text
stringlengths 2.5k
6.39M
| kind
stringclasses 3
values |
|---|---|
<i>Copyright (c) Microsoft Corporation. All rights reserved.</i>
<i>Licensed under the MIT License.</i>
# Evaluation
Evaluation with offline metrics is pivotal to assess the quality of a recommender before it goes into production. Usually, evaluation metrics are carefully chosen based on the actual application scenario of a recommendation system. It is hence important to data scientists and AI developers that build recommendation systems to understand how each evaluation metric is calculated and what it is for.
This notebook deep dives into several commonly used evaluation metrics, and illustrates how these metrics are used in practice. The metrics covered in this notebook are merely for off-line evaluations.
## 0 Global settings
Most of the functions used in the notebook can be found in the `reco_utils` directory.
```
# set the environment path to find Recommenders
import sys
sys.path.append("../../")
import pandas as pd
import pyspark
from sklearn.preprocessing import minmax_scale
from reco_utils.common.spark_utils import start_or_get_spark
from reco_utils.evaluation.spark_evaluation import SparkRankingEvaluation, SparkRatingEvaluation
from reco_utils.evaluation.python_evaluation import auc, logloss
from reco_utils.recommender.sar.sar_singlenode import SARSingleNode
from reco_utils.dataset.download_utils import maybe_download
from reco_utils.dataset.python_splitters import python_random_split
print("System version: {}".format(sys.version))
print("Pandas version: {}".format(pd.__version__))
print("PySpark version: {}".format(pyspark.__version__))
```
Note to successfully run Spark codes with the Jupyter kernel, one needs to correctly set the environment variables of `PYSPARK_PYTHON` and `PYSPARK_DRIVER_PYTHON` that point to Python executables with the desired version. Detailed information can be found in the setup instruction document [SETUP.md](../../SETUP.md).
```
COL_USER = "UserId"
COL_ITEM = "MovieId"
COL_RATING = "Rating"
COL_PREDICTION = "Rating"
HEADER = {
"col_user": COL_USER,
"col_item": COL_ITEM,
"col_rating": COL_RATING,
"col_prediction": COL_PREDICTION,
}
```
## 1 Prepare data
### 1.1 Prepare dummy data
For illustration purpose, a dummy data set is created for demonstrating how different evaluation metrics work.
The data has the schema that can be frequently found in a recommendation problem, that is, each row in the dataset is a (user, item, rating) tuple, where "rating" can be an ordinal rating score (e.g., discrete integers of 1, 2, 3, etc.) or an numerical float number that quantitatively indicates the preference of the user towards that item.
For simplicity reason, the column of rating in the dummy dataset we use in the example represent some ordinal ratings.
```
df_true = pd.DataFrame(
{
COL_USER: [1, 1, 1, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3],
COL_ITEM: [1, 2, 3, 1, 4, 5, 6, 7, 2, 5, 6, 8, 9, 10, 11, 12, 13, 14],
COL_RATING: [5, 4, 3, 5, 5, 3, 3, 1, 5, 5, 5, 4, 4, 3, 3, 3, 2, 1],
}
)
df_pred = pd.DataFrame(
{
COL_USER: [1, 1, 1, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3],
COL_ITEM: [3, 10, 12, 10, 3, 5, 11, 13, 4, 10, 7, 13, 1, 3, 5, 2, 11, 14],
COL_PREDICTION: [14, 13, 12, 14, 13, 12, 11, 10, 14, 13, 12, 11, 10, 9, 8, 7, 6, 5]
}
)
```
Take a look at ratings of the user with ID "1" in the dummy dataset.
```
df_true[df_true[COL_USER] == 1]
df_pred[df_pred[COL_USER] == 1]
```
### 1.2 Prepare Spark data
Spark framework is sometimes used to evaluate metrics given datasets that are hard to fit into memory. In our example, Spark DataFrames can be created from the Python dummy dataset.
```
spark = start_or_get_spark("EvaluationTesting", "local")
dfs_true = spark.createDataFrame(df_true)
dfs_pred = spark.createDataFrame(df_pred)
dfs_true.filter(dfs_true[COL_USER] == 1).show()
dfs_pred.filter(dfs_pred[COL_USER] == 1).show()
```
## 2 Evaluation metrics
### 2.1 Rating metrics
Rating metrics are similar to regression metrics used for evaluating a regression model that predicts numerical values given input observations. In the context of recommendation system, rating metrics are to evaluate how accurate a recommender is to predict ratings that users may give to items. Therefore, the metrics are **calculated exactly on the same group of (user, item) pairs that exist in both ground-truth dataset and prediction dataset** and **averaged by the total number of users**.
#### 2.1.1 Use cases
Rating metrics are effective in measuring the model accuracy. However, in some cases, the rating metrics are limited if
* **the recommender is to predict ranking instead of explicit rating**. For example, if the consumer of the recommender cares about the ranked recommended items, rating metrics do not apply directly. Usually a relevancy function such as top-k will be applied to generate the ranked list from predicted ratings in order to evaluate the recommender with other metrics.
* **the recommender is to generate recommendation scores that have different scales with the original ratings (e.g., the SAR algorithm)**. In this case, the difference between the generated scores and the original scores (or, ratings) is not valid for measuring accuracy of the model.
#### 2.1.2 How-to with the evaluation utilities
A few notes about the interface of the Rating evaluator class:
1. The columns of user, item, and rating (prediction) should be present in the ground-truth DataFrame (prediction DataFrame).
2. There should be no duplicates of (user, item) pairs in the ground-truth and the prediction DataFrames, othewise there may be unexpected behavior in calculating certain metrics.
3. Default column names for user, item, rating, and prediction are "UserId", "ItemId", "Rating", and "Prediciton", respectively.
In our examples below, to calculate rating metrics for input data frames in Spark, a Spark object, `SparkRatingEvaluation` is initialized. The input data schemas for the ground-truth dataset and the prediction dataset are
* Ground-truth dataset.
|Column|Data type|Description|
|-------------|------------|-------------|
|`COL_USER`|<int\>|User ID|
|`COL_ITEM`|<int\>|Item ID|
|`COL_RATING`|<float\>|Rating or numerical value of user preference.|
* Prediction dataset.
|Column|Data type|Description|
|-------------|------------|-------------|
|`COL_USER`|<int\>|User ID|
|`COL_ITEM`|<int\>|Item ID|
|`COL_RATING`|<float\>|Predicted rating or numerical value of user preference.|
```
spark_rate_eval = SparkRatingEvaluation(dfs_true, dfs_pred, **HEADER)
```
#### 2.1.3 Root Mean Square Error (RMSE)
RMSE is for evaluating the accuracy of prediction on ratings. RMSE is the most widely used metric to evaluate a recommendation algorithm that predicts missing ratings. The benefit is that RMSE is easy to explain and calculate.
```
print("The RMSE is {}".format(spark_rate_eval.rmse()))
```
#### 2.1.4 R Squared (R2)
R2 is also called "coefficient of determination" in some context. It is a metric that evaluates how well a regression model performs, based on the proportion of total variations of the observed results.
```
print("The R2 is {}".format(spark_rate_eval.rsquared()))
```
#### 2.1.5 Mean Absolute Error (MAE)
MAE evaluates accuracy of prediction. It computes the metric value from ground truths and prediction in the same scale. Compared to RMSE, MAE is more explainable.
```
print("The MAE is {}".format(spark_rate_eval.mae()))
```
#### 2.1.6 Explained Variance
Explained variance is usually used to measure how well a model performs with regard to the impact from the variation of the dataset.
```
print("The explained variance is {}".format(spark_rate_eval.exp_var()))
```
#### 2.1.7 Summary
|Metric|Range|Selection criteria|Limitation|Reference|
|------|-------------------------------|---------|----------|---------|
|RMSE|$> 0$|The smaller the better.|May be biased, and less explainable than MSE|[link](https://en.wikipedia.org/wiki/Root-mean-square_deviation)|
|R2|$\leq 1$|The closer to $1$ the better.|Depend on variable distributions.|[link](https://en.wikipedia.org/wiki/Coefficient_of_determination)|
|MSE|$\geq 0$|The smaller the better.|Dependent on variable scale.|[link](https://en.wikipedia.org/wiki/Mean_absolute_error)|
|Explained variance|$\leq 1$|The closer to $1$ the better.|Depend on variable distributions.|[link](https://en.wikipedia.org/wiki/Explained_variation)|
### 2.2 Ranking metrics
"Beyond-accuray evaluation" was proposed to evaluate how relevant recommendations are for users. In this case, a recommendation system is a treated as a ranking system. Given a relency definition, recommendation system outputs a list of recommended items to each user, which is ordered by relevance. The evaluation part takes ground-truth data, the actual items that users interact with (e.g., liked, purchased, etc.), and the recommendation data, as inputs, to calculate ranking evaluation metrics.
#### 2.2.1 Use cases
Ranking metrics are often used when hit and/or ranking of the items are considered:
* **Hit** - defined by relevancy, a hit usually means whether the recommended "k" items hit the "relevant" items by the user. For example, a user may have clicked, viewed, or purchased an item for many times, and a hit in the recommended items indicate that the recommender performs well. Metrics like "precision", "recall", etc. measure the performance of such hitting accuracy.
* **Ranking** - ranking metrics give more explanations about, for the hitted items, whether they are ranked in a way that is preferred by the users whom the items will be recommended to. Metrics like "mean average precision", "ndcg", etc., evaluate whether the relevant items are ranked higher than the less-relevant or irrelevant items.
#### 2.2.2 How-to with evaluation utilities
A few notes about the interface of the Rating evaluator class:
1. The columns of user, item, and rating (prediction) should be present in the ground-truth DataFrame (prediction DataFrame). The column of timestamp is optional, but it is required if certain relevanc function is used. For example, timestamps will be used if the most recent items are defined as the relevant one.
2. There should be no duplicates of (user, item) pairs in the ground-truth and the prediction DataFrames, othewise there may be unexpected behavior in calculating certain metrics.
3. Default column names for user, item, rating, and prediction are "UserId", "ItemId", "Rating", and "Prediciton", respectively.
#### 2.2.1 Relevancy of recommendation
Relevancy of recommendation can be measured in different ways:
* **By ranking** - In this case, relevant items in the recommendations are defined as the top ranked items, i.e., top k items, which are taken from the list of the recommended items that is ordered by the predicted ratings (or other numerical scores that indicate preference of a user to an item).
* **By timestamp** - Relevant items are defined as the most recently viewed k items, which are obtained from the recommended items ranked by timestamps.
* **By rating** - Relevant items are defined as items with ratings (or other numerical scores that indicate preference of a user to an item) that are above a given threshold.
Similarly, a ranking metric object can be initialized as below. The input data schema is
* Ground-truth dataset.
|Column|Data type|Description|
|-------------|------------|-------------|
|`COL_USER`|<int\>|User ID|
|`COL_ITEM`|<int\>|Item ID|
|`COL_RATING`|<float\>|Rating or numerical value of user preference.|
|`COL_TIMESTAMP`|<string\>|Timestamps.|
* Prediction dataset.
|Column|Data type|Description|
|-------------|------------|-------------|
|`COL_USER`|<int\>|User ID|
|`COL_ITEM`|<int\>|Item ID|
|`COL_RATING`|<float\>|Predicted rating or numerical value of user preference.|
|`COL_TIMESTAM`|<string\>|Timestamps.|
In this case, in addition to the input datasets, there are also other arguments used for calculating the ranking metrics:
|Argument|Data type|Description|
|------------|------------|--------------|
|`k`|<int\>|Number of items recommended to user.|
|`revelancy_method`|<string\>|Methonds that extract relevant items from the recommendation list|
For example, the following code initializes a ranking metric object that calculates the metrics.
```
spark_rank_eval = SparkRankingEvaluation(dfs_true, dfs_pred, k=3, relevancy_method="top_k", **HEADER)
```
A few ranking metrics can then be calculated.
#### 2.2.1 Precision
Precision@k is a metric that evaluates how many items in the recommendation list are relevant (hit) in the ground-truth data. For each user the precision score is normalized by `k` and then the overall precision scores are averaged by the total number of users.
Note it is apparent that the precision@k metric grows with the number of `k`.
```
print("The precision at k is {}".format(spark_rank_eval.precision_at_k()))
```
#### 2.2.2 Recall
Recall@k is a metric that evaluates how many relevant items in the ground-truth data are in the recommendation list. For each user the recall score is normalized by the total number of ground-truth items and then the overall recall scores are averaged by the total number of users.
```
print("The recall at k is {}".format(spark_rank_eval.recall_at_k()))
```
#### 2.2.3 Normalized Discounted Cumulative Gain (NDCG)
NDCG is a metric that evaluates how well the recommender performs in recommending ranked items to users. Therefore both hit of relevant items and correctness in ranking of these items matter to the NDCG evaluation. The total NDCG score is normalized by the total number of users.
```
print("The ndcg at k is {}".format(spark_rank_eval.ndcg_at_k()))
```
#### 2.2.4 Mean Average Precision (MAP)
MAP is a metric that evaluates the average precision for each user in the datasets. It also penalizes ranking correctness of the recommended items. The overall MAP score is normalized by the total number of users.
```
print("The map at k is {}".format(spark_rank_eval.map_at_k()))
```
#### 2.2.5 ROC and AUC
ROC, as well as AUC, is a well known metric that is used for evaluating binary classification problem. It is similar in the case of binary rating typed recommendation algorithm where the "hit" accuracy on the relevant items is used for measuring the recommender's performance.
To demonstrate the evaluation method, the original data for testing is manipuldated in a way that the ratings in the testing data are arranged as binary scores, whilst the ones in the prediction are scaled in 0 to 1.
```
# Convert the original rating to 0 and 1.
df_true_bin = df_true.copy()
df_true_bin[COL_RATING] = df_true_bin[COL_RATING].apply(lambda x: 1 if x > 3 else 0)
df_true_bin
# Convert the predicted ratings into a [0, 1] scale.
df_pred_bin = df_pred.copy()
df_pred_bin[COL_PREDICTION] = minmax_scale(df_pred_bin[COL_PREDICTION].astype(float))
df_pred_bin
# Calculate the AUC metric
auc_score = auc(
df_true_bin,
df_pred_bin,
col_user = COL_USER,
col_item = COL_ITEM,
col_rating = COL_RATING,
col_prediction = COL_RATING
)
print("The auc score is {}".format(auc_score))
```
It is worth mentioning that in some literature there are variants of the original AUC metric, that considers the effect of **the number of the recommended items (k)**, **grouping effect of users (compute AUC for each user group, and take the average across different groups)**. These variants are applicable to various different scenarios, and choosing an appropriate one depends on the context of the use case itself.
#### 2.3.2 Logistic loss
Logistic loss (sometimes it is called simply logloss, or cross-entropy loss) is another useful metric to evaluate the hit accuracy. It is defined as the negative log-likelihood of the true labels given the predictions of a classifier.
```
# Calculate the logloss metric
logloss_score = logloss(
df_true_bin,
df_pred_bin,
col_user = COL_USER,
col_item = COL_ITEM,
col_rating = COL_RATING,
col_prediction = COL_RATING
)
print("The logloss score is {}".format(logloss_score))
```
It is worth noting that logloss may be sensitive to the class balance of datasets, as it penalizes heavily classifiers that are confident about incorrect classifications. To demonstrate, the ground truth data set for testing is manipulated purposely to unbalance the binary labels. For example, the following binarizes the original rating data by using a lower threshold, i.e., 2, to create more positive feedback from the user.
```
df_true_bin_pos = df_true.copy()
df_true_bin_pos[COL_RATING] = df_true_bin_pos[COL_RATING].apply(lambda x: 1 if x > 2 else 0)
df_true_bin_pos
```
By using threshold of 2, the labels in the ground truth data is not balanced, and the ratio of 1 over 0 is
```
one_zero_ratio = df_true_bin_pos[COL_PREDICTION].sum() / (df_true_bin_pos.shape[0] - df_true_bin_pos[COL_PREDICTION].sum())
print('The ratio between label 1 and label 0 is {}'.format(one_zero_ratio))
```
Another prediction data is also created, where the probabilities for label 1 and label 0 are fixed. Without loss of generity, the probability of predicting 1 is 0.6. The data set is purposely created to make the precision to be 100% given an presumption of cut-off equal to 0.5.
```
prob_true = 0.6
df_pred_bin_pos = df_true_bin_pos.copy()
df_pred_bin_pos[COL_PREDICTION] = df_pred_bin_pos[COL_PREDICTION].apply(lambda x: prob_true if x==1 else 1-prob_true)
df_pred_bin_pos
```
Then the logloss is calculated as follows.
```
# Calculate the logloss metric
logloss_score_pos = logloss(
df_true_bin_pos,
df_pred_bin_pos,
col_user = COL_USER,
col_item = COL_ITEM,
col_rating = COL_RATING,
col_prediction = COL_RATING
)
print("The logloss score is {}".format(logloss_score))
```
For comparison, a similar process is used with a threshold value of 3 to create a more balanced dataset. Another prediction dataset is also created by using the balanced dataset. Again, the probabilities of predicting label 1 and label 0 are fixed as 0.6 and 0.4, respectively. **NOTE**, same as above, in this case, the prediction also gives us a 100% precision. The only difference is the proportion of binary labels.
```
prob_true = 0.6
df_pred_bin_balanced = df_true_bin.copy()
df_pred_bin_balanced[COL_PREDICTION] = df_pred_bin_balanced[COL_PREDICTION].apply(lambda x: prob_true if x==1 else 1-prob_true)
df_pred_bin_balanced
```
The ratio of label 1 and label 0 is
```
one_zero_ratio = df_true_bin[COL_PREDICTION].sum() / (df_true_bin.shape[0] - df_true_bin[COL_PREDICTION].sum())
print('The ratio between label 1 and label 0 is {}'.format(one_zero_ratio))
```
It is perfectly balanced.
Applying the logloss function to calculate the metric gives us a more promising result, as shown below.
```
# Calculate the logloss metric
logloss_score = logloss(
df_true_bin,
df_pred_bin_balanced,
col_user = COL_USER,
col_item = COL_ITEM,
col_rating = COL_RATING,
col_prediction = COL_RATING
)
print("The logloss score is {}".format(logloss_score))
```
It can be seen that the score is more close to 0, and, by definition, it means that the predictions are generating better results than the one before where binary labels are more biased.
#### 2.2.5 Summary
|Metric|Range|Selection criteria|Limitation|Reference|
|------|-------------------------------|---------|----------|---------|
|Precision|$\geq 0$ and $\leq 1$|The closer to $1$ the better.|Only for hits in recommendations.|[link](https://spark.apache.org/docs/2.3.0/mllib-evaluation-metrics.html#ranking-systems)|
|Recall|$\geq 0$ and $\leq 1$|The closer to $1$ the better.|Only for hits in the ground truth.|[link](https://en.wikipedia.org/wiki/Precision_and_recall)|
|NDCG|$\geq 0$ and $\leq 1$|The closer to $1$ the better.|Does not penalize for bad/missing items, and does not perform for several equally good items.|[link](https://spark.apache.org/docs/2.3.0/mllib-evaluation-metrics.html#ranking-systems)|
|MAP|$\geq 0$ and $\leq 1$|The closer to $1$ the better.|Depend on variable distributions.|[link](https://spark.apache.org/docs/2.3.0/mllib-evaluation-metrics.html#ranking-systems)|
|AUC|$\geq 0$ and $\leq 1$|The closer to $1$ the better. 0.5 indicates an uninformative classifier|Depend on the number of recommended items (k).|[link](https://en.wikipedia.org/wiki/Receiver_operating_characteristic#Area_under_the_curve)|
|Logloss|$0$ to $\infty$|The closer to $0$ the better.|Logloss can be sensitive to imbalanced datasets.|[link](https://en.wikipedia.org/wiki/Cross_entropy#Relation_to_log-likelihood)|
## References
1. Guy Shani and Asela Gunawardana, "Evaluating Recommendation Systems", Recommender Systems Handbook, Springer, 2015.
2. PySpark MLlib evaluation metrics, url: https://spark.apache.org/docs/2.3.0/mllib-evaluation-metrics.html.
3. Dimitris Paraschakis et al, "Comparative Evaluation of Top-N Recommenders in e-Commerce: An Industrial Perspective", IEEE ICMLA, 2015, Miami, FL, USA.
4. Yehuda Koren and Robert Bell, "Advances in Collaborative Filtering", Recommender Systems Handbook, Springer, 2015.
5. Chris Bishop, "Pattern Recognition and Machine Learning", Springer, 2006.
|
github_jupyter
|
## <div style="text-align: center"> 20 ML Algorithms from start to Finish for Iris</div>
<div style="text-align: center"> I want to solve<b> iris problem</b> a popular machine learning Dataset as a comprehensive workflow with python packages.
After reading, you can use this workflow to solve other real problems and use it as a template to deal with <b>machine learning</b> problems.</div>

<div style="text-align:center">last update: <b>10/28/2018</b></div>
>###### you may be interested have a look at it: [**10-steps-to-become-a-data-scientist**](https://github.com/mjbahmani/10-steps-to-become-a-data-scientist)
---------------------------------------------------------------------
you can Fork and Run this kernel on Github:
> ###### [ GitHub](https://github.com/mjbahmani/Machine-Learning-Workflow-with-Python)
-------------------------------------------------------------------------------------------------------------
**I hope you find this kernel helpful and some <font color="red"><b>UPVOTES</b></font> would be very much appreciated**
-----------
## Notebook Content
* 1- [Introduction](#1)
* 2- [Machine learning workflow](#2)
* 2-1 [Real world Application Vs Competitions](#2)
* 3- [Problem Definition](#3)
* 3-1 [Problem feature](#4)
* 3-2 [Aim](#5)
* 3-3 [Variables](#6)
* 4-[ Inputs & Outputs](#7)
* 4-1 [Inputs ](#8)
* 4-2 [Outputs](#9)
* 5- [Installation](#10)
* 5-1 [ jupyter notebook](#11)
* 5-2[ kaggle kernel](#12)
* 5-3 [Colab notebook](#13)
* 5-4 [install python & packages](#14)
* 5-5 [Loading Packages](#15)
* 6- [Exploratory data analysis](#16)
* 6-1 [Data Collection](#17)
* 6-2 [Visualization](#18)
* 6-2-1 [Scatter plot](#19)
* 6-2-2 [Box](#20)
* 6-2-3 [Histogram](#21)
* 6-2-4 [Multivariate Plots](#22)
* 6-2-5 [Violinplots](#23)
* 6-2-6 [Pair plot](#24)
* 6-2-7 [Kde plot](#25)
* 6-2-8 [Joint plot](#26)
* 6-2-9 [Andrews curves](#27)
* 6-2-10 [Heatmap](#28)
* 6-2-11 [Radviz](#29)
* 6-3 [Data Preprocessing](#30)
* 6-4 [Data Cleaning](#31)
* 7- [Model Deployment](#32)
* 7-1[ KNN](#33)
* 7-2 [Radius Neighbors Classifier](#34)
* 7-3 [Logistic Regression](#35)
* 7-4 [Passive Aggressive Classifier](#36)
* 7-5 [Naive Bayes](#37)
* 7-6 [MultinomialNB](#38)
* 7-7 [BernoulliNB](#39)
* 7-8 [SVM](#40)
* 7-9 [Nu-Support Vector Classification](#41)
* 7-10 [Linear Support Vector Classification](#42)
* 7-11 [Decision Tree](#43)
* 7-12 [ExtraTreeClassifier](#44)
* 7-13 [Neural network](#45)
* 7-13-1 [What is a Perceptron?](#45)
* 7-14 [RandomForest](#46)
* 7-15 [Bagging classifier ](#47)
* 7-16 [AdaBoost classifier](#48)
* 7-17 [Gradient Boosting Classifier](#49)
* 7-18 [Linear Discriminant Analysis](#50)
* 7-19 [Quadratic Discriminant Analysis](#51)
* 7-20 [Kmeans](#52)
* 7-21 [Backpropagation](#53)
* 8- [Conclusion](#54)
* 10- [References](#55)
<a id="1"></a> <br>
## 1- Introduction
This is a **comprehensive ML techniques with python** , that I have spent for more than two months to complete it.
it is clear that everyone in this community is familiar with IRIS dataset but if you need to review your information about the dataset please visit this [link](https://archive.ics.uci.edu/ml/datasets/iris).
I have tried to help **beginners** in Kaggle how to face machine learning problems. and I think it is a great opportunity for who want to learn machine learning workflow with python completely.
I have covered most of the methods that are implemented for iris until **2018**, you can start to learn and review your knowledge about ML with a simple dataset and try to learn and memorize the workflow for your journey in Data science world.
## 1-1 Courses
There are alot of Online courses that can help you develop your knowledge, here I have just listed some of them:
1. [Machine Learning Certification by Stanford University (Coursera)](https://www.coursera.org/learn/machine-learning/)
2. [Machine Learning A-Z™: Hands-On Python & R In Data Science (Udemy)](https://www.udemy.com/machinelearning/)
3. [Deep Learning Certification by Andrew Ng from deeplearning.ai (Coursera)](https://www.coursera.org/specializations/deep-learning)
4. [Python for Data Science and Machine Learning Bootcamp (Udemy)](Python for Data Science and Machine Learning Bootcamp (Udemy))
5. [Mathematics for Machine Learning by Imperial College London](https://www.coursera.org/specializations/mathematics-machine-learning)
6. [Deep Learning A-Z™: Hands-On Artificial Neural Networks](https://www.udemy.com/deeplearning/)
7. [Complete Guide to TensorFlow for Deep Learning Tutorial with Python](https://www.udemy.com/complete-guide-to-tensorflow-for-deep-learning-with-python/)
8. [Data Science and Machine Learning Tutorial with Python – Hands On](https://www.udemy.com/data-science-and-machine-learning-with-python-hands-on/)
9. [Machine Learning Certification by University of Washington](https://www.coursera.org/specializations/machine-learning)
10. [Data Science and Machine Learning Bootcamp with R](https://www.udemy.com/data-science-and-machine-learning-bootcamp-with-r/)
5- [https://www.kaggle.com/startupsci/titanic-data-science-solutions](https://www.kaggle.com/startupsci/titanic-data-science-solutions)
I am open to getting your feedback for improving this **kernel**
<a id="2"></a> <br>
## 2- Machine Learning Workflow
Field of study that gives computers the ability to learn without being
explicitly programmed.
Arthur Samuel, 1959
If you have already read some [machine learning books](https://towardsdatascience.com/list-of-free-must-read-machine-learning-books-89576749d2ff). You have noticed that there are different ways to stream data into machine learning.
most of these books share the following steps (checklist):
* Define the Problem(Look at the big picture)
* Specify Inputs & Outputs
* Data Collection
* Exploratory data analysis
* Data Preprocessing
* Model Design, Training, and Offline Evaluation
* Model Deployment, Online Evaluation, and Monitoring
* Model Maintenance, Diagnosis, and Retraining
**You can see my workflow in the below image** :
<img src="http://s9.picofile.com/file/8338227634/workflow.png" />
**you should feel free to adapt this checklist to your needs**
## 2-1 Real world Application Vs Competitions
<img src="http://s9.picofile.com/file/8339956300/reallife.png" height="600" width="500" />
<a id="3"></a> <br>
## 3- Problem Definition
I think one of the important things when you start a new machine learning project is Defining your problem. that means you should understand business problem.( **Problem Formalization**)
Problem Definition has four steps that have illustrated in the picture below:
<img src="http://s8.picofile.com/file/8338227734/ProblemDefination.png">
<a id="4"></a> <br>
### 3-1 Problem Feature
we will use the classic Iris data set. This dataset contains information about three different types of Iris flowers:
* Iris Versicolor
* Iris Virginica
* Iris Setosa
The data set contains measurements of four variables :
* sepal length
* sepal width
* petal length
* petal width
The Iris data set has a number of interesting features:
1. One of the classes (Iris Setosa) is linearly separable from the other two. However, the other two classes are not linearly separable.
2. There is some overlap between the Versicolor and Virginica classes, so it is unlikely to achieve a perfect classification rate.
3. There is some redundancy in the four input variables, so it is possible to achieve a good solution with only three of them, or even (with difficulty) from two, but the precise choice of best variables is not obvious.
**Why am I using iris dataset:**
1- This is a good project because it is so well understood.
2- Attributes are numeric so you have to figure out how to load and handle data.
3- It is a classification problem, allowing you to practice with perhaps an easier type of supervised learning algorithm.
4- It is a multi-class classification problem (multi-nominal) that may require some specialized handling.
5- It only has 4 attributes and 150 rows, meaning it is small and easily fits into memory (and a screen or A4 page).
6- All of the numeric attributes are in the same units and the same scale, not requiring any special scaling or transforms to get started.[5]
7- we can define problem as clustering(unsupervised algorithm) project too.
<a id="5"></a> <br>
### 3-2 Aim
The aim is to classify iris flowers among three species (setosa, versicolor or virginica) from measurements of length and width of sepals and petals
<a id="6"></a> <br>
### 3-3 Variables
The variables are :
**sepal_length**: Sepal length, in centimeters, used as input.
**sepal_width**: Sepal width, in centimeters, used as input.
**petal_length**: Petal length, in centimeters, used as input.
**petal_width**: Petal width, in centimeters, used as input.
**setosa**: Iris setosa, true or false, used as target.
**versicolour**: Iris versicolour, true or false, used as target.
**virginica**: Iris virginica, true or false, used as target.
**<< Note >>**
> You must answer the following question:
How does your company expact to use and benfit from your model.
<a id="7"></a> <br>
## 4- Inputs & Outputs
<a id="8"></a> <br>
### 4-1 Inputs
**Iris** is a very popular **classification** and **clustering** problem in machine learning and it is such as "Hello world" program when you start learning a new programming language. then I decided to apply Iris on 20 machine learning method on it.
The Iris flower data set or Fisher's Iris data set is a **multivariate data set** introduced by the British statistician and biologist Ronald Fisher in his 1936 paper The use of multiple measurements in taxonomic problems as an example of linear discriminant analysis. It is sometimes called Anderson's Iris data set because Edgar Anderson collected the data to quantify the morphologic variation of Iris flowers in three related species. Two of the three species were collected in the Gaspé Peninsula "all from the same pasture, and picked on the same day and measured at the same time by the same person with the same apparatus".
The data set consists of 50 samples from each of three species of Iris (Iris setosa, Iris virginica, and Iris versicolor). Four features were measured from each sample: the length and the width of the sepals and petals, in centimeters. Based on the combination of these four features, Fisher developed a linear discriminant model to distinguish the species from each other.
As a result, **iris dataset is used as the input of all algorithms**.
<a id="9"></a> <br>
### 4-2 Outputs
the outputs for our algorithms totally depend on the type of classification or clustering algorithms.
the outputs can be the number of clusters or predict for new input.
**setosa**: Iris setosa, true or false, used as target.
**versicolour**: Iris versicolour, true or false, used as target.
**virginica**: Iris virginica, true or false, used as a target.
<a id="10"></a> <br>
## 5-Installation
#### Windows:
* Anaconda (from https://www.continuum.io) is a free Python distribution for SciPy stack. It is also available for Linux and Mac.
* Canopy (https://www.enthought.com/products/canopy/) is available as free as well as commercial distribution with full SciPy stack for Windows, Linux and Mac.
* Python (x,y) is a free Python distribution with SciPy stack and Spyder IDE for Windows OS. (Downloadable from http://python-xy.github.io/)
#### Linux
Package managers of respective Linux distributions are used to install one or more packages in SciPy stack.
For Ubuntu Users:
sudo apt-get install python-numpy python-scipy python-matplotlibipythonipythonnotebook
python-pandas python-sympy python-nose
<a id="11"></a> <br>
## 5-1 Jupyter notebook
I strongly recommend installing **Python** and **Jupyter** using the **[Anaconda Distribution](https://www.anaconda.com/download/)**, which includes Python, the Jupyter Notebook, and other commonly used packages for scientific computing and data science.
First, download Anaconda. We recommend downloading Anaconda’s latest Python 3 version.
Second, install the version of Anaconda which you downloaded, following the instructions on the download page.
Congratulations, you have installed Jupyter Notebook! To run the notebook, run the following command at the Terminal (Mac/Linux) or Command Prompt (Windows):
> jupyter notebook
>
<a id="12"></a> <br>
## 5-2 Kaggle Kernel
Kaggle kernel is an environment just like you use jupyter notebook, it's an **extension** of the where in you are able to carry out all the functions of jupyter notebooks plus it has some added tools like forking et al.
<a id="13"></a> <br>
## 5-3 Colab notebook
**Colaboratory** is a research tool for machine learning education and research. It’s a Jupyter notebook environment that requires no setup to use.
### 5-3-1 What browsers are supported?
Colaboratory works with most major browsers, and is most thoroughly tested with desktop versions of Chrome and Firefox.
### 5-3-2 Is it free to use?
Yes. Colaboratory is a research project that is free to use.
### 5-3-3 What is the difference between Jupyter and Colaboratory?
Jupyter is the open source project on which Colaboratory is based. Colaboratory allows you to use and share Jupyter notebooks with others without having to download, install, or run anything on your own computer other than a browser.
<a id="15"></a> <br>
## 5-5 Loading Packages
In this kernel we are using the following packages:
<img src="http://s8.picofile.com/file/8338227868/packages.png">
### 5-5-1 Import
```
from sklearn.cross_validation import train_test_split
from sklearn.metrics import classification_report
from sklearn.metrics import confusion_matrix
from sklearn.metrics import accuracy_score
from sklearn.decomposition import PCA
import matplotlib.pyplot as plt
from pandas import get_dummies
import plotly.graph_objs as go
from sklearn import datasets
import plotly.plotly as py
import seaborn as sns
import pandas as pd
import numpy as np
import matplotlib
import warnings
import sklearn
import scipy
import numpy
import json
import sys
import csv
import os
```
### 5-5-2 Print
```
print('matplotlib: {}'.format(matplotlib.__version__))
print('sklearn: {}'.format(sklearn.__version__))
print('scipy: {}'.format(scipy.__version__))
print('seaborn: {}'.format(sns.__version__))
print('pandas: {}'.format(pd.__version__))
print('numpy: {}'.format(np.__version__))
print('Python: {}'.format(sys.version))
#show plot inline
%matplotlib inline
```
<a id="16"></a> <br>
## 6- Exploratory Data Analysis(EDA)
In this section, you'll learn how to use graphical and numerical techniques to begin uncovering the structure of your data.
* Which variables suggest interesting relationships?
* Which observations are unusual?
By the end of the section, you'll be able to answer these questions and more, while generating graphics that are both insightful and beautiful. then We will review analytical and statistical operations:
* 5-1 Data Collection
* 5-2 Visualization
* 5-3 Data Preprocessing
* 5-4 Data Cleaning
<img src="http://s9.picofile.com/file/8338476134/EDA.png">
<a id="17"></a> <br>
## 6-1 Data Collection
**Data collection** is the process of gathering and measuring data, information or any variables of interest in a standardized and established manner that enables the collector to answer or test hypothesis and evaluate outcomes of the particular collection.[techopedia]
**Iris dataset** consists of 3 different types of irises’ (Setosa, Versicolour, and Virginica) petal and sepal length, stored in a 150x4 numpy.ndarray
The rows being the samples and the columns being: Sepal Length, Sepal Width, Petal Length and Petal Width.[6]
```
# import Dataset to play with it
dataset = pd.read_csv('../input/Iris.csv')
```
**<< Note 1 >>**
* Each row is an observation (also known as : sample, example, instance, record)
* Each column is a feature (also known as: Predictor, attribute, Independent Variable, input, regressor, Covariate)
After loading the data via **pandas**, we should checkout what the content is, description and via the following:
```
type(dataset)
```
<a id="18"></a> <br>
## 6-2 Visualization
**Data visualization** is the presentation of data in a pictorial or graphical format. It enables decision makers to see analytics presented visually, so they can grasp difficult concepts or identify new patterns.
With interactive visualization, you can take the concept a step further by using technology to drill down into charts and graphs for more detail, interactively changing what data you see and how it’s processed.[SAS]
In this section I show you **11 plots** with **matplotlib** and **seaborn** that is listed in the blew picture:
<img src="http://s8.picofile.com/file/8338475500/visualization.jpg" />
<a id="19"></a> <br>
### 6-2-1 Scatter plot
Scatter plot Purpose To identify the type of relationship (if any) between two quantitative variables
```
# Modify the graph above by assigning each species an individual color.
sns.FacetGrid(dataset, hue="Species", size=5) \
.map(plt.scatter, "SepalLengthCm", "SepalWidthCm") \
.add_legend()
plt.show()
```
<a id="20"></a> <br>
### 6-2-2 Box
In descriptive statistics, a **box plot** or boxplot is a method for graphically depicting groups of numerical data through their quartiles. Box plots may also have lines extending vertically from the boxes (whiskers) indicating variability outside the upper and lower quartiles, hence the terms box-and-whisker plot and box-and-whisker diagram.[wikipedia]
```
dataset.plot(kind='box', subplots=True, layout=(2,3), sharex=False, sharey=False)
plt.figure()
#This gives us a much clearer idea of the distribution of the input attributes:
# To plot the species data using a box plot:
sns.boxplot(x="Species", y="PetalLengthCm", data=dataset )
plt.show()
# Use Seaborn's striplot to add data points on top of the box plot
# Insert jitter=True so that the data points remain scattered and not piled into a verticle line.
# Assign ax to each axis, so that each plot is ontop of the previous axis.
ax= sns.boxplot(x="Species", y="PetalLengthCm", data=dataset)
ax= sns.stripplot(x="Species", y="PetalLengthCm", data=dataset, jitter=True, edgecolor="gray")
plt.show()
# Tweek the plot above to change fill and border color color using ax.artists.
# Assing ax.artists a variable name, and insert the box number into the corresponding brackets
ax= sns.boxplot(x="Species", y="PetalLengthCm", data=dataset)
ax= sns.stripplot(x="Species", y="PetalLengthCm", data=dataset, jitter=True, edgecolor="gray")
boxtwo = ax.artists[2]
boxtwo.set_facecolor('red')
boxtwo.set_edgecolor('black')
boxthree=ax.artists[1]
boxthree.set_facecolor('yellow')
boxthree.set_edgecolor('black')
plt.show()
```
<a id="21"></a> <br>
### 6-2-3 Histogram
We can also create a **histogram** of each input variable to get an idea of the distribution.
```
# histograms
dataset.hist(figsize=(15,20))
plt.figure()
```
It looks like perhaps two of the input variables have a Gaussian distribution. This is useful to note as we can use algorithms that can exploit this assumption.
```
dataset["PetalLengthCm"].hist();
```
<a id="22"></a> <br>
### 6-2-4 Multivariate Plots
Now we can look at the interactions between the variables.
First, let’s look at scatterplots of all pairs of attributes. This can be helpful to spot structured relationships between input variables.
```
# scatter plot matrix
pd.plotting.scatter_matrix(dataset,figsize=(10,10))
plt.figure()
```
Note the diagonal grouping of some pairs of attributes. This suggests a high correlation and a predictable relationship.
<a id="23"></a> <br>
### 6-2-5 violinplots
```
# violinplots on petal-length for each species
sns.violinplot(data=dataset,x="Species", y="PetalLengthCm")
```
<a id="24"></a> <br>
### 6-2-6 pairplot
```
# Using seaborn pairplot to see the bivariate relation between each pair of features
sns.pairplot(dataset, hue="Species")
```
From the plot, we can see that the species setosa is separataed from the other two across all feature combinations
We can also replace the histograms shown in the diagonal of the pairplot by kde.
```
# updating the diagonal elements in a pairplot to show a kde
sns.pairplot(dataset, hue="Species",diag_kind="kde")
```
<a id="25"></a> <br>
### 6-2-7 kdeplot
```
# seaborn's kdeplot, plots univariate or bivariate density estimates.
#Size can be changed by tweeking the value used
sns.FacetGrid(dataset, hue="Species", size=5).map(sns.kdeplot, "PetalLengthCm").add_legend()
plt.show()
```
<a id="26"></a> <br>
### 6-2-8 jointplot
```
# Use seaborn's jointplot to make a hexagonal bin plot
#Set desired size and ratio and choose a color.
sns.jointplot(x="SepalLengthCm", y="SepalWidthCm", data=dataset, size=10,ratio=10, kind='hex',color='green')
plt.show()
```
<a id="27"></a> <br>
### 6-2-9 andrews_curves
```
#In Pandas use Andrews Curves to plot and visualize data structure.
#Each multivariate observation is transformed into a curve and represents the coefficients of a Fourier series.
#This useful for detecting outliers in times series data.
#Use colormap to change the color of the curves
from pandas.tools.plotting import andrews_curves
andrews_curves(dataset.drop("Id", axis=1), "Species",colormap='rainbow')
plt.show()
# we will use seaborn jointplot shows bivariate scatterplots and univariate histograms with Kernel density
# estimation in the same figure
sns.jointplot(x="SepalLengthCm", y="SepalWidthCm", data=dataset, size=6, kind='kde', color='#800000', space=0)
```
<a id="28"></a> <br>
### 6-2-10 Heatmap
```
plt.figure(figsize=(7,4))
sns.heatmap(dataset.corr(),annot=True,cmap='cubehelix_r') #draws heatmap with input as the correlation matrix calculted by(iris.corr())
plt.show()
```
<a id="29"></a> <br>
### 6-2-11 radviz
```
# A final multivariate visualization technique pandas has is radviz
# Which puts each feature as a point on a 2D plane, and then simulates
# having each sample attached to those points through a spring weighted
# by the relative value for that feature
from pandas.tools.plotting import radviz
radviz(dataset.drop("Id", axis=1), "Species")
```
### 6-2-12 Bar Plot
```
dataset['Species'].value_counts().plot(kind="bar");
```
### 6-2-14 visualization with Plotly
```
import plotly.offline as py
import plotly.graph_objs as go
py.init_notebook_mode(connected=True)
from plotly import tools
import plotly.figure_factory as ff
iris = datasets.load_iris()
X = iris.data[:, :2] # we only take the first two features.
Y = iris.target
x_min, x_max = X[:, 0].min() - .5, X[:, 0].max() + .5
y_min, y_max = X[:, 1].min() - .5, X[:, 1].max() + .5
trace = go.Scatter(x=X[:, 0],
y=X[:, 1],
mode='markers',
marker=dict(color=np.random.randn(150),
size=10,
colorscale='Viridis',
showscale=False))
layout = go.Layout(title='Training Points',
xaxis=dict(title='Sepal length',
showgrid=False),
yaxis=dict(title='Sepal width',
showgrid=False),
)
fig = go.Figure(data=[trace], layout=layout)
py.iplot(fig)
```
**<< Note >>**
**Yellowbrick** is a suite of visual diagnostic tools called “Visualizers” that extend the Scikit-Learn API to allow human steering of the model selection process. In a nutshell, Yellowbrick combines scikit-learn with matplotlib in the best tradition of the scikit-learn documentation, but to produce visualizations for your models!
### 6-2-13 Conclusion
we have used Python to apply data visualization tools to the Iris dataset. Color and size changes were made to the data points in scatterplots. I changed the border and fill color of the boxplot and violin, respectively.
<a id="30"></a> <br>
## 6-3 Data Preprocessing
**Data preprocessing** refers to the transformations applied to our data before feeding it to the algorithm.
Data Preprocessing is a technique that is used to convert the raw data into a clean data set. In other words, whenever the data is gathered from different sources it is collected in raw format which is not feasible for the analysis.
there are plenty of steps for data preprocessing and we just listed some of them :
* removing Target column (id)
* Sampling (without replacement)
* Making part of iris unbalanced and balancing (with undersampling and SMOTE)
* Introducing missing values and treating them (replacing by average values)
* Noise filtering
* Data discretization
* Normalization and standardization
* PCA analysis
* Feature selection (filter, embedded, wrapper)
## 6-3-1 Features
Features:
* numeric
* categorical
* ordinal
* datetime
* coordinates
find the type of features in titanic dataset
<img src="http://s9.picofile.com/file/8339959442/titanic.png" height="700" width="600" />
### 6-3-2 Explorer Dataset
1- Dimensions of the dataset.
2- Peek at the data itself.
3- Statistical summary of all attributes.
4- Breakdown of the data by the class variable.[7]
Don’t worry, each look at the data is **one command**. These are useful commands that you can use again and again on future projects.
```
# shape
print(dataset.shape)
#columns*rows
dataset.size
```
how many NA elements in every column
```
dataset.isnull().sum()
# remove rows that have NA's
dataset = dataset.dropna()
```
We can get a quick idea of how many instances (rows) and how many attributes (columns) the data contains with the shape property.
You should see 150 instances and 5 attributes:
for getting some information about the dataset you can use **info()** command
```
print(dataset.info())
```
you see number of unique item for Species with command below:
```
dataset['Species'].unique()
dataset["Species"].value_counts()
```
to check the first 5 rows of the data set, we can use head(5).
```
dataset.head(5)
```
to check out last 5 row of the data set, we use tail() function
```
dataset.tail()
```
to pop up 5 random rows from the data set, we can use **sample(5)** function
```
dataset.sample(5)
```
to give a statistical summary about the dataset, we can use **describe()
```
dataset.describe()
```
to check out how many null info are on the dataset, we can use **isnull().sum()
```
dataset.isnull().sum()
dataset.groupby('Species').count()
```
to print dataset **columns**, we can use columns atribute
```
dataset.columns
```
**<< Note 2 >>**
in pandas's data frame you can perform some query such as "where"
```
dataset.where(dataset ['Species']=='Iris-setosa')
```
as you can see in the below in python, it is so easy perform some query on the dataframe:
```
dataset[dataset['SepalLengthCm']>7.2]
# Seperating the data into dependent and independent variables
X = dataset.iloc[:, :-1].values
y = dataset.iloc[:, -1].values
```
**<< Note >>**
>**Preprocessing and generation pipelines depend on a model type**
<a id="31"></a> <br>
## 6-4 Data Cleaning
When dealing with real-world data, dirty data is the norm rather than the exception. We continuously need to predict correct values, impute missing ones, and find links between various data artefacts such as schemas and records. We need to stop treating data cleaning as a piecemeal exercise (resolving different types of errors in isolation), and instead leverage all signals and resources (such as constraints, available statistics, and dictionaries) to accurately predict corrective actions.
The primary goal of data cleaning is to detect and remove errors and **anomalies** to increase the value of data in analytics and decision making. While it has been the focus of many researchers for several years, individual problems have been addressed separately. These include missing value imputation, outliers detection, transformations, integrity constraints violations detection and repair, consistent query answering, deduplication, and many other related problems such as profiling and constraints mining.[8]
```
cols = dataset.columns
features = cols[0:4]
labels = cols[4]
print(features)
print(labels)
#Well conditioned data will have zero mean and equal variance
#We get this automattically when we calculate the Z Scores for the data
data_norm = pd.DataFrame(dataset)
for feature in features:
dataset[feature] = (dataset[feature] - dataset[feature].mean())/dataset[feature].std()
#Show that should now have zero mean
print("Averages")
print(dataset.mean())
print("\n Deviations")
#Show that we have equal variance
print(pow(dataset.std(),2))
#Shuffle The data
indices = data_norm.index.tolist()
indices = np.array(indices)
np.random.shuffle(indices)
# One Hot Encode as a dataframe
from sklearn.cross_validation import train_test_split
y = get_dummies(y)
# Generate Training and Validation Sets
X_train, X_test, y_train, y_test = train_test_split(X,y, test_size=.3)
# Convert to np arrays so that we can use with TensorFlow
X_train = np.array(X_train).astype(np.float32)
X_test = np.array(X_test).astype(np.float32)
y_train = np.array(y_train).astype(np.float32)
y_test = np.array(y_test).astype(np.float32)
#Check to make sure split still has 4 features and 3 labels
print(X_train.shape, y_train.shape)
print(X_test.shape, y_test.shape)
```
<a id="32"></a> <br>
## 7- Model Deployment
In this section have been applied more than **20 learning algorithms** that play an important rule in your experiences and improve your knowledge in case of ML technique.
> **<< Note 3 >>** : The results shown here may be slightly different for your analysis because, for example, the neural network algorithms use random number generators for fixing the initial value of the weights (starting points) of the neural networks, which often result in obtaining slightly different (local minima) solutions each time you run the analysis. Also note that changing the seed for the random number generator used to create the train, test, and validation samples can change your results.
## Families of ML algorithms
There are several categories for machine learning algorithms, below are some of these categories:
* Linear
* Linear Regression
* Logistic Regression
* Support Vector Machines
* Tree-Based
* Decision Tree
* Random Forest
* GBDT
* KNN
* Neural Networks
-----------------------------
And if we want to categorize ML algorithms with the type of learning, there are below type:
* Classification
* k-Nearest Neighbors
* LinearRegression
* SVM
* DT
* NN
* clustering
* K-means
* HCA
* Expectation Maximization
* Visualization and dimensionality reduction:
* Principal Component Analysis(PCA)
* Kernel PCA
* Locally -Linear Embedding (LLE)
* t-distributed Stochastic Neighbor Embedding (t-SNE)
* Association rule learning
* Apriori
* Eclat
* Semisupervised learning
* Reinforcement Learning
* Q-learning
* Batch learning & Online learning
* Ensemble Learning
**<< Note >>**
> Here is no method which outperforms all others for all tasks
<a id="33"></a> <br>
## Prepare Features & Targets
First of all seperating the data into dependent(Feature) and independent(Target) variables.
**<< Note 4 >>**
* X==>>Feature
* y==>>Target
```
X = dataset.iloc[:, :-1].values
y = dataset.iloc[:, -1].values
# Splitting the dataset into the Training set and Test set
from sklearn.cross_validation import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state = 0)
```
## Accuracy and precision
* **precision** :
In pattern recognition, information retrieval and binary classification, precision (also called positive predictive value) is the fraction of relevant instances among the retrieved instances,
* **recall** :
recall is the fraction of relevant instances that have been retrieved over the total amount of relevant instances.
* **F-score** :
the F1 score is a measure of a test's accuracy. It considers both the precision p and the recall r of the test to compute the score: p is the number of correct positive results divided by the number of all positive results returned by the classifier, and r is the number of correct positive results divided by the number of all relevant samples (all samples that should have been identified as positive). The F1 score is the harmonic average of the precision and recall, where an F1 score reaches its best value at 1 (perfect precision and recall) and worst at 0.
**What is the difference between accuracy and precision?
"Accuracy" and "precision" are general terms throughout science. A good way to internalize the difference are the common "bullseye diagrams". In machine learning/statistics as a whole, accuracy vs. precision is analogous to bias vs. variance.
<a id="33"></a> <br>
## 7-1 K-Nearest Neighbours
In **Machine Learning**, the **k-nearest neighbors algorithm** (k-NN) is a non-parametric method used for classification and regression. In both cases, the input consists of the k closest training examples in the feature space. The output depends on whether k-NN is used for classification or regression:
In k-NN classification, the output is a class membership. An object is classified by a majority vote of its neighbors, with the object being assigned to the class most common among its k nearest neighbors (k is a positive integer, typically small). If k = 1, then the object is simply assigned to the class of that single nearest neighbor.
In k-NN regression, the output is the property value for the object. This value is the average of the values of its k nearest neighbors.
k-NN is a type of instance-based learning, or lazy learning, where the function is only approximated locally and all computation is deferred until classification. The k-NN algorithm is among the simplest of all machine learning algorithms.
```
# K-Nearest Neighbours
from sklearn.neighbors import KNeighborsClassifier
Model = KNeighborsClassifier(n_neighbors=8)
Model.fit(X_train, y_train)
y_pred = Model.predict(X_test)
# Summary of the predictions made by the classifier
print(classification_report(y_test, y_pred))
print(confusion_matrix(y_test, y_pred))
# Accuracy score
print('accuracy is',accuracy_score(y_pred,y_test))
```
<a id="34"></a> <br>
## 7-2 Radius Neighbors Classifier
Classifier implementing a **vote** among neighbors within a given **radius**
In scikit-learn **RadiusNeighborsClassifier** is very similar to **KNeighborsClassifier** with the exception of two parameters. First, in RadiusNeighborsClassifier we need to specify the radius of the fixed area used to determine if an observation is a neighbor using radius. Unless there is some substantive reason for setting radius to some value, it is best to treat it like any other hyperparameter and tune it during model selection. The second useful parameter is outlier_label, which indicates what label to give an observation that has no observations within the radius - which itself can often be a useful tool for identifying outliers.
```
from sklearn.neighbors import RadiusNeighborsClassifier
Model=RadiusNeighborsClassifier(radius=8.0)
Model.fit(X_train,y_train)
y_pred=Model.predict(X_test)
#summary of the predictions made by the classifier
print(classification_report(y_test,y_pred))
print(confusion_matrix(y_test,y_pred))
#Accouracy score
print('accuracy is ', accuracy_score(y_test,y_pred))
```
<a id="35"></a> <br>
## 7-3 Logistic Regression
Logistic regression is the appropriate regression analysis to conduct when the dependent variable is **dichotomous** (binary). Like all regression analyses, the logistic regression is a **predictive analysis**.
In statistics, the logistic model (or logit model) is a widely used statistical model that, in its basic form, uses a logistic function to model a binary dependent variable; many more complex extensions exist. In regression analysis, logistic regression (or logit regression) is estimating the parameters of a logistic model; it is a form of binomial regression. Mathematically, a binary logistic model has a dependent variable with two possible values, such as pass/fail, win/lose, alive/dead or healthy/sick; these are represented by an indicator variable, where the two values are labeled "0" and "1"
```
# LogisticRegression
from sklearn.linear_model import LogisticRegression
Model = LogisticRegression()
Model.fit(X_train, y_train)
y_pred = Model.predict(X_test)
# Summary of the predictions made by the classifier
print(classification_report(y_test, y_pred))
print(confusion_matrix(y_test, y_pred))
# Accuracy score
print('accuracy is',accuracy_score(y_pred,y_test))
```
<a id="36"></a> <br>
## 7-4 Passive Aggressive Classifier
```
from sklearn.linear_model import PassiveAggressiveClassifier
Model = PassiveAggressiveClassifier()
Model.fit(X_train, y_train)
y_pred = Model.predict(X_test)
# Summary of the predictions made by the classifier
print(classification_report(y_test, y_pred))
print(confusion_matrix(y_test, y_pred))
# Accuracy score
print('accuracy is',accuracy_score(y_pred,y_test))
```
<a id="37"></a> <br>
## 7-5 Naive Bayes
In machine learning, naive Bayes classifiers are a family of simple "**probabilistic classifiers**" based on applying Bayes' theorem with strong (naive) independence assumptions between the features.
```
# Naive Bayes
from sklearn.naive_bayes import GaussianNB
Model = GaussianNB()
Model.fit(X_train, y_train)
y_pred = Model.predict(X_test)
# Summary of the predictions made by the classifier
print(classification_report(y_test, y_pred))
print(confusion_matrix(y_test, y_pred))
# Accuracy score
print('accuracy is',accuracy_score(y_pred,y_test))
```
<a id="39"></a> <br>
## 7-7 BernoulliNB
Like MultinomialNB, this classifier is suitable for **discrete data**. The difference is that while MultinomialNB works with occurrence counts, BernoulliNB is designed for binary/boolean features.
```
# BernoulliNB
from sklearn.naive_bayes import BernoulliNB
Model = BernoulliNB()
Model.fit(X_train, y_train)
y_pred = Model.predict(X_test)
# Summary of the predictions made by the classifier
print(classification_report(y_test, y_pred))
print(confusion_matrix(y_test, y_pred))
# Accuracy score
print('accuracy is',accuracy_score(y_pred,y_test))
```
<a id="40"></a> <br>
## 7-8 SVM
The advantages of support vector machines are:
* Effective in high dimensional spaces.
* Still effective in cases where number of dimensions is greater than the number of samples.
* Uses a subset of training points in the decision function (called support vectors), so it is also memory efficient.
* Versatile: different Kernel functions can be specified for the decision function. Common kernels are provided, but it is also possible to specify custom kernels.
The disadvantages of support vector machines include:
* If the number of features is much greater than the number of samples, avoid over-fitting in choosing Kernel functions and regularization term is crucial.
* SVMs do not directly provide probability estimates, these are calculated using an expensive five-fold cross-validation
```
# Support Vector Machine
from sklearn.svm import SVC
Model = SVC()
Model.fit(X_train, y_train)
y_pred = Model.predict(X_test)
# Summary of the predictions made by the classifier
print(classification_report(y_test, y_pred))
print(confusion_matrix(y_test, y_pred))
# Accuracy score
print('accuracy is',accuracy_score(y_pred,y_test))
```
<a id="41"></a> <br>
## 7-9 Nu-Support Vector Classification
> Similar to SVC but uses a parameter to control the number of support vectors.
```
# Support Vector Machine's
from sklearn.svm import NuSVC
Model = NuSVC()
Model.fit(X_train, y_train)
y_pred = Model.predict(X_test)
# Summary of the predictions made by the classifier
print(classification_report(y_test, y_pred))
print(confusion_matrix(y_test, y_pred))
# Accuracy score
print('accuracy is',accuracy_score(y_pred,y_test))
```
<a id="42"></a> <br>
## 7-10 Linear Support Vector Classification
Similar to **SVC** with parameter kernel=’linear’, but implemented in terms of liblinear rather than libsvm, so it has more flexibility in the choice of penalties and loss functions and should scale better to large numbers of samples.
```
# Linear Support Vector Classification
from sklearn.svm import LinearSVC
Model = LinearSVC()
Model.fit(X_train, y_train)
y_pred = Model.predict(X_test)
# Summary of the predictions made by the classifier
print(classification_report(y_test, y_pred))
print(confusion_matrix(y_test, y_pred))
# Accuracy score
print('accuracy is',accuracy_score(y_pred,y_test))
```
<a id="43"></a> <br>
## 7-11 Decision Tree
Decision Trees (DTs) are a non-parametric supervised learning method used for **classification** and **regression**. The goal is to create a model that predicts the value of a target variable by learning simple **decision rules** inferred from the data features.
```
# Decision Tree's
from sklearn.tree import DecisionTreeClassifier
Model = DecisionTreeClassifier()
Model.fit(X_train, y_train)
y_pred = Model.predict(X_test)
# Summary of the predictions made by the classifier
print(classification_report(y_test, y_pred))
print(confusion_matrix(y_test, y_pred))
# Accuracy score
print('accuracy is',accuracy_score(y_pred,y_test))
```
<a id="44"></a> <br>
## 7-12 ExtraTreeClassifier
An extremely randomized tree classifier.
Extra-trees differ from classic decision trees in the way they are built. When looking for the best split to separate the samples of a node into two groups, random splits are drawn for each of the **max_features** randomly selected features and the best split among those is chosen. When max_features is set 1, this amounts to building a totally random decision tree.
**Warning**: Extra-trees should only be used within ensemble methods.
```
# ExtraTreeClassifier
from sklearn.tree import ExtraTreeClassifier
Model = ExtraTreeClassifier()
Model.fit(X_train, y_train)
y_pred = Model.predict(X_test)
# Summary of the predictions made by the classifier
print(classification_report(y_test, y_pred))
print(confusion_matrix(y_test, y_pred))
# Accuracy score
print('accuracy is',accuracy_score(y_pred,y_test))
```
<a id="45"></a> <br>
## 7-13 Neural network
I have used multi-layer Perceptron classifier.
This model optimizes the log-loss function using **LBFGS** or **stochastic gradient descent**.
## 7-13-1 What is a Perceptron?
There are many online examples and tutorials on perceptrons and learning. Here is a list of some articles:
- [Wikipedia on Perceptrons](https://en.wikipedia.org/wiki/Perceptron)
- Jurafsky and Martin (ed. 3), Chapter 8
This is an example that I have taken from a draft of the 3rd edition of Jurafsky and Martin, with slight modifications:
We import *numpy* and use its *exp* function. We could use the same function from the *math* module, or some other module like *scipy*. The *sigmoid* function is defined as in the textbook:
```
import numpy as np
def sigmoid(z):
return 1 / (1 + np.exp(-z))
```
Our example data, **weights** $w$, **bias** $b$, and **input** $x$ are defined as:
```
w = np.array([0.2, 0.3, 0.8])
b = 0.5
x = np.array([0.5, 0.6, 0.1])
```
Our neural unit would compute $z$ as the **dot-product** $w \cdot x$ and add the **bias** $b$ to it. The sigmoid function defined above will convert this $z$ value to the **activation value** $a$ of the unit:
```
z = w.dot(x) + b
print("z:", z)
print("a:", sigmoid(z))
```
### The XOR Problem
The power of neural units comes from combining them into larger networks. Minsky and Papert (1969): A single neural unit cannot compute the simple logical function XOR.
The task is to implement a simple **perceptron** to compute logical operations like AND, OR, and XOR.
- Input: $x_1$ and $x_2$
- Bias: $b = -1$ for AND; $b = 0$ for OR
- Weights: $w = [1, 1]$
with the following activation function:
$$
y = \begin{cases}
\ 0 & \quad \text{if } w \cdot x + b \leq 0\\
\ 1 & \quad \text{if } w \cdot x + b > 0
\end{cases}
$$
We can define this activation function in Python as:
```
def activation(z):
if z > 0:
return 1
return 0
```
For AND we could implement a perceptron as:
```
w = np.array([1, 1])
b = -1
x = np.array([0, 0])
print("0 AND 0:", activation(w.dot(x) + b))
x = np.array([1, 0])
print("1 AND 0:", activation(w.dot(x) + b))
x = np.array([0, 1])
print("0 AND 1:", activation(w.dot(x) + b))
x = np.array([1, 1])
print("1 AND 1:", activation(w.dot(x) + b))
```
For OR we could implement a perceptron as:
```
w = np.array([1, 1])
b = 0
x = np.array([0, 0])
print("0 OR 0:", activation(w.dot(x) + b))
x = np.array([1, 0])
print("1 OR 0:", activation(w.dot(x) + b))
x = np.array([0, 1])
print("0 OR 1:", activation(w.dot(x) + b))
x = np.array([1, 1])
print("1 OR 1:", activation(w.dot(x) + b))
```
There is no way to implement a perceptron for XOR this way.
no see our prediction for iris
```
from sklearn.neural_network import MLPClassifier
Model=MLPClassifier()
Model.fit(X_train,y_train)
y_pred=Model.predict(X_test)
# Summary of the predictions
print(classification_report(y_test,y_pred))
print(confusion_matrix(y_test,y_pred))
#Accuracy Score
print('accuracy is ',accuracy_score(y_pred,y_test))
```
<a id="46"></a> <br>
## 7-14 RandomForest
A random forest is a meta estimator that **fits a number of decision tree classifiers** on various sub-samples of the dataset and uses averaging to improve the predictive accuracy and control over-fitting.
The sub-sample size is always the same as the original input sample size but the samples are drawn with replacement if bootstrap=True (default).
```
from sklearn.ensemble import RandomForestClassifier
Model=RandomForestClassifier(max_depth=2)
Model.fit(X_train,y_train)
y_pred=Model.predict(X_test)
print(classification_report(y_test,y_pred))
print(confusion_matrix(y_pred,y_test))
#Accuracy Score
print('accuracy is ',accuracy_score(y_pred,y_test))
```
<a id="47"></a> <br>
## 7-15 Bagging classifier
A Bagging classifier is an ensemble **meta-estimator** that fits base classifiers each on random subsets of the original dataset and then aggregate their individual predictions (either by voting or by averaging) to form a final prediction. Such a meta-estimator can typically be used as a way to reduce the variance of a black-box estimator (e.g., a decision tree), by introducing randomization into its construction procedure and then making an ensemble out of it.
This algorithm encompasses several works from the literature. When random subsets of the dataset are drawn as random subsets of the samples, then this algorithm is known as Pasting . If samples are drawn with replacement, then the method is known as Bagging . When random subsets of the dataset are drawn as random subsets of the features, then the method is known as Random Subspaces . Finally, when base estimators are built on subsets of both samples and features, then the method is known as Random Patches .[http://scikit-learn.org]
```
from sklearn.ensemble import BaggingClassifier
Model=BaggingClassifier()
Model.fit(X_train,y_train)
y_pred=Model.predict(X_test)
print(classification_report(y_test,y_pred))
print(confusion_matrix(y_pred,y_test))
#Accuracy Score
print('accuracy is ',accuracy_score(y_pred,y_test))
```
<a id="48"></a> <br>
## 7-16 AdaBoost classifier
An AdaBoost classifier is a meta-estimator that begins by fitting a classifier on the original dataset and then fits additional copies of the classifier on the same dataset but where the weights of incorrectly classified instances are adjusted such that subsequent classifiers focus more on difficult cases.
This class implements the algorithm known as **AdaBoost-SAMME** .
```
from sklearn.ensemble import AdaBoostClassifier
Model=AdaBoostClassifier()
Model.fit(X_train,y_train)
y_pred=Model.predict(X_test)
print(classification_report(y_test,y_pred))
print(confusion_matrix(y_pred,y_test))
#Accuracy Score
print('accuracy is ',accuracy_score(y_pred,y_test))
```
<a id="49"></a> <br>
## 7-17 Gradient Boosting Classifier
GB builds an additive model in a forward stage-wise fashion; it allows for the optimization of arbitrary differentiable loss functions.
```
from sklearn.ensemble import GradientBoostingClassifier
Model=GradientBoostingClassifier()
Model.fit(X_train,y_train)
y_pred=Model.predict(X_test)
print(classification_report(y_test,y_pred))
print(confusion_matrix(y_pred,y_test))
#Accuracy Score
print('accuracy is ',accuracy_score(y_pred,y_test))
```
<a id="50"></a> <br>
## 7-18 Linear Discriminant Analysis
Linear Discriminant Analysis (discriminant_analysis.LinearDiscriminantAnalysis) and Quadratic Discriminant Analysis (discriminant_analysis.QuadraticDiscriminantAnalysis) are two classic classifiers, with, as their names suggest, a **linear and a quadratic decision surface**, respectively.
These classifiers are attractive because they have closed-form solutions that can be easily computed, are inherently multiclass, have proven to work well in practice, and have no **hyperparameters** to tune.
```
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
Model=LinearDiscriminantAnalysis()
Model.fit(X_train,y_train)
y_pred=Model.predict(X_test)
print(classification_report(y_test,y_pred))
print(confusion_matrix(y_pred,y_test))
#Accuracy Score
print('accuracy is ',accuracy_score(y_pred,y_test))
```
<a id="51"></a> <br>
## 7-19 Quadratic Discriminant Analysis
A classifier with a quadratic decision boundary, generated by fitting class conditional densities to the data and using Bayes’ rule.
The model fits a **Gaussian** density to each class.
```
from sklearn.discriminant_analysis import QuadraticDiscriminantAnalysis
Model=QuadraticDiscriminantAnalysis()
Model.fit(X_train,y_train)
y_pred=Model.predict(X_test)
print(classification_report(y_test,y_pred))
print(confusion_matrix(y_pred,y_test))
#Accuracy Score
print('accuracy is ',accuracy_score(y_pred,y_test))
```
<a id="52"></a> <br>
## 7-20 Kmeans
K-means clustering is a type of unsupervised learning, which is used when you have unlabeled data (i.e., data without defined categories or groups).
The goal of this algorithm is **to find groups in the data**, with the number of groups represented by the variable K. The algorithm works iteratively to assign each data point to one of K groups based on the features that are provided.
```
from sklearn.cluster import KMeans
iris_SP = dataset[['SepalLengthCm','SepalWidthCm','PetalLengthCm','PetalWidthCm']]
# k-means cluster analysis for 1-15 clusters
from scipy.spatial.distance import cdist
clusters=range(1,15)
meandist=[]
# loop through each cluster and fit the model to the train set
# generate the predicted cluster assingment and append the mean
# distance my taking the sum divided by the shape
for k in clusters:
model=KMeans(n_clusters=k)
model.fit(iris_SP)
clusassign=model.predict(iris_SP)
meandist.append(sum(np.min(cdist(iris_SP, model.cluster_centers_, 'euclidean'), axis=1))
/ iris_SP.shape[0])
"""
Plot average distance from observations from the cluster centroid
to use the Elbow Method to identify number of clusters to choose
"""
plt.plot(clusters, meandist)
plt.xlabel('Number of clusters')
plt.ylabel('Average distance')
plt.title('Selecting k with the Elbow Method')
# pick the fewest number of clusters that reduces the average distance
# If you observe after 3 we can see graph is almost linear
```
<a id="53"></a> <br>
## 7-21- Backpropagation
Backpropagation is a method used in artificial neural networks to calculate a gradient that is needed in the calculation of the weights to be used in the network.It is commonly used to train deep neural networks,a term referring to neural networks with more than one hidden layer.
In this example we will use a very simple network to start with. The network will only have one input and one output layer. We want to make the following predictions from the input:
| Input | Output |
| ------ |:------:|
| 0 0 1 | 0 |
| 1 1 1 | 1 |
| 1 0 1 | 1 |
| 0 1 1 | 0 |
We will use **Numpy** to compute the network parameters, weights, activation, and outputs:
We will use the *[Sigmoid](http://ml-cheatsheet.readthedocs.io/en/latest/activation_functions.html#sigmoid)* activation function:
```
def sigmoid(z):
"""The sigmoid activation function."""
return 1 / (1 + np.exp(-z))
```
We could use the [ReLU](http://ml-cheatsheet.readthedocs.io/en/latest/activation_functions.html#activation-relu) activation function instead:
```
def relu(z):
"""The ReLU activation function."""
return max(0, z)
```
The [Sigmoid](http://ml-cheatsheet.readthedocs.io/en/latest/activation_functions.html#sigmoid) activation function introduces non-linearity to the computation. It maps the input value to an output value between $0$ and $1$.
<img src="http://s8.picofile.com/file/8339774900/SigmoidFunction1.png" style="max-width:100%; width: 30%; max-width: none">
The derivative of the sigmoid function is maximal at $x=0$ and minimal for lower or higher values of $x$:
<img src="http://s9.picofile.com/file/8339770650/sigmoid_prime.png" style="max-width:100%; width: 25%; max-width: none">
The *sigmoid_prime* function returns the derivative of the sigmoid for any given $z$. The derivative of the sigmoid is $z * (1 - z)$. This is basically the slope of the sigmoid function at any given point:
```
def sigmoid_prime(z):
"""The derivative of sigmoid for z."""
return z * (1 - z)
```
We define the inputs as rows in *X*. There are three input nodes (three columns per vector in $X$. Each row is one trainig example:
```
X = np.array([ [ 0, 0, 1 ],
[ 0, 1, 1 ],
[ 1, 0, 1 ],
[ 1, 1, 1 ] ])
print(X)
```
The outputs are stored in *y*, where each row represents the output for the corresponding input vector (row) in *X*. The vector is initiated as a single row vector and with four columns and transposed (using the $.T$ method) into a column vector with four rows:
```
y = np.array([[0,0,1,1]]).T
print(y)
```
To make the outputs deterministic, we seed the random number generator with a constant. This will guarantee that every time you run the code, you will get the same random distribution:
```
np.random.seed(1)
```
We create a weight matrix ($Wo$) with randomly initialized weights:
```
n_inputs = 3
n_outputs = 1
#Wo = 2 * np.random.random( (n_inputs, n_outputs) ) - 1
Wo = np.random.random( (n_inputs, n_outputs) ) * np.sqrt(2.0/n_inputs)
print(Wo)
```
The reason for the output weight matrix ($Wo$) to have 3 rows and 1 column is that it represents the weights of the connections from the three input neurons to the single output neuron. The initialization of the weight matrix is random with a mean of $0$ and a variance of $1$. There is a good reason for chosing a mean of zero in the weight initialization. See for details the section on Weight Initialization in the [Stanford course CS231n on Convolutional Neural Networks for Visual Recognition](https://cs231n.github.io/neural-networks-2/#init).
The core representation of this network is basically the weight matrix *Wo*. The rest, input matrix, output vector and so on are components that we need to learning and evaluation. The leraning result is stored in the *Wo* weight matrix.
We loop in the optimization and learning cycle 10,000 times. In the *forward propagation* line we process the entire input matrix for training. This is called **full batch** training. I do not use an alternative variable name to represent the input layer, instead I use the input matrix $X$ directly here. Think of this as the different inputs to the input neurons computed at once. In principle the input or training data could have many more training examples, the code would stay the same.
```
for n in range(10000):
# forward propagation
l1 = sigmoid(np.dot(X, Wo))
# compute the loss
l1_error = y - l1
#print("l1_error:\n", l1_error)
# multiply the loss by the slope of the sigmoid at l1
l1_delta = l1_error * sigmoid_prime(l1)
#print("l1_delta:\n", l1_delta)
#print("error:", l1_error, "\nderivative:", sigmoid(l1, True), "\ndelta:", l1_delta, "\n", "-"*10, "\n")
# update weights
Wo += np.dot(X.T, l1_delta)
print("l1:\n", l1)
```
The dots in $l1$ represent the lines in the graphic below. The lines represent the slope of the sigmoid in the particular position. The slope is highest with a value $x = 0$ (blue dot). It is rather shallow with $x = 2$ (green dot), and not so shallow and not as high with $x = -1$. All derivatives are between $0$ and $1$, of course, that is, no slope or a maximal slope of $1$. There is no negative slope in a sigmoid function.
<img src="http://s8.picofile.com/file/8339770734/sigmoid_deriv_2.png" style="max-width:100%; width: 50%; max-width: none">
The matrix $l1\_error$ is a 4 by 1 matrix (4 rows, 1 column). The derivative matrix $sigmoid\_prime(l1)$ is also a 4 by one matrix. The returned matrix of the element-wise product $l1\_delta$ is also the 4 by 1 matrix.
The product of the error and the slopes **reduces the error of high confidence predictions**. When the sigmoid slope is very shallow, the network had a very high or a very low value, that is, it was rather confident. If the network guessed something close to $x=0, y=0.5$, it was not very confident. Such predictions without confidence are updated most significantly. The other peripheral scores are multiplied with a number closer to $0$.
In the prediction line $l1 = sigmoid(np.dot(X, Wo))$ we compute the dot-product of the input vectors with the weights and compute the sigmoid on the sums.
The result of the dot-product is the number of rows of the first matrix ($X$) and the number of columns of the second matrix ($Wo$).
In the computation of the difference between the true (or gold) values in $y$ and the "guessed" values in $l1$ we have an estimate of the miss.
An example computation for the input $[ 1, 0, 1 ]$ and the weights $[ 9.5, 0.2, -0.1 ]$ and an output of $0.99$: If $y = 1$, the $l1\_error = y - l2 = 0.01$, and $l1\_delta = 0.01 * tiny\_deriv$:
<img src="http://s8.picofile.com/file/8339770792/toy_network_deriv.png" style="max-width:100%; width: 40%; max-width: none">
## 7-21-1 More Complex Example with Backpropagation
Consider now a more complicated example where no column has a correlation with the output:
| Input | Output |
| ------ |:------:|
| 0 0 1 | 0 |
| 0 1 1 | 1 |
| 1 0 1 | 1 |
| 1 1 1 | 0 |
The pattern here is our XOR pattern or problem: If there is a $1$ in either column $1$ or $2$, but not in both, the output is $1$ (XOR over column $1$ and $2$).
From our discussion of the XOR problem we remember that this is a *non-linear pattern*, a **one-to-one relationship between a combination of inputs**.
To cope with this problem, we need a network with another layer, that is a layer that will combine and transform the input, and an additional layer will map it to the output. We will add a *hidden layer* with randomized weights and then train those to optimize the output probabilities of the table above.
We will define a new $X$ input matrix that reflects the above table:
```
X = np.array([[0, 0, 1],
[0, 1, 1],
[1, 0, 1],
[1, 1, 1]])
print(X)
```
We also define a new output matrix $y$:
```
y = np.array([[ 0, 1, 1, 0]]).T
print(y)
```
We initialize the random number generator with a constant again:
```
np.random.seed(1)
```
Assume that our 3 inputs are mapped to 4 hidden layer ($Wh$) neurons, we have to initialize the hidden layer weights in a 3 by 4 matrix. The outout layer ($Wo$) is a single neuron that is connected to the hidden layer, thus the output layer is a 4 by 1 matrix:
```
n_inputs = 3
n_hidden_neurons = 4
n_output_neurons = 1
Wh = np.random.random( (n_inputs, n_hidden_neurons) ) * np.sqrt(2.0/n_inputs)
Wo = np.random.random( (n_hidden_neurons, n_output_neurons) ) * np.sqrt(2.0/n_hidden_neurons)
print("Wh:\n", Wh)
print("Wo:\n", Wo)
```
We will loop now 60,000 times to optimize the weights:
```
for i in range(100000):
l1 = sigmoid(np.dot(X, Wh))
l2 = sigmoid(np.dot(l1, Wo))
l2_error = y - l2
if (i % 10000) == 0:
print("Error:", np.mean(np.abs(l2_error)))
# gradient, changing towards the target value
l2_delta = l2_error * sigmoid_prime(l2)
# compute the l1 contribution by value to the l2 error, given the output weights
l1_error = l2_delta.dot(Wo.T)
# direction of the l1 target:
# in what direction is the target l1?
l1_delta = l1_error * sigmoid_prime(l1)
Wo += np.dot(l1.T, l2_delta)
Wh += np.dot(X.T, l1_delta)
print("Wo:\n", Wo)
print("Wh:\n", Wh)
```
The new computation in this new loop is $l1\_error = l2\_delta.dot(Wo.T)$, a **confidence weighted error** from $l2$ to compute an error for $l1$. The computation sends the error across the weights from $l2$ to $l1$. The result is a **contribution weighted error**, because we learn how much each node value in $l1$ **contributed** to the error in $l2$. This step is called **backpropagation**. We update $Wh$ using the same steps we did in the 2 layer implementation.
```
from sklearn import datasets
iris = datasets.load_iris()
X_iris = iris.data
y_iris = iris.target
plt.figure('sepal')
colormarkers = [ ['red','s'], ['greenyellow','o'], ['blue','x']]
for i in range(len(colormarkers)):
px = X_iris[:, 0][y_iris == i]
py = X_iris[:, 1][y_iris == i]
plt.scatter(px, py, c=colormarkers[i][0], marker=colormarkers[i][1])
plt.title('Iris Dataset: Sepal width vs sepal length')
plt.legend(iris.target_names)
plt.xlabel('Sepal length')
plt.ylabel('Sepal width')
plt.figure('petal')
for i in range(len(colormarkers)):
px = X_iris[:, 2][y_iris == i]
py = X_iris[:, 3][y_iris == i]
plt.scatter(px, py, c=colormarkers[i][0], marker=colormarkers[i][1])
plt.title('Iris Dataset: petal width vs petal length')
plt.legend(iris.target_names)
plt.xlabel('Petal length')
plt.ylabel('Petal width')
plt.show()
```
-----------------
<a id="54"></a> <br>
# 8- Conclusion
In this kernel, I have tried to cover all the parts related to the process of **Machine Learning** with a variety of Python packages and I know that there are still some problems then I hope to get your feedback to improve it.
you can follow me on:
> #### [ GitHub](https://github.com/mjbahmani)
--------------------------------------
**I hope you find this kernel helpful and some <font color="red"><b>UPVOTES</b></font> would be very much appreciated**
<a id="55"></a> <br>
-----------
# 9- References
* [1] [Iris image](https://rpubs.com/wjholst/322258)
* [2] [IRIS](https://archive.ics.uci.edu/ml/datasets/iris)
* [3] [https://skymind.ai/wiki/machine-learning-workflow](https://skymind.ai/wiki/machine-learning-workflow)
* [4] [IRIS-wiki](https://archive.ics.uci.edu/ml/datasets/iris)
* [5] [Problem-define](https://machinelearningmastery.com/machine-learning-in-python-step-by-step/)
* [6] [Sklearn](http://scikit-learn.org/)
* [7] [machine-learning-in-python-step-by-step](https://machinelearningmastery.com/machine-learning-in-python-step-by-step/)
* [8] [Data Cleaning](http://wp.sigmod.org/?p=2288)
* [9] [competitive data science](https://www.coursera.org/learn/competitive-data-science/)
-------------
|
github_jupyter
|
# Writing Low-Level TensorFlow Code
**Learning Objectives**
1. Practice defining and performing basic operations on constant Tensors
2. Use Tensorflow's automatic differentiation capability
3. Learn how to train a linear regression from scratch with TensorFLow
## Introduction
In this notebook, we will start by reviewing the main operations on Tensors in TensorFlow and understand how to manipulate TensorFlow Variables. We explain how these are compatible with python built-in list and numpy arrays.
Then we will jump to the problem of training a linear regression from scratch with gradient descent. The first order of business will be to understand how to compute the gradients of a function (the loss here) with respect to some of its arguments (the model weights here). The TensorFlow construct allowing us to do that is `tf.GradientTape`, which we will describe.
At last we will create a simple training loop to learn the weights of a 1-dim linear regression using synthetic data generated from a linear model.
As a bonus exercise, we will do the same for data generated from a non linear model, forcing us to manual engineer non-linear features to improve our linear model performance.
Each learning objective will correspond to a #TODO in the [student lab notebook](https://github.com/GoogleCloudPlatform/training-data-analyst/blob/master/courses/machine_learning/deepdive2/introduction_to_tensorflow/labs/write_low_level_code.ipynb) -- try to complete that notebook first before reviewing this solution notebook.
```
!sudo chown -R jupyter:jupyter /home/jupyter/training-data-analyst
# Ensure the right version of Tensorflow is installed.
!pip freeze | grep tensorflow==2.1 || pip install tensorflow==2.1
import numpy as np
import tensorflow as tf
from matplotlib import pyplot as plt
print(tf.__version__)
```
## Operations on Tensors
### Variables and Constants
Tensors in TensorFlow are either contant (`tf.constant`) or variables (`tf.Variable`).
Constant values can not be changed, while variables values can be.
The main difference is that instances of `tf.Variable` have methods allowing us to change
their values while tensors constructed with `tf.constant` don't have these methods, and
therefore their values can not be changed. When you want to change the value of a `tf.Variable`
`x` use one of the following method:
* `x.assign(new_value)`
* `x.assign_add(value_to_be_added)`
* `x.assign_sub(value_to_be_subtracted`
```
x = tf.constant([2, 3, 4])
x
x = tf.Variable(2.0, dtype=tf.float32, name="my_variable")
x.assign(45.8)
x
x.assign_add(4)
x
x.assign_sub(3)
x
```
### Point-wise operations
Tensorflow offers similar point-wise tensor operations as numpy does:
* `tf.add` allows to add the components of a tensor
* `tf.multiply` allows us to multiply the components of a tensor
* `tf.subtract` allow us to substract the components of a tensor
* `tf.math.*` contains the usual math operations to be applied on the components of a tensor
* and many more...
Most of the standard aritmetic operations (`tf.add`, `tf.substrac`, etc.) are overloaded by the usual corresponding arithmetic symbols (`+`, `-`, etc.)
**Lab Task #1:** Performing basic operations on Tensors
1. Compute the sum of the constants `a` and `b` below using `tf.add` and `+` and verify both operations produce the same values.
2. Compute the product of the constants `a` and `b` below using `tf.multiply` and `*` and verify both operations produce the same values.
3. Compute the exponential of the constant `a` using `tf.math.exp`. Note, you'll need to specify the type for this operation.
```
# TODO 1a
a = # TODO -- Your code here.
b = # TODO -- Your code here.
c = # TODO -- Your code here.
d = # TODO -- Your code here.
print("c:", c)
print("d:", d)
# TODO 1b
a = # TODO -- Your code here.
b = # TODO -- Your code here.
c = # TODO -- Your code here.
d = # TODO -- Your code here.
print("c:", c)
print("d:", d)
# TODO 1c
# tf.math.exp expects floats so we need to explicitly give the type
a = # TODO -- Your code here.
b = # TODO -- Your code here.
print("b:", b)
```
### NumPy Interoperability
In addition to native TF tensors, tensorflow operations can take native python types and NumPy arrays as operands.
```
# native python list
a_py = [1, 2]
b_py = [3, 4]
tf.add(a_py, b_py)
# numpy arrays
a_np = np.array([1, 2])
b_np = np.array([3, 4])
tf.add(a_np, b_np)
# native TF tensor
a_tf = tf.constant([1, 2])
b_tf = tf.constant([3, 4])
tf.add(a_tf, b_tf)
```
You can convert a native TF tensor to a NumPy array using .numpy()
```
a_tf.numpy()
```
## Linear Regression
Now let's use low level tensorflow operations to implement linear regression.
Later in the course you'll see abstracted ways to do this using high level TensorFlow.
### Toy Dataset
We'll model the following function:
\begin{equation}
y= 2x + 10
\end{equation}
```
X = tf.constant(range(10), dtype=tf.float32)
Y = 2 * X + 10
print(f"X:{X}")
print(f"Y:{Y}")
```
Let's also create a test dataset to evaluate our models:
```
X_test = tf.constant(range(10, 20), dtype=tf.float32)
Y_test = 2 * X_test + 10
print(f"X_test:{X_test}")
print(f"Y_test:{Y_test}")
```
#### Loss Function
The simplest model we can build is a model that for each value of x returns the sample mean of the training set:
```
y_mean = Y.numpy().mean()
def predict_mean(X):
y_hat = [y_mean] * len(X)
return y_hat
Y_hat = predict_mean(X_test)
```
Using mean squared error, our loss is:
\begin{equation}
MSE = \frac{1}{m}\sum_{i=1}^{m}(\hat{Y}_i-Y_i)^2
\end{equation}
For this simple model the loss is then:
```
errors = (Y_hat - Y) ** 2
loss = tf.reduce_mean(errors)
loss.numpy()
```
This values for the MSE loss above will give us a baseline to compare how a more complex model is doing.
Now, if $\hat{Y}$ represents the vector containing our model's predictions when we use a linear regression model
\begin{equation}
\hat{Y} = w_0X + w_1
\end{equation}
we can write a loss function taking as arguments the coefficients of the model:
```
def loss_mse(X, Y, w0, w1):
Y_hat = w0 * X + w1
errors = (Y_hat - Y) ** 2
return tf.reduce_mean(errors)
```
### Gradient Function
To use gradient descent we need to take the partial derivatives of the loss function with respect to each of the weights. We could manually compute the derivatives, but with Tensorflow's automatic differentiation capabilities we don't have to!
During gradient descent we think of the loss as a function of the parameters $w_0$ and $w_1$. Thus, we want to compute the partial derivative with respect to these variables.
For that we need to wrap our loss computation within the context of `tf.GradientTape` instance which will reccord gradient information:
```python
with tf.GradientTape() as tape:
loss = # computation
```
This will allow us to later compute the gradients of any tensor computed within the `tf.GradientTape` context with respect to instances of `tf.Variable`:
```python
gradients = tape.gradient(loss, [w0, w1])
```
We illustrate this procedure with by computing the loss gradients with respect to the model weights:
**Lab Task #2:** Complete the function below to compute the loss gradients with respect to the model weights `w0` and `w1`.
```
# TODO 2
def compute_gradients(X, Y, w0, w1):
# TODO -- Your code here.
w0 = tf.Variable(0.0)
w1 = tf.Variable(0.0)
dw0, dw1 = compute_gradients(X, Y, w0, w1)
print("dw0:", dw0.numpy())
print("dw1", dw1.numpy())
```
### Training Loop
Here we have a very simple training loop that converges. Note we are ignoring best practices like batching, creating a separate test set, and random weight initialization for the sake of simplicity.
**Lab Task #3:** Complete the `for` loop below to train a linear regression.
1. Use `compute_gradients` to compute `dw0` and `dw1`.
2. Then, re-assign the value of `w0` and `w1` using the `.assign_sub(...)` method with the computed gradient values and the `LEARNING_RATE`.
3. Finally, for every 100th step , we'll compute and print the `loss`. Use the `loss_mse` function we created above to compute the `loss`.
```
# TODO 3
STEPS = 1000
LEARNING_RATE = .02
MSG = "STEP {step} - loss: {loss}, w0: {w0}, w1: {w1}\n"
w0 = tf.Variable(0.0)
w1 = tf.Variable(0.0)
for step in range(0, STEPS + 1):
dw0, dw1 = # TODO -- Your code here.
if step % 100 == 0:
loss = # TODO -- Your code here.
print(MSG.format(step=step, loss=loss, w0=w0.numpy(), w1=w1.numpy()))
```
Now let's compare the test loss for this linear regression to the test loss from the baseline model that outputs always the mean of the training set:
```
loss = loss_mse(X_test, Y_test, w0, w1)
loss.numpy()
```
This is indeed much better!
## Bonus
Try modelling a non-linear function such as: $y=xe^{-x^2}$
```
X = tf.constant(np.linspace(0, 2, 1000), dtype=tf.float32)
Y = X * tf.exp(-(X ** 2))
%matplotlib inline
plt.plot(X, Y)
def make_features(X):
f1 = tf.ones_like(X) # Bias.
f2 = X
f3 = tf.square(X)
f4 = tf.sqrt(X)
f5 = tf.exp(X)
return tf.stack([f1, f2, f3, f4, f5], axis=1)
def predict(X, W):
return tf.squeeze(X @ W, -1)
def loss_mse(X, Y, W):
Y_hat = predict(X, W)
errors = (Y_hat - Y) ** 2
return tf.reduce_mean(errors)
def compute_gradients(X, Y, W):
with tf.GradientTape() as tape:
loss = loss_mse(Xf, Y, W)
return tape.gradient(loss, W)
STEPS = 2000
LEARNING_RATE = 0.02
Xf = make_features(X)
n_weights = Xf.shape[1]
W = tf.Variable(np.zeros((n_weights, 1)), dtype=tf.float32)
# For plotting
steps, losses = [], []
plt.figure()
for step in range(1, STEPS + 1):
dW = compute_gradients(X, Y, W)
W.assign_sub(dW * LEARNING_RATE)
if step % 100 == 0:
loss = loss_mse(Xf, Y, W)
steps.append(step)
losses.append(loss)
plt.clf()
plt.plot(steps, losses)
print(f"STEP: {STEPS} MSE: {loss_mse(Xf, Y, W)}")
plt.figure()
plt.plot(X, Y, label="actual")
plt.plot(X, predict(Xf, W), label="predicted")
plt.legend()
```
Copyright 2020 Google Inc. Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0 Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License
|
github_jupyter
|
# Simple Test between NumPy and Numba
$$
x = \exp(-\Gamma_s d)
$$
```
import numba
import cython
import numexpr
import numpy as np
%load_ext cython
from empymod import filters
from scipy.constants import mu_0 # Magn. permeability of free space [H/m]
from scipy.constants import epsilon_0 # Elec. permittivity of free space [F/m]
res = np.array([2e14, 0.3, 1, 50, 1]) # nlay
freq = np.arange(1, 201)/20. # nfre
off = np.arange(1, 101)*1000 # noff
lambd = filters.key_201_2009().base/off[:, None] # nwav
aniso = np.array([1, 1, 1.5, 2, 1])
epermH = np.array([1, 80, 9, 20, 1])
epermV = np.array([1, 40, 9, 10, 1])
mpermH = np.array([1, 1, 3, 5, 1])
etaH = 1/res + np.outer(2j*np.pi*freq, epermH*epsilon_0)
etaV = 1/(res*aniso*aniso) + np.outer(2j*np.pi*freq, epermV*epsilon_0)
zetaH = np.outer(2j*np.pi*freq, mpermH*mu_0)
Gam = np.sqrt((etaH/etaV)[:, None, :, None] * (lambd*lambd)[None, :, None, :] + (zetaH*etaH)[:, None, :, None])
```
## NumPy
Numpy version to check result and compare times
```
def test_numpy(lGam, d):
return np.exp(-lGam*d)
```
## Numba @vectorize
This is exactly the same function as with NumPy, just added the @vectorize decorater.
```
@numba.vectorize('c16(c16, f8)')
def test_numba_vnp(lGam, d):
return np.exp(-lGam*d)
@numba.vectorize('c16(c16, f8)', target='parallel')
def test_numba_v(lGam, d):
return np.exp(-lGam*d)
```
## Numba @njit
```
@numba.njit
def test_numba_nnp(lGam, d):
out = np.empty_like(lGam)
for nf in numba.prange(lGam.shape[0]):
for no in numba.prange(lGam.shape[1]):
for ni in numba.prange(lGam.shape[2]):
out[nf, no, ni] = np.exp(-lGam[nf, no, ni] * d)
return out
@numba.njit(nogil=True, parallel=True)
def test_numba_n(lGam, d):
out = np.empty_like(lGam)
for nf in numba.prange(lGam.shape[0]):
for no in numba.prange(lGam.shape[1]):
for ni in numba.prange(lGam.shape[2]):
out[nf, no, ni] = np.exp(-lGam[nf, no, ni] * d)
return out
```
## Run comparison for a small and a big matrix
```
lGam = Gam[:, :, 1, :]
d = 100
# Output shape
out_shape = (freq.size, off.size, filters.key_201_2009().base.size)
print(' Shape Test Matrix ::', out_shape, '; total # elements:: '+str(freq.size*off.size*filters.key_201_2009().base.size))
print('------------------------------------------------------------------------------------------')
print(' NumPy :: ', end='')
# Get NumPy result for comparison
numpy_result = test_numpy(lGam, d)
# Get runtime
%timeit test_numpy(lGam, d)
print(' Numba @vectorize :: ', end='')
# Ensure it agrees with NumPy
numba_vnp_result = test_numba_vnp(lGam, d)
if not np.allclose(numpy_result, numba_vnp_result, atol=0, rtol=1e-10):
print('\n * FAIL, DOES NOT AGREE WITH NumPy RESULT!')
# Get runtime
%timeit test_numba_vnp(lGam, d)
print(' Numba @vectorize par :: ', end='')
# Ensure it agrees with NumPy
numba_v_result = test_numba_v(lGam, d)
if not np.allclose(numpy_result, numba_v_result, atol=0, rtol=1e-10):
print('\n * FAIL, DOES NOT AGREE WITH NumPy RESULT!')
# Get runtime
%timeit test_numba_v(lGam, d)
print(' Numba @njit :: ', end='')
# Ensure it agrees with NumPy
numba_nnp_result = test_numba_nnp(lGam, d)
if not np.allclose(numpy_result, numba_nnp_result, atol=0, rtol=1e-10):
print('\n * FAIL, DOES NOT AGREE WITH NumPy RESULT!')
# Get runtime
%timeit test_numba_nnp(lGam, d)
print(' Numba @njit par :: ', end='')
# Ensure it agrees with NumPy
numba_n_result = test_numba_n(lGam, d)
if not np.allclose(numpy_result, numba_n_result, atol=0, rtol=1e-10):
print('\n * FAIL, DOES NOT AGREE WITH NumPy RESULT!')
# Get runtime
%timeit test_numba_n(lGam, d)
from empymod import versions
versions('HTML', add_pckg=[cython, numba], ncol=5)
```
|
github_jupyter
|
```
import argparse
import copy
import os
import os.path as osp
import pprint
import sys
import time
from pathlib import Path
import open3d.ml as _ml3d
import open3d.ml.tf as ml3d
import yaml
from open3d.ml.datasets import S3DIS, SemanticKITTI, SmartLab
from open3d.ml.tf.models import RandLANet
from open3d.ml.tf.pipelines import SemanticSegmentation
from open3d.ml.utils import Config, get_module
randlanet_smartlab_cfg = "/home/threedee/repos/Open3D-ML/ml3d/configs/randlanet_smartlab.yml"
randlanet_semantickitti_cfg = "/home/threedee/repos/Open3D-ML/ml3d/configs/randlanet_semantickitti.yml"
randlanet_s3dis_cfg = "/home/threedee/repos/Open3D-ML/ml3d/configs/randlanet_s3dis.yml"
cfg = _ml3d.utils.Config.load_from_file(randlanet_smartlab_cfg)
# construct a dataset by specifying dataset_path
dataset = ml3d.datasets.SmartLab(**cfg.dataset)
# get the 'all' split that combines training, validation and test set
split = dataset.get_split("training")
# print the attributes of the first datum
print(split.get_attr(0))
# print the shape of the first point cloud
print(split.get_data(0)["point"].shape)
# for idx in range(split.__len__()):
# print(split.get_data(idx)["point"].shape[0])
# show the first 100 frames using the visualizer
vis = ml3d.vis.Visualizer()
vis.visualize_dataset(dataset, "training") # , indices=range(100)
cfg = _ml3d.utils.Config.load_from_file(randlanet_s3dis_cfg)
dataset = S3DIS("/home/charith/datasets/S3DIS/", use_cache=True)
model = RandLANet(**cfg.model)
pipeline = SemanticSegmentation(model=model, dataset=dataset, max_epoch=100)
pipeline.cfg_tb = {
"readme": "readme",
"cmd_line": "cmd_line",
"dataset": pprint.pformat("S3DIS", indent=2),
"model": pprint.pformat("RandLANet", indent=2),
"pipeline": pprint.pformat("SemanticSegmentation", indent=2),
}
pipeline.run_train()
# Inference and test example
from open3d.ml.tf.models import RandLANet
from open3d.ml.tf.pipelines import SemanticSegmentation
Pipeline = get_module("pipeline", "SemanticSegmentation", "tf")
Model = get_module("model", "RandLANet", "tf")
Dataset = get_module("dataset", "SemanticKITTI")
RandLANet = Model(ckpt_path=args.path_ckpt_randlanet)
# Initialize by specifying config file path
SemanticKITTI = Dataset(args.path_semantickitti, use_cache=False)
pipeline = Pipeline(model=RandLANet, dataset=SemanticKITTI)
# inference
# get data
train_split = SemanticKITTI.get_split("train")
data = train_split.get_data(0)
# restore weights
# run inference
results = pipeline.run_inference(data)
print(results)
# test
pipeline.run_test()
```
|
github_jupyter
|
```
import numpy as np
import pandas as pd
import scipy
import pickle
import matplotlib.pyplot as plt
import seaborn as sns
import ipdb
```
# generate data
## 4 types of GalSim images
```
#### 1000 training images
with open("data/galsim_simulated_2500gals_lambda0.4_theta3.14159_2021-05-20-17-01.pkl", 'rb') as handle:
group1 = pickle.load(handle)
with open("data/galsim_simulated_2500gals_lambda0.4_theta2.3562_2021-05-20-17-42.pkl", 'rb') as handle:
group2 = pickle.load(handle)
with open("data/galsim_simulated_2500gals_lambda0.4_theta1.5708_2021-05-20-17-08.pkl", 'rb') as handle:
group3 = pickle.load(handle)
with open("data/galsim_simulated_2500gals_lambda0.4_theta0.7854_2021-05-20-17-44.pkl", 'rb') as handle:
group4 = pickle.load(handle)
sns.heatmap(group1['galaxies_generated'][0])
plt.show()
sns.heatmap(group2['galaxies_generated'][0])
plt.show()
sns.heatmap(group3['galaxies_generated'][0])
plt.show()
sns.heatmap(group4['galaxies_generated'][0])
plt.show()
#### 1000 test images
with open("data/galsim_simulated_250gals_lambda0.4_theta3.14159_2021-05-20-18-14.pkl", 'rb') as handle:#
test1 = pickle.load(handle)
with open("data/galsim_simulated_250gals_lambda0.4_theta2.3562_2021-05-20-18-14.pkl", 'rb') as handle:
test2 = pickle.load(handle)
with open("data/galsim_simulated_250gals_lambda0.4_theta1.5708_2021-05-20-18-14.pkl", 'rb') as handle:
test3 = pickle.load(handle)
with open("data/galsim_simulated_250gals_lambda0.4_theta0.7854_2021-05-20-18-14.pkl", 'rb') as handle:
test4 = pickle.load(handle)
gal_img1 = group1['galaxies_generated']
gal_img2 = group2['galaxies_generated']
gal_img3 = group3['galaxies_generated']
gal_img4 = group4['galaxies_generated']
all_gal_imgs = np.vstack([gal_img1, gal_img2, gal_img3, gal_img4])
all_gal_imgs.shape
test_img1 = test1['galaxies_generated']
test_img2 = test2['galaxies_generated']
test_img3 = test3['galaxies_generated']
test_img4 = test4['galaxies_generated']
all_test_imgs = np.vstack([test_img1, test_img2, test_img3, test_img4])
all_test_imgs.shape
all_train_test_imgs = np.vstack([all_gal_imgs, all_test_imgs])
all_train_test_imgs.shape
#with open('galsim_conformal_imgs_20210520.pkl', 'wb') as handle:
# pickle.dump(all_train_test_imgs, handle, protocol=pickle.HIGHEST_PROTOCOL)
```
## 4 distributions with same mean and variance (gaussian, uniform, exponential, bimodal)
```
# N(1,1)
z1 = np.random.normal(1, 1, size=2500)
# Unif(1-sqrt(3),1+sqrt(3))
z2 = np.random.uniform(1-np.sqrt(3), 1+np.sqrt(3), size=2500)
# Expo(1)
z3 = np.random.exponential(1, size=2500)
# 0.5N(0.25,0.4375) + 0.5N(1.75,0.4375)
z4_ind = np.random.binomial(n=1, p=0.5, size=2500)
z4 = z4_ind*np.random.normal(0.25, 0.4375, size=2500) + (1-z4_ind)*np.random.normal(1.75, 0.4375, size=2500)
fig, ax = plt.subplots(figsize=(7,6))
sns.distplot(z1, color='green', label='N(1,1)', ax=ax)
sns.distplot(z2, label='Uniform(-0.732,2.732)', ax=ax)
sns.distplot(z3, label='Expo(1)', ax=ax)
sns.distplot(z4, color='purple', label='0.5N(0.25,0.4375) + 0.5N(1.75,0.4375)', bins=50, ax=ax)
plt.legend(fontsize=13)
plt.xlabel('Y', fontsize=14)
plt.ylabel('Density', fontsize=14)
plt.tick_params(axis='both', which='major', labelsize=12)
plt.savefig('z_dists_v1.pdf')
all_zs = np.hstack([z1, z2, z3, z4])
test_z1 = np.random.normal(1, 1, size=250)
test_z2 = np.random.uniform(1-np.sqrt(3), 1+np.sqrt(3), size=250)
test_z3 = np.random.exponential(1, size=250)
test_z4_ind = np.random.binomial(n=1, p=0.5, size=250)
test_z4 = test_z4_ind*np.random.normal(0.25, 0.4375, size=250) + (1-test_z4_ind)*np.random.normal(1.75, 0.4375, size=250)
all_test_zs = np.hstack([test_z1, test_z2, test_z3, test_z4])
all_train_test_zs = np.hstack([all_zs, all_test_zs])
#with open('z_conformal_20210520.pkl', 'wb') as handle:
# pickle.dump(all_train_test_zs, handle, protocol=pickle.HIGHEST_PROTOCOL)
```
# fit neural density model
# run CDE diagnostics
# conformal approach
|
github_jupyter
|
# Tracking Callbacks
```
from fastai.gen_doc.nbdoc import *
from fastai.vision import *
from fastai.callbacks import *
```
This module regroups the callbacks that track one of the metrics computed at the end of each epoch to take some decision about training. To show examples of use, we'll use our sample of MNIST and a simple cnn model.
```
path = untar_data(URLs.MNIST_SAMPLE)
data = ImageDataBunch.from_folder(path)
show_doc(TerminateOnNaNCallback)
```
Sometimes, training diverges and the loss goes to nan. In that case, there's no point continuing, so this callback stops the training.
```
model = simple_cnn((3,16,16,2))
learn = Learner(data, model, metrics=[accuracy])
learn.fit_one_cycle(1,1e4)
```
Using it prevents that situation to happen.
```
model = simple_cnn((3,16,16,2))
learn = Learner(data, model, metrics=[accuracy], callbacks=[TerminateOnNaNCallback()])
learn.fit(2,1e4)
```
### Callback methods
You don't call these yourself - they're called by fastai's [`Callback`](/callback.html#Callback) system automatically to enable the class's functionality.
```
show_doc(TerminateOnNaNCallback.on_batch_end)
show_doc(TerminateOnNaNCallback.on_epoch_end)
show_doc(EarlyStoppingCallback)
```
This callback tracks the quantity in `monitor` during the training of `learn`. `mode` can be forced to 'min' or 'max' but will automatically try to determine if the quantity should be the lowest possible (validation loss) or the highest possible (accuracy). Will stop training after `patience` epochs if the quantity hasn't improved by `min_delta`.
```
model = simple_cnn((3,16,16,2))
learn = Learner(data, model, metrics=[accuracy],
callback_fns=[partial(EarlyStoppingCallback, monitor='accuracy', min_delta=0.01, patience=3)])
learn.fit(50,1e-42)
```
### Callback methods
You don't call these yourself - they're called by fastai's [`Callback`](/callback.html#Callback) system automatically to enable the class's functionality.
```
show_doc(EarlyStoppingCallback.on_train_begin)
show_doc(EarlyStoppingCallback.on_epoch_end)
show_doc(SaveModelCallback)
```
This callback tracks the quantity in `monitor` during the training of `learn`. `mode` can be forced to 'min' or 'max' but will automatically try to determine if the quantity should be the lowest possible (validation loss) or the highest possible (accuracy). Will save the model in `name` whenever determined by `every` ('improvement' or 'epoch'). Loads the best model at the end of training is `every='improvement'`.
### Callback methods
You don't call these yourself - they're called by fastai's [`Callback`](/callback.html#Callback) system automatically to enable the class's functionality.
```
show_doc(SaveModelCallback.on_epoch_end)
show_doc(SaveModelCallback.on_train_end)
show_doc(ReduceLROnPlateauCallback)
```
This callback tracks the quantity in `monitor` during the training of `learn`. `mode` can be forced to 'min' or 'max' but will automatically try to determine if the quantity should be the lowest possible (validation loss) or the highest possible (accuracy). Will reduce the learning rate by `factor` after `patience` epochs if the quantity hasn't improved by `min_delta`.
### Callback methods
You don't call these yourself - they're called by fastai's [`Callback`](/callback.html#Callback) system automatically to enable the class's functionality.
```
show_doc(ReduceLROnPlateauCallback.on_train_begin)
show_doc(ReduceLROnPlateauCallback.on_epoch_end)
show_doc(TrackerCallback)
show_doc(TrackerCallback.get_monitor_value)
```
### Callback methods
You don't call these yourself - they're called by fastai's [`Callback`](/callback.html#Callback) system automatically to enable the class's functionality.
```
show_doc(TrackerCallback.on_train_begin)
```
## Undocumented Methods - Methods moved below this line will intentionally be hidden
## New Methods - Please document or move to the undocumented section
|
github_jupyter
|
## Современные библиотеки градиентного бустинга
Ранее мы использовали наивную версию градиентного бустинга из scikit-learn, [придуманную](https://projecteuclid.org/download/pdf_1/euclid.aos/1013203451) в 1999 году Фридманом. С тех пор было предложено много реализаций, которые оказываются лучше на практике. На сегодняшний день популярны три библиотеки, реализующие градиентный бустинг:
* **XGBoost**. После выхода быстро набрала популярность и оставалась стандартом до конца 2016 года. Одними из основных особенностей имплементации были оптимизированность построения деревьев, а также различные регуляризации модели.
* **LightGBM**. Отличительной чертой является быстрота построения композиции. Например, используется следующий трюк для ускорения обучения: при построении вершины дерева вместо перебора по всем значениям признака производится перебор значений гистограммы этого признака. Таким образом, вместо $O(\ell)$ требуется $O(\text{#bins})$. Кроме того, в отличие от других библиотек, которые строят дерево по уровням, LightGBM использует стратегию best-first, т.е. на каждом шаге строит вершину, дающую наибольшее уменьшение функционала. Таким образом, каждое дерево является цепочкой с прикрепленными листьями.
* **CatBoost**. Библиотека от компании Яндекс. Позволяет автоматически обрабатывать категориальные признаки (даже если их значения представлены в виде строк). Кроме того, алгоритм является менее чувствительным к выбору конкретных гиперпараметров. За счёт этого уменьшается время, которое тратит человек на подбор оптимальных гиперпараметров.
### Основные параметры
(lightgbm/catboost)
* `objective` – функционал, на который будет настраиваться композиция
* `eta` / `learning_rate` – темп (скорость) обучения
* `num_iterations` / `n_estimators` – число итераций бустинга
#### Параметры, отвечающие за сложность деревьев
* `max_depth` – максимальная глубина
* `max_leaves` / num_leaves – максимальное число вершин в дереве
* `gamma` / `min_gain_to_split` – порог на уменьшение функции ошибки при расщеплении в дереве
* `min_data_in_leaf` – минимальное число объектов в листе
* `min_sum_hessian_in_leaf` – минимальная сумма весов объектов в листе, минимальное число объектов, при котором делается расщепление
* `lambda` – коэффициент регуляризации (L2)
* `subsample` / `bagging_fraction` – какую часть объектов обучения использовать для построения одного дерева
* `colsample_bytree` / `feature_fraction` – какую часть признаков использовать для построения одного дерева
Подбор всех этих параметров — настоящее искусство. Но начать их настройку можно с самых главных параметров: `learning_rate` и `n_estimators`. Обычно один из них фиксируют, а оставшийся из этих двух параметров подбирают (например, фиксируют `n_estimators=1000` и подбирают `learning_rate`). Следующим по важности является `max_depth`. В силу того, что мы заинтересованы в неглубоких деревьях, обычно его перебирают из диапазона [3; 7].
```
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
plt.style.use('seaborn')
%matplotlib inline
plt.rcParams['figure.figsize'] = (8, 5)
# !pip install catboost
# !pip install lightgbm
# !pip install xgboost
!pip install mlxtend
from sklearn.datasets import make_classification
from sklearn.model_selection import train_test_split
X, y = make_classification(n_samples=500, n_features=2, n_informative=2,
n_redundant=0, n_repeated=0,
n_classes=2, n_clusters_per_class=2,
flip_y=0.05, class_sep=0.8, random_state=241)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=241)
plt.scatter(X[:, 0], X[:, 1], c=y, cmap='YlGn');
```
## Catboost
```
from catboost import CatBoostClassifier
??CatBoostClassifier
```
#### Задание 1.
- Обучите CatBoostClassifier с дефолтными параметрами, используя 300 деревьев.
- Нарисуйте decision boundary
- Посчитайте roc_auc_score
```
from sklearn.metrics import roc_auc_score
from mlxtend.plotting import plot_decision_regions
fig, ax = plt.subplots(1,1)
clf = CatBoostClassifier(iterations=200, logging_level='Silent')
clf.fit(X_train, y_train)
plot_decision_regions(X_test, y_test, clf, ax=ax)
print(roc_auc_score(y_test, clf.predict_proba(X_test)[:,1]))
y_test_pred = clf.predict_proba(X_test)[:, 1]
roc_auc_score(y_test, y_test_pred)
```
### Learning rate
Default is 0.03
#### Задание 2.
- Обучите CatBoostClassifier с разными значениями `learning_rate`.
- Посчитайте roc_auc_score на тестовой и тренировочной выборках
- Написуйте график зависимости roc_auc от скорости обучения (learning_rate)
```
lrs = np.arange(0.001, 1.1, 0.005)
quals_train = [] # to store roc auc on trian
quals_test = [] # to store roc auc on test
for l in lrs:
clf = CatBoostClassifier(iterations=150, logging_level='Silent',
learning_rate=l)
clf.fit(X_train, y_train)
q_train = roc_auc_score(y_train, clf.predict_proba(X_train)[:,1])
q_test = roc_auc_score(y_test, clf.predict_proba(X_test)[:,1])
quals_train.append(q_train)
quals_test.append(q_test)
# YOUR CODE HERE
plt.plot(lrs, quals_train, marker='.', label='train')
plt.plot(lrs, quals_test, marker='.', label='test')
plt.xlabel('LR')
plt.ylabel('AUC-ROC')
plt.legend()
# YOUR CODE HERE (make the plot)
```
### Number of trees
Важно также подобрать количество деревьев
#### Задание 3.
- Обучите CatBoostClassifier с разными значениями `iterations`.
- Посчитайте roc_auc_score на тестовой и тренировочной выборках
- Написуйте график зависимости roc_auc от размера копозиции
```
%%timeit
n_trees = [1, 5, 10, 100, 200, 300, 400, 500, 600, 700]
quals_train = []
quals_test = []
for n in n_trees:
clf = CatBoostClassifier(iterations=n, logging_level='Silent', learning_rate=0.02)
clf.fit(X_train, y_train)
q_train = roc_auc_score(y_train, clf.predict_proba(X_train)[:,1])
q_test = roc_auc_score(y_test, clf.predict_proba(X_test)[:,1])
quals_train.append(q_train)
quals_test.append(q_test)
# YOUR CODE HERE
plt.plot(n_trees, quals_train, marker='.', label='train')
plt.plot(n_trees, quals_test, marker='.', label='test')
plt.xlabel('N trees')
plt.ylabel('AUC-ROC')
plt.legend()
plt.plot(n_trees, quals_train, marker='.', label='train')
plt.plot(n_trees, quals_test, marker='.', label='test')
plt.xlabel('Number of trees')
plt.ylabel('AUC-ROC')
plt.legend()
plt.show()
```
### Staged prediction
Как сделать то же самое, но быстрее. Для этого в библиотеке CatBoost есть метод `staged_predict_proba`
```
%%timeit
# train the model with max trees
clf = CatBoostClassifier(iterations=700,
logging_level='Silent',
learning_rate = 0.01)
clf.fit(X_train, y_train)
# obtain staged predictiond on test
predictions_test = clf.staged_predict_proba(
data=X_test,
ntree_start=0,
ntree_end=700,
eval_period=25
)
# obtain staged predictiond on train
predictions_train = clf.staged_predict_proba(
data=X_train,
ntree_start=0,
ntree_end=700,
eval_period=25
)
# calculate roc_auc
quals_train = []
quals_test = []
n_trees = []
for iteration, (test_pred, train_pred) in enumerate(zip(predictions_test, predictions_train)):
n_trees.append((iteration+1)*25)
quals_test.append(roc_auc_score(y_test, test_pred[:, 1]))
quals_train.append(roc_auc_score(y_train, train_pred[:, 1]))
plt.plot(n_trees, quals_train, marker='.', label='train')
plt.plot(n_trees, quals_test, marker='.', label='test')
plt.xlabel('Number of trees')
plt.ylabel('AUC-ROC')
plt.legend()
plt.show()
```
## LightGBM
```
from lightgbm import LGBMClassifier
??LGBMClassifier
```
#### Задание 4.
- Обучите LGBMClassifier с дефолтными параметрами, используя 300 деревьев.
- Нарисуйте decision boundary
- Посчитайте roc_auc_score
```
clf = LGBMClassifier(n_estimators=200)
clf.fit(X_train, y_train)
plot_decision_regions(X_test, y_test, clf)
print(roc_auc_score(y_test, clf.predict_proba(X_test)[:,1]))
n_trees = [1, 5, 10, 100, 200, 300, 400, 500, 600, 700]
quals_train = []
quals_test = []
for n in n_trees:
clf = LGBMClassifier(n_estimators=n)
clf.fit(X_train, y_train)
q_train = roc_auc_score(y_train, clf.predict_proba(X_train)[:, 1])
q_test = roc_auc_score(y_test, clf.predict_proba(X_test)[:, 1])
quals_train.append(q_train)
quals_test.append(q_test)
plt.plot(n_trees, quals_train, marker='.', label='train')
plt.plot(n_trees, quals_test, marker='.', label='test')
plt.xlabel('Number of trees')
plt.ylabel('AUC-ROC')
plt.legend()
```
Теперь попробуем взять фиксированное количество деревьев, но будем менять максимальнyю глубину
```
depth = list(range(1, 17, 2))
quals_train = []
quals_test = []
for d in depth:
lgb = LGBMClassifier(n_estimators=100, max_depth=d)
lgb.fit(X_train, y_train)
q_train = roc_auc_score(y_train, lgb.predict_proba(X_train)[:, 1])
q_test = roc_auc_score(y_test, lgb.predict_proba(X_test)[:, 1])
quals_train.append(q_train)
quals_test.append(q_test)
plt.plot(depth, quals_train, marker='.', label='train')
plt.plot(depth, quals_test, marker='.', label='test')
plt.xlabel('Depth of trees')
plt.ylabel('AUC-ROC')
plt.legend()
```
И сравним с Catboost:
#### Задание 5.
- Обучите CatBoostClassifier с разной глубиной
- Посчитайте roc_auc_score,
- Сравните лучший результат с LGBM
```
depth = list(range(1, 17, 2))
quals_train = []
quals_test = []
```
Теперь, когда у нас получились отличные модели, нужно их сохранить!
```
clf = CatBoostClassifier(n_estimators=200, learning_rate=0.01,
max_depth=5, logging_level="Silent")
clf.fit(X_train, y_train)
clf.save_model('catboost.cbm', format='cbm');
lgb = LGBMClassifier(n_estimators=100, max_depth=3)
lgb.fit(X_train, y_train)
lgb.booster_.save_model('lightgbm.txt')
```
И загрузим обратно, когда понадобится их применить
```
lgb = LGBMClassifier(model_file='lightgbm.txt')
clf = clf.load_model('catboost.cbm')
```
## Блендинг и Стекинг
Блендинг представляет из себя "мета-алгоритм", предсказание которого строится как взвешенная сумма базовых алгоритмов.
Рассмотрим простой пример блендинга бустинга и линейной регрессии.
```
from sklearn.datasets import load_boston
from sklearn.metrics import mean_squared_error
data = load_boston()
X = pd.DataFrame(data.data, columns=data.feature_names)
y = data.target
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=10)
```
#### Задание 6.
- Обучите CatBoostRegressor со следующими гиперпараметрами:
`iterations=100, max_depth=4, learning_rate=0.01, loss_function='RMSE'`
- Посчитайте предсказание и RMSE на тестовой и тренировочной выборках
```
from catboost import CatBoostRegressor
cbm = CatBoostRegressor(iterations=100, max_depth=5, learning_rate=0.02,
loss_function='RMSE', logging_level='Silent')
cbm.fit(X_train, y_train)
y_pred_cbm = cbm.predict(X_test)
y_train_pred_cbm = cbm.predict(X_train)
print("Train RMSE = %.4f" % mean_squared_error(y_train, y_train_pred_cbm))
print("Test RMSE = %.4f" % mean_squared_error(y_test, y_pred_cbm))
```
#### Задание 7.
- Отмасштабируйте данные (StandardScaler) и обучите линейную регрессию
- Посчитайте предсказание и RMSE на тестовой и тренировочной выборках
```
from sklearn.linear_model import LinearRegression
from sklearn.preprocessing import StandardScaler
lr = LinearRegression(normalize=True)
lr.fit(X_train, y_train)
y_pred_lr = lr.predict(X_test)
y_train_lr = lr.predict(X_train)
print("Train RMSE = %.4f" % mean_squared_error(y_train, y_train_lr))
print("Test RMSE = %.4f" % mean_squared_error(y_test, y_pred_lr))
```
#### Блендинг
Будем считать, что новый алгоритм $a(x)$ представим как
$$
a(x)
=
\sum_{n = 1}^{N}
w_n b_n(x),
$$
где $\sum\limits_{n=1}^N w_n =1$
Нам нужно обучить линейную регрессию на предсказаниях двух обченных выше алгоритмов
#### Задание 8.
```
predictions_train = pd.DataFrame([y_train_lr, y_train_pred_cbm]).T
predictions_test = pd.DataFrame([y_pred_lr, y_pred_cbm]).T
lr_blend = LinearRegression()
lr_blend.fit(predictions_train, y_train)
y_pred_blend = lr_blend.predict(predictions_test)
y_train_blend = lr_blend.predict(predictions_train)
print("Train RMSE = %.4f" % mean_squared_error(y_train, y_train_blend))
print("Test RMSE = %.4f" % mean_squared_error(y_test, y_pred_blend))
```
#### Стекинг
Теперь обучим более сложную функцию композиции
$$
a(x) = f(b_1(x), b_2(x))
$$
где $f()$ это обученная модель градиентного бустинга
#### Задание 9.
```
from lightgbm import LGBMRegressor
lgb_stack = LGBMRegressor(n_estimators=100, max_depth=2)
lgb_stack.fit(predictions_train, y_train)
y_pred_stack = lgb_stack.predict(predictions_test)
mean_squared_error(y_test, y_pred_stack)
```
В итоге получаем качество на тестовой выборке лучше, чем у каждого алгоритма в отдельности.
Полезные ссылки:
* [Видео про стекинг](https://www.coursera.org/lecture/competitive-data-science/stacking-Qdtt6)
## XGBoost
```
# based on https://www.analyticsvidhya.com/blog/2016/03/complete-guide-parameter-tuning-xgboost-with-codes-python/
import pandas as pd
import numpy as np
from sklearn.model_selection import GridSearchCV
from sklearn import metrics
titanic = pd.read_csv('titanic.csv')
X = titanic[['Pclass', 'Age', 'SibSp', 'Fare']]
y = titanic.Survived.values
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
X_train.shape, y_train.shape, X_test.shape, y_test.shape
from xgboost.sklearn import XGBClassifier
??XGBClassifier
def modelfit(alg, dtrain, y, X_test=None, y_test=None, test=True):
#Fit the algorithm on the data
alg.fit(dtrain, y, eval_metric='auc')
#Predict training set:
dtrain_predictions = alg.predict(dtrain)
dtrain_predprob = alg.predict_proba(dtrain)[:,1]
#Print model report:
print ("\nModel Report")
print ("Accuracy (Train): %.4g" % metrics.accuracy_score(y, dtrain_predictions))
print ("AUC Score (Train): %f" % metrics.roc_auc_score(y, dtrain_predprob))
if test:
dtest_predictions = alg.predict(X_test)
dtest_predprob = alg.predict_proba(X_test)[:,1]
print ("Accuracy (Test): %.4g" % metrics.accuracy_score(y_test, dtest_predictions))
print ("AUC Score (Test): %f" % metrics.roc_auc_score(y_test, dtest_predprob))
# plot feature importance
feat_imp = pd.Series(alg.get_booster().get_fscore()).sort_values(ascending=False)
feat_imp.plot(kind='bar', title='Feature Importances')
plt.ylabel('Feature Importance Score')
```
These parameters are used to define the optimization objective the metric to be calculated at each step.
<table><tr>
<td> <img src="https://github.com/AKuzina/ml_dpo/blob/main/practicals/xgb.png?raw=1" alt="Drawing" style="width: 700px;"/> </td>
</tr></table>
```
xgb1 = XGBClassifier(objective='binary:logistic',
eval_metric='auc',
learning_rate =0.1,
n_estimators=1000,
booster='gbtree',
seed=27)
modelfit(xgb1, X_train, y_train, X_test, y_test)
```
#### Задание 10.
- Задайте сетку для перечисленных ниже параметров
`max_depth` - Maximum tree depth for base learners.
`gamma` - Minimum loss reduction required to make a further partition on a leaf node of the tree.
`subsample` - Subsample ratio of the training instance.
`colsample_bytree` - Subsample ratio of columns when constructing each tree.
`reg_alpha` - L1 regularization term on weights
- Запустите поиск, используя `GridSearchCV` c 5 фолдами. Используйте смесь из 100 деревьев.
```
param_grid = {
# YOUR CODE HERE
}
gsearch1 = # YOUR CODE HERE
gsearch1.best_params_, gsearch1.best_score_
```
Теперь можем взять больше деревьев, но меньше lr
```
xgb_best = XGBClassifier(objective='binary:logistic',
eval_metric='auc',
learning_rate =0.01,
n_estimators=1000,
booster='gbtree',
seed=27,
max_depth = gsearch1.best_params_['max_depth'],
gamma = gsearch1.best_params_['gamma'],
subsample = gsearch1.best_params_['subsample'],
colsample_bytree = gsearch1.best_params_['colsample_bytree'],
reg_alpha = gsearch1.best_params_['reg_alpha']
)
modelfit(xgb_best, X_train, y_train, X_test, y_test)
```
## Важность признаков
В курсе мы подробно обсуждаем, как добиваться хорошего качества решения задачи: имея выборку $X, y$, построить алгоритм с наименьшей ошибкой. Однако заказчику часто важно понимать, как работает алгоритм, почему он делает такие предсказания. Обсудим несколько мотиваций.
#### Доверие алгоритму
Например, в банках на основе решений, принятых алгоритмом, выполняются финансовые операции, и менеджер, ответственный за эти операции, будет готов использовать алгоритм, только если он понимает, что его решения обоснованы. По этой причине в банках очень часто используют простые линейные алгоритмы. Другой пример из области медицины: поскольку цена ошибки может быть очень велика, врачи готовы использовать только интерпретируемые алгоритмы.
#### Отсутствие дискриминации (fairness)
Вновь пример с банком: алгоритм кредитного скоринга не должен учитывать расовую принадлежность (racial bias) заемщика или его пол (gender bias). Между тем, такие зависимости часто могут присутствовать в датасете (исторические данные), на котором обучался алгоритм. Еще один пример: известно, что нейросетевые векторы слов содержат gender bias. Если эти вектора использовались при построении системы поиска по резюме для рекрутера, то, например, по запросу `technical skill` он может видеть женские резюме в конце ранжированного списка.
#### Учет контекста
Данные, на которых обучается алгоритм, не отображают всю предметную область. Интерпретация алгоритма позволит оценить, насколько найденные зависимости связаны с реальной жизнью. Если предсказания интерпретируемы, это также говорит о высокой обобщающей способности алгоритма.
Теперь обсудим несколько вариантов, как можно оценивать важность признаков.
### Веса линейной модели
Самый простой способ, который уже был рассмотрен на семинаре про линейные модели: после построения модели каждому признаку будет соответствовать свой вес - если признаки масштабированы, то чем он больше по модулю, тем важнее признак, а знак будет говорить о положительном или отрицательном влиянии на величину целевой переменной.
```
from sklearn.datasets import load_boston
from sklearn.metrics import mean_squared_error
data = load_boston()
X = pd.DataFrame(data.data, columns=data.feature_names)
y = data.target
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=10)
```
### FSTR (Feature strength)
[Fstr](https://catboost.ai/docs/concepts/fstr.html) говорит, что важность признака — это то, насколько в среднем меняется ответ модели при изменении значения данного признака (изменении значения разбиения).
Рассчитать его можно так:
$$feature\_importance_{F} = \sum_{tree, leaves_F} (v_1 - avr)^2\cdot c_1 +(v_2 - avr)^2\cdot c_2 = \left(v_1 - v_2\right)^2\frac{c_1c_2}{c_1 + c_2}\\
\qquad avr = \frac{v_1 \cdot c_1 + v_2 \cdot c_2}{c_1 + c_2}.$$
Мы сравниваем листы, отличающиеся значением сплита в узле на пути к ним: если условие сплита выполняется, объект попадает в левое поддерево, иначе — в правое.
$c_1, c_2$ - число объектов обучающего датасета, попавших в левое и правое поддерево соответственно, либо суммарный вес этих объектов, если используются веса; $v_1, v_2$ - значение модели в левом и правом поддереве (например, среднее)
Далее значения $feature\_importance$ нормируются, и получаются величины, которые суммируются в 100.
```
clf = CatBoostClassifier(n_estimators=200, learning_rate=0.01,
max_depth=5, logging_level="Silent")
# load the trained catboost model
clf = clf.load_model('catboost.cbm')
for val, name in sorted(zip(cbm.feature_importances_, data.feature_names))[::-1]:
print(name, val)
feature_importances = pd.DataFrame({'importance':cbm.feature_importances_}, index=data.feature_names)
feature_importances.sort_values('importance').plot.bar();
print(data.DESCR)
```
### Impurity-based feature importances
Важность признака рассчитывается как (нормированное) общее снижение критерия информативности за счет этого признака.
Приведем простейший пример, как можно получить такую оценку в sklearn-реализации RandomForest
```
from sklearn.ensemble import RandomForestRegressor
clf = RandomForestRegressor(n_estimators=100, oob_score=True)
clf.fit(X_train, y_train)
clf.feature_importances_
feature_importances = pd.DataFrame({'importance':clf.feature_importances_}, index=X_train.columns)
feature_importances.sort_values('importance').plot.bar();
```
|
github_jupyter
|
```
%cd ../
from torchsignal.datasets import OPENBMI
from torchsignal.datasets.multiplesubjects import MultipleSubjects
from torchsignal.trainer.multitask import Multitask_Trainer
from torchsignal.model import MultitaskSSVEP
import numpy as np
import torch
import matplotlib.pyplot as plt
from matplotlib.pyplot import figure
config = {
"exp_name": "multitask-run1",
"seed": 12,
"segment_config": {
"window_len": 1,
"shift_len": 1000,
"sample_rate": 1000,
"add_segment_axis": True
},
"bandpass_config": {
"sample_rate": 1000,
"lowcut": 1,
"highcut": 40,
"order": 6
},
"train_subject_ids": {
"low": 1,
"high": 54
},
"test_subject_ids": {
"low": 1,
"high": 54
},
"root": "../data/openbmi",
"selected_channels": ['P7', 'P3', 'Pz', 'P4', 'P8', 'PO9', 'O1', 'Oz', 'O2', 'PO10'],
"sessions": [1,2],
"tsdata": False,
"num_classes": 4,
"num_channel": 10,
"batchsize": 256,
"learning_rate": 0.001,
"epochs": 100,
"patience": 5,
"early_stopping": 10,
"model": {
"n1": 4,
"kernel_window_ssvep": 59,
"kernel_window": 19,
"conv_3_dilation": 4,
"conv_4_dilation": 4
},
"gpu": 0,
"multitask": True,
"runkfold": 4,
"check_model": True
}
device = torch.device("cuda:"+str(config['gpu']) if torch.cuda.is_available() else "cpu")
print('device', device)
```
# Load Data - OPENBMI
```
subject_ids = list(np.arange(config['train_subject_ids']['low'], config['train_subject_ids']['high']+1, dtype=int))
openbmi_data = MultipleSubjects(
dataset=OPENBMI,
root=config['root'],
subject_ids=subject_ids,
sessions=config['sessions'],
selected_channels=config['selected_channels'],
segment_config=config['segment_config'],
bandpass_config=config['bandpass_config'],
one_hot_labels=True,
)
```
# Train-Test model - leave one subject out
```
train_loader, val_loader, test_loader = openbmi_data.leave_one_subject_out(selected_subject_id=1)
dataloaders_dict = {
'train': train_loader,
'val': val_loader
}
check_model = config['check_model'] if 'check_model' in config else False
if check_model:
x = torch.ones((20, 10, 1000)).to(device)
if config['tsdata'] == True:
x = torch.ones((40, config['num_channel'], config['segment_config']['window_len'] * config['bandpass_config']['sample_rate'])).to(device)
model = MultitaskSSVEP(num_channel=config['num_channel'],
num_classes=config['num_classes'],
signal_length=config['segment_config']['window_len'] * config['bandpass_config']['sample_rate'],
filters_n1= config['model']['n1'],
kernel_window_ssvep= config['model']['kernel_window_ssvep'],
kernel_window= config['model']['kernel_window'],
conv_3_dilation= config['model']['conv_3_dilation'],
conv_4_dilation= config['model']['conv_4_dilation'],
).to(device)
out = model(x)
print('output',out.shape)
def count_params(model):
return sum(p.numel() for p in model.parameters() if p.requires_grad)
print('model size',count_params(model))
del model
del out
model = MultitaskSSVEP(num_channel=config['num_channel'],
num_classes=config['num_classes'],
signal_length=config['segment_config']['window_len'] * config['bandpass_config']['sample_rate'],
filters_n1= config['model']['n1'],
kernel_window_ssvep= config['model']['kernel_window_ssvep'],
kernel_window= config['model']['kernel_window'],
conv_3_dilation= config['model']['conv_3_dilation'],
conv_4_dilation= config['model']['conv_4_dilation'],
).to(device)
epochs=config['epochs'] if 'epochs' in config else 50
patience=config['patience'] if 'patience' in config else 20
early_stopping=config['early_stopping'] if 'early_stopping' in config else 40
trainer = Multitask_Trainer(model, model_name="multitask", device=device, num_classes=config['num_classes'], multitask_learning=True, patience=patience, verbose=True)
trainer.fit(dataloaders_dict, num_epochs=epochs, early_stopping=early_stopping, topk_accuracy=1, save_model=False)
test_loss, test_acc, test_metric = trainer.validate(test_loader, 1)
print('test: {:.5f}, {:.5f}, {:.5f}'.format(test_loss, test_acc, test_metric))
```
# Train-Test model - k-fold and leave one subject out
```
subject_kfold_acc = {}
subject_kfold_f1 = {}
test_subject_ids = list(np.arange(config['test_subject_ids']['low'], config['test_subject_ids']['high']+1, dtype=int))
for subject_id in test_subject_ids:
print('Subject', subject_id)
kfold_acc = []
kfold_f1 = []
for k in range(config['runkfold']):
openbmi_data.split_by_kfold(kfold_k=k, kfold_split=config['runkfold'])
train_loader, val_loader, test_loader = openbmi_data.leave_one_subject_out(selected_subject_id=subject_id, dataloader_batchsize=config['batchsize'])
dataloaders_dict = {
'train': train_loader,
'val': val_loader
}
model = MultitaskSSVEP(num_channel=config['num_channel'],
num_classes=config['num_classes'],
signal_length=config['segment_config']['window_len'] * config['bandpass_config']['sample_rate'],
filters_n1= config['model']['n1'],
kernel_window_ssvep= config['model']['kernel_window_ssvep'],
kernel_window= config['model']['kernel_window'],
conv_3_dilation= config['model']['conv_3_dilation'],
conv_4_dilation= config['model']['conv_4_dilation'],
).to(device)
epochs=config['epochs'] if 'epochs' in config else 50
patience=config['patience'] if 'patience' in config else 20
early_stopping=config['early_stopping'] if 'early_stopping' in config else 40
trainer = Multitask_Trainer(model, model_name="Network064b_1-8sub", device=device, num_classes=config['num_classes'], multitask_learning=True, patience=patience, verbose=False)
trainer.fit(dataloaders_dict, num_epochs=epochs, early_stopping=early_stopping, topk_accuracy=1, save_model=True)
test_loss, test_acc, test_metric = trainer.validate(test_loader, 1)
# print('test: {:.5f}, {:.5f}, {:.5f}'.format(test_loss, test_acc, test_metric))
kfold_acc.append(test_acc)
kfold_f1.append(test_metric)
subject_kfold_acc[subject_id] = kfold_acc
subject_kfold_f1[subject_id] = kfold_f1
print('results')
print('subject_kfold_acc', subject_kfold_acc)
print('subject_kfold_f1', subject_kfold_f1)
# acc
subjects = []
acc = []
acc_min = 1.0
acc_max = 0.0
for subject_id in subject_kfold_acc:
subjects.append(subject_id)
avg_acc = np.mean(subject_kfold_acc[subject_id])
if avg_acc < acc_min:
acc_min = avg_acc
if avg_acc > acc_max:
acc_max = avg_acc
acc.append(avg_acc)
x_pos = [i for i, _ in enumerate(subjects)]
figure(num=None, figsize=(15, 3), dpi=80, facecolor='w', edgecolor='k')
plt.bar(x_pos, acc, color='skyblue')
plt.xlabel("Subject")
plt.ylabel("Accuracies")
plt.title("Average k-fold Accuracies by subjects")
plt.xticks(x_pos, subjects)
plt.ylim([acc_min-0.02, acc_max+0.02])
plt.show()
# f1
subjects = []
f1 = []
f1_min = 1.0
f1_max = 0.0
for subject_id in subject_kfold_f1:
subjects.append(subject_id)
avg_f1 = np.mean(subject_kfold_f1[subject_id])
if avg_f1 < f1_min:
f1_min = avg_f1
if avg_f1 > f1_max:
f1_max = avg_f1
f1.append(avg_f1)
x_pos = [i for i, _ in enumerate(subjects)]
figure(num=None, figsize=(15, 3), dpi=80, facecolor='w', edgecolor='k')
plt.bar(x_pos, f1, color='skyblue')
plt.xlabel("Subject")
plt.ylabel("Accuracies")
plt.title("Average k-fold F1 by subjects")
plt.xticks(x_pos, subjects)
plt.ylim([f1_min-0.02, f1_max+0.02])
plt.show()
print('Average acc:', np.mean(acc))
print('Average f1:', np.mean(f1))
```
|
github_jupyter
|
```
# TODO
# 1. # of words
# 2. # of sensor types
# 3. how bag of words clustering works
# 4. how data feature classification works on sensor types
# 5. how data feature classification works on tag classification
# 6. # of unique sentence structure
import json
from functools import reduce
import os.path
import os
import random
import pandas as pd
from scipy.stats import entropy
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import AdaBoostClassifier
from sklearn.neural_network import MLPClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC
from sklearn.naive_bayes import GaussianNB
from sklearn.tree import DecisionTreeClassifier
from sklearn.discriminant_analysis import QuadraticDiscriminantAnalysis
from sklearn.gaussian_process.kernels import RBF
from sklearn.gaussian_process import GaussianProcessClassifier
nae_dict = {
'bonner': ['607', '608', '609', '557', '610'],
'ap_m': ['514', '513','604'],
'bsb': ['519', '568', '567', '566', '564', '565'],
'ebu3b': ['505', '506']
}
def counterize_feature(feat):
indexList = [not np.isnan(val) for val in feat]
maxVal = max(feat.loc[indexList])
minVal = min(feat.loc[indexList])
gran = 100
interval = (maxVal-minVal)/100.0
keys = np.arange(minVal,maxVal,interval)
resultDict = defaultdict(int)
for key, val in feat.iteritems():
try:
if np.isnan(val):
resultDict[None] += 1
continue
diffList = [abs(key-val) for key in keys]
minVal = min(diffList)
minIdx = diffList.index(minVal)
minKey = keys[minIdx]
resultDict[minKey] += 1
except:
print key, val
return resultDict
true_df.loc[true_df['Unique Identifier']=='505_0_3000003']['Schema Label'].ravel()[0]
#sklearn.ensemble.RandomForestClassifier
building_list = ['ebu3b']
for building_name in building_list:
print("============ %s ==========="%building_name)
with open('metadata/%s_sentence_dict.json'%building_name, 'r') as fp:
sentence_dict = json.load(fp)
srcid_list = list(sentence_dict.keys())
# 1. Number of unique words
adder = lambda x,y:x+y
num_remover = lambda xlist: ["number" if x.isdigit() else x for x in xlist]
total_word_set = set(reduce(adder, map(num_remover,sentence_dict.values()), []))
print("# of unique words: %d"%(len(total_word_set)))
# 2. of sensor types
labeled_metadata_filename = 'metadata/%s_sensor_types_location.csv'%building_name
if os.path.isfile(labeled_metadata_filename):
true_df = pd.read_csv(labeled_metadata_filename)
else:
true_df = None
if isinstance(true_df, pd.DataFrame):
sensor_type_set = set(true_df['Schema Label'].ravel())
print("# of unique sensor types: %d"%(len(sensor_type_set)))
else:
sensor_type_set = None
# 3. how bag of words clustering works
with open('model/%s_word_clustering.json'%building_name, 'r') as fp:
cluster_dict = json.load(fp)
print("# of word clusterings: %d"%(len(cluster_dict)))
small_cluster_num = 0
large_cluster_num = 0
for cluster_id, srcids in cluster_dict.items():
if len(srcids)<5:
small_cluster_num +=1
else:
large_cluster_num +=1
print("# of word small (<5)clusterings: %d"%small_cluster_num)
print("# of word large (>=5)clusterings: %d"%large_cluster_num)
# 4. how data feature classification works on sensor types
with open('model/fe_%s.json'%building_name, 'r') as fp:
#data_feature_dict = json.load(fp)
pass
with open('model/fe_%s_normalized.json'%building_name, 'r') as fp:
data_feature_dict = json.load(fp)
pass
feature_num = len(list(data_feature_dict.values())[0])
data_available_srcid_list = list(data_feature_dict.keys())
if isinstance(true_df, pd.DataFrame):
sample_num = 500
sample_idx_list = random.sample(range(0,len(data_feature_dict)), sample_num)
learning_srcid_list = [data_available_srcid_list[sample_idx]
for sample_idx in sample_idx_list]
learning_x = [data_feature_dict[srcid] for srcid in learning_srcid_list]
learning_y = [true_df.loc[true_df['Unique Identifier']==srcid]
['Schema Label'].ravel()[0]
for srcid in learning_srcid_list]
test_srcid_list = [srcid for srcid in data_available_srcid_list
if srcid not in learning_srcid_list]
test_x = [data_feature_dict[srcid] for srcid in test_srcid_list]
classifier_list = [RandomForestClassifier(),
AdaBoostClassifier(),
MLPClassifier(),
KNeighborsClassifier(),
SVC(),
GaussianNB(),
DecisionTreeClassifier()
]
for classifier in classifier_list:
classifier.fit(learning_x, learning_y)
test_y = classifier.predict(test_x)
precision = calc_accuracy(test_srcid_list, test_y)
print(type(classifier).__name__, precision)
# 5. How entropy varies in clusters
entropy_dict = dict()
for cluster_id, cluster in cluster_dict.items():
entropy_list = list()
for feature_idx in range(0,feature_num):
entropy_list.append(\
entropy([data_feature_dict[srcid][feature_idx] + 0.01
for srcid in cluster #random_sample_srcid_list \
if srcid in data_available_srcid_list]))
entropy_dict[cluster_id] = entropy_list
# 5. how data feature classification works on tag classification
#if isinstance()
# 6. # of unique sentence structure
def feature_check(data_feature_dict):
for srcid, features in data_feature_dict.items():
for feat in features:
#if np.isnan(feat):
if feat < -100:
print(srcid, features)
correct_cnt = 0
for i, srcid in enumerate(test_srcid_list):
schema_label = true_df.loc[true_df['Unique Identifier']==srcid]['Schema Label'].ravel()[0]
if schema_label==test_y[i]:
correct_cnt += 1
print(correct_cnt)
print(correct_cnt/len(test_srcid_list))
```
|
github_jupyter
|
##### Copyright 2019 The TensorFlow Authors.
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# 케라스를 사용한 분산 훈련
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://www.tensorflow.org/tutorials/distribute/keras"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />TensorFlow.org에서 보기</a>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/docs/blob/master/site/ko/tutorials/distribute/keras.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />구글 코랩(Colab)에서 실행하기</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/docs/blob/master/site/ko/tutorials/distribute/keras.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />깃허브(GitHub) 소스 보기</a>
</td>
</table>
Note: 이 문서는 텐서플로 커뮤니티에서 번역했습니다. 커뮤니티 번역 활동의 특성상 정확한 번역과 최신 내용을 반영하기 위해 노력함에도 불구하고 [공식 영문 문서](https://github.com/tensorflow/docs/blob/master/site/en/tutorials/distribute/keras.ipynb)의 내용과 일치하지 않을 수 있습니다. 이 번역에 개선할 부분이 있다면 [tensorflow/docs](https://github.com/tensorflow/docs) 깃허브 저장소로 풀 리퀘스트를 보내주시기 바랍니다. 문서 번역이나 리뷰에 참여하려면 [[email protected]](https://groups.google.com/a/tensorflow.org/forum/#!forum/docs-ko)로 메일을 보내주시기 바랍니다.
## 개요
`tf.distribute.Strategy` API는 훈련을 여러 처리 장치들로 분산시키는 것을 추상화한 것입니다. 기존의 모델이나 훈련 코드를 조금만 바꾸어 분산 훈련을 할 수 있게 하는 것이 분산 전략 API의 목표입니다.
이 튜토리얼에서는 `tf.distribute.MirroredStrategy`를 사용합니다. 이 전략은 동기화된 훈련 방식을 활용하여 한 장비에 있는 여러 개의 GPU로 그래프 내 복제를 수행합니다. 다시 말하자면, 모델의 모든 변수를 각 프로세서에 복사합니다. 그리고 각 프로세서의 그래디언트(gradient)를 [올 리듀스(all-reduce)](http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/)를 사용하여 모읍니다. 그다음 모아서 계산한 값을 각 프로세서의 모델 복사본에 적용합니다.
`MirroredStategy`는 텐서플로에서 기본으로 제공하는 몇 가지 분산 전략 중 하나입니다. 다른 전략들에 대해서는 [분산 전략 가이드](../../guide/distribute_strategy.ipynb)를 참고하십시오.
### 케라스 API
이 예는 모델과 훈련 루프를 만들기 위해 `tf.keras` API를 사용합니다. 직접 훈련 코드를 작성하는 방법은 [사용자 정의 훈련 루프로 분산 훈련하기](training_loops.ipynb) 튜토리얼을 참고하십시오.
## 필요한 패키지 가져오기
```
from __future__ import absolute_import, division, print_function, unicode_literals
# 텐서플로와 텐서플로 데이터셋 패키지 가져오기
!pip install tensorflow-gpu==2.0.0-beta1
import tensorflow_datasets as tfds
import tensorflow as tf
tfds.disable_progress_bar()
import os
```
## 데이터셋 다운로드
MNIST 데이터셋을 [TensorFlow Datasets](https://www.tensorflow.org/datasets)에서 다운로드받은 후 불러옵니다. 이 함수는 `tf.data` 형식을 반환합니다.
`with_info`를 `True`로 설정하면 전체 데이터에 대한 메타 정보도 함께 불러옵니다. 이 정보는 `info` 변수에 저장됩니다. 여기에는 훈련과 테스트 샘플 수를 비롯한 여러가지 정보들이 들어있습니다.
```
datasets, info = tfds.load(name='mnist', with_info=True, as_supervised=True)
mnist_train, mnist_test = datasets['train'], datasets['test']
```
## 분산 전략 정의하기
분산과 관련된 처리를 하는 `MirroredStrategy` 객체를 만듭니다. 이 객체가 컨텍스트 관리자(`tf.distribute.MirroredStrategy.scope`)도 제공하는데, 이 안에서 모델을 만들어야 합니다.
```
strategy = tf.distribute.MirroredStrategy()
print('장치의 수: {}'.format(strategy.num_replicas_in_sync))
```
## 입력 파이프라인 구성하기
다중 GPU로 모델을 훈련할 때는 배치 크기를 늘려야 컴퓨팅 자원을 효과적으로 사용할 수 있습니다. 기본적으로는 GPU 메모리에 맞추어 가능한 가장 큰 배치 크기를 사용하십시오. 이에 맞게 학습률도 조정해야 합니다.
```
# 데이터셋 내 샘플의 수는 info.splits.total_num_examples 로도
# 얻을 수 있습니다.
num_train_examples = info.splits['train'].num_examples
num_test_examples = info.splits['test'].num_examples
BUFFER_SIZE = 10000
BATCH_SIZE_PER_REPLICA = 64
BATCH_SIZE = BATCH_SIZE_PER_REPLICA * strategy.num_replicas_in_sync
```
픽셀의 값은 0~255 사이이므로 [0-1 범위로 정규화](https://en.wikipedia.org/wiki/Feature_scaling)해야 합니다. 정규화 함수를 정의합니다.
```
def scale(image, label):
image = tf.cast(image, tf.float32)
image /= 255
return image, label
```
이 함수를 훈련과 테스트 데이터에 적용합니다. 훈련 데이터 순서를 섞고, [훈련을 위해 배치로 묶습니다](https://www.tensorflow.org/api_docs/python/tf/data/Dataset#batch).
```
train_dataset = mnist_train.map(scale).shuffle(BUFFER_SIZE).batch(BATCH_SIZE)
eval_dataset = mnist_test.map(scale).batch(BATCH_SIZE)
```
## 모델 만들기
`strategy.scope` 컨텍스트 안에서 케라스 모델을 만들고 컴파일합니다.
```
with strategy.scope():
model = tf.keras.Sequential([
tf.keras.layers.Conv2D(32, 3, activation='relu', input_shape=(28, 28, 1)),
tf.keras.layers.MaxPooling2D(),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(64, activation='relu'),
tf.keras.layers.Dense(10, activation='softmax')
])
model.compile(loss='sparse_categorical_crossentropy',
optimizer=tf.keras.optimizers.Adam(),
metrics=['accuracy'])
```
## 콜백 정의하기
여기서 사용하는 콜백은 다음과 같습니다.
* *텐서보드(TensorBoard)*: 이 콜백은 텐서보드용 로그를 남겨서, 텐서보드에서 그래프를 그릴 수 있게 해줍니다.
* *모델 체크포인트(Checkpoint)*: 이 콜백은 매 에포크(epoch)가 끝난 후 모델을 저장합니다.
* *학습률 스케줄러*: 이 콜백을 사용하면 매 에포크 혹은 배치가 끝난 후 학습률을 바꿀 수 있습니다.
콜백을 추가하는 방법을 보여드리기 위하여 노트북에 *학습률*을 표시하는 콜백도 추가하겠습니다.
```
# 체크포인트를 저장할 체크포인트 디렉터리를 지정합니다.
checkpoint_dir = './training_checkpoints'
# 체크포인트 파일의 이름
checkpoint_prefix = os.path.join(checkpoint_dir, "ckpt_{epoch}")
# 학습률을 점점 줄이기 위한 함수
# 필요한 함수를 직접 정의하여 사용할 수 있습니다.
def decay(epoch):
if epoch < 3:
return 1e-3
elif epoch >= 3 and epoch < 7:
return 1e-4
else:
return 1e-5
# 에포크가 끝날 때마다 학습률을 출력하는 콜백.
class PrintLR(tf.keras.callbacks.Callback):
def on_epoch_end(self, epoch, logs=None):
print('\n에포크 {}의 학습률은 {}입니다.'.format(epoch + 1,
model.optimizer.lr.numpy()))
callbacks = [
tf.keras.callbacks.TensorBoard(log_dir='./logs'),
tf.keras.callbacks.ModelCheckpoint(filepath=checkpoint_prefix,
save_weights_only=True),
tf.keras.callbacks.LearningRateScheduler(decay),
PrintLR()
]
```
## 훈련과 평가
이제 평소처럼 모델을 학습합시다. 모델의 `fit` 함수를 호출하고 튜토리얼의 시작 부분에서 만든 데이터셋을 넘깁니다. 이 단계는 분산 훈련 여부와 상관없이 동일합니다.
```
model.fit(train_dataset, epochs=12, callbacks=callbacks)
```
아래에서 볼 수 있듯이 체크포인트가 저장되고 있습니다.
```
# 체크포인트 디렉터리 확인하기
!ls {checkpoint_dir}
```
모델의 성능이 어떤지 확인하기 위하여, 가장 최근 체크포인트를 불러온 후 테스트 데이터에 대하여 `evaluate`를 호출합니다.
평소와 마찬가지로 적절한 데이터셋과 함께 `evaluate`를 호출하면 됩니다.
```
model.load_weights(tf.train.latest_checkpoint(checkpoint_dir))
eval_loss, eval_acc = model.evaluate(eval_dataset)
print('평가 손실: {}, 평가 정확도: {}'.format(eval_loss, eval_acc))
```
텐서보드 로그를 다운로드받은 후 터미널에서 다음과 같이 텐서보드를 실행하여 훈련 결과를 확인할 수 있습니다.
```
$ tensorboard --logdir=path/to/log-directory
```
```
!ls -sh ./logs
```
## SavedModel로 내보내기
플랫폼에 무관한 SavedModel 형식으로 그래프와 변수들을 내보냅니다. 모델을 내보낸 후에는, 전략 범위(scope) 없이 불러올 수도 있고, 전략 범위와 함께 불러올 수도 있습니다.
```
path = 'saved_model/'
tf.keras.experimental.export_saved_model(model, path)
```
`strategy.scope` 없이 모델 불러오기.
```
unreplicated_model = tf.keras.experimental.load_from_saved_model(path)
unreplicated_model.compile(
loss='sparse_categorical_crossentropy',
optimizer=tf.keras.optimizers.Adam(),
metrics=['accuracy'])
eval_loss, eval_acc = unreplicated_model.evaluate(eval_dataset)
print('평가 손실: {}, 평가 정확도: {}'.format(eval_loss, eval_acc))
```
`strategy.scope`와 함께 모델 불러오기.
```
with strategy.scope():
replicated_model = tf.keras.experimental.load_from_saved_model(path)
replicated_model.compile(loss='sparse_categorical_crossentropy',
optimizer=tf.keras.optimizers.Adam(),
metrics=['accuracy'])
eval_loss, eval_acc = replicated_model.evaluate(eval_dataset)
print ('평가 손실: {}, 평가 정확도: {}'.format(eval_loss, eval_acc))
```
### 예제와 튜토리얼
케라스 적합/컴파일과 함께 분산 전략을 쓰는 예제들이 더 있습니다.
1. `tf.distribute.MirroredStrategy`를 사용하여 학습한 [Transformer](https://github.com/tensorflow/models/blob/master/official/transformer/v2/transformer_main.py) 예제.
2. `tf.distribute.MirroredStrategy`를 사용하여 학습한 [NCF](https://github.com/tensorflow/models/blob/master/official/recommendation/ncf_keras_main.py) 예제.
[분산 전략 가이드](../../guide/distribute_strategy.ipynb#examples_and_tutorials)에 더 많은 예제 목록이 있습니다.
## 다음 단계
* [분산 전략 가이드](../../guide/distribute_strategy.ipynb)를 읽어보세요.
* [사용자 정의 훈련 루프를 사용한 분산 훈련](training_loops.ipynb) 튜토리얼을 읽어보세요.
Note: `tf.distribute.Strategy`은 현재 활발히 개발 중입니다. 근시일내에 예제나 튜토리얼이 더 추가될 수 있습니다. 한 번 사용해 보세요. [깃허브 이슈](https://github.com/tensorflow/tensorflow/issues/new)를 통하여 피드백을 주시면 감사하겠습니다.
|
github_jupyter
|
Lambda School Data Science, Unit 2: Predictive Modeling
# Applied Modeling, Module 1
You will use your portfolio project dataset for all assignments this sprint.
## Assignment
Complete these tasks for your project, and document your decisions.
- [ ] Choose your target. Which column in your tabular dataset will you predict?
- [ ] Choose which observations you will use to train, validate, and test your model. And which observations, if any, to exclude.
- [ ] Determine whether your problem is regression or classification.
- [ ] Choose your evaluation metric.
- [ ] Begin with baselines: majority class baseline for classification, or mean baseline for regression, with your metric of choice.
- [ ] Begin to clean and explore your data.
- [ ] Choose which features, if any, to exclude. Would some features "leak" information from the future?
## Reading
- [Attacking discrimination with smarter machine learning](https://research.google.com/bigpicture/attacking-discrimination-in-ml/), by Google Research, with interactive visualizations. _"A threshold classifier essentially makes a yes/no decision, putting things in one category or another. We look at how these classifiers work, ways they can potentially be unfair, and how you might turn an unfair classifier into a fairer one. As an illustrative example, we focus on loan granting scenarios where a bank may grant or deny a loan based on a single, automatically computed number such as a credit score."_
- [How Shopify Capital Uses Quantile Regression To Help Merchants Succeed](https://engineering.shopify.com/blogs/engineering/how-shopify-uses-machine-learning-to-help-our-merchants-grow-their-business)
- [Maximizing Scarce Maintenance Resources with Data: Applying predictive modeling, precision at k, and clustering to optimize impact](https://towardsdatascience.com/maximizing-scarce-maintenance-resources-with-data-8f3491133050), **by Lambda DS3 student** Michael Brady. His blog post extends the Tanzania Waterpumps scenario, far beyond what's in the lecture notebook.
- [Notebook about how to calculate expected value from a confusion matrix by treating it as a cost-benefit matrix](https://github.com/podopie/DAT18NYC/blob/master/classes/13-expected_value_cost_benefit_analysis.ipynb)
- [Simple guide to confusion matrix terminology](https://www.dataschool.io/simple-guide-to-confusion-matrix-terminology/) by Kevin Markham, with video
- [Visualizing Machine Learning Thresholds to Make Better Business Decisions](https://blog.insightdatascience.com/visualizing-machine-learning-thresholds-to-make-better-business-decisions-4ab07f823415)
## Overview of Intent
## Targets
**Original Feature-Targets**
* BMI
* HealthScore
**Generated Feature-Targets**
* BMI -> Underweight, Average Weight, Overweight, Obese
* health_trajectory (matched polynomial regression of BMI & health scores - still in development)
## Features
**Original Features**
From Activity Response:
* Case ID only (for join)
From Child Response:
* Case ID only (for join)
From General Response (In Development):
* (In Development)
**Transformed Features**
From Activity Response:
* Total Secondary Eating (drinking not associated with primary meals)
* Total Secondary Drinking (drinking not associated with primary meals)
From Child Response:
* Total Assisted Meals
* Number Children Under 19 in Household
From General Response (In Development):
* (In Development)
## Evaluation Metrics
**Bifurcated design**
One phase will use regression forms to estimate future BMI & BMI Trajectory Over Time (coefficients of line)
Another will use classification in an attempt to model reported health status (1 thru 5; already encoded in data)
**Useful Metrics**
Accuracy scores and confusion matrices for health status. Balanced accuracy may be considered if target distribution is skewed.
r^2 and t-statistics for BMI prediction. Will also look at explained variance to see how much the model is capturing.
## Data Cleaning and Exploration
**For work on combining datasets and first pass feature engineering, see stitch.ipynb in this folder**
|
github_jupyter
|
# Python Basics
## Variables
Python variables are untyped, i.e. no datatype is required to define a variable
```
x=10 # static allocation
print(x) # to print a variable
```
Sometimes variables are allocated dynamically during runtime by user input. Python not only creates a new variable on-demand, also, it assigns corresponding type.
```
y=input('Enter something : ')
print(y)
```
To check the type of a variable use `type()` method.
```
type(x)
type(y)
```
Any input given is Python is by default of string type. You may use different __typecasting__ constructors to change it.
```
# without typeasting
y=input('Enter a number... ')
print(f'type of y is {type(y)}') # string formatting
# with typeasting into integer
y=int(input('Enter a number... '))
print(f'type of y is {type(y)}') # string formatting
```
How to check if an existing variable is of a given type ?
```
x=10
print(isinstance(x,int))
print(isinstance(x,float))
```
## Control Flow Structure
### if-else-elif
```
name = input ('Enter name...')
age = int(input('Enter age... '))
if age in range(0,150):
if age < 18 :
print(f'{name} is a minor')
elif age >= 18 and age < 60:
print(f'{name} is a young person')
else:
print(f'{name} is an elderly person')
else:
print('Invalid age')
```
### For-loop
```
print('Table Generator\n************************')
num = int(input('Enter a number... '))
for i in range(1,11):
print(f'{num} x {i} \t = {num*i}')
```
### While-loop
```
print('Table Generator\n************************')
num = int(input('Enter a number...'))
i = 1
while i<11:
print(f'{num} x {i} \t = {num * i}')
i += 1
```
## Primitive Data-structures
### List
List is a heterogenous linked-list stucture in python
```
lst = [1,2,'a','b'] #creating a list
lst
type(lst)
loc=2
print(f'item at location {loc} is {lst[loc]}') # reading item by location
lst[2]='abc' # updating an item in a list
lst
lst.insert(2,'a') # inserting into a specific location
lst
lst.pop(2) # deleting from a specific location
lst
len(lst) # length of a list
lst.reverse() # reversing a list
lst
test_list=[1,5,7,8,10] # sorting a list
test_list.sort(reverse=False)
test_list
```
### Set
```
P = {2,3,5,7} # Set of single digit prime numbers
O = {1,3,5,7} # Set of single digit odd numbers
E = {0,2,4,6,8} # Set of ingle digit even numbers
type(P)
P.union(O) # odd or prime
P.intersection(E) # even and prime
P-E # Prime but not even
```
Finding distinct numbers from a list of numbers by typecasting into set
```
lst = [1,2,4,5,6,2,1,4,5,6,1]
print(lst)
lst = list(set(lst)) # List --> Set --> List
print(lst)
```
### Dictionarry
Unordered named list, i.e. values are index by alphanumeric indices called key.
```
import random as rnd
test_d = {
'name' : 'Something', #kay : value
'age' : rnd.randint(18,60),
'marks' : {
'Physics' : rnd.randint(0,100),
'Chemistry' : rnd.randint(0,100),
'Mathematics' : rnd.randint(0,100),
'Biology' : rnd.randint(0,100),
}
}
test_d
```
A list of dictionarry forms a tabular structure. Each key becomes a column and the corresponding value becomes the value that specific row at that coloumn.
```
test_d['marks'] # reading a value by key
test_d['name'] = 'anything' # updating a value by its key
test_d
for k in test_d.keys(): # reading values iteratively by its key
print(f'value at key {k} is {test_d[k]} of type {type(test_d[k])}')
```
### Tuples
Immutable ordered collection of heterogenous data.
```
tup1 = ('a',1,2)
tup1
type(tup1)
tup1[1] # reading from index
tup1[1] = 3 # immutable collection, updation is not possible
lst1 = list(tup1) #typecast into list
lst1
```
## Serialization
### Theory
Computer networks are defined as a collection interconnected autonomous systems. The connections (edges) between netwrok devices (nodes) are descibed by its Topology which is modeled by Graph Theoretic principles and the computing modeles i.e. Algorithms are designed based on Distributed Systems. The connetions are inheritly FIFO (Sequential) in nature, thus it cannot carry any non-linear data-structures. However, duting RPC communication limiting the procedures to only linear structures are not realistic, especially while using Objects, as Objects are stored in memory Heap. Therefore Data stored in a Non-Linear DS must be converted into a linear format (Byte-Stream) before transmitting in a way that the receiver must reconstruct the source DS and retrive the original data. This transformation is called Serialization. All Modern programming languages such as Java and Python support Serializtion.
### Serializing primitive ADTs
```
test_d = {
'name' : 'Something', #kay : value
'age' : rnd.randint(18,60),
'marks' : {
'Physics' : rnd.randint(0,100),
'Chemistry' : rnd.randint(0,100),
'Mathematics' : rnd.randint(0,100),
'Biology' : rnd.randint(0,100),
},
'optionals' : ['music', 'Mechanics']
}
test_d
import json # default serialization library commonly used in RESTFul APIs
# Step 1
ser_dat = json.dumps(test_d) # Serialization
print(ser_dat)
print(type(ser_dat))
# Step 2
bs_data = ser_dat.encode() # Encoding into ByteStream
print(bs_data)
print(type(bs_data))
# Step 3
ser_data2 = bytes.decode(bs_data) # Decoding strings from ByteStream
print(ser_data2)
print(type(ser_data2))
# Step 4
json.loads(ser_data2) # Deserializing
```
### Serializing Objects
```
class MyClass: # defining class
# member variables
name
age
# member functions
def __init__(self,name, age): #__init__() = Constructor
self.name = name #'self' is like 'this' in java
self.age = age
def get_info(self): # returns a dictionary
return {'name' : self.name , 'age' : self.age}
obj1 = MyClass('abc',20) # crates an object
obj1.get_info() #invoke functions from object
json.dumps(obj1) # object can't be serializable in string
import pickle as pkl # pickle library is used to serialize objects
bs_data = pkl.dumps(obj1) # serialization + encoding
print(bs_data)
print(type(bs_data))
obj2 = pkl.loads(bs_data) # Decoding + Deserialization
obj2.get_info()
```
# Interfacing with the Operating System
In this section we will discuss various methods a Python script may use to interface with an Operating Systems. We'll fist understand the Local interfacing i.e. the script runs on top of the OS. Later, We'll see how it communicates with a remote computer using networking protocols such as Telnet and SSH.
## Local interfacing
```
import os
cmd = 'dir *.exe' # command to be executed
for i in os.popen(cmd).readlines():
print(i)
```
To run a command without any outputs
```
import os
# write a batch of commnad
cmds = ['md test_dir' ,
'cd test_dir' ,
'fsutil file createnew test1.txt 0',
'fsutil file createnew test2.txt 0',
'fsutil file createnew test3.txt 0',
'cd..'
]
# call commands from the batch
for c in cmds:
os.system(c)
# verify
for i in os.popen('dir test*.txt').readlines():
print(i)
```
## Remote Interfacing
* Install Telnet daemon on the Linux host : `sudo apt -y install telnetd`
* Verify installation using : `namp localhiost`
```
import telnetlib as tn
import getpass
host = '192.168.1.84'
user = input("Enter your remote account: ")
password = getpass.getpass()
tn_session = tn.Telnet(host)
tn_session.read_until(b"login: ")
tn_session.write(user.encode('ascii') + b"\n")
if password:
tn_session.read_until(b"Password: ")
tn_session.write(password.encode('ascii') + b"\n")
tn_session.write(b"ls\n")
print(tn_session.read_all().decode('ascii'))
```

