
def get_common_unigrams(unigram_question1, unigram_question2):
"""method to find and return the common unigrams found in both question1 and question2"""
return len(set(unigram_question1).intersection(set(unigram_question2)))
def get_common_unigram_ratio(unigram_question1, unigram_question2):
"method to find and return common unigram ratio for question1 and question"
count = get_common_unigrams(unigram_question1, unigram_question2)
return float(count) / max(len(set(get_unigrams(unigram_question1)).union(set(get_unigrams(unigram_question2)))), 1)
def get_similarity(vector1, vector2):
"""method to compute the cosine similarity between question1 and question2"""
intersection = set(vector1.keys()) & set(vector2.keys())
numerator = sum([vector1[x] * vector2[x] for x in intersection])
sum1 = sum([vector1[x] ** 2 for x in vector1.keys()])
sum2 = sum([vector2[x] ** 2 for x in vector2.keys()])
denominator = math.sqrt(sum1) * math.sqrt(sum2)
if not denominator:
return 0.0
else:
return float(numerator) / denominator
def text_to_vector(sentence):
if sentence is np.NAN:
sentence = ""
words = WORD.findall(sentence)
return Counter(words)
# Finding the number of words in the question
train_df['q1_total_words'] = train_df['question1'].apply(lambda x: find_total_words(x))
train_df['q1_total_words'].value_counts().plot(kind='bar')
train_df['q2_total_words'] = train_df['question2'].apply(lambda x: find_total_words(x))
train_df['q2_total_words'].value_counts().plot(kind='bar')
# Finding the length of the question
train_df['q1_length'] = train_df['question1'].apply(lambda x: find_length(x))
train_df['q1_length'].value_counts().plot(kind='bar')
train_df['q2_length'] = train_df['question2'].apply(lambda x: find_length(x))
train_df['q2_length'].value_counts().plot(kind='bar')
# finding the total words distribution on both question 1 and question 2
train_df['all_question_length'] = train_df.apply(lambda row: find_length(row['question1']) +
find_length(row['question2']), axis=1)
train_df['all_question_length'].value_counts().plot(kind='bar')
# Finding the total sentences of the question
train_df['q1_sentence_length'] = train_df['question1'].apply(lambda x: find_total_sentences(x))
train_df['q1_sentence_length'].value_counts().plot(kind='bar')
train_df['q2_sentence_length'] = train_df['question2'].apply(lambda x: find_total_sentences(x))
train_df['q2_sentence_length'].value_counts().plot(kind='bar')
# Finding the number of stop words in the question
train_df['q1_total_stopwords'] = train_df['question1'].apply(lambda x: find_total_stopwords(x))
train_df['q1_total_stopwords'].value_counts().plot(kind='bar')
train_df['q2_total_stopwords'] = train_df['question2'].apply(lambda x: find_total_stopwords(x))
train_df['q2_total_stopwords'].value_counts().plot(kind='bar')
# finding the unigrams in both question 1 and question 2
train_df['unigrams_question1'] = train_df['question1'].apply(lambda x: get_unigrams(x))
train_df['unigrams_question2'] = train_df['question2'].apply(lambda x: get_unigrams(x))
train_df.head()
# finding the common unigram count
train_df['common_unigram_count'] = train_df.apply(lambda row: get_common_unigrams(row['unigrams_question1'],
row['unigrams_question2']), axis=1)
train_df['common_unigram_count'].value_counts().plot(kind='bar')
# finding the common unigram ratio
train_df['common_unigram_ratio'] = train_df.apply(lambda row: get_common_unigram_ratio(row['unigrams_question1'],
row['unigrams_question2']), axis=1)
train_df['common_unigram_ratio'].head()
# comparision between common unigram count and is_duplicate
plt.figure(figsize=(8, 6))
sns.boxplot(x='is_duplicate', y='common_unigram_count', data=train_df)
plt.xlabel('Is Duplicate', fontsize=12)
plt.ylabel('Common Unigram Count', fontsize=12)
plt.show()
# comparision between common unigram ratio and is_duplicate
plt.figure(figsize=(8, 6))
sns.boxplot(x='is_duplicate', y='common_unigram_ratio', data=train_df)
plt.xlabel('Is Duplicate', fontsize=12)
plt.ylabel('Common Unigram Ratio', fontsize=12)
plt.show()
# calculating weighted word match share
train_df['word_share'] = train_df.apply(lambda row: get_similarity(text_to_vector(row['question1']),
text_to_vector(row['question2'])), axis=1)
train_df['word_share'].head()
# getting all the data
train_df.head()
train_df.info()
train_df.describe()
# comparision on is_duplicate and word share
plt.figure(figsize=(12, 6))
plt.subplot(1, 2, 1)
sns.violinplot(x='is_duplicate', y='word_share', data=train_df[0:5000])
plt.subplot(1, 2, 2)
sns.distplot(train_df[train_df['is_duplicate'] == 1.0]['word_share'][0:10000], color='green')
sns.distplot(train_df[train_df['is_duplicate'] == 0.0]['word_share'][0:10000], color='red')
plt.figure(figsize=(15, 5))
plt.hist(train_df['word_share'][train_df['is_duplicate'] == 0], bins=20, normed=True, label='Not Duplicated')
plt.hist(train_df['word_share'][train_df['is_duplicate'] == 1], bins=20, normed=True, alpha=0.7, label='Duplicated')
plt.legend()
plt.title('Label distribution over word share', fontsize=15)
plt.xlabel('Word share', fontsize=15)
# checking the correlation amoung columns
corr = train_df.corr()
corr
plt.figure(figsize=(16, 10))
sns.heatmap(corr, annot=True, xticklabels=corr.columns.values, yticklabels=corr.columns.values)
plt.show()
numeric_training_df = train_df[['q1_length', 'q2_length', 'q1_total_words', 'q2_total_words', 'is_duplicate']]
sns.pairplot(data=numeric_training_df[0:10000], hue='is_duplicate')
```
# Model Building
```
# Generating basic model using Logistic Regression
# We can treat this model as a base line model - for this model we're using only numerical variable
# importing model building libraries
import sklearn
from sklearn import preprocessing
from sklearn import model_selection
from sklearn import linear_model
from sklearn import metrics
basic_train_df = train_df[['q1_length', 'q2_length', 'q1_total_words', 'q2_total_words', 'word_share']]
basic_train_df.head()
# creating scalar object
scalar = preprocessing.StandardScaler()
X = scalar.fit_transform(basic_train_df)
y = train_df['is_duplicate']
# Splitting the data into training and test
X_train, X_test, y_train, y_test = model_selection.train_test_split(X, y, test_size=0.30, random_state=42)
print(X_train.shape)
print(y_train.shape)
print(X_test.shape)
print(y_train.shape)
# Creating logistic regression instance
lr_clf = linear_model.LogisticRegression()
# creating grid parameters
n_folds = 5
parameters = {
'C': [0.001, 0.01, 1, 10, 100, 1000],
'penalty': ['l1', 'l2']
}
model_cv = model_selection.GridSearchCV(estimator=lr_clf,
param_grid=parameters,
scoring='neg_log_loss',
cv=n_folds,
verbose=1,
n_jobs=1,
return_train_score=1)
model_cv.fit(X_train, y_train)
cv_results = pd.DataFrame(model_cv.cv_results_)
cv_results.head()
# plot of C versus train and test scores
plt.figure(figsize=(8, 6))
plt.plot(cv_results['param_C'], cv_results['mean_test_score'])
plt.plot(cv_results['param_C'], cv_results['mean_train_score'])
plt.xlabel('C')
plt.ylabel('Neg Log Loss')
plt.legend(['test accuracy', 'train accuracy'], loc='bottom right')
plt.xscale('log')
plt.show()
best_score = model_cv.best_score_
best_params = model_cv.best_params_
best_estimator = model_cv.best_estimator_
print('Best score: ', best_score)
print('Best parameters: ', best_params)
print('Best estimator: ', best_estimator)
lr = linear_model.LogisticRegression(C=best_params.get('C'), penalty=best_params.get('penalty'))
lr.fit(X_train, y_train)
# prediction
y_pred = lr.predict(X_test)
print('Model accuracy: ', metrics.accuracy_score(y_true=y_test, y_pred=y_pred))
print('Confusion matrix')
print(metrics.confusion_matrix(y_true=y_test, y_pred=y_pred))
print('Classification Report')
print(metrics.classification_report(y_true=y_test, y_pred=y_pred))
print('Accuracy of the logistic regression model for training set: {}'.format(lr.score(X=X, y=y)))
# including graph based features
all_questions = pd.concat([train_df[['question1', 'question2']],
test_df[['question1', 'question2']]], axis=0).reset_index(drop='index')
all_questions.head()
from collections import defaultdict
question_dict = defaultdict(set)
for i in range(all_questions.shape[0]):
question_dict[all_questions.question1[i]].add(all_questions.question2[i])
question_dict[all_questions.question2[i]].add(all_questions.question1[i])
set(question_dict[train_df['question1'][15]])
# utiiltiy functions to calculate word frequencies in both question 1 and question 2
def question1_frequency(row):
return len(question_dict[row['question1']])
def question2_frequency(row):
return len(question_dict[row['question2']])
def questions_intersection(row):
return len(set(question_dict[row['question1']]).intersection(set(question_dict[row['question2']])))
train_df['question1_freq'] = train_df.apply(lambda row: question1_frequency(row), axis=1, raw=True)
train_df['question2_freq'] = train_df.apply(lambda row: question2_frequency(row), axis=1, raw=True)
train_df['q1_q2_intersect'] = train_df.apply(lambda row: questions_intersection(row), axis=1, raw=True)
cnt_srs = train_df['q1_q2_intersect'].value_counts()
plt.figure(figsize=(12, 6))
sns.barplot(cnt_srs.index, np.log(cnt_srs.values), alpha=0.8)
plt.xlabel('Q1-Q2 neighbor intersection count', fontsize=12)
plt.ylabel('Log of number of occurences', fontsize=12)
plt.xticks(rotation='vertical')
plt.show()
grouped_df = train_df.groupby('q1_q2_intersect')['is_duplicate'].aggregate(np.mean).reset_index()
plt.figure(figsize=(12, 8))
sns.pointplot(grouped_df['q1_q2_intersect'].values,
grouped_df['is_duplicate'].values,
alpha=0.8)
plt.xlabel('Q1-Q2 neighbor intersection count', fontsize=12)
plt.ylabel('Mean is_duplicate', fontsize=12)
plt.xticks(rotation='vertical')
plt.show()
pvt_df = train_df.pivot_table(index='question1_freq',
columns='question2_freq',
values='is_duplicate')
plt.figure(figsize=(12, 12))
sns.heatmap(pvt_df)
plt.title('Mean is_duplicate across question1 and question2 frequency')
plt.show()
columns = ['question1_freq', 'question2_freq', 'q1_q2_intersect', 'is_duplicate']
temp_df = train_df[columns]
corr = temp_df.corr(method='spearman')
corr
# plotting the correlation matrix between different frequencies
sns.heatmap(corr, vmax=1, square=True, annot=True)
plt.title('Leaky variables correlation map', fontsize=15)
plt.show()
```
**Advanced Feature Engineering**
*below are the utility functions which calcuate some advanced features in our data set*
```
# method to calculate cosine similarity between two sentences
def stem_tokens(sentence):
"""method to stem the given list of words and return the same"""
if sentence is np.NAN:
sentence = ""
else:
sentence = str(sentence).lower()
stemmer = nltk.stem.porter.PorterStemmer()
tokens = nltk.word_tokenize(sentence)
stemmed_tokens = [stemmer.stem(item) for item in tokens]
return " ".join(stemmed_tokens)
# calculating the cosine similiarity
train_df['q1_q2_cosine_similarity'] = train_df.apply(lambda row: get_similarity(text_to_vector(stem_tokens(row['question1'])), text_to_vector(stem_tokens(row['question2']))), axis=1)
train_df.head()
# calculating Jaccard distance
def get_jaccard_distance(sentence1, sentence2):
"""method to calculate the jaccard distance between two strings and return the same"""
sentence1_lwr = str(sentence1).lower()
sentence2_lwr = str(sentence2).lower()
sentence1_tokens = set(nltk.word_tokenize(sentence1_lwr))
sentence2_tokens = set(nltk.word_tokenize(sentence2_lwr))
return nltk.jaccard_distance(sentence1_tokens, sentence2_tokens)
# calculating the jaccard distance
train_df['q1_q2_jaccard_distance'] = train_df.apply(lambda row: get_jaccard_distance(row['question1'], row['question2']), axis=1)
train_df.head()
# calculating similarity using sequence matcher
import difflib
def get_similarity_sm(sentence1, sentence2):
"""method to get the similarity ratio using python difflib.SequenceMatcher"""
sentence1_lwr = str(sentence1).lower()
sentence2_lwr = str(sentence2).lower()
return difflib.SequenceMatcher(None, sentence1_lwr, sentence2_lwr).ratio() * 100
# calculating the similarity measure using sequence matcher
train_df['similarity'] = train_df.apply(lambda row: get_similarity_sm(row['question1'], row['question2']), axis=1)
train_df.head()
# calculate the alphabet count from the given sentence
def get_alphabet_count(sentence):
"""method to get alphabet count from removing all the numbers, spl., characters etc., and return the same"""
sentence_lwr = str(sentence).lower()
return len(re.sub('[^a-z]+', '', sentence_lwr))
train_df['q1_alphabet_count'] = train_df['question1'].apply(lambda x: get_alphabet_count(x))
train_df['q2_alphabet_count'] = train_df['question2'].apply(lambda x: get_alphabet_count(x))
train_df.head()
# method for decision maker
def is_equal(sentence1, sentence2):
"""method to check whether the given sentence are exactly match or not"""
sentence1_lwr = str(sentence1).lower()
sentence2_lwr = str(sentence2).lower()
if sentence1_lwr == sentence2_lwr:
return 1
else:
return 0
train_df['is_equal'] = train_df.apply(lambda row: is_equal(row['question1'], row['question2']), axis=1)
train_df.head()
# get_common_unigrams(get_unigrams('Programming in Java, 3rd edition'), get_unigrams('Programming in scala, 2nd edition'))
def get_matched_token_count(tokens, match_words):
"""methdo to compute total number of matching words in the given string"""
counter = 0
for token in list(tokens):
if token in match_words:
counter += 1
return counter
def get_word_match_share(sentence1, sentence2):
"""method to compute the word match share in the given sentences and return the same."""
sentence1_lwr = str(sentence1).lower()
sentence2_lwr = str(sentence2).lower()
sentence1_unigrams = get_unigrams(sentence1_lwr)
sentence2_unigrams = get_unigrams(sentence2_lwr)
match_words = set(sentence1_unigrams).intersection(set(sentence2_unigrams))
sentence1_tokens = nltk.word_tokenize(sentence1_lwr)
sentence2_tokens = nltk.word_tokenize(sentence2_lwr)
sentence1_matched_length = get_matched_token_count(sentence1_tokens, match_words)
sentence2_matched_length = get_matched_token_count(sentence2_tokens, match_words)
question1_word_length = len(sentence1_unigrams)
question2_word_length = len(sentence2_unigrams)
return ((sentence1_matched_length + sentence2_matched_length)* 1.0) / (question1_word_length + question2_word_length)
train_df['word_match_shre'] = train_df.apply(lambda row: get_word_match_share(row['question1'], row['question2']), axis=1)
train_df.head()
# computing the tf-idf word match share for the given sentences
def computeTF(wordDict, bow):
tfDict = {}
bowCount = len(bow)
for word, count in wordDict.items():
tfDict[word] = count / float(bowCount)
return tfDict
def computeIDF(docList):
import math
idfDict = {}
N = len(docList)
idfDict = dict.fromkeys(docList[0].keys(), 0)
for doc in docList:
for word, val in doc.items():
if val > 0:
idfDict[word] += 1
for word, val in idfDict.items():
idfDict[word] = math.log10(N / float(val))
return idfDict
def computeTFIDF(tfBow, idfs):
tfidf = {}
for word, val in tfBow.items():
tfidf[word] = val * idfs[word]
return tfidf
def get_score(sentence1, sentence2):
sentence1_lwr = ""
sentence2_lwr = ""
if sentence1 is not np.NaN:
sentence1_lwr = str(sentence1).lower()
else:
sentence1_lwr = "?"
if sentence2 is not np.NaN:
sentence2_lwr = str(sentence2).lower()
else:
sentence2_lwr = "?"
sentence1_words = sentence1_lwr.split()
sentence2_words = sentence2_lwr.split()
word_set = set(sentence1_words).union(set(sentence2_words))
sentence1_dictionary = dict.fromkeys(word_set, 0)
sentence2_dictionary = dict.fromkeys(word_set, 0)
for word in sentence1_words:
sentence1_dictionary[word] += 1
for word in sentence2_words:
sentence2_dictionary[word] += 1
sentence1_tf = computeTF(sentence1_dictionary, sentence1_words)
sentence2_tf = computeTF(sentence2_dictionary, sentence2_words)
idf = computeIDF([sentence1_dictionary, sentence2_dictionary])
return computeTFIDF(sentence1_tf, idf), computeTFIDF(sentence2_tf, idf)
# computing tf-idf metrics for question1
def get_result_list(dictionary):
result = []
for d in dictionary:
result.append(int(d))
return result
score = train_df.apply(lambda row: get_score(row['question1'], row['question2']), axis=1)
train_df['question1_tf_idf_sum'] = score.apply(lambda x: np.sum(get_result_list(x[0].values())))
train_df['question1_tf_idf_mean'] = score.apply(lambda x: np.mean(get_result_list(x[0].values())))
train_df['question1_tf_idf_min'] = score.apply(lambda x: np.min(get_result_list(x[0].values())))
train_df['question1_tf_idf_max'] = score.apply(lambda x: np.max(get_result_list(x[0].values())))
train_df.head()
train_df['question2_tf_idf_sum'] = score.apply(lambda x: np.sum(get_result_list(x[1].values())))
train_df['question2_tf_idf_mean'] = score.apply(lambda x: np.mean(get_result_list(x[1].values())))
train_df['question2_tf_idf_min'] = score.apply(lambda x: np.min(get_result_list(x[1].values())))
train_df['question2_tf_idf_max'] = score.apply(lambda x: np.max(get_result_list(x[1].values())))
train_df.head()
# computing the bigrams
from nltk.util import ngrams
def find_ngrams(sentence, weight):
"""method to find the bigrams in the given sentence"""
tokens = nltk.word_tokenize(sentence)
return set(ngrams(tokens, weight))
def get_shared_ngrams(sentence1, sentence2, weight):
"""method to find the common bigrams in the given sentence and return the same"""
if sentence1 is not np.NAN:
sentence1 = str(sentence1).lower()
else:
sentence1 = ""
if sentence2 is not np.NAN:
sentence2 = str(sentence2).lower()
else:
sentence2 = ""
sentence1_bigrams = find_ngrams(sentence1, weight)
sentence2_bigrams = find_ngrams(sentence2, weight)
shared_bigrams = sentence1_bigrams.intersection(sentence2_bigrams)
if len(shared_bigrams) == 0:
return 0
else:
return (len(shared_bigrams) * 1.0) / (len(sentence1_bigrams) + len(sentence2_bigrams))
# computing the common bigrams in the question1 & question2
train_df['shared_bigrams'] = train_df.apply(lambda row: get_shared_ngrams(row['question1'], row['question2'], 2), axis=1)
train_df.head()
# computing the common trigrams in the question1 & question2
train_df['shared_trigrams'] = train_df.apply(lambda row: get_shared_ngrams(row['question1'], row['question2'], 3), axis=1)
train_df.head()
# finding the avg_word_length_diff
def get_average_word_length_difference(sentence1, sentence2):
"""method to find the difference between the average length of given sentences"""
if sentence1 is np.NAN or sentence2 is np.NAN:
return 0
else:
sentence1_avg_word_length = find_length(sentence1) / find_total_words(sentence1)
sentence2_avg_word_length = find_length(sentence2) / find_total_words(sentence2)
return sentence1_avg_word_length - sentence2_avg_word_length
# calculating average lenght of question1 and question2
train_df['avg_word_length_diff'] = train_df.apply(lambda row: get_average_word_length_difference(row['question1'], row['question2']), axis=1)
train_df.head()
# method to find the average stopwords in the given sentences
def avg_stop_words(sentence1, sentence2):
"""method to find the average stopwords in given sentences and return the same"""
sentence1_lwr = ""
sentence2_lwr = ""
if sentence1 is np.NAN or sentence2 is np.NAN:
return 0
else:
sentence1_lwr = str(sentence1).lower()
sentence2_lwr = str(sentence2).lower()
sentence1_stop_words = find_total_stopwords(sentence1_lwr)
sentence2_stop_words = find_total_stopwords(sentence2_lwr)
return (((sentence1_stop_words * 1.0) / find_total_words(sentence1_lwr)) - ((sentence2_stop_words * 1.0) / find_total_words(sentence2_lwr)))
# calculating average lenght of stop words of question1 and question2
train_df['avg_stop_word_length_diff'] = train_df.apply(lambda row: avg_stop_words(row['question1'], row['question2']), axis=1)
train_df.head()
# method to calculate the noun count difference between two sentences
from textblob import TextBlob
def find_noun_count_difference(sentence1, sentence2):
"""method to find the noun count difference between given sentences and return the same"""
sentence1_lwr = ""
sentence2_lwr = ""
if sentence1 is np.NAN or sentence2 is np.NAN:
return 0
else:
sentence1_lwr = str(sentence1).lower()
sentence2_lwr = str(sentence2).lower()
sentence1_blob = TextBlob(sentence1_lwr)
sentence2_blob = TextBlob(sentence2_lwr)
sentence1_noun_count = len(sentence1_blob.noun_phrases)
sentence2_noun_count = len(sentence2_blob.noun_phrases)
return sentence1_noun_count - sentence2_noun_count
# calculating noun count difference between given question1 and question2
train_df['noun_count_diff'] = train_df.apply(lambda row: find_noun_count_difference(row['question1'], row['question2']), axis=1)
train_df.head()
# finding capital letters ratio for two given strings
def find_capital_letters_diff(sentence1, sentence2):
"""method to find the capital letters count difference for the given sentences and return the same"""
if sentence1 is np.NAN:
sentence1 = ""
if sentence2 is np.NAN:
sentence2 = ""
sentence1_tokens = nltk.word_tokenize(sentence1)
sentence2_tokens = nltk.word_tokenize(sentence2)
sentence1_capital_count = len([token for token in sentence1_tokens if token.istitle()])
sentence2_capital_count = len([token for token in sentence2_tokens if token.istitle()])
return sentence1_capital_count - sentence2_capital_count
# calculating capital letter count difference between given question1 and question2
train_df['capital_count_diff'] = train_df.apply(lambda row: find_capital_letters_diff(row['question1'], row['question2']), axis=1)
train_df.head()
```
**Model Building - using new data**
```
# creating the model using newly created variables
train_df.info()
X = train_df.drop(["question1", "question2", "unigrams_question1", "unigrams_question2","is_duplicate"], axis=1)
y = train_df['is_duplicate']
print(X.shape)
print(y.shape)
# standardising the data
scalar = preprocessing.StandardScaler()
X = scalar.fit_transform(X)
# Splitting the data into training and test
X_train, X_test, y_train, y_test = model_selection.train_test_split(X, y, test_size=0.30, random_state=42)
print(X_train.shape)
print(y_train.shape)
print(X_test.shape)
print(y_train.shape)
lr_clf = linear_model.LogisticRegression()
# creating grid parameters
n_folds = 5
parameters = {
'C': [0.001, 0.01, 1, 10, 100, 1000],
'penalty': ['l1', 'l2']
}
model_cv = model_selection.GridSearchCV(estimator=lr_clf,
param_grid=parameters,
scoring='neg_log_loss',
cv=n_folds,
verbose=1,
n_jobs=1,
return_train_score=1)
model_cv.fit(X_train, y_train)
cv_results = pd.DataFrame(model_cv.cv_results_)
cv_results.head()
# plot of C versus train and test scores
plt.figure(figsize=(8, 6))
plt.plot(cv_results['param_C'], cv_results['mean_test_score'])
plt.plot(cv_results['param_C'], cv_results['mean_train_score'])
plt.xlabel('C')
plt.ylabel('Neg Log Loss')
plt.legend(['test accuracy', 'train accuracy'], loc='bottom right')
plt.xscale('log')
plt.show()
best_score = model_cv.best_score_
best_params = model_cv.best_params_
best_estimator = model_cv.best_estimator_
print('Best score: ', best_score)
print('Best parameters: ', best_params)
print('Best estimator: ', best_estimator)
lr = linear_model.LogisticRegression(C=best_params.get('C'), penalty=best_params.get('penalty'))
lr.fit(X_train, y_train)
# prediction
y_pred = lr.predict(X_test)
print('Model accuracy: ', metrics.accuracy_score(y_true=y_test, y_pred=y_pred))
print('Confusion matrix')
print(metrics.confusion_matrix(y_true=y_test, y_pred=y_pred))
print('Classification Report')
print(metrics.classification_report(y_true=y_test, y_pred=y_pred))
print('Accuracy of the logistic regression model for training set: {}'.format(lr.score(X=X, y=y)))
```
|
github_jupyter
|
# This Python 3 environment comes with many helpful analytics libraries installed
# It is defined by the kaggle/python docker image: https://github.com/kaggle/docker-python
# For example, here's several helpful packages to load in
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory
import os
print(os.listdir("../input"))
# Any results you write to the current directory are saved as output.
# importing packages
import warnings
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
warnings.filterwarnings('ignore')
train_df = pd.read_csv("../input/train.csv")
train_df.head()
test_df = pd.read_csv("../input/test.csv")
test_df.head()
# checking the summary of the data
train_df.info()
# checking the basic statistics on numeric column
train_df.describe()
# checking the null columns
train_df.isnull().sum()
# sum it up to check how many rows have all missing values
train_df.isnull().all(axis = 1).sum()
# summing up the missing values (column-wise) in percentage
round(100 * (train_df.isnull().sum() / len(train_df.index)), 2)
# Basic preprocessing, Feature extraction and EDA
# Some basic EDA
# looking at target variable
is_duplicate = train_df['is_duplicate'].value_counts()
is_duplicate.plot(kind='bar')
# is_duplicate ratio / checking the data is balance or not
data_ratio = is_duplicate / is_duplicate.sum()
data_ratio * 100
# importing nltk packages
import math
import re
import nltk
from collections import Counter
from nltk.corpus import stopwords
from nltk import (word_tokenize, ngrams)
WORD = re.compile(r'\w+')
english_stopwords = set(stopwords.words('english'))
# checking the total words in the question
def find_total_words(element):
"""method to find the number of words in the question"""
return len(str(element).split())
def find_length(element):
"method to find the length of question in the question"
return len(str(element))
def find_total_sentences(element):
"method to find the total number of sentences in the question"
return len(str(element).split("."))
def find_total_stopwords(element):
"method to find total number of stopwords in the question"
total_words = str(element).split()
return len([word for word in total_words if word in english_stopwords])
def get_unigrams(element):
"""method to find and return available unigrams in the question"""
lower_str = str(element).lower()
total_words = word_tokenize(lower_str)
total_unigrams = [word for word in total_words if word not in english_stopwords]
return total_unigrams
def get_common_unigrams(unigram_question1, unigram_question2):
"""method to find and return the common unigrams found in both question1 and question2"""
return len(set(unigram_question1).intersection(set(unigram_question2)))
def get_common_unigram_ratio(unigram_question1, unigram_question2):
"method to find and return common unigram ratio for question1 and question"
count = get_common_unigrams(unigram_question1, unigram_question2)
return float(count) / max(len(set(get_unigrams(unigram_question1)).union(set(get_unigrams(unigram_question2)))), 1)
def get_similarity(vector1, vector2):
"""method to compute the cosine similarity between question1 and question2"""
intersection = set(vector1.keys()) & set(vector2.keys())
numerator = sum([vector1[x] * vector2[x] for x in intersection])
sum1 = sum([vector1[x] ** 2 for x in vector1.keys()])
sum2 = sum([vector2[x] ** 2 for x in vector2.keys()])
denominator = math.sqrt(sum1) * math.sqrt(sum2)
if not denominator:
return 0.0
else:
return float(numerator) / denominator
def text_to_vector(sentence):
if sentence is np.NAN:
sentence = ""
words = WORD.findall(sentence)
return Counter(words)
# Finding the number of words in the question
train_df['q1_total_words'] = train_df['question1'].apply(lambda x: find_total_words(x))
train_df['q1_total_words'].value_counts().plot(kind='bar')
train_df['q2_total_words'] = train_df['question2'].apply(lambda x: find_total_words(x))
train_df['q2_total_words'].value_counts().plot(kind='bar')
# Finding the length of the question
train_df['q1_length'] = train_df['question1'].apply(lambda x: find_length(x))
train_df['q1_length'].value_counts().plot(kind='bar')
train_df['q2_length'] = train_df['question2'].apply(lambda x: find_length(x))
train_df['q2_length'].value_counts().plot(kind='bar')
# finding the total words distribution on both question 1 and question 2
train_df['all_question_length'] = train_df.apply(lambda row: find_length(row['question1']) +
find_length(row['question2']), axis=1)
train_df['all_question_length'].value_counts().plot(kind='bar')
# Finding the total sentences of the question
train_df['q1_sentence_length'] = train_df['question1'].apply(lambda x: find_total_sentences(x))
train_df['q1_sentence_length'].value_counts().plot(kind='bar')
train_df['q2_sentence_length'] = train_df['question2'].apply(lambda x: find_total_sentences(x))
train_df['q2_sentence_length'].value_counts().plot(kind='bar')
# Finding the number of stop words in the question
train_df['q1_total_stopwords'] = train_df['question1'].apply(lambda x: find_total_stopwords(x))
train_df['q1_total_stopwords'].value_counts().plot(kind='bar')
train_df['q2_total_stopwords'] = train_df['question2'].apply(lambda x: find_total_stopwords(x))
train_df['q2_total_stopwords'].value_counts().plot(kind='bar')
# finding the unigrams in both question 1 and question 2
train_df['unigrams_question1'] = train_df['question1'].apply(lambda x: get_unigrams(x))
train_df['unigrams_question2'] = train_df['question2'].apply(lambda x: get_unigrams(x))
train_df.head()
# finding the common unigram count
train_df['common_unigram_count'] = train_df.apply(lambda row: get_common_unigrams(row['unigrams_question1'],
row['unigrams_question2']), axis=1)
train_df['common_unigram_count'].value_counts().plot(kind='bar')
# finding the common unigram ratio
train_df['common_unigram_ratio'] = train_df.apply(lambda row: get_common_unigram_ratio(row['unigrams_question1'],
row['unigrams_question2']), axis=1)
train_df['common_unigram_ratio'].head()
# comparision between common unigram count and is_duplicate
plt.figure(figsize=(8, 6))
sns.boxplot(x='is_duplicate', y='common_unigram_count', data=train_df)
plt.xlabel('Is Duplicate', fontsize=12)
plt.ylabel('Common Unigram Count', fontsize=12)
plt.show()
# comparision between common unigram ratio and is_duplicate
plt.figure(figsize=(8, 6))
sns.boxplot(x='is_duplicate', y='common_unigram_ratio', data=train_df)
plt.xlabel('Is Duplicate', fontsize=12)
plt.ylabel('Common Unigram Ratio', fontsize=12)
plt.show()
# calculating weighted word match share
train_df['word_share'] = train_df.apply(lambda row: get_similarity(text_to_vector(row['question1']),
text_to_vector(row['question2'])), axis=1)
train_df['word_share'].head()
# getting all the data
train_df.head()
train_df.info()
train_df.describe()
# comparision on is_duplicate and word share
plt.figure(figsize=(12, 6))
plt.subplot(1, 2, 1)
sns.violinplot(x='is_duplicate', y='word_share', data=train_df[0:5000])
plt.subplot(1, 2, 2)
sns.distplot(train_df[train_df['is_duplicate'] == 1.0]['word_share'][0:10000], color='green')
sns.distplot(train_df[train_df['is_duplicate'] == 0.0]['word_share'][0:10000], color='red')
plt.figure(figsize=(15, 5))
plt.hist(train_df['word_share'][train_df['is_duplicate'] == 0], bins=20, normed=True, label='Not Duplicated')
plt.hist(train_df['word_share'][train_df['is_duplicate'] == 1], bins=20, normed=True, alpha=0.7, label='Duplicated')
plt.legend()
plt.title('Label distribution over word share', fontsize=15)
plt.xlabel('Word share', fontsize=15)
# checking the correlation amoung columns
corr = train_df.corr()
corr
plt.figure(figsize=(16, 10))
sns.heatmap(corr, annot=True, xticklabels=corr.columns.values, yticklabels=corr.columns.values)
plt.show()
numeric_training_df = train_df[['q1_length', 'q2_length', 'q1_total_words', 'q2_total_words', 'is_duplicate']]
sns.pairplot(data=numeric_training_df[0:10000], hue='is_duplicate')
# Generating basic model using Logistic Regression
# We can treat this model as a base line model - for this model we're using only numerical variable
# importing model building libraries
import sklearn
from sklearn import preprocessing
from sklearn import model_selection
from sklearn import linear_model
from sklearn import metrics
basic_train_df = train_df[['q1_length', 'q2_length', 'q1_total_words', 'q2_total_words', 'word_share']]
basic_train_df.head()
# creating scalar object
scalar = preprocessing.StandardScaler()
X = scalar.fit_transform(basic_train_df)
y = train_df['is_duplicate']
# Splitting the data into training and test
X_train, X_test, y_train, y_test = model_selection.train_test_split(X, y, test_size=0.30, random_state=42)
print(X_train.shape)
print(y_train.shape)
print(X_test.shape)
print(y_train.shape)
# Creating logistic regression instance
lr_clf = linear_model.LogisticRegression()
# creating grid parameters
n_folds = 5
parameters = {
'C': [0.001, 0.01, 1, 10, 100, 1000],
'penalty': ['l1', 'l2']
}
model_cv = model_selection.GridSearchCV(estimator=lr_clf,
param_grid=parameters,
scoring='neg_log_loss',
cv=n_folds,
verbose=1,
n_jobs=1,
return_train_score=1)
model_cv.fit(X_train, y_train)
cv_results = pd.DataFrame(model_cv.cv_results_)
cv_results.head()
# plot of C versus train and test scores
plt.figure(figsize=(8, 6))
plt.plot(cv_results['param_C'], cv_results['mean_test_score'])
plt.plot(cv_results['param_C'], cv_results['mean_train_score'])
plt.xlabel('C')
plt.ylabel('Neg Log Loss')
plt.legend(['test accuracy', 'train accuracy'], loc='bottom right')
plt.xscale('log')
plt.show()
best_score = model_cv.best_score_
best_params = model_cv.best_params_
best_estimator = model_cv.best_estimator_
print('Best score: ', best_score)
print('Best parameters: ', best_params)
print('Best estimator: ', best_estimator)
lr = linear_model.LogisticRegression(C=best_params.get('C'), penalty=best_params.get('penalty'))
lr.fit(X_train, y_train)
# prediction
y_pred = lr.predict(X_test)
print('Model accuracy: ', metrics.accuracy_score(y_true=y_test, y_pred=y_pred))
print('Confusion matrix')
print(metrics.confusion_matrix(y_true=y_test, y_pred=y_pred))
print('Classification Report')
print(metrics.classification_report(y_true=y_test, y_pred=y_pred))
print('Accuracy of the logistic regression model for training set: {}'.format(lr.score(X=X, y=y)))
# including graph based features
all_questions = pd.concat([train_df[['question1', 'question2']],
test_df[['question1', 'question2']]], axis=0).reset_index(drop='index')
all_questions.head()
from collections import defaultdict
question_dict = defaultdict(set)
for i in range(all_questions.shape[0]):
question_dict[all_questions.question1[i]].add(all_questions.question2[i])
question_dict[all_questions.question2[i]].add(all_questions.question1[i])
set(question_dict[train_df['question1'][15]])
# utiiltiy functions to calculate word frequencies in both question 1 and question 2
def question1_frequency(row):
return len(question_dict[row['question1']])
def question2_frequency(row):
return len(question_dict[row['question2']])
def questions_intersection(row):
return len(set(question_dict[row['question1']]).intersection(set(question_dict[row['question2']])))
train_df['question1_freq'] = train_df.apply(lambda row: question1_frequency(row), axis=1, raw=True)
train_df['question2_freq'] = train_df.apply(lambda row: question2_frequency(row), axis=1, raw=True)
train_df['q1_q2_intersect'] = train_df.apply(lambda row: questions_intersection(row), axis=1, raw=True)
cnt_srs = train_df['q1_q2_intersect'].value_counts()
plt.figure(figsize=(12, 6))
sns.barplot(cnt_srs.index, np.log(cnt_srs.values), alpha=0.8)
plt.xlabel('Q1-Q2 neighbor intersection count', fontsize=12)
plt.ylabel('Log of number of occurences', fontsize=12)
plt.xticks(rotation='vertical')
plt.show()
grouped_df = train_df.groupby('q1_q2_intersect')['is_duplicate'].aggregate(np.mean).reset_index()
plt.figure(figsize=(12, 8))
sns.pointplot(grouped_df['q1_q2_intersect'].values,
grouped_df['is_duplicate'].values,
alpha=0.8)
plt.xlabel('Q1-Q2 neighbor intersection count', fontsize=12)
plt.ylabel('Mean is_duplicate', fontsize=12)
plt.xticks(rotation='vertical')
plt.show()
pvt_df = train_df.pivot_table(index='question1_freq',
columns='question2_freq',
values='is_duplicate')
plt.figure(figsize=(12, 12))
sns.heatmap(pvt_df)
plt.title('Mean is_duplicate across question1 and question2 frequency')
plt.show()
columns = ['question1_freq', 'question2_freq', 'q1_q2_intersect', 'is_duplicate']
temp_df = train_df[columns]
corr = temp_df.corr(method='spearman')
corr
# plotting the correlation matrix between different frequencies
sns.heatmap(corr, vmax=1, square=True, annot=True)
plt.title('Leaky variables correlation map', fontsize=15)
plt.show()
# method to calculate cosine similarity between two sentences
def stem_tokens(sentence):
"""method to stem the given list of words and return the same"""
if sentence is np.NAN:
sentence = ""
else:
sentence = str(sentence).lower()
stemmer = nltk.stem.porter.PorterStemmer()
tokens = nltk.word_tokenize(sentence)
stemmed_tokens = [stemmer.stem(item) for item in tokens]
return " ".join(stemmed_tokens)
# calculating the cosine similiarity
train_df['q1_q2_cosine_similarity'] = train_df.apply(lambda row: get_similarity(text_to_vector(stem_tokens(row['question1'])), text_to_vector(stem_tokens(row['question2']))), axis=1)
train_df.head()
# calculating Jaccard distance
def get_jaccard_distance(sentence1, sentence2):
"""method to calculate the jaccard distance between two strings and return the same"""
sentence1_lwr = str(sentence1).lower()
sentence2_lwr = str(sentence2).lower()
sentence1_tokens = set(nltk.word_tokenize(sentence1_lwr))
sentence2_tokens = set(nltk.word_tokenize(sentence2_lwr))
return nltk.jaccard_distance(sentence1_tokens, sentence2_tokens)
# calculating the jaccard distance
train_df['q1_q2_jaccard_distance'] = train_df.apply(lambda row: get_jaccard_distance(row['question1'], row['question2']), axis=1)
train_df.head()
# calculating similarity using sequence matcher
import difflib
def get_similarity_sm(sentence1, sentence2):
"""method to get the similarity ratio using python difflib.SequenceMatcher"""
sentence1_lwr = str(sentence1).lower()
sentence2_lwr = str(sentence2).lower()
return difflib.SequenceMatcher(None, sentence1_lwr, sentence2_lwr).ratio() * 100
# calculating the similarity measure using sequence matcher
train_df['similarity'] = train_df.apply(lambda row: get_similarity_sm(row['question1'], row['question2']), axis=1)
train_df.head()
# calculate the alphabet count from the given sentence
def get_alphabet_count(sentence):
"""method to get alphabet count from removing all the numbers, spl., characters etc., and return the same"""
sentence_lwr = str(sentence).lower()
return len(re.sub('[^a-z]+', '', sentence_lwr))
train_df['q1_alphabet_count'] = train_df['question1'].apply(lambda x: get_alphabet_count(x))
train_df['q2_alphabet_count'] = train_df['question2'].apply(lambda x: get_alphabet_count(x))
train_df.head()
# method for decision maker
def is_equal(sentence1, sentence2):
"""method to check whether the given sentence are exactly match or not"""
sentence1_lwr = str(sentence1).lower()
sentence2_lwr = str(sentence2).lower()
if sentence1_lwr == sentence2_lwr:
return 1
else:
return 0
train_df['is_equal'] = train_df.apply(lambda row: is_equal(row['question1'], row['question2']), axis=1)
train_df.head()
# get_common_unigrams(get_unigrams('Programming in Java, 3rd edition'), get_unigrams('Programming in scala, 2nd edition'))
def get_matched_token_count(tokens, match_words):
"""methdo to compute total number of matching words in the given string"""
counter = 0
for token in list(tokens):
if token in match_words:
counter += 1
return counter
def get_word_match_share(sentence1, sentence2):
"""method to compute the word match share in the given sentences and return the same."""
sentence1_lwr = str(sentence1).lower()
sentence2_lwr = str(sentence2).lower()
sentence1_unigrams = get_unigrams(sentence1_lwr)
sentence2_unigrams = get_unigrams(sentence2_lwr)
match_words = set(sentence1_unigrams).intersection(set(sentence2_unigrams))
sentence1_tokens = nltk.word_tokenize(sentence1_lwr)
sentence2_tokens = nltk.word_tokenize(sentence2_lwr)
sentence1_matched_length = get_matched_token_count(sentence1_tokens, match_words)
sentence2_matched_length = get_matched_token_count(sentence2_tokens, match_words)
question1_word_length = len(sentence1_unigrams)
question2_word_length = len(sentence2_unigrams)
return ((sentence1_matched_length + sentence2_matched_length)* 1.0) / (question1_word_length + question2_word_length)
train_df['word_match_shre'] = train_df.apply(lambda row: get_word_match_share(row['question1'], row['question2']), axis=1)
train_df.head()
# computing the tf-idf word match share for the given sentences
def computeTF(wordDict, bow):
tfDict = {}
bowCount = len(bow)
for word, count in wordDict.items():
tfDict[word] = count / float(bowCount)
return tfDict
def computeIDF(docList):
import math
idfDict = {}
N = len(docList)
idfDict = dict.fromkeys(docList[0].keys(), 0)
for doc in docList:
for word, val in doc.items():
if val > 0:
idfDict[word] += 1
for word, val in idfDict.items():
idfDict[word] = math.log10(N / float(val))
return idfDict
def computeTFIDF(tfBow, idfs):
tfidf = {}
for word, val in tfBow.items():
tfidf[word] = val * idfs[word]
return tfidf
def get_score(sentence1, sentence2):
sentence1_lwr = ""
sentence2_lwr = ""
if sentence1 is not np.NaN:
sentence1_lwr = str(sentence1).lower()
else:
sentence1_lwr = "?"
if sentence2 is not np.NaN:
sentence2_lwr = str(sentence2).lower()
else:
sentence2_lwr = "?"
sentence1_words = sentence1_lwr.split()
sentence2_words = sentence2_lwr.split()
word_set = set(sentence1_words).union(set(sentence2_words))
sentence1_dictionary = dict.fromkeys(word_set, 0)
sentence2_dictionary = dict.fromkeys(word_set, 0)
for word in sentence1_words:
sentence1_dictionary[word] += 1
for word in sentence2_words:
sentence2_dictionary[word] += 1
sentence1_tf = computeTF(sentence1_dictionary, sentence1_words)
sentence2_tf = computeTF(sentence2_dictionary, sentence2_words)
idf = computeIDF([sentence1_dictionary, sentence2_dictionary])
return computeTFIDF(sentence1_tf, idf), computeTFIDF(sentence2_tf, idf)
# computing tf-idf metrics for question1
def get_result_list(dictionary):
result = []
for d in dictionary:
result.append(int(d))
return result
score = train_df.apply(lambda row: get_score(row['question1'], row['question2']), axis=1)
train_df['question1_tf_idf_sum'] = score.apply(lambda x: np.sum(get_result_list(x[0].values())))
train_df['question1_tf_idf_mean'] = score.apply(lambda x: np.mean(get_result_list(x[0].values())))
train_df['question1_tf_idf_min'] = score.apply(lambda x: np.min(get_result_list(x[0].values())))
train_df['question1_tf_idf_max'] = score.apply(lambda x: np.max(get_result_list(x[0].values())))
train_df.head()
train_df['question2_tf_idf_sum'] = score.apply(lambda x: np.sum(get_result_list(x[1].values())))
train_df['question2_tf_idf_mean'] = score.apply(lambda x: np.mean(get_result_list(x[1].values())))
train_df['question2_tf_idf_min'] = score.apply(lambda x: np.min(get_result_list(x[1].values())))
train_df['question2_tf_idf_max'] = score.apply(lambda x: np.max(get_result_list(x[1].values())))
train_df.head()
# computing the bigrams
from nltk.util import ngrams
def find_ngrams(sentence, weight):
"""method to find the bigrams in the given sentence"""
tokens = nltk.word_tokenize(sentence)
return set(ngrams(tokens, weight))
def get_shared_ngrams(sentence1, sentence2, weight):
"""method to find the common bigrams in the given sentence and return the same"""
if sentence1 is not np.NAN:
sentence1 = str(sentence1).lower()
else:
sentence1 = ""
if sentence2 is not np.NAN:
sentence2 = str(sentence2).lower()
else:
sentence2 = ""
sentence1_bigrams = find_ngrams(sentence1, weight)
sentence2_bigrams = find_ngrams(sentence2, weight)
shared_bigrams = sentence1_bigrams.intersection(sentence2_bigrams)
if len(shared_bigrams) == 0:
return 0
else:
return (len(shared_bigrams) * 1.0) / (len(sentence1_bigrams) + len(sentence2_bigrams))
# computing the common bigrams in the question1 & question2
train_df['shared_bigrams'] = train_df.apply(lambda row: get_shared_ngrams(row['question1'], row['question2'], 2), axis=1)
train_df.head()
# computing the common trigrams in the question1 & question2
train_df['shared_trigrams'] = train_df.apply(lambda row: get_shared_ngrams(row['question1'], row['question2'], 3), axis=1)
train_df.head()
# finding the avg_word_length_diff
def get_average_word_length_difference(sentence1, sentence2):
"""method to find the difference between the average length of given sentences"""
if sentence1 is np.NAN or sentence2 is np.NAN:
return 0
else:
sentence1_avg_word_length = find_length(sentence1) / find_total_words(sentence1)
sentence2_avg_word_length = find_length(sentence2) / find_total_words(sentence2)
return sentence1_avg_word_length - sentence2_avg_word_length
# calculating average lenght of question1 and question2
train_df['avg_word_length_diff'] = train_df.apply(lambda row: get_average_word_length_difference(row['question1'], row['question2']), axis=1)
train_df.head()
# method to find the average stopwords in the given sentences
def avg_stop_words(sentence1, sentence2):
"""method to find the average stopwords in given sentences and return the same"""
sentence1_lwr = ""
sentence2_lwr = ""
if sentence1 is np.NAN or sentence2 is np.NAN:
return 0
else:
sentence1_lwr = str(sentence1).lower()
sentence2_lwr = str(sentence2).lower()
sentence1_stop_words = find_total_stopwords(sentence1_lwr)
sentence2_stop_words = find_total_stopwords(sentence2_lwr)
return (((sentence1_stop_words * 1.0) / find_total_words(sentence1_lwr)) - ((sentence2_stop_words * 1.0) / find_total_words(sentence2_lwr)))
# calculating average lenght of stop words of question1 and question2
train_df['avg_stop_word_length_diff'] = train_df.apply(lambda row: avg_stop_words(row['question1'], row['question2']), axis=1)
train_df.head()
# method to calculate the noun count difference between two sentences
from textblob import TextBlob
def find_noun_count_difference(sentence1, sentence2):
"""method to find the noun count difference between given sentences and return the same"""
sentence1_lwr = ""
sentence2_lwr = ""
if sentence1 is np.NAN or sentence2 is np.NAN:
return 0
else:
sentence1_lwr = str(sentence1).lower()
sentence2_lwr = str(sentence2).lower()
sentence1_blob = TextBlob(sentence1_lwr)
sentence2_blob = TextBlob(sentence2_lwr)
sentence1_noun_count = len(sentence1_blob.noun_phrases)
sentence2_noun_count = len(sentence2_blob.noun_phrases)
return sentence1_noun_count - sentence2_noun_count
# calculating noun count difference between given question1 and question2
train_df['noun_count_diff'] = train_df.apply(lambda row: find_noun_count_difference(row['question1'], row['question2']), axis=1)
train_df.head()
# finding capital letters ratio for two given strings
def find_capital_letters_diff(sentence1, sentence2):
"""method to find the capital letters count difference for the given sentences and return the same"""
if sentence1 is np.NAN:
sentence1 = ""
if sentence2 is np.NAN:
sentence2 = ""
sentence1_tokens = nltk.word_tokenize(sentence1)
sentence2_tokens = nltk.word_tokenize(sentence2)
sentence1_capital_count = len([token for token in sentence1_tokens if token.istitle()])
sentence2_capital_count = len([token for token in sentence2_tokens if token.istitle()])
return sentence1_capital_count - sentence2_capital_count
# calculating capital letter count difference between given question1 and question2
train_df['capital_count_diff'] = train_df.apply(lambda row: find_capital_letters_diff(row['question1'], row['question2']), axis=1)
train_df.head()
# creating the model using newly created variables
train_df.info()
X = train_df.drop(["question1", "question2", "unigrams_question1", "unigrams_question2","is_duplicate"], axis=1)
y = train_df['is_duplicate']
print(X.shape)
print(y.shape)
# standardising the data
scalar = preprocessing.StandardScaler()
X = scalar.fit_transform(X)
# Splitting the data into training and test
X_train, X_test, y_train, y_test = model_selection.train_test_split(X, y, test_size=0.30, random_state=42)
print(X_train.shape)
print(y_train.shape)
print(X_test.shape)
print(y_train.shape)
lr_clf = linear_model.LogisticRegression()
# creating grid parameters
n_folds = 5
parameters = {
'C': [0.001, 0.01, 1, 10, 100, 1000],
'penalty': ['l1', 'l2']
}
model_cv = model_selection.GridSearchCV(estimator=lr_clf,
param_grid=parameters,
scoring='neg_log_loss',
cv=n_folds,
verbose=1,
n_jobs=1,
return_train_score=1)
model_cv.fit(X_train, y_train)
cv_results = pd.DataFrame(model_cv.cv_results_)
cv_results.head()
# plot of C versus train and test scores
plt.figure(figsize=(8, 6))
plt.plot(cv_results['param_C'], cv_results['mean_test_score'])
plt.plot(cv_results['param_C'], cv_results['mean_train_score'])
plt.xlabel('C')
plt.ylabel('Neg Log Loss')
plt.legend(['test accuracy', 'train accuracy'], loc='bottom right')
plt.xscale('log')
plt.show()
best_score = model_cv.best_score_
best_params = model_cv.best_params_
best_estimator = model_cv.best_estimator_
print('Best score: ', best_score)
print('Best parameters: ', best_params)
print('Best estimator: ', best_estimator)
lr = linear_model.LogisticRegression(C=best_params.get('C'), penalty=best_params.get('penalty'))
lr.fit(X_train, y_train)
# prediction
y_pred = lr.predict(X_test)
print('Model accuracy: ', metrics.accuracy_score(y_true=y_test, y_pred=y_pred))
print('Confusion matrix')
print(metrics.confusion_matrix(y_true=y_test, y_pred=y_pred))
print('Classification Report')
print(metrics.classification_report(y_true=y_test, y_pred=y_pred))
print('Accuracy of the logistic regression model for training set: {}'.format(lr.score(X=X, y=y)))
| 0.628407 | 0.400867 |
# Practical Deep Learning for Coders, v3
# 00_notebook_tutorial
**Important note:** You should always work on a duplicate of the course notebook. On the page you used to open this, tick the box next to the name of the notebook and click duplicate to easily create a new version of this notebook.<br>
You will get errors each time you try to update your course repository if you don't do this, and your changes will end up being erased by the original course version.<br>
**重要提示:** 你应该在课程notebook的副本上工作。在你打开notebook的页面上,勾选notebook名称旁的选择框,然后点击复制就能轻松创建一个新的notebook副本了。<br>
如果你不这样做,那么当你尝试更新课程资源库时就会报错,你的改动会被课程的原始内容所覆盖。
# Welcome to Jupyter Notebooks!
# 欢迎来到Jupyter Notebooks!
If you want to learn how to use this tool you've come to the right place. This article will teach you all you need to know to use Jupyter Notebooks effectively. You only need to go through Section 1 to learn the basics and you can go into Section 2 if you want to further increase your productivity.<br>
如果你想学习如何使用这个工具,你来对地方了。这篇文章将教你高效使用jupyter notebook的所有应知应会的内容。你只需完成第1节的内容,就能学到基础知识,如果你想进一步生提高生产率,可以去学习第2节的内容。
You might be reading this tutorial in a web page (maybe Github or the course's webpage). We strongly suggest to read this tutorial in a (yes, you guessed it) Jupyter Notebook. This way you will be able to actually *try* the different commands we will introduce here.<br>
你可能正在通过网页来阅读这篇教程(可能是github网站,或课程的网页上)。我们强烈建议你(没错,你猜对了)在jupyter notebook中来阅读本教程。这种方式可以让你实际*尝试*在本文中介绍到的不同命令。
## Section 1: Need to Know
## 第1节:需知
### Introduction 简介
Let's build up from the basics, what is a Jupyter Notebook? Well, you are reading one. It is a document made of cells. You can write like I am writing now (markdown cells) or you can perform calculations in Python (code cells) and run them like this:<br>
让我们从最基础的部分开始说起,Jupyter Notebook是什么? 你现在看到的就是一个notebook。它是由一些单元格(cells)组成的文档。你可以像我这样(使用markdown cells)写入内容,你也可以(使用code cells)执行Python中的计算程序并且像下面这样来运行它们:
```
1+1
```
Cool huh? This combination of prose and code makes Jupyter Notebook ideal for experimentation: we can see the rationale for each experiment, the code and the results in one comprehensive document. In fast.ai, each lesson is documented in a notebook and you can later use that notebook to experiment yourself. <br>
是不是很cool?这种将普通文本和代码结合起来的模式,使得Jupyter Notebook成为做实验的绝佳选择:在一篇综合性文档中,我们可以既可以看到每个实验的原理讲解,又可以看到对应代码,甚至还有代码运行后的结果。在fast.ai课程里,每一节课的内容都以notebook方式来呈现,随后你也可以自己使用对应的notebook来做实验。
Other renowned institutions in academy and industry use Jupyter Notebook: Google, Microsoft, IBM, Bloomberg, Berkeley and NASA among others. Even Nobel-winning economists [use Jupyter Notebooks](https://paulromer.net/jupyter-mathematica-and-the-future-of-the-research-paper/) for their experiments and some suggest that Jupyter Notebooks will be the [new format for research papers](https://www.theatlantic.com/science/archive/2018/04/the-scientific-paper-is-obsolete/556676/).<br>
许多在学术界和工业界久负盛名的机构也在使用Jupyter Notebook,比如Google, Microsoft,IBM,Bloomberg,Berkeley以及NASA等,甚至诺贝尔经济学奖得主也在[使用Jupyter Notebooks](https://paulromer.net/jupyter-mathematica-and-the-future-of-the-research-paper/) 来进行实验,其中有一些经济学奖得主认为Jupyter Notebook将成为[新的学术论文格式](https://www.theatlantic.com/science/archive/2018/04/the-scientific-paper-is-obsolete/556676/)。
### Writing 写作
A type of cell in which you can write like this is called _Markdown_. [_Markdown_](https://en.wikipedia.org/wiki/Markdown) is a very popular markup language. To specify that a cell is _Markdown_ you need to click in the drop-down menu in the toolbar and select _Markdown_.<br>
_Markdown_ 是jupyter notebook里单元格的一种类型,它可以让你进行本文写作。[Markdown](https://en.wikipedia.org/wiki/Markdown) 是一种非常流行的标记语言。为了指定一个单元格为*_Markdown_*,你需要点击工具栏中的下拉菜单并且选择*_Markdown_*。
Click on the the '+' button on the left and select _Markdown_ from the toolbar.<br>
点击左边的“+”按钮,从工具栏中选择*_Markdown_*。
Now you can type your first _Markdown_ cell. Write 'My first markdown cell' and press run.<br>
现在你可以创建你的第一个*_Markdown_*单元格了。在单元格中输入“My first markdown cell”并点击run。

You should see something like this: <br>
你将看到下面的内容:
My first markdown cell
Now try making your first _Code_ cell: follow the same steps as before but don't change the cell type (when you add a cell its default type is _Code_). Type something like 3/2. You should see '1.5' as output.<br>
现在试着创建你的第一个*_Code_*单元格:遵循前面介绍的步骤,但是不要修改单元格的类型(当你添加一个单元格时,它的默认类型就是*_Code_*)。输入一些代码,比如3/2,那么你的输出为“1.5”。
```
3/2
```
### Modes 模式
If you made a mistake in your *Markdown* cell and you have already ran it, you will notice that you cannot edit it just by clicking on it. This is because you are in **Command Mode**. Jupyter Notebooks have two distinct modes:<br>
如果你在*Markdown*单元格中犯了错误并且已经运行过此单元格,你会发现不能仅通过点击它来进行编辑。这是因为你处于**命令模式**。Jupyter notebooks有两种不同的工作模式:<br>
1. **Edit Mode**: Allows you to edit a cell's content.<br>
**编辑模式**:允许你对单个单元格的内容进行编辑。
2. **Command Mode**: Allows you to edit the notebook as a whole and use keyboard shortcuts but not edit a cell's content. <br>
**命令模式**:允许你使用键盘快捷键,将notebook作为一个整体进行编辑,但不能对单个单元格的内容进行编辑。
You can toggle between these two by either pressing <kbd>ESC</kbd> and <kbd>Enter</kbd> or clicking outside a cell or inside it (you need to
double click if its a Markdown cell). You can always know which mode you're on since the current cell has a green border if in **Edit Mode** and a blue border in **Command Mode**. Try it!<br>
你可以在这两种模式间转换,方法是同时按下<kbd>ESC</kbd>和<kbd>Enter</kbd>键,或者通过点击一个单元格的外面或者里面来切换(如果是Markdown单元格,你需要双击实现模式切换)。你总是可以通过观察当前单元格的边框颜色来判断当前单元格处于什么模式:如果边框是绿色则表示处在**编辑模式**,如果是蓝色边框则表示处在**命令模式**。试一试吧!
### Other Important Considerations 其他重要考虑因素
1. Your notebook is autosaved every 120 seconds. If you want to manually save it you can just press the save button on the upper left corner or press <kbd>s</kbd> in **Command Mode**.<br>
你的notebook每过120秒就将自动保存。如果你希望手工保存,只需点击左上角的save按钮即可,或者在**命令模式**下按下<kbd>s</kbd>键。

2. To know if your kernel is computing or not you can check the dot in your upper right corner. If the dot is full, it means that the kernel is working. If not, it is idle. You can place the mouse on it and see the state of the kernel be displayed.<br>
如果你想知道你的kernel是否在运行中,你可以检查右上角的圆点。如果是实心的,表示kernel正在工作中,如果是空心的,则表示kernel空闲。你也可以将鼠标悬浮于圆点上,来查看kernel的状态。

3. There are a couple of shortcuts you must know about which we use **all** the time (always in **Command Mode**). These are:<br>
(处于**命令模式**时)有一些我们**总是**要用的键盘快捷键,你必须掌握。如下所示:
<kbd>Shift</kbd>+<kbd>Enter</kbd>: Runs the code or markdown on a cell<br>
<kbd>Shift</kbd>+<kbd>Enter</kbd>:运行一个单元格中的代码或者格式化文本
<kbd>Up Arrow</kbd>+<kbd>Down Arrow</kbd>: Toggle across cells<br>
<kbd>Up Arrow</kbd>+<kbd>Down Arrow</kbd>:在单元格之间切换选择
<kbd>b</kbd>: Create new cell<br>
<kbd>b</kbd>: 创建一个新的单元格
<kbd>0</kbd>+<kbd>0</kbd>: Reset Kernel<br>
<kbd>0</kbd>+<kbd>0</kbd>: 重置 Kernel
You can find more shortcuts in the Shortcuts section below.<br>
在下面的章节,你还会看到更多快捷键的说明。
4. You may need to use a terminal in a Jupyter Notebook environment (for example to git pull on a repository). That is very easy to do, just press 'New' in your Home directory and 'Terminal'. Don't know how to use the Terminal? We made a tutorial for that as well. You can find it [here](https://course.fast.ai/terminal_tutorial.html).<br>
你可能需要在Jupyter Notebook的环境中使用terminal(比如通过git pull指令拉取一个repo)。这也非常简单,只需要在你的首页点击“New”,再选择“Terminal”即可。不知道具体怎么用Terminal?我们准备了一篇教程,你可以在 [这里](https://course.fast.ai/terminal_tutorial.html) 找到。

That's it. This is all you need to know to use Jupyter Notebooks. That said, we have more tips and tricks below ↓↓↓<br>
好了,这就是使用Jupyter Notebooks时,你需要知道的知识点。当然了,下面还会介绍更多小技巧↓↓↓
## Section 2: Going deeper
## 第2节:更进一步
### Markdown formatting 设定markdown的格式
#### Italics, Bold, Strikethrough, Inline, Blockquotes and Links
#### 斜体,粗体,删除线,内联,引用和链接
The five most important concepts to format your code appropriately when using markdown are:<br>
当你使用markdown时,有五种最重要的格式设定,它们的作用如下:
1. *Italics*: Surround your text with '\_' or '\*'
*斜体*: 在文本两边包裹上“\_”或者“\*” <br>
2. **Bold**: Surround your text with '\__' or '\**'
**粗体**: 在文本两边包裹上“\__”或者“**”<br>
3. `inline`: Surround your text with '\`'
`内联`: 文本两边包裹上“\`”<br>
4. > blockquote: Place '\>' before your text.
> 引用:在文本前加上前缀“\>”<br>
5. [Links](https://course.fast.ai/): Surround the text you want to link with '\[\]' and place the link adjacent to the text, surrounded with '()' <br>
[链接](https://course.fast.ai/): 在文本两边包裹上 “\[\]”(这里是方括号),并且紧跟着将链接文本放在“()”中
#### Headings 标题
Notice that including a hashtag before the text in a markdown cell makes the text a heading. The number of hashtags you include will determine the priority of the header ('#' is level one, '##' is level two, '###' is level three and '####' is level four). We will add three new cells with the '+' button on the left to see how every level of heading looks.<br>
在一个markdown单元格的文本前添加一个“#”,就可将该文本设定为标题了。“#”的个数决定了文本的优先级别。(“#”表示一级标题,“##”表示二级标题,“###”表示三级标题,“####”表示四级标题)。我们通过点击“+”来添加三个新的单元格来演示各个级别的标题都是什么样子的。
Double click on some headings and find out what level they are!<br>
双击下面的标题,看看他们都是什么级别的吧!
#### Lists 列表
There are three types of lists in markdown.<br>
在markdown中有三种类型的列表。
Ordered list: 有序列表
1. Step 1<br>A.Step 1B
2. Step 3
Unordered list 无序列表
* learning rate 学习速率
* cycle length 周期长度
* weight decay 权重衰减
Task list 任务列表
- [x] Learn Jupyter Notebooks 学习Jupyter Notebooks
- [x] Writing 写作
- [x] Modes 模式
- [x] Other Considerations 其他考虑因素
- [ ] Change the world 改变世界
Double click on each to see how they are built!
双击查看这些列表是怎么构建出来的!
### Code Capabilities 代码能力
**Code** cells are different than **Markdown** cells in that they have an output cell. This means that we can *keep* the results of our code within the notebook and share them. Let's say we want to show a graph that explains the result of an experiment. We can just run the necessary cells and save the notebook. The output will be there when we open it again! Try it out by running the next four cells.<br>
**Code**单元格和**Markdown**单元格是不同类型的单元格,因为**Code**单元格中有一个输出单元格。这意味着我们可以在notebook中 *保留* 代码执行结果,并分享它们。当我们想要展示实验结果的图表时,我们只需要运行必要的单元格并保存notebook。运行结果会在我们再次打开时显示出来!试试看运行接下来的4个单元格吧!
```
# Import necessary libraries
from fastai.vision import *
import matplotlib.pyplot as plt
from PIL import Image
a = 1
b = a + 1
c = b + a + 1
d = c + b + a + 1
a, b, c ,d
plt.plot([a,b,c,d])
plt.show()
```
We can also print images while experimenting. I am watching you.<br>
我们也可以在做实验过程中显示一些图片。(这只猫的图片就像在说)“我在看着你哦”。
```
Image.open('images/notebook_tutorial/cat_example.jpg')
```
### Running the app locally 本地运行app
You may be running Jupyter Notebook from an interactive coding environment like Gradient, Sagemaker or Salamander. You can also run a Jupyter Notebook server from your local computer. What's more, if you have installed Anaconda you don't even need to install Jupyter (if not, just `pip install jupyter`).<br>
你可能在Gradient, Sagemaker或者Salamander,这样的交互式编码环境中运行Jupyter Notebook。你也可以在本地计算机上运行一个Jupyter Notebook服务器。此外,如果你安装了Anaconda,你甚至不用单独安装Jupyter(如果没有安装的话,只要运行一下`pip install jupyter`就可以了)。
You just need to run `jupyter notebook` in your terminal. Remember to run it from a folder that contains all the folders/files you will want to access. You will be able to open, view and edit files located within the directory in which you run this command but not files in parent directories.<br>
你只需要在你的terminal上运行`jupyter notebook`命令即可。记住在包含你希望访问的文件夹/文件的总文件夹那里来运行这条命令。这样你就可以打开,查看和编辑,运行了`jupyter notebook`命令的文件夹中的文件了,但是记在父目录里面的文件是不能打开查看或者编辑的。<br>
If a browser tab does not open automatically once you run the command, you should CTRL+CLICK the link starting with 'https://localhost:' and this will open a new tab in your default browser.<br>
如果你运行了上面的命令,却没有自动打开浏览器,你也可以按住CTRL键,然后点击以 “https://localhost:” 开头的链接,这样你的默认浏览器中就会打开一个新的标签页。
### Creating a notebook 创建一个notebook
Click on 'New' in the upper left corner and 'Python 3' in the drop-down list (we are going to use a [Python kernel](https://github.com/ipython/ipython) for all our experiments).<br>
点击左上角的“New”按钮,随后在下拉列表中选择“Python 3”(我们将在我们的所有实验中使用一个[Python内核](https://github.com/ipython/ipython))

Note: You will sometimes hear people talking about the Notebook 'kernel'. The 'kernel' is just the Python engine that performs the computations for you. <br>
注意:你有时可能听到人们谈论Notebook “kernel”,“kernel”就是替你执行计算的Python引擎。
### Shortcuts and tricks 快捷键和技巧
#### Command Mode Shortcuts 命令模式下的快捷键
There are a couple of useful keyboard shortcuts in `Command Mode` that you can leverage to make Jupyter Notebook faster to use. Remember that to switch back and forth between `Command Mode` and `Edit Mode` with <kbd>Esc</kbd> and <kbd>Enter</kbd>.<br>
在`命令模式`下有一些可以提高效率的快捷键。记住在`命令模式`和`编辑`模式间来回切换的快捷键是<kbd>Esc</kbd> 和 <kbd>Enter</kbd>。<br><br>
<kbd>m</kbd>: Convert cell to Markdown 将单元格转换为Markdown单元格
<kbd>y</kbd>: Convert cell to Code 将单元格转换为Code代码单元格
<kbd>D</kbd>+<kbd>D</kbd>: Delete cell 删除单元格
<kbd>o</kbd>: Toggle between hide or show output 切换显示或者隐藏输出信息
<kbd>Shift</kbd>+<kbd>Arrow up上箭头/Arrow down下箭头</kbd>: Selects multiple cells. Once you have selected them you can operate on them like a batch (run, copy, paste etc).
用于选择多个单元格。一旦你选中了多个单元格,你就可以批量操作他们(比如运行,复制,粘贴等操作)。
<kbd>Shift</kbd>+<kbd>M</kbd>: Merge selected cells. 合并选中的单元格为一个单元格
<kbd>Shift</kbd>+<kbd>Tab</kbd>: [press once] Tells you which parameters to pass on a function
[按键一次]提示函数有哪些参数
<kbd>Shift</kbd>+<kbd>Tab</kbd>: [press three times] Gives additional information on the method
[按键三次] 提示这个方法的更多信息
#### Cell Tricks 单元格小技巧
```
from fastai import*
from fastai.vision import *
```
There are also some tricks that you can code into a cell.<br>
这里还有一些在单元格编码的一些小技巧。
`?function-name`: Shows the definition and docstring for that function <br>
`?function-name`:显示该函数的定义和文档信息
```
?ImageDataBunch
```
`??function-name`: Shows the source code for that function<br>
`??function-name`:显示函数的源代码
```
??ImageDataBunch
```
`doc(function-name)`: Shows the definition, docstring **and links to the documentation** of the function
(only works with fastai library imported)<br>
`doc(function-name)`:显示定义、文档信息以及**详细文档的链接**(只有在import导入了fastai库之后才能工作)
```
doc(ImageDataBunch)
```
#### Line Magics
Line magics are functions that you can run on cells and take as an argument the rest of the line from where they are called. You call them by placing a '%' sign before the command. The most useful ones are:<br>
Line magics是可以在单元格中运行并且将该行的其他信息作为参数的函数。通过在命令之前添加一个“%”来调用他们。最有用的是以下几个:
`%matplotlib inline`: This command ensures that all matplotlib plots will be plotted in the output cell within the notebook and will be kept in the notebook when saved.<br>
`%matplotlib inline`:该命令确保所有的matplotlib图表都将绘制在notebook的输出单元格中,并且在保存时一并保留在notebook中。
`%reload_ext autoreload`, `%autoreload 2`: Reload all modules before executing a new line. If a module is edited, it is not necessary to rerun the import commands, the modules will be reloaded automatically.<br>
`%reload_ext autoreload`,`%autoreload 2`:这两条命令指示在执行新的行代码时重新加载所有的模块。如果一个模块修改过了,没有必要再次运行import命令,模块将自动重新加载。
These three commands are always called together at the beginning of every notebook. <br>
通常这3条命令在每一个notebook的起始部分被一起调用。
```
%matplotlib inline
%reload_ext autoreload
%autoreload 2
```
`%timeit`: Runs a line a ten thousand times and displays the average time it took to run it.<br>
`%timeit`:这个命令将会运行一行代码1000次并且显示平均运行的时间。
```
%timeit [i+1 for i in range(1000)]
```
`%debug`: Allows to inspect a function which is showing an error using the [Python debugger](https://docs.python.org/3/library/pdb.html).<br>
`%debug`:允许你使用[Python调试器](https://docs.python.org/3/library/pdb.html)来检查报错的函数。
```
for i in range(1000):
a = i+1
b = 'string'
c = b+1
%debug
```
|
github_jupyter
|
1+1
3/2
# Import necessary libraries
from fastai.vision import *
import matplotlib.pyplot as plt
from PIL import Image
a = 1
b = a + 1
c = b + a + 1
d = c + b + a + 1
a, b, c ,d
plt.plot([a,b,c,d])
plt.show()
Image.open('images/notebook_tutorial/cat_example.jpg')
from fastai import*
from fastai.vision import *
?ImageDataBunch
??ImageDataBunch
doc(ImageDataBunch)
%matplotlib inline
%reload_ext autoreload
%autoreload 2
%timeit [i+1 for i in range(1000)]
for i in range(1000):
a = i+1
b = 'string'
c = b+1
%debug
| 0.243642 | 0.916969 |
```
import numpy
%matplotlib notebook
import matplotlib.pyplot
import scipy.interpolate
import scipy.integrate
import pynverse
```
# Arc Length Reparameterization
## Overview
To have control over the speed and acceleration of an object along a path, the path should be parameterized on distance. It is much easier to express a parameterized curve in terms of time, so we need to convert it from time-parameterized to arc-length-parameterized.
## Time-Parameterized
The original equation we wish to convert to arc-length-parameterized is a spline curve connecting two roads. The details of the curve are irrelevant so we will focus on a specific example without loss of generality. Consider the following curves:
$$ \vec{\ell}_a(t) = \langle 3, t \rangle \quad \forall t \in [0, 1] $$
$$ \vec{\ell}_b(t) = \langle 1 - t, 3 \rangle \quad \forall t \in [0, 1] $$
```
l_a = lambda t: numpy.array([[3 + 0 * t], [0 + 1 * t]])
l_b = lambda t: numpy.array([[1 - 1 * t], [3 + 0 * t]])
```
To get a better idea of these curves, here they are plotted.
```
fig, ax = matplotlib.pyplot.subplots()
t_a = t_b = numpy.linspace(0, 1, 100)
ax.plot(l_a(t_a)[0].flatten(), l_a(t_a)[1].flatten(), 'r-')
ax.plot(l_b(t_b)[0].flatten(), l_b(t_b)[1].flatten(), 'r-')
ax.axis('equal')
matplotlib.pyplot.show()
```
Now we can connect them with a spline interpolation. We will call it `time_path` to differentiate from the arc-length parameterized expression later. This path will now be referenced using the following symbols
$$ \ell(t) = \langle x(t), y(t) \rangle \quad \forall t \in [0, 1] $$
```
t_a = t_b = numpy.linspace(0, 1, 100)
x = numpy.hstack([l_a(t_a)[0].flatten(), l_b(t_b)[0].flatten()])
y = numpy.hstack([l_a(t_a)[1].flatten(), l_b(t_b)[1].flatten()])
points = [x, y]
tck, u = scipy.interpolate.splprep(points, s=0)
time_path = lambda t: scipy.interpolate.splev(t, tck, der=0)
```
To confirm the interpolation is correct, we can plot the result.
```
fig, ax = matplotlib.pyplot.subplots()
t = numpy.linspace(0, 1, 20)
ax.plot(time_path(t)[0], time_path(t)[1], 'r-')
ax.axis('equal')
matplotlib.pyplot.show()
```
## Speed
Before we reparameterize the curve, we will investigate why time-parameterization is insufficient. First we calculate the velocity and speed of the curve.
$$ \vec{v}(t) = \left\langle \frac{\partial x}{\partial t}(t), \frac{\partial y}{\partial t}(t) \right\rangle \quad \forall t \in [0, 1] $$
$$ v(t) = \left\lVert \left\langle \frac{\partial x}{\partial t}(t), \frac{\partial y}{\partial t}(t) \right\rangle \right\rVert \quad \forall t \in [0, 1] $$
SciPy has built in functions for calculating the derivatives of spline curves.
```
velocity = lambda t: scipy.interpolate.splev(t, tck, der=1)
speed = lambda t: numpy.linalg.norm(velocity(t), axis=0)
```
Over time, we want constant speed along the curve. Let's plot the curve to see if this is the case.
```
fig, ax = matplotlib.pyplot.subplots()
t = numpy.linspace(0, 1, 1000)
ax.plot(t, speed(t), 'r-')
matplotlib.pyplot.show()
```
Unfortunately, the speed doesn't remain constant over the turn. Granted, it is a small difference, but it is still there and could cause trouble.
## Arc-Length-Parameterized
The first task is to determine the length of the curve from starting position at a specific time.
$$ L(t) = \int_0^t \left\lVert \left\langle \frac{\partial x}{\partial t}(t), \frac{\partial y}{\partial t}(t) \right\rangle \right\rVert dt = \int_0^t v(t) dt $$
We can use SciPy's integration `quad` technique and the speed function calculated above to evaluate this expression.
```
length = lambda t: scipy.integrate.quad(speed, 0, t)[0]
```
The total length of the curve is given by
$$ L_{\mathrm{total}} = \int_0^1 \left\lVert \left\langle \frac{\partial x}{\partial t}(t), \frac{\partial y}{\partial t}(t) \right\rangle \right\rVert dt = \int_0^1 v(t) dt $$
Using the total length, we can normalized the length function so $L^*(t) \in [0, 1]$.
$$ L^*(t) = \frac{L(t)}{L_{\mathrm{total}}} = \frac{1}{L_{\mathrm{total}}} \int_0^t \left\lVert \left\langle \frac{\partial x}{\partial t}(t), \frac{\partial y}{\partial t}(t) \right\rangle \right\rVert dt = \frac{1}{L_{\mathrm{total}}} \int_0^t v(t) dt $$
```
length_total = length(1)
length_star = lambda t: length(t) / length_total
```
We can now plot the length with respect to time. If the speed were constant, this would be a perfectly linear relationship. Since we know it is not, there should be a slight divergence.
```
fig, ax = matplotlib.pyplot.subplots()
t = numpy.linspace(0, 1, 100)
ax.plot(t, t, 'b-', alpha=0.66)
ax.plot(t, numpy.vectorize(length_star)(t), 'r-')
matplotlib.pyplot.show()
```
Notice that this plot took a long time to compute. We naively `vectorize` the length expression to allow for array inputs. This is super slow and, if you think about it for a second, very wasteful. Using the `cumtrapz` function, this can be significantly optimized as we can compute the integration values within a range cumulatively without iteration, but the intuition might be less clear.
```
fig, ax = matplotlib.pyplot.subplots()
t = numpy.linspace(0, 1, 100)
ax.plot(t, t, 'b-', alpha=0.66)
ax.plot(t, scipy.integrate.cumtrapz(speed(t) / length_total, t, initial=0), 'r-')
matplotlib.pyplot.show()
```
The next step to reparameterize with respect to arc-length is to invert the normalized length function. This allows us to define the curve in the following way
$$ \ell(s^*) = \left\langle x\left(L^{-1}(s^*)\right), y\left(L^{-1}(s^*)\right) \right\rangle \quad \forall s^* \in [0, 1] $$
Unfortunately, our length function doesn't have a known analytical form. Therefore, we have to compute the inverse numerically. Because our functions are normalized, we can compute several points in the range $[0, 1]$ and fit a spline curve to the inverted data. This will approximate our inverse.
