text
stringlengths 2.5k
6.39M
| kind
stringclasses 3
values |
|---|---|
** ----- IMPORTANT ------ **
The code presented here assumes that you're running TensorFlow v1.3.0 or higher, this was not released yet so the easiet way to run this is update your TensorFlow version to TensorFlow's master.
To do that go [here](https://github.com/tensorflow/tensorflow#installation) and then execute:
`pip install --ignore-installed --upgrade <URL for the right binary for your machine>`.
For example, considering a Linux CPU-only running python2:
`pip install --upgrade https://ci.tensorflow.org/view/Nightly/job/nightly-matrix-cpu/TF_BUILD_IS_OPT=OPT,TF_BUILD_IS_PIP=PIP,TF_BUILD_PYTHON_VERSION=PYTHON2,label=cpu-slave/lastSuccessfulBuild/artifact/pip_test/whl/tensorflow-1.2.1-cp27-none-linux_x86_64.whl`
## Here is walk-through to help getting started with tensorflow
1) Simple Linear Regression with low-level TensorFlow
2) Simple Linear Regression with a canned estimator
3) Playing with real data: linear regressor and DNN
4) Building a custom estimator to classify handwritten digits (MNIST)
### [What's next?](https://goo.gl/hZaLPA)
## Dependencies
```
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import collections
# tensorflow
import tensorflow as tf
print('Expected TensorFlow version is v1.3.0 or higher')
print('Your TensorFlow version:', tf.__version__)
# data manipulation
import numpy as np
import pandas as pd
# visualization
import matplotlib
import matplotlib.pyplot as plt
%matplotlib inline
matplotlib.rcParams['figure.figsize'] = [12,8]
```
## 1) Simple Linear Regression with low-level TensorFlow
### Generating data
This function creates a noisy dataset that's roughly linear, according to the equation y = mx + b + noise.
Notice that the expected value for m is 0.1 and for b is 0.3. This is the values we expect the model to predict.
```
def make_noisy_data(m=0.1, b=0.3, n=100):
x = np.random.randn(n)
noise = np.random.normal(scale=0.01, size=len(x))
y = m * x + b + noise
return x, y
```
Create training data
```
x_train, y_train = make_noisy_data()
```
Plot the training data
```
plt.plot(x_train, y_train, 'b.')
```
### The Model
```
# input and output
x = tf.placeholder(shape=[None], dtype=tf.float32, name='x')
y_label = tf.placeholder(shape=[None], dtype=tf.float32, name='y_label')
# variables
W = tf.Variable(tf.random_normal([1], name="W")) # weight
b = tf.Variable(tf.random_normal([1], name="b")) # bias
# actual model
y = W * x + b
```
### The Loss and Optimizer
Define a loss function (here, squared error) and an optimizer (here, gradient descent).
```
loss = tf.reduce_mean(tf.square(y - y_label))
optimizer = tf.train.GradientDescentOptimizer(learning_rate=0.1)
train = optimizer.minimize(loss)
```
### The Training Loop and generating predictions
```
init = tf.global_variables_initializer()
with tf.Session() as sess:
sess.run(init) # initialize variables
for i in range(100): # train for 100 steps
sess.run(train, feed_dict={x: x_train, y_label:y_train})
x_plot = np.linspace(-3, 3, 101) # return evenly spaced numbers over a specified interval
# using the trained model to predict values for the training data
y_plot = sess.run(y, feed_dict={x: x_plot})
# saving final weight and bias
final_W = sess.run(W)
final_b = sess.run(b)
```
### Visualizing predictions
```
plt.scatter(x_train, y_train)
plt.plot(x_plot, y_plot, 'g')
```
### What is the final weight and bias?
```
print('W:', final_W, 'expected: 0.1')
print('b:', final_b, 'expected: 0.3')
```
## 2) Simple Linear Regression with a canned estimator
### Input Pipeline
```
x_dict = {'x': x_train}
train_input = tf.estimator.inputs.numpy_input_fn(x_dict, y_train,
shuffle=True,
num_epochs=None) # repeat forever
```
### Describe input feature usage
```
features = [tf.feature_column.numeric_column('x')] # because x is a real number
```
### Build and train the model
```
estimator = tf.estimator.LinearRegressor(features)
estimator.train(train_input, steps = 1000)
```
### Generating and visualizing predictions
```
x_test_dict = {'x': np.linspace(-5, 5, 11)}
data_source = tf.estimator.inputs.numpy_input_fn(x_test_dict, shuffle=False)
predictions = list(estimator.predict(data_source))
preds = [p['predictions'][0] for p in predictions]
for y in predictions:
print(y['predictions'])
plt.scatter(x_train, y_train)
plt.plot(x_test_dict['x'], preds, 'g')
```
## 3) Playing with real data: linear regressor and DNN
### Get the data
The Adult dataset is from the Census bureau and the task is to predict whether a given adult makes more than $50,000 a year based attributes such as education, hours of work per week, etc.
But the code here presented can be easilly aplicable to any csv dataset that fits in memory.
More about the data [here](https://archive.ics.uci.edu/ml/machine-learning-databases/adult/old.adult.names)
```
census_train_url = 'https://archive.ics.uci.edu/ml/machine-learning-databases/adult/adult.data'
census_train_path = tf.contrib.keras.utils.get_file('census.train', census_train_url)
census_test_url = 'https://archive.ics.uci.edu/ml/machine-learning-databases/adult/adult.test'
census_test_path = tf.contrib.keras.utils.get_file('census.test', census_test_url)
```
### Load the data
```
column_names = [
'age', 'workclass', 'fnlwgt', 'education', 'education-num',
'marital-status', 'occupation', 'relationship', 'race', 'sex',
'capital-gain', 'capital-loss', 'hours-per-week', 'native-country',
'income'
]
census_train = pd.read_csv(census_train_path, index_col=False, names=column_names)
census_test = pd.read_csv(census_train_path, index_col=False, names=column_names)
census_train_label = census_train.pop('income') == " >50K"
census_test_label = census_test.pop('income') == " >50K"
census_train.head(10)
census_train_label[:20]
```
### Input pipeline
```
train_input = tf.estimator.inputs.pandas_input_fn(
census_train,
census_train_label,
shuffle=True,
batch_size = 32, # process 32 examples at a time
num_epochs=None,
)
test_input = tf.estimator.inputs.pandas_input_fn(
census_test,
census_test_label,
shuffle=True,
num_epochs=1)
features, labels = train_input()
features
```
### Feature description
```
features = [
tf.feature_column.numeric_column('hours-per-week'),
tf.feature_column.bucketized_column(tf.feature_column.numeric_column('education-num'), list(range(25))),
tf.feature_column.categorical_column_with_vocabulary_list('sex', ['male','female']),
tf.feature_column.categorical_column_with_hash_bucket('native-country', 1000),
]
estimator = tf.estimator.LinearClassifier(features, model_dir='census/linear',n_classes=2)
estimator.train(train_input, steps=5000)
```
### Evaluate the model
```
estimator.evaluate(test_input)
```
## DNN model
### Update input pre-processing
```
features = [
tf.feature_column.numeric_column('education-num'),
tf.feature_column.numeric_column('hours-per-week'),
tf.feature_column.numeric_column('age'),
tf.feature_column.indicator_column(
tf.feature_column.categorical_column_with_vocabulary_list('sex',['male','female'])),
tf.feature_column.embedding_column( # now using embedding!
tf.feature_column.categorical_column_with_hash_bucket('native-country', 1000), 10)
]
estimator = tf.estimator.DNNClassifier(hidden_units=[20,20],
feature_columns=features,
n_classes=2,
model_dir='census/dnn')
estimator.train(train_input, steps=5000)
estimator.evaluate(test_input)
```
## Custom Input Pipeline using Datasets API
### Read the data
```
def census_input_fn(path):
def input_fn():
dataset = (
tf.contrib.data.TextLineDataset(path)
.map(csv_decoder)
.shuffle(buffer_size=100)
.batch(32)
.repeat())
columns = dataset.make_one_shot_iterator().get_next()
income = tf.equal(columns.pop('income')," >50K")
return columns, income
return input_fn
csv_defaults = collections.OrderedDict([
('age',[0]),
('workclass',['']),
('fnlwgt',[0]),
('education',['']),
('education-num',[0]),
('marital-status',['']),
('occupation',['']),
('relationship',['']),
('race',['']),
('sex',['']),
('capital-gain',[0]),
('capital-loss',[0]),
('hours-per-week',[0]),
('native-country',['']),
('income',['']),
])
def csv_decoder(line):
parsed = tf.decode_csv(line, csv_defaults.values())
return dict(zip(csv_defaults.keys(), parsed))
```
### Try the input function
```
tf.reset_default_graph()
census_input = census_input_fn(census_train_path)
training_batch = census_input()
with tf.Session() as sess:
features, high_income = sess.run(training_batch)
print(features['education'])
print(features['age'])
print(high_income)
```
## 4) Building a custom estimator to classify handwritten digits (MNIST)
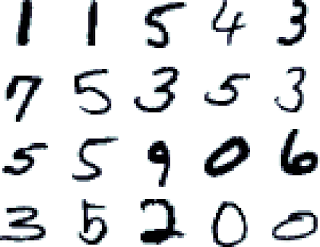
Image from: http://rodrigob.github.io/are_we_there_yet/build/images/mnist.png?1363085077
```
train,test = tf.contrib.keras.datasets.mnist.load_data()
x_train,y_train = train
x_test,y_test = test
mnist_train_input = tf.estimator.inputs.numpy_input_fn({'x':np.array(x_train, dtype=np.float32)},
np.array(y_train,dtype=np.int32),
shuffle=True,
num_epochs=None)
mnist_test_input = tf.estimator.inputs.numpy_input_fn({'x':np.array(x_test, dtype=np.float32)},
np.array(y_test,dtype=np.int32),
shuffle=True,
num_epochs=1)
```
### tf.estimator.LinearClassifier
```
estimator = tf.estimator.LinearClassifier([tf.feature_column.numeric_column('x',shape=784)],
n_classes=10,
model_dir="mnist/linear")
estimator.train(mnist_train_input, steps = 10000)
estimator.evaluate(mnist_test_input)
```
### Examine the results with [TensorBoard](http://0.0.0.0:6006)
$> tensorboard --logdir mnnist/DNN
```
estimator = tf.estimator.DNNClassifier(hidden_units=[256],
feature_columns=[tf.feature_column.numeric_column('x',shape=784)],
n_classes=10,
model_dir="mnist/DNN")
estimator.train(mnist_train_input, steps = 10000)
estimator.evaluate(mnist_test_input)
# Parameters
BATCH_SIZE = 128
STEPS = 10000
```
## A Custom Model
```
def build_cnn(input_layer, mode):
with tf.name_scope("conv1"):
conv1 = tf.layers.conv2d(inputs=input_layer,filters=32, kernel_size=[5, 5],
padding='same', activation=tf.nn.relu)
with tf.name_scope("pool1"):
pool1 = tf.layers.max_pooling2d(inputs=conv1, pool_size=[2, 2], strides=2)
with tf.name_scope("conv2"):
conv2 = tf.layers.conv2d(inputs=pool1,filters=64, kernel_size=[5, 5],
padding='same', activation=tf.nn.relu)
with tf.name_scope("pool2"):
pool2 = tf.layers.max_pooling2d(inputs=conv2, pool_size=[2, 2], strides=2)
with tf.name_scope("dense"):
pool2_flat = tf.reshape(pool2, [-1, 7 * 7 * 64])
dense = tf.layers.dense(inputs=pool2_flat, units=1024, activation=tf.nn.relu)
with tf.name_scope("dropout"):
is_training_mode = mode == tf.estimator.ModeKeys.TRAIN
dropout = tf.layers.dropout(inputs=dense, rate=0.4, training=is_training_mode)
logits = tf.layers.dense(inputs=dropout, units=10)
return logits
def model_fn(features, labels, mode):
# Describing the model
input_layer = tf.reshape(features['x'], [-1, 28, 28, 1])
tf.summary.image('mnist_input',input_layer)
logits = build_cnn(input_layer, mode)
# Generate Predictions
classes = tf.argmax(input=logits, axis=1)
predictions = {
'classes': classes,
'probabilities': tf.nn.softmax(logits, name='softmax_tensor')
}
if mode == tf.estimator.ModeKeys.PREDICT:
# Return an EstimatorSpec object
return tf.estimator.EstimatorSpec(mode=mode, predictions=predictions)
with tf.name_scope('loss'):
loss = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=labels, logits=logits)
loss = tf.reduce_sum(loss)
tf.summary.scalar('loss', loss)
with tf.name_scope('accuracy'):
accuracy = tf.cast(tf.equal(tf.cast(classes,tf.int32),labels),tf.float32)
accuracy = tf.reduce_mean(accuracy)
tf.summary.scalar('accuracy', accuracy)
# Configure the Training Op (for TRAIN mode)
if mode == tf.estimator.ModeKeys.TRAIN:
train_op = tf.contrib.layers.optimize_loss(
loss=loss,
global_step=tf.train.get_global_step(),
learning_rate=1e-4,
optimizer='Adam')
return tf.estimator.EstimatorSpec(mode=mode, predictions=predictions,
loss=loss, train_op=train_op)
# Configure the accuracy metric for evaluation
eval_metric_ops = {
'accuracy': tf.metrics.accuracy(
classes,
input=labels)
}
return tf.estimator.EstimatorSpec(mode=mode, predictions=predictions,
loss=loss, eval_metric_ops=eval_metric_ops)
```
## Runs estimator
```
# create estimator
run_config = tf.contrib.learn.RunConfig(model_dir='mnist/CNN')
estimator = tf.estimator.Estimator(model_fn=model_fn, config=run_config)
# train for 10000 steps
estimator.train(input_fn=mnist_train_input, steps=10000)
# evaluate
estimator.evaluate(input_fn=mnist_test_input)
# predict
preds = estimator.predict(input_fn=test_input_fn)
```
## Distributed tensorflow: using experiments
```
# Run an experiment
from tensorflow.contrib.learn.python.learn import learn_runner
# Enable TensorFlow logs
tf.logging.set_verbosity(tf.logging.INFO)
# create experiment
def experiment_fn(run_config, hparams):
# create estimator
estimator = tf.estimator.Estimator(model_fn=model_fn,
config=run_config)
return tf.contrib.learn.Experiment(
estimator,
train_input_fn=train_input_fn,
eval_input_fn=test_input_fn,
train_steps=STEPS
)
# run experiment
learn_runner.run(experiment_fn,
run_config=run_config)
```
### Examine the results with [TensorBoard](http://0.0.0.0:6006)
$> tensorboard --logdir mnist/CNN
|
github_jupyter
|
```
from autoreduce import *
import numpy as np
from sympy import symbols
# Post conservation law and other approximations phenomenological model at the RNA level
n = 4 # Number of states
nouts = 2 # Number of outputs
# Inputs by user
x_init = np.zeros(n)
n = 4 # Number of states
timepoints_ode = np.linspace(0, 100, 100)
C = [[0, 0, 1, 0], [0, 0, 0, 1]]
nstates_tol = 3
error_tol = 0.3
# System dynamics symbolically
# params = [100, 50, 10, 5, 5, 0.02, 0.02, 0.01, 0.01]
# params = [1, 1, 5, 0.1, 0.2, 1, 1, 100, 100] # Parameter set for which reduction doesn't work
# K,b_t,b_l,d_t,d_l,del_t,del_l,beta_t,beta_l = params
x0 = symbols('x0')
x1 = symbols('x1')
x2 = symbols('x2')
x3 = symbols('x3')
x = [x0, x1, x2, x3]
K = symbols('K')
b_t = symbols('b_t')
b_l = symbols('b_l')
d_t = symbols('d_t')
d_l = symbols('d_l')
del_t = symbols('del_t')
del_l = symbols('del_l')
beta_t = symbols('beta_t')
beta_l = symbols('beta_l')
params = [K,b_t,b_l,d_t,d_l,del_t,del_l,beta_t,beta_l]
f0 = K * b_t**2/(b_t**2 + x[3]**2) - d_t * x[0]
f1 = K * b_l**2/(b_l**2 + x[2]**2) - d_l * x[1]
f2 = beta_t * x[0] - del_t * x[2]
f3 = beta_l * x[1] - del_l * x[3]
f = [f0,f1,f2,f3]
# parameter values
params_values = [100, 50, 10, 5, 5, 0.02, 0.02, 0.01, 0.01]
sys = System(x, f, params = params, params_values = params_values, C = C, x_init = x_init)
from autoreduce.utils import get_ODE
sys_ode = get_ODE(sys, timepoints_ode)
sol = sys_ode.solve_system().T
try:
import matplotlib.pyplot as plt
plt.plot(timepoints_ode, np.transpose(np.array(C)@sol))
plt.xlabel('Time')
plt.ylabel('[Outputs]')
plt.show()
except:
print('Plotting libraries missing.')
from autoreduce.utils import get_SSM
timepoints_ssm = np.linspace(0,100,100)
sys_ssm = get_SSM(sys, timepoints_ssm)
Ss = sys_ssm.compute_SSM() # len(timepoints) x len(params) x len(states)
out_Ss = []
for i in range(len(params)):
out_Ss.append((np.array(C)@(Ss[:,i,:].T)))
out_Ss = np.reshape(np.array(out_Ss), (len(timepoints_ssm), len(params), nouts))
try:
import seaborn as sn
import matplotlib.pyplot as plt
for j in range(nouts):
sn.heatmap(out_Ss[:,:,j].T)
plt.xlabel('Time')
plt.ylabel('Parameters')
plt.title('Sensitivity of output[{0}] with respect to all parameters'.format(j))
plt.show()
except:
print('Plotting libraries missing.')
from autoreduce.utils import get_reducible
timepoints_ssm = np.linspace(0,100,10)
timepoints_ode = np.linspace(0, 100, 100)
sys_reduce = get_reducible(sys, timepoints_ode, timepoints_ssm)
results = sys_reduce.reduce_simple()
list(results.keys())[0].f[1]
reduced_system, collapsed_system = sys_reduce.solve_timescale_separation([x0,x1], fast_states = [x3, x2])
reduced_system.f[1]
```
|
github_jupyter
|
# Understanding the FFT Algorithm
Copy from http://jakevdp.github.io/blog/2013/08/28/understanding-the-fft/
*This notebook first appeared as a post by Jake Vanderplas on [Pythonic Perambulations](http://jakevdp.github.io/blog/2013/08/28/understanding-the-fft/). The notebook content is BSD-licensed.*
<!-- PELICAN_BEGIN_SUMMARY -->
The Fast Fourier Transform (FFT) is one of the most important algorithms in signal processing and data analysis. I've used it for years, but having no formal computer science background, It occurred to me this week that I've never thought to ask *how* the FFT computes the discrete Fourier transform so quickly. I dusted off an old algorithms book and looked into it, and enjoyed reading about the deceptively simple computational trick that JW Cooley and John Tukey outlined in their classic [1965 paper](http://www.ams.org/journals/mcom/1965-19-090/S0025-5718-1965-0178586-1/) introducing the subject.
The goal of this post is to dive into the Cooley-Tukey FFT algorithm, explaining the symmetries that lead to it, and to show some straightforward Python implementations putting the theory into practice. My hope is that this exploration will give data scientists like myself a more complete picture of what's going on in the background of the algorithms we use.
<!-- PELICAN_END_SUMMARY -->
## The Discrete Fourier Transform
The FFT is a fast, $\mathcal{O}[N\log N]$ algorithm to compute the Discrete Fourier Transform (DFT), which
naively is an $\mathcal{O}[N^2]$ computation. The DFT, like the more familiar continuous version of the Fourier transform, has a forward and inverse form which are defined as follows:
**Forward Discrete Fourier Transform (DFT):**
$$X_k = \sum_{n=0}^{N-1} x_n \cdot e^{-i~2\pi~k~n~/~N}$$
**Inverse Discrete Fourier Transform (IDFT):**
$$x_n = \frac{1}{N}\sum_{k=0}^{N-1} X_k e^{i~2\pi~k~n~/~N}$$
The transformation from $x_n \to X_k$ is a translation from configuration space to frequency space, and can be very useful in both exploring the power spectrum of a signal, and also for transforming certain problems for more efficient computation. For some examples of this in action, you can check out Chapter 10 of our upcoming Astronomy/Statistics book, with figures and Python source code available [here](http://www.astroml.org/book_figures/chapter10/). For an example of the FFT being used to simplify an otherwise difficult differential equation integration, see my post on [Solving the Schrodinger Equation in Python](http://jakevdp.github.io/blog/2012/09/05/quantum-python/).
Because of the importance of the FFT in so many fields, Python contains many standard tools and wrappers to compute this. Both NumPy and SciPy have wrappers of the extremely well-tested FFTPACK library, found in the submodules ``numpy.fft`` and ``scipy.fftpack`` respectively. The fastest FFT I am aware of is in the [FFTW](http://www.fftw.org/) package, which is also available in Python via the [PyFFTW](https://pypi.python.org/pypi/pyFFTW) package.
For the moment, though, let's leave these implementations aside and ask how we might compute the FFT in Python from scratch.
## Computing the Discrete Fourier Transform
For simplicity, we'll concern ourself only with the forward transform, as the inverse transform can be implemented in a very similar manner. Taking a look at the DFT expression above, we see that it is nothing more than a straightforward linear operation: a matrix-vector multiplication of $\vec{x}$,
$$\vec{X} = M \cdot \vec{x}$$
with the matrix $M$ given by
$$M_{kn} = e^{-i~2\pi~k~n~/~N}.$$
With this in mind, we can compute the DFT using simple matrix multiplication as follows:
```
import numpy as np
def DFT_slow(x):
"""Compute the discrete Fourier Transform of the 1D array x"""
x = np.asarray(x, dtype=float)
N = x.shape[0]
n = np.arange(N)
k = n.reshape((N, 1))
M = np.exp(-2j * np.pi * k * n / N)
return np.dot(M, x)
```
We can double-check the result by comparing to numpy's built-in FFT function:
```
x = np.random.random(1024)
np.allclose(DFT_slow(x), np.fft.fft(x))
```
Just to confirm the sluggishness of our algorithm, we can compare the execution times
of these two approaches:
```
%timeit DFT_slow(x)
%timeit np.fft.fft(x)
```
We are over 1000 times slower, which is to be expected for such a simplistic implementation. But that's not the worst of it. For an input vector of length $N$, the FFT algorithm scales as $\mathcal{O}[N\log N]$, while our slow algorithm scales as $\mathcal{O}[N^2]$. That means that for $N=10^6$ elements, we'd expect the FFT to complete in somewhere around 50 ms, while our slow algorithm would take nearly 20 hours!
So how does the FFT accomplish this speedup? The answer lies in exploiting symmetry.
## Symmetries in the Discrete Fourier Transform
One of the most important tools in the belt of an algorithm-builder is to exploit symmetries of a problem. If you can show analytically that one piece of a problem is simply related to another, you can compute the subresult
only once and save that computational cost. Cooley and Tukey used exactly this approach in deriving the FFT.
We'll start by asking what the value of $X_{N+k}$ is. From our above expression:
$$
\begin{align*}
X_{N + k} &= \sum_{n=0}^{N-1} x_n \cdot e^{-i~2\pi~(N + k)~n~/~N}\\
&= \sum_{n=0}^{N-1} x_n \cdot e^{- i~2\pi~n} \cdot e^{-i~2\pi~k~n~/~N}\\
&= \sum_{n=0}^{N-1} x_n \cdot e^{-i~2\pi~k~n~/~N}
\end{align*}
$$
where we've used the identity $\exp[2\pi~i~n] = 1$ which holds for any integer $n$.
The last line shows a nice symmetry property of the DFT:
$$X_{N+k} = X_k.$$
By a simple extension,
$$X_{k + i \cdot N} = X_k$$
for any integer $i$. As we'll see below, this symmetry can be exploited to compute the DFT much more quickly.
## DFT to FFT: Exploiting Symmetry
Cooley and Tukey showed that it's possible to divide the DFT computation into two smaller parts. From
the definition of the DFT we have:
$$
\begin{align}
X_k &= \sum_{n=0}^{N-1} x_n \cdot e^{-i~2\pi~k~n~/~N} \\
&= \sum_{m=0}^{N/2 - 1} x_{2m} \cdot e^{-i~2\pi~k~(2m)~/~N} + \sum_{m=0}^{N/2 - 1} x_{2m + 1} \cdot e^{-i~2\pi~k~(2m + 1)~/~N} \\
&= \sum_{m=0}^{N/2 - 1} x_{2m} \cdot e^{-i~2\pi~k~m~/~(N/2)} + e^{-i~2\pi~k~/~N} \sum_{m=0}^{N/2 - 1} x_{2m + 1} \cdot e^{-i~2\pi~k~m~/~(N/2)}
\end{align}
$$
We've split the single Discrete Fourier transform into two terms which themselves look very similar to smaller Discrete Fourier Transforms, one on the odd-numbered values, and one on the even-numbered values. So far, however, we haven't saved any computational cycles. Each term consists of $(N/2)*N$ computations, for a total of $N^2$.
The trick comes in making use of symmetries in each of these terms. Because the range of $k$ is $0 \le k < N$, while the range of $n$ is $0 \le n < M \equiv N/2$, we see from the symmetry properties above that we need only perform half the computations for each sub-problem. Our $\mathcal{O}[N^2]$ computation has become $\mathcal{O}[M^2]$, with $M$ half the size of $N$.
But there's no reason to stop there: as long as our smaller Fourier transforms have an even-valued $M$, we can reapply this divide-and-conquer approach, halving the computational cost each time, until our arrays are small enough that the strategy is no longer beneficial. In the asymptotic limit, this recursive approach scales as $\mathcal{O}[N\log N]$.
This recursive algorithm can be implemented very quickly in Python, falling-back on our slow DFT code when the size of the sub-problem becomes suitably small:
```
def FFT(x):
"""A recursive implementation of the 1D Cooley-Tukey FFT"""
x = np.asarray(x, dtype=float)
N = x.shape[0]
if N % 2 > 0:
raise ValueError("size of x must be a power of 2")
elif N <= 32: # this cutoff should be optimized
return DFT_slow(x)
else:
X_even = FFT(x[::2])
X_odd = FFT(x[1::2])
factor = np.exp(-2j * np.pi * np.arange(N) / N)
return np.concatenate([X_even + factor[:N / 2] * X_odd,
X_even + factor[N / 2:] * X_odd])
```
Here we'll do a quick check that our algorithm produces the correct result:
```
x = np.random.random(1024)
np.allclose(FFT(x), np.fft.fft(x))
```
And we'll time this algorithm against our slow version:
```
%timeit DFT_slow(x)
%timeit FFT(x)
%timeit np.fft.fft(x)
```
Our calculation is faster than the naive version by over an order of magnitude! What's more, our recursive algorithm is asymptotically $\mathcal{O}[N\log N]$: we've implemented the Fast Fourier Transform.
Note that we still haven't come close to the speed of the built-in FFT algorithm in numpy, and this is to be expected. The FFTPACK algorithm behind numpy's ``fft`` is a Fortran implementation which has received years of tweaks and optimizations. Furthermore, our NumPy solution involves both Python-stack recursions and the allocation of many temporary arrays, which adds significant computation time.
A good strategy to speed up code when working with Python/NumPy is to vectorize repeated computations where possible. We can do this, and in the process remove our recursive function calls, and make our Python FFT even more efficient.
## Vectorized Numpy Version
Notice that in the above recursive FFT implementation, at the lowest recursion level we perform $N~/~32$ identical matrix-vector products. The efficiency of our algorithm would benefit by computing these matrix-vector products all at once as a single matrix-matrix product. At each subsequent level of recursion, we also perform duplicate operations which can be vectorized. NumPy excels at this sort of operation, and we can make use of that fact to create this vectorized version of the Fast Fourier Transform:
```
def FFT_vectorized(x):
"""A vectorized, non-recursive version of the Cooley-Tukey FFT"""
x = np.asarray(x, dtype=float)
N = x.shape[0]
if np.log2(N) % 1 > 0:
raise ValueError("size of x must be a power of 2")
# N_min here is equivalent to the stopping condition above,
# and should be a power of 2
N_min = min(N, 32)
# Perform an O[N^2] DFT on all length-N_min sub-problems at once
n = np.arange(N_min)
k = n[:, None]
M = np.exp(-2j * np.pi * n * k / N_min)
X = np.dot(M, x.reshape((N_min, -1)))
# build-up each level of the recursive calculation all at once
while X.shape[0] < N:
X_even = X[:, :X.shape[1] / 2]
X_odd = X[:, X.shape[1] / 2:]
factor = np.exp(-1j * np.pi * np.arange(X.shape[0])
/ X.shape[0])[:, None]
X = np.vstack([X_even + factor * X_odd,
X_even - factor * X_odd])
return X.ravel()
```
Though the algorithm is a bit more opaque, it is simply a rearrangement of the operations used in the recursive version with one exception: we exploit a symmetry in the ``factor`` computation and construct only half of the array. Again, we'll confirm that our function yields the correct result:
```
x = np.random.random(1024)
np.allclose(FFT_vectorized(x), np.fft.fft(x))
```
Because our algorithms are becoming much more efficient, we can use a larger array to compare the timings,
leaving out ``DFT_slow``:
```
x = np.random.random(1024 * 16)
%timeit FFT(x)
%timeit FFT_vectorized(x)
%timeit np.fft.fft(x)
```
We've improved our implementation by another order of magnitude! We're now within about a factor of 10 of the FFTPACK benchmark, using only a couple dozen lines of pure Python + NumPy. Though it's still no match computationally speaking, readibility-wise the Python version is far superior to the FFTPACK source, which you can browse [here](http://www.netlib.org/fftpack/fft.c).
So how does FFTPACK attain this last bit of speedup? Well, mainly it's just a matter of detailed bookkeeping. FFTPACK spends a lot of time making sure to reuse any sub-computation that can be reused. Our numpy version still involves an excess of memory allocation and copying; in a low-level language like Fortran it's easier to control and minimize memory use. In addition, the Cooley-Tukey algorithm can be extended to use splits of size other than 2 (what we've implemented here is known as the *radix-2* Cooley-Tukey FFT). Also, other more sophisticated FFT algorithms may be used, including fundamentally distinct approaches based on convolutions (see, e.g. Bluestein's algorithm and Rader's algorithm). The combination of the above extensions and techniques can lead to very fast FFTs even on arrays whose size is not a power of two.
Though the pure-Python functions are probably not useful in practice, I hope they've provided a bit of an intuition into what's going on in the background of FFT-based data analysis. As data scientists, we can make-do with black-box implementations of fundamental tools constructed by our more algorithmically-minded colleagues, but I am a firm believer that the more understanding we have about the low-level algorithms we're applying to our data, the better practitioners we'll be.
*This blog post was written entirely in the IPython Notebook. The full notebook can be downloaded
[here](http://jakevdp.github.io/downloads/notebooks/UnderstandingTheFFT.ipynb),
or viewed statically
[here](http://nbviewer.ipython.org/url/jakevdp.github.io/downloads/notebooks/UnderstandingTheFFT.ipynb).*
|
github_jupyter
|
```
# default_exp label
```
# Label
> A collection of functions to do label-based quantification
```
#hide
from nbdev.showdoc import *
```
## Label search
The label search is implemented based on the compare_frags from the search.
We have a fixed number of reporter channels and check if we find a respective peak within the search tolerance.
Useful resources:
- [IsobaricAnalyzer](https://abibuilder.informatik.uni-tuebingen.de/archive/openms/Documentation/nightly/html/TOPP_IsobaricAnalyzer.html)
- [TMT Talk from Hupo 2015](https://assets.thermofisher.com/TFS-Assets/CMD/Reference-Materials/PP-TMT-Multiplexed-Protein-Quantification-HUPO2015-EN.pdf)
```
#export
from numba import njit
from alphapept.search import compare_frags
import numpy as np
@njit
def label_search(query_frag: np.ndarray, query_int: np.ndarray, label: np.ndarray, reporter_frag_tol:float, ppm:bool)-> (np.ndarray, np.ndarray):
"""Function to search for a label for a given spectrum.
Args:
query_frag (np.ndarray): Array with query fragments.
query_int (np.ndarray): Array with query intensities.
label (np.ndarray): Array with label masses.
reporter_frag_tol (float): Fragment tolerance for search.
ppm (bool): Flag to use ppm instead of Dalton.
Returns:
np.ndarray: Array with intensities for the respective label channel.
np.ndarray: Array with offset masses.
"""
report = np.zeros(len(label))
off_mass = np.zeros_like(label)
hits = compare_frags(query_frag, label, reporter_frag_tol, ppm)
for idx, _ in enumerate(hits):
if _ > 0:
report[idx] = query_int[_-1]
off_mass[idx] = query_frag[_-1] - label[idx]
if ppm:
off_mass[idx] = off_mass[idx] / (query_frag[_-1] + label[idx]) *2 * 1e6
return report, off_mass
def test_label_search():
query_frag = np.array([1,2,3,4,5])
query_int = np.array([1,2,3,4,5])
label = np.array([1.0, 2.0, 3.0, 4.0, 5.0])
frag_tolerance = 0.1
ppm= False
assert np.allclose(label_search(query_frag, query_int, label, frag_tolerance, ppm)[0], query_int)
query_frag = np.array([1,2,3,4,6])
query_int = np.array([1,2,3,4,5])
assert np.allclose(label_search(query_frag, query_int, label, frag_tolerance, ppm)[0], np.array([1,2,3,4,0]))
query_frag = np.array([1,2,3,4,6])
query_int = np.array([5,4,3,2,1])
assert np.allclose(label_search(query_frag, query_int, label, frag_tolerance, ppm)[0], np.array([5,4,3,2,0]))
query_frag = np.array([1.1, 2.2, 3.3, 4.4, 6.6])
query_int = np.array([1,2,3,4,5])
frag_tolerance = 0.5
ppm= False
assert np.allclose(label_search(query_frag, query_int, label, frag_tolerance, ppm)[1], np.array([0.1, 0.2, 0.3, 0.4, 0.0]))
test_label_search()
#Example usage
query_frag = np.array([127, 128, 129.1, 132])
query_int = np.array([100, 200, 300, 400, 500])
label = np.array([127.0, 128.0, 129.0, 130.0])
frag_tolerance = 0.1
ppm = False
report, offset = label_search(query_frag, query_int, label, frag_tolerance, ppm)
print(f'Reported intensities {report}, Offset {offset}')
```
## MS2 Search
```
#export
from typing import NamedTuple
import alphapept.io
def search_label_on_ms_file(file_name:str, label:NamedTuple, reporter_frag_tol:float, ppm:bool):
"""Wrapper function to search labels on an ms_file and write results to the peptide_fdr of the file.
Args:
file_name (str): Path to ms_file:
label (NamedTuple): Label with channels, mod_name and masses.
reporter_frag_tol (float): Fragment tolerance for search.
ppm (bool): Flag to use ppm instead of Dalton.
"""
ms_file = alphapept.io.MS_Data_File(file_name, is_read_only = False)
df = ms_file.read(dataset_name='peptide_fdr')
label_intensities = np.zeros((len(df), len(label.channels)))
off_masses = np.zeros((len(df), len(label.channels)))
labeled = df['sequence'].str.startswith(label.mod_name).values
query_data = ms_file.read_DDA_query_data()
query_indices = query_data["indices_ms2"]
query_frags = query_data['mass_list_ms2']
query_ints = query_data['int_list_ms2']
for idx, query_idx in enumerate(df['raw_idx']):
query_idx_start = query_indices[query_idx]
query_idx_end = query_indices[query_idx + 1]
query_frag = query_frags[query_idx_start:query_idx_end]
query_int = query_ints[query_idx_start:query_idx_end]
query_frag_idx = query_frag < label.masses[-1]+1
query_frag = query_frag[query_frag_idx]
query_int = query_int[query_frag_idx]
if labeled[idx]:
label_int, off_mass = label_search(query_frag, query_int, label.masses, reporter_frag_tol, ppm)
label_intensities[idx, :] = label_int
off_masses[idx, :] = off_mass
df[label.channels] = label_intensities
df[[_+'_off_ppm' for _ in label.channels]] = off_masses
ms_file.write(df, dataset_name="peptide_fdr", overwrite=True) #Overwrite dataframe with label information
#export
import logging
import os
from alphapept.constants import label_dict
def find_labels(
to_process: dict,
callback: callable = None,
parallel:bool = False
) -> bool:
"""Wrapper function to search for labels.
Args:
to_process (dict): A dictionary with settings indicating which files are to be processed and how.
callback (callable): A function that accepts a float between 0 and 1 as progress. Defaults to None.
parallel (bool): If True, process multiple files in parallel.
This is not implemented yet!
Defaults to False.
Returns:
bool: True if and only if the label finding was succesful.
"""
index, settings = to_process
raw_file = settings['experiment']['file_paths'][index]
try:
base, ext = os.path.splitext(raw_file)
file_name = base+'.ms_data.hdf'
label = label_dict[settings['isobaric_label']['label']]
reporter_frag_tol = settings['isobaric_label']['reporter_frag_tolerance']
ppm = settings['isobaric_label']['reporter_frag_tolerance_ppm']
search_label_on_ms_file(file_name, label, reporter_frag_tol, ppm)
logging.info(f'Tag finding of file {file_name} complete.')
return True
except Exception as e:
logging.error(f'Tag finding of file {file_name} failed. Exception {e}')
return f"{e}" #Can't return exception object, cast as string
return True
#hide
from nbdev.export import *
notebook2script()
```
|
github_jupyter
|
```
import numpy as np
import pandas as pd
import warnings
warnings.filterwarnings('ignore')
import seaborn as sns
sns.set_palette('Set2')
import matplotlib.pyplot as plt
%matplotlib inline
from sklearn.metrics import confusion_matrix, mean_squared_error
from sklearn.preprocessing import LabelEncoder, MinMaxScaler, StandardScaler
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.linear_model import LinearRegression, Lasso, Ridge, SGDRegressor
from sklearn.preprocessing import KBinsDiscretizer
from sklearn.ensemble import RandomForestRegressor
from sklearn.svm import LinearSVC, SVC, LinearSVR, SVR
from sklearn.tree import DecisionTreeRegressor
from sklearn.model_selection import GridSearchCV
from scipy.stats import zscore
from sklearn.metrics import mean_squared_error
import requests
import json
from datetime import datetime
import time
import os
from pandas.tseries.holiday import USFederalHolidayCalendar as calendar
from config import yelp_api_key
from config import darksky_api_key
```
## Set Up
```
# Analysis Dates
start_date = '2017-01-01' # Start Date Inclusive
end_date = '2019-06-19' # End Date Exclusive
search_business = 'Jupiter Disco'
location = 'Brooklyn, NY'
```
## Pull Weather Data
### Latitude + Longitude from Yelp API
```
host = 'https://api.yelp.com'
path = '/v3/businesses/search'
search_limit = 10
# Yelp Authorization Header with API Key
headers = {
'Authorization': 'Bearer {}'.format(yelp_api_key)
}
# Build Requests Syntax with Yelp Host and Path and URL Paramaters
# Return JSON response
def request(host, path, url_params=None):
url_params = url_params or {}
url = '{}{}'.format(host, path)
response = requests.get(url, headers=headers, params=url_params)
return response.json()
# Build URL Params for the Request and provide the host and path
def search(term, location):
url_params = {
'term': term.replace(' ', '+'),
'location': location.replace(' ', '+'),
'limit': search_limit
}
return request(host, path, url_params=url_params)
# Return Coordinates if Exact Match Found
def yelp_lat_long(business, location):
# Call search function here with business name and location
response = search(business, location)
# Set state to 'No Match' in case no Yelp match found
state = 'No Match'
possible_matches = []
# Check search returns for match wtith business
for i in range(len(response['businesses'])):
# If match found:
if response['businesses'][i]['name'] == business:
# Local variables to help navigate JSON return
response_ = response['businesses'][0]
name_ = response_['name']
print(f'Weather Location: {name_}')
state = 'Match Found'
#print(response['businesses'][0])
return response_['coordinates']['latitude'], response_['coordinates']['longitude']
else:
# If no exact match, append all search returns to list
possible_matches.append(response['businesses'][i]['name'])
# If no match, show user potential matches
if state == 'No Match':
print('Exact match not found, did you mean one of the following? \n')
for possible_match in possible_matches:
print(possible_match)
return None, None
lat, long = yelp_lat_long(search_business, location)
#print(f'Latitude: {lat}\nLongitude: {long}')
```
### Darksky API Call
```
# Create List of Dates of target Weather Data
def find_dates(start_date, end_date):
list_of_days = []
daterange = pd.date_range(start_date, end_date)
for single_date in daterange:
list_of_days.append(single_date.strftime("%Y-%m-%d"))
return list_of_days
# Concatenate URL to make API Call
def build_url(api_key, lat, long, day):
_base_url = 'https://api.darksky.net/forecast/'
_time = 'T20:00:00'
_url = f'{_base_url}{api_key}/{lat},{long},{day + _time}?America/New_York&exclude=flags'
return _url
def make_api_call(url):
r = requests.get(url)
return r.json()
# Try / Except Helper Function for Handling JSON API Output
def find_val(dictionary, *keys):
level = dictionary
for key in keys:
try:
level = level[key]
except:
return np.NAN
return level
# Parse API Call Data using Try / Except Helper Function
def parse_data(data):
time = datetime.fromtimestamp(data['currently']['time']).strftime('%Y-%m-%d')
try:
precip_max_time = datetime.fromtimestamp(find_val(data, 'daily', 'data', 0, 'precipIntensityMaxTime')).strftime('%I:%M%p')
except:
precip_max_time = datetime(1900,1,1,5,1).strftime('%I:%M%p')
entry = {'date': time,
'temperature': float(find_val(data, 'currently', 'temperature')),
'apparent_temperature': float(find_val(data, 'currently', 'apparentTemperature')),
'humidity': float(find_val(data, 'currently', 'humidity')),
'precip_intensity_max': float(find_val(data,'daily','data', 0, 'precipIntensityMax')),
'precip_type': find_val(data, 'daily', 'data', 0, 'precipType'),
'precip_prob': float(find_val(data, 'currently', 'precipProbability')),
'pressure': float(find_val(data, 'currently', 'pressure')),
'summary': find_val(data, 'currently', 'icon'),
'precip_max_time': precip_max_time}
return entry
# Create List of Weather Data Dictionaries & Input Target Dates
def weather_call(start_date, end_date, _lat, _long):
weather = []
list_of_days = find_dates(start_date, end_date)
for day in list_of_days:
data = make_api_call(build_url(darksky_api_key, _lat, _long, day))
weather.append(parse_data(data))
return weather
result = weather_call(start_date, end_date, lat, long)
# Build DataFrame from List of Dictionaries
def build_weather_df(api_call_results):
df = pd.DataFrame(api_call_results)
# Add day of week to DataFrame + Set Index as date
df['date'] = pd.to_datetime(df['date'])
df['day_of_week'] = df['date'].dt.weekday
df['month'] = df['date'].dt.month
df.set_index('date', inplace=True)
df['apparent_temperature'].fillna(method='ffill',inplace=True)
df['temperature'].fillna(method='ffill',inplace=True)
df['humidity'].fillna(method='ffill',inplace=True)
df['precip_prob'].fillna(method='ffill', inplace=True)
df['pressure'].fillna(method='ffill', inplace=True)
df['precip_type'].fillna(value='none', inplace=True)
return df
weather_df = build_weather_df(result);
weather_df.to_csv(f'weather_{start_date}_to_{end_date}.csv')
weather_csv_file = f'weather_{start_date}_to_{end_date}.csv'
```
## Import / Clean / Prep File
```
# Import Sales Data
bar_sales_file = 'bar_x_sales_export.csv'
rest_1_file = 'rest_1_dinner_sales_w_covers_061819.csv'
# Set File
current_file = rest_1_file
weather_csv_file = 'weather_2017-01-01_to_2019-06-19.csv'
# HELPER FUNCTION
def filter_df(df, start_date, end_date):
return df[(df.index > start_date) & (df.index < end_date)]
# HELPER FUNCTION
def import_parse(file):
data = pd.read_csv(file, index_col = 'date', parse_dates=True)
df = pd.DataFrame(data)
# Rename Column to 'sales'
df = df.rename(columns={df.columns[0]: 'sales',
'dinner_covers': 'covers'})
# Drop NaN
#df = df.query('sales > 0').copy()
df.fillna(0, inplace=True)
print(f'"{file}" has been imported + parsed. The file has {len(df)} rows.')
return df
# HELPER FUNCTION
def prepare_data(current_file, weather_file):
df = filter_df(import_parse(current_file), start_date, end_date)
weather_df_csv = pd.read_csv(weather_csv_file, parse_dates=True, index_col='date')
weather_df_csv['summary'].fillna(value='none', inplace=True)
df = pd.merge(df, weather_df_csv, how='left', on='date')
return df
```
### Encode Closed Days
```
# Set Closed Dates using Sales
## REST 1 CLOSED DATES
additional_closed_dates = ['2018-12-24', '2017-12-24', '2017-02-05', '2017-03-14', '2018-01-01', '2018-02-04', '2019-02-03']
## BAR CLOSED DATES
#additional_closed_dates = ['2018-12-24', '2017-12-24', '2017-10-22']
closed_dates = [pd.to_datetime(date) for date in additional_closed_dates]
# Drop or Encode Closed Days
def encode_closed_days(df):
# CLOSED FEATURE
cal = calendar()
# Local list of days with zero sales
potential_closed_dates = df[df['sales'] == 0].index
# Enocodes closed days with 1
df['closed'] = np.where((((df.index.isin(potential_closed_dates)) & \
(df.index.isin(cal.holidays(start_date, end_date)))) | df.index.isin(closed_dates)), 1, 0)
df['sales'] = np.where(df['closed'] == 1, 0, df['sales'])
return df
baseline_df = encode_closed_days(prepare_data(current_file, weather_csv_file))
baseline_df = baseline_df[['sales', 'outside', 'day_of_week', 'month', 'closed']]
baseline_df = add_dummies(add_clusters(baseline_df))
mod_baseline = target_trend_engineering(add_cal_features(impute_outliers(baseline_df)))
mod_baseline = mod_baseline.drop(['month'], axis=1)
```
### Replace Outliers in Training Data
```
# Replace Outliers with Medians
## Targets for Outliers
z_thresh = 3
def impute_outliers(df, *col):
# Check for Outliers in Sales + Covers
for c in col:
# Impute Median for Sales & Covers Based on Day of Week Outiers
for d in df['day_of_week'].unique():
# Median / Mean / STD for each day of the week
daily_median = np.median(df[df['day_of_week'] == d][c])
daily_mean = np.mean(df[df['day_of_week'] == d][c])
daily_std = np.std(df[df['day_of_week'] ==d ][c])
# Temporary column encoded if Target Columns have an Outlier
df['temp_col'] = np.where((df['day_of_week'] == d) & (df['closed'] == 0) & ((np.abs(df[c] - daily_mean)) > (daily_std * z_thresh)), 1, 0)
# Replace Outlier with Median
df[c] = np.where(df['temp_col'] == 1, daily_median, df[c])
df = df.drop(['temp_col'], axis=1)
return df
def add_ppa(df):
df['ppa'] = np.where(df['covers'] > 0, df['sales'] / df['covers'], 0)
return df
```
## Clean File Here
```
data = add_ppa(impute_outliers(encode_closed_days(prepare_data(current_file, weather_csv_file)), 'sales', 'covers'))
```
### Download CSV for EDA
```
data.to_csv('CSV_for_EDA.csv')
df_outside = data['outside']
```
## CHOOSE TARGET --> SALES OR COVERS
```
target = 'sales'
def daily_average_matrix_ann(df, target):
matrix = df.groupby([df.index.dayofweek, df.index.month, df.index.year]).agg({target: 'mean'})
matrix = matrix.rename_axis(['day', 'month', 'year'])
return matrix.unstack(level=1)
daily_average_matrix_ann(data, target)
```
### Create Month Clusters
```
from sklearn.cluster import KMeans
day_k = 7
mo_k = 3
def create_clusters(df, target, col, k):
# MAKE DATAFRAME USING CENTRAL TENDENCIES AS FEATURES
describe = df.groupby(col)[target].aggregate(['median', 'std', 'max'])
df = describe.reset_index()
# SCALE TEMPORARY DF
scaler = MinMaxScaler()
f = scaler.fit_transform(df)
# INSTANTIATE MODEL
km = KMeans(n_clusters=k, random_state=0).fit(f)
# GET KMEANS CLUSTER PREDICTIONS
labels = km.predict(f)
# MAKE SERIES FROM PREDICTIONS
temp = pd.DataFrame(labels, columns = ['cluster'], index=df.index)
# CONCAT CLUSTERS TO DATAFRAME
df = pd.concat([df, temp], axis=1)
# CREATE CLUSTER DICTIONARY
temp_dict = {}
for i in list(df[col]):
temp_dict[i] = df.loc[df[col] == i, 'cluster'].iloc[0]
return temp_dict
# Create Global Dictionaries to Categorize Day / Month
#day_dict = create_clusters(data, 'day_of_week', day_k)
month_dict = create_clusters(data, target, 'month', mo_k)
# Print Clusters
#print('Day Clusters: ', day_dict, '\n', 'Total Clusters: ', len(set(day_dict.values())), '\n')
print('Month Clusters: ', month_dict, '\n', 'Total Clusters: ', len(set(month_dict.values())))
```
### Add Temperature Onehot Categories
```
def encode_temp(df):
temp_enc = KBinsDiscretizer(n_bins=5, encode='onehot', strategy='kmeans')
temp_enc.fit(df[['apparent_temperature']])
return temp_enc
def one_hot_temp(df, temp_enc):
binned_transform = temp_enc.transform(df[['apparent_temperature']])
binned_df = pd.DataFrame(binned_transform.toarray(), index=df.index, columns=['temp_very_cold', 'temp_cold', 'temp_warm', 'temp_hot', 'temp_very_hot'])
df = df.merge(binned_df, how='left', on='date')
df.drop(['apparent_temperature', 'temperature'], axis=1, inplace=True)
return df, temp_enc
```
## Feature Engineering
```
# Add Clusters to DataFrame to use as Features
def add_clusters(df):
#df['day_cluster'] = df['day_of_week'].apply(lambda x: day_dict[x]).astype('category')
df['month_cluster'] = df['month'].apply(lambda x: month_dict[x]).astype('category')
return df
```
### Add Weather Features
```
hours_start = '05:00PM'
hours_end = '11:59PM'
hs_dt = datetime.strptime(hours_start, "%I:%M%p")
he_dt = datetime.strptime(hours_end, "%I:%M%p")
def between_time(check_time):
if hs_dt <= datetime.strptime(check_time, "%I:%M%p") <= he_dt:
return 1
else:
return 0
add_weather = True
temp_delta_window = 1
def add_weather_features(df):
if add_weather:
# POOR WEATHER FEATURES
df['precip_while_open'] = df['precip_max_time'].apply(lambda x: between_time(x))
# DROP FEATURES
features_to_drop = ['precip_max_time']
df.drop(features_to_drop, axis=1, inplace=True)
return df
```
### Add Calendar Features
```
def add_cal_features(df):
cal = calendar()
# THREE DAY WEEKEND FEATURE
sunday_three_days = [date + pd.DateOffset(-1) for date in cal.holidays(start_date, end_date) if date.dayofweek == 0]
df['sunday_three_day'] = np.where(df.index.isin(sunday_three_days), 1, 0)
return df
```
### Add Dummies
```
def add_dummies(df):
df['day_of_week'] = df['day_of_week'].astype('category')
df = pd.get_dummies(data=df, columns=['day_of_week', 'month_cluster'])
return df
```
### Add Interactions
```
def add_interactions(df):
apply_this_interaction = False
if apply_this_interaction:
for d in [col for col in df.columns if col.startswith('day_cluster')]:
for m in [col for col in df.columns if col.startswith('month_cluster')]:
col_name = d + '_X_' + m
df[col_name] = df[d] * df[m]
df.drop([d], axis=1, inplace=True)
df.drop([col for col in df.columns if col.startswith('month_cluster')], axis=1, inplace=True)
return df
else:
return df
def add_weather_interactions(df):
apply_this_interaction = True
if apply_this_interaction:
try:
df['outside_X_precip_open'] = df['outside'] * df['precip_while_open']
for w in [col for col in df.columns if col.startswith('temp_')]:
col_name = w + '_X_' + 'outside'
df[col_name] = df[w] * df['outside']
df.drop(['outside'], axis=1, inplace=True)
except:
pass
return df
else:
return df
```
### Feature Selection
```
def feature_selection(df):
try:
target_list = ['sales', 'covers', 'ppa']
target_to_drop = [t for t in target_list if t != target]
df = df.drop(target_to_drop, axis=1)
except:
pass
# Feature Selection / Drop unnecessary or correlated columns
cols_to_drop = ['month', 'precip_type', 'summary', 'pressure', 'precip_intensity_max', 'day_of_week_0']
df = df.drop(cols_to_drop, axis=1)
return df
```
### Add Target Trend Feature Engineering
```
trend_days_rolling = 31
trend_days_shift = 7
days_fwd = trend_days_rolling + trend_days_shift + 1
def target_trend_engineering(df):
df['target_trend'] = df[target].rolling(trend_days_rolling).mean() / df[target].shift(trend_days_shift).rolling(trend_days_rolling).mean()
#df['target_delta'] = df[target].shift(7) + df[target].shift(14) - df[target].shift(21) - df[target].shift(28)
return df
```
## Start Here
```
# IMPORT & PARSE CLEAN TRAINING SET
data = add_ppa(impute_outliers(encode_closed_days(prepare_data(current_file, weather_csv_file)), 'sales', 'covers'));
# One Hot Encode Temperature Data
data, temp_enc = one_hot_temp(data, encode_temp(data))
# Create CSV
data.to_csv('csv_before_features.csv')
def feature_engineering(df):
df.columns = df.columns.map(str)
# Add day & Month Clusters // Dicts with data held in Global Variable
df = add_clusters(df)
# Add Engineered Features for Weather & Calendar
df = add_weather_features(df)
df = add_cal_features(df)
# Create Dummies
df = add_dummies(df)
# Add Interactions
df = add_interactions(df)
df = add_weather_interactions(df)
# Drop Selected Columns
df = feature_selection(df)
return df
dfx = feature_engineering(data)
dfx = target_trend_engineering(dfx)
def corr_chart(df):
corr = df.corr()
mask = np.zeros_like(corr, dtype=np.bool)
mask[np.triu_indices_from(mask)] = True
# Set up the matplotlib figure
sns.set_style('whitegrid')
f, ax = plt.subplots(figsize=(16, 12))
# Generate a custom diverging colormap
cmap = sns.diverging_palette(220, 10, as_cmap=True)
# Draw the heatmap with the mask and correct aspect ratio
sns.heatmap(corr, mask=mask, cmap=cmap, vmax=1, vmin=-1, center=0,
square=True, linewidths=.75, annot=False, cbar_kws={"shrink": .75});
corr_chart(dfx)
```
## Update Sales
```
# # File from Start based on Target Variable
# current_sales_df = import_parse(rest_1_file)
# date = '2019-06-13'
# sales = 15209.75
# covers = 207
# outside = 1
# closed = 0
# def add_sales_row(date, sales):
# df = pd.DataFrame({'sales': sales,
# 'covers': covers,
# 'outside': outside,
# 'closed': closed},
# index=[date])
# return df
# temp = add_sales_row(date, sales)
# def build_sales_df(df, temp):
# df = df.append(temp)
# return df
# current_sales_df = build_sales_df(current_sales_df, temp)
# # Download Current DataFrame to CSV
# current_sales_df.to_csv(f'rest_1_clean_updated_{start_date}_to_{end_date}.csv')
# df_import = pd.read_csv('rest_1_clean_updated_2017-01-01_to_2019-06-17.csv', parse_dates=True, index_col='Unnamed: 0')
# def import_current(df):
# df.index = pd.to_datetime(df.index)
# df = add_ppa(df)
# target_list = ['sales', 'covers', 'ppa']
# target_to_drop = [t for t in target_list if t != target]
# df = df.drop(target_to_drop, axis=1)
# return df
# current_df = import_current(df_import)
```
### Add Recent Sales Data
```
# # Import Most Recent DataFrame
# df_before_features = pd.read_csv('csv_before_features.csv', index_col='date', parse_dates=True)
# # Create New Weather DataFrame with Updated Data
# new_date_start = '2019-06-15'
# new_date_end = '2019-06-17'
# def update_current_df(sales_df, df_before_features, new_date_start, new_end_date):
# sales_df = sales_df[new_date_start:]
# sales_df = sales_df.rename_axis(index = 'date')
# sales_df.index = pd.to_datetime(sales_df.index)
# ## Find Lat Long for Business
# lat, long = yelp_lat_long(search_business, location)
# ## Pull Weather Data / Forecast
# weather_df = build_weather_df(weather_call(new_date_start, new_date_end, lat, long))
# ## Parse, Clean, Engineer
# df = pd.merge(sales_df, weather_df, how='left', on='date')
# df, _ = one_hot_temp(df, temp_enc)
# df = pd.concat([df_before_features, df])
# df = target_trend_engineering(feature_engineering(df))
# return df
# current_df = update_current_df(current_df, df_before_features, new_date_start, new_date_end)
```
## Test / Train / Split
### Drop Closed Days?
```
drop_all_closed = False
if drop_all_closed:
current_df = current_df[current_df['closed'] == 0]
dfx.columns
def drop_weather(df):
no_weather = False
if no_weather:
df = df.drop(['humidity', 'precip_prob', 'temp_very_cold', 'temp_cold', 'temp_hot', 'temp_very_hot', 'precip_while_open', \
'temp_very_cold_X_outside', 'temp_cold_X_outside', 'temp_hot_X_outside','temp_very_hot_X_outside', 'outside_X_precip_open'], axis=1)
df = df.merge(df_outside, on='date', how='left')
return df
else:
return df
dfx = drop_weather(dfx)
dfx.head()
def cv_split(df):
features = dfx.drop([target], axis=1)[days_fwd:]
y = dfx[target][days_fwd:]
return features, y
cv_features, cv_y = cv_split(dfx)
baseline_cv_x, baseline_cv_y = cv_split(mod_baseline)
def train_test_split(df):
# Separate Target & Features
y = df[target]
features = df.drop([target], axis=1)
# Test / Train / Split
train_date_start = '2017-01-01'
train_date_end = '2018-12-31'
X_train = features[pd.to_datetime(train_date_start) + pd.DateOffset(days_fwd):train_date_end]
X_test = features[pd.to_datetime(train_date_end) + pd.DateOffset(1): ]
y_train = y[pd.to_datetime(train_date_start) + pd.DateOffset(days_fwd):train_date_end]
y_test = y[pd.to_datetime(train_date_end) + pd.DateOffset(1): ]
# Scale
scaler = MinMaxScaler()
X_train_scaled = scaler.fit_transform(X_train)
X_test_scaled = scaler.transform(X_test)
X_train = pd.DataFrame(X_train_scaled, columns=X_train.columns)
X_test = pd.DataFrame(X_test_scaled, columns=X_train.columns)
print('Train set: ', len(X_train))
print('Test set: ', len(X_test))
return X_train, X_test, y_train, y_test, scaler
X_train, X_test, y_train, y_test, scaler = train_test_split(dfx)
baseline_X_train, baseline_X_test, baseline_y_train, baseline_y_test, baseline_scaler = train_test_split(mod_baseline)
```
### Linear Regression
```
def linear_regression_model(X_train, y_train):
lr = LinearRegression(fit_intercept=True)
lr_rgr = lr.fit(X_train, y_train)
return lr_rgr
lr_rgr = linear_regression_model(X_train, y_train)
baseline_lr_rgr = linear_regression_model(baseline_X_train, baseline_y_train)
def rgr_score(rgr, X_train, y_train, X_test, y_test, cv_features, cv_y):
y_hat = rgr.predict(X_test)
sum_squares_residual = sum((y_test - y_hat)**2)
sum_squares_total = sum((y_test - np.mean(y_test))**2)
r_squared = 1 - (float(sum_squares_residual))/sum_squares_total
adjusted_r_squared = 1 - (1-r_squared)*(len(y_test)-1)/(len(y_test)-X_test.shape[1]-1)
print('Formula Scores - R-Squared: ', r_squared, 'Adjusted R-Squared: ', adjusted_r_squared, '\n')
train_score = rgr.score(X_train, y_train)
test_score = rgr.score(X_test, y_test)
y_pred = rgr.predict(X_test)
rmse = np.sqrt(mean_squared_error(y_test, y_pred))
pred_df = pd.DataFrame(y_pred, index=y_test.index)
pred_df = pred_df.rename(columns={0: target})
print('Train R-Squared: ', train_score)
print('Test R-Squared: ', test_score, '\n')
print('Root Mean Squared Error: ', rmse, '\n')
print('Cross Val Avg R-Squared: ', \
np.mean(cross_val_score(rgr, cv_features, cv_y, cv=10, scoring='r2')), '\n')
print('Intercept: ', rgr.intercept_, '\n')
print('Coefficients: \n')
for index, col_name in enumerate(X_test.columns):
print(col_name, ' --> ', rgr.coef_[index])
return pred_df
pred_df = rgr_score(lr_rgr, X_train, y_train, X_test, y_test, cv_features, cv_y)
baseline_preds = rgr_score(baseline_lr_rgr, baseline_X_train, baseline_y_train, baseline_X_test, baseline_y_test, baseline_cv_x, baseline_cv_y)
```
### Prediction Function
```
outside = 1
def predict_df(clf, scaler, X_train, current_df, date_1, date_2):
# Find Lat Long for Business
lat, long = yelp_lat_long(search_business, location)
# Pull Weather Data / Forecast
weather_df = build_weather_df(weather_call(date_1, date_2, lat, long))
day_of_week, apparent_temperature = weather_df['day_of_week'], weather_df['apparent_temperature']
weather_df['outside'] = outside
# One Hot Encode Temperature Using Fitted Encoder
df, _ = one_hot_temp(weather_df, temp_enc)
df['closed'] = 0
# Add Feature Engineering
df = feature_engineering(df)
# Add Sales Data for Sales Trend Engineering
current_df = current_df[target]
df = pd.merge(df, current_df, on='date', how='left')
df[target] = df[target].fillna(method='ffill')
df = target_trend_engineering(df)
df = df.drop([target], axis=1)
# Ensure Column Parity
missing_cols = set(X_train.columns) - set(df.columns)
for c in missing_cols:
df[c] = 0
df = df[X_train.columns][-2:]
# Scale Transform
df_scaled = scaler.transform(df)
df = pd.DataFrame(df_scaled, columns=df.columns, index=df.index)
# Predict and Build Prediction DataFrame for Review
pred_array = pd.DataFrame(clf.predict(df), index=df.index, columns=[target])
pred_df = df[df.columns[(df != 0).any()]]
pred_df = pd.concat([pred_df, day_of_week, apparent_temperature], axis=1)
final_predict = pd.concat([pred_array, pred_df], axis=1)
return final_predict
tonight = predict_df(lr_rgr, scaler, X_train, dfx, pd.datetime.now().date() + pd.DateOffset(-days_fwd), pd.datetime.now().date())
tonight[-2:]
```
## Lasso
```
def lasso_model(X_train, y_train):
lassoReg = Lasso(fit_intercept=True, alpha=.05)
lasso_rgr = lassoReg.fit(X_train,y_train)
return lasso_rgr
lasso_rgr = lasso_model(X_train, y_train)
baseline_lasso = lasso_model(baseline_X_train, baseline_y_train)
pred_df_ppa_lasso = rgr_score(lasso_rgr, X_train, y_train, X_test, y_test, cv_features, cv_y)
baseline_lasso = rgr_score(baseline_lasso, baseline_X_train, baseline_y_train, baseline_X_test, baseline_y_test, baseline_cv_x, baseline_cv_y)
tonight = predict_df(lasso_rgr, scaler, X_train, dfx, pd.datetime.now().date() + pd.DateOffset(-days_fwd), pd.datetime.now().date())
tonight[-2:]
from yellowbrick.regressor import ResidualsPlot
from yellowbrick.features.importances import FeatureImportances
plt.figure(figsize=(12,8))
visualizer = ResidualsPlot(lasso_rgr, hist=False)
visualizer.fit(X_train, y_train)
visualizer.score(X_test, y_test)
visualizer.poof()
features = list(X_train.columns)
fig = plt.figure(figsize=(12,8))
ax = fig.add_subplot()
labels = list(map(lambda x: x.title(), features))
visualizer = FeatureImportances(lasso_rgr, ax=ax, labels=labels, relative=False)
visualizer.fit(X_train, y_train)
visualizer.poof()
```
### Random Forest Regression
```
(1/6)**20
8543 * 8543
def rf_regression_model(X_train, y_train):
rfr = RandomForestRegressor(max_depth= 11,
max_features= 0.60,
min_impurity_decrease= 0.005,
n_estimators= 300,
min_samples_leaf = 2,
min_samples_split = 2,
random_state = 0)
rfr_rgr = rfr.fit(X_train, y_train)
return rfr_rgr
rfr_rgr = rf_regression_model(X_train, y_train)
def rfr_score(rgr, X_test, y_test, cv_features, cv_y):
y_hat = rgr.predict(X_test)
sum_squares_residual = sum((y_test - y_hat)**2)
sum_squares_total = sum((y_test - np.mean(y_test))**2)
r_squared = 1 - (float(sum_squares_residual))/sum_squares_total
adjusted_r_squared = 1 - (1-r_squared)*(len(y_test)-1)/(len(y_test)-X_test.shape[1]-1)
print('Formula Scores - R-Squared: ', r_squared, 'Adjusted R-Squared: ', adjusted_r_squared, '\n')
train_score = rgr.score(X_train, y_train)
test_score = rgr.score(X_test, y_test)
y_pred = rgr.predict(X_test)
rmse = np.sqrt(mean_squared_error(y_test, y_pred))
pred_df = pd.DataFrame(y_pred, index=y_test.index)
pred_df = pred_df.rename(columns={0: target})
print('Train R-Squared: ', train_score)
print('Test R-Squared: ', test_score, '\n')
print('Root Mean Squared Error: ', rmse, '\n')
print('Cross Val Avg R-Squared: ', \
np.mean(cross_val_score(rgr, cv_features, cv_y, cv=10, scoring='r2')), '\n')
return pred_df
pred_df = rfr_score(rfr_rgr, X_test, y_test, cv_features, cv_y)
```
# Random Forest Regression Prediction
```
tonight = predict_df(rfr_rgr, scaler, X_train, dfx, pd.datetime.now().date() + pd.DateOffset(-days_fwd), pd.datetime.now().date())
tonight[-2:]
```
### Grid Search Helper Function
```
def run_grid_search(rgr, params, X_train, y_train):
cv = 5
n_jobs = -1
scoring = 'neg_mean_squared_error'
grid = GridSearchCV(rgr, params, cv=cv, n_jobs=n_jobs, scoring=scoring, verbose=10)
grid = grid.fit(X_train, y_train)
best_grid_rgr = grid.best_estimator_
print('Grid Search: ', rgr.__class__.__name__, '\n')
print('Grid Search Best Score: ', grid.best_score_)
print('Grid Search Best Params: ', grid.best_params_)
print('Grid Search Best Estimator: ', grid.best_estimator_)
return best_grid_rgr
params = {
'n_estimators': [250, 275, 300, 350, 400,500],
'max_depth': [5, 7, 9, 11, 13, 15],
'min_impurity_decrease': [0.005, 0.001, 0.0001],
'max_features': ['auto', 0.65, 0.75, 0.85, 0.95]
}
best_grid_rgr = run_grid_search(rfr_rgr, params, X_train, y_train)
```
### OLS Model
```
import statsmodels.api as sm
from statsmodels.formula.api import ols
f = ''
for c in dfx.columns:
f += c + '+'
x = f[6:-1]
f= target + '~' + x
model = ols(formula=f, data=dfx).fit()
model.summary()
```
## XGB Regressor
```
from xgboost import XGBRegressor
def xgb_model(X_train, y_train):
objective = 'reg:linear'
booster = 'gbtree'
nthread = 4
learning_rate = 0.02
max_depth = 3
colsample_bytree = 0.75
n_estimators = 450
min_child_weight = 2
xgb_rgr= XGBRegressor(booster=booster, objective=objective, colsample_bytree=colsample_bytree, learning_rate=learning_rate, \
max_depth=max_depth, nthread=nthread, n_estimators=n_estimators, min_child_weight=min_child_weight, random_state = 0)
xgb_rgr = xgb_rgr.fit(X_train, y_train)
return xgb_rgr
xgb_rgr = xgb_model(X_train, y_train)
# Convert column names back to original
X_test = X_test[X_train.columns]
pred_df_covers_xgb = rfr_score(xgb_rgr, X_test, y_test, cv_features, cv_y)
```
## HYBRID
```
filex = 'predicted_ppa_timeseries.csv'
df_ts = pd.read_csv(filex,index_col='date',parse_dates=True)
pred_df = pred_df_covers_xgb.merge(df_ts, on='date', how='left')
pred_df.head()
pred_df['sales'] = pred_df['covers'] * pred_df['pred_ppa']
pred_df = pred_df[['sales']]
r2 = metrics.r2_score(y_test, pred_df)
r2
rmse = np.sqrt(mean_squared_error(y_test, pred_df))
rmse
tonight = predict_df(xgb_rgr, scaler, X_train, dfx, pd.datetime.now().date() + pd.DateOffset(-days_fwd), pd.datetime.now().date())
tonight[-2:]
xgb1 = XGBRegressor()
parameters = {'nthread':[4], #when use hyperthread, xgboost may become slower
'objective':['reg:linear'],
'learning_rate': [.015, 0.02, .025], #so called `eta` value
'max_depth': [3, 4, 5],
'min_child_weight': [1, 2, 3],
'silent': [1],
'subsample': [0.7],
'colsample_bytree': [0.55, 0.60, 0.65],
'n_estimators': [400, 500, 600]}
xgb_grid = GridSearchCV(xgb1,
parameters,
cv = 3,
n_jobs = 5,
verbose=True,
scoring = 'neg_mean_squared_error')
xgb_grid.fit(X_train,
y_train)
print(xgb_grid.best_score_)
print(xgb_grid.best_params_)
```
## CLEAN RUN
|
github_jupyter
|
# Numpy
### GitHub repository: https://github.com/jorgemauricio/curso_itesm
### Instructor: Jorge Mauricio
```
# librerías
import numpy as np
```
# Crear Numpy Arrays
## De una lista de python
Creamos el arreglo directamente de una lista o listas de python
```
my_list = [1,2,3]
my_list
np.array(my_list)
my_matrix = [[1,2,3],[4,5,6],[7,8,9]]
my_matrix
```
## Métodos
### arange
```
np.arange(0,10)
np.arange(0,11,2)
```
### ceros y unos
Generar arreglos de ceros y unos
```
np.zeros(3)
np.zeros((5,5))
np.ones(3)
np.ones((3,3))
```
### linspace
Generar un arreglo especificando un intervalo
```
np.linspace(0,10,3)
np.linspace(0,10,50)
```
### eye
Generar matrices de identidad
```
np.eye(4)
```
### Random
### rand
Generar un arreglo con una forma determinada de numeros con una distribución uniforme [0,1]
```
np.random.rand(2)
np.random.rand(5,5)
```
### randn
Generar un arreglo con una distribucion estandar a diferencia de rand que es uniforme
```
np.random.randn(2)
np.random.randn(5,5)
```
### randint
Generar numeros aleatorios en un rango determinado
```
np.random.randint(1,100)
np.random.randint(1,100,10)
```
### Métodos y atributos de arreglos
```
arr = np.arange(25)
ranarr = np.random.randint(0,50,10)
arr
ranarr
```
### Reshape
Regresa el mismo arreglo pero en diferente forma
```
arr.reshape(5,5)
```
### max, min, argmax, argmin
Metodos para encontrar máximos y mínimos de los valores y sus indices
```
ranarr
ranarr.max()
ranarr.argmax()
ranarr.min()
ranarr.argmin()
```
### Shape
Atributo para desplegar la forma que tienen el arreglo
```
# Vector
arr.shape
# Tomar en cuenta que se implementan dos corchetes
arr.reshape(1,25)
arr.reshape(1,25).shape
arr.reshape(25,1)
arr.reshape(25,1).shape
```
### dtype
Despliega el tipo de dato de los objetos del arreglo
```
arr.dtype
```
# Selección e indices en Numpy
```
# crear un arreglo
arr = np.arange(0,11)
# desplegar el arreglo
arr
```
# Selección utilizando corchetes
```
# obtener el valor del indice 8
arr[8]
# obtener los valores de un rango
arr[1:5]
#obtener los valores de otro rango
arr[2:6]
```
# Reemplazar valores
```
# reemplazar valores en un rango determinado
arr[0:5]=100
# desplegar el arreglo
arr
# Generar nuevamente el arreglo
arr = np.arange(0,11)
# desplegar
arr
# corte de un arreglo
slice_of_arr = arr[0:6]
# desplegar el corte
slice_of_arr
# cambiar valores del corte
slice_of_arr[:]=99
# desplegar los valores del corte
slice_of_arr
# desplegar arreglo
arr
# para obtener una copia se debe hacer explicitamente
arr_copy = arr.copy()
# desplegar el arreglo copia
arr_copy
```
## Indices en un arreglo 2D (matrices)
La forma general de un arreglo 2d es la siguiente **arr_2d[row][col]** o **arr_2d[row,col]**
```
# generar un arreglo 2D
arr_2d = np.array(([5,10,15],[20,25,30],[35,40,45]))
#Show
arr_2d
# indices de filas
arr_2d[1]
# Formato es arr_2d[row][col] o arr_2d[row,col]
# Seleccionar un solo elemento
arr_2d[1][0]
# Seleccionar un solo elemento
arr_2d[1,0]
# Cortes en 2D
# forma (2,2) desde la esquina superior derecha
arr_2d[:2,1:]
#forma desde la ultima fila
arr_2d[2]
# forma desde la ultima fila
arr_2d[2,:]
# longitud de un arreglo
arr_length = arr_2d.shape[1]
arr_length
```
# Selección
```
arr = np.arange(1,11)
arr
arr > 4
bool_arr = arr>4
bool_arr
arr[bool_arr]
arr[arr>2]
x = 2
arr[arr>x]
```
|
github_jupyter
|
```
import os
import numpy as np
import pandas as pd
import glob
from prediction_utils.util import yaml_read, df_dict_concat
table_path = '../figures/hyperparameters/'
os.makedirs(table_path, exist_ok = True)
param_grid_base = {
"lr": [1e-3, 1e-4, 1e-5],
"batch_size": [128, 256, 512],
"drop_prob": [0.0, 0.25, 0.5, 0.75],
"num_hidden": [1, 2, 3],
"hidden_dim": [128, 256],
}
the_dict = {'hyperparameter': [], 'Grid': []}
for key, value in param_grid_base.items():
the_dict['hyperparameter'].append(key)
the_dict['Grid'].append(value)
the_df = pd.DataFrame(the_dict)
rename_grid = {
'hyperparameter': ['lr', 'batch_size', 'drop_prob', 'num_hidden', 'hidden_dim'],
'Hyperparameter': ['Learning Rate', 'Batch Size', 'Dropout Probability', 'Number of Hidden Layers', 'Hidden Dimension']
}
rename_df = pd.DataFrame(rename_grid)
the_df = the_df.merge(rename_df)[['Hyperparameter', 'Grid']].sort_values('Hyperparameter')
the_df
the_df.to_latex(os.path.join(table_path, 'param_grid.txt'), index=False)
selected_models_path = '/share/pi/nigam/projects/spfohl/cohorts/admissions/optum/experiments/baseline_tuning_fold_1/config/selected_models'
selected_models_path_dict = {
'starr': '/share/pi/nigam/projects/spfohl/cohorts/admissions/starr_20200523/experiments/baseline_tuning_fold_1_10/config/selected_models',
'mimic': '/share/pi/nigam/projects/spfohl/cohorts/admissions/mimic_omop/experiments/baseline_tuning_fold_1_10/config/selected_models',
'optum': '/share/pi/nigam/projects/spfohl/cohorts/admissions/optum/experiments/baseline_tuning_fold_1/config/selected_models',
}
selected_param_dict = {
db: {
task: yaml_read(glob.glob(os.path.join(db_path, task, '*.yaml'), recursive=True)[0]) for task in os.listdir(db_path)
}
for db, db_path in selected_models_path_dict.items()
}
col_order = {
'starr': ['hospital_mortality', 'LOS_7', 'readmission_30'],
'mimic': ['los_icu_3days', 'los_icu_7days', 'mortality_hospital', 'mortality_icu'],
'optum': ['readmission_30', 'LOS_7'],
}
for db in selected_param_dict.keys():
db_params = selected_param_dict[db]
db_df = (
pd.concat({key: pd.DataFrame(value, index= [0]) for key, value in db_params.items()})
.reset_index(level=1,drop=True)
.rename_axis('task')
.transpose()
.rename_axis('hyperparameter')
.reset_index()
.merge(rename_df, how ='right')
)
db_df = db_df[['Hyperparameter'] + list(set(db_df.columns) - set(['hyperparameter', 'Hyperparameter']))].sort_values('Hyperparameter')
db_df = db_df[['Hyperparameter'] + col_order[db]].sort_values('Hyperparameter')
selected_param_path = os.path.join(table_path, db)
os.makedirs(selected_param_path, exist_ok=True)
db_df.to_latex(os.path.join(selected_param_path, 'selected_param_table.txt'), index=False)
```
|
github_jupyter
|
# 时间序列预测
时间序列是随着时间的推移定期收集的数据。时间序列预测是指根据历史数据预测未来数据点的任务。时间序列预测用途很广泛,包括天气预报、零售和销量预测、股市预测,以及行为预测(例如预测一天的车流量)。时间序列数据有很多,识别此类数据中的模式是很活跃的机器学习研究领域。
<img src='notebook_ims/time_series_examples.png' width=80% />
在此 notebook 中,我们将学习寻找时间规律的一种方法,即使用 SageMaker 的监督式学习模型 [DeepAR](https://docs.aws.amazon.com/sagemaker/latest/dg/deepar.html)。
### DeepAR
DeepAR 使用循环神经网络 (RNN),它会接受序列数据点作为历史输入,并生成预测序列数据点。这种模型如何学习?
在训练过程中,你需要向 DeepAR estimator 提供训练数据集(由多个时间序列组成)。该 estimator 会查看所有的训练时间序列并尝试发现它们之间的相似性。它通过从训练时间序列中随机抽取**训练样本**进行训练。
* 每个训练样本都由相邻的**上下文**和**预测**窗口(长度已提前固定好)对组成。
* `context_length` 参数会控制模型能看到过去多久的数据。
* `prediction_length` 参数会控制模型可以对未来多久做出预测。
* 详情请参阅[此文档](https://docs.aws.amazon.com/sagemaker/latest/dg/deepar_how-it-works.html)。
<img src='notebook_ims/context_prediction_windows.png' width=50% />
> 因为 DeepAR 用多个时间序列进行训练,所以很适合有**循环规律**的数据。
在任何预测任务中,选择的上下文窗口都应该能向模型提供足够的**相关**信息,这样才能生成准确的预测。通常,最接近预测时间帧的数据包含的信息对确定预测结果的影响最大。在很多预测应用中,例如预测月销量,上下文和预测窗口大小一样,但有时候有必要设置更大的上下文窗口,从而发现数据中的更长期规律。
### 能耗数据
在此 notebook 中,我们将使用的数据是全球的家庭耗电量数据。数据集来自 [Kaggle](https://www.kaggle.com/uciml/electric-power-consumption-data-set),表示从 2006 年到 2010 年的耗电量数据。对于这么庞大的数据集,我们可以预测很长时间的耗电量,例如几天、几周或几个月。预测能耗有很多用途,例如确定耗电量的季节性价格,以及根据预测用量有效地向居民供电。
**趣味阅读**:Google DeepMind 最近展开了一项逆相关项目,他们使用机器学习预测风力发电机产生的电量,并有效地将电力输送给电网。你可以在[这篇帖子](https://deepmind.com/blog/machine-learning-can-boost-value-wind-energy/)中详细了解这项研究。
### 机器学习工作流程
此 notebook 将时间序列预测分成了以下几个步骤:
* 加载和探索数据
* 创建时间序列训练集和测试集
* 将数据变成 JSON 文件并上传到 S3
* 实例化和训练 DeepAR estimator
* 部署模型并创建预测器
* 评估预测器
---
首先加载常规资源。
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
```
# 加载和探索数据
我们将收集在几年内收集的全球能耗数据。以下单元格将加载并解压缩此数据,并为你提供一个文本数据文件 `household_power_consumption.txt`。
```
! wget https://s3.amazonaws.com/video.udacity-data.com/topher/2019/March/5c88a3f1_household-electric-power-consumption/household-electric-power-consumption.zip
! unzip household-electric-power-consumption
```
### 读取 `.txt` 文件
下个单元格显示了文本文件里的前几行,我们可以看看数据格式。
```
# display first ten lines of text data
n_lines = 10
with open('household_power_consumption.txt') as file:
head = [next(file) for line in range(n_lines)]
display(head)
```
## 预处理数据
household_power_consumption.txt 文件具有以下属性:
* 每个数据点都具有日期和时间记录 (时:分:秒)
* 各个数据特征用分号 (;) 分隔
* 某些数据为“nan”或“?”,我们将它们都当做 `NaN` 值
### 处理 `NaN` 值
此 DataFrame 包含一些缺失值的数据点。到目前为止,我们只是丢弃这些值,但是还有其他处理 `NaN` 值的方式。一种技巧是用缺失值所在列的**均值**填充;这样填充的值可能比较符合实际。
我在 `txt_preprocessing.py` 中提供了一些辅助函数,可以帮助你将原始文本文件加载为 DataFrame,并且用各列的平均特征值填充 `NaN` 值。这种技巧对于长期预测来说是可行的,如果是按小时分析和预测,则最好丢弃这些 `NaN` 值或对很小的滑动窗口求平均值,而不是采用整个数据列的平均值。
**在下面的单元格中,我将文件读取为 DataFrame 并用特征级平均值填充 `NaN` 值。**
```
import txt_preprocessing as pprocess
# create df from text file
initial_df = pprocess.create_df('household_power_consumption.txt', sep=';')
# fill NaN column values with *average* column value
df = pprocess.fill_nan_with_mean(initial_df)
# print some stats about the data
print('Data shape: ', df.shape)
df.head()
```
## 全球有效能耗
在此示例中,我们想要预测全球有效能耗,即全球的家庭每分钟平均有效能耗(千瓦)。在下面获取这列数据并显示生成的图形。
```
# Select Global active power data
power_df = df['Global_active_power'].copy()
print(power_df.shape)
# display the data
plt.figure(figsize=(12,6))
# all data points
power_df.plot(title='Global active power', color='blue')
plt.show()
```
因为数据是每分钟记录的,上图包含很多值。所以我只在下面显示了一小部分数据。
```
# can plot a slice of hourly data
end_mins = 1440 # 1440 mins = 1 day
plt.figure(figsize=(12,6))
power_df[0:end_mins].plot(title='Global active power, over one day', color='blue')
plt.show()
```
### 每小时与每天
每分钟收集了很多数据,我可以通过以下两种方式之一分析数据:
1. 创建很多简短的时间序列,例如一周左右,并且每小时都记录一次能耗,尝试预测接下来的几小时或几天的能耗。
2. 创建更少的很长时间序列,数据每天记录一次,并使用这些数据预测未来几周或几个月的能耗。
两种任务都很有意思。具体取决于你是要预测一天/一周还是更长时间(例如一个月)的规律。鉴于我拥有的数据量,我认为可以查看在多个月或一年内发生的更长重复性趋势。所以我将重采样“全球有效能耗”,将**每日**数据点记录为 24 小时的平均值。
> 我们可以使用 pandas [时间序列工具](https://pandas.pydata.org/pandas-docs/stable/user_guide/timeseries.html)根据特定的频率重采样数据,例如按照每小时 ('H') 或每天 ('D') 重采样数据点。
```
# resample over day (D)
freq = 'D'
# calculate the mean active power for a day
mean_power_df = power_df.resample(freq).mean()
# display the mean values
plt.figure(figsize=(15,8))
mean_power_df.plot(title='Global active power, mean per day', color='blue')
plt.tight_layout()
plt.show()
```
在此图形中,可以看到每年都出现了有趣的趋势。每年初和每年末都会出现能耗高峰,这时候是冬季,供暖和照明使用量都更高。在 8 月份左右也出现小高峰,这时候全球的温度通常更高。
数据依然不够平滑,但是展现了明显的趋势,所以适合用机器学习模型识别这些规律。
---
## 创建时间序列
我的目标是看看能否根据从 2007-2009 的整年数据,准确地预测 2010 年多个月的平均全球有效能耗。
接下来为每个完整的年份数据创建一个时间序列。这只是一种设计决策,我决定使用一整年的数据,从 2007 年 1 月开始,因为 2006 年的数据点不太多,并且这种划分更容易处理闰年。我还可以从第一个收集的数据点开始构建时间序列,只需在下面的函数中更改 `t_start` 和 `t_end` 即可。
函数 `make_time_series` 将为传入的每个年份 `['2007', '2008', '2009']` 创建 pandas `Series`。
* 所有的时间序列将从相同的时间点 `t_start`(或 t0)开始。
* 在准备数据时,需要为每个时间序列使用一致的起始点;DeepAR 将此时间点作为参考帧,从而学习循环规律,例如工作日的行为与周末不一样,或者夏天与冬天不一样。
* 你可以更改起始和结束索引,并定义你创建的任何时间序列。
* 在创建时间序列时,我们应该考虑到闰年,例如 2008 年。
* 通常,我们通过从 DataFrame 获取相关的全球能耗数据和日期索引创建 `Series`。
```
# get global consumption data
data = mean_power_df[start_idx:end_idx]
# create time series for the year
index = pd.DatetimeIndex(start=t_start, end=t_end, freq='D')
time_series.append(pd.Series(data=data, index=index))
```
```
def make_time_series(mean_power_df, years, freq='D', start_idx=16):
'''Creates as many time series as there are complete years. This code
accounts for the leap year, 2008.
:param mean_power_df: A dataframe of global power consumption, averaged by day.
This dataframe should also be indexed by a datetime.
:param years: A list of years to make time series out of, ex. ['2007', '2008'].
:param freq: The frequency of data recording (D = daily)
:param start_idx: The starting dataframe index of the first point in the first time series.
The default, 16, points to '2017-01-01'.
:return: A list of pd.Series(), time series data.
'''
# store time series
time_series = []
# store leap year in this dataset
leap = '2008'
# create time series for each year in years
for i in range(len(years)):
year = years[i]
if(year == leap):
end_idx = start_idx+366
else:
end_idx = start_idx+365
# create start and end datetimes
t_start = year + '-01-01' # Jan 1st of each year = t_start
t_end = year + '-12-31' # Dec 31st = t_end
# get global consumption data
data = mean_power_df[start_idx:end_idx]
# create time series for the year
index = pd.DatetimeIndex(start=t_start, end=t_end, freq=freq)
time_series.append(pd.Series(data=data, index=index))
start_idx = end_idx
# return list of time series
return time_series
```
## 测试结果
下面为每个完整的年份创建一个时间序列,并显示结果。
```
# test out the code above
# yearly time series for our three complete years
full_years = ['2007', '2008', '2009']
freq='D' # daily recordings
# make time series
time_series = make_time_series(mean_power_df, full_years, freq=freq)
# display first time series
time_series_idx = 0
plt.figure(figsize=(12,6))
time_series[time_series_idx].plot()
plt.show()
```
---
# 按时间拆分数据
我们将用测试数据集评估模型。对于分类等机器学习任务,我们通常随机将样本拆分成不同的数据集,创建训练/测试数据。对于未来预测来说,一定要按照**时间**拆分训练/测试数据,不能按照单个数据点拆分。
> 通常,在创建训练数据时,我们从每个完整的时间序列中去除最后 `prediction_length` 个数据点,并创建训练时间序列。
### 练习:创建训练时间序列
请完成 `create_training_series` 函数,它应该接受完整时间序列数据列表,并返回截断的训练时间序列列表。
* 在此例中,我们想预测一个月的数据,将 `prediction_length` 设为 30 天。
* 为了创建训练数据集,我们将从生成的每个时间序列中去除最后 30 个数据点,所以仅使用第一部分作为训练数据。
* **测试集包含每个时间序列的完整范围**。
```
# create truncated, training time series
def create_training_series(complete_time_series, prediction_length):
'''Given a complete list of time series data, create training time series.
:param complete_time_series: A list of all complete time series.
:param prediction_length: The number of points we want to predict.
:return: A list of training time series.
'''
# your code here
pass
# test your code!
# set prediction length
prediction_length = 30 # 30 days ~ a month
time_series_training = create_training_series(time_series, prediction_length)
```
### 训练和测试序列
我们可以将训练/测试序列绘制到同一个坐标轴上,可视化这些序列。我们应该看到测试序列包含一年的所有数据,训练序列包含最后 `prediction_length` 个数据点之外的数据。
```
# display train/test time series
time_series_idx = 0
plt.figure(figsize=(15,8))
# test data is the whole time series
time_series[time_series_idx].plot(label='test', lw=3)
# train data is all but the last prediction pts
time_series_training[time_series_idx].plot(label='train', ls=':', lw=3)
plt.legend()
plt.show()
```
## 转换为 JSON
根据 [DeepAR 文档](https://docs.aws.amazon.com/sagemaker/latest/dg/deepar.html),DeepAR 要求输入训练数据是 JSON 格式,并包含以下字段:
* **start**:定义时间序列开始日期的字符串,格式为“YYYY-MM-DD HH:MM:SS”。
* **target**:表示时间序列的数值数组。
* **cat**(可选):类别特征数值数组,可以用于表示记录所属的组。这个字段适合按照项目类别寻找模型,例如对于零售销量,可以将 {'shoes', 'jackets', 'pants'} 表示成类别 {0, 1, 2}。
输入数据的格式应该为,在 JSON 文件中每行一个时间序列。每行看起来像字典,例如:
```
{"start":'2007-01-01 00:00:00', "target": [2.54, 6.3, ...], "cat": [1]}
{"start": "2012-01-30 00:00:00", "target": [1.0, -5.0, ...], "cat": [0]}
...
```
在上述示例中,每个时间序列都有一个相关的类别特征和一个时间序列特征。
### 练习:格式化能耗数据
对于我们的数据来说:
* 开始日期“start”将为时间序列中第一行的索引,即这一年的 1 月 1 日。
* “target”将为时间序列存储的所有能耗值。
* 我们将不使用可选“cat”字段。
请完成以下实用函数,它应该将 `pandas.Series` 对象转换成 DeepAR 可以使用的相应 JSON 字符串。
```
def series_to_json_obj(ts):
'''Returns a dictionary of values in DeepAR, JSON format.
:param ts: A single time series.
:return: A dictionary of values with "start" and "target" keys.
'''
# your code here
pass
# test out the code
ts = time_series[0]
json_obj = series_to_json_obj(ts)
print(json_obj)
```
### 将数据保存到本地
下面的辅助函数会将一个序列放入 JSON 文件的一行中,并使用换行符“\n”分隔。数据还会编码并写入我们指定的文件名中。
```
# import json for formatting data
import json
import os # and os for saving
def write_json_dataset(time_series, filename):
with open(filename, 'wb') as f:
# for each of our times series, there is one JSON line
for ts in time_series:
json_line = json.dumps(series_to_json_obj(ts)) + '\n'
json_line = json_line.encode('utf-8')
f.write(json_line)
print(filename + ' saved.')
# save this data to a local directory
data_dir = 'json_energy_data'
# make data dir, if it does not exist
if not os.path.exists(data_dir):
os.makedirs(data_dir)
# directories to save train/test data
train_key = os.path.join(data_dir, 'train.json')
test_key = os.path.join(data_dir, 'test.json')
# write train/test JSON files
write_json_dataset(time_series_training, train_key)
write_json_dataset(time_series, test_key)
```
---
## 将数据上传到 S3
接下来,为了使 estimator 能够访问此数据,我将数据上传到 S3。
### Sagemaker 资源
首先指定:
* 训练模型用到的 sagemaker 角色和会话。
* 默认 S3 存储桶,我们可以在其中存储训练、测试和模型数据。
```
import boto3
import sagemaker
from sagemaker import get_execution_role
# session, role, bucket
sagemaker_session = sagemaker.Session()
role = get_execution_role()
bucket = sagemaker_session.default_bucket()
```
### 练习:将训练和测试 JSON 文件上传到 S3
指定唯一的训练和测试 prefix,它们定义了数据在 S3 中的位置。
* 将训练数据上传到 S3 中的某个位置,并将该位置保存到 `train_path`
* 将测试数据上传到 S3 中的某个位置,并将该位置保存到 `test_path`
```
# suggested that you set prefixes for directories in S3
# upload data to S3, and save unique locations
train_path = None
test_path = None
# check locations
print('Training data is stored in: '+ train_path)
print('Test data is stored in: '+ test_path)
```
---
# 训练 DeepAR Estimator
某些 estimator 具有特定的 SageMaker 构造函数,但是并非都有。你可以创建一个基本 `Estimator` 并传入保存特定模型的特定镜像(或容器)。
接下来,配置要在我们运行模型所在的区域使用的容器镜像。
```
from sagemaker.amazon.amazon_estimator import get_image_uri
image_name = get_image_uri(boto3.Session().region_name, # get the region
'forecasting-deepar') # specify image
```
### 练习:实例化 Estimator
现在可以定义将启动训练作业的 estimator 了。一般的 Estimator 将由普通的构造函数参数和 `image_name` 进行定义。
> 你可以查看 [estimator 源代码](https://github.com/aws/sagemaker-python-sdk/blob/master/src/sagemaker/estimator.py#L595)了解详情。
```
from sagemaker.estimator import Estimator
# instantiate a DeepAR estimator
estimator = None
```
## 设置超参数
接下来,我们需要定义一些 DeepAR 超参数,这些超参数定义了模型大小和训练行为。需要定义周期数评论、预测时长和上下文时长。
* **epochs**:在训练时遍历数据的最大次数。
* **time_freq**:数据集中的时间序列频率(“D”表示每天)。
* **prediction_length**:一个字符串,表示训练模型预测的时间步数量(基于频率单位)。
* **context_length**:模型在做出预测之前可以看到的时间点数量。
### 上下文长度
通常,建议从 `context_length`=`prediction_length` 开始。这是因为 DeepAR 模型还会从目标时间序列那接收“延迟”输入,使模型能够捕获长期依赖关系。例如,每日时间序列可以具有每年季节效应,DeepAR 会自动包含一年延迟。所以上下文长度可以短于一年,模型依然能够捕获这种季节效应。
模型选择的延迟值取决于时间序列的频率。例如,每日频率的延迟值是上一周、上两周、上三周、上四周和一年。详情请参阅 [DeepAR 工作原理文档](https://docs.aws.amazon.com/sagemaker/latest/dg/deepar_how-it-works.html)。
### 可选超参数
你还可以配置可选超参数,以进一步优化模型。包括 RNN 模型的层数、每层的单元格数量、似然率函数,以及训练选项,例如批次大小和学习速率。
要了解所有不同 DeepAR 超参数的详尽列表,请参阅 DeepAR [超参数文档](https://docs.aws.amazon.com/sagemaker/latest/dg/deepar_hyperparameters.html)。
```
freq='D'
context_length=30 # same as prediction_length
hyperparameters = {
"epochs": "50",
"time_freq": freq,
"prediction_length": str(prediction_length),
"context_length": str(context_length),
"num_cells": "50",
"num_layers": "2",
"mini_batch_size": "128",
"learning_rate": "0.001",
"early_stopping_patience": "10"
}
# set the hyperparams
estimator.set_hyperparameters(**hyperparameters)
```
## 训练作业
现在我们可以启动训练作业了。SageMaker 将启动 EC2 实例、从 S3 下载数据、开始训练模型并保存训练过的模型。
如果你提供了 `test` 数据通道(就像在示例中一样),DeepAR 还会计算训练过的模型在此测试数据集上的准确率指标。计算方法是预测测试集中每个时间序列的最后 `prediction_length` 个点,并将它们与时间序列的实际值进行比较。计算的误差指标将包含在日志输出中。
下个单元格可能需要几分钟才能完成,取决于数据大小、模型复杂度和训练选项。
```
%%time
# train and test channels
data_channels = {
"train": train_path,
"test": test_path
}
# fit the estimator
estimator.fit(inputs=data_channels)
```
## 部署和创建预测器
训练模型后,我们可以将模型部署到预测器端点上,并使用模型做出预测。
在此 notebook 结束时,记得**删除端点**。我们将在此 notebook 的最后提供一个删除单元格,但是建议提前记住。
```
%%time
# create a predictor
predictor = estimator.deploy(
initial_instance_count=1,
instance_type='ml.t2.medium',
content_type="application/json" # specify that it will accept/produce JSON
)
```
---
# 生成预测
根据 DeepAR 的[推理格式](https://docs.aws.amazon.com/sagemaker/latest/dg/deepar-in-formats.html),`predictor` 要求输入数据是 JSON 格式,并具有以下键:
* **instances**:一个 JSON 格式的时间序列列表,模型应该预测这些时间序列。
* **configuration**(可选):一个配置信息字典,定义了请求希望的响应类型。
在 configuration 中,可以配置以下键:
* **num_samples**:一个整数,指定了模型在做出概率预测时,生成的样本数。
* **output_types**:一个指定响应类型的列表。我们需要 **quantiles**,它会查看模型生成的 num_samples 列表,并根据这些值为每个时间点生成[分位数估值](https://en.wikipedia.org/wiki/Quantile)。
* **quantiles**:一个列表,指定生成哪些分位数估值并在响应中返回这些估值。
下面是向 DeepAR 模型端点发出 JSON 查询的示例。
```
{
"instances": [
{ "start": "2009-11-01 00:00:00", "target": [4.0, 10.0, 50.0, 100.0, 113.0] },
{ "start": "1999-01-30", "target": [2.0, 1.0] }
],
"configuration": {
"num_samples": 50,
"output_types": ["quantiles"],
"quantiles": ["0.5", "0.9"]
}
}
```
## JSON 预测请求
以下代码接受时间序列**列表**作为输入并接受一些配置参数。然后将该序列变成 JSON 实例格式,并将输入转换成相应格式的 JSON_input。
```
def json_predictor_input(input_ts, num_samples=50, quantiles=['0.1', '0.5', '0.9']):
'''Accepts a list of input time series and produces a formatted input.
:input_ts: An list of input time series.
:num_samples: Number of samples to calculate metrics with.
:quantiles: A list of quantiles to return in the predicted output.
:return: The JSON-formatted input.
'''
# request data is made of JSON objects (instances)
# and an output configuration that details the type of data/quantiles we want
instances = []
for k in range(len(input_ts)):
# get JSON objects for input time series
instances.append(series_to_json_obj(input_ts[k]))
# specify the output quantiles and samples
configuration = {"num_samples": num_samples,
"output_types": ["quantiles"],
"quantiles": quantiles}
request_data = {"instances": instances,
"configuration": configuration}
json_request = json.dumps(request_data).encode('utf-8')
return json_request
```
### 获得预测
然后,我们可以使用该函数获得格式化时间序列的预测。
在下个单元格中,我获得了时间序列输入和已知目标,并将格式化输入传入预测器端点中,以获得预测。
```
# get all input and target (test) time series
input_ts = time_series_training
target_ts = time_series
# get formatted input time series
json_input_ts = json_predictor_input(input_ts)
# get the prediction from the predictor
json_prediction = predictor.predict(json_input_ts)
print(json_prediction)
```
## 解码预测
预测器返回 predictor returns JSON 格式的预测,所以我们需要提取可视化结果所需的预测和分位数数据。以下函数会读取 JSON 格式的预测并生成每个分位数中的预测列表。
```
# helper function to decode JSON prediction
def decode_prediction(prediction, encoding='utf-8'):
'''Accepts a JSON prediction and returns a list of prediction data.
'''
prediction_data = json.loads(prediction.decode(encoding))
prediction_list = []
for k in range(len(prediction_data['predictions'])):
prediction_list.append(pd.DataFrame(data=prediction_data['predictions'][k]['quantiles']))
return prediction_list
# get quantiles/predictions
prediction_list = decode_prediction(json_prediction)
# should get a list of 30 predictions
# with corresponding quantile values
print(prediction_list[0])
```
## 显示结果
分位数数据可以提供查看预测结果所需的所有信息。
* 分位数 0.1 和 0.9 表示预测值的上下限。
* 分位数 0.5 表示所有样本预测的中位数。
```
# display the prediction median against the actual data
def display_quantiles(prediction_list, target_ts=None):
# show predictions for all input ts
for k in range(len(prediction_list)):
plt.figure(figsize=(12,6))
# get the target month of data
if target_ts is not None:
target = target_ts[k][-prediction_length:]
plt.plot(range(len(target)), target, label='target')
# get the quantile values at 10 and 90%
p10 = prediction_list[k]['0.1']
p90 = prediction_list[k]['0.9']
# fill the 80% confidence interval
plt.fill_between(p10.index, p10, p90, color='y', alpha=0.5, label='80% confidence interval')
# plot the median prediction line
prediction_list[k]['0.5'].plot(label='prediction median')
plt.legend()
plt.show()
# display predictions
display_quantiles(prediction_list, target_ts)
```
## 预测未来
我们没有向模型提供任何 2010 年数据,但是我们看看如果只有已知开始日期,**没有目标**,模型能否预测能耗。
### 练习:为“未来”预测设定请求
请创建一个格式化输入并传入部署的 `predictor`,同时传入常规“configuration”参数。这里的“instances”只有 1 个实例,定义如下:
* **start**:开始时间将为你指定的时间戳。要预测 2010 年的前 30 天,从 1 月 1 日“2010-01-01”开始。
* **target**:目标将为空列表,因为这一年没有完整的相关时间序列。我们特意从模型中去除了该信息,以便测试模型。
```
{"start": start_time, "target": []} # empty target
```
```
# Starting my prediction at the beginning of 2010
start_date = '2010-01-01'
timestamp = '00:00:00'
# formatting start_date
start_time = start_date +' '+ timestamp
# format the request_data
# with "instances" and "configuration"
request_data = None
# create JSON input
json_input = json.dumps(request_data).encode('utf-8')
print('Requesting prediction for '+start_time)
```
然后正常地获取和解码预测响应。
```
# get prediction response
json_prediction = predictor.predict(json_input)
prediction_2010 = decode_prediction(json_prediction)
```
最后,将预测与已知目标序列进行比较。此目标将来自 2010 年的时间序列,我在下面创建了该序列。
```
# create 2010 time series
ts_2010 = []
# get global consumption data
# index 1112 is where the 2011 data starts
data_2010 = mean_power_df.values[1112:]
index = pd.DatetimeIndex(start=start_date, periods=len(data_2010), freq='D')
ts_2010.append(pd.Series(data=data_2010, index=index))
# range of actual data to compare
start_idx=0 # days since Jan 1st 2010
end_idx=start_idx+prediction_length
# get target data
target_2010_ts = [ts_2010[0][start_idx:end_idx]]
# display predictions
display_quantiles(prediction_2010, target_2010_ts)
```
## 删除端点
请用不同的时间序列尝试你的代码。建议调节 DeepAR 超参数,看看能否改进此预测器的性能。
评估完预测器(任何预测器)后,记得删除端点。
```
## TODO: delete the endpoint
predictor.delete_endpoint()
```
## 总结
你已经见过一个复杂但是应用广泛的时间序列预测方法,并且掌握了将 DeepAR 模型应用到你感兴趣的数据上所需的技能。
|
github_jupyter
|
### Netflix Scrapper
The purpose of the code is to get details of all the Categories on Netflix and then to gather information about Sub-Categories and movies under each Sub-Category.