Remote config with SSH (Secure Communication)
```
import paramiko
import getpass
host = input('Enter host IP')
port = 22
username = input("Enter your remote account: ")
password = getpass.getpass()
command = "ls"
ssh = paramiko.SSHClient()
ssh.set_missing_host_key_policy(paramiko.AutoAddPolicy())
ssh.connect(host, port, username, password)
stdin, stdout, stderr = ssh.exec_command(command)
for l in stdout.readlines():
print(l)
```

# Home Tasks
1. Write a python API that runs shell scripts on demand. The shell scripts must be present on the system. The API must take the name of the script as input and display output from the script. Create at least 3 shell scripts of your choice to demonstrate.
2. Write a python API that automatically calls DHCP request for dynamic IP allocation on a given interface, if it doesnt have any IP address.
3. Write a python API that organises files.
* The API first takes a directory as input on which it will run the organization
* Thereafter, it asks for a list of pairs (filetype, destination_folder).
* For example, [('mp3','music'),('png','images'),('jpg','images'),('mov','videos')] means all '.mp3' files will be moved to 'Music' directory likewise for images and Videos. In case the directories do not exist, the API must create them.
4. Write a python API that remotely monitors number of processes running on a system over a given period.
# Course Suggestion
https://www.linkedin.com/learning/python-essential-training-2/
|
github_jupyter
|
```
import scipy.io as io
import matplotlib.pyplot as plt
import matplotlib.pylab as pylab
#Set up parameters for figure display
params = {'legend.fontsize': 'x-large',
'figure.figsize': (8, 10),
'axes.labelsize': 'x-large',
'axes.titlesize':'x-large',
'axes.labelweight': 'bold',
'xtick.labelsize':'x-large',
'ytick.labelsize':'x-large'}
pylab.rcParams.update(params)
pylab.rcParams["font.family"] = "serif"
pylab.rcParams["font.weight"] = "heavy"
#Load the hori data from some samples..
mat_hori = io.loadmat('/work/imagingQ/SpatialAttention_Drowsiness/Jagannathan_Neuroimage2018/'
'Scripts/mat_files/horigraphics.mat')
data_hori = mat_hori['Hori_graphics']
#take the data for different scales..
y_hori1 = data_hori[0,]
y_hori2 = data_hori[3,]
y_hori3 = data_hori[6,]
y_hori4 = data_hori[9,]
y_hori5 = data_hori[12,]
y_hori6 = data_hori[13,]
y_hori7 = data_hori[15,]
y_hori8 = data_hori[18,]
y_hori9 = data_hori[21,]
y_hori10 = data_hori[23,]
#Set the bolding range..
x = list(range(0, 1001))
bold_hori1a = slice(0, 500)
bold_hori1b = slice(500, 1000)
bold_hori2a = slice(50, 460)
bold_hori2b = slice(625, 835)
bold_hori3a = slice(825, 1000)
bold_hori4a = slice(0, 1000)
bold_hori6a = slice(800, 875)
bold_hori7a = slice(200, 250)
bold_hori7b = slice(280, 350)
bold_hori7c = slice(450, 525)
bold_hori7d = slice(550, 620)
bold_hori7e = slice(750, 800)
bold_hori8a = slice(650, 750)
bold_hori8b = slice(750, 795)
bold_hori9a = slice(200, 325)
bold_hori10a = slice(720, 855)
#Set the main figure of the Hori scale..
plt.style.use('ggplot')
ax1 = plt.subplot2grid((60, 1), (0, 0), rowspan=6)
ax2 = plt.subplot2grid((60, 1), (6, 0), rowspan=6)
ax3 = plt.subplot2grid((60, 1), (12, 0), rowspan=6)
ax4 = plt.subplot2grid((60, 1), (18, 0), rowspan=6)
ax5 = plt.subplot2grid((60, 1), (24, 0), rowspan=6)
ax6 = plt.subplot2grid((60, 1), (30, 0), rowspan=6)
ax7 = plt.subplot2grid((60, 1), (36, 0), rowspan=6)
ax8 = plt.subplot2grid((60, 1), (42, 0), rowspan=6)
ax9 = plt.subplot2grid((60, 1), (48, 0), rowspan=6)
ax10 = plt.subplot2grid((60, 1), (54, 0), rowspan=6)
plt.setp(ax1, xticks=[0, 250, 500, 750, 999], xticklabels=['0','1', '2', '3', '4'])
plt.setp(ax2, xticks=[0, 250, 500, 750, 999], xticklabels=['0','1', '2', '3', '4'])
plt.setp(ax3, xticks=[0, 250, 500, 750, 999], xticklabels=['0','1', '2', '3', '4'])
plt.setp(ax4, xticks=[0, 250, 500, 750, 999], xticklabels=['0','1', '2', '3', '4'])
plt.setp(ax5, xticks=[0, 250, 500, 750, 999], xticklabels=['0','1', '2', '3', '4'])
plt.setp(ax6, xticks=[0, 250, 500, 750, 999], xticklabels=['0','1', '2', '3', '4'])
plt.setp(ax7, xticks=[0, 250, 500, 750, 999], xticklabels=['0','1', '2', '3', '4'])
plt.setp(ax8, xticks=[0, 250, 500, 750, 999], xticklabels=['0','1', '2', '3', '4'])
plt.setp(ax9, xticks=[0, 250, 500, 750, 999], xticklabels=['0','1', '2', '3', '4'])
plt.setp(ax10, xticks=[0, 250, 500, 750, 999], xticklabels=['0','1', '2', '3', '4'])
plt.subplots_adjust(wspace=0, hspace=0)
ax1.plot(x, y_hori1, 'k-', alpha=0.5, linewidth=2.0)
ax1.plot(x[bold_hori1a], y_hori1[bold_hori1a], 'b-', alpha=0.75)
ax1.plot(x[bold_hori1b], y_hori1[bold_hori1b], 'b-', alpha=0.75)
ax1.set_ylim([-150, 150])
ax1.axes.xaxis.set_ticklabels([])
ax1.set_ylabel('1: Alpha wave \ntrain', rotation=0,ha='right',va='center', fontsize=20, labelpad=10)
ax2.plot(x, y_hori2, 'k-', alpha=0.5, linewidth=2.0)
ax2.plot(x[bold_hori2a], y_hori2[bold_hori2a], 'b-', alpha=0.75)
ax2.plot(x[bold_hori2b], y_hori2[bold_hori2b], 'b-', alpha=0.75)
ax2.set_ylim([-150, 150])
ax2.axes.xaxis.set_ticklabels([])
ax2.set_ylabel('2: Alpha wave \nintermittent(>50%)', rotation=0,ha='right',va='center',
fontsize=20, labelpad=10)
ax3.plot(x, y_hori3, 'k-', alpha=0.5, linewidth=2.0)
ax3.plot(x[bold_hori3a], y_hori3[bold_hori3a], 'b-', alpha=0.75)
ax3.set_ylim([-150, 150])
ax3.axes.xaxis.set_ticklabels([])
ax3.set_ylabel('3: Alpha wave \nintermittent(<50%)', rotation=0,ha='right',va='center',
fontsize=20, labelpad=10)
ax4.plot(x, y_hori4, 'g-', alpha=0.5, linewidth=2.0)
ax4.plot(x[bold_hori4a], y_hori4[bold_hori4a], 'g-', alpha=0.75)
ax4.set_ylim([-150, 150])
ax4.axes.xaxis.set_ticklabels([])
ax4.set_ylabel('4: EEG flattening', rotation=0,ha='right',va='center', fontsize=20, labelpad=10)
ax5.plot(x, y_hori5, 'g-', alpha=0.5, linewidth=2.0)
ax5.plot(x[bold_hori4a], y_hori5[bold_hori4a], 'g-', alpha=0.75)
ax5.set_ylim([-150, 150])
ax5.axes.xaxis.set_ticklabels([])
ax5.set_ylabel('5: Ripples', rotation=0,ha='right',va='center', fontsize=20, labelpad=10)
ax6.plot(x, y_hori6, 'k-', alpha=0.5, linewidth=2.0)
ax6.plot(x[bold_hori6a], y_hori6[bold_hori6a], 'r-', alpha=0.75)
ax6.set_ylim([-150, 150])
ax6.axes.xaxis.set_ticklabels([])
ax6.set_ylabel('6: Vertex sharp wave \nsolitary', rotation=0,ha='right',va='center',
fontsize=20, labelpad=10)
ax7.plot(x, y_hori7, 'k-', alpha=0.5, linewidth=2.0)
ax7.plot(x[bold_hori7a], y_hori7[bold_hori7a], 'r-', alpha=0.75)
ax7.plot(x[bold_hori7b], y_hori7[bold_hori7b], 'r-', alpha=0.75)
ax7.plot(x[bold_hori7c], y_hori7[bold_hori7c], 'r-', alpha=0.75)
ax7.plot(x[bold_hori7d], y_hori7[bold_hori7d], 'r-', alpha=0.75)
ax7.plot(x[bold_hori7e], y_hori7[bold_hori7e], 'r-', alpha=0.75)
ax7.set_ylim([-150, 150])
ax7.set_ylabel('7: Vertex sharp wave \nbursts', rotation=0,ha='right',va='center',
fontsize=20, labelpad=10)
ax7.axes.xaxis.set_ticklabels([])
ax8.plot(x, y_hori8, 'k-', alpha=0.5, linewidth=2.0)
ax8.plot(x[bold_hori8a], y_hori8[bold_hori8a], 'r-', alpha=0.75)
ax8.plot(x[bold_hori8b], y_hori8[bold_hori8b], 'm-', alpha=0.75)
ax8.set_ylim([-150, 150])
ax8.set_ylabel('8: Vertex sharp wave \nand incomplete spindles', rotation=0,ha='right',va='center',
fontsize=20, labelpad=10)
ax8.axes.xaxis.set_ticklabels([])
ax9.plot(x, y_hori9, 'k-', alpha=0.5, linewidth=2.0)
ax9.plot(x[bold_hori9a], y_hori9[bold_hori9a], 'm-', alpha=0.75)
ax9.set_ylim([-40, 40])
ax9.set_ylabel('9: Spindles', rotation=0,ha='right',va='center', fontsize=20, labelpad=10)
ax9.axes.xaxis.set_ticklabels([])
ax10.plot(x, y_hori10, 'k-', alpha=0.5, linewidth=2.0)
ax10.plot(x[bold_hori10a], y_hori10[bold_hori10a], 'c-', alpha=0.75)
ax10.set_ylim([-175, 175])
ax10.set_ylabel('10: K-complexes', rotation=0,ha='right',va='center', fontsize=20, labelpad=10)
ax10.set_xlabel('Time(seconds)', rotation=0,ha='center',va='center', fontsize=20, labelpad=10)
ax1.axes.yaxis.set_ticklabels([' ',' ',''])
ax2.axes.yaxis.set_ticklabels([' ',' ',''])
ax3.axes.yaxis.set_ticklabels([' ',' ',''])
ax4.axes.yaxis.set_ticklabels([' ',' ',''])
ax5.axes.yaxis.set_ticklabels([' ',' ',''])
ax6.axes.yaxis.set_ticklabels([' ',' ',''])
ax7.axes.yaxis.set_ticklabels([' ',' ',''])
ax8.axes.yaxis.set_ticklabels([' ',' ',''])
ax9.axes.yaxis.set_ticklabels([' ',' ',''])
ax10.axes.yaxis.set_ticklabels(['-100(uV)','','100(uV)'])
ax10.axes.yaxis.tick_right()
ax1.axes.yaxis.set_ticks([-100, 0, 100])
ax2.axes.yaxis.set_ticks([-100, 0, 100])
ax3.axes.yaxis.set_ticks([-100, 0, 100])
ax4.axes.yaxis.set_ticks([-100, 0, 100])
ax5.axes.yaxis.set_ticks([-100, 0, 100])
ax6.axes.yaxis.set_ticks([-100, 0, 100])
ax7.axes.yaxis.set_ticks([-100, 0, 100])
ax8.axes.yaxis.set_ticks([-100, 0, 100])
ax9.axes.yaxis.set_ticks([-100, 0, 100])
ax10.axes.yaxis.set_ticks([-100, 0, 100])
# Here is the label of interest
ax2.annotate('Wake', xy=(-0.85, 0.90), xytext=(-0.85, 1.00), xycoords='axes fraction',rotation='vertical',
fontsize=20, ha='center', va='center')
ax6.annotate('N1', xy=(-0.85, 1), xytext=(-0.85, 1), xycoords='axes fraction', rotation='vertical',
fontsize=20, ha='center', va='center')
ax10.annotate('N2', xy=(-0.85, 0.90), xytext=(-0.85, 1.00), xycoords='axes fraction', rotation='vertical',
fontsize=20, ha='center', va='center')
#Set up the vertex element now..
params = {'figure.figsize': (3, 6)}
pylab.rcParams.update(params)
y_hori6 = data_hori[13,]
y_hori7 = data_hori[15,]
x = list(range(0, 101))
x_spin = list(range(0, 301))
x_kcomp = list(range(0, 301))
y_hori6 = y_hori6[800:901]
y_hori7 = y_hori7[281:382]
#Vertex
bold_biphasic = slice(8, 75)
bold_monophasic = slice(8, 65)
plt.style.use('ggplot')
f, axarr = plt.subplots(2, sharey=True) # makes the 2 subplots share an axis.
f.suptitle('Vertex element', size=12, fontweight='bold')
plt.setp(axarr, xticks=[0, 50,100], xticklabels=['0', '0.5', '1'],
yticks=[-150,0, 150])
axarr[0].plot(x, y_hori6, 'k-', alpha=0.5, linewidth=2.0)
axarr[0].plot(x[bold_biphasic], y_hori6[bold_biphasic], 'r-', alpha=0.75)
axarr[0].set_title('Biphasic', fontsize=10, fontweight='bold')
axarr[0].set_ylim([-150, 150])
axarr[1].plot(x, y_hori7, 'k-', alpha=0.5, linewidth=2.0)
axarr[1].plot(x[bold_monophasic], y_hori7[bold_monophasic], 'r-', alpha=0.75)
axarr[1].set_title('Monophasic', fontsize=10, fontweight='bold')
axarr[1].set_xlabel('Time(s)')
f.text(-0.2, 0.5, 'Amp(uV)', va='center', rotation='vertical', fontsize=20)
f.subplots_adjust(hspace=0.3)
#Set up the Spindle element now..
params = {'figure.figsize': (3, 1.5)}
pylab.rcParams.update(params)
bold_spindle = slice(95, 205)
y_hori8 = data_hori[21,]
y_hori8 = y_hori8[101:402]
fspin, axarrspin = plt.subplots(1, sharey=False) # makes the 2 subplots share an axis.
plt.setp(axarrspin, xticks=[0, 150,300], xticklabels=['0', '1.5', '3'],
yticks=[-100,0, 100])
axarrspin.plot(x_spin, y_hori8, 'k-', alpha=0.5, linewidth=2.0)
axarrspin.plot(x_spin[bold_spindle], y_hori8[bold_spindle], 'r-', alpha=0.75)
axarrspin.set_title('', fontsize=10, fontweight='bold')
axarrspin.set_ylim([-100, 100])
axarrspin.set_xlabel('Time(s)')
fspin.text(0.3, 1.5, 'Spindle element', va='center', rotation='horizontal', fontsize=12)
fspin.subplots_adjust(hspace=0.3)
#Set up the K-complex element now..
bold_kcomp = slice(20, 150)
y_hori10 = data_hori[23,]
y_hori10 = y_hori10[700:1007]
fkcomp, axarrkcomp = plt.subplots(1, sharey=False) # makes the 2 subplots share an axis.
plt.setp(axarrkcomp, xticks=[0, 150,300], xticklabels=['0', '1.5', '3'],
yticks=[-200,0, 200])
axarrkcomp.plot(x_kcomp, y_hori10, 'k-', alpha=0.5, linewidth=2.0)
axarrkcomp.plot(x_kcomp[bold_kcomp], y_hori10[bold_kcomp], 'r-', alpha=0.75)
axarrkcomp.set_title('', fontsize=10, fontweight='bold')
axarrkcomp.set_ylim([-200, 200])
axarrkcomp.set_xlabel('Time(s)')
fkcomp.text(0.3, 1.5, 'K-complex element', va='center', rotation='horizontal', fontsize=12)
fkcomp.subplots_adjust(hspace=0.3)
```
|
github_jupyter
|
# sift down
```
# python3
class HeapBuilder:
def __init__(self):
self._swaps = [] #array of tuples or arrays
self._data = []
def ReadData(self):
n = int(input())
self._data = [int(s) for s in input().split()]
assert n == len(self._data)
def WriteResponse(self):
print(len(self._swaps))
for swap in self._swaps:
print(swap[0], swap[1])
def swapdown(self,i):
n = len(self._data)
min_index = i
l = 2*i+1 if (2*i+1<n) else -1
r = 2*i+2 if (2*i+2<n) else -1
if l != -1 and self._data[l] < self._data[min_index]:
min_index = l
if r != - 1 and self._data[r] < self._data[min_index]:
min_index = r
if i != min_index:
self._swaps.append((i, min_index))
self._data[i], self._data[min_index] = \
self._data[min_index], self._data[i]
self.swapdown(min_index)
def GenerateSwaps(self):
for i in range(len(self._data)//2 ,-1,-1):
self.swapdown(i)
def Solve(self):
self.ReadData()
self.GenerateSwaps()
self.WriteResponse()
if __name__ == '__main__':
heap_builder = HeapBuilder()
heap_builder.Solve()
```
# sift up initialized swap array
```
%%time
# python3
class HeapBuilder:
# index =0
# global index
def __init__(self,n):
self._swaps = [[None ,None]]*4*n #array of tuples or arrays
self._data = []
self.n = n
self.index = 0
def ReadData(self):
# n = int(input())
self._data = [int(s) for s in input().split()]
assert self.n == len(self._data)
def WriteResponse(self):
print(self.index)
for _ in range(self.index):
print(self._swaps[_][0],self._swaps[_][1])
# print(len(self._swaps))
# for swap in self._swaps:
# print(swap[0], swap[1])
def swapup(self,i):
if i !=0:
# print(self._data[int((i-1)/2)], self._data[i])
# print(self.index)
# print(i)
if self._data[int((i-1)/2)]> self._data[i]:
# print('2')
# self._swaps.append(((int((i-1)/2)),i))
self._swaps[self.index] = ((int((i-1)/2)),i)
# print(((int((i-1)/2)),i))
self.index+=1
# print(self.index)
self._data[int((i-1)/2)], self._data[i] = self._data[i],self._data[int((i-1)/2)]
self.swapup(int((i-1)/2))
def GenerateSwaps(self):
# The following naive implementation just sorts
# the given sequence using selection sort algorithm
# and saves the resulting sequence of swaps.
# This turns the given array into a heap,
# but in the worst case gives a quadratic number of swaps.
#
# TODO: replace by a more efficient implementation
# efficient implementation is complete binary tree. but here you're not getting data 1 by 1, instead everything at once
# so for i in range(0,n), implement swap up ai < a2i+1 ai < a2i+2
for i in range(self.n-1,0,-1):
# print(i)
self.swapup(i)
# print('1')
# for j in range(i + 1, len(self._data)):
# if self._data[i] > self._data[j]:
# self._swaps.append((i, j))
# self._data[i], self._data[j] = self._data[j], self._data[i]
def Solve(self):
self.ReadData()
self.GenerateSwaps()
self.WriteResponse()
if __name__ == '__main__':
n = int(input())
heap_builder = HeapBuilder(n)
heap_builder.Solve()
assert(len(heap_builder._swaps)<=4*len(heap_builder._data))
a = [None]*4
for i in range(4):
a[i] = (i,i+1000000)
print(a)
k = 100
for i in range(k,0,-1):
print(i,end = ' ')
%%time
# python3
class HeapBuilder:
def __init__(self):
self._swaps = [] #array of tuples or arrays
self._data = []
def ReadData(self):
n = int(input())
self._data = [int(s) for s in input().split()]
assert n == len(self._data)
def WriteResponse(self):
print(len(self._swaps))
for swap in self._swaps:
print(swap[0], swap[1])
def swapup(self,i):
if i !=0:
if self._data[int((i-1)/2)]> self._data[i]:
self._swaps.append(((int((i-1)/2)),i))
self._data[int((i-1)/2)], self._data[i] = self._data[i],self._data[int((i-1)/2)]
self.swapup(int((i-1)/2))
def GenerateSwaps(self):
for i in range(len(self._data)-1,0,-1):
self.swapup(i)
def Solve(self):
self.ReadData()
self.GenerateSwaps()
self.WriteResponse()
if __name__ == '__main__':
heap_builder = HeapBuilder()
heap_builder.Solve()
26148864/536870912
0.30/3.00
# python3
class HeapBuilder:
"""Converts an array of integers into a min-heap.
A binary heap is a complete binary tree which satisfies the heap ordering
property: the value of each node is greater than or equal to the value of
its parent, with the minimum-value element at the root.
Samples:
>>> heap = HeapBuilder()
>>> heap.array = [5, 4, 3, 2, 1]
>>> heap.generate_swaps()
>>> heap.swaps
[(1, 4), (0, 1), (1, 3)]
>>> # Explanation: After swapping elements 4 in position 1 and 1 in position
>>> # 4 the array becomes 5 1 3 2 4. After swapping elements 5 in position 0
>>> # and 1 in position 1 the array becomes 1 5 3 2 4. After swapping
>>> # elements 5 in position 1 and 2 in position 3 the array becomes
>>> # 1 2 3 5 4, which is already a heap, because a[0] = 1 < 2 = a[1],
>>> # a[0] = 1 < 3 = a[2], a[1] = 2 < 5 = a[3], a[1] = 2 < 4 = a[4].
>>> heap = HeapBuilder()
>>> heap.array = [1, 2, 3, 4, 5]
>>> heap.generate_swaps()
>>> heap.swaps
[]
>>> # Explanation: The input array is already a heap, because it is sorted
>>> # in increasing order.
"""
def __init__(self):
self.swaps = []
self.array = []
@property
def size(self):
return len(self.array)
def read_data(self):
"""Reads data from standard input."""
n = int(input())
self.array = [int(s) for s in input().split()]
assert n == self.size
def write_response(self):
"""Writes the response to standard output."""
print(len(self.swaps))
for swap in self.swaps:
print(swap[0], swap[1])
def l_child_index(self, index):
"""Returns the index of left child.
If there's no left child, returns -1.
"""
l_child_index = 2 * index + 1
if l_child_index >= self.size:
return -1
return l_child_index
def r_child_index(self, index):
"""Returns the index of right child.
If there's no right child, returns -1.
"""
r_child_index = 2 * index + 2
if r_child_index >= self.size:
return -1
return r_child_index
def sift_down(self, i):
"""Sifts i-th node down until both of its children have bigger value.
At each step of swapping, indices of swapped nodes are appended
to HeapBuilder.swaps attribute.
"""
min_index = i
l = self.l_child_index(i)
r = self.r_child_index(i)
print(i,l,r)
if l != -1 and self.array[l] < self.array[min_index]:
min_index = l
if r != - 1 and self.array[r] < self.array[min_index]:
min_index = r
if i != min_index:
self.swaps.append((i, min_index))
self.array[i], self.array[min_index] = \
self.array[min_index], self.array[i]
self.sift_down(min_index)
def generate_swaps(self):
"""Heapify procedure.
It calls sift down procedure 'size // 2' times. It's enough to make
the heap completed.
"""
for i in range(self.size // 2, -1, -1):
self.sift_down(i)
def solve(self):
self.read_data()
self.generate_swaps()
self.write_response()
if __name__ == "__main__":
heap_builder = HeapBuilder()
heap_builder.solve()
```
|
github_jupyter
|
If you're opening this Notebook on colab, you will probably need to install 🤗 Transformers and 🤗 Datasets as well as other dependencies. Uncomment the following cell and run it.
```
#! pip install datasets transformers rouge-score nltk
```
If you're opening this notebook locally, make sure your environment has the last version of those libraries installed.
You can find a script version of this notebook to fine-tune your model in a distributed fashion using multiple GPUs or TPUs [here](https://github.com/huggingface/transformers/tree/master/examples/seq2seq).
# Fine-tuning a model on a summarization task
In this notebook, we will see how to fine-tune one of the [🤗 Transformers](https://github.com/huggingface/transformers) model for a summarization task. We will use the [XSum dataset](https://arxiv.org/pdf/1808.08745.pdf) (for extreme summarization) which contains BBC articles accompanied with single-sentence summaries.

We will see how to easily load the dataset for this task using 🤗 Datasets and how to fine-tune a model on it using the `Trainer` API.
```
model_checkpoint = "t5-small"
```
This notebook is built to run with any model checkpoint from the [Model Hub](https://huggingface.co/models) as long as that model has a sequence-to-sequence version in the Transformers library. Here we picked the [`t5-small`](https://huggingface.co/t5-small) checkpoint.
## Loading the dataset
We will use the [🤗 Datasets](https://github.com/huggingface/datasets) library to download the data and get the metric we need to use for evaluation (to compare our model to the benchmark). This can be easily done with the functions `load_dataset` and `load_metric`.
```
from datasets import load_dataset, load_metric
raw_datasets = load_dataset("xsum")
metric = load_metric("rouge")
```
The `dataset` object itself is [`DatasetDict`](https://huggingface.co/docs/datasets/package_reference/main_classes.html#datasetdict), which contains one key for the training, validation and test set:
```
raw_datasets
```
To access an actual element, you need to select a split first, then give an index:
```
raw_datasets["train"][0]
```
To get a sense of what the data looks like, the following function will show some examples picked randomly in the dataset.
```
import datasets
import random
import pandas as pd
from IPython.display import display, HTML
def show_random_elements(dataset, num_examples=5):
assert num_examples <= len(dataset), "Can't pick more elements than there are in the dataset."
picks = []
for _ in range(num_examples):
pick = random.randint(0, len(dataset)-1)
while pick in picks:
pick = random.randint(0, len(dataset)-1)
picks.append(pick)
df = pd.DataFrame(dataset[picks])
for column, typ in dataset.features.items():
if isinstance(typ, datasets.ClassLabel):
df[column] = df[column].transform(lambda i: typ.names[i])
display(HTML(df.to_html()))
show_random_elements(raw_datasets["train"])
```
The metric is an instance of [`datasets.Metric`](https://huggingface.co/docs/datasets/package_reference/main_classes.html#datasets.Metric):
```
metric
```
You can call its `compute` method with your predictions and labels, which need to be list of decoded strings:
```
fake_preds = ["hello there", "general kenobi"]
fake_labels = ["hello there", "general kenobi"]
metric.compute(predictions=fake_preds, references=fake_labels)
```
## Preprocessing the data
Before we can feed those texts to our model, we need to preprocess them. This is done by a 🤗 Transformers `Tokenizer` which will (as the name indicates) tokenize the inputs (including converting the tokens to their corresponding IDs in the pretrained vocabulary) and put it in a format the model expects, as well as generate the other inputs that the model requires.
To do all of this, we instantiate our tokenizer with the `AutoTokenizer.from_pretrained` method, which will ensure:
- we get a tokenizer that corresponds to the model architecture we want to use,
- we download the vocabulary used when pretraining this specific checkpoint.
That vocabulary will be cached, so it's not downloaded again the next time we run the cell.
```
from transformers import AutoTokenizer
tokenizer = AutoTokenizer.from_pretrained(model_checkpoint)
```
By default, the call above will use one of the fast tokenizers (backed by Rust) from the 🤗 Tokenizers library.
You can directly call this tokenizer on one sentence or a pair of sentences:
```
tokenizer("Hello, this one sentence!")
```
Depending on the model you selected, you will see different keys in the dictionary returned by the cell above. They don't matter much for what we're doing here (just know they are required by the model we will instantiate later), you can learn more about them in [this tutorial](https://huggingface.co/transformers/preprocessing.html) if you're interested.
Instead of one sentence, we can pass along a list of sentences:
```
tokenizer(["Hello, this one sentence!", "This is another sentence."])
```
To prepare the targets for our model, we need to tokenize them inside the `as_target_tokenizer` context manager. This will make sure the tokenizer uses the special tokens corresponding to the targets:
```
with tokenizer.as_target_tokenizer():
print(tokenizer(["Hello, this one sentence!", "This is another sentence."]))
```
If you are using one of the five T5 checkpoints we have to prefix the inputs with "summarize:" (the model can also translate and it needs the prefix to know which task it has to perform).
```
if model_checkpoint in ["t5-small", "t5-base", "t5-larg", "t5-3b", "t5-11b"]:
prefix = "summarize: "
else:
prefix = ""
```
We can then write the function that will preprocess our samples. We just feed them to the `tokenizer` with the argument `truncation=True`. This will ensure that an input longer that what the model selected can handle will be truncated to the maximum length accepted by the model. The padding will be dealt with later on (in a data collator) so we pad examples to the longest length in the batch and not the whole dataset.
```
max_input_length = 1024
max_target_length = 128
def preprocess_function(examples):
inputs = [prefix + doc for doc in examples["document"]]
model_inputs = tokenizer(inputs, max_length=max_input_length, truncation=True)
# Setup the tokenizer for targets
with tokenizer.as_target_tokenizer():
labels = tokenizer(examples["summary"], max_length=max_target_length, truncation=True)
model_inputs["labels"] = labels["input_ids"]
return model_inputs
```
This function works with one or several examples. In the case of several examples, the tokenizer will return a list of lists for each key:
```
preprocess_function(raw_datasets['train'][:2])
```
To apply this function on all the pairs of sentences in our dataset, we just use the `map` method of our `dataset` object we created earlier. This will apply the function on all the elements of all the splits in `dataset`, so our training, validation and testing data will be preprocessed in one single command.
```
tokenized_datasets = raw_datasets.map(preprocess_function, batched=True)
```
Even better, the results are automatically cached by the 🤗 Datasets library to avoid spending time on this step the next time you run your notebook. The 🤗 Datasets library is normally smart enough to detect when the function you pass to map has changed (and thus requires to not use the cache data). For instance, it will properly detect if you change the task in the first cell and rerun the notebook. 🤗 Datasets warns you when it uses cached files, you can pass `load_from_cache_file=False` in the call to `map` to not use the cached files and force the preprocessing to be applied again.
Note that we passed `batched=True` to encode the texts by batches together. This is to leverage the full benefit of the fast tokenizer we loaded earlier, which will use multi-threading to treat the texts in a batch concurrently.
## Fine-tuning the model
Now that our data is ready, we can download the pretrained model and fine-tune it. Since our task is of the sequence-to-sequence kind, we use the `AutoModelForSeq2SeqLM` class. Like with the tokenizer, the `from_pretrained` method will download and cache the model for us.
```
from transformers import AutoModelForSeq2SeqLM, DataCollatorForSeq2Seq, Seq2SeqTrainingArguments, Seq2SeqTrainer
model = AutoModelForSeq2SeqLM.from_pretrained(model_checkpoint)
```
Note that we don't get a warning like in our classification example. This means we used all the weights of the pretrained model and there is no randomly initialized head in this case.
To instantiate a `Seq2SeqTrainer`, we will need to define three more things. The most important is the [`Seq2SeqTrainingArguments`](https://huggingface.co/transformers/main_classes/trainer.html#transformers.Seq2SeqTrainingArguments), which is a class that contains all the attributes to customize the training. It requires one folder name, which will be used to save the checkpoints of the model, and all other arguments are optional:
```
batch_size = 16
args = Seq2SeqTrainingArguments(
"test-summarization",
evaluation_strategy = "epoch",
learning_rate=2e-5,
per_device_train_batch_size=batch_size,
per_device_eval_batch_size=batch_size,
weight_decay=0.01,
save_total_limit=3,
num_train_epochs=1,
predict_with_generate=True,
fp16=True,
)
```
Here we set the evaluation to be done at the end of each epoch, tweak the learning rate, use the `batch_size` defined at the top of the cell and customize the weight decay. Since the `Seq2SeqTrainer` will save the model regularly and our dataset is quite large, we tell it to make three saves maximum. Lastly, we use the `predict_with_generate` option (to properly generate summaries) and activate mixed precision training (to go a bit faster).
Then, we need a special kind of data collator, which will not only pad the inputs to the maximum length in the batch, but also the labels:
```
data_collator = DataCollatorForSeq2Seq(tokenizer, model=model)
```
The last thing to define for our `Seq2SeqTrainer` is how to compute the metrics from the predictions. We need to define a function for this, which will just use the `metric` we loaded earlier, and we have to do a bit of pre-processing to decode the predictions into texts:
```
import nltk
import numpy as np
def compute_metrics(eval_pred):
predictions, labels = eval_pred
decoded_preds = tokenizer.batch_decode(predictions, skip_special_tokens=True)
# Replace -100 in the labels as we can't decode them.
labels = np.where(labels != -100, labels, tokenizer.pad_token_id)
decoded_labels = tokenizer.batch_decode(labels, skip_special_tokens=True)
# Rouge expects a newline after each sentence
decoded_preds = ["\n".join(nltk.sent_tokenize(pred.strip())) for pred in decoded_preds]
decoded_labels = ["\n".join(nltk.sent_tokenize(label.strip())) for label in decoded_labels]
result = metric.compute(predictions=decoded_preds, references=decoded_labels, use_stemmer=True)
# Extract a few results
result = {key: value.mid.fmeasure * 100 for key, value in result.items()}
# Add mean generated length
prediction_lens = [np.count_nonzero(pred != tokenizer.pad_token_id) for pred in predictions]
result["gen_len"] = np.mean(prediction_lens)
return {k: round(v, 4) for k, v in result.items()}
```
Then we just need to pass all of this along with our datasets to the `Seq2SeqTrainer`:
```
trainer = Seq2SeqTrainer(
model,
args,
train_dataset=tokenized_datasets["train"],
eval_dataset=tokenized_datasets["validation"],
data_collator=data_collator,
tokenizer=tokenizer,
compute_metrics=compute_metrics
)
```
We can now finetune our model by just calling the `train` method:
```
trainer.train()
```
Don't forget to [upload your model](https://huggingface.co/transformers/model_sharing.html) on the [🤗 Model Hub](https://huggingface.co/models). You can then use it only to generate results like the one shown in the first picture of this notebook!
|
github_jupyter
|
# Welcome to Python 101
<a href="http://pyladies.org"><img align="right" src="http://www.pyladies.com/assets/images/pylady_geek.png" alt="Pyladies" style="position:relative;top:-80px;right:30px;height:50px;" /></a>
Welcome! This notebook is appropriate for people who have never programmed before. A few tips:
- To execute a cell, click in it and then type `[shift]` + `[enter]`
- This notebook's kernel will restart if the page becomes idle for 10 minutes, meaning you'll have to rerun steps again
- Try.jupyter.org is awesome, and <a href="http://rackspace.com">Rackspace</a> is awesome for hosting this, but you will want your own Python on your computer too. Hopefully you are in a class and someone helped you install. If not:
+ [Anaconda][anaconda-download] is great if you use Windows
or will only use Python for data analysis.
+ If you want to contribute to open source code, you want the standard
Python release. (Follow
the [Hitchhiker's Guide to Python][python-guide].)
## Outline
- Operators and functions
- Data and container types
- Control structures
- I/O, including basic web APIs
- How to write and run a Python script
[anaconda-download]: http://continuum.io/downloads
[python-guide]: http://docs.python-guide.org/
### First, try Python as a calculator.
Python can be used as a shell interpreter. After you install Python, you can open a command line terminal (*e.g.* powershell or bash), type `python3` or `python`, and a Python shell will open.
For now, we are using the notebook.
Here is simple math. Go to town!
```
1 + 1
3 / 4 # caution: in Python 2 the result will be an integer
7 ** 3
```
## Challenge for you
The arithmetic operators in Python are:
```python
+ - * / ** % //
```
Use the Python interpreter to calculate:
- 16 times 26515
- 1835 [modulo][wiki-modulo] 163
<p style="font-size:smaller">(psst...)
If you're stuck, try</p>
```python
help()
```
<p style="font-size:smaller">and then in the interactive box, type <tt>symbols</tt>
</p>
[wiki-modulo]: https://en.wikipedia.org/wiki/Modulo_operation
## More math requires the math module
```
import math
print("The square root of 3 is:", math.sqrt(3))
print("pi is:", math.pi)
print("The sin of 90 degrees is:", math.sin(math.radians(90)))
```
- The `import` statement imports the module into the namespace
- Then access functions (or constants) by using:
```python
<module>.<function>
```
- And get help on what is in the module by using:
```python
help(<module>)
```
## Challenge for you
Hint: `help(math)` will show all the functions...
- What is the arc cosine of `0.743144` in degrees?
```
from math import acos, degrees # use 'from' sparingly
int(degrees(acos(0.743144))) # 'int' to make an integer
```
## Math takeaways
- Operators are what you think
- Be careful of unintended integer math
- the `math` module has the remaining functions
# Strings
(Easier in Python than in any other language ever. Even Perl.)
## Strings
Use `help(str)` to see available functions for string objects. For help on a particular function from the class, type the class name and the function name: `help(str.join)`
String operations are easy:
```
s = "foobar"
"bar" in s
s.find("bar")
index = s.find("bar")
s[:index]
s[index:] + " this is intuitive! Hooray!"
s[-1] # The last element in the list or string
```
Strings are **immutable**, meaning they cannot be modified, only copied or replaced. (This is related to memory use, and interesting for experienced programmers ... don't worry if you don't get what this means.)
```
# Here's to start.
s = "foobar"
"bar" in s
# You try out the other ones!
s.find("bar")
```
## Challenge for you
Using only string addition (concatenation) and the function `str.join`, combine `declaration` and `sayings` :
```python
declaration = "We are the knights who say:\n"
sayings = ['"icky"'] * 3 + ['"p\'tang"']
# the (\') escapes the quote
```
to a variable, `sentence`, that when printed does this:
```python
>>> print(sentence)
We are the knights who say:
"icky", "icky", "icky", "p'tang"!
```
```
help(str.join)
declaration = "We are now the knights who say:\n"
sayings = ['"icky"'] * 3 + ['"p\'tang"']
# You do the rest -- fix the below :-)
print(sayings)
```
### Don't peek until you're done with your own code!
```
sentence = declaration + ", ".join(sayings) + "!"
print(sentence)
print() # empty 'print' makes a newline
# By the way, you use 'split' to split a string...
# (note what happens to the commas):
print(" - ".join(['ni'] * 12))
print("\n".join("icky, icky, icky, p'tang!".split(", ")))
```
## String formatting
There are a bunch of ways to do string formatting:
- C-style:
```python
"%s is: %.3f (or %d in Indiana)" % \
("Pi", math.pi, math.pi)
# %s = string
# %0.3f = floating point number, 3 places to the left of the decimal
# %d = decimal number
#
# Style notes:
# Line continuation with '\' works but
# is frowned upon. Indent twice
# (8 spaces) so it doesn't look
# like a control statement
```
```
print("%s is: %.3f (well, %d in Indiana)" % ("Pi", math.pi, math.pi))
```
- New in Python 2.6, `str.format` doesn't require types:
```python
"{0} is: {1} ({1:3.2} truncated)".format(
"Pi", math.pi)
# More style notes:
# Line continuation in square or curly
# braces or parenthesis is better.
```
```
# Use a colon and then decimals to control the
# number of decimals that print out.
#
# Also note the number {1} appears twice, so that
# the argument `math.pi` is reused.
print("{0} is: {1} ({1:.3} truncated)".format("Pi", math.pi))
```
- And Python 2.7+ allows named specifications:
```python
"{pi} is {pie:05.3}".format(
pi="Pi", pie=math.pi)
# 05.3 = zero-padded number, with
# 5 total characters, and
# 3 significant digits.
```
```
# Go to town -- change the decimal places!
print("{pi} is: {pie:05.2}".format(pi="Pi", pie=math.pi))
```
## String takeaways
- `str.split` and `str.join`, plus the **regex** module (pattern matching tools for strings), make Python my language of choice for data manipulation
- There are many ways to format a string
- `help(str)` for more
# Quick look at other types
```
# Boolean
x = True
type(x)
```
## Python has containers built in...
Lists, dictionaries, sets. We will talk about them later.
There is also a library [`collections`][collections] with additional specialized container types.
[collections]: https://docs.python.org/3/library/collections.html
```
# Lists can contain multiple types
x = [True, 1, 1.2, 'hi', [1], (1,2,3), {}, None]
type(x)
# (the underscores are for special internal variables)
# List access. Try other numbers!
x[1]
print("x[0] is:", x[0], "... and x[1] is:", x[1]) # Python is zero-indexed
x.append(set(["a", "b", "c"]))
for item in x:
print(item, "... type =", type(item))
```
If you need to check an object's type, do this:
```python
isinstance(x, list)
isinstance(x[1], bool)
```
```
# You do it!
isinstance(x, tuple)
```
## Caveat
Lists, when copied, are copied by pointer. What that means is every symbol that points to a list, points to that same list.
Same with dictionaries and sets.
### Example:
```python
fifth_element = x[4]
fifth_element.append("Both!")
print(fifth_element)
print(x)
```
Why? The assignment (`=`) operator copies the pointer to the place on the computer where the list (or dictionary or set) is: it does not copy the actual contents of the whole object, just the address where the data is in the computer. This is efficent because the object could be megabytes big.
```
# You do it!
fifth_element = x[4]
print(fifth_element)
fifth_element.append("Both!")
print(fifth_element)
# and see, the original list is changed too!
print(x)
```
### To make a duplicate copy you must do it explicitly
[The copy module ] [copy]
Example:
```python
import copy
# -------------------- A shallow copy
x[4] = ["list"]
shallow_copy_of_x = copy.copy(x)
shallow_copy_of_x[0] = "Shallow copy"
fifth_element = x[4]
fifth_element.append("Both?")
def print_list(l):
print("-" * 10)
for elem in l:
print(elem)
print()
# look at them
print_list(shallow_copy_of_x)
print_list(x)
fifth_element
```
[copy]: https://docs.python.org/3/library/copy.html
```
import copy
# -------------------- A shallow copy
x[4] = ["list"]
shallow_copy_of_x = copy.copy(x)
shallow_copy_of_x[0] = "Shallow copy"
fifth_element = x[4]
fifth_element.append("Both?")
# look at them
def print_list(l):
print("-" * 8, "the list, element-by-element", "-" * 8)
for elem in l:
print(elem)
print()
print_list(shallow_copy_of_x)
print_list(x)
```
## And here is a deep copy
```python
# -------------------- A deep copy
x[4] = ["list"]
deep_copy_of_x = copy.deepcopy(x)
deep_copy_of_x[0] = "Deep copy"
fifth_element = deep_copy_of_x[4]
fifth_element.append("Both?")
# look at them
print_list(deep_copy_of_x)
print_list(x)
fifth_element
```
```
# -------------------- A deep copy
x[4] = ["list"]
deep_copy_of_x = copy.deepcopy(x)
deep_copy_of_x[0] = "Deep copy"
fifth_element = deep_copy_of_x[4]
fifth_element.append("Both? -- no, just this one got it!")
# look at them
print(fifth_element)
print("\nand...the fifth element in the original list:")
print(x[4])
```
## Common atomic types
<table style="border:3px solid white;"><tr>
<td> boolean</td>
<td> integer </td>
<td> float </td>
<td>string</td>
<td>None</td>
</tr><tr>
<td><tt>True</tt></td>
<td><tt>42</tt></td>
<td><tt>42.0</tt></td>
<td><tt>"hello"</tt></td>
<td><tt>None</tt></td>
</tr></table>
## Common container types
<table style="border:3px solid white;"><tr>
<td> list </td>
<td> tuple </td>
<td> set </td>
<td>dictionary</td>
</tr><tr style="font-size:smaller;">
<td><ul style="margin:5px 2px 0px 15px;"><li>Iterable</li><li>Mutable</li>
<li>No restriction on elements</li>
<li>Elements are ordered</li></ul></td>
<td><ul style="margin:5px 2px 0px 15px;"><li>Iterable</li><li>Immutable</li>
<li>Elements must be hashable</li>
<li>Elements are ordered</li></ul></td>
<td><ul style="margin:5px 2px 0px 15px;"><li>Iterable</li><li>Mutable</li>
<li>Elements are<br/>
unique and must<br/>
be hashable</li>
<li>Elements are not ordered</li></ul></td>
<td><ul style="margin:5px 2px 0px 15px;"><li>Iterable</li><li>Mutable</li>
<li>Key, value pairs.<br/>
Keys are unique and<br/>
must be hashable</li>
<li>Keys are not ordered</li></ul></td>
</tr></table>
### Iterable
You can loop over it
### Mutable
You can change it
### Hashable
A hash function converts an object to a number that will always be the same for the object. They help with identifying the object. A better explanation kind of has to go into the guts of the code...
# Container examples
## List
- To make a list, use square braces.
```
l = ["a", 0, [1, 2] ]
l[1] = "second element"
type(l)
print(l)
```
- Items in a list can be anything: <br/>
sets, other lists, dictionaries, atoms
```
indices = range(len(l))
print(indices)
# Iterate over the indices using i=0, i=1, i=2
for i in indices:
print(l[i])
# Or iterate over the items in `x` directly
for x in l:
print(x)
```
## Tuple
To make a tuple, use parenthesis.
```
t = ("a", 0, "tuple")
type(t)
for x in t:
print x
```
## Set
To make a set, wrap a list with the function `set()`.
- Items in a set are unique
- Lists, dictionaries, and sets cannot be in a set
```
s = set(['a', 0])
if 'b' in s:
print("has b")
s.add("b")
s.remove("a")
if 'b' in s:
print("has b")
l = [1,2,3]
try:
s.add(l)
except TypeError:
print("Could not add the list")
#raise # uncomment this to raise an error
```
## Dictionary
To make a dictionary, use curly braces.
- A dictionary is a set of key,value pairs where the keys
are unique.
- Lists, dictionaries, and sets cannot be dictionary keys
- To iterate over a dictionary use `items`
```
# two ways to do the same thing
d = {"mother":"hamster",
"father":"elderberries"}
d = dict(mother="hamster",
father="elderberries")
d['mother']
print("the dictionary keys:", d.keys())
print()
print("the dictionary values:", d.values())
# When iterating over a dictionary, use items() and two variables:
for k, v in d.items():
print("key: ", k, end=" ... ")
print("val: ", v)
# If you don't you will just get the keys:
for k in d:
print(k)
```
## Type takeaways
- Lists, tuples, dictionaries, sets all are base Python objects
- Be careful of duck typing
- Remember about copy / deepcopy
```python
# For more information, use help(object)
help(tuple)
help(set)
help()
```
## Function definition and punctuation
The syntax for creating a function is:
```python
def function_name(arg1, arg2, kwarg1=default1):
"""Docstring goes here -- triple quoted."""
pass # the 'pass' keyword means 'do nothing'
# The next thing unindented statement is outside
# of the function. Leave a blank line between the
# end of the function and the next statement.
```
- The **def** keyword begins a function declaration.
- The colon (`:`) finishes the signature.
- The body must be indented. The indentation must be exactly the same.
- There are no curly braces for function bodies in Python — white space at the beginning of a line tells Python that this line is "inside" the body of whatever came before it.
Also, at the end of a function, leave at least one blank line to separate the thought from the next thing in the script.
```
def function_name(arg1, arg2, kwarg1="my_default_value"):
"""Docstring goes here -- triple quoted."""
pass # the 'pass' keyword means 'do nothing'
# See the docstring appear when using `help`
help(function_name)
```
## Whitespace matters
The 'tab' character **'\t'** counts as one single character even if it looks like multiple characters in your editor.
**But indentation is how you denote nesting!**
So, this can seriously mess up your coding. The [Python style guide][pep8] recommends configuring your editor to make the tab keypress type four spaces automatically.
To set the spacing for Python code in Sublime, go to **Sublime Text** → **Preferences** → **Settings - More** → **Syntax Specific - User**
It will open up the file **Python.sublime-settings**. Please put this inside, then save and close.
```
{
"tab_size": 4,
"translate_tabs_to_spaces": true
}
```
[pep8]: https://www.python.org/dev/peps/pep-0008/
## Your first function
Copy this and paste it in the cell below
```python
def greet_person(person):
"""Greet the named person.
usage:
>>> greet_person("world")
hello world
"""
print('hello', person)
```
```
# Paste the function definition below:
# Here's the help statement
help(greet_person)
# And here's the function in action!
greet_person("world")
```
## Duck typing
Python's philosophy for handling data types is called **duck typing** (If it walks like a duck, and quacks like a duck, it's a duck). Functions do no type checking — they happily process an argument until something breaks. This is great for fast coding but can sometimes make for odd errors. (If you care to specify types, there is a [standard way to do it][pep484], but don't worry about this if you're a beginner.)
[pep484]: https://www.python.org/dev/peps/pep-0484/
## Challenge for you
Create another function named `greet_people` that takes a list of people and greets them all one by one. Hint: you can call the function `greet_person`.
```
# your function
def greet_people(list_of_people)
"""Documentation string goes here."""
# You do it here!
pass
```
### don't peek...
```
def greet_people(list_of_people):
for person in list_of_people:
greet_person(person)
greet_people(["world", "awesome python user!", "rockstar!!!"])
```
## Quack quack
Make a list of all of the people in your group and use your function to greet them:
```python
people = ["King Arthur",
"Sir Galahad",
"Sir Robin"]
greet_people(people)
# What do you think will happen if I do:
greet_people("pyladies")
```
```
# Try it!
```
## WTW?
Remember strings are iterable...
<div style="text-align:center;">quack!</div>
<div style="text-align:right;">quack!</div>
## Whitespace / duck typing takeways
- Indentation is how to denote nesting in Python
- Do not use tabs; expand them to spaces
- If it walks like a duck and quacks like a duck, it's a duck
# Control structures
### Common comparison operators
<table style="border:3px solid white;"><tr>
<td><tt>==</tt></td>
<td><tt>!=</tt></td>
<td><tt><=</tt> or <tt><</tt><br/>
<tt>>=</tt> or <tt>></tt></td>
<td><tt>x in (1, 2)</tt></td>
<td><tt>x is None<br/>
x is not None</tt></td>
</tr><tr style="font-size:smaller;">
<td>equals</td>
<td>not equals</td>
<td>less or<br/>equal, etc.</td>
<td>works for sets,<br/>
lists, tuples,<br/>
dictionary keys,<br/>
strings</td>
<td>just for <tt>None</tt></td>
</tr></table>
### If statement
The `if` statement checks whether the condition after `if` is true.
Note the placement of colons (`:`) and the indentation. These are not optional.
- If it is, it does the thing below it.
- Otherwise it goes to the next comparison.
- You do not need any `elif` or `else` statements if you only
want to do something if your test condition is true.
Advanced users, there is no switch statement in Python.
```
# Standard if / then / else statement.
#
# Go ahead and change `i`
i = 1
if i is None:
print("None!")
elif i % 2 == 0:
print("`i` is an even number!")
else:
print("`i` is neither None nor even")
# This format is for very short one-line if / then / else.
# It is called a `ternary` statement.
#
"Y" if i==1 else "N"
```
### While loop
The `while` loop requires you to set up something first. Then it
tests whether the statement after the `while` is true.
Again note the colon (`:`) and the indentation.
- If the condition is true, then the body of
the `while` loop will execute
- Otherwise it will break out of the loop and go on
to the next code underneath the `while` block
```
i = 0
while i < 3:
print("i is:", i)
i += 1
print("We exited the loop, and now i is:", i)
```
### For loop
The `for` loop iterates over the items after the `for`,
executing the body of the loop once per item.
```
for i in range(3):
print("in the for loop. `i` is:", i)
print()
print("outside the for loop. `i` is:", i)
# or loop directly over a list or tuple
for element in ("one", 2, "three"):
print("in the for loop. `element` is:", element)
print()
print("outside the for loop. `element` is:", element)
```
## Challenge for you
Please look at this code and think of what will happen, then copy it and run it. We introduce `break` and `continue`...can you tell what they do?
- When will it stop?
- What will it print out?
- What will `i` be at the end?
```python
for i in range(20):
if i == 15:
break
elif i % 2 == 0:
continue
for j in range(5):
print(i + j, end="...")
print() # newline
```
```
# Paste it here, and run!
```
# You are done, welcome to Python!
## ... and you rock!
### Now join (or start!) a friendly PyLadies group near you ...
[PyLadies locations][locations]
[locations]: http://www.pyladies.com/locations/
<div style="font-size:80%;color:#333333;text-align:center;">
<h4>Psst...contribute to this repo!</h4>
<span style="font-size:70%;">
Here is the
<a href="https://github.com/jupyter/docker-demo-images">
link to the github repo that hosts these
</a>.
Make them better!
</span>
</div>
|
github_jupyter
|
# Part 4: Create an approximate nearest neighbor index for the item embeddings
This notebook is the fourth of five notebooks that guide you through running the [Real-time Item-to-item Recommendation with BigQuery ML Matrix Factorization and ScaNN](https://github.com/GoogleCloudPlatform/analytics-componentized-patterns/tree/master/retail/recommendation-system/bqml-scann) solution.
Use this notebook to create an approximate nearest neighbor (ANN) index for the item embeddings by using the [ScaNN](https://github.com/google-research/google-research/tree/master/scann) framework. You create the index as a model, train the model on AI Platform Training, then export the index to Cloud Storage so that it can serve ANN information.
Before starting this notebook, you must run the [03_create_embedding_lookup_model](03_create_embedding_lookup_model.ipynb) notebook to process the item embeddings data and export it to Cloud Storage.
After completing this notebook, run the [05_deploy_lookup_and_scann_caip](05_deploy_lookup_and_scann_caip.ipynb) notebook to deploy the solution. Once deployed, you can submit song IDs to the solution and get similar song recommendations in return, based on the ANN index.
## Setup
Import the required libraries, configure the environment variables, and authenticate your GCP account.
```
!pip install -q scann
```
### Import libraries
```
import tensorflow as tf
import numpy as np
from datetime import datetime
```
### Configure GCP environment settings
Update the following variables to reflect the values for your GCP environment:
+ `PROJECT_ID`: The ID of the Google Cloud project you are using to implement this solution.
+ `BUCKET`: The name of the Cloud Storage bucket you created to use with this solution. The `BUCKET` value should be just the bucket name, so `myBucket` rather than `gs://myBucket`.
+ `REGION`: The region to use for the AI Platform Training job.
```
PROJECT_ID = 'yourProject' # Change to your project.
BUCKET = 'yourBucketName' # Change to the bucket you created.
REGION = 'yourTrainingRegion' # Change to your AI Platform Training region.
EMBEDDING_FILES_PREFIX = f'gs://{BUCKET}/bqml/item_embeddings/embeddings-*'
OUTPUT_INDEX_DIR = f'gs://{BUCKET}/bqml/scann_index'
```
### Authenticate your GCP account
This is required if you run the notebook in Colab. If you use an AI Platform notebook, you should already be authenticated.
```
try:
from google.colab import auth
auth.authenticate_user()
print("Colab user is authenticated.")
except: pass
```
## Build the ANN index
Use the `build` method implemented in the [indexer.py](index_builder/builder/indexer.py) module to load the embeddings from the CSV files, create the ANN index model and train it on the embedding data, and save the SavedModel file to Cloud Storage. You pass the following three parameters to this method:
+ `embedding_files_path`, which specifies the Cloud Storage location from which to load the embedding vectors.
+ `num_leaves`, which provides the value for a hyperparameter that tunes the model based on the trade-off between retrieval latency and recall. A higher `num_leaves` value will use more data and provide better recall, but will also increase latency. If `num_leaves` is set to `None` or `0`, the `num_leaves` value is the square root of the number of items.
+ `output_dir`, which specifies the Cloud Storage location to write the ANN index SavedModel file to.
Other configuration options for the model are set based on the [rules-of-thumb](https://github.com/google-research/google-research/blob/master/scann/docs/algorithms.md#rules-of-thumb) provided by ScaNN.
### Build the index locally
```
from index_builder.builder import indexer
indexer.build(EMBEDDING_FILES_PREFIX, OUTPUT_INDEX_DIR)
```
### Build the index using AI Platform Training
Submit an AI Platform Training job to build the ScaNN index at scale. The [index_builder](index_builder) directory contains the expected [training application packaging structure](https://cloud.google.com/ai-platform/training/docs/packaging-trainer) for submitting the AI Platform Training job.
```
if tf.io.gfile.exists(OUTPUT_INDEX_DIR):
print("Removing {} contents...".format(OUTPUT_INDEX_DIR))
tf.io.gfile.rmtree(OUTPUT_INDEX_DIR)
print("Creating output: {}".format(OUTPUT_INDEX_DIR))
tf.io.gfile.makedirs(OUTPUT_INDEX_DIR)
timestamp = datetime.utcnow().strftime('%y%m%d%H%M%S')
job_name = f'ks_bqml_build_scann_index_{timestamp}'
!gcloud ai-platform jobs submit training {job_name} \
--project={PROJECT_ID} \
--region={REGION} \
--job-dir={OUTPUT_INDEX_DIR}/jobs/ \
--package-path=index_builder/builder \
--module-name=builder.task \
--config='index_builder/config.yaml' \
--runtime-version=2.2 \
--python-version=3.7 \
--\
--embedding-files-path={EMBEDDING_FILES_PREFIX} \
--output-dir={OUTPUT_INDEX_DIR} \
--num-leaves=500
```
After the AI Platform Training job finishes, check that the `scann_index` folder has been created in your Cloud Storage bucket:
```
!gsutil ls {OUTPUT_INDEX_DIR}
```
## Test the ANN index
Test the ANN index by using the `ScaNNMatcher` class implemented in the [index_server/matching.py](index_server/matching.py) module.
Run the following code snippets to create an item embedding from random generated values and pass it to `scann_matcher`, which returns the items IDs for the five items that are the approximate nearest neighbors of the embedding you submitted.
```
from index_server.matching import ScaNNMatcher
scann_matcher = ScaNNMatcher(OUTPUT_INDEX_DIR)
vector = np.random.rand(50)
scann_matcher.match(vector, 5)
```
## License
Copyright 2020 Google LLC
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License. You may obtain a copy of the License at: http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and limitations under the License.
**This is not an official Google product but sample code provided for an educational purpose**
|
github_jupyter
|
# Day 24 - Cellular automaton
We are back to [cellar automatons](https://en.wikipedia.org/wiki/Cellular_automaton), in a finite 2D grid, just like [day 18 of 2018](../2018/Day%2018.ipynb). I'll use similar techniques, with [`scipy.signal.convolve2d()`](https://docs.scipy.org/doc/scipy-0.18.1/reference/generated/scipy.signal.convolve2d.html) to turn neighbor counts into the next state. Our state is simpler, a simple on or off so we can use simple boolean selections here.
```
from __future__ import annotations
from typing import Set, Sequence, Tuple
import numpy as np
from scipy.signal import convolve2d
def readmap(maplines: Sequence[str]) -> np.array:
return np.array([
c == "#" for line in maplines for c in line
]).reshape((5, -1))
def biodiversity_rating(matrix: np.array) -> int:
# booleans -> single int by multiplying with powers of 2, then summing
return (
matrix.reshape((-1)) *
np.logspace(0, matrix.size - 1, num=matrix.size, base=2, dtype=np.uint)
).sum()
def find_repeat(matrix: np.array) -> int:
# the four adjacent tiles matter, not the diagonals
kernel = np.array([[0, 1, 0], [1, 0, 1], [0, 1, 0]])
# previous states seen (matrix flattened to a tuple)
seen: Set[Tuple] = set()
while True:
counts = convolve2d(matrix, kernel, mode='same')
matrix = (
# A bug dies (becoming an empty space) unless there is exactly one bug adjacent to it.
(matrix & (counts == 1)) |
# An empty space becomes infested with a bug if exactly one or two bugs are adjacent to it.
(~matrix & ((counts == 1) | (counts == 2)))
)
key = tuple(matrix.flatten())
if key in seen:
return biodiversity_rating(matrix)
seen.add(key)
test_matrix = readmap("""\
....#
#..#.
#..##
..#..
#....""".splitlines())
assert find_repeat(test_matrix) == 2129920
import aocd
data = aocd.get_data(day=24, year=2019)
erismap = readmap(data.splitlines())
print("Part 1:", find_repeat(erismap))
# how fast is this?
%timeit find_repeat(erismap)
```
## Part 2, adding a 3rd dimension
I'm not sure if we might be able to use [`scipy.signal.convolve()`](https://docs.scipy.org/doc/scipy-0.18.1/reference/generated/scipy.signal.convolve.html#scipy.signal.convolve) (the N-dimensional variant of `convolve2d()`) to count neighbours across multiple layers in one go. It works for counting neighbours across a single layer however, and for 200 steps, the additional 8 computations are not exactly strenuous.
I'm creating all layers needed to fit all the steps. An empty layer is filled across 2 steps; first the inner ring, then the outer ring, at which point another layer is needed. So for 200 steps we need 100 layers below and a 100 layers above, ending up with 201 layers. These are added by using [np.pad()](https://docs.scipy.org/doc/numpy/reference/generated/numpy.pad.html).
Then use `convolve()` to count neighbours on the same level, and a few sums for additional counts from the levels above and below.
```
from scipy.signal import convolve
def run_multidimensional(matrix: np.array, steps: int = 200) -> int:
# 3d kernel; only those on the same level, not above or below
kernel = np.array([
[[0, 0, 0], [0, 0, 0], [0, 0, 0]],
[[0, 1, 0], [1, 0, 1], [0, 1, 0]],
[[0, 0, 0], [0, 0, 0], [0, 0, 0]],
])
matrix = np.pad(matrix[None], [((steps + 1) // 2,), (0,), (0,)])
for _ in range(steps):
# count neighbours on the same layer, then clear the hole
counts = convolve(matrix, kernel, mode='same')
counts[:, 2, 2] = 0
# layer below, counts[:-1, ...] are updated from kernel[1:, ...].sum()s
counts[:-1, 1, 2] += matrix[1:, 0, :].sum(axis=1) # cell above hole += top row next level
counts[:-1, 3, 2] += matrix[1:, -1, :].sum(axis=1) # cell below hole += bottom row next level
counts[:-1, 2, 1] += matrix[1:, :, 0].sum(axis=1) # cell left of hole += left column next level
counts[:-1, 2, 3] += matrix[1:, :, -1].sum(axis=1) # cell right of hole += right column next level
# layer above, counts[1-:, ...] slices are updated from kernel[:-1, ...] indices (true -> 1)
counts[1:, 0, :] += matrix[:-1, 1, 2, None] # top row += cell above hole next level
counts[1:, -1, :] += matrix[:-1, 3, 2, None] # bottom row += cell below hole next level
counts[1:, :, 0] += matrix[:-1, 2, 1, None] # left column += cell left of hole next level
counts[1:, :, -1] += matrix[:-1, 2, 3, None] # right column += cell right of hole next level
# next step is the same as part 1:
matrix = (
# A bug dies (becoming an empty space) unless there is exactly one bug adjacent to it.
(matrix & (counts == 1)) |
# An empty space becomes infested with a bug if exactly one or two bugs are adjacent to it.
(~matrix & ((counts == 1) | (counts == 2)))
)
return matrix.sum()
assert run_multidimensional(test_matrix, 10) == 99
print("Part 2:", run_multidimensional(erismap))
# how fast is this?
%timeit run_multidimensional(erismap)
```
|
github_jupyter
|
```
"""
Please run notebook locally (if you have all the dependencies and a GPU).
Technically you can run this notebook on Google Colab but you need to set up microphone for Colab.
Instructions for setting up Colab are as follows:
1. Open a new Python 3 notebook.
2. Import this notebook from GitHub (File -> Upload Notebook -> "GITHUB" tab -> copy/paste GitHub URL)
3. Connect to an instance with a GPU (Runtime -> Change runtime type -> select "GPU" for hardware accelerator)
4. Run this cell to set up dependencies.
5. Set up microphone for Colab
"""
# If you're using Google Colab and not running locally, run this cell.
## Install dependencies
!pip install wget
!apt-get install sox libsndfile1 ffmpeg portaudio19-dev
!pip install unidecode
!pip install pyaudio
# ## Install NeMo
BRANCH = 'main'
!python -m pip install git+https://github.com/NVIDIA/NeMo.git@$BRANCH#egg=nemo_toolkit[asr]
## Install TorchAudio
!pip install torchaudio>=0.10.0 -f https://download.pytorch.org/whl/torch_stable.html
```
This notebook demonstrates offline and online (from a microphone's stream in NeMo) speech commands recognition
The notebook requires PyAudio library to get a signal from an audio device.
For Ubuntu, please run the following commands to install it:
```
sudo apt-get install -y portaudio19-dev
pip install pyaudio
```
This notebook requires the `torchaudio` library to be installed for MatchboxNet. Please follow the instructions available at the [torchaudio Github page](https://github.com/pytorch/audio#installation) to install the appropriate version of torchaudio.
If you would like to install the latest version, please run the following command to install it:
```
conda install -c pytorch torchaudio
```
```
import numpy as np
import pyaudio as pa
import os, time
import librosa
import IPython.display as ipd
import matplotlib.pyplot as plt
%matplotlib inline
import nemo
import nemo.collections.asr as nemo_asr
# sample rate, Hz
SAMPLE_RATE = 16000
```
## Restore the model from NGC
```
mbn_model = nemo_asr.models.EncDecClassificationModel.from_pretrained("commandrecognition_en_matchboxnet3x1x64_v2")
```
Since speech commands model MatchBoxNet doesn't consider non-speech scenario,
here we use a Voice Activity Detection (VAD) model to help reduce false alarm for background noise/silence. When there is speech activity detected, the speech command inference will be activated.
**Please note the VAD model is not perfect for various microphone input and you might need to finetune on your input and play with different parameters.**
```
vad_model = nemo_asr.models.EncDecClassificationModel.from_pretrained('vad_marblenet')
```
## Observing the config of the model
```
from omegaconf import OmegaConf
import copy
# Preserve a copy of the full config
vad_cfg = copy.deepcopy(vad_model._cfg)
mbn_cfg = copy.deepcopy(mbn_model._cfg)
print(OmegaConf.to_yaml(mbn_cfg))
```
## What classes can this model recognize?
Before we begin inference on the actual audio stream, let's look at what are the classes this model was trained to recognize.
**MatchBoxNet model is not designed to recognize words out of vocabulary (OOV).**
```
labels = mbn_cfg.labels
for i in range(len(labels)):
print('%-10s' % (labels[i]), end=' ')
```
## Setup preprocessor with these settings
```
# Set model to inference mode
mbn_model.eval();
vad_model.eval();
```
## Setting up data for Streaming Inference
```
from nemo.core.classes import IterableDataset
from nemo.core.neural_types import NeuralType, AudioSignal, LengthsType
import torch
from torch.utils.data import DataLoader
# simple data layer to pass audio signal
class AudioDataLayer(IterableDataset):
@property
def output_types(self):
return {
'audio_signal': NeuralType(('B', 'T'), AudioSignal(freq=self._sample_rate)),
'a_sig_length': NeuralType(tuple('B'), LengthsType()),
}
def __init__(self, sample_rate):
super().__init__()
self._sample_rate = sample_rate
self.output = True
def __iter__(self):
return self
def __next__(self):
if not self.output:
raise StopIteration
self.output = False
return torch.as_tensor(self.signal, dtype=torch.float32), \
torch.as_tensor(self.signal_shape, dtype=torch.int64)
def set_signal(self, signal):
self.signal = signal.astype(np.float32)/32768.
self.signal_shape = self.signal.size
self.output = True
def __len__(self):
return 1
data_layer = AudioDataLayer(sample_rate=mbn_cfg.train_ds.sample_rate)
data_loader = DataLoader(data_layer, batch_size=1, collate_fn=data_layer.collate_fn)
```
## inference method for audio signal (single instance)
```
def infer_signal(model, signal):
data_layer.set_signal(signal)
batch = next(iter(data_loader))
audio_signal, audio_signal_len = batch
audio_signal, audio_signal_len = audio_signal.to(model.device), audio_signal_len.to(model.device)
logits = model.forward(input_signal=audio_signal, input_signal_length=audio_signal_len)
return logits
```
we don't include postprocessing techniques here.
```
# class for streaming frame-based ASR
# 1) use reset() method to reset FrameASR's state
# 2) call transcribe(frame) to do ASR on
# contiguous signal's frames
class FrameASR:
def __init__(self, model_definition,
frame_len=2, frame_overlap=2.5,
offset=0):
'''
Args:
frame_len (seconds): Frame's duration
frame_overlap (seconds): Duration of overlaps before and after current frame.
offset: Number of symbols to drop for smooth streaming.
'''
self.task = model_definition['task']
self.vocab = list(model_definition['labels'])
self.sr = model_definition['sample_rate']
self.frame_len = frame_len
self.n_frame_len = int(frame_len * self.sr)
self.frame_overlap = frame_overlap
self.n_frame_overlap = int(frame_overlap * self.sr)
timestep_duration = model_definition['AudioToMFCCPreprocessor']['window_stride']
for block in model_definition['JasperEncoder']['jasper']:
timestep_duration *= block['stride'][0] ** block['repeat']
self.buffer = np.zeros(shape=2*self.n_frame_overlap + self.n_frame_len,
dtype=np.float32)
self.offset = offset
self.reset()
@torch.no_grad()
def _decode(self, frame, offset=0):
assert len(frame)==self.n_frame_len
self.buffer[:-self.n_frame_len] = self.buffer[self.n_frame_len:]
self.buffer[-self.n_frame_len:] = frame
if self.task == 'mbn':
logits = infer_signal(mbn_model, self.buffer).to('cpu').numpy()[0]
decoded = self._mbn_greedy_decoder(logits, self.vocab)
elif self.task == 'vad':
logits = infer_signal(vad_model, self.buffer).to('cpu').numpy()[0]
decoded = self._vad_greedy_decoder(logits, self.vocab)
else:
raise("Task should either be of mbn or vad!")
return decoded[:len(decoded)-offset]
def transcribe(self, frame=None,merge=False):
if frame is None:
frame = np.zeros(shape=self.n_frame_len, dtype=np.float32)
if len(frame) < self.n_frame_len:
frame = np.pad(frame, [0, self.n_frame_len - len(frame)], 'constant')
unmerged = self._decode(frame, self.offset)
return unmerged
def reset(self):
'''
Reset frame_history and decoder's state
'''
self.buffer=np.zeros(shape=self.buffer.shape, dtype=np.float32)
self.mbn_s = []
self.vad_s = []
@staticmethod
def _mbn_greedy_decoder(logits, vocab):
mbn_s = []
if logits.shape[0]:
class_idx = np.argmax(logits)
class_label = vocab[class_idx]
mbn_s.append(class_label)
return mbn_s
@staticmethod
def _vad_greedy_decoder(logits, vocab):
vad_s = []
if logits.shape[0]:
probs = torch.softmax(torch.as_tensor(logits), dim=-1)
probas, preds = torch.max(probs, dim=-1)
vad_s = [preds.item(), str(vocab[preds]), probs[0].item(), probs[1].item(), str(logits)]
return vad_s
```
# Streaming Inference
## offline inference
Here we show an example of offline streaming inference. you can use your file or download the provided demo audio file.
Streaming inference depends on a few factors, such as the frame length (STEP) and buffer size (WINDOW SIZE). Experiment with a few values to see their effects in the below cells.
```
STEP = 0.25
WINDOW_SIZE = 1.28 # input segment length for NN we used for training
import wave
def offline_inference(wave_file, STEP = 0.25, WINDOW_SIZE = 0.31):
"""
Arg:
wav_file: wave file to be performed inference on.
STEP: infer every STEP seconds
WINDOW_SIZE : lenght of audio to be sent to NN.
"""
FRAME_LEN = STEP
CHANNELS = 1 # number of audio channels (expect mono signal)
RATE = SAMPLE_RATE # sample rate, 16000 Hz
CHUNK_SIZE = int(FRAME_LEN * SAMPLE_RATE)
mbn = FrameASR(model_definition = {
'task': 'mbn',
'sample_rate': SAMPLE_RATE,
'AudioToMFCCPreprocessor': mbn_cfg.preprocessor,
'JasperEncoder': mbn_cfg.encoder,
'labels': mbn_cfg.labels
},
frame_len=FRAME_LEN, frame_overlap = (WINDOW_SIZE - FRAME_LEN)/2,
offset=0)
wf = wave.open(wave_file, 'rb')
data = wf.readframes(CHUNK_SIZE)
while len(data) > 0:
data = wf.readframes(CHUNK_SIZE)
signal = np.frombuffer(data, dtype=np.int16)
mbn_result = mbn.transcribe(signal)
if len(mbn_result):
print(mbn_result)
mbn.reset()
demo_wave = 'SpeechCommands_demo.wav'
if not os.path.exists(demo_wave):
!wget "https://dldata-public.s3.us-east-2.amazonaws.com/SpeechCommands_demo.wav"
wave_file = demo_wave
CHANNELS = 1
audio, sample_rate = librosa.load(wave_file, sr=SAMPLE_RATE)
dur = librosa.get_duration(audio)
print(dur)
ipd.Audio(audio, rate=sample_rate)
# Ground-truth is Yes No
offline_inference(wave_file, STEP, WINDOW_SIZE)
```
## Online inference through microphone
Please note MatchBoxNet and VAD model are not perfect for various microphone input and you might need to finetune on your input and play with different parameter. \
**We also recommend to use a headphone.**
```
vad_threshold = 0.8
STEP = 0.1
WINDOW_SIZE = 0.15
mbn_WINDOW_SIZE = 1
CHANNELS = 1
RATE = SAMPLE_RATE
FRAME_LEN = STEP # use step of vad inference as frame len
CHUNK_SIZE = int(STEP * RATE)
vad = FrameASR(model_definition = {
'task': 'vad',
'sample_rate': SAMPLE_RATE,
'AudioToMFCCPreprocessor': vad_cfg.preprocessor,
'JasperEncoder': vad_cfg.encoder,
'labels': vad_cfg.labels
},
frame_len=FRAME_LEN, frame_overlap=(WINDOW_SIZE - FRAME_LEN) / 2,
offset=0)
mbn = FrameASR(model_definition = {
'task': 'mbn',
'sample_rate': SAMPLE_RATE,
'AudioToMFCCPreprocessor': mbn_cfg.preprocessor,
'JasperEncoder': mbn_cfg.encoder,
'labels': mbn_cfg.labels
},
frame_len=FRAME_LEN, frame_overlap = (mbn_WINDOW_SIZE-FRAME_LEN)/2,
offset=0)
vad.reset()
mbn.reset()
# Setup input device
p = pa.PyAudio()
print('Available audio input devices:')
input_devices = []
for i in range(p.get_device_count()):
dev = p.get_device_info_by_index(i)
if dev.get('maxInputChannels'):
input_devices.append(i)
print(i, dev.get('name'))
if len(input_devices):
dev_idx = -2
while dev_idx not in input_devices:
print('Please type input device ID:')
dev_idx = int(input())
def callback(in_data, frame_count, time_info, status):
"""
callback function for streaming audio and performing inference
"""
signal = np.frombuffer(in_data, dtype=np.int16)
vad_result = vad.transcribe(signal)
mbn_result = mbn.transcribe(signal)
if len(vad_result):
# if speech prob is higher than threshold, we decide it contains speech utterance
# and activate MatchBoxNet
if vad_result[3] >= vad_threshold:
print(mbn_result) # print mbn result when speech present
else:
print("no-speech")
return (in_data, pa.paContinue)
# streaming
stream = p.open(format=pa.paInt16,
channels=CHANNELS,
rate=SAMPLE_RATE,
input=True,
input_device_index=dev_idx,
stream_callback=callback,
frames_per_buffer=CHUNK_SIZE)
print('Listening...')
stream.start_stream()
# Interrupt kernel and then speak for a few more words to exit the pyaudio loop !
try:
while stream.is_active():
time.sleep(0.1)
finally:
stream.stop_stream()
stream.close()
p.terminate()
print()
print("PyAudio stopped")
else:
print('ERROR: No audio input device found.')
```
## ONNX Deployment
You can also export the model to ONNX file and deploy it to TensorRT or MS ONNX Runtime inference engines. If you don't have one installed yet, please run:
```
!pip install --upgrade onnxruntime # for gpu, use onnxruntime-gpu
# !mkdir -p ort
# %cd ort
# !git clone --depth 1 --branch v1.8.0 https://github.com/microsoft/onnxruntime.git .
# !./build.sh --skip_tests --config Release --build_shared_lib --parallel --use_cuda --cuda_home /usr/local/cuda --cudnn_home /usr/lib/x86_64-linux-gnu --build_wheel
# !pip install ./build/Linux/Release/dist/onnxruntime*.whl
# %cd ..
```
Then just replace `infer_signal` implementation with this code:
```
import onnxruntime
mbn_model.export('mbn.onnx')
ort_session = onnxruntime.InferenceSession('mbn.onnx')
def to_numpy(tensor):
return tensor.detach().cpu().numpy() if tensor.requires_grad else tensor.cpu().numpy()
def infer_signal(signal):
data_layer.set_signal(signal)
batch = next(iter(data_loader))
audio_signal, audio_signal_len = batch
audio_signal, audio_signal_len = audio_signal.to(mbn_model.device), audio_signal_len.to(mbn_model.device)
processed_signal, processed_signal_len = mbn_model.preprocessor(
input_signal=audio_signal, length=audio_signal_len,
)
ort_inputs = {ort_session.get_inputs()[0].name: to_numpy(processed_signal), }
ologits = ort_session.run(None, ort_inputs)
alogits = np.asarray(ologits)
logits = torch.from_numpy(alogits[0])
return logits
```
|
github_jupyter
|
# Quantum Counting
To understand this algorithm, it is important that you first understand both Grover’s algorithm and the quantum phase estimation algorithm. Whereas Grover’s algorithm attempts to find a solution to the Oracle, the quantum counting algorithm tells us how many of these solutions there are. This algorithm is interesting as it combines both quantum search and quantum phase estimation.
## Contents
1. [Overview](#overview)
1.1 [Intuition](#intuition)
1.2 [A Closer Look](#closer_look)
2. [The Code](#code)
2.1 [Initialising our Code](#init_code)
2.2 [The Controlled-Grover Iteration](#cont_grover)
2.3 [The Inverse QFT](#inv_qft)
2.4 [Putting it Together](#putting_together)
3. [Simulating](#simulating)
4. [Finding the Number of Solutions](#finding_m)
5. [Exercises](#exercises)
6. [References](#references)
## 1. Overview <a id='overview'></a>
### 1.1 Intuition <a id='intuition'></a>
In quantum counting, we simply use the quantum phase estimation algorithm to find an eigenvalue of a Grover search iteration. You will remember that an iteration of Grover’s algorithm, $G$, rotates the state vector by $\theta$ in the $|\omega\rangle$, $|s’\rangle$ basis:

The percentage number of solutions in our search space affects the difference between $|s\rangle$ and $|s’\rangle$. For example, if there are not many solutions, $|s\rangle$ will be very close to $|s’\rangle$ and $\theta$ will be very small. It turns out that the eigenvalues of the Grover iterator are $e^{\pm i\theta}$, and we can extract this using quantum phase estimation (QPE) to estimate the number of solutions ($M$).
### 1.2 A Closer Look <a id='closer_look'></a>
In the $|\omega\rangle$,$|s’\rangle$ basis we can write the Grover iterator as the matrix:
$$
G =
\begin{pmatrix}
\cos{\theta} && -\sin{\theta}\\
\sin{\theta} && \cos{\theta}
\end{pmatrix}
$$
The matrix $G$ has eigenvectors:
$$
\begin{pmatrix}
-i\\
1
\end{pmatrix}
,
\begin{pmatrix}
i\\
1
\end{pmatrix}
$$
With the aforementioned eigenvalues $e^{\pm i\theta}$. Fortunately, we do not need to prepare our register in either of these states, the state $|s\rangle$ is in the space spanned by $|\omega\rangle$, $|s’\rangle$, and thus is a superposition of the two vectors.
$$
|s\rangle = \alpha |\omega\rangle + \beta|s'\rangle
$$
As a result, the output of the QPE algorithm will be a superposition of the two phases, and when we measure the register we will obtain one of these two values! We can then use some simple maths to get our estimate of $M$.

## 2. The Code <a id='code'></a>
### 2.1 Initialising our Code <a id='init_code'></a>
First, let’s import everything we’re going to need:
```
import matplotlib.pyplot as plt
import numpy as np
import math
# importing Qiskit
import qiskit
from qiskit import IBMQ, Aer
from qiskit import QuantumCircuit, execute
# import basic plot tools
from qiskit.visualization import plot_histogram
```
In this guide will choose to ‘count’ on the first 4 qubits on our circuit (we call the number of counting qubits $t$, so $t = 4$), and to 'search' through the last 4 qubits ($n = 4$). With this in mind, we can start creating the building blocks of our circuit.
### 2.2 The Controlled-Grover Iteration <a id='cont_grover'></a>
We have already covered Grover iterations in the Grover’s algorithm section. Here is an example with an Oracle we know has 5 solutions ($M = 5$) of 16 states ($N = 2^n = 16$), combined with a diffusion operator:
```
def example_grover_iteration():
"""Small circuit with 5/16 solutions"""
# Do circuit
qc = QuantumCircuit(4)
# Oracle
qc.h([2,3])
qc.ccx(0,1,2)
qc.h(2)
qc.x(2)
qc.ccx(0,2,3)
qc.x(2)
qc.h(3)
qc.x([1,3])
qc.h(2)
qc.mct([0,1,3],2)
qc.x([1,3])
qc.h(2)
# Diffuser
qc.h(range(3))
qc.x(range(3))
qc.z(3)
qc.mct([0,1,2],3)
qc.x(range(3))
qc.h(range(3))
qc.z(3)
return qc
```
Notice the python function takes no input and returns a `QuantumCircuit` object with 4 qubits. In the past the functions you created might have modified an existing circuit, but a function like this allows us to turn the `QuantmCircuit` object into a single gate we can then control.
We can use `.to_gate()` and `.control()` to create a controlled gate from a circuit. We will call our Grover iterator `grit` and the controlled Grover iterator `cgrit`:
```
# Create controlled-Grover
grit = example_grover_iteration().to_gate()
cgrit = grit.control()
cgrit.label = "Grover"
```
### 2.3 The Inverse QFT <a id='inv_qft'></a>
We now need to create an inverse QFT. This code implements the QFT on n qubits:
```
def qft(n):
"""Creates an n-qubit QFT circuit"""
circuit = QuantumCircuit(4)
def swap_registers(circuit, n):
for qubit in range(n//2):
circuit.swap(qubit, n-qubit-1)
return circuit
def qft_rotations(circuit, n):
"""Performs qft on the first n qubits in circuit (without swaps)"""
if n == 0:
return circuit
n -= 1
circuit.h(n)
for qubit in range(n):
circuit.cu1(np.pi/2**(n-qubit), qubit, n)
qft_rotations(circuit, n)
qft_rotations(circuit, n)
swap_registers(circuit, n)
return circuit
```
Again, note we have chosen to return another `QuantumCircuit` object, this is so we can easily invert the gate. We create the gate with t = 4 qubits as this is the number of counting qubits we have chosen in this guide:
```
qft_dagger = qft(4).to_gate().inverse()
qft_dagger.label = "QFT†"
```
### 2.4 Putting it Together <a id='putting_together'></a>
We now have everything we need to complete our circuit! Let’s put it together.
First we need to put all qubits in the $|+\rangle$ state:
```
# Create QuantumCircuit
t = 4 # no. of counting qubits
n = 4 # no. of searching qubits
qc = QuantumCircuit(n+t, t) # Circuit with n+t qubits and t classical bits
# Initialise all qubits to |+>
for qubit in range(t+n):
qc.h(qubit)
# Begin controlled Grover iterations
iterations = 1
for qubit in range(t):
for i in range(iterations):
qc.append(cgrit, [qubit] + [*range(t, n+t)])
iterations *= 2
# Do inverse QFT on counting qubits
qc.append(qft_dagger, range(t))
# Measure counting qubits
qc.measure(range(t), range(t))
# Display the circuit
qc.draw()
```
Great! Now let’s see some results.
## 3. Simulating <a id='simulating'></a>
```
# Execute and see results
emulator = Aer.get_backend('qasm_simulator')
job = execute(qc, emulator, shots=2048 )
hist = job.result().get_counts()
plot_histogram(hist)
```
We can see two values stand out, having a much higher probability of measurement than the rest. These two values correspond to $e^{i\theta}$ and $e^{-i\theta}$, but we can’t see the number of solutions yet. We need to little more processing to get this information, so first let us get our output into something we can work with (an `int`).
We will get the string of the most probable result from our output data:
```
measured_str = max(hist, key=hist.get)
```
Let us now store this as an integer:
```
measured_int = int(measured_str,2)
print("Register Output = %i" % measured_int)
```
## 4. Finding the Number of Solutions (M) <a id='finding_m'></a>
We will create a function, `calculate_M()` that takes as input the decimal integer output of our register, the number of counting qubits ($t$) and the number of searching qubits ($n$).
First we want to get $\theta$ from `measured_int`. You will remember that QPE gives us a measured $\text{value} = 2^n \phi$ from the eigenvalue $e^{2\pi i\phi}$, so to get $\theta$ we need to do:
$$
\theta = \text{value}\times\frac{2\pi}{2^t}
$$
Or, in code:
```
theta = (measured_int/(2**t))*math.pi*2
print("Theta = %.5f" % theta)
```
You may remember that we can get the angle $\theta/2$ can from the inner product of $|s\rangle$ and $|s’\rangle$:

$$
\langle s'|s\rangle = \cos{\tfrac{\theta}{2}}
$$
And that the inner product of these vectors is:
$$
\langle s'|s\rangle = \sqrt{\frac{N-M}{N}}
$$
We can combine these equations, then use some trigonometry and algebra to show:
$$
N\sin^2{\frac{\theta}{2}} = M
$$
From the [Grover's algorithm](https://qiskit.org/textbook/ch-algorithms/grover.html) chapter, you will remember that a common way to create a diffusion operator, $U_s$, is actually to implement $-U_s$. This implementation is used in the Grover iteration provided in this chapter. In a normal Grover search, this phase is global and can be ignored, but now we are controlling our Grover iterations, this phase does have an effect. The result is that we have effectively searched for the states that are _not_ solutions, and our quantum counting algorithm will tell us how many states are _not_ solutions. To fix this, we simply calculate $N-M$.
And in code:
```
N = 2**n
M = N * (math.sin(theta/2)**2)
print("No. of Solutions = %.1f" % (N-M))
```
And we can see we have (approximately) the correct answer! We can approximately calculate the error in this answer using:
```
m = t - 1 # Upper bound: Will be less than this
err = (math.sqrt(2*M*N) + N/(2**(m-1)))*(2**(-m))
print("Error < %.2f" % err)
```
Explaining the error calculation is outside the scope of this article, but an explanation can be found in [1].
Finally, here is the finished function `calculate_M()`:
```
def calculate_M(measured_int, t, n):
"""For Processing Output of Quantum Counting"""
# Calculate Theta
theta = (measured_int/(2**t))*math.pi*2
print("Theta = %.5f" % theta)
# Calculate No. of Solutions
N = 2**n
M = N * (math.sin(theta/2)**2)
print("No. of Solutions = %.1f" % (N-M))
# Calculate Upper Error Bound
m = t - 1 #Will be less than this (out of scope)
err = (math.sqrt(2*M*N) + N/(2**(m-1)))*(2**(-m))
print("Error < %.2f" % err)
```
## 5. Exercises <a id='exercises'></a>
1. Can you create an oracle with a different number of solutions? How does the accuracy of the quantum counting algorithm change?
2. Can you adapt the circuit to use more or less counting qubits to get a different precision in your result?
## 6. References <a id='references'></a>
[1] Michael A. Nielsen and Isaac L. Chuang. 2011. Quantum Computation and Quantum Information: 10th Anniversary Edition (10th ed.). Cambridge University Press, New York, NY, USA.
```
import qiskit
qiskit.__qiskit_version__
```
|
github_jupyter
|
# CORD-19 overview
In this notebook, we provide an overview of publication medatata for CORD-19.
```
%matplotlib inline
import matplotlib.pyplot as plt
# magics and warnings
%load_ext autoreload
%autoreload 2
import warnings; warnings.simplefilter('ignore')
import os, random, codecs, json
import pandas as pd
import numpy as np
seed = 99
random.seed(seed)
np.random.seed(seed)
import nltk, sklearn
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(style="white")
sns.set_context("notebook", font_scale=1.2, rc={"lines.linewidth": 2.5})
# load metadata
df_meta = pd.read_csv("datasets_output/df_pub.csv",compression="gzip")
df_datasource = pd.read_csv("datasets_output/sql_tables/datasource.csv",sep="\t",header=None,names=['datasource_metadata_id', 'datasource', 'url'])
df_pub_datasource = pd.read_csv("datasets_output/sql_tables/pub_datasource.csv",sep="\t",header=None,names=['pub_id','datasource_metadata_id'])
df_cord_meta = pd.read_csv("datasets_output/sql_tables/cord19_metadata.csv",sep="\t",header=None,names=[ 'cord19_metadata_id', 'source', 'license', 'ms_academic_id',
'who_covidence', 'sha', 'full_text', 'pub_id'])
df_meta.head()
df_meta.columns
df_datasource
df_pub_datasource.head()
df_cord_meta.head()
```
#### Select just CORD-19
```
df_meta = df_meta.merge(df_pub_datasource, how="inner", left_on="pub_id", right_on="pub_id")
df_meta = df_meta.merge(df_datasource, how="inner", left_on="datasource_metadata_id", right_on="datasource_metadata_id")
df_cord19 = df_meta[df_meta.datasource_metadata_id==0]
df_cord19 = df_cord19.merge(df_cord_meta, how="inner", left_on="pub_id", right_on="pub_id")
df_meta.shape
df_cord19.shape
df_cord19.head()
```
#### Publication years
```
import re
def clean_year(s):
if pd.isna(s):
return np.nan
if not (s>1900):
return np.nan
elif s>2020:
return 2020
return s
df_cord19["publication_year"] = df_cord19["publication_year"].apply(clean_year)
df_cord19.publication_year.describe()
sns.distplot(df_cord19.publication_year.tolist(), bins=60, kde=False)
plt.xlabel("Publication year", fontsize=15)
plt.ylabel("Publication count", fontsize=15)
plt.tight_layout()
plt.savefig("figures/publication_year_all.pdf")
sns.distplot(df_cord19[(pd.notnull(df_cord19.publication_year)) & (df_cord19.publication_year > 2000)].publication_year.tolist(), bins=20, hist=True, kde=False)
plt.xlabel("Publication year", fontsize=15)
plt.ylabel("Publication count", fontsize=15)
plt.tight_layout()
plt.savefig("figures/publication_year_2000.pdf")
which = "PMC"
sns.distplot(df_cord19[(pd.notnull(df_cord19.publication_year)) & (df_cord19.publication_year > 2000) & (df_cord19.source == which)].publication_year.tolist(), bins=20, hist=True, kde=False)
plt.xlabel("Publication year", fontsize=15)
plt.ylabel("Publication count", fontsize=15)
plt.tight_layout()
# recent uptake
df_cord19[df_cord19.publication_year>2018].groupby([(df_cord19.publication_year),(df_cord19.publication_month)]).count().pub_id
```
#### Null values
```
df_cord19.shape
df_cord19["abstract_length"] = df_cord19.abstract.str.len()
df_cord19[df_cord19.abstract_length>0].shape
sum(pd.notnull(df_cord19.abstract))
sum(pd.notnull(df_cord19.doi))
sum(pd.notnull(df_cord19.pmcid))
sum(pd.notnull(df_cord19.pmid))
sum(pd.notnull(df_cord19.journal))
```
#### Journals
```
df_cord19.journal.value_counts()[:30]
df_sub = df_cord19[df_cord19.journal.isin(df_cord19.journal.value_counts()[:20].index.tolist())]
b = sns.countplot(y="journal", data=df_sub, order=df_sub['journal'].value_counts().index)
#b.axes.set_title("Title",fontsize=50)
b.set_xlabel("Publication count",fontsize=15)
b.set_ylabel("Journal",fontsize=15)
b.tick_params(labelsize=12)
plt.tight_layout()
plt.savefig("figures/journals.pdf")
```
#### Sources and licenses
```
# source
df_sub = df_cord19[df_cord19.source.isin(df_cord19.source.value_counts()[:10].index.tolist())]
b = sns.countplot(y="source", data=df_sub, order=df_sub['source'].value_counts().index)
#b.axes.set_title("Title",fontsize=50)
b.set_xlabel("Publication count",fontsize=15)
b.set_ylabel("Source",fontsize=15)
b.tick_params(labelsize=12)
plt.tight_layout()
plt.savefig("figures/sources.pdf")
# license
df_sub = df_cord19[df_cord19.license.isin(df_cord19.license.value_counts()[:30].index.tolist())]
b = sns.countplot(y="license", data=df_sub, order=df_sub['license'].value_counts().index)
#b.axes.set_title("Title",fontsize=50)
b.set_xlabel("Publication count",fontsize=15)
b.set_ylabel("License",fontsize=15)
b.tick_params(labelsize=12)
plt.tight_layout()
plt.savefig("figures/licenses.pdf")
```
#### Full text availability
```
df_cord19["has_full_text"] = pd.notnull(df_cord19.full_text)
df_cord19["has_full_text"].sum()
# full text x source
df_plot = df_cord19.groupby(['has_full_text', 'source']).size().reset_index().pivot(columns='has_full_text', index='source', values=0)
df_plot.plot(kind='bar', stacked=True)
plt.xlabel("Source", fontsize=15)
plt.ylabel("Publication count", fontsize=15)
#plt.tight_layout()
plt.savefig("figures/source_ft.pdf")
# full text x journal
df_sub = df_cord19[df_cord19.journal.isin(df_cord19.journal.value_counts()[:20].index.tolist())]
df_plot = df_sub.groupby(['has_full_text', 'journal']).size().reset_index().pivot(columns='has_full_text', index='journal', values=0)
df_plot.plot(kind='bar', stacked=True)
plt.xlabel("Source", fontsize=15)
plt.ylabel("Publication count", fontsize=15)
#plt.tight_layout()
plt.savefig("figures/journal_ft.pdf")
# full text x year
df_sub = df_cord19[(pd.notnull(df_cord19.publication_year)) & (df_cord19.publication_year > 2000)]
df_plot = df_sub.groupby(['has_full_text', 'publication_year']).size().reset_index().pivot(columns='has_full_text', index='publication_year', values=0)
df_plot.plot(kind='bar', stacked=True)
plt.xticks(np.arange(20), [int(x) for x in df_plot.index.values], rotation=45)
plt.xlabel("Publication year", fontsize=15)
plt.ylabel("Publication count", fontsize=15)
plt.tight_layout()
plt.savefig("figures/year_ft.pdf")
```
## Dimensions
```
# load Dimensions data (you will need to download it on your own!)
directory_name = "datasets_output/json_dimensions_cwts"
all_dimensions = list()
for root, dirs, files in os.walk(directory_name):
for file in files:
if ".json" in file:
all_data = codecs.open(os.path.join(root,file)).read()
for record in all_data.split("\n"):
if record:
all_dimensions.append(json.loads(record))
df_dimensions = pd.DataFrame.from_dict({
"id":[r["id"] for r in all_dimensions],
"publication_type":[r["publication_type"] for r in all_dimensions],
"doi":[r["doi"] for r in all_dimensions],
"pmid":[r["pmid"] for r in all_dimensions],
"issn":[r["journal"]["issn"] for r in all_dimensions],
"times_cited":[r["times_cited"] for r in all_dimensions],
"relative_citation_ratio":[r["relative_citation_ratio"] for r in all_dimensions],
"for_top":[r["for"][0]["first_level"]["name"] if len(r["for"])>0 else "" for r in all_dimensions],
"for_bottom":[r["for"][0]["second_level"]["name"] if len(r["for"])>0 else "" for r in all_dimensions],
"open_access_versions":[r["open_access_versions"] for r in all_dimensions]
})
df_dimensions.head()
df_dimensions.pmid = df_dimensions.pmid.astype(float)
df_dimensions.shape
df_joined_doi = df_cord19[pd.notnull(df_cord19.doi)].merge(df_dimensions[pd.notnull(df_dimensions.doi)], how="inner", left_on="doi", right_on="doi")
df_joined_doi.shape
df_joined_pmid = df_cord19[pd.isnull(df_cord19.doi) & pd.notnull(df_cord19.pmid)].merge(df_dimensions[pd.isnull(df_dimensions.doi) & pd.notnull(df_dimensions.pmid)], how="inner", left_on="pmid", right_on="pmid")
df_joined_pmid.shape
df_joined = pd.concat([df_joined_doi,df_joined_pmid])
# nearly all publications from CORD-19 are in Dimensions
df_joined.shape
df_cord19.shape
# publication type
df_sub = df_joined[df_joined.publication_type.isin(df_joined.publication_type.value_counts()[:10].index.tolist())]
b = sns.countplot(y="publication_type", data=df_sub, order=df_sub['publication_type'].value_counts().index)
#b.axes.set_title("Title",fontsize=50)
b.set_xlabel("Publication count",fontsize=15)
b.set_ylabel("Publication type",fontsize=15)
b.tick_params(labelsize=12)
plt.tight_layout()
plt.savefig("figures/dim_pub_type.pdf")
```
#### Citation counts
```
# scatter of citations vs time of publication
sns.scatterplot(df_joined.publication_year.to_list(),df_joined.times_cited.to_list())
plt.xlabel("Publication year", fontsize=15)
plt.ylabel("Citation count", fontsize=15)
plt.tight_layout()
plt.savefig("figures/dim_citations_year.png")
# most cited papers
df_joined[["title","times_cited","relative_citation_ratio","journal","publication_year","doi"]].sort_values("times_cited",ascending=False).head(20)
# same but in 2020; note that duplicates are due to SI or pre-prints with different PMIDs
df_joined[df_joined.publication_year>2019][["title","times_cited","relative_citation_ratio","journal","publication_year","doi"]].sort_values("times_cited",ascending=False).head(10)
# most cited journals
df_joined[['journal','times_cited']].groupby('journal').sum().sort_values('times_cited',ascending=False).head(20)
```
#### Categories
```
# FOR jeywords distribution, TOP
df_sub = df_joined[df_joined.for_top.isin(df_joined.for_top.value_counts()[:10].index.tolist())]
b = sns.countplot(y="for_top", data=df_sub, order=df_sub['for_top'].value_counts().index)
#b.axes.set_title("Title",fontsize=50)
b.set_xlabel("Publication count",fontsize=15)
b.set_ylabel("FOR first level",fontsize=15)
b.tick_params(labelsize=12)
plt.tight_layout()
plt.savefig("figures/dim_for_top.pdf")
# FOR jeywords distribution, TOP
df_sub = df_joined[df_joined.for_bottom.isin(df_joined.for_bottom.value_counts()[:10].index.tolist())]
b = sns.countplot(y="for_bottom", data=df_sub, order=df_sub['for_bottom'].value_counts().index)
#b.axes.set_title("Title",fontsize=50)
b.set_xlabel("Publication count",fontsize=15)
b.set_ylabel("FOR second level",fontsize=15)
b.tick_params(labelsize=12)
plt.tight_layout()
plt.savefig("figures/dim_for_bottom.pdf")
```
|
github_jupyter
|
### 2.2 CNN Models - Test Cases
The trained CNN model was performed to a hold-out test set with 10,873 images.
The network obtained 0.743 and 0.997 AUC-PRC on the hold-out test set for cored plaque and diffuse plaque respectively.
```
import time, os
import torch
torch.manual_seed(42)
from torch.autograd import Variable
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from torch.optim import lr_scheduler
import torchvision
from torchvision import transforms
from matplotlib import pyplot as plt
import numpy as np
import pandas as pd
CSV_DIR = 'data/CSVs/test.csv'
MODEL_DIR = 'models/CNN_model_parameters.pkl'
IMG_DIR = 'data/tiles/hold-out/'
NEGATIVE_DIR = 'data/seg/negatives/'
SAVE_DIR = 'data/outputs/'
if not os.path.exists(SAVE_DIR):
os.makedirs(SAVE_DIR)
batch_size = 32
num_workers = 8
norm = np.load('utils/normalization.npy', allow_pickle=True).item()
from torch.utils.data import Dataset
from PIL import Image
class MultilabelDataset(Dataset):
def __init__(self, csv_path, img_path, transform=None):
"""
Args:
csv_path (string): path to csv file
img_path (string): path to the folder where images are
transform: pytorch transforms for transforms and tensor conversion
"""
self.data_info = pd.read_csv(csv_path)
self.img_path = img_path
self.transform = transform
c=torch.Tensor(self.data_info.loc[:,'cored'])
d=torch.Tensor(self.data_info.loc[:,'diffuse'])
a=torch.Tensor(self.data_info.loc[:,'CAA'])
c=c.view(c.shape[0],1)
d=d.view(d.shape[0],1)
a=a.view(a.shape[0],1)
self.raw_labels = torch.cat([c,d,a], dim=1)
self.labels = (torch.cat([c,d,a], dim=1)>0.99).type(torch.FloatTensor)
def __getitem__(self, index):
# Get label(class) of the image based on the cropped pandas column
single_image_label = self.labels[index]
raw_label = self.raw_labels[index]
# Get image name from the pandas df
single_image_name = str(self.data_info.loc[index,'imagename'])
# Open image
try:
img_as_img = Image.open(self.img_path + single_image_name)
except:
img_as_img = Image.open(NEGATIVE_DIR + single_image_name)
# Transform image to tensor
if self.transform is not None:
img_as_img = self.transform(img_as_img)
# Return image and the label
return (img_as_img, single_image_label, raw_label, single_image_name)
def __len__(self):
return len(self.data_info.index)
data_transforms = {
'test' : transforms.Compose([
transforms.ToTensor(),
transforms.Normalize(norm['mean'], norm['std'])
])
}
image_datasets = {'test': MultilabelDataset(CSV_DIR, IMG_DIR,
data_transforms['test'])}
dataloaders = {x: torch.utils.data.DataLoader(image_datasets[x],
batch_size=batch_size,
shuffle=False,
num_workers=num_workers)
for x in ['test']}
dataset_sizes = {x: len(image_datasets[x]) for x in ['test']}
image_classes = ['cored','diffuse','CAA']
use_gpu = torch.cuda.is_available()
def imshow(inp, title=None):
"""Imshow for Tensor."""
inp = inp.numpy().transpose((1, 2, 0))
mean = np.array(norm['mean'])
std = np.array(norm['std'])
inp = std * inp + mean
inp = np.clip(inp, 0, 1)
plt.figure()
plt.imshow(inp)
if title is not None:
plt.title(title)
plt.pause(0.001) # pause a bit so that plots are updated
# Get a batch of training data
inputs, labels, raw_labels, names = next(iter(dataloaders['test']))
# Make a grid from batch
out = torchvision.utils.make_grid(inputs)
imshow(out)
class Net(nn.Module):
def __init__(self, fc_nodes=512, num_classes=3, dropout=0.5):
super(Net, self).__init__()
def forward(self, x):
x = self.features(x)
x = x.view(x.size(0), -1)
x = self.classifier(x)
return x
def dev_model(model, criterion, phase='test', gpu_id=None):
phase = phase
since = time.time()
dataloaders = {x: torch.utils.data.DataLoader(image_datasets[x], batch_size=batch_size,
shuffle=False, num_workers=num_workers)
for x in [phase]}
model.train(False)
running_loss = 0.0
running_corrects = torch.zeros(len(image_classes))
running_preds = torch.Tensor(0)
running_predictions = torch.Tensor(0)
running_labels = torch.Tensor(0)
running_raw_labels = torch.Tensor(0)
# Iterate over data.
step = 0
for data in dataloaders[phase]:
step += 1
# get the inputs
inputs, labels, raw_labels, names = data
running_labels = torch.cat([running_labels, labels])
running_raw_labels = torch.cat([running_raw_labels, raw_labels])
# wrap them in Variable
if use_gpu:
inputs = Variable(inputs.cuda(gpu_id))
labels = Variable(labels.cuda(gpu_id))
else:
inputs, labels = Variable(inputs), Variable(labels)
# forward
outputs = model(inputs)
preds = F.sigmoid(outputs) #posibility for each class
#print(preds)
if use_gpu:
predictions = (preds>0.5).type(torch.cuda.FloatTensor)
else:
predictions = (preds>0.5).type(torch.FloatTensor)
loss = criterion(outputs, labels)
preds = preds.data.cpu()
predictions = predictions.data.cpu()
labels = labels.data.cpu()
# statistics
running_loss += loss.data[0]
running_corrects += torch.sum(predictions==labels, 0).type(torch.FloatTensor)
running_preds = torch.cat([running_preds, preds])
running_predictions = torch.cat([running_predictions, predictions])
epoch_loss = running_loss / dataset_sizes[phase]
epoch_acc = running_corrects / dataset_sizes[phase]
print('{} Loss: {:.4f}\n Cored: {:.4f} Diffuse: {:.4f} CAA: {:.4f}'.format(
phase, epoch_loss, epoch_acc[0], epoch_acc[1], epoch_acc[2]))
print()
time_elapsed = time.time() - since
print('Prediction complete in {:.0f}m {:.0f}s'.format(
time_elapsed // 60, time_elapsed % 60))
return epoch_acc, running_preds, running_predictions, running_labels
from sklearn.metrics import roc_curve, auc, precision_recall_curve
def plot_roc(preds, label, image_classes, size=20, path=None):
colors = ['pink','c','deeppink', 'b', 'g', 'm', 'y', 'r', 'k']
fig = plt.figure(figsize=(1.2*size, size))
ax = plt.axes()
for i in range(preds.shape[1]):
fpr, tpr, _ = roc_curve(label[:,i].ravel(), preds[:,i].ravel())
lw = 0.2*size
# Plot all ROC curves
ax.plot([0, 1], [0, 1], 'k--', lw=lw, label='random')
ax.plot(fpr, tpr,
label='ROC-curve of {}'.format(image_classes[i])+ '( area = {0:0.3f})'
''.format(auc(fpr, tpr)),
color=colors[(i+preds.shape[1])%len(colors)], linewidth=lw)
ax.set_xlim([0.0, 1.0])
ax.set_ylim([0.0, 1.05])
ax.set_xlabel('False Positive Rate', fontsize=1.8*size)
ax.set_ylabel('True Positive Rate', fontsize=1.8*size)
ax.set_title('Receiver operating characteristic Curve', fontsize=1.8*size, y=1.01)
ax.legend(loc=0, fontsize=1.5*size)
ax.xaxis.set_tick_params(labelsize=1.6*size, size=size/2, width=0.2*size)
ax.yaxis.set_tick_params(labelsize=1.6*size, size=size/2, width=0.2*size)
if path != None:
fig.savefig(path)
# plt.close(fig)
print('saved')
def plot_prc(preds, label, image_classes, size=20, path=None):
colors = ['pink','c','deeppink', 'b', 'g', 'm', 'y', 'r', 'k']
fig = plt.figure(figsize=(1.2*size,size))
ax = plt.axes()
for i in range(preds.shape[1]):
rp = (label[:,i]>0).sum()/len(label)
precision, recall, _ = precision_recall_curve(label[:,i].ravel(), preds[:,i].ravel())
lw=0.2*size
ax.plot(recall, precision,
label='PR-curve of {}'.format(image_classes[i])+ '( area = {0:0.3f})'
''.format(auc(recall, precision)),
color=colors[(i+preds.shape[1])%len(colors)], linewidth=lw)
ax.plot([0, 1], [rp, rp], 'k--', color=colors[(i+preds.shape[1])%len(colors)], lw=lw, label='random')
ax.set_xlim([0.0, 1.0])
ax.set_ylim([0.0, 1.05])
ax.set_xlabel('Recall', fontsize=1.8*size)
ax.set_ylabel('Precision', fontsize=1.8*size)
ax.set_title('Precision-Recall curve', fontsize=1.8*size, y=1.01)
ax.legend(loc="lower left", bbox_to_anchor=(0.01, 0.1), fontsize=1.5*size)
ax.xaxis.set_tick_params(labelsize=1.6*size, size=size/2, width=0.2*size)
ax.yaxis.set_tick_params(labelsize=1.6*size, size=size/2, width=0.2*size)
if path != None:
fig.savefig(path)
# plt.close(fig)
print('saved')
def auc_roc(preds, label):
aucroc = []
for i in range(preds.shape[1]):
fpr, tpr, _ = roc_curve(label[:,i].ravel(), preds[:,i].ravel())
aucroc.append(auc(fpr, tpr))
return aucroc
def auc_prc(preds, label):
aucprc = []
for i in range(preds.shape[1]):
precision, recall, _ = precision_recall_curve(label[:,i].ravel(), preds[:,i].ravel())
aucprc.append(auc(recall, precision))
return aucprc
criterion = nn.MultiLabelSoftMarginLoss(size_average=False)
model = torch.load(MODEL_DIR, map_location=lambda storage, loc: storage)
if use_gpu:
model = model.module.cuda()
# take 10s running on single GPU
try:
acc, pred, prediction, target = dev_model(model.module, criterion, phase='test', gpu_id=None)
except:
acc, pred, prediction, target = dev_model(model, criterion, phase='test', gpu_id=None)
label = target.numpy()
preds = pred.numpy()
output = {}
for i in range(3):
fpr, tpr, _ = roc_curve(label[:,i].ravel(), preds[:,i].ravel())
precision, recall, _ = precision_recall_curve(label[:,i].ravel(), preds[:,i].ravel())
output['{} fpr'.format(image_classes[i])] = fpr
output['{} tpr'.format(image_classes[i])] = tpr
output['{} precision'.format(image_classes[i])] = precision
output['{} recall'.format(image_classes[i])] = recall
outcsv = pd.DataFrame(dict([ (k,pd.Series(v)) for k,v in output.items() ]))
outcsv.to_csv(SAVE_DIR+'CNN_test_output.csv', index=False)
plot_roc(pred.numpy(), target.numpy(), image_classes, size=30)
plot_prc(pred.numpy(), target.numpy(), image_classes, size=30)
```
|
github_jupyter
|
TSG023 - Get all BDC objects (Kubernetes)
=========================================
Description
-----------
Get a summary of all Kubernetes resources for the system namespace and
the Big Data Cluster namespace
Steps
-----
### Common functions
Define helper functions used in this notebook.
```
# Define `run` function for transient fault handling, suggestions on error, and scrolling updates on Windows
import sys
import os
import re
import json
import platform
import shlex
import shutil
import datetime
from subprocess import Popen, PIPE
from IPython.display import Markdown
retry_hints = {} # Output in stderr known to be transient, therefore automatically retry
error_hints = {} # Output in stderr where a known SOP/TSG exists which will be HINTed for further help
install_hint = {} # The SOP to help install the executable if it cannot be found
first_run = True
rules = None
debug_logging = False
def run(cmd, return_output=False, no_output=False, retry_count=0, base64_decode=False, return_as_json=False):
"""Run shell command, stream stdout, print stderr and optionally return output
NOTES:
1. Commands that need this kind of ' quoting on Windows e.g.:
kubectl get nodes -o jsonpath={.items[?(@.metadata.annotations.pv-candidate=='data-pool')].metadata.name}
Need to actually pass in as '"':
kubectl get nodes -o jsonpath={.items[?(@.metadata.annotations.pv-candidate=='"'data-pool'"')].metadata.name}
The ' quote approach, although correct when pasting into Windows cmd, will hang at the line:
`iter(p.stdout.readline, b'')`
The shlex.split call does the right thing for each platform, just use the '"' pattern for a '
"""
MAX_RETRIES = 5
output = ""
retry = False
global first_run
global rules
if first_run:
first_run = False
rules = load_rules()
# When running `azdata sql query` on Windows, replace any \n in """ strings, with " ", otherwise we see:
#
# ('HY090', '[HY090] [Microsoft][ODBC Driver Manager] Invalid string or buffer length (0) (SQLExecDirectW)')
#
if platform.system() == "Windows" and cmd.startswith("azdata sql query"):
cmd = cmd.replace("\n", " ")
# shlex.split is required on bash and for Windows paths with spaces
#
cmd_actual = shlex.split(cmd)
# Store this (i.e. kubectl, python etc.) to support binary context aware error_hints and retries
#
user_provided_exe_name = cmd_actual[0].lower()
# When running python, use the python in the ADS sandbox ({sys.executable})
#
if cmd.startswith("python "):
cmd_actual[0] = cmd_actual[0].replace("python", sys.executable)
# On Mac, when ADS is not launched from terminal, LC_ALL may not be set, which causes pip installs to fail
# with:
#
# UnicodeDecodeError: 'ascii' codec can't decode byte 0xc5 in position 4969: ordinal not in range(128)
#
# Setting it to a default value of "en_US.UTF-8" enables pip install to complete
#
if platform.system() == "Darwin" and "LC_ALL" not in os.environ:
os.environ["LC_ALL"] = "en_US.UTF-8"
# When running `kubectl`, if AZDATA_OPENSHIFT is set, use `oc`
#
if cmd.startswith("kubectl ") and "AZDATA_OPENSHIFT" in os.environ:
cmd_actual[0] = cmd_actual[0].replace("kubectl", "oc")
# To aid supportability, determine which binary file will actually be executed on the machine
#
which_binary = None
# Special case for CURL on Windows. The version of CURL in Windows System32 does not work to
# get JWT tokens, it returns "(56) Failure when receiving data from the peer". If another instance
# of CURL exists on the machine use that one. (Unfortunately the curl.exe in System32 is almost
# always the first curl.exe in the path, and it can't be uninstalled from System32, so here we
# look for the 2nd installation of CURL in the path)
if platform.system() == "Windows" and cmd.startswith("curl "):
path = os.getenv('PATH')
for p in path.split(os.path.pathsep):
p = os.path.join(p, "curl.exe")
if os.path.exists(p) and os.access(p, os.X_OK):
if p.lower().find("system32") == -1:
cmd_actual[0] = p
which_binary = p
break
# Find the path based location (shutil.which) of the executable that will be run (and display it to aid supportability), this
# seems to be required for .msi installs of azdata.cmd/az.cmd. (otherwise Popen returns FileNotFound)
#
# NOTE: Bash needs cmd to be the list of the space separated values hence shlex.split.
#
if which_binary == None:
which_binary = shutil.which(cmd_actual[0])
# Display an install HINT, so the user can click on a SOP to install the missing binary
#
if which_binary == None:
print(f"The path used to search for '{cmd_actual[0]}' was:")
print(sys.path)
if user_provided_exe_name in install_hint and install_hint[user_provided_exe_name] is not None:
display(Markdown(f'HINT: Use [{install_hint[user_provided_exe_name][0]}]({install_hint[user_provided_exe_name][1]}) to resolve this issue.'))
raise FileNotFoundError(f"Executable '{cmd_actual[0]}' not found in path (where/which)")
else:
cmd_actual[0] = which_binary
start_time = datetime.datetime.now().replace(microsecond=0)
print(f"START: {cmd} @ {start_time} ({datetime.datetime.utcnow().replace(microsecond=0)} UTC)")
print(f" using: {which_binary} ({platform.system()} {platform.release()} on {platform.machine()})")
print(f" cwd: {os.getcwd()}")
# Command-line tools such as CURL and AZDATA HDFS commands output
# scrolling progress bars, which causes Jupyter to hang forever, to
# workaround this, use no_output=True
#
# Work around a infinite hang when a notebook generates a non-zero return code, break out, and do not wait
#
wait = True
try:
if no_output:
p = Popen(cmd_actual)
else:
p = Popen(cmd_actual, stdout=PIPE, stderr=PIPE, bufsize=1)
with p.stdout:
for line in iter(p.stdout.readline, b''):
line = line.decode()
if return_output:
output = output + line
else:
if cmd.startswith("azdata notebook run"): # Hyperlink the .ipynb file
regex = re.compile(' "(.*)"\: "(.*)"')
match = regex.match(line)
if match:
if match.group(1).find("HTML") != -1:
display(Markdown(f' - "{match.group(1)}": "{match.group(2)}"'))
else:
display(Markdown(f' - "{match.group(1)}": "[{match.group(2)}]({match.group(2)})"'))
wait = False
break # otherwise infinite hang, have not worked out why yet.
else:
print(line, end='')
if rules is not None:
apply_expert_rules(line)
if wait:
p.wait()
except FileNotFoundError as e:
if install_hint is not None:
display(Markdown(f'HINT: Use {install_hint} to resolve this issue.'))
raise FileNotFoundError(f"Executable '{cmd_actual[0]}' not found in path (where/which)") from e
exit_code_workaround = 0 # WORKAROUND: azdata hangs on exception from notebook on p.wait()
if not no_output:
for line in iter(p.stderr.readline, b''):
try:
line_decoded = line.decode()
except UnicodeDecodeError:
# NOTE: Sometimes we get characters back that cannot be decoded(), e.g.
#
# \xa0
#
# For example see this in the response from `az group create`:
#
# ERROR: Get Token request returned http error: 400 and server
# response: {"error":"invalid_grant",# "error_description":"AADSTS700082:
# The refresh token has expired due to inactivity.\xa0The token was
# issued on 2018-10-25T23:35:11.9832872Z
#
# which generates the exception:
#
# UnicodeDecodeError: 'utf-8' codec can't decode byte 0xa0 in position 179: invalid start byte
#
print("WARNING: Unable to decode stderr line, printing raw bytes:")
print(line)
line_decoded = ""
pass
else:
# azdata emits a single empty line to stderr when doing an hdfs cp, don't
# print this empty "ERR:" as it confuses.
#
if line_decoded == "":
continue
print(f"STDERR: {line_decoded}", end='')
if line_decoded.startswith("An exception has occurred") or line_decoded.startswith("ERROR: An error occurred while executing the following cell"):
exit_code_workaround = 1
# inject HINTs to next TSG/SOP based on output in stderr
#
if user_provided_exe_name in error_hints:
for error_hint in error_hints[user_provided_exe_name]:
if line_decoded.find(error_hint[0]) != -1:
display(Markdown(f'HINT: Use [{error_hint[1]}]({error_hint[2]}) to resolve this issue.'))
# apply expert rules (to run follow-on notebooks), based on output
#
if rules is not None:
apply_expert_rules(line_decoded)
# Verify if a transient error, if so automatically retry (recursive)
#
if user_provided_exe_name in retry_hints:
for retry_hint in retry_hints[user_provided_exe_name]:
if line_decoded.find(retry_hint) != -1:
if retry_count < MAX_RETRIES:
print(f"RETRY: {retry_count} (due to: {retry_hint})")
retry_count = retry_count + 1
output = run(cmd, return_output=return_output, retry_count=retry_count)
if return_output:
if base64_decode:
import base64
return base64.b64decode(output).decode('utf-8')
else:
return output
elapsed = datetime.datetime.now().replace(microsecond=0) - start_time
# WORKAROUND: We avoid infinite hang above in the `azdata notebook run` failure case, by inferring success (from stdout output), so
# don't wait here, if success known above
#
if wait:
if p.returncode != 0:
raise SystemExit(f'Shell command:\n\n\t{cmd} ({elapsed}s elapsed)\n\nreturned non-zero exit code: {str(p.returncode)}.\n')
else:
if exit_code_workaround !=0 :
raise SystemExit(f'Shell command:\n\n\t{cmd} ({elapsed}s elapsed)\n\nreturned non-zero exit code: {str(exit_code_workaround)}.\n')
print(f'\nSUCCESS: {elapsed}s elapsed.\n')
if return_output:
if base64_decode:
import base64
return base64.b64decode(output).decode('utf-8')
else:
return output
def load_json(filename):
"""Load a json file from disk and return the contents"""
with open(filename, encoding="utf8") as json_file:
return json.load(json_file)
def load_rules():
"""Load any 'expert rules' from the metadata of this notebook (.ipynb) that should be applied to the stderr of the running executable"""
# Load this notebook as json to get access to the expert rules in the notebook metadata.
#
try:
j = load_json("tsg023-run-kubectl-get-all.ipynb")
except:
pass # If the user has renamed the book, we can't load ourself. NOTE: Is there a way in Jupyter, to know your own filename?
else:
if "metadata" in j and \
"azdata" in j["metadata"] and \
"expert" in j["metadata"]["azdata"] and \
"expanded_rules" in j["metadata"]["azdata"]["expert"]:
rules = j["metadata"]["azdata"]["expert"]["expanded_rules"]
rules.sort() # Sort rules, so they run in priority order (the [0] element). Lowest value first.
# print (f"EXPERT: There are {len(rules)} rules to evaluate.")
return rules
def apply_expert_rules(line):
"""Determine if the stderr line passed in, matches the regular expressions for any of the 'expert rules', if so
inject a 'HINT' to the follow-on SOP/TSG to run"""
global rules
for rule in rules:
notebook = rule[1]
cell_type = rule[2]
output_type = rule[3] # i.e. stream or error
output_type_name = rule[4] # i.e. ename or name
output_type_value = rule[5] # i.e. SystemExit or stdout
details_name = rule[6] # i.e. evalue or text
expression = rule[7].replace("\\*", "*") # Something escaped *, and put a \ in front of it!
if debug_logging:
print(f"EXPERT: If rule '{expression}' satisfied', run '{notebook}'.")
if re.match(expression, line, re.DOTALL):
if debug_logging:
print("EXPERT: MATCH: name = value: '{0}' = '{1}' matched expression '{2}', therefore HINT '{4}'".format(output_type_name, output_type_value, expression, notebook))
match_found = True
display(Markdown(f'HINT: Use [{notebook}]({notebook}) to resolve this issue.'))
print('Common functions defined successfully.')
# Hints for binary (transient fault) retry, (known) error and install guide
#
retry_hints = {'kubectl': ['A connection attempt failed because the connected party did not properly respond after a period of time, or established connection failed because connected host has failed to respond']}
error_hints = {'kubectl': [['no such host', 'TSG010 - Get configuration contexts', '../monitor-k8s/tsg010-get-kubernetes-contexts.ipynb'], ['No connection could be made because the target machine actively refused it', 'TSG056 - Kubectl fails with No connection could be made because the target machine actively refused it', '../repair/tsg056-kubectl-no-connection-could-be-made.ipynb']]}
install_hint = {'kubectl': ['SOP036 - Install kubectl command line interface', '../install/sop036-install-kubectl.ipynb']}
```
### Run kubectl get all for the system namespace
```
run("kubectl get all")
```
### Get the Kubernetes namespace for the big data cluster
Get the namespace of the Big Data Cluster use the kubectl command line
interface .
**NOTE:**
If there is more than one Big Data Cluster in the target Kubernetes
cluster, then either:
- set \[0\] to the correct value for the big data cluster.
- set the environment variable AZDATA\_NAMESPACE, before starting
Azure Data Studio.
```
# Place Kubernetes namespace name for BDC into 'namespace' variable
if "AZDATA_NAMESPACE" in os.environ:
namespace = os.environ["AZDATA_NAMESPACE"]
else:
try:
namespace = run(f'kubectl get namespace --selector=MSSQL_CLUSTER -o jsonpath={{.items[0].metadata.name}}', return_output=True)
except:
from IPython.display import Markdown
print(f"ERROR: Unable to find a Kubernetes namespace with label 'MSSQL_CLUSTER'. SQL Server Big Data Cluster Kubernetes namespaces contain the label 'MSSQL_CLUSTER'.")
display(Markdown(f'HINT: Use [TSG081 - Get namespaces (Kubernetes)](../monitor-k8s/tsg081-get-kubernetes-namespaces.ipynb) to resolve this issue.'))
display(Markdown(f'HINT: Use [TSG010 - Get configuration contexts](../monitor-k8s/tsg010-get-kubernetes-contexts.ipynb) to resolve this issue.'))
display(Markdown(f'HINT: Use [SOP011 - Set kubernetes configuration context](../common/sop011-set-kubernetes-context.ipynb) to resolve this issue.'))
raise
print(f'The SQL Server Big Data Cluster Kubernetes namespace is: {namespace}')
```
### Run kubectl get all for the Big Data Cluster namespace
```
run(f"kubectl get all -n {namespace}")
print('Notebook execution complete.')
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/araffin/rl-tutorial-jnrr19/blob/master/1_getting_started.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Stable Baselines Tutorial - Getting Started
Github repo: https://github.com/araffin/rl-tutorial-jnrr19
Stable-Baselines: https://github.com/hill-a/stable-baselines
Documentation: https://stable-baselines.readthedocs.io/en/master/
RL Baselines zoo: https://github.com/araffin/rl-baselines-zoo
Medium article: [https://medium.com/@araffin/stable-baselines-a-fork-of-openai-baselines-df87c4b2fc82](https://medium.com/@araffin/stable-baselines-a-fork-of-openai-baselines-df87c4b2fc82)
[RL Baselines Zoo](https://github.com/araffin/rl-baselines-zoo) is a collection of pre-trained Reinforcement Learning agents using Stable-Baselines.
It also provides basic scripts for training, evaluating agents, tuning hyperparameters and recording videos.
## Introduction
In this notebook, you will learn the basics for using stable baselines library: how to create a RL model, train it and evaluate it. Because all algorithms share the same interface, we will see how simple it is to switch from one algorithm to another.
## Install Dependencies and Stable Baselines Using Pip
List of full dependencies can be found in the [README](https://github.com/hill-a/stable-baselines).
```
sudo apt-get update && sudo apt-get install cmake libopenmpi-dev zlib1g-dev
```
```
pip install stable-baselines[mpi]
```
```
# Stable Baselines only supports tensorflow 1.x for now
%tensorflow_version 1.x
!apt-get install ffmpeg freeglut3-dev xvfb # For visualization
!pip install stable-baselines[mpi]==2.10.0
```
## Imports
Stable-Baselines works on environments that follow the [gym interface](https://stable-baselines.readthedocs.io/en/master/guide/custom_env.html).
You can find a list of available environment [here](https://gym.openai.com/envs/#classic_control).
It is also recommended to check the [source code](https://github.com/openai/gym) to learn more about the observation and action space of each env, as gym does not have a proper documentation.
Not all algorithms can work with all action spaces, you can find more in this [recap table](https://stable-baselines.readthedocs.io/en/master/guide/algos.html)
```
import gym
import numpy as np
```
The first thing you need to import is the RL model, check the documentation to know what you can use on which problem
```
from stable_baselines import PPO2
```
The next thing you need to import is the policy class that will be used to create the networks (for the policy/value functions).
This step is optional as you can directly use strings in the constructor:
```PPO2('MlpPolicy', env)``` instead of ```PPO2(MlpPolicy, env)```
Note that some algorithms like `SAC` have their own `MlpPolicy` (different from `stable_baselines.common.policies.MlpPolicy`), that's why using string for the policy is the recommened option.
```
from stable_baselines.common.policies import MlpPolicy
```
## Create the Gym env and instantiate the agent
For this example, we will use CartPole environment, a classic control problem.
"A pole is attached by an un-actuated joint to a cart, which moves along a frictionless track. The system is controlled by applying a force of +1 or -1 to the cart. The pendulum starts upright, and the goal is to prevent it from falling over. A reward of +1 is provided for every timestep that the pole remains upright. "
Cartpole environment: [https://gym.openai.com/envs/CartPole-v1/](https://gym.openai.com/envs/CartPole-v1/)
We chose the MlpPolicy because the observation of the CartPole task is a feature vector, not images.
The type of action to use (discrete/continuous) will be automatically deduced from the environment action space
Here we are using the [Proximal Policy Optimization](https://stable-baselines.readthedocs.io/en/master/modules/ppo2.html) algorithm (PPO2 is the version optimized for GPU), which is an Actor-Critic method: it uses a value function to improve the policy gradient descent (by reducing the variance).
It combines ideas from [A2C](https://stable-baselines.readthedocs.io/en/master/modules/a2c.html) (having multiple workers and using an entropy bonus for exploration) and [TRPO](https://stable-baselines.readthedocs.io/en/master/modules/trpo.html) (it uses a trust region to improve stability and avoid catastrophic drops in performance).
PPO is an on-policy algorithm, which means that the trajectories used to update the networks must be collected using the latest policy.
It is usually less sample efficient than off-policy alorithms like [DQN](https://stable-baselines.readthedocs.io/en/master/modules/dqn.html), [SAC](https://stable-baselines.readthedocs.io/en/master/modules/sac.html) or [TD3](https://stable-baselines.readthedocs.io/en/master/modules/td3.html), but is much faster regarding wall-clock time.
```
env = gym.make('CartPole-v1')
model = PPO2(MlpPolicy, env, verbose=0)
```
We create a helper function to evaluate the agent:
```
def evaluate(model, num_episodes=100):
"""
Evaluate a RL agent
:param model: (BaseRLModel object) the RL Agent
:param num_episodes: (int) number of episodes to evaluate it
:return: (float) Mean reward for the last num_episodes
"""
# This function will only work for a single Environment
env = model.get_env()
all_episode_rewards = []
for i in range(num_episodes):
episode_rewards = []
done = False
obs = env.reset()
while not done:
# _states are only useful when using LSTM policies
action, _states = model.predict(obs)
# here, action, rewards and dones are arrays
# because we are using vectorized env
obs, reward, done, info = env.step(action)
episode_rewards.append(reward)
all_episode_rewards.append(sum(episode_rewards))
mean_episode_reward = np.mean(all_episode_rewards)
print("Mean reward:", mean_episode_reward, "Num episodes:", num_episodes)
return mean_episode_reward
```
Let's evaluate the un-trained agent, this should be a random agent.
```
# Random Agent, before training
mean_reward_before_train = evaluate(model, num_episodes=100)
```
Stable-Baselines already provides you with that helper:
```
from stable_baselines.common.evaluation import evaluate_policy
mean_reward, std_reward = evaluate_policy(model, env, n_eval_episodes=100)
print(f"mean_reward:{mean_reward:.2f} +/- {std_reward:.2f}")
```
## Train the agent and evaluate it
```
# Train the agent for 10000 steps
model.learn(total_timesteps=10000)
# Evaluate the trained agent
mean_reward, std_reward = evaluate_policy(model, env, n_eval_episodes=100)
print(f"mean_reward:{mean_reward:.2f} +/- {std_reward:.2f}")
```
Apparently the training went well, the mean reward increased a lot !
### Prepare video recording
```
# Set up fake display; otherwise rendering will fail
import os
os.system("Xvfb :1 -screen 0 1024x768x24 &")
os.environ['DISPLAY'] = ':1'
import base64
from pathlib import Path
from IPython import display as ipythondisplay
def show_videos(video_path='', prefix=''):
"""
Taken from https://github.com/eleurent/highway-env
:param video_path: (str) Path to the folder containing videos
:param prefix: (str) Filter the video, showing only the only starting with this prefix
"""
html = []
for mp4 in Path(video_path).glob("{}*.mp4".format(prefix)):
video_b64 = base64.b64encode(mp4.read_bytes())
html.append('''<video alt="{}" autoplay
loop controls style="height: 400px;">
<source src="data:video/mp4;base64,{}" type="video/mp4" />
</video>'''.format(mp4, video_b64.decode('ascii')))
ipythondisplay.display(ipythondisplay.HTML(data="<br>".join(html)))
```
We will record a video using the [VecVideoRecorder](https://stable-baselines.readthedocs.io/en/master/guide/vec_envs.html#vecvideorecorder) wrapper, you will learn about those wrapper in the next notebook.
```
from stable_baselines.common.vec_env import VecVideoRecorder, DummyVecEnv
def record_video(env_id, model, video_length=500, prefix='', video_folder='videos/'):
"""
:param env_id: (str)
:param model: (RL model)
:param video_length: (int)
:param prefix: (str)
:param video_folder: (str)
"""
eval_env = DummyVecEnv([lambda: gym.make(env_id)])
# Start the video at step=0 and record 500 steps
eval_env = VecVideoRecorder(eval_env, video_folder=video_folder,
record_video_trigger=lambda step: step == 0, video_length=video_length,
name_prefix=prefix)
obs = eval_env.reset()
for _ in range(video_length):
action, _ = model.predict(obs)
obs, _, _, _ = eval_env.step(action)
# Close the video recorder
eval_env.close()
```
### Visualize trained agent
```
record_video('CartPole-v1', model, video_length=500, prefix='ppo2-cartpole')
show_videos('videos', prefix='ppo2')
```
## Bonus: Train a RL Model in One Line
The policy class to use will be inferred and the environment will be automatically created. This works because both are [registered](https://stable-baselines.readthedocs.io/en/master/guide/quickstart.html).
```
model = PPO2('MlpPolicy', "CartPole-v1", verbose=1).learn(1000)
```
## Train a DQN agent
In the previous example, we have used PPO, which one of the many algorithms provided by stable-baselines.
In the next example, we are going train a [Deep Q-Network agent (DQN)](https://stable-baselines.readthedocs.io/en/master/modules/dqn.html), and try to see possible improvements provided by its extensions (Double-DQN, Dueling-DQN, Prioritized Experience Replay).
The essential point of this section is to show you how simple it is to tweak hyperparameters.
The main advantage of stable-baselines is that it provides a common interface to use the algorithms, so the code will be quite similar.
DQN paper: https://arxiv.org/abs/1312.5602
Dueling DQN: https://arxiv.org/abs/1511.06581
Double-Q Learning: https://arxiv.org/abs/1509.06461
Prioritized Experience Replay: https://arxiv.org/abs/1511.05952
### Vanilla DQN: DQN without extensions
```
# Same as before we instantiate the agent along with the environment
from stable_baselines import DQN
# Deactivate all the DQN extensions to have the original version
# In practice, it is recommend to have them activated
kwargs = {'double_q': False, 'prioritized_replay': False, 'policy_kwargs': dict(dueling=False)}
# Note that the MlpPolicy of DQN is different from the one of PPO
# but stable-baselines handles that automatically if you pass a string
dqn_model = DQN('MlpPolicy', 'CartPole-v1', verbose=1, **kwargs)
# Random Agent, before training
mean_reward_before_train = evaluate(dqn_model, num_episodes=100)
# Train the agent for 10000 steps
dqn_model.learn(total_timesteps=10000, log_interval=10)
# Evaluate the trained agent
mean_reward = evaluate(dqn_model, num_episodes=100)
```
### DQN + Prioritized Replay
```
# Activate only the prioritized replay
kwargs = {'double_q': False, 'prioritized_replay': True, 'policy_kwargs': dict(dueling=False)}
dqn_per_model = DQN('MlpPolicy', 'CartPole-v1', verbose=1, **kwargs)
dqn_per_model.learn(total_timesteps=10000, log_interval=10)
# Evaluate the trained agent
mean_reward = evaluate(dqn_per_model, num_episodes=100)
```
### DQN + Prioritized Experience Replay + Double Q-Learning + Dueling
```
# Activate all extensions
kwargs = {'double_q': True, 'prioritized_replay': True, 'policy_kwargs': dict(dueling=True)}
dqn_full_model = DQN('MlpPolicy', 'CartPole-v1', verbose=1, **kwargs)
dqn_full_model.learn(total_timesteps=10000, log_interval=10)
mean_reward = evaluate(dqn_per_model, num_episodes=100)
```
In this particular example, the extensions does not seem to give any improvement compared to the simple DQN version.
They are several reasons for that:
1. `CartPole-v1` is a pretty simple environment
2. We trained DQN for very few timesteps, not enough to see any difference
3. The default hyperparameters for DQN are tuned for atari games, where the number of training timesteps is much larger (10^6) and input observations are images
4. We have only compared one random seed per experiment
## Conclusion
In this notebook we have seen:
- how to define and train a RL model using stable baselines, it takes only one line of code ;)
- how to use different RL algorithms and change some hyperparameters
```
```
|
github_jupyter
|
<img src="https://github.com/pmservice/ai-openscale-tutorials/raw/master/notebooks/images/banner.png" align="left" alt="banner">
# Working with Watson OpenScale - Custom Machine Learning Provider
This notebook should be run using with **Python 3.7.x** runtime environment. **If you are viewing this in Watson Studio and do not see Python 3.7.x in the upper right corner of your screen, please update the runtime now.** It requires service credentials for the following services:
* Watson OpenScale
* A Custom ML provider which is hosted in a VM that can be accessible from CPD PODs, specifically OpenScale PODs namely ML Gateway fairness, quality, drift, and explain.
* DB2 - as part of this notebook, we make use of an existing data mart.
The notebook will configure a OpenScale data mart subscription for Custom ML Provider deployment. We configure and execute the fairness, explain, quality and drift monitors.
## Custom Machine Learning Provider Setup
Following code can be used to start a gunicorn/flask application that can be hosted in a VM, such that it can be accessable from CPD system.
This code does the following:
* It wraps a Watson Machine Learning model that is deployed to a space.
* So the hosting application URL should contain the SPACE ID and the DEPLOYMENT ID. Then, the same can be used to talk to the target WML model/deployment.
* Having said that, this is only for this tutorial purpose, and you can define your Custom ML provider endpoint in any fashion you want, such that it wraps your own custom ML engine.
* The scoring request and response payload should confirm to the schema as described here at: https://dataplatform.cloud.ibm.com/docs/content/wsj/model/wos-frameworks-custom.html
* To start the application using the below code, make sure you install following python packages in your VM:
python -m pip install gunicorn
python -m pip install flask
python -m pip install numpy
python -m pip install pandas
python -m pip install requests
python -m pip install joblib==0.11
python -m pip install scipy==0.19.1
python -m pip install --user numpy scipy matplotlib ipython jupyter pandas sympy nose
python -m pip install ibm_watson_machine_learning
-----------------
```
from flask import Flask, request, abort, jsonify
import json
import base64
import requests, io
import pandas as pd
from ibm_watson_machine_learning import APIClient
app = Flask(__name__)
WML_CREDENTIALS = {
"url": "https://namespace1-cpd-namespace1.apps.xxxxx.os.fyre.ibm.com",
"username": "admin",
"password" : "xxxx",
"instance_id": "wml_local",
"version" : "3.5"
}
@app.route('/spaces/<space_id>/deployments/<deployment_id>/predictions', methods=['POST'])
def wml_scoring(space_id, deployment_id):
if not request.json:
abort(400)
wml_credentials = WML_CREDENTIALS
payload_scoring = {
"input_data": [
request.json
]
}
wml_client = APIClient(wml_credentials)
wml_client.set.default_space(space_id)
records_list=[]
scoring_response = wml_client.deployments.score(deployment_id, payload_scoring)
return jsonify(scoring_response["predictions"][0])
if __name__ == '__main__':
app.run(host='xxxx.fyre.ibm.com', port=9443, debug=True)
```
-----------------
# Setup <a name="setup"></a>
## Package installation
```
import warnings
warnings.filterwarnings('ignore')
!pip install --upgrade pyspark==2.4 --no-cache | tail -n 1
!pip install --upgrade pandas==0.25.3 --no-cache | tail -n 1
!pip install --upgrade requests==2.23 --no-cache | tail -n 1
!pip install numpy==1.16.4 --no-cache | tail -n 1
!pip install scikit-learn==0.20 --no-cache | tail -n 1
!pip install SciPy --no-cache | tail -n 1
!pip install lime --no-cache | tail -n 1
!pip install --upgrade ibm-watson-machine-learning --user | tail -n 1
!pip install --upgrade ibm-watson-openscale --no-cache | tail -n 1
!pip install --upgrade ibm-wos-utils --no-cache | tail -n 1
```
### Action: restart the kernel!
## Configure credentials
- WOS_CREDENTIALS (CP4D)
- WML_CREDENTIALS (CP4D)
- DATABASE_CREDENTIALS (DB2 on CP4D or Cloud Object Storage (COS))
- SCHEMA_NAME
```
#masked
WOS_CREDENTIALS = {
"url": "https://namespace1-cpd-namespace1.apps.xxxxx.os.fyre.ibm.com",
"username": "admin",
"password": "xxxxx",
"version": "3.5"
}
CUSTOM_ML_PROVIDER_SCORING_URL = 'https://xxxxx.fyre.ibm.com:9443/spaces/$SPACE_ID/deployments/$DEPLOYMENT_ID/predictions'
scoring_url = CUSTOM_ML_PROVIDER_SCORING_URL
label_column="Risk"
model_type = "binary"
import os
import base64
import json
import requests
from requests.auth import HTTPBasicAuth
```
## Save training data to Cloud Object Storage
### Cloud object storage details¶
In next cells, you will need to paste some credentials to Cloud Object Storage. If you haven't worked with COS yet please visit getting started with COS tutorial. You can find COS_API_KEY_ID and COS_RESOURCE_CRN variables in Service Credentials in menu of your COS instance. Used COS Service Credentials must be created with Role parameter set as Writer. Later training data file will be loaded to the bucket of your instance and used as training refecence in subsription. COS_ENDPOINT variable can be found in Endpoint field of the menu.
```
IAM_URL="https://iam.ng.bluemix.net/oidc/token"
# masked
COS_API_KEY_ID = "*****"
COS_RESOURCE_CRN = "*****"
COS_ENDPOINT = "https://s3.us.cloud-object-storage.appdomain.cloud" # Current list avaiable at https://control.cloud-object-storage.cloud.ibm.com/v2/endpoints
BUCKET_NAME = "*****"
FILE_NAME = "german_credit_data_biased_training.csv"
```
# Load and explore data
```
!rm german_credit_data_biased_training.csv
!wget https://raw.githubusercontent.com/pmservice/ai-openscale-tutorials/master/assets/historical_data/german_credit_risk/wml/german_credit_data_biased_training.csv
```
## Explore data
```
training_data_references = [
{
"id": "Credit Risk",
"type": "s3",
"connection": {
"access_key_id": COS_API_KEY_ID,
"endpoint_url": COS_ENDPOINT,
"resource_instance_id":COS_RESOURCE_CRN
},
"location": {
"bucket": BUCKET_NAME,
"path": FILE_NAME,
}
}
]
```
## Construct the scoring payload
```
import pandas as pd
df = pd.read_csv("german_credit_data_biased_training.csv")
df.head()
cols_to_remove = [label_column]
def get_scoring_payload(no_of_records_to_score = 1):
for col in cols_to_remove:
if col in df.columns:
del df[col]
fields = df.columns.tolist()
values = df[fields].values.tolist()
payload_scoring ={"fields": fields, "values": values[:no_of_records_to_score]}
return payload_scoring
#debug
payload_scoring = get_scoring_payload(1)
payload_scoring
```
## Method to perform scoring
```
def custom_ml_scoring():
header = {"Content-Type": "application/json", "x":"y"}
print(scoring_url)
scoring_response = requests.post(scoring_url, json=payload_scoring, headers=header, verify=False)
jsonify_scoring_response = scoring_response.json()
return jsonify_scoring_response
```
## Method to perform payload logging
```
import uuid
scoring_id = None
from ibm_watson_openscale.supporting_classes.payload_record import PayloadRecord
def payload_logging(payload_scoring, scoring_response):
scoring_id = str(uuid.uuid4())
records_list=[]
#manual PL logging for custom ml provider
pl_record = PayloadRecord(scoring_id=scoring_id, request=payload_scoring, response=scoring_response, response_time=int(460))
records_list.append(pl_record)
wos_client.data_sets.store_records(data_set_id = payload_data_set_id, request_body=records_list)
time.sleep(5)
pl_records_count = wos_client.data_sets.get_records_count(payload_data_set_id)
print("Number of records in the payload logging table: {}".format(pl_records_count))
return scoring_id
```
## Score the model and print the scoring response
### Sample Scoring
```
custom_ml_scoring()
```
# Configure OpenScale
The notebook will now import the necessary libraries and set up a Python OpenScale client.
```
from ibm_watson_openscale import APIClient
from ibm_watson_openscale.utils import *
from ibm_watson_openscale.supporting_classes import *
from ibm_watson_openscale.supporting_classes.enums import *
from ibm_watson_openscale.base_classes.watson_open_scale_v2 import *
from ibm_cloud_sdk_core.authenticators import CloudPakForDataAuthenticator
import json
import requests
import base64
from requests.auth import HTTPBasicAuth
import time
```
## Get a instance of the OpenScale SDK client
```
authenticator = CloudPakForDataAuthenticator(
url=WOS_CREDENTIALS['url'],
username=WOS_CREDENTIALS['username'],
password=WOS_CREDENTIALS['password'],
disable_ssl_verification=True
)
wos_client = APIClient(service_url=WOS_CREDENTIALS['url'],authenticator=authenticator)
wos_client.version
```
## Set up datamart
Watson OpenScale uses a database to store payload logs and calculated metrics. If database credentials were not supplied above, the notebook will use the free, internal lite database. If database credentials were supplied, the datamart will be created there unless there is an existing datamart and the KEEP_MY_INTERNAL_POSTGRES variable is set to True. If an OpenScale datamart exists in Db2 or PostgreSQL, the existing datamart will be used and no data will be overwritten.
Prior instances of the model will be removed from OpenScale monitoring.
```
wos_client.data_marts.show()
data_marts = wos_client.data_marts.list().result.data_marts
if len(data_marts) == 0:
raise Exception("Missing data mart.")
data_mart_id=data_marts[0].metadata.id
print('Using existing datamart {}'.format(data_mart_id))
data_mart_details = wos_client.data_marts.list().result.data_marts[0]
data_mart_details.to_dict()
wos_client.service_providers.show()
```
## Remove existing service provider connected with used WML instance.
Multiple service providers for the same engine instance are avaiable in Watson OpenScale. To avoid multiple service providers of used WML instance in the tutorial notebook the following code deletes existing service provder(s) and then adds new one.
```
SERVICE_PROVIDER_NAME = "Custom ML Provider Demo - All Monitors"
SERVICE_PROVIDER_DESCRIPTION = "Added by tutorial WOS notebook to showcase monitoring Fairness, Quality, Drift and Explainability against a Custom ML provider."
service_providers = wos_client.service_providers.list().result.service_providers
for service_provider in service_providers:
service_instance_name = service_provider.entity.name
if service_instance_name == SERVICE_PROVIDER_NAME:
service_provider_id = service_provider.metadata.id
wos_client.service_providers.delete(service_provider_id)
print("Deleted existing service_provider for WML instance: {}".format(service_provider_id))
```
## Add service provider
Watson OpenScale needs to be bound to the Watson Machine Learning instance to capture payload data into and out of the model.
Note: You can bind more than one engine instance if needed by calling wos_client.service_providers.add method. Next, you can refer to particular service provider using service_provider_id.
```
request_headers = {"Content-Type": "application/json", "Custom_header_X": "Custom_header_X_value_Y"}
MLCredentials = {}
added_service_provider_result = wos_client.service_providers.add(
name=SERVICE_PROVIDER_NAME,
description=SERVICE_PROVIDER_DESCRIPTION,
service_type=ServiceTypes.CUSTOM_MACHINE_LEARNING,
request_headers=request_headers,
operational_space_id = "production",
credentials=MLCredentials,
background_mode=False
).result
service_provider_id = added_service_provider_result.metadata.id
print(wos_client.service_providers.get(service_provider_id).result)
print('Data Mart ID : ' + data_mart_id)
print('Service Provider ID : ' + service_provider_id)
```
## Subscriptions
Remove existing credit risk subscriptions
This code removes previous subscriptions to the model to refresh the monitors with the new model and new data.
```
wos_client.subscriptions.show()
```
## Remove the existing subscription
```
SUBSCRIPTION_NAME = "Custom ML Subscription - All Monitors"
subscriptions = wos_client.subscriptions.list().result.subscriptions
for subscription in subscriptions:
if subscription.entity.asset.name == "[asset] " + SUBSCRIPTION_NAME:
sub_model_id = subscription.metadata.id
wos_client.subscriptions.delete(subscription.metadata.id)
print('Deleted existing subscription for model', sub_model_id)
```
This code creates the model subscription in OpenScale using the Python client API. Note that we need to provide the model unique identifier, and some information about the model itself.
```
feature_columns=["CheckingStatus","LoanDuration","CreditHistory","LoanPurpose","LoanAmount","ExistingSavings","EmploymentDuration","InstallmentPercent","Sex","OthersOnLoan","CurrentResidenceDuration","OwnsProperty","Age","InstallmentPlans","Housing","ExistingCreditsCount","Job","Dependents","Telephone","ForeignWorker"]
cat_features=["CheckingStatus","CreditHistory","LoanPurpose","ExistingSavings","EmploymentDuration","Sex","OthersOnLoan","OwnsProperty","InstallmentPlans","Housing","Job","Telephone","ForeignWorker"]
import uuid
asset_id = str(uuid.uuid4())
asset_name = '[asset] ' + SUBSCRIPTION_NAME
url = ''
asset_deployment_id = str(uuid.uuid4())
asset_deployment_name = asset_name
asset_deployment_scoring_url = scoring_url
scoring_endpoint_url = scoring_url
scoring_request_headers = {
"Content-Type": "application/json",
"Custom_header_X": "Custom_header_X_value_Y"
}
subscription_details = wos_client.subscriptions.add(
data_mart_id=data_mart_id,
service_provider_id=service_provider_id,
asset=Asset(
asset_id=asset_id,
name=asset_name,
url=url,
asset_type=AssetTypes.MODEL,
input_data_type=InputDataType.STRUCTURED,
problem_type=ProblemType.BINARY_CLASSIFICATION
),
deployment=AssetDeploymentRequest(
deployment_id=asset_deployment_id,
name=asset_deployment_name,
deployment_type= DeploymentTypes.ONLINE,
scoring_endpoint=ScoringEndpointRequest(
url=scoring_endpoint_url,
request_headers=scoring_request_headers
)
),
asset_properties=AssetPropertiesRequest(
label_column=label_column,
probability_fields=["probability"],
prediction_field="predictedLabel",
feature_fields = feature_columns,
categorical_fields = cat_features,
training_data_reference=TrainingDataReference(type="cos",
location=COSTrainingDataReferenceLocation(bucket = BUCKET_NAME,
file_name = FILE_NAME),
connection=COSTrainingDataReferenceConnection.from_dict({
"resource_instance_id": COS_RESOURCE_CRN,
"url": COS_ENDPOINT,
"api_key": COS_API_KEY_ID,
"iam_url": IAM_URL}))
)
).result
subscription_id = subscription_details.metadata.id
print('Subscription ID: ' + subscription_id)
import time
time.sleep(5)
payload_data_set_id = None
payload_data_set_id = wos_client.data_sets.list(type=DataSetTypes.PAYLOAD_LOGGING,
target_target_id=subscription_id,
target_target_type=TargetTypes.SUBSCRIPTION).result.data_sets[0].metadata.id
if payload_data_set_id is None:
print("Payload data set not found. Please check subscription status.")
else:
print("Payload data set id:", payload_data_set_id)
```
### Before the payload logging
wos_client.subscriptions.get(subscription_id).result.to_dict()
# Score the model so we can configure monitors
Now that the WML service has been bound and the subscription has been created, we need to send a request to the model before we configure OpenScale. This allows OpenScale to create a payload log in the datamart with the correct schema, so it can capture data coming into and out of the model.
```
no_of_records_to_score = 100
```
### Construct the scoring payload
```
payload_scoring = get_scoring_payload(no_of_records_to_score)
```
### Perform the scoring against the Custom ML Provider
```
scoring_response = custom_ml_scoring()
```
### Perform payload logging by passing the scoring payload and scoring response
```
scoring_id = payload_logging(payload_scoring, scoring_response)
```
### The scoring id, which would be later used for explanation of the randomly picked transactions
```
print('scoring_id: ' + str(scoring_id))
```
# Fairness configuration <a name="Fairness"></a>
The code below configures fairness monitoring for our model. It turns on monitoring for two features, sex and age. In each case, we must specify:
Which model feature to monitor One or more majority groups, which are values of that feature that we expect to receive a higher percentage of favorable outcomes One or more minority groups, which are values of that feature that we expect to receive a higher percentage of unfavorable outcomes The threshold at which we would like OpenScale to display an alert if the fairness measurement falls below (in this case, 80%) Additionally, we must specify which outcomes from the model are favourable outcomes, and which are unfavourable. We must also provide the number of records OpenScale will use to calculate the fairness score. In this case, OpenScale's fairness monitor will run hourly, but will not calculate a new fairness rating until at least 100 records have been added. Finally, to calculate fairness, OpenScale must perform some calculations on the training data, so we provide the dataframe containing the data.
### Create Fairness Monitor Instance
```
target = Target(
target_type=TargetTypes.SUBSCRIPTION,
target_id=subscription_id
)
parameters = {
"features": [
{"feature": "Sex",
"majority": ['male'],
"minority": ['female']
},
{"feature": "Age",
"majority": [[26, 75]],
"minority": [[18, 25]]
}
],
"favourable_class": ["No Risk"],
"unfavourable_class": ["Risk"],
"min_records": 100
}
thresholds = [{
"metric_id": "fairness_value",
"specific_values": [{
"applies_to": [{
"key": "feature",
"type": "tag",
"value": "Age"
}],
"value": 95
},
{
"applies_to": [{
"key": "feature",
"type": "tag",
"value": "Sex"
}],
"value": 95
}
],
"type": "lower_limit",
"value": 80.0
}]
fairness_monitor_details = wos_client.monitor_instances.create(
data_mart_id=data_mart_id,
background_mode=False,
monitor_definition_id=wos_client.monitor_definitions.MONITORS.FAIRNESS.ID,
target=target,
parameters=parameters,
thresholds=thresholds).result
fairness_monitor_instance_id = fairness_monitor_details.metadata.id
```
### Get Fairness Monitor Instance
```
wos_client.monitor_instances.show()
```
### Get run details
In case of production subscription, initial monitoring run is triggered internally. Checking its status
```
runs = wos_client.monitor_instances.list_runs(fairness_monitor_instance_id, limit=1).result.to_dict()
fairness_monitoring_run_id = runs["runs"][0]["metadata"]["id"]
run_status = None
while(run_status not in ["finished", "error"]):
run_details = wos_client.monitor_instances.get_run_details(fairness_monitor_instance_id, fairness_monitoring_run_id).result.to_dict()
run_status = run_details["entity"]["status"]["state"]
print('run_status: ', run_status)
if run_status in ["finished", "error"]:
break
time.sleep(10)
```
### Fairness run output
```
wos_client.monitor_instances.get_run_details(fairness_monitor_instance_id, fairness_monitoring_run_id).result.to_dict()
wos_client.monitor_instances.show_metrics(monitor_instance_id=fairness_monitor_instance_id)
```
# Configure Explainability <a name="explain"></a>
We provide OpenScale with the training data to enable and configure the explainability features.
```
target = Target(
target_type=TargetTypes.SUBSCRIPTION,
target_id=subscription_id
)
parameters = {
"enabled": True
}
explain_monitor_details = wos_client.monitor_instances.create(
data_mart_id=data_mart_id,
background_mode=False,
monitor_definition_id=wos_client.monitor_definitions.MONITORS.EXPLAINABILITY.ID,
target=target,
parameters=parameters
).result
explain_monitor_details.metadata.id
scoring_ids = []
sample_size = 2
import random
for i in range(0, sample_size):
n = random.randint(1,100)
scoring_ids.append(scoring_id + '-' + str(n))
print("Running explanations on scoring IDs: {}".format(scoring_ids))
explanation_types = ["lime", "contrastive"]
result = wos_client.monitor_instances.explanation_tasks(scoring_ids=scoring_ids, explanation_types=explanation_types).result
print(result)
```
### Explanation tasks
```
explanation_task_ids=result.metadata.explanation_task_ids
explanation_task_ids
```
### Wait for the explanation tasks to complete - all of them
```
import time
def finish_explanation_tasks():
finished_explanations = []
finished_explanation_task_ids = []
# Check for the explanation task status for finished status.
# If it is in-progress state, then sleep for some time and check again.
# Perform the same for couple of times, so that all tasks get into finished state.
for i in range(0, 5):
# for each explanation
print('iteration ' + str(i))
#check status for all explanation tasks
for explanation_task_id in explanation_task_ids:
if explanation_task_id not in finished_explanation_task_ids:
result = wos_client.monitor_instances.get_explanation_tasks(explanation_task_id=explanation_task_id).result
print(explanation_task_id + ' : ' + result.entity.status.state)
if (result.entity.status.state == 'finished' or result.entity.status.state == 'error') and explanation_task_id not in finished_explanation_task_ids:
finished_explanation_task_ids.append(explanation_task_id)
finished_explanations.append(result)
# if there is altest one explanation task that is not yet completed, then sleep for sometime,
# and check for all those tasks, for which explanation is not yet completeed.
if len(finished_explanation_task_ids) != sample_size:
print('sleeping for some time..')
time.sleep(10)
else:
break
return finished_explanations
```
### You may have to run the below multiple times till all explanation tasks are either finished or error'ed.
```
finished_explanations = finish_explanation_tasks()
len(finished_explanations)
def construct_explanation_features_map(feature_name, feature_weight):
if feature_name in explanation_features_map:
explanation_features_map[feature_name].append(feature_weight)
else:
explanation_features_map[feature_name] = [feature_weight]
explanation_features_map = {}
for result in finished_explanations:
print('\n>>>>>>>>>>>>>>>>>>>>>>\n')
print('explanation task: ' + str(result.metadata.explanation_task_id) + ', perturbed:' + str(result.entity.perturbed))
if result.entity.explanations is not None:
explanations = result.entity.explanations
for explanation in explanations:
if 'predictions' in explanation:
predictions = explanation['predictions']
for prediction in predictions:
predicted_value = prediction['value']
probability = prediction['probability']
print('prediction : ' + str(predicted_value) + ', probability : ' + str(probability))
if 'explanation_features' in prediction:
explanation_features = prediction['explanation_features']
for explanation_feature in explanation_features:
feature_name = explanation_feature['feature_name']
feature_weight = explanation_feature['weight']
if (feature_weight >= 0 ):
feature_weight_percent = round(feature_weight * 100, 2)
print(str(feature_name) + ' : ' + str(feature_weight_percent))
task_feature_weight_map = {}
task_feature_weight_map[result.metadata.explanation_task_id] = feature_weight_percent
construct_explanation_features_map(feature_name, feature_weight_percent)
print('\n>>>>>>>>>>>>>>>>>>>>>>\n')
explanation_features_map
import matplotlib.pyplot as plt
for key in explanation_features_map.keys():
#plot_graph(key, explanation_features_map[key])
values = explanation_features_map[key]
plt.title(key)
plt.ylabel('Weight')
plt.bar(range(len(values)), values)
plt.show()
```
# Quality monitoring and feedback logging <a name="quality"></a>
## Enable quality monitoring
The code below waits ten seconds to allow the payload logging table to be set up before it begins enabling monitors. First, it turns on the quality (accuracy) monitor and sets an alert threshold of 70%. OpenScale will show an alert on the dashboard if the model accuracy measurement (area under the curve, in the case of a binary classifier) falls below this threshold.
The second paramater supplied, min_records, specifies the minimum number of feedback records OpenScale needs before it calculates a new measurement. The quality monitor runs hourly, but the accuracy reading in the dashboard will not change until an additional 50 feedback records have been added, via the user interface, the Python client, or the supplied feedback endpoint.
```
import time
#time.sleep(10)
target = Target(
target_type=TargetTypes.SUBSCRIPTION,
target_id=subscription_id
)
parameters = {
"min_feedback_data_size": 90
}
thresholds = [
{
"metric_id": "area_under_roc",
"type": "lower_limit",
"value": .80
}
]
quality_monitor_details = wos_client.monitor_instances.create(
data_mart_id=data_mart_id,
background_mode=False,
monitor_definition_id=wos_client.monitor_definitions.MONITORS.QUALITY.ID,
target=target,
parameters=parameters,
thresholds=thresholds
).result
quality_monitor_instance_id = quality_monitor_details.metadata.id
quality_monitor_instance_id
```
## Feedback logging
The code below downloads and stores enough feedback data to meet the minimum threshold so that OpenScale can calculate a new accuracy measurement. It then kicks off the accuracy monitor. The monitors run hourly, or can be initiated via the Python API, the REST API, or the graphical user interface.
```
!rm additional_feedback_data_v2.json
!wget https://raw.githubusercontent.com/IBM/watson-openscale-samples/main/IBM%20Cloud/WML/assets/data/credit_risk/additional_feedback_data_v2.json
```
## Get feedback logging dataset ID
```
feedback_dataset_id = None
feedback_dataset = wos_client.data_sets.list(type=DataSetTypes.FEEDBACK,
target_target_id=subscription_id,
target_target_type=TargetTypes.SUBSCRIPTION).result
feedback_dataset_id = feedback_dataset.data_sets[0].metadata.id
if feedback_dataset_id is None:
print("Feedback data set not found. Please check quality monitor status.")
with open('additional_feedback_data_v2.json') as feedback_file:
additional_feedback_data = json.load(feedback_file)
wos_client.data_sets.store_records(feedback_dataset_id, request_body=additional_feedback_data, background_mode=False)
wos_client.data_sets.get_records_count(data_set_id=feedback_dataset_id)
run_details = wos_client.monitor_instances.run(monitor_instance_id=quality_monitor_instance_id, background_mode=False).result
wos_client.monitor_instances.show_metrics(monitor_instance_id=quality_monitor_instance_id)
```
# Drift configuration <a name="drift"></a>
# Drift detection model generation
Please update the score function which will be used forgenerating drift detection model which will used for drift detection . This might take sometime to generate model and time taken depends on the training dataset size. The output of the score function should be a 2 arrays 1. Array of model prediction 2. Array of probabilities
- User is expected to make sure that the data type of the "class label" column selected and the prediction column are same . For eg : If class label is numeric , the prediction array should also be numeric
- Each entry of a probability array should have all the probabities of the unique class lable .
For eg: If the model_type=multiclass and unique class labels are A, B, C, D . Each entry in the probability array should be a array of size 4 . Eg : [ [50,30,10,10] ,[40,20,30,10]...]
**Note:**
- *User is expected to add "score" method , which should output prediction column array and probability column array.*
- *The data type of the label column and prediction column should be same . User needs to make sure that label column and prediction column array should have the same unique class labels*
- **Please update the score function below with the help of templates documented [here](https://github.com/IBM-Watson/aios-data-distribution/blob/master/Score%20function%20templates%20for%20drift%20detection.md)**
```
import pandas as pd
df = pd.read_csv("german_credit_data_biased_training.csv")
df.head()
def score(training_data_frame):
#The data type of the label column and prediction column should be same .
#User needs to make sure that label column and prediction column array should have the same unique class labels
prediction_column_name = "predictedLabel"
probability_column_name = "probability"
feature_columns = list(training_data_frame.columns)
training_data_rows = training_data_frame[feature_columns].values.tolist()
payload_scoring_records = {
"fields": feature_columns,
"values": [x for x in training_data_rows]
}
header = {"Content-Type": "application/json", "x":"y"}
scoring_response_raw = requests.post(scoring_url, json=payload_scoring_records, headers=header, verify=False)
scoring_response = scoring_response_raw.json()
probability_array = None
prediction_vector = None
prob_col_index = list(scoring_response.get('fields')).index(probability_column_name)
predict_col_index = list(scoring_response.get('fields')).index(prediction_column_name)
if prob_col_index < 0 or predict_col_index < 0:
raise Exception("Missing prediction/probability column in the scoring response")
import numpy as np
probability_array = np.array([value[prob_col_index] for value in scoring_response.get('values')])
prediction_vector = np.array([value[predict_col_index] for value in scoring_response.get('values')])
return probability_array, prediction_vector
```
### Define the drift detection input
```
drift_detection_input = {
"feature_columns": feature_columns,
"categorical_columns": cat_features,
"label_column": label_column,
"problem_type": model_type
}
print(drift_detection_input)
```
### Generate drift detection model
```
!rm drift_detection_model.tar.gz
from ibm_wos_utils.drift.drift_trainer import DriftTrainer
drift_trainer = DriftTrainer(df,drift_detection_input)
if model_type != "regression":
#Note: batch_size can be customized by user as per the training data size
drift_trainer.generate_drift_detection_model(score,batch_size=df.shape[0])
#Note: Two column constraints are not computed beyond two_column_learner_limit(default set to 200)
#User can adjust the value depending on the requirement
drift_trainer.learn_constraints(two_column_learner_limit=200)
drift_trainer.create_archive()
!ls -al
filename = 'drift_detection_model.tar.gz'
```
### Upload the drift detection model to OpenScale subscription
```
wos_client.monitor_instances.upload_drift_model(
model_path=filename,
archive_name=filename,
data_mart_id=data_mart_id,
subscription_id=subscription_id,
enable_data_drift=True,
enable_model_drift=True
)
```
### Delete the existing drift monitor instance for the subscription
```
monitor_instances = wos_client.monitor_instances.list().result.monitor_instances
for monitor_instance in monitor_instances:
monitor_def_id=monitor_instance.entity.monitor_definition_id
if monitor_def_id == "drift" and monitor_instance.entity.target.target_id == subscription_id:
wos_client.monitor_instances.delete(monitor_instance.metadata.id)
print('Deleted existing drift monitor instance with id: ', monitor_instance.metadata.id)
target = Target(
target_type=TargetTypes.SUBSCRIPTION,
target_id=subscription_id
)
parameters = {
"min_samples": 100,
"drift_threshold": 0.1,
"train_drift_model": False,
"enable_model_drift": True,
"enable_data_drift": True
}
drift_monitor_details = wos_client.monitor_instances.create(
data_mart_id=data_mart_id,
background_mode=False,
monitor_definition_id=wos_client.monitor_definitions.MONITORS.DRIFT.ID,
target=target,
parameters=parameters
).result
drift_monitor_instance_id = drift_monitor_details.metadata.id
drift_monitor_instance_id
```
### Drift run
```
drift_run_details = wos_client.monitor_instances.run(monitor_instance_id=drift_monitor_instance_id, background_mode=False)
time.sleep(5)
wos_client.monitor_instances.show_metrics(monitor_instance_id=drift_monitor_instance_id)
```
## Summary
As part of this notebook, we have performed the following:
* Create a subscription to an custom ML end point
* Scored the custom ML provider with 100 records
* With the scored payload and also the scored response, we called the DataSets SDK method to store the payload logging records into the data mart. While doing so, we have set the scoring_id attribute.
* Configured the fairness monitor and executed it and viewed the fairness metrics output.
* Configured explainabilty monitor
* Randomly selected 5 transactions for which we want to get the prediction explanation.
* Submitted explainability tasks for the selected scoring ids, and waited for their completion.
* In the end, we composed a weight map of feature and its weight across transactions. And plotted the same.
* For example:
```
{'ForeignWorker': [33.29, 5.23],
'OthersOnLoan': [15.96, 19.97, 12.76],
'OwnsProperty': [15.43, 3.92, 4.44, 10.36],
'Dependents': [9.06],
'InstallmentPercent': [9.05],
'CurrentResidenceDuration': [8.74, 13.15, 12.1, 10.83],
'Sex': [2.96, 12.76],
'InstallmentPlans': [2.4, 5.67, 6.57],
'Age': [2.28, 8.6, 11.26],
'Job': [0.84],
'LoanDuration': [15.02, 10.87, 18.91, 12.72],
'EmploymentDuration': [14.02, 14.05, 12.1],
'LoanAmount': [9.28, 12.42, 7.85],
'Housing': [4.35],
'CreditHistory': [6.5]}
```
The understanding of the above map is like this:
* LoanDuration, CurrentResidenceDuration, OwnsProperty are the most contributing features across transactions for their respective prediction. Their weights for the respective prediction can also be seen.
* And the low contributing features are CreditHistory, Housing, Job, InstallmentPercent and Dependents, with their respective weights can also be seen as printed.
* We configured quality monitor and uploaded feedback data, and thereby ran the quality monitor
* For drift monitoring purposes, we created the drift detection model and uploaded to the OpenScale subscription.
* Executed the drift monitor.
Thank You! for working on tutorial notebook.
Author: Ravi Chamarthy ([email protected])
|
github_jupyter
|
# ETS models
The ETS models are a family of time series models with an underlying state space model consisting of a level component, a trend component (T), a seasonal component (S), and an error term (E).
This notebook shows how they can be used with `statsmodels`. For a more thorough treatment we refer to [1], chapter 8 (free online resource), on which the implementation in statsmodels and the examples used in this notebook are based.
`statmodels` implements all combinations of:
- additive and multiplicative error model
- additive and multiplicative trend, possibly dampened
- additive and multiplicative seasonality
However, not all of these methods are stable. Refer to [1] and references therein for more info about model stability.
[1] Hyndman, Rob J., and George Athanasopoulos. *Forecasting: principles and practice*, 3rd edition, OTexts, 2019. https://www.otexts.org/fpp3/7
```
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
%matplotlib inline
from statsmodels.tsa.exponential_smoothing.ets import ETSModel
plt.rcParams['figure.figsize'] = (12, 8)
```
## Simple exponential smoothing
The simplest of the ETS models is also known as *simple exponential smoothing*. In ETS terms, it corresponds to the (A, N, N) model, that is, a model with additive errors, no trend, and no seasonality. The state space formulation of Holt's method is:
\begin{align}
y_{t} &= y_{t-1} + e_t\\
l_{t} &= l_{t-1} + \alpha e_t\\
\end{align}
This state space formulation can be turned into a different formulation, a forecast and a smoothing equation (as can be done with all ETS models):
\begin{align}
\hat{y}_{t|t-1} &= l_{t-1}\\
l_{t} &= \alpha y_{t-1} + (1 - \alpha) l_{t-1}
\end{align}
Here, $\hat{y}_{t|t-1}$ is the forecast/expectation of $y_t$ given the information of the previous step. In the simple exponential smoothing model, the forecast corresponds to the previous level. The second equation (smoothing equation) calculates the next level as weighted average of the previous level and the previous observation.
```
oildata = [
111.0091, 130.8284, 141.2871, 154.2278,
162.7409, 192.1665, 240.7997, 304.2174,
384.0046, 429.6622, 359.3169, 437.2519,
468.4008, 424.4353, 487.9794, 509.8284,
506.3473, 340.1842, 240.2589, 219.0328,
172.0747, 252.5901, 221.0711, 276.5188,
271.1480, 342.6186, 428.3558, 442.3946,
432.7851, 437.2497, 437.2092, 445.3641,
453.1950, 454.4096, 422.3789, 456.0371,
440.3866, 425.1944, 486.2052, 500.4291,
521.2759, 508.9476, 488.8889, 509.8706,
456.7229, 473.8166, 525.9509, 549.8338,
542.3405
]
oil = pd.Series(oildata, index=pd.date_range('1965', '2013', freq='AS'))
oil.plot()
plt.ylabel("Annual oil production in Saudi Arabia (Mt)");
```
The plot above shows annual oil production in Saudi Arabia in million tonnes. The data are taken from the R package `fpp2` (companion package to prior version [1]).
Below you can see how to fit a simple exponential smoothing model using statsmodels's ETS implementation to this data. Additionally, the fit using `forecast` in R is shown as comparison.
```
model = ETSModel(oil, error='add', trend='add', damped_trend=True)
fit = model.fit(maxiter=10000)
oil.plot(label='data')
fit.fittedvalues.plot(label='statsmodels fit')
plt.ylabel("Annual oil production in Saudi Arabia (Mt)");
# obtained from R
params_R = [0.99989969, 0.11888177503085334, 0.80000197, 36.46466837, 34.72584983]
yhat = model.smooth(params_R).fittedvalues
yhat.plot(label='R fit', linestyle='--')
plt.legend();
```
By default the initial states are considered to be fitting parameters and are estimated by maximizing log-likelihood. Additionally it is possible to only use a heuristic for the initial values. In this case this leads to better agreement with the R implementation.
```
model_heuristic = ETSModel(oil, error='add', trend='add', damped_trend=True,
initialization_method='heuristic')
fit_heuristic = model_heuristic.fit()
oil.plot(label='data')
fit.fittedvalues.plot(label='estimated')
fit_heuristic.fittedvalues.plot(label='heuristic', linestyle='--')
plt.ylabel("Annual oil production in Saudi Arabia (Mt)");
# obtained from R
params = [0.99989969, 0.11888177503085334, 0.80000197, 36.46466837, 34.72584983]
yhat = model.smooth(params).fittedvalues
yhat.plot(label='with R params', linestyle=':')
plt.legend();
```
The fitted parameters and some other measures are shown using `fit.summary()`. Here we can see that the log-likelihood of the model using fitted initial states is a bit lower than the one using a heuristic for the initial states.
Additionally, we see that $\beta$ (`smoothing_trend`) is at the boundary of the default parameter bounds, and therefore it's not possible to estimate confidence intervals for $\beta$.
```
fit.summary()
fit_heuristic.summary()
```
## Holt-Winters' seasonal method
The exponential smoothing method can be modified to incorporate a trend and a seasonal component. In the additive Holt-Winters' method, the seasonal component is added to the rest. This model corresponds to the ETS(A, A, A) model, and has the following state space formulation:
\begin{align}
y_t &= l_{t-1} + b_{t-1} + s_{t-m} + e_t\\
l_{t} &= l_{t-1} + b_{t-1} + \alpha e_t\\
b_{t} &= b_{t-1} + \beta e_t\\
s_{t} &= s_{t-m} + \gamma e_t
\end{align}
```
austourists_data = [
30.05251300, 19.14849600, 25.31769200, 27.59143700,
32.07645600, 23.48796100, 28.47594000, 35.12375300,
36.83848500, 25.00701700, 30.72223000, 28.69375900,
36.64098600, 23.82460900, 29.31168300, 31.77030900,
35.17787700, 19.77524400, 29.60175000, 34.53884200,
41.27359900, 26.65586200, 28.27985900, 35.19115300,
42.20566386, 24.64917133, 32.66733514, 37.25735401,
45.24246027, 29.35048127, 36.34420728, 41.78208136,
49.27659843, 31.27540139, 37.85062549, 38.83704413,
51.23690034, 31.83855162, 41.32342126, 42.79900337,
55.70835836, 33.40714492, 42.31663797, 45.15712257,
59.57607996, 34.83733016, 44.84168072, 46.97124960,
60.01903094, 38.37117851, 46.97586413, 50.73379646,
61.64687319, 39.29956937, 52.67120908, 54.33231689,
66.83435838, 40.87118847, 51.82853579, 57.49190993,
65.25146985, 43.06120822, 54.76075713, 59.83447494,
73.25702747, 47.69662373, 61.09776802, 66.05576122,
]
index = pd.date_range("1999-03-01", "2015-12-01", freq="3MS")
austourists = pd.Series(austourists_data, index=index)
austourists.plot()
plt.ylabel('Australian Tourists');
# fit in statsmodels
model = ETSModel(austourists, error="add", trend="add", seasonal="add",
damped_trend=True, seasonal_periods=4)
fit = model.fit()
# fit with R params
params_R = [
0.35445427, 0.03200749, 0.39993387, 0.97999997, 24.01278357,
0.97770147, 1.76951063, -0.50735902, -6.61171798, 5.34956637
]
fit_R = model.smooth(params_R)
austourists.plot(label='data')
plt.ylabel('Australian Tourists')
fit.fittedvalues.plot(label='statsmodels fit')
fit_R.fittedvalues.plot(label='R fit', linestyle='--')
plt.legend();
fit.summary()
```
## Predictions
The ETS model can also be used for predicting. There are several different methods available:
- `forecast`: makes out of sample predictions
- `predict`: in sample and out of sample predictions
- `simulate`: runs simulations of the statespace model
- `get_prediction`: in sample and out of sample predictions, as well as prediction intervals
We can use them on our previously fitted model to predict from 2014 to 2020.
```
pred = fit.get_prediction(start='2014', end='2020')
df = pred.summary_frame(alpha=0.05)
df
```
In this case the prediction intervals were calculated using an analytical formula. This is not available for all models. For these other models, prediction intervals are calculated by performing multiple simulations (1000 by default) and using the percentiles of the simulation results. This is done internally by the `get_prediction` method.
We can also manually run simulations, e.g. to plot them. Since the data ranges until end of 2015, we have to simulate from the first quarter of 2016 to the first quarter of 2020, which means 17 steps.
```
simulated = fit.simulate(anchor="end", nsimulations=17, repetitions=100)
for i in range(simulated.shape[1]):
simulated.iloc[:,i].plot(label='_', color='gray', alpha=0.1)
df["mean"].plot(label='mean prediction')
df["pi_lower"].plot(linestyle='--', color='tab:blue', label='95% interval')
df["pi_upper"].plot(linestyle='--', color='tab:blue', label='_')
pred.endog.plot(label='data')
plt.legend()
```
In this case, we chose "end" as simulation anchor, which means that the first simulated value will be the first out of sample value. It is also possible to choose other anchor inside the sample.
|
github_jupyter
|
## **Bootstrap Your Own Latent A New Approach to Self-Supervised Learning:** https://arxiv.org/pdf/2006.07733.pdf
```
# !pip install torch==1.6.0+cu101 torchvision==0.7.0+cu101 -f https://download.pytorch.org/whl/torch_stable.html
# !pip install -qqU fastai fastcore
# !pip install nbdev
import fastai, fastcore, torch
fastai.__version__ , fastcore.__version__, torch.__version__
from fastai.vision.all import *
```
### Sizes
Resize -> RandomCrop
320 -> 256 | 224 -> 192 | 160 -> 128
```
resize = 320
size = 256
```
## 1. Implementation Details (Section 3.2 from the paper)
### 1.1 Image Augmentations
Same as SimCLR with optional grayscale
```
import kornia
def get_aug_pipe(size, stats=imagenet_stats, s=.6):
"SimCLR augmentations"
rrc = kornia.augmentation.RandomResizedCrop((size, size), scale=(0.2, 1.0), ratio=(3/4, 4/3))
rhf = kornia.augmentation.RandomHorizontalFlip()
rcj = kornia.augmentation.ColorJitter(0.8*s, 0.8*s, 0.8*s, 0.2*s)
rgs = kornia.augmentation.RandomGrayscale(p=0.2)
tfms = [rrc, rhf, rcj, rgs, Normalize.from_stats(*stats)]
pipe = Pipeline(tfms)
pipe.split_idx = 0
return pipe
```
### 1.2 Architecture
```
def create_encoder(arch, n_in=3, pretrained=True, cut=None, concat_pool=True):
"Create encoder from a given arch backbone"
encoder = create_body(arch, n_in, pretrained, cut)
pool = AdaptiveConcatPool2d() if concat_pool else nn.AdaptiveAvgPool2d(1)
return nn.Sequential(*encoder, pool, Flatten())
class MLP(Module):
"MLP module as described in paper"
def __init__(self, dim, projection_size=256, hidden_size=2048):
self.net = nn.Sequential(
nn.Linear(dim, hidden_size),
nn.BatchNorm1d(hidden_size),
nn.ReLU(inplace=True),
nn.Linear(hidden_size, projection_size)
)
def forward(self, x):
return self.net(x)
class BYOLModel(Module):
"Compute predictions of v1 and v2"
def __init__(self,encoder,projector,predictor):
self.encoder,self.projector,self.predictor = encoder,projector,predictor
def forward(self,v1,v2):
q1 = self.predictor(self.projector(self.encoder(v1)))
q2 = self.predictor(self.projector(self.encoder(v2)))
return (q1,q2)
def create_byol_model(arch=resnet50, hidden_size=4096, pretrained=True, projection_size=256, concat_pool=False):
encoder = create_encoder(arch, pretrained=pretrained, concat_pool=concat_pool)
with torch.no_grad():
x = torch.randn((2,3,128,128))
representation = encoder(x)
projector = MLP(representation.size(1), projection_size, hidden_size=hidden_size)
predictor = MLP(projection_size, projection_size, hidden_size=hidden_size)
apply_init(projector)
apply_init(predictor)
return BYOLModel(encoder, projector, predictor)
```
### 1.3 BYOLCallback
```
def _mse_loss(x, y):
x = F.normalize(x, dim=-1, p=2)
y = F.normalize(y, dim=-1, p=2)
return 2 - 2 * (x * y).sum(dim=-1)
def symmetric_mse_loss(pred, *yb):
(q1,q2),z1,z2 = pred,*yb
return (_mse_loss(q1,z2) + _mse_loss(q2,z1)).mean()
x = torch.randn((64,256))
y = torch.randn((64,256))
test_close(symmetric_mse_loss((x,y),y,x), 0) # perfect
test_close(symmetric_mse_loss((x,y),x,y), 4, 1e-1) # random
```
Useful Discussions and Supportive Material:
- https://www.reddit.com/r/MachineLearning/comments/hju274/d_byol_bootstrap_your_own_latent_cheating/fwohtky/
- https://untitled-ai.github.io/understanding-self-supervised-contrastive-learning.html
```
import copy
class BYOLCallback(Callback):
"Implementation of https://arxiv.org/pdf/2006.07733.pdf"
def __init__(self, T=0.99, debug=True, size=224, **aug_kwargs):
self.T, self.debug = T, debug
self.aug1 = get_aug_pipe(size, **aug_kwargs)
self.aug2 = get_aug_pipe(size, **aug_kwargs)
def before_fit(self):
"Create target model"
self.target_model = copy.deepcopy(self.learn.model).to(self.dls.device)
self.T_sched = SchedCos(self.T, 1) # used in paper
# self.T_sched = SchedNo(self.T, 1) # used in open source implementation
def before_batch(self):
"Generate 2 views of the same image and calculate target projections for these views"
if self.debug: print(f"self.x[0]: {self.x[0]}")
v1,v2 = self.aug1(self.x), self.aug2(self.x.clone())
self.learn.xb = (v1,v2)
if self.debug:
print(f"v1[0]: {v1[0]}\nv2[0]: {v2[0]}")
self.show_one()
assert not torch.equal(*self.learn.xb)
with torch.no_grad():
z1 = self.target_model.projector(self.target_model.encoder(v1))
z2 = self.target_model.projector(self.target_model.encoder(v2))
self.learn.yb = (z1,z2)
def after_step(self):
"Update target model and T"
self.T = self.T_sched(self.pct_train)
with torch.no_grad():
for param_k, param_q in zip(self.target_model.parameters(), self.model.parameters()):
param_k.data = param_k.data * self.T + param_q.data * (1. - self.T)
def show_one(self):
b1 = self.aug1.normalize.decode(to_detach(self.learn.xb[0]))
b2 = self.aug1.normalize.decode(to_detach(self.learn.xb[1]))
i = np.random.choice(len(b1))
show_images([b1[i],b2[i]], nrows=1, ncols=2)
def after_train(self):
if self.debug: self.show_one()
def after_validate(self):
if self.debug: self.show_one()
```
## 2. Pretext Training
```
sqrmom=0.99
mom=0.95
beta=0.
eps=1e-4
opt_func = partial(ranger, mom=mom, sqr_mom=sqrmom, eps=eps, beta=beta)
bs=128
def get_dls(size, bs, workers=None):
path = URLs.IMAGEWANG_160 if size <= 160 else URLs.IMAGEWANG
source = untar_data(path)
files = get_image_files(source)
tfms = [[PILImage.create, ToTensor, RandomResizedCrop(size, min_scale=0.9)],
[parent_label, Categorize()]]
dsets = Datasets(files, tfms=tfms, splits=RandomSplitter(valid_pct=0.1)(files))
batch_tfms = [IntToFloatTensor]
dls = dsets.dataloaders(bs=bs, num_workers=workers, after_batch=batch_tfms)
return dls
dls = get_dls(resize, bs)
model = create_byol_model(arch=xresnet34, pretrained=False)
learn = Learner(dls, model, symmetric_mse_loss, opt_func=opt_func,
cbs=[BYOLCallback(T=0.99, size=size, debug=False), TerminateOnNaNCallback()])
learn.to_fp16();
learn.lr_find()
lr=1e-3
wd=1e-2
epochs=100
learn.unfreeze()
learn.fit_flat_cos(epochs, lr, wd=wd, pct_start=0.5)
save_name = f'byol_iwang_sz{size}_epc{epochs}'
learn.save(save_name)
torch.save(learn.model.encoder.state_dict(), learn.path/learn.model_dir/f'{save_name}_encoder.pth')
learn.load(save_name);
lr=1e-4
wd=1e-2
epochs=100
learn.unfreeze()
learn.fit_flat_cos(epochs, lr, wd=wd, pct_start=0.5)
save_name = f'byol_iwang_sz{size}_epc200'
learn.save(save_name)
torch.save(learn.model.encoder.state_dict(), learn.path/learn.model_dir/f'{save_name}_encoder.pth')
lr=1e-4
wd=1e-2
epochs=30
learn.unfreeze()
learn.fit_flat_cos(epochs, lr, wd=wd, pct_start=0.5)
save_name = f'byol_iwang_sz{size}_epc230'
learn.save(save_name)
torch.save(learn.model.encoder.state_dict(), learn.path/learn.model_dir/f'{save_name}_encoder.pth')
lr=5e-5
wd=1e-2
epochs=30
learn.unfreeze()
learn.fit_flat_cos(epochs, lr, wd=wd, pct_start=0.5)
save_name = f'byol_iwang_sz{size}_epc260'
learn.save(save_name)
torch.save(learn.model.encoder.state_dict(), learn.path/learn.model_dir/f'{save_name}_encoder.pth')
learn.recorder.plot_loss()
save_name
```
## 3. Downstream Task - Image Classification
```
def get_dls(size, bs, workers=None):
path = URLs.IMAGEWANG_160 if size <= 160 else URLs.IMAGEWANG
source = untar_data(path)
files = get_image_files(source, folders=['train', 'val'])
splits = GrandparentSplitter(valid_name='val')(files)
item_aug = [RandomResizedCrop(size, min_scale=0.35), FlipItem(0.5)]
tfms = [[PILImage.create, ToTensor, *item_aug],
[parent_label, Categorize()]]
dsets = Datasets(files, tfms=tfms, splits=splits)
batch_tfms = [IntToFloatTensor, Normalize.from_stats(*imagenet_stats)]
dls = dsets.dataloaders(bs=bs, num_workers=workers, after_batch=batch_tfms)
return dls
def do_train(epochs=5, runs=5, lr=2e-2, size=size, bs=bs, save_name=None):
dls = get_dls(size, bs)
for run in range(runs):
print(f'Run: {run}')
learn = cnn_learner(dls, xresnet34, opt_func=opt_func, normalize=False,
metrics=[accuracy,top_k_accuracy], loss_func=LabelSmoothingCrossEntropy(),
pretrained=False)
# learn.to_fp16()
if save_name is not None:
state_dict = torch.load(learn.path/learn.model_dir/f'{save_name}_encoder.pth')
learn.model[0].load_state_dict(state_dict)
print("Model loaded...")
learn.unfreeze()
learn.fit_flat_cos(epochs, lr, wd=wd)
```
### ImageWang Leaderboard
**sz-256**
**Contrastive Learning**
- 5 epochs: 67.70%
- 20 epochs: 70.03%
- 80 epochs: 70.71%
- 200 epochs: 71.78%
**BYOL**
- 5 epochs: 64.74%
- 20 epochs: **71.01%**
- 80 epochs: **72.58%**
- 200 epochs: **72.13%**
### 5 epochs
```
# we are using old pretrained model with size 192 for transfer learning
# link: https://github.com/KeremTurgutlu/self_supervised/blob/252269827da41b41091cf0db533b65c0d1312f85/nbs/byol_iwang_192.ipynb
save_name = 'byol_iwang_sz192_epc230'
lr = 1e-2
wd=1e-2
bs=128
epochs = 5
runs = 5
do_train(epochs, runs, lr=lr, bs=bs, save_name=save_name)
np.mean([0.657165,0.637312,0.631967,0.646729,0.664291])
```
### 20 epochs
```
lr=2e-2
epochs = 20
runs = 3
do_train(epochs, runs, lr=lr, save_name=save_name)
np.mean([0.711631, 0.705269, 0.713413])
```
### 80 epochs
```
epochs = 80
runs = 1
do_train(epochs, runs, save_name=save_name)
```
### 200 epochs
```
epochs = 200
runs = 1
do_train(epochs, runs, save_name=save_name)
```
|
github_jupyter
|
# Fitbit Data Analysis
## About Fitbit Data Analysis
This project provides some high-level data analysis of steps, sleep, heart rate and weight data from Fitbit tracking.
Please using fitbit_downloader file to first collect and export your data.
-------
### Dependencies and Libraries
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt, matplotlib.font_manager as fm
from datetime import datetime
import seaborn
%matplotlib inline
```
-------
# Steps
```
daily_steps = pd.read_csv('data/daily_steps.csv', encoding='utf-8')
daily_steps['Date'] = pd.to_datetime(daily_steps['Date'])
daily_steps['dow'] = daily_steps['Date'].dt.weekday
daily_steps['day_of_week'] = daily_steps['Date'].dt.weekday_name
daily_steps.tail()
len(daily_steps)
# drop days with now steps
daily_steps = daily_steps[daily_steps.Steps > 0]
len(daily_steps)
daily_steps.Steps.max()
daily_steps.Steps.min()
daily_steps.Steps.max()
```
### Step Charts
```
daily_steps['RollingMeanSteps'] = daily_steps.Steps.rolling(window=10, center=True).mean()
daily_steps.plot(x='Date', y='RollingMeanSteps', title= 'Daily step counts rolling mean over 10 days')
daily_steps.groupby(['dow'])['Steps'].mean()
ax = daily_steps.groupby(['dow'])['Steps'].mean().plot(kind='bar', x='day_of_week')
plt.suptitle('Average Steps by Day of the Week', fontsize=16)
plt.xlabel('Day of Week: 0 = Monday, 6 = Sunday', fontsize=12, color='red')
```
# Sleep
```
daily_sleep = pd.read_csv('data/daily_sleep.csv', encoding='utf-8')
daily_inbed = pd.read_csv('data/daily_inbed.csv', encoding='utf-8')
len(daily_sleep)
sleep_data = pd.merge(daily_sleep, daily_inbed, how='inner', on='Date')
sleep_data['Date'] = pd.to_datetime(sleep_data['Date'])
sleep_data['dow'] = sleep_data['Date'].dt.weekday
sleep_data['day_of_week'] = sleep_data['Date'].dt.weekday_name
sleep_data['day_of_week'] = sleep_data["day_of_week"].astype('category')
sleep_data['InBedHours'] = round((sleep_data.InBed / 60), 2)
sleep_data = sleep_data[sleep_data.Sleep > 0]
len(daily_sleep)
sleep_data.info()
sleep_data.tail()
sleep_data.describe()
sleep_data.plot(x='Date', y='Hours')
sleep_data['RollingMeanSleep'] = sleep_data.Sleep.rolling(window=10, center=True).mean()
sleep_data.plot(x='Date', y='RollingMeanSleep', title= 'Daily sleep counts rolling mean over 10 days')
sleep_data.groupby(['dow'])['Hours'].mean()
ax = sleep_data.groupby(['dow'])['Hours'].mean().plot(kind='bar', x='day_of_week')
plt.suptitle('Average Sleep by Night of the Week', fontsize=16)
plt.xlabel('Day of Week: 0 = Monday, 6 = Sunday', fontsize=12, color='red')
```
|
github_jupyter
|
# R API Serving Examples
In this example, we demonstrate how to quickly compare the runtimes of three methods for serving a model from an R hosted REST API. The following SageMaker examples discuss each method in detail:
* **Plumber**
* Website: [https://www.rplumber.io/](https://www.rplumber.io)
* SageMaker Example: [r_serving_with_plumber](../r_serving_with_plumber)
* **RestRServe**
* Website: [https://restrserve.org](https://restrserve.org)
* SageMaker Example: [r_serving_with_restrserve](../r_serving_with_restrserve)
* **FastAPI** (reticulated from Python)
* Website: [https://fastapi.tiangolo.com](https://fastapi.tiangolo.com)
* SageMaker Example: [r_serving_with_fastapi](../r_serving_with_fastapi)
We will reuse the docker images from each of these examples. Each one is configured to serve a small XGBoost model which has already been trained on the classical Iris dataset.
## Building Docker Images for Serving
First, we will build each docker image from the provided SageMaker Examples.
### Plumber Serving Image
```
!cd .. && docker build -t r-plumber -f r_serving_with_plumber/Dockerfile r_serving_with_plumber
```
### RestRServe Serving Image
```
!cd .. && docker build -t r-restrserve -f r_serving_with_restrserve/Dockerfile r_serving_with_restrserve
```
### FastAPI Serving Image
```
!cd .. && docker build -t r-fastapi -f r_serving_with_fastapi/Dockerfile r_serving_with_fastapi
```
## Launch Serving Containers
Next, we will launch each search container. The containers will be launch on the following ports to avoid port collisions on your local machine or SageMaker Notebook instance:
```
ports = {
"plumber": 5000,
"restrserve": 5001,
"fastapi": 5002,
}
!bash launch.sh
!docker container list
```
## Define Simple Client
```
import requests
from tqdm import tqdm
import pandas as pd
def get_predictions(examples, instance=requests, port=5000):
payload = {"features": examples}
return instance.post(f"http://127.0.0.1:{port}/invocations", json=payload)
def get_health(instance=requests, port=5000):
instance.get(f"http://127.0.0.1:{port}/ping")
```
## Define Example Inputs
Next, we define a example inputs from the classical [Iris](https://archive.ics.uci.edu/ml/datasets/iris) dataset.
* Dua, D. and Graff, C. (2019). UCI Machine Learning Repository [http://archive.ics.uci.edu/ml]. Irvine, CA: University of California, School of Information and Computer Science.
```
column_names = ["Sepal.Length", "Sepal.Width", "Petal.Length", "Petal.Width", "Label"]
iris = pd.read_csv(
"s3://sagemaker-sample-files/datasets/tabular/iris/iris.data", names=column_names
)
iris_features = iris[["Sepal.Length", "Sepal.Width", "Petal.Length", "Petal.Width"]]
example = iris_features.values[:1].tolist()
many_examples = iris_features.values[:100].tolist()
```
## Testing
Now it's time to test how each API server performs under stress.
We will test two use cases:
* **New Requests**: In this scenario, we test how quickly the server can respond with predictions when each client request establishes a new connection with the server. This simulates the server's ability to handle real-time requests. We could make this more realistic by creating an asynchronous environment that tests the server's ability to fulfill concurrent rather than sequential requests.
* **Keep Alive / Reuse Session**: In this scenario, we test how quickly the server can respond with predictions when each client request uses a session to keep its connection to the server alive between requests. This simulates the server's ability to handle sequential batch requests from the same client.
For each of the two use cases, we will test the performance on following situations:
* 1000 requests of a single example
* 1000 requests of 100 examples
* 1000 pings for health status
## New Requests
### Plumber
```
# verify the prediction output
get_predictions(example, port=ports["plumber"]).json()
for i in tqdm(range(1000)):
_ = get_predictions(example, port=ports["plumber"])
for i in tqdm(range(1000)):
_ = get_predictions(many_examples, port=ports["plumber"])
for i in tqdm(range(1000)):
get_health(port=ports["plumber"])
```
### RestRserve
```
# verify the prediction output
get_predictions(example, port=ports["restrserve"]).json()
for i in tqdm(range(1000)):
_ = get_predictions(example, port=ports["restrserve"])
for i in tqdm(range(1000)):
_ = get_predictions(many_examples, port=ports["restrserve"])
for i in tqdm(range(1000)):
get_health(port=ports["restrserve"])
```
### FastAPI
```
# verify the prediction output
get_predictions(example, port=ports["fastapi"]).json()
for i in tqdm(range(1000)):
_ = get_predictions(example, port=ports["fastapi"])
for i in tqdm(range(1000)):
_ = get_predictions(many_examples, port=ports["fastapi"])
for i in tqdm(range(1000)):
get_health(port=ports["fastapi"])
```
## Keep Alive (Reuse Session)
Now, let's test how each one performs when each request reuses a session connection.
```
# reuse the session for each post and get request
instance = requests.Session()
```
### Plumber
```
for i in tqdm(range(1000)):
_ = get_predictions(example, instance=instance, port=ports["plumber"])
for i in tqdm(range(1000)):
_ = get_predictions(many_examples, instance=instance, port=ports["plumber"])
for i in tqdm(range(1000)):
get_health(instance=instance, port=ports["plumber"])
```
### RestRserve
```
for i in tqdm(range(1000)):
_ = get_predictions(example, instance=instance, port=ports["restrserve"])
for i in tqdm(range(1000)):
_ = get_predictions(many_examples, instance=instance, port=ports["restrserve"])
for i in tqdm(range(1000)):
get_health(instance=instance, port=ports["restrserve"])
```
### FastAPI
```
for i in tqdm(range(1000)):
_ = get_predictions(example, instance=instance, port=ports["fastapi"])
for i in tqdm(range(1000)):
_ = get_predictions(many_examples, instance=instance, port=ports["fastapi"])
for i in tqdm(range(1000)):
get_health(instance=instance, port=ports["fastapi"])
```
### Stop All Serving Containers
Finally, we will shut down the serving containers we launched for the tests.
```
!docker kill $(docker ps -q)
```
## Conclusion
In this example, we demonstrated how to conduct a simple performance benchmark across three R model serving solutions. We leave the choice of serving solution up to the reader since in some cases it might be appropriate to customize the benchmark in the following ways:
* Update the serving example to serve a specific model
* Perform the tests across multiple instances types
* Modify the serving example and client to test asynchronous requests.
* Deploy the serving examples to SageMaker Endpoints to test within an autoscaling environment.
For more information on serving your models in custom containers on SageMaker, please see our [support documentation](https://docs.aws.amazon.com/sagemaker/latest/dg/your-algorithms-inference-main.html) for the latest updates and best practices.
|
github_jupyter
|
** Build Adjacency Matrix **
**Note:** You must put the generated JSON file into a zip file. We probably should code this in too.
```
import sqlite3
import json
# Progress Bar I found on the internet.
# https://github.com/alexanderkuk/log-progress
from progress_bar import log_progress
PLOS_PMC_DB = 'sqlite_data/data.plos-pmc.sqlite'
ALL_DB = 'sqlite_data/data.all.sqlite'
PLOS_PMC_MATRIX = 'json_data/plos-pmc/adjacency_matrix.json'
ALL_MATRIX = 'json_data/all/adjacency_matrix.json'
conn_plos_pmc = sqlite3.connect(PLOS_PMC_DB)
cursor_plos_pmc = conn_plos_pmc.cursor()
conn_all = sqlite3.connect(ALL_DB)
cursor_all = conn_all.cursor()
```
Queries
```
# For getting the maximum row id
QUERY_MAX_ID = "SELECT id FROM interactions ORDER BY id DESC LIMIT 1"
# Get interaction data
QUERY_INTERACTION = "SELECT geneids1, geneids2, probability FROM interactions WHERE id = {}"
# Get all at once
QUERY_ALL_INTERACTION = "SELECT geneids1, geneids2, probability FROM interactions"
actions = [
# {
# "db":PLOS_PMC_DB,
# "matrix" : PLOS_PMC_MATRIX,
# "conn": conn_plos_pmc,
# "cursor": cursor_plos_pmc,
# },
{
"db":ALL_DB,
"matrix" : ALL_MATRIX,
"conn": conn_all,
"cursor": cursor_all,
},
]
```
Step through every interaction.
1. If geneids1 not in matrix - insert it as dict.
2. If geneids2 not in matrix[geneids1] - insert it as []
3. If probability not in matrix[geneids1][geneids2] - insert it.
4. Perform the reverse.
```
# for action in actions:
for action in log_progress(actions, every=1, name="Matrix"):
print("Executing SQL query. May take a minute.")
matrix = {}
cursor = action["cursor"].execute(QUERY_ALL_INTERACTION)
interactions = cursor.fetchall()
print("Query complete")
for row in log_progress(interactions, every=10000, name=action["matrix"]+" rows"):
if row == None:
continue
id1 = row[0]
id2 = row[1]
try:
prob = int(round(row[2],2) * 1000)
except Exception:
continue
# Forward
if id1 not in matrix:
matrix[id1] = {}
if id2 not in matrix[id1]:
matrix[id1][id2] = []
if prob not in matrix[id1][id2]:
matrix[id1][id2].append(prob)
# Backwards
if id2 not in matrix:
matrix[id2] = {}
if id1 not in matrix[id2]:
matrix[id2][id1] = []
if prob not in matrix[id2][id1]:
matrix[id2][id1].append(prob)
with open(action["matrix"], "w+") as file:
file.write(json.dumps( matrix ))
print("All Matrices generated")
action["conn"].close()
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/skredenmathias/DS-Unit-2-Applied-Modeling/blob/master/module4/assignment_applied_modeling_1.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
Lambda School Data Science
*Unit 2, Sprint 3, Module 1*
---
# Define ML problems
You will use your portfolio project dataset for all assignments this sprint.
## Assignment
Complete these tasks for your project, and document your decisions.
- [ ] Choose your target. Which column in your tabular dataset will you predict?
- [ ] Is your problem regression or classification?
- [ ] How is your target distributed?
- Classification: How many classes? Are the classes imbalanced?
- Regression: Is the target right-skewed? If so, you may want to log transform the target.
- [ ] Choose your evaluation metric(s).
- Classification: Is your majority class frequency >= 50% and < 70% ? If so, you can just use accuracy if you want. Outside that range, accuracy could be misleading. What evaluation metric will you choose, in addition to or instead of accuracy?
- Regression: Will you use mean absolute error, root mean squared error, R^2, or other regression metrics?
- [ ] Choose which observations you will use to train, validate, and test your model.
- Are some observations outliers? Will you exclude them?
- Will you do a random split or a time-based split?
- [ ] Begin to clean and explore your data.
- [ ] Begin to choose which features, if any, to exclude. Would some features "leak" future information?
If you haven't found a dataset yet, do that today. [Review requirements for your portfolio project](https://lambdaschool.github.io/ds/unit2) and choose your dataset.
```
# I will use the worlds_2019 dataset for now.
import pandas as pd
!git clone https://github.com/skredenmathias/DS-Unit-1-Build.git
path = '/content/DS-Unit-1-Build/'
worlds_2019 = pd.read_excel(path+'2019-summer-match-data-OraclesElixir-2019-11-10.xlsx')
(print(worlds_2019.shape))
worlds_2019.head()
```
Choose your target. Which column in your tabular dataset will you predict?
```
df = worlds_2019
target = df['result']
# Initially I seek if I can predict if a team will win or lose.
# The goal is to see how much variance is explained by each factor.
# / see how much certain factors contribute to the result.
# From here I might look at questions such as:
# How do win conditions change per patch?
# What are the win percentages for red / blue side? Are different objectives
# more important to one side?
# See how different positions have different degrees of impact based on teams.
#
```
Is your problem regression or classification?
```
# Classification.
```
How is your target distributed?
Classification: How many classes? Are the classes imbalanced?
```
target.value_counts(normalize=True)
```
Choose your evaluation metric(s).
Classification: Is your majority class frequency >= 50% and < 70% ?
If so, you can just use accuracy if you want. Outside that range, accuracy could be misleading.
What evaluation metric will you choose, in addition to or instead of accuracy?
```
# Accuracy. What others could I choose?
```
Choose which observations you will use to train, validate, and test your model.
Are some observations outliers? Will you exclude them?
Will you do a random split or a time-based split?
```
# Depends on ceteris paribus, other things equal:
# I might have to keep it on the same patch.
# Feature importances will differ across regions & tournaments & patches.
# Gamelength might be a leak?
# Should I also make a separate df with all 5 players grouped as a team w/
# most of the stats retained?
# Outliers:
# Gamelength beyond 50 minutes. I can filter these out if needed.
# Leaks / uninteresting columns:
# gameid, url, (league), (split), date, week, game, (patchno), playerid,
# (position), (team), gamelength?, total gold?, firsttothreetowers?,
# teamtowerkills?, opptowerkills?,
df.head()
df.columns
```
Begin to clean and explore your data.
```
# Lot's of cleaning done in the unit 1 build notebook.
# Will focus on exploration here for now.
df['gamelength'].plot.hist() # We see a small outlier here.
df['gamelength'].describe()
import seaborn as sns
# Note: Will be other outliers in the big dataset.
# Couldn't upload full dataset to Git, 80mb is too big.
# Why is it so big, it's just a text file?
sns.distplot(df['gamelength']);
```
# Fast first model
```
from sklearn.model_selection import train_test_split
train, val = train_test_split(df, test_size=.25)
!pip install category_encoders
import category_encoders as ce
from sklearn.pipeline import make_pipeline
from sklearn.ensemble import RandomForestClassifier
from sklearn.impute import SimpleImputer
from sklearn.metrics import accuracy_score
target = 'result'
X_train = train.drop(columns=target)
y_train = train[target]
X_val = val.drop(columns=target)
y_val = val[target]
# X_test = test.drop(columns=target)
# y_test = test[target]
X_train.shape, X_val.shape
pipeline = make_pipeline(
ce.OrdinalEncoder(),
SimpleImputer(),
RandomForestClassifier(random_state=42, n_jobs=-1)
)
pipeline.fit(X_train, y_train)
# Get validation accuracy
y_pred = pipeline.predict(X_val)
print('Validation Accuracy:', pipeline.score(X_val, y_val)) # We've got leakage!
print('X_train shape before encoding', X_train.shape)
encoder = pipeline.named_steps['ordinalencoder']
encoded = encoder.transform(X_train)
print('X_train shape after encoding', encoded.shape)
# Plot feature importances to find leak
%matplotlib inline
import matplotlib.pyplot as plt
# Get feature importances
rf = pipeline.named_steps['randomforestclassifier']
importances = pd.Series(rf.feature_importances_, encoded.columns)
# Plot top n feature importances
n = 20
plt.figure(figsize=(10,n/2))
plt.title(f'Top {n} features')
importances.sort_values()[-n:].plot.barh(color='grey')
```
# XGBoost
```
from xgboost import XGBClassifier
pipeline = make_pipeline(
ce.OrdinalEncoder(),
XGBClassifier(n_estimators=100, random_state=42, n_jobs=-1)
)
pipeline.fit(X_train, y_train)
from sklearn.metrics import accuracy_score
y_pred = pipeline.predict(X_val)
print('Validation Accuracy', accuracy_score(y_val, y_pred))
```
# Partial dependence plots
```
import matplotlib.pyplot as plt
# plt.rcParams['figure.dpi] = 72
!pip install pdpbox
!pip install shap
from pdpbox.pdp import pdp_isolate, pdp_plot
feature = 'teamtowerkills'
isolated = pdp_isolate(
model=pipeline,
dataset=X_val,
model_features=X_val.columns,
feature=feature,
num_grid_points=50
)
pdp_plot(isolated, feature_name=feature, plot_lines=True,
frac_to_plot=0.1) # leakage
plt.xlim(5, 12);
```
# Permutation importances
```
!pip install eli5
import eli5
from eli5.sklearn import PermutationImportance
transformers = make_pipeline(
ce.OrdinalEncoder(),
SimpleImputer(strategy='median')
)
X_train_transformed = transformers.fit_transform(X_train)
X_val_transformed = transformers.transform(X_val)
model = RandomForestClassifier(n_estimators=100, random_state=42, n_jobs=-1)
model.fit(X_train_transformed, y_train)
permuter = PermutationImportance(
model,
scoring='accuracy',
n_iter=10,
random_state=42
)
permuter.fit(X_val_transformed, y_val)
feature_names = X_val.columns.tolist()
pd.Series(permuter.feature_importances_, feature_names).sort_values()
eli5.show_weights(
permuter,
top=None,
feature_names=feature_names
)
```
# Dropping 'teamtowerkills' & 'opptowerkills'
```
def wrangle(X):
X = X.copy()
# Drop teamtowerkills & opptowerkills
model_breakers = ['teamtowerkills','opptowerkills']
X = X.drop(columns = model_breakers)
return X
# train = wrangle(train)
val = wrangle(val)
val.columns
train.shape, val.shape
```
# Running XGBoost again
```
X_train = train.drop(columns=target)
y_train = train[target]
X_val = val.drop(columns=target)
y_val = val[target]
pipeline = make_pipeline(
ce.OrdinalEncoder(),
XGBClassifier(n_estimators=100, random_state=42, n_jobs=-1)
)
pipeline.fit(X_train, y_train)
from sklearn.metrics import accuracy_score
y_pred = pipeline.predict(X_val)
print('Validation Accuracy', accuracy_score(y_val, y_pred))
```
# Feature importances, again
```
transformers = make_pipeline(
ce.OrdinalEncoder(),
SimpleImputer(strategy='median')
)
X_train_transformed = transformers.fit_transform(X_train)
X_val_transformed = transformers.transform(X_val)
model = RandomForestClassifier(n_estimators=100, random_state=42, n_jobs=-1)
model.fit(X_train_transformed, y_train)
permuter = PermutationImportance(
model,
scoring='accuracy',
n_iter=10,
random_state=42
)
permuter.fit(X_val_transformed, y_val)
feature_names = X_val.columns.tolist()
pd.Series(permuter.feature_importances_, feature_names).sort_values()
eli5.show_weights(
permuter,
top=None,
feature_names=feature_names
)
```
# Dropping a fuckton of columns, then repeat
```
# Leaks / uninteresting columns:
# gameid, url, (league), (split), date, week, game, (patchno), playerid,
# (position), (team), gamelength?, total gold?, firsttothreetowers?,
# teamtowerkills?, opptowerkills?,
def wrangle2(X):
X = X.copy()
# Drops
low_importance = ['gameid', 'url', 'league', 'split', 'date', 'week',
'patchno', 'position', 'gamelength']
X = X.drop(columns = low_importance)
return X
# train = wrangle(train)
train = wrangle2(train)
val = wrangle2(val)
train.columns
X_train = train.drop(columns=target)
y_train = train[target]
X_val = val.drop(columns=target)
y_val = val[target]
pipeline = make_pipeline(
ce.OrdinalEncoder(),
XGBClassifier(n_estimators=100, random_state=42, n_jobs=-1)
)
pipeline.fit(X_train, y_train)
from sklearn.metrics import accuracy_score
y_pred = pipeline.predict(X_val)
print('Validation Accuracy', accuracy_score(y_val, y_pred))
```
# Feature importances, iterations
```
transformers = make_pipeline(
ce.OrdinalEncoder(),
SimpleImputer(strategy='median')
)
X_train_transformed = transformers.fit_transform(X_train)
X_val_transformed = transformers.transform(X_val)
model = RandomForestClassifier(n_estimators=100, random_state=42, n_jobs=-1)
model.fit(X_train_transformed, y_train)
permuter = PermutationImportance(
model,
scoring='accuracy',
n_iter=10,
random_state=42
)
permuter.fit(X_val_transformed, y_val)
feature_names = X_val.columns.tolist()
pd.Series(permuter.feature_importances_, feature_names).sort_values()
eli5.show_weights(
permuter,
top=None,
feature_names=feature_names
)
```
# Shapley plot
```
row = X_val.iloc[[0]]
y_val.iloc[[0]]
row #model.predict(row)
import shap
explainer = shap.TreeExplainer(model)
shap_values = explainer.shap_values(row)
shap.initjs()
shap.force_plot(
base_value = explainer.expected_value,
shap_values = shap_values,
features=row
)
```
# Using importances for feature selection
```
X_train.shape
minimum_importance = 0
mask = permuter.feature_importances_ > minimum_importance
```
|
github_jupyter
|
# KNN
Importing required python modules
---------------------------------
```
import matplotlib.pyplot as plt
from sklearn.neighbors import KNeighborsClassifier
from sklearn.cross_validation import train_test_split
from sklearn import metrics
from sklearn.preprocessing import normalize,scale
from sklearn.cross_validation import cross_val_score
import numpy as np
import pandas as pd
```
The following libraries have been used :
* **Pandas** : pandas is an open source, BSD-licensed library providing high-performance, easy-to-use data structures and data analysis tools for the Python programming language.
* **Numpy** : NumPy is the fundamental package for scientific computing with Python.
* **Matplotlib** : matplotlib is a python 2D plotting library which produces publication quality figures in a variety of hardcopy formats and interactive environments .
* **Sklearn** : It features various classification, regression and clustering algorithms including support vector machines, random forests, gradient boosting, k-means and DBSCAN, and is designed to interoperate with the Python numerical and scientific libraries NumPy and SciPy.
Retrieving the dataset
----------------------
```
data = pd.read_csv('heart.csv', header=None)
df = pd.DataFrame(data)
x = df.iloc[:, 0:5]
x = x.drop(x.columns[1:3], axis=1)
x = pd.DataFrame(scale(x))
y = df.iloc[:, 13]
y = y-1
```
1. Dataset is imported.
2. The imported dataset is converted into a pandas DataFrame.
3. Attributes(x) and labels(y) are extracted.
```
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.4)
```
Train/Test split is 0.4
Plotting the dataset
--------------------
```
fig = plt.figure()
ax1 = fig.add_subplot(1,2,1)
ax1.scatter(x[1],x[2], c=y)
ax1.set_title("Original Data")
```
Matplotlib is used to plot the loaded pandas DataFrame.
Learning from the data
----------------------
```
model = KNeighborsClassifier(n_neighbors=5)
scores = cross_val_score(model, x, y, scoring='accuracy', cv=10)
print ("10-Fold Accuracy : ", scores.mean()*100)
model.fit(x_train,y_train)
print ("Testing Accuracy : ",model.score(x_test, y_test)*100)
predicted = model.predict(x)
```
Here **model** is an instance of KNeighborsClassifier method from sklearn.neighbors. 10 Fold Cross Validation is used to verify the results.
```
ax2 = fig.add_subplot(1,2,2)
ax2.scatter(x[1],x[2], c=predicted)
ax2.set_title("KNearestNeighbours")
```
The learned data is plotted.
```
cm = metrics.confusion_matrix(y, predicted)
print (cm/len(y))
print (metrics.classification_report(y, predicted))
plt.show()
```
Compute confusion matrix to evaluate the accuracy of a classification and build a text report showing the main classification metrics.
|
github_jupyter
|
# Jupyter Superpower - Extend SQL analysis with Python
> Making collboration with Notebook possible and share perfect SQL analysis with Notebook.
- toc: true
- badges: true
- comments: true
- author: noklam
- categories: ["python", "reviewnb", "sql"]
- hide: false
- canonical_url: https://blog.reviewnb.com/jupyter-sql-notebook/
If you have ever written SQL queries to extract data from a database, chances are you are familiar with an IDE like the screenshot below. The IDE offers features like auto-completion, visualize the query output, display the table schema and the ER diagram. Whenever you need to write a query, this is your go-to tool. However, you may want to add `Jupyter Notebook` into your toolkit. It improves my productivity by complementing some missing features in IDE.
")
```
#collapse-hide
# !pip install ipython_sql
%load_ext sql
%config SqlMagic.displaycon = False
%config SqlMagic.feedback = False
# Download the file from https://github.com/cwoodruff/ChinookDatabase/blob/master/Scripts/Chinook_Sqlite.sqlite
%sql sqlite:///sales.sqlite.db
from pathlib import Path
DATA_DIR = Path('../_demo/sql_notebook')
%%sql
select ProductId, Sum(Unit) from Sales group by ProductId;
```
## Notebook as a self-contained report
As a data scientist/data analyst, you write SQL queries for ad-hoc analyses all the time. After getting the right data, you make nice-looking charts and put them in a PowerPoint and you are ready to present your findings. Unlike a well-defined ETL job, you are exploring the data and testing your hypotheses all the time. You make assumptions, which is often wrong but you only realized it after a few weeks. But all you got is a CSV that you cannot recall how it was generated in the first place.
Data is not stationary, why should your analysis be? I have seen many screenshots, fragmented scripts flying around in organizations. As a data scientist, I learned that you need to be cautious about what you heard. Don't trust peoples' words easily, verify the result! To achieve that, we need to know exactly how the data was extracted, what kind of assumptions have been made? Unfortunately, this information usually is not available. As a result, people are redoing the same analysis over and over. You will be surprised that this is very common in organizations. In fact, numbers often do not align because every department has its own definition for a given metric. It is not shared among the organization, and verbal communication is inaccurate and error-prone. It would be really nice if anyone in the organization can reproduce the same result with just a single click. Jupyter Notebook can achieve that reproducibility and keep your entire analysis (documentation, data, and code) in the same place.
## Notebook as an extension of IDE
Writing SQL queries in a notebook gives you extra flexibility of a full programming language alongside SQL.
For example:
* Write complex processing logic that is not easy in pure SQL
* Create visualizations directly from SQL results without exporting to an intermediate CSV
For instance, you can pipe your `SQL` query with `pandas` and then make a plot. It allows you to generate analysis with richer content. If you find bugs in your code, you can modify the code and re-run the analysis. This reduces the hustles to reproduce an analysis greatly. In contrast, if your analysis is reading data from an anonymous exported CSV, it is almost guaranteed that the definition of the data will be lost. No one will be able to reproduce the dataset.
You can make use of the `ipython_sql` library to make queries in a notebook. To do this, you need to use the **magic** function with the inline magic `%` or cell magic `%%`.
```
sales = %sql SELECT * from sales LIMIT 3
sales
```
To make it fancier, you can even parameterize your query with variables. Tools like [papermill](https://www.bing.com/search?q=github+paramter+notebook&cvid=5b17218ec803438fb1ca41212d53d90a&FORM=ANAB01&PC=U531) allows you to parameterize your notebook. If you execute the notebook regularly with a scheduler, you can get a updated dashboard. To reference the python variable, the `$` sign is used.
```
table = "sales"
query = f"SELECT * from {table} LIMIT 3"
sales = %sql $query
sales
```
With a little bit of python code, you can make a nice plot to summarize your finding. You can even make an interactive plot if you want. This is a very powerful way to extend your analysis.
```
import seaborn as sns
sales = %sql SELECT * FROM SALES
sales_df = sales.DataFrame()
sales_df = sales_df.groupby('ProductId', as_index=False).sum()
ax = sns.barplot(x='ProductId', y='Unit', data=sales_df)
ax.set_title('Sales by ProductId');
```
## Notebook as a collaboration tool
Jupyter Notebook is flexible and it fits extremely well with exploratory data analysis. To share to a non-coder, you can share the notebook or export it as an HTML file. They can read the report or any cached executed result. If they need to verify the data or add some extra plots, they can do it easily themselves.
It is true that Jupyter Notebook has an infamous reputation. It is not friendly to version control, it's hard to collaborate with notebooks. Luckily, there are efforts that make collaboration in notebook a lot easier now.
Here what I did not show you is that the table has an `isDeleted` column. Some of the records are invalid and we should exclude them. In reality, this happens frequently when you are dealing with hundreds of tables that you are not familiar with. These tables are made for applications, transactions, and they do not have analytic in mind. Data Analytic is usually an afterthought. Therefore, you need to consult the SME or the maintainer of that tables. It takes many iterations to get the correct data that can be used to produce useful insight.
With [ReviewNB](https://www.reviewnb.com/), you can publish your result and invite some domain expert to review your analysis. This is where notebook shine, this kind of workflow is not possible with just the SQL script or a screenshot of your finding. The notebook itself is a useful documentation and collaboration tool.
### Step 1 - Review PR online

You can view your notebook and add comments on a particular cell on [ReviewNB](https://www.reviewnb.com/). This lowers the technical barrier as your analysts do not have to understand Git. He can review changes and make comments on the web without the need to pull code at all. As soon as your analyst makes a suggestion, you can make changes.
### Step 2 - Review Changes

Once you have made changes to the notebook, you can review it side by side. This is very trivial to do it in your local machine. Without ReviewNB, you have to pull both notebooks separately. As Git tracks line-level changes, you can't really read the changes as it consists of a lot of confusing noise. It would also be impossible to view changes about the chart with git.
### Step 3 - Resolve Discussion