```
t = numpy.linspace(0, 1, 100)
tck2 = scipy.interpolate.splrep(scipy.integrate.cumtrapz(speed(t) / length_total, t, initial=0), t, s=0)
length_star_inverse = lambda t: scipy.interpolate.splev(t, tck2)
```
Let's plot the inverse to make sure the output makes sense.
```
fig, ax = matplotlib.pyplot.subplots()
t = numpy.linspace(0, 1, 100)
ax.plot(t, length_star_inverse(t), 'r-')
ax.plot(t, t, 'b-', alpha=0.66)
matplotlib.pyplot.show()
```
Now we have all the necessary components to reparameterized the curve with respect to arc-length.
```
arc_length_path = lambda s: time_path(length_star_inverse(s))
```
First we can plot the paths to make sure they align.
```
fig, ax = matplotlib.pyplot.subplots()
s = numpy.linspace(0, 1, 100)
t = numpy.linspace(0, 1, 100)
ax.plot(arc_length_path(s)[0], arc_length_path(s)[1], 'r-')
ax.plot(time_path(t)[0], time_path(t)[1], 'b-')
ax.axis('equal')
matplotlib.pyplot.show()
```
And we can compare the speed versus time graphs. The reparameterized graph should be constant.
```
fig, ax = matplotlib.pyplot.subplots()
s = numpy.linspace(0, 1, 100)
t = numpy.linspace(0, 1, 100)
ax.plot(s, numpy.linalg.norm(numpy.gradient(arc_length_path(s), axis=1), axis=0), 'r-')
ax.plot(t, numpy.linalg.norm(numpy.gradient(time_path(t), axis=1), axis=0), 'b-')
matplotlib.pyplot.show()
```
Although there is a slight jag in the red curve where the turn begins and ends, it is a vast improvement over the previous curve definition. I attribute the jag to approximations made with numerical solutions to inverses and gradients as this jag was nonexistant using the slower vectorized solution that I replaced with this much faster solution.
|
github_jupyter
|
import numpy
%matplotlib notebook
import matplotlib.pyplot
import scipy.interpolate
import scipy.integrate
import pynverse
l_a = lambda t: numpy.array([[3 + 0 * t], [0 + 1 * t]])
l_b = lambda t: numpy.array([[1 - 1 * t], [3 + 0 * t]])
fig, ax = matplotlib.pyplot.subplots()
t_a = t_b = numpy.linspace(0, 1, 100)
ax.plot(l_a(t_a)[0].flatten(), l_a(t_a)[1].flatten(), 'r-')
ax.plot(l_b(t_b)[0].flatten(), l_b(t_b)[1].flatten(), 'r-')
ax.axis('equal')
matplotlib.pyplot.show()
t_a = t_b = numpy.linspace(0, 1, 100)
x = numpy.hstack([l_a(t_a)[0].flatten(), l_b(t_b)[0].flatten()])
y = numpy.hstack([l_a(t_a)[1].flatten(), l_b(t_b)[1].flatten()])
points = [x, y]
tck, u = scipy.interpolate.splprep(points, s=0)
time_path = lambda t: scipy.interpolate.splev(t, tck, der=0)
fig, ax = matplotlib.pyplot.subplots()
t = numpy.linspace(0, 1, 20)
ax.plot(time_path(t)[0], time_path(t)[1], 'r-')
ax.axis('equal')
matplotlib.pyplot.show()
velocity = lambda t: scipy.interpolate.splev(t, tck, der=1)
speed = lambda t: numpy.linalg.norm(velocity(t), axis=0)
fig, ax = matplotlib.pyplot.subplots()
t = numpy.linspace(0, 1, 1000)
ax.plot(t, speed(t), 'r-')
matplotlib.pyplot.show()
length = lambda t: scipy.integrate.quad(speed, 0, t)[0]
length_total = length(1)
length_star = lambda t: length(t) / length_total
fig, ax = matplotlib.pyplot.subplots()
t = numpy.linspace(0, 1, 100)
ax.plot(t, t, 'b-', alpha=0.66)
ax.plot(t, numpy.vectorize(length_star)(t), 'r-')
matplotlib.pyplot.show()
fig, ax = matplotlib.pyplot.subplots()
t = numpy.linspace(0, 1, 100)
ax.plot(t, t, 'b-', alpha=0.66)
ax.plot(t, scipy.integrate.cumtrapz(speed(t) / length_total, t, initial=0), 'r-')
matplotlib.pyplot.show()
t = numpy.linspace(0, 1, 100)
tck2 = scipy.interpolate.splrep(scipy.integrate.cumtrapz(speed(t) / length_total, t, initial=0), t, s=0)
length_star_inverse = lambda t: scipy.interpolate.splev(t, tck2)
fig, ax = matplotlib.pyplot.subplots()
t = numpy.linspace(0, 1, 100)
ax.plot(t, length_star_inverse(t), 'r-')
ax.plot(t, t, 'b-', alpha=0.66)
matplotlib.pyplot.show()
arc_length_path = lambda s: time_path(length_star_inverse(s))
fig, ax = matplotlib.pyplot.subplots()
s = numpy.linspace(0, 1, 100)
t = numpy.linspace(0, 1, 100)
ax.plot(arc_length_path(s)[0], arc_length_path(s)[1], 'r-')
ax.plot(time_path(t)[0], time_path(t)[1], 'b-')
ax.axis('equal')
matplotlib.pyplot.show()
fig, ax = matplotlib.pyplot.subplots()
s = numpy.linspace(0, 1, 100)
t = numpy.linspace(0, 1, 100)
ax.plot(s, numpy.linalg.norm(numpy.gradient(arc_length_path(s), axis=1), axis=0), 'r-')
ax.plot(t, numpy.linalg.norm(numpy.gradient(time_path(t), axis=1), axis=0), 'b-')
matplotlib.pyplot.show()
| 0.518059 | 0.981275 |
```
import os
import pickle
import pandas as pd
import numpy as np
from PIL import Image
from keras.applications.inception_resnet_v2 import InceptionResNetV2, preprocess_input
from keras.utils import to_categorical
from keras.preprocessing import image
EMOTIONS = [
"angry",
"calm",
"disgust",
"fear",
"sad",
"happy",
"neutral",
"surprise"
]
DATA_PATH = 'data/'
IMG_WIDTH, IMG_HEIGHT = 100, 100
SEQ_LENGTH = 2
OVERLAP_IDX = int(0.9 * SEQ_LENGTH)
SEQUENCE_PATH = 'sequence/'
model = InceptionResNetV2(include_top=False, weights='imagenet')
def extract_feature_sequence():
X, y = [], []
for emotion in EMOTIONS:
print(emotion)
video_list = [f for f in os.listdir(DATA_PATH + emotion)]
for video in video_list:
video_path = DATA_PATH + emotion + '/' + video + '/' + video + '_aligned'
frames = [f for f in os.listdir(video_path) if os.path.isfile(os.path.join(video_path, f))]
if len(frames) >= SEQ_LENGTH:
X, y = process_frames(frames, video_path, emotion, X, y)
print('{} sequences extracted'.format(emotion))
# use onehot encoding for LSTM
y = to_categorical(y, num_classes=len(EMOTIONS))
# save to binary files
print('Saving sequence')
np.save(SEQUENCE_PATH + 'X_InceptionResNetV2', X)
np.save(SEQUENCE_PATH + 'y_InceptionResNetV2', y)
print('Saving finished')
def process_frames(frames, video_path, emotion, X, y):
sequence = []
for frame in frames:
frame = video_path + '/' + frame
features = extract_features(model, frame)
sequence.append(features)
if len(sequence) == SEQ_LENGTH:
X.append(sequence)
y.append(EMOTIONS.index(emotion))
sequence = sequence[OVERLAP_IDX:]
return X, y
def extract_features(model, image_path):
# load and preprocess the frame
img = image.load_img(image_path, target_size=(IMG_WIDTH, IMG_HEIGHT))
x = image.img_to_array(img)
x = np.expand_dims(x, axis=0)
x = preprocess_input(x)
# Get the prediction.
features = model.predict(x)
features = features[0]
return features
extract_feature_sequence()
from sklearn.model_selection import train_test_split
def split_dataset(X, y, test_size=0.2, val_split=True):
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=test_size, random_state=42, stratify=y)
if val_split:
X_val, X_test, y_val, y_test = train_test_split(X_test, y_test, test_size=0.5, random_state=42, stratify=y_test)
return X_train, y_train, X_val, y_val, X_test, y_test
else:
return X_train, y_train, X_test, y_test
def load_sequence():
X = np.load(SEQUENCE_PATH + 'X_InceptionResNetV2.npy')
X = X.reshape(X.shape[0], X.shape[1], X.shape[2] * X.shape[3] * X.shape[4])
y = np.load(SEQUENCE_PATH + 'y_InceptionResNetV2.npy')
X_train, y_train, X_val, y_val, X_test, y_test = split_dataset(X, y, test_size=0.2)
return X_train, y_train, X_val, y_val, X_test, y_test
X_train, y_train, X_val, y_val, X_test, y_test = load_sequence()
import matplotlib.pyplot as plt
import time
from keras.models import Sequential, load_model
from keras.layers import LSTM, Dense
from keras.callbacks import TensorBoard, ModelCheckpoint
from sklearn.metrics import confusion_matrix
import itertools
def plot_confusion_matrix(cm, title='Confusion matrix', float_display='.4f', cmap=plt.cm.Greens, class_names=None):
# create confusion matrix plot
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = np.arange(cm.shape[1])
plt.xticks(tick_marks)
ax = plt.gca()
ax.set_xticklabels(class_names)
plt.yticks(tick_marks)
ax.set_yticklabels(class_names)
thresh = cm.max() / 2.
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, format(cm[i, j], float_display),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.tight_layout()
plt.ylabel('Actual')
plt.xlabel('Predicted')
def get_predictions_and_labels(model, X, y):
predictions = model.predict(X)
y_true = []
y_pred = []
for i in range(len(y)):
label = list(y[i]).index(1)
pred = list(predictions[i])
max_value = max(pred)
max_index = pred.index(max_value)
p = max_index
y_true.append(label)
y_pred.append(p)
return y_true, y_pred
class LSTMNetwork:
def __init__(self, n_layer, lstm_unit, input_shape, feature, data_type):
self.EMOTIONS = [
"angry",
"calm",
"disgust",
"fear",
"sad",
"happy",
"neutral",
"surprise"
]
self.model = Sequential()
if n_layer > 1:
self.model.add(LSTM(lstm_unit, return_sequences=True, input_shape=input_shape,
dropout=0.2))
layer_count = 1
while layer_count < n_layer:
if layer_count == n_layer - 1:
self.model.add(LSTM(lstm_unit, return_sequences=False, dropout=0.2))
else:
self.model.add(LSTM(lstm_unit, return_sequences=True, dropout=0.2))
layer_count += 1
else:
self.model.add(LSTM(lstm_unit, return_sequences=False, input_shape=input_shape,
dropout=0.2))
nb_class = len(self.EMOTIONS)
self.model.add(Dense(nb_class, activation='softmax'))
current_time = time.strftime("%Y%m%d-%H%M%S")
self.base_dir = 'LSTM/' + data_type + '/' + feature + '/'
self.model_dir = 'LSTM_' + str(n_layer) + '_' + str(lstm_unit) + '_' + current_time + '/'
filename = 'LSTM.h5'
self.model_file = self.base_dir + self.model_dir + filename
def train(self, X_train, y_train, X_val, y_val, epochs, batch_size):
# compile and train the model
if not os.path.exists(self.base_dir + self.model_dir):
os.makedirs(self.base_dir + self.model_dir)
log_dir = self.base_dir + self.model_dir + 'log/'
os.mkdir(log_dir)
self.model.compile(optimizer='adam',
loss='categorical_crossentropy',
metrics=['accuracy'])
callbacks = [ModelCheckpoint(self.model_file, monitor='val_loss', save_best_only=True, verbose=0),
TensorBoard(log_dir=log_dir, write_graph=True)]
self.model.fit(X_train, y_train,
epochs=epochs,
batch_size=batch_size,
validation_data=(X_val, y_val),
callbacks=callbacks)
def evaluate(self, X_val, y_val):
# evaluate_vgg16 the model with validation set
model = load_model(self.model_file)
scores = model.evaluate(X_val, y_val)
print('val_loss: {}, val_acc: {}'.format(scores[0], scores[1]))
y_true, y_pred = get_predictions_and_labels(model, X_val, y_val)
cm = confusion_matrix(y_true, y_pred)
cm_percent = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
df = pd.DataFrame(cm_percent, index=self.EMOTIONS, columns=self.EMOTIONS)
df.index.name = 'Actual'
df.columns.name = 'Predicted'
df.to_csv(self.base_dir + self.model_dir + 'cm_val.csv', float_format='%.4f')
# plot percentage confusion matrix
fig1, ax1 = plt.subplots()
plot_confusion_matrix(cm_percent, class_names=self.EMOTIONS)
plt.savefig(self.base_dir + self.model_dir + 'cm_percent_val.png', format='png')
# plot normal confusion matrix
fig2, ax2 = plt.subplots()
plot_confusion_matrix(cm, float_display='.0f', class_names=self.EMOTIONS)
plt.savefig(self.base_dir + self.model_dir + 'cm_val.png', format='png')
plt.show()
def compare_model(self, X_val, y_val):
folder_list = [model_dir for model_dir in os.listdir(self.base_dir) if 'LSTM' in model_dir]
for folder in folder_list:
filename = 'LSTM.h5'
path = os.path.join(self.base_dir, folder, filename)
model = load_model(path)
scores = model.evaluate(X_val, y_val)
print('model: {}, val_loss: {}, val_acc: {}'.format(folder, scores[0], scores[1]))
y_true, y_pred = get_predictions_and_labels(model, X_val, y_val)
cm = confusion_matrix(y_true, y_pred)
cm_percent = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
# plot percentage confusion matrix
fig1, ax1 = plt.subplots()
plot_confusion_matrix(cm_percent, class_names=self.EMOTIONS)
plt.savefig(os.path.join(self.base_dir, folder, 'cm_percent_test.png'), format='png')
# plot normal confusion matrix
fig2, ax2 = plt.subplots()
plot_confusion_matrix(cm, float_display='.0f', class_names=self.EMOTIONS)
plt.savefig(os.path.join(self.base_dir, folder, 'cm_test.png'), format='png')
feature = 'InceptionResNetV2'
data_type = 'Basic'
n_layer = 1
lstm_unit = 32
batch_size = 256
epochs = 250
lstm_net = LSTMNetwork(n_layer, lstm_unit, X_train.shape[1:], feature, data_type)
lstm_net.train(X_train, y_train, X_test, y_test, epochs, batch_size)
lstm_net.evaluate(X_val, y_val)
```
|
github_jupyter
|
import os
import pickle
import pandas as pd
import numpy as np
from PIL import Image
from keras.applications.inception_resnet_v2 import InceptionResNetV2, preprocess_input
from keras.utils import to_categorical
from keras.preprocessing import image
EMOTIONS = [
"angry",
"calm",
"disgust",
"fear",
"sad",
"happy",
"neutral",
"surprise"
]
DATA_PATH = 'data/'
IMG_WIDTH, IMG_HEIGHT = 100, 100
SEQ_LENGTH = 2
OVERLAP_IDX = int(0.9 * SEQ_LENGTH)
SEQUENCE_PATH = 'sequence/'
model = InceptionResNetV2(include_top=False, weights='imagenet')
def extract_feature_sequence():
X, y = [], []
for emotion in EMOTIONS:
print(emotion)
video_list = [f for f in os.listdir(DATA_PATH + emotion)]
for video in video_list:
video_path = DATA_PATH + emotion + '/' + video + '/' + video + '_aligned'
frames = [f for f in os.listdir(video_path) if os.path.isfile(os.path.join(video_path, f))]
if len(frames) >= SEQ_LENGTH:
X, y = process_frames(frames, video_path, emotion, X, y)
print('{} sequences extracted'.format(emotion))
# use onehot encoding for LSTM
y = to_categorical(y, num_classes=len(EMOTIONS))
# save to binary files
print('Saving sequence')
np.save(SEQUENCE_PATH + 'X_InceptionResNetV2', X)
np.save(SEQUENCE_PATH + 'y_InceptionResNetV2', y)
print('Saving finished')
def process_frames(frames, video_path, emotion, X, y):
sequence = []
for frame in frames:
frame = video_path + '/' + frame
features = extract_features(model, frame)
sequence.append(features)
if len(sequence) == SEQ_LENGTH:
X.append(sequence)
y.append(EMOTIONS.index(emotion))
sequence = sequence[OVERLAP_IDX:]
return X, y
def extract_features(model, image_path):
# load and preprocess the frame
img = image.load_img(image_path, target_size=(IMG_WIDTH, IMG_HEIGHT))
x = image.img_to_array(img)
x = np.expand_dims(x, axis=0)
x = preprocess_input(x)
# Get the prediction.
features = model.predict(x)
features = features[0]
return features
extract_feature_sequence()
from sklearn.model_selection import train_test_split
def split_dataset(X, y, test_size=0.2, val_split=True):
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=test_size, random_state=42, stratify=y)
if val_split:
X_val, X_test, y_val, y_test = train_test_split(X_test, y_test, test_size=0.5, random_state=42, stratify=y_test)
return X_train, y_train, X_val, y_val, X_test, y_test
else:
return X_train, y_train, X_test, y_test
def load_sequence():
X = np.load(SEQUENCE_PATH + 'X_InceptionResNetV2.npy')
X = X.reshape(X.shape[0], X.shape[1], X.shape[2] * X.shape[3] * X.shape[4])
y = np.load(SEQUENCE_PATH + 'y_InceptionResNetV2.npy')
X_train, y_train, X_val, y_val, X_test, y_test = split_dataset(X, y, test_size=0.2)
return X_train, y_train, X_val, y_val, X_test, y_test
X_train, y_train, X_val, y_val, X_test, y_test = load_sequence()
import matplotlib.pyplot as plt
import time
from keras.models import Sequential, load_model
from keras.layers import LSTM, Dense
from keras.callbacks import TensorBoard, ModelCheckpoint
from sklearn.metrics import confusion_matrix
import itertools
def plot_confusion_matrix(cm, title='Confusion matrix', float_display='.4f', cmap=plt.cm.Greens, class_names=None):
# create confusion matrix plot
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = np.arange(cm.shape[1])
plt.xticks(tick_marks)
ax = plt.gca()
ax.set_xticklabels(class_names)
plt.yticks(tick_marks)
ax.set_yticklabels(class_names)
thresh = cm.max() / 2.
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, format(cm[i, j], float_display),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.tight_layout()
plt.ylabel('Actual')
plt.xlabel('Predicted')
def get_predictions_and_labels(model, X, y):
predictions = model.predict(X)
y_true = []
y_pred = []
for i in range(len(y)):
label = list(y[i]).index(1)
pred = list(predictions[i])
max_value = max(pred)
max_index = pred.index(max_value)
p = max_index
y_true.append(label)
y_pred.append(p)
return y_true, y_pred
class LSTMNetwork:
def __init__(self, n_layer, lstm_unit, input_shape, feature, data_type):
self.EMOTIONS = [
"angry",
"calm",
"disgust",
"fear",
"sad",
"happy",
"neutral",
"surprise"
]
self.model = Sequential()
if n_layer > 1:
self.model.add(LSTM(lstm_unit, return_sequences=True, input_shape=input_shape,
dropout=0.2))
layer_count = 1
while layer_count < n_layer:
if layer_count == n_layer - 1:
self.model.add(LSTM(lstm_unit, return_sequences=False, dropout=0.2))
else:
self.model.add(LSTM(lstm_unit, return_sequences=True, dropout=0.2))
layer_count += 1
else:
self.model.add(LSTM(lstm_unit, return_sequences=False, input_shape=input_shape,
dropout=0.2))
nb_class = len(self.EMOTIONS)
self.model.add(Dense(nb_class, activation='softmax'))
current_time = time.strftime("%Y%m%d-%H%M%S")
self.base_dir = 'LSTM/' + data_type + '/' + feature + '/'
self.model_dir = 'LSTM_' + str(n_layer) + '_' + str(lstm_unit) + '_' + current_time + '/'
filename = 'LSTM.h5'
self.model_file = self.base_dir + self.model_dir + filename
def train(self, X_train, y_train, X_val, y_val, epochs, batch_size):
# compile and train the model
if not os.path.exists(self.base_dir + self.model_dir):
os.makedirs(self.base_dir + self.model_dir)
log_dir = self.base_dir + self.model_dir + 'log/'
os.mkdir(log_dir)
self.model.compile(optimizer='adam',
loss='categorical_crossentropy',
metrics=['accuracy'])
callbacks = [ModelCheckpoint(self.model_file, monitor='val_loss', save_best_only=True, verbose=0),
TensorBoard(log_dir=log_dir, write_graph=True)]
self.model.fit(X_train, y_train,
epochs=epochs,
batch_size=batch_size,
validation_data=(X_val, y_val),
callbacks=callbacks)
def evaluate(self, X_val, y_val):
# evaluate_vgg16 the model with validation set
model = load_model(self.model_file)
scores = model.evaluate(X_val, y_val)
print('val_loss: {}, val_acc: {}'.format(scores[0], scores[1]))
y_true, y_pred = get_predictions_and_labels(model, X_val, y_val)
cm = confusion_matrix(y_true, y_pred)
cm_percent = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
df = pd.DataFrame(cm_percent, index=self.EMOTIONS, columns=self.EMOTIONS)
df.index.name = 'Actual'
df.columns.name = 'Predicted'
df.to_csv(self.base_dir + self.model_dir + 'cm_val.csv', float_format='%.4f')
# plot percentage confusion matrix
fig1, ax1 = plt.subplots()
plot_confusion_matrix(cm_percent, class_names=self.EMOTIONS)
plt.savefig(self.base_dir + self.model_dir + 'cm_percent_val.png', format='png')
# plot normal confusion matrix
fig2, ax2 = plt.subplots()
plot_confusion_matrix(cm, float_display='.0f', class_names=self.EMOTIONS)
plt.savefig(self.base_dir + self.model_dir + 'cm_val.png', format='png')
plt.show()
def compare_model(self, X_val, y_val):
folder_list = [model_dir for model_dir in os.listdir(self.base_dir) if 'LSTM' in model_dir]
for folder in folder_list:
filename = 'LSTM.h5'
path = os.path.join(self.base_dir, folder, filename)
model = load_model(path)
scores = model.evaluate(X_val, y_val)
print('model: {}, val_loss: {}, val_acc: {}'.format(folder, scores[0], scores[1]))
y_true, y_pred = get_predictions_and_labels(model, X_val, y_val)
cm = confusion_matrix(y_true, y_pred)
cm_percent = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
# plot percentage confusion matrix
fig1, ax1 = plt.subplots()
plot_confusion_matrix(cm_percent, class_names=self.EMOTIONS)
plt.savefig(os.path.join(self.base_dir, folder, 'cm_percent_test.png'), format='png')
# plot normal confusion matrix
fig2, ax2 = plt.subplots()
plot_confusion_matrix(cm, float_display='.0f', class_names=self.EMOTIONS)
plt.savefig(os.path.join(self.base_dir, folder, 'cm_test.png'), format='png')
feature = 'InceptionResNetV2'
data_type = 'Basic'
n_layer = 1
lstm_unit = 32
batch_size = 256
epochs = 250
lstm_net = LSTMNetwork(n_layer, lstm_unit, X_train.shape[1:], feature, data_type)
lstm_net.train(X_train, y_train, X_test, y_test, epochs, batch_size)
lstm_net.evaluate(X_val, y_val)
| 0.50952 | 0.379148 |
# XGBoost
Ranklib is a relatively old library and doesn't have the wide spread use that XGBoost does. Ranklib is still under active development, but the fork of the project OSC created reflects an older version.
The ES-LTR plugin is designed to work with XGBoost model format. This notebook starts with the `classic` training data generated in `hello-ltr.py` and shows how you could use XGBoost instead of Ranklib to create a model and use it with the plugin.
### Input Data
Gather the data generated for our `classic` model in `hello-ltr.ipynb`. If this file doesn't exist yet, rerun that notebook!
```
import ltr.judgments as judge
df = [j for j in judge.judgments_from_file(open('data/classic-training.txt'))]
df = judge.judgments_to_dataframe(df)
df
```
### Libraries for xgboost-ing
Just the dependencies we need to train and visualize out model trained with XG-Boost instead of Ranklib.
```
import pandas as pd
import xgboost as xgb
from matplotlib.pylab import rcParams
rcParams['figure.figsize'] = 50,150
```
### Set up our training Matrix
XGBoost has it's data specficiations so we need to get out features into that format to use it.
```
df = df[['grade', 'features0']]
features = df[['features0']]
labels = df[['grade']]
dmx = xgb.DMatrix(features, labels)
```
### Train the first XGBoost model
Using the demo parameters for our model, we will train a standard regression tree
```
param = {'max_depth':2, 'eta':1, 'silent':1}
num_round = 2
model = xgb.train(param, dmx, num_round)
```
### Inspect as dataframe
Looking at the model as a dataframe can tell you which splits helped the most
```
model.trees_to_dataframe()
xgb.plot_tree(model)
```
### Adjust the objective for LTR
Really we don't want the regression as our objective function. In LTR we take advantage of a new pairwise loss function to find the optimal splits for a regression tree.
This doesn't make a massive difference for the model that is generated because it is still a regression tree at the end of the day, but we are not longer using residual sqared error.
```
param2 = {'max_depth':2, 'eta':1, 'silent':1, 'objective':'rank:pairwise'}
ranking_model = xgb.train(param2, dmx, num_round)
ranking_model.trees_to_dataframe()
xgb.plot_tree(ranking_model)
```
### Uploading an XGBoost model to the plugin
Since the model can be represented with JSON, the plugin can parse it. But we need to make sure the plugin gets the proper feature value names in order for it to parse properly.
These are supplied via a mapping `txt` file, `fmap.txt`.
The first step is to dump the model with the feature mapping to the features already stored in the plugin.
```
model_dump = ranking_model.get_dump(fmap='fmap.txt', dump_format='json')
```
### Massage the JSON
Manipulate the XGBoost output format to clean it up for posting to the plugin.
```
import json
clean_model = []
for line in model_dump:
clean_model.append(json.loads(line))
```
### Post it to the plugin
Still referencing the index and feature set the model will be associated with.
```
import ltr.client as client
client = client.ElasticClient()
client.submit_xgboost_model('release', 'tmdb', 'xgb', clean_model)
```
### Confirm it works
```
from ltr.release_date_plot import search
search(client, 'batman', 'xgb')
```
### Compare it to the classic Ranklib model
```
from ltr.release_date_plot import plot
plot(client, "batman", models = ['classic', 'xgb'])
```
|
github_jupyter
|
import ltr.judgments as judge
df = [j for j in judge.judgments_from_file(open('data/classic-training.txt'))]
df = judge.judgments_to_dataframe(df)
df
import pandas as pd
import xgboost as xgb
from matplotlib.pylab import rcParams
rcParams['figure.figsize'] = 50,150
df = df[['grade', 'features0']]
features = df[['features0']]
labels = df[['grade']]
dmx = xgb.DMatrix(features, labels)
param = {'max_depth':2, 'eta':1, 'silent':1}
num_round = 2
model = xgb.train(param, dmx, num_round)
model.trees_to_dataframe()
xgb.plot_tree(model)
param2 = {'max_depth':2, 'eta':1, 'silent':1, 'objective':'rank:pairwise'}
ranking_model = xgb.train(param2, dmx, num_round)
ranking_model.trees_to_dataframe()
xgb.plot_tree(ranking_model)
model_dump = ranking_model.get_dump(fmap='fmap.txt', dump_format='json')
import json
clean_model = []
for line in model_dump:
clean_model.append(json.loads(line))
import ltr.client as client
client = client.ElasticClient()
client.submit_xgboost_model('release', 'tmdb', 'xgb', clean_model)
from ltr.release_date_plot import search
search(client, 'batman', 'xgb')
from ltr.release_date_plot import plot
plot(client, "batman", models = ['classic', 'xgb'])
| 0.26341 | 0.924005 |
# Survival Analysis in Python
Chapter 1
Allen B. Downey
[MIT License](https://en.wikipedia.org/wiki/MIT_License)
```
# Configure Jupyter so figures appear in the notebook
%matplotlib inline
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(style='white')
import utils
from utils import decorate
from empyrical_dist import Pmf, Cdf
```
Data from [https://gist.github.com/epogrebnyak/7933e16c0ad215742c4c104be4fbdeb1]
```
Dataset from:
V.J. Menon and D.C. Agrawal, Renewal Rate of Filament Lamps:
Theory and Experiment. Journal of Failure Analysis and Prevention.
December 2007, p. 421, Table 2/
DOI: 10.1007/s11668-007-9074-9
Description:
An assembly of 50 new Philips (India) lamps with the
rating 40 W, 220 V (AC) was taken and installed in the horizontal
orientation and uniformly distributed over a lab area 11 m 9 7 m.
The assembly was monitored at regular intervals of 12 h to
look for failures. The instants of recorded failures were
called t‘ and a total of 32 data points were obtained such
that even the last bulb failed.
Variables:
i - observation number
h - time in hours since experiment start
f - number of failed lamps at particular time h
K - number of surviving lamps at particular time h
```
```
df = pd.read_csv("data/lamps.csv")
df.head()
pmf = Pmf(df.f.values, index=df.h)
pmf.normalize()
pmf.head()
pmf.bar(width=30)
decorate(xlabel='Lifetime (hours)',
ylabel='PMF',
title='PMF of lightbulb lifetimes')
cdf = pmf.make_cdf()
cdf.head()
cdf.step()
decorate(xlabel='Lifetime (hours)',
ylabel='CDF',
title='CDF of lightbulb lifetimes')
def make_surv(cdf):
return Surv(1-cdf.ps, index=cdf.qs)
Cdf.make_surv = make_surv
def underride(d, **options):
"""Add key-value pairs to d only if key is not in d.
d: dictionary
options: keyword args to add to d
:return: modified d
"""
for key, val in options.items():
d.setdefault(key, val)
return d
from scipy.interpolate import interp1d
class Surv(pd.Series):
"""Represents a survival function (complementary CDF)."""
def __init__(self, *args, **kwargs):
"""Initialize a survival function.
Note: this cleans up a weird Series behavior, which is
that Series() and Series([]) yield different results.
See: https://github.com/pandas-dev/pandas/issues/16737
"""
if args:
super().__init__(*args, **kwargs)
else:
underride(kwargs, dtype=np.float64)
super().__init__([], **kwargs)
def copy(self, deep=True):
"""Make a copy.
:return: new Surv
"""
return Surv(self, copy=deep)
@staticmethod
def from_seq(seq, normalize=True, sort=True, **options):
"""Make a Surv from a sequence of values.
seq: any kind of sequence
normalize: whether to normalize the Surv, default True
sort: whether to sort the Surv by values, default True
options: passed to the pd.Series constructor
:return: Surv object
"""
pmf = Pmf.from_seq(seq, normalize=False, sort=sort, **options)
cdf = pmf.make_cdf(normalize=normalize)
return cdf.make_surv()
@property
def qs(self):
"""Get the quantities.
:return: NumPy array
"""
return self.index.values
@property
def ps(self):
"""Get the probabilities.
:return: NumPy array
"""
return self.values
def _repr_html_(self):
"""Returns an HTML representation of the series.
Mostly used for Jupyter notebooks.
"""
df = pd.DataFrame(dict(probs=self))
return df._repr_html_()
def plot(self, **options):
"""Plot the Cdf as a line.
:param options: passed to plt.plot
:return:
"""
underride(options, label=self.name)
plt.plot(self.qs, self.ps, **options)
def step(self, **options):
"""Plot the Cdf as a step function.
:param options: passed to plt.step
:return:
"""
underride(options, label=self.name, where='post')
plt.step(self.qs, self.ps, **options)
def normalize(self):
"""Make the probabilities add up to 1 (modifies self).
:return: normalizing constant
"""
total = self.ps[-1]
self /= total
return total
@property
def forward(self, **kwargs):
"""Compute the forward Cdf
:param kwargs: keyword arguments passed to interp1d
:return array of probabilities
"""
underride(kwargs, kind='previous',
copy=False,
assume_sorted=True,
bounds_error=False,
fill_value=(1, 0))
interp = interp1d(self.qs, self.ps, **kwargs)
return interp
@property
def inverse(self, **kwargs):
"""Compute the inverse Cdf
:param kwargs: keyword arguments passed to interp1d
:return array of quantities
"""
interp = self.make_cdf().inverse
return lambda ps: interp(1-ps, **kwargs)
# calling a Cdf like a function does forward lookup
__call__ = forward
# quantile is the same as an inverse lookup
quantile = inverse
def make_cdf(self, normalize=False):
"""Make a Cdf from the Surv.
:return: Cdf
"""
cdf = Cdf(1-self.ps, index=self.qs)
if normalize:
cdf.normalize()
return cdf
def make_pmf(self, normalize=False):
"""Make a Pmf from the Surv.
:return: Pmf
"""
cdf = self.make_cdf(normalize=False)
pmf = cdf.make_pmf(normalize=normalize)
return pmf
def make_hazard(self, **kwargs):
"""Make a Hazard object from the Surv.
:return: Hazard object
"""
# TODO: Get the Pandas-idiomatic version of this
lams = pd.Series(index=self.qs)
prev = 1.0
for q, p in self.iteritems():
lams[q] = (prev - p) / prev
prev = p
return Hazard(lams, **kwargs)
def choice(self, *args, **kwargs):
"""Makes a random sample.
Uses the probabilities as weights unless `p` is provided.
args: same as np.random.choice
options: same as np.random.choice
:return: NumPy array
"""
# TODO: Make this more efficient by implementing the inverse CDF method.
pmf = self.make_pmf()
return pmf.choice(*args, **kwargs)
def sample(self, *args, **kwargs):
"""Makes a random sample.
Uses the probabilities as weights unless `weights` is provided.
This function returns an array containing a sample of the quantities in this Pmf,
which is different from Series.sample, which returns a Series with a sample of
the rows in the original Series.
args: same as Series.sample
options: same as Series.sample
:return: NumPy array
"""
# TODO: Make this more efficient by implementing the inverse CDF method.
pmf = self.make_pmf()
return pmf.sample(*args, **kwargs)
def mean(self):
"""Expected value.
:return: float
"""
return self.make_pmf().mean()
def var(self):
"""Variance.
:return: float
"""
return self.make_pmf().var()
def std(self):
"""Standard deviation.
:return: float
"""
return self.make_pmf().std()
def median(self):
"""Median (50th percentile).
:return: float
"""
return self.quantile(0.5)
surv = cdf.make_surv()
surv.head()
cdf.step(color='gray', alpha=0.3)
surv.step()
decorate(xlabel='Lifetime (hours)',
ylabel='Prob(lifetime>t)',
title='Survival function of lightbulb lifetimes')
surv(-1)
surv(3000)
surv.median()
surv.mean()
class Hazard(pd.Series):
"""Represents a Hazard function."""
def __init__(self, *args, **kwargs):
"""Initialize a Hazard.
Note: this cleans up a weird Series behavior, which is
that Series() and Series([]) yield different results.
See: https://github.com/pandas-dev/pandas/issues/16737
"""
if args:
super().__init__(*args, **kwargs)
else:
underride(kwargs, dtype=np.float64)
super().__init__([], **kwargs)
def copy(self, deep=True):
"""Make a copy.
:return: new Pmf
"""
return Hazard(self, copy=deep)
def __getitem__(self, qs):
"""Look up qs and return ps."""
try:
return super().__getitem__(qs)
except (KeyError, ValueError, IndexError):
return 0
@property
def qs(self):
"""Get the quantities.
:return: NumPy array
"""
return self.index.values
@property
def ps(self):
"""Get the probabilities.
:return: NumPy array
"""
return self.values
def _repr_html_(self):
"""Returns an HTML representation of the series.
Mostly used for Jupyter notebooks.
"""
df = pd.DataFrame(dict(probs=self))
return df._repr_html_()
def mean(self):
"""Computes expected value.
:return: float
"""
raise ValueError()
def median(self):
"""Median (50th percentile).
:return: float
"""
raise ValueError()
def quantile(self, ps, **kwargs):
"""Quantiles.
Computes the inverse CDF of ps, that is,
the values that correspond to the given probabilities.
:return: float
"""
raise ValueError()
def var(self):
"""Variance of a PMF.
:return: float
"""
raise ValueError()
def std(self):
"""Standard deviation of a PMF.
:return: float
"""
raise ValueError()
def choice(self, *args, **kwargs):
"""Makes a random sample.
Uses the probabilities as weights unless `p` is provided.
args: same as np.random.choice
kwargs: same as np.random.choice
:return: NumPy array
"""
raise ValueError()
def sample(self, *args, **kwargs):
"""Makes a random sample.
Uses the probabilities as weights unless `weights` is provided.
This function returns an array containing a sample of the quantities,
which is different from Series.sample, which returns a Series with a sample of
the rows in the original Series.
args: same as Series.sample
options: same as Series.sample
:return: NumPy array
"""
raise ValueError()
def plot(self, **options):
"""Plot the Pmf as a line.
:param options: passed to plt.plot
:return:
"""
underride(options, label=self.name)
plt.plot(self.qs, self.ps, **options)
def bar(self, **options):
"""Makes a bar plot.
options: passed to plt.bar
"""
underride(options, label=self.name)
plt.bar(self.qs, self.ps, **options)
def make_cdf(self, normalize=True):
"""Make a Cdf from the Hazard.
It can be good to normalize the cdf even if the Pmf was normalized,
to guarantee that the last element of `ps` is 1.
:return: Cdf
"""
cdf = self.make_surv().make_cdf()
return cdf
def make_surv(self, normalize=True):
"""Make a Surv from the Hazard.
:return: Surv
"""
ps = (1 - self.ps).cumprod()
return Surv(ps, index=self.qs)
@staticmethod
def from_seq(seq, **options):
"""Make a PMF from a sequence of values.
seq: any kind of sequence
normalize: whether to normalize the Pmf, default True
sort: whether to sort the Pmf by values, default True
options: passed to the pd.Series constructor
:return: Pmf object
"""
cdf = Cdf.from_seq(seq, **options)
surv = cdf.make_surv()
return surv.make_hazard()
haz = surv.make_hazard()
haz.bar(width=30)
decorate(xlabel='Lifetime (hours)',
ylabel='Hazard rate',
title='Hazard function of lightbulb lifetimes')
surv2 = haz.make_surv()
max(abs(surv - surv2))
cdf2 = surv2.make_cdf()
max(abs(cdf - cdf2))
pmf2 = cdf2.make_pmf()
max(abs(pmf.ps - pmf2.ps))
from scipy.stats import gaussian_kde
def estimate_smooth_hazard(pmf):
xs = np.linspace(min(pmf.qs), max(pmf.qs))
kde = gaussian_kde(pmf.qs, weights=pmf.ps)
pdf = Pmf(kde(xs), index=xs)
pdf.normalize()
ps = pdf.make_cdf().make_surv().forward(xs)
surv = Surv(ps, index=xs)
lams = pdf.ps / surv
haz = Hazard(lams)
return pdf, surv, haz
pdf, surv, haz = estimate_smooth_hazard(pmf)
pdf.plot()
decorate(xlabel='Lifetime (hours)',
ylabel='Probability density',
title='Estimated PDF of lightbulb lifetimes')
surv.plot()
decorate(xlabel='Lifetime (hours)',
ylabel='Prob(lifetime > t)',
title='Estimated survival function of lightbulb lifetimes')
haz.plot()
decorate(xlabel='Lifetime (hours)',
ylabel='Hazard rate',
title='Estimated hazard function of lightbulb lifetimes')
pmf = Pmf(df.f.values, index=df.h)
kde = gaussian_kde(pmf.qs, weights=pmf.ps)
size = pmf.sum()
for i in range(100):
sample = kde.resample(size).flatten()
pdf, surv, haz = estimate_smooth_hazard(Pmf.from_seq(sample))
surv.plot(color='gray', alpha=0.1)
decorate(xlabel='Lifetime (hours)',
ylabel='Prob(lifetime > t)',
title='Estimated survival function of lightbulb lifetimes')
for i in range(100):
sample = kde.resample(size).flatten()
pdf, surv, haz = estimate_smooth_hazard(Pmf.from_seq(sample))
haz.plot(color='gray', alpha=0.1)
decorate(xlabel='Lifetime (hours)',
ylabel='Prob(lifetime > t)',
title='Estimated survival function of lightbulb lifetimes')
from collections import Counter
pmf = Pmf(df.f.values, index=df.h)
counter = Counter(dict(pmf.iteritems()))
data = np.fromiter(counter.elements(), dtype=np.int64)
len(data), pmf.sum()
from scipy.stats import exponweib
a, c, loc, scale = exponweib.fit(data)
qs = pmf.qs
ps = exponweib(a, c, loc, scale).sf(qs)
plt.plot(qs, ps)
pmf.make_cdf().make_surv().plot()
decorate(xlabel='Lifetime (hours)',
ylabel='Prob(lifetime > t)',
title='Survival function of lightbulb lifetimes')
```
### Popcorn
Description of the experiment here:
https://www.reddit.com/r/dataisbeautiful/comments/8uo1q9/popcorn_analytics_oc/
Data downloaded from here
https://textuploader.com/dzqpd
```
df = pd.read_csv('data/popcorn.csv', sep=';')
df.head()
data = df['time of the pop in seconds']
pmf = Pmf.from_seq(data)
pdf, surv, haz = estimate_smooth_hazard(pmf)
pdf.plot()
decorate(xlabel='Time until pop (seconds)',
ylabel='Probability density',
title='Estimated PDF of popcorn popping time')
surv.plot()
decorate(xlabel='Time until pop (seconds)',
ylabel='Prob(lifetime>t)',
title='Estimated survival function of popcorn popping time')
haz.plot()
decorate(xlabel='Time until pop (seconds)',
ylabel='Hazard rate',
title='Estimated hazard function of popcorn popping time')
kde = gaussian_kde(data)
size = len(data)
for i in range(20):
sample = kde.resample(size).flatten()
pdf, surv, haz = estimate_smooth_hazard(Pmf.from_seq(sample))
surv.plot(color='gray', alpha=0.1)
pmf.make_cdf().make_surv().plot()
decorate(xlabel='Time until pop (seconds)',
ylabel='Prob(lifetime>t)',
title='Estimated survival function of popcorn popping time')
for i in range(20):
sample = kde.resample(size).flatten()
pdf, surv, haz = estimate_smooth_hazard(Pmf.from_seq(sample))
haz.plot(color='gray', alpha=0.1)
haz.plot()
decorate(xlabel='Time until pop (seconds)',
ylabel='Hazard rate',
title='Estimated hazard function of popcorn popping time')
from scipy.stats import exponweib
a, c, loc, scale = exponweib.fit(data)
a, c, loc, scale
qs = pmf.qs
ps = exponweib(a, c, loc, scale).sf(qs)
plt.plot(qs, ps)
pmf.make_cdf().make_surv().plot()
cdf = pmf.make_cdf()
cdf.plot()
decorate(xlabel='Time until pop (seconds)',
ylabel='Prob(lifetime>t)',
title='Survival function of popcorn popping time')
from scipy.stats import norm
rv = norm(pmf.mean(), pmf.std())
qs = np.linspace(data.min(), data.max())
ps = rv.cdf(qs)
model_cdf = Cdf(ps, index=qs)
model_cdf.plot(color='gray', alpha=0.5)
cdf.plot()
decorate(xlabel='Time until pop (seconds)',
ylabel='Prob(lifetime>t)',
title='Survival function of popcorn popping time')
model_haz = model_cdf.make_surv().make_hazard()
model_haz.plot(color='gray')
pdf, surv, haz = estimate_smooth_hazard(pmf)
haz.plot()
decorate(xlabel='Time until pop (seconds)',
ylabel='Hazard rate',
title='Hazard function of popcorn popping time')
```
Why estimating the right side of the hazard function is nearly impossible:
1) If we take a data-intensive approach, we are trying to estimate the probabilities of rare events, which is hard.
2) The right side of the survival curve is noisy enough, but then we amplify the noise twice, first by taking a derivative, and then by dividing through by the survival function as it goes to zero. In particular, the last point is always bogus.
3) If we take a model-intensive approach, we are depending on the validity of estimating a model based on the bulk of the distribution and then extrapolating into the tail. But when reality deviates from a model, the tail is where it happens. In the light-bulb example, the left tail is probably populated by manufacturing defects. The right tail might include procedural errors (wrong kind of light bulb, operated under non-compliant conditions), measurement errors, recording errors,etc.
If we plot confidence intervals, we are at least aware of where the estimate is not reliable. But in general it is probably unwise to base any conclusion on the right half of an estimated hazard function.
|
github_jupyter
|
# Configure Jupyter so figures appear in the notebook
%matplotlib inline
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(style='white')
import utils
from utils import decorate
from empyrical_dist import Pmf, Cdf
Dataset from:
V.J. Menon and D.C. Agrawal, Renewal Rate of Filament Lamps:
Theory and Experiment. Journal of Failure Analysis and Prevention.
December 2007, p. 421, Table 2/
DOI: 10.1007/s11668-007-9074-9
Description:
An assembly of 50 new Philips (India) lamps with the
rating 40 W, 220 V (AC) was taken and installed in the horizontal
orientation and uniformly distributed over a lab area 11 m 9 7 m.
The assembly was monitored at regular intervals of 12 h to
look for failures. The instants of recorded failures were
called t‘ and a total of 32 data points were obtained such
that even the last bulb failed.
Variables:
i - observation number
h - time in hours since experiment start
f - number of failed lamps at particular time h
K - number of surviving lamps at particular time h
df = pd.read_csv("data/lamps.csv")
df.head()
pmf = Pmf(df.f.values, index=df.h)
pmf.normalize()
pmf.head()
pmf.bar(width=30)
decorate(xlabel='Lifetime (hours)',
ylabel='PMF',
title='PMF of lightbulb lifetimes')
cdf = pmf.make_cdf()
cdf.head()
cdf.step()
decorate(xlabel='Lifetime (hours)',
ylabel='CDF',
title='CDF of lightbulb lifetimes')
def make_surv(cdf):
return Surv(1-cdf.ps, index=cdf.qs)
Cdf.make_surv = make_surv
def underride(d, **options):
"""Add key-value pairs to d only if key is not in d.
d: dictionary
options: keyword args to add to d
:return: modified d
"""
for key, val in options.items():
d.setdefault(key, val)
return d
from scipy.interpolate import interp1d
class Surv(pd.Series):
"""Represents a survival function (complementary CDF)."""
def __init__(self, *args, **kwargs):
"""Initialize a survival function.
Note: this cleans up a weird Series behavior, which is
that Series() and Series([]) yield different results.
See: https://github.com/pandas-dev/pandas/issues/16737
"""
if args:
super().__init__(*args, **kwargs)
else:
underride(kwargs, dtype=np.float64)
super().__init__([], **kwargs)
def copy(self, deep=True):
"""Make a copy.
:return: new Surv
"""
return Surv(self, copy=deep)
@staticmethod
def from_seq(seq, normalize=True, sort=True, **options):
"""Make a Surv from a sequence of values.
seq: any kind of sequence
normalize: whether to normalize the Surv, default True
sort: whether to sort the Surv by values, default True
options: passed to the pd.Series constructor
:return: Surv object
"""
pmf = Pmf.from_seq(seq, normalize=False, sort=sort, **options)
cdf = pmf.make_cdf(normalize=normalize)
return cdf.make_surv()
@property
def qs(self):
"""Get the quantities.
:return: NumPy array
"""
return self.index.values
@property
def ps(self):
"""Get the probabilities.
:return: NumPy array
"""
return self.values
def _repr_html_(self):
"""Returns an HTML representation of the series.
Mostly used for Jupyter notebooks.
"""
df = pd.DataFrame(dict(probs=self))
return df._repr_html_()
def plot(self, **options):
"""Plot the Cdf as a line.
:param options: passed to plt.plot
:return:
"""
underride(options, label=self.name)
plt.plot(self.qs, self.ps, **options)
def step(self, **options):
"""Plot the Cdf as a step function.
:param options: passed to plt.step
:return:
"""
underride(options, label=self.name, where='post')
plt.step(self.qs, self.ps, **options)
def normalize(self):
"""Make the probabilities add up to 1 (modifies self).
:return: normalizing constant
"""
total = self.ps[-1]
self /= total
return total
@property
def forward(self, **kwargs):
"""Compute the forward Cdf
:param kwargs: keyword arguments passed to interp1d
:return array of probabilities
"""
underride(kwargs, kind='previous',
copy=False,
assume_sorted=True,
bounds_error=False,
fill_value=(1, 0))
interp = interp1d(self.qs, self.ps, **kwargs)
return interp
@property
def inverse(self, **kwargs):
"""Compute the inverse Cdf
:param kwargs: keyword arguments passed to interp1d
:return array of quantities
"""
interp = self.make_cdf().inverse
return lambda ps: interp(1-ps, **kwargs)
# calling a Cdf like a function does forward lookup
__call__ = forward
# quantile is the same as an inverse lookup
quantile = inverse
def make_cdf(self, normalize=False):
"""Make a Cdf from the Surv.
:return: Cdf
"""
cdf = Cdf(1-self.ps, index=self.qs)
if normalize:
cdf.normalize()
return cdf
def make_pmf(self, normalize=False):
"""Make a Pmf from the Surv.
:return: Pmf
"""
cdf = self.make_cdf(normalize=False)
pmf = cdf.make_pmf(normalize=normalize)
return pmf
def make_hazard(self, **kwargs):
"""Make a Hazard object from the Surv.
:return: Hazard object
"""
# TODO: Get the Pandas-idiomatic version of this
lams = pd.Series(index=self.qs)
prev = 1.0
for q, p in self.iteritems():
lams[q] = (prev - p) / prev
prev = p
return Hazard(lams, **kwargs)
def choice(self, *args, **kwargs):
"""Makes a random sample.
Uses the probabilities as weights unless `p` is provided.
args: same as np.random.choice
options: same as np.random.choice
:return: NumPy array
"""
# TODO: Make this more efficient by implementing the inverse CDF method.
pmf = self.make_pmf()
return pmf.choice(*args, **kwargs)
def sample(self, *args, **kwargs):
"""Makes a random sample.
Uses the probabilities as weights unless `weights` is provided.
This function returns an array containing a sample of the quantities in this Pmf,
which is different from Series.sample, which returns a Series with a sample of
the rows in the original Series.
args: same as Series.sample
options: same as Series.sample
:return: NumPy array
"""
# TODO: Make this more efficient by implementing the inverse CDF method.
pmf = self.make_pmf()
return pmf.sample(*args, **kwargs)
def mean(self):
"""Expected value.
:return: float
"""
return self.make_pmf().mean()
def var(self):
"""Variance.
:return: float
"""
return self.make_pmf().var()
def std(self):
"""Standard deviation.
:return: float
"""
return self.make_pmf().std()
def median(self):
"""Median (50th percentile).
:return: float
"""
return self.quantile(0.5)
surv = cdf.make_surv()
surv.head()
cdf.step(color='gray', alpha=0.3)
surv.step()
decorate(xlabel='Lifetime (hours)',
ylabel='Prob(lifetime>t)',
title='Survival function of lightbulb lifetimes')
surv(-1)
surv(3000)
surv.median()
surv.mean()
class Hazard(pd.Series):
"""Represents a Hazard function."""
def __init__(self, *args, **kwargs):
"""Initialize a Hazard.
Note: this cleans up a weird Series behavior, which is
that Series() and Series([]) yield different results.
See: https://github.com/pandas-dev/pandas/issues/16737
"""
if args:
super().__init__(*args, **kwargs)
else:
underride(kwargs, dtype=np.float64)
super().__init__([], **kwargs)
def copy(self, deep=True):
"""Make a copy.
:return: new Pmf
"""
return Hazard(self, copy=deep)
def __getitem__(self, qs):
"""Look up qs and return ps."""
try:
return super().__getitem__(qs)
except (KeyError, ValueError, IndexError):
return 0
@property
def qs(self):
"""Get the quantities.
:return: NumPy array
"""
return self.index.values
@property
def ps(self):
"""Get the probabilities.
:return: NumPy array
"""
return self.values
def _repr_html_(self):
"""Returns an HTML representation of the series.
Mostly used for Jupyter notebooks.
"""
df = pd.DataFrame(dict(probs=self))
return df._repr_html_()
def mean(self):
"""Computes expected value.
:return: float
"""
raise ValueError()
def median(self):
"""Median (50th percentile).
:return: float
"""
raise ValueError()
def quantile(self, ps, **kwargs):
"""Quantiles.
Computes the inverse CDF of ps, that is,
the values that correspond to the given probabilities.
:return: float
"""
raise ValueError()
def var(self):
"""Variance of a PMF.
:return: float
"""
raise ValueError()
def std(self):
"""Standard deviation of a PMF.
:return: float
"""
raise ValueError()
def choice(self, *args, **kwargs):
"""Makes a random sample.
Uses the probabilities as weights unless `p` is provided.
args: same as np.random.choice
kwargs: same as np.random.choice
:return: NumPy array
"""
raise ValueError()
def sample(self, *args, **kwargs):
"""Makes a random sample.
Uses the probabilities as weights unless `weights` is provided.
This function returns an array containing a sample of the quantities,
which is different from Series.sample, which returns a Series with a sample of
the rows in the original Series.
args: same as Series.sample
options: same as Series.sample
:return: NumPy array
"""
raise ValueError()
def plot(self, **options):
"""Plot the Pmf as a line.
:param options: passed to plt.plot
:return:
"""
underride(options, label=self.name)
plt.plot(self.qs, self.ps, **options)
def bar(self, **options):
"""Makes a bar plot.
options: passed to plt.bar
"""
underride(options, label=self.name)
plt.bar(self.qs, self.ps, **options)
def make_cdf(self, normalize=True):
"""Make a Cdf from the Hazard.
It can be good to normalize the cdf even if the Pmf was normalized,
to guarantee that the last element of `ps` is 1.
:return: Cdf
"""
cdf = self.make_surv().make_cdf()
return cdf
def make_surv(self, normalize=True):
"""Make a Surv from the Hazard.
:return: Surv
"""
ps = (1 - self.ps).cumprod()
return Surv(ps, index=self.qs)
@staticmethod
def from_seq(seq, **options):
"""Make a PMF from a sequence of values.
seq: any kind of sequence
normalize: whether to normalize the Pmf, default True
sort: whether to sort the Pmf by values, default True
options: passed to the pd.Series constructor
:return: Pmf object
"""
cdf = Cdf.from_seq(seq, **options)
surv = cdf.make_surv()
return surv.make_hazard()
haz = surv.make_hazard()
haz.bar(width=30)
decorate(xlabel='Lifetime (hours)',
ylabel='Hazard rate',
title='Hazard function of lightbulb lifetimes')
surv2 = haz.make_surv()
max(abs(surv - surv2))
cdf2 = surv2.make_cdf()
max(abs(cdf - cdf2))
pmf2 = cdf2.make_pmf()
max(abs(pmf.ps - pmf2.ps))
from scipy.stats import gaussian_kde
def estimate_smooth_hazard(pmf):
xs = np.linspace(min(pmf.qs), max(pmf.qs))
kde = gaussian_kde(pmf.qs, weights=pmf.ps)
pdf = Pmf(kde(xs), index=xs)
pdf.normalize()
ps = pdf.make_cdf().make_surv().forward(xs)
surv = Surv(ps, index=xs)
lams = pdf.ps / surv
haz = Hazard(lams)
return pdf, surv, haz
pdf, surv, haz = estimate_smooth_hazard(pmf)
pdf.plot()
decorate(xlabel='Lifetime (hours)',
ylabel='Probability density',
title='Estimated PDF of lightbulb lifetimes')
surv.plot()
decorate(xlabel='Lifetime (hours)',
ylabel='Prob(lifetime > t)',
title='Estimated survival function of lightbulb lifetimes')
haz.plot()
decorate(xlabel='Lifetime (hours)',
ylabel='Hazard rate',
title='Estimated hazard function of lightbulb lifetimes')
pmf = Pmf(df.f.values, index=df.h)
kde = gaussian_kde(pmf.qs, weights=pmf.ps)
size = pmf.sum()
for i in range(100):
sample = kde.resample(size).flatten()
pdf, surv, haz = estimate_smooth_hazard(Pmf.from_seq(sample))
surv.plot(color='gray', alpha=0.1)
decorate(xlabel='Lifetime (hours)',
ylabel='Prob(lifetime > t)',
title='Estimated survival function of lightbulb lifetimes')
for i in range(100):
sample = kde.resample(size).flatten()
pdf, surv, haz = estimate_smooth_hazard(Pmf.from_seq(sample))
haz.plot(color='gray', alpha=0.1)
decorate(xlabel='Lifetime (hours)',
ylabel='Prob(lifetime > t)',
title='Estimated survival function of lightbulb lifetimes')
from collections import Counter
pmf = Pmf(df.f.values, index=df.h)
counter = Counter(dict(pmf.iteritems()))
data = np.fromiter(counter.elements(), dtype=np.int64)
len(data), pmf.sum()
from scipy.stats import exponweib
a, c, loc, scale = exponweib.fit(data)
qs = pmf.qs
ps = exponweib(a, c, loc, scale).sf(qs)
plt.plot(qs, ps)
pmf.make_cdf().make_surv().plot()
decorate(xlabel='Lifetime (hours)',
ylabel='Prob(lifetime > t)',
title='Survival function of lightbulb lifetimes')
df = pd.read_csv('data/popcorn.csv', sep=';')
df.head()
data = df['time of the pop in seconds']
pmf = Pmf.from_seq(data)
pdf, surv, haz = estimate_smooth_hazard(pmf)
pdf.plot()
decorate(xlabel='Time until pop (seconds)',
ylabel='Probability density',
title='Estimated PDF of popcorn popping time')
surv.plot()
decorate(xlabel='Time until pop (seconds)',
ylabel='Prob(lifetime>t)',
title='Estimated survival function of popcorn popping time')
haz.plot()
decorate(xlabel='Time until pop (seconds)',
ylabel='Hazard rate',
title='Estimated hazard function of popcorn popping time')
kde = gaussian_kde(data)
size = len(data)
for i in range(20):
sample = kde.resample(size).flatten()
pdf, surv, haz = estimate_smooth_hazard(Pmf.from_seq(sample))
surv.plot(color='gray', alpha=0.1)
pmf.make_cdf().make_surv().plot()
decorate(xlabel='Time until pop (seconds)',
ylabel='Prob(lifetime>t)',
title='Estimated survival function of popcorn popping time')
for i in range(20):
sample = kde.resample(size).flatten()
pdf, surv, haz = estimate_smooth_hazard(Pmf.from_seq(sample))
haz.plot(color='gray', alpha=0.1)
haz.plot()
decorate(xlabel='Time until pop (seconds)',
ylabel='Hazard rate',
title='Estimated hazard function of popcorn popping time')
from scipy.stats import exponweib
a, c, loc, scale = exponweib.fit(data)
a, c, loc, scale
qs = pmf.qs
ps = exponweib(a, c, loc, scale).sf(qs)
plt.plot(qs, ps)
pmf.make_cdf().make_surv().plot()
cdf = pmf.make_cdf()
cdf.plot()
decorate(xlabel='Time until pop (seconds)',
ylabel='Prob(lifetime>t)',
title='Survival function of popcorn popping time')
from scipy.stats import norm
rv = norm(pmf.mean(), pmf.std())
qs = np.linspace(data.min(), data.max())
ps = rv.cdf(qs)
model_cdf = Cdf(ps, index=qs)
model_cdf.plot(color='gray', alpha=0.5)
cdf.plot()
decorate(xlabel='Time until pop (seconds)',
ylabel='Prob(lifetime>t)',
title='Survival function of popcorn popping time')
model_haz = model_cdf.make_surv().make_hazard()
model_haz.plot(color='gray')
pdf, surv, haz = estimate_smooth_hazard(pmf)
haz.plot()
decorate(xlabel='Time until pop (seconds)',
ylabel='Hazard rate',
title='Hazard function of popcorn popping time')
| 0.796055 | 0.892609 |
<a href="https://colab.research.google.com/github/jaya-shankar/education-impact/blob/jaya-shankar/randomForest.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
!rm -rf education-impact
!rm education-impact
!git clone https://github.com/jaya-shankar/education-impact.git
!pip install tensorflow_decision_forests
!pip install wurlitzer
root = "education-impact/"
datasets_path = {
"infant_mortality" : root+ "datasets/Infant_Mortality_Rate.csv",
"child_mortality" : root+ "datasets/child_mortality_0_5_year_olds_dying_per_1000_born.csv",
"children_per_woman" : root+ "datasets/children_per_woman_total_fertility.csv",
"co2_emissions" : root+"datasets/co2_emissions_tonnes_per_person.csv",
"population" : root+ "datasets/converted_pop.csv",
"food_supply" : root+ "datasets/food_supply_kilocalories_per_person_and_day.csv",
"gdp_per_captia" : root+ "datasets/gdp_per_capita_yearly_growth.csv",
"gini_index" : root+ "datasets/gini.csv",
"life_expectancy" : root+ "datasets/life_expectancy_years.csv",
"malnutrition" : root+ "datasets/malnutrition_weight_for_age_percent_of_children_under_5.csv",
"poverty_index" : root+ "datasets/mincpcap_cppp.csv",
"maternal_mortality" : root+ "datasets/mmr_who.csv",
"people_in_poverty" : root+ "datasets/number_of_people_in_poverty.csv",
"primary_completion" : root+ "datasets/primary_school_completion_percent_of_girls.csv",
"ratio_b/g_in_primary" : root+ "datasets/ratio_of_girls_to_boys_in_primary_and_secondary_education_perc.csv",
"wcde-15--24" : root+ "datasets/wcde-15--24.csv",
"wcde-25--34" : root+ "datasets/wcde-25--34.csv",
"wcde-35--44" : root+ "datasets/wcde-35--44.csv",
"wcde-45--54" : root+ "datasets/wcde-45--54.csv",
"wcde-55--64" : root+ "datasets/wcde-55--64.csv",
"wcde-65--74" : root+ "datasets/wcde-65--74.csv",
"wcde-75--84" : root+ "datasets/wcde-75--84.csv",
"wcde-85--94" : root+ "datasets/wcde-85--94.csv",
"wcde-95--" : root+ "datasets/wcde-95--.csv",
}
import pandas as pd
import numpy as np
import math
import tensorflow_decision_forests as tfdf
from sklearn.model_selection import train_test_split
from wurlitzer import sys_pipes
# to find out how many countries each dataset has
countries_arr = []
for path in datasets_path:
df = pd.read_csv(datasets_path[path])
print(f"{'Factor: ' + path:<30} count: {len(set(df.Country.unique()))}")
```
from the above output
- **malnutrition & people in povery** have least no of countries
- **infant mortality & gdp per captia** have highest no of countries
*Doubt:* Does having more data for one factor will make the decision tree bias?
###Steps
1. create a csv file such that each row contains all values of particular year & country present
2. the output for each row is year + 40 years corresponding value
1. **outputs** - life expectany, education level, gdp
```
PREDICT_FUTURE = 40
OUTPUTS = ['life_expectancy', 'gdp_per_captia', 'primary_completion' ]
# creating a list of all countries & years
countries = list(pd.read_csv('education-impact/datasets/Infant_Mortality_Rate.csv').Country.unique())
years = [y for y in range(1960,2015-PREDICT_FUTURE+1)]
keys=[]
for y in years:
for c in countries:
keys.append((c,str(y)))
big_dic = {k : [] for k in keys}
for path in datasets_path:
df = pd.read_csv(datasets_path[path])
df.set_index("Country", inplace=True)
for k in keys:
try:
big_dic[k].append(df.loc[k[0]][k[1]])
except:
big_dic[k].append(np.NaN)
for output_path in OUTPUTS:
df = pd.read_csv(datasets_path[output_path])
df.set_index("Country", inplace=True)
for k in keys:
try:
big_dic[k].append(df.loc[k[0]][str(int(k[1])+PREDICT_FUTURE)])
except:
big_dic[k].append(np.NaN)
columns = [k for k in datasets_path ]
output_columns = ["o_"+o for o in OUTPUTS]
columns.extend(output_columns)
input_df = pd.DataFrame.from_dict(big_dic,orient='index', columns = columns)
output_df = input_df[["o_"+o for o in OUTPUTS]]
input_df.drop(labels=["o_"+o for o in OUTPUTS], axis = 1, inplace=True)
```
From above output
- if we dont drop any rows our table size = 4256 entries
- if we drop rows containing any if all of outputs missing then our table size = 3039 entries
- if we drop rows containing any one of output missing then our table size = 1745 entries
so, I think its is better to go with second choice and build different models, but not sure it will not effect performance of the model
now we have the dataframe containing both inputs and ouputs,our next step is
1. split the data into train & test data
1. try to split data based on continents to reduce bias
2. build DF model using tensorflow
3. check the accuracy of the model
```
X_train, X_test, y_train, y_test = train_test_split(input_df, output_df, test_size=0.30, random_state=43)
frames = [X_train,y_train['o_life_expectancy']]
le_model_df = pd.concat(frames,axis=1)
le_model_df.dropna(subset=['o_life_expectancy'],inplace=True)
le_model_df
train_ds = tfdf.keras.pd_dataframe_to_tf_dataset(le_model_df, label='o_life_expectancy', task=tfdf.keras.Task.REGRESSION)
model = tfdf.keras.RandomForestModel(task = tfdf.keras.Task.REGRESSION)
with sys_pipes():
model.fit(x=train_ds)
frames = [X_test,y_test['o_life_expectancy']]
le_model_test_df = pd.concat(frames,axis=1)
le_model_test_df.dropna(subset=['o_life_expectancy'],inplace=True)
le_model_test_df
# Convert it to a TensorFlow dataset
test_ds = tfdf.keras.pd_dataframe_to_tf_dataset(le_model_test_df, label='o_life_expectancy', task=tfdf.keras.Task.REGRESSION)
# Evaluate the model
model.compile(metrics=["mse"])
# Evaluate the model on the test dataset.
evaluation = model.evaluate(test_ds, return_dict=True)
print(evaluation)
print()
print(f"MSE: {evaluation['mse']}")
print(f"RMSE: {math.sqrt(evaluation['mse'])}")
tfdf.model_plotter.plot_model_in_colab(model, tree_idx=0)
```
|
github_jupyter
|
!rm -rf education-impact
!rm education-impact
!git clone https://github.com/jaya-shankar/education-impact.git
!pip install tensorflow_decision_forests
!pip install wurlitzer
root = "education-impact/"
datasets_path = {
"infant_mortality" : root+ "datasets/Infant_Mortality_Rate.csv",
"child_mortality" : root+ "datasets/child_mortality_0_5_year_olds_dying_per_1000_born.csv",
"children_per_woman" : root+ "datasets/children_per_woman_total_fertility.csv",
"co2_emissions" : root+"datasets/co2_emissions_tonnes_per_person.csv",
"population" : root+ "datasets/converted_pop.csv",
"food_supply" : root+ "datasets/food_supply_kilocalories_per_person_and_day.csv",
"gdp_per_captia" : root+ "datasets/gdp_per_capita_yearly_growth.csv",
"gini_index" : root+ "datasets/gini.csv",
"life_expectancy" : root+ "datasets/life_expectancy_years.csv",
"malnutrition" : root+ "datasets/malnutrition_weight_for_age_percent_of_children_under_5.csv",
"poverty_index" : root+ "datasets/mincpcap_cppp.csv",
"maternal_mortality" : root+ "datasets/mmr_who.csv",
"people_in_poverty" : root+ "datasets/number_of_people_in_poverty.csv",
"primary_completion" : root+ "datasets/primary_school_completion_percent_of_girls.csv",
"ratio_b/g_in_primary" : root+ "datasets/ratio_of_girls_to_boys_in_primary_and_secondary_education_perc.csv",
"wcde-15--24" : root+ "datasets/wcde-15--24.csv",
"wcde-25--34" : root+ "datasets/wcde-25--34.csv",
"wcde-35--44" : root+ "datasets/wcde-35--44.csv",
"wcde-45--54" : root+ "datasets/wcde-45--54.csv",
"wcde-55--64" : root+ "datasets/wcde-55--64.csv",
"wcde-65--74" : root+ "datasets/wcde-65--74.csv",
"wcde-75--84" : root+ "datasets/wcde-75--84.csv",
"wcde-85--94" : root+ "datasets/wcde-85--94.csv",
"wcde-95--" : root+ "datasets/wcde-95--.csv",
}
import pandas as pd
import numpy as np
import math
import tensorflow_decision_forests as tfdf
from sklearn.model_selection import train_test_split
from wurlitzer import sys_pipes
# to find out how many countries each dataset has
countries_arr = []
for path in datasets_path:
df = pd.read_csv(datasets_path[path])
print(f"{'Factor: ' + path:<30} count: {len(set(df.Country.unique()))}")
PREDICT_FUTURE = 40
OUTPUTS = ['life_expectancy', 'gdp_per_captia', 'primary_completion' ]
# creating a list of all countries & years
countries = list(pd.read_csv('education-impact/datasets/Infant_Mortality_Rate.csv').Country.unique())
years = [y for y in range(1960,2015-PREDICT_FUTURE+1)]
keys=[]
for y in years:
for c in countries:
keys.append((c,str(y)))
big_dic = {k : [] for k in keys}
for path in datasets_path:
df = pd.read_csv(datasets_path[path])
df.set_index("Country", inplace=True)
for k in keys:
try:
big_dic[k].append(df.loc[k[0]][k[1]])
except:
big_dic[k].append(np.NaN)
for output_path in OUTPUTS:
df = pd.read_csv(datasets_path[output_path])
df.set_index("Country", inplace=True)
for k in keys:
try:
big_dic[k].append(df.loc[k[0]][str(int(k[1])+PREDICT_FUTURE)])
except:
big_dic[k].append(np.NaN)
columns = [k for k in datasets_path ]
output_columns = ["o_"+o for o in OUTPUTS]
columns.extend(output_columns)
input_df = pd.DataFrame.from_dict(big_dic,orient='index', columns = columns)
output_df = input_df[["o_"+o for o in OUTPUTS]]
input_df.drop(labels=["o_"+o for o in OUTPUTS], axis = 1, inplace=True)
X_train, X_test, y_train, y_test = train_test_split(input_df, output_df, test_size=0.30, random_state=43)
frames = [X_train,y_train['o_life_expectancy']]
le_model_df = pd.concat(frames,axis=1)
le_model_df.dropna(subset=['o_life_expectancy'],inplace=True)
le_model_df
train_ds = tfdf.keras.pd_dataframe_to_tf_dataset(le_model_df, label='o_life_expectancy', task=tfdf.keras.Task.REGRESSION)
model = tfdf.keras.RandomForestModel(task = tfdf.keras.Task.REGRESSION)
with sys_pipes():
model.fit(x=train_ds)
frames = [X_test,y_test['o_life_expectancy']]
le_model_test_df = pd.concat(frames,axis=1)
le_model_test_df.dropna(subset=['o_life_expectancy'],inplace=True)
le_model_test_df
# Convert it to a TensorFlow dataset
test_ds = tfdf.keras.pd_dataframe_to_tf_dataset(le_model_test_df, label='o_life_expectancy', task=tfdf.keras.Task.REGRESSION)
# Evaluate the model
model.compile(metrics=["mse"])