```
from bs4 import BeautifulSoup
import requests
import pandas as pd
import numpy as np
def make_soup(url):
return BeautifulSoup(requests.get(url).text, 'html.parser')
def browseCategory(category, data):
category_url = data[category-1][2]
category = data[category-1][1]
subCategory_details = []
count = 1
subCategories = []
soup = make_soup(category_url)
cards_list = soup.find_all('section',{'class':'nm-collections-row'})
for card in cards_list:
try:
subCategory = card.find('h1').text
movie_list = []
movies = card.find_all('li')
movie_count = 1
for movie in movies:
try:
movie_title = movie.find('span',{'class':'nm-collections-title-name'}).text
movie_link = movie.find('a').get('href')
movie_list.append([movie_count, movie_title , movie_link])
movie_count += 1
except AttributeError:
pass
subCategories.append(subCategory)
subCategory_details.append(movie_list)
count += 1
except AttributeError:
pass
return subCategories, subCategory_details, count-1
def getCategories(base_url):
category_soup = make_soup(base_url)
categories = category_soup.find_all('section',{'class':'nm-collections-row'})
result=[]
count = 1
for category in categories:
try:
Title = category.find('span', {'class':'nm-collections-row-name'}).text
url = category.find('a').get('href')
result.append([count, Title, url])
count += 1
except AttributeError:
pass
#print(result)
return result
def main():
netflix_url = "https://www.netflix.com/in/browse/genre/839338"
categories = getCategories(netflix_url)
print("Please select one of the category")
df = pd.DataFrame(np.array(categories), columns=['Sr.No', 'Title', 'link'])
print(df.to_string(index=False))
choice = int(input('\n\n Please Enter your Choice: \n'))
subCategories, movieList, count = browseCategory(choice, categories)
for i in range(0, count):
print(subCategories[i],'\n\n')
subCategory_df = pd.DataFrame(np.array(movieList[i]), columns=['Sr.No', 'Title', 'link'])
print(subCategory_df.to_string(index=False))
print("\n\n\n")
if __name__ == '__main__':
main()
```
|
github_jupyter
|
# Explain Attacking BERT models using CAptum
Captum is a PyTorch library to explain neural networks
Here we show a minimal example using Captum to explain BERT models from TextAttack
[](https://colab.research.google.com/github/QData/TextAttack/blob/master/docs/2notebook/Example_5_Explain_BERT.ipynb)
[](https://github.com/QData/TextAttack/blob/master/docs/2notebook/Example_5_Explain_BERT.ipynb)
```
import torch
from copy import deepcopy
from textattack.datasets import HuggingFaceDataset
from textattack.models.tokenizers import AutoTokenizer
from textattack.models.wrappers import HuggingFaceModelWrapper
from textattack.models.wrappers import ModelWrapper
from transformers import AutoModelForSequenceClassification
from captum.attr import IntegratedGradients, LayerConductance, LayerIntegratedGradients, LayerDeepLiftShap, InternalInfluence, LayerGradientXActivation
from captum.attr import visualization as viz
device = torch.device("cuda:2" if torch.cuda.is_available() else "cpu")
print(device)
torch.cuda.set_device(device)
dataset = HuggingFaceDataset("ag_news", None, "train")
original_model = AutoModelForSequenceClassification.from_pretrained("textattack/bert-base-uncased-ag-news")
original_tokenizer = AutoTokenizer("textattack/bert-base-uncased-ag-news")
model = HuggingFaceModelWrapper(original_model,original_tokenizer)
def captum_form(encoded):
input_dict = {k: [_dict[k] for _dict in encoded] for k in encoded[0]}
batch_encoded = { k: torch.tensor(v).to(device) for k, v in input_dict.items()}
return batch_encoded
def get_text(tokenizer,input_ids,token_type_ids,attention_mask):
list_of_text = []
number = input_ids.size()[0]
for i in range(number):
ii = input_ids[i,].cpu().numpy()
tt = token_type_ids[i,]
am = attention_mask[i,]
txt = tokenizer.decode(ii, skip_special_tokens=True)
list_of_text.append(txt)
return list_of_text
sel =2
encoded = model.tokenizer.batch_encode([dataset[i][0]['text'] for i in range(sel)])
labels = [dataset[i][1] for i in range(sel)]
batch_encoded = captum_form(encoded)
clone = deepcopy(model)
clone.model.to(device)
def calculate(input_ids,token_type_ids,attention_mask):
#convert back to list of text
return clone.model(input_ids,token_type_ids,attention_mask)[0]
# x = calculate(**batch_encoded)
lig = LayerIntegratedGradients(calculate, clone.model.bert.embeddings)
# lig = InternalInfluence(calculate, clone.model.bert.embeddings)
# lig = LayerGradientXActivation(calculate, clone.model.bert.embeddings)
bsl = torch.zeros(batch_encoded['input_ids'].size()).type(torch.LongTensor).to(device)
labels = torch.tensor(labels).to(device)
attributions,delta = lig.attribute(inputs=batch_encoded['input_ids'],
baselines=bsl,
additional_forward_args=(batch_encoded['token_type_ids'], batch_encoded['attention_mask']),
n_steps = 10,
target = labels,
return_convergence_delta=True
)
atts = attributions.sum(dim=-1).squeeze(0)
atts = atts / torch.norm(atts)
# print(attributions.size())
atts = attributions.sum(dim=-1).squeeze(0)
atts = atts / torch.norm(atts)
from textattack.attack_recipes import PWWSRen2019
attack = PWWSRen2019.build(model)
results_iterable = attack.attack_dataset(dataset, indices=range(10))
viz_list = []
for n,result in enumerate(results_iterable):
orig = result.original_text()
pert = result.perturbed_text()
encoded = model.tokenizer.batch_encode([orig])
batch_encoded = captum_form(encoded)
x = calculate(**batch_encoded)
print(x)
print(dataset[n][1])
pert_encoded = model.tokenizer.batch_encode([pert])
pert_batch_encoded = captum_form(pert_encoded)
x_pert = calculate(**pert_batch_encoded)
attributions,delta = lig.attribute(inputs=batch_encoded['input_ids'],
# baselines=bsl,
additional_forward_args=(batch_encoded['token_type_ids'], batch_encoded['attention_mask']),
n_steps = 10,
target = torch.argmax(calculate(**batch_encoded)).item(),
return_convergence_delta=True
)
attributions_pert,delta_pert = lig.attribute(inputs=pert_batch_encoded['input_ids'],
# baselines=bsl,
additional_forward_args=(pert_batch_encoded['token_type_ids'], pert_batch_encoded['attention_mask']),
n_steps = 10,
target = torch.argmax(calculate(**pert_batch_encoded)).item(),
return_convergence_delta=True
)
orig = original_tokenizer.tokenizer.tokenize(orig)
pert = original_tokenizer.tokenizer.tokenize(pert)
atts = attributions.sum(dim=-1).squeeze(0)
atts = atts / torch.norm(atts)
atts_pert = attributions_pert.sum(dim=-1).squeeze(0)
atts_pert = atts_pert / torch.norm(atts)
all_tokens = original_tokenizer.tokenizer.convert_ids_to_tokens(batch_encoded['input_ids'][0])
all_tokens_pert = original_tokenizer.tokenizer.convert_ids_to_tokens(pert_batch_encoded['input_ids'][0])
v = viz.VisualizationDataRecord(
atts[:45].detach().cpu(),
torch.max(x).item(),
torch.argmax(x,dim=1).item(),
dataset[n][1],
2,
atts.sum().detach(),
all_tokens[:45],
delta)
v_pert = viz.VisualizationDataRecord(
atts_pert[:45].detach().cpu(),
torch.max(x_pert).item(),
torch.argmax(x_pert,dim=1).item(),
dataset[n][1],
2,
atts_pert.sum().detach(),
all_tokens_pert[:45],
delta_pert)
viz_list.append(v)
viz_list.append(v_pert)
# print(result.perturbed_text())
print(result.__str__(color_method='ansi'))
print('\033[1m', 'Visualizations For AG NEWS', '\033[0m')
viz.visualize_text(viz_list)
# reference for viz datarecord
# def __init__(
# self,
# word_attributions,
# pred_prob,
# pred_class,
# true_class,
# attr_class,
# attr_score,
# raw_input,
# convergence_score,
# ):
```
|
github_jupyter
|
```
import pandas as pd
import datetime
import vk_api
import os
import requests
import json
import random
%matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sns
import sys
token = '4e6e771d37dbcbcfcc3b53d291a274d3ae21560a2e81f058a7c177aff044b5141941e89aff1fead50be4f'
vk_session = vk_api.VkApi(token=token)
vk = vk_session.get_api()
vk.messages.send(
chat_id=1,
random_id=2,
message='Matrix has you ...')
df = pd.read_csv('/home/jupyter-an.karpov/shared/ads_data.csv.zip', compression='zip')
ad_data = df.groupby(['ad_id', 'event'], as_index=False) \
.agg({'user_id': 'count'})
ad_data = ad_data.pivot(index='ad_id', columns='event', values='user_id').reset_index()
ad_data = ad_data.fillna(0).assign(ctr=ad_data.click / ad_data.view)
top_ctr = ad_data.query('click > 20 & view > 100').sort_values('ctr').tail(10)
top_ctr.to_csv('top_ctr_data.csv', index=False)
path = '/home/jupyter-an.karpov/lesson_7/top_ctr_data.csv'
file_name = 'top_ctr_data.csv'
path_to_file = path
upload_url = vk.docs.getMessagesUploadServer(peer_id=2000000001)["upload_url"]
file = {'file': (file_name, open(path_to_file, 'rb'))}
response = requests.post(upload_url, files=file)
json_data = json.loads(response.text)
json_data
saved_file = vk.docs.save(file=json_data['file'], title=file_name)
saved_file
attachment = 'doc{}_{}'.format(saved_file['doc']['owner_id'], saved_file['doc']['id'])
attachment
vk.messages.send(
chat_id=1,
random_id=3,
message='Топ объявлений по CTR',
attachment = attachment
)
import gspread
from df2gspread import df2gspread as d2g
from oauth2client.service_account import ServiceAccountCredentials
scope = ['https://spreadsheets.google.com/feeds',
'https://www.googleapis.com/auth/drive']
my_mail = '[email protected]'
# Authorization
credentials = ServiceAccountCredentials.from_json_keyfile_name('heroic-venture-268009-1dfbcc34e5fa.json', scope)
gs = gspread.authorize(credentials)
# Name of the table in google sheets,
# can be url for open_by_url
# or id (key) part for open_by_key
table_name = 'to_sequence' # Your table
# Get this table
work_sheet = gs.open(table_name)
# Select 1st sheet
sheet1 = work_sheet.sheet1
# Get data in python lists format
data = sheet1.get_all_values()
data
headers = data.pop(0)
df = pd.DataFrame(data, columns=headers)
df
df.sort_values('price', ascending=False)
sheet1.append_row([500, 'group_4'])
# Looks like spreadsheet should be already present at the dist (so, run code in create table section)
spreadsheet_name = 'A new spreadsheet'
sheet = 'KarpovCorses2'
d2g.upload(df, spreadsheet_name, sheet, credentials=credentials, row_names=True)
url = 'https://api-metrika.yandex.net/stat/v1/data?'
visits = 'metrics=ym:s:visits&dimensions=ym:s:date&id=44147844'
vistis_url = url + visits
vistis_request = requests.get(vistis_url)
vistis_request
json_data = json.loads(vistis_request.text)
json_data['data']
y_df = pd.DataFrame([(i['dimensions'][0]['name'],
i['metrics'][0]) for i in json_data['data']], columns=['date',
'visits'])
spreadsheet_name = 'A new spreadsheet'
sheet = 'Yandex_visits'
d2g.upload(y_df, spreadsheet_name, sheet, credentials=credentials, row_names=True)
```
|
github_jupyter
|
# Software Analytics Mini Tutorial Part I: Jupyter Notebook and Python basics
## Introduction
This series of notebooks are a simple mini tutorial to introduce you to the basic functionality of Jupyter, Python, pandas and matplotlib. The comprehensive explanations should guide you to be able to analyze software data on your own. Therefore, the examples is chosen in such a way that we come across the typical methods in a data analysis. Have fun!
*This is part I: The basics of Jupyter Notebook and Python. For the data analysis framework pandas and the visualization library matplotlib, go to [the other tutorial](10%20pandas%20and%20matplotlib%20basics.ipynb).*
## The Jupyter Notebook System
First, we'll take a closer look at Jupyter Notebook. What you see here is Jupyter, the interactive notebook environment for programming. Jupyter Notebook allows us us to write code and documentation in executable **cells**.
We see below a cell in which we can enter Python code.
#### Execute a cell
1. select the next cell (mouse click or arrow keys).
1. type in for example `"Hello World!`.
1. execute the cell with a `Ctrl`+`Enter`.
1. click on the cell again
1. execute the cell with `Shift`+`Enter`.
##### Discussion
* What's the difference between the two different ways of executing cells?
#### Create a new cell
We will use the built-in keyboard shortcuts for this:
1. if not happened yet, click on this cell.
1. enter **command mode**, selectable with `Esc` key.
1. create a **new cell** after this text by pressing the key `b`.
1. change the **cell type** to **Markdown** with key `m`.
1. switch to **edit mode** with `Enter`
1. write a text
1. execute cell with `Ctrl` + `Enter`.
*Additional information:*
* We've seen an important feature of Jupyter Notebook: The distinction between **command mode** (accessible via the `Esc` key) and **edit mode** (accessible via the `Enter` key). Note the differences:
* In command mode, the border of the current cell is blue. This mode allows you to manipulate the **notebook**'s content.
* In edit mode, the border turns green. This allows you to manipulate the **cell**'s content.
* **Markdown** is a simple markup language that can be used to write and format texts. This allows us to directly document the steps we have taken.
## Python Basics
Let's take a look at some basic Python programming constructs that we will need later when working with the pandas data analysis framework.
We look at very basic functions:
* variable assignments
* value range accesses
* method calls
#### Assign text to a variable
1. **assign** the text **value** "Hello World" to the **variable** `text` by using the syntax `<variable> = <value>`.
1. type the variable `text` in the next line and execute the cell.
1. execute the cell (this will be necessary for each upcoming cell, so we won't mention it from now on)
#### Accessing slices of information
By using the array notation with the square brackets `[` and `]` (the slice operators), we can access the first letter of our text with a 0-based index (this also works for other types like lists).
1. access the first letter in `text` with `[0]`.
#### Select last character
1. access the last letter in `text` with `[-1]`.
#### Select ranges
1. access a range of `text` with the **slice** `[2:5]`.
#### Select open ranges
1. access an open range with the slice `[:5]` (which is an abbreviation for a 0-based slice `[0:5]`)
#### Reverse a list of values
1. reverse the text (or a list) by using the `::` notation with an following `-1`.
#### Use auto completion and execute a method
1. append a `.` to `text` and look at the functions with the `Tab` key.
1. find and execute the **method** `upper()` of the `text` object (Tip: Type a `u` when using auto completion).
#### Execute a method with parameters
...and find out how this works by using the integrated, interactive documentation:
1. select the `split` method of `text`.
1. press `Shift`+`Tab`.
1. press `Shift`+`Tab` twice in quick succession.
1. press `Shift`+`Tab` three times in quick succession (and then `ESC` to hide).
1. read the documentation of `split`
1. split the text in `text` with `split` exactly once (**parameter** `maxsplit`) apart by using an `l` (a lower case "L") as separator (parameter `sep`).
*Note: What we are using here additionally is Python's feature to swap the methods parameter. This works, if you assign the inputs directly to the method argument's name (e.g. `maxsplit=1`).
## Summary
OK, we were just warming up! Proceed to [the next section](10%20pandas%20and%20matplotlib%20basics.ipynb)
If you want to dive deeper into this topic, take a look at my [blog posts on that topic](http://www.feststelltaste.de/category/software-analytics/) or my microsite [softwareanalytics.de](https://softwareanalytics.de/). I'm looking forward to your comments and feedback on [GitHub](https://github.com/feststelltaste/software-analytics-workshop/issues) or on [Twitter](https://www.twitter.com/feststelltaste)!
|
github_jupyter
|
```
## plot the histogram showing the modeled and labeled result
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
# for loop version
def read_comp(file):
Pwave = {}
Pwave['correct'] = []
Pwave['wrongphase'] = []
Pwave['miss'] = 0
Pwave['multiphase'] = []
with open(file, 'r') as comp:
for line in comp:
pickline = line.split()
if pickline[0].strip()[-1] == pickline[1].strip() and len(pickline)==3:
Pwave['correct'].append(float(pickline[2][:-2]))
if pickline[0].strip()[-1] != pickline[1].strip() and pickline[1].strip() != 'N':
Pwave['wrongphase'].append (float(pickline[2][:-2]))
if pickline[0].strip()[-1] == pickline[1].strip() and len(pickline)>3:
Pwave['multiphase'].append(len(pickline)-2)
if pickline[1].strip() == 'N':
Pwave['miss'] +=1
return Pwave
# run all the output file
ty = ['EQS','EQP','SUS','SUP','THS','THP','SNS','SNP','PXS','PXP']
wave = {}
for name in ty:
wave[name] = read_comp('../comparison_out/comp.'+name+'.out')
# plot histogram of the correct plot
def plotfig(name):
fig, ax = plt.subplots(figsize = (10,6))
# filename = wave['EQP']['correct']
fig = plt.hist(name,bins= 10)
ax.set_ylabel('number', fontsize=15)
ax.set_xlabel('time difference (s)', fontsize=15)
ax.set_title('Phase with time difference')
plt.xticks(fontsize=15)
#plt.xticks( rotation='vertical')
plt.xticks(np.arange(-10, 10, step=1))
plt.yticks(fontsize=15)
plt.legend(fontsize=15)
#plt.savefig('test.jpg')
for k in wave.keys():
for t in wave[k].keys():
# print (k, t)
fig, ax = plt.subplots(figsize = (10,6))
# filename = wave['EQP']['correct']
fig = plt.hist(wave[k][t],bins= 10)
ax.set_ylabel('number', fontsize=15)
ax.set_xlabel('time difference (s)', fontsize=15)
ax.set_title('{} Phase with time difference for {}'.format(t,k))
plt.xticks(fontsize=15)
#plt.xticks( rotation='vertical')
plt.xticks(np.arange(-10, 10, step=1))
plt.yticks(fontsize=15)
#plt.legend(fontsize=15)
#plt.savefig('{}_{}.jpg'.format(k,t))
plt.hist(wave['EQS']['correct'])
#plt.xlim(0, 50)
plt.savefig('time_diff.jpg')
plotfig(wave['EQS']['wrongphase'])
plotfig(wave['EQS']['multiphase'])
plotfig(wave['EQS']['correct'])
# plot histogram of the wrongphase plot
fig, ax = plt.subplots(figsize = (10,6))
#k = np.random.normal(float(test['EQS'][ID]['time']), 3, 1000)
fig = plt.hist(wave['EQP']['wrongphase'])
ax.set_ylabel('number', fontsize=15)
ax.set_xlabel('time difference (S)', fontsize=15)
ax.set_title('Correct Phase with time difference')
plt.xticks(fontsize=15)
plt.yticks(fontsize=15)
plt.xticks(np.arange(-10, 10, step=1))
plt.legend(fontsize=15)
# plot histogram of the wrongphase plot
fig, ax = plt.subplots(figsize = (10,6))
#k = np.random.normal(float(test['EQS'][ID]['time']), 3, 1000)
fig = plt.hist(test['multiphase'])
ax.set_ylabel('number', fontsize=15)
ax.set_xlabel('time difference (S)', fontsize=15)
ax.set_title('Correct Phase with time difference')
plt.xticks(fontsize=15)
plt.yticks(fontsize=15)
#plt.xticks(np.arange(-10, 10, step=1))
plt.legend(fontsize=15)
comp = pd. read_csv('../comparison.out', names=['type','mod_type','time'],sep = ' ')
comp.head()
comp['mod_type'].value_counts()
comp['time'][comp['type']=='THP'].describe()
comp = pd. read_csv('../comparison_out/comp.EQP.out', sep = '/s+')
comp.head()
with open('../comparison_out/comp.EQP.out', 'r') as comp:
leng = []
for line in comp:
pickline = line.split(' ')
leng.append(len(pickline))
max(leng)
```
|
github_jupyter
|
```
%matplotlib inline
```
torchaudio Tutorial
===================
PyTorch is an open source deep learning platform that provides a
seamless path from research prototyping to production deployment with
GPU support.
Significant effort in solving machine learning problems goes into data
preparation. ``torchaudio`` leverages PyTorch’s GPU support, and provides
many tools to make data loading easy and more readable. In this
tutorial, we will see how to load and preprocess data from a simple
dataset.
For this tutorial, please make sure the ``matplotlib`` package is
installed for easier visualization.
```
import torch
import torchaudio
import matplotlib.pyplot as plt
```
Opening a file
-----------------
``torchaudio`` also supports loading sound files in the wav and mp3 format. We
call waveform the resulting raw audio signal.
```
filename = "../_static/img/steam-train-whistle-daniel_simon-converted-from-mp3.wav"
waveform, sample_rate = torchaudio.load(filename)
print("Shape of waveform: {}".format(waveform.size()))
print("Sample rate of waveform: {}".format(sample_rate))
plt.figure()
plt.plot(waveform.t().numpy())
```
When you load a file in ``torchaudio``, you can optionally specify the backend to use either
`SoX <https://pypi.org/project/sox/>`_ or `SoundFile <https://pypi.org/project/SoundFile/>`_
via ``torchaudio.set_audio_backend``. These backends are loaded lazily when needed.
``torchaudio`` also makes JIT compilation optional for functions, and uses ``nn.Module`` where possible.
Transformations
---------------
``torchaudio`` supports a growing list of
`transformations <https://pytorch.org/audio/transforms.html>`_.
- **Resample**: Resample waveform to a different sample rate.
- **Spectrogram**: Create a spectrogram from a waveform.
- **GriffinLim**: Compute waveform from a linear scale magnitude spectrogram using
the Griffin-Lim transformation.
- **ComputeDeltas**: Compute delta coefficients of a tensor, usually a spectrogram.
- **ComplexNorm**: Compute the norm of a complex tensor.
- **MelScale**: This turns a normal STFT into a Mel-frequency STFT,
using a conversion matrix.
- **AmplitudeToDB**: This turns a spectrogram from the
power/amplitude scale to the decibel scale.
- **MFCC**: Create the Mel-frequency cepstrum coefficients from a
waveform.
- **MelSpectrogram**: Create MEL Spectrograms from a waveform using the
STFT function in PyTorch.
- **MuLawEncoding**: Encode waveform based on mu-law companding.
- **MuLawDecoding**: Decode mu-law encoded waveform.
- **TimeStretch**: Stretch a spectrogram in time without modifying pitch for a given rate.
- **FrequencyMasking**: Apply masking to a spectrogram in the frequency domain.
- **TimeMasking**: Apply masking to a spectrogram in the time domain.
Each transform supports batching: you can perform a transform on a single raw
audio signal or spectrogram, or many of the same shape.
Since all transforms are ``nn.Modules`` or ``jit.ScriptModules``, they can be
used as part of a neural network at any point.
To start, we can look at the log of the spectrogram on a log scale.
```
specgram = torchaudio.transforms.Spectrogram()(waveform)
print("Shape of spectrogram: {}".format(specgram.size()))
plt.figure()
plt.imshow(specgram.log2()[0,:,:].numpy(), cmap='gray')
```
Or we can look at the Mel Spectrogram on a log scale.
```
specgram = torchaudio.transforms.MelSpectrogram()(waveform)
print("Shape of spectrogram: {}".format(specgram.size()))
plt.figure()
p = plt.imshow(specgram.log2()[0,:,:].detach().numpy(), cmap='gray')
```
We can resample the waveform, one channel at a time.
```
new_sample_rate = sample_rate/10
# Since Resample applies to a single channel, we resample first channel here
channel = 0
transformed = torchaudio.transforms.Resample(sample_rate, new_sample_rate)(waveform[channel,:].view(1,-1))
print("Shape of transformed waveform: {}".format(transformed.size()))
plt.figure()
plt.plot(transformed[0,:].numpy())
```
As another example of transformations, we can encode the signal based on
Mu-Law enconding. But to do so, we need the signal to be between -1 and
1. Since the tensor is just a regular PyTorch tensor, we can apply
standard operators on it.
```
# Let's check if the tensor is in the interval [-1,1]
print("Min of waveform: {}\nMax of waveform: {}\nMean of waveform: {}".format(waveform.min(), waveform.max(), waveform.mean()))
```
Since the waveform is already between -1 and 1, we do not need to
normalize it.
```
def normalize(tensor):
# Subtract the mean, and scale to the interval [-1,1]
tensor_minusmean = tensor - tensor.mean()
return tensor_minusmean/tensor_minusmean.abs().max()
# Let's normalize to the full interval [-1,1]
# waveform = normalize(waveform)
```
Let’s apply encode the waveform.
```
transformed = torchaudio.transforms.MuLawEncoding()(waveform)
print("Shape of transformed waveform: {}".format(transformed.size()))
plt.figure()
plt.plot(transformed[0,:].numpy())
```
And now decode.
```
reconstructed = torchaudio.transforms.MuLawDecoding()(transformed)
print("Shape of recovered waveform: {}".format(reconstructed.size()))
plt.figure()
plt.plot(reconstructed[0,:].numpy())
```
We can finally compare the original waveform with its reconstructed
version.
```
# Compute median relative difference
err = ((waveform-reconstructed).abs() / waveform.abs()).median()
print("Median relative difference between original and MuLaw reconstucted signals: {:.2%}".format(err))
```
Functional
---------------
The transformations seen above rely on lower level stateless functions for their computations.
These functions are available under ``torchaudio.functional``. The complete list is available
`here <https://pytorch.org/audio/functional.html>`_ and includes:
- **istft**: Inverse short time Fourier Transform.
- **gain**: Applies amplification or attenuation to the whole waveform.
- **dither**: Increases the perceived dynamic range of audio stored at a
particular bit-depth.
- **compute_deltas**: Compute delta coefficients of a tensor.
- **equalizer_biquad**: Design biquad peaking equalizer filter and perform filtering.
- **lowpass_biquad**: Design biquad lowpass filter and perform filtering.
- **highpass_biquad**:Design biquad highpass filter and perform filtering.
For example, let's try the `mu_law_encoding` functional:
```
mu_law_encoding_waveform = torchaudio.functional.mu_law_encoding(waveform, quantization_channels=256)
print("Shape of transformed waveform: {}".format(mu_law_encoding_waveform.size()))
plt.figure()
plt.plot(mu_law_encoding_waveform[0,:].numpy())
```
You can see how the output fron ``torchaudio.functional.mu_law_encoding`` is the same as
the output from ``torchaudio.transforms.MuLawEncoding``.
Now let's experiment with a few of the other functionals and visualize their output. Taking our
spectogram, we can compute it's deltas:
```
computed = torchaudio.functional.compute_deltas(specgram, win_length=3)
print("Shape of computed deltas: {}".format(computed.shape))
plt.figure()
plt.imshow(computed.log2()[0,:,:].detach().numpy(), cmap='gray')
```
We can take the original waveform and apply different effects to it.
```
gain_waveform = torchaudio.functional.gain(waveform, gain_db=5.0)
print("Min of gain_waveform: {}\nMax of gain_waveform: {}\nMean of gain_waveform: {}".format(gain_waveform.min(), gain_waveform.max(), gain_waveform.mean()))
dither_waveform = torchaudio.functional.dither(waveform)
print("Min of dither_waveform: {}\nMax of dither_waveform: {}\nMean of dither_waveform: {}".format(dither_waveform.min(), dither_waveform.max(), dither_waveform.mean()))
```
Another example of the capabilities in ``torchaudio.functional`` are applying filters to our
waveform. Applying the lowpass biquad filter to our waveform will output a new waveform with
the signal of the frequency modified.
```
lowpass_waveform = torchaudio.functional.lowpass_biquad(waveform, sample_rate, cutoff_freq=3000)
print("Min of lowpass_waveform: {}\nMax of lowpass_waveform: {}\nMean of lowpass_waveform: {}".format(lowpass_waveform.min(), lowpass_waveform.max(), lowpass_waveform.mean()))
plt.figure()
plt.plot(lowpass_waveform.t().numpy())
```
We can also visualize a waveform with the highpass biquad filter.
```
highpass_waveform = torchaudio.functional.highpass_biquad(waveform, sample_rate, cutoff_freq=2000)
print("Min of highpass_waveform: {}\nMax of highpass_waveform: {}\nMean of highpass_waveform: {}".format(highpass_waveform.min(), highpass_waveform.max(), highpass_waveform.mean()))
plt.figure()
plt.plot(highpass_waveform.t().numpy())
```
Migrating to torchaudio from Kaldi
----------------------------------
Users may be familiar with
`Kaldi <http://github.com/kaldi-asr/kaldi>`_, a toolkit for speech
recognition. ``torchaudio`` offers compatibility with it in
``torchaudio.kaldi_io``. It can indeed read from kaldi scp, or ark file
or streams with:
- read_vec_int_ark
- read_vec_flt_scp
- read_vec_flt_arkfile/stream
- read_mat_scp
- read_mat_ark
``torchaudio`` provides Kaldi-compatible transforms for ``spectrogram``,
``fbank``, ``mfcc``, and ``resample_waveform with the benefit of GPU support, see
`here <compliance.kaldi.html>`__ for more information.
```
n_fft = 400.0
frame_length = n_fft / sample_rate * 1000.0
frame_shift = frame_length / 2.0
params = {
"channel": 0,
"dither": 0.0,
"window_type": "hanning",
"frame_length": frame_length,
"frame_shift": frame_shift,
"remove_dc_offset": False,
"round_to_power_of_two": False,
"sample_frequency": sample_rate,
}
specgram = torchaudio.compliance.kaldi.spectrogram(waveform, **params)
print("Shape of spectrogram: {}".format(specgram.size()))
plt.figure()
plt.imshow(specgram.t().numpy(), cmap='gray')
```
We also support computing the filterbank features from waveforms,
matching Kaldi’s implementation.
```
fbank = torchaudio.compliance.kaldi.fbank(waveform, **params)
print("Shape of fbank: {}".format(fbank.size()))
plt.figure()
plt.imshow(fbank.t().numpy(), cmap='gray')
```
You can create mel frequency cepstral coefficients from a raw audio signal
This matches the input/output of Kaldi’s compute-mfcc-feats.
```
mfcc = torchaudio.compliance.kaldi.mfcc(waveform, **params)
print("Shape of mfcc: {}".format(mfcc.size()))
plt.figure()
plt.imshow(mfcc.t().numpy(), cmap='gray')
```
Available Datasets
-----------------
If you do not want to create your own dataset to train your model, ``torchaudio`` offers a
unified dataset interface. This interface supports lazy-loading of files to memory, download
and extract functions, and datasets to build models.
The datasets ``torchaudio`` currently supports are:
- **VCTK**: Speech data uttered by 109 native speakers of English with various accents
(`Read more here <https://homepages.inf.ed.ac.uk/jyamagis/page3/page58/page58.html>`_).
- **Yesno**: Sixty recordings of one individual saying yes or no in Hebrew; each
recording is eight words long (`Read more here <https://www.openslr.org/1/>`_).
- **Common Voice**: An open source, multi-language dataset of voices that anyone can use
to train speech-enabled applications (`Read more here <https://voice.mozilla.org/en/datasets>`_).
- **LibriSpeech**: Large-scale (1000 hours) corpus of read English speech (`Read more here <http://www.openslr.org/12>`_).
```
yesno_data = torchaudio.datasets.YESNO('./', download=True)
# A data point in Yesno is a tuple (waveform, sample_rate, labels) where labels is a list of integers with 1 for yes and 0 for no.
# Pick data point number 3 to see an example of the the yesno_data:
n = 3
waveform, sample_rate, labels = yesno_data[n]
print("Waveform: {}\nSample rate: {}\nLabels: {}".format(waveform, sample_rate, labels))
plt.figure()
plt.plot(waveform.t().numpy())
```
Now, whenever you ask for a sound file from the dataset, it is loaded in memory only when you ask for it.
Meaning, the dataset only loads and keeps in memory the items that you want and use, saving on memory.
Conclusion
----------
We used an example raw audio signal, or waveform, to illustrate how to
open an audio file using ``torchaudio``, and how to pre-process,
transform, and apply functions to such waveform. We also demonstrated how
to use familiar Kaldi functions, as well as utilize built-in datasets to
construct our models. Given that ``torchaudio`` is built on PyTorch,
these techniques can be used as building blocks for more advanced audio
applications, such as speech recognition, while leveraging GPUs.
|
github_jupyter
|
# EEP/IAS 118 - Section 6
## Fixed Effects Regression
### August 1, 2019
Today we will practice with fixed effects regressions in __R__. We have two different ways to estimate the model, and we will see how to do both and the situations in which we might favor one versus the other.
Let's give this a try using the dataset `wateruse.dta`, a panel of residential water use for residents in Alameda and Contra Costa Counties. The subset of households are high water users, people who used over 1,000 gallons per billing cycle. We have information on their water use, weather during the period, as well as information on the city and zipcode of where the home is located, and information on the size and value of the house.
Suppose we are interested in running the following panel regression of residential water use:
$$ GPD_{it} = \beta_0 + \beta_1 degree\_days_{it} + \beta_2 precip_{it} ~~~~~~~~~~~~~~~~~~~~~~~(1)$$
Where $GPD$ is the gallons used per day by household $i$ in billing cycle $t$, $degree\_days$ the count of degree days experienced by the household in that billing cycle (degree days are a measure of cumulative time spent above a certain temperature threshold), and $precip$ the amount of precipitation in millimeters.
```
library(tidyverse)
library(haven)
library(lfe)
waterdata <- read_dta("wateruse.dta") %>%
mutate(gpd = (unit*748)/num_days)
waterdata <- mutate(waterdata,
n = 1:nrow(waterdata))
head(waterdata)
dim(waterdata)
reg1 <- lm(gpd ~ degree_days + precip, data = waterdata)
summary(reg1)
```
Here we obtain an estimate of $\hat\beta_1 = 0.777$, telling us that an additional degree day per billing cycle is associated with an additional $0.7769$ gallon used per day. These billing cycles are roughly two months long, so this suggests an increase of roughly 47 gallons per billing cycle. Our estimate is statistically significant at all conventional levels, suggesting residential water use does respond to increased exposure to high heat.
We estimate a statistically insignificant coefficient on additional precipitation, which tells us that on average household water use in our sample doesn't adjust to how much it rains.
We might think that characteristics of the home impact how much water is used there, so we add in some home controls:
$$ GPD_{it} = \beta_0 + \beta_1 degree\_days_{it} + \beta_2 precip_{it} + \beta_3 lotsize_{i} + \beta_4 homesize_i + \beta_5 num\_baths_i + \beta_6 num\_beds_i + \beta_7 homeval_i~~~~~~~~~~~~~~~~~~~~~~~(2)$$
```
reg2 <- lm(gpd ~ degree_days + precip + lotsize + homesize + num_baths + num_beds + homeval, data = waterdata)
summary(reg2)
```
Our coefficient on $degree\_days$ remains statistically significant and doesn't change much, so we find that $\hat\beta_1$ is robust to the addition of home characteristics. Of these characteristics, we obtain statistically significant coefficients on the size of the lot (in acres), the size of the home ($ft^2$), and the number of bedrooms in the home.
We get a curious result for $\hat\beta_6$: for each additional bedroom in the home we predict that water use will _fall_ by 48 gallons per day.
### Discussion: what might be driving this effect?
```
waterdata %>%
filter( city <= 9) %>%
ggplot(aes(x=num_beds, y=gpd)) +
geom_point() +
facet_grid(. ~ city)
waterdata %>%
filter(city> 9 & city <= 18) %>%
ggplot(aes(x=num_beds, y=gpd)) +
geom_point() +
facet_grid(. ~ city)
waterdata %>%
filter( city> 18) %>%
ggplot(aes(x=num_beds, y=gpd)) +
geom_point() +
facet_grid(. ~ city)
```
Since there are likely a number of sources of omitted variable bias in the previous model, we think it might be worth including some fixed effects in our model.
## Method 1: Fixed Effects with lm()
Up to this point we have been running our regressions using the `lm()` function. We can still use `lm()` for our fixed effects models, but it takes some more work.
Recall that we can write our general panel fixed effects model as
$$ y_{it} = \beta x_{it} + \mathbf{a}_i + {d}_t + u_{it} $$
* $y$ our outcome of interest, which varies in both the time and cross-sectional dimensions
* $x_{it}$ our set of time-varying unit characteristics
* $\mathbf{a}_i$ our set of unit fixed effects
* $d_t$ our time fixed effects
We can estimate this model in `lm()` provided we have variables in our dataframe that correspond to $a_i$ and $d_t$. This means we'll have to generate them before we can run any regression.
### Generating Dummy Variables
In order to include fixed effects for our regression, we have to first generate the set of dummy variables that we want. For example, if we want to include a set of city fixed effects in our model, we need to generate them.
We can do this in a few ways.
1. First, we can use `mutate()` and add a separate line for each individual city:
```
fe_1 <- waterdata %>%
mutate(city_1 = as.numeric((city==1)),
city_2 = as.numeric((city ==2)),
city_3 = as.numeric((city ==3))) %>%
select(n, city, city_1, city_2, city_3)
head(fe_1)
```
This can be super tedious though when we have a bunch of different levels of our variable that we want to make fixed effects for. In this case, we have 27 different cities.
2. Alternatively, we can use the `spread()` function to help us out. Here we add in a constant variable `v` that is equal to one in all rows, and a copy of city that adds "city_" to the front of the city number. Then we pass the data to `spread`, telling it to split the variable `cty` into dummy variables for all its levels, with all the "false" cases filled with zeros.
```
fe_2 <- waterdata %>%
select(n, city)
head(fe_2)
fe_2 %>%
mutate(v = 1, cty = paste0("city_", city)) %>%
spread(cty, v, fill = 0)
```
That is much easier! Let's now do that so that they'll be in `waterdata`:
```
waterdata <- waterdata %>%
mutate(v = 1, cty = paste0("city_", city)) %>%
spread(cty, v, fill = 0)
head(waterdata)
names(waterdata)
```
Note that both of the variables we used in `spread` are no longer in our dataset.
While we're at it, let's also add in a set of billing cycle fixed effects.
```
waterdata <- waterdata %>%
mutate(v = 1, cyc = paste0("cycle_", billingcycle)) %>%
spread(cyc, v, fill = 0)
head(waterdata)
```
Now we have all our variables to run the regression
$$ GPD_{it} = \beta_0 + \beta_1 degree\_days_{it} + \beta_2 precip_{it} + \mathbf{a}_i + \mathbf{d}_t~~~~~~~~~~~~~~~~~~~~~~~(2)$$
Where $\mathbf{a}_i$ are our city fixed effects, and $\mathbf{d}_t$ our billing cycle fixed effects.
Now we can run our model! Well, now we can _write out_ our model. The challenge here is that we need to specify all of the dummy variables in our formula. We could do this all by hand, but when we end up with a bunch of fixed effects it's easier to use the following trick: we can write `y ~ .` to tell __R__ we want it to put every variable in our dataset other than $y$ on the right hand side of our regression. That means we can create a version of our dataset with only $gpd$, $degree\_days$, $precip$, and our fixed effects and won't have to write out all those fixed effects by hand!
Note that we can use `select()` and `-` to remove variables from our dataframe. If there is a list of variables in order that we want to get rid of, we can do it easily by passing a vector through! For instance, we want to get rid of the first 12 variables in our data, so we can add `-unit:-hh` in select to get rid of them all at once. if we separate with a comma, we can drop other sections of our data too!
```
fe_data <- waterdata %>%
select(-unit:-hh, -city, -n)
head(fe_data)
fe_reg1 <- lm(gpd ~ ., data = fe_data)
summary(fe_reg1)
```
Since I specified it this way, __R__ chose the last dummy variable for each set of fixed effect to leave out as our omitted group.
Now that we account for which billing cycle we're in (i.e. whether we're in the winter or whether we're in the summer), we find that the coefficient on $degree\_days$ is now much smaller and statistically insignificant. This makes sense, as we were falsely attributing the extra water use that comes from seasonality to temperature on its own. Now that we control for the season we're in via billing cycle fixed effects, we find that deviations in temperature exposure during a billing cycle doesn't result in dramatically higher water use within the sample.
### Discussion: Why did we drop the home characteristics from our model?
## Method 2: Fixed Effects with felm()
Alternatively, we could do everything way faster using the `felm()` function from the package __lfe__. This package doesn't require us to produce all the dummy variables by hand. Further, it performs the background math way faster so will be much quicker to estimate models using large datasets and many variables.
The syntax we use is now
`felm(y ~ x1 + x2 + ... + xk | FE_1 + FE_2, data = df)`
* The first section $y \sim x1 + x2 +... xk$ is our formula, written the same way as with `lm()`
* We now add a `|` and in the second section we specify our fixed effects. Here we say $FE\_1 + FE\_2$ which tells __R__ to include fixed effects for each level of $FE\_1$ and $FE\_2$.
* Note that our fixed effect variables must be of class "factor" - we can force our variables to take this class by adding them as `as.factor(FE_1) + as.factor(FE_2)`.
* we add the data source after the comma, as before.
Let's go ahead and try this now with our water data model:
```
fe_reg2 <- felm(gpd ~ degree_days + precip | as.factor(city) + as.factor(billingcycle), data = waterdata)
summary(fe_reg2)
```
And we estimate the exact same coefficients on $degree\_days$ and $precip$ as in the case where we specified everything by hand! We didn't have to mutate our data or add any variables. The one potential downside is that this approach doesn't report the fixed effects themselves. However, the tradeoff is that `felm` runs a lot faster than `lm`. To see this, we can compare run times:
```
lm_start <- Sys.time()
fe_data <- waterdata %>%
mutate(v = 1, cyc = paste0("cycle_", billingcycle)) %>%
spread(cyc, v, fill = 0) %>%
mutate(v = 1, cty = paste0("city_", city)) %>%
spread(cty, v, fill = 0) %>%
select(-unit:-hh, -city, -n)
lm(gpd ~ ., data = fe_data)
lm_end <- Sys.time()
lm_dur <- lm_end - lm_start
felm_start <- Sys.time()
felm(gpd ~ degree_days + precip | as.factor(city) + as.factor(billingcycle), data = waterdata)
felm_end <- Sys.time()
felm_dur <- felm_end - felm_start
print(paste0("lm() duration is ", lm_dur, " seconds, while felm() duration is ", felm_dur, " seconds."))
```
Okay, neither of these models took very long, but that's because we only have two covariates other than our fixed effects and only around 2300 observations. If we have hundreds of covariates and millions of observations, this time difference becomes massive.
# Regression Discontinuity
Let's practice running a regression discontinuity model. Suppose we were interested in exploring the weird relationship we saw earlier between water use and number of bedrooms in a home. Let's take a look at that relationship a bit more closely.
```
waterdata %>%
ggplot(aes(x = num_beds, y = gpd)) +
geom_point( alpha = 0.4, colour = "royalblue")
```
We see that average water use appears to rise as we add bedrooms to a house from a low number, peaks when households have five bedrooms, then begins to fall with larger and larger houses... though there are a few high outliers in the 6-9 bedroom cases as well, which might overshadow that trend.
Is there something else that's correlated with the number of bedrooms in a home that may also be driving this?
```
waterdata %>%
ggplot(aes(x = num_beds, y = lotsize)) +
geom_point( alpha = 0.4, colour = "royalblue")
```
It looks like lotsize and the number of bedrooms share a similar relationship - lot size increasing in \# bedrooms up until 5, then declining from there.
Given that it looks like 5 bedrooms is where the relationship changes, let's use this as our running variable and allow the relationship for the number of bedrooms to differ around a threshold of five bedrooms. We can write an RD model as
$$ GPD_{it} = \beta_0 + \beta_1 T_i + \beta_2 (num\_beds - 5) + \beta_3\left( T_i \times (num\_beds - 5) \right) + u_{it} $$
```
rd_data <- waterdata %>%
select(gpd, num_beds, lotsize) %>%
mutate(treat = (num_beds > 5),
beds_below = num_beds - 5,
beds_above = treat * (num_beds - 5))
rd_reg <- lm(gpd ~ treat + beds_below + beds_above, data = rd_data)
summary(rd_reg)
```
What if we limit our comparison closer to around the threshold? Right now we're using data from the entire sample, but this might not be a valid comparison. Let's see what happens when we reduce our bandwidth to 3 and look at only homes with between 3 and 8 bedrooms.
```
rd_data_trim <- rd_data %>%
filter(!(num_beds < 2)) %>%
filter(!(num_beds > 8))
rd_reg2 <- lm(gpd ~ treat + beds_below + beds_above, data = rd_data_trim)
summary(rd_reg2)
```
We now estimate a treatment effect at the discontinuity! Our model finds a discontinuity, estimating a jump down of 284 gallons per day right around the 5 bedroom threshold.
However, we saw earlier that it appears that lotsize is correlated with the number of bedrooms in the home, and is definitely a factor correlated with residential water use. What happens to our LATE estimate when we control for lot size in our regression?
```
rd_reg3 <- lm(gpd ~ treat + beds_below + beds_above + lotsize, data = rd_data_trim)
summary(rd_reg3)
```
Once we control for lot size, our interpretation changes. Now we estimate a coefficient on treatment nearly half the magnitude as before and without statistical significance.
### Discussion Q: What does this tell us about the covariance between lot size and the number of bedrooms?
# Fixed Effects Practice Question #1
#### From a random sample of agricultural yields Y (1000 dollars per acre) for region $i$ in year $t$ for the US, we have estimated the following eqation:
\begin{align*} \widehat{\log(Y)}_{it} &= 0.49 + .01 GE_{it} ~~~~ R^2 = .32\\
&~~~~~(.11) ~~~~ (.01) ~~~~ n = 1526 \end{align*}
#### (a) Interpret the results on the Genetically engineered ($GE$) technology on yields. (follow SSS= Sign Size Significance)
#### (b) Suppose $GE$ is used more on the West Coast, where crop yields are also higher. How would the estimated effect of GE change if we include a West Coast region dummy variable in the equation? Justify your answer.
#### (c) If we include region fixed effects, would they control for the factors in (b)? Justify your answer.
#### (d) If yields have been generally improving over time and GE adoption was only recently introduced in the USA, what would happen to the coefficient of GE if we included year fixed effects?
# Fixed Effects Practice Question #2
#### A recent paper investigates whether advertisement for Viagra causes increases in birth rates in the USA. Apparently, advertising for products, including Viagra, happens on TV and reaches households that have a TV within a Marketing region and does not happen in areas outside a designated marketing region. What the authors do is look at hospital birth rates in regions inside and near the advertising region border and collect data on dollars per 100 people (Ads) for a certain time, and compare those to the birth rates in hospitals located outside and near the advertising region designated border. They conduct a panel data analysis and estimate the following model:
$$ Births_{it} = \beta_0 + \beta_1 Ads + \beta_2 Ads^2 + Z_i + M_t + u_{it}$$
#### Where $Z_i$ are zipcode fixed effects and $M_t$ monthly fixed effects.
#### (a) Why do the authors include Zip Code Fixed Effects? In particular, what would be a variable that they are controlling for when adding Zip Code fixed effects that could cause a problem when interpreting the marginal effect of ad spending on birth rates? What would that (solved) problem be?
#### (b) Why do they add month fixed effects?
|
github_jupyter
|
# SVM Classification Using Individual Replicas
This notebook analyzes the quality of the classifiers resulting from training on individual replicas of read counts rather than averaged values. Data are adjusted for library size and gene length.
Training data
1. Uses individual replicas (not averaged)
1. Uses all genes
1. Includes time T1 (normoxia is not combined with resuscitation)
```
import init
from common import constants as cn
from common.trinary_data import TrinaryData
from common.data_provider import DataProvider
from common_python.plots import util_plots
from plots import util_plots as xutil_plots
from common_python.classifier import classifier_ensemble
from common_python.classifier import util_classifier
from common_python.classifier import classifier_collection
from common_python.classifier.classifier_ensemble_random_forest import ClassifierEnsembleRandomForest
from common_python.plots import util_plots as common_plots
import collections
import os
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import sklearn
from sklearn.decomposition import PCA
from sklearn import svm
from sklearn.model_selection import cross_val_score
%matplotlib inline
```
## Analyze Replica Data
The following shows the extent to which replicas agree with the tranary values that are assigned.
```
def compareDFValues(df1, df2, title):
RANGE = [-8, 8]
plt.figure()
arr1 = df1.values.flatten()
arr2 = df2.values.flatten()
plt.scatter(arr1, arr2)
# Define region of 0 values
plt.plot([-1, -1], [-1, 1], color="b")
plt.plot([1, 1], [-1, 1], color="b")
plt.plot([-1, 1], [-1, -1], color="b")
plt.plot([-1, 1], [1, 1], color="b")
# Define region of 1 values
plt.plot([1, 1], [1, RANGE[1]], color="b")
plt.plot([1, RANGE[1]], [1, 1], color="b")
# Define region of -1 values
plt.plot([-1, -1], [-1, RANGE[0]], color="b")
plt.plot([-1, RANGE[0]], [-1, -1], color="b")
plt.plot(RANGE, RANGE, color="r")
plt.title(title)
provider = DataProvider()
provider.do()
dfs = []
for idx in range(3):
dfs.append(provider.dfs_adjusted_read_count_wrtT0_log2[idx])
compareDFValues(dfs[0], dfs[1], "0 vs 1")
compareDFValues(dfs[0], dfs[2], "0 vs 2")
compareDFValues(dfs[1], dfs[2], "1 vs 2")
dfs[0].values.flatten()
```
## Accuracy With Replicas
Compares accuracies with replicas (is_averaged=False) vs. with replicas as the data.
```
def clfEval(is_averaged, high_rank=15, ensemble_size=50, is_randomize=False, num_iterations=10):
trinary = TrinaryData(is_averaged=is_averaged, is_dropT1=is_averaged)
df_X = trinary.df_X.copy()
df_X.columns = trinary.features
ser_y = trinary.ser_y.copy()
if is_randomize:
# Randomize the relationship between features and state
df_X = df_X.sample(frac=1)
ser_y = ser_y.sample(frac=1)
#
svm_ensemble = classifier_ensemble.ClassifierEnsemble(
classifier_ensemble.ClassifierDescriptorSVM(), size=ensemble_size,
filter_high_rank=high_rank)
return classifier_ensemble.ClassifierEnsemble.crossValidateByState(
svm_ensemble, df_X, ser_y, num_iterations)
clfEval(True, ensemble_size=50, is_randomize=True)
clfEval(False, ensemble_size=50, is_randomize=True)
clfEval(True)
clfEval(False)
```
## Analysis of Classifier Accuracy by State
```
# Plot values by state
def plotValuesByState(states, values, stds=None, ylabel="percent"):
if stds is None:
plt.bar(states, values)
else:
plt.bar(states, values, yerr=stds, alpha=0.5)
plt.xticks(rotation=45)
plt.xlabel("state")
plt.ylabel(ylabel)
# State statistics
def plotStateDistributions():
PERCENT = "percent"
VALUE = "value"
NAME = "name"
trinary = TrinaryData(is_averaged=False, is_dropT1=False)
df = pd.DataFrame(trinary.ser_y)
df[VALUE] = list(np.repeat(1, len(df)))
df_group = pd.DataFrame(df.groupby(NAME).count())
dct = {v: k for k, v in trinary.state_dict.items()}
df_group.index = [dct[s] for s in df_group.index]
df_group[PERCENT] = 100*df_group[VALUE] / len(df)
plotValuesByState(df_group.index, df_group[PERCENT])
plotStateDistributions()
# Classification accuracy by state
def stateClassificationAccuracy(state, num_iterations=10, is_averaged=False):
NUM_HOLDOUTS = 1
is_dropT1 = is_averaged
trinary = TrinaryData(is_averaged=is_averaged, is_dropT1=is_dropT1)
df_X = trinary.df_X.copy()
df_X.columns = trinary.features
ser_y = trinary.ser_y
results = []
for _ in range(num_iterations):
test_indices = []
ser_sample = ser_y[ser_y == state].sample(n=NUM_HOLDOUTS)
test_indices.extend(list(ser_sample.index))
train_indices = list(set(df_X.index).difference(test_indices))
svm_ensemble = classifier_ensemble.ClassifierEnsemble(
classifier_ensemble.ClassifierDescriptorSVM(), size=30,
filter_high_rank=1500,
classes=list(ser_y.values))
svm_ensemble.fit(df_X.loc[train_indices, :], ser_y.loc[train_indices])
results.append(svm_ensemble.score(df_X.loc[test_indices, :], ser_y[test_indices]))
return results
def plotStateAccuracies(is_averaged=True):
is_dropT1 = is_averaged
trinary = TrinaryData(is_averaged=is_averaged, is_dropT1=is_dropT1)
states = list(trinary.state_dict.values())
avgs = []
stds = []
for state in states:
values = stateClassificationAccuracy(state, is_averaged=is_averaged)
avgs.append(np.mean(values))
stds.append(np.std(values))
plotValuesByState(list(trinary.state_dict.keys()), avgs, stds=stds, ylabel="accuracy")
plotStateAccuracies(is_averaged=True)
plt.figure()
plotStateAccuracies(is_averaged=False)
if False:
is_averaged = False
df = df_confuse.applymap(lambda v: np.nan if v <= 0.2 else 1)
df.columns = [c - 0.5 for c in df.columns]
trinary = TrinaryData(is_averaged=is_averaged, is_dropT1=is_averaged)
states = trinary.ser_y.values
state_colors = ["grey", "orange", "green", "pink", "peru", "greenyellow"]
heatmap = plt.pcolor(df.T, cmap='jet')
#fig = heatmap.get_figure()
#axes = fig.get_axes()[0]
#yaxis = axes.get_yaxis()
#xv = [x + 0.5 for x in range(len(df.T.columns))]
#yv = [y + 0.5 for y in range(len(df.T))]
#plt.xticks(xv)
#plt.yticks(yv)
positions = [p - 0.5 for p in range(-1, len(states))]
labels = [str(int(c-.5)) if c >= 0 else "" for c in positions]
plt.yticks(positions, labels)
for idx, state in enumerate(states):
color = state_colors[state]
plt.scatter(idx, [-1], color=color)
#plt.colorbar(heatmap)
is_averaged = False
trinary = TrinaryData(is_averaged=is_averaged, is_dropT1=is_averaged)
svm_ensemble = classifier_ensemble.ClassifierEnsemble(
classifier_ensemble.ClassifierDescriptorSVM(), size=50,
filter_high_rank=None)
ser_pred = svm_ensemble.makeInstancePredictionDF(trinary.df_X, trinary.ser_y)
util_classifier.plotInstancePredictions(trinary.ser_y, ser_pred, is_plot=True)
```
|
github_jupyter
|
# Inference in Google Earth Engine + Colab
> Scaling up machine learning with GEE and Google Colab.
- toc: true
- badges: true
- author: Drew Bollinger
- comments: false
- hide: false
- sticky_rank: 11
# Inference in Google Earth Engine + Colab
Here we demonstrate how to take a trained model and apply to to imagery with Google Earth Engine + Colab + Tensorflow. This is adapted from an [Earth Engine <> TensorFlow demonstration notebook](https://developers.google.com/earth-engine/guides/tf_examples). We'll be taking the trained model from the [Deep Learning Crop Type Segmentation Model Example](https://developmentseed.org/sat-ml-training/DeepLearning_CropType_Segmentation).
# Setup software libraries
Authenticate and import as necessary.
```
# Import, authenticate and initialize the Earth Engine library.
import ee
ee.Authenticate()
ee.Initialize()
# Mount our Google Drive
from google.colab import drive
drive.mount('/content/drive')
# Add necessary libraries.
!pip install -q focal-loss
import os
from os import path as op
import tensorflow as tf
import folium
from focal_loss import SparseCategoricalFocalLoss
```
# Variables
Declare the variables that will be in use throughout the notebook.
```
# Specify names locations for outputs in Google Drive.
FOLDER = 'servir-inference-demo'
ROOT_DIR = '/content/drive/My Drive/'
# Specify inputs (Sentinel indexes) to the model.
BANDS = ['NDVI', 'WDRVI', 'SAVI']
# Specify the size and shape of patches expected by the model.
KERNEL_SIZE = 224
KERNEL_SHAPE = [KERNEL_SIZE, KERNEL_SIZE]
```
# Imagery
Gather and setup the imagery to use for inputs. It's important that we match the index inputs from the earlier analysis. This is a three-month Sentinel-2 composite. Display it in the notebook for a sanity check.
```
# Use Sentinel-2 data.
def add_indexes(img):
ndvi = img.expression(
'(nir - red) / (nir + red + a)', {
'a': 1e-5,
'nir': img.select('B8'),
'red': img.select('B4')
}
).rename('NDVI')
wdrvi = img.expression(
'(a * nir - red) / (a * nir + red)', {
'a': 0.2,
'nir': img.select('B8'),
'red': img.select('B4')
}
).rename('WDRVI')
savi = img.expression(
'1.5 * (nir - red) / (nir + red + 0.5)', {
'nir': img.select('B8'),
'red': img.select('B4')
}
).rename('SAVI')
return ee.Image.cat([ndvi, wdrvi, savi])
image = ee.ImageCollection('COPERNICUS/S2') \
.filterDate('2018-01-01', '2018-04-01') \
.filter(ee.Filter.lt('CLOUDY_PIXEL_PERCENTAGE', 20)) \
.map(add_indexes) \
.median()
# Use folium to visualize the imagery.
mapid = image.getMapId({'bands': BANDS, 'min': -1, 'max': 1})
map = folium.Map(location=[
-29.177943749121233,
30.55984497070313,
])
folium.TileLayer(
tiles=mapid['tile_fetcher'].url_format,
attr='Map Data © <a href="https://earthengine.google.com/">Google Earth Engine</a>',
overlay=True,
name='median composite',
).add_to(map)
map.add_child(folium.LayerControl())
map
```
# Load our saved model
```
# Load a trained model.
MODEL_DIR = '/content/drive/Shared drives/servir-sat-ml/data/model_out/10062020/'
model = tf.keras.models.load_model(MODEL_DIR)
model.summary()
```
# Prediction
The prediction pipeline is:
1. Export imagery on which to do predictions from Earth Engine in TFRecord format to Google Drive.
2. Use the trained model to make the predictions.
3. Write the predictions to a TFRecord file in Google Drive.
4. Manually upload the predictions TFRecord file to Earth Engine.
The following functions handle this process. It's useful to separate the export from the predictions so that you can experiment with different models without running the export every time.
```
def doExport(out_image_base, shape, region):
"""Run the image export task. Block until complete.
"""
task = ee.batch.Export.image.toDrive(
image = image.select(BANDS),
description = out_image_base,
fileNamePrefix = out_image_base,
folder = FOLDER,
region = region.getInfo()['coordinates'],
scale = 30,
fileFormat = 'TFRecord',
maxPixels = 1e10,
formatOptions = {
'patchDimensions': shape,
'compressed': True,
'maxFileSize': 104857600
}
)
task.start()
# Block until the task completes.
print('Running image export to Google Drive...')
import time
while task.active():
time.sleep(30)
# Error condition
if task.status()['state'] != 'COMPLETED':
print('Error with image export.')
else:
print('Image export completed.')
def doPrediction(out_image_base, kernel_shape, region):
"""Perform inference on exported imagery.
"""
print('Looking for TFRecord files...')
# Get a list of all the files in the output bucket.
filesList = os.listdir(op.join(ROOT_DIR, FOLDER))
# Get only the files generated by the image export.
exportFilesList = [s for s in filesList if out_image_base in s]
# Get the list of image files and the JSON mixer file.
imageFilesList = []
jsonFile = None
for f in exportFilesList:
if f.endswith('.tfrecord.gz'):
imageFilesList.append(op.join(ROOT_DIR, FOLDER, f))
elif f.endswith('.json'):
jsonFile = f
# Make sure the files are in the right order.
imageFilesList.sort()
from pprint import pprint
pprint(imageFilesList)
print(jsonFile)
import json
# Load the contents of the mixer file to a JSON object.
with open(op.join(ROOT_DIR, FOLDER, jsonFile), 'r') as f:
mixer = json.load(f)
pprint(mixer)
patches = mixer['totalPatches']
# Get set up for prediction.
imageColumns = [
tf.io.FixedLenFeature(shape=kernel_shape, dtype=tf.float32)
for k in BANDS
]
imageFeaturesDict = dict(zip(BANDS, imageColumns))
def parse_image(example_proto):
return tf.io.parse_single_example(example_proto, imageFeaturesDict)
def toTupleImage(inputs):
inputsList = [inputs.get(key) for key in BANDS]
stacked = tf.stack(inputsList, axis=0)
stacked = tf.transpose(stacked, [1, 2, 0])
return stacked
# Create a dataset from the TFRecord file(s) in Cloud Storage.
imageDataset = tf.data.TFRecordDataset(imageFilesList, compression_type='GZIP')
imageDataset = imageDataset.map(parse_image, num_parallel_calls=5)
imageDataset = imageDataset.map(toTupleImage).batch(1)
# Perform inference.
print('Running predictions...')
predictions = model.predict(imageDataset, steps=patches, verbose=1)
# print(predictions[0])
print('Writing predictions...')
out_image_file = op.join(ROOT_DIR, FOLDER, f'{out_image_base}pred.TFRecord')
writer = tf.io.TFRecordWriter(out_image_file)
patches = 0
for predictionPatch in predictions:
print('Writing patch ' + str(patches) + '...')
predictionPatch = tf.argmax(predictionPatch, axis=2)
# Create an example.
example = tf.train.Example(
features=tf.train.Features(
feature={
'class': tf.train.Feature(
float_list=tf.train.FloatList(
value=predictionPatch.numpy().flatten()))
}
)
)
# Write the example.
writer.write(example.SerializeToString())
patches += 1
writer.close()
```
Now there's all the code needed to run the prediction pipeline, all that remains is to specify the output region in which to do the prediction, the names of the output files, where to put them, and the shape of the outputs.
```
# Base file name to use for TFRecord files and assets.
image_base = 'servir_inference_demo_'
# South Africa (near training data)
region = ee.Geometry.Polygon(
[[[
30.55984497070313,
-29.177943749121233
],
[
30.843429565429684,
-29.177943749121233
],
[
30.843429565429684,
-28.994928377910732
],
[
30.55984497070313,
-28.994928377910732
]]], None, False)
# Run the export.
doExport(image_base, KERNEL_SHAPE, region)
# Run the prediction.
doPrediction(image_base, KERNEL_SHAPE, region)
```
# Display the output
One the data has been exported, the model has made predictions and the predictions have been written to a file, we need to [manually import the TFRecord to Earth Engine](https://developers.google.com/earth-engine/guides/tfrecord#uploading-tfrecords-to-earth-engine). Then we can display our crop type predictions as an image asset
```
out_image = ee.Image('users/drew/servir_inference_demo_-mixer')
mapid = out_image.getMapId({'min': 0, 'max': 10, 'palette': ['00A600','63C600','E6E600','E9BD3A','ECB176','EFC2B3','F2F2F2']})
map = folium.Map(location=[
-29.177943749121233,
30.55984497070313,
])
folium.TileLayer(
tiles=mapid['tile_fetcher'].url_format,
attr='Map Data © <a href="https://earthengine.google.com/">Google Earth Engine</a>',
overlay=True,
name='predicted crop type',
).add_to(map)
map.add_child(folium.LayerControl())
map
```
|
github_jupyter
|
<a href="https://cognitiveclass.ai"><img src = "https://ibm.box.com/shared/static/9gegpsmnsoo25ikkbl4qzlvlyjbgxs5x.png" width = 400> </a>
<h1 align=center><font size = 5>From Understanding to Preparation</font></h1>
## Introduction
In this lab, we will continue learning about the data science methodology, and focus on the **Data Understanding** and the **Data Preparation** stages.
## Table of Contents
<div class="alert alert-block alert-info" style="margin-top: 20px">
1. [Recap](#0)<br>
2. [Data Understanding](#2)<br>
3. [Data Preparation](#4)<br>
</div>
<hr>
# Recap <a id="0"></a>
In Lab **From Requirements to Collection**, we learned that the data we need to answer the question developed in the business understanding stage, namely *can we automate the process of determining the cuisine of a given recipe?*, is readily available. A researcher named Yong-Yeol Ahn scraped tens of thousands of food recipes (cuisines and ingredients) from three different websites, namely:
<img src = "https://ibm.box.com/shared/static/4fruwan7wmjov3gywiz3swlojw0srv54.png" width=500>
www.allrecipes.com
<img src = "https://ibm.box.com/shared/static/cebfdbr22fjxa47lltp0bs533r103g0z.png" width=500>
www.epicurious.com
<img src = "https://ibm.box.com/shared/static/epk727njg7xrz49pbkpkzd05cm5ywqmu.png" width=500>
www.menupan.com
For more information on Yong-Yeol Ahn and his research, you can read his paper on [Flavor Network and the Principles of Food Pairing](http://yongyeol.com/papers/ahn-flavornet-2011.pdf).
We also collected the data and placed it on an IBM server for your convenience.
------------
# Data Understanding <a id="2"></a>
<img src="https://ibm.box.com/shared/static/89geb3m0ge1z73s92hl8o8wdcpcrggtz.png" width=500>
<strong> Important note:</strong> Please note that you are not expected to know how to program in python. The following code is meant to illustrate the stages of data understanding and data preparation, so it is totally fine if you do not understand the individual lines of code. We have a full course on programming in python, <a href="http://cocl.us/PY0101EN_DS0103EN_LAB3_PYTHON">Python for Data Science</a>, so please feel free to complete the course if you are interested in learning how to program in python.
### Using this notebook:
To run any of the following cells of code, you can type **Shift + Enter** to excute the code in a cell.
Get the version of Python installed.
```
# check Python version
!python -V
```
Download the library and dependencies that we will need to run this lab.
```
import pandas as pd # import library to read data into dataframe
pd.set_option('display.max_columns', None)
import numpy as np # import numpy library
import re # import library for regular expression
```
Download the data from the IBM server and read it into a *pandas* dataframe.
```
recipes = pd.read_csv("https://ibm.box.com/shared/static/5wah9atr5o1akuuavl2z9tkjzdinr1lv.csv")
print("Data read into dataframe!") # takes about 30 seconds
```
Show the first few rows.
```
recipes.head()
```
Get the dimensions of the dataframe.
```
recipes.shape
```
So our dataset consists of 57,691 recipes. Each row represents a recipe, and for each recipe, the corresponding cuisine is documented as well as whether 384 ingredients exist in the recipe or not, beginning with almond and ending with zucchini.
We know that a basic sushi recipe includes the ingredients:
* rice
* soy sauce
* wasabi
* some fish/vegetables
Let's check that these ingredients exist in our dataframe:
```
ingredients = list(recipes.columns.values)
print([match.group(0) for ingredient in ingredients for match in [(re.compile(".*(rice).*")).search(ingredient)] if match])
print([match.group(0) for ingredient in ingredients for match in [(re.compile(".*(wasabi).*")).search(ingredient)] if match])
print([match.group(0) for ingredient in ingredients for match in [(re.compile(".*(soy).*")).search(ingredient)] if match])
```
Yes, they do!
* rice exists as rice.
* wasabi exists as wasabi.
* soy exists as soy_sauce.
So maybe if a recipe contains all three ingredients: rice, wasabi, and soy_sauce, then we can confidently say that the recipe is a **Japanese** cuisine! Let's keep this in mind!
----------------
# Data Preparation <a id="4"></a>
<img src="https://ibm.box.com/shared/static/lqc2j3r0ndhokh77mubohwjqybzf8dhk.png" width=500>
In this section, we will prepare data for the next stage in the data science methodology, which is modeling. This stage involves exploring the data further and making sure that it is in the right format for the machine learning algorithm that we selected in the analytic approach stage, which is decision trees.
First, look at the data to see if it needs cleaning.
```
recipes["country"].value_counts() # frequency table
```
By looking at the above table, we can make the following observations:
1. Cuisine column is labeled as Country, which is inaccurate.
2. Cuisine names are not consistent as not all of them start with an uppercase first letter.
3. Some cuisines are duplicated as variation of the country name, such as Vietnam and Vietnamese.
4. Some cuisines have very few recipes.
#### Let's fixes these problems.
Fix the name of the column showing the cuisine.
```
column_names = recipes.columns.values
column_names[0] = "cuisine"
recipes.columns = column_names
recipes
```
Make all the cuisine names lowercase.
```
recipes["cuisine"] = recipes["cuisine"].str.lower()
```
Make the cuisine names consistent.
```
recipes.loc[recipes["cuisine"] == "austria", "cuisine"] = "austrian"
recipes.loc[recipes["cuisine"] == "belgium", "cuisine"] = "belgian"
recipes.loc[recipes["cuisine"] == "china", "cuisine"] = "chinese"
recipes.loc[recipes["cuisine"] == "canada", "cuisine"] = "canadian"
recipes.loc[recipes["cuisine"] == "netherlands", "cuisine"] = "dutch"
recipes.loc[recipes["cuisine"] == "france", "cuisine"] = "french"
recipes.loc[recipes["cuisine"] == "germany", "cuisine"] = "german"
recipes.loc[recipes["cuisine"] == "india", "cuisine"] = "indian"
recipes.loc[recipes["cuisine"] == "indonesia", "cuisine"] = "indonesian"
recipes.loc[recipes["cuisine"] == "iran", "cuisine"] = "iranian"
recipes.loc[recipes["cuisine"] == "italy", "cuisine"] = "italian"
recipes.loc[recipes["cuisine"] == "japan", "cuisine"] = "japanese"
recipes.loc[recipes["cuisine"] == "israel", "cuisine"] = "jewish"
recipes.loc[recipes["cuisine"] == "korea", "cuisine"] = "korean"
recipes.loc[recipes["cuisine"] == "lebanon", "cuisine"] = "lebanese"
recipes.loc[recipes["cuisine"] == "malaysia", "cuisine"] = "malaysian"
recipes.loc[recipes["cuisine"] == "mexico", "cuisine"] = "mexican"
recipes.loc[recipes["cuisine"] == "pakistan", "cuisine"] = "pakistani"
recipes.loc[recipes["cuisine"] == "philippines", "cuisine"] = "philippine"
recipes.loc[recipes["cuisine"] == "scandinavia", "cuisine"] = "scandinavian"
recipes.loc[recipes["cuisine"] == "spain", "cuisine"] = "spanish_portuguese"
recipes.loc[recipes["cuisine"] == "portugal", "cuisine"] = "spanish_portuguese"
recipes.loc[recipes["cuisine"] == "switzerland", "cuisine"] = "swiss"
recipes.loc[recipes["cuisine"] == "thailand", "cuisine"] = "thai"
recipes.loc[recipes["cuisine"] == "turkey", "cuisine"] = "turkish"
recipes.loc[recipes["cuisine"] == "vietnam", "cuisine"] = "vietnamese"
recipes.loc[recipes["cuisine"] == "uk-and-ireland", "cuisine"] = "uk-and-irish"
recipes.loc[recipes["cuisine"] == "irish", "cuisine"] = "uk-and-irish"
recipes
```
Remove cuisines with < 50 recipes.
```
# get list of cuisines to keep
recipes_counts = recipes["cuisine"].value_counts()
cuisines_indices = recipes_counts > 50
cuisines_to_keep = list(np.array(recipes_counts.index.values)[np.array(cuisines_indices)])
rows_before = recipes.shape[0] # number of rows of original dataframe
print("Number of rows of original dataframe is {}.".format(rows_before))
recipes = recipes.loc[recipes['cuisine'].isin(cuisines_to_keep)]
rows_after = recipes.shape[0] # number of rows of processed dataframe
print("Number of rows of processed dataframe is {}.".format(rows_after))
print("{} rows removed!".format(rows_before - rows_after))
```
Convert all Yes's to 1's and the No's to 0's
```
recipes = recipes.replace(to_replace="Yes", value=1)
recipes = recipes.replace(to_replace="No", value=0)
```
#### Let's analyze the data a little more in order to learn the data better and note any interesting preliminary observations.
Run the following cell to get the recipes that contain **rice** *and* **soy** *and* **wasabi** *and* **seaweed**.
```
recipes.head()
check_recipes = recipes.loc[
(recipes["rice"] == 1) &
(recipes["soy_sauce"] == 1) &
(recipes["wasabi"] == 1) &
(recipes["seaweed"] == 1)
]
check_recipes
```
Based on the results of the above code, can we classify all recipes that contain **rice** *and* **soy** *and* **wasabi** *and* **seaweed** as **Japanese** recipes? Why?
Double-click __here__ for the solution.
<!-- The correct answer is:
No, because other recipes such as Asian and East_Asian recipes also contain these ingredients.
-->
Let's count the ingredients across all recipes.
```
# sum each column
ing = recipes.iloc[:, 1:].sum(axis=0)
# define each column as a pandas series
ingredient = pd.Series(ing.index.values, index = np.arange(len(ing)))
count = pd.Series(list(ing), index = np.arange(len(ing)))
# create the dataframe
ing_df = pd.DataFrame(dict(ingredient = ingredient, count = count))
ing_df = ing_df[["ingredient", "count"]]
print(ing_df.to_string())
```
Now we have a dataframe of ingredients and their total counts across all recipes. Let's sort this dataframe in descending order.
```
ing_df.sort_values(["count"], ascending=False, inplace=True)
ing_df.reset_index(inplace=True, drop=True)
print(ing_df)
```
#### What are the 3 most popular ingredients?
Double-click __here__ for the solution.
<!-- The correct answer is:
// 1. Egg with <strong>21,025</strong> occurrences.
// 2. Wheat with <strong>20,781</strong> occurrences.
// 3. Butter with <strong>20,719</strong> occurrences.
-->
However, note that there is a problem with the above table. There are ~40,000 American recipes in our dataset, which means that the data is biased towards American ingredients.
**Therefore**, let's compute a more objective summary of the ingredients by looking at the ingredients per cuisine.
#### Let's create a *profile* for each cuisine.
In other words, let's try to find out what ingredients Chinese people typically use, and what is **Canadian** food for example.
```
cuisines = recipes.groupby("cuisine").mean()
cuisines.head()
```
As shown above, we have just created a dataframe where each row is a cuisine and each column (except for the first column) is an ingredient, and the row values represent the percentage of each ingredient in the corresponding cuisine.
**For example**:
* *almond* is present across 15.65% of all of the **African** recipes.
* *butter* is present across 38.11% of all of the **Canadian** recipes.
Let's print out the profile for each cuisine by displaying the top four ingredients in each cuisine.
```
num_ingredients = 4 # define number of top ingredients to print
# define a function that prints the top ingredients for each cuisine
def print_top_ingredients(row):
print(row.name.upper())
row_sorted = row.sort_values(ascending=False)*100
top_ingredients = list(row_sorted.index.values)[0:num_ingredients]
row_sorted = list(row_sorted)[0:num_ingredients]
for ind, ingredient in enumerate(top_ingredients):
print("%s (%d%%)" % (ingredient, row_sorted[ind]), end=' ')
print("\n")
# apply function to cuisines dataframe
create_cuisines_profiles = cuisines.apply(print_top_ingredients, axis=1)
```
At this point, we feel that we have understood the data well and the data is ready and is in the right format for modeling!
-----------
### Thank you for completing this lab!
This notebook was created by [Alex Aklson](https://www.linkedin.com/in/aklson/). We hope you found this lab session interesting. Feel free to contact us if you have any questions!
This notebook is part of the free course on **Cognitive Class** called *Data Science Methodology*. If you accessed this notebook outside the course, you can take this free self-paced course, online by clicking [here](https://cocl.us/DS0103EN_LAB3_PYTHON).
<hr>
Copyright © 2018 [Cognitive Class](https://cognitiveclass.ai/?utm_source=bducopyrightlink&utm_medium=dswb&utm_campaign=bdu). This notebook and its source code are released under the terms of the [MIT License](https://bigdatauniversity.com/mit-license/).
|
github_jupyter
|
# Autoencoder (Semi-supervised)
```
%load_ext autoreload
%autoreload 2
# Seed value
# Apparently you may use different seed values at each stage
seed_value= 0
# 1. Set the `PYTHONHASHSEED` environment variable at a fixed value
import os
os.environ['PYTHONHASHSEED']=str(seed_value)
# 2. Set the `python` built-in pseudo-random generator at a fixed value
import random
random.seed(seed_value)
# 3. Set the `numpy` pseudo-random generator at a fixed value
import numpy as np
np.random.seed(seed_value)
# 4. Set the `tensorflow` pseudo-random generator at a fixed value
import tensorflow as tf
tf.set_random_seed(seed_value)
# 5. Configure a new global `tensorflow` session
from keras import backend as K
session_conf = tf.ConfigProto(intra_op_parallelism_threads=1, inter_op_parallelism_threads=1)
sess = tf.Session(graph=tf.get_default_graph(), config=session_conf)
K.set_session(sess)
import keras
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import sklearn
# plt.style.use('fivethirtyeight')
sns.set_style("whitegrid")
sns.set_context("notebook")
DATA_PATH = '../data/'
VAL_SPLITS = 4
from plot_utils import plot_confusion_matrix
from cv_utils import run_cv_f1
from cv_utils import plot_cv_roc
from cv_utils import plot_cv_roc_prc
from sklearn.model_selection import StratifiedShuffleSplit
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import cross_validate
```
For this part of the project, we will only work with the training set, that we will split again into train and validation to perform the hyperparameter tuning.
We will save the test set for the final part, when we have already tuned our hyperparameters.
```
df = pd.read_csv(os.path.join(DATA_PATH,'df_train.csv'))
df.drop(columns= df.columns[0:2],inplace=True)
df.head()
```
## Preprocessing the data
Although we are always using cross validation with `VAL_SPLITS` folds, (in general, 4), here we are gonna set only one split in order to explore how the Autoencoder works and get intuition.
```
cv = StratifiedShuffleSplit(n_splits=1,test_size=0.15,random_state=0)
# In case we want to select a subset of features
# df_ = df[['Class','V9','V14','V16','V2','V3','V17']]
df_ = df[['Class','V4','V14','V16','V12','V3','V17']]
X = df_.drop(columns='Class').to_numpy()
y = df_['Class'].to_numpy()
for idx_t, idx_v in cv.split(X,y):
X_train = X[idx_t]
y_train = y[idx_t]
X_val = X[idx_v]
y_val = y[idx_v]
# Now we need to erase the FRAUD cases on the TRAINING set
X_train_normal = X_train[y_train==0]
```
## Defining the model
```
# this is the size of our encoded representations
ENCODED_DIM = 2
INPUT_DIM = X.shape[1]
from keras.layers import Input, Dense
from keras.models import Model
from keras.models import Sequential
from keras.layers import Dense, Activation
from keras.layers import LeakyReLU
def create_encoder(input_dim, encoded_dim):
encoder = Sequential([
Dense(32, input_shape=(input_dim,)),
LeakyReLU(),
Dense(16),
LeakyReLU(),
Dense(8),
LeakyReLU(),
Dense(encoded_dim)
], name='encoder')
return encoder
def create_decoder(input_dim, encoded_dim):
decoder = Sequential([
Dense(8, input_shape=(encoded_dim,) ),
LeakyReLU(),
Dense(16),
LeakyReLU(),
Dense(8),
LeakyReLU(),
Dense(input_dim)
],name='decoder')
return decoder
def create_autoencoder(input_dim, encoded_dim, return_encoder = True):
encoder = create_encoder(input_dim,encoded_dim)
decoder = create_decoder(input_dim,encoded_dim)
inp = Input(shape=(INPUT_DIM,),name='Input_Layer')
# a layer instance is callable on a tensor, and returns a tensor
x_enc = encoder(inp)
x_out = decoder(x_enc)
# This creates a model that includes
# the Input layer and three Dense layers
autoencoder = Model(inputs=inp, outputs=x_out)
if return_encoder:
return autoencoder, encoder
else:
return autoencoder
autoencoder, encoder = create_autoencoder(INPUT_DIM,ENCODED_DIM)
print('ENCODER SUMMARY\n')
print(encoder.summary())
print('AUTOENCODER SUMMARY\n')
print(autoencoder.summary())
autoencoder.compile(optimizer='adam',
loss='mean_squared_error')
```
## Training the model
```
autoencoder.fit(x=X_train_normal, y= X_train_normal,
batch_size=512,epochs=40, validation_split=0.1) # starts training
```
## Testing
```
X_enc = encoder.predict(X_val)
X_enc_normal = X_enc[y_val==0]
X_enc_fraud = X_enc[y_val==1]
sns.scatterplot(x = X_enc_normal[:,0], y = X_enc_normal[:,1] ,label='Normal', alpha=0.5)
sns.scatterplot(x = X_enc_fraud[:,0], y = X_enc_fraud[:,1] ,label='Fraud')
X_out = autoencoder.predict(X_val)
print(X_out.shape)
X_val.shape
distances = np.sum((X_out-X_val)**2,axis=1)
bins = np.linspace(0,np.max(distances),40)
sns.distplot(distances[y_val==0],label='Normal',kde=False,
bins=bins, norm_hist=True, axlabel='Distance')
sns.distplot(distances[y_val==1],label='Fraud',kde=False, bins=bins, norm_hist=True)
bins = np.linspace(0,100,40)
sns.distplot(distances[y_val==0],label='Normal',kde=False,
bins=bins, norm_hist=True, axlabel='Distance')
sns.distplot(distances[y_val==1],label='Fraud',kde=False, bins=bins, norm_hist=True)
plt.xlim((0,100))
```
## Validating the model
```
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import cross_validate
from sklearn.metrics import f1_score
def clf_autoencoder(X,autoencoder,threshold):
"""
Classifier based in the autoencoder.
A datapoint is a nomaly if the distance of the original points
and the result of the autoencoder is greater than the threshold.
"""
X_out = autoencoder.predict(X)
distances = np.sum((X_out-X)**2,axis=1).reshape((-1,1))
# y_pred = 1 if it is anomaly
y_pred = 1.*(distances > threshold )
return y_pred
cv = StratifiedShuffleSplit(n_splits=VAL_SPLITS,test_size=0.15,random_state=0)
# Thresholds to validate
thresholds = np.linspace(0,100,100)
# List with the f1 of all the thresholds at each validation fold
f1_all = []
for i,(idx_t, idx_v) in enumerate(cv.split(X,y)):
X_train = X[idx_t]
y_train = y[idx_t]
X_val = X[idx_v]
y_val = y[idx_v]
# Now we need to erase the FRAUD cases on the TRAINING set
X_train_normal = X_train[y_train==0]
# Train the autoencoder
autoencoder, encoder = create_autoencoder(INPUT_DIM,ENCODED_DIM)
autoencoder.compile(optimizer='adam',
loss='mean_squared_error')
autoencoder.fit(x=X_train_normal, y= X_train_normal,
batch_size=512,epochs=30, shuffle=True,
verbose=0) # starts training
# Plot of the validation set in the embedding space
X_enc = encoder.predict(X_val)
X_enc_normal = X_enc[y_val==0]
X_enc_fraud = X_enc[y_val==1]
sns.scatterplot(x = X_enc_normal[:,0], y = X_enc_normal[:,1] ,label='Normal', alpha=0.5)
sns.scatterplot(x = X_enc_fraud[:,0], y = X_enc_fraud[:,1] ,label='Fraud')
plt.show()
# Transformation of the points through the autoencoder
# and calculate the predictions
y_preds=clf_autoencoder(X_val,autoencoder,thresholds)
metrics_f1 = np.array([ f1_score(y_val,y_pred) for y_pred in y_preds.T ])
f1_all.append(metrics_f1)
# Save the models into files for future use
autoencoder.save('models_autoencoder/autoencoder_fold_'+str(i+1)+'.h5')
encoder.save('models_autoencoder/encoder_fold_'+str(i+1)+'.h5')
del(autoencoder,encoder)
f1_mean = np.mean(f1_all,axis=0)
# Plot of F1-Threshold curves
for i,f1_fold in enumerate(f1_all):
sns.lineplot(thresholds,f1_fold, label='Fold '+str(i+1))
sns.scatterplot(thresholds,f1_mean,label='Mean')
plt.show()
f1_opt = f1_mean.max()
threshold_opt = thresholds[np.argmax(f1_mean)]
print('F1 max = {:.3f} at threshold = {:.3f}'.format(f1_opt,threshold_opt))
```
|
github_jupyter
|
# Facial Keypoint Detection
This project will be all about defining and training a convolutional neural network to perform facial keypoint detection, and using computer vision techniques to transform images of faces. The first step in any challenge like this will be to load and visualize the data you'll be working with.
Let's take a look at some examples of images and corresponding facial keypoints.
<img src='images/key_pts_example.png' width=50% height=50%/>
Facial keypoints (also called facial landmarks) are the small magenta dots shown on each of the faces in the image above. In each training and test image, there is a single face and **68 keypoints, with coordinates (x, y), for that face**. These keypoints mark important areas of the face: the eyes, corners of the mouth, the nose, etc. These keypoints are relevant for a variety of tasks, such as face filters, emotion recognition, pose recognition, and so on. Here they are, numbered, and you can see that specific ranges of points match different portions of the face.
<img src='images/landmarks_numbered.jpg' width=30% height=30%/>
---
## Load and Visualize Data
The first step in working with any dataset is to become familiar with your data; you'll need to load in the images of faces and their keypoints and visualize them! This set of image data has been extracted from the [YouTube Faces Dataset](https://www.cs.tau.ac.il/~wolf/ytfaces/), which includes videos of people in YouTube videos. These videos have been fed through some processing steps and turned into sets of image frames containing one face and the associated keypoints.
#### Training and Testing Data
This facial keypoints dataset consists of 5770 color images. All of these images are separated into either a training or a test set of data.
* 3462 of these images are training images, for you to use as you create a model to predict keypoints.
* 2308 are test images, which will be used to test the accuracy of your model.
The information about the images and keypoints in this dataset are summarized in CSV files, which we can read in using `pandas`. Let's read the training CSV and get the annotations in an (N, 2) array where N is the number of keypoints and 2 is the dimension of the keypoint coordinates (x, y).
---
First, before we do anything, we have to load in our image data. This data is stored in a zip file and in the below cell, we access it by it's URL and unzip the data in a `/data/` directory that is separate from the workspace home directory.
```
# -- DO NOT CHANGE THIS CELL -- #
!mkdir /data
!wget -P /data/ https://s3.amazonaws.com/video.udacity-data.com/topher/2018/May/5aea1b91_train-test-data/train-test-data.zip
!unzip -n /data/train-test-data.zip -d /data
# import the required libraries
import glob
import os
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import cv2
```
Then, let's load in our training data and display some stats about that dat ato make sure it's been loaded in correctly!
```
key_pts_frame = pd.read_csv('/data/training_frames_keypoints.csv')
n = 0
image_name = key_pts_frame.iloc[n, 0]
key_pts = key_pts_frame.iloc[n, 1:].as_matrix()
key_pts = key_pts.astype('float').reshape(-1, 2)
print('Image name: ', image_name)
print('Landmarks shape: ', key_pts.shape)
print('First 4 key pts: {}'.format(key_pts[:4]))
# print out some stats about the data
print('Number of images: ', key_pts_frame.shape[0])
img2 = cv2.imread('/data/training/Nicolas_Sarkozy_02.jpg')
width, height, channels = img2.shape
npixel = img2.size
print("npixel", npixel)
print("width {}, height {}, channels {}".format(width, height, channels))
```
## Look at some images
Below, is a function `show_keypoints` that takes in an image and keypoints and displays them. As you look at this data, **note that these images are not all of the same size**, and neither are the faces! To eventually train a neural network on these images, we'll need to standardize their shape.
```
def show_keypoints(image, key_pts):
"""Show image with keypoints"""
plt.imshow(image)
plt.scatter(key_pts[:, 0], key_pts[:, 1], s=20, marker='.', c='m')
# Display a few different types of images by changing the index n
# select an image by index in our data frame
n = 0
image_name = key_pts_frame.iloc[n, 0]
key_pts = key_pts_frame.iloc[n, 1:].as_matrix()
key_pts = key_pts.astype('float').reshape(-1, 2)
plt.figure(figsize=(5, 5))
show_keypoints(mpimg.imread(os.path.join('/data/training/', image_name)), key_pts)
plt.show()
```
## Dataset class and Transformations
To prepare our data for training, we'll be using PyTorch's Dataset class. Much of this this code is a modified version of what can be found in the [PyTorch data loading tutorial](http://pytorch.org/tutorials/beginner/data_loading_tutorial.html).
#### Dataset class
``torch.utils.data.Dataset`` is an abstract class representing a
dataset. This class will allow us to load batches of image/keypoint data, and uniformly apply transformations to our data, such as rescaling and normalizing images for training a neural network.
Your custom dataset should inherit ``Dataset`` and override the following
methods:
- ``__len__`` so that ``len(dataset)`` returns the size of the dataset.
- ``__getitem__`` to support the indexing such that ``dataset[i]`` can
be used to get the i-th sample of image/keypoint data.
Let's create a dataset class for our face keypoints dataset. We will
read the CSV file in ``__init__`` but leave the reading of images to
``__getitem__``. This is memory efficient because all the images are not
stored in the memory at once but read as required.
A sample of our dataset will be a dictionary
``{'image': image, 'keypoints': key_pts}``. Our dataset will take an
optional argument ``transform`` so that any required processing can be
applied on the sample. We will see the usefulness of ``transform`` in the
next section.
```
from torch.utils.data import Dataset, DataLoader
class FacialKeypointsDataset(Dataset):
"""Face Landmarks dataset."""
def __init__(self, csv_file, root_dir, transform=None):
"""
Args:
csv_file (string): Path to the csv file with annotations.
root_dir (string): Directory with all the images.
transform (callable, optional): Optional transform to be applied
on a sample.
"""
self.key_pts_frame = pd.read_csv(csv_file)
self.root_dir = root_dir
self.transform = transform
def __len__(self):
return len(self.key_pts_frame)
def __getitem__(self, idx):
image_name = os.path.join(self.root_dir,
self.key_pts_frame.iloc[idx, 0])
image = mpimg.imread(image_name)
# if image has an alpha color channel, get rid of it
if(image.shape[2] == 4):
image = image[:,:,0:3]
key_pts = self.key_pts_frame.iloc[idx, 1:].as_matrix()
key_pts = key_pts.astype('float').reshape(-1, 2)
sample = {'image': image, 'keypoints': key_pts}
if self.transform:
sample = self.transform(sample)
return sample
```
Now that we've defined this class, let's instantiate the dataset and display some images.
```
# Construct the dataset
face_dataset = FacialKeypointsDataset(csv_file='/data/training_frames_keypoints.csv',
root_dir='/data/training/')
# print some stats about the dataset
print('Length of dataset: ', len(face_dataset))
# Display a few of the images from the dataset
num_to_display = 3
for i in range(num_to_display):
# define the size of images
fig = plt.figure(figsize=(20,10))
# randomly select a sample
rand_i = np.random.randint(0, len(face_dataset))
sample = face_dataset[rand_i]
# print the shape of the image and keypoints
print(i, sample['image'].shape, sample['keypoints'].shape)
ax = plt.subplot(1, num_to_display, i + 1)
ax.set_title('Sample #{}'.format(i))
# Using the same display function, defined earlier
show_keypoints(sample['image'], sample['keypoints'])
```
## Transforms
Now, the images above are not of the same size, and neural networks often expect images that are standardized; a fixed size, with a normalized range for color ranges and coordinates, and (for PyTorch) converted from numpy lists and arrays to Tensors.
Therefore, we will need to write some pre-processing code.
Let's create four transforms:
- ``Normalize``: to convert a color image to grayscale values with a range of [0,1] and normalize the keypoints to be in a range of about [-1, 1]
- ``Rescale``: to rescale an image to a desired size.
- ``RandomCrop``: to crop an image randomly.
- ``ToTensor``: to convert numpy images to torch images.
We will write them as callable classes instead of simple functions so
that parameters of the transform need not be passed everytime it's
called. For this, we just need to implement ``__call__`` method and
(if we require parameters to be passed in), the ``__init__`` method.
We can then use a transform like this:
tx = Transform(params)
transformed_sample = tx(sample)
Observe below how these transforms are generally applied to both the image and its keypoints.
```
import torch
from torchvision import transforms, utils
# tranforms
class Normalize(object):
"""Convert a color image to grayscale and normalize the color range to [0,1]."""
def __call__(self, sample):
image, key_pts = sample['image'], sample['keypoints']
image_copy = np.copy(image)
key_pts_copy = np.copy(key_pts)
# convert image to grayscale
image_copy = cv2.cvtColor(image, cv2.COLOR_RGB2GRAY)
# scale color range from [0, 255] to [0, 1]
image_copy= image_copy/255.0
# scale keypoints to be centered around 0 with a range of [-1, 1]
# mean = 100, sqrt = 50, so, pts should be (pts - 100)/50
key_pts_copy = (key_pts_copy - 100)/50.0
return {'image': image_copy, 'keypoints': key_pts_copy}
class Rescale(object):
"""Rescale the image in a sample to a given size.
Args:
output_size (tuple or int): Desired output size. If tuple, output is
matched to output_size. If int, smaller of image edges is matched
to output_size keeping aspect ratio the same.
"""
def __init__(self, output_size):
assert isinstance(output_size, (int, tuple))
self.output_size = output_size
def __call__(self, sample):
image, key_pts = sample['image'], sample['keypoints']
h, w = image.shape[:2]
if isinstance(self.output_size, int):
if h > w:
new_h, new_w = self.output_size * h / w, self.output_size
else:
new_h, new_w = self.output_size, self.output_size * w / h
else:
new_h, new_w = self.output_size
new_h, new_w = int(new_h), int(new_w)
img = cv2.resize(image, (new_w, new_h))
# scale the pts, too
key_pts = key_pts * [new_w / w, new_h / h]
return {'image': img, 'keypoints': key_pts}
class RandomCrop(object):
"""Crop randomly the image in a sample.
Args:
output_size (tuple or int): Desired output size. If int, square crop
is made.
"""
def __init__(self, output_size):
assert isinstance(output_size, (int, tuple))
if isinstance(output_size, int):
self.output_size = (output_size, output_size)
else:
assert len(output_size) == 2
self.output_size = output_size
def __call__(self, sample):
image, key_pts = sample['image'], sample['keypoints']
h, w = image.shape[:2]
new_h, new_w = self.output_size
top = np.random.randint(0, h - new_h)
left = np.random.randint(0, w - new_w)
image = image[top: top + new_h,
left: left + new_w]
key_pts = key_pts - [left, top]
return {'image': image, 'keypoints': key_pts}
class ToTensor(object):
"""Convert ndarrays in sample to Tensors."""
def __call__(self, sample):
image, key_pts = sample['image'], sample['keypoints']
# if image has no grayscale color channel, add one
if(len(image.shape) == 2):
# add that third color dim
image = image.reshape(image.shape[0], image.shape[1], 1)
# swap color axis because
# numpy image: H x W x C
# torch image: C X H X W
image = image.transpose((2, 0, 1))
return {'image': torch.from_numpy(image),
'keypoints': torch.from_numpy(key_pts)}
```
## Test out the transforms
Let's test these transforms out to make sure they behave as expected. As you look at each transform, note that, in this case, **order does matter**. For example, you cannot crop a image using a value smaller than the original image (and the orginal images vary in size!), but, if you first rescale the original image, you can then crop it to any size smaller than the rescaled size.
```
# test out some of these transforms
rescale = Rescale(100)
crop = RandomCrop(50)
composed = transforms.Compose([Rescale(250),
RandomCrop(224)])
# apply the transforms to a sample image
test_num = 500
sample = face_dataset[test_num]
fig = plt.figure()
for i, tx in enumerate([rescale, crop, composed]):
transformed_sample = tx(sample)
ax = plt.subplot(1, 3, i + 1)
plt.tight_layout()
ax.set_title(type(tx).__name__)
show_keypoints(transformed_sample['image'], transformed_sample['keypoints'])
plt.show()
```
## Create the transformed dataset
Apply the transforms in order to get grayscale images of the same shape. Verify that your transform works by printing out the shape of the resulting data (printing out a few examples should show you a consistent tensor size).
```
# define the data tranform
# order matters! i.e. rescaling should come before a smaller crop
data_transform = transforms.Compose([Rescale(250),
RandomCrop(224),
Normalize(),
ToTensor()])
# create the transformed dataset
transformed_dataset = FacialKeypointsDataset(csv_file='/data/training_frames_keypoints.csv',
root_dir='/data/training/',
transform=data_transform)
# print some stats about the transformed data
print('Number of images: ', len(transformed_dataset))
# make sure the sample tensors are the expected size
for i in range(5):
sample = transformed_dataset[i]
print(i, sample['image'].size(), sample['keypoints'].size())
```
## Data Iteration and Batching
Right now, we are iterating over this data using a ``for`` loop, but we are missing out on a lot of PyTorch's dataset capabilities, specifically the abilities to:
- Batch the data
- Shuffle the data
- Load the data in parallel using ``multiprocessing`` workers.
``torch.utils.data.DataLoader`` is an iterator which provides all these
features, and we'll see this in use in the *next* notebook, Notebook 2, when we load data in batches to train a neural network!
---
## Ready to Train!
Now that you've seen how to load and transform our data, you're ready to build a neural network to train on this data.
In the next notebook, you'll be tasked with creating a CNN for facial keypoint detection.
|
github_jupyter
|
```
import os
import pickle
import re
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sn
import numpy as np
import re
import xgboost as xgb
import shap
from sklearn import ensemble
from sklearn import dummy
from sklearn import linear_model
from sklearn import svm
from sklearn import neural_network
from sklearn import metrics
from sklearn import preprocessing
from sklearn.model_selection import train_test_split
from sklearn.utils.fixes import loguniform
import scipy
from misc import save_model, load_model, regression_results, grid_search_cv
# Options of settings with different Xs and Ys
options = ["../data/Train_Compound_Viral_interactions_for_Supervised_Learning_with_LS_LS.csv",
"../data/Train_Compound_Viral_interactions_for_Supervised_Learning_with_MFP_LS.csv",
".."] #(to be continued)
data_type_options = ["LS_Compound_LS_Protein",
"MFP_Compound_LS_Protein",
".."
]
# input option is also used to control the model parameters below
input_option = 0
classification_task = False
classification_th = 85
data_type=data_type_options[input_option]
filename = options[input_option]
with open(filename, "rb") as file:
print("Loading ", filename)
big_df = pd.read_csv(filename, header='infer', delimiter=",")
total_length = len(big_df.columns)
X = big_df.iloc[:,range(5,total_length)]
Y = big_df[['pchembl_value']].to_numpy().flatten()
meta_X = big_df.iloc[:,[0,1,2,3]]
print("Lengths --> X = %d, Y = %d" % (len(X), len(Y)))
print(X.columns)
n_samples = len(X)
indices = np.arange(n_samples)
X_train = X
y_train = Y
print(X_train[:10])
print(X_train.shape,y_train.shape)
print(X_train.columns)
print(big_df.isnull().sum().sum())
def calculate_classification_metrics(labels, predictions):
predictions = predictions.round()
fpr, tpr, thresholds = metrics.roc_curve(labels, predictions)
auc = metrics.auc(fpr, tpr)
aupr = metrics.average_precision_score(labels,predictions)
return metrics.accuracy_score(labels, predictions),\
metrics.f1_score(labels, predictions, average='binary'),\
auc,\
aupr
def calculate_regression_metrics(labels, predictions):
return metrics.mean_absolute_error(labels, predictions),\
metrics.mean_squared_error(labels, predictions),\
metrics.r2_score(labels, predictions),\
scipy.stats.pearsonr(np.array(labels).flatten(),np.array(predictions.flatten()))[0],\
scipy.stats.spearmanr(np.array(labels).flatten(),np.array(predictions.flatten()))[0]
def supervised_learning_steps(method,scoring,data_type,task,model,params,X_train,y_train,n_iter):
gs = grid_search_cv(model, params, X_train, y_train, scoring=scoring, n_iter = n_iter)
y_pred = gs.predict(X_train)
y_pred[y_pred < 0] = 0
if task:
results=calculate_classification_metrics(y_train, y_pred)
print("Acc: %.3f, F1: %.3f, AUC: %.3f, AUPR: %.3f" % (results[0], results[1], results[2], results[3]))
else:
results=calculate_regression_metrics(y_train,y_pred)
print("MAE: %.3f, MSE: %.3f, R2: %.3f, Pearson R: %.3f, Spearman R: %.3f" % (results[0], results[1], results[2], results[3], results[4]))
print('Parameters')
print('----------')
for p,v in gs.best_estimator_.get_params().items():
print(p, ":", v)
print('-' * 80)
if task:
save_model(gs, "%s_models/%s_%s_classifier_gs.pk" % (method,method,data_type))
save_model(gs.best_estimator_, "%s_models/%s_%s_classifier_best_estimator.pk" %(method,method,data_type))
else:
save_model(gs, "%s_models/%s_%s_regressor_gs.pk" % (method,method,data_type))
save_model(gs.best_estimator_, "%s_models/%s_%s_regressor_best_estimator.pk" %(method,method,data_type))
return(gs)
if classification_task:
model = ensemble.RandomForestRegressor(n_estimators=100, criterion='auc',
max_depth=None, min_samples_split=2,
min_samples_leaf=1, min_weight_fraction_leaf=0.0,
max_features='auto', max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
bootstrap=True, oob_score=False,
n_jobs=-1, random_state=328, verbose=1,
warm_start=False, ccp_alpha=0.0, max_samples=None)
else:
model = ensemble.RandomForestRegressor(n_estimators=100, criterion='mse',
max_depth=None, min_samples_split=2,
min_samples_leaf=1, min_weight_fraction_leaf=0.0,
max_features='auto', max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
bootstrap=True, oob_score=False,
n_jobs=-1, random_state=328, verbose=1,
warm_start=False, ccp_alpha=0.0, max_samples=None)
# Grid parameters
param_rf = {"n_estimators": scipy.stats.randint(20, 500),
"max_depth": scipy.stats.randint(1, 9),
"min_samples_leaf": scipy.stats.randint(1, 10),
"max_features": scipy.stats.uniform.ppf([0.1,0.7])
}
n_iter=200
if classification_task:
rf_gs=supervised_learning_steps("rf","roc_auc",data_type,classification_task,model,param_rf,X_train,y_train,n_iter)
else:
rf_gs=supervised_learning_steps("rf","r2",data_type,classification_task,model,param_rf,X_train,y_train,n_iter)
rf_gs.cv_results_
rf_gs = load_model("rf_models/rf__LS_Drug_LS_Protein_regressor_gs.pk")
np.max(rf_gs.cv_results_["mean_test_score"])
file_list = ["../data/Test_Compound_Viral_interactions_for_Supervised_Learning_with_LS_LS.csv",
"../data/Test_Compound_Viral_interactions_for_Supervised_Learning_with_MFP_LS.csv"]
filename = file_list[input_option]
with open(filename, "rb") as file:
print("Loading ", filename)
big_df = pd.read_csv(filename, header='infer', delimiter=",")
total_length = len(big_df.columns)
X = big_df.iloc[:,range(5,total_length)]
Y = big_df[['pchembl_value']].to_numpy().flatten()
meta_X = big_df.iloc[:,[0,1,2,3]]
print("Lengths --> X = %d, Y = %d" % (len(X), len(Y)))
print(X.columns)
n_samples = len(X)
indices = np.arange(n_samples)
X_test = X
y_test = Y
rf_best = rf_gs.best_estimator_
y_pred_rf=rf_best.predict(X_test)
print(calculate_regression_metrics(y_test,y_pred_rf))
#Write the output in the results folder
meta_X["predictions"]=y_pred_xgb
meta_X["labels"]=y_test
rev_output_df = meta_X.iloc[:,[0,2,4,5]].copy()
rev_output_df.to_csv("../results/RF_"+data_type_options[input_option]+"supervised_test_predictions.csv",index=False)
## load JS visualization code to notebook (Doesn't work for random forest)
#shap.initjs()
## explain the model's predictions using SHAP values
#explainer = shap.TreeExplainer(xgb_gs.best_estimator_)
#shap_values = explainer.shap_values(X_train)
#shap.summary_plot(shap_values, X_train)
##Get results for SARS-COV-2
#big_X_test = pd.read_csv("../data/COVID-19/sars_cov_2_additional_compound_viral_interactions_to_predict_with_LS_v2.csv",header='infer',sep=",")
#total_length = len(big_X_test.columns)
#X_test = big_X_test.iloc[:,range(8,total_length)]
#rf_best = load_model("../models/rf_models/rf__LS_Drug_LS_Protein_regressor_best_estimator.pk")
#y_pred = rf_best.predict(X_test)
#meta_X_test = big_X_test.iloc[:,[0,2]].copy()
#meta_X_test.loc[:,'predictions']=y_pred
#meta_X_test.loc[:,'labels']=0
#meta_X_test.to_csv("../results/RF_supervised_sars_cov2_additional_test_predictions.csv",index=False)
```
|
github_jupyter
|
# 2.4 Deep Taylor Decomposition Part 2.
## Tensorflow Walkthrough
### 1. Import Dependencies
I made a custom `Taylor` class for Deep Taylor Decomposition. If you are interested in the details, check out `models_3_2.py` in the models directory.
```
import os
import re
from tensorflow.examples.tutorials.mnist import input_data
from tensorflow.python.ops import nn_ops, gen_nn_ops
import matplotlib.pyplot as plt
import tensorflow as tf
import numpy as np
from models.models_2_4 import MNIST_CNN, Taylor
%matplotlib inline
mnist = input_data.read_data_sets("MNIST_data/", one_hot=True)
images = mnist.train.images
labels = mnist.train.labels
logdir = './tf_logs/2_4_DTD/'
ckptdir = logdir + 'model'
if not os.path.exists(logdir):
os.mkdir(logdir)
```
### 2. Building Graph
```
with tf.name_scope('Classifier'):
# Initialize neural network
DNN = MNIST_CNN('CNN')
# Setup training process
X = tf.placeholder(tf.float32, [None, 784], name='X')
Y = tf.placeholder(tf.float32, [None, 10], name='Y')
activations, logits = DNN(X)
tf.add_to_collection('DTD', X)
for activation in activations:
tf.add_to_collection('DTD', activation)
cost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits=logits, labels=Y))
optimizer = tf.train.AdamOptimizer().minimize(cost, var_list=DNN.vars)
correct_prediction = tf.equal(tf.argmax(logits, 1), tf.argmax(Y, 1))
accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))
cost_summary = tf.summary.scalar('Cost', cost)
accuray_summary = tf.summary.scalar('Accuracy', accuracy)
summary = tf.summary.merge_all()
```
### 3. Training Network
This is the step where the DNN is trained to classify the 10 digits of the MNIST images. Summaries are written into the logdir and you can visualize the statistics using tensorboard by typing this command: `tensorboard --lodir=./tf_logs`
```
sess = tf.InteractiveSession()
sess.run(tf.global_variables_initializer())
saver = tf.train.Saver()
file_writer = tf.summary.FileWriter(logdir, tf.get_default_graph())
# Hyper parameters
training_epochs = 15
batch_size = 100
for epoch in range(training_epochs):
total_batch = int(mnist.train.num_examples / batch_size)
avg_cost = 0
avg_acc = 0
for i in range(total_batch):
batch_xs, batch_ys = mnist.train.next_batch(batch_size)
_, c, a, summary_str = sess.run([optimizer, cost, accuracy, summary], feed_dict={X: batch_xs, Y: batch_ys})
avg_cost += c / total_batch
avg_acc += a / total_batch
file_writer.add_summary(summary_str, epoch * total_batch + i)
print('Epoch:', '%04d' % (epoch + 1), 'cost =', '{:.9f}'.format(avg_cost), 'accuracy =', '{:.9f}'.format(avg_acc))
saver.save(sess, ckptdir)
print('Accuracy:', sess.run(accuracy, feed_dict={X: mnist.test.images, Y: mnist.test.labels}))
sess.close()
```
### 4. Restoring Subgraph
Here we first rebuild the DNN graph from metagraph, restore DNN parameters from the checkpoint and then gather the necessary weight and biases for Deep Taylor Decomposition using the `tf.get_collection()` function.