Once the changes are reviewed, you can resolve the discussion and share your insight with the team. You can publish the notebook to internal sharing platform like [knowledge-repo](https://github.com/airbnb/knowledge-repo) to organize the analysis.
I hope this convince you that Notebook is a good choice for adhoc analytics. It is possible to collaborate with notebook with proper software in place. Regarless if you use notebook or not, you should try your best to document the process. Let's make more reproducible analyses!
|
github_jupyter
|
```
#Import Required Packages
import requests
import time
import schedule
import os
import json
import newspaper
from bs4 import BeautifulSoup
from datetime import datetime
from newspaper import fulltext
import newspaper
import pandas as pd
import numpy as np
import pickle
#Set Today's Date
#dates = [datetime.today().strftime('%m-%d-%y')]
dates = [datetime.today().strftime('%m-%d')]
```
### Define Urls for Newsapi
```
#Define urls for newsapi
urls=[
'https://newsapi.org/v2/top-headlines?sources=associated-press&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=independent&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=bbc-news&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=reuters&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=the-wall-street-journal&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=the-washington-post&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=national-geographic&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=usa-today&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=cnn&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=fox-news&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=al-jazeera-english&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=bloomberg&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=business-insider&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=cnbc&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=the-new-york-times&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=new-scientist&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=news-com-au&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=newsweek&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=the-economist&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=the-hill&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=the-huffington-post&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=the-next-web&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=the-telegraph&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=the-washington-times&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=time&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=the-jerusalem-post&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=the-irish-times&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=the-globe-and-mail&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=the-american-conservative&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=techcrunch-cn&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4',
'https://newsapi.org/v2/top-headlines?sources=recode&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4'
]
```
### Develop news site folder structure and write top 10 headline urls to API file
```
for date in dates:
print('saving {} ...'.format(date))
for url in urls:
r = requests.get(url)
source = url.replace('https://newsapi.org/v2/top-headlines?sources=','').replace('&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4','')
print(source)
filename = './data/Credible/{0}/articles/{1}/api.txt'.format(source, date)
os.makedirs(os.path.dirname(filename), exist_ok=True)
with open(filename, 'w') as f:
json.dump(json.loads(r.text), f)
print('Finished')
```
### From individual API files, download news source link and extract text using newspaper python package
```
def saving_json():
print('saving ...')
for url in urls:
url = url.strip()
for date in dates:
source = url.replace('https://newsapi.org/v2/top-headlines?sources=','').replace('&apiKey=3fb3c0c1e622430b8df3f9693c7a55b4','')
print(source)
sourcename = './data/Credible/{0}/articles/{1}/api.txt'.format(source, date)
os.makedirs(os.path.dirname(sourcename), exist_ok=True)
with open (sourcename) as f:
jdata = json.load(f)
jdata2=jdata['articles']
for i in range(0,len(jdata2)):
r=jdata2[i]['url']
print(r)
link = newspaper.Article(r)
link.download()
html = link.html
if 'video' in r:
pass
elif link:
try:
link.parse()
text = fulltext(html)
date_longform = dates[0]
article = {}
article["html"] = html
article["title"] = link.title
article["url"] = link.url
article["date"] = date_longform
article["source"] = source
article["text"] = link.text
article["images"] = list(link.images)
article["videos"] = link.movies
count=i+1
filename = './data/Credible/{0}/articles/{1}/article_{2}.txt'.format(source, date, count)
os.makedirs(os.path.dirname(filename), exist_ok=True)
with open(filename, 'w',encoding="utf8",newline='') as file:
json.dump(article,file)
except:
pass
else:
pass
print('Finished')
return None
saving_json()
# #Create initial modeling DataFrame - Only Ran Once then commented out
# modeling = pd.DataFrame(columns=('label', 'text', 'title'))
# #Save initial DataFrame - Only Ran Once - Only Ran Once then commented out
# with open('./data/credible_news_df.pickle', 'wb') as file:
# pickle.dump(modeling, file)
#Open Corpus of News Article Text
with open('./data/credible_news_df.pickle', 'rb') as file:
credible_news_df = pickle.load(file)
i = credible_news_df.shape[0] #Will start adding at the last row of the dataframe
for source in os.listdir("./data/Credible/"):
for file in os.listdir('./data/Credible/'+source+'/articles/'+dates[0]):
if file.endswith(".txt") and 'api' not in file:
curr_file = os.path.join('./data/Credible/'+source+'/articles/'+dates[0], file)
#print curr_file
with open(curr_file) as json_file:
try:
data = json.load(json_file)
credible_news_df.loc[i] = [0,data["text"],data["title"]]
i = i + 1
except ValueError:
continue
#Will Increase Daily
credible_news_df.shape
#Save Updated Data Frame
with open('./data/credible_news_df.pickle', 'wb') as file:
pickle.dump(credible_news_df, file)
```
|
github_jupyter
|
Universidade Federal do Rio Grande do Sul (UFRGS)
Programa de Pós-Graduação em Engenharia Civil (PPGEC)
# PEC00144: Experimental Methods in Civil Engineering
### Reading the serial port of an Arduino device
---
_Prof. Marcelo M. Rocha, Dr.techn._ [(ORCID)](https://orcid.org/0000-0001-5640-1020)
_Porto Alegre, RS, Brazil_
```
# Importing Python modules required for this notebook
# (this cell must be executed with "shift+enter" before any other Python cell)
import sys
import time
import serial
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from MRPy import MRPy
```
### 1. Setup serial communication
In order to run this notebook, the Python module ``pyserial`` must be installed.
To ensure the module availability, open a conda terminal and issue the command:
conda install -c anaconda pyserial
Before openning the serial port, verify with Arduino IDE which USB identifier the
board has be assigned (in Windows it has the form "COMxx", while in Linux it
it is something like "/dev/ttyXXXX").
```
#port = '/dev/ttyUSB0'
#baud = 9600
port = 'COM5' # change this address according to your computer
baud = 9600 # match this number with the Arduino's output baud rate
Ardn = serial.Serial(port, baud, timeout=1)
time.sleep(3) # this is important to give time for serial settling
```
### 2. Define function for reading one incoming line
```
def ReadSerial(nchar, nvar, nlines=1):
Ardn.write(str(nlines).encode())
data = np.zeros((nlines,nvar))
for k in range(nlines):
wait = True
while(wait):
if (Ardn.inWaiting() >= nchar):
wait = False
bdat = Ardn.readline()
sdat = bdat.decode()
sdat = sdat.replace('\n',' ').split()
data[k, :] = np.array(sdat[0:nvar], dtype='int')
return data
```
### 3. Acquire data lines from serial port
```
try:
data = ReadSerial(16, 2, nlines=64)
t = data[:,0]
LC = data[:,1]
Ardn.close()
print('Acquisition ok!')
except:
Ardn.close()
sys.exit('Acquisition failure!')
```
### 4. Create ``MRPy`` instance and save to file
```
ti = (t - t[0])/1000
LC = (LC + 1270)/2**23
data = MRPy.resampling(ti, LC)
data.to_file('read_HX711', form='excel')
print('Average sampling rate is {0:5.1f}Hz.'.format(data.fs))
print('Total record duration is {0:5.1f}Hz.'.format(data.Td))
print((2**23)*data.mean())
```
### 5. Data visualization
```
fig1 = data.plot_time(fig=1, figsize=(12,8), axis_t=[0, data.Td, -0.01, 0.01])
```
|
github_jupyter
|
```
%reload_ext autoreload
%autoreload 2
%matplotlib inline
from fastai.text import *
path = Path('./WikiTextTR')
path.ls()
LANG_FILENAMES = [str(f) for f in path.rglob("*/*")]
print(len(LANG_FILENAMES))
print(LANG_FILENAMES[:5])
LANG_TEXT = []
for i in LANG_FILENAMES:
try:
for line in open(i, encoding="utf-8"):
LANG_TEXT.append(json.loads(line))
except:
break
LANG_TEXT = pd.DataFrame(LANG_TEXT)
LANG_TEXT.head()
LANG_TEXT.to_csv(f"{path}/Wiki_Turkish_Corpus.csv", index=False)
LANG_TEXT = pd.read_csv(f"{path}/Wiki_Turkish_Corpus.csv")
LANG_TEXT.head()
LANG_TEXT.drop(["id","url","title"],axis=1,inplace=True)
LANG_TEXT = (LANG_TEXT.assign(labels = 0)
.pipe(lambda x: x[['labels', 'text']])
.to_csv(f"{path}/Wiki_Turkish_Corpus2.csv", index=False))
LANG_TEXT.head()
LANG_TEXT = pd.read_csv(f"{path}/Wiki_Turkish_Corpus2.csv")
LANG_TEXT.head()
def split_title_from_text(text):
words = text.split("\n\n")
if len(words) >= 2:
return ''.join(words[1:])
else:
return ''.join(words)
LANG_TEXT['text'] = LANG_TEXT['text'].apply(lambda x: split_title_from_text(x))
LANG_TEXT.isna().any()
LANG_TEXT.shape
LANG_TEXT['text'].apply(lambda x: len(x.split(" "))).sum()
re1 = re.compile(r' +')
def fixup(x):
x = x.replace('ü', "u").replace('Ü', 'U').replace('ı', "i").replace(
'ğ', 'g').replace('İ', 'I').replace('Ğ', "G").replace('ö', "o").replace(
'Ö', "o").replace('\n\n', ' ').replace("\'",' ').replace('\n\nSection::::',' ').replace(
'\n',' ').replace('\\', ' \\ ').replace('ç', 'c').replace('Ç', 'C').replace('ş', 's').replace('Ş', 'S')
return re1.sub(' ', html.unescape(x))
LANG_TEXT.to_csv(f"{path}/Wiki_Turkish_Corpus3.csv", index=False)
LANG_TEXT = pd.read_csv(f"{path}/Wiki_Turkish_Corpus3.csv")#, chunksize=5000)
LANG_TEXT.head()
import torch
torch.cuda.device(0)
torch.cuda.get_device_name(0)
LANG_TEXT.dropna(axis=0, inplace=True)
df = LANG_TEXT.iloc[np.random.permutation(len(LANG_TEXT))]
cut1 = int(0.8 * len(df)) + 1
cut1
df_train, df_valid = df[:cut1], df[cut1:]
df = LANG_TEXT.iloc[np.random.permutation(len(LANG_TEXT))]
cut1 = int(0.8 * len(df)) + 1
df_train, df_valid = df[:cut1], df[cut1:]
df_train.shape, df_valid.shape
df_train.head()
data_lm = TextLMDataBunch.from_df(path, train_df=df_train, valid_df= df_valid, label_cols="labels", text_cols="text")
data_lm.save('data_lm.pkl')
bs=16
data_lm = load_data(path, 'data_lm.pkl', bs=bs)
data_lm.show_batch()
learner = language_model_learner(data_lm, AWD_LSTM, pretrained=False, drop_mult=0.5)
learner.lr_find()
learner.recorder.plot()
learner.fit_one_cycle(1,1e-2)
learner.save("model1")
learner.load("model1")
TEXT = "Birinci dünya savaşında"
N_WORDS = 40
N_SENTENCES = 2
print("\n".join(learner.predict(TEXT, N_WORDS, temperature=0.75) for _ in range(N_SENTENCES)))
file = open("itos.pkl","wb")
pickle.dump(data_lm.vocab.itos, file)
file.close()
```
|
github_jupyter
|
```
import numpy as np
import numpy as np
import matplotlib.pyplot as plt
import matplotlib
%matplotlib notebook
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
font = {'weight' : 'medium',
'size' : 13}
matplotlib.rc('font', **font)
import time
import concurrent.futures as cf
import warnings
warnings.filterwarnings("ignore")
import scipy.constants
mec2 = scipy.constants.value('electron mass energy equivalent in MeV')*1e6
c_light = scipy.constants.c
e_charge = scipy.constants.e
r_e = scipy.constants.value('classical electron radius')
```
### Parameters
```
gamma = 5000
rho = 1.5 # Bend radius in m
beta = (1-1/gamma**2)**(1/2)
sigma_x = 50e-6
sigma_z = 50e-6
# Entrance angle
phi = 0.1/rho
```
## code
```
from csr2d.core2 import psi_s, psi_x0_hat
import numpy as np
gamma = 5000
rho = 1.5 # Bend radius in m
beta = (1-1/gamma**2)**(1/2)
sigma_x = 50e-6
sigma_z = 50e-6
nz = 100
nx = 100
dz = (10*sigma_z) / (nz - 1)
dx = (10*sigma_x) / (nx - 1)
zvec = np.linspace(-5*sigma_z, 5*sigma_z, nz)
xvec = np.linspace(-5*sigma_x, 5*sigma_x, nx)
zm, xm = np.meshgrid(zvec, xvec, indexing='ij')
psi_s_grid = psi_s(zm, xm, beta)
psi_x_grid = psi_x0_hat(zm, xm, beta, dx)
from csr2d.core2 import psi_s, psi_x_hat, psi_x0_hat
from scipy.interpolate import RectBivariateSpline
from numba import njit, vectorize, float64
from csr2d.kick2 import green_meshes_hat, green_meshes
# Bypassing the beam, use smooth Gaussian distribution for testing
def lamb_2d(z,x):
return 1/(2*np.pi*sigma_x*sigma_z)* np.exp(-z**2 / 2 / sigma_z**2 - x**2 / 2 / sigma_x**2)
def lamb_2d_prime(z,x):
return 1/(2*np.pi*sigma_x*sigma_z)* np.exp(-z**2 / 2 / sigma_z**2 - x**2 / 2 / sigma_x**2) * (-z / sigma_z**2)
nz = 100
nx = 100
zvec = np.linspace(-5*sigma_z, 5*sigma_z, nz)
xvec = np.linspace(-5*sigma_x, 5*sigma_x, nx)
zm, xm = np.meshgrid(zvec, xvec, indexing='ij')
lambda_grid_filtered = lamb_2d(zm,xm)
lambda_grid_filtered_prime = lamb_2d_prime(zm,xm)
dz = (10*sigma_z) / (nz - 1)
dx = (10*sigma_x) / (nx - 1)
psi_s_grid = psi_s(zm, xm, beta)
psi_s_grid, psi_x_grid, zvec2, xvec2 = green_meshes_hat(nz, nx, dz, dx, rho=rho, beta=beta)
```
# Integral term code development
```
# Convolution for a specific observatino point only
@njit
def my_2d_convolve2(g1, g2, ix1, ix2):
d1, d2 = g1.shape
g2_flip = np.flip(g2)
g2_cut = g2_flip[d1-ix1:2*d1-ix1, d2-ix2:2*d2-ix2]
sums = 0
for i in range(d1):
for j in range(d2):
sums+= g1[i,j]*g2_cut[i,j]
return sums
#@njit
# njit doesn't like the condition grid and interpolation....
def transient_calc_lambda(phi, z_observe, x_observe, zvec, xvec, dz, dx, lambda_grid_filtered_prime, psi_s_grid, psi_x_grid):
x_observe_index = np.argmin(np.abs(xvec - x_observe))
#print('x_observe_index :', x_observe_index )
z_observe_index = np.argmin(np.abs(zvec - z_observe))
#print('z_observe_index :', z_observe_index )
# Boundary condition
temp = (x_observe - xvec)/rho
zi_vec = rho*( phi - beta*np.sqrt(temp**2 + 4*(1 + temp)*np.sin(phi/2)**2))
zo_vec = -beta*np.abs(x_observe - xvec)
condition_grid = np.array([(zvec > z_observe - zo_vec[i]) | (zvec < z_observe - zi_vec[i]) for i in range(len(xvec))])
lambda_grid_filtered_prime_bounded = np.where(condition_grid.T, 0, lambda_grid_filtered_prime)
conv_s = my_2d_convolve2(lambda_grid_filtered_prime_bounded, psi_s_grid, z_observe_index, x_observe_index)
conv_x = my_2d_convolve2(lambda_grid_filtered_prime_bounded, psi_x_grid, z_observe_index, x_observe_index)
##conv_s, conv_x = fftconvolve2(lambda_grid_filtered_prime_bounded, psi_s_grid, psi_x_grid)
#Ws_grid = (beta**2 / abs(rho)) * (conv_s) * (dz * dx)
#Wx_grid = (beta**2 / abs(rho)) * (conv_x) * (dz * dx)
#lambda_interp = RectBivariateSpline(zvec, xvec, lambda_grid_filtered) # lambda lives in the observation grid
#lambda_zi_vec = lambda_interp.ev( z_observe - zi_vec, xvec )
#psi_x_zi_vec = psi_x0(zi_vec/2/rho, temp, beta, dx)
#Wx_zi = (beta**2 / rho) * np.dot(psi_x_zi_vec, lambda_zi_vec)*dx
#lambda_zo_vec = lambda_interp.ev( z_observe - zo_vec, xvec )
#psi_x_zo_vec = psi_x0(zo_vec/2/rho, temp, beta, dx)
#Wx_zo = (beta**2 / rho) * np.dot(psi_x_zo_vec, lambda_zo_vec)*dx
#return Wx_grid[ z_observe_index ][ x_observe_index ], Wx_zi, Wx_zo
#return conv_x, Wx_zi, Wx_zo
return conv_x
#return condition_grid
@njit
def transient_calc_lambda_2(phi, z_observe, x_observe, zvec, xvec, dz, dx, lambda_grid_filtered_prime, psi_s_grid, psi_x_grid):
x_observe_index = np.argmin(np.abs(xvec - x_observe))
#print('x_observe_index :', x_observe_index )
z_observe_index = np.argmin(np.abs(zvec - z_observe))
#print('z_observe_index :', z_observe_index )
# Boundary condition
temp = (x_observe - xvec)/rho
zi_vec = rho*( phi - beta*np.sqrt(temp**2 + 4*(1 + temp)*np.sin(phi/2)**2))
zo_vec = -beta*np.abs(x_observe - xvec)
nz = len(zvec)
nx = len(xvec)
# Allocate array for histogrammed data
cond = np.zeros( (nz,nx) )
for i in range(nx):
cond[:,i] = (zvec > z_observe - zo_vec[i]) | (zvec < z_observe - zi_vec[i])
#condition_grid = np.array([(zvec < z_observe - zi_vec[i]) for i in range(len(xvec))])
#condition_grid = np.array([(zvec > z_observe - zo_vec[i]) | (zvec < z_observe - zi_vec[i]) for i in range(len(xvec))])
lambda_grid_filtered_prime_bounded = np.where(cond, 0, lambda_grid_filtered_prime)
conv_s = my_2d_convolve2(lambda_grid_filtered_prime_bounded, psi_s_grid, z_observe_index, x_observe_index)
conv_x = my_2d_convolve2(lambda_grid_filtered_prime_bounded, psi_x_grid, z_observe_index, x_observe_index)
return conv_x
```
# Applying the codes
### Note that numba-jitted code are slower the FIRST time
```
t1 = time.time()
r1 = transient_calc_lambda(phi, 2*sigma_z, sigma_x, zvec, xvec, dz, dx,lambda_grid_filtered_prime, psi_s_grid, psi_x_grid)
print(r1)
t2 = time.time()
print('Mapping takes:', t2-t1, 'sec')
t1 = time.time()
r1 = transient_calc_lambda_2(phi, 2*sigma_z, sigma_x, zvec, xvec, dz, dx,lambda_grid_filtered_prime, psi_s_grid, psi_x_grid)
print(r1)
t2 = time.time()
print('Mapping takes:', t2-t1, 'sec')
```
## super version for parallelism
```
def transient_calc_lambda_super(z_observe, x_observe):
return transient_calc_lambda(phi, z_observe, x_observe, zvec, xvec, dz, dx,lambda_grid_filtered_prime, psi_s_grid, psi_x_grid)
#@njit
@vectorize([float64(float64,float64)], target='parallel')
def transient_calc_lambda_2_super(z_observe, x_observe):
return transient_calc_lambda_2(phi, z_observe, x_observe, zvec, xvec, dz, dx,lambda_grid_filtered_prime, psi_s_grid, psi_x_grid)
t1 = time.time()
with cf.ProcessPoolExecutor(max_workers=20) as executor:
result = executor.map(transient_calc_lambda_super, zm.flatten(), xm.flatten())
g1 = np.array(list(result)).reshape(zm.shape)
t2 = time.time()
print('Mapping takes:', t2-t1, 'sec')
t1 = time.time()
g4 = transient_calc_lambda_boundary_super_new(zm,xm)
t2 = time.time()
print('Mapping takes:', t2-t1, 'sec')
fig, ax = plt.subplots(figsize=(8,8))
ax = plt.axes(projection='3d')
ax.plot_surface(zm*1e5, xm*1e5, yaya , cmap='inferno', zorder=1)
ax.set_xlabel(r'z $(10^{-5}m)$')
ax.set_ylabel(r'x $(10^{-5}m)$')
ax.set_zlabel(r'$W_x$ $(\times 10^3/m^2)$ ')
ax.zaxis.labelpad = 10
ax.set_title(r'$W_x$ benchmarking')
# To be fixed
from scipy.integrate import quad
def transient_calc_lambda_boundary_quad(phi, z_observe, x_observe, dx):
def integrand_zi(xp):
temp = (x_observe - xp)/rho
zi = rho*( phi - beta*np.sqrt(temp**2 + 4*(1 + temp)*np.sin(phi/2)**2))
#return psi_x_hat(zi/2/rho, temp, beta)*lamb_2d(z_observe - zi, xp)
return psi_x0_hat(zi/2/rho, temp, beta, dx)*lamb_2d(z_observe - zi, xp)
def integrand_zo(xp):
zo = -beta*np.abs(x_observe - xp)
#return psi_x_hat(zo/2/rho, temp, beta)*lamb_2d(z_observe - zo, xp)
return psi_x0_hat(zo/2/rho, temp, beta, dx)*lamb_2d(z_observe - zo, xp)
return quad(integrand_zi, -5*sigma_x, 5*sigma_x)[0]/dx
factor = (beta**2 / rho)*dx
diff = np.abs((g4.reshape(zm.shape) - g3.reshape(zm.shape))/g3.reshape(zm.shape) )* 100
diff = np.abs((g0 - g3.reshape(zm.shape))/g3.reshape(zm.shape)) * 100
g3.shape
fig, ax = plt.subplots(figsize=(8,8))
ax = plt.axes(projection='3d')
ax.plot_surface(zm*1e5, xm*1e5, factor*g3, cmap='inferno', zorder=1)
ax.set_xlabel(r'z $(10^{-5}m)$')
ax.set_ylabel(r'x $(10^{-5}m)$')
ax.set_zlabel(r'$W_x$ $(m^{-2}$) ')
ax.zaxis.labelpad = 10
ax.set_title(r'$W_x$ benchmarking')
fig, ax = plt.subplots(figsize=(8,8))
ax = plt.axes(projection='3d')
ax.plot_surface(zm*1e5, xm*1e5, diff, cmap='inferno', zorder=1)
ax.set_xlabel(r'z $(10^{-5}m)$')
ax.set_ylabel(r'x $(10^{-5}m)$')
ax.set_zlabel(r'$W_x$ $(\times 10^3/m^2)$ ')
ax.zaxis.labelpad = 10
ax.set_title(r'$W_x$ benchmarking')
ax.zaxis.set_scale('log')
plt.plot(diff[30:100,100])
```
|
github_jupyter
|
# Data Analysis
# FINM September Launch
# Homework Solution 5
## Imports
```
import pandas as pd
import numpy as np
import statsmodels.api as sm
from sklearn.linear_model import LinearRegression
from sklearn.decomposition import PCA
from sklearn.cross_decomposition import PLSRegression
from numpy.linalg import svd
import matplotlib.pyplot as plt
import seaborn as sns
sns.set()
import warnings
warnings.filterwarnings("ignore")
```
## Data
```
data = pd.read_excel("../data/single_name_return_data.xlsx", sheet_name="total returns").set_index("Date")
data.head()
equities = data.drop(columns=['SPY', 'SHV'])
equities.head()
```
## 1 Principal Components
#### 1.1
**Calculate the principal components of the return series.**
Using linear algebra:
```
clean = equities - equities.mean()
u, s, vh = svd(clean)
factors = clean @ vh.T
factors.columns = np.arange(1,23)
factors
```
Using a package:
```
pca = PCA(svd_solver='full')
pca.fit(equities.values)
pca_factors = pd.DataFrame(pca.transform(equities.values),
columns=['Factor {}'.format(i+1) for i in range(pca.n_components_)],
index=equities.index)
pca_factors
```
#### 1.2
**Report the eigenvalues associated with these principal components. Report each eigenvalue as a percentage of the sum of all the eigenvalues. This is the total variation each PCA explains.**
Using linear algebra:
```
PCA_eigenvals = pd.DataFrame(index=factors.columns, columns=['Eigen Value', 'Percentage Explained'])
PCA_eigenvals['Eigen Value'] = s**2
PCA_eigenvals['Percentage Explained'] = s**2 / (s**2).sum()
PCA_eigenvals
```
Using package (no eignvalues method):
```
pkg_explained_var = pd.DataFrame(data = pca.explained_variance_ratio_,
index = factors.columns,
columns = ['Explained Variance'])
pkg_explained_var
```
#### 1.3
**How many PCs are needed to explain 75% of the variation?**
```
pkg_explained_var.cumsum()
```
We need the first 5 PCs to explain at least 75% of the variation
#### 1.4
**Calculate the correlation between the first (largest eigenvalue) principal component with each of the 22 single-name equities. Which correlation is highest?**
```
corr_4 = equities.copy()
corr_4['factor 1'] = factors[1]
corr_equities = corr_4.corr()['factor 1'].to_frame('Correlation to factor 1')
corr_equities.iloc[:len(equities.columns)]
```
#### 1.5
**Calculate the correlation between the SPY and the first, second, third principal components.**
```
fac_corr = factors[[1,2,3]]
fac_corr['SPY'] = data['SPY']
SPY_corr = fac_corr.corr()['SPY'].to_frame('Correlation to SPY').iloc[:3]
SPY_corr
```
## 2 PCR and PLS
#### 2.1
**Principal Component Regression (PCR) refers to using PCA for dimension reduction, and then
utilizing the principal components in a regression. Try this by regressing SPY on the first 3 PCs
calculated in the previous section. Report the r-squared.**
```
y_PCR = data['SPY']
X_PCR = factors[[1,2,3]]
model_PCR = LinearRegression().fit(X_PCR,y_PCR)
print('PCR R-squared: ' + str(round(model_PCR.score(X_PCR, y_PCR),3)))
```
#### 2.2
**Calculate the Partial Least Squares estimation of SPY on the 22 single-name equities. Model it for 3 factors. Report the r-squared.**
```
X_PLS = equities
y_PLS = data['SPY']
model_PLS = PLSRegression(n_components=3).fit(X_PLS, y_PLS)
print('PLS R-squared: ' + str(round(model_PLS.score(X_PLS, y_PLS),3)))
```
#### 2.3
**Compare the results between these two approaches and against penalized regression seen in the past homework.**
PCR and PLS both seek to maximize the ability to explain the variation in y variable, and therefore they will have high $R^{2}$ in-sample. When using LASSO or Ridge as our model, we are conservatively forming factors, and penalizing for additional factors. This makes in-sample $R^{2}$ lower as we saw in Homework #4, but may make more robust OOS predictions.
|
github_jupyter
|
# Training and Evaluating ACGAN Model
*by Marvin Bertin*
<img src="../../images/keras-tensorflow-logo.jpg" width="400">
# Imports
```
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import tensorflow as tf
import numpy as np
from collections import defaultdict
import cPickle as pickle
import matplotlib.pyplot as plt
import pandas as pd
from PIL import Image
from six.moves import range
from glob import glob
models = tf.contrib.keras.models
layers = tf.contrib.keras.layers
utils = tf.contrib.keras.utils
losses = tf.contrib.keras.losses
optimizers = tf.contrib.keras.optimizers
metrics = tf.contrib.keras.metrics
preprocessing_image = tf.contrib.keras.preprocessing.image
datasets = tf.contrib.keras.datasets
```
# Construct Generator
```
def generator(latent_size, classes=10):
def up_sampling_block(x, filter_size):
x = layers.UpSampling2D(size=(2, 2))(x)
x = layers.Conv2D(filter_size, (5,5), padding='same', activation='relu')(x)
return x
# Input 1
# image class label
image_class = layers.Input(shape=(1,), dtype='int32', name='image_class')
# class embeddings
emb = layers.Embedding(classes, latent_size,
embeddings_initializer='glorot_normal')(image_class)
# 10 classes in MNIST
cls = layers.Flatten()(emb)
# Input 2
# latent noise vector
latent_input = layers.Input(shape=(latent_size,), name='latent_noise')
# hadamard product between latent embedding and a class conditional embedding
h = layers.multiply([latent_input, cls])
# Conv generator
x = layers.Dense(1024, activation='relu')(h)
x = layers.Dense(128 * 7 * 7, activation='relu')(x)
x = layers.Reshape((7, 7, 128))(x)
# upsample to (14, 14, 128)
x = up_sampling_block(x, 256)
# upsample to (28, 28, 256)
x = up_sampling_block(x, 128)
# reduce channel into binary image (28, 28, 1)
generated_img = layers.Conv2D(1, (2,2), padding='same', activation='tanh')(x)
return models.Model(inputs=[latent_input, image_class],
outputs=generated_img,
name='generator')
```
# Construct Discriminator
```
def discriminator(input_shape=(28, 28, 1)):
def conv_block(x, filter_size, stride):
x = layers.Conv2D(filter_size, (3,3), padding='same', strides=stride)(x)
x = layers.LeakyReLU()(x)
x = layers.Dropout(0.3)(x)
return x
input_img = layers.Input(shape=input_shape)
x = conv_block(input_img, 32, (2,2))
x = conv_block(x, 64, (1,1))
x = conv_block(x, 128, (2,2))
x = conv_block(x, 256, (1,1))
features = layers.Flatten()(x)
# binary classifier, image fake or real
fake = layers.Dense(1, activation='sigmoid', name='generation')(features)
# multi-class classifier, image digit class
aux = layers.Dense(10, activation='softmax', name='auxiliary')(features)
return models.Model(inputs=input_img, outputs=[fake, aux], name='discriminator')
```
# Combine Generator with Discriminator
```
# Adam parameters suggested in paper
adam_lr = 0.0002
adam_beta_1 = 0.5
def ACGAN(latent_size = 100):
# build the discriminator
dis = discriminator()
dis.compile(
optimizer=optimizers.Adam(lr=adam_lr, beta_1=adam_beta_1),
loss=['binary_crossentropy', 'sparse_categorical_crossentropy']
)
# build the generator
gen = generator(latent_size)
gen.compile(optimizer=optimizers.Adam(lr=adam_lr, beta_1=adam_beta_1),
loss='binary_crossentropy')
# Inputs
latent = layers.Input(shape=(latent_size, ), name='latent_noise')
image_class = layers.Input(shape=(1,), dtype='int32', name='image_class')
# Get a fake image
fake_img = gen([latent, image_class])
# Only train generator in combined model
dis.trainable = False
fake, aux = dis(fake_img)
combined = models.Model(inputs=[latent, image_class],
outputs=[fake, aux],
name='ACGAN')
combined.compile(
optimizer=optimizers.Adam(lr=adam_lr, beta_1=adam_beta_1),
loss=['binary_crossentropy', 'sparse_categorical_crossentropy']
)
return combined, dis, gen
```
# Load and Normalize MNIST Dataset
```
# reshape to (..., 28, 28, 1)
# normalize dataset with range [-1, 1]
(X_train, y_train), (X_test, y_test) = datasets.mnist.load_data()
# normalize and reshape train set
X_train = (X_train.astype(np.float32) - 127.5) / 127.5
X_train = np.expand_dims(X_train, axis=-1)
# normalize and reshape test set
X_test = (X_test.astype(np.float32) - 127.5) / 127.5
X_test = np.expand_dims(X_test, axis=-1)
nb_train, nb_test = X_train.shape[0], X_test.shape[0]
```
# Training Helper Functions
```
def print_logs(metrics_names, train_history, test_history):
print('{0:<22s} | {1:4s} | {2:15s} | {3:5s}'.format(
'component', *metrics_names))
print('-' * 65)
ROW_FMT = '{0:<22s} | {1:<4.2f} | {2:<15.2f} | {3:<5.2f}'
print(ROW_FMT.format('generator (train)',
*train_history['generator'][-1]))
print(ROW_FMT.format('generator (test)',
*test_history['generator'][-1]))
print(ROW_FMT.format('discriminator (train)',
*train_history['discriminator'][-1]))
print(ROW_FMT.format('discriminator (test)',
*test_history['discriminator'][-1]))
def generate_batch_noise_and_labels(batch_size, latent_size):
# generate a new batch of noise
noise = np.random.uniform(-1, 1, (batch_size, latent_size))
# sample some labels
sampled_labels = np.random.randint(0, 10, batch_size)
return noise, sampled_labels
```
# Train and Evaluate ACGAN on MNIST
```
nb_epochs = 50
batch_size = 100
train_history = defaultdict(list)
test_history = defaultdict(list)
combined, dis, gen = ACGAN(latent_size = 100)
for epoch in range(nb_epochs):
print('Epoch {} of {}'.format(epoch + 1, nb_epochs))
nb_batches = int(X_train.shape[0] / batch_size)
progress_bar = utils.Progbar(target=nb_batches)
epoch_gen_loss = []
epoch_disc_loss = []
for index in range(nb_batches):
progress_bar.update(index)
### Train Discriminator ###
# generate noise and labels
noise, sampled_labels = generate_batch_noise_and_labels(batch_size, latent_size)
# generate a batch of fake images, using the generated labels as a conditioner
generated_images = gen.predict([noise, sampled_labels.reshape((-1, 1))], verbose=0)
# get a batch of real images
image_batch = X_train[index * batch_size:(index + 1) * batch_size]
label_batch = y_train[index * batch_size:(index + 1) * batch_size]
# construct discriminator dataset
X = np.concatenate((image_batch, generated_images))
y = np.array([1] * batch_size + [0] * batch_size)
aux_y = np.concatenate((label_batch, sampled_labels), axis=0)
# train discriminator
epoch_disc_loss.append(dis.train_on_batch(X, [y, aux_y]))
### Train Generator ###
# generate 2 * batch size here such that we have
# the generator optimize over an identical number of images as the
# discriminator
noise, sampled_labels = generate_batch_noise_and_labels(2 * batch_size, latent_size)
# we want to train the generator to trick the discriminator
# so all the labels should be not-fake (1)
trick = np.ones(2 * batch_size)
epoch_gen_loss.append(combined.train_on_batch(
[noise, sampled_labels.reshape((-1, 1))], [trick, sampled_labels]))
print('\nTesting for epoch {}:'.format(epoch + 1))
### Evaluate Discriminator ###
# generate a new batch of noise
noise, sampled_labels = generate_batch_noise_and_labels(nb_test, latent_size)
# generate images
generated_images = gen.predict(
[noise, sampled_labels.reshape((-1, 1))], verbose=False)
# construct discriminator evaluation dataset
X = np.concatenate((X_test, generated_images))
y = np.array([1] * nb_test + [0] * nb_test)
aux_y = np.concatenate((y_test, sampled_labels), axis=0)
# evaluate discriminator
# test loss
discriminator_test_loss = dis.evaluate(X, [y, aux_y], verbose=False)
# train loss
discriminator_train_loss = np.mean(np.array(epoch_disc_loss), axis=0)
### Evaluate Generator ###
# make new noise
noise, sampled_labels = generate_batch_noise_and_labels(2 * nb_test, latent_size)
# create labels
trick = np.ones(2 * nb_test)
# evaluate generator
# test loss
generator_test_loss = combined.evaluate(
[noise, sampled_labels.reshape((-1, 1))],
[trick, sampled_labels], verbose=False)
# train loss
generator_train_loss = np.mean(np.array(epoch_gen_loss), axis=0)
### Save Losses per Epoch ###
# append train losses
train_history['generator'].append(generator_train_loss)
train_history['discriminator'].append(discriminator_train_loss)
# append test losses
test_history['generator'].append(generator_test_loss)
test_history['discriminator'].append(discriminator_test_loss)
# print training and test losses
print_logs(dis.metrics_names, train_history, test_history)
# save weights every epoch
gen.save_weights(
'../logs/params_generator_epoch_{0:03d}.hdf5'.format(epoch), True)
dis.save_weights(
'../logs/params_discriminator_epoch_{0:03d}.hdf5'.format(epoch), True)
# Save train test loss history
pickle.dump({'train': train_history, 'test': test_history},
open('../logs/acgan-history.pkl', 'wb'))
```
# Generator Loss History
```
hist = pickle.load(open('../logs/acgan-history.pkl'))
for p in ['train', 'test']:
for g in ['discriminator', 'generator']:
hist[p][g] = pd.DataFrame(hist[p][g], columns=['loss', 'generation_loss', 'auxiliary_loss'])
plt.plot(hist[p][g]['generation_loss'], label='{} ({})'.format(g, p))
# get the NE and show as an equilibrium point
plt.hlines(-np.log(0.5), 0, hist[p][g]['generation_loss'].shape[0], label='Nash Equilibrium')
plt.legend()
plt.title(r'$L_s$ (generation loss) per Epoch')
plt.xlabel('Epoch')
plt.ylabel(r'$L_s$')
plt.show()
```
<img src="../../images/gen-loss.png" width="500">
** Generator Loss: **
- loss associated with tricking the discriminator
- training losses converges to the Nash Equilibrium point
- shakiness comes from the generator and the discriminator competing at the equilibrium.
# Label Classification Loss History
```
for g in ['discriminator', 'generator']:
for p in ['train', 'test']:
plt.plot(hist[p][g]['auxiliary_loss'], label='{} ({})'.format(g, p))
plt.legend()
plt.title(r'$L_c$ (classification loss) per Epoch')
plt.xlabel('Epoch')
plt.ylabel(r'$L_c$')
plt.semilogy()
plt.show()
```
<img src="../../images/class-loss.png" width="500">
** Label classification loss: **
- loss associated with the discriminator getting the correct label
- discriminator and generator loss reach stable congerence point
# Generate Digits Conditioned on Class Label
```
# load the weights from the last epoch
gen.load_weights(sorted(glob('../logs/params_generator*'))[-1])
# construct batch of noise and labels
noise = np.tile(np.random.uniform(-1, 1, (10, latent_size)), (10, 1))
sampled_labels = np.array([[i] * 10 for i in range(10)]).reshape(-1, 1)
# generate digits
generated_images = gen.predict([noise, sampled_labels], verbose=0)
# arrange them into a grid and un-normalize the pixels
img = (np.concatenate([r.reshape(-1, 28)
for r in np.split(generated_images, 10)
], axis=-1) * 127.5 + 127.5).astype(np.uint8)
# plot images
plt.imshow(img, cmap='gray')
_ = plt.axis('off')
```
<img src="../../images/generated-digits.png" width="500">
## End of Section
<img src="../../images/divider.png" width="100">
|
github_jupyter
|
# 自动微分
:label:`sec_autograd`
正如我们在 :numref:`sec_calculus`中所说的那样,求导是几乎所有深度学习优化算法的关键步骤。
虽然求导的计算很简单,只需要一些基本的微积分。
但对于复杂的模型,手工进行更新是一件很痛苦的事情(而且经常容易出错)。
深度学习框架通过自动计算导数,即*自动微分*(automatic differentiation)来加快求导。
实际中,根据我们设计的模型,系统会构建一个*计算图*(computational graph),
来跟踪计算是哪些数据通过哪些操作组合起来产生输出。
自动微分使系统能够随后反向传播梯度。
这里,*反向传播*(backpropagate)意味着跟踪整个计算图,填充关于每个参数的偏导数。
## 一个简单的例子
作为一个演示例子,(**假设我们想对函数$y=2\mathbf{x}^{\top}\mathbf{x}$关于列向量$\mathbf{x}$求导**)。
首先,我们创建变量`x`并为其分配一个初始值。
```
import tensorflow as tf
x = tf.range(4, dtype=tf.float32)
x
```
[**在我们计算$y$关于$\mathbf{x}$的梯度之前,我们需要一个地方来存储梯度。**]
重要的是,我们不会在每次对一个参数求导时都分配新的内存。
因为我们经常会成千上万次地更新相同的参数,每次都分配新的内存可能很快就会将内存耗尽。
注意,一个标量函数关于向量$\mathbf{x}$的梯度是向量,并且与$\mathbf{x}$具有相同的形状。
```
x = tf.Variable(x)
```
(**现在让我们计算$y$。**)
```
# 把所有计算记录在磁带上
with tf.GradientTape() as t:
y = 2 * tf.tensordot(x, x, axes=1)
y
```
`x`是一个长度为4的向量,计算`x`和`x`的点积,得到了我们赋值给`y`的标量输出。
接下来,我们[**通过调用反向传播函数来自动计算`y`关于`x`每个分量的梯度**],并打印这些梯度。
```
x_grad = t.gradient(y, x)
x_grad
```
函数$y=2\mathbf{x}^{\top}\mathbf{x}$关于$\mathbf{x}$的梯度应为$4\mathbf{x}$。
让我们快速验证这个梯度是否计算正确。
```
x_grad == 4 * x
```
[**现在让我们计算`x`的另一个函数。**]
```
with tf.GradientTape() as t:
y = tf.reduce_sum(x)
t.gradient(y, x) # 被新计算的梯度覆盖
```
## 非标量变量的反向传播
当`y`不是标量时,向量`y`关于向量`x`的导数的最自然解释是一个矩阵。
对于高阶和高维的`y`和`x`,求导的结果可以是一个高阶张量。
然而,虽然这些更奇特的对象确实出现在高级机器学习中(包括[**深度学习中**]),
但当我们调用向量的反向计算时,我们通常会试图计算一批训练样本中每个组成部分的损失函数的导数。
这里(**,我们的目的不是计算微分矩阵,而是单独计算批量中每个样本的偏导数之和。**)
```
with tf.GradientTape() as t:
y = x * x
t.gradient(y, x) # 等价于y=tf.reduce_sum(x*x)
```
## 分离计算
有时,我们希望[**将某些计算移动到记录的计算图之外**]。
例如,假设`y`是作为`x`的函数计算的,而`z`则是作为`y`和`x`的函数计算的。
想象一下,我们想计算`z`关于`x`的梯度,但由于某种原因,我们希望将`y`视为一个常数,
并且只考虑到`x`在`y`被计算后发挥的作用。
在这里,我们可以分离`y`来返回一个新变量`u`,该变量与`y`具有相同的值,
但丢弃计算图中如何计算`y`的任何信息。
换句话说,梯度不会向后流经`u`到`x`。
因此,下面的反向传播函数计算`z=u*x`关于`x`的偏导数,同时将`u`作为常数处理,
而不是`z=x*x*x`关于`x`的偏导数。
```
# 设置persistent=True来运行t.gradient多次
with tf.GradientTape(persistent=True) as t:
y = x * x
u = tf.stop_gradient(y)
z = u * x
x_grad = t.gradient(z, x)
x_grad == u
```
由于记录了`y`的计算结果,我们可以随后在`y`上调用反向传播,
得到`y=x*x`关于的`x`的导数,即`2*x`。
```
t.gradient(y, x) == 2 * x
```
## Python控制流的梯度计算
使用自动微分的一个好处是:
[**即使构建函数的计算图需要通过Python控制流(例如,条件、循环或任意函数调用),我们仍然可以计算得到的变量的梯度**]。
在下面的代码中,`while`循环的迭代次数和`if`语句的结果都取决于输入`a`的值。
```
def f(a):
b = a * 2
while tf.norm(b) < 1000:
b = b * 2
if tf.reduce_sum(b) > 0:
c = b
else:
c = 100 * b
return c
```
让我们计算梯度。
```
a = tf.Variable(tf.random.normal(shape=()))
with tf.GradientTape() as t:
d = f(a)
d_grad = t.gradient(d, a)
d_grad
```
我们现在可以分析上面定义的`f`函数。
请注意,它在其输入`a`中是分段线性的。
换言之,对于任何`a`,存在某个常量标量`k`,使得`f(a)=k*a`,其中`k`的值取决于输入`a`。
因此,我们可以用`d/a`验证梯度是否正确。
```
d_grad == d / a
```
## 小结
* 深度学习框架可以自动计算导数:我们首先将梯度附加到想要对其计算偏导数的变量上。然后我们记录目标值的计算,执行它的反向传播函数,并访问得到的梯度。
## 练习
1. 为什么计算二阶导数比一阶导数的开销要更大?
1. 在运行反向传播函数之后,立即再次运行它,看看会发生什么。
1. 在控制流的例子中,我们计算`d`关于`a`的导数,如果我们将变量`a`更改为随机向量或矩阵,会发生什么?
1. 重新设计一个求控制流梯度的例子,运行并分析结果。
1. 使$f(x)=\sin(x)$,绘制$f(x)$和$\frac{df(x)}{dx}$的图像,其中后者不使用$f'(x)=\cos(x)$。
[Discussions](https://discuss.d2l.ai/t/1757)
|
github_jupyter
|
# Rossman data preparation
To illustrate the techniques we need to apply before feeding all the data to a Deep Learning model, we are going to take the example of the [Rossmann sales Kaggle competition](https://www.kaggle.com/c/rossmann-store-sales). Given a wide range of information about a store, we are going to try predict their sale number on a given day. This is very useful to be able to manage stock properly and be able to properly satisfy the demand without wasting anything. The official training set was giving a lot of informations about various stores in Germany, but it was also allowed to use additional data, as long as it was made public and available to all participants.
We are going to reproduce most of the steps of one of the winning teams that they highlighted in [Entity Embeddings of Categorical Variables](https://arxiv.org/pdf/1604.06737.pdf). In addition to the official data, teams in the top of the leaderboard also used information about the weather, the states of the stores or the Google trends of those days. We have assembled all that additional data in one file available for download [here](http://files.fast.ai/part2/lesson14/rossmann.tgz) if you want to replicate those steps.
### A first look at the data
First things first, let's import everything we will need.
```
from fastai.tabular.all import *
```
If you have download the previous file and decompressed it in a folder named rossmann in the fastai data folder, you should see the following list of files with this instruction:
```
path = Config().data/'rossmann'
path.ls()
```
The data that comes from Kaggle is in 'train.csv', 'test.csv', 'store.csv' and 'sample_submission.csv'. The other files are the additional data we were talking about. Let's start by loading everything using pandas.
```
table_names = ['train', 'store', 'store_states', 'state_names', 'googletrend', 'weather', 'test']
tables = [pd.read_csv(path/f'{fname}.csv', low_memory=False) for fname in table_names]
train, store, store_states, state_names, googletrend, weather, test = tables
```
To get an idea of the amount of data available, let's just look at the length of the training and test tables.
```
len(train), len(test)
```
So we have more than one million records available. Let's have a look at what's inside:
```
train.head()
```
The `Store` column contains the id of the stores, then we are given the id of the day of the week, the exact date, if the store was open on that day, if there were any promotion in that store during that day, and if it was a state or school holiday. The `Customers` column is given as an indication, and the `Sales` column is what we will try to predict.
If we look at the test table, we have the same columns, minus `Sales` and `Customers`, and it looks like we will have to predict on dates that are after the ones of the train table.
```
test.head()
```
The other table given by Kaggle contains some information specific to the stores: their type, what the competition looks like, if they are engaged in a permanent promotion program, and if so since then.
```
store.head().T
```
Now let's have a quick look at our four additional dataframes. `store_states` just gives us the abbreviated name of the sate of each store.
```
store_states.head()
```
We can match them to their real names with `state_names`.
```
state_names.head()
```
Which is going to be necessary if we want to use the `weather` table:
```
weather.head().T
```
Lastly the googletrend table gives us the trend of the brand in each state and in the whole of Germany.
```
googletrend.head()
```
Before we apply the fastai preprocessing, we will need to join the store table and the additional ones with our training and test table. Then, as we saw in our first example in chapter 1, we will need to split our variables between categorical and continuous. Before we do that, though, there is one type of variable that is a bit different from the others: dates.
We could turn each particular day in a category but there are cyclical information in dates we would miss if we did that. We already have the day of the week in our tables, but maybe the day of the month also bears some significance. People might be more inclined to go shopping at the beggining or the end of the month. The number of the week/month is also important to detect seasonal influences.
Then we will try to exctract meaningful information from those dates. For instance promotions on their own are important inputs, but maybe the number of running weeks with promotion is another useful information as it will influence customers. A state holiday in itself is important, but it's more significant to know if we are the day before or after such a holiday as it will impact sales. All of those might seem very specific to this dataset, but you can actually apply them in any tabular data containing time information.
This first step is called feature-engineering and is extremely important: your model will try to extract useful information from your data but any extra help you can give it in advance is going to make training easier, and the final result better. In Kaggle Competitions using tabular data, it's often the way people prepared their data that makes the difference in the final leaderboard, not the exact model used.
### Feature Engineering
#### Merging tables
To merge tables together, we will use this little helper function that relies on the pandas library. It will merge the tables `left` and `right` by looking at the column(s) which names are in `left_on` and `right_on`: the information in `right` will be added to the rows of the tables in `left` when the data in `left_on` inside `left` is the same as the data in `right_on` inside `right`. If `left_on` and `right_on` are the same, we don't have to pass `right_on`. We keep the fields in `right` that have the same names as fields in `left` and add a `_y` suffix (by default) to those field names.
```
def join_df(left, right, left_on, right_on=None, suffix='_y'):
if right_on is None: right_on = left_on
return left.merge(right, how='left', left_on=left_on, right_on=right_on,
suffixes=("", suffix))
```
First, let's replace the state names in the weather table by the abbreviations, since that's what is used in the other tables.
```
weather = join_df(weather, state_names, "file", "StateName")
weather[['file', 'Date', 'State', 'StateName']].head()
```
To double-check the merge happened without incident, we can check that every row has a `State` with this line:
```
len(weather[weather.State.isnull()])
```
We can now safely remove the columns with the state names (`file` and `StateName`) since they we'll use the short codes.
```
weather.drop(columns=['file', 'StateName'], inplace=True)
```
To add the weather informations to our `store` table, we first use the table `store_states` to match a store code with the corresponding state, then we merge with our weather table.
```
store = join_df(store, store_states, 'Store')
store = join_df(store, weather, 'State')
```
And again, we can check if the merge went well by looking if new NaNs where introduced.
```
len(store[store.Mean_TemperatureC.isnull()])
```
Next, we want to join the `googletrend` table to this `store` table. If you remember from our previous look at it, it's not exactly in the same format:
```
googletrend.head()
```
We will need to change the column with the states and the columns with the dates:
- in the column `fil`, the state names contain `Rossmann_DE_XX` with `XX` being the code of the state, so we want to remove `Rossmann_DE`. We will do this by creating a new column containing the last part of a split of the string by '\_'.
- in the column `week`, we will extract the date corresponding to the beginning of the week in a new column by taking the last part of a split on ' - '.
In pandas, creating a new column is very easy: you just have to define them.
```
googletrend['Date'] = googletrend.week.str.split(' - ', expand=True)[0]
googletrend['State'] = googletrend.file.str.split('_', expand=True)[2]
googletrend.head()
```
Let's check everything went well by looking at the values in the new `State` column of our `googletrend` table.
```
store['State'].unique(),googletrend['State'].unique()
```
We have two additional values in the second (`None` and 'SL') but this isn't a problem since they'll be ignored when we join. One problem however is that 'HB,NI' in the first table is named 'NI' in the second one, so we need to change that.
```
googletrend.loc[googletrend.State=='NI', "State"] = 'HB,NI'
```
Why do we have a `None` in state? As we said before, there is a global trend for Germany that corresponds to `Rosmann_DE` in the field `file`. For those, the previous split failed which gave the `None` value. We will keep this global trend and put it in a new column.
```
trend_de = googletrend[googletrend.file == 'Rossmann_DE'][['Date', 'trend']]
```
Then we can merge it with the rest of our trends, by adding the suffix '\_DE' to know it's the general trend.
```
googletrend = join_df(googletrend, trend_de, 'Date', suffix='_DE')
```
Then at this stage, we can remove the columns `file` and `week`since they won't be useful anymore, as well as the rows where `State` is `None` (since they correspond to the global trend that we saved in another column).
```
googletrend.drop(columns=['file', 'week'], axis=1, inplace=True)
googletrend = googletrend[~googletrend['State'].isnull()]
```
The last thing missing to be able to join this with or store table is to extract the week from the date in this table and in the store table: we need to join them on week values since each trend is given for the full week that starts on the indicated date. This is linked to the next topic in feature engineering: extracting dateparts.
#### Adding dateparts
If your table contains dates, you will need to split the information there in several column for your Deep Learning model to be able to train properly. There is the basic stuff, such as the day number, week number, month number or year number, but anything that can be relevant to your problem is also useful. Is it the beginning or the end of the month? Is it a holiday?
To help with this, the fastai library as a convenience function called `add_datepart`. It will take a dataframe and a column you indicate, try to read it as a date, then add all those new columns. If we go back to our `googletrend` table, we now have gour columns.
```
googletrend.head()
```
If we add the dateparts, we will gain a lot more
```
googletrend = add_datepart(googletrend, 'Date', drop=False)
googletrend.head().T
```
We chose the option `drop=False` as we want to keep the `Date` column for now. Another option is to add the `time` part of the date, but it's not relevant to our problem here.
Now we can join our Google trends with the information in the `store` table, it's just a join on \['Week', 'Year'\] once we apply `add_datepart` to that table. Note that we only keep the initial columns of `googletrend` with `Week` and `Year` to avoid all the duplicates.
```
googletrend = googletrend[['trend', 'State', 'trend_DE', 'Week', 'Year']]
store = add_datepart(store, 'Date', drop=False)
store = join_df(store, googletrend, ['Week', 'Year', 'State'])
```
At this stage, `store` contains all the information about the stores, the weather on that day and the Google trends applicable. We only have to join it with our training and test table. We have to use `make_date` before being able to execute that merge, to convert the `Date` column of `train` and `test` to proper date format.
```
make_date(train, 'Date')
make_date(test, 'Date')
train_fe = join_df(train, store, ['Store', 'Date'])
test_fe = join_df(test, store, ['Store', 'Date'])
```
#### Elapsed times
Another feature that can be useful is the elapsed time before/after a certain event occurs. For instance the number of days since the last promotion or before the next school holiday. Like for the date parts, there is a fastai convenience function that will automatically add them.
One thing to take into account here is that you will need to use that function on the whole time series you have, even the test data: there might be a school holiday that takes place during the training data and it's going to impact those new features in the test data.
```
all_ftrs = train_fe.append(test_fe, sort=False)
```
We will consider the elapsed times for three events: 'Promo', 'StateHoliday' and 'SchoolHoliday'. Note that those must correspondon to booleans in your dataframe. 'Promo' and 'SchoolHoliday' already are (only 0s and 1s) but 'StateHoliday' has multiple values.
```
all_ftrs['StateHoliday'].unique()
```
If we refer to the explanation on Kaggle, 'b' is for Easter, 'c' for Christmas and 'a' for the other holidays. We will just converts this into a boolean that flags any holiday.
```
all_ftrs.StateHoliday = all_ftrs.StateHoliday!='0'
```
Now we can add, for each store, the number of days since or until the next promotion, state or school holiday. This will take a little while since the whole table is big.
```
all_ftrs = add_elapsed_times(all_ftrs, ['Promo', 'StateHoliday', 'SchoolHoliday'],
date_field='Date', base_field='Store')
```
It added a four new features. If we look at 'StateHoliday' for instance:
```
[c for c in all_ftrs.columns if 'StateHoliday' in c]
```
The column 'AfterStateHoliday' contains the number of days since the last state holiday, 'BeforeStateHoliday' the number of days until the next one. As for 'StateHoliday_bw' and 'StateHoliday_fw', they contain the number of state holidays in the past or future seven days respectively. The same four columns have been added for 'Promo' and 'SchoolHoliday'.
Now that we have added those features, we can split again our tables between the training and the test one.
```
train_df = all_ftrs.iloc[:len(train_fe)]
test_df = all_ftrs.iloc[len(train_fe):]
```
One last thing the authors of this winning solution did was to remove the rows with no sales, which correspond to exceptional closures of the stores. This might not have been a good idea since even if we don't have access to the same features in the test data, it can explain why we have some spikes in the training data.
```
train_df = train_df[train_df.Sales != 0.]
```
We will use those for training but since all those steps took a bit of time, it's a good idea to save our progress until now. We will just pickle those tables on the hard drive.
```
train_df.to_pickle(path/'train_clean')
test_df.to_pickle(path/'test_clean')
```
|
github_jupyter
|
* [1.0 - Introduction](#1.0---Introduction)
- [1.1 - Library imports and loading the data from SQL to pandas](#1.1---Library-imports-and-loading-the-data-from-SQL-to-pandas)
* [2.0 - Data Cleaning](#2.0---Data-Cleaning)
- [2.1 - Pre-cleaning, investigating data types](#2.1---Pre-cleaning,-investigating-data-types)
- [2.2 - Dealing with non-numerical values](#2.2---Dealing-with-non-numerical-values)
* [3.0 - Creating New Features](#)
- [3.1 - Creating the 'gender' column](#3.1---Creating-the-'gender'-column)
- [3.2 - Categorizing job titles](#3.2---Categorizing-job-titles)
* [4.0 - Data Analysis and Visualizations](#4.0---Data-Analysis-and-Visualizations)
- [4.1 - Overview of the gender gap](#4.1---Overview-of-the-gender-gap)
- [4.2 - Exploring the year column](#4.2---Exploring-the-year-column)
- [4.3 - Full time vs. part time employees](#4.3---Full-time-vs.-part-time-employees)
- [4.4 - Breaking down the total pay](#4.4---Breaking-down-the-total-pay)
- [4.5 - Breaking down the base pay by job category](#4.5---Breaking-down-the-base-pay-by-job-category)
- [4.6 - Gender representation by job category](#4.6---Gender-representation-by-job-category)
- [4.7 - Significance testing by exact job title](#4.7---Significance-testing-by-exact-job-title)
* [5.0 - San Francisco vs. Newport Beach](#5.0---San Francisco-vs.-Newport-Beach)
- [5.1 - Part time vs. full time workers](#5.1---Part-time-vs.-full-time-workers)
- [5.2 - Comparisons by job cateogry](#5.2---Comparisons-by-job-cateogry)
- [5.3 - Gender representation by job category](#5.3---Gender-representation-by-job-category)
* [6.0 - Conclusion](#6.0---Conclusion)
### 1.0 - Introduction
In this notebook, I will focus on data analysis and preprocessing for the gender wage gap. Specifically, I am going to focus on public jobs in the city of San Francisco and Newport Beach. This data set is publically available on [Kaggle](https://www.kaggle.com/kaggle/sf-salaries) and [Transparent California](https://transparentcalifornia.com/).
I also created a web application based on this dataset. You can play arround with it [here](https://gendergapvisualization.herokuapp.com/). For a complete list of requirements and files used for my web app, check out my GitHub repository [here](https://github.com/sengkchu/gendergapvisualization).
In this notebook following questions will be explored:
+ Is there an overall gender wage gap for public jobs in San Francisco?
+ Is the gender gap really 78 cents on the dollar?
+ Is there a gender wage gap for full time employees?
+ Is there a gender wage gap for part time employees?
+ Is there a gender wage gap if the employees were grouped by job categories?
+ Is there a gender wage gap if the employees were grouped by exact job title?
+ If the gender wage gap exists, is the data statistically significant?
+ If the gender wage gap exists, how does the gender wage gap in San Francisco compare with more conservative cities in California?
Lastly, I want to mention that I am not affiliated with any political group, everything I write in this project is based on my perspective of the data alone.
#### 1.1 - Library imports and loading the data from SQL to pandas
The SQL database is about 18 megabytes, which is small enough for my computer to handle. So I've decided to just load the entire database into memory using pandas. However, I created a function that takes in a SQL query and returns the result as a pandas dataframe just in case I need to use SQL queries.
```
import pandas as pd
import numpy as np
import sqlite3
import matplotlib.pyplot as plt
import seaborn as sns
import gender_guesser.detector as gender
import time
import collections
%matplotlib inline
sns.set(font_scale=1.5)
def run_query(query):
with sqlite3.connect('database.sqlite') as conn:
return pd.read_sql(query, conn)
#Read the data from SQL->Pandas
q1 = '''
SELECT * FROM Salaries
'''
data = run_query(q1)
data.head()
```
### 2.0 - Data Cleaning
Fortunately, this data set is already very clean. However, we should still look into every column. Specifically, we are interested in the data types of each column, and check for null values within the rows.
#### 2.1 - Pre-cleaning, investigating data types
Before we do anything to the dataframe, we are going to simply explore the data a little bit.
```
data.dtypes
data['JobTitle'].nunique()
```
There is no gender column, so we'll have to create one. In addition, we'll need to reduce the number of unique values in the `'JobTitle'` column. `'BasePay'`, `'OvertimePay'`, `'OtherPay'`, and `'Benefits'` are all object columns. We'll need to find a way to covert these into numeric values.
Let's take a look at the rest of the columns using the `.value_counts()` method.
```
data['Year'].value_counts()
data['Notes'].value_counts()
data['Agency'].value_counts()
data['Status'].value_counts()
```
It looks like the data is split into 4 years. The `'Notes'` column is empty for 148654 rows, so we should just remove it. The `'Agency'` column is also not useful, because we already know the data is for San Francisco.
The `'Status'` column shows a separation for full time employees and part time employees. We should leave that alone for now.
#### 2.2 - Dealing with non-numerical values
Let's tackle the object columns first, we are going to convert everything into integers using the `pandas.to_numeric()` function. If we run into any errors, the returned value will be NaN.
```
def process_pay(df):
cols = ['BasePay','OvertimePay', 'OtherPay', 'Benefits']
print('Checking for nulls:')
for col in cols:
df[col] = pd.to_numeric(df[col], errors ='coerce')
print(len(col)*'-')
print(col)
print(len(col)*'-')
print(df[col].isnull().value_counts())
return df
data = process_pay(data.copy())
```
Looking at our results above, we found 609 null values in `BasePay` and 36163 null values in `Benefits`. We are going to drop the rows with null values in `BasePay`. Not everyone will recieve benefits for their job, so it makes more sense to fill in the null values for `Benefits` with zeroes.
```
def process_pay2(df):
df['Benefits'] = df['Benefits'].fillna(0)
df = df.dropna()
print(df['BasePay'].isnull().value_counts())
return df
data = process_pay2(data)
```
Lastly, let's drop the `Agency` and `Notes` columns as they do not provide any information.
```
data = data.drop(columns=['Agency', 'Notes'])
```
### 3.0 - Creating New Features
Unfortunately, this data set does not include demographic information. Since this project is focused on investigating the gender wage gap, we need a way to classify a person's gender. Furthermore, the `JobTitle` column has 2159 unique values. We'll need to simplify this column.
#### 3.1 - Creating the 'gender' column
Due to the limitations of this data set. We'll have to assume the gender of the employee by using their first name. The `gender_guesser` library is very useful for this.
```
#Create the 'Gender' column based on employee's first name.
d = gender.Detector(case_sensitive=False)
data['FirstName'] = data['EmployeeName'].str.split().apply(lambda x: x[0])
data['Gender'] = data['FirstName'].apply(lambda x: d.get_gender(x))
data['Gender'].value_counts()
```
We are just going to remove employees with ambiguous or gender neutral first names from our analysis.
```
#Retain data with 'male' and 'female' names.
male_female_only = data[(data['Gender'] == 'male') | (data['Gender'] == 'female')].copy()
male_female_only['Gender'].value_counts()
```
#### 3.2 - Categorizing job titles
Next, we'll have to simplify the `JobTitles` column. To do this, we'll use the brute force method. I created an ordered dictionary with keywords and their associated job category. The generic titles are at the bottom of the dictionary, and the more specific titles are at the top of the dictionary. Then we are going to use a for loop in conjunction with the `.map()` method on the column.
I used the same labels as this [kernel](https://www.kaggle.com/mevanoff24/data-exploration-predicting-salaries) on Kaggle, but I heavily modified the code for readability.
```
def find_job_title2(row):
#Prioritize specific titles on top
titles = collections.OrderedDict([
('Police',['police', 'sherif', 'probation', 'sergeant', 'officer', 'lieutenant']),
('Fire', ['fire']),
('Transit',['mta', 'transit']),
('Medical',['anesth', 'medical', 'nurs', 'health', 'physician', 'orthopedic', 'pharm', 'care']),
('Architect', ['architect']),
('Court',['court', 'legal']),
('Mayor Office', ['mayoral']),
('Library', ['librar']),
('Public Works', ['public']),
('Attorney', ['attorney']),
('Custodian', ['custodian']),
('Gardener', ['garden']),
('Recreation Leader', ['recreation']),
('Automotive',['automotive', 'mechanic', 'truck']),
('Engineer',['engineer', 'engr', 'eng', 'program']),
('General Laborer',['general laborer', 'painter', 'inspector', 'carpenter', 'electrician', 'plumber', 'maintenance']),
('Food Services', ['food serv']),
('Clerk', ['clerk']),
('Porter', ['porter']),
('Airport Staff', ['airport']),
('Social Worker',['worker']),
('Guard', ['guard']),
('Assistant',['aide', 'assistant', 'secretary', 'attendant']),
('Analyst', ['analy']),
('Manager', ['manager'])
])
#Loops through the dictionaries
for group, keywords in titles.items():
for keyword in keywords:
if keyword in row.lower():
return group
return 'Other'
start_time = time.time()
male_female_only["Job_Group"] = male_female_only["JobTitle"].map(find_job_title2)
print("--- Run Time: %s seconds ---" % (time.time() - start_time))
male_female_only['Job_Group'].value_counts()
```
### 4.0 - Data Analysis and Visualizations
In this section, we are going to use the data to answer the questions stated in the [introduction section](#1.0---Introduction).
#### 4.1 - Overview of the gender gap
Let's begin by splitting the data set in half, one for females and one for males. Then we'll plot the overall income distribution using kernel density estimation based on the gausian function.
```
fig = plt.figure(figsize=(10, 5))
male_only = male_female_only[male_female_only['Gender'] == 'male']
female_only = male_female_only[male_female_only['Gender'] == 'female']
ax = sns.kdeplot(male_only['TotalPayBenefits'], color ='Blue', label='Male', shade=True)
ax = sns.kdeplot(female_only['TotalPayBenefits'], color='Red', label='Female', shade=True)
plt.yticks([])
plt.title('Overall Income Distribution')
plt.ylabel('Density of Employees')
plt.xlabel('Total Pay + Benefits ($)')
plt.xlim(0, 350000)
plt.show()
```
The income distribution plot is bimodal. In addition, we see a gender wage gap in favor of males in between the ~110000 and the ~275000 region. But, this plot doesn't capture the whole story. We need to break down the data some more. But first, let's explore the percentage of employees based on gender.
```
fig = plt.figure(figsize=(5, 5))
colors = ['#AFAFF5', '#EFAFB5']
labels = ['Male', 'Female']
sizes = [len(male_only), len(female_only)]
explode = (0.05, 0)
sns.set(font_scale=1.5)
ax = plt.pie(sizes, labels=labels, explode=explode, colors=colors, shadow=True, startangle=90, autopct='%1.f%%')
plt.title('Estimated Percentages of Employees: Overall')
plt.show()
```
Another key factor we have to consider is the number of employees. How do we know if there are simply more men working at higher paying jobs? How can we determine if social injustice has occured?
The chart above only tells us the total percentage of employees across all job categories, but it does give us an overview of the data.
#### 4.2 - Exploring the year column
The data set contain information on employees between 2011-2014. Let's take a look at an overview of the income based on the `Year` column regardless of gender.
```
data_2011 = male_female_only[male_female_only['Year'] == 2011]
data_2012 = male_female_only[male_female_only['Year'] == 2012]
data_2013 = male_female_only[male_female_only['Year'] == 2013]
data_2014 = male_female_only[male_female_only['Year'] == 2014]
plt.figure(figsize=(10,7.5))
ax = plt.boxplot([data_2011['TotalPayBenefits'].values, data_2012['TotalPayBenefits'].values, \
data_2013['TotalPayBenefits'].values, data_2014['TotalPayBenefits'].values])
plt.ylim(0, 350000)
plt.xticks([1, 2, 3, 4], ['2011', '2012', '2013', '2014'])
plt.xlabel('Year')
plt.ylabel('Total Pay + Benefits ($)')
plt.tight_layout()
```
From the boxplots, we see that the total pay is increasing for every year. We'll have to consider inflation in our analysis. In addition, it is very possible for an employee to stay at their job for multiple years. We don't want to double sample on these employees.
To simplify the data for the purpose of investigating the gender gap. It makes more sense to only choose only one year for our analysis. From our data exploration, we noticed that the majority of the `status` column was blank. Let's break the data down by year using the `.value_counts()` method.
```
years = ['2011', '2012', '2013', '2014']
all_data = [data_2011, data_2012, data_2013, data_2014]
for i in range(4):
print(len(years[i])*'-')
print(years[i])
print(len(years[i])*'-')
print(all_data[i]['Status'].value_counts())
```
The status of the employee is critical to our analysis, only year 2014 has this information. So it makes sense to focus on analysis on 2014.
```
data_2014_FT = data_2014[data_2014['Status'] == 'FT']
data_2014_PT = data_2014[data_2014['Status'] == 'PT']
```
#### 4.3 - Full time vs. part time employees
Let's take a look at the kernal density estimation plot for part time and full time employees.
```
fig = plt.figure(figsize=(10, 5))
ax = sns.kdeplot(data_2014_PT['TotalPayBenefits'], color = 'Orange', label='Part Time Workers', shade=True)
ax = sns.kdeplot(data_2014_FT['TotalPayBenefits'], color = 'Green', label='Full Time Workers', shade=True)
plt.yticks([])
plt.title('Part Time Workers vs. Full Time Workers')
plt.ylabel('Density of Employees')
plt.xlabel('Total Pay + Benefits ($)')
plt.xlim(0, 350000)
plt.show()
```
If we split the data by employment status, we can see that the kernal distribution plot is no longer bimodal. Next, let's see how these two plots look if we seperate the data by gender.
```
fig = plt.figure(figsize=(10, 10))
fig.subplots_adjust(hspace=.5)
#Generate the top plot
male_only = data_2014_FT[data_2014_FT['Gender'] == 'male']
female_only = data_2014_FT[data_2014_FT['Gender'] == 'female']
ax = fig.add_subplot(2, 1, 1)
ax = sns.kdeplot(male_only['TotalPayBenefits'], color ='Blue', label='Male', shade=True)
ax = sns.kdeplot(female_only['TotalPayBenefits'], color='Red', label='Female', shade=True)
plt.title('Full Time Workers')
plt.ylabel('Density of Employees')
plt.xlabel('Total Pay & Benefits ($)')
plt.xlim(0, 350000)
plt.yticks([])
#Generate the bottom plot
male_only = data_2014_PT[data_2014_PT['Gender'] == 'male']
female_only = data_2014_PT[data_2014_PT['Gender'] == 'female']
ax2 = fig.add_subplot(2, 1, 2)
ax2 = sns.kdeplot(male_only['TotalPayBenefits'], color ='Blue', label='Male', shade=True)
ax2 = sns.kdeplot(female_only['TotalPayBenefits'], color='Red', label='Female', shade=True)
plt.title('Part Time Workers')
plt.ylabel('Density of Employees')
plt.xlabel('Total Pay & Benefits ($)')
plt.xlim(0, 350000)
plt.yticks([])
plt.show()
```
For part time workers, the KDE plot is nearly identical for both males and females.
For full time workers, we still see a gender gap. We'll need to break down the data some more.
#### 4.4 - Breaking down the total pay
We used total pay including benefits for the x-axis for the KDE plot in the previous section. Is this a fair way to analyze the data? What if men work more overtime hours than women? Can we break down the data some more?
```
male_only = data_2014_FT[data_2014_FT['Gender'] == 'male']
female_only = data_2014_FT[data_2014_FT['Gender'] == 'female']
fig = plt.figure(figsize=(10, 15))
fig.subplots_adjust(hspace=.5)
#Generate the top plot
ax = fig.add_subplot(3, 1, 1)
ax = sns.kdeplot(male_only['OvertimePay'], color ='Blue', label='Male', shade=True)
ax = sns.kdeplot(female_only['OvertimePay'], color='Red', label='Female', shade=True)
plt.title('Full Time Workers')
plt.ylabel('Density of Employees')
plt.xlabel('Overtime Pay ($)')
plt.xlim(0, 60000)
plt.yticks([])
#Generate the middle plot
ax2 = fig.add_subplot(3, 1, 2)
ax2 = sns.kdeplot(male_only['Benefits'], color ='Blue', label='Male', shade=True)
ax2 = sns.kdeplot(female_only['Benefits'], color='Red', label='Female', shade=True)
plt.ylabel('Density of Employees')
plt.xlabel('Benefits Only ($)')
plt.xlim(0, 75000)
plt.yticks([])
#Generate the bottom plot
ax3 = fig.add_subplot(3, 1, 3)
ax3 = sns.kdeplot(male_only['BasePay'], color ='Blue', label='Male', shade=True)
ax3 = sns.kdeplot(female_only['BasePay'], color='Red', label='Female', shade=True)
plt.ylabel('Density of Employees')
plt.xlabel('Base Pay Only ($)')
plt.xlim(0, 300000)
plt.yticks([])
plt.show()
```
We see a gender gap for all three plots above. Looks like we'll have to dig even deeper and analyze the data by job cateogries.
But first, let's take a look at the overall correlation for the data set.
```
data_2014_FT.corr()
```
The correlation table above uses Pearson's R to determine the values. The `BasePay` and `Benefits` column are very closely related. We can visualize this relationship using a scatter plot.
```
fig = plt.figure(figsize=(10, 5))
ax = plt.scatter(data_2014_FT['BasePay'], data_2014_FT['Benefits'])
plt.ylabel('Benefits ($)')
plt.xlabel('Base Pay ($)')
plt.show()
```
This makes a lot of sense because an employee's benefits is based on a percentage of their base pay. The San Francisco Human Resources department includes this information on their website [here](http://sfdhr.org/benefits-overview).
As we move further into our analysis of the data, it makes the most sense to focus on the `BasePay` column. Both `Benefits` and `OvertimePay` are dependent of the `BasePay`.
#### 4.5 - Breaking down the base pay by job category
Next we'll analyze the base pay of full time workers by job category.
```
pal = sns.diverging_palette(0, 255, n=2)
ax = sns.factorplot(x='BasePay', y='Job_Group', hue='Gender', data=data_2014_FT,
size=10, kind="bar", palette=pal, ci=None)
plt.title('Full Time Workers')
plt.xlabel('Base Pay ($)')
plt.ylabel('Job Group')
plt.show()
```
At a glance, we can't really draw any conclusive statements about the gender wage gap. Some job categories favor females, some favor males. It really depends on what job group the employee is actually in. Maybe it makes more sense to calculate the the difference between these two bars.
```
salaries_by_group = pd.pivot_table(data = data_2014_FT,
values = 'BasePay',
columns = 'Job_Group', index='Gender',
aggfunc = np.mean)
count_by_group = pd.pivot_table(data = data_2014_FT,
values = 'Id',
columns = 'Job_Group', index='Gender',
aggfunc = len)
salaries_by_group
fig = plt.figure(figsize=(10, 15))
sns.set(font_scale=1.5)
differences = (salaries_by_group.loc['female'] - salaries_by_group.loc['male'])*100/salaries_by_group.loc['male']
labels = differences.sort_values().index
x = differences.sort_values()
y = [i for i in range(len(differences))]
palette = sns.diverging_palette(240, 10, n=28, center ='dark')
ax = sns.barplot(x, y, orient = 'h', palette = palette)
#Draws the two arrows
bbox_props = dict(boxstyle="rarrow,pad=0.3", fc="white", ec="black", lw=1)
t = plt.text(5.5, 12, "Higher pay for females", ha="center", va="center", rotation=0,
size=15,
bbox=bbox_props)
bbox_props2 = dict(boxstyle="larrow,pad=0.3", fc="white", ec="black", lw=1)
t = plt.text(-5.5, 12, "Higher pay for males", ha="center", va="center", rotation=0,
size=15,
bbox=bbox_props2)
#Labels each bar with the percentage of females
percent_labels = count_by_group[labels].iloc[0]*100 \
/(count_by_group[labels].iloc[0] + count_by_group[labels].iloc[1])
for i in range(len(ax.patches)):
p = ax.patches[i]
width = p.get_width()*1+1
ax.text(15,
p.get_y()+p.get_height()/2+0.3,
'{:1.0f}'.format(percent_labels[i])+' %',
ha="center")
ax.text(15, -1+0.3, 'Female Representation',
ha="center", fontname='Arial', rotation = 0)
plt.yticks(range(len(differences)), labels)
plt.title('Full Time Workers (Base Pay)')
plt.xlabel('Mean Percent Difference in Pay (Females - Males)')
plt.xlim(-11, 11)
plt.show()
```
I believe this is a better way to represent the gender wage gap. I calculated the mean difference between female and male pay based on job categories. Then I converted the values into a percentage by using this formula:
$$ \text{Mean Percent Difference} = \frac{\text{(Female Mean Pay - Male Mean Pay)*100}} {\text{Male Mean Pay}} $$
The theory stating that women makes 78 cents for every dollar men makes implies a 22% pay difference. None of these percentages were more than 10%, and not all of these percentage values showed favoritism towards males. However, we should keep in mind that this data set only applies to San Francisco public jobs. We should also keep in mind that we do not have access to job experience data which would directly correlate with base pay.
In addition, I included a short table of female representation for each job group on the right side of the graph. We'll dig further into this on the next section.
#### 4.6 - Gender representation by job category
```
contingency_table = pd.crosstab(
data_2014_FT['Gender'],
data_2014_FT['Job_Group'],
margins = True
)
contingency_table
#Assigns the frequency values
femalecount = contingency_table.iloc[0][0:-1].values
malecount = contingency_table.iloc[1][0:-1].values
totals = contingency_table.iloc[2][0:-1]
femalepercentages = femalecount*100/totals
malepercentages = malecount*100/totals
malepercentages=malepercentages.sort_values(ascending=True)
femalepercentages=femalepercentages.sort_values(ascending=False)
length = range(len(femalepercentages))
#Plots the bar chart
fig = plt.figure(figsize=(10, 12))
sns.set(font_scale=1.5)
p1 = plt.barh(length, malepercentages.values, 0.55, label='Male', color='#AFAFF5')
p2 = plt.barh(length, femalepercentages, 0.55, left=malepercentages, color='#EFAFB5', label='Female')
labels = malepercentages.index
plt.yticks(range(len(malepercentages)), labels)
plt.xticks([0, 25, 50, 75, 100], ['0 %', '25 %', '50 %', '75 %', '100 %'])
plt.xlabel('Percentage of Males')
plt.title('Gender Representation by Job Group')
plt.legend(bbox_to_anchor=(0, 1, 1, 0), loc=3,
ncol=2, mode="expand", borderaxespad=0)
plt.show()
```
The chart above does not include any information based on pay. I wanted to show an overview of gender representation based on job category. It is safe to say, women don't like working with automotives with <1% female representation. Where as female representation is highest for medical jobs at 73%.
#### 4.7 - Significance testing by exact job title
So what if breaking down the wage gap by job category is not good enough? Should we break down the gender gap by exact job title? Afterall, the argument is for equal pay for equal work. We can assume equal work if the job titles are exactly the same.
We can use hypothesis testing using the Welch's t-test to determine if there is a statistically significant result between male and female wages. The Welch's t-test is very robust as it doesn't assume equal variance and equal sample size. It does however, assume a normal distrbution which is well represented by the KDE plots. I talk about this in detail in my blog post [here](https://codingdisciple.com/hypothesis-testing-welch-python.html).
Let's state our null and alternative hypothesis:
$ H_0 : \text{There is no statistically significant relationship between gender and pay.} $
$ H_a : \text{There is a statistically significant relationship between gender and pay.} $
We are going to use only job titles with more than 100 employees, and job titles with more than 30 females and 30 males for this t-test. Using a for loop, we'll perform the Welch's t-test on every job title tat matches our criteria.
```
from scipy import stats
#Significance testing by job title
job_titles = data_2014['JobTitle'].value_counts(dropna=True)
job_titles_over_100 = job_titles[job_titles > 100 ]
t_scores = {}
for title,count in job_titles_over_100.iteritems():
male_pay = pd.to_numeric(male_only[male_only['JobTitle'] == title]['BasePay'])
female_pay = pd.to_numeric(female_only[female_only['JobTitle'] == title]['BasePay'])
if female_pay.shape[0] < 30:
continue
if male_pay.shape[0] < 30:
continue
t_scores[title] = stats.ttest_ind_from_stats(
mean1=male_pay.mean(), std1=(male_pay.std()), nobs1= male_pay.shape[0], \
mean2=female_pay.mean(), std2=(female_pay.std()), nobs2=female_pay.shape[0], \
equal_var=False)
for key, value in t_scores.items():
if value[1] < 0.05:
print(len(key)*'-')
print(key)
print(len(key)*'-')
print(t_scores[key])
print(' ')
print('Male: {}'.format((male_only[male_only['JobTitle'] == key]['BasePay']).mean()))
print('sample size: {}'.format(male_only[male_only['JobTitle'] == key].shape[0]))
print(' ')
print('Female: {}'.format((female_only[female_only['JobTitle'] == key]['BasePay']).mean()))
print('sample size: {}'.format(female_only[female_only['JobTitle'] == key].shape[0]))
len(t_scores)
```
Out of the 25 jobs that were tested using the Welch's t-test, 5 jobs resulted in a p-value of less than 0.05. However, not all jobs showed favoritism towards males. 'Registered Nurse' and 'Senior Clerk' both showed an average pay in favor of females. However, we should take the Welch's t-test results with a grain of salt. We do not have data on the work experience of the employees. Maybe female nurses have more work experience over males. Maybe male transit operators have more work experience over females. We don't actually know. Since `BasePay` is a function of work experience, without this critical piece of information, we can not make any conclusions based on the t-test alone. All we know is that a statistically significant difference exists.
### 5.0 - San Francisco vs. Newport Beach
Let's take a look at more a more conservative city such as Newport Beach. This data can be downloaded at Transparent California [here](https://transparentcalifornia.com/salaries/2016/newport-beach/).
We can process the data similar to the San Francisco data set. The following code performs the following:
+ Read the data using pandas
+ Create the `Job_Group` column
+ Create the `Gender` column
+ Create two new dataframes: one for part time workers and one for full time workers
```
#Reads in the data
nb_data = pd.read_csv('newport-beach-2016.csv')
#Creates job groups
def find_job_title_nb(row):
titles = collections.OrderedDict([
('Police',['police', 'sherif', 'probation', 'sergeant', 'officer', 'lieutenant']),
('Fire', ['fire']),
('Transit',['mta', 'transit']),
('Medical',['anesth', 'medical', 'nurs', 'health', 'physician', 'orthopedic', 'pharm', 'care']),
('Architect', ['architect']),
('Court',['court', 'legal']),
('Mayor Office', ['mayoral']),
('Library', ['librar']),
('Public Works', ['public']),
('Attorney', ['attorney']),
('Custodian', ['custodian']),
('Gardener', ['garden']),
('Recreation Leader', ['recreation']),
('Automotive',['automotive', 'mechanic', 'truck']),
('Engineer',['engineer', 'engr', 'eng', 'program']),
('General Laborer',['general laborer', 'painter', 'inspector', 'carpenter', 'electrician', 'plumber', 'maintenance']),
('Food Services', ['food serv']),
('Clerk', ['clerk']),
('Porter', ['porter']),
('Airport Staff', ['airport']),
('Social Worker',['worker']),
('Guard', ['guard']),
('Assistant',['aide', 'assistant', 'secretary', 'attendant']),
('Analyst', ['analy']),
('Manager', ['manager'])
])
#Loops through the dictionaries
for group, keywords in titles.items():
for keyword in keywords:
if keyword in row.lower():
return group
return 'Other'
start_time = time.time()
nb_data["Job_Group"]=data["JobTitle"].map(find_job_title_nb)
#Create the 'Gender' column based on employee's first name.
d = gender.Detector(case_sensitive=False)
nb_data['FirstName'] = nb_data['Employee Name'].str.split().apply(lambda x: x[0])
nb_data['Gender'] = nb_data['FirstName'].apply(lambda x: d.get_gender(x))
nb_data['Gender'].value_counts()
#Retain data with 'male' and 'female' names.
nb_male_female_only = nb_data[(nb_data['Gender'] == 'male') | (nb_data['Gender'] == 'female')]
nb_male_female_only['Gender'].value_counts()
#Seperates full time/part time data
nb_data_FT = nb_male_female_only[nb_male_female_only['Status'] == 'FT']
nb_data_PT = nb_male_female_only[nb_male_female_only['Status'] == 'PT']
nb_data_FT.head()
```
#### 5.1 - Part time vs. full time workers
```
fig = plt.figure(figsize=(10, 5))
nb_male_only = nb_data_PT[nb_data_PT['Gender'] == 'male']
nb_female_only = nb_data_PT[nb_data_PT['Gender'] == 'female']
ax = fig.add_subplot(1, 1, 1)
ax = sns.kdeplot(nb_male_only['Total Pay & Benefits'], color ='Blue', label='Male', shade=True)
ax = sns.kdeplot(nb_female_only['Total Pay & Benefits'], color='Red', label='Female', shade=True)
plt.title('Newport Beach: Part Time Workers')
plt.ylabel('Density of Employees')
plt.xlabel('Total Pay + Benefits ($)')
plt.xlim(0, 400000)
plt.yticks([])
plt.show()
```
Similar to the KDE plot for San Francisco, the KDE plot is nearly identical for both males and females for part time workers.
Let's take a look at the full time workers.
```
fig = plt.figure(figsize=(10, 10))
fig.subplots_adjust(hspace=.5)
#Generate the top chart
nb_male_only = nb_data_FT[nb_data_FT['Gender'] == 'male']
nb_female_only = nb_data_FT[nb_data_FT['Gender'] == 'female']
ax = fig.add_subplot(2, 1, 1)
ax = sns.kdeplot(nb_male_only['Total Pay & Benefits'], color ='Blue', label='Male', shade=True)
ax = sns.kdeplot(nb_female_only['Total Pay & Benefits'], color='Red', label='Female', shade=True)
plt.title('Newport Beach: Full Time Workers')
plt.ylabel('Density of Employees')
plt.xlabel('Total Pay + Benefits ($)')
plt.xlim(0, 400000)
plt.yticks([])
#Generate the bottom chart
male_only = data_2014_FT[data_2014_FT['Gender'] == 'male']
female_only = data_2014_FT[data_2014_FT['Gender'] == 'female']
ax2 = fig.add_subplot(2, 1, 2)
ax2 = sns.kdeplot(male_only['TotalPayBenefits'], color ='Blue', label='Male', shade=True)
ax2 = sns.kdeplot(female_only['TotalPayBenefits'], color='Red', label='Female', shade=True)
plt.title('San Francisco: Full Time Workers')
plt.ylabel('Density of Employees')
plt.xlabel('Total Pay + Benefits ($)')
plt.xlim(0, 400000)
plt.yticks([])
plt.show()
```
The kurtosis of the KDE plot for Newport Beach full time workers is lower than KDE plot for San Francisco full time workers. We can see a higher gender wage gap for Newport beach workers than San Francisco workers. However, these two plots do not tell us the full story. We need to break down the data by job category.
#### 5.2 - Comparisons by job cateogry
```
nb_salaries_by_group = pd.pivot_table(data = nb_data_FT,
values = 'Base Pay',
columns = 'Job_Group', index='Gender',
aggfunc = np.mean,)
nb_salaries_by_group
fig = plt.figure(figsize=(10, 7.5))
sns.set(font_scale=1.5)
differences = (nb_salaries_by_group.loc['female'] - nb_salaries_by_group.loc['male'])*100/nb_salaries_by_group.loc['male']
nb_labels = differences.sort_values().index
x = differences.sort_values()
y = [i for i in range(len(differences))]
nb_palette = sns.diverging_palette(240, 10, n=9, center ='dark')
ax = sns.barplot(x, y, orient = 'h', palette = nb_palette)
plt.yticks(range(len(differences)), nb_labels)
plt.title('Newport Beach: Full Time Workers (Base Pay)')
plt.xlabel('Mean Percent Difference in Pay (Females - Males)')
plt.xlim(-25, 25)
plt.show()
```
Most of these jobs shows a higher average pay for males. The only job category where females were paid higher on average was 'Manager'. Some of these job categories do not even have a single female within the category, so the difference cannot be calculated. We should create a contingency table to check the sample size of our data.
#### 5.3 - Gender representation by job category
```
nb_contingency_table = pd.crosstab(
nb_data_FT['Gender'],
nb_data_FT['Job_Group'],
margins = True
)
nb_contingency_table
```
The number of public jobs is much lower in Newport Beach compared to San Francisco. With only 3 female managers working full time in Newport Beach, we can't really say female managers make more money on average than male managers.
```
#Assigns the frequency values
nb_femalecount = nb_contingency_table.iloc[0][0:-1].values
nb_malecount = nb_contingency_table.iloc[1][0:-1].values
nb_totals = nb_contingency_table.iloc[2][0:-1]
nb_femalepercentages = nb_femalecount*100/nb_totals
nb_malepercentages = nb_malecount*100/nb_totals
nb_malepercentages=nb_malepercentages.sort_values(ascending=True)
nb_femalepercentages=nb_femalepercentages.sort_values(ascending=False)
nb_length = range(len(nb_malepercentages))
#Plots the bar chart
fig = plt.figure(figsize=(10, 10))
sns.set(font_scale=1.5)
p1 = plt.barh(nb_length, nb_malepercentages.values, 0.55, label='Male', color='#AFAFF5')
p2 = plt.barh(nb_length, nb_femalepercentages, 0.55, left=nb_malepercentages, color='#EFAFB5', label='Female')
labels = nb_malepercentages.index
plt.yticks(range(len(nb_malepercentages)), labels)
plt.xticks([0, 25, 50, 75, 100], ['0 %', '25 %', '50 %', '75 %', '100 %'])
plt.xlabel('Percentage of Males')
plt.title('Gender Representation by Job Group')
plt.legend(bbox_to_anchor=(0, 1, 1, 0), loc=3,
ncol=2, mode="expand", borderaxespad=0)
plt.show()
fig = plt.figure(figsize=(10, 5))
colors = ['#AFAFF5', '#EFAFB5']
labels = ['Male', 'Female']
sizes = [len(nb_male_only), len(nb_female_only)]
explode = (0.05, 0)
sns.set(font_scale=1.5)
ax = fig.add_subplot(1, 2, 1)
ax = plt.pie(sizes, labels=labels, explode=explode, colors=colors, shadow=True, startangle=90, autopct='%1.f%%')
plt.title('Newport Beach: Full Time')
sizes = [len(male_only), len(female_only)]
explode = (0.05, 0)
sns.set(font_scale=1.5)
ax2 = fig.add_subplot(1, 2, 2)
ax2 = plt.pie(sizes, labels=labels, explode=explode, colors=colors, shadow=True, startangle=90, autopct='%1.f%%')
plt.title('San Francisco: Full Time')
plt.show()
```
Looking at the plots above. There are fewer females working full time public jobs in Newport Beach compared to San Francisco.
### 6.0 - Conclusion
It is very easy for people to say there is a gender wage gap and make general statements about it. But the real concern is whether if there is social injustice and discrimination involved. Yes, there is an overall gender wage gap for both San Francisco and Newport Beach. In both cases, the income distribution for part time employees were nearly identical for both males and females.
For full time public positions in San Francisco, an overall gender wage gap can be observed. When the full time positions were broken down to job categories, the gender wage gap went both ways. Some jobs favored men, some favored women. For full time public positions in Newport Beach, the majority of the jobs favored men.
However, we were missing a critical piece of information in this entire analysis. We don't have any information on the job experience of the employees. Maybe the men just had more job experience in Newport Beach, we don't actually know. For San Francisco, we assumed equal experience by comparing employees with the same exact job titles. Only job titles with a size greater than 100 were chosen. Out of the 25 job titles that were selected, 5 of them showed a statistically significant result with the Welch's t-test. Two of those jobs showed an average base pay in favor of females.
Overall, I do not believe the '78 cents to a dollar' is a fair statement. It generalizes the data and oversimplifies the problem. There are many hidden factors that is not shown by the data. Maybe women are less likely to ask for a promotion. Maybe women perform really well in the medical world. Maybe the men's body is more suitable for the police officer role. Maybe women are more organized than men and make better libarians. The list goes on and on, the point is, we should always be skeptical of what the numbers tell us. The truth is, men and women are different on a fundamental level. Social injustices and gender discrimination should be analyzed on a case by case basis.
|
github_jupyter
|
```
import tensorflow as tf
import keras
import keras.backend as K
from sklearn.utils import shuffle
from sklearn.metrics import classification_report, confusion_matrix, accuracy_score, f1_score
from collections import Counter
from keras import regularizers
from keras.models import Sequential, Model, load_model, model_from_json
from keras.utils import to_categorical
from keras.layers import Input, Dense, Flatten, Reshape, Concatenate, Dropout
from keras.layers import Conv2D, MaxPooling2D, UpSampling2D, Conv2DTranspose
from keras.layers.normalization import BatchNormalization
from keras.callbacks import ModelCheckpoint
from keras.utils import np_utils
from keras.layers.advanced_activations import LeakyReLU
def get_class_weights(y):
counter = Counter(y)
majority = max(counter.values())
return {cls: float(majority/count) for cls, count in counter.items()}
class Estimator:
l2p = 0.001
@staticmethod
def early_layers(inp, fm = (1,3), hid_act_func="relu"):
# Start
x = Conv2D(64, fm, padding="same", kernel_regularizer=regularizers.l2(Estimator.l2p), activation=hid_act_func)(inp)
x = BatchNormalization()(x)
x = MaxPooling2D(pool_size=(1, 2))(x)
x = Dropout(0.25)(x)
# 1
x = Conv2D(64, fm, padding="same", kernel_regularizer=regularizers.l2(Estimator.l2p), activation=hid_act_func)(x)
x = BatchNormalization()(x)
x = MaxPooling2D(pool_size=(1, 2))(x)
x = Dropout(0.25)(x)
return x
@staticmethod
def late_layers(inp, num_classes, fm = (1,3), act_func="softmax", hid_act_func="relu", b_name="Identifier"):
# 2
x = Conv2D(32, fm, padding="same", kernel_regularizer=regularizers.l2(Estimator.l2p), activation=hid_act_func)(inp)
x = BatchNormalization()(x)
x = MaxPooling2D(pool_size=(1, 2))(x)
x = Dropout(0.25)(x)
# 3
x = Conv2D(32, fm, padding="same", kernel_regularizer=regularizers.l2(Estimator.l2p), activation=hid_act_func)(x)
x = BatchNormalization()(x)
x = MaxPooling2D(pool_size=(1, 2))(x)
x = Dropout(0.25)(x)
# End
x = Flatten()(x)
x = Dense(256, kernel_regularizer=regularizers.l2(Estimator.l2p), activation=hid_act_func)(x)
x = BatchNormalization()(x)
x = Dropout(0.5)(x)
x = Dense(64, kernel_regularizer=regularizers.l2(Estimator.l2p), activation=hid_act_func)(x)
x = BatchNormalization()(x)
x = Dropout(0.5)(x)
x = Dense(num_classes, activation=act_func, name = b_name)(x)
return x
@staticmethod
def build(height, width, num_classes, name, fm = (1,3), act_func="softmax",hid_act_func="relu"):
inp = Input(shape=(height, width, 1))
early = Estimator.early_layers(inp, fm, hid_act_func=hid_act_func)
late = Estimator.late_layers(early, num_classes, fm, act_func=act_func, hid_act_func=hid_act_func)
model = Model(inputs=inp, outputs=late ,name=name)
return model
import numpy as np
import pandas as pd
from pandas.plotting import autocorrelation_plot
import matplotlib.pyplot as plt
def get_ds_infos():
"""
Read the file includes data subject information.
Data Columns:
0: code [1-24]
1: weight [kg]
2: height [cm]
3: age [years]
4: gender [0:Female, 1:Male]
Returns:
A pandas DataFrame that contains inforamtion about data subjects' attributes
"""
dss = pd.read_csv("data_subjects_info.csv")
print("[INFO] -- Data subjects' information is imported.")
return dss
def set_data_types(data_types=["userAcceleration"]):
"""
Select the sensors and the mode to shape the final dataset.
Args:
data_types: A list of sensor data type from this list: [attitude, gravity, rotationRate, userAcceleration]
Returns:
It returns a list of columns to use for creating time-series from files.
"""
dt_list = []
for t in data_types:
if t != "attitude":
dt_list.append([t+".x",t+".y",t+".z"])
else:
dt_list.append([t+".roll", t+".pitch", t+".yaw"])
return dt_list
def creat_time_series(dt_list, act_labels, trial_codes, mode="mag", labeled=True, combine_grav_acc=False):
"""
Args:
dt_list: A list of columns that shows the type of data we want.
act_labels: list of activites
trial_codes: list of trials
mode: It can be "raw" which means you want raw data
for every dimention of each data type,
[attitude(roll, pitch, yaw); gravity(x, y, z); rotationRate(x, y, z); userAcceleration(x,y,z)].
or it can be "mag" which means you only want the magnitude for each data type: (x^2+y^2+z^2)^(1/2)
labeled: True, if we want a labeld dataset. False, if we only want sensor values.
combine_grav_acc: True, means adding each axis of gravity to corresponding axis of userAcceleration.
Returns:
It returns a time-series of sensor data.
"""
num_data_cols = len(dt_list) if mode == "mag" else len(dt_list*3)
if labeled:
dataset = np.zeros((0,num_data_cols+7)) # "7" --> [act, code, weight, height, age, gender, trial]
else:
dataset = np.zeros((0,num_data_cols))
ds_list = get_ds_infos()
print("[INFO] -- Creating Time-Series")
for sub_id in ds_list["code"]:
for act_id, act in enumerate(act_labels):
for trial in trial_codes[act_id]:
fname = 'A_DeviceMotion_data/'+act+'_'+str(trial)+'/sub_'+str(int(sub_id))+'.csv'
raw_data = pd.read_csv(fname)
raw_data = raw_data.drop(['Unnamed: 0'], axis=1)
vals = np.zeros((len(raw_data), num_data_cols))
if combine_grav_acc:
raw_data["userAcceleration.x"] = raw_data["userAcceleration.x"].add(raw_data["gravity.x"])
raw_data["userAcceleration.y"] = raw_data["userAcceleration.y"].add(raw_data["gravity.y"])
raw_data["userAcceleration.z"] = raw_data["userAcceleration.z"].add(raw_data["gravity.z"])
for x_id, axes in enumerate(dt_list):
if mode == "mag":
vals[:,x_id] = (raw_data[axes]**2).sum(axis=1)**0.5
else:
vals[:,x_id*3:(x_id+1)*3] = raw_data[axes].values
vals = vals[:,:num_data_cols]
if labeled:
lbls = np.array([[act_id,
sub_id-1,
ds_list["weight"][sub_id-1],
ds_list["height"][sub_id-1],
ds_list["age"][sub_id-1],
ds_list["gender"][sub_id-1],
trial
]]*len(raw_data))
vals = np.concatenate((vals, lbls), axis=1)
dataset = np.append(dataset,vals, axis=0)
cols = []
for axes in dt_list:
if mode == "raw":
cols += axes
else:
cols += [str(axes[0][:-2])]
if labeled:
cols += ["act", "id", "weight", "height", "age", "gender", "trial"]
dataset = pd.DataFrame(data=dataset, columns=cols)
return dataset
#________________________________
#________________________________
def ts_to_secs(dataset, w, s, standardize = False, **options):
data = dataset[dataset.columns[:-7]].values
act_labels = dataset["act"].values
id_labels = dataset["id"].values
trial_labels = dataset["trial"].values
mean = 0
std = 1
if standardize:
## Standardize each sensor’s data to have a zero mean and unity standard deviation.
## As usual, we normalize test dataset by training dataset's parameters
if options:
mean = options.get("mean")
std = options.get("std")
print("[INFO] -- Test/Val Data has been standardized")
else:
mean = data.mean(axis=0)
std = data.std(axis=0)
print("[INFO] -- Training Data has been standardized: the mean is = "+str(mean)+" ; and the std is = "+str(std))
data -= mean
data /= std
else:
print("[INFO] -- Without Standardization.....")
## We want the Rows of matrices show each Feature and the Columns show time points.
data = data.T
m = data.shape[0] # Data Dimension
ttp = data.shape[1] # Total Time Points
number_of_secs = int(round(((ttp - w)/s)))
## Create a 3D matrix for Storing Sections
secs_data = np.zeros((number_of_secs , m , w ))
act_secs_labels = np.zeros(number_of_secs)
id_secs_labels = np.zeros(number_of_secs)
k=0
for i in range(0 , ttp-w, s):
j = i // s
if j >= number_of_secs:
break
if id_labels[i] != id_labels[i+w-1]:
continue
if act_labels[i] != act_labels[i+w-1]:
continue
if trial_labels[i] != trial_labels[i+w-1]:
continue
secs_data[k] = data[:, i:i+w]
act_secs_labels[k] = act_labels[i].astype(int)
id_secs_labels[k] = id_labels[i].astype(int)
k = k+1
secs_data = secs_data[0:k]
act_secs_labels = act_secs_labels[0:k]
id_secs_labels = id_secs_labels[0:k]
return secs_data, act_secs_labels, id_secs_labels, mean, std
##________________________________________________________________
ACT_LABELS = ["dws","ups", "wlk", "jog", "std", "sit"]
TRIAL_CODES = {
ACT_LABELS[0]:[1,2,11],
ACT_LABELS[1]:[3,4,12],
ACT_LABELS[2]:[7,8,15],
ACT_LABELS[3]:[9,16],
ACT_LABELS[4]:[6,14],
ACT_LABELS[5]:[5,13],
}
#https://stackoverflow.com/a/45305384/5210098
def f1_metric(y_true, y_pred):
def recall(y_true, y_pred):
"""Recall metric.
Only computes a batch-wise average of recall.
Computes the recall, a metric for multi-label classification of
how many relevant items are selected.
"""
true_positives = K.sum(K.round(K.clip(y_true * y_pred, 0, 1)))
possible_positives = K.sum(K.round(K.clip(y_true, 0, 1)))
recall = true_positives / (possible_positives + K.epsilon())
return recall
def precision(y_true, y_pred):
"""Precision metric.
Only computes a batch-wise average of precision.
Computes the precision, a metric for multi-label classification of
how many selected items are relevant.
"""
true_positives = K.sum(K.round(K.clip(y_true * y_pred, 0, 1)))
predicted_positives = K.sum(K.round(K.clip(y_pred, 0, 1)))
precision = true_positives / (predicted_positives + K.epsilon())
return precision
precision = precision(y_true, y_pred)
recall = recall(y_true, y_pred)
return 2*((precision*recall)/(precision+recall+K.epsilon()))
def eval_id(sdt, mode, ep, cga):
print("[INFO] -- Selected sensor data types: "+str(sdt)+" -- Mode: "+str(mode)+" -- Grav+Acc: "+str(cga))
act_labels = ACT_LABELS [0:4]
print("[INFO] -- Selected activites: "+str(act_labels))
trial_codes = [TRIAL_CODES[act] for act in act_labels]
dt_list = set_data_types(sdt)
dataset = creat_time_series(dt_list, act_labels, trial_codes, mode=mode, labeled=True, combine_grav_acc = cga)
print("[INFO] -- Shape of time-Series dataset:"+str(dataset.shape))
#*****************
TRAIN_TEST_TYPE = "trial" # "subject" or "trial"
#*****************
if TRAIN_TEST_TYPE == "subject":
test_ids = [4,9,11,21]
print("[INFO] -- Test IDs: "+str(test_ids))
test_ts = dataset.loc[(dataset['id'].isin(test_ids))]
train_ts = dataset.loc[~(dataset['id'].isin(test_ids))]
else:
test_trail = [11,12,13,14,15,16]
print("[INFO] -- Test Trials: "+str(test_trail))
test_ts = dataset.loc[(dataset['trial'].isin(test_trail))]
train_ts = dataset.loc[~(dataset['trial'].isin(test_trail))]
print("[INFO] -- Shape of Train Time-Series :"+str(train_ts.shape))
print("[INFO] -- Shape of Test Time-Series :"+str(test_ts.shape))
print("___________________________________________________")
## This Variable Defines the Size of Sliding Window
## ( e.g. 100 means in each snapshot we just consider 100 consecutive observations of each sensor)
w = 128 # 50 Equals to 1 second for MotionSense Dataset (it is on 50Hz samplig rate)
## Here We Choose Step Size for Building Diffrent Snapshots from Time-Series Data
## ( smaller step size will increase the amount of the instances and higher computational cost may be incurred )
s = 10
train_data, act_train, id_train, train_mean, train_std = ts_to_secs(train_ts.copy(),
w,
s,
standardize = True)
s = 10
test_data, act_test, id_test, test_mean, test_std = ts_to_secs(test_ts.copy(),
w,
s,
standardize = True,
mean = train_mean,
std = train_std)
print("[INFO] -- Training Sections: "+str(train_data.shape))
print("[INFO] -- Test Sections: "+str(test_data.shape))
id_train_labels = to_categorical(id_train)
id_test_labels = to_categorical(id_test)
act_train_labels = to_categorical(act_train)
act_test_labels = to_categorical(act_test)
## Here we add an extra dimension to the datasets just to be ready for using with Convolution2D
train_data = np.expand_dims(train_data,axis=3)
print("[INFO] -- Training Sections:"+str(train_data.shape))
test_data = np.expand_dims(test_data,axis=3)
print("[INFO] -- Test Sections:"+str(test_data.shape))
height = train_data.shape[1]
width = train_data.shape[2]
id_class_numbers = 24
act_class_numbers = 4
fm = (1,5)
print("___________________________________________________")
## Callbacks
#eval_metric= "val_acc"
eval_metric= "val_f1_metric"
early_stop = keras.callbacks.EarlyStopping(monitor=eval_metric, mode='max', patience = 7)
filepath="MID.best.hdf5"
checkpoint = ModelCheckpoint(filepath, monitor=eval_metric, verbose=0, save_best_only=True, mode='max')
callbacks_list = [early_stop,
checkpoint
]
## Callbacks
eval_id = Estimator.build(height, width, id_class_numbers, name ="EVAL_ID", fm=fm, act_func="softmax",hid_act_func="relu")
eval_id.compile( loss="categorical_crossentropy", optimizer='adam', metrics=['acc', f1_metric])
print("Model Size = "+str(eval_id.count_params()))
eval_id.fit(train_data, id_train_labels,
validation_data = (test_data, id_test_labels),
epochs = ep,
batch_size = 128,
verbose = 0,
class_weight = get_class_weights(np.argmax(id_train_labels,axis=1)),
callbacks = callbacks_list
)
eval_id.load_weights("MID.best.hdf5")
eval_id.compile( loss="categorical_crossentropy", optimizer='adam', metrics=['acc',f1_metric])
result1 = eval_id.evaluate(test_data, id_test_labels, verbose = 2)
id_acc = result1[1]
print("***[RESULT]*** ID Accuracy: "+str(id_acc))
rf1 = result1[2].round(4)*100
print("***[RESULT]*** ID F1: "+str(rf1))
preds = eval_id.predict(test_data)
preds = np.argmax(preds, axis=1)
conf_mat = confusion_matrix(np.argmax(id_test_labels, axis=1), preds)
conf_mat = conf_mat.astype('float') / conf_mat.sum(axis=1)[:, np.newaxis]
print("***[RESULT]*** ID Confusion Matrix")
print((np.array(conf_mat).diagonal()).round(3)*100)
d_test_ids = [4,9,11,21]
to_avg = 0
for i in range(len(d_test_ids)):
true_positive = conf_mat[d_test_ids[i],d_test_ids[i]]
print("True Positive Rate for "+str(d_test_ids[i])+" : "+str(true_positive*100))
to_avg+=true_positive
atp = to_avg/len(d_test_ids)
print("Average TP:"+str(atp*100))
f1id = f1_score(np.argmax(id_test_labels, axis=1), preds, average=None).mean()
print("***[RESULT]*** ID Averaged F-1 Score : "+str(f1id))
return [round(id_acc,4), round(f1id,4), round(atp,4)]
results ={}
## Here we set parameter to build labeld time-series from dataset of "(A)DeviceMotion_data"
## attitude(roll, pitch, yaw); gravity(x, y, z); rotationRate(x, y, z); userAcceleration(x,y,z)
sdt = ["rotationRate"]
mode = "mag"
ep = 40
cga = False # Add gravity to acceleration or not
for i in range(5):
results[str(sdt)+"--"+str(mode)+"--"+str(cga)+"--"+str(i)] = eval_id(sdt, mode, ep, cga)
## Here we set parameter to build labeld time-series from dataset of "(A)DeviceMotion_data"
## attitude(roll, pitch, yaw); gravity(x, y, z); rotationRate(x, y, z); userAcceleration(x,y,z)
sdt = ["rotationRate"]
mode = "raw"
ep = 40
cga = False # Add gravity to acceleration or not
for i in range(5):
results[str(sdt)+"--"+str(mode)+"--"+str(cga)+"--"+str(i)] = eval_id(sdt, mode, ep, cga)
results
## Here we set parameter to build labeld time-series from dataset of "(A)DeviceMotion_data"
## attitude(roll, pitch, yaw); gravity(x, y, z); rotationRate(x, y, z); userAcceleration(x,y,z)
sdt = ["userAcceleration"]
mode = "mag"
ep = 40
cga = True # Add gravity to acceleration or not
for i in range(5):
results[str(sdt)+"--"+str(mode)+"--"+str(cga)+"--"+str(i)] = eval_id(sdt, mode, ep, cga)
results
## Here we set parameter to build labeld time-series from dataset of "(A)DeviceMotion_data"
## attitude(roll, pitch, yaw); gravity(x, y, z); rotationRate(x, y, z); userAcceleration(x,y,z)
sdt = ["userAcceleration"]
mode = "raw"
ep = 40
cga = True # Add gravity to acceleration or not
for i in range(5):
results[str(sdt)+"--"+str(mode)+"--"+str(cga)+"--"+str(i)] = eval_id(sdt, mode, ep, cga)
results
## Here we set parameter to build labeld time-series from dataset of "(A)DeviceMotion_data"
## attitude(roll, pitch, yaw); gravity(x, y, z); rotationRate(x, y, z); userAcceleration(x,y,z)
sdt = ["rotationRate","userAcceleration"]
mode = "mag"
ep = 40
cga = True # Add gravity to acceleration or not
for i in range(5):
results[str(sdt)+"--"+str(mode)+"--"+str(cga)+"--"+str(i)] = eval_id(sdt, mode, ep, cga)
results
## Here we set parameter to build labeld time-series from dataset of "(A)DeviceMotion_data"
## attitude(roll, pitch, yaw); gravity(x, y, z); rotationRate(x, y, z); userAcceleration(x,y,z)
sdt = ["rotationRate","userAcceleration"]
mode = "raw"
ep = 40
cga = True # Add gravity to acceleration or not
for i in range(5):
results[str(sdt)+"--"+str(mode)+"--"+str(cga)+"--"+str(i)] = eval_id(sdt, mode, ep, cga)
results
#https://stackoverflow.com/a/45305384/5210098
def f1_metric(y_true, y_pred):
def recall(y_true, y_pred):
"""Recall metric.
Only computes a batch-wise average of recall.
Computes the recall, a metric for multi-label classification of
how many relevant items are selected.
"""
true_positives = K.sum(K.round(K.clip(y_true * y_pred, 0, 1)))
possible_positives = K.sum(K.round(K.clip(y_true, 0, 1)))
recall = true_positives / (possible_positives + K.epsilon())
return recall
def precision(y_true, y_pred):
"""Precision metric.
Only computes a batch-wise average of precision.
Computes the precision, a metric for multi-label classification of
how many selected items are relevant.
"""
true_positives = K.sum(K.round(K.clip(y_true * y_pred, 0, 1)))
predicted_positives = K.sum(K.round(K.clip(y_pred, 0, 1)))
precision = true_positives / (predicted_positives + K.epsilon())
return precision
precision = precision(y_true, y_pred)
recall = recall(y_true, y_pred)
return 2*((precision*recall)/(precision+recall+K.epsilon()))
def eval_id(sdt, mode, ep, cga):
print("[INFO] -- Selected sensor data types: "+str(sdt)+" -- Mode: "+str(mode)+" -- Grav+Acc: "+str(cga))
act_labels = ACT_LABELS [0:4]
print("[INFO] -- Selected activites: "+str(act_labels))
trial_codes = [TRIAL_CODES[act] for act in act_labels]
dt_list = set_data_types(sdt)
dataset = creat_time_series(dt_list, act_labels, trial_codes, mode=mode, labeled=True, combine_grav_acc = cga)
print("[INFO] -- Shape of time-Series dataset:"+str(dataset.shape))
#*****************
TRAIN_TEST_TYPE = "trial" # "subject" or "trial"
#*****************
if TRAIN_TEST_TYPE == "subject":
test_ids = [4,9,11,21]
print("[INFO] -- Test IDs: "+str(test_ids))
test_ts = dataset.loc[(dataset['id'].isin(test_ids))]
train_ts = dataset.loc[~(dataset['id'].isin(test_ids))]
else:
test_trail = [11,12,13,14,15,16]
print("[INFO] -- Test Trials: "+str(test_trail))
test_ts = dataset.loc[(dataset['trial'].isin(test_trail))]
train_ts = dataset.loc[~(dataset['trial'].isin(test_trail))]
print("[INFO] -- Shape of Train Time-Series :"+str(train_ts.shape))
print("[INFO] -- Shape of Test Time-Series :"+str(test_ts.shape))
# print("___________Train_VAL____________")
# val_trail = [11,12,13,14,15,16]
# val_ts = train_ts.loc[(train_ts['trial'].isin(val_trail))]
# train_ts = train_ts.loc[~(train_ts['trial'].isin(val_trail))]
# print("[INFO] -- Training Time-Series :"+str(train_ts.shape))
# print("[INFO] -- Validation Time-Series :"+str(val_ts.shape))
print("___________________________________________________")
## This Variable Defines the Size of Sliding Window
## ( e.g. 100 means in each snapshot we just consider 100 consecutive observations of each sensor)
w = 128 # 50 Equals to 1 second for MotionSense Dataset (it is on 50Hz samplig rate)
## Here We Choose Step Size for Building Diffrent Snapshots from Time-Series Data
## ( smaller step size will increase the amount of the instances and higher computational cost may be incurred )
s = 10
train_data, act_train, id_train, train_mean, train_std = ts_to_secs(train_ts.copy(),
w,
s,
standardize = True)
s = 10
test_data, act_test, id_test, test_mean, test_std = ts_to_secs(test_ts.copy(),
w,
s,
standardize = True,
mean = train_mean,
std = train_std)
print("[INFO] -- Training Sections: "+str(train_data.shape))
print("[INFO] -- Test Sections: "+str(test_data.shape))
id_train_labels = to_categorical(id_train)
id_test_labels = to_categorical(id_test)
act_train_labels = to_categorical(act_train)
act_test_labels = to_categorical(act_test)
## Here we add an extra dimension to the datasets just to be ready for using with Convolution2D
train_data = np.expand_dims(train_data,axis=3)
print("[INFO] -- Training Sections:"+str(train_data.shape))
test_data = np.expand_dims(test_data,axis=3)
print("[INFO] -- Test Sections:"+str(test_data.shape))
height = train_data.shape[1]
width = train_data.shape[2]
id_class_numbers = 24
act_class_numbers = 4
fm = (2,5)
print("___________________________________________________")
## Callbacks
#eval_metric= "val_acc"
eval_metric= "val_f1_metric"
early_stop = keras.callbacks.EarlyStopping(monitor=eval_metric, mode='max', patience = 7)
filepath="MID.best.hdf5"
checkpoint = ModelCheckpoint(filepath, monitor=eval_metric, verbose=0, save_best_only=True, mode='max')
callbacks_list = [early_stop,
checkpoint
]
## Callbacks
eval_id = Estimator.build(height, width, id_class_numbers, name ="EVAL_ID", fm=fm, act_func="softmax",hid_act_func="relu")
eval_id.compile( loss="categorical_crossentropy", optimizer='adam', metrics=['acc', f1_metric])
print("Model Size = "+str(eval_id.count_params()))
eval_id.fit(train_data, id_train_labels,
validation_data = (test_data, id_test_labels),
epochs = ep,
batch_size = 128,
verbose = 0,
class_weight = get_class_weights(np.argmax(id_train_labels,axis=1)),
callbacks = callbacks_list
)
eval_id.load_weights("MID.best.hdf5")
eval_id.compile( loss="categorical_crossentropy", optimizer='adam', metrics=['acc',f1_metric])
result1 = eval_id.evaluate(test_data, id_test_labels, verbose = 2)
id_acc = result1[1]
print("***[RESULT]*** ID Accuracy: "+str(id_acc))
rf1 = result1[2].round(4)*100
print("***[RESULT]*** ID F1: "+str(rf1))
preds = eval_id.predict(test_data)
preds = np.argmax(preds, axis=1)
conf_mat = confusion_matrix(np.argmax(id_test_labels, axis=1), preds)
conf_mat = conf_mat.astype('float') / conf_mat.sum(axis=1)[:, np.newaxis]
print("***[RESULT]*** ID Confusion Matrix")
print((np.array(conf_mat).diagonal()).round(3)*100)
d_test_ids = [4,9,11,21]
to_avg = 0
for i in range(len(d_test_ids)):
true_positive = conf_mat[d_test_ids[i],d_test_ids[i]]
print("True Positive Rate for "+str(d_test_ids[i])+" : "+str(true_positive*100))
to_avg+=true_positive
atp = to_avg/len(d_test_ids)
print("Average TP:"+str(atp*100))
f1id = f1_score(np.argmax(id_test_labels, axis=1), preds, average=None).mean()
print("***[RESULT]*** ID Averaged F-1 Score : "+str(f1id))
return [round(id_acc,4), round(f1id,4), round(atp,4)]
## Here we set parameter to build labeld time-series from dataset of "(A)DeviceMotion_data"
## attitude(roll, pitch, yaw); gravity(x, y, z); rotationRate(x, y, z); userAcceleration(x,y,z)
sdt = ["rotationRate","userAcceleration"]
mode = "mag"
ep = 40
cga = True # Add gravity to acceleration or not
for i in range(5):
results[str(sdt)+"-2D-"+str(mode)+"--"+str(cga)+"--"+str(i)] = eval_id(sdt, mode, ep, cga)
```
|
github_jupyter
|
<figure>
<IMG SRC="https://raw.githubusercontent.com/mbakker7/exploratory_computing_with_python/master/tudelft_logo.png" WIDTH=250 ALIGN="right">
</figure>
# Exploratory Computing with Python
*Developed by Mark Bakker*
## Notebook 9: Discrete random variables
In this Notebook you learn how to deal with discrete random variables. Many of the functions we will use are included in the `random` subpackage of `numpy`. We will import this package and call it `rnd` so that we don't have to type `np.random.` all the time.
```
import numpy as np
import matplotlib.pyplot as plt
import numpy.random as rnd
%matplotlib inline
```
### Random numbers
A random number generator lets you draw, at random, a number from a specified distribution. Several random number generators are included in the `random` subpackage of `numpy`. For example, the `ranint(low, high, size)` function returns an integer array of shape `size` at random from `low` up to (but not including) `high`. For example, let's flip a coin 10 times and assign a 0 to heads and a 1 to tails. Note that the `high` is specified as `1 + 1`, which means it is `1` higher than the value we want.
```
rnd.randint(0, 1 + 1, 10)
```
If we call the `ran_int` function again, we get a different sequence of heads (zeros) and tails (ones):
```
rnd.randint(0, 1 + 1, 10)
```
Internally, the random number generator starts with what is called a *seed*. The seed is a number and is generated automatically (and supposedly at random) when you call the random number generator. The value of the seed exactly defines the sequence of random numbers that you get (so some people may argue that the generated sequence is at best pseudo-random, and you may not want to use the sequence for any serious cryptographic use, but for our purposes they are random enough). For example, let's set `seed` equal to 10
```
rnd.seed(10)
rnd.randint(0, 1 + 1, 10)
```
If we now specify the seed again as 10, we can generate the exact same sequence
```
rnd.seed(10)
rnd.randint(0, 1 + 1, 10)
```
The ability to generate the exact same sequence is useful during code development. For example, by seeding the random number generator, you can compare your output to output of others trying to solve the same problem.
### Flipping a coin
Enough for now about random number generators. Let's flip a coin 100 times and count the number of heads (0-s) and the number of tails (1-s):
```
flip = rnd.randint(0, 1 + 1, 100)
headcount = 0
tailcount = 0
for i in range(100):
if flip[i] == 0:
headcount += 1
else:
tailcount += 1
print('number of heads:', headcount)
print('number of tails:', tailcount)
```
First of all, note that the number of heads and the number of tails add up to 100. Also, note how we counted the heads and tails. We created counters `headcount` and `tailcount`, looped through all flips, and added 1 to the appropriate counter. Instead of a loop, we could have used a condition for the indices combined with a summation as follows
```
headcount = np.count_nonzero(flip == 0)
tailcount = np.count_nonzero(flip == 1)
print('headcount', headcount)
print('tailcount', tailcount)
```
How does that work? You may recall that the `flip == 0` statement returns an array with length 100 (equal to the lenght of `flip`) with the value `True` when the condition is met, and `False` when the condition is not met. The boolean `True` has the value 1, and the boolean `False` has the value 0. So we simply need to count the nonzero values using the `np.count_nonzero` function to find out how many items are `True`.
The code above is easy, but if we do an experiment with more than two outcomes, it may be cumbersome to count the non-zero items for every possible outcome. So let's try to rewrite this part of the code using a loop. For this specific case the number of lines of code doesn't decrease, but when we have an experiment with many different outcomes this will be much more efficient. Note that `dtype='int'` sets the array to integers.
```
outcomes = np.zeros(2, dtype='int') # Two outcomes. heads are stored in outcome[0], tails in outcome[1]
for i in range (2):
outcomes[i] = np.count_nonzero(flip == i)
print('outcome ', i, ' is ', outcomes[i])
```
### Exercise 1. <a name="back1"></a>Throwing a dice
Throw a dice 100 times and report how many times you throw 1, 2, 3, 4, 5, and 6. Use a seed of 33. Make sure that the reported values add up to 100. Make sure you use a loop in your code as we did in the previous code cell.
<a href="#ex1answer">Answers to Exercise 1</a>
### Flipping a coin twice
Next we are going to flip a coin twice for 100 times and count the number of tails. We generate a random array of 0-s (heads) and 1-s (tails) with two rows (representing two coin flips) and 100 colums. The sum of the two rows represents the number of tails. The `np.sum` function takes an array and by default sums all the values in the array and returns one number. In this case we want to sum the rows. For that, the `sum` function has a keyword argument called `axis`, where `axis=0` sums over index 0 of the array (the rows), `axis=1` sums over the index 1 of the array (the columns), etc.
```
rnd.seed(55)
flips = rnd.randint(low=0, high=1 + 1, size=(2, 100))
tails = np.sum(flips, axis=0)
number_of_tails = np.zeros(3, dtype='int')
for i in range(3):
number_of_tails[i] = np.count_nonzero(tails == i)
print('number of 0, 1, 2 tails:', number_of_tails)
```
Another way to simulate flipping a coin twice, is to draw a number at random from a set of 2 numbers (0 and 1). You need to replace the number after every draw, of course. The `numpy` function to draw a random number from a given array is called `choice`. The `choice` function has a keyword to specify whether values are replaced or not. Hence the following two ways to generate 5 flips are identical.
```
rnd.seed(55)
flips1 = rnd.randint(low=0, high=1 + 1, size=5)
rnd.seed(55)
flips2 = rnd.choice(range(2), size=5, replace=True)
np.alltrue(flips1 == flips2) # Check whether all values in the two arrays are equal
```
### Bar graph
The outcome of the experiment may also be plotted with a bar graph
```
plt.bar(range(0, 3), number_of_tails)
plt.xticks(range(0, 3))
plt.xlabel('number of tails')
plt.ylabel('occurence in 100 trials');
```
### Cumulative Probability
Next we compute the experimental probability of 0 tails, 1 tail, and 2 tails through division by the total number of trials (one trial is two coin flips). The three probabilities add up to 1. The cumulative probability distribution is obtained by cumulatively summing the probabilities using the `cumsum` function of `numpy`. The first value is the probability of throwing 0 tails. The second value is the probability of 1 or fewer tails, and the third value it the probability of 2 or fewer tails. The probability is computed as the number of tails divided by the total number of trials.
```
prob = number_of_tails / 100 # number_of_tails was computed two code cells back
cum_prob = np.cumsum(prob) # So cum_prob[0] = prob[0], cum_prob[1] = prob[0] + prob[1], etc.
print('cum_prob ', cum_prob)
```
The cumulative probability distribution is plotted with a bar graph, making sure that all the bars touch each other (by setting the width to 1, in the case below)
```
plt.bar(range(0, 3), cum_prob, width=1)
plt.xticks(range(0, 3))
plt.xlabel('number of tails in two flips')
plt.ylabel('cumulative probability');
```
### Exercise 2. <a name="back2"></a>Flip a coin five times
Flip a coin five times in a row and record how many times you obtain tails (varying from 0-5). Perform the exeriment 1000 times. Make a bar graph with the total number of tails on the horizontal axis and the emperically computed probability to get that many tails, on the vertical axis. Execute your code several times (hit [shift]-[enter]) and see that the graph changes a bit every time, as the sequence of random numbers changes every time.
Compute the cumulative probability. Print the values to the screen and make a plot of the cumulative probability function using a bar graph.
<a href="#ex2answer">Answers to Exercise 2</a>
### Probability of a Bernouilli variable
In the previous exercise, we computed the probability of a certain number of heads in five flips experimentally. But we can, of course, compute the value exactly by using a few simple formulas. Consider the random variable $Y$, which is the outcome of an experiment with two possible values 0 and 1. Let $p$ be the probability of success, $p=P(Y=1)$.
Then $Y$ is said to be a Bernoulli variable. The experiment is repeated $n$ times and we define $X$ as the number of successes in the experiment. The variable $X$ has a Binomial Distribution with parameters $n$ and $p$. The probability that $X$ takes value $k$ can be computed as (see for example [here](http://en.wikipedia.org/wiki/Binomial_distribution))
$$P(X=k) = \binom{n}{k}p^k(1-p)^{n-k}$$
The term $\binom{n}{k}$ may be computed with the `comb` function, which needs to be imported from the `scipy.misc` package.
### Exercise 3. <a name="back3"></a>Flip a coin 5 times revisited
Go back to the experiment where we flip a coin five times in a row and record how many times we obtain tails.
Compute the theoretical probability for 0, 1, 2, 3, 4, and 5 tails and compare your answer to the probability computed from 1000 trials, 10000 trials, and 100000 trials (use a loop for these three sets of trials). Do you approach the theoretical value with more trials?
<a href="#ex3answer">Answers to Exercise 3</a>
### Exercise 4. <a name="back4"></a>Maximum value of two dice throws
Throw a dice two times and record the maximum value of the two throws. Use the `np.max` function to compute the maximum value. Like the `np.sum` function, the `np.max` function takes an array as input argument and an optional keyword argument named `axis`. Perform the experiment 1000 times and compute the probability that the highest value is 1, 2, 3, 4, 5, or 6. Make a graph of the cumulative probability distribution function using a step graph.
<a href="#ex4answer">Answers to Exercise 4</a>
### Exercise 5. <a name="back5"></a>Maximum value of two dice throws revisited
Refer back to Exercise 4.
Compute the theoretical value of the probability of the highest dice when throwing the dice twice (the throws are labeled T1 and T2, respectively). There are 36 possible outcomes for this experiment. Let $M$ denote the random variable corresponding to this experiment (this means for instance that $M=3$ when your first throw is a 2, and the second throw is a 3). All outcomes of $M$ can easily be written down, as shown in the following Table:
| T1$\downarrow$ T2$\to$ | 1 | 2 | 3 | 4 | 5 | 6 |
|-----------:|------------:|:------------:|
| 1 | 1 | 2 | 3 | 4 | 5 | 6 |
| 2 | 2 | 2 | 3 | 4 | 5 | 6 |
| 3 | 3 | 3 | 3 | 4 | 5 | 6 |
| 4 | 4 | 4 | 4 | 4 | 5 | 6 |
| 5 | 5 | 5 | 5 | 5 | 5 | 6 |
| 6 | 6 | 6 | 6 | 6 | 6 | 6 |
Use the 36 possible outcomes shown in the Table to compute the theoretical probability of $M$ being 1, 2, 3, 4, 5, or 6. Compare the theoretical outcome with the experimental outcome for 100, 1000, and 10000 dice throws.
<a href="#ex5answer">Answers to Exercise 5</a>
### Generate random integers with non-equal probabilities
So far, we have generated random numbers of which the probability of each outcome was the same (heads or tails, or the numbers on a dice, considering the throwing device was "fair"). What now if we want to generate outcomes that don't have the same probability? For example, consider the case that we have a bucket with 4 blue balls and 6 red balls. When you draw a ball at random, the probability of a blue ball is 0.4 and the probability of a red ball is 0.6. A sequence of drawing ten balls, with replacement, may be generated as follows
```
balls = np.zeros(10, dtype='int') # zero is blue
balls[4:] = 1 # one is red
print('balls:', balls)
drawing = rnd.choice(balls, 10, replace=True)
print('drawing:', drawing)
print('blue balls:', np.count_nonzero(drawing == 0))
print('red balls:', np.count_nonzero(drawing == 1))
```
### Exercise 6. <a name="back6"></a>Election poll
Consider an election where one million people will vote. 490,000 people will vote for candidate $A$ and 510,000 people will vote for candidate $B$. One day before the election, the company of 'Maurice the Dog' conducts a pole among 1000 randomly chosen voters. Compute whether the Dog will predict the winner correctly using the approach explained above and a seed of 2.
Perform the pole 1000 times. Count how many times the outcome of the pole is that candidate $A$ wins and how many times the outcome of the pole is that candidate $B$ wins. What is the probability that the Dog will predict the correct winner based on these 1000 poles of 1000 people?
Compute the probability that the Dog will predict the correct winner based on 1000 poles of 5000 people? Does the probability that The Dog predicts the correct winner increase significantly when he poles 5000 people?
<a href="#ex6answer">Answers to Exercise 6</a>
### Answers to the exercises
<a name="ex1answer">Answers to Exercise 1</a>
```
rnd.seed(33)
dicethrow = rnd.randint(1, 6 + 1, 100)
side = np.zeros(6, dtype='int')
for i in range(6):
side[i] = np.count_nonzero(dicethrow == i + 1)
print('number of times', i + 1, 'is', side[i])
print('total number of throws ', sum(side))
```
<a href="#back1">Back to Exercise 1</a>
<a name="ex2answer">Answers to Exercise 2</a>
```
N = 1000
tails = np.sum(rnd.randint(0, 1 + 1, (5, 1000)), axis=0)
counttails = np.zeros(6, dtype='int')
for i in range(6):
counttails[i] = np.count_nonzero(tails == i)
plt.bar(range(0, 6), counttails / N)
plt.xlabel('number of tails in five flips')
plt.ylabel('probability');
cumprob = np.cumsum(counttails / N)
print('cumprob:', cumprob)
plt.bar(range(0, 6), cumprob, width=1)
plt.xlabel('number of tails in five flips')
plt.ylabel('cumulative probability');
```
<a href="#back2">Back to Exercise 2</a>
<a name="ex3answer">Answers to Exercise 3</a>
```
from scipy.misc import comb
print('Theoretical probabilities:')
for k in range(6):
print(k, ' tails ', comb(5, k) * 0.5 ** k * 0.5 ** (5 - k))
for N in (1000, 10000, 100000):
tails = np.sum(rnd.randint(0, 1 + 1, (5, N)), axis=0)
counttails = np.zeros(6)
for i in range(6):
counttails[i] = np.count_nonzero(tails==i)
print('Probability with', N, 'trials: ', counttails / float(N))
```
<a href="#back3">Back to Exercise 3</a>
<a name="ex4answer">Answers to Exercise 4</a>
```
dice = rnd.randint(1, 6 + 1, (2, 1000))
highest_dice = np.max(dice, 0)
outcome = np.zeros(6)
for i in range(6):
outcome[i] = np.sum(highest_dice == i + 1) / 1000
plt.bar(left=np.arange(1, 7), height=outcome, width=1)
plt.xlabel('highest dice in two throws')
plt.ylabel('probability');
```
<a href="#back4">Back to Exercise 4</a>
<a name="ex5answer">Answers to Exercise 5</a>
```
for N in [100, 1000, 10000]:
dice = rnd.randint(1, 6 + 1, (2, N))
highest_dice = np.max(dice, axis=0)
outcome = np.zeros(6)
for i in range(6):
outcome[i] = np.sum(highest_dice == i + 1) / N
print('Outcome for', N, 'throws: ', outcome)
# Exact values
exact = np.zeros(6)
for i, j in enumerate(range(1, 12, 2)):
exact[i] = j / 36
print('Exact probabilities: ',exact)
```
<a href="#back5">Back to Exercise 5</a>
<a name="ex6answer">Answers to Exercise 6</a>
```
rnd.seed(2)
people = np.zeros(1000000, dtype='int') # candidate A is 0
people[490000:] = 1 # candidate B is 1
pole = rnd.choice(people, 1000)
poled_for_A = np.count_nonzero(pole == 0)
print('poled for A:', poled_for_A)
if poled_for_A > 500:
print('The Dog will predict the wrong winner')
else:
print('The Dog will predict the correct winner')
Awins = 0
Bwins = 0
for i in range(1000):
people = np.zeros(1000000, dtype='int') # candidate A is 0
people[490000:] = 1 # candidate B is 1
pole = rnd.choice(people, 1000)
poled_for_A = np.count_nonzero(pole == 0)
if poled_for_A > 500:
Awins += 1
else:
Bwins += 1
print('1000 poles of 1000 people')
print('Probability that The Dog predicts candidate A to win:', Awins / 1000)
Awins = 0
Bwins = 0
for i in range(1000):
people = np.zeros(1000000, dtype='int') # candidate A is 0
people[490000:] = 1 # candidate B is 1
pole = rnd.choice(people, 5000)
poled_for_A = np.count_nonzero(pole == 0)
if poled_for_A > 2500:
Awins += 1
else:
Bwins += 1
print('1000 poles of 5000 people')
print('Probability that The Dog predicts candidate A to win:', Awins / 5000)
```
<a href="#back6">Back to Exercise 6</a>
|
github_jupyter
|
Lambda School Data Science
*Unit 2, Sprint 3, Module 3*
---
# Permutation & Boosting
- Get **permutation importances** for model interpretation and feature selection
- Use xgboost for **gradient boosting**
### Setup
Run the code cell below. You can work locally (follow the [local setup instructions](https://lambdaschool.github.io/ds/unit2/local/)) or on Colab.
Libraries:
- category_encoders
- [**eli5**](https://eli5.readthedocs.io/en/latest/)
- matplotlib
- numpy
- pandas
- scikit-learn
- [**xgboost**](https://xgboost.readthedocs.io/en/latest/)
```
%%capture
import sys
# If you're on Colab:
if 'google.colab' in sys.modules:
DATA_PATH = 'https://raw.githubusercontent.com/LambdaSchool/DS-Unit-2-Applied-Modeling/master/data/'
!pip install category_encoders==2.*
!pip install eli5
# If you're working locally:
else:
DATA_PATH = '../data/'
```
We'll go back to Tanzania Waterpumps for this lesson.
```
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
# Merge train_features.csv & train_labels.csv
train = pd.merge(pd.read_csv(DATA_PATH+'waterpumps/train_features.csv'),
pd.read_csv(DATA_PATH+'waterpumps/train_labels.csv'))
# Read test_features.csv & sample_submission.csv
test = pd.read_csv(DATA_PATH+'waterpumps/test_features.csv')
sample_submission = pd.read_csv(DATA_PATH+'waterpumps/sample_submission.csv')
# Split train into train & val
train, val = train_test_split(train, train_size=0.80, test_size=0.20,
stratify=train['status_group'], random_state=42)
def wrangle(X):
"""Wrangle train, validate, and test sets in the same way"""
# Prevent SettingWithCopyWarning
X = X.copy()
# About 3% of the time, latitude has small values near zero,
# outside Tanzania, so we'll treat these values like zero.
X['latitude'] = X['latitude'].replace(-2e-08, 0)
# When columns have zeros and shouldn't, they are like null values.
# So we will replace the zeros with nulls, and impute missing values later.
# Also create a "missing indicator" column, because the fact that
# values are missing may be a predictive signal.
cols_with_zeros = ['longitude', 'latitude', 'construction_year',
'gps_height', 'population']
for col in cols_with_zeros:
X[col] = X[col].replace(0, np.nan)
X[col+'_MISSING'] = X[col].isnull()
# Drop duplicate columns
duplicates = ['quantity_group', 'payment_type']
X = X.drop(columns=duplicates)
# Drop recorded_by (never varies) and id (always varies, random)
unusable_variance = ['recorded_by', 'id']
X = X.drop(columns=unusable_variance)
# Convert date_recorded to datetime
X['date_recorded'] = pd.to_datetime(X['date_recorded'], infer_datetime_format=True)
# Extract components from date_recorded, then drop the original column
X['year_recorded'] = X['date_recorded'].dt.year
X['month_recorded'] = X['date_recorded'].dt.month
X['day_recorded'] = X['date_recorded'].dt.day
X = X.drop(columns='date_recorded')
# Engineer feature: how many years from construction_year to date_recorded
X['years'] = X['year_recorded'] - X['construction_year']
X['years_MISSING'] = X['years'].isnull()
# return the wrangled dataframe
return X
train = wrangle(train)
val = wrangle(val)
test = wrangle(test)
# Arrange data into X features matrix and y target vector
target = 'status_group'
X_train = train.drop(columns=target)
y_train = train[target]
X_val = val.drop(columns=target)
y_val = val[target]
X_test = test
import category_encoders as ce
from sklearn.impute import SimpleImputer
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn.pipeline import make_pipeline
pipeline = make_pipeline(
ce.OrdinalEncoder(),
SimpleImputer(strategy='median'),
RandomForestClassifier(n_estimators=100, random_state=42, n_jobs=-1)
)
# Fit on train, score on val
pipeline.fit(X_train, y_train)
print('Validation Accuracy', pipeline.score(X_val, y_val))
```
# Get permutation importances for model interpretation and feature selection
## Overview
Default Feature Importances are fast, but Permutation Importances may be more accurate.
These links go deeper with explanations and examples:
- Permutation Importances
- [Kaggle / Dan Becker: Machine Learning Explainability](https://www.kaggle.com/dansbecker/permutation-importance)
- [Christoph Molnar: Interpretable Machine Learning](https://christophm.github.io/interpretable-ml-book/feature-importance.html)
- (Default) Feature Importances
- [Ando Saabas: Selecting good features, Part 3, Random Forests](https://blog.datadive.net/selecting-good-features-part-iii-random-forests/)
- [Terence Parr, et al: Beware Default Random Forest Importances](https://explained.ai/rf-importance/index.html)
There are three types of feature importances:
### 1. (Default) Feature Importances
Fastest, good for first estimates, but be aware:
>**When the dataset has two (or more) correlated features, then from the point of view of the model, any of these correlated features can be used as the predictor, with no concrete preference of one over the others.** But once one of them is used, the importance of others is significantly reduced since effectively the impurity they can remove is already removed by the first feature. As a consequence, they will have a lower reported importance. This is not an issue when we want to use feature selection to reduce overfitting, since it makes sense to remove features that are mostly duplicated by other features. But when interpreting the data, it can lead to the incorrect conclusion that one of the variables is a strong predictor while the others in the same group are unimportant, while actually they are very close in terms of their relationship with the response variable. — [Selecting good features – Part III: random forests](https://blog.datadive.net/selecting-good-features-part-iii-random-forests/)
> **The scikit-learn Random Forest feature importance ... tends to inflate the importance of continuous or high-cardinality categorical variables.** ... Breiman and Cutler, the inventors of Random Forests, indicate that this method of “adding up the gini decreases for each individual variable over all trees in the forest gives a **fast** variable importance that is often very consistent with the permutation importance measure.” — [Beware Default Random Forest Importances](https://explained.ai/rf-importance/index.html)
```
# Get feature importances
rf = pipeline.named_steps['randomforestclassifier']
importances = pd.Series(rf.feature_importances_, X_train.columns)
# Plot feature importances
%matplotlib inline
import matplotlib.pyplot as plt
n = 20
plt.figure(figsize=(10,n/2))
plt.title(f'Top {n} features')
importances.sort_values()[-n:].plot.barh(color='grey');
```
### 2. Drop-Column Importance
The best in theory, but too slow in practice
```
column = 'quantity'
# Fit without column
pipeline = make_pipeline(
ce.OrdinalEncoder(),
SimpleImputer(strategy='median'),
RandomForestClassifier(n_estimators=100, random_state=42, n_jobs=-1)
)
pipeline.fit(X_train.drop(columns=column), y_train)
score_without = pipeline.score(X_val.drop(columns=column), y_val)
print(f'Validation Accuracy without {column}: {score_without}')
# Fit with column
pipeline = make_pipeline(
ce.OrdinalEncoder(),
SimpleImputer(strategy='median'),
RandomForestClassifier(n_estimators=100, random_state=42, n_jobs=-1)
)
pipeline.fit(X_train, y_train)
score_with = pipeline.score(X_val, y_val)
print(f'Validation Accuracy with {column}: {score_with}')
# Compare the error with & without column
print(f'Drop-Column Importance for {column}: {score_with - score_without}')
```
### 3. Permutation Importance
Permutation Importance is a good compromise between Feature Importance based on impurity reduction (which is the fastest) and Drop Column Importance (which is the "best.")
[The ELI5 library documentation explains,](https://eli5.readthedocs.io/en/latest/blackbox/permutation_importance.html)
> Importance can be measured by looking at how much the score (accuracy, F1, R^2, etc. - any score we’re interested in) decreases when a feature is not available.
>
> To do that one can remove feature from the dataset, re-train the estimator and check the score. But it requires re-training an estimator for each feature, which can be computationally intensive. ...
>
>To avoid re-training the estimator we can remove a feature only from the test part of the dataset, and compute score without using this feature. It doesn’t work as-is, because estimators expect feature to be present. So instead of removing a feature we can replace it with random noise - feature column is still there, but it no longer contains useful information. This method works if noise is drawn from the same distribution as original feature values (as otherwise estimator may fail). The simplest way to get such noise is to shuffle values for a feature, i.e. use other examples’ feature values - this is how permutation importance is computed.
>
>The method is most suitable for computing feature importances when a number of columns (features) is not huge; it can be resource-intensive otherwise.
### Do-It-Yourself way, for intuition
### With eli5 library
For more documentation on using this library, see:
- [eli5.sklearn.PermutationImportance](https://eli5.readthedocs.io/en/latest/autodocs/sklearn.html#eli5.sklearn.permutation_importance.PermutationImportance)
- [eli5.show_weights](https://eli5.readthedocs.io/en/latest/autodocs/eli5.html#eli5.show_weights)
- [scikit-learn user guide, `scoring` parameter](https://scikit-learn.org/stable/modules/model_evaluation.html#the-scoring-parameter-defining-model-evaluation-rules)
eli5 doesn't work with pipelines.
```
# Ignore warnings
```
### We can use importances for feature selection
For example, we can remove features with zero importance. The model trains faster and the score does not decrease.
# Use xgboost for gradient boosting
## Overview
In the Random Forest lesson, you learned this advice:
#### Try Tree Ensembles when you do machine learning with labeled, tabular data
- "Tree Ensembles" means Random Forest or **Gradient Boosting** models.
- [Tree Ensembles often have the best predictive accuracy](https://arxiv.org/abs/1708.05070) with labeled, tabular data.
- Why? Because trees can fit non-linear, non-[monotonic](https://en.wikipedia.org/wiki/Monotonic_function) relationships, and [interactions](https://christophm.github.io/interpretable-ml-book/interaction.html) between features.
- A single decision tree, grown to unlimited depth, will [overfit](http://www.r2d3.us/visual-intro-to-machine-learning-part-1/). We solve this problem by ensembling trees, with bagging (Random Forest) or **[boosting](https://www.youtube.com/watch?v=GM3CDQfQ4sw)** (Gradient Boosting).
- Random Forest's advantage: may be less sensitive to hyperparameters. **Gradient Boosting's advantage:** may get better predictive accuracy.
Like Random Forest, Gradient Boosting uses ensembles of trees. But the details of the ensembling technique are different:
### Understand the difference between boosting & bagging
Boosting (used by Gradient Boosting) is different than Bagging (used by Random Forests).
Here's an excerpt from [_An Introduction to Statistical Learning_](http://www-bcf.usc.edu/~gareth/ISL/ISLR%20Seventh%20Printing.pdf) Chapter 8.2.3, Boosting:
>Recall that bagging involves creating multiple copies of the original training data set using the bootstrap, fitting a separate decision tree to each copy, and then combining all of the trees in order to create a single predictive model.
>
>**Boosting works in a similar way, except that the trees are grown _sequentially_: each tree is grown using information from previously grown trees.**
>
>Unlike fitting a single large decision tree to the data, which amounts to _fitting the data hard_ and potentially overfitting, the boosting approach instead _learns slowly._ Given the current model, we fit a decision tree to the residuals from the model.
>
>We then add this new decision tree into the fitted function in order to update the residuals. Each of these trees can be rather small, with just a few terminal nodes. **By fitting small trees to the residuals, we slowly improve fˆ in areas where it does not perform well.**
>
>Note that in boosting, unlike in bagging, the construction of each tree depends strongly on the trees that have already been grown.
This high-level overview is all you need to know for now. If you want to go deeper, we recommend you watch the StatQuest videos on gradient boosting!
Let's write some code. We have lots of options for which libraries to use:
#### Python libraries for Gradient Boosting
- [scikit-learn Gradient Tree Boosting](https://scikit-learn.org/stable/modules/ensemble.html#gradient-boosting) — slower than other libraries, but [the new version may be better](https://twitter.com/amuellerml/status/1129443826945396737)
- Anaconda: already installed
- Google Colab: already installed
- [xgboost](https://xgboost.readthedocs.io/en/latest/) — can accept missing values and enforce [monotonic constraints](https://xiaoxiaowang87.github.io/monotonicity_constraint/)
- Anaconda, Mac/Linux: `conda install -c conda-forge xgboost`
- Windows: `conda install -c anaconda py-xgboost`
- Google Colab: already installed
- [LightGBM](https://lightgbm.readthedocs.io/en/latest/) — can accept missing values and enforce [monotonic constraints](https://blog.datadive.net/monotonicity-constraints-in-machine-learning/)
- Anaconda: `conda install -c conda-forge lightgbm`
- Google Colab: already installed
- [CatBoost](https://catboost.ai/) — can accept missing values and use [categorical features](https://catboost.ai/docs/concepts/algorithm-main-stages_cat-to-numberic.html) without preprocessing
- Anaconda: `conda install -c conda-forge catboost`
- Google Colab: `pip install catboost`
In this lesson, you'll use a new library, xgboost — But it has an API that's almost the same as scikit-learn, so it won't be a hard adjustment!
#### [XGBoost Python API Reference: Scikit-Learn API](https://xgboost.readthedocs.io/en/latest/python/python_api.html#module-xgboost.sklearn)
#### [Avoid Overfitting By Early Stopping With XGBoost In Python](https://machinelearningmastery.com/avoid-overfitting-by-early-stopping-with-xgboost-in-python/)
Why is early stopping better than a For loop, or GridSearchCV, to optimize `n_estimators`?
With early stopping, if `n_iterations` is our number of iterations, then we fit `n_iterations` decision trees.
With a for loop, or GridSearchCV, we'd fit `sum(range(1,n_rounds+1))` trees.
But it doesn't work well with pipelines. You may need to re-run multiple times with different values of other parameters such as `max_depth` and `learning_rate`.
#### XGBoost parameters
- [Notes on parameter tuning](https://xgboost.readthedocs.io/en/latest/tutorials/param_tuning.html)
- [Parameters documentation](https://xgboost.readthedocs.io/en/latest/parameter.html)
### Try adjusting these hyperparameters
#### Random Forest
- class_weight (for imbalanced classes)
- max_depth (usually high, can try decreasing)
- n_estimators (too low underfits, too high wastes time)
- min_samples_leaf (increase if overfitting)
- max_features (decrease for more diverse trees)
#### Xgboost
- scale_pos_weight (for imbalanced classes)
- max_depth (usually low, can try increasing)
- n_estimators (too low underfits, too high wastes time/overfits) — Use Early Stopping!
- learning_rate (too low underfits, too high overfits)
For more ideas, see [Notes on Parameter Tuning](https://xgboost.readthedocs.io/en/latest/tutorials/param_tuning.html) and [DART booster](https://xgboost.readthedocs.io/en/latest/tutorials/dart.html).
## Challenge
You will use your portfolio project dataset for all assignments this sprint. Complete these tasks for your project, and document your work.
- Continue to clean and explore your data. Make exploratory visualizations.
- Fit a model. Does it beat your baseline?
- Try xgboost.
- Get your model's permutation importances.
You should try to complete an initial model today, because the rest of the week, we're making model interpretation visualizations.
But, if you aren't ready to try xgboost and permutation importances with your dataset today, you can practice with another dataset instead. You may choose any dataset you've worked with previously.
|
github_jupyter
|
# Partial Correlation
The purpose of this notebook is to understand how to compute the [partial correlation](https://en.wikipedia.org/wiki/Partial_correlation) between two variables, $X$ and $Y$, given a third $Z$. In particular, these variables are assumed to be guassians (or, in general, multivariate gaussians).
Why is it important to estimate partial correlations? The primary reason for estimating a partial correlation is to use it to detect for [confounding](https://en.wikipedia.org/wiki/Confounding_variable) variables during causal analysis.
## Simulation
Let's start out by simulating 3 data sets. Graphically, these data sets comes from graphs represented by the following.
* $X \rightarrow Z \rightarrow Y$ (serial)
* $X \leftarrow Z \rightarrow Y$ (diverging)
* $X \rightarrow Z \leftarrow Y$ (converging)
```
%matplotlib inline
import matplotlib.pyplot as plt
import networkx as nx
import warnings
warnings.filterwarnings('ignore')
plt.style.use('ggplot')
def get_serial_graph():
g = nx.DiGraph()
g.add_node('X')
g.add_node('Y')
g.add_node('Z')
g.add_edge('X', 'Z')
g.add_edge('Z', 'Y')
return g
def get_diverging_graph():
g = nx.DiGraph()
g.add_node('X')
g.add_node('Y')
g.add_node('Z')
g.add_edge('Z', 'X')
g.add_edge('Z', 'Y')
return g
def get_converging_graph():
g = nx.DiGraph()
g.add_node('X')
g.add_node('Y')
g.add_node('Z')
g.add_edge('X', 'Z')
g.add_edge('Y', 'Z')
return g
g_serial = get_serial_graph()
g_diverging = get_diverging_graph()
g_converging = get_converging_graph()
p_serial = nx.nx_agraph.graphviz_layout(g_serial, prog='dot', args='-Kcirco')
p_diverging = nx.nx_agraph.graphviz_layout(g_diverging, prog='dot', args='-Kcirco')
p_converging = nx.nx_agraph.graphviz_layout(g_converging, prog='dot', args='-Kcirco')
fig, ax = plt.subplots(3, 1, figsize=(5, 5))
nx.draw(g_serial, pos=p_serial, with_labels=True, node_color='#e0e0e0', node_size=800, arrowsize=20, ax=ax[0])
nx.draw(g_diverging, pos=p_diverging, with_labels=True, node_color='#e0e0e0', node_size=800, arrowsize=20, ax=ax[1])
nx.draw(g_converging, pos=p_converging, with_labels=True, node_color='#e0e0e0', node_size=800, arrowsize=20, ax=ax[2])
ax[0].set_title('Serial')
ax[1].set_title('Diverging')
ax[2].set_title('Converging')
plt.tight_layout()
```
In the serial graph, `X` causes `Z` and `Z` causes `Y`. In the diverging graph, `Z` causes both `X` and `Y`. In the converging graph, `X` and `Y` cause `Z`. Below, the serial, diverging, and converging data sets are named S, D, and C, correspondingly.
Note that in the serial graph, the data is sampled as follows.
* $X \sim \mathcal{N}(0, 1)$
* $Z \sim 2 + 1.8 \times X$
* $Y \sim 5 + 2.7 \times Z$
In the diverging graph, the data is sampled as follows.
* $Z \sim \mathcal{N}(0, 1)$
* $X \sim 4.3 + 3.3 \times Z$
* $Y \sim 5.0 + 2.7 \times Z$
Lastly, in the converging graph, the data is sampled as follows.
* $X \sim \mathcal{N}(0, 1)$
* $Y \sim \mathcal{N}(5.5, 1)$
* $Z \sim 2.0 + 0.8 \times X + 1.2 \times Y$
Note the ordering of the sampling with the variables follows the structure of the corresponding graph.
```
import numpy as np
np.random.seed(37)
def get_error(N=10000, mu=0.0, std=0.2):
return np.random.normal(mu, std, N)
def to_matrix(X, Z, Y):
return np.concatenate([
X.reshape(-1, 1),
Z.reshape(-1, 1),
Y.reshape(-1, 1)], axis=1)
def get_serial(N=10000, e_mu=0.0, e_std=0.2):
X = np.random.normal(0, 1, N) + get_error(N, e_mu, e_std)
Z = 2 + 1.8 * X + get_error(N, e_mu, e_std)
Y = 5 + 2.7 * Z + get_error(N, e_mu, e_std)
return to_matrix(X, Z, Y)
def get_diverging(N=10000, e_mu=0.0, e_std=0.2):
Z = np.random.normal(0, 1, N) + get_error(N, e_mu, e_std)
X = 4.3 + 3.3 * Z + get_error(N, e_mu, e_std)
Y = 5 + 2.7 * Z + get_error(N, e_mu, e_std)
return to_matrix(X, Z, Y)
def get_converging(N=10000, e_mu=0.0, e_std=0.2):
X = np.random.normal(0, 1, N) + get_error(N, e_mu, e_std)
Y = np.random.normal(5.5, 1, N) + get_error(N, e_mu, e_std)
Z = 2 + 0.8 * X + 1.2 * Y + get_error(N, e_mu, e_std)
return to_matrix(X, Z, Y)
S = get_serial()
D = get_diverging()
C = get_converging()
```
## Computation
For the three datasets, `S`, `D`, and `C`, we want to compute the partial correlation between $X$ and $Y$ given $Z$. The way to do this is as follows.
* Regress $X$ on $Z$ and also $Y$ on $Z$
* $X = b_X + w_X * Z$
* $Y = b_Y + w_Y * Z$
* With the new weights $(b_X, w_X)$ and $(b_Y, w_Y)$, predict $X$ and $Y$.
* $\hat{X} = b_X + w_X * Z$
* $\hat{Y} = b_Y + w_Y * Z$
* Now compute the residuals between the true and predicted values.
* $R_X = X - \hat{X}$
* $R_Y = Y - \hat{Y}$
* Finally, compute the Pearson correlation between $R_X$ and $R_Y$.
The correlation between the residuals is the partial correlation and runs from -1 to +1. More interesting is the test of significance. If $p > \alpha$, where $\alpha \in [0.1, 0.05, 0.01]$, then assume independence. For example, assume $\alpha = 0.01$ and $p = 0.002$, then $X$ is conditionally independent of $Y$ given $Z$.
```
from sklearn.linear_model import LinearRegression
from sklearn.metrics import r2_score
from scipy.stats import pearsonr
from scipy import stats
def get_cond_indep_test(c_xy_z, N=10000, alpha=0.01):
point = stats.norm.ppf(1 - (alpha / 2.0))
z_transform = np.sqrt(N - 3) * np.abs(0.5 * np.log((1 + c_xy_z) / (1 - c_xy_z)))
return z_transform, point, z_transform > point
def get_partial_corr(M):
X = M[:, 0]
Z = M[:, 1].reshape(-1, 1)
Y = M[:, 2]
mXZ = LinearRegression()
mXZ.fit(Z, X)
pXZ = mXZ.predict(Z)
rXZ = X - pXZ
mYZ = LinearRegression()
mYZ.fit(Z, Y)
pYZ = mYZ.predict(Z)
rYZ = Y - pYZ
c_xy, p_xy = pearsonr(X, Y)
c_xy_z, p_xy_z = pearsonr(rXZ, rYZ)
return c_xy, p_xy, c_xy_z, p_xy_z
```
## Serial graph data
For $X \rightarrow Z \rightarrow Y$, note that the marginal correlation is high (0.99) and the correlation is significant (p < 0.01). However, the correlation between X and Y vanishes given Z to -0.01 (p > 0.01). Note the conditional independence test fails to reject the null hypothesis.
```
c_xy, p_xy, c_xy_z, p_xy_z = get_partial_corr(S)
print(f'corr_xy={c_xy:.5f}, p_xy={p_xy:.5f}')
print(f'corr_xy_z={c_xy_z:.5f}, p_xy_z={p_xy_z:.5f}')
print(get_cond_indep_test(c_xy_z))
```
## Diverging graph data
For $X \leftarrow Z \rightarrow Y$, note that the marginal correlation is high (0.99) and the correlation is significant (p < 0.01). However, the correlation between X and Y vanishes given Z to 0.01 (p > 0.01). Note the conditional independence test fails to reject the null hypothesis.
```
c_xy, p_xy, c_xy_z, p_xy_z = get_partial_corr(D)
print(f'corr_xy={c_xy:.5f}, p_xy={p_xy:.5f}')
print(f'corr_xy_z={c_xy_z:.5f}, p_xy_z={p_xy_z:.5f}')
print(get_cond_indep_test(c_xy_z))
```
## Converging graph data
For $X \rightarrow Z \leftarrow Y$, note that the correlation is low (-0.00) and the correlation is insignficiant (p > 0.01). However, the correlation between X and Y increases to -0.96 and becomes significant (p < 0.01)! Note the conditional independence test rejects the null hypothesis.
```
c_xy, p_xy, c_xy_z, p_xy_z = get_partial_corr(C)
print(f'corr_xy={c_xy:.5f}, p_xy={p_xy:.5f}')
print(f'corr_xy_z={c_xy_z:.5f}, p_xy_z={p_xy_z:.5f}')
print(get_cond_indep_test(c_xy_z))
```
## Statistically Distinguishable
The `serial` and `diverging` graphs are said to be `statistically indistingishable` since $X$ and $Y$ are both `conditionally independent` given $Z$. However, the `converging` graph is `statistically distinguishable` since it is the only graph where $X$ and $Y$ are `conditionally dependent` given $Z$.
|
github_jupyter
|
```
# Purpose: Analyze results from Predictions Files created by Models
# Inputs: Prediction files from Random Forest, Elastic Net, XGBoost, and Team Ensembles
# Outputs: Figures (some included in the paper, some in SI)
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import astropy.stats as AS
from scipy.stats.stats import pearsonr
from os import listdir
from sklearn.decomposition import PCA
%matplotlib inline
```
## Reading Data, generating In-Sample Scores
```
name_dict = {'lassoRF_prediction': 'Lasso RF','elastic_prediction': 'Elastic Net','RF_prediction': 'Ensemble RF',
'LR_prediction': 'Ensemble LR','weighted_multiRF_prediction': 'Nested RF',
'weighted_avrg_prediction': 'Weighted Team Avg', 'avrg_prediction': 'Team Avg',
'xgboost_prediction': 'Gradient Boosted Tree'}
training=pd.read_csv('../data/train.csv',index_col = 'challengeID')
baseline=np.mean(training, axis=0)
BL_CV_scores = pd.DataFrame(columns = ['outcome','type','model','score_avg'])
for outcome in training.columns.values:
y = training[outcome].dropna()
y_hat = baseline[outcome]
partition_scores = list()
for i in range(10,110,10):
bools = y.index<np.percentile(y.index,i)
y_curr=y[bools]
partition_scores.append(np.linalg.norm(y_curr-y_hat)/len(y_curr))
bootstrapped_means = AS.bootstrap(np.array(partition_scores),samples = 10, bootnum = 100, bootfunc = np.mean)
to_add = pd.DataFrame({'outcome':list(len(bootstrapped_means)*[outcome]),'type':len(bootstrapped_means)*['In-Sample Error'],'model':len(bootstrapped_means)*['Baseline'],'score_avg':bootstrapped_means})
BL_CV_scores = BL_CV_scores.append(to_add, ignore_index = True)
name_dict
bootstrapped_scores_all = {}
for name in list(name_dict.keys()):
model_name = name_dict[name]
data=pd.read_csv(str('../output/final_pred/'+name+'.csv'), index_col = 'challengeID')
CV_scores = pd.DataFrame(columns = ['outcome','type','model','score_avg'])
for outcome in training.columns.values:
y = training[outcome].dropna()
y_hat = data[outcome][np.in1d(data.index,y.index)]
partition_scores = list()
for i in range(10,110,10):
bools = y.index<np.percentile(y.index,i)
y_curr=y[bools]
y_hat_curr = y_hat[bools]
partition_scores.append(np.linalg.norm(y_curr-y_hat_curr)/len(y_curr))
bootstrapped_means = AS.bootstrap(np.array(partition_scores),samples = 10, bootnum = 100, bootfunc = np.mean)
bootstrapped_means = (1-np.divide(bootstrapped_means,BL_CV_scores.score_avg[BL_CV_scores.outcome==outcome]))*100
to_add = pd.DataFrame({'outcome':list(len(bootstrapped_means)*[outcome]),'type':len(bootstrapped_means)*['In-Sample Error'],'model':len(bootstrapped_means)*[model_name],'score_avg':bootstrapped_means})
CV_scores = CV_scores.append(to_add, ignore_index = True)
bootstrapped_scores_all[name] = CV_scores
```
## Individual Model Scores
```
GBT_CV = bootstrapped_scores_all['xgboost_prediction']
GBT_leaderboard = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Leaderboard'],'model':6*['Gradient Boosted Tree'],'score_avg':[0.37543,0.22008,0.02437,0.05453,0.17406,0.19676]})
GBT_holdout = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Holdout'],'model':6*['Gradient Boosted Tree'],'score_avg':[0.34379983,0.238180899,0.019950074,0.056877623,0.167392429,0.177202581]})
GBT_scores = GBT_CV.append(GBT_leaderboard.append(GBT_holdout,ignore_index = True),ignore_index = True)
avrg_CV = bootstrapped_scores_all['avrg_prediction']
avrg_leaderboard = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Leaderboard'],'model':6*['Team Avg'],'score_avg':[0.36587,0.21287,0.02313,0.05025,0.17467,0.20058]})
avrg_holdout = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Holdout'],'model':6*['Team Avg'],'score_avg':[0.352115776,0.241462042,0.019888218,0.053480264,0.169287396,0.181767792]})
avrg_scores = avrg_CV.append(avrg_leaderboard.append(avrg_holdout,ignore_index = True),ignore_index = True)
weighted_avrg_CV = bootstrapped_scores_all['weighted_avrg_prediction']
weighted_avrg_leaderboard = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Leaderboard'],'model':6*['Weighted Team Avg'],'score_avg':[0.36587,0.21287,0.02301,0.04917,0.1696,0.19782]})
weighted_avrg_holdout = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Holdout'],'model':6*['Weighted Team Avg'],'score_avg':[0.352115776,0.241462042,0.020189616,0.053818827,0.162462938,0.178098036]})
weighted_avrg_scores = weighted_avrg_CV.append(weighted_avrg_leaderboard.append(weighted_avrg_holdout,ignore_index = True),ignore_index = True)
multi_RF_CV = bootstrapped_scores_all['weighted_multiRF_prediction']
multi_RF_leaderboard = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Leaderboard'],'model':6*['Nested RF'],'score_avg':[0.38766,0.22353,0.02542,0.05446,0.20228,0.22092]})
multi_RF_holdout = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Holdout'],'model':6*['Nested RF'],'score_avg':[0.365114483,0.248124154,0.021174361,0.063930882,0.207400541,0.191352482]})
multi_RF_scores = multi_RF_CV.append(multi_RF_leaderboard.append(multi_RF_holdout,ignore_index = True),ignore_index = True)
LR_CV = bootstrapped_scores_all['LR_prediction']
LR_leaderboard = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Leaderboard'],'model':6*['Ensemble LR'],'score_avg':[0.37674,0.2244,0.02715,0.05092,0.18341,0.22311]})
LR_holdout = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Holdout'],'model':6*['Ensemble LR'],'score_avg':[0.364780108,0.247382526,0.021359837,0.058200047,0.181441591,0.194502527]})
LR_scores = LR_CV.append(LR_leaderboard.append(LR_holdout,ignore_index = True),ignore_index = True)
RF_CV = bootstrapped_scores_all['RF_prediction']
RF_leaderboard = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Leaderboard'],'model':6*['Ensemble RF'],'score_avg':[0.38615,0.22342,0.02547,0.05475,0.20346,0.22135]})
RF_holdout = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Holdout'],'model':6*['Ensemble RF'],'score_avg':[0.364609923,0.247940405,0.021135379,0.064494339,0.208869867,0.191742726]})
RF_scores = RF_CV.append(RF_leaderboard.append(RF_holdout,ignore_index = True),ignore_index = True)
lasso_RF_CV = bootstrapped_scores_all['lassoRF_prediction']
lasso_RF_leaderboard = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Leaderboard'],'model':6*['Lasso RF'],'score_avg':[0.37483,0.21686,0.02519,0.05226,0.17223,0.20028]})
lasso_RF_holdout = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Holdout'],'model':6*['Lasso RF'],'score_avg':[0.361450643,0.243745261,0.020491841,0.054397319,0.165154165,0.180446409]})
lasso_scores = lasso_RF_CV.append(lasso_RF_leaderboard.append(lasso_RF_holdout,ignore_index = True),ignore_index = True)
eNet_CV = bootstrapped_scores_all['elastic_prediction']
eNet_leaderboard = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Leaderboard'],'model':6*['Elastic Net'],'score_avg':[0.36477,0.21252,0.02353,0.05341,0.17435,0.20224]})
eNet_holdout = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Holdout'],'model':6*['Elastic Net'],'score_avg':[0.350083,0.239361,0.019791,0.055458,0.167224,0.185329]})
eNet_scores = eNet_CV.append(eNet_leaderboard.append(eNet_holdout,ignore_index = True),ignore_index = True)
#bools = np.in1d(eNet_scores.outcome,['gpa','grit','materialHardship'])
#eNet_scores = eNet_scores.loc[bools]
```
## Score Aggregation and Plotting
```
## Baseline Scores:
BL_LB = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Leaderboard'],'model':6*['Baseline'],'score_avg':[0.39273,0.21997,0.02880,0.05341,0.17435,0.20224]})
BL_HO = pd.DataFrame({'outcome':['gpa','grit','materialHardship','eviction','layoff','jobTraining'],'type':6*['Holdout'],'model':6*['Baseline'],'score_avg':[0.425148881,0.252983596,0.024905617,0.055457913,0.167223718,0.185329492]})
scores_all = eNet_scores.append(lasso_scores.append(RF_scores.append(LR_scores.append(multi_RF_scores.append(weighted_avrg_scores.append(avrg_scores.append(GBT_scores,ignore_index = True),ignore_index = True),ignore_index = True),ignore_index = True),ignore_index = True),ignore_index = True), ignore_index = True)
scores_ADJ = scores_all
scores = scores_all.loc[scores_all.type != 'In-Sample Error']
for OUTCOME in training.columns.values:
f, ax = plt.subplots(nrows=1, ncols=1, figsize=(12, 7), sharex=True)
temp=scores.loc[scores.outcome==OUTCOME]
temp.score_avg.loc[temp.type=='Leaderboard']=(1-np.divide(temp.score_avg.loc[temp.type=='Leaderboard'],BL_LB.score_avg.loc[BL_LB.outcome==OUTCOME]))*100
temp.score_avg.loc[temp.type=='Holdout']=(1-np.divide(temp.score_avg.loc[temp.type=='Holdout'],BL_HO.score_avg.loc[BL_HO.outcome==OUTCOME]))*100
scores_ADJ.score_avg.loc[(scores_ADJ.outcome==OUTCOME) & (scores_ADJ.type=='Leaderboard')] = (1-np.divide(scores_ADJ.score_avg.loc[(scores_ADJ.outcome==OUTCOME) & (scores_ADJ.type=='Leaderboard')],BL_LB.score_avg.loc[BL_LB.outcome==OUTCOME]))*100
scores_ADJ.score_avg.loc[(scores_ADJ.outcome==OUTCOME) & (scores_ADJ.type=='Holdout')] = (1-np.divide(scores_ADJ.score_avg.loc[(scores_ADJ.outcome==OUTCOME) & (scores_ADJ.type=='Holdout')],BL_HO.score_avg.loc[BL_HO.outcome==OUTCOME]))*100
sns.barplot('model','score_avg',hue = 'type', data = temp, ci = 'sd', ax=ax)
ax.set_title(str(OUTCOME))
ax.set_xlabel('Model')
ax.set_ylabel('Accuracy Improvement over Baseline (%)')
plt.setp( ax.xaxis.get_majorticklabels(), rotation=30)
ax.tick_params(labelsize=18)
plt.savefig(str('../output/fig/'+OUTCOME+'.pdf'))
bools_L = (scores.type=='Leaderboard') & (scores.outcome==OUTCOME)
bools_H = (scores.type=='Holdout') & (scores.outcome==OUTCOME)
print(OUTCOME)
print('Best Leaderboard Model: ',scores.loc[(bools_L)&(scores.loc[bools_L].score_avg==max(scores.loc[bools_L].score_avg))].model)
print('Best Holdout Model: ',scores.loc[(bools_H)&(scores.loc[bools_H].score_avg==max(scores.loc[bools_H].score_avg))].model)
print()
scores = scores_all.loc[scores_all.type=='In-Sample Error']
f, ax = plt.subplots(nrows=1, ncols=1, figsize=(24, 7), sharex=True)
sns.barplot('model','score_avg', hue = 'outcome', data = scores, ci = 'sd', ax=ax)
ax.set_title('In-Sample Model Performance Improvement')
ax.set_xlabel('Model')
ax.set_ylabel('Accuracy Improvement over Baseline (%)')
plt.setp( ax.xaxis.get_majorticklabels(), rotation=30)
plt.ylim([-20,100])
ax.tick_params(labelsize=18)
plt.savefig(str('../output/fig/ALL_IS.pdf'))
scores = scores_all.loc[scores_all.type=='In-Sample Error']
for OUTCOME in training.columns.values:
f, ax = plt.subplots(nrows=1, ncols=1, figsize=(12, 7), sharex=True)
temp=scores.loc[scores.outcome==OUTCOME]
sns.barplot('model','score_avg', data = temp, ci = 'sd', ax=ax, color = 'red')
ax.set_title(str(OUTCOME))
ax.set_xlabel('Model')
ax.set_ylabel('Accuracy Improvement over Baseline (%)')
plt.setp( ax.xaxis.get_majorticklabels(), rotation=30)
ax.tick_params(labelsize=18)
plt.savefig(str('../output/fig/'+OUTCOME+'_IS.pdf'))
bools_L = (scores.type=='Leaderboard') & (scores.outcome==OUTCOME)
bools_H = (scores.type=='Holdout') & (scores.outcome==OUTCOME)
```
# Data Partition Performance
```
scores_PLT = scores_ADJ
scores_PLT = scores_PLT.loc[~((scores_ADJ.model=='Elastic Net') & np.in1d(scores_ADJ.outcome,['eviction','layoff','jobTraining']))]
scores_PLT['color'] = [-1]*np.shape(scores_PLT)[0]
for i,OUTCOME in enumerate(['gpa', 'grit', 'materialHardship', 'eviction', 'layoff', 'jobTraining']):
scores_PLT.color.loc[scores_PLT.outcome==OUTCOME] = i
# LEADERBOARD vs HOLDOUT
scores_X = scores_PLT.loc[scores_PLT.type=='Leaderboard']
scores_Y = scores_PLT.loc[scores_PLT.type=='Holdout']
txt = [str(a) for a,b in zip(scores_X.model,scores_X.outcome)]
f, ax = plt.subplots(nrows=1, ncols=1, figsize=(12, 12), sharex=True)
colors = ['red','blue','green','black','yellow','orange']
for i in range(6):
corr_temp = np.round(pearsonr(scores_X.score_avg.loc[scores_X.color==i],
scores_Y.score_avg.loc[scores_Y.color==i]),decimals = 3)
plt.scatter(x = scores_X.score_avg.loc[scores_X.color==i],
s=20, y = scores_Y.score_avg.loc[scores_Y.color==i],
c = colors[i],label=str(scores_X.outcome.loc[scores_X.color==i].iloc[0])+': r^2='+str(corr_temp[0])+' p='+str(corr_temp[1]))
print(i)
print(len(scores_X.score_avg.loc[scores_X.color==i]),
len(scores_Y.score_avg.loc[scores_Y.color==i]))
ax.set_xlabel('Leaderboard Improvement Over Baseline (%)')
ax.set_ylabel('Holdout Improvement Over Baseline (%)')
ax.tick_params(labelsize=18)
plt.ylim([-26, 22])
plt.xlim([-26, 22])
ax.plot([-26,22],[-26,22], 'k-')
ax.legend()
for i,n in enumerate(txt):
ax.annotate(n,(scores_X.score_avg.iloc[i],scores_Y.score_avg.iloc[i]),
size = 10,textcoords='data')
plt.savefig(str('../output/fig/LB_vs_HO.pdf'))
# LEADERBOARD VS IN-SAMPLE
scores_X = scores_PLT.loc[scores_PLT.type=='Leaderboard']
scores_Y = scores_PLT.loc[scores_PLT.type=='In-Sample Error']
scores_Y = pd.DataFrame(scores_Y.groupby([scores_Y.model,scores_Y.outcome]).mean())
txt = [str(a) for a,b in zip(scores_X.model,scores_X.outcome)]
f, ax = plt.subplots(nrows=1, ncols=1, figsize=(12, 12), sharex=True)
colors = ['red','blue','green','black','yellow','orange']
for i in range(6):
corr_temp = np.round(pearsonr(scores_X.score_avg.loc[scores_X.color==i],
scores_Y.score_avg.loc[scores_Y.color==i]),decimals = 3)
plt.scatter(x = scores_X.score_avg.loc[scores_X.color==i],
s=20, y = scores_Y.score_avg.loc[scores_Y.color==i],
c = colors[i],label=str(scores_X.outcome.loc[scores_X.color==i].iloc[0])+': r^2='+str(corr_temp[0])+' p='+str(corr_temp[1]))
print(i)
print(len(scores_X.score_avg.loc[scores_X.color==i]),
len(scores_Y.score_avg.loc[scores_Y.color==i]))
ax.set_xlabel('Leaderboard Improvement Over Baseline (%)')
ax.set_ylabel('In-Sample Error Improvement Over Baseline (%)')
ax.tick_params(labelsize=18)
#plt.ylim([-26, 22])
#plt.xlim([-26, 22])
#ax.plot([-26,22],[-26,22], 'k-')
ax.legend()
for i,n in enumerate(txt):
ax.annotate(n,(scores_X.score_avg.iloc[i],scores_Y.score_avg.iloc[i]),
size = 10,textcoords='data')
plt.savefig(str('../output/fig/LB_vs_IS.pdf'))
# HOLDOUT VS IN-SAMPLE
scores_X = scores_PLT.loc[scores_PLT.type=='Holdout']
scores_Y = scores_PLT.loc[scores_PLT.type=='In-Sample Error']
scores_Y = scores_Y.groupby([scores_Y.model,scores_Y.outcome]).mean().reset_index()
# UNCOMMENT if STD
#scores_Y.color = [0, 1, 2, 3, 0, 1, 5, 4, 2, 3, 0, 1, 5, 4, 2, 3, 0, 1, 5, 4, 2, 3, 0,
# 1, 5, 4, 2, 3, 0, 1, 5, 4, 2, 3, 0, 1, 5, 4, 2, 3, 0, 1, 5, 4, 2]
txt = [str(a) for a,b in zip(scores_X.model,scores_X.outcome)]
f, ax = plt.subplots(nrows=1, ncols=1, figsize=(12, 12), sharex=True)
colors = ['red','blue','green','black','yellow','orange']
for i in range(6):
corr_temp = np.round(pearsonr(scores_X.score_avg.loc[scores_X.color==i],
scores_Y.score_avg.loc[scores_Y.color==i]),decimals = 3)
plt.scatter(x = scores_X.score_avg.loc[scores_X.color==i],
s=20, y = scores_Y.score_avg.loc[scores_Y.color==i],
c = colors[i],label=str(scores_X.outcome.loc[scores_X.color==i].iloc[0])+': r^2='+str(corr_temp[0])+' p='+str(corr_temp[1]))
print(i)
print(len(scores_X.score_avg.loc[scores_X.color==i]),
len(scores_Y.score_avg.loc[scores_Y.color==i]))
ax.set_xlabel('Holdout Improvement Over Baseline (%)')
ax.set_ylabel('In-Sample Error Improvement Over Baseline (%)')
ax.tick_params(labelsize=18)
#plt.ylim([-26, 22])
#plt.xlim([-26, 22])
#ax.plot([-26,22],[-26,22], 'k-')
ax.legend()
for i,n in enumerate(txt):
ax.annotate(n,(scores_X.score_avg.iloc[i],scores_Y.score_avg.iloc[i]),
size = 10,textcoords='data')
plt.savefig(str('../output/fig/HO_vs_IS.pdf'))
```
### Bootstrapping Correlation Values
```
bootnum = 10000
all_keys_boot = ['gpa']*bootnum
temp = ['grit']*bootnum
all_keys_boot.extend(temp)
temp = ['materialHardship']*bootnum
all_keys_boot.extend(temp)
temp = ['eviction']*bootnum
all_keys_boot.extend(temp)
temp = ['layoff']*bootnum
all_keys_boot.extend(temp)
temp = ['jobTraining']*bootnum
all_keys_boot.extend(temp)
temp = ['overall']*bootnum
all_keys_boot.extend(temp)
scores_ADJ = scores_all
keys = ['gpa', 'grit', 'materialHardship', 'eviction', 'layoff', 'jobTraining','overall']
t1 = ['In-Sample Error']*14
temp = ['Leaderboard']*7
t1.extend(temp)
t2 = ['Leaderboard']*7
temp = ['Holdout']*14
t2.extend(temp)
all_keys_boot = ['gpa']*bootnum
temp = ['grit']*bootnum
all_keys_boot.extend(temp)
temp = ['materialHardship']*bootnum
all_keys_boot.extend(temp)
temp = ['eviction']*bootnum
all_keys_boot.extend(temp)
temp = ['layoff']*bootnum
all_keys_boot.extend(temp)
temp = ['jobTraining']*bootnum
all_keys_boot.extend(temp)
temp = ['overall']*bootnum
all_keys_boot.extend(temp)
df_full = pd.DataFrame(columns = ['T1-T2', 'condition', 'avg_corr','sd_corr'])
for [T1,T2] in [['In-Sample Error','Leaderboard'],['In-Sample Error','Holdout'],['Leaderboard','Holdout']]:
X_type = scores_ADJ.loc[scores_ADJ.type==T1]
Y_type = scores_ADJ.loc[scores_ADJ.type==T2]
avg_corr = list([])
# For Ind. Outcomes
for OUTCOME in ['gpa', 'grit', 'materialHardship', 'eviction', 'layoff', 'jobTraining']:
corr = np.zeros(bootnum)
X_OC = X_type.loc[X_type.outcome==OUTCOME]
Y_OC = Y_type.loc[Y_type.outcome==OUTCOME]
X_curr = X_OC.groupby(X_OC.model).score_avg.mean()
Y_curr = Y_OC.groupby(Y_OC.model).score_avg.mean()
for i in range(bootnum):
index = np.random.choice(list(range(len(X_curr))),len(X_curr))
avg_corr.append(pearsonr(X_curr[index].values,Y_curr[index].values)[0])
# For Overall
X_curr = X_type.groupby([X_type.model,X_type.outcome]).score_avg.mean()
Y_curr = Y_type.groupby([Y_type.model,Y_type.outcome]).score_avg.mean()
corr = np.zeros(bootnum)
for i in range(bootnum):
index = np.random.choice(list(range(len(X_curr))),len(X_curr))
avg_corr.append(pearsonr(X_curr[index].values,Y_curr[index].values)[0])
to_add = pd.DataFrame({'T1-T2':7*bootnum*[str(T1)+' w/ '+str(T2)], 'condition': all_keys_boot,
'avg_corr':avg_corr})
df_full = df_full.append(to_add)
f, ax = plt.subplots(nrows=1, ncols=1, figsize=(12, 7), sharex=True)
sns.barplot('T1-T2','avg_corr', hue = 'condition', data = df_full, ci = 'sd', ax=ax)
ax.set_title('Correlation Comparison')
ax.set_xlabel('Data Partitions Compared')
ax.set_ylabel('Avg. Correlation')
plt.setp( ax.xaxis.get_majorticklabels(), rotation=30)
plt.ylim([-1.3,1.2])
ax.tick_params(labelsize=18)
plt.savefig(str('../output/fig/Correlation_Comparison.pdf'))
f, ax = plt.subplots(nrows=1, ncols=1, figsize=(12, 7), sharex=True)
sns.barplot('T1-T2','avg_corr', hue = 'condition', data = df_full.loc[df_full.condition=='overall'], ci = 'sd', ax=ax)
ax.set_title('Correlation Comparison')
ax.set_xlabel('Data Partitions Compared')
ax.set_ylabel('Avg. Correlation')
plt.setp( ax.xaxis.get_majorticklabels(), rotation=30)
plt.ylim([0,1])
ax.tick_params(labelsize=18)
plt.savefig(str('../output/fig/Correlations_Overall.pdf'))
```
## Feature Importance XGBoost
```
father = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],
'characteristic': 6*['Father'],'score': [0.199531305,0.140893472,0.221546773,0.1923971,0.130434782,0.27181208]})
homevisit = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],
'characteristic': 6*['Home Visit'],'score': [0.203213929,0.209621994,0.189125295,0.112949541,0.036789297,0.187919463]})
child = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],
'characteristic': 6*['Child'],'score': [0.044861065,0.003436426,0.082404594,0.01572542,0.006688963,0.023489933]})
kinder = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],
'characteristic': 6*['Kindergarden'],'score': [0.003347841,0.003436426,0.00810537,0.008432472,0.003344482,0.006711409]})
mother = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],
'characteristic': 6*['Mother'],'score': [0.349849352,0.515463913,0.360351229,0.569032313,0.66889632,0.395973155]})
other = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],
'characteristic': 6*['Other'],'score': [0.016069635,0.01718213,0.003377237,0.0097999,0.006688963,0.016778523]})
care = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],
'characteristic': 6*['Caregiver'],'score': [0.085369937,0.048109966,0.10570753,0.060713797,0.140468227,0.080536912]})
teacher = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],
'characteristic': 6*['Teacher'],'score': [0.087378641,0.058419244,0.023302938,0.02306395,0.006688963,0.016778524]})
wav1 = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],
'characteristic': 6*['Wave 1'],'score': [0.109809175,0.048109966,0.101654846,0.317288843,0.046822742,0.104026846]})
wav2 = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],
'characteristic': 6*['Wave 2'],'score': [0.126548378,0.085910654,0.125295507,0.122612698,0.117056855,0.073825504]})
wav3 = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],
'characteristic': 6*['Wave 3'],'score': [0.189822567,0.206185568,0.173252278,0.162496011,0.143812707,0.271812079]})
wav4 = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],
'characteristic': 6*['Wave 4'],'score': [0.172079012,0.230240552,0.205336034,0.166826199,0.217391305,0.241610739]})
wav5 = pd.DataFrame({'outcome': ['gpa','eviction','grit','materialHardship','jobTraining','layoff'],
'characteristic': 6*['Wave 5'],'score': [0.388014734,0.422680407,0.380276931,0.214458269,0.471571907,0.302013422]})
who_df = pd.concat([mother,father,care,homevisit,child,teacher,kinder,other],ignore_index = True)
when_df = pd.concat([wav1,wav2,wav3,wav4,wav5],ignore_index = True)
f, ax = plt.subplots(nrows=1, ncols=1, figsize=(12, 5), sharex=True)
sns.barplot('characteristic','score', hue = 'outcome', data = who_df,
ci = None,ax=ax)
ax.set_ylabel('Feature Importance (Sum)')
ax.tick_params(labelsize=13)
ax.set_ylim(0,0.7)
plt.savefig('../output/fig/Who_Feature_Importance.pdf')
f, ax = plt.subplots(nrows=1, ncols=1, figsize=(12, 5), sharex=True)
sns.barplot('characteristic','score', hue = 'outcome', data = when_df,
ci = None,ax=ax)
ax.set_ylabel('Feature Importance (Sum)')
ax.tick_params(labelsize=13)
ax.set_ylim(0,0.7)
plt.savefig('../output/fig/When_Feature_Importance.pdf')
```
## Comparison of Feature Selection Methods
```
LASSO_files = listdir('../output/LASSO_ALL/')
MI_files = ['data_univariate_feature_selection_5.csv','data_univariate_feature_selection_15.csv','data_univariate_feature_selection_50.csv','data_univariate_feature_selection_100.csv','data_univariate_feature_selection_200.csv','data_univariate_feature_selection_300.csv','data_univariate_feature_selection_500.csv','data_univariate_feature_selection_700.csv','data_univariate_feature_selection_1000.csv','data_univariate_feature_selection_1500.csv','data_univariate_feature_selection_2000.csv','data_univariate_feature_selection_3000.csv','data_univariate_feature_selection_4000.csv']
msk = [i!='.DS_Store' for i in LASSO_files]
LASSO_files = [i for i,j in zip(LASSO_files,msk) if j]
LASSO_files = np.sort(LASSO_files)
MI_file = MI_files[0]
L_file = LASSO_files[0]
perc_similar = np.zeros((len(LASSO_files),len(MI_files)))
PC1_corr = np.zeros((len(LASSO_files),len(MI_files)))
L_names = []
MI_names = []
for i,L_file in enumerate(LASSO_files):
temp_L = pd.read_csv(('../output/LASSO_ALL/'+L_file))
L_names.append(np.shape(temp_L.columns.values)[0])
L_PC = PCA(n_components=2).fit_transform(temp_L)
for j,MI_file in enumerate(MI_files):
temp_M = pd.read_csv(('../output/MI/'+MI_file))
MI_names.append(np.shape(temp_M.columns.values)[0])
MI_PC = PCA(n_components=2).fit_transform(temp_M)
PC1_corr[i,j] = pearsonr(L_PC[:,0],MI_PC[:,0])[0]
perc_similar[i,j]= sum(np.in1d(temp_L.columns.values,temp_M.columns.values))
data_named = pd.DataFrame(perc_similar,index = L_names, columns = np.unique(MI_names))
columns = data_named.columns.tolist()
columns = columns[::-1]
data_named = data_named[columns]
f, ax = plt.subplots(nrows=1, ncols=1, figsize=(15, 15), sharex=True)
sns.heatmap(data_named, annot = True)
plt.savefig('../output/fig/feature_heatmap.png')
L_names = [str('r^2='+str(i)) for i in np.linspace(0.1,0.9,9)]
MI_names = [str('K='+str(i)) for i in [5,15,50,100,200,300,500,700,1000,1500,2000,3000,4000]]
data_PC = pd.DataFrame(PC1_corr,index = L_names, columns = MI_names)
columns = data_named.columns.tolist()
columns = columns[::-1]
data_named = data_named[columns]
f, ax = plt.subplots(nrows=1, ncols=1, figsize=(15, 15), sharex=True)
sns.heatmap(data_PC, annot = True)
plt.savefig('../output/fig/PC1_heatmap.png')
```
|
github_jupyter
|
# Benchmarking the Permanent
This tutorial shows how to use the permanent function using The Walrus, which calculates the permanent using Ryser's algorithm
### The Permanent
The permanent of an $n$-by-$n$ matrix A = $a_{i,j}$ is defined as
$\text{perm}(A)=\sum_{\sigma\in S_n}\prod_{i=1}^n a_{i,\sigma(i)}.$
The sum here extends over all elements $\sigma$ of the symmetric group $S_n$; i.e. over all permutations of the numbers $1, 2, \ldots, n$. ([see Wikipedia](https://en.wikipedia.org/wiki/Permanent)).
The function `thewalrus.perm` implements [Ryser's algorithm](https://en.wikipedia.org/wiki/Computing_the_permanent#Ryser_formula) to calculate the permanent of an arbitrary matrix using [Gray code](https://en.wikipedia.org/wiki/Gray_code) ordering.
## Using the library
Once installed or compiled, one imports the library in the usual way:
```
from thewalrus import perm
```
To use it we need to pass square numpy arrays thus we also import NumPy:
```
import numpy as np
import time
```
The library provides functions to compute permanents of real and complex matrices. The functions take as arguments the matrix; the number of threads to be used to do the computation are determined using OpenMP.
```
size = 20
matrix = np.ones([size,size])
perm(matrix)
size = 20
matrix = np.ones([size,size], dtype=np.complex128)
perm(matrix)
```
Not surprisingly, the permanent of a matrix containing only ones equals the factorial of the dimension of the matrix, in our case $20!$.
```
from math import factorial
factorial(20)
```
### Benchmarking the performance of the code
For sizes $n=1,28$ we will generate random unitary matrices and measure the (average) amount of time it takes to calculate their permanent. The number of samples for each will be geometrically distirbuted with a 1000 samples for size $n=1$ and 10 samples for $n=28$. The unitaries will be random Haar distributed.
```
a0 = 1000.
anm1 = 10.
n = 28
r = (anm1/a0)**(1./(n-1))
nreps = [(int)(a0*(r**((i)))) for i in range(n)]
nreps
```
The following function generates random Haar unitaries of dimensions $n$
```
from scipy import diagonal, randn
from scipy.linalg import qr
def haar_measure(n):
'''A Random matrix distributed with Haar measure
See https://arxiv.org/abs/math-ph/0609050
How to generate random matrices from the classical compact groups
by Francesco Mezzadri '''
z = (randn(n,n) + 1j*randn(n,n))/np.sqrt(2.0)
q,r = qr(z)
d = diagonal(r)
ph = d/np.abs(d)
q = np.multiply(q,ph,q)
return q
```
Now let's bench mark the scaling of the calculation with the matrix size:
```
times = np.empty(n)
for ind, reps in enumerate(nreps):
#print(ind+1,reps)
start = time.time()
for i in range(reps):
size = ind+1
nth = 1
matrix = haar_measure(size)
res = perm(matrix)
end = time.time()
times[ind] = (end - start)/reps
print(ind+1, times[ind])
```
We can now plot the (average) time it takes to calculate the permanent vs. the size of the matrix:
```
import matplotlib.pyplot as plt
%matplotlib inline
%config InlineBackend.figure_formats=['svg']
plt.semilogy(np.arange(1,n+1),times,"+")
plt.xlabel(r"Matrix size $n$")
plt.ylabel(r"Time in seconds for 4 threads")
```
We can also fit to the theoretical scaling of $ c n 2^n$ and use it to extrapolate for larger sizes:
```
def fit(n,c):
return c*n*2**n
from scipy.optimize import curve_fit
popt, pcov = curve_fit(fit, np.arange(1,n+1)[15:-1],times[15:-1])
```
The scaling prefactor is
```
popt[0]
```
And we can use it to extrapolate the time it takes to calculate permanents of bigger dimensions
```
flags = [3600,3600*24*7, 3600*24*365, 3600*24*365*1000]
labels = ["1 hour", "1 week", "1 year", "1000 years"]
plt.semilogy(np.arange(1,n+1), times, "+", np.arange(1,61), fit(np.arange(1,61),popt[0]))
plt.xlabel(r"Matrix size $n$")
plt.ylabel(r"Time in seconds for single thread")
plt.hlines(flags,0,60,label="1 hr",linestyles=u'dotted')
for i in range(len(flags)):
plt.text(0,2*flags[i], labels[i])
```
The specs of the computer on which this benchmark was performed are:
```
!cat /proc/cpuinfo|head -19
```
|
github_jupyter
|
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# Import all the necessary files!
import os
import tensorflow as tf
from tensorflow.keras import layers
from tensorflow.keras import Model
# Download the inception v3 weights
!wget --no-check-certificate \
https://storage.googleapis.com/mledu-datasets/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5 \
-O /tmp/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5
# Import the inception model
from tensorflow.keras.applications.inception_v3 import InceptionV3
# Create an instance of the inception model from the local pre-trained weights
local_weights_file = '/tmp/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5'
# Your Code Here
pre_trained_model = InceptionV3(input_shape=(150,150,3),
include_top = False,
weights = None)
pre_trained_model.load_weights(local_weights_file)
# Make all the layers in the pre-trained model non-trainable
for layer in pre_trained_model.layers:
# Your Code Here
layer.trainable = False
# Print the model summary
pre_trained_model.summary()
# Expected Output is extremely large, but should end with:
#batch_normalization_v1_281 (Bat (None, 3, 3, 192) 576 conv2d_281[0][0]
#__________________________________________________________________________________________________
#activation_273 (Activation) (None, 3, 3, 320) 0 batch_normalization_v1_273[0][0]
#__________________________________________________________________________________________________
#mixed9_1 (Concatenate) (None, 3, 3, 768) 0 activation_275[0][0]
# activation_276[0][0]
#__________________________________________________________________________________________________
#concatenate_5 (Concatenate) (None, 3, 3, 768) 0 activation_279[0][0]
# activation_280[0][0]
#__________________________________________________________________________________________________
#activation_281 (Activation) (None, 3, 3, 192) 0 batch_normalization_v1_281[0][0]
#__________________________________________________________________________________________________
#mixed10 (Concatenate) (None, 3, 3, 2048) 0 activation_273[0][0]
# mixed9_1[0][0]
# concatenate_5[0][0]
# activation_281[0][0]
#==================================================================================================
#Total params: 21,802,784
#Trainable params: 0
#Non-trainable params: 21,802,784
last_layer = pre_trained_model.get_layer('mixed7')
print('last layer output shape: ', last_layer.output_shape)
last_output = last_layer.output# Your Code Here
# Expected Output:
# ('last layer output shape: ', (None, 7, 7, 768))
# Define a Callback class that stops training once accuracy reaches 99.9%
class myCallback(tf.keras.callbacks.Callback):
def on_epoch_end(self, epoch, logs={}):
if logs.get('accuracy'):
if(logs.get('accuracy')>0.999):
print("\nReached 99.9% accuracy so cancelling training!")
self.model.stop_training = True
from tensorflow.keras.optimizers import RMSprop
# Flatten the output layer to 1 dimension
x = layers.Flatten()(last_output)
# Add a fully connected layer with 1,024 hidden units and ReLU activation
x = layers.Dense(1024, activation='relu')(x)
# Add a dropout rate of 0.2
x = layers.Dropout(0.2)(x)
# Add a final sigmoid layer for classification
x = layers.Dense (1, activation='sigmoid')(x)
model = Model(pre_trained_model.input, x)
model.compile(optimizer = RMSprop(lr=0.0001),
loss = 'binary_crossentropy',
metrics = ['acc'])
model.summary()
# Expected output will be large. Last few lines should be:
# mixed7 (Concatenate) (None, 7, 7, 768) 0 activation_248[0][0]
# activation_251[0][0]
# activation_256[0][0]
# activation_257[0][0]
# __________________________________________________________________________________________________
# flatten_4 (Flatten) (None, 37632) 0 mixed7[0][0]
# __________________________________________________________________________________________________
# dense_8 (Dense) (None, 1024) 38536192 flatten_4[0][0]
# __________________________________________________________________________________________________
# dropout_4 (Dropout) (None, 1024) 0 dense_8[0][0]
# __________________________________________________________________________________________________
# dense_9 (Dense) (None, 1) 1025 dropout_4[0][0]
# ==================================================================================================
# Total params: 47,512,481
# Trainable params: 38,537,217
# Non-trainable params: 8,975,264
# Get the Horse or Human dataset
!wget --no-check-certificate https://storage.googleapis.com/laurencemoroney-blog.appspot.com/horse-or-human.zip -O /tmp/horse-or-human.zip
# Get the Horse or Human Validation dataset
!wget --no-check-certificate https://storage.googleapis.com/laurencemoroney-blog.appspot.com/validation-horse-or-human.zip -O /tmp/validation-horse-or-human.zip
from tensorflow.keras.preprocessing.image import ImageDataGenerator
import os
import zipfile
local_zip = '//tmp/horse-or-human.zip'
zip_ref = zipfile.ZipFile(local_zip, 'r')
zip_ref.extractall('/tmp/training')
zip_ref.close()
local_zip = '//tmp/validation-horse-or-human.zip'
zip_ref = zipfile.ZipFile(local_zip, 'r')
zip_ref.extractall('/tmp/validation')
zip_ref.close()
train_horses_dir = "/tmp/training/horses/" # Your Code Here
train_humans_dir = "/tmp/training/humans" # Your Code Here
validation_horses_dir = "/tmp/validation/horses" # Your Code Here
validation_humans_dir = "/tmp/validation/humans" # Your Code Here
train_horses_fnames = len(os.listdir(train_horses_dir))
train_humans_fnames = len(os.listdir(train_humans_dir)) # Your Code Here
validation_horses_fnames = len(os.listdir(validation_horses_dir)) # Your Code Here
validation_humans_fnames = len(os.listdir(validation_humans_dir)) # Your Code Here
print(train_horses_fnames)
print(train_humans_fnames)
print(validation_horses_fnames)
print(validation_humans_fnames)
# Expected Output:
# 500
# 527
# 128
# 128
# Define our example directories and files
train_dir = '/tmp/training'
validation_dir = '/tmp/validation'
train_datagen = ImageDataGenerator(rescale=1./255,
rotation_range=40,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
fill_mode='nearest')
# Note that the validation data should not be augmented!
test_datagen = ImageDataGenerator( rescale = 1.0/255.)
# Flow training images in batches of 20 using train_datagen generator
train_generator = train_datagen.flow_from_directory(train_dir,
batch_size=20,
class_mode='binary',
target_size=(150, 150))
# Flow validation images in batches of 20 using test_datagen generator
validation_generator = test_datagen.flow_from_directory(validation_dir,
batch_size=20,
class_mode = 'binary',
target_size = (150, 150))
# Expected Output:
# Found 1027 images belonging to 2 classes.
# Found 256 images belonging to 2 classes.
# Run this and see how many epochs it should take before the callback
# fires, and stops training at 99.9% accuracy
# (It should take less than 100 epochs)
callbacks = myCallback()# Your Code Here
history = model.fit_generator(train_generator,
epochs=3,
verbose=1,
validation_data=validation_generator,
callbacks=[callbacks])
import matplotlib.pyplot as plt
acc = history.history['acc']
val_acc = history.history['val_acc']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(len(acc))
plt.plot(epochs, acc, 'r', label='Training accuracy')
plt.plot(epochs, val_acc, 'b', label='Validation accuracy')
plt.title('Training and validation accuracy')
plt.legend(loc=0)
plt.figure()
plt.show()
```
|
github_jupyter
|
```
from keras import applications
# python image_scraper.py "yellow labrador retriever" --count 500 --label labrador
from keras.preprocessing.image import ImageDataGenerator
from keras_tqdm import TQDMNotebookCallback
from keras import optimizers
from keras.models import Sequential, Model
from keras.layers import (Dropout, Flatten, Dense, Conv2D,
Activation, MaxPooling2D)
from keras.applications.inception_v3 import InceptionV3
from keras.layers import GlobalAveragePooling2D
from keras import backend as K
from sklearn.cross_validation import train_test_split
import os, glob
from tqdm import tqdm
from collections import Counter
import pandas as pd
from sklearn.utils import shuffle
import numpy as np
import shutil
more_im = glob.glob("collie_lab/*/*.jpg")
more_im = shuffle(more_im)
collie = [x for x in more_im if "coll" in x.split("\\")[-2]]
lab = [x for x in shuffle(more_im) if "lab" in x.split("\\")[-2]]
print(len(collie))
print(len(lab))
for_labeling = collie + lab
for_labeling = shuffle(for_labeling)
Counter([x.split("\\")[-2] for x in more_im]).most_common()
import shutil
from tqdm import tqdm
%mkdir collie_lab_train
%mkdir collie_lab_valid
%mkdir collie_lab_train\\collie
%mkdir collie_lab_train\\lab
%mkdir collie_lab_valid\\collie
%mkdir collie_lab_valid\\lab
for index, image in tqdm(enumerate(for_labeling)):
if index < 1000:
label = image.split("\\")[-2]
image_name = image.split("\\")[-1]
if "coll" in label:
shutil.copy(image, 'collie_lab_train\\collie\\{}'.format(image_name))
if "lab" in label:
shutil.copy(image, 'collie_lab_train\\lab\\{}'.format(image_name))
if index > 1000:
label = image.split("\\")[-2]
image_name = image.split("\\")[-1]
if "coll" in label:
shutil.copy(image, 'collie_lab_valid\\collie\\{}'.format(image_name))
if "lab" in label:
shutil.copy(image, 'collie_lab_valid\\lab\\{}'.format(image_name))
train_datagen = ImageDataGenerator(
rescale=1. / 255,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=False)
test_datagen = ImageDataGenerator(rescale=1. / 255)
train_generator = train_datagen.flow_from_directory(
'collie_lab_train/',
target_size=(150, 150),
batch_size=32,
shuffle=True,
class_mode='binary')
validation_generator = test_datagen.flow_from_directory(
'collie_lab_valid/',
target_size=(150, 150),
batch_size=32,
class_mode='binary')
model = Sequential()
model.add(Conv2D(32, (3, 3), input_shape=(150, 150, 3)))
model.add(Activation('relu')) #tanh
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.5))
model.add(Conv2D(32, (3, 3)))
model.add(Activation('relu')) #tanh
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.5))
model.add(Conv2D(64, (3, 3)))
model.add(Activation('relu'))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.5))
model.add(Flatten())
model.add(Dense(96))
model.add(Activation('relu'))
model.add(Dropout(0.5))
model.add(Dense(1)) # binary
model.add(Activation('sigmoid'))
model.compile(loss='binary_crossentropy',
optimizer='adam',
metrics=['accuracy'])
model.fit_generator(
train_generator,
steps_per_epoch= 3000 // 32, # give me more data
epochs=30,
callbacks=[TQDMNotebookCallback()],
verbose=0,
validation_data=validation_generator,
validation_steps= 300 // 32)
```
|
github_jupyter
|
# Applying GrandPrix on the cell cycle single cell nCounter data of PC3 human prostate cancer
_Sumon Ahmed_, 2017, 2018
This notebooks describes how GrandPrix with informative prior over the latent space can be used to infer the cell cycle stages from the single cell nCounter data of the PC3 human prostate cancer cell line.
```
import pandas as pd
import numpy as np
from GrandPrix import GrandPrix
```
# Data decription
<a href="https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4102402/" terget="_blank">McDavid et al. (2014)</a> assayed the expression profiles of the PC3 human prostate cancer cell line. They identified the cells in G0/G1, S and G2/M cell cycle stages. The cells identified as G0/G1, S and G2/M have been mapped to the capture times of 1, 2 and 3, respectively. Due to the additional challenge of optimizing pseudotime parameters for periodic data, random pseudotimes having the largest log likelihood to estimate cell cycle peak time points have been used to initilize the prior.
The __McDavidtrainingData.csv__ file contains the expression profiles of the top __56__ differentially expressed genes in __361__ cells from the PC3 human prostate cancer cell line which have been used in the inference.
The __McDavidCellMeta.csv__ file contains the additional information of the data such as capture time of each cells, different initializations of pseudotimes, etc.
```
Y = pd.read_csv('../data/McDavid/McDavidtrainingData.csv', index_col=[0]).T
mData = pd.read_csv('../data/McDavid/McDavidCellMeta.csv', index_col=[0])
N, D = Y.shape
print('Time Points: %s, Genes: %s'%(N, D))
mData.head()
```
## Model with Informative prior
Capture time points have been used as the informative prior information over pseudotime. Following arguments have been passed to initialize the model.
<!--
- __data__: _array-like, shape N x D_. Observed data, where N is the number of time points and D is the number of genes.
- __latent_prior_mean__: _array-like, shape N_ x 1, _optional (default:_ __0__). > Mean of the prior distribution over pseudotime.
- __latent_prior_var__: _array-like, shape N_ x 1, _optional (default:_ __1.__). Variance of the prior distribution over pseudotime.
- __latent_mean__: _array-like, shape N_ x 1, _optional (default:_ __1.__). Initial mean values of the approximate posterior distribution over pseudotime.
- __latent_var__: _array-like, shape N_ x 1, _optional (default:_ __1.__). Initial variance of the approximate posterior distribution over pseudotime.
- __kernel:__ _optional (default: RBF kernel with lengthscale and variance set to 1.0)_. Covariance function to define the mapping from the latent space to the data space in Gaussian process prior.
-->
- __data__: _array-like, shape N x D_. Observed data, where N is the number of time points and D is the number of genes.
- __latent_prior_mean__: _array-like, shape N_ x 1. Mean of the prior distribution over pseudotime.
- __latent_prior_mean__: _array-like, shape N_ x 1. Mean of the prior distribution over pseudotime.
- __latent_prior_var__: _array-like, shape N_ x 1. Variance of the prior distribution over pseudotime.
- __latent_mean__: _array-like, shape N_ x 1. Initial mean values of the approximate posterior distribution over pseudotime.
<!--
- __latent_var__: _array-like, shape N_ x 1. Initial variance of the approximate posterior distribution over pseudotime.
-->
- __kernel__: Covariance function to define the mapping from the latent space to the data space in Gaussian process prior. Here we have used the standard periodic covariance function <a href="http://www.ics.uci.edu/~welling/teaching/KernelsICS273B/gpB.pdf" terget="_blank">(MacKay, 1998)</a>, to restrict the Gaussian Process (GP) prior to periodic functions only.
- __predict__: _int_. The number of new points. The mean of the expression level and associated variance of these new data points will be predicted.
```
np.random.seed(10)
sigma_t = .5
prior_mean = mData['prior'].values[:, None]
init_mean = mData['capture.orig'].values[:, None]
X_mean = [init_mean[i, 0] + sigma_t * np.random.randn(1) for i in range(0, N)] # initialisation of latent_mean
mp = GrandPrix.fit_model(data=Y.values, n_inducing_points = 20, latent_prior_mean=prior_mean, latent_prior_var=np.square(sigma_t),
latent_mean=np.asarray(X_mean), kernel={'name':'Periodic', 'ls':5.0, 'var':1.0}, predict=100)
pseudotimes = mp[0]
posterior_var = mp[1]
mean = mp[2] # mean of predictive distribution
var = mp[3] # variance of predictive distribution
Xnew = np.linspace(min(pseudotimes), max(pseudotimes), 100)[:, None]
```
# Visualize the results
The expression profile of some interesting genes have been plotted against the estimated pseudotime. Each point corresponds to a particular gene expression in a cell.
The points are coloured based on cell cycle stages according to <a href="https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4102402/" terget="_blank" style="text-decoration:none;">McDavid et al. (2014)</a>. The circular horizontal axis (where both first and last labels are G2/M) represents the periodicity realized by the method in pseudotime inference.
The solid black line is the posterior predicted mean of expression profiles while the grey ribbon depicts the 95% confidence interval.
The vertical dotted lines are the CycleBase peak times for the selected genes.
To see the expression profiles of a different set of genes a list containing gene names shound be passed to the function `plot_genes`.
```
selectedGenes = ['CDC6', 'MKI67', 'NUF2', 'PRR11', 'PTTG1', 'TPX2']
geneProfiles = pd.DataFrame({selectedGenes[i]: Y[selectedGenes[i]] for i in range(len(selectedGenes))})
```
## Binding gene names with predictive mean and variations
```
geneNames = Y.columns.values
name = [_ for _ in geneNames]
posterior_mean = pd.DataFrame(mean, columns=name)
posterior_var = pd.DataFrame(var, columns=name)
```
## geneData description
The __"McDavidgene.csv"__ file contains gene specific information such as peak time, etc. for the top 56 differentially expressed genes.
```
geneData = pd.read_csv('../data/McDavid/McDavid_gene.csv', index_col=0).T
geneData.head()
%matplotlib inline
from utils import plot_genes
cpt = mData['capture.orig'].values
plot_genes(pseudotimes, geneProfiles, geneData, cpt, prediction=(Xnew, posterior_mean, posterior_var))
```
|
github_jupyter
|
# Sequences
## `sequence.DNA`
`coral.DNA` is the core data structure of `coral`. If you are already familiar with core python data structures, it mostly acts like a container similar to lists or strings, but also provides further object-oriented methods for DNA-specific tasks, like reverse complementation. Most design functions in `coral` return a `coral.DNA` object or something that contains a `coral.DNA` object (like `coral.Primer`). In addition, there are related `coral.RNA` and `coral.Peptide` objects for representing RNA and peptide sequences and methods for converting between them.
To get started with `coral.DNA`, import `coral`:
```
import coral as cor
```
### Your first sequence
Let's jump right into things. Let's make a sequence that's the first 30 bases of gfp from *A. victoria*. To initialize a sequence, you feed it a string of DNA characters.
```
example_dna = cor.DNA('atgagtaaaggagaagaacttttcactgga')
display(example_dna)
```
A few things just happened behind the scenes. First, the input was checked to make sure it's DNA (A, T, G, and C). For now, it supports only unambiguous letters - no N, Y, R, etc. Second, the internal representation is converted to an uppercase string - this way, DNA is displayed uniformly and functional elements (like annealing and overhang regions of primers) can be delineated using case. If you input a non-DNA sequence, a `ValueError` is raised.
For the most part, a `sequence.DNA` instance acts like a python container and many string-like operations work.
```
# Extract the first three bases
display(example_dna[0:3])
# Extract the last seven bases
display(example_dna[-7:])
# Reverse a sequence
display(example_dna[::-1])
# Grab every other base starting at index 0
display(example_dna[::2])
# Is the sequence 'AT' in our sequence? How about 'AC'?
print "'AT' is in our sequence: {}.".format("AT" in example_dna)
print "'ATT' is in our sequence: {}.".format("ATT" in example_dna)
```
Several other common special methods and operators are defined for sequences - you can concatenate DNA (so long as it isn't circular) using `+`, repeat linear sequences using `*` with an integer, check for equality with `==` and `!=` (note: features, not just sequences, must be identical), check the length with `len(dna_object)`, etc.
### Simple sequences - methods
In addition to slicing, `sequence.DNA` provides methods for common molecular manipulations. For example, reverse complementing a sequence is a single call:
```
example_dna.reverse_complement()
```
An extremely important method is the `.copy()` method. It may seem redundant to have an entire function for copying a sequence - why not just assign a `sequence.DNA` object to a new variable? As in most high-level languages, python does not actually copy entire objects in memory when assignment happens - it just adds another reference to the same data. The short of it is that the very common operation of generating a lot of new variants to a sequence, or copying a sequence, requires the use of a `.copy()` method. For example, if you want to generate a new list of variants where an 'a' is substituted one at a time at each part of the sequence, using `.copy()` returns the correct result (the first example) while directly accessing example_dna has horrible consequences (the edits build up, as they all modify the same piece of data sequentially):
```
example_dna.copy()
# Incorrect way (editing shared + mutable sequence):
example_dna = cor.DNA('atgagtaaaggagaagaacttttcactgga')
variant_list = []
for i, base in enumerate(example_dna):
variant = example_dna
variant.top[i] = 'A'
variant.bottom[i] = 'T'
variant_list.append(variant)
print [str(x) for x in variant_list]
print
# Correct way (copy mutable sequence, then edit):
example_dna = cor.DNA('atgagtaaaggagaagaacttttcactgga')
variant_list = []
for i, base in enumerate(example_dna):
variant = example_dna.copy()
variant.top[i] = 'A'
variant.bottom[i] = 'T'
variant_list.append(variant)
print [str(x) for x in variant_list]
```
An important fact about `sequence.DNA` methods and slicing is that none of the operations modify the object directly (they don't mutate their parent) - if we look at example_dna, it has not been reverse-complemented itself. Running `example_dna.reverse_complement()` outputs a new sequence, so if you want to save your chance you need to assign a variable:
```
revcomp_dna = example_dna.reverse_complement()
display(example_dna)
display(revcomp_dna)
```
You also have direct access important attributes of a `sequence.DNA` object. The following are examples of how to get important sequences or information about a sequence.
```
# The top strand - a simple python string in the 5' -> 3' orientation.
example_dna.top
# The bottom strand - another python string, also in the 5' -> 3' orientation.
example_dna.bottom
# Sequences are double stranded, or 'ds' by default.
# This is a directly accessible attribute, not a method, so () is not required.
example_dna.ds
# DNA can be linear or circular - check the boolean `circular` attribute.
example_dna.circular
# You can switch between topologies using the .circularize and .linearize methods.
# Circular DNA has different properties:
# 1) it can't be concatenated to
# 2) sequence searches using .locate will search over the current origin (e.g. from -10 to +10 for a 20-base sequence).
circular_dna = example_dna.circularize()
circular_dna.circular
# Linearization is more complex - you can choose the index at which to linearize a circular sequence.
# This simulates a precise double stranded break at the index of your choosing.
# The following example shows the difference between linearizing at index 0 (default) versus index 2
# (python 0-indexes, so index 2 = 3rd base, i.e. 'g' in 'atg')
print circular_dna.linearize()
print
print circular_dna.linearize(2)
# Sometimes you just want to rotate the sequence around - i.e. switch the top and bottom strands.
# For this, use the .flip() method
example_dna.flip()
```
|
github_jupyter
|
```
import os
os.chdir('C:\\Users\\SHAILESH TIWARI\\Downloads\\Classification\\hr')
%matplotlib inline
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
train = pd.read_csv('train.csv')
# getting their shapes
print("Shape of train :", train.shape)
#print("Shape of test :", test.shape)
train.shape
train.head()
train.columns
train.isna().sum()
#calculation of percentage of missing data
total = train.isnull().sum().sort_values(ascending=False)
percent_1 = train.isnull().sum()/train.isnull().count()*100
percent_2 = (round(percent_1, 1)).sort_values(ascending=False)
missing_data = pd.concat([total, percent_2], axis=1, keys=['Total', '%'])
missing_data.head(5)
train['is_promoted'].value_counts() #unbalanced
train.shape
# finding the %age of people promoted
promoted = (4668/54808)*100
print("Percentage of Promoted Employees is {:.2f}%".format(promoted))
#plotting a scatter plot
plt.hist(train['is_promoted'])
plt.title('plot to show the gap in Promoted and Non-Promoted Employees', fontsize = 30)
plt.xlabel('0 -No Promotion and 1- Promotion', fontsize = 20)
plt.ylabel('count')
plt.show()
s1=train.dtypes
s1.groupby(s1).count()
train.dtypes
corr_matrix = train.corr(method='pearson')
corr_matrix['is_promoted'].sort_values(kind="quicksort")
#dropping the column
train.drop(['employee_id','region'], axis = 1, inplace = True)
train.head()
train.columns.values
#check for missing value, unique etc
FileNameDesc = pd.DataFrame(columns = ['column_name','missing_count','percent_missing','unique_count'])
for col in list(train.columns.values):
sum_missing = train[col].isnull().sum()
percent_missing = sum_missing/len(train)*100
uniq_count = (train.groupby([col])[col].count()).count()
FileNameDesc = FileNameDesc.append({'column_name':col,'missing_count':sum_missing,
'percent_missing':percent_missing,'unique_count':uniq_count},
ignore_index = True)
FileNameDesc
#Apply Mode strategy to populate the categorical data
train.groupby('education').agg({'education': np.size})
train["education"]=train["education"].fillna('Attchd')
train["education"]=train["education"].astype('category')
train["education"] = train["education"].cat.codes
train.isnull().sum()
train['previous_year_rating'].unique()
train['previous_year_rating'].mode()
train['previous_year_rating'].fillna(1, inplace = True)
train.isnull().sum()
train.dtypes
data=pd.get_dummies(train,columns=['department','gender','recruitment_channel','previous_year_rating'],drop_first=True)
data
df1=data['is_promoted']
data.drop(['is_promoted'], axis = 1, inplace = True)
data=pd.concat([data,df1],axis=1)
#Key data analysis
len(data)
data.head()
data.isnull().any()
data.isnull().sum()
data.corr()
sns.heatmap(data.corr(),annot=False)
data.columns
x = data.iloc[:,0:22].values
y = data.iloc[:,-1:].values
x
y
from sklearn.preprocessing import StandardScaler # to make the data in standard format to read
sc = StandardScaler() # feature scaling because salary and age are both in different scale
x=sc.fit_transform(x)
pd.DataFrame(x)
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test= train_test_split(x,y,test_size=0.20, random_state=0)
# applying logistic regression
from sklearn.linear_model import LogisticRegression
logmodel = LogisticRegression()
logmodel.fit(x_train,y_train)
# prediction for x_test
y_pred = logmodel.predict(x_test)
y_pred
y_test
# concept of confusion matrix
from sklearn.metrics import confusion_matrix
confusion_matrix(y_test,y_pred)
from sklearn.metrics import accuracy_score
accuracy_score(y_test,y_pred)
len(y_test)
sns.pairplot(train)
# applying cross validation on top of algo
from sklearn.model_selection import cross_val_score
accuracies = cross_val_score(estimator=logmodel, X=x_train,y=y_train,cv=10)
accuracies
accuracies.mean()
# k nearest neighbour algo applying
from sklearn.neighbors import KNeighborsClassifier
classifier_knn =KNeighborsClassifier(n_neighbors=11,metric='euclidean',p=2)
classifier_knn.fit(x_train,y_train)
y_pred_knn = classifier_knn.predict(x_test)
y_pred_knn
y_test
from sklearn.metrics import confusion_matrix
confusion_matrix(y_test,y_pred_knn)
from sklearn.metrics import accuracy_score
accuracy_score(y_test,y_pred_knn)
from sklearn.model_selection import cross_val_score
accuracies = cross_val_score(estimator=classifier_knn, X=x_train,y=y_train,cv=10)
accuracies
accuracies.mean()
# naiye baise algo application
from sklearn.naive_bayes import GaussianNB
classifier_nb =GaussianNB()
classifier_nb.fit(x_train,y_train)
y_pred_nb = classifier_nb.predict(x_test)
y_pred_nb
y_test
from sklearn.metrics import confusion_matrix
confusion_matrix(y_test,y_pred_nb)
from sklearn.metrics import accuracy_score
accuracy_score(y_test,y_pred_nb)
from sklearn.model_selection import cross_val_score
accuracies = cross_val_score(estimator=classifier_nb, X=x_train,y=y_train,cv=10)
accuracies
accuracies.mean()
# support vector machine application through sigmoid kernel
from sklearn.svm import SVC
classifier_svm_sig = SVC(kernel='sigmoid')
classifier_svm_sig.fit(x_train,y_train)
pred_svm_sig = classifier_svm_sig.predict(x_test)
pred_svm_sig
from sklearn.metrics import confusion_matrix
confusion_matrix(y_test,pred_svm_sig)
from sklearn.metrics import accuracy_score
accuracy_score(y_test,pred_svm_sig)
# support vector machine application through linear kernel
from sklearn.svm import SVC
classifier_svm_lin = SVC(kernel='linear')
classifier_svm_lin.fit(x_train,y_train)
y_pred_svm_lin = classifier_svm_lin.predict(x_test)
y_pred_svm_lin
from sklearn.metrics import confusion_matrix
confusion_matrix(y_test,y_pred_svm_lin)
from sklearn.metrics import accuracy_score
accuracy_score(y_test,y_pred_svm_lin)
# support vector machine application through polynomial kernel
from sklearn.svm import SVC
classifier_svm_poly = SVC(kernel='poly')
classifier_svm_poly.fit(x_train,y_train)
y_pred_svm_poly = classifier_svm_poly.predict(x_test)
y_pred_svm_poly
from sklearn.metrics import confusion_matrix
confusion_matrix(y_test,y_pred_svm_poly)
from sklearn.metrics import accuracy_score
accuracy_score(y_test,y_pred_svm_poly)
# support vector machine application through rbf kernel
from sklearn.svm import SVC
classifier_svm_rbf = SVC(kernel='rbf')
classifier_svm_rbf.fit(x_train,y_train)
y_pred_svm_rbf = classifier_svm_rbf.predict(x_test)
y_pred_svm_rbf
from sklearn.metrics import confusion_matrix
confusion_matrix(y_test,y_pred_svm_rbf)
#accuracy score
from sklearn.metrics import accuracy_score
accuracy_score(y_test,y_pred_svm_rbf)
#running decision tree algo
from sklearn.tree import DecisionTreeClassifier
classifier_dt =DecisionTreeClassifier(criterion='entropy') # also can use gini
classifier_dt.fit(x_train,y_train)
y_pred_dt =classifier_dt.predict(x_test)
y_pred_dt
from sklearn.metrics import confusion_matrix
confusion_matrix(y_test,y_pred_dt)
# accuracy score calculation
from sklearn.metrics import accuracy_score
accuracy_score(y_test,y_pred_dt)
# running random forest algorithm
from sklearn.ensemble import RandomForestClassifier
classifier_rf =RandomForestClassifier(n_estimators=3, criterion='entropy')
classifier_rf.fit(x_train,y_train)
y_pred_rf =classifier_rf.predict(x_test)
y_pred_rf
from sklearn.metrics import confusion_matrix
confusion_matrix(y_test,y_pred_rf)
from sklearn.metrics import accuracy_score
accuracy_score(y_test,y_pred_rf)
```
|
github_jupyter
|
# 3. Image-Similar-FCNN-Binary
For landmark-recognition-2019 algorithm validation
## Run name
```
import time
project_name = 'Dog-Breed'
step_name = '3-Image-Similar-FCNN-Binary'
time_str = time.strftime("%Y%m%d-%H%M%S", time.localtime())
run_name = project_name + '_' + step_name + '_' + time_str
print('run_name: ' + run_name)
t0 = time.time()
```
## Important params
```
import multiprocessing
cpu_amount = multiprocessing.cpu_count()
print('cpu_amount: ', cpu_amount)
```
## Import PKGs
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
%matplotlib inline
from IPython.display import display
import os
import sys
import gc
import math
import shutil
import zipfile
import pickle
import h5py
from tqdm import tqdm
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix, accuracy_score
import keras
from keras.utils import Sequence
from keras.layers import *
from keras.models import *
from keras.applications import *
from keras.optimizers import *
from keras.regularizers import *
from keras.preprocessing.image import *
from keras.applications.inception_v3 import preprocess_input
```
## Project folders
```
cwd = os.getcwd()
feature_folder = os.path.join(cwd, 'feature')
input_folder = os.path.join(cwd, 'input')
output_folder = os.path.join(cwd, 'output')
model_folder = os.path.join(cwd, 'model')
org_train_folder = os.path.join(input_folder, 'org_train')
org_test_folder = os.path.join(input_folder, 'org_test')
train_folder = os.path.join(input_folder, 'data_train')
val_folder = os.path.join(input_folder, 'data_val')
test_folder = os.path.join(input_folder, 'data_test')
test_sub_folder = os.path.join(test_folder, 'test')
vgg16_feature_file = os.path.join(feature_folder, 'feature_wrapper_171023.h5')
train_csv_file = os.path.join(input_folder, 'train.csv')
test_csv_file = os.path.join(input_folder, 'test.csv')
sample_submission_folder = os.path.join(input_folder, 'sample_submission.csv')
print(vgg16_feature_file)
print(train_csv_file)
print(test_csv_file)
print(sample_submission_folder)
```
## Load feature
```
with h5py.File(vgg16_feature_file, 'r') as h:
x_train = np.array(h['train'])
y_train = np.array(h['train_label'])
x_val = np.array(h['val'])
y_val = np.array(h['val_label'])
print(x_train.shape)
print(y_train.shape)
print(x_val.shape)
print(y_val.shape)
import random
random.choice(list(range(10)))
import copy
a = list(range(10, 20))
print(a)
a.remove(13)
print(a)
```
## ImageSequence
```
# class ImageSequence(Sequence):
# def __init__(self, x, y, batch_size, times_for_1_image, positive_rate):
# self.x = x
# self.y = y
# self.batch_size = batch_size
# self.times_for_1_image = times_for_1_image
# self.positive_rate = positive_rate
# self.len_x = self.x.shape[0]
# self.index = list(range(self.len_x))
# self.group = {}
# self.classes = list(set(self.y))
# self.classes.sort()
# for c in self.classes:
# self.group[c] = []
# for i, y_i in enumerate(self.y):
# temp_arr = self.group[y_i]
# temp_arr.append(i)
# self.group[y_i] = temp_arr
# def __len__(self):
# # times_for_1_image: the times to train one image
# # 2: positive example and negative example
# return self.times_for_1_image * 2 * (math.ceil(self.len_x/self.batch_size))
# def __getitem__(self, idx):
# batch_main_x = []
# batch_libary_x = []
# batch_x = {}
# batch_y = [] # 0 or 1
# for i in range(self.batch_size):
# # prepare main image
# item_main_image_idx = random.choice(self.index) # random choice one image from all train images
# item_main_image_y = self.y[item_main_image_idx]
# # prepare libary image
# is_positive = random.random() < self.positive_rate
# if is_positive: # chioce a positive image as libary_x
# # choice one image from itself group
# item_libary_image_idx = random.choice(self.group[item_main_image_y]) # don't exclude item_main_image_idx, so it could choice a idx same to item_main_image_idx.
# else: # chioce a negative image as libary_x
# # choice group
# new_class = copy.deepcopy(self.classes)
# new_class.remove(item_main_image_y)
# item_libary_image_group_num = random.choice(new_class)
# # choice one image from group
# item_libary_image_idx = random.choice(self.group[item_libary_image_group_num])
# # add item data to batch
# batch_main_x.append(self.x[item_main_image_idx])
# batch_libary_x.append(self.x[item_libary_image_idx])
# batch_y.append(int(is_positive))
# # concatenate array to np.array
# batch_x = {
# 'main_input': np.array(batch_main_x),
# 'library_input': np.array(batch_libary_x)
# }
# batch_y = np.array(batch_y)
# return batch_x, batch_y
# demo_sequence = ImageSequence(x_train[:200], y_train[:200], 128, 3, 0.1)
# print(len(demo_sequence))
# print(type(demo_sequence))
# batch_index = 0
# demo_batch = demo_sequence[batch_index]
# demo_batch_x = demo_batch[0]
# demo_batch_y = demo_batch[1]
# print(type(demo_batch_x))
# print(type(demo_batch_y))
# demo_main_input = demo_batch_x['main_input']
# demo_library_input = demo_batch_x['library_input']
# print(demo_main_input.shape)
# print(demo_library_input.shape)
# print(demo_batch_y.shape)
# # print(demo_main_input[0])
# print(demo_batch_y)
class ImageSequence(Sequence):
def __init__(self, x, y, batch_size, times_for_1_image, positive_rate):
self.x = x
self.y = y
self.batch_size = batch_size
self.times_for_1_image = times_for_1_image
self.positive_rate = positive_rate
self.len_x = self.x.shape[0]
self.index = list(range(self.len_x))
self.group = {}
self.classes = list(set(self.y))
self.classes.sort()
for c in self.classes:
self.group[c] = []
for i, y_i in enumerate(self.y):
temp_arr = self.group[y_i]
temp_arr.append(i)
self.group[y_i] = temp_arr
def __len__(self):
# times_for_1_image: the times to train one image
# 2: positive example and negative example
return self.times_for_1_image * 2 * (math.ceil(self.len_x/self.batch_size))
def __getitem__(self, idx):
batch_main_x = np.zeros((self.batch_size, self.x.shape[1]))
batch_libary_x = np.zeros((self.batch_size, self.x.shape[1]))
batch_x = {}
batch_y = [] # 0 or 1
for i in range(self.batch_size):
# prepare main image
item_main_image_idx = random.choice(self.index) # random choice one image from all train images
item_main_image_y = self.y[item_main_image_idx]
# prepare libary image
is_positive = random.random() < self.positive_rate
if is_positive: # chioce a positive image as libary_x
# choice one image from itself group
item_libary_image_idx = random.choice(self.group[item_main_image_y]) # don't exclude item_main_image_idx, so it could choice a idx same to item_main_image_idx.
else: # chioce a negative image as libary_x
# choice group
new_class = copy.deepcopy(self.classes)
new_class.remove(item_main_image_y)
item_libary_image_group_num = random.choice(new_class)
# choice one image from group
item_libary_image_idx = random.choice(self.group[item_libary_image_group_num])
# add item data to batch
batch_main_x[i] = self.x[item_main_image_idx]
batch_libary_x[i] = self.x[item_libary_image_idx]
batch_y.append(int(is_positive))
# concatenate array to np.array
batch_x = {
'main_input': batch_main_x,
'library_input': batch_libary_x
}
batch_y = np.array(batch_y)
return batch_x, batch_y
demo_sequence = ImageSequence(x_train[:200], y_train[:200], 128, 3, 0.1)
print(len(demo_sequence))
print(type(demo_sequence))
batch_index = 0
demo_batch = demo_sequence[batch_index]
demo_batch_x = demo_batch[0]
demo_batch_y = demo_batch[1]
print(type(demo_batch_x))
print(type(demo_batch_y))
demo_main_input = demo_batch_x['main_input']
demo_library_input = demo_batch_x['library_input']
print(demo_main_input.shape)
print(demo_library_input.shape)
print(demo_batch_y.shape)
# print(demo_main_input[0])
print(demo_batch_y)
train_sequence = ImageSequence(x_train, y_train, 32, 3, 0.5)
val_sequence = ImageSequence(x_val, y_val, 32, 3, 0.5)
```
## Model
```
main_input = Input((x_train.shape[1],), dtype='float32', name='main_input')
library_input = Input((x_train.shape[1],), dtype='float32', name='library_input')
x = keras.layers.concatenate([main_input, library_input])
x = Dense(x_train.shape[1]*2, activation='sigmoid')(x)
x = Dense(1024, activation='sigmoid')(x)
x = Dense(1024, activation='sigmoid')(x)
output = Dense(1, activation='sigmoid')(x)
model = Model(inputs=[main_input, library_input], outputs=[output])
model.compile(optimizer=Adam(lr=1e-4), loss='binary_crossentropy', metrics=['accuracy'])
model.summary()
hist = model.fit_generator(
train_sequence,
steps_per_epoch=128,
epochs=300,
verbose=1,
callbacks=None,
validation_data=val_sequence,
validation_steps=128,
class_weight=None,
max_queue_size=10,
workers=1,
use_multiprocessing=False,
shuffle=True,
initial_epoch=0
)
import matplotlib.pyplot as plt
%matplotlib inline
plt.plot(hist.history['loss'], color='b')
plt.plot(hist.history['val_loss'], color='r')
plt.show()
plt.plot(hist.history['acc'], color='b')
plt.plot(hist.history['val_acc'], color='r')
plt.show()
def saveModel(model, run_name):
cwd = os.getcwd()
modelPath = os.path.join(cwd, 'model')
if not os.path.isdir(modelPath):
os.mkdir(modelPath)
weigths_file = os.path.join(modelPath, run_name + '.h5')
print(weigths_file)
model.save(weigths_file)
saveModel(model, run_name)
print('Time elapsed: %.1fs' % (time.time() - t0))
print(run_name)
```
|
github_jupyter
|
# Support Vector Machine
```
from PIL import Image
import numpy as np
%matplotlib inline
import matplotlib
import matplotlib.pyplot as plt
from sklearn import datasets, svm, linear_model
matplotlib.style.use('bmh')
matplotlib.rcParams['figure.figsize']=(10,10)
```
### 2D Linear
```
# Random 2d X
X0 = np.random.normal(-2, size=(30,2))
X1 = np.random.normal(2, size=(30,2))
X = np.concatenate([X0,X1], axis=0)
y = X @ [1,1] > 0
clf=svm.SVC(kernel='linear', C=1000)
clf.fit(X, y)
# 邊界
x_min, y_min = X.min(axis=0)-1
x_max, y_max = X.max(axis=0)+1
# 座標點
grid = np.mgrid[x_min:x_max:200j, y_min:y_max:200j]
# grid.shape = (2, 200, 200)
# 在座標點 算出 svm 的判斷函數
Z = clf.decision_function(grid.reshape(2, -1).T)
Z = Z.reshape(grid.shape[1:])
# 畫出顏色和邊界
plt.pcolormesh(grid[0], grid[1], Z > 0, cmap=plt.cm.rainbow, alpha=0.1)
plt.contour(grid[0], grid[1], Z, colors=['k', 'k', 'k'], linestyles=['--', '-', '--'],
levels=[-1, 0, 1])
# 標出 sample 點
plt.scatter(X[:,0], X[:, 1], c=y, cmap=plt.cm.rainbow, zorder=10, s=50);
```
3D view
```
from mpl_toolkits.mplot3d import Axes3D
ax = plt.gca(projection='3d')
ax.plot_surface(grid[0], grid[1], Z, cmap=plt.cm.rainbow, alpha=0.2)
ax.plot_wireframe(grid[0], grid[1], Z, alpha=0.2, rstride=20, cstride=20)
ax.scatter(X[:, 0], X[:, 1], y, c=y, cmap=plt.cm.rainbow, s=30);
ax.set_zlim3d(-2,2)
ax.set_xlim3d(-3,3)
ax.set_ylim3d(-3,3)
ax.view_init(15, -75)
```
Linear Nonseparable
```
# Random 2d X
X = np.random.uniform(-1.5, 1.5, size=(100,2))
y = (X**2).sum(axis=1) > 1
clf=svm.SVC(kernel='linear', C=1000)
clf.fit(X, y)
# 邊界
x_min, y_min = X.min(axis=0)-1
x_max, y_max = X.max(axis=0)+1
# 座標點
grid = np.mgrid[x_min:x_max:200j, y_min:y_max:200j]
# grid.shape = (2, 200, 200)
# 在座標點 算出 svm 的判斷函數
Z = clf.decision_function(grid.reshape(2, -1).T)
Z = Z.reshape(grid.shape[1:])
# 畫出顏色和邊界
plt.pcolormesh(grid[0], grid[1], Z > 0, cmap=plt.cm.rainbow, alpha=0.1)
plt.contour(grid[0], grid[1], Z, colors=['k', 'k', 'k'], linestyles=['--', '-', '--'],
levels=[-1, 0, 1])
# 標出 sample 點
plt.scatter(X[:,0], X[:, 1], c=y, cmap=plt.cm.rainbow, zorder=10, s=20);
(np.linspace(-1.5,1.5, 10)[:, None] @ np.linspace(-1.5,1.5, 10)[None, :]).shape
# Random 2d X
X = np.random.uniform(-1.5, 1.5, size=(100,2))
# more feature (x**2, y**2, x*y)
X2 = np.concatenate([X, X**2, (X[:, 0]*X[:, 1])[:, None]], axis=1)
y = (X**2).sum(axis=1) > 1
clf=svm.SVC(kernel='linear', C=1000)
clf.fit(X2, y)
# 邊界
x_min, y_min = X.min(axis=0)-1
x_max, y_max = X.max(axis=0)+1
# 座標點
grid = np.mgrid[x_min:x_max:200j, y_min:y_max:200j]
# grid.shape = (2, 200, 200)
G = grid.reshape(2, -1).T
G = np.concatenate([G, G**2, (G[:, 0]*G[:, 1])[:, None]], axis=1)
# 在座標點 算出 svm 的判斷函數
Z = clf.decision_function(G)
Z = Z.reshape(grid.shape[1:])
# 畫出顏色和邊界
plt.pcolormesh(grid[0], grid[1], Z > 0, cmap=plt.cm.rainbow, alpha=0.1)
plt.contour(grid[0], grid[1], Z, colors=['k', 'k', 'k'], linestyles=['--', '-', '--'],
levels=[-1, 0, 1])
# 標出 sample 點
plt.scatter(X[:,0], X[:, 1], c=y, cmap=plt.cm.rainbow, zorder=10, s=20);
#%matplotlib qt
ax = plt.gca(projection='3d')
ax.plot_surface(grid[0], grid[1], Z, cmap=plt.cm.rainbow, alpha=0.2)
ax.plot_wireframe(grid[0], grid[1], Z, alpha=0.2, rstride=20, cstride=20)
ax.scatter(X[:, 0], X[:, 1], y, c=y, cmap=plt.cm.rainbow, s=30);
#plt.show()
```
With kernel
```
%matplotlib inline
matplotlib.rcParams['figure.figsize']=(10,10)
# Random 2d X
X = np.random.uniform(-1.5, 1.5, size=(100,2))
# more feature (x**2, y**2, x*y)
X2 = np.concatenate([X, X**2, (X[:, 0]*X[:, 1])[:, None]], axis=1)
y = (X**2).sum(axis=1) > 1
clf=svm.SVC(kernel='rbf', C=1000)
clf.fit(X2, y)
# 邊界
x_min, y_min = X.min(axis=0)-1
x_max, y_max = X.max(axis=0)+1
# 座標點
grid = np.mgrid[x_min:x_max:200j, y_min:y_max:200j]
# grid.shape = (2, 200, 200)
G = grid.reshape(2, -1).T
G = np.concatenate([G, G**2, (G[:, 0]*G[:, 1])[:, None]], axis=1)
# 在座標點 算出 svm 的判斷函數
Z = clf.decision_function(G)
Z = Z.reshape(grid.shape[1:])
# 畫出顏色和邊界
plt.pcolormesh(grid[0], grid[1], Z > 0, cmap=plt.cm.rainbow, alpha=0.1)
plt.contour(grid[0], grid[1], Z, colors=['k', 'k', 'k'], linestyles=['--', '-', '--'],
levels=[-1, 0, 1])
# 標出 sample 點
plt.scatter(X[:,0], X[:, 1], c=y, cmap=plt.cm.rainbow, zorder=10, s=20);
#%matplotlib qt
ax = plt.gca(projection='3d')
ax.plot_surface(grid[0], grid[1], Z, cmap=plt.cm.rainbow, alpha=0.2)
ax.plot_wireframe(grid[0], grid[1], Z, alpha=0.2, rstride=20, cstride=20)
ax.scatter(X[:, 0], X[:, 1], y, c=y, cmap=plt.cm.rainbow, s=30);
#plt.show()
```
|
github_jupyter
|
```
import sys
import numpy as np
```
# Numpy
Numpy proporciona un nuevo contenedor de datos a Python, los `ndarray`s, además de funcionalidad especializada para poder manipularlos de forma eficiente.
Hablar de manipulación de datos en Python es sinónimo de Numpy y prácticamente todo el ecosistema científico de Python está construido sobre Numpy. Digamos que Numpy es el ladrillo que ha permitido levantar edificios tan sólidos como Pandas, Matplotlib, Scipy, scikit-learn,...
**Índice**
* [¿Por qué un nuevo contenedor de datos?](#%C2%BFPor-qu%C3%A9-un-nuevo-contenedor-de-datos?)
* [Tipos de datos](#Tipos-de-datos)
* [Creación de `numpy` arrays](#Creaci%C3%B3n-de-numpy-arrays)
* [Operaciones disponibles más típicas](#Operaciones-disponibles-m%C3%A1s-t%C3%ADpicas)
* [Metadatos y anatomía de un `ndarray`](#Metadatos-y-anatom%C3%ADa-de-un-ndarray)
* [Indexación](#Indexaci%C3%B3n)
* [Manejo de valores especiales](#Manejo-de-valores-especiales)
* [Subarrays, vistas y copias](#Subarrays,-vistas-y-copias)
* [¿Cómo funcionan los ejes de un `ndarray`?](#%C2%BFC%C3%B3mo-funcionan-los-ejes-en-un-ndarray?)
* [Reformateo de `ndarray`s](#Reformateo-de-ndarrays)
* [Broadcasting](#Broadcasting)
* [`ndarrays` estructurados y `recarray`s](#ndarrays-estructurados-y-recarrays)
* [Concatenación y partición de `ndarray`s](#Concatenaci%C3%B3n-y-partici%C3%B3n-de-ndarrays)
* [Funciones matemáticas, funciones universales *ufuncs* y vectorización](#Funciones-matem%C3%A1ticas,-funciones-universales-ufuncs-y-vectorizaci%C3%B3n)
* [Estadística](#Estad%C3%ADstica)
* [Ordenando, buscando y contando](#Ordenando,-buscando-y-contando)
* [Polinomios](#Polinomios)
* [Álgebra lineal](#%C3%81lgebra-lineal)
* [Manipulación de `ndarray`s](#Manipulaci%C3%B3n-de-ndarrays)
* [Módulos de interés dentro de numpy](#M%C3%B3dulos-de-inter%C3%A9s-dentro-de-numpy)
* [Cálculo matricial](#C%C3%A1lculo-matricial)
## ¿Por qué un nuevo contenedor de datos?
En Python, disponemos, de partida, de diversos contenedores de datos, listas, tuplas, diccionarios, conjuntos,..., ¿por qué añadir uno más?.
¡Por conveniencia!, a pesar de la pérdida de flexibilidad. Es una solución de compromiso.
* Uso de memoria más eficiente: Por ejemplo, una lista puede contener distintos tipos de objetos lo que provoca que Python deba guardar información del tipo de cada elemento contenido en la lista. Por otra parte, un `ndarray` contiene tipos homogéneos, es decir, todos los elementos son del mismo tipo, por lo que la información del tipo solo debe guardarse una vez independientemente del número de elementos que tenga el `ndarray`.