# Evaluate the model on the test dataset.
evaluation = model.evaluate(test_ds, return_dict=True)
print(evaluation)
print()
print(f"MSE: {evaluation['mse']}")
print(f"RMSE: {math.sqrt(evaluation['mse'])}")
tfdf.model_plotter.plot_model_in_colab(model, tree_idx=0)
| 0.455199 | 0.904566 |
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = (20,20)
import os,time
from glob import glob
from PIL import Image
from sklearn import neighbors
import re
df = pd.read_csv('flags_url.csv')
def read_flag(countrycode='IN',file='', res=(128,64)):
countrycode = countrycode.upper()
#url = df[df['alpha-2']==countrycode].image_url
path = f'flags/{countrycode}.png'
if file!='':
path = file
flag = Image.open(path).convert('RGB').resize(res,)
flag = np.array(flag)
return flag
re.match('set\d+','set10')
name2code = lambda x: df[df.country==x]['alpha-2'].to_list()[0]
set1 = ['Venezuela', 'Ecuador', 'Colombia']
set2 = ['Slovenia', 'Russia', 'Slovakia']
set3 = ['Luxembourg','Netherlands']
set4 = ['Norway','Iceland']
set5 = ['New Zealand', 'Australia']
set6 = ['Indonesia', 'Monaco']
set7 = ['Senegal','Mali']
set8= ['India','Niger']
set9= ['Yemen','Syria']
set10 = ['Mexico','Italy']
sets=[]
local_vars = locals()
for var in local_vars:
if re.match('set\d+',var):
sets += [eval(var)]
#print(sets)
codesets = [ list(map(name2code,s)) for s in sets ]
CATEGORIES = list(map(lambda x: x[0], sets))
allcodes = []
for cs in codesets: allcodes+=cs
CATEGORIES
# Similar Flags Plot
fig,axes = plt.subplots(len(codesets),max(map(len,codesets)))
plt.axis('off')
for idx,similars in enumerate(codesets):
#similars = similars+[[]] if len(similars)==2 else similars
for idy,s in enumerate(similars):
if len(s): axes[idx,idy].imshow(read_flag(s))
#axes[idx,idy].axis('off')
X = np.zeros((len(allcodes),read_flag(allcodes[0]).size))
Y = np.zeros(len(allcodes))
for idx,code in enumerate(allcodes):
# find the category
category = 0
for i,cset in enumerate(codesets):
if code in cset:
category = i
break
x = read_flag(code)
X[idx,:] = x.flatten()
Y[idx] = category
clf = neighbors.KNeighborsClassifier(n_neighbors=len(CATEGORIES), weights='distance')
clf.fit(X,Y)
clf.predict([read_flag(codesets[3][1]).flatten()])
print(df[df['alpha-2']==codesets[9][-1]].image_url.to_list())
alltests = glob('test_flags/*.png')
fig,axes = plt.subplots(len(alltests),2,figsize=(20,40))
for idx,f in enumerate(glob('test_flags/*.png')):
#print(f)
test = read_flag(file=f)
test_big = read_flag(file=f,res=(521,256))
p = clf.predict([test.flatten()])[0]
pname = CATEGORIES[int(p)]
pimg = read_flag(name2code(pname), res=(512,256))
axes[idx][0].imshow(test_big)
axes[idx][1].imshow(pimg)
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = (20,20)
import os,time
from glob import glob
from PIL import Image
from sklearn import neighbors
import re
df = pd.read_csv('flags_url.csv')
def read_flag(countrycode='IN',file='', res=(128,64)):
countrycode = countrycode.upper()
#url = df[df['alpha-2']==countrycode].image_url
path = f'flags/{countrycode}.png'
if file!='':
path = file
flag = Image.open(path).convert('RGB').resize(res,)
flag = np.array(flag)
return flag
re.match('set\d+','set10')
name2code = lambda x: df[df.country==x]['alpha-2'].to_list()[0]
set1 = ['Venezuela', 'Ecuador', 'Colombia']
set2 = ['Slovenia', 'Russia', 'Slovakia']
set3 = ['Luxembourg','Netherlands']
set4 = ['Norway','Iceland']
set5 = ['New Zealand', 'Australia']
set6 = ['Indonesia', 'Monaco']
set7 = ['Senegal','Mali']
set8= ['India','Niger']
set9= ['Yemen','Syria']
set10 = ['Mexico','Italy']
sets=[]
local_vars = locals()
for var in local_vars:
if re.match('set\d+',var):
sets += [eval(var)]
#print(sets)
codesets = [ list(map(name2code,s)) for s in sets ]
CATEGORIES = list(map(lambda x: x[0], sets))
allcodes = []
for cs in codesets: allcodes+=cs
CATEGORIES
# Similar Flags Plot
fig,axes = plt.subplots(len(codesets),max(map(len,codesets)))
plt.axis('off')
for idx,similars in enumerate(codesets):
#similars = similars+[[]] if len(similars)==2 else similars
for idy,s in enumerate(similars):
if len(s): axes[idx,idy].imshow(read_flag(s))
#axes[idx,idy].axis('off')
X = np.zeros((len(allcodes),read_flag(allcodes[0]).size))
Y = np.zeros(len(allcodes))
for idx,code in enumerate(allcodes):
# find the category
category = 0
for i,cset in enumerate(codesets):
if code in cset:
category = i
break
x = read_flag(code)
X[idx,:] = x.flatten()
Y[idx] = category
clf = neighbors.KNeighborsClassifier(n_neighbors=len(CATEGORIES), weights='distance')
clf.fit(X,Y)
clf.predict([read_flag(codesets[3][1]).flatten()])
print(df[df['alpha-2']==codesets[9][-1]].image_url.to_list())
alltests = glob('test_flags/*.png')
fig,axes = plt.subplots(len(alltests),2,figsize=(20,40))
for idx,f in enumerate(glob('test_flags/*.png')):
#print(f)
test = read_flag(file=f)
test_big = read_flag(file=f,res=(521,256))
p = clf.predict([test.flatten()])[0]
pname = CATEGORIES[int(p)]
pimg = read_flag(name2code(pname), res=(512,256))
axes[idx][0].imshow(test_big)
axes[idx][1].imshow(pimg)
| 0.158891 | 0.365145 |
# Aggregated model - class - test set: 3 domain features
## Table of contents
1. [Linear Regression](#LinearRegression)
2. [MLP (Dense)](#MLP)
3. [AE combined latent](#AE_combined)
4. [AE OTU latent](#AE_latentOTU)
```
import sys
sys.path.append('../../Src/')
from data import *
from train_2 import *
from transfer_learning import *
from test_functions import *
from layers import *
from utils import *
from loss import *
from metric import *
from results import *
import tensorflow as tf
import tensorflow.keras as keras
from tensorflow.keras import layers
df_microbioma_train, df_microbioma_test, _, _, \
df_domain_train, df_domain_test, _, _, otu_columns, domain_columns = \
read_df_with_transfer_learning_subset_fewerDomainFeatures( \
metadata_names=['age','Temperature','Precipitation3Days'],
otu_filename='../../Datasets/Aggregated/otu_table_Class.csv',
metadata_filename='../../Datasets/Aggregated/metadata_table_all_80.csv')
print(df_microbioma_train.shape[1])
print(df_microbioma_test.shape)
print('TRAIN:')
print('age:' + str(df_domain_train.loc[:,'age'].mean()))
print('rain:' + str(df_domain_train.loc[:,'Precipitation3Days'].mean()))
print('Tª:' + str(df_domain_train.loc[:,'Temperature'].mean()))
print('TEST:')
print('age:' + str(df_domain_test.loc[:,'age'].mean()))
print('rain:' + str(df_domain_test.loc[:,'Precipitation3Days'].mean()))
print('Tª:' + str(df_domain_test.loc[:,'Temperature'].mean()))
```
### Get numpy objects
```
data_microbioma_train = df_microbioma_train.to_numpy(dtype=np.float32)
data_microbioma_test = df_microbioma_test.to_numpy(dtype=np.float32)
data_domain_train = df_domain_train.to_numpy(dtype=np.float32)
data_domain_test = df_domain_test.to_numpy(dtype=np.float32)
```
### Compute default error
```
# Absolute abundance transformed to TSS (with epsilon=1E-6)
def transform_to_rel_abundance(dataset):
epsilon=1E-6
sum_per_sample = dataset.sum(axis=1)
num_samples = sum_per_sample.shape
num_OTUs = np.shape(dataset)[-1]
sum_per_sample = sum_per_sample + (num_OTUs * epsilon)
dividend=dataset+epsilon
dataset_rel_abund = np.divide(dividend,sum_per_sample[:,None])
#display(Markdown("{}</p>".format(np.array2string(actual_array,precision=6,floatmode='fixed'))))
#actual_array.sum(axis=1)
return dataset_rel_abund
```
#### Pearson correlation
```
data_microbioma_rel = transform_to_rel_abundance(data_microbioma_train)
random_seed=347
folds=5
tf.random.set_seed(random_seed) # BGJ
kf = KFold(n_splits=folds, random_state=random_seed, shuffle=True)
tf.random.set_seed(random_seed)
tot_cv_r = 0.0
for train_index, test_index in kf.split(data_microbioma_rel):
m_train, m_test = data_microbioma_rel[train_index], data_microbioma_rel[test_index]
# prediction = average training samples
pred = data_microbioma_rel[train_index].mean(axis=0)
tot = 0.0
count = 0
for i,actual in enumerate(data_microbioma_rel[test_index]):
r, _ = scipy.stats.pearsonr(actual,pred)
if not np.isnan(r):
count += 1
tot += r
r_cv = tot/count
#print(r_cv)
tot_cv_r += r_cv
tot_cv_r/folds
```
#### Bray-Curtis
```
from skbio.diversity import beta_diversity
data_microbioma_rel = transform_to_rel_abundance(data_microbioma_train)
random_seed=347
folds=5
tf.random.set_seed(random_seed) # BGJ
kf = KFold(n_splits=folds, random_state=random_seed, shuffle=True)
tf.random.set_seed(random_seed)
tot_cv = 0.0
for train_index, test_index in kf.split(data_microbioma_rel):
m_train, m_test = data_microbioma_rel[train_index], data_microbioma_rel[test_index]
# prediction = average training samples
pred = data_microbioma_rel[train_index].mean(axis=0)
tot_bc = 0.0
for i,actual in enumerate(data_microbioma_rel[test_index]):
bc_dm = beta_diversity("braycurtis", [actual,pred]) # Source: http://scikit-bio.org/docs/0.4.2/diversity.html
bc = bc_dm[0,1]
tot_bc += bc
bc_cv = tot_bc/(test_index.shape[0])
#print(bc_cv)
tot_cv += bc_cv
tot_cv/folds
```
# 1. Linear regression <a name="LinearRegression"></a>
```
def model(shape_in, shape_out, output_transform):
in_layer = layers.Input(shape=(shape_in,))
net = in_layer
net = layers.Dense(shape_out, activation='linear')(net)
if output_transform is not None:
net = output_transform(net)
out_layer = net
model = keras.Model(inputs=[in_layer], outputs=[out_layer], name='model')
return model
def compile_model(model, optimizer, reconstruction_error, input_transform, output_transform):
metrics = get_experiment_metrics(input_transform, output_transform)[0][3:]
model.compile(optimizer=optimizer, loss=reconstruction_error, metrics=metrics)
def model_fn():
m = model(shape_in=data_domain_train.shape[1],
shape_out=data_microbioma_train.shape[1],
output_transform=None)
compile_model(model=m,
optimizer=optimizers.Adam(lr=0.001),
reconstruction_error=LossMeanSquaredErrorWrapper(CenterLogRatio(), None),
input_transform=CenterLogRatio(),
output_transform=None)
return m, None, m, None
latent_space = 0
results, modelsLR = train(model_fn,
data_microbioma_train,
data_domain_train,
latent_space=latent_space,
folds=5,
epochs=100,
batch_size=64,
learning_rate_scheduler=None,
verbose=-1)
print_results(results)
predictions = test_model(modelsLR, CenterLogRatio, None, otu_columns, data_microbioma_test, data_domain_test)
#save_predictions(predictions, 'experiment_testSet_linear_regresion_3var.txt')
```
# 2. MLP (Dense) <a name="MLP"></a>
```
def model(shape_in, shape_out, output_transform, layers_list, activation_fn):
in_layer = layers.Input(shape=(shape_in,))
net = in_layer
for s in layers_list:
net = layers.Dense(s, activation=activation_fn)(net)
net = layers.Dense(shape_out, activation='linear')(net)
if output_transform is not None:
net = output_transform(net)
out_layer = net
model = keras.Model(inputs=[in_layer], outputs=[out_layer], name='model')
return model
def compile_model(model, optimizer, reconstruction_error, input_transform, output_transform):
metrics = get_experiment_metrics(input_transform, output_transform)[0][3:]
model.compile(optimizer=optimizer, loss=reconstruction_error, metrics=metrics)
def model_fn():
m = model(shape_in=data_domain_train.shape[1],
shape_out=data_microbioma_train.shape[1],
output_transform=None,
layers_list=[128,512],
activation_fn='tanh')
compile_model(model=m,
optimizer=optimizers.Adam(lr=0.01),
reconstruction_error=LossMeanSquaredErrorWrapper(CenterLogRatio(), None),
input_transform=CenterLogRatio(),
output_transform=None)
return m, None, m, None
latent_space=0
results, modelsMLP = train(model_fn,
data_microbioma_train,
data_domain_train,
latent_space=latent_space,
folds=5,
epochs=100,
batch_size=64,
learning_rate_scheduler=None,
verbose=-1)
print_results(results)
predictions = test_model(modelsMLP, CenterLogRatio, None, otu_columns, data_microbioma_test, data_domain_test)
#save_predictions(predictions, 'experiment_testSet_MLP_3var.txt')
```
# 3. Auto-encoder combined latent <a name="AE_combined"></a>
### To create auto-encoder combined model
```
# Train the selected model (the best one from those with the smallest latent space (10)): no.351
experiment_metrics, models, results = perform_experiment_2(cv_folds=5,
epochs=100,
batch_size=64,
learning_rate=0.001,
optimizer=optimizers.Adam,
learning_rate_scheduler=None,
input_transform=Percentage,
output_transform=tf.keras.layers.Softmax,
reconstruction_loss=MakeLoss(LossBrayCurtis, Percentage, None),
latent_space=10,
layers=[512,256],
#layers=[128],
activation='tanh',
activation_latent='tanh',
data_microbioma_train=data_microbioma_train,
data_domain_train=data_domain_train,
show_results=True,
device='/CPU:0')
predictions = test_model_cv_predictions(models, Percentage, tf.keras.layers.Softmax, otu_columns, data_microbioma_test, data_domain_test)
#save_predictions(predictions, 'experiment_aggregated_phylum_testSet_AE_combinedLatent_5CV_3var.txt')
```
# 4. Auto-encoder OTU latent <a name="AE_latentOTU"></a>
```
# Train the selected model (the best one from those with the smallest latent space (10)): no.351
experiment_metrics, models, results = perform_experiment_2(cv_folds=0,
epochs=100,
batch_size=64,
learning_rate=0.001,
optimizer=optimizers.Adam,
learning_rate_scheduler=None,
input_transform=Percentage,
output_transform=tf.keras.layers.Softmax,
reconstruction_loss=MakeLoss(LossBrayCurtis, Percentage, None),
latent_space=10,
layers=[512,256],
#layers=[128],
activation='tanh',
activation_latent='tanh',
data_microbioma_train=data_microbioma_train,
data_domain_train=None,
show_results=True,
device='/CPU:0')
```
### To get encoders and decoders to use in domain->latent model
```
model, encoder, _ ,decoder = models[0]
df_domain_train.shape
```
### To predict latent space for samples in domain->latent model
```
latent_train = encoder.predict(data_microbioma_train)
latent_test = encoder.predict(data_microbioma_test)
```
### To build model to predict latent space
```
def model_fn_latent():
in_layer = layers.Input(shape=(data_domain_train.shape[1],))
net = layers.Dense(128, activation='tanh')(in_layer)
net = layers.Dense(64, activation='tanh')(net)
net = layers.Dense(32, activation='tanh')(net)
net = layers.Dense(16, activation='tanh')(net)
out_layer = layers.Dense(latent_train.shape[1], activation=None)(net) # 'tanh already'
model = keras.Model(inputs=[in_layer], outputs=[out_layer], name='model')
model.compile(optimizer=optimizers.Adam(lr=0.001), loss=tf.keras.losses.MeanSquaredError(),
metrics=[tf.keras.metrics.MeanSquaredError()])
return model
result_latent, model_latent = train_tl_noEnsemble(model_fn_latent,
latent_train,
latent_train,
data_domain_train,
data_domain_train,
epochs=100,
batch_size=16,
verbose=-1)
print_results_noEnsemble(result_latent)
# Test only Dense(domain->latent)
predictions = test_model_tl_latent(model_latent, latent_test, data_domain_test)
#save_predictions(predictions, 'experiment_testSet_domain-latent_AE_OTUlatent_3var.txt)
```
### Domain -> latent -> microbiome. Test set
```
predictions = test_model_tl_noEnsemble(model_latent, decoder, Percentage, tf.keras.layers.Softmax, data_microbioma_test, data_domain_test)
```
|
github_jupyter
|
import sys
sys.path.append('../../Src/')
from data import *
from train_2 import *
from transfer_learning import *
from test_functions import *
from layers import *
from utils import *
from loss import *
from metric import *
from results import *
import tensorflow as tf
import tensorflow.keras as keras
from tensorflow.keras import layers
df_microbioma_train, df_microbioma_test, _, _, \
df_domain_train, df_domain_test, _, _, otu_columns, domain_columns = \
read_df_with_transfer_learning_subset_fewerDomainFeatures( \
metadata_names=['age','Temperature','Precipitation3Days'],
otu_filename='../../Datasets/Aggregated/otu_table_Class.csv',
metadata_filename='../../Datasets/Aggregated/metadata_table_all_80.csv')
print(df_microbioma_train.shape[1])
print(df_microbioma_test.shape)
print('TRAIN:')
print('age:' + str(df_domain_train.loc[:,'age'].mean()))
print('rain:' + str(df_domain_train.loc[:,'Precipitation3Days'].mean()))
print('Tª:' + str(df_domain_train.loc[:,'Temperature'].mean()))
print('TEST:')
print('age:' + str(df_domain_test.loc[:,'age'].mean()))
print('rain:' + str(df_domain_test.loc[:,'Precipitation3Days'].mean()))
print('Tª:' + str(df_domain_test.loc[:,'Temperature'].mean()))
data_microbioma_train = df_microbioma_train.to_numpy(dtype=np.float32)
data_microbioma_test = df_microbioma_test.to_numpy(dtype=np.float32)
data_domain_train = df_domain_train.to_numpy(dtype=np.float32)
data_domain_test = df_domain_test.to_numpy(dtype=np.float32)
# Absolute abundance transformed to TSS (with epsilon=1E-6)
def transform_to_rel_abundance(dataset):
epsilon=1E-6
sum_per_sample = dataset.sum(axis=1)
num_samples = sum_per_sample.shape
num_OTUs = np.shape(dataset)[-1]
sum_per_sample = sum_per_sample + (num_OTUs * epsilon)
dividend=dataset+epsilon
dataset_rel_abund = np.divide(dividend,sum_per_sample[:,None])
#display(Markdown("{}</p>".format(np.array2string(actual_array,precision=6,floatmode='fixed'))))
#actual_array.sum(axis=1)
return dataset_rel_abund
data_microbioma_rel = transform_to_rel_abundance(data_microbioma_train)
random_seed=347
folds=5
tf.random.set_seed(random_seed) # BGJ
kf = KFold(n_splits=folds, random_state=random_seed, shuffle=True)
tf.random.set_seed(random_seed)
tot_cv_r = 0.0
for train_index, test_index in kf.split(data_microbioma_rel):
m_train, m_test = data_microbioma_rel[train_index], data_microbioma_rel[test_index]
# prediction = average training samples
pred = data_microbioma_rel[train_index].mean(axis=0)
tot = 0.0
count = 0
for i,actual in enumerate(data_microbioma_rel[test_index]):
r, _ = scipy.stats.pearsonr(actual,pred)
if not np.isnan(r):
count += 1
tot += r
r_cv = tot/count
#print(r_cv)
tot_cv_r += r_cv
tot_cv_r/folds
from skbio.diversity import beta_diversity
data_microbioma_rel = transform_to_rel_abundance(data_microbioma_train)
random_seed=347
folds=5
tf.random.set_seed(random_seed) # BGJ
kf = KFold(n_splits=folds, random_state=random_seed, shuffle=True)
tf.random.set_seed(random_seed)
tot_cv = 0.0
for train_index, test_index in kf.split(data_microbioma_rel):
m_train, m_test = data_microbioma_rel[train_index], data_microbioma_rel[test_index]
# prediction = average training samples
pred = data_microbioma_rel[train_index].mean(axis=0)
tot_bc = 0.0
for i,actual in enumerate(data_microbioma_rel[test_index]):
bc_dm = beta_diversity("braycurtis", [actual,pred]) # Source: http://scikit-bio.org/docs/0.4.2/diversity.html
bc = bc_dm[0,1]
tot_bc += bc
bc_cv = tot_bc/(test_index.shape[0])
#print(bc_cv)
tot_cv += bc_cv
tot_cv/folds
def model(shape_in, shape_out, output_transform):
in_layer = layers.Input(shape=(shape_in,))
net = in_layer
net = layers.Dense(shape_out, activation='linear')(net)
if output_transform is not None:
net = output_transform(net)
out_layer = net
model = keras.Model(inputs=[in_layer], outputs=[out_layer], name='model')
return model
def compile_model(model, optimizer, reconstruction_error, input_transform, output_transform):
metrics = get_experiment_metrics(input_transform, output_transform)[0][3:]
model.compile(optimizer=optimizer, loss=reconstruction_error, metrics=metrics)
def model_fn():
m = model(shape_in=data_domain_train.shape[1],
shape_out=data_microbioma_train.shape[1],
output_transform=None)
compile_model(model=m,
optimizer=optimizers.Adam(lr=0.001),
reconstruction_error=LossMeanSquaredErrorWrapper(CenterLogRatio(), None),
input_transform=CenterLogRatio(),
output_transform=None)
return m, None, m, None
latent_space = 0
results, modelsLR = train(model_fn,
data_microbioma_train,
data_domain_train,
latent_space=latent_space,
folds=5,
epochs=100,
batch_size=64,
learning_rate_scheduler=None,
verbose=-1)
print_results(results)
predictions = test_model(modelsLR, CenterLogRatio, None, otu_columns, data_microbioma_test, data_domain_test)
#save_predictions(predictions, 'experiment_testSet_linear_regresion_3var.txt')
def model(shape_in, shape_out, output_transform, layers_list, activation_fn):
in_layer = layers.Input(shape=(shape_in,))
net = in_layer
for s in layers_list:
net = layers.Dense(s, activation=activation_fn)(net)
net = layers.Dense(shape_out, activation='linear')(net)
if output_transform is not None:
net = output_transform(net)
out_layer = net
model = keras.Model(inputs=[in_layer], outputs=[out_layer], name='model')
return model
def compile_model(model, optimizer, reconstruction_error, input_transform, output_transform):
metrics = get_experiment_metrics(input_transform, output_transform)[0][3:]
model.compile(optimizer=optimizer, loss=reconstruction_error, metrics=metrics)
def model_fn():
m = model(shape_in=data_domain_train.shape[1],
shape_out=data_microbioma_train.shape[1],
output_transform=None,
layers_list=[128,512],
activation_fn='tanh')
compile_model(model=m,
optimizer=optimizers.Adam(lr=0.01),
reconstruction_error=LossMeanSquaredErrorWrapper(CenterLogRatio(), None),
input_transform=CenterLogRatio(),
output_transform=None)
return m, None, m, None
latent_space=0
results, modelsMLP = train(model_fn,
data_microbioma_train,
data_domain_train,
latent_space=latent_space,
folds=5,
epochs=100,
batch_size=64,
learning_rate_scheduler=None,
verbose=-1)
print_results(results)
predictions = test_model(modelsMLP, CenterLogRatio, None, otu_columns, data_microbioma_test, data_domain_test)
#save_predictions(predictions, 'experiment_testSet_MLP_3var.txt')
# Train the selected model (the best one from those with the smallest latent space (10)): no.351
experiment_metrics, models, results = perform_experiment_2(cv_folds=5,
epochs=100,
batch_size=64,
learning_rate=0.001,
optimizer=optimizers.Adam,
learning_rate_scheduler=None,
input_transform=Percentage,
output_transform=tf.keras.layers.Softmax,
reconstruction_loss=MakeLoss(LossBrayCurtis, Percentage, None),
latent_space=10,
layers=[512,256],
#layers=[128],
activation='tanh',
activation_latent='tanh',
data_microbioma_train=data_microbioma_train,
data_domain_train=data_domain_train,
show_results=True,
device='/CPU:0')
predictions = test_model_cv_predictions(models, Percentage, tf.keras.layers.Softmax, otu_columns, data_microbioma_test, data_domain_test)
#save_predictions(predictions, 'experiment_aggregated_phylum_testSet_AE_combinedLatent_5CV_3var.txt')
# Train the selected model (the best one from those with the smallest latent space (10)): no.351
experiment_metrics, models, results = perform_experiment_2(cv_folds=0,
epochs=100,
batch_size=64,
learning_rate=0.001,
optimizer=optimizers.Adam,
learning_rate_scheduler=None,
input_transform=Percentage,
output_transform=tf.keras.layers.Softmax,
reconstruction_loss=MakeLoss(LossBrayCurtis, Percentage, None),
latent_space=10,
layers=[512,256],
#layers=[128],
activation='tanh',
activation_latent='tanh',
data_microbioma_train=data_microbioma_train,
data_domain_train=None,
show_results=True,
device='/CPU:0')
model, encoder, _ ,decoder = models[0]
df_domain_train.shape
latent_train = encoder.predict(data_microbioma_train)
latent_test = encoder.predict(data_microbioma_test)
def model_fn_latent():
in_layer = layers.Input(shape=(data_domain_train.shape[1],))
net = layers.Dense(128, activation='tanh')(in_layer)
net = layers.Dense(64, activation='tanh')(net)
net = layers.Dense(32, activation='tanh')(net)
net = layers.Dense(16, activation='tanh')(net)
out_layer = layers.Dense(latent_train.shape[1], activation=None)(net) # 'tanh already'
model = keras.Model(inputs=[in_layer], outputs=[out_layer], name='model')
model.compile(optimizer=optimizers.Adam(lr=0.001), loss=tf.keras.losses.MeanSquaredError(),
metrics=[tf.keras.metrics.MeanSquaredError()])
return model
result_latent, model_latent = train_tl_noEnsemble(model_fn_latent,
latent_train,
latent_train,
data_domain_train,
data_domain_train,
epochs=100,
batch_size=16,
verbose=-1)
print_results_noEnsemble(result_latent)
# Test only Dense(domain->latent)
predictions = test_model_tl_latent(model_latent, latent_test, data_domain_test)
#save_predictions(predictions, 'experiment_testSet_domain-latent_AE_OTUlatent_3var.txt)
predictions = test_model_tl_noEnsemble(model_latent, decoder, Percentage, tf.keras.layers.Softmax, data_microbioma_test, data_domain_test)
| 0.361277 | 0.873215 |
```
class ContosoSIS(BaseOEAModule):
def __init__(self, oea, source_folder='contoso_sis', pseudonymize = True):
BaseOEAModule.__init__(self, oea, source_folder, pseudonymize)
self.schemas['studentattendance'] = [['id', 'string', 'no-op'],
['student_id', 'string', 'hash-no-lookup'],
['school_year', 'integer', 'no-op'],
['school_id', 'string', 'no-op'],
['attendance_date', 'timestamp', 'no-op'],
['all_day', 'string', 'no-op'],
['Period', 'short', 'no-op'],
['section_id', 'string', 'no-op'],
['AttendanceCode', 'string', 'no-op'],
['PresenceFlag', 'boolean', 'no-op'],
['attendance_status', 'string', 'no-op'],
['attendance_type', 'string', 'no-op'],
['attendance_sequence', 'short', 'no-op']]
self.schemas['studentsectionmark'] = [['id', 'string', 'no-op'],
['student_id', 'string', 'hash-no-lookup'],
['section_id', 'string', 'no-op'],
['school_year', 'string', 'no-op'],
['term_id', 'string', 'no-op'],
['numeric_grade_earned', 'short', 'no-op'],
['alpha_grade_earned', 'string', 'no-op'],
['is_final_grade', 'string', 'no-op'],
['credits_attempted', 'short', 'no-op'],
['credits_earned', 'short', 'no-op'],
['grad_credit_type', 'string', 'no-op']]
def process_latest_from_stage1(self):
latest = oea.get_latest_folder(self.stage1np)
self._process_entity_from_stage1(latest, 'studentattendance', 'csv', 'overwrite', 'true')
self._process_entity_from_stage1(latest, 'studentsectionmark', 'csv', 'overwrite', 'true')
def process_data_from_stage1(self):
self._process_entity_from_stage1('studentattendance', 'csv', 'overwrite', 'true')
self._process_entity_from_stage1('studentsectionmark', 'csv', 'overwrite', 'true')
def copy_test_data_to_stage1(self):
mssparkutils.fs.cp(self.module_path + '/test_data/studentattendance.csv', self.stage1np + '/studentattendance/studentattendance.csv', True)
mssparkutils.fs.cp(self.module_path + '/test_data/studentsectionmark.csv', self.stage1np + '/studentsectionmark/studentsectionmark.csv', True)
class M365(BaseOEAModule):
"""
Provides data processing methods for MS Insights data v0.2 format.
"""
def __init__(self, oea, source_folder='m365', pseudonymize = False):
BaseOEAModule.__init__(self, oea, source_folder, pseudonymize)
self.schemas['Activity'] = [['SignalType', 'string', 'no-op'],
['StartTime', 'timestamp', 'no-op'],
['UserAgent', 'string', 'no-op'],
['SignalId', 'string', 'no-op'],
['SISClassId', 'string', 'no-op'],
['OfficeClassId', 'string', 'no-op'],
['ChannelId', 'string', 'no-op'],
['AppName', 'string', 'no-op'],
['ActorId', 'string', 'hash-no-lookup'],
['ActorRole', 'string', 'no-op'],
['SchemaVersion', 'string', 'no-op'],
['AssignmentId', 'string', 'no-op'],
['SubmissionId', 'string', 'no-op'],
['Action', 'string', 'no-op'],
['AssginmentDueDate', 'string', 'no-op'],
['ClassCreationDate', 'string', 'no-op'],
['Grade', 'string', 'no-op'],
['SourceFileExtension', 'string', 'no-op'],
['MeetingDuration', 'string', 'no-op']]
self.schemas['Calendar'] = [['Id', 'string', 'no-op'],
['Name', 'string', 'no-op'],
['Description', 'string', 'no-op'],
['SchoolYear', 'integer', 'no-op'],
['IsCurrent', 'boolean', 'no-op'],
['ExternalId', 'string', 'no-op'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['OrgId', 'string', 'no-op']]
self.schemas['Course'] = [['Id', 'string', 'no-op'],
['Name', 'string', 'no-op'],
['Code', 'string', 'no-op'],
['Description', 'string', 'no-op'],
['ExternalId', 'string', 'no-op'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['CalendarId', 'string', 'no-op']]
self.schemas['Org'] = [['Id', 'string', 'no-op'],
['Name', 'string', 'no-op'],
['Identifier', 'string', 'no-op'],
['ExternalId', 'string', 'no-op'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['ParentOrgId', 'string', 'no-op'],
['RefOrgTypeId', 'string', 'no-op'],
['SourceSystemId', 'string', 'no-op']]
self.schemas['Person'] = [['Id', 'string', 'hash'],
['FirstName', 'string', 'mask'],
['MiddleName', 'string', 'mask'],
['LastName', 'string', 'mask'],
['GenerationCode', 'string', 'no-op'],
['Prefix', 'string', 'no-op'],
['EnabledUser', 'string', 'no-op'],
['ExternalId', 'string', 'hash'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['SourceSystemId', 'string', 'no-op']]
self.schemas['PersonIdentifier'] = [['Id', 'string', 'hash'],
['Identifier', 'string', 'hash'],
['Description', 'string', 'no-op'],
['RefIdentifierTypeId', 'string', 'no-op'],
['ExternalId', 'string', 'hash'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['PersonId', 'string', 'hash'],
['SourceSystemId', 'string', 'no-op']]
self.schemas['RefDefinition'] = [['Id', 'string', 'no-op'],
['RefType', 'string', 'no-op'],
['Namespace', 'string', 'no-op'],
['Code', 'string', 'no-op'],
['SortOrder', 'integer', 'no-op'],
['Description', 'string', 'no-op'],
['IsActive', 'boolean', 'no-op']]
self.schemas['Section'] = [['Id', 'string', 'no-op'],
['Name', 'string', 'no-op'],
['Code', 'string', 'no-op'],
['Location', 'string', 'no-op'],
['ExternalId', 'string', 'no-op'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['CourseId', 'string', 'no-op'],
['RefSectionTypeId', 'string', 'no-op'],
['SessionId', 'string', 'no-op'],
['OrgId', 'string', 'no-op']]
self.schemas['Session'] = [['Id', 'string', 'no-op'],
['Name', 'string', 'no-op'],
['BeginDate', 'timestamp', 'no-op'],
['EndDate', 'timestamp', 'no-op'],
['ExternalId', 'string', 'no-op'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['CalendarId', 'string', 'no-op'],
['ParentSessionId', 'string', 'no-op'],
['RefSessionTypeId', 'string', 'no-op']]
self.schemas['StaffOrgAffiliation'] = [['Id', 'string', 'no-op'],
['IsPrimary', 'boolean', 'no-op'],
['EntryDate', 'timestamp', 'no-op'],
['ExitDate', 'timestamp', 'no-op'],
['ExternalId', 'string', 'no-op'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['OrgId', 'string', 'no-op'],
['PersonId', 'string', 'hash'],
['RefStaffOrgRoleId', 'string', 'no-op']]
self.schemas['StaffSectionMembership'] = [['Id', 'string', 'no-op'],
['IsPrimaryStaffForSection', 'boolean', 'no-op'],
['EntryDate', 'timestamp', 'no-op'],
['ExitDate', 'timestamp', 'no-op'],
['ExternalId', 'string', 'no-op'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['PersonId', 'string', 'hash'],
['RefStaffSectionRoleId', 'string', 'no-op'],
['SectionId', 'string', 'no-op']]
self.schemas['StudentOrgAffiliation'] = [['Id', 'string', 'no-op'],
['IsPrimary', 'boolean', 'no-op'],
['EntryDate', 'timestamp', 'no-op'],
['ExitDate', 'timestamp', 'no-op'],
['ExternalId', 'string', 'no-op'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['OrgId', 'string', 'no-op'],
['PersonId', 'string', 'hash'],
['RefGradeLevelId', 'string', 'no-op'],
['RefStudentOrgRoleId', 'string', 'no-op'],
['RefEnrollmentStatusId', 'string', 'no-op']]
self.schemas['StudentSectionMembership'] = [['Id', 'string', 'no-op'],
['EntryDate', 'timestamp', 'no-op'],
['ExitDate', 'timestamp', 'no-op'],
['ExternalId', 'string', 'no-op'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['PersonId', 'string', 'hash'],
['RefGradeLevelWhenCourseTakenId', 'string', 'no-op'],
['RefStudentSectionRoleId', 'string', 'no-op'],
['SectionId', 'string', 'no-op']]
def process_activity_data_from_stage1(self):
""" Processes activity data from stage1 into stage2 using structured streaming.
https://spark.apache.org/docs/latest/structured-streaming-programming-guide.html
"""
latest = oea.get_latest_folder(self.stage1np)
logger.info("Processing activity data from: " + self.stage1np + '/' + latest)
spark_schema = self.oea.to_spark_schema(self.schemas['Activity'])
df = spark.read.csv(self.stage1np + '/' + latest + '/Activity/*.csv', header='false', schema=spark_schema)
sqlContext.registerDataFrameAsTable(df, 'Activity')
sqlContext.registerDataFrameAsTable(spark.read.format('parquet').load(self.oea.stage2np + '/m365/PersonIdentifier'), 'PersonIdentifier')
sqlContext.registerDataFrameAsTable(spark.read.format('parquet').load(self.oea.stage2np + '/m365/RefDefinition'), 'RefDefinition')
df = spark.sql(
"select act.SignalType, act.StartTime, act.UserAgent, act.SignalId, act.SISClassId, act.OfficeClassId, act.ChannelId, \
act.AppName, act.ActorId, act.ActorRole, act.SchemaVersion, act.AssignmentId, act.SubmissionId, act.Action, act.AssginmentDueDate, \
act.ClassCreationDate, act.Grade, act.SourceFileExtension, act.MeetingDuration, pi.PersonId \
from PersonIdentifier pi, RefDefinition rd, Activity act \
where \
pi.RefIdentifierTypeId = rd.Id \
and rd.RefType = 'RefIdentifierType' \
and rd.Code = 'ActiveDirectoryId' \
and pi.Identifier = act.ActorId")
df = df.dropDuplicates(['SignalId'])
df = df.withColumn('year', F.year(F.col('StartTime'))).withColumn('month', F.month(F.col('StartTime')))
df = self.oea.fix_column_names(df)
df.write.format('parquet').mode('overwrite').option("mergeSchema", "true").save(self.stage2np + '/TechActivity')
def reset_activity_processing(self):
""" Resets all TechActivity processing. This is intended for use during initial testing - use with caution. """
self.oea.rm_if_exists(self.stage2p + '/TechActivity')
self.oea.rm_if_exists(self.stage2np + '/TechActivity')
logger.info(f"Deleted TechActivity from stage2")
def _process_roster_entity(self, path):
try:
base_path, filename = self.oea.pop_from_path(path)
entity = filename[:-4]
logger.debug(f"Processing roster entity: path={path}, entity={entity}")
spark_schema = self.oea.to_spark_schema(self.schemas[entity])
df = spark.read.csv(path, header='false', schema=spark_schema)
df = self.oea.fix_column_names(df)
df.write.format('parquet').mode('overwrite').option("mergeSchema", "true").save(self.stage2np + '/' + entity)
except (AnalysisException) as error:
logger.exception(str(error))
def process_latest_roster_from_stage1(self):
latest = oea.get_latest_folder(self.stage1np)
items = mssparkutils.fs.ls(self.stage1np + '/' + latest)
for item in items:
if item.name != 'Activity':
self._process_entity_from_stage1(latest, item.name, 'csv', 'overwrite', 'false')
def xprocess_roster_data_from_stage1(self):
""" Processes all roster data in stage1 and writes out to stage2 and stage2p """
latest = oea.get_latest_folder(self.stage1np)
logger.info("Processing ms_insights roster data from: " + self.stage1np + '/' + latest)
items = mssparkutils.fs.ls(self.stage1np + '/' + latest)
for item in items:
if item.name != 'Activity':
self._process_roster_entity(item.path)
def reset_roster_processing(self):
""" Resets all stage1 to stage2 processing of roster data. """
# cleanup stage2np
if self.oea.path_exists(self.stage2np):
# Delete roster tables (everything other than TechActivity)
items = mssparkutils.fs.ls(self.stage2np)
#print(file.name, file.isDir, file.isFile, file.path, file.size)
for item in items:
if item.name != 'TechActivity':
mssparkutils.fs.rm(item.path, True)
# cleanup stage2p
if self.oea.path_exists(self.stage2p):
# Delete roster tables (everything other than TechActivity)
items = mssparkutils.fs.ls(self.stage2p)
#print(file.name, file.isDir, file.isFile, file.path, file.size)
for item in items:
if item.name != 'TechActivity':
mssparkutils.fs.rm(item.path, True)
```
|
github_jupyter
|
class ContosoSIS(BaseOEAModule):
def __init__(self, oea, source_folder='contoso_sis', pseudonymize = True):
BaseOEAModule.__init__(self, oea, source_folder, pseudonymize)
self.schemas['studentattendance'] = [['id', 'string', 'no-op'],
['student_id', 'string', 'hash-no-lookup'],
['school_year', 'integer', 'no-op'],
['school_id', 'string', 'no-op'],
['attendance_date', 'timestamp', 'no-op'],
['all_day', 'string', 'no-op'],
['Period', 'short', 'no-op'],
['section_id', 'string', 'no-op'],
['AttendanceCode', 'string', 'no-op'],
['PresenceFlag', 'boolean', 'no-op'],
['attendance_status', 'string', 'no-op'],
['attendance_type', 'string', 'no-op'],
['attendance_sequence', 'short', 'no-op']]
self.schemas['studentsectionmark'] = [['id', 'string', 'no-op'],
['student_id', 'string', 'hash-no-lookup'],
['section_id', 'string', 'no-op'],
['school_year', 'string', 'no-op'],
['term_id', 'string', 'no-op'],
['numeric_grade_earned', 'short', 'no-op'],
['alpha_grade_earned', 'string', 'no-op'],
['is_final_grade', 'string', 'no-op'],
['credits_attempted', 'short', 'no-op'],
['credits_earned', 'short', 'no-op'],
['grad_credit_type', 'string', 'no-op']]
def process_latest_from_stage1(self):
latest = oea.get_latest_folder(self.stage1np)
self._process_entity_from_stage1(latest, 'studentattendance', 'csv', 'overwrite', 'true')
self._process_entity_from_stage1(latest, 'studentsectionmark', 'csv', 'overwrite', 'true')
def process_data_from_stage1(self):
self._process_entity_from_stage1('studentattendance', 'csv', 'overwrite', 'true')
self._process_entity_from_stage1('studentsectionmark', 'csv', 'overwrite', 'true')
def copy_test_data_to_stage1(self):
mssparkutils.fs.cp(self.module_path + '/test_data/studentattendance.csv', self.stage1np + '/studentattendance/studentattendance.csv', True)
mssparkutils.fs.cp(self.module_path + '/test_data/studentsectionmark.csv', self.stage1np + '/studentsectionmark/studentsectionmark.csv', True)
class M365(BaseOEAModule):
"""
Provides data processing methods for MS Insights data v0.2 format.
"""
def __init__(self, oea, source_folder='m365', pseudonymize = False):
BaseOEAModule.__init__(self, oea, source_folder, pseudonymize)
self.schemas['Activity'] = [['SignalType', 'string', 'no-op'],
['StartTime', 'timestamp', 'no-op'],
['UserAgent', 'string', 'no-op'],
['SignalId', 'string', 'no-op'],
['SISClassId', 'string', 'no-op'],
['OfficeClassId', 'string', 'no-op'],
['ChannelId', 'string', 'no-op'],
['AppName', 'string', 'no-op'],
['ActorId', 'string', 'hash-no-lookup'],
['ActorRole', 'string', 'no-op'],
['SchemaVersion', 'string', 'no-op'],
['AssignmentId', 'string', 'no-op'],
['SubmissionId', 'string', 'no-op'],
['Action', 'string', 'no-op'],
['AssginmentDueDate', 'string', 'no-op'],
['ClassCreationDate', 'string', 'no-op'],
['Grade', 'string', 'no-op'],
['SourceFileExtension', 'string', 'no-op'],
['MeetingDuration', 'string', 'no-op']]
self.schemas['Calendar'] = [['Id', 'string', 'no-op'],
['Name', 'string', 'no-op'],
['Description', 'string', 'no-op'],
['SchoolYear', 'integer', 'no-op'],
['IsCurrent', 'boolean', 'no-op'],
['ExternalId', 'string', 'no-op'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['OrgId', 'string', 'no-op']]
self.schemas['Course'] = [['Id', 'string', 'no-op'],
['Name', 'string', 'no-op'],
['Code', 'string', 'no-op'],
['Description', 'string', 'no-op'],
['ExternalId', 'string', 'no-op'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['CalendarId', 'string', 'no-op']]
self.schemas['Org'] = [['Id', 'string', 'no-op'],
['Name', 'string', 'no-op'],
['Identifier', 'string', 'no-op'],
['ExternalId', 'string', 'no-op'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['ParentOrgId', 'string', 'no-op'],
['RefOrgTypeId', 'string', 'no-op'],
['SourceSystemId', 'string', 'no-op']]
self.schemas['Person'] = [['Id', 'string', 'hash'],
['FirstName', 'string', 'mask'],
['MiddleName', 'string', 'mask'],
['LastName', 'string', 'mask'],
['GenerationCode', 'string', 'no-op'],
['Prefix', 'string', 'no-op'],
['EnabledUser', 'string', 'no-op'],
['ExternalId', 'string', 'hash'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['SourceSystemId', 'string', 'no-op']]
self.schemas['PersonIdentifier'] = [['Id', 'string', 'hash'],
['Identifier', 'string', 'hash'],
['Description', 'string', 'no-op'],
['RefIdentifierTypeId', 'string', 'no-op'],
['ExternalId', 'string', 'hash'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['PersonId', 'string', 'hash'],
['SourceSystemId', 'string', 'no-op']]
self.schemas['RefDefinition'] = [['Id', 'string', 'no-op'],
['RefType', 'string', 'no-op'],
['Namespace', 'string', 'no-op'],
['Code', 'string', 'no-op'],
['SortOrder', 'integer', 'no-op'],
['Description', 'string', 'no-op'],
['IsActive', 'boolean', 'no-op']]
self.schemas['Section'] = [['Id', 'string', 'no-op'],
['Name', 'string', 'no-op'],
['Code', 'string', 'no-op'],
['Location', 'string', 'no-op'],
['ExternalId', 'string', 'no-op'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['CourseId', 'string', 'no-op'],
['RefSectionTypeId', 'string', 'no-op'],
['SessionId', 'string', 'no-op'],
['OrgId', 'string', 'no-op']]
self.schemas['Session'] = [['Id', 'string', 'no-op'],
['Name', 'string', 'no-op'],
['BeginDate', 'timestamp', 'no-op'],
['EndDate', 'timestamp', 'no-op'],
['ExternalId', 'string', 'no-op'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['CalendarId', 'string', 'no-op'],
['ParentSessionId', 'string', 'no-op'],
['RefSessionTypeId', 'string', 'no-op']]
self.schemas['StaffOrgAffiliation'] = [['Id', 'string', 'no-op'],
['IsPrimary', 'boolean', 'no-op'],
['EntryDate', 'timestamp', 'no-op'],
['ExitDate', 'timestamp', 'no-op'],
['ExternalId', 'string', 'no-op'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['OrgId', 'string', 'no-op'],
['PersonId', 'string', 'hash'],
['RefStaffOrgRoleId', 'string', 'no-op']]
self.schemas['StaffSectionMembership'] = [['Id', 'string', 'no-op'],
['IsPrimaryStaffForSection', 'boolean', 'no-op'],
['EntryDate', 'timestamp', 'no-op'],
['ExitDate', 'timestamp', 'no-op'],
['ExternalId', 'string', 'no-op'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['PersonId', 'string', 'hash'],
['RefStaffSectionRoleId', 'string', 'no-op'],
['SectionId', 'string', 'no-op']]
self.schemas['StudentOrgAffiliation'] = [['Id', 'string', 'no-op'],
['IsPrimary', 'boolean', 'no-op'],
['EntryDate', 'timestamp', 'no-op'],
['ExitDate', 'timestamp', 'no-op'],
['ExternalId', 'string', 'no-op'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['OrgId', 'string', 'no-op'],
['PersonId', 'string', 'hash'],
['RefGradeLevelId', 'string', 'no-op'],
['RefStudentOrgRoleId', 'string', 'no-op'],
['RefEnrollmentStatusId', 'string', 'no-op']]
self.schemas['StudentSectionMembership'] = [['Id', 'string', 'no-op'],
['EntryDate', 'timestamp', 'no-op'],
['ExitDate', 'timestamp', 'no-op'],
['ExternalId', 'string', 'no-op'],
['CreateDate', 'timestamp', 'no-op'],
['LastModifiedDate', 'timestamp', 'no-op'],
['IsActive', 'boolean', 'no-op'],
['PersonId', 'string', 'hash'],
['RefGradeLevelWhenCourseTakenId', 'string', 'no-op'],
['RefStudentSectionRoleId', 'string', 'no-op'],
['SectionId', 'string', 'no-op']]
def process_activity_data_from_stage1(self):
""" Processes activity data from stage1 into stage2 using structured streaming.
https://spark.apache.org/docs/latest/structured-streaming-programming-guide.html
"""
latest = oea.get_latest_folder(self.stage1np)
logger.info("Processing activity data from: " + self.stage1np + '/' + latest)
spark_schema = self.oea.to_spark_schema(self.schemas['Activity'])
df = spark.read.csv(self.stage1np + '/' + latest + '/Activity/*.csv', header='false', schema=spark_schema)
sqlContext.registerDataFrameAsTable(df, 'Activity')
sqlContext.registerDataFrameAsTable(spark.read.format('parquet').load(self.oea.stage2np + '/m365/PersonIdentifier'), 'PersonIdentifier')
sqlContext.registerDataFrameAsTable(spark.read.format('parquet').load(self.oea.stage2np + '/m365/RefDefinition'), 'RefDefinition')
df = spark.sql(
"select act.SignalType, act.StartTime, act.UserAgent, act.SignalId, act.SISClassId, act.OfficeClassId, act.ChannelId, \
act.AppName, act.ActorId, act.ActorRole, act.SchemaVersion, act.AssignmentId, act.SubmissionId, act.Action, act.AssginmentDueDate, \
act.ClassCreationDate, act.Grade, act.SourceFileExtension, act.MeetingDuration, pi.PersonId \
from PersonIdentifier pi, RefDefinition rd, Activity act \
where \
pi.RefIdentifierTypeId = rd.Id \
and rd.RefType = 'RefIdentifierType' \
and rd.Code = 'ActiveDirectoryId' \
and pi.Identifier = act.ActorId")
df = df.dropDuplicates(['SignalId'])
df = df.withColumn('year', F.year(F.col('StartTime'))).withColumn('month', F.month(F.col('StartTime')))
df = self.oea.fix_column_names(df)
df.write.format('parquet').mode('overwrite').option("mergeSchema", "true").save(self.stage2np + '/TechActivity')
def reset_activity_processing(self):
""" Resets all TechActivity processing. This is intended for use during initial testing - use with caution. """
self.oea.rm_if_exists(self.stage2p + '/TechActivity')
self.oea.rm_if_exists(self.stage2np + '/TechActivity')
logger.info(f"Deleted TechActivity from stage2")
def _process_roster_entity(self, path):
try:
base_path, filename = self.oea.pop_from_path(path)
entity = filename[:-4]
logger.debug(f"Processing roster entity: path={path}, entity={entity}")
spark_schema = self.oea.to_spark_schema(self.schemas[entity])
df = spark.read.csv(path, header='false', schema=spark_schema)
df = self.oea.fix_column_names(df)
df.write.format('parquet').mode('overwrite').option("mergeSchema", "true").save(self.stage2np + '/' + entity)
except (AnalysisException) as error:
logger.exception(str(error))
def process_latest_roster_from_stage1(self):
latest = oea.get_latest_folder(self.stage1np)
items = mssparkutils.fs.ls(self.stage1np + '/' + latest)
for item in items:
if item.name != 'Activity':
self._process_entity_from_stage1(latest, item.name, 'csv', 'overwrite', 'false')
def xprocess_roster_data_from_stage1(self):
""" Processes all roster data in stage1 and writes out to stage2 and stage2p """
latest = oea.get_latest_folder(self.stage1np)
logger.info("Processing ms_insights roster data from: " + self.stage1np + '/' + latest)
items = mssparkutils.fs.ls(self.stage1np + '/' + latest)
for item in items:
if item.name != 'Activity':
self._process_roster_entity(item.path)
def reset_roster_processing(self):
""" Resets all stage1 to stage2 processing of roster data. """
# cleanup stage2np
if self.oea.path_exists(self.stage2np):
# Delete roster tables (everything other than TechActivity)
items = mssparkutils.fs.ls(self.stage2np)
#print(file.name, file.isDir, file.isFile, file.path, file.size)
for item in items:
if item.name != 'TechActivity':
mssparkutils.fs.rm(item.path, True)
# cleanup stage2p
if self.oea.path_exists(self.stage2p):
# Delete roster tables (everything other than TechActivity)
items = mssparkutils.fs.ls(self.stage2p)
#print(file.name, file.isDir, file.isFile, file.path, file.size)
for item in items:
if item.name != 'TechActivity':
mssparkutils.fs.rm(item.path, True)
| 0.278453 | 0.238107 |
# Solve a Generalized Assignment Problem using Lagrangian relaxation
This tutorial includes data and information that you need to set up decision optimization engines and build mathematical programming models to solve a Generalized Assignment Problem using Lagrangian relaxation.
When you finish this tutorial, you'll have a foundational knowledge of _Prescriptive Analytics_.
>This notebook is part of the [Prescriptive Analytics for Python](https://rawgit.com/IBMDecisionOptimization/docplex-doc/master/docs/index.html)
>It requires an [installation of CPLEX Optimizers](http://ibmdecisionoptimization.github.io/docplex-doc/getting_started.html)
Discover us [here](https://developer.ibm.com/docloud)
Some familiarity with Python is recommended. This notebook runs on Python 2.
## Table of contents
* [Describe the business problem](#describe-problem)
* [How Decision Optimization can help](#do-help)
* [Use Decision Optimization to create and solve the model](#do-model-create-solve)
* [Summary](#summary)<br>
## Describe the business problem
This notebook illustrates how to solve an optimization model using Lagrangian relaxation technics.
It solves a generalized assignment problem (GAP), as defined by Wolsey, using this relaxation technic.
The main aim is to show multiple optimization through modifications of different models existing in a single environment, not to show how to solve a GAP problem.
In the field of Mathematical Programming, this technic consists in the approximation of a difficult constrained problem by a simpler problem: you remove difficult constraints by integrating them in the objective function, penalizing it if the constraint is not respected.
The method penalizes violations of inequality constraints using a Lagrange multiplier, which imposes a cost on violations. These added costs are used instead of the strict inequality constraints in the optimization. In practice, this relaxed problem can often be solved more easily than the original problem.
For more information, see the following Wikipedia articles: [Generalized assignment problem](https://en.wikipedia.org/wiki/Generalized_assignment_problem) and [Lagrangian relaxation](https://en.wikipedia.org/wiki/Lagrangian_relaxation).
This notebook first solves the standard problem (which is not important here), then shows how to reformulate it to meet the Lagrangian Relaxation features.
## How decision optimization can help
Prescriptive analytics (decision optimization) technology recommends actions that are based on desired outcomes. It considers specific scenarios, resources, and knowledge of past and current events. With this insight, your organization can make better decisions and have greater control over business outcomes.
Prescriptive analytics is the next step on the path to insight-based actions. It creates value through synergy with predictive analytics, which analyzes data to predict future outcomes. Prescriptive analytics takes that insight to the next level by suggesting the optimal way to handle a future situation. Organizations that act fast in dynamic conditions and make superior decisions in uncertain environments gain a strong competitive advantage.
With prescriptive analytics, you can:
* Automate the complex decisions and trade-offs to better manage your limited resources.
* Take advantage of a future opportunity or mitigate a future risk.
* Proactively update recommendations based on changing events.
* Meet operational goals, increase customer loyalty, prevent threats and fraud, and optimize business processes.
## Use Decision Optimization
Perform the following steps to create and solve the model.
1. [Import the library](#Step-1:-Import-the-library)<br>
2. [Model the Data](#Step-2:-Model-the-data)<br>
3. [Set up the prescriptive model](#Step-3:-Set-up-the-prescriptive-model)<br>
3.1 [Define the decision variables](#Define-the-decision-variables)<br>
3.2 [Express the business constraints](#Express-the-business-constraints)<br>
3.3 [Express the objective](#Express-the-objective)<br>
3.4 [Solve the model](#3.4.-Solve-the-model)<br>
3.5 [Solve the model with Lagrangian Relaxation](#3.5.-Solve-the-model-with-Lagrangian-Relaxation-method)<br>
4. [Investigate the solution and run an example analysis](#Step-4:-Investigate-the-solution-and-then-run-an-)<br>
### 1. Import the library
Run the following code to import the Decision Optimization CPLEX Modeling library. The *DOcplex* library contains two modeling packages, mathematical programming (docplex.mp) package and constraint programming (docplex.cp) package.
```
import sys
try:
import docplex.mp
except:
raise Exception('Please install docplex. See https://pypi.org/project/docplex/')
```
If *CPLEX* is not installed, install CPLEX Community edition.
```
try:
import cplex
except:
raise Exception('Please install CPLEX. See https://pypi.org/project/cplex/')
```
### 2. Model the data
In this scenario, the data is simple. It is delivered as 3 input arrays: A, B, and C. The data does not need changing or refactoring.
```
B = [15, 15, 15]
C = [
[ 6, 10, 1],
[12, 12, 5],
[15, 4, 3],
[10, 3, 9],
[8, 9, 5]
]
A = [
[ 5, 7, 2],
[14, 8, 7],
[10, 6, 12],
[ 8, 4, 15],
[ 6, 12, 5]
]
```
### 3. Set up the prescriptive model
Start with viewing the environment information. This information should be updated when you run the notebook.
```
from docplex.mp.environment import Environment
env = Environment()
env.print_information()
```
We will firt create an optimization problem, composed of 2 basic constraints blocks, then we will resolve it using Lagrangian Relaxation on 1 of the constraints block.
#### 3.1 Create the DOcplex model
The model contains the business constraints and the objective.
```
from docplex.mp.model import Model
mdl = Model("GAP per Wolsey")
```
#### 3.2 Define the decision variables
```
print("#As={}, #Bs={}, #Cs={}".format(len(A), len(B), len(C)))
number_of_cs = len(C)
# variables
x_vars = [mdl.binary_var_list(c, name=None) for c in C]
```
#### 3.3 Define the business constraints
```
# constraints
cts = mdl.add_constraints(mdl.sum(xv) <= 1 for xv in x_vars)
mdl.add_constraints(mdl.sum(x_vars[ii][j] * A[ii][j] for ii in range(number_of_cs)) <= bs for j, bs in enumerate(B))
# objective
total_profit = mdl.sum(mdl.scal_prod(x_i, c_i) for c_i, x_i in zip(C, x_vars))
mdl.maximize(total_profit)
mdl.print_information()
```
#### 3.4. Solve the model
Use the Decision Optimization to solve the model.
```
s = mdl.solve()
assert s is not None
obj = s.objective_value
print("* GAP with no relaxation run OK, best objective is: {:g}".format(obj))
```
#### 3.5. Solve the model with Lagrangian Relaxation method
Let's consider for the demonstration of the Lagrangian Relaxation that this model was hard to solve for CPLEX.
We will approximate this problem by doing an iterative model, where the objective is modified at each iteration.
(Wait a few seconds for the solution, due to a time limit parameter.)
We first remove the culprit constraints from the model
```
for ct in cts:
mdl.remove_constraint(ct)
#p_vars are the penalties attached to violating the constraints
p_vars = mdl.continuous_var_list(C, name='p') # new for relaxation
# new version of the approximated constraint where we apply the penalties
mdl.add_constraints(mdl.sum(xv) == 1 - pv for xv, pv in zip(x_vars, p_vars))
;
#Define the maximum number of iterations
max_iters = 10
number_of_cs = len(C)
c_range = range(number_of_cs)
# Langrangian relaxation loop
eps = 1e-6
loop_count = 0
best = 0
initial_multiplier = 1
multipliers = [initial_multiplier] * len(C)
# Objective function
# I'd write the key perfromance indicator (kpi) as
# total_profit = mdl.sum(mdl.sum(x_vars[task][worker] * C[task][worker]) for task, worker in zip(tasks, workers))
total_profit = mdl.sum(mdl.scal_prod(x_i, c_i) for c_i, x_i in zip(C, x_vars))
mdl.add_kpi(total_profit, "Total profit")
print("starting the loop")
while loop_count <= max_iters:
loop_count += 1
# Rebuilt at each loop iteration
total_penalty = mdl.scal_prod(p_vars, multipliers)
mdl.maximize(total_profit + total_penalty)
s = mdl.solve()
if not s:
print("*** solve fails, stopping at iteration: %d" % loop_count)
break
best = s.objective_value
penalties = [pv.solution_value for pv in p_vars]
print('%d> new lagrangian iteration:\n\t obj=%g, m=%s, p=%s' % (loop_count, best, str(multipliers), str(penalties)))
do_stop = True
justifier = 0
for k in c_range:
penalized_violation = penalties[k] * multipliers[k]
if penalized_violation >= eps:
do_stop = False
justifier = penalized_violation
break
if do_stop:
print("* Lagrangian relaxation succeeds, best={:g}, penalty={:g}, #iterations={}"
.format(best, total_penalty.solution_value, loop_count))
break
else:
# Update multipliers and start the loop again.
scale_factor = 1.0 / float(loop_count)
multipliers = [max(multipliers[i] - scale_factor * penalties[i], 0.) for i in c_range]
print('{0}> -- loop continues, m={1!s}, justifier={2:g}'.format(loop_count, multipliers, justifier))
print(best)
```
### 4. Investigate the solution and run an example analysis
You can see that with this relaxation method applied to this simple model, we find the same solution to the problem.
## Summary
You learned how to set up and use IBM Decision Optimization CPLEX Modeling for Python to formulate a Constraint Programming model and solve it with IBM Decision Optimization on Cloud.
## References
* [CPLEX Modeling for Python documentation](https://rawgit.com/IBMDecisionOptimization/docplex-doc/master/docs/index.html)
* [Decision Optimization on Cloud](https://developer.ibm.com/docloud/)
* [Decision Optimization documentation](https://datascience.ibm.com/docs/content/DO/DOinDSX.html)
* For help with DOcplex, or to report a defect, go [here](https://developer.ibm.com/answers/smartspace/docloud).
* Contact us at [email protected]
<hr>
Copyright © IBM Corp. 2017-2018. Released as licensed Sample Materials.
|
github_jupyter
|
import sys
try:
import docplex.mp
except:
raise Exception('Please install docplex. See https://pypi.org/project/docplex/')
try:
import cplex
except:
raise Exception('Please install CPLEX. See https://pypi.org/project/cplex/')
B = [15, 15, 15]
C = [
[ 6, 10, 1],
[12, 12, 5],
[15, 4, 3],
[10, 3, 9],
[8, 9, 5]
]
A = [
[ 5, 7, 2],
[14, 8, 7],
[10, 6, 12],
[ 8, 4, 15],
[ 6, 12, 5]
]
from docplex.mp.environment import Environment
env = Environment()
env.print_information()
from docplex.mp.model import Model
mdl = Model("GAP per Wolsey")
print("#As={}, #Bs={}, #Cs={}".format(len(A), len(B), len(C)))
number_of_cs = len(C)
# variables
x_vars = [mdl.binary_var_list(c, name=None) for c in C]
# constraints
cts = mdl.add_constraints(mdl.sum(xv) <= 1 for xv in x_vars)
mdl.add_constraints(mdl.sum(x_vars[ii][j] * A[ii][j] for ii in range(number_of_cs)) <= bs for j, bs in enumerate(B))
# objective
total_profit = mdl.sum(mdl.scal_prod(x_i, c_i) for c_i, x_i in zip(C, x_vars))
mdl.maximize(total_profit)
mdl.print_information()
s = mdl.solve()
assert s is not None
obj = s.objective_value
print("* GAP with no relaxation run OK, best objective is: {:g}".format(obj))
for ct in cts:
mdl.remove_constraint(ct)
#p_vars are the penalties attached to violating the constraints
p_vars = mdl.continuous_var_list(C, name='p') # new for relaxation
# new version of the approximated constraint where we apply the penalties
mdl.add_constraints(mdl.sum(xv) == 1 - pv for xv, pv in zip(x_vars, p_vars))
;
#Define the maximum number of iterations
max_iters = 10
number_of_cs = len(C)
c_range = range(number_of_cs)
# Langrangian relaxation loop
eps = 1e-6
loop_count = 0
best = 0
initial_multiplier = 1
multipliers = [initial_multiplier] * len(C)
# Objective function
# I'd write the key perfromance indicator (kpi) as
# total_profit = mdl.sum(mdl.sum(x_vars[task][worker] * C[task][worker]) for task, worker in zip(tasks, workers))
total_profit = mdl.sum(mdl.scal_prod(x_i, c_i) for c_i, x_i in zip(C, x_vars))
mdl.add_kpi(total_profit, "Total profit")
print("starting the loop")
while loop_count <= max_iters:
loop_count += 1
# Rebuilt at each loop iteration
total_penalty = mdl.scal_prod(p_vars, multipliers)
mdl.maximize(total_profit + total_penalty)
s = mdl.solve()
if not s:
print("*** solve fails, stopping at iteration: %d" % loop_count)
break
best = s.objective_value
penalties = [pv.solution_value for pv in p_vars]
print('%d> new lagrangian iteration:\n\t obj=%g, m=%s, p=%s' % (loop_count, best, str(multipliers), str(penalties)))
do_stop = True
justifier = 0
for k in c_range:
penalized_violation = penalties[k] * multipliers[k]
if penalized_violation >= eps:
do_stop = False
justifier = penalized_violation
break
if do_stop:
print("* Lagrangian relaxation succeeds, best={:g}, penalty={:g}, #iterations={}"
.format(best, total_penalty.solution_value, loop_count))
break
else:
# Update multipliers and start the loop again.
scale_factor = 1.0 / float(loop_count)
multipliers = [max(multipliers[i] - scale_factor * penalties[i], 0.) for i in c_range]
print('{0}> -- loop continues, m={1!s}, justifier={2:g}'.format(loop_count, multipliers, justifier))
print(best)
| 0.342022 | 0.985454 |
# 项目:全美枪支数据分析
## 目录
<ul>
<li><a href="#intro">简介</a></li>
<li><a href="#wrangling">数据整理</a></li>
<li><a href="#eda">探索性数据分析</a></li>
<li><a href="#conclusions">结论</a></li>
</ul>
<a id='intro'></a>
## 简介
> 该数据来自联邦调查局 (FBI) 的全国即时犯罪背景调查系统 (NICS)。NICS 用于确定潜在买家是否有资格购买枪支或爆炸物。枪支店可以进入这个系统,以确保每位客户没有犯罪记录或符合资格购买。该数据已经收纳了来自 census.gov 的州级数据作为补充数据。NICS 数据在一个 xlsx 文件格式的一个表格中,它包含了按照月份 (month)、州 (state) 、类型 (type) 统计的武器调查数量 (the number of firearm checks) ;美国的人口普查数据 (U.S. census data) 储存在一个 csv 文件中。它包含了州级的几个变量,每个州的大多数变量在 2016 年只有一个数据点,但一些变量有一年以上的数据。
> 通过以上数据,我们将提出以下问题:
> 1. 各个州的枪支拥有数量
> 2. 各个州的人均拥有枪支的数量
> 3. 各个州的合法枪支的比例
> 4. 各个州白人比例于合法枪支比例的关系
> 5. 各个州人口增加率和武器增长率的关系(2011-2016)
```
# 用这个框对你计划使用的所有数据包进行设置
# 导入语句。
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
```
<a id='wrangling'></a>
## 数据整理
> 读取<kbd>gun_data.xlsx</kbd>枪支持有数据 以及<kbd>U.S. Census Data.csv</kbd> 人口数据。分析并且清理数据
### 数据读取
```
# 加载数据并打印几行。进行这几项操作,来检查数据
# 类型,以及是否有缺失数据或错误数据的情况。
df_gun = pd.read_excel('gun_data.xlsx')
df_census = pd.read_csv('U.S. Census Data.csv')
```
### 数据分析
```
#分析枪支持有数据
df_gun.info()
```
> 通过信息分析,可以看到数据有很多缺失。查看原生表格我们知道,有些数据,尤其是早期数据是没有相关的枪支种类的。那么,我们可以用0填充缺失的数据,表示没有该枪支。另外,我们也发现日期格式不对。所以在以下清理工作中,要修正日期格式。
### 数据清理
> 填充枪支数据中缺失的数据
```
df_gun.fillna(0, inplace=True)
df_gun.info()
```
> 查看是否有重复数据
```
df_gun[df_gun.duplicated(['month', 'state']) == True].shape
```
> 没有重复数据,无需去重
> 查看所有州的条目数
```
df_gun['state'].value_counts()
```
### 分析人口数据集
```
df_census.info()
```
> 通过信息,可以知道。大多数数据只有65行。而整个表格是85行。通过查看表格可以知道,65行之后只是一些统计的无效数据。所以需要删除无效数据
```
df_census.drop('Fact Note', axis=1, inplace=True)
df_census.drop(np.arange(64,85), inplace=True)
df_census.info()
```
> 我们通过时间来匹配对应的数据,在人口表中。比较齐全的是2016年的数据,所以我们以2016的数据作为统计的依据。
```
df_census_2016 = df_census[df_census.Fact.str.contains('2016')].set_index('Fact')
df_census_2016 = df_census_2016.transpose()
df_gun_2016 = df_gun[df_gun['month'].str.contains('2016')].groupby('state').mean()
df_census_2016.head()
```
> 通过输出人口表的数据,我们发现数据包含一些特殊符号。如逗号和百分号。我们需要把这些字段转化成float
```
# df_census_2016.info()
df_census_2016.replace({',': ''}, regex=True, inplace=True) # 处理带逗号的数值,去掉逗号
df_census_2016.replace({'Z': 0}, inplace=True) #将数据中的 Z 替换为 0
df_census_2016 = df_census_2016.applymap(lambda x: float(x[:-2])/100 if str(x)[-1]=='%' else x) #转换带 % 的数据
df_census_2016 = df_census_2016.applymap(lambda x: str(x)[1:] if str(x)[0]=='$' else x) #转换带 $ 的数据
df_census_2016 = df_census_2016.apply(pd.to_numeric, errors='coerce') # 将数据集的列转换为数字格式
df_census_2016.head()
```
> 判断是否有重复州的数据
```
df_census_2016[df_census_2016.duplicated() == True].shape
```
> 无重复数据需要处理 接下来 要合并枪支数据和人口数据
```
df_2016 = df_gun_2016.join(df_census_2016)
df_2016.head()
```
<a id='eda'></a>
## 探索性数据分析
> 到这里,基本的数据清理工作就完成了。得到一张2016年的枪支和人口之间的关系表。通过这张表,可以做出以下分析。
### 研究问题1:每个州的武器持有数量对比
```
width = 1
plt.figure(figsize=(20,10))
plt.bar(df_2016.index, df_2016['totals'], width)
plt.title("USA Guns Number")
plt.xlabel("States")
plt.ylabel("Guns Number")
plt.xticks(rotation = 90);
```
### 以上图表是当前每个州的枪支持有的数量对比,我们可以看到kentuchy和California的持枪总量比较高
> 研究 每个州的持枪率
```
df_2016['Population estimates, July 1, 2016, (V2016)'] = df_2016['Population estimates, July 1, 2016, (V2016)'].astype(float)
width = 1
plt.figure(figsize=(20,10))
plt.bar(df_2016.index, df_2016['totals']/df_2016['Population estimates, July 1, 2016, (V2016)'], width)
plt.title("USA Guns Number Percent")
plt.xlabel("States")
plt.ylabel("Guns Number Percent")
plt.xticks(rotation = 90);
```
### 以上图表是表示每个州的人均持枪的数量,其中kentuchy一枝独秀。人均持枪数接近0.07
> 研究 每个周的合法枪支的比率
```
df_2016['Population estimates, July 1, 2016, (V2016)'] = df_2016['Population estimates, July 1, 2016, (V2016)'].astype(float)
width = 1
plt.figure(figsize=(20,10))
plt.bar(df_2016.index, (df_2016['permit'] + df_2016['permit_recheck'])/df_2016['totals'], width)
plt.title("USA Guns Permit Percent")
plt.xlabel("States")
plt.ylabel("Guns Permit Percent")
plt.xticks(rotation = 90);
```
### 以上图标显示每个州的合法持枪的比较。其中Hawaii合法枪支比例最高。接近100%
### 研究问题 2:白人比例和合法枪支比率的关系
```
plt.scatter(df_2016['White alone, percent, July 1, 2016, (V2016)'],(df_2016['permit'] + df_2016['permit_recheck'])/df_2016['totals'])
plt.title("Guns Permit By White Alone Percent")
plt.xlabel("White Alone Percent")
plt.ylabel("Guns Permit Percent")
```
### 当前白人的比例和合法枪支比例的关系,通过图表可以知道。并没有非常明显的线性关系。
> 2011 到 2016年的武器增长率
```
# 2011 年的枪支数量的平均值
df_gun_2011= df_gun[df_gun['month'].str.contains('2011')].groupby('state').mean()
df_gun_2011.head()
width = 1
plt.figure(figsize=(20,10))
plt.bar(df_gun_2016.index, (df_gun_2016['totals'] - df_gun_2011['totals'])/df_gun_2011['totals'], width)
plt.title("USA Guns Incr Percent")
plt.xlabel("States")
plt.ylabel("Guns Incr Percent")
plt.ylim(-10, 20)
plt.xticks(rotation = 90);
```
### 当前每个州2011-2016武器的增长率。当前Mariana Islands 增长率最高。但是通过表格数据,发现枪支数较低。参考意义不大。
> 人口增长率和武器增长率之间的关系
```
plt.scatter(df_2016['Population, percent change - April 1, 2010 (estimates base) to July 1, 2016, (V2016)'],(df_gun_2016['totals'] - df_gun_2011['totals'])/df_gun_2011['totals'])
plt.title("Relation Between Polulation And Guns Number Incr")
plt.xlabel("Polulation Incr Percent")
plt.ylabel("Guns Incr Percent")
```
### 武器增长率和人口增长率的关系。通过图表可以看出,武器增长率比较稳定。和人口增加率没有直接的正向关系。
<a id='conclusions'></a>
## 结论
> 通过以上数据分析图表,得出以下结论。
> 1. Kentucky 人均持枪数最高。
> 2. 2011到2016 Mariana Islands 增加率最高,但是由于基数太低,无参考意义。
> 3. Hawaii 武器的合法率最高。
> 4. 人口增加和武器增长,并没有直接的线性关系。
>
### 不足
实际上,当前的数据是以2016年为标准,样本数量不足。判断比较武断。 而且通过查看表格可以看到,有些武器数据出现的异常数量,会影响数据的准确性。
当前对于数据的分析大多是基于人口数量做出的评定。没有对人口构成,房屋拥有情况进行分析。存在限制。
|
github_jupyter
|
# 用这个框对你计划使用的所有数据包进行设置
# 导入语句。
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# 加载数据并打印几行。进行这几项操作,来检查数据
# 类型,以及是否有缺失数据或错误数据的情况。
df_gun = pd.read_excel('gun_data.xlsx')
df_census = pd.read_csv('U.S. Census Data.csv')
#分析枪支持有数据
df_gun.info()
df_gun.fillna(0, inplace=True)
df_gun.info()
df_gun[df_gun.duplicated(['month', 'state']) == True].shape
df_gun['state'].value_counts()
df_census.info()
df_census.drop('Fact Note', axis=1, inplace=True)
df_census.drop(np.arange(64,85), inplace=True)
df_census.info()
df_census_2016 = df_census[df_census.Fact.str.contains('2016')].set_index('Fact')
df_census_2016 = df_census_2016.transpose()
df_gun_2016 = df_gun[df_gun['month'].str.contains('2016')].groupby('state').mean()
df_census_2016.head()
# df_census_2016.info()
df_census_2016.replace({',': ''}, regex=True, inplace=True) # 处理带逗号的数值,去掉逗号
df_census_2016.replace({'Z': 0}, inplace=True) #将数据中的 Z 替换为 0
df_census_2016 = df_census_2016.applymap(lambda x: float(x[:-2])/100 if str(x)[-1]=='%' else x) #转换带 % 的数据
df_census_2016 = df_census_2016.applymap(lambda x: str(x)[1:] if str(x)[0]=='$' else x) #转换带 $ 的数据
df_census_2016 = df_census_2016.apply(pd.to_numeric, errors='coerce') # 将数据集的列转换为数字格式
df_census_2016.head()
df_census_2016[df_census_2016.duplicated() == True].shape
df_2016 = df_gun_2016.join(df_census_2016)
df_2016.head()
width = 1
plt.figure(figsize=(20,10))
plt.bar(df_2016.index, df_2016['totals'], width)
plt.title("USA Guns Number")
plt.xlabel("States")
plt.ylabel("Guns Number")
plt.xticks(rotation = 90);
df_2016['Population estimates, July 1, 2016, (V2016)'] = df_2016['Population estimates, July 1, 2016, (V2016)'].astype(float)
width = 1
plt.figure(figsize=(20,10))
plt.bar(df_2016.index, df_2016['totals']/df_2016['Population estimates, July 1, 2016, (V2016)'], width)
plt.title("USA Guns Number Percent")
plt.xlabel("States")
plt.ylabel("Guns Number Percent")
plt.xticks(rotation = 90);
df_2016['Population estimates, July 1, 2016, (V2016)'] = df_2016['Population estimates, July 1, 2016, (V2016)'].astype(float)
width = 1
plt.figure(figsize=(20,10))
plt.bar(df_2016.index, (df_2016['permit'] + df_2016['permit_recheck'])/df_2016['totals'], width)
plt.title("USA Guns Permit Percent")
plt.xlabel("States")
plt.ylabel("Guns Permit Percent")
plt.xticks(rotation = 90);
plt.scatter(df_2016['White alone, percent, July 1, 2016, (V2016)'],(df_2016['permit'] + df_2016['permit_recheck'])/df_2016['totals'])
plt.title("Guns Permit By White Alone Percent")
plt.xlabel("White Alone Percent")
plt.ylabel("Guns Permit Percent")
# 2011 年的枪支数量的平均值
df_gun_2011= df_gun[df_gun['month'].str.contains('2011')].groupby('state').mean()
df_gun_2011.head()
width = 1
plt.figure(figsize=(20,10))
plt.bar(df_gun_2016.index, (df_gun_2016['totals'] - df_gun_2011['totals'])/df_gun_2011['totals'], width)
plt.title("USA Guns Incr Percent")
plt.xlabel("States")
plt.ylabel("Guns Incr Percent")
plt.ylim(-10, 20)
plt.xticks(rotation = 90);
plt.scatter(df_2016['Population, percent change - April 1, 2010 (estimates base) to July 1, 2016, (V2016)'],(df_gun_2016['totals'] - df_gun_2011['totals'])/df_gun_2011['totals'])
plt.title("Relation Between Polulation And Guns Number Incr")
plt.xlabel("Polulation Incr Percent")
plt.ylabel("Guns Incr Percent")
| 0.282394 | 0.842831 |
# Covid-19 status in Chile
> Covid-19 overview in Chile
- toc: true
- badges: true
- comments: true
- author: Alonso Silva Allende
- categories: [jupyter]
- image: images/Chile-total-confirmed-cases.png
```
#hide
import numpy as np
import pandas as pd
import altair as alt
#hide
from IPython.display import display_html, HTML
#hide
update_date = pd.to_datetime('today') - pd.offsets.Hour(19)
today = update_date.strftime('%Y-%m-%d')
today
#hide
date_one_week_ago = (update_date - pd.offsets.Day(7)).strftime('%Y-%m-%d')
date_one_week_ago
#hide_input
print(f"Updated on {update_date.strftime('%B %d, %Y')}.")
#hide
casos_raw = pd.read_csv(
"https://raw.githubusercontent.com/MinCiencia/Datos-COVID19/master/output/producto3/CasosTotalesCumulativo.csv",
index_col='Region')
#hide
s = "https://raw.githubusercontent.com/MinCiencia/Datos-COVID19/master/output/producto4/" \
+ today + "-CasosConfirmados-totalRegional.csv"
deaths_raw = pd.read_csv(s, index_col='Region')
#hide
s = "https://raw.githubusercontent.com/MinCiencia/Datos-COVID19/master/output/producto4/" \
+ date_one_week_ago + "-CasosConfirmados-totalRegional.csv"
deaths_one_week_ago = pd.read_csv(s, index_col='Region')
#hide
tests_raw = pd.read_csv(
'https://raw.githubusercontent.com/MinCiencia/Datos-COVID19/master/output/producto7/PCR.csv',
index_col='Region')
#hide
data_table = pd.DataFrame()
data_table["Region"] = casos_raw[today].drop("Total").reset_index()["Region"]
data_table["Confirmed cases"] = casos_raw[today].drop("Total").reset_index()[today]
data_table["Confirmed cases per 100,000 people"] = np.round(100000*(casos_raw[today]/tests_raw['Poblacion']).drop('Total'), decimals=1).values
data_table["Confirmed deaths"] = deaths_raw["Fallecidos"].drop("Total").values
data_table["Confirmed deaths per 100.000 people"] = \
np.round(100000*deaths_raw['Fallecidos'].drop('Total')/(tests_raw['Poblacion'].values), decimals=1).values
#hide
total_confirmed = casos_raw.iloc[-1,-1]
total_deaths = deaths_raw.iloc[-1,-1]
#hide
total_confirmed_one_week_ago = casos_raw.iloc[-1,-8]
total_deaths_one_week_ago = deaths_one_week_ago.loc["Total", "Fallecidos"]
#hide
diff_cases = total_confirmed-total_confirmed_one_week_ago
diff_deaths = total_deaths-total_deaths_one_week_ago
#hide
total_confirmed = '{:,}'.format(total_confirmed) #.replace(',', '.')
total_deaths = '{:,}'.format(total_deaths) #.replace(',', '.')
#hide
diff_cases = '{:,}'.format(diff_cases) #.replace(',', '.')
diff_deaths = '{:,}'.format(diff_deaths) #.replace(',', '.')
#hide_input
HTML(f'<p style="color:#3361ff;"><span style="font-weight:bold;">Confirmed cases:</span> {total_confirmed} (+{diff_cases} since 7 days ago)</p><p style="color:#FF3F3F;"><span style="font-weight:bold;">Confirmed deaths:</span> {total_deaths} (+{diff_deaths} since 7 days ago)</p>')
#hide_input
(data_table.style.set_properties(**{'text-align': 'right'}).background_gradient(cmap='Reds').hide_index()).set_caption(
'Statistics by region: Confirmed cases and confirmed deaths')
```
# Evolution of total confirmed cases by region
```
#hide
data_raw = pd.read_csv(
"https://raw.githubusercontent.com/MinCiencia/Datos-COVID19/master/output/producto3/CasosTotalesCumulativo.csv",
index_col='Region')
#hide
data_raw = data_raw.drop("Total")
#hide
data = pd.DataFrame()
i = 0
for date in data_raw.keys():
for n, region in enumerate(data_raw.index):
data[i] = date, region, data_raw[date].loc[region], n
i += 1
#hide
data_raw = data_raw.reset_index()
regiones = data_raw['Region'].values
data = data.T
data = data.rename(columns={0: "date", 1: "region", 2: "casos", 3: "codigo region"})
#hide
data["casos"] = data["casos"].astype(int)
#hide_input
input_dropdown = alt.binding_select(options=data['region'].unique())
selection1 = alt.selection_single(fields=['region'], bind=input_dropdown, name=' ')
selection2 = alt.selection_multi(fields=['region'], on='mouseover')
color = alt.condition(selection1 | selection2,
alt.Color('region:N', scale=alt.Scale(scheme='tableau20'), legend=None),
alt.value('lightgray'))
chart = alt.Chart(data).mark_bar().encode(
x=alt.X('date:O', axis=alt.Axis(title='Date')),
y=alt.Y('casos', axis=alt.Axis(title='Confirmed cases')),
color=color,
tooltip=['date', 'region', 'casos'],
order=alt.Order(
# Sort the segments of the bars by this field
'codigo region',
sort='descending'
)
).properties(
title='COVID-19 in Chile: Total confirmed cases by región'
).add_selection(
selection1, selection2
).transform_filter(
selection1
)
legend = alt.Chart(data).mark_point().encode(
y=alt.Y('region:N', axis=alt.Axis(orient='right'), sort=regiones),
color=color
).add_selection(
selection1, selection2
)
chart.properties(width=600, height=400) | legend
```
# Evolution of total confirmed deaths by region
```
#hide
data = pd.DataFrame()
data_raw = pd.read_csv(
"https://raw.githubusercontent.com/MinCiencia/Datos-COVID19/master/output/producto4/2020-03-24-CasosConfirmados-totalRegional.csv",
index_col='Region')
data['2020-03-24'] = data_raw['Fallecidos']
#hide
first_death_date = '2020-03-24'
total_days = (pd.to_datetime(today)-pd.to_datetime(first_death_date)).days
#hide
for i in np.arange(total_days+1):
date = (pd.to_datetime(first_death_date)+pd.DateOffset(i)).strftime('%Y-%m-%d')
s = "https://raw.githubusercontent.com/MinCiencia/Datos-COVID19/master/output/producto4/" + date + "-CasosConfirmados-totalRegional.csv"
data_by_date = pd.read_csv(s)
if 'Fallecidos' in data_by_date.columns:
data[date] = data_by_date["Fallecidos"].values
elif 'Casos fallecidos' in data_by_date.columns:
data[date] = data_by_date["Casos fallecidos"].values
else:
data[date] = data_by_date[" Casos fallecidos"].values
#hide
data = data.drop("Total")
#hide
new_data = pd.DataFrame()
i = 0
for date in data.keys():
for n, region in enumerate(data.index):
new_data[i] = date, region, data[date].loc[region], n
i += 1
#hide
data = data.reset_index()
regiones = data['Region'].values
new_data = new_data.T
new_data = new_data.rename(columns={0: "date", 1: "region", 2: "fallecidos", 3: "codigo region"})
#hide
new_data["fallecidos"] = new_data["fallecidos"].astype(int)
#hide_input
input_dropdown = alt.binding_select(options=new_data['region'].unique())
selection1 = alt.selection_single(fields=['region'], bind=input_dropdown, name=' ')
selection2 = alt.selection_multi(fields=['region'], on='mouseover')
color = alt.condition(selection1 | selection2,
alt.Color('region:N', scale=alt.Scale(scheme='tableau20'), legend=None),
alt.value('lightgray'))
bars = alt.Chart(new_data).mark_bar().encode(
x=alt.X('date:O', axis=alt.Axis(title='Date')),
y=alt.Y('fallecidos', axis=alt.Axis(title='Confirmed deaths')),
color=color,
tooltip=['date', 'region', 'fallecidos'],
order=alt.Order(
# Sort the segments of the bars by this field
'codigo region',
sort='descending'
)
).properties(
title='COVID-19 in Chile: Total confirmed deaths by region'
).add_selection(
selection1, selection2
).transform_filter(
selection1
)
legend = alt.Chart(new_data).mark_point().encode(
y=alt.Y('region:N', axis=alt.Axis(orient='right'), sort=regiones),
color=color
).add_selection(
selection1, selection2
)
bars.properties(width=600, height=400) | legend
#hide
# Window size cases
WS_cases = 7
#hide
def my_cases_plot(region):
aux1 = casos_raw.loc[region]
aux2 = [t - s for s, t in zip(aux1, aux1.loc['2020-03-04':])]
# Create the dataframe
new_cases = pd.DataFrame()
new_cases["Date"] = casos_raw.loc[region].reset_index()["index"].iloc[1:].values
new_cases["Confirmed cases"] = aux2
new_cases["Rolling"] = new_cases["Confirmed cases"].rolling(window=WS_cases).mean()
new_cases["Daily confirmed cases"] = len(new_cases) * ["Daily confirmed cases"]
new_cases["7-day rolling average"] = len(new_cases) * ["7-day rolling average"]
# Make the plot
bars = alt.Chart(new_cases).mark_bar(opacity=0.7).encode(
x = alt.X('Date:N', axis=alt.Axis(title='Date')),
y = alt.Y('Confirmed cases:Q', axis=alt.Axis(title='Daily confirmed cases')),
tooltip = ['Date', 'Confirmed cases'],
opacity=alt.Opacity('Daily confirmed cases', legend=alt.Legend(title=""))
)
line = alt.Chart(new_cases).mark_line(point={
"filled": True,
"fill": "#1f77b4"
}, color="#1f77b4").encode(
x=alt.X('Date:N', axis=alt.Axis(title='Date')),
y = alt.Y('Rolling:Q'),
shape=alt.Shape('7-day rolling average', legend=alt.Legend(title=""))
)
return bars, line
```
Data source: [Science Ministry](https://github.com/MinCiencia/Datos-COVID19).