```
tf.reset_default_graph()
sess = tf.InteractiveSession()
new_saver = tf.train.import_meta_graph(ckptdir + '.meta')
new_saver.restore(sess, tf.train.latest_checkpoint(logdir))
weights = tf.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES, scope='CNN')
activations = tf.get_collection('DTD')
X = activations[0]
```
### 5. Attaching Subgraph for Calculating Relevance Scores
```
conv_ksize = [1, 3, 3, 1]
pool_ksize = [1, 2, 2, 1]
conv_strides = [1, 1, 1, 1]
pool_strides = [1, 2, 2, 1]
weights.reverse()
activations.reverse()
taylor = Taylor(activations, weights, conv_ksize, pool_ksize, conv_strides, pool_strides, 'Taylor')
Rs = []
for i in range(10):
Rs.append(taylor(i))
```
### 6. Calculating Relevance Scores $R(x_i)$
```
sample_imgs = []
for i in range(10):
sample_imgs.append(images[np.argmax(labels, axis=1) == i][3])
imgs = []
for i in range(10):
imgs.append(sess.run(Rs[i], feed_dict={X: sample_imgs[i][None,:]}))
```
### 7. Displaying Images
The relevance scores are visualized as heat maps. You can see which features/data points influenced the DNN most its decision making.
```
plt.figure(figsize=(15,15))
for i in range(5):
plt.subplot(5, 2, 2 * i + 1)
plt.imshow(np.reshape(imgs[2 * i], [28, 28]), cmap='hot_r')
plt.title('Digit: {}'.format(2 * i))
plt.colorbar()
plt.subplot(5, 2, 2 * i + 2)
plt.imshow(np.reshape(imgs[2 * i + 1], [28, 28]), cmap='hot_r')
plt.title('Digit: {}'.format(2 * i + 1))
plt.colorbar()
plt.tight_layout()
```
|
github_jupyter
|
# Initialization
Welcome to the first assignment of "Improving Deep Neural Networks".
Training your neural network requires specifying an initial value of the weights. A well chosen initialization method will help learning.
If you completed the previous course of this specialization, you probably followed our instructions for weight initialization, and it has worked out so far. But how do you choose the initialization for a new neural network? In this notebook, you will see how different initializations lead to different results.
A well chosen initialization can:
- Speed up the convergence of gradient descent
- Increase the odds of gradient descent converging to a lower training (and generalization) error
To get started, run the following cell to load the packages and the planar dataset you will try to classify.
```
import numpy as np
import matplotlib.pyplot as plt
import sklearn
import sklearn.datasets
from init_utils import sigmoid, relu, compute_loss, forward_propagation, backward_propagation
from init_utils import update_parameters, predict, load_dataset, plot_decision_boundary, predict_dec
%matplotlib inline
plt.rcParams['figure.figsize'] = (7.0, 4.0) # set default size of plots
plt.rcParams['image.interpolation'] = 'nearest'
plt.rcParams['image.cmap'] = 'gray'
# load image dataset: blue/red dots in circles
train_X, train_Y, test_X, test_Y = load_dataset()
```
You would like a classifier to separate the blue dots from the red dots.
## 1 - Neural Network model
You will use a 3-layer neural network (already implemented for you). Here are the initialization methods you will experiment with:
- *Zeros initialization* -- setting `initialization = "zeros"` in the input argument.
- *Random initialization* -- setting `initialization = "random"` in the input argument. This initializes the weights to large random values.
- *He initialization* -- setting `initialization = "he"` in the input argument. This initializes the weights to random values scaled according to a paper by He et al., 2015.
**Instructions**: Please quickly read over the code below, and run it. In the next part you will implement the three initialization methods that this `model()` calls.
```
def model(X, Y, learning_rate = 0.01, num_iterations = 15000, print_cost = True, initialization = "he"):
"""
Implements a three-layer neural network: LINEAR->RELU->LINEAR->RELU->LINEAR->SIGMOID.
Arguments:
X -- input data, of shape (2, number of examples)
Y -- true "label" vector (containing 0 for red dots; 1 for blue dots), of shape (1, number of examples)
learning_rate -- learning rate for gradient descent
num_iterations -- number of iterations to run gradient descent
print_cost -- if True, print the cost every 1000 iterations
initialization -- flag to choose which initialization to use ("zeros","random" or "he")
Returns:
parameters -- parameters learnt by the model
"""
grads = {}
costs = [] # to keep track of the loss
m = X.shape[1] # number of examples
layers_dims = [X.shape[0], 10, 5, 1]
# Initialize parameters dictionary.
if initialization == "zeros":
parameters = initialize_parameters_zeros(layers_dims)
elif initialization == "random":
parameters = initialize_parameters_random(layers_dims)
elif initialization == "he":
parameters = initialize_parameters_he(layers_dims)
# Loop (gradient descent)
for i in range(0, num_iterations):
# Forward propagation: LINEAR -> RELU -> LINEAR -> RELU -> LINEAR -> SIGMOID.
a3, cache = forward_propagation(X, parameters)
# Loss
cost = compute_loss(a3, Y)
# Backward propagation.
grads = backward_propagation(X, Y, cache)
# Update parameters.
parameters = update_parameters(parameters, grads, learning_rate)
# Print the loss every 1000 iterations
if print_cost and i % 1000 == 0:
print("Cost after iteration {}: {}".format(i, cost))
costs.append(cost)
# plot the loss
plt.plot(costs)
plt.ylabel('cost')
plt.xlabel('iterations (per hundreds)')
plt.title("Learning rate =" + str(learning_rate))
plt.show()
return parameters
```
## 2 - Zero initialization
There are two types of parameters to initialize in a neural network:
- the weight matrices $(W^{[1]}, W^{[2]}, W^{[3]}, ..., W^{[L-1]}, W^{[L]})$
- the bias vectors $(b^{[1]}, b^{[2]}, b^{[3]}, ..., b^{[L-1]}, b^{[L]})$
**Exercise**: Implement the following function to initialize all parameters to zeros. You'll see later that this does not work well since it fails to "break symmetry", but lets try it anyway and see what happens. Use np.zeros((..,..)) with the correct shapes.
```
# GRADED FUNCTION: initialize_parameters_zeros
def initialize_parameters_zeros(layers_dims):
"""
Arguments:
layer_dims -- python array (list) containing the size of each layer.
Returns:
parameters -- python dictionary containing your parameters "W1", "b1", ..., "WL", "bL":
W1 -- weight matrix of shape (layers_dims[1], layers_dims[0])
b1 -- bias vector of shape (layers_dims[1], 1)
...
WL -- weight matrix of shape (layers_dims[L], layers_dims[L-1])
bL -- bias vector of shape (layers_dims[L], 1)
"""
parameters = {}
L = len(layers_dims) # number of layers in the network
for l in range(1, L):
### START CODE HERE ### (≈ 2 lines of code)
parameters['W' + str(l)] = np.zeros((layers_dims[l], layers_dims[l-1]))
parameters['b' + str(l)] = np.zeros((layers_dims[l],1))
### END CODE HERE ###
return parameters
parameters = initialize_parameters_zeros([3,2,1])
print("W1 = " + str(parameters["W1"]))
print("b1 = " + str(parameters["b1"]))
print("W2 = " + str(parameters["W2"]))
print("b2 = " + str(parameters["b2"]))
```
**Expected Output**:
<table>
<tr>
<td>
**W1**
</td>
<td>
[[ 0. 0. 0.]
[ 0. 0. 0.]]
</td>
</tr>
<tr>
<td>
**b1**
</td>
<td>
[[ 0.]
[ 0.]]
</td>
</tr>
<tr>
<td>
**W2**
</td>
<td>
[[ 0. 0.]]
</td>
</tr>
<tr>
<td>
**b2**
</td>
<td>
[[ 0.]]
</td>
</tr>
</table>
Run the following code to train your model on 15,000 iterations using zeros initialization.
```
parameters = model(train_X, train_Y, initialization = "zeros")
print ("On the train set:")
predictions_train = predict(train_X, train_Y, parameters)
print ("On the test set:")
predictions_test = predict(test_X, test_Y, parameters)
```
The performance is really bad, and the cost does not really decrease, and the algorithm performs no better than random guessing. Why? Lets look at the details of the predictions and the decision boundary:
```
print ("predictions_train = " + str(predictions_train))
print ("predictions_test = " + str(predictions_test))
plt.title("Model with Zeros initialization")
axes = plt.gca()
axes.set_xlim([-1.5,1.5])
axes.set_ylim([-1.5,1.5])
plot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y)
```
The model is predicting 0 for every example.
In general, initializing all the weights to zero results in the network failing to break symmetry. This means that every neuron in each layer will learn the same thing, and you might as well be training a neural network with $n^{[l]}=1$ for every layer, and the network is no more powerful than a linear classifier such as logistic regression.
<font color='blue'>
**What you should remember**:
- The weights $W^{[l]}$ should be initialized randomly to break symmetry.
- It is however okay to initialize the biases $b^{[l]}$ to zeros. Symmetry is still broken so long as $W^{[l]}$ is initialized randomly.
## 3 - Random initialization
To break symmetry, lets intialize the weights randomly. Following random initialization, each neuron can then proceed to learn a different function of its inputs. In this exercise, you will see what happens if the weights are intialized randomly, but to very large values.
**Exercise**: Implement the following function to initialize your weights to large random values (scaled by \*10) and your biases to zeros. Use `np.random.randn(..,..) * 10` for weights and `np.zeros((.., ..))` for biases. We are using a fixed `np.random.seed(..)` to make sure your "random" weights match ours, so don't worry if running several times your code gives you always the same initial values for the parameters.
```
# GRADED FUNCTION: initialize_parameters_random
def initialize_parameters_random(layers_dims):
"""
Arguments:
layer_dims -- python array (list) containing the size of each layer.
Returns:
parameters -- python dictionary containing your parameters "W1", "b1", ..., "WL", "bL":
W1 -- weight matrix of shape (layers_dims[1], layers_dims[0])
b1 -- bias vector of shape (layers_dims[1], 1)
...
WL -- weight matrix of shape (layers_dims[L], layers_dims[L-1])
bL -- bias vector of shape (layers_dims[L], 1)
"""
np.random.seed(3) # This seed makes sure your "random" numbers will be the as ours
parameters = {}
L = len(layers_dims) # integer representing the number of layers
for l in range(1, L):
### START CODE HERE ### (≈ 2 lines of code)
parameters['W' + str(l)] = np.random.randn(layers_dims[l], layers_dims[l-1]) * 10
parameters['b' + str(l)] = np.zeros((layers_dims[l], 1))
### END CODE HERE ###
return parameters
parameters = initialize_parameters_random([3, 2, 1])
print("W1 = " + str(parameters["W1"]))
print("b1 = " + str(parameters["b1"]))
print("W2 = " + str(parameters["W2"]))
print("b2 = " + str(parameters["b2"]))
```
**Expected Output**:
<table>
<tr>
<td>
**W1**
</td>
<td>
[[ 17.88628473 4.36509851 0.96497468]
[-18.63492703 -2.77388203 -3.54758979]]
</td>
</tr>
<tr>
<td>
**b1**
</td>
<td>
[[ 0.]
[ 0.]]
</td>
</tr>
<tr>
<td>
**W2**
</td>
<td>
[[-0.82741481 -6.27000677]]
</td>
</tr>
<tr>
<td>
**b2**
</td>
<td>
[[ 0.]]
</td>
</tr>
</table>
Run the following code to train your model on 15,000 iterations using random initialization.
```
parameters = model(train_X, train_Y, initialization = "random")
print ("On the train set:")
predictions_train = predict(train_X, train_Y, parameters)
print ("On the test set:")
predictions_test = predict(test_X, test_Y, parameters)
```
If you see "inf" as the cost after the iteration 0, this is because of numerical roundoff; a more numerically sophisticated implementation would fix this. But this isn't worth worrying about for our purposes.
Anyway, it looks like you have broken symmetry, and this gives better results. than before. The model is no longer outputting all 0s.
```
print (predictions_train)
print (predictions_test)
plt.title("Model with large random initialization")
axes = plt.gca()
axes.set_xlim([-1.5,1.5])
axes.set_ylim([-1.5,1.5])
plot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y)
```
**Observations**:
- The cost starts very high. This is because with large random-valued weights, the last activation (sigmoid) outputs results that are very close to 0 or 1 for some examples, and when it gets that example wrong it incurs a very high loss for that example. Indeed, when $\log(a^{[3]}) = \log(0)$, the loss goes to infinity.
- Poor initialization can lead to vanishing/exploding gradients, which also slows down the optimization algorithm.
- If you train this network longer you will see better results, but initializing with overly large random numbers slows down the optimization.
<font color='blue'>
**In summary**:
- Initializing weights to very large random values does not work well.
- Hopefully intializing with small random values does better. The important question is: how small should be these random values be? Lets find out in the next part!
## 4 - He initialization
Finally, try "He Initialization"; this is named for the first author of He et al., 2015. (If you have heard of "Xavier initialization", this is similar except Xavier initialization uses a scaling factor for the weights $W^{[l]}$ of `sqrt(1./layers_dims[l-1])` where He initialization would use `sqrt(2./layers_dims[l-1])`.)
**Exercise**: Implement the following function to initialize your parameters with He initialization.
**Hint**: This function is similar to the previous `initialize_parameters_random(...)`. The only difference is that instead of multiplying `np.random.randn(..,..)` by 10, you will multiply it by $\sqrt{\frac{2}{\text{dimension of the previous layer}}}$, which is what He initialization recommends for layers with a ReLU activation.
```
# GRADED FUNCTION: initialize_parameters_he
def initialize_parameters_he(layers_dims):
"""
Arguments:
layer_dims -- python array (list) containing the size of each layer.
Returns:
parameters -- python dictionary containing your parameters "W1", "b1", ..., "WL", "bL":
W1 -- weight matrix of shape (layers_dims[1], layers_dims[0])
b1 -- bias vector of shape (layers_dims[1], 1)
...
WL -- weight matrix of shape (layers_dims[L], layers_dims[L-1])
bL -- bias vector of shape (layers_dims[L], 1)
"""
np.random.seed(3)
parameters = {}
L = len(layers_dims) - 1 # integer representing the number of layers
for l in range(1, L + 1):
### START CODE HERE ### (≈ 2 lines of code)
parameters['W' + str(l)] = np.random.randn(layers_dims[l], layers_dims[l-1]) * np.sqrt(2./layers_dims[l-1])
parameters['b' + str(l)] = np.zeros((layers_dims[l], 1))
### END CODE HERE ###
return parameters
parameters = initialize_parameters_he([2, 4, 1])
print("W1 = " + str(parameters["W1"]))
print("b1 = " + str(parameters["b1"]))
print("W2 = " + str(parameters["W2"]))
print("b2 = " + str(parameters["b2"]))
```
**Expected Output**:
<table>
<tr>
<td>
**W1**
</td>
<td>
[[ 1.78862847 0.43650985]
[ 0.09649747 -1.8634927 ]
[-0.2773882 -0.35475898]
[-0.08274148 -0.62700068]]
</td>
</tr>
<tr>
<td>
**b1**
</td>
<td>
[[ 0.]
[ 0.]
[ 0.]
[ 0.]]
</td>
</tr>
<tr>
<td>
**W2**
</td>
<td>
[[-0.03098412 -0.33744411 -0.92904268 0.62552248]]
</td>
</tr>
<tr>
<td>
**b2**
</td>
<td>
[[ 0.]]
</td>
</tr>
</table>
Run the following code to train your model on 15,000 iterations using He initialization.
```
parameters = model(train_X, train_Y, initialization = "he")
print ("On the train set:")
predictions_train = predict(train_X, train_Y, parameters)
print ("On the test set:")
predictions_test = predict(test_X, test_Y, parameters)
plt.title("Model with He initialization")
axes = plt.gca()
axes.set_xlim([-1.5,1.5])
axes.set_ylim([-1.5,1.5])
plot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y)
```
**Observations**:
- The model with He initialization separates the blue and the red dots very well in a small number of iterations.
## 5 - Conclusions
You have seen three different types of initializations. For the same number of iterations and same hyperparameters the comparison is:
<table>
<tr>
<td>
**Model**
</td>
<td>
**Train accuracy**
</td>
<td>
**Problem/Comment**
</td>
</tr>
<td>
3-layer NN with zeros initialization
</td>
<td>
50%
</td>
<td>
fails to break symmetry
</td>
<tr>
<td>
3-layer NN with large random initialization
</td>
<td>
83%
</td>
<td>
too large weights
</td>
</tr>
<tr>
<td>
3-layer NN with He initialization
</td>
<td>
99%
</td>
<td>
recommended method
</td>
</tr>
</table>
<font color='blue'>
**What you should remember from this notebook**:
- Different initializations lead to different results
- Random initialization is used to break symmetry and make sure different hidden units can learn different things
- Don't intialize to values that are too large
- He initialization works well for networks with ReLU activations.
```
!tar cvfz notebook.tar.gz *
!tar cvfz notebook.tar.gz *
!tar cvfz notebook.tar.gz *
```
|
github_jupyter
|
```
# The usual preamble
%matplotlib inline
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
# Make the graphs a bit prettier, and bigger
plt.style.use('ggplot')
plt.rcParams['figure.figsize'] = (15, 5)
plt.rcParams['font.family'] = 'sans-serif'
# This is necessary to show lots of columns in pandas 0.12.
# Not necessary in pandas 0.13.
pd.set_option('display.width', 5000)
pd.set_option('display.max_columns', 60)
```
One of the main problems with messy data is: how do you know if it's messy or not?
We're going to use the NYC 311 service request dataset again here, since it's big and a bit unwieldy.
```
requests = pd.read_csv('data/311-service-requests.csv', low_memory=False)
```
# 7.1 How do we know if it's messy?
We're going to look at a few columns here. I know already that there are some problems with the zip code, so let's look at that first.
To get a sense for whether a column has problems, I usually use `.unique()` to look at all its values. If it's a numeric column, I'll instead plot a histogram to get a sense of the distribution.
When we look at the unique values in "Incident Zip", it quickly becomes clear that this is a mess.
Some of the problems:
* Some have been parsed as strings, and some as floats
* There are `nan`s
* Some of the zip codes are `29616-0759` or `83`
* There are some N/A values that pandas didn't recognize, like 'N/A' and 'NO CLUE'
What we can do:
* Normalize 'N/A' and 'NO CLUE' into regular nan values
* Look at what's up with the 83, and decide what to do
* Make everything strings
```
requests['Incident Zip'].unique()
```
# 7.2 Fixing the nan values and string/float confusion
We can pass a `na_values` option to `pd.read_csv` to clean this up a little bit. We can also specify that the type of Incident Zip is a string, not a float.
```
na_values = ['NO CLUE', 'N/A', '0']
requests = pd.read_csv('data/311-service-requests.csv', na_values=na_values, dtype={'Incident Zip': str})
requests['Incident Zip'].unique()
```
# 7.3 What's up with the dashes?
```
rows_with_dashes = requests['Incident Zip'].str.contains('-').fillna(False)
len(requests[rows_with_dashes])
requests[rows_with_dashes]
```
I thought these were missing data and originally deleted them like this:
`requests['Incident Zip'][rows_with_dashes] = np.nan`
But then my friend Dave pointed out that 9-digit zip codes are normal. Let's look at all the zip codes with more than 5 digits, make sure they're okay, and then truncate them.
```
long_zip_codes = requests['Incident Zip'].str.len() > 5
requests['Incident Zip'][long_zip_codes].unique()
```
Those all look okay to truncate to me.
```
requests['Incident Zip'] = requests['Incident Zip'].str.slice(0, 5)
```
Done.
Earlier I thought 00083 was a broken zip code, but turns out Central Park's zip code 00083! Shows what I know. I'm still concerned about the 00000 zip codes, though: let's look at that.
```
requests[requests['Incident Zip'] == '00000']
```
This looks bad to me. Let's set these to nan.
```
zero_zips = requests['Incident Zip'] == '00000'
requests.loc[zero_zips, 'Incident Zip'] = np.nan
```
Great. Let's see where we are now:
```
unique_zips = requests['Incident Zip'].unique()
unique_zips
```
Amazing! This is much cleaner. There's something a bit weird here, though -- I looked up 77056 on Google maps, and that's in Texas.
Let's take a closer look:
```
zips = requests['Incident Zip']
# Let's say the zips starting with '0' and '1' are okay, for now. (this isn't actually true -- 13221 is in Syracuse, and why?)
is_close = zips.str.startswith('0') | zips.str.startswith('1')
# There are a bunch of NaNs, but we're not interested in them right now, so we'll say they're False
is_far = ~(is_close) & zips.notnull()
zips[is_far]
requests[is_far][['Incident Zip', 'Descriptor', 'City']].sort_values(by='Incident Zip')
```
Okay, there really are requests coming from LA and Houston! Good to know. Filtering by zip code is probably a bad way to handle this -- we should really be looking at the city instead.
```
requests['City'].str.upper().value_counts()
```
It looks like these are legitimate complaints, so we'll just leave them alone.
# 7.4 Putting it together
Here's what we ended up doing to clean up our zip codes, all together:
```
na_values = ['NO CLUE', 'N/A', '0']
requests = pd.read_csv('data/311-service-requests.csv',
na_values=na_values,
dtype={'Incident Zip': str})
def fix_zip_codes(zips):
# Truncate everything to length 5
zips = zips.str.slice(0, 5)
# Set 00000 zip codes to nan
zero_zips = zips == '00000'
zips[zero_zips] = np.nan
return zips
requests['Incident Zip'] = fix_zip_codes(requests['Incident Zip'])
requests['Incident Zip'].unique()
```
<style>
@font-face {
font-family: "Computer Modern";
src: url('http://mirrors.ctan.org/fonts/cm-unicode/fonts/otf/cmunss.otf');
}
div.cell{
width:800px;
margin-left:16% !important;
margin-right:auto;
}
h1 {
font-family: Helvetica, serif;
}
h4{
margin-top:12px;
margin-bottom: 3px;
}
div.text_cell_render{
font-family: Computer Modern, "Helvetica Neue", Arial, Helvetica, Geneva, sans-serif;
line-height: 145%;
font-size: 130%;
width:800px;
margin-left:auto;
margin-right:auto;
}
.CodeMirror{
font-family: "Source Code Pro", source-code-pro,Consolas, monospace;
}
.text_cell_render h5 {
font-weight: 300;
font-size: 22pt;
color: #4057A1;
font-style: italic;
margin-bottom: .5em;
margin-top: 0.5em;
display: block;
}
.warning{
color: rgb( 240, 20, 20 )
}
|
github_jupyter
|
```
import boto3
import sagemaker
import time
import pandas as pd
import numpy as np
role = sagemaker.get_execution_role()
region = boto3.Session().region_name
sagemaker_session = sagemaker.Session()
bucket_name = sagemaker_session.default_bucket()
prefix = 'endtoendmlsm'
print(region)
print(role)
print(bucket_name)
s3 = boto3.resource('s3')
file_key = 'data/raw/windturbine_raw_data.csv'
copy_source = {
'Bucket': 'gianpo-public',
'Key': 'endtoendml/{0}'.format(file_key)
}
s3.Bucket(bucket_name).copy(copy_source, '{0}/'.format(prefix) + file_key)
#sagemaker_session.upload_data('/home/ec2-user/SageMaker/windturbine_raw_data_2.csv', bucket=bucket_name, key_prefix='endtoendmlsm/data/raw')
sagemaker_session.download_data(path='.', bucket=bucket_name, key_prefix='endtoendmlsm/data/raw/windturbine_raw_data.csv', extra_args=None)
original_ds = pd.read_csv('./windturbine_raw_data.csv', names = ['turbine_id','turbine_type','wind_speed','rpm_blade','oil_temperature','oil_level','temperature','humidity','vibrations_frequency','pressure','wind_direction','breakdown'])
original_ds.head()
original_ds.describe(include='all')
%matplotlib inline
import matplotlib.pyplot as plt
plt.scatter(original_ds.wind_speed, original_ds.rpm_blade)
plt.hist(original_ds.wind_speed, bins=70)
#wind_speed: gaussian, mean=50, std=30
ws = abs(np.random.normal(50, 30, 1000000)).astype(int)
#temperature: gaussian, mean=20, std=18
temp = abs(np.random.normal(20, 18, 1000000)).astype(int)
#humidity: gaussian, mean=50, std=5
hum = abs(np.random.normal(50, 5, 1000000)).astype(int)
#pressure: gaussian, mean=40, std=25
press = abs(np.random.normal(40, 25, 1000000)).astype(int)
#oil_level: uniform, min=5, max=35
oil_lev = np.random.uniform(5,35,1000000).astype(int)
#rpm_blade: alpha*wind_speed + error
alpha = 5
rpm_blade = abs(alpha*ws + np.random.normal(0,30,1000000)).astype(int)
#vibration_freq: beta*rpm_blade + gamma*pressure + error
beta = 3.5
gamma = 2
vibration_freq = abs(beta*rpm_blade + gamma*press + np.random.normal(0,50,1000000)).astype(int)
#oil_temp: delta*temp + error
#delta = 4.5
#oil_temperature = abs(delta*temp + np.random.normal(0,50,1000000)).astype(int)
#breakdown: k1*rpm_blade + k2*vibration_freq + k3*oil_temp + error
new_dataset = pd.DataFrame()
new_dataset['turbine_id'] = original_ds['turbine_id']
new_dataset['turbine_type'] = original_ds['turbine_type']
new_dataset['wind_direction'] = original_ds['wind_direction']
new_dataset['wind_speed'] = ws
new_dataset['temperature'] = temp
new_dataset['humidity'] = hum
new_dataset['pressure'] = press
new_dataset['oil_level'] = oil_lev
new_dataset['rpm_blade'] = rpm_blade
new_dataset['vibrations_frequency'] = vibration_freq
new_dataset['oil_temperature'] = original_ds['oil_temperature']
new_dataset.describe()
plt.scatter(new_dataset['wind_speed'][:10000], new_dataset['rpm_blade'][:10000])
plt.hist(new_dataset['rpm_blade'][:10000])
from scipy.special import expit
k1=0.0003
k2=0.0005
k3=0.0033
breakdown = k1*rpm_blade + k2*vibration_freq + k3*oil_lev + np.random.normal(0,0.1,1000000)
new_dataset['breakdown_num'] = breakdown
new_dataset.loc[new_dataset['breakdown_num'] <= 0.9, 'breakdown'] = 'no'
new_dataset.loc[new_dataset['breakdown_num'] > 0.9, 'breakdown'] = 'yes'
new_dataset.describe(include='all')
plt.scatter(new_dataset['breakdown'][:10000], new_dataset['rpm_blade'][:10000])
plt.scatter(new_dataset['breakdown'][:10000], new_dataset['vibrations_frequency'][:10000])
final_dataset = new_dataset
final_dataset = final_dataset.drop(columns=['breakdown_num'])
final_dataset.to_csv('windturbine_raw_data.csv', index=False, columns = ['turbine_id','turbine_type','wind_speed','rpm_blade','oil_temperature','oil_level','temperature','humidity','vibrations_frequency','pressure','wind_direction','breakdown'])
sagemaker_session.upload_data('windturbine_raw_data.csv', bucket=bucket_name, key_prefix='endtoendmlsm/data/raw')
```
|
github_jupyter
|
---
title: "Create empty feature groups for Online Feature Store"
date: 2021-04-25
type: technical_note
draft: false
---
```
import json
from pyspark.sql.types import StructField, StructType, StringType, DoubleType, TimestampType, LongType, IntegerType
```
# Create empty feature groups
In this demo example we are expecting to recieve data from Kafka topic, read using spark streaming, do streamig aggregations and ingest aggregated data to feature groups. Thus we will create empy feature groups where we will ingest streaming data.
### Define schema for feature groups
```
card_schema = StructType([StructField('tid', StringType(), True),
StructField('datetime', StringType(), True),
StructField('cc_num', LongType(), True),
StructField('amount', DoubleType(), True)])
schema_10m = StructType([StructField('cc_num', LongType(), True),
StructField('num_trans_per_10m', LongType(), True),
StructField('avg_amt_per_10m', DoubleType(), True),
StructField('stdev_amt_per_10m', DoubleType(), True)])
schema_1h = StructType([StructField('cc_num', LongType(), True),
StructField('num_trans_per_1h', LongType(), True),
StructField('avg_amt_per_1h', DoubleType(), True),
StructField('stdev_amt_per_1h', DoubleType(), True)])
schema_12h = StructType([StructField('cc_num', LongType(), True),
StructField('num_trans_per_12h', LongType(), True),
StructField('avg_amt_per_12h', DoubleType(), True),
StructField('stdev_amt_per_12h', DoubleType(), True)])
```
### Create empty spark dataframes
```
empty_card_df = sqlContext.createDataFrame(sc.emptyRDD(), card_schema)
empty_10m_agg_df = sqlContext.createDataFrame(sc.emptyRDD(), schema_10m)
empty_1h_agg_df = sqlContext.createDataFrame(sc.emptyRDD(), schema_1h)
empty_12h_agg_df = sqlContext.createDataFrame(sc.emptyRDD(), schema_12h)
```
### Establish a connection with your Hopsworks feature store.
```
import hsfs
connection = hsfs.connection()
# get a reference to the feature store, you can access also shared feature stores by providing the feature store name
fs = connection.get_feature_store()
```
### Create feature group metadata objects and save empty spark dataframes to materialise them in hopsworks feature store.
Now We will create each feature group and enable online feature store. Since we are plannig to use these feature groups durring online model serving primary key(s) are required to retrieve feature vector from online feature store.
```
card_transactions = fs.create_feature_group("card_transactions",
version = 1,
online_enabled=False,
statistics_config=False,
primary_key=["tid"])
card_transactions.save(empty_card_df)
card_transactions_10m_agg = fs.create_feature_group("card_transactions_10m_agg",
version = 1,
online_enabled=True,
statistics_config=False,
primary_key=["cc_num"])
card_transactions_10m_agg.save(empty_10m_agg_df)
card_transactions_1h_agg = fs.create_feature_group("card_transactions_1h_agg",
version = 1,
online_enabled=True,
statistics_config=False,
primary_key=["cc_num"])
card_transactions_1h_agg.save(empty_1h_agg_df)
card_transactions_12h_agg = fs.create_feature_group("card_transactions_12h_agg",
version = 1,
online_enabled=True,
statistics_config=False,
primary_key=["cc_num"])
card_transactions_12h_agg.save(empty_12h_agg_df)
```
|
github_jupyter
|
# Freesurfer space to native space using `mri_vol2vol`
BMED360-2021: `freesurfer-to-native-space.ipynb`
```
%matplotlib inline
import os
import pathlib
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import nibabel as nib
from os.path import expanduser, join, basename, split
import sys
sys.path.append('.') # path to utils.py
import utils
import time
import shutil # copy files
cwd = os.getcwd()
```
### We will use the `fs711_subjects` Freesurfer tree previously run for the `bids_bg_bmed360` sample
```
fs711_home = '/usr/local/freesurfer'
working_dir = join(cwd, 'data')
bids_dir = '%s/bids_bg_bmed360' % (working_dir)
fs711_subj = '%s/fs711_subjects' % (working_dir)
dmri_res = '%s/dmri_results' % (working_dir)
if not os.path.exists(dmri_res):
os.makedirs(dmri_res)
else:
print('subdirectory dmri_results already exists')
```
The Freesurfer environment:
```
%%bash -s '/usr/local/freesurfer' './data/fs711_subjects'
echo $1
echo $2
FREESURFER_HOME=${1}; export FREESURFER_HOME
PATH=${FREESURFER_HOME}/bin:${PATH}; export PATH
SUBJECTS_DIR=${2}; export SUBJECTS_DIR
FSLDIR=/usr/local/fsl; export FSLDIR
PATH=${FSLDIR}/bin:${PATH}; export PATH
. ${FSLDIR}/etc/fslconf/fsl.sh
source ${FREESURFER_HOME}/SetUpFreeSurfer.sh
```
## How to Convert from FreeSurfer Space Back to Native Anatomical Space
See: https://surfer.nmr.mgh.harvard.edu/fswiki/FsAnat-to-NativeAnat
Question: I have successfully run a subject's data through [FreeSurfer](https://surfer.nmr.mgh.harvard.edu/fswiki/FreeSurfer).
FreeSurfer creates volumes in 1 mm$^3$, 256$^3$ space, but I want the FreeSurfer results in the space of my original anatomical. How do I do this?<br>
The exact command you use depends on what you want to convert, an image (like brain.mgz) or a segmentation (like aseg.mgz).
For an image:
```
# cd $SUBJECTS_DIR/<subjid>/mri
# mri_vol2vol --mov brain.mgz --targ rawavg.mgz --regheader --o brain-in-rawavg.mgz --no-save-reg
```
For a segmentation (aseg.mgz, aparc+aseg.mgz, wmparc.mgz, etc):
```
# cd $SUBJECTS_DIR/<subjid>/mri
# mri_label2vol --seg aseg.mgz --temp rawavg.mgz --o aseg-in-rawavg.mgz --regheader aseg.mgz
```
Map the surface to the native space:
```
# mri_surf2surf --sval-xyz pial --reg register.native.dat rawavg.mgz --tval lh.pial.native --tval-xyz rawavg.mgz --hemi lh --s subjectname
```
The output will be stored in $SUBJECTS_DIR/subjectname/surf/lh.pial.native and can be viewed with freeview rawavg.mgz -f ../surf/lh.pial.native<br>
To verify that this worked, run
```
# freeview -v rawavg.mgz -f lh.pial.native
MRI_VOL2VOL = '%s/bin/mri_vol2vol' % (fs711_home)
print(os.popen(MRI_VOL2VOL).read())
def my_mri_vol2vol(subj_dir, sub, ses, inp_image, targ_image, targ_name, out_dir, interptype):
"""
Ex.
cd $SUBJECTS_DIR/<subjid>/mri
mri_vol2vol --mov brain.mgz --targ rawavg.mgz --regheader --o brain-in-rawavg.mgz --no-save-reg
--interp interptype : interpolation cubic, trilin, nearest (def is trilin)
"""
fs_mri = join('%s' % (subj_dir), 'sub_%d_tp%d/mri' % (sub, ses))
cmd = [
MRI_VOL2VOL,
'--mov', '%s/%s.mgz' % (fs_mri, inp_image),
'--targ', '%s' % (targ_image),
'--regheader',
'--interp', '%s' % (interptype),
'--o', '%s/sub_%d_tp%d_%s_in_%s.nii.gz' % (out_dir, sub, ses, inp_image, targ_name),
'--no-save-reg']
# ' 2>', error_output_log,'>', output_log]
cmd_str = " ".join(cmd)
#print('cmd_str = \n%s\n' % cmd_str)
# EXECUTE
os.system(cmd_str)
```
### Testing the native space conversion on one subject (sub_102_tp1)
**using the `sub-102_ses-1_T1w.nii.gz` in the `bids_bg_bmed360` tree as target image**
```
subj_dir = fs711_subj
out_dir = dmri_res
sub = 102
ses = 1
targ_image = '%s/sub-%d/ses-%d/anat/sub-%d_ses-%d_T1w.nii.gz' % (bids_dir, sub, ses, sub, ses)
targ_name = 'native_space'
```
**Use the `my_mri_vol2vol()`function on different source images and masks using approriate interpolation ('trilinear' and 'nearest neighbour')**
```
%%time
shutil.copy2(targ_image,out_dir) # copy the original anatomy file in bids tree to out_dir
interptype = 'trilin'
inp_image = 'orig'
my_mri_vol2vol(subj_dir, sub, ses, inp_image, targ_image, targ_name, out_dir, interptype)
interptype = 'trilin'
inp_image = 'brain'
my_mri_vol2vol(subj_dir, sub, ses, inp_image, targ_image, targ_name, out_dir, interptype)
interptype = 'nearest'
inp_image = 'ribbon'
my_mri_vol2vol(subj_dir, sub, ses, inp_image, targ_image, targ_name, out_dir, interptype)
interptype = 'nearest'
inp_image = 'aseg'
my_mri_vol2vol(subj_dir, sub, ses, inp_image, targ_image, targ_name, out_dir, interptype)
interptype = 'nearest'
inp_image = 'wmparc'
my_mri_vol2vol(subj_dir, sub, ses, inp_image, targ_image, targ_name, out_dir, interptype)
interptype = 'nearest'
inp_image = 'aparc+aseg'
my_mri_vol2vol(subj_dir, sub, ses, inp_image, targ_image, targ_name, out_dir, interptype)
```
#### Run the native space conversion on all subjects and sessions using the `_T1_biascorr_brain.nii.gz` image obtained from `03-fsl-anat.ipynb` as target image.
```
%%time
subj_dir = fs711_subj
bids_dir = bids_dir
out_dir = dmri_res
targ_name = 'native_space'
for sub in [102, 103, 111, 123]:
for ses in [1, 2]:
print(f'Computing sub:{sub} ses:{ses}')
targ_image = join(bids_dir,'sub-%d/ses-%d/anat/sub-%d_ses-%d_T1w.nii.gz' % (sub, ses, sub, ses))
shutil.copy2(targ_image,out_dir) # copy the original anatomy file in bids tree to out_dir
inp_image = 'orig'
interptype = 'trilin'
my_mri_vol2vol(subj_dir, sub, ses, inp_image, targ_image, targ_name, out_dir, interptype)
inp_image = 'brain'
interptype = 'trilin'
my_mri_vol2vol(subj_dir, sub, ses, inp_image, targ_image, targ_name, out_dir, interptype)
inp_image = 'brainmask'
interptype = 'nearest'
my_mri_vol2vol(subj_dir, sub, ses, inp_image, targ_image, targ_name, out_dir, interptype)
inp_image = 'ribbon'
interptype = 'nearest'
my_mri_vol2vol(subj_dir, sub, ses, inp_image, targ_image, targ_name, out_dir, interptype)
inp_image = 'aseg'
interptype = 'nearest'
my_mri_vol2vol(subj_dir, sub, ses, inp_image, targ_image, targ_name, out_dir, interptype)
inp_image = 'wmparc'
interptype = 'nearest'
my_mri_vol2vol(subj_dir, sub, ses, inp_image, targ_image, targ_name, out_dir, interptype)
inp_image = 'aparc+aseg'
interptype = 'nearest'
my_mri_vol2vol(subj_dir, sub, ses, inp_image, targ_image, targ_name, out_dir, interptype)
```
|
github_jupyter
|
# Tokenizers
```
! pipenv install nltk
import nltk
from nltk import tokenize
s1 = """Why wase time say lot word when few word do trick?"""
s2 = """Hickory dickory dock, the mouse ran up the clock."""
from nltk.tokenize import word_tokenize
! df -h /home/christangrant/nltk_data
# nltk.download('punkt') # Download the model
word_tokenize(s1)
word_tokenize(s2)
paragraph = [s1,s2]
paragraph
' '.join(paragraph)
from nltk.tokenize import sent_tokenize
sent_tokenize(' '.join(paragraph))
[ word_tokenize(sent) for sent in sent_tokenize(' '.join(paragraph))]
```
# Vectorization
Given a corpus C, normalize C, then vectorize C.
Vectorizing C gives us a **vocabulary** and it gives us **weights**.
Get vocabulary => vocabulary is a list of terms that appear in the corpus.
Create a vector where each entry represents a voculary item.
If our vocabulary size is 10K, our vector size if 10K. (Good Normalization shrinks this vector size.)
If V is the vocabulary, V + 5 => vocabulary size.
- OOV (Out of Vocabulary) /Unknown terms
- Redacted terms
- grouped terms
-
Corpus =
>Old macdonald had a farm, on his farm he had a cow.
>Old macdonald had a farm, on his farm he had a pig.
>Old macdonald had a farm, on his farm he had a goat.
Normalize(Corpus)
>Old, macdonald, farm, pig, cow, goat
```pre
Old, macdonald, farm, pig, cow, goat
[1, 1, 1, 0, 1 ,0]
[1, 1, 1, 1 ,0, 0]
[1, 1, 1, 0, 0, 1]
```
**One-hot encoding** of text.
How can we get some positional information in this one-hot encoded format?
> Use weights to represent positions?
> Use nggrams to group terms together
### N-gram encoding
Vocabulary size grows with the size of the ngram
```pre
Old, macdonald, farm, pig, cow, goat
<s> Old, Old macdonald, macdonald farm, farm pig, pig cow, cow goat, goat </s>
[1,1,1,0,0,0,0]
[1,1,1,0,0,0,1]
[1,1,1,0,0,0,0]
```
Useful to have a range of n-grams when vectorizing.
## Bag of words model
Unordered bag representation of the vocabulary.
>Old macdonald had a farm, on his farm he had a cow.
>Old macdonald had a farm, on his farm he had a pig.
>Old macdonald had a farm, on his farm he had a goat.
```pre
bow =
Old, macdonald, farm, pig, cow, goat
[1, 1, 2, 0, 1 ,0]
[1, 1, 2, 1 ,0, 0]
[1, 1, 2, 0, 0, 1]
```
Unique words may be important!
## Term frequency
The raw frequency value of a term in a particular document.
$$
tf(word,document) = \sum_{v \in D} 1 (if v == w)
$$
## Document frequency
The number of documents that contain a word w.
## TF*IDF
idf = 1/df = log(N/(df+epslilon)) + epsilon
Term frequency * Inverse document freqency
## Smoothing.
Adding a small term to help handle out of vocabulary errors floating point issues.
```
! pipenv install sklearn
```
Sklearn has an api to documents.
>Transformers take a document and processes it using the function .fit_transform
>Vectorizers take a document and process it using .fit()
>fit create an internal model using the document
```
corpus = [ word_tokenize(sent) for sent in sent_tokenize(' '.join(paragraph))]
corpus
import sklearn
from sklearn.feature_extraction.text import CountVectorizer
cv = CountVectorizer(min_df=0., max_df=1.)
cv_matrix = cv.fit_transform([s1, s2])
print(cv_matrix)
cv_matrix.toarray()
vocab = cv.get_feature_names_out()
vocab
import pandas as pd
df = pd.DataFrame(cv_matrix.toarray(), columns=vocab)
df
bv = CountVectorizer(ngram_range=(2,2))
bv_matrix = bv.fit_transform([s1, s2])
vocab = bv.get_feature_names_out()
df1 = pd.DataFrame(bv_matrix.toarray(), columns=vocab)
df1
```
|
github_jupyter
|
```
%load_ext autoreload
%autoreload 2
from IPython.display import Markdown, display
def printmd(string):
display(Markdown(string))
def colorize(string,color="red"):
return f"<span style=\"color:{color}\">{string}</span>"
```
# Problem description
### Subtask2: Detecting antecedent and consequence
Indicating causal insight is an inherent characteristic of counterfactual. To further detect the causal knowledge conveyed in counterfactual statements, subtask 2 aims to locate antecedent and consequent in counterfactuals.
According to (Nelson Goodman, 1947. The problem of counterfactual conditionals), a counterfactual statement can be converted to a contrapositive with a true antecedent and consequent. Consider example “Her post-traumatic stress could have been avoided if a combination of paroxetine and exposure therapy had been prescribed two months earlier”; it can be transposed into “because her post-traumatic stress was not avoided, (we know) a combination of paroxetine and exposure therapy was not prescribed”. Such knowledge can be not only used for analyzing the specific statement but also be accumulated across corpora to develop domain causal knowledge (e.g., a combination of paroxetine and exposure may help cure post-traumatic stress).
Please note that __in some cases there is only an antecedent part while without a consequent part in a counterfactual statement__. For example, "Frankly, I wish he had issued this order two years ago instead of this year", in this sentence we could only get the antecedent part. In our subtask2, when locating the antecedent and consequent part, please set '-1' as consequent starting index (character index) and ending index (character index) to refer that there is no consequent part in this sentence. For details, please refer to the 'Evaluation' on this website.
```
!ls
import pandas as pd
!pwd
df = pd.read_csv('../../.data/semeval2020_5/train_task2.csv')
```
We have this amount of data:
```
len(df)
import random
i = random.randint(0,len(df))
print(df.iloc[i])
print("-"*50)
print(df["sentence"].iloc[i])
print("-"*50)
print(df["antecedent"].iloc[i])
print("-"*50)
print(df["consequent"].iloc[i])
import random
i = random.randint(0,len(df))
s = df.loc[df["sentenceID"]==203483]
#print(s)
print("-"*50)
print(s["sentence"].iloc[0])
print("-"*50)
print(s["antecedent"].iloc[0])
print("-"*50)
print(s["consequent"].iloc[0])
df["antecedent"].iloc[0]
df["consequent"].iloc[0]
df["sentence"].iloc[0][df["consequent_startid"].iloc[0]:df["consequent_endid"].iloc[0]]
```
Check whether all indices fit the annotation
_Note: annotation indices are inclusive!_
```
for i in range(len(df)):
assert df["sentence"].iloc[i][df["antecedent_startid"].iloc[i]:df["antecedent_endid"].iloc[i]+1] \
== df["antecedent"].iloc[i]
if df["consequent_startid"].iloc[i]>0:
assert df["sentence"].iloc[i][df["consequent_startid"].iloc[i]:df["consequent_endid"].iloc[i]+1] \
== df["consequent"].iloc[i]
```
__Consequent part might not always exist!__
```
df.loc[df['consequent_startid'] == -1]
```
It does not exist in this number of cases
```
df_without_conseq = df.loc[df['consequent_startid'] == -1]
print(f"{len(df_without_conseq)} / {len(df)}")
```
Lets check what are the lengths of sentences, and how much sentences without consequent correlate with length.
```
all_lens = [len(s.split()) for s in df["sentence"].values.tolist()]
no_conseq_lens = [len(s.split()) for s in df_without_conseq["sentence"].values.tolist()]
all_lens
import matplotlib.pyplot as plt
values1 = all_lens
values2= no_conseq_lens
bins=100
_range=(0,max(all_lens))
fig = plt.figure(figsize=(8,6))
ax = fig.add_subplot(111)
ax.hist(values1, alpha=0.5, bins=bins, range=_range, color= 'b', label='All sentences')
ax.hist(values2, alpha=0.5, bins=bins, range=_range, color= 'r', label='Sentences without consequent')
ax.legend(loc='upper right', prop={'size':14})
plt.show()
```
Distribution is skewed a little bit toward smaller values, but there does not seem to be any big correlation here...
|
github_jupyter
|
```
import numpy as np
import scipy.io as sio
from sklearn import svm
from sklearn.model_selection import cross_val_score
from sklearn.metrics.pairwise import pairwise_distances
from sklearn import manifold
import matplotlib.pyplot as plt
from sklearn.decomposition import PCA
from sklearn.svm import SVC
from graph_kernels_lib import WeisfeilerLehmanKernel, fit_n_components
ppi = sio.loadmat("PPI.mat")
ppi_graphs = ppi['G'][0]
ppi_labels = ppi['labels'].ravel()
n = ppi_labels.shape[0]
wl_kernel = WeisfeilerLehmanKernel()
K = wl_kernel.eval_similarities(ppi_graphs[:]['am'], 2)
D = pairwise_distances(K, metric='euclidean')
plt.imshow(D, zorder=2, cmap='Blues', interpolation='nearest')
plt.colorbar();
plt.style.use("ggplot")
plt.show()
```
# SVM Linear Classifier
```
from sklearn.model_selection import StratifiedKFold
strat_k_fold = StratifiedKFold(n_splits = 10, shuffle = True) #10
clf = svm.SVC(kernel="linear", C = 1.0)
scores_ln = cross_val_score(clf, D, ppi_labels, cv = strat_k_fold)
print(str(np.min(scores_ln)) +" - "+str(np.mean(scores_ln))+ " - " + str(np.max(scores_ln)) + " - "+ str(np.std(scores_ln)))
PCA_D = PCA(n_components = 2).fit_transform(D)
plt.plot(np.cumsum(PCA().fit(D).explained_variance_ratio_))
plt.show()
np.cumsum(PCA().fit(D).explained_variance_ratio_)[:3]
acidovorax = PCA_D[ppi_labels == 1]
acidobacteria = PCA_D[ppi_labels == 2]
clf = clf.fit(PCA_D, ppi_labels)
w = clf.coef_[0]
a = -w[0] / w[1]
xx = np.linspace(np.min(PCA_D), np.max(PCA_D))
yy = a * xx - (clf.intercept_[0]) / w[1]
plt.figure(figsize=(10,5))
ax_av = plt.scatter(acidovorax[:, 0], acidovorax[:, 1], color = "xkcd:red", marker = "^",label = "Acidovorax", s = 455, alpha = 0.65)
ax_ab = plt.scatter(acidobacteria[:, 0], acidobacteria[:, 1], color = "green", label = "Acidobacteria", s = 250, alpha = 0.75)
svm_line = plt.plot(xx, yy, color = "xkcd:sky blue", linestyle = "--", linewidth = 3.0)
plt.axis('tight');
#plt.grid(True)
plt.legend(prop={'size': 15})
ax_av.set_facecolor('xkcd:salmon')
ax_ab.set_facecolor('xkcd:pale green')
plt.show()
from mpl_toolkits.mplot3d import Axes3D
fig = plt.figure(figsize=(10,8))
ax = fig.add_subplot(111, projection='3d')
PCA_D = PCA(n_components = 3).fit_transform(D)
acidovorax = PCA_D[ppi_labels == 1]
acidobacteria = PCA_D[ppi_labels == 2]
clf = clf.fit(PCA_D, ppi_labels)
w = clf.coef_[0]
a = -w[0] / w[1]
xx = np.linspace(np.min(PCA_D), np.max(PCA_D))
yy = a * xx - (clf.intercept_[0]) / w[1]
#plt.figure(figsize=(10,5))
ax_av = ax.scatter(acidovorax[:, 0], acidovorax[:, 1], acidovorax[:, 2],c = "xkcd:red", marker = "^",label = "Acidovorax", s = 455, alpha = 0.65)
ax_ab = ax.scatter(acidobacteria[:, 0], acidobacteria[:, 1], acidobacteria[:, 2], c = "green", label = "Acidobacteria", s = 250, alpha = 0.75)
#svm_line = plt.plot(xx, yy, color = "xkcd:sky blue", linestyle = "--", linewidth = 3.0)
plt.axis('tight');
#plt.grid(True)
plt.legend(prop={'size': 15})
ax_av.set_facecolor('xkcd:salmon')
ax_ab.set_facecolor('xkcd:pale green')
ax.view_init(azim = 30, elev = 25)
plt.show()
```
# Manifold Learning Isomap
```
n_neighbors = 14#15
n_components = 2
iso_prj_D = manifold.Isomap(n_neighbors, n_components).fit_transform(D)
scores_ln = cross_val_score(clf, iso_prj_D, ppi_labels, cv = strat_k_fold, n_jobs= 8)
np.mean(scores_ln)
```
It seems that manifold learning with Isomap does not improve the performance of our linear svm classifier
### Plots for Isomap
```
acidovorax = iso_prj_D[ppi_labels == 1]
acidobacteria = iso_prj_D[ppi_labels == 2]
clf = clf.fit(iso_prj_D, ppi_labels)
w = clf.coef_[0]
a = -w[0] / w[1]
xx = np.linspace(np.min(iso_prj_D), np.max(iso_prj_D))
yy = a * xx - (clf.intercept_[0]) / w[1]
plt.figure(figsize=(10,5))
ax_av = plt.scatter(acidovorax[:, 0], acidovorax[:, 1], color = "xkcd:red", marker = "^",label = "Acidovorax", s = 455, alpha = 0.65)
ax_ab = plt.scatter(acidobacteria[:, 0], acidobacteria[:, 1], color = "green", label = "Acidobacteria", s = 250, alpha = 0.75)
svm_line = plt.plot(xx, yy, color = "xkcd:sky blue", linestyle = "--", linewidth = 3.0)
plt.axis('tight');
#plt.grid(True)
plt.legend(prop={'size': 15})
ax_av.set_facecolor('xkcd:salmon')
ax_ab.set_facecolor('xkcd:pale green')
plt.show()
```
#### Fit with best n of components
```
opt_n_components = fit_n_components(D, ppi_labels, manifold.Isomap, n_iteration= 10)
opt_iso_prj_D = manifold.Isomap(n_neighbors, opt_n_components).fit_transform(D)
scores_ln = cross_val_score(clf, opt_iso_prj_D, ppi_labels, cv = strat_k_fold, n_jobs= 8)
np.mean(scores_ln)
```
# Manifold Learning LocalLinearEmbedding
```
n_neighbors = 13#15
n_components = 15
lle_prj_D = manifold.LocallyLinearEmbedding(n_neighbors, n_components).fit_transform(D)
scores_ln = cross_val_score(clf, lle_prj_D, ppi_labels, cv = strat_k_fold, n_jobs= 8)
np.mean(scores_ln)
```
It seems that also manifold learning with LocalLinearEmbedding does not improve the performance of our linear svm classifier
### Plots for LLE
```
acidovorax = lle_prj_D[ppi_labels == 1]
acidobacteria = lle_prj_D[ppi_labels == 2]
clf = clf.fit(lle_prj_D, ppi_labels)
w = clf.coef_[0]
a = -w[0] / w[1]
xx = np.linspace(-0.2,0.25)
yy = a * xx - (clf.intercept_[0]) / w[1]
plt.figure(figsize=(10,5))
ax_av = plt.scatter(acidovorax[:, 0], acidovorax[:, 1], color = "xkcd:red", marker = "^",label = "Acidovorax", s = 455, alpha = 0.65)
ax_ab = plt.scatter(acidobacteria[:, 0], acidobacteria[:, 1], color = "green", label = "Acidobacteria", s = 250, alpha = 0.75)
svm_line = plt.plot(xx, yy, color = "xkcd:sky blue", linestyle = "--", linewidth = 3.0)
plt.axis('tight');
#plt.grid(True)
plt.legend(prop={'size': 15})
ax_av.set_facecolor('xkcd:salmon')
ax_ab.set_facecolor('xkcd:pale green')
plt.show()
```
#### Fit with best n of components
```
opt_n_components = fit_n_components(D, ppi_labels, manifold.LocallyLinearEmbedding, n_neighbors=13, n_iteration= 10)
opt_n_components
opt_lle_prj_D = manifold.LocallyLinearEmbedding(13, opt_n_components).fit_transform(D)
scores_ln = cross_val_score(clf, opt_lle_prj_D, ppi_labels, cv = strat_k_fold, n_jobs= 8)
np.mean(scores_ln)
```
# Graphs plots
```
import networkx as nx
G = nx.from_numpy_matrix(ppi_graphs[10]['am'])
#pos=nx.spring_layout(G) # positions for all nodes
pos = nx.spring_layout(G, k = 0.9, iterations = 1000)
nx.draw_networkx_nodes(G, pos, with_labels= False, node_color = "green", node_size = 300, alpha = 0.8)
nx.draw_networkx_edges(G, pos, width = 2, alpha=0.5,edge_color='r')
plt.axis('off')
#plt.savefig("acidovorax_graph_10.png") # save as png
plt.show() # display
G = nx.from_numpy_matrix(ppi_graphs[59]['am'])
#pos=nx.spring_layout(G) # positions for all nodes
pos = nx.spring_layout(G, k = 0.9, iterations = 1000)
nx.draw_networkx_nodes(G, pos, with_labels= False, node_color = "green", node_size = 300, alpha = 0.8)
nx.draw_networkx_edges(G, pos, width = 2, alpha=0.5,edge_color='r')
plt.axis('off')
#plt.savefig("Acidobacteria_graph_59.png") # save as png
plt.show() # display
G = nx.from_numpy_matrix(ppi_graphs[6]['am'])
#pos=nx.spring_layout(G) # positions for all nodes
pos = nx.spring_layout(G, k = 0.9, iterations = 1000)
nx.draw_networkx_nodes(G, pos, with_labels= False, node_color = "green", node_size = 300, alpha = 0.8)
nx.draw_networkx_edges(G, pos, width = 2, alpha=0.5,edge_color='r')
plt.axis('off')
#plt.savefig("acidovorax_graph_2.png") # save as png
plt.show() # display
G = nx.from_numpy_matrix(ppi_graphs[48]['am'])
#pos=nx.spring_layout(G) # positions for all nodes
pos = nx.spring_layout(G, k = 0.9, iterations = 1000)
nx.draw_networkx_nodes(G, pos, with_labels= False, node_color = "green", node_size = 300, alpha = 0.8)
nx.draw_networkx_edges(G, pos, width = 2, alpha=0.5,edge_color='r')
plt.axis('off')
#plt.savefig("Acidobacteria_graph_48.png") # save as png
plt.show() # display
node_labels = wl_kernel.extract_graphs_labels(ppi_graphs[:]['am'])
size = int(np.max(np.concatenate(node_labels)))
degree_component = np.zeros((n, size))
for i in range(len(node_labels)):
for j in node_labels[i]:
degree_component[i,int(j)-1] += 1
degree_component[0]
```
|
github_jupyter
|
# Phase 2 Review
```
import pandas as pd
import numpy as np
from sklearn.preprocessing import StandardScaler
from statsmodels.formula.api import ols
pd.set_option('display.max_columns', 100)
```
### Check Your Data … Quickly
The first thing you want to do when you get a new dataset, is to quickly to verify the contents with the .head() method.
```
df = pd.read_csv('movie_metadata.csv')
print(df.shape)
df.head()
```
## Question 1
A Hollywood executive wants to know how much an R-rated movie released after 2000 will earn. The data above is a sample of some of the movies with that rating during that timeframe, as well as other movies. How would you go about answering her question? Talk through it theoretically and then do it in code.
What is the 95% confidence interval for a post-2000 R-rated movie's box office gross?
```
df.isna().sum()
# talk through your answer here
'''
Drop null values.
Filter dataframe for movies after 2000, content rating is 'R'.
Calculate mean, standard deviation, sample size, and plug those into the confidence interval formula to
find the lower and upper bounds of what the executive can expect such a movie to make.
'''
# do it in code here
df.dropna(subset=['gross'], inplace=True)
df_2000R = df[(df['title_year'] > 2000) & (df['content_rating'] == 'R')]
mean = df_2000R.gross.mean()
sd = df_2000R.gross.std()
n = df_2000R.gross.count()
mean, sd, n
se = sd/n**.5
# 95% confidence interval
mean - 1.96 * (sd / n**.5), mean + 1.96 * (sd / n**.5)
```
## Question 2a
Your ability to answer the first question has the executive excited and now she has many other questions about the types of movies being made and the differences in those movies budgets and gross amounts.
Read through the questions below and **determine what type of statistical test you should use** for each question and **write down the null and alternative hypothesis for those tests**.
- Is there a relationship between the number of Facebook likes for a cast and the box office gross of the movie?
- Do foreign films perform differently at the box office than non-foreign films?
- Of all movies created are 40% rated R?
- Is there a relationship between the language of a film and the content rating (G, PG, PG-13, R) of that film?
- Is there a relationship between the content rating of a film and its budget?
# your answer here
### Facebook Likes (cast) and Box Office Gross
'''
Correlation/simple linear regression
'''
Ho: Beta =0
Ha: Beta != 0
### Domestic vs. Foreign and Box Office Gross
'''
Two-sample T-Test
'''
Ho: mu_domestic = mu_foreign
Ha: mu_domestic != mu_foreign
### Rated R
'''
One-sample Z-Test of proportion
'''
Ho: P = 0.4
Ha: P != 0.4
### Language and Content rating
'''
Chi-square
'''
Ho: distributions are equal
Ha: distributions are not equal
### Content rating and budget
'''
ANOVA
'''
Ho: mu_r = mu_PG13 = mu_PG = mu_G
Ha: They are not all equal
## Question 2b
Calculate the answer for the second question:
- Do foreign films perform differently at the box office than non-foreign films?
```
df.head()
import scipy
import numpy as np
USA_array = np.array(df[df.country == "USA"].gross)
Foreign_array = np.array(df[df.country != "USA"].gross)
scipy.stats.ttest_ind(USA_array,Foreign_array, nan_policy = 'omit')
# your answer here
df_foreign = df[df.country != 'USA'].dropna(subset=['country'])
df_domestic = df[df.country == 'USA']
df_foreign.shape, df_domestic.shape
from scipy.stats import ttest_ind
ttest_ind(df_foreign.gross, df_domestic.gross)
'''
Yes! There is a statistically significant difference between the box office gross of foreign and domestic films.
'''
```
## Question 3
Now that you have answered all of those questions, the executive wants you to create a model that predicts the money a movie will make if it is released next year in the US. She wants to use this to evaluate different scripts and then decide which one has the largest revenue potential.
Below is a list of potential features you could use in the model. Create a new frame containing only those variables.
Would you use all of these features in the model?
Identify which features you might drop and why.
*Remember you want to be able to use this model to predict the box office gross of a film **before** anyone has seen it.*
- **budget**: The amount of money spent to make the movie
- **title_year**: The year the movie first came out in the box office
- **years_old**: How long has it been since the movie was released
- **genres**: Each movie is assigned one genre category like action, horror, comedy
- **imdb_score**: This rating is taken from Rotten tomatoes, and is the average rating given to the movie by the audience
- **actor_1_facebook_likes**: The number of likes that the most popular actor in the movie has
- **cast_total_facebook_likes**: The sum of likes for the three most popular actors in the movie
- **language**: the original spoken language of the film
```
df.loc[0, 'genres'].split('|')
df['genres'] = df.genres.apply(lambda x: x.split('|')[0])
df.genre.head()
df.columns
# your answer here
model_data = df[[
'gross', 'budget', 'actor_1_facebook_likes', 'cast_total_facebook_likes',
'title_year', 'content_rating', 'genres'
]]
model_data.corr()
# '''
# drop either `cast_total_facebook_likes` or `actor_1_facebook_likes` due to multicollinearity
# '''
'''
`num_critic_for_reviews` and `imdb_score` can't be known before the movie is released.
we'll drop them from the model.
drop either `cast_total_facebook_likes` or `actor_1_facebook_likes` due to multicollinearity.
'''
```
## Question 4a
Create the following variables:
- `years_old`: The number of years since the film was released.
- Dummy categories for each of the following ratings:
- `G`
- `PG`
- `R`
Once you have those variables, create a summary output for the following OLS model:
`gross~cast_total_facebook_likes+budget+years_old+G+PG+R`
```
import pandas as pd
model_data['years_old'] = 2020 - model_data.title_year
model_data = pd.get_dummies(model_data, columns=['content_rating']).drop(columns='content_rating_PG-13')
model_data.columns
from statsmodels.formula.api import ols writing out the formula
from statsmodels.api import OLS using x,y
# your answer here
lr_model = ols(formula='gross~cast_total_facebook_likes+budget+years_old+G+PG+R', data=model_data).fit()
lr_model.summary()
```
## Question 4b
Below is the summary output you should have gotten above. Identify any key takeaways from it.
- How ‘good’ is this model?
- Which features help to explain the variance in the target variable?
- Which do not?
<img src="ols_summary.png" style="withd:300px;">
```
'''
The model is not very good in that it only explains about 7.9% (13.9% in mine) of the variation
in the data around the mean. (based on R-squared value)
In the photo, Total Facebook likes, budget, age, PG rating, and R rating help to explain the variance,
whereas G rating does not. (based on p-values)
In mine, everything other than years old helps to explain the variance.
'''
```
## Question 5
**Bayes Theorem**
An advertising executive is studying television viewing habits of married men and women during prime time hours. Based on the past viewing records he has determined that during prime time wives are watching television 60% of the time. It has also been determined that when the wife is watching television, 40% of the time the husband is also watching. When the wife is not watching the television, 30% of the time the husband is watching the television. Find the probability that if the husband is watching the television, the wife is also watching the television.
```
# your answer here
'''
P(A) = Probability wife is watching tv
P(B) = Probability husband is watching tv
P(A|B) = Probability wife is watching tv given husband is
P(B|A) = Probability husband is watching tv given wife is
'''
p_A = 0.6
p_notA = 1 - p_A
p_B_given_A = 0.4
p_B_given_notA = 0.3
p_A_given_B = (p_B_given_A * p_A) / (p_B_given_A * p_A + p_B_given_notA * p_notA)
p_A_given_B
```
## Question 6
Explain what a Type I error is and how it relates to the significance level when doing a statistical test.
```
# your answer here
'''
A Type I error occurs when you reject the null hypothesis even though the null hypothesis is True.
The likelihood of a Type I error is directly related to changes in the significance level. If you
increase the significance level, the likelihood of a Type I error also increases and vice versa.
If our significane lecel is 95%, that means we have a 5% chance of making a type one error.
'''
```
## Question 7
How is the confidence interval for a sample related to a one sample t-test?
The range of a confidence interval sets the limits of the values for which you would reject a null hypothesis. For example, if a confidence interval for a population mean was 100 to 105, we would reject any null hypothesis where the proposed population mean is outside of that range.
|
github_jupyter
|
# Stirlingの公式(対数近似)
* $\log n! \sim n\log n - n$
* $n!$はおおよそ$\left(\frac{n}{e}\right)^n$になる
* 参考: [スターリングの公式(対数近似)の導出](https://starpentagon.net/analytics/stirling_log_formula/)
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
```
## $\log n!$の上からの評価
```
MIN_X = 0.5
MAX_X = 10
x = np.linspace(MIN_X, MAX_X, 100)
y = np.log(x)
p = plt.plot(x, y, label='$\log x$')
p = plt.hlines([0], MIN_X, MAX_X)
p = plt.xlim(MIN_X, MAX_X-0.5)
p = plt.xticks(range(1, MAX_X+1))
p = plt.ylim([-0.2, 2.3])
# 面積log kの矩形を描画
for k in range(2, MAX_X):
p = plt.vlines(k, 0, np.log(k), linestyles='dashed')
p = plt.hlines(np.log(k), k, k+1, linestyles='dashed')
p = plt.legend()
plt.show(p)
```
## $\log n!$の下からの評価
```
MIN_X = 0.5
MAX_X = 10
x = np.linspace(MIN_X, MAX_X, 100)
y = np.log(x)
p = plt.plot(x, y, label='$\log x$')
p = plt.hlines([0], MIN_X, MAX_X)
p = plt.xlim(MIN_X, MAX_X-0.5)
p = plt.xticks(range(1, MAX_X+1))
p = plt.ylim([-0.2, 2.3])
# 面積log kの矩形を描画
for k in range(2, MAX_X):
p = plt.vlines(k-1, 0, np.log(k), linestyles='dashed')
p = plt.hlines(np.log(k), k-1, k, linestyles='dashed')
p = plt.vlines(MAX_X-1, 0, np.log(MAX_X), linestyles='dashed')
p = plt.legend()
plt.show(p)
```
## $n \log n - n$の近似精度
```
def log_factorial(n):
'''log n!を返す'''
val = 0.0
for i in range(1, n+1):
val += np.log(i)
return val
# test of log_factorial
eps = 10**-5
assert abs(log_factorial(1) - 0.0) < eps
assert abs(log_factorial(2) - np.log(2)) < eps
assert abs(log_factorial(5) - np.log(120)) < eps
def log_factorial_approx(n):
'''log n!の近似: n log n - nを返す'''
return n * np.log(n) - n
# test of log_factorial_approx
assert abs(log_factorial_approx(1) - (-1)) < eps
assert abs(log_factorial_approx(2) - (2 * np.log(2) - 2)) < eps
# log_factorial, log_factorial_approxをplot
n_list = range(1, 50+1)
y_fact = [log_factorial(n) for n in n_list]
y_approx = [log_factorial_approx(n) for n in n_list]
p = plt.plot(n_list, y_fact, label='$\log n!$')
p = plt.plot(n_list, y_approx, label='$n \log n - n$')
p = plt.legend()
plt.show(p)
# 近似精度を評価
n_list = [5, 10, 20, 50, 100, 1000]
approx_df = pd.DataFrame()
approx_df['n'] = n_list
approx_df['log n!'] = [log_factorial(n) for n in n_list]
approx_df['n log(n)-n'] = [log_factorial_approx(n) for n in n_list]
approx_df['error(%)'] = 100 * (approx_df['log n!'] - approx_df['n log(n)-n']) / approx_df['log n!']
pd.options.display.float_format = '{:.1f}'.format
approx_df
```
## $n!$と$\left(\frac{n}{e}\right)^n$の比較
```
n_list = [5, 10, 20, 50, 100]
approx_df = pd.DataFrame()
approx_df['n'] = n_list
approx_df['n!'] = [np.exp(log_factorial(n)) for n in n_list]
approx_df['(n/e)^n'] = [np.exp(log_factorial_approx(n)) for n in n_list]
approx_df['error(%)'] = 100 * (approx_df['n!'] - approx_df['(n/e)^n']) / approx_df['n!']
pd.options.display.float_format = None
pd.options.display.precision = 2
approx_df
```
|
github_jupyter
|
# Part 1: Extracting a Journal's Publications+Researchers Datasets
In this notebook we are going to
* extract all publications data for a given journal
* have a quick look at the publications' authors and affiliations
* review how many authors have been disambiguated with a Dimensions Researcher ID
* produce a dataset of non-disambiguated authors that can be used for manual disambiguation
## Prerequisites: Installing the Dimensions Library and Logging in
```
# @markdown # Get the API library and login
# @markdown Click the 'play' button on the left (or shift+enter) after entering your API credentials
username = "" #@param {type: "string"}
password = "" #@param {type: "string"}
endpoint = "https://app.dimensions.ai" #@param {type: "string"}
!pip install dimcli plotly tqdm -U --quiet
import dimcli
from dimcli.shortcuts import *
dimcli.login(username, password, endpoint)
dsl = dimcli.Dsl()
#
# load common libraries
import time
import sys
import json
import os
import pandas as pd
from pandas.io.json import json_normalize
from tqdm.notebook import tqdm as progress
#
# charts libs
# import plotly_express as px
import plotly.express as px
if not 'google.colab' in sys.modules:
# make js dependecies local / needed by html exports
from plotly.offline import init_notebook_mode
init_notebook_mode(connected=True)
#
# create output data folder
if not(os.path.exists("data")):
os.mkdir("data")
```
## Selecting a Journal and Extracting All Publications Metadata
```
#@title Select a journal from the dropdown
#@markdown If the journal isn't there, you can try type in the exact name instead.
journal_title = "Nature Genetics" #@param ['Nature', 'The Science of Nature', 'Nature Communications', 'Nature Biotechnology', 'Nature Medicine', 'Nature Genetics', 'Nature Neuroscience', 'Nature Structural & Molecular Biology', 'Nature Methods', 'Nature Cell Biology', 'Nature Immunology', 'Nature Reviews Drug Discovery', 'Nature Materials', 'Nature Physics', 'Nature Reviews Neuroscience', 'Nature Nanotechnology', 'Nature Reviews Genetics', 'Nature Reviews Urology', 'Nature Reviews Molecular Cell Biology', 'Nature Precedings', 'Nature Reviews Cancer', 'Nature Photonics', 'Nature Reviews Immunology', 'Nature Reviews Cardiology', 'Nature Reviews Gastroenterology & Hepatology', 'Nature Reviews Clinical Oncology', 'Nature Reviews Endocrinology', 'Nature Reviews Neurology', 'Nature Chemical Biology', 'Nature Reviews Microbiology', 'Nature Geoscience', 'Nature Reviews Rheumatology', 'Nature Climate Change', 'Nature Reviews Nephrology', 'Nature Chemistry', 'Nature Digest', 'Nature Protocols', 'Nature Middle East', 'Nature India', 'Nature China', 'Nature Plants', 'Nature Microbiology', 'Nature Ecology & Evolution', 'Nature Astronomy', 'Nature Energy', 'Nature Human Behaviour', 'AfCS-Nature Molecule Pages', 'Human Nature', 'Nature Reviews Disease Primers', 'Nature Biomedical Engineering', 'Nature Reports Stem Cells', 'Nature Reviews Materials', 'Nature Sustainability', 'Nature Catalysis', 'Nature Electronics', 'Nature Reviews Chemistry', 'Nature Metabolism', 'Nature Reviews Physics', 'Nature Machine Intelligence', 'NCI Nature Pathway Interaction Database', 'Nature Reports: Climate Change'] {allow-input: true}
start_year = 2015 #@param {type: "number"}
#@markdown ---
# PS
# To get titles from the API one can do this:
# > %dsldf search publications where journal.title~"Nature" and publisher="Springer Nature" return journal limit 100
# > ", ".join([f"'{x}'" for x in list(dsl_last_results.title)])
#
q_template = """search publications where
journal.title="{}" and
year>={}
return publications[basics+altmetric+times_cited]"""
q = q_template.format(journal_title, start_year)
print("DSL Query:\n----\n", q, "\n----")
pubs = dsl.query_iterative(q.format(journal_title, start_year), limit=500)
```
Save the data as a CSV file in case we want to reuse it later
```
dfpubs = pubs.as_dataframe()
dfpubs.to_csv("data/1.pubs_metadata_with_metrics.csv")
# preview the publications
dfpubs.head(10)
```
Extract the authors data
```
# preview the authors data
authors = pubs.as_dataframe_authors()
authors.to_csv("data/1.publications_authors.csv", index=False)
authors.head(10)
```
Extract the affiliations data
```
affiliations = pubs.as_dataframe_authors_affiliations()
affiliations.to_csv("data/1.publications_authors_affiliations.csv", index=False)
affiliations.head(10)
```
## Some stats about authors
* count how many authors in total
* count how many authors have a researcher ID
* count how many unique researchers IDs we have in total
```
researchers = authors.query("researcher_id!=''")
#
df = pd.DataFrame({
'measure' : ['Authors in total (non unique)', 'Authors with a researcher ID', 'Authors with a researcher ID (unique)'],
'count' : [len(authors), len(researchers), researchers['researcher_id'].nunique()],
})
px.bar(df, x="measure", y="count", title=f"Author stats for {journal_title} (from {start_year})")
# save the researchers data to a file
researchers.to_csv("data/1.authors_with_researchers_id.csv")
```
## Apprendix: A quick look at authors *without a Researcher ID*
We're not going to try to disambiguate them here, but still it's good to have a quick look at them...
Looks like the most common surname is `Wang`, while the most common first name is an empty value
```
authors_without_id = authors.query("researcher_id==''")
authors_without_id[['first_name', 'last_name']].describe()
```
Top Ten surnames seem all Chinese..
```
authors_without_id['last_name'].value_counts()[:10]
```
### Any common patterns?
If we try to group the data by name+surname we can see some interesting patterns
* some entries are things which are not persons (presumably the results of bad source data in Dimensions, eg from the publisher)
* there are some apparently meaningful name+surname combinations with a lot of hits
* not many Chinese names in the top ones
```
test = authors_without_id.groupby(["first_name", "last_name"]).size()
test.sort_values(ascending=False, inplace=True)
test.head(50)
```
## Conclusion and next steps
For the next tasks, we will focus on the disambiguated authors as the Researcher ID links will let us carry out useful analyses.
Still, we can **save the authors with missing IDs** results and try to do some manual disambiguation later. To this end, adding a simple google-search URL can help in making sense of these data quickly.
```
from urllib.parse import quote
out = []
for index, value in test.items():
# compose a simple URL of the form 'https://www.google.com/search?q=tonu+esko'
if index[0] or index[1]:
n, s = quote(index[0]), quote(index[1])
url = f"https://www.google.com/search?q={n}+{s}"
else:
url = ""
d = {'name': index[0] , 'surname' : index[1] , 'frequency' : value , 'search_url' : url }
out.append(d)
dftest = pd.DataFrame.from_dict(out)
# set order of columns
dftest = dftest[['name', 'surname', 'frequency', 'search_url']]
dftest.head(20)
# save the data
#
dftest.to_csv("data/1.authors_not_disambiguated_frequency.csv", header=True)
if COLAB_ENV:
files.download("data/1.authors_not_disambiguated_frequency.csv")
files.download("data/1.authors_with_researchers_id.csv")
files.download("data/1.publications_authors.csv")
files.download("data/1.publications_authors_affiliations.csv")
files.download("data/1.pubs_metadata_with_metrics.csv")
```
That's it!
Now let's go and open this in [Google Sheets](https://docs.google.com/spreadsheets/)...
|
github_jupyter
|
Your name here.
Your section number here.
# Workshop 1: Python basics, and a little plotting
**Submit this notebook to bCourses to receive a grade for this Workshop.**
Please complete workshop activities in code cells in this iPython notebook. The activities titled **Practice** are purely for you to explore Python, and no particular output is expected. Some of them have some code written, and you should try to modify it in different ways to understand how it works. Although no particular output is expected at submission time, it is _highly_ recommended that you read and work through the practice activities before or alongside the exercises. However, the activities titled **Exercise** have specific tasks and specific outputs expected. Include comments in your code when necessary. Enter your name in the cell at the top of the notebook. The workshop should be submitted on bCourses under the Assignments tab.
To submit the assignment, click File->Download As->Notebook (.ipynb). Then upload the completed (.ipynb) file to the corresponding bCourses assignment.
## Practice: Writing Python code
### The iPython Interpreter
Time to write your first python code! In Jupyter, the code is written in "Cells". Click on the "+" button above to create a new cell and type in "2+2" (without the quotes ... or with them!) and see what happens! To execute, click "Run" button or press "Shift-Enter". Also try switching the type of the cell from "Code" to "Markdown" and see what happens
```
2+2
```
## Practice: Peforming arithmetic in Python
If you get bored of using WolframAlpha to help with physics homework, Python can also be used as a "glorified calculator". Python code must follow certain syntax in order to run properly--it tends to be a bit more picky than Wolfram. However, once you get used to the Python language, you have the freedom to calculate pretty much anything you can think of.
To start, let's see how to perform the basic arithmetic operations. The syntax is
<h3><center><i>number</i> operator <i>number</i></center></h3>
Run the cells below and take a look at several of the different operators that you can use in Python (text after "#" are non-executable comments).
```
3+2 #addition
3-2 #subtraction
3*2 #multiplication
3/2 #division
3%2 #modulus (remainder after division) see https://en.wikipedia.org/wiki/Modulo_operation
3**2 #exponentiation, note: 3^2 means something different in Python
```
Python cares __*a lot*__ about the spaces, tabs, and enters you type (this is known as whitespace in programming). Many of your errors this semester will involve improper indentation. However, in this case, you are free to put a lot of space between numbers and operators as long as you keep everything in one line.
```
5 * 3 #This is valid code
```
You are not limited to just 2 numbers and a single operator; you can put a whole bunch of operations on one line.
```
5 * 4 + 3 / 2
```
Python follows the standard order of operations (PEMDAS) : Parentheses -> Exponentiation -> Multiplication/Division -> Addition/Subtraction. If you use parentheses, make sure every ```(``` has a corresponding ```)```
```
5 * (4 + 3) / 2
```
## Practice: Strings vs numbers
If you're familiar with programming in other languages, you are probably aware that different [_types_](https://realpython.com/python-data-types/) of things exist--you can do more than work with numbers (and not all numbers are the same type). If you'd like to work with letters, words, or sentences in Python, then you'll be using something called a string. To input a string, simply put single `' '` or double `" "` quotes around your desired phrase.
```
"Hello world"
```
Some (but not all) of the arithmetic operations also work with strings; you can add two of them together.
```
"Phys" + "ics"
```
You can multiply a string by a number.
```
"ha"*3
```
This one doesn't work; try reading the error message and see if you understand what it's saying (this is a useful skill to develop).
```
"error"/3
```
## Practice: Printing
Up until this point, we've just been typing a single line of code in each Jupyter cell and running it. Most Python interpreters will display the result of the final thing you typed, but occassionally you want to display the results of many things in a single Python script.
```
"These are some numbers:"
3*2
3*3
3*4
```
In the cell above, there are several multiplications happening but only the final result is displayed. To display everything, we simply use a "print statement" on each line.
```
print("These are some numbers:")
print(3*2)
print(3*3)
print(3*4)
```
If you'd like to print multiple things on one line, you can separate them by commas within the print statement.
```
print("These are some numbers:", 3*2, 3*3, 3*4)
```
## Exercise 1: Four Fours
[Inspired by Harvey Mudd College's CS5 course] Here's an arithmetic game to try your hand at. Your task is to compute each of the numbers, from 1 through 11, using exactly four 4's and simple math operations. You're allowed to use `+` (addition), `-` (subtraction), `*` (multiplication), `/` (division), `sqrt()` (square root), `factorial()` (factorial), and `%` (modulus). You're also allowed to use `.4` (that's one 4) or `44` (that's two 4's) if you'd like. Just remember, you must use exactly four 4 digits total!
As a reminder, four factorial (denoted by $!$ in mathematics) is $4! = 4 \cdot 3 \cdot 2 \cdot 1$, and the modulus operator (usually denoted by $\text{mod}$ in mathematics) is the remainder after division. For instance, $\ 5\ \text{mod}\ 2 = 1$, $\ 13\ \text{mod}\ 7 = 6$, and $\ 14\ \text{mod}\ 7 = 0$.
We've given you `zero` for free, as `4 - 4 + 4 - 4`. Of course, we could have also done `44 * (.4 - .4)` or `factorial(4) - 4 * (4 + sqrt(4))`, since both of those also yield `0` (or rather, `0.0`. Why is that?) and use exactly four 4's.
```
### Exercise 1
from math import factorial, sqrt
print('Zero:', 4 - 4 + 4 - 4)
print('One:')
print('Two:')
print('Three:')
print('Four:')
print('Five:')
print('Six:')
print('Seven:')
print('Eight:')
print('Nine:')
print('Ten:')
print('Eleven:')
```
Your final source code will be full of four fours formulas, but your final output should look like this:
Zero: 0
One: 1
Two: 2
Three: 3
Four: 4
Five: 5
Six: 6
Seven: 7
Eight: 8
Nine: 9
Ten: 10
Eleven: 11
It's ok if some of these have a trailing `.0` (`0.0`, for instance), but make sure you understand why they do!
## Practice: Variables, functions, namespaces
### Variables
Suppose you calculate something in Python and would like to use the result later in your program (instead of just printing it and immediately throwing it away). One big difference between a calculator and a computer language is an ability to store the values in memory, give that memory block a name, and use the value in later calculations. Such named memory block is called a _variable_. To create a variable, use an _assignment_ opperator = . Once you have created the variable, you can use it in the calculations.
```
x = "Phys"
y = "ics!"
z = x + y # Put 'em together
z # See what we got!
y + x # Backwards!
len(z) # 8 characters in total ...
len(z)**2 # Computing the area?
z[0] # Grab the first character
z[1:3] # Grab the next two characters
z[:4]
z[:4] == x # Test a match!
z[4:] == y
z[:] # The whole string
z[::-1] # The whole string, right to left
z[1::3] # Start at the second character and take every third character from there
z*3 + 5*z[-1] # Woo!
```
### Namespaces
This notebook and interpreter are a great place to test things out and mess around. Some interpreters (like Canopy) comes preloaded with a couple libraries (like numpy and matplotlib) that we will use a lot in this course. In Jupyter, you have to pre-load each package before using it. This is a good python practice anyway ! Here is an example.
```
log(e)
```
Both the function `log` and the number `e` are from the `numpy` library, which needs to be loaded into Jupyter. "pylab" adds `matplotlib` (the standard plotting tool) to `numpy`, so we will use that.
```
from pylab import *
log(e)
```
Or type `pie([1,2,3])`, since `pie` is defined by matplotlib!
```
pie([1,2,3])
matplotlib.pyplot.show() #This line is needed so matplotlib actually displays the plot
```
Note that we imported all library definitions from `pylab` into the default <i>namespace</i>, and can use the functions directly instead of having to add the name or alias of the package:
```
import numpy as np
np.log(np.e)
```
Loading into the default namespace can be convenient, but also confusing since many names and variables are already used in ways you might not expect. When writing scripts you'll have to manually import any library you want to use. This little inconvenience is greatly worth the confusion it can save.
### Functions (looking a bit ahead)
You'll often find yourself performing the same operations on several different variables. For example, we might want to convert heights from feet to meters.
```
burj_khalifa = 2717 #height in feet
shanghai_tower = 2073 #height in feet
print(burj_khalifa / 3.281) #height in meters
print(shanghai_tower / 3.281) #height in meters
```
You could just type the same thing over and over (or copy and paste), but this becomes tedious as your operations become more complex. To simplify things, you can define a function in Python (above, you were able to use the `log()` function from the `numpy` library).
```
'''A function definition starts with the 'def' keyword,
followed by the function name. The input variables are then
placed in parentheses after the function name. The first line
ends with a colon'''
def feet_to_meters(height):
#The operations to be performed by the function are now written out at the first indentation level
#You can indent with tabs or a constant number of spaces; just be consistent
converted_height = height / 3.281
print("Your height is being converted to meters.")
return converted_height #To return a value from a function, use the 'return' keyword
```
To use a function, simply type its name with the appropriate input variables in parentheses.
```
feet_to_meters(burj_khalifa)
```
If you'd like a function with multiple input variables, simply separate them with commas in the function declaration.
```
def difference_in_meters(height1, height2):
converted_height1 = height1 / 3.281
converted_height2 = height2 / 3.281
return converted_height1 - converted_height2
difference_in_meters(burj_khalifa, shanghai_tower)
```
## Practice: Formatted output
Usually the data you manipulate has finate precision. You do not know it absolutely precisely, and therefore you should not report it with an arbitrary number of digits. One of the cardinal rules of a good science paper: round off all your numbers to the precision you know them (or care about) -- and no more !
#### Examples:
```
x = 20.0 # I only know 3 digits
print(x) # OK, let Python handle it
```
That's actually pretty good -- Python remembered stored precision !
What happens if you now use x in a calculation ?
```
print(sqrt(x))
```
Do we really know the output to 10 significant digits ? No ! So let's truncate it
```
print('sqrt(x) = {0:5.3f}'.format(sqrt(x)))
```
There are several formatting options available to you, but the basic idea is this:
place `{:.#f}` wherever you'd like to insert a variable into your string (where `#` is
the number of digits you'd like after the decimal point). Then type `.format()` after
the string and place the variable names within the parentheses.
```
from math import e
print("Euler's number with 5 decimal places is {:.5f} and with 3 decimal places is {:.3f}".format(e,e))
```
For more formatting options, see https://pyformat.info/
### Practice
Using what you just learned, try writing program to print only 4 decimal places of $\pi$ and $\log\pi$. The result should look like:
Hello world! Have some pie! 3.1416
And some pie from a log! 1.1447
```
from math import pi
#Your print statement here
```
## Exercise 2: Coulomb force
Write a function that calculates the magnitude of the force between two charged particles. The function should take the charge of each particle ($q_1$ and $q_2$) and the distance between them, $r$, as input (three input variables total). The electrostatic force between two particles is given by:
$ F = k\frac{q_1 q_2}{r^2}$
```
k = 8.99e9 #Coulomb constant, units: N * m**2 / C**2
def calculate_force(q1, q2, r):
#calculate (and return) the force between the two particles
```
Now call the function with random input values (of your choosing) and print the result with 3 decimal places. What happens if you call the function with the value $r=0$ ?
## Practice: Simple plotting
In order to do some plotting, we'll need the tools from two commonly used Python libraries: `matplotlib` (similar to Matlab plotting) and `numpy` (NUMerical PYthon). You've seen importing at work before with `from math import sqrt`; we can also import an entire library (or a large part of it) with the following syntax:
```
import numpy as np
import matplotlib.pyplot as plt
```
You could have also typed `import numpy`, but programmers are lazy when it comes to typing. By including `as np`, you now only have to type the two-letter word `np` when you'd like to use functions from the library. The `np` and `plt` part of the import statements can be whatever you like--these are just the standard names.
Numpy has a lot of the same functions as the `math` library; for example we have `sqrt`, `log`, and `exp`:
```
np.sqrt(4)
np.log(4)
np.exp(3)
np.log(np.exp(5))
```
We could have just gotten these functions from the `math` library, so why bother with `numpy`? There's another variable type in Python known as a *__list__*, which is exactly like it sounds--just a list of some things (numbers, strings, more lists, etc.). We'll talk about these more at some point, but the important thing is that `numpy` has a way better alternative: the `numpy` array. Usually anything you'd want to do with a list can also be done with a `numpy` array, but faster.
Let's just demonstrate by example. Suppose we want to plot the function `x**2`. To do this, we'll plot a collection of (x,y) points and connect them with lines. If the points are spaced closely enough, the plot will look nice and smooth on the screen.
```
x_values = np.linspace(-5, 5, 11)
print(x_values)
```
The `linspace` function from `numpy` gave us an array of 11 numbers, evenly spaced between -5 and 5. We'll want our points a bit closer, so let's change 11 to something larger.
```
x_values = np.linspace(-5, 5 , 1000)
y_values = x_values**2
```
To get the corresponding y values, we can just perform operations on the entire array of x values. Now, we can plot these using the `matplotlib` library.
```
plt.plot(x_values, y_values)
```
There's a ton of stuff you can do with `matplotlib.pyplot` or the `matplotlib` library as a whole, but here are a few basics to get you started.
```
plt.plot(x_values, x_values**3) #As before, this plots the (x,y) points and connects them with lines
plt.show() #This forces matplotlib to display the current figure
plt.figure() #This creates a new, empty figure
plt.plot(x_values, np.exp(x_values), 'g--') #There are lots of optional arguments that do cool things
plt.title(r'$e^x$') #Creates a title; you can use LaTeX formatting in matplotlib as shown here
plt.xlabel('y values') #Label for x-axis
plt.ylabel('exp(x)') #Label for y-axis
plt.show()
```
## Exercise 3: Plotting Radioactivity Data
[Adapted from Ayars, Problem 0-2]
The file Ba137.txt contains two columns. The first is counts from a Geiger counter, the second is time in seconds. If you opened this Workshop notebook using the Interact Link (from the bCourses page), then you should already have Ba137.txt in your datahub directory.
If not, it's available [here](https://raw.githubusercontent.com/celegante/code_chapter_0-_github/master/Ba137.txt). Open the link, right-click and save as a .txt file. Then upload to datahub.berkeley.edu or move it to whichever folder you're keeping this notebook.
1. Make a useful graph of this data, with axes labels and a title.
2. If this data follows an exponential curve, then plotting the natural log of the data (or plotting the raw data on a logarithmic scale) will result in a straight line. Determine whether this is the case, and explain your conclusion with---you guessed it---an appropriate graph.
Be sure to add comments throughout your code so it's clear what each section of the code is doing! It may help to refer to the lecture notes or Ayars Chapter 0.
Try using `'x'` or `'^'` as the marker type in your `plt.plot()` functions (instead of `'g-'`, for instance), to get a single x or triangle for each data point instead of a connected line. Google if you'd like to learn more options!
Once you're through, your code should produce two graphs, one with the data, another with the natural log of the data, both labelled appropriately. It should also print out a clear answer to the question in part 2 (e.g., `Yes, the data follows an exponential curve`, or `No, the data does not follow an exponential curve`).
```
### Exercise 3
import numpy as np
import matplotlib.pyplot as plt
### Load the data here
counts, times = np.loadtxt('Ba137.txt', unpack = True)
plt.figure() # Start a clean figure for your first plot
### Your code for the first plot here!
plt.figure() # Start a clean figure for your second plot
### Your code for the second plot here!
plt.show() # This tells python to display the plots you've made
```
#### Hints
Put the file in the same directory as your python file, and use numpy's `loadtxt` or `genfromtxt` function to load each column into an array for use in your plots.
If your file isn't loading correctly, it might be because your IPython working directory isn't the same as the directory your script and Ba137.txt are in.
If you'd like to learn more about what `loadtxt` does (or why the `unpack = True` option is important), type `loadtxt?` or `help(loadtxt)` into the python interpreter for documentation. Press `q` to get out of the documentation.
## Practice: Debugging
[Adapted from Langtangen, Exercise 1.16] Working with a partner, type these statements into your python interpreter. Figure out why some statements fail and correct the errors.
*Hint: Try testing the left- and right-hand sides seperately before you put them together in statements. It's ok if you don't understand yet why some of the expressions evaluate to the results they do (like the last one).*
1a = 2
a1 = b
x = 2
y = X + 4 # is it 6?
5 = 5 # is it True?
4/5 == 4.0/5.0 # is it True? (this depends on which version of Python you're using)
type(10/2) == type(10/2.) # is it True? (again, this depends on the Python version)
from Math import factorial
print factorial(pi)
discount = 12%
## You're done!
Congratulations, you've finished this week's workshop! You're welcome to leave early or get started on this week's homework.
|
github_jupyter
|
## Recursive Functions
A recursive function is a function that makes calls to itself. It works like the loops we described before, but sometimes it the situation is better to use recursion than loops.
Every recursive function has two components: a base case and a recursive step. The base case is usually the smallest input and has an easily verifiable solution. This is also the mechanism that stops the function from calling itself forever. The recursive step is the set of all cases where a recursive call, or a function call to itself, is made.
Consider the example of computing the factorial of a number. For example, the factorial of a number $n$ is given by $f(n) = 1 \ \times \ 2 \ \times \ 3 \ \times \ \dots \ \times \ (n-1) \ \times \ n$.
The recursive from of a factorial is
$$
f(n) = \left\{ \begin{array}{ll} 1 & if \ n=1 \\
n \ \times \ f(n-1) & otherwise\end{array} \right.
$$
which can be expressed in code as
```
def factorial_n(n):
assert type(n) == int, 'Input must be an integer'
if n == 1: #this is the base case
return 1
else: #this is the recursive step
return n * factorial_n(n-1)
factorial_n(1)
factorial_n(2)
factorial_n(5)
1*2*3*4*5
#We can use debbuging tools to understand the code
from pdb import set_trace
def factorial_n(n):
assert type(n) == int, 'Input must be an integer'
set_trace()
if n == 1: #this is the base case
return 1
else: #this is the recursive step
return n * factorial_n(n-1)
factorial_n(1)
factorial_n(3)
```
## mini challenge 1
Fibonacci numbers were originally developed to model the idealized population growth of rabbits. Since then, they have been found to be significant in any naturally occurring phenomena.
Use recursivity to compute the Fibonacci numbers.
The recursive form of the Fibonacci numbers.
$$
f(n) = \left\{ \begin{array}{ll} 1 & if \ n=1 \\
1 & if \ n=2 \\
f(n-1) + f(n-2) & otherwise\end{array} \right.
$$
```
#examples
fibonacci(1) = 1
fibonacci(2) = 1
fibonacci(3) = 2
fibonacci(4) = 3
fibonacci(5) = 5
fibonacci(35) = 9227465
def fibonacci(n) :
assert type(n) == int, 'Input must be an integer'
if n == 1:
return 1
if n == 2:
return 1
else:
return fibonacci(n-1) + fibonacci(n-2)
```
## mini challenge 2
An integer number $n$ is said to be **prime** if is divisible only by itself and one. If $n$ is divisible by any other number between $1$ and $n$, the the number is not prime.
Write a recursive function to verify if a number n is prime.
```
def prime(N, div = 2):
if N == 1:
return True
else:
if N == 2:
return True
elif (N%div) == 0 :
return False
else:
prime(N,div+1)
return True
prime(7)
```
|
github_jupyter
|
```
from astropy.constants import G
import astropy.coordinates as coord
import astropy.table as at
import astropy.units as u
from astropy.time import Time
import matplotlib as mpl
import matplotlib.pyplot as plt
%matplotlib inline
import numpy as np
from gala.units import galactic, UnitSystem
from twobody import TwoBodyKeplerElements, KeplerOrbit
from twobody.anomaly import mean_anomaly_from_eccentric_anomaly
usys = UnitSystem(1e12*u.Msun, u.kpc, u.Gyr, u.radian)
true_m31_sky_c = coord.SkyCoord(
10.64628564*u.deg,
41.23456631*u.deg
)
```
## Simulate some Keplerian data
```
M1 = 1.4e12 * u.Msun
M2 = 2.4e12 * u.Msun
M = M1 + M2
a = 511 * u.kpc
eta = 4.3 * u.rad
e = 0.981
mean_anomaly = mean_anomaly_from_eccentric_anomaly(eta, e)
elem = TwoBodyKeplerElements(
a=a, m1=M1, m2=M2, e=e,
omega=0*u.rad, i=90*u.deg,
M0=0.*u.rad, t0=0. * u.Gyr,
units=galactic
)
orb1 = KeplerOrbit(elem.primary)
orb2 = KeplerOrbit(elem.secondary)
Romega = coord.matrix_utilities.rotation_matrix(elem.secondary.omega, 'z')
xyz1 = orb1.orbital_plane(0. * u.Gyr)
xyz2 = orb2.orbital_plane(0. * u.Gyr).transform(Romega)
xyz1, xyz2
time = (elem.P * (mean_anomaly / (2*np.pi*u.rad))).to(u.Gyr)
xyz1 = orb1.orbital_plane(time)
xyz2 = orb2.orbital_plane(time).transform(Romega)
(xyz1.without_differentials()
- xyz2.without_differentials()).norm().to(u.kpc)
a * (1 - e * np.cos(eta))
times = np.linspace(0, 1, 1024) * elem.P
xyzs1 = orb1.orbital_plane(times)
xyzs2 = orb2.orbital_plane(times).transform(Romega)
rs = (xyzs1.without_differentials()
- xyzs2.without_differentials()).norm().to(u.kpc)
fig, ax = plt.subplots(1, 1, figsize=(8, 8))
ax.plot(xyzs1.x, xyzs1.y, marker='')
ax.plot(xyzs2.x, xyzs2.y, marker='')
ax.plot(xyz1.x, xyz1.y, zorder=100, ms=10, color='tab:orange')
ax.plot(xyz2.x, xyz2.y, zorder=100, ms=10, color='tab:red')
ax.set_xlim(-2*a.value, 2*a.value)
ax.set_ylim(-2*a.value, 2*a.value)
plt.plot(times.value, rs.value)
dxs = xyzs1.without_differentials() - xyzs2.without_differentials()
dvs = xyzs1.differentials['s'] - xyzs2.differentials['s']
dx_cyl = dxs.represent_as(coord.CylindricalRepresentation)
dv_cyl = dvs.represent_as(coord.CylindricalDifferential, dxs)
vrads = dv_cyl.d_rho
vtans = (dx_cyl.rho * dv_cyl.d_phi).to(u.km/u.s, u.dimensionless_angles())
etas = np.linspace(0, 2*np.pi, 1024) * u.rad
mean_anoms = mean_anomaly_from_eccentric_anomaly(etas, e)
eq_times = elem.P * (mean_anoms / (2*np.pi*u.rad))
eq_vrad = np.sqrt(G * M / a) * (e * np.sin(etas)) / (1 - e * np.cos(etas))
eq_vtan = np.sqrt(G * M / a) * np.sqrt(1 - e**2) / (1 - e * np.cos(etas))
plt.plot(times.value, vrads.to_value(u.km/u.s))
plt.plot(times.value, vtans.to_value(u.km/u.s))
plt.plot(eq_times.value, eq_vrad.to_value(u.km/u.s))
plt.plot(eq_times.value, eq_vtan.to_value(u.km/u.s))
plt.ylim(-500, 500)
```
### Transform to ICRS
```
dx = xyz1.without_differentials() - xyz2.without_differentials()
dv = xyz1.differentials['s'] - xyz2.differentials['s']
dx_cyl = dx.represent_as(coord.CylindricalRepresentation)
dv_cyl = dv.represent_as(coord.CylindricalDifferential, dx)
vrad = dv_cyl.d_rho.to(u.km/u.s)
vtan = (dx_cyl.rho * dv_cyl.d_phi).to(u.km/u.s, u.dimensionless_angles())
r = dx.norm()
sun_galcen_dist = coord.Galactocentric().galcen_distance
gamma = coord.Galactocentric().galcen_coord.separation(true_m31_sky_c)
sun_m31_dist = (sun_galcen_dist * np.cos(gamma)) + np.sqrt(
r**2 - sun_galcen_dist**2 * np.sin(gamma)**2
)
r, sun_m31_dist
vscale = np.sqrt(G * M / a)
print(vscale.decompose(usys).value,
vrad.decompose(usys).value,
vtan.decompose(usys).value)
alpha = 32.4 * u.deg
galcen_pos = coord.SkyCoord(true_m31_sky_c.ra,
true_m31_sky_c.dec,
distance=sun_m31_dist)
galcen_pos = galcen_pos.transform_to(coord.Galactocentric())
# galcen_pos = coord.CartesianRepresentation(
# -375 * u.kpc, 605 * u.kpc, -279 * u.kpc)
# galcen_pos = coord.Galactocentric(galcen_pos / galcen_pos.norm() * r)
galcen_sph = galcen_pos.represent_as('spherical')
gc_Rz = coord.matrix_utilities.rotation_matrix(-galcen_sph.lon, 'z')
gc_Ry = coord.matrix_utilities.rotation_matrix(galcen_sph.lat, 'y')
gc_Rx = coord.matrix_utilities.rotation_matrix(alpha, 'x')
R = gc_Rz @ gc_Ry @ gc_Rx
fake_X = R @ [r.value, 0, 0] * r.unit
fake_V = R @ [vrad.to_value(u.km/u.s), vtan.to_value(u.km/u.s), 0.] * u.km/u.s
fake_galcen = coord.Galactocentric(*fake_X, *fake_V)
fake_icrs = fake_galcen.transform_to(coord.ICRS())
fake_icrs
```
## Check roundtripping
```
def tt_sph_to_xyz(r, lon, lat):
return [
r * np.cos(lon) * np.cos(lat),
r * np.sin(lon) * np.cos(lat),
r * np.sin(lat)
]
def tt_cross(a, b):
return np.array([
a[1]*b[2] - a[2]*b[1],
a[2]*b[0] - a[0]*b[2],
a[0]*b[1] - a[1]*b[0]
])
def tt_rotation_matrix(angle_rad, axis):
s = np.sin(angle_rad)
c = np.cos(angle_rad)
if axis == 'x':
R = np.array([
1., 0, 0,
0, c, s,
0, -s, c
])
elif axis == 'y':
R = np.array([
c, 0, -s,
0, 1., 0,
s, 0, c
])
elif axis == 'z':
R = np.array([
c, s, 0,
-s, c, 0,
0, 0, 1.