***(imagen por Jake VanderPlas y extraída [de GitHub](https://github.com/jakevdp/PythonDataScienceHandbook)).***
* Más rápido: Por ejemplo, en una lista que consta de elementos con diferentes tipos Python debe realizar trabajos extra para saber si los tipos son compatibles con las operaciones que estamos realizando. Cuando trabajamos con un `ndarray` ya podemos saber eso de partida y podemos tener operaciones más eficientes (además de que mucha funcionalidad está programada en C, C++, Cython, Fortran).
* Operaciones vectorizadas
* Funcionalidad extra: Muchas operaciones de álgebra lineal, transformadas rápidas de Fourier, estadística básica, histogramas,...
* Acceso a los elementos más conveniente: Indexación más avanzada que con los tipos normales de Python
* ...
Uso de memoria
```
# AVISO: SYS.GETSYZEOF NO ES FIABLE
lista = list(range(5_000_000))
arr = np.array(lista, dtype=np.uint32)
print("5 millones de elementos")
print(sys.getsizeof(lista))
print(sys.getsizeof(arr))
print()
lista = list(range(100))
arr = np.array(lista, dtype=np.uint8)
print("100 elementos")
print(sys.getsizeof(lista))
print(sys.getsizeof(arr))
```
Velocidad de operaciones
```
a = list(range(1000000))
%timeit sum(a)
print(sum(a))
a = np.array(a)
%timeit np.sum(a)
print(np.sum(a))
```
Operaciones vectorizadas
```
# Suma de dos vectores elemento a elemento
a = [1, 1, 1]
b = [3, 4, 3]
print(a + b)
print('Fail')
# Suma de dos vectores elemento a elemento
a = np.array([1, 1, 1])
b = np.array([3, 4, 3])
print(a + b)
print('\o/')
```
Funcionalidad más conveniente
```
# suma acumulada
a = list(range(100))
print([sum(a[:i+1]) for i in a])
a = np.array(a)
print(a.cumsum())
```
Acceso a elementos más conveniente
```
a = [[11, 12, 13],
[21, 22, 23],
[31, 32, 33]]
print('acceso a la primera fila: ', a[0])
print('acceso a la primera columna: ', a[:][0], ' Fail!!!')
a = np.array(a)
print('acceso a la primera fila: ', a[0])
print('acceso a la primera columna: ', a[:,0], ' \o/')
```
...
Recapitulando un poco.
***Los `ndarray`s son contenedores multidimensionales, homogéneos con elementos de tamaño fijo, de dimensión predefinida.***
## Tipos de datos
Como los arrays deben ser homogéneos tenemos tipos de datos. Algunos de ellos se pueden ver en la siguiente tabla:
| Data type | Descripción |
|---------------|-------------|
| ``bool_`` | Booleano (True o False) almacenado como un Byte |
| ``int_`` | El tipo entero por defecto (igual que el `long` de C; normalmente será `int64` o `int32`)|
| ``intc`` | Idéntico al ``int`` de C (normalmente `int32` o `int64`)|
| ``intp`` | Entero usado para indexación (igual que `ssize_t` en C; normalmente `int32` o `int64`)|
| ``int8`` | Byte (de -128 a 127)|
| ``int16`` | Entero (de -32768 a 32767)|
| ``int32`` | Entero (de -2147483648 a 2147483647)|
| ``int64`` | Entero (de -9223372036854775808 a 9223372036854775807)|
| ``uint8`` | Entero sin signo (de 0 a 255)|
| ``uint16`` | Entero sin signo (de 0 a 65535)|
| ``uint32`` | Entero sin signo (de 0 a 4294967295)|
| ``uint64`` | Entero sin signo (de 0 a 18446744073709551615)|
| ``float_`` | Atajo para ``float64``.|
| ``float16`` | Half precision float: un bit para el signo, 5 bits para el exponente, 10 bits para la mantissa|
| ``float32`` | Single precision float: un bit para el signo, 8 bits para el exponente, 23 bits para la mantissa|
| ``float64`` | Double precision float: un bit para el signo, 11 bits para el exponente, 52 bits para la mantissa|
| ``complex_`` | Atajo para `complex128`.|
| ``complex64`` | Número complejo, represantedo por dos *floats* de 32-bits|
| ``complex128``| Número complejo, represantedo por dos *floats* de 64-bits|
Es posible tener una especificación de tipos más detallada, pudiendo especificar números con *big endian* o *little endian*. No vamos a ver esto en este momento.
El tipo por defecto que usa `numpy` al crear un *ndarray* es `np.float_`, siempre que no específiquemos explícitamente el tipo a usar.
Por ejemplo, un array de tipo `np.uint8` puede tener los siguientes valores:
```
import itertools
for i, bits in enumerate(itertools.product((0, 1), repeat=8)):
print(i, bits)
```
Es decir, puede contener valores que van de 0 a 255 ($2^8$).
¿Cuántos bytes tendrá un `ndarray` de 10 elementos cuyo tipo de datos es un `np.int8`?
```
a = np.arange(10, dtype=np.int8)
print(a.nbytes)
print(sys.getsizeof(a))
a = np.repeat(1, 100000).astype(np.int8)
print(a.nbytes)
print(sys.getsizeof(a))
```
## Creación de numpy arrays
Podemos crear numpy arrays de muchas formas.
* Rangos numéricos
`np.arange`, `np.linspace`, `np.logspace`
* Datos homogéneos
`np.zeros`, `np.ones`
* Elementos diagonales
`np.diag`, `np.eye`
* A partir de otras estructuras de datos ya creadas
`np.array`
* A partir de otros numpy arrays
`np.empty_like`
* A partir de ficheros
`np.loadtxt`, `np.genfromtxt`,...
* A partir de un escalar
`np.full`, `np.tile`,...
* A partir de valores aleatorios
`np.random.randint`, `np.random.randint`, `np.random.randn`,...
...
```
a = np.arange(10) # similar a range pero devuelve un ndarray en lugar de un objeto range
print(a)
a = np.linspace(0, 1, 101)
print(a)
a_i = np.zeros((2, 3), dtype=np.int)
a_f = np.zeros((2, 3))
print(a_i)
print(a_f)
a = np.eye(3)
print(a)
a = np.array(
(
(1, 2, 3, 4, 5, 6),
(10, 20, 30, 40, 50, 60)
),
dtype=np.float
)
print(a)
np.full((5, 5), -999)
np.random.randint(0, 50, 15)
```
<div class="alert alert-success">
<p>Referencias:</p>
<p><a href="https://docs.scipy.org/doc/numpy/user/basics.creation.html#arrays-creation">array creation</a></p>
<p><a href="https://docs.scipy.org/doc/numpy/reference/routines.array-creation.html#routines-array-creation">routines for array creation</a></p>
</div>
**Practicando**
Recordad que siempre podéis usar `help`, `?`, `np.lookfor`,..., para obtener más información.
```
help(np.sum)
np.rad2deg?
np.lookfor("create array")
```
Ved un poco como funciona `np.repeat`, `np.empty_like`,...
```
# Play area
%load ../../solutions/03_01_np_array_creacion.py
```
## Operaciones disponibles más típicas
```
a = np.random.rand(5, 2)
print(a)
a.sum()
a.sum(axis=0)
a.sum(axis=1)
a.ravel()
a.reshape(2, 5)
a.T
a.transpose()
a.mean()
a.mean(axis=1)
a.cumsum(axis=1)
```
<div class="alert alert-success">
<p>Referencias:</p>
<p><a href="https://docs.scipy.org/doc/numpy/user/quickstart.html">Quick start tutorial</a></p>
</div>
**Practicando**
Mirad más métodos de un `ndarray` y toquetead. Si no entendéis algo, preguntad:
```
dir(a)
# Play area
%load ../../solutions/03_02_np_operaciones_tipicas.py
```
## Metadatos y anatomía de un `ndarray`
En realidad, un `ndarray` es un bloque de memoria con información extra sobre como interpretar su contenido. La memoria dinámica (RAM) se puede considerar como un 'churro' lineal y es por ello que necesitamos esa información extra para saber como formar ese `ndarray`, sobre todo la información de `shape` y `strides`.
Esta parte va a ser un poco más esotérica para los no iniciados pero considero que es necesaria para poder entender mejor nuestra nueva estructura de datos y poder sacarle mejor partido.
```
a = np.random.randn(5000, 5000)
```
El número de dimensiones del `ndarray`
```
a.ndim
```
El número de elementos en cada una de las dimensiones
```
a.shape
```
El número de elementos
```
a.size
```
El tipo de datos de los elementos
```
a.dtype
```
El número de bytes de cada elemento
```
a.itemsize
```
El número de bytes que ocupa el `ndarray` (es lo mismo que `size` por `itemsize`)
```
a.nbytes
```
El *buffer* que contiene los elementos del `ndarray`
```
a.data
```
Pasos a dar en cada dimensión cuando nos movemos entre elementos
```
a.strides
```

***(imagen extraída [de GitHub](https://github.com/btel/2016-erlangen-euroscipy-advanced-numpy)).***
Más cosas
```
a.flags
```
Pequeño ejercicio, ¿por qué tarda menos en sumar elementos en una dimensión que en otra si es un array regular?
```
%timeit a.sum(axis=0)
%timeit a.sum(axis=1)
```
Pequeño ejercicio, ¿por qué ahora el resultado es diferente?
```
aT = a.T
%timeit aT.sum(axis=0)
%timeit aT.sum(axis=1)
print(aT.strides)
print(aT.flags)
print(np.repeat((1,2,3), 3))
print()
a = np.repeat((1,2,3), 3).reshape(3, 3)
print(a)
print()
print(a.sum(axis=0))
print()
print(a.sum(axis=1))
```
<div class="alert alert-success">
<p>Referencias:</p>
<p><a href="https://docs.scipy.org/doc/numpy/reference/arrays.ndarray.html#internal-memory-layout-of-an-ndarray">Internal memory layout of an ndarray</a></p>
<p><a href="https://docs.scipy.org/doc/numpy/reference/internals.html#multidimensional-array-indexing-order-issues">multidimensional array indexing order issues</a></p>
</div>
## Indexación
Si ya has trabajado con indexación en estructuras de Python, como listas, tuplas o strings, la indexación en Numpy te resultará muy familiar.
Por ejemplo, por hacer las cosas sencillas, vamos a crear un `ndarray` de 1D:
```
a = np.arange(10, dtype=np.uint8)
print(a)
print(a[:]) # para acceder a todos los elementos
print(a[:-1]) # todos los elementos menos el último
print(a[1:]) # todos los elementos menos el primero
print(a[::2]) # el primer, el tercer, el quinto,..., elemento
print(a[3]) # el cuarto elemento
print(a[-1:-5:-1]) # ¿?
# Practicad vosotros
```
Para *ndarrays* de una dimensión es exactamente igual que si usásemos listas o tuplas de Python:
* Primer elemento tiene índice 0
* Los índices negativos empiezan a contar desde el final
* slices/rebanadas con `[start:stop:step]`
Con un `ndarray` de más dimensiones las cosas ya cambian con respecto a Python puro:
```
a = np.random.randn(10, 2)
print(a)
a[1] # ¿Qué nos dará esto?
a[1, 1] # Si queremos acceder a un elemento específico hay que dar su posición completa en el ndarray
a[::3, 1]
```
Si tenemos dimensiones mayores a 1 es parecido a las listas pero los índices se separan por comas para las nuevas dimensiones.
<img src="../../images/03_03_arraygraphics_0.png" width=400px />
(imagen extraída de [aquí](https://github.com/gertingold/euroscipy-numpy-tutorial))
```
a = np.arange(40).reshape(5, 8)
print(a)
a[2, -3]
```
Para obtener más de un elemento hacemos *slicing* para cada eje:
<img src="../../images/03_04_arraygraphics_1.png" width=400px />
(imagen extraída de [aquí](https://github.com/gertingold/euroscipy-numpy-tutorial))
```
a[:3, :5]
```
¿Cómo podemos conseguir los elementos señalados en esta imagen?
<img src="../../images/03_06_arraygraphics_2_wo.png" width=400px />
(imagen extraída de [aquí](https://github.com/gertingold/euroscipy-numpy-tutorial))
```
a[x:x ,x:x]
```
¿Cómo podemos conseguir los elementos señalados en esta imagen?
<img src="../../images/03_08_arraygraphics_3_wo.png" width=400px />
(imagen extraída de [aquí](https://github.com/gertingold/euroscipy-numpy-tutorial))
```
a[x:x ,x:x]
```
¿Cómo podemos conseguir los elementos señalados en esta imagen?
<img src="../../images/03_10_arraygraphics_4_wo.png" width=400px />
(imagen extraída de [aquí](https://github.com/gertingold/euroscipy-numpy-tutorial))
```
a[x:x ,x:x]
```
¿Cómo podemos conseguir los elementos señalados en esta imagen?
<img src="../../images/03_12_arraygraphics_5_wo.png" width=400px />
(imagen extraída de [aquí](https://github.com/gertingold/euroscipy-numpy-tutorial))
```
a[x:x ,x:x]
```
Soluciones a lo anterior:
<img src="../../images/03_05_arraygraphics_2.png" width=200px />
<img src="../../images/03_07_arraygraphics_3.png" width=200px />
<img src="../../images/03_09_arraygraphics_4.png" width=200px />
<img src="../../images/03_11_arraygraphics_5.png" width=200px />
(imágenes extraídas de [aquí](https://github.com/gertingold/euroscipy-numpy-tutorial))
**Fancy indexing**
Con *fancy indexing* podemos hacer cosas tan variopintas como:
<img src="../../images/03_13_arraygraphics_6.png" width=300px />
<img src="../../images/03_14_arraygraphics_7.png" width=300px />
(imágenes extraídas de [aquí](https://github.com/gertingold/euroscipy-numpy-tutorial))
Es decir, podemos indexar usando `ndarray`s de booleanos ó usando listas de índices para extraer elementos concretos de una sola vez.
**WARNING: En el momento que usamos *fancy indexing* nos devuelve un nuevo *ndarray* que no tiene porque conservar la estructura original.**
Por ejemplo, en el siguiente caso no devuelve un *ndarray* de dos dimensiones porque la máscara no tiene porqué ser regular y, por tanto, devuelve solo los valores que cumplen el criterio en un vector (*ndarray* de una dimensión).
```
a = np.arange(10).reshape(2, 5)
print(a)
bool_indexes = (a % 2 == 0)
print(bool_indexes)
a[bool_indexes]
```
Sin embargo, sí que lo podríamos usar para modificar el *ndarray* original en base a un criterio y seguir manteniendo la misma forma.
```
a[bool_indexes] = 999
print(a)
```
<div class="alert alert-success">
<p>Referencias:</p>
<p><a href="https://docs.scipy.org/doc/numpy/reference/arrays.indexing.html#arrays-indexing">array indexing</a></p>
<p><a href="https://docs.scipy.org/doc/numpy/reference/arrays.ndarray.html#indexing-arrays">indexing arrays</a></p>
</div>
## Manejo de valores especiales
`numpy` provee de varios valores especiales: `np.nan`, `np.Inf`, `np.Infinity`, `np.inf`, `np.infty`,...
```
a = 1 / np.arange(10)
print(a)
a[0] == np.inf
a.max() # Esto no es lo que queremos
a.mean() # Esto no es lo que queremos
a[np.isfinite(a)].max()
a[-1] = np.nan
print(a)
a.mean()
np.isnan(a)
np.isfinite(a)
np.isinf(a) # podéis mirar también np.isneginf, np.isposinf
```
`numpy` usa el estándar IEEE de números flotantes para aritmética (IEEE 754). Esto significa que *Not a
Number* no es equivalente a *infinity*. También, *positive infinity* no es equivalente a *negative infinity*. Pero *infinity* es equivalente a *positive infinity*.
```
1 < np.inf
1 < -np.inf
1 > -np.inf
1 == np.inf
1 < np.nan
1 > np.nan
1 == np.nan
```
## Subarrays, vistas y copias
**¡IMPORTANTE!**
Vistas y copias: `numpy`, por defecto, siempre devuelve vistas para evitar incrementos innecesarios de memoria. Este comportamiento difiere del de Python puro donde una rebanada (*slicing*) de una lista devuelve una copia. Si queremos una copia de un `ndarray` debemos obtenerla de forma explícita:
```
a = np.arange(10)
b = a[2:5]
print(a)
print(b)
b[0] = 222
print(a)
print(b)
```
Este comportamiento por defecto es realmente muy útil, significa que, trabajando con grandes conjuntos de datos, podemos acceder y procesar piezas de estos conjuntos de datos sin necesidad de copiar el buffer de datos original.
A veces, es necesario crear una copia. Esto se puede realizar fácilmente usando el método `copy` de los *ndarrays*. El ejemplo anterior usando una copia en lugar de una vista:
```
a = np.arange(10)
b = a[2:5].copy()
print(a)
print(b)
b[0] = 222
print(a)
print(b)
```
## ¿Cómo funcionan los ejes en un `ndarray`?
Por ejemplo, cuando hacemos `a.sum()`, `a.sum(axis=0)`, `a.sum(axis=1)`.
¿Qué pasa si tenemos más de dos dimensiones?
Vamos a ver ejemplos:
```
a = np.arange(10).reshape(5,2)
a.shape
a.sum()
a.sum(axis=0)
a.sum(axis=1)
```

(imagen extraída de [aquí](https://github.com/gertingold/euroscipy-numpy-tutorial))
```
a = np.arange(9).reshape(3, 3)
print(a)
print(a.sum(axis=0))
print(a.sum(axis=1))
```

(imagen extraída de [aquí](https://github.com/gertingold/euroscipy-numpy-tutorial))
```
a = np.arange(24).reshape(2, 3, 4)
print(a)
print(a.sum(axis=0))
print(a.sum(axis=1))
print(a.sum(axis=2))
```
Por ejemplo, en el primer caso, `axis=0`, lo que sucede es que cogemos todos los elementos del primer índice y aplicamos la operación para cada uno de los elementos de los otros dos ejes. Hecho de uno en uno sería lo siguiente:
```
print(a[:,0,0].sum(), a[:,0,1].sum(), a[:,0,2].sum(), a[:,0,3].sum())
print(a[:,1,0].sum(), a[:,1,1].sum(), a[:,1,2].sum(), a[:,1,3].sum())
print(a[:,2,0].sum(), a[:,2,1].sum(), a[:,2,2].sum(), a[:,2,3].sum())
```
Sin contar el eje que estamos usando, las dimensiones que quedan son 3 x 4 (segunda y tercera dimensiones) por lo que el resultado son 12 elementos.
Para el caso de `axis=1`:
```
print(a[0,:,0].sum(), a[0,:,1].sum(), a[0,:,2].sum(), a[0,:,3].sum())
print(a[1,:,0].sum(), a[1,:,1].sum(), a[1,:,2].sum(), a[1,:,3].sum())
```
Sin contar el eje que estamos usando, las dimensiones que quedan son 2 x 4 (primera y tercera dimensiones) por lo que el resultado son 8 elementos.
Para el caso de `axis=2`:
```
print(a[0,0,:].sum(), a[0,1,:].sum(), a[0,2,:].sum())
print(a[1,0,:].sum(), a[1,1,:].sum(), a[1,2,:].sum())
```
Sin contar el eje que estamos usando, las dimensiones que quedan son 2 x 3 (primera y segunda dimensiones) por lo que el resultado son 3 elementos.
## Reformateo de `ndarray`s
Podemos cambiar la forma de los `ndarray`s usando el método `reshape`. Por ejemplo, si queremos colocar los números del 1 al 9 en un grid $3 \times 3$ lo podemos hacer de la siguiente forma:
```
a = np.arange(1, 10).reshape(3, 3)
```
Para que el cambio de forma no dé errores hemos de tener cuidado en que los tamaños del `ndarray` inicial y del `ndarray` final sean compatibles.
```
# Por ejemplo, lo siguiente dará error?
a = np.arange(1, 10). reshape(5, 2)
```
Otro patrón común de cambio de forma sería la conversion de un `ndarray` de 1D en uno de 2D añadiendo un nuevo eje. Lo podemos hacer usando, nuevamente, el método `reshape` o usando `numpy.newaxis`.
```
# Por ejemplo un array 2D de una fila
a = np.arange(3)
a1_2D = a.reshape(1,3)
a2_2D = a[np.newaxis, :]
print(a1_2D)
print(a1_2D.shape)
print(a2_2D)
print(a2_2D.shape)
# Por ejemplo un array 2D de una columna
a = np.arange(3)
a1_2D = a.reshape(3,1)
a2_2D = a[:, np.newaxis]
print(a1_2D)
print(a1_2D.shape)
print(a2_2D)
print(a2_2D.shape)
```
## Broadcasting
Es poible realizar operaciones en *ndarrays* de diferentes tamaños. En algunos casos `numpy` puede transformar estos *ndarrays* automáticamente de forma que todos tienen la misma forma. Esta conversión automática se llama **broadcasting**.
Normas del Broadcasting
Para determinar la interacción entre dos `ndarray`s en Numpy se sigue un conjunto de reglas estrictas:
* Regla 1: Si dos `ndarray`s difieren en su número de dimensiones la forma de aquel con menos dimensiones se rellena con 1's a su derecha.
- Regla 2: Si la forma de dos `ndarray`s no es la misma en ninguna de sus dimensiones, el `ndarry` con forma igual a 1 en esa dimensión se 'alarga' para tener simulares dimensiones que los del otros `ndarray`.
- Regla 3: Si en cualquier dimensión el tamaño no es igual y ninguno de ellos es igual a 1 entonces obtendremos un error.
Resumiendo, cuando se opera en dos *ndarrays*, `numpy` compara sus formas (*shapes*) elemento a elemento. Empieza por las dimensiones más a la izquierda y trabaja hacia las siguientes dimensiones. Dos dimensiones son compatibles cuando
ambas son iguales o
una de ellas es 1
Si estas condiciones no se cumplen se lanzará una excepción `ValueError: frames are not aligned` indicando que los *ndarrays* tienen formas incompatibles. El tamaño del *ndarray* resultante es el tamaño máximo a lo largo de cada dimensión de los *ndarrays* de partida.
De forma más gráfica:

(imagen extraída de [aquí](https://github.com/btel/2016-erlangen-euroscipy-advanced-numpy))
```
a: 4 x 3 a: 4 x 3 a: 4 x 1
b: 4 x 3 b: 3 b: 3
result: 4 x 3 result: 4 x 3 result: 4 x 3
```
Intentemos reproducir los esquemas de la imagen anterior.
```
a = np.repeat((0, 10, 20, 30), 3).reshape(4, 3)
b = np.repeat((0, 1, 2), 4).reshape(3,4).T
print(a)
print(b)
print(a + b)
a = np.repeat((0, 10, 20, 30), 3).reshape(4, 3)
b = np.array((0, 1, 2))
print(a)
print(b)
print(a + b)
a = np.array((0, 10, 20, 30)).reshape(4,1)
b = np.array((0, 1, 2))
print(a)
print(b)
print(a + b)
```
<div class="alert alert-success">
<p>Referencias:</p>
<p><a href="https://docs.scipy.org/doc/numpy/user/basics.broadcasting.html">Basic broadcasting</a></p>
<p><a href="http://scipy.github.io/old-wiki/pages/EricsBroadcastingDoc">Broadcasting more in depth</a></p>
</div>
## `ndarrays` estructurados y `recarray`s
Antes hemos comentado que los `ndarray`s deben ser homogéneos pero era un poco inexacto, en realidad, podemos tener `ndarray`s que tengan diferentes tipos. Estos se llaman `ndarray`s estructurados y `recarray`s.
Veamos ejemplos:
```
nombre = ['paca', 'pancracio', 'nemesia', 'eulogio']
edad = [72, 68, 86, 91]
a = np.array(np.zeros(4), dtype=[('name', '<S10'), ('age', np.int)])
a['name'] = nombre
a['age'] = edad
print(a)
```
Podemos acceder a las columnas por nombre
```
a['name']
```
A todos los elementos menos el primero
```
a['age'][1:]
```
Un `recarray` es similar pero podemos acceder a los campos con notación de punto (*dot notation*).
```
ra = a.view(np.recarray)
ra.name
```
Esto introduce un poco de *overhead* para acceder ya que se realizan algunas operaciones de más.
## Concatenación y partición de `ndarrays`
Podemos combinar múltiples *ndarrays* en uno o separar uno en varios.
Para concatenar podemos usar `np.concatenate`, `np.hstack`, `np.vstack`, `np.dstack`. Ejemplos:
```
a = np.array([1, 1, 1, 1])
b = np.array([2, 2, 2, 2])
```
Podemos concatenar esos dos arrays usando `np.concatenate`:
```
np.concatenate([a, b])
```
No solo podemos concatenar *ndarrays* de una sola dimensión:
```
np.concatenate([a.reshape(2, 2), b.reshape(2, 2)])
```
Podemos elegir sobre qué eje concatenamos:
```
np.concatenate([a.reshape(2, 2), b.reshape(2, 2)], axis=1)
```
Podemos concatenar más de dos arrays:
```
c = [3, 3, 3, 3]
np.concatenate([a, b, c])
```
Si queremos ser más explícitos podemos usar `np.hstack` o `np.vstack`. La `h` y la `v` son para horizontal y vertical, respectivamente.
```
np.hstack([a, b])
np.vstack([a, b])
```
Podemos concatenar en la tercera dimensión usamos `np.dstack`.
De la misma forma que podemos concatenar, podemos partir *ndarrays* usando `np.split`, `np.hsplit`, `np.vsplit`, `np.dsplit`.
```
# Intentamos entender como funciona la partición probando...
```
## Funciones matemáticas, funciones universales *ufuncs* y vectorización
¿Qué es eso de *ufunc*?
De la [documentación oficial de Numpy](http://docs.scipy.org/doc/numpy/reference/ufuncs.html):
> A universal function (or ufunc for short) is a function that operates on ndarrays in an element-by-element fashion, supporting array broadcasting, type casting, and several other standard features. That is, a ufunc is a “**vectorized**” wrapper for a function that takes a **fixed number of scalar inputs** and produces a **fixed number of scalar outputs**.
Una *ufunc* es una *Universal function* o función universal que actúa sobre todos los elementos de un `ndarray`, es decir aplica la funcionalidad sobre cada uno de los elementos del `ndarray`. Esto se conoce como vectorización.
Por ejemplo, veamos la operación de elevar al cuadrado una lista en python puro o en `numpy`:
```
# En Python puro
a_list = list(range(10000))
%timeit [i ** 2 for i in a_list]
# En numpy
an_arr = np.arange(10000)
%timeit np.power(an_arr, 2)
a = np.arange(10)
np.power(a, 2)
```
La función anterior eleva al cuadrado cada uno de los elementos del `ndarray` anterior.
Dentro de `numpy` hay muchísimas *ufuncs* y `scipy` (no lo vamos a ver) dispone de muchas más *ufuns* mucho más especializadas.
En `numpy` tenemos, por ejemplo:
* Funciones trigonométricas: `sin`, `cos`, `tan`, `arcsin`, `arccos`, `arctan`, `hypot`, `arctan2`, `degrees`, `radians`, `unwrap`, `deg2rad`, `rad2deg`
```
# juguemos un poco con ellas
```
* Funciones hiperbólicas: `sinh`, `cosh`, `tanh`, `arcsinh`, `arccosh`, `arctanh`
```
# juguemos un poco con ellas
```
* Redondeo: `around`, `round_`, `rint`, `fix`, `floor`, `ceil`, `trunc`
```
# juguemos un poco con ellas
```
* Sumas, productos, diferencias: `prod`, `sum`, `nansum`, `cumprod`, `cumsum`, `diff`, `ediff1d`, `gradient`, `cross`, `trapz`
```
# juguemos un poco con ellas
```
* Exponentes y logaritmos: `exp`, `expm1`, `exp2`, `log`, `log10`, `log2`, `log1p`, `logaddexp`, `logaddexp2`
```
# juguemos un poco con ellas
```
* Otras funciones especiales: `i0`, `sinc`
```
# juguemos un poco con ellas
```
* Trabajo con decimales: `signbit`, `copysign`, `frexp`, `ldexp`
```
# juguemos un poco con ellas
```
* Operaciones aritméticas: `add`, `reciprocal`, `negative`, `multiply`, `divide`, `power`, `subtract`, `true_divide`, `floor_divide`, `fmod`, `mod`, `modf`, `remainder`
```
# juguemos un poco con ellas
```
* Manejo de números complejos: `angle`, `real`, `imag`, `conj`
```
# juguemos un poco con ellas
```
* Miscelanea: `convolve`, `clip`, `sqrt`, `square`, `absolute`, `fabs`, `sign`, `maximum`, `minimum`, `fmax`, `fmin`, `nan_to_num`, `real_if_close`, `interp`
...
```
# juguemos un poco con ellas
```
<div class="alert alert-success">
<p>Referencias:</p>
<p><a href="http://docs.scipy.org/doc/numpy/reference/ufuncs.html">Ufuncs</a></p>
</div>
## Estadística
* Orden: `amin`, `amax`, `nanmin`, `nanmax`, `ptp`, `percentile`, `nanpercentile`
* Medias y varianzas: `median`, `average`, `mean`, `std`, `var`, `nanmedian`, `nanmean`, `nanstd`, `nanvar`
* Correlacionando: `corrcoef`, `correlate`, `cov`
* Histogramas: `histogram`, `histogram2d`, `histogramdd`, `bincount`, `digitize`
...
```
# juguemos un poco con ellas
```
## Ordenando, buscando y contando
* Ordenando: `sort`, `lexsort`, `argsort`, `ndarray.sort`, `msort`, `sort_complex`, `partition`, `argpartition`
* Buscando: `argmax`, `nanargmax`, `argmin`, `nanargmin`, `argwhere`, `nonzero`, `flatnonzero`, `where`, `searchsorted`, `extract`
* Contando: `count_nonzero`
...
```
# juguemos un poco con ellas
```
## Polinomios
* Series de potencias: `numpy.polynomial.polynomial`
* Clase Polynomial: `np.polynomial.Polynomial`
* Básicos: `polyval`, `polyval2d`, `polyval3d`, `polygrid2d`, `polygrid3d`, `polyroots`, `polyfromroots`
* Ajuste: `polyfit`, `polyvander`, `polyvander2d`, `polyvander3d`
* Cálculo: `polyder`, `polyint`
* Álgebra: `polyadd`, `polysub`, `polymul`, `polymulx`, `polydiv`, `polypow`
* Miscelánea: `polycompanion`, `polydomain`, `polyzero`, `polyone`, `polyx`, `polytrim`, `polyline`
* Otras funciones polinómicas: `Chebyshev`, `Legendre`, `Laguerre`, `Hermite`
...
```
# juguemos un poco con ellas
```
## Álgebra lineal
Lo siguiente que se encuentra dentro de `numpy.linalg` vendrá precedido por `LA`.
* Productos para vectores y matrices: `dot`, `vdot`, `inner`, `outer`, `matmul`, `tensordot`, `einsum`, `LA.matrix_power`, `kron`
* Descomposiciones: `LA.cholesky`, `LA.qr`, `LA.svd`
* Eigenvalores: `LA.eig`, `LA.eigh`, `LA.eigvals`, `LA.eigvalsh`
* Normas y otros números: `LA.norm`, `LA.cond`, `LA.det`, `LA.matrix_rank`, `LA.slogdet`, `trace`
* Resolución de ecuaciones e inversión de matrices: `LA.solve`, `LA.tensorsolve`, `LA.lstsq`, `LA.inv`, `LA.pinv`, `LA.tensorinv`
Dentro de `scipy` tenemos más cosas relacionadas.
```
# juguemos un poco con ellas
```
## Manipulación de `ndarrays`
`tile`, `hstack`, `vstack`, `dstack`, `hsplit`, `vsplit`, `dsplit`, `repeat`, `reshape`, `ravel`, `resize`,...
```
# juguemos un poco con ellas
```
## Módulos de interés dentro de `numpy`
Dentro de `numpy` podemos encontrar módulos para:
* Usar números aleatorios: `np.random`
* Usar FFT: `np.fft`
* Usar *masked arrays*: `np.ma`
* Usar polinomios: `np.polynomial`
* Usar álgebra lineal: `np.linalg`
* Usar matrices: `np.matlib`
* ...
Toda esta funcionalidad se puede ampliar y mejorar usando `scipy`.
## Cálculo matricial
```
a1 = np.repeat(2, 9).reshape(3, 3)
a2 = np.tile(2, (3, 3))
a3 = np.ones((3, 3), dtype=np.int) * 2
print(a1)
print(a2)
print(a3)
b = np.arange(1,4)
print(b)
print(a1.dot(b))
print(np.dot(a2, b))
print(a3 @ b) # only python version >= 3.5
```
Lo anterior lo hemos hecho usando *ndarrays* pero `numpy` también ofrece una estructura de datos `matrix`.
```
a_mat = np.matrix(a1)
a_mat
b_mat = np.matrix(b)
a_mat @ b_mat
a_mat @ b_mat.T
```
Como vemos, con los *ndarrays* no hace falta que seamos rigurosos con las dimensiones, en cambio, si usamos `np.matrix` como tipos hemos de realizar operaciones matriciales válidas (por ejemplo, que las dimensiones sean correctas).
A efectos prácticos, en general, los *ndarrays* se pueden usar como `matrix` conociendo estas pequeñas cosas.
|
github_jupyter
|
### Processing Echosounder Data from Ocean Observatories Initiative with `echopype`.
Downloading a file from the OOI website. We pick August 21, 2017 since this was the day of the solar eclipse which affected the traditional patterns of the marine life.
```
# downloading the file
!wget https://rawdata.oceanobservatories.org/files/CE04OSPS/PC01B/ZPLSCB102_10.33.10.143/OOI-D20170821-T163049.raw
filename = 'OOI-D20170821-T163049.raw'
```
**Converting from Raw to Standartized Netcdf Format**
```
# import as part of a submodule
from echopype.convert import ConvertEK60
data_tmp = ConvertEK60(filename)
data_tmp.raw2nc()
os.remove(filename)
```
**Calibrating, Denoising, Mean Volume Backscatter Strength**
```
from echopype.model import EchoData
data = EchoData(filename[:-4]+'.nc')
data.calibrate() # Calibration and echo-integration
data.remove_noise(save=True) # Save denoised Sv to FILENAME_Sv_clean.nc
data.get_MVBS(save=True)
```
**Visualizing the Result**
```
%matplotlib inline
data.MVBS.MVBS.sel(frequency=200000).plot(x='ping_time',cmap = 'jet')
```
**Processing Multiple Files**
To process multiple file from the OOI website we need to scrape the names of the existing files there. We will use the `Beautiful Soup`
package for that.
```
!conda install --yes beautifulsoup4
from bs4 import BeautifulSoup
from urllib.request import urlopen
path = 'https://rawdata.oceanobservatories.org/files/CE04OSPS/PC01B/ZPLSCB102_10.33.10.143/'
response = urlopen(path)
soup = BeautifulSoup(response.read(), "html.parser")
# urls = []
# for item in soup.find_all(text=True):
# if '.raw' in item:
# urls.append(path+'/'+item)
urls = [path+'/'+item for item in soup.find_all(text=True) if '.raw' in item]
# urls
from datetime import datetime
```
Specify range:
```
start_time = '20170821-T000000'
end_time = '20170822-T235959'
# convert the times to datetime format
start_datetime = datetime.strptime(start_time,'%Y%m%d-T%H%M%S')
end_datetime = datetime.strptime(end_time,'%Y%m%d-T%H%M%S')
# function to check if a date is in the date range
def in_range(date_str, start_time, end_time):
date_str = datetime.strptime(date_str,'%Y%m%d-T%H%M%S')
true = date_str >= start_datetime and date_str <= end_datetime
return(true)
# identify the list of urls in range
range_urls = []
for url in urls:
date_str = url[-20:-4]
if in_range(date_str, start_time, end_time):
range_urls.append(url)
range_urls
rawnames = [url.split('//')[-1] for url in range_urls]
ls
import os
```
**Downloading the Files**
```
# Download the files
import requests
rawnames = []
for url in range_urls:
r = requests.get(url, allow_redirects=True)
rawnames.append(url.split('//')[-1])
open(url.split('//')[-1], 'wb').write(r.content)
!pip install echopype
ls
```
**Converting from Raw to Standartized Netcdf Format**
```
# import as part of a submodule
from echopype.convert import ConvertEK60
for filename in rawnames:
data_tmp = ConvertEK60(filename)
data_tmp.raw2nc()
os.remove(filename)
#ls
```
**Calibrating, Denoising, Mean Volume Backscatter Strength**
```
# calibrate and denoise
from echopype.model import EchoData
for filename in rawnames:
data = EchoData(filename[:-4]+'.nc')
data.calibrate() # Calibration and echo-integration
data.remove_noise(save=False) # Save denoised Sv to FILENAME_Sv_clean.nc
data.get_MVBS(save=True)
os.remove(filename[:-4]+'.nc')
os.remove(filename[:-4]+'_Sv.nc')
```
**Opening and Visualizing the Results in Parallel**
No that all files are in an appropriate format, we can open them and visualize them in parallel. For that we will need to install the `dask` parallelization library.
```
!conda install --yes dask
import xarray as xr
res = xr.open_mfdataset('*MVBS.nc')
import matplotlib.pyplot as plt
plt.figure(figsize = (15,5))
res.MVBS.sel(frequency=200000).plot(x='ping_time',cmap = 'jet')
```
|
github_jupyter
|
Variables with more than one value
==================================
You have already seen ordinary variables that store a single value. However other variable types can hold more than one value. The simplest type is called a list. Here is a example of a list being used:
```
which_one = int(input("What month (1-12)? "))
months = ['January', 'February', 'March', 'April', 'May', 'June', 'July',\
'August', 'September', 'October', 'November', 'December']
if 1 <= which_one <= 12:
print("The month is", months[which_one - 1])
```
and an output example:
In this example the months is a list. months is defined with the lines months = ['January', 'February', 'March', 'April', 'May', 'June', 'July', 'August', 'September', 'October', 'November', 'December'] (Note that a `\` can be used to split a long line). The `[` and `]` start and end the list with comma's (`,`) separating the list items. The list is used in `months[which_one - 1]`. A list consists of items that are numbered starting at 0. In other words if you wanted January you would type in 1 and that would have 1 subtracted off to use `months[0]`. Give a list a number and it will return the value that is stored at that location.
The statement `if 1 <= which_one <= 12:` will only be true if `which_one` is between one and twelve inclusive (in other words it is what you would expect if you have seen that in algebra). Since 1 is subtracted from `which_one` we get list locations from 0 to 11.
Lists can be thought of as a series of boxes. For example, the boxes created by demolist = ['life', 42, 'the', 'universe', 6, 'and', 7] would look like this:
| | | | | | | | |
| --- | --- | --- | --- | --- | --- | --- | --- |
| box number | 0 | 1 | 2 | 3 | 4 | 5 | 6 |
| demolist | 'life' | 42 | 'the' | 'universe' | 6 | 'and' | 7 |
Each box is referenced by its number so the statement `demolist[0]` would get 'life', `demolist[1]` would get 42 and so on up to `demolist[6]` getting 7.
More features of lists
======================
The next example is just to show a lot of other stuff lists can do (for once, I don't expect you to type it in, but you should probably play around with lists until you are comfortable with them. Also, there will be another program that uses most of these features soon.). Here goes:
```
demolist = ['life', 42, 'the', 'universe', 6, 'and', 7]
print('demolist = ', demolist)
demolist.append('everything')
print("after 'everything' was appended demolist is now:")
print(demolist)
print('len(demolist) =', len(demolist))
print('demolist.index(42) =', demolist.index(42))
print('demolist[1] =', demolist[1])
#Next we will loop through the list
c = 0
while c < len(demolist):
print('demolist[', c, ']=', demolist[c])
c = c + 1
del demolist[2]
print("After 'the universe' was removed demolist is now:")
print(demolist)
if 'life' in demolist:
print("'life' was found in demolist")
else:
print("'life' was not found in demolist")
if 'amoeba' in demolist:
print("'amoeba' was found in demolist")
if 'amoeba' not in demolist:
print("'amoeba' was not found in demolist")
int_list = []
c = 0
while c < len(demolist):
if type(0) == type(demolist[c]):
int_list.append(demolist[c])
c = c + 1
print('int_list is', int_list)
int_list.sort()
print('The sorted int_list is ', int_list)
```
The output is:
This example uses a whole bunch of new functions. Notice that you can
just print a whole list. Next the append function is used
to add a new item to the end of the list. `len` returns how many
items are in a list. The valid indexes (as in numbers that can be
used inside of the []) of a list range from 0 to len - 1. The
index function tell where the first location of an item is
located in a list. Notice how `demolist.index(42)` returns 1 and
when `demolist[1]` is run it returns 42. The line
`#Next we will loop through the list` is a just a reminder to the
programmer (also called a comment). Python will ignore any lines that
start with a `#`. Next the lines:
```
c = 0
while c < len(demolist):
print('demolist[', c, ']=', demolist[c])
c = c + 1
```
This creates a variable c which starts at 0 and is incremented until it reaches the last index of the list. Meanwhile the print function prints out each element of the list.
The `del` command can be used to remove a given element in a list. The next few lines use the in operator to test if a element is in or is not in a list.
The `sort` function sorts the list. This is useful if you need a
list in order from smallest number to largest or alphabetical. Note
that this rearranges the list. Note also that the numbers were put in
a new list, and that was sorted, instead of trying to sort a mixed
list. Sorting numbers and strings does not really make sense and results
in an error.
In summary for a list the following operations exist:
| | | | |
| --- | --- | --- | --- |
| example | explanation | | |
| list[2] | accesses the element at index 2 | | |
| list[2] = 3 | sets the element at index 2 to be 3 | | |
| del list[2] | removes the element at index 2 | | |
| len(list) | returns the length of list | | |
| "value" in list | is true if "value" is an element in list | | |
| "value" not in list | is true if "value" is not an element in list | | |
| list.sort() | sorts list | | |
| list.index("value") | returns the index of the first place that "value" occurs | | |
| list.append("value") | adds an element "value" at the end of the list | | |
This next example uses these features in a more useful way:
```
menu_item = 0
list = []
while menu_item != 9:
print("--------------------")
print("1. Print the list")
print("2. Add a name to the list")
print("3. Remove a name from the list")
print("4. Change an item in the list")
print("9. Quit")
menu_item = int(input("Pick an item from the menu: "))
if menu_item == 1:
current = 0
if len(list) > 0:
while current < len(list):
print(current, ". ", list[current])
current = current + 1
else:
print("List is empty")
elif menu_item == 2:
name = input("Type in a name to add: ")
list.append(name)
elif menu_item == 3:
del_name = input("What name would you like to remove: ")
if del_name in list:
item_number = list.index(del_name)
del list[item_number]
#The code above only removes the first occurance of
# the name. The code below from Gerald removes all.
#while del_name in list:
# item_number = list.index(del_name)
# del list[item_number]
else:
print(del_name, " was not found")
elif menu_item == 4:
old_name = input("What name would you like to change: ")
if old_name in list:
item_number = list.index(old_name)
new_name = input("What is the new name: ")
list[item_number] = new_name
else:
print(old_name, " was not found")
print("Goodbye")
```
And here is part of the output:
That was a long program. Let's take a look at the source code. The line `list = []` makes the variable list a list with no items (or elements). The next important line is `while menu_item != 9:` . This line starts a loop that allows the menu system for this program. The next few lines display a menu and decide which part of the program to run.
The section:
goes through the list and prints each name. `len(list_name)` tell how many items are in a list. If len returns `0` then the list is empty.
Then a few lines later the statement `list.append(name)` appears. It uses the append function to add a item to the end of the list. Jump down another two lines and notice this section of code:
Here the index function is used to find the index value that will be used later to remove the item. `del list[item_number]` is used to remove a element of the list.
The next section
uses index to find the `item_number` and then puts `new_name` where the `old_name` was.
Congratulations, with lists under your belt, you now know enough of the language
that you could do any computations that a computer can do (this is technically known as Turing-Completeness). Of course, there are still many features that
are used to make your life easier.
Examples
========
test.py
```
## This program runs a test of knowledge
# First get the test questions
# Later this will be modified to use file io.
def get_questions():
# notice how the data is stored as a list of lists
return [["What color is the daytime sky on a clear day?", "blue"],\
["What is the answer to life, the universe and everything?", "42"],\
["What is a three letter word for mouse trap?", "cat"]]
# This will test a single question
# it takes a single question in
# it returns true if the user typed the correct answer, otherwise false
def check_question(question_and_answer):
#extract the question and the answer from the list
question = question_and_answer[0]
answer = question_and_answer[1]
# give the question to the user
given_answer = input(question)
# compare the user's answer to the tester's answer
if answer == given_answer:
print("Correct")
return True
else:
print("Incorrect, correct was:", answer)
return False
# This will run through all the questions
def run_test(questions):
if len(questions) == 0:
print("No questions were given.")
# the return exits the function
return
index = 0
right = 0
while index < len(questions):
#Check the question
if check_question(questions[index]):
right = right + 1
#go to the next question
index = index + 1
#notice the order of the computation, first multiply, then divide
print("You got ", right*100//len(questions), "% right out of", len(questions))
#now lets run the questions
run_test(get_questions())
```
Sample Output:
Exercises
=========
Expand the test.py program so it has menu giving the option of taking
the test, viewing the list of questions and answers, and an option to
Quit. Also, add a new question to ask, “What noise does a truly
advanced machine make?” with the answer of “ping”.
|
github_jupyter
|
# Control of a hydropower dam
Consider a hydropower plant with a dam. We want to control the flow through the dam gates in order to keep the amount of water at a desired level.
<p><img src="hydropowerdam-wikipedia.png" alt="Hydro power from Wikipedia" width="400"></p>
The system is a typical integrator, and is given by the difference equation
$$ y(k+1) = y(k) + b_uu(k) - b_vv(k), $$
where $x$ is the deviation of the water level from a reference level, $u$ is the change in the flow through the dam gates. A positive value of $u$ corresponds to less flow through the gates, relative to an operating point. The flow $v$ corresponds to changes in the flow in (from river) or out (through power plant).
The pulse transfer function of the dam is thus $$H(z) = \frac{b_u}{z-1}.$$
We want to control the system using a two-degree-of-freedom controller, including an anti-aliasing filter modelled as a delay of one sampling period. This gives the block diagram <p><img src="2dof-block-integrator.png" alt="Block diagram" width="700"></p>
The desired closed-loop system from the command signal $u_c$ to the output $y$ should have poles in $z=0.7$, and any observer poles should be chosen faster than the closed-loop poles, say in $z=0.5$.
## The closed-loop pulse-transfer functions
With $F_b(z) = \frac{S(z)}{R(z)}$ and $F_f(z) = \frac{T(z)}{R(z)}$, and using Mason's rule, we get that the closed-loop pulse-transfer function from command signal $u_c$ to output $y$ becomes
$$G_c(z) = \frac{\frac{T(z)}{R(z)}\frac{b_u}{z-1}}{1 + \frac{S(z)}{R(z)} \frac{b_u}{(z-1)z}} = \frac{b_uzT(z)}{z(z-1)R(z) + b_uS(z)}.$$
The closed-loop transfer function from disturbance to output becomes
$$G_{cv}(z) = \frac{\frac{b_v}{z-1}}{1 + \frac{S(z)}{R(z)} \frac{b_u}{(z-1)z}} = \frac{b_vzR(z)}{z(z-1)R(z) + b_uS(z)}.$$
## The Diophantine equation
The diophantine equation becomes
$$z(z-1)R(z) + b_uS(z) = A_c(z)A_o(z)$$ We want to find the smallest order controller that can satisfy the Diophantine equation. Since the feedback controller is given by
$$ F_b(z) = \frac{s_0z^n + s_1z^{n-1} + \cdots + s_n}{z^n + r_1z^{n-1} + \cdots + r_n}$$ and has $2\deg R + 1$ unknown parameters, and since we should choose the order of the Diphantine equation to be the same as the number of unknown parameters, we get
$$ \deg \big((z(z-1)R(z) + b_uS(z)\big) = \deg R + 2 = 2\deg R + 1 \quad \Rightarrow \quad \deg R = n = 1.$$
The Diophantine equation thus becomes
$$ z(z-1)(z+r_1) + b_u(s_0z+s_1) = (z-0.7)^2(z-0.5), $$
where $A_o(z) = z-0.5$ is the observer polynomial. Working out the expressions on both sides gives
$$ z^3-(1-r_1)z^2 -r_1 z + b_us_0z + b_us_1 = (z^2 - 1.4z + 0.49)(z-0.5)$$
$$ z^3 -(1-r_1)z^2 +(b_us_0-r_1)z + b_us_1 = z^3 - (1.4+0.5)z^2 + (0.49+0.7)z -0.245$$
From the Diophantine equation we get the following equations in the unknowns
\begin{align}
z^2: &\quad 1-r_1 = 1.9\\
z^1: &\quad b_us_0 - r_1 = 1.19\\
z^0: &\quad b_us_1 = -0.245
\end{align}
This is a linear system of equations in the unknown, and can be solved in many different ways. Here we see that with simple substitution we find
\begin{align}
r_1 &= 1-1.9 = -0.9\\
s_0 &= \frac{1}{b_u}(1.19+r_1) = \frac{0.29}{b_u}\\
s_1 &= -\frac{0.245}{b_u}
\end{align}
## The feedforward
We set $T(z) = t_0A_o(z)$ which gives the closed-loop pulse-transfer function
$$G_c(z) = \frac{b_uzT(z)}{z(z-1)R(z) + b_uS(z)}= \frac{b_ut_0zA_o(z)}{A_c(z)A_o(z)} = \frac{b_u t_0z}{A_c(z)}$$
In order for this pulse-transfer function to have unit DC-gain (static gain) we must have $G_c(1) = 1$, or
$$ \frac{b_ut_0}{A_c(1)} = 1. $$
The solution is
$$ t_0 = \frac{A_c(1)}{b_u} = \frac{(1-0.7)^2}{b_u} = \frac{0.3^2}{b_u}. $$
## Verify by symbolic computer algebra
```
import numpy as np
import sympy as sy
z = sy.symbols('z', real=False)
bu,r1,s0,s1 = sy.symbols('bu,r1,s0,s1', real=True)
pc,po = sy.symbols('pc,po', real=True) # Closed-loop pole and observer pole
# The polynomials
Ap = sy.Poly(z*(z-1), z)
Bp = sy.Poly(bu,z)
Rp = sy.Poly(z+r1, z)
Sp = sy.Poly(s0*z+s1, z)
Ac = sy.Poly((z-pc)**2, z)
Ao = sy.Poly(z-po, z)
# The diophantine eqn
dioph = Ap*Rp + Bp*Sp - Ac*Ao
# Form system of eqs from coefficients, then solve
dioph_coeffs = dioph.all_coeffs()
# Solve for r1, s0 and s1,
sol = sy.solve(dioph_coeffs, (r1,s0,s1))
print('r_1 = %s' % sol[r1])
print('s_0 = %s' % sol[s0])
print('s_1 = %s' % sol[s1])
# Substitute values for the desired closed-loop pole and observer pole
substitutions = [(pc, 0.7), (po, 0.5)]
print('r_1 = %s' % sol[r1].subs(substitutions))
print('s_0 = %s' % sol[s0].subs(substitutions))
print('s_1 = %s' % sol[s1].subs(substitutions))
# The forward controller
t0 = (Ac.eval(1)/Bp.eval(1))
print('t_0 = %s' % t0)
print('t_0 = %s' % t0.subs(substitutions))
```
## Requirements on the closed-loop poles and observer poles in order to obtain stable controller
Notice the solution for the controller denominator
$$ R(z) = z+r_1 = z -2p_c -p_o + 1, $$
where $0\ge p_c<1$ is the desired closed-loop pole and $0 \ge p_o<1$ is the observer pole. Sketch in the $(p_c, p_o)$-plane the region which will give a stable controller $F_b(z) = \frac{S(z)}{R(z)}$!
## Simulate a particular case
Let $b_u=1$, $p_c = p_o = \frac{2}{3}$. Analyze the closed-loop system by simulation
```
import control
import control.matlab as cm
import matplotlib.pyplot as plt
sbs = [(bu, 1), (pc, 2.0/3.0), (po, 2.0/3.0)]
Rcoeffs = [1, float(sol[r1].subs(sbs))]
Scoeffs = [float(sol[s0].subs(sbs)), float(sol[s1].subs(sbs))]
Tcoeffs = float(t0.subs(sbs))*np.array([1, float(pc.subs(sbs))])
Acoeffs = [1, -1]
H = cm.tf(float(bu.subs(sbs)), Acoeffs, 1)
Ff = cm.tf(Tcoeffs, Rcoeffs, 1)
Fb = cm.tf(Scoeffs, Rcoeffs, 1)
Haa = cm.tf(1, [1, 0], 1) # The delay due to the anti-aliasing filter
Gc = cm.minreal(Ff*cm.feedback(H, Haa*Fb ))
Gcv = cm.feedback(H, Haa*Fb)
# Pulse trf fcn from command signal to control signal
Gcu = Ff*cm.feedback(1, H*Haa*Fb)
cm.pzmap(Fb)
tvec = np.arange(40)
(t1, y1) = control.step_response(Gc,tvec)
plt.figure(figsize=(14,4))
plt.step(t1, y1[0])
plt.xlabel('k')
plt.ylabel('y')
plt.title('Output')
(t1, y1) = control.step_response(Gcv,tvec)
plt.figure(figsize=(14,4))
plt.step(t1, y1[0])
plt.xlabel('k')
plt.ylabel('y')
plt.title('Output')
```
|
github_jupyter
|
```
import warnings
warnings.filterwarnings('ignore')
import nltk
nltk.download('stopwords')
nltk.download('punkt')
nltk.download('wordnet')
from nltk.corpus import stopwords
import pandas as pd
import numpy as np
from glove import Glove
from sklearn.preprocessing import LabelEncoder
from sklearn import metrics
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import OneHotEncoder
from keras.models import Sequential
from keras.layers import Dense
import geopandas as gpd
import os
import json
import h5py
labelEncoder = LabelEncoder()
one_enc = OneHotEncoder()
lemma = nltk.WordNetLemmatizer()
```
## Manual Classification
```
#Dir = '/mnt/d/Dropbox/Ranee_Joshi_PhD_Local/04_PythonCodes/dh2loop_old/shp_NSW'
#DF=litho_Dataframe(Dir)
#DF.to_csv('export.csv')
DF = pd.read_csv('/mnt/d/Dropbox/Ranee_Joshi_PhD_Local/04_PythonCodes/dh2loop/notebooks/Upscaled_Litho_Test2.csv')
DF['FromDepth'] = pd.to_numeric(DF.FromDepth)
DF['ToDepth'] = pd.to_numeric(DF.ToDepth)
DF['TopElev'] = pd.to_numeric(DF.TopElev)
DF['BottomElev'] = pd.to_numeric(DF.BottomElev)
DF['x'] = pd.to_numeric(DF.x)
DF['y'] = pd.to_numeric(DF.y)
print('number of original litho classes:', len(DF.MajorLithCode.unique()))
print('number of litho classes :',
len(DF['reclass'].unique()))
print('unclassified descriptions:',
len(DF[DF['reclass'].isnull()]))
def save_file(DF, name):
'''Function to save manually reclassified dataframe
Inputs:
-DF: reclassified pandas dataframe
-name: name (string) to save dataframe file
'''
DF.to_pickle('{}.pkl'.format(name))
save_file(DF, 'manualTest_ygsb')
```
## MLP Classification
```
def load_geovec(path):
instance = Glove()
with h5py.File(path, 'r') as f:
v = np.zeros(f['vectors'].shape, f['vectors'].dtype)
f['vectors'].read_direct(v)
dct = f['dct'][()].tostring().decode('utf-8')
dct = json.loads(dct)
instance.word_vectors = v
instance.no_components = v.shape[1]
instance.word_biases = np.zeros(v.shape[0])
instance.add_dictionary(dct)
return instance
# Stopwords
extra_stopwords = [
'also',
]
stop = stopwords.words('english') + extra_stopwords
def tokenize(text, min_len=1):
'''Function that tokenize a set of strings
Input:
-text: set of strings
-min_len: tokens length
Output:
-list containing set of tokens'''
tokens = [word.lower() for sent in nltk.sent_tokenize(text)
for word in nltk.word_tokenize(sent)]
filtered_tokens = []
for token in tokens:
if token.isalpha() and len(token) >= min_len:
filtered_tokens.append(token)
return [x.lower() for x in filtered_tokens if x not in stop]
def tokenize_and_lemma(text, min_len=0):
'''Function that retrieves lemmatised tokens
Inputs:
-text: set of strings
-min_len: length of text
Outputs:
-list containing lemmatised tokens'''
filtered_tokens = tokenize(text, min_len=min_len)
lemmas = [lemma.lemmatize(t) for t in filtered_tokens]
return lemmas
def get_vector(word, model, return_zero=False):
'''Function that retrieves word embeddings (vector)
Inputs:
-word: token (string)
-model: trained MLP model
-return_zero: boolean variable
Outputs:
-wv: numpy array (vector)'''
epsilon = 1.e-10
unk_idx = model.dictionary['unk']
idx = model.dictionary.get(word, unk_idx)
wv = model.word_vectors[idx].copy()
if return_zero and word not in model.dictionary:
n_comp = model.word_vectors.shape[1]
wv = np.zeros(n_comp) + epsilon
return wv
def mean_embeddings(dataframe_file, model):
'''Function to retrieve sentence embeddings from dataframe with
lithological descriptions.
Inputs:
-dataframe_file: pandas dataframe containing lithological descriptions
and reclassified lithologies
-model: word embeddings model generated using GloVe
Outputs:
-DF: pandas dataframe including sentence embeddings'''
DF = pd.read_pickle(dataframe_file)
DF = DF.drop_duplicates(subset=['x', 'y', 'z'])
DF['tokens'] = DF['Description'].apply(lambda x: tokenize_and_lemma(x))
DF['length'] = DF['tokens'].apply(lambda x: len(x))
DF = DF.loc[DF['length']> 0]
DF['vectors'] = DF['tokens'].apply(lambda x: np.asarray([get_vector(n, model) for n in x]))
DF['mean'] = DF['vectors'].apply(lambda x: np.mean(x[~np.all(x == 1.e-10, axis=1)], axis=0))
DF['reclass'] = pd.Categorical(DF.reclass)
DF['code'] = DF.reclass.cat.codes
DF['drop'] = DF['mean'].apply(lambda x: (~np.isnan(x).any()))
DF = DF[DF['drop']]
return DF
# loading word embeddings model
# (This can be obtained from https://github.com/spadarian/GeoVec )
#modelEmb = Glove.load('/home/ignacio/Documents/chapter2/best_glove_300_317413_w10_lemma.pkl')
modelEmb = load_geovec('geovec_300d_v1.h5')
# getting the mean embeddings of descriptions
DF = mean_embeddings('manualTest_ygsb.pkl', modelEmb)
DF2 = DF[DF['code'].isin(DF['code'].value_counts()[DF['code'].value_counts()>2].index)]
print(DF2)
def split_stratified_dataset(Dataframe, test_size, validation_size):
'''Function that split dataset into test, training and validation subsets
Inputs:
-Dataframe: pandas dataframe with sentence mean_embeddings
-test_size: decimal number to generate the test subset
-validation_size: decimal number to generate the validation subset
Outputs:
-X: numpy array with embeddings
-Y: numpy array with lithological classes
-X_test: numpy array with embeddings for test subset
-Y_test: numpy array with lithological classes for test subset
-Xt: numpy array with embeddings for training subset
-yt: numpy array with lithological classes for training subset
-Xv: numpy array with embeddings for validation subset
-yv: numpy array with lithological classes for validation subset
'''
#df2 = Dataframe[Dataframe['code'].isin(Dataframe['code'].value_counts()[Dataframe['code'].value_counts()>2].index)]
#X = np.vstack(df2['mean'].values)
#Y = df2.code.values.reshape(len(df2.code), 1)
X = np.vstack(Dataframe['mean'].values)
Y = Dataframe.code.values.reshape(len(Dataframe.code), 1)
#print(X.shape)
#print (Dataframe.code.values.shape)
#print (len(Dataframe.code))
#print (Y.shape)
X_train, X_test, y_train, y_test = train_test_split(X,
Y, stratify=Y,
test_size=test_size,
random_state=42)
#print(X_train.shape)
#print(Y_train.shape)
Xt, Xv, yt, yv = train_test_split(X_train,
y_train,
test_size=validation_size,
stratify=None,
random_state=1)
return X, Y, X_test, y_test, Xt, yt, Xv, yv
# subseting dataset for training classifier
X, Y, X_test, Y_test, X_train, Y_train, X_validation, Y_validation = split_stratified_dataset(DF2, 0.1, 0.1)
# encoding lithological classes
encodes = one_enc.fit_transform(Y_train).toarray()
# MLP model generation
model = Sequential()
model.add(Dense(100, input_dim=300, activation='relu'))
model.add(Dense(100, activation='relu'))
model.add(Dense(100, activation='relu'))
model.add(Dense(units=len(DF2.code.unique()), activation='softmax'))
model.compile(loss='categorical_crossentropy',
optimizer='adam',
metrics=['accuracy'])
# training MLP model
model.fit(X_train, encodes, epochs=30, batch_size=100, verbose=2)
# saving MLP model
model.save('mlp_prob_model.h5')
def retrieve_predictions(classifier, x):
'''Function that retrieves lithological classes using the trained classifier
Inputs:
-classifier: trained MLP classifier
-x: numpy array containing embbedings
Outputs:
-codes_pred: numpy array containing lithological classes predicted'''
preds = classifier.predict(x, verbose=0)
new_onehot = np.zeros((x.shape[0], 72))
new_onehot[np.arange(len(preds)), preds.argmax(axis=1)] = 1
codes_pred = one_enc.inverse_transform(new_onehot)
return codes_pred
def classifier_assess(classifier, x, y):
'''Function that prints the performance of the classifier
Inputs:
-classifier: trained MLP classifier
-x: numpy array with embeddings
-y: numpy array with lithological classes predicted'''
Y2 = retrieve_predictions(classifier, x)
print('f1 score: ', metrics.f1_score(y, Y2, average='macro'),
'accuracy: ', metrics.accuracy_score(y, Y2),
'balanced_accuracy:', metrics.balanced_accuracy_score(y, Y2))
def save_predictions(Dataframe, classifier, x, name):
'''Function that saves dataframe predictions as a pickle file
Inputs:
-Dataframe: pandas dataframe with mean_embeddings
-classifier: trained MLP model,
-x: numpy array with embeddings,
-name: string name to save dataframe
Outputs:
-save dataframe'''
preds = classifier.predict(x, verbose=0)
Dataframe['predicted_probabilities'] = preds.tolist()
Dataframe['pred'] = retrieve_predictions(classifier, x).astype(np.int32)
Dataframe[['x', 'y', 'FromDepth', 'ToDepth', 'TopElev', 'BottomElev',
'mean', 'predicted_probabilities', 'pred', 'reclass', 'code']].to_pickle('{}.pkl'.format(name))
# assessment of model performance
classifier_assess(model, X_validation, Y_validation)
# save lithological prediction likelihoods dataframe
save_predictions(DF2, model, X, 'YGSBpredictions')
import pickle
with open('YGSBpredictions.pkl', 'rb') as f:
data = pickle.load(f)
print(data)
len(data)
data.head()
tmp = data['predicted_probabilities'][0]
len(tmp)
data.to_csv('YGSBpredictions.csv')
import base64
with open('a.csv', 'a', encoding='utf8') as csv_file:
wr = csv.writer(csv_file, delimiter='|')
pickle_bytes = pickle.dumps(obj) # unsafe to write
b64_bytes = base64.b64encode(pickle_bytes) # safe to write but still bytes
b64_str = b64_bytes.decode('utf8') # safe and in utf8
wr.writerow(['col1', 'col2', b64_str])
# the file contains
# col1|col2|gANdcQAu
with open('a.csv', 'r') as csv_file:
for line in csv_file:
line = line.strip('\n')
b64_str = line.split('|')[2] # take the pickled obj
obj = pickle.loads(base64.b64decode(b64_str)) #
```
|
github_jupyter
|
```
# https://community.plotly.com/t/different-colors-for-bars-in-barchart-by-their-value/6527/7
%reset
# Run this app with `python app.py` ando
# visit http://127.0.0.1:8050/ in your web browser.
import dash
import dash_core_components as dcc
import dash_html_components as html
import plotly.express as px
import jupyter_dash
import pandas as pd
from dash.dependencies import Input, Output
import plotly.graph_objects as go
from plotly.subplots import make_subplots
external_stylesheets = ['https://codepen.io/chriddyp/pen/bWLwgP.css']
top_artists_df = pd.read_csv('~/qs/lastfm/data/lastfm_top_artists.csv')
top_tracks_df = pd.read_csv('~/qs/lastfm/data/lastfm_top_tracks.csv')
top_albums_df = pd.read_csv('~/qs/lastfm/data/lastfm_top_albums.csv')
new_top_artists_df = pd.read_csv('~/qs/lastfm/data/lastfm_top_artists_with_tags.csv', usecols=[1, 2, 3])
new_top_artists_df
top_artists_df = new_top_artists_df
top_artists_df
print('Top Artists')
print(f"{top_artists_df.head(5)} \n")
# print('Top Tracks')
# print(f"{top_tracks_df.head(5)} \n")
# print('Top Albums')
# print(top_albums_df.head(5))
top_artists_df.tail(5)
df = top_artists_df
# total_songs = top_artists_df['play_count'].sum()
# total_songs
# total_artists = len(top_artists_df['artist'])
# total_artists
# def num_unique_tags(df):
# unique_tags = []
# for tags, num_artists_in_tags in df.groupby('tags'):
# unique_tags.append(len(num_artists_in_tags))
# unique_tags.sort(reverse=True)
# return len(unique_tags)
# num_unique_tags(top_artists_df)
metal = 'metal|core|sludge'
rock = 'rock'
len(df.loc[df['tags'].str.contains(metal)])
# if you want custom colors for certain words in the tags
#https://stackoverflow.com/questions/23400743/pandas-modify-column-values-in-place-based-on-boolean-array
#px.colors.qualitative.Plotly
custom_colors = ['#EF553B'] * 200
df['colors'] = custom_colors
metal = 'metal|core|sludge'
df.loc[df['tags'].str.contains(metal), 'colors'] = '#636EFA'
rock = 'rock|blues'
df.loc[df['tags'].str.contains(rock), 'colors'] = '#00CC96'
punk = 'punk'
df.loc[df['tags'].str.contains(punk), 'colors'] = '#AB63FA'
alternative = 'alternative'
df.loc[df['tags'].str.contains(alternative), 'colors'] = '#FFA15A'
indie = 'indie'
df.loc[df['tags'].str.contains(indie), 'colors'] = '#19D3F3'
billy = 'billy'
df.loc[df['tags'].str.contains(billy), 'colors'] = '#FF6692'
rap = 'rap|hip|rnb'
df.loc[df['tags'].str.contains(rap), 'colors'] = '#B6E880'
pop = 'pop|soul'
df.loc[df['tags'].str.contains(pop), 'colors'] = '#FF97FF'
electronic = 'electronic|synthwave'
df.loc[df['tags'].str.contains(electronic), 'colors'] = '#FECB52'
df
title = f'Last.fm Dashboard'
#https://stackoverflow.com/questions/22291395/sorting-the-grouped-data-as-per-group-size-in-pandas
df_grouped = sorted(df.groupby('tags'), key=lambda x: len(x[1]), reverse=True)
artist = df['artist']
play_count = df['play_count']
tags = df['tags']
total_songs = df['play_count'].sum()
total_artists = len(df['artist'])
num_unique_tags = len(df.groupby('tags'))
fig = go.Figure()
def make_traces(df):
for tags, df in df_grouped:
num_tags = len(df)
fig.add_trace(go.Bar(y=df['artist'],
x=df['play_count'],
orientation='h',
text=df['play_count'],
textposition='outside',
name=f"{tags} ({num_tags})",
customdata = df['tags'],
hovertemplate =
"Artist: %{y}<br>" +
"Play Count: %{x}<br>" +
"Tag: %{customdata}" +
"<extra></extra>",
# for custom colors
marker_color=df['colors'],
#https://community.plotly.com/t/different-colors-for-bars-in-barchart-by-their-value/6527/4
#marker={'color': colors[tags]},
showlegend=True,
))
make_traces(top_artists_df)
fig.update_layout(title=dict(text=title,
yanchor="top",
y=.95,
xanchor="left",
x=.075),
legend_title=f'Unique Tags: ({num_unique_tags})',
legend_itemclick='toggle',
legend_itemdoubleclick='toggleothers',
margin_l = 240,
xaxis=dict(fixedrange=True),
# https://towardsdatascience.com/4-ways-to-improve-your-plotly-graphs-517c75947f7e
yaxis=dict(categoryorder='total descending'),
dragmode='pan',
annotations=[
#https://plotly.com/python/text-and-annotations/#adding-annotations-with-xref-and-yref-as-paper
#https://community.plotly.com/t/how-to-add-a-text-area-with-custom-text-as-a-kind-of-legend-in-a-plotly-plot/24349/3
go.layout.Annotation(
text=f'Total Songs Tracked: {total_songs}<br>Total Artists Tracked: {total_artists}',
align='left',
showarrow=False,
xref='paper',
yref='paper',
yanchor="bottom",
y=1.02,
xanchor="right",
x=1,
)])
fig.update_yaxes(title_text="Artist",
type='category',
range=[25.5, -.5],
# https://plotly.com/python/setting-graph-size/#adjusting-height-width--margins
automargin=False
)
fig.update_xaxes(title_text="Play Count ",
range=[0, 700],
dtick=100,
)
app = jupyter_dash.JupyterDash(__name__,
external_stylesheets=external_stylesheets,
title=f"{title}")
def make_footer():
return html.Div(html.Footer([
'Matthew Waage',
html.A('github.com/mcwaage1',
href='http://www.github.com/mcwaage1',
target='_blank',
style = {'margin': '.5em'}),
html.A('[email protected]',
href="mailto:[email protected]",
target='_blank',
style = {'margin': '.5em'}),
html.A('waage.dev',
href='http://www.waage.dev',
target='_blank',
style = {'margin': '.5em'})
], style={'position': 'fixed',
'text-align': 'right',
'left': '0px',
'bottom': '0px',
'margin-right': '10%',
'color': 'black',
'display': 'inline-block',
'background': '#f2f2f2',
'border-top': 'solid 2px #e4e4e4',
'width': '100%'}))
app.layout = html.Div([
dcc.Graph(
figure=fig,
#https://plotly.com/python/setting-graph-size/
#https://stackoverflow.com/questions/46287189/how-can-i-change-the-size-of-my-dash-graph
style={'height': '95vh'}
),
make_footer(),
])
if __name__ == '__main__':
app.run_server(mode ='external', port=8070, debug=True)
# title = '2020 Last.fm Dashboard'
# fig = go.Figure()
# fig.add_trace(go.Bar(
# x=top_artists_df['artist'],
# y=top_artists_df['play_count'],
# text=top_artists_df['play_count'],
# ))
# fig.update_traces(textposition='outside')
# fig.update_layout(title=dict(text=title,
# yanchor="top",
# y=.95,
# xanchor="left",
# x=.075),
# dragmode='pan',
# annotations=[
# #https://plotly.com/python/text-and-annotations/#adding-annotations-with-xref-and-yref-as-paper
# #https://community.plotly.com/t/how-to-add-a-text-area-with-custom-text-as-a-kind-of-legend-in-a-plotly-plot/24349/3
# go.layout.Annotation(
# text=f'Total Songs Played: {total_songs}',
# align='left',
# showarrow=False,
# xref='paper',
# yref='paper',
# yanchor="bottom",
# y=1.02,
# xanchor="right",
# x=1,
# )])
# # https://stackoverflow.com/questions/61782622/plotly-how-to-add-a-horizontal-scrollbar-to-a-plotly-express-figure
# # https://community.plotly.com/t/freeze-y-axis-while-scrolling-along-x-axis/4898/5
# fig.update_layout(
# xaxis=dict(
# rangeslider=dict(
# visible=True,
# )))
# fig.update_xaxes(title_text="Artist",
# type='category',
# range=[-.5, 25.5],
# )
# fig.update_yaxes(title_text="Play Count ",
# range=[0, 750],
# dtick=100,
# )
# app = jupyter_dash.JupyterDash(__name__,
# external_stylesheets=external_stylesheets,
# title=f"{title}")
# app.layout = html.Div([
# dcc.Graph(
# figure=fig,
# #https://plotly.com/python/setting-graph-size/
# #https://stackoverflow.com/questions/46287189/how-can-i-change-the-size-of-my-dash-graph
# style={'height': '95vh'}
# )
# ])
# if __name__ == '__main__':
# app.run_server(mode ='external', debug=True, port=8080)
import requests
import json
headers = {
'user-agent': 'mcwaage1'
}
with open("data/credentials.json", "r") as file:
credentials = json.load(file)
last_fm_cr = credentials['last_fm']
key = last_fm_cr['KEY']
username = last_fm_cr['USERNAME']
limit = 20 #api lets you retrieve up to 200 records per call
extended = 0 #api lets you retrieve extended data for each track, 0=no, 1=yes
page = 1 #page of results to start retrieving at
```
### Testing out api calls of artists
```
artist = "death from above 1979"
artist = artist.replace(' ', '+')
artist
method = 'artist.gettoptags'
request_url = f'http://ws.audioscrobbler.com/2.0/?method={method}&artist={artist}&api_key={key}&limit={limit}&extended={extended}&page={page}&format=json'
response = requests.get(request_url, headers=headers)
response.status_code
artist_tags = [tag['name'] for tag in response.json()['toptags']['tag'][:3]]
artist_tags
method = 'tag.gettoptags'
request_url = f'http://ws.audioscrobbler.com/2.0/?method={method}&api_key={key}&limit={limit}&extended={extended}&page={page}&format=json'
response = requests.get(request_url, headers=headers)
response.status_code
top_tags = artist_tags
top_tags
method = 'user.getinfo'
request_url = f'http://ws.audioscrobbler.com/2.0/?method={method}&user={username}&api_key={key}&limit={limit}&extended={extended}&page={page}&format=json'
response = requests.get(request_url, headers=headers)
response.status_code
response.json()
user_info = [response.json()['user']['name']]
user_info
user_info.append(response.json()['user']['url'])
user_info.append(response.json()['user']['image'][0]['#text'])
user_info
# def make_user_info(user_info):
# return html.Div(children=[html.Img(src=f'{user_info[2]}'),
# html.A(f'{user_info[0]}',
# href=f'{user_info[1]}',
# target='_blank',
# style={'margin': '.5em'}
# ),
# ])
```
### End of testing
```
artists = []
artists
def get_artists():
artists = []
for artist in top_artists_df['artist']:
artist = artist.replace(' ', '+')
artists.append(artist)
return artists
artists_to_parse = get_artists()
artists_to_parse
```
# To start the api calling process and get new data
```
# replace the [:1] with [:3] or whatever for more tags of artist
artist_genre = []
for artist in artists_to_parse:
request_url = f'http://ws.audioscrobbler.com/2.0/?method=artist.gettoptags&artist={artist}&api_key={key}&limit={limit}&extended={extended}&page={page}&format=json'
response = requests.get(request_url, headers=headers)
artist_tags = [tag['name'] for tag in response.json()['toptags']['tag'][:1]]
artist_genre.append(artist_tags)
artist_genre
# https://stackoverflow.com/questions/12555323/adding-new-column-to-existing-dataframe-in-python-pandas
top_artists_df['tags'] = artist_genre
top_artists_df.head(5)
top_artists_df['tags'] = top_artists_df['tags'].astype(str)
top_artists_df['tags'] = top_artists_df['tags'].str.strip("['']")
top_artists_df['tags'] = top_artists_df['tags'].str.lower()
top_artists_df.head(5)
```
### To replace tags that you don't want
```
tags_to_replace = 'seen live|vocalists'
def get_new_artists(tags_to_replace):
artists_to_replace = []
for artist in df.loc[df['tags'].str.contains(tags_to_replace)]['artist']:
artists_to_replace.append(artist)
return artists_to_replace
get_new_artists(tags_to_replace)
tags_to_replace = 'seen live|vocalists'
def get_artists_to_replace(tags_to_replace):
artists_to_replace = []
for artist in df.loc[df['tags'].str.contains(tags_to_replace)]['artist']:
artist = artist.replace(' ', '+')
artists_to_replace.append(artist)
return artists_to_replace
get_artists_to_replace(tags_to_replace)
new_artists_to_parse = get_artists_to_replace(tags_to_replace)
new_artists_tags = []
for artist in new_artists_to_parse:
request_url = f'http://ws.audioscrobbler.com/2.0/?method=artist.gettoptags&artist={artist}&api_key={key}&limit={limit}&extended={extended}&page={page}&format=json'
response = requests.get(request_url, headers=headers)
artist_tags = [tag['name'] for tag in response.json()['toptags']['tag'][1:2]]
new_artists_tags.append(artist_tags)
new_artists_tags
new_artists_tags = [str(x) for x in new_artists_tags]
new_artists_tags = [x.strip("['']") for x in new_artists_tags]
new_artists_tags = [x.lower() for x in new_artists_tags]
new_artists_tags
for artist in get_new_artists(tags_to_replace):
print(artist)
for k, v in zip(get_new_artists(tags_to_replace), new_artists_tags):
df.loc[df['artist'].str.contains(k), 'tags'] = v
```
### End of replacing tags
```
top_artists_df.to_csv('~/qs/lastfm/data/lastfm_top_artists_with_tags.csv')
from IPython.display import display
with pd.option_context('display.max_rows', 205, 'display.max_columns', 5):
display(top_artists_df)
# unique_tags = top_artists_df['tags'].unique()
# unique_tags = pd.Series(unique_tags)
# print("Type: ", type(unique_tags))
# print('')
# for tag in unique_tags:
# print(tag)
# len(unique_tags)
# def get_sorted_tags(df):
# unique_tags = df['tags'].unique()
# unique_tags = pd.Series(unique_tags)
# sorted_tags = []
# for tag in unique_tags:
# #sorted_tags.append((top_artists_df['tags'].str.count(tag).sum(), tag))
# #sorted_tags.append(top_artists_df['tags'].str.count(tag).sum())
# sorted_tags.sort(reverse=True)
# return sorted_tags
# get_sorted_tags(top_artists_df)
# unique_tags = unique_tags.str.split()
# type(unique_tags)
# unique_tags
# for tag in unique_tags:
# print(tag, unique_tags.str.count(tag).sum())
# px.colors.qualitative.Plotly
# https://plotly.com/python/discrete-color/
#fig = px.colors.qualitative.swatches()
#https://plotly.com/python/renderers/
#fig.show(renderer='iframe')
# One way to replace value in one series based on another, better version below
# top_artists_df['colors'][top_artists_df['tags'].str.contains('metal')] = '#636EFA'
# top_tags
#https://stackoverflow.com/questions/23400743/pandas-modify-column-values-in-place-based-on-boolean-array
px.colors.qualitative.Plotly
# custom_colors = ['#EF553B'] * 200
# df['colors'] = custom_colors
# df.loc[df['tags'].str.contains('metal'), 'colors'] = '#636EFA'
# df.loc[df['tags'].str.contains('rock'), 'colors'] = '#00CC96'
# df.loc[df['tags'].str.contains('punk'), 'colors'] = '#AB63FA'
# df.loc[df['tags'].str.contains('alternative'), 'colors'] = '#FFA15A'
# df.loc[df['tags'].str.contains('indie'), 'colors'] = '#19D3F3'
# df.loc[df['tags'].str.contains('billy'), 'colors'] = '#FF6692'
# df
# colors = {'Ugly': 'red',
# 'Bad': 'orange',
# 'Good': 'lightgreen',
# 'Great': 'darkgreen'
# }
# from IPython.display import display
# with pd.option_context('display.max_rows', 265, 'display.max_columns', 5):
# display(top_artists_df)
# for tag, artist in top_artists_df.groupby('tags'):
# print(tag, len(artist))
# print(artist)
# print('')
# top_artists_df.loc[top_artists_df['tags'].str.contains('metal')]
# type(top_artists_df.loc[top_artists_df['tags'].str.contains('metal')])
# len(top_artists_df.loc[top_artists_df['tags'].str.contains('metal')])
# def print_tags(df):
# printed_tags = []
# for tags, top_artists in top_artists_df.groupby('tags'):
# printed_tags.append([len(top_artists), tags])
# printed_tags.sort(reverse=True)
# return printed_tags
# print_tags(top_artists_df)
```
|
github_jupyter
|
# Hawaii - A Climate Analysis And Exploration
### For data between August 23, 2016 - August 23, 2017
---
```
# Import dependencies
%matplotlib inline
from matplotlib import style
style.use('fivethirtyeight')
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import datetime as dt
# Python SQL toolkit and Object Relational Mapper
import sqlalchemy
from sqlalchemy.ext.automap import automap_base
from sqlalchemy.orm import Session
from sqlalchemy import create_engine, func, inspect
```
## Reflect Tables into SQLAlchemy ORM
```
# Set up query engine. 'echo=True is the default - will keep a log of activities'
engine = create_engine("sqlite:///Resources/hawaii.sqlite")
# Reflect an existing database into a new model
Base = automap_base()
# Reflect the tables
Base.prepare(engine, reflect=True)
# We can view all of the classes that automap found
Base.classes.keys()
# Another way to get table names from SQL-lite
inspector = inspect(engine)
inspector.get_table_names()
```
## Exploratory Climate Analysis
```
# Save references to each table
Measurement = Base.classes.measurement
Station = Base.classes.station
# Create our session (link) from Python to the DB
session = Session(engine)
# Display details of 'measurement' table
columns = inspector.get_columns('measurement')
for c in columns:
print(c['name'], c['type'])
# DISPLY number of line items measurement, and remove tuple form
result, = engine.execute('SELECT COUNT(*) FROM measurement').fetchall()[0]
print(result,)
# Display details of 'station' table
columns = inspector.get_columns('station')
for c in columns:
print(c['name'], c['type'])
# DISPLY number of line items station, and remove tuple form
result, = engine.execute('SELECT COUNT(*) FROM station').fetchall()[0]
print(result,)
# FULL INNTER JOIN BOTH THE MEASUREMENT AND STATION TABLE
# engine.execute('SELECT measurement.*, station.name, station.latitude FROM measurement INNER JOIN station ON measurement.station = station.station;').fetchall()
join_result = engine.execute('SELECT * FROM measurement INNER JOIN station ON measurement.station = station.station;').fetchall()
join_result
# Another way to PERFORM AN INNER JOIN ON THE MEASUREMENT AND STATION TABLES
engine.execute('SELECT measurement.*, station.* FROM measurement, station WHERE measurement.station=station.station;').fetchall()
# Query last date of the measurement file
last_date = session.query(Measurement.date).order_by(Measurement.date.desc()).first()[0]
print(last_date)
last_date_measurement = dt.date(2017, 8 ,23)
# Calculate the date 1 year delta of the "last date measurement"
one_year_ago = last_date_measurement - dt.timedelta(days=365)
print(one_year_ago)
# Plotting precipitation data from 1 year ago
date = dt.date(2016, 8, 23)
#sel = [Measurement.id, Measurement.station, Measurement.date, Measurement.prcp, Measurement.tobs]
sel = [Measurement.date, Measurement.prcp]
print(date)
# date = "2016-08-23"
result = session.query(Measurement.date, Measurement.prcp).\
filter(Measurement.date >= date).all()
# get the count / length of the list of tuples
print(len(result))
# Created a line plot and saved the figure
df = pd.DataFrame(result, columns=['Date', 'Precipitation'])
df.sort_values(by=['Date'])
df.set_index('Date', inplace=True)
s = df['Precipitation']
ax = s.plot(figsize=(8,6), use_index=True, title='Precipitation Data Between 8/23/2016 - 8/23/2017')
fig = ax.get_figure()
fig.savefig('./Images/precipitation_line.png')
# Use Pandas to calcualte the summary statistics for the precipitation data
df.describe()
# Design a query to show how many stations are available in this dataset?
session.query(Measurement.station).\
group_by(Measurement.station).count()
# Querying for the most active stations (i.e. what stations have the most rows)?
# List the stations and the counts in descending order.
engine.execute('SELECT DISTINCT station, COUNT(id) FROM measurement GROUP BY station ORDER BY COUNT(id) DESC').fetchall()
# Query for stations from the measurement table
session.query(Measurement.station).\
group_by(Measurement.station).all()
# Using the station id from the previous query, calculate the lowest temperature recorded,
# highest temperature recorded, and average temperature most active station?
sel = [func.min(Measurement.tobs), func.max(Measurement.tobs), func.avg(Measurement.tobs)]
session.query(*sel).\
filter(Measurement.station == 'USC00519281').all()
# Query the dates of the last 12 months of the most active station
last_date = session.query(Measurement.date).\
filter(Measurement.station == 'USC00519281').\
order_by(Measurement.date.desc()).first()[0]
print(last_date)
last_date_USC00519281 = dt.date(2017, 8 ,18)
last_year_USC00519281 = last_date_USC00519281 - dt.timedelta(days=365)
print(last_year_USC00519281)
# SET UP HISTOGRAM QUERY AND PLOT
sel_two = [Measurement.tobs]
results_tobs_hist = session.query(*sel_two).\
filter(Measurement.date >= last_year_USC00519281).\
filter(Measurement.station == 'USC00519281').all()
# HISTOGRAM Plot
df = pd.DataFrame(results_tobs_hist, columns=['tobs'])
ax = df.plot.hist(figsize=(8,6), bins=12, use_index=False, title='Hawaii - Temperature Histogram Between 8/23/2016 - 8/23/2017')
fig = ax.get_figure()
fig.savefig('./Images/temperature_histogram.png')
# Created a function called `calc_temps` that accepts a 'start date' and 'end date' in the format 'YYYY-MM-DD'
# and return the minimum, average, and maximum temperatures for that range of dates
def calc_temps(start_date, end_date):
"""Temp MIN,Temp AVG, and Temp MAX for a list of dates.
Args are:
start_date (string): A date string in the format YYYY-MM-DD
end_date (string): A date string in the format YYYY-MM-DD
Returns:
T-MIN, T-AVG, and T-MAX
"""
return session.query(func.min(Measurement.tobs), func.avg(Measurement.tobs), func.max(Measurement.tobs)).\
filter(Measurement.date >= start_date).filter(Measurement.date <= end_date).all()
print(calc_temps('2017-08-01', '2017-08-07'))
```
|
github_jupyter
|
```
import os
import glob
base_dir = os.path.join('F:/0Sem 7/B.TECH PROJECT/0Image data/cell_images')
infected_dir = os.path.join(base_dir,'Parasitized')
healthy_dir = os.path.join(base_dir,'Uninfected')
infected_files = glob.glob(infected_dir+'/*.png')
healthy_files = glob.glob(healthy_dir+'/*.png')
print("Infected samples:",len(infected_files))
print("Uninfected samples:",len(healthy_files))
import numpy as np
import pandas as pd
np.random.seed(42)
files_df = pd.DataFrame({
'filename': infected_files + healthy_files,
'label': ['malaria'] * len(infected_files) + ['healthy'] * len(healthy_files)
}).sample(frac=1, random_state=42).reset_index(drop=True)
files_df.head()
from sklearn.model_selection import train_test_split
from collections import Counter
train_files, test_files, train_labels, test_labels = train_test_split(files_df['filename'].values,
files_df['label'].values,
test_size=0.3, random_state=42)
train_files, val_files, train_labels, val_labels = train_test_split(train_files,
train_labels,
test_size=0.1, random_state=42)
print(train_files.shape, val_files.shape, test_files.shape)
print('Train:', Counter(train_labels), '\nVal:', Counter(val_labels), '\nTest:', Counter(test_labels))
import cv2
from concurrent import futures
import threading
def get_img_shape_parallel(idx, img, total_imgs):
if idx % 5000 == 0 or idx == (total_imgs - 1):
print('{}: working on img num: {}'.format(threading.current_thread().name,
idx))
return cv2.imread(img).shape
ex = futures.ThreadPoolExecutor(max_workers=None)
data_inp = [(idx, img, len(train_files)) for idx, img in enumerate(train_files)]
print('Starting Img shape computation:')
train_img_dims_map = ex.map(get_img_shape_parallel,
[record[0] for record in data_inp],
[record[1] for record in data_inp],
[record[2] for record in data_inp])
train_img_dims = list(train_img_dims_map)
print('Min Dimensions:', np.min(train_img_dims, axis=0))
print('Avg Dimensions:', np.mean(train_img_dims, axis=0))
print('Median Dimensions:', np.median(train_img_dims, axis=0))
print('Max Dimensions:', np.max(train_img_dims, axis=0))
IMG_DIMS = (32, 32)
def get_img_data_parallel(idx, img, total_imgs):
if idx % 5000 == 0 or idx == (total_imgs - 1):
print('{}: working on img num: {}'.format(threading.current_thread().name,
idx))
img = cv2.imread(img)
img = cv2.resize(img, dsize=IMG_DIMS,
interpolation=cv2.INTER_CUBIC)
img = np.array(img, dtype=np.float32)
return img
ex = futures.ThreadPoolExecutor(max_workers=None)
train_data_inp = [(idx, img, len(train_files)) for idx, img in enumerate(train_files)]
val_data_inp = [(idx, img, len(val_files)) for idx, img in enumerate(val_files)]
test_data_inp = [(idx, img, len(test_files)) for idx, img in enumerate(test_files)]
print('Loading Train Images:')
train_data_map = ex.map(get_img_data_parallel,
[record[0] for record in train_data_inp],
[record[1] for record in train_data_inp],
[record[2] for record in train_data_inp])
train_data = np.array(list(train_data_map))
print('\nLoading Validation Images:')
val_data_map = ex.map(get_img_data_parallel,
[record[0] for record in val_data_inp],
[record[1] for record in val_data_inp],
[record[2] for record in val_data_inp])
val_data = np.array(list(val_data_map))
print('\nLoading Test Images:')
test_data_map = ex.map(get_img_data_parallel,
[record[0] for record in test_data_inp],
[record[1] for record in test_data_inp],
[record[2] for record in test_data_inp])
test_data = np.array(list(test_data_map))
train_data.shape, val_data.shape, test_data.shape
import matplotlib.pyplot as plt
%matplotlib inline
plt.figure(1 , figsize = (8 , 8))
n = 0
for i in range(16):
n += 1
r = np.random.randint(0 , train_data.shape[0] , 1)
plt.subplot(4 , 4 , n)
plt.subplots_adjust(hspace = 0.5 , wspace = 0.5)
plt.imshow(train_data[r[0]]/255.)
plt.title('{}'.format(train_labels[r[0]]))
plt.xticks([]) , plt.yticks([])
BATCH_SIZE = 32
NUM_CLASSES = 2
EPOCHS = 25
INPUT_SHAPE = (32, 32, 3)
train_imgs_scaled = train_data / 255.
val_imgs_scaled = val_data / 255.
# encode text category labels
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
le.fit(train_labels)
train_labels_enc = le.transform(train_labels)
val_labels_enc = le.transform(val_labels)
print(train_labels[:6], train_labels_enc[:6])
import tensorflow as tf
vgg = tf.keras.applications.mobilenet.MobileNet(include_top=False, alpha=1.0, weights='imagenet',
input_shape=INPUT_SHAPE)
# Freeze the layers
vgg.trainable = True
set_trainable = False
for layer in vgg.layers:
layer.trainable = False
base_vgg = vgg
base_out = base_vgg.output
pool_out = tf.keras.layers.Flatten()(base_out)
hidden1 = tf.keras.layers.Dense(512, activation='relu')(pool_out)
drop1 = tf.keras.layers.Dropout(rate=0.3)(hidden1)
hidden2 = tf.keras.layers.Dense(512, activation='relu')(drop1)
drop2 = tf.keras.layers.Dropout(rate=0.3)(hidden2)
out = tf.keras.layers.Dense(1, activation='sigmoid')(drop2)
model = tf.keras.Model(inputs=base_vgg.input, outputs=out)
from tensorflow.keras.optimizers import Adam
adam = Adam(lr=0.0001)
model.compile(optimizer=adam,
loss='binary_crossentropy',
metrics=['accuracy'])
print("Total Layers:", len(model.layers))
print("Total trainable layers:", sum([1 for l in model.layers if l.trainable]))
print(model.summary())
history = model.fit(x=train_imgs_scaled, y=train_labels_enc,
batch_size=BATCH_SIZE,
epochs=EPOCHS,
validation_data=(val_imgs_scaled, val_labels_enc),
verbose=1)
f, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 4))
t = f.suptitle('Basic CNN Performance', fontsize=12)
f.subplots_adjust(top=0.85, wspace=0.3)
max_epoch = len(history.history['accuracy'])+1
epoch_list = list(range(1,max_epoch))
ax1.plot(epoch_list, history.history['accuracy'], label='Train Accuracy')
ax1.plot(epoch_list, history.history['val_accuracy'], label='Validation Accuracy')
ax1.set_xticks(np.arange(1, max_epoch, 5))
ax1.set_ylabel('Accuracy Value')
ax1.set_xlabel('Epoch')
ax1.set_title('Accuracy')
l1 = ax1.legend(loc="best")
ax2.plot(epoch_list, history.history['loss'], label='Train Loss')
ax2.plot(epoch_list, history.history['val_loss'], label='Validation Loss')
ax2.set_xticks(np.arange(1, max_epoch, 5))
ax2.set_ylabel('Loss Value')
ax2.set_xlabel('Epoch')
ax2.set_title('Loss')
l2 = ax2.legend(loc="best")
test_imgs_scaled = test_data/255.
test_labels_enc = le.transform(test_labels)
# evaluate the model
_, train_acc = model.evaluate(train_imgs_scaled, train_labels_enc, verbose=0)
_, test_acc = model.evaluate(test_imgs_scaled, test_labels_enc, verbose=0)
print('Train: %.3f, Test: %.3f' % (train_acc, test_acc))
print(model.summary())
```
|
github_jupyter
|
##### Copyright 2020 The TensorFlow Authors.
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# Barren plateaus
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://www.tensorflow.org/quantum/tutorials/barren_plateaus"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on TensorFlow.org</a>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/quantum/blob/master/docs/tutorials/barren_plateaus.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/quantum/blob/master/docs/tutorials/barren_plateaus.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
</td>
<td>
<a href="https://storage.googleapis.com/tensorflow_docs/quantum/docs/tutorials/barren_plateaus.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png" />Download notebook</a>
</td>
</table>
In this example you will explore the result of <a href="https://www.nature.com/articles/s41467-018-07090-4" class="external">McClean, 2019</a> that says not just any quantum neural network structure will do well when it comes to learning. In particular you will see that a certain large family of random quantum circuits do not serve as good quantum neural networks, because they have gradients that vanish almost everywhere. In this example you won't be training any models for a specific learning problem, but instead focusing on the simpler problem of understanding the behaviors of gradients.
## Setup
```
try:
%tensorflow_version 2.x
except Exception:
pass
```
Install TensorFlow Quantum:
```
!pip install tensorflow-quantum
```
Now import TensorFlow and the module dependencies:
```
import tensorflow as tf
import tensorflow_quantum as tfq
import cirq
import sympy
import numpy as np
# visualization tools
%matplotlib inline
import matplotlib.pyplot as plt
from cirq.contrib.svg import SVGCircuit
np.random.seed(1234)
```
## 1. Summary
Random quantum circuits with many blocks that look like this ($R_{P}(\theta)$ is a random Pauli rotation):<br/>
<img src="./images/barren_2.png" width=700>
Where if $f(x)$ is defined as the expectation value w.r.t. $Z_{a}Z_{b}$ for any qubits $a$ and $b$, then there is a problem that $f'(x)$ has a mean very close to 0 and does not vary much. You will see this below:
## 2. Generating random circuits
The construction from the paper is straightforward to follow. The following implements a simple function that generates a random quantum circuit—sometimes referred to as a *quantum neural network* (QNN)—with the given depth on a set of qubits:
```
def generate_random_qnn(qubits, symbol, depth):
"""Generate random QNN's with the same structure from McClean et al."""
circuit = cirq.Circuit()
for qubit in qubits:
circuit += cirq.Ry(np.pi / 4.0)(qubit)
for d in range(depth):
# Add a series of single qubit rotations.
for i, qubit in enumerate(qubits):
random_n = np.random.uniform()
random_rot = np.random.uniform(
) * 2.0 * np.pi if i != 0 or d != 0 else symbol
if random_n > 2. / 3.:
# Add a Z.
circuit += cirq.Rz(random_rot)(qubit)
elif random_n > 1. / 3.:
# Add a Y.
circuit += cirq.Ry(random_rot)(qubit)
else:
# Add a X.
circuit += cirq.Rx(random_rot)(qubit)
# Add CZ ladder.
for src, dest in zip(qubits, qubits[1:]):
circuit += cirq.CZ(src, dest)
return circuit
generate_random_qnn(cirq.GridQubit.rect(1, 3), sympy.Symbol('theta'), 2)
```
The authors investigate the gradient of a single parameter $\theta_{1,1}$. Let's follow along by placing a `sympy.Symbol` in the circuit where $\theta_{1,1}$ would be. Since the authors do not analyze the statistics for any other symbols in the circuit, let's replace them with random values now instead of later.
## 3. Running the circuits
Generate a few of these circuits along with an observable to test the claim that the gradients don't vary much. First, generate a batch of random circuits. Choose a random *ZZ* observable and batch calculate the gradients and variance using TensorFlow Quantum.
### 3.1 Batch variance computation
Let's write a helper function that computes the variance of the gradient of a given observable over a batch of circuits:
```
def process_batch(circuits, symbol, op):
"""Compute the variance of a batch of expectations w.r.t. op on each circuit that
contains `symbol`. Note that this method sets up a new compute graph every time it is
called so it isn't as performant as possible."""
# Setup a simple layer to batch compute the expectation gradients.
expectation = tfq.layers.Expectation()
# Prep the inputs as tensors
circuit_tensor = tfq.convert_to_tensor(circuits)
values_tensor = tf.convert_to_tensor(
np.random.uniform(0, 2 * np.pi, (n_circuits, 1)).astype(np.float32))
# Use TensorFlow GradientTape to track gradients.
with tf.GradientTape() as g:
g.watch(values_tensor)
forward = expectation(circuit_tensor,
operators=op,
symbol_names=[symbol],
symbol_values=values_tensor)
# Return variance of gradients across all circuits.
grads = g.gradient(forward, values_tensor)
grad_var = tf.math.reduce_std(grads, axis=0)
return grad_var.numpy()[0]
```
### 3.1 Set up and run
Choose the number of random circuits to generate along with their depth and the amount of qubits they should act on. Then plot the results.
```
n_qubits = [2 * i for i in range(2, 7)
] # Ranges studied in paper are between 2 and 24.
depth = 50 # Ranges studied in paper are between 50 and 500.
n_circuits = 200
theta_var = []
for n in n_qubits:
# Generate the random circuits and observable for the given n.
qubits = cirq.GridQubit.rect(1, n)
symbol = sympy.Symbol('theta')
circuits = [
generate_random_qnn(qubits, symbol, depth) for _ in range(n_circuits)
]
op = cirq.Z(qubits[0]) * cirq.Z(qubits[1])
theta_var.append(process_batch(circuits, symbol, op))
plt.semilogy(n_qubits, theta_var)
plt.title('Gradient Variance in QNNs')
plt.xlabel('n_qubits')
plt.ylabel('$\\partial \\theta$ variance')
plt.show()
```
This plot shows that for quantum machine learning problems, you can't simply guess a random QNN ansatz and hope for the best. Some structure must be present in the model circuit in order for gradients to vary to the point where learning can happen.
## 4. Heuristics
An interesting heuristic by <a href="https://arxiv.org/pdf/1903.05076.pdf" class="external">Grant, 2019</a> allows one to start very close to random, but not quite. Using the same circuits as McClean et al., the authors propose a different initialization technique for the classical control parameters to avoid barren plateaus. The initialization technique starts some layers with totally random control parameters—but, in the layers immediately following, choose parameters such that the initial transformation made by the first few layers is undone. The authors call this an *identity block*.
The advantage of this heuristic is that by changing just a single parameter, all other blocks outside of the current block will remain the identity—and the gradient signal comes through much stronger than before. This allows the user to pick and choose which variables and blocks to modify to get a strong gradient signal. This heuristic does not prevent the user from falling in to a barren plateau during the training phase (and restricts a fully simultaneous update), it just guarantees that you can start outside of a plateau.
### 4.1 New QNN construction
Now construct a function to generate identity block QNNs. This implementation is slightly different than the one from the paper. For now, look at the behavior of the gradient of a single parameter so it is consistent with McClean et al, so some simplifications can be made.
To generate an identity block and train the model, generally you need $U1(\theta_{1a}) U1(\theta_{1b})^{\dagger}$ and not $U1(\theta_1) U1(\theta_1)^{\dagger}$. Initially $\theta_{1a}$ and $\theta_{1b}$ are the same angles but they are learned independently. Otherwise, you will always get the identity even after training. The choice for the number of identity blocks is empirical. The deeper the block, the smaller the variance in the middle of the block. But at the start and end of the block, the variance of the parameter gradients should be large.
```
def generate_identity_qnn(qubits, symbol, block_depth, total_depth):
"""Generate random QNN's with the same structure from Grant et al."""
circuit = cirq.Circuit()
# Generate initial block with symbol.
prep_and_U = generate_random_qnn(qubits, symbol, block_depth)
circuit += prep_and_U
# Generate dagger of initial block without symbol.
U_dagger = (prep_and_U[1:])**-1
circuit += cirq.resolve_parameters(
U_dagger, param_resolver={symbol: np.random.uniform() * 2 * np.pi})
for d in range(total_depth - 1):
# Get a random QNN.
prep_and_U_circuit = generate_random_qnn(
qubits,
np.random.uniform() * 2 * np.pi, block_depth)
# Remove the state-prep component
U_circuit = prep_and_U_circuit[1:]
# Add U
circuit += U_circuit
# Add U^dagger
circuit += U_circuit**-1
return circuit
generate_identity_qnn(cirq.GridQubit.rect(1, 3), sympy.Symbol('theta'), 2, 2)
```
### 4.2 Comparison
Here you can see that the heuristic does help to keep the variance of the gradient from vanishing as quickly:
```
block_depth = 10
total_depth = 5
heuristic_theta_var = []
for n in n_qubits:
# Generate the identity block circuits and observable for the given n.
qubits = cirq.GridQubit.rect(1, n)
symbol = sympy.Symbol('theta')
circuits = [
generate_identity_qnn(qubits, symbol, block_depth, total_depth)
for _ in range(n_circuits)
]
op = cirq.Z(qubits[0]) * cirq.Z(qubits[1])
heuristic_theta_var.append(process_batch(circuits, symbol, op))
plt.semilogy(n_qubits, theta_var)
plt.semilogy(n_qubits, heuristic_theta_var)
plt.title('Heuristic vs. Random')
plt.xlabel('n_qubits')
plt.ylabel('$\\partial \\theta$ variance')
plt.show()
```
This is a great improvement in getting stronger gradient signals from (near) random QNNs.
|
github_jupyter
|
```
import pandas as pd
import numpy as np
import h5py
import matplotlib.pyplot as plt
import scipy
from PIL import Image
from scipy import ndimage
#from dnn_app_utils_v2 import *
import pandas as pd
%matplotlib inline
from pandas import ExcelWriter
from pandas import ExcelFile
%load_ext autoreload
%autoreload 2
from sklearn.utils import resample
import tensorflow as tf
from tensorflow.python.framework import ops
import openpyxl
import keras
import xlsxwriter
from keras.layers import Dense, Dropout
from keras import optimizers
import pandas as pd
import numpy as np
import h5py
import matplotlib.pyplot as plt
import scipy
from PIL import Image
from scipy import ndimage
#from dnn_app_utils_v2 import *
import pandas as pd
%matplotlib inline
from pandas import ExcelWriter
from pandas import ExcelFile
%load_ext autoreload
%autoreload 2
from sklearn.utils import resample
import tensorflow as tf
from tensorflow.python.framework import ops
import openpyxl
import keras
import xlsxwriter
from keras.models import Sequential
from keras.layers import Dense
from keras.wrappers.scikit_learn import KerasClassifier
from sklearn.model_selection import cross_val_score
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import StratifiedKFold
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import Pipeline
print(" All the necessary Libraries have been loaded")
print(" ")
print(" ")
print(" The code after this is for loading your data into train and test. Make sure you load the correct features")
xls = pd.ExcelFile("test_selected.xlsx")
test_selected_x = pd.read_excel(xls, 'test_selected_x')
test_selected_y = pd.read_excel(xls, 'test_selected_y')
print(" The selected important features data for spesific model is loaded into train, and test")
print(" ")
test_selected_x=test_selected_x.values
test_selected_y=test_selected_y.values
print("##################################################################################################")
print("Now you load the model but with correct model name")
print(" loading the trained model ")
print(" ")
from keras.models import model_from_json
# load json and create model
json_file = open('1_model.json', 'r')
loaded_model_json = json_file.read()
json_file.close()
loaded_model_1 = model_from_json(loaded_model_json)
# load weights into new model
loaded_model_1.load_weights("1_model.h5")
print("Loaded model from disk")
print(" ")
json_file = open('2_model.json', 'r')
loaded_model_json = json_file.read()
json_file.close()
loaded_model_2 = model_from_json(loaded_model_json)
# load weights into new model
loaded_model_2.load_weights("2_model.h5")
print("Loaded model from disk")
print(" ")
json_file = open('3_model.json', 'r')
loaded_model_json = json_file.read()
json_file.close()
loaded_model_3 = model_from_json(loaded_model_json)
# load weights into new model
loaded_model_3.load_weights("3_model.h5")
print("Loaded model from disk")
print(" ")
json_file = open('4_model.json', 'r')
loaded_model_json = json_file.read()
json_file.close()
loaded_model_4 = model_from_json(loaded_model_json)
# load weights into new model
loaded_model_4.load_weights("4_model.h5")
print("Loaded model from disk")
print(" ")
print(" Computing the AUCROC using the loded model for checking ")
print(" ")
from sklearn.metrics import roc_auc_score, roc_curve
pred_test_1 = loaded_model_1.predict(test_selected_x)
pred_test_2 = loaded_model_2.predict(test_selected_x)
pred_test_3 = loaded_model_3.predict(test_selected_x)
pred_test_4 = loaded_model_4.predict(test_selected_x)
pred_test=(pred_test_1+pred_test_2+pred_test_3+pred_test_4)/4
auc_test = roc_auc_score(test_selected_y, pred_test)
print ("AUROC_test: " + str(auc_test))
```
|
github_jupyter
|
# LKJ Cholesky Covariance Priors for Multivariate Normal Models
While the [inverse-Wishart distribution](https://en.wikipedia.org/wiki/Inverse-Wishart_distribution) is the conjugate prior for the covariance matrix of a multivariate normal distribution, it is [not very well-suited](https://github.com/pymc-devs/pymc3/issues/538#issuecomment-94153586) to modern Bayesian computational methods. For this reason, the [LKJ prior](http://www.sciencedirect.com/science/article/pii/S0047259X09000876) is recommended when modeling the covariance matrix of a multivariate normal distribution.
To illustrate modelling covariance with the LKJ distribution, we first generate a two-dimensional normally-distributed sample data set.
```
import arviz as az
import numpy as np
import pymc3 as pm
import seaborn as sns
import warnings
from matplotlib.patches import Ellipse
from matplotlib import pyplot as plt
az.style.use("arviz-darkgrid")
warnings.simplefilter(action="ignore", category=FutureWarning)
RANDOM_SEED = 8924
np.random.seed(3264602) # from random.org
N = 10000
μ_actual = np.array([1.0, -2.0])
sigmas_actual = np.array([0.7, 1.5])
Rho_actual = np.matrix([[1.0, -0.4], [-0.4, 1.0]])
Σ_actual = np.diag(sigmas_actual) * Rho_actual * np.diag(sigmas_actual)
x = np.random.multivariate_normal(μ_actual, Σ_actual, size=N)
Σ_actual
var, U = np.linalg.eig(Σ_actual)
angle = 180.0 / np.pi * np.arccos(np.abs(U[0, 0]))
fig, ax = plt.subplots(figsize=(8, 6))
blue, _, red, *_ = sns.color_palette()
e = Ellipse(
μ_actual, 2 * np.sqrt(5.991 * var[0]), 2 * np.sqrt(5.991 * var[1]), angle=angle
)
e.set_alpha(0.5)
e.set_facecolor(blue)
e.set_zorder(10)
ax.add_artist(e)
ax.scatter(x[:, 0], x[:, 1], c="k", alpha=0.05, zorder=11)
rect = plt.Rectangle((0, 0), 1, 1, fc=blue, alpha=0.5)
ax.legend([rect], ["95% density region"], loc=2);
```
The sampling distribution for the multivariate normal model is $\mathbf{x} \sim N(\mu, \Sigma)$, where $\Sigma$ is the covariance matrix of the sampling distribution, with $\Sigma_{ij} = \textrm{Cov}(x_i, x_j)$. The density of this distribution is
$$f(\mathbf{x}\ |\ \mu, \Sigma^{-1}) = (2 \pi)^{-\frac{k}{2}} |\Sigma|^{-\frac{1}{2}} \exp\left(-\frac{1}{2} (\mathbf{x} - \mu)^{\top} \Sigma^{-1} (\mathbf{x} - \mu)\right).$$
The LKJ distribution provides a prior on the correlation matrix, $\mathbf{C} = \textrm{Corr}(x_i, x_j)$, which, combined with priors on the standard deviations of each component, [induces](http://www3.stat.sinica.edu.tw/statistica/oldpdf/A10n416.pdf) a prior on the covariance matrix, $\Sigma$. Since inverting $\Sigma$ is numerically unstable and inefficient, it is computationally advantageous to use the [Cholesky decompositon](https://en.wikipedia.org/wiki/Cholesky_decomposition) of $\Sigma$, $\Sigma = \mathbf{L} \mathbf{L}^{\top}$, where $\mathbf{L}$ is a lower-triangular matrix. This decompositon allows computation of the term $(\mathbf{x} - \mu)^{\top} \Sigma^{-1} (\mathbf{x} - \mu)$ using back-substitution, which is more numerically stable and efficient than direct matrix inversion.
PyMC3 supports LKJ priors for the Cholesky decomposition of the covariance matrix via the [LKJCholeskyCov](../api/distributions/multivariate.rst) distribution. This distribution has parameters `n` and `sd_dist`, which are the dimension of the observations, $\mathbf{x}$, and the PyMC3 distribution of the component standard deviations, respectively. It also has a hyperparamter `eta`, which controls the amount of correlation between components of $\mathbf{x}$. The LKJ distribution has the density $f(\mathbf{C}\ |\ \eta) \propto |\mathbf{C}|^{\eta - 1}$, so $\eta = 1$ leads to a uniform distribution on correlation matrices, while the magnitude of correlations between components decreases as $\eta \to \infty$.
In this example, we model the standard deviations with $\textrm{Exponential}(1.0)$ priors, and the correlation matrix as $\mathbf{C} \sim \textrm{LKJ}(\eta = 2)$.
```
with pm.Model() as m:
packed_L = pm.LKJCholeskyCov(
"packed_L", n=2, eta=2.0, sd_dist=pm.Exponential.dist(1.0)
)
```
Since the Cholesky decompositon of $\Sigma$ is lower triangular, `LKJCholeskyCov` only stores the diagonal and sub-diagonal entries, for efficiency:
```
packed_L.tag.test_value.shape
```
We use [expand_packed_triangular](../api/math.rst) to transform this vector into the lower triangular matrix $\mathbf{L}$, which appears in the Cholesky decomposition $\Sigma = \mathbf{L} \mathbf{L}^{\top}$.
```
with m:
L = pm.expand_packed_triangular(2, packed_L)
Σ = L.dot(L.T)
L.tag.test_value.shape
```
Often however, you'll be interested in the posterior distribution of the correlations matrix and of the standard deviations, not in the posterior Cholesky covariance matrix *per se*. Why? Because the correlations and standard deviations are easier to interpret and often have a scientific meaning in the model. As of PyMC 3.9, there is a way to tell PyMC to automatically do these computations and store the posteriors in the trace. You just have to specify `compute_corr=True` in `pm.LKJCholeskyCov`:
```
with pm.Model() as model:
chol, corr, stds = pm.LKJCholeskyCov(
"chol", n=2, eta=2.0, sd_dist=pm.Exponential.dist(1.0), compute_corr=True
)
cov = pm.Deterministic("cov", chol.dot(chol.T))
```
To complete our model, we place independent, weakly regularizing priors, $N(0, 1.5),$ on $\mu$:
```
with model:
μ = pm.Normal("μ", 0.0, 1.5, shape=2, testval=x.mean(axis=0))
obs = pm.MvNormal("obs", μ, chol=chol, observed=x)
```
We sample from this model using NUTS and give the trace to [ArviZ](https://arviz-devs.github.io/arviz/):
```
with model:
trace = pm.sample(random_seed=RANDOM_SEED, init="adapt_diag")
idata = az.from_pymc3(trace)
az.summary(idata, var_names=["~chol"], round_to=2)
```
Sampling went smoothly: no divergences and good r-hats. You can also see that the sampler recovered the true means, correlations and standard deviations. As often, that will be clearer in a graph:
```
az.plot_trace(
idata,
var_names=["~chol"],
compact=True,
lines=[
("μ", {}, μ_actual),
("cov", {}, Σ_actual),
("chol_stds", {}, sigmas_actual),
("chol_corr", {}, Rho_actual),
],
);
```
The posterior expected values are very close to the true value of each component! How close exactly? Let's compute the percentage of closeness for $\mu$ and $\Sigma$:
```
μ_post = trace["μ"].mean(axis=0)
(1 - μ_post / μ_actual).round(2)
Σ_post = trace["cov"].mean(axis=0)
(1 - Σ_post / Σ_actual).round(2)
```
So the posterior means are within 3% of the true values of $\mu$ and $\Sigma$.
Now let's replicate the plot we did at the beginning, but let's overlay the posterior distribution on top of the true distribution -- you'll see there is excellent visual agreement between both:
```
var_post, U_post = np.linalg.eig(Σ_post)
angle_post = 180.0 / np.pi * np.arccos(np.abs(U_post[0, 0]))
fig, ax = plt.subplots(figsize=(8, 6))
e = Ellipse(
μ_actual, 2 * np.sqrt(5.991 * var[0]), 2 * np.sqrt(5.991 * var[1]), angle=angle
)
e.set_alpha(0.5)
e.set_facecolor(blue)
e.set_zorder(10)
ax.add_artist(e)
e_post = Ellipse(
μ_post,
2 * np.sqrt(5.991 * var_post[0]),
2 * np.sqrt(5.991 * var_post[1]),
angle=angle_post,
)
e_post.set_alpha(0.5)
e_post.set_facecolor(red)
e_post.set_zorder(10)
ax.add_artist(e_post)
ax.scatter(x[:, 0], x[:, 1], c="k", alpha=0.05, zorder=11)
rect = plt.Rectangle((0, 0), 1, 1, fc=blue, alpha=0.5)
rect_post = plt.Rectangle((0, 0), 1, 1, fc=red, alpha=0.5)
ax.legend(
[rect, rect_post],
["True 95% density region", "Estimated 95% density region"],
loc=2,
);
%load_ext watermark
%watermark -n -u -v -iv -w
```
|
github_jupyter
|
# Forecasting on Contraceptive Use - A Multi-step Ensemble Approach¶
Update: 09/07/2020
Github Repository: https://github.com/herbsh/USAID_Forecast_submit
## key idea
- The goal is to forecast on site_code & product_code level demand.
- The site_code & product_code level demand fluctuates too much and doesn't have any obvious pattern.
- The aggregate level is easier to forecast. The noise cancels out.
- We don't know what is the best level to aggregate. It's possible that it varies regarding each site too.
- we aggregate on various levels, and use "Ensemble Learning" to to determine the final result
## Aggregate
How to aggregate? Here is the structure of the supply chain and products. We can use this guide our aggregation:
- site_code -> district -> region
- product_code -> product_type
After we aggregate on some level, we get a time series of stock_distributed, {Y}.
We supplement some external data and match/aggregate to the same level, {X}.
we use a time-series model to forecast {Y} with {Y} and {X}.
## Forecast the Aggregate : Time Series Modeling, Auto_SARIMAX
- We use a SARIMAX stucture (ARIMA (AR, MA with a trend) with Seasonality and external data )
- The specific order of SARIMAX is determined within each time series with an Auto_ARIMA function.
- Use BIC criteria to pick the optimal order for ARIMA (p, d, q) and seasonality (P,D,Q) (BIC : best for forecast power) (range of p,d,q and P,D,Q - small, less than 4)
- use the time series model to make aggregate level forecast. Store the results for later use.
## De-aggregate (Distribute) : Machine Learning Modeling
- We use machine learning to learn the variable "share", the share of the specific stock_distributed as a fraction of the aggregate sum.
- training data:
- all the data, excluding the last 3 month. Encode the year, month, region, district, product type, code, plus all available external data, matched to site_code+product level.
- target: actual shares
- model: RandomForest Decision Regression Tree
- use the fitted model to make prediction on shares. "Distribute" aggregate stocks to individual forecasts.
## Emsemble
- From Aggregate and Distribute, we arrive at X different forecast numbers for each site+product_code. (We also have a lot of intermediary forecasts that could be of interests to various parties).
- We introduce another model to perform the ensemble
- For each training observation, we have multiple fourcasts and one actual realization, denote them as F1, F2, F3, F4 and Y (ommitting site, product, t time subscripts). We also have all the features X ( temperature, roads, etc).
- The emsemble part estimate another model to take inputs (F1..F4, and features X) to arrive at an estimated Y_hat that minimizes its MSE to Y ( actual stock_distributed).
- We used XGBoost to perform the ensemble learning part.
## Key Takeaway of this approach
- Combines traditional forecast methods (SARIMAX) and Machine Learning (XGBoost)
- It's very transferable to other scenarios
- It uses external data and it's easy to plug in more external data to improve the forecasts.
- The ensemble piece makes adding model possible and easy
```
# suppress warning to make cleaner output
import warnings
warnings.filterwarnings("ignore")
```
# Outline
## Step 1: Data Cleaning
### Upsample (fill in gaps in time series)
- notebook: datacleaning_upsample.ipynb
```
%run datacleaning_upsample.ipynb
```
- inputs: "..\0_data\contraceptive_logistics_data.csv"
- output: "..\2_pipeline\df_upsample.csv"
- steps:
- upsample - make sure all individual product-site series has no gaps in time even though they may differ in length
- fill in 0 for NA in stock_distributed
### Supplement
(very import thing about supplement data - if we are to use any supplement data, the value should exist for the 3 months that are to be forecasted) ( 10, 11, 12) Must be careful when constructing supplement dataset
- notebook: datacleaning_prep_supplement.ipynb
- input:
- "..\0_data\supplement_data_raw.csv"
- steps:
- time invariant supplement data:
- identifiers: site_code product_code region district
- information: (currently) road condition, product type
- output: "../0_data/time_invariant_supplement.dta"
- time variant supplement data: (include rows for time that need to be forecasted)
- identifiers: temp_timeindex year month site_code product_code region district
- information: maxtemp temp pressure relative rain visibility windspeed maxsus* storm fog
- output: "../0_data/time_variant_supplement.dta"
### Combine
- notebook: datacleaning_combine.ipynb
```
%run datacleaning_combine.ipynb
```
- input:
- "../2_pipeline/df_upsample.csv"
- '../0_data/submission_format.csv'
- "../0_data/time_invariant_supplement.dta"
- "../0_data/time_variant_supplement.dta"
- '../0_data/service_delivery_site_data.csv'
- output: a site_code & product_code & date level logistics data with time variant and time invariant exogenous features :
- for development: '../0_data/df_training.csv'
- for final prediction(contained 3 last month exog vars and space holder) '../0_data/df_combined_fullsample.csv'
## Step 2: Multiple Agg-Forecast-Distribute Models in parallel
### Region Level
- notebook: model_SARIMAX_Distribute_region.ipynb
%run model_SARIMAX_Distribute_region.ipynb
- input:
- '../0_data/df_combined_fullsample.csv'
- output:
1. ../2_pipeline/final_pred_region_lev.csv
2. ./2_pipeline/final_distribute_regionlev.csv'
### District Level
- notebook: model_SARIMAX_Distribute_District.ipynb
```
%run model_SARIMAX_Distribute_District.ipynb
```
- input:
- '../0_data/df_combined_fullsample.csv'
- output::
1. '../2_pipeline/final_pred_district_lev.csv'
2. '../2_pipeline/final_distribute_districtlev.csv'
### Region-Product_type level
- notebook: model_SARIMAX_Distribute_regionproducttype.ipynb
```
%run model_SARIMAX_Distribute_regionproducttype.ipynb
```
- input:
- '../0_data/df_combined_fullsample.csv'
- output:
1. ../2_pipeline/final_pred_region_producttype_lev.csv
2. '../2_pipeline/final_distribute_regionproducttypelev.csv'
### Individual Level, with raw data, winsorized data, and rolling smoothed data
- notebook: model_SARIMAX_individual.ipyn
```
%run model_SARIMAX_individual.ipynb
```
- input:
- '../0_data/df_combined_fullsample.csv'
- output:
- '../2_pipeline/final_pred_ind_lev.csv'
- notebook: model_SARIMAX_individual_winsorized.ipynb
```
%run model_SARIMAX_individual_winsorized.ipynb
```
- input:
- '../0_data/df_combined_fullsample.csv'
- output:
- '../2_pipeline/final_pred_ind_winsorized_lev.csv'
- notebook: model_SARIMAX_individual_rollingsmoothed.ipynb
```
%run model_SARIMAX_individual_rollingsmoothed.ipynb
```
- input:
- '../0_data/df_combined_fullsample.csv'
- output:
- '../2_pipeline/final_pred_ind_rollingsmoothed_lev.csv'
## Step 3: Ensemble, learn the ensemble model, make final prediction
- notebook: ensemble.ipynb
```
%run ensemble.ipynb
```
- input:
- Distribution model results: glob.glob('../2_pipeline/final_distribute_*.csv')
- SARIMAX results: glob.glob('../2_pipeline/final_pred_ind*.csv')
- output:
# Ensemble Model Details
## Import : data with actual stock distributed and exogenous variables
```
import pandas as pd
df_combined=pd.read_csv('../0_data/df_combined_fullsample.csv')
```
## Import results from distribution(shares) models
```
import glob
temp=glob.glob('../2_pipeline/final_distribute_*.csv')
print('\n importing results from the distribution stage of various aggregation levels \n')
print(temp)
distribute_districtlev=pd.read_csv('../2_pipeline\\final_distribute_districtlev.csv').drop(columns=['Unnamed: 0'])
distribute_regionlev=pd.read_csv('../2_pipeline\\final_distribute_regionlev.csv').drop(columns=['Unnamed: 0'])
distribute_regionproducttypelev=pd.read_csv('../2_pipeline\\final_distribute_regionproducttypelev.csv').drop(columns=['Unnamed: 0'])
```
## Import SARIMAX_agg model results and merge with distribute
```
print('Import SARIMAX_agg model results and merge with predicted distribute values')
sarimax_pred_region=pd.read_csv('../2_pipeline/final_pred_region_lev.csv').rename(columns={'Unnamed: 0':'date',}).rename(columns={'stock_distributed_forecasted':'stock_forecast_agg_region'})
sarimax_pred_regionproducttype=pd.read_csv('../2_pipeline/final_pred_region_producttype_lev.csv').rename(columns={'Unnamed: 0':'date'}).rename(columns={'stock_distributed_forecasted':'stock_forecast_agg_regionproducttype'})
sarimax_pred_district=pd.read_csv('../2_pipeline/final_pred_district_lev.csv').rename(columns={'Unnamed: 0':'date'}).rename(columns={'stock_distributed_forecasted':'stock_forecast_agg_district'})
```
- merge sarimax_pred_region with distribute_region
```
pred_agg_region=pd.merge(left=sarimax_pred_region,right=distribute_regionlev,on=['date','region','product_code'],how='right')
pred_agg_region.describe()
```
- merge sarimax_pred_regionproducttype with distribute_regionproducttype
```
pred_agg_regionproducttype=pd.merge(left=sarimax_pred_regionproducttype,right=distribute_regionproducttypelev,on=['date','region','product_type'],how='right')
pred_agg_regionproducttype.describe()
```
- merge sarimax_pred_district with distribute_districtlev
```
pred_agg_district=pd.merge(left=sarimax_pred_district,right=distribute_districtlev,on=['date','district','product_code'],how='right')
pred_agg_district.describe()
```
## Import three individual level sarimax results
```
import glob
temp=glob.glob('../2_pipeline/final_pred_ind*.csv')
print('\n Import three individual level sarimax results \n')
print(temp)
sarimax_ind=pd.read_csv('../2_pipeline/final_pred_ind_lev.csv').rename(columns={'Unnamed: 0':'date','stock_distributed_forecasted':'stock_forecast_agg_ind'})
sarimax_ind.head(2)
sarimax_ind_smooth=pd.read_csv('../2_pipeline/final_pred_ind_rollingsmoothed_lev.csv').rename(columns={'Unnamed: 0':'date','stock_distributed_forecasted':'stock_forecast_agg_ind_smooth'})
sarimax_ind_smooth.head(2)
sarimax_ind_winsorized=pd.read_csv('../2_pipeline/final_pred_ind_winsorized_lev.csv').rename(columns={'Unnamed: 0':'date','stock_distributed_forecasted':'stock_forecast_agg_ind_winsorized'})
sarimax_ind_winsorized.head(2)
df_ensemble=pd.merge(left=df_combined,right=pred_agg_region.drop(columns=['agg_level']),on=['date','region','product_code','site_code'],how='left')
len(df_ensemble)
df_ensemble=pd.merge(left=df_ensemble,right=pred_agg_regionproducttype.drop(columns=['agg_level']),on=['date','region','product_type','site_code','product_code'],how='left')
len(df_ensemble)
df_ensemble=pd.merge(left=df_ensemble,right=pred_agg_district.drop(columns=['agg_level']),on=['date','district','site_code','product_code'],how='left')
df_ensemble=pd.merge(left=df_ensemble,right=sarimax_ind,on=['date','site_code','product_code'],how='left')
df_ensemble=pd.merge(left=df_ensemble,right=sarimax_ind_smooth,on=['date','site_code','product_code'],how='left')
df_ensemble=pd.merge(left=df_ensemble,right=sarimax_ind_winsorized,on=['date','site_code','product_code'],how='left')
df_ensemble['date']=pd.to_datetime(df_ensemble['date'])
df_ensemble.set_index('date')['2019-10':].describe()
df_ensemble=df_ensemble.fillna(0)
df_ensemble.head()
```
## Sort df_ensemble dataframe by date to ensure the train-test data are set up correctly
```
df_ensemble.sort_values(by='date',inplace=True)
```
## Feature Engineering
### Add a few interactions
```
df_ensemble['interaction_1']=df_ensemble['pred_share_regionlev']*df_ensemble['stock_forecast_agg_regionproducttype']
df_ensemble['interaction_2']=df_ensemble['pred_share_districtlev']*df_ensemble['stock_forecast_agg_regionproducttype']
df_ensemble['weather_interaction']=df_ensemble['maxtemp']*df_ensemble['rainfallsnowmelt']
columns_to_encode=['site_code', 'product_code', 'year', 'month',
'region', 'district', 'product_type','site_type']
columns_continuous_exog=['regionroads',
'regionasphaltroads', 'regionearthroads', 'regionsurfacetreatmentroads',
'regionpoorroads', 'poorroads', 'earthroads', 'asphaltroads', 'temp',
'maxtemp', 'pressure', 'relativehumidity', 'rainfallsnowmelt',
'visibility', 'windspeed', 'maxsustainedwindspeed', 'rainordrizzle',
'storm', 'fog','weather_interaction']
columns_continuous_frommodel=['stock_forecast_agg_region', 'pred_share_regionlev',
'stock_forecast_agg_regionproducttype',
'pred_share_regionproducttype_tlev', 'stock_forecast_agg_district',
'pred_share_districtlev', 'stock_forecast_agg_ind',
'stock_forecast_agg_ind_smooth', 'stock_forecast_agg_ind_winsorized','interaction_1','interaction_2']
```
## Setting up target
- the y vector should have all 0s for the last 3 months worth of data
```
y=df_ensemble.stock_distributed
```
## Setting up features
```
# Import libraries and download example data
from sklearn.preprocessing import OneHotEncoder
ohe = OneHotEncoder(sparse=False,categories='auto')
encoded_columns = ohe.fit_transform(df_ensemble[columns_to_encode])
import numpy as np
np.shape(encoded_columns)
```
- produce one-hot encoding for categorical values
```
features=pd.DataFrame(data=encoded_columns,columns=ohe.get_feature_names(columns_to_encode))
features.describe()
```
- add continuous values. Put everything to a X matrix
```
X=features
X[columns_continuous_exog]=df_ensemble[columns_continuous_exog]
X[columns_continuous_frommodel]=df_ensemble[columns_continuous_frommodel]
X.to_csv('x.csv')
```
## Scale X
```
from sklearn.preprocessing import scale
Xs = scale(X)
```
# Use XGboost to make final prediction
## XGBoost
```
import xgboost as xgb
from sklearn.metrics import mean_squared_error
import pandas as pd
import numpy as np
df_train=df_ensemble.set_index('date')[:'2019-9']
df_pred=df_ensemble.set_index('date')['2019-10':'2019-12']
Xs_train=Xs[:df_train.shape[0]]
y_train=y[:df_train.shape[0]]
Xs_pred=Xs[-df_pred.shape[0]:]
data_dmatrix = xgb.DMatrix(data=Xs,label=y)
xg_reg = xgb.XGBRegressor(objective ='reg:squarederror', colsample_bytree = 0.3, learning_rate = 0.1,
max_depth = 30, alpha = 10, n_estimators = 20)
xg_reg.fit(Xs_train,y_train)
preds = xg_reg.predict(Xs_pred)
len(preds)
import matplotlib.pyplot as plt
%matplotlib inline
xgb.plot_importance(xg_reg)
plt.rcParams['figure.figsize'] = [8,10]
plt.savefig('../2_pipeline/xgboost_plot_importance.jpg')
plt.show()
```
## Collect Results
```
temp=df_pred[['year','month','site_code','product_code']].copy()
temp['predicted_value']=preds
temp=temp.reset_index()
temp=temp.drop(columns='date')
submission_format=pd.read_csv('../0_data/submission_format.csv')
submission=pd.merge(left=submission_format.drop(columns='predicted_value'),right=temp,on=['year','month','site_code','product_code'],how='left')
submission.describe()
submission['predicted_value']=submission['predicted_value'].apply(lambda x: max(x,0))
submission.describe()
submission.head()
submission[['year','month','site_code','product_code','predicted_value']].to_csv('../submission.csv')
```
|
github_jupyter
|
# Data Visualization With Safas
This notebook demonstrates plotting the results from Safas video analysis.
## Import modules and data
Import safas and other components for display and analysis. safas has several example images in the safas/data directory. These images are accessible as attributes of the data module because the __init__ function of safas/data also acts as a loader.
```
import sys
from matplotlib import pyplot as plt
from matplotlib.ticker import ScalarFormatter
%matplotlib inline
import pandas as pd
import cv2
from safas import filters
from safas import data
from safas.filters.sobel_focus import imfilter as sobel_filter
from safas.filters.imfilters_module import add_contours
```
## Object properties
Users may interactively select and link objects in safas. When the data is saved, the data is written in tabular form where the properties of each oject are stored.
The object properties are calculated with the Scikit-Image function [regionprops.](https://scikit-image.org/docs/dev/api/skimage.measure.html#skimage.measure.regionprops). A complete description of these properties may be found in the [regionprops.](https://scikit-image.org/docs/dev/api/skimage.measure.html#skimage.measure.regionprops) documentation. At this time, the following properties are stored in a .xlsx file:
property | unit |
--- | --- |
area | $\mu m^2$ |
equivalent_diameter | $\mu m$ |
perimeter | $\mu m$ |
euler_number | -- |
minor_axis_length | $\mu m$ |
major_axis_length | $\mu m$ |
extent | -- |
If a selected object is linked to an object in the next frame, the instantaneous velocity will be calculated based on the displacement of the object centroid and the frame rate of the video.
property | unit | description
--- | --- | ---|
vel_mean | [mm/s] | velocity |
vel_N | [--] | number of objects linked|
vel_std | [mm/s] | standard deviation of velocity |
## Plot a settling velocity versus floc size
```
# load the excel file as a Pandas DataFrame
df = pd.read_excel('data/floc_props.xlsx')
# see the keys
print(df.keys())
# plot velocity vs major_axis_length
f, ax = plt.subplots(1,1, figsize=(3.5, 2.2), dpi=250)
# note: remove *10 factor if floc_props.xlsx file is updated: previous version was output in [cm/s]
ax.plot(df.major_axis_length, df.vel_mean*10, marker='o', linestyle='None')
ax.set_xlabel('Floc size [$\mu m$]')
ax.set_ylabel('Settling velocity [mm/s]')
# convert to log-log
ax.loglog()
ax.axis([100, 5000, 0.1, 100])
for axis in [ax.xaxis, ax.yaxis]:
axis.set_major_formatter(ScalarFormatter())
plt.tight_layout()
save = True
if save:
plt.savefig('png/vel_size.png', dpi=900)
```
|
github_jupyter
|
```
from sys import modules
IN_COLAB = 'google.colab' in modules
if IN_COLAB:
!pip install -q ir_axioms[examples] python-terrier
# Start/initialize PyTerrier.
from pyterrier import started, init
if not started():
init(tqdm="auto", no_download=True)
from pyterrier.datasets import get_dataset, Dataset
# Load dataset.
dataset_name = "msmarco-passage"
dataset: Dataset = get_dataset(f"irds:{dataset_name}")
dataset_train: Dataset = get_dataset(f"irds:{dataset_name}/trec-dl-2019/judged")
dataset_test: Dataset = get_dataset(f"irds:{dataset_name}/trec-dl-2020/judged")
from pathlib import Path
cache_dir = Path("cache/")
index_dir = cache_dir / "indices" / dataset_name.split("/")[0]
from pyterrier.index import IterDictIndexer
if not index_dir.exists():
indexer = IterDictIndexer(str(index_dir.absolute()))
indexer.index(
dataset.get_corpus_iter(),
fields=["text"]
)
from pyterrier.batchretrieve import BatchRetrieve
# BM25 baseline retrieval.
bm25 = BatchRetrieve(str(index_dir.absolute()), wmodel="BM25")
from ir_axioms.axiom import (
ArgUC, QTArg, QTPArg, aSL, PROX1, PROX2, PROX3, PROX4, PROX5, TFC1, TFC3, RS_TF, RS_TF_IDF, RS_BM25, RS_PL2, RS_QL,
AND, LEN_AND, M_AND, LEN_M_AND, DIV, LEN_DIV, M_TDC, LEN_M_TDC, STMC1, STMC1_f, STMC2, STMC2_f, LNC1, TF_LNC, LB1,
REG, ANTI_REG, REG_f, ANTI_REG_f, ASPECT_REG, ASPECT_REG_f, ORIG
)
axioms = [
~ArgUC(), ~QTArg(), ~QTPArg(), ~aSL(),
~LNC1(), ~TF_LNC(), ~LB1(),
~PROX1(), ~PROX2(), ~PROX3(), ~PROX4(), ~PROX5(),
~REG(), ~REG_f(), ~ANTI_REG(), ~ANTI_REG_f(), ~ASPECT_REG(), ~ASPECT_REG_f(),
~AND(), ~LEN_AND(), ~M_AND(), ~LEN_M_AND(), ~DIV(), ~LEN_DIV(),
~RS_TF(), ~RS_TF_IDF(), ~RS_BM25(), ~RS_PL2(), ~RS_QL(),
~TFC1(), ~TFC3(), ~M_TDC(), ~LEN_M_TDC(),
~STMC1(), ~STMC1_f(), ~STMC2(), ~STMC2_f(),
ORIG()
]
from sklearn.ensemble import RandomForestClassifier
from ir_axioms.modules.pivot import MiddlePivotSelection
from ir_axioms.backend.pyterrier.estimator import EstimatorKwikSortReranker
random_forest = RandomForestClassifier(
max_depth=3,
)
kwiksort_random_forest = bm25 % 20 >> EstimatorKwikSortReranker(
axioms=axioms,
estimator=random_forest,
index=index_dir,
dataset=dataset_name,
pivot_selection=MiddlePivotSelection(),
cache_dir=cache_dir,
verbose=True,
)
kwiksort_random_forest.fit(dataset_train.get_topics(), dataset_train.get_qrels())
from pyterrier.pipelines import Experiment
from ir_measures import nDCG, MAP, RR
experiment = Experiment(
[bm25, kwiksort_random_forest ^ bm25],
dataset_test.get_topics(),
dataset_test.get_qrels(),
[nDCG @ 10, RR, MAP],
["BM25", "KwikSort Random Forest"],
verbose=True,
)
experiment.sort_values(by="nDCG@10", ascending=False, inplace=True)
experiment
random_forest.feature_importances_
```
|
github_jupyter
|
```
# -*- coding: utf-8 -*-
"""
EVCのためのEV-GMMを構築します. そして, 適応学習する.
詳細 : https://pdfs.semanticscholar.org/cbfe/71798ded05fb8bf8674580aabf534c4dbb8bc.pdf
This program make EV-GMM for EVC. Then, it make adaptation learning.
Check detail : https://pdfs.semanticscholar.org/cbfe/71798ded05fb8bf8674580abf534c4dbb8bc.pdf
"""
from __future__ import division, print_function
import os
from shutil import rmtree
import argparse
import glob
import pickle
import time
import numpy as np
from numpy.linalg import norm
from sklearn.decomposition import PCA
from sklearn.mixture import GMM # sklearn 0.20.0から使えない
from sklearn.preprocessing import StandardScaler
import scipy.signal
import scipy.sparse
%matplotlib inline
import matplotlib.pyplot as plt
import IPython
from IPython.display import Audio
import soundfile as sf
import wave
import pyworld as pw
import librosa.display
from dtw import dtw
import warnings
warnings.filterwarnings('ignore')
"""
Parameters
__Mixtured : GMM混合数
__versions : 実験セット
__convert_source : 変換元話者のパス
__convert_target : 変換先話者のパス
"""
# parameters
__Mixtured = 40
__versions = 'pre-stored0.1.1'
__convert_source = 'input/EJM10/V01/T01/TIMIT/000/*.wav'
__convert_target = 'adaptation/EJM04/V01/T01/ATR503/A/*.wav'
# settings
__same_path = './utterance/' + __versions + '/'
__output_path = __same_path + 'output/EJM04/' # EJF01, EJF07, EJM04, EJM05
Mixtured = __Mixtured
pre_stored_pickle = __same_path + __versions + '.pickle'
pre_stored_source_list = __same_path + 'pre-source/**/V01/T01/**/*.wav'
pre_stored_list = __same_path + "pre/**/V01/T01/**/*.wav"
#pre_stored_target_list = "" (not yet)
pre_stored_gmm_init_pickle = __same_path + __versions + '_init-gmm.pickle'
pre_stored_sv_npy = __same_path + __versions + '_sv.npy'
save_for_evgmm_covarXX = __output_path + __versions + '_covarXX.npy'
save_for_evgmm_covarYX = __output_path + __versions + '_covarYX.npy'
save_for_evgmm_fitted_source = __output_path + __versions + '_fitted_source.npy'
save_for_evgmm_fitted_target = __output_path + __versions + '_fitted_target.npy'
save_for_evgmm_weights = __output_path + __versions + '_weights.npy'
save_for_evgmm_source_means = __output_path + __versions + '_source_means.npy'
for_convert_source = __same_path + __convert_source
for_convert_target = __same_path + __convert_target
converted_voice_npy = __output_path + 'sp_converted_' + __versions
converted_voice_wav = __output_path + 'sp_converted_' + __versions
mfcc_save_fig_png = __output_path + 'mfcc3dim_' + __versions
f0_save_fig_png = __output_path + 'f0_converted' + __versions
converted_voice_with_f0_wav = __output_path + 'sp_f0_converted' + __versions
EPSILON = 1e-8
class MFCC:
"""
MFCC() : メル周波数ケプストラム係数(MFCC)を求めたり、MFCCからスペクトルに変換したりするクラス.
動的特徴量(delta)が実装途中.
ref : http://aidiary.hatenablog.com/entry/20120225/1330179868
"""
def __init__(self, frequency, nfft=1026, dimension=24, channels=24):
"""
各種パラメータのセット
nfft : FFTのサンプル点数
frequency : サンプリング周波数
dimension : MFCC次元数
channles : メルフィルタバンクのチャンネル数(dimensionに依存)
fscale : 周波数スケール軸
filterbankl, fcenters : フィルタバンク行列, フィルタバンクの頂点(?)
"""
self.nfft = nfft
self.frequency = frequency
self.dimension = dimension
self.channels = channels
self.fscale = np.fft.fftfreq(self.nfft, d = 1.0 / self.frequency)[: int(self.nfft / 2)]
self.filterbank, self.fcenters = self.melFilterBank()
def hz2mel(self, f):
"""
周波数からメル周波数に変換
"""
return 1127.01048 * np.log(f / 700.0 + 1.0)
def mel2hz(self, m):
"""
メル周波数から周波数に変換
"""
return 700.0 * (np.exp(m / 1127.01048) - 1.0)
def melFilterBank(self):
"""
メルフィルタバンクを生成する
"""
fmax = self.frequency / 2
melmax = self.hz2mel(fmax)
nmax = int(self.nfft / 2)
df = self.frequency / self.nfft
dmel = melmax / (self.channels + 1)
melcenters = np.arange(1, self.channels + 1) * dmel
fcenters = self.mel2hz(melcenters)
indexcenter = np.round(fcenters / df)
indexstart = np.hstack(([0], indexcenter[0:self.channels - 1]))
indexstop = np.hstack((indexcenter[1:self.channels], [nmax]))
filterbank = np.zeros((self.channels, nmax))
for c in np.arange(0, self.channels):
increment = 1.0 / (indexcenter[c] - indexstart[c])
# np,int_ は np.arangeが[0. 1. 2. ..]となるのをintにする
for i in np.int_(np.arange(indexstart[c], indexcenter[c])):
filterbank[c, i] = (i - indexstart[c]) * increment
decrement = 1.0 / (indexstop[c] - indexcenter[c])
# np,int_ は np.arangeが[0. 1. 2. ..]となるのをintにする
for i in np.int_(np.arange(indexcenter[c], indexstop[c])):
filterbank[c, i] = 1.0 - ((i - indexcenter[c]) * decrement)
return filterbank, fcenters
def mfcc(self, spectrum):
"""
スペクトルからMFCCを求める.
"""
mspec = []
mspec = np.log10(np.dot(spectrum, self.filterbank.T))
mspec = np.array(mspec)
return scipy.fftpack.realtransforms.dct(mspec, type=2, norm="ortho", axis=-1)
def delta(self, mfcc):
"""
MFCCから動的特徴量を求める.
現在は,求める特徴量フレームtをt-1とt+1の平均としている.
"""
mfcc = np.concatenate([
[mfcc[0]],
mfcc,
[mfcc[-1]]