# Daily confirmed cases
## Chile
```
#hide
region = "Total"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in Chile',
width=600
)
```
## Arica y Parinacota
```
#hide
region = "Arica y Parinacota"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
```
## Tarapacá
```
#hide
region = "Tarapacá"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
```
## Antofagasta
```
#hide
region = "Antofagasta"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
```
## Atacama
```
#hide
region = "Atacama"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
```
## Coquimbo
```
#hide
region = "Coquimbo"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
```
## Valparaíso
```
#hide
region = "Valparaíso"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
```
## Metropolitana
```
#hide
region = "Metropolitana"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
```
## O'Higgins
```
#hide
region = "O’Higgins"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
```
## Maule
```
#hide
region = "Maule"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
```
## Ñuble
```
#hide
region = "Ñuble"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
```
## Biobío
```
#hide
region = "Biobío"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
```
## Araucanía
```
#hide
region = "Araucanía"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
```
## Los Ríos
```
#hide
region = "Los Ríos"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
```
## Los Lagos
```
#hide
region = "Los Lagos"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
```
## Aysén
```
#hide
region = "Aysén"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
```
## Magallanes
```
#hide
region = "Magallanes"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
```
# Daily confirmed deaths
```
#hide
data = pd.DataFrame()
data_raw = pd.read_csv(
"https://raw.githubusercontent.com/MinCiencia/Datos-COVID19/master/output/producto4/2020-03-24-CasosConfirmados-totalRegional.csv",
index_col='Region')
data['2020-03-24'] = data_raw['Fallecidos']
#hide
first_death_date = '2020-03-24'
total_days = (pd.to_datetime(today)-pd.to_datetime(first_death_date)).days
#hide
for i in np.arange(total_days+1):
date = (pd.to_datetime(first_death_date)+pd.DateOffset(i)).strftime('%Y-%m-%d')
s = "https://raw.githubusercontent.com/MinCiencia/Datos-COVID19/master/output/producto4/" + date + "-CasosConfirmados-totalRegional.csv"
data_by_date = pd.read_csv(s)
if 'Fallecidos' in data_by_date.columns:
data[date] = data_by_date["Fallecidos"].values
elif 'Casos fallecidos' in data_by_date.columns:
data[date] = data_by_date["Casos fallecidos"].values
else:
data[date] = data_by_date[" Casos fallecidos"].values
#hide
WS_deaths = 7
#hide
def my_deaths_plot(region):
aux1 = data.loc[region]
aux2 = [t - s for s, t in zip(aux1, aux1.loc['2020-03-25':])]
new_deaths = pd.DataFrame()
new_deaths["Dates"] = data.loc["Total"].reset_index()["index"].iloc[1:].values
new_deaths["New_deaths"] = aux2
new_deaths["Rolling"] = new_deaths["New_deaths"].rolling(window=WS_deaths).mean()
new_deaths["Daily confirmed deaths"] = len(new_deaths) * ["Daily confirmed deaths"]
new_deaths["7-day rolling average"] = len(new_deaths) * ["7-day rolling average"]
bars = alt.Chart(new_deaths).mark_bar(opacity=0.7, color='firebrick').encode(
x = alt.X('Dates:N', axis=alt.Axis(title='Date')),
y = alt.Y('New_deaths:Q', axis=alt.Axis(title='Daily confirmed deaths')),
tooltip = ['Dates', 'New_deaths'],
opacity=alt.Opacity('Daily confirmed deaths', legend=alt.Legend(title=""))
)
line = alt.Chart(new_deaths).mark_line(point={
"filled": True,
"fill": "firebrick"
}, color='firebrick').encode(
x=alt.X('Dates:N', axis=alt.Axis(title='Date')),
y = alt.Y('Rolling:Q'),
shape=alt.Shape('7-day rolling average', legend=alt.Legend(title=""))
)
return bars, line
```
## Chile
```
#hide
region = "Total"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in Chile',
width=600
)
```
## Arica y Parinacota
```
#hide
region = "Arica y Parinacota"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
```
## Tarapacá
```
#hide
region = "Tarapacá"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
```
## Antofagasta
```
#hide
region = "Antofagasta"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
```
## Atacama
```
#hide
region = "Atacama"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
```
## Coquimbo
```
#hide
region = "Coquimbo"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
```
## Valparaíso
```
#hide
region = "Valparaíso"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
```
## Metropolitana
```
#hide
region = "Metropolitana"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
```
## O’Higgins
```
#hide
region = "O’Higgins"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
```
## Maule
```
#hide
region = "Maule"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
```
## Ñuble
```
#hide
region = "Ñuble"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
```
## Biobío
```
#hide
region = "Biobío"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
```
## Araucanía
```
#hide
region = "Araucanía"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
```
## Los Ríos
```
#hide
region = "Los Ríos"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
```
## Los Lagos
```
#hide
region = "Los Lagos"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
```
## Aysén
```
#hide
region = "Aysén"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
```
## Magallanes
```
#hide
region = "Magallanes"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
```
|
github_jupyter
|
#hide
import numpy as np
import pandas as pd
import altair as alt
#hide
from IPython.display import display_html, HTML
#hide
update_date = pd.to_datetime('today') - pd.offsets.Hour(19)
today = update_date.strftime('%Y-%m-%d')
today
#hide
date_one_week_ago = (update_date - pd.offsets.Day(7)).strftime('%Y-%m-%d')
date_one_week_ago
#hide_input
print(f"Updated on {update_date.strftime('%B %d, %Y')}.")
#hide
casos_raw = pd.read_csv(
"https://raw.githubusercontent.com/MinCiencia/Datos-COVID19/master/output/producto3/CasosTotalesCumulativo.csv",
index_col='Region')
#hide
s = "https://raw.githubusercontent.com/MinCiencia/Datos-COVID19/master/output/producto4/" \
+ today + "-CasosConfirmados-totalRegional.csv"
deaths_raw = pd.read_csv(s, index_col='Region')
#hide
s = "https://raw.githubusercontent.com/MinCiencia/Datos-COVID19/master/output/producto4/" \
+ date_one_week_ago + "-CasosConfirmados-totalRegional.csv"
deaths_one_week_ago = pd.read_csv(s, index_col='Region')
#hide
tests_raw = pd.read_csv(
'https://raw.githubusercontent.com/MinCiencia/Datos-COVID19/master/output/producto7/PCR.csv',
index_col='Region')
#hide
data_table = pd.DataFrame()
data_table["Region"] = casos_raw[today].drop("Total").reset_index()["Region"]
data_table["Confirmed cases"] = casos_raw[today].drop("Total").reset_index()[today]
data_table["Confirmed cases per 100,000 people"] = np.round(100000*(casos_raw[today]/tests_raw['Poblacion']).drop('Total'), decimals=1).values
data_table["Confirmed deaths"] = deaths_raw["Fallecidos"].drop("Total").values
data_table["Confirmed deaths per 100.000 people"] = \
np.round(100000*deaths_raw['Fallecidos'].drop('Total')/(tests_raw['Poblacion'].values), decimals=1).values
#hide
total_confirmed = casos_raw.iloc[-1,-1]
total_deaths = deaths_raw.iloc[-1,-1]
#hide
total_confirmed_one_week_ago = casos_raw.iloc[-1,-8]
total_deaths_one_week_ago = deaths_one_week_ago.loc["Total", "Fallecidos"]
#hide
diff_cases = total_confirmed-total_confirmed_one_week_ago
diff_deaths = total_deaths-total_deaths_one_week_ago
#hide
total_confirmed = '{:,}'.format(total_confirmed) #.replace(',', '.')
total_deaths = '{:,}'.format(total_deaths) #.replace(',', '.')
#hide
diff_cases = '{:,}'.format(diff_cases) #.replace(',', '.')
diff_deaths = '{:,}'.format(diff_deaths) #.replace(',', '.')
#hide_input
HTML(f'<p style="color:#3361ff;"><span style="font-weight:bold;">Confirmed cases:</span> {total_confirmed} (+{diff_cases} since 7 days ago)</p><p style="color:#FF3F3F;"><span style="font-weight:bold;">Confirmed deaths:</span> {total_deaths} (+{diff_deaths} since 7 days ago)</p>')
#hide_input
(data_table.style.set_properties(**{'text-align': 'right'}).background_gradient(cmap='Reds').hide_index()).set_caption(
'Statistics by region: Confirmed cases and confirmed deaths')
#hide
data_raw = pd.read_csv(
"https://raw.githubusercontent.com/MinCiencia/Datos-COVID19/master/output/producto3/CasosTotalesCumulativo.csv",
index_col='Region')
#hide
data_raw = data_raw.drop("Total")
#hide
data = pd.DataFrame()
i = 0
for date in data_raw.keys():
for n, region in enumerate(data_raw.index):
data[i] = date, region, data_raw[date].loc[region], n
i += 1
#hide
data_raw = data_raw.reset_index()
regiones = data_raw['Region'].values
data = data.T
data = data.rename(columns={0: "date", 1: "region", 2: "casos", 3: "codigo region"})
#hide
data["casos"] = data["casos"].astype(int)
#hide_input
input_dropdown = alt.binding_select(options=data['region'].unique())
selection1 = alt.selection_single(fields=['region'], bind=input_dropdown, name=' ')
selection2 = alt.selection_multi(fields=['region'], on='mouseover')
color = alt.condition(selection1 | selection2,
alt.Color('region:N', scale=alt.Scale(scheme='tableau20'), legend=None),
alt.value('lightgray'))
chart = alt.Chart(data).mark_bar().encode(
x=alt.X('date:O', axis=alt.Axis(title='Date')),
y=alt.Y('casos', axis=alt.Axis(title='Confirmed cases')),
color=color,
tooltip=['date', 'region', 'casos'],
order=alt.Order(
# Sort the segments of the bars by this field
'codigo region',
sort='descending'
)
).properties(
title='COVID-19 in Chile: Total confirmed cases by región'
).add_selection(
selection1, selection2
).transform_filter(
selection1
)
legend = alt.Chart(data).mark_point().encode(
y=alt.Y('region:N', axis=alt.Axis(orient='right'), sort=regiones),
color=color
).add_selection(
selection1, selection2
)
chart.properties(width=600, height=400) | legend
#hide
data = pd.DataFrame()
data_raw = pd.read_csv(
"https://raw.githubusercontent.com/MinCiencia/Datos-COVID19/master/output/producto4/2020-03-24-CasosConfirmados-totalRegional.csv",
index_col='Region')
data['2020-03-24'] = data_raw['Fallecidos']
#hide
first_death_date = '2020-03-24'
total_days = (pd.to_datetime(today)-pd.to_datetime(first_death_date)).days
#hide
for i in np.arange(total_days+1):
date = (pd.to_datetime(first_death_date)+pd.DateOffset(i)).strftime('%Y-%m-%d')
s = "https://raw.githubusercontent.com/MinCiencia/Datos-COVID19/master/output/producto4/" + date + "-CasosConfirmados-totalRegional.csv"
data_by_date = pd.read_csv(s)
if 'Fallecidos' in data_by_date.columns:
data[date] = data_by_date["Fallecidos"].values
elif 'Casos fallecidos' in data_by_date.columns:
data[date] = data_by_date["Casos fallecidos"].values
else:
data[date] = data_by_date[" Casos fallecidos"].values
#hide
data = data.drop("Total")
#hide
new_data = pd.DataFrame()
i = 0
for date in data.keys():
for n, region in enumerate(data.index):
new_data[i] = date, region, data[date].loc[region], n
i += 1
#hide
data = data.reset_index()
regiones = data['Region'].values
new_data = new_data.T
new_data = new_data.rename(columns={0: "date", 1: "region", 2: "fallecidos", 3: "codigo region"})
#hide
new_data["fallecidos"] = new_data["fallecidos"].astype(int)
#hide_input
input_dropdown = alt.binding_select(options=new_data['region'].unique())
selection1 = alt.selection_single(fields=['region'], bind=input_dropdown, name=' ')
selection2 = alt.selection_multi(fields=['region'], on='mouseover')
color = alt.condition(selection1 | selection2,
alt.Color('region:N', scale=alt.Scale(scheme='tableau20'), legend=None),
alt.value('lightgray'))
bars = alt.Chart(new_data).mark_bar().encode(
x=alt.X('date:O', axis=alt.Axis(title='Date')),
y=alt.Y('fallecidos', axis=alt.Axis(title='Confirmed deaths')),
color=color,
tooltip=['date', 'region', 'fallecidos'],
order=alt.Order(
# Sort the segments of the bars by this field
'codigo region',
sort='descending'
)
).properties(
title='COVID-19 in Chile: Total confirmed deaths by region'
).add_selection(
selection1, selection2
).transform_filter(
selection1
)
legend = alt.Chart(new_data).mark_point().encode(
y=alt.Y('region:N', axis=alt.Axis(orient='right'), sort=regiones),
color=color
).add_selection(
selection1, selection2
)
bars.properties(width=600, height=400) | legend
#hide
# Window size cases
WS_cases = 7
#hide
def my_cases_plot(region):
aux1 = casos_raw.loc[region]
aux2 = [t - s for s, t in zip(aux1, aux1.loc['2020-03-04':])]
# Create the dataframe
new_cases = pd.DataFrame()
new_cases["Date"] = casos_raw.loc[region].reset_index()["index"].iloc[1:].values
new_cases["Confirmed cases"] = aux2
new_cases["Rolling"] = new_cases["Confirmed cases"].rolling(window=WS_cases).mean()
new_cases["Daily confirmed cases"] = len(new_cases) * ["Daily confirmed cases"]
new_cases["7-day rolling average"] = len(new_cases) * ["7-day rolling average"]
# Make the plot
bars = alt.Chart(new_cases).mark_bar(opacity=0.7).encode(
x = alt.X('Date:N', axis=alt.Axis(title='Date')),
y = alt.Y('Confirmed cases:Q', axis=alt.Axis(title='Daily confirmed cases')),
tooltip = ['Date', 'Confirmed cases'],
opacity=alt.Opacity('Daily confirmed cases', legend=alt.Legend(title=""))
)
line = alt.Chart(new_cases).mark_line(point={
"filled": True,
"fill": "#1f77b4"
}, color="#1f77b4").encode(
x=alt.X('Date:N', axis=alt.Axis(title='Date')),
y = alt.Y('Rolling:Q'),
shape=alt.Shape('7-day rolling average', legend=alt.Legend(title=""))
)
return bars, line
#hide
region = "Total"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in Chile',
width=600
)
#hide
region = "Arica y Parinacota"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
#hide
region = "Tarapacá"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
#hide
region = "Antofagasta"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
#hide
region = "Atacama"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
#hide
region = "Coquimbo"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
#hide
region = "Valparaíso"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
#hide
region = "Metropolitana"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
#hide
region = "O’Higgins"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
#hide
region = "Maule"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
#hide
region = "Ñuble"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
#hide
region = "Biobío"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
#hide
region = "Araucanía"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
#hide
region = "Los Ríos"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
#hide
region = "Los Lagos"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
#hide
region = "Aysén"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
#hide
region = "Magallanes"
#hide_input
bars, line = my_cases_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed cases in {region}',
width=600
)
#hide
data = pd.DataFrame()
data_raw = pd.read_csv(
"https://raw.githubusercontent.com/MinCiencia/Datos-COVID19/master/output/producto4/2020-03-24-CasosConfirmados-totalRegional.csv",
index_col='Region')
data['2020-03-24'] = data_raw['Fallecidos']
#hide
first_death_date = '2020-03-24'
total_days = (pd.to_datetime(today)-pd.to_datetime(first_death_date)).days
#hide
for i in np.arange(total_days+1):
date = (pd.to_datetime(first_death_date)+pd.DateOffset(i)).strftime('%Y-%m-%d')
s = "https://raw.githubusercontent.com/MinCiencia/Datos-COVID19/master/output/producto4/" + date + "-CasosConfirmados-totalRegional.csv"
data_by_date = pd.read_csv(s)
if 'Fallecidos' in data_by_date.columns:
data[date] = data_by_date["Fallecidos"].values
elif 'Casos fallecidos' in data_by_date.columns:
data[date] = data_by_date["Casos fallecidos"].values
else:
data[date] = data_by_date[" Casos fallecidos"].values
#hide
WS_deaths = 7
#hide
def my_deaths_plot(region):
aux1 = data.loc[region]
aux2 = [t - s for s, t in zip(aux1, aux1.loc['2020-03-25':])]
new_deaths = pd.DataFrame()
new_deaths["Dates"] = data.loc["Total"].reset_index()["index"].iloc[1:].values
new_deaths["New_deaths"] = aux2
new_deaths["Rolling"] = new_deaths["New_deaths"].rolling(window=WS_deaths).mean()
new_deaths["Daily confirmed deaths"] = len(new_deaths) * ["Daily confirmed deaths"]
new_deaths["7-day rolling average"] = len(new_deaths) * ["7-day rolling average"]
bars = alt.Chart(new_deaths).mark_bar(opacity=0.7, color='firebrick').encode(
x = alt.X('Dates:N', axis=alt.Axis(title='Date')),
y = alt.Y('New_deaths:Q', axis=alt.Axis(title='Daily confirmed deaths')),
tooltip = ['Dates', 'New_deaths'],
opacity=alt.Opacity('Daily confirmed deaths', legend=alt.Legend(title=""))
)
line = alt.Chart(new_deaths).mark_line(point={
"filled": True,
"fill": "firebrick"
}, color='firebrick').encode(
x=alt.X('Dates:N', axis=alt.Axis(title='Date')),
y = alt.Y('Rolling:Q'),
shape=alt.Shape('7-day rolling average', legend=alt.Legend(title=""))
)
return bars, line
#hide
region = "Total"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in Chile',
width=600
)
#hide
region = "Arica y Parinacota"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
#hide
region = "Tarapacá"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
#hide
region = "Antofagasta"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
#hide
region = "Atacama"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
#hide
region = "Coquimbo"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
#hide
region = "Valparaíso"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
#hide
region = "Metropolitana"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
#hide
region = "O’Higgins"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
#hide
region = "Maule"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
#hide
region = "Ñuble"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
#hide
region = "Biobío"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
#hide
region = "Araucanía"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
#hide
region = "Los Ríos"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
#hide
region = "Los Lagos"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
#hide
region = "Aysén"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
#hide
region = "Magallanes"
#hide_input
bars, line = my_deaths_plot(region)
(bars + line).properties(
title=f'Covid-19 in Chile: Daily confirmed deaths in {region}',
width=600
)
| 0.236957 | 0.645176 |
```
%load_ext autoreload
%autoreload 2
from GPy.models import GPRegression
from GPy.kern import Matern52, Exponential
from summit.utils.models import GPyModel, ModelGroup
from summit.utils.dataset import DataSet
from GPy.inference.optimization import Adam, RProp, Optimizer
from scipydirect import minimize as direct
import numpy as np
ds = DataSet.read_csv(f'data/python/20200604/experiment_0.csv')
```
## LBSG Optimization Algorithm
```
models = ModelGroup({f'y_{i}': GPyModel(input_dim = 5) for i in range(2)})
lhs_data = ds[ds['strategy']=="LHS"]
inputs = [f'x_{i}' for i in range(5)]
models.fit(lhs_data[inputs], lhs_data[['y_0', 'y_1']], num_restarts=100, parallel=True)
print("Log Likelihood:", models['y_1']._model.log_likelihood())
models['y_1']._model
print("Log Likelihood:", models['y_0']._model.log_likelihood())
models['y_0']._model
print("Log Likelihood:", models['y_0']._model.log_likelihood())
models['y_0']._model
print("Log Likelihood:", models['y_1']._model.log_likelihood())
models['y_1']._model
```
## Direct Optimization Algorithm
```
class Direct(Optimizer):
def __init__(self, **kwargs):
super().__init__(**kwargs)
self.opt_name = 'DIRECT'
def opt(self, x_init, f_fp=None, f=None, fp=None):
assert f is not None
if self.max_iters > 6000:
raise ValueError("Maximum iterations must be less than 6000 for DIRECT algorithm")
# bounds = [[np.log(np.sqrt(1e-3)), np.log(np.sqrt(1e3))] for _ in range(len(x_init))]
# bounds[-1] = [-6, ]
bounds = [[np.sqrt(1e-3), np.sqrt(1e3)] for _ in range(len(x_init))]
print(x_init)
bounds[-1] = [np.exp(-6), 1]
res = direct(f, bounds, maxT=self.max_iters)
self.f_opt = res.fun
self.x_opt = res.x
self.status = 'maximum number of function evaluations exceeded'
inputs = [f'x_{i}' for i in range(5)]
outputs = ['y_0', 'y_1']
X = ds[inputs].standardize()
y = ds[outputs].standardize()
kern = Matern52(input_dim =5, ARD=True)
model = GPRegression(X,y, kern)
```
Long story short, this really doesn't work well.
## Adam
```
class Adam(Optimizer):
# We want the optimizer to know some things in the Optimizer implementation:
def __init__(self, step_rate=.0002,
decay=0,
decay_mom1=0.1,
decay_mom2=0.001,
momentum=0,
offset=1e-8, *args, **kwargs):
super(Adam, self).__init__(*args, **kwargs)
self.opt_name = 'Adam (climin)'
self.step_rate = step_rate
self.decay = 1-1e-8
self.decay_mom1 = decay_mom1
self.decay_mom2 = decay_mom2
self.momentum = momentum
self.offset = offset
# _check_for_climin()
def opt(self, x_init, f_fp=None, f=None, fp=None):
# We only need the gradient of the
assert not fp is None
import climin
# Do the optimization, giving previously stored parameters
opt = climin.adam.Adam(x_init, fp,
step_rate=self.step_rate, decay=self.decay,
decay_mom1=self.decay_mom1, decay_mom2=self.decay_mom2,
momentum=self.momentum,offset=self.offset)
# Get the optimized state and transform it into Paramz readable format by setting
# values on this object:
# Important ones are x_opt and status:
for info in opt:
if info['n_iter']>=self.max_iters:
self.x_opt = opt.wrt
self.f_opt = fp(opt.wrt)
self.status = 'maximum number of function evaluations exceeded'
break
else: # pragma: no cover
pass
inputs = [f'x_{i}' for i in range(5)]
outputs = ['y_0', 'y_1']
X = ds[inputs].standardize()
y = ds[outputs].standardize()
kern = Matern52(input_dim =5, ARD=True)
model = GPRegression(X,y, kern)
model.optimize_restarts(num_restarts=10, optimizer=Adam(max_iters=1e4))
model.log_likelihood()
```
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
from GPy.models import GPRegression
from GPy.kern import Matern52, Exponential
from summit.utils.models import GPyModel, ModelGroup
from summit.utils.dataset import DataSet
from GPy.inference.optimization import Adam, RProp, Optimizer
from scipydirect import minimize as direct
import numpy as np
ds = DataSet.read_csv(f'data/python/20200604/experiment_0.csv')
models = ModelGroup({f'y_{i}': GPyModel(input_dim = 5) for i in range(2)})
lhs_data = ds[ds['strategy']=="LHS"]
inputs = [f'x_{i}' for i in range(5)]
models.fit(lhs_data[inputs], lhs_data[['y_0', 'y_1']], num_restarts=100, parallel=True)
print("Log Likelihood:", models['y_1']._model.log_likelihood())
models['y_1']._model
print("Log Likelihood:", models['y_0']._model.log_likelihood())
models['y_0']._model
print("Log Likelihood:", models['y_0']._model.log_likelihood())
models['y_0']._model
print("Log Likelihood:", models['y_1']._model.log_likelihood())
models['y_1']._model
class Direct(Optimizer):
def __init__(self, **kwargs):
super().__init__(**kwargs)
self.opt_name = 'DIRECT'
def opt(self, x_init, f_fp=None, f=None, fp=None):
assert f is not None
if self.max_iters > 6000:
raise ValueError("Maximum iterations must be less than 6000 for DIRECT algorithm")
# bounds = [[np.log(np.sqrt(1e-3)), np.log(np.sqrt(1e3))] for _ in range(len(x_init))]
# bounds[-1] = [-6, ]
bounds = [[np.sqrt(1e-3), np.sqrt(1e3)] for _ in range(len(x_init))]
print(x_init)
bounds[-1] = [np.exp(-6), 1]
res = direct(f, bounds, maxT=self.max_iters)
self.f_opt = res.fun
self.x_opt = res.x
self.status = 'maximum number of function evaluations exceeded'
inputs = [f'x_{i}' for i in range(5)]
outputs = ['y_0', 'y_1']
X = ds[inputs].standardize()
y = ds[outputs].standardize()
kern = Matern52(input_dim =5, ARD=True)
model = GPRegression(X,y, kern)
class Adam(Optimizer):
# We want the optimizer to know some things in the Optimizer implementation:
def __init__(self, step_rate=.0002,
decay=0,
decay_mom1=0.1,
decay_mom2=0.001,
momentum=0,
offset=1e-8, *args, **kwargs):
super(Adam, self).__init__(*args, **kwargs)
self.opt_name = 'Adam (climin)'
self.step_rate = step_rate
self.decay = 1-1e-8
self.decay_mom1 = decay_mom1
self.decay_mom2 = decay_mom2
self.momentum = momentum
self.offset = offset
# _check_for_climin()
def opt(self, x_init, f_fp=None, f=None, fp=None):
# We only need the gradient of the
assert not fp is None
import climin
# Do the optimization, giving previously stored parameters
opt = climin.adam.Adam(x_init, fp,
step_rate=self.step_rate, decay=self.decay,
decay_mom1=self.decay_mom1, decay_mom2=self.decay_mom2,
momentum=self.momentum,offset=self.offset)
# Get the optimized state and transform it into Paramz readable format by setting
# values on this object:
# Important ones are x_opt and status:
for info in opt:
if info['n_iter']>=self.max_iters:
self.x_opt = opt.wrt
self.f_opt = fp(opt.wrt)
self.status = 'maximum number of function evaluations exceeded'
break
else: # pragma: no cover
pass
inputs = [f'x_{i}' for i in range(5)]
outputs = ['y_0', 'y_1']
X = ds[inputs].standardize()
y = ds[outputs].standardize()
kern = Matern52(input_dim =5, ARD=True)
model = GPRegression(X,y, kern)
model.optimize_restarts(num_restarts=10, optimizer=Adam(max_iters=1e4))
model.log_likelihood()
| 0.735642 | 0.689155 |
# The Inference Button: Bayesian GLMs made easy with PyMC3
Author: Thomas Wiecki
This tutorial appeared as a post in a small series on Bayesian GLMs on my blog:
1. [The Inference Button: Bayesian GLMs made easy with PyMC3](http://twiecki.github.com/blog/2013/08/12/bayesian-glms-1/)
2. [This world is far from Normal(ly distributed): Robust Regression in PyMC3](http://twiecki.github.io/blog/2013/08/27/bayesian-glms-2/)
3. [The Best Of Both Worlds: Hierarchical Linear Regression in PyMC3](http://twiecki.github.io/blog/2014/03/17/bayesian-glms-3/)
In this blog post I will talk about:
- How the Bayesian Revolution in many scientific disciplines is hindered by poor usability of current Probabilistic Programming languages.
- A gentle introduction to Bayesian linear regression and how it differs from the frequentist approach.
- A preview of [PyMC3](https://github.com/pymc-devs/pymc/tree/pymc3) (currently in alpha) and its new GLM submodule I wrote to allow creation and estimation of Bayesian GLMs as easy as frequentist GLMs in R.
Ready? Lets get started!
There is a huge paradigm shift underway in many scientific disciplines: The Bayesian Revolution.
While the theoretical benefits of Bayesian over Frequentist stats have been discussed at length elsewhere (see *Further Reading* below), there is a major obstacle that hinders wider adoption -- *usability* (this is one of the reasons DARPA wrote out a huge grant to [improve Probabilistic Programming](http://www.darpa.mil/Our_Work/I2O/Programs/Probabilistic_Programming_for_Advanced_Machine_Learning_%28PPAML%29.aspx)).
This is mildly ironic because the beauty of Bayesian statistics is their generality. Frequentist stats have a bazillion different tests for every different scenario. In Bayesian land you define your model exactly as you think is appropriate and hit the *Inference Button(TM)* (i.e. running the magical MCMC sampling algorithm).
Yet when I ask my colleagues why they use frequentist stats (even though they would like to use Bayesian stats) the answer is that software packages like SPSS or R make it very easy to run all those individuals tests with a single command (and more often then not, they don't know the exact model and inference method being used).
While there are great Bayesian software packages like [JAGS](http://mcmc-jags.sourceforge.net/), [BUGS](http://www.mrc-bsu.cam.ac.uk/bugs/), [Stan](http://mc-stan.org/) and [PyMC](http://pymc-devs.github.io/pymc/), they are written for Bayesians statisticians who know very well what model they want to build.
Unfortunately, ["the vast majority of statistical analysis is not performed by statisticians"](http://simplystatistics.org/2013/06/14/the-vast-majority-of-statistical-analysis-is-not-performed-by-statisticians/) -- so what we really need are tools for *scientists* and not for statisticians.
In the interest of putting my code where my mouth is I wrote a submodule for the upcoming [PyMC3](https://github.com/pymc-devs/pymc/tree/pymc3) that makes construction of Bayesian Generalized Linear Models (GLMs) as easy as Frequentist ones in R.
Linear Regression
-----------------
While future blog posts will explore more complex models, I will start here with the simplest GLM -- linear regression.
In general, frequentists think about Linear Regression as follows:
$$ Y = X\beta + \epsilon $$
where $Y$ is the output we want to predict (or *dependent* variable), $X$ is our predictor (or *independent* variable), and $\beta$ are the coefficients (or parameters) of the model we want to estimate. $\epsilon$ is an error term which is assumed to be normally distributed.
We can then use Ordinary Least Squares or Maximum Likelihood to find the best fitting $\beta$.
Probabilistic Reformulation
---------------------------
Bayesians take a probabilistic view of the world and express this model in terms of probability distributions. Our above linear regression can be rewritten to yield:
$$ Y \sim \mathcal{N}(X \beta, \sigma^2) $$
In words, we view $Y$ as a random variable (or random vector) of which each element (data point) is distributed according to a Normal distribution. The mean of this normal distribution is provided by our linear predictor with variance $\sigma^2$.
While this is essentially the same model, there are two critical advantages of Bayesian estimation:
- Priors: We can quantify any prior knowledge we might have by placing priors on the paramters. For example, if we think that $\sigma$ is likely to be small we would choose a prior with more probability mass on low values.
- Quantifying uncertainty: We do not get a single estimate of $\beta$ as above but instead a complete posterior distribution about how likely different values of $\beta$ are. For example, with few data points our uncertainty in $\beta$ will be very high and we'd be getting very wide posteriors.
Bayesian GLMs in PyMC3
----------------------
With the new GLM module in PyMC3 it is very easy to build this and much more complex models.
First, lets import the required modules.
```
%matplotlib inline
from pymc3 import *
import numpy as np
import matplotlib.pyplot as plt
```
### Generating data
Create some toy data to play around with and scatter-plot it.
Essentially we are creating a regression line defined by intercept and slope and add data points by sampling from a Normal with the mean set to the regression line.
```
size = 200
true_intercept = 1
true_slope = 2
x = np.linspace(0, 1, size)
# y = a + b*x
true_regression_line = true_intercept + true_slope * x
# add noise
y = true_regression_line + np.random.normal(scale=.5, size=size)
data = dict(x=x, y=y)
fig = plt.figure(figsize=(7, 7))
ax = fig.add_subplot(111, xlabel='x', ylabel='y', title='Generated data and underlying model')
ax.plot(x, y, 'x', label='sampled data')
ax.plot(x, true_regression_line, label='true regression line', lw=2.)
plt.legend(loc=0);
```
### Estimating the model
Lets fit a Bayesian linear regression model to this data. As you can see, model specifications in `PyMC3` are wrapped in a `with` statement.
Here we use the awesome new [NUTS sampler](http://arxiv.org/abs/1111.4246) (our Inference Button) to draw 2000 posterior samples.
```
with Model() as model: # model specifications in PyMC3 are wrapped in a with-statement
# Define priors
sigma = HalfCauchy('sigma', beta=10, testval=1.)
intercept = Normal('Intercept', 0, sd=20)
x_coeff = Normal('x', 0, sd=20)
# Define likelihood
likelihood = Normal('y', mu=intercept + x_coeff * x,
sd=sigma, observed=y)
# Inference!
start = find_MAP() # Find starting value by optimization
step = NUTS(scaling=start) # Instantiate MCMC sampling algorithm
trace = sample(2000, step, start=start, progressbar=False) # draw 2000 posterior samples using NUTS sampling
```
This should be fairly readable for people who know probabilistic programming. However, would my non-statistican friend know what all this does? Moreover, recall that this is an extremely simple model that would be one line in R. Having multiple, potentially transformed regressors, interaction terms or link-functions would also make this much more complex and error prone.
The new `glm()` function instead takes a [Patsy](http://patsy.readthedocs.org/en/latest/quickstart.html) linear model specifier from which it creates a design matrix. `glm()` then adds random variables for each of the coefficients and an appopriate likelihood to the model.
```
with Model() as model:
# specify glm and pass in data. The resulting linear model, its likelihood and
# and all its parameters are automatically added to our model.
glm.glm('y ~ x', data)
start = find_MAP()
step = NUTS(scaling=start) # Instantiate MCMC sampling algorithm
trace = sample(2000, step, progressbar=False) # draw 2000 posterior samples using NUTS sampling
```
Much shorter, but this code does the exact same thing as the above model specification (you can change priors and everything else too if we wanted). `glm()` parses the `Patsy` model string, adds random variables for each regressor (`Intercept` and slope `x` in this case), adds a likelihood (by default, a Normal is chosen), and all other variables (`sigma`). Finally, `glm()` then initializes the parameters to a good starting point by estimating a frequentist linear model using [statsmodels](http://statsmodels.sourceforge.net/devel/).
If you are not familiar with R's syntax, `'y ~ x'` specifies that we have an output variable `y` that we want to estimate as a linear function of `x`.
### Analyzing the model
Bayesian inference does not give us only one best fitting line (as maximum likelihood does) but rather a whole posterior distribution of likely parameters. Lets plot the posterior distribution of our parameters and the individual samples we drew.
```
plt.figure(figsize=(7, 7))
traceplot(trace[100:])
plt.tight_layout();
```
The left side shows our marginal posterior -- for each parameter value on the x-axis we get a probability on the y-axis that tells us how likely that parameter value is.
There are a couple of things to see here. The first is that our sampling chains for the individual parameters (left side) seem well converged and stationary (there are no large drifts or other odd patterns).
Secondly, the maximum posterior estimate of each variable (the peak in the left side distributions) is very close to the true parameters used to generate the data (`x` is the regression coefficient and `sigma` is the standard deviation of our normal).
In the GLM we thus do not only have one best fitting regression line, but many. A posterior predictive plot takes multiple samples from the posterior (intercepts and slopes) and plots a regression line for each of them. Here we are using the `glm.plot_posterior_predictive()` convenience function for this.
```
plt.figure(figsize=(7, 7))
plt.plot(x, y, 'x', label='data')
glm.plot_posterior_predictive(trace, samples=100,
label='posterior predictive regression lines')
plt.plot(x, true_regression_line, label='true regression line', lw=3., c='y')
plt.title('Posterior predictive regression lines')
plt.legend(loc=0)
plt.xlabel('x')
plt.ylabel('y');
```
As you can see, our estimated regression lines are very similar to the true regression line. But since we only have limited data we have *uncertainty* in our estimates, here expressed by the variability of the lines.
## Summary
- Usability is currently a huge hurdle for wider adoption of Bayesian statistics.
- `PyMC3` allows GLM specification with convenient syntax borrowed from R.
- Posterior predictive plots allow us to evaluate fit and our uncertainty in it.
### Further reading
This is the first post of a small series on Bayesian GLMs I am preparing. Next week I will describe how the Student T distribution can be used to perform robust linear regression.
Then there are also other good resources on Bayesian statistics:
- The excellent book [Doing Bayesian Data Analysis by John Kruschke](http://www.indiana.edu/~kruschke/DoingBayesianDataAnalysis/).
- [Andrew Gelman's blog](http://andrewgelman.com/)
- [Baeu Cronins blog post on Probabilistic Programming](https://plus.google.com/u/0/107971134877020469960/posts/KpeRdJKR6Z1)
|
github_jupyter
|
%matplotlib inline
from pymc3 import *
import numpy as np
import matplotlib.pyplot as plt
size = 200
true_intercept = 1
true_slope = 2
x = np.linspace(0, 1, size)
# y = a + b*x
true_regression_line = true_intercept + true_slope * x
# add noise
y = true_regression_line + np.random.normal(scale=.5, size=size)
data = dict(x=x, y=y)
fig = plt.figure(figsize=(7, 7))
ax = fig.add_subplot(111, xlabel='x', ylabel='y', title='Generated data and underlying model')
ax.plot(x, y, 'x', label='sampled data')
ax.plot(x, true_regression_line, label='true regression line', lw=2.)
plt.legend(loc=0);
with Model() as model: # model specifications in PyMC3 are wrapped in a with-statement
# Define priors
sigma = HalfCauchy('sigma', beta=10, testval=1.)
intercept = Normal('Intercept', 0, sd=20)
x_coeff = Normal('x', 0, sd=20)
# Define likelihood
likelihood = Normal('y', mu=intercept + x_coeff * x,
sd=sigma, observed=y)
# Inference!
start = find_MAP() # Find starting value by optimization
step = NUTS(scaling=start) # Instantiate MCMC sampling algorithm
trace = sample(2000, step, start=start, progressbar=False) # draw 2000 posterior samples using NUTS sampling
with Model() as model:
# specify glm and pass in data. The resulting linear model, its likelihood and
# and all its parameters are automatically added to our model.
glm.glm('y ~ x', data)
start = find_MAP()
step = NUTS(scaling=start) # Instantiate MCMC sampling algorithm
trace = sample(2000, step, progressbar=False) # draw 2000 posterior samples using NUTS sampling
plt.figure(figsize=(7, 7))
traceplot(trace[100:])
plt.tight_layout();
plt.figure(figsize=(7, 7))
plt.plot(x, y, 'x', label='data')
glm.plot_posterior_predictive(trace, samples=100,
label='posterior predictive regression lines')
plt.plot(x, true_regression_line, label='true regression line', lw=3., c='y')
plt.title('Posterior predictive regression lines')
plt.legend(loc=0)
plt.xlabel('x')
plt.ylabel('y');
| 0.77373 | 0.974067 |
# Simulations with Model Violations: Aperiodic
In this set of simulations, we will test power spectrum parameterization performance across power spectra which violate model assumptions, specifically in the aperiodic component.
In particular, we will explore the influence of simulating data and fitting with aperiodic modes that do or do not match the properties of the data.
```
%matplotlib inline
from os.path import join as pjoin
import numpy as np
import matplotlib.pyplot as plt
from scipy.stats import spearmanr, mode
from fooof import FOOOF, FOOOFGroup, fit_fooof_3d
from fooof.plts import plot_spectrum
from fooof.sim import gen_power_spectrum, gen_group_power_spectra
from fooof.sim.utils import set_random_seed
# Import project specific (local) custom code
import sys
sys.path.append('../code')
from plts import *
from sims import *
from utils import *
from analysis import *
from settings import *
```
## Settings
```
# Set random seed
set_random_seed(303)
# Set plotting style from seaborn
import seaborn as sns
sns.set_style('white')
# Set a folder name (for saving data & figures)
FOLDER = '04_mv-ap/'
# Data & Model Setting
GEN_SIMS = False
SAVE_SIMS = False
FIT_MODELS = False
SAVE_MODELS = False
# Run Settings
SAVE_FIG = False
PLT_LOG = True
# Check the conditions to simulate across: knee values
print('Knees: ', KNEES)
# Set the number of power spectra - this is per noise condition
n_psds = N_PSDS
```
## Example Model Violation Simulation
In this example, we will simulate a power spectrum with a knee, but fit a model in the mode without a knee, to see how much this impacts fitting.
```
# Test generate a power spectrum
off_val = 1
kne_val = 10
exp_val = 2.0
ap = [off_val, kne_val, exp_val]
peak = [[10, 0.3, 1], [75, 0.2, 1]]
nlv = 0.0
freqs, pows = gen_power_spectrum(F_RANGE_LONG, ap, peak, nlv, F_RES)
# Plot example simulated power spectrum
plot_spectrum(freqs, pows, True, True, ax=get_ax())
# Initialize FOOOF models, with different aperiodic modes
fm1 = FOOOF(aperiodic_mode='fixed')
fm2 = FOOOF(aperiodic_mode='knee')
# Fit models to example simulated spectrum
fm1.fit(freqs, pows)
fm2.fit(freqs, pows)
# Plot comparison of fitting with and without a knee
fig, axes = plt.subplots(1, 2, figsize=[15, 5])
fm1.plot(plt_log=True, add_legend=False, ax=axes[0])
fm2.plot(plt_log=True, add_legend=False, ax=axes[1])
plt.subplots_adjust(wspace=0.3)
# Replot individually, for saving out, without displaying
fm1.plot(plt_log=True, add_legend=True,
save_fig=SAVE_FIG, file_path=pjoin(FIGS_PATH, FOLDER), file_name='example_fixed' + SAVE_EXT)
plt.close()
fm2.plot(plt_log=True, add_legend=True,
save_fig=SAVE_FIG, file_path=pjoin(FIGS_PATH, FOLDER), file_name='example_knee' + SAVE_EXT)
plt.close()
# Check how well our example was fit, focusing on the aperiodic exponent
string = ("With a simulated exponent value of {:1.1f}:\n\n"
" The fixed mode fit value was:\t {:1.3f}\n"
" The knee mode fit value was: \t {:1.3f}")
print(string.format(exp_val,
fm1.get_params('aperiodic_params', 'exponent'),
fm2.get_params('aperiodic_params', 'exponent')))
```
As we can see above, the estimation of the exponent, when there is a knee in the data, but no knee term in the model, is not a very good estimation.
This example suggests that it is quite important for the model to be set with the appropriate mode for aperiodic fitting.
Next we can simulate an expanded set of data, to further explore model fitting in this situation.
## Simulate Power Spectra
In this set of power spectra, we will simulate across a fixed set of knee values.
For each simulation, we will sample the other aperiodic components, and also add one peak.
We will then parameterize these data in 'fixed' mode - that is, an aperiodic mode inconsistent with the mode that simulated the data - to see how sensitive fit quality is to model assumptions.
```
# Use generators to sample peak & aperiodic parameters
peaks = gen_peaks_both()
# Get data sizes
n_conds = len(KNEES)
n_freqs = int((F_RANGE_LONG[1] - F_RANGE_LONG[0]) / F_RES + 1)
# Generate or load power spectra
data_name = 'mvap_kne_sims'
if GEN_SIMS:
# Initialize data stores
psds = np.empty(shape=[n_conds, n_psds, n_freqs])
sim_params = [None] * n_conds
# Generate simulated power spectra
for n_ind, knee in enumerate(KNEES):
aps = gen_ap_knee_def(knee=knee)
freqs, psds[n_ind, :, :], sim_params[n_ind] = \
gen_group_power_spectra(n_psds, F_RANGE_LONG, aps, peaks, NLV,
F_RES, return_params=True)
# Save out generated simulated data & parameter definitions
if SAVE_SIMS:
save_sim_data(data_name, FOLDER, freqs, psds, sim_params)
else:
# Reload simulated data and parameter definitions
freqs, psds, sim_params = load_sim_data(data_name, FOLDER)
# Check shape
print('n_conds, n_spectra, n_freqs : ', psds.shape)
# Extract ground truth values
peak_truths, ap_truths = get_ground_truth(sim_params)
```
### Fit Power Spectra with FOOOF
```
# Initialize FOOOFGroup to test with
fg = FOOOFGroup(*FOOOF_SETTINGS, verbose=False)
# Print out settings used for fitting simulated power spectra
fg.print_settings()
# Fit power spectra
if FIT_MODELS:
fgs = fit_fooof_3d(fg, freqs, psds)
if SAVE_MODELS:
save_model_data(data_name, FOLDER, fgs)
else:
# Reload model fit data
fgs = load_model_data(data_name, FOLDER, n_conds)
```
### Extract FOOOF fit Data
```
# Extract data of interest from FOOOF fits
peak_fits, ap_fits, err_fits, r2_fits, n_fit_peaks = get_fit_data(fgs)
# Calculate errors of the aperiodic parameters that were fit
off_errors = calc_errors(ap_truths[:, :, 0], ap_fits[:, :, 0])
exp_errors = calc_errors(ap_truths[:, :, -1], ap_fits[:, :, -1])
# Get count of number of fit peaks as related to simulated knee value
n_peak_counter = count_peak_conditions(n_fit_peaks, KNEES)
```
### Check Average Errors & Stats
```
# Check overall fit quality
temp = r2_fits.flatten()
print('Min/Max R^2: \t{:1.4f} \t{:1.4f}'.format(np.nanmin(temp), np.nanmax(temp)))
print('Median R^2: \t{:1.4f}'.format(np.nanmedian(temp)))
# Collect data together
datas = {
'OFF' : off_errors,
'EXP' : exp_errors,
'ERR' : err_fits,
'R^2' : r2_fits
}
# Print out the average error for each parameter, across number of peaks
# Also prints out the average model error and R^2 per peak count
with np.printoptions(precision=4, suppress=True):
for label, data in datas.items():
print(label, '\n\t', np.nanmedian(data, 1))
```
### Create Plots
#### Aperiodic Components
First, lets check how well we do reconstructing the aperiodic parameters that we did fit.
In the plots below we can see that the presence of a knee does impact the fit error of out aperiodic components, and that the degree of this effect scales with the value of the knee.
```
# Multi-peak fitting: plot error of aperiodic offset reconstruction across number of peaks
plot_errors_violin(off_errors, 'Offset', plt_log=PLT_LOG,
x_axis='knees', ylim=YLIMS_AP2, #ylim=[-1, 0.5],
save_fig=SAVE_FIG, save_name=pjoin(FOLDER, 'off_error'))
# Multi-peak fitting: plot error of aperiodic offset reconstruction across number of peaks
plot_errors_violin(exp_errors, 'Exponent', plt_log=PLT_LOG,
x_axis='knees', ylim=[-3.5, 0.25], # ylim=YLIMS_AP2,
save_fig=SAVE_FIG, save_name=pjoin(FOLDER, 'exp_error'))
```
#### Periodic Components
Here, we will check how the peak fitting went, checking in particular how many peaks are fit.
Note that all spectra were created with two peaks.
In the plot below, we can see that in all cases with a knee (knee value != 0), the model fits too many peaks.
```
# Plot the correspondance between number of simulated peaks & number of fit peaks
plot_n_peaks_bubbles(n_peak_counter, x_label='knee', ms_val=12,
save_fig=SAVE_FIG, save_name=pjoin(FOLDER, 'number_of_peaks'))
```
#### Goodness of Fit Measures
```
# Multi-peak fitting: plot error levels across knee values
plot_errors_violin(err_fits, 'Fit Error', plt_log=False, x_axis='knees',
save_fig=SAVE_FIG, save_name=pjoin(FOLDER, 'model_error'))
# Multi-peak fitting: plot R^2 levels across knee values
plot_errors_violin(r2_fits, 'R2', plt_log=False, x_axis='knees',
save_fig=SAVE_FIG, save_name=pjoin(FOLDER, 'model_r_squared'))
```
### Example FOOOF Fits
```
# Grab example case of no knee
ind = 0
fm1 = fgs[0].get_fooof(ind, True)
print(sim_params[0][ind])
fm1.print_results()
fm1.plot(plt_log=True, add_legend=True, save_fig=SAVE_FIG,
file_path=pjoin(FIGS_PATH, FOLDER), file_name='example_knee0' + SAVE_EXT)
# Grab example case of highest value knee
ind = 0
fm2 = fgs[-1].get_fooof(ind, True)
print(sim_params[-1][ind])
fm2.print_results()
fm2.plot(plt_log=True, add_legend=True, save_fig=SAVE_FIG,
file_path=pjoin(FIGS_PATH, FOLDER), file_name='example_knee5' + SAVE_EXT)
```
## Fitting a knee model with no knee
Note that so far we have investigated fitting data with a knee, with a model without a knee.
Next, let's check if there is an impact of fitting a model with a knee parameter to data without a knee.
To do so, we will run a model fits in 'knee' mode on our set of simulation with a knee value of 0.
```
# Initialize a new FOOOFGroup, in 'knee mode'
nk_fg = FOOOFGroup(*FOOOF_SETTINGS_KNEE, verbose=False)
# Fit the first set of data, where knee value is 0
nk_fg.fit(freqs, psds[0, :, :])
# Get the fit data
nk_peak_fits, nk_ap_fits, nk_err_fits, nk_r2_fits, nk_n_fit_peaks = get_fit_data([nk_fg])
# Squeeze because we only have one 'condition' for these sims
nk_peak_fits = np.squeeze(nk_peak_fits)
nk_ap_fits = np.squeeze(nk_ap_fits)
nk_err_fits = np.squeeze(nk_err_fits)
nk_r2_fits = np.squeeze(nk_r2_fits)
nk_n_fit_peaks = np.squeeze(nk_n_fit_peaks)
# Extract the true simulated parameters for the no-knee data condition, used here
nk_ap_truths = ap_truths[0, :, :]
# Calculate errors of the aperiodic parameters that were fit
nk_off_errors = calc_errors(nk_ap_truths[:, 0], nk_ap_fits[:, 0])
nk_exp_errors = calc_errors(nk_ap_truths[:, -1], nk_ap_fits[:, -1])
# Check the goodness of fit measures
print('Average fit error: \t {:1.3f}'.format(np.mean(nk_err_fits)))
print('Average fit R^2: \t {:1.3f}'.format(np.mean(nk_r2_fits)))
# Check the error of fit offset and exponent as individual values
_, axes = plt.subplots(1, 2, figsize=[7, 5])
plt.subplots_adjust(wspace=0.5)
plot_single_data(nk_off_errors, 'Offset', ax=axes[0])
plot_single_data(nk_exp_errors, 'Exponent', ax=axes[1])
# Check the error of fit offset and exponent as violinplots
_, axes = plt.subplots(1, 2, figsize=[7, 5])
plt.subplots_adjust(wspace=0.5)
plot_errors_violin(nk_off_errors[np.newaxis, :], title='Offset', plt_log=PLT_LOG,
x_axis=None, ylim=YLIMS_AP, ax=axes[0])
plot_errors_violin(nk_exp_errors[np.newaxis, :], title='Exponent', plt_log=PLT_LOG,
x_axis=None, ylim=YLIMS_AP, ax=axes[1])
# Check the actual fit knee values in the knee-less data
print('Mean value for knee value: {:1.3f}'.format(
np.mean(nk_fg.get_params('aperiodic_params', 'knee'))))
plot_single_data(nk_fg.get_params('aperiodic_params', 'knee'), ylabel='Knee Value')
# Check the average number of fit peaks
print('The modal number of fit peaks is:\t {:1.0f}'.format(mode(nk_n_fit_peaks).mode[0]))
print('The mean number of fit peaks is:\t {:1.2f}'.format(np.mean(nk_n_fit_peaks)))
```
#### Knee Model Fit Conclusion
For fitting a model with a knee parameter on data that does not have a knee:
- the fit knee value tends to be fit as approximately 0
- since there is no knee, exactly 0 is the true parameter value
- the model fit overall does well: with low overall error
- aperiodic parameters are well fit
- there is no obvious bias in the number of fit peaks
This suggests fitting a model with a knee parameter to data that does not a knee is generally fine.
|
github_jupyter
|
%matplotlib inline
from os.path import join as pjoin
import numpy as np
import matplotlib.pyplot as plt
from scipy.stats import spearmanr, mode
from fooof import FOOOF, FOOOFGroup, fit_fooof_3d
from fooof.plts import plot_spectrum
from fooof.sim import gen_power_spectrum, gen_group_power_spectra
from fooof.sim.utils import set_random_seed
# Import project specific (local) custom code
import sys
sys.path.append('../code')
from plts import *
from sims import *
from utils import *
from analysis import *
from settings import *
# Set random seed
set_random_seed(303)
# Set plotting style from seaborn
import seaborn as sns
sns.set_style('white')
# Set a folder name (for saving data & figures)
FOLDER = '04_mv-ap/'
# Data & Model Setting
GEN_SIMS = False
SAVE_SIMS = False
FIT_MODELS = False
SAVE_MODELS = False
# Run Settings
SAVE_FIG = False
PLT_LOG = True
# Check the conditions to simulate across: knee values
print('Knees: ', KNEES)
# Set the number of power spectra - this is per noise condition
n_psds = N_PSDS
# Test generate a power spectrum
off_val = 1
kne_val = 10
exp_val = 2.0
ap = [off_val, kne_val, exp_val]
peak = [[10, 0.3, 1], [75, 0.2, 1]]
nlv = 0.0
freqs, pows = gen_power_spectrum(F_RANGE_LONG, ap, peak, nlv, F_RES)
# Plot example simulated power spectrum
plot_spectrum(freqs, pows, True, True, ax=get_ax())
# Initialize FOOOF models, with different aperiodic modes
fm1 = FOOOF(aperiodic_mode='fixed')
fm2 = FOOOF(aperiodic_mode='knee')
# Fit models to example simulated spectrum
fm1.fit(freqs, pows)
fm2.fit(freqs, pows)
# Plot comparison of fitting with and without a knee
fig, axes = plt.subplots(1, 2, figsize=[15, 5])
fm1.plot(plt_log=True, add_legend=False, ax=axes[0])
fm2.plot(plt_log=True, add_legend=False, ax=axes[1])
plt.subplots_adjust(wspace=0.3)
# Replot individually, for saving out, without displaying
fm1.plot(plt_log=True, add_legend=True,
save_fig=SAVE_FIG, file_path=pjoin(FIGS_PATH, FOLDER), file_name='example_fixed' + SAVE_EXT)
plt.close()
fm2.plot(plt_log=True, add_legend=True,
save_fig=SAVE_FIG, file_path=pjoin(FIGS_PATH, FOLDER), file_name='example_knee' + SAVE_EXT)
plt.close()
# Check how well our example was fit, focusing on the aperiodic exponent
string = ("With a simulated exponent value of {:1.1f}:\n\n"
" The fixed mode fit value was:\t {:1.3f}\n"
" The knee mode fit value was: \t {:1.3f}")
print(string.format(exp_val,
fm1.get_params('aperiodic_params', 'exponent'),
fm2.get_params('aperiodic_params', 'exponent')))
# Use generators to sample peak & aperiodic parameters
peaks = gen_peaks_both()
# Get data sizes
n_conds = len(KNEES)
n_freqs = int((F_RANGE_LONG[1] - F_RANGE_LONG[0]) / F_RES + 1)
# Generate or load power spectra
data_name = 'mvap_kne_sims'
if GEN_SIMS:
# Initialize data stores
psds = np.empty(shape=[n_conds, n_psds, n_freqs])
sim_params = [None] * n_conds
# Generate simulated power spectra
for n_ind, knee in enumerate(KNEES):
aps = gen_ap_knee_def(knee=knee)
freqs, psds[n_ind, :, :], sim_params[n_ind] = \
gen_group_power_spectra(n_psds, F_RANGE_LONG, aps, peaks, NLV,
F_RES, return_params=True)
# Save out generated simulated data & parameter definitions
if SAVE_SIMS:
save_sim_data(data_name, FOLDER, freqs, psds, sim_params)
else:
# Reload simulated data and parameter definitions
freqs, psds, sim_params = load_sim_data(data_name, FOLDER)
# Check shape
print('n_conds, n_spectra, n_freqs : ', psds.shape)
# Extract ground truth values
peak_truths, ap_truths = get_ground_truth(sim_params)
# Initialize FOOOFGroup to test with
fg = FOOOFGroup(*FOOOF_SETTINGS, verbose=False)
# Print out settings used for fitting simulated power spectra
fg.print_settings()
# Fit power spectra
if FIT_MODELS:
fgs = fit_fooof_3d(fg, freqs, psds)
if SAVE_MODELS:
save_model_data(data_name, FOLDER, fgs)
else:
# Reload model fit data
fgs = load_model_data(data_name, FOLDER, n_conds)
# Extract data of interest from FOOOF fits
peak_fits, ap_fits, err_fits, r2_fits, n_fit_peaks = get_fit_data(fgs)
# Calculate errors of the aperiodic parameters that were fit
off_errors = calc_errors(ap_truths[:, :, 0], ap_fits[:, :, 0])
exp_errors = calc_errors(ap_truths[:, :, -1], ap_fits[:, :, -1])
# Get count of number of fit peaks as related to simulated knee value
n_peak_counter = count_peak_conditions(n_fit_peaks, KNEES)
# Check overall fit quality
temp = r2_fits.flatten()
print('Min/Max R^2: \t{:1.4f} \t{:1.4f}'.format(np.nanmin(temp), np.nanmax(temp)))
print('Median R^2: \t{:1.4f}'.format(np.nanmedian(temp)))
# Collect data together
datas = {
'OFF' : off_errors,
'EXP' : exp_errors,
'ERR' : err_fits,
'R^2' : r2_fits
}
# Print out the average error for each parameter, across number of peaks
# Also prints out the average model error and R^2 per peak count
with np.printoptions(precision=4, suppress=True):
for label, data in datas.items():
print(label, '\n\t', np.nanmedian(data, 1))
# Multi-peak fitting: plot error of aperiodic offset reconstruction across number of peaks
plot_errors_violin(off_errors, 'Offset', plt_log=PLT_LOG,
x_axis='knees', ylim=YLIMS_AP2, #ylim=[-1, 0.5],
save_fig=SAVE_FIG, save_name=pjoin(FOLDER, 'off_error'))
# Multi-peak fitting: plot error of aperiodic offset reconstruction across number of peaks
plot_errors_violin(exp_errors, 'Exponent', plt_log=PLT_LOG,
x_axis='knees', ylim=[-3.5, 0.25], # ylim=YLIMS_AP2,
save_fig=SAVE_FIG, save_name=pjoin(FOLDER, 'exp_error'))
# Plot the correspondance between number of simulated peaks & number of fit peaks
plot_n_peaks_bubbles(n_peak_counter, x_label='knee', ms_val=12,
save_fig=SAVE_FIG, save_name=pjoin(FOLDER, 'number_of_peaks'))
# Multi-peak fitting: plot error levels across knee values
plot_errors_violin(err_fits, 'Fit Error', plt_log=False, x_axis='knees',
save_fig=SAVE_FIG, save_name=pjoin(FOLDER, 'model_error'))
# Multi-peak fitting: plot R^2 levels across knee values
plot_errors_violin(r2_fits, 'R2', plt_log=False, x_axis='knees',
save_fig=SAVE_FIG, save_name=pjoin(FOLDER, 'model_r_squared'))
# Grab example case of no knee
ind = 0
fm1 = fgs[0].get_fooof(ind, True)
print(sim_params[0][ind])
fm1.print_results()
fm1.plot(plt_log=True, add_legend=True, save_fig=SAVE_FIG,
file_path=pjoin(FIGS_PATH, FOLDER), file_name='example_knee0' + SAVE_EXT)
# Grab example case of highest value knee
ind = 0
fm2 = fgs[-1].get_fooof(ind, True)
print(sim_params[-1][ind])
fm2.print_results()
fm2.plot(plt_log=True, add_legend=True, save_fig=SAVE_FIG,
file_path=pjoin(FIGS_PATH, FOLDER), file_name='example_knee5' + SAVE_EXT)
# Initialize a new FOOOFGroup, in 'knee mode'
nk_fg = FOOOFGroup(*FOOOF_SETTINGS_KNEE, verbose=False)
# Fit the first set of data, where knee value is 0
nk_fg.fit(freqs, psds[0, :, :])
# Get the fit data
nk_peak_fits, nk_ap_fits, nk_err_fits, nk_r2_fits, nk_n_fit_peaks = get_fit_data([nk_fg])
# Squeeze because we only have one 'condition' for these sims
nk_peak_fits = np.squeeze(nk_peak_fits)
nk_ap_fits = np.squeeze(nk_ap_fits)
nk_err_fits = np.squeeze(nk_err_fits)
nk_r2_fits = np.squeeze(nk_r2_fits)
nk_n_fit_peaks = np.squeeze(nk_n_fit_peaks)
# Extract the true simulated parameters for the no-knee data condition, used here
nk_ap_truths = ap_truths[0, :, :]
# Calculate errors of the aperiodic parameters that were fit
nk_off_errors = calc_errors(nk_ap_truths[:, 0], nk_ap_fits[:, 0])
nk_exp_errors = calc_errors(nk_ap_truths[:, -1], nk_ap_fits[:, -1])
# Check the goodness of fit measures
print('Average fit error: \t {:1.3f}'.format(np.mean(nk_err_fits)))
print('Average fit R^2: \t {:1.3f}'.format(np.mean(nk_r2_fits)))
# Check the error of fit offset and exponent as individual values
_, axes = plt.subplots(1, 2, figsize=[7, 5])
plt.subplots_adjust(wspace=0.5)
plot_single_data(nk_off_errors, 'Offset', ax=axes[0])
plot_single_data(nk_exp_errors, 'Exponent', ax=axes[1])
# Check the error of fit offset and exponent as violinplots
_, axes = plt.subplots(1, 2, figsize=[7, 5])
plt.subplots_adjust(wspace=0.5)
plot_errors_violin(nk_off_errors[np.newaxis, :], title='Offset', plt_log=PLT_LOG,
x_axis=None, ylim=YLIMS_AP, ax=axes[0])
plot_errors_violin(nk_exp_errors[np.newaxis, :], title='Exponent', plt_log=PLT_LOG,
x_axis=None, ylim=YLIMS_AP, ax=axes[1])
# Check the actual fit knee values in the knee-less data
print('Mean value for knee value: {:1.3f}'.format(
np.mean(nk_fg.get_params('aperiodic_params', 'knee'))))
plot_single_data(nk_fg.get_params('aperiodic_params', 'knee'), ylabel='Knee Value')
# Check the average number of fit peaks
print('The modal number of fit peaks is:\t {:1.0f}'.format(mode(nk_n_fit_peaks).mode[0]))
print('The mean number of fit peaks is:\t {:1.2f}'.format(np.mean(nk_n_fit_peaks)))
| 0.687 | 0.98652 |
```
import pandas as pd
import itertools
from sklearn.metrics import confusion_matrix
from tqdm import tqdm
tqdm.pandas()
```
# Summary
ABCD Face recognition models are regular convolutional neural networks models. They represent face photos as vectors. We find the distance between these two vectors to compare two faces. Finally, we classify two faces as same person whose distance is less than a threshold value.
The question is that how to determine the threshold. In this notebook, we will find the best split point for a threshold.