])
else:
raise ValueError('bork')
return np.reshape(R, (3, 3))
def ugh(m31_ra_rad, m31_dec_rad, m31_distance_kpc, r, vrad, vtan):
galcen_frame = coord.Galactocentric()
# tangent bases: ra, dec, r
M = np.array([
[-np.sin(m31_ra_rad), np.cos(m31_ra_rad), 0.],
[-np.sin(m31_dec_rad)*np.cos(m31_ra_rad), -np.sin(m31_dec_rad)*np.sin(m31_ra_rad), np.cos(m31_dec_rad)],
[np.cos(m31_dec_rad)*np.cos(m31_ra_rad), np.cos(m31_dec_rad)*np.sin(m31_ra_rad), np.sin(m31_dec_rad)]
])
# Matrix to go from ICRS to Galactocentric
R_I2G, offset_I2G = coord.builtin_frames.galactocentric.get_matrix_vectors(
galcen_frame, inverse=False)
dxyz_I2G = offset_I2G.xyz.to_value(usys['length'])
dvxyz_I2G = offset_I2G.differentials['s'].d_xyz.to_value(usys['velocity'])
# Matrix to go from Galactocentric to ICRS
R_G2I, offset_G2I = coord.builtin_frames.galactocentric.get_matrix_vectors(
galcen_frame, inverse=True)
dxyz_G2I = offset_G2I.xyz.to_value(usys['length'])
dvxyz_G2I = offset_G2I.differentials['s'].d_xyz.to_value(usys['velocity'])
m31_icrs_xyz = tt_sph_to_xyz(m31_distance_kpc,
m31_ra_rad, m31_dec_rad)
m31_galcen_xyz = np.dot(R_I2G, m31_icrs_xyz) + dxyz_I2G
m31_galcen_lon = np.arctan2(m31_galcen_xyz[1], m31_galcen_xyz[0])
m31_galcen_lat = np.arcsin(m31_galcen_xyz[2] / r)
xhat = m31_galcen_xyz / r
Rz = tt_rotation_matrix(-m31_galcen_lon, 'z')
print(gc_Ry)
Ry = tt_rotation_matrix(m31_galcen_lat, 'y')
print(Ry)
Rx = tt_rotation_matrix(alpha, 'x')
yhat = np.dot(np.dot(Rz, np.dot(Ry, Rx)), [0, 1, 0.])
zhat = tt_cross(xhat, yhat)
R_LGtoG = np.stack((xhat, yhat, zhat), axis=1)
print(R_LGtoG - R)
x_LG = np.array([r, 0., 0.])
v_LG = np.array([vrad, vtan, 0.])
x_I = np.dot(R_G2I, np.dot(R_LGtoG, x_LG)) + dxyz_G2I
v_I = np.dot(R_G2I, np.dot(R_LGtoG, v_LG)) + dvxyz_G2I
v_I_tangent_plane = np.dot(M, v_I) # alpha, delta, radial
shit1 = coord.CartesianRepresentation(*((R @ x_LG) * usys['length']))
shit2 = coord.CartesianDifferential(*((R @ v_LG) * usys['velocity']))
shit = coord.SkyCoord(shit1.with_differentials(shit2), frame=coord.Galactocentric())
return x_I, v_I, shit.transform_to(coord.ICRS()).velocity
ugh(fake_icrs.ra.radian, fake_icrs.dec.radian, fake_icrs.distance.to_value(u.kpc),
r.decompose(usys).value, vrad.decompose(usys).value, vtan.decompose(usys).value)
fake_icrs.velocity
def ugh2():
galcen_frame = coord.Galactocentric()
# Matrix to go from ICRS to Galactocentric
R_I2G, offset_I2G = coord.builtin_frames.galactocentric.get_matrix_vectors(
galcen_frame, inverse=False)
dxyz_I2G = offset_I2G.xyz.to_value(usys['length'])
dvxyz_I2G = offset_I2G.differentials['s'].d_xyz.to_value(usys['velocity'])
# Matrix to go from Galactocentric to ICRS
R_G2I, offset_G2I = coord.builtin_frames.galactocentric.get_matrix_vectors(
galcen_frame, inverse=True)
dxyz_G2I = offset_G2I.xyz.to_value(usys['length'])
dvxyz_G2I = offset_G2I.differentials['s'].d_xyz.to_value(usys['velocity'])
m31_icrs_xyz = tt_sph_to_xyz(m31_distance_kpc,
m31_ra_rad, m31_dec_rad)
m31_galcen_xyz = np.dot(R_I2G, m31_icrs_xyz) + dxyz_I2G
m31_galcen_lon = np.arctan2(m31_galcen_xyz[1], m31_galcen_xyz[0])
m31_galcen_lat = np.arcsin(m31_galcen_xyz[2] / r)
xhat = m31_galcen_xyz / r
Rz = tt_rotation_matrix(-m31_galcen_lon, 'z')
Ry = tt_rotation_matrix(m31_galcen_lat, 'y')
Rx = tt_rotation_matrix(alpha, 'x')
yhat = np.dot(np.dot(Rz, np.dot(Ry, Rx)), [0, 1, 0.])
zhat = tt_cross(xhat, yhat)
R_LGtoG = np.stack((xhat, yhat, zhat), axis=1)
x_LG = np.array([r, 0., 0.])
v_LG = np.array([vrad, vtan, 0.])
x_I = np.dot(R_G2I, np.dot(R_LGtoG, x_LG)) + dxyz_G2I
v_I = np.dot(R_G2I, np.dot(R_LGtoG, v_LG)) + dvxyz_G2I
v_I_tangent_plane = np.dot(M, v_I) # alpha, delta, radial
shit1 = coord.CartesianRepresentation(*((R @ x_LG) * usys['length']))
shit2 = coord.CartesianDifferential(*((R @ v_LG) * usys['velocity']))
shit = coord.SkyCoord(shit1.with_differentials(shit2), frame=coord.Galactocentric())
return x_I, v_I, shit.transform_to(coord.ICRS()).velocity
```
## Write data to files:
```
rng = np.random.default_rng(seed=42)
dist_err = 11. * u.kpc
pmra_err = 3 * u.microarcsecond / u.yr
pmdec_err = 4 * u.microarcsecond / u.yr
rv_err = 2. * u.km/u.s
t_err = 0.11 * u.Gyr
tbl = {}
tbl['ra'] = u.Quantity(fake_icrs.ra)
tbl['dec'] = u.Quantity(fake_icrs.dec)
tbl['distance'] = rng.normal(fake_icrs.distance.to_value(u.kpc),
dist_err.to_value(u.kpc)) * u.kpc
tbl['distance_err'] = dist_err
tbl['pm_ra_cosdec'] = rng.normal(
fake_icrs.pm_ra_cosdec.to_value(pmra_err.unit),
pmra_err.value) * pmra_err.unit
tbl['pm_ra_cosdec_err'] = pmra_err
tbl['pm_dec'] = rng.normal(
fake_icrs.pm_dec.to_value(pmdec_err.unit),
pmdec_err.value) * pmdec_err.unit
tbl['pm_dec_err'] = pmdec_err
tbl['radial_velocity'] = rng.normal(
fake_icrs.radial_velocity.to_value(rv_err.unit),
rv_err.value) * rv_err.unit
tbl['radial_velocity_err'] = rv_err
tbl['tperi'] = rng.normal(
time.to_value(t_err.unit),
t_err.value) * t_err.unit
tbl['tperi_err'] = t_err
t = at.QTable({k: [] * tbl[k].unit for k in tbl})
t.add_row(tbl)
t.meta['title'] = 'Simulated Two-body'
t.write('../datasets/apw-simulated.ecsv', overwrite=True)
rng = np.random.default_rng(seed=42)
dist_err = 1. * u.kpc
pmra_err = 0.1 * u.microarcsecond / u.yr
pmdec_err = 0.1 * u.microarcsecond / u.yr
rv_err = 0.1 * u.km/u.s
t_err = 0.02 * u.Gyr
tbl = {}
tbl['ra'] = u.Quantity(fake_icrs.ra)
tbl['dec'] = u.Quantity(fake_icrs.dec)
tbl['distance'] = rng.normal(fake_icrs.distance.to_value(u.kpc),
dist_err.to_value(u.kpc)) * u.kpc
tbl['distance_err'] = dist_err
tbl['pm_ra_cosdec'] = rng.normal(
fake_icrs.pm_ra_cosdec.to_value(pmra_err.unit),
pmra_err.value) * pmra_err.unit
tbl['pm_ra_cosdec_err'] = pmra_err
tbl['pm_dec'] = rng.normal(
fake_icrs.pm_dec.to_value(pmdec_err.unit),
pmdec_err.value) * pmdec_err.unit
tbl['pm_dec_err'] = pmdec_err
tbl['radial_velocity'] = rng.normal(
fake_icrs.radial_velocity.to_value(rv_err.unit),
rv_err.value) * rv_err.unit
tbl['radial_velocity_err'] = rv_err
tbl['tperi'] = rng.normal(
time.to_value(t_err.unit),
t_err.value) * t_err.unit
tbl['tperi_err'] = t_err
t = at.QTable({k: [] * tbl[k].unit for k in tbl})
t.add_row(tbl)
t.meta['title'] = 'Simulated Two-body - precise'
t.write('../datasets/apw-simulated-precise.ecsv', overwrite=True)
rng = np.random.default_rng(42)
tbl = {}
vrad_err = 1 * u.km/u.s
vtan_err = 1 * u.km/u.s
t_err = 0.1 * u.Gyr
r_err = 1 * u.kpc
tbl['vrad'] = rng.normal(
vrad.to_value(vrad_err.unit),
vrad_err.value) * vrad_err.unit
tbl['vrad_err'] = vrad_err
tbl['vtan'] = rng.normal(
vtan.to_value(vtan_err.unit),
vtan_err.value) * vtan_err.unit
tbl['vtan_err'] = vtan_err
tbl['r'] = rng.normal(
r.to_value(r_err.unit),
r_err.value) * r_err.unit
tbl['r_err'] = r_err
tbl['tperi'] = rng.normal(
time.to_value(t_err.unit),
t_err.value) * t_err.unit
tbl['tperi_err'] = t_err
t = at.QTable({k: [] * tbl[k].unit for k in tbl})
t.add_row(tbl)
t.meta['title'] = 'Simulated Two-body - simple vrad, vtan'
t.write('../datasets/apw-simulated-simple.ecsv', overwrite=True)
```
|
github_jupyter
|
Peakcalling Bam Stats and Filtering Report - Insert Sizes
================================================================
This notebook is for the analysis of outputs from the peakcalling pipeline
There are severals stats that you want collected and graphed (topics covered in this notebook in bold).
These are:
- how many reads input
- how many reads removed at each step (numbers and percentages)
- how many reads left after filtering
- inset size distribution pre filtering for PE reads
- how many reads mapping to each chromosome before filtering?
- how many reads mapping to each chromosome after filtering?
- X:Y reads ratio
- **inset size distribution after filtering for PE reads**
- samtools flags - check how many reads are in categories they shouldn't be
- picard stats - check how many reads are in categories they shouldn't be
This notebook takes the sqlite3 database created by cgat peakcalling_pipeline.py and uses it for plotting the above statistics
It assumes a file directory of:
location of database = project_folder/csvdb
location of this notebook = project_folder/notebooks.dir/
Firstly lets load all the things that might be needed
Insert size distribution
------------------------
This section get the size distribution of the fragements that have been sequeced in paired-end sequencing. The pipeline calculates the size distribution by caluculating the distance between the most 5' possition of both reads, for those mapping to the + stand this is the left-post possition, for those mapping to the - strand is the rightmost coordinate.
This plot is especially useful for ATAC-Seq experiments as good samples should show peaks with a period approximately equivelent to the length of a nucleosome (~ 146bp) a lack of this phasing might indicate poor quality samples and either over (if lots of small fragments) or under intergration (if an excess of large fragments) of the topoisomerase.
```
import sqlite3
import pandas as pd
import numpy as np
%matplotlib inline
import matplotlib
import numpy as np
import matplotlib.pyplot as plt
#import cgatcore.pipeline as P
import os
import statistics
#import collections
#load R and the R packages required
#%load_ext rpy2.ipython
#%R require(ggplot2)
# use these functions to display tables nicely as html
from IPython.display import display, HTML
plt.style.use('ggplot')
#plt.style.available
```
This is where we are and when the notebook was run
```
!pwd
!date
```
First lets set the output path for where we want our plots to be saved and the database path and see what tables it contains
```
database_path = '../csvdb'
output_path = '.'
#database_path= "/ifs/projects/charlotteg/pipeline_peakcalling/csvdb"
```
This code adds a button to see/hide code in html
```
HTML('''<script>
code_show=true;
function code_toggle() {
if (code_show){
$('div.input').hide();
} else {
$('div.input').show();
}
code_show = !code_show
}
$( document ).ready(code_toggle);
</script>
<form action="javascript:code_toggle()"><input type="submit" value="Click here to toggle on/off the raw code."></form>''')
```
The code below provides functions for accessing the project database and extract a table names so you can see what tables have been loaded into the database and are available for plotting. It also has a function for geting table from the database and indexing the table with the track name
```
def getTableNamesFromDB(database_path):
# Create a SQL connection to our SQLite database
con = sqlite3.connect(database_path)
cur = con.cursor()
# the result of a "cursor.execute" can be iterated over by row
cur.execute("SELECT name FROM sqlite_master WHERE type='table' ORDER BY name;")
available_tables = (cur.fetchall())
#Be sure to close the connection.
con.close()
return available_tables
db_tables = getTableNamesFromDB(database_path)
print('Tables contained by the database:')
for x in db_tables:
print('\t\t%s' % x[0])
#This function retrieves a table from sql database and indexes it with track name
def getTableFromDB(statement,database_path):
'''gets table from sql database depending on statement
and set track as index if contains track in column names'''
conn = sqlite3.connect(database_path)
df = pd.read_sql_query(statement,conn)
if 'track' in df.columns:
df.index = df['track']
return df
```
Insert Size Summary
====================
1) lets getthe insert_sizes table from database
Firsly lets look at the summary statistics that us the mean fragment size, sequencing type and mean read length. This table is produced using macs2 for PE data, or bamtools for SE data
If IDR has been run the insert_size table will contain entries for the pooled and pseudo replicates too - we don't really want this as it will duplicate the data from the origional samples so we subset this out
```
insert_df = getTableFromDB('select * from insert_sizes;',database_path)
insert_df = insert_df[insert_df["filename"].str.contains('pseudo')==False].copy()
insert_df = insert_df[insert_df["filename"].str.contains('pooled')==False].copy()
def add_expt_to_insertdf(dataframe):
''' splits track name for example HsTh1-RATotal-R1.star into expt
featues, expt, sample_treatment and replicate and adds these as
collumns to the dataframe'''
expt = []
treatment = []
replicate = []
for value in dataframe.filename:
x = value.split('/')[-1]
x = x.split('_insert')[0]
# split into design features
y = x.split('-')
expt.append(y[-3])
treatment.append(y[-2])
replicate.append(y[-1])
if len(expt) == len(treatment) and len(expt)== len(replicate):
print ('all values in list correctly')
else:
print ('error in loading values into lists')
#add collums to dataframe
dataframe['expt_name'] = expt
dataframe['sample_treatment'] = treatment
dataframe['replicate'] = replicate
return dataframe
insert_df = add_expt_to_insertdf(insert_df)
insert_df
```
lets graph the fragment length mean and tag size grouped by sample so we can see if they are much different
```
ax = insert_df.boxplot(column='fragmentsize_mean', by='sample_treatment')
ax.set_title('for mean fragment size',size=10)
ax.set_ylabel('mean fragment length')
ax.set_xlabel('sample treatment')
ax = insert_df.boxplot(column='tagsize', by='sample_treatment')
ax.set_title('for tag size',size=10)
ax.set_ylabel('tag size')
ax.set_xlabel('sample treatment')
ax.set_ylim(((insert_df.tagsize.min()-2),(insert_df.tagsize.max()+2)))
```
Ok now get get the fragment length distributiions for each sample and plot them
```
def getFraglengthTables(database_path):
'''Takes path to sqlite3 database and retrieves fraglengths tables for individual samples
, returns a dictionary where keys = sample table names, values = fraglengths dataframe'''
frag_tabs = []
db_tables = getTableNamesFromDB(database_path)
for table_name in db_tables:
if 'fraglengths' in str(table_name[0]):
tab_name = str(table_name[0])
statement ='select * from %s;' % tab_name
df = getTableFromDB(statement,database_path)
frag_tabs.append((tab_name,df))
print('detected fragment length distribution tables for %s files: \n' % len(frag_tabs))
for val in frag_tabs:
print(val[0])
return frag_tabs
def getDFofFragLengths(database_path):
''' this takes a path to database and gets a dataframe where length of fragments is the index,
each column is a sample and values are the number of reads that have that fragment length in that
sample
'''
fraglength_dfs_list = getFraglengthTables(database_path)
dfs=[]
for item in fraglength_dfs_list:
track = item[0].split('_filtered_fraglengths')[0]
df = item[1]
#rename collumns so that they are correct - correct this in the pipeline then delete this
#df.rename(columns={'frequency':'frag_length', 'frag_length':'frequency'}, inplace=True)
df.index = df.frag_length
df.drop('frag_length',axis=1,inplace=True)
df.rename(columns={'frequency':track},inplace=True)
dfs.append(df)
frag_length_df = pd.concat(dfs,axis=1)
frag_length_df.fillna(0, inplace=True)
return frag_length_df
#Note the frequency and fragment lengths are around the wrong way!
#frequency is actually fragment length, and fragement length is the frequency
#This gets the tables from db and makes master df of all fragment length frequencies
frag_length_df = getDFofFragLengths(database_path)
#plot fragment length frequencies
ax = frag_length_df.divide(1000).plot()
ax.set_ylabel('Number of fragments\n(thousands)')
ax.legend(loc=2,bbox_to_anchor=(1.05, 1),borderaxespad=0. )
ax.set_title('fragment length distribution')
ax.set_xlabel('fragment length (bp)')
ax.set_xlim()
```
Now lets zoom in on the interesting region of the plot (the default in the code looks at fragment lengths from 0 to 800bp - you can change this below by setting the tuple in the ax.set_xlim() function
```
ax = frag_length_df.divide(1000).plot(figsize=(9,9))
ax.set_ylabel('Number of fragments\n(thousands)')
ax.legend(loc=2,bbox_to_anchor=(1.05, 1),borderaxespad=0. )
ax.set_title('fragment length distribution')
ax.set_xlabel('fragment length (bp)')
ax.set_xlim((0,800))
```
it is a bit trickly to see differences between samples of different library sizes so lets look and see if the reads for each fragment length is similar
```
percent_frag_length_df = pd.DataFrame(index=frag_length_df.index)
for column in frag_length_df:
total_frags = frag_length_df[column].sum()
percent_frag_length_df[column] = frag_length_df[column].divide(total_frags)*100
ax = percent_frag_length_df.plot(figsize=(9,9))
ax.set_ylabel('Percentage of fragments')
ax.legend(loc=2,bbox_to_anchor=(1.05, 1),borderaxespad=0. )
ax.set_title('percentage fragment length distribution')
ax.set_xlabel('fragment length (bp)')
ax.set_xlim((0,800))
```
SUMMARISE HERE
==============
From these plots you should be able to tell wether there are any distinctive patterns in the size of the fragment lengths,this is especially important for ATAC-Seq data as in successful experiments you should be able to detect nucleosome phasing - it can also indicate over fragmentation or biases in cutting.
Lets looks at the picard insert size metrics also
```
insert_df = getTableFromDB('select * from picard_stats_insert_size_metrics;',database_path)
for c in insert_df.columns:
print (c)
insert_df
```
These metrics are actually quite different to the ones we calculate themselves - for some reason it seems to split the files into 2 and dives a distribution for smaller fragments and for larger fragments- not sure why at the moment
|
github_jupyter
|
```
import keras
from keras.models import Sequential, Model, load_model
from keras.layers import Dense, Dropout, Flatten, Input, Lambda, Concatenate
from keras.layers import Conv1D, MaxPooling1D
from keras.callbacks import ModelCheckpoint, EarlyStopping
from keras import backend as K
import keras.losses
import tensorflow as tf
import pandas as pd
import os
import numpy as np
import scipy.sparse as sp
import scipy.io as spio
import matplotlib.pyplot as plt
import matplotlib.cm as cm
import isolearn.io as isoio
import isolearn.keras as iso
from scipy.stats import pearsonr
```
<h2>Load 5' Alternative Splicing Data</h2>
- Load a Pandas DataFrame + Matlab Matrix of measured Splicing Sequences<br/>
- isolearn.io loads all .csv and .mat files of a directory into memory as a dictionary<br/>
- The DataFrame has one column - padded_sequence - containing the splice donor sequence<br/>
- The Matrix contains RNA-Seq counts of measured splicing at each position across the sequence<br/>
```
#Load Splicing Data
splicing_dict = isoio.load('data/processed_data/splicing_5ss_data/splicing_5ss_data')
```
<h2>Create a Training and Test Set</h2>
- We create an index containing row numbers corresponding to training and test sequences<br/>
- Notice that we do not alter the underlying DataFrame, we only make lists of pointers to rows<br/>
```
#Generate training, validation and test set indexes
valid_set_size = 0.10
test_set_size = 0.10
data_index = np.arange(len(splicing_dict['df']), dtype=np.int)
train_index = data_index[:-int(len(data_index) * (valid_set_size + test_set_size))]
valid_index = data_index[train_index.shape[0]:-int(len(data_index) * test_set_size)]
test_index = data_index[train_index.shape[0] + valid_index.shape[0]:]
print('Training set size = ' + str(train_index.shape[0]))
print('Validation set size = ' + str(valid_index.shape[0]))
print('Test set size = ' + str(test_index.shape[0]))
```
<h2>Create Data Generators</h2>
- In Isolearn, we always build data generators that will encode and feed us the data on the fly<br/>
- Here, for example, we create a training and test generator separately (using list comprehension)<br/>
- First argument: The list of row indices (of data points) for this generator<br/>
- Second argument: Dictionary or data sources<br/>
- Third argument: Batch size for the data generator
- Fourth argument: List of inputs, where each input is specified as a dictionary of attributes<br/>
- Fifth argument: List of outputs<br/>
- Sixth argument: List of any randomizers (see description below)<br/>
- Seventh argument: Shuffle the dataset or not<br/>
- Eight argument: True if some data source matrices are in sparse format<br/>
- Ninth argument: In Keras, we typically want to specfiy the Outputs as Inputs when training. <br/>This argument achieves this by moving the outputs over to the input list and replaces the output with a dummy encoder.<br/>
In this example, we specify a One-Hot encoder as the input encoder for the entire splice donor sequence (centered on the splice donor).<br/>
We also specify the target output as the normalized RNA-Seq count at position 120 in the count matrix for each cell line (4 outputs).<br/>
Besides the canonical splice donor at position 120 in the sequence, there are many other splice donors inserted randomly at neighboring positions. If we wanted to learn a general model of splicing, it would be a lot better if we could stochastically "align" sequences on any of the possible splice donors, perturbing both the input sequence and the RNA-Seq count matrix that we estimate splice donor usage from.<br/>
This is achieved using the built-in CutAlignSampler class, which allows us to randomly sample a position in the sequence with supporting splice junction counts, and shift both the sequence and splice count vector to be centered around that position. In this example, we specfiy the sampling rate of splice donors to be 0.5 (p_pos) and the rate of sampling some other, non-splice-site, position at a rate of 0.5 (p_neg).<br/>
```
#Create a One-Hot data generator, to be used for a convolutional net to regress SD1 Usage
total_cuts = splicing_dict['hek_count'] + splicing_dict['hela_count'] + splicing_dict['mcf7_count'] + splicing_dict['cho_count']
shifter = iso.CutAlignSampler(total_cuts, 240, 120, [], 0.0, p_pos=0.5, p_neg=0.5, sparse_source=True)
splicing_gens = {
gen_id : iso.DataGenerator(
idx,
{
'df' : splicing_dict['df'],
'hek_count' : splicing_dict['hek_count'],
'hela_count' : splicing_dict['hela_count'],
'mcf7_count' : splicing_dict['mcf7_count'],
'cho_count' : splicing_dict['cho_count'],
},
batch_size=32,
inputs = [
{
'id' : 'seq',
'source_type' : 'dataframe',
'source' : 'df',
'extractor' : iso.SequenceExtractor('padded_sequence', start_pos=0, end_pos=240, shifter=shifter if gen_id == 'train' else None),
'encoder' : iso.OneHotEncoder(seq_length=240),
'dim' : (240, 4),
'sparsify' : False
}
],
outputs = [
{
'id' : cell_type + '_sd1_usage',
'source_type' : 'matrix',
'source' : cell_type + '_count',
'extractor' : iso.CountExtractor(start_pos=0, end_pos=240, static_poses=[-1], shifter=shifter if gen_id == 'train' else None, sparse_source=False),
'transformer' : lambda t: t[120] / np.sum(t)
} for cell_type in ['hek', 'hela', 'mcf7', 'cho']
],
randomizers = [shifter] if gen_id in ['train'] else [],
shuffle = True if gen_id in ['train'] else False,
densify_batch_matrices=True,
move_outputs_to_inputs=True if gen_id in ['train', 'valid'] else False
) for gen_id, idx in [('train', train_index), ('valid', valid_index), ('test', test_index)]
}
```
<h2>Keras Loss Functions</h2>
Here we specfiy a few loss function (Cross-Entropy and KL-divergence) to be used when optimizing our Splicing CNN.<br/>
```
#Keras loss functions
def sigmoid_entropy(inputs) :
y_true, y_pred = inputs
y_pred = K.clip(y_pred, K.epsilon(), 1. - K.epsilon())
return -K.sum(y_true * K.log(y_pred) + (1.0 - y_true) * K.log(1.0 - y_pred), axis=-1)
def mean_sigmoid_entropy(inputs) :
y_true, y_pred = inputs
y_pred = K.clip(y_pred, K.epsilon(), 1. - K.epsilon())
return -K.mean(y_true * K.log(y_pred) + (1.0 - y_true) * K.log(1.0 - y_pred), axis=-1)
def sigmoid_kl_divergence(inputs) :
y_true, y_pred = inputs
y_pred = K.clip(y_pred, K.epsilon(), 1. - K.epsilon())
y_true = K.clip(y_true, K.epsilon(), 1. - K.epsilon())
return K.sum(y_true * K.log(y_true / y_pred) + (1.0 - y_true) * K.log((1.0 - y_true) / (1.0 - y_pred)), axis=-1)
def mean_sigmoid_kl_divergence(inputs) :
y_true, y_pred = inputs
y_pred = K.clip(y_pred, K.epsilon(), 1. - K.epsilon())
y_true = K.clip(y_true, K.epsilon(), 1. - K.epsilon())
return K.mean(y_true * K.log(y_true / y_pred) + (1.0 - y_true) * K.log((1.0 - y_true) / (1.0 - y_pred)), axis=-1)
```
<h2>Splicing Model Definition</h2>
Here we specfiy the Keras Inputs that we expect to receive from the data generators.<br/>
We also define the model architecture (2 convolutional-layer CNN with MaxPooling).<br/>
```
#Splicing Model Definition (CNN)
#Inputs
seq_input = Input(shape=(240, 4))
#Outputs
true_usage_hek = Input(shape=(1,))
true_usage_hela = Input(shape=(1,))
true_usage_mcf7 = Input(shape=(1,))
true_usage_cho = Input(shape=(1,))
#Shared Model Definition (Applied to each randomized sequence region)
layer_1 = Conv1D(64, 8, padding='valid', activation='relu')
layer_1_pool = MaxPooling1D(pool_size=2)
layer_2 = Conv1D(128, 6, padding='valid', activation='relu')
def shared_model(seq_input) :
return Flatten()(
layer_2(
layer_1_pool(
layer_1(
seq_input
)
)
)
)
shared_out = shared_model(seq_input)
#Layers applied to the concatenated hidden representation
layer_dense = Dense(256, activation='relu')
layer_drop = Dropout(0.2)
dropped_dense_out = layer_drop(layer_dense(shared_out))
#Final cell-line specific regression layers
layer_usage_hek = Dense(1, activation='sigmoid', kernel_initializer='zeros')
layer_usage_hela = Dense(1, activation='sigmoid', kernel_initializer='zeros')
layer_usage_mcf7 = Dense(1, activation='sigmoid', kernel_initializer='zeros')
layer_usage_cho = Dense(1, activation='sigmoid', kernel_initializer='zeros')
pred_usage_hek = layer_usage_hek(dropped_dense_out)
pred_usage_hela = layer_usage_hela(dropped_dense_out)
pred_usage_mcf7 = layer_usage_mcf7(dropped_dense_out)
pred_usage_cho = layer_usage_cho(dropped_dense_out)
#Compile Splicing Model
splicing_model = Model(
inputs=[
seq_input
],
outputs=[
pred_usage_hek,
pred_usage_hela,
pred_usage_mcf7,
pred_usage_cho
]
)
```
<h2>Loss Model Definition</h2>
Here we specfiy our loss function, and we build it as a separate Keras Model.<br/>
In our case, our loss model averages the KL-divergence of predicted vs. true Splice Donor Usage across the 4 different cell types.<br/>
```
#Loss Model Definition
loss_hek = Lambda(sigmoid_kl_divergence, output_shape = (1,))([true_usage_hek, pred_usage_hek])
loss_hela = Lambda(sigmoid_kl_divergence, output_shape = (1,))([true_usage_hela, pred_usage_hela])
loss_mcf7 = Lambda(sigmoid_kl_divergence, output_shape = (1,))([true_usage_mcf7, pred_usage_mcf7])
loss_cho = Lambda(sigmoid_kl_divergence, output_shape = (1,))([true_usage_cho, pred_usage_cho])
total_loss = Lambda(
lambda l: (l[0] + l[1] + l[2] + l[3]) / 4.,
output_shape = (1,)
)(
[
loss_hek,
loss_hela,
loss_mcf7,
loss_cho
]
)
loss_model = Model([
#Inputs
seq_input,
#Target SD Usages
true_usage_hek,
true_usage_hela,
true_usage_mcf7,
true_usage_cho
], total_loss)
```
<h2>Optimize the Loss Model</h2>
Here we use SGD to optimize the Loss Model (defined in the previous notebook cell).<br/>
Since our Loss Model indirectly depends on predicted outputs from our CNN Splicing Model, SGD will optimize the weights of our CNN<br/>
<br/>
Note that we very easily pass the data generators, and run them in parallel, by simply calling Keras fit_generator.<br/>
```
#Optimize CNN with Keras using the Data Generators to stream genomic data features
opt = keras.optimizers.SGD(lr=0.1, decay=1e-6, momentum=0.9, nesterov=True)
loss_model.compile(loss=lambda true, pred: pred, optimizer=opt)
callbacks =[
EarlyStopping(monitor='val_loss', min_delta=0.001, patience=2, verbose=0, mode='auto')
]
loss_model.fit_generator(
generator=splicing_gens['train'],
validation_data=splicing_gens['valid'],
epochs=10,
use_multiprocessing=True,
workers=4,
callbacks=callbacks
)
#Save model
save_dir = os.path.join(os.getcwd(), 'saved_models')
if not os.path.isdir(save_dir):
os.makedirs(save_dir)
model_name = 'splicing_cnn_perturbed_multicell.h5'
model_path = os.path.join(save_dir, model_name)
splicing_model.save(model_path)
print('Saved trained model at %s ' % model_path)
#Load model
save_dir = os.path.join(os.getcwd(), 'saved_models')
model_name = 'splicing_cnn_perturbed_multicell.h5'
model_path = os.path.join(save_dir, model_name)
splicing_model = load_model(model_path)
```
<h2>Evaluate the Splicing CNN</h2>
Here we run our Splicing CNN on the Test set data generator (using Keras predict_generator).<br/>
We then compare our predictions of splice donor usage against the true RNA-Seq measurements.<br/>
```
#Evaluate predictions on test set
predictions = splicing_model.predict_generator(splicing_gens['test'], workers=4, use_multiprocessing=True)
pred_usage_hek, pred_usage_hela, pred_usage_mcf7, pred_usage_cho = [np.ravel(prediction) for prediction in predictions]
targets = zip(*[splicing_gens['test'][i][1] for i in range(len(splicing_gens['test']))])
true_usage_hek, true_usage_hela, true_usage_mcf7, true_usage_cho = [np.concatenate(list(target)) for target in targets]
cell_lines = [
('hek', (pred_usage_hek, true_usage_hek)),
('hela', (pred_usage_hela, true_usage_hela)),
('mcf7', (pred_usage_mcf7, true_usage_mcf7)),
('cho', (pred_usage_cho, true_usage_cho))
]
for cell_name, [y_true, y_pred] in cell_lines :
r_val, p_val = pearsonr(y_pred, y_true)
print("Test set R^2 = " + str(round(r_val * r_val, 2)) + ", p = " + str(p_val))
#Plot test set scatter
f = plt.figure(figsize=(4, 4))
plt.scatter(y_pred, y_true, color='black', s=5, alpha=0.05)
plt.xticks([0.0, 0.25, 0.5, 0.75, 1.0], fontsize=14)
plt.yticks([0.0, 0.25, 0.5, 0.75, 1.0], fontsize=14)
plt.xlabel('Predicted SD1 Usage', fontsize=14)
plt.ylabel('True SD1 Usage', fontsize=14)
plt.title(str(cell_name), fontsize=16)
plt.xlim(0, 1)
plt.ylim(0, 1)
plt.tight_layout()
plt.show()
```
|
github_jupyter
|
# Working with MODFLOW-NWT v 1.1 option blocks
In MODFLOW-NWT an option block is present for the WEL file, UZF file, and SFR file. This block takes keyword arguments that are supplied in an option line in other versions of MODFLOW.
The `OptionBlock` class was created to provide combatibility with the MODFLOW-NWT option block and allow the user to easily edit values within the option block
```
import os
import sys
import platform
try:
import flopy
except:
fpth = os.path.abspath(os.path.join("..", ".."))
sys.path.append(fpth)
import flopy
from flopy.utils import OptionBlock
print(sys.version)
print("flopy version: {}".format(flopy.__version__))
load_ws = os.path.join("..", "data", "options", "sagehen")
model_ws = os.path.join("temp", "nwt_options", "output")
```
## Loading a MODFLOW-NWT model that has option block options
It is critical to set the `version` flag in `flopy.modflow.Modflow.load()` to `version='mfnwt'`
We are going to load a modified version of the Sagehen test problem from GSFLOW to illustrate compatibility
```
mfexe = "mfnwt"
if platform.system() == "Windows":
mfexe += ".exe"
ml = flopy.modflow.Modflow.load(
"sagehen.nam", model_ws=load_ws, exe_name=mfexe, version="mfnwt"
)
ml.change_model_ws(new_pth=model_ws)
ml.write_input()
success, buff = ml.run_model(silent=True)
if not success:
print("Something bad happened.")
```
## Let's look at the options attribute of the UZF object
The `uzf.options` attribute is an `OptionBlock` object. The representation of this object is the option block that will be written to output, which allows the user to easily check to make sure the block has the options they want.
```
uzf = ml.get_package("UZF")
uzf.options
```
The `OptionBlock` object also has attributes which correspond to the option names listed in the online guide to modflow
The user can call and edit the options within the option block
```
print(uzf.options.nosurfleak)
print(uzf.options.savefinf)
uzf.options.etsquare = False
uzf.options
uzf.options.etsquare = True
uzf.options
```
### The user can also see the single line representation of the options
```
uzf.options.single_line_options
```
### And the user can easily change to single line options writing
```
uzf.options.block = False
# write out only the uzf file
uzf_name = "uzf_opt.uzf"
uzf.write_file(os.path.join(model_ws, uzf_name))
```
Now let's examine the first few lines of the new UZF file
```
f = open(os.path.join(model_ws, uzf_name))
for ix, line in enumerate(f):
if ix == 3:
break
else:
print(line)
```
And let's load the new UZF file
```
uzf2 = flopy.modflow.ModflowUzf1.load(
os.path.join(model_ws, uzf_name), ml, check=False
)
```
### Now we can look at the options object, and check if it's block or line format
`block=False` indicates that options will be written as line format
```
print(uzf2.options)
print(uzf2.options.block)
```
### Finally we can convert back to block format
```
uzf2.options.block = True
uzf2.write_file(os.path.join(model_ws, uzf_name))
ml.remove_package("UZF")
uzf3 = flopy.modflow.ModflowUzf1.load(
os.path.join(model_ws, uzf_name), ml, check=False
)
print("\n")
print(uzf3.options)
print(uzf3.options.block)
```
## We can also look at the WEL object
```
wel = ml.get_package("WEL")
wel.options
```
Let's write this out as a single line option block and examine the first few lines
```
wel_name = "wel_opt.wel"
wel.options.block = False
wel.write_file(os.path.join(model_ws, wel_name))
f = open(os.path.join(model_ws, wel_name))
for ix, line in enumerate(f):
if ix == 4:
break
else:
print(line)
```
And we can load the new single line options WEL file and confirm that it is being read as an option line
```
ml.remove_package("WEL")
wel2 = flopy.modflow.ModflowWel.load(
os.path.join(model_ws, wel_name), ml, nper=ml.nper, check=False
)
wel2.options
wel2.options.block
```
# Building an OptionBlock from scratch
The user can also build an `OptionBlock` object from scratch to add to a `ModflowSfr2`, `ModflowUzf1`, or `ModflowWel` file.
The `OptionBlock` class has two required parameters and one optional parameter
`option_line`: a one line, string based representation of the options
`package`: a modflow package object
`block`: boolean flag for line based or block based options
```
opt_line = "specify 0.1 20"
options = OptionBlock(opt_line, flopy.modflow.ModflowWel, block=True)
options
```
from here we can set the noprint flag by using `options.noprint`
```
options.noprint = True
```
and the user can also add auxillary variables by using `options.auxillary`
```
options.auxillary = ["aux", "iface"]
```
### Now we can create a new wel file using this `OptionBlock`
and write it to output
```
wel3 = flopy.modflow.ModflowWel(
ml,
stress_period_data=wel.stress_period_data,
options=options,
unitnumber=99,
)
wel3.write_file(os.path.join(model_ws, wel_name))
```
And now let's examine the first few lines of the file
```
f = open(os.path.join(model_ws, wel_name))
for ix, line in enumerate(f):
if ix == 8:
break
else:
print(line)
```
We can see that everything that the OptionBlock class writes out options in the correct location.
### The user can also switch the options over to option line style and write out the output too!
```
wel3.options.block = False
wel3.write_file(os.path.join(model_ws, wel_name))
f = open(os.path.join(model_ws, wel_name))
for ix, line in enumerate(f):
if ix == 6:
break
else:
print(line)
```
|
github_jupyter
|
<h1>Table of Contents<span class="tocSkip"></span></h1>
<div class="toc"><ul class="toc-item"><li><span><a href="#Matplotlib" data-toc-modified-id="Matplotlib-1"><span class="toc-item-num">1 </span>Matplotlib</a></span><ul class="toc-item"><li><span><a href="#Customization" data-toc-modified-id="Customization-1.1"><span class="toc-item-num">1.1 </span>Customization</a></span></li></ul></li><li><span><a href="#subplot" data-toc-modified-id="subplot-2"><span class="toc-item-num">2 </span>subplot</a></span></li></ul></div>
# Intermediate Python for Data Science
## Matplotlib
- source: https://www.datacamp.com/courses/intermediate-python-for-data-science
- color code: https://matplotlib.org/examples/color/named_colors.html
```
# Quick cheat sheet
# to change plot size
plt.figure(figsize=(20,8))
'''Line Plot'''
# Print the last item from years and populations
print(year[-1])
print(pop[-1])
# Import matplotlib.pyplot as plt
import matplotlib.pyplot as plt
# Make a line plot: year on the x-axis, pop on the y-axis
plt.plot(year, pop)
# Display the plot with plt.show()
plt.show()
# Print the last item of gdp_cap and life_exp
print(gdp_cap[-1])
print(life_exp[-1])
# Make a line plot, gdp_cap on the x-axis, life_exp on the y-axis
plt.plot(gdp_cap, life_exp)
# Display the plot
plt.show()
'''Scatter Plot'''
# Change the line plot below to a scatter plot
plt.scatter(gdp_cap, life_exp)
# Put the x-axis on a logarithmic scale
plt.xscale('log')
# Show plot
plt.show()
'''Scatter Plot'''
# Import package
import matplotlib.pyplot as plt
# Build Scatter plot
plt.scatter(pop, life_exp)
# Show plot
plt.show()
'''Histogram'''
# Create histogram of life_exp data
plt.hist(life_exp)
# Display histogram
plt.show()
'''Histogram bins'''
# Build histogram with 5 bins
plt.hist(life_exp, bins = 5)
# Show and clear plot
plt.show()
plt.clf() # cleans it up again so you can start afresh.
# Build histogram with 20 bins
plt.hist(life_exp, bins = 20)
# Show and clear plot again
plt.show()
plt.clf()
'''Histogram compare'''
# Histogram of life_exp, 15 bins
plt.hist(life_exp, bins = 15)
# Show and clear plot
plt.show()
plt.clf()
# Histogram of life_exp1950, 15 bins
plt.hist(life_exp1950, bins = 15)
# Show and clear plot again
plt.show()
plt.clf()
```
### Customization
```
'''Label'''
# Basic scatter plot, log scale
plt.scatter(gdp_cap, life_exp)
plt.xscale('log')
# Strings
xlab = 'GDP per Capita [in USD]'
ylab = 'Life Expectancy [in years]'
title = 'World Development in 2007'
# Add axis labels
plt.xlabel(xlab)
plt.ylabel(ylab)
# Add title
plt.title(title)
# After customizing, display the plot
plt.show()
'''Ticks'''
# Scatter plot
plt.scatter(gdp_cap, life_exp)
# Previous customizations
plt.xscale('log')
plt.xlabel('GDP per Capita [in USD]')
plt.ylabel('Life Expectancy [in years]')
plt.title('World Development in 2007')
# Definition of tick_val and tick_lab
tick_val = [1000,10000,100000]
tick_lab = ['1k','10k','100k']
# Adapt the ticks on the x-axis
plt.xticks(tick_val, tick_lab)
# After customizing, display the plot
plt.show()
'''Sizes
Wouldn't it be nice if the size of the dots corresponds to the population?
'''
# Import numpy as np
import numpy as np
# Store pop as a numpy array: np_pop
np_pop = np.array(pop)
# Double np_pop
np_pop = np_pop * 2
# Update: set s argument to np_pop # s is size
plt.scatter(gdp_cap, life_exp, s = np_pop)
# Previous customizations
plt.xscale('log')
plt.xlabel('GDP per Capita [in USD]')
plt.ylabel('Life Expectancy [in years]')
plt.title('World Development in 2007')
plt.xticks([1000, 10000, 100000],['1k', '10k', '100k'])
# Display the plot
plt.show()
'''Colors
The next step is making the plot more colorful! To do this, a list col has been created for you. It's a list with a color for each corresponding country, depending on the continent the country is part of.
How did we make the list col you ask? The Gapminder data contains a list continent with the continent each country belongs to. A dictionary is constructed that maps continents onto colors:
'''
dict = {
'Asia':'red',
'Europe':'green',
'Africa':'blue',
'Americas':'yellow',
'Oceania':'black'
}
# c = color, alpha = opacity
# Specify c and alpha inside plt.scatter()
plt.scatter(x = gdp_cap, y = life_exp, s = np.array(pop) * 2, c = col, alpha = 0.8)
# Previous customizations
plt.xscale('log')
plt.xlabel('GDP per Capita [in USD]')
plt.ylabel('Life Expectancy [in years]')
plt.title('World Development in 2007')
plt.xticks([1000,10000,100000], ['1k','10k','100k'])
# Show the plot
plt.show()
'''Additional Customizations'''
# Scatter plot
plt.scatter(x = gdp_cap, y = life_exp, s = np.array(pop) * 2, c = col, alpha = 0.8)
# Previous customizations
plt.xscale('log')
plt.xlabel('GDP per Capita [in USD]')
plt.ylabel('Life Expectancy [in years]')
plt.title('World Development in 2007')
plt.xticks([1000,10000,100000], ['1k','10k','100k'])
# Additional customizations
plt.text(1550, 71, 'India')
plt.text(5700, 80, 'China')
# Add grid() call
plt.grid(True)
# Show the plot
plt.show()
from sklearn.datasets import load_iris
data = load_iris()
data.target[[10, 25, 50]]
list(data.target_names)
```
## subplot
source: https://matplotlib.org/examples/pylab_examples/subplot_demo.html
```
# subplot(nrows, ncols, plot_number)
import numpy as np
import matplotlib.pyplot as plt
x1 = np.linspace(0.0, 5.0)
x2 = np.linspace(0.0, 2.0)
y1 = np.cos(2 * np.pi * x1) * np.exp(-x1)
y2 = np.cos(2 * np.pi * x2)
plt.subplot(2, 1, 1)
plt.plot(x1, y1, 'ko-')
plt.title('A tale of 2 subplots')
plt.ylabel('Damped oscillation')
plt.subplot(2, 1, 2)
plt.plot(x2, y2, 'r.-')
plt.xlabel('time (s)')
plt.ylabel('Undamped')
plt.show()
plt.subplot(2, 1, 1)
plt.plot(x1, y1, 'ko-')
plt.title('A tale of 2 subplots')
plt.ylabel('Damped oscillation')
plt.subplot(2, 1, 2)
plt.plot(x2, y2, 'r.-')
plt.xlabel('time (s)')
plt.ylabel('Undamped')
plt.show()
plt.subplots(2, 2, sharex=True, sharey=True)
plt.show()
x = np.linspace(0, 2*np.pi, 400)
y = np.sin(x**2)
fig, axes = plt.subplots(1,2, sharey=True)
axes[0].plot(x, y)
axes[1].scatter(x, y)
plt.show()
# Two subplots, unpack the axes array immediately
f, (ax1, ax2) = plt.subplots(1, 2, sharey=True)
ax1.plot(x, y)
ax1.set_title('Sharing Y axis')
ax2.scatter(x, y)
plt.show()
fig, axes = plt.subplots(1,3, sharey=True, sharex=True)
for i in range(3):
axes[i].scatter(center[i],xn)
axes[i].set_title('Cluster ' + str(i+1))
axes[i].grid(True)
plt.yticks(xn,var)
plt.subplots_adjust(wspace=0, hspace=0)
#plt.grid(True)
plt.show()
```
|
github_jupyter
|
```
import seaborn as sns
import pandas as pd
import numpy as np
import altair as alt
from markdown import markdown
from IPython.display import Markdown
from ipywidgets.widgets import HTML, Tab
from ipywidgets import widgets
from datetime import timedelta
from matplotlib import pyplot as plt
import os.path as op
from mod import load_data, alt_theme
def author_url(author):
return f"https://github.com/{author}"
# Parameters
fmt_date = "{:%Y-%m-%d}"
n_days = 90
start_date = fmt_date.format(pd.datetime.today() - timedelta(days=n_days))
end_date = fmt_date.format(pd.datetime.today())
renderer = "jupyterlab"
github_orgs = ["jupyterhub", "jupyter", "jupyterlab", "jupyter-widgets", "ipython", "binder-examples", "nteract"]
# Parameters
renderer = "kaggle"
start_date = "2019-02-01"
end_date = "2019-03-01"
comments, issues, prs = load_data('../data/')
bot_names = pd.read_csv('bot_names.csv')['names'].tolist()
comments = comments.query('author not in @bot_names').drop_duplicates()
issues = issues.query('author not in @bot_names').drop_duplicates()
prs = prs.query('author not in @bot_names').drop_duplicates()
# Only keep the dates we want
comments = comments.query('updatedAt > @start_date and updatedAt < @end_date')
issues = issues.query('updatedAt > @start_date and updatedAt < @end_date')
prs = prs.query('updatedAt > @start_date and updatedAt < @end_date')
alt.renderers.enable(renderer);
alt.themes.register('my_theme', alt_theme)
alt.themes.enable("my_theme")
# Information about out time window
time_delta = pd.to_datetime(end_date) - pd.to_datetime(start_date)
n_days = time_delta.days
# Information about the data we loaded
github_orgs = comments['org'].unique()
```
# GitHub activity
Jupyter also has lots of activity across GitHub repositories. The following sections contain
overviews of recent activity across the following GitHub organizations:
```
# Define colors we'll use for GitHub membership
author_types = ['MEMBER', 'CONTRIBUTOR', 'COLLABORATOR', "NONE"]
author_palette = sns.palettes.blend_palette(["lightgrey", "lightgreen", "darkgreen"], 4)
author_colors = ["rgb({}, {}, {}, {})".format(*(ii*256)) for ii in author_palette]
author_color_dict = {key: val for key, val in zip(author_types, author_palette)}
orgs_md = []
for org in github_orgs:
orgs_md.append(f'* [github.com/{org}](https://github.com/{org})')
Markdown('\n'.join(orgs_md))
Markdown(f"Showing GitHub activity from **{start_date}** to **{end_date}**")
```
## List of all contributors per organization
First, we'll list each contributor that has contributed to each organization in the last several days.
Contributions to open source projects are diverse, and involve much more than just contributing code and
code review. Thanks to everybody in the Jupyter communities for all that they do.
```
n_plot = 5
tabs = widgets.Tab(children=[])
for ii, org in enumerate(github_orgs):
authors_comments = comments.query('org == @org')['author']
authors_prs = prs.query('org == @org')['author']
unique_participants = np.unique(np.hstack([authors_comments.values, authors_prs.values]).astype(str)).tolist()
unique_participants.sort(key=lambda a: a.lower())
all_participants = [f"[{participant}](https://github.com/{participant})" for participant in unique_participants]
participants_md = " | ".join(all_participants)
md_html = HTML("<center>{}</center>".format(markdown(participants_md)))
children = list(tabs.children)
children.append(md_html)
tabs.children = tuple(children)
tabs.set_title(ii, org)
display(Markdown(f"All participants across issues and pull requests in each org in the last {n_days} days"))
display(tabs)
```
## Merged Pull requests
Here's an analysis of **merged pull requests** across each of the repositories in the Jupyter
ecosystem.
```
merged = prs.query('state == "MERGED" and closedAt > @start_date and closedAt < @end_date')
prs_by_repo = merged.groupby(['org', 'repo']).count()['author'].reset_index().sort_values(['org', 'author'], ascending=False)
alt.Chart(data=prs_by_repo, title=f"Merged PRs in the last {n_days} days").mark_bar().encode(
x=alt.X('repo', sort=prs_by_repo['repo'].values.tolist()),
y='author',
color='org'
)
```
### A list of merged PRs by project
Below is a tabbed readout of recently-merged PRs. Check out the title to get an idea for what they
implemented, and be sure to thank the PR author for their hard work!
```
tabs = widgets.Tab(children=[])
merged_by = {}
pr_by = {}
for ii, (org, idata) in enumerate(merged.groupby('org')):
issue_md = []
issue_md.append(f"#### Closed PRs for org: `{org}`")
issue_md.append("")
for (org, repo), prs in idata.groupby(['org', 'repo']):
issue_md.append(f"##### [{org}/{repo}](https://github.com/{org}/{repo})")
for _, pr in prs.iterrows():
user_name = pr['author']
user_url = author_url(user_name)
pr_number = pr['number']
pr_html = pr['url']
pr_title = pr['title']
pr_closedby = pr['mergedBy']
pr_closedby_url = f"https://github.com/{pr_closedby}"
if user_name not in pr_by:
pr_by[user_name] = 1
else:
pr_by[user_name] += 1
if pr_closedby not in merged_by:
merged_by[pr_closedby] = 1
else:
merged_by[pr_closedby] += 1
text = f"* [(#{pr_number})]({pr_html}): _{pr_title}_ by **[@{user_name}]({user_url})** merged by **[@{pr_closedby}]({pr_closedby_url})**"
issue_md.append(text)
issue_md.append('')
markdown_html = markdown('\n'.join(issue_md))
children = list(tabs.children)
children.append(HTML(markdown_html))
tabs.children = tuple(children)
tabs.set_title(ii, org)
tabs
```
### Authoring and merging stats by repository
Let's see who has been doing most of the PR authoring and merging. The PR author is generally the
person that implemented a change in the repository (code, documentation, etc). The PR merger is
the person that "pressed the green button" and got the change into the main codebase.
```
# Prep our merging DF
merged_by_repo = merged.groupby(['org', 'repo', 'author'], as_index=False).agg({'id': 'count', 'authorAssociation': 'first'}).rename(columns={'id': "authored", 'author': 'username'})
closed_by_repo = merged.groupby(['org', 'repo', 'mergedBy']).count()['id'].reset_index().rename(columns={'id': "closed", "mergedBy": "username"})
n_plot = 50
charts = []
for ii, (iorg, idata) in enumerate(merged_by_repo.replace(np.nan, 0).groupby(['org'])):
title = f"PR authors for {iorg} in the last {n_days} days"
idata = idata.groupby('username', as_index=False).agg({'authored': 'sum', 'authorAssociation': 'first'})
idata = idata.sort_values('authored', ascending=False).head(n_plot)
ch = alt.Chart(data=idata, width=1000, title=title).mark_bar().encode(
x='username',
y='authored',
color=alt.Color('authorAssociation', scale=alt.Scale(domain=author_types, range=author_colors))
)
charts.append(ch)
alt.hconcat(*charts)
charts = []
for ii, (iorg, idata) in enumerate(closed_by_repo.replace(np.nan, 0).groupby(['org'])):
title = f"Merges for {iorg} in the last {n_days} days"
ch = alt.Chart(data=idata, width=1000, title=title).mark_bar().encode(
x='username',
y='closed',
)
charts.append(ch)
alt.hconcat(*charts)
```
## Issues
Issues are **conversations** that happen on our GitHub repositories. Here's an
analysis of issues across the Jupyter organizations.
```
created = issues.query('state == "OPEN" and createdAt > @start_date and createdAt < @end_date')
closed = issues.query('state == "CLOSED" and closedAt > @start_date and closedAt < @end_date')
created_counts = created.groupby(['org', 'repo']).count()['number'].reset_index()
created_counts['org/repo'] = created_counts.apply(lambda a: a['org'] + '/' + a['repo'], axis=1)
sorted_vals = created_counts.sort_values(['org', 'number'], ascending=False)['repo'].values
alt.Chart(data=created_counts, title=f"Issues created in the last {n_days} days").mark_bar().encode(
x=alt.X('repo', sort=alt.Sort(sorted_vals.tolist())),
y='number',
color='org',
)
closed_counts = closed.groupby(['org', 'repo']).count()['number'].reset_index()
closed_counts['org/repo'] = closed_counts.apply(lambda a: a['org'] + '/' + a['repo'], axis=1)
sorted_vals = closed_counts.sort_values(['org', 'number'], ascending=False)['repo'].values
alt.Chart(data=closed_counts, title=f"Issues closed in the last {n_days} days").mark_bar().encode(
x=alt.X('repo', sort=alt.Sort(sorted_vals.tolist())),
y='number',
color='org',
)
created_closed = pd.merge(created_counts.rename(columns={'number': 'created'}).drop(columns='org/repo'),
closed_counts.rename(columns={'number': 'closed'}).drop(columns='org/repo'),
on=['org', 'repo'], how='outer')
created_closed = pd.melt(created_closed, id_vars=['org', 'repo'], var_name="kind", value_name="count").replace(np.nan, 0)
charts = []
for org in github_orgs:
# Pick the top 10 repositories
this_issues = created_closed.query('org == @org')
top_repos = this_issues.groupby(['repo']).sum().sort_values(by='count', ascending=False).head(10).index
ch = alt.Chart(this_issues.query('repo in @top_repos'), width=120).mark_bar().encode(
x=alt.X("kind", axis=alt.Axis(labelFontSize=15, title="")),
y=alt.Y('count', axis=alt.Axis(titleFontSize=15, labelFontSize=12)),
color='kind',
column=alt.Column("repo", header=alt.Header(title=f"Issue activity, last {n_days} days for {org}", titleFontSize=15, labelFontSize=12))
)
charts.append(ch)
alt.hconcat(*charts)
# Set to datetime
for kind in ['createdAt', 'closedAt']:
closed.loc[:, kind] = pd.to_datetime(closed[kind])
closed.loc[:, 'time_open'] = closed['closedAt'] - closed['createdAt']
closed.loc[:, 'time_open'] = closed['time_open'].dt.total_seconds()
time_open = closed.groupby(['org', 'repo']).agg({'time_open': 'median'}).reset_index()
time_open['time_open'] = time_open['time_open'] / (60 * 60 * 24)
time_open['org/repo'] = time_open.apply(lambda a: a['org'] + '/' + a['repo'], axis=1)
sorted_vals = time_open.sort_values(['org', 'time_open'], ascending=False)['repo'].values
alt.Chart(data=time_open, title=f"Time to close for issues closed in the last {n_days} days").mark_bar().encode(
x=alt.X('repo', sort=alt.Sort(sorted_vals.tolist())),
y=alt.Y('time_open', title="Median Days Open"),
color='org',
)
```
### A list of recent issues
Below is a list of issues with recent activity in each repository. If they seem of interest
to you, click on their links and jump in to participate!
```
# Add comment count data to issues and PRs
comment_counts = (
comments
.query("createdAt > @start_date and createdAt < @end_date")
.groupby(['org', 'repo', 'issue_id'])
.count().iloc[:, 0].to_frame()
)
comment_counts.columns = ['n_comments']
comment_counts = comment_counts.reset_index()
n_plot = 5
tabs = widgets.Tab(children=[])
for ii, (org, idata) in enumerate(comment_counts.groupby('org')):
issue_md = []
issue_md.append(f"#### {org}")
issue_md.append("")
for repo, i_issues in idata.groupby('repo'):
issue_md.append(f"##### [{org}/{repo}](https://github.com/{org}/{repo})")
top_issues = i_issues.sort_values('n_comments', ascending=False).head(n_plot)
top_issue_list = pd.merge(issues, top_issues, left_on=['org', 'repo', 'number'], right_on=['org', 'repo', 'issue_id'])
for _, issue in top_issue_list.sort_values('n_comments', ascending=False).head(n_plot).iterrows():
user_name = issue['author']
user_url = author_url(user_name)
issue_number = issue['number']
issue_html = issue['url']
issue_title = issue['title']
text = f"* [(#{issue_number})]({issue_html}): _{issue_title}_ by **[@{user_name}]({user_url})**"
issue_md.append(text)
issue_md.append('')
md_html = HTML(markdown('\n'.join(issue_md)))
children = list(tabs.children)
children.append(HTML(markdown('\n'.join(issue_md))))
tabs.children = tuple(children)
tabs.set_title(ii, org)
display(Markdown(f"Here are the top {n_plot} active issues in each repository in the last {n_days} days"))
display(tabs)
```
## Commenters across repositories
These are commenters across all issues and pull requests in the last several days.
These are colored by the commenter's association with the organization. For information
about what these associations mean, [see this StackOverflow post](https://stackoverflow.com/a/28866914/1927102).
```
commentors = (
comments
.query("createdAt > @start_date and createdAt < @end_date")
.groupby(['org', 'repo', 'author', 'authorAssociation'])
.count().rename(columns={'id': 'count'})['count']
.reset_index()
.sort_values(['org', 'count'], ascending=False)
)
n_plot = 50
charts = []
for ii, (iorg, idata) in enumerate(commentors.groupby(['org'])):
title = f"Top {n_plot} commentors for {iorg} in the last {n_days} days"
idata = idata.groupby('author', as_index=False).agg({'count': 'sum', 'authorAssociation': 'first'})
idata = idata.sort_values('count', ascending=False).head(n_plot)
ch = alt.Chart(data=idata.head(n_plot), width=1000, title=title).mark_bar().encode(
x='author',
y='count',
color=alt.Color('authorAssociation', scale=alt.Scale(domain=author_types, range=author_colors))
)
charts.append(ch)
alt.hconcat(*charts)
```
## First responders
First responders are the first people to respond to a new issue in one of the repositories.
The following plots show first responders for recently-created issues.
```
first_comments = []
for (org, repo, issue_id), i_comments in comments.groupby(['org', 'repo', 'issue_id']):
ix_min = pd.to_datetime(i_comments['createdAt']).idxmin()
first_comment = i_comments.loc[ix_min]
if isinstance(first_comment, pd.DataFrame):
first_comment = first_comment.iloc[0]
first_comments.append(first_comment)
first_comments = pd.concat(first_comments, axis=1).T
first_responder_counts = first_comments.groupby(['org', 'author', 'authorAssociation'], as_index=False).\
count().rename(columns={'id': 'n_first_responses'}).sort_values(['org', 'n_first_responses'], ascending=False)
n_plot = 50
charts = []
for ii, (iorg, idata) in enumerate(first_responder_counts.groupby(['org'])):
title = f"Top {n_plot} first responders for {iorg} in the last {n_days} days"
idata = idata.groupby('author', as_index=False).agg({'n_first_responses': 'sum', 'authorAssociation': 'first'})
idata = idata.sort_values('n_first_responses', ascending=False).head(n_plot)
ch = alt.Chart(data=idata.head(n_plot), width=1000, title=title).mark_bar().encode(
x='author',
y='n_first_responses',
color=alt.Color('authorAssociation', scale=alt.Scale(domain=author_types, range=author_colors))
)
charts.append(ch)
alt.hconcat(*charts)
%%html
<script src="https://cdn.rawgit.com/parente/4c3e6936d0d7a46fd071/raw/65b816fb9bdd3c28b4ddf3af602bfd6015486383/code_toggle.js"></script>
```
|
github_jupyter
|
# Introduction to Logistic Regression
## Learning Objectives
1. Create Seaborn plots for Exploratory Data Analysis
2. Train a Logistic Regression Model using Scikit-Learn
## Introduction
This lab is an introduction to logistic regression using Python and Scikit-Learn. This lab serves as a foundation for more complex algorithms and machine learning models that you will encounter in the course. In this lab, we will use a synthetic advertising data set, indicating whether or not a particular internet user clicked on an Advertisement on a company website. We will try to create a model that will predict whether or not they will click on an ad based off the features of that user.
Each learning objective will correspond to a __#TODO__ in this student lab notebook -- try to complete this notebook first and then review the [solution notebook](../solutions/intro_logistic_regression.ipynb).
### Import Libraries
```
import os
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
```
### Load the Dataset
We will use a synthetic [advertising](https://www.kaggle.com/fayomi/advertising) dataset. This data set contains the following features:
* 'Daily Time Spent on Site': consumer time on site in minutes
* 'Age': customer age in years
* 'Area Income': Avg. Income of geographical area of consumer
* 'Daily Internet Usage': Avg. minutes a day consumer is on the internet
* 'Ad Topic Line': Headline of the advertisement
* 'City': City of consumer
* 'Male': Whether or not consumer was male
* 'Country': Country of consumer
* 'Timestamp': Time at which consumer clicked on Ad or closed window
* 'Clicked on Ad': 0 or 1 indicated clicking on Ad
```
# TODO 1: Read in the advertising.csv file and set it to a data frame called ad_data.
# TODO: Your code goes here
```
**Check the head of ad_data**
```
ad_data.head()
```
**Use info and describe() on ad_data**
```
ad_data.info()
ad_data.describe()
```
Let's check for any null values.
```
ad_data.isnull().sum()
```
## Exploratory Data Analysis (EDA)
Let's use seaborn to explore the data! Try recreating the plots shown below!
TODO 1: **Create a histogram of the Age**
```
# TODO: Your code goes here
```
TODO 1: **Create a jointplot showing Area Income versus Age.**
```
# TODO: Your code goes here
```
TODO 2: **Create a jointplot showing the kde distributions of Daily Time spent on site vs. Age.**
```
# TODO: Your code goes here
```
TODO 1: **Create a jointplot of 'Daily Time Spent on Site' vs. 'Daily Internet Usage'**
```
# TODO: Your code goes here
```
# Logistic Regression
Logistic regression is a supervised machine learning process. It is similar to linear regression, but rather than predict a continuous value, we try to estimate probabilities by using a logistic function. Note that even though it has regression in the name, it is for classification.
While linear regression is acceptable for estimating values, logistic regression is best for predicting the class of an observation
Now it's time to do a train test split, and train our model! You'll have the freedom here to choose columns that you want to train on!
```
from sklearn.model_selection import train_test_split
```
Next, let's define the features and label. Briefly, feature is input; label is output. This applies to both classification and regression problems.
```
X = ad_data[['Daily Time Spent on Site', 'Age', 'Area Income','Daily Internet Usage', 'Male']]
y = ad_data['Clicked on Ad']
```
TODO 2: **Split the data into training set and testing set using train_test_split**
```
# TODO: Your code goes here
```
**Train and fit a logistic regression model on the training set.**
```
from sklearn.linear_model import LogisticRegression
logmodel = LogisticRegression()
logmodel.fit(X_train,y_train)
```
## Predictions and Evaluations
**Now predict values for the testing data.**
```
predictions = logmodel.predict(X_test)
```
**Create a classification report for the model.**
```
from sklearn.metrics import classification_report
print(classification_report(y_test,predictions))
```
Copyright 2021 Google Inc. Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
|
github_jupyter
|
# Generative Adversarial Network
In this notebook, we'll be building a generative adversarial network (GAN) trained on the MNIST dataset. From this, we'll be able to generate new handwritten digits!
GANs were [first reported on](https://arxiv.org/abs/1406.2661) in 2014 from Ian Goodfellow and others in Yoshua Bengio's lab. Since then, GANs have exploded in popularity. Here are a few examples to check out:
* [Pix2Pix](https://affinelayer.com/pixsrv/)
* [CycleGAN & Pix2Pix in PyTorch, Jun-Yan Zhu](https://github.com/junyanz/pytorch-CycleGAN-and-pix2pix)
* [A list of generative models](https://github.com/wiseodd/generative-models)
The idea behind GANs is that you have two networks, a generator $G$ and a discriminator $D$, competing against each other. The generator makes "fake" data to pass to the discriminator. The discriminator also sees real training data and predicts if the data it's received is real or fake.
> * The generator is trained to fool the discriminator, it wants to output data that looks _as close as possible_ to real, training data.
* The discriminator is a classifier that is trained to figure out which data is real and which is fake.
What ends up happening is that the generator learns to make data that is indistinguishable from real data to the discriminator.
<img src='assets/gan_pipeline.png' width=70% />
The general structure of a GAN is shown in the diagram above, using MNIST images as data. The latent sample is a random vector that the generator uses to construct its fake images. This is often called a **latent vector** and that vector space is called **latent space**. As the generator trains, it figures out how to map latent vectors to recognizable images that can fool the discriminator.
If you're interested in generating only new images, you can throw out the discriminator after training. In this notebook, I'll show you how to define and train these adversarial networks in PyTorch and generate new images!
```
%matplotlib inline
import numpy as np
import torch
import matplotlib.pyplot as plt
from torchvision import datasets
import torchvision.transforms as transforms
# number of subprocesses to use for data loading
num_workers = 0
# how many samples per batch to load
batch_size = 64
# convert data to torch.FloatTensor
transform = transforms.ToTensor()
# get the training datasets
train_data = datasets.MNIST(root='data', train=True,
download=True, transform=transform)
# prepare data loader
train_loader = torch.utils.data.DataLoader(train_data, batch_size=batch_size,
num_workers=num_workers)
```
### Visualize the data
```
# obtain one batch of training images
dataiter = iter(train_loader)
images, labels = dataiter.next()
images = images.numpy()
# get one image from the batch
img = np.squeeze(images[0])
fig = plt.figure(figsize = (3,3))
ax = fig.add_subplot(111)
ax.imshow(img, cmap='gray')
```
---
# Define the Model
A GAN is comprised of two adversarial networks, a discriminator and a generator.
## Discriminator
The discriminator network is going to be a pretty typical linear classifier. To make this network a universal function approximator, we'll need at least one hidden layer, and these hidden layers should have one key attribute:
> All hidden layers will have a [Leaky ReLu](https://pytorch.org/docs/stable/nn.html#torch.nn.LeakyReLU) activation function applied to their outputs.
<img src='assets/gan_network.png' width=70% />
#### Leaky ReLu
We should use a leaky ReLU to allow gradients to flow backwards through the layer unimpeded. A leaky ReLU is like a normal ReLU, except that there is a small non-zero output for negative input values.
<img src='assets/leaky_relu.png' width=40% />
#### Sigmoid Output
We'll also take the approach of using a more numerically stable loss function on the outputs. Recall that we want the discriminator to output a value 0-1 indicating whether an image is _real or fake_.
> We will ultimately use [BCEWithLogitsLoss](https://pytorch.org/docs/stable/nn.html#bcewithlogitsloss), which combines a `sigmoid` activation function **and** and binary cross entropy loss in one function.
So, our final output layer should not have any activation function applied to it.
```
import torch.nn as nn
import torch.nn.functional as F
class Discriminator(nn.Module):
def __init__(self, input_size, hidden_dim, output_size):
super(Discriminator, self).__init__()
# define all layers
self.fc1 = nn.Linear(input_size, hidden_dim * 4)
self.fc2 = nn.Linear(hidden_dim * 4, hidden_dim * 2)
self.fc3 = nn.Linear(hidden_dim * 2, hidden_dim)
self.fc4 = nn.Linear(hidden_dim, output_size)
self.dropout = nn.Dropout(0.3)
def forward(self, x):
# flatten image
x = x.view(-1, 28 * 28)
# pass x through all layers
# apply leaky relu activation to all hidden layers
x = F.leaky_relu(self.fc1(x), 0.2)
x = self.dropout(x)
x = F.leaky_relu(self.fc2(x), 0.2)
x = self.dropout(x)
x = F.leaky_relu(self.fc3(x), 0.2)
x = self.dropout(x)
out = self.fc4(x)
return out
```
## Generator
The generator network will be almost exactly the same as the discriminator network, except that we're applying a [tanh activation function](https://pytorch.org/docs/stable/nn.html#tanh) to our output layer.
#### tanh Output
The generator has been found to perform the best with $tanh$ for the generator output, which scales the output to be between -1 and 1, instead of 0 and 1.
<img src='assets/tanh_fn.png' width=40% />
Recall that we also want these outputs to be comparable to the *real* input pixel values, which are read in as normalized values between 0 and 1.
> So, we'll also have to **scale our real input images to have pixel values between -1 and 1** when we train the discriminator.
I'll do this in the training loop, later on.
```
class Generator(nn.Module):
def __init__(self, input_size, hidden_dim, output_size):
super(Generator, self).__init__()
# define all layers
self.fc1 = nn.Linear(input_size, hidden_dim)
self.fc2 = nn.Linear(hidden_dim, hidden_dim * 2)
self.fc3 = nn.Linear(hidden_dim * 2, hidden_dim * 4)
self.fc4 = nn.Linear(hidden_dim * 4, output_size)
self.dropout = nn.Dropout(0.2)
def forward(self, x):
# pass x through all layers
x = F.leaky_relu(self.fc1(x), 0.2)
x = self.dropout(x)
x = F.leaky_relu(self.fc2(x), 0.2)
x = self.dropout(x)
x = F.leaky_relu(self.fc3(x), 0.2)
x = self.dropout(x)
# final layer should have tanh applied
out = F.tanh(self.fc4(x))
return out
```
## Model hyperparameters
```
# Discriminator hyperparams
# Size of input image to discriminator (28*28)
input_size = 784
# Size of discriminator output (real or fake)
d_output_size = 1
# Size of *last* hidden layer in the discriminator
d_hidden_size = 32
# Generator hyperparams
# Size of latent vector to give to generator
z_size = 100
# Size of discriminator output (generated image)
g_output_size = 784
# Size of *first* hidden layer in the generator
g_hidden_size = 32
```
## Build complete network
Now we're instantiating the discriminator and generator from the classes defined above. Make sure you've passed in the correct input arguments.
```
# instantiate discriminator and generator
D = Discriminator(input_size, d_hidden_size, d_output_size)
G = Generator(z_size, g_hidden_size, g_output_size)
# check that they are as you expect
print(D)
print()
print(G)
```
---
## Discriminator and Generator Losses
Now we need to calculate the losses.
### Discriminator Losses
> * For the discriminator, the total loss is the sum of the losses for real and fake images, `d_loss = d_real_loss + d_fake_loss`.
* Remember that we want the discriminator to output 1 for real images and 0 for fake images, so we need to set up the losses to reflect that.
<img src='assets/gan_pipeline.png' width=70% />
The losses will by binary cross entropy loss with logits, which we can get with [BCEWithLogitsLoss](https://pytorch.org/docs/stable/nn.html#bcewithlogitsloss). This combines a `sigmoid` activation function **and** and binary cross entropy loss in one function.
For the real images, we want `D(real_images) = 1`. That is, we want the discriminator to classify the the real images with a label = 1, indicating that these are real. To help the discriminator generalize better, the labels are **reduced a bit from 1.0 to 0.9**. For this, we'll use the parameter `smooth`; if True, then we should smooth our labels. In PyTorch, this looks like `labels = torch.ones(size) * 0.9`
The discriminator loss for the fake data is similar. We want `D(fake_images) = 0`, where the fake images are the _generator output_, `fake_images = G(z)`.
### Generator Loss
The generator loss will look similar only with flipped labels. The generator's goal is to get `D(fake_images) = 1`. In this case, the labels are **flipped** to represent that the generator is trying to fool the discriminator into thinking that the images it generates (fakes) are real!
```
# Calculate losses
def real_loss(D_out, smooth=False):
# compare logits to real labels
# smooth labels if smooth=True
batch_size = D_out.size(0)
if smooth:
labels = torch.ones(batch_size) * 0.9
else:
labels = torch.ones(batch_size)
criterion = nn.BCEWithLogitsLoss()
loss = criterion(D_out.squeeze(), labels)
return loss
def fake_loss(D_out):
# compare logits to fake labels
batch_size = D_out.size(0)
labels = torch.zeros(batch_size)
criterion = nn.BCEWithLogitsLoss()
loss = criterion(D_out.squeeze(), labels)
return loss
```
## Optimizers
We want to update the generator and discriminator variables separately. So, we'll define two separate Adam optimizers.
```
import torch.optim as optim
# learning rate for optimizers
lr = 0.002
# Create optimizers for the discriminator and generator
d_optimizer = optim.Adam(D.parameters(), lr)
g_optimizer = optim.Adam(G.parameters(), lr)
```
---
## Training
Training will involve alternating between training the discriminator and the generator. We'll use our functions `real_loss` and `fake_loss` to help us calculate the discriminator losses in all of the following cases.
### Discriminator training
1. Compute the discriminator loss on real, training images
2. Generate fake images
3. Compute the discriminator loss on fake, generated images
4. Add up real and fake loss
5. Perform backpropagation + an optimization step to update the discriminator's weights
### Generator training
1. Generate fake images
2. Compute the discriminator loss on fake images, using **flipped** labels!
3. Perform backpropagation + an optimization step to update the generator's weights
#### Saving Samples
As we train, we'll also print out some loss statistics and save some generated "fake" samples.
```
import pickle as pkl
# training hyperparams
num_epochs = 40
# keep track of loss and generated, "fake" samples
samples = []
losses = []
print_every = 400
# Get some fixed data for sampling. These are images that are held
# constant throughout training, and allow us to inspect the model's performance
sample_size=16
fixed_z = np.random.uniform(-1, 1, size=(sample_size, z_size))
fixed_z = torch.from_numpy(fixed_z).float()
# train the network
D.train()
G.train()
for epoch in range(num_epochs):
for batch_i, (real_images, _) in enumerate(train_loader):
batch_size = real_images.size(0)
## Important rescaling step ##
real_images = real_images*2 - 1 # rescale input images from [0,1) to [-1, 1)
# ============================================
# TRAIN THE DISCRIMINATOR
# ============================================
d_optimizer.zero_grad()
# 1. Train with real images
# Compute the discriminator losses on real images
# use smoothed labels
D_real = D(real_images)
d_real_loss = real_loss(D_real, smooth=True)
# 2. Train with fake images
# Generate fake images
z = np.random.uniform(-1, 1, size=(batch_size, z_size))
z = torch.from_numpy(z).float()
fake_images = G(z)
# Compute the discriminator losses on fake images
D_fake = D(fake_images)
d_fake_loss = fake_loss(D_fake)
# add up real and fake losses and perform backprop
d_loss = d_real_loss + d_fake_loss
d_loss.backward()
d_optimizer.step()
# =========================================
# TRAIN THE GENERATOR
# =========================================
g_optimizer.zero_grad()
# 1. Train with fake images and flipped labels
# Generate fake images
z = np.random.uniform(-1, 1, size=(batch_size, z_size))
z = torch.from_numpy(z).float()
fake_images = G(z)
# Compute the discriminator losses on fake images
# using flipped labels!
D_fake = D(fake_images)
g_loss = real_loss(D_fake)
# perform backprop
g_loss.backward()
g_optimizer.step()
# Print some loss stats
if batch_i % print_every == 0:
# print discriminator and generator loss
print('Epoch [{:5d}/{:5d}] | d_loss: {:6.4f} | g_loss: {:6.4f}'.format(
epoch+1, num_epochs, d_loss.item(), g_loss.item()))
## AFTER EACH EPOCH##
# append discriminator loss and generator loss
losses.append((d_loss.item(), g_loss.item()))
# generate and save sample, fake images
G.eval() # eval mode for generating samples
samples_z = G(fixed_z)
samples.append(samples_z)
G.train() # back to train mode
# Save training generator samples
with open('train_samples.pkl', 'wb') as f:
pkl.dump(samples, f)
```
## Training loss
Here we'll plot the training losses for the generator and discriminator, recorded after each epoch.
```
fig, ax = plt.subplots()
losses = np.array(losses)
plt.plot(losses.T[0], label='Discriminator')
plt.plot(losses.T[1], label='Generator')
plt.title("Training Losses")
plt.legend()
```
## Generator samples from training
Here we can view samples of images from the generator. First we'll look at the images we saved during training.
```
# helper function for viewing a list of passed in sample images
def view_samples(epoch, samples):
fig, axes = plt.subplots(figsize=(7,7), nrows=4, ncols=4, sharey=True, sharex=True)
for ax, img in zip(axes.flatten(), samples[epoch]):
img = img.detach()
ax.xaxis.set_visible(False)
ax.yaxis.set_visible(False)
im = ax.imshow(img.reshape((28,28)), cmap='Greys_r')
# Load samples from generator, taken while training
with open('train_samples.pkl', 'rb') as f:
samples = pkl.load(f)
```
These are samples from the final training epoch. You can see the generator is able to reproduce numbers like 1, 7, 3, 2. Since this is just a sample, it isn't representative of the full range of images this generator can make.
```
# -1 indicates final epoch's samples (the last in the list)
view_samples(-1, samples)
```
Below I'm showing the generated images as the network was training, every 10 epochs.
```
rows = 10 # split epochs into 10, so 100/10 = every 10 epochs
cols = 6
fig, axes = plt.subplots(figsize=(7,12), nrows=rows, ncols=cols, sharex=True, sharey=True)
for sample, ax_row in zip(samples[::int(len(samples)/rows)], axes):
for img, ax in zip(sample[::int(len(sample)/cols)], ax_row):
img = img.detach()
ax.imshow(img.reshape((28,28)), cmap='Greys_r')
ax.xaxis.set_visible(False)
ax.yaxis.set_visible(False)
```
It starts out as all noise. Then it learns to make only the center white and the rest black. You can start to see some number like structures appear out of the noise like 1s and 9s.
## Sampling from the generator
We can also get completely new images from the generator by using the checkpoint we saved after training. **We just need to pass in a new latent vector $z$ and we'll get new samples**!
```
# randomly generated, new latent vectors
sample_size=16
rand_z = np.random.uniform(-1, 1, size=(sample_size, z_size))
rand_z = torch.from_numpy(rand_z).float()
G.eval() # eval mode
# generated samples
rand_images = G(rand_z)
# 0 indicates the first set of samples in the passed in list
# and we only have one batch of samples, here
view_samples(0, [rand_images])
```
|
github_jupyter
|
```
"""
We use following lines because we are running on Google Colab
If you are running notebook on a local computer, you don't need this cell
"""
from google.colab import drive
drive.mount('/content/gdrive')
import os
os.chdir('/content/gdrive/My Drive/finch/tensorflow1/free_chat/chinese/main')
%tensorflow_version 1.x
!pip install texar
import tensorflow as tf
import texar.tf as tx
import numpy as np
import copy
from texar.tf.modules import TransformerEncoder, TransformerDecoder
print("TensorFlow Version", tf.__version__)
print('GPU Enabled:', tf.test.is_gpu_available())
def forward(features, labels, mode):
if isinstance(features, dict):
words = features['words']
else:
words = features
words_len = tf.count_nonzero(words, 1, dtype=tf.int32)
is_training = (mode == tf.estimator.ModeKeys.TRAIN)
batch_sz = tf.shape(words)[0]
with tf.variable_scope('Embedding'):
embedding = tf.Variable(np.load('../vocab/char.npy'),
dtype=tf.float32,
name='fasttext_vectors')
embedding = tf.concat([tf.zeros(shape=[1, params['embed_dim']]), embedding[1:, :]], axis=0)
x = tf.nn.embedding_lookup(embedding, words)
pos_embedder = tx.modules.SinusoidsPositionEmbedder(
position_size = 2*params['max_len'],
hparams = config_model.position_embedder_hparams)
x = (x * config_model.hidden_dim ** 0.5) + pos_embedder(sequence_length=words_len)
with tf.variable_scope('Encoder'):
encoder = TransformerEncoder(hparams=config_model.encoder)
enc_out = encoder(inputs=x, sequence_length=words_len)
with tf.variable_scope('Decoder'):
decoder = TransformerDecoder(vocab_size=len(params['char2idx'])+1,
output_layer=tf.transpose(embedding, (1, 0)),
hparams=config_model.decoder)
start_tokens = tf.fill([batch_sz], 1)
def _embedding_fn(x, y):
x_w_embed = tf.nn.embedding_lookup(embedding, x)
y_p_embed = pos_embedder(y)
return x_w_embed * config_model.hidden_dim ** 0.5 + y_p_embed
predictions = decoder(
memory=enc_out,
memory_sequence_length=words_len,
beam_width=params['beam_width'],
length_penalty=params['length_penalty'],
start_tokens=start_tokens,
end_token=2,
embedding=_embedding_fn,
max_decoding_length=params['max_len'],
mode=tf.estimator.ModeKeys.PREDICT)
return predictions['sample_id'][:, :, :params['top_k']]
def model_fn(features, labels, mode, params):
logits_or_ids = forward(features, labels, mode)
if mode == tf.estimator.ModeKeys.PREDICT:
return tf.estimator.EstimatorSpec(mode, predictions=logits_or_ids)
class config_model:
hidden_dim = 300
num_heads = 8
dropout_rate = .2
num_blocks = 6
position_embedder_hparams = {
'dim': hidden_dim
}
encoder = {
'dim': hidden_dim,
'embedding_dropout': dropout_rate,
'residual_dropout': dropout_rate,
'num_blocks': num_blocks,
'initializer': {
'type': 'variance_scaling_initializer',
'kwargs': {
'scale': 1.0,
'mode': 'fan_avg',
'distribution': 'uniform',
},
},
'multihead_attention': {
'dropout_rate': dropout_rate,
'num_heads': num_heads,
'output_dim': hidden_dim,
'use_bias': True,
},
'poswise_feedforward': {
'name': 'fnn',
'layers': [
{
'type': 'Dense',
'kwargs': {
'name': 'conv1',
'units': hidden_dim * 2,
'activation': 'gelu',
'use_bias': True,
},
},
{
'type': 'Dropout',
'kwargs': {
'rate': dropout_rate,
}
},
{
'type': 'Dense',
'kwargs': {
'name': 'conv2',
'units': hidden_dim,
'use_bias': True,
}
}
],
},
}
decoder = copy.deepcopy(encoder)
decoder['output_layer_bias'] = True
params = {
'model_dir': '../model/transformer',
'export_dir': '../model/transformer_export',
'vocab_path': '../vocab/char.txt',
'max_len': 15,
'embed_dim': config_model.hidden_dim,
'beam_width': 5,
'top_k': 3,
'length_penalty': .6,
}
def serving_input_receiver_fn():
words = tf.placeholder(tf.int32, [None, None], 'words')
features = {'words': words}
receiver_tensors = features
return tf.estimator.export.ServingInputReceiver(features, receiver_tensors)
def get_vocab(f_path):
word2idx = {}
with open(f_path) as f:
for i, line in enumerate(f):
line = line.rstrip('\n')
word2idx[line] = i
return word2idx
params['char2idx'] = get_vocab(params['vocab_path'])
params['idx2char'] = {idx: char for char, idx in params['char2idx'].items()}
estimator = tf.estimator.Estimator(model_fn, params['model_dir'])
estimator.export_saved_model(params['export_dir'], serving_input_receiver_fn)
```
|
github_jupyter
|
# PyIndMach012: an example of user-model using DSS Python
This example runs a modified example from the OpenDSS distribution for the induction machine model with a sample PyIndMach012 implementation, written in Python, and the original, built-in IndMach012.
Check the `PyIndMach012.py` file for more comments. Comparing it to [the Pascal code for IndMach012](https://github.com/dss-extensions/dss_capi/blob/master/Version7/Source/PCElements/IndMach012.pas) can be useful to understand some of the inner workings of OpenDSS.
The user-model code in DSS Python is not stable yet but can be used to develop new ideas before commiting the final model in a traditional DLL user-model. Particularly, I (@PMeira) found some issues with callbacks with newer Version 8 COM DLLs, so changes related to that are expected.
```
%matplotlib inline
import os
import numpy as np
from matplotlib import pyplot as plt
from dss.UserModels import GenUserModel # used to get the DLL path
import PyIndMach012 # we need to import the model so it gets registered
```
## The model class
```
??PyIndMach012
```
## OpenDSS setup
For this example, we can use either COM or DSS Python (DSS C-API). The IndMach012 model in DSS C-API seems to have a bug somewhere though -- this is being tracked in [dss_capi#62](https://github.com/dss-extensions/dss_capi/issues/62).
```
original_dir = os.getcwd() # same the original working directory since the COM module messes with it
USE_COM = True # toggle this value to run with DSS C-API
if USE_COM:
from dss import patch_dss_com
import win32com.client
DSS = patch_dss_com(win32com.client.gencache.EnsureDispatch('OpenDSSengine.DSS'))
DSS.DataPath = original_dir
os.chdir(original_dir)
else:
from dss import DSS
DSS.Version
Text = DSS.Text
Monitors = DSS.ActiveCircuit.Monitors
```
## Using the model
To use a Python model for generators:
- the model class needs to be registered in advance
- create a generator with `model=6`
- pass a `usermodel="{dll_path}"` as in the following DSS command in the `run` function
- pass a `"pymodel=MODELNAME"` parameter in the userdata property, where MODELNAME is the name of the model class in Python
```
def run(pymodel):
Text.Command = 'redirect "master.dss"'
if pymodel:
# This uses our custom user-model in Python
Text.Command = 'New "Generator.Motor1" bus1=Bg2 kW=1200 conn=delta kVA=1500.000 H=6 model=6 kv=0.48 usermodel="{dll_path}" userdata=(pymodel=PyIndMach012 purs=0.048 puxs=0.075 purr=0.018 puxr=0.12 puxm=3.8 slip=0.02 SlipOption=variableslip)'.format(
dll_path=GenUserModel.dll_path,
)
Text.Command = 'New "Monitor.mfr2" element=Generator.Motor1 terminal=1 mode=3'
else:
# This uses the built-in model for comparison
Text.Command = 'New "IndMach012.Motor1" bus1=Bg2 kW=1200 conn=delta kVA=1500.000 H=6 purs=0.048 puxs=0.075 purr=0.018 puxr=0.12 puxm=3.8 slip=0.02 SlipOption=variableslip kv=0.48'
Text.Command = 'New "Monitor.mfr2" element=IndMach012.Motor1 terminal=1 mode=3'
# This will run a power-flow solution
Text.Command = 'Solve'
# This will toggle to the dynamics mode
Text.Command = 'Set mode=dynamics number=1 h=0.000166667'
# And finally run 5000 steps for the dynamic simulation
Text.Command = f'Solve number=5000'
# There are the channels from the Pascal/built-in IndMach012
channels_pas = (' Frequency', 'Theta (deg)', 'E1', 'dSpeed (deg/sec)', 'dTheta (deg)', 'Slip', 'Is1', 'Is2', 'Ir1', 'Ir2', 'Stator Losses', 'Rotor Losses', 'Shaft Power (hp)', 'Power Factor', 'Efficiency (%)')
# There are the channels from the Python module -- we define part of these and part come from the generator model itself
channels_py = (' Frequency', 'Theta (Deg)', 'E1_pu', 'dSpeed (Deg/sec)', 'dTheta (Deg)', 'Slip', 'Is1', 'Is2', 'Ir1', 'Ir2', 'StatorLosses', 'RotorLosses', 'ShaftPower_hp', 'PowerFactor', 'Efficiency_pct')
```
## Running and saving the outputs
Let's run the Pascal/built-in version of IndMach012 and our custom Python version for comparison:
```
run(False)
Monitors.Name = 'mfr2'
outputs_pas = {channel: Monitors.Channel(Monitors.Header.index(channel) + 1) for channel in channels_pas}
run(True)
Monitors.Name = 'mfr2'
outputs_py = {channel: Monitors.Channel(Monitors.Header.index(channel) + 1) for channel in channels_py}
time = np.arange(1, 5000 + 1) * 0.000166667
offset = int(0.1 / 0.000166667)
```
## Plotting the various output channels
The example circuit applies a fault at 0.3 s, isolating the machine at 0.4s (check `master.dss` for more details).
As we can see from the figures below, the outputs match very closely. After the induction machine is isolated, the efficiency and power factor values can misbehave as the power goes to zero, seem especially in the Pascal version.
```
for ch_pas, ch_py in zip(channels_pas, channels_py):
plt.figure(figsize=(8,4))
plt.plot(time, outputs_pas[ch_pas], label='IndMach012', lw=3)
plt.plot(time, outputs_py[ch_py], label='PyIndMach012', ls='--', lw=2)
plt.axvline(0.3, linestyle=':', color='k', alpha=0.5, label='Fault occurs')
plt.axvline(0.4, linestyle='--', color='r', alpha=0.5, label='Relays operate')
plt.legend()
plt.xlabel('Time (s)')
plt.ylabel(ch_pas)
if ch_pas == 'Efficiency (%)':
# Limit efficiency to 0-100
plt.ylim(0, 100)
plt.xlim(0, time[-1])
plt.tight_layout()
```
|
github_jupyter
|
# A Two-Level, Six-Factor Full Factorial Design
<br />
<br />
<br />
### Table of Contents
* [Introduction](#intro)
* Factorial Experimental Design:
* [Two-Level Six-Factor Full Factorial Design](#fullfactorial)
* [Variables and Variable Labels](#varlabels)
* [Computing Main and Interaction Effects](#computing_effects)
* Analysis of results:
* [Analyzing Effects](#analyzing_effects)
* [Quantile-Quantile Effects Plot](#quantile_effects)
* [Utilizing Degrees of Freedom](#dof)
* [Ordinary Least Squares Regression Model](#ols)
* [Goodness of Fit](#goodness_of_fit)
* [Distribution of Error](#distribution_of_error)
* [Aggregating Results](#aggregating)
* [Distribution of Variance](#dist_variance)
* [Residual vs. Response Plots](#residual)
<br />
<br />
<br />
<a name="intro"></a>
## Introduction
This notebook roughly follows content from Box and Draper's _Empirical Model-Building and Response Surfaces_ (Wiley, 1984). This content is covered by Chapter 4 of Box and Draper.
In this notebook, we'll carry out an anaylsis of a full factorial design, and show how we can obtain inforomation about a system and its responses, and a quantifiable range of certainty about those values. This is the fundamental idea behind empirical model-building and allows us to construct cheap and simple models to represent complex, nonlinear systems.
```
%matplotlib inline
import pandas as pd
import numpy as np
from numpy.random import rand, seed
import seaborn as sns
import scipy.stats as stats
from matplotlib.pyplot import *
seed(10)
```
<a name="fullfactorial"></a>
## Two-Level Six-Factor Full Factorial Design
Let's start with our six-factor factorial design example. Six factors means there are six input variables; this is still a two-level experiment, so this is now a $2^6$-factorial experiment.
Additionally, there are now three response variables, $(y_1, y_2, y_3)$.
To generate a table of the 64 experiments to be run at each factor level, we will use the ```itertools.product``` function below. This is all put into a DataFrame.
This example generates some random response data, by multiplying a vector of random numbers by the vector of input variable values. (Nothing too complicated.)
```
import itertools
# Create the inputs:
encoded_inputs = list( itertools.product([-1,1],[-1,1],[-1,1],[-1,1],[-1,1],[-1,1]) )
# Create the experiment design table:
doe = pd.DataFrame(encoded_inputs,columns=['x%d'%(i+1) for i in range(6)])
# "Manufacture" observed data y
doe['y1'] = doe.apply( lambda z : sum([ rand()*z["x%d"%(i)]+0.01*(0.5-rand()) for i in range(1,7) ]), axis=1)
doe['y2'] = doe.apply( lambda z : sum([ 5*rand()*z["x%d"%(i)]+0.01*(0.5-rand()) for i in range(1,7) ]), axis=1)
doe['y3'] = doe.apply( lambda z : sum([ 100*rand()*z["x%d"%(i)]+0.01*(0.5-rand()) for i in range(1,7) ]), axis=1)
print(doe[['y1','y2','y3']])
```
<a name="varlablels"></a>
## Defining Variables and Variable Labels
Next we'll define some containers for input variable labels, output variable labels, and any interaction terms that we'll be computing:
```
labels = {}
labels[1] = ['x1','x2','x3','x4','x5','x6']
for i in [2,3,4,5,6]:
labels[i] = list(itertools.combinations(labels[1], i))
obs_list = ['y1','y2','y3']
for k in labels.keys():
print(str(k) + " : " + str(labels[k]))
```
Now that we have variable labels for each main effect and interaction effect, we can actually compute those effects.
<a name="computing_effects"></a>
## Computing Main and Interaction Effects
We'll start by finding the constant effect, which is the mean of each response:
```
effects = {}
# Start with the constant effect: this is $\overline{y}$
effects[0] = {'x0' : [doe['y1'].mean(),doe['y2'].mean(),doe['y3'].mean()]}
print(effects[0])
```
Next, compute the main effect of each variable, which quantifies the amount the response changes by when the input variable is changed from the -1 to +1 level. That is, it computes the average effect of an input variable $x_i$ on each of the three response variables $y_1, y_2, y_3$.
```
effects[1] = {}
for key in labels[1]:
effects_result = []
for obs in obs_list:
effects_df = doe.groupby(key)[obs].mean()
result = sum([ zz*effects_df.ix[zz] for zz in effects_df.index ])
effects_result.append(result)
effects[1][key] = effects_result
effects[1]
```
Our next step is to crank through each variable interaction level: two-variable, three-variable, and on up to six-variable interaction effects. We compute interaction effects for each two-variable combination, three-variable combination, etc.
```
for c in [2,3,4,5,6]:
effects[c] = {}
for key in labels[c]:
effects_result = []
for obs in obs_list:
effects_df = doe.groupby(key)[obs].mean()
result = sum([ np.prod(zz)*effects_df.ix[zz]/(2**(len(zz)-1)) for zz in effects_df.index ])
effects_result.append(result)
effects[c][key] = effects_result
def printd(d):
for k in d.keys():
print("%25s : %s"%(k,d[k]))
for i in range(1,7):
printd(effects[i])
```
We've computed the main and interaction effects for every variable combination (whew!), but now we're at a point where we want to start doing things with these quantities.
<a name="analyzing_effects"></a>
## Analyzing Effects
The first and most important question is, what variable, or combination of variables, has the strongest effect on the three responses $y_1$? $y_2$? $y_3$?
To figure this out, we'll need to use the data we computed above. Python makes it easy to slice and dice data. In this case, we've constructed a nested dictionary, with the outer keys mapping to the number of variables and inner keys mapping to particular combinations of input variables. Its pretty easy to convert this to a flat data structure that we can use to sort by variable effects. We've got six "levels" of variable combinations, so we'll flatten ```effects``` by looping through all six dictionaries of variable combinations (from main effects to six-variable interaction effects), and adding each entry to a master dictionary.
The master dictionary will be a flat dictionary, and once we've populated it, we can use it to make a DataFrame for easier sorting, printing, manipulating, aggregating, and so on.
```
print(len(effects))
master_dict = {}
for nvars in effects.keys():
effect = effects[nvars]
for k in effect.keys():
v = effect[k]
master_dict[k] = v
master_df = pd.DataFrame(master_dict).T
master_df.columns = obs_list
y1 = master_df['y1'].copy()
y1.sort_values(inplace=True,ascending=False)
print("Top 10 effects for observable y1:")
print(y1[:10])
y2 = master_df['y2'].copy()
y2.sort_values(inplace=True,ascending=False)
print("Top 10 effects for observable y2:")
print(y2[:10])
y3 = master_df['y3'].copy()
y3.sort_values(inplace=True,ascending=False)
print("Top 10 effects for observable y3:")
print(y3[:10])
```
If we were only to look at the list of rankings of each variable, we would see that each response is affected by different input variables, listed below in order of descending importance:
* $y_1$: 136254
* $y_2$: 561234
* $y_3$: 453216
This is a somewhat mixed message that's hard to interpret - can we get rid of variable 2? We can't eliminate 1, 4, or 5, and probably not 3 or 6 either.
However, looking at the quantile-quantile plot of the effects answers the question in a more visual way.
<a name="quantile_effects"></a>
## Quantile-Quantile Effects Plot
We can examine the distribution of the various input variable effects using a quantile-quantile plot of the effects. Quantile-quantile plots arrange the effects in order from least to greatest, and can be applied in several contexts (as we'll see below, when assessing model fits). If the quantities plotted on a quantile-qantile plot are normally distributed, they will fall on a straight line; data that do not fall on the straight line indicate significant deviations from normal behavior.
In the case of a quantile-quantile plot of effects, non-normal behavior means the effect is paticularly strong. By identifying the outlier points on thse quantile-quantile plots (they're ranked in order, so they correspond to the lists printed above), we can identify the input variables most likely to have a strong impact on the responses.
We need to look both at the top (the variables that have the largest overall positive effect) and the bottom (the variables that have the largest overall negative effect) for significant outliers. When we find outliers, we can add them to a list of variabls that we have decided are important and will keep in our analysis.
```
# Quantify which effects are not normally distributed,
# to assist in identifying important variables
fig = figure(figsize=(14,4))
ax1 = fig.add_subplot(131)
ax2 = fig.add_subplot(132)
ax3 = fig.add_subplot(133)
stats.probplot(y1, dist="norm", plot=ax1)
ax1.set_title('y1')
stats.probplot(y2, dist="norm", plot=ax2)
ax2.set_title('y2')
stats.probplot(y3, dist="norm", plot=ax3)
ax3.set_title('y3')
```
Normally, we would use the main effects that were computed, and their rankings, to eliminate any variables that don't have a strong effect on any of our variables. However, this analysis shows that sometimes we can't eliminate any variables.
All six input variables are depicted as the effects that fall far from the red line - indicating all have a statistically meaningful (i.e., not normally distributed) effect on all three response variables. This means we should keep all six factors in our analysis.
There is also a point on the $y_3$ graph that appears significant on the bottom. Examining the output of the lists above, this point represents the effect for the six-way interaction of all input variables. High-order interactions are highly unlikely (and in this case it is a numerical artifact of the way the responses were generated), so we'll keep things simple and stick to a linear model.
Let's continue our analysis without eliminating any of the six factors, since they are important to all of our responses.
<a name="dof"></a>
## Utilizing Degrees of Freedom
Our very expensive, 64-experiment full factorial design (the data for which maps $(x_1,x_2,\dots,x_6)$ to $(y_1,y_2,y_3)$) gives us 64 data points, and 64 degrees of freedom. What we do with those 64 degrees of freedom is up to us.
We _could_ fit an empirical model, or response surface, that has 64 independent parameters, and account for many of the high-order interaction terms - all the way up to six-variable interaction effects. However, high-order effects are rarely important, and are a waste of our degrees of freedom.
Alternatively, we can fit an empirical model with fewer coefficients, using up fewer degrees of freedom, and use the remaining degrees of freedom to characterize the error introduced by our approximate model.
To describe a model with the 6 variables listed above and no other variable interaction effects would use only 6 degrees of freedom, plus 1 degree of freedom for the constant term, leaving 57 degrees of freedom available to quantify error, attribute variance, etc.
Our goal is to use least squares to compute model equations for $(y_1,y_2,y_3)$ as functions of $(x_1,x_2,x_3,x_4,x_5,x_6)$.
```
xlabs = ['x1','x2','x3','x4','x5','x6']
ylabs = ['y1','y2','y3']
ls_data = doe[xlabs+ylabs]
import statsmodels.api as sm
import numpy as np
x = ls_data[xlabs]
x = sm.add_constant(x)
```
The first ordinary least squares linear model is created to predict values of the first variable, $y_1$, as a function of each of our input variables, the list of which are contained in the ```xlabs``` variable. When we perform the linear regression fitting, we see much of the same information that we found in the prior two-level three-factor full factorial design, but here, everything is done automatically.
The model is linear, meaning it's fitting the coefficients of the function:
$$
\hat{y} = a_0 + a_1 x_1 + a_2 x_2 + a_3 + x_3 + a_4 x_4 + a_5 x_5 + a_6 x_6
$$
(here, the variables $y$ and $x$ are vectors, with one component for each response; in our case, they are three-dimensional vectors.)
Because there are 64 observations and 7 coefficients, the 57 extra observations give us extra degrees of freedom with which to assess how good the model is. That analysis can be done with an ordinary least squares (OLS) model, available through the statsmodel library in Python.
<a name="ols"></a>
## Ordinary Least Squares Regression Model
This built-in OLS model will fit an input vector $(x_1,x_2,x_3,x_4,x_5,x_6)$ to an output vector $(y_1,y_2,y_3)$ using a linear model; the OLS model is designed to fit the model with more observations than coefficients, and utilize the remaining data to quantify the fit of the model.
Let's run through one of these, and analyze the results:
```
y1 = ls_data['y1']
est1 = sm.OLS(y1,x).fit()
print(est1.summary())
```
The StatsModel OLS object prints out quite a bit of useful information, in a nicely-formatted table. Starting at the top, we see a couple of important pieces of information: specifically, the name of the dependent variable (the response) that we're looking at, the number of observations, and the number of degrees of freedom.
We can see an $R^2$ statistic, which indicates how well this data is fit with our linear model, and an adjusted $R^2$ statistic, which accounts for the large nubmer of degrees of freedom. While an adjusted $R^2$ of 0.73 is not great, we have to remember that this linear model is trying to capture a wealth of complexity in six coefficients. Furthermore, the adjusted $R^2$ value is too broad to sum up how good our model actually is.
The table in the middle is where the most useful information is located. The `coef` column shows the coefficients $a_0, a_1, a_2, \dots$ for the model equation:
$$
\hat{y} = a_0 + a_1 x_1 + a_2 x_2 + a_3 + x_3 + a_4 x_4 + a_5 x_5 + a_6 x_6
$$
Using the extra degrees of freedom, an estime $s^2$ of the variance in the regression coefficients is also computed, and reported in the the `std err` column. Each linear term is attributed the same amount of variance, $\pm 0.082$.
```
y2 = ls_data['y2']
est2 = sm.OLS(y2,x).fit()
print(est2.summary())
y3 = ls_data['y3']
est3 = sm.OLS(y3,x).fit()
print(est3.summary())
```
<a name="goodness_of_fit"></a>
## Quantifying Model Goodness-of-Fit
We can now use these linear models to evaluate each set of inputs and compare the model response $\hat{y}$ to the actual observed response $y$. What we would expect to see, if our model does an adequate job of representing the underlying behavior of the model, is that in each of the 64 experiments, the difference between the model prediction $M$ and the measured data $d$, defined as the residual $r$,
$$
r = \left| d - M \right|
$$
should be comparable across all experiments. If the residuals appear to have functional dependence on the input variables, it is an indication that our model is missing important effects and needs more or different terms. The way we determine this, mathematically, is by looking at a quantile-quantile plot of our errors (that is, a ranked plot of our error magnitudes).
If the residuals are normally distributed, they will follow a straight line; if the plot shows the data have significant wiggle and do not follow a line, it is an indication that the errors are not normally distributed, and are therefore skewed (indicating terms missing from our OLS model).
```
%matplotlib inline
import seaborn as sns
import scipy.stats as stats
from matplotlib.pyplot import *
# Quantify goodness of fit
fig = figure(figsize=(14,4))
ax1 = fig.add_subplot(131)
ax2 = fig.add_subplot(132)
ax3 = fig.add_subplot(133)
r1 = y1 - est1.predict(x)
r2 = y2 - est2.predict(x)
r3 = y3 - est3.predict(x)
stats.probplot(r1, dist="norm", plot=ax1)
ax1.set_title('Residuals, y1')
stats.probplot(r2, dist="norm", plot=ax2)
ax2.set_title('Residuals, y2')
stats.probplot(r3, dist="norm", plot=ax3)
ax3.set_title('Residuals, y3')
```
Determining whether significant trends are being missed by the model depends on how many points deviate from the red line, and how significantly. If there is a single point that deviates, it does not necessarily indicate a problem; but if there is significant wiggle and most points deviate significantly from the red line, it means that there is something about the relationship between the inputs and the outputs that our model is missing.
There are only a few points deviating from the red line. We saw from the effect quantile for $y_3$ that there was an interaction variable that was important to modeling the response $y_3$, and it is likely this interaction that is leading to noise at the tail end of these residuals. This indicates residual errors (deviations of the model from data) that do not follow a natural, normal distribution, which indicates there is a _pattern_ in the deviations - namely, the interaction effect.
The conclusion about the error from the quantile plots above is that there are only a few points deviation from the line, and no particularly significant outliers. Our model can use some improvement, but it's a pretty good first-pass model.
<a name="distribution_of_error"></a>
## Distribution of Error
Another thing we can look at is the normalized error: what are the residual errors (differences between our model prediction and our data)? How are their values distributed?
A kernel density estimate (KDE) plot, which is a smoothed histogram, shows the probability distribution of the normalized residual errors. As expected, they're bunched pretty close to zero. There are some bumps far from zero, corresponding to the outliers on the quantile-quantile plot of the errors above. However, they're pretty close to randomly distributed, and therefore it doesn't look like there is any systemic bias there.
```
fig = figure(figsize=(10,12))
ax1 = fig.add_subplot(311)
ax2 = fig.add_subplot(312)
ax3 = fig.add_subplot(313)
axes = [ax1,ax2,ax3]
colors = sns.xkcd_palette(["windows blue", "amber", "faded green", "dusty purple","aqua blue"])
#resids = [r1, r2, r3]
normed_resids = [r1/y1, r2/y2, r3/y3]
for (dataa, axx, colorr) in zip(normed_resids,axes,colors):
sns.kdeplot(dataa, bw=1.0, ax=axx, color=colorr, shade=True, alpha=0.5);
ax1.set_title('Probability Distribution: Normalized Residual Error, y1')
ax2.set_title('Normalized Residual Error, y2')
ax3.set_title('Normalized Residual Error, y3')
```
Note that in these figures, the bumps at extreme value are caused by the fact that the interval containing the responses includes 0 and values close to 0, so the normalization factor is very tiny, leading to large values.
<a name="aggregating"></a>
## Aggregating Results
Let's next aggregate experimental results, by taking the mean over various variables to compute the mean effect for regressed varables. For example, we may want to look at the effects of variables 2, 3, and 4, and take the mean over the other three variables.
This is simple to do with Pandas, by grouping the data by each variable, and applying the mean function on all of the results. The code looks like this:
```
# Our original regression variables
xlabs = ['x2','x3','x4']
doe.groupby(xlabs)[ylabs].mean()
# If we decided to go for a different variable set
xlabs = ['x2','x3','x4','x6']
doe.groupby(xlabs)[ylabs].mean()
```
This functionality can also be used to determine the variance in all of the experimental observations being aggregated. For example, here we aggregate over $x_3 \dots x_6$ and show the variance broken down by $x_1, x_2$ vs $y_1, y_2, y_3$.
```
xlabs = ['x1','x2']
doe.groupby(xlabs)[ylabs].var()
```
Or even the number of experimental observations being aggregated!