]) # 最初のフレームを最初に、最後のフレームを最後に付け足す
delta = None
for i in range(1, mfcc.shape[0] - 1):
slope = (mfcc[i+1] - mfcc[i-1]) / 2
if delta is None:
delta = slope
else:
delta = np.vstack([delta, slope])
return delta
def imfcc(self, mfcc, spectrogram):
"""
MFCCからスペクトルを求める.
"""
im_sp = np.array([])
for i in range(mfcc.shape[0]):
mfcc_s = np.hstack([mfcc[i], [0] * (self.channels - self.dimension)])
mspectrum = scipy.fftpack.idct(mfcc_s, norm='ortho')
# splrep はスプライン補間のための補間関数を求める
tck = scipy.interpolate.splrep(self.fcenters, np.power(10, mspectrum))
# splev は指定座標での補間値を求める
im_spectrogram = scipy.interpolate.splev(self.fscale, tck)
im_sp = np.concatenate((im_sp, im_spectrogram), axis=0)
return im_sp.reshape(spectrogram.shape)
def trim_zeros_frames(x, eps=1e-7):
"""
無音区間を取り除く.
"""
T, D = x.shape
s = np.sum(np.abs(x), axis=1)
s[s < 1e-7] = 0.
return x[s > eps]
def analyse_by_world_with_harverst(x, fs):
"""
WORLD音声分析合成器で基本周波数F0,スペクトル包絡,非周期成分を求める.
基本周波数F0についてはharvest法により,より精度良く求める.
"""
# 4 Harvest with F0 refinement (using Stonemask)
frame_period = 5
_f0_h, t_h = pw.harvest(x, fs, frame_period=frame_period)
f0_h = pw.stonemask(x, _f0_h, t_h, fs)
sp_h = pw.cheaptrick(x, f0_h, t_h, fs)
ap_h = pw.d4c(x, f0_h, t_h, fs)
return f0_h, sp_h, ap_h
def wavread(file):
"""
wavファイルから音声トラックとサンプリング周波数を抽出する.
"""
wf = wave.open(file, "r")
fs = wf.getframerate()
x = wf.readframes(wf.getnframes())
x = np.frombuffer(x, dtype= "int16") / 32768.0
wf.close()
return x, float(fs)
def preEmphasis(signal, p=0.97):
"""
MFCC抽出のための高域強調フィルタ.
波形を通すことで,高域成分が強調される.
"""
return scipy.signal.lfilter([1.0, -p], 1, signal)
def alignment(source, target, path):
"""
タイムアライメントを取る.
target音声をsource音声の長さに合うように調整する.
"""
# ここでは814に合わせよう(targetに合わせる)
# p_p = 0 if source.shape[0] > target.shape[0] else 1
#shapes = source.shape if source.shape[0] > target.shape[0] else target.shape
shapes = source.shape
align = np.array([])
for (i, p) in enumerate(path[0]):
if i != 0:
if j != p:
temp = np.array(target[path[1][i]])
align = np.concatenate((align, temp), axis=0)
else:
temp = np.array(target[path[1][i]])
align = np.concatenate((align, temp), axis=0)
j = p
return align.reshape(shapes)
"""
pre-stored学習のためのパラレル学習データを作る。
時間がかかるため、利用できるlearn-data.pickleがある場合はそれを利用する。
それがない場合は一から作り直す。
"""
timer_start = time.time()
if os.path.exists(pre_stored_pickle):
print("exist, ", pre_stored_pickle)
with open(pre_stored_pickle, mode='rb') as f:
total_data = pickle.load(f)
print("open, ", pre_stored_pickle)
print("Load pre-stored time = ", time.time() - timer_start , "[sec]")
else:
source_mfcc = []
#source_data_sets = []
for name in sorted(glob.iglob(pre_stored_source_list, recursive=True)):
print(name)
x, fs = sf.read(name)
f0, sp, ap = analyse_by_world_with_harverst(x, fs)
mfcc = MFCC(fs)
source_mfcc_temp = mfcc.mfcc(sp)
#source_data = np.hstack([source_mfcc_temp, mfcc.delta(source_mfcc_temp)]) # static & dynamic featuers
source_mfcc.append(source_mfcc_temp)
#source_data_sets.append(source_data)
total_data = []
i = 0
_s_len = len(source_mfcc)
for name in sorted(glob.iglob(pre_stored_list, recursive=True)):
print(name, len(total_data))
x, fs = sf.read(name)
f0, sp, ap = analyse_by_world_with_harverst(x, fs)
mfcc = MFCC(fs)
target_mfcc = mfcc.mfcc(sp)
dist, cost, acc, path = dtw(source_mfcc[i%_s_len], target_mfcc, dist=lambda x, y: norm(x - y, ord=1))
#print('Normalized distance between the two sounds:' + str(dist))
#print("target_mfcc = {0}".format(target_mfcc.shape))
aligned = alignment(source_mfcc[i%_s_len], target_mfcc, path)
#target_data_sets = np.hstack([aligned, mfcc.delta(aligned)]) # static & dynamic features
#learn_data = np.hstack((source_data_sets[i], target_data_sets))
learn_data = np.hstack([source_mfcc[i%_s_len], aligned])
total_data.append(learn_data)
i += 1
with open(pre_stored_pickle, 'wb') as output:
pickle.dump(total_data, output)
print("Make, ", pre_stored_pickle)
print("Make pre-stored time = ", time.time() - timer_start , "[sec]")
"""
全事前学習出力話者からラムダを推定する.
ラムダは適応学習で変容する.
"""
S = len(total_data)
D = int(total_data[0].shape[1] / 2)
print("total_data[0].shape = ", total_data[0].shape)
print("S = ", S)
print("D = ", D)
timer_start = time.time()
if os.path.exists(pre_stored_gmm_init_pickle):
print("exist, ", pre_stored_gmm_init_pickle)
with open(pre_stored_gmm_init_pickle, mode='rb') as f:
initial_gmm = pickle.load(f)
print("open, ", pre_stored_gmm_init_pickle)
print("Load initial_gmm time = ", time.time() - timer_start , "[sec]")
else:
initial_gmm = GMM(n_components = Mixtured, covariance_type = 'full')
initial_gmm.fit(np.vstack(total_data))
with open(pre_stored_gmm_init_pickle, 'wb') as output:
pickle.dump(initial_gmm, output)
print("Make, ", initial_gmm)
print("Make initial_gmm time = ", time.time() - timer_start , "[sec]")
weights = initial_gmm.weights_
source_means = initial_gmm.means_[:, :D]
target_means = initial_gmm.means_[:, D:]
covarXX = initial_gmm.covars_[:, :D, :D]
covarXY = initial_gmm.covars_[:, :D, D:]
covarYX = initial_gmm.covars_[:, D:, :D]
covarYY = initial_gmm.covars_[:, D:, D:]
fitted_source = source_means
fitted_target = target_means
"""
SVはGMMスーパーベクトルで、各pre-stored学習における出力話者について平均ベクトルを推定する。
GMMの学習を見てみる必要があるか?
"""
timer_start = time.time()
if os.path.exists(pre_stored_sv_npy):
print("exist, ", pre_stored_sv_npy)
sv = np.load(pre_stored_sv_npy)
print("open, ", pre_stored_sv_npy)
print("Load pre_stored_sv time = ", time.time() - timer_start , "[sec]")
else:
sv = []
for i in range(S):
gmm = GMM(n_components = Mixtured, params = 'm', init_params = '', covariance_type = 'full')
gmm.weights_ = initial_gmm.weights_
gmm.means_ = initial_gmm.means_
gmm.covars_ = initial_gmm.covars_
gmm.fit(total_data[i])
sv.append(gmm.means_)
sv = np.array(sv)
np.save(pre_stored_sv_npy, sv)
print("Make pre_stored_sv time = ", time.time() - timer_start , "[sec]")
"""
各事前学習出力話者のGMM平均ベクトルに対して主成分分析(PCA)を行う.
PCAで求めた固有値と固有ベクトルからeigenvectorsとbiasvectorsを作る.
"""
timer_start = time.time()
#source_pca
source_n_component, source_n_features = sv[:, :, :D].reshape(S, Mixtured*D).shape
# 標準化(分散を1、平均を0にする)
source_stdsc = StandardScaler()
# 共分散行列を求める
source_X_std = source_stdsc.fit_transform(sv[:, :, :D].reshape(S, Mixtured*D))
# PCAを行う
source_cov = source_X_std.T @ source_X_std / (source_n_component - 1)
source_W, source_V_pca = np.linalg.eig(source_cov)
print(source_W.shape)
print(source_V_pca.shape)
# データを主成分の空間に変換する
source_X_pca = source_X_std @ source_V_pca
print(source_X_pca.shape)
#target_pca
target_n_component, target_n_features = sv[:, :, D:].reshape(S, Mixtured*D).shape
# 標準化(分散を1、平均を0にする)
target_stdsc = StandardScaler()
#共分散行列を求める
target_X_std = target_stdsc.fit_transform(sv[:, :, D:].reshape(S, Mixtured*D))
#PCAを行う
target_cov = target_X_std.T @ target_X_std / (target_n_component - 1)
target_W, target_V_pca = np.linalg.eig(target_cov)
print(target_W.shape)
print(target_V_pca.shape)
# データを主成分の空間に変換する
target_X_pca = target_X_std @ target_V_pca
print(target_X_pca.shape)
eigenvectors = source_X_pca.reshape((Mixtured, D, S)), target_X_pca.reshape((Mixtured, D, S))
source_bias = np.mean(sv[:, :, :D], axis=0)
target_bias = np.mean(sv[:, :, D:], axis=0)
biasvectors = source_bias.reshape((Mixtured, D)), target_bias.reshape((Mixtured, D))
print("Do PCA time = ", time.time() - timer_start , "[sec]")
"""
声質変換に用いる変換元音声と目標音声を読み込む.
"""
timer_start = time.time()
source_mfcc_for_convert = []
source_sp_for_convert = []
source_f0_for_convert = []
source_ap_for_convert = []
fs_source = None
for name in sorted(glob.iglob(for_convert_source, recursive=True)):
print("source = ", name)
x_source, fs_source = sf.read(name)
f0_source, sp_source, ap_source = analyse_by_world_with_harverst(x_source, fs_source)
mfcc_source = MFCC(fs_source)
#mfcc_s_tmp = mfcc_s.mfcc(sp)
#source_mfcc_for_convert = np.hstack([mfcc_s_tmp, mfcc_s.delta(mfcc_s_tmp)])
source_mfcc_for_convert.append(mfcc_source.mfcc(sp_source))
source_sp_for_convert.append(sp_source)
source_f0_for_convert.append(f0_source)
source_ap_for_convert.append(ap_source)
target_mfcc_for_fit = []
target_f0_for_fit = []
target_ap_for_fit = []
for name in sorted(glob.iglob(for_convert_target, recursive=True)):
print("target = ", name)
x_target, fs_target = sf.read(name)
f0_target, sp_target, ap_target = analyse_by_world_with_harverst(x_target, fs_target)
mfcc_target = MFCC(fs_target)
#mfcc_target_tmp = mfcc_target.mfcc(sp_target)
#target_mfcc_for_fit = np.hstack([mfcc_t_tmp, mfcc_t.delta(mfcc_t_tmp)])
target_mfcc_for_fit.append(mfcc_target.mfcc(sp_target))
target_f0_for_fit.append(f0_target)
target_ap_for_fit.append(ap_target)
# 全部numpy.arrrayにしておく
source_data_mfcc = np.array(source_mfcc_for_convert)
source_data_sp = np.array(source_sp_for_convert)
source_data_f0 = np.array(source_f0_for_convert)
source_data_ap = np.array(source_ap_for_convert)
target_mfcc = np.array(target_mfcc_for_fit)
target_f0 = np.array(target_f0_for_fit)
target_ap = np.array(target_ap_for_fit)
print("Load Input and Target Voice time = ", time.time() - timer_start , "[sec]")
"""
適応話者学習を行う.
つまり,事前学習出力話者から目標話者の空間を作りだす.
適応話者文数ごとにfitted_targetを集めるのは未実装.
"""
timer_start = time.time()
epoch=100
py = GMM(n_components = Mixtured, covariance_type = 'full')
py.weights_ = weights
py.means_ = target_means
py.covars_ = covarYY
fitted_target = None
for i in range(len(target_mfcc)):
print("adaptation = ", i+1, "/", len(target_mfcc))
target = target_mfcc[i]
for x in range(epoch):
print("epoch = ", x)
predict = py.predict_proba(np.atleast_2d(target))
y = np.sum([predict[:, i: i + 1] * (target - biasvectors[1][i])
for i in range(Mixtured)], axis = 1)
gamma = np.sum(predict, axis = 0)
left = np.sum([gamma[i] * np.dot(eigenvectors[1][i].T,
np.linalg.solve(py.covars_, eigenvectors[1])[i])
for i in range(Mixtured)], axis=0)
right = np.sum([np.dot(eigenvectors[1][i].T,
np.linalg.solve(py.covars_, y)[i])
for i in range(Mixtured)], axis = 0)
weight = np.linalg.solve(left, right)
fitted_target = np.dot(eigenvectors[1], weight) + biasvectors[1]
py.means_ = fitted_target
print("Load Input and Target Voice time = ", time.time() - timer_start , "[sec]")
"""
変換に必要なものを残しておく.
"""
np.save(save_for_evgmm_covarXX, covarXX)
np.save(save_for_evgmm_covarYX, covarYX)
np.save(save_for_evgmm_fitted_source, fitted_source)
np.save(save_for_evgmm_fitted_target, fitted_target)
np.save(save_for_evgmm_weights, weights)
np.save(save_for_evgmm_source_means, source_means)
```
|
github_jupyter
|
Code:<a href="https://github.com/lotapp/BaseCode" target="_blank">https://github.com/lotapp/BaseCode</a>
多图旧排版:<a href="https://www.cnblogs.com/dunitian/p/9119986.html" target="_blank">https://www.cnblogs.com/dunitian/p/9119986.html</a>
在线编程:<a href="https://mybinder.org/v2/gh/lotapp/BaseCode/master" target="_blank">https://mybinder.org/v2/gh/lotapp/BaseCode/master</a>
Python设计的目的就是 ==> **让程序员解放出来,不要过于关注代码本身**
步入正题:**欢迎提出更简单或者效率更高的方法**
**基础系列**:(这边重点说说`Python`,上次讲过的东西我就一笔带过了)
## 1.基础回顾
### 1.1.输出+类型转换
```
user_num1=input("输入第一个数:")
user_num2=input("输入第二个数:")
print("两数之和:%d"%(int(user_num1)+int(user_num2)))
```
### 1.2.字符串拼接+拼接输出方式
```
user_name=input("输入昵称:")
user_pass=input("输入密码:")
user_url="192.168.1.121"
#拼接输出方式一:
print("ftp://"+user_name+":"+user_pass+"@"+user_url)
#拼接输出方式二:
print("ftp://%s:%s@%s"%(user_name,user_pass,user_url))
```
## 2.字符串遍历、下标、切片
### 2.1.Python
重点说下`python`的 **下标**,有点意思,最后一个元素,我们一般都是`len(str)-1`,他可以直接用`-1`,倒2自然就是`-2`了
**最后一个元素:`user_str[-1]`**
user_str[-1]
user_str[len(user_str)-1] #其他编程语言写法
**倒数第二个元素:`user_str[-2]`**
user_str[-1]
user_str[len(user_str)-2] #其他编程语言写法
```
user_str="七大姑曰:工作了吗?八大姨问:买房了吗?异性说:结婚了吗?"
#遍历
for item in user_str:
print(item,end=" ") # 不换行,以“ ”方式拼接
#长度:len(user_str)
len(user_str)
# #第一个元素:user_str[0]
user_str[0]
# 最后一个元素:user_str[-1]
print(user_str[-1])
print(user_str[len(user_str)-1])#其他编程语言写法
#倒数第二个元素:user_str[-2]
print(user_str[-2])
print(user_str[len(user_str)-2])#其他编程语言写法
```
**python切片语法**:`[start_index:end_index:step]` (**end_index取不到**)
eg:`str[1:4]` 取str[1]、str[2]、str[3]
eg:`str[2:]` 取下标为2开始到最后的元素
eg:`str[2:-1]` 取下标为2~到倒数第二个元素(end_index取不到)
eg:`str[1:6:2]` 隔着取~str[1]、str[3]、str[5](案例会详细说)
eg:`str[::-1]` 逆向输出(案例会详细说)
```
it_str="我爱编程,编程爱它,它是程序,程序是谁?"
# eg:取“编程爱它” it_str[5:9]
print(it_str[5:9])
print(it_str[5:-11]) # end_index用-xx也一样
print(it_str[-15:-11])# start_index用-xx也可以
# eg:取“编程爱它,它是程序,程序是谁?” it_str[5:]
print(it_str[5:])# 不写默认取到最后一个
# eg:一个隔一个跳着取("我编,程它它程,序谁") it_str[0::2]
print(it_str[0::2])# step=△index(eg:0,1,2,3。这里的step=> 2-0 => 间隔1)
# eg:倒序输出 it_str[::-1]
# end_index不写默认是取到最后一个,是正取(从左往右)还是逆取(从右往左),就看step是正是负
print(it_str[::-1])
print(it_str[-1::-1])# 等价于上一个
```
### 2.2.CSharp
这次为了更加形象对比,一句一句翻译成C#
有没有发现规律,`user_str[user_str.Length-1]`==> -1是最后一个
`user_str[user_str.Length-2]`==> -2是最后第二个
python的切片其实就是在这方面简化了
```
%%script csharp
//# # 字符串遍历、下标、切片
//# user_str="七大姑曰:工作了吗?八大姨问:买房了吗?异性说:结婚了吗?"
var user_str = "七大姑曰:工作了吗?八大姨问:买房了吗?异性说:结婚了吗?";
//# #遍历
//# for item in user_str:
//# print(item,end=" ")
foreach (var item in user_str)
{
Console.Write(item);
}
//# #长度:len(user_str)
//# print(len(user_str))
Console.WriteLine(user_str.Length);
//# #第一个元素:user_str[0]
//# print(user_str[0])
Console.WriteLine(user_str[0]);
//# #最后一个元素:user_str[-1]
//# print(user_str[-1])
//# print(user_str[len(user_str)-1])#其他编程语言写法
Console.WriteLine(user_str[user_str.Length - 1]);
//
//# #倒数第二个元素:user_str[-2]
//# print(user_str[-2])
Console.WriteLine(user_str[user_str.Length - 2]);
```
其实你用`Pytho`n跟其他语言对比反差更大,`net`真的很强大了。
补充(对比看就清楚`Python`的`step`为什么是2了,i+=2==>2)
```
%%script csharp
//# 切片:[start_index:end_index:step] (end_index取不到)
//# eg:str[1:4] 取str[1]、str[2]、str[3]
//# eg:str[2:] 取下标为2开始到最后的元素
//# eg:str[2:-1] 取下标为2~到倒数第二个元素(end_index取不到)
//# eg:str[1:6:2] 隔着取~str[1]、str[3]、str[5](案例会详细说)
//# eg:str[::-1] 逆向输出(案例会详细说,)
//
var it_str = "我爱编程,编程爱它,它是程序,程序是谁?";
//
//#eg:取“编程爱它” it_str[5:9]
// print(it_str[5:9])
// print(it_str[5:-11]) #end_index用-xx也一样
// print(it_str[-15:-11])#start_index用-xx也可以
//Substring(int startIndex, int length)
Console.WriteLine(it_str.Substring(5, 4));//第二个参数是长度
//
//#eg:取“编程爱它,它是程序,程序是谁?” it_str[5:]
// print(it_str[5:])#不写默认取到最后一个
Console.WriteLine(it_str.Substring(5));//不写默认取到最后一个
//#eg:一个隔一个跳着取("我编,程它它程,序谁") it_str[0::2]
// print(it_str[0::2])#step=△index(eg:0,1,2,3。这里的step=> 2-0 => 间隔1)
//这个我第一反应是用linq ^_^
for (int i = 0; i < it_str.Length; i += 2)//对比看就清除Python的step为什么是2了,i+=2==》2
{
Console.Write(it_str[i]);
}
Console.WriteLine("\n倒序:");
//#eg:倒序输出 it_str[::-1]
//# end_index不写默认是取到最后一个,是正取(从左往右)还是逆取(从右往左),就看step是正是负
// print(it_str[::-1])
// print(it_str[-1::-1])#等价于上一个
for (int i = it_str.Length - 1; i >= 0; i--)
{
Console.Write(it_str[i]);
}
//其实可以用Linq:Console.WriteLine(new string(it_str.ToCharArray().Reverse().ToArray()));
```
## 3.Python字符串方法系列
### 3.1.Python查找
`find`,`rfind`,`index`,`rindex`
Python查找 **推荐**你用`find`和`rfind`
```
test_str = "ABCDabcdefacddbdf"
# 查找:find,rfind,index,rindex
# xxx.find(str, start, end)
print(test_str.find("cd"))#从左往右
print(test_str.rfind("cd"))#从右往左
print(test_str.find("dnt"))#find和rfind找不到就返回-1
# index和rindex用法和find一样,只是找不到会报错(以后用find系即可)
print(test_str.index("dnt"))
```
### 3.2.Python计数
python:`xxx.count(str, start, end)`
```
# 计数:count
# xxx.count(str, start, end)
print(test_str.count("d"))#4
print(test_str.count("cd"))#2
```
### 3.3.Python替换
Python:`xxx.replace(str1, str2, 替换次数)`
```
# 替换:replace
# xxx.replace(str1, str2, 替换次数)
print(test_str)
print(test_str.replace("b","B"))#并没有改变原字符串,只是生成了一个新的字符串
print(test_str)
# replace可以指定替换几次
print(test_str.replace("b","B",1))#ABCDaBcdefacddbdf
```
### 3.4.Python分割
`split`(按指定字符分割),`splitlines`(按行分割)
`partition`(以str分割成三部分,str前,str和str后),`rpartition`(从右边开始)
说下 **split的切片用法**:`print(test_input.split(" ",3))` 在第三个空格处切片,后面的不切了
```
# 分割:split(按指定字符分割),splitlines(按行分割),partition(以str分割成三部分,str前,str和str后),rpartition
test_list=test_str.split("a")#a有两个,按照a分割,那么会分成三段,返回类型是列表(List),并且返回结果中没有a
print(test_list)
test_input="hi my name is dnt"
print(test_input.split(" ")) #返回列表格式(后面会说)['hi', 'my', 'name', 'is', 'dnt']
print(test_input.split(" ",3))#在第三个空格处切片,后面的不管了
```
继续说说`splitlines`(按行分割),和`split("\n")`的区别:
```
# splitlines()按行分割,返回类型为List
test_line_str="abc\nbca\ncab\n"
print(test_line_str.splitlines())#['abc', 'bca', 'cab']
print(test_line_str.split("\n"))#看出区别了吧:['abc', 'bca', 'cab', '']
# splitlines(按行分割),和split("\n")的区别没看出来就再来个案例
test_line_str2="abc\nbca\ncab\nLLL"
print(test_line_str2.splitlines())#['abc', 'bca', 'cab', 'LLL']
print(test_line_str2.split("\n"))#再提示一下,最后不是\n就和上面一样效果
```
扩展:`split()`,默认按 **空字符**切割(`空格、\t、\n`等等,不用担心返回`''`)
```
# 扩展:split(),默认按空字符切割(空格、\t、\n等等,不用担心返回'')
print("hi my name is dnt\t\n m\n\t\n".split())
```
最后说一下`partition`和`rpartition`: 返回是元祖类型(后面会说的)
方式和find一样,找到第一个匹配的就罢工了【**注意一下没找到的情况**】
```
# partition(以str分割成三部分,str前,str和str后)
# 返回是元祖类型(后面会说的),方式和find一样,找到第一个匹配的就罢工了【注意一下没找到的情况】
print(test_str.partition("cd"))#('ABCDab', 'cd', 'efacddbdf')
print(test_str.rpartition("cd"))#('ABCDabcdefa', 'cd', 'dbdf')
print(test_str.partition("感觉自己萌萌哒"))#没找到:('ABCDabcdefacddbdf', '', '')
```
### 3.5.Python字符串连接
**join** :`"-".join(test_list)`
```
# 连接:join
# separat.join(xxx)
# 错误用法:xxx.join("-")
print("-".join(test_list))
```
### 3.6.Python头尾判断
`startswith`(以。。。开头),`endswith`(以。。。结尾)
```
# 头尾判断:startswith(以。。。开头),endswith(以。。。结尾)
# test_str.startswith(以。。。开头)
start_end_str="http://www.baidu.net"
print(start_end_str.startswith("https://") or start_end_str.startswith("http://"))
print(start_end_str.endswith(".com"))
```
### 3.7.Python大小写系
`lower`(字符串转换为小写),`upper`(字符串转换为大写)
`title`(单词首字母大写),`capitalize`(第一个字符大写,其他变小写)
```
# 大小写系:lower(字符串转换为小写),upper(字符串转换为大写)
# title(单词首字母大写),capitalize(第一个字符大写,其他变小写)
print(test_str)
print(test_str.upper())#ABCDABCDEFACDDBDF
print(test_str.lower())#abcdabcdefacddbdf
print(test_str.capitalize())#第一个字符大写,其他变小写
```
### 3.8.Python格式系列
`lstrip`(去除左边空格),`rstrip`(去除右边空格)
**`strip`** (去除两边空格)美化输出系列:`ljust`,`rjust`,`center`
`ljust,rjust,center`这些就不说了,python经常在linux终端中输出,所以这几个用的比较多
```
# 格式系列:lstrip(去除左边空格),rstrip(去除右边空格),strip(去除两边空格)美化输出系列:ljust,rjust,center
strip_str=" I Have a Dream "
print(strip_str.strip()+"|")#我加 | 是为了看清后面空格,没有别的用处
print(strip_str.lstrip()+"|")
print(strip_str.rstrip()+"|")
#这个就是格式化输出,就不讲了
print(test_str.ljust(50))
print(test_str.rjust(50))
print(test_str.center(50))
```
### 3.9.Python验证系列
`isalpha`(是否是纯字母),`isalnum`(是否是数字|字母)
`isdigit`(是否是纯数字),`isspace`(是否是纯空格)
注意~ `test_str5=" \t \n "` # **isspace() ==>true**
```
# 验证系列:isalpha(是否是纯字母),isalnum(是否是数字|字母),isdigit(是否是纯数字),isspace(是否是纯空格)
# 注意哦~ test_str5=" \t \n " #isspace() ==>true
test_str2="Abcd123"
test_str3="123456"
test_str4=" \t" #isspace() ==>true
test_str5=" \t \n " #isspace() ==>true
test_str.isalpha() #是否是纯字母
test_str.isalnum() #是否是数字|字母
test_str.isdigit() #是否是纯数字
test_str.isspace() #是否是纯空格
test_str2.isalnum() #是否是数字和字母组成
test_str2.isdigit() #是否是纯数字
test_str3.isdigit() #是否是纯数字
test_str5.isspace() #是否是纯空格
test_str4.isspace() #是否是纯空格
```
### Python补充
像这些方法练习用`ipython3`就好了(`sudo apt-get install ipython3`)
code的话需要一个个的print,比较麻烦(我这边因为需要写文章,所以只能一个个code)
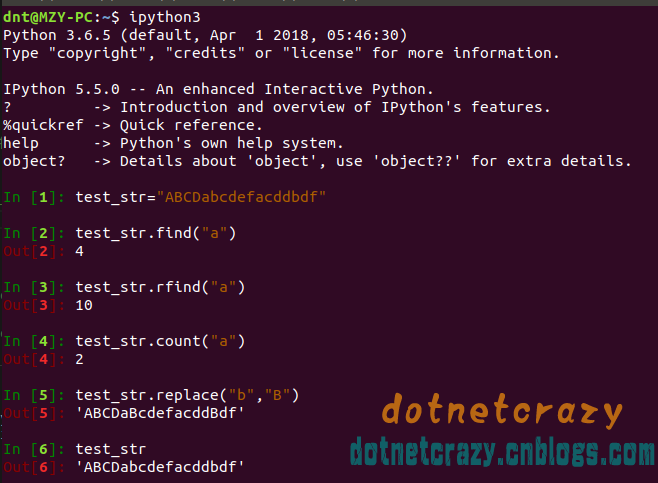
## 4.CSharp字符串方法系列
### 4.1.查找
`index0f`就相当于python里面的`find`
`LastIndexOf` ==> `rfind`
```
%%script csharp
var test_str = "ABCDabcdefacddbdf";
//# # 查找:find,rfind,index,rindex
//# # xxx.find(str, start, end)
//# print(test_str.find("cd"))#从左往右
Console.WriteLine(test_str.IndexOf('a'));//4
Console.WriteLine(test_str.IndexOf("cd"));//6
//# print(test_str.rfind("cd"))#从右往左
Console.WriteLine(test_str.LastIndexOf("cd"));//11
//# print(test_str.find("dnt"))#find和rfind找不到就返回-1
Console.WriteLine(test_str.IndexOf("dnt"));//-1
```
### 4.2.计数
这个真用基础来解决的话,两种方法:
第一种自己变形一下:(原字符串长度 - 替换后的长度) / 字符串长度
```csharp
//# # 计数:count
//# # xxx.count(str, start, end)
// print(test_str.count("d"))#4
// print(test_str.count("cd"))#2
// 第一反应,字典、正则、linq,后来想怎么用基础知识解决,于是有了这个~(原字符串长度-替换后的长度)/字符串长度
Console.WriteLine(test_str.Length - test_str.Replace("d", "").Length);//统计单个字符就简单了
Console.WriteLine((test_str.Length - test_str.Replace("cd", "").Length) / "cd".Length);
Console.WriteLine(test_str);//不用担心原字符串改变(python和C#都是有字符串不可变性的)
```
字符串统计另一种方法(<a href="https://github.com/dunitian/LoTCodeBase/tree/master/NetCode/2.面向对象/4.字符串" target="_blank">就用index</a>)
```csharp
int count = 0;
int index = input.IndexOf("abc");
while (index != -1)
{
count++;
index = input.IndexOf("abc", index + 3);//index指向abc的后一位
}
```
### 4.3.替换
替换指定次数的功能有点业余,就不说了,你可以自行思考哦~
```
%%script csharp
var test_str = "ABCDabcdefacddbdf";
Console.WriteLine(test_str.Replace("b", "B"));
```
### 4.4.分割
`split`里面很多重载方法,可以自己去查看下
eg:`Split("\n",StringSplitOptions.RemoveEmptyEntries)`
再说一下这个:`test_str.Split('a');` //返回数组
如果要和Python一样返回列表==》`test_str.Split('a').ToList();` 【需要引用linq的命名空间哦】
```csharp
var test_array = test_str.Split('a');//返回数组(如果要返回列表==》test_str.Split('a').ToList();)
var test_input = "hi my name is dnt";
//# print(test_input.split(" ")) #返回列表格式(后面会说)['hi', 'my', 'name', 'is', 'dnt']
test_input.Split(" ");
//# 按行分割,返回类型为List
var test_line_str = "abc\nbca\ncab\n";
//# print(test_line_str.splitlines())#['abc', 'bca', 'cab']
test_line_str.Split("\n", StringSplitOptions.RemoveEmptyEntries);
```
### 4.5.连接
**`string.Join(分隔符,数组)`**
```csharp
Console.WriteLine(string.Join("-", test_array));//test_array是数组 ABCD-bcdef-cddbdf
```
### 4.6.头尾判断
`StartsWith`(以。。。开头),`EndsWith`(以。。。结尾)
```
%%script csharp
var start_end_str = "http://www.baidu.net";
//# print(start_end_str.startswith("https://") or start_end_str.startswith("http://"))
System.Console.WriteLine(start_end_str.StartsWith("https://") || start_end_str.StartsWith("http://"));
//# print(start_end_str.endswith(".com"))
System.Console.WriteLine(start_end_str.EndsWith(".com"));
```
### 4.7.大小写系
```csharp
//# print(test_str.upper())#ABCDABCDEFACDDBDF
Console.WriteLine(test_str.ToUpper());
//# print(test_str.lower())#abcdabcdefacddbdf
Console.WriteLine(test_str.ToLower());
```
### 4.8.格式化系
`Tirm`很强大,除了去空格还可以去除你想去除的任意字符
net里面`string.Format`各种格式化输出,可以参考,这边就不讲了
```
%%script csharp
var strip_str = " I Have a Dream ";
//# print(strip_str.strip()+"|")#我加 | 是为了看清后面空格,没有别的用处
Console.WriteLine(strip_str.Trim() + "|");
//# print(strip_str.lstrip()+"|")
Console.WriteLine(strip_str.TrimStart() + "|");
//# print(strip_str.rstrip()+"|")
Console.WriteLine(strip_str.TrimEnd() + "|");
```
### 4.9.验证系列
`string.IsNullOrEmpty` 和 `string.IsNullOrWhiteSpace` 是系统自带的
```
%%script csharp
var test_str4 = " \t";
var test_str5 = " \t \n "; //#isspace() ==>true
// string.IsNullOrEmpty 和 string.IsNullOrWhiteSpace 是系统自带的,其他的你需要自己封装一个扩展类
Console.WriteLine(string.IsNullOrEmpty(test_str4)); //false
Console.WriteLine(string.IsNullOrWhiteSpace(test_str4));//true
Console.WriteLine(string.IsNullOrEmpty(test_str5));//false
Console.WriteLine(string.IsNullOrWhiteSpace(test_str5));//true
```
其他的你需要自己封装一个扩展类(eg:<a href="https://github.com/dunitian/LoTCodeBase/blob/master/NetCode/5.逆天类库/LoTLibrary/Validation/ValidationHelper.cs" target="_blank">简单封装</a>)
```csharp
using System;
using System.Collections.Generic;
using System.Linq;
using System.Text.RegularExpressions;
public static partial class ValidationHelper
{
#region 常用验证
#region 集合系列
/// <summary>
/// 判断集合是否有数据
/// </summary>
/// <typeparam name="T"></typeparam>
/// <param name="list"></param>
/// <returns></returns>
public static bool ExistsData<T>(this IEnumerable<T> list)
{
bool b = false;
if (list != null && list.Count() > 0)
{
b = true;
}
return b;
}
#endregion
#region Null判断系列
/// <summary>
/// 判断是否为空或Null
/// </summary>
/// <param name="objStr"></param>
/// <returns></returns>
public static bool IsNullOrWhiteSpace(this string objStr)
{
if (string.IsNullOrWhiteSpace(objStr))
{
return true;
}
else
{
return false;
}
}
/// <summary>
/// 判断类型是否为可空类型
/// </summary>
/// <param name="theType"></param>
/// <returns></returns>
public static bool IsNullableType(Type theType)
{
return (theType.IsGenericType && theType.GetGenericTypeDefinition().Equals(typeof(Nullable<>)));
}
#endregion
#region 数字字符串检查
/// <summary>
/// 是否数字字符串(包括小数)
/// </summary>
/// <param name="objStr">输入字符串</param>
/// <returns></returns>
public static bool IsNumber(this string objStr)
{
try
{
return Regex.IsMatch(objStr, @"^\d+(\.\d+)?$");
}
catch
{
return false;
}
}
/// <summary>
/// 是否是浮点数
/// </summary>
/// <param name="objStr">输入字符串</param>
/// <returns></returns>
public static bool IsDecimal(this string objStr)
{
try
{
return Regex.IsMatch(objStr, @"^(-?\d+)(\.\d+)?$");
}
catch
{
return false;
}
}
#endregion
#endregion
#region 业务常用
#region 中文检测
/// <summary>
/// 检测是否有中文字符
/// </summary>
/// <param name="objStr"></param>
/// <returns></returns>
public static bool IsZhCN(this string objStr)
{
try
{
return Regex.IsMatch(objStr, "[\u4e00-\u9fa5]");
}
catch
{
return false;
}
}
#endregion
#region 邮箱验证
/// <summary>
/// 判断邮箱地址是否正确
/// </summary>
/// <param name="objStr"></param>
/// <returns></returns>
public static bool IsEmail(this string objStr)
{
try
{
return Regex.IsMatch(objStr, @"^([\w-\.]+)@((\[[0-9]{1,3}\.[0-9]{1,3}\.[0-9]{1,3}\.)|(([\w-]+\.)+))([a-zA-Z]{2,4}|[0-9]{1,3})(\]?)$");
}
catch
{
return false;
}
}
#endregion
#region IP系列验证
/// <summary>
/// 是否为ip
/// </summary>
/// <param name="objStr"></param>
/// <returns></returns>
public static bool IsIP(this string objStr)
{
return Regex.IsMatch(objStr, @"^((2[0-4]\d|25[0-5]|[01]?\d\d?)\.){3}(2[0-4]\d|25[0-5]|[01]?\d\d?)$");
}
/// <summary>
/// 判断输入的字符串是否是表示一个IP地址
/// </summary>
/// <param name="objStr">被比较的字符串</param>
/// <returns>是IP地址则为True</returns>
public static bool IsIPv4(this string objStr)
{
string[] IPs = objStr.Split('.');
for (int i = 0; i < IPs.Length; i++)
{
if (!Regex.IsMatch(IPs[i], @"^\d+$"))
{
return false;
}
if (Convert.ToUInt16(IPs[i]) > 255)
{
return false;
}
}
return true;
}
/// <summary>
/// 判断输入的字符串是否是合法的IPV6 地址
/// </summary>
/// <param name="input"></param>
/// <returns></returns>
public static bool IsIPV6(string input)
{
string temp = input;
string[] strs = temp.Split(':');
if (strs.Length > 8)
{
return false;
}
int count = input.GetStrCount("::");
if (count > 1)
{
return false;
}
else if (count == 0)
{
return Regex.IsMatch(input, @"^([\da-f]{1,4}:){7}[\da-f]{1,4}$");
}
else
{
return Regex.IsMatch(input, @"^([\da-f]{1,4}:){0,5}::([\da-f]{1,4}:){0,5}[\da-f]{1,4}$");
}
}
#endregion
#region 网址系列验证
/// <summary>
/// 验证网址是否正确(http:或者https:)【后期添加 // 的情况】
/// </summary>
/// <param name="objStr">地址</param>
/// <returns></returns>
public static bool IsWebUrl(this string objStr)
{
try
{
return Regex.IsMatch(objStr, @"http://([\w-]+\.)+[\w-]+(/[\w- ./?%&=]*)?|https://([\w-]+\.)+[\w-]+(/[\w- ./?%&=]*)?");
}
catch
{
return false;
}
}
/// <summary>
/// 判断输入的字符串是否是一个超链接
/// </summary>
/// <param name="objStr"></param>
/// <returns></returns>
public static bool IsURL(this string objStr)
{
string pattern = @"^[a-zA-Z]+://(\w+(-\w+)*)(\.(\w+(-\w+)*))*(\?\S*)?$";
return Regex.IsMatch(objStr, pattern);
}
#endregion
#region 邮政编码验证
/// <summary>
/// 验证邮政编码是否正确
/// </summary>
/// <param name="objStr">输入字符串</param>
/// <returns></returns>
public static bool IsZipCode(this string objStr)
{
try
{
return Regex.IsMatch(objStr, @"\d{6}");
}
catch
{
return false;
}
}
#endregion
#region 电话+手机验证
/// <summary>
/// 验证手机号是否正确
/// </summary>
/// <param name="objStr">手机号</param>
/// <returns></returns>
public static bool IsMobile(this string objStr)
{
try
{
return Regex.IsMatch(objStr, @"^13[0-9]{9}|15[012356789][0-9]{8}|18[0123456789][0-9]{8}|147[0-9]{8}$");
}
catch
{
return false;
}
}
/// <summary>
/// 匹配3位或4位区号的电话号码,其中区号可以用小括号括起来,也可以不用,区号与本地号间可以用连字号或空格间隔,也可以没有间隔
/// </summary>
/// <param name="objStr"></param>
/// <returns></returns>
public static bool IsPhone(this string objStr)
{
try
{
return Regex.IsMatch(objStr, "^\\(0\\d{2}\\)[- ]?\\d{8}$|^0\\d{2}[- ]?\\d{8}$|^\\(0\\d{3}\\)[- ]?\\d{7}$|^0\\d{3}[- ]?\\d{7}$");
}
catch
{
return false;
}
}
#endregion
#region 字母或数字验证
/// <summary>
/// 是否只是字母或数字
/// </summary>
/// <param name="objStr"></param>
/// <returns></returns>
public static bool IsAbcOr123(this string objStr)
{
try
{
return Regex.IsMatch(objStr, @"^[0-9a-zA-Z\$]+$");
}
catch
{
return false;
}
}
#endregion
#endregion
}
```
|
github_jupyter
|
## Importing and mapping netCDF data with xarray and cartopy
- Read data from a netCDF file with xarray
- Select (index) and modify variables using xarray
- Create user-defined functions
- Set up map features with cartopy (lat/lon tickmarks, continents, country/state borders); create a function to automate these steps
- Overlay various plot types: contour lines, filled contours, vectors, and barbs
- Customize plot elements such as the colorbar and titles
- Save figure
```
## Imports
import os, sys
import numpy as np
import xarray as xr
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import cartopy.feature as cfeature
from cartopy.mpl.geoaxes import GeoAxes
from cartopy.mpl.ticker import LongitudeFormatter, LatitudeFormatter
```
### Load netcdf data with Xarray
This example demonstrates importing and mapping ERA5 reanalysis data for an AR-Thunderstorm event that occurred in Santa Barbara County on 6 March 2019. The data file created for this example can be found in the `sample-data` folder that 6-hourly ERA5 Reanalysis on a 0.5 x 0.5 deg lat-lon grid 4-8 March. ERA5 data was retrieved from the Climate Data Store and subset to a regional domain over the Western US/N. Pacific.
The xarray package provides an easy interface for importing and analyzing multidimensional data. Because xarray was designed around the netCDF data model, it is an exceptionally powerful tool for working with weather and climate data.
Xarray has two fundamental **data structures**:
**1)** a **`DataArray`**, which holds a single n-dimensional variable. Elements of a DataArray include:
- `values`: numpy array of data values
- `dims`: list of named dimensions (for example, `['time','lat','lon']`)
- `coords`: coordinate arrays (e.g., vectors of lat/lon values or datetime data)
- `atts`: variable attributes such as `units` and `standard_name`
**2)** a **`Dataset`**, which holds multiple n-dimensional variables (shared coordinates). Elements of a Dataset: data variables, dimensions, coordinates, and attributes.
In the cell below, we will load the ERA5 data (netcdf file) into an xarray dataset.
```
# Path to ERA5 data
filepath = "../sample-data/era5.6hr.AR-thunderstorm.20190304_08.nc"
# Read nc file into xarray dataset
ds = xr.open_dataset(filepath)
# Print dataset contents
print(ds)
```
### Selecting/Indexing data with xarray
We can always use regular numpy indexing and slicing on DataArrays and Datasets; however,
it is often more powerful and easier to use xarray’s `.sel()` method of label-based indexing.
```
# Select a single time
ds.sel(time='2019-03-05T18:00:00') # 5 March 2019 at 18 UTC
# Select all times within a single day
ds.sel(time='2019-03-06')
# Select times at 06 UTC
idx = (ds['time.hour'] == 6) # selection uses boolean indexing
hr06 = ds.sel(time=idx) # statements could be combined into a single line
print(hr06)
print(hr06.time.values) # check time coordinates in new dataset
```
In the previous block, we used the `ds['time.hour']` to access the 'hour' component of a datetime object. Other datetime components include 'year', 'month', 'day', 'dayofyear', and 'season'. The 'season' component is unqiue to xarray; valid seasons include 'DJF', 'MAM', 'JJA', and 'SON'.
```
# Select a single grid point
ds.sel(latitude=40, longitude=-120)
# Select the grid point nearest to 34.4208° N, 119.6982°W;
ds.sel(latitude=34.4208, longitude=-119.6982, method='nearest')
# Select range of lats (30-40 N)
# because ERA5 data latitudes are listed from 90N to 90S
# you have to slice from latmax to latmin
latmin=30
latmax=40
ds.sel(latitude=slice(latmax,latmin))
```
Select data at the peak of the AR-Thunderstorm event (06-Mar-2019, 18UTC).
```
# Select the date/time of the AR event (~06 March 2019 at 06 UTC);
# assign subset selection to new dataset `dsAR`
dsAR = ds.sel(time='2019-03-06T06:00:00')
print(dsAR)
# Select data on a single pressure level `plev`
plev = '250'
dsAR = dsAR.sel(level=plev)
```
In the following code block, we select the data and coordinate variables needed to create a map of 250-hPa heights and winds at the time of the AR-Thunderstorm event.
```
# coordinate arrays
lats = dsAR['latitude'].values # .values extracts var as numpy array
lons = dsAR['longitude'].values
#print(lats.shape, lons.shape)
#print(lats)
# data variables
uwnd = dsAR['u'].values
vwnd = dsAR['v'].values
hgts = dsAR['zg'].values
# check the shape and values of
print(hgts.shape)
print(hgts)
```
### Simple arithmetic
Calculate the magnitude of horizontal wind (wind speed) from its u and v components.
Convert wspd data from m/s to knots.
```
# Define a function to calculate wind speed from u and v wind components
def calc_wspd(u, v):
"""Computes wind speed from u and v components"""
wspd = np.sqrt(u**2 + v**2)
return wspd
# Use calc_wspd() function on uwnd & vwnd
wspd = calc_wspd(uwnd, vwnd)
# Define a function to convert m/s to knots
# Hint: 1 m/s = 1.9438445 knots
def to_knots(x):
x_kt = x * 1.9438445
return x_kt
# Convert wspd data to knots, save as separate array
wspd_kt = to_knots(wspd)
print(wspd_kt)
```
### Plotting with Cartopy
Map 250-hPa height lines, isotachs (in knots), and wind vectors or barbs.
```
# Set up map properties
# Projection/Coordinate systems
datacrs = ccrs.PlateCarree() # data/source
mapcrs = ccrs.PlateCarree() # map/destination
# Map extent
lonmin = lons.min()
lonmax = lons.max()
latmin = lats.min()
latmax = lats.max()
# Tickmark Locations
dx = 10; dy = 10
xticks = np.arange(lonmin, lonmax+1, dx) # np.arange(start, stop, interval) returns 1d array
yticks = np.arange(latmin, latmax+1, dy) # that ranges from `start` to `stop-1` by `interval`
print('xticks:', xticks)
print('yticks:', yticks)
```
First, we need to create a basemap to plot our data on. In creating the basemap, we will set the map extent, draw lat/lon tickmarks, and add/customize map features such as coastlines and country borders. Next, use the `contour()` function to draw lines of 250-hPa geopotential heights.
```
# Create figure
fig = plt.figure(figsize=(11,8))
# Add plot axes
ax = fig.add_subplot(111, projection=mapcrs)
ax.set_extent([lonmin,lonmax,latmin,latmax], crs=mapcrs)
# xticks (longitude tickmarks)
ax.set_xticks(xticks, crs=mapcrs)
lon_formatter = LongitudeFormatter()
ax.xaxis.set_major_formatter(lon_formatter)
# yticks (latitude tickmarks)
ax.set_yticks(yticks, crs=mapcrs)
lat_formatter = LatitudeFormatter()
ax.yaxis.set_major_formatter(lat_formatter)
# format tickmarks
ax.tick_params(direction='out', # draws ticks outside of plot (`out`,`in`,`inout)
labelsize=8.5, # font size of ticklabel,
length=5, # lenght of tickmark in points
pad=2, # points between tickmark anmd label
color='black')
# Add map features
ax.add_feature(cfeature.LAND, facecolor='0.9') # color fill land gray
ax.add_feature(cfeature.COASTLINE, edgecolor='k', linewidth=1.0) # coastlines
ax.add_feature(cfeature.BORDERS, edgecolor='0.1', linewidth=0.7) # country borders
ax.add_feature(cfeature.STATES, edgecolor='0.1', linewidth=0.7) # state borders
# Create arr of contour levels using np.arange(start,stop,interval)
clevs_hgts = np.arange(8400,12800,120)
#print(clevs_hgts)
# Draw contour lines for geop heights
cs = ax.contour(lons, lats, hgts, transform=datacrs, # first line= required
levels=clevs_hgts, # contour levels
colors='blue', # line color
linewidths=1.2) # line thickness (default=1.0)
# Add labels to contour lines
plt.clabel(cs, fmt='%d', fontsize=9, inline_spacing=5)
# # Show
plt.show()
```
Create a function that will create and return a figure with a background map. This saves us from having to copy/paste lines 1-27 in the previous block each time we create a new map.
```
def draw_basemap():
# Create figure
fig = plt.figure(figsize=(11,9))
# Add plot axes and draw basemap
ax = fig.add_subplot(111, projection=mapcrs)
ax.set_extent([lonmin,lonmax,latmin,latmax], crs=mapcrs)
# xticks
ax.set_xticks(xticks, crs=mapcrs)
lon_formatter = LongitudeFormatter()
ax.xaxis.set_major_formatter(lon_formatter)
# yticks
ax.set_yticks(yticks, crs=mapcrs)
lat_formatter = LatitudeFormatter()
ax.yaxis.set_major_formatter(lat_formatter)
# tick params
ax.tick_params(direction='out', labelsize=8.5, length=5, pad=2, color='black')
# Map features
ax.add_feature(cfeature.LAND, facecolor='0.9')
ax.add_feature(cfeature.COASTLINE, edgecolor='k', linewidth=1.0)
ax.add_feature(cfeature.BORDERS, edgecolor='0.1', linewidth=0.7)
ax.add_feature(cfeature.STATES, edgecolor='0.1', linewidth=0.7)
return fig, ax
```
Use your `draw_basemap` function to create a new figure and background map. Plot height contours, then use `contourf()` to plot filled contours for wind speed (knots).
```
# Draw basemap
fig, ax = draw_basemap()
# Geopotential Heights (contour lines)
clevs_hgts = np.arange(8400,12800,120)
cs = ax.contour(lons, lats, hgts, transform=datacrs,
levels=clevs_hgts, # contour levels
colors='b', # line color
linewidths=1.2) # line thickness
# Add labels to contour lines
plt.clabel(cs, fmt='%d',fontsize=8.5, inline_spacing=5)
# Wind speed - contour fill
clevs_wspd = np.arange(70,121,10)
cf = ax.contourf(lons, lats, wspd_kt, transform=datacrs,
levels=clevs_wspd,
cmap='BuPu', # colormap
extend='max',
alpha=0.8) # transparency (0=transparent, 1=opaque)
# show
plt.show()
```
Add wind vectors using `quiver()`
```
# Draw basemap
fig, ax = draw_basemap()
# Geopotenital height lines
clevs_hgts = np.arange(840,1280,12)
cs = ax.contour(lons, lats, hgts/10., transform=datacrs,
levels=clevs_hgts,
colors='b', # line color
linewidths=1.2) # line thickness
# Add labels to contour lines
plt.clabel(cs, fmt='%d',fontsize=9, inline_spacing=5)
# Wind speed - contour fill
clevs_wspd = np.arange(70,131,10)
cf = ax.contourf(lons, lats, wspd_kt, transform=datacrs,
levels=clevs_wspd,
cmap='BuPu',
extend='max', # use if max data value
alpha=0.8) # transparency (0=transparent, 1=opaque)
# Wind vectors
ax.quiver(lons, lats, uwnd, vwnd, transform=datacrs,
color='k',
pivot='middle',
regrid_shape=12) # increasing regrid_shape increases the number/density of vectors
# show
plt.show()
```
Plot barbs instead of vectors using `barbs()`
```
# Draw basemap
fig, ax = draw_basemap()
# Geopotenital height lines
clevs_hgts = np.arange(840,1280,12)
cs = ax.contour(lons, lats, hgts/10., transform=datacrs,
levels=clevs_hgts,
colors='b', # line color
linewidths=1.25) # line thickness
# Add labels to contour lines
plt.clabel(cs, fmt='%d',fontsize=9, inline_spacing=5)
# Wind speed - contour fill
clevs_wspd = np.arange(70,131,10)
cf = ax.contourf(lons, lats, wspd_kt, transform=datacrs,
levels=clevs_wspd,
cmap='BuPu',
extend='max',
alpha=0.8) # transparency (0=transparent, 1=opaque)
# Wind barbs
ax.barbs(lons, lats, uwnd, vwnd, transform=datacrs, # uses the same args as quiver
color='k', regrid_shape=12, pivot='middle')
# show
plt.show()
```
Add plot elements such as a colorbar and title. Option to save figure.
```
# Draw basemap
fig, ax = draw_basemap()
# Geopotenital height lines
clevs_hgts = np.arange(8400,12800,120)
cs = ax.contour(lons, lats, hgts, transform=datacrs,
levels=clevs_hgts,
colors='b', # line color
linewidths=1.2) # line thickness
# Add labels to contour lines
plt.clabel(cs, fmt='%d',fontsize=8.5, inline_spacing=5)
# Wind speed - contour fill
clevs_wspd = np.arange(70,131,10)
cf = ax.contourf(lons, lats, wspd_kt, transform=datacrs,
levels=clevs_wspd,
cmap='BuPu',
extend='max',
alpha=0.8) # transparency (0=transparent, 1=opaque)
# Wind barbs
ax.barbs(lons, lats, uwnd, vwnd, transform=datacrs, # uses the same args as quiver
color='k', regrid_shape=12, pivot='middle')
# Add colorbar
cb = plt.colorbar(cf, orientation='vertical', # 'horizontal' or 'vertical'
shrink=0.7, pad=0.03) # fraction to shrink cb by; pad= space between cb and plot
cb.set_label('knots')
# Plot title
titlestring = f"{plev}-hPa Hgts/Wind" # uses new f-string formatting
ax.set_title(titlestring, loc='center',fontsize=13) # loc: {'center','right','left'}
# Save figure
outfile = 'map-250hPa.png'
plt.savefig(outfile,
bbox_inches='tight', # trims excess whitespace from around figure
dpi=300) # resolution in dots per inch
# show
plt.show()
```
|
github_jupyter
|
# Add external catalog for source matching: allWISE catalog
This notebook will create a dabase containing the allWISE all-sky mid-infrared catalog. As the catalogs grows (the allWISE catalog we are inserting contains of the order of hundreds of millions sources), using an index on the geoJSON corrdinate type to support the queries becomes unpractical, as such an index does not compress well. In this case, and healpix based indexing offers a good compromise. We will use an healpix grid of order 16, which has a resolution of ~ 3 arcseconds, simlar to the FWHM of ZTF images.
References, data access, and documentation on the catalog can be found at:
http://wise2.ipac.caltech.edu/docs/release/allwise/
http://irsa.ipac.caltech.edu/data/download/wise-allwise/
This notebook is straight to the point, more like an actual piece of code than a demo. For an explanation of the various steps needed in the see the 'insert_example' notebook in this same folder.
## 1) Inserting:
```
import numpy as np
from healpy import ang2pix
from extcats import CatalogPusher
# build the pusher object and point it to the raw files.
wisep = CatalogPusher.CatalogPusher(
catalog_name = 'wise',
data_source = '../testdata/AllWISE/',
file_type = ".bz2")
# read column names and types from schema file
schema_file = "../testdata/AllWISE/wise-allwise-cat-schema.txt"
names, types = [], {}
with open(schema_file) as schema:
for l in schema:
if "#" in l or (not l.strip()):
continue
name, dtype = zip(
[p.strip() for p in l.strip().split(" ") if not p in [""]])
name, dtype = name[0], dtype[0]
#print (name, dtype)
names.append(name)
# convert the data type
if "char" in dtype:
types[name] = str
elif "decimal" in dtype:
types[name] = np.float64
elif "serial" in dtype or "integer" in dtype:
types[name] = int
elif "smallfloat" in dtype:
types[name] = np.float16
elif "smallint" in dtype:
types[name] = np.int16
elif dtype == "int8":
types[name] = np.int8
else:
print("unknown data type: %s"%dtype)
# select the columns you want to use.
use_cols = []
select = ["Basic Position and Identification Information",
"Primary Photometric Information",
"Measurement Quality and Source Reliability Information",
"2MASS PSC Association Information"]
with open(schema_file) as schema:
blocks = schema.read().split("#")
for block in blocks:
if any([k in block for k in select]):
for l in block.split("\n")[1:]:
if "#" in l or (not l.strip()):
continue
name, dtype = zip(
[p.strip() for p in l.strip().split(" ") if not p in [""]])
use_cols.append(name[0])
print("we will be using %d columns out of %d"%(len(use_cols), len(names)))
# now assign the reader to the catalog pusher object
import pandas as pd
wisep.assign_file_reader(
reader_func = pd.read_csv,
read_chunks = True,
names = names,
usecols = lambda x : x in use_cols,
#dtype = types, #this mess up with NaN values
chunksize=5000,
header=None,
engine='c',
sep='|',
na_values = 'nnnn')
# define the dictionary modifier that will act on the single entries
def modifier(srcdict):
srcdict['hpxid_16'] = int(
ang2pix(2**16, srcdict['ra'], srcdict['dec'], lonlat = True, nest = True))
#srcdict['_id'] = srcdict.pop('source_id') doesn't work, seems it is not unique
return srcdict
wisep.assign_dict_modifier(modifier)
# finally push it in the databse
wisep.push_to_db(
coll_name = 'srcs',
index_on = "hpxid_16",
overwrite_coll = True,
append_to_coll = False)
# if needed print extensive info on database
#wisep.info()
```
## 2) Testing the catalog
At this stage, a simple test is run on the database, consisting in crossmatching with a set of randomly distributed points.
```
# now test the database for query performances. We use
# a sample of randomly distributed points on a sphere
# as targets.
# define the funtion to test coordinate based queries:
from healpy import ang2pix, get_all_neighbours
from astropy.table import Table
from astropy.coordinates import SkyCoord
return_fields = ['designation', 'ra', 'dec']
project = {}
for field in return_fields: project[field] = 1
print (project)
hp_order, rs_arcsec = 16, 30.
def test_query(ra, dec, coll):
"""query collection for points within rs of target ra, dec.
The results as returned as an astropy Table."""
# find the index of the target pixel and its neighbours
target_pix = int( ang2pix(2**hp_order, ra, dec, nest = True, lonlat = True) )
neighbs = get_all_neighbours(2**hp_order, ra, dec, nest = True, lonlat = True)
# remove non-existing neigbours (in case of E/W/N/S) and add center pixel
pix_group = [int(pix_id) for pix_id in neighbs if pix_id != -1] + [target_pix]
# query the database for sources in these pixels
qfilter = { 'hpxid_%d'%hp_order: { '$in': pix_group } }
qresults = [o for o in coll.find(qfilter)]
if len(qresults)==0:
return None
# then use astropy to find the closest match
tab = Table(qresults)
target = SkyCoord(ra, dec, unit = 'deg')
matches_pos = SkyCoord(tab['ra'], tab['dec'], unit = 'deg')
d2t = target.separation(matches_pos).arcsecond
match_id = np.argmin(d2t)
# if it's too far away don't use it
if d2t[match_id]>rs_arcsec:
return None
return tab[match_id]
# run the test
wisep.run_test(test_query, npoints = 10000)
```
# 3) Adding metadata
Once the database is set up and the query performance are satisfactory, metadata describing the catalog content, contact person, and query strategies have to be added to the catalog database. If presents, the keys and parameters for the healpix partitioning of the sources are also to be given, as well as the name of the compound geoJSON/legacy pair entry in the documents.
This information will be added into the 'metadata' collection of the database which will be accessed by the CatalogQuery. The metadata will be stored in a dedicated collection so that the database containig a given catalog will have two collections:
- db['srcs'] : contains the sources.
- db['meta'] : describes the catalog.
```
mqp.healpix_meta(healpix_id_key = 'hpxid_16', order = 16, is_indexed = True, nest = True)
mqp.science_meta(
contact = 'C. Norris',
email = '[email protected]',
description = 'allWISE infrared catalog',
reference = 'http://wise2.ipac.caltech.edu/docs/release/allwise/')
```
|
github_jupyter
|
<a id="title_ID"></a>
# JWST Pipeline Validation Testing Notebook: spec2, extract_2d step
<span style="color:red"> **Instruments Affected**</span>: NIRSpec
Tested on CV3 data
### Table of Contents
<div style="text-align: left">
<br> [Imports](#imports_ID) <br> [Introduction](#intro_ID) <br> [Testing Data Set](#data_ID) <br> [Run the JWST pipeline and assign_wcs validation tests](#pipeline_ID): [FS Full-Frame test](#FULLFRAME), [FS ALLSLITS test](#ALLSLITS), [MOS test](#MOS) <br> [About This Notebook](#about_ID)<br> [Results](#results) <br>
</div>
<a id="imports_ID"></a>
# Imports
The library imports relevant to this notebook are aready taken care of by importing PTT.
* astropy.io for opening fits files
* jwst.module.PipelineStep is the pipeline step being tested
* matplotlib.pyplot.plt to generate plot
NOTE: This notebook assumes that the pipeline version to be tested is already installed and its environment is activated.
To be able to run this notebook you need to install nptt.
If all goes well you will be able to import PTT.
[Top of Page](#title_ID)
```
# Create a temporary directory to hold notebook output, and change the working directory to that directory.
from tempfile import TemporaryDirectory
import os
import shutil
data_dir = TemporaryDirectory()
os.chdir(data_dir.name)
# Choose CRDS cache location
use_local_crds_cache = False
crds_cache_tempdir = False
crds_cache_notebook_dir = True
crds_cache_home = False
crds_cache_custom_dir = False
crds_cache_dir_name = ""
if use_local_crds_cache:
if crds_cache_tempdir:
os.environ['CRDS_PATH'] = os.path.join(os.getcwd(), "crds")
elif crds_cache_notebook_dir:
try:
os.environ['CRDS_PATH'] = os.path.join(orig_dir, "crds")
except Exception as e:
os.environ['CRDS_PATH'] = os.path.join(os.getcwd(), "crds")
elif crds_cache_home:
os.environ['CRDS_PATH'] = os.path.join(os.environ['HOME'], 'crds', 'cache')
elif crds_cache_custom_dir:
os.environ['CRDS_PATH'] = crds_cache_dir_name
import warnings
import psutil
from astropy.io import fits
# Only print a DeprecationWarning the first time it shows up, not every time.
with warnings.catch_warnings():
warnings.simplefilter("once", category=DeprecationWarning)
import jwst
from jwst.pipeline.calwebb_detector1 import Detector1Pipeline
from jwst.assign_wcs.assign_wcs_step import AssignWcsStep
from jwst.msaflagopen.msaflagopen_step import MSAFlagOpenStep
from jwst.extract_2d.extract_2d_step import Extract2dStep
# The latest version of NPTT is installed in the requirements text file at:
# /jwst_validation_notebooks/environment.yml
# import NPTT
import nirspec_pipe_testing_tool as nptt
# To get data from Artifactory
from ci_watson.artifactory_helpers import get_bigdata
# Print the versions used for the pipeline and NPTT
pipeline_version = jwst.__version__
nptt_version = nptt.__version__
print("Using jwst pipeline version: ", pipeline_version)
print("Using NPTT version: ", nptt_version)
```
<a id="intro_ID"></a>
# Test Description
We compared Institute's pipeline product of the assign_wcs step with our benchmark files, or with the intermediary products from the ESA pipeline, which is completely independent from the Institute's. The comparison file is referred to as 'truth'. We calculated the relative difference and expected it to be equal to or less than computer precision: relative_difference = absolute_value( (Truth - ST)/Truth ) <= 1x10^-7.
For the test to be considered PASSED, every single slit (for FS data), slitlet (for MOS data) or slice (for IFU data) in the input file has to pass. If there is any failure, the whole test will be considered as FAILED.
The code for this test can be obtained at: https://github.com/spacetelescope/nirspec_pipe_testing_tool/blob/master/nirspec_pipe_testing_tool/calwebb_spec2_pytests/auxiliary_code/check_corners_extract2d.py. Multi Object Spectroscopy (MOS), the code is in the same repository but is named ```compare_wcs_mos.py```, and for Integral Field Unit (IFU) data, the test is named ```compare_wcs_ifu.py```.
The input file is defined in the variable ```input_file``` (see section [Testing Data Set and Variable Setup](#data_ID)).
Step description: https://jwst-pipeline.readthedocs.io/en/latest/jwst/extract_2d/main.html
Pipeline code: https://github.com/spacetelescope/jwst/tree/master/jwst/extract_2d
### Results
If the test **PASSED** this means that all slits, slitlets, or slices individually passed the test. However, if ony one individual slit (for FS data), slitlet (for MOS data) or slice (for IFU data) test failed, the whole test will be reported as **FAILED**.
### Calibration WG Requested Algorithm:
A short description and link to the page:
https://outerspace.stsci.edu/display/JWSTCC/Vanilla+Path-Loss+Correction
### Defining Term
Acronymns used un this notebook:
pipeline: calibration pipeline
spec2: spectroscopic calibration pipeline level 2b
PTT: NIRSpec pipeline testing tool (https://github.com/spacetelescope/nirspec_pipe_testing_tool)
[Top of Page](#title_ID)
<a id="pipeline_ID"></a>
# Run the JWST pipeline and extract_2d validation tests
The pipeline can be run from the command line in two variants: full or per step.
Tu run the spec2 pipeline in full use the command:
$ strun jwst.pipeline.Spec2Pipeline jwtest_rate.fits
Tu only run the extract_2d step, use the command:
$ strun jwst.extract_2d.Extract2dStep jwtest_previous_step_output.fits
These options are also callable from a script with the testing environment active. The Python call for running the pipeline in full or by step are:
$\gt$ from jwst.pipeline.calwebb_spec2 import Spec2Pipeline
$\gt$ Spec2Pipeline.call(jwtest_rate.fits)
or
$\gt$ from jwst.extract_2d import Extract2dStep
$\gt$ Extract2dStep.call(jwtest_previous_step_output.fits)
PTT can run the spec2 pipeline either in full or per step, as well as the imaging pipeline in full. In this notebook we will use PTT to run the pipeline and the validation tests. To run PTT, follow the directions in the corresponding repo page.
[Top of Page](#title_ID)
<a id="data_ID"></a>
# Testing Data Set
All testing data is from the CV3 campaign. We chose these files because this is our most complete data set, i.e. all modes and filter-grating combinations.
Data used was for testing was only FS and MOS, since extract_2d is skipped for IFU. Data sets are:
- FS_PRISM_CLEAR
- FS_FULLFRAME_G395H_F290LP
- FS_ALLSLITS_G140H_F100LP
- MOS_G140M_LINE1
- MOS_PRISM_CLEAR
[Top of Page](#title_ID)
```
testing_data = {'fs_prism_clear':{
'uncal_file_nrs1': 'fs_prism_nrs1_uncal.fits',
'uncal_file_nrs2': 'fs_prism_nrs2_uncal.fits',
'truth_file_nrs1': 'fs_prism_nrs1_extract_2d_truth.fits',
'truth_file_nrs2': 'fs_prism_nrs2_extract_2d_truth.fits',
'msa_shutter_config': None },
'fs_fullframe_g395h_f290lp':{
'uncal_file_nrs1': 'fs_fullframe_g35h_f290lp_nrs1_uncal.fits',
'uncal_file_nrs2': 'fs_fullframe_g35h_f290lp_nrs2_uncal.fits',
'truth_file_nrs1': 'fs_fullframe_g35h_f290lp_nrs1_extract_2d_truth.fits',
'truth_file_nrs2': 'fs_fullframe_g35h_f290lp_nrs2_extract_2d_truth.fits',
'msa_shutter_config': None },
'fs_allslits_g140h_f100lp':{
'uncal_file_nrs1': 'fs_allslits_g140h_f100lp_nrs1_uncal.fits',
'uncal_file_nrs2': 'fs_allslits_g140h_f100lp_nrs2_uncal.fits',
'truth_file_nrs1': 'fs_allslits_g140h_f100lp_nrs1_extract_2d_truth.fits',
'truth_file_nrs2': 'fs_allslits_g140h_f100lp_nrs2_extract_2d_truth.fits',
'msa_shutter_config': None },
# Commented out because the pipeline is failing with this file
#'bots_g235h_f170lp':{
# 'uncal_file_nrs1': 'bots_g235h_f170lp_nrs1_uncal.fits',
# 'uncal_file_nrs2': 'bots_g235h_f170lp_nrs2_uncal.fits',
# 'truth_file_nrs1': 'bots_g235h_f170lp_nrs1_extract_2d_truth.fits',
# 'truth_file_nrs2': 'bots_g235h_f170lp_nrs2_extract_2d_truth.fits',
# 'msa_shutter_config': None },
'mos_prism_clear':{
'uncal_file_nrs1': 'mos_prism_nrs1_uncal.fits',
'uncal_file_nrs2': 'mos_prism_nrs2_uncal.fits',
'truth_file_nrs1': 'mos_prism_nrs1_extract_2d_truth.fits',
'truth_file_nrs2': None,
'msa_shutter_config': 'V0030006000104_msa.fits' },
'mos_g140m_f100lp':{
'uncal_file_nrs1': 'mos_g140m_line1_NRS1_uncal.fits',
'uncal_file_nrs2': 'mos_g140m_line1_NRS2_uncal.fits',
'truth_file_nrs1': 'mos_g140m_line1_nrs1_extract_2d_truth.fits',
'truth_file_nrs2': 'mos_g140m_line1_nrs2_extract_2d_truth.fits',
'msa_shutter_config': 'V8460001000101_msa.fits' },
}
# define function to pull data from Artifactory
def get_artifactory_file(data_set_dict, detector):
"""This function creates a list with all the files needed per detector to run the test.
Args:
data_set_dict: dictionary, contains inputs for a specific mode and configuration
detector: string, either nrs1 or nrs2
Returns:
data: list, contains all files needed to run test
"""
files2obtain = ['uncal_file_nrs1', 'truth_file_nrs1', 'msa_shutter_config']
data = []
for file in files2obtain:
data_file = None
try:
if '_nrs' in file and '2' in detector:
file = file.replace('_nrs1', '_nrs2')
data_file = get_bigdata('jwst_validation_notebooks',
'validation_data',
'nirspec_data',
data_set_dict[file])
except TypeError:
data.append(None)
continue
data.append(data_file)
return data
# Set common NPTT switches for NPTT and run the test for both detectors in each data set
# define benchmark (or 'truth') file
compare_assign_wcs_and_extract_2d_with_esa = False
# accepted threshold difference with respect to benchmark files
extract_2d_threshold_diff = 4
# define benchmark (or 'truth') file
esa_files_path, raw_data_root_file = None, None
compare_assign_wcs_and_extract_2d_with_esa = False
# Get the data
results_dict = {}
detectors = ['nrs1', 'nrs2']
for mode_config, data_set_dict in testing_data.items():
for det in detectors:
print('Testing files for detector: ', det)
data = get_artifactory_file(data_set_dict, det)
uncal_file, truth_file, msa_shutter_config = data
print('Working with uncal_file: ', uncal_file)
uncal_basename = os.path.basename(uncal_file)
# Make sure that there is an assign_wcs truth product to compare to, else skip this data set
if truth_file is None:
print('No truth file to compare to for this detector, skipping this file. \n')
skip_file = True
else:
skip_file = False
if not skip_file:
# Run the stage 1 pipeline
rate_object = Detector1Pipeline.call(uncal_file)
# Make sure the MSA shutter configuration file is set up correctly
if msa_shutter_config is not None:
msa_metadata = rate_object.meta.instrument.msa_metadata_file
print(msa_metadata)
if msa_metadata is None or msa_metadata == 'N/A':
rate_object.meta.instrument.msa_metadata_file = msa_shutter_config
# Run the stage 2 pipeline steps
pipe_object = AssignWcsStep.call(rate_object)
if 'mos' in uncal_basename.lower():
pipe_object = MSAFlagOpenStep.call(pipe_object)
extract_2d_object = Extract2dStep.call(pipe_object)
# Run the validation test
%matplotlib inline
if 'fs' in uncal_file.lower():
print('Running test for FS...')
result, _ = nptt.calwebb_spec2_pytests.auxiliary_code.check_corners_extract2d.find_FSwindowcorners(
extract_2d_object,
truth_file=truth_file,
esa_files_path=esa_files_path,
extract_2d_threshold_diff=extract_2d_threshold_diff)
if 'mos' in uncal_file.lower():
print('Running test for MOS...')
result, _ = nptt.calwebb_spec2_pytests.auxiliary_code.check_corners_extract2d.find_MOSwindowcorners(
extract_2d_object,
msa_shutter_config,
truth_file=truth_file,
esa_files_path=esa_files_path,
extract_2d_threshold_diff= extract_2d_threshold_diff)
else:
result = 'skipped'
# Did the test passed
print("Did assign_wcs validation test passed? ", result, "\n\n")
rd = {uncal_basename: result}
results_dict.update(rd)
# close all open files
psutil.Process().open_files()
closing_files = []
for fd in psutil.Process().open_files():
if data_dir.name in fd.path:
closing_files.append(fd)
for fd in closing_files:
try:
print('Closing file: ', fd)
open(fd.fd).close()
except:
print('File already closed: ', fd)
# Quickly see if the test passed
print('These are the final results of the tests: ')
for key, val in results_dict.items():
print(key, val)
```
<a id="about_ID"></a>
## About this Notebook
**Author:** Maria A. Pena-Guerrero, Staff Scientist II - Systems Science Support, NIRSpec
<br>**Updated On:** Mar/24/2021
[Top of Page](#title_ID)
<img style="float: right;" src="./stsci_pri_combo_mark_horizonal_white_bkgd.png" alt="stsci_pri_combo_mark_horizonal_white_bkgd" width="200px"/>
|
github_jupyter
|
# Pump Calculations
```
import numpy as np
```
## Power Input
```
#Constants and inputs
g = 32.174; #gravitational acceleration, ft/s^2
rho_LOx = 71.27; #Density of Liquid Oxygen- lbm/ft^3
rho_LCH4 = 26.3; #Density of Liquid Methane- lbm/ft^3
Differential = #Desired pressure differential (psi)
mLOx = #Mass flow of Liquid Oxygen (lb/s)
mLCH4 = #Mass Flow of Liquid Methane (lb/s)
#Head Calculations
HLOx = (((Differential)*144)/(rho_LOx * g))*32.174 #Head of Liquid Oxygen - ft
HLCH4 = (((Differential)*144)/(rho_LCH4 * g))*32.174 #Head of Liquid Methane - ft
#Power Calculations - Assume a 75% effiency (Minimum value that we can reach)
Power_LOx = (((mLOx * g * HLOx)/0.75)/32.174) * 1.36; #Output is in Watts
Power_LCH4 = (((mLCH4 * g * HLCH4)/0.75)/32.174) * 1.36; #Output is in Watts
```
## Impeller Calculations
### Constants
```
QLOx = mLOx/rho_LOx #Volumetric flow rate of Liquid Oxygen in ft^3/s
QLCH4 = mLCH4/rho_LCH4 #Volumetric flow rate of Liquid Methane in ft^3/s
Eff_Vol = #Volumetric effiency is a measure of how much fluid is lost due to leakages, estimate the value
QImp_LOx = QLox/Eff_Vol #Impeller flow rate of Liquid oxygen in ft^3/s
QImp_LCH4 = QLCH4/Eff_Vol #Impeller flow rate of Liquid methane in ft^3/s
n = #RPM of impeller, pick such that nq_LOx is low but not too low
nq_LOx = n * (QImp_LOx ** 0.5)/(HLOx ** 0.75) #Specific speed of Liquid Oxygen
nq_LCH4 = n * (QImp_LCH4 ** 0.5)/(HLCH4 ** 0.75) #Specific speed of Liquid Methane
omegas_LOx = nq_LOx/52.9 #Universal specific speed
omegas_LCH4 = nq_LCH4/52.9 #Universal specific speed
tau = #Shear stress of desired metal (Pa)
fq = 1 #Number of impeller inlets, either 1 or 2
f_t = 1.1 #Given earlier in the text
PC_LOx = 1.21*f_t*(np.exp(-0.408*omegas_LOx))* nq #Pressure coefficient of static pressure rise in impeller of Liquid Oxygen, the equation given uses nq_ref, but I just use nq because I didn't define an nq_ref
PC_LCH4 = 1.21*f_t*(np.exp(-0.408*omegas_LCH4))* nq #Pressure coefficienct of static pressure rise in impeller of Liquid Methane
```
#### Shaft diameter
```
dw_LOx = 3.65(Power_LOx)/(rpm*tau) #Shaft diameter of Liquid Oxygen Impeller
dw_LCH4 = 3.65(Power_LCH4)/(rpm*tau) #Shaft diameter of Liquid Methane Impeller
```
#### Specific Speed
```
q_LOx = QLOx * 3600 * (.3048 ** 3) #converts ft^3/s to m^3/h
q_LCH4 = QLCH4 * 3600 * (.3048 ** 3) #converts ft^3/s to m^3/h
ps = 200 #static pressure in fluid close to impeller in psi
pv_LOX =
pv_LCH4 =
A_LOx = #see two lines below to see what to do
A_LCH4 = #see two lines below to see what to do
v_LOx = (mLOx / rho_LOx) / A_LOx #Define A above as the area of the inlet pipe in ft^2
v_LCH4 = (mLCH4 / rho_LCH4) / A_LCH4 #Define A above as the area of the inlet pipe in ft^2
NPSH_LOx = ps/rho_LOx * (v_LOx ** 2)/(2*9.81) - pv/rho_LOx #substitue pv as Vapor Pressure of Oxygen at temperature in psi above
NPSH_LCH4 = ps/rho_LOx * (v_LOx ** 2)/(2*9.81) - pv/rho_LOx #substitue pv as Vapor Pressure of Methane at temperature in psi above
nss_LOx = n*(q_LOx ** 0.5)/(NPSH_LOx ** 0.75)
nss_LCH4 = n*(q_LCH4 ** 0.5)/(NPSH_LCH4 ** 0.75)
```
#### Inlet diameter
```
#Note: The equation given in the book uses a (1+tan(Beta1)/tan(alpha1)) term, but since the impeller is radial, alpha1 is 90 so the term goes to infinity and therefore results in a multiplication by 1
#Beta1 is determined by finding the specific suction speed** and reading off of the graph, or using:
#kn = 1 - (dn ** 2)/(d1 ** 2); Just choose a value (I assumed inlet diameter ~ 1.15x the size of dn, the hub diameter) since d1 depends on the value of kn and vice versa
tan_Beta1_LOx = (kn) ** 1.1 * (125/nss_LOx) ** 2.2 * (nq_LOx/27) ** 0.418 #Calculates Beta with a 40% std deviation, so a large amount of values is determined with this formula
tan_Beta1_LCH4 = (kn) ** 1.1 * (125/nss_LCH4) ** 2.2 * (nq_LCH4/27) ** 0.418 #Calculates Beta with a 40% std deviation, so a large amount of values is determined with this formula
d1_LOx = 2.9 * (QImp_LOx/(fq*n*kn*tan_Beta1_LOx))^(1/3)
d1_LCH4 = 2.9 * (QImp_LCH4/(fq*n*kn*tan_Beta1_LCH4))^(1/3)
```
#### Exit Diameter
```
d2_LOx = 60/(np.pi * n) * (2 * 9.81 * (HLOx * 0.3048)/(PC_LOX)) ** 0.5
d2_LCH4 = 60/(np.pi * n) * (2 * 9.81 * (HLCH4 * 0.3048)/(PC_LCH4)) ** 0.5
```
#### Blade Thickness
```
e_LOx = 0.022 * d2_LOx #Blade thickness for LOx, this number may have to go up for manufacturing purposes
e_LCH4 = 0.022 * d2_LCH4 #Blade thickness for LCH4, this number may have to go up for manufacturing purposes
```
#### Leading and Trailing Edge Profiles
```
cp_min_sf = 0.155
Lp1_LOx = (2 + (4 + 4 * ((cp_min_sf/0.373)/e_LOx)*(0.373 * e_LOx)) ** 0.5)/ (2 * (cp_min_sf/0.373)/e_LOx) #Leading Edge profile, simplification of formula in Centrifugal Pumps in terms of Quadratic formula
Lp2_LOx = (2 - (4 + 4 * ((cp_min_sf/0.373)/e_LOx)*(0.373 * e_LOx)) ** 0.5)/ (2 * (cp_min_sf/0.373)/e_LOx) #Leading Edge profile, simplification of formula in Centrifugal Pumps in terms of Quadratic formula
Lp1_LCH4 = (2 + (4 + 4 * ((cp_min_sf/0.373)/e_LCH4)*(0.373 * e_LCH4)) ** 0.5)/ (2 * (cp_min_sf/0.373)/e_LCH4) #Leading Edge profile, simplification of formula in Centrifugal Pumps in terms of Quadratic formula
Lp2_LCH4 = (2 - (4 + 4 * ((cp_min_sf/0.373)/e_LCH4)*(0.373 * e_LCH4)) ** 0.5)/ (2 * (cp_min_sf/0.373)/e_LCH4) #Leading Edge profile, simplification of formula in Centrifugal Pumps in terms of Quadratic formula
#Take whichever value above comes out positive, assumed an elliptical profile where cp,min,sf was given as 0.155. Formula changes if cp_min_sf changes
TE_LOx = e_LOx/2 #Trailing edge for Liquid Oxygen using the most simple formula given
TE_LCH4 = e_LCH4/2 #Trailing edge for Liquid Methane using the most simple formula given
```
# Impeller Calcuations
```
#Reference values given on page 667 of Centrifugal Pumps and then converted to imperial from metric
nq_ref = 40 #unitless
Href = 3280.84 #meters to feet
rho_ref = 62.428 #lb/ft^3
tau3 = 1 #given
epsilon_sp = np.pi #Radians. Guessed from the fact that doube volutes are generally at 180
QLe_LOx = QImp_LOx/0.95 * 0.0283168 #m^3/s. Assume that the leakages due to the volute are really low
QLe_LCH4 = QImp_LCH4/0.95 * 0.0283168 #m^3/s
b3_LOx = 1 #Guess; Width of the diffuser inlet (cm)
b3_LCH4 = 1 #Guess; Width of the diffuser inlet (cm)
u2_LOX = (np.pi*d2_LOx*n)/60 #Circumferential speed at the outer diameter of the impeller for Liquid Oxygen
u2_LCH4 = (np.pi*d2_LCH4*n)/60 #Circumferential speed at the outer diameter of the impeller for Liquid Methane
u1m_LOx = (np.pi*d1_LOx*n)/60 #Circumferential speed at the inner diameter of the impeller for Liquid Oxygen
u1m_LCH4 = (np.pi*d1_LOx*n)/60 #Circumferential speed at the inner diameter of the impeller for Liquid Methane
c1u = 1 #Formula is c1m/tan(alpha1) but alpha1 is 90 degrees, so it simplifies to 1
Qref = 1 #Since Volumetric Flow was calculated absolutely, the "reference" value is 1
a = 1 #Taken from book for Q less than or equal to 1 m^3/s
m_LOx = 0.08 * a * (Qref/QImp_LOx) ** 0.15 * (45/nq_LOx) ** 0.06 #Exponential to find hydraulic efficiency
m_LCH4 = 0.08 * a * (Qref/QImp_LCH4) ** 0.15 * (45/nq_LCH4) ** 0.06 #Expoential to find hydraulic efficiency
Eff_Hyd_LOx = 1 - 0.055 * (Qref/QImp_LOx) ** m_LOx - 0.2 * (0.26 - np.log10(nq_LOx/25)) ** 2 #Hydraulic Efficiency of LOx Pump
Eff_Hyd_LCH4 = 1 - 0.055 * (Qref/QImp_LCH4) ** m_LCH4 - 0.2 * (0.26 - np.log10(nq_LCH4/25)) ** 2 #Hydraulic Efficiency of LCH4 Pump
c2u_LOx = (g*HLOx)/(Eff_Hyd_LOx*u2_LOx)+(u1m_LOx*c1u)/u2_LOx #Circumferential component of absolute velocity at impeller outlet for Liquid Oxygen
c2u_LCH4 = (g*HLCH4)/(Eff_Hyd_LCH4*u2_LCH4)+(u1m_LCH4*c1u)/u2_LCH4 #Circumferential component of absolute velocity at impeller outlet for Liquid Methane
d3_LOx = d2_LOx * (1.03 + 0.1*(nq_LOx/nq_ref)*0.07(rho_LOx * HLOX)/(rho_ref*Href)) #distance of the gap bewteen the impeller and volute for Liquid Oxygen
d3_LCH4 = d2_LCH4 * (1.03 + 0.1*(nq_LCH4/nq_ref)*0.07(rho_LCH4 * HLCH4)/(rho_ref*Href)) #distance of the gap bewteen the impeller and volute for Liquid Methane
c3u_LOx = d2_LOx * c2u_LOx / d3_LOx #Circumferential component of absolute velocity at diffuser inlet for Liquid Oxygen
c3u_LCH4 = d2_LCH4 * c2u_LCH4 / d3_LCH4 #Circumferential component of absolute velocity at diffuser inlet for Liquid Methane
c3m_LOx = QLe_LOx*tau3/(np.pi*d3_LOx*b3_LOx) #Meridional component of absolute velocity at diffuser inlet for Liquid Oxygen
c3m_LCH4 = QLe_LCH4 * tau3/(np.pi*d3_LCH4*b3_LCH4) #Meridional component of absolute velocity at diffuser inlet for Liquid Methane
tan_alpha3_LOx = c3m_LOx/c3u_LOx #Flow angle at diffuser inlet with blockage for Liquid Oxygen
tan_alpha3_LCH4 = c3m_LCH4/c3u_LCH4 #Flow angle at diffuser inlet with blockage for Liquid Methane
alpha3b_LOx = np.degrees(np.arctan(tan_alpha3_LOx)) + 3 #Degrees. Diffuser vane inlet, can change the scalar 3 anywhere in the realm of real numbers of [-3,3] for Liquid Oxygen
alpha3b_LCH4 = np.degrees(np.arctan(tan_alpha3_LCH4)) + 3 #Degrees. Diffuser vane inlet, can change the scalar 3 anywhere in the realm of real numbers of [-3,3] for Liquid Methane
r2_LOx = d2_LOx/2 #Radius of the impeller outlet for Liquid Oxygen
r2_LCH4 = d2_LCH4/2 #Radius of the impeller outlet for Liquid Methane
#Throat area calculations, many variables are used that aren't entirely explained
Xsp_LOx = (QLe_LOx * epsilon_sp)/(np.pi*c2u_LOx*r2_LOx * 2 * np.pi)
Xsp_LCH4 = (QLe_LCH4 * epsilon_sp)/(np.pi*c2u_LCH4*r2_LCH4 * 2 * np.pi)
d3q_LOx = Xsp_LOx + (2*d3_LOx*Xsp_LOx) ** 0.5
d3q_LCH4 = Xsp_LCH4 + (2*d3_LCH4*Xsp_LCH4) ** 0.5
A3q_LOx = np.pi*((d3q_LOx) ** 2)/4
A3q_LCH4 = np.pi*((d3q_LCH4) ** 2)/4
```
|
github_jupyter
|
```
import numpy as np
import tensorflow as tf
from tensorflow.examples.tutorials.mnist import input_data
from functools import partial
n_inputs = 28*28
n_hidden1 = 100
n_hidden2 = 100
n_hidden3 = 100
n_hidden4 = 100
n_hidden5 = 100
n_outputs = 5
# Let's define the placeholders for the inputs and the targets
X = tf.placeholder(tf.float32, shape=(None, n_inputs), name="X")
y = tf.placeholder(tf.int64, shape=(None), name="y")
# Let's create the DNN
he_init = tf.contrib.layers.variance_scaling_initializer()
my_dense_layer = partial(
tf.layers.dense, activation=tf.nn.elu,
kernel_initializer=he_init)
with tf.name_scope("dnn"):
hidden1 = my_dense_layer(X, n_hidden1, name="hidden1")
hidden2 = my_dense_layer(hidden1, n_hidden2, name="hidden2")
hidden3 = my_dense_layer(hidden2, n_hidden3, name="hidden3")
hidden4 = my_dense_layer(hidden3, n_hidden4, name="hidden4")
hidden5 = my_dense_layer(hidden4, n_hidden5, name="hidden5")
logits = my_dense_layer(hidden5, n_outputs, activation=None, name="outputs")
Y_proba = tf.nn.softmax(logits, name="Y_proba")
learning_rate = 0.01
with tf.name_scope("loss"):
xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)
loss = tf.reduce_mean(xentropy, name="loss")
with tf.name_scope("train"):
optimizer = tf.train.AdamOptimizer(learning_rate)
training_op = optimizer.minimize(loss, name="training_op")
with tf.name_scope("eval"):
correct = tf.nn.in_top_k(logits, y , 1)
accuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name="accuracy")
init = tf.global_variables_initializer()
saver = tf.train.Saver()
mnist = input_data.read_data_sets("/tmp/data/")
X_train1 = mnist.train.images[mnist.train.labels < 5]
y_train1 = mnist.train.labels[mnist.train.labels < 5]
X_valid1 = mnist.validation.images[mnist.validation.labels < 5]
y_valid1 = mnist.validation.labels[mnist.validation.labels < 5]
X_test1 = mnist.test.images[mnist.test.labels < 5]
y_test1 = mnist.test.labels[mnist.test.labels < 5]
n_epochs = 1000
batch_size = 20
max_checks_without_progress = 20
checks_without_progress = 0
best_loss = np.infty
with tf.Session() as sess:
init.run()
for epoch in range(n_epochs):
rnd_idx = np.random.permutation(len(X_train1))
for rnd_indices in np.array_split(rnd_idx, len(X_train1) // batch_size):
X_batch, y_batch = X_train1[rnd_indices], y_train1[rnd_indices]
sess.run(training_op, feed_dict={X: X_batch, y: y_batch})
# Calculate loss and acc on the validation set to do early stopping
loss_val, acc_val = sess.run([loss, accuracy], feed_dict={X: X_valid1, y: y_valid1})
if loss_val < best_loss:
save_path = saver.save(sess, "./my_mnist_model_0_to_4.ckpt")
best_loss = loss_val
checks_without_progress = 0
else:
checks_without_progress += 1
if checks_without_progress > max_checks_without_progress:
print("Early stopping!")
break
print("{}\tValidation loss: {:.6f}\tBest loss: {:.6f}\tAccuracy: {:.2f}%".format(
epoch, loss_val, best_loss, acc_val * 100))
with tf.Session() as sess:
saver.restore(sess, "./my_mnist_model_0_to_4.ckpt")
acc_test = accuracy.eval(feed_dict={X: X_test1, y: y_test1})
print("Final test accuracy: {:.2f}%".format(acc_test * 100))
```
<h1>DNNClassifier</h1>
```
import numpy as np
import tensorflow as tf
from sklearn.base import BaseEstimator, ClassifierMixin
from sklearn.exceptions import NotFittedError
class DNNClassifier(BaseEstimator, ClassifierMixin):
def __init__(self, n_hidden_layers=5, n_neurons=100, optimizer_class=tf.train.AdamOptimizer,
learning_rate=0.01, batch_size=20, activation=tf.nn.elu, initializer=he_init,
batch_norm_momentum=None, dropout_rate=None, random_state=None):
"""Initialize the DNNClassifier by simply storing all the hyperparameters."""
self.n_hidden_layers = n_hidden_layers
self.n_neurons = n_neurons
self.optimizer_class = optimizer_class
self.learning_rate = learning_rate
self.batch_size = batch_size
self.activation = activation
self.initializer = initializer
self.batch_norm_momentum = batch_norm_momentum
self.dropout_rate = dropout_rate
self.random_state = random_state
self._session = None
def _dnn(self, inputs):
"""Build the hidden layers, with support for batch normalization and dropout."""
for layer in range(self.n_hidden_layers):
if self.dropout_rate:
inputs = tf.layers.dropout(inputs, self.dropout_rate, training=self._training)
inputs = tf.layers.dense(inputs, self.n_neurons,
kernel_initializer=self.initializer,
name="hidden%d" % (layer + 1))
if self.batch_norm_momentum:
inputs = tf.layers.batch_normalization(inputs, momentum=self.batch_norm_momentum,
training=self._training)
inputs = self.activation(inputs, name="hidden%d_out" % (layer + 1))
return inputs
def _build_graph(self, n_inputs, n_outputs):
"""Build the same model as earlier"""
if self.random_state is not None:
tf.set_random_seed(self.random_state)
np.random.seed(self.random_state)
X = tf.placeholder(tf.float32, shape=(None, n_inputs), name="X")
y = tf.placeholder(tf.int32, shape=(None), name="y")
if self.batch_norm_momentum or self.dropout_rate:
self._training = tf.placeholder_with_default(False, shape=(), name='training')
else:
self._training = None
dnn_outputs = self._dnn(X)
logits = tf.layers.dense(dnn_outputs, n_outputs, kernel_initializer=he_init, name="logits")
Y_proba = tf.nn.softmax(logits, name="Y_proba")
xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y,
logits=logits)
loss = tf.reduce_mean(xentropy, name="loss")
optimizer = self.optimizer_class(learning_rate=self.learning_rate)
training_op = optimizer.minimize(loss)
correct = tf.nn.in_top_k(logits, y, 1)
accuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name="accuracy")
init = tf.global_variables_initializer()
saver = tf.train.Saver()
# Make the important operations available easily through instance variables
self._X, self._y = X, y
self._Y_proba, self._loss = Y_proba, loss
self._training_op, self._accuracy = training_op, accuracy
self._init, self._saver = init, saver
def close_session(self):
if self._session:
self._session.close()
def _get_model_params(self):
"""Get all variable values (used for early stopping, faster than saving to disk)"""
with self._graph.as_default():
gvars = tf.get_collection(tf.GraphKeys.GLOBAL_VARIABLES)
return {gvar.op.name: value for gvar, value in zip(gvars, self._session.run(gvars))}
def _restore_model_params(self, model_params):
"""Set all variables to the given values (for early stopping, faster than loading from disk)"""
gvar_names = list(model_params.keys())
assign_ops = {gvar_name: self._graph.get_operation_by_name(gvar_name + "/Assign")
for gvar_name in gvar_names}
init_values = {gvar_name: assign_op.inputs[1] for gvar_name, assign_op in assign_ops.items()}
feed_dict = {init_values[gvar_name]: model_params[gvar_name] for gvar_name in gvar_names}
self._session.run(assign_ops, feed_dict=feed_dict)
def fit(self, X, y, n_epochs=100, X_valid=None, y_valid=None):
"""Fit the model to the training set. If X_valid and y_valid are provided, use early stopping."""
self.close_session()
# infer n_inputs and n_outputs from the training set.
n_inputs = X.shape[1]
self.classes_ = np.unique(y)
n_outputs = len(self.classes_)
# Translate the labels vector to a vector of sorted class indices, containing
# integers from 0 to n_outputs - 1.
# For example, if y is equal to [8, 8, 9, 5, 7, 6, 6, 6], then the sorted class
# labels (self.classes_) will be equal to [5, 6, 7, 8, 9], and the labels vector
# will be translated to [3, 3, 4, 0, 2, 1, 1, 1]
self.class_to_index_ = {label: index
for index, label in enumerate(self.classes_)}
y = np.array([self.class_to_index_[label]
for label in y], dtype=np.int32)
self._graph = tf.Graph()
with self._graph.as_default():
self._build_graph(n_inputs, n_outputs)
# extra ops for batch normalization
extra_update_ops = tf.get_collection(tf.GraphKeys.UPDATE_OPS)
# needed in case of early stopping
max_checks_without_progress = 20
checks_without_progress = 0
best_loss = np.infty
best_params = None
# Now train the model!
self._session = tf.Session(graph=self._graph)
with self._session.as_default() as sess:
self._init.run()
for epoch in range(n_epochs):
rnd_idx = np.random.permutation(len(X))
for rnd_indices in np.array_split(rnd_idx, len(X) // self.batch_size):
X_batch, y_batch = X[rnd_indices], y[rnd_indices]
feed_dict = {self._X: X_batch, self._y: y_batch}
if self._training is not None:
feed_dict[self._training] = True
sess.run(self._training_op, feed_dict=feed_dict)
if extra_update_ops:
sess.run(extra_update_ops, feed_dict=feed_dict)
if X_valid is not None and y_valid is not None:
loss_val, acc_val = sess.run([self._loss, self._accuracy],
feed_dict={self._X: X_valid,
self._y: y_valid})
if loss_val < best_loss:
best_params = self._get_model_params()
best_loss = loss_val
checks_without_progress = 0
else:
checks_without_progress += 1
print("{}\tValidation loss: {:.6f}\tBest loss: {:.6f}\tAccuracy: {:.2f}%".format(
epoch, loss_val, best_loss, acc_val * 100))
if checks_without_progress > max_checks_without_progress:
print("Early stopping!")
break
else:
loss_train, acc_train = sess.run([self._loss, self._accuracy],
feed_dict={self._X: X_batch,
self._y: y_batch})
print("{}\tLast training batch loss: {:.6f}\tAccuracy: {:.2f}%".format(
epoch, loss_train, acc_train * 100))
# If we used early stopping then rollback to the best model found
if best_params:
self._restore_model_params(best_params)
return self
def predict_proba(self, X):
if not self._session:
raise NotFittedError("This %s instance is not fitted yet" % self.__class__.__name__)
with self._session.as_default() as sess:
return self._Y_proba.eval(feed_dict={self._X: X})
def predict(self, X):
class_indices = np.argmax(self.predict_proba(X), axis=1)
return np.array([[self.classes_[class_index]]
for class_index in class_indices], np.int32)
def save(self, path):
self._saver.save(self._session, path)
dnn_clf = DNNClassifier(random_state=42)
dnn_clf.fit(X_train1, y_train1, n_epochs=1000, X_valid=X_valid1, y_valid=y_valid1)
from sklearn.metrics import accuracy_score
y_pred = dnn_clf.predict(X_test1)
accuracy_score(y_test1, y_pred)
from sklearn.model_selection import RandomizedSearchCV
def leaky_relu(alpha=0.01):
def parametrized_leaky_relu(z, name=None):
return tf.maximum(alpha * z, z, name=name)
return parametrized_leaky_relu
param_distribs = {
"n_neurons": [10, 30, 50, 70, 90, 100, 120, 140, 160],
"batch_size": [10, 50, 100, 500],
"learning_rate": [0.01, 0.02, 0.05, 0.1],
"activation": [tf.nn.relu, tf.nn.elu, leaky_relu(alpha=0.01), leaky_relu(alpha=0.1)],
# you could also try exploring different numbers of hidden layers, different optimizers, etc.
#"n_hidden_layers": [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10],
#"optimizer_class": [tf.train.AdamOptimizer, partial(tf.train.MomentumOptimizer, momentum=0.95)],
}
rnd_search = RandomizedSearchCV(DNNClassifier(random_state=42), param_distribs, n_iter=50,
fit_params={"X_valid": X_valid1, "y_valid": y_valid1, "n_epochs": 1000},
random_state=42, verbose=2)
rnd_search.fit(X_train1, y_train1)
rnd_search.best_params_
y_pred = rnd_search.predict(X_test1)
accuracy_score(y_test1, y_pred)
rnd_search.best_estimator_.save("./my_best_mnist_model_0_to_4")
# Let's train the best model found, once again, to see how fast it converges
dnn_clf = DNNClassifier(activation=leaky_relu(alpha=0.1), batch_size=500, learning_rate=0.01,
n_neurons=50, random_state=42)
dnn_clf.fit(X_train1, y_train1, n_epochs=1000, X_valid=X_valid1, y_valid=y_valid1)
y_pred = dnn_clf.predict(X_test1)
accuracy_score(y_test1, y_pred)
# Here the accuracy is different because I put leaky_relu in the training instead of relu as the rnd_search_best_params says.
# However, the accuracy is better...Wtf?!
# Let's try to add Batch Normalization
dnn_clf_bn = DNNClassifier(activation=leaky_relu(alpha=0.1), batch_size=500, learning_rate=0.01,
n_neurons=50, random_state=42,
batch_norm_momentum=0.95)
dnn_clf_bn.fit(X_train1, y_train1, n_epochs=1000, X_valid=X_valid1, y_valid=y_valid1)
y_pred = dnn_clf_bn.predict(X_test1)
accuracy_score(y_test1, y_pred)
# Mmm, Batch Normalization did not improve the accuracy. We should try to do another tuning for hyperparameters with BN
# and try again.
# ...
# Now let's go back to our previous model and see how well perform on the training set
y_pred = dnn_clf.predict(X_train1)
accuracy_score(y_train1, y_pred)
# Much better than the test set, so probably it is overfitting the training set. Let's try using dropout
dnn_clf_dropout = DNNClassifier(activation=leaky_relu(alpha=0.1), batch_size=500, learning_rate=0.01,
n_neurons=50, random_state=42,
dropout_rate=0.5)
dnn_clf_dropout.fit(X_train1, y_train1, n_epochs=1000, X_valid=X_valid1, y_valid=y_valid1)
y_pred = dnn_clf_dropout.predict(X_test1)
accuracy_score(y_test1, y_pred)
# Dropout doesn't seem to help. As said before, we could try to tune the network with dropout and see what we got.
# ...
```
<h1>Transfer Learning</h1>
<p>Let's try to reuse the previous model on digits from 5 to 9, using only 100 images per digit!</p>
```
restore_saver = tf.train.import_meta_graph("./my_best_mnist_model_0_to_4.meta")
X = tf.get_default_graph().get_tensor_by_name("X:0")
y = tf.get_default_graph().get_tensor_by_name("y:0")
loss = tf.get_default_graph().get_tensor_by_name("loss:0")
Y_proba = tf.get_default_graph().get_tensor_by_name("Y_proba:0")
logits = Y_proba.op.inputs[0]
accuracy = tf.get_default_graph().get_tensor_by_name("accuracy:0")
learning_rate = 0.01
output_layer_vars = tf.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES, scope="logits")
optimizer = tf.train.AdamOptimizer(learning_rate, name="Adam2")
# Freeze all the hidden layers
training_op = optimizer.minimize(loss, var_list=output_layer_vars)
correct = tf.nn.in_top_k(logits, y, 1)
accuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name="accuracy")
init = tf.global_variables_initializer()
five_frozen_saver = tf.train.Saver()
X_train2_full = mnist.train.images[mnist.train.labels >= 5]
y_train2_full = mnist.train.labels[mnist.train.labels >= 5] - 5
X_valid2_full = mnist.validation.images[mnist.validation.labels >= 5]
y_valid2_full = mnist.validation.labels[mnist.validation.labels >= 5] - 5
X_test2 = mnist.test.images[mnist.test.labels >= 5]
y_test2 = mnist.test.labels[mnist.test.labels >= 5] - 5
def sample_n_instances_per_class(X, y, n=100):
Xs, ys = [], []
for label in np.unique(y):
idx = (y == label)
Xc = X[idx][:n]
yc = y[idx][:n]
Xs.append(Xc)
ys.append(yc)
return np.concatenate(Xs), np.concatenate(ys)
X_train2, y_train2 = sample_n_instances_per_class(X_train2_full, y_train2_full, n=100)
X_valid2, y_valid2 = sample_n_instances_per_class(X_valid2_full, y_valid2_full, n=30)
import time
n_epochs = 1000
batch_size = 20
max_checks_without_progress = 20
checks_without_progress = 0
best_loss = np.infty
with tf.Session() as sess:
init.run()
restore_saver.restore(sess, "./my_best_mnist_model_0_to_4")
for var in output_layer_vars:
var.initializer.run()
t0 = time.time()
for epoch in range(n_epochs):
rnd_idx = np.random.permutation(len(X_train2))
for rnd_indices in np.array_split(rnd_idx, len(X_train2) // batch_size):
X_batch, y_batch = X_train2[rnd_indices], y_train2[rnd_indices]
sess.run(training_op, feed_dict={X: X_batch, y: y_batch})
loss_val, acc_val = sess.run([loss, accuracy], feed_dict={X: X_valid2, y: y_valid2})
if loss_val < best_loss:
save_path = five_frozen_saver.save(sess, "./my_mnist_model_5_to_9_five_frozen")
best_loss = loss_val
checks_without_progress = 0
else:
checks_without_progress += 1
if checks_without_progress > max_checks_without_progress:
print("Early stopping!")
break
print("{}\tValidation loss: {:.6f}\tBest loss: {:.6f}\tAccuracy: {:.2f}%".format(
epoch, loss_val, best_loss, acc_val * 100))
t1 = time.time()
print("Total training time: {:.1f}s".format(t1 - t0))
with tf.Session() as sess:
five_frozen_saver.restore(sess, "./my_mnist_model_5_to_9_five_frozen")
acc_test = accuracy.eval(feed_dict={X: X_test2, y: y_test2})
print("Final test accuracy: {:.2f}%".format(acc_test * 100))
```
<p>As we can see, not so good...But of course, we're using 100 images per digit and we only changed the output layer.</p>
```
# Let's try to reuse only 4 hidden layers instead of 5
n_outputs = 5
restore_saver = tf.train.import_meta_graph("./my_best_mnist_model_0_to_4.meta")
X = tf.get_default_graph().get_tensor_by_name("X:0")
y = tf.get_default_graph().get_tensor_by_name("y:0")
hidden4_out = tf.get_default_graph().get_tensor_by_name("hidden4_out:0")
logits = tf.layers.dense(hidden4_out, n_outputs, kernel_initializer=he_init, name="new_logits")
Y_proba = tf.nn.softmax(logits)
xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)
loss = tf.reduce_mean(xentropy)
correct = tf.nn.in_top_k(logits, y, 1)
accuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name="accuracy")
learning_rate = 0.01
output_layer_vars = tf.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES, scope="new_logits")
optimizer = tf.train.AdamOptimizer(learning_rate, name="Adam2")
training_op = optimizer.minimize(loss, var_list=output_layer_vars)
init = tf.global_variables_initializer()
four_frozen_saver = tf.train.Saver()
n_epochs = 1000
batch_size = 20
max_checks_without_progress = 20
checks_without_progress = 0
best_loss = np.infty
with tf.Session() as sess:
init.run()
restore_saver.restore(sess, "./my_best_mnist_model_0_to_4")
for epoch in range(n_epochs):
rnd_idx = np.random.permutation(len(X_train2))
for rnd_indices in np.array_split(rnd_idx, len(X_train2) // batch_size):
X_batch, y_batch = X_train2[rnd_indices], y_train2[rnd_indices]
sess.run(training_op, feed_dict={X: X_batch, y: y_batch})
loss_val, acc_val = sess.run([loss, accuracy], feed_dict={X: X_valid2, y: y_valid2})
if loss_val < best_loss:
save_path = four_frozen_saver.save(sess, "./my_mnist_model_5_to_9_four_frozen")
best_loss = loss_val
checks_without_progress = 0
else:
checks_without_progress += 1
if checks_without_progress > max_checks_without_progress:
print("Early stopping!")
break
print("{}\tValidation loss: {:.6f}\tBest loss: {:.6f}\tAccuracy: {:.2f}%".format(
epoch, loss_val, best_loss, acc_val * 100))
with tf.Session() as sess:
four_frozen_saver.restore(sess, "./my_mnist_model_5_to_9_four_frozen")
acc_test = accuracy.eval(feed_dict={X: X_test2, y: y_test2})
print("Final test accuracy: {:.2f}%".format(acc_test * 100))
```
<p>Well, a bit better...</p>
```
# Let's try now to unfreeze the last two layers
learning_rate = 0.01
unfrozen_vars = tf.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES, scope="hidden[34]|new_logits")
optimizer = tf.train.AdamOptimizer(learning_rate, name="Adam3")
training_op = optimizer.minimize(loss, var_list=unfrozen_vars)
init = tf.global_variables_initializer()
two_frozen_saver = tf.train.Saver()
n_epochs = 1000
batch_size = 20
max_checks_without_progress = 20
checks_without_progress = 0
best_loss = np.infty
with tf.Session() as sess:
init.run()
four_frozen_saver.restore(sess, "./my_mnist_model_5_to_9_four_frozen")
for epoch in range(n_epochs):
rnd_idx = np.random.permutation(len(X_train2))
for rnd_indices in np.array_split(rnd_idx, len(X_train2) // batch_size):
X_batch, y_batch = X_train2[rnd_indices], y_train2[rnd_indices]
sess.run(training_op, feed_dict={X: X_batch, y: y_batch})
loss_val, acc_val = sess.run([loss, accuracy], feed_dict={X: X_valid2, y: y_valid2})
if loss_val < best_loss:
save_path = two_frozen_saver.save(sess, "./my_mnist_model_5_to_9_two_frozen")
best_loss = loss_val
checks_without_progress = 0
else:
checks_without_progress += 1
if checks_without_progress > max_checks_without_progress:
print("Early stopping!")
break
print("{}\tValidation loss: {:.6f}\tBest loss: {:.6f}\tAccuracy: {:.2f}%".format(
epoch, loss_val, best_loss, acc_val * 100))
with tf.Session() as sess:
two_frozen_saver.restore(sess, "./my_mnist_model_5_to_9_two_frozen")
acc_test = accuracy.eval(feed_dict={X: X_test2, y: y_test2})
print("Final test accuracy: {:.2f}%".format(acc_test * 100))
```
<p>Not bad...And what if we unfreeze all the layers?</p>
```
learning_rate = 0.01
optimizer = tf.train.AdamOptimizer(learning_rate, name="Adam4")
training_op = optimizer.minimize(loss)
init = tf.global_variables_initializer()
no_frozen_saver = tf.train.Saver()
n_epochs = 1000
batch_size = 20
max_checks_without_progress = 20
checks_without_progress = 0
best_loss = np.infty
with tf.Session() as sess:
init.run()
two_frozen_saver.restore(sess, "./my_mnist_model_5_to_9_two_frozen")
for epoch in range(n_epochs):
rnd_idx = np.random.permutation(len(X_train2))
for rnd_indices in np.array_split(rnd_idx, len(X_train2) // batch_size):
X_batch, y_batch = X_train2[rnd_indices], y_train2[rnd_indices]
sess.run(training_op, feed_dict={X: X_batch, y: y_batch})
loss_val, acc_val = sess.run([loss, accuracy], feed_dict={X: X_valid2, y: y_valid2})
if loss_val < best_loss:
save_path = no_frozen_saver.save(sess, "./my_mnist_model_5_to_9_no_frozen")
best_loss = loss_val
checks_without_progress = 0
else:
checks_without_progress += 1
if checks_without_progress > max_checks_without_progress:
print("Early stopping!")
break
print("{}\tValidation loss: {:.6f}\tBest loss: {:.6f}\tAccuracy: {:.2f}%".format(
epoch, loss_val, best_loss, acc_val * 100))
with tf.Session() as sess:
no_frozen_saver.restore(sess, "./my_mnist_model_5_to_9_no_frozen")
acc_test = accuracy.eval(feed_dict={X: X_test2, y: y_test2})
print("Final test accuracy: {:.2f}%".format(acc_test * 100))
# Let's compare this result with a DNN trained from scratch
dnn_clf_5_to_9 = DNNClassifier(n_hidden_layers=4, random_state=42)
dnn_clf_5_to_9.fit(X_train2, y_train2, n_epochs=1000, X_valid=X_valid2, y_valid=y_valid2)
from sklearn.metrics import accuracy_score
y_pred = dnn_clf_5_to_9.predict(X_test2)
accuracy_score(y_test2, y_pred)
```
<p>Unfortunately in this case transfer learning did not help too much.</p>
|
github_jupyter
|
# LB-Colloids Colloid particle tracking
LB-Colloids allows the user to perform colloid and nanoparticle tracking simulations on Computational Fluid Dynamics domains. As the user, you supply the chemical and physical properties, and the code performs the mathematics and particle tracking!
Let's set up our workspace to begin. And we will use the Synthetic5 example problem to parameterize run LB-Colloids
```
%matplotlib inline
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import os
from lb_colloids import LBImage, LB2DModel
from lb_colloids import ColloidModel, cIO
workspace = os.path.join("..", "data")
domain = "Synth100_5.png"
lb_name = "s5.hdf5"
endpoint = "s5.endpoint"
```
First thing, let's run a lattice Boltzmann model to get our fluid domain. For more details see the LB2D Notebook.
```
lbi = LBImage.Images(os.path.join(workspace, domain))
bc = LBImage.BoundaryCondition(lbi.arr, fluidvx=[253], solidvx=[0], nlayers=5)
lbm = LB2DModel(bc.binarized)
lbm.niters = 1000
lbm.run(output=os.path.join(workspace, lb_name), verbose=1000)
```
## Setting up a Colloids particle tracking model
We can begin setting up a Colloids model by using the `ColloidsConfig()` class. This class ensures that valid values are supplied to particle tracking variables and allows the user to write an external particle tracking configuration file for documentation and later use if wanted.
Let's generate an empty `ColloidsConfig` instance
```
io = cIO.ColloidsConfig()
```
`ColloidsConfig()` uses dictionary keys to be parameterized. Common parameterization variables include
`lbmodel`: required parameter that points to the CFD fluid domain
`ncols`: required parameter that describes the number of colloids released
`iters`: number of time steps to simulate transport
`lbres`: the lattice Boltzmann simulation resolution in meters
`gridref`: optional grid refinement parameter, uses bi-linear interpolation
`ac`: colloid radius in meters
`timestep`: the timestep length in seconds. Recommend very small timesteps!
`continuous`: flag for continuous release. If 0 one release of colloids occurs, if > 0 a release of colloids occurs at continuous number of timesteps
`i`: fluid ionic strength in M
`print_time`: how often iteration progress prints to the screen
`endpoint`: endpoint file name to store breakthrough information
`store_time`: internal function that can be used to reduce memory requirements, a higher store_time equals less memory devoted to storing colloid positions (old positions are striped every store_time timesteps).
`zeta_colloid`: zeta potential of the colloid in V
`zeta_solid`: zeta potential of the solid in V
`plot`: boolean flag that generates a plot at the end of the model run
`showfig`: boolean flag that determines weather to show the figure or save it to disk
A complete listing of these are available in the user guide.
```
# model parameters
io["lbmodel"] = os.path.join(workspace, lb_name)
io['ncols'] = 2000
io['iters'] = 50000
io['lbres'] = 1e-6
io['gridref'] = 10
io['ac'] = 1e-06
io['timestep'] = 1e-06 # should be less than or equal to colloid radius!
io['continuous'] = 0
# chemical parameters
io['i'] = 1e-03 # Molar ionic strength of solution
io['zeta_colloid'] = -49.11e-3 # zeta potential of Na-Kaolinite at 1e-03 M NaCl
io['zeta_solid'] = -61.76e-3 # zeta potential of Glass Beads at 1e-03 M NaCl
# output control
io['print_time'] = 10000
io['endpoint'] = os.path.join(workspace, endpoint)
io['store_time'] = 100
io['plot'] = True
io['showfig'] = True
```
We can now look at the parameter dictionaries `ColloidConfig` creates!
```
io.model_parameters, io.chemical_parameters, io.physical_parameters, io.output_control_parameters
```
we can also write a config file for documentation and later runs! And see the information that will be written to the config file by using the `io.config` call
```
io.write(os.path.join(workspace, "s2.config"))
io.config
```
the `ColloidsConfig` can be directly be used with the `Config` reader to instanstiate a LB-Colloids model
```
config = cIO.Config(io.config)
```
and we can run the model using the `ColloidModel.run()` call
```
ColloidModel.run(config)
```
The output image shows the path of colloids which haven't yet broke through the model domain!
### For ColloidModel outputs please see the LB_Colloids_output_contol notebook
|
github_jupyter
|
```
import os
path = '/home/yash/Desktop/tensorflow-adversarial/tf_example'
os.chdir(path)
# supress tensorflow logging other than errors
os.environ['TF_CPP_MIN_LOG_LEVEL'] = '3'
import numpy as np
import tensorflow as tf
from tensorflow.contrib.learn import ModeKeys, Estimator
import matplotlib
matplotlib.use('Agg')
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
from fgsm4 import fgsm
import mnist
img_rows = 28
img_cols = 28
img_chas = 1
input_shape = (img_rows, img_cols, img_chas)
n_classes = 10
print('\nLoading mnist')
(X_train, y_train), (X_test, y_test) = mnist.load_data()
X_train = X_train.astype('float32') / 255.
X_test = X_test.astype('float32') / 255.
X_train = X_train.reshape(-1, img_rows, img_cols, img_chas)
X_test = X_test.reshape(-1, img_rows, img_cols, img_chas)
# one hot encoding, basically creates hte si
def _to_categorical(x, n_classes):
x = np.array(x, dtype=int).ravel()
n = x.shape[0]
ret = np.zeros((n, n_classes))
ret[np.arange(n), x] = 1
return ret
def find_l2(X_test, X_adv):
a=X_test.reshape(-1,28*28)
b=X_adv.reshape(-1,28*28)
l2_unsquared = np.sum(np.square(a-b),axis=1)
return l2_unsquared
y_train = _to_categorical(y_train, n_classes)
y_test = _to_categorical(y_test, n_classes)
print('\nShuffling training data')
ind = np.random.permutation(X_train.shape[0])
X_train, y_train = X_train[ind], y_train[ind]
# X_train = X_train[:1000]
# y_train = y_train[:1000]
# split training/validation dataset
validation_split = 0.1
n_train = int(X_train.shape[0]*(1-validation_split))
X_valid = X_train[n_train:]
X_train = X_train[:n_train]
y_valid = y_train[n_train:]
y_train = y_train[:n_train]
class Dummy:
pass
env = Dummy()
def model(x, logits=False, training=False):
conv0 = tf.layers.conv2d(x, filters=32, kernel_size=[3, 3],
padding='same', name='conv0',
activation=tf.nn.relu)
pool0 = tf.layers.max_pooling2d(conv0, pool_size=[2, 2],
strides=2, name='pool0')
conv1 = tf.layers.conv2d(pool0, filters=64,
kernel_size=[3, 3], padding='same',
name='conv1', activation=tf.nn.relu)
pool1 = tf.layers.max_pooling2d(conv1, pool_size=[2, 2],
strides=2, name='pool1')
flat = tf.reshape(pool1, [-1, 7*7*64], name='flatten')
dense1 = tf.layers.dense(flat, units=1024, activation=tf.nn.relu,
name='dense1')
dense2 = tf.layers.dense(dense1, units=128, activation=tf.nn.relu,
name='dense2')
logits_ = tf.layers.dense(dense2, units=10, name='logits') #removed dropout
y = tf.nn.softmax(logits_, name='ybar')
if logits:
return y, logits_
return y
# We need a scope since the inference graph will be reused later
with tf.variable_scope('model'):
env.x = tf.placeholder(tf.float32, (None, img_rows, img_cols,
img_chas), name='x')
env.y = tf.placeholder(tf.float32, (None, n_classes), name='y')
env.training = tf.placeholder(bool, (), name='mode')
env.ybar, logits = model(env.x, logits=True,
training=env.training)
z = tf.argmax(env.y, axis=1)
zbar = tf.argmax(env.ybar, axis=1)
env.count = tf.cast(tf.equal(z, zbar), tf.float32)
env.acc = tf.reduce_mean(env.count, name='acc')
xent = tf.nn.softmax_cross_entropy_with_logits(labels=env.y,
logits=logits)
env.loss = tf.reduce_mean(xent, name='loss')
extra_update_ops = tf.get_collection(tf.GraphKeys.UPDATE_OPS)
with tf.control_dependencies(extra_update_ops):
env.optim = tf.train.AdamOptimizer(beta1=0.9, beta2=0.999, epsilon=1e-08,).minimize(env.loss)
with tf.variable_scope('model', reuse=True):
env.x_adv, env.all_flipped = fgsm(model, env.x, step_size=.05, bbox_semi_side=10) #epochs is redundant now!
sess = tf.InteractiveSession()
sess.run(tf.global_variables_initializer())
sess.run(tf.local_variables_initializer())
def save_model(label):
saver = tf.train.Saver()
saver.save(sess, './models/mnist/' + label)
def restore_model(label):
saver = tf.train.Saver()
saver.restore(sess, './models/mnist/' + label)
def _evaluate(X_data, y_data, env):
print('\nEvaluating')
n_sample = X_data.shape[0]
batch_size = 128
n_batch = int(np.ceil(n_sample/batch_size))
loss, acc = 0, 0
ns = 0
for ind in range(n_batch):
print(' batch {0}/{1}'.format(ind+1, n_batch), end='\r')
start = ind*batch_size
end = min(n_sample, start+batch_size)
batch_loss, batch_count, batch_acc = sess.run(
[env.loss, env.count, env.acc],
feed_dict={env.x: X_data[start:end],
env.y: y_data[start:end],
env.training: False})
loss += batch_loss*batch_size
# print('batch count: {0}'.format(np.sum(batch_count)))
ns+=batch_size
acc += batch_acc*batch_size
loss /= ns
acc /= ns
# print (ns)
# print (n_sample)
print(' loss: {0:.4f} acc: {1:.4f}'.format(loss, acc))
return loss, acc
def _predict(X_data, env):
print('\nPredicting')
n_sample = X_data.shape[0]
batch_size = 128
n_batch = int(np.ceil(n_sample/batch_size))
yval = np.empty((X_data.shape[0], n_classes))
for ind in range(n_batch):
print(' batch {0}/{1}'.format(ind+1, n_batch), end='\r')
start = ind*batch_size
end = min(n_sample, start+batch_size)
batch_y = sess.run(env.ybar, feed_dict={
env.x: X_data[start:end], env.training: False})
yval[start:end] = batch_y
return yval
def train(label):
print('\nTraining')
n_sample = X_train.shape[0]
batch_size = 128
n_batch = int(np.ceil(n_sample/batch_size))
n_epoch = 50
for epoch in range(n_epoch):
print('Epoch {0}/{1}'.format(epoch+1, n_epoch))
for ind in range(n_batch):
print(' batch {0}/{1}'.format(ind+1, n_batch), end='\r')
start = ind*batch_size
end = min(n_sample, start+batch_size)
sess.run(env.optim, feed_dict={env.x: X_train[start:end],
env.y: y_train[start:end],
env.training: True})
if(epoch%5 == 0):
model_label = label+ '{0}'.format(epoch)
print("saving model " + model_label)
save_model(model_label)
save_model(label)
def create_adv(X, Y, label):
print('\nCrafting adversarial')
n_sample = X.shape[0]
batch_size = 1
n_batch = int(np.ceil(n_sample/batch_size))
n_epoch = 20
X_adv = np.empty_like(X)
for ind in range(n_batch):
print(' batch {0}/{1}'.format(ind+1, n_batch), end='\r')
start = ind*batch_size
end = min(n_sample, start+batch_size)
tmp, all_flipped = sess.run([env.x_adv, env.all_flipped], feed_dict={env.x: X[start:end],
env.y: Y[start:end],
env.training: False})
# _evaluate(tmp, Y[start:end],env)
X_adv[start:end] = tmp
# print(all_flipped)
print('\nSaving adversarial')
os.makedirs('data', exist_ok=True)
np.save('data/mnist/' + label + '.npy', X_adv)
return X_adv
label = "mnist_with_cnn"
# train(label) # else
#Assuming that you've started a session already else do that first!
restore_model(label + '5')
# restore_model(label + '10')
# restore_model(label + '50')
# restore_model(label + '100')
_evaluate(X_train, y_train, env)
def random_normal_func(X):
X=X.reshape(-1,28*28)
print(X.shape)
mean, std = np.mean(X, axis=0), np.std(X,axis=0)
randomX = np.zeros([10000,X[0].size])
print(randomX.shape)
for i in range(X[0].size):
randomX[:,i] = np.random.normal(mean[i],std[i],10000)
randomX = randomX.reshape(-1,28,28,1)
ans = sess.run(env.ybar, feed_dict={env.x: randomX,env.training: False})
labels = _to_categorical(np.argmax(ans,axis=1), n_classes)
return randomX,labels
test = "test_fs_exp1_0"
train = "train_fs_exp1_0"
random = "random_fs_exp1_0"
random_normal= "random_normal_fs_exp1_0"
X_train_sub = X_train[:10000]
y_train_sub = sess.run(env.ybar, feed_dict={env.x: X_train_sub,env.training: False})
y_train_sub = _to_categorical(np.argmax(y_train_sub, axis=1), n_classes)
y_test_sub = sess.run(env.ybar, feed_dict={env.x: X_test,env.training: False})
y_test_sub = _to_categorical(np.argmax(y_test_sub, axis=1), n_classes)
X_random = np.random.rand(10000,28,28,1)
X_random = X_random[:10000]
y_random = sess.run(env.ybar, feed_dict={env.x: X_random,env.training: False})
y_random = _to_categorical(np.argmax(y_random, axis=1), n_classes)
X_random_normal, y_random_normal = random_normal_func(X_train)
X_adv_test = create_adv(X_test, y_test_sub, test)
X_adv_train = create_adv(X_train_sub, y_train_sub, train)
X_adv_random = create_adv(X_random,y_random, random)
X_adv_random_normal = create_adv(X_random_normal, y_random_normal, random_normal)
# X_adv_test = np.load('data/mnist/' + test + '.npy')
# X_adv_train = np.load('data/mnist/' + train + '.npy')
# X_adv_random = np.load('data/mnist/' + random + '.npy')
# X_adv_random_normal = np.load('data/mnist/' + random_normal + '.npy')
l2_test = find_l2(X_adv_test,X_test)
l2_train = find_l2(X_adv_train, X_train_sub)
l2_random = find_l2(X_adv_random,X_random)
l2_random_normal = find_l2(X_adv_random_normal,X_random_normal)
print(l2_train)
print(X_adv_random_normal[0][3])
%matplotlib inline
# evenly sampled time at 200ms intervals
t = np.arange(1,10001, 1)
# red dashes, blue squares and green triangles
plt.plot(t, l2_test, 'r--', t, l2_train, 'b--', t, l2_random, 'y--', l2_random_normal, 'g--')
plt.show()
import matplotlib.patches as mpatches
%matplotlib inline
# evenly sampled time at 200ms intervals
t = np.arange(1,101, 1)
# red dashes, blue squares and green triangles
plt.plot(t, l2_test[:100], 'r--', t, l2_train[:100], 'b--',t, l2_random[:100], 'y--',l2_random_normal[:100], 'g--')
blue_patch = mpatches.Patch(color='blue', label='Train Data')
plt.legend(handles=[blue_patch])
plt.show()
%matplotlib inline
plt.hist(l2_test,100)
plt.title("L2 distance of test data")
plt.xlabel("Distance")
plt.ylabel("Frequency")
plt.show()
%matplotlib inline
plt.hist(l2_train,100)
plt.title("L2 distance of train data")
plt.xlabel("Distance")
plt.ylabel("Frequency")
plt.show()
%matplotlib inline
plt.hist(l2_random,100)
plt.title("L2 distance of random data")
plt.xlabel("Distance")
plt.ylabel("Frequency")
plt.show()
%matplotlib inline
plt.hist(l2_random_normal,100)
plt.title("L2 distance of random normal data")
plt.xlabel("Distance")
plt.ylabel("Frequency")
plt.show()
```
|
github_jupyter
|
```
import ipywidgets
tabs = ipywidgets.Tab()
tabs.children = [ipywidgets.Label(value='tab1'), ipywidgets.Label(value='tab2'), ipywidgets.Label(value='tab3'), ipywidgets.Label(value='tab4')]
tabs.observe(lambda change: print(f"selected index: {change['new']}") , names='selected_index')
def change_children(_):
id = tabs.selected_index
tabs.selected_index = None # Warning : this will emit a change event
tabs.children = [ipywidgets.Label(value='tab1'), ipywidgets.Label(value='tab2'), ipywidgets.Label(value='tab3'), ipywidgets.Label(value='tab4')]
tabs.selected_index = id
btn = ipywidgets.Button(description='change_children')
btn.on_click(change_children)
ipywidgets.VBox([tabs, btn])
import ipywidgets as widgets
tab_contents = ['P0', 'P1']
children = [widgets.Text(description=name) for name in tab_contents]
tab = widgets.Tab()
tab.children = children
for i in range(len(children)):
tab.set_title(i, str(i))
def tab_toggle_var(*args):
global vartest
if tab.selected_index ==0:
vartest = 0
else:
vartest = 1
tab.observe(tab_toggle_var)
tab_toggle_var()
print(children)
metadata={}
def _observe_test(change):
print(change)
def _observe_config(change):
print('_observe_config')
metadata[ widget_elec_config.description] = widget_elec_config.value
metadata_json_raw = json.dumps(metadata, indent=4)
export.value = "<pre>{}</pre>".format(
html.escape(metadata_json_raw))
export = widgets.HTML()
vbox_metadata = widgets.VBox(
[
widgets.HTML('''
<h4>Preview of metadata export:</h4>
<hr style="height:1px;border-width:0;color:black;background-color:gray">
'''),
export
]
)
for child in tab.children:
print(child)
child.observe(_observe_test)
display(tab)
w = widgets.Dropdown(
options=['Addition', 'Multiplication', 'Subtraction', 'Division'],
value='Addition',
description='Task:',
)
def on_change(change):
if change['type'] == 'change' and change['name'] == 'value':
print("changed to %s" % change['new'])
w.observe(on_change)
display(w)
from IPython.display import display
import ipywidgets as widgets
int_range0_slider = widgets.IntSlider()
int_range1_slider = widgets.IntSlider()
output = widgets.Output()
def interactive_function(inp0,inp1):
with output:
print('ie changed. int_range0_slider: '+str(inp0)+' int_range1_slider: '+str(inp1))
return
def report_int_range0_change(change):
with output:
print('int_range0 change observed'+str(change))
return
def report_ie_change(change):
with output:
print('ie change observed'+str(change))
return
ie = widgets.interactive(interactive_function, inp0=int_range0_slider,inp1=int_range1_slider)
# print(int_range0_slider.observe)
# print(ie.observe)
# int_range0_slider.observe(report_int_range0_change, names='value')
for child in ie.children:
child.observe(report_ie_change)
display(int_range0_slider,int_range1_slider,output)
```
|
github_jupyter
|
# Home Credit Default Risk
Can you predict how capable each applicant is of repaying a loan?
Many people struggle to get loans due to **insufficient or non-existent credit histories**. And, unfortunately, this population is often taken advantage of by untrustworthy lenders.
Home Credit strives to broaden financial inclusion for the **unbanked population by providing a positive and safe borrowing experience**. In order to make sure this underserved population has a positive loan experience, Home Credit makes use of a variety of alternative data--including telco and transactional information--to predict their clients' repayment abilities.
While Home Credit is currently using various statistical and machine learning methods to make these predictions, they're challenging Kagglers to help them unlock the full potential of their data. Doing so will ensure that clients capable of repayment are not rejected and that loans are given with a principal, maturity, and repayment calendar that will empower their clients to be successful.
**Submissions are evaluated on area under the ROC curve between the predicted probability and the observed target.**
# Dataset
```
# #Python Libraries
import numpy as np
import scipy as sp
import pandas as pd
import statsmodels
import pandas_profiling
%matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sns
import os
import sys
import time
import requests
import datetime
import missingno as msno
import math
import sys
import gc
import os
# #sklearn
from sklearn.model_selection import train_test_split
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RandomizedSearchCV
from sklearn.model_selection import GridSearchCV
from sklearn.ensemble import RandomForestRegressor
from sklearn import preprocessing
# #sklearn - metrics
from sklearn.metrics import mean_squared_error
from sklearn.metrics import mean_absolute_error
from sklearn.metrics import r2_score
# #XGBoost & LightGBM
import xgboost as xgb
import lightgbm as lgb
# #Missing value imputation
from fancyimpute import KNN, MICE
```
## Data Dictionary
```
!ls -l ../data/
```
- application_{train|test}.csv
This is the main table, broken into two files for Train (**with TARGET**) and Test (without TARGET).
Static data for all applications. **One row represents one loan in our data sample.**
- bureau.csv
All client's previous credits provided by other financial institutions that were reported to Credit Bureau (for clients who have a loan in our sample).
For every loan in our sample, there are as many rows as number of credits the client had in Credit Bureau before the application date.
- bureau_balance.csv
Monthly balances of previous credits in Credit Bureau.
This table has one row for each month of history of every previous credit reported to Credit Bureau – i.e the table has (#loans in sample * # of relative previous credits * # of months where we have some history observable for the previous credits) rows.
- POS_CASH_balance.csv
Monthly balance snapshots of previous POS (point of sales) and cash loans that the applicant had with Home Credit.
This table has one row for each month of history of every previous credit in Home Credit (consumer credit and cash loans) related to loans in our sample – i.e. the table has (#loans in sample * # of relative previous credits * # of months in which we have some history observable for the previous credits) rows.
- credit_card_balance.csv
Monthly balance snapshots of previous credit cards that the applicant has with Home Credit.
This table has one row for each month of history of every previous credit in Home Credit (consumer credit and cash loans) related to loans in our sample – i.e. the table has (#loans in sample * # of relative previous credit cards * # of months where we have some history observable for the previous credit card) rows.
- previous_application.csv
All previous applications for Home Credit loans of clients who have loans in our sample.
There is one row for each previous application related to loans in our data sample.
- installments_payments.csv
Repayment history for the previously disbursed credits in Home Credit related to the loans in our sample.
There is a) one row for every payment that was made plus b) one row each for missed payment.
One row is equivalent to one payment of one installment OR one installment corresponding to one payment of one previous Home Credit credit related to loans in our sample.
- HomeCredit_columns_description.csv
This file contains descriptions for the columns in the various data files.