# Data set
```
# Ref: https://github.com/serengil/deepface/tree/master/tests/dataset
idendities = {
"Angelina": ["img1.jpg", "img2.jpg", "img4.jpg", "img5.jpg", "img6.jpg", "img7.jpg", "img10.jpg", "img11.jpg"],
"Scarlett": ["img8.jpg", "img9.jpg"],
"Jennifer": ["img3.jpg", "img12.jpg"],
"Mark": ["img13.jpg", "img14.jpg", "img15.jpg"],
"Jack": ["img16.jpg", "img17.jpg"],
"Elon": ["img18.jpg", "img19.jpg"],
"Jeff": ["img20.jpg", "img21.jpg"],
"Marissa": ["img22.jpg", "img23.jpg"],
"Sundar": ["img24.jpg", "img25.jpg"]
}
```
# Positive samples
Find different photos of same people
```
positives = []
for key, values in idendities.items():
#print(key)
for i in range(0, len(values)-1):
for j in range(i+1, len(values)):
#print(values[i], " and ", values[j])
positive = []
positive.append(values[i])
positive.append(values[j])
positives.append(positive)
positives = pd.DataFrame(positives, columns = ["file_x", "file_y"])
positives["decision"] = "Yes"
```
# Negative samples
Compare photos of different people
```
samples_list = list(idendities.values())
negatives = []
for i in range(0, len(idendities) - 1):
for j in range(i+1, len(idendities)):
#print(samples_list[i], " vs ",samples_list[j])
cross_product = itertools.product(samples_list[i], samples_list[j])
cross_product = list(cross_product)
#print(cross_product)
for cross_sample in cross_product:
#print(cross_sample[0], " vs ", cross_sample[1])
negative = []
negative.append(cross_sample[0])
negative.append(cross_sample[1])
negatives.append(negative)
negatives = pd.DataFrame(negatives, columns = ["file_x", "file_y"])
negatives["decision"] = "No"
```
# Merge Positives and Negative Samples
```
df = pd.concat([positives, negatives]).reset_index(drop = True)
df.shape
df.decision.value_counts()
df.file_x = "deepface/tests/dataset/"+df.file_x
df.file_y = "deepface/tests/dataset/"+df.file_y
```
# DeepFace
```
from deepface import DeepFace
instances = df[["file_x", "file_y"]].values.tolist()
model_name = "VGG-Face"
distance_metric = "cosine"
resp_obj = DeepFace.verify(instances, model_name = model_name, distance_metric = distance_metric)
distances = []
for i in range(0, len(instances)):
distance = round(resp_obj["pair_%s" % (i+1)]["distance"], 4)
distances.append(distance)
df["distance"] = distances
```
# Analyzing Distances
```
tp_mean = round(df[df.decision == "Yes"].mean().values[0], 4)
tp_std = round(df[df.decision == "Yes"].std().values[0], 4)
fp_mean = round(df[df.decision == "No"].mean().values[0], 4)
fp_std = round(df[df.decision == "No"].std().values[0], 4)
print("Mean of true positives: ", tp_mean)
print("Std of true positives: ", tp_std)
print("Mean of false positives: ", fp_mean)
print("Std of false positives: ", fp_std)
```
# Distribution
```
df[df.decision == "Yes"].distance.plot.kde()
df[df.decision == "No"].distance.plot.kde()
```
# Best Split Point
```
from chefboost import Chefboost as chef
config = {'algorithm': 'C4.5'}
tmp_df = df[['distance', 'decision']].rename(columns = {"decision": "Decision"}).copy()
model = chef.fit(tmp_df, config)
```
# Sigma
```
sigma = 2
#2 sigma corresponds 95.45% confidence, and 3 sigma corresponds 99.73% confidence
#threshold = round(tp_mean + sigma * tp_std, 4)
threshold = 0.3147 #comes from c4.5 algorithm
print("threshold: ", threshold)
df[df.decision == 'Yes'].distance.max()
df[df.decision == 'No'].distance.min()
```
# Evaluation
```
df["prediction"] = "No"
idx = df[df.distance <= threshold].index
df.loc[idx, 'prediction'] = 'Yes'
df.sample(5)
cm = confusion_matrix(df.decision.values, df.prediction.values)
cm
tn, fp, fn, tp = cm.ravel()
tn, fp, fn, tp
recall = tp / (tp + fn)
precision = tp / (tp + fp)
accuracy = (tp + tn)/(tn + fp + fn + tp)
f1 = 2 * (precision * recall) / (precision + recall)
print("Precision: ", 100*precision,"%")
print("Recall: ", 100*recall,"%")
print("F1 score ",100*f1, "%")
print("Accuracy: ", 100*accuracy,"%")
df.to_csv("threshold_pivot.csv", index = False)
```
## Test results
### Threshold = 0.3147 (C4.5 best split point)
Precision: 100.0 %
Recall: 89.47368421052632 %
F1 score 94.44444444444444%
Accuracy: 98.66666666666667 %
### Threshold = 0.3751 (2 sigma)
Precision: 90.47619047619048 %
Recall: 100.0 %
F1 score 95.0 %
Accuracy: 98.66666666666667 %
|
github_jupyter
|
import pandas as pd
import itertools
from sklearn.metrics import confusion_matrix
from tqdm import tqdm
tqdm.pandas()
# Ref: https://github.com/serengil/deepface/tree/master/tests/dataset
idendities = {
"Angelina": ["img1.jpg", "img2.jpg", "img4.jpg", "img5.jpg", "img6.jpg", "img7.jpg", "img10.jpg", "img11.jpg"],
"Scarlett": ["img8.jpg", "img9.jpg"],
"Jennifer": ["img3.jpg", "img12.jpg"],
"Mark": ["img13.jpg", "img14.jpg", "img15.jpg"],
"Jack": ["img16.jpg", "img17.jpg"],
"Elon": ["img18.jpg", "img19.jpg"],
"Jeff": ["img20.jpg", "img21.jpg"],
"Marissa": ["img22.jpg", "img23.jpg"],
"Sundar": ["img24.jpg", "img25.jpg"]
}
positives = []
for key, values in idendities.items():
#print(key)
for i in range(0, len(values)-1):
for j in range(i+1, len(values)):
#print(values[i], " and ", values[j])
positive = []
positive.append(values[i])
positive.append(values[j])
positives.append(positive)
positives = pd.DataFrame(positives, columns = ["file_x", "file_y"])
positives["decision"] = "Yes"
samples_list = list(idendities.values())
negatives = []
for i in range(0, len(idendities) - 1):
for j in range(i+1, len(idendities)):
#print(samples_list[i], " vs ",samples_list[j])
cross_product = itertools.product(samples_list[i], samples_list[j])
cross_product = list(cross_product)
#print(cross_product)
for cross_sample in cross_product:
#print(cross_sample[0], " vs ", cross_sample[1])
negative = []
negative.append(cross_sample[0])
negative.append(cross_sample[1])
negatives.append(negative)
negatives = pd.DataFrame(negatives, columns = ["file_x", "file_y"])
negatives["decision"] = "No"
df = pd.concat([positives, negatives]).reset_index(drop = True)
df.shape
df.decision.value_counts()
df.file_x = "deepface/tests/dataset/"+df.file_x
df.file_y = "deepface/tests/dataset/"+df.file_y
from deepface import DeepFace
instances = df[["file_x", "file_y"]].values.tolist()
model_name = "VGG-Face"
distance_metric = "cosine"
resp_obj = DeepFace.verify(instances, model_name = model_name, distance_metric = distance_metric)
distances = []
for i in range(0, len(instances)):
distance = round(resp_obj["pair_%s" % (i+1)]["distance"], 4)
distances.append(distance)
df["distance"] = distances
tp_mean = round(df[df.decision == "Yes"].mean().values[0], 4)
tp_std = round(df[df.decision == "Yes"].std().values[0], 4)
fp_mean = round(df[df.decision == "No"].mean().values[0], 4)
fp_std = round(df[df.decision == "No"].std().values[0], 4)
print("Mean of true positives: ", tp_mean)
print("Std of true positives: ", tp_std)
print("Mean of false positives: ", fp_mean)
print("Std of false positives: ", fp_std)
df[df.decision == "Yes"].distance.plot.kde()
df[df.decision == "No"].distance.plot.kde()
from chefboost import Chefboost as chef
config = {'algorithm': 'C4.5'}
tmp_df = df[['distance', 'decision']].rename(columns = {"decision": "Decision"}).copy()
model = chef.fit(tmp_df, config)
sigma = 2
#2 sigma corresponds 95.45% confidence, and 3 sigma corresponds 99.73% confidence
#threshold = round(tp_mean + sigma * tp_std, 4)
threshold = 0.3147 #comes from c4.5 algorithm
print("threshold: ", threshold)
df[df.decision == 'Yes'].distance.max()
df[df.decision == 'No'].distance.min()
df["prediction"] = "No"
idx = df[df.distance <= threshold].index
df.loc[idx, 'prediction'] = 'Yes'
df.sample(5)
cm = confusion_matrix(df.decision.values, df.prediction.values)
cm
tn, fp, fn, tp = cm.ravel()
tn, fp, fn, tp
recall = tp / (tp + fn)
precision = tp / (tp + fp)
accuracy = (tp + tn)/(tn + fp + fn + tp)
f1 = 2 * (precision * recall) / (precision + recall)
print("Precision: ", 100*precision,"%")
print("Recall: ", 100*recall,"%")
print("F1 score ",100*f1, "%")
print("Accuracy: ", 100*accuracy,"%")
df.to_csv("threshold_pivot.csv", index = False)
| 0.324985 | 0.878419 |
## Dependencies
```
!pip install --quiet /kaggle/input/kerasapplications
!pip install --quiet /kaggle/input/efficientnet-git
import warnings, glob
from tensorflow.keras import Sequential, Model
import efficientnet.tfkeras as efn
from cassava_scripts import *
seed = 0
seed_everything(seed)
warnings.filterwarnings('ignore')
```
### Hardware configuration
```
# TPU or GPU detection
# Detect hardware, return appropriate distribution strategy
strategy, tpu = set_up_strategy()
AUTO = tf.data.experimental.AUTOTUNE
REPLICAS = strategy.num_replicas_in_sync
print(f'REPLICAS: {REPLICAS}')
```
# Model parameters
```
BATCH_SIZE = 8 * REPLICAS
HEIGHT = 512
WIDTH = 512
CHANNELS = 3
N_CLASSES = 5
TTA_STEPS = 0 # Do TTA if > 0
```
# Augmentation
```
def data_augment(image, label):
p_spatial = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_rotate = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_pixel_1 = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_pixel_2 = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_pixel_3 = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_crop = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
# Flips
image = tf.image.random_flip_left_right(image)
image = tf.image.random_flip_up_down(image)
if p_spatial > .75:
image = tf.image.transpose(image)
# Rotates
if p_rotate > .75:
image = tf.image.rot90(image, k=3) # rotate 270º
elif p_rotate > .5:
image = tf.image.rot90(image, k=2) # rotate 180º
elif p_rotate > .25:
image = tf.image.rot90(image, k=1) # rotate 90º
# Pixel-level transforms
if p_pixel_1 >= .4:
image = tf.image.random_saturation(image, lower=.7, upper=1.3)
if p_pixel_2 >= .4:
image = tf.image.random_contrast(image, lower=.8, upper=1.2)
if p_pixel_3 >= .4:
image = tf.image.random_brightness(image, max_delta=.1)
# Crops
if p_crop > .7:
if p_crop > .9:
image = tf.image.central_crop(image, central_fraction=.7)
elif p_crop > .8:
image = tf.image.central_crop(image, central_fraction=.8)
else:
image = tf.image.central_crop(image, central_fraction=.9)
elif p_crop > .4:
crop_size = tf.random.uniform([], int(HEIGHT*.8), HEIGHT, dtype=tf.int32)
image = tf.image.random_crop(image, size=[crop_size, crop_size, CHANNELS])
# # Crops
# if p_crop > .6:
# if p_crop > .9:
# image = tf.image.central_crop(image, central_fraction=.5)
# elif p_crop > .8:
# image = tf.image.central_crop(image, central_fraction=.6)
# elif p_crop > .7:
# image = tf.image.central_crop(image, central_fraction=.7)
# else:
# image = tf.image.central_crop(image, central_fraction=.8)
# elif p_crop > .3:
# crop_size = tf.random.uniform([], int(HEIGHT*.6), HEIGHT, dtype=tf.int32)
# image = tf.image.random_crop(image, size=[crop_size, crop_size, CHANNELS])
return image, label
```
## Auxiliary functions
```
# Datasets utility functions
def resize_image(image, label):
image = tf.image.resize(image, [HEIGHT, WIDTH])
image = tf.reshape(image, [HEIGHT, WIDTH, CHANNELS])
return image, label
def process_path(file_path):
name = get_name(file_path)
img = tf.io.read_file(file_path)
img = decode_image(img)
img, _ = scale_image(img, None)
# img = center_crop(img, HEIGHT, WIDTH)
return img, name
def get_dataset(files_path, shuffled=False, tta=False, extension='jpg'):
dataset = tf.data.Dataset.list_files(f'{files_path}*{extension}', shuffle=shuffled)
dataset = dataset.map(process_path, num_parallel_calls=AUTO)
if tta:
dataset = dataset.map(data_augment, num_parallel_calls=AUTO)
dataset = dataset.map(resize_image, num_parallel_calls=AUTO)
dataset = dataset.batch(BATCH_SIZE)
dataset = dataset.prefetch(AUTO)
return dataset
```
# Load data
```
database_base_path = '/kaggle/input/cassava-leaf-disease-classification/'
submission = pd.read_csv(f'{database_base_path}sample_submission.csv')
display(submission.head())
TEST_FILENAMES = tf.io.gfile.glob(f'{database_base_path}test_tfrecords/ld_test*.tfrec')
NUM_TEST_IMAGES = count_data_items(TEST_FILENAMES)
print(f'GCS: test: {NUM_TEST_IMAGES}')
model_path_list = glob.glob('/kaggle/input/39-cassava-leaf-effnetb5-better-crop-512x512/*.h5')
model_path_list.sort()
print('Models to predict:')
print(*model_path_list, sep='\n')
```
# Model
```
def model_fn(input_shape, N_CLASSES):
inputs = L.Input(shape=input_shape, name='input_image')
base_model = efn.EfficientNetB5(input_tensor=inputs,
include_top=False,
weights=None,
pooling='avg')
x = L.Dropout(.25)(base_model.output)
output = L.Dense(N_CLASSES, activation='softmax', name='output')(x)
model = Model(inputs=inputs, outputs=output)
return model
with strategy.scope():
model = model_fn((None, None, CHANNELS), N_CLASSES)
model.summary()
```
# Test set predictions
```
files_path = f'{database_base_path}test_images/'
test_size = len(os.listdir(files_path))
test_preds = np.zeros((test_size, N_CLASSES))
for model_path in model_path_list:
print(model_path)
K.clear_session()
model.load_weights(model_path)
if TTA_STEPS > 0:
test_ds = get_dataset(files_path, tta=True).repeat()
ct_steps = TTA_STEPS * ((test_size/BATCH_SIZE) + 1)
preds = model.predict(test_ds, steps=ct_steps, verbose=1)[:(test_size * TTA_STEPS)]
preds = np.mean(preds.reshape(test_size, TTA_STEPS, N_CLASSES, order='F'), axis=1)
test_preds += preds / len(model_path_list)
else:
test_ds = get_dataset(files_path, tta=False)
x_test = test_ds.map(lambda image, image_name: image)
test_preds += model.predict(x_test) / len(model_path_list)
test_preds = np.argmax(test_preds, axis=-1)
test_names_ds = get_dataset(files_path)
image_names = [img_name.numpy().decode('utf-8') for img, img_name in iter(test_names_ds.unbatch())]
submission = pd.DataFrame({'image_id': image_names, 'label': test_preds})
submission.to_csv('submission.csv', index=False)
display(submission.head())
```
|
github_jupyter
|
!pip install --quiet /kaggle/input/kerasapplications
!pip install --quiet /kaggle/input/efficientnet-git
import warnings, glob
from tensorflow.keras import Sequential, Model
import efficientnet.tfkeras as efn
from cassava_scripts import *
seed = 0
seed_everything(seed)
warnings.filterwarnings('ignore')
# TPU or GPU detection
# Detect hardware, return appropriate distribution strategy
strategy, tpu = set_up_strategy()
AUTO = tf.data.experimental.AUTOTUNE
REPLICAS = strategy.num_replicas_in_sync
print(f'REPLICAS: {REPLICAS}')
BATCH_SIZE = 8 * REPLICAS
HEIGHT = 512
WIDTH = 512
CHANNELS = 3
N_CLASSES = 5
TTA_STEPS = 0 # Do TTA if > 0
def data_augment(image, label):
p_spatial = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_rotate = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_pixel_1 = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_pixel_2 = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_pixel_3 = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_crop = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
# Flips
image = tf.image.random_flip_left_right(image)
image = tf.image.random_flip_up_down(image)
if p_spatial > .75:
image = tf.image.transpose(image)
# Rotates
if p_rotate > .75:
image = tf.image.rot90(image, k=3) # rotate 270º
elif p_rotate > .5:
image = tf.image.rot90(image, k=2) # rotate 180º
elif p_rotate > .25:
image = tf.image.rot90(image, k=1) # rotate 90º
# Pixel-level transforms
if p_pixel_1 >= .4:
image = tf.image.random_saturation(image, lower=.7, upper=1.3)
if p_pixel_2 >= .4:
image = tf.image.random_contrast(image, lower=.8, upper=1.2)
if p_pixel_3 >= .4:
image = tf.image.random_brightness(image, max_delta=.1)
# Crops
if p_crop > .7:
if p_crop > .9:
image = tf.image.central_crop(image, central_fraction=.7)
elif p_crop > .8:
image = tf.image.central_crop(image, central_fraction=.8)
else:
image = tf.image.central_crop(image, central_fraction=.9)
elif p_crop > .4:
crop_size = tf.random.uniform([], int(HEIGHT*.8), HEIGHT, dtype=tf.int32)
image = tf.image.random_crop(image, size=[crop_size, crop_size, CHANNELS])
# # Crops
# if p_crop > .6:
# if p_crop > .9:
# image = tf.image.central_crop(image, central_fraction=.5)
# elif p_crop > .8:
# image = tf.image.central_crop(image, central_fraction=.6)
# elif p_crop > .7:
# image = tf.image.central_crop(image, central_fraction=.7)
# else:
# image = tf.image.central_crop(image, central_fraction=.8)
# elif p_crop > .3:
# crop_size = tf.random.uniform([], int(HEIGHT*.6), HEIGHT, dtype=tf.int32)
# image = tf.image.random_crop(image, size=[crop_size, crop_size, CHANNELS])
return image, label
# Datasets utility functions
def resize_image(image, label):
image = tf.image.resize(image, [HEIGHT, WIDTH])
image = tf.reshape(image, [HEIGHT, WIDTH, CHANNELS])
return image, label
def process_path(file_path):
name = get_name(file_path)
img = tf.io.read_file(file_path)
img = decode_image(img)
img, _ = scale_image(img, None)
# img = center_crop(img, HEIGHT, WIDTH)
return img, name
def get_dataset(files_path, shuffled=False, tta=False, extension='jpg'):
dataset = tf.data.Dataset.list_files(f'{files_path}*{extension}', shuffle=shuffled)
dataset = dataset.map(process_path, num_parallel_calls=AUTO)
if tta:
dataset = dataset.map(data_augment, num_parallel_calls=AUTO)
dataset = dataset.map(resize_image, num_parallel_calls=AUTO)
dataset = dataset.batch(BATCH_SIZE)
dataset = dataset.prefetch(AUTO)
return dataset
database_base_path = '/kaggle/input/cassava-leaf-disease-classification/'
submission = pd.read_csv(f'{database_base_path}sample_submission.csv')
display(submission.head())
TEST_FILENAMES = tf.io.gfile.glob(f'{database_base_path}test_tfrecords/ld_test*.tfrec')
NUM_TEST_IMAGES = count_data_items(TEST_FILENAMES)
print(f'GCS: test: {NUM_TEST_IMAGES}')
model_path_list = glob.glob('/kaggle/input/39-cassava-leaf-effnetb5-better-crop-512x512/*.h5')
model_path_list.sort()
print('Models to predict:')
print(*model_path_list, sep='\n')
def model_fn(input_shape, N_CLASSES):
inputs = L.Input(shape=input_shape, name='input_image')
base_model = efn.EfficientNetB5(input_tensor=inputs,
include_top=False,
weights=None,
pooling='avg')
x = L.Dropout(.25)(base_model.output)
output = L.Dense(N_CLASSES, activation='softmax', name='output')(x)
model = Model(inputs=inputs, outputs=output)
return model
with strategy.scope():
model = model_fn((None, None, CHANNELS), N_CLASSES)
model.summary()
files_path = f'{database_base_path}test_images/'
test_size = len(os.listdir(files_path))
test_preds = np.zeros((test_size, N_CLASSES))
for model_path in model_path_list:
print(model_path)
K.clear_session()
model.load_weights(model_path)
if TTA_STEPS > 0:
test_ds = get_dataset(files_path, tta=True).repeat()
ct_steps = TTA_STEPS * ((test_size/BATCH_SIZE) + 1)
preds = model.predict(test_ds, steps=ct_steps, verbose=1)[:(test_size * TTA_STEPS)]
preds = np.mean(preds.reshape(test_size, TTA_STEPS, N_CLASSES, order='F'), axis=1)
test_preds += preds / len(model_path_list)
else:
test_ds = get_dataset(files_path, tta=False)
x_test = test_ds.map(lambda image, image_name: image)
test_preds += model.predict(x_test) / len(model_path_list)
test_preds = np.argmax(test_preds, axis=-1)
test_names_ds = get_dataset(files_path)
image_names = [img_name.numpy().decode('utf-8') for img, img_name in iter(test_names_ds.unbatch())]
submission = pd.DataFrame({'image_id': image_names, 'label': test_preds})
submission.to_csv('submission.csv', index=False)
display(submission.head())
| 0.553264 | 0.547343 |
<a href="https://colab.research.google.com/github/nsstnaka/machine_learning_handson/blob/master/stock_price_prediction_with_rnn.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# ディープラーニングによる株価予測
直近50営業日の4本値+出来高をもとに、終値を予想します。
## 前準備
ライブラリのimport
```
import pandas as pd
import pandas_datareader as pdr
import numpy as np
import tensorflow as tf
import seaborn as sns
import matplotlib.pyplot as plt
```
TensorFlowのバージョン確認
```
tf.__version__
```
## 株価取得
```
df = pdr.data.DataReader('^DJI', 'yahoo', '2017-04-01', '2020-03-31') # '^DJI'の部分を変えると違う株価を拾える(例:AAPL, GOOG)
df.reset_index(inplace=True) # 後続処理のためインデックスを振りなおす
df.head(10)
```
可視化
```
df.plot(x='Date', y=['Close'])
```
統計情報
```
df.describe()
```
## 前処理
4本値+出来高の抽出
```
features = df[['High', 'Low', 'Open', 'Close', 'Volume']].values
features
```
正規化
```
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
features_scaled = scaler.fit_transform(features)
features_scaled
```
時系列データおよびラベルの作成
```
N = 50
all_xs = []
all_ys = []
for idx in range(len(features_scaled)):
if idx < N:
continue
all_xs.append(features_scaled[idx-N:idx, :]) # 直近N営業日のHigh, Low, Open, Close, Volume
all_ys.append(features_scaled[idx, 3]) # 当日のClose
all_xs = np.array(all_xs)
all_ys = np.array(all_ys)
```
できたデータのshapeを確認
```
all_xs.shape
all_ys.shape
```
訓練データとテストデータに分割
(最新の100営業日分をテストデータにし、残りを訓練データにする)
```
test_num = 100
train_xs = all_xs[:-test_num]
train_ys = all_ys[:-test_num]
test_xs = all_xs[-test_num:]
test_ys = all_ys[-test_num:]
print(train_xs.shape, train_ys.shape, test_xs.shape, test_ys.shape)
```
## 学習と評価
### (1) 1層のRNN
モデル構築
```
model = tf.keras.models.Sequential([
tf.keras.layers.LSTM(128, input_shape=(N, train_xs.shape[2])),
# tf.keras.layers.GRU(128, input_shape=(N, 1)),
# tf.keras.layers.SimpleRNN(128, input_shape=(N, 1)),
tf.keras.layers.Dense(1)
])
model.compile(optimizer='adam', loss='mean_squared_error')
```
モデル確認
```
model.summary()
```
学習
```
model.fit(x=train_xs, y=train_ys, batch_size=8, epochs=50)
```
評価
```
pred = model.predict(x=test_xs)
pred = pred / scaler.scale_[3] + scaler.data_min_[3]
test_df = pd.DataFrame(pred, columns=['predict'])
test_df['actual'] = test_ys / scaler.scale_[3] + scaler.data_min_[3]
test_df.plot()
```
### 2層のRNN(ドロップアウト付)
モデル構築
```
model = tf.keras.models.Sequential([
tf.keras.layers.LSTM(128, input_shape=(N, train_xs.shape[2]), return_sequences=True),
# tf.keras.layers.GRU(128, input_shape=(N, 1)),
# tf.keras.layers.SimpleRNN(128, input_shape=(N, 1)),
tf.keras.layers.Dropout(0.5),
tf.keras.layers.LSTM(128),
tf.keras.layers.Dropout(0.5),
tf.keras.layers.Dense(1)
])
model.compile(optimizer='adam', loss='mean_squared_error')
```
モデル確認
```
model.summary()
```
学習
```
model.fit(x=train_xs, y=train_ys, batch_size=8, epochs=50)
```
評価
```
pred = model.predict(x=test_xs)
pred = pred / scaler.scale_[3] + scaler.data_min_[3]
test_df = pd.DataFrame(pred, columns=['predict'])
test_df['actual'] = test_ys / scaler.scale_[3] + scaler.data_min_[3]
test_df.plot()
```
|
github_jupyter
|
import pandas as pd
import pandas_datareader as pdr
import numpy as np
import tensorflow as tf
import seaborn as sns
import matplotlib.pyplot as plt
tf.__version__
df = pdr.data.DataReader('^DJI', 'yahoo', '2017-04-01', '2020-03-31') # '^DJI'の部分を変えると違う株価を拾える(例:AAPL, GOOG)
df.reset_index(inplace=True) # 後続処理のためインデックスを振りなおす
df.head(10)
df.plot(x='Date', y=['Close'])
df.describe()
features = df[['High', 'Low', 'Open', 'Close', 'Volume']].values
features
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
features_scaled = scaler.fit_transform(features)
features_scaled
N = 50
all_xs = []
all_ys = []
for idx in range(len(features_scaled)):
if idx < N:
continue
all_xs.append(features_scaled[idx-N:idx, :]) # 直近N営業日のHigh, Low, Open, Close, Volume
all_ys.append(features_scaled[idx, 3]) # 当日のClose
all_xs = np.array(all_xs)
all_ys = np.array(all_ys)
all_xs.shape
all_ys.shape
test_num = 100
train_xs = all_xs[:-test_num]
train_ys = all_ys[:-test_num]
test_xs = all_xs[-test_num:]
test_ys = all_ys[-test_num:]
print(train_xs.shape, train_ys.shape, test_xs.shape, test_ys.shape)
model = tf.keras.models.Sequential([
tf.keras.layers.LSTM(128, input_shape=(N, train_xs.shape[2])),
# tf.keras.layers.GRU(128, input_shape=(N, 1)),
# tf.keras.layers.SimpleRNN(128, input_shape=(N, 1)),
tf.keras.layers.Dense(1)
])
model.compile(optimizer='adam', loss='mean_squared_error')
model.summary()
model.fit(x=train_xs, y=train_ys, batch_size=8, epochs=50)
pred = model.predict(x=test_xs)
pred = pred / scaler.scale_[3] + scaler.data_min_[3]
test_df = pd.DataFrame(pred, columns=['predict'])
test_df['actual'] = test_ys / scaler.scale_[3] + scaler.data_min_[3]
test_df.plot()
model = tf.keras.models.Sequential([
tf.keras.layers.LSTM(128, input_shape=(N, train_xs.shape[2]), return_sequences=True),
# tf.keras.layers.GRU(128, input_shape=(N, 1)),
# tf.keras.layers.SimpleRNN(128, input_shape=(N, 1)),
tf.keras.layers.Dropout(0.5),
tf.keras.layers.LSTM(128),
tf.keras.layers.Dropout(0.5),
tf.keras.layers.Dense(1)
])
model.compile(optimizer='adam', loss='mean_squared_error')
model.summary()
model.fit(x=train_xs, y=train_ys, batch_size=8, epochs=50)
pred = model.predict(x=test_xs)
pred = pred / scaler.scale_[3] + scaler.data_min_[3]
test_df = pd.DataFrame(pred, columns=['predict'])
test_df['actual'] = test_ys / scaler.scale_[3] + scaler.data_min_[3]
test_df.plot()
| 0.558809 | 0.976625 |
# Preliminary XGBoost
This notebook outlines preliminary work done to tune the XGBoost classifier. **The results in this notebook are superceeded by those in `xgb_tuning.ipynb` for the purposes of the report.** This notebook does however show comparable results for gradient boosting and explores a broader design space. The results from this notebook were used in part as the basis of understanding in order to set the initial hyperparameter values for the final tuning process.
```
import matplotlib.pyplot as plt
from sklearn.metrics import roc_curve
from sklearn.metrics import auc
from itertools import cycle
from sklearn.metrics import RocCurveDisplay
classes = ['anger', 'fear', 'joy', 'love', 'sadness', 'surprise']
def plot_mc_roc(Y_test_bin, Y_test_proba, n_classes, title='ROC Curve'):
ax = plt.axes()
leg = []
auc_sum = 0
for i in range(n_classes):
y_score = Y_test_proba[:, i]
fpr, tpr, _ = roc_curve(Y_test_bin[:, i], y_score)
roc_auc = auc(fpr, tpr)
auc_sum += roc_auc
leg.append(classes[i] + ' (auc = {:.4})'.format(roc_auc))
RocCurveDisplay(fpr=fpr, tpr=tpr).plot(ax=ax)
plt.xlim((0, .4))
plt.ylim((.6, 1))
plt.legend(leg, loc='lower right')
plt.title(title)
print('Unweighted Average AUC: {:.4}'.format(auc_sum/n_classes))
import pandas as pd
# read in all data
test = pd.read_csv('../data/test.txt', delimiter=';', names=['text', 'target'])
train = pd.read_csv('../data/train.txt', delimiter=';',
names=['text', 'target'])
val = pd.read_csv('../data/val.txt', delimiter=';', names=['text', 'target'])
trainval = pd.concat([train,val])
testval = pd.concat([test,val])
import numpy as np
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.preprocessing import LabelEncoder
# create vectorier for BoW
vectorizer = CountVectorizer(max_df=0.5, min_df=5, stop_words='english', ngram_range=(1, 1))
BoW = vectorizer.fit_transform(trainval.text)
print('Number of Features in BoW: ', len(vectorizer.get_feature_names_out()))
enc = LabelEncoder().fit(['anger', 'fear', 'joy', 'love', 'sadness', 'surprise'])
# transform all the data
X_trainval = vectorizer.transform(trainval.text).toarray()
Y_trainval = enc.transform(trainval.target)
X_train = vectorizer.transform(train.text).toarray()
Y_train = enc.transform(train.target)
X_val = vectorizer.transform(val.text).toarray()
Y_val = enc.transform(val.target)
X_test = vectorizer.transform(test.text).toarray()
Y_test = enc.transform(test.target)
```
## Coarse Hyperparameter Search
Performing a course randomized search cross-validation to get an estimate of where the best model resides in the design space. Further tuning and optimization will be performed based on the results of the randomized cross-validation.
```
from xgboost import XGBClassifier
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import RandomizedSearchCV
clf = XGBClassifier(random_state=0, eval_metric='mlogloss', use_label_encoder=False)
params = {'n_estimators':[10, 50, 100, 250, 500],
'max_depth':[1, 5, 10, 25, 50],
'reg_alpha':[0, .1, 1, 10, 100],
'learning_rate':[.01, .1, .25, .5, 1]
}
#grid_cv = GridSearchCV(clf,param_grid=params, scoring='f1_macro', verbose=3, n_jobs=1, cv=5)
rand_cv = RandomizedSearchCV(clf, param_distributions=params, n_iter=25, n_jobs=-1, verbose=1, scoring='f1_macro', cv=2, random_state=0)
rand_cv.fit(X_trainval, Y_trainval)
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
import numpy as np
print('Best Parameters: ', rand_cv.best_params_)
print('Best Score: ', rand_cv.best_score_)
best_model = rand_cv.best_estimator_
res = rand_cv.cv_results_
res.pop('params',None)
res.pop('split0_test_score',None)
res.pop('split1_test_score',None)
df = pd.DataFrame(res).sort_values('rank_test_score')
df['param_reg_alpha'] = df['param_reg_alpha'].astype(np.float64)
df['param_n_estimators'] = df['param_n_estimators'].astype(np.float64)
df['param_learning_rate'] = df['param_learning_rate'].astype(np.float64)
df['param_max_depth'] = df['param_max_depth'].astype(np.float64)
display(df)
# display corr heat map
fig, ax = plt.subplots(figsize=(8, 8))
sns.heatmap(df.corr(), center=0, annot=True)
from sklearn.preprocessing import label_binarize
from sklearn.metrics import classification_report
from sklearn.metrics import accuracy_score
Y_test_pred = best_model.predict(X_test)
Y_trainval_pred = best_model.predict(X_trainval)
print('Coarse Estimation Best Model Train Accuracy: {:.4}'.format(accuracy_score(Y_trainval, Y_trainval_pred)))
print('Coarse Estimation Best Model Test Accuracy: ', accuracy_score(Y_test, Y_test_pred))
print(classification_report(Y_test, Y_test_pred))
Y_test_bin = label_binarize(Y_test, classes=[0, 1, 2, 3, 4, 5])
Y_test_proba = best_model.predict_proba(X_test)
plot_mc_roc(Y_test_bin, Y_test_proba, n_classes=6,
title='ROC for Coarse CV XGBoost (one-vs-all)')
```
### Analysis of Coarse Hyperparameter Estimation
The coarse estimation of hyperparameter performed provides an initial assessment of how the design responds to changes in different parameters. We keep the search space coarse to reduce computation time, however this reduction inherently makes the cross-validation sparse meaning we can only initially infer some general trends. Additional fine-tuning will be performed based on these results.
The primary takeaways from initial coarse estimation are as follows:
- Regularization has a strong negative correlation with the the F1 validation score meaning that alpha regularization should be kept small or not used at all
- Max tree depth and the learning rate both have significant positive correlation with the F1 validation score. This means that deeper trees and higher learning rates tend to increase the accuracy of the model. This is supported by the fact that most top scoring runs in the table have a high value of one or both of those parameters.
- The number of trees used had no correlation to the validation scores. This indicates that the number of trees is a less important parameter than the the others used. We know that higher number of estimators will tend to perform better, however these results show that the impact is less significant than that of other variables.
The performance of the best model selected exceeds that of the random forrest model previously analysed.
**Note that these takeaways are specific to the models tested, and will not necessarily hold true for all design spaces.** For example, while learning rate had a positive correlation, that will not always be the case depending on the number of estimators used.
## Hyperparameter Fine-Tuning
Now that we have a general sense for where the optimal model lies in the design space, we can perform more fine-tuning via additional cross-validation. For the sake of reducing computational requirements we wil tune a subset of parameters at a time. Regularization showed no evidence of benefit in the coarse estimation so it is not included in the tuning.
### Learning Rate and N-Estimators Tuning
The learning rate proved to have a positive correlation with accuracy. Although we observed that the number of estimators (trees) had lower impact on the score, we know that the number of trees and the optimal learning rate are closely connected, with fewer estimators requiring a higher learning rate and visa-versa. From coarse estimation we know higher depths of trees tend to perform better. For the sake of training speed we use a relatively low depth in order to evaluate the best learning rate and n-estimators, and then in a future step evaluate the best value for the depth of the trees.
```
from xgboost import XGBClassifier
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import RandomizedSearchCV
clf = XGBClassifier(random_state=0, eval_metric='mlogloss',use_label_encoder=False)
params = {'n_estimators': [50, 250, 500, 750],
'max_depth': [5],
'learning_rate': [.1, .5, 1, 1.5]#[.5, 2, 5]
}
grid_cv = GridSearchCV(clf,param_grid=params, scoring='f1_macro', verbose=1, n_jobs=-1, cv=2)
grid_cv.fit(X_trainval, Y_trainval)
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
import numpy as np
print('Best Parameters: ', grid_cv.best_params_)
print('Best Score: ', grid_cv.best_score_)
best_model = grid_cv.best_estimator_
res = grid_cv.cv_results_
res.pop('params', None)
res.pop('split0_test_score', None)
res.pop('split1_test_score', None)
df = pd.DataFrame(res).sort_values('rank_test_score')
display(df)
from sklearn.preprocessing import label_binarize
from sklearn.metrics import classification_report
from sklearn.metrics import accuracy_score
Y_test_pred = best_model.predict(X_test)
Y_trainval_pred = best_model.predict(X_trainval)
print('Best Model Train Accuracy: {:.4}'.format(accuracy_score(Y_trainval, Y_trainval_pred)))
print('Best Model Test Accuracy: ', accuracy_score(Y_test, Y_test_pred))
print(classification_report(Y_test, Y_test_pred))
Y_test_bin = label_binarize(Y_test, classes=[0, 1, 2, 3, 4, 5])
Y_test_proba = best_model.predict_proba(X_test)
plot_mc_roc(Y_test_bin, Y_test_proba, n_classes=6,
title='ROC for Coarse CV XGBoost (one-vs-all)')
from xgboost import XGBClassifier
best_try_clf = XGBClassifier(n_estimators=250, learning_rate=.5, max_depth=25, n_jobs=-1, random_state=0, eval_metric='mlogloss',use_label_encoder=False)
best_try_clf.fit(X_trainval, Y_trainval)
from sklearn.preprocessing import label_binarize
from sklearn.metrics import classification_report
from sklearn.metrics import accuracy_score
Y_test_pred = best_try_clf.predict(X_test)
Y_trainval_pred = best_try_clf.predict(X_trainval)
print('Coarse Estimation Best Model Train Accuracy: {:.4}'.format(accuracy_score(Y_trainval, Y_trainval_pred)))
print('Coarse Estimation Best Model Test Accuracy: ', accuracy_score(Y_test, Y_test_pred))
print(classification_report(Y_test, Y_test_pred))
Y_test_bin = label_binarize(Y_test, classes=[0, 1, 2, 3, 4, 5])
Y_test_proba = best_try_clf.predict_proba(X_test)
plot_mc_roc(Y_test_bin, Y_test_proba, n_classes=6,
title='ROC for Coarse CV XGBoost (one-vs-all)')
from xgboost import XGBClassifier
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import RandomizedSearchCV
clf = XGBClassifier(random_state=0, eval_metric='mlogloss',
use_label_encoder=False)
params = {'n_estimators': [250],
'max_depth': [1, 5, 10, 17, 25, 20],
'learning_rate': [.5]
}
grid_cv = GridSearchCV(clf,param_grid=params, scoring='f1_macro', verbose=1, n_jobs=-1, cv=2)
grid_cv.fit(X_trainval, Y_trainval)
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
import numpy as np
print('Best Parameters: ', grid_cv.best_params_)
print('Best Score: ', grid_cv.best_score_)
best_model = grid_cv.best_estimator_
res = grid_cv.cv_results_
res.pop('params', None)
res.pop('split0_test_score', None)
res.pop('split1_test_score', None)
df = pd.DataFrame(res).sort_values('rank_test_score')
display(df)
```
|
github_jupyter
|
import matplotlib.pyplot as plt
from sklearn.metrics import roc_curve
from sklearn.metrics import auc
from itertools import cycle
from sklearn.metrics import RocCurveDisplay
classes = ['anger', 'fear', 'joy', 'love', 'sadness', 'surprise']
def plot_mc_roc(Y_test_bin, Y_test_proba, n_classes, title='ROC Curve'):
ax = plt.axes()
leg = []
auc_sum = 0
for i in range(n_classes):
y_score = Y_test_proba[:, i]
fpr, tpr, _ = roc_curve(Y_test_bin[:, i], y_score)
roc_auc = auc(fpr, tpr)
auc_sum += roc_auc
leg.append(classes[i] + ' (auc = {:.4})'.format(roc_auc))
RocCurveDisplay(fpr=fpr, tpr=tpr).plot(ax=ax)
plt.xlim((0, .4))
plt.ylim((.6, 1))
plt.legend(leg, loc='lower right')
plt.title(title)
print('Unweighted Average AUC: {:.4}'.format(auc_sum/n_classes))
import pandas as pd
# read in all data
test = pd.read_csv('../data/test.txt', delimiter=';', names=['text', 'target'])
train = pd.read_csv('../data/train.txt', delimiter=';',
names=['text', 'target'])
val = pd.read_csv('../data/val.txt', delimiter=';', names=['text', 'target'])
trainval = pd.concat([train,val])
testval = pd.concat([test,val])
import numpy as np
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.preprocessing import LabelEncoder
# create vectorier for BoW
vectorizer = CountVectorizer(max_df=0.5, min_df=5, stop_words='english', ngram_range=(1, 1))
BoW = vectorizer.fit_transform(trainval.text)
print('Number of Features in BoW: ', len(vectorizer.get_feature_names_out()))
enc = LabelEncoder().fit(['anger', 'fear', 'joy', 'love', 'sadness', 'surprise'])
# transform all the data
X_trainval = vectorizer.transform(trainval.text).toarray()
Y_trainval = enc.transform(trainval.target)
X_train = vectorizer.transform(train.text).toarray()
Y_train = enc.transform(train.target)
X_val = vectorizer.transform(val.text).toarray()
Y_val = enc.transform(val.target)
X_test = vectorizer.transform(test.text).toarray()
Y_test = enc.transform(test.target)
from xgboost import XGBClassifier
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import RandomizedSearchCV
clf = XGBClassifier(random_state=0, eval_metric='mlogloss', use_label_encoder=False)
params = {'n_estimators':[10, 50, 100, 250, 500],
'max_depth':[1, 5, 10, 25, 50],
'reg_alpha':[0, .1, 1, 10, 100],
'learning_rate':[.01, .1, .25, .5, 1]
}
#grid_cv = GridSearchCV(clf,param_grid=params, scoring='f1_macro', verbose=3, n_jobs=1, cv=5)
rand_cv = RandomizedSearchCV(clf, param_distributions=params, n_iter=25, n_jobs=-1, verbose=1, scoring='f1_macro', cv=2, random_state=0)
rand_cv.fit(X_trainval, Y_trainval)
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
import numpy as np
print('Best Parameters: ', rand_cv.best_params_)
print('Best Score: ', rand_cv.best_score_)
best_model = rand_cv.best_estimator_
res = rand_cv.cv_results_
res.pop('params',None)
res.pop('split0_test_score',None)
res.pop('split1_test_score',None)
df = pd.DataFrame(res).sort_values('rank_test_score')
df['param_reg_alpha'] = df['param_reg_alpha'].astype(np.float64)
df['param_n_estimators'] = df['param_n_estimators'].astype(np.float64)
df['param_learning_rate'] = df['param_learning_rate'].astype(np.float64)
df['param_max_depth'] = df['param_max_depth'].astype(np.float64)
display(df)
# display corr heat map
fig, ax = plt.subplots(figsize=(8, 8))
sns.heatmap(df.corr(), center=0, annot=True)
from sklearn.preprocessing import label_binarize
from sklearn.metrics import classification_report
from sklearn.metrics import accuracy_score
Y_test_pred = best_model.predict(X_test)
Y_trainval_pred = best_model.predict(X_trainval)
print('Coarse Estimation Best Model Train Accuracy: {:.4}'.format(accuracy_score(Y_trainval, Y_trainval_pred)))
print('Coarse Estimation Best Model Test Accuracy: ', accuracy_score(Y_test, Y_test_pred))
print(classification_report(Y_test, Y_test_pred))
Y_test_bin = label_binarize(Y_test, classes=[0, 1, 2, 3, 4, 5])
Y_test_proba = best_model.predict_proba(X_test)
plot_mc_roc(Y_test_bin, Y_test_proba, n_classes=6,
title='ROC for Coarse CV XGBoost (one-vs-all)')
from xgboost import XGBClassifier
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import RandomizedSearchCV
clf = XGBClassifier(random_state=0, eval_metric='mlogloss',use_label_encoder=False)
params = {'n_estimators': [50, 250, 500, 750],
'max_depth': [5],
'learning_rate': [.1, .5, 1, 1.5]#[.5, 2, 5]
}
grid_cv = GridSearchCV(clf,param_grid=params, scoring='f1_macro', verbose=1, n_jobs=-1, cv=2)
grid_cv.fit(X_trainval, Y_trainval)
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
import numpy as np
print('Best Parameters: ', grid_cv.best_params_)
print('Best Score: ', grid_cv.best_score_)
best_model = grid_cv.best_estimator_
res = grid_cv.cv_results_
res.pop('params', None)
res.pop('split0_test_score', None)
res.pop('split1_test_score', None)
df = pd.DataFrame(res).sort_values('rank_test_score')
display(df)
from sklearn.preprocessing import label_binarize
from sklearn.metrics import classification_report
from sklearn.metrics import accuracy_score
Y_test_pred = best_model.predict(X_test)
Y_trainval_pred = best_model.predict(X_trainval)
print('Best Model Train Accuracy: {:.4}'.format(accuracy_score(Y_trainval, Y_trainval_pred)))
print('Best Model Test Accuracy: ', accuracy_score(Y_test, Y_test_pred))
print(classification_report(Y_test, Y_test_pred))
Y_test_bin = label_binarize(Y_test, classes=[0, 1, 2, 3, 4, 5])
Y_test_proba = best_model.predict_proba(X_test)
plot_mc_roc(Y_test_bin, Y_test_proba, n_classes=6,
title='ROC for Coarse CV XGBoost (one-vs-all)')
from xgboost import XGBClassifier
best_try_clf = XGBClassifier(n_estimators=250, learning_rate=.5, max_depth=25, n_jobs=-1, random_state=0, eval_metric='mlogloss',use_label_encoder=False)
best_try_clf.fit(X_trainval, Y_trainval)
from sklearn.preprocessing import label_binarize
from sklearn.metrics import classification_report
from sklearn.metrics import accuracy_score
Y_test_pred = best_try_clf.predict(X_test)
Y_trainval_pred = best_try_clf.predict(X_trainval)
print('Coarse Estimation Best Model Train Accuracy: {:.4}'.format(accuracy_score(Y_trainval, Y_trainval_pred)))
print('Coarse Estimation Best Model Test Accuracy: ', accuracy_score(Y_test, Y_test_pred))
print(classification_report(Y_test, Y_test_pred))
Y_test_bin = label_binarize(Y_test, classes=[0, 1, 2, 3, 4, 5])
Y_test_proba = best_try_clf.predict_proba(X_test)
plot_mc_roc(Y_test_bin, Y_test_proba, n_classes=6,
title='ROC for Coarse CV XGBoost (one-vs-all)')
from xgboost import XGBClassifier
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import RandomizedSearchCV
clf = XGBClassifier(random_state=0, eval_metric='mlogloss',
use_label_encoder=False)
params = {'n_estimators': [250],
'max_depth': [1, 5, 10, 17, 25, 20],
'learning_rate': [.5]
}
grid_cv = GridSearchCV(clf,param_grid=params, scoring='f1_macro', verbose=1, n_jobs=-1, cv=2)
grid_cv.fit(X_trainval, Y_trainval)
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
import numpy as np
print('Best Parameters: ', grid_cv.best_params_)
print('Best Score: ', grid_cv.best_score_)
best_model = grid_cv.best_estimator_
res = grid_cv.cv_results_
res.pop('params', None)
res.pop('split0_test_score', None)
res.pop('split1_test_score', None)
df = pd.DataFrame(res).sort_values('rank_test_score')
display(df)
| 0.63375 | 0.917635 |
# Modern Data Science
**(Module 11: Data Analytics (IV))**
---
- Materials in this module include resources collected from various open-source online repositories.
- You are free to use, change and distribute this package.
Prepared by and for
**Student Members** |
2006-2018 [TULIP Lab](http://www.tulip.org.au), Australia
---
## Session 11A - Case Study: Prediction
The purpose of this session is to demonstrate different coefficient and linear regression.
### Content
### Part 1 Linear Regression
1.1 [Linear Regression Package](#lrp)
1.2 [Evaluation](#eva)
### Part 2 Classificiation
2.1 [Skulls Dataset](#data)
2.2 [Data Preprocessing](#datapre)
2.3 [KNN](#knn)
2.4 [Decision Tree](#dt)
2.5 [Random Forest](#rf)
---
## <span style="color:#0b486b">1. Linear Regression</span>
<a id = "lrp"></a>
### <span style="color:#0b486b">1.1 Linear Regression Package</span>
We will learn how to use sklearn package to do linear regression
```
from sklearn.datasets import load_diabetes
from sklearn.linear_model import LinearRegression
import matplotlib.pyplot as plt
%matplotlib inline
```
Now create an instance of the diabetes data set by using the <b>load_diabetes</b> function as a variable called <b>diabetes</b>.
```
diabetes = load_diabetes()
```
We will work with one feature only.
```
diabetes_X = diabetes.data[:, None, 2]
```
Now create an instance of the LinearRegression called LinReg.
```
LinReg = LinearRegression()
```
Now to perform <b>train/test split</b> we have to split the <b>X</b> and <b>y</b> into two different sets: The <b>training</b> and <b>testing</b> set. Luckily there is a sklearn function for just that!
Import the <b>train_test_split</b> from <b>sklearn.cross_validation</b>
```
from sklearn.model_selection import train_test_split
```
Now <b>train_test_split</b> will return <b>4</b> different parameters. We will name this <b>X_trainset</b>, <b>X_testset</b>, <b>y_trainset</b>, <b>y_testset</b>.
Now let's use <b>diabetes_X</b> as the <b>Feature Matrix</b> and <b>diabetes.target</b> as the <b>response vector</b> and split it up using <b>train_test_split</b> function we imported earlier (<i>If you haven't, please import it</i>). The <b>train_test_split</b> function should have <b>test_size = 0.3</b> and a <b>random state = 7</b>.
The <b>train_test_split</b> will need the parameters <b>X</b>, <b>y</b>, <b>test_size=0.3</b>, and <b>random_state=7</b>. The <b>X</b> and <b>y</b> are the arrays required before the split, the <b>test_size</b> represents the ratio of the testing dataset, and the <b>random_state</b> ensures we obtain the same splits.
```
X_trainset, X_testset, y_trainset, y_testset = train_test_split(diabetes_X, diabetes.target, test_size=0.3, random_state=7)
```
Train the <b>LinReg</b> model using <b>X_trainset</b> and <b>y_trainset</b>
```
LinReg.fit(X_trainset, y_trainset)
```
Now let's <i>plot</i> the graph.
<p> Use plt's <b>scatter</b> function to plot all the datapoints of <b>X_testset</b> and <b>y_testset</b> and color it <b>black</b> </p>
<p> Use plt's <b>plot</b> function to plot the line of best fit with <b>X_testset</b> and <b>LinReg.predict(X_testset)</b>. Color it <b>blue</b> with a <b>linewidth</b> of <b>3</b>. </p> <br>
<b>Note</b>: Please ignore the FutureWarning.
```
plt.scatter(X_testset, y_testset, color='black')
plt.plot(X_testset, LinReg.predict(X_testset), color='blue', linewidth=3)
```
<a id = "eva"></a>
### <span style="color:#0b486b">1.2 Evaluation</span>
In this part, you will learn the about the different evaluation models and metrics. You will be able to identify the strengths and weaknesses of each model and how to incorporate underfitting or overfilling them also referred to as the Bias-Variance trade-off.
```
import numpy as np
```
<br><b> Here's a list of useful functions: </b><br>
mean -> np.mean()<br>
exponent -> **<br>
absolute value -> abs()
We use three evaluation metrics:
$$ MAE = \frac{\sum_{j=1}^n|y_i-\hat y_i|}{n} $$
$$ MSE = \frac{\sum_{j=1}^n (y_i-\hat y_i)^2}{n} $$
$$ RMSE = \sqrt{\frac{\sum_{j=1}^n (y_i-\hat y_i)^2}{n}} $$
```
print(np.mean(abs(LinReg.predict(X_testset) - y_testset)))
print(np.mean((LinReg.predict(X_testset) - y_testset) ** 2) )
print(np.mean((LinReg.predict(X_testset) - y_testset) ** 2) ** (0.5) )
```
---
## <span style="color:#0b486b">2. Classification</span>
<a id = "cls"></a>
<a id = "data"></a>
### <span style="color:#0b486b">2.1 Skulls dataset</span>
In this section, we will take a closer look at a data set.
Everything starts off with how the data is stored. We will be working with .csv files, or comma separated value files. As the name implies, each attribute (or column) in the data is separated by commas.
Next, a little information about the dataset. We are using a dataset called skulls.csv, which contains the measurements made on Egyptian skulls from five epochs.
#### The attributes of the data are as follows:
<b>epoch</b> - The epoch the skull as assigned to, a factor with levels c4000BC c3300BC, c1850BC, c200BC, and cAD150, where the years are only given approximately.
<b>mb</b> - Maximal Breadth of the skull.
<b>bh</b> - Basiregmatic Heights of the skull.
<b>bl</b> - Basilveolar Length of the skull.
<b>nh</b> - Nasal Heights of the skull.
#### Importing Libraries
Before we begin, we need to import some libraries, as they have useful functions that will be used later on.<br>
If you look at the imports below, you will notice the return of **numpy**! Remember that numpy is homogeneous multidimensional array (ndarray).
```
import numpy as np
import pandas
```
---
We need the **pandas** library for a function to read .csv files
<ul>
<li> <b>pandas.read_csv</b> - Reads data into DataFrame </li>
<li> The read_csv function takes in <i>2 parameters</i>: </li>
<ul>
<li> The .csv file as the first parameter </li>
<li> The delimiter as the second parameter </li>
</ul>
</ul>
-----------------------------
Save the "<b> skulls.csv </b>" data file into a variable called <b> my_data </b>
```
!pip install wget
import wget
link_to_data = 'https://github.com/tuliplab/mds/raw/master/Jupyter/data/skulls.csv'
DataSet = wget.download(link_to_data)
my_data = pandas.read_csv("skulls.csv", delimiter=",")
my_data.describe()
```
Print out the data in <b> my_data </b>
```
print(my_data)
```
Check the type of <b> my_data </b>
```
print (type(my_data))
```
-----------
There are various functions that the **pandas** library has to look at the data
<ul>
<li> <font color = "red"> [DataFrame Data].columns </font> - Displays the Header of the Data </li>
<ul>
<li> Type: pandas.indexes.base.Index </li>
</ul>
</ul>
<ul>
<li> <font color = "red"> [DataFrame Data].values </font> (or <font color = "red"> [DataFrame Data].as_matrix() </font>) - Displays the values of the data (without headers) </li>
<ul>
<li> Type: numpy.ndarray </li>
</ul>
</ul>
<ul>
<li> <font color = "red"> [DataFrame Data].shape </font> - Displays the dimensions of the data (rows x columns) </li>
<ul>
<li> Type: tuple </li>
</ul>
</ul>
----------
Using the <b> my_data </b> variable containing the DataFrame data, retrieve the <b> header </b> data, data <b> values </b>, and <b> shape </b> of the data.
```
print( my_data.columns)
print (my_data.values)
print (my_data.shape)
```
<a id = "datapre"></a>
### <span style="color:#0b486b">2.2 Data Preprocessing</span>
When we train a model, the model requires two inputs, X and y
<ul>
<li> X: Feature Matrix, or array that contains the data. </li>
<li> y: Response Vector, or 1-D array that contains the classification categories </li>
</ul>
------------
There are some problems with the data in my_data:
<ul>
<li> There is a header on the data (Unnamed: 0 epoch mb bh bl nh) </li>
<li> The data needs to be in numpy.ndarray format in order to use it in the machine learning model </li>
<li> There is non-numeric data within the dataset </li>
<li> There are row numbers associated with each row that affect the model </li>
</ul>
To resolve these problems, I have created a function that fixes these for us:
<b> removeColumns(pandasArray, column) </b>
This function produces one output and requires two inputs.
<ul>
<li> 1st Input: A pandas array. The pandas array we have been using is my_data </li>
<li> 2nd Input: Any number of integer values (order doesn't matter) that represent the columns that we want to remove. (Look at the data again and find which column contains the non-numeric values). We also want to remove the first column because that only contains the row number, which is irrelevant to our analysis.</li>
<ul>
<li> Note: Remember that Python is zero-indexed, therefore the first column would be 0. </li>
</ul>
</ul>
```
# Remove the column containing the target name since it doesn't contain numeric values.
# Also remove the column that contains the row number
# axis=1 means we are removing columns instead of rows.
# Function takes in a pandas array and column numbers and returns a numpy array without
# the stated columns
def removeColumns(pandasArray, *column):
return pandasArray.drop(pandasArray.columns[[column]], axis=1).values
```
---------
Using the function, store the values from the DataFrame data into a variable called new_data.
```
new_data = removeColumns(my_data, 0, 1)
```
Print out the data in <b> new_data </b>
```
print(new_data)
```
-------
Now, we have one half of the required data to fit a model, which is X or new_data
Next, we need to get the response vector y. Since we cannot use .target and .target_names, I have created a function that will do this for us.
<b> targetAndtargetNames(numpyArray, targetColumnIndex) </b>
This function produces two outputs, and requires two inputs.
<ul>
<li> <font size = 3.5><b><i>1st Input</i></b></font>: A numpy array. The numpy array you will use is my_data.values (or my_data.as_matrix())</li>
<ul>
<li> Note: DO NOT USE <b> new_data </b> here. We need the original .csv data file without the headers </li>
</ul>
</ul>
<ul>
<li> <font size = 3.5><b><i>2nd Input</i></b></font>: An integer value that represents the target column . (Look at the data again and find which column contains the non-numeric values. This is the target column)</li>
<ul>
<li> Note: Remember that Python is zero-indexed, therefore the first column would be 0. </li>
</ul>
</ul>
<ul>
<li> <font size = 3.5><b><i>1st Output</i></b></font>: The response vector (target) </li>
<li> <font size = 3.5><b><i>2nd Output</i></b></font>: The target names (target_names) </li>
</ul>
```
def targetAndtargetNames(numpyArray, targetColumnIndex):
target_dict = dict()
target = list()
target_names = list()
count = -1
for i in range(len(my_data.values)):
if my_data.values[i][targetColumnIndex] not in target_dict:
count += 1
target_dict[my_data.values[i][targetColumnIndex]] = count
target.append(target_dict[my_data.values[i][targetColumnIndex]])
# Since a dictionary is not ordered, we need to order it and output it to a list so the
# target names will match the target.
for targetName in sorted(target_dict, key=target_dict.get):
target_names.append(targetName)
return np.asarray(target), target_names
```
Using the targetAndtargetNames function, create two variables called <b>target</b> and <b>target_names</b>
```
y, targetNames = targetAndtargetNames(my_data, 1)
```
Print out the <b>y</b> and <b>targetNames</b> variables you created.
```
print(y)
print(targetNames)
```
Now that we have the two required variables to fit the data, a sneak peak at how to fit data will be shown in the cell below.
<a id = "knn"></a>
### <span style="color:#0b486b">2.3 KNN</span>
**K-Nearest Neighbors** is an algorithm for supervised learning. Where the data is 'trained' with data points corresponding to their classification. Once a point is to be predicted, it takes into account the 'K' nearest points to it to determine it's classification.
#### Here's an visualization of the K-Nearest Neighbors algorithm.
<img src = "https://raw.githubusercontent.com/tuliplab/mds/master/Jupyter/image/KNN.png">
In this case, we have data points of Class A and B. We want to predict what the star (test data point) is. If we consider a k value of 3 (3 nearest data points) we will obtain a prediction of Class B. Yet if we consider a k value of 6, we will obtain a prediction of Class A.
In this sense, it is important to consider the value of k. But hopefully from this diagram, you should get a sense of what the K-Nearest Neighbors algorithm is. It considers the 'K' Nearest Neighbors (points) when it predicts the classification of the test point.
```
# X = removeColumns(my_data, 0, 1)
# y = target(my_data, 1)
X = new_data
print( X.shape)
print (y.shape)
```
Now to perform <b>train/test split</b> we have to split the <b>X</b> and <b>y</b> into two different sets: The <b>training</b> and <b>testing</b> set. Luckily there is a sklearn function for just that!
Import the <b>train_test_split</b> from <b>sklearn.cross_validation</b>
Now <b>train_test_split</b> will return <b>4</b> different parameters. We will name this <b>X_trainset</b>, <b>X_testset</b>, <b>y_trainset</b>, <b>y_testset</b>. The <b>train_test_split</b> will need the parameters <b>X</b>, <b>y</b>, <b>test_size=0.3</b>, and <b>random_state=7</b>. The <b>X</b> and <b>y</b> are the arrays required before the split, the <b>test_size</b> represents the ratio of the testing dataset, and the <b>random_state</b> ensures we obtain the same splits.
```
X_trainset, X_testset, y_trainset, y_testset = train_test_split(X, y, test_size=0.3, random_state=7)
```
Now let's print the shape of the training sets to see if they match.
```
print (X_trainset.shape)
print (y_trainset.shape)
```
Let's check the same with the testing sets! They should both match up!
```
print (X_testset.shape)
print (y_testset.shape)
```
Now similarly with the last lab, let's create declarations of KNeighborsClassifier. Except we will create 3 different ones:
- neigh -> n_neighbors = 1
- neigh23 -> n_neighbors = 23
- neigh90 -> n_neighbors = 90
```
from sklearn.neighbors import KNeighborsClassifier
neigh = KNeighborsClassifier(n_neighbors = 1)
neigh23 = KNeighborsClassifier(n_neighbors = 23)
neigh90 = KNeighborsClassifier(n_neighbors = 90)
```
Now we will fit each instance of <b>KNeighborsClassifier</b> with the <b>X_trainset</b> and <b>y_trainset</b>
```
neigh.fit(X_trainset, y_trainset)
neigh23.fit(X_trainset, y_trainset)
neigh90.fit(X_trainset, y_trainset)
```
Now you are able to predict with <b>multiple</b> datapoints. We can do this by just passing in the <b>y_testset</b> which contains multiple test points into a <b>predict</b> function of <b>KNeighborsClassifier</b>.
Let's pass the <b>y_testset</b> in the <b>predict</b> function each instance of <b>KNeighborsClassifier</b> but store it's returned value into <b>pred</b>, <b>pred23</b>, <b>pred90</b> (corresponding to each of their names)
```
pred = neigh.predict(X_testset)
pred23 = neigh23.predict(X_testset)
pred90 = neigh90.predict(X_testset)
```
Awesome! Now let's compute neigh's <b>prediction accuracy</b>. We can do this by using the <b>metrics.accuracy_score</b> function
```
from sklearn import metrics
print("Neigh's Accuracy: "), metrics.accuracy_score(y_testset, pred)
```
Interesting! Let's do the same for the other instances of KNeighborsClassifier.
```
print("Neigh23's Accuracy: "), metrics.accuracy_score(y_testset, pred23)
print("Neigh90's Accuracy: "), metrics.accuracy_score(y_testset, pred90)
```
As shown, the accuracy of <b>neigh23</b> is the highest. When <b>n_neighbors = 1</b>, the model was <b>overfit</b> to the training data (<i>too specific</i>) and when <b>n_neighbors = 90</b>, the model was <b>underfit</b> (<i>too generalized</i>). In comparison, <b>n_neighbors = 23</b> had a <b>good balance</b> between <b>Bias</b> and <b>Variance</b>, creating a generalized model that neither <b>underfit</b> the data nor <b>overfit</b> it.
<a id = "dt"></a>
### <span style="color:#0b486b">2.4 Decision Tree</span>
In this section, you will learn <b>decision trees</b> and <b>random forests</b>.
The <b> getFeatureNames </b> is a function made to get the attribute names for specific columns
This function produces one output and requires two inputs:
<ul>
<li> <b>1st Input</b>: A pandas array. The pandas array we have been using is <b>my_data</b>. </li>
<li> <b>2nd Input</b>: Any number of integer values (order doesn't matter) that represent the columns that we want to include. In our case we want <b>columns 2-5</b>. </li>
<ul> <li> Note: Remember that Python is zero-indexed, therefore the first column would be 0. </li> </ul>
```
def getFeatureNames(pandasArray, *column):
actualColumns = list()
allColumns = list(pandasArray.columns.values)
for i in sorted(column):
actualColumns.append(allColumns[i])
return actualColumns
```
Now we prepare the data for decision tree construction.
```
#X = removeColumns(my_data, 0, 1)
#y, targetNames = targetAndtargetNames(my_data, 1)
featureNames = getFeatureNames(my_data, 2,3,4,5)
```
Print out <b>y</b>, <b>targetNames</b>, and <b>featureNames</b> to use in the next example. Remember that the numbers correspond to the names, 0 being the first name,1 being the second name, and so on.
```
print( y )
print (targetNames )
print (featureNames)
```
We will first create an instance of the <b>DecisionTreeClassifier</b> called <b>skullsTree</b>.<br>
Inside of the classifier, specify <i> criterion="entropy" </i> so we can see the information gain of each node.
```
from sklearn.tree import DecisionTreeClassifier
skullsTree = DecisionTreeClassifier(criterion="entropy")
skullsTree.fit(X_trainset,y_trainset)
```
Let's make some <b>predictions</b> on the testing dataset and store it into a variable called <b>predTree</b>.
```
predTree = skullsTree.predict(X_testset)
```
You can print out <b>predTree</b> and <b>y_testset</b> if you want to visually compare the prediction to the actual values.
```
print (predTree)
print (y_testset)
```
Next, let's import metrics from sklearn and check the accuracy of our model.
```
from sklearn import metrics
print("DecisionTrees's Accuracy: "), metrics.accuracy_score(y_testset, predTree)
```
Now we can visualize the tree constructed.
However, it should be noted that the following code may not work in all Python2 environment. You can try to see packages like <b>pydot</b>, <b>pydot2</b>, <b>pydot2</b>, <b>pydotplus</b>, etc., and see which one works in your platform.
```
!pip install pydotplus
#!pip install pydot2
#!pip install pyparsing==2.2.0
!pip install pydot
!conda install sklearn
from IPython.display import Image
from sklearn.externals.six import StringIO
from sklearn import tree
import pydot
import pydotplus
import pandas as pd
dot_data = StringIO()
tree.export_graphviz(skullsTree, out_file=dot_data,
feature_names=featureNames,
class_names=targetNames,
filled=True, rounded=True,
special_characters=True,
leaves_parallel=True)
graph = pydotplus.graph_from_dot_data(dot_data.getvalue())
Image(graph.create_png())
```
<a id = "rf"></a>
### <span style="color:#0b486b">2.5 Random Forest</span>
Import the <b>RandomForestClassifier</b> class from <b>sklearn.ensemble</b>
```
from sklearn.ensemble import RandomForestClassifier
```
Create an instance of the <b>RandomForestClassifier()</b> called <b>skullsForest</b>, where the forest has <b>10 decision tree estimators</b> (<i>n_estimators=10</i>) and the <b>criterion is entropy</b> (<i>criterion="entropy"</i>)
```
skullsForest=RandomForestClassifier(n_estimators=10, criterion="entropy")
```
Let's use the same <b>X_trainset</b>, <b>y_trainset</b> datasets that we made when dealing with the <b>Decision Trees</b> above to fit <b>skullsForest</b>.
<br> <br>
<b>Note</b>: Make sure you have ran through the Decision Trees section.
```
skullsForest.fit(X_trainset, y_trainset)
```
Let's now create a variable called <b>predForest</b> using a predict on <b>X_testset</b> with <b>skullsForest</b>.
```
predForest = skullsForest.predict(X_testset)
```
You can print out <b>predForest</b> and <b>y_testset</b> if you want to visually compare the prediction to the actual values.
```
print (predForest )
print (y_testset)
```
Let's check the accuracy of our model. <br>
Note: Make sure you have metrics imported from sklearn
```
print("RandomForests's Accuracy: "), metrics.accuracy_score(y_testset, predForest)
```
We can also see what trees are in our <b> skullsForest </b> variable by using the <b> .estimators_ </b> attribute. This attribute is indexable, so we can look at any individual tree we want.
```
print(skullsForest.estimators_)
```
You can choose to view any tree by using the code below. Replace the <i>"&"</i> in <b>skullsForest[&]</b> with the tree you want to see.
The following block may not work in your Python enrionment.
```
from IPython.display import Image
from sklearn.externals.six import StringIO
import pydot
dot_data = StringIO()
#Replace the '&' below with the tree number
tree.export_graphviz(skullsForest[&], out_file=dot_data,
feature_names=featureNames,
class_names=targetNames,
filled=True, rounded=True,
special_characters=True,
leaves_parallel=True)
graph = pydotplus.graph_from_dot_data(dot_data.getvalue())
Image(graph.create_png())
```
|
github_jupyter
|
from sklearn.datasets import load_diabetes
from sklearn.linear_model import LinearRegression
import matplotlib.pyplot as plt
%matplotlib inline
diabetes = load_diabetes()
diabetes_X = diabetes.data[:, None, 2]
LinReg = LinearRegression()
from sklearn.model_selection import train_test_split
X_trainset, X_testset, y_trainset, y_testset = train_test_split(diabetes_X, diabetes.target, test_size=0.3, random_state=7)
LinReg.fit(X_trainset, y_trainset)
plt.scatter(X_testset, y_testset, color='black')
plt.plot(X_testset, LinReg.predict(X_testset), color='blue', linewidth=3)
import numpy as np
print(np.mean(abs(LinReg.predict(X_testset) - y_testset)))
print(np.mean((LinReg.predict(X_testset) - y_testset) ** 2) )
print(np.mean((LinReg.predict(X_testset) - y_testset) ** 2) ** (0.5) )
import numpy as np
import pandas
!pip install wget
import wget
link_to_data = 'https://github.com/tuliplab/mds/raw/master/Jupyter/data/skulls.csv'
DataSet = wget.download(link_to_data)
my_data = pandas.read_csv("skulls.csv", delimiter=",")
my_data.describe()
print(my_data)
print (type(my_data))
print( my_data.columns)
print (my_data.values)
print (my_data.shape)
# Remove the column containing the target name since it doesn't contain numeric values.
# Also remove the column that contains the row number
# axis=1 means we are removing columns instead of rows.
# Function takes in a pandas array and column numbers and returns a numpy array without
# the stated columns
def removeColumns(pandasArray, *column):
return pandasArray.drop(pandasArray.columns[[column]], axis=1).values
new_data = removeColumns(my_data, 0, 1)
print(new_data)
def targetAndtargetNames(numpyArray, targetColumnIndex):
target_dict = dict()
target = list()
target_names = list()
count = -1
for i in range(len(my_data.values)):
if my_data.values[i][targetColumnIndex] not in target_dict:
count += 1
target_dict[my_data.values[i][targetColumnIndex]] = count
target.append(target_dict[my_data.values[i][targetColumnIndex]])
# Since a dictionary is not ordered, we need to order it and output it to a list so the
# target names will match the target.
for targetName in sorted(target_dict, key=target_dict.get):
target_names.append(targetName)
return np.asarray(target), target_names
y, targetNames = targetAndtargetNames(my_data, 1)
print(y)
print(targetNames)
# X = removeColumns(my_data, 0, 1)
# y = target(my_data, 1)
X = new_data
print( X.shape)
print (y.shape)
X_trainset, X_testset, y_trainset, y_testset = train_test_split(X, y, test_size=0.3, random_state=7)
print (X_trainset.shape)
print (y_trainset.shape)
print (X_testset.shape)
print (y_testset.shape)
from sklearn.neighbors import KNeighborsClassifier
neigh = KNeighborsClassifier(n_neighbors = 1)
neigh23 = KNeighborsClassifier(n_neighbors = 23)
neigh90 = KNeighborsClassifier(n_neighbors = 90)
neigh.fit(X_trainset, y_trainset)
neigh23.fit(X_trainset, y_trainset)
neigh90.fit(X_trainset, y_trainset)
pred = neigh.predict(X_testset)
pred23 = neigh23.predict(X_testset)
pred90 = neigh90.predict(X_testset)
from sklearn import metrics
print("Neigh's Accuracy: "), metrics.accuracy_score(y_testset, pred)
print("Neigh23's Accuracy: "), metrics.accuracy_score(y_testset, pred23)
print("Neigh90's Accuracy: "), metrics.accuracy_score(y_testset, pred90)
def getFeatureNames(pandasArray, *column):
actualColumns = list()
allColumns = list(pandasArray.columns.values)
for i in sorted(column):
actualColumns.append(allColumns[i])
return actualColumns
#X = removeColumns(my_data, 0, 1)
#y, targetNames = targetAndtargetNames(my_data, 1)
featureNames = getFeatureNames(my_data, 2,3,4,5)
print( y )
print (targetNames )
print (featureNames)
from sklearn.tree import DecisionTreeClassifier
skullsTree = DecisionTreeClassifier(criterion="entropy")
skullsTree.fit(X_trainset,y_trainset)
predTree = skullsTree.predict(X_testset)
print (predTree)
print (y_testset)
from sklearn import metrics
print("DecisionTrees's Accuracy: "), metrics.accuracy_score(y_testset, predTree)
!pip install pydotplus
#!pip install pydot2
#!pip install pyparsing==2.2.0
!pip install pydot
!conda install sklearn
from IPython.display import Image
from sklearn.externals.six import StringIO
from sklearn import tree
import pydot
import pydotplus
import pandas as pd
dot_data = StringIO()
tree.export_graphviz(skullsTree, out_file=dot_data,
feature_names=featureNames,
class_names=targetNames,
filled=True, rounded=True,
special_characters=True,
leaves_parallel=True)
graph = pydotplus.graph_from_dot_data(dot_data.getvalue())
Image(graph.create_png())
from sklearn.ensemble import RandomForestClassifier
skullsForest=RandomForestClassifier(n_estimators=10, criterion="entropy")
skullsForest.fit(X_trainset, y_trainset)
predForest = skullsForest.predict(X_testset)
print (predForest )
print (y_testset)
print("RandomForests's Accuracy: "), metrics.accuracy_score(y_testset, predForest)
print(skullsForest.estimators_)
from IPython.display import Image
from sklearn.externals.six import StringIO
import pydot
dot_data = StringIO()
#Replace the '&' below with the tree number
tree.export_graphviz(skullsForest[&], out_file=dot_data,
feature_names=featureNames,
class_names=targetNames,
filled=True, rounded=True,
special_characters=True,
leaves_parallel=True)
graph = pydotplus.graph_from_dot_data(dot_data.getvalue())
Image(graph.create_png())
| 0.708313 | 0.991546 |
# Single Layer Perceptron
```
import numpy as np
class Perceptron(object):
def __init__(self, input_size, lr = 1, epochs = 10):
self.W = np.zeros(input_size + 1)
self.epochs = epochs
self.lr = lr
def activation_fn(self, x):
return 1 if x >= 0 else 0
def predict(self, x):
z = self.W.T.dot(x)
a = self.activation_fn(z)
return a
def fit(self, X, d):
for _ in range(self.epochs):
for i in range(d.shape[0]):
x = np.insert(X[i], 0, 1)
y = self.predict(x)
e = d[i] - y
self.W = self.W + self.lr * e * x
def test(self, X, d):
p = np.zeros(d.shape[0])
print("Input\tExpected\tPredicted")
for i in range(d.shape[0]):
x = np.insert(X[i], 0, 1)
t = self.predict(x)
print(X[i], " ", d[i], "\t\t", t)
p[i] = t
return p
```
### AND Gate
```
# AND Gate
if __name__ == '__main__':
X = np.array([
[0, 0],
[0, 1],
[1, 0],
[1, 1]
])
d = np.array([0, 0, 0, 1])
print("AND Gate with Single Layer Perceptron")
perceptron = Perceptron(input_size = 2)
perceptron.fit(X, d)
print("Trained Weights: ", perceptron.W)
p = perceptron.test(X, d)
```
### OR Gate
```
# OR Gate
if __name__ == '__main__':
X = np.array([
[0, 0],
[0, 1],
[1, 0],
[1, 1]
])
d = np.array([0, 1, 1, 1])
print("OR Gate with Single Layer Perceptron")
perceptron = Perceptron(input_size = 2)
perceptron.fit(X, d)
print("Trained Weights: ", perceptron.W)
p = perceptron.test(X, d)
```
# Multi Level Perceptron
```
class mlp(object):
def __init__(self, input_dim, hidden_dim, output_dim, epochs = 10000, lr = 1.0):
self.W1 = np.random.random((input_dim, hidden_dim))
self.W2 = np.random.random((hidden_dim, output_dim))
self.epochs = epochs
self.lr = lr
def sigmoid(self, x):
return 1 / (1 + np.exp(-x))
def sigmoid_derivative(self, sx):
return sx * (1 - sx)
def cost(self, predicted, truth):
return truth - predicted
def fit(self, X, Y):
for epoch_n in range(self.epochs):
layer0 = X
# forward propagation
layer1 = self.sigmoid(np.dot(layer0, self.W1))
layer2 = self.sigmoid(np.dot(layer1, self.W2))
# back propagation (Y -> Layer2)
layer2_error = self.cost(layer2, Y)
layer2_delta = layer2_error * self.sigmoid_derivative(layer2)
# back propagation (Layer2 -> Layer1)
layer1_error = np.dot(layer2_delta, self.W2.T)
layer1_delta = layer1_error * self.sigmoid_derivative(layer1)
# update weights
self.W2 += self.lr * np.dot(layer1.T, layer2_delta)
self.W1 += self.lr * np.dot(layer0.T, layer1_delta)
def predict(self, X, Y):
p = []
q = []
for x, y in zip(X, Y):
layer1_prediction = self.sigmoid(np.dot(self.W1.T, x))
prediction = layer2_prediction = self.sigmoid(np.dot(self.W2.T, layer1_prediction))
print(prediction, "\t", int(prediction > 0.5), "\t\t\t", y)
p.append(prediction)
return p
def weights(self):
print("W1 = ", self.W1)
print("W2 = ", self.W2)
```
### XOR Gate
```
xor_input = np.array([
[0, 0],
[0, 1],
[1, 0],
[1, 1]
])
xor_output = np.array([[0, 1, 1, 0]]).T
X = xor_input
Y = xor_output
print("Multi Layer Perceptron for XOR Gate")
perceptron = mlp(input_dim = 2, hidden_dim = 5, output_dim = 1)
perceptron.fit(X, Y)
print("Fitted Weights: ")
perceptron.weights()
print("Model's Prediction")
print("Output Value\tRounded Up Output\tExpected Value")
p = perceptron.predict(X, Y)
```
### Using dataset
```
class mlp(object):
def __init__(self, input_dim, hidden_dim, output_dim, epochs = 10000, lr = 1.0):
self.W1 = np.random.random((input_dim, hidden_dim))
self.W2 = np.random.random((hidden_dim, output_dim))
self.epochs = epochs
self.lr = lr
def sigmoid(self, x):
return 1 / (1 + np.exp(-x))
def sigmoid_derivative(self, sx):
return sx * (1 - sx)
def cost(self, predicted, truth):
for i in range(len(predicted)):
return 0 if truth[i] == predicted[i] else 1
def fit(self, X, Y):
for epoch_n in range(self.epochs):
layer0 = X
# forward propagation
layer1 = self.sigmoid(np.dot(layer0, self.W1))
layer2 = self.sigmoid(np.dot(layer1, self.W2))
# back propagation (Y -> Layer2)
layer2_error = self.cost(layer2, Y)
layer2_delta = layer2_error * self.sigmoid_derivative(layer2)
# back propagation (Layer2 -> Layer1)
layer1_error = np.dot(layer2_delta, self.W2.T)
layer1_delta = layer1_error * self.sigmoid_derivative(layer1)
# update weights
self.W2 += self.lr * np.dot(layer1.T, layer2_delta)
self.W1 += self.lr * np.dot(layer0.T, layer1_delta)
def predict(self, X, Y):
p = []
q = []
for x, y in zip(X, Y):
layer1_prediction = self.sigmoid(np.dot(self.W1.T, x))
prediction = layer2_prediction = self.sigmoid(np.dot(self.W2.T, layer1_prediction))
print(prediction, "\t", int(prediction > 0.5), "\t\t\t", y)
p.append(prediction)
return p
def weights(self):
print("W1 = ", self.W1)
print("W2 = ", self.W2)
%cd "/home/mona/3074 ML Lab/Datasets"
import pandas as pd
# read data
data = pd.read_csv('iris_data.csv')
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
x_train, x_test, y_train, y_test = train_test_split(np.array(data)[:, :-1], np.array(data)[:, -1], test_size = 0.20)
scaler = StandardScaler()
scaler.fit(x_train)
x_train = scaler.transform(x_train)
x_test = scaler.transform(x_test)
print("Multi Layer Perceptron for Iris dataset")
perceptron = mlp(input_dim = 4, hidden_dim = 5, output_dim = 1)
perceptron.fit(x_train, y_train)
print("Fitted Weights: ")
perceptron.weights()
print("Model's Prediction")
print("Output Value\tRounded Up Output\tExpected Value")
p = perceptron.predict(x_test, y_test)
```
# Spot
Train a single layer perceptronto classify whether a fruit presented to the perceptron is going to be liked by a certain person or not, based on three features attributed to the presented fruit: its taste (whether it is sweet or not), its seeds (whether they are edible or not) and its skin (whether it is edible or not). Consider the following table for the inputs and the target output of the perceptron. Since there are three (binary) input values (taste, seeds and skin) and one (binary) target output, construct a single-layer perceptronwith three inputs and one output.
```
import numpy as np
class Perceptron(object):
def __init__(self, input_size, lr = 1, epochs = 20):
self.W = np.zeros(input_size + 1)
self.epochs = epochs
self.lr = lr
def activation_fn(self, x):
return 1 if x >= 0 else 0
def predict(self, x):
z = self.W.T.dot(x)
a = self.activation_fn(z)
return a
def fit(self, X, d):
for _ in range(self.epochs):
for i in range(d.shape[0]):
x = np.insert(X[i], 0, 1)
y = self.predict(x)
e = d[i] - y
self.W = self.W + self.lr * e * x
def test(self, X, d):
p = np.zeros(d.shape[0])
print("Input\tExpected\tPredicted")
for i in range(d.shape[0]):
x = np.insert(X[i], 0, 1)
t = self.predict(x)
print(X[i], " ", d[i], "\t\t", t)
p[i] = t
return p
%cd "/home/mona/3074 ML Lab/Datasets"
import pandas as pd
# read data
data = pd.read_csv('fruits.csv')
data
# classifying if fruits would be liked
if __name__ == '__main__':
X = np.array(data.iloc[:, 1:4])
d = np.array(data.iloc[:, -1])
print("Classifying Fruit Preferences with Single Layer Perceptron")
perceptron = Perceptron(input_size = 3)
perceptron.fit(X, d)
print("Trained Weights: ", perceptron.W)
p = perceptron.test(X, d)
```
|
github_jupyter
|
import numpy as np
class Perceptron(object):
def __init__(self, input_size, lr = 1, epochs = 10):
self.W = np.zeros(input_size + 1)
self.epochs = epochs
self.lr = lr
def activation_fn(self, x):
return 1 if x >= 0 else 0
def predict(self, x):
z = self.W.T.dot(x)
a = self.activation_fn(z)
return a
def fit(self, X, d):
for _ in range(self.epochs):
for i in range(d.shape[0]):
x = np.insert(X[i], 0, 1)
y = self.predict(x)
e = d[i] - y
self.W = self.W + self.lr * e * x
def test(self, X, d):
p = np.zeros(d.shape[0])
print("Input\tExpected\tPredicted")
for i in range(d.shape[0]):
x = np.insert(X[i], 0, 1)
t = self.predict(x)
print(X[i], " ", d[i], "\t\t", t)
p[i] = t
return p
# AND Gate
if __name__ == '__main__':
X = np.array([
[0, 0],
[0, 1],
[1, 0],
[1, 1]
])
d = np.array([0, 0, 0, 1])
print("AND Gate with Single Layer Perceptron")
perceptron = Perceptron(input_size = 2)
perceptron.fit(X, d)
print("Trained Weights: ", perceptron.W)
p = perceptron.test(X, d)
# OR Gate
if __name__ == '__main__':
X = np.array([
[0, 0],
[0, 1],
[1, 0],
[1, 1]
])
d = np.array([0, 1, 1, 1])
print("OR Gate with Single Layer Perceptron")
perceptron = Perceptron(input_size = 2)
perceptron.fit(X, d)
print("Trained Weights: ", perceptron.W)
p = perceptron.test(X, d)
class mlp(object):
def __init__(self, input_dim, hidden_dim, output_dim, epochs = 10000, lr = 1.0):
self.W1 = np.random.random((input_dim, hidden_dim))
self.W2 = np.random.random((hidden_dim, output_dim))
self.epochs = epochs
self.lr = lr
def sigmoid(self, x):
return 1 / (1 + np.exp(-x))
def sigmoid_derivative(self, sx):
return sx * (1 - sx)
def cost(self, predicted, truth):
return truth - predicted
def fit(self, X, Y):
for epoch_n in range(self.epochs):
layer0 = X
# forward propagation
layer1 = self.sigmoid(np.dot(layer0, self.W1))
layer2 = self.sigmoid(np.dot(layer1, self.W2))
# back propagation (Y -> Layer2)
layer2_error = self.cost(layer2, Y)
layer2_delta = layer2_error * self.sigmoid_derivative(layer2)
# back propagation (Layer2 -> Layer1)
layer1_error = np.dot(layer2_delta, self.W2.T)
layer1_delta = layer1_error * self.sigmoid_derivative(layer1)
# update weights
self.W2 += self.lr * np.dot(layer1.T, layer2_delta)
self.W1 += self.lr * np.dot(layer0.T, layer1_delta)
def predict(self, X, Y):
p = []
q = []
for x, y in zip(X, Y):
layer1_prediction = self.sigmoid(np.dot(self.W1.T, x))
prediction = layer2_prediction = self.sigmoid(np.dot(self.W2.T, layer1_prediction))
print(prediction, "\t", int(prediction > 0.5), "\t\t\t", y)
p.append(prediction)
return p
def weights(self):
print("W1 = ", self.W1)
print("W2 = ", self.W2)
xor_input = np.array([
[0, 0],
[0, 1],
[1, 0],
[1, 1]
])
xor_output = np.array([[0, 1, 1, 0]]).T
X = xor_input
Y = xor_output
print("Multi Layer Perceptron for XOR Gate")
perceptron = mlp(input_dim = 2, hidden_dim = 5, output_dim = 1)
perceptron.fit(X, Y)
print("Fitted Weights: ")
perceptron.weights()
print("Model's Prediction")
print("Output Value\tRounded Up Output\tExpected Value")
p = perceptron.predict(X, Y)
class mlp(object):
def __init__(self, input_dim, hidden_dim, output_dim, epochs = 10000, lr = 1.0):
self.W1 = np.random.random((input_dim, hidden_dim))
self.W2 = np.random.random((hidden_dim, output_dim))
self.epochs = epochs
self.lr = lr
def sigmoid(self, x):
return 1 / (1 + np.exp(-x))
def sigmoid_derivative(self, sx):
return sx * (1 - sx)
def cost(self, predicted, truth):
for i in range(len(predicted)):
return 0 if truth[i] == predicted[i] else 1
def fit(self, X, Y):
for epoch_n in range(self.epochs):
layer0 = X
# forward propagation
layer1 = self.sigmoid(np.dot(layer0, self.W1))
layer2 = self.sigmoid(np.dot(layer1, self.W2))
# back propagation (Y -> Layer2)
layer2_error = self.cost(layer2, Y)
layer2_delta = layer2_error * self.sigmoid_derivative(layer2)
# back propagation (Layer2 -> Layer1)
layer1_error = np.dot(layer2_delta, self.W2.T)
layer1_delta = layer1_error * self.sigmoid_derivative(layer1)
# update weights
self.W2 += self.lr * np.dot(layer1.T, layer2_delta)
self.W1 += self.lr * np.dot(layer0.T, layer1_delta)
def predict(self, X, Y):
p = []
q = []
for x, y in zip(X, Y):
layer1_prediction = self.sigmoid(np.dot(self.W1.T, x))
prediction = layer2_prediction = self.sigmoid(np.dot(self.W2.T, layer1_prediction))
print(prediction, "\t", int(prediction > 0.5), "\t\t\t", y)
p.append(prediction)
return p
def weights(self):
print("W1 = ", self.W1)
print("W2 = ", self.W2)
%cd "/home/mona/3074 ML Lab/Datasets"
import pandas as pd
# read data
data = pd.read_csv('iris_data.csv')
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
x_train, x_test, y_train, y_test = train_test_split(np.array(data)[:, :-1], np.array(data)[:, -1], test_size = 0.20)
scaler = StandardScaler()
scaler.fit(x_train)
x_train = scaler.transform(x_train)
x_test = scaler.transform(x_test)
print("Multi Layer Perceptron for Iris dataset")
perceptron = mlp(input_dim = 4, hidden_dim = 5, output_dim = 1)
perceptron.fit(x_train, y_train)
print("Fitted Weights: ")
perceptron.weights()
print("Model's Prediction")
print("Output Value\tRounded Up Output\tExpected Value")
p = perceptron.predict(x_test, y_test)
import numpy as np
class Perceptron(object):
def __init__(self, input_size, lr = 1, epochs = 20):
self.W = np.zeros(input_size + 1)
self.epochs = epochs
self.lr = lr
def activation_fn(self, x):
return 1 if x >= 0 else 0
def predict(self, x):
z = self.W.T.dot(x)
a = self.activation_fn(z)
return a
def fit(self, X, d):
for _ in range(self.epochs):
for i in range(d.shape[0]):
x = np.insert(X[i], 0, 1)
y = self.predict(x)
e = d[i] - y
self.W = self.W + self.lr * e * x
def test(self, X, d):
p = np.zeros(d.shape[0])
print("Input\tExpected\tPredicted")
for i in range(d.shape[0]):
x = np.insert(X[i], 0, 1)
t = self.predict(x)
print(X[i], " ", d[i], "\t\t", t)
p[i] = t
return p
%cd "/home/mona/3074 ML Lab/Datasets"
import pandas as pd
# read data
data = pd.read_csv('fruits.csv')
data
# classifying if fruits would be liked
if __name__ == '__main__':
X = np.array(data.iloc[:, 1:4])
d = np.array(data.iloc[:, -1])
print("Classifying Fruit Preferences with Single Layer Perceptron")
perceptron = Perceptron(input_size = 3)
perceptron.fit(X, d)
print("Trained Weights: ", perceptron.W)
p = perceptron.test(X, d)
| 0.70416 | 0.913754 |
## Evaluating HPO Space of SVD algorithm
This notebook contains evaluation of RMSE of SVD models at Movielens datasets
using different numbers of factors and regularization constants.
Initial setup: imports and working dir
```
import os
while not os.path.exists('.gitmodules'):
os.chdir('..')
from typing import Dict
import matplotlib.pyplot as plt
import pandas as pd
from parameters import get_env_parameters
from util.hpo_space_eval_utils import eval_svd_hpo_space, visualize_hpo_space
from util.datasets import MOVIELENS_100K, MOVIELENS_1M, MOVIELENS_10M
from util.docker.eigen3_svd import Eigen3SVDService
```
Start the docker container for native SVD/SVD++ models.