```
doe.groupby(xlabs)[ylabs].count()
```
<a name="dist_variance"></a>
## Distributions of Variance
We can convert these dataframes of averages, variances, and counts into data for plotting. For example, if we want to make a histogram of every value in the groupby dataframe, we can use the ```.values``` method, so that this:
doe.gorupby(xlabs)[ylabs].mean()
becomes this:
doe.groupby(xlabs)[ylabs].mean().values
This $M \times N$ array can then be flattened into a vector using the ```ravel()``` method from numpy:
np.ravel( doe.groupby(xlabs)[ylabs].mean().values )
The resulting data can be used to generate histograms, as shown below:
```
# Histogram of means of response values, grouped by xlabs
xlabs = ['x1','x2','x3','x4']
print("Grouping responses by %s"%( "-".join(xlabs) ))
dat = np.ravel(doe.groupby(xlabs)[ylabs].mean().values) / np.ravel(doe.groupby(xlabs)[ylabs].var().values)
hist(dat, 10, normed=False, color=colors[3]);
xlabel(r'Relative Variance ($\mu$/$\sigma^2$)')
show()
# Histogram of variances of response values, grouped by xlabs
print("Grouping responses by %s"%( "-".join(xlabs) ))
dat = np.ravel(doe.groupby(xlabs)['y1'].var().values)
hist(dat, normed=True, color=colors[4])
xlabel(r'Variance in $y_{1}$ Response')
ylabel(r'Frequency')
show()
```
The distribution of variance looks _mostly_ normal, with some outliers. These are the same outliers that showed up in our quantile-quantile plot, and they'll show up in the plots below as well.
<a name="residual"></a>
## Residual vs. Response Plots
Another thing we can do, to look for uncaptured effects, is to look at our residuals vs. $\hat{y}$. This is a further effort to look for underlying functional relationships between $\hat{y}$ and the residuals, which would indicate that our system exhibits behavior not captured by our linear model.
```
# normal plot of residuals
fig = figure(figsize=(14,4))
ax1 = fig.add_subplot(131)
ax2 = fig.add_subplot(132)
ax3 = fig.add_subplot(133)
ax1.plot(y1,r1,'o',color=colors[0])
ax1.set_xlabel('Response value $y_1$')
ax1.set_ylabel('Residual $r_1$')
ax2.plot(y2,r2,'o',color=colors[1])
ax2.set_xlabel('Response value $y_2$')
ax2.set_ylabel('Residual $r_2$')
ax2.set_title('Response vs. Residual Plots')
ax3.plot(y1,r1,'o',color=colors[2])
ax3.set_xlabel('Response value $y_3$')
ax3.set_ylabel('Residual $r_3$')
show()
```
Notice that each plot is trending up and to the right - indicative of an underlying trend that our model $\hat{y}$ is not capturing. The trend is relatively weak, however, indicating that our linear model does a good job of capturing _most_ of the relevant effects of this system.
# Discussion
The analysis shows that there are some higher-order or nonlinear effects in the system that a purely linear model does not account for. Next steps would involve adding higher order points for a quadratic or higher order polynomial model to gather additional data to fit the higher-degree models.
|
github_jupyter
|
<a href="https://colab.research.google.com/github/NikolaZubic/AppliedGameTheoryHomeworkSolutions/blob/main/domaci3.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# TREĆI DOMAĆI ZADATAK iz predmeta "Primenjena teorija igara" (Applied Game Theory)
Razvoj bota za igranje igre Ajnc (BlackJack) koristeći "Q-learning" pristup.
# Potrebni import-i
```
import gym
import numpy as np
import matplotlib.pyplot as plt
from gym import spaces
import seaborn as sns
```
# Definisanje Ajnc okruženja koristeći "Open AI Gym" toolkit
```
class BlackJackEnvironment(gym.Env):
# Because of human-friendly output
metadata = {'render.modes':['human']}
def __init__(self):
"""
We will define possible number of states with observation_space.
Player's sum can go from 4 to 32: Now when the sum is 22, and the player chooses to hit, he may get a card with value 10, resulting in a sum of 32, and thus loosing the game.
Dealer's card can be from 1 to 10 and we have 2 actions.
Total number of states: 29 * 10 * 2 = 580
Total number of actions = 2 = len( {"HIT", "STAND"} )
"""
self.observation_space = spaces.Discrete(580)
self.action_space = spaces.Discrete(2)
self.step_count = 0 # at the beginning of the game we have 0 actions taken
def check_usable_ace(self,hand):
"""
If someone has an usable ace, we will replace that ace (1) with 11.
:param hand: player's or dealer's card
:return: True if we have usable ace, False otherwise
"""
temp_hand = hand.copy()
# Check if there is ace in hand
if np.any(temp_hand == 1):
# If we have any ace then replace it with 11, but if we have more than one ace replace the first one with 11
temp_hand[np.where(temp_hand == 1)[0][0]] = 11
# If the sum is less or equal than 21 then we can use it
if temp_hand.sum() <= 21:
return True
return False
def use_ace(self,hand):
"""
If there is usable ace in function above, then replace 1 with 11.
:param hand: player's or dealer's hand
:return: new hand where 1 is replaced with 11
"""
temp_hand = hand.copy()
temp_hand[np.where(temp_hand == 1)[0][0]] = 11
return temp_hand
def reset(self):
# Resets the environment after one game.
# Initialize player's hand
self.current_hand = np.random.choice(range(1,11),2)
# Initialize usable Ace to False, since we don't have it at the very beginning
self.usable_ace = False
self.dealer_stand, self.player_stand = False, False
# Replace usable ace in the player's hand
if self.check_usable_ace(self.current_hand):
self.usable_ace = True
self.current_hand = self.use_ace(self.current_hand)
# Player's current sum
self.current_sum = self.current_hand.sum()
# Dealer's hand
self.dealer_hand = np.random.choice(range(1,11),2)
# Dealer's sum
self.dealer_sum = self.dealer_hand.sum()
# First element of self.dealer_hand is the current showing card of dealer
self.dealer_showing_card = self.dealer_hand[0]
# Replace usable ace in the dealer's hand
if self.check_usable_ace(self.dealer_hand):
temp_dealer_hand = self.use_ace(self.dealer_hand)
self.dealer_sum = temp_dealer_hand.sum()
def take_turn(self, current_player):
"""
Play one turn for the player. This function will be called from step() function, directly depending on the game state.
We will take new random card, add it to the current_player hand.
:param player: {"player", "dealer"}
:return: None
"""
if current_player == 'dealer':
# Take new random card
new_card = np.random.choice(range(1,11))
# Add new card to the current_player hand
new_dealer_hand = np.array(self.dealer_hand.tolist() + [new_card])
# Check for usable ace and replace if found
if self.check_usable_ace(new_dealer_hand):
new_dealer_hand = self.use_ace(new_dealer_hand)
self.dealer_hand = new_dealer_hand
# Update his sum
self.dealer_sum = self.dealer_hand.sum()
if current_player == 'player':
new_card = np.random.choice(range(1,11))
new_player_hand = np.array(self.current_hand.tolist()+ [new_card])
if self.check_usable_ace(new_player_hand):
self.usable_ace = True
new_player_hand = self.use_ace(new_player_hand)
self.current_hand = new_player_hand
self.current_sum = self.current_hand.sum()
def check_game_status(self, mode = 'normal'):
"""
Check the current status of the game.
During the 'normal' we check after each turn whether we got in the terminal state.
In the 'compare' mode we compare the totals of both players (player vs dealer) in order to pronounce the winner.
:param mode: {'normal', 'compare'}
:return: dictionary with the winner, whether the game is finished and the reward of the game
"""
result = {'winner':'',
'is_done': False,
'reward':0}
if mode == 'normal':
if self.current_sum > 21:
result['winner'] = 'dealer'
result['is_done'] = True
result['reward'] = -1
elif self.dealer_sum > 21:
result['winner'] = 'player'
result['is_done'] = True
result['reward'] = 1
elif self.current_sum == 21:
result['winner'] = 'player'
result['is_done'] = True
result['reward'] = 1
elif self.dealer_sum == 21:
result['winner'] = 'dealer'
result['is_done'] = True
result['reward'] = -1
elif mode == 'compare':
result['is_done'] = True
diff_21_player = 21 - self.current_sum
diff_21_dealer = 21 - self.dealer_sum
if diff_21_player > diff_21_dealer:
result['reward'] = -1
result['winner'] = 'dealer'
elif diff_21_player < diff_21_dealer:
result['reward'] = 1
result['winner'] = 'player'
else:
result['reward'] = 0
result['winner'] = 'draw'
return result
return result
def step(self,action):
"""
Performs one action.
:param action:
:return: dictionary with the winner, whether the game is finished and the reward of the game
"""
# Increase number of actions that are taken during the game.
self.step_count += 1
result = {'winner':'',
'is_done': False,
'reward':0}
"""
Before taking the first step of the game, we need to ensure that there is no winning condition.
Check if the initial two cards of the players are 21. If anyone has 21, then that player wins.
If both players have 21, then the game is DRAW. Otherwise, we will continue with the game.
"""
if self.step_count == 1:
if self.check_usable_ace(self.current_hand):
self.current_hand = self.use_ace(self.current_hand)
if self.check_usable_ace(self.dealer_hand):
self.current_hand = self.use_ace(self.dealer_hand)
if self.current_sum == 21 and self.dealer_sum == 21:
result['is_done'] = True
result['reward'] = 0
result['winner'] = 'draw'
return result
elif self.current_sum == 21 and self.dealer_sum < 21:
result['is_done'] = True
result['reward'] = 1
result['winner'] = 'player'
return result
elif self.dealer_sum == 21 and self.current_sum < 21:
result['is_done'] = True
result['reward'] = -1
result['winner'] = 'dealer'
return result
if self.dealer_sum >= 17:
self.dealer_stand = True
# action = 0 means "HIT"
if action == 0:
self.take_turn('player')
result = self.check_game_status()
if result['is_done'] == True:
return result
# action = 1 means "STAND"
if action == 1:
if self.dealer_stand == True:
return self.check_game_status(mode = 'compare')
"""
If the dealer hasn't stand, he will hit unless his sum is greater than or equal to 17.
After that, he will stand.
"""
while self.dealer_sum < 17:
self.take_turn('dealer')
result = self.check_game_status()
# After dealer stands, check the game status.
if result['is_done'] == True:
return result
# If the game hasn't finished yet, we set dealer_stand to True, so the player will either HIT or STAND
self.dealer_stand = True
return result
def get_current_state(self):
"""
Get current state which is comprised of current player's sum, dealer's showing card and usable ace presence.
:return: return current state variables
"""
current_state = {}
current_state['current_sum'] = self.current_sum
current_state['dealer_showing_card'] = self.dealer_showing_card
current_state['usable_ace'] = self.usable_ace
return current_state
def render(self):
print("OBSERVABLE STATES")
print("Current player's sum: {}".format(self.current_sum))
print("Dealer's showing card: {}".format(self.dealer_showing_card))
print("Player has usable Ace: {}".format(self.usable_ace))
print("INFORMATION ABOUT CARDS AND DEALER'S SUM")
print("Player's hand: {}".format(self.current_hand))
print("Dealer's hand: {}".format(self.dealer_hand))
print("Dealer's sum: {}".format(self.dealer_sum))
```
# Pomoćne funkcije za Q-learning
```
# dictionaries used for converting the state values to indexes in the Q table
current_sum_to_index = dict(zip(np.arange(4,33),np.arange(29)))
dealer_showing_card_to_index = dict(zip(np.arange(1,11),np.arange(10)))
usable_ace_index = dict(zip([False,True],[0,1]))
action_index = dict(zip(['HIT','STAND'],[0,1]))
def get_state_q_indices(current_state):
"""
Get indexes of Q table for any given state.
:param current_state: comprised of current player's sum, dealer's showing card and usable ace presence.
:return: get table indexes for a state
"""
current_sum_idx = current_sum_to_index[current_state['current_sum']]
dealer_showing_card_idx = dealer_showing_card_to_index[current_state['dealer_showing_card']]
usable_ace_idx = usable_ace_index[current_state['usable_ace']]
return [current_sum_idx,dealer_showing_card_idx,usable_ace_idx]
def get_max_action(Q_sa, current_state):
"""
Get the action with the max Q-value for the given current state and the Q table.
:param Q_sa: given Q table
:param current_state: current state
:return: best action for given state and Q table
"""
state_q_idxs = get_state_q_indices(current_state)
action = Q_sa[state_q_idxs[0],state_q_idxs[1],state_q_idxs[2],:].argmax()
return action
def get_q_value(Q_sa, state, action):
"""
Get Q(s,a) value for state and action in certain Q table.
:param Q_sa: given Q table
:param state: given state
:param action: given action
:return: Q(s, a)
"""
state_q_idxs = get_state_q_indices(state)
q_value = Q_sa[state_q_idxs[0],state_q_idxs[1],state_q_idxs[2],action]
return q_value
```
# Q-learning
Inicijalizacija Q tabele.
```
"""
Player's current sum is ranging from 4 to 32 => 32 - 4 + 1 = 29
Dealer's showing card can be one from the following set {1, 2, 3, 4, 5, 6, 7, 8, 9, 10} => 10 values
Ace can be usable or not => 2
Actions are from the following set {"HIT", "STAND"} => 2
"""
Q = np.zeros((29,10,2,2))
```
Proces treniranja.
```
episode_count = 0
total_episodes = 2000000
# Discounting factor
gamma = 0.9
# Used for filtering q-values, learning rate
LAMBDA = 0.1
# Defined Black Jack Environment
environment = BlackJackEnvironment()
while episode_count < total_episodes:
environment.reset()
current_state = environment.get_current_state()
current_action = get_max_action(Q, current_state)
# Take action
step_result = environment.step(current_action)
# Get into next state and get the reward
next_state = environment.get_current_state()
next_max_action = get_max_action(Q, next_state)
immediate_reward = step_result['reward']
next_state_q_idxs = get_state_q_indices(next_state)
# Get the q-value for the next state and max action in the next state
q_max_s_a = get_q_value(Q, next_state, next_max_action)
td_target = immediate_reward + gamma * q_max_s_a
# Get the q-value for the current state and action
q_current_s_a = get_q_value(Q, current_state, current_action)
td_error = td_target - q_current_s_a
state_q_idxs = get_state_q_indices(current_state)
# Update the current Q(s, a)
Q[state_q_idxs[0],state_q_idxs[1],state_q_idxs[2],current_action] = q_current_s_a + LAMBDA * td_error
# get into the next state
current_state = next_state
if step_result['is_done']:
episode_count += 1
if episode_count % 100000 == 0:
print("Episode number: {}".format(episode_count))
```
# Diskusija rezultata
```
fig, ax = plt.subplots(ncols= 2,figsize=(16,8))
sns.heatmap(Q[:,:,0,0],cmap = sns.light_palette((210, 90, 60), input="husl"), ax = ax[0],
xticklabels=np.arange(1,11),yticklabels=np.arange(4,33))
ax[0].set_title("Usable Ace = False, Action = HIT")
ax[0].set_xlabel("Dealer's Showing Card")
ax[0].set_ylabel("Current Player's Sum")
sns.heatmap(Q[:,:,0,1],cmap = sns.light_palette((210, 90, 60), input="husl"), ax = ax[1],
xticklabels=np.arange(1,11),yticklabels=np.arange(4,33))
ax[1].set_title("Usable Ace = False, Action = STAND")
ax[1].set_xlabel("Dealer's Showing Card")
ax[1].set_ylabel("Current Player's Sum")
```
Na osnovu gornjih heatmapa možemo uočiti koje je to akcije dobro izvršiti u kojem stanju.
**Zaključak sa lijeve heatmape**: kada je ukupna suma igrača manja od 12, 13 onda je najbolje da se izvršava akcija "HIT".
**Zaključak sa desne heatmape**: Za veće vrijednosti otkrivene karte djelitelja i veće vrijednosti ukupne sume igrača bolje je izvršiti akciju "STAND".
```
fig, ax = plt.subplots(ncols = 2, figsize=(16,8))
sns.heatmap(Q[:,:,1,0],cmap = sns.light_palette((210, 90, 60), input="husl"), ax = ax[0],
xticklabels=np.arange(1,11),yticklabels=np.arange(4,33))
ax[0].set_title("Usable Ace = True, Action = HIT")
ax[0].set_xlabel("Dealer's Showing Card")
ax[0].set_ylabel("Current Player's Sum")
sns.heatmap(Q[:,:,1,1],cmap = sns.light_palette((210, 90, 60), input="husl"), ax = ax[1],
xticklabels=np.arange(1,11),yticklabels=np.arange(4,33))
ax[1].set_title("Usable Ace = True, Action = STAND")
ax[1].set_xlabel("Dealer's Showing Card")
ax[1].set_ylabel("Current Player's Sum")
```
U slučaju kad imamo iskoristiv kec, broj semplova je znatno manji, tako da paterni Q-vrijednosti nisu baš potpuno jasni, ali može se zaključiti da je najbolje izvršiti akciju **"HIT" u slučajevima kad je suma igrača oko 12**, dok se akcija **"STAND" izvršava u slučaju kada je igra pri kraju po pitanju sume igrača**.
Sada ćemo pogledati naučene politike (za slučaj pohlepne politike, jer želimo da naš igrač uvijek bira onako da najbolje igra).
**Sa crnim blokovima označeno je kada treba izvršiti akciju "HIT"**, a imamo 2 heatmape za slučaj kad nemamo i imamo iskoristiv kec.
```
fig, ax = plt.subplots(ncols= 1,figsize=(8,6))
sns.heatmap(np.argmax(Q[:17,:,0,:],axis=2),cmap = sns.cubehelix_palette(8, start=2, rot=0, dark=0, light=.95, reverse=True)\
,linewidths=1,xticklabels=np.arange(1,11),yticklabels=np.arange(4,33))
ax.set_title("Usable Ace = False")
ax.set_xlabel("Dealer's Showing Card")
ax.set_ylabel("Current Player's Sum")
fig, ax = plt.subplots(ncols= 1,figsize=(8,6))
sns.heatmap(np.argmax(Q[:17,:,1,:],axis=2),cmap = sns.cubehelix_palette(8, start=2, rot=0, dark=0, light=.95, reverse=True)\
,linewidths=1,xticklabels=np.arange(1,11),yticklabels=np.arange(4,33))
ax.set_title("Usable Ace = True")
ax.set_xlabel("Dealer's Showing Card")
ax.set_ylabel("Current Player's Sum")
```
# Na kraju, nakon 2 miliona iteracija treniranja, testiraćemo algoritam na 10 000 partija.
```
player_wins = 0
dealer_wins = 0
NUMBER_OF_GAMES = 10000
for i in range(NUMBER_OF_GAMES):
environment.reset()
while True:
current_state = environment.get_current_state()
current_action = get_max_action(Q, current_state)
# Take action
step_result = environment.step(current_action)
#environment.render()
next_state = environment.get_current_state()
current_state = next_state
if step_result['is_done']:
break
if step_result['winner'] == 'player':
player_wins += 1
elif step_result['winner'] == 'dealer':
dealer_wins += 1
print("Player wins: " + str(player_wins))
print("Dealer wins: " + str(dealer_wins))
print("Player wins percentage = " + str(round(100 * (player_wins / (player_wins + dealer_wins)), 2)) + "%")
```
|
github_jupyter
|
```
# Copyright 2021 NVIDIA Corporation. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# Accelerate BERT encoder with TF-TRT
## Introduction
The NVIDIA TensorRT is a C++ library that facilitates high performance inference on NVIDIA graphics processing units (GPUs). TensorFlow™ integration with TensorRT™ (TF-TRT) optimizes TensorRT compatible parts of your computation graph, allowing TensorFlow to execute the remaining graph. While you can use TensorFlow's wide and flexible feature set, TensorRT will produce a highly optimized runtime engine for the TensorRT compatible subgraphs of your network.
In this notebook, we demonstrate accelerating BERT inference using TF-TRT. We focus on the encoder.
## Requirements
This notebook requires at least TF 2.5 and TRT 7.1.3.
## 1. Download the model
We will download a bert base model from [TF-Hub](https://tfhub.dev/tensorflow/bert_en_uncased_L-12_H-768_A-12/3).
```
!pip install -q tf-models-official
import tensorflow as tf
import tensorflow_hub as hub
tfhub_handle_encoder = 'https://tfhub.dev/tensorflow/bert_en_uncased_L-12_H-768_A-12/3'
bert_saved_model_path = 'bert_base'
bert_model = hub.load(tfhub_handle_encoder)
tf.saved_model.save(bert_model, bert_saved_model_path)
```
## 2. Inference
In this section we will convert the model using TF-TRT and run inference.
```
import matplotlib.pyplot as plt
import numpy as np
from tensorflow.python.saved_model import signature_constants
from tensorflow.python.saved_model import tag_constants
from tensorflow.python.compiler.tensorrt import trt_convert as trt
from timeit import default_timer as timer
tf.get_logger().setLevel('ERROR')
```
### 2.1 Helper functions
```
def get_func_from_saved_model(saved_model_dir):
saved_model_loaded = tf.saved_model.load(
saved_model_dir, tags=[tag_constants.SERVING])
graph_func = saved_model_loaded.signatures[
signature_constants.DEFAULT_SERVING_SIGNATURE_DEF_KEY]
return graph_func, saved_model_loaded
def predict_and_benchmark_throughput(input_dict, model, N_warmup_run=50, N_run=500,
result_key='predictions', batch_size=None):
elapsed_time = []
for val in input_dict.values():
input_batch_size = val.shape[0]
break
if batch_size is None or batch_size > input_batch_size:
batch_size = input_batch_size
print('Benchmarking with batch size', batch_size)
elapsed_time = np.zeros(N_run)
for i in range(N_warmup_run):
preds = model(**input_dict)
# Force device synchronization with .numpy()
tmp = preds[result_key][0].numpy()
for i in range(N_run):
start_time = timer()
preds = model(**input_dict)
# Synchronize
tmp += preds[result_key][0].numpy()
end_time = timer()
elapsed_time[i] = end_time - start_time
if i>=50 and i % 50 == 0:
print('Steps {}-{} average: {:4.1f}ms'.format(i-50, i, (elapsed_time[i-50:i].mean()) * 1000))
latency = elapsed_time.mean() * 1000
print('Latency: {:5.2f}+/-{:4.2f}ms'.format(latency, elapsed_time.std() * 1000))
print('Throughput: {:.0f} samples/s'.format(N_run * batch_size / elapsed_time.sum()))
return latency
def trt_convert(input_path, output_path, input_shapes, explicit_batch=False,
dtype=np.float32, precision='FP32', prof_strategy='Optimal'):
conv_params=trt.TrtConversionParams(
precision_mode=precision, minimum_segment_size=50,
max_workspace_size_bytes=12*1<<30, maximum_cached_engines=1)
converter = trt.TrtGraphConverterV2(
input_saved_model_dir=input_path, conversion_params=conv_params,
use_dynamic_shape=explicit_batch,
dynamic_shape_profile_strategy=prof_strategy)
converter.convert()
def input_fn():
for shapes in input_shapes:
# return a list of input tensors
yield [np.ones(shape=x).astype(dtype) for x in shapes]
converter.build(input_fn)
converter.save(output_path)
def random_input(batch_size, seq_length):
# Generate random input data
mask = tf.convert_to_tensor(np.ones((batch_size, seq_length), dtype=np.int32))
type_id = tf.convert_to_tensor(np.zeros((batch_size, seq_length), dtype=np.int32))
word_id = tf.convert_to_tensor(np.random.randint(0, 1000, size=[batch_size, seq_length], dtype=np.int32))
return {'input_mask':mask, 'input_type_ids': type_id, 'input_word_ids':word_id}
```
### 2.2 Convert the model with TF-TRT
```
bert_trt_path = bert_saved_model_path + '_trt'
input_shapes = [[(1, 128), (1, 128), (1, 128)]]
trt_convert(bert_saved_model_path, bert_trt_path, input_shapes, True, np.int32, precision='FP16')
```
### 2.3 Run inference with converted model
```
trt_func, _ = get_func_from_saved_model(bert_trt_path)
input_dict = random_input(1, 128)
result_key = 'bert_encoder_1' # 'classifier'
res = predict_and_benchmark_throughput(input_dict, trt_func, result_key=result_key)
```
### Compare to the original function
```
func, model = get_func_from_saved_model(bert_saved_model_path)
res = predict_and_benchmark_throughput(input_dict, func, result_key=result_key)
```
## 3. Dynamic sequence length
The sequence length for the encoder is dynamic, we can use different input sequence lengths. Here we call the original model for two sequences.
```
seq1 = random_input(1, 128)
res1 = func(**seq1)
seq2 = random_input(1, 180)
res2 = func(**seq2)
```
The converted model is optimized for a sequnce length of 128 (and batch size 8). If we infer the converted model using a different sequence length, then two things can happen:
1. If `TrtConversionParams.allow_build_at_runtime` == False: native TF model is inferred
2. if `TrtConversionParams.allow_build_at_runtime` == True a new TRT engine is created which is optimized for the new sequence length.
The first option do not provide TRT accelaration while the second one creates a large overhead while the new engine is constructed. In the next section we convert the model to handle multiple sequence lengths.
### 3.1 TRT Conversion with dynamic sequence length
```
bert_trt_path = bert_saved_model_path + '_trt2'
input_shapes = [[(1, 128), (1, 128), (1, 128)], [(1, 180), (1, 180), (1, 180)]]
trt_convert(bert_saved_model_path, bert_trt_path, input_shapes, True, np.int32, precision='FP16',
prof_strategy='Range')
trt_func_dynamic, _ = get_func_from_saved_model(bert_trt_path)
trt_res = trt_func_dynamic(**seq1)
result_key = 'bert_encoder_1' # 'classifier'
res = predict_and_benchmark_throughput(seq1, trt_func_dynamic, result_key=result_key)
res = predict_and_benchmark_throughput(seq2, trt_func_dynamic, result_key=result_key)
```
|
github_jupyter
|
```
import sys
sys.path.append('..') # for import src
import os
import cloudpickle
import lzma
import pandas as pd
import numpy as np
from sklearn.linear_model import Ridge
from sklearn.model_selection import cross_val_predict
from scipy.stats import pearsonr
import matplotlib.pyplot as plt
import lightgbm as lgb
import talib
import src
from src.ml_utils import (
fetch_ohlcv,
visualize_result,
normalize_position,
calc_position_cv,
get_feature_columns,
get_symbols,
unbiased_rank,
ewm_finite,
)
cloudpickle.register_pickle_by_value(src) # for model portability
# symbols = 'BTC,ETH'.split(',')
symbols = os.getenv('ALPHASEA_SYMBOLS').split(',') # 売買代金が多く、古いもの
df = fetch_ohlcv(symbols=symbols, with_target=True)
df.to_pickle('/tmp/df.pkl')
display(df)
class ExampleModelRank:
def __init__(self):
self._model = Ridge(fit_intercept=False, alpha=1e5)
self.max_data_sec = 7 * 24 * 60 * 60 # for predict script
def fit(self, df):
df = self._calc_features(df)
features = get_feature_columns(df)
df['ret_rank'] = unbiased_rank(df.groupby('timestamp')['ret']) - 0.5
df = df.dropna()
self.symbols = get_symbols(df) # for predict script
return self._model.fit(df[features], df['ret_rank'])
def predict(self, df):
df = self._calc_features(df)
features = get_feature_columns(df)
y_pred = self._model.predict(df[features])
df['position'] = np.sign(y_pred)
normalize_position(df)
return df['position']
def _calc_features(self, df):
df = df.copy()
for i in [2, 4, 8, 24, 48, 72]:
df['feature_momentum_{}'.format(i)] = (df['cl'] / df.groupby('symbol')['cl'].shift(i) - 1).fillna(0)
for i in [2, 4, 8, 24, 48, 72]:
df['feature_rsi_{}'.format(i)] = df.groupby('symbol')['cl'].transform(lambda x: talib.RSI(x, timeperiod=i).fillna(50))
for col in get_feature_columns(df):
df[col] = unbiased_rank(df.groupby('timestamp')[col]) - 0.5
return df
df = pd.read_pickle('/tmp/df.pkl')
model = ExampleModelRank()
# cv
calc_position_cv(model, df)
visualize_result(df.dropna())
# fit and save model as portable format
model.fit(df)
data = cloudpickle.dumps(model)
data = lzma.compress(data)
with open('/home/jovyan/data/example_model_rank.xz', 'wb') as f:
f.write(data)
# model validation (Just run this cell in the new kernel to make sure you saved it in a portable format.)
import os
import joblib
import pandas as pd
model = joblib.load('/home/jovyan/data/example_model_rank.xz')
df = pd.read_pickle('/tmp/df.pkl')
df = df[['op', 'hi', 'lo', 'cl']]
max_timestamp = df.index.get_level_values('timestamp').max()
df = df.loc[max_timestamp - pd.to_timedelta(model.max_data_sec, unit='S') <= df.index.get_level_values('timestamp')]
print(model.predict(df))
print(model.symbols)
```
|
github_jupyter
|
# Packages
```
#!/usr/bin/env python
# coding: utf-8
import requests
import numpy as np
import json
import os
import time as tm
import pandas as pd
import http.client
import io
import boto3
import zipfile
from threading import Thread
import logging
from datetime import datetime
import time
from operator import itemgetter
import xlsxwriter
```
# Define API Call Variables
In the following codeblock you'll need to input data for the following variables:
***app_group_api_key*** - This is the string that allows you to access the API calls for an App Group. The App Group API Key can be found in the Braze Dashboard under Settings -> Developer Console -> Rest API Keys. An App Group API key that does not have sufficient access grants may result in an error.
***API_URL*** - This is the URL used to make the Rest API Call.The current value of 'https://rest.iad-01.braze.com' may need to be updated to match the Cluster of your Braze instance. For example, your the cluster of your Braze instance may be 02, and you would update the url to 'https://rest.iad-02.braze.com'. You can find the integer value for the API URL by checking the same value next to "dashboard-0" in the URL you use to access the Braze Dashboard.
***EXPORT_DATE*** - This field is optional, only if you have run the segment export on a prior date to the same S3 bucket. It can be left blank and will export the most recent user profile data for the selected SEGMENT_ID. If not, enter a date when the export was run previously in the following format: 'YYYY-MM-DD'. All other date formats will fail to return results
***SEGMENT_ID*** - This is the Segment API Identifier used to return user data from the segment for the API call. This script can only return results for one segment at a time, and it is recommmended that the segment have no more than 200k users due to hardware limitations that were verified during testing. The Segment API Identifier can be found in the Braze Dashboard under Settings -> Developer Console -> Additional API Identifiers. Under the dropdown menu select 'Segments' and then click the 'Search for a value' dropdown to see a list of segments. Select the segement name that you wish to return results for and copy the value listed under "API Identifier".
***The App Group API Key and Segment ID should never be shared outside of your organization, or be saved in a publically accessible workspace.***
```
app_group_api_key =
now = datetime.now().strftime("%Y-%m-%d")
API_URL = "https://rest.iad-01.braze.com"
EXPORT_DATE = []
SEGMENT_ID =
REQUEST_HEADERS = {
'Content-Type': 'application/json',
'Authorization': 'Bearer ' + app_group_api_key
}
FIELDS_TO_EXPORT = ["braze_id", "custom_attributes", "country", "total_revenue", "push_subscribe",
"email_subscribe", "custom_events", "purchases", "devices", "created_at", "apps",
"campaigns_received", "canvases_received", "cards_clicked", "push_tokens"]
```
# Define S3 Client Variables & Initializing the S3 Client
The codeblock below will initialize the client for Amazon S3 once the following values for the following variables have been added:
***access_key*** - Listed under "AWS Access ID"
***secret_key*** - Listed under "AWS Secret Access Key"
***region_name*** - The region that your S3 bucket is listed under
***user_export_bucket_name*** - The name of the S3 storage bucket that you would like to store the User Profile Export in.
All of these values, with the exception of the user_export_bucket_name can be found in the Braze Dashboard under "Integrations" -> "Technology Partners" -> "AWS Cloud Storage" -> "Data Export Using AWS Credentials".
If there are no values currently listed in this section of the Braze Dashboard, you will need to work with your System Admin to either create them for the first time, or access them. In the event that you are using MFA for AWS S3, you will need to create an account that does not require the use of MFA, as otherwise the export will fail.
***This script will not function without the proper integration between Braze and Amazon S3. While it is possible to modify the script so that the files are returned to your local machine, that functionality requires additional development.***
*You can test your credentials by entering the proper values under 'AWS Access Key', 'AWS Secret Access Key' and 'AWS S3 Bucket Name' and then press 'Test Credentials'. If you see a success message, press save. If you do not see the success message, you'll need to work with your System Admin. to create an account and S3 bucket with the correct access controls.*
**Necessary to point out**: Keep in mind costs related to a high amount of `GET` requests for the user profiles. While these costs are minimal, S3 storage is not free, so keep that in mind before making a high volume of API requests.
Once the S3 credentials have been tested and verified via the Braze Dashboard, you should be all set to store files from the `POST` request for the [User Profiles by Semgent endpoint](https://www.braze.com/docs/api/endpoints/export/user_data/post_users_segment/).
After the variables have been entered, the S3 client will be initialized, and functions will be created so that the ZIP files returned from the API request to the S3 bucket can be processed and transformed into a pandas dataframe.
```
access_key =
secret_key =
region_name =
user_export_bucket_name =
s3 = boto3.resource(
service_name='s3',
aws_access_key_id=access_key,
aws_secret_access_key=secret_key,
region_name=region_name
)
user_export_bucket = s3.Bucket(user_export_bucket_name)
```
# Segment List Endpoint
Here we'll call the [Segment List API Endpoint](https://www.braze.com/docs/api/endpoints/export/segments/get_segment/) in order to return some data needed to build the dataframe and later to return user data from that segment.
```
page, finished = 0, False
braze_segments = []
while True:
endpoint = f"{API_URL}/segments/list?page={page}&sort_direction=desc"
results = requests.get(endpoint, headers=REQUEST_HEADERS).json()['segments']
if not results:
break
braze_segments.extend(results)
page += 1
braze_segments_df = pd.DataFrame(braze_segments)
braze_segments_df.columns = ['segment_id', 'segment_name',
'segment_analytics_tracking_enabled', 'segment_tags']
braze_segments_df = braze_segments_df[braze_segments_df['segment_id'] == SEGMENT_ID]
```
# Defining Functions to Process User Profiles Stored in S3
```
def process_s3_profiles_to_dataframe(objects):
"""Build a DataFrame chunk by chunk and return it.
Temporary function for testing efficiency of building a DataFrame as we go.
There are number of great hosting solutions for us but most come with memory limit. Wanted to leave this function
for troubleshooting potential memory issues there.
Parameters
----------
objects: s3.ObjectSummary
S3 object iterator returned from `bucket.objects.filter` method
Returns
-------
pd.DataFrame
New dataframe with exported user profile from the selected objects
"""
frame_chunks = []
for obj in objects:
segment_id = obj.key.split('/')[1]
with io.BytesIO(obj.get()["Body"].read()) as zip_bytes:
user_chunk = process_s3_zip_object(zip_bytes, segment_id)
frame_chunk = pd.DataFrame(user_chunk)
frame_chunks.append(frame_chunk)
return pd.concat(frame_chunks, ignore_index=True)
def process_s3_profiles(objects, user_data):
"""Extract and process zip user profiles obtained from user segment export.
Parameters
----------
objects : s3.ObjectSummary
S3 object iterator returned from `bucket.objects.filter` method
user_data : list
Store for the extracted profile objects
"""
for obj in objects:
segment_id = obj.key.split('/')[1]
with io.BytesIO(obj.get()["Body"].read()) as zip_bytes:
user_chunk = process_s3_zip_object(zip_bytes, segment_id)
user_data.extend(user_chunk)
def process_s3_zip_object(zip_bytes, segment_id):
"""Extract the zip file contents and process each text file within that zip file.
Text files extracted contain user data JSONs, separated by new line.
Parameters
----------
zip_bytes : io.BytesIO
segment_id : string
Returns
-------
list
Extracted user profile dictionaries from the zip file
"""
profiles = []
with zipfile.ZipFile(zip_bytes) as open_zip:
for user_file in open_zip.namelist():
with open_zip.open(user_file) as users:
for line in users:
user_profile = json.loads(line.decode('utf-8'))
user_profile['segment_id'] = segment_id
profiles.append(user_profile)
return profiles
```
# Define Functions for Processing Campaign Data
The below codeblock:defines functions to enable `GET` requests from the [Campaign Details Endpoint](https://www.braze.com/docs/api/endpoints/export/campaigns/get_campaign_details/) for one or many campaign_ids. It also creates functions to enable the creation of the Channel Combo and Custom Events used in Campaigns.
The URL may need to be updated in the same manner as above following 'iad-0' depending on the cluster of your Braze Instance. For example, you may need to update the string "https://rest.iad-01.braze.com/campaigns/" to "https://rest.iad-02.braze.com/campaigns/" if the Cluster for your Braze instance is 02.
The MAX_RETRIES variable is the number of times that the script will attempt to make a request to the the API Endpoint. If the number is increased the script will take longer to return results from the Campaign Details Endpoint.
```
MAX_RETRIES=3
def process_campaign_id(campaign_id, endpoint):
requests_made = 0
while requests_made < MAX_RETRIES:
try:
response = requests.get("https://rest.iad-01.braze.com/campaigns/"+endpoint+"?campaign_id="+campaign_id, headers=REQUEST_HEADERS
)
return response.json()
except requests.exceptions.HTTPError:
requests_made += 1
tm.sleep(0.5)
if requests_made >= MAX_RETRIES:
raise
### processes a range of ids
def process_campaign_id_range(campaign_id_range, endpoint, store=None):
"""process a number of ids, storing the results in a dict"""
if store is None:
store = {}
for campaign_id in campaign_id_range:
store[campaign_id] = process_campaign_id(campaign_id, endpoint)
return store
def threaded_process_campaigns(nthreads, campaign_id_range, endpoint):
"""process the id range in a specified number of threads"""
try:
store = {}
threads = []
for i in range(nthreads):
campaign_ids = campaign_id_range[i::nthreads]
t = Thread(target=process_campaign_id_range,
args=(campaign_ids, endpoint, store))
threads.append(t)
for t in threads:
t.start()
for t in threads:
t.join()
return store
except Exception as e:
logging.error("Threading exception: "+str(e))
tm.sleep(30)
def get_campaign_id(df_column):
try:
return df_column.get('api_campaign_id')
except AttributeError:
return float('NaN')
def get_message_variation_id(df_column):
try:
return df_column.get('variation_api_id')
except AttributeError:
return float('NaN')
def parse_channel(row):
if row.num_channels > 0:
return row.channel
elif type(row.campaigns_received) != dict:
return "No Messages Received"
else:
return "Unable to Retrieve Campaign Details"
def parse_channel_combo(row):
if type(row.channel_combo) != float:
return row.channel_combo
elif row.channel == "No Messages Received":
return "No Messages Received"
else:
return "Unable to Retrieve Campaign Details"
def get_campaign_custom_event(df_column):
try:
return df_column.get('custom_event_name')
except AttributeError:
return float('NaN')
```
# Define Field Getters to Enable Segment Analytics
The functions defined in the codeblocks below will get the corresponding fields from nested dictionaries stored in dataframes columns that are returned from the User Profiles Endpoint.
```
def get_email_open_engagement(df_column):
try:
return df_column.get('opened_email')
except AttributeError:
return False
def get_email_click_engagement(df_column):
try:
return df_column.get('clicked_email')
except AttributeError:
return False
def get_push_engagement(df_column):
try:
return df_column.get('opened_push')
except AttributeError:
return False
def get_iam_engagement(df_column):
try:
return df_column.get('clicked_in_app_message')
except AttributeError:
return False
def get_conversions(df_column):
try:
return df_column.get('converted')
except AttributeError:
return False
### Create get engagement
def calc_engagement(series):
return series.sum()/series.count()
def get_cards_clicked(row):
if row.channel == 'No Messages Received':
return 0
else:
return len(row.cards_clicked)
def days_between(d1, d2):
d1 = datetime.datetime.strptime(str(d1), '%Y-%m-%dT%H:%M:%S.%f%z')
d2 = datetime.datetime.strptime(str(d2), '%Y-%m-%dT%H:%M:%S.%f%z')
return (d2 - d1).days
def get_custom_event_name(df_column):
try:
return df_column.get('name')
except AttributeError:
return float('NaN')
def get_custom_event_count(df_column):
try:
return df_column.get('count')
except AttributeError:
return float('NaN')
def get_custom_event_first_date(df_column):
try:
return df_column.get('first')
except AttributeError:
return float('NaN')
def get_custom_event_last_date(df_column):
try:
return df_column.get('last')
except AttributeError:
return float('NaN')
def get_notifications_enabled(df_column):
try:
return df_column.get('notifications_enabled')
except AttributeError:
return False
def get_token(df_column):
try:
return df_column.get('token')
except AttributeError:
return 'None'
def get_platform(df_column):
try:
return df_column.get('platform')
except AttributeError:
return 'No platform token'
```
# Export Data from S3 for either Today or a Prior Date
The below codeblock will do the following depending on the value entered above for the EXPORT_DATE variable:
***If the EXPORT_DATE is left blank***:
- Make a request to the [Users by Segment Endpoint](https://www.braze.com/docs/api/endpoints/export/user_data/post_users_segment/)
- Process the user profile data returned to S3 following the successful request for the selected SEGMENT_ID
- Displays the number of user profiles that have been returned.
If the value returned is 0 it is likely that some of the above variables were not configured properly. You'll need to double check and try again.
If the number of user profiles exported is low, it could be because of latency between the Braze API and S3. Try running the code block again and see if the number of users returned increases
***If the EXPORT_DATE is a properly formatted data from a prior export***
- Process the user profile data returned to S3 following the successful request for the selected SEGMENT_ID
- Displays the number of user profiles that have been returned.
If the EXPORT_DATE is not formatted 'YYYY-MM-DD' the below codeblock will fail and you will be asked to try again.
If completed successfully, the segment_df data should return a dataframe for all user profiles from the segment, along with data from the fields listed out in the *FIELDS_TO_EXPORT* variable. Each row in the dataframe corresponds to one user profile within the selected segment.
```
if len(EXPORT_DATE) == 0:
object_prefix_by_segment_id = []
payload = {
"segment_id": SEGMENT_ID,
"fields_to_export": FIELDS_TO_EXPORT
}
res = requests.post(f"{API_URL}/users/export/segment",
headers=REQUEST_HEADERS, json=payload)
res_data = res.json()
print(res_data)
EXPORT_DATE = datetime.today().strftime('%Y-%m-%d')
objects = user_export_bucket.objects.filter(
Prefix=f"segment-export/{SEGMENT_ID}/{EXPORT_DATE}")
tm.sleep(300)
print("Waiting for data to be returned from the Users by Segment Endpoint.")
start = time.time()
user_data = []
print("Reading exported user data from S3")
process_s3_profiles(objects, user_data)
print(f"Took {(time.time() - start):.2f}s")
print(len(user_data))
elif len(EXPORT_DATE) == 10 and EXPORT_DATE.count('-') == 2 and len(EXPORT_DATE) > 0:
year, month, day = EXPORT_DATE.split('-')
isValidDate = True
try:
datetime(int(year), int(month), int(day))
except ValueError:
print("Input date is not the valid YYYY-MM-DD format. Please return to the Define Variables cell and try again enter a properly formatted Date.")
isValidDate = False
if(isValidDate):
objects = user_export_bucket.objects.filter(
Prefix=f"segment-export/{SEGMENT_ID}/{EXPORT_DATE}")
start = time.time()
user_data = []
print("Reading exported user data from S3")
process_s3_profiles(objects, user_data)
print(f"Took {(time.time() - start):.2f}s")
print(len(user_data))
else:
print("This is the text that will display if export date is neither blank nor properly formatted.")
segment_df_raw = pd.DataFrame(user_data)\
.dropna(subset=['braze_id'])
segment_df = pd.merge(segment_df_raw, braze_segments_df,
how='left',
left_on=['segment_id'],
right_on=['segment_id'],
suffixes=['_from_user_segment_endpoint', '_from_segment_list'])
```
# Creating Separate Dataframes for Each KPI
The below codeblock will split the segment_df into the appropriate dataframes so that the following analytical outputs can be viewed from the selected Segment:
1. Rolling Retention
2. Purchasing Rates &
3. Purchase Retention
4. Session Engagement Metrics
5. Custom Event Metrics
5. Message Engagement Rates
6. Custom Events used in Campaigns
7. Opt-In Rates for Push and Email
1-5 will also be crossed by the following dimensions from Message Engagement so that the impact of different messaging strategies can be viewed at the segement level:
- Channel
- Channel Combo
- Campaign Tag
In the event that a segment fails one of the checks below, you can skip those sections in the script. For example, say you are tracking session data, but not purchasing data. Skip the purchasing codeblocks and comment out the final outputs associated with those metrics.
```
rolling_retention_columns = ['braze_id', 'segment_id',
'apps', 'segment_name', 'segment_tags']
purchasing_stats_columns = ['braze_id',
'segment_id', 'apps', 'segment_name', 'segment_tags', 'purchases', 'total_revenue']
sessions_stats_columns = ['braze_id', 'segment_id',
'apps', 'segment_name', 'segment_tags']
custom_events_stats_columns = ['braze_id', 'segment_id', 'apps',
'segment_name', 'segment_tags', 'custom_events']
engagement_stats_columns_all = ['braze_id', 'segment_id', 'country', 'apps', 'segment_name',
'segment_tags', 'campaigns_received', 'canvases_received', 'cards_clicked']
engagement_stats_columns_canvas = ['braze_id', 'segment_id', 'country', 'apps',
'segment_name', 'segment_tags', 'canvases_received', 'cards_clicked']
engagement_stats_columns_campaigns = ['braze_id', 'segment_id', 'country', 'apps',
'segment_name', 'segment_tags', 'campaigns_received', 'cards_clicked']
opt_ins_columns_all = ['braze_id', 'segment_id', 'segment_name', 'apps', 'push_tokens', 'email_subscribe', 'email_opted_in_at', 'push_subscribe',
'push_opted_in_at', 'email_unsubscribed_at', 'push_unsubscribed_at']
opt_ins_columns_email = ['braze_id', 'segment_id', 'segment_name', 'apps', 'push_tokens',
'email_subscribe', 'email_opted_in_at', 'email_unsubscribed_at']
opt_ins_columns_push = ['braze_id', 'segment_id', 'segment_name', 'apps', 'push_tokens',
'push_subscribe', 'push_opted_in_at', 'push_unsubscribed_at']
users_have_sessions = "apps" in segment_df
users_have_purchases = "purchases" in segment_df
users_have_custom_events = "custom_events" in segment_df
users_received_campaigns = "campaigns_received" in segment_df
users_received_canvas = "canvases_received" in segment_df
users_subscribed_email = "email_subscribe" in segment_df
users_subscribed_push = "push_subscribe" in segment_df
if users_have_sessions == True:
segment_rolling_retention_pre_apps = segment_df[rolling_retention_columns]
segment_rolling_retention_pre_apps = segment_rolling_retention_pre_apps.reset_index()
else:
print("Users in these Segments do not have Retention Data")
if users_have_purchases == True:
segment_purchasing_stats_pre_apps = segment_df[purchasing_stats_columns]
segment_purchasing_stats_pre_apps = segment_purchasing_stats_pre_apps.reset_index()
else:
print("Users in these Segments do not have Purchasing Data")
if users_have_sessions == True:
segment_sessions_stats_pre_apps = segment_df[sessions_stats_columns]
segment_sessions_stats_pre_apps = segment_sessions_stats_pre_apps.reset_index()
else:
print("Users in these Segments do not have Session Data")
if users_have_custom_events == True:
segment_custom_event_stats_pre_custom_event = segment_df[custom_events_stats_columns]
segment_custom_event_stats_pre_custom_event = segment_custom_event_stats_pre_custom_event.reset_index()
else:
print("Users in these Segments do not have Custom Event Data")
if (users_received_campaigns == True and users_received_canvas == True):
segment_engagement_stats_pre_apps = segment_df[engagement_stats_columns_all]
elif (users_received_campaigns == False and users_received_canvas == True):
segment_engagement_stats_pre_apps = segment_df[engagement_stats_columns_canvas]
elif (users_received_campaigns == True and users_received_canvas == False):
segment_engagement_stats_pre_apps = segment_df[engagement_stats_columns_campaigns]
elif (users_received_campaigns == False and users_received_canvas == False):
print("Users in these Segments do not have Engagement Data")
if (users_subscribed_email == True and users_subscribed_push == True):
segment_opt_in_stats_pre_apps = segment_df[opt_ins_columns_all]
elif (users_subscribed_email == False and users_subscribed_push == True):
segment_opt_in_stats_pre_apps = segment_df[users_subscribed_push]
elif (users_subscribed_email == True and users_subscribed_push == False):
segment_opt_in_stats_pre_apps = segment_df[users_subscribed_email]
elif (users_subscribed_email == False and users_subscribed_push == False):
print("Users in these Segments do not have Opt-In Data")
```
# Campaign & Engagement Data
The below codeblocks will complete the following tasks:
- Return all of the campaign ids received by the exported Segment
- Send `GET` results from the [Campaign Details API](https://www.braze.com/docs/api/endpoints/export/campaigns/get_campaign_details/#campaign-details-endpoint-api-response) and process the data that is returned.
- Users that received messages from campaign_ids that do not have details returned will be assigned the 'Unable to Retrieve Campaign Details' value for both Channel and Channel Combo.
- Create the Channel_Combo dimension. Please note that the Channel Combo is being created at the Campaign Level and not the User Level.
- Removing Users in the control_group for multivariate campaigns
- Cleaning the Channel names and Channel Combo names
- Creating the dataframe used to caclulate Message Engagement Metrics
- Creating dataframes used to cross other metrics with Channel, Channel Combo, and Campaign Tag
```
segment_engagement_temp = segment_engagement_stats_pre_apps.explode(
'campaigns_received')
segment_engagement_temp['campaign_id'] = list(
map(get_campaign_id, segment_engagement_temp['campaigns_received']))
braze_campaigns = segment_engagement_temp[segment_engagement_temp['campaign_id'].isnull(
) == False]['campaign_id']
braze_campaigns = list(set(braze_campaigns))
campaign_dict = threaded_process_campaigns(
10, braze_campaigns, 'details')
campaign_details_df = pd.DataFrame.from_dict(campaign_dict, orient='index')
campaign_details_df = campaign_details_df.reset_index()
campaign_details_df.rename(columns={"index": "campaign_id"},
inplace=True)
campaign_details_df = campaign_details_df[campaign_details_df['message'] == 'success']
campaign_details_df['num_channels'] = campaign_details_df.channels.apply(len)
campaign_details_df = campaign_details_df[campaign_details_df['num_channels'] > 0]
joined_campaign = pd.merge(segment_engagement_temp, campaign_details_df,
how='left',
left_on=['campaign_id'],
right_on=['campaign_id'],
suffixes=['_from_segments', '_from_campaigns'])
segment_data_engagement_stats_temp = joined_campaign
segment_data_engagement_stats_temp.rename(columns={"channels": "channel"},
inplace=True)
segment_data_engagement_stats_temp['in_control']=segment_data_engagement_stats_temp.campaigns_received.apply(
lambda x: x.get('in_control') if type(x) != float else x)
segment_data_engagement_stats_temp=segment_data_engagement_stats_temp[segment_data_engagement_stats_temp['in_control']!=True]
segment_data_engagement_stats_temp.loc[:, 'channel'] = segment_data_engagement_stats_temp.apply(
parse_channel, axis=1)
segment_data_engagement_stats_temp = segment_data_engagement_stats_temp.explode(
'channel')
segment_data_engagement_stats_temp['channel'] = segment_data_engagement_stats_temp.channel.apply(
lambda x: 'mobile_push' if x == 'android_push' or x == 'ios_push' else x)
segment_data_engagement_stats_temp['channel'] = segment_data_engagement_stats_temp.channel.apply(
lambda x: 'in_app_message' if x == 'legacy_in_app_message' or x == 'trigger_in_app_message ' or x == 'trigger_in_app_message' else x)
segment_data_engagement_stats_temp['channel'] = segment_data_engagement_stats_temp.channel.apply(
lambda x: x.replace("_", " "))
segment_data_engagement_stats_temp['channel'] = segment_data_engagement_stats_temp.channel.apply(
lambda x: x.title())
segment_data_channel_combo = segment_data_engagement_stats_temp[(segment_data_engagement_stats_temp['channel'] != 'No Messages Received')]
segment_data_channel_combo = segment_data_engagement_stats_temp[segment_data_engagement_stats_temp['channel'] != 'Unable To Retrieve Campaign Details']
segment_data_channel_combo = segment_data_channel_combo[[
'braze_id', 'channel']].drop_duplicates()
segment_data_channel_combo = segment_data_channel_combo.dropna(subset=[
'channel'])
segment_data_channel_combo = segment_data_channel_combo.groupby('braze_id')
segment_data_channel_combo = segment_data_channel_combo.apply(
lambda x: x['channel'].unique()).reset_index()
segment_data_channel_combo.columns = ['braze_id', 'channel_combo']
segment_data_channel_combo['channel_combo'] = segment_data_channel_combo.channel_combo.apply(
lambda x: np.ndarray.tolist(x))
segment_data_channel_combo['channel_combo'] = segment_data_channel_combo['channel_combo'].apply(
lambda x: list(set(x)))
segment_data_channel_combo['channel_combo'] = segment_data_channel_combo.channel_combo.apply(
sorted)
segment_data_channel_combo['channel_combo'] = [
', '.join(map(str, l)) for l in segment_data_channel_combo['channel_combo']]
segment_data_channel_combo = segment_data_channel_combo.drop_duplicates()
segment_data_engagement_stats = pd.merge(segment_data_engagement_stats_temp, segment_data_channel_combo,
how='left',
left_on=['braze_id'],
right_on=['braze_id'],
suffixes=['_from_engagement', '_from_channel_combo'])
segment_data_engagement_stats.loc[:, 'channel_combo'] = segment_data_engagement_stats.apply(
parse_channel_combo, axis=1)
users_per_channel_df = segment_data_engagement_stats.groupby(
['segment_name', 'segment_id','channel']).agg(num_users=('braze_id', 'nunique'))
users_per_channel_df = users_per_channel_df.reset_index(level=[0, 1, 2])
users_per_channel_combo_df = segment_data_engagement_stats.groupby(
['segment_name', 'segment_id','channel_combo']).agg(num_users=('braze_id', 'nunique'))
users_per_channel_combo_df = users_per_channel_combo_df.reset_index(level=[
0, 1, 2])
users_per_campaign_tags_df = segment_data_engagement_stats.explode('tags')
users_per_campaign_tags_df['tags'] = users_per_campaign_tags_df.tags.fillna(
'No Messages')
users_per_campaign_tags_df = users_per_campaign_tags_df.groupby(
['segment_name', 'segment_id','tags']).agg(num_users=('braze_id', 'nunique'))
users_per_campaign_tags_df = users_per_campaign_tags_df.reset_index(level=[
0, 1, 2])
```
# Calculate Engagement
The below codeblocks will return Messge Engagement rates for all channels. If the segment did not receive a channel it will simply return a value of zero under the engagement metric.
The following Message Engagement Rates will be returned:
- Number of Users
- Email Open Rate
- Email Click Rate
- Push Open Rate
- In-App Message Click Rate
- Message Conversion Rates (of all Conversion Criteria)
- Content Card Click Rate
Message Engagement Rates will be returned by:
- Segment
- Channel
- Channel Combo
- Campaign Tag
```
segment_data_engagement_stats['campaign_engaged'] = segment_data_engagement_stats.campaigns_received.apply(
lambda x: x.get('engaged') if type(x) != float else x)
segment_data_engagement_stats['opened_email'] = list(
map(get_email_open_engagement, segment_data_engagement_stats['campaign_engaged']))
segment_data_engagement_stats['clicked_email'] = list(map(
get_email_click_engagement, segment_data_engagement_stats['campaign_engaged']))
segment_data_engagement_stats['opened_push'] = list(
map(get_push_engagement, segment_data_engagement_stats['campaign_engaged']))
segment_data_engagement_stats['clicked_iam'] = list(
map(get_iam_engagement, segment_data_engagement_stats['campaign_engaged']))
segment_data_engagement_stats['converted'] = list(
map(get_conversions, segment_data_engagement_stats['campaigns_received']))
segment_data_engagement_stats['converted'] = segment_data_engagement_stats.converted.fillna(
value=False)
segment_data_engagement_stats['cards_clicked'] = segment_data_engagement_stats.cards_clicked.fillna(
value='')
segment_data_engagement_stats.loc[:, 'cards_clicked'] = segment_data_engagement_stats.apply(
get_cards_clicked, axis=1)
engagement_by_segment_preagg = segment_data_engagement_stats.groupby(
['segment_name', 'segment_id'])
engagement_by_segment = engagement_by_segment_preagg.agg(
num_users=('braze_id', 'nunique'), email_open_rate=('opened_email', calc_engagement), email_click_rate=('clicked_email', calc_engagement),
push_open_rate=('opened_push', calc_engagement), iam_click_rate=('clicked_iam', calc_engagement), conversion_rate=('converted', calc_engagement), content_card_click_rate=('cards_clicked', calc_engagement))
engagement_by_segment_and_channel_preagg = segment_data_engagement_stats.groupby(
['segment_name', 'segment_id', 'channel'])
engagement_by_segment_and_channel = engagement_by_segment_and_channel_preagg.agg(
num_users=('braze_id', 'nunique'), email_open_rate=('opened_email', calc_engagement), email_click_rate=('clicked_email', calc_engagement),
push_open_rate=('opened_push', calc_engagement), iam_click_rate=('clicked_iam', calc_engagement), conversion_rate=('converted', calc_engagement), content_card_click_rate=('cards_clicked', calc_engagement))
engagement_by_segment_and_channel_combo_preagg = segment_data_engagement_stats.groupby(
['segment_name', 'segment_id', 'channel_combo'])
engagement_by_segment_and_channel_combo = engagement_by_segment_and_channel_combo_preagg.agg(
num_users=('braze_id', 'nunique'), email_open_rate=('opened_email', calc_engagement), email_click_rate=('clicked_email', calc_engagement),
push_open_rate=('opened_push', calc_engagement), iam_click_rate=('clicked_iam', calc_engagement), conversion_rate=('converted', calc_engagement), content_card_click_rate=('cards_clicked', calc_engagement))
engagement_by_segment_and_channel_combo = engagement_by_segment_and_channel_combo
segment_data_engagement_stats_by_campaign_tags = segment_data_engagement_stats.explode(
'tags')
engagement_by_segment_and_campaign_tag_preagg = segment_data_engagement_stats_by_campaign_tags.groupby([
'segment_name', 'segment_id', 'tags'])
engagement_by_segment_and_campaign_tag = engagement_by_segment_and_campaign_tag_preagg.agg(
num_users=('braze_id', 'nunique'), email_open_rate=('opened_email', calc_engagement), email_click_rate=('clicked_email', calc_engagement),
push_open_rate=('opened_push', calc_engagement), iam_click_rate=('clicked_iam', calc_engagement), conversion_rate=('converted', calc_engagement), content_card_click_rate=('cards_clicked', calc_engagement))
```
# Rolling Retention
The below codeblocks will return Rolling Retetion Rates. You can view the Rolling Retention Methodology [here](https://www.braze.com/resources/articles/calculate-retention-rate).
Rolling Retention Rates will be returned by:
- Segment
- Channel
- Channel Combo
- Campaign Tag
```
segment_rolling_retention_temp = segment_rolling_retention_pre_apps.explode(
'apps')
segment_rolling_retention_temp = segment_rolling_retention_temp.dropna(subset=[
'apps'])
segment_rolling_retention_temp['first_used'] = segment_rolling_retention_temp['apps'].apply(
lambda x: x.get('first_used'))
segment_rolling_retention_temp['last_used'] = segment_rolling_retention_temp['apps'].apply(
lambda x: x.get('last_used'))
segment_rolling_retention_temp['platform'] = segment_rolling_retention_temp['apps'].apply(
lambda x: x.get('platform'))
segment_rolling_retention_temp[['first_used', 'last_used']] = segment_rolling_retention_temp[[
'first_used', 'last_used']].apply(pd.to_datetime)
segment_rolling_retention_temp['day_num'] = (
segment_rolling_retention_temp['last_used'] - segment_rolling_retention_temp['first_used']).dt.days
segment_rolling_retention_temp['day_num'] = segment_rolling_retention_temp['day_num'].astype(
'int')
segment_rolling_retention_raw = pd.pivot_table(segment_rolling_retention_temp,
values=("braze_id"),
index=("segment_name", 'segment_id',
"platform"),
columns="day_num",
aggfunc='nunique')\
.fillna(0)
segment_rolling_retention_raw = segment_rolling_retention_raw[segment_rolling_retention_raw
.columns[::-1]].cumsum(axis=1)
segment_rolling_retention_raw = segment_rolling_retention_raw[
segment_rolling_retention_raw.columns[::-1]]
segment_rolling_retention_raw["num_users"] = segment_rolling_retention_raw[0]
segment_rolling_retention_raw = segment_rolling_retention_raw.groupby(
['segment_name', 'segment_id', 'platform']).sum()
segment_rolling_retention = pd.concat([segment_rolling_retention_raw["num_users"],
segment_rolling_retention_raw
.drop(["num_users"], axis=1)
.div(segment_rolling_retention_raw["num_users"], axis=0)],
axis=1).fillna(0)
segment_rolling_retention=segment_rolling_retention.drop(0,axis=1)
segment_engagement_user_data = segment_data_engagement_stats[[
'braze_id', 'segment_id', 'segment_name', 'apps', 'channel', 'tags', 'channel_combo']]
segment_engagement_data_for_retention = segment_engagement_user_data.explode(
'apps')
segment_engagement_data_for_retention = segment_engagement_data_for_retention.dropna(subset=[
'apps'])
segment_engagement_data_for_retention['platform'] = segment_engagement_data_for_retention['apps'].apply(
lambda x: x.get('platform'))
segment_rolling_retention_by_engagement_temp = pd.merge(segment_rolling_retention_temp.reset_index(), segment_engagement_data_for_retention.reset_index(),
how='left',
left_on=[
'braze_id', 'platform', 'segment_id', 'segment_name'],
right_on=[
'braze_id', 'platform', 'segment_id', 'segment_name'],
suffixes=['_from_retention', '_from_engagement'])
segment_rolling_retention_by_engagement_temp['day_num'] = segment_rolling_retention_by_engagement_temp['day_num'].astype(
'int')
segment_rolling_retention_by_engagement_raw = pd.pivot_table(segment_rolling_retention_by_engagement_temp,
values=(
"braze_id"),
index=(
"segment_name", "segment_id", "platform", "channel"),
columns="day_num",
aggfunc='nunique')\
.fillna(0)
segment_rolling_retention_by_engagement_raw = segment_rolling_retention_by_engagement_raw[segment_rolling_retention_by_engagement_raw
.columns[::-1]].cumsum(axis=1)
segment_rolling_retention_by_engagement_raw = segment_rolling_retention_by_engagement_raw[
segment_rolling_retention_by_engagement_raw.columns[::-1]]
segment_rolling_retention_by_engagement_raw["num_users"] = segment_rolling_retention_by_engagement_raw[0]
segment_rolling_retention_by_engagement_raw = segment_rolling_retention_by_engagement_raw.groupby(
['segment_name', 'segment_id', 'platform', "channel"]).sum()
segment_rolling_retention_by_engagement = pd.concat([segment_rolling_retention_by_engagement_raw["num_users"],
segment_rolling_retention_by_engagement_raw
.drop(["num_users"], axis=1)
.div(segment_rolling_retention_by_engagement_raw["num_users"], axis=0)],
axis=1).fillna(0)
segment_rolling_retention_by_engagement=segment_rolling_retention_by_engagement.drop(0,axis=1)
segment_rolling_retention_by_engagement_temp['day_num'] = segment_rolling_retention_by_engagement_temp['day_num'].astype(
'int')
segment_campaign_tag_data_for_retention_temp = segment_rolling_retention_by_engagement_temp.explode(
'tags')
segment_campaign_tag_data_for_retention_temp = segment_campaign_tag_data_for_retention_temp.dropna(subset=[
'tags'])
segment_rolling_retention_by_campaign_tag_raw = pd.pivot_table(segment_campaign_tag_data_for_retention_temp,
values=(
"braze_id"),
index=(
"segment_name", "segment_id", "platform", "tags"),
columns="day_num",
aggfunc='nunique')\
.fillna(0)
segment_rolling_retention_by_campaign_tag_raw = segment_rolling_retention_by_campaign_tag_raw[segment_rolling_retention_by_campaign_tag_raw
.columns[::-1]].cumsum(axis=1)
segment_rolling_retention_by_campaign_tag_raw = segment_rolling_retention_by_campaign_tag_raw[
segment_rolling_retention_by_campaign_tag_raw.columns[::-1]]
segment_rolling_retention_by_campaign_tag_raw["num_users"] = segment_rolling_retention_by_campaign_tag_raw[0]
segment_rolling_retention_by_campaign_tag_raw = segment_rolling_retention_by_campaign_tag_raw.groupby(
['segment_name', 'segment_id', 'platform', "tags"]).sum()
segment_rolling_retention_by_campaign_tag = pd.concat([segment_rolling_retention_by_campaign_tag_raw["num_users"],
segment_rolling_retention_by_campaign_tag_raw
.drop(["num_users"], axis=1)
.div(segment_rolling_retention_by_campaign_tag_raw["num_users"], axis=0)],
axis=1).fillna(0)
segment_rolling_retention_by_campaign_tag =segment_rolling_retention_by_campaign_tag.drop(0,axis=1)
segment_rolling_retention_by_engagement_temp['day_num'] = segment_rolling_retention_by_engagement_temp['day_num'].astype(
'int')
segment_rolling_retention_by_channel_combo_raw = pd.pivot_table(segment_rolling_retention_by_engagement_temp,
values=(
"braze_id"),
index=(
"segment_name", "segment_id", "platform", "channel_combo"),
columns="day_num",
aggfunc='nunique')\
.fillna(0)
segment_rolling_retention_by_channel_combo_raw = segment_rolling_retention_by_channel_combo_raw[segment_rolling_retention_by_channel_combo_raw
.columns[::-1]].cumsum(axis=1)
segment_rolling_retention_by_channel_combo_raw = segment_rolling_retention_by_channel_combo_raw[
segment_rolling_retention_by_channel_combo_raw.columns[::-1]]
segment_rolling_retention_by_channel_combo_raw["num_users"] = segment_rolling_retention_by_channel_combo_raw[0]
segment_rolling_retention_by_channel_combo_raw = segment_rolling_retention_by_channel_combo_raw.groupby(
['segment_name', 'segment_id', 'platform', "channel_combo"]).sum()
segment_rolling_retention_by_channel_combo = pd.concat([segment_rolling_retention_by_channel_combo_raw["num_users"],
segment_rolling_retention_by_channel_combo_raw
.drop(["num_users"], axis=1)
.div(segment_rolling_retention_by_channel_combo_raw["num_users"], axis=0)],
axis=1).fillna(0)
segment_rolling_retention_by_channel_combo=segment_rolling_retention_by_channel_combo.drop(0,axis=1)
```
# Purchasing Stats
The following purchasing metrics will be returned in the first purchasing stats dataframe:
- Number of Buyers
- Number of Repeat Buyers
- % Buyers
- % Repeat Buyers
- Number of Purchases
- Total Revenue
- Average Revenue per Buyer
- Average time to Purchase
- Purchases per Buyer
The second purchasing stats dataframe will return purchase retention rates.
Both purchasing stats dataframes will returned by:
- Segment
- Channel
- Channel Combo
- Campaign Tag
```
num_users = segment_df.braze_id.nunique()
segment_purchasing_stats_temp = segment_purchasing_stats_pre_apps.dropna(
subset=['apps', 'purchases'])
segment_purchasing_dates = segment_purchasing_stats_pre_apps.dropna(
subset=['apps', 'purchases'])
segment_purchasing_dates = segment_purchasing_dates.explode(
'purchases')
segment_purchasing_dates = segment_purchasing_dates.explode(
'apps')
segment_purchasing_stats_temp['num_purchases'] = segment_purchasing_stats_temp['purchases'].apply(
lambda x: sum(map(itemgetter('count'), x)))
segment_purchasing_dates['first_purchase'] = segment_purchasing_dates['purchases'].apply(
lambda x: x.get('first'))
segment_purchasing_dates['last_purchase'] = segment_purchasing_dates['purchases'].apply(
lambda x: x.get('last'))
segment_purchasing_dates['first_session'] = segment_purchasing_dates['apps'].apply(
lambda x: x.get('first_used'))
segment_purchasing_dates['first_purchase'] = pd.to_datetime(
segment_purchasing_dates['first_purchase'])
segment_purchasing_dates['last_purchase'] = pd.to_datetime(
segment_purchasing_dates['last_purchase'])
segment_purchasing_dates['first_session'] = pd.to_datetime(
segment_purchasing_dates['first_session'])
segment_purchasing_dates_temp = segment_purchasing_dates.groupby(['segment_name', 'segment_id', 'braze_id']).agg(first_purchase_date=(
'first_purchase', 'min'), last_purchase_date=('last_purchase', 'max'), first_session_date=('first_session', 'min'))
segment_purchasing_dates_temp = segment_purchasing_dates_temp.reset_index(level=[
0, 1, 2])
segment_purchasing_stats_temp = pd.merge(segment_purchasing_stats_temp, segment_purchasing_dates_temp,
how='left',
left_on=[
'braze_id', 'segment_id', 'segment_name'],
right_on=[
'braze_id', 'segment_id', 'segment_name'])
segment_purchasing_stats_temp['repeat_buyer'] = segment_purchasing_stats_temp[
'first_purchase_date'] != segment_purchasing_stats_temp['last_purchase_date']
segment_purchasing_stats_temp['repeat_buyer_id'] = segment_purchasing_stats_temp.apply(
lambda row: row.braze_id if row.repeat_buyer == True else 'NaN', axis=1)
segment_purchasing_stats_temp['days_to_purchase'] = (
segment_purchasing_stats_temp['first_purchase_date'] - segment_purchasing_stats_temp['first_session_date']).dt.seconds
segment_purchasing_stats_temp['days_to_purchase'] = segment_purchasing_stats_temp['days_to_purchase']/86400
segment_purchase_retention_temp = segment_purchasing_stats_temp
segment_purchase_data = segment_purchasing_stats_temp
segment_purchasing_stats_temp = segment_purchasing_stats_temp.groupby(['segment_name', 'segment_id']).agg(buyers=('braze_id', 'nunique'), repeat_buyers=('repeat_buyer_id', 'nunique'), num_purchases=(
'num_purchases', 'sum'), total_revenue=('total_revenue', 'sum'), avg_revenue_per_buyer=('total_revenue', 'mean'), avg_time_to_purchase=('days_to_purchase', 'mean'))
segment_purchasing_stats_temp['pct_repeat_buyers'] = round(
segment_purchasing_stats_temp.repeat_buyers/segment_purchasing_stats_temp.buyers, 2)
segment_purchasing_stats_temp['purchases_per_buyer'] = round(
segment_purchasing_stats_temp.num_purchases/segment_purchasing_stats_temp.buyers, 2)
segment_purchasing_stats_temp['revenue_per_item_purchased'] = round(
segment_purchasing_stats_temp.total_revenue/segment_purchasing_stats_temp.num_purchases, 2)
segment_purchasing_stats_temp['purchases_per_user'] = round(
segment_purchasing_stats_temp.num_purchases/num_users, 2)
segment_purchasing_stats_temp['pct_buyer'] = round(
segment_purchasing_stats_temp.buyers/num_users, 2)
segment_purchasing_stats = segment_purchasing_stats_temp
segment_purchase_retention_temp['day_num'] = (
segment_purchase_retention_temp['last_purchase_date'] - segment_purchase_retention_temp['first_purchase_date']).dt.days
segment_purchase_retention_temp['day_num'] = segment_purchase_retention_temp['day_num'].astype(
'int')
segment_purchase_retention_raw = pd.pivot_table(segment_purchase_retention_temp,
values=("braze_id"),
index=("segment_name",
"segment_id"),
columns="day_num",
aggfunc='nunique')\
.fillna(0)
segment_purchase_retention_raw = segment_purchase_retention_raw[segment_purchase_retention_raw
.columns[::-1]].cumsum(axis=1)
segment_purchase_retention_raw = segment_purchase_retention_raw[
segment_purchase_retention_raw.columns[::-1]]
segment_purchase_retention_raw["num_users"] = segment_purchase_retention_raw[0]
segment_purchase_retention_raw = segment_purchase_retention_raw.groupby(
['segment_name', 'segment_id']).sum()
segment_purchase_retention = pd.concat([segment_purchase_retention_raw["num_users"],
segment_purchase_retention_raw
.drop(["num_users"], axis=1)
.div(segment_purchase_retention_raw["num_users"], axis=0)],
axis=1).fillna(0)
segment_purchase_retention=segment_purchase_retention.drop(0,axis=1)
segment_purchase_stats_by_engagement_temp = pd.merge(segment_purchase_data, segment_engagement_user_data,
how='left',
left_on=[
'braze_id', 'segment_id', 'segment_name'],
right_on=[
'braze_id', 'segment_id', 'segment_name'],
suffixes=['_from_retention', '_from_engagement'])
segment_purchase_stats_by_engagement_temp['day_num'] = (
segment_purchase_stats_by_engagement_temp['last_purchase_date'] - segment_purchase_stats_by_engagement_temp['first_purchase_date']).dt.days
segment_purchase_stats_by_engagement_temp['channel'] = segment_purchase_stats_by_engagement_temp.channel.fillna(
'No Messages')
segment_purchase_stats_by_engagement_temp['channel_combo'] = segment_purchase_stats_by_engagement_temp.channel_combo.fillna(
'No Messages')
segment_purchase_stats_by_channel_temp = segment_purchase_stats_by_engagement_temp.groupby(['segment_name', 'segment_id', 'channel']).agg(buyers=('braze_id', 'nunique'), repeat_buyers=('repeat_buyer_id', 'nunique'), num_purchases=(
'num_purchases', 'sum'), total_revenue=('total_revenue', 'sum'), avg_revenue_per_buyer=('total_revenue', 'mean'), avg_time_to_purchase=('days_to_purchase', 'mean'), total_buyers=('braze_id', 'count'), total_repeat_buyers=('repeat_buyer_id', 'count'))
segment_purchase_stats_by_channel_temp['pct_repeat_buyers'] = round(
segment_purchase_stats_by_channel_temp.repeat_buyers/segment_purchase_stats_by_channel_temp.buyers, 2)
segment_purchase_stats_by_channel_temp['purchases_per_buyer'] = round(
segment_purchase_stats_by_channel_temp.num_purchases/segment_purchase_stats_by_channel_temp.total_buyers, 2)
segment_purchase_stats_by_channel_temp['revenue_per_item_purchased'] = round(
segment_purchase_stats_by_channel_temp.total_revenue/segment_purchase_stats_by_channel_temp.num_purchases, 2)
segment_purchase_stats_by_channel = pd.merge(segment_purchase_stats_by_channel_temp, users_per_channel_df,
how='left',
left_on=[
'segment_name', 'segment_id','channel'],
right_on=['segment_name', 'segment_id','channel'])
segment_purchase_stats_by_channel['pct_buyers'] = round(
segment_purchase_stats_by_channel.buyers/segment_purchase_stats_by_channel.num_users, 2)
segment_purchase_stats_by_channel = segment_purchase_stats_by_channel[['segment_name', 'segment_id', 'channel', 'buyers', 'repeat_buyers', 'num_users', 'pct_buyers',
'pct_repeat_buyers', 'purchases_per_buyer', 'avg_revenue_per_buyer', 'avg_time_to_purchase', 'revenue_per_item_purchased']].set_index(['segment_name', 'segment_id', 'channel'])
segment_purchase_stats_by_channel_combo_temp = segment_purchase_stats_by_engagement_temp.groupby(['segment_name', 'segment_id', 'channel_combo']).agg(buyers=('braze_id', 'nunique'), repeat_buyers=('repeat_buyer_id', 'nunique'), num_purchases=(
'num_purchases', 'sum'), total_revenue=('total_revenue', 'sum'), avg_revenue_per_buyer=('total_revenue', 'mean'), avg_time_to_purchase=('days_to_purchase', 'mean'), total_buyers=('braze_id', 'count'), total_repeat_buyers=('repeat_buyer_id', 'count'))
segment_purchase_stats_by_channel_combo_temp['pct_repeat_buyers'] = round(
segment_purchase_stats_by_channel_combo_temp.repeat_buyers/segment_purchase_stats_by_channel_combo_temp.buyers, 2)
segment_purchase_stats_by_channel_combo_temp['purchases_per_buyer'] = round(
segment_purchase_stats_by_channel_combo_temp.num_purchases/segment_purchase_stats_by_channel_combo_temp.total_buyers, 2)
segment_purchase_stats_by_channel_combo_temp['revenue_per_item_purchased'] = round(
segment_purchase_stats_by_channel_combo_temp.total_revenue/segment_purchase_stats_by_channel_combo_temp.num_purchases, 2)
segment_purchase_stats_by_channel_combo = pd.merge(segment_purchase_stats_by_channel_combo_temp, users_per_channel_combo_df,
how='left',
left_on=[
'segment_name', 'segment_id','channel_combo'],
right_on=['segment_name', 'segment_id','channel_combo'])
segment_purchase_stats_by_channel_combo['pct_buyers'] = round(
segment_purchase_stats_by_channel_combo.buyers/segment_purchase_stats_by_channel_combo.num_users, 2)
segment_purchase_stats_by_channel_combo = segment_purchase_stats_by_channel_combo[['segment_name', 'segment_id', 'channel_combo', 'buyers', 'repeat_buyers', 'num_users', 'pct_buyers',
'pct_repeat_buyers', 'purchases_per_buyer', 'avg_revenue_per_buyer', 'avg_time_to_purchase', 'revenue_per_item_purchased']].set_index(['segment_name', 'segment_id', 'channel_combo'])
segment_purchase_stats_by_campaign_tag_temp = segment_purchase_stats_by_engagement_temp.explode(
'tags')
segment_purchase_stats_by_campaign_tag_temp = segment_purchase_stats_by_campaign_tag_temp.groupby(['segment_name', 'segment_id', 'tags']).agg(buyers=('braze_id', 'nunique'), repeat_buyers=('repeat_buyer_id', 'nunique'), num_purchases=(
'num_purchases', 'sum'), total_revenue=('total_revenue', 'sum'), avg_revenue_per_buyer=('total_revenue', 'mean'), avg_time_to_purchase=('days_to_purchase', 'mean'), total_buyers=('braze_id', 'count'), total_repeat_buyers=('repeat_buyer_id', 'count'))
segment_purchase_stats_by_campaign_tag_temp['pct_repeat_buyers'] = round(
segment_purchase_stats_by_campaign_tag_temp.repeat_buyers/segment_purchase_stats_by_campaign_tag_temp.repeat_buyers, 2)
segment_purchase_stats_by_campaign_tag_temp['purchases_per_buyer'] = round(
segment_purchase_stats_by_campaign_tag_temp.num_purchases/segment_purchase_stats_by_campaign_tag_temp.total_buyers, 2)
segment_purchase_stats_by_campaign_tag_temp['revenue_per_item_purchased'] = round(
segment_purchase_stats_by_campaign_tag_temp.total_revenue/segment_purchase_stats_by_campaign_tag_temp.num_purchases, 2)
segment_purchase_stats_by_campaign_tag = pd.merge(segment_purchase_stats_by_campaign_tag_temp, users_per_campaign_tags_df,
how='left',
left_on=[
'segment_name', 'tags'],
right_on=['segment_name', 'tags'])
segment_purchase_stats_by_campaign_tag['pct_buyers'] = round(
segment_purchase_stats_by_campaign_tag.buyers/segment_purchase_stats_by_campaign_tag.num_users, 2)
segment_purchase_stats_by_campaign_tag = segment_purchase_stats_by_campaign_tag[['segment_name', 'segment_id', 'tags', 'buyers', 'repeat_buyers', 'num_users', 'pct_buyers',
'pct_repeat_buyers', 'purchases_per_buyer', 'avg_revenue_per_buyer', 'avg_time_to_purchase', 'revenue_per_item_purchased']].set_index(['segment_name', 'segment_id', 'tags'])
segment_purchase_stats_by_engagement_temp['day_num'] = segment_purchase_stats_by_engagement_temp['day_num'].astype(
'int')
segment_purchase_retention_by_channel_raw = pd.pivot_table(segment_purchase_stats_by_engagement_temp,
values=("braze_id"),
index=(
"segment_name", "segment_id", "channel"),
columns="day_num",
aggfunc='nunique')\
.fillna(0)
segment_purchase_retention_by_channel_raw = segment_purchase_retention_by_channel_raw[segment_purchase_retention_by_channel_raw
.columns[::-1]].cumsum(axis=1)
segment_purchase_retention_by_channel_raw = segment_purchase_retention_by_channel_raw[
segment_purchase_retention_by_channel_raw.columns[::-1]]
segment_purchase_retention_by_channel_raw["num_users"] = segment_purchase_retention_by_channel_raw[0]
segment_purchase_retention_by_channel_raw = segment_purchase_retention_by_channel_raw.groupby(
['segment_name', 'segment_id', "channel"]).sum()
segment_purchase_retention_by_channel = pd.concat([segment_purchase_retention_by_channel_raw["num_users"],
segment_purchase_retention_by_channel_raw
.drop(["num_users"], axis=1)
.div(segment_purchase_retention_by_channel_raw["num_users"], axis=0)],
axis=1).fillna(0)
segment_purchase_retention_by_channel=segment_purchase_retention_by_channel.drop(0, axis=1)
segment_purchase_stats_by_engagement_temp['day_num'] = segment_purchase_stats_by_engagement_temp['day_num'].astype(
'int')
segment_purchase_retention_by_channel_combo_raw = pd.pivot_table(segment_purchase_stats_by_engagement_temp,
values=(
"braze_id"),
index=(
"segment_name", "segment_id", "channel_combo"),
columns="day_num",
aggfunc='nunique')\
.fillna(0)
segment_purchase_retention_by_channel_combo_raw = segment_purchase_retention_by_channel_combo_raw[segment_purchase_retention_by_channel_combo_raw
.columns[::-1]].cumsum(axis=1)
segment_purchase_retention_by_channel_combo_raw = segment_purchase_retention_by_channel_combo_raw[
segment_purchase_retention_by_channel_combo_raw.columns[::-1]]
segment_purchase_retention_by_channel_combo_raw[
"num_users"] = segment_purchase_retention_by_channel_combo_raw[0]
segment_purchase_retention_by_channel_combo_raw = segment_purchase_retention_by_channel_combo_raw.groupby(
['segment_name', 'segment_id', "channel_combo"]).sum()
segment_purchase_retention_by_channel_combo = pd.concat([segment_purchase_retention_by_channel_combo_raw["num_users"],
segment_purchase_retention_by_channel_combo_raw
.drop(["num_users"], axis=1)
.div(segment_purchase_retention_by_channel_combo_raw["num_users"], axis=0)],
axis=1).fillna(0)
segment_purchase_retention_by_channel_combo=segment_purchase_retention_by_channel_combo.drop(0,axis=1)
segment_purchase_stats_by_engagement_temp['day_num'] = segment_purchase_stats_by_engagement_temp['day_num'].astype(
'int')
segment_purchase_stats_by_campaign_tag_temp = segment_purchase_stats_by_engagement_temp.explode(
'tags')
segment_purchase_retention_by_campaign_tags_raw = pd.pivot_table(segment_purchase_stats_by_campaign_tag_temp,
values=(
"braze_id"),
index=(
"segment_name", "segment_id", "tags"),
columns="day_num",
aggfunc='nunique')\
.fillna(0)
### Get the cumulative sum of users based on "last day"
segment_purchase_retention_by_campaign_tags_raw = segment_purchase_retention_by_campaign_tags_raw[segment_purchase_retention_by_campaign_tags_raw
.columns[::-1]].cumsum(axis=1)
segment_purchase_retention_by_campaign_tags_raw = segment_purchase_retention_by_campaign_tags_raw[
segment_purchase_retention_by_campaign_tags_raw.columns[::-1]]
segment_purchase_retention_by_campaign_tags_raw[
"num_users"] = segment_purchase_retention_by_campaign_tags_raw[0]
segment_purchase_retention_by_campaign_tags_raw = segment_purchase_retention_by_campaign_tags_raw.groupby(
['segment_name', 'segment_id', "tags"]).sum()
segment_purchase_retention_by_campaign_tags = pd.concat([segment_purchase_retention_by_campaign_tags_raw["num_users"],
segment_purchase_retention_by_campaign_tags_raw
.drop(["num_users"], axis=1)
.div(segment_purchase_retention_by_campaign_tags_raw["num_users"], axis=0)],
axis=1).fillna(0)
segment_purchase_retention_by_campaign_tags=segment_purchase_retention_by_campaign_tags.drop(0,axis=1)
```
# Session Stats
The following Session Engagement Metrics will be returned by the codeblocks below:
- Number of Users
- Sessions per User
Session Engagement Metrics will returned by:
- Segment
- Channel
- Channel Combo
- Campaign Tag
```
segment_sessions_stats_temp = segment_sessions_stats_pre_apps.explode('apps')
segment_sessions_stats_temp = segment_sessions_stats_temp.dropna(subset=[
'apps'])
segment_sessions_stats_temp['sessions'] = segment_sessions_stats_temp['apps'].apply(
lambda x: x.get('sessions'))
segment_sessions_stats_temp['platform'] = segment_sessions_stats_temp['apps'].apply(
lambda x: x.get('platform'))
segment_sessions_stats_temp = segment_sessions_stats_temp.groupby(['segment_name', 'segment_id']).agg(
num_users=("braze_id", 'nunique'), total_sessions=('sessions', 'sum'))
segment_sessions_stats_temp['sessions_per_user'] = segment_sessions_stats_temp.total_sessions / \
segment_sessions_stats_temp.num_users
segment_sessions_stats = segment_sessions_stats_temp
segment_sessions_stats_by_engagement_temp = segment_engagement_user_data.explode(
'apps')
segment_sessions_stats_by_engagement_temp = segment_sessions_stats_by_engagement_temp.dropna(subset=[
'apps'])
segment_sessions_stats_by_engagement_temp['sessions'] = segment_sessions_stats_by_engagement_temp['apps'].apply(
lambda x: x.get('sessions'))
segment_sessions_stats_by_engagement_temp['platform'] = segment_sessions_stats_by_engagement_temp['apps'].apply(
lambda x: x.get('platform'))
segment_sessions_stats_by_channel_temp = segment_sessions_stats_by_engagement_temp.groupby(['segment_name', 'segment_id', 'channel']).agg(
total_users=("braze_id", 'count'), total_sessions=('sessions', 'sum'), num_users=("braze_id", 'nunique'))
segment_sessions_stats_by_channel_temp = segment_sessions_stats_by_channel_temp.reset_index()
segment_sessions_stats_by_channel_temp['sessions_per_user'] = segment_sessions_stats_by_channel_temp.total_sessions / \
segment_sessions_stats_by_channel_temp.total_users
segment_sessions_stats_by_channel = segment_sessions_stats_by_channel_temp[[
'segment_name', 'segment_id', 'channel', 'num_users', 'sessions_per_user']].set_index(['segment_name', 'segment_id', 'channel'])
segment_sessions_stats_by_channel_combo_temp = segment_sessions_stats_by_engagement_temp.groupby(['segment_name', 'segment_id', 'channel_combo']).agg(
total_users=("braze_id", 'count'), total_sessions=('sessions', 'sum'), num_users=("braze_id", 'nunique'))
segment_sessions_stats_by_channel_combo_temp = segment_sessions_stats_by_channel_combo_temp.reset_index()
segment_sessions_stats_by_channel_combo_temp['sessions_per_user'] = segment_sessions_stats_by_channel_combo_temp.total_sessions / \
segment_sessions_stats_by_channel_combo_temp.total_users
segment_sessions_stats_by_channel_combo = segment_sessions_stats_by_channel_combo_temp[[
'segment_name', 'segment_id', 'channel_combo', 'num_users', 'sessions_per_user']].set_index(['segment_name', 'segment_id', 'channel_combo'])
segment_sessions_stats_by_campaign_tag_temp = segment_sessions_stats_by_engagement_temp.explode(
'tags')
segment_sessions_stats_by_campaign_tag_temp = segment_sessions_stats_by_campaign_tag_temp.groupby(['segment_name', 'segment_id', 'tags']).agg(
total_users=("braze_id", 'count'), total_sessions=('sessions', 'sum'), num_users=("braze_id", 'nunique'))
segment_sessions_stats_by_campaign_tag_temp = segment_sessions_stats_by_campaign_tag_temp.reset_index()
segment_sessions_stats_by_campaign_tag_temp['sessions_per_user'] = segment_sessions_stats_by_campaign_tag_temp.total_sessions / \
segment_sessions_stats_by_campaign_tag_temp.total_users
segment_sessions_stats_by_campaign_tag = segment_sessions_stats_by_campaign_tag_temp[[
'segment_name', 'segment_id', 'tags', 'num_users', 'sessions_per_user']].set_index(['segment_name', 'segment_id', 'tags'])
```
# Custom Event Stats
The following Custom Events Stats will be calculated:
- Number of Users Completing the Custom Event
- Number of Users
- Total Count of Custom Event
- % of Users Completing Custom Events
- Custom Events per User
- Avg. Days between each occurence of a Custom Event
- Avg. Custom Event Completion per Day
Custom Event stats dataframes will returned by:
- Segment
- Channel
- Channel Combo
- Campaign Tag
```
segment_custom_event_stats_temp = segment_custom_event_stats_pre_custom_event.explode(
'custom_events')
segment_custom_event_stats_temp['custom_event_name'] = list(
map(get_custom_event_name, segment_custom_event_stats_temp['custom_events']))
segment_custom_event_stats_temp['custom_event_count'] = list(
map(get_custom_event_count, segment_custom_event_stats_temp['custom_events']))
segment_custom_event_stats_temp['custom_event_first_date'] = list(map(
get_custom_event_first_date, segment_custom_event_stats_temp['custom_events']))
segment_custom_event_stats_temp['custom_event_last_date'] = list(
map(get_custom_event_last_date, segment_custom_event_stats_temp['custom_events']))
segment_custom_event_stats_temp[['custom_event_first_date', 'custom_event_last_date']] = segment_custom_event_stats_temp[[
'custom_event_first_date', 'custom_event_last_date']].apply(pd.to_datetime)
segment_custom_event_stats_temp['days_between_events'] = (
segment_custom_event_stats_temp['custom_event_last_date'] - segment_custom_event_stats_temp['custom_event_first_date']).dt.days
segment_custom_event_stats_temp['custom_event_per_day'] = np.round(np.where(segment_custom_event_stats_temp['days_between_events'] > 0,
segment_custom_event_stats_temp.custom_event_count/segment_custom_event_stats_temp.days_between_events, segment_custom_event_stats_temp.custom_event_count), 1)
total_segment_users_custom_event = segment_custom_event_stats_temp.braze_id.nunique()
segment_custom_event_stats_by_segment = segment_custom_event_stats_temp.groupby(
['segment_name', 'segment_id', 'custom_event_name']).agg(num_users_completing_custom_event=(
'braze_id', 'nunique'), total_custom_events=('custom_event_count', 'sum'), avg_days_between_events=('days_between_events', 'mean'), avg_custom_event_per_day=('custom_event_per_day', 'mean'))
segment_custom_event_stats_by_segment['custom_event_per_user'] = segment_custom_event_stats_by_segment.total_custom_events / \
total_segment_users_custom_event
segment_custom_event_stats_by_segment['pct_users_completing_custom_event'] = segment_custom_event_stats_by_segment.num_users_completing_custom_event / \
total_segment_users_custom_event
segment_custom_event_stats_by_segment['num_users'] = total_segment_users_custom_event
segment_custom_event_stats_by_segment = segment_custom_event_stats_by_segment[[
'num_users_completing_custom_event', 'num_users', 'total_custom_events', 'pct_users_completing_custom_event', 'custom_event_per_user', 'avg_days_between_events', 'avg_custom_event_per_day']]
segment_custom_event_stats_by_engagement_temp = pd.merge(segment_custom_event_stats_temp, segment_engagement_user_data,
how='left',
left_on=[
'braze_id', 'segment_id', 'segment_name'],
right_on=[
'braze_id', 'segment_id', 'segment_name'],
suffixes=['_from_custom_events', '_from_engagement'])
segment_custom_event_stats_by_engagement_temp['channel'] = segment_custom_event_stats_by_engagement_temp.channel.fillna(
'No Messages')
segment_custom_event_stats_by_engagement_temp['channel_combo'] = segment_custom_event_stats_by_engagement_temp.channel_combo.fillna(
'No Messages')
segment_custom_event_stats_by_engagement_temp['tags'] = segment_custom_event_stats_by_engagement_temp.tags.fillna(
'No Messages')
segment_custom_event_stats_by_segment_and_channel_temp = segment_custom_event_stats_by_engagement_temp.groupby(['segment_name', 'segment_id', 'channel', 'custom_event_name']).agg(num_users_completing_custom_event=(
'braze_id', 'nunique'), total_custom_events=('custom_event_count', 'sum'), avg_days_between_events=('days_between_events', 'mean'), avg_custom_event_per_day=('custom_event_per_day', 'mean'))
segment_custom_event_stats_by_segment_and_channel_temp = segment_custom_event_stats_by_segment_and_channel_temp.reset_index()
segment_custom_event_stats_by_segment_and_channel = pd.merge(segment_custom_event_stats_by_segment_and_channel_temp, users_per_channel_df,
how='left',
left_on=[
'segment_name', 'segment_id','channel'],
right_on=['segment_name', 'segment_id','channel'])
segment_custom_event_stats_by_segment_and_channel['custom_event_per_user'] = segment_custom_event_stats_by_segment_and_channel.total_custom_events / \
segment_custom_event_stats_by_segment_and_channel.num_users
segment_custom_event_stats_by_segment_and_channel['pct_users_completing_custom_event'] = segment_custom_event_stats_by_segment_and_channel.num_users_completing_custom_event / \
segment_custom_event_stats_by_segment_and_channel.num_users
segment_custom_event_stats_by_segment_and_channel = segment_custom_event_stats_by_segment_and_channel[['segment_name', 'segment_id', 'channel','custom_event_name', 'num_users_completing_custom_event', 'num_users',
'total_custom_events', 'pct_users_completing_custom_event', 'custom_event_per_user', 'avg_days_between_events', 'avg_custom_event_per_day']].set_index(['segment_name', 'segment_id', 'channel'])
segment_custom_event_stats_by_segment_and_channel_combo_temp = segment_custom_event_stats_by_engagement_temp.groupby(['segment_name', 'segment_id', 'channel_combo', 'custom_event_name']).agg(num_users_completing_custom_event=(
'braze_id', 'nunique'), total_custom_events=('custom_event_count', 'sum'), avg_days_between_events=('days_between_events', 'mean'), avg_custom_event_per_day=('custom_event_per_day', 'mean'))
segment_custom_event_stats_by_segment_and_channel_combo_temp = segment_custom_event_stats_by_segment_and_channel_combo_temp.reset_index()
segment_custom_event_stats_by_segment_and_channel_combo = pd.merge(segment_custom_event_stats_by_segment_and_channel_combo_temp, users_per_channel_combo_df,
how='left',
left_on=[
'segment_name', 'segment_id','channel_combo'],
right_on=['segment_name', 'segment_id','channel_combo'])
segment_custom_event_stats_by_segment_and_channel_combo['custom_event_per_user'] = segment_custom_event_stats_by_segment_and_channel_combo.total_custom_events / \
segment_custom_event_stats_by_segment_and_channel_combo.num_users
segment_custom_event_stats_by_segment_and_channel_combo['pct_users_completing_custom_event'] = segment_custom_event_stats_by_segment_and_channel_combo.num_users_completing_custom_event / \
segment_custom_event_stats_by_segment_and_channel_combo.num_users
segment_custom_event_stats_by_segment_and_channel_combo = segment_custom_event_stats_by_segment_and_channel_combo[['segment_name', 'segment_id', 'channel_combo', 'custom_event_name','num_users_completing_custom_event', 'num_users',
'total_custom_events', 'pct_users_completing_custom_event', 'custom_event_per_user', 'avg_days_between_events', 'avg_custom_event_per_day']].set_index(['segment_name', 'segment_id', 'channel_combo'])
segment_custom_event_stats_by_segment_and_campaign_tags_df = segment_custom_event_stats_by_engagement_temp.explode(
'tags')
segment_custom_event_stats_by_segment_and_campaign_tags_temp = segment_custom_event_stats_by_segment_and_campaign_tags_df.groupby(['segment_name', 'segment_id', 'tags', 'custom_event_name']).agg(num_users_completing_custom_event=(
'braze_id', 'nunique'), total_custom_events=('custom_event_count', 'sum'), avg_days_between_events=('days_between_events', 'mean'), avg_custom_event_per_day=('custom_event_per_day', 'mean'))
segment_custom_event_stats_by_segment_and_campaign_tags_temp = segment_custom_event_stats_by_segment_and_campaign_tags_temp.reset_index()
segment_custom_event_stats_by_segment_and_campaign_tags = pd.merge(segment_custom_event_stats_by_segment_and_campaign_tags_temp, users_per_campaign_tags_df,
how='left',
left_on=[
'segment_name', 'segment_id','tags'],
right_on=['segment_name', 'segment_id','tags'])
segment_custom_event_stats_by_segment_and_campaign_tags['custom_event_per_user'] = segment_custom_event_stats_by_segment_and_campaign_tags.total_custom_events / \
segment_custom_event_stats_by_segment_and_campaign_tags.num_users
segment_custom_event_stats_by_segment_and_campaign_tags['pct_users_completing_custom_event'] = segment_custom_event_stats_by_segment_and_campaign_tags.num_users_completing_custom_event / \
segment_custom_event_stats_by_segment_and_campaign_tags.num_users
segment_custom_event_stats_by_segment_and_campaign_tags = segment_custom_event_stats_by_segment_and_campaign_tags[[
'segment_name', 'segment_id', 'tags', 'custom_event_name','num_users_completing_custom_event', 'num_users', 'total_custom_events', 'pct_users_completing_custom_event', 'custom_event_per_user', 'avg_days_between_events', 'avg_custom_event_per_day']].set_index(['segment_name', 'segment_id', 'tags'])
```
## Custom Events Used in Campaigns
The codeblock below will return all custom events that used in campaigns received by the selected segment.
```
campaign_details_custom_event_temp = campaign_details_df[[
'campaign_id', 'conversion_behaviors']]
campaign_details_custom_event_temp = campaign_details_custom_event_temp.dropna(
subset=['conversion_behaviors'])
campaign_details_custom_event_temp = campaign_details_custom_event_temp.explode(
'conversion_behaviors')
campaign_details_custom_event_temp['custom_event_conversion_behavior'] = list(map(
get_campaign_custom_event, campaign_details_custom_event_temp['conversion_behaviors']))
campaign_details_custom_event_temp = campaign_details_custom_event_temp.dropna(
subset=['custom_event_conversion_behavior'])
campaign_details_custom_event = campaign_details_custom_event_temp[[
'campaign_id', 'custom_event_conversion_behavior']].drop_duplicates()
campaign_details_custom_event = campaign_details_custom_event.set_index(
'campaign_id')
```
# Segment Opt-In Rates
The codeblock below will return the opt-in rates for Push and Email for all users across the following platforms:
- iOS
- Android
- Web
```
segment_opt_ins_temp = segment_opt_in_stats_pre_apps.explode('apps')
segment_opt_ins_temp = segment_opt_ins_temp.dropna(subset=['apps'])
segment_opt_ins_temp = segment_opt_ins_temp.explode('push_tokens')
segment_opt_ins_temp['notifications_enabled'] = list(
map(get_notifications_enabled, segment_opt_ins_temp['push_tokens']))
segment_opt_ins_temp['token'] = list(
map(get_token, segment_opt_ins_temp['push_tokens']))
segment_opt_ins_temp['push_token_platform'] = list(
map(get_platform, segment_opt_ins_temp['push_tokens']))
segment_opt_ins_temp['app_platform'] = segment_opt_ins_temp['apps'].apply(
lambda x: x.get('platform'))
segment_opt_ins_temp_android = segment_opt_ins_temp[segment_opt_ins_temp['app_platform'] == 'Android'].copy()
segment_opt_ins_temp_android['push_opted_in'] = segment_opt_ins_temp.apply(lambda x: True
if x['notifications_enabled'] == True and x['token'] != "None" else False, axis=1)
segment_opt_ins_temp_android['email_opted_in'] = segment_opt_ins_temp.apply(lambda x: True
if x['email_subscribe'] == 'opted_in' else False, axis=1)
segment_opt_ins_temp_ios = segment_opt_ins_temp[segment_opt_ins_temp['app_platform'] == 'iOS'].copy()
segment_opt_ins_temp_ios['push_opted_in'] = segment_opt_ins_temp.apply(lambda x: True
if x['notifications_enabled'] == True and x['token'] != "None" else False, axis=1)
segment_opt_ins_temp_ios['email_opted_in'] = segment_opt_ins_temp.apply(lambda x: True
if x['email_subscribe'] == 'opted_in' else False, axis=1)
segment_opt_ins_temp_web = segment_opt_ins_temp[segment_opt_ins_temp['app_platform'] == 'Web'].copy()
segment_opt_ins_temp_web['push_opted_in'] = segment_opt_ins_temp.apply(lambda x: True
if x['notifications_enabled'] == True and x['token'] != "None" else False, axis=1)
segment_opt_ins_temp_web['email_opted_in'] = segment_opt_ins_temp.apply(lambda x: True
if x['email_subscribe'] == 'opted_in' else False, axis=1)
segment_opt_ins_android_pre_agg = segment_opt_ins_temp_android.groupby(
['segment_id', 'segment_name', 'app_platform'])
opt_ins_aggregator = {'push_opted_in': calc_engagement,
'email_opted_in': calc_engagement}
segment_opt_ins_android = segment_opt_ins_android_pre_agg.agg(
opt_ins_aggregator)
segment_opt_ins_ios_pre_agg = segment_opt_ins_temp_ios.groupby(
['segment_id', 'segment_name', 'app_platform'])
segment_opt_ins_ios = segment_opt_ins_ios_pre_agg.agg(opt_ins_aggregator)
segment_opt_ins_web_pre_agg = segment_opt_ins_temp_web.groupby(
['segment_id', 'segment_name', 'app_platform'])
segment_opt_ins_web = segment_opt_ins_web_pre_agg.agg(opt_ins_aggregator)
segment_opt_ins = pd.concat(
[segment_opt_ins_android, segment_opt_ins_ios, segment_opt_ins_web])
```
## Exporting Outputs to Excel
Please note that attempting to export dataframes that were not created will result in an error.
```
file_name = "Segment Analytics {date}.xlsx".format(date = datetime.now().date())
writer = pd.ExcelWriter(file_name, engine='xlsxwriter')
engagement_by_segment.to_excel(writer, sheet_name='Eng. by Segment')
engagement_by_segment_and_channel.to_excel(
writer, sheet_name='Eng. by Channel')
engagement_by_segment_and_channel_combo.to_excel(
writer, sheet_name='Eng. by Channel Combo')
engagement_by_segment_and_campaign_tag.to_excel(
writer, sheet_name='Eng. by Campaign Tag')
segment_rolling_retention.to_excel(writer, sheet_name='Ret. by Segment')
segment_rolling_retention_by_engagement.to_excel(
writer, sheet_name='Ret. by Channel')
segment_rolling_retention_by_channel_combo.to_excel(
writer, sheet_name='Ret. by Channel Combo')
segment_rolling_retention_by_campaign_tag.to_excel(
writer, sheet_name='Ret. by Campaign Tag')
segment_purchasing_stats.to_excel(writer, sheet_name='Purch. Stats by Segment')
segment_purchase_stats_by_channel.to_excel(
writer, sheet_name='Purch. Stats by Channel')
segment_purchase_stats_by_channel_combo.to_excel(
writer, sheet_name='Purch. Stats by Combo')
segment_purchase_stats_by_campaign_tag.to_excel(
writer, sheet_name='Purch. Stats by Campaign Tag')
segment_purchase_retention.to_excel(writer, sheet_name='Purch. Ret by Segment')
segment_purchase_retention_by_channel.to_excel(
writer, sheet_name='Purch. Ret by Channel')
segment_purchase_retention_by_channel_combo.to_excel(
writer, sheet_name='Purch. Ret by Combo')
segment_purchase_retention_by_campaign_tags.to_excel(
writer, sheet_name='Purch. Ret by Campaign Tag')
segment_sessions_stats.to_excel(writer, sheet_name='Sess. Stats by Segment')
segment_sessions_stats_by_channel.to_excel(
writer, sheet_name='Sess. Stats by Channel')
segment_sessions_stats_by_channel_combo.to_excel(
writer, sheet_name='Sess. Stats by Combo')
segment_sessions_stats_by_campaign_tag.to_excel(
writer, sheet_name='Sess. Stats by Campaign Tag')
segment_custom_event_stats_by_segment.to_excel(
writer, sheet_name='CE Stats by Segment')
segment_custom_event_stats_by_segment_and_channel.to_excel(
writer, sheet_name='CE Stats by Channel')
segment_custom_event_stats_by_segment_and_channel_combo.to_excel(
writer, sheet_name='CE Stats by Combo')
segment_custom_event_stats_by_segment_and_campaign_tags.to_excel(
writer, sheet_name='CE Stats by Campaign Tag')
campaign_details_custom_event.to_excel(
writer, sheet_name='CE Used in Campaigns')
segment_opt_ins.to_excel(writer, sheet_name='Opt-Ins by Segment')
writer.save()
```
|
github_jupyter
|
# DJL BERT Inference Demo
## Introduction
In this tutorial, you walk through running inference using DJL on a [BERT](https://towardsdatascience.com/bert-explained-state-of-the-art-language-model-for-nlp-f8b21a9b6270) QA model trained with MXNet.
You can provide a question and a paragraph containing the answer to the model. The model is then able to find the best answer from the answer paragraph.