# Data Pre-processing
```
df_application_train = pd.read_csv("../data/application_train.csv")
df_application_train.head()
df_application_test = pd.read_csv("../data/application_test.csv")
df_application_test.head()
```
## Missing Value Imputation
```
df_application_train_imputed = pd.read_csv("../transformed_data/application_train_imputed.csv")
df_application_test_imputed = pd.read_csv("../transformed_data/application_test_imputed.csv")
df_application_train.shape, df_application_test.shape
df_application_train_imputed.shape, df_application_test_imputed.shape
df_application_train.isnull().sum(axis = 0).sum(), df_application_test.isnull().sum(axis = 0).sum()
df_application_train_imputed.isnull().sum(axis = 0).sum(), df_application_test_imputed.isnull().sum(axis = 0).sum()
```
# Model Building
## Encode categorical columns
```
# arr_categorical_columns = df_application_train.select_dtypes(['object']).columns
# for var_col in arr_categorical_columns:
# df_application_train[var_col] = df_application_train[var_col].astype('category').cat.codes
# arr_categorical_columns = df_application_test.select_dtypes(['object']).columns
# for var_col in arr_categorical_columns:
# df_application_test[var_col] = df_application_test[var_col].astype('category').cat.codes
```
## Train-Validation Split
```
input_columns = df_application_train_imputed.columns
input_columns = input_columns[input_columns != 'TARGET']
target_column = 'TARGET'
X = df_application_train_imputed[input_columns]
y = df_application_train_imputed[target_column]
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)
xgb_params = {
'seed': 0,
'colsample_bytree': 0.8,
'silent': 1,
'subsample': 0.6,
'learning_rate': 0.01,
'objective': 'binary:logistic',
'eval_metric': 'auc',
'max_depth': 6,
'num_parallel_tree': 1,
'min_child_weight': 5,
}
watchlist = [(xgb.DMatrix(X_train, y_train), 'train'), (xgb.DMatrix(X_test, y_test), 'valid')]
model = xgb.train(xgb_params, xgb.DMatrix(X_train, y_train), 270, watchlist, maximize=True, verbose_eval=100)
df_predict = model.predict(xgb.DMatrix(df_application_test_imputed), ntree_limit=model.best_ntree_limit)
submission = pd.DataFrame()
submission["SK_ID_CURR"] = df_application_test["SK_ID_CURR"]
submission["TARGET"] = df_predict
submission.to_csv("../submissions/model_1_xgbstarter_missingdata_MICE_imputed.csv", index=False)
submission.shape
input_columns = df_application_train.columns
input_columns = input_columns[input_columns != 'TARGET']
target_column = 'TARGET'
X = df_application_train[input_columns]
y = df_application_train[target_column]
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)
watchlist = [(xgb.DMatrix(X_train, y_train), 'train'), (xgb.DMatrix(X_test, y_test), 'valid')]
model = xgb.train(xgb_params, xgb.DMatrix(X_train, y_train), 270, watchlist, maximize=True, verbose_eval=100)
df_predict = model.predict(xgb.DMatrix(df_application_test), ntree_limit=model.best_ntree_limit)
submission = pd.DataFrame()
submission["SK_ID_CURR"] = df_application_test["SK_ID_CURR"]
submission["TARGET"] = df_predict
submission.to_csv("../submissions/model_1_xgbstarter_missingdata_MICE_nonimputed_hypothesis.csv", index=False)
submission.shape
```
|
github_jupyter
|
```
library(repr) ; options(repr.plot.width = 5, repr.plot.height = 6) # Change plot sizes (in cm)
```
# Bootstrapping using rTPC package
## Introduction
In this Chapter we will work through an example of model fitting using the rTPC package in R. This references the previous chapters' work, especially [Model Fitting the Bayesian way](https://www.youtube.com/watch?v=dQw4w9WgXcQ).
Lets start with the requirements!
```
require('ggplot2')
require('nls.multstart')
require('broom')
require('tidyverse')
require('rTPC')
require('dplyr')
require('data.table')
require('car')
require('boot')
require('patchwork')
require('minpack.lm')
require("tidyr")
require('purrr')
# update.packages(ask = FALSE)
rm(list=ls())
graphics.off()
setwd("/home/primuser/Documents/VByte/VecMismatchPaper1/code/")
```
Now that we have the background requirements going, we can start using the rTPC package. Lets look through the different models available!
```
#take a look at the different models available
get_model_names()
```
There are 24 models to choose from. For our purposes in this chapter we will be using the sharpesschoolhigh_1981 model. More information on the model can be found [here](https://padpadpadpad.github.io/rTPC/reference/sharpeschoolhigh_1981.html).
From here lets load in our data from the overall repository. This will be called '../data/Final_Traitofinterest.csv'.
```
#read in the trait data
final_trait_data <- read.csv('../data/Final_Traitofinterest.csv')
```
Lets reduce this to a single trait. This data comes from the [VectorBiTE database](https://legacy.vectorbyte.org/) and so has unique IDs. We will use this to get our species and trait of interest isolated from the larger dataset. In this example we will be looking at Development Rate across temperatures for Aedes albopictus, which we can find an example of in csm7I.
```
df1 <- final_trait_data %>%
dplyr::select('originalid', 'originaltraitname', 'originaltraitunit', 'originaltraitvalue', 'interactor1', 'ambienttemp', 'citation')
#filter to single species and trait
df2 <- dplyr::filter(df1, originalid == 'csm7I')
```
Now lets visualize our data in ggplot.
```
#visualize
ggplot(df2, aes(ambienttemp, originaltraitvalue))+
geom_point()+
theme_bw(base_size = 12) +
labs(x = 'Temperature (ºC)',
y = 'Development Rate',
title = 'Development Rate across temperatures for Aedes albopictus')
```
We will need to write which model we are using (sharpschoolhigh_1981). From here we can actually build our fit. We will use ''nls_multstart'' to automatically find our starting values. This lets us skip the [starting value problem](https://mhasoba.github.io/TheMulQuaBio/notebooks/20-ModelFitting-NLLS.html#the-starting-values-problem). From here we build our predicted line.
```
# choose model
mod = 'sharpschoolhigh_1981'
d<- df2 %>%
rename(temp = ambienttemp,
rate = originaltraitvalue)
# fit Sharpe-Schoolfield model
d_fit <- nest(d, data = c(temp, rate)) %>%
mutate(sharpeschoolhigh = map(data, ~nls_multstart(rate~sharpeschoolhigh_1981(temp = temp, r_tref,e,eh,th, tref = 15),
data = .x,
iter = c(3,3,3,3),
start_lower = get_start_vals(.x$temp, .x$rate, model_name = 'sharpeschoolhigh_1981') - 10,
start_upper = get_start_vals(.x$temp, .x$rate, model_name = 'sharpeschoolhigh_1981') + 10,
lower = get_lower_lims(.x$temp, .x$rate, model_name = 'sharpeschoolhigh_1981'),
upper = get_upper_lims(.x$temp, .x$rate, model_name = 'sharpeschoolhigh_1981'),
supp_errors = 'Y',
convergence_count = FALSE)),
# create new temperature data
new_data = map(data, ~tibble(temp = seq(min(.x$temp), max(.x$temp), length.out = 100))),
# predict over that data,
preds = map2(sharpeschoolhigh, new_data, ~augment(.x, newdata = .y)))
# unnest predictions
d_preds <- select(d_fit, preds) %>%
unnest(preds)
```
Lets visualize the line:
```
# plot data and predictions
ggplot() +
geom_line(aes(temp, .fitted), d_preds, col = 'blue') +
geom_point(aes(temp, rate), d, size = 2, alpha = 0.5) +
theme_bw(base_size = 12) +
labs(x = 'Temperature (ºC)',
y = 'Growth rate',
title = 'Growth rate across temperatures')
```
This looks like a good fit! We can start exploring using bootstrapping. Lets start with refitting the model using nlsLM.
```
# refit model using nlsLM
fit_nlsLM <- minpack.lm::nlsLM(rate~sharpeschoolhigh_1981(temp = temp, r_tref,e,eh,th, tref = 15),
data = d,
start = coef(d_fit$sharpeschoolhigh[[1]]),
lower = get_lower_lims(d$temp, d$rate, model_name = 'sharpeschoolhigh_1981'),
upper = get_upper_lims(d$temp, d$rate, model_name = 'sharpeschoolhigh_1981'),
weights = rep(1, times = nrow(d)))
```
Now we can actually bootstrap.
```
# bootstrap using case resampling
boot1 <- Boot(fit_nlsLM, method = 'case')
```
It is a good idea to explore the data again now.
```
# look at the data
head(boot1$t)
hist(boot1, layout = c(2,2))
```
Now we use the bootstrapped model to build predictions which we can explore visually.
```
# create predictions of each bootstrapped model
boot1_preds <- boot1$t %>%
as.data.frame() %>%
drop_na() %>%
mutate(iter = 1:n()) %>%
group_by_all() %>%
do(data.frame(temp = seq(min(d$temp), max(d$temp), length.out = 100))) %>%
ungroup() %>%
mutate(pred = sharpeschoolhigh_1981(temp, r_tref, e, eh, th, tref = 15))
# calculate bootstrapped confidence intervals
boot1_conf_preds <- group_by(boot1_preds, temp) %>%
summarise(conf_lower = quantile(pred, 0.025),
conf_upper = quantile(pred, 0.975)) %>%
ungroup()
# plot bootstrapped CIs
p1 <- ggplot() +
geom_line(aes(temp, .fitted), d_preds, col = 'blue') +
geom_ribbon(aes(temp, ymin = conf_lower, ymax = conf_upper), boot1_conf_preds, fill = 'blue', alpha = 0.3) +
geom_point(aes(temp, rate), d, size = 2, alpha = 0.5) +
theme_bw(base_size = 12) +
labs(x = 'Temperature (ºC)',
y = 'Growth rate',
title = 'Growth rate across temperatures')
# plot bootstrapped predictions
p2 <- ggplot() +
geom_line(aes(temp, .fitted), d_preds, col = 'blue') +
geom_line(aes(temp, pred, group = iter), boot1_preds, col = 'blue', alpha = 0.007) +
geom_point(aes(temp, rate), d, size = 2, alpha = 0.5) +
theme_bw(base_size = 12) +
labs(x = 'Temperature (ºC)',
y = 'Growth rate',
title = 'Growth rate across temperatures')
p1 + p2
```
We can see here that when we bootstrap this data, the fit is not as good as we would expect from the initial exploration. We do not necessarily get a good thermal optima from this data. However, this does show how to use this function in the future. Please see Daniel Padfields [git](https://padpadpadpad.github.io/rTPC/articles/rTPC.html) for more information on using the rTPC package.
# Please go to the [landing page](https://www.youtube.com/watch?v=YddwkMJG1Jo) and proceed on to the next stage of the training!
|
github_jupyter
|
# Targeting Direct Marketing with Amazon SageMaker XGBoost
_**Supervised Learning with Gradient Boosted Trees: A Binary Prediction Problem With Unbalanced Classes**_
---
## Background
Direct marketing, either through mail, email, phone, etc., is a common tactic to acquire customers. Because resources and a customer's attention is limited, the goal is to only target the subset of prospects who are likely to engage with a specific offer. Predicting those potential customers based on readily available information like demographics, past interactions, and environmental factors is a common machine learning problem.
This notebook presents an example problem to predict if a customer will enroll for a term deposit at a bank, after one or more phone calls. The steps include:
* Preparing your Amazon SageMaker notebook
* Downloading data from the internet into Amazon SageMaker
* Investigating and transforming the data so that it can be fed to Amazon SageMaker algorithms
* Estimating a model using the Gradient Boosting algorithm
* Evaluating the effectiveness of the model
* Setting the model up to make on-going predictions
---
## Preparation
_This notebook was created and tested on an ml.m4.xlarge notebook instance._
Let's start by specifying:
- The S3 bucket and prefix that you want to use for training and model data. This should be within the same region as the Notebook Instance, training, and hosting.
- The IAM role arn used to give training and hosting access to your data. See the documentation for how to create these. Note, if more than one role is required for notebook instances, training, and/or hosting, please replace the boto regexp with a the appropriate full IAM role arn string(s).
```
# cell 01
import sagemaker
bucket=sagemaker.Session().default_bucket()
prefix = 'sagemaker/DEMO-xgboost-dm'
# Define IAM role
import boto3
import re
from sagemaker import get_execution_role
role = get_execution_role()
```
Now let's bring in the Python libraries that we'll use throughout the analysis
```
# cell 02
import numpy as np # For matrix operations and numerical processing
import pandas as pd # For munging tabular data
import matplotlib.pyplot as plt # For charts and visualizations
from IPython.display import Image # For displaying images in the notebook
from IPython.display import display # For displaying outputs in the notebook
from time import gmtime, strftime # For labeling SageMaker models, endpoints, etc.
import sys # For writing outputs to notebook
import math # For ceiling function
import json # For parsing hosting outputs
import os # For manipulating filepath names
import sagemaker
import zipfile # Amazon SageMaker's Python SDK provides many helper functions
```
---
## Data
Let's start by downloading the [direct marketing dataset](https://sagemaker-sample-data-us-west-2.s3-us-west-2.amazonaws.com/autopilot/direct_marketing/bank-additional.zip) from the sample data s3 bucket.
\[Moro et al., 2014\] S. Moro, P. Cortez and P. Rita. A Data-Driven Approach to Predict the Success of Bank Telemarketing. Decision Support Systems, Elsevier, 62:22-31, June 2014
```
# cell 03
!wget https://sagemaker-sample-data-us-west-2.s3-us-west-2.amazonaws.com/autopilot/direct_marketing/bank-additional.zip
with zipfile.ZipFile('bank-additional.zip', 'r') as zip_ref:
zip_ref.extractall('.')
```
Now lets read this into a Pandas data frame and take a look.
```
# cell 04
data = pd.read_csv('./bank-additional/bank-additional-full.csv')
pd.set_option('display.max_columns', 500) # Make sure we can see all of the columns
pd.set_option('display.max_rows', 20) # Keep the output on one page
data
```
We will store this natively in S3 to then process it with SageMaker Processing.
```
# cell 05
from sagemaker import Session
sess = Session()
input_source = sess.upload_data('./bank-additional/bank-additional-full.csv', bucket=bucket, key_prefix=f'{prefix}/input_data')
input_source
```
# Feature Engineering with Amazon SageMaker Processing
Amazon SageMaker Processing allows you to run steps for data pre- or post-processing, feature engineering, data validation, or model evaluation workloads on Amazon SageMaker. Processing jobs accept data from Amazon S3 as input and store data into Amazon S3 as output.
Here, we'll import the dataset and transform it with SageMaker Processing, which can be used to process terabytes of data in a SageMaker-managed cluster separate from the instance running your notebook server. In a typical SageMaker workflow, notebooks are only used for prototyping and can be run on relatively inexpensive and less powerful instances, while processing, training and model hosting tasks are run on separate, more powerful SageMaker-managed instances. SageMaker Processing includes off-the-shelf support for Scikit-learn, as well as a Bring Your Own Container option, so it can be used with many different data transformation technologies and tasks.
To use SageMaker Processing, simply supply a Python data preprocessing script as shown below. For this example, we're using a SageMaker prebuilt Scikit-learn container, which includes many common functions for processing data. There are few limitations on what kinds of code and operations you can run, and only a minimal contract: input and output data must be placed in specified directories. If this is done, SageMaker Processing automatically loads the input data from S3 and uploads transformed data back to S3 when the job is complete.
```
# cell 06
%%writefile preprocessing.py
import pandas as pd
import numpy as np
import argparse
import os
from sklearn.preprocessing import OrdinalEncoder
def _parse_args():
parser = argparse.ArgumentParser()
# Data, model, and output directories
# model_dir is always passed in from SageMaker. By default this is a S3 path under the default bucket.
parser.add_argument('--filepath', type=str, default='/opt/ml/processing/input/')
parser.add_argument('--filename', type=str, default='bank-additional-full.csv')
parser.add_argument('--outputpath', type=str, default='/opt/ml/processing/output/')
parser.add_argument('--categorical_features', type=str, default='y, job, marital, education, default, housing, loan, contact, month, day_of_week, poutcome')
return parser.parse_known_args()
if __name__=="__main__":
# Process arguments
args, _ = _parse_args()
# Load data
df = pd.read_csv(os.path.join(args.filepath, args.filename))
# Change the value . into _
df = df.replace(regex=r'\.', value='_')
df = df.replace(regex=r'\_$', value='')
# Add two new indicators
df["no_previous_contact"] = (df["pdays"] == 999).astype(int)
df["not_working"] = df["job"].isin(["student", "retired", "unemployed"]).astype(int)
df = df.drop(['duration', 'emp.var.rate', 'cons.price.idx', 'cons.conf.idx', 'euribor3m', 'nr.employed'], axis=1)
# Encode the categorical features
df = pd.get_dummies(df)
# Train, test, validation split
train_data, validation_data, test_data = np.split(df.sample(frac=1, random_state=42), [int(0.7 * len(df)), int(0.9 * len(df))]) # Randomly sort the data then split out first 70%, second 20%, and last 10%
# Local store
pd.concat([train_data['y_yes'], train_data.drop(['y_yes','y_no'], axis=1)], axis=1).to_csv(os.path.join(args.outputpath, 'train/train.csv'), index=False, header=False)
pd.concat([validation_data['y_yes'], validation_data.drop(['y_yes','y_no'], axis=1)], axis=1).to_csv(os.path.join(args.outputpath, 'validation/validation.csv'), index=False, header=False)
test_data['y_yes'].to_csv(os.path.join(args.outputpath, 'test/test_y.csv'), index=False, header=False)
test_data.drop(['y_yes','y_no'], axis=1).to_csv(os.path.join(args.outputpath, 'test/test_x.csv'), index=False, header=False)
print("## Processing complete. Exiting.")
```
Before starting the SageMaker Processing job, we instantiate a `SKLearnProcessor` object. This object allows you to specify the instance type to use in the job, as well as how many instances.
```
# cell 07
train_path = f"s3://{bucket}/{prefix}/train"
validation_path = f"s3://{bucket}/{prefix}/validation"
test_path = f"s3://{bucket}/{prefix}/test"
# cell 08
from sagemaker.sklearn.processing import SKLearnProcessor
from sagemaker.processing import ProcessingInput, ProcessingOutput
from sagemaker import get_execution_role
sklearn_processor = SKLearnProcessor(
framework_version="0.23-1",
role=get_execution_role(),
instance_type="ml.m5.large",
instance_count=1,
base_job_name='sm-immday-skprocessing'
)
sklearn_processor.run(
code='preprocessing.py',
# arguments = ['arg1', 'arg2'],
inputs=[
ProcessingInput(
source=input_source,
destination="/opt/ml/processing/input",
s3_input_mode="File",
s3_data_distribution_type="ShardedByS3Key"
)
],
outputs=[
ProcessingOutput(
output_name="train_data",
source="/opt/ml/processing/output/train",
destination=train_path,
),
ProcessingOutput(output_name="validation_data", source="/opt/ml/processing/output/validation", destination=validation_path),
ProcessingOutput(output_name="test_data", source="/opt/ml/processing/output/test", destination=test_path),
]
)
# cell 09
!aws s3 ls $train_path/
```
---
## End of Lab 1
---
## Training
Now we know that most of our features have skewed distributions, some are highly correlated with one another, and some appear to have non-linear relationships with our target variable. Also, for targeting future prospects, good predictive accuracy is preferred to being able to explain why that prospect was targeted. Taken together, these aspects make gradient boosted trees a good candidate algorithm.
There are several intricacies to understanding the algorithm, but at a high level, gradient boosted trees works by combining predictions from many simple models, each of which tries to address the weaknesses of the previous models. By doing this the collection of simple models can actually outperform large, complex models. Other Amazon SageMaker notebooks elaborate on gradient boosting trees further and how they differ from similar algorithms.
`xgboost` is an extremely popular, open-source package for gradient boosted trees. It is computationally powerful, fully featured, and has been successfully used in many machine learning competitions. Let's start with a simple `xgboost` model, trained using Amazon SageMaker's managed, distributed training framework.
First we'll need to specify the ECR container location for Amazon SageMaker's implementation of XGBoost.
```
# cell 10
container = sagemaker.image_uris.retrieve(region=boto3.Session().region_name, framework='xgboost', version='latest')
```
Then, because we're training with the CSV file format, we'll create `s3_input`s that our training function can use as a pointer to the files in S3, which also specify that the content type is CSV.
```
# cell 11
s3_input_train = sagemaker.inputs.TrainingInput(s3_data=train_path.format(bucket, prefix), content_type='csv')
s3_input_validation = sagemaker.inputs.TrainingInput(s3_data=validation_path.format(bucket, prefix), content_type='csv')
```
First we'll need to specify training parameters to the estimator. This includes:
1. The `xgboost` algorithm container
1. The IAM role to use
1. Training instance type and count
1. S3 location for output data
1. Algorithm hyperparameters
And then a `.fit()` function which specifies:
1. S3 location for output data. In this case we have both a training and validation set which are passed in.
```
# cell 12
sess = sagemaker.Session()
xgb = sagemaker.estimator.Estimator(container,
role,
instance_count=1,
instance_type='ml.m4.xlarge',
output_path='s3://{}/{}/output'.format(bucket, prefix),
sagemaker_session=sess)
xgb.set_hyperparameters(max_depth=5,
eta=0.2,
gamma=4,
min_child_weight=6,
subsample=0.8,
silent=0,
objective='binary:logistic',
num_round=100)
xgb.fit({'train': s3_input_train, 'validation': s3_input_validation})
```
---
## Hosting
Now that we've trained the `xgboost` algorithm on our data, let's deploy a model that's hosted behind a real-time endpoint.
```
# cell 13
xgb_predictor = xgb.deploy(initial_instance_count=1,
instance_type='ml.m4.xlarge')
```
---
## Evaluation
There are many ways to compare the performance of a machine learning model, but let's start by simply comparing actual to predicted values. In this case, we're simply predicting whether the customer subscribed to a term deposit (`1`) or not (`0`), which produces a simple confusion matrix.
First we'll need to determine how we pass data into and receive data from our endpoint. Our data is currently stored as NumPy arrays in memory of our notebook instance. To send it in an HTTP POST request, we'll serialize it as a CSV string and then decode the resulting CSV.
*Note: For inference with CSV format, SageMaker XGBoost requires that the data does NOT include the target variable.*
```
# cell 14
xgb_predictor.serializer = sagemaker.serializers.CSVSerializer()
```
Now, we'll use a simple function to:
1. Loop over our test dataset
1. Split it into mini-batches of rows
1. Convert those mini-batches to CSV string payloads (notice, we drop the target variable from our dataset first)
1. Retrieve mini-batch predictions by invoking the XGBoost endpoint
1. Collect predictions and convert from the CSV output our model provides into a NumPy array
```
# cell 15
!aws s3 cp $test_path/test_x.csv /tmp/test_x.csv
!aws s3 cp $test_path/test_y.csv /tmp/test_y.csv
# cell 16
def predict(data, predictor, rows=500 ):
split_array = np.array_split(data, int(data.shape[0] / float(rows) + 1))
predictions = ''
for array in split_array:
predictions = ','.join([predictions, predictor.predict(array).decode('utf-8')])
return np.fromstring(predictions[1:], sep=',')
test_x = pd.read_csv('/tmp/test_x.csv', names=[f'{i}' for i in range(59)])
test_y = pd.read_csv('/tmp/test_y.csv', names=['y'])
predictions = predict(test_x.drop(test_x.columns[0], axis=1).to_numpy(), xgb_predictor)
```
Now we'll check our confusion matrix to see how well we predicted versus actuals.
```
# cell 17
pd.crosstab(index=test_y['y'].values, columns=np.round(predictions), rownames=['actuals'], colnames=['predictions'])
```
So, of the ~4000 potential customers, we predicted 136 would subscribe and 94 of them actually did. We also had 389 subscribers who subscribed that we did not predict would. This is less than desirable, but the model can (and should) be tuned to improve this. Most importantly, note that with minimal effort, our model produced accuracies similar to those published [here](http://media.salford-systems.com/video/tutorial/2015/targeted_marketing.pdf).
_Note that because there is some element of randomness in the algorithm's subsample, your results may differ slightly from the text written above._
## Automatic model Tuning (optional)
Amazon SageMaker automatic model tuning, also known as hyperparameter tuning, finds the best version of a model by running many training jobs on your dataset using the algorithm and ranges of hyperparameters that you specify. It then chooses the hyperparameter values that result in a model that performs the best, as measured by a metric that you choose.
For example, suppose that you want to solve a binary classification problem on this marketing dataset. Your goal is to maximize the area under the curve (auc) metric of the algorithm by training an XGBoost Algorithm model. You don't know which values of the eta, alpha, min_child_weight, and max_depth hyperparameters to use to train the best model. To find the best values for these hyperparameters, you can specify ranges of values that Amazon SageMaker hyperparameter tuning searches to find the combination of values that results in the training job that performs the best as measured by the objective metric that you chose. Hyperparameter tuning launches training jobs that use hyperparameter values in the ranges that you specified, and returns the training job with highest auc.
```
# cell 18
from sagemaker.tuner import IntegerParameter, CategoricalParameter, ContinuousParameter, HyperparameterTuner
hyperparameter_ranges = {'eta': ContinuousParameter(0, 1),
'min_child_weight': ContinuousParameter(1, 10),
'alpha': ContinuousParameter(0, 2),
'max_depth': IntegerParameter(1, 10)}
# cell 19
objective_metric_name = 'validation:auc'
# cell 20
tuner = HyperparameterTuner(xgb,
objective_metric_name,
hyperparameter_ranges,
max_jobs=20,
max_parallel_jobs=3)
# cell 21
tuner.fit({'train': s3_input_train, 'validation': s3_input_validation})
# cell 22
boto3.client('sagemaker').describe_hyper_parameter_tuning_job(
HyperParameterTuningJobName=tuner.latest_tuning_job.job_name)['HyperParameterTuningJobStatus']
# cell 23
# return the best training job name
tuner.best_training_job()
# cell 24
# Deploy the best trained or user specified model to an Amazon SageMaker endpoint
tuner_predictor = tuner.deploy(initial_instance_count=1,
instance_type='ml.m4.xlarge')
# cell 25
# Create a serializer
tuner_predictor.serializer = sagemaker.serializers.CSVSerializer()
# cell 26
# Predict
predictions = predict(test_x.to_numpy(),tuner_predictor)
# cell 27
# Collect predictions and convert from the CSV output our model provides into a NumPy array
pd.crosstab(index=test_y['y'].values, columns=np.round(predictions), rownames=['actuals'], colnames=['predictions'])
```
---
## Extensions
This example analyzed a relatively small dataset, but utilized Amazon SageMaker features such as distributed, managed training and real-time model hosting, which could easily be applied to much larger problems. In order to improve predictive accuracy further, we could tweak value we threshold our predictions at to alter the mix of false-positives and false-negatives, or we could explore techniques like hyperparameter tuning. In a real-world scenario, we would also spend more time engineering features by hand and would likely look for additional datasets to include which contain customer information not available in our initial dataset.
### (Optional) Clean-up
If you are done with this notebook, please run the cell below. This will remove the hosted endpoint you created and avoid any charges from a stray instance being left on.
```
# cell 28
xgb_predictor.delete_endpoint(delete_endpoint_config=True)
# cell 29
tuner_predictor.delete_endpoint(delete_endpoint_config=True)
```
|
github_jupyter
|
# Machine Learning
## Types of learning
- Whether or not they are trained with human supervision (supervised, unsupervised, semisupervised, and Reinforcement Learning)
- Whether or not they can learn incrementally on the fly (online versus batch learning)
- Whether they work by simply comparing new data points to known data points, or instead detect patterns in the training data and build a predictive model, much like scientists do (instance-based versus model-based learning)
# Types Of Machine Learning Systems
## Supervised VS Unsupervised learning
### Supervised Learning
- In supervised learning, the training data you feed to the algorithm includes the desired solutions, called labels
- k-Nearest Neighbors
- Linear Regression
- Logistic Regression
- Support Vector Machines (SVMs)
- Decision Trees and Random Forests
- Neural networks
### Unsupervised Learning
- In unsupervised learning, as you might guess, the training data is unlabeled. The system tries to learn without a teacher
- Clustering
- k-Means
- Hierarchical Cluster Analysis (HCA)
- Expectation Maximization
- DBSCAN
- Visualization and dimensionality reduction
- Principal Component Analysis (PCA)
- Kernel PCA
- Locally-Linear Embedding (LLE)
- t-distributed Stochastic Neighbor Embedding (t-SNE)
- Association rule learning
- Apriori
- Eclat
- Anamoly detection and novelty detection
- One-Class SVM
- Isolation Forest
## Semisupervised Learning
- Some algorithms can deal with partially labeled training data, usually a lot of unla beled data and a little bit of labeled data. This is called semisupervised learning
- Most semisupervised learning algorithms are combinations of unsupervised and supervised algorithms. For example, deep belief networks (DBNs) are based on unsu‐ pervised components called restricted Boltzmann machines (RBMs) stacked on top of one another. RBMs are trained sequentially in an unsupervised manner, and then the whole system is fine-tuned using supervised learning techniques.
## Reinforcement Learning
- Reinforcement Learning is a very different beast. The learning system, called an agent in this context, can observe the environment, select and perform actions, and get rewards in return (or penalties in the form of negative rewards. It must then learn by itself what is the best strategy, called a policy, to get the most reward over time. A policy defines what action the agent should choose when it is in a given situation.
## Batch And Online Learning
### Batch Learning
- In batch learning, the system is incapable of learning incrementally: it must be trained using all the available data. This will generally take a lot of time and computing resources, so it is typically done offline. First the system is trained, and then it is launched into production and runs without learning anymore; it just applies what it has learned. This is called offline learning.
- If you want a batch learning system to know about new data (such as a new type of spam), you need to train a new version of the system from scratch on the full dataset (not just the new data, but also the old data), then stop the old system and replace it with the new one.
### Online Learning
- In online learning, you train the system incrementally by feeding it data instances sequentially, either individually or by small groups called mini-batches. Each learning step is fast and cheap, so the system can learn about new data on the fly, as it arrives
- Online learning is great for systems that receive data as a continuous flow (e.g., stock prices) and need to adapt to change rapidly or autonomously. It is also a good option if you have limited computing resources: once an online learning system has learned about new data instances, it does not need them anymore, so you can discard them (unless you want to be able to roll back to a previous state and “replay” the data). This can save a huge amount of space.
- Online learning algorithms can also be used to train systems on huge datasets that cannot fit in one machine’s main memory (this is called out-of-core learning). The algorithm loads part of the data, runs a training step on that data, and repeats the process until it has run on all of the data
- This whole process is usually done offline (i.e., not on the live system), so online learning can be a confusing name. Think of it as incremental learning
- Importance of Learning Rate in Online Learning
## Instance-Based Vs Model-Based Learning
### Instance-Based Learning
- the system learns the examples by heart, then generalizes to new cases using a similarity measure
### Model-Based Learning
- Another way to generalize from a set of examples is to build a model of these examples, then use that model to make predictions. This is called model-based learning
# Challenges of Machine Learning
- Insuffcient Quantity of Training Data
- Nonrepresentative Training Data
- Poor-Quality Data
- Irrelevant Features
- Overfitting the Training Data
- Underfitting the Training Data
Most Common Supervised Learning Tasks are Classification(predicting classes) And Regression(predicting Values)
|
github_jupyter
|
<img src="https://raw.githubusercontent.com/Qiskit/qiskit-tutorials/master/images/qiskit-heading.png" alt="Note: In order for images to show up in this jupyter notebook you need to select File => Trusted Notebook" width="500 px" align="left">
## _*Superposition*_
The latest version of this notebook is available on https://github.com/qiskit/qiskit-tutorial.
***
### Contributors
Jay Gambetta, Antonio Córcoles, Andrew Cross, Anna Phan
### Qiskit Package Versions
```
import qiskit
qiskit.__qiskit_version__
```
## Introduction
Many people tend to think quantum physics is hard math, but this is not actually true. Quantum concepts are very similar to those seen in the linear algebra classes you may have taken as a freshman in college, or even in high school. The challenge of quantum physics is the necessity to accept counter-intuitive ideas, and its lack of a simple underlying theory. We believe that if you can grasp the following two Principles, you will have a good start:
1. A physical system in a definite state can still behave randomly.
2. Two systems that are too far apart to influence each other can nevertheless behave in ways that, though individually random, are somehow strongly correlated.
In this tutorial, we will be discussing the first of these Principles, the second is discussed in [this other tutorial](entanglement_introduction.ipynb).
```
# useful additional packages
import matplotlib.pyplot as plt
%matplotlib inline
import numpy as np
# importing Qiskit
from qiskit import QuantumCircuit, QuantumRegister, ClassicalRegister, execute
from qiskit import BasicAer, IBMQ
# import basic plot tools
from qiskit.tools.visualization import plot_histogram
backend = BasicAer.get_backend('qasm_simulator') # run on local simulator by default
# Uncomment the following lines to run on a real device
#IBMQ.load_accounts()
#from qiskit.providers.ibmq import least_busy
#backend = least_busy(IBMQ.backends(operational=True, simulator=False))
#print("the best backend is " + backend.name())
```
## Quantum States - Basis States and Superpositions<a id='section1'></a>
The first Principle above tells us that the results of measuring a quantum state may be random or deterministic, depending on what basis is used. To demonstrate, we will first introduce the computational (or standard) basis for a qubit.
The computational basis is the set containing the ground and excited state $\{|0\rangle,|1\rangle\}$, which also corresponds to the following vectors:
$$|0\rangle =\begin{pmatrix} 1 \\ 0 \end{pmatrix}$$
$$|1\rangle =\begin{pmatrix} 0 \\ 1 \end{pmatrix}$$
In Python these are represented by
```
zero = np.array([[1],[0]])
one = np.array([[0],[1]])
```
In our quantum processor system (and many other physical quantum processors) it is natural for all qubits to start in the $|0\rangle$ state, known as the ground state. To make the $|1\rangle$ (or excited) state, we use the operator
$$ X =\begin{pmatrix} 0 & 1 \\ 1 & 0 \end{pmatrix}.$$
This $X$ operator is often called a bit-flip because it exactly implements the following:
$$X: |0\rangle \rightarrow |1\rangle$$
$$X: |1\rangle \rightarrow |0\rangle.$$
In Python this can be represented by the following:
```
X = np.array([[0,1],[1,0]])
print(np.dot(X,zero))
print(np.dot(X,one))
```
Next, we give the two quantum circuits for preparing and measuring a single qubit in the ground and excited states using Qiskit.
```
# Creating registers
qr = QuantumRegister(1)
cr = ClassicalRegister(1)
# Quantum circuit ground
qc_ground = QuantumCircuit(qr, cr)
qc_ground.measure(qr[0], cr[0])
# Quantum circuit excited
qc_excited = QuantumCircuit(qr, cr)
qc_excited.x(qr)
qc_excited.measure(qr[0], cr[0])
qc_ground.draw(output='mpl')
qc_excited.draw(output='mpl')
```
Here we have created two jobs with different quantum circuits; the first to prepare the ground state, and the second to prepare the excited state. Now we can run the prepared jobs.
```
circuits = [qc_ground, qc_excited]
job = execute(circuits, backend)
result = job.result()
```
After the run has been completed, the data can be extracted from the API output and plotted.
```
plot_histogram(result.get_counts(qc_ground))
plot_histogram(result.get_counts(qc_excited))
```
Here we see that the qubit is in the $|0\rangle$ state with 100% probability for the first circuit and in the $|1\rangle$ state with 100% probability for the second circuit. If we had run on a quantum processor rather than the simulator, there would be a difference from the ideal perfect answer due to a combination of measurement error, preparation error, and gate error (for the $|1\rangle$ state).
Up to this point, nothing is different from a classical system of a bit. To go beyond, we must explore what it means to make a superposition. The operation in the quantum circuit language for generating a superposition is the Hadamard gate, $H$. Let's assume for now that this gate is like flipping a fair coin. The result of a flip has two possible outcomes, heads or tails, each occurring with equal probability. If we repeat this simple thought experiment many times, we would expect that on average we will measure as many heads as we do tails. Let heads be $|0\rangle$ and tails be $|1\rangle$.
Let's run the quantum version of this experiment. First we prepare the qubit in the ground state $|0\rangle$. We then apply the Hadamard gate (coin flip). Finally, we measure the state of the qubit. Repeat the experiment 1024 times (shots). As you likely predicted, half the outcomes will be in the $|0\rangle$ state and half will be in the $|1\rangle$ state.
Try the program below.
```
# Quantum circuit superposition
qc_superposition = QuantumCircuit(qr, cr)
qc_superposition.h(qr)
qc_superposition.measure(qr[0], cr[0])
qc_superposition.draw()
job = execute(qc_superposition, backend, shots = 1024)
result = job.result()
plot_histogram(result.get_counts(qc_superposition))
```
Indeed, much like a coin flip, the results are close to 50/50 with some non-ideality due to errors (again due to state preparation, measurement, and gate errors). So far, this is still not unexpected. Let's run the experiment again, but this time with two $H$ gates in succession. If we consider the $H$ gate to be analog to a coin flip, here we would be flipping it twice, and still expecting a 50/50 distribution.
```
# Quantum circuit two Hadamards
qc_twohadamard = QuantumCircuit(qr, cr)
qc_twohadamard.h(qr)
qc_twohadamard.barrier()
qc_twohadamard.h(qr)
qc_twohadamard.measure(qr[0], cr[0])
qc_twohadamard.draw(output='mpl')
job = execute(qc_twohadamard, backend)
result = job.result()
plot_histogram(result.get_counts(qc_twohadamard))
```
This time, the results are surprising. Unlike the classical case, with high probability the outcome is not random, but in the $|0\rangle$ state. *Quantum randomness* is not simply like a classical random coin flip. In both of the above experiments, the system (without noise) is in a definite state, but only in the first case does it behave randomly. This is because, in the first case, via the $H$ gate, we make a uniform superposition of the ground and excited state, $(|0\rangle+|1\rangle)/\sqrt{2}$, but then follow it with a measurement in the computational basis. The act of measurement in the computational basis forces the system to be in either the $|0\rangle$ state or the $|1\rangle$ state with an equal probability (due to the uniformity of the superposition). In the second case, we can think of the second $H$ gate as being a part of the final measurement operation; it changes the measurement basis from the computational basis to a *superposition* basis. The following equations illustrate the action of the $H$ gate on the computational basis states:
$$H: |0\rangle \rightarrow |+\rangle=\frac{|0\rangle+|1\rangle}{\sqrt{2}}$$
$$H: |1\rangle \rightarrow |-\rangle=\frac{|0\rangle-|1\rangle}{\sqrt{2}}.$$
We can redefine this new transformed basis, the superposition basis, as the set {$|+\rangle$, $|-\rangle$}. We now have a different way of looking at the second experiment above. The first $H$ gate prepares the system into a superposition state, namely the $|+\rangle$ state. The second $H$ gate followed by the standard measurement changes it into a measurement in the superposition basis. If the measurement gives 0, we can conclude that the system was in the $|+\rangle$ state before the second $H$ gate, and if we obtain 1, it means the system was in the $|-\rangle$ state. In the above experiment we see that the outcome is mainly 0, suggesting that our system was in the $|+\rangle$ superposition state before the second $H$ gate.
The math is best understood if we represent the quantum superposition state $|+\rangle$ and $|-\rangle$ by:
$$|+\rangle =\frac{1}{\sqrt{2}}\begin{pmatrix} 1 \\ 1 \end{pmatrix}$$
$$|-\rangle =\frac{1}{\sqrt{2}}\begin{pmatrix} 1 \\ -1 \end{pmatrix}$$
A standard measurement, known in quantum mechanics as a projective or von Neumann measurement, takes any superposition state of the qubit and projects it to either the state $|0\rangle$ or the state $|1\rangle$ with a probability determined by:
$$P(i|\psi) = |\langle i|\psi\rangle|^2$$
where $P(i|\psi)$ is the probability of measuring the system in state $i$ given preparation $\psi$.
We have written the Python function ```state_overlap``` to return this:
```
state_overlap = lambda state1, state2: np.absolute(np.dot(state1.conj().T,state2))**2
```
Now that we have a simple way of going from a state to the probability distribution of a standard measurement, we can go back to the case of a superposition made from the Hadamard gate. The Hadamard gate is defined by the matrix:
$$ H =\frac{1}{\sqrt{2}}\begin{pmatrix} 1 & 1 \\ 1 & -1 \end{pmatrix}$$
The $H$ gate acting on the state $|0\rangle$ gives:
```
Hadamard = np.array([[1,1],[1,-1]],dtype=complex)/np.sqrt(2)
psi1 = np.dot(Hadamard,zero)
P0 = state_overlap(zero,psi1)
P1 = state_overlap(one,psi1)
plot_histogram({'0' : P0.item(0), '1' : P1.item(0)})
```
which is the ideal version of the first superposition experiment.
The second experiment involves applying the Hadamard gate twice. While matrix multiplication shows that the product of two Hadamards is the identity operator (meaning that the state $|0\rangle$ remains unchanged), here (as previously mentioned) we prefer to interpret this as doing a measurement in the superposition basis. Using the above definitions, you can show that $H$ transforms the computational basis to the superposition basis.
```
print(np.dot(Hadamard,zero))
print(np.dot(Hadamard,one))
```
This is just the beginning of how a quantum state differs from a classical state. Please continue to [Amplitude and Phase](amplitude_and_phase.ipynb) to explore further!
|
github_jupyter
|
### Image Classification - Conv Nets -Pytorch
> Classifying if an image is a `bee` of an `ant` using `ConvNets` in pytorch
### Imports
```
import cv2
import matplotlib.pyplot as plt
import numpy as np
from sklearn.model_selection import train_test_split
import torch
from torch import nn
import torch.nn.functional as F
import os
```
### Data Preparation
```
class Insect:
BEE = 'BEE'
ANT = "ANT"
BEES_IMAGES_PATH = 'data/colored/rgb/bees'
ANTS_IMAGES_PATH = 'data/colored/rgb/ants'
classes = {'bee': 0, 'ant' : 1}
classes =dict([(i, j) for (j, i) in classes.items()])
classes
os.path.exists(Insect.BEES_IMAGES_PATH)
insects = []
for path in os.listdir(Insect.BEES_IMAGES_PATH):
img_path = os.path.join(Insect.BEES_IMAGES_PATH, path)
image = np.array(cv2.imread(img_path, cv2.IMREAD_UNCHANGED), dtype='float32')
image = image / 255
insects.append([image, 0])
for path in os.listdir(Insect.ANTS_IMAGES_PATH):
img_path = os.path.join(Insect.ANTS_IMAGES_PATH, path)
image = np.array(cv2.imread(img_path, cv2.IMREAD_UNCHANGED), dtype='float32')
image = image / 255
insects.append([image, 1])
insects = np.array(insects)
np.random.shuffle(insects)
```
### Visualization
```
plt.imshow(insects[7][0], cmap="gray"), insects[10][0].shape
```
> Seperating Labels and features
```
X = np.array([insect[0] for insect in insects])
y = np.array([insect[1] for insect in insects])
X[0].shape
```
> Splitting the data into training and test.
```
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=33, test_size=.2)
X_train.shape, y_train.shape, y_test.shape, X_test.shape
```
> Converting the data into `torch` tensor.
```
X_train = torch.from_numpy(X_train.astype('float32'))
X_test = torch.from_numpy(X_test.astype('float32'))
y_train = torch.Tensor(y_train)
y_test = torch.Tensor(y_test)
```
### Model Creation
```
class Net(nn.Module):
def __init__(self):
super().__init__()
self.conv1 = nn.Conv2d(in_channels=3, out_channels= 32, kernel_size=(3, 3))
self.conv2 = nn.Conv2d(32, 64, (3, 3))
self.conv3 = nn.Conv2d(64, 64, (3, 3))
self._to_linear = None # protected variable
self.x = torch.randn(3, 200, 200).view(-1, 3, 200, 200)
self.conv(self.x)
self.fc1 = nn.Linear(self._to_linear, 64)
self.fc2 = nn.Linear(64, 2)
def conv(self, x):
x = F.max_pool2d(F.relu(self.conv1(x)), (2, 2))
x = F.max_pool2d(F.relu(self.conv2(x)), (2, 2))
x = F.max_pool2d(F.relu(self.conv3(x)), (2, 2))
if self._to_linear is None:
self._to_linear = x.shape[1] * x.shape[2] * x.shape[3]
return x
def forward(self, x):
x = self.conv(x)
x = x.view(-1, self._to_linear)
x = F.relu(self.fc1(x))
return x
net = Net()
net
optimizer = torch.optim.SGD(net.parameters(), lr=1e-3)
loss_function = nn.CrossEntropyLoss()
EPOCHS = 10
BATCH_SIZE = 5
for epoch in range(EPOCHS):
print(f'Epochs: {epoch+1}/{EPOCHS}')
for i in range(0, len(y_train), BATCH_SIZE):
X_batch = X_train[i: i+BATCH_SIZE].view(-1, 3, 200, 200)
y_batch = y_train[i: i+BATCH_SIZE].long()
net.zero_grad() ## or you can say optimizer.zero_grad()
outputs = net(X_batch)
loss = loss_function(outputs, y_batch)
loss.backward()
optimizer.step()
print("Loss", loss)
```
### Evaluating the model
### Test set
```
total, correct = 0, 0
with torch.no_grad():
for i in range(len(X_test)):
correct_label = torch.argmax(y_test[i])
prediction = torch.argmax(net(X_test[i].view(-1, 3, 200, 200))[0])
if prediction == correct_label:
correct+=1
total +=1
print(f"Accuracy: {correct/total}")
torch.argmax(net(X_test[1].view(-1, 3, 200, 200))), y_test[0]
```
### Train set
```
total, correct = 0, 0
with torch.no_grad():
for i in range(len(X_train)):
correct_label = torch.argmax(y_train[i])
prediction = torch.argmax(net(X_train[i].view(-1, 3, 200, 200))[0])
if prediction == correct_label:
correct+=1
total +=1
print(f"Accuracy: {correct/total}")
```
### Making Predictions
```
plt.imshow(X_test[12])
plt.title(classes[torch.argmax(net(X_test[12].view(-1, 3, 200, 200))).item()].title(), fontsize=16)
plt.show()
fig, ax = plt.subplots(nrows=3, ncols=3, figsize=(10, 10))
for row in ax:
for col in row:
col.imshow(X_test[2])
plt.show()
```
|
github_jupyter
|
# Using Models as Layers in Another Model
In this notebook, we show how you can use Keras models as Layers within a larger model and still perform pruning on that model.
```
# Import required packages
import tensorflow as tf
import mann
from sklearn.metrics import confusion_matrix, classification_report
# Load the data
(x_train, y_train), (x_test, y_test) = tf.keras.datasets.fashion_mnist.load_data()
# Convert images from grayscale to RGB
x_train = tf.image.grayscale_to_rgb(tf.Variable(x_train.reshape(-1, 28, 28, 1)))
x_test = tf.image.grayscale_to_rgb(tf.Variable(x_test.reshape(-1, 28, 28, 1)))
```
## Model Creation
In the following cells, we create two models and put them together to create a larger model. The first model, called the `preprocess_model`, takes in images, divides the pixel values by 255 to ensure all values are between 0 and 1, resized the image to a height and width of 40 pixels. It then performs training data augmentation by randomly flips some images across the y-axis, randomly rotates images, and randomly translates the images.
The second model, called the `true_model`, contains the logic for performing prediction on images. It contains blocks of convolutional layers followed by max pooling and dropout layers. The output of these blocks is flattened and passed through fully-connected layers to output predicted class probabilities.
These two models are combined in the `training_model` to be trained.
```
preprocess_model = tf.keras.models.Sequential()
preprocess_model.add(tf.keras.layers.Rescaling(1./255))
preprocess_model.add(tf.keras.layers.Resizing(40, 40, input_shape = (None, None, 3)))
preprocess_model.add(tf.keras.layers.RandomFlip('horizontal'))
preprocess_model.add(tf.keras.layers.RandomRotation(0.1))
preprocess_model.add(tf.keras.layers.RandomTranslation(0.1, 0.1))
true_model = tf.keras.models.Sequential()
true_model.add(mann.layers.MaskedConv2D(16, padding = 'same', input_shape = (40, 40, 3)))
true_model.add(mann.layers.MaskedConv2D(16, padding = 'same'))
true_model.add(tf.keras.layers.MaxPool2D())
true_model.add(tf.keras.layers.Dropout(0.2))
true_model.add(mann.layers.MaskedConv2D(32, padding = 'same', activation = 'relu'))
true_model.add(mann.layers.MaskedConv2D(32, padding = 'same', activation = 'relu'))
true_model.add(tf.keras.layers.MaxPool2D())
true_model.add(tf.keras.layers.Dropout(0.2))
true_model.add(mann.layers.MaskedConv2D(64, padding = 'same', activation = 'relu'))
true_model.add(mann.layers.MaskedConv2D(64, padding = 'same', activation = 'relu'))
true_model.add(tf.keras.layers.MaxPool2D())
true_model.add(tf.keras.layers.Dropout(0.2))
true_model.add(tf.keras.layers.Flatten())
true_model.add(mann.layers.MaskedDense(256, activation = 'relu'))
true_model.add(mann.layers.MaskedDense(256, activation = 'relu'))
true_model.add(mann.layers.MaskedDense(10, activation = 'softmax'))
training_input = tf.keras.layers.Input((None, None, 3))
training_x = preprocess_model(training_input)
training_output = true_model(training_x)
training_model = tf.keras.models.Model(
training_input,
training_output
)
training_model.compile(
loss = 'sparse_categorical_crossentropy',
metrics = ['accuracy'],
optimizer = 'adam'
)
training_model.summary()
```
## Model Training
In this cell, we create the `ActiveSparsification` object to continually sparsify the model as it trains, and train the model.
```
callback = mann.utils.ActiveSparsification(
0.80,
sparsification_rate = 5
)
training_model.fit(
x_train,
y_train,
epochs = 200,
batch_size = 512,
validation_split = 0.2,
callbacks = [callback]
)
```
## Convert the model to not have masking layers
In the following cell, we configure the model to remove masking layers and replace them with non-masking native TensorFlow layers. We then perform prediction on the resulting model and present the results.
```
model = mann.utils.remove_layer_masks(training_model)
preds = model.predict(x_test).argmax(axis = 1)
print(confusion_matrix(y_test, preds))
print(classification_report(y_test, preds))
```
## Save only the model that performs prediction
Lastly, save only the part of the model that performs prediction
```
model.layers[2].save('ModelLayer.h5')
```
|
github_jupyter
|
```
import pathlib
import lzma
import re
import os
import datetime
import copy
import functools
import numpy as np
import pandas as pd
# Makes it so any changes in pymedphys is automatically
# propagated into the notebook without needing a kernel reset.
from IPython.lib.deepreload import reload
%load_ext autoreload
%autoreload 2
import pymedphys._icom.extract
import pymedphys.mudensity
import pymedphys
patient_id = '008566'
patients_dir = pathlib.Path(r'\\physics-server\iComLogFiles\patients')
patient_data_paths = list(patients_dir.glob(f'{patient_id}_*/*.xz'))
patient_data_paths
patient_data_path = patient_data_paths[0]
with lzma.open(patient_data_path, 'r') as f:
patient_data = f.read()
DATE_PATTERN = re.compile(rb"\d\d\d\d-\d\d-\d\d\d\d:\d\d:\d\d.")
def get_data_points(data):
date_index = [m.span() for m in DATE_PATTERN.finditer(data)]
start_points = [span[0] - 8 for span in date_index]
end_points = start_points[1::] + [None]
data_points = [data[start:end] for start, end in zip(start_points, end_points)]
return data_points
patient_data_list = get_data_points(patient_data)
len(patient_data_list)
@functools.lru_cache()
def get_coll_regex(label, number):
header = rb"0\xb8\x00DS\x00R.\x00\x00\x00" + label + b"\n"
item = rb"0\x1c\x01DS\x00R.\x00\x00\x00(-?\d+\.\d+)"
regex = re.compile(header + b"\n".join([item] * number))
return regex
def extract_coll(data, label, number):
regex = get_coll_regex(label, number)
match = regex.search(data)
span = match.span()
data = data[0 : span[0]] + data[span[1] + 1 : :]
items = np.array([float(item) for item in match.groups()])
return data, items
def get_delivery_data_items(single_icom_stream):
shrunk_stream, mu = pymedphys._icom.extract.extract(single_icom_stream, "Delivery MU")
shrunk_stream, gantry = pymedphys._icom.extract.extract(shrunk_stream, "Gantry")
shrunk_stream, collimator = pymedphys._icom.extract.extract(shrunk_stream, "Collimator")
shrunk_stream, mlc = extract_coll(shrunk_stream, b"MLCX", 160)
mlc = mlc.reshape((80,2))
mlc = np.fliplr(np.flipud(mlc * 10))
mlc[:,1] = -mlc[:,1]
mlc = np.round(mlc,10)
# shrunk_stream, result["ASYMX"] = extract_coll(shrunk_stream, b"ASYMX", 2)
shrunk_stream, jaw = extract_coll(shrunk_stream, b"ASYMY", 2)
jaw = np.round(np.array(jaw) * 10, 10)
jaw = np.flipud(jaw)
return mu, gantry, collimator, mlc, jaw
mu, gantry, collimator, mlc, jaw = get_delivery_data_items(patient_data_list[250])
gantry
collimator
mu
jaw
len(patient_data_list)
delivery_raw = [
get_delivery_data_items(single_icom_stream)
for single_icom_stream in patient_data_list
]
mu = np.array([item[0] for item in delivery_raw])
diff_mu = np.concatenate([[0], np.diff(mu)])
diff_mu[diff_mu<0] = 0
mu = np.cumsum(diff_mu)
gantry = np.array([item[1] for item in delivery_raw])
collimator = np.array([item[2] for item in delivery_raw])
mlc = np.array([item[3] for item in delivery_raw])
jaw = np.array([item[4] for item in delivery_raw])
icom_delivery = pymedphys.Delivery(mu, gantry, collimator, mlc, jaw)
icom_delivery = icom_delivery._filter_cps()
monaco_directory = pathlib.Path(r'\\monacoda\FocalData\RCCC\1~Clinical')
tel_path = list(monaco_directory.glob(f'*~{patient_id}/plan/*/tel.1'))[-1]
tel_path
GRID = pymedphys.mudensity.grid()
delivery_tel = pymedphys.Delivery.from_monaco(tel_path)
mudensity_tel = delivery_tel.mudensity()
pymedphys.mudensity.display(GRID, mudensity_tel)
mudensity_icom = icom_delivery.mudensity()
pymedphys.mudensity.display(GRID, mudensity_icom)
icom_delivery.mu[16]
delivery_tel.mu[1]
delivery_tel.mlc[1]
icom_delivery.mlc[16]
delivery_tel.jaw[1]
icom_delivery.jaw[16]
# new_mlc = np.fliplr(np.flipud(np.array(icom_delivery.mlc[16]) * 10))
# new_mlc[:,1] = -new_mlc[:,1]
# new_mlc
```
|
github_jupyter
|
# Transform JD text files into an LDA model and pyLDAvis visualization
### Steps:
1. Use spaCy phrase matching to identify skills
2. Parse the job descriptions. A full, readable job description gets turned into a bunch of newline-delimited skills.
3. Create a Gensim corpus and dictionary from the parsed skills
4. Train an LDA model using the corpus and dictionary
5. Visualize the LDA model
6. Compare user input to the LDA model; get out a list of relevant skills
```
# Modeling and visualization
import gensim
from gensim.corpora import Dictionary, MmCorpus
from gensim.models.ldamodel import LdaModel
import pyLDAvis
import pyLDAvis.gensim
# Utilities
import codecs
import pickle
import os
import warnings
# Black magic
import spacy
from spacy.matcher import Matcher
from spacy.attrs import *
nlp = spacy.load('en')
```
### 1. Use spaCy phrase matching to ID skills in job descriptions
**First, we read in a pickled dictionary that contains the word patterns we'll use to extract skills from JDs. Here's what the first few patterns look like:**
``` Python
{
0 : [{"lower": "after"}, {"lower": "effects"}],
1 : [{"lower": "amazon"}, {"lower": "web"}, {"lower": "services"}],
2 : [{"lower": "angular"}, {"lower": "js"}],
3 : [{"lower": "ansible"}],
4 : [{"lower": "bash"}, {"lower": "shell"}],
5 : [{"lower": "business"}, {"lower": "intelligence"}]
}
```
**We generated the pickled dictionary through some (rather heavy) preprocessing steps:**
1. Train a word2vec model on all of the job descriptions. Cluster the word embeddings, identify clusters associated with hard skills, and annotate all of the words in those clusters. Save those words as a "skill repository" (a text document that we'll use as the canonical list of hard tech skills).
2. Clean the skill repository. Inevitably, terms that are not hard skills made it into the word2vec "skill" clusters. Remove them. In this case, we defined a "skill" as "a tool, platform, or language that would make sense as a skill to learn or improve."
3. Use the skill repository to train an Named Entity Recognition model (in our case, using Prodigy). Use the training process to identify hard skills that we previously did not have in our repository. Add the new skills to the repository.
4. Create a Python dictionary of the skills. Format the dictionary so that the values can be ingested as spaCy language patterns.
See spaCy's [matcher documentation](https://spacy.io/api/matcher#init) for more details.
```
# read pickled dict() object
with open('skill_dict.pkl', 'rb') as f:
skill_dict = pickle.load(f)
%%time
# Read JDs into memory
import os
directory = os.fsencode('../local_data/')
jds = []
for file in os.listdir(directory):
filename = os.fsdecode(file)
path = '../local_data/' + filename
with open(path, 'r') as infile:
jds.append(infile.read())
print(len(jds), "JDs")
import sys
print(sys.getsizeof(jds)/1000000, "Megabytes")
```
### 2. Parse job descriptions
From each JD, generate a list of skills.
```
%%time
# Write skill-parsed JDs to file.
# This took about three hours for 106k jobs.
for idx, jd in enumerate(jds):
out_path = '../skill_parsed/'+ str(idx+1) + '.txt'
with open(out_path, 'w') as outfile:
# Creating a matcher object
doc = nlp(jd)
matcher = Matcher(nlp.vocab)
for label, pattern in skill_dict.items():
matcher.add(label, None, pattern)
matches = matcher(doc)
for match in matches:
# match object returns a tuple with (id, startpos, endpos)
output = str(doc[match[1]:match[2]]).replace(' ', '_').lower()
outfile.write(output)
outfile.write('\n')
```
### 3. Generate a Gensim corpus and dictionary from the parsed skill documents
```
%%time
# Load parsed items back into memory
directory = os.fsencode('skill_parsed//')
parsed_jds = []
for file in os.listdir(directory):
filename = os.fsdecode(file)
path = 'skill_parsed/' + filename
# Ran into an encoding issue; changing to latin-1 fixed it
with codecs.open(path, 'r', encoding='latin-1') as infile:
parsed_jds.append(infile.read())
%%time
'''
Gensim needs documents to be formatted as a list-of-lists, where the inner
lists are simply lists including the tokens (skills) from a given document.
It's important to note that any bigram or trigram skills are already tokenized
with underscores instead of spaces to preserve them as tokens.
'''
nested_dict_corpus = [text.split() for text in parsed_jds]
print(nested_dict_corpus[222:226])
from gensim.corpora import Dictionary, MmCorpus
gensim_skills_dict = Dictionary(nested_dict_corpus)
# save the dict
gensim_skills_dict.save('gensim_skills.dict')
corpus = [gensim_skills_dict.doc2bow(text) for text in nested_dict_corpus]
# Save the corpus
gensim.corpora.MmCorpus.serialize('skill_bow_corpus.mm', corpus, id2word=gensim_skills_dict)
# Load up the dictionary
gensim_skills_dict = Dictionary.load('gensim_skills.dict')
# Load the corpus
bow_corpus = MmCorpus('skill_bow_corpus.mm')
```
### 4. Create the LDA model using Gensim
```
%%time
with warnings.catch_warnings():
warnings.simplefilter('ignore')
lda_alpha_auto = LdaModel(bow_corpus,
id2word=gensim_skills_dict,
num_topics=20)
lda_alpha_auto.save('lda/skills_lda')
# load the finished LDA model from disk
lda = LdaModel.load('lda/skills_lda')
```
### 5. Visualize using pyLDAvis
```
LDAvis_data_filepath = 'lda/ldavis/ldavis'
%%time
LDAvis_prepared = pyLDAvis.gensim.prepare(lda, bow_corpus,
gensim_skills_dict)
with open(LDAvis_data_filepath, 'wb') as f:
pickle.dump(LDAvis_prepared, f)
# load the pre-prepared pyLDAvis data from disk
with open(LDAvis_data_filepath, 'rb') as f:
LDAvis_prepared = pickle.load(f)
pyLDAvis.display(LDAvis_prepared)
# Save the file as HTML
pyLDAvis.save_html(LDAvis_prepared, 'lda/html/lda.html')
```
### 6. Compare user input to the LDA model
Output the skills a user has and does not have from various topics.
```
# Look at the topics
def explore_topic(topic_number, topn=20):
"""
accept a topic number and print out a
formatted list of the top terms
"""
print(u'{:20} {}'.format(u'term', u'frequency') + u'')
for term, frequency in lda.show_topic(topic_number, topn=40):
print(u'{:20} {:.3f}'.format(term, round(frequency, 3)))
for i in range(20): # Same number as the types of jobs we scraped initially
print("\n\nTopic %s" % i)
explore_topic(topic_number=i)
# A stab at naming the topics
topic_names = {1: u'Data Engineering (Big Data Focus)',
2: u'Microsoft OOP Engineering (C, C++, .NET)',
3: u'Web Application Development (Ruby, Rails, JS, Databases)',
4: u'Linux/Unix, Software Engineering, and Scripting',
5: u'Database Administration',
6: u'Project Management (Agile Focus)',
7: u'Project Management (General Software)',
8: u'Product Management',
9: u'General Management & Productivity (Microsoft Office Focus)',
10: u'Software Program Management',
11: u'Project and Program Management',
12: u'DevOps and Cloud Computing/Infrastructure',
13: u'Frontend Software Engineering and Design',
14: u'Business Intelligence',
15: u'Analytics',
16: u'Quality Engineering, Version Control, & Build',
17: u'Big Data Analytics; Hardware & Scientific Computing',
18: u'Software Engineering',
19: u'Data Science, Machine Learning, and AI',
20: u'Design'}
```
#### Ingest user input & transform into list of skills
```
matcher = Matcher(nlp.vocab)
user_input = '''
My skills are Postgresql, and Python.
Experience with Chef Puppet and Docker required.
I also happen to know Blastoise and Charzard. Also NeuRal neTwOrk.
I use Git, Github, svn, Subversion, but not git, github or subversion.
Additionally, I can program using Perl, Java, and Haskell. But not perl, java, or haskell.'''
# Construct matcher object
doc = nlp(user_input)
for label, pattern in skill_dict.items():
matcher.add(label, None, pattern)
# Compare input to pre-defined skill patterns
user_skills = []
matches = matcher(doc)
for match in matches:
if match is not None:
# match object returns a tuple with (id, startpos, endpos)
output = str(doc[match[1]:match[2]]).lower()
user_skills.append(output)
print("*** User skills: *** ")
for skill in user_skills:
print(skill)
```
#### Compare user skills to the LDA model
```
def top_match_items(input_doc, lda_model, input_dictionary, num_terms=20):
"""
(1) parse input doc with spaCy, apply text pre-proccessing steps,
(3) create a bag-of-words representation (4) create an LDA representation
"""
doc_bow = gensim_skills_dict.doc2bow(input_doc)
# create an LDA representation
document_lda = lda_model[doc_bow]
# Sort in descending order
sorted_doc_lda = sorted(document_lda, key=lambda review_lda: -review_lda[1])
topic_number, freq = sorted_doc_lda[0][0], sorted_doc_lda[0][1]
highest_probability_topic = topic_names[topic_number+1]
top_topic_skills = []
for term, term_freq in lda.show_topic(topic_number, topn=num_terms):
top_topic_skills.append(term)
return highest_probability_topic, round(freq, 3), top_topic_skills
matched_topic, matched_freq, top_topic_skills = top_match_items(user_skills, lda, gensim_skills_dict)
def common_skills(top_topic_skills, user_skills):
return [item for item in top_topic_skills if item in user_skills]
def non_common_skills(top_topic_skills, user_skills):
return [item for item in top_topic_skills if item not in user_skills]
print("**** User's matched topic and percent match:")
print(matched_topic, matched_freq)
print("\n**** Skills user has in common with topic:")
for skill in common_skills(top_topic_skills, user_skills):
print(skill)
print("\n**** Skills user does NOT have in common with topic:")
for skill in non_common_skills(top_topic_skills, user_skills):
print(skill)
```
|
github_jupyter
|
# Unconstrainted optimization with NN models
In this tutorial we will go over type 1 optimization problem which entails nn.Module rerpesented cost function and __no constarint__ at all. This type of problem is often written as follows:
$$ \min_{x} f_{\theta}(x) $$
we can find Type1 problems quite easily. For instance assuming you are the manager of some manufactoring facilities, then your primary objective would be to maximize the yield of the manufactoring process. In industrial grade of manufactoring process the model of process is often __unknown__. hence we may need to learn the model through your favorite differentiable models such as neural networks and perform the graident based optimization to find the (local) optimums that minimize (or maximize) the yield.
### General problem solving tricks; Cast your problem into QP, approximately.
As far as I know, Convex optimization is the most general class of optmization problems where we have algorithms that can solve the problem optimally. Qudartic progamming (QP) is a type of convex optimization problems which is well developed in the side of theory and computations. We will heavily utilize QPs to solve the optimziation problems that have dependency with `torch` models.
Our general problem solving tricks are as follows:
1. Construct the cost or constraint models from the data
2. By utilizting `torch` automatic differentiation functionality, compute the jacobian or hessians of the moodels.
3. solve (possibley many times) QP with the estimated jacobian and hessians.
> It is noteworthy that even we locally cast the problem into QP, that doesn't mean our original problem is convex. Therefore, we cannot say that this approahces we will look over can find the global optimum.
```
import torch
import numpy as np
import matplotlib.pyplot as plt
from torch.utils.data import TensorDataset, DataLoader
from src.utils import generate_y
from src.nn.MLP import MLP
```
## Generate training dataset
```
x_min, x_max = -4.0, 4.0
xs_linspace = torch.linspace(-4, 4, 2000).view(-1, 1)
ys_linspace = generate_y(xs_linspace)
# samples to construct training dataset
x_dist = torch.distributions.uniform.Uniform(-4.0, 4.0)
xs = x_dist.sample(sample_shape=(500, 1))
ys = generate_y(xs)
BS = 64 # Batch size
ds = TensorDataset(xs, ys)
loader = DataLoader(ds, batch_size=BS, shuffle=True)
input_dim, output_dim = 1, 1
m = MLP(input_dim, output_dim, num_neurons=[128, 128])
mse_criteria = torch.nn.MSELoss()
opt = torch.optim.Adam(m.parameters(), lr=1e-3)
n_update = 0
print_every = 500
epochs = 200
for _ in range(epochs):
for x, y in loader:
y_pred = m(x)
loss = mse_criteria(y_pred, y)
opt.zero_grad()
loss.backward()
opt.step()
n_update += 1
if n_update % print_every == 0:
print(n_update, loss.item())
# save model for the later usages
torch.save(m.state_dict(), './model.pt')
```
## Solve the unconstraint optimization problem
Let's solve the unconstraint optimization problem with torch estmiated graidents and simple gradient descent method.
```
def minimize_y(x_init, model, num_steps=15, step_size=1e-1):
def _grad(model, x):
return torch.autograd.functional.jacobian(model, x).squeeze()
x = x_init
xs = [x]
ys = [model(x)]
gs = [_grad(model, x)]
for _ in range(num_steps):
grad = _grad(model, x)
x = (x- step_size * grad).clone()
y = model(x)
xs.append(x)
ys.append(y)
gs.append(grad)
xs = torch.stack(xs).detach().numpy()
ys = torch.stack(ys).detach().numpy()
gs = torch.stack(gs).detach().numpy()
return xs, ys, gs
x_min, x_max = -4.0, 4.0
n_steps = 40
x_init = torch.tensor(np.random.uniform(x_min, x_max, 1)).float()
opt_xs, opt_ys, grad = minimize_y(x_init, m, n_steps)
pred_ys = m(xs_linspace).detach()
fig, axes = plt.subplots(1, 1, figsize=(10, 5))
axes.grid()
axes.plot(xs_linspace, ys_linspace, label='Ground truth')
axes.plot(xs_linspace, pred_ys, label='Model prediction')
axes.scatter(opt_xs[0], opt_ys[0], label='Opt start',
c='green', marker='*', s=100.0)
axes.scatter(opt_xs[1:], opt_ys[1:], label='NN opt', c='green')
_ = axes.legend()
```
|
github_jupyter
|
# Function Practice Exercises
Problems are arranged in increasing difficulty:
* Warmup - these can be solved using basic comparisons and methods
* Level 1 - these may involve if/then conditional statements and simple methods
* Level 2 - these may require iterating over sequences, usually with some kind of loop
* Challenging - these will take some creativity to solve
## WARMUP SECTION:
#### LESSER OF TWO EVENS: Write a function that returns the lesser of two given numbers *if* both numbers are even, but returns the greater if one or both numbers are odd
lesser_of_two_evens(2,4) --> 2
lesser_of_two_evens(2,5) --> 5
```
def lesser_of_two_evens(a,b):
pass
# Check
lesser_of_two_evens(2,4)
# Check
lesser_of_two_evens(2,5)
```
#### ANIMAL CRACKERS: Write a function takes a two-word string and returns True if both words begin with same letter
animal_crackers('Levelheaded Llama') --> True
animal_crackers('Crazy Kangaroo') --> False
```
def animal_crackers(text):
pass
# Check
animal_crackers('Levelheaded Llama')
# Check
animal_crackers('Crazy Kangaroo')
```
#### MAKES TWENTY: Given two integers, return True if the sum of the integers is 20 *or* if one of the integers is 20. If not, return False
makes_twenty(20,10) --> True
makes_twenty(12,8) --> True
makes_twenty(2,3) --> False
```
def makes_twenty(n1,n2):
pass
# Check
makes_twenty(20,10)
# Check
makes_twenty(2,3)
```
# LEVEL 1 PROBLEMS
#### OLD MACDONALD: Write a function that capitalizes the first and fourth letters of a name
old_macdonald('macdonald') --> MacDonald
Note: `'macdonald'.capitalize()` returns `'Macdonald'`
```
def old_macdonald(name):
pass
# Check
old_macdonald('macdonald')
```
#### MASTER YODA: Given a sentence, return a sentence with the words reversed
master_yoda('I am home') --> 'home am I'
master_yoda('We are ready') --> 'ready are We'
Note: The .join() method may be useful here. The .join() method allows you to join together strings in a list with some connector string. For example, some uses of the .join() method:
>>> "--".join(['a','b','c'])
>>> 'a--b--c'
This means if you had a list of words you wanted to turn back into a sentence, you could just join them with a single space string:
>>> " ".join(['Hello','world'])
>>> "Hello world"
```
def master_yoda(text):
pass
# Check
master_yoda('I am home')
# Check
master_yoda('We are ready')
```
#### ALMOST THERE: Given an integer n, return True if n is within 10 of either 100 or 200
almost_there(90) --> True
almost_there(104) --> True
almost_there(150) --> False
almost_there(209) --> True
NOTE: `abs(num)` returns the absolute value of a number
```
def almost_there(n):
pass
# Check
almost_there(104)
# Check
almost_there(150)
# Check
almost_there(209)
```
# LEVEL 2 PROBLEMS
#### FIND 33:
Given a list of ints, return True if the array contains a 3 next to a 3 somewhere.
has_33([1, 3, 3]) → True
has_33([1, 3, 1, 3]) → False
has_33([3, 1, 3]) → False
```
def has_33(nums):
pass
# Check
has_33([1, 3, 3])
# Check
has_33([1, 3, 1, 3])
# Check
has_33([3, 1, 3])
```
#### PAPER DOLL: Given a string, return a string where for every character in the original there are three characters
paper_doll('Hello') --> 'HHHeeellllllooo'
paper_doll('Mississippi') --> 'MMMiiissssssiiippppppiii'
```
def paper_doll(text):
pass
# Check
paper_doll('Hello')
# Check
paper_doll('Mississippi')
```
#### BLACKJACK: Given three integers between 1 and 11, if their sum is less than or equal to 21, return their sum. If their sum exceeds 21 *and* there's an eleven, reduce the total sum by 10. Finally, if the sum (even after adjustment) exceeds 21, return 'BUST'
blackjack(5,6,7) --> 18
blackjack(9,9,9) --> 'BUST'
blackjack(9,9,11) --> 19
```
def blackjack(a,b,c):
pass
# Check
blackjack(5,6,7)
# Check
blackjack(9,9,9)
# Check
blackjack(9,9,11)
```
#### SUMMER OF '69: Return the sum of the numbers in the array, except ignore sections of numbers starting with a 6 and extending to the next 9 (every 6 will be followed by at least one 9). Return 0 for no numbers.
summer_69([1, 3, 5]) --> 9
summer_69([4, 5, 6, 7, 8, 9]) --> 9
summer_69([2, 1, 6, 9, 11]) --> 14
```
def summer_69(arr):
pass
# Check
summer_69([1, 3, 5])
# Check
summer_69([4, 5, 6, 7, 8, 9])
# Check
summer_69([2, 1, 6, 9, 11])
```
# CHALLENGING PROBLEMS
#### SPY GAME: Write a function that takes in a list of integers and returns True if it contains 007 in order
spy_game([1,2,4,0,0,7,5]) --> True
spy_game([1,0,2,4,0,5,7]) --> True
spy_game([1,7,2,0,4,5,0]) --> False
```
def spy_game(nums):
pass
# Check
spy_game([1,2,4,0,0,7,5])
# Check
spy_game([1,0,2,4,0,5,7])
# Check
spy_game([1,7,2,0,4,5,0])
```
#### COUNT PRIMES: Write a function that returns the *number* of prime numbers that exist up to and including a given number
count_primes(100) --> 25
By convention, 0 and 1 are not prime.
```
def count_primes(num):
pass
# Check
count_primes(100)
```
### Just for fun:
#### PRINT BIG: Write a function that takes in a single letter, and returns a 5x5 representation of that letter
print_big('a')
out: *
* *
*****
* *
* *
HINT: Consider making a dictionary of possible patterns, and mapping the alphabet to specific 5-line combinations of patterns. <br>For purposes of this exercise, it's ok if your dictionary stops at "E".
```
def print_big(letter):
pass
print_big('a')
```
## Great Job!
|
github_jupyter
|
*This notebook contains an excerpt from the [Whirlwind Tour of Python](http://www.oreilly.com/programming/free/a-whirlwind-tour-of-python.csp) by Jake VanderPlas; the content is available [on GitHub](https://github.com/jakevdp/WhirlwindTourOfPython).*
*The text and code are released under the [CC0](https://github.com/jakevdp/WhirlwindTourOfPython/blob/master/LICENSE) license; see also the companion project, the [Python Data Science Handbook](https://github.com/jakevdp/PythonDataScienceHandbook).*
# A Quick Tour of Python Language Syntax
```
x = 1
y = 4
z = x + y
z
# set the midpoint
midpoint = 5
# make two empty lists
lower = []; upper = []
# split the numbers into lower and upper
for i in range(10):
if (i < midpoint):
lower.append(i)
else:
upper.append(i)
print("lower:", lower)
print("upper:", upper)
```
## Comments Are Marked by ``#``
The script starts with a comment:
``` python
# set the midpoint
```
Comments in Python are indicated by a pound sign (``#``), and anything on the line following the pound sign is ignored by the interpreter.
This means, for example, that you can have stand-alone comments like the one just shown, as well as inline comments that follow a statement. For example:
``` python
x += 2 # shorthand for x = x + 2
```
Python does not have any syntax for multi-line comments, such as the ``/* ... */`` syntax used in C and C++, though multi-line strings are often used as a replacement for multi-line comments (more on this in [String Manipulation and Regular Expressions](14-Strings-and-Regular-Expressions.ipynb)).
## End-of-Line Terminates a Statement
The next line in the script is
``` python
midpoint = 5
```
This is an assignment operation, where we've created a variable named ``midpoint`` and assigned it the value ``5``.
Notice that the end of this statement is simply marked by the end of the line.
This is in contrast to languages like C and C++, where every statement must end with a semicolon (``;``).
In Python, if you'd like a statement to continue to the next line, it is possible to use the "``\``" marker to indicate this:
## Semicolon Can Optionally Terminate a Statement
Sometimes it can be useful to put multiple statements on a single line.
The next portion of the script is
``` python
lower = []; upper = []
```
This shows the example of how the semicolon (``;``) familiar in C can be used optionally in Python to put two statements on a single line.
Functionally, this is entirely equivalent to writing
``` python
lower = []
upper = []
```
Using a semicolon to put multiple statements on a single line is generally discouraged by most Python style guides, though occasionally it proves convenient.
## Indentation: Whitespace Matters!
Next, we get to the main block of code:
``` Python
for i in range(10):
if i < midpoint:
lower.append(i)
else:
upper.append(i)
```
This is a compound control-flow statement including a loop and a conditional – we'll look at these types of statements in a moment.
For now, consider that this demonstrates what is perhaps the most controversial feature of Python's syntax: whitespace is meaningful!
## Parentheses Are for Grouping or Calling
In the previous code snippet, we see two uses of parentheses.
First, they can be used in the typical way to group statements or mathematical operations:
```
2 * (3 + 4)
```
They can also be used to indicate that a *function* is being called.
In the next snippet, the ``print()`` function is used to display the contents of a variable (see the sidebar).
The function call is indicated by a pair of opening and closing parentheses, with the *arguments* to the function contained within:
```
print('first value:', 1)
```
# Basic Python Semantics: Variables and Objects
## Python Variables Are Pointers
Assigning variables in Python is as easy as putting a variable name to the left of the equals (``=``) sign:
```python
# assign 4 to the variable x
x = 4
```
```
x = 1 # x is an integer
x = 'hello' # now x is a string
x = [1, 2, 3] # now x is a list
x = [1, 2, 3]
y = x
```
We've created two variables ``x`` and ``y`` which both point to the same object.
Because of this, if we modify the list via one of its names, we'll see that the "other" list will be modified as well:
```
print(y)
x.append(4) # append 4 to the list pointed to by x
print(y) # y's list is modified as well!
print(y)
x = 10
y = x
x += 5 # add 5 to x's value, and assign it to x
print("x =", x)
print("y =", y)
```
When we call ``x += 5``, we are not modifying the value of the ``10`` object pointed to by ``x``; we are rather changing the variable ``x`` so that it points to a new integer object with value ``15``.
For this reason, the value of ``y`` is not affected by the operation.
## Everything Is an Object
Python is an object-oriented programming language, and in Python everything is an object.
Let's flesh-out what this means. Earlier we saw that variables are simply pointers, and the variable names themselves have no attached type information.
This leads some to claim erroneously that Python is a type-free language. But this is not the case!
Consider the following:
```
x = 4
type(x)
x.bit_length()
x.real
x = 'hello'
type(x)
x.upper()
x = 3.14159
type(x)
x.as_integer_ratio()
```
Python has types; however, the types are linked not to the variable names but *to the objects themselves*.
In object-oriented programming languages like Python, an *object* is an entity that contains data along with associated metadata and/or functionality.
```
L = [1, 2, 3]
L.append(100)
print(L)
```
While it might be expected for compound objects like lists to have attributes and methods, what is sometimes unexpected is that in Python even simple types have attached attributes and methods.
For example, numerical types have a ``real`` and ``imag`` attribute that returns the real and imaginary part of the value, if viewed as a complex number:
```
x = 4.5
print(x.real, "+", x.imag, 'i')
```
Methods are like attributes, except they are functions that you can call using opening and closing parentheses.
For example, floating point numbers have a method called ``is_integer`` that checks whether the value is an integer:
```
x = 4.5
x.is_integer()
x = 4.0
x.is_integer()
```
|
github_jupyter
|
<!-- :Author: Arthur Goldberg <[email protected]> -->
<!-- :Date: 2020-08-02 -->
<!-- :Copyright: 2020, Karr Lab -->
<!-- :License: MIT -->
# DE-Sim: Ordering simultaneous events
DE-Sim makes it easy to build and simulate discrete-event models.
This notebook discusses DE-Sim's methods for controlling the execution order of simultaneous messages.
## Installation
Use `pip` to install `de_sim`.
## Scheduling events with equal simulation times
A discrete-event simulation may execute multiple events simultaneously, that is, at a particular simulation time.
To ensure that simulation runs are reproducible and deterministic, a simulator must provide mechanisms that deterministically control the execution order of simultaneous events.
Two types of situations arise, *local* and *global*.
A local situation arises when a simulation object receives multiple event messages simultaneously, while a global situation arises when multiple simulation objects execute events simultaneously.
Separate *local* and *global* mechanisms ensure that these situations are simulated deterministically.
The local mechanism ensures that simultaneous events are handled deterministically at a single simulation object, while the global mechanism ensures that simultaneous events are handled deterministically across all objects in a simulation.
### Local mechanism: simultaneous event messages at a simulation object
The local mechanism, called *event superposition* after the [physics concept of superposition](https://en.wikipedia.org/wiki/Superposition_principle), involves two components:
1. When a simulation object receives multiple event messages at the same time, the simulator passes all of the event messages to the object's event handler in a list.
(However, if simultaneous event messages have different handlers then the simulator raises a `SimulatorError` exception.)
2. The simulator sorts the events in the list so that any given list of events will always be arranged in the same order.
Event messages are sorted by the pair (event message priority, event message content).
Sorting costs O(n log n), but since simultaneous events are usually rare, sorting event lists is unlikely to slow down simulations.
```
""" This example illustrates the local mechanism that handles simultaneous
event messages received by a simulation object
"""
import random
import de_sim
from de_sim.event import Event
class Double(de_sim.EventMessage):
'Double value'
class Increment(de_sim.EventMessage):
'Increment value'
class IncrementThenDoubleSimObject(de_sim.SimulationObject):
""" Execute Increment before Double, demonstrating superposition """
def __init__(self, name):
super().__init__(name)
self.value = 0
def init_before_run(self):
self.send_events()
def handle_superposed_events(self, event_list):
""" Process superposed events in an event list
Each Increment message increments value, and each Double message doubles value.
Assumes that `event_list` contains an Increment event followed by a Double event.
Args:
event_list (:obj:`event_list` of :obj:`de_sim.Event`): list of events
"""
for event in event_list:
if isinstance(event.message, Increment):
self.value += 1
elif isinstance(event.message, Double):
self.value *= 2
self.send_events()
# The order of the message types in event_handlers, (Increment, Double), determines
# the sort order of messages in `event_list` received by `handle_superposed_events`
event_handlers = [(Increment, 'handle_superposed_events'),
(Double, 'handle_superposed_events')]
def send_events(self):
# To show that the simulator delivers event messages to `handle_superposed_events`
# sorted into the order (Increment, Double), send them in a random order.
if random.randrange(2):
self.send_event(1, self, Double())
self.send_event(1, self, Increment())
else:
self.send_event(1, self, Increment())
self.send_event(1, self, Double())
# Register the message types sent
messages_sent = (Increment, Double)
class TestSuperposition(object):
def increment_then_double_from_0(self, iterations):
v = 0
for _ in range(iterations):
v += 1
v *= 2
return v
def test_superposition(self, max_time):
simulator = de_sim.Simulator()
simulator.add_object(IncrementThenDoubleSimObject('name'))
simulator.initialize()
simulator.simulate(max_time)
for sim_obj in simulator.get_objects():
assert sim_obj.value, self.increment_then_double_from_0(max_time)
print(f'Simulation to {max_time} executed all messages in the order (Increment, Double).')
TestSuperposition().test_superposition(20)
```
This example shows how event superposition handles simultaneous events.
An `IncrementThenDoubleSimObject` simulation object stores an integer value.
It receives two events every time unit, one carrying an `Increment` message and another containing a `Double` message.
Executing an `Increment` event increments the value, while executing a `Double` message event doubles the value.
The design for `IncrementThenDoubleSimObject` requires that it increments before doubling.
Several features of DE-Sim and `IncrementThenDoubleSimObject` ensure this behavior:
1. The mapping between event message types and event handlers, stored in the list `event_handlers`, contains `Increment` before `Double`. This gives events containing an `Increment` message a higher priority than events containing `Double`.
2. Under the covers, when DE-Sim passes superposed events to a subclass of [`SimulationObject`](https://docs.karrlab.org/de_sim/master/source/de_sim.html#de_sim.simulation_object.SimulationObject), it sorts the messages by their (event message priority, event message content), which sorts events with higher priority message types earlier.
3. The message handler `handle_superposed_events` receives a list of events and executes them in order.
To challenge and test this superposition mechanism, the `send_events()` method in `IncrementThenDoubleSimObject` randomizes the order in which it sends `Increment` and `Double` events.
Finally, `TestSuperposition().test_superposition()` runs a simulation of `IncrementThenDoubleSimObject` and asserts that the value it computes equals the correct value for a sequence of increment and double operations.
### Global mechanism: simultaneous event messages at multiple simulation objects
A *global* mechanism is needed to ensure that simultaneous events which occur at distinct objects in a simulation are executed in a deterministic order.
Otherwise, the discrete-event simulator might execute simultaneous events at distinct simulation objects in a different order in different simulation runs that use the same input.
When using a simulator that allows 0-delay event messages or global state shared between simulation objects -- both of which DE-Sim supports -- this can alter the simulation's predictions and thereby imperil debugging efforts, statistical analyses of predictions and other essential uses of simulation results.
The global mechanism employed by DE-Sim conceives of the simulation time as a pair -- the event time, and a *sub-time* which breaks event time ties.
Sub-time values within a particular simulation time must be distinct.
Given that constraint, many approaches for selecting the sub-time would achieve the objective.
DE-Sim creates a distinct sub-time from the state of the simulation object receiving an event.
The sub-time is a pair composed of a priority assigned to the simulation class and a unique identifier for each class instance.
Each simulation class defines a `class_priority` attribute that determines the relative execution order of simultaneous events by different simulation classes.
Among multiple instances of a simulation class, the attribute `event_time_tiebreaker`, which defaults to a simulation instance's unique name, breaks ties.
All classes have the same default priority of `LOW`. If class priorities are not set and `event_time_tiebreaker`s are not set for individual simulation objects, then an object's global priority is given by its name.
```
from de_sim.simulation_object import SimObjClassPriority
class ExampleMsg(de_sim.EventMessage):
'Example message'
class NoPrioritySimObj(de_sim.SimulationObject):
def init_before_run(self):
self.send_event(0., self, ExampleMsg())
# register the message types sent
messages_sent = (ExampleMsg, )
class LowPrioritySimObj(NoPrioritySimObj):
def handler(self, event):
print(f"{self.time}: LowPrioritySimObj {self.name} running")
self.send_event(1., self, ExampleMsg())
event_handlers = [(ExampleMsg, 'handler')]
# have `LowPrioritySimObj`s execute at low priority
class_priority = SimObjClassPriority.LOW
class MediumPrioritySimObj(NoPrioritySimObj):
def handler(self, event):
print(f"{self.time}: MediumPrioritySimObj {self.name} running")
self.send_event(1., self, ExampleMsg())
event_handlers = [(ExampleMsg, 'handler')]
# have `MediumPrioritySimObj`s execute at medium priority
class_priority = SimObjClassPriority.MEDIUM
simulator = de_sim.Simulator()
simulator.add_object(LowPrioritySimObj('A'))
simulator.add_object(MediumPrioritySimObj('B'))
simulator.initialize()
print(simulator.simulate(2).num_events, 'events executed')
```
This example illustrates the scheduling of simultaneous event messages.
`SimObjClassPriority` is an `IntEnum` that provides simulation object class priorities, including `LOW`, `MEDIUM`, and `HIGH`.
We create two classes, `LowPrioritySimObj` and `MediumPrioritySimObj`, with `LOW` and `MEDIUM` priorities, respectively, and execute them simultaneously at simulation times 0, 1, 2, ...
At each time, the `MediumPrioritySimObj` object runs before the `LowPrioritySimObj` one.
#### Execution order of objects without an assigned `class_priority`
The next example shows the ordering of simultaneous events executed by objects that don't have assigned priorities.
```
class DefaultPrioritySimObj(NoPrioritySimObj):
def handler(self, event):
print(f"{self.time}: DefaultPrioritySimObj {self.name} running")
self.send_event(1., self, ExampleMsg())
event_handlers = [(ExampleMsg, 'handler')]
simulator = de_sim.Simulator()
for name in random.sample(range(10), k=3):
sim_obj = DefaultPrioritySimObj(str(name))
print(f"{sim_obj.name} priority: {sim_obj.class_event_priority.name}")
simulator.add_object(sim_obj)
simulator.initialize()
print(simulator.simulate(2).num_events, 'events executed')
```
In this example, the [`SimulationObject`s](https://docs.karrlab.org/de_sim/master/source/de_sim.html#de_sim.simulation_object.SimulationObject) have no priorities assigned, so their default priorities are `LOW`. (The `class_event_priority` attribute of a simulation object is a `SimObjClassPriority`)
Three objects with names randomly selected from '0', '1', ..., '9', are created.
When they execute simultaneously, events are ordered by the sort order of the objects' names.
#### Execution order of instances of simulation object classes with relative priorities
Often, a modeler wants to control the *relative* simultaneous priorities of simulation objects, but does not care about their absolute priorities.
The next example shows how to specify relative priorities.
```
class FirstNoPrioritySimObj(NoPrioritySimObj):
def handler(self, event):
print(f"{self.time}: FirstNoPrioritySimObj {self.name} running")
self.send_event(1., self, ExampleMsg())
event_handlers = [(ExampleMsg, 'handler')]
class SecondNoPrioritySimObj(NoPrioritySimObj):
def handler(self, event):
print(f"{self.time}: SecondNoPrioritySimObj {self.name} running")
self.send_event(1., self, ExampleMsg())
event_handlers = [(ExampleMsg, 'handler')]
# Assign decreasing priorities to classes in [FirstNoPrioritySimObj, SecondNoPrioritySimObj]
SimObjClassPriority.assign_decreasing_priority([FirstNoPrioritySimObj,
SecondNoPrioritySimObj])
simulator = de_sim.Simulator()
simulator.add_object(SecondNoPrioritySimObj('A'))
simulator.add_object(FirstNoPrioritySimObj('B'))
for sim_obj in simulator.simulation_objects.values():
print(f"{type(sim_obj).__name__}: {sim_obj.name}; "
f"priority: {sim_obj.class_event_priority.name}")
simulator.initialize()
print(simulator.simulate(2).num_events, 'events executed')
```
The `assign_decreasing_priority` method of `SimObjClassPriority` takes an iterator over `SimulationObject` subclasses, and assigns them decreasing simultaneous event priorities.
The `FirstNoPrioritySimObj` instance therefore executes before the `SecondNoPrioritySimObj` instance at each discrete simulation time.
|
github_jupyter
|
```
%matplotlib inline
import numpy as np
from matplotlib import pyplot as plt
from matplotlib import cm
import pandas as pd
import matplotlib as mpl
mpl.rcParams['text.usetex'] = True
mpl.rcParams['text.latex.unicode'] = True
blues = cm.get_cmap(plt.get_cmap('Blues'))
greens = cm.get_cmap(plt.get_cmap('Greens'))
reds = cm.get_cmap(plt.get_cmap('Reds'))
oranges = cm.get_cmap(plt.get_cmap('Oranges'))
purples = cm.get_cmap(plt.get_cmap('Purples'))
greys = cm.get_cmap(plt.get_cmap('Greys'))
from IPython.core.display import display, HTML
display(HTML("<style>.container { width:100% !important; }</style>"))
import warnings
warnings.filterwarnings('ignore')
des2a = pd.read_csv('../Data/design2a.csv')
des2b = pd.read_csv('../Data/design2b.csv')
fig, axis = plt.subplots(nrows=2,ncols=4,figsize=(11,5),sharex='row',sharey='row')
x1 = np.arange(1)
x2 = np.arange(1)
_ = axis[0,0].bar(x1,des2a['CpuMeanTime'][0],yerr=des2a['CpuStdTime'][0], color=blues(150))
_ = axis[0,0].bar(x2+1,des2a['GpuMeanTime'][0],yerr=des2a['GpuStdTime'][0], color=reds(150))
_ = axis[0,0].set_xticks([0,1])
_ = axis[0,0].grid('on', linestyle=':', linewidth=2)
_ = axis[0,0].set_xticklabels(['CPUs', 'GPUs'], fontsize=22)
_ = axis[0,1].bar(x1,des2a['CpuMeanTime'][1],yerr=des2a['CpuStdTime'][1], color=blues(150))
_ = axis[0,1].bar(x2+1,des2a['GpuMeanTime'][1],yerr=des2a['GpuStdTime'][1], color=reds(150))
_ = axis[0,1].set_xticks([0,1])
_ = axis[0,1].grid('on', linestyle=':', linewidth=2)
_ = axis[0,1].set_xticklabels(['CPUs', 'GPUs'], fontsize=22)
_ = axis[0,2].bar(x1,des2a['CpuMeanTime'][2],yerr=des2a['CpuStdTime'][2], color=blues(150))
_ = axis[0,2].bar(x2+1,des2a['GpuMeanTime'][2],yerr=des2a['GpuStdTime'][2], color=reds(150))
_ = axis[0,2].set_xticks([0,1])
_ = axis[0,2].grid('on', linestyle=':', linewidth=2)
_ = axis[0,2].set_xticklabels(['CPUs', 'GPUs'], fontsize=22)
_ = axis[0,3].bar(x1,des2a['CpuMeanTime'][3],yerr=des2a['CpuStdTime'][3], color=blues(150))
_ = axis[0,3].bar(x2+1,des2a['GpuMeanTime'][3],yerr=des2a['GpuStdTime'][3], color=reds(150))
_ = axis[0,3].set_xticks([0,1])
_ = axis[0,3].grid('on', linestyle=':', linewidth=2)
_ = axis[0,3].set_xticklabels(['CPUs', 'GPUs'], fontsize=22)
_ = axis[0,0].set_yticklabels(axis[0,0].get_yticks().astype('int').tolist(),fontsize=22)
_ = axis[1,0].hist(eval(des2a['Images'][0]),bins=50)
_ = axis[1,1].hist(eval(des2a['Images'][1]),bins=50)
_ = axis[1,2].hist(eval(des2a['Images'][2]),bins=50)
_ = axis[1,3].hist(eval(des2a['Images'][3]),bins=50)
_ = axis[0,0].set_title('Node 1',fontsize=24)
_ = axis[0,1].set_title('Node 2',fontsize=24)
_ = axis[0,2].set_title('Node 3',fontsize=24)
_ = axis[0,3].set_title('Node 4',fontsize=24)
_ = axis[1,0].set_xlabel('Image Size\nin MBs', fontsize=24)
_ = axis[1,1].set_xlabel('Image Size\nin MBs', fontsize=24)
_ = axis[1,2].set_xlabel('Image Size\nin MBs', fontsize=24)
_ = axis[1,3].set_xlabel('Image Size\nin MBs', fontsize=24)
_ = axis[0,0].set_ylabel('Time', fontsize=24)
_ = axis[1,0].set_yticklabels(axis[1,0].get_yticks().astype('int').tolist(),fontsize=22)
_ = axis[1,0].set_xticklabels(axis[1,0].get_xticks().astype('int').tolist(),fontsize=22)
_ = axis[1,1].set_xticklabels(axis[1,1].get_xticks().astype('int').tolist(),fontsize=22)
_ = axis[1,2].set_xticklabels(axis[1,2].get_xticks().astype('int').tolist(),fontsize=22)
_ = axis[1,3].set_xticklabels(axis[1,3].get_xticks().astype('int').tolist(),fontsize=22)
_ = axis[1,0].grid('on', linestyle=':', linewidth=2)
_ = axis[1,1].grid('on', linestyle=':', linewidth=2)
_ = axis[1,2].grid('on', linestyle=':', linewidth=2)
_ = axis[1,3].grid('on', linestyle=':', linewidth=2)
_ = axis[1,0].set_ylabel('Number of\nImages', fontsize=24)
_ = axis[0,0].set_ylabel('Time', fontsize=24)
_ = axis[1,0].set_yticklabels(axis[1,0].get_yticks().astype('int').tolist(),fontsize=22)
#fig.savefig('design2_timelines.pdf',dpi=800,bbox_inches='tight')
level0 = axis[0,0].get_ylim()
level1 = axis[1,0].get_ylim()
fig, axis = plt.subplots(nrows=2,ncols=4,figsize=(11,5),sharex='row',sharey='row')
x1 = np.arange(1)
x2 = np.arange(1)
_ = axis[0,0].set_ylim(level0)
_ = axis[1,0].set_ylim(level1)
_ = axis[0,0].bar(x1,des2b['CpuMeanTime'][0],yerr=des2b['CpuStdTime'][0], color=blues(150))
_ = axis[0,0].bar(x2+1,des2b['GpuMeanTime'][0],yerr=des2b['GpuStdTime'][0], color=reds(150))
_ = axis[0,0].set_xticks([0,1])
_ = axis[0,0].grid('on', linestyle=':', linewidth=2)
_ = axis[0,0].set_xticklabels(['CPUs', 'GPUs'], fontsize=22)
_ = axis[0,1].bar(x1,des2b['CpuMeanTime'][1],yerr=des2b['CpuStdTime'][1], color=blues(150))
_ = axis[0,1].bar(x2+1,des2b['GpuMeanTime'][1],yerr=des2b['GpuStdTime'][1], color=reds(150))
_ = axis[0,1].set_xticks([0,1])
_ = axis[0,1].grid('on', linestyle=':', linewidth=2)
_ = axis[0,1].set_xticklabels(['CPUs', 'GPUs'], fontsize=22)
_ = axis[0,2].bar(x1,des2b['CpuMeanTime'][2],yerr=des2b['CpuStdTime'][2], color=blues(150))
_ = axis[0,2].bar(x2+1,des2b['GpuMeanTime'][2],yerr=des2b['GpuStdTime'][2], color=reds(150))
_ = axis[0,2].set_xticks([0,1])
_ = axis[0,2].grid('on', linestyle=':', linewidth=2)
_ = axis[0,2].set_xticklabels(['CPUs', 'GPUs'], fontsize=22)
_ = axis[0,3].bar(x1,des2b['CpuMeanTime'][3],yerr=des2b['CpuStdTime'][3], color=blues(150))
_ = axis[0,3].bar(x2+1,des2b['GpuMeanTime'][3],yerr=des2b['GpuStdTime'][3], color=reds(150))
_ = axis[0,3].set_xticks([0,1])
_ = axis[0,3].grid(axis='both', linestyle=':', linewidth=2)
_ = axis[0,3].set_xticklabels(['CPUs', 'GPUs'], fontsize=22)
_ = axis[0,0].set_yticklabels(axis[0,0].get_yticks().astype('int').tolist(),fontsize=22)
_ = axis[1,0].grid('on', linestyle=':', linewidth=2)
_ = axis[1,1].grid('on', linestyle=':', linewidth=2)
_ = axis[1,2].grid('on', linestyle=':', linewidth=2)
_ = axis[1,3].grid('on', linestyle=':', linewidth=2)
_ = axis[1,0].hist(eval(des2b['Images'][0]),bins=50)
_ = axis[1,1].hist(eval(des2b['Images'][1]),bins=50)
_ = axis[1,2].hist(eval(des2b['Images'][2]),bins=50)
_ = axis[1,3].hist(eval(des2b['Images'][3]),bins=50)
_ = axis[0,0].set_title('Node 1',fontsize=24)
_ = axis[0,1].set_title('Node 2',fontsize=24)
_ = axis[0,2].set_title('Node 3',fontsize=24)
_ = axis[0,3].set_title('Node 4',fontsize=24)
_ = axis[1,0].set_xlabel('Image Size\nin MBs', fontsize=24)
_ = axis[1,1].set_xlabel('Image Size\nin MBs', fontsize=24)
_ = axis[1,2].set_xlabel('Image Size\nin MBs', fontsize=24)
_ = axis[1,3].set_xlabel('Image Size\nin MBs', fontsize=24)
_ = axis[1,0].set_ylabel('Number of\nImages', fontsize=24)
_ = axis[0,0].set_ylabel('Time', fontsize=24)
_ = axis[1,0].set_yticklabels(axis[1,0].get_yticks().astype('int').tolist(),fontsize=22)
_ = axis[1,0].set_xticklabels(axis[1,0].get_xticks().astype('int').tolist(),fontsize=22)
_ = axis[1,1].set_xticklabels(axis[1,1].get_xticks().astype('int').tolist(),fontsize=22)
_ = axis[1,2].set_xticklabels(axis[1,2].get_xticks().astype('int').tolist(),fontsize=22)
_ = axis[1,3].set_xticklabels(axis[1,3].get_xticks().astype('int').tolist(),fontsize=22)
#fig.savefig('design2a_timelines.pdf',dpi=800,bbox_inches='tight')
```
|
github_jupyter
|
# HandGestureDetection using OpenCV
This code template is for Hand Gesture detection in a video using OpenCV Library.
### Required Packages
```
!pip install opencv-python
!pip install mediapipe
import cv2
import mediapipe as mp
import time
```
### Hand Detection
For detecting hands in the image, we use the detectMultiScale() function.
It detects objects of different sizes in the input image. The detected objects are returned as a list of rectangles which can be then plotted around the hands.
####Tuning Parameters:
**image** - Matrix of the type CV_8U containing an image where objects are detected.
**objects** - Vector of rectangles where each rectangle contains the detected object, the rectangles may be partially outside the original image.
**scaleFactor** - Parameter specifying how much the image size is reduced at each image scale.
**minNeighbors** - Parameter specifying how many neighbors each candidate rectangle should have to retain it.
```
class handDetector():
def __init__(self, mode = False, maxHands = 2, detectionCon = 0.5, trackCon = 0.5):
self.mode = mode
self.maxHands = maxHands
self.detectionCon = detectionCon
self.trackCon = trackCon
self.mpHands = mp.solutions.hands
self.hands = self.mpHands.Hands(self.mode, self.maxHands, self.detectionCon, self.trackCon)
self.mpDraw = mp.solutions.drawing_utils
def findHands(self,img, draw = True):
imgRGB = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
self.results = self.hands.process(imgRGB)
# print(results.multi_hand_landmarks)
if self.results.multi_hand_landmarks:
for handLms in self.results.multi_hand_landmarks:
if draw:
self.mpDraw.draw_landmarks(img, handLms, self.mpHands.HAND_CONNECTIONS)
return img
def findPosition(self, img, handNo = 0, draw = True):
lmlist = []
if self.results.multi_hand_landmarks:
myHand = self.results.multi_hand_landmarks[handNo]
for id, lm in enumerate(myHand.landmark):
h, w, c = img.shape
cx, cy = int(lm.x * w), int(lm.y * h)
lmlist.append([id, cx, cy])
if draw:
cv2.circle(img, (cx, cy), 3, (255, 0, 255), cv2.FILLED)
return lmlist
```
To run the handDetector(), save this file with .py extension and allow your webcam to take a video. The co-ordinates will be outputted at the terminal.
```
pTime = 0
cTime = 0
cap = cv2.VideoCapture(0)
detector = handDetector()
while True:
success, img = cap.read()
img = detector.findHands(img)
lmlist = detector.findPosition(img)
if len(lmlist) != 0:
print(lmlist[4])
cTime = time.time()
fps = 1 / (cTime - pTime)
pTime = cTime
cv2.putText(img, str(int(fps)), (10, 70), cv2.FONT_HERSHEY_PLAIN, 3, (255, 0, 255), 3)
cv2.imshow("Image", img)
cv2.waitKey(1)
```
#### Creator: Ayush Gupta , Github: [Profile](https://github.com/guptayush179)
|
github_jupyter
|
The purpose of this notebook is to convert the wide-format car data to long-format. The car data comes from the mlogit package. The data description is reproduced below. Note the data originally comes from McFadden and Train (2000).
#### Description
- Cross-Sectional Dataset
- Number of Observations: 4,654
- Unit of Observation: Individual
- Country: United States
#### Format
A dataframe containing :
- choice: choice of a vehicule amoung 6 propositions
- college: college education?
- hsg2: size of household greater than 2?
- coml5: commulte lower than 5 miles a day?
- typez: body type, one of regcar (regular car), sportuv (sport utility vehicule), sportcar, stwagon (station wagon), truck, van, for each proposition z from 1 to 6
- fuelz: fuel for proposition z, one of gasoline, methanol, cng (compressed natural gas), electric. pricez price of vehicule divided by the logarithme of income
- rangez: hundreds of miles vehicule can travel between refuelings/rechargings
- accz: acceleration, tens of seconds required to reach 30 mph from stop
- speedz: highest attainable speed in hundreds of mph
- pollutionz: tailpipe emissions as fraction of those for new gas vehicule
- sizez: 0 for a mini, 1 for a subcompact, 2 for a compact and 3 for a mid–size or large vehicule
- spacez: fraction of luggage space in comparable new gas vehicule
- costz: cost per mile of travel(tens of cents). Either cost of home recharging for electric vehicule or the cost of station refueling otherwise
- stationz: fraction of stations that can refuel/recharge vehicule
#### Source
McFadden, Daniel and Kenneth Train (2000) “Mixed MNL models for discrete response”, Journal of Applied Econometrics, 15(5), 447–470.
Journal of Applied Econometrics data archive : http://jae.wiley.com/jae/
```
import pandas as pd
import numpy as np
import pylogit as pl
```
# Load the Car data
```
wide_car = pd.read_csv("../data/raw/car_wide_format.csv")
wide_car.head().T
```
# Convert the Car dataset to long-format
```
# Look at the columns of the car data
print(wide_car.columns.tolist())
# Create the list of individual specific variables
ind_variables = wide_car.columns.tolist()[1:4]
# Specify the variables that vary across individuals and some or all alternatives
# The keys are the column names that will be used in the long format dataframe.
# The values are dictionaries whose key-value pairs are the alternative id and
# the column name of the corresponding column that encodes that variable for
# the given alternative. Examples below.
new_name_to_old_base = {'body_type': 'type{}',
'fuel_type': 'fuel{}',
'price_over_log_income': 'price{}',
'range': 'range{}',
'acceleration': 'acc{}',
'top_speed': 'speed{}',
'pollution': 'pollution{}',
'vehicle_size': 'size{}',
'luggage_space': 'space{}',
'cents_per_mile': 'cost{}',
'station_availability': 'station{}'}
alt_varying_variables =\
{k: dict([(x, v.format(x)) for x in range(1, 7)])
for k, v in list(new_name_to_old_base.items())}
# Specify the availability variables
# Note that the keys of the dictionary are the alternative id's.
# The values are the columns denoting the availability for the
# given mode in the dataset.
availability_variables =\
{x: 'avail_{}'.format(x) for x in range(1, 7)}
for col in availability_variables.values():
wide_car[col] = 1
##########
# Determine the columns for: alternative ids, the observation ids and the choice
##########
# The 'custom_alt_id' is the name of a column to be created in the long-format data
# It will identify the alternative associated with each row.
custom_alt_id = "alt_id"
# Create a custom id column that ignores the fact that this is a
# panel/repeated-observations dataset. Note the +1 ensures the id's start at one.
obs_id_column = "obs_id"
wide_car[obs_id_column] =\
np.arange(1, wide_car.shape[0] + 1, dtype=int)
# Create a variable recording the choice column
choice_column = "choice"
# Store the original choice column in a new variable
wide_car['orig_choices'] = wide_car['choice'].values
# Alter the original choice column
choice_str_to_value = {'choice{}'.format(x): x for x in range(1, 7)}
wide_car[choice_column] =\
wide_car[choice_column].map(choice_str_to_value)
# Convert the wide-format data to long format
long_car =\
pl.convert_wide_to_long(wide_data=wide_car,
ind_vars=ind_variables,
alt_specific_vars=alt_varying_variables,
availability_vars=availability_variables,
obs_id_col=obs_id_column,
choice_col=choice_column,
new_alt_id_name=custom_alt_id)
long_car.head().T
# Save the long-format data
long_car.to_csv("../data/interim/car_long_format.csv",
index=False)
```
|
github_jupyter
|
# Score for the Fed's dual mandate
The U.S. Congress established three key objectives for monetary policy
in the Federal Reserve Act: *Maximum employment, stable prices*, and
moderate long-term interest rates. The first two objectives are
sometimes referred to as the Federal Reserve's **dual mandate**.
Here we examine unemployment and inflation data to construct
a time-series which gives a numerical score to the
Fed's performance on the dual mandate.
(This notebook could be extended to studies of
the **Phillips curve**, see Appendix 1).
The key is to find comparable units to measure performance
and a suitable scalar measure to show deviation
from the dual mandate.
Our visualization features *time-sequential* scatter plots using color *heat* map.
Short URL: https://git.io/phillips
*Dependencies:*
- fecon235 repository https://github.com/rsvp/fecon235
- Python: matplotlib, pandas
*CHANGE LOG*
2016-11-14 Fix #2 by v5 and PREAMBLE-p6.16.0428 upgrades.
Switch from fecon to fecon235 for main import module.
Minor edits given additional year of data.
2015-12-15 Switch to yi_0sys dependencies. Phillips curve.
2015-11-18 First version.
```
from fecon235.fecon235 import *
# PREAMBLE-p6.16.0428 :: Settings and system details
from __future__ import absolute_import, print_function
system.specs()
pwd = system.getpwd() # present working directory as variable.
print(" :: $pwd:", pwd)
# If a module is modified, automatically reload it:
%load_ext autoreload
%autoreload 2
# Use 0 to disable this feature.
# Notebook DISPLAY options:
# Represent pandas DataFrames as text; not HTML representation:
import pandas as pd
pd.set_option( 'display.notebook_repr_html', False )
from IPython.display import HTML # useful for snippets
# e.g. HTML('<iframe src=http://en.mobile.wikipedia.org/?useformat=mobile width=700 height=350></iframe>')
from IPython.display import Image
# e.g. Image(filename='holt-winters-equations.png', embed=True) # url= also works
from IPython.display import YouTubeVideo
# e.g. YouTubeVideo('1j_HxD4iLn8', start='43', width=600, height=400)
from IPython.core import page
get_ipython().set_hook('show_in_pager', page.as_hook(page.display_page), 0)
# Or equivalently in config file: "InteractiveShell.display_page = True",
# which will display results in secondary notebook pager frame in a cell.
# Generate PLOTS inside notebook, "inline" generates static png:
%matplotlib inline
# "notebook" argument allows interactive zoom and resize.
```
## Comparable unit for comparison
A 1% change in inflation may have different economic significance
than a 1% change in unemployment.
We retrieve historical data, then let one standard deviation
represent one unit for scoring purposes.
Note that the past scores will thus be represented *ex-post*,
which is to say, influenced by recent data.
The dual mandate is not required to be explicitly
stated numerically by the Federal Reserve,
so we assign explicit target values
for unemployment and inflation
(those frequently mentioned in Congressional testimonies
and used as benchmarks by the market).
These targets can be finely reset below
so that the Fed performance can be re-evaluated.
Sidenote: "The natural rate of unemployment
[**NAIRU**, *non-accelerating inflation rate of unemployment*]
is the rate of unemployment
arising from all sources except fluctuations in aggregate demand.
Estimates of potential GDP are based on the long-term natural rate.
The short-term natural rate is used to gauge the amount of current
and projected slack in labor markets."
See https://research.stlouisfed.org/fred2/series/NROU
and Appendix 1 for details.
```
# Set DUAL MANDATE, assumed throughout this notebook:
unem_target = 5.0
infl_target = 2.0
# The Fed varies the targets over time,
# sometimes only implicitly. So for example,
# there may be disagreement among Board members
# regarding NAIRU -- but we set it to what
# seems to be the prevalent market assumption.
```
## Unemployment rate
```
unem = get( m4unemp )
# m4 implies monthly frequency.
# # Starts 1948-01-01, uncomment to view:
# stats( unem )
# Standard deviation for unemployment rate:
unem_std = unem.std()
unem_std
# Uncomment to plot raw unemployment rate:
# plot( unem )
# Score unemployment as standard deviations from target:
unem_score = todf( (unem - unem_target) / unem_std )
```
## Inflation
```
# Use synthetic inflation
# which averages CPI and PCE for both headline and core versions:
infl_level = get( m4infl )
# Get the YoY inflation rate:
infl = todf(pcent( infl_level, 12 ))
# # Starts 1960-01-01, uncomment to view:
# stats(infl)
infl_std = infl.std()
infl_std
# # Uncomment to plot inflation rate:
# plot( infl )
# Score inflation as standard deviations from target:
infl_score = todf( (infl - infl_target) / infl_std )
```
## Expressing duality as complex number
We encode each joint score for unemployment and inflation into a single complex number.
Let *u* be the unemployment score and *i* the inflation score.
(Note: we follow the Python/engineering convention by letting **j**
be the imaginary number $\sqrt -1$.)
So let *z* be our dual encoding as follows:
$ z = u + i \mathbf{j} $
(In the history of mathematics, this was the precursor
to the idea of a *vector*.)
```
# Let's start constructing our 4-column dataframe:
scores = paste( [unem_score, infl_score, infl_score, infl_score ] )
# Third and fouth columns are dummy placeholders to be replaced later.
# Give names to the scores columns:
scores.columns = ['z_unem', 'z_infl', 'z', 'z_norm']
# Fill in THIRD column z as complex number per our discussion:
scores.z = scores.z_unem + (scores.z_infl * 1j)
# The imaginary number in itself in Python is represented as 1j,
# since j may be a variable elsewhere.
```
## Computing the Fed score
Each dual score can be interpreted as a vector in the
complex plane. Its component parts, real for unemployment
and imaginary for inflation, measure deviation from
respective targets in units expressed as standard deviations.
*Our **key idea** is to use the length of this vector (from
the origin, 0+0**j**, representing the dual mandate) as
the Fed score* = |z|.
Python, which natively handles complex numbers, can
compute the *norm* of such a vector using abs(z).
Later we shall visualize the trajectory of the component
parts using a color heat map.
```
# Finally fill-in the FOURTH placeholder column:
scores.z_norm = abs( scores.z )
# Tail end of recent scores:
tail( scores )
# ... nicely revealing the data structure:
```
## Visualizing Fed scores
z_norm is expressed in standard deviation units, thus
it truly represents deviation from the dual mandate
on a Gaussian scale.
```
# Define descriptive dataframe from our mathematical construct:
fed_score = todf( scores.z_norm )
# FED SCORE
plot( fed_score )
```
*We can say that a score greater than 2 is definitively cause for concern.
For example, between 1974 and 1984, there are two peaks
extending into 4 standard deviations mainly due to high inflation.
Fed score during the Great Recession hit 3 mainly due to
severe unemployment.*
```
stats( fed_score )
```
## Remarks on fed_score
Our fed_score *handles both positive and negative deviations from the Fed's dual mandate*,
moreover, it handles them jointly using a historical fair measuring unit:
the standard deviation. This avoids using ad-hoc weights to balance
the importance between unemployment and inflation.
The **fed_score is always a positve real number (since it is a norm)
which is zero if and only if the Fed has achieved targets**
(which we have explicitly specified).
*That score can be interpreted as the number of standard deviations
away from the dual mandate.*
Our **fed_score** can also be simply interpreted
as an economic **crisis level** indicator
(much like a *n-alarm* fire) when Fed monetary
policy becomes crucial for the US economy.
Since 1960, ex-post fed_score averages around 1.31 where
the mid-50% percentile range is approximately [0.47, 1.79].
But keep in mind that this computation relies on our
current fixed targets, so fed_score is most
useful in accessing recent performance
from the perspective of historical variance.
This means that the fed_score for a particular
point in time may change as new incoming data arrives.
2015-12-15 notes: The current fed_score is 0.46
gravitating towards zero as the Fed is preparing
its first rate hike in a decade.
2016-11-14 notes: The current fed_score is 0.14
as the Fed is expected to announce
the second rate hike since the Great Recession.
The labor market has exhibited sustained improvement.
## CONCLUSION: visualize fed_score components over time
When FOMC makes its releases, the market becomes
fixated on the so-called blue dot chart
of expected future interest rates by its members.
The next scatter plot helps us to understand
the motivations and constraints behind
the Federal Reserve's monetary policy.
We see that the key components of the U.S. economy,
unemployment and inflation, came back
"full circle" to target from 2005 to 2016,
or more accurately "full figure eight"
with *major* (3 standard) deviations seen in the unemployment component.
```
# Scatter plot of recent data using color heat map:
scatter( scores['2005':], col=[0, 1] )
# (Ignore FutureWarning from matplotlib/collections.py:590
# regarding elementwise comparison due to upstream numpy.)
```
In this scatter plot: z_unem is shown along the x-axis,
z_infl along the y-axis,
such that the color *heat* map depicts chronological movement
from *blue to green to red*.
The coordinate (0, 0) represents our Fed dual mandate,
so it is easy to see deviations from target.
Geometrically a point's distance from the origin is what
we have computed as fed_score (= z_norm, in the complex plane).
- - - -
## Appendix 1: Phillips curve
The Phillips curve purports to explain the relationship between
inflation and unemployment, however, as it turns out the
relationship is one of mere correlation during certain periods
in a given country. It is too simplistic to assert that
decreased unemployment (i.e. increased levels of employment)
will cause with the inflation rate to increase.
"Most economists no longer use the Phillips curve in its original
form because it was shown to be too simplistic. This can be seen
in a cursory analysis of US inflation and unemployment data from
1953-92. There is no single curve that will fit the data, but
there are three rough aggregations—1955–71, 1974–84, and 1985–92 —
each of which shows a general, downwards slope, but at three
very different levels with the shifts occurring abruptly.
The data for 1953-54 and 1972-73 do not group easily,
and a more formal analysis posits up to five groups/curves
over the period.
Modern versions distinguish between short-run and long-run
effects on unemployment. The "short-run Phillips curve" is also called
the "expectations-augmented Phillips curve," since it shifts up when
inflationary expectations rise. In the long run, this implies
that monetary policy cannot affect unemployment,
which adjusts back to its "natural rate", also called the "NAIRU" or
"long-run Phillips curve". However, this long-run "neutrality" of
monetary policy does allow for short run fluctuations and the ability
of the monetary authority to temporarily decrease unemployment
by increasing permanent inflation, and vice versa.
In many recent stochastic general equilibrium models, with sticky prices,
there is a positive relation between the rate of inflation and
the level of demand, and therefore a negative relation between
the rate of inflation and the rate of unemployment. This relationship is
often called the "New Keynesian Phillips curve." Like the
expectations-augmented Phillips curve, the New Keynesian Phillips curve
implies that increased inflation can lower unemployment temporarily,
but cannot lower it permanently."
For more details, see https://en.wikipedia.org/wiki/Phillips_curve
Curiously, according to Edmund Phelps who won the 2006 Nobel Prize in Economics,
the **long-run Phillips Curve is *vertical* **
such that the rate of inflation has no effect on unemployment at its NAIRU.
The name "NAIRU" arises because with actual unemployment below it,
inflation accelerates, while with unemployment above it,
inflation decelerates. With the actual rate equal to it,
inflation is stable, neither accelerating nor decelerating.
```
Image(url="https://upload.wikimedia.org/wikipedia/commons/e/e3/NAIRU-SR-and-LR.svg", embed=False)
```
We know from our data that economic reality is far more
complicated than the ideal diagram above.
For example, in the late 1990s, the unemployment rate
fell below 4%, much lower than almost all estimates of the NAIRU.
But inflation stayed very moderate rather than accelerating.
Since our z data captures the normalized version of
unemployment vs inflation rates, it is easy
to visualize the actual paths using our scatter plots.
```
# Uncomment for # Scatter plot of selected data using color heat map:
# scatter( scores['1995':'2000'], col=[0, 1] )
```
|
github_jupyter
|
# <center>Welcome to Supervised Learning</center>
## <center>Part 2: How to prepare your data for supervised machine learning</center>
## <center>Instructor: Andras Zsom</center>
### <center>https://github.com/azsom/Supervised-Learning<center>
## The topic of the course series: supervised Machine Learning (ML)
- how to build an ML pipeline from beginning to deployment
- we assume you already performed data cleaning
- this is the first course out of 6 courses
- Part 1: Introduction to machine learning and the bias-variance tradeoff
- **Part 2: How to prepare your data for supervised machine learning**
- Part 3: Evaluation metrics in supervised machine learning
- Part 4: SVMs, Random Forests, XGBoost
- Part 5: Missing data in supervised ML
- Part 6: Interpretability
- you can complete the courses in sequence or complete individual courses based on your interest
### Structured data
| X|feature_1|feature_2|...|feature_j|...|feature_m|<font color='red'>Y</font>|
|-|:-:|:-:|:-:|:-:|:-:|:-:|:-:|
|__data_point_1__|x_11|x_12|...|x_1j|...|x_1m|__<font color='red'>y_1</font>__|
|__data_point_2__|x_21|x_22|...|x_2j|...|x_2m|__<font color='red'>y_2</font>__|
|__...__|...|...|...|...|...|...|__<font color='red'>...</font>__|
|__data_point_i__|x_i1|x_i2|...|x_ij|...|x_im|__<font color='red'>y_i</font>__|
|__...__|...|...|...|...|...|...|__<font color='red'>...</font>__|
|__data_point_n__|x_n1|x_n2|...|x_nj|...|x_nm|__<font color='red'>y_n</font>__|
We focus on the feature matrix (X) in this course.
### Learning objectives of this course
By the end of the course, you will be able to
- describe why data splitting is necessary in machine learning
- summarize the properties of IID data
- list examples of non-IID datasets
- apply IID splitting techniques
- apply non-IID splitting techniques
- identify when a custom splitting strategy is necessary
- describe the two motivating concepts behind preprocessing
- apply various preprocessors to categorical and continuous features
- perform preprocessing with a sklearn pipeline and ColumnTransformer
# Module 1: Split IID data
### Learning objectives of this module:
- describe why data splitting is necessary in machine learning
- summarize the properties of IID data
- apply IID splitting techniques
## Why do we split the data?
- we want to find the best hyper-parameters of our ML algorithms
- fit models to training data
- evaluate each model on validation set
- we find hyper-parameter values that optimize the validation score
- we want to know how the model will perform on previously unseen data - the generalization error
- apply our final model on the test set
### We need to split the data into three parts!
## Ask yourself these questions!
- What is the intended use of the model? What is it supposed to do/predict?
- What data/info do you have available at the time of prediction?
- Your split must mimic the intended use of the model only then will you accurately estimate how well the model will perform on previously unseen points (generalization error).
- two examples:
- if you want to predict the outcome of a new patient's visit to the ER:
- your test score must be based on patients not included in training and validation
- your validation score must be based on patients not included in training
- points of one patient should not be distributed over multiple sets because your generalization error will be off
- predict stocks price
- it is a time series data
- if you predict the stocks price at a certain time in development, make sure that you only use information predating that time
## How should we split the data into train/validation/test?
- data is **Independent and Identically Distributed** (iid)
- all samples stem from the same generative process and the generative process is assumed to have no memory of past generated samples
- identify cats and dogs on images
- predict the house price
- predict if someone's salary is above or below 50k
- examples of not iid data:
- data generated by time-dependent processes
- data has group structure (samples collected from e.g., different subjects, experiments, measurement devices)
## Splitting strategies for iid data: basic approach
- 60% train, 20% validation, 20% test for small datasets
- 98% train, 1% validation, 1% test for large datasets
- if you have 1 million points, you still have 10000 points in validation and test which is plenty to assess model performance
### Let's work with the adult data!
https://archive.ics.uci.edu/ml/datasets/adult
```
import pandas as pd
from sklearn.model_selection import train_test_split
df = pd.read_csv('data/adult_data.csv')
# let's separate the feature matrix X, and target variable y
y = df['gross-income'] # remember, we want to predict who earns more than 50k or less than 50k
X = df.loc[:, df.columns != 'gross-income'] # all other columns are features
print(y)
print(X.head())
help(train_test_split)
random_state = 42
# first split to separate out the training set
X_train, X_other, y_train, y_other = train_test_split(X,y,train_size = 0.6,random_state=random_state)
print('training set:',X_train.shape, y_train.shape) # 60% of points are in train
print(X_other.shape, y_other.shape) # 40% of points are in other
print(X_train.head())
# second split to separate out the validation and test sets
X_val, X_test, y_val, y_test = train_test_split(X_other,y_other,train_size = 0.5,random_state=random_state)
print('validation set:',X_val.shape, y_val.shape) # 20% of points are in validation
print('test set:',X_test.shape, y_test.shape) # 20% of points are in test
print(X_val.head())
print(X_test.head())
```
## Randomness due to splitting
- the model performance, validation and test scores will change depending on which points are in train, val, test
- inherent randomness or uncertainty of the ML pipeline
- change the random state a couple of times and repeat the whole ML pipeline to assess how much the random splitting affects your test score
- you would expect a similar uncertainty when the model is deployed
## Splitting strategies for iid data: k-fold splitting
<center><img src="figures/grid_search_cross_validation.png" width="600"></center>
```
from sklearn.model_selection import KFold
help(KFold)
random_state = 42
# first split to separate out the test set
X_other, X_test, y_other, y_test = train_test_split(X,y,test_size = 0.2,random_state=random_state)
print(X_other.shape,y_other.shape)
print('test set:',X_test.shape,y_test.shape)
# do KFold split on other
kf = KFold(n_splits=5,shuffle=True,random_state=random_state)
for train_index, val_index in kf.split(X_other,y_other):
X_train = X_other.iloc[train_index]
y_train = y_other.iloc[train_index]
X_val = X_other.iloc[val_index]
y_val = y_other.iloc[val_index]
print(' training set:',X_train.shape, y_train.shape)
print(' validation set:',X_val.shape, y_val.shape)
# the validation set contains different points in each iteration
print(X_val[['age','workclass','education']].head())
```
## How many splits should I create?
- tough question, 3-5 is most common
- if you do $n$ splits, $n$ models will be trained, so the larger the $n$, the most computationally intensive it will be to train the models
- KFold is usually better suited for small datasets
- KFold is good to estimate uncertainty due to random splitting of train and val, but it is not perfect
- the test set remains the same
### Why shuffling iid data is important?
- by default, data is not shuffled by Kfold which can introduce errors!
<center><img src="figures/kfold.png" width="600"></center>
## Imbalanced data
- imbalanced data: only a small fraction of the points are in one of the classes, usually ~5% or less but there is no hard limit here
- examples:
- people visit a bank's website. do they sign up for a new credit card?
- most customers just browse and leave the page
- usually 1% or less of the customers get a credit card (class 1), the rest leaves the page without signing up (class 0).
- fraud detection
- only a tiny fraction of credit card payments are fraudulent
- rare disease diagnosis
- the issue with imbalanced data:
- if you apply train_test_split or KFold, you might not have class 1 points in one of your sets by chance
- this is what we need to fix
## Solution: stratified splits
```
random_state = 42
X_train, X_other, y_train, y_other = train_test_split(X,y,train_size = 0.6,random_state=random_state)
X_val, X_test, y_val, y_test = train_test_split(X_other,y_other,train_size = 0.5,random_state=random_state)
print('**balance without stratification:**')
# a variation on the order of 1% which would be too much for imbalanced data!
print(y_train.value_counts(normalize=True))
print(y_val.value_counts(normalize=True))
print(y_test.value_counts(normalize=True))
X_train, X_other, y_train, y_other = train_test_split(X,y,train_size = 0.6,stratify=y,random_state=random_state)
X_val, X_test, y_val, y_test = train_test_split(X_other,y_other,train_size = 0.5,stratify=y_other,random_state=random_state)
print('**balance with stratification:**')
# very little variation (in the 4th decimal point only) which is important if the problem is imbalanced
print(y_train.value_counts(normalize=True))
print(y_val.value_counts(normalize=True))
print(y_test.value_counts(normalize=True))
```
## Stratified folds
<center><img src="figures/stratified_kfold.png" width="600"></center>
```
from sklearn.model_selection import StratifiedKFold
help(StratifiedKFold)
# what we did before: variance in balance on the order of 1%
random_state = 42
X_other, X_test, y_other, y_test = train_test_split(X,y,test_size = 0.2,random_state=random_state)
print('test balance:',y_test.value_counts(normalize=True))
# do KFold split on other
kf = KFold(n_splits=5,shuffle=True,random_state=random_state)
for train_index, val_index in kf.split(X_other,y_other):
X_train = X_other.iloc[train_index]
y_train = y_other.iloc[train_index]
X_val = X_other.iloc[val_index]
y_val = y_other.iloc[val_index]
print('train balance:')
print(y_train.value_counts(normalize=True))
print('val balance:')
print(y_val.value_counts(normalize=True))
# stratified K Fold: variation in balance is very small (4th decimal point)
random_state = 42
# stratified train-test split
X_other, X_test, y_other, y_test = train_test_split(X,y,test_size = 0.2,stratify=y,random_state=random_state)
print('test balance:',y_test.value_counts(normalize=True))
# do StratifiedKFold split on other
kf = StratifiedKFold(n_splits=5,shuffle=True,random_state=random_state)
for train_index, val_index in kf.split(X_other,y_other):
X_train = X_other.iloc[train_index]
y_train = y_other.iloc[train_index]
X_val = X_other.iloc[val_index]
y_val = y_other.iloc[val_index]
print('train balance:')
print(y_train.value_counts(normalize=True))
print('val balance:')
print(y_val.value_counts(normalize=True))
```
# Module 2: Split non-IID data
### Learning objectives of this module:
- list examples of non-IID datasets
- apply non-IID splitting techniques
- identify when a custom splitting strategy is necessary
## Examples of non-iid data
- if there is any sort of time or group structure in your data, it is likely non-iid
- group structure:
- each point is someone's visit to the ER and some people visited the ER multiple times
- each point is a customer's visit to website and customers tend to return regularly
- time structure
- each point is the stocks price at a given time
- eahc point is a person's health or activity status
## Group-based split: GroupShuffleSplit
<center><img src="figures/groupshufflesplit.png" width="600"></center>
```
import numpy as np
from sklearn.model_selection import GroupShuffleSplit
X = np.ones(shape=(8, 2))
y = np.ones(shape=(8, 1))
groups = np.array([1, 1, 2, 2, 2, 3, 3, 3])
gss = GroupShuffleSplit(n_splits=10, train_size=.8, random_state=42)
for train_idx, test_idx in gss.split(X, y, groups):
print("TRAIN:", train_idx, "TEST:", test_idx)
```
## Group-based split: GroupKFold
<center><img src="figures/groupkfold.png" width="600"></center>
```
from sklearn.model_selection import GroupKFold
group_kfold = GroupKFold(n_splits=3)
for train_index, test_index in group_kfold.split(X, y, groups):
print("TRAIN:", train_index, "TEST:", test_index)
help(GroupKFold)
```
## Data leakage in time series data is similar!
- do NOT use information in validation or test which will not be available once your model is deployed
- don't use future information!
<center><img src="figures/timeseriessplit.png" width="600"></center>
```
import numpy as np
from sklearn.model_selection import TimeSeriesSplit
X = np.array([[1, 2], [3, 4], [1, 2], [3, 4], [1, 2], [3, 4]])
y = np.array([1, 2, 3, 4, 5, 6])
tscv = TimeSeriesSplit()
for train_index, test_index in tscv.split(X):
print("TRAIN:", train_index, "TEST:", test_index)
X_train, X_test = X[train_index], X[test_index]
y_train, y_test = y[train_index], y[test_index]
```
## When should you develop your own splitting function?
- there are certain splitting strategies sklearn can't handle at the moment
- time series data with group structure is one example
- if you want certain groups to be in certain sets
- group structure in classification where all points in a group belong to a certain class
- you might want a roughly equal number of groups of each class to be in each set
- check out the [model selection](https://scikit-learn.org/stable/modules/classes.html#module-sklearn.model_selection) part of sklearn
- if the splitting stragey you want to follow is not there, implement your own function
# Module 3: Preprocess continuous and categorical features
### Learning objectives of this module:
- describe the two motivating concepts behind preprocessing
- apply various preprocessors to categorical and continuous features
- perform preprocessing with a sklearn pipeline and ColumnTransformer
### Data almost never comes in a format that's directly usable in ML
- ML works with numerical data but some columns are text (e.g., home country, educational level, gender, race)
- some ML algorithms accept (and prefer) a non-numerical feature matrix (like [CatBoost](https://catboost.ai/) ) but that's not standard
- sklearn throws an error message if the feature matrix contains non-numerical elements
- the order of magnitude of numerical features can vary greatly which is not good for most ML algorithms (e.g., salary in USD, age in years, time spent on the site in sec)
- many ML algorithms are distance-based and they perform better and converge faster if the features are standardized (features have a mean of 0 and the same standard deviation, usually 1)
- Lasso and Ridge regression because of the penalty term, K Nearest Neightbors, SVM, linear models if you want to use the coefficients to measure feature importance (more on this in part 6), neural networks
- tree-based methods don't require standardization
- check out part 1 to learn more about linear and logistic regression, Lasso and Ridge
- check out part 4 to learn more about SVMs, tree-based methods, and K Nearest Neighbors
### scikit-learn transformers to the rescue!
Preprocessing is done with various transformers. All transformes have three methods:
- **fit** method: estimates parameters necessary to do the transformation,
- **transform** method: transforms the data based on the estimated parameters,
- **fit_transform** method: both steps are performed at once, this can be faster than doing the steps separately.
### Transformers we cover
- **OrdinalEncoder** - converts categorical features into an integer array
- **OneHotEncoder** - converts categorical features into dummy arrays
- **StandardScaler** - standardizes continuous features by removing the mean and scaling to unit variance
## Ordered categorical data: OrdinalEncoder
Let's assume we have a categorical feature and training and test sets
The cateogies can be ordered or ranked
E.g., educational level in the adult dataset
```
import pandas as pd
train_edu = {'educational level':['Bachelors','Masters','Bachelors','Doctorate','HS-grad','Masters']}
test_edu = {'educational level':['HS-grad','Masters','Masters','College','Bachelors']}
X_train = pd.DataFrame(train_edu)
X_test = pd.DataFrame(test_edu)
from sklearn.preprocessing import OrdinalEncoder
help(OrdinalEncoder)
# initialize the encoder
cats = ['HS-grad','Bachelors','Masters','Doctorate']
enc = OrdinalEncoder(categories = [cats]) # The ordered list of
# categories need to be provided. By default, the categories are alphabetically ordered!
# fit the training data
enc.fit(X_train)
# print the categories - not really important because we manually gave the ordered list of categories
print(enc.categories_)
# transform X_train. We could have used enc.fit_transform(X_train) to combine fit and transform
X_train_oe = enc.transform(X_train)
print(X_train_oe)
# transform X_test
X_test_oe = enc.transform(X_test) # OrdinalEncoder always throws an error message if
# it encounters an unknown category in test
print(X_test_oe)
```
## Unordered categorical data: one-hot encoder
some categories cannot be ordered. e.g., workclass, relationship status
first feature: gender (male, female, unknown)
second feature: browser used
these categories cannot be ordered
```
train = {'gender':['Male','Female','Unknown','Male','Female','Female'],\
'browser':['Safari','Safari','Internet Explorer','Chrome','Chrome','Internet Explorer']}
test = {'gender':['Female','Male','Unknown','Female'],'browser':['Chrome','Firefox','Internet Explorer','Safari']}
X_train = pd.DataFrame(train)
X_test = pd.DataFrame(test)
# How do we convert this to numerical features?
from sklearn.preprocessing import OneHotEncoder
help(OneHotEncoder)
# initialize the encoder
enc = OneHotEncoder(sparse=False) # by default, OneHotEncoder returns a sparse matrix. sparse=False returns a 2D array
# fit the training data
enc.fit(X_train)
print('categories:',enc.categories_)
print('feature names:',enc.get_feature_names())
# transform X_train
X_train_ohe = enc.transform(X_train)
#print(X_train_ohe)
# do all of this in one step
X_train_ohe = enc.fit_transform(X_train)
print(X_train_ohe)
# transform X_test
X_test_ohe = enc.transform(X_test)
print('X_test transformed')
print(X_test_ohe)
```
## Continuous features: StandardScaler
```
train = {'salary':[50_000,75_000,40_000,1_000_000,30_000,250_000,35_000,45_000]}
test = {'salary':[25_000,55_000,1_500_000,60_000]}
X_train = pd.DataFrame(train)
X_test = pd.DataFrame(test)
from sklearn.preprocessing import StandardScaler
help(StandardScaler)
scaler = StandardScaler()
print(scaler.fit_transform(X_train))
print(scaler.transform(X_test))
```
## How and when to do preprocessing in the ML pipeline?
- **SPLIT YOUR DATA FIRST!**
- **APPLY TRANSFORMER.FIT ONLY ON YOUR TRAINING DATA!** Then transform the validation and test sets.
- One of the most common mistake practitioners make is leaking statistics!
- fit_transform is applied to the whole dataset, then the data is split into train/validation/test
- this is wrong because the test set statistics impacts how the training and validation sets are transformed
- but the test set must be separated from train and val, and val must be separated from train
- or fit_transform is applied to the train, then fit_transform is applied to the validation set, and fit_transform is applied to the test set
- this is wrong because the relative position of the points change
<center><img src="figures/no_separate_scaling.png" width="1200"></center>
## Scikit-learn's pipelines
- Preprocessing and model training (not the splitting) can be chained together into a scikit-learn pipeline which consists of transformers and one final estimator which is usually your classifier or regression model.
- It neatly combines the preprocessing steps and it helps to avoid leaking statistics.
https://scikit-learn.org/stable/auto_examples/compose/plot_column_transformer_mixed_types.html
```
import pandas as pd
import numpy as np
from sklearn.compose import ColumnTransformer
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler, OneHotEncoder, OrdinalEncoder, MinMaxScaler
from sklearn.model_selection import train_test_split
np.random.seed(0)
df = pd.read_csv('data/adult_data.csv')
# let's separate the feature matrix X, and target variable y
y = df['gross-income'] # remember, we want to predict who earns more than 50k or less than 50k
X = df.loc[:, df.columns != 'gross-income'] # all other columns are features
random_state = 42
# first split to separate out the training set
X_train, X_other, y_train, y_other = train_test_split(X,y,train_size = 0.6,random_state=random_state)
# second split to separate out the validation and test sets
X_val, X_test, y_val, y_test = train_test_split(X_other,y_other,train_size = 0.5,random_state=random_state)
# collect which encoder to use on each feature
# needs to be done manually
ordinal_ftrs = ['education']
ordinal_cats = [[' Preschool',' 1st-4th',' 5th-6th',' 7th-8th',' 9th',' 10th',' 11th',' 12th',' HS-grad',\
' Some-college',' Assoc-voc',' Assoc-acdm',' Bachelors',' Masters',' Prof-school',' Doctorate']]
onehot_ftrs = ['workclass','marital-status','occupation','relationship','race','sex','native-country']
std_ftrs = ['capital-gain','capital-loss','age','hours-per-week']
# collect all the encoders
preprocessor = ColumnTransformer(
transformers=[
('ord', OrdinalEncoder(categories = ordinal_cats), ordinal_ftrs),
('onehot', OneHotEncoder(sparse=False,handle_unknown='ignore'), onehot_ftrs),
('std', StandardScaler(), std_ftrs)])
# for now we only preprocess, later on we will add other steps here
# note the final scaler which is a standard scaler
# the ordinal and one hot encoded features do not have a mean of 0 and an std of 1
# the final scaler standardizes those features
clf = Pipeline(steps=[('preprocessor', preprocessor),('final scaler',StandardScaler())])
X_train_prep = clf.fit_transform(X_train)
X_val_prep = clf.transform(X_val)
X_test_prep = clf.transform(X_test)
print(X_train.shape)
print(X_train_prep.shape)
print(np.mean(X_train_prep,axis=0))
print(np.std(X_train_prep,axis=0))
print(np.mean(X_val_prep,axis=0))
print(np.std(X_val_prep,axis=0))
print(np.mean(X_test_prep,axis=0))
print(np.std(X_test_prep,axis=0))
```
|
github_jupyter
|
## 2. Random Forest
### a)
```
import pandas as pd
headers = ["Number of times pregnant",
"Plasma glucose concentration a 2 hours in an oral glucose tolerance test",
"Diastolic blood pressure (mm Hg)",
"Triceps skinfold thickness (mm)",
"2-Hour serum insulin (mu U/ml)",
"Body mass index (weight in kg/(height in m)^2)",
"Diabetes pedigree function",
"Age (years)",
"Class variable(0 or 1)"]
raw_df = pd.read_csv('Diabetes.csv', names= headers)
print(raw_df.shape)
raw_df.head(6)
```
<hr style = "border-top: 3px solid #000000 ; border-radius: 3px;">
<p style =" direction:rtl;text-align:right;">
ابتدا یک لیست از اسامی هدر ها ایجاد کرده و سپس فایل csv را به کمک ان فراخوانی می کنیم
</p>
<hr style = "border-top: 3px solid #000000 ; border-radius: 3px;">
### b)
#### Handling Missing Values
```
raw_df.isnull().sum()
raw_df['Class variable(0 or 1)'].value_counts()
```
<hr style = "border-top: 3px solid #000000 ; border-radius: 3px;">
<p style =" direction:rtl;text-align:right;">
همانطور که مشخص است داده missing در دیتاست وجود ندارد.
همینطور با توجه به محموع تعداد هرکدام از مقادیر یکتا که برابر است به تعداد کل رکوردها می توان فهمید که در ستون کلاس null نداریم.
این تحلیل را برای سایر ستون ها هم می توان انجام داد.
</p>
<hr style = "border-top: 3px solid #000000 ; border-radius: 3px;">
#### Normalizing Numerical Columns and Encoding Categorical Ones
```
raw_df.dtypes
```
<hr style = "border-top: 3px solid #000000 ; border-radius: 3px;">
<p style =" direction:rtl;text-align:right;">
متغیر کتگوریکال نداریم پس نیازی به انکود نیست
</p>
<hr style = "border-top: 3px solid #000000 ; border-radius: 3px;">
```
from sklearn.preprocessing import normalize
numericals = pd.DataFrame(raw_df.loc[:, raw_df.columns != 'Class variable(0 or 1)'])
categoricals = pd.DataFrame(raw_df['Class variable(0 or 1)'])
#remove outlires
print("Before Removing Outliers: ",raw_df.shape)
Q1 = raw_df.loc[:, raw_df.columns != 'Class variable(0 or 1)'].quantile(0.25)
Q3 = raw_df.loc[:, raw_df.columns != 'Class variable(0 or 1)'].quantile(0.75)
IQR = Q3 - Q1
mask = ~((raw_df < (Q1 - 1.5 * IQR)) | (raw_df > (Q3 + 1.5 * IQR))).any(axis=1)
print("#Outliers = ",raw_df[~mask].dropna().shape[0])
print("#Not outliers = ",raw_df.shape[0]-raw_df[~mask].dropna().shape[0])
raw_df= raw_df[mask]
print("After Removing Outliers: ",raw_df.shape)
raw_df.head()
numericals = pd.DataFrame(raw_df.loc[:, raw_df.columns != 'Class variable(0 or 1)'])
categoricals = pd.DataFrame(raw_df['Class variable(0 or 1)'])
#normalize
raw_df.loc[:, raw_df.columns != 'Class variable(0 or 1)'] = normalize(numericals, norm='l2')
df.loc[:,:]=raw_df
df.head()
```
<hr style = "border-top: 3px solid #000000 ; border-radius: 3px;">
<p style =" direction:rtl;text-align:right;">
با جزئیاتی کاملا مشابه سوال قبل حذف داده های پرت و نرمال سازی انجام شده است.</p>
<hr style = "border-top: 3px solid #000000 ; border-radius: 3px;">
### c)
```
Y = df['Class variable(0 or 1)']
X = df.drop('Class variable(0 or 1)', axis=1)
```
### d)
```
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, Y, test_size=0.2)
print('Distribution of defferent classes in training data:(%) ')
y = pd.DataFrame(y_train)
((y.groupby('Class variable(0 or 1)')['Class variable(0 or 1)'].count())/y_train.shape[0])*100
print('Distribution of defferent classes in test data:(%) ')
y = pd.DataFrame(y_test)
((y.groupby('Class variable(0 or 1)')['Class variable(0 or 1)'].count())/y_test.shape[0])*100
```
<hr style = "border-top: 3px solid #000000 ; border-radius: 3px;">
<p style =" direction:rtl;text-align:right;">
به کمک تابع train_test_split داده ها را به دو بخش آموزشی و تست با نسبت خواسته شده تقسیم می کنیم.سپس درصد وجود هر کلاس را در داده های آموزش و تست محاسبه می کنیم.
همانطور که مشحص است هر کدام از این دسته ها درصدهای نزدیک به همی در دو نوع داده دارند
</p>
<hr style = "border-top: 3px solid #000000 ; border-radius: 3px;">
### e)
```
from sklearn.ensemble import RandomForestClassifier
myclassifier = RandomForestClassifier(max_depth=3, criterion='entropy').fit(X_train, y_train)
```
<hr style = "border-top: 3px solid #000000 ; border-radius: 3px;">
<p style =" direction:rtl;text-align:right;">
کلاسیفایر جنگل تصادفی را با پارامترهای گفته شده در سوال می سازیم .
سپس داده ها را برای آموزش به مدل دادیم.
</p>
<hr style = "border-top: 3px solid #000000 ; border-radius: 3px;">
### f)
```
from sklearn.metrics import classification_report, confusion_matrix, accuracy_score
y_predicted = myclassifier.predict(X_test)
print("[+] confusion matrix\n")
print(confusion_matrix(y_test, y_predicted))
print("\n[+] classification report\n")
print(classification_report(y_test, y_predicted))
print("Mean Accuracy = ",myclassifier.score(X_test,y_test,y_predicted))
print("Accuracy Score = " ,accuracy_score(y_test, y_predicted))
```
<hr style = "border-top: 3px solid #000000 ; border-radius: 3px;">
<p style =" direction:rtl;text-align:right;">
توضیح مفصل confusion matrix را در بخش های بعد ذکر خواهم کرد.
اما در این مرحله خطای mse و دقت را به صورت فوق گزارش کرده ام که دقت حوالی 70 است.
</p>
<hr style = "border-top: 3px solid #000000 ; border-radius: 3px;">
### g)
```
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.legend_handler import HandlerLine2D
# ranging from 1 to 32
max_depths = np.linspace(1, 32, 32, endpoint=True)
train_results = []
test_results = []
for max_depth in max_depths:
rf = RandomForestClassifier(max_depth=max_depth, criterion='entropy')
rf.fit(X_train, y_train)
train_pred = rf.predict(X_train)
ac = accuracy_score(y_train, train_pred)
train_results.append(ac)
y_pred = rf.predict(X_test)
acc = accuracy_score(y_test, y_pred)
test_results.append(acc)
fig, axes = plt.subplots(1,1,figsize=(9,9))
trains, = plt.plot(max_depths, train_results, 'b', label='Train Accuracy')
tests, = plt.plot(max_depths, test_results, 'r', label='Test Accuracy')
plt.legend(handler_map={trains: HandlerLine2D(numpoints=2)})
plt.ylabel('Acccuracy Score')
plt.xlabel('max_depth')
plt.show()
from sklearn.metrics import roc_curve, auc
max_depths = np.linspace(1, 32, 32, endpoint=True)
train_results = []
test_results = []
for max_depth in max_depths:
dt = RandomForestClassifier(max_depth=max_depth, criterion='entropy')
dt.fit(X_train, y_train)
train_pred = dt.predict(X_train)
false_positive_rate, true_positive_rate, thresholds = roc_curve(y_train, train_pred)
roc_auc = auc(false_positive_rate, true_positive_rate)
train_results.append(roc_auc)
y_pred = dt.predict(X_test)
false_positive_rate, true_positive_rate, thresholds = roc_curve(y_test, y_pred)
roc_auc = auc(false_positive_rate, true_positive_rate)
test_results.append(roc_auc)
fig, axes = plt.subplots(1,1,figsize=(9,9))
trains, = plt.plot(max_depths, train_results, 'b', label='Train Accuracy')
tests, = plt.plot(max_depths, test_results, 'r', label='Test Accuracy')
plt.legend(handler_map={trains: HandlerLine2D(numpoints=2)})
plt.ylabel('Acccuracy Score')
plt.xlabel('max_depth')
plt.show()
myclassifier = RandomForestClassifier(max_depth=5, criterion='entropy').fit(X_train, y_train)
y_predicted = myclassifier.predict(X_test)
print("[+] confusion matrix\n")
print(confusion_matrix(y_test, y_predicted))
print("\n[+] classification report\n")
print(classification_report(y_test, y_predicted))
print("Mean Accuracy = ",myclassifier.score(X_test,y_test,y_predicted))
print("Accuracy Score = " ,accuracy_score(y_test, y_predicted))
```
<hr style = "border-top: 3px solid #000000 ; border-radius: 3px;">
<p style =" direction:rtl;text-align:right;">
این پارامتر بیشینه عمق درخت را نمایش می دهد. اگر ست نشود مدل تا جایی ادامه می دهد تا به نودهای تماما خالص برسد یا برگها کمتر ازmin_samples_split باشند
همانطور که در نمودار هایی که برای دقت در دو بخش train و test رسم کرده ایم مشخص است بعد از مدتی ( حوالی 10 )
مدل ما با خطر overfit شدن مواجه می شود.
یعنی برای داده های آموزش نتایج بسیار خوبی حاصل می شود اما قادر به پیش بینی داده های جدید مخواهد بود.
به همین علت باید این پارامتر را به صورتی مقداررهی کنیم که نه این اتفاق رخ بدهد نه اینکه مدل به علت مقدار کم آن به خوبی آموزش نبیند.
پس این مقدار را برابر 5 قرارداده ام که قبل از بیش برازش است و دقت تست خوبی هم دارد.
</p>
<hr style = "border-top: 3px solid #000000 ; border-radius: 3px;">
|
github_jupyter
|
# Similarity Encoders with Keras
## using the model definition from `simec.py`
```
from __future__ import unicode_literals, division, print_function, absolute_import
from builtins import range
import numpy as np
np.random.seed(28)
import matplotlib.pyplot as plt
from sklearn.manifold import Isomap
from sklearn.decomposition import KernelPCA
from sklearn.preprocessing import StandardScaler
from sklearn.datasets import fetch_mldata, fetch_20newsgroups
import tensorflow as tf
tf.set_random_seed(28)
import keras
from keras.models import Sequential, Model
from keras.layers import Input, Dense, Activation
# https://github.com/cod3licious/nlputils
from nlputils.features import FeatureTransform, features2mat
from simec import SimilarityEncoder
from utils import center_K, check_embed_match, check_similarity_match
from utils_plotting import plot_mnist, plot_20news
%matplotlib inline
%load_ext autoreload
%autoreload 2
```
### MNIST with Linear Kernel
```
# load digits
mnist = fetch_mldata('MNIST original', data_home='data')
X = mnist.data/255. # normalize to 0-1
y = np.array(mnist.target, dtype=int)
# subsample 10000 random data points
np.random.seed(42)
n_samples = 10000
n_test = 2000
rnd_idx = np.random.permutation(X.shape[0])[:n_samples]
X_test, y_test = X[rnd_idx[:n_test],:], y[rnd_idx[:n_test]]
X, y = X[rnd_idx[n_test:],:], y[rnd_idx[n_test:]]
ss = StandardScaler(with_std=False)
X = ss.fit_transform(X)
X_test = ss.transform(X_test)
n_train, n_features = X.shape
# centered linear kernel matrix
K_lin = center_K(np.dot(X, X.T))
# linear kPCA
kpca = KernelPCA(n_components=2, kernel='linear')
X_embed = kpca.fit_transform(X)
X_embed_test = kpca.transform(X_test)
plot_mnist(X_embed, y, X_embed_test, y_test, title='MNIST - linear Kernel PCA')
print("error similarity match: msqe: %.10f ; r^2: %.10f ; rho: %.10f" % check_similarity_match(X_embed, K_lin))
# on how many target similarities you want to train - faster and works equally well than training on all
n_targets = 1000 # K_lin.shape[1]
# initialize the model
simec = SimilarityEncoder(X.shape[1], 2, n_targets, s_ll_reg=0.5, S_ll=K_lin[:n_targets,:n_targets])
# train the model to get an embedding with which the target similarities
# can be linearly approximated
simec.fit(X, K_lin[:,:n_targets], epochs=25)
# get the embeddings
X_embeds = simec.transform(X)
X_embed_tests = simec.transform(X_test)
plot_mnist(X_embeds, y, X_embed_tests, y_test, title='MNIST - SimEc (lin. kernel, linear)')
# correlation with the embedding produced by the spectral method should be high
print("correlation with lin kPCA : %f" % check_embed_match(X_embed, X_embeds)[1])
print("correlation with lin kPCA (test): %f" % check_embed_match(X_embed_test, X_embed_tests)[1])
# similarity match error should be similar to the one from kpca
print("error similarity match: msqe: %.10f ; r^2: %.10f ; rho: %.10f" % check_similarity_match(X_embeds, K_lin))
```
### Non-linear MNIST embedding with isomap
```
# isomap
isomap = Isomap(n_neighbors=10, n_components=2)
X_embed = isomap.fit_transform(X)
X_embed_test = isomap.transform(X_test)
plot_mnist(X_embed, y, X_embed_test, y_test, title='MNIST - isomap')
# non-linear SimEc to approximate isomap solution
K_geod = center_K(-0.5*(isomap.dist_matrix_**2))
n_targets = 1000
# initialize the model
simec = SimilarityEncoder(X.shape[1], 2, n_targets, hidden_layers=[(20, 'tanh')], s_ll_reg=0.5,
S_ll=K_geod[:n_targets,:n_targets], opt=keras.optimizers.Adamax(lr=0.01))
# train the model to get an embedding with which the target similarities
# can be linearly approximated
simec.fit(X, K_geod[:,:n_targets], epochs=25)
# get the embeddings
X_embeds = simec.transform(X)
X_embed_tests = simec.transform(X_test)
plot_mnist(X_embeds, y, X_embed_tests, y_test, title='MNIST - SimEc (isomap, 1 h.l.)')
print("correlation with isomap : %f" % check_embed_match(X_embed, X_embeds)[1])
print("correlation with isomap (test): %f" % check_embed_match(X_embed_test, X_embed_tests)[1])
```
## 20newsgroups embedding
```
## load the data and transform it into a tf-idf representation
categories = [
"comp.graphics",
"rec.autos",
"rec.sport.baseball",
"sci.med",
"sci.space",
"soc.religion.christian",
"talk.politics.guns"
]
newsgroups_train = fetch_20newsgroups(subset='train', remove=(
'headers', 'footers', 'quotes'), data_home='data', categories=categories, random_state=42)
newsgroups_test = fetch_20newsgroups(subset='test', remove=(
'headers', 'footers', 'quotes'), data_home='data', categories=categories, random_state=42)
# store in dicts (if the text contains more than 3 words)
textdict = {i: t for i, t in enumerate(newsgroups_train.data) if len(t.split()) > 3}
textdict.update({i: t for i, t in enumerate(newsgroups_test.data, len(newsgroups_train.data)) if len(t.split()) > 3})
train_ids = [i for i in range(len(newsgroups_train.data)) if i in textdict]
test_ids = [i for i in range(len(newsgroups_train.data), len(textdict)) if i in textdict]
print("%i training and %i test samples" % (len(train_ids), len(test_ids)))
# transform into tf-idf features
ft = FeatureTransform(norm='max', weight=True, renorm='max')
docfeats = ft.texts2features(textdict, fit_ids=train_ids)
# organize in feature matrix
X, featurenames = features2mat(docfeats, train_ids)
X_test, _ = features2mat(docfeats, test_ids, featurenames)
print("%i features" % len(featurenames))
targets = np.hstack([newsgroups_train.target,newsgroups_test.target])
y = targets[train_ids]
y_test = targets[test_ids]
target_names = newsgroups_train.target_names
n_targets = 1000
# linear kPCA
kpca = KernelPCA(n_components=2, kernel='linear')
X_embed = kpca.fit_transform(X)
X_embed_test = kpca.transform(X_test)
plot_20news(X_embed, y, target_names, X_embed_test, y_test,
title='20newsgroups - linear Kernel PCA', legend=True)
# compute linear kernel and center
K_lin = center_K(X.dot(X.T).A)
K_lin_test = center_K(X_test.dot(X_test.T).A)
print("similarity approximation : msqe: %.10f ; r^2: %.10f ; rho: %.10f" % check_similarity_match(X_embed, K_lin))
print("similarity approximation (test): msqe: %.10f ; r^2: %.10f ; rho: %.10f" % check_similarity_match(X_embed_test, K_lin_test))
# project to 2d with linear similarity encoder
# careful: our input is sparse!!!
simec = SimilarityEncoder(X.shape[1], 2, n_targets, sparse_inputs=True, opt=keras.optimizers.SGD(lr=50.))
# train the model to get an embedding with which the target similarities
# can be linearly approximated
simec.fit(X, K_lin[:,:n_targets], epochs=25)
# get the embeddings
X_embeds = simec.transform(X)
X_embed_tests = simec.transform(X_test)
plot_20news(X_embeds, y, target_names, X_embed_tests, y_test,
title='20 newsgroups - SimEc (lin. kernel, linear)', legend=True)
print("correlation with lin kPCA : %f" % check_embed_match(X_embed, X_embeds)[1])
print("correlation with lin kPCA (test): %f" % check_embed_match(X_embed_test, X_embed_tests)[1])
print("similarity approximation : msqe: %.10f ; r^2: %.10f ; rho: %.10f" % check_similarity_match(X_embeds, K_lin))
print("similarity approximation (test): msqe: %.10f ; r^2: %.10f ; rho: %.10f" % check_similarity_match(X_embed_tests, K_lin_test))
```
|
github_jupyter
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.