*If either the container or needed image does not exist, they will be created programmatically.*
This behavior is controlled by service constructor flags.
```
env_params = get_env_parameters()
eigen3_svd_service = Eigen3SVDService(env_params=env_params,
do_init_container=True,
allow_auto_build_image=True,
allow_auto_create_container=True,
allow_auto_run_container=True)
```
Define experiment settings: hyperparameter values to evaluate for needed datasets.
```
eval_params_map = {
MOVIELENS_100K.id: {
'n_factors_list': [(i + 1) * 10 for i in range(10)],
'reg_weight_list': [round(i * 0.01, 2) for i in range(10)]
},
MOVIELENS_1M.id: {
'n_factors_list': [(i + 1) * 10 for i in range(10)],
'reg_weight_list': [round(i * 0.01, 2) for i in range(10)]
},
MOVIELENS_10M.id: {
'n_factors_list': [(i + 1) * 10 for i in range(10)],
'reg_weight_list': [round(i * 0.01, 2) for i in range(10)]
}
}
```
Define map with existing experiment results.
This allows taking previous RMSE evaluations from disk without running model pipeline.
```
existing_results_files_map = {
MOVIELENS_100K.id: [
'ml100k-svd-f10-100-regw0.0-0.09.csv'
],
MOVIELENS_1M.id: [
'ml1m-svd-f10-100-regw0.0-0.09.csv'
],
MOVIELENS_10M.id: [
'ml10m-svd-f10-100-regw0.0-0.09.csv'
]
}
```
Run the experiments.
Function ```eval_svd_hpo_space``` returns ```pd.DataFrame``` and saves CSV results file on disk.
```
results_map: Dict[str, pd.DataFrame] = dict()
for dd in [MOVIELENS_100K, MOVIELENS_1M, MOVIELENS_10M]:
results_map[dd.id] = eval_svd_hpo_space(
eval_params_map=eval_params_map,
existing_results_files_map=existing_results_files_map,
dataset_descriptor=dd,
eigen3_svd_service=eigen3_svd_service
)
```
Visualize model loss on given hyperparameter configuration space.
```
fig, ax = plt.subplots(nrows=3, ncols=1, figsize=(5, 5), facecolor='white')
for i, dd in enumerate([MOVIELENS_100K, MOVIELENS_1M, MOVIELENS_10M]):
visualize_hpo_space(results_map[dd.id], fig, ax[i])
ax[i].set_title(dd.name)
fig.tight_layout()
```
|
github_jupyter
|
import os
while not os.path.exists('.gitmodules'):
os.chdir('..')
from typing import Dict
import matplotlib.pyplot as plt
import pandas as pd
from parameters import get_env_parameters
from util.hpo_space_eval_utils import eval_svd_hpo_space, visualize_hpo_space
from util.datasets import MOVIELENS_100K, MOVIELENS_1M, MOVIELENS_10M
from util.docker.eigen3_svd import Eigen3SVDService
env_params = get_env_parameters()
eigen3_svd_service = Eigen3SVDService(env_params=env_params,
do_init_container=True,
allow_auto_build_image=True,
allow_auto_create_container=True,
allow_auto_run_container=True)
eval_params_map = {
MOVIELENS_100K.id: {
'n_factors_list': [(i + 1) * 10 for i in range(10)],
'reg_weight_list': [round(i * 0.01, 2) for i in range(10)]
},
MOVIELENS_1M.id: {
'n_factors_list': [(i + 1) * 10 for i in range(10)],
'reg_weight_list': [round(i * 0.01, 2) for i in range(10)]
},
MOVIELENS_10M.id: {
'n_factors_list': [(i + 1) * 10 for i in range(10)],
'reg_weight_list': [round(i * 0.01, 2) for i in range(10)]
}
}
existing_results_files_map = {
MOVIELENS_100K.id: [
'ml100k-svd-f10-100-regw0.0-0.09.csv'
],
MOVIELENS_1M.id: [
'ml1m-svd-f10-100-regw0.0-0.09.csv'
],
MOVIELENS_10M.id: [
'ml10m-svd-f10-100-regw0.0-0.09.csv'
]
}
results_map: Dict[str, pd.DataFrame] = dict()
for dd in [MOVIELENS_100K, MOVIELENS_1M, MOVIELENS_10M]:
results_map[dd.id] = eval_svd_hpo_space(
eval_params_map=eval_params_map,
existing_results_files_map=existing_results_files_map,
dataset_descriptor=dd,
eigen3_svd_service=eigen3_svd_service
)
fig, ax = plt.subplots(nrows=3, ncols=1, figsize=(5, 5), facecolor='white')
for i, dd in enumerate([MOVIELENS_100K, MOVIELENS_1M, MOVIELENS_10M]):
visualize_hpo_space(results_map[dd.id], fig, ax[i])
ax[i].set_title(dd.name)
fig.tight_layout()
| 0.5144 | 0.832134 |
```
%matplotlib inline
```
ImageContainer object
=====================
This tutorial shows how to use `squidpy.im.ImageContainer` to interact
with image structured data.
The ImageContainer is the central object in Squidpy containing the high
resolution images. It wraps `xarray.Dataset` and provides different
cropping, processing, and feature extraction functions.
::: {.seealso}
For more details on specific `squidpy.im.ImageContainer` functions, have
a look at the following examples:
> - [Interactive visualization with
> Napari](../external_tutorials/tutorial_napari.ipynb).
> - `sphx_glr_auto_tutorials_tutorial_image_container_zstacks.py`.
> - `sphx_glr_auto_examples_image_compute_crops.py`.
> - `sphx_glr_auto_examples_image_compute_show.py`.
:::
```
import squidpy as sq
import numpy as np
```
Initialize ImageContainer
=========================
The `squidpy.im.ImageContainer` constructor can read in memory
`numpy.ndarray`/`xarray.DataArray` or on-disk image files. The
[ImageContainer]{.title-ref} can store multiple image layers (for
example an image and a matching segmentation mask).
Images are expected to have at least a [x]{.title-ref} and
[y]{.title-ref} dimension, with optional [channel]{.title-ref} and
[z]{.title-ref} dimensions. Here, we will focus on 2D images without at
[z]{.title-ref} dimension, see
`sphx_glr_auto_tutorials_tutorial_image_container_zstacks.py` for a
tutorial on how to use z-stacks with [ImageContainer]{.title-ref}.
Most important arguments upon initialization are:
> - [img]{.title-ref} - the image.
> - [layer]{.title-ref} - the name of the image layer.
> - [dims]{.title-ref} - to specify the dimensions names of
> [img]{.title-ref}.
> - [lazy]{.title-ref} - set to True to allow lazy computations.
> - [scale]{.title-ref} - set this to the scaling factor between the
> image and the coordinates saved.
Let us see these arguments in action with a toy example.
```
arr = np.ones((100, 100, 3))
arr[40:60, 40:60] = [0, 0.7, 1]
print(arr.shape)
img = sq.im.ImageContainer(arr, layer="img1")
img
```
[img]{.title-ref} now contains one layer, [img1]{.title-ref}. The
default value of [dims]{.title-ref} expects the image to have dimensions
`y, x, channels` or `y, x, z, channels`. If the image has different
dimensions, you can specify another strategy or a tuple of dimension
names:
```
arr1 = arr.transpose(2, 0, 1)
print(arr1.shape)
img = sq.im.ImageContainer(arr1, dims=("channels", "y", "x"), layer="img1")
img
```
Add layers to ImageContainer
============================
You can add image layers into the ImageContainer using
`squidpy.im.ImageContainer.add_img`.
The new layer has to share [x]{.title-ref}, [y]{.title-ref} (and
[z]{.title-ref}) dimensions with the already existing image. It can have
different channel dimensions. This is useful for add e.g., segmentation
masks.
By default, unique layer and channel dimension names are chosen, you can
specify them using the [layer]{.title-ref} and [dims]{.title-ref}
arguments.
```
arr_seg = np.zeros((100, 100))
arr_seg[40:60, 40:60] = 1
img.add_img(arr_seg, layer="seg1")
img
```
For convenience, you can also assign image layers directly using the new
layer name:
```
img["seg2"] = arr_seg
img
```
You can get a list of layers contained in an ImageContainer, and access
specific image-structured arrays using their names:
```
print(list(img))
img["img1"]
```
Renaming of image layers is also possible using
`squidpy.im.ImageContainer.rename`:
```
img.rename("seg2", "new-name")
```
Visualization
=============
Use `squidpy.im.ImageContainer.show` to visualize (small) images
statically. See `sphx_glr_auto_examples_image_compute_show.py` for more
details.
For large images and for interactive visualization of
`squidpy.im.ImageContainer` together with spatial \'omics data, we
recommend using `squidpy.im.ImageContainer.interactive`, which uses
Napari. See [Interactive visualization with
Napari](../external_tutorials/tutorial_napari.ipynb) for more details.
```
img.show(layer="img1")
```
Crop and scale images
=====================
Images can be cropped and scaled using
`squidpy.im.ImageContainer.crop_corner` and
`squidpy.im.ImageContainer.crop_center`. See
`sphx_glr_auto_examples_image_compute_crops.py` for more details.
```
crop1 = img.crop_corner(30, 40, size=(30, 30), scale=1)
crop1.show(layer="img1")
crop2 = crop1.crop_corner(0, 0, size=(40, 40), scale=0.5)
crop2.show(layer="img1")
```
Internally, the [ImageContainer]{.title-ref} keeps track of the crop
coordinates in the dataset attributes. This enables mapping from cropped
[ImageContainers]{.title-ref} to observations in [adata]{.title-ref} for
interactive visualization and feature extraction.
Using `squidpy.im.ImageContainer.uncrop`, we can reconstruct the
original image. Even when chaining multiple calls to `crop`, `uncrop`
correctly places the crop in the image. Note that `uncrop` only undoes
the cropping, not the scaling.
```
print(crop1.data.attrs)
print(crop2.data.attrs)
sq.im.ImageContainer.uncrop([crop1], shape=img.shape).show(layer="img1")
sq.im.ImageContainer.uncrop([crop2], shape=(50, 50)).show(layer="img1")
```
After cropping the ImageContainer, you can subset the associated
[adata]{.title-ref} to the cropped image using
`squidpy.im.ImageContainer.subset`. See
`sphx_glr_auto_examples_image_compute_crops.py` for an example.
Processing images and extracting features
=========================================
The main purpose of ImageContainer is to allow efficient image
processing, segmentation and features extraction.
For details on each of these steps, have a look a the following examples
using the high-level API:
> - `sphx_glr_auto_examples_image_compute_process_hires.py` for
> `sq.im.process`.
> - `sphx_glr_auto_examples_image_compute_segment_fluo.py` for
> `sq.im.segment`.
> - `sphx_glr_auto_examples_image_compute_features.py` for
> `sq.im.extract_features`.
These functions are build to be general and flexible. All of them allow
you to pass custom processing and feature extraction functions for easy
use of external packages with Squidpy.
For even more control, you can also use low-level functions provided by
\`ImageContainer\`:
> - `sq.im.ImageContainer.apply` for custom processing functions that
> should be applied to a specific image layer.
> - `sq.im.ImageContainner.feature_custom` for extracting features.
There are two generators, that allow you to iterate over a sequence of
image crops and apply processing functions to smaller crops (e.g. to
allow parallelization or processing images that won\'t fit in memory:
> - `sq.im.ImageContainer.generate_equal_crops`, for evenly
> decomposing the image into equally sized crops.
> - `sq.im.ImageContainer.generate_spot_crops`, for extracting image
> crops for each observation in the associated [adata]{.title-ref}.
Internal representation of images
=================================
Internally, the images are represented in a `xarray.Dataset`. You can
access this dataset using `img.data`.
```
img.data
```
Whenever possible, images are represented as lazy `dask` arrays. This
allows lazy computations, which only load and compute the data when it
is required.
Let us load an on-disk image that is provided by the `squidpy.datasets`
module: By default, the [lazy]{.title-ref} argument is
[True]{.title-ref}, therefore resulting in a `dask.array.Array`.
```
img_on_disk = sq.datasets.visium_hne_image()
print(type(img_on_disk["image"].data))
```
We can use `squidpy.im.ImageContainer.compute` to force loading of the
data:
```
img_on_disk.compute()
print(type(img_on_disk["image"].data))
```
ImageContainers can be saved and loaded from a *Zarr* store, using
`squidpy.im.ImageContainer.save` and `squidpy.im.ImageContainer.load`.
|
github_jupyter
|
%matplotlib inline
import squidpy as sq
import numpy as np
arr = np.ones((100, 100, 3))
arr[40:60, 40:60] = [0, 0.7, 1]
print(arr.shape)
img = sq.im.ImageContainer(arr, layer="img1")
img
arr1 = arr.transpose(2, 0, 1)
print(arr1.shape)
img = sq.im.ImageContainer(arr1, dims=("channels", "y", "x"), layer="img1")
img
arr_seg = np.zeros((100, 100))
arr_seg[40:60, 40:60] = 1
img.add_img(arr_seg, layer="seg1")
img
img["seg2"] = arr_seg
img
print(list(img))
img["img1"]
img.rename("seg2", "new-name")
img.show(layer="img1")
crop1 = img.crop_corner(30, 40, size=(30, 30), scale=1)
crop1.show(layer="img1")
crop2 = crop1.crop_corner(0, 0, size=(40, 40), scale=0.5)
crop2.show(layer="img1")
print(crop1.data.attrs)
print(crop2.data.attrs)
sq.im.ImageContainer.uncrop([crop1], shape=img.shape).show(layer="img1")
sq.im.ImageContainer.uncrop([crop2], shape=(50, 50)).show(layer="img1")
img.data
img_on_disk = sq.datasets.visium_hne_image()
print(type(img_on_disk["image"].data))
img_on_disk.compute()
print(type(img_on_disk["image"].data))
| 0.272508 | 0.990642 |
## 1. Import Libraries
```
import psycopg2
import pandas as pd
import numpy as np
import xgboost as xgb
import tensorflow as tf
from functools import reduce
from sklearn import metrics
from sklearn.model_selection import train_test_split
from sklearn.pipeline import Pipeline
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import LogisticRegression
from sklearn.neural_network import MLPClassifier
from sklearn.metrics import confusion_matrix, ConfusionMatrixDisplay
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.layers.experimental import preprocessing
from keras.models import Sequential
from matplotlib import pyplot as plt
```
## 2. Connect to DB
```
# information used to create a database connection
sqluser = 'postgres'
dbname = 'mimic4'
hostname = 'localhost'
port_number = 5434
schema_name = 'mimiciv'
# Connect to postgres with a copy of the MIMIC-III database
con = psycopg2.connect(dbname=dbname, user=sqluser, host=hostname, port=port_number, password='mysecretpassword')
# the below statement is prepended to queries to ensure they select from the right schema
query_schema = 'set search_path to ' + schema_name + ';'
```
## 3. Extract Features
### 3.1 Static Features
```
query = query_schema + \
"""
WITH ht AS
(
SELECT
c.subject_id, c.stay_id, c.charttime,
-- Ensure that all heights are in centimeters, and fix data as needed
CASE
-- rule for neonates
WHEN pt.anchor_age = 0
AND (c.valuenum * 2.54) < 80
THEN c.valuenum * 2.54
-- rule for adults
WHEN pt.anchor_age > 0
AND (c.valuenum * 2.54) > 120
AND (c.valuenum * 2.54) < 230
THEN c.valuenum * 2.54
-- set bad data to NULL
ELSE NULL
END AS height
, ROW_NUMBER() OVER (PARTITION BY stay_id ORDER BY charttime DESC) AS rn
FROM mimiciv.chartevents c
INNER JOIN mimiciv.patients pt
ON c.subject_id = pt.subject_id
WHERE c.valuenum IS NOT NULL
AND c.valuenum != 0
AND c.itemid IN
(
226707 -- Height (measured in inches)
-- note we intentionally ignore the below ITEMID in metavision
-- these are duplicate data in a different unit
-- , 226730 -- Height (cm)
)
)
, wt AS
(
SELECT
c.stay_id
, c.charttime
-- TODO: eliminate obvious outliers if there is a reasonable weight
, c.valuenum as weight
, ROW_NUMBER() OVER (PARTITION BY stay_id ORDER BY charttime DESC) AS rn
FROM mimiciv.chartevents c
WHERE c.valuenum IS NOT NULL
AND c.itemid = 226512 -- Admit Wt
AND c.stay_id IS NOT NULL
AND c.valuenum > 0
)
SELECT
ie.subject_id, ie.hadm_id, ie.stay_id
, CASE WHEN pat.gender = 'M' THEN '1' ELSE '0' END AS is_male
, FLOOR(DATE_PART('day', adm.admittime - make_timestamp(pat.anchor_year, 1, 1, 0, 0, 0))/365.0) + pat.anchor_age as age
, CASE WHEN adm.ethnicity LIKE '%WHITE%' THEN '1' ELSE '0' END AS race_white
, CASE WHEN adm.ethnicity LIKE '%BLACK%' THEN '1' ELSE '0' END AS race_black
, CASE WHEN adm.ethnicity LIKE '%HISPANIC%' THEN '1' ELSE '0' END AS race_hispanic
, CASE WHEN adm.ethnicity LIKE '%ASIAN%' THEN '1' ELSE '0' END AS race_asian
, CASE WHEN adm.ethnicity LIKE '%OTHER%' THEN '1' ELSE '0' END AS race_other
, CASE WHEN adm.admission_type LIKE '%EMER%' THEN '1' ELSE '0' END AS emergency_admission
, CASE
WHEN ht.height IS NOT null AND wt.weight IS NOT null
THEN (wt.weight / (ht.height/100*ht.height/100))
ELSE null
END AS bmi
, ht.height as height
, wt.weight as weight
, (
SELECT
CASE WHEN COUNT(*) = 0 THEN 0 ELSE 1 END
FROM mimiciv.transfers car_trs
WHERE car_trs.hadm_id = adm.hadm_id
AND lower(car_trs.careunit) LIKE '%card%'
AND lower(car_trs.careunit) LIKE '%surg%'
) AS service_any_card_surg
, (
SELECT
CASE WHEN COUNT(*) = 0 THEN 0 ELSE 1 END
FROM mimiciv.transfers car_trs
WHERE car_trs.hadm_id = adm.hadm_id
AND lower(car_trs.careunit) NOT LIKE '%card%'
AND lower(car_trs.careunit) LIKE '%surg%'
) AS service_any_noncard_surg
, (
SELECT
CASE WHEN COUNT(*) = 0 THEN 0 ELSE 1 END
FROM mimiciv.transfers car_trs
WHERE car_trs.hadm_id = adm.hadm_id
AND lower(car_trs.careunit) LIKE '%trauma%'
) AS service_trauma
-- , adm.hospital_expire_flag
FROM mimiciv.icustays ie
INNER JOIN mimiciv.admissions adm
ON ie.hadm_id = adm.hadm_id
INNER JOIN mimiciv.patients pat
ON ie.subject_id = pat.subject_id
LEFT JOIN ht
ON ie.stay_id = ht.stay_id AND ht.rn = 1
LEFT JOIN wt
ON ie.stay_id = wt.stay_id AND wt.rn = 1
"""
static = pd.read_sql_query(query, con)
static
```
### 3.2 First Lab Measurement Attributes
```
query = query_schema + \
"""
WITH labs_preceeding AS
(
SELECT icu.stay_id, l.valuenum, l.charttime
, CASE
WHEN itemid = 51006 THEN 'BUN'
WHEN itemid = 50806 THEN 'CHLORIDE'
WHEN itemid = 50902 THEN 'CHLORIDE'
WHEN itemid = 50912 THEN 'CREATININE'
WHEN itemid = 50811 THEN 'HEMOGLOBIN'
WHEN itemid = 51222 THEN 'HEMOGLOBIN'
WHEN itemid = 51265 THEN 'PLATELET'
WHEN itemid = 50822 THEN 'POTASSIUM'
WHEN itemid = 50971 THEN 'POTASSIUM'
WHEN itemid = 50824 THEN 'SODIUM'
WHEN itemid = 50983 THEN 'SODIUM'
WHEN itemid = 50803 THEN 'BICARBONATE'
WHEN itemid = 50882 THEN 'BICARBONATE'
WHEN itemid = 50804 THEN 'TOTALCO2'
WHEN itemid = 50821 THEN 'PO2'
WHEN itemid = 52042 THEN 'PO2'
WHEN itemid = 50832 THEN 'PO2'
WHEN itemid = 50818 THEN 'PCO2'
WHEN itemid = 52040 THEN 'PCO2'
WHEN itemid = 50830 THEN 'PCO2'
WHEN itemid = 50820 THEN 'PH'
WHEN itemid = 52041 THEN 'PH'
WHEN itemid = 50831 THEN 'PH'
WHEN itemid = 51300 THEN 'WBC'
WHEN itemid = 51301 THEN 'WBC'
WHEN itemid = 50802 THEN 'BASEEXCESS'
WHEN itemid = 52038 THEN 'BASEEXCESS'
WHEN itemid = 50805 THEN 'CARBOXYHEMOGLOBIN'
WHEN itemid = 50814 THEN 'METHEMOGLOBIN'
WHEN itemid = 50868 THEN 'ANIONGAP'
WHEN itemid = 52500 THEN 'ANIONGAP'
WHEN itemid = 50862 THEN 'ALBUMIN'
WHEN itemid = 51144 THEN 'BANDS'
WHEN itemid = 50885 THEN 'BILRUBIN'
WHEN itemid = 51478 THEN 'GLUCOSE'
WHEN itemid = 50931 THEN 'GLUCOSE'
WHEN itemid = 51221 THEN 'HEMATOCRIT'
WHEN itemid = 50813 THEN 'LACTATE'
WHEN itemid = 51275 THEN 'PTT'
WHEN itemid = 51237 THEN 'INR'
ELSE null
END AS LABEL
FROM mimiciv.icustays icu
INNER JOIN mimiciv.admissions adm
ON icu.hadm_id = adm.hadm_id
INNER JOIN mimiciv.patients pat
ON icu.subject_id = pat.subject_id
INNER JOIN mimiciv.labevents l
ON l.hadm_id = icu.hadm_id
AND l.charttime >= icu.intime - interval '8 hour'
AND l.charttime <= icu.intime + interval '24 hour'
WHERE l.itemid IN
(
51300,51301 -- wbc
, 50811,51222 -- hgb
, 51265 -- platelet
, 50824, 50983 -- sodium
, 50822, 50971 -- potassium
, 50804 -- Total CO2 or ...
, 50803, 50882 -- bicarbonate
, 50806, 50902 -- chloride
, 51006 -- bun
, 50912 -- creatinine
, 50821, 52042, 50832 -- po2
, 50818, 52040, 50830 -- pco2
, 50820, 52041, 50831 -- ph
, 50802, 52038 -- Base Excess
, 50805 -- carboxyhemoglobin
, 50814 -- methemoglobin
, 50868, 52500 -- aniongap
, 50862 -- albumin
, 51144 -- bands
, 50885 -- bilrubin
, 51478, 50931 -- glucose
, 51221 -- hematocrit
, 50813 -- lactate
, 51275 -- ptt
, 51237 -- inr
)
AND valuenum IS NOT null
)
, labs_rn AS
(
SELECT
stay_id, valuenum, label
, ROW_NUMBER() OVER (PARTITION BY stay_id, label ORDER BY charttime) AS rn
FROM labs_preceeding
)
, labs_grp AS
(
SELECT
stay_id
, COALESCE(MAX(CASE WHEN label = 'BUN' THEN valuenum ELSE null END)) AS BUN
, COALESCE(MAX(CASE WHEN label = 'CHLORIDE' THEN valuenum ELSE null END)) AS CHLORIDE
, COALESCE(MAX(CASE WHEN label = 'CREATININE' THEN valuenum ELSE null END)) AS CREATININE
, COALESCE(MAX(CASE WHEN label = 'HEMOGLOBIN' THEN valuenum ELSE null END)) AS HEMOGLOBIN
, COALESCE(MAX(CASE WHEN label = 'PLATELET' THEN valuenum ELSE null END)) AS PLATELET
, COALESCE(MAX(CASE WHEN label = 'POTASSIUM' THEN valuenum ELSE null END)) AS POTASSIUM
, COALESCE(MAX(CASE WHEN label = 'SODIUM' THEN valuenum ELSE null END)) AS SODIUM
, COALESCE(MAX(CASE WHEN label = 'TOTALCO2' THEN valuenum ELSE null END)) AS TOTALCO2
, COALESCE(MAX(CASE WHEN label = 'WBC' THEN valuenum ELSE null END)) AS WBC
, COALESCE(MAX(CASE WHEN label = 'PO2' THEN valuenum ELSE null END)) AS PO2
, COALESCE(MAX(CASE WHEN label = 'PCO2' THEN valuenum ELSE null END)) AS PCO2
, COALESCE(MAX(CASE WHEN label = 'PH' THEN valuenum ELSE null END)) AS PH
, COALESCE(MAX(CASE WHEN label = 'BASEEXCESS' THEN valuenum ELSE null END)) AS BASEEXCESS
, COALESCE(MAX(CASE WHEN label = 'CARBOXYHEMOGLOBIN' THEN valuenum ELSE null END)) AS CARBOXYHEMOGLOBIN
, COALESCE(MAX(CASE WHEN label = 'METHEMOGLOBIN' THEN valuenum ELSE null END)) AS METHEMOGLOBIN
, COALESCE(MAX(CASE WHEN label = 'ANIONGAP' THEN valuenum ELSE null END)) AS ANIONGAP
, COALESCE(MAX(CASE WHEN label = 'ALBUMIN' THEN valuenum ELSE null END)) AS ALBUMIN
, COALESCE(MAX(CASE WHEN label = 'BANDS' THEN valuenum ELSE null END)) AS BANDS
, COALESCE(MAX(CASE WHEN label = 'BICARBONATE' THEN valuenum ELSE null END)) AS BICARBONATE
, COALESCE(MAX(CASE WHEN label = 'BILRUBIN' THEN valuenum ELSE null END)) AS BILRUBIN
, COALESCE(MAX(CASE WHEN label = 'GLUCOSE' THEN valuenum ELSE null END)) AS GLUCOSE
, COALESCE(MAX(CASE WHEN label = 'HEMATOCRIT' THEN valuenum ELSE null END)) AS HEMATOCRIT
, COALESCE(MAX(CASE WHEN label = 'LACTATE' THEN valuenum ELSE null END)) AS LACTATE
, COALESCE(MAX(CASE WHEN label = 'PTT' THEN valuenum ELSE null END)) AS PTT
, COALESCE(MAX(CASE WHEN label = 'INR' THEN valuenum ELSE null END)) AS INR
FROM labs_rn
WHERE rn = 1
GROUP BY stay_id
)
SELECT icu.stay_id
, lg.bun AS bun_first
, lg.chloride AS chloride_first
, lg.creatinine AS creatinine_first
, lg.HEMOGLOBIN AS hgb_first
, lg.platelet AS platelet_first
, lg.potassium AS potassium_first
, lg.sodium AS sodium_first
, lg.TOTALCO2 AS tco2_first
, lg.wbc AS wbc_first
, lg.po2 AS bg_po2_first
, lg.pco2 AS bg_pco2_first
, lg.ph AS bg_ph_first
, lg.BASEEXCESS AS bg_baseexcess_first
, lg.CARBOXYHEMOGLOBIN AS bg_carboxyhemoglobin_first
, lg.METHEMOGLOBIN AS bg_methemomoglobin_first
, lg.ANIONGAP AS aniongap_first
, lg.ALBUMIN AS albumin_first
, lg.BANDS AS bands_first
, lg.BICARBONATE AS bicarbonate_first
, lg.BILRUBIN AS bilrubin_first
, lg.GLUCOSE AS glucose_first
, lg.HEMATOCRIT AS hematocrit_first
, lg.LACTATE AS lactate_first
, lg.PTT AS ptt_first
, lg.INR AS inr_first
FROM mimiciv.icustays icu
LEFT JOIN labs_grp lg
ON icu.stay_id = lg.stay_id
"""
first_lab = pd.read_sql_query(query, con)
first_lab
```
### 3.3 Last Lab Measurement Attributes
```
query = query_schema + \
"""
WITH labs_preceeding AS
(
SELECT icu.stay_id, l.valuenum, l.charttime
, CASE
WHEN itemid = 51006 THEN 'BUN'
WHEN itemid = 50806 THEN 'CHLORIDE'
WHEN itemid = 50902 THEN 'CHLORIDE'
WHEN itemid = 50912 THEN 'CREATININE'
WHEN itemid = 50811 THEN 'HEMOGLOBIN'
WHEN itemid = 51222 THEN 'HEMOGLOBIN'
WHEN itemid = 51265 THEN 'PLATELET'
WHEN itemid = 50822 THEN 'POTASSIUM'
WHEN itemid = 50971 THEN 'POTASSIUM'
WHEN itemid = 50824 THEN 'SODIUM'
WHEN itemid = 50983 THEN 'SODIUM'
WHEN itemid = 50803 THEN 'BICARBONATE'
WHEN itemid = 50882 THEN 'BICARBONATE'
WHEN itemid = 50804 THEN 'TOTALCO2'
WHEN itemid = 50821 THEN 'PO2'
WHEN itemid = 52042 THEN 'PO2'
WHEN itemid = 50832 THEN 'PO2'
WHEN itemid = 50818 THEN 'PCO2'
WHEN itemid = 52040 THEN 'PCO2'
WHEN itemid = 50830 THEN 'PCO2'
WHEN itemid = 50820 THEN 'PH'
WHEN itemid = 52041 THEN 'PH'
WHEN itemid = 50831 THEN 'PH'
WHEN itemid = 51300 THEN 'WBC'
WHEN itemid = 51301 THEN 'WBC'
WHEN itemid = 50802 THEN 'BASEEXCESS'
WHEN itemid = 52038 THEN 'BASEEXCESS'
WHEN itemid = 50805 THEN 'CARBOXYHEMOGLOBIN'
WHEN itemid = 50814 THEN 'METHEMOGLOBIN'
WHEN itemid = 50868 THEN 'ANIONGAP'
WHEN itemid = 52500 THEN 'ANIONGAP'
WHEN itemid = 50862 THEN 'ALBUMIN'
WHEN itemid = 51144 THEN 'BANDS'
WHEN itemid = 50885 THEN 'BILRUBIN'
WHEN itemid = 51478 THEN 'GLUCOSE'
WHEN itemid = 50931 THEN 'GLUCOSE'
WHEN itemid = 51221 THEN 'HEMATOCRIT'
WHEN itemid = 50813 THEN 'LACTATE'
WHEN itemid = 51275 THEN 'PTT'
WHEN itemid = 51237 THEN 'INR'
ELSE null
END AS LABEL
FROM mimiciv.icustays icu
INNER JOIN mimiciv.admissions adm
ON icu.hadm_id = adm.hadm_id
INNER JOIN mimiciv.patients pat
ON icu.subject_id = pat.subject_id
INNER JOIN mimiciv.labevents l
ON l.hadm_id = icu.hadm_id
AND l.charttime >= icu.intime - interval '8 hour'
AND l.charttime <= icu.intime + interval '24 hour'
WHERE l.itemid IN
(
51300,51301 -- wbc
, 50811,51222 -- hgb
, 51265 -- platelet
, 50824, 50983 -- sodium
, 50822, 50971 -- potassium
, 50804 -- Total CO2 or ...
, 50803, 50882 -- bicarbonate
, 50806, 50902 -- chloride
, 51006 -- bun
, 50912 -- creatinine
, 50821, 52042, 50832 -- po2
, 50818, 52040, 50830 -- pco2
, 50820, 52041, 50831 -- ph
, 50802, 52038 -- Base Excess
, 50805 -- carboxyhemoglobin
, 50814 -- methemoglobin
, 50868, 52500 -- aniongap
, 50862 -- albumin
, 51144 -- bands
, 50885 -- bilrubin
, 51478, 50931 -- glucose
, 51221 -- hematocrit
, 50813 -- lactate
, 51275 -- ptt
, 51237 -- inr
)
AND valuenum IS NOT null
)
, labs_rn AS
(
SELECT
stay_id, valuenum, label
, ROW_NUMBER() OVER (PARTITION BY stay_id, label ORDER BY charttime DESC) AS rn
FROM labs_preceeding
)
, labs_grp AS
(
SELECT
stay_id
, COALESCE(MAX(CASE WHEN label = 'BUN' THEN valuenum ELSE null END)) AS BUN
, COALESCE(MAX(CASE WHEN label = 'CHLORIDE' THEN valuenum ELSE null END)) AS CHLORIDE
, COALESCE(MAX(CASE WHEN label = 'CREATININE' THEN valuenum ELSE null END)) AS CREATININE
, COALESCE(MAX(CASE WHEN label = 'HEMOGLOBIN' THEN valuenum ELSE null END)) AS HEMOGLOBIN
, COALESCE(MAX(CASE WHEN label = 'PLATELET' THEN valuenum ELSE null END)) AS PLATELET
, COALESCE(MAX(CASE WHEN label = 'POTASSIUM' THEN valuenum ELSE null END)) AS POTASSIUM
, COALESCE(MAX(CASE WHEN label = 'SODIUM' THEN valuenum ELSE null END)) AS SODIUM
, COALESCE(MAX(CASE WHEN label = 'TOTALCO2' THEN valuenum ELSE null END)) AS TOTALCO2
, COALESCE(MAX(CASE WHEN label = 'WBC' THEN valuenum ELSE null END)) AS WBC
, COALESCE(MAX(CASE WHEN label = 'PO2' THEN valuenum ELSE null END)) AS PO2
, COALESCE(MAX(CASE WHEN label = 'PCO2' THEN valuenum ELSE null END)) AS PCO2
, COALESCE(MAX(CASE WHEN label = 'PH' THEN valuenum ELSE null END)) AS PH
, COALESCE(MAX(CASE WHEN label = 'BASEEXCESS' THEN valuenum ELSE null END)) AS BASEEXCESS
, COALESCE(MAX(CASE WHEN label = 'CARBOXYHEMOGLOBIN' THEN valuenum ELSE null END)) AS CARBOXYHEMOGLOBIN
, COALESCE(MAX(CASE WHEN label = 'METHEMOGLOBIN' THEN valuenum ELSE null END)) AS METHEMOGLOBIN
, COALESCE(MAX(CASE WHEN label = 'ANIONGAP' THEN valuenum ELSE null END)) AS ANIONGAP
, COALESCE(MAX(CASE WHEN label = 'ALBUMIN' THEN valuenum ELSE null END)) AS ALBUMIN
, COALESCE(MAX(CASE WHEN label = 'BANDS' THEN valuenum ELSE null END)) AS BANDS
, COALESCE(MAX(CASE WHEN label = 'BICARBONATE' THEN valuenum ELSE null END)) AS BICARBONATE
, COALESCE(MAX(CASE WHEN label = 'BILRUBIN' THEN valuenum ELSE null END)) AS BILRUBIN
, COALESCE(MAX(CASE WHEN label = 'GLUCOSE' THEN valuenum ELSE null END)) AS GLUCOSE
, COALESCE(MAX(CASE WHEN label = 'HEMATOCRIT' THEN valuenum ELSE null END)) AS HEMATOCRIT
, COALESCE(MAX(CASE WHEN label = 'LACTATE' THEN valuenum ELSE null END)) AS LACTATE
, COALESCE(MAX(CASE WHEN label = 'PTT' THEN valuenum ELSE null END)) AS PTT
, COALESCE(MAX(CASE WHEN label = 'INR' THEN valuenum ELSE null END)) AS INR
FROM labs_rn
WHERE rn = 1
GROUP BY stay_id
)
SELECT icu.stay_id
, lg.bun AS bun_last
, lg.chloride AS chloride_last
, lg.creatinine AS creatinine_last
, lg.HEMOGLOBIN AS hgb_last
, lg.platelet AS platelet_last
, lg.potassium AS potassium_last
, lg.sodium AS sodium_last
, lg.TOTALCO2 AS tco2_last
, lg.wbc AS wbc_last
, lg.po2 AS bg_po2_last
, lg.pco2 AS bg_pco2_last
, lg.ph AS bg_ph_last
, lg.BASEEXCESS AS bg_baseexcess_last
, lg.CARBOXYHEMOGLOBIN AS bg_carboxyhemoglobin_last
, lg.METHEMOGLOBIN AS bg_methemomoglobin_last
, lg.ANIONGAP AS aniongap_last
, lg.ALBUMIN AS albumin_last
, lg.BANDS AS bands_last
, lg.BICARBONATE AS bicarbonate_last
, lg.BILRUBIN AS bilrubin_last
, lg.GLUCOSE AS glucose_last
, lg.HEMATOCRIT AS hematocrit_last
, lg.LACTATE AS lactate_last
, lg.PTT AS ptt_last
, lg.INR AS inr_last
FROM mimiciv.icustays icu
LEFT JOIN labs_grp lg
ON icu.stay_id = lg.stay_id
"""
last_lab = pd.read_sql_query(query, con)
last_lab
```
### 3.4 First Vitals Measurement Attributes
```
query = query_schema + \
"""
WITH vitals_stg_1 AS
(
SELECT icu.stay_id, cev.charttime
, CASE
WHEN itemid = 223761 THEN (cev.valuenum-32)/1.8
ELSE cev.valuenum
END AS valuenum
, CASE
WHEN itemid = 220045 THEN 'HEARTRATE'
WHEN itemid = 220050 THEN 'SYSBP'
WHEN itemid = 220179 THEN 'SYSBP'
WHEN itemid = 220051 THEN 'DIASBP'
WHEN itemid = 220180 THEN 'DIASBP'
WHEN itemid = 220052 THEN 'MEANBP'
WHEN itemid = 220181 THEN 'MEANBP'
WHEN itemid = 225312 THEN 'MEANBP'
WHEN itemid = 220210 THEN 'RESPRATE'
WHEN itemid = 224688 THEN 'RESPRATE'
WHEN itemid = 224689 THEN 'RESPRATE'
WHEN itemid = 224690 THEN 'RESPRATE'
WHEN itemid = 223761 THEN 'TEMPC'
WHEN itemid = 223762 THEN 'TEMPC'
WHEN itemid = 220277 THEN 'SPO2'
WHEN itemid = 220739 THEN 'GCSEYE'
WHEN itemid = 223900 THEN 'GCSVERBAL'
WHEN itemid = 223901 THEN 'GCSMOTOR'
ELSE null
END AS label
FROM mimiciv.icustays icu
INNER JOIN mimiciv.chartevents cev
ON cev.stay_id = icu.stay_id
AND cev.charttime >= icu.intime
AND cev.charttime <= icu.intime + interval '24 hour'
WHERE cev.itemid IN
(
220045 -- heartrate
, 220050, 220179 -- sysbp
, 220051, 220180 -- diasbp
, 220052, 220181, 225312 -- meanbp
, 220210, 224688, 224689, 224690 -- resprate
, 223761, 223762 -- tempc
, 220277 -- SpO2
, 220739 -- gcseye
, 223900 -- gcsverbal
, 223901 -- gscmotor
)
AND valuenum IS NOT null
)
, vitals_stg_2 AS
(
SELECT
stay_id, valuenum, label
, ROW_NUMBER() OVER (PARTITION BY stay_id, label ORDER BY charttime) AS rn
FROM vitals_stg_1
)
, vitals_stg_3 AS
(
SELECT
stay_id
, COALESCE(MAX(CASE WHEN label = 'HEARTRATE' THEN valuenum ELSE null END)) AS heartrate_first
, COALESCE(MAX(CASE WHEN label = 'SYSBP' THEN valuenum ELSE null END)) AS sysbp_first
, COALESCE(MAX(CASE WHEN label = 'DIASBP' THEN valuenum ELSE null END)) AS diabp_first
, COALESCE(MAX(CASE WHEN label = 'MEANBP' THEN valuenum ELSE null END)) AS meanbp_first
, COALESCE(MAX(CASE WHEN label = 'RESPRATE' THEN valuenum ELSE null END)) AS resprate_first
, COALESCE(MAX(CASE WHEN label = 'TEMPC' THEN valuenum ELSE null END)) AS tempc_first
, COALESCE(MAX(CASE WHEN label = 'SPO2' THEN valuenum ELSE null END)) AS spo2_first
, COALESCE(MAX(CASE WHEN label = 'GCSEYE' THEN valuenum ELSE null END)) AS gcseye_first
, COALESCE(MAX(CASE WHEN label = 'GCSVERBAL' THEN valuenum ELSE null END)) AS gcsverbal_first
, COALESCE(MAX(CASE WHEN label = 'GCSMOTOR' THEN valuenum ELSE null END)) AS gcsmotor_first
FROM vitals_stg_2
WHERE rn = 1
GROUP BY stay_id
)
SELECT * FROM vitals_stg_3
"""
first_vitals = pd.read_sql_query(query, con)
first_vitals
```
### 3.5 Last Vitals Measurement Attributes
```
query = query_schema + \
"""
WITH vitals_stg_1 AS
(
SELECT icu.stay_id, cev.charttime
, CASE
WHEN itemid = 223761 THEN (cev.valuenum-32)/1.8
ELSE cev.valuenum
END AS valuenum
, CASE
WHEN itemid = 220045 THEN 'HEARTRATE'
WHEN itemid = 220050 THEN 'SYSBP'
WHEN itemid = 220179 THEN 'SYSBP'
WHEN itemid = 220051 THEN 'DIASBP'
WHEN itemid = 220180 THEN 'DIASBP'
WHEN itemid = 220052 THEN 'MEANBP'
WHEN itemid = 220181 THEN 'MEANBP'
WHEN itemid = 225312 THEN 'MEANBP'
WHEN itemid = 220210 THEN 'RESPRATE'
WHEN itemid = 224688 THEN 'RESPRATE'
WHEN itemid = 224689 THEN 'RESPRATE'
WHEN itemid = 224690 THEN 'RESPRATE'
WHEN itemid = 223761 THEN 'TEMPC'
WHEN itemid = 223762 THEN 'TEMPC'
WHEN itemid = 220277 THEN 'SPO2'
WHEN itemid = 220739 THEN 'GCSEYE'
WHEN itemid = 223900 THEN 'GCSVERBAL'
WHEN itemid = 223901 THEN 'GCSMOTOR'
ELSE null
END AS label
FROM mimiciv.icustays icu
INNER JOIN mimiciv.chartevents cev
ON cev.stay_id = icu.stay_id
AND cev.charttime >= icu.intime
AND cev.charttime <= icu.intime + interval '24 hour'
WHERE cev.itemid IN
(
220045 -- heartrate
, 220050, 220179 -- sysbp
, 220051, 220180 -- diasbp
, 220052, 220181, 225312 -- meanbp
, 220210, 224688, 224689, 224690 -- resprate
, 223761, 223762 -- tempc
, 220277 -- SpO2
, 220739 -- gcseye
, 223900 -- gcsverbal
, 223901 -- gscmotor
)
AND valuenum IS NOT null
)
, vitals_stg_2 AS
(
SELECT
stay_id, valuenum, label
, ROW_NUMBER() OVER (PARTITION BY stay_id, label ORDER BY charttime DESC) AS rn
FROM vitals_stg_1
)
, vitals_stg_3 AS
(
SELECT
stay_id
, COALESCE(MAX(CASE WHEN label = 'HEARTRATE' THEN valuenum ELSE null END)) AS heartrate_last
, COALESCE(MAX(CASE WHEN label = 'SYSBP' THEN valuenum ELSE null END)) AS sysbp_last
, COALESCE(MAX(CASE WHEN label = 'DIASBP' THEN valuenum ELSE null END)) AS diabp_last
, COALESCE(MAX(CASE WHEN label = 'MEANBP' THEN valuenum ELSE null END)) AS meanbp_last
, COALESCE(MAX(CASE WHEN label = 'RESPRATE' THEN valuenum ELSE null END)) AS resprate_last
, COALESCE(MAX(CASE WHEN label = 'TEMPC' THEN valuenum ELSE null END)) AS tempc_last
, COALESCE(MAX(CASE WHEN label = 'SPO2' THEN valuenum ELSE null END)) AS spo2_last
, COALESCE(MAX(CASE WHEN label = 'GCSEYE' THEN valuenum ELSE null END)) AS gcseye_last
, COALESCE(MAX(CASE WHEN label = 'GCSVERBAL' THEN valuenum ELSE null END)) AS gcsverbal_last
, COALESCE(MAX(CASE WHEN label = 'GCSMOTOR' THEN valuenum ELSE null END)) AS gcsmotor_last
FROM vitals_stg_2
WHERE rn = 1
GROUP BY stay_id
)
SELECT * FROM vitals_stg_3
"""
last_vitals = pd.read_sql_query(query, con)
last_vitals
```
### 3.6 Max Vitals Measurement Attributes
```
query = query_schema + \
"""
WITH vitals_stg_1 AS
(
SELECT icu.stay_id, cev.charttime
, CASE
WHEN itemid = 223761 THEN (cev.valuenum-32)/1.8
ELSE cev.valuenum
END AS valuenum
, CASE
WHEN itemid = 220045 THEN 'HEARTRATE'
WHEN itemid = 220050 THEN 'SYSBP'
WHEN itemid = 220179 THEN 'SYSBP'
WHEN itemid = 220051 THEN 'DIASBP'
WHEN itemid = 220180 THEN 'DIASBP'
WHEN itemid = 220052 THEN 'MEANBP'
WHEN itemid = 220181 THEN 'MEANBP'
WHEN itemid = 225312 THEN 'MEANBP'
WHEN itemid = 220210 THEN 'RESPRATE'
WHEN itemid = 224688 THEN 'RESPRATE'
WHEN itemid = 224689 THEN 'RESPRATE'
WHEN itemid = 224690 THEN 'RESPRATE'
WHEN itemid = 223761 THEN 'TEMPC'
WHEN itemid = 223762 THEN 'TEMPC'
WHEN itemid = 220277 THEN 'SPO2'
WHEN itemid = 220739 THEN 'GCSEYE'
WHEN itemid = 223900 THEN 'GCSVERBAL'
WHEN itemid = 223901 THEN 'GCSMOTOR'
ELSE null
END AS label
FROM mimiciv.icustays icu
INNER JOIN mimiciv.chartevents cev
ON cev.stay_id = icu.stay_id
AND cev.charttime >= icu.intime
AND cev.charttime <= icu.intime + interval '24 hour'
WHERE cev.itemid IN
(
220045 -- heartrate
, 220050, 220179 -- sysbp
, 220051, 220180 -- diasbp
, 220052, 220181, 225312 -- meanbp
, 220210, 224688, 224689, 224690 -- resprate
, 223761, 223762 -- tempc
, 220277 -- SpO2
, 220739 -- gcseye
, 223900 -- gcsverbal
, 223901 -- gscmotor
)
AND valuenum IS NOT null
)
, vitals_stg_2 AS
(
SELECT
stay_id, valuenum, label
, ROW_NUMBER() OVER (PARTITION BY stay_id, label ORDER BY charttime) AS rn
FROM vitals_stg_1
)
, vitals_stg_3 AS
(
SELECT
stay_id
, rn
, COALESCE(MAX(CASE WHEN label = 'HEARTRATE' THEN valuenum ELSE null END)) AS heartrate
, COALESCE(MAX(CASE WHEN label = 'SYSBP' THEN valuenum ELSE null END)) AS sysbp
, COALESCE(MAX(CASE WHEN label = 'DIASBP' THEN valuenum ELSE null END)) AS diabp
, COALESCE(MAX(CASE WHEN label = 'MEANBP' THEN valuenum ELSE null END)) AS meanbp
, COALESCE(MAX(CASE WHEN label = 'RESPRATE' THEN valuenum ELSE null END)) AS resprate
, COALESCE(MAX(CASE WHEN label = 'TEMPC' THEN valuenum ELSE null END)) AS tempc
, COALESCE(MAX(CASE WHEN label = 'SPO2' THEN valuenum ELSE null END)) AS spo2
, COALESCE(MAX(CASE WHEN label = 'GCSEYE' THEN valuenum ELSE null END)) AS gcseye
, COALESCE(MAX(CASE WHEN label = 'GCSVERBAL' THEN valuenum ELSE null END)) AS gcsverbal
, COALESCE(MAX(CASE WHEN label = 'GCSMOTOR' THEN valuenum ELSE null END)) AS gcsmotor
FROM vitals_stg_2
GROUP BY stay_id, rn
)
, vitals_stg_4 AS
(
SELECT
stay_id,
MAX(heartrate) AS heartrate_max
, MAX(sysbp) AS sysbp_max
, MAX(diabp) AS diabp_max
, MAX(meanbp) AS meanbp_max
, MAX(resprate) AS resprate_max
, MAX(tempc) AS tempc_max
, MAX(spo2) AS spo2_max
, MAX(gcseye) AS gcseye_max
, MAX(gcsverbal) AS gcsverbal_max
, MAX(gcsmotor) AS gcsmotor_max
FROM vitals_stg_3
GROUP BY stay_id
)
SELECT * FROM vitals_stg_4
"""
max_vitals = pd.read_sql_query(query, con)
max_vitals
```
### 3.7 Min Vitals Measurement Attributes
```
query = query_schema + \
"""
WITH vitals_stg_1 AS
(
SELECT icu.stay_id, cev.charttime
, CASE
WHEN itemid = 223761 THEN (cev.valuenum-32)/1.8
ELSE cev.valuenum
END AS valuenum
, CASE
WHEN itemid = 220045 THEN 'HEARTRATE'
WHEN itemid = 220050 THEN 'SYSBP'
WHEN itemid = 220179 THEN 'SYSBP'
WHEN itemid = 220051 THEN 'DIASBP'
WHEN itemid = 220180 THEN 'DIASBP'
WHEN itemid = 220052 THEN 'MEANBP'
WHEN itemid = 220181 THEN 'MEANBP'
WHEN itemid = 225312 THEN 'MEANBP'
WHEN itemid = 220210 THEN 'RESPRATE'
WHEN itemid = 224688 THEN 'RESPRATE'
WHEN itemid = 224689 THEN 'RESPRATE'
WHEN itemid = 224690 THEN 'RESPRATE'
WHEN itemid = 223761 THEN 'TEMPC'
WHEN itemid = 223762 THEN 'TEMPC'
WHEN itemid = 220277 THEN 'SPO2'
WHEN itemid = 220739 THEN 'GCSEYE'
WHEN itemid = 223900 THEN 'GCSVERBAL'
WHEN itemid = 223901 THEN 'GCSMOTOR'
ELSE null
END AS label
FROM mimiciv.icustays icu
INNER JOIN mimiciv.chartevents cev
ON cev.stay_id = icu.stay_id
AND cev.charttime >= icu.intime
AND cev.charttime <= icu.intime + interval '24 hour'
WHERE cev.itemid IN
(
220045 -- heartrate
, 220050, 220179 -- sysbp
, 220051, 220180 -- diasbp
, 220052, 220181, 225312 -- meanbp
, 220210, 224688, 224689, 224690 -- resprate
, 223761, 223762 -- tempc
, 220277 -- SpO2
, 220739 -- gcseye
, 223900 -- gcsverbal
, 223901 -- gscmotor
)
AND valuenum IS NOT null
)
, vitals_stg_2 AS
(
SELECT
stay_id, valuenum, label
, ROW_NUMBER() OVER (PARTITION BY stay_id, label ORDER BY charttime) AS rn
FROM vitals_stg_1
)
, vitals_stg_3 AS
(
SELECT
stay_id
, rn
, COALESCE(MAX(CASE WHEN label = 'HEARTRATE' THEN valuenum ELSE null END)) AS heartrate
, COALESCE(MAX(CASE WHEN label = 'SYSBP' THEN valuenum ELSE null END)) AS sysbp
, COALESCE(MAX(CASE WHEN label = 'DIASBP' THEN valuenum ELSE null END)) AS diabp
, COALESCE(MAX(CASE WHEN label = 'MEANBP' THEN valuenum ELSE null END)) AS meanbp
, COALESCE(MAX(CASE WHEN label = 'RESPRATE' THEN valuenum ELSE null END)) AS resprate
, COALESCE(MAX(CASE WHEN label = 'TEMPC' THEN valuenum ELSE null END)) AS tempc
, COALESCE(MAX(CASE WHEN label = 'SPO2' THEN valuenum ELSE null END)) AS spo2
, COALESCE(MAX(CASE WHEN label = 'GCSEYE' THEN valuenum ELSE null END)) AS gcseye
, COALESCE(MAX(CASE WHEN label = 'GCSVERBAL' THEN valuenum ELSE null END)) AS gcsverbal
, COALESCE(MAX(CASE WHEN label = 'GCSMOTOR' THEN valuenum ELSE null END)) AS gcsmotor
FROM vitals_stg_2
GROUP BY stay_id, rn
)
, vitals_stg_4 AS
(
SELECT
stay_id,
MIN(heartrate) AS heartrate_min
, MIN(sysbp) AS sysbp_min
, MIN(diabp) AS diabp_min
, MIN(meanbp) AS meanbp_min
, MIN(resprate) AS resprate_min
, MIN(tempc) AS tempc_min
, MIN(spo2) AS spo2_min
, MIN(gcseye) AS gcseye_min
, MIN(gcsverbal) AS gcsverbal_min
, MIN(gcsmotor) AS gcsmotor_min
FROM vitals_stg_3
GROUP BY stay_id
)
SELECT * FROM vitals_stg_4
"""
min_vitals = pd.read_sql_query(query, con)
min_vitals
```
### 3.8 Average Vitals Measurement Attributes
```
query = query_schema + \
"""
WITH vitals_stg_1 AS
(
SELECT icu.stay_id, cev.charttime
, CASE
WHEN itemid = 223761 THEN (cev.valuenum-32)/1.8
ELSE cev.valuenum
END AS valuenum
, CASE
WHEN itemid = 220045 THEN 'HEARTRATE'
WHEN itemid = 220050 THEN 'SYSBP'
WHEN itemid = 220179 THEN 'SYSBP'
WHEN itemid = 220051 THEN 'DIASBP'
WHEN itemid = 220180 THEN 'DIASBP'
WHEN itemid = 220052 THEN 'MEANBP'
WHEN itemid = 220181 THEN 'MEANBP'
WHEN itemid = 225312 THEN 'MEANBP'
WHEN itemid = 220210 THEN 'RESPRATE'
WHEN itemid = 224688 THEN 'RESPRATE'
WHEN itemid = 224689 THEN 'RESPRATE'
WHEN itemid = 224690 THEN 'RESPRATE'
WHEN itemid = 223761 THEN 'TEMPC'
WHEN itemid = 223762 THEN 'TEMPC'
WHEN itemid = 220277 THEN 'SPO2'
WHEN itemid = 220739 THEN 'GCSEYE'
WHEN itemid = 223900 THEN 'GCSVERBAL'
WHEN itemid = 223901 THEN 'GCSMOTOR'
ELSE null
END AS label
FROM mimiciv.icustays icu
INNER JOIN mimiciv.chartevents cev
ON cev.stay_id = icu.stay_id
AND cev.charttime >= icu.intime
AND cev.charttime <= icu.intime + interval '24 hour'
WHERE cev.itemid IN
(
220045 -- heartrate
, 220050, 220179 -- sysbp
, 220051, 220180 -- diasbp
, 220052, 220181, 225312 -- meanbp
, 220210, 224688, 224689, 224690 -- resprate
, 223761, 223762 -- tempc
, 220277 -- SpO2
, 220739 -- gcseye
, 223900 -- gcsverbal
, 223901 -- gscmotor
)
AND valuenum IS NOT null
)
, vitals_stg_2 AS
(
SELECT
stay_id, valuenum, label
, ROW_NUMBER() OVER (PARTITION BY stay_id, label ORDER BY charttime) AS rn
FROM vitals_stg_1
)
, vitals_stg_3 AS
(
SELECT
stay_id
, rn
, COALESCE(MAX(CASE WHEN label = 'HEARTRATE' THEN valuenum ELSE null END)) AS heartrate
, COALESCE(MAX(CASE WHEN label = 'SYSBP' THEN valuenum ELSE null END)) AS sysbp
, COALESCE(MAX(CASE WHEN label = 'DIASBP' THEN valuenum ELSE null END)) AS diabp
, COALESCE(MAX(CASE WHEN label = 'MEANBP' THEN valuenum ELSE null END)) AS meanbp
, COALESCE(MAX(CASE WHEN label = 'RESPRATE' THEN valuenum ELSE null END)) AS resprate
, COALESCE(MAX(CASE WHEN label = 'TEMPC' THEN valuenum ELSE null END)) AS tempc
, COALESCE(MAX(CASE WHEN label = 'SPO2' THEN valuenum ELSE null END)) AS spo2
, COALESCE(MAX(CASE WHEN label = 'GCSEYE' THEN valuenum ELSE null END)) AS gcseye
, COALESCE(MAX(CASE WHEN label = 'GCSVERBAL' THEN valuenum ELSE null END)) AS gcsverbal
, COALESCE(MAX(CASE WHEN label = 'GCSMOTOR' THEN valuenum ELSE null END)) AS gcsmotor
FROM vitals_stg_2
GROUP BY stay_id, rn
)
, vitals_stg_4 AS
(
SELECT
stay_id,
AVG(heartrate) AS heartrate_avg
, AVG(sysbp) AS sysbp_avg
, AVG(diabp) AS diabp_avg
, AVG(meanbp) AS meanbp_avg
, AVG(resprate) AS resprate_avg
, AVG(tempc) AS tempc_avg
, AVG(spo2) AS spo2_avg
, AVG(gcseye) AS gcseye_avg
, AVG(gcsverbal) AS gcsverbal_avg
, AVG(gcsmotor) AS gcsmotor_avg
FROM vitals_stg_3
GROUP BY stay_id
)
SELECT * FROM vitals_stg_4
"""
avg_vitals = pd.read_sql_query(query, con)
avg_vitals
```
### 3.9 Inhospital Mortality
```
query = query_schema + \
"""
SELECT
icu.stay_id, adm.hospital_expire_flag
FROM
mimiciv.icustays icu
INNER JOIN mimiciv.admissions adm
ON adm.hadm_id = icu.hadm_id
"""
mortality = pd.read_sql_query(query, con)
mortality
```
### 3.10 Filter
```
query = query_schema + \
"""
SELECT
icu.stay_id
FROM
mimiciv.icustays icu
INNER JOIN mimiciv.admissions adm
ON adm.hadm_id = icu.hadm_id
INNER JOIN mimiciv.patients pat
ON pat.subject_id = adm.subject_id
AND icu.intime = (
SELECT MAX(icu_max.intime) FROM mimiciv.icustays icu_max WHERE icu_max.hadm_id = icu.hadm_id
)
WHERE (icu.outtime >= icu.intime + interval '24 hour')
AND (FLOOR(DATE_PART('day', adm.admittime - make_timestamp(pat.anchor_year, 1, 1, 0, 0, 0))/365.0) + pat.anchor_age) > 18
"""
filtered = pd.read_sql_query(query, con)
filtered
```
## 4. Combine
```
static.shape, first_lab.shape, last_lab.shape, first_vitals.shape, last_vitals.shape, max_vitals.shape, min_vitals.shape, avg_vitals.shape, mortality.shape, filtered.shape
dfs = [filtered, static, first_lab, last_lab, first_vitals, last_vitals, max_vitals, min_vitals, avg_vitals, mortality]
data = reduce(lambda left, right: pd.merge(left, right, on=['stay_id'], how='inner'), dfs)
```
## 5. Sanity Check
### 5.1 Missing Data
```
na_counts = data.isna().sum().reset_index(name="n")
na_counts = na_counts.sort_values(by=['n'], ascending=[False])
plt.figure(figsize=(10,40))
plt.tick_params(top="on")
plt.barh(y=na_counts['index'], width=na_counts.n)
```
### 5.2 Data Distributions
### 5.3 Outliers
## 6. Format Data
```
data.drop(columns=['subject_id', 'hadm_id', 'stay_id'], inplace=True)
data.shape
# convert to numpy data (assumes target, death, is the last column)
X = data.values
y = X[:,-1]
y = y.astype('int')
X = X[:,0:-1]
X_header = [x for x in data.columns.values]
X_header = X_header[0:-1]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
```
## 7. Machine Learning
### 7.1 Logistic Regression
```
estimator = Pipeline([("imputer", SimpleImputer(missing_values=np.nan, strategy="mean")),
("scaler", StandardScaler()),
("logreg" , LogisticRegression(fit_intercept=True))])
mdl = estimator.fit(X_train, y_train)
probs = mdl.predict_proba(X_test)
probs = probs[:,1]
preds = mdl.predict(X_test)
# calculate score (AUROC)
roc_score = metrics.roc_auc_score(y_test, probs)
roc_score
# calculate score (AUROC)
classification_report = metrics.classification_report(y_test, preds)
print(classification_report)
lrDf = pd.DataFrame(data=list(zip(X_header, mdl.named_steps['logreg'].coef_[0])), columns=['feature', 'weight'])
lrDf.sort_values('weight', inplace=True)
plt.figure(figsize=(10,40))
plt.barh(y=lrDf.feature, width=lrDf.weight)
```
### 7.2 XGB Classifier
```
estimator = Pipeline([("imputer", SimpleImputer(missing_values=np.nan, strategy="mean")),
("scaler", StandardScaler()),
("xgb" , xgb.XGBClassifier(max_depth=3, n_estimators=300, learning_rate=0.05))])
mdl = estimator.fit(X_train, y_train)
probs = mdl.predict_proba(X_test)
probs = probs[:,1]
preds = mdl.predict(X_test)
# calculate score (AUROC)
roc_score = metrics.roc_auc_score(y_test, probs)
roc_score
# calculate score (AUROC)
classification_report = metrics.classification_report(y_test, preds)
print(classification_report)
feature_importance_list = []
feature_imp_dict = mdl.named_steps['xgb'].get_booster().get_score(importance_type="gain")
for feature_imp in feature_imp_dict:
feature_importance_list.append([X_header[int(feature_imp[1:])], feature_imp_dict[feature_imp]])
xgbDf = pd.DataFrame(feature_importance_list, columns = ['feature', 'weight'])
xgbDf.sort_values('weight', inplace=True)
plt.figure(figsize=(10,40))
plt.barh(y=xgbDf.feature, width=xgbDf.weight)
```
### 7.3 Neural Network
#### 7.3.1 Using SKLearn
```
estimator = Pipeline([("imputer", SimpleImputer(missing_values=np.nan, strategy="mean")),
("scaler", StandardScaler()),
("mlp" , MLPClassifier(hidden_layer_sizes=10))])
mdl = estimator.fit(X_train, y_train)
probs = mdl.predict_proba(X_test)
probs = probs[:,1]
preds = mdl.predict(X_test)
# calculate score (AUROC)
roc_score = metrics.roc_auc_score(y_test, probs)
roc_score
# calculate score (AUROC)
classification_report = metrics.classification_report(y_test, preds)
print(classification_report)
```
#### 7.3.2 Using Keras
```
X_train_array = np.asarray(X_train).astype('float32')
X_test_array = np.asarray(X_test).astype('float32')
imp_mean = SimpleImputer(missing_values=np.nan, strategy='mean')
imp_mean.fit(X_train_array)
X_train_imputed = imp_mean.transform(X_train_array)
X_test_imputed = imp_mean.transform(X_test_array)
normalizer = preprocessing.Normalization()
normalizer.adapt(X_train_imputed)
X_train_noralized = normalizer(X_train_imputed)
X_test_noralized = normalizer(X_test_imputed)
model = Sequential()
model.add(layers.Dense(X_train.shape[1], activation=tf.nn.relu, kernel_initializer='he_normal', bias_initializer='zeros'))
model.add(layers.Dense(40, activation=tf.nn.relu, kernel_initializer='he_normal', bias_initializer='zeros'))
model.add(layers.Dense(1, activation=tf.nn.sigmoid, kernel_initializer='he_normal', bias_initializer='zeros'))
model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
model.fit(X_train_noralized, y_train, validation_data=(X_test_noralized, y_test), epochs=5, batch_size=256)
model.summary()
probs = model.predict_proba(X_test_noralized)
preds = np.around(probs)
# calculate score (AUROC)
roc_score = metrics.roc_auc_score(y_test, probs)
roc_score
# calculate score (AUROC)
classification_report = metrics.classification_report(y_test, preds)
print(classification_report)
cm = confusion_matrix(y_test, preds, labels=[0, 1])
disp = ConfusionMatrixDisplay(confusion_matrix=cm, display_labels=['survided', 'not-survived'])
disp.plot()
```
|
github_jupyter
|
import psycopg2
import pandas as pd
import numpy as np
import xgboost as xgb
import tensorflow as tf
from functools import reduce
from sklearn import metrics
from sklearn.model_selection import train_test_split
from sklearn.pipeline import Pipeline
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import LogisticRegression
from sklearn.neural_network import MLPClassifier
from sklearn.metrics import confusion_matrix, ConfusionMatrixDisplay
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.layers.experimental import preprocessing
from keras.models import Sequential
from matplotlib import pyplot as plt
# information used to create a database connection
sqluser = 'postgres'
dbname = 'mimic4'
hostname = 'localhost'
port_number = 5434
schema_name = 'mimiciv'
# Connect to postgres with a copy of the MIMIC-III database
con = psycopg2.connect(dbname=dbname, user=sqluser, host=hostname, port=port_number, password='mysecretpassword')
# the below statement is prepended to queries to ensure they select from the right schema
query_schema = 'set search_path to ' + schema_name + ';'
query = query_schema + \
"""
WITH ht AS
(
SELECT
c.subject_id, c.stay_id, c.charttime,
-- Ensure that all heights are in centimeters, and fix data as needed
CASE
-- rule for neonates
WHEN pt.anchor_age = 0
AND (c.valuenum * 2.54) < 80
THEN c.valuenum * 2.54
-- rule for adults
WHEN pt.anchor_age > 0
AND (c.valuenum * 2.54) > 120
AND (c.valuenum * 2.54) < 230
THEN c.valuenum * 2.54
-- set bad data to NULL
ELSE NULL
END AS height
, ROW_NUMBER() OVER (PARTITION BY stay_id ORDER BY charttime DESC) AS rn
FROM mimiciv.chartevents c
INNER JOIN mimiciv.patients pt
ON c.subject_id = pt.subject_id
WHERE c.valuenum IS NOT NULL
AND c.valuenum != 0
AND c.itemid IN
(
226707 -- Height (measured in inches)
-- note we intentionally ignore the below ITEMID in metavision
-- these are duplicate data in a different unit
-- , 226730 -- Height (cm)
)
)
, wt AS
(
SELECT
c.stay_id
, c.charttime
-- TODO: eliminate obvious outliers if there is a reasonable weight
, c.valuenum as weight
, ROW_NUMBER() OVER (PARTITION BY stay_id ORDER BY charttime DESC) AS rn
FROM mimiciv.chartevents c
WHERE c.valuenum IS NOT NULL
AND c.itemid = 226512 -- Admit Wt
AND c.stay_id IS NOT NULL
AND c.valuenum > 0
)
SELECT
ie.subject_id, ie.hadm_id, ie.stay_id
, CASE WHEN pat.gender = 'M' THEN '1' ELSE '0' END AS is_male
, FLOOR(DATE_PART('day', adm.admittime - make_timestamp(pat.anchor_year, 1, 1, 0, 0, 0))/365.0) + pat.anchor_age as age
, CASE WHEN adm.ethnicity LIKE '%WHITE%' THEN '1' ELSE '0' END AS race_white
, CASE WHEN adm.ethnicity LIKE '%BLACK%' THEN '1' ELSE '0' END AS race_black
, CASE WHEN adm.ethnicity LIKE '%HISPANIC%' THEN '1' ELSE '0' END AS race_hispanic
, CASE WHEN adm.ethnicity LIKE '%ASIAN%' THEN '1' ELSE '0' END AS race_asian
, CASE WHEN adm.ethnicity LIKE '%OTHER%' THEN '1' ELSE '0' END AS race_other
, CASE WHEN adm.admission_type LIKE '%EMER%' THEN '1' ELSE '0' END AS emergency_admission
, CASE
WHEN ht.height IS NOT null AND wt.weight IS NOT null
THEN (wt.weight / (ht.height/100*ht.height/100))
ELSE null
END AS bmi
, ht.height as height
, wt.weight as weight
, (
SELECT
CASE WHEN COUNT(*) = 0 THEN 0 ELSE 1 END
FROM mimiciv.transfers car_trs
WHERE car_trs.hadm_id = adm.hadm_id
AND lower(car_trs.careunit) LIKE '%card%'
AND lower(car_trs.careunit) LIKE '%surg%'
) AS service_any_card_surg
, (
SELECT
CASE WHEN COUNT(*) = 0 THEN 0 ELSE 1 END
FROM mimiciv.transfers car_trs
WHERE car_trs.hadm_id = adm.hadm_id
AND lower(car_trs.careunit) NOT LIKE '%card%'
AND lower(car_trs.careunit) LIKE '%surg%'
) AS service_any_noncard_surg
, (
SELECT
CASE WHEN COUNT(*) = 0 THEN 0 ELSE 1 END
FROM mimiciv.transfers car_trs
WHERE car_trs.hadm_id = adm.hadm_id
AND lower(car_trs.careunit) LIKE '%trauma%'
) AS service_trauma
-- , adm.hospital_expire_flag
FROM mimiciv.icustays ie
INNER JOIN mimiciv.admissions adm
ON ie.hadm_id = adm.hadm_id
INNER JOIN mimiciv.patients pat
ON ie.subject_id = pat.subject_id
LEFT JOIN ht
ON ie.stay_id = ht.stay_id AND ht.rn = 1
LEFT JOIN wt
ON ie.stay_id = wt.stay_id AND wt.rn = 1
"""
static = pd.read_sql_query(query, con)
static
query = query_schema + \
"""
WITH labs_preceeding AS
(
SELECT icu.stay_id, l.valuenum, l.charttime
, CASE
WHEN itemid = 51006 THEN 'BUN'
WHEN itemid = 50806 THEN 'CHLORIDE'
WHEN itemid = 50902 THEN 'CHLORIDE'
WHEN itemid = 50912 THEN 'CREATININE'
WHEN itemid = 50811 THEN 'HEMOGLOBIN'
WHEN itemid = 51222 THEN 'HEMOGLOBIN'
WHEN itemid = 51265 THEN 'PLATELET'
WHEN itemid = 50822 THEN 'POTASSIUM'
WHEN itemid = 50971 THEN 'POTASSIUM'
WHEN itemid = 50824 THEN 'SODIUM'
WHEN itemid = 50983 THEN 'SODIUM'
WHEN itemid = 50803 THEN 'BICARBONATE'
WHEN itemid = 50882 THEN 'BICARBONATE'
WHEN itemid = 50804 THEN 'TOTALCO2'
WHEN itemid = 50821 THEN 'PO2'
WHEN itemid = 52042 THEN 'PO2'
WHEN itemid = 50832 THEN 'PO2'
WHEN itemid = 50818 THEN 'PCO2'
WHEN itemid = 52040 THEN 'PCO2'
WHEN itemid = 50830 THEN 'PCO2'
WHEN itemid = 50820 THEN 'PH'
WHEN itemid = 52041 THEN 'PH'
WHEN itemid = 50831 THEN 'PH'
WHEN itemid = 51300 THEN 'WBC'
WHEN itemid = 51301 THEN 'WBC'
WHEN itemid = 50802 THEN 'BASEEXCESS'
WHEN itemid = 52038 THEN 'BASEEXCESS'
WHEN itemid = 50805 THEN 'CARBOXYHEMOGLOBIN'
WHEN itemid = 50814 THEN 'METHEMOGLOBIN'
WHEN itemid = 50868 THEN 'ANIONGAP'
WHEN itemid = 52500 THEN 'ANIONGAP'
WHEN itemid = 50862 THEN 'ALBUMIN'
WHEN itemid = 51144 THEN 'BANDS'
WHEN itemid = 50885 THEN 'BILRUBIN'
WHEN itemid = 51478 THEN 'GLUCOSE'
WHEN itemid = 50931 THEN 'GLUCOSE'
WHEN itemid = 51221 THEN 'HEMATOCRIT'
WHEN itemid = 50813 THEN 'LACTATE'
WHEN itemid = 51275 THEN 'PTT'
WHEN itemid = 51237 THEN 'INR'
ELSE null
END AS LABEL
FROM mimiciv.icustays icu
INNER JOIN mimiciv.admissions adm
ON icu.hadm_id = adm.hadm_id
INNER JOIN mimiciv.patients pat
ON icu.subject_id = pat.subject_id
INNER JOIN mimiciv.labevents l
ON l.hadm_id = icu.hadm_id
AND l.charttime >= icu.intime - interval '8 hour'
AND l.charttime <= icu.intime + interval '24 hour'
WHERE l.itemid IN
(
51300,51301 -- wbc
, 50811,51222 -- hgb
, 51265 -- platelet
, 50824, 50983 -- sodium
, 50822, 50971 -- potassium
, 50804 -- Total CO2 or ...
, 50803, 50882 -- bicarbonate
, 50806, 50902 -- chloride
, 51006 -- bun
, 50912 -- creatinine
, 50821, 52042, 50832 -- po2
, 50818, 52040, 50830 -- pco2
, 50820, 52041, 50831 -- ph
, 50802, 52038 -- Base Excess
, 50805 -- carboxyhemoglobin
, 50814 -- methemoglobin
, 50868, 52500 -- aniongap
, 50862 -- albumin
, 51144 -- bands
, 50885 -- bilrubin
, 51478, 50931 -- glucose
, 51221 -- hematocrit
, 50813 -- lactate
, 51275 -- ptt
, 51237 -- inr
)
AND valuenum IS NOT null
)
, labs_rn AS
(
SELECT
stay_id, valuenum, label
, ROW_NUMBER() OVER (PARTITION BY stay_id, label ORDER BY charttime) AS rn
FROM labs_preceeding
)
, labs_grp AS
(
SELECT
stay_id
, COALESCE(MAX(CASE WHEN label = 'BUN' THEN valuenum ELSE null END)) AS BUN
, COALESCE(MAX(CASE WHEN label = 'CHLORIDE' THEN valuenum ELSE null END)) AS CHLORIDE
, COALESCE(MAX(CASE WHEN label = 'CREATININE' THEN valuenum ELSE null END)) AS CREATININE
, COALESCE(MAX(CASE WHEN label = 'HEMOGLOBIN' THEN valuenum ELSE null END)) AS HEMOGLOBIN
, COALESCE(MAX(CASE WHEN label = 'PLATELET' THEN valuenum ELSE null END)) AS PLATELET
, COALESCE(MAX(CASE WHEN label = 'POTASSIUM' THEN valuenum ELSE null END)) AS POTASSIUM
, COALESCE(MAX(CASE WHEN label = 'SODIUM' THEN valuenum ELSE null END)) AS SODIUM
, COALESCE(MAX(CASE WHEN label = 'TOTALCO2' THEN valuenum ELSE null END)) AS TOTALCO2
, COALESCE(MAX(CASE WHEN label = 'WBC' THEN valuenum ELSE null END)) AS WBC
, COALESCE(MAX(CASE WHEN label = 'PO2' THEN valuenum ELSE null END)) AS PO2
, COALESCE(MAX(CASE WHEN label = 'PCO2' THEN valuenum ELSE null END)) AS PCO2
, COALESCE(MAX(CASE WHEN label = 'PH' THEN valuenum ELSE null END)) AS PH
, COALESCE(MAX(CASE WHEN label = 'BASEEXCESS' THEN valuenum ELSE null END)) AS BASEEXCESS
, COALESCE(MAX(CASE WHEN label = 'CARBOXYHEMOGLOBIN' THEN valuenum ELSE null END)) AS CARBOXYHEMOGLOBIN
, COALESCE(MAX(CASE WHEN label = 'METHEMOGLOBIN' THEN valuenum ELSE null END)) AS METHEMOGLOBIN
, COALESCE(MAX(CASE WHEN label = 'ANIONGAP' THEN valuenum ELSE null END)) AS ANIONGAP
, COALESCE(MAX(CASE WHEN label = 'ALBUMIN' THEN valuenum ELSE null END)) AS ALBUMIN
, COALESCE(MAX(CASE WHEN label = 'BANDS' THEN valuenum ELSE null END)) AS BANDS
, COALESCE(MAX(CASE WHEN label = 'BICARBONATE' THEN valuenum ELSE null END)) AS BICARBONATE
, COALESCE(MAX(CASE WHEN label = 'BILRUBIN' THEN valuenum ELSE null END)) AS BILRUBIN
, COALESCE(MAX(CASE WHEN label = 'GLUCOSE' THEN valuenum ELSE null END)) AS GLUCOSE
, COALESCE(MAX(CASE WHEN label = 'HEMATOCRIT' THEN valuenum ELSE null END)) AS HEMATOCRIT
, COALESCE(MAX(CASE WHEN label = 'LACTATE' THEN valuenum ELSE null END)) AS LACTATE
, COALESCE(MAX(CASE WHEN label = 'PTT' THEN valuenum ELSE null END)) AS PTT
, COALESCE(MAX(CASE WHEN label = 'INR' THEN valuenum ELSE null END)) AS INR
FROM labs_rn
WHERE rn = 1
GROUP BY stay_id
)
SELECT icu.stay_id
, lg.bun AS bun_first
, lg.chloride AS chloride_first
, lg.creatinine AS creatinine_first
, lg.HEMOGLOBIN AS hgb_first
, lg.platelet AS platelet_first
, lg.potassium AS potassium_first
, lg.sodium AS sodium_first
, lg.TOTALCO2 AS tco2_first
, lg.wbc AS wbc_first
, lg.po2 AS bg_po2_first
, lg.pco2 AS bg_pco2_first
, lg.ph AS bg_ph_first
, lg.BASEEXCESS AS bg_baseexcess_first
, lg.CARBOXYHEMOGLOBIN AS bg_carboxyhemoglobin_first
, lg.METHEMOGLOBIN AS bg_methemomoglobin_first
, lg.ANIONGAP AS aniongap_first
, lg.ALBUMIN AS albumin_first
, lg.BANDS AS bands_first
, lg.BICARBONATE AS bicarbonate_first
, lg.BILRUBIN AS bilrubin_first
, lg.GLUCOSE AS glucose_first
, lg.HEMATOCRIT AS hematocrit_first
, lg.LACTATE AS lactate_first
, lg.PTT AS ptt_first
, lg.INR AS inr_first
FROM mimiciv.icustays icu
LEFT JOIN labs_grp lg
ON icu.stay_id = lg.stay_id
"""
first_lab = pd.read_sql_query(query, con)
first_lab
query = query_schema + \
"""
WITH labs_preceeding AS
(
SELECT icu.stay_id, l.valuenum, l.charttime
, CASE
WHEN itemid = 51006 THEN 'BUN'
WHEN itemid = 50806 THEN 'CHLORIDE'
WHEN itemid = 50902 THEN 'CHLORIDE'
WHEN itemid = 50912 THEN 'CREATININE'
WHEN itemid = 50811 THEN 'HEMOGLOBIN'
WHEN itemid = 51222 THEN 'HEMOGLOBIN'
WHEN itemid = 51265 THEN 'PLATELET'
WHEN itemid = 50822 THEN 'POTASSIUM'
WHEN itemid = 50971 THEN 'POTASSIUM'
WHEN itemid = 50824 THEN 'SODIUM'
WHEN itemid = 50983 THEN 'SODIUM'
WHEN itemid = 50803 THEN 'BICARBONATE'
WHEN itemid = 50882 THEN 'BICARBONATE'
WHEN itemid = 50804 THEN 'TOTALCO2'
WHEN itemid = 50821 THEN 'PO2'
WHEN itemid = 52042 THEN 'PO2'
WHEN itemid = 50832 THEN 'PO2'
WHEN itemid = 50818 THEN 'PCO2'
WHEN itemid = 52040 THEN 'PCO2'
WHEN itemid = 50830 THEN 'PCO2'
WHEN itemid = 50820 THEN 'PH'
WHEN itemid = 52041 THEN 'PH'
WHEN itemid = 50831 THEN 'PH'
WHEN itemid = 51300 THEN 'WBC'
WHEN itemid = 51301 THEN 'WBC'
WHEN itemid = 50802 THEN 'BASEEXCESS'
WHEN itemid = 52038 THEN 'BASEEXCESS'
WHEN itemid = 50805 THEN 'CARBOXYHEMOGLOBIN'
WHEN itemid = 50814 THEN 'METHEMOGLOBIN'
WHEN itemid = 50868 THEN 'ANIONGAP'
WHEN itemid = 52500 THEN 'ANIONGAP'
WHEN itemid = 50862 THEN 'ALBUMIN'
WHEN itemid = 51144 THEN 'BANDS'
WHEN itemid = 50885 THEN 'BILRUBIN'
WHEN itemid = 51478 THEN 'GLUCOSE'
WHEN itemid = 50931 THEN 'GLUCOSE'
WHEN itemid = 51221 THEN 'HEMATOCRIT'
WHEN itemid = 50813 THEN 'LACTATE'
WHEN itemid = 51275 THEN 'PTT'
WHEN itemid = 51237 THEN 'INR'
ELSE null
END AS LABEL
FROM mimiciv.icustays icu
INNER JOIN mimiciv.admissions adm
ON icu.hadm_id = adm.hadm_id
INNER JOIN mimiciv.patients pat
ON icu.subject_id = pat.subject_id
INNER JOIN mimiciv.labevents l
ON l.hadm_id = icu.hadm_id
AND l.charttime >= icu.intime - interval '8 hour'
AND l.charttime <= icu.intime + interval '24 hour'
WHERE l.itemid IN
(
51300,51301 -- wbc
, 50811,51222 -- hgb
, 51265 -- platelet
, 50824, 50983 -- sodium
, 50822, 50971 -- potassium
, 50804 -- Total CO2 or ...