Example:
```text
Q: When did BBC Japan start broadcasting?
```
Answer paragraph:
```text
BBC Japan was a general entertainment channel, which operated between December 2004 and April 2006.
It ceased operations after its Japanese distributor folded.
```
And it picked the right answer:
```text
A: December 2004
```
## Preparation
This tutorial requires the installation of Java Kernel. To install the Java Kernel, see the [README](https://github.com/awslabs/djl/blob/master/jupyter/README.md).
```
%maven ai.djl:api:0.2.0
%maven ai.djl.mxnet:mxnet-engine:0.2.0
%maven ai.djl:repository:0.2.0
%maven ai.djl.mxnet:mxnet-model-zoo:0.2.0
%maven org.slf4j:slf4j-api:1.7.26
%maven org.slf4j:slf4j-simple:1.7.26
%maven net.java.dev.jna:jna:5.3.0
```
### Include MXNet engine dependency
This tutorial uses MXNet engine as its backend. MXNet has different [build flavor](https://mxnet.apache.org/get_started?version=v1.5.1&platform=linux&language=python&environ=pip&processor=cpu) and it is platform specific.
Please read [here](https://github.com/awslabs/djl/blob/master/examples/README.md#engine-selection) for how to select MXNet engine flavor.
```
String classifier = System.getProperty("os.name").startsWith("Mac") ? "osx-x86_64" : "linux-x86_64";
%maven ai.djl.mxnet:mxnet-native-mkl:jar:${classifier}:1.6.0-a
```
### Import java packages by running the following:
```
import java.io.*;
import java.nio.charset.*;
import java.nio.file.*;
import java.util.*;
import com.google.gson.*;
import com.google.gson.annotations.*;
import ai.djl.*;
import ai.djl.inference.*;
import ai.djl.metric.*;
import ai.djl.mxnet.zoo.*;
import ai.djl.mxnet.zoo.nlp.qa.*;
import ai.djl.repository.zoo.*;
import ai.djl.ndarray.*;
import ai.djl.ndarray.types.*;
import ai.djl.training.util.*;
import ai.djl.translate.*;
import ai.djl.util.*;
```
Now that all of the prerequisites are complete, start writing code to run inference with this example.
## Load the model and input
The model requires three inputs:
- word indices: The index of each word in a sentence
- word types: The type index of the word. All Questions will be labelled with 0 and all Answers will be labelled with 1.
- sequence length: You need to limit the length of the input. In this case, the length is 384
- valid length: The actual length of the question and answer tokens
**First, load the input**
```
var question = "When did BBC Japan start broadcasting?";
var resourceDocument = "BBC Japan was a general entertainment Channel.\n" +
"Which operated between December 2004 and April 2006.\n" +
"It ceased operations after its Japanese distributor folded.";
QAInput input = new QAInput(question, resourceDocument, 384);
```
Then load the model and vocabulary. Create a variable `model` by using the `ModelZoo` as shown in the following code.
```
Map<String, String> criteria = new ConcurrentHashMap<>();
criteria.put("backbone", "bert");
criteria.put("dataset", "book_corpus_wiki_en_uncased");
ZooModel<QAInput, String> model = MxModelZoo.BERT_QA.loadModel(criteria, new ProgressBar());
```
## Run inference
Once the model is loaded, you can call `Predictor` and run inference as follows
```
Predictor<QAInput, String> predictor = model.newPredictor();
String answer = predictor.predict(input);
answer
```
Running inference on DJL is that easy. In the example, you use a model from the `ModelZoo`. However, you can also load the model on your own and use custom classes as the input and output. The process for that is illustrated in greater detail later in this tutorial.
## Dive deep into Translator
Inference in deep learning is the process of predicting the output for a given input based on a pre-defined model.
DJL abstracts away the whole process for ease of use. It can load the model, perform inference on the input, and provide
output. DJL also allows you to provide user-defined inputs. The workflow looks like the following:
The red block ("Images") in the workflow is the input that DJL expects from you. The green block ("Images
bounding box") is the output that you expect. Because DJL does not know which input to expect and which output format that you prefer, DJL provides the `Translator` interface so you can define your own
input and output.
The `Translator` interface encompasses the two white blocks: Pre-processing and Post-processing. The pre-processing
component converts the user-defined input objects into an NDList, so that the `Predictor` in DJL can understand the
input and make its prediction. Similarly, the post-processing block receives an NDList as the output from the
`Predictor`. The post-processing block allows you to convert the output from the `Predictor` to the desired output
format.
### Pre-processing
Now, you need to convert the sentences into tokens. You can use `BertDataParser.tokenizer` to convert questions and answers into tokens. Then, use `BertDataParser.formTokens` to create Bert-Formatted tokens. Once you have properly formatted tokens, use `parser.token2idx` to create the indices.
The following code block converts the question and answer defined earlier into bert-formatted tokens and creates word types for the tokens.
```
// Create token lists for question and answer
List<String> tokenQ = BertDataParser.tokenizer(question.toLowerCase());
List<String> tokenA = BertDataParser.tokenizer(resourceDocument.toLowerCase());
int validLength = tokenQ.size() + tokenA.size();
System.out.println("Question Token: " + tokenQ);
System.out.println("Answer Token: " + tokenA);
System.out.println("Valid length: " + validLength);
```
Normally, words/sentences are represented as indices instead of Strings for training. They typically work like a vector in a n-dimensional space. In this case, you need to map them into indices. The form tokens also pad the sentence to the required length.
```
// Create Bert-formatted tokens
List<String> tokens = BertDataParser.formTokens(tokenQ, tokenA, 384);
// Convert tokens into indices in the vocabulary
BertDataParser parser = model.getArtifact("vocab.json", BertDataParser::parse);
List<Integer> indices = parser.token2idx(tokens);
```
Finally, the model needs to understand which part is the Question and which part is the Answer. Mask the tokens as follows:
```
[Question tokens...AnswerTokens...padding tokens] => [000000...11111....0000]
```
```
// Get token types
List<Float> tokenTypes = BertDataParser.getTokenTypes(tokenQ, tokenA, 384);
```
To properly convert them into `float[]` for `NDArray` creation, here is the helper function:
```
/**
* Convert a List of Number to float array.
*
* @param list the list to be converted
* @return float array
*/
public static float[] toFloatArray(List<? extends Number> list) {
float[] ret = new float[list.size()];
int idx = 0;
for (Number n : list) {
ret[idx++] = n.floatValue();
}
return ret;
}
float[] indicesFloat = toFloatArray(indices);
float[] types = toFloatArray(tokenTypes);
```
Now that you have everything you need, you can create an NDList and populate all of the inputs you formatted earlier. You're done with pre-processing!
#### Construct `Translator`
You need to do this processing within an implementation of the `Translator` interface. `Translator` is designed to do pre-processing and post-processing. You must define the input and output objects. It contains the following two override classes:
- `public NDList processInput(TranslatorContext ctx, I)`
- `public String processOutput(TranslatorContext ctx, O)`
Every translator takes in input and returns output in the form of generic objects. In this case, the translator takes input in the form of `QAInput` (I) and returns output as a `String` (O). `QAInput` is just an object that holds questions and answer; We have prepared the Input class for you.
Armed with the needed knowledge, you can write an implementation of the `Translator` interface. `BertTranslator` uses the code snippets explained previously to implement the `processInput`method. For more information, see [`NDManager`](https://javadoc.djl.ai/api/0.2.0/ai/djl/ndarray/NDManager.html).
```
manager.create(Number[] data, Shape)
manager.create(Number[] data)
```
The `Shape` for `data0` and `data1` is (num_of_batches, sequence_length). For `data2` is just 1.
```
public class BertTranslator implements Translator<QAInput, String> {
private BertDataParser parser;
private List<String> tokens;
private int seqLength;
BertTranslator(BertDataParser parser) {
this.parser = parser;
this.seqLength = 384;
}
@Override
public Batchifier getBatchifier() {
return null;
}
@Override
public NDList processInput(TranslatorContext ctx, QAInput input) throws IOException {
BertDataParser parser = ctx.getModel().getArtifact("vocab.json", BertDataParser::parse);
// Pre-processing - tokenize sentence
// Create token lists for question and answer
List<String> tokenQ = BertDataParser.tokenizer(question.toLowerCase());
List<String> tokenA = BertDataParser.tokenizer(resourceDocument.toLowerCase());
// Calculate valid length (length(Question tokens) + length(resourceDocument tokens))
var validLength = tokenQ.size() + tokenA.size();
// Create Bert-formatted tokens
tokens = BertDataParser.formTokens(tokenQ, tokenA, 384);
if (tokens == null) {
throw new IllegalStateException("tokens is not defined");
}
// Convert tokens into indices in the vocabulary
List<Integer> indices = parser.token2idx(tokens);
// Get token types
List<Float> tokenTypes = BertDataParser.getTokenTypes(tokenQ, tokenA, 384);
NDManager manager = ctx.getNDManager();
// Using the manager created, create NDArrays for the indices, types, and valid length.
// in that order. The type of the NDArray should all be float
NDArray indicesNd = manager.create(toFloatArray(indices), new Shape(1, 384));
indicesNd.setName("data0");
NDArray typesNd = manager.create(toFloatArray(tokenTypes), new Shape(1, 384));
typesNd.setName("data1");
NDArray validLengthNd = manager.create(new float[]{validLength});
validLengthNd.setName("data2");
NDList list = new NDList(3);
list.add(indicesNd);
list.add(typesNd);
list.add(validLengthNd);
return list;
}
@Override
public String processOutput(TranslatorContext ctx, NDList list) {
NDArray array = list.singletonOrThrow();
NDList output = array.split(2, 2);
// Get the formatted logits result
NDArray startLogits = output.get(0).reshape(new Shape(1, -1));
NDArray endLogits = output.get(1).reshape(new Shape(1, -1));
// Get Probability distribution
NDArray startProb = startLogits.softmax(-1);
NDArray endProb = endLogits.softmax(-1);
int startIdx = (int) startProb.argMax(1).getFloat();
int endIdx = (int) endProb.argMax(1).getFloat();
return tokens.subList(startIdx, endIdx + 1).toString();
}
}
```
Congrats! You have created your first Translator! We have pre-filled the `processOutput()` that will process the `NDList` and return it in a desired format. `processInput()` and `processOutput()` offer the flexibility to get the predictions from the model in any format you desire.
With the Translator implemented, you need to bring up the predictor that uses your `Translator` to start making predictions. You can find the usage for `Predictor` in the [Predictor Javadoc](https://javadoc.djl.ai/api/0.2.0/ai/djl/inference/Predictor.html). Create a translator and use the `question` and `resourceDocument` provided previously.
```
String predictResult = null;
QAInput input = new QAInput(question, resourceDocument, 384);
BertTranslator translator = new BertTranslator(parser);
// Create a Predictor and use it to predict the output
try (Predictor<QAInput, String> predictor = model.newPredictor(translator)) {
predictResult = predictor.predict(input);
}
System.out.println(question);
System.out.println(predictResult);
```
Based on the input, the following result will be shown:
```
[december, 2004]
```
That's it!
You can try with more questions and answers. Here are the samples:
**Answer Material**
The Normans (Norman: Nourmands; French: Normands; Latin: Normanni) were the people who in the 10th and 11th centuries gave their name to Normandy, a region in France. They were descended from Norse ("Norman" comes from "Norseman") raiders and pirates from Denmark, Iceland and Norway who, under their leader Rollo, agreed to swear fealty to King Charles III of West Francia. Through generations of assimilation and mixing with the native Frankish and Roman-Gaulish populations, their descendants would gradually merge with the Carolingian-based cultures of West Francia. The distinct cultural and ethnic identity of the Normans emerged initially in the first half of the 10th century, and it continued to evolve over the succeeding centuries.
**Question**
Q: When were the Normans in Normandy?
A: 10th and 11th centuries
Q: In what country is Normandy located?
A: france
|
github_jupyter
|
# Run Train of Bubble-Agent (DQN)
- Team: TToBoT
- Member: { Sejun, Steve, Victor } @kaist
## Objective
- run training simultaneously w/ notebook
- to compare the performance of traing
## For Competition
1. prepare the final trained IQN Model (checkpoint w/ 100 iteration)
2. need to customize of env.step()
- it should work only 1 live. (later, we can use 3 lives)
- need to enumerate all stage (1~99) level w/ at least 10 (250,000 x 10) (8hr x 100 = 800/33d) iteration. (model should be same)
- using origin step(). do train w/ random level, do loop (iteration) forever! (final training)
3. in final competion, it will load the latest checkpoint for initial Model paramters.
4. win the competition!!
```
import os, sys, gin
# use parent folder as shared lib path..
if "../" not in sys.path:
sys.path.append("../")
# major libraries
import gin.tf
import seaborn as sns
import matplotlib.pyplot as plt
from absl import flags
import numpy as np
import tensorflow as tf
# show tf version.
print('! tf.ver = {}'.format(tf.__version__))
# Globals
# BASE_PATH = './!experimental_results_bubble/run3'
# let Dopamine .py files to be imported as modules in Jupiter notebook
module_path = os.path.abspath(os.path.join('../dopamine'))
if module_path not in sys.path:
sys.path.append(module_path)
print(module_path)
# try to load `Dopamine` libraries
import bubble
from dopamine.colab import utils as colab_utils
```
## Train Bubble w/ DQN
```
# @title Load the configuration for DQN.
# DQN_PATH = os.path.join(BASE_PATH, 'rainbow')
# Modified from dopamine/agents/dqn/config/dqn_cartpole.gin
# CONFIG FOR DQN (see @bubble/dqn_nature.gin)
gin_config = '''
# run_experiment
# python -um dopamine.discrete_domains.train --base_dir=/tmp/bubble --gin_files='bubble/dqn_nature.gin'
# python -um dopamine.discrete_domains.train --base_dir=/tmp/bubble --gin_files='bubble/dqn_nature.gin' --gin_bindings='DQNAgent.tf_device="/cpu:*"'
# Hyperparameters used in Mnih et al. (2015).
import dopamine.discrete_domains.atari_lib
import dopamine.discrete_domains.run_experiment
import dopamine.agents.dqn.dqn_agent
import dopamine.replay_memory.circular_replay_buffer
import gin.tf.external_configurables
import bubble.retro_lib
import bubble.bubble_agent
retro_lib.create_retro_environment.game_name = 'BubbleBobble'
retro_lib.create_retro_environment.level = 1
Runner.create_environment_fn = @retro_lib.create_retro_environment
create_agent.agent_name = 'dqn'
RetroPreprocessing.wall_offset = 0
DQNAgent.gamma = 0.99
DQNAgent.update_horizon = 1
DQNAgent.min_replay_history = 50000 # agent steps
DQNAgent.update_period = 4
DQNAgent.target_update_period = 10000 # agent steps
DQNAgent.epsilon_train = 0.1
DQNAgent.epsilon_eval = 0.05
DQNAgent.epsilon_decay_period = 1000000 # agent steps
DQNAgent.tf_device = '/gpu:1' # use '/cpu:*' for non-GPU version
DQNAgent.optimizer = @tf.train.RMSPropOptimizer()
tf.train.RMSPropOptimizer.learning_rate = 0.00025
tf.train.RMSPropOptimizer.decay = 0.95
tf.train.RMSPropOptimizer.momentum = 0.0
tf.train.RMSPropOptimizer.epsilon = 0.00001
tf.train.RMSPropOptimizer.centered = True
# atari_lib.create_atari_environment.game_name = 'Pong'
# Deterministic ALE version used in the DQN Nature paper (Mnih et al., 2015).
# atari_lib.create_atari_environment.sticky_actions = False
# create_agent.agent_name = 'dqn'
Runner.num_iterations = 200
Runner.training_steps = 250000 # agent steps
Runner.evaluation_steps = 125000 # agent steps
Runner.max_steps_per_episode = 27000 # agent steps
AtariPreprocessing.terminal_on_life_loss = True
WrappedReplayBuffer.replay_capacity = 1000000
WrappedReplayBuffer.batch_size = 32
'''
# parse this config
gin.parse_config(gin_config, skip_unknown=False)
# Train DQN on Cartpole
#dqn_runner = create_runner(DQN_PATH, schedule='continuous_train')
#print('\n\n\nStart Training...\n\n\n')
#dqn_runner.run_experiment()
#print('\n\n\nDone training\n\n\n')
#dqn4 (5/28) - reward := -0.01 + 1*K - 3*D + log(S,100) + 5*L
#dqn5 (6/02) - same reward, but wall_offset = 0
#dqn7 (6/04) - final reward
DQN_PATH = '/tmp/bubble_dqn7'
# import main run()
from dopamine.discrete_domains import run_experiment
# config main file
gin_files = []
# bindings.....
gin_bindings = ['Runner.evaluation_steps=0']
# # code from train.main()
# tf.logging.set_verbosity(tf.logging.INFO)
# run_experiment.load_gin_configs(gin_files, gin_bindings)
# runner = run_experiment.create_runner(DQN_PATH)
# # start run
# runner.run_experiment()
```
## Thread for updating status
```
# Thread for update canvas
import threading, time
def get_ioloop():
import IPython, zmq
ipython = IPython.get_ipython()
if ipython and hasattr(ipython, 'kernel'):
return zmq.eventloop.ioloop.IOLoop.instance()
# The IOloop is shared
ioloop = get_ioloop()
# Main Thread
class MyThread(threading.Thread):
'''Thread for drawing into canvas in live'''
def __init__(self, sleep = 0.5, name = 'my'):
super().__init__()
self._quit = threading.Event()
self.sleep = 0.5
self.name = name
self.start()
def run(self):
while not self._quit.isSet():
def update_progress():
if self._quit.isSet():
return
self.display()
time.sleep(self.sleep)
ioloop.add_callback(update_progress)
print("! T[{}].Quit()".format(self.name))
def quit(self):
self._quit.set()
def display(self):
pass
# display basic
from ipycanvas import Canvas
canvas = Canvas(width=640, height=480)
if canvas:
canvas.stroke_text('hello canvas! -------------', 0, 10)
# show canvas in here.
canvas
# Helper for Canvas
#canvas.fill_style = 'green'
#canvas.fill_rect(25, 25, 100, 100)
#canvas.clear_rect(45, 45, 60, 60)
def drawPlot2Canvas(fig = None, x=0, y=0):
'''draw current plt to canvas at (x,y)'''
fig = plt.gcf() if fig is None else fig
plt.close() # not to update on screen.
fig.canvas.draw() # draw fig to canvas
arr = np.array(fig.canvas.renderer._renderer)
print('! arr = {}'.format(np.shape(arr)))
h, w, d = np.shape(arr)
print('! w,h,d = {}'.format(w))
cv = Canvas(width=w, height=h)
cv.put_image_data(arr, 0, 0)
cv.stroke_rect(x, y, x+w-1, y+h-1)
canvas.clear_rect(x,y,x+w,y+h)
canvas.draw_image(cv, x, y)
def drawText2Canvas(txt='msg!', x=10, y=10):
w,h,o = 200,10,10
#canvas.fill_style = 'green'
#canvas.fill_rect(x, y-o, x+w, y+h-o)
canvas.clear_rect(x, y-o, x+w, y+h-o)
canvas.stroke_text(txt, x, y)
# draw plot....
fig = plt.figure(1)
plt.plot([[1,3],[3,3],[7,1]])
# draw plot-to-canvas
drawPlot2Canvas(fig, x=0)
drawText2Canvas('hello world')
#drawText2Canvas('......................')
```
### support Multi-Processing
```
from multiprocessing import Process, Queue
# process list
proc_list = []
proc_queue = None
# train function
def processTrain(name = 'train', Q = None):
global gin_files, gin_bindings, DQN_PATH
from dopamine.discrete_domains import run_experiment
Q.put('init!') if Q else None
tf.logging.set_verbosity(tf.logging.INFO)
run_experiment.load_gin_configs(gin_files, gin_bindings)
runner = run_experiment.create_runner(DQN_PATH)
# access to env
env = runner._environment
o = env.reset()
Q.put('! o({}) = {}'.format(type(o), o[0:10,0,]))
Q.put('start!') if Q else None
runner.run_experiment()
Q.put('! P[{}].stop()'.format(name))
# train thread
def startProcessTrain(target = None):
global proc_queue, proc_list
target = target if target is not None else processTrain
proc_queue = Queue() if proc_queue is None else proc_queue
proc = Process(target = target, args = ('T0', proc_queue))
proc_list.append(proc)
proc.start()
return proc
# stop(or kill) processes
def stopProcess():
global proc_list
for proc in proc_list:
proc.terminate()
proc.join()
# trainer = startProcessTrain()
# stop
# stopProcess()
# show process
# !ps -ax | grep python
# proc_queue
```
### MyTrainer and MyThread
```
from dopamine.discrete_domains import run_experiment
# MyRunner for Train
# - report every episode status.
class MyRunner(run_experiment.Runner):
def __init__(self, base_dir, create_agent_fn):
'''initialize runner'''
super(MyRunner, self).__init__(base_dir, create_agent_fn)
self._load_logger()
def _run_one_episode(self):
'''override to post episode status'''
global proc_queue
episode_length, episode_return = super(MyRunner, self)._run_one_episode()
data = {'episode':{'length': episode_length, 'return': episode_return }}
#proc_queue.put('! epsode[len,ret] = {},{}'.format(episode_length, episode_return))
proc_queue.put(data)
return episode_length, episode_return
def _load_logger(self):
'''load logger to save into file'''
import logging, os
# get TF logger
log = logging.getLogger('tensorflow')
log.setLevel(logging.DEBUG)
# create file handler which logs even debug messages
formatter = logging.Formatter('%(asctime)s - %(name)s - %(levelname)s - %(message)s')
fh = logging.FileHandler(os.path.join(DQN_PATH, 'tensorflow.log'))
fh.setLevel(logging.INFO)
fh.setFormatter(formatter)
log.addHandler(fh)
#! start runner
def startMyRunner(name = 'train', Q = None):
global gin_files, gin_bindings, DQN_PATH
from dopamine.discrete_domains import run_experiment
Q.put('! start: my-runner') if Q else None
tf.logging.set_verbosity(tf.logging.INFO)
run_experiment.load_gin_configs(gin_files, gin_bindings)
runner = MyRunner(DQN_PATH, run_experiment.create_agent)
runner.run_experiment()
Q.put('! P[{}].stop()'.format(name)) if Q else None
#! start process of runner
startProcessTrain(target = startMyRunner)
```
#### Train Results (01/Jun/2020)
```pre
INFO:tensorflow:Starting iteration 87
INFO:tensorflow:Average undiscounted return per training episode: 19.29
INFO:tensorflow:Average training steps per second: 98.36
INFO:tensorflow:Starting iteration 88
INFO:tensorflow:Average undiscounted return per training episode: 17.34
INFO:tensorflow:Starting iteration 89
INFO:tensorflow:Average undiscounted return per training episode: 18.19
INFO:tensorflow:Starting iteration 90
INFO:tensorflow:Average undiscounted return per training episode: 16.46
INFO:tensorflow:Starting iteration 91
INFO:tensorflow:Average undiscounted return per training episode: 18.53
INFO:tensorflow:Starting iteration 92
INFO:tensorflow:Average undiscounted return per training episode: 18.22
INFO:tensorflow:Starting iteration 99
INFO:tensorflow:Average undiscounted return per training episode: 17.893
INFO:tensorflow:Starting iteration 100
INFO:tensorflow:Average undiscounted return per training episode: 18.24
INFO:tensorflow:Starting iteration 101
INFO:tensorflow:Average undiscounted return per training episode: 19.01
INFO:tensorflow:Starting iteration 102
INFO:tensorflow:Average undiscounted return per training episode: 19.94
INFO:tensorflow:Starting iteration 103
INFO:tensorflow:Average undiscounted return per training episode: 17.44
INFO:tensorflow:Starting iteration 104
INFO:tensorflow:Average undiscounted return per training episode: 17.876
INFO:tensorflow:Starting iteration 105
INFO:tensorflow:Average undiscounted return per training episode: 17.42
INFO:tensorflow:Starting iteration 106
INFO:tensorflow:Average undiscounted return per training episode: 17.595
INFO:tensorflow:Starting iteration 107
INFO:tensorflow:Average undiscounted return per training episode: 17.779
```
```
# MyThread for status display
class MyTrainStatus(MyThread):
def __init__(self):
super().__init__(name='status')
self.episodes = np.array([[0,0]])
print('! MyTrainStatus({})'.format(self.name))
def display(self):
global canvas, proc_queue, plt
episodes = []
# pop all queue...
while not proc_queue.empty():
msg = proc_queue.get()
if msg and 'episode' in msg:
E = msg['episode']
episodes.append([E['length'], E['return']])
# print('>> episodes = {}'.format(episodes))
# draw plot if len > 0
if len(episodes) > 0:
arr = np.array(episodes)
print('>> arr = {}'.format(arr))
# draw plot...
if 1>0:
self.episodes = np.vstack((self.episodes, arr))
#print('>> self.episodes = {}'.format(self.episodes))
#fig = plt.figure(1)
#plt.plot(self.episodes)
fig, ax1 = plt.subplots()
ax2 = ax1.twinx()
ax1.plot(self.episodes[:,0], 'g-')
ax2.plot(self.episodes[:,1], 'b-')
ax1.set_xlabel('episode count')
ax1.set_ylabel('length', color='g')
ax2.set_ylabel('return', color='b')
drawPlot2Canvas(fig)
#! start thread for status
tstatus = MyTrainStatus()
episode_length, episode_return = 1,3
msg = {'episode':{'length': episode_length, 'return': episode_return }}
proc_queue.put(msg)
print('> msg.org = {}'.format(msg))
# stop - thread of status
tstatus.quit() if tstatus else None
# stop - process of train
stopProcess()
```
|
github_jupyter
|
```
import sys, os; sys.path.append('..')
import pyzx as zx
import random
import math
from fractions import Fraction
%config InlineBackend.figure_format = 'svg'
c = zx.qasm("""
qreg q[3];
cx q[0], q[1];
""")
zx.d3.draw(c)
c = zx.qasm("""
qreg q[2];
rx(0.5*pi) q[1];
t q[0];
cx q[0], q[1];
cx q[1], q[0];
cx q[0], q[1];
tdg q[1];
rx(-0.5*pi) q[0];
""")
zx.d3.draw(c)
c.gates
g = c.to_graph()
g
zx.d3.draw(g)
zx.simplify.spider_simp(g)
zx.d3.draw(g)
zx.full_reduce(g)
zx.d3.draw(g)
g = zx.sqasm("""
qreg S[1];
qreg q[3];
t S[0];
cx q[0], S[0];
cx q[1], S[0];
cx q[2], S[0];
""")
zx.d3.draw(g)
g = zx.sqasm("""
qreg S[2];
qreg q[3];
t S[0];
cx q[0], S[0];
cx q[1], S[0];
cx q[2], S[0];
tdg S[1];
cx q[0], S[1];
cx q[1], S[1];
cx q[2], S[1];
""")
zx.d3.draw(g)
zx.clifford_simp(g)
zx.d3.draw(g)
zx.full_reduce(g)
zx.d3.draw(g)
g = zx.Circuit.load("test.qsim").to_graph()
zx.d3.draw(g)
g1 = g.copy()
g1.map_qubits([
(0,0), (1, 0), (2, 0), (3, 0),
(0,1), (1, 1), (2, 1), (3, 1),
(0,2), (1, 2), (2, 2), (3, 2),
(0,3), (1, 3), (2, 3), (3, 3)
])
zx.d3.draw(g1)
zx.full_reduce(g1)
zx.d3.draw(g1)
def t_optimiser(c):
g = c.to_graph()
g = zx.simplify.teleport_reduce(g)
c_opt = zx.Circuit.from_graph(g).split_phase_gates().to_basic_gates()
return zx.optimize.basic_optimization(c_opt).to_basic_gates()
c = zx.Circuit.load('../circuits/Fast/grover_5.qc')
zx.d3.draw(c.to_graph())
print(zx.tcount(c))
c1 = t_optimiser(c)
zx.d3.draw(c1.to_graph())
print(zx.tcount(c1))
c.verify_equality(c1)
c2 = c1.copy()
c2.add_circuit(c.adjoint())
g = c2.to_graph()
zx.simplify.full_reduce(g)
zx.d3.draw(g)
c1.gates[10] = zx.gates.T(6, adjoint=True)
c.verify_equality(c1)
c2 = c1.copy()
c2.add_circuit(c.adjoint())
g = c2.to_graph()
zx.simplify.full_reduce(g)
zx.d3.draw(g)
g = zx.Circuit.load('../circuits/Fast/hwb6.qc').to_graph()
zx.d3.draw(g)
print(zx.tcount(g))
zx.simplify.full_reduce(g)
zx.d3.draw(g)
print(zx.tcount(g))
g.apply_state("++---+-")
g.apply_effect("+011-1-")
zx.simplify.full_reduce(g)
print(zx.tcount(g))
zx.drawing.arrange_scalar_diagram(g)
zx.d3.draw(g)
def compute_decomp(g):
if zx.tcount(g) >= 6:
gsum = zx.simulate.replace_magic_states(g)
gsum.reduce_scalar()
terms = 0
vals = 0
for g1 in gsum.graphs:
t,v = compute_decomp(g1)
terms += t
vals += v
return (terms, vals)
else:
return (2 ** math.ceil(zx.tcount(g)/2), g.to_matrix())
math.ceil(2**(0.468 * zx.tcount(g)))
compute_decomp(g)
zx.simulate.calculate_path_sum(g)
```
|
github_jupyter
|
# PDOS data analysis and plotting
---
### Import Modules
```
import os
print(os.getcwd())
import sys
import plotly.graph_objs as go
import matplotlib.pyplot as plt
from scipy import stats
# #########################################################
from methods import get_df_features_targets
from proj_data import scatter_marker_props, layout_shared
```
### Read Data
```
df_features_targets = get_df_features_targets()
```
```
# df_features_targets.columns.tolist()
pband_indices = df_features_targets[[
(
'features',
# 'oh',
'o',
'p_band_center',
)
]].dropna().index.tolist()
df_i = df_features_targets.loc[
pband_indices
][[
("targets", "g_oh", ""),
("targets", "g_o", ""),
("targets", "g_o_m_oh", ""),
("targets", "e_oh", ""),
("targets", "e_o", ""),
("targets", "e_o_m_oh", ""),
("features", "o", "p_band_center"),
]]
# pband_indices =
df_features_targets[[
(
'features',
# 'oh',
'o',
'p_band_center',
)
]]
# ]].dropna().index.tolist()
# assert False
# df_features_targets.shape
# (288, 7)
# (311, 7)
# (312, 7)
# (316, 7)
# df_i.shape
# assert False
# df_i[""]
df = df_i
df = df[
(df["features", "o", "p_band_center"] > -3.5) &
(df["features", "o", "p_band_center"] < -2.) &
# (df[""] == "") &
# (df[""] == "") &
[True for i in range(len(df))]
]
df_i = df
x = df_i["features", "o", "p_band_center"]
# y = df_i["targets", "g_oh", ""]
# y = df_i["targets", "g_o", ""]
y = df_i["targets", "g_o_m_oh", ""]
# y = df_i["targets", "e_o_m_oh", ""]
res = stats.linregress(x, y)
y_new_fit = res.intercept + res.slope * x
def colin_fit(p_i):
g_o_m_oh_i = 0.94 * p_i + 3.58
return(g_o_m_oh_i)
trace_colin_fit = go.Scatter(
x=[-6, 0],
y=[colin_fit(-6), colin_fit(0)],
mode="lines",
name="Colin fit (G_OmOH = 0.94 * p_i + 3.58)",
)
trace_my_fit = go.Scatter(
x=x,
y=y_new_fit,
mode="lines",
name="Colin fit (G_OmOH = 0.94 * p_i + 3.58)",
)
y_new_fit
trace = go.Scatter(
x=x, y=y,
mode="markers",
name="My DFT data",
)
x_i = x.to_numpy()
X = x_i.reshape(-1, 1)
import numpy as np
from sklearn.linear_model import LinearRegression
# X = np.array([[1, 1], [1, 2], [2, 2], [2, 3]])
# y = 1 * x_0 + 2 * x_1 + 3
# y = np.dot(X, np.array([1, 2])) + 3
reg = LinearRegression().fit(X, y)
reg.score(X, y)
print(
reg.coef_,
reg.intercept_,
)
# reg.predict(np.array([[3, 5]]))
y_pred_mine = reg.predict(
[[-6], [2]],
)
trace_my_fit = go.Scatter(
x=[-6, 2],
y=y_pred_mine,
mode="lines",
name="My fit (G_OmOH = 0.75 * p_i + 3.55)",
)
data = [trace, trace_colin_fit, trace_my_fit]
# data = [trace, trace_colin_fit, trace_my_fit]
layout_mine = go.Layout(
showlegend=False,
xaxis=go.layout.XAxis(
title=go.layout.xaxis.Title(
text="ε<sub>2p</sub>",
),
range=[-6, 0, ]
),
yaxis=go.layout.YAxis(
title=go.layout.yaxis.Title(
text="ΔE<sub>O-OH</sub>",
),
range=[-3, 4, ]
),
)
# #########################################################
layout_shared_i = layout_shared.update(layout_mine)
fig = go.Figure(data=data, layout=layout_shared_i)
fig.show()
```
```
# df_i
# df_features_targets
# (0.94 * 0 + 3.58) - (0.94 * 3 + 3.58)
# 0.94 * 0.3
# res.intercept
# res.slope
# layout = go.Layout(
# xaxis=go.layout.XAxis(
# title=go.layout.xaxis.Title(
# text="ε<sub>2p</sub>",
# ),
# ),
# yaxis=go.layout.YAxis(
# title=go.layout.yaxis.Title(
# text="ΔE<sub>O-OH</sub>",
# ),
# ),
# )
```
|
github_jupyter
|
### Basic Functions for Interactively Exploring the CORTX Metrics Stored in Pickles
```
%cd /home/johnbent/cortx/metrics
import cortx_community
import cortx_graphing
import os
from github import Github
gh = Github(os.environ.get('GH_OATH'))
stx = gh.get_organization('Seagate')
repos = cortx_community.get_repos()
ps = cortx_community.PersistentStats()
# a function which can test the progress of a running scrape_metrics.py process
def check_scan_progress(date,ps):
done=0
for repo in ps.get_repos():
(a,b)=ps.get_latest(repo)
if b == date:
done+=1
#print("Last report for %s is %s" % (repo,b))
print("%d out of %d repos have been scanned" % (done,len(ps.get_repos())))
# a function for comparing a field in a repo over time
# for example, if you want to see the change in innersource_committers over time, you can use this function
def compare_fields(ps,repo,field,verbose=False):
last = None
first = None
for date in sorted(ps.stats[repo].keys()):
try:
last = ps.stats[repo][date][field]
except KeyError:
pass # many fields didn't always exist so might not appear in old stats
if first is None:
first = last
if verbose:
print("%s -> %s" % (date, last))
print("Difference between first and last is: %s" % (first-last))
print("Difference between last and first is: %s" % (last-first))
compare_fields(ps,'GLOBAL','external_committers',verbose=True)
#print(ps.get_values('GLOBAL','external_committers'))
# manually add some values so they can be in executive report
repo='GLOBAL'
slack_members_key='slack_total_members'
slack_wau_key='slack_weekly_ave_active'
newsletter_key='newsletter_subscribers'
webinar_key='webinar_attendees'
integrations_key='integrations'
dates=ps.get_dates(repo)
ec_key='external_committers'
ep_key='external_participants'
hc_key='hackathon_committers'
hp_key='hackathon_participants'
eup_key='eu_r&d_participants'
# the helper function to load them
def add_stats(List,Key):
for item in List:
date=item[0]
value=item[1]
print("Repo",repo,"Date",date,"Stat",Key,"Value",value)
ps.add_stat(date=date,repo=repo,stat=Key,value=value)
# the date strings to use
jun_date='2020-07-06'
sep_date='2020-10-07'
oct_date='2020-11-03'
nov_date='2020-12-21'
dec_date='2020-12-30'
last_date='2021-01-06' # must add metrics for the latest date or else they don't show up in plots . . .
def update_slack():
# adding the slack metrics for executive report
slack_members =[(jun_date,97), (sep_date,212), (nov_date,329), (dec_date,355),(last_date,355)]
slack_weekly_average_active=[(jun_date,11.3),(sep_date,53.0),(nov_date,69.9),(dec_date,59.9),(last_date,59.9)]
add_stats(slack_members,slack_members_key)
add_stats(slack_weekly_average_active,slack_wau_key)
print(ps.get_values(repo,slack_members_key))
print(ps.get_values(repo,slack_wau_key))
def update_newsletter():
# adding the newsletter metrics for executive report
dec_date='2020-12-30'
feb_date='2021-02-13'
newsletter_feb=set(["[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]"," [email protected]"," [email protected]"," [email protected]"," [email protected]"," [email protected]"," [email protected]"," [email protected]"," [email protected]"," [email protected]"," [email protected]"," [email protected]"," [email protected]"," [email protected]"," [email protected]"," [email protected]"," [email protected]"," [email protected]"," [email protected]"," [email protected]"," [email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]"])
newsletter_members_dec=set(["[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]","[email protected]"])
print(len(newsletter_feb))
newsletter = [(sep_date,429),(oct_date,459),(nov_date,477),(dec_date,newsletter_members_dec),(last_date,newsletter_members_dec)]
newsletter = [(feb_date,newsletter_feb)]
add_stats(newsletter,newsletter_key)
print(ps.get_values_as_numbers(repo,newsletter_key))
def update_webinar():
# adding the webinar metrics for executive report
webinar_nov=set(["Andrei Zheregelia","Andrew List","Andriy Tkachuk","Anitoliy Bilenko","Anthony Toccco","Ben Wason","Charles Kunkel","Chetan Deshmukh","Chetan Kumar","Clay Curry","Daniar Kurniawan","Dima.c","Dmitri Sandler","Dmytro Podgornyi","Eduard Aleksandrov","Gary Phillips","Guy Carbonneau","Hanesan Umanesan","Igor Pivovarov","Iman Anvari","Ivan Alekhin","Jason Sliger-Sparks","Jjohn Carrier","Julia Rubtsov","Kalpesh Chhajed","Kaustubh Suresh Deorukhkar","Ken Haugen","Ketan Anil Arlulkar","Konstatin Nekrasov","Lance Blumberg","Madhavrao Vemuri","Mark Jedraszek","Maxim Malezhin","Max Medved","Mehul Joshi","Nicholas Krauter","Nigel Hart","Nikita Danilov","Patrick Raaf","Paul Woods","Philippe Nicolas","Phil Ruff","Raydon Gordon","Ricardo Alvarez-Miranda","Sachin Punadikar","Sailesh Manjrekar","Sai Narasimhamurthy","Sarang Sawant","Serkay Olmez","Shankar More","Shiji Zhang","Swapnil Khandare","Taro Iwata","Ujjwal Lanjewar"])
webinar_dec=set(["Andriy Tkachuk","Anthony Tocco","Charles Kunkel","Daniar Kurniawan","Dan Olster","Ganesan Umanesan","Gary Phillips","Guy Carbonneau","Ivan Poddubnyy","Justin Rackowski","Nicolas Krauter","Nigel Hart","Paul Benn","Praveen Viraraghavan","Rajesh Nambiar","Ricardo Alvarez-miranda","Sachin Punadikar","Sarang Sawant","Shankar More","Shiji Zhang","Swapnil Khandare","Trend Geerdes","Ujjwal Lanjewar","Walter Lopatka",])
webinar_jan=set(["Unknown1","Anatoliy Bilenko","Andrea Chamorro","Andriy Tkachuk","Anthony Tocco","Unknown2","Charles Kunkel","Chetan Kumar","Chirs Cramer","Erin Foley","Gabe Wham","Gary Grider","Gregory Touretsky","Iman Anvari","Joseph Rebovich","Justin Rackowski","Keith Pine","Ken Haugen","Ketan Anil Arlulkar","Madhavrao Vemuri","Amandar Sawant","Mark Jedraszek","Mark Sprouse","Matthew Halcomb","Matthew L Curry Sandia","Max","Mehul Joshi","Meng Wang","Mike Sevilla","Muhul Malhotra","Nedko Amaudov","Oded Kellner","Paul Kusbel","Pedro Fernandez","Pritesh Pawar","Priyanka Borawake","Quinn D Mitchell","Rajesh Bhalerao","Ricardo Alvarez-Miranda","Robert Pechman","Rohan Puri","Sachin Punadikar","Sai Narasimhamurthy","Sarang Sawant","Shailesh","Shankar More","Sharad Mehrotra","Shlomi Avihou","Shreya Karmakar","Shrihari Waskar","Unknown","Sridbar Dubhaka","Stephane Thiell","Swapril Khandare","Tong Shi","Ujjwal Lanjewar","Venky P","Vijay Nanjunda Swamy","Vikram","Vojtech Juranek","Walkter Lopatka","Ziv","Theodore Omtzigt","Rajkumar Patel","Anjinkya Deshpande","Anatoliy Bilenko","Chetan Deshmukh","Henry Newman","Paul Benn","Paul Woods","Kyle Lamb"])
webinar_feb=set(["Ashwin Agrawal","Jean Luca Bez","Rex Tanakit","Samuel Spencer","Shailesh Vaidya","Tripti Srivastava","Abraham Checkoway","Abhijeet Dhumal","Anatoliy Bilenko","Anthony Tocco","Antoine Le Bideau","Basavaraj Kirunge","BK Singh","Branislav Radovanovic","Charles Kunkel","Chetan Deshmukh","Carlos Thomaz","Dan Olster","Debashish Pal","Geert Wenes","Gary Grider","Gary Lowell","Jason Sliger-Sparks","Jean-Thomas","Justin Rackowski","Justin Woo","Kalpesh Chhajed","Keith Pine","Ken Haugen","Ketan Anil Arlulkar","Kiran Mangalore","Liang Gan","Madhavrao Vemuri","Mandar Sawant","Mark Jedraszek","Mehul Joshi","Mukul Malhotra","Nicolau Manubens","Nigel Hart","Nilesh Navale","Parag Joshi","Parks Fields","Paul Benn","Paul Woods","Peyton McNully","Prudence Huang","Philippe Nicolas","Pranali Ramdas Tirkhunde","Ryan Cassidy","Rob Wilson","Robert Read","Rohan Puri","Ryan Tyler","Sarang Sawant","Serkay Olmez","Shankar More","Seth Kindley","Swarajya Pendharkar","Sumedh Kulkarni","Sven Breuner","Sven Breuner","Theodore Omtzigt","Tim Coullter","Ravi Tripathi","Tushar Tarkas","Ujjwal Lanjewar","Venky P","Walter Lopatka","Earl Dodd","Wendell Wenjen","Weikuan Yu","George Zhi Qiao",])
jan_date='2021-01-06'
feb_date='2021-02-06'
dec_date='2020-12-21'
nov_date='2020-11-03'
#print(len(webinar_nov),len(webinar_dec),len(webinar_jan))
#webinar = [(nov_date,webinar_nov),(dec_date,webinar_dec),(jan_date,webinar_jan)]
webinar = [(feb_date,webinar_feb),(dec_date,webinar_dec),(jan_date,webinar_jan),(nov_date,webinar_nov)]
add_stats(webinar,webinar_key)
print(ps.get_values_as_numbers(repo,webinar_key))
def update_integrations():
# add the integrations metric for executive report
integrations = [(sep_date,0),(oct_date,1),(nov_date,1),(dec_date,6),(last_date,6)]
add_stats(integrations,integrations_key)
print(ps.get_values_as_numbers(repo,integrations_key))
def update_external(repo):
def update_external_single(repo,date,original,removals,key):
if original is None:
return
for r in removals:
try:
original -= r
except TypeError:
pass # can't subtract None from a set
ps.add_stat(repo=repo,date=date,stat=key,value=original)
# so we used to double count hackathon folks as external folks
# we fixed that but now the external counts suddenly unexpectedly dropped
# let's fix that in the historical record
# actually this should be easy
# just iterate through every date and subtract hackathon folks from external folks and resave the difference as external
#external participants
for date in dates:
ec = ps.get_values(repo, ec_key,[date])[0]
hc = ps.get_values(repo, hc_key,[date])[0]
ep = ps.get_values(repo, ep_key,[date])[0]
hp = ps.get_values(repo, hp_key,[date])[0]
eup = {'u-u-h', 'jayeshbadwaik'}
update_external_single(repo=repo,date=date,original=ec,removals=[hc], key=ec_key)
update_external_single(repo=repo,date=date,original=ep,removals=[hp,eup], key=ep_key)
def manually_add_historical_committers():
# for external committers also go through and manually add them in the early months
sep_committers=set(['cotigao','daniarherikurniawan','jan--f'])
oct_committers=sep_committers | set(['raikrahul'])
committers = [(jun_date,set()),(sep_date,sep_committers),(oct_date,oct_committers),('2020-12-20',oct_committers),('2020-12-21',oct_committers),('2020-12-23',oct_committers)]
add_stats(committers,ec_key)
#print(ps.get_values_as_numbers(repo, 'external_participants'))
def clean_external():
print(dates)
for r in ps.get_repos():
print("Will clean external committers and participants for %s" % r)
update_external(r)
print(ps.get_values_as_numbers(r, ec_key))
print(ps.get_values_as_numbers(r, ep_key))
print(ps.get_values(repo, ec_key))
update_webinar()
#print(ps.get_dates(repo))
update_newsletter()
ps.persist()
#manually_add_historical_committers()
#update_slack()
#update_webinar()
#update_integrations()
#for i in [3,4,5,10,11,12,20]:
# print(ps.get_dates('GLOBAL')[i], '->', ps.get_values_as_numbers('GLOBAL',webinar_key)[i])
def clean_external_participants():
ep1=ps.stats[repo][dates[4]][ep_key]
ep2=ps.stats[repo][dates[5]][ep_key]
print(len(ep1),len(ep2))
print(dates[4],ep1)
print(dates[5],ep2)
print(ep1-ep2)
ps.stats[repo][dates[5]][ep_key]=ep2|{'jan--f'}
ps.persist()
print(ps.get_values_as_numbers(repo, ep_key))
ep3=ps.stats[repo][dates[-1]][ep_key]
ep4=ps.stats[repo][dates[-2]][ep_key]
print(ep4-ep3)
#clean_external_participants()
def get_logins(Type):
folks=set()
people=cortx_community.CortxCommunity()
for p in people.values():
if p.type == Type:
folks.add(p.login)
return folks
# we once mistakenly characterized a few team folks as innersource folks
def clean_innersource(repo,Type,folks):
key='innersource_%s'%Type
for d in ps.get_dates(repo):
try:
values=ps.stats[repo][d][key]
except KeyError:
continue # some keys didn't always exist
if isinstance(values,set):
bad = [v for v in values if v not in folks]
if len(bad)>0:
print("Need to remove",bad,"from %s:%s on %s" % (repo,key,d))
new_values = values - set(bad)
print("Will reduce from %d to %d" % (len(values),len(new_values)))
ps.stats[repo][d][key]=new_values
def print_values(repo,key,dates=None):
for v in ps.get_values(repo,key,dates):
try:
print(len(v),sorted(v))
except:
print(v)
def bulk_clean():
for r in ps.get_repos():
for t in ('participants','committers'):
clean_innersource(repo=r,Type=t,folks=folks)
print(r,t,ps.get_values_as_numbers(r,'innersource_%s'%t))
folks=get_logins('Innersource')
print(folks)
#bulk_clean()
#ps.persist()
#print("all cleaned?!?!")
def clean_ip():
repo='GLOBAL'
key='innersource_participants'
dates=ps.get_dates(repo)
good_values=ps.get_values(repo=repo,key=key,dates=[dates[5]])[0]
for d in [dates[2],dates[3],dates[4]]:
ps.add_stat(date=d,repo=repo,stat=key,value=good_values)
print_values(repo,key,dates)
print(len(values),values)
def clean_ic():
repo='GLOBAL'
key='innersource_committers'
clean_innersource(repo,'committers',folks)
#clean_ip()
def print_innersource(Key):
for r in ps.get_repos():
print(r, ps.get_values_as_numbers(repo=r,key=Key))
clean_ic()
print_innersource('innersource_committers')
ps.persist()
compare_fields(ps=ps,repo='cortx',field='innersource_committers',verbose=False)
compare_fields(ps=ps,repo='cortx',field='external_email_addresses',verbose=False)
targets=['issues_closed_ave_age_in_s','issues_closed']
for target in targets:
for r in ['GLOBAL','cortx-ha','cortx-hare']:
print("%s %s -> %d " % (r, target, ps.stats[r]['2020-12-29'][target]))
check_scan_progress('2021-01-06',ps)
(a,b)=ps.get_latest('cortx-hare')
a['issues_open']
b
ps.stats['GLOBAL']['2021-01-02']['stars']
# this block is a one-time thing to add historical data from before we automated the scraping
d1={'innersource_participants' : 5, 'pull_requests_external' : 0,
'external_participants' : 0,
'watchers' : 34, 'stars' : 19, 'forks' : 13, 'views_unique_14_days' : 106,
'clones_count_14_days' : 38, 'clones_unique_14_days' : 4,
'seagate_blog_referrer_uniques' : 0, 'seagate_referrer_uniques' : 0,
'downloads_vms' : 0}
d1_date='2020-05-19'
d2={'innersource_participants' : 8, 'pull_requests_external' : 0,
'external_participants' : 0,
'watchers' : 69, 'stars' : 52, 'forks' : 42,
'views_unique_14_days' : 86,
'clones_count_14_days' : 15, 'clones_unique_14_days' : 6,
'seagate_blog_referrer_uniques' : 0, 'seagate_referrer_uniques' : 0,
'downloads_vms' : 0}
d2_date='2020-07-06'
d3={'innersource_participants' : 18, 'pull_requests_external' : 1,
'external_participants' : 0,
'watchers' : 62, 'stars' : 116, 'forks' : 31,
'views_unique_14_days' : 1817,
'clones_count_14_days' : 468, 'clones_unique_14_days' : 224,
'seagate_blog_referrer_uniques' : 0, 'seagate_referrer_uniques' : 0,
'downloads_vms' : 130}
d3_date='2020-10-07'
d4={'innersource_participants' : 18, 'pull_requests_external' : 4,
'external_participants' : 0,
'watchers' : 65, 'stars' : 159, 'forks' : 45,
'views_unique_14_days' : 817,
'clones_count_14_days' : 1851, 'clones_unique_14_days' : 259,
'seagate_blog_referrer_uniques' : 0, 'seagate_referrer_uniques' : 0,
'downloads_vms' : 363}
d4_date='2020-11-03'
print(d1)
#ps.add_stats(date=d1_date,repo='GLOBAL',stats=d1)
#ps.add_stats(date=d2_date,repo='GLOBAL',stats=d2)
#ps.add_stats(date=d3_date,repo='GLOBAL',stats=d3)
#ps.add_stats(date=d4_date,repo='GLOBAL',stats=d4)
ps.get_dates('GLOBAL')
```
|
github_jupyter
|
# Furniture Rearrangement - How to setup a new interaction task in Habitat-Lab
This tutorial demonstrates how to setup a new task in Habitat that utilizes interaction capabilities in Habitat Simulator.

## Task Definition:
The working example in this demo will be the task of **Furniture Rearrangement** - The agent will be randomly spawned in an environment in which the furniture are initially displaced from their desired position. The agent is tasked with navigating the environment, picking furniture and putting them in the desired position. To keep the tutorial simple and easy to follow, we will rearrange just a single object.
To setup this task, we will build on top of existing API in Habitat-Simulator and Habitat-Lab. Here is a summary of all the steps involved in setting up this task:
1. **Setup the Simulator**: Using existing functionalities of the Habitat-Sim, we can add or remove objects from the scene. We will use these methods to spawn the agent and the objects at some pre-defined initial configuration.
2. **Create a New Dataset**: We will define a new dataset class to save / load a list of episodes for the agent to train and evaluate on.
3. **Grab / Release Action**: We will add the "grab/release" action to the agent's action space to allow the agent to pickup / drop an object under a crosshair.
4. **Extend the Simulator Class**: We will extend the Simulator Class to add support for new actions implemented in previous step and add other additional utility functions
5. **Create a New Task**: Create a new task definition, implement new *sensors* and *metrics*.
6. **Train an RL agent**: We will define rewards for this task and utilize it to train an RL agent using the PPO algorithm.
Let's get started!
```
# @title Installation { display-mode: "form" }
# @markdown (double click to show code).
!curl -L https://raw.githubusercontent.com/facebookresearch/habitat-sim/master/examples/colab_utils/colab_install.sh | NIGHTLY=true bash -s
%cd /content
!gdown --id 1Pc-J6pZzXEd8RSeLM94t3iwO8q_RQ853
!unzip -o /content/coda.zip -d /content/habitat-sim/data/scene_datasets
# reload the cffi version
import sys
if "google.colab" in sys.modules:
import importlib
import cffi
importlib.reload(cffi)
# @title Path Setup and Imports { display-mode: "form" }
# @markdown (double click to show code).
%cd /content/habitat-lab
## [setup]
import gzip
import json
import os
import sys
from typing import Any, Dict, List, Optional, Type
import attr
import cv2
import git
import magnum as mn
import numpy as np
%matplotlib inline
from matplotlib import pyplot as plt
from PIL import Image
import habitat
import habitat_sim
from habitat.config import Config
from habitat.core.registry import registry
from habitat_sim.utils import viz_utils as vut
if "google.colab" in sys.modules:
os.environ["IMAGEIO_FFMPEG_EXE"] = "/usr/bin/ffmpeg"
repo = git.Repo(".", search_parent_directories=True)
dir_path = repo.working_tree_dir
%cd $dir_path
data_path = os.path.join(dir_path, "data")
output_directory = "data/tutorials/output/" # @param {type:"string"}
output_path = os.path.join(dir_path, output_directory)
if __name__ == "__main__":
import argparse
parser = argparse.ArgumentParser()
parser.add_argument("--no-display", dest="display", action="store_false")
parser.add_argument(
"--no-make-video", dest="make_video", action="store_false"
)
parser.set_defaults(show_video=True, make_video=True)
args, _ = parser.parse_known_args()
show_video = args.display
display = args.display
make_video = args.make_video
else:
show_video = False
make_video = False
display = False
if make_video and not os.path.exists(output_path):
os.makedirs(output_path)
# @title Util functions to visualize observations
# @markdown - `make_video_cv2`: Renders a video from a list of observations
# @markdown - `simulate`: Runs simulation for a given amount of time at 60Hz
# @markdown - `simulate_and_make_vid` Runs simulation and creates video
def make_video_cv2(
observations, cross_hair=None, prefix="", open_vid=True, fps=60
):
sensor_keys = list(observations[0])
videodims = observations[0][sensor_keys[0]].shape
videodims = (videodims[1], videodims[0]) # flip to w,h order
print(videodims)
video_file = output_path + prefix + ".mp4"
print("Encoding the video: %s " % video_file)
writer = vut.get_fast_video_writer(video_file, fps=fps)
for ob in observations:
# If in RGB/RGBA format, remove the alpha channel
rgb_im_1st_person = cv2.cvtColor(ob["rgb"], cv2.COLOR_RGBA2RGB)
if cross_hair is not None:
rgb_im_1st_person[
cross_hair[0] - 2 : cross_hair[0] + 2,
cross_hair[1] - 2 : cross_hair[1] + 2,
] = [255, 0, 0]
if rgb_im_1st_person.shape[:2] != videodims:
rgb_im_1st_person = cv2.resize(
rgb_im_1st_person, videodims, interpolation=cv2.INTER_AREA
)
# write the 1st person observation to video
writer.append_data(rgb_im_1st_person)
writer.close()
if open_vid:
print("Displaying video")
vut.display_video(video_file)
def simulate(sim, dt=1.0, get_frames=True):
# simulate dt seconds at 60Hz to the nearest fixed timestep
print("Simulating " + str(dt) + " world seconds.")
observations = []
start_time = sim.get_world_time()
while sim.get_world_time() < start_time + dt:
sim.step_physics(1.0 / 60.0)
if get_frames:
observations.append(sim.get_sensor_observations())
return observations
# convenience wrapper for simulate and make_video_cv2
def simulate_and_make_vid(sim, crosshair, prefix, dt=1.0, open_vid=True):
observations = simulate(sim, dt)
make_video_cv2(observations, crosshair, prefix=prefix, open_vid=open_vid)
def display_sample(
rgb_obs,
semantic_obs=np.array([]),
depth_obs=np.array([]),
key_points=None, # noqa: B006
):
from habitat_sim.utils.common import d3_40_colors_rgb
rgb_img = Image.fromarray(rgb_obs, mode="RGB")
arr = [rgb_img]
titles = ["rgb"]
if semantic_obs.size != 0:
semantic_img = Image.new(
"P", (semantic_obs.shape[1], semantic_obs.shape[0])
)
semantic_img.putpalette(d3_40_colors_rgb.flatten())
semantic_img.putdata((semantic_obs.flatten() % 40).astype(np.uint8))
semantic_img = semantic_img.convert("RGBA")
arr.append(semantic_img)
titles.append("semantic")
if depth_obs.size != 0:
depth_img = Image.fromarray(
(depth_obs / 10 * 255).astype(np.uint8), mode="L"
)
arr.append(depth_img)
titles.append("depth")
plt.figure(figsize=(12, 8))
for i, data in enumerate(arr):
ax = plt.subplot(1, 3, i + 1)
ax.axis("off")
ax.set_title(titles[i])
# plot points on images
if key_points is not None:
for point in key_points:
plt.plot(
point[0], point[1], marker="o", markersize=10, alpha=0.8
)
plt.imshow(data)
plt.show(block=False)
```
## 1. Setup the Simulator
---
```
# @title Setup simulator configuration
# @markdown We'll start with setting up simulator with the following configurations
# @markdown - The simulator will render both RGB, Depth observations of 256x256 resolution.
# @markdown - The actions available will be `move_forward`, `turn_left`, `turn_right`.
def make_cfg(settings):
sim_cfg = habitat_sim.SimulatorConfiguration()
sim_cfg.gpu_device_id = 0
sim_cfg.default_agent_id = settings["default_agent_id"]
sim_cfg.scene.id = settings["scene"]
sim_cfg.enable_physics = settings["enable_physics"]
sim_cfg.physics_config_file = settings["physics_config_file"]
# Note: all sensors must have the same resolution
sensors = {
"rgb": {
"sensor_type": habitat_sim.SensorType.COLOR,
"resolution": [settings["height"], settings["width"]],
"position": [0.0, settings["sensor_height"], 0.0],
},
"depth": {
"sensor_type": habitat_sim.SensorType.DEPTH,
"resolution": [settings["height"], settings["width"]],
"position": [0.0, settings["sensor_height"], 0.0],
},
}
sensor_specs = []
for sensor_uuid, sensor_params in sensors.items():
if settings[sensor_uuid]:
sensor_spec = habitat_sim.SensorSpec()
sensor_spec.uuid = sensor_uuid
sensor_spec.sensor_type = sensor_params["sensor_type"]
sensor_spec.resolution = sensor_params["resolution"]
sensor_spec.position = sensor_params["position"]
sensor_specs.append(sensor_spec)
# Here you can specify the amount of displacement in a forward action and the turn angle
agent_cfg = habitat_sim.agent.AgentConfiguration()
agent_cfg.sensor_specifications = sensor_specs
agent_cfg.action_space = {
"move_forward": habitat_sim.agent.ActionSpec(
"move_forward", habitat_sim.agent.ActuationSpec(amount=0.1)
),
"turn_left": habitat_sim.agent.ActionSpec(
"turn_left", habitat_sim.agent.ActuationSpec(amount=10.0)
),
"turn_right": habitat_sim.agent.ActionSpec(
"turn_right", habitat_sim.agent.ActuationSpec(amount=10.0)
),
}
return habitat_sim.Configuration(sim_cfg, [agent_cfg])
settings = {
"max_frames": 10,
"width": 256,
"height": 256,
"scene": "data/scene_datasets/coda/coda.glb",
"default_agent_id": 0,
"sensor_height": 1.5, # Height of sensors in meters
"rgb": True, # RGB sensor
"depth": True, # Depth sensor
"seed": 1,
"enable_physics": True,
"physics_config_file": "data/default.phys_scene_config.json",
"silent": False,
"compute_shortest_path": False,
"compute_action_shortest_path": False,
"save_png": True,
}
cfg = make_cfg(settings)
# @title Spawn the agent at a pre-defined location
def init_agent(sim):
agent_pos = np.array([-0.15776923, 0.18244143, 0.2988735])
# Place the agent
sim.agents[0].scene_node.translation = agent_pos
agent_orientation_y = -40
sim.agents[0].scene_node.rotation = mn.Quaternion.rotation(
mn.Deg(agent_orientation_y), mn.Vector3(0, 1.0, 0)
)
cfg.sim_cfg.default_agent_id = 0
with habitat_sim.Simulator(cfg) as sim:
init_agent(sim)
if make_video:
# Visualize the agent's initial position
simulate_and_make_vid(
sim, None, "sim-init", dt=1.0, open_vid=show_video
)
# @title Set the object's initial and final position
# @markdown Defines two utility functions:
# @markdown - `remove_all_objects`: This will remove all objects from the scene
# @markdown - `set_object_in_front_of_agent`: This will add an object in the scene in front of the agent at the specified distance.
# @markdown Here we add a chair *3.0m* away from the agent and the task is to place the agent at the desired final position which is *7.0m* in front of the agent.
def remove_all_objects(sim):
for id in sim.get_existing_object_ids():
sim.remove_object(id)
def set_object_in_front_of_agent(sim, obj_id, z_offset=-1.5):
r"""
Adds an object in front of the agent at some distance.
"""
agent_transform = sim.agents[0].scene_node.transformation_matrix()
obj_translation = agent_transform.transform_point(
np.array([0, 0, z_offset])
)
sim.set_translation(obj_translation, obj_id)
obj_node = sim.get_object_scene_node(obj_id)
xform_bb = habitat_sim.geo.get_transformed_bb(
obj_node.cumulative_bb, obj_node.transformation
)
# also account for collision margin of the scene
scene_collision_margin = 0.04
y_translation = mn.Vector3(
0, xform_bb.size_y() / 2.0 + scene_collision_margin, 0
)
sim.set_translation(y_translation + sim.get_translation(obj_id), obj_id)
def init_objects(sim):
# Manager of Object Attributes Templates
obj_attr_mgr = sim.get_object_template_manager()
# Add a chair into the scene.
obj_path = "test_assets/objects/chair"
chair_template_id = obj_attr_mgr.load_object_configs(
str(os.path.join(data_path, obj_path))
)[0]
chair_attr = obj_attr_mgr.get_template_by_ID(chair_template_id)
obj_attr_mgr.register_template(chair_attr)
# Object's initial position 3m away from the agent.
object_id = sim.add_object_by_handle(chair_attr.handle)
set_object_in_front_of_agent(sim, object_id, -3.0)
sim.set_object_motion_type(
habitat_sim.physics.MotionType.STATIC, object_id
)
# Object's final position 7m away from the agent
goal_id = sim.add_object_by_handle(chair_attr.handle)
set_object_in_front_of_agent(sim, goal_id, -7.0)
sim.set_object_motion_type(habitat_sim.physics.MotionType.STATIC, goal_id)
return object_id, goal_id
with habitat_sim.Simulator(cfg) as sim:
init_agent(sim)
init_objects(sim)
# Visualize the scene after the chair is added into the scene.
if make_video:
simulate_and_make_vid(
sim, None, "object-init", dt=1.0, open_vid=show_video
)
```
## Rearrangement Dataset

In the previous section, we created a single episode of the rearrangement task. Let's define a format to store all the necessary information about a single episode. It should store the *scene* the episode belongs to, *initial spawn position and orientation* of the agent, *object type*, object's *initial position and orientation* as well as *final position and orientation*.
The format will be as follows:
```
{
'episode_id': 0,
'scene_id': 'data/scene_datasets/coda/coda.glb',
'goals': {
'position': [4.34, 0.67, -5.06],
'rotation': [0.0, 0.0, 0.0, 1.0]
},
'objects': {
'object_id': 0,
'object_template': 'data/test_assets/objects/chair',
'position': [1.77, 0.67, -1.99],
'rotation': [0.0, 0.0, 0.0, 1.0]
},
'start_position': [-0.15, 0.18, 0.29],
'start_rotation': [-0.0, -0.34, -0.0, 0.93]}
}
```
Once an episode is defined, a dataset will just be a collection of such episodes. For simplicity, in this notebook, the dataset will only contain one episode defined above.
```
# @title Create a new dataset
# @markdown Utility functions to define and save the dataset for the rearrangement task
def get_rotation(sim, object_id):
quat = sim.get_rotation(object_id)
return np.array(quat.vector).tolist() + [quat.scalar]
def init_episode_dict(episode_id, scene_id, agent_pos, agent_rot):
episode_dict = {
"episode_id": episode_id,
"scene_id": "data/scene_datasets/coda/coda.glb",
"start_position": agent_pos,
"start_rotation": agent_rot,
"info": {},
}
return episode_dict
def add_object_details(sim, episode_dict, id, object_template, object_id):
object_template = {
"object_id": id,
"object_template": object_template,
"position": np.array(sim.get_translation(object_id)).tolist(),
"rotation": get_rotation(sim, object_id),
}
episode_dict["objects"] = object_template
return episode_dict
def add_goal_details(sim, episode_dict, object_id):
goal_template = {
"position": np.array(sim.get_translation(object_id)).tolist(),
"rotation": get_rotation(sim, object_id),
}
episode_dict["goals"] = goal_template
return episode_dict
# set the number of objects to 1 always for now.
def build_episode(sim, episode_num, object_id, goal_id):
episodes = {"episodes": []}
for episode in range(episode_num):
agent_state = sim.get_agent(0).get_state()
agent_pos = np.array(agent_state.position).tolist()
agent_quat = agent_state.rotation
agent_rot = np.array(agent_quat.vec).tolist() + [agent_quat.real]
episode_dict = init_episode_dict(
episode, settings["scene"], agent_pos, agent_rot
)
object_attr = sim.get_object_initialization_template(object_id)
object_path = os.path.relpath(
os.path.splitext(object_attr.render_asset_handle)[0]
)
episode_dict = add_object_details(
sim, episode_dict, 0, object_path, object_id
)
episode_dict = add_goal_details(sim, episode_dict, goal_id)
episodes["episodes"].append(episode_dict)
return episodes
with habitat_sim.Simulator(cfg) as sim:
init_agent(sim)
object_id, goal_id = init_objects(sim)
episodes = build_episode(sim, 1, object_id, goal_id)
dataset_content_path = "data/datasets/rearrangement/coda/v1/train/"
if not os.path.exists(dataset_content_path):
os.makedirs(dataset_content_path)
with gzip.open(
os.path.join(dataset_content_path, "train.json.gz"), "wt"
) as f:
json.dump(episodes, f)
print(
"Dataset written to {}".format(
os.path.join(dataset_content_path, "train.json.gz")
)
)
# @title Dataset class to read the saved dataset in Habitat-Lab.
# @markdown To read the saved episodes in Habitat-Lab, we will extend the `Dataset` class and the `Episode` base class. It will help provide all the relevant details about the episode through a consistent API to all downstream tasks.
# @markdown - We will first create a `RearrangementEpisode` by extending the `NavigationEpisode` to include additional information about object's initial configuration and desired final configuration.
# @markdown - We will then define a `RearrangementDatasetV0` class that builds on top of `PointNavDatasetV1` class to read the JSON file stored earlier and initialize a list of `RearrangementEpisode`.
from habitat.core.utils import DatasetFloatJSONEncoder, not_none_validator
from habitat.datasets.pointnav.pointnav_dataset import (
CONTENT_SCENES_PATH_FIELD,
DEFAULT_SCENE_PATH_PREFIX,
PointNavDatasetV1,
)
from habitat.tasks.nav.nav import NavigationEpisode
@attr.s(auto_attribs=True, kw_only=True)
class RearrangementSpec:
r"""Specifications that capture a particular position of final position
or initial position of the object.
"""
position: List[float] = attr.ib(default=None, validator=not_none_validator)
rotation: List[float] = attr.ib(default=None, validator=not_none_validator)
info: Optional[Dict[str, str]] = attr.ib(default=None)
@attr.s(auto_attribs=True, kw_only=True)
class RearrangementObjectSpec(RearrangementSpec):
r"""Object specifications that capture position of each object in the scene,
the associated object template.
"""
object_id: str = attr.ib(default=None, validator=not_none_validator)
object_template: Optional[str] = attr.ib(
default="data/test_assets/objects/chair"
)
@attr.s(auto_attribs=True, kw_only=True)
class RearrangementEpisode(NavigationEpisode):
r"""Specification of episode that includes initial position and rotation
of agent, all goal specifications, all object specifications
Args:
episode_id: id of episode in the dataset
scene_id: id of scene inside the simulator.
start_position: numpy ndarray containing 3 entries for (x, y, z).
start_rotation: numpy ndarray with 4 entries for (x, y, z, w)
elements of unit quaternion (versor) representing agent 3D
orientation.
goal: object's goal position and rotation
object: object's start specification defined with object type,
position, and rotation.
"""
objects: RearrangementObjectSpec = attr.ib(
default=None, validator=not_none_validator
)
goals: RearrangementSpec = attr.ib(
default=None, validator=not_none_validator
)
@registry.register_dataset(name="RearrangementDataset-v0")
class RearrangementDatasetV0(PointNavDatasetV1):
r"""Class inherited from PointNavDataset that loads Rearrangement dataset."""
episodes: List[RearrangementEpisode]
content_scenes_path: str = "{data_path}/content/{scene}.json.gz"
def to_json(self) -> str:
result = DatasetFloatJSONEncoder().encode(self)
return result
def __init__(self, config: Optional[Config] = None) -> None:
super().__init__(config)
def from_json(
self, json_str: str, scenes_dir: Optional[str] = None
) -> None:
deserialized = json.loads(json_str)
if CONTENT_SCENES_PATH_FIELD in deserialized:
self.content_scenes_path = deserialized[CONTENT_SCENES_PATH_FIELD]
for i, episode in enumerate(deserialized["episodes"]):
rearrangement_episode = RearrangementEpisode(**episode)
rearrangement_episode.episode_id = str(i)
if scenes_dir is not None:
if rearrangement_episode.scene_id.startswith(
DEFAULT_SCENE_PATH_PREFIX
):
rearrangement_episode.scene_id = (
rearrangement_episode.scene_id[
len(DEFAULT_SCENE_PATH_PREFIX) :
]
)
rearrangement_episode.scene_id = os.path.join(
scenes_dir, rearrangement_episode.scene_id
)
rearrangement_episode.objects = RearrangementObjectSpec(
**rearrangement_episode.objects
)
rearrangement_episode.goals = RearrangementSpec(
**rearrangement_episode.goals
)
self.episodes.append(rearrangement_episode)
# @title Load the saved dataset using the Dataset class
config = habitat.get_config("configs/datasets/pointnav/habitat_test.yaml")
config.defrost()
config.DATASET.DATA_PATH = (
"data/datasets/rearrangement/coda/v1/{split}/{split}.json.gz"
)
config.DATASET.TYPE = "RearrangementDataset-v0"
config.freeze()
dataset = RearrangementDatasetV0(config.DATASET)
# check if the dataset got correctly deserialized
assert len(dataset.episodes) == 1
assert dataset.episodes[0].objects.position == [
1.770593523979187,
0.6726829409599304,
-1.9992598295211792,
]
assert dataset.episodes[0].objects.rotation == [0.0, 0.0, 0.0, 1.0]
assert (
dataset.episodes[0].objects.object_template
== "data/test_assets/objects/chair"
)
assert dataset.episodes[0].goals.position == [
4.3417439460754395,
0.6726829409599304,
-5.0634379386901855,
]
assert dataset.episodes[0].goals.rotation == [0.0, 0.0, 0.0, 1.0]
```
## Implement Grab/Release Action
```
# @title RayCast utility to implement Grab/Release Under Cross-Hair Action
# @markdown Cast a ray in the direction of crosshair from the camera and check if it collides with another object within a certain distance threshold
def raycast(sim, sensor_name, crosshair_pos=(128, 128), max_distance=2.0):
r"""Cast a ray in the direction of crosshair and check if it collides
with another object within a certain distance threshold
:param sim: Simulator object
:param sensor_name: name of the visual sensor to be used for raycasting
:param crosshair_pos: 2D coordiante in the viewport towards which the
ray will be cast
:param max_distance: distance threshold beyond which objects won't
be considered
"""
visual_sensor = sim._sensors[sensor_name]
scene_graph = sim.get_active_scene_graph()
scene_graph.set_default_render_camera_parameters(
visual_sensor._sensor_object
)
render_camera = scene_graph.get_default_render_camera()
center_ray = render_camera.unproject(mn.Vector2i(crosshair_pos))
raycast_results = sim.cast_ray(center_ray, max_distance=max_distance)
closest_object = -1
closest_dist = 1000.0
if raycast_results.has_hits():
for hit in raycast_results.hits:
if hit.ray_distance < closest_dist:
closest_dist = hit.ray_distance
closest_object = hit.object_id
return closest_object
# Test the raycast utility.
with habitat_sim.Simulator(cfg) as sim:
init_agent(sim)
obj_attr_mgr = sim.get_object_template_manager()
obj_path = "test_assets/objects/chair"
chair_template_id = obj_attr_mgr.load_object_configs(
str(os.path.join(data_path, obj_path))
)[0]
chair_attr = obj_attr_mgr.get_template_by_ID(chair_template_id)
obj_attr_mgr.register_template(chair_attr)
object_id = sim.add_object_by_handle(chair_attr.handle)
print(f"Chair's object id is {object_id}")
set_object_in_front_of_agent(sim, object_id, -1.5)
sim.set_object_motion_type(
habitat_sim.physics.MotionType.STATIC, object_id
)
if make_video:
# Visualize the agent's initial position
simulate_and_make_vid(
sim, [190, 128], "sim-before-grab", dt=1.0, open_vid=show_video
)
# Distance threshold=2 is greater than agent-to-chair distance.
# Should return chair's object id
closest_object = raycast(
sim, "rgb", crosshair_pos=[128, 190], max_distance=2.0
)
print(f"Closest Object ID: {closest_object} using 2.0 threshold")
assert (
closest_object == object_id
), f"Could not pick chair with ID: {object_id}"
# Distance threshold=1 is smaller than agent-to-chair distance .
# Should return -1
closest_object = raycast(
sim, "rgb", crosshair_pos=[128, 190], max_distance=1.0
)
print(f"Closest Object ID: {closest_object} using 1.0 threshold")
assert closest_object == -1, "Agent shoud not be able to pick any object"
# @title Define a Grab/Release action and create a new action space.
# @markdown Each new action is defined by a `ActionSpec` and an `ActuationSpec`. `ActionSpec` is mapping between the action name and its corresponding `ActuationSpec`. `ActuationSpec` contains all the necessary specifications required to define the action.
from habitat.config.default import _C, CN
from habitat.core.embodied_task import SimulatorTaskAction
from habitat.sims.habitat_simulator.actions import (
HabitatSimActions,
HabitatSimV1ActionSpaceConfiguration,
)
from habitat_sim.agent.controls.controls import ActuationSpec
from habitat_sim.physics import MotionType
# @markdown For instance, `GrabReleaseActuationSpec` contains the following:
# @markdown - `visual_sensor_name` defines which viewport (rgb, depth, etc) to to use to cast the ray.
# @markdown - `crosshair_pos` stores the position in the viewport through which the ray passes. Any object which intersects with this ray can be grabbed by the agent.
# @markdown - `amount` defines a distance threshold. Objects which are farther than the treshold cannot be picked up by the agent.
@attr.s(auto_attribs=True, slots=True)
class GrabReleaseActuationSpec(ActuationSpec):
visual_sensor_name: str = "rgb"
crosshair_pos: List[int] = [128, 128]
amount: float = 2.0
# @markdown Then, we extend the `HabitatSimV1ActionSpaceConfiguration` to add the above action into the agent's action space. `ActionSpaceConfiguration` is a mapping between action name and the corresponding `ActionSpec`
@registry.register_action_space_configuration(name="RearrangementActions-v0")
class RearrangementSimV0ActionSpaceConfiguration(
HabitatSimV1ActionSpaceConfiguration
):
def __init__(self, config):
super().__init__(config)
if not HabitatSimActions.has_action("GRAB_RELEASE"):
HabitatSimActions.extend_action_space("GRAB_RELEASE")
def get(self):
config = super().get()
new_config = {
HabitatSimActions.GRAB_RELEASE: habitat_sim.ActionSpec(
"grab_or_release_object_under_crosshair",
GrabReleaseActuationSpec(
visual_sensor_name=self.config.VISUAL_SENSOR,
crosshair_pos=self.config.CROSSHAIR_POS,
amount=self.config.GRAB_DISTANCE,
),
)
}
config.update(new_config)
return config
# @markdown Finally, we extend `SimualtorTaskAction` which tells the simulator which action to call when a named action ('GRAB_RELEASE' in this case) is predicte by the agent's policy.
@registry.register_task_action
class GrabOrReleaseAction(SimulatorTaskAction):
def step(self, *args: Any, **kwargs: Any):
r"""This method is called from ``Env`` on each ``step``."""
return self._sim.step(HabitatSimActions.GRAB_RELEASE)
_C.TASK.ACTIONS.GRAB_RELEASE = CN()
_C.TASK.ACTIONS.GRAB_RELEASE.TYPE = "GrabOrReleaseAction"
_C.SIMULATOR.CROSSHAIR_POS = [128, 160]
_C.SIMULATOR.GRAB_DISTANCE = 2.0
_C.SIMULATOR.VISUAL_SENSOR = "rgb"
```
##Setup Simulator Class for Rearrangement Task

```
# @title RearrangementSim Class
# @markdown Here we will extend the `HabitatSim` class for the rearrangement task. We will make the following changes:
# @markdown - define a new `_initialize_objects` function which will load the object in its initial configuration as defined by the episode.
# @markdown - define a `gripped_object_id` property that stores whether the agent is holding any object or not.
# @markdown - modify the `step` function of the simulator to use the `grab/release` action we define earlier.
# @markdown #### Writing the `step` function:
# @markdown Since we added a new action for this task, we have to modify the `step` function to define what happens when `grab/release` action is called. If a simple navigation action (`move_forward`, `turn_left`, `turn_right`) is called, we pass it forward to `act` function of the agent which already defines the behavior of these actions.
# @markdown For the `grab/release` action, if the agent is not already holding an object, we first call the `raycast` function using the values from the `ActuationSpec` to see if any object is grippable. If it returns a valid object id, we put the object in a "invisible" inventory and remove it from the scene.
# @markdown If the agent was already holding an object, `grab/release` action will try release the object at the same relative position as it was grabbed. If the object can be placed without any collision, then the `release` action is successful.
from habitat.sims.habitat_simulator.habitat_simulator import HabitatSim
from habitat_sim.nav import NavMeshSettings
from habitat_sim.utils.common import quat_from_coeffs, quat_to_magnum
@registry.register_simulator(name="RearrangementSim-v0")
class RearrangementSim(HabitatSim):
r"""Simulator wrapper over habitat-sim with
object rearrangement functionalities.
"""
def __init__(self, config: Config) -> None:
self.did_reset = False
super().__init__(config=config)
self.grip_offset = np.eye(4)
agent_id = self.habitat_config.DEFAULT_AGENT_ID
agent_config = self._get_agent_config(agent_id)
self.navmesh_settings = NavMeshSettings()
self.navmesh_settings.set_defaults()
self.navmesh_settings.agent_radius = agent_config.RADIUS
self.navmesh_settings.agent_height = agent_config.HEIGHT
def reconfigure(self, config: Config) -> None:
super().reconfigure(config)
self._initialize_objects()
def reset(self):
sim_obs = super().reset()
if self._update_agents_state():
sim_obs = self.get_sensor_observations()
self._prev_sim_obs = sim_obs
self.did_reset = True
self.grip_offset = np.eye(4)
return self._sensor_suite.get_observations(sim_obs)
def _initialize_objects(self):
objects = self.habitat_config.objects[0]
obj_attr_mgr = self.get_object_template_manager()
# first remove all existing objects
existing_object_ids = self.get_existing_object_ids()
if len(existing_object_ids) > 0:
for obj_id in existing_object_ids:
self.remove_object(obj_id)
self.sim_object_to_objid_mapping = {}
self.objid_to_sim_object_mapping = {}
if objects is not None:
object_template = objects["object_template"]
object_pos = objects["position"]
object_rot = objects["rotation"]
object_template_id = obj_attr_mgr.load_object_configs(
object_template
)[0]
object_attr = obj_attr_mgr.get_template_by_ID(object_template_id)
obj_attr_mgr.register_template(object_attr)
object_id = self.add_object_by_handle(object_attr.handle)
self.sim_object_to_objid_mapping[object_id] = objects["object_id"]
self.objid_to_sim_object_mapping[objects["object_id"]] = object_id
self.set_translation(object_pos, object_id)
if isinstance(object_rot, list):
object_rot = quat_from_coeffs(object_rot)
object_rot = quat_to_magnum(object_rot)
self.set_rotation(object_rot, object_id)
self.set_object_motion_type(MotionType.STATIC, object_id)
# Recompute the navmesh after placing all the objects.
self.recompute_navmesh(self.pathfinder, self.navmesh_settings, True)
def _sync_gripped_object(self, gripped_object_id):
r"""
Sync the gripped object with the object associated with the agent.
"""
if gripped_object_id != -1:
agent_body_transformation = (
self._default_agent.scene_node.transformation
)
self.set_transformation(
agent_body_transformation, gripped_object_id
)
translation = agent_body_transformation.transform_point(
np.array([0, 2.0, 0])
)
self.set_translation(translation, gripped_object_id)
@property
def gripped_object_id(self):
return self._prev_sim_obs.get("gripped_object_id", -1)
def step(self, action: int):
dt = 1 / 60.0
self._num_total_frames += 1
collided = False
gripped_object_id = self.gripped_object_id
agent_config = self._default_agent.agent_config
action_spec = agent_config.action_space[action]
if action_spec.name == "grab_or_release_object_under_crosshair":
# If already holding an agent
if gripped_object_id != -1:
agent_body_transformation = (
self._default_agent.scene_node.transformation
)
T = np.dot(agent_body_transformation, self.grip_offset)
self.set_transformation(T, gripped_object_id)
position = self.get_translation(gripped_object_id)
if self.pathfinder.is_navigable(position):
self.set_object_motion_type(
MotionType.STATIC, gripped_object_id
)
gripped_object_id = -1
self.recompute_navmesh(
self.pathfinder, self.navmesh_settings, True
)
# if not holding an object, then try to grab
else:
gripped_object_id = raycast(
self,
action_spec.actuation.visual_sensor_name,
crosshair_pos=action_spec.actuation.crosshair_pos,
max_distance=action_spec.actuation.amount,
)
# found a grabbable object.
if gripped_object_id != -1:
agent_body_transformation = (
self._default_agent.scene_node.transformation
)
self.grip_offset = np.dot(
np.array(agent_body_transformation.inverted()),
np.array(self.get_transformation(gripped_object_id)),
)
self.set_object_motion_type(
MotionType.KINEMATIC, gripped_object_id
)
self.recompute_navmesh(
self.pathfinder, self.navmesh_settings, True
)
else:
collided = self._default_agent.act(action)
self._last_state = self._default_agent.get_state()
# step physics by dt
super().step_world(dt)
# Sync the gripped object after the agent moves.
self._sync_gripped_object(gripped_object_id)
# obtain observations
self._prev_sim_obs = self.get_sensor_observations()
self._prev_sim_obs["collided"] = collided
self._prev_sim_obs["gripped_object_id"] = gripped_object_id
observations = self._sensor_suite.get_observations(self._prev_sim_obs)
return observations
```
## Create the Rearrangement Task

```
# @title Implement new sensors and measurements
# @markdown After defining the dataset, action space and simulator functions for the rearrangement task, we are one step closer to training agents to solve this task.
# @markdown Here we define inputs to the policy and other measurements required to design reward functions.
# @markdown **Sensors**: These define various part of the simulator state that's visible to the agent. For simplicity, we'll assume that agent knows the object's current position, object's final goal position relative to the agent's current position.
# @markdown - Object's current position will be made given by the `ObjectPosition` sensor
# @markdown - Object's goal position will be available through the `ObjectGoal` sensor.
# @markdown - Finally, we will also use `GrippedObject` sensor to tell the agent if it's holding any object or not.
# @markdown **Measures**: These define various metrics about the task which can be used to measure task progress and define rewards. Note that measurements are *privileged* information not accessible to the agent as part of the observation space. We will need the following measurements:
# @markdown - `AgentToObjectDistance` which measure the euclidean distance between the agent and the object.
# @markdown - `ObjectToGoalDistance` which measures the euclidean distance between the object and the goal.
from gym import spaces
import habitat_sim
from habitat.config.default import CN, Config
from habitat.core.dataset import Episode
from habitat.core.embodied_task import Measure
from habitat.core.simulator import Observations, Sensor, SensorTypes, Simulator
from habitat.tasks.nav.nav import PointGoalSensor
@registry.register_sensor
class GrippedObjectSensor(Sensor):
cls_uuid = "gripped_object_id"
def __init__(
self, *args: Any, sim: RearrangementSim, config: Config, **kwargs: Any
):
self._sim = sim
super().__init__(config=config)
def _get_uuid(self, *args: Any, **kwargs: Any) -> str:
return self.cls_uuid
def _get_observation_space(self, *args: Any, **kwargs: Any):
return spaces.Discrete(self._sim.get_existing_object_ids())
def _get_sensor_type(self, *args: Any, **kwargs: Any):
return SensorTypes.MEASUREMENT
def get_observation(
self,
observations: Dict[str, Observations],
episode: Episode,
*args: Any,
**kwargs: Any,
):
obj_id = self._sim.sim_object_to_objid_mapping.get(
self._sim.gripped_object_id, -1
)
return obj_id
@registry.register_sensor
class ObjectPosition(PointGoalSensor):
cls_uuid: str = "object_position"
def _get_observation_space(self, *args: Any, **kwargs: Any):
sensor_shape = (self._dimensionality,)
return spaces.Box(
low=np.finfo(np.float32).min,
high=np.finfo(np.float32).max,
shape=sensor_shape,
dtype=np.float32,
)
def get_observation(
self, *args: Any, observations, episode, **kwargs: Any
):
agent_state = self._sim.get_agent_state()
agent_position = agent_state.position
rotation_world_agent = agent_state.rotation
object_id = self._sim.get_existing_object_ids()[0]
object_position = self._sim.get_translation(object_id)
pointgoal = self._compute_pointgoal(
agent_position, rotation_world_agent, object_position
)
return pointgoal
@registry.register_sensor
class ObjectGoal(PointGoalSensor):
cls_uuid: str = "object_goal"
def _get_observation_space(self, *args: Any, **kwargs: Any):
sensor_shape = (self._dimensionality,)
return spaces.Box(
low=np.finfo(np.float32).min,
high=np.finfo(np.float32).max,
shape=sensor_shape,
dtype=np.float32,
)
def get_observation(
self, *args: Any, observations, episode, **kwargs: Any
):
agent_state = self._sim.get_agent_state()
agent_position = agent_state.position
rotation_world_agent = agent_state.rotation
goal_position = np.array(episode.goals.position, dtype=np.float32)
point_goal = self._compute_pointgoal(
agent_position, rotation_world_agent, goal_position
)
return point_goal
@registry.register_measure
class ObjectToGoalDistance(Measure):
"""The measure calculates distance of object towards the goal."""
cls_uuid: str = "object_to_goal_distance"
def __init__(
self, sim: Simulator, config: Config, *args: Any, **kwargs: Any
):
self._sim = sim
self._config = config
super().__init__(**kwargs)
@staticmethod
def _get_uuid(*args: Any, **kwargs: Any):
return ObjectToGoalDistance.cls_uuid
def reset_metric(self, episode, *args: Any, **kwargs: Any):
self.update_metric(*args, episode=episode, **kwargs)
def _geo_dist(self, src_pos, goal_pos: np.array) -> float:
return self._sim.geodesic_distance(src_pos, [goal_pos])
def _euclidean_distance(self, position_a, position_b):
return np.linalg.norm(
np.array(position_b) - np.array(position_a), ord=2
)
def update_metric(self, episode, *args: Any, **kwargs: Any):
sim_obj_id = self._sim.get_existing_object_ids()[0]
previous_position = np.array(
self._sim.get_translation(sim_obj_id)
).tolist()
goal_position = episode.goals.position
self._metric = self._euclidean_distance(
previous_position, goal_position
)
@registry.register_measure
class AgentToObjectDistance(Measure):
"""The measure calculates the distance of objects from the agent"""
cls_uuid: str = "agent_to_object_distance"
def __init__(
self, sim: Simulator, config: Config, *args: Any, **kwargs: Any
):
self._sim = sim
self._config = config
super().__init__(**kwargs)
@staticmethod
def _get_uuid(*args: Any, **kwargs: Any):
return AgentToObjectDistance.cls_uuid
def reset_metric(self, episode, *args: Any, **kwargs: Any):
self.update_metric(*args, episode=episode, **kwargs)
def _euclidean_distance(self, position_a, position_b):
return np.linalg.norm(
np.array(position_b) - np.array(position_a), ord=2
)
def update_metric(self, episode, *args: Any, **kwargs: Any):
sim_obj_id = self._sim.get_existing_object_ids()[0]
previous_position = np.array(
self._sim.get_translation(sim_obj_id)
).tolist()
agent_state = self._sim.get_agent_state()
agent_position = agent_state.position
self._metric = self._euclidean_distance(
previous_position, agent_position
)
# -----------------------------------------------------------------------------
# # REARRANGEMENT TASK GRIPPED OBJECT SENSOR
# -----------------------------------------------------------------------------
_C.TASK.GRIPPED_OBJECT_SENSOR = CN()
_C.TASK.GRIPPED_OBJECT_SENSOR.TYPE = "GrippedObjectSensor"
# -----------------------------------------------------------------------------
# # REARRANGEMENT TASK ALL OBJECT POSITIONS SENSOR
# -----------------------------------------------------------------------------
_C.TASK.OBJECT_POSITION = CN()
_C.TASK.OBJECT_POSITION.TYPE = "ObjectPosition"
_C.TASK.OBJECT_POSITION.GOAL_FORMAT = "POLAR"
_C.TASK.OBJECT_POSITION.DIMENSIONALITY = 2
# -----------------------------------------------------------------------------
# # REARRANGEMENT TASK ALL OBJECT GOALS SENSOR
# -----------------------------------------------------------------------------
_C.TASK.OBJECT_GOAL = CN()
_C.TASK.OBJECT_GOAL.TYPE = "ObjectGoal"
_C.TASK.OBJECT_GOAL.GOAL_FORMAT = "POLAR"
_C.TASK.OBJECT_GOAL.DIMENSIONALITY = 2
# -----------------------------------------------------------------------------
# # OBJECT_DISTANCE_TO_GOAL MEASUREMENT
# -----------------------------------------------------------------------------
_C.TASK.OBJECT_TO_GOAL_DISTANCE = CN()
_C.TASK.OBJECT_TO_GOAL_DISTANCE.TYPE = "ObjectToGoalDistance"
# -----------------------------------------------------------------------------
# # OBJECT_DISTANCE_FROM_AGENT MEASUREMENT
# -----------------------------------------------------------------------------
_C.TASK.AGENT_TO_OBJECT_DISTANCE = CN()
_C.TASK.AGENT_TO_OBJECT_DISTANCE.TYPE = "AgentToObjectDistance"
from habitat.config.default import CN, Config
# @title Define `RearrangementTask` by extending `NavigationTask`
from habitat.tasks.nav.nav import NavigationTask, merge_sim_episode_config
def merge_sim_episode_with_object_config(
sim_config: Config, episode: Type[Episode]
) -> Any:
sim_config = merge_sim_episode_config(sim_config, episode)
sim_config.defrost()
sim_config.objects = [episode.objects.__dict__]
sim_config.freeze()
return sim_config
@registry.register_task(name="RearrangementTask-v0")
class RearrangementTask(NavigationTask):
r"""Embodied Rearrangement Task
Goal: An agent must place objects at their corresponding goal position.
"""
def __init__(self, **kwargs) -> None:
super().__init__(**kwargs)
def overwrite_sim_config(self, sim_config, episode):
return merge_sim_episode_with_object_config(sim_config, episode)
```
## Implement a hard-coded and an RL agent
```
# @title Load the `RearrangementTask` in Habitat-Lab and run a hard-coded agent
import habitat
config = habitat.get_config("configs/tasks/pointnav.yaml")
config.defrost()
config.ENVIRONMENT.MAX_EPISODE_STEPS = 50
config.SIMULATOR.TYPE = "RearrangementSim-v0"
config.SIMULATOR.ACTION_SPACE_CONFIG = "RearrangementActions-v0"
config.SIMULATOR.GRAB_DISTANCE = 2.0
config.SIMULATOR.HABITAT_SIM_V0.ENABLE_PHYSICS = True
config.TASK.TYPE = "RearrangementTask-v0"
config.TASK.SUCCESS_DISTANCE = 1.0
config.TASK.SENSORS = [
"GRIPPED_OBJECT_SENSOR",
"OBJECT_POSITION",
"OBJECT_GOAL",
]
config.TASK.GOAL_SENSOR_UUID = "object_goal"
config.TASK.MEASUREMENTS = [
"OBJECT_TO_GOAL_DISTANCE",
"AGENT_TO_OBJECT_DISTANCE",
]
config.TASK.POSSIBLE_ACTIONS = ["STOP", "MOVE_FORWARD", "GRAB_RELEASE"]
config.DATASET.TYPE = "RearrangementDataset-v0"
config.DATASET.SPLIT = "train"
config.DATASET.DATA_PATH = (
"data/datasets/rearrangement/coda/v1/{split}/{split}.json.gz"
)
config.freeze()
def print_info(obs, metrics):
print(
"Gripped Object: {}, Distance To Object: {}, Distance To Goal: {}".format(
obs["gripped_object_id"],
metrics["agent_to_object_distance"],
metrics["object_to_goal_distance"],
)
)
try: # Got to make initialization idiot proof
sim.close()
except NameError:
pass
with habitat.Env(config) as env:
obs = env.reset()
obs_list = []
# Get closer to the object
while True:
obs = env.step(1)
obs_list.append(obs)
metrics = env.get_metrics()
print_info(obs, metrics)
if metrics["agent_to_object_distance"] < 2.0:
break
# Grab the object
obs = env.step(2)
obs_list.append(obs)
metrics = env.get_metrics()
print_info(obs, metrics)
assert obs["gripped_object_id"] != -1
# Get closer to the goal
while True:
obs = env.step(1)
obs_list.append(obs)
metrics = env.get_metrics()
print_info(obs, metrics)
if metrics["object_to_goal_distance"] < 2.0:
break
# Release the object
obs = env.step(2)
obs_list.append(obs)
metrics = env.get_metrics()
print_info(obs, metrics)
assert obs["gripped_object_id"] == -1
if make_video:
make_video_cv2(
obs_list,
[190, 128],
"hard-coded-agent",
fps=5.0,
open_vid=show_video,
)
# @title Create a task specific RL Environment with a new reward definition.
# @markdown We create a `RearragenmentRLEnv` class and modify the `get_reward()` function.
# @markdown The reward sturcture is as follows:
# @markdown - The agent gets a positive reward if the agent gets closer to the object otherwise a negative reward.
# @markdown - The agent gets a positive reward if it moves the object closer to goal otherwise a negative reward.
# @markdown - The agent gets a positive reward when the agent "picks" up an object for the first time. For all other "grab/release" action, it gets a negative reward.
# @markdown - The agent gets a slack penalty of -0.01 for every action it takes in the environment.
# @markdown - Finally the agent gets a large success reward when the episode is completed successfully.
from typing import Optional, Type
import numpy as np
import habitat
from habitat import Config, Dataset
from habitat_baselines.common.baseline_registry import baseline_registry
from habitat_baselines.common.environments import NavRLEnv
@baseline_registry.register_env(name="RearrangementRLEnv")
class RearrangementRLEnv(NavRLEnv):
def __init__(self, config: Config, dataset: Optional[Dataset] = None):
self._prev_measure = {
"agent_to_object_distance": 0.0,
"object_to_goal_distance": 0.0,
"gripped_object_id": -1,
"gripped_object_count": 0,
}
super().__init__(config, dataset)
self._success_distance = self._core_env_config.TASK.SUCCESS_DISTANCE
def reset(self):
self._previous_action = None
observations = super().reset()
self._prev_measure.update(self.habitat_env.get_metrics())
self._prev_measure["gripped_object_id"] = -1
self._prev_measure["gripped_object_count"] = 0
return observations
def step(self, *args, **kwargs):
self._previous_action = kwargs["action"]
return super().step(*args, **kwargs)
def get_reward_range(self):
return (
self._rl_config.SLACK_REWARD - 1.0,
self._rl_config.SUCCESS_REWARD + 1.0,
)
def get_reward(self, observations):
reward = self._rl_config.SLACK_REWARD
gripped_success_reward = 0.0
episode_success_reward = 0.0
agent_to_object_dist_reward = 0.0
object_to_goal_dist_reward = 0.0
action_name = self._env.task.get_action_name(
self._previous_action["action"]
)
# If object grabbed, add a success reward
# The reward gets awarded only once for an object.
if (
action_name == "GRAB_RELEASE"
and observations["gripped_object_id"] >= 0
):
obj_id = observations["gripped_object_id"]
self._prev_measure["gripped_object_count"] += 1
gripped_success_reward = (
self._rl_config.GRIPPED_SUCCESS_REWARD
if self._prev_measure["gripped_object_count"] == 1
else 0.0
)
# add a penalty everytime grab/action is called and doesn't do anything
elif action_name == "GRAB_RELEASE":
gripped_success_reward += -0.1
self._prev_measure["gripped_object_id"] = observations[
"gripped_object_id"
]
# If the action is not a grab/release action, and the agent
# has not picked up an object, then give reward based on agent to
# object distance.
if (
action_name != "GRAB_RELEASE"
and self._prev_measure["gripped_object_id"] == -1
):
agent_to_object_dist_reward = self.get_agent_to_object_dist_reward(
observations
)
# If the action is not a grab/release action, and the agent
# has picked up an object, then give reward based on object to
# to goal distance.
if (
action_name != "GRAB_RELEASE"
and self._prev_measure["gripped_object_id"] != -1
):
object_to_goal_dist_reward = self.get_object_to_goal_dist_reward()
if (
self._episode_success(observations)
and self._prev_measure["gripped_object_id"] == -1
and action_name == "STOP"
):
episode_success_reward = self._rl_config.SUCCESS_REWARD
reward += (
agent_to_object_dist_reward
+ object_to_goal_dist_reward
+ gripped_success_reward
+ episode_success_reward
)
return reward
def get_agent_to_object_dist_reward(self, observations):
"""
Encourage the agent to move towards the closest object which is not already in place.
"""
curr_metric = self._env.get_metrics()["agent_to_object_distance"]
prev_metric = self._prev_measure["agent_to_object_distance"]
dist_reward = prev_metric - curr_metric
self._prev_measure["agent_to_object_distance"] = curr_metric
return dist_reward
def get_object_to_goal_dist_reward(self):
curr_metric = self._env.get_metrics()["object_to_goal_distance"]
prev_metric = self._prev_measure["object_to_goal_distance"]
dist_reward = prev_metric - curr_metric
self._prev_measure["object_to_goal_distance"] = curr_metric
return dist_reward
def _episode_success(self, observations):
r"""Returns True if object is within distance threshold of the goal."""
dist = self._env.get_metrics()["object_to_goal_distance"]
if (
abs(dist) > self._success_distance
or observations["gripped_object_id"] != -1
):
return False
return True
def _gripped_success(self, observations):
if (
observations["gripped_object_id"] >= 0
and observations["gripped_object_id"]
!= self._prev_measure["gripped_object_id"]
):
return True
return False
def get_done(self, observations):
done = False
action_name = self._env.task.get_action_name(
self._previous_action["action"]
)
if self._env.episode_over or (
self._episode_success(observations)
and self._prev_measure["gripped_object_id"] == -1
and action_name == "STOP"
):
done = True
return done
def get_info(self, observations):
info = self.habitat_env.get_metrics()
info["episode_success"] = self._episode_success(observations)
return info
import os
import time
from collections import defaultdict, deque
from typing import Any, Dict, List, Optional
import numpy as np
from torch.optim.lr_scheduler import LambdaLR
from habitat import Config, logger
from habitat.utils.visualizations.utils import observations_to_image
from habitat_baselines.common.baseline_registry import baseline_registry
from habitat_baselines.common.environments import get_env_class
from habitat_baselines.common.rollout_storage import RolloutStorage
from habitat_baselines.common.tensorboard_utils import TensorboardWriter
from habitat_baselines.rl.models.rnn_state_encoder import RNNStateEncoder
from habitat_baselines.rl.ppo import PPO
from habitat_baselines.rl.ppo.policy import Net, Policy
from habitat_baselines.rl.ppo.ppo_trainer import PPOTrainer
from habitat_baselines.utils.common import (
batch_obs,
generate_video,
linear_decay,
)
from habitat_baselines.utils.env_utils import make_env_fn
def construct_envs(
config,
env_class,
workers_ignore_signals=False,
):
r"""Create VectorEnv object with specified config and env class type.
To allow better performance, dataset are split into small ones for
each individual env, grouped by scenes.
:param config: configs that contain num_processes as well as information
:param necessary to create individual environments.
:param env_class: class type of the envs to be created.
:param workers_ignore_signals: Passed to :ref:`habitat.VectorEnv`'s constructor
:return: VectorEnv object created according to specification.
"""
num_processes = config.NUM_PROCESSES
configs = []
env_classes = [env_class for _ in range(num_processes)]
dataset = habitat.datasets.make_dataset(config.TASK_CONFIG.DATASET.TYPE)
scenes = config.TASK_CONFIG.DATASET.CONTENT_SCENES
if "*" in config.TASK_CONFIG.DATASET.CONTENT_SCENES:
scenes = dataset.get_scenes_to_load(config.TASK_CONFIG.DATASET)
if num_processes > 1:
if len(scenes) == 0:
raise RuntimeError(
"No scenes to load, multiple process logic relies on being able to split scenes uniquely between processes"
)
if len(scenes) < num_processes:
scenes = scenes * num_processes
random.shuffle(scenes)
scene_splits = [[] for _ in range(num_processes)]
for idx, scene in enumerate(scenes):
scene_splits[idx % len(scene_splits)].append(scene)
assert sum(map(len, scene_splits)) == len(scenes)
for i in range(num_processes):
proc_config = config.clone()
proc_config.defrost()
task_config = proc_config.TASK_CONFIG
task_config.SEED = task_config.SEED + i
if len(scenes) > 0:
task_config.DATASET.CONTENT_SCENES = scene_splits[i]
task_config.SIMULATOR.HABITAT_SIM_V0.GPU_DEVICE_ID = (
config.SIMULATOR_GPU_ID
)
task_config.SIMULATOR.AGENT_0.SENSORS = config.SENSORS
proc_config.freeze()
configs.append(proc_config)
envs = habitat.ThreadedVectorEnv(
make_env_fn=make_env_fn,
env_fn_args=tuple(zip(configs, env_classes)),
workers_ignore_signals=workers_ignore_signals,
)
return envs
class RearrangementBaselinePolicy(Policy):
def __init__(self, observation_space, action_space, hidden_size=512):
super().__init__(
RearrangementBaselineNet(
observation_space=observation_space, hidden_size=hidden_size
),
action_space.n,
)
def from_config(cls, config, envs):
pass
class RearrangementBaselineNet(Net):
r"""Network which passes the input image through CNN and concatenates
goal vector with CNN's output and passes that through RNN.
"""
def __init__(self, observation_space, hidden_size):
super().__init__()
self._n_input_goal = observation_space.spaces[
ObjectGoal.cls_uuid
].shape[0]
self._hidden_size = hidden_size
self.state_encoder = RNNStateEncoder(
2 * self._n_input_goal,
self._hidden_size,
)
self.train()
@property
def output_size(self):
return self._hidden_size
@property
def is_blind(self):
return False
@property
def num_recurrent_layers(self):
return self.state_encoder.num_recurrent_layers
def forward(self, observations, rnn_hidden_states, prev_actions, masks):
object_goal_encoding = observations[ObjectGoal.cls_uuid]
object_pos_encoding = observations[ObjectPosition.cls_uuid]
x = [object_goal_encoding, object_pos_encoding]
x = torch.cat(x, dim=1)
x, rnn_hidden_states = self.state_encoder(x, rnn_hidden_states, masks)
return x, rnn_hidden_states
@baseline_registry.register_trainer(name="ppo-rearrangement")
class RearrangementTrainer(PPOTrainer):
supported_tasks = ["RearrangementTask-v0"]
def _setup_actor_critic_agent(self, ppo_cfg: Config) -> None:
r"""Sets up actor critic and agent for PPO.
Args:
ppo_cfg: config node with relevant params
Returns:
None
"""
logger.add_filehandler(self.config.LOG_FILE)
self.actor_critic = RearrangementBaselinePolicy(
observation_space=self.envs.observation_spaces[0],
action_space=self.envs.action_spaces[0],
hidden_size=ppo_cfg.hidden_size,
)
self.actor_critic.to(self.device)
self.agent = PPO(
actor_critic=self.actor_critic,
clip_param=ppo_cfg.clip_param,
ppo_epoch=ppo_cfg.ppo_epoch,
num_mini_batch=ppo_cfg.num_mini_batch,
value_loss_coef=ppo_cfg.value_loss_coef,
entropy_coef=ppo_cfg.entropy_coef,
lr=ppo_cfg.lr,
eps=ppo_cfg.eps,
max_grad_norm=ppo_cfg.max_grad_norm,
use_normalized_advantage=ppo_cfg.use_normalized_advantage,
)
def train(self) -> None:
r"""Main method for training PPO.