, 50803, 50882 -- bicarbonate
, 50806, 50902 -- chloride
, 51006 -- bun
, 50912 -- creatinine
, 50821, 52042, 50832 -- po2
, 50818, 52040, 50830 -- pco2
, 50820, 52041, 50831 -- ph
, 50802, 52038 -- Base Excess
, 50805 -- carboxyhemoglobin
, 50814 -- methemoglobin
, 50868, 52500 -- aniongap
, 50862 -- albumin
, 51144 -- bands
, 50885 -- bilrubin
, 51478, 50931 -- glucose
, 51221 -- hematocrit
, 50813 -- lactate
, 51275 -- ptt
, 51237 -- inr
)
AND valuenum IS NOT null
)
, labs_rn AS
(
SELECT
stay_id, valuenum, label
, ROW_NUMBER() OVER (PARTITION BY stay_id, label ORDER BY charttime DESC) AS rn
FROM labs_preceeding
)
, labs_grp AS
(
SELECT
stay_id
, COALESCE(MAX(CASE WHEN label = 'BUN' THEN valuenum ELSE null END)) AS BUN
, COALESCE(MAX(CASE WHEN label = 'CHLORIDE' THEN valuenum ELSE null END)) AS CHLORIDE
, COALESCE(MAX(CASE WHEN label = 'CREATININE' THEN valuenum ELSE null END)) AS CREATININE
, COALESCE(MAX(CASE WHEN label = 'HEMOGLOBIN' THEN valuenum ELSE null END)) AS HEMOGLOBIN
, COALESCE(MAX(CASE WHEN label = 'PLATELET' THEN valuenum ELSE null END)) AS PLATELET
, COALESCE(MAX(CASE WHEN label = 'POTASSIUM' THEN valuenum ELSE null END)) AS POTASSIUM
, COALESCE(MAX(CASE WHEN label = 'SODIUM' THEN valuenum ELSE null END)) AS SODIUM
, COALESCE(MAX(CASE WHEN label = 'TOTALCO2' THEN valuenum ELSE null END)) AS TOTALCO2
, COALESCE(MAX(CASE WHEN label = 'WBC' THEN valuenum ELSE null END)) AS WBC
, COALESCE(MAX(CASE WHEN label = 'PO2' THEN valuenum ELSE null END)) AS PO2
, COALESCE(MAX(CASE WHEN label = 'PCO2' THEN valuenum ELSE null END)) AS PCO2
, COALESCE(MAX(CASE WHEN label = 'PH' THEN valuenum ELSE null END)) AS PH
, COALESCE(MAX(CASE WHEN label = 'BASEEXCESS' THEN valuenum ELSE null END)) AS BASEEXCESS
, COALESCE(MAX(CASE WHEN label = 'CARBOXYHEMOGLOBIN' THEN valuenum ELSE null END)) AS CARBOXYHEMOGLOBIN
, COALESCE(MAX(CASE WHEN label = 'METHEMOGLOBIN' THEN valuenum ELSE null END)) AS METHEMOGLOBIN
, COALESCE(MAX(CASE WHEN label = 'ANIONGAP' THEN valuenum ELSE null END)) AS ANIONGAP
, COALESCE(MAX(CASE WHEN label = 'ALBUMIN' THEN valuenum ELSE null END)) AS ALBUMIN
, COALESCE(MAX(CASE WHEN label = 'BANDS' THEN valuenum ELSE null END)) AS BANDS
, COALESCE(MAX(CASE WHEN label = 'BICARBONATE' THEN valuenum ELSE null END)) AS BICARBONATE
, COALESCE(MAX(CASE WHEN label = 'BILRUBIN' THEN valuenum ELSE null END)) AS BILRUBIN
, COALESCE(MAX(CASE WHEN label = 'GLUCOSE' THEN valuenum ELSE null END)) AS GLUCOSE
, COALESCE(MAX(CASE WHEN label = 'HEMATOCRIT' THEN valuenum ELSE null END)) AS HEMATOCRIT
, COALESCE(MAX(CASE WHEN label = 'LACTATE' THEN valuenum ELSE null END)) AS LACTATE
, COALESCE(MAX(CASE WHEN label = 'PTT' THEN valuenum ELSE null END)) AS PTT
, COALESCE(MAX(CASE WHEN label = 'INR' THEN valuenum ELSE null END)) AS INR
FROM labs_rn
WHERE rn = 1
GROUP BY stay_id
)
SELECT icu.stay_id
, lg.bun AS bun_last
, lg.chloride AS chloride_last
, lg.creatinine AS creatinine_last
, lg.HEMOGLOBIN AS hgb_last
, lg.platelet AS platelet_last
, lg.potassium AS potassium_last
, lg.sodium AS sodium_last
, lg.TOTALCO2 AS tco2_last
, lg.wbc AS wbc_last
, lg.po2 AS bg_po2_last
, lg.pco2 AS bg_pco2_last
, lg.ph AS bg_ph_last
, lg.BASEEXCESS AS bg_baseexcess_last
, lg.CARBOXYHEMOGLOBIN AS bg_carboxyhemoglobin_last
, lg.METHEMOGLOBIN AS bg_methemomoglobin_last
, lg.ANIONGAP AS aniongap_last
, lg.ALBUMIN AS albumin_last
, lg.BANDS AS bands_last
, lg.BICARBONATE AS bicarbonate_last
, lg.BILRUBIN AS bilrubin_last
, lg.GLUCOSE AS glucose_last
, lg.HEMATOCRIT AS hematocrit_last
, lg.LACTATE AS lactate_last
, lg.PTT AS ptt_last
, lg.INR AS inr_last
FROM mimiciv.icustays icu
LEFT JOIN labs_grp lg
ON icu.stay_id = lg.stay_id
"""
last_lab = pd.read_sql_query(query, con)
last_lab
query = query_schema + \
"""
WITH vitals_stg_1 AS
(
SELECT icu.stay_id, cev.charttime
, CASE
WHEN itemid = 223761 THEN (cev.valuenum-32)/1.8
ELSE cev.valuenum
END AS valuenum
, CASE
WHEN itemid = 220045 THEN 'HEARTRATE'
WHEN itemid = 220050 THEN 'SYSBP'
WHEN itemid = 220179 THEN 'SYSBP'
WHEN itemid = 220051 THEN 'DIASBP'
WHEN itemid = 220180 THEN 'DIASBP'
WHEN itemid = 220052 THEN 'MEANBP'
WHEN itemid = 220181 THEN 'MEANBP'
WHEN itemid = 225312 THEN 'MEANBP'
WHEN itemid = 220210 THEN 'RESPRATE'
WHEN itemid = 224688 THEN 'RESPRATE'
WHEN itemid = 224689 THEN 'RESPRATE'
WHEN itemid = 224690 THEN 'RESPRATE'
WHEN itemid = 223761 THEN 'TEMPC'
WHEN itemid = 223762 THEN 'TEMPC'
WHEN itemid = 220277 THEN 'SPO2'
WHEN itemid = 220739 THEN 'GCSEYE'
WHEN itemid = 223900 THEN 'GCSVERBAL'
WHEN itemid = 223901 THEN 'GCSMOTOR'
ELSE null
END AS label
FROM mimiciv.icustays icu
INNER JOIN mimiciv.chartevents cev
ON cev.stay_id = icu.stay_id
AND cev.charttime >= icu.intime
AND cev.charttime <= icu.intime + interval '24 hour'
WHERE cev.itemid IN
(
220045 -- heartrate
, 220050, 220179 -- sysbp
, 220051, 220180 -- diasbp
, 220052, 220181, 225312 -- meanbp
, 220210, 224688, 224689, 224690 -- resprate
, 223761, 223762 -- tempc
, 220277 -- SpO2
, 220739 -- gcseye
, 223900 -- gcsverbal
, 223901 -- gscmotor
)
AND valuenum IS NOT null
)
, vitals_stg_2 AS
(
SELECT
stay_id, valuenum, label
, ROW_NUMBER() OVER (PARTITION BY stay_id, label ORDER BY charttime) AS rn
FROM vitals_stg_1
)
, vitals_stg_3 AS
(
SELECT
stay_id
, COALESCE(MAX(CASE WHEN label = 'HEARTRATE' THEN valuenum ELSE null END)) AS heartrate_first
, COALESCE(MAX(CASE WHEN label = 'SYSBP' THEN valuenum ELSE null END)) AS sysbp_first
, COALESCE(MAX(CASE WHEN label = 'DIASBP' THEN valuenum ELSE null END)) AS diabp_first
, COALESCE(MAX(CASE WHEN label = 'MEANBP' THEN valuenum ELSE null END)) AS meanbp_first
, COALESCE(MAX(CASE WHEN label = 'RESPRATE' THEN valuenum ELSE null END)) AS resprate_first
, COALESCE(MAX(CASE WHEN label = 'TEMPC' THEN valuenum ELSE null END)) AS tempc_first
, COALESCE(MAX(CASE WHEN label = 'SPO2' THEN valuenum ELSE null END)) AS spo2_first
, COALESCE(MAX(CASE WHEN label = 'GCSEYE' THEN valuenum ELSE null END)) AS gcseye_first
, COALESCE(MAX(CASE WHEN label = 'GCSVERBAL' THEN valuenum ELSE null END)) AS gcsverbal_first
, COALESCE(MAX(CASE WHEN label = 'GCSMOTOR' THEN valuenum ELSE null END)) AS gcsmotor_first
FROM vitals_stg_2
WHERE rn = 1
GROUP BY stay_id
)
SELECT * FROM vitals_stg_3
"""
first_vitals = pd.read_sql_query(query, con)
first_vitals
query = query_schema + \
"""
WITH vitals_stg_1 AS
(
SELECT icu.stay_id, cev.charttime
, CASE
WHEN itemid = 223761 THEN (cev.valuenum-32)/1.8
ELSE cev.valuenum
END AS valuenum
, CASE
WHEN itemid = 220045 THEN 'HEARTRATE'
WHEN itemid = 220050 THEN 'SYSBP'
WHEN itemid = 220179 THEN 'SYSBP'
WHEN itemid = 220051 THEN 'DIASBP'
WHEN itemid = 220180 THEN 'DIASBP'
WHEN itemid = 220052 THEN 'MEANBP'
WHEN itemid = 220181 THEN 'MEANBP'
WHEN itemid = 225312 THEN 'MEANBP'
WHEN itemid = 220210 THEN 'RESPRATE'
WHEN itemid = 224688 THEN 'RESPRATE'
WHEN itemid = 224689 THEN 'RESPRATE'
WHEN itemid = 224690 THEN 'RESPRATE'
WHEN itemid = 223761 THEN 'TEMPC'
WHEN itemid = 223762 THEN 'TEMPC'
WHEN itemid = 220277 THEN 'SPO2'
WHEN itemid = 220739 THEN 'GCSEYE'
WHEN itemid = 223900 THEN 'GCSVERBAL'
WHEN itemid = 223901 THEN 'GCSMOTOR'
ELSE null
END AS label
FROM mimiciv.icustays icu
INNER JOIN mimiciv.chartevents cev
ON cev.stay_id = icu.stay_id
AND cev.charttime >= icu.intime
AND cev.charttime <= icu.intime + interval '24 hour'
WHERE cev.itemid IN
(
220045 -- heartrate
, 220050, 220179 -- sysbp
, 220051, 220180 -- diasbp
, 220052, 220181, 225312 -- meanbp
, 220210, 224688, 224689, 224690 -- resprate
, 223761, 223762 -- tempc
, 220277 -- SpO2
, 220739 -- gcseye
, 223900 -- gcsverbal
, 223901 -- gscmotor
)
AND valuenum IS NOT null
)
, vitals_stg_2 AS
(
SELECT
stay_id, valuenum, label
, ROW_NUMBER() OVER (PARTITION BY stay_id, label ORDER BY charttime DESC) AS rn
FROM vitals_stg_1
)
, vitals_stg_3 AS
(
SELECT
stay_id
, COALESCE(MAX(CASE WHEN label = 'HEARTRATE' THEN valuenum ELSE null END)) AS heartrate_last
, COALESCE(MAX(CASE WHEN label = 'SYSBP' THEN valuenum ELSE null END)) AS sysbp_last
, COALESCE(MAX(CASE WHEN label = 'DIASBP' THEN valuenum ELSE null END)) AS diabp_last
, COALESCE(MAX(CASE WHEN label = 'MEANBP' THEN valuenum ELSE null END)) AS meanbp_last
, COALESCE(MAX(CASE WHEN label = 'RESPRATE' THEN valuenum ELSE null END)) AS resprate_last
, COALESCE(MAX(CASE WHEN label = 'TEMPC' THEN valuenum ELSE null END)) AS tempc_last
, COALESCE(MAX(CASE WHEN label = 'SPO2' THEN valuenum ELSE null END)) AS spo2_last
, COALESCE(MAX(CASE WHEN label = 'GCSEYE' THEN valuenum ELSE null END)) AS gcseye_last
, COALESCE(MAX(CASE WHEN label = 'GCSVERBAL' THEN valuenum ELSE null END)) AS gcsverbal_last
, COALESCE(MAX(CASE WHEN label = 'GCSMOTOR' THEN valuenum ELSE null END)) AS gcsmotor_last
FROM vitals_stg_2
WHERE rn = 1
GROUP BY stay_id
)
SELECT * FROM vitals_stg_3
"""
last_vitals = pd.read_sql_query(query, con)
last_vitals
query = query_schema + \
"""
WITH vitals_stg_1 AS
(
SELECT icu.stay_id, cev.charttime
, CASE
WHEN itemid = 223761 THEN (cev.valuenum-32)/1.8
ELSE cev.valuenum
END AS valuenum
, CASE
WHEN itemid = 220045 THEN 'HEARTRATE'
WHEN itemid = 220050 THEN 'SYSBP'
WHEN itemid = 220179 THEN 'SYSBP'
WHEN itemid = 220051 THEN 'DIASBP'
WHEN itemid = 220180 THEN 'DIASBP'
WHEN itemid = 220052 THEN 'MEANBP'
WHEN itemid = 220181 THEN 'MEANBP'
WHEN itemid = 225312 THEN 'MEANBP'
WHEN itemid = 220210 THEN 'RESPRATE'
WHEN itemid = 224688 THEN 'RESPRATE'
WHEN itemid = 224689 THEN 'RESPRATE'
WHEN itemid = 224690 THEN 'RESPRATE'
WHEN itemid = 223761 THEN 'TEMPC'
WHEN itemid = 223762 THEN 'TEMPC'
WHEN itemid = 220277 THEN 'SPO2'
WHEN itemid = 220739 THEN 'GCSEYE'
WHEN itemid = 223900 THEN 'GCSVERBAL'
WHEN itemid = 223901 THEN 'GCSMOTOR'
ELSE null
END AS label
FROM mimiciv.icustays icu
INNER JOIN mimiciv.chartevents cev
ON cev.stay_id = icu.stay_id
AND cev.charttime >= icu.intime
AND cev.charttime <= icu.intime + interval '24 hour'
WHERE cev.itemid IN
(
220045 -- heartrate
, 220050, 220179 -- sysbp
, 220051, 220180 -- diasbp
, 220052, 220181, 225312 -- meanbp
, 220210, 224688, 224689, 224690 -- resprate
, 223761, 223762 -- tempc
, 220277 -- SpO2
, 220739 -- gcseye
, 223900 -- gcsverbal
, 223901 -- gscmotor
)
AND valuenum IS NOT null
)
, vitals_stg_2 AS
(
SELECT
stay_id, valuenum, label
, ROW_NUMBER() OVER (PARTITION BY stay_id, label ORDER BY charttime) AS rn
FROM vitals_stg_1
)
, vitals_stg_3 AS
(
SELECT
stay_id
, rn
, COALESCE(MAX(CASE WHEN label = 'HEARTRATE' THEN valuenum ELSE null END)) AS heartrate
, COALESCE(MAX(CASE WHEN label = 'SYSBP' THEN valuenum ELSE null END)) AS sysbp
, COALESCE(MAX(CASE WHEN label = 'DIASBP' THEN valuenum ELSE null END)) AS diabp
, COALESCE(MAX(CASE WHEN label = 'MEANBP' THEN valuenum ELSE null END)) AS meanbp
, COALESCE(MAX(CASE WHEN label = 'RESPRATE' THEN valuenum ELSE null END)) AS resprate
, COALESCE(MAX(CASE WHEN label = 'TEMPC' THEN valuenum ELSE null END)) AS tempc
, COALESCE(MAX(CASE WHEN label = 'SPO2' THEN valuenum ELSE null END)) AS spo2
, COALESCE(MAX(CASE WHEN label = 'GCSEYE' THEN valuenum ELSE null END)) AS gcseye
, COALESCE(MAX(CASE WHEN label = 'GCSVERBAL' THEN valuenum ELSE null END)) AS gcsverbal
, COALESCE(MAX(CASE WHEN label = 'GCSMOTOR' THEN valuenum ELSE null END)) AS gcsmotor
FROM vitals_stg_2
GROUP BY stay_id, rn
)
, vitals_stg_4 AS
(
SELECT
stay_id,
MAX(heartrate) AS heartrate_max
, MAX(sysbp) AS sysbp_max
, MAX(diabp) AS diabp_max
, MAX(meanbp) AS meanbp_max
, MAX(resprate) AS resprate_max
, MAX(tempc) AS tempc_max
, MAX(spo2) AS spo2_max
, MAX(gcseye) AS gcseye_max
, MAX(gcsverbal) AS gcsverbal_max
, MAX(gcsmotor) AS gcsmotor_max
FROM vitals_stg_3
GROUP BY stay_id
)
SELECT * FROM vitals_stg_4
"""
max_vitals = pd.read_sql_query(query, con)
max_vitals
query = query_schema + \
"""
WITH vitals_stg_1 AS
(
SELECT icu.stay_id, cev.charttime
, CASE
WHEN itemid = 223761 THEN (cev.valuenum-32)/1.8
ELSE cev.valuenum
END AS valuenum
, CASE
WHEN itemid = 220045 THEN 'HEARTRATE'
WHEN itemid = 220050 THEN 'SYSBP'
WHEN itemid = 220179 THEN 'SYSBP'
WHEN itemid = 220051 THEN 'DIASBP'
WHEN itemid = 220180 THEN 'DIASBP'
WHEN itemid = 220052 THEN 'MEANBP'
WHEN itemid = 220181 THEN 'MEANBP'
WHEN itemid = 225312 THEN 'MEANBP'
WHEN itemid = 220210 THEN 'RESPRATE'
WHEN itemid = 224688 THEN 'RESPRATE'
WHEN itemid = 224689 THEN 'RESPRATE'
WHEN itemid = 224690 THEN 'RESPRATE'
WHEN itemid = 223761 THEN 'TEMPC'
WHEN itemid = 223762 THEN 'TEMPC'
WHEN itemid = 220277 THEN 'SPO2'
WHEN itemid = 220739 THEN 'GCSEYE'
WHEN itemid = 223900 THEN 'GCSVERBAL'
WHEN itemid = 223901 THEN 'GCSMOTOR'
ELSE null
END AS label
FROM mimiciv.icustays icu
INNER JOIN mimiciv.chartevents cev
ON cev.stay_id = icu.stay_id
AND cev.charttime >= icu.intime
AND cev.charttime <= icu.intime + interval '24 hour'
WHERE cev.itemid IN
(
220045 -- heartrate
, 220050, 220179 -- sysbp
, 220051, 220180 -- diasbp
, 220052, 220181, 225312 -- meanbp
, 220210, 224688, 224689, 224690 -- resprate
, 223761, 223762 -- tempc
, 220277 -- SpO2
, 220739 -- gcseye
, 223900 -- gcsverbal
, 223901 -- gscmotor
)
AND valuenum IS NOT null
)
, vitals_stg_2 AS
(
SELECT
stay_id, valuenum, label
, ROW_NUMBER() OVER (PARTITION BY stay_id, label ORDER BY charttime) AS rn
FROM vitals_stg_1
)
, vitals_stg_3 AS
(
SELECT
stay_id
, rn
, COALESCE(MAX(CASE WHEN label = 'HEARTRATE' THEN valuenum ELSE null END)) AS heartrate
, COALESCE(MAX(CASE WHEN label = 'SYSBP' THEN valuenum ELSE null END)) AS sysbp
, COALESCE(MAX(CASE WHEN label = 'DIASBP' THEN valuenum ELSE null END)) AS diabp
, COALESCE(MAX(CASE WHEN label = 'MEANBP' THEN valuenum ELSE null END)) AS meanbp
, COALESCE(MAX(CASE WHEN label = 'RESPRATE' THEN valuenum ELSE null END)) AS resprate
, COALESCE(MAX(CASE WHEN label = 'TEMPC' THEN valuenum ELSE null END)) AS tempc
, COALESCE(MAX(CASE WHEN label = 'SPO2' THEN valuenum ELSE null END)) AS spo2
, COALESCE(MAX(CASE WHEN label = 'GCSEYE' THEN valuenum ELSE null END)) AS gcseye
, COALESCE(MAX(CASE WHEN label = 'GCSVERBAL' THEN valuenum ELSE null END)) AS gcsverbal
, COALESCE(MAX(CASE WHEN label = 'GCSMOTOR' THEN valuenum ELSE null END)) AS gcsmotor
FROM vitals_stg_2
GROUP BY stay_id, rn
)
, vitals_stg_4 AS
(
SELECT
stay_id,
MIN(heartrate) AS heartrate_min
, MIN(sysbp) AS sysbp_min
, MIN(diabp) AS diabp_min
, MIN(meanbp) AS meanbp_min
, MIN(resprate) AS resprate_min
, MIN(tempc) AS tempc_min
, MIN(spo2) AS spo2_min
, MIN(gcseye) AS gcseye_min
, MIN(gcsverbal) AS gcsverbal_min
, MIN(gcsmotor) AS gcsmotor_min
FROM vitals_stg_3
GROUP BY stay_id
)
SELECT * FROM vitals_stg_4
"""
min_vitals = pd.read_sql_query(query, con)
min_vitals
query = query_schema + \
"""
WITH vitals_stg_1 AS
(
SELECT icu.stay_id, cev.charttime
, CASE
WHEN itemid = 223761 THEN (cev.valuenum-32)/1.8
ELSE cev.valuenum
END AS valuenum
, CASE
WHEN itemid = 220045 THEN 'HEARTRATE'
WHEN itemid = 220050 THEN 'SYSBP'
WHEN itemid = 220179 THEN 'SYSBP'
WHEN itemid = 220051 THEN 'DIASBP'
WHEN itemid = 220180 THEN 'DIASBP'
WHEN itemid = 220052 THEN 'MEANBP'
WHEN itemid = 220181 THEN 'MEANBP'
WHEN itemid = 225312 THEN 'MEANBP'
WHEN itemid = 220210 THEN 'RESPRATE'
WHEN itemid = 224688 THEN 'RESPRATE'
WHEN itemid = 224689 THEN 'RESPRATE'
WHEN itemid = 224690 THEN 'RESPRATE'
WHEN itemid = 223761 THEN 'TEMPC'
WHEN itemid = 223762 THEN 'TEMPC'
WHEN itemid = 220277 THEN 'SPO2'
WHEN itemid = 220739 THEN 'GCSEYE'
WHEN itemid = 223900 THEN 'GCSVERBAL'
WHEN itemid = 223901 THEN 'GCSMOTOR'
ELSE null
END AS label
FROM mimiciv.icustays icu
INNER JOIN mimiciv.chartevents cev
ON cev.stay_id = icu.stay_id
AND cev.charttime >= icu.intime
AND cev.charttime <= icu.intime + interval '24 hour'
WHERE cev.itemid IN
(
220045 -- heartrate
, 220050, 220179 -- sysbp
, 220051, 220180 -- diasbp
, 220052, 220181, 225312 -- meanbp
, 220210, 224688, 224689, 224690 -- resprate
, 223761, 223762 -- tempc
, 220277 -- SpO2
, 220739 -- gcseye
, 223900 -- gcsverbal
, 223901 -- gscmotor
)
AND valuenum IS NOT null
)
, vitals_stg_2 AS
(
SELECT
stay_id, valuenum, label
, ROW_NUMBER() OVER (PARTITION BY stay_id, label ORDER BY charttime) AS rn
FROM vitals_stg_1
)
, vitals_stg_3 AS
(
SELECT
stay_id
, rn
, COALESCE(MAX(CASE WHEN label = 'HEARTRATE' THEN valuenum ELSE null END)) AS heartrate
, COALESCE(MAX(CASE WHEN label = 'SYSBP' THEN valuenum ELSE null END)) AS sysbp
, COALESCE(MAX(CASE WHEN label = 'DIASBP' THEN valuenum ELSE null END)) AS diabp
, COALESCE(MAX(CASE WHEN label = 'MEANBP' THEN valuenum ELSE null END)) AS meanbp
, COALESCE(MAX(CASE WHEN label = 'RESPRATE' THEN valuenum ELSE null END)) AS resprate
, COALESCE(MAX(CASE WHEN label = 'TEMPC' THEN valuenum ELSE null END)) AS tempc
, COALESCE(MAX(CASE WHEN label = 'SPO2' THEN valuenum ELSE null END)) AS spo2
, COALESCE(MAX(CASE WHEN label = 'GCSEYE' THEN valuenum ELSE null END)) AS gcseye
, COALESCE(MAX(CASE WHEN label = 'GCSVERBAL' THEN valuenum ELSE null END)) AS gcsverbal
, COALESCE(MAX(CASE WHEN label = 'GCSMOTOR' THEN valuenum ELSE null END)) AS gcsmotor
FROM vitals_stg_2
GROUP BY stay_id, rn
)
, vitals_stg_4 AS
(
SELECT
stay_id,
AVG(heartrate) AS heartrate_avg
, AVG(sysbp) AS sysbp_avg
, AVG(diabp) AS diabp_avg
, AVG(meanbp) AS meanbp_avg
, AVG(resprate) AS resprate_avg
, AVG(tempc) AS tempc_avg
, AVG(spo2) AS spo2_avg
, AVG(gcseye) AS gcseye_avg
, AVG(gcsverbal) AS gcsverbal_avg
, AVG(gcsmotor) AS gcsmotor_avg
FROM vitals_stg_3
GROUP BY stay_id
)
SELECT * FROM vitals_stg_4
"""
avg_vitals = pd.read_sql_query(query, con)
avg_vitals
query = query_schema + \
"""
SELECT
icu.stay_id, adm.hospital_expire_flag
FROM
mimiciv.icustays icu
INNER JOIN mimiciv.admissions adm
ON adm.hadm_id = icu.hadm_id
"""
mortality = pd.read_sql_query(query, con)
mortality
query = query_schema + \
"""
SELECT
icu.stay_id
FROM
mimiciv.icustays icu
INNER JOIN mimiciv.admissions adm
ON adm.hadm_id = icu.hadm_id
INNER JOIN mimiciv.patients pat
ON pat.subject_id = adm.subject_id
AND icu.intime = (
SELECT MAX(icu_max.intime) FROM mimiciv.icustays icu_max WHERE icu_max.hadm_id = icu.hadm_id
)
WHERE (icu.outtime >= icu.intime + interval '24 hour')
AND (FLOOR(DATE_PART('day', adm.admittime - make_timestamp(pat.anchor_year, 1, 1, 0, 0, 0))/365.0) + pat.anchor_age) > 18
"""
filtered = pd.read_sql_query(query, con)
filtered
static.shape, first_lab.shape, last_lab.shape, first_vitals.shape, last_vitals.shape, max_vitals.shape, min_vitals.shape, avg_vitals.shape, mortality.shape, filtered.shape
dfs = [filtered, static, first_lab, last_lab, first_vitals, last_vitals, max_vitals, min_vitals, avg_vitals, mortality]
data = reduce(lambda left, right: pd.merge(left, right, on=['stay_id'], how='inner'), dfs)
na_counts = data.isna().sum().reset_index(name="n")
na_counts = na_counts.sort_values(by=['n'], ascending=[False])
plt.figure(figsize=(10,40))
plt.tick_params(top="on")
plt.barh(y=na_counts['index'], width=na_counts.n)
data.drop(columns=['subject_id', 'hadm_id', 'stay_id'], inplace=True)
data.shape
# convert to numpy data (assumes target, death, is the last column)
X = data.values
y = X[:,-1]
y = y.astype('int')
X = X[:,0:-1]
X_header = [x for x in data.columns.values]
X_header = X_header[0:-1]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
estimator = Pipeline([("imputer", SimpleImputer(missing_values=np.nan, strategy="mean")),
("scaler", StandardScaler()),
("logreg" , LogisticRegression(fit_intercept=True))])
mdl = estimator.fit(X_train, y_train)
probs = mdl.predict_proba(X_test)
probs = probs[:,1]
preds = mdl.predict(X_test)
# calculate score (AUROC)
roc_score = metrics.roc_auc_score(y_test, probs)
roc_score
# calculate score (AUROC)
classification_report = metrics.classification_report(y_test, preds)
print(classification_report)
lrDf = pd.DataFrame(data=list(zip(X_header, mdl.named_steps['logreg'].coef_[0])), columns=['feature', 'weight'])
lrDf.sort_values('weight', inplace=True)
plt.figure(figsize=(10,40))
plt.barh(y=lrDf.feature, width=lrDf.weight)
estimator = Pipeline([("imputer", SimpleImputer(missing_values=np.nan, strategy="mean")),
("scaler", StandardScaler()),
("xgb" , xgb.XGBClassifier(max_depth=3, n_estimators=300, learning_rate=0.05))])
mdl = estimator.fit(X_train, y_train)
probs = mdl.predict_proba(X_test)
probs = probs[:,1]
preds = mdl.predict(X_test)
# calculate score (AUROC)
roc_score = metrics.roc_auc_score(y_test, probs)
roc_score
# calculate score (AUROC)
classification_report = metrics.classification_report(y_test, preds)
print(classification_report)
feature_importance_list = []
feature_imp_dict = mdl.named_steps['xgb'].get_booster().get_score(importance_type="gain")
for feature_imp in feature_imp_dict:
feature_importance_list.append([X_header[int(feature_imp[1:])], feature_imp_dict[feature_imp]])
xgbDf = pd.DataFrame(feature_importance_list, columns = ['feature', 'weight'])
xgbDf.sort_values('weight', inplace=True)
plt.figure(figsize=(10,40))
plt.barh(y=xgbDf.feature, width=xgbDf.weight)
estimator = Pipeline([("imputer", SimpleImputer(missing_values=np.nan, strategy="mean")),
("scaler", StandardScaler()),
("mlp" , MLPClassifier(hidden_layer_sizes=10))])
mdl = estimator.fit(X_train, y_train)
probs = mdl.predict_proba(X_test)
probs = probs[:,1]
preds = mdl.predict(X_test)
# calculate score (AUROC)
roc_score = metrics.roc_auc_score(y_test, probs)
roc_score
# calculate score (AUROC)
classification_report = metrics.classification_report(y_test, preds)
print(classification_report)
X_train_array = np.asarray(X_train).astype('float32')
X_test_array = np.asarray(X_test).astype('float32')
imp_mean = SimpleImputer(missing_values=np.nan, strategy='mean')
imp_mean.fit(X_train_array)
X_train_imputed = imp_mean.transform(X_train_array)
X_test_imputed = imp_mean.transform(X_test_array)
normalizer = preprocessing.Normalization()
normalizer.adapt(X_train_imputed)
X_train_noralized = normalizer(X_train_imputed)
X_test_noralized = normalizer(X_test_imputed)
model = Sequential()
model.add(layers.Dense(X_train.shape[1], activation=tf.nn.relu, kernel_initializer='he_normal', bias_initializer='zeros'))
model.add(layers.Dense(40, activation=tf.nn.relu, kernel_initializer='he_normal', bias_initializer='zeros'))
model.add(layers.Dense(1, activation=tf.nn.sigmoid, kernel_initializer='he_normal', bias_initializer='zeros'))
model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
model.fit(X_train_noralized, y_train, validation_data=(X_test_noralized, y_test), epochs=5, batch_size=256)
model.summary()
probs = model.predict_proba(X_test_noralized)
preds = np.around(probs)
# calculate score (AUROC)
roc_score = metrics.roc_auc_score(y_test, probs)
roc_score
# calculate score (AUROC)
classification_report = metrics.classification_report(y_test, preds)
print(classification_report)
cm = confusion_matrix(y_test, preds, labels=[0, 1])
disp = ConfusionMatrixDisplay(confusion_matrix=cm, display_labels=['survided', 'not-survived'])
disp.plot()
| 0.234582 | 0.616878 |
<a href="https://colab.research.google.com/github/yukinaga/ai_programming/blob/main/lecture_05/01_gradient_decent.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# 勾配降下法
勾配降下法では、関数の傾き(勾配)に基づき関数を最小化します。
ディープラーニングにおいて、出力と正解の誤差を最小化するために使われます。
## 勾配降下法とは?
「勾配降下法」は勾配法の一種で、勾配に基づき最小値を探索します。
こちらの多変数関数、$y=f(X)$の最小値を勾配降下法により探索する例を考えます。
$$f(X) = f(x_1,x_2,\cdots, x_i,\cdots, x_n)$$
このとき、$X$の初期値を適当に決めた上で、以下のような式に基づき$X$の全ての要素を更新します。
(式 1)
$$ x_i \leftarrow x_i-\eta\frac{\partial f(X)}{\partial x_i} $$
ここで$\eta$は学習係数と呼ばれる定数で、$x_i$の更新速度を決めます。
この式により、勾配$\frac{\partial f(X)}{\partial x_i} $が大きいほど(傾きが大きいほど)、$x_i$の値は大きく変更されることになります。
これを$f(X)$が変化しなくなるまで(勾配が0になるまで)繰り返すことで、$f(X)$の最小値を求めます。
## 勾配降下法の実装
以下の簡単な一変数関数$f(x)$の最小値を、勾配降下法を使って求めます。
$$f(x) = x^2 - 2x$$
この関数は、$x$の値が1のときに最小値$f(1) = -1$をとります。また、この関数を$x$で微分すると次の通りです。
$$\frac{d f(x)}{d x} = 2x-2$$
一変数なので、偏微分ではなく通常の微分を使っています。
以下のコードは、上記の関数の最小値を、勾配降下法により求めます。
(式1)を使って20回$x$を更新し、その過程を最後にグラフで表示します。
```
import numpy as np
import matplotlib.pyplot as plt
def my_func(x): # 最小値を求める関数
return x**2 - 2*x
def grad_func(x): # 導関数
return 2*x - 2
eta = 0.1 # 学習係数
x = 4.0 # xに初期値を設定
record_x = [] # xの記録
record_y = [] # yの記録
for i in range(20): # 20回xを更新する
y = my_func(x)
record_x.append(x)
record_y.append(y)
x -= eta * grad_func(x) # (式1)
x_f = np.linspace(-2, 4) # 表示範囲
y_f = my_func(x_f)
plt.plot(x_f, y_f, linestyle="dashed") # 関数を点線で描画
plt.scatter(record_x, record_y) # xとyの記録を点で表示
plt.xlabel("x", size=14)
plt.ylabel("y", size=14)
plt.grid()
plt.show()
```
$x$の初期値は4ですが、そこから関数を滑り落ちるようにして最小値付近に到達しました。
次第に$x$の間隔は狭くなっており、勾配が小さくなるとともに$x$の更新量が小さくなることが確認できます。
勾配降下法により求められる最小値は厳密な最小値ではありませんが、現実の問題を扱う際は関数の形状さえ分からないことが多いので、勾配降下法により最小値を少しずつ探索するアプローチは有効です。
## 局所的な最小値
最小値には、全体の最小値と局所的な最小値があります。
先程の例では関数が比較的単純なので、全体の最小値にあっさりとたどり着くことができました。
しかしながら、ニューラルネットワークは複雑な関数なので、局所的な最小値に捕獲されて全体の最小値にたどり着けないことがあります。
以下では、局所的な最小値の例を見ていきます。以下の関数$f(x)$の最小値を、勾配降下法を使って求めます。
$$f(x) = x^4 + 2x^3 -3x^2 - 2x$$
この関数を$x$で微分すると次のようになります。
$$\frac{d f(x)}{d x} = 4x^3 + 6x^2 - 6x - 2$$
以下のコードでは、上記の関数に勾配降下法を適用しています。
```
import numpy as np
import matplotlib.pyplot as plt
def my_func(x): # 最小値を求める関数
return x**4 + 2*x**3 - 3*x**2 - 2*x
def grad_func(x): # 導関数
return 4*x**3 + 6*x**2 - 6*x - 2
eta = 0.01 # 学習係数
x = 1.6 # xに初期値を設定
record_x = [] # xの記録
record_y = [] # yの記録
for i in range(20): # 20回xを更新する
y = my_func(x)
record_x.append(x)
record_y.append(y)
x -= eta * grad_func(x) # (式1)
x_f = np.linspace(-2.8, 1.6) # 表示範囲
y_f = my_func(x_f)
plt.plot(x_f, y_f, linestyle="dashed") # 関数を点線で描画
plt.scatter(record_x, record_y) # xとyの記録を点で表示
plt.xlabel("x", size=14)
plt.ylabel("y", size=14)
plt.grid()
plt.show()
```
関数の曲線が点線で表されていますが、左側の谷が全体の最小値で、右側の谷が局所的な最小値です。
上記のコードでは`x = 1.6`を初期値としましたが、右側の谷に捕獲されて抜け出せなくなってしまいます。
ディープラーニングにおいて、このような局所的な最小値に捕らわれてしまうのは深刻な問題です。
適切に初期値を設定したり、あるいはランダムな動きを導入したりして、このような問題への対策が行われています。
今回のケースでは、初期値が適切に設定されれば全体の最小値にたどり着くことができます。
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
def my_func(x): # 最小値を求める関数
return x**2 - 2*x
def grad_func(x): # 導関数
return 2*x - 2
eta = 0.1 # 学習係数
x = 4.0 # xに初期値を設定
record_x = [] # xの記録
record_y = [] # yの記録
for i in range(20): # 20回xを更新する
y = my_func(x)
record_x.append(x)
record_y.append(y)
x -= eta * grad_func(x) # (式1)
x_f = np.linspace(-2, 4) # 表示範囲
y_f = my_func(x_f)
plt.plot(x_f, y_f, linestyle="dashed") # 関数を点線で描画
plt.scatter(record_x, record_y) # xとyの記録を点で表示
plt.xlabel("x", size=14)
plt.ylabel("y", size=14)
plt.grid()
plt.show()
import numpy as np
import matplotlib.pyplot as plt
def my_func(x): # 最小値を求める関数
return x**4 + 2*x**3 - 3*x**2 - 2*x
def grad_func(x): # 導関数
return 4*x**3 + 6*x**2 - 6*x - 2
eta = 0.01 # 学習係数
x = 1.6 # xに初期値を設定
record_x = [] # xの記録
record_y = [] # yの記録
for i in range(20): # 20回xを更新する
y = my_func(x)
record_x.append(x)
record_y.append(y)
x -= eta * grad_func(x) # (式1)
x_f = np.linspace(-2.8, 1.6) # 表示範囲
y_f = my_func(x_f)
plt.plot(x_f, y_f, linestyle="dashed") # 関数を点線で描画
plt.scatter(record_x, record_y) # xとyの記録を点で表示
plt.xlabel("x", size=14)
plt.ylabel("y", size=14)
plt.grid()
plt.show()
| 0.209551 | 0.976669 |
<a href="https://colab.research.google.com/github/amita-kapoor/UO-Artificial-Intelligence-Cloud-and-Edge-Implementations/blob/master/Excercise_Classification.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
## Classification Exercises
For these exercises use the GPU in Google Colab. To enable GPU go to top menu bar in **EDIT** menu go to **NoteBook Settings**. Once you click it a window opens, in the hardware accelerator dropdown menu choose GPU.

### Introduction
We have already learned about Neural Networks and discussed Multilayered Perceptrons in depth. In this exercise, we will be testing our understanding of the underlying concepts with special emphasis to [Hyperparameter tuning](https://towardsdatascience.com/understanding-hyperparameters-and-its-optimisation-techniques-f0debba07568).
After doing these exercises, you would be able to better understand:
* The architecture of a neural network
* The parameters (training) of a neural network and how they change with changing architecture.
* Hyperparameter tuning: batch size, number of hidden units and optimizers.
We encourage you to work with other hyperparameters as well like learning rate, number of layers, activation functions etc. And in the end there is an optional exercise, where you can see if what you observe for the MNIST dataset is true for other dataset as well.
The Notebook is divided in three parts: Building the Model, Reading the dataset and Hyperparameters. It contains five exercises in total and one additional optional exercise:
* [Exercise 1](#ex_1)
* [Exercise 2](#ex_2)
* [Exercise 3](#ex_3)
* [Exercise 4](#ex_4)
* [Exercise 5](#ex_5)
* [Optional Exercise](#ex_O)
You have to do all the five exercises. Run the code given with each exercise and write down your answer just below each exercise. Wish you all the best.
### Part 1: Building the model
Below we define a function to built a neural network model using TensorFlow Keras.
```
import tensorflow as tf
import numpy as np
from tensorflow import keras
def built_model(input_shape, n_hidden, nb_classes, optimizer='SGD'):
'''
The function builds a fully connected neural network with two hidden layers
Arguments:
input_shape: The number of inputs to the neural network
n_hidden: Number of hidden neurons in the hidden layers
nb_classes: Number of neurons in the output layer
optimizer: The optimizer used to train the model.
By default we use Stochastic Gradient Descent.
Returns:
The function returns A model with loss and optimizer defined
'''
model = tf.keras.models.Sequential()
## First Hidden layer
model.add(keras.layers.Dense(n_hidden,
input_shape=(input_shape,),
name='dense_layer', activation='relu'))
## Second Hidden Layer
model.add(keras.layers.Dense(n_hidden,
name='dense_layer_2', activation='relu'))
## Output Layer
model.add(keras.layers.Dense(nb_classes,
name='dense_layer_3', activation='softmax'))
## Define loss and optimizer
model.compile(optimizer=optimizer,
loss='categorical_crossentropy',
metrics=['accuracy'])
return model
```
<a id='ex_1'></a>
**Exercise 1** What should be the values of the arguments `INPUT_SHAPE`: the number of input units, `N_HIDDEN`: the number of hidden units, and `NB_CLASSES`: the number of output units, if we want to build a model using `built_model` function with the specifications given in the figure:

To build this network we used TensorFlow Keras `plot_model` function available in `utils` model. You can learn more about the function from [TensorFlow docs](https://www.tensorflow.org/api_docs/python/tf/keras/utils/plot_model).
```
# Task to do
INPUT_SHAPE = #?
N_HIDDEN = #?
NB_CLASSES = #?
## Do not change anything below
assert(INPUT_SHAPE == 5), "Input shape incorrect"
assert(N_HIDDEN == 10), "Number of hidden neurons incorrect"
assert(NB_CLASSES == 2), "Number of output units incorrect"
model = built_model(INPUT_SHAPE, N_HIDDEN,NB_CLASSES)
```
<a id='ex_2'></a>
**Exercise 2** Based on the input, hidden and output units what are the total number of trainable parameters in this model?
```
# Task to do
trainable_parameters = #?
## Do not change anything below
assert trainable_parameters==model.count_params(), "Your answer is incorrect"
print("Number of trainable parameters in the model are", trainable_parameters)
```
Good work! Let us now visualize the summary of the model created.
```
model.summary()
```
### Part 2: Reading the dataset
We will continue with the MNIST dataset.
###### Just run the cells in this part of the notebook. Do not change anything.
```
mnist = keras.datasets.mnist
(X_train, Y_train), (X_test, Y_test) = mnist.load_data()
# Processing the data
assert(len(X_train.shape)==3), "The input data is not of the right shape"
RESHAPED = X_train.shape[1]*X_train.shape[2]
X_train = X_train.reshape(60000, RESHAPED)
X_test = X_test.reshape(10000, RESHAPED)
X_train = X_train.astype('float32')
X_test = X_test.astype('float32')
# Data Normalization
X_train, X_test = X_train / 255.0, X_test / 255.0
print(X_train.shape[0], 'train samples')
print(X_test.shape[0], 'test samples')
```
For the MNIST dataset the number of input and number of output units are fixed. However we can choose different values of hidden units.
```
INPUT_SHAPE = RESHAPED
NB_CLASSES = len(set(Y_train))
# one-hot encode
Y_train = tf.keras.utils.to_categorical(Y_train, NB_CLASSES)
Y_test = tf.keras.utils.to_categorical(Y_test, NB_CLASSES)
```
### Part 3: Hyperparameters
<a id='ex_3'></a>
**Exercise 3:** The aim of this exercise is to understand the affect of changing number of hidden units on the model performance. Change the number of hidden units, and train the model. Compare the model performance in terms of accuracy. What do you understand from this?
**Answer** Please type your answer here (Double click to edit)
```
# Task to do choose different values for number of hidden units (minimum five different values)
N_HIDDEN = #? Choose a value
## Do not change anything below
model = built_model(INPUT_SHAPE,N_HIDDEN, NB_CLASSES)
history = model.fit(X_train, Y_train,
batch_size=128, epochs=50,
verbose=1, validation_split=0.2)
# Evaluate the model
test_loss, test_acc = model.evaluate(X_test, Y_test)
print('Test accuracy: {:.2f} %'.format(test_acc*100))
```
<a id='ex_4'></a>
**Exercise 4:** Let us now repeat the same after changing the batch size (minimum 5 different values). Compare the model performance in terms of accuracy. What do you understand from this?
**Answer** Please type your answer here (Double click to edit)
```
# Task to do choose different values for batch size (minimum five different values)
BATCH_SIZE = #? Choose a value
## Do not change anything below
model = built_model(INPUT_SHAPE,128, NB_CLASSES)
history = model.fit(X_train, Y_train,
batch_size=BATCH_SIZE, epochs=50,
verbose=1, validation_split=0.2)
# Evaluate the model
test_loss, test_acc = model.evaluate(X_test, Y_test)
print('Test accuracy: {:.2f} %'.format(test_acc*100))
```
<a id='ex_5'></a>
**Exercise 5:** And now we do the same with different [optimizers](https://www.tensorflow.org/api_docs/python/tf/keras/optimizers) available in TensorFlow. Change the optimizers and compare the model performance in terms of accuracy. What do you understand from this?
**Answer** Please type your answer here (Double click to edit)
```
# Task to do choose different optimizers
opt = #? Choose from available optimizers
## Do not change anything below
N_HIDDEN = 128
model = built_model(INPUT_SHAPE,N_HIDDEN, NB_CLASSES, opt)
history = model.fit(X_train, Y_train,
batch_size=128, epochs=50,
verbose=1, validation_split=0.2)
# Evaluate the model
test_loss, test_acc = model.evaluate(X_test, Y_test)
print('Test accuracy: {:.2f} %'.format(test_acc*100))
```
<a id='ex_O'></a>
### Optional Exercise: Fashion MNIST
Repeat the above exercises (3-5) with different dataset. You can use Fashion MNIST another popular ML dataset. Are the results same? Comment.
To download fashion mnist you can use the following code:
```
fashion_mnist = keras.datasets.fashion_mnist
(X_train, Y_train), (X_test, Y_test) = fashion_mnist.load_data()
```
|
github_jupyter
|
import tensorflow as tf
import numpy as np
from tensorflow import keras
def built_model(input_shape, n_hidden, nb_classes, optimizer='SGD'):
'''
The function builds a fully connected neural network with two hidden layers
Arguments:
input_shape: The number of inputs to the neural network
n_hidden: Number of hidden neurons in the hidden layers
nb_classes: Number of neurons in the output layer
optimizer: The optimizer used to train the model.
By default we use Stochastic Gradient Descent.
Returns:
The function returns A model with loss and optimizer defined
'''
model = tf.keras.models.Sequential()
## First Hidden layer
model.add(keras.layers.Dense(n_hidden,
input_shape=(input_shape,),
name='dense_layer', activation='relu'))
## Second Hidden Layer
model.add(keras.layers.Dense(n_hidden,
name='dense_layer_2', activation='relu'))
## Output Layer
model.add(keras.layers.Dense(nb_classes,
name='dense_layer_3', activation='softmax'))
## Define loss and optimizer
model.compile(optimizer=optimizer,
loss='categorical_crossentropy',
metrics=['accuracy'])
return model
# Task to do
INPUT_SHAPE = #?
N_HIDDEN = #?
NB_CLASSES = #?
## Do not change anything below
assert(INPUT_SHAPE == 5), "Input shape incorrect"
assert(N_HIDDEN == 10), "Number of hidden neurons incorrect"
assert(NB_CLASSES == 2), "Number of output units incorrect"
model = built_model(INPUT_SHAPE, N_HIDDEN,NB_CLASSES)
# Task to do
trainable_parameters = #?
## Do not change anything below
assert trainable_parameters==model.count_params(), "Your answer is incorrect"
print("Number of trainable parameters in the model are", trainable_parameters)
model.summary()
mnist = keras.datasets.mnist
(X_train, Y_train), (X_test, Y_test) = mnist.load_data()
# Processing the data
assert(len(X_train.shape)==3), "The input data is not of the right shape"
RESHAPED = X_train.shape[1]*X_train.shape[2]
X_train = X_train.reshape(60000, RESHAPED)
X_test = X_test.reshape(10000, RESHAPED)
X_train = X_train.astype('float32')
X_test = X_test.astype('float32')
# Data Normalization
X_train, X_test = X_train / 255.0, X_test / 255.0
print(X_train.shape[0], 'train samples')
print(X_test.shape[0], 'test samples')
INPUT_SHAPE = RESHAPED
NB_CLASSES = len(set(Y_train))
# one-hot encode
Y_train = tf.keras.utils.to_categorical(Y_train, NB_CLASSES)
Y_test = tf.keras.utils.to_categorical(Y_test, NB_CLASSES)
# Task to do choose different values for number of hidden units (minimum five different values)
N_HIDDEN = #? Choose a value
## Do not change anything below
model = built_model(INPUT_SHAPE,N_HIDDEN, NB_CLASSES)
history = model.fit(X_train, Y_train,
batch_size=128, epochs=50,
verbose=1, validation_split=0.2)
# Evaluate the model
test_loss, test_acc = model.evaluate(X_test, Y_test)
print('Test accuracy: {:.2f} %'.format(test_acc*100))
# Task to do choose different values for batch size (minimum five different values)
BATCH_SIZE = #? Choose a value
## Do not change anything below
model = built_model(INPUT_SHAPE,128, NB_CLASSES)
history = model.fit(X_train, Y_train,
batch_size=BATCH_SIZE, epochs=50,
verbose=1, validation_split=0.2)
# Evaluate the model
test_loss, test_acc = model.evaluate(X_test, Y_test)
print('Test accuracy: {:.2f} %'.format(test_acc*100))
# Task to do choose different optimizers
opt = #? Choose from available optimizers
## Do not change anything below
N_HIDDEN = 128
model = built_model(INPUT_SHAPE,N_HIDDEN, NB_CLASSES, opt)
history = model.fit(X_train, Y_train,
batch_size=128, epochs=50,
verbose=1, validation_split=0.2)
# Evaluate the model
test_loss, test_acc = model.evaluate(X_test, Y_test)
print('Test accuracy: {:.2f} %'.format(test_acc*100))
fashion_mnist = keras.datasets.fashion_mnist
(X_train, Y_train), (X_test, Y_test) = fashion_mnist.load_data()
| 0.886972 | 0.993029 |
<a href="https://colab.research.google.com/github/aletcher/impossibility-global-convergence/blob/master/impossibility_global_convergence.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
Accompanying code for the paper: [On the Impossibility of Global Convergence in Differentiable Games](https://arxiv.org/pdf/2005.12649.pdf). Implements a number of multi-loss optimization methods that are shown to enter limit cycles instead of converging in a two-parameter zero-sum game, *despite* being weakly-coercive, analytic and nondegenerate. This includes (simultaneous) GD, [AGD](https://arxiv.org/pdf/1907.04392.pdf), [EG](https://arxiv.org/pdf/1906.05945.pdf), [OMD](https://arxiv.org/pdf/1711.00141.pdf), [CO](https://arxiv.org/pdf/1705.10461.pdf), [SGA](https://arxiv.org/pdf/1802.05642.pdf), [LA](https://openreview.net/pdf?id=SyGjjsC5tQ), [LOLA](https://arxiv.org/pdf/1709.04326.pdf), [SOS](https://openreview.net/pdf?id=SyGjjsC5tQ), and [CGD](https://arxiv.org/pdf/1905.12103.pdf). The notebook runs in ~5 minutes with GPU accelerator enabled in colab.
This undesirable phenomenom is shown to apply more generally to *any* 'reasonable' algorithm in the paper, ruling out the possibility of global convergence guarantees in multi-loss optimization.
```
import numpy as np
import torch
import matplotlib.pyplot as plt
import seaborn as sns
plt.style.use('seaborn-darkgrid')
#@markdown Plotting function.
def plot_param(th, algo, start=0):
fig, ax = plt.subplots(nrows=2, ncols=5, figsize=(20, 8))
ax = ax.flatten()
for i, algo in enumerate(algos):
ax[i].set_xlim(-1.45, 1.45)
ax[i].set_ylim(-1.45, 1.45)
ax[i].plot(th_out[i, start:, 0], th_out[i, start:, 1],
sns.xkcd_rgb["prussian blue"], lw=1.5)
ax[i].set_title(algo.upper())
if i < 5:
ax[i].get_xaxis().set_ticklabels([])
if not (i == 0 or i == 5 or i == 4 or i == 9):
ax[i].get_yaxis().set_ticklabels([])
if i == 4 or i == 9:
ax[i].yaxis.tick_right()
plt.show()
#@markdown Gradient computations for each algorithm.
def init_th(dims, std=1):
th = []
for i in range(len(dims)):
if std > 0:
init = torch.nn.init.normal_(torch.empty(dims[i], requires_grad=True), std=std)
else:
init = torch.zeros(dims[i], requires_grad=True)
th.append(init)
return th
def get_gradient(function, param):
try:
grad = torch.autograd.grad(function, param, create_graph=True)[0]
except:
grad = torch.zeros(param.shape)
return grad
def get_hessian(th, grad_L, diag=True, off_diag=True):
n = len(th)
H = []
for i in range(n):
row_block = []
for j in range(n):
if (i == j and diag) or (i != j and off_diag):
block = [torch.unsqueeze(get_gradient(grad_L[i][i][k], th[j]), dim=0)
for k in range(len(th[i]))]
row_block.append(torch.cat(block, dim=0))
else:
row_block.append(torch.zeros(len(th[i]), len(th[j])))
H.append(torch.cat(row_block, dim=1))
return torch.cat(H, dim=0)
def update_th(th, Ls, alpha, algo, sos_a=0.5, sos_b=0.5, co_gam=0.01, omd_th=None):
n = len(th)
losses = Ls(th)
# Compute gradients
grad_L = [[get_gradient(losses[j], th[i]) for j in range(n)] for i in range(n)]
if algo == 'la':
terms = [sum([torch.dot(grad_L[j][i], grad_L[j][j].detach())
for j in range(n) if j != i]) for i in range(n)]
grads = [grad_L[i][i]-alpha*get_gradient(terms[i], th[i]) for i in range(n)]
elif algo == 'lola':
terms = [sum([torch.dot(grad_L[j][i], grad_L[j][j])
for j in range(n) if j != i]) for i in range(n)]
grads = [grad_L[i][i]-alpha*get_gradient(terms[i], th[i]) for i in range(n)]
elif algo == 'sos':
terms = [sum([torch.dot(grad_L[j][i], grad_L[j][j].detach())
for j in range(n) if j != i]) for i in range(n)]
xi_0 = [grad_L[i][i]-alpha*get_gradient(terms[i], th[i]) for i in range(n)]
chi = [get_gradient(sum([torch.dot(grad_L[j][i].detach(), grad_L[j][j])
for j in range(n) if j != i]), th[i]) for i in range(n)]
# Compute p
dot = torch.dot(-alpha*torch.cat(chi), torch.cat(xi_0))
p1 = 1 if dot >= 0 else min(1, -sos_a*torch.norm(torch.cat(xi_0))**2/dot)
xi = torch.cat([grad_L[i][i] for i in range(n)])
xi_norm = torch.norm(xi)
p2 = xi_norm**2 if xi_norm < sos_b else 1
p = min(p1, p2)
grads = [xi_0[i]-p*alpha*chi[i] for i in range(n)]
elif algo == 'sga':
xi = torch.cat([grad_L[i][i] for i in range(n)])
norm = torch.dot(xi, xi.detach())
H_t_xi = [get_gradient(norm, th[i]) for i in range(n)]
H_xi = [get_gradient(sum([torch.dot(grad_L[j][i], grad_L[j][j].detach())
for j in range(n)]), th[i]) for i in range(n)]
A_t_xi = [H_t_xi[i]/2-H_xi[i]/2 for i in range(n)]
# Compute lambda (sga with alignment)
dot_xi = torch.dot(xi, torch.cat(H_t_xi))
dot_A = torch.dot(torch.cat(A_t_xi), torch.cat(H_t_xi))
d = sum([len(th[i]) for i in range(n)])
lam = torch.sign(dot_xi*dot_A/d)
grads = [grad_L[i][i]+lam*A_t_xi[i] for i in range(n)]
elif algo == 'co':
xi = torch.cat([grad_L[i][i] for i in range(n)])
norm = torch.dot(xi, xi.detach())
grads = [grad_L[i][i]+co_gam*get_gradient(norm, th[i]) for i in range(n)]
elif algo == 'eg':
th_eg = [th[i]-alpha*grad_L[i][i] for i in range(n)]
losses_eg = Ls(th_eg)
grads = [get_gradient(losses_eg[i], th_eg[i]) for i in range(n)]
elif algo == 'agd':
th_agd = [th[i] for i in range(n)]
grads = []
for i in range(n):
losses_agd = Ls(th_agd)
grad = get_gradient(losses_agd[i], th_agd[i])
grads.append(grad)
th_agd[i] = th_agd[i]-alpha*grad
elif algo == 'omd':
past_grad = [get_gradient(Ls(omd_th)[i], omd_th[i]) for i in range(n)]
grads = [2*grad_L[i][i]-past_grad[i] for i in range(n)]
elif algo == 'cgd': # Slow implementation (exact matrix inversion)
dims = [len(th[i]) for i in range(n)]
xi = torch.cat([grad_L[i][i] for i in range(n)])
H_o = get_hessian(th, grad_L, diag=False)
grad = torch.matmul(torch.inverse(torch.eye(sum(dims))+alpha*H_o), xi)
grads = [grad[sum(dims[:i]):sum(dims[:i+1])] for i in range(n)]
else: # (simultaneous) GD
grads = [grad_L[i][i] for i in range(n)]
# Update theta
past_th = [th[i].clone() for i in range(n)]
with torch.no_grad():
for i in range(n):
th[i] -= alpha*grads[i]
return th, losses, past_th
#@markdown Game definitions.
def market():
dims = [1, 1]
def Ls(th):
x, y = th
g = y**4/(1+x**2)-x**4/(1+y**2)
L_1 = x*y-x**2/2+x**6/6+g/4
L_2 = -x*y-y**2/2+y**6/6-g/4
return [L_1, L_2]
return dims, Ls
def zerosum():
dims = [1, 1]
def Ls(th):
x, y = th
L_1 = x*y-x**2/2+y**2/2+x**4/4-y**4/4
return [L_1, -L_1]
return dims, Ls
def market_with_min(sig=0.01):
dims = [1, 1]
def Ls(th):
x, y = th
if x**2+y**2 >= sig**2:
f = (x**2+y**2-sig**2)/2
else:
f = (y**2-3*x**2)*(x**2+y**2-sig**2)/(2*sig**2)
g = y**4/(1+x**2)-x**4/(1+y**2)
L_1 = x**6/6-x**2+f+x*y+g/4
L_2 = y**6/6-f-x*y-g/4
return [L_1, L_2]
return dims, Ls
# Run each algorithm on a chosen game (market, zerosum or market_with_min)
dims, Ls = zerosum()
algos = ['gd', 'agd', 'eg', 'omd', 'sga', 'co', 'cgd', 'la', 'lola', 'sos']
alpha = 0.01
num_epochs = 3000
th_out = np.zeros((len(algos), num_epochs, sum(dims)))
for i, algo in enumerate(algos):
th = init_th(dims, std=1)
past_th = th.copy()
losses_out = np.zeros((num_epochs, len(dims)))
for k in range(num_epochs):
th_out[i, k] = [theta.clone().data.numpy() for theta in th]
th, losses, past_th = update_th(th, Ls, alpha, algo, co_gam=0.01, omd_th=past_th)
losses_out[k] = [loss for loss in losses]
# Plot results from epoch=start onwards (for visual clarity)
plot_param(th_out, algos, start=0)
```
|
github_jupyter
|
import numpy as np
import torch
import matplotlib.pyplot as plt
import seaborn as sns
plt.style.use('seaborn-darkgrid')
#@markdown Plotting function.
def plot_param(th, algo, start=0):
fig, ax = plt.subplots(nrows=2, ncols=5, figsize=(20, 8))
ax = ax.flatten()
for i, algo in enumerate(algos):
ax[i].set_xlim(-1.45, 1.45)
ax[i].set_ylim(-1.45, 1.45)
ax[i].plot(th_out[i, start:, 0], th_out[i, start:, 1],
sns.xkcd_rgb["prussian blue"], lw=1.5)
ax[i].set_title(algo.upper())
if i < 5:
ax[i].get_xaxis().set_ticklabels([])
if not (i == 0 or i == 5 or i == 4 or i == 9):
ax[i].get_yaxis().set_ticklabels([])
if i == 4 or i == 9:
ax[i].yaxis.tick_right()
plt.show()
#@markdown Gradient computations for each algorithm.
def init_th(dims, std=1):
th = []
for i in range(len(dims)):
if std > 0:
init = torch.nn.init.normal_(torch.empty(dims[i], requires_grad=True), std=std)
else:
init = torch.zeros(dims[i], requires_grad=True)
th.append(init)
return th
def get_gradient(function, param):
try:
grad = torch.autograd.grad(function, param, create_graph=True)[0]
except:
grad = torch.zeros(param.shape)
return grad
def get_hessian(th, grad_L, diag=True, off_diag=True):
n = len(th)
H = []
for i in range(n):
row_block = []
for j in range(n):
if (i == j and diag) or (i != j and off_diag):
block = [torch.unsqueeze(get_gradient(grad_L[i][i][k], th[j]), dim=0)
for k in range(len(th[i]))]
row_block.append(torch.cat(block, dim=0))
else:
row_block.append(torch.zeros(len(th[i]), len(th[j])))
H.append(torch.cat(row_block, dim=1))
return torch.cat(H, dim=0)
def update_th(th, Ls, alpha, algo, sos_a=0.5, sos_b=0.5, co_gam=0.01, omd_th=None):
n = len(th)
losses = Ls(th)
# Compute gradients
grad_L = [[get_gradient(losses[j], th[i]) for j in range(n)] for i in range(n)]
if algo == 'la':
terms = [sum([torch.dot(grad_L[j][i], grad_L[j][j].detach())
for j in range(n) if j != i]) for i in range(n)]
grads = [grad_L[i][i]-alpha*get_gradient(terms[i], th[i]) for i in range(n)]
elif algo == 'lola':
terms = [sum([torch.dot(grad_L[j][i], grad_L[j][j])
for j in range(n) if j != i]) for i in range(n)]
grads = [grad_L[i][i]-alpha*get_gradient(terms[i], th[i]) for i in range(n)]
elif algo == 'sos':
terms = [sum([torch.dot(grad_L[j][i], grad_L[j][j].detach())
for j in range(n) if j != i]) for i in range(n)]
xi_0 = [grad_L[i][i]-alpha*get_gradient(terms[i], th[i]) for i in range(n)]
chi = [get_gradient(sum([torch.dot(grad_L[j][i].detach(), grad_L[j][j])
for j in range(n) if j != i]), th[i]) for i in range(n)]
# Compute p
dot = torch.dot(-alpha*torch.cat(chi), torch.cat(xi_0))
p1 = 1 if dot >= 0 else min(1, -sos_a*torch.norm(torch.cat(xi_0))**2/dot)
xi = torch.cat([grad_L[i][i] for i in range(n)])
xi_norm = torch.norm(xi)
p2 = xi_norm**2 if xi_norm < sos_b else 1
p = min(p1, p2)
grads = [xi_0[i]-p*alpha*chi[i] for i in range(n)]
elif algo == 'sga':
xi = torch.cat([grad_L[i][i] for i in range(n)])
norm = torch.dot(xi, xi.detach())
H_t_xi = [get_gradient(norm, th[i]) for i in range(n)]
H_xi = [get_gradient(sum([torch.dot(grad_L[j][i], grad_L[j][j].detach())
for j in range(n)]), th[i]) for i in range(n)]
A_t_xi = [H_t_xi[i]/2-H_xi[i]/2 for i in range(n)]
# Compute lambda (sga with alignment)
dot_xi = torch.dot(xi, torch.cat(H_t_xi))
dot_A = torch.dot(torch.cat(A_t_xi), torch.cat(H_t_xi))
d = sum([len(th[i]) for i in range(n)])
lam = torch.sign(dot_xi*dot_A/d)
grads = [grad_L[i][i]+lam*A_t_xi[i] for i in range(n)]
elif algo == 'co':
xi = torch.cat([grad_L[i][i] for i in range(n)])
norm = torch.dot(xi, xi.detach())
grads = [grad_L[i][i]+co_gam*get_gradient(norm, th[i]) for i in range(n)]
elif algo == 'eg':
th_eg = [th[i]-alpha*grad_L[i][i] for i in range(n)]
losses_eg = Ls(th_eg)
grads = [get_gradient(losses_eg[i], th_eg[i]) for i in range(n)]
elif algo == 'agd':
th_agd = [th[i] for i in range(n)]
grads = []
for i in range(n):
losses_agd = Ls(th_agd)
grad = get_gradient(losses_agd[i], th_agd[i])
grads.append(grad)
th_agd[i] = th_agd[i]-alpha*grad
elif algo == 'omd':
past_grad = [get_gradient(Ls(omd_th)[i], omd_th[i]) for i in range(n)]
grads = [2*grad_L[i][i]-past_grad[i] for i in range(n)]
elif algo == 'cgd': # Slow implementation (exact matrix inversion)
dims = [len(th[i]) for i in range(n)]
xi = torch.cat([grad_L[i][i] for i in range(n)])
H_o = get_hessian(th, grad_L, diag=False)
grad = torch.matmul(torch.inverse(torch.eye(sum(dims))+alpha*H_o), xi)
grads = [grad[sum(dims[:i]):sum(dims[:i+1])] for i in range(n)]
else: # (simultaneous) GD
grads = [grad_L[i][i] for i in range(n)]
# Update theta
past_th = [th[i].clone() for i in range(n)]
with torch.no_grad():
for i in range(n):
th[i] -= alpha*grads[i]
return th, losses, past_th
#@markdown Game definitions.
def market():
dims = [1, 1]
def Ls(th):
x, y = th
g = y**4/(1+x**2)-x**4/(1+y**2)
L_1 = x*y-x**2/2+x**6/6+g/4
L_2 = -x*y-y**2/2+y**6/6-g/4
return [L_1, L_2]
return dims, Ls
def zerosum():
dims = [1, 1]
def Ls(th):
x, y = th
L_1 = x*y-x**2/2+y**2/2+x**4/4-y**4/4
return [L_1, -L_1]
return dims, Ls
def market_with_min(sig=0.01):
dims = [1, 1]
def Ls(th):
x, y = th
if x**2+y**2 >= sig**2:
f = (x**2+y**2-sig**2)/2
else:
f = (y**2-3*x**2)*(x**2+y**2-sig**2)/(2*sig**2)
g = y**4/(1+x**2)-x**4/(1+y**2)
L_1 = x**6/6-x**2+f+x*y+g/4
L_2 = y**6/6-f-x*y-g/4
return [L_1, L_2]
return dims, Ls
# Run each algorithm on a chosen game (market, zerosum or market_with_min)
dims, Ls = zerosum()
algos = ['gd', 'agd', 'eg', 'omd', 'sga', 'co', 'cgd', 'la', 'lola', 'sos']
alpha = 0.01
num_epochs = 3000
th_out = np.zeros((len(algos), num_epochs, sum(dims)))
for i, algo in enumerate(algos):
th = init_th(dims, std=1)
past_th = th.copy()
losses_out = np.zeros((num_epochs, len(dims)))
for k in range(num_epochs):
th_out[i, k] = [theta.clone().data.numpy() for theta in th]
th, losses, past_th = update_th(th, Ls, alpha, algo, co_gam=0.01, omd_th=past_th)
losses_out[k] = [loss for loss in losses]
# Plot results from epoch=start onwards (for visual clarity)
plot_param(th_out, algos, start=0)
| 0.550849 | 0.945951 |
```
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
from statsmodels.graphics.tsaplots import plot_pacf
from statsmodels.graphics.tsaplots import plot_acf
from matplotlib.pyplot import figure
from sklearn.metrics import f1_score
from sklearn.metrics import confusion_matrix
def visualize_prediction(confidence, first_f, last_f, no_features, name_time_column, dataframe, title, location_line, line=False):
fig, axes = plt.subplots(2, 2, figsize=(15,15), dpi=200)
ax = axes.ravel()
columns = dataframe.columns
plt.subplots_adjust(hspace=0.6, wspace=0.4)
for i in range(first_f,last_f):
# Target values (actual sensor values)
sns.scatterplot(data=dataframe,
x=name_time_column,
y=columns[i],
ax=ax[i-first_f],
label="target",
palette="Greys")
# Predicted mu
sns.lineplot(data=dataframe,
x=name_time_column,
y=columns[i+no_features],
ax=ax[i-first_f],
label="predicted",
palette="Blues")
# Confidence bound (+- 2 sigma)
ax[i-first_f].fill_between(dataframe[i+no_features].values - confidence * dataframe[i+2*no_features].values,
dataframe[i+no_features].values + confidence * dataframe[i+2*no_features].values,
alpha=0.2,
color='blue')
if line:
ax[i-first_f].axvline(location_line, 0,2, color="r", linestyle ="--", linewidth=1, label="train-validation")
ax[i-first_f].set(xlabel='up time', ylabel='sensor value')
ax[i-first_f].set(title=columns[i])
plt.setp(ax[i-first_f].get_xticklabels(), rotation=45)
fig.suptitle(title)
plt.subplots_adjust(wspace=0.3, hspace=0.3)
```
# Visualisation of Prediction - Artifical Signal
```
artifical_data = pd.read_csv("../../files/prediction/MLE/artifical_2_signals.csv", sep=";")
subset = artifical_data.iloc[10000:,:]
fig, axes = plt.subplots(2, 1, figsize=(30,15))
ax = axes.ravel()
columns = subset.columns
t = subset.iloc[:,0]
for i in range(1,3):
#t = range(10000,artifical_data.shape[0]+10000)
target = subset.iloc[:,i].values
mu = subset.iloc[:,i+2].values
sigma = subset.iloc[:,i+4].values
lower_bound = mu - 2*sigma
upper_bound = mu + 2* sigma
sns.lineplot(data=subset,
x=t,
y=columns[i],
ax=ax[i-1],
color="black")
sns.scatterplot(data=subset,
x=t,
y=columns[i+2],
ax=ax[i-1],
hue=columns[i+9],
palette=["blue", "red"])
ax[i-1].fill_between(t, lower_bound, upper_bound, color="grey", alpha=0.5)
if i == 1:
ax[i-1].axvspan(10251,10301, alpha=0.2, color='red')
ax[i-1].axvspan(10450,10500, alpha=0.2, color='red')
ax[i-1].axvspan(10750,10850, alpha=0.2, color='red')
ax[i-1].axvspan(11150,11300, alpha=0.2, color='red')
ax[i-1].axvspan(11500,11650, alpha=0.2, color='red')
ax[i-1].axvspan(11700,11800, alpha=0.2, color='red')
plt.show()
```
## Evaluation Metrics
### F1-score
Exact labels are provided, thus F1-score can be computed
```
anomaly_label_ground_truth = artifical_data.anomaly
anomaly_label_by_model = artifical_data["Anomaly Sensor_1"]
print(f1_score(anomaly_label_ground_truth, anomaly_label_by_model, average='macro'))
```
### Confusion Matrix
```
tn, fp, fn, tp = confusion_matrix(anomaly_label_ground_truth, anomaly_label_by_model).ravel()
print("Positive --> Anomaly")
print("Negative --> Normal Behaviour")
print("--"*15)
print("True negative: {}".format(tn))
print("False positive: {}".format(fp))
print("False negative: {}".format(fn))
print("True positive: {}".format(tp))
```
|
github_jupyter
|
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
from statsmodels.graphics.tsaplots import plot_pacf
from statsmodels.graphics.tsaplots import plot_acf
from matplotlib.pyplot import figure
from sklearn.metrics import f1_score
from sklearn.metrics import confusion_matrix
def visualize_prediction(confidence, first_f, last_f, no_features, name_time_column, dataframe, title, location_line, line=False):
fig, axes = plt.subplots(2, 2, figsize=(15,15), dpi=200)
ax = axes.ravel()
columns = dataframe.columns
plt.subplots_adjust(hspace=0.6, wspace=0.4)
for i in range(first_f,last_f):
# Target values (actual sensor values)
sns.scatterplot(data=dataframe,
x=name_time_column,
y=columns[i],
ax=ax[i-first_f],
label="target",
palette="Greys")
# Predicted mu
sns.lineplot(data=dataframe,
x=name_time_column,
y=columns[i+no_features],
ax=ax[i-first_f],
label="predicted",
palette="Blues")
# Confidence bound (+- 2 sigma)
ax[i-first_f].fill_between(dataframe[i+no_features].values - confidence * dataframe[i+2*no_features].values,
dataframe[i+no_features].values + confidence * dataframe[i+2*no_features].values,
alpha=0.2,
color='blue')
if line:
ax[i-first_f].axvline(location_line, 0,2, color="r", linestyle ="--", linewidth=1, label="train-validation")
ax[i-first_f].set(xlabel='up time', ylabel='sensor value')
ax[i-first_f].set(title=columns[i])
plt.setp(ax[i-first_f].get_xticklabels(), rotation=45)
fig.suptitle(title)
plt.subplots_adjust(wspace=0.3, hspace=0.3)
artifical_data = pd.read_csv("../../files/prediction/MLE/artifical_2_signals.csv", sep=";")
subset = artifical_data.iloc[10000:,:]
fig, axes = plt.subplots(2, 1, figsize=(30,15))
ax = axes.ravel()
columns = subset.columns
t = subset.iloc[:,0]
for i in range(1,3):
#t = range(10000,artifical_data.shape[0]+10000)
target = subset.iloc[:,i].values
mu = subset.iloc[:,i+2].values
sigma = subset.iloc[:,i+4].values
lower_bound = mu - 2*sigma
upper_bound = mu + 2* sigma
sns.lineplot(data=subset,
x=t,
y=columns[i],
ax=ax[i-1],
color="black")
sns.scatterplot(data=subset,
x=t,
y=columns[i+2],
ax=ax[i-1],
hue=columns[i+9],
palette=["blue", "red"])
ax[i-1].fill_between(t, lower_bound, upper_bound, color="grey", alpha=0.5)
if i == 1:
ax[i-1].axvspan(10251,10301, alpha=0.2, color='red')
ax[i-1].axvspan(10450,10500, alpha=0.2, color='red')
ax[i-1].axvspan(10750,10850, alpha=0.2, color='red')
ax[i-1].axvspan(11150,11300, alpha=0.2, color='red')
ax[i-1].axvspan(11500,11650, alpha=0.2, color='red')
ax[i-1].axvspan(11700,11800, alpha=0.2, color='red')
plt.show()
anomaly_label_ground_truth = artifical_data.anomaly
anomaly_label_by_model = artifical_data["Anomaly Sensor_1"]
print(f1_score(anomaly_label_ground_truth, anomaly_label_by_model, average='macro'))
tn, fp, fn, tp = confusion_matrix(anomaly_label_ground_truth, anomaly_label_by_model).ravel()
print("Positive --> Anomaly")
print("Negative --> Normal Behaviour")
print("--"*15)
print("True negative: {}".format(tn))
print("False positive: {}".format(fp))
print("False negative: {}".format(fn))
print("True positive: {}".format(tp))
| 0.527803 | 0.800926 |
<img src="../img/logo_amds.png" alt="Logo" style="width: 128px;"/>
# AmsterdamUMCdb - Freely Accessible ICU Database
version 1.0.2 March 2020
Copyright © 2003-2020 Amsterdam UMC - Amsterdam Medical Data Science
# <a id='freetextitems'></a>freetextitems table
The *freetextitems* table contains all observations, including laboratory results, that store text (non-numeric) data. Personal data, including references to hospital staff have been removed. All items have an associated admissionid from the [admissions](admissions.ipynb#admissions) table.