Returns:
None
"""
self.envs = construct_envs(
self.config, get_env_class(self.config.ENV_NAME)
)
ppo_cfg = self.config.RL.PPO
self.device = (
torch.device("cuda", self.config.TORCH_GPU_ID)
if torch.cuda.is_available()
else torch.device("cpu")
)
if not os.path.isdir(self.config.CHECKPOINT_FOLDER):
os.makedirs(self.config.CHECKPOINT_FOLDER)
self._setup_actor_critic_agent(ppo_cfg)
logger.info(
"agent number of parameters: {}".format(
sum(param.numel() for param in self.agent.parameters())
)
)
rollouts = RolloutStorage(
ppo_cfg.num_steps,
self.envs.num_envs,
self.envs.observation_spaces[0],
self.envs.action_spaces[0],
ppo_cfg.hidden_size,
)
rollouts.to(self.device)
observations = self.envs.reset()
batch = batch_obs(observations, device=self.device)
for sensor in rollouts.observations:
rollouts.observations[sensor][0].copy_(batch[sensor])
# batch and observations may contain shared PyTorch CUDA
# tensors. We must explicitly clear them here otherwise
# they will be kept in memory for the entire duration of training!
batch = None
observations = None
current_episode_reward = torch.zeros(self.envs.num_envs, 1)
running_episode_stats = dict(
count=torch.zeros(self.envs.num_envs, 1),
reward=torch.zeros(self.envs.num_envs, 1),
)
window_episode_stats = defaultdict(
lambda: deque(maxlen=ppo_cfg.reward_window_size)
)
t_start = time.time()
env_time = 0
pth_time = 0
count_steps = 0
count_checkpoints = 0
lr_scheduler = LambdaLR(
optimizer=self.agent.optimizer,
lr_lambda=lambda x: linear_decay(x, self.config.NUM_UPDATES),
)
with TensorboardWriter(
self.config.TENSORBOARD_DIR, flush_secs=self.flush_secs
) as writer:
for update in range(self.config.NUM_UPDATES):
if ppo_cfg.use_linear_lr_decay:
lr_scheduler.step()
if ppo_cfg.use_linear_clip_decay:
self.agent.clip_param = ppo_cfg.clip_param * linear_decay(
update, self.config.NUM_UPDATES
)
for _step in range(ppo_cfg.num_steps):
(
delta_pth_time,
delta_env_time,
delta_steps,
) = self._collect_rollout_step(
rollouts, current_episode_reward, running_episode_stats
)
pth_time += delta_pth_time
env_time += delta_env_time
count_steps += delta_steps
(
delta_pth_time,
value_loss,
action_loss,
dist_entropy,
) = self._update_agent(ppo_cfg, rollouts)
pth_time += delta_pth_time
for k, v in running_episode_stats.items():
window_episode_stats[k].append(v.clone())
deltas = {
k: (
(v[-1] - v[0]).sum().item()
if len(v) > 1
else v[0].sum().item()
)
for k, v in window_episode_stats.items()
}
deltas["count"] = max(deltas["count"], 1.0)
writer.add_scalar(
"reward", deltas["reward"] / deltas["count"], count_steps
)
# Check to see if there are any metrics
# that haven't been logged yet
for k, v in deltas.items():
if k not in {"reward", "count"}:
writer.add_scalar(
"metric/" + k, v / deltas["count"], count_steps
)
losses = [value_loss, action_loss]
for l, k in zip(losses, ["value, policy"]):
writer.add_scalar("losses/" + k, l, count_steps)
# log stats
if update > 0 and update % self.config.LOG_INTERVAL == 0:
logger.info(
"update: {}\tfps: {:.3f}\t".format(
update, count_steps / (time.time() - t_start)
)
)
logger.info(
"update: {}\tenv-time: {:.3f}s\tpth-time: {:.3f}s\t"
"frames: {}".format(
update, env_time, pth_time, count_steps
)
)
logger.info(
"Average window size: {} {}".format(
len(window_episode_stats["count"]),
" ".join(
"{}: {:.3f}".format(k, v / deltas["count"])
for k, v in deltas.items()
if k != "count"
),
)
)
# checkpoint model
if update % self.config.CHECKPOINT_INTERVAL == 0:
self.save_checkpoint(
f"ckpt.{count_checkpoints}.pth", dict(step=count_steps)
)
count_checkpoints += 1
self.envs.close()
def eval(self) -> None:
r"""Evaluates the current model
Returns:
None
"""
config = self.config.clone()
if len(self.config.VIDEO_OPTION) > 0:
config.defrost()
config.NUM_PROCESSES = 1
config.freeze()
logger.info(f"env config: {config}")
with construct_envs(config, get_env_class(config.ENV_NAME)) as envs:
observations = envs.reset()
batch = batch_obs(observations, device=self.device)
current_episode_reward = torch.zeros(
envs.num_envs, 1, device=self.device
)
ppo_cfg = self.config.RL.PPO
test_recurrent_hidden_states = torch.zeros(
self.actor_critic.net.num_recurrent_layers,
config.NUM_PROCESSES,
ppo_cfg.hidden_size,
device=self.device,
)
prev_actions = torch.zeros(
config.NUM_PROCESSES, 1, device=self.device, dtype=torch.long
)
not_done_masks = torch.zeros(
config.NUM_PROCESSES, 1, device=self.device
)
rgb_frames = [
[] for _ in range(self.config.NUM_PROCESSES)
] # type: List[List[np.ndarray]]
if len(config.VIDEO_OPTION) > 0:
os.makedirs(config.VIDEO_DIR, exist_ok=True)
self.actor_critic.eval()
for _i in range(config.TASK_CONFIG.ENVIRONMENT.MAX_EPISODE_STEPS):
current_episodes = envs.current_episodes()
with torch.no_grad():
(
_,
actions,
_,
test_recurrent_hidden_states,
) = self.actor_critic.act(
batch,
test_recurrent_hidden_states,
prev_actions,
not_done_masks,
deterministic=False,
)
prev_actions.copy_(actions)
outputs = envs.step([a[0].item() for a in actions])
observations, rewards, dones, infos = [
list(x) for x in zip(*outputs)
]
batch = batch_obs(observations, device=self.device)
not_done_masks = torch.tensor(
[[0.0] if done else [1.0] for done in dones],
dtype=torch.float,
device=self.device,
)
rewards = torch.tensor(
rewards, dtype=torch.float, device=self.device
).unsqueeze(1)
current_episode_reward += rewards
# episode ended
if not_done_masks[0].item() == 0:
generate_video(
video_option=self.config.VIDEO_OPTION,
video_dir=self.config.VIDEO_DIR,
images=rgb_frames[0],
episode_id=current_episodes[0].episode_id,
checkpoint_idx=0,
metrics=self._extract_scalars_from_info(infos[0]),
tb_writer=None,
)
print("Evaluation Finished.")
print("Success: {}".format(infos[0]["episode_success"]))
print(
"Reward: {}".format(current_episode_reward[0].item())
)
print(
"Distance To Goal: {}".format(
infos[0]["object_to_goal_distance"]
)
)
return
# episode continues
elif len(self.config.VIDEO_OPTION) > 0:
frame = observations_to_image(observations[0], infos[0])
rgb_frames[0].append(frame)
%load_ext tensorboard
%tensorboard --logdir data/tb
# @title Train an RL agent on a single episode
!if [ -d "data/tb" ]; then rm -r data/tb; fi
import random
import numpy as np
import torch
import habitat
from habitat import Config
from habitat_baselines.config.default import get_config as get_baseline_config
baseline_config = get_baseline_config(
"habitat_baselines/config/pointnav/ppo_pointnav.yaml"
)
baseline_config.defrost()
baseline_config.TASK_CONFIG = config
baseline_config.TRAINER_NAME = "ddppo"
baseline_config.ENV_NAME = "RearrangementRLEnv"
baseline_config.SIMULATOR_GPU_ID = 0
baseline_config.TORCH_GPU_ID = 0
baseline_config.VIDEO_OPTION = ["disk"]
baseline_config.TENSORBOARD_DIR = "data/tb"
baseline_config.VIDEO_DIR = "data/videos"
baseline_config.NUM_PROCESSES = 2
baseline_config.SENSORS = ["RGB_SENSOR", "DEPTH_SENSOR"]
baseline_config.CHECKPOINT_FOLDER = "data/checkpoints"
if vut.is_notebook():
baseline_config.NUM_UPDATES = 400 # @param {type:"number"}
else:
baseline_config.NUM_UPDATES = 1
baseline_config.LOG_INTERVAL = 10
baseline_config.CHECKPOINT_INTERVAL = 50
baseline_config.LOG_FILE = "data/checkpoints/train.log"
baseline_config.EVAL.SPLIT = "train"
baseline_config.RL.SUCCESS_REWARD = 2.5 # @param {type:"number"}
baseline_config.RL.SUCCESS_MEASURE = "object_to_goal_distance"
baseline_config.RL.REWARD_MEASURE = "object_to_goal_distance"
baseline_config.RL.GRIPPED_SUCCESS_REWARD = 2.5 # @param {type:"number"}
baseline_config.freeze()
random.seed(baseline_config.TASK_CONFIG.SEED)
np.random.seed(baseline_config.TASK_CONFIG.SEED)
torch.manual_seed(baseline_config.TASK_CONFIG.SEED)
if __name__ == "__main__":
trainer = RearrangementTrainer(baseline_config)
trainer.train()
trainer.eval()
if make_video:
video_file = os.listdir("data/videos")[0]
vut.display_video(os.path.join("data/videos", video_file))
```
|
github_jupyter
|
# Language Translation
In this project, you’re going to take a peek into the realm of neural network machine translation. You’ll be training a sequence to sequence model on a dataset of English and French sentences that can translate new sentences from English to French.
## Get the Data
Since translating the whole language of English to French will take lots of time to train, we have provided you with a small portion of the English corpus.
```
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import helper
import problem_unittests as tests
source_path = 'data/small_vocab_en'
target_path = 'data/small_vocab_fr'
source_text = helper.load_data(source_path)
target_text = helper.load_data(target_path)
```
## Explore the Data
Play around with view_sentence_range to view different parts of the data.
```
view_sentence_range = (0, 10)
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import numpy as np
print('Dataset Stats')
print('Roughly the number of unique words: {}'.format(len({word: None for word in source_text.split()})))
sentences = source_text.split('\n')
word_counts = [len(sentence.split()) for sentence in sentences]
print('Number of sentences: {}'.format(len(sentences)))
print('Average number of words in a sentence: {}'.format(np.average(word_counts)))
print()
print('English sentences {} to {}:'.format(*view_sentence_range))
print('\n'.join(source_text.split('\n')[view_sentence_range[0]:view_sentence_range[1]]))
print()
print('French sentences {} to {}:'.format(*view_sentence_range))
print('\n'.join(target_text.split('\n')[view_sentence_range[0]:view_sentence_range[1]]))
```
## Implement Preprocessing Function
### Text to Word Ids
As you did with other RNNs, you must turn the text into a number so the computer can understand it. In the function `text_to_ids()`, you'll turn `source_text` and `target_text` from words to ids. However, you need to add the `<EOS>` word id at the end of `target_text`. This will help the neural network predict when the sentence should end.
You can get the `<EOS>` word id by doing:
```python
target_vocab_to_int['<EOS>']
```
You can get other word ids using `source_vocab_to_int` and `target_vocab_to_int`.
```
def text_to_ids(source_text, target_text, source_vocab_to_int, target_vocab_to_int):
"""
Convert source and target text to proper word ids
:param source_text: String that contains all the source text.
:param target_text: String that contains all the target text.
:param source_vocab_to_int: Dictionary to go from the source words to an id
:param target_vocab_to_int: Dictionary to go from the target words to an id
:return: A tuple of lists (source_id_text, target_id_text)
"""
# Split text into sentences
source_text_lst = source_text.split('\n')
target_text_lst = target_text.split('\n')
# Append <EOS> at the end of each sententence
target_text_lst = [sentence + ' <EOS>' for sentence in target_text_lst]
# Make lists using vocab to int mapping
source_id_text = [[source_vocab_to_int[word] for word in sentence.split()] for sentence in source_text_lst]
target_id_text = [[target_vocab_to_int[word] for word in sentence.split()] for sentence in target_text_lst]
return source_id_text, target_id_text
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_text_to_ids(text_to_ids)
```
### Preprocess all the data and save it
Running the code cell below will preprocess all the data and save it to file.
```
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
helper.preprocess_and_save_data(source_path, target_path, text_to_ids)
```
# Check Point
This is your first checkpoint. If you ever decide to come back to this notebook or have to restart the notebook, you can start from here. The preprocessed data has been saved to disk.
```
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import numpy as np
import helper
(source_int_text, target_int_text), (source_vocab_to_int, target_vocab_to_int), _ = helper.load_preprocess()
```
### Check the Version of TensorFlow and Access to GPU
This will check to make sure you have the correct version of TensorFlow and access to a GPU
```
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
from distutils.version import LooseVersion
import warnings
import tensorflow as tf
from tensorflow.python.layers.core import Dense
# Check TensorFlow Version
assert LooseVersion(tf.__version__) >= LooseVersion('1.1'), 'Please use TensorFlow version 1.1 or newer'
print('TensorFlow Version: {}'.format(tf.__version__))
# Check for a GPU
if not tf.test.gpu_device_name():
warnings.warn('No GPU found. Please use a GPU to train your neural network.')
else:
print('Default GPU Device: {}'.format(tf.test.gpu_device_name()))
```
## Build the Neural Network
You'll build the components necessary to build a Sequence-to-Sequence model by implementing the following functions below:
- `model_inputs`
- `process_decoder_input`
- `encoding_layer`
- `decoding_layer_train`
- `decoding_layer_infer`
- `decoding_layer`
- `seq2seq_model`
### Input
Implement the `model_inputs()` function to create TF Placeholders for the Neural Network. It should create the following placeholders:
- Input text placeholder named "input" using the TF Placeholder name parameter with rank 2.
- Targets placeholder with rank 2.
- Learning rate placeholder with rank 0.
- Keep probability placeholder named "keep_prob" using the TF Placeholder name parameter with rank 0.
- Target sequence length placeholder named "target_sequence_length" with rank 1
- Max target sequence length tensor named "max_target_len" getting its value from applying tf.reduce_max on the target_sequence_length placeholder. Rank 0.
- Source sequence length placeholder named "source_sequence_length" with rank 1
Return the placeholders in the following the tuple (input, targets, learning rate, keep probability, target sequence length, max target sequence length, source sequence length)
```
def model_inputs():
"""
Create TF Placeholders for input, targets, learning rate, and lengths of source and target sequences.
:return: Tuple (input, targets, learning rate, keep probability, target sequence length,
max target sequence length, source sequence length)
"""
inputs = tf.placeholder(tf.int32, shape=(None, None), name='input')
targets = tf.placeholder(tf.int32, shape=(None, None))
learn_rate = tf.placeholder(tf.float32, shape=None)
keep_prob = tf.placeholder(tf.float32, shape=None, name='keep_prob')
target_sequence_length = tf.placeholder(tf.int32, (None,), name='target_sequence_length')
max_target_sequence_length = tf.reduce_max(target_sequence_length, name='max_target_len')
source_sequence_length = tf.placeholder(tf.int32, (None,), name='source_sequence_length')
return inputs, targets, learn_rate, keep_prob, target_sequence_length, max_target_sequence_length, source_sequence_length
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_model_inputs(model_inputs)
```
### Process Decoder Input
Implement `process_decoder_input` by removing the last word id from each batch in `target_data` and concat the GO ID to the begining of each batch.
```
def process_decoder_input(target_data, target_vocab_to_int, batch_size):
"""
Preprocess target data for encoding
:param target_data: Target Placehoder
:param target_vocab_to_int: Dictionary to go from the target words to an id
:param batch_size: Batch Size
:return: Preprocessed target data
"""
ending = tf.strided_slice(target_data, [0, 0], [batch_size, -1], [1, 1])
dec_input = tf.concat([tf.fill([batch_size, 1], target_vocab_to_int['<GO>']), ending], 1)
return dec_input
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_process_encoding_input(process_decoder_input)
```
### Encoding
Implement `encoding_layer()` to create a Encoder RNN layer:
* Embed the encoder input using [`tf.contrib.layers.embed_sequence`](https://www.tensorflow.org/api_docs/python/tf/contrib/layers/embed_sequence)
* Construct a [stacked](https://github.com/tensorflow/tensorflow/blob/6947f65a374ebf29e74bb71e36fd82760056d82c/tensorflow/docs_src/tutorials/recurrent.md#stacking-multiple-lstms) [`tf.contrib.rnn.LSTMCell`](https://www.tensorflow.org/api_docs/python/tf/contrib/rnn/LSTMCell) wrapped in a [`tf.contrib.rnn.DropoutWrapper`](https://www.tensorflow.org/api_docs/python/tf/contrib/rnn/DropoutWrapper)
* Pass cell and embedded input to [`tf.nn.dynamic_rnn()`](https://www.tensorflow.org/api_docs/python/tf/nn/dynamic_rnn)
```
from imp import reload
reload(tests)
def encoding_layer(rnn_inputs, rnn_size, num_layers, keep_prob,
source_sequence_length, source_vocab_size,
encoding_embedding_size):
"""
Create encoding layer
:param rnn_inputs: Inputs for the RNN
:param rnn_size: RNN Size
:param num_layers: Number of layers
:param keep_prob: Dropout keep probability
:param source_sequence_length: a list of the lengths of each sequence in the batch
:param source_vocab_size: vocabulary size of source data
:param encoding_embedding_size: embedding size of source data
:return: tuple (RNN output, RNN state)
"""
# Encoder embedding
enc_embed_input = tf.contrib.layers.embed_sequence(rnn_inputs, source_vocab_size, encoding_embedding_size)
# RNN cell
def make_cell(rnn_size):
enc_cell = tf.contrib.rnn.LSTMCell(rnn_size,
initializer=tf.random_uniform_initializer(-0.1, 0.1, seed=2))
return enc_cell
enc_cell = tf.contrib.rnn.MultiRNNCell([make_cell(rnn_size) for _ in range(num_layers)])
drop = tf.contrib.rnn.DropoutWrapper(enc_cell, output_keep_prob=keep_prob)
enc_output, enc_state = tf.nn.dynamic_rnn(drop, enc_embed_input, sequence_length=source_sequence_length, dtype=tf.float32)
return enc_output, enc_state
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_encoding_layer(encoding_layer)
```
### Decoding - Training
Create a training decoding layer:
* Create a [`tf.contrib.seq2seq.TrainingHelper`](https://www.tensorflow.org/api_docs/python/tf/contrib/seq2seq/TrainingHelper)
* Create a [`tf.contrib.seq2seq.BasicDecoder`](https://www.tensorflow.org/api_docs/python/tf/contrib/seq2seq/BasicDecoder)
* Obtain the decoder outputs from [`tf.contrib.seq2seq.dynamic_decode`](https://www.tensorflow.org/api_docs/python/tf/contrib/seq2seq/dynamic_decode)
```
def decoding_layer_train(encoder_state, dec_cell, dec_embed_input,
target_sequence_length, max_summary_length,
output_layer, keep_prob):
"""
Create a decoding layer for training
:param encoder_state: Encoder State
:param dec_cell: Decoder RNN Cell
:param dec_embed_input: Decoder embedded input
:param target_sequence_length: The lengths of each sequence in the target batch
:param max_summary_length: The length of the longest sequence in the batch
:param output_layer: Function to apply the output layer
:param keep_prob: Dropout keep probability
:return: BasicDecoderOutput containing training logits and sample_id
"""
# Helper for the training process. Used by BasicDecoder to read inputs.
training_helper = tf.contrib.seq2seq.TrainingHelper(inputs=dec_embed_input,
sequence_length=target_sequence_length,
time_major=False)
# Basic decoder
training_decoder = tf.contrib.seq2seq.BasicDecoder(dec_cell,
training_helper,
encoder_state,
output_layer)
# Perform dynamic decoding using the decoder
training_decoder_output, _ = tf.contrib.seq2seq.dynamic_decode(training_decoder,
impute_finished=True,
maximum_iterations=max_summary_length)
return training_decoder_output
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_decoding_layer_train(decoding_layer_train)
```
### Decoding - Inference
Create inference decoder:
* Create a [`tf.contrib.seq2seq.GreedyEmbeddingHelper`](https://www.tensorflow.org/api_docs/python/tf/contrib/seq2seq/GreedyEmbeddingHelper)
* Create a [`tf.contrib.seq2seq.BasicDecoder`](https://www.tensorflow.org/api_docs/python/tf/contrib/seq2seq/BasicDecoder)
* Obtain the decoder outputs from [`tf.contrib.seq2seq.dynamic_decode`](https://www.tensorflow.org/api_docs/python/tf/contrib/seq2seq/dynamic_decode)
```
def decoding_layer_infer(encoder_state, dec_cell, dec_embeddings, start_of_sequence_id,
end_of_sequence_id, max_target_sequence_length,
vocab_size, output_layer, batch_size, keep_prob):
"""
Create a decoding layer for inference
:param encoder_state: Encoder state
:param dec_cell: Decoder RNN Cell
:param dec_embeddings: Decoder embeddings
:param start_of_sequence_id: GO ID
:param end_of_sequence_id: EOS Id
:param max_target_sequence_length: Maximum length of target sequences
:param vocab_size: Size of decoder/target vocabulary
:param decoding_scope: TenorFlow Variable Scope for decoding
:param output_layer: Function to apply the output layer
:param batch_size: Batch size
:param keep_prob: Dropout keep probability
:return: BasicDecoderOutput containing inference logits and sample_id
"""
# Reuses the same parameters trained by the training process
start_tokens = tf.tile(tf.constant([start_of_sequence_id], dtype=tf.int32), [batch_size], name='start_tokens')
# Helper for the inference process.
inference_helper = tf.contrib.seq2seq.GreedyEmbeddingHelper(dec_embeddings,
start_tokens,
end_of_sequence_id)
# Basic decoder
inference_decoder = tf.contrib.seq2seq.BasicDecoder(dec_cell,
inference_helper,
encoder_state,
output_layer)
# Perform dynamic decoding using the decoder
inference_decoder_output, _ = tf.contrib.seq2seq.dynamic_decode(inference_decoder,
impute_finished=True,
maximum_iterations=max_target_sequence_length)
return inference_decoder_output
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_decoding_layer_infer(decoding_layer_infer)
```
### Build the Decoding Layer
Implement `decoding_layer()` to create a Decoder RNN layer.
* Embed the target sequences
* Construct the decoder LSTM cell (just like you constructed the encoder cell above)
* Create an output layer to map the outputs of the decoder to the elements of our vocabulary
* Use the your `decoding_layer_train(encoder_state, dec_cell, dec_embed_input, target_sequence_length, max_target_sequence_length, output_layer, keep_prob)` function to get the training logits.
* Use your `decoding_layer_infer(encoder_state, dec_cell, dec_embeddings, start_of_sequence_id, end_of_sequence_id, max_target_sequence_length, vocab_size, output_layer, batch_size, keep_prob)` function to get the inference logits.
Note: You'll need to use [tf.variable_scope](https://www.tensorflow.org/api_docs/python/tf/variable_scope) to share variables between training and inference.
```
def decoding_layer(dec_input, encoder_state,
target_sequence_length, max_target_sequence_length,
rnn_size,
num_layers, target_vocab_to_int, target_vocab_size,
batch_size, keep_prob, decoding_embedding_size):
"""
Create decoding layer
:param dec_input: Decoder input
:param encoder_state: Encoder state
:param target_sequence_length: The lengths of each sequence in the target batch
:param max_target_sequence_length: Maximum length of target sequences
:param rnn_size: RNN Size
:param num_layers: Number of layers
:param target_vocab_to_int: Dictionary to go from the target words to an id
:param target_vocab_size: Size of target vocabulary
:param batch_size: The size of the batch
:param keep_prob: Dropout keep probability
:param decoding_embedding_size: Decoding embedding size
:return: Tuple of (Training BasicDecoderOutput, Inference BasicDecoderOutput)
"""
# Embed the target sequences
dec_embeddings = tf.Variable(tf.random_uniform([target_vocab_size, decoding_embedding_size]))
dec_embed_inputs = tf.nn.embedding_lookup(dec_embeddings, dec_input)
# Construct the decoder LSTM cell (just like you constructed the encoder cell above)
def make_cell(rnn_size):
dec_cell = tf.contrib.rnn.LSTMCell(rnn_size, initializer=tf.random_uniform_initializer(-0.1, 0.1, seed=2))
return dec_cell
dec_cell = tf.contrib.rnn.MultiRNNCell([make_cell(rnn_size) for _ in range(num_layers)])
# Create an output layer to map the outputs of the decoder to the elements of our vocabulary
output_layer = Dense(target_vocab_size, kernel_initializer = tf.truncated_normal_initializer(mean = 0.0, stddev=0.1))
with tf.variable_scope("decode"):
# Helper for the training process. Used by BasicDecoder to read inputs.
training_helper = tf.contrib.seq2seq.TrainingHelper(inputs=dec_embed_inputs,
sequence_length=target_sequence_length,
time_major=False)
# Basic decoder
training_decoder = tf.contrib.seq2seq.BasicDecoder(dec_cell,
training_helper,
encoder_state,
output_layer)
# Perform dynamic decoding using the decoder
training_decoder_output, _ = tf.contrib.seq2seq.dynamic_decode(training_decoder,
impute_finished=True,
maximum_iterations=max_target_sequence_length)
# 5. Inference Decoder
# Reuses the same parameters trained by the training process
with tf.variable_scope("decode", reuse=True):
start_tokens = tf.tile(tf.constant([target_vocab_to_int['<GO>']], dtype=tf.int32), [batch_size], name='start_tokens')
# Helper for the inference process.
inference_helper = tf.contrib.seq2seq.GreedyEmbeddingHelper(dec_embeddings,
start_tokens,
target_vocab_to_int['<EOS>'])
# Basic decoder
inference_decoder = tf.contrib.seq2seq.BasicDecoder(dec_cell,
inference_helper,
encoder_state,
output_layer)
# Perform dynamic decoding using the decoder
inference_decoder_output, _ = tf.contrib.seq2seq.dynamic_decode(inference_decoder,
impute_finished=True,
maximum_iterations=max_target_sequence_length)
return training_decoder_output, inference_decoder_output
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_decoding_layer(decoding_layer)
```
### Build the Neural Network
Apply the functions you implemented above to:
- Encode the input using your `encoding_layer(rnn_inputs, rnn_size, num_layers, keep_prob, source_sequence_length, source_vocab_size, encoding_embedding_size)`.
- Process target data using your `process_decoder_input(target_data, target_vocab_to_int, batch_size)` function.
- Decode the encoded input using your `decoding_layer(dec_input, enc_state, target_sequence_length, max_target_sentence_length, rnn_size, num_layers, target_vocab_to_int, target_vocab_size, batch_size, keep_prob, dec_embedding_size)` function.
```
def seq2seq_model(input_data, target_data, keep_prob, batch_size,
source_sequence_length, target_sequence_length,
max_target_sentence_length,
source_vocab_size, target_vocab_size,
enc_embedding_size, dec_embedding_size,
rnn_size, num_layers, target_vocab_to_int):
"""
Build the Sequence-to-Sequence part of the neural network
:param input_data: Input placeholder
:param target_data: Target placeholder
:param keep_prob: Dropout keep probability placeholder
:param batch_size: Batch Size
:param source_sequence_length: Sequence Lengths of source sequences in the batch
:param target_sequence_length: Sequence Lengths of target sequences in the batch
:param source_vocab_size: Source vocabulary size
:param target_vocab_size: Target vocabulary size
:param enc_embedding_size: Decoder embedding size
:param dec_embedding_size: Encoder embedding size
:param rnn_size: RNN Size
:param num_layers: Number of layers
:param target_vocab_to_int: Dictionary to go from the target words to an id
:return: Tuple of (Training BasicDecoderOutput, Inference BasicDecoderOutput)
"""
# Pass the input data through the encoder. We'll ignore the encoder output, but use the state
_, enc_state = encoding_layer(input_data,
rnn_size,
num_layers,
keep_prob,
source_sequence_length,
source_vocab_size,
enc_embedding_size)
# Prepare the target sequences we'll feed to the decoder in training mode
dec_input = process_decoder_input(target_data, target_vocab_to_int, batch_size)
# Pass encoder state and decoder inputs to the decoders
training_decoder_output, inference_decoder_output = decoding_layer(dec_input,
enc_state,
target_sequence_length,
max_target_sentence_length,
rnn_size,
num_layers,
target_vocab_to_int,
target_vocab_size,
batch_size,
keep_prob,
dec_embedding_size)
return training_decoder_output, inference_decoder_output
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_seq2seq_model(seq2seq_model)
```
## Neural Network Training
### Hyperparameters
Tune the following parameters:
- Set `epochs` to the number of epochs.
- Set `batch_size` to the batch size.
- Set `rnn_size` to the size of the RNNs.
- Set `num_layers` to the number of layers.
- Set `encoding_embedding_size` to the size of the embedding for the encoder.
- Set `decoding_embedding_size` to the size of the embedding for the decoder.
- Set `learning_rate` to the learning rate.
- Set `keep_probability` to the Dropout keep probability
- Set `display_step` to state how many steps between each debug output statement
```
# Number of Epochs
epochs = 30
# Batch Size
batch_size = 128
# RNN Size
rnn_size = 50
# Number of Layers
num_layers = 2
# Embedding Size
encoding_embedding_size = 13
decoding_embedding_size = 13
# Learning Rate
learning_rate = 0.001
# Dropout Keep Probability
keep_probability = 0.8
display_step = 10
```
### Build the Graph
Build the graph using the neural network you implemented.
```
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
save_path = 'checkpoints/dev'
(source_int_text, target_int_text), (source_vocab_to_int, target_vocab_to_int), _ = helper.load_preprocess()
max_target_sentence_length = max([len(sentence) for sentence in source_int_text])
train_graph = tf.Graph()
with train_graph.as_default():
input_data, targets, lr, keep_prob, target_sequence_length, max_target_sequence_length, source_sequence_length = model_inputs()
#sequence_length = tf.placeholder_with_default(max_target_sentence_length, None, name='sequence_length')
input_shape = tf.shape(input_data)
train_logits, inference_logits = seq2seq_model(tf.reverse(input_data, [-1]),
targets,
keep_prob,
batch_size,
source_sequence_length,
target_sequence_length,
max_target_sequence_length,
len(source_vocab_to_int),
len(target_vocab_to_int),
encoding_embedding_size,
decoding_embedding_size,
rnn_size,
num_layers,
target_vocab_to_int)
training_logits = tf.identity(train_logits.rnn_output, name='logits')
inference_logits = tf.identity(inference_logits.sample_id, name='predictions')
masks = tf.sequence_mask(target_sequence_length, max_target_sequence_length, dtype=tf.float32, name='masks')
with tf.name_scope("optimization"):
# Loss function
cost = tf.contrib.seq2seq.sequence_loss(
training_logits,
targets,
masks)
# Optimizer
optimizer = tf.train.AdamOptimizer(lr)
# Gradient Clipping
gradients = optimizer.compute_gradients(cost)
capped_gradients = [(tf.clip_by_value(grad, -1., 1.), var) for grad, var in gradients if grad is not None]
train_op = optimizer.apply_gradients(capped_gradients)
```
Batch and pad the source and target sequences
```
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
def pad_sentence_batch(sentence_batch, pad_int):
"""Pad sentences with <PAD> so that each sentence of a batch has the same length"""
max_sentence = max([len(sentence) for sentence in sentence_batch])
return [sentence + [pad_int] * (max_sentence - len(sentence)) for sentence in sentence_batch]
def get_batches(sources, targets, batch_size, source_pad_int, target_pad_int):
"""Batch targets, sources, and the lengths of their sentences together"""
for batch_i in range(0, len(sources)//batch_size):
start_i = batch_i * batch_size
# Slice the right amount for the batch
sources_batch = sources[start_i:start_i + batch_size]
targets_batch = targets[start_i:start_i + batch_size]
# Pad
pad_sources_batch = np.array(pad_sentence_batch(sources_batch, source_pad_int))
pad_targets_batch = np.array(pad_sentence_batch(targets_batch, target_pad_int))
# Need the lengths for the _lengths parameters
pad_targets_lengths = []
for target in pad_targets_batch:
pad_targets_lengths.append(len(target))
pad_source_lengths = []
for source in pad_sources_batch:
pad_source_lengths.append(len(source))
yield pad_sources_batch, pad_targets_batch, pad_source_lengths, pad_targets_lengths
```
### Train
Train the neural network on the preprocessed data. If you have a hard time getting a good loss, check the forms to see if anyone is having the same problem.
```
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
def get_accuracy(target, logits):
"""
Calculate accuracy
"""
max_seq = max(target.shape[1], logits.shape[1])
if max_seq - target.shape[1]:
target = np.pad(
target,
[(0,0),(0,max_seq - target.shape[1])],
'constant')
if max_seq - logits.shape[1]:
logits = np.pad(
logits,
[(0,0),(0,max_seq - logits.shape[1])],
'constant')
return np.mean(np.equal(target, logits))
# Split data to training and validation sets
train_source = source_int_text[batch_size:]
train_target = target_int_text[batch_size:]
valid_source = source_int_text[:batch_size]
valid_target = target_int_text[:batch_size]
(valid_sources_batch, valid_targets_batch, valid_sources_lengths, valid_targets_lengths ) = next(get_batches(valid_source,
valid_target,
batch_size,
source_vocab_to_int['<PAD>'],
target_vocab_to_int['<PAD>']))
with tf.Session(graph=train_graph) as sess:
sess.run(tf.global_variables_initializer())
for epoch_i in range(epochs):
for batch_i, (source_batch, target_batch, sources_lengths, targets_lengths) in enumerate(
get_batches(train_source, train_target, batch_size,
source_vocab_to_int['<PAD>'],
target_vocab_to_int['<PAD>'])):
_, loss = sess.run(
[train_op, cost],
{input_data: source_batch,
targets: target_batch,
lr: learning_rate,
target_sequence_length: targets_lengths,
source_sequence_length: sources_lengths,
keep_prob: keep_probability})
if batch_i % display_step == 0 and batch_i > 0:
batch_train_logits = sess.run(
inference_logits,
{input_data: source_batch,
source_sequence_length: sources_lengths,
target_sequence_length: targets_lengths,
keep_prob: 1.0})
batch_valid_logits = sess.run(
inference_logits,
{input_data: valid_sources_batch,
source_sequence_length: valid_sources_lengths,
target_sequence_length: valid_targets_lengths,
keep_prob: 1.0})
train_acc = get_accuracy(target_batch, batch_train_logits)
valid_acc = get_accuracy(valid_targets_batch, batch_valid_logits)
print('Epoch {:>3} Batch {:>4}/{} - Train Accuracy: {:>6.4f}, Validation Accuracy: {:>6.4f}, Loss: {:>6.4f}'
.format(epoch_i, batch_i, len(source_int_text) // batch_size, train_acc, valid_acc, loss))
# Save Model
saver = tf.train.Saver()
saver.save(sess, save_path)
print('Model Trained and Saved')
```
### Save Parameters
Save the `batch_size` and `save_path` parameters for inference.
```
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
# Save parameters for checkpoint
helper.save_params(save_path)
```
# Checkpoint
```
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import tensorflow as tf
import numpy as np
import helper
import problem_unittests as tests
_, (source_vocab_to_int, target_vocab_to_int), (source_int_to_vocab, target_int_to_vocab) = helper.load_preprocess()
load_path = helper.load_params()
```
## Sentence to Sequence
To feed a sentence into the model for translation, you first need to preprocess it. Implement the function `sentence_to_seq()` to preprocess new sentences.
- Convert the sentence to lowercase
- Convert words into ids using `vocab_to_int`
- Convert words not in the vocabulary, to the `<UNK>` word id.
```
def sentence_to_seq(sentence, vocab_to_int):
"""
Convert a sentence to a sequence of ids
:param sentence: String
:param vocab_to_int: Dictionary to go from the words to an id
:return: List of word ids
"""
# TODO: Implement Function
return None
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_sentence_to_seq(sentence_to_seq)
```
## Translate
This will translate `translate_sentence` from English to French.
```
translate_sentence = 'he saw a old yellow truck .'
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
translate_sentence = sentence_to_seq(translate_sentence, source_vocab_to_int)
loaded_graph = tf.Graph()
with tf.Session(graph=loaded_graph) as sess:
# Load saved model
loader = tf.train.import_meta_graph(load_path + '.meta')
loader.restore(sess, load_path)
input_data = loaded_graph.get_tensor_by_name('input:0')
logits = loaded_graph.get_tensor_by_name('predictions:0')
target_sequence_length = loaded_graph.get_tensor_by_name('target_sequence_length:0')
source_sequence_length = loaded_graph.get_tensor_by_name('source_sequence_length:0')
keep_prob = loaded_graph.get_tensor_by_name('keep_prob:0')
translate_logits = sess.run(logits, {input_data: [translate_sentence]*batch_size,
target_sequence_length: [len(translate_sentence)*2]*batch_size,
source_sequence_length: [len(translate_sentence)]*batch_size,
keep_prob: 1.0})[0]
print('Input')
print(' Word Ids: {}'.format([i for i in translate_sentence]))
print(' English Words: {}'.format([source_int_to_vocab[i] for i in translate_sentence]))
print('\nPrediction')
print(' Word Ids: {}'.format([i for i in translate_logits]))
print(' French Words: {}'.format(" ".join([target_int_to_vocab[i] for i in translate_logits])))
```
## Imperfect Translation
You might notice that some sentences translate better than others. Since the dataset you're using only has a vocabulary of 227 English words of the thousands that you use, you're only going to see good results using these words. For this project, you don't need a perfect translation. However, if you want to create a better translation model, you'll need better data.
You can train on the [WMT10 French-English corpus](http://www.statmt.org/wmt10/training-giga-fren.tar). This dataset has more vocabulary and richer in topics discussed. However, this will take you days to train, so make sure you've a GPU and the neural network is performing well on dataset we provided. Just make sure you play with the WMT10 corpus after you've submitted this project.
## Submitting This Project
When submitting this project, make sure to run all the cells before saving the notebook. Save the notebook file as "dlnd_language_translation.ipynb" and save it as a HTML file under "File" -> "Download as". Include the "helper.py" and "problem_unittests.py" files in your submission.
|
github_jupyter
|
<a href="https://colab.research.google.com/github/yohanesnuwara/machine-learning/blob/master/06_simple_linear_regression/simple_linear_reg_algorithm.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# **Simple Linear Regression**
```
import numpy as np
import matplotlib.pyplot as plt
```
## Method 1 ("Traditional")
Calculate bias (or intercept $B_0$) and slope ($B_1$) using:
$$B_1 = \frac{\sum_{i=1}^{n}(x_i-mean(x))(y_i-mean(y))}{\sum_{i=1}^{n}(x_i-mean(x))^2}$$
$$B_0 = mean(y) - B_1 \cdot mean(x)$$
to construct simple linear regression model: $$y = B_0 + B_1 \cdot x$$
```
x = [1, 2, 4, 3, 5]
y = [1, 3, 3, 2, 5]
# visualize our data
plt.plot(x, y, 'o')
```
Calculate mean of data
```
mean_x = np.mean(x)
mean_y = np.mean(y)
print(mean_x, mean_y)
```
Calculate error
```
err_x = x - mean_x
err_y = y - mean_y
print(err_x)
print(err_y)
```
Multiply error of x and error of y
```
err_mult = err_x * err_y
print(err_mult)
```
Calculate numerator by summing up the errors
```
numerator = np.sum(err_mult)
numerator
```
Calculate denominator by squaring the x error and summing them up
```
err_x_squared = err_x**2
denominator = np.sum(err_x_squared)
print(denominator)
```
Calculate the **slope (B1)** !
```
B1 = numerator / denominator
print(B1)
```
And we can calculate the **intercept (c)** !
```
B0 = mean_y - B1 * mean_x
print(B0)
```
We now have the coefficents for our simple linear regression equation.
$$y = B_0 + B_1 x = 0.4 + 0.8 x$$
### Test the model to our training data
```
x_test = np.array([1, 2, 3, 4, 5])
y_predicted = B0 + B1 * x_test
p1 = plt.plot(x, y, 'o')
p2 = plt.plot(x_test, y_predicted, 'o-', color='r')
plt.legend((p1[0], p2[0]), (['y data', 'predicted y']))
```
### Estimating Error (Root Mean Squared Error)
$$RMSE = \sqrt{\frac{\sum_{i=1}^{n} (p_i - y_i)^2}{n}}$$
```
numerator = np.sum((y_predicted - y)**2)
denominator = len(y)
rmse = np.sqrt(numerator / denominator)
rmse
```
### Wrap all up
```
def simple_linear_regression_traditional(x, y, x_test):
import numpy as np
x = np.array(x); y = np.array(y); x_test = np.array(x_test)
mean_x = np.mean(x)
mean_y = np.mean(y)
err_x = x - mean_x
err_y = y - mean_y
err_mult = err_x * err_y
numerator = np.sum(err_mult)
err_x_squared = err_x**2
denominator = np.sum(err_x_squared)
B1 = numerator / denominator
B0 = mean_y - B1 * mean_x
y_predicted = B0 + B1 * x_test
return(B0, B1, y_predicted)
def linreg_error(y, y_predicted):
import numpy as np
y = np.array(y); y_predicted = np.array(y_predicted)
numerator = np.sum((y_predicted - y)**2)
denominator = len(y)
rmse = np.sqrt(numerator / denominator)
return(rmse)
```
## Method 2 ("Advanced")
Calculate bias (or intercept $B_0$) and slope ($B_1$) using:
$$B_1 = corr(x, y) \cdot \frac{stdev(y)}{stdev(x)}$$
Then, similar to **Method 1**.
$$B_0 = mean(y) - B_1 \cdot mean(x)$$
to construct simple linear regression model: $$y = B_0 + B_1 \cdot x$$
Calculate the **pearson's correlation coefficient $corr(x,y)$**. First, calculate mean and standard deviation.
```
import statistics as stat
mean_x = np.mean(x)
mean_y = np.mean(y)
stdev_x = stat.stdev(x)
stdev_y = stat.stdev(y)
print(stdev_x, stdev_y)
```
Calculate **covariance**. Covariance is the relationship that can be summarized between two variables. The sign of the covariance can be interpreted as whether the two variables change in the same direction (positive) or change in different directions (negative). A covariance value of zero indicates that both variables are completely independent.
```
cov_x_y = (np.sum((x - mean_x) * (y - mean_y))) * (1 / (len(x) - 1))
cov_x_y
```
Calculate **Pearson's Correlation Coefficient**. It summarizes the strength of the linear relationship between two data samples. It is the normalization of the covariance between the two variables. The coefficient returns a value between -1 and 1 that represents the limits of correlation from a full negative correlation to a full positive correlation. A value of 0 means no correlation. The value must be interpreted, where often a value below -0.5 or above 0.5 indicates a notable correlation, and values below those values suggests a less notable correlation.
```
corr_x_y = cov_x_y / (stdev_x * stdev_y)
corr_x_y
```
Calculate slope $B_1$
```
B1 = corr_x_y * (stdev_y / stdev_x)
B1
```
Next, is similar to **Method 1**.
```
B0 = mean_y - B1 * mean_x
x_test = np.array([1, 2, 3, 4, 5])
y_predicted = B0 + B1 * x_test
p1 = plt.plot(x, y, 'o')
p2 = plt.plot(x_test, y_predicted, 'o-', color='r')
plt.legend((p1[0], p2[0]), (['y data', 'predicted y']))
```
Calculate RMSE
```
rmse = linreg_error(y, y_predicted)
rmse
```
### Wrap all up
```
def simple_linear_regression_advanced(x, y, x_test):
import numpy as np
import statistics as stat
x = np.array(x); y = np.array(y); x_test = np.array(x_test)
mean_x = np.mean(x)
mean_y = np.mean(y)
stdev_x = stat.stdev(x)
stdev_y = stat.stdev(y)
cov_x_y = (np.sum((x - mean_x) * (y - mean_y))) * (1 / (len(x) - 1))
corr_x_y = cov_x_y / (stdev_x * stdev_y)
B1 = corr_x_y * (stdev_y / stdev_x)
B0 = mean_y - B1 * mean_x
y_predicted = B0 + B1 * x_test
return(B0, B1, y_predicted)
```
## Implement to Real Dataset
Simple linear regression to WTI and Brent Daily Oil Price (1980-2020)
```
!git clone https://www.github.com/yohanesnuwara/machine-learning
import pandas as pd
brent = pd.read_csv('/content/machine-learning/datasets/brent-daily_csv.csv')
wti = pd.read_csv('/content/machine-learning/datasets/wti-daily_csv.csv')
# Converting to Panda datetime
brent['Date'] = pd.to_datetime(brent['Date'], format='%Y-%m-%d') # depends on the data, format check web: https://strftime.org/
wti['Date'] = pd.to_datetime(wti['Date'], format='%Y-%m-%d') # depends on the data, format check web: https://strftime.org/
brent.head(10)
```
Visualize data
```
from pandas.plotting import register_matplotlib_converters
register_matplotlib_converters()
plt.figure(figsize=(15, 6))
plt.plot(brent.Date, brent.Price, '.', color='blue')
plt.plot(wti.Date, wti.Price, '.', color='red')
plt.title('Daily Oil Price')
plt.xlabel('Year'); plt.ylabel('Price ($/bbl)')
# convert datetime to ordinal
import datetime as dt
brent_date = np.array(brent['Date'].map(dt.datetime.toordinal))
brent_price = brent.Price
brent_test = brent_date
B0_brent, B1_brent, brent_price_predicted = simple_linear_regression_advanced(brent_date, brent_price, brent_test)
wti_date = np.array(wti['Date'].map(dt.datetime.toordinal))
wti_price = wti.Price
wti_test = wti_date
B0_wti, B1_wti, wti_price_predicted = simple_linear_regression_advanced(wti_date, wti_price, wti_test)
plt.figure(figsize=(15, 6))
p1 = plt.plot(brent.Date, brent.Price, '.', color='blue')
p2 = plt.plot(wti.Date, wti.Price, '.', color='red')
p3 = plt.plot(brent_test, brent_price_predicted, color='blue')
p4 = plt.plot(wti_test, wti_price_predicted, color='red')
plt.legend((p1[0], p2[0], p3[0], p4[0]), (['Brent data', 'WTI data', 'Brent predicted', 'WTI predicted']))
plt.title('Daily Oil Price')
plt.xlabel('Year'); plt.ylabel('Price ($/bbl)')
```
|
github_jupyter
|
```
import matplotlib.pyplot as plt
%matplotlib inline
import pickle
import numpy as np
from scipy.spatial.distance import pdist, squareform
with open('exp_features.p', 'rb') as f:
data = pickle.load(f)
```
## visualize
```
def get_continuous_quantile(x, y, n_interval=100, q=1):
"""
Take continuous x and y, bin the data according to the intervals of x
and then calculate the quantiles of y within this bin
Args:
x (list): array of x values
y (list): array of y values
n_interval (int): number of intervals on x
q (float): quantile value [0, 1]
"""
ind = np.argsort(x)
x = x[ind]
y = y[ind]
boundaries = np.linspace(x[0], x[-1], n_interval+1)
dx = boundaries[1] - boundaries[0]
x_center = np.linspace(x[0]+dx/2, x[-1]-dx/2, n_interval)
y_q = []
for x_min, x_max in zip(boundaries[:-1], boundaries[1:]):
ind = (x>=x_min) & (x<x_max)
ys = y[ind]
if len(ys) > 0:
y_q.append(np.quantile(ys, q))
else:
y_q.append(y_q[-1])
y_q = np.array(y_q)
return x_center, y_q
def visualize(key, n_interval=100, interval=5, alpha=0.5, data_file="100_0.xlsx"):
"""
Visualize the data specified by key.
Args:
key (str): key in data
n_interval (int): number of intervals for drawing the quantile bounds
interval (int): subsamping of the data. Sometimes the input data is too large for visualization
we just subsample the data
"""
keys = list(data['band_gap'].keys())
f = np.concatenate([data[key][i] for i in keys], axis=0)
values = np.array([data['band_gap'][i] for i in keys])
sort_index = np.argsort(values)
fscale = (f-np.min(f, axis=0)) / (np.max(f, axis=0) - np.min(f, axis=0))
d = pdist(fscale)
v_dist = pdist(values.reshape((-1, 1)))
ind = (d>0) & (d<1)
d_ = d[ind]
v_ = v_dist[ind]
#print(d_.shape, v_.shape)
x_center, y_q = get_continuous_quantile(d_, v_, n_interval=n_interval, q=1)
plt.rcParams['font.size'] = 22
plt.rcParams['font.family'] = 'Arial'
plt.figure(figsize=(5.7, 5.0 ))
d_ = d_[::interval]
v_ = v_[::interval]
print(v_.shape)
plt.plot(d_, v_, 'o', alpha=alpha, c='#21c277')
plt.plot(x_center, y_q, '--', c='#21c277', lw=2, alpha=0.5)
import pandas as pd
x = np.round(np.concatenate([d_, x_center]), 3)
y = np.round(np.concatenate([v_, y_q]), 3)
df = pd.DataFrame({"dF": x, "dEg": y})
with pd.ExcelWriter(data_file) as writer:
df.to_excel(writer)
plt.xlim([0, 1])
plt.ylim([0, 13])
plt.xticks(np.linspace(0, 1, 5))
plt.yticks(np.linspace(0, 12.5, 6))
plt.xlabel('$d_{F}$ (a.u.)')
plt.ylabel("$\Delta E_{g}$ (eV)")
plt.tight_layout()
visualize('100_0', n_interval=100, interval=15, alpha=0.08, data_file='100_0.xlsx')
plt.savefig("100_0.pdf")
visualize('100_41000', n_interval=100, interval=15, alpha=0.08, data_file='100_41000.xlsx')
plt.savefig("100_41000.pdf")
```
|
github_jupyter
|
```
print("Hello world!")
a=10
a
b=5
b
#addition demo
sum=a+b
print("the sum of a and b is:",sum)
x=2**3
x
y=5/2
y
y=5//2
y
input("Enter some variable")
a=int(input("enter the first number"))
b=int(input("enter the second number"))
int("The sum of first number and second number is:",a+b)
int("The difference of the first and second number is:",a-b)
int("The product of the first and second numberis:",a*b)
int("The quotient of the first and second number is:",a//b)
new=("String demo")
new
#Slice operator
#slice operator works like this
# string variable[x::y::z]
#[x::y::z]
#[start_index=:end index 1:skip index:2]
new[0:5:2]
new[0:]
new[5:]
new*2
new*5
#repetition operator
new*3
#concatenation operator
new +" Test"
new
#short way a=10
new += " Extra"
listt= [ "Kar",'']
listt
#length method to find the value of n(the number of elements in list)
#len()
len(listt)
students=["Akhil","Akila","John","sonum","khushi"]
students
#append function- add a new element to the existing list
#append9()
students.append("jyoti")
students
#Insert function- add a new eleent at the end of index position given
#insert (index,value)
students.insert(3,"papa")
students
students
#if else in python
age=int(input("enter your age:"))
if age>=18:
print("You are eligible")
else:
print("You are not eligible")
#No switch case in python
#if-elif-lse block
a=200
b=33
if b>a:
print("B is greater than A")
elif a==b :
print("A is equal to B")
else:
print("A is greater than B")
```
### Nested if example
age=18
if age>=18:
print("Allow inside club")
if age>=21:
print("Drinking allowed")
else:
print("Drinking not allowed")
else :
print("Not allowed inside club")
```
# For loop
#range function = for var in range(int,int,int):
#range(value)= start from 0 upto value
#range(value 1,value2)= start from value 1 and go upto value2
#range(v1,v2,v3)= start from v1 and go upto v2 and skip every v3
#for x in range(2,9,2):
#x=[2,4,6,8]
#print all value of variable one by one
for x in range(2,9,2):
print("The value of x is:",x)
# i=[1,2,3,4,5]
# i=5
for i in range(1,6):
print(i*"*")
# while loop
# while condition:
# statement
# incrementer
x=0
while x<4:
print(x)
x+=1
for x in range(1,10):
if x==6:
print("Existing loop")
break
else:
print("The value of x is :",x)
def add(a,b):
print("a=",a)
print("b=",b)
return a+b
c= add(5,2)
print(c)
```
###### for i=[1,2,3,4,5,6]
#fori=5
for i in range(1,6):
print("i=1,i<=6,i++")
```
i=1
for i in range(1,6):
j=1
for j in range(i,i+1):
print(j,end=" ")
print()
class student:
def_init_(self,sname,sage,spercent):
listt=['kar','abcd',706,2.33,'johny',70.2,36.3,755]
listt
type(listt[0])
type(listt[3])
int i2;
double d2;
char s2[100]; // this is not scalable for input of unknown size
// Read inputs from stdin
scan("%d", &i2);
scan("%lf", &d2);
scan("%*[\n] %[^\n]", s2);
// Print outputs to stdout
print("%d\n", i + i2);
print("%.01lf\n", d + d2);
print("%s%s", s, s2);
i=int(input(4))
d=int(input(4.0))
s=int(input(Hackerank))
i2 = int(input(12)) # read int
d2 = float(input()) # read double
s2 = input() # read string
# print summed and concatenated values
print(i + i2)
print(d + d2)
print(s + s2)
>>>str="hello"
>>>str[:2]
>>>
import maths
def add(a,b):
return a+b
def sub(a,b):
return a-b
def mul(a,b):
return a*b
def div(a,b):
return a/b
def sqrt(a)
return (math,sqrt.(a))
def powa
return (a**a)
while true:
enter = input("enter function name:")
if function== 'addition'
try:
a =int(input("enter first number"))
b=int(input("enter second number"))
print(add(a,b))
except Value error:
print("please provide valid numbers")
if function == 'subtraction'
try:
a= int(input("enter first number"))
b= int(input("enter second number"))
print(sub(a,b))
except Value error:
print("please provide valid numbers")
if function == 'multiplication'
try:
a=int(input("enter first number"))
b= int(input("enter second number"))
print(mul(a,b))
except Value error:
print("please provide valid numbers")
if function ==;'division'
try :
a=int(input("enter first number"))
b= int(input("enter second number"))
print(div(a,b))
except Value error:
print("please")
import numpy as np
a= np.arange(30).reshape(2,15)
a
```
|
github_jupyter
|
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from IPython.core.display import display, HTML
plt.style.use('fivethirtyeight')
plt.rc('figure', figsize=(5.0, 2.0))
pokemon=pd.read_csv("../dataset/pokemon.csv")
# Which pokémon is the most difficult to catch?
pokemon['capture_rate']=pd.to_numeric(pokemon['capture_rate'])
pokemon['name'][pokemon['capture_rate']==min(pokemon['capture_rate'])]
#Which no-legendary pokémon is the most diffucult to catch? </b>
no_legendary=pokemon[pokemon['is_legendary']==False]
no_legendary['name'][no_legendary['capture_rate']==min(no_legendary['capture_rate'])]
display(HTML("<img src='../img/beldum.png' width='200px' height='200px'>"))
rate=pokemon[pokemon['name']=='Beldum']['capture_rate'].values
beldum_rate=num = "{0:.2f}".format((rate[0]*100)/255)
print("Name: Beldum\n"+"Percentage of catch: " + beldum_rate + " %")
```
<div class="alert alert-info">
The min value for attack/special-attack and defense/special-defense statistics was calculated from the subsets of the pokemon which have the highest physical/special statistic compared to the special/physical equivalent. In this way the results acquire greater relevance.
</div>
```
#Speed
pokemon['name'][pokemon['speed']==max(pokemon['speed'])]
pokemon['name'][pokemon['speed']==min(pokemon['speed'])]
```
<b> Atk </b>
```
pokemon['name'][pokemon['attack']==max(pokemon['attack'])]
physical_atk=pokemon[pokemon['attack']>=pokemon['sp_attack']]
physical_atk['name'][physical_atk['attack']==min(physical_atk['attack'])]
```
<b>Def</b>
```
pokemon['name'][pokemon['defense']==max(pokemon['defense'])]
physical_def=pokemon[pokemon['defense']>=pokemon['sp_defense']]
physical_def['name'][physical_def['defense']==min(physical_def['defense'])]
```
<b> Sp.Atk</b>
```
pokemon['name'][pokemon['sp_attack']==max(pokemon['sp_attack'])]
special_atk=pokemon[pokemon['sp_attack']>=pokemon['attack']]
special_atk['name'][special_atk['sp_attack']==min(special_atk['sp_attack'])]
```
<b>Sp.Def</b>
```
pokemon['name'][pokemon['sp_defense']==max(pokemon['sp_defense'])]
special_def=pokemon[pokemon['sp_defense']>=pokemon['defense']]
special_def['name'][special_def['sp_defense']==min(special_def['sp_defense'])]
```
<b>Hp</b>
```
pokemon['name'][pokemon['hp']==max(pokemon['hp'])]
pokemon['name'][pokemon['hp']==min(pokemon['hp'])]
```
Combining all the information we can see how <code>Shuckle</code> is a pokémon with <b>very particular statistics</b>. Look at them:
```
display(HTML("<img src='../img/shuckle.png' width='200px' height='200px'>"))
pokemon.iloc[212][['name','hp','attack','sp_attack','defense','sp_defense','speed']]
```
# Which type is the most common?
To answer this question, I think it's more interesting seeing the <b>absolute frequencies</b> for each type of pokémon in a <b>bar chart</b>.
```
types_abs_freq=(pokemon['type1'].value_counts()+pokemon['type2'].value_counts()).sort_values(ascending=False)
x=types_abs_freq.index
y=types_abs_freq.values
types_abs_freq.plot.bar()
plt.show()
```
<div class="alert alert-info">
Absolute frequencies were calculated from a set constructed as the union between the set of types 1 and 2 of each pokémon.
</div>
The result obtained shows us a subdivision of the pokémon by type rather conform to reality: the pokémon closest to have an animal correspondent in the real world are the most widespread.<br>
<b>The most common type is water</b> but the most interesting data is that the psychic type is the fifth most common type, even exceeding the bug type.
# Which ability is the most common?
We answer this question by printing the top 10 most common abilities.
```
ser_abilities=pokemon['abilities']
abilities=[]
for i in range(0,801):
arr_ab=ser_abilities[i].split(',')
for j in range(0,len(arr_ab)):
ability=arr_ab[j].replace("[","").replace("'","").replace("]","")
abilities.append(ability)
abilities_freq=pd.Series(abilities).value_counts().sort_values(ascending=False)
abilities_freq.head(10)
```
<b> Be very careful to do earthquake! </b>
# Correlation
```
import seaborn as sns
plt.figure(figsize=(20,20))
sns.heatmap(pokemon.corr(), linewidths=.5)
plt.show()
```
There is a strong positive correlation between:
- generation and pokédex number (good information for building generation clusters),
- against_ghost and against_dark (thanks to ghost type),
- base_egg_steps and is_legendary (good information for building legendary classifier).
There is a good positive correlation between:
- single stats and base_total,
- height and weight.
There is a strong negative correlation between:
- capture_rate and base_total,
- single stats and capture_rate,
- against_fight and against_ghost (thanks to normal type),
- against_psychic and against_bug (thanks to dark type),
- against_ground and against_ice (Why?),
- base_happiness and base_egg_steps.
There is a good negative correlation between:
- base_happiness and weight,
- base_happiness and is_legendary.
And so on.
|
github_jupyter
|
```
import random
import gym
#import math
import numpy as np
from collections import deque
import tensorflow as tf
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Conv2D, MaxPooling2D, Flatten
from tensorflow.keras.optimizers import Adam
EPOCHS = 1000
THRESHOLD = 10
MONITOR = True
class DQN():
def __init__(self, env_string,batch_size=64, IM_SIZE = 84, m = 4):
self.memory = deque(maxlen=5000)
self.env = gym.make(env_string)
input_size = self.env.observation_space.shape[0]
action_size = self.env.action_space.n
self.batch_size = batch_size
self.gamma = 1.0
self.epsilon = 1.0
self.epsilon_min = 0.01
self.epsilon_decay = 0.995
self.IM_SIZE = IM_SIZE
self.m = m
alpha=0.01
alpha_decay=0.01
if MONITOR: self.env = gym.wrappers.Monitor(self.env, '../data/'+env_string, force=True)
# Init model
self.model = Sequential()
self.model.add( Conv2D(32, 8, (4,4), activation='relu',padding='valid', input_shape=(IM_SIZE, IM_SIZE, m)))
#self.model.add(MaxPooling2D())
self.model.add( Conv2D(64, 4, (2,2), activation='relu',padding='valid'))
self.model.add(MaxPooling2D())
self.model.add( Conv2D(64, 3, (1,1), activation='relu',padding='valid'))
self.model.add(MaxPooling2D())
self.model.add(Flatten())
self.model.add(Dense(256, activation='elu'))
self.model.add(Dense(action_size, activation='linear'))
self.model.compile(loss='mse', optimizer=Adam(lr=alpha, decay=alpha_decay))
self.model_target = tf.keras.models.clone_model(self.model)
def remember(self, state, action, reward, next_state, done):
self.memory.append((state, action, reward, next_state, done))
def choose_action(self, state, epsilon):
if np.random.random() <= epsilon:
return self.env.action_space.sample()
else:
return np.argmax(self.model.predict(state))
def preprocess_state(self, img):
img_temp = img[31:195] # Choose the important area of the image
img_temp = tf.image.rgb_to_grayscale(img_temp)
img_temp = tf.image.resize(img_temp, [self.IM_SIZE, self.IM_SIZE],
method=tf.image.ResizeMethod.NEAREST_NEIGHBOR)
img_temp = tf.cast(img_temp, tf.float32)
return img_temp[:,:,0]
def combine_images(self, img1, img2):
if len(img1.shape) == 3 and img1.shape[0] == self.m:
im = np.append(img1[1:,:, :],np.expand_dims(img2,0), axis=2)
return tf.expand_dims(im, 0)
else:
im = np.stack([img1]*self.m, axis = 2)
return tf.expand_dims(im, 0)
#return np.reshape(state, [1, 4])
def replay(self, batch_size):
x_batch, y_batch = [], []
minibatch = random.sample(self.memory, min(len(self.memory), batch_size))
for state, action, reward, next_state, done in minibatch:
y_target = self.model_target.predict(state)
y_target[0][action] = reward if done else reward + self.gamma * np.max(self.model.predict(next_state)[0])
x_batch.append(state[0])
y_batch.append(y_target[0])
self.model.fit(np.array(x_batch), np.array(y_batch), batch_size=len(x_batch), verbose=0)
#epsilon = max(epsilon_min, epsilon_decay*epsilon) # decrease epsilon
def train(self):
scores = deque(maxlen=100)
avg_scores = []
for e in range(EPOCHS):
state = self.env.reset()
state = self.preprocess_state(state)
state = self.combine_images(state, state)
done = False
i = 0
while not done:
action = self.choose_action(state,self.epsilon)
next_state, reward, done, _ = self.env.step(action)
next_state = self.preprocess_state(next_state)
next_state = self.combine_images(next_state, state)
#print(next_state.shape)
self.remember(state, action, reward, next_state, done)
state = next_state
self.epsilon = max(self.epsilon_min, self.epsilon_decay*self.epsilon) # decrease epsilon
i += reward
scores.append(i)
mean_score = np.mean(scores)
avg_scores.append(mean_score)
if mean_score >= THRESHOLD:
print('Solved after {} trials ✔'.format(e))
return avg_scores
if e % 10 == 0:
print('[Episode {}] - Average Score: {}.'.format(e, mean_score))
self.model_target.set_weights(self.model.get_weights())
self.replay(self.batch_size)
print('Did not solve after {} episodes 😞'.format(e))
return avg_scores
env_string = 'BreakoutDeterministic-v4'
agent = DQN(env_string)
agent.model.summary()
agent.model_target.summary()
scores = agent.train()
import matplotlib.pyplot as plt
plt.plot(scores)
plt.show()
agent.env.close()
```
|
github_jupyter
|
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import glob
import sys
import argparse as argp
change_50_dat = pd.read_csv('/Users/leg2015/workspace/Aagos/Data/Mut_Treat_Change_50_CleanedDataStatFit.csv', index_col="update", float_precision="high")
change_0_dat = pd.read_csv('/Users/leg2015/workspace/Aagos/Data/Mut_Treat_Change_0_CleanedDataStatFit.csv', index_col="update", float_precision="high")
mut_dat = pd.read_csv('/Users/leg2015/workspace/Aagos/Data/Change_Treat_f_.003_CleanedDataStatFit.csv', index_col="update", float_precision="high")
# max_gen_data = all_dat.loc[49000]
# early_gen_data = all_dat.loc[10000]
change_50_max = change_50_dat.loc[50000]
change_0_max = change_0_dat.loc[50000]
mut_max = mut_dat.loc[50000]
change_50_plot = sns.boxplot(y=max_gen_change_data["mean_coding_sites"], x="change", data=max_gen_change_data)
plt.suptitle("gen 49000 mean coding sites ")
plt.savefig("Change_m_.003_c_.01_f_.001_mean_coding_2.pdf")
max_gen_fitness = max_gen_data.loc[:,[ 'max_fitness', 'c', 'm', 'f', 'replicate']]
max_gen_gene_len = max_gen_data.loc[:, ['max_gene_length', 'c', 'm', 'f', 'replicate']]
max_gen_overlap = max_gen_data.loc[:,[ 'max_overlap', 'c', 'm', 'f', 'replicate']]
max_gen_coding = max_gen_data.loc[:,[ 'max_coding_sites', 'c', 'm', 'f', 'replicate']]
max_gen_neutral = max_gen_data.loc[:,[ 'max_neutral_sites', 'c', 'm', 'f', 'replicate']]
max_gen_neighbor = max_gen_data.loc[:,[ 'max_neighbor_genes', 'c', 'm', 'f', 'replicate']]
facet = sns.FacetGrid(max_gen_gene_len, col="c", row="m",)
facet.map_dataframe(sns.boxplot, x="f", y="max_gene_length")
for axis in facet.axes.flat:
axis.set_xlabel("f")
plt.subplots_adjust(top=0.95)
facet.fig.suptitle('Gen 49,000 max gene length') # can also get
facet.savefig("Mut_Rate_Low_max_genlen.pdf")
# TODO: figure early_gen_neighbor how to save pdfs to figure directory
# also way so don't have to boilerplate would be nice
facet = sns.FacetGrid(early_gen_rep_gene_len, col="c", row="m",)
facet.map_dataframe(sns.boxplot, x="f", y="genome_size")
for axis in facet.axes.flat:
axis.set_xlabel("f")
plt.subplots_adjust(top=0.95)
facet.fig.suptitle('Gen 10,000 genome size of representative org') # can also get
facet.savefig("early_gen_rep_gene_len.pdf")
facet = sns.FacetGrid(max_gen_overlap, col="c", row="m",)
facet.map_dataframe(sns.boxplot, x="f", y="mean_Overlap")
for axis in facet.axes.flat:
axis.set_xlabel("f")
plt.subplots_adjust(top=0.95)
facet.fig.suptitle('Gen 49,000 mean overlap') # can also get
facet.savefig("max_gen_mean_overlap.pdf")
facet = sns.FacetGrid(early_gen_overlap, col="c", row="m",)
facet.map_dataframe(sns.boxplot, x="f", y="mean_Overlap")
for axis in facet.axes.flat:
axis.set_xlabel("f")
plt.subplots_adjust(top=0.95)
facet.fig.suptitle('Gen 10,000 mean overlap') # can also get
facet.savefig("early_gen_mean_overlap.pdf")
facet = sns.FacetGrid(early_gen_fitness, col="c", row="m",)
facet.map_dataframe(sns.boxplot, x="f", y="mean_fitness")
for axis in facet.axes.flat:
axis.set_xlabel("f")
plt.subplots_adjust(top=0.95)
facet.fig.suptitle('Gen 10,000 mean fitness') # can also get
facet.savefig("early_gen_mean_fitness.pdf")
facet = sns.FacetGrid(max_gen_fitness, col="c", row="m",)
facet.map_dataframe(sns.boxplot, x="f", y="mean_fitness")
# for plot in plt.subplots():
# print(plot.AxesSubplot)
# print(facet.axes[4,0].get_yticklabels())
# print(facet.axes[4,0].get_xticklabels())
# print("\n")
# labels = ["meme1", "meme2"]
for axis in facet.axes.flat:
# locs, labels = axis.xticks()
# axis.set_yticklabels(labels)
axis.set_xlabel("f")
# _ = plt.setp(axis.get_yticklabels(), visible=True)
# _ = plt.setp(axis.get_xticklabels(), visible=True)
# axis.set_yticklabels(axis.get_yticklabels(), visible=True)
# print(axis.get_xticklabels())
# axis.set_xticklabels(axis.get_xticklabels(), visible=True)
# axis.set_yticklabels(facet.axes[4,0].get_yticklabels(), visible=True)
# axis.set_xticklabels(facet.axes[4,0].get_xticklabels(), visible=True)
plt.subplots_adjust(hspace=0.3)
# yticklabels = facet.axes[4,0].get_yticklabels()
# xticklabels = facet.axes[4,0].get_xticklabels()
# meme = []
# mema = []
# for ax in facet.axes[-1,:]:
# xlabel = ax.get_xticklabels()
# print("x lab ", xlabel)
# meme.append(xlabel)
# for ax in facet.axes[:,0]:
# ylabel = ax.get_xticklabels()
# print("y lab", ylabel)
# mema.append(ylabel)
# for i in range(len(meme)):
# for j in range(len(mema)):
# facet.axes[j,i].set_xticklabels("meme")
# facet.axes[j,i].set_yticklabels("mema")
# for ax in facet.axes:
# _ = plt.setp(ax.get_yticklabels(), visible=True)
# _ = plt.setp(ax.get_xticklabels(), visible=True)
# for ax in facet.axes:
# plt.show()
plt.subplots_adjust(top=0.95)
facet.fig.suptitle('Gen 49,000 mean fitness') # can also get
facet.savefig("max_gen_mean_fitness.pdf")
facet = sns.FacetGrid(max_gen_fitness, col="c", row="m",)
facet.map_dataframe(sns.boxplot, x="f", y="mean_fitness")
# for plot in plt.subplots():
# print(plot.AxesSubplot)
# print(facet.axes[4,0].get_yticklabels())
# print(facet.axes[4,0].get_xticklabels())
# print("\n")
# labels = ["meme1", "meme2"]
for axis in facet.axes.flat:
# locs, labels = axis.xticks()
# axis.set_yticklabels(labels)
axis.set_xlabel("f")
# _ = plt.setp(axis.get_yticklabels(), visible=True)
# _ = plt.setp(axis.get_xticklabels(), visible=True)
# axis.set_yticklabels(axis.get_yticklabels(), visible=True)
# print(axis.get_xticklabels())
# axis.set_xticklabels(axis.get_xticklabels(), visible=True)
# axis.set_yticklabels(facet.axes[4,0].get_yticklabels(), visible=True)
# axis.set_xticklabels(facet.axes[4,0].get_xticklabels(), visible=True)
plt.subplots_adjust(hspace=0.3)
# yticklabels = facet.axes[4,0].get_yticklabels()
# xticklabels = facet.axes[4,0].get_xticklabels()
# meme = []
# mema = []
# for ax in facet.axes[-1,:]:
# xlabel = ax.get_xticklabels()
# print("x lab ", xlabel)
# meme.append(xlabel)
# for ax in facet.axes[:,0]:
# ylabel = ax.get_xticklabels()
# print("y lab", ylabel)
# mema.append(ylabel)
# for i in range(len(meme)):
# for j in range(len(mema)):
# facet.axes[j,i].set_xticklabels("meme")
# facet.axes[j,i].set_yticklabels("mema")
# for ax in facet.axes:
# _ = plt.setp(ax.get_yticklabels(), visible=True)
# _ = plt.setp(ax.get_xticklabels(), visible=True)
# for ax in facet.axes:
plt.show()
facet.savefig("max_gen__mean_fitness.pdf")
for curr in group:
plt.scatter((curr[1].m + curr[1].f + curr[1].c), curr[1].mean_fitness)
plt.show()
playData = max_gen_fitness.iloc[0:5]
playData
memes = max_gen_fitness.iloc[20:25]
memes
plt.boxplot(playData.mean_fitness)
plt.show()
# plt.boxplot(playData.mean_fitness)
# plt.boxplot(memes.mean_fitness)
superData = [playData.mean_fitness, memes.mean_fitness]
plt.boxplot(superData)
plt.show()
```
|
github_jupyter
|
# Deploy model
**Important**: Change the kernel to *PROJECT_NAME local*. You can do this from the *Kernel* menu under *Change kernel*. You cannot deploy the model using the *PROJECT_NAME docker* kernel.
```
from azureml.api.schema.dataTypes import DataTypes
from azureml.api.schema.sampleDefinition import SampleDefinition
from azureml.api.realtime.services import generate_schema
import pandas as pd
import numpy as np
import imp
import pickle
import os
import sys
import json
from azureml.logging import get_azureml_logger
run_logger = get_azureml_logger()
run_logger.log('amlrealworld.timeseries.deploy-model','true')
```
Enter the name of the model to deploy.
```
model_name = "linear_regression"
```
Load the test dataset and retain just one row. This record will be used to create and input schema for the web service. It will also allow us to simulate invoking the web service with features for one hour period and generating a demand forecast for this hour.
```
aml_dir = os.environ['AZUREML_NATIVE_SHARE_DIRECTORY']
test_df = pd.read_csv(os.path.join(aml_dir, 'nyc_demand_test.csv'), parse_dates=['timeStamp'])
test_df = test_df.drop(['demand', 'timeStamp'], axis=1).copy().iloc[[0]]
test_df
```
Load model from disk and transfer it to the working directory.
```
with open(os.path.join(aml_dir, model_name + '.pkl'), 'rb') as f:
mod = pickle.load(f)
with open('model_deploy.pkl', 'wb') as f:
pickle.dump(mod, f)
```
Check model object has loaded as expected.
```
mod
```
Apply model to predict test record
```
np.asscalar(mod.predict(test_df))
```
### Author a realtime web service
Create a score.py script which implements the scoring function to run inside the web service. Change model_name variable as required.
```
%%writefile score.py
# The init and run functions will load and score the input using the saved model.
# The score.py file will be included in the web service deployment package.
def init():
import pickle
import os
global model
with open('model_deploy.pkl', 'rb') as f:
model = pickle.load(f)
def run(input_df):
input_df = input_df[['precip', 'temp', 'hour', 'month', 'dayofweek',
'temp_lag1', 'temp_lag2', 'temp_lag3', 'temp_lag4', 'temp_lag5',
'temp_lag6', 'demand_lag1', 'demand_lag2', 'demand_lag3',
'demand_lag4', 'demand_lag5', 'demand_lag6']]
try:
if (input_df.shape != (1,17)):
return 'Bad imput: Expecting dataframe of shape (1,17)'
else:
pred = model.predict(input_df)
return int(pred)
except Exception as e:
return(str(e))
```
This script will be written to your current working directory:
```
os.getcwd()
```
#### Test the *init* and *run* functions
```
import score
imp.reload(score)
score.init()
score.run(test_df)
```
#### Create web service schema
The web service schema provides details on the required structure of the input data as well as the data types of each column.
```
inputs = {"input_df": SampleDefinition(DataTypes.PANDAS, test_df)}
generate_schema(run_func=score.run, inputs=inputs, filepath='service_schema.json')
```
#### Deploy the web service
The command below deploys a web service names "demandforecast", with input schema defined by "service_schema.json". The web service runs "score.py" which scores the input data using the model "model_deploy.pkl". This may take a few minutes.
```
!az ml service create realtime -f score.py -m model_deploy.pkl -s service_schema.json -n demandforecast -r python
```
Check web service is running.
```
!az ml service show realtime -i demandforecast
```
Test the web service is working by invoking it with a test record.
```
!az ml service run realtime -i demandforecast -d "{\"input_df\": [{\"hour\": 0, \"month\": 6, \"demand_lag3\": 7576.558, \"temp_lag5\": 77.36, \"temp\": 74.63, \"demand_lag1\": 6912.7, \"demand_lag5\": 7788.292, \"temp_lag6\": 80.92, \"temp_lag3\": 76.72, \"demand_lag6\": 8102.142, \"temp_lag4\": 75.85, \"precip\": 0.0, \"temp_lag2\": 75.72, \"demand_lag2\": 7332.625, \"temp_lag1\": 75.1, \"demand_lag4\": 7603.008, \"dayofweek\": 4}]}"
```
#### Delete the web service
```
!az ml service delete realtime --id=demandforecast
```
|
github_jupyter
|
# DiFuMo (Dictionaries of Functional Modes)
<div class="alert alert-block alert-danger">
<b>NEW:</b> New in release 0.7.1
</div>
## Outline
- <a href="#descr">Description</a>
- <a href="#howto">Description</a>
- <a href="#closer">Coser look on the object</a>
- <a href="#visualize">Visualize</a>
<span id="descr"></span>
## Description
- New atlas fetcher :func:`nilearn.datasets.fetch_atlas_difumo`
- Download statistical maps which can serve as atlases to extract functional signals with different dimensionalities (64, 128, 256, 512, and 1024)
- These modes are optimized to represent well raw BOLD timeseries, over a with range of experimental conditions.
<span id="howto"></span>
## How to use it?
First of all, make sure you have nilearn >= 0.7.1 installed:
```
import nilearn
print(nilearn.__version__)
```
If this is verified, we should be able to export the difumo fetcher from the `datasets` module:
```
from nilearn.datasets import fetch_atlas_difumo
```
The documentation for this function can be seen on the website [here](http://nilearn.github.io/modules/generated/nilearn.datasets.fetch_atlas_difumo.html#nilearn.datasets.fetch_atlas_difumo) or thanks to the Jupyter magic command:
```
?fetch_atlas_difumo
```
Looking at the docstring, it looks like there are mainly two parameters to control the data we wish to donwload:
- dimension: this will be the number of functional maps of the atlas. It must be 64, 128, 256, 512, or 1024
- resolution: this enables to download atlas sampled either at 2mm or 3mm resolution
Let's try it:
```
difumo_64 = fetch_atlas_difumo(dimension=64, # Feel free to change these parameters!
resolution_mm=2)
```
This should have either downloaded the 64 component atlas sampled at 2mm from osf, or simply grabed the data in the nilearn cache if you have downloaded it already.
<span id="closer"></span>
## Closer look on the object
Like for any dataset in nilearn, the resulting object is a scikit-learn Bunch object with the following keys:
- description: string describing the dataset
- maps: the actual data
- labels: label information for the maps
```
type(difumo_64)
difumo_64.keys()
```
Reading the description before usage is always recommanded:
```
print(difumo_64.description.decode()) # Note that description strings will be soon shipped as Python strings,
# avoiding the anoying call to decode...
```
Label information is directly available:
```
assert len(difumo_64.labels) == 64 # We have one label information tuple per component
difumo_64.labels[:6] # Print the first 6 label information
```
We can see that each component has:
- a label index going from 1 to 64
- a name
- a network (todo: explain)
- a network (todo: explain)
- coordinates (todo: explain)
Finally, the actual data is a simple path to a nifti image on disk, which is the usual way to represent niimg in Nilearn:
```
difumo_64.maps
```
If you wan to have a look at the actual data, you can open this image using usual nilearn loading utilities:
```
from nilearn.image import get_data
raw_maps = get_data(difumo_64.maps) # raw_maps is a 4D numpy array holding the
raw_maps.shape # coefficients of the functional modes
```
<span id="visualize"></span>
## Visualize it
**Method 1**
Looking at probabilitic atlases can be done with the function `plot_prob_atlas` of the `plotting` module:
```
from nilearn.plotting import plot_prob_atlas
plot_prob_atlas(difumo_64.maps, title='DiFuMo 64')
```
**Method 2**
Another way to visualize the atlas is through the report of the `NiftiMapsMasker` object.
<div class="alert alert-block alert-danger">
<b>Danger:</b> This feature is under development and still not available in 0.7.1. I might remove this section if I don't submit my PR in time.
</div>
```
from nilearn.input_data import NiftiMapsMasker
masker = NiftiMapsMasker(difumo_64.maps)
masker
```
|
github_jupyter
|
# Tutorial 06: Networks from OpenStreetMap
In this tutorial, we discuss how networks that have been imported from OpenStreetMap can be integrated and run in Flow. This will all be presented via the Bay Bridge network, seen in the figure below. Networks from OpenStreetMap are commonly used in many traffic simulators for the purposes of replicating traffic in realistic traffic geometries. This is true in both SUMO and Aimsun (which are both supported in Flow), with each supporting several techniques for importing such network files. This process is further simplified and abstracted in Flow, with users simply required to specify the path to the osm file in order to simulate traffic in the network.
<img src="img/bay_bridge_osm.png" width=750>
<center> **Figure 1**: Snapshot of the Bay Bridge from OpenStreetMap </center>
Before we begin, let us import all relevant Flow parameters as we have done for previous tutorials. If you are unfamiliar with these parameters, you are encouraged to review tutorial 1.
```
# the TestEnv environment is used to simply simulate the network
from flow.envs import TestEnv
# the Experiment class is used for running simulations
from flow.core.experiment import Experiment
# all other imports are standard
from flow.core.params import VehicleParams
from flow.core.params import NetParams
from flow.core.params import InitialConfig
from flow.core.params import EnvParams
from flow.core.params import SumoParams
```
## 1. Running a Default Simulation
In order to create a network object in Flow with network features depicted from OpenStreetMap, we will use the base `Network` class. This class can sufficiently support the generation of any .osm file.
```
from flow.networks import Network
```
In order to recreate the network features of a specific osm file, the path to the osm file must be specified in `NetParams`. For this example, we will use an osm file extracted from the section of the Bay Bridge as depicted in Figure 1.
In order to specify the path to the osm file, simply fill in the `osm_path` attribute with the path to the .osm file as follows:
```
net_params = NetParams(
osm_path='networks/bay_bridge.osm'
)
```
Next, we create all other parameters as we have in tutorials 1 and 2. For this example, we will assume a total of 1000 are uniformly spread across the Bay Bridge. Once again, if the choice of parameters is unclear, you are encouraged to review Tutorial 1.
```
# create the remainding parameters
env_params = EnvParams()
sim_params = SumoParams(render=True)
initial_config = InitialConfig()
vehicles = VehicleParams()
vehicles.add('human', num_vehicles=100)
# create the network
network = Network(
name='bay_bridge',
net_params=net_params,
initial_config=initial_config,
vehicles=vehicles
)
```
We are finally ready to test our network in simulation. In order to do so, we create an `Experiment` object and run the simulation for a number of steps. This is done in the cell below.
```
# create the environment
env = TestEnv(
env_params=env_params,
sim_params=sim_params,
network=network
)
# run the simulation for 1000 steps
exp = Experiment(env=env)
exp.run(1, 1000)
```
## 2. Customizing the Network
While the above example does allow you to view the network within Flow, the simulation is limited for two reasons. For one, vehicles are placed on all edges within the network; if we wished to simulate traffic solely on the on the bridge and do not care about the artireols, for instance, this would result in unnecessary computational burdens. Next, as you may have noticed if you ran the above example to completion, routes in the base network class are defaulted to consist of the vehicles' current edges only, meaning that vehicles exit the network as soon as they reach the end of the edge they are originated on. In the next subsections, we discuss how the network can be modified to resolve these issues.
### 2.1 Specifying Traversable Edges
In order to limit the edges vehicles are placed on to the road sections edges corresponding to the westbound Bay Bridge, we define an `EDGES_DISTRIBUTION` variable. This variable specifies the names of the edges within the network that vehicles are permitted to originated in, and is assigned to the network via the `edges_distribution` component of the `InitialConfig` input parameter, as seen in the code snippet below. Note that the names of the edges can be identified from the .osm file or by right clicking on specific edges from the SUMO gui (see the figure below).
<img src="img/osm_edge_name.png" width=600>
<center> **Figure 2**: Name of an edge from SUMO </center>
```
# we define an EDGES_DISTRIBUTION variable with the edges within
# the westbound Bay Bridge
EDGES_DISTRIBUTION = [
"11197898",
"123741311",
"123741303",
"90077193#0",
"90077193#1",
"340686922",
"236348366",
"340686911#0",
"340686911#1",
"340686911#2",
"340686911#3",
"236348361",
"236348360#0",
"236348360#1"
]
# the above variable is added to initial_config
new_initial_config = InitialConfig(
edges_distribution=EDGES_DISTRIBUTION
)
```
### 2.2 Creating Custom Routes
Next, we choose to specify the routes of vehicles so that they can traverse the entire Bay Bridge, instead of the only the edge they are currently on. In order to this, we create a new network class that inherits all its properties from `Network` and simply redefine the routes by modifying the `specify_routes` variable. This method was originally introduced in Tutorial 07: Creating Custom Network. The new network class looks as follows:
```
# we create a new network class to specify the expected routes
class BayBridgeOSMNetwork(Network):
def specify_routes(self, net_params):
return {
"11197898": [
"11197898", "123741311", "123741303", "90077193#0", "90077193#1",
"340686922", "236348366", "340686911#0", "340686911#1",
"340686911#2", "340686911#3", "236348361", "236348360#0", "236348360#1",
],
"123741311": [
"123741311", "123741303", "90077193#0", "90077193#1", "340686922",
"236348366", "340686911#0", "340686911#1", "340686911#2",
"340686911#3", "236348361", "236348360#0", "236348360#1"
],
"123741303": [
"123741303", "90077193#0", "90077193#1", "340686922", "236348366",
"340686911#0", "340686911#1", "340686911#2", "340686911#3", "236348361",
"236348360#0", "236348360#1"
],
"90077193#0": [
"90077193#0", "90077193#1", "340686922", "236348366", "340686911#0",
"340686911#1", "340686911#2", "340686911#3", "236348361", "236348360#0",
"236348360#1"
],
"90077193#1": [
"90077193#1", "340686922", "236348366", "340686911#0", "340686911#1",
"340686911#2", "340686911#3", "236348361", "236348360#0", "236348360#1"
],
"340686922": [
"340686922", "236348366", "340686911#0", "340686911#1", "340686911#2",
"340686911#3", "236348361", "236348360#0", "236348360#1"
],
"236348366": [
"236348366", "340686911#0", "340686911#1", "340686911#2", "340686911#3",
"236348361", "236348360#0", "236348360#1"
],
"340686911#0": [
"340686911#0", "340686911#1", "340686911#2", "340686911#3", "236348361",
"236348360#0", "236348360#1"
],
"340686911#1": [
"340686911#1", "340686911#2", "340686911#3", "236348361", "236348360#0",
"236348360#1"
],
"340686911#2": [
"340686911#2", "340686911#3", "236348361", "236348360#0", "236348360#1"
],
"340686911#3": [
"340686911#3", "236348361", "236348360#0", "236348360#1"
],
"236348361": [
"236348361", "236348360#0", "236348360#1"
],
"236348360#0": [
"236348360#0", "236348360#1"
],
"236348360#1": [
"236348360#1"
]
}
```
### 2.3 Rerunning the SImulation
We are now ready to rerun the simulation with fully defined vehicle routes and a limited number of traversable edges. If we run the cell below, we can see the new simulation in action.
```
# create the network
new_network = BayBridgeOSMNetwork(
name='bay_bridge',
net_params=net_params,
initial_config=new_initial_config,
vehicles=vehicles,
)
# create the environment
env = TestEnv(
env_params=env_params,
sim_params=sim_params,
network=new_network
)
# run the simulation for 1000 steps
exp = Experiment(env=env)
exp.run(1, 10000)
```
## 3. Other Tips
This tutorial introduces how to incorporate OpenStreetMap files in Flow. This feature, however, does not negate other features that are introduced in other tutorials and documentation. For example, if you would like to not have vehicles be originated side-by-side within a network, this can still be done by specifying a "random" spacing for vehicles as follows:
initial_config = InitialConfig(
spacing="random",
edges_distribution=EDGES_DISTRIBUTION
)
In addition, inflows of vehicles can be added to networks imported from OpenStreetMap as they are for any other network (see the tutorial on adding inflows for more on this).
|
github_jupyter
|
<table width="100%"> <tr>
<td style="background-color:#ffffff;">
<a href="http://qworld.lu.lv" target="_blank"><img src="../images/qworld.jpg" width="35%" align="left"> </a></td>
<td style="background-color:#ffffff;vertical-align:bottom;text-align:right;">
prepared by Abuzer Yakaryilmaz (<a href="http://qworld.lu.lv/index.php/qlatvia/" target="_blank">QLatvia</a>)
<br>
Özlem Salehi | July 6, 2019 (updated)
</td>
</tr></table>
<table width="100%"><tr><td style="color:#bbbbbb;background-color:#ffffff;font-size:11px;font-style:italic;text-align:right;">This cell contains some macros. If there is a problem with displaying mathematical formulas, please run this cell to load these macros. </td></tr></table>
$ \newcommand{\bra}[1]{\langle #1|} $
$ \newcommand{\ket}[1]{|#1\rangle} $
$ \newcommand{\braket}[2]{\langle #1|#2\rangle} $
$ \newcommand{\dot}[2]{ #1 \cdot #2} $
$ \newcommand{\biginner}[2]{\left\langle #1,#2\right\rangle} $
$ \newcommand{\mymatrix}[2]{\left( \begin{array}{#1} #2\end{array} \right)} $
$ \newcommand{\myvector}[1]{\mymatrix{c}{#1}} $
$ \newcommand{\myrvector}[1]{\mymatrix{r}{#1}} $
$ \newcommand{\mypar}[1]{\left( #1 \right)} $
$ \newcommand{\mybigpar}[1]{ \Big( #1 \Big)} $
$ \newcommand{\sqrttwo}{\frac{1}{\sqrt{2}}} $
$ \newcommand{\dsqrttwo}{\dfrac{1}{\sqrt{2}}} $
$ \newcommand{\onehalf}{\frac{1}{2}} $
$ \newcommand{\donehalf}{\dfrac{1}{2}} $
$ \newcommand{\hadamard}{ \mymatrix{rr}{ \sqrttwo & \sqrttwo \\ \sqrttwo & -\sqrttwo }} $
$ \newcommand{\vzero}{\myvector{1\\0}} $
$ \newcommand{\vone}{\myvector{0\\1}} $
$ \newcommand{\vhadamardzero}{\myvector{ \sqrttwo \\ \sqrttwo } } $
$ \newcommand{\vhadamardone}{ \myrvector{ \sqrttwo \\ -\sqrttwo } } $
$ \newcommand{\myarray}[2]{ \begin{array}{#1}#2\end{array}} $
$ \newcommand{\X}{ \mymatrix{cc}{0 & 1 \\ 1 & 0} } $
$ \newcommand{\Z}{ \mymatrix{rr}{1 & 0 \\ 0 & -1} } $
$ \newcommand{\Htwo}{ \mymatrix{rrrr}{ \frac{1}{2} & \frac{1}{2} & \frac{1}{2} & \frac{1}{2} \\ \frac{1}{2} & -\frac{1}{2} & \frac{1}{2} & -\frac{1}{2} \\ \frac{1}{2} & \frac{1}{2} & -\frac{1}{2} & -\frac{1}{2} \\ \frac{1}{2} & -\frac{1}{2} & -\frac{1}{2} & \frac{1}{2} } } $
$ \newcommand{\CNOT}{ \mymatrix{cccc}{1 & 0 & 0 & 0 \\ 0 & 1 & 0 & 0 \\ 0 & 0 & 0 & 1 \\ 0 & 0 & 1 & 0} } $
$ \newcommand{\norm}[1]{ \left\lVert #1 \right\rVert } $
<h2> <font color="blue"> Solutions for </font>Rotation Automata</h2>
<a id="task1"></a>
<h3> Task 1 </h3>
Do the same task given above by using different angles.
Test at least three different angles.
Please modify the code above.
<h3>Solution</h3>
Any odd multiple of $ \frac{\pi}{16} $ works: $ i \frac{\pi}{16} $, where $ i \in \{1,3,5,7,\ldots\} $
<a id="task2"></a>
<h3> Task 2 </h3>
Let $ \mathsf{p} = 11 $.
Determine an angle of rotation such that when the length of stream is a multiple of $ \sf p $, then we observe only state $ 0 $, and we can also observe state $ 1 $, otherwise.
Test your rotation by using a quantum circuit. Execute the circuit for all streams of lengths from 1 to 11.
<h3>Solution</h3>
We can pick any angle $ k\frac{2\pi}{11} $ for $ k \in \{1,\ldots,10\} $.
```
from qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer
from math import pi
from random import randrange
# the angle of rotation
r = randrange(1,11)
print("the picked angle is",r,"times of 2pi/11")
print()
theta = r*pi/11
# we read streams of length from 1 to 11
for i in range(1,12):
# quantum circuit with one qubit and one bit
qreg = QuantumRegister(1)
creg = ClassicalRegister(1)
mycircuit = QuantumCircuit(qreg,creg)
# the stream of length i
for j in range(i):
mycircuit.ry(2*theta,qreg[0]) # apply one rotation for each symbol
# we measure after reading the whole stream
#mycircuit.measure(qreg[0],creg[0])
# execute the circuit 1000 times
job = execute(mycircuit,Aer.get_backend('unitary_simulator'))
u=job.result().get_unitary(mycircuit,decimals=3)
# we print the unitary matrix in nice format
for i in range(len(u)):
s=""
for j in range(len(u)):
val = str(u[i][j].real)
while(len(val)<8): val = " "+val
s = s + val
print(s)
```
<a id="task3"></a>
<h3> Task 3 </h3>
List down 10 possible different angles for Task 2, where each angle should be between 0 and $2\pi$.
<h3>Solution</h3>
Any angle $ k\frac{2\pi}{11} $ for $ k \in \{1,\ldots,10\} $.
<h3>Task 4</h3>
For each stream of length from 1 to 10, experimentially determine the best angle of rotation (we observe state $\ket{1}$ the most) by using your circuit.
<h3>Solution</h3>
```
from qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer
from math import pi
from random import randrange
# for each stream of length from 1 to 10
for i in range(1,11):
# we try each angle of the form k*2*pi/11 for k=1,...,10
# we try to find the best k for which we observe 1 the most
number_of_one_state = 0
best_k = 1
all_outcomes_for_i = "length "+str(i)+"-> "
for k in range(1,11):
theta = k*2*pi/11
# quantum circuit with one qubit and one bit
qreg = QuantumRegister(1)
creg = ClassicalRegister(1)
mycircuit = QuantumCircuit(qreg,creg)
# the stream of length i
for j in range(i):
mycircuit.ry(2*theta,qreg[0]) # apply one rotation for each symbol
# we measure after reading the whole stream
mycircuit.measure(qreg[0],creg[0])
# execute the circuit 10000 times
job = execute(mycircuit,Aer.get_backend('qasm_simulator'),shots=10000)
counts = job.result().get_counts(mycircuit)
all_outcomes_for_i = all_outcomes_for_i + str(k)+ ":" + str(counts['1']) + " "
if int(counts['1']) > number_of_one_state:
number_of_one_state = counts['1']
best_k = k
print(all_outcomes_for_i)
print("for length",i,", the best k is",best_k)
print()
```
<a id="task5"></a>
<h3> Task 5 </h3>
Let $ \mathsf{p} = 31 $.
Create a circuit with three quantum bits and three classical bits.
Rotate the qubits with angles $ 3\frac{2\pi}{31} $, $ 7\frac{2\pi}{31} $, and $ 11\frac{2\pi}{31} $, respectively.
Execute your circuit for all streams of lengths from 1 to 30. Check whether the number of state $ \ket{000} $ is less than half or not.
<i>Note that whether a key is in dictionary or not can be checked as follows:</i>
```python
if '000' in counts.keys():
c = counts['000']
else:
c = 0
```
<h3>Solution</h3>
```
from qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer
from math import pi
from random import randrange
# the angles of rotations
theta1 = 3*2*pi/31
theta2 = 7*2*pi/31
theta3 = 11*2*pi/31
# we read streams of length from 1 to 30
for i in range(1,32):
# quantum circuit with three qubits and three bits
qreg = QuantumRegister(3)
creg = ClassicalRegister(3)
mycircuit = QuantumCircuit(qreg,creg)
# the stream of length i
for j in range(i):
# apply rotations for each symbol
mycircuit.ry(2*theta1,qreg[0])
mycircuit.ry(2*theta2,qreg[1])
mycircuit.ry(2*theta3,qreg[2])
# we measure after reading the whole stream
mycircuit.measure(qreg,creg)
# execute the circuit N times
N = 1000
job = execute(mycircuit,Aer.get_backend('qasm_simulator'),shots=N)
counts = job.result().get_counts(mycircuit)
print(counts)
if '000' in counts.keys():
c = counts['000']
else:
c = 0
print('000 is observed',c,'times out of',N)
percentange = round(c/N*100,1)
print("the ratio of 000 is ",percentange,"%")
print()
```
<a id="task6"></a>
<h3> Task 6 </h3>
Let $ \mathsf{p} = 31 $.
Create a circuit with three quantum bits and three classical bits.
Rotate the qubits with random angles of the form $ k\frac{2\pi}{31}, $ where $ k
\in \{1,\ldots,30\}.$
Execute your circuit for all streams of lengths from 1 to 30.
Calculate the maximum percentage of observing the state $ \ket{000} $.
Repeat this task for a few times.
<i>Note that whether a key is in dictionary or not can be checked as follows:</i>
```python
if '000' in counts.keys():
c = counts['000']
else:
c = 0
```
<h3>Solution</h3>
```
from qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer
from math import pi
from random import randrange
# randomly picked angles of rotations
k1 = randrange(1,31)
theta1 = k1*2*pi/31
k2 = randrange(1,31)
theta2 = k2*2*pi/31
k3 = randrange(1,31)
theta3 = k3*2*pi/31
print("k1 =",k1,"k2 =",k2,"k3 =",k3)
print()
max_percentange = 0
# we read streams of length from 1 to 30
for i in range(1,31):
k1 = randrange(1,31)
theta1 = k1*2*pi/31
k2 = randrange(1,31)
theta2 = k2*2*pi/31
k3 = randrange(1,31)
theta3 = k3*2*pi/31
# quantum circuit with three qubits and three bits
qreg = QuantumRegister(3)
creg = ClassicalRegister(3)
mycircuit = QuantumCircuit(qreg,creg)
# the stream of length i
for j in range(i):
# apply rotations for each symbol
mycircuit.ry(2*theta1,qreg[0])
mycircuit.ry(2*theta2,qreg[1])
mycircuit.ry(2*theta3,qreg[2])
# we measure after reading the whole stream
mycircuit.measure(qreg,creg)
# execute the circuit N times
N = 1000
job = execute(mycircuit,Aer.get_backend('qasm_simulator'),shots=N)
counts = job.result().get_counts(mycircuit)
# print(counts)
if '000' in counts.keys():
c = counts['000']
else:
c = 0
# print('000 is observed',c,'times out of',N)
percentange = round(c/N*100,1)
if max_percentange < percentange: max_percentange = percentange
# print("the ration of 000 is ",percentange,"%")
# print()
print("max percentage is",max_percentange)
```
<a id="task7"></a>
<h3> Task 7 </h3>
Repeat Task 6 by using four and five qubits.