## Fields
|Name|Type|Description|
|:---|:---|:---|
|admissionid|integer|links the items with the admissionid in the [admissions](admissions.ipynb#admissions) table|
|itemid|integer|id to identify the type of result|
|[item](#item)|string|type of result|
|[value](#value)|string|value of this text result|
|[comment](#comment)|string|comment associated with this result|
|measuredat|integer|time associated with this result in milliseconds since the first admission. Negative results imply that the sample was drawn before the first ICU admission|
|registeredat|integer|time the result was saved to the database in milliseconds since the first admission. Negative results imply that the sample was drawn before the first ICU admission.|
|registeredby|string|user group that updated the result ('System')|
|updatedat|integer|time the result was updated, in milliseconds since the first admission|
|updatedby|string|user group that updated the result ('System')|
|islabresult|bit|determines wheter the observation is a labresult. Currently, only freetext labresults have been released|
<h2>Example Python and SQL scripts</h2>
## Imports
```
%matplotlib inline
import amsterdamumcdb
import psycopg2
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
import matplotlib as mpl
import io
from IPython.display import display, HTML, Markdown
```
## Display settings
```
#matplotlib settings for image size
#needs to be in a different cell from %matplotlib inline
plt.style.use('seaborn-darkgrid')
plt.rcParams["figure.dpi"] = 288
plt.rcParams["figure.figsize"] = [8, 6]
plt.rcParams["font.size"] = 12
pd.options.display.max_columns = None
pd.options.display.max_rows = None
pd.options.display.max_colwidth = 1000
```
## Connection settings
```
#Modify config.ini in the root folder of the repository to change the settings to connect to your postgreSQL database
import configparser
import os
config = configparser.ConfigParser()
if os.path.isfile('../config.ini'):
config.read('../config.ini')
else:
config.read('../config.SAMPLE.ini')
#Open a connection to the postgres database:
con = psycopg2.connect(database=config['psycopg2']['database'],
user=config['psycopg2']['username'], password=config['psycopg2']['password'],
host=config['psycopg2']['host'], port=config['psycopg2']['port'])
con.set_client_encoding('WIN1252') #Uses code page for Dutch accented characters.
con.set_session(autocommit=True)
cursor = con.cursor()
cursor.execute('SET SCHEMA \'amsterdamumcdb\''); #set search_path to amsterdamumcdb schema
```
## Overview of all fields
```
sql = """
SELECT * FROM freetextitems
"""
display(Markdown("``` mysql\n" + sql + "\n```"))
freetexitems = pd.read_sql(sql,con)
freetexitems.head(10)
freetexitems.describe()
```
## <a id='item'></a> item
```
sql = """
SELECT item, value, comment
FROM freetextitems
WHERE item ILIKE '%coombs%'
"""
display(Markdown("``` mysql\n" + sql + "\n```"))
df = pd.read_sql(sql,con)
df.head()
```
## <a id='value'></a> value
```
sql = """
SELECT DISTINCT value
FROM freetextitems
"""
display(Markdown("``` mysql\n" + sql + "\n```"))
df = pd.read_sql(sql,con)
df.head(10)
sql = """
SELECT item, COUNT(item) AS "Number of results"
FROM freetextitems
WHERE NOT itemid = 11646 -- not a real result: location of bloodsample: arterial, venous, etc.
GROUP BY item
ORDER BY "Number of results" DESC
LIMIT 20
"""
display(Markdown("``` mysql\n" + sql + "\n```"))
df = pd.read_sql(sql,con)
cm = plt.get_cmap('RdPu')
color_step = int(-255/len(df.index))
ax = df.plot.barh(x=df.columns[0],y=df.columns[1], legend=False,color=cm(range(255,1,color_step)))
ax.set(xlabel=df.columns[1])
ax.xaxis.set_major_formatter(mpl.ticker.StrMethodFormatter('{x:,.0f}'))
sql = """
SELECT DISTINCT value, COUNT(item) AS "Number of results"
FROM freetextitems
WHERE itemid = 11646 -- location of bloodsample: arterial, venous, etc.
GROUP BY value
ORDER BY "Number of results" DESC
"""
display(Markdown("``` mysql\n" + sql + "\n```"))
df = pd.read_sql(sql,con)
df.head(10)
sql = """
SELECT DISTINCT value, COUNT(item) AS "Number of results"
FROM freetextitems
WHERE itemid = 11646 -- location of bloodsample: arterial, venous, etc.
GROUP BY value
ORDER BY "Number of results" DESC
LIMIT 5
"""
display(Markdown("``` mysql\n" + sql + "\n```"))
df = pd.read_sql(sql,con)
cm = plt.get_cmap('RdPu')
color_step = int(-255/len(df.index))
ax = df.plot.bar(x=df.columns[0],y=df.columns[1], legend=False,color=cm(range(255,1,color_step)))
ax.set(ylabel=df.columns[1])
ax.yaxis.set_major_formatter(mpl.ticker.StrMethodFormatter('{x:,.0f}'))
```
## <a id='comment'></a> comment
```
sql = """
SELECT DISTINCT comment
FROM freetextitems
"""
display(Markdown("``` mysql\n" + sql + "\n```"))
df = pd.read_sql(sql,con)
df.head()
```
|
github_jupyter
|
%matplotlib inline
import amsterdamumcdb
import psycopg2
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
import matplotlib as mpl
import io
from IPython.display import display, HTML, Markdown
#matplotlib settings for image size
#needs to be in a different cell from %matplotlib inline
plt.style.use('seaborn-darkgrid')
plt.rcParams["figure.dpi"] = 288
plt.rcParams["figure.figsize"] = [8, 6]
plt.rcParams["font.size"] = 12
pd.options.display.max_columns = None
pd.options.display.max_rows = None
pd.options.display.max_colwidth = 1000
#Modify config.ini in the root folder of the repository to change the settings to connect to your postgreSQL database
import configparser
import os
config = configparser.ConfigParser()
if os.path.isfile('../config.ini'):
config.read('../config.ini')
else:
config.read('../config.SAMPLE.ini')
#Open a connection to the postgres database:
con = psycopg2.connect(database=config['psycopg2']['database'],
user=config['psycopg2']['username'], password=config['psycopg2']['password'],
host=config['psycopg2']['host'], port=config['psycopg2']['port'])
con.set_client_encoding('WIN1252') #Uses code page for Dutch accented characters.
con.set_session(autocommit=True)
cursor = con.cursor()
cursor.execute('SET SCHEMA \'amsterdamumcdb\''); #set search_path to amsterdamumcdb schema
sql = """
SELECT * FROM freetextitems
"""
display(Markdown("``` mysql\n" + sql + "\n```"))
freetexitems = pd.read_sql(sql,con)
freetexitems.head(10)
freetexitems.describe()
sql = """
SELECT item, value, comment
FROM freetextitems
WHERE item ILIKE '%coombs%'
"""
display(Markdown("``` mysql\n" + sql + "\n```"))
df = pd.read_sql(sql,con)
df.head()
sql = """
SELECT DISTINCT value
FROM freetextitems
"""
display(Markdown("``` mysql\n" + sql + "\n```"))
df = pd.read_sql(sql,con)
df.head(10)
sql = """
SELECT item, COUNT(item) AS "Number of results"
FROM freetextitems
WHERE NOT itemid = 11646 -- not a real result: location of bloodsample: arterial, venous, etc.
GROUP BY item
ORDER BY "Number of results" DESC
LIMIT 20
"""
display(Markdown("``` mysql\n" + sql + "\n```"))
df = pd.read_sql(sql,con)
cm = plt.get_cmap('RdPu')
color_step = int(-255/len(df.index))
ax = df.plot.barh(x=df.columns[0],y=df.columns[1], legend=False,color=cm(range(255,1,color_step)))
ax.set(xlabel=df.columns[1])
ax.xaxis.set_major_formatter(mpl.ticker.StrMethodFormatter('{x:,.0f}'))
sql = """
SELECT DISTINCT value, COUNT(item) AS "Number of results"
FROM freetextitems
WHERE itemid = 11646 -- location of bloodsample: arterial, venous, etc.
GROUP BY value
ORDER BY "Number of results" DESC
"""
display(Markdown("``` mysql\n" + sql + "\n```"))
df = pd.read_sql(sql,con)
df.head(10)
sql = """
SELECT DISTINCT value, COUNT(item) AS "Number of results"
FROM freetextitems
WHERE itemid = 11646 -- location of bloodsample: arterial, venous, etc.
GROUP BY value
ORDER BY "Number of results" DESC
LIMIT 5
"""
display(Markdown("``` mysql\n" + sql + "\n```"))
df = pd.read_sql(sql,con)
cm = plt.get_cmap('RdPu')
color_step = int(-255/len(df.index))
ax = df.plot.bar(x=df.columns[0],y=df.columns[1], legend=False,color=cm(range(255,1,color_step)))
ax.set(ylabel=df.columns[1])
ax.yaxis.set_major_formatter(mpl.ticker.StrMethodFormatter('{x:,.0f}'))
sql = """
SELECT DISTINCT comment
FROM freetextitems
"""
display(Markdown("``` mysql\n" + sql + "\n```"))
df = pd.read_sql(sql,con)
df.head()
| 0.36557 | 0.848282 |
<a href="https://colab.research.google.com/github/maxigaarp/Gestion-De-Datos-en-R/blob/main/Tarea1_V2.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
En las clases de gestión de datos hemos aprendido acerca de la eficiencia de bases de datos, modelos relacionales, visualizar información y unir diferentes bases de datos para obtener la mayor información posible. Esta tarea consistirá de dos preguntas las cuales deberá responder de forma completa (código, texto e imágenes explicativas). Puede entregar la tarea en el formato, que estime conveniente (doc, pdf, script de R, url de Colab) pero tenga en cuenta que debe adjuntar el código utilizado. Se recomiendo el uso de Colab R.
```
#rendim2017 <- read.csv("http://datos.mineduc.cl/datasets/180324-rendimiento-escolar-ano-2017.download/",row.names=NULL, sep=";")
#rendim2018 <- read.csv("http://datos.mineduc.cl/datasets/189328-rendimiento-escolar-ano-2018.download/",row.names=NULL, sep=";")
## Hay un problema descargando directamente los datos desde la pagina de mineduc asi que subi los datos a mi google drive,
## si es que les fallara pueden subirlo a su propio google drive o cargar los datos de manera directa a Google Colab R
system("gdown https://drive.google.com/uc?id=1aARNSlskNDlnp95AMIfAnTQJticwxzVD")
system("gdown https://drive.google.com/uc?id=1mdr2SpmggrOQAOUDIUjRyE1hul_7YyY6")
rendim2017 <- read.csv("/content/20180213_Rendimiento_2017_20180131_PUBL.csv", sep=";")
rendim2018 <- read.csv("/content/20190220_Rendimiento_2018_20190131_PUBL.csv", sep=";")
columnas2017 <- paste(names(rendim2017), "2017", sep = "_")
columnas2018 <- paste(names(rendim2018), "2018", sep = "_")
rendim_17_18 <- data.frame(matrix(ncol = 71, nrow = 0))
nombres_columnas <- c(columnas2017, columnas2018)
colnames(rendim_17_18) <- nombres_columnas
```
# Problema 1 (1.5 Pts)
En el taller, revisamos el siguiente código para hacer un join
de dos bases de datos que compartieran un identificador.
```
ciclo1 <- 3000
ciclo2 <- 3000
n <- 1
for(i in 1:ciclo1){
mrun <- rendim2017$mrun[i]
for(j in 1:ciclo2){
mrun2 <- rendim2018$MRUN[j]
if(mrun == mrun2){
fila <- c(as.character(rendim2017[i,]), as.character(rendim2018[j,]))
rendim_17_18[n, ] <- fila
n <- n+1
}
}
}
```
Suponga que usted tiene un disco duro con dos bases de datos con 3000 filas cada una que ocupan 300 paginas de disco cada una, en otras palabras cada pagina tiene 10 tuplas. Por otro lado usted tiene una memoria RAM de 101 páginas de almacenamiento.
(a) (0.5 pts) Dibuje el diagrama de flujo del algoritmo.
```
```
(b) (0.5 pts) Cuente la cantidad de paginas que se deben ir a buscar desde el disco duro para almacenarlas en memoria de la forma que esta escrito actualmente el algoritmo.
```
```
(c) (0.5 pts) Plantee y programe una forma para reducir las lecturas a disco a al menos 1300 páginas. El algoritmo que requiera menos consultas del curso tendrá un bonus de 0.5 puntos.
```
```
#Problema 2 (1.5 pts)
(a) (0.7 pts) Descarga la base de datos de Cargos Docentes del año 2015 en R. Crea dos tablas distintas: una para el establecimiento y la otra para los docentes, además agrega con los 4 atributos más importantes en cada relación. Además crea una tabla ''trabaja\_en'' que vincule el docente con el establecimiento. Notese que buscamos reducir el tamaño de la información por lo que en las tablas no deberían aparecer valores duplicados.
```
#docentes2015 <- read.csv("http://datos.mineduc.cl/datasets/176034-directorio-oficial-de-docentes-2015.download/")
## Hay un problema descargando directamente los datos desde la pagina de mineduc asi que subi los datos a mi google drive,
## si es que les fallara pueden subirlo a su propio google drive o cargar los datos de manera directa a Google Colab R
docentes2015 <- read.csv("https://drive.google.com/uc?id=1_cXZSUur89uZi49hSwI8DNyx1xrB0G7L", sep=";")
docentes2015
```
(b) (0.4 pts) Utilizando los operadores de álgebra relacional escribe la siguiente consulta para las tablas que definiste en el punto anterior:
```
Liste los MRUN de los profesores que trabajan en más de un colegio.
```
Hint: no se debe contar para esta pregunta, solo se debe hacer el producto cruz de la tabla ''trabaja\_en'' de la pregunta (a) con sigo misma y luego filtrar de manera adecuada
```
```
(c) (0.4 pts) Implementa esta consulta en R (puedes usar el procedimiento dado por el álgebra relacional pero no es obligatorio).
```
```
#Problema 3 (3 pts)
En esta pregunta usted deberá trabajar con las bases de datos disponibles dentro del portal de datos para investigación del MINEDUC datosabiertos.mineduc.cl. En esta pregunta el objetivo es comprobar alguna hipótesis basándose en visualización de información, por lo que las elecciones de cada paso deben ser concordantes.
(a) (0.5 Pts) Revise la página web del MINEDUC y formule a lo menos tres distintas hipótesis que puedan responderse con los estos datos. Estas deben cumplir que:
* Deben resolverse mediante el cruce de información entre 2 o más tablas (no es valido usar la misma base de datos para distintos años).
* No deben ser triviales pues se evaluará también la calidad de la pregunta.
(b) Seleccione tres bases de datos que ayuden a resolver una o más hipótesis planteadas. Explique su elección y con sus propias palabras que contiene cada base de datos seleccionada.
(c) (1 Pt)Compruebe usando R que las bases de datos se pueden mezclar entre si. Es decir, pueden hacer un join entre ellas cuyo resultado es distinto de vacío. Si es que este proceso no entrega la cantidad de tuplas suficiente para estudiar ninguna de las tres hipótesis (o al menos tener confianza del resultado) deberá rehacer los pasos (a), (b) y (c).
(d) (1 Pt) Seleccione una de las tres hipótesis y a través de la visualización de información determine si esta pareciese estar acertada o no. Entregue una pequeña conclusión y análisis sobre este mini-estudio.
|
github_jupyter
|
#rendim2017 <- read.csv("http://datos.mineduc.cl/datasets/180324-rendimiento-escolar-ano-2017.download/",row.names=NULL, sep=";")
#rendim2018 <- read.csv("http://datos.mineduc.cl/datasets/189328-rendimiento-escolar-ano-2018.download/",row.names=NULL, sep=";")
## Hay un problema descargando directamente los datos desde la pagina de mineduc asi que subi los datos a mi google drive,
## si es que les fallara pueden subirlo a su propio google drive o cargar los datos de manera directa a Google Colab R
system("gdown https://drive.google.com/uc?id=1aARNSlskNDlnp95AMIfAnTQJticwxzVD")
system("gdown https://drive.google.com/uc?id=1mdr2SpmggrOQAOUDIUjRyE1hul_7YyY6")
rendim2017 <- read.csv("/content/20180213_Rendimiento_2017_20180131_PUBL.csv", sep=";")
rendim2018 <- read.csv("/content/20190220_Rendimiento_2018_20190131_PUBL.csv", sep=";")
columnas2017 <- paste(names(rendim2017), "2017", sep = "_")
columnas2018 <- paste(names(rendim2018), "2018", sep = "_")
rendim_17_18 <- data.frame(matrix(ncol = 71, nrow = 0))
nombres_columnas <- c(columnas2017, columnas2018)
colnames(rendim_17_18) <- nombres_columnas
ciclo1 <- 3000
ciclo2 <- 3000
n <- 1
for(i in 1:ciclo1){
mrun <- rendim2017$mrun[i]
for(j in 1:ciclo2){
mrun2 <- rendim2018$MRUN[j]
if(mrun == mrun2){
fila <- c(as.character(rendim2017[i,]), as.character(rendim2018[j,]))
rendim_17_18[n, ] <- fila
n <- n+1
}
}
}
```
(b) (0.5 pts) Cuente la cantidad de paginas que se deben ir a buscar desde el disco duro para almacenarlas en memoria de la forma que esta escrito actualmente el algoritmo.
(c) (0.5 pts) Plantee y programe una forma para reducir las lecturas a disco a al menos 1300 páginas. El algoritmo que requiera menos consultas del curso tendrá un bonus de 0.5 puntos.
#Problema 2 (1.5 pts)
(a) (0.7 pts) Descarga la base de datos de Cargos Docentes del año 2015 en R. Crea dos tablas distintas: una para el establecimiento y la otra para los docentes, además agrega con los 4 atributos más importantes en cada relación. Además crea una tabla ''trabaja\_en'' que vincule el docente con el establecimiento. Notese que buscamos reducir el tamaño de la información por lo que en las tablas no deberían aparecer valores duplicados.
(b) (0.4 pts) Utilizando los operadores de álgebra relacional escribe la siguiente consulta para las tablas que definiste en el punto anterior:
Hint: no se debe contar para esta pregunta, solo se debe hacer el producto cruz de la tabla ''trabaja\_en'' de la pregunta (a) con sigo misma y luego filtrar de manera adecuada
(c) (0.4 pts) Implementa esta consulta en R (puedes usar el procedimiento dado por el álgebra relacional pero no es obligatorio).
| 0.258981 | 0.934455 |
```
%matplotlib inline
%run ../setup/nb_setup
```
# Orbits 3: Orbits in Triaxial Potentials
Author(s): Adrian Price-Whelan
## Learning goals
In this tutorial, we will introduce triaxial potential models, and explore the additional complexity that this brings to the landscape of orbits, as compared to orbits in axisymmetric potential models.
## Introduction
In the last tutorial, we saw that reducing the symmetry of a potential from spherical to axisymmetric reduced the number if isolating integrals of motion from four to three. Will removing another potential symmetry — going from axisymmetric to triaxial — further reduce the number of isolating integrals? Luckily no: It turns out that regular orbits in triaxial potentials still have three isolating integrals. However, the orbit structure of triaxial potentials is more complex: A new type of orbit family can exist in triaxial potentials known as "box orbits," and tube orbits (what we are familiar with from axisymmetric models) can only exist around either the short or long axis of the potential. Triaxial potentials also generally have larger regions of chaotic orbits, making chaos more relevant in triaxial mass models.
In this tutorial, we will introduce some triaxial gravitational potential models, demonstrate the different types of orbits that can exist in triaxial models, and compare chaotic and regular orbits.
## Terminology and Notation
- (See Orbits tutorials 1 and 2)
### Notebook Setup and Package Imports
```
from astropy.constants import G
import astropy.units as u
from IPython.display import HTML
import matplotlib as mpl
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
import numpy as np
import gala.dynamics as gd
import gala.integrate as gi
import gala.potential as gp
from gala.units import galactic
```
## Triaxial Potential Models
To recap the last tutorial, we found that we could define axisymmetric potential–density pairs by replacing the spherical radius with a elliptical radius that scales the $z$ axis differently. Axisymmetric models are useful for defining potentials for flattened mass distributions, such as from galaxy disks. Triaxial potential models are models in which the isopotential or isodensity contours coincide with ellipsoidal surfaces. These models are useful for representing the outskirts of dark matter halos (which have been found to be triaxial, to first order) and for representing the stellar distributions in elliptical galaxies.
To create a triaxial potential, we can instead replace the spherical radius with an ellipsoidal radius defined as
$$
r^2_{\rm ell} = \frac{x^2}{a^2} + \frac{y^2}{b^2} + \frac{z^2}{c^2}
$$
where often by convention, $a$ is set to $a=1$ so that $b$ is the $y/x$ *axis ratio* and $c$ is the $z/x$ axis ratio. As with the axisymmetric models, we can either replace the radius in the expression of a spherical potential, or replace the radius in the expression of the density function, but these have different effects. Ideally, we would want to replace the radius in the form of a spherical density with the ellipsoidal radius, because then the density would be guaranteed to be positive and physical at all positions. However, there are very few triaxial solutions to Poisson's equation known, so most triaxial density distributions do not have analytic expressions for the potential (or gradients, which we need to compute orbits). It is therefore common to instead replace that radius in a potential expression with an ellipsoidal radius, but this has the same caveat as we saw with axisymmetric models that for some choices of $b, c$ the density can become negative.
### Example: The Triaxial Log Potential
One example of such a potential model is the triaxial extension of the flattened log potential model discussed in the previous tutorial. The form of the potential is given by
$$
\Phi_L(\boldsymbol{x}) = \frac{1}{2}\,v_0^2 \, \ln\left(\frac{x^2}{q_1^2} + \frac{y^2}{q_2^2} +
\frac{z^2}{q_3^2} + r_h^2 \right)
$$
where the axis ratios are given by the parameters $(q_1, q_2, q_3)$, and by convention we typically set $q_1=1$.
With Gala, we can define a triaxial log potential by specifying axis ratio parameters `q1`, `q2`, `q3`.
### Exercise: Define a Triaxial Log Potential with Gala
Define a Gala `LogarithmicPotential` object with:
* $v_c=230~\textrm{km}~\textrm{s}^{-1}$
* $r_h=15~\textrm{kpc}$
* $q_1 = 1$
* $q_2 = 0.9$
* $q_3 = 0.8$
```
# Define the potential object here
# triaxial_log =
```
## Integrals of Motion and Orbit Classes in Triaxial Models
Regular orbits in triaxial potential models still have three integrals of motion, however, like in the axisymmetric case, these are again not easily associated with classical integrals or analytic expressions. Regular orbits do have actions, but the interpretation of the actions often depends on the type of orbit being studied. To understand this a bit more, we need to look at the different types of orbit families that exist in triaxial potential models. Recall that in spherical models, all orbits are planar, and generic orbits form rosette patterns because of radial and azimuthal oscillations that often have an irrational frequency ratio. In axisymmetric models, orbits conserve the $z$ component of their angular momentum and so preserve their circulation around the symmetry axis of the potential, forming vertically-thickened analogs to the planar rosette patterns called "tube" orbits. In triaxial models, there are four general classes of orbits: short-axis tube orbits, "inner" long-axis tube orbits, "outer" long-axis tube orbits, and box orbits.
## Tube Orbits
Tube orbits in triaxial models can only exist around the long and short axes of the potential, but around these axes the orbits look a lot like their axisymmetric counterparts. In the exercise below, we will compute a few tube orbits and take a closer look at their properties.
### Exercise: Long- and short-axis tube orbits
Define a `PhaseSpacePosition` object to represent two initial conditions:
* At $\boldsymbol{x} = (10, 1, 3)~\textrm{kpc}$, with $v_y = v_c$
* At $\boldsymbol{x} = (3, 10, 1)~\textrm{kpc}$, with $v_z = v_c$
(other velocity components set to 0)
```
# Define the initial conditions here
# tube_w0s = ...
```
Integrate these orbits in the `triaxial_log` potential defined above for a total integration time of 100 Gyr with a timestep of 2 Myr:
```
# Integrate the orbits here:
# tube_orbits = ...
```
Plot the two orbits in all 2D projections of the 3D positions (x-y, x-z, y-z) on separate figures:
Compute and plot the angular momentum components for the two orbits as a function of time:
```
# tube_orbits_L = ...
```
What differences do you see in the time-series angular momentum components as compared to orbits in an axisymmetric model?
Answer here: ...
**Bonus plots**: Some animations showing the 3D structure of these orbits
```
azims = np.linspace(0, 180, 128)
half_elevs = np.linspace(-80, 80, len(azims) // 2)
elevs = np.concatenate((half_elevs, half_elevs[::-1]))
fig, anim = animate_3d(tube_orbits[:, 0], azims=azims, elevs=elevs, total_time=8)
plt.close(fig)
HTML(anim.to_html5_video())
fig, anim = animate_3d(tube_orbits[:, 1], azims=azims, elevs=elevs, total_time=8)
plt.close(fig)
HTML(anim.to_html5_video())
```
### Exercise: Tube orbits around the intermediate axis?
Set up initial conditions to compute a tube orbit around the intermediate axis, starting from the position $\boldsymbol{x} = (10, 0.5, 0)~\textrm{kpc}$.
```
# y_tube_w0 = ...
```
Integrate this orbit for the same time array as the tube orbits we computed above:
```
# y_tube_orbit = ...
```
Plot the orbit in projections:
Compute the angular momentum components and plot them:
What is different about the angular momentum component time series for this orbit?
## Box Orbits
As mentioned above, triaxial potentials host another class of orbits that are distinct from tube orbits: *Box orbits*. These orbits do not maintain a fixed sense of rotation or circulation around an axis and so they fill a boxy volume in position space over time. Box orbits tend to have initial conditions without much tangential velocity, for example:
```
box_orbit1_w0 = gd.PhaseSpacePosition(
pos=[12, 4.0, 4.0] * u.kpc, vel=[0, 0, 0.0] * u.km / u.s
)
box_orbit1 = triaxial_log.integrate_orbit(
box_orbit1_w0, t=tube_orbits.t, Integrator=gi.DOPRI853Integrator
)
_ = box_orbit1.plot_3d()
```
Because they do not maintain a sense of circulation around any of the axes, we can guess that none of the angular momentum components are even approximately conserved: We expect that all three components likely change sign over the course of the orbit. For the orbit we computed above, we can see that this is true:
```
plt.plot(box_orbit1.t.value, box_orbit1.angular_momentum().T)
plt.xlim(0, 10000)
```
Because these orbits look nothing like tube orbits, and so there is no sense in which the epicycle approximation makes sense here, the actions for box orbits have a different interpretation than the actions for tube orbits (which can be understood as radial, azimuthal, and vertical actions like in the axisymmetric case). For box orbits, the actions do not have an exact physical interpretation: they will be quantifications of the amplitudes of independent oscillations. However, to first order, if the potential model is aligned along the coordinate axes, conceptually they will be related to the amplitude of oscillation in the $x$, $y$, and $z$ directions. Actions for box orbits are typically referred to as
$$
\boldsymbol{J} = (J_1, J_2, J_3)
$$
to emphasize generality, i.e. that there is no direct or precise correspondence to coordinates.
### Exercise: A grid of orbits with equal energy
In this exercise, we are going to compute a grid of orbits started with the same total energy to map out the orbit structure of a portion of phase-space. How should we choose the initial conditions for our grid of orbits? We need to set the 6 phase-space coordinates for each orbit. Requiring that they have the same energy gives us 1 constraint. To further reduce the dimensionality, we will assume $y=v_x=v_z=0$ (we now have 4 constraints). We will then choose a grid in $x, z$ to set the final two coordinates. At any location in our $x,z$ grid, we will use the energy to determine the value of $v_y$ from:
$$
E = \frac{1}{2}(v_x^2 + v_y^2 + v_z^2) + \Phi(x,y,z)\\
v_y = \sqrt{2\,(E - \Phi(x, 0, z))}
$$
For the energy, we will use $E = 0.195~\textrm{kpc}^2~\textrm{Myr}^{-2}$:
```
# (No modifications needed here)
grid_E = 0.195 * (u.kpc / u.Myr) ** 2
```
Generate a 1D grid of 41 $x$ values between $(15, 25)~\textrm{kpc}$, and a 1D grid of 41 $z$ values between $(0, 20)~\textrm{kpc}$. Use these 1D grids to construct a 2D grid with all 1,681 pairs of coordinates (*Hint: use `numpy.meshgrid()`*). Store an array of all $x,y,z$ values (all $y$ values are 0) in the variable `grid_pos`:
```
_x_grid = np.linspace(15, 25, 41)
_z_grid = np.linspace(0, 20, 41)
grid_shape = (len(_x_grid), len(_z_grid))
# x_grid, z_grid = ...
```
Compute the potential energy at all locations in the grid, and use the difference of the grid energy `grid_E` and the potential energy to compute the initial $v_y$:
```
# grid_Phi = ...
# vy_grid = ...
```
(Some of the $v_y$ values may come out as NaN: that is ok, you can ignore those - there are some values of our $x,z$ grid that are outside of the iso-potential-energy surface)
Plot the grid of $x,y$ positions colored by the value of $v_y$ at each location (i.e. the following cell should execute)
```
# (No modifications needed here)
plt.figure(figsize=(7, 6))
plt.pcolormesh(x_grid, z_grid, vy_grid.reshape(grid_shape).to_value(u.km / u.s))
plt.xlabel("$x_0$")
plt.ylabel("$z_0$")
cb = plt.colorbar()
cb.set_label(r"$v_y$")
```
Set up the full grid of initial conditions as a `PhaseSpacePosition` object named `grid_w0`:
```
# (No modifications needed here)
grid_vel = np.zeros(grid_pos.shape) * u.km / u.s
grid_vel[1] = vy_grid
grid_w0 = gd.PhaseSpacePosition(pos=grid_pos * u.kpc, vel=grid_vel)
```
Compute the orbits for all of the initial conditions in the grid using the default `LeapfrogIntegrator`, using a timestep of 2 Myr, and integrate for 10 Gyr
```
# grid_orbits = ...
```
Compute the angular momentum components for all orbits, and then compute the peak-to-peak spread in each angular momentum component for each orbit (i.e. compute $\textrm{max}(L_i) - \textrm{min}(L_i)$ for each component $i$ for each orbit)
```
# grid_orbits_L = ...
# ptp_L = ...
```
Make a 3 panel plot (panels corresponding to the 3 angular momentum components) showing a 2D image of the peak-to-peak spread in each component (i.e. the plot commands below should execute)
* What structure do you see in this diagram?
* What do you think causes the structure we see in this diagram?
* Can you identify the transition from tube to box orbits?
## Irregular and Chaotic Orbits
A final class of orbits that are found in very flattened axisymmetric potentials and more frequently in triaxial potentials are known as *chaotic* orbits. There is a lot to say about chaos, but one way of thinking of chaotic orbits are that they are special orbits in which the actions do not exist: only energy is conserved (in time-independent potentials with chaotic orbits). The fact that chaotic orbits no longer have the three actions as isolating integrals of motion means that they are free to explore a much larger volume in phase-space. Recall that regular orbits exist in 6D phase-space, but the existence of three actions limits orbits (with particular values for their actions) to exploring a 3D manifold embedded within the 6D phase-space. Orbits that only conserve energy as an integral of motion can instead explore a 5D manifold known as the *energy hypersurface*.
Chaotic orbits also look more erratic: Regular orbits tend to have a smooth, well-behaved shape to their trajectories.
### Example: Comparing the morphology of regular and chaotic orbits
As a demonstration of the visual differences between regular and chaotic orbits, we will use a triaxial Navarro-Frenk-White potential model to compute two orbits:
```
triaxial_nfw = gp.NFWPotential.from_circular_velocity(
v_c=230 * u.km / u.s, r_s=5 * u.kpc, a=1, b=0.9, c=0.8, units=galactic
)
```
Compare the two orbits below -- which one do you think is chaotic?
```
regular_chaotic_w0 = gd.PhaseSpacePosition(
pos=([[0, 20, 0], [0.1, 15, 0]] * u.kpc).T,
vel=([[15, 0, 120], [5.0, 0.0, 230]] * u.km / u.s).T,
)
regular_chaotic_orbits = triaxial_nfw.integrate_orbit(
regular_chaotic_w0, dt=0.5, t1=0, t2=10 * u.Gyr
)
for n in range(regular_chaotic_orbits.norbits):
_ = regular_chaotic_orbits[:, n].plot()
```
### Chaotic Timescales
Not all chaotic orbits obviously appear chaotic from their morphologies. This is often because some orbits are only *weakly chaotic* whereas others are *strongly chaotic*. There is no global definition to strong and weak chaos because it depends on the relevant timescales of whatever system you are looking at: Weakly chaotic orbits have a chaotic timescale much smaller than the dynamical time, whereas strongly chaotic orbits have comparable timescales. In the case of weakly chaotic orbits, the orbits may look and behave like nearby regular orbits for a long time (i.e. for all astronomically-relevant timescales). In the case of strongly chaotic orbits, the morphology of the orbit changes on the order of an orbital timescale, like in the example above.
One way of quantifying the strength of chaos for an orbit is with the *Maximum Lyapunov Exponent* or the inverse, sometimes called the *Lyapunov Time*. The MLE is a measure of how quickly two infinitesimally-close orbits diverge from one another. We will not go through the math to derive the MLE here (see section 3.7.3 c in Binney and Tremaine). However, I will note that you can compute the MLE in Gala using the `gala.dynamics.fast_lyapunov_max()` function. For example, for the two orbits shown above:
```
lyap1 = gd.fast_lyapunov_max(
regular_chaotic_w0[0], triaxial_nfw, dt=2.0, n_steps=200_000, return_orbit=False
)
lyap2 = gd.fast_lyapunov_max(
regular_chaotic_w0[1], triaxial_nfw, dt=2.0, n_steps=200_000, return_orbit=False
)
```
For regular orbits, the estimate of the Lyapunov exponent continues decreasing as the integration time increases. However for chaotic orbits, the value eventually saturates to a number: This is the estimate of the MLE.
Let's plot the time series estimate of the LE for the two orbits above:
```
plt.loglog(lyap1[:, 0])
plt.loglog(lyap2[:, 0])
plt.xlabel("timestep")
plt.ylabel("Lyapunov exponent")
```
We can see that the first orbit, which visually looks erratic, has a finite MLE that saturates to a value around $4\times 10^{-4}~\textrm{Myr^{-1}}$, corresponding to a Lyapunov timescale of about $2.5~\textrm{Gyr}$.
There are many other interesting aspects of chaotic orbits, but we will leave that to be covered in discussions (if you get to this point, ask the workshop coordinators about chaos :-)).
|
github_jupyter
|
%matplotlib inline
%run ../setup/nb_setup
from astropy.constants import G
import astropy.units as u
from IPython.display import HTML
import matplotlib as mpl
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
import numpy as np
import gala.dynamics as gd
import gala.integrate as gi
import gala.potential as gp
from gala.units import galactic
# Define the potential object here
# triaxial_log =
# Define the initial conditions here
# tube_w0s = ...
# Integrate the orbits here:
# tube_orbits = ...
# tube_orbits_L = ...
azims = np.linspace(0, 180, 128)
half_elevs = np.linspace(-80, 80, len(azims) // 2)
elevs = np.concatenate((half_elevs, half_elevs[::-1]))
fig, anim = animate_3d(tube_orbits[:, 0], azims=azims, elevs=elevs, total_time=8)
plt.close(fig)
HTML(anim.to_html5_video())
fig, anim = animate_3d(tube_orbits[:, 1], azims=azims, elevs=elevs, total_time=8)
plt.close(fig)
HTML(anim.to_html5_video())
# y_tube_w0 = ...
# y_tube_orbit = ...
box_orbit1_w0 = gd.PhaseSpacePosition(
pos=[12, 4.0, 4.0] * u.kpc, vel=[0, 0, 0.0] * u.km / u.s
)
box_orbit1 = triaxial_log.integrate_orbit(
box_orbit1_w0, t=tube_orbits.t, Integrator=gi.DOPRI853Integrator
)
_ = box_orbit1.plot_3d()
plt.plot(box_orbit1.t.value, box_orbit1.angular_momentum().T)
plt.xlim(0, 10000)
# (No modifications needed here)
grid_E = 0.195 * (u.kpc / u.Myr) ** 2
_x_grid = np.linspace(15, 25, 41)
_z_grid = np.linspace(0, 20, 41)
grid_shape = (len(_x_grid), len(_z_grid))
# x_grid, z_grid = ...
# grid_Phi = ...
# vy_grid = ...
# (No modifications needed here)
plt.figure(figsize=(7, 6))
plt.pcolormesh(x_grid, z_grid, vy_grid.reshape(grid_shape).to_value(u.km / u.s))
plt.xlabel("$x_0$")
plt.ylabel("$z_0$")
cb = plt.colorbar()
cb.set_label(r"$v_y$")
# (No modifications needed here)
grid_vel = np.zeros(grid_pos.shape) * u.km / u.s
grid_vel[1] = vy_grid
grid_w0 = gd.PhaseSpacePosition(pos=grid_pos * u.kpc, vel=grid_vel)
# grid_orbits = ...
# grid_orbits_L = ...
# ptp_L = ...
triaxial_nfw = gp.NFWPotential.from_circular_velocity(
v_c=230 * u.km / u.s, r_s=5 * u.kpc, a=1, b=0.9, c=0.8, units=galactic
)
regular_chaotic_w0 = gd.PhaseSpacePosition(
pos=([[0, 20, 0], [0.1, 15, 0]] * u.kpc).T,
vel=([[15, 0, 120], [5.0, 0.0, 230]] * u.km / u.s).T,
)
regular_chaotic_orbits = triaxial_nfw.integrate_orbit(
regular_chaotic_w0, dt=0.5, t1=0, t2=10 * u.Gyr
)
for n in range(regular_chaotic_orbits.norbits):
_ = regular_chaotic_orbits[:, n].plot()
lyap1 = gd.fast_lyapunov_max(
regular_chaotic_w0[0], triaxial_nfw, dt=2.0, n_steps=200_000, return_orbit=False
)
lyap2 = gd.fast_lyapunov_max(
regular_chaotic_w0[1], triaxial_nfw, dt=2.0, n_steps=200_000, return_orbit=False
)
plt.loglog(lyap1[:, 0])
plt.loglog(lyap2[:, 0])
plt.xlabel("timestep")
plt.ylabel("Lyapunov exponent")
| 0.601477 | 0.993235 |
<a href="https://qworld.net" target="_blank" align="left"><img src="../qworld/images/header.jpg" align="left"></a>
$ \newcommand{\bra}[1]{\langle #1|} $
$ \newcommand{\ket}[1]{|#1\rangle} $
$ \newcommand{\braket}[2]{\langle #1|#2\rangle} $
$ \newcommand{\dot}[2]{ #1 \cdot #2} $
$ \newcommand{\biginner}[2]{\left\langle #1,#2\right\rangle} $
$ \newcommand{\mymatrix}[2]{\left( \begin{array}{#1} #2\end{array} \right)} $
$ \newcommand{\myvector}[1]{\mymatrix{c}{#1}} $
$ \newcommand{\myrvector}[1]{\mymatrix{r}{#1}} $
$ \newcommand{\mypar}[1]{\left( #1 \right)} $
$ \newcommand{\mybigpar}[1]{ \Big( #1 \Big)} $
$ \newcommand{\sqrttwo}{\frac{1}{\sqrt{2}}} $
$ \newcommand{\dsqrttwo}{\dfrac{1}{\sqrt{2}}} $
$ \newcommand{\onehalf}{\frac{1}{2}} $
$ \newcommand{\donehalf}{\dfrac{1}{2}} $
$ \newcommand{\hadamard}{ \mymatrix{rr}{ \sqrttwo & \sqrttwo \\ \sqrttwo & -\sqrttwo }} $
$ \newcommand{\vzero}{\myvector{1\\0}} $
$ \newcommand{\vone}{\myvector{0\\1}} $
$ \newcommand{\stateplus}{\myvector{ \sqrttwo \\ \sqrttwo } } $
$ \newcommand{\stateminus}{ \myrvector{ \sqrttwo \\ -\sqrttwo } } $
$ \newcommand{\myarray}[2]{ \begin{array}{#1}#2\end{array}} $
$ \newcommand{\X}{ \mymatrix{cc}{0 & 1 \\ 1 & 0} } $
$ \newcommand{\I}{ \mymatrix{rr}{1 & 0 \\ 0 & 1} } $
$ \newcommand{\Z}{ \mymatrix{rr}{1 & 0 \\ 0 & -1} } $
$ \newcommand{\Htwo}{ \mymatrix{rrrr}{ \frac{1}{2} & \frac{1}{2} & \frac{1}{2} & \frac{1}{2} \\ \frac{1}{2} & -\frac{1}{2} & \frac{1}{2} & -\frac{1}{2} \\ \frac{1}{2} & \frac{1}{2} & -\frac{1}{2} & -\frac{1}{2} \\ \frac{1}{2} & -\frac{1}{2} & -\frac{1}{2} & \frac{1}{2} } } $
$ \newcommand{\CNOT}{ \mymatrix{cccc}{1 & 0 & 0 & 0 \\ 0 & 1 & 0 & 0 \\ 0 & 0 & 0 & 1 \\ 0 & 0 & 1 & 0} } $
$ \newcommand{\norm}[1]{ \left\lVert #1 \right\rVert } $
$ \newcommand{\pstate}[1]{ \lceil \mspace{-1mu} #1 \mspace{-1.5mu} \rfloor } $
$ \newcommand{\greenbit}[1] {\mathbf{{\color{green}#1}}} $
$ \newcommand{\bluebit}[1] {\mathbf{{\color{blue}#1}}} $
$ \newcommand{\redbit}[1] {\mathbf{{\color{red}#1}}} $
$ \newcommand{\brownbit}[1] {\mathbf{{\color{brown}#1}}} $
$ \newcommand{\blackbit}[1] {\mathbf{{\color{black}#1}}} $
<font style="font-size:28px;" align="left"><b> <font color="blue"> Solution for </font>Operations on the Unit Circle </b></font>
<br>
_prepared by Abuzer Yakaryilmaz_
<br><br>
<a id="task3"></a>
<h3> Task 3</h3>
Randomly pick an angle $ \theta \in [0,2\pi) $.
Suppose that we have 1000 copies of quantum state $ \ket{v} = \myvector{ \cos \theta \\ \sin \theta } $ and we measure each of them.
What are the expected numbers of observing the states 0 and 1?
Implement the above experiment by designing a quantum circuit and set the quantum state by using ry-gate.
Compare your experimental and analytic results.
Repeat the task a couple of times.
<h3> Solution </h3>
**Analytical results**
```
from random import randrange
from math import sin,cos, pi
# randomly pick an angle
random_angle = randrange(360)
print("random angle is",random_angle)
# pick angle in radian
rotation_angle = random_angle/360*2*pi
# the quantum state
quantum_state = [ cos(rotation_angle) , sin (rotation_angle) ]
the_expected_number_of_zeros = 1000*cos(rotation_angle)**2
the_expected_number_of_ones = 1000*sin(rotation_angle)**2
# expected results
print("The expected value of observing '0' is",round(the_expected_number_of_zeros,4))
print("The expected value of observing '1' is",round(the_expected_number_of_ones,4))
# draw the quantum state
%run quantum.py
draw_qubit()
draw_quantum_state(quantum_state[0],quantum_state[1],"|v>")
show_plt()
```
**Experimental results**
```
from qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer
from qiskit.visualization import plot_histogram
# we define a quantum circuit with one qubit and one bit
q = QuantumRegister(1) # quantum register with a single qubit
c = ClassicalRegister(1) # classical register with a single bit
qc = QuantumCircuit(q,c) # quantum circuit with quantum and classical registers
# rotate the qubit with rotation_angle
qc.ry(2*rotation_angle,q[0])
# measure the qubit
qc.measure(q,c)
# draw the circuit
qc.draw(output='mpl')
# execute the program 1000 times
job = execute(qc,Aer.get_backend('qasm_simulator'),shots=1000)
# print the results
counts = job.result().get_counts(qc)
print(counts)
the_observed_number_of_ones = 0
if '1' in counts:
the_observed_number_of_ones= counts['1']
# draw the histogram
plot_histogram(counts)
```
**Compare the results**
```
difference = abs(the_expected_number_of_ones - the_observed_number_of_ones)
print("The expected number of ones is",the_expected_number_of_ones)
print("The observed number of ones is",the_observed_number_of_ones)
print("The difference is",difference)
print("The difference in percentage is",difference/100,"%")
```
|
github_jupyter
|
from random import randrange
from math import sin,cos, pi
# randomly pick an angle
random_angle = randrange(360)
print("random angle is",random_angle)
# pick angle in radian
rotation_angle = random_angle/360*2*pi
# the quantum state
quantum_state = [ cos(rotation_angle) , sin (rotation_angle) ]
the_expected_number_of_zeros = 1000*cos(rotation_angle)**2
the_expected_number_of_ones = 1000*sin(rotation_angle)**2
# expected results
print("The expected value of observing '0' is",round(the_expected_number_of_zeros,4))
print("The expected value of observing '1' is",round(the_expected_number_of_ones,4))
# draw the quantum state
%run quantum.py
draw_qubit()
draw_quantum_state(quantum_state[0],quantum_state[1],"|v>")
show_plt()
from qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer
from qiskit.visualization import plot_histogram
# we define a quantum circuit with one qubit and one bit
q = QuantumRegister(1) # quantum register with a single qubit
c = ClassicalRegister(1) # classical register with a single bit
qc = QuantumCircuit(q,c) # quantum circuit with quantum and classical registers
# rotate the qubit with rotation_angle
qc.ry(2*rotation_angle,q[0])
# measure the qubit
qc.measure(q,c)
# draw the circuit
qc.draw(output='mpl')
# execute the program 1000 times
job = execute(qc,Aer.get_backend('qasm_simulator'),shots=1000)
# print the results
counts = job.result().get_counts(qc)
print(counts)
the_observed_number_of_ones = 0
if '1' in counts:
the_observed_number_of_ones= counts['1']
# draw the histogram
plot_histogram(counts)
difference = abs(the_expected_number_of_ones - the_observed_number_of_ones)
print("The expected number of ones is",the_expected_number_of_ones)
print("The observed number of ones is",the_observed_number_of_ones)
print("The difference is",difference)
print("The difference in percentage is",difference/100,"%")
| 0.705278 | 0.988734 |
```
import os
import sys
module_path = os.path.abspath(os.path.join('..'))
if module_path not in sys.path:
sys.path.append(module_path)
import pandas as pd
import numpy as np
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import GridSearchCV
from joblib import dump
from src.models import eval_model as evm
from src.models import eval_baseline as evb
from sklearn.utils import resample
%load_ext autoreload
%autoreload 2
```
# Case for Upsample
```
from sklearn.model_selection import train_test_split
def read_and_split_data(file):
df = pd.read_csv(file)
x=df.drop(['TARGET_5Yrs','TARGET_5Yrs_Inv'],axis=1)
y=df['TARGET_5Yrs_Inv']
x_data , x_test ,y_data, y_test = train_test_split(x, y, test_size=0.2, random_state = 8, stratify=y)
x_train , x_val , y_train, y_val = train_test_split(x_data, y_data, test_size=0.2, random_state = 8, stratify=y_data)
print('y',y.value_counts())
print('y_train',y_train.value_counts())
print('y_val', y_val.value_counts())
print('y_test',y_test.value_counts())
return x_train , x_val , y_train, y_val, x_test, y_test
x_train , x_val , y_train, y_val, x_test, y_test = read_and_split_data("../data/processed/df_cleaned_upsampled_nba_prediction.csv")
evb.eval_baseline(x_train,y_train)
evm.eval_model(RandomForestClassifier(random_state=8),x_train,y_train,x_val,y_val)
rf_data = [
['n_estimators', 5],
['n_estimators', 25],
['n_estimators',50],
['n_estimators', 150],
['n_estimators',250],
['n_estimators',350],
['max_depth',2],
['max_depth',6],
['max_depth',12],
['max_depth',25],
['max_depth',50],
['min_samples_leaf',1],
['min_samples_leaf',2],
['min_samples_leaf',3],
['min_samples_leaf',5],
['min_samples_leaf',10],
['min_samples_leaf',15],
['min_samples_leaf',50],
['max_features',2],
['max_features',5],
['max_features',10],
['max_features',15]
]
rf_data
rf_param = pd.DataFrame(rf_data, columns = ['param', 'value'])
rf_param
```
# Training is overfitting. Tunning parameter starting with n_estimators
```
for _,row in rf_param[rf_param['param']=='n_estimators'].iterrows():
evm.eval_model(RandomForestClassifier(random_state=8,n_estimators = int(row.value)) ,x_train,y_train,x_val,y_val)
```
# Taking n_estimators = 50, tunning max_depth
```
nestimator = 50
for _,row in rf_param[rf_param['param']=='max_depth'].iterrows():
evm.eval_model(RandomForestClassifier(random_state=8,n_estimators = nestimator, max_depth= int(row.value)) ,x_train,y_train,x_val,y_val)
```
# Taking max_depth = 25, tunning min_samples_leaf
```
maxdepth = 25
for _,row in rf_param[rf_param['param']=='min_samples_leaf'].iterrows():
evm.eval_model(RandomForestClassifier(random_state=8,n_estimators =nestimator, max_depth=maxdepth,min_samples_leaf = int(row.value)) ,x_train,y_train,x_val,y_val)
```
# Taking min_samples_leaf = 1, Tunning max_features
```
minsamplesleaf=1
for _,row in rf_param[rf_param['param']=='max_features'].iterrows():
evm.eval_model(RandomForestClassifier(random_state=8,n_estimators=nestimator,max_depth=maxdepth,min_samples_leaf=minsamplesleaf,max_features = int(row.value)) ,x_train,y_train,x_val,y_val)
```
# Taking max_feature = 2, now using hyperopt to see if it can get better
```
maxfeatures=2
print('max_depth=',maxdepth,' max_features=',maxfeatures, ' min_samples_leaf=',minsamplesleaf,' n_estimators=',nestimator)
rf_upsample_mod0 = RandomForestClassifier(random_state=8,n_estimators=nestimator,max_depth=maxdepth,min_samples_leaf=minsamplesleaf,max_features = maxfeatures)
evm.get_performance(rf_upsample_mod, x_test, y_test, "Test", True)
from hyperopt import Trials, STATUS_OK, tpe, hp, fmin
from sklearn.metrics import accuracy_score, confusion_matrix,roc_curve, roc_auc_score, precision_score, recall_score, precision_recall_curve, f1_score, plot_confusion_matrix
from sklearn.model_selection import cross_val_score , cross_validate
space = {
'max_depth': hp.choice('max_depth',range(1,100,1)),
'max_features': hp.choice('max_features',range(1,15,1)),
'min_samples_leaf': hp.choice('min_samples_leaf',range(1,14,1)),
'n_estimators': hp.choice('n_estimators',range(10,500,1))
}
space
def objective(space):
rf = RandomForestClassifier(max_depth = space['max_depth'],max_features = space['max_features'],min_samples_leaf = space['min_samples_leaf'],n_estimators = space['n_estimators'])
acc=cross_val_score(rf, x_train,y_train,cv=50, scoring='roc_auc').mean()
return{'loss': 1-acc, 'status': STATUS_OK }
best = fmin(
fn=objective,
space=space,
algo=tpe.suggest,
max_evals=5
)
best
```
# Validate Results form Hyperopt
```
rf_upsample_mod = RandomForestClassifier(random_state=8,n_estimators = best['n_estimators'], max_depth = best['max_depth'], max_features = best['max_features'])
evm.eval_model(rf_upsample_mod,x_train,y_train,x_val,y_val)
evm.get_performance(rf_upsample_mod, x_test, y_test, "Test", True)
#max_depth= 25 max_features= 2 min_samples_leaf= 1 n_estimators= 50
space = {
'max_depth': hp.choice('max_depth',range(1,30,1)),
'max_features': hp.choice('max_features',range(1,15,1)),
'min_samples_leaf': hp.choice('min_samples_leaf',range(1,14,1)),
'n_estimators': hp.choice('n_estimators',range(10,250,1))
}
space
def objective(space):
rf = RandomForestClassifier(max_depth = space['max_depth'],max_features = space['max_features'],min_samples_leaf = space['min_samples_leaf'],n_estimators = space['n_estimators'])
acc=cross_val_score(rf, x_train,y_train,cv=50, scoring='roc_auc').mean()
return{'loss': 1-acc, 'status': STATUS_OK }
best = fmin(
fn=objective,
space=space,
algo=tpe.suggest,
max_evals=5
)
best
rf_upsample_mod1 = RandomForestClassifier(random_state=8,n_estimators = best['n_estimators'], max_depth = best['max_depth'], max_features = best['max_features'], min_samples_leaf = best['min_samples_leaf'])
evm.eval_model(rf_upsample_mod1,x_train,y_train,x_val,y_val)
evm.get_performance(rf_upsample_mod1, x_test, y_test, "Test", True)
```
# Predict for Upsample
```
def read_and_split_data(file):
df = pd.read_csv(file)
df_cleaned = df.copy()
print('Before Data Clean')
for cols in df_cleaned.columns:
chk_rows = df_cleaned[df_cleaned[cols]<0].shape[0]
if chk_rows > 0 :
print(f'Column Name {cols},\tRows with Negative Value {chk_rows},\tPercentage {chk_rows/len(df)*100}')
df_cleaned[ df_cleaned<0 ] = 0
df_cleaned.loc[df_cleaned['3P Made'] <= 0, ['3P Made', '3PA', 'CALC3P%']] = 0, 0, 0
df_cleaned.loc[df_cleaned['FGM'] <= 0, ['FGM', 'FGA', 'CALCFG%']] = 0, 0, 0
df_cleaned.loc[df_cleaned['FTM'] <= 0, ['FTM', 'FTA', 'CALCFT%']] = 0, 0, 0
df_cleaned.loc[df_cleaned['3P Made'] > df_cleaned['3PA'], ['3P Made' , '3PA', 'CALC3P%']] = 0, 0, 0
df_cleaned.loc[df_cleaned['FGM'] > df_cleaned['FGA'], ['FGM', 'FGA', 'CALCFG%']] = 0, 0, 0
df_cleaned.loc[df_cleaned['FTM'] > df_cleaned['FTA'], ['FTM', 'FTA', 'CALCFT%']] = 0, 0, 0
df_cleaned.loc[df_cleaned['3P Made'] > 0, ['CALC3P%']] = df_cleaned['3P Made']/df_cleaned['3PA']*100
df_cleaned.loc[df_cleaned['FGM'] > 0, ['CALCFG%']] =df_cleaned['FGM']/df_cleaned['FGA']*100
df_cleaned.loc[df_cleaned['FTM'] > 0, ['CALCFT%']] = df_cleaned['FTM']/df_cleaned['FTA']*100
print(df_cleaned.head(5))
print('After Data Clean')
for cols in df_cleaned.columns:
chk_rows = df_cleaned[df_cleaned[cols]<0].shape[0]
if chk_rows > 0 :
print(f'Column Name {cols},\tRows with Negative Value {chk_rows},\tPercentage {chk_rows/len(df)*100}')
x = df_cleaned.drop(['3P%','FT%','FG%','Id_old','Id'],axis=1)
print(df_cleaned.columns)
return x , df_cleaned
x, df_cleaned =read_and_split_data('../data/raw/test.csv')
y_upsample_pred_proba=rf_upsample_mod.predict_proba(x)
y_upsample_pred=rf_upsample_mod.predict(x)
print(np.unique(y_upsample_pred_proba,return_counts=True))
print(np.unique(y_upsample_pred,return_counts=True))
print(rf_upsample_mod.classes_)
df_cleaned_upsample_result = df_cleaned.copy()
df_cleaned_upsample_result['TARGET_5Yrs'] = y_upsample_pred_proba[:,0]
print(df_cleaned_upsample_result['TARGET_5Yrs'].round().value_counts())
df_cleaned_upsample_result.to_csv('../data/processed/TestResult_RF_UpSampleUpdated_Result.csv',index=False,columns=['Id', 'TARGET_5Yrs'])
y_upsample_pred_proba=rf_upsample_mod1.predict_proba(x)
y_upsample_pred=rf_upsample_mod.predict(x)
print(np.unique(y_upsample_pred,return_counts=True))
df_cleaned_upsample_result = df_cleaned.copy()
df_cleaned_upsample_result['TARGET_5Yrs'] = y_upsample_pred_proba[:,0]
print(df_cleaned_upsample_result['TARGET_5Yrs'].round().value_counts())
df_cleaned_upsample_result.to_csv('../data/processed/TestResult_RF_UpSampleUpdated_Result.csv',index=False,columns=['Id', 'TARGET_5Yrs'])
```
|
github_jupyter
|
import os
import sys
module_path = os.path.abspath(os.path.join('..'))
if module_path not in sys.path:
sys.path.append(module_path)
import pandas as pd
import numpy as np
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import GridSearchCV
from joblib import dump
from src.models import eval_model as evm
from src.models import eval_baseline as evb
from sklearn.utils import resample
%load_ext autoreload
%autoreload 2
from sklearn.model_selection import train_test_split
def read_and_split_data(file):
df = pd.read_csv(file)
x=df.drop(['TARGET_5Yrs','TARGET_5Yrs_Inv'],axis=1)
y=df['TARGET_5Yrs_Inv']
x_data , x_test ,y_data, y_test = train_test_split(x, y, test_size=0.2, random_state = 8, stratify=y)
x_train , x_val , y_train, y_val = train_test_split(x_data, y_data, test_size=0.2, random_state = 8, stratify=y_data)
print('y',y.value_counts())
print('y_train',y_train.value_counts())
print('y_val', y_val.value_counts())
print('y_test',y_test.value_counts())
return x_train , x_val , y_train, y_val, x_test, y_test
x_train , x_val , y_train, y_val, x_test, y_test = read_and_split_data("../data/processed/df_cleaned_upsampled_nba_prediction.csv")
evb.eval_baseline(x_train,y_train)
evm.eval_model(RandomForestClassifier(random_state=8),x_train,y_train,x_val,y_val)
rf_data = [
['n_estimators', 5],
['n_estimators', 25],
['n_estimators',50],
['n_estimators', 150],
['n_estimators',250],
['n_estimators',350],
['max_depth',2],
['max_depth',6],
['max_depth',12],
['max_depth',25],
['max_depth',50],
['min_samples_leaf',1],
['min_samples_leaf',2],
['min_samples_leaf',3],
['min_samples_leaf',5],
['min_samples_leaf',10],
['min_samples_leaf',15],
['min_samples_leaf',50],
['max_features',2],
['max_features',5],
['max_features',10],
['max_features',15]
]
rf_data
rf_param = pd.DataFrame(rf_data, columns = ['param', 'value'])
rf_param
for _,row in rf_param[rf_param['param']=='n_estimators'].iterrows():
evm.eval_model(RandomForestClassifier(random_state=8,n_estimators = int(row.value)) ,x_train,y_train,x_val,y_val)
nestimator = 50
for _,row in rf_param[rf_param['param']=='max_depth'].iterrows():
evm.eval_model(RandomForestClassifier(random_state=8,n_estimators = nestimator, max_depth= int(row.value)) ,x_train,y_train,x_val,y_val)
maxdepth = 25
for _,row in rf_param[rf_param['param']=='min_samples_leaf'].iterrows():
evm.eval_model(RandomForestClassifier(random_state=8,n_estimators =nestimator, max_depth=maxdepth,min_samples_leaf = int(row.value)) ,x_train,y_train,x_val,y_val)
minsamplesleaf=1
for _,row in rf_param[rf_param['param']=='max_features'].iterrows():
evm.eval_model(RandomForestClassifier(random_state=8,n_estimators=nestimator,max_depth=maxdepth,min_samples_leaf=minsamplesleaf,max_features = int(row.value)) ,x_train,y_train,x_val,y_val)
maxfeatures=2
print('max_depth=',maxdepth,' max_features=',maxfeatures, ' min_samples_leaf=',minsamplesleaf,' n_estimators=',nestimator)
rf_upsample_mod0 = RandomForestClassifier(random_state=8,n_estimators=nestimator,max_depth=maxdepth,min_samples_leaf=minsamplesleaf,max_features = maxfeatures)
evm.get_performance(rf_upsample_mod, x_test, y_test, "Test", True)
from hyperopt import Trials, STATUS_OK, tpe, hp, fmin
from sklearn.metrics import accuracy_score, confusion_matrix,roc_curve, roc_auc_score, precision_score, recall_score, precision_recall_curve, f1_score, plot_confusion_matrix
from sklearn.model_selection import cross_val_score , cross_validate
space = {
'max_depth': hp.choice('max_depth',range(1,100,1)),
'max_features': hp.choice('max_features',range(1,15,1)),
'min_samples_leaf': hp.choice('min_samples_leaf',range(1,14,1)),
'n_estimators': hp.choice('n_estimators',range(10,500,1))
}
space
def objective(space):
rf = RandomForestClassifier(max_depth = space['max_depth'],max_features = space['max_features'],min_samples_leaf = space['min_samples_leaf'],n_estimators = space['n_estimators'])
acc=cross_val_score(rf, x_train,y_train,cv=50, scoring='roc_auc').mean()
return{'loss': 1-acc, 'status': STATUS_OK }
best = fmin(
fn=objective,
space=space,
algo=tpe.suggest,
max_evals=5
)
best
rf_upsample_mod = RandomForestClassifier(random_state=8,n_estimators = best['n_estimators'], max_depth = best['max_depth'], max_features = best['max_features'])
evm.eval_model(rf_upsample_mod,x_train,y_train,x_val,y_val)
evm.get_performance(rf_upsample_mod, x_test, y_test, "Test", True)
#max_depth= 25 max_features= 2 min_samples_leaf= 1 n_estimators= 50
space = {
'max_depth': hp.choice('max_depth',range(1,30,1)),
'max_features': hp.choice('max_features',range(1,15,1)),
'min_samples_leaf': hp.choice('min_samples_leaf',range(1,14,1)),
'n_estimators': hp.choice('n_estimators',range(10,250,1))
}
space
def objective(space):
rf = RandomForestClassifier(max_depth = space['max_depth'],max_features = space['max_features'],min_samples_leaf = space['min_samples_leaf'],n_estimators = space['n_estimators'])
acc=cross_val_score(rf, x_train,y_train,cv=50, scoring='roc_auc').mean()
return{'loss': 1-acc, 'status': STATUS_OK }
best = fmin(
fn=objective,
space=space,
algo=tpe.suggest,
max_evals=5
)
best
rf_upsample_mod1 = RandomForestClassifier(random_state=8,n_estimators = best['n_estimators'], max_depth = best['max_depth'], max_features = best['max_features'], min_samples_leaf = best['min_samples_leaf'])
evm.eval_model(rf_upsample_mod1,x_train,y_train,x_val,y_val)
evm.get_performance(rf_upsample_mod1, x_test, y_test, "Test", True)
def read_and_split_data(file):
df = pd.read_csv(file)
df_cleaned = df.copy()
print('Before Data Clean')
for cols in df_cleaned.columns:
chk_rows = df_cleaned[df_cleaned[cols]<0].shape[0]
if chk_rows > 0 :
print(f'Column Name {cols},\tRows with Negative Value {chk_rows},\tPercentage {chk_rows/len(df)*100}')
df_cleaned[ df_cleaned<0 ] = 0
df_cleaned.loc[df_cleaned['3P Made'] <= 0, ['3P Made', '3PA', 'CALC3P%']] = 0, 0, 0
df_cleaned.loc[df_cleaned['FGM'] <= 0, ['FGM', 'FGA', 'CALCFG%']] = 0, 0, 0
df_cleaned.loc[df_cleaned['FTM'] <= 0, ['FTM', 'FTA', 'CALCFT%']] = 0, 0, 0
df_cleaned.loc[df_cleaned['3P Made'] > df_cleaned['3PA'], ['3P Made' , '3PA', 'CALC3P%']] = 0, 0, 0
df_cleaned.loc[df_cleaned['FGM'] > df_cleaned['FGA'], ['FGM', 'FGA', 'CALCFG%']] = 0, 0, 0
df_cleaned.loc[df_cleaned['FTM'] > df_cleaned['FTA'], ['FTM', 'FTA', 'CALCFT%']] = 0, 0, 0
df_cleaned.loc[df_cleaned['3P Made'] > 0, ['CALC3P%']] = df_cleaned['3P Made']/df_cleaned['3PA']*100
df_cleaned.loc[df_cleaned['FGM'] > 0, ['CALCFG%']] =df_cleaned['FGM']/df_cleaned['FGA']*100
df_cleaned.loc[df_cleaned['FTM'] > 0, ['CALCFT%']] = df_cleaned['FTM']/df_cleaned['FTA']*100
print(df_cleaned.head(5))
print('After Data Clean')
for cols in df_cleaned.columns:
chk_rows = df_cleaned[df_cleaned[cols]<0].shape[0]
if chk_rows > 0 :
print(f'Column Name {cols},\tRows with Negative Value {chk_rows},\tPercentage {chk_rows/len(df)*100}')
x = df_cleaned.drop(['3P%','FT%','FG%','Id_old','Id'],axis=1)
print(df_cleaned.columns)
return x , df_cleaned
x, df_cleaned =read_and_split_data('../data/raw/test.csv')
y_upsample_pred_proba=rf_upsample_mod.predict_proba(x)
y_upsample_pred=rf_upsample_mod.predict(x)
print(np.unique(y_upsample_pred_proba,return_counts=True))
print(np.unique(y_upsample_pred,return_counts=True))
print(rf_upsample_mod.classes_)
df_cleaned_upsample_result = df_cleaned.copy()
df_cleaned_upsample_result['TARGET_5Yrs'] = y_upsample_pred_proba[:,0]
print(df_cleaned_upsample_result['TARGET_5Yrs'].round().value_counts())
df_cleaned_upsample_result.to_csv('../data/processed/TestResult_RF_UpSampleUpdated_Result.csv',index=False,columns=['Id', 'TARGET_5Yrs'])
y_upsample_pred_proba=rf_upsample_mod1.predict_proba(x)
y_upsample_pred=rf_upsample_mod.predict(x)
print(np.unique(y_upsample_pred,return_counts=True))
df_cleaned_upsample_result = df_cleaned.copy()
df_cleaned_upsample_result['TARGET_5Yrs'] = y_upsample_pred_proba[:,0]
print(df_cleaned_upsample_result['TARGET_5Yrs'].round().value_counts())
df_cleaned_upsample_result.to_csv('../data/processed/TestResult_RF_UpSampleUpdated_Result.csv',index=False,columns=['Id', 'TARGET_5Yrs'])
| 0.312055 | 0.685818 |
# Create your own fake fMRI results
With this short jupyter notebook, you can create your own fake fMRI results. The only thing that you have to do is to specify the fake clusters that you want to create under **Targets**. After that you can run the whole notebook, either by using SHIFT+ENTER for each cell, or by selecting "Kernel > Restart & Run all" from the menu.
The notebook will create the fake dataset and save it in a output file called ``res.nii.gz``. It will also create different visualizations, plus extract the peak location of each cluster, its extend and the probability of anatomy, according to two different atlases.
# Targets
To create your fake fMRI results, you need to specify the following parameters:
- where in the brain do you want to have your significant cluster (x,y,z coordination)
- size of the cluster (specified by a radius of a sphere)
- intensity of the cluster (best chose about a value of 1, and add the directionality with '+' or '-')
For example, a cluster at 150,105,80, with a radius of 20, and an intensity of +1, is defined as:
target = [([150, 105, 80], 20, 1.)]
```
# Target: Location, radius, intensity
target = [([150, 105, 80], 20, 1.),
([30, 105, 80], 25, -1.),
([65, 30, 75], 30, -1.2),
([115, 30, 75], 30, 1.2)]
```
# Code to run the notebook
### Import python modules
```
%pylab inline
import numpy as np
import nibabel as nb
from nilearn.plotting import plot_stat_map, plot_glass_brain, cm
from nilearn.image import smooth_img
from scipy.stats import zscore
```
### Create dataset, insert target spheres and add some noise
```
# Load MNI152 template mask
mask = nb.load('templates/MNI152_T1_1mm_brain_mask.nii.gz')
# Create empty dataset
data = np.array(mask.get_data().copy() * 0, dtype='float')
# Add noise to dataset
data += np.random.normal(loc=1., scale=2., size=data.shape)
# Go through all the targets
for t in target:
# Create noisy sphere
radius = t[1]
r2 = np.arange(-radius, radius + 1)**2
r2 = -(r2 - r2.max())
dist = np.asarray(r2[:, None, None] + r2[:, None] + r2, dtype='float')
dist -= dist[0].max()
dist[dist <= 0] = 0
dist /= dist.max()
dist *= np.random.normal(loc=2.0, scale=2.5, size=dist.shape) * t[2]
# Add noisy sphere to dataset
c = t[0]
data[c[0] - radius:c[0] + radius + 1,
c[1] - radius:c[1] + radius + 1,
c[2] - radius:c[2] + radius + 1] += dist
# zscore data
data = zscore(data)
# Create NIfTI dataset
img = nb.Nifti1Image(data, mask.affine, mask.header)
# Smooth dataset
fwhm = 6
img = smooth_img(img, fwhm)
# Mask brain
data = img.get_data()
data *= mask.get_data()
# Threshold data and rescale it
data /= np.abs(data).max()
data[np.abs(data) <= np.percentile(np.abs(data), 99.5)] = 0
tmp = np.abs(data) - np.abs(data[data != 0]).min()
tmp[tmp <= 0] = 0
tmp /= tmp.max()
data = tmp * np.sign(data)
# Save Dataset
img = nb.Nifti1Image(data, img.affine, img.header)
nb.save(img, 'res.nii.gz')
```
# Visualize Results
## Visualize results on a glassbrain
There are multiple parameters that you can change, if you want:
- `display_mode`: changes which views you will see (i.e. left, right, front, top)
- `black_bg`: change if you want a black or white background
- `cmap`: what color scheme you want to use
```
plot_glass_brain('res.nii.gz', threshold=0.2, plot_abs=False, colorbar=True,
display_mode='lyrz', black_bg=False, cmap=cm.cold_hot)
```
## Visualize results on multiple slices
You can also visualize your results on multiple x, y or z sclises, with the MNI152 template as a background. In this approach you can change the following parameters:
- `display_mode`: use 'x', 'y', or 'z' do specify the plane to cut through
- `cut_coords`: specify which slices to cut, i.e. visualize
- `cmap`: what color scheme you want to use
```
anatimg = 'templates/MNI152_T1_1mm.nii.gz'
```
### Cut through z-plane
```
plot_stat_map('res.nii.gz', bg_img=anatimg, threshold=0.2,
cut_coords=(-5, 0, 5, 10, 15), display_mode='z', cmap=cm.cold_hot)
```
### Cut through y-plane
```
plot_stat_map('res.nii.gz', bg_img=anatimg, threshold=0.2,
cut_coords=(-100, -90, -22, -20, -18), display_mode='y', cmap=cm.cold_hot)
```
### Cut through x-plane
```
plot_stat_map('res.nii.gz', bg_img=anatimg, threshold=0.2,
cut_coords=(-65, -63, -61, -59), display_mode='x', cmap=cm.cold_hot)
plot_stat_map('res.nii.gz', bg_img=anatimg, threshold=0.2,
cut_coords=(59, 61, 63, 65), display_mode='x', cmap=cm.cold_hot)
```
### Show all planes at a specific location
```
plot_stat_map('res.nii.gz', bg_img=anatimg, threshold=0.2, display_mode='ortho',
cut_coords=(24, -92, 3), draw_cross=False, cmap=cm.cold_hot)
```
# Extract Cluster Information
To extract information about the clusters, we can run the following command. This command will extract for each cluster, the size (in voxels), the intensity at its peak (arbitrary value around 1) and the anatomical location of the peak, according to two different atlases. More information about the atlases can be found [here](https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/Atlases).
```
from scipy.ndimage import label
labels, nlabels = label(img.get_data())
for i in range(nlabels):
cSize = (labels == i + 1).sum()
maxcoord = np.abs(data) == np.abs(data[labels == i + 1]).max()
cPeak = np.round(data[maxcoord][0], 3)
print('Cluster %.2d\n==========' % (i + 1))
print(' Size = %s voxels\n Peak Value: %s\n' % (cSize, cPeak))
coord = np.dot(img.affine, np.hstack(
(np.ravel(np.where(maxcoord)), 1)))[:3].tolist()
coordStr = ','.join([str(coord[0]), str(coord[1]), str(coord[2])])
%run atlas_reader.py all $coordStr 0 1
```
|
github_jupyter
|
# Target: Location, radius, intensity
target = [([150, 105, 80], 20, 1.),
([30, 105, 80], 25, -1.),
([65, 30, 75], 30, -1.2),
([115, 30, 75], 30, 1.2)]
%pylab inline
import numpy as np
import nibabel as nb
from nilearn.plotting import plot_stat_map, plot_glass_brain, cm
from nilearn.image import smooth_img
from scipy.stats import zscore
# Load MNI152 template mask
mask = nb.load('templates/MNI152_T1_1mm_brain_mask.nii.gz')
# Create empty dataset
data = np.array(mask.get_data().copy() * 0, dtype='float')
# Add noise to dataset
data += np.random.normal(loc=1., scale=2., size=data.shape)
# Go through all the targets
for t in target:
# Create noisy sphere
radius = t[1]
r2 = np.arange(-radius, radius + 1)**2
r2 = -(r2 - r2.max())
dist = np.asarray(r2[:, None, None] + r2[:, None] + r2, dtype='float')
dist -= dist[0].max()
dist[dist <= 0] = 0
dist /= dist.max()
dist *= np.random.normal(loc=2.0, scale=2.5, size=dist.shape) * t[2]
# Add noisy sphere to dataset
c = t[0]
data[c[0] - radius:c[0] + radius + 1,
c[1] - radius:c[1] + radius + 1,
c[2] - radius:c[2] + radius + 1] += dist
# zscore data
data = zscore(data)
# Create NIfTI dataset
img = nb.Nifti1Image(data, mask.affine, mask.header)
# Smooth dataset
fwhm = 6
img = smooth_img(img, fwhm)
# Mask brain
data = img.get_data()
data *= mask.get_data()
# Threshold data and rescale it
data /= np.abs(data).max()
data[np.abs(data) <= np.percentile(np.abs(data), 99.5)] = 0
tmp = np.abs(data) - np.abs(data[data != 0]).min()
tmp[tmp <= 0] = 0
tmp /= tmp.max()
data = tmp * np.sign(data)
# Save Dataset
img = nb.Nifti1Image(data, img.affine, img.header)
nb.save(img, 'res.nii.gz')
plot_glass_brain('res.nii.gz', threshold=0.2, plot_abs=False, colorbar=True,
display_mode='lyrz', black_bg=False, cmap=cm.cold_hot)
anatimg = 'templates/MNI152_T1_1mm.nii.gz'
plot_stat_map('res.nii.gz', bg_img=anatimg, threshold=0.2,
cut_coords=(-5, 0, 5, 10, 15), display_mode='z', cmap=cm.cold_hot)
plot_stat_map('res.nii.gz', bg_img=anatimg, threshold=0.2,
cut_coords=(-100, -90, -22, -20, -18), display_mode='y', cmap=cm.cold_hot)
plot_stat_map('res.nii.gz', bg_img=anatimg, threshold=0.2,
cut_coords=(-65, -63, -61, -59), display_mode='x', cmap=cm.cold_hot)
plot_stat_map('res.nii.gz', bg_img=anatimg, threshold=0.2,
cut_coords=(59, 61, 63, 65), display_mode='x', cmap=cm.cold_hot)
plot_stat_map('res.nii.gz', bg_img=anatimg, threshold=0.2, display_mode='ortho',
cut_coords=(24, -92, 3), draw_cross=False, cmap=cm.cold_hot)
from scipy.ndimage import label
labels, nlabels = label(img.get_data())
for i in range(nlabels):
cSize = (labels == i + 1).sum()
maxcoord = np.abs(data) == np.abs(data[labels == i + 1]).max()
cPeak = np.round(data[maxcoord][0], 3)
print('Cluster %.2d\n==========' % (i + 1))
print(' Size = %s voxels\n Peak Value: %s\n' % (cSize, cPeak))
coord = np.dot(img.affine, np.hstack(
(np.ravel(np.where(maxcoord)), 1)))[:3].tolist()
coordStr = ','.join([str(coord[0]), str(coord[1]), str(coord[2])])
%run atlas_reader.py all $coordStr 0 1
| 0.761361 | 0.873107 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.