<h3>Solution</h3>
```
from qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer
from math import pi
from random import randrange
number_of_qubits = 4
#number_of_qubits = 5
# randomly picked angles of rotations
theta = []
for i in range(number_of_qubits):
k = randrange(1,31)
print("k",str(i),"=",k)
theta += [k*2*pi/31]
# print(theta)
# we count the number of zeros
zeros = ''
for i in range(number_of_qubits):
zeros = zeros + '0'
print("zeros = ",zeros)
print()
max_percentange = 0
# we read streams of length from 1 to 30
for i in range(1,31):
# quantum circuit with qubits and bits
qreg = QuantumRegister(number_of_qubits)
creg = ClassicalRegister(number_of_qubits)
mycircuit = QuantumCircuit(qreg,creg)
# the stream of length i
for j in range(i):
# apply rotations for each symbol
for k in range(number_of_qubits):
mycircuit.ry(2*theta[k],qreg[k])
# we measure after reading the whole stream
mycircuit.measure(qreg,creg)
# execute the circuit N times
N = 1000
job = execute(mycircuit,Aer.get_backend('qasm_simulator'),shots=N)
counts = job.result().get_counts(mycircuit)
# print(counts)
if zeros in counts.keys():
c = counts[zeros]
else:
c = 0
# print('000 is observed',c,'times out of',N)
percentange = round(c/N*100,1)
if max_percentange < percentange: max_percentange = percentange
# print("the ration of 000 is ",percentange,"%")
# print()
print("max percentage is",max_percentange)
```
|
github_jupyter
|
```
# Execute this code block to install dependencies when running on colab
try:
import torch
except:
from os.path import exists
from wheel.pep425tags import get_abbr_impl, get_impl_ver, get_abi_tag
platform = '{}{}-{}'.format(get_abbr_impl(), get_impl_ver(), get_abi_tag())
cuda_output = !ldconfig -p|grep cudart.so|sed -e 's/.*\.\([0-9]*\)\.\([0-9]*\)$/cu\1\2/'
accelerator = cuda_output[0] if exists('/dev/nvidia0') else 'cpu'
!pip install -q http://download.pytorch.org/whl/{accelerator}/torch-1.0.0-{platform}-linux_x86_64.whl torchvision
! pip install --upgrade git+https://github.com/sovrasov/flops-counter.pytorch.git
try:
import torchtext
except:
!pip install torchtext
try:
import spacy
except:
!pip install spacy
try:
spacy.load('en')
except:
!python -m spacy download en
```
# Data loading and preprocessing
```
import torch
from torchtext import data
import torch.nn.functional as F
from torch import nn
from torch import optim
from torch.distributions import Categorical
from torch.distributions import Binomial
from torchtext import datasets
import os.path
import random
import numpy as np
TEXT = data.Field(tokenize='spacy', lower=True, include_lengths=True)
LABEL = data.LabelField(dtype=torch.float)
_train_data, test_data = datasets.IMDB.splits(TEXT, LABEL)
train_data, valid_data = _train_data.split(0.8)
print(f'Number of training examples: {len(train_data)}')
print(f'Number of validation examples: {len(valid_data)}')
print(f'Number of testing examples: {len(test_data)}')
TEXT.build_vocab(train_data, max_size=100000, vectors="glove.6B.100d")
LABEL.build_vocab(train_data)
print(f"Unique tokens in TEXT vocabulary: {len(TEXT.vocab)}")
print(f"Unique tokens in LABEL vocabulary: {len(LABEL.vocab)}")
device = torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')
device = 'cpu'
# Assume that we are on a CUDA machine, then this should print a CUDA device:
print(device)
```
# Model and training
```
BATCH_SIZE = 50
train_iterator, valid_iterator, test_iterator = data.BucketIterator.splits(
(train_data, valid_data, test_data),
batch_size=BATCH_SIZE,
device=device,
shuffle = False,
sort_key=lambda x: len(x.text),
sort_within_batch=True)
eps = torch.tensor(1e-9)
temp = []
R = 20 # chunk of words read or skipped
def reward_function(prob, true_label):
"""
Returns 1 if correct prediction, -1 otherwise
"""
# print("true_label", "prob", true_label, prob)
if prob>0.5 and true_label>0.5:
return torch.tensor(1.0, requires_grad=True)
if prob<0.5 and true_label<0.5:
return torch.tensor(1.0, requires_grad=True)
return torch.tensor(-1.0, requires_grad=True)
def sample_binary(prob):
if prob>random.random:
return torch.tensor(1)
return torch.tensor(0)
class SkipReadingModel(nn.Module):
def __init__(self, input_dim, embedding_dim, hidden_dim, output_dim, gamma=0.99, train_mode=True, K=4):
super().__init__()
# store dimensions and constants
self.input_dim = input_dim
self.embedding_dim = embedding_dim
self.hidden_dim = hidden_dim
self.output_dim = output_dim
self.gamma = torch.tensor(gamma)
self.train_mode = train_mode
self.K = K
# create layers
self.embedding = nn.Embedding(input_dim, embedding_dim)
self.lstm_cell = nn.LSTMCell(input_size = embedding_dim, hidden_size = hidden_dim, bias = True)
self.stop_linear_1 = nn.Linear(hidden_dim, hidden_dim)
self.stop_linear_2 = nn.Linear(hidden_dim, hidden_dim)
self.stop_linear_3 = nn.Linear(hidden_dim, 1)
self.jumping_linear_1 = nn.Linear(hidden_dim, hidden_dim)
self.jumping_linear_2 = nn.Linear(hidden_dim, hidden_dim)
self.jumping_linear_3 = nn.Linear(hidden_dim, K)
self.output_linear_1 = nn.Linear(hidden_dim, hidden_dim)
self.output_linear_2 = nn.Linear(hidden_dim, output_dim)
self.value_head = nn.Linear(hidden_dim, 1)
# Baseline weight
self.wb = nn.Parameter(data=torch.zeros(self.hidden_dim), requires_grad=True)
self.cb = nn.Parameter(data=torch.tensor((0.0)), requires_grad=True)
# Initialize lstm_cell states
self.initialize_lstm_cell_states()
# Initalize episode number and time number
self.initialize_for_new_batch()
self.initialize_time_number()
# Overall reward and loss history
self.reward_history = []
self.loss_history = []
self.training_accuracies = []
self.validation_accuracies = []
# torch.tensor((0.0), requires_grad=True)
def initialize_lstm_cell_states(self):
self.c = torch.zeros(1, self.hidden_dim, requires_grad=True)
self.h = torch.zeros(1, self.hidden_dim, requires_grad=True)
def initialize_episode_number(self):
self.ep = 0
def initialize_time_number(self):
self.t = 0
def clear_batch_lists(self):
del self.saved_log_probs_s[:]
del self.saved_log_probs_n[:]
del self.saved_log_probs_o[:]
del self.reward_baselines[:]
del self.rewards[:]
del self.label_targets[:]
del self.label_predictions[:]
del self.state_values[:]
self.initialize_episode_number()
self.training_accuracy = 0.0
def initialize_for_new_batch(self):
"""
Cleans history of log probabilities, rewards, targets etc for the last
batch
"""
self.initialize_episode_number()
# Episode policy and reward history
self.saved_log_probs_s = [] # log probabilities for each time step t in each episode in batch
self.saved_log_probs_n = [] # log probs for jump
self.saved_log_probs_o = [] # log_prob for class
self.rewards = [] # reward at final time step of each episode in batch
self.reward_baselines = [] # reward baselines for each time step t in each episode in batch
self.state_values = []
# Predictions and targets history (for cross entropy loss calculation)
self.label_predictions = [] # 1 probability for each episode
self.label_targets = []# 1 label for each episode
self.training_accuracy = 0.0
def classify(self):
# global temp
# temp.append(self.c[0])
out = self.output_linear_1(self.c[0])
out = self.output_linear_2(out)
self.label_predictions.append(out)
prob_o = torch.sigmoid(out)
class_categ = Binomial(probs=prob_o)
_class = class_categ.sample()
if self.train_mode:
self.rewards.append(reward_function(_class, self.label_targets[-1]))
self.saved_log_probs_o.append((class_categ.log_prob(_class), ))
# return torch.sigmoid(out)
def get_baseline(self):
return torch.dot(self.wb, self.c[0].detach()) + self.cb
def save_training_accuracy(self):
correct = 0
for _r in self.rewards:
if _r > 0:
correct += 1
self.training_accuracy = correct/len(self.rewards)
self.training_accuracies.append(self.training_accuracy)
def forward(self, pack):
texts, lengths, labels = pack
embeddeds = self.embedding(texts)
# embeddeds = nn.utils.rnn.pack_padded_sequence(embeddeds, lengths)
self.initialize_for_new_batch()
for episode_number in range(embeddeds.shape[1]):
# load episode data
self.ep = episode_number
embedded = embeddeds[:, self.ep, :]
#print(texts.shape, embeddeds.shape, embedded.shape)
#print(label)
# initialize counters and index
tokens_read = 0
jumps_made = 0
word_index = 0
words_len = embedded.shape[0]
self.initialize_lstm_cell_states()
self.initialize_time_number()
self.saved_log_probs_s.append([])
self.saved_log_probs_n.append([])
self.state_values.append([])
self.reward_baselines.append([])
if self.train_mode:
label = labels[self.ep].reshape(1)
self.label_targets.append(label)
# start iterating through sequence, while skipping some words
while word_index<words_len and word_index<400:
self.t += 1
#print("embedded_word", embedded_word.shape)
# generate next lstm cell state
for _r in range(min(R, words_len-word_index)):
embedded_word = embedded[word_index]
self.h, self.c = self.lstm_cell(torch.reshape(embedded_word, (1, -1)), (self.h, self.c))
word_index += 1
# print('word_index', word_index, 'tokens_read', tokens_read, 'jumps_made', jumps_made)
# print(self.c)
_state_value = self.value_head(self.c[0])
self.state_values.append(_state_value)
_s = self.stop_linear_1(self.c[0])
_s = F.relu(_s)
_s = self.stop_linear_2(_s)
_s = F.relu(_s)
_s = self.stop_linear_3(_s)
probs_s = torch.sigmoid(_s)
try:
stop_categ = Binomial(probs=probs_s)
stop = stop_categ.sample()
except:
print("_c", self.c)
#temp = (self.c, _s, probs_s, self.stop_linear_1, self.stop_linear_2, self.stop_linear_3, stop_categ)
raise ValueError('got the expected error')
# Add log probability of our chosen action to our history
self.saved_log_probs_s[-1].append(stop_categ.log_prob(stop))
self.reward_baselines[-1].append(self.get_baseline())
if stop > 0.5:
self.classify()
break
else:
_n = self.jumping_linear_1(self.c[0])
_n = F.relu(_n)
_n = self.jumping_linear_2(_n)
_n = F.relu(_n)
_n = self.jumping_linear_3(_n)
_n = F.softmax(_n)
n_categ = Categorical(_n)
n = n_categ.sample()
self.saved_log_probs_n[-1].append(n_categ.log_prob(n))
word_index += n * R
else:
# print("Finished while loop")
# raise ValueError('Finshed ')
self.classify()
if self.train_mode:
self.save_training_accuracy()
return self.label_predictions
INPUT_DIM = len(TEXT.vocab)
EMBEDDING_DIM = 100
HIDDEN_DIM = 128
OUTPUT_DIM = 1
FLOP_COST = 0.0001
seed = 7
torch.manual_seed(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
np.random.seed(seed)
bce = nn.BCEWithLogitsLoss(reduction='mean')
policy_model = SkipReadingModel(INPUT_DIM, EMBEDDING_DIM, HIDDEN_DIM, OUTPUT_DIM).to(device)
pretrained_embeddings = TEXT.vocab.vectors
policy_model.embedding.weight.data.copy_(pretrained_embeddings)
if os.path.exists('fast_text_model.weights'):
policy_model.load_state_dict(torch.load('fast_text_model.weights'))
# define the optimiser
optimizer = optim.Adam(policy_model.parameters(), lr=0.01)
def update_policy():
#print(len(policy_model.rewards), len(policy_model.label_predictions), len(policy_model.label_targets))
#print(len(policy_model.saved_log_probs_o), len(policy_model.saved_log_probs_n), len(policy_model.saved_log_probs_s))
#print(len(policy_model.reward_baselines))
policy_loss_sum = torch.tensor(0.0, requires_grad=True)
reward_sum = torch.tensor(0.0, requires_grad=True)
baseline_loss_sum = torch.tensor(0.0, requires_grad=True)
value_loss_sum = torch.tensor(0.0, requires_grad=True)
for reward, prediction, target, log_probs_o, log_probs_n, log_probs_s, baselines, svs in zip(
policy_model.rewards, policy_model.label_predictions,
policy_model.label_targets, policy_model.saved_log_probs_o,
policy_model.saved_log_probs_n, policy_model.saved_log_probs_s,
policy_model.reward_baselines, policy_model.state_values):
for lpn in log_probs_n:
policy_loss_sum = policy_loss_sum + lpn
for i, (lps, b, sv) in enumerate(zip(log_probs_s, baselines, svs)):
policy_loss_sum = policy_loss_sum + lps
r = torch.pow(policy_model.gamma, i) * (-FLOP_COST)
if i == len(svs)-1:
r = r + torch.pow(policy_model.gamma, i) * reward
adv = r - sv.item()
reward_sum = reward_sum + adv
value_loss_sum = value_loss_sum + F.smooth_l1_loss(sv, torch.tensor([r]))
# baseline_loss_sum = baseline_loss_sum + torch.pow(rew, 2)
# baseline_loss_sum = baseline_loss_sum - torch.pow(rew, 2) + torch.pow(torch.pow(policy_model.gamma, i) * reward + rew, 2)
policy_loss_sum = policy_loss_sum + log_probs_o[0]
# print("reward sum", reward_sum)
# print("policy_loss_sum", policy_loss_sum)
loss = policy_loss_sum * reward_sum + value_loss_sum
optimizer.zero_grad()
# print('policy_loss', policy_loss)
loss.backward(retain_graph=True)
optimizer.step()
policy_model.clear_batch_lists()
def test_model():
policy_model.train_mode = True
correct = 0
total=0
for _data in test_iterator:
# get the inputs
texts, text_lengths, labels = _data.text[0], _data.text[1], _data.label
# print("Input review texts, text_lengths, labels", texts.shape, text_lengths.shape, labels.shape)
predictions = policy_model((texts.to(device), text_lengths.to(device), labels.to(device)))
for (prediction, label) in zip(predictions, labels):
if reward_function(label, prediction) > 0:
correct += 1
total += 1
if total%1000 == 0:
print(total)
if total%5000 == 0:
break
print("Test accuracy :", correct/total)
policy_model.train_mode = True
return correct/total
def validate_model():
policy_model.train_mode = True
correct = 0
total=0
for _data in valid_iterator:
# get the inputs
texts, text_lengths, labels = _data.text[0], _data.text[1], _data.label
# print("Input review texts, text_lengths, labels", texts.shape, text_lengths.shape, labels.shape)
predictions = policy_model((texts.to(device), text_lengths.to(device), labels.to(device)))
for (prediction, label) in zip(predictions, labels):
if reward_function(label, prediction) > 0:
correct += 1
total += 1
if total%1000 == 0:
break
print("Validation accuracy :", correct/total)
policy_model.train_mode = True
policy_model.validation_accuracies.append(correct/total)
return correct/total
# test_model()
# the epoch loop
with torch.enable_grad():
validate_model()
for epoch in range(10):
running_reward = 10
t = 0
for _data in train_iterator:
# get the inputs
texts, text_lengths, labels = _data.text[0], _data.text[1], _data.label
# print("Input review texts, text_lengths, labels", texts.shape, text_lengths.shape, labels.shape)
prediction = policy_model((texts.to(device), text_lengths.to(device), labels.to(device)))
# print("Prediction", prediction.item())
# raise ValueError('Not done')
t += 1
if t%2 == 0:
print("batch no. %d, training accuracy %4.2f" % (t, policy_model.training_accuracy))
if t%10 == 0:
validate_model()
#print("wb", policy_model.wb)
# print("lstm hh hi", policy_model.lstm_cell.weight_hh[0][::10], policy_model.lstm_cell.weight_ih[0][::10])
#print("lstm hh hi", policy_model.lstm_cell.weight_hh, policy_model.lstm_cell.weight_ih)
#print("emb", policy_model.embedding.weight)
#print("jmp", policy_model.jumping_linear.weight)
#print("out", policy_model.output_linear.weight)
if t%1000 == 0:
break
update_policy()
#running_reward = 0.05 * policy_model.reward_episode + (1 - 0.05) * running_reward
#print("Epoch %d, reward %4.2f" % (epoch, running_reward))
print("Epoch %d" % (epoch))
print('**** Finished Training ****')
# test_model()
torch.save(policy_model.state_dict(), 'fast_text_model.weights')
```
# Evaluation
```
import timeit
import spacy
import matplotlib.pyplot as plt
nlp = spacy.load('en')
def predict_sentiment(model, sentence):
model.train_mode = False
tokenized = [tok.text for tok in nlp.tokenizer(sentence)]
indexed = [TEXT.vocab.stoi[t] for t in tokenized]
# tensor = torch.LongTensor(indexed).to(device)
tensor = torch.LongTensor(indexed).to('cpu')
tensor = tensor.unsqueeze(1)
model((tensor, torch.tensor([tensor.shape[0]]), None))
res = torch.sigmoid(model.label_predictions[0])
model.train_mode = False
return res
times = []
lengths=[]
train_iterator, valid_iterator, test_iterator = data.BucketIterator.splits(
(train_data, valid_data, test_data),
batch_size=1,
device=device,
shuffle = True,
# sort_key=lambda x: len(x.text),
sort_within_batch=False)
i = 0
for _data in test_iterator:
if i%100 == 0:
# get the inputs
texts, text_lengths, labels = _data.text[0], _data.text[1], _data.label
#print(texts.shape, text_lengths.shape, labels.shape)
start_time = timeit.default_timer()
predictions = policy_model((texts.to(device), text_lengths.to(device), labels.to(device)))
elapsed = timeit.default_timer() - start_time
lengths.append(texts.shape[0])
times.append(elapsed)
# print("Input review texts, text_lengths, labels", texts.shape, text_lengths.shape, labels.shape)
if i>20000:
break
i += 1
import pickle
pickle_out = open("test_times_1.pickle","wb")
pickle.dump((lengths, times), pickle_out)
pickle_out.close()
plt.scatter(lengths, times, label='skip-model')
plt.xlabel('Lengths of sentences')
plt.ylabel('Time taken for prediction')
plt.show()
predict_sentiment(policy_model, "This film is terrible what can I say")
import pickle
pickle_out = open("training_epochs_1.pickle","wb")
pickle.dump((policy_model.training_accuracies, policy_model.validation_accuracies), pickle_out)
pickle_out.close()
```
|
github_jupyter
|
```
from crystal_toolkit.helpers.layouts import Columns, Column
from crystal_toolkit.settings import SETTINGS
from jupyter_dash import JupyterDash
from pydefect.analyzer.calc_results import CalcResults
from pydefect.analyzer.dash_components.cpd_energy_dash import CpdEnergy2D3DComponent, CpdEnergyOtherComponent
from pydefect.chem_pot_diag.chem_pot_diag import ChemPotDiag, CpdPlotInfo, \
CompositionEnergy
from pydefect.corrections.manual_correction import ManualCorrection
from pydefect.input_maker.defect_entry import DefectEntry
from pymatgen import Composition, Structure, Lattice, Element
import dash_html_components as html
import crystal_toolkit.components as ctc
from dash.dependencies import Input, Output, State
import json
app = JupyterDash(suppress_callback_exceptions=True,
assets_folder=SETTINGS.ASSETS_PATH)
from vise.analyzer.band_edge_properties import BandEdge
comp_energies = [
CompositionEnergy(Composition("Mg"), 0.0, "a"),
CompositionEnergy(Composition("Ca"), 0.0, "a"),
CompositionEnergy(Composition("Sr"), 0.0, "a"),
CompositionEnergy(Composition("O"), 0.0, "a"),
CompositionEnergy(Composition("H"), 0.0, "a"),
# CompositionEnergy(Composition("MgCaO3"), -100.0, "a"),
CompositionEnergy(Composition("MgCaSrO3"), -100.0, "a"),
]
#cpd = ChemPotDiag(comp_energies, target=Composition("MgCaO3"))
cpd = ChemPotDiag(comp_energies, target=Composition("MgCaSrO3"))
cpd_plot_info = CpdPlotInfo(cpd)
print(cpd.target.elements)
print(cpd.dim)
print(cpd.target_vertices)
print(cpd.all_compounds)
print(cpd.impurity_abs_energy(Element.H, label="A"))
structure = Structure(Lattice.cubic(1), species=["O"] * 2, coords=[[0]*3]*2)
defect_structure = Structure(Lattice.cubic(1), species=["O"] * 1, coords=[[0]*3])
common = dict(site_symmetry="1",
magnetization=0.0,
kpoint_coords=[[0]*3],
kpoint_weights=[1.0],
potentials=[0.0],
vbm_info=BandEdge(0.0),
cbm_info=BandEdge(1.0),
fermi_level=0.0)
perfect = CalcResults(structure=structure,energy=0, **common)
defects = [CalcResults(structure=defect_structure, energy=1.0, **common),
CalcResults(structure=defect_structure, energy=0.5, **common)]
de_common = dict(name="Va_O1",
structure=defect_structure, site_symmetry="1",
perturbed_structure=defect_structure, defect_center=[[0]*3])
defect_entries = [DefectEntry(charge=0, **de_common),
DefectEntry(charge=1, **de_common)]
corrections = [ManualCorrection(correction_energy=1.0),
ManualCorrection(correction_energy=1.0)]
cpd_e_component = CpdEnergyOtherComponent(cpd_plot_info,
perfect,
defects,
defect_entries,
corrections)
my_layout = html.Div([Column(cpd_e_component.layout)])
ctc.register_crystal_toolkit(app=app, layout=my_layout, cache=None)
app.run_server(port=8097)
#app.run_server(mode='inline', port=8094)
```
|
github_jupyter
|
# V2: SCF optimization with VAMPyR
## V2.1: Hydrogen atom
In order to solve the one-electron Schr\"{o}dinger equation in MWs we reformulate them in an integral form [1].
\begin{equation}
\phi = -2\hat{G}_{\mu}\hat{V}\phi
\end{equation}
Where $\hat{V}$ is the potential acting on the system, $\phi$ is the wavefunction, $\hat{G}$ is the Helmholtz integral operator, where its kernel is defined as $G_\mu(r - r') = \frac{\exp(-\mu |r - r'|)}{4\pi |r - r'|}$
and $\mu$ is a parameter defined above through the energy.
The Helmholtz operator is already implemented in vampyr, therefore the only things you need are the integral KS equation and the definition of $\mu$
\begin{equation}
\mu = \sqrt{-2E}
\end{equation}
The way you initialize the Helmholtz operator is as follows
```
H = vp.HelmholtzOperator( mra, exp=mu, prec=eps )
```
where `mu` is the $\mu$ is the parameter defined above, mra you have seen before, and `eps` is the desired threshold precision. This operator is applied the same way you applied the vp.ScalingProjector earlier.
In this exercise you will be solving the KS equation iteratively for a simple system, the Hydrogen atom. This means that you only have the nuclear potential to take into account for the potential term in the KS equation.
$$ V_{nuc}(\mathbf{r}) = -\frac{1}{|\mathbf{r}|}$$
We will also be working with a single orbital, of which the initial guess is
$$ \phi_0(\mathbf{r}) = e^{-|\mathbf{r}|^2} $$
where
$$ |\mathbf{r}| = \sqrt{x^2 + y^2 + z^2}$$
The orbital update is defined as follows
\begin{align}
\Delta\tilde{\phi}^n &= -2\hat{G}[V_{nuc}\phi^n] - \phi^n \\
\Delta\tilde{\phi}^n &= \tilde{\phi}^{n+1} - \phi^n
\end{align}
where we use \~ to denote a function that is **not** normalized, and $n$ is the iteration index.
#### Implementation exercise:
1. Make a nuclear potential as a python function `f_nuc(r)`
2. Make an initial guess for the orbital as a python function `f_phi(r)` (hint use `np.exp` to get an exponetial function)
3. Create a Helmholtz operator $G_\mu$ with $\mu$ as shown above, use the exact value of $E = -0.5 a.u.$ for a hydrogen atom
4. Project both nuclear potential ($V$) and orbital ($\phi_n$) to the MW basis using a `vp.ScalingProjector` with precision $\epsilon=1.0e-3$
5. Compute new orbital through application of the Helmholtz operator
6. Compute the size of the orbital update $||\tilde{\phi}^{n+1} - \phi^n||$
7. Normalize the orbital $\phi^{n+1} = \tilde{\phi}^{n+1}/||\tilde{\phi}^{n+1}||$
8. Update orbital $\phi^{n+1} \rightarrow \phi^{n}$ for next iteration
9. Repeat steps 5-8 until your wavefunction has converged
The convergence criterion is the norm of $\Delta \phi^n$, but you should start by looping a set amount of times before trying the threshold.
```
from vampyr import vampyr3d as vp
import numpy as np
import matplotlib.pyplot as plt
r_x = np.linspace(-0.99, 0.99, 1000) # create an evenly spaced set of points between -0.99 and 0.99
r_y = np.zeros(1000)
r_z = np.zeros(1000)
r = [r_x, r_y, r_z]
# Analytic nuclear potential
def f_nuc(r):
# TODO: implement the nuclear potential
return
# Analytic guess for solution
def f_phi(r):
# TODO: implement the initial guess for the orbital
return
# Prepare Helmholtz operator
E = -0.5
mu = np.sqrt(-2*E)
G_mu = # TODO: Construct BSH operator from mu)
V = # TODO: Project nuclear potential V from f_nuc
phi_n = # TODO: Project starting guess phi_n from f_phi
phi_n.normalize()
# Optimization loop
thrs = 1.0e-3
update = 1.0
i = 0
while (i < 3): # switch to (update > thrs) later
# TODO:
# Compute product of potential V and wavefunction phi_n
# Apply Helmholtz operator to obtain phi_np1
# Compute norm = ||phi^{n+1}||
# Compute update = ||phi^{n+1} - phi^{n}||
# this will plot the wavefunction at each iteration
phi_n_plt = [phi_n([x, 0.0, 0.0]) for x in r_x]
plt.plot(r_x, phi_n_plt)
# this will print some info, you need to compute in the loop:
print("iteration: {} Norm: {} Update: {}".format(i, norm, update))
i += 1
plt.show()
```
## V2.2 Extension to Helium
A few things change when you go from Hydrogen to Helium:
1. The energy is no longer known exactly, and thus will have to be computed from the wavefunction
2. The Helmholtz operator which depends on the energy through $\mu = \sqrt{-2E}$ needs to be updated in every iteration
3. The potential operator $V$ depends on the wavefunction and must be updated in every iteration
In this example we will use the Hartree-Fock model, which for a single-orbital system like Helium, reduces to the following potential operator:
\begin{align}
\hat{V} &= \hat{V}_{nuc} + 2\hat{J} - \hat{K}\\
&= \hat{V}_{nuc} + \hat{J}
\end{align}
since $\hat{K} = \hat{J}$ for a doubly occupied single orbital.
The Coulomb potential $\hat{J}$ can be computed by application of the Poisson operator $P$:
\begin{equation}
\hat{J}(r) = P\left[4\pi\rho\right]
\end{equation}
Where $\rho$ is the square of the orbital
\begin{equation}
\rho = \phi*\phi
\end{equation}
#### Pen and paper exercise:
Use the fact that
\begin{equation}
\tilde{\phi}^{n+1} = -\Big[\hat{T} - E^n\Big]^{-1} V^n\phi^n \end{equation}
to show that
\begin{equation}
E^{n+1} = \frac{\langle\tilde{\phi}^{n+1}|\hat{T} +
\hat{V}^{n+1}|\tilde{\phi}^{n+1}\rangle}
{||\tilde{\phi}^{n+1}||^2}
\end{equation}
can be written as a pure update $dE^n$ involving only the potentials $\hat{V}^{n+1}$, $\hat{V}^n$ as well as the orbitals $\tilde{\phi}^{n+1}$ and $\phi^n$
\begin{equation}
E^{n+1} = E^{n} + dE^n
\end{equation}
#### Implementation exercise:
1. Make a nuclear potential function `f_nuc(r)` for the Helium atom
2. Make an initial guess for the orbital as a python function `f_phi(r)` (hint use `np.exp` to get an exponetial function)
3. Project both nuclear potential ($V$) and orbital ($\phi_n$) to the MW basis using a `vp.ScalingProjector` with precision $\epsilon=1.0e-3$
4. Create a Helmholtz operator $G^n$ with $\mu^n$ using the current energy $E^n$
5. Compute total potential $\hat{V^n} = \hat{V}_{nuc} + \hat{J^n}$, where the Coulomb potential is computed using the `vp.PoissonOperator` on the current squared orbital $\rho^n = ||\phi^n||^2$
6. Compute new orbital through application of the Helmholtz operator on $\phi^{n+1} = -2\hat{G}^n\hat{V}^n\phi^n$
7. Compute the size of the orbital update $||\tilde{\phi}^{n+1} - \phi^n||$
8. Normalize the orbital $\phi^{n+1} = \tilde{\phi}^{n+1}/||\tilde{\phi}^{n+1}||$
9. Update orbital $\phi^{n+1} \rightarrow \phi^{n}$ for next iteration
10. Repeat steps 4-9 until your wavefunction has converged
The convergence criterion is the norm of $\Delta \phi^n$, but you should start by looping a set amount of times before trying the threshold.
```
from vampyr import vampyr3d as vp
import numpy as np
import matplotlib.pyplot as plt
r_x = np.linspace(-0.99, 0.99, 1000) # create an evenly spaced set of points between -0.99 and 0.99
r_y = np.zeros(1000)
r_z = np.zeros(1000)
r = [r_x, r_y, r_z]
# Analytic nuclear potential Helium
def f_nuc(r):
#implement the nuclear potential
return
# Analytic guess for solution (same as for Hydrogen)
def f_phi(r):
# implement the initial guess for the orbital
return
# TODO:
# Project nuclear potential V_nuc from f_nuc
# Project starting guess phi_n from f_phi
# Set a starting guess E_n for the energy
# Optimization loop
thrs = 1.0e-3
update = 1.0
i = 0
while (i < 3): # switch to (update > thrs) later
# Prepare Helmholtz operator from current energy
mu_n = np.sqrt(-2*E_n)
G_n = # TODO: Construct BSH operator from mu_n)
# TODO:
# Compute rho
# Initialize vp.PoissonOperator and compute J
# Compute total potential V = V_nuc + J
# Iterate Helmholtz operator to get new orbital phi^{n+1}
dE_n = # TODO: insert energy expression from above
# Prepare for next iteration
E_n += dE_n
phi_n += dPhi_n
# This will plot the wavefunction at each iteration
phi_n_plt = [phi_n([x, 0.0, 0.0]) for x in r_x]
plt.plot(r_x, phi_n_plt)
# this will print some info, you need to compute in the loop:
# norm = ||phi^{n+1}||
# update = ||phi^{n+1} - phi^{n}||
print("iteration: {} Energy: {} Norm: {} Update: {}".format(i, E_n, norm, update))
i += 1
plt.show()
```
You should expect the orbital energy to converge towards
$E_n \approx -0.918$.
#### Bonus exercise:
The total energy can be computed after convergence as
$E_{tot} = 2E_n - \langle\rho|J\rangle$, should be around $E_{tot} \approx -2.86$.
## Sources
[1] Stig Rune Jensen, Santanu Saha, José A. Flores-Livas, William Huhn, Volker Blum, Stefan Goedecker, and Luca Frediani The Elephant in the Room of Density Functional Theory Calculations. The Journal of Physical Chemistry Letters 2017 8 (7), 1449-1457
DOI: 10.1021/acs.jpclett.7b00255
|
github_jupyter
|
##### Copyright 2018 The TensorFlow Authors.
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# Using TFRecords and `tf.Example`
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://www.tensorflow.org/tutorials/load_data/tf-records"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on TensorFlow.org</a>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/tutorials/load_data/tf-records.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/docs/blob/master/site/en/tutorials/load_data/tf-records.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
</td>
</table>
To read data efficiently it can be helpful to serialize your data and store it in a set of files (100-200MB each) that can each be read linearly. This is especially true if the data is being streamed over a network. This can also be useful for caching any data-preprocessing.
The TFRecord format is a simple format for storing a sequence of binary records.
[Protocol buffers](https://developers.google.com/protocol-buffers/) are a cross-platform, cross-language library for efficient serialization of structured data.
Protocol messages are defined by `.proto` files, these are often the easiest way to understand a message type.
The `tf.Example` message (or protobuf) is a flexible message type that represents a `{"string": value}` mapping. It is designed for use with TensorFlow and is used throughout the higher-level APIs such as [TFX](https://www.tensorflow.org/tfx/).
This notebook will demonstrate how to create, parse, and use the `tf.Example` message, and then serialize, write, and read `tf.Example` messages to and from `.tfrecord` files.
Note: While useful, these structures are optional. There is no need to convert existing code to use TFRecords, unless you are using [`tf.data`](https://www.tensorflow.org/guide/datasets) and reading data is still the bottleneck to training. See [Data Input Pipeline Performance](https://www.tensorflow.org/guide/performance/datasets) for dataset performance tips.
## Setup
```
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import tensorflow as tf
tf.enable_eager_execution()
import numpy as np
import IPython.display as display
```
## `tf.Example`
### Data types for `tf.Example`
Fundamentally a `tf.Example` is a `{"string": tf.train.Feature}` mapping.
The `tf.train.Feature` message type can accept one of the following three types (See the [`.proto` file]((https://github.com/tensorflow/tensorflow/blob/master/tensorflow/core/example/feature.proto) for reference). Most other generic types can be coerced into one of these.
1. `tf.train.BytesList` (the following types can be coerced)
- `string`
- `byte`
1. `tf.train.FloatList` (the following types can be coerced)
- `float` (`float32`)
- `double` (`float64`)
1. `tf.train.Int64List` (the following types can be coerced)
- `bool`
- `enum`
- `int32`
- `uint32`
- `int64`
- `uint64`
In order to convert a standard TensorFlow type to a `tf.Example`-compatible `tf.train.Feature`, you can use the following shortcut functions:
Each function takes a scalar input value and returns a `tf.train.Feature` containing one of the three `list` types above.
```
# The following functions can be used to convert a value to a type compatible
# with tf.Example.
def _bytes_feature(value):
"""Returns a bytes_list from a string / byte."""
return tf.train.Feature(bytes_list=tf.train.BytesList(value=[value]))
def _float_feature(value):
"""Returns a float_list from a float / double."""
return tf.train.Feature(float_list=tf.train.FloatList(value=[value]))
def _int64_feature(value):
"""Returns an int64_list from a bool / enum / int / uint."""
return tf.train.Feature(int64_list=tf.train.Int64List(value=[value]))
```
Note: To stay simple, this example only uses scalar inputs. The simplest way to handle non-scalar features is to use `tf.serialize_tensor` to convert tensors to binary-strings. Strings are scalars in tensorflow. Use `tf.parse_tensor` to convert the binary-string back to a tensor.
Below are some examples of how these functions work. Note the varying input types and the standardizes output types. If the input type for a function does not match one of the coercible types stated above, the function will raise an exception (e.g. `_int64_feature(1.0)` will error out, since `1.0` is a float, so should be used with the `_float_feature` function instead).
```
print(_bytes_feature(b'test_string'))
print(_bytes_feature(u'test_bytes'.encode('utf-8')))
print(_float_feature(np.exp(1)))
print(_int64_feature(True))
print(_int64_feature(1))
```
All proto messages can be serialized to a binary-string using the `.SerializeToString` method.
```
feature = _float_feature(np.exp(1))
feature.SerializeToString()
```
### Creating a `tf.Example` message
Suppose you want to create a `tf.Example` message from existing data. In practice, the dataset may come from anywhere, but the procedure of creating the `tf.Example` message from a single observation will be the same.
1. Within each observation, each value needs to be converted to a `tf.train.Feature` containing one of the 3 compatible types, using one of the functions above.
1. We create a map (dictionary) from the feature name string to the encoded feature value produced in #1.
1. The map produced in #2 is converted to a [`Features` message](https://github.com/tensorflow/tensorflow/blob/master/tensorflow/core/example/feature.proto#L85).
In this notebook, we will create a dataset using NumPy.
This dataset will have 4 features.
- a boolean feature, `False` or `True` with equal probability
- a random bytes feature, uniform across the entire support
- an integer feature uniformly randomly chosen from `[-10000, 10000)`
- a float feature from a standard normal distribution
Consider a sample consisting of 10,000 independently and identically distributed observations from each of the above distributions.
```
# the number of observations in the dataset
n_observations = int(1e4)
# boolean feature, encoded as False or True
feature0 = np.random.choice([False, True], n_observations)
# integer feature, random between -10000 and 10000
feature1 = np.random.randint(0, 5, n_observations)
# bytes feature
strings = np.array([b'cat', b'dog', b'chicken', b'horse', b'goat'])
feature2 = strings[feature1]
# float feature, from a standard normal distribution
feature3 = np.random.randn(n_observations)
```
Each of these features can be coerced into a `tf.Example`-compatible type using one of `_bytes_feature`, `_float_feature`, `_int64_feature`. We can then create a `tf.Example` message from these encoded features.
```
def serialize_example(feature0, feature1, feature2, feature3):
"""
Creates a tf.Example message ready to be written to a file.
"""
# Create a dictionary mapping the feature name to the tf.Example-compatible
# data type.
feature = {
'feature0': _int64_feature(feature0),
'feature1': _int64_feature(feature1),
'feature2': _bytes_feature(feature2),
'feature3': _float_feature(feature3),
}
# Create a Features message using tf.train.Example.
example_proto = tf.train.Example(features=tf.train.Features(feature=feature))
return example_proto.SerializeToString()
```
For example, suppose we have a single observation from the dataset, `[False, 4, bytes('goat'), 0.9876]`. We can create and print the `tf.Example` message for this observation using `create_message()`. Each single observation will be written as a `Features` message as per the above. Note that the `tf.Example` [message](https://github.com/tensorflow/tensorflow/blob/master/tensorflow/core/example/example.proto#L88) is just a wrapper around the `Features` message.
```
# This is an example observation from the dataset.
example_observation = []
serialized_example = serialize_example(False, 4, b'goat', 0.9876)
serialized_example
```
To decode the message use the `tf.train.Example.FromString` method.
```
example_proto = tf.train.Example.FromString(serialized_example)
example_proto
```
## TFRecord files using `tf.data`
The `tf.data` module also provides tools for reading and writing data in tensorflow.
### Writing a TFRecord file
The easiest way to get the data into a dataset is to use the `from_tensor_slices` method.
Applied to an array, it returns a dataset of scalars.
```
tf.data.Dataset.from_tensor_slices(feature1)
```
Applies to a tuple of arrays, it returns a dataset of tuples:
```
features_dataset = tf.data.Dataset.from_tensor_slices((feature0, feature1, feature2, feature3))
features_dataset
# Use `take(1)` to only pull one example from the dataset.
for f0,f1,f2,f3 in features_dataset.take(1):
print(f0)
print(f1)
print(f2)
print(f3)
```
Use the `tf.data.Dataset.map` method to apply a function to each element of a `Dataset`.
The mapped function must operate in TensorFlow graph mode: It must operate on and return `tf.Tensors`. A non-tensor function, like `create_example`, can be wrapped with `tf.py_func` to make it compatible.
Using `tf.py_func` requires that you specify the shape and type information that is otherwise unavailable:
```
def tf_serialize_example(f0,f1,f2,f3):
tf_string = tf.py_func(
serialize_example,
(f0,f1,f2,f3), # pass these args to the above function.
tf.string) # the return type is `tf.string`.
return tf.reshape(tf_string, ()) # The result is a scalar
```
Apply this function to each element in the dataset:
```
serialized_features_dataset = features_dataset.map(tf_serialize_example)
serialized_features_dataset
```
And write them to a TFRecord file:
```
filename = 'test.tfrecord'
writer = tf.data.experimental.TFRecordWriter(filename)
writer.write(serialized_features_dataset)
```
### Reading a TFRecord file
We can also read the TFRecord file using the `tf.data.TFRecordDataset` class.
More information on consuming TFRecord files using `tf.data` can be found [here](https://www.tensorflow.org/guide/datasets#consuming_tfrecord_data).
Using `TFRecordDataset`s can be useful for standardizing input data and optimizing performance.
```
filenames = [filename]
raw_dataset = tf.data.TFRecordDataset(filenames)
raw_dataset
```
At this point the dataset contains serialized `tf.train.Example` messages. When iterated over it returns these as scalar string tensors.
Use the `.take` method to only show the first 10 records.
Note: iterating over a `tf.data.Dataset` only works with eager execution enabled.
```
for raw_record in raw_dataset.take(10):
print(repr(raw_record))
```
These tensors can be parsed using the function below.
Note: The `feature_description` is necessary here because datasets use graph-execution, and need this description to build their shape and type signature.
```
# Create a description of the features.
feature_description = {
'feature0': tf.FixedLenFeature([], tf.int64, default_value=0),
'feature1': tf.FixedLenFeature([], tf.int64, default_value=0),
'feature2': tf.FixedLenFeature([], tf.string, default_value=''),
'feature3': tf.FixedLenFeature([], tf.float32, default_value=0.0),
}
def _parse_function(example_proto):
# Parse the input tf.Example proto using the dictionary above.
return tf.parse_single_example(example_proto, feature_description)
```
Or use `tf.parse example` to parse a whole batch at once.
Apply this finction to each item in the dataset using the `tf.data.Dataset.map` method:
```
parsed_dataset = raw_dataset.map(_parse_function)
parsed_dataset
```
Use eager execution to display the observations in the dataset. There are 10,000 observations in this dataset, but we only display the first 10. The data is displayed as a dictionary of features. Each item is a `tf.Tensor`, and the `numpy` element of this tensor displays the value of the feature.
```
for parsed_record in parsed_dataset.take(10):
print(repr(raw_record))
```
Here, the `tf.parse_example` function unpacks the `tf.Example` fields into standard tensors.
## TFRecord files using tf.python_io
The `tf.python_io` module also contains pure-Python functions for reading and writing TFRecord files.
### Writing a TFRecord file
Now write the 10,000 observations to the file `test.tfrecords`. Each observation is converted to a `tf.Example` message, then written to file. We can then verify that the file `test.tfrecords` has been created.
```
# Write the `tf.Example` observations to the file.
with tf.python_io.TFRecordWriter(filename) as writer:
for i in range(n_observations):
example = serialize_example(feature0[i], feature1[i], feature2[i], feature3[i])
writer.write(example)
!ls
```
### Reading a TFRecord file
Suppose we now want to read this data back, to be input as data into a model.
The following example imports the data as is, as a `tf.Example` message. This can be useful to verify that a the file contains the data that we expect. This can also be useful if the input data is stored as TFRecords but you would prefer to input NumPy data (or some other input data type), for example [here](https://www.tensorflow.org/guide/datasets#consuming_numpy_arrays), since this example allows us to read the values themselves.
We iterate through the TFRecords in the infile, extract the `tf.Example` message, and can read/store the values within.
```
record_iterator = tf.python_io.tf_record_iterator(path=filename)
for string_record in record_iterator:
example = tf.train.Example()
example.ParseFromString(string_record)
print(example)
# Exit after 1 iteration as this is purely demonstrative.
break
```
The features of the `example` object (created above of type `tf.Example`) can be accessed using its getters (similarly to any protocol buffer message). `example.features` returns a `repeated feature` message, then getting the `feature` message returns a map of feature name to feature value (stored in Python as a dictionary).
```
print(dict(example.features.feature))
```
From this dictionary, you can get any given value as with a dictionary.
```
print(example.features.feature['feature3'])
```
Now, we can access the value using the getters again.
```
print(example.features.feature['feature3'].float_list.value)
```
## Walkthrough: Reading/Writing Image Data
This is an example of how to read and write image data using TFRecords. The purpose of this is to show how, end to end, input data (in this case an image) and write the data as a TFRecord file, then read the file back and display the image.
This can be useful if, for example, you want to use several models on the same input dataset. Instead of storing the image data raw, it can be preprocessed into the TFRecords format, and that can be used in all further processing and modelling.
First, let's download [this image](https://commons.wikimedia.org/wiki/File:Felis_catus-cat_on_snow.jpg) of a cat in the snow and [this photo](https://upload.wikimedia.org/wikipedia/commons/f/fe/New_East_River_Bridge_from_Brooklyn_det.4a09796u.jpg) of the Williamsburg Bridge, NYC under construction.
### Fetch the images
```
cat_in_snow = tf.keras.utils.get_file('320px-Felis_catus-cat_on_snow.jpg', 'https://storage.googleapis.com/download.tensorflow.org/example_images/320px-Felis_catus-cat_on_snow.jpg')
williamsburg_bridge = tf.keras.utils.get_file('194px-New_East_River_Bridge_from_Brooklyn_det.4a09796u.jpg','https://storage.googleapis.com/download.tensorflow.org/example_images/194px-New_East_River_Bridge_from_Brooklyn_det.4a09796u.jpg')
display.display(display.Image(filename=cat_in_snow))
display.display(display.HTML('Image cc-by: <a "href=https://commons.wikimedia.org/wiki/File:Felis_catus-cat_on_snow.jpg">Von.grzanka</a>'))
display.display(display.Image(filename=williamsburg_bridge))
display.display(display.HTML('<a "href=https://commons.wikimedia.org/wiki/File:New_East_River_Bridge_from_Brooklyn_det.4a09796u.jpg">source</a>'))
```
### Write the TFRecord file
As we did earlier, we can now encode the features as types compatible with `tf.Example`. In this case, we will not only store the raw image string as a feature, but we will store the height, width, depth, and an arbitrary `label` feature, which is used when we write the file to distinguish between the cat image and the bridge image. We will use `0` for the cat image, and `1` for the bridge image.
```
image_labels = {
cat_in_snow : 0,
williamsburg_bridge : 1,
}
# This is an example, just using the cat image.
image_string = open(cat_in_snow, 'rb').read()
label = image_labels[cat_in_snow]
# Create a dictionary with features that may be relevant.
def image_example(image_string, label):
image_shape = tf.image.decode_jpeg(image_string).shape
feature = {
'height': _int64_feature(image_shape[0]),
'width': _int64_feature(image_shape[1]),
'depth': _int64_feature(image_shape[2]),
'label': _int64_feature(label),
'image_raw': _bytes_feature(image_string),
}
return tf.train.Example(features=tf.train.Features(feature=feature))
for line in str(image_example(image_string, label)).split('\n')[:15]:
print(line)
print('...')
```
We see that all of the features are now stores in the `tf.Example` message. Now, we functionalize the code above and write the example messages to a file, `images.tfrecords`.
```
# Write the raw image files to images.tfrecords.
# First, process the two images into tf.Example messages.
# Then, write to a .tfrecords file.
with tf.python_io.TFRecordWriter('images.tfrecords') as writer:
for filename, label in image_labels.items():
image_string = open(filename, 'rb').read()
tf_example = image_example(image_string, label)
writer.write(tf_example.SerializeToString())
!ls
```
### Read the TFRecord file
We now have the file `images.tfrecords`. We can now iterate over the records in the file to read back what we wrote. Since, for our use case we will just reproduce the image, the only feature we need is the raw image string. We can extract that using the getters described above, namely `example.features.feature['image_raw'].bytes_list.value[0]`. We also use the labels to determine which record is the cat as opposed to the bridge.
```
raw_image_dataset = tf.data.TFRecordDataset('images.tfrecords')
# Create a dictionary describing the features.
image_feature_description = {
'height': tf.FixedLenFeature([], tf.int64),
'width': tf.FixedLenFeature([], tf.int64),
'depth': tf.FixedLenFeature([], tf.int64),
'label': tf.FixedLenFeature([], tf.int64),
'image_raw': tf.FixedLenFeature([], tf.string),
}
def _parse_image_function(example_proto):
# Parse the input tf.Example proto using the dictionary above.
return tf.parse_single_example(example_proto, image_feature_description)
parsed_image_dataset = raw_image_dataset.map(_parse_image_function)
parsed_image_dataset
```
Recover the images from the TFRecord file:
```
for image_features in parsed_image_dataset:
image_raw = image_features['image_raw'].numpy()
display.display(display.Image(data=image_raw))
```
|
github_jupyter
|
# Módulo 2 - Modelos preditivos e séries temporais
# Desafio do Módulo 2
```
import pandas as pd
import numpy as np
base = pd.read_csv('https://pycourse.s3.amazonaws.com/banknote_authentication.txt', header=None)
base.head()
#labels:
#variance, skewness, curtosis e entropy)
base.columns=['variance', 'skewness', 'curtosis', 'entropy', 'class']
base.head()
#Qual o tamanho desse dataset (número de linhas, número de colunas)?
base.shape
#Qual variável possui o maior range (diferença entre valor máximo e mínimo)?
#Qual a média da coluna skewness?
#Qual a média da coluna entropy?
#Qual a desvio padrão da coluna curtosis?
base.describe()
#Qual a mediana da coluna variance?
base.median()
#Qual a porcentagem de exemplos do dataset que são cédulas falsas (class=1)?
falsas = (base['class'] == 1).sum()
total = base.shape[0]
falsas / total * 100
#Qual o valor da correlação de Pearson entre as variáveis skewness e curtosis?
import scipy as sp
from scipy import stats
sp.stats.pearsonr(base['skewness'], base['curtosis'])
#Qual a acurácia do KNN no conjunto de teste?
from sklearn.model_selection import train_test_split
x = base.drop('class', axis=1)
y = base['class']
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.30, random_state=1)
import sklearn
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.svm import SVC
from sklearn.neural_network import MLPClassifier
from sklearn.metrics import accuracy_score #Medir acurária abaixo
#a. Algoritmo KNN:
clf_KNN = KNeighborsClassifier(n_neighbors=5)
#b. Algoritmo Árvore de Decisão (Decision Tree):
clf_arvore = DecisionTreeClassifier(random_state=1)
#c. Algoritmo Floresta Aleatória (Random Forest):
clf_floresta = RandomForestClassifier(max_depth=8, random_state=1)
#d. Algoritmo SVM:
clf_svm = SVC(gamma='auto',kernel='rbf', random_state=1)
#e. Algoritmo Rede MLP:
clf_mlp = MLPClassifier(hidden_layer_sizes=(2,), solver='lbfgs', random_state=1)
#Qual a acurácia do KNN no conjunto de teste?
clf_KNN.fit(x_train, y_train)
knn_predict = clf_KNN.predict(x_test)
accuracy_score(y_test, knn_predict)
#Qual a acurácia da Árvore de Decisão no conjunto de teste?
clf_arvore.fit(x_train, y_train)
arvore_predict = clf_arvore.predict(x_test)
accuracy_score(y_test, arvore_predict)
#Qual a acurácia do Random Forest no conjunto de teste?
clf_floresta.fit(x_train, y_train)
floresta_redict = clf_floresta.predict(x_test)
accuracy_score(y_test, floresta_redict)
#Qual a acurácia do SVM no conjunto de teste?
clf_svm.fit(x_train, y_train)
svm_predict = clf_svm.predict(x_test)
accuracy_score(y_test, svm_predict)
#Qual a acurácia da rede MLP no conjunto de teste?
clf_mlp.fit(x_train, y_train)
mlp_predict = clf_mlp.predict(x_test)
accuracy_score(y_test, mlp_predict)
#Analisando o valor da importância relativa das features do Random Forest (atributo feature_importances_),
#qual feature melhor contribuiu para a predição de class?
#Qual o valor da importância relativa da feature skewness?
relativos = clf_floresta.feature_importances_
base.head()
relativos
```
# Fim
# Visite o meu [github](https://github.com/k3ybladewielder) <3
|
github_jupyter
|
<img src="../../images/banners/python-advanced.png" width="600"/>
# <img src="../../images/logos/python.png" width="23"/> Python's property(): Add Managed Attributes to Your Classes
## <img src="../../images/logos/toc.png" width="20"/> Table of Contents
* [Managing Attributes in Your Classes](#managing_attributes_in_your_classes)
* [The Getter and Setter Approach in Python](#the_getter_and_setter_approach_in_python)
* [The Pythonic Approach](#the_pythonic_approach)
* [Getting Started With Python’s `property()`](#getting_started_with_python’s_`property()`)
* [Creating Attributes With `property()`](#creating_attributes_with_`property()`)
* [Using `property()` as a Decorator](#using_`property()`_as_a_decorator)
* [Providing Read-Only Attributes](#providing_read-only_attributes)
* [Creating Read-Write Attributes](#creating_read-write_attributes)
* [Providing Write-Only Attributes](#providing_write-only_attributes)
* [Putting Python’s `property()` Into Action](#putting_python’s_`property()`_into_action)
* [Validating Input Values](#validating_input_values)
* [Providing Computed Attributes](#providing_computed_attributes)
* [Caching Computed Attributes](#caching_computed_attributes)
* [Logging Attribute Access and Mutation](#logging_attribute_access_and_mutation)
* [Managing Attribute Deletion](#managing_attribute_deletion)
* [Creating Backward-Compatible Class APIs](#creating_backward-compatible_class_apis)
* [Overriding Properties in Subclasses](#overriding_properties_in_subclasses)
* [Conclusion](#conclusion)
---
With Python’s [`property()`](https://docs.python.org/3/library/functions.html#property), you can create **managed attributes** in your classes. You can use managed attributes, also known as **properties**, when you need to modify their internal implementation without changing the public [API](https://en.wikipedia.org/wiki/API) of the class. Providing stable APIs can help you avoid breaking your users’ code when they rely on your classes and objects.
Properties are arguably the most popular way to create managed attributes quickly and in the purest [Pythonic](https://realpython.com/learning-paths/writing-pythonic-code/) style.
**In this tutorial, you’ll learn how to:**
- Create **managed attributes** or **properties** in your classes
- Perform **lazy attribute evaluation** and provide **computed attributes**
- Avoid **setter** and **getter** methods to make your classes more Pythonic
- Create **read-only**, **read-write**, and **write-only** properties
- Create consistent and **backward-compatible APIs** for your classes
You’ll also write a few practical examples that use `property()` for validating input data, computing attribute values dynamically, logging your code, and more. To get the most out of this tutorial, you should know the basics of [object-oriented](https://realpython.com/python3-object-oriented-programming/) programming and [decorators](https://realpython.com/primer-on-python-decorators/) in Python.
<a class="anchor" id="managing_attributes_in_your_classes"></a>
## Managing Attributes in Your Classes
When you define a class in an [object-oriented](https://en.wikipedia.org/wiki/Object-oriented_programming) programming language, you’ll probably end up with some instance and class [attributes](https://realpython.com/python3-object-oriented-programming/#class-and-instance-attributes). In other words, you’ll end up with variables that are accessible through the instance, class, or even both, depending on the language. Attributes represent or hold the internal [state](https://en.wikipedia.org/wiki/State_(computer_science)) of a given object, which you’ll often need to access and mutate.
Typically, you have at least two ways to manage an attribute. Either you can access and mutate the attribute directly or you can use **methods**. Methods are functions attached to a given class. They provide the behaviors and actions that an object can perform with its internal data and attributes.
If you expose your attributes to the user, then they become part of the public [API](https://en.wikipedia.org/wiki/API) of your classes. Your user will access and mutate them directly in their code. The problem comes when you need to change the internal implementation of a given attribute.
Say you’re working on a `Circle` class. The initial implementation has a single attribute called `.radius`. You finish coding the class and make it available to your end users. They start using `Circle` in their code to create a lot of awesome projects and applications. Good job!
Now suppose that you have an important user that comes to you with a new requirement. They don’t want `Circle` to store the radius any longer. They need a public `.diameter` attribute.
At this point, removing `.radius` to start using `.diameter` could break the code of some of your end users. You need to manage this situation in a way other than removing `.radius`.
Programming languages such as [Java](https://realpython.com/oop-in-python-vs-java/) and [C++](https://en.wikipedia.org/wiki/C%2B%2B) encourage you to never expose your attributes to avoid this kind of problem. Instead, you should provide **getter** and **setter** methods, also known as [accessors](https://en.wikipedia.org/wiki/Accessor_method) and [mutators](https://en.wikipedia.org/wiki/Mutator_method), respectively. These methods offer a way to change the internal implementation of your attributes without changing your public API.
In the end, these languages need getter and setter methods because they don’t provide a suitable way to change the internal implementation of an attribute if a given requirement changes. Changing the internal implementation would require an API modification, which can break your end users’ code.
<a class="anchor" id="the_getter_and_setter_approach_in_python"></a>
### The Getter and Setter Approach in Python
Technically, there’s nothing that stops you from using getter and setter [methods](https://realpython.com/python3-object-oriented-programming/#instance-methods) in Python. Here’s how this approach would look:
```
# point.py
class Point:
def __init__(self, x, y):
self._x = x
self._y = y
def get_x(self):
return self._x
def set_x(self, value):
self._x = value
def get_y(self):
return self._y
def set_y(self, value):
self._y = value
```
In this example, you create `Point` with two **non-public attributes** `._x` and `._y` to hold the [Cartesian coordinates](https://en.wikipedia.org/wiki/Cartesian_coordinate_system) of the point at hand.
To access and mutate the value of either `._x` or `._y`, you can use the corresponding getter and setter methods. Go ahead and save the above definition of `Point` in a Python [module](https://realpython.com/python-modules-packages/) and [import](https://realpython.com/python-import/) the class into your [interactive shell](https://realpython.com/interacting-with-python/).
Here’s how you can work with `Point` in your code:
```
point = Point(12, 5)
point.get_x()
point.get_y()
point.set_x(42)
point.get_x()
# Non-public attributes are still accessible
point._x
point._y
```
With `.get_x()` and `.get_y()`, you can access the current values of `._x` and `._y`. You can use the setter method to store a new value in the corresponding managed attribute. From this code, you can confirm that Python doesn’t restrict access to non-public attributes. Whether or not you do so is up to you.
<a class="anchor" id="the_pythonic_approach"></a>
### The Pythonic Approach
Even though the example you just saw uses the Python coding style, it doesn’t look Pythonic. In the example, the getter and setter methods don’t perform any further processing with `._x` and `._y`. You can rewrite `Point` in a more concise and Pythonic way:
```
class Point:
def __init__(self, x, y):
self.x = x
self.y = y
point = Point(12, 5)
point.x
point.y
point.x = 42
point.x
```
This code uncovers a fundamental principle. Exposing attributes to the end user is normal and common in Python. You don’t need to clutter your classes with getter and setter methods all the time, which sounds pretty cool! However, how can you handle requirement changes that would seem to involve API changes?
Unlike Java and C++, Python provides handy tools that allow you to change the underlying implementation of your attributes without changing your public API. The most popular approach is to turn your attributes into **properties**.
[Properties](https://en.wikipedia.org/wiki/Property_(programming)) represent an intermediate functionality between a plain attribute (or field) and a method. In other words, they allow you to create methods that behave like attributes. With properties, you can change how you compute the target attribute whenever you need to do so.
For example, you can turn both `.x` and `.y` into properties. With this change, you can continue accessing them as attributes. You’ll also have an underlying method holding `.x` and `.y` that will allow you to modify their internal implementation and perform actions on them right before your users access and mutate them.
The main advantage of Python properties is that they allow you to expose your attributes as part of your public API. If you ever need to change the underlying implementation, then you can turn the attribute into a property at any time without much pain.
In the following sections, you’ll learn how to create properties in Python.
<a class="anchor" id="getting_started_with_python’s_`property()`"></a>
## Getting Started With Python’s `property()`
Python’s [`property()`](https://docs.python.org/3/library/functions.html#property) is the Pythonic way to avoid formal getter and setter methods in your code. This function allows you to turn [class attributes](https://realpython.com/python3-object-oriented-programming/#class-and-instance-attributes) into **properties** or **managed attributes**. Since `property()` is a built-in function, you can use it without importing anything. Additionally, `property()` was [implemented in C](https://github.com/python/cpython/blob/main/Objects/descrobject.c#L1460) to ensure optimal performance.
With `property()`, you can attach getter and setter methods to given class attributes. This way, you can handle the internal implementation for that attribute without exposing getter and setter methods in your API. You can also specify a way to handle attribute deletion and provide an appropriate [docstring](https://realpython.com/documenting-python-code/) for your properties.
Here’s the full signature of `property()`:
```
property(fget=None, fset=None, fdel=None, doc=None)
```
The first two arguments take function objects that will play the role of getter (`fget`) and setter (`fset`) methods. Here’s a summary of what each argument does:
|Argument|Description|
|:--|:--|
|`fget`|Function that returns the value of the managed attribute|
|`fset`|Function that allows you to set the value of the managed attribute|
|`fdel`|Function to define how the managed attribute handles deletion|
|`doc`|String representing the property’s docstring|
The [return](https://realpython.com/python-return-statement/) value of `property()` is the managed attribute itself. If you access the managed attribute, as in `obj.attr`, then Python automatically calls `fget()`. If you assign a new value to the attribute, as in `obj.attr = value`, then Python calls `fset()` using the input `value` as an argument. Finally, if you run a `del obj.attr` statement, then Python automatically calls `fdel()`.
You can use `doc` to provide an appropriate docstring for your properties. You and your fellow programmers will be able to read that docstring using Python’s [`help()`](https://docs.python.org/3/library/functions.html#help). The `doc` argument is also useful when you’re working with [code editors and IDEs](https://realpython.com/python-ides-code-editors-guide/) that support docstring access.
You can use `property()` either as a [function](https://realpython.com/defining-your-own-python-function/) or a [decorator](https://realpython.com/primer-on-python-decorators/) to build your properties. In the following two sections, you’ll learn how to use both approaches. However, you should know up front that the decorator approach is more popular in the Python community.
<a class="anchor" id="creating_attributes_with_`property()`"></a>
### Creating Attributes With `property()`
You can create a property by calling `property()` with an appropriate set of arguments and assigning its return value to a class attribute. All the arguments to `property()` are optional. However, you typically provide at least a **setter function**.
The following example shows how to create a `Circle` class with a handy property to manage its radius:
```
# circle.py
class Circle:
def __init__(self, radius):
self._radius = radius
def _get_radius(self):
print("Get radius")
return self._radius
def _set_radius(self, value):
print("Set radius")
self._radius = value
def _del_radius(self):
print("Delete radius")
del self._radius
radius = property(
fget=_get_radius,
fset=_set_radius,
fdel=_del_radius,
doc="The radius property."
)
```
In this code snippet, you create `Circle`. The class initializer, `.__init__()`, takes `radius` as an argument and stores it in a non-public attribute called `._radius`. Then you define three non-public methods:
1. **`._get_radius()`** returns the current value of `._radius`
2. **`._set_radius()`** takes `value` as an argument and assigns it to `._radius`
3. **`._del_radius()`** deletes the instance attribute `._radius`
Once you have these three methods in place, you create a class attribute called `.radius` to store the property object. To initialize the property, you pass the three methods as arguments to `property()`. You also pass a suitable docstring for your property.
In this example, you use [keyword arguments](https://realpython.com/defining-your-own-python-function/#keyword-arguments) to improve the code readability and prevent confusion. That way, you know exactly which method goes into each argument.
To give `Circle` a try, run the following code:
```
circle = Circle(42.0)
circle.radius
circle.radius = 100.0
circle.radius
del circle.radius
circle.radius
help(circle)
```
The `.radius` property hides the non-public instance attribute `._radius`, which is now your managed attribute in this example. You can access and assign `.radius` directly. Internally, Python automatically calls `._get_radius()` and `._set_radius()` when needed. When you execute `del circle.radius`, Python calls `._del_radius()`, which deletes the underlying `._radius`.
<div class="alert alert-success" role="alert">
Properties are <strong>class attributes</strong> that manage <strong>instance attributes</strong>.
</div>
You can think of a property as a collection of methods bundled together. If you inspect `.radius` carefully, then you can find the raw methods you provided as the `fget`, `fset`, and `fdel` arguments:
```
Circle.radius.fget
Circle.radius.fset
Circle.radius.fdel
dir(Circle.radius)
```
You can access the getter, setter, and deleter methods in a given property through the corresponding `.fget`, `.fset`, and `.fdel`.
Properties are also **overriding descriptors**. If you use [`dir()`](https://realpython.com/python-scope-legb-rule/#dir) to check the internal members of a given property, then you’ll find `.__set__()` and `.__get__()` in the list. These methods provide a default implementation of the [descriptor protocol](https://docs.python.org/3/howto/descriptor.html#descriptor-protocol).
The default implementation of `.__set__()`, for example, runs when you don’t provide a custom setter method. In this case, you get an `AttributeError` because there’s no way to set the underlying property.
<a class="anchor" id="using_`property()`_as_a_decorator"></a>
### Using `property()` as a Decorator
Decorators are everywhere in Python. They’re functions that take another function as an argument and return a new function with added functionality. With a decorator, you can attach pre- and post-processing operations to an existing function.
When [Python 2.2](https://docs.python.org/3/whatsnew/2.2.html#attribute-access) introduced `property()`, the decorator syntax wasn’t available. The only way to define properties was to pass getter, setter, and deleter methods, as you learned before. The decorator syntax was added in [Python 2.4](https://docs.python.org/3/whatsnew/2.4.html#pep-318-decorators-for-functions-and-methods), and nowadays, using `property()` as a decorator is the most popular practice in the Python community.
The decorator syntax consists of placing the name of the decorator function with a leading `@` symbol right before the definition of the function you want to decorate:
```
@decorator
def func(a):
return a
```
In this code fragment, `@decorator` can be a function or class intended to decorate `func()`. This syntax is equivalent to the following:
```
def func(a):
return a
func = decorator(func)
```
The final line of code reassigns the name `func` to hold the result of calling `decorator(func)`. Note that this is the same syntax you used to create a property in the section above.
Python’s `property()` can also work as a decorator, so you can use the `@property` syntax to create your properties quickly:
```
# circle.py
class Circle:
def __init__(self, radius):
self._radius = radius
@property
def radius(self):
"""The radius property."""
print("Get radius")
return self._radius
@radius.setter
def radius(self, value):
print("Set radius")
self._radius = value
@radius.deleter
def radius(self):
print("Delete radius")
del self._radius
```
This code looks pretty different from the getter and setter methods approach. `Circle` now looks more Pythonic and clean. You don’t need to use method names such as `._get_radius()`, `._set_radius()`, and `._del_radius()` anymore. Now you have three methods with the same clean and descriptive attribute-like name. How is that possible?
The decorator approach for creating properties requires defining a first method using the public name for the underlying managed attribute, which is `.radius` in this case. This method should implement the getter logic.
Then you define the setter method for `.radius`. In this case, the syntax is fairly different. Instead of using `@property` again, you use `@radius.setter`. Why do you need to do that? Take another look at the `dir()` output:
```
dir(Circle.radius)
```
Besides `.fget`, `.fset`, `.fdel`, and a bunch of other special attributes and methods, `property` also provides `.deleter()`, `.getter()`, and `.setter()`. These three methods each return a new property.
When you decorate the second `.radius()` method with `@radius.setter`, you create a new property and reassign the class-level name `.radius` (line 8) to hold it. This new property contains the same set of methods of the initial property at line 8 with the addition of the new setter method provided on line 14. Finally, the decorator syntax reassigns the new property to the `.radius` class-level name.
The mechanism to define the deleter method is similar. This time, you need to use the `@radius.deleter` decorator. At the end of the process, you get a full-fledged property with the getter, setter, and deleter methods.
Finally, how can you provide suitable docstrings for your properties when you use the decorator approach? If you check `Circle` again, you’ll note that you already did so by adding a docstring to the getter method on line 9.
The new `Circle` implementation works the same as the example in the section above:
```
circle = Circle(42.0)
circle.radius
circle.radius = 100.0
circle.radius
del circle.radius
# This should raise AttributeError cause we deleted circle.radius above
circle.radius
help(circle)
```
You don’t need to use a pair of parentheses for calling `.radius()` as a method. Instead, you can access `.radius` as you would access a regular attribute, which is the primary use of properties. They allow you to treat methods as attributes, and they take care of calling the underlying set of methods automatically.
Here’s a recap of some important points to remember when you’re creating properties with the decorator approach:
- The `@property` decorator must decorate the **getter method**.
- The docstring must go in the **getter method**.
- The **setter and deleter methods** must be decorated with the name of the getter method plus `.setter` and `.deleter`, respectively.
Up to this point, you’ve created managed attributes using `property()` as a function and as a decorator. If you check your `Circle` implementations so far, then you’ll note that their getter and setter methods don’t add any real extra processing on top of your attributes.
In general, you should avoid turning attributes that don’t require extra processing into properties. Using properties in those situations can make your code:
- Unnecessarily verbose
- Confusing to other developers
- Slower than code based on regular attributes
Unless you need something more than bare attribute access, don’t write properties. They’re a waste of [CPU](https://en.wikipedia.org/wiki/Central_processing_unit) time, and more importantly, they’re a waste of *your* time. Finally, you should avoid writing explicit getter and setter methods and then wrapping them in a property. Instead, use the `@property` decorator. That’s currently the most Pythonic way to go.
<a class="anchor" id="providing_read-only_attributes"></a>
## Providing Read-Only Attributes
Probably the most elementary use case of `property()` is to provide **read-only attributes** in your classes. Say you need an [immutable](https://docs.python.org/3/glossary.html#term-immutable) `Point` class that doesn’t allow the user to mutate the original value of its coordinates, `x` and `y`. To achieve this goal, you can create `Point` like in the following example:
```
# point.py
class Point:
def __init__(self, x, y):
self._x = x
self._y = y
@property
def x(self):
return self._x
@property
def y(self):
return self._y
```
Here, you store the input arguments in the attributes `._x` and `._y`. As you already learned, using the leading underscore (`_`) in names tells other developers that they’re non-public attributes and shouldn’t be accessed using dot notation, such as in `point._x`. Finally, you define two getter methods and decorate them with `@property`.
Now you have two read-only properties, `.x` and `.y`, as your coordinates:
```
point = Point(12, 5)
# Read coordinates
point.x
point.y
# Write coordinates
point.x = 42
```
Here, `point.x` and `point.y` are bare-bone examples of read-only properties. Their behavior relies on the underlying descriptor that `property` provides. As you already saw, the default `.__set__()` implementation raises an `AttributeError` when you don’t define a proper setter method.
You can take this implementation of `Point` a little bit further and provide explicit setter methods that raise a custom exception with more elaborate and specific messages:
```
# point.py
class WriteCoordinateError(Exception):
pass
class Point:
def __init__(self, x, y):
self._x = x
self._y = y
@property
def x(self):
return self._x
@x.setter
def x(self, value):
raise WriteCoordinateError("x coordinate is read-only")
@property
def y(self):
return self._y
@y.setter
def y(self, value):
raise WriteCoordinateError("y coordinate is read-only")
```
In this example, you define a custom exception called `WriteCoordinateError`. This exception allows you to customize the way you implement your immutable `Point` class. Now, both setter methods raise your custom exception with a more explicit message. Go ahead and give your improved `Point` a try!
<a class="anchor" id="creating_read-write_attributes"></a>
## Creating Read-Write Attributes
You can also use `property()` to provide managed attributes with **read-write** capabilities. In practice, you just need to provide the appropriate getter method (“read”) and setter method (“write”) to your properties in order to create read-write managed attributes.
Say you want your `Circle` class to have a `.diameter` attribute. However, taking the radius and the diameter in the class initializer seems unnecessary because you can compute the one using the other. Here’s a `Circle` that manages `.radius` and `.diameter` as read-write attributes:
```
# circle.py
import math
class Circle:
def __init__(self, radius):
self.radius = radius
@property
def radius(self):
return self._radius
@radius.setter
def radius(self, value):
self._radius = float(value)
@property
def diameter(self):
return self.radius * 2
@diameter.setter
def diameter(self, value):
self.radius = value / 2
```
Here, you create a `Circle` class with a read-write `.radius`. In this case, the getter method just returns the radius value. The setter method converts the input value for the radius and assigns it to the non-public `._radius`, which is the variable you use to store the final data.
> **Note:** There is a subtle detail to note in this new implementation of `Circle` and its `.radius` attribute. In this case, the class initializer assigns the input value to the `.radius` property directly instead of storing it in a dedicated non-public attribute, such as `._radius`.
Why? Because you need to make sure that every value provided as a radius, including the initialization value, goes through the setter method and gets converted to a floating-point number.
`Circle` also implements a `.diameter` attribute as a property. The getter method computes the diameter using the radius. The setter method does something curious. Instead of storing the input diameter `value` in a dedicated attribute, it calculates the radius and writes the result into `.radius`.
Here’s how your `Circle` works:
```
circle = Circle(42)
circle.radius
circle.diameter
circle.diameter = 100
circle.diameter
circle.radius
```
Both `.radius` and `.diameter` work as normal attributes in these examples, providing a clean and Pythonic public API for your `Circle` class.
<a class="anchor" id="providing_write-only_attributes"></a>
## Providing Write-Only Attributes
You can also create **write-only** attributes by tweaking how you implement the getter method of your properties. For example, you can make your getter method raise an exception every time a user accesses the underlying attribute value.
Here’s an example of handling passwords with a write-only property:
```
# users.py
import hashlib
import os
class User:
def __init__(self, name, password):
self.name = name
self.password = password
@property
def password(self):
raise AttributeError("Password is write-only")
@password.setter
def password(self, plaintext):
salt = os.urandom(32)
self._hashed_password = hashlib.pbkdf2_hmac(
"sha256", plaintext.encode("utf-8"), salt, 100_000
)
```
The initializer of `User` takes a username and a password as arguments and stores them in `.name` and `.password`, respectively. You use a property to manage how your class processes the input password. The getter method raises an `AttributeError` whenever a user tries to retrieve the current password. This turns `.password` into a write-only attribute:
```
john = User("John", "secret")
john._hashed_password
john.password
john.password = "supersecret"
john._hashed_password
```
In this example, you create `john` as a `User` instance with an initial password. The setter method hashes the password and stores it in `._hashed_password`. Note that when you try to access `.password` directly, you get an `AttributeError`. Finally, assigning a new value to `.password` triggers the setter method and creates a new hashed password.
In the setter method of `.password`, you use `os.urandom()` to generate a 32-byte random [string](https://realpython.com/python-strings/) as your hashing function’s [salt](https://en.wikipedia.org/wiki/Salt_(cryptography)). To generate the hashed password, you use [`hashlib.pbkdf2_hmac()`](https://docs.python.org/3/library/hashlib.html#hashlib.pbkdf2_hmac). Then you store the resulting hashed password in the non-public attribute `._hashed_password`. Doing so ensures that you never save the plaintext password in any retrievable attribute.
<a class="anchor" id="putting_python’s_`property()`_into_action"></a>
## Putting Python’s `property()` Into Action
So far, you’ve learned how to use Python’s `property()` built-in function to create managed attributes in your classes. You used `property()` as a function and as a decorator and learned about the differences between these two approaches. You also learned how to create read-only, read-write, and write-only attributes.
In the following sections, you’ll code a few examples that will help you get a better practical understanding of common use cases of `property()`.
<a class="anchor" id="validating_input_values"></a>
### Validating Input Values
One of the most common use cases of `property()` is building managed attributes that validate the input data before storing or even accepting it as a secure input. [Data validation](https://en.wikipedia.org/wiki/Data_validation) is a common requirement in code that takes input from users or other information sources that you consider untrusted.
Python’s `property()` provides a quick and reliable tool for dealing with input data validation. For example, thinking back to the `Point` example, you may require the values of `.x` and `.y` to be valid [numbers](https://realpython.com/python-numbers/). Since your users are free to enter any type of data, you need to make sure that your point only accepts numbers.
Here’s an implementation of `Point` that manages this requirement:
```
# point.py
class Point:
def __init__(self, x, y):
self.x = x
self.y = y
@property
def x(self):
return self._x
@x.setter
def x(self, value):
try:
self._x = float(value)
print("Validated!")
except ValueError:
raise ValueError('"x" must be a number') from None
@property
def y(self):
return self._y
@y.setter
def y(self, value):
try:
self._y = float(value)
print("Validated!")
except ValueError:
raise ValueError('"y" must be a number') from None
```
The setter methods of `.x` and `.y` use [`try` … `except`](https://realpython.com/python-exceptions/#the-try-and-except-block-handling-exceptions) blocks that validate input data using the Python [EAFP](https://docs.python.org/3/glossary.html#term-eafp) style. If the call to `float()` succeeds, then the input data is valid, and you get `Validated!` on your screen. If `float()` raises a `ValueError`, then the user gets a `ValueError` with a more specific message.
It’s important to note that assigning the `.x` and `.y` properties directly in `.__init__()` ensures that the validation also occurs during object initialization. Not doing so is a common mistake when using `property()` for data validation.
Here’s how your `Point` class works now:
```
point = Point(12, 5)
point.x
point.y
point.x = 42
point.x
point.y = 100.0
point.y
point.x = "one"
point.y = "1o"
```
If you assign `.x` and `.y` values that `float()` can turn into floating-point numbers, then the validation is successful, and the value is accepted. Otherwise, you get a `ValueError`.
This implementation of `Point` uncovers a fundamental weakness of `property()`. Did you spot it?
That’s it! You have repetitive code that follows specific patterns. This repetition breaks the [DRY (Don’t Repeat Yourself)](https://en.wikipedia.org/wiki/Don%27t_repeat_yourself) principle, so you would want to [refactor](https://realpython.com/python-refactoring/) this code to avoid it. To do so, you can abstract out the repetitive logic using a descriptor:
```
# point.py
class Coordinate:
def __set_name__(self, owner, name):
self._name = name
def __get__(self, instance, owner):
return instance.__dict__[self._name]
def __set__(self, instance, value):
try:
instance.__dict__[self._name] = float(value)
print("Validated!")
except ValueError:
raise ValueError(f'"{self._name}" must be a number') from None
class Point:
x = Coordinate()
y = Coordinate()
def __init__(self, x, y):
self.x = x
self.y = y
```
Now your code is a bit shorter. You managed to remove repetitive code by defining `Coordinate` as a [descriptor](https://realpython.com/python-descriptors/) that manages your data validation in a single place. The code works just like your earlier implementation. Go ahead and give it a try!
In general, if you find yourself copying and pasting property definitions all around your code or if you spot repetitive code like in the example above, then you should consider using a proper descriptor.
<a class="anchor" id="providing_computed_attributes"></a>
### Providing Computed Attributes
If you need an attribute that builds its value dynamically whenever you access it, then `property()` is the way to go. These kinds of attributes are commonly known as **computed attributes**. They’re handy when you need them to look like [eager](https://en.wikipedia.org/wiki/Eager_evaluation) attributes, but you want them to be [lazy](https://en.wikipedia.org/wiki/Lazy_evaluation).
The main reason for creating eager attributes is to optimize computation costs when you access the attribute often. On the other hand, if you rarely use a given attribute, then a lazy property can postpone its computation until needed, which can make your programs more efficient.
Here’s an example of how to use `property()` to create a computed attribute `.area` in a `Rectangle` class:
```
class Rectangle:
def __init__(self, width, height):
self.width = width
self.height = height
@property
def area(self):
return self.width * self.height
```
In this example, the `Rectangle` initializer takes `width` and `height` as arguments and stores them in regular instance attributes. The read-only property `.area` computes and returns the area of the current rectangle every time you access it.
Another common use case of properties is to provide an auto-formatted value for a given attribute:
```
class Product:
def __init__(self, name, price):
self._name = name
self._price = float(price)
@property
def price(self):
return f"${self._price:,.2f}"
```
In this example, `.price` is a property that formats and returns the price of a particular product. To provide a currency-like format, you use an [f-string](https://realpython.com/python-f-strings/) with appropriate formatting options.
As a final example of computed attributes, say you have a `Point` class that uses `.x` and `.y` as Cartesian coordinates. You want to provide [polar coordinates](https://en.wikipedia.org/wiki/Polar_coordinate_system) for your point so that you can use them in a few computations. The polar coordinate system represents each point using the distance to the origin and the angle with the horizontal coordinate axis.
Here’s a Cartesian coordinates `Point` class that also provides computed polar coordinates:
```
# point.py
import math
class Point:
def __init__(self, x, y):
self.x = x
self.y = y
@property
def distance(self):
return round(math.dist((0, 0), (self.x, self.y)))
@property
def angle(self):
return round(math.degrees(math.atan(self.y / self.x)), 1)
def as_cartesian(self):
return self.x, self.y
def as_polar(self):
return self.distance, self.angle
```
This example shows how to compute the distance and angle of a given `Point` object using its `.x` and `.y` Cartesian coordinates. Here’s how this code works in practice:
```
point = Point(12, 5)
point.x
point.y
point.distance
point.angle
point.as_cartesian()
point.as_polar()
```
When it comes to providing computed or lazy attributes, `property()` is a pretty handy tool. However, if you’re creating an attribute that you use frequently, then computing it every time can be costly and wasteful. A good strategy is to [cache](https://realpython.com/lru-cache-python/) them once the computation is done.
<a class="anchor" id="caching_computed_attributes"></a>
### Caching Computed Attributes
Sometimes you have a given computed attribute that you use frequently. Constantly repeating the same computation may be unnecessary and expensive. To work around this problem, you can cache the computed value and save it in a non-public dedicated attribute for further reuse.
To prevent unexpected behaviors, you need to think of the mutability of the input data. If you have a property that computes its value from constant input values, then the result will never change. In that case, you can compute the value just once:
```
# circle.py
from time import sleep
class Circle:
def __init__(self, radius):
self.radius = radius
self._diameter = None
@property
def diameter(self):
if self._diameter is None:
sleep(0.5) # Simulate a costly computation
self._diameter = self.radius * 2
return self._diameter
```
Even though this implementation of `Circle` properly caches the computed diameter, it has the drawback that if you ever change the value of `.radius`, then `.diameter` won’t return a correct value:
```
circle = Circle(42.0)
circle.radius
circle.diameter # With delay
circle.diameter # Without delay
circle.radius = 100.0
circle.diameter # Wrong diameter
```
In these examples, you create a circle with a radius equal to `42.0`. The `.diameter` property computes its value only the first time you access it. That’s why you see a delay in the first execution and no delay in the second. Note that even though you change the value of the radius, the diameter stays the same.
If the input data for a computed attribute mutates, then you need to recalculate the attribute:
```
# circle.py
from time import sleep
class Circle:
def __init__(self, radius):
self.radius = radius
@property
def radius(self):
return self._radius
@radius.setter
def radius(self, value):
self._diameter = None
self._radius = value
@property
def diameter(self):
if self._diameter is None:
sleep(0.5) # Simulate a costly computation
self._diameter = self._radius * 2
return self._diameter
```
The setter method of the `.radius` property resets `._diameter` to [`None`](https://realpython.com/null-in-python/) every time you change the value of the radius. With this little update, `.diameter` recalculates its value the first time you access it after every mutation of `.radius`:
```
circle = Circle(42.0)
circle.radius
circle.diameter # With delay
circle.diameter # Without delay
circle.radius = 100.0
circle.diameter # With delay
circle.diameter # Without delay
```
Cool! `Circle` works correctly now! It computes the diameter the first time you access it and also every time you change the radius.
Another option to create cached properties is to use [`functools.cached_property()`](https://docs.python.org/3/library/functools.html#functools.cached_property) from the standard library. This function works as a decorator that allows you to transform a method into a cached property. The property computes its value only once and caches it as a normal attribute during the lifetime of the instance:
```
# circle.py
from functools import cached_property
from time import sleep
class Circle:
def __init__(self, radius):
self.radius = radius
@cached_property
def diameter(self):
sleep(0.5) # Simulate a costly computation
return self.radius * 2
```
Here, `.diameter` computes and caches its value the first time you access it. This kind of implementation is suitable for those computations in which the input values don’t mutate. Here’s how it works:
```
circle = Circle(42.0)
circle.diameter # With delay
circle.diameter # Without delay
circle.radius = 100
circle.diameter # Wrong diameter
# Allow direct assignment
circle.diameter = 200
circle.diameter # Cached value
```
When you access `.diameter`, you get its computed value. That value remains the same from this point on. However, unlike `property()`, `cached_property()` doesn’t block attribute mutations unless you provide a proper setter method. That’s why you can update the diameter to `200` in the last couple of lines.
If you want to create a cached property that doesn’t allow modification, then you can use `property()` and [`functools.cache()`](https://docs.python.org/3/library/functools.html#functools.cache) like in the following example:
```
# circle.py
# this import works in python 3.9+
from functools import cache
from time import sleep
class Circle:
def __init__(self, radius):
self.radius = radius
@property
@cache
def diameter(self):
sleep(0.5) # Simulate a costly computation
return self.radius * 2
```
This code stacks `@property` on top of `@cache`. The combination of both decorators builds a cached property that prevents mutations:
```
circle = Circle(42.0)
circle.diameter # With delay
circle.diameter # Without delay
circle.radius = 100
circle.diameter
circle.diameter = 200
```
In these examples, when you try to assign a new value to `.diameter`, you get an `AttributeError` because the setter functionality comes from the internal descriptor of `property`.
<a class="anchor" id="logging_attribute_access_and_mutation"></a>
### Logging Attribute Access and Mutation
Sometimes you need to keep track of what your code does and how your programs flow. A way to do that in Python is to use [`logging`](https://realpython.com/python-logging/). This module provides all the functionality you would require for logging your code. It’ll allow you to constantly watch the code and generate useful information about how it works.
If you ever need to keep track of how and when you access and mutate a given attribute, then you can take advantage of `property()` for that, too:
```
# circle.py
import logging
logging.basicConfig(
format="%(asctime)s: %(message)s",
level=logging.INFO,
datefmt="%H:%M:%S"
)
class Circle:
def __init__(self, radius):
self._msg = '"radius" was {state}. Current value: {value}'
self.radius = radius
@property
def radius(self):
"""The radius property."""
logging.info(self._msg.format(state="accessed", value=str(self._radius)))
return self._radius
@radius.setter
def radius(self, value):
try:
self._radius = float(value)
logging.info(self._msg.format(state="mutated", value=str(self._radius)))
except ValueError:
logging.info('validation error while mutating "radius"')
```
Here, you first import `logging` and define a basic configuration. Then you implement `Circle` with a managed attribute `.radius`. The getter method generates log information every time you access `.radius` in your code. The setter method logs each mutation that you perform on `.radius`. It also logs those situations in which you get an error because of bad input data.
Here’s how you can use `Circle` in your code:
```
circle = Circle(42.0)
circle.radius
circle.radius = 100
circle.radius
circle.radius = "value"
```
Logging useful data from attribute access and mutation can help you debug your code. Logging can also help you identify sources of problematic data input, analyze the performance of your code, spot usage patterns, and more.
<a class="anchor" id="managing_attribute_deletion"></a>
### Managing Attribute Deletion
You can also create properties that implement deleting functionality. This might be a rare use case of `property()`, but having a way to delete an attribute can be handy in some situations.
Say you’re implementing your own [tree](https://en.wikipedia.org/wiki/Tree_(data_structure)) data type. A tree is an [abstract data type](https://en.wikipedia.org/wiki/Abstract_data_type) that stores elements in a hierarchy. The tree components are commonly known as **nodes**. Each node in a tree has a parent node, except for the root node. Nodes can have zero or more children.
Now suppose you need to provide a way to delete or clear the list of children of a given node. Here’s an example that implements a tree node that uses `property()` to provide most of its functionality, including the ability to clear the list of children of the node at hand:
```
# tree.py
class TreeNode:
def __init__(self, data):
self._data = data
self._children = []
@property
def children(self):
return self._children
@children.setter
def children(self, value):
if isinstance(value, list):
self._children = value
else:
del self.children
self._children.append(value)
@children.deleter
def children(self):
self._children.clear()
def __repr__(self):
return f'{self.__class__.__name__}("{self._data}")'
```
In this example, `TreeNode` represents a node in your custom tree data type. Each node stores its children in a Python [list](https://realpython.com/python-lists-tuples/). Then you implement `.children` as a property to manage the underlying list of children. The deleter method calls `.clear()` on the list of children to remove them all:
```
root = TreeNode("root")
child1 = TreeNode("child 1")
child2 = TreeNode("child 2")
root.children = [child1, child2]
root.children
del root.children
root.children
```
Here, you first create a `root` node to start populating the tree. Then you create two new nodes and assign them to `.children` using a list. The [`del`](https://realpython.com/python-keywords/#the-del-keyword) statement triggers the internal deleter method of `.children` and clears the list.
<a class="anchor" id="creating_backward-compatible_class_apis"></a>
### Creating Backward-Compatible Class APIs
As you already know, properties turn method calls into direct attribute lookups. This feature allows you to create clean and Pythonic APIs for your classes. You can expose your attributes publicly without the need for getter and setter methods.
If you ever need to modify how you compute a given public attribute, then you can turn it into a property. Properties make it possible to perform extra processing, such as data validation, without having to modify your public APIs.
Suppose you’re creating an accounting application and you need a base class to manage currencies. To this end, you create a `Currency` class that exposes two attributes, `.units` and `.cents`:
```
class Currency:
def __init__(self, units, cents):
self.units = units
self.cents = cents
# Currency implementation...
```
This class looks clean and Pythonic. Now say that your requirements change, and you decide to store the total number of cents instead of the units and cents. Removing `.units` and `.cents` from your public API to use something like `.total_cents` would break more than one client’s code.
In this situation, `property()` can be an excellent option to keep your current API unchanged. Here’s how you can work around the problem and avoid breaking your clients’ code:
```
# currency.py
CENTS_PER_UNIT = 100
class Currency:
def __init__(self, units, cents):
self._total_cents = units * CENTS_PER_UNIT + cents
@property
def units(self):
return self._total_cents // CENTS_PER_UNIT
@units.setter
def units(self, value):
self._total_cents = self.cents + value * CENTS_PER_UNIT
@property
def cents(self):
return self._total_cents % CENTS_PER_UNIT
@cents.setter
def cents(self, value):
self._total_cents = self.units * CENTS_PER_UNIT + value
# Currency implementation...
```
Now your class stores the total number of cents instead of independent units and cents. However, your users can still access and mutate `.units` and `.cents` in their code and get the same result as before. Go ahead and give it a try!
When you write something upon which many people are going to build, you need to guarantee that modifications to the internal implementation don’t affect how end users work with your classes.
<a class="anchor" id="overriding_properties_in_subclasses"></a>
## Overriding Properties in Subclasses
When you create Python classes that include properties and release them in a package or library, you should expect your users to do a lot of different things with them. One of those things could be **subclassing** them to customize their functionalities. In these cases, your users have to be careful and be aware of a subtle gotcha. If you partially override a property, then you lose the non-overridden functionality.
For example, suppose you’re coding an `Employee` class to manage employee information in your company’s internal accounting system. You already have a class called `Person`, and you think about subclassing it to reuse its functionalities.
`Person` has a `.name` attribute implemented as a property. The current implementation of `.name` doesn’t meet the requirement of returning the name in uppercase letters. Here’s how you end up solving this:
```
# persons.py
class Person:
def __init__(self, name):
self._name = name
@property
def name(self):
return self._name
@name.setter
def name(self, value):
self._name = value
# Person implementation...
class Employee(Person):
@property
def name(self):
return super().name.upper()
# Employee implementation...
```
In `Employee`, you override `.name` to make sure that when you access the attribute, you get the employee name in uppercase:
```
person = Person("John")
person.name
person.name = "John Doe"
person.name
employee = Employee("John")
employee.name
```
Great! `Employee` works as you need! It returns the name using uppercase letters. However, subsequent tests uncover an unexpected behavior:
```
# this should raise AttributeError
employee.name = "John Doe"
```
What happened? Well, when you override an existing property from a parent class, you override the whole functionality of that property. In this example, you reimplemented the getter method only. Because of that, `.name` lost the rest of the functionality from the base class. You don’t have a setter method any longer.
The idea is that if you ever need to override a property in a subclass, then you should provide all the functionality you need in the new version of the property at hand.
<a class="anchor" id="conclusion"></a>
## Conclusion
A **property** is a special type of class member that provides functionality that’s somewhere in between regular attributes and methods. Properties allow you to modify the implementation of instance attributes without changing the public API of the class. Being able to keep your APIs unchanged helps you avoid breaking code your users wrote on top of older versions of your classes.
Properties are the [Pythonic](https://realpython.com/learning-paths/writing-pythonic-code/) way to create **managed attributes** in your classes. They have several use cases in real-world programming, making them a great addition to your skill set as a Python developer.
**In this tutorial, you learned how to:**
- Create **managed attributes** with Python’s `property()`
- Perform **lazy attribute evaluation** and provide **computed attributes**
- Avoid **setter** and **getter** methods with properties
- Create **read-only**, **read-write**, and **write-only** attributes
- Create consistent and **backward-compatible APIs** for your classes
You also wrote several practical examples that walked you through the most common use cases of `property()`. Those examples include input [data validation](#validating-input-values), computed attributes, [logging](https://realpython.com/python-logging/) your code, and more.
|
github_jupyter
|
# iMCSpec (iSpec+emcee)
iMCSpec is a tool which combines iSpec(https://www.blancocuaresma.com/s/iSpec) and emcee(https://emcee.readthedocs.io/en/stable/) into a single unit to perform Bayesian analysis of spectroscopic data to estimate stellar parameters. For more details on the individual code please refer to the links above. This code have been tested on Syntehtic dataset as well as GAIA BENCHMARK stars (https://www.blancocuaresma.com/s/benchmarkstars). The example shown here is for the grid genarated MARCS.GES_atom_hfs. If you want to use any other grid, just download it from the https://www.cfa.harvard.edu/~sblancoc/iSpec/grid/ and make the necessary changes in the line_regions.
Let us import all the necessary packages that are required for this analysis.
```
import os
import sys
import numpy as np
import pandas as pd
import emcee
from multiprocessing import Pool
import matplotlib.pyplot as plt
os.environ["OMP_NUM_THREADS"] = "1"
os.environ['QT_QPA_PLATFORM']='offscreen'
os.environ["NUMEXPR_MAX_THREADS"] = "8" #CHECK NUMBER OF CORES ON YOUR MACHINE AND CHOOSE APPROPRIATELY
ispec_dir = '/home/swastik/iSpec' #MENTION YOUR DIRECTORY WHERE iSPEC is present
sys.path.insert(0, os.path.abspath(ispec_dir))
import ispec
#np.seterr(all="ignore") #FOR MCMC THE WARNING COMES FOR RED BLUE MOVES WHEN ANY PARTICULAR WALKER VALUE DONOT LIE IN THE PARAMETER SPACE
```
Let us read the input spectra. Here I have the input spectrum in .txt format for reading the spectra. You can use the .fits format also for reading the spectra using Astropy (https://docs.astropy.org/en/stable/io/fits/). Please note that my input spectra is normalized and radial velocity (RV) corrected. For normalization and RV correction you can used iSpec or iraf.
```
df = pd.read_csv('/home/swastik/Downloads/test/HPArcturus.txt', sep ='\s+') #ENTER YOUR INPUT SPECTRA
df = df[df.flux != 0] #FOR SOME SPECTROGRAPH PARTS OF SPECTRA ARE MISSING AND THE CORRESPONDING FLUX VALUES ARE LABELLED AS ZEROS. WE WANT TO IGNORE SUCH POINTS
x = df['waveobs'].values
y = df['flux'].values
yerr = df['err'].values
df = np.array(df,dtype=[('waveobs', '<f8'), ('flux', '<f8'), ('err', '<f8')])
```
You can perform the analysis on the entire spectrum or choose specific regions/segments for which you want to perform the analysis for.
```
#--- Read lines with atomic data ------------------------------------------------
# line_regions = ispec.read_line_regions(ispec_dir + "/input/regions/47000_GES/grid_synth_good_for_params_all.txt") #CHANGE THIS ACCORDINGLY FOR THE INPUT GRID
# line_regions = ispec.adjust_linemasks(df, line_regions, max_margin=0.5)
# segments = ispec.create_segments_around_lines(line_regions, margin=0.5)
# ### Add also regions from the wings of strong lines:
# ## H beta
# hbeta_segments = ispec.read_segment_regions(ispec_dir + "/input/regions/wings_Hbeta_segments.txt")
# #segments = hbeta_segments
# segments = np.hstack((segments, hbeta_segments))
# ## H ALPHA
# halpha_segments = ispec.read_segment_regions(ispec_dir + "/input/regions/wings_Halpha_segments.txt")
# segments = np.hstack((segments, halpha_segments))
# ## MG TRIPLET
# mgtriplet_segments = ispec.read_segment_regions(ispec_dir + "/input/regions/wings_MgTriplet_segments.txt")
# segments = np.hstack((segments, mgtriplet_segments))
##IRON
# fe_segments = ispec.read_segment_regions(ispec_dir + "/input/regions/fe_lines_segments.txt")
# segments = np.hstack((segments, fe_segments))
##CALCIUM TRIPLET
# catriplet_segments = ispec.read_segment_regions(ispec_dir + "/input/regions/Calcium_Triplet_segments.txt")
# segments = np.hstack((segments, catriplet_segments))
##Na doublet
# NaDoublet_segments = ispec.read_segment_regions(ispec_dir + "/input/regions/Calcium_Triplet_segments.txt")
# segments = np.hstack((segments, NaDoublet_segments_segments))
# for j in range(len(segments)):
# segments[j][0] = segments[j][0]+0.05
# segments[j][1] = segments[j][1]-0.05
#YOU CAN CHANGE THE STARTING AND ENDING POINTS OF THE SEGEMENT
```
I will create a mask all false values with the same dimension as my original spectra in 1D. I will keep only those values of wavelength and flux for which the value falls in the segments (i.e, Mask is True).
```
# mask =np.zeros(x.shape,dtype =bool)
# for i in range(len(segments)):
# mask|= (x>segments[i][0])&(x<segments[i][1])
# x = x[mask] #SELECTING THOSE VALUES ONLY FOR WHICH MASK VALUE IS TRUE
# y = y[mask]
# #yerr = yerr[mask]
yerr = y*0.0015 #IF ERROR IS NOT SPECIFIED YOU CAN CHOOSE ACCORDINGLY
```
Now let us interpolate the spectrum using iSpec. Here for simplicity I have considered only Teff, log g and [M/H] as free parameters. Vmic and Vmac are obtained from emperical relations by Jofre et al.2013 and Maria Bergemann
```
def synthesize_spectrum(theta):
teff ,logg ,MH = theta
# alpha = ispec.determine_abundance_enchancements(MH)
alpha =0.0
microturbulence_vel = ispec.estimate_vmic(teff, logg, MH)
macroturbulence = ispec.estimate_vmac(teff, logg, MH)
limb_darkening_coeff = 0.6
resolution = 47000
vsini = 1.6 #CHANGE HERE
code = "grid"
precomputed_grid_dir = ispec_dir + "/input/grid/SPECTRUM_MARCS.GES_GESv6_atom_hfs_iso.480_680nm/"
# precomputed_grid_dir = ispec_dir + "/input/grid/SPECTRUM_MARCS.GES_GESv6_atom_hfs_iso.480_680nm_light/"
# The light grid comes bundled with iSpec. It is just for testing purpose. Donot use it for Scientific purpose.
grid = ispec.load_spectral_grid(precomputed_grid_dir)
atomic_linelist = None
isotopes = None
modeled_layers_pack = None
solar_abundances = None
fixed_abundances = None
abundances = None
atmosphere_layers = None
regions = None
if not ispec.valid_interpolated_spectrum_target(grid, {'teff':teff, 'logg':logg, 'MH':MH, 'alpha':alpha, 'vmic': microturbulence_vel}):
msg = "The specified effective temperature, gravity (log g) and metallicity [M/H] \
fall out of the spectral grid limits."
print(msg)
# Interpolation
synth_spectrum = ispec.create_spectrum_structure(x)
synth_spectrum['flux'] = ispec.generate_spectrum(synth_spectrum['waveobs'], \
atmosphere_layers, teff, logg, MH, alpha, atomic_linelist, isotopes, abundances, \
fixed_abundances, microturbulence_vel = microturbulence_vel, \
macroturbulence=macroturbulence, vsini=vsini, limb_darkening_coeff=limb_darkening_coeff, \
R=resolution, regions=regions, verbose=1,
code=code, grid=grid)
return synth_spectrum
```
You can also synthesize the spectrum directly from various atmospheric models. A skeleton of the code taken from iSpec is shown below. For more details check example.py in iSpec.
```
# def synthesize_spectrum(theta,code="spectrum"):
# teff ,logg ,MH = theta
# resolution = 47000
# alpha = ispec.determine_abundance_enchancements(MH)
# microturbulence_vel = ispec.estimate_vmic(teff, logg, MH)
# macroturbulence = ispec.estimate_vmac(teff, logg, MH)
# limb_darkening_coeff = 0.6
# regions = None
# # Selected model amtosphere, linelist and solar abundances
# #model = ispec_dir + "/input/atmospheres/MARCS/"
# #model = ispec_dir + "/input/atmospheres/MARCS.GES/"
# #model = ispec_dir + "/input/atmospheres/MARCS.APOGEE/"
# #model = ispec_dir + "/input/atmospheres/ATLAS9.APOGEE/"
# model = ispec_dir + "/input/atmospheres/ATLAS9.Castelli/"
# #model = ispec_dir + "/input/atmospheres/ATLAS9.Kurucz/"
# #model = ispec_dir + "/input/atmospheres/ATLAS9.Kirby/"
# #atomic_linelist_file = ispec_dir + "/input/linelists/transitions/VALD.300_1100nm/atomic_lines.tsv"
# #atomic_linelist_file = ispec_dir + "/input/linelists/transitions/VALD.1100_2400nm/atomic_lines.tsv"
# atomic_linelist_file = ispec_dir + "/input/linelists/transitions/GESv6_atom_hfs_iso.420_920nm/atomic_lines.tsv"
# #atomic_linelist_file = ispec_dir + "/input/linelists/transitions/GESv6_atom_nohfs_noiso.420_920nm/atomic_lines.tsv"
# isotope_file = ispec_dir + "/input/isotopes/SPECTRUM.lst"
# atomic_linelist = ispec.read_atomic_linelist(atomic_linelist_file, wave_base=wave_base, wave_top=wave_top)
# atomic_linelist = atomic_linelist[atomic_linelist['theoretical_depth'] >= 0.01]
# isotopes = ispec.read_isotope_data(isotope_file)
# if "ATLAS" in model:
# solar_abundances_file = ispec_dir + "/input/abundances/Grevesse.1998/stdatom.dat"
# else:
# # MARCS
# solar_abundances_file = ispec_dir + "/input/abundances/Grevesse.2007/stdatom.dat"
# #solar_abundances_file = ispec_dir + "/input/abundances/Asplund.2005/stdatom.dat"
# #solar_abundances_file = ispec_dir + "/input/abundances/Asplund.2009/stdatom.dat"
# #solar_abundances_file = ispec_dir + "/input/abundances/Anders.1989/stdatom.dat"
# modeled_layers_pack = ispec.load_modeled_layers_pack(model)
# solar_abundances = ispec.read_solar_abundances(solar_abundances_file)
# ## Custom fixed abundances
# #fixed_abundances = ispec.create_free_abundances_structure(["C", "N", "O"], chemical_elements, solar_abundances)
# #fixed_abundances['Abund'] = [-3.49, -3.71, -3.54] # Abundances in SPECTRUM scale (i.e., x - 12.0 - 0.036) and in the same order ["C", "N", "O"]
# ## No fixed abundances
# fixed_abundances = None
# atmosphere_layers = ispec.interpolate_atmosphere_layers(modeled_layers_pack, {'teff':teff, 'logg':logg, 'MH':MH, 'alpha':alpha}, code=code)
# synth_spectrum = ispec.create_spectrum_structure(x)
# synth_spectrum['flux'] = ispec.generate_spectrum(synth_spectrum['waveobs'],
# atmosphere_layers, teff, logg, MH, alpha, atomic_linelist, isotopes, solar_abundances,
# fixed_abundances, microturbulence_vel = microturbulence_vel,
# macroturbulence=macroturbulence, vsini=vsini, limb_darkening_coeff=limb_darkening_coeff,
# R=resolution, regions=regions, verbose=0,
# code=code)
# return synth_spectrum
```
So far we have discussed about reading the input original spectra and interpolating the synthetic spectra from iSpec. Now the important part that comes into picture is to compare the original spectra and the interpolated spectra. For this we will use Montecarlo Markhov chain method to compare both the spectrums. For this we have used the emcee package by
Dan Foreman-Mackey.
```
walkers = eval(input("Enter Walkers: ")) #WALKER IMPLIES THE INDEPENDENT RANDOMLY SELECTED PARAMETER SETS. NOTE IT SHOULD HAVE ATLEAST TWICE THE VALUE OF AVAILABLE FREE PARAMETERS
Iter = eval(input("Enter Iterations: ")) #ITERATION IMPLIES NUMBER OF RUNS THE PARAMETERS WILL BE CHECKED FOR CONVERGENCE. FOR MOST CASES 250-300 SHOULD DO.
```
We will be creating four functions for this MCMC run. The first is straightforward, and is known as the model. The model function should take as an argument a list representing our θ vector, and return the model evaluated at that θ. For completion, your model function should also have your parameter array as an input. The form of this function comes from the Gaussian probability distribution P(x)dx.
```
def log_likelihood(theta):
model = synthesize_spectrum(theta) #GENARATING THE SPECTRUM FOR A GIVEN VALUE OF THETA
sigma2 = yerr ** 2 # FINDING THE Variance
return -0.5 * np.sum((y - (model['flux'])) ** 2/ sigma2) # returns the -chi^2/2 value
```
There is no unique way to set up your prior function. For the simplistic case we have choosen the log prior function returns zero if the input values genarated randomly lies wittin the specified ranges and -infinity if it doesnt(atleast one vale should satisty this criterion). You can choose your own prior function as well.
```
def log_prior(theta):
teff, logg, MH = theta
if 3200 < teff < 6900 and 1.1 < logg < 4.8 and -2.49 < MH <= 0.49 : #CHANGE HERE
return 0.0
return -np.inf
```
The last function we need to define is lnprob(). This function combines the steps above by running the lnprior function, and if the function returned -np.inf, passing that through as a return, and if not (if all priors are good), returning the lnlike for that model (by convention we say it’s the lnprior output + lnlike output, since lnprior’s output should be zero if the priors are good). lnprob needs to take as arguments theta,x,y,and yerr, since these get passed through to lnlike.
```
def log_probability(theta):
lp = log_prior(theta)
if not np.isfinite(lp):
return -np.inf
return lp + log_likelihood(theta)
```
Select input guess values and create intial set of stellar parameters RANDOMLY
```
initial = np.array([4650,1.8,-0.7]) #INPUT GUESS VALUES
pos = initial + np.array([100,0.1,0.1])*np.random.randn(walkers, 3) # YOU CAN CHOOSE UNIFORM RANDOM FUNCTION OR GAUSSIAUN RANDOM NUMBER GENARATOR
nwalkers, ndim = pos.shape
```
Now we will run the EMCEE sampler to run the code. This will take some time depending on the your system. But don't worry :)
```
sampler = emcee.EnsembleSampler(nwalkers, ndim, log_probability)
sampler.run_mcmc(pos,Iter, progress=True)
```
Let us plot the Walkers and Iterations. Check out for convergence in this plot. If you see the convergence you are good to go.
```
fig, axes = plt.subplots(3, figsize=(10, 7), sharex=True)
samples = sampler.get_chain()
accepted = sampler.backend.accepted.astype(bool) #Here accepted indicated that the lines for each parameter below have converged/moved at least one time.
labels = ["teff","logg","MH"]
for i in range(ndim):
ax = axes[i]
ax.plot(samples[:, :, i], "k", alpha=0.3)
ax.set_ylabel(labels[i])
ax.yaxis.set_label_coords(-0.1, 0.5)
axes[-1].set_xlabel("step number");
```
Let us check how good is the fitting.....
```
fig, ax = plt.subplots(1, figsize=(10, 7), sharex=True)
samples = sampler.flatchain
theta_max = samples[np.argmax(sampler.flatlnprobability)]
best_fit_model = synthesize_spectrum(theta_max)
ax.plot(x,y,alpha=0.3)
ax.plot(x,best_fit_model['flux'],alpha =0.3)
ax.plot(x,y-best_fit_model['flux'],alpha =0.3)
plt.savefig('t2.pdf') #CHANGE HERE
print(('Theta max: ',theta_max)) # Genarating the spectrum for the Maximum likelyhood function.
#NOTE THE SPIKES IN THE PLOT BELOW. THESE ARE DUE TO THE FACT THAT END POINTS OF THE SPECTRUMS ARE EXTRAPOLATED
```
Since the first few runs the walkers are exploring the parameter space and convergence have not yet been achived. We will ignore such runs. This is also known as "BURN-IN".
```
new_samples = sampler.get_chain(discard=100, thin=1, flat=False)
new_samples = new_samples[:,accepted,:] # WE ARE ONLY CHOOSING THE VALUES FOR WHICH THE WALKER HAVE MOVED ATLEAST ONCE DURING THE ENTIRE ITERATION. Stagnent walkers indicates that the prior function might have returned -inf.
```
Checking the Convergence after the BURN-IN... If it seems to be converged then it is DONE.
```
fig, axes = plt.subplots(3, figsize=(10, 7), sharex=True)
for i in range(ndim):
ax = axes[i]
ax.plot(new_samples[:, :, i], "k", alpha=0.3)
ax.set_ylabel(labels[i])
ax.yaxis.set_label_coords(-0.1, 0.5)
axes[-1].set_xlabel("step number")
plt.savefig('t3.pdf') #CHANGE HERE
flat_samples = new_samples.reshape(-1,new_samples.shape[2])
np.savetxt("RNtesto.txt",flat_samples,delimiter='\t') #CHANGE HERE
```
# DATA VISUALIZATION
Now after the final list of stellar parameters it is important to visualise the stellar parameter distribution. Also it is important to check for any correlation among the stellar parameters. Here I have shown two medhods by which you can do this. Note: I have taken very few points for analysis and for a proper plot you actually need a much larger dataset (40x300:150 Burns at minimum)
```
import corner
from pandas.plotting import scatter_matrix
df = pd.read_csv('/home/swastik/RNtesto.txt',delimiter='\t',header = None)
df.columns = ["$T_{eff}$", "logg", "[M/H]"]
df.hist() #Plotting Histogram for each individual stellar parameters. THIS NEED NOT BE A GAUSSIAN ONE
#df = df[df.logg < 4.451 ] #REMOVE ANY OUTLIER DISTRIBUTION
scatter_matrix(df, alpha=0.2, figsize=(6, 6), diagonal='kde') #PLOTTING THE SCATTER MATRIX. I HAVE USED A VERY LIGHT DATASET FOR TEST PURPOSE> YOU CAN USE A MORE WALKER X ITERATION FOR A BETTER RESULT
samples = np.vstack([df]) #IT IS NECESSARY TO STACK THE DATA VERTICALLY TO OBTAIN THE DISTRIBUTION FROM THE DATA FRAME
value2 = np.mean(samples, axis=0)
plt.rcParams["font.size"] = "10" #THIS CHANGES THE FONT SIZE OF THE LABELS(NOT LEGEND)
#FINALLY... MAKING THE CORNER PLOT>>>>
#fig = corner.corner(df,show_titles=True,plot_datapoints=True,quantiles=[0.16, 0.5, 0.84],color ='black',levels=(1-np.exp(-0.5),),label_kwargs=dict(fontsize=20,color = 'black'),hist_kwargs=dict(fill = True,color = 'dodgerblue'),alpha =0.2)
fig = corner.corner(df,show_titles=True,plot_datapoints=True,quantiles=[0.16, 0.5, 0.84],color ='black',label_kwargs=dict(fontsize=20,color = 'black'),hist_kwargs=dict(fill = True,color = 'dodgerblue'),alpha =0.2)
axes = np.array(fig.axes).reshape((3, 3))
for i in range(3):
ax = axes[i, i]
ax.axvline(value2[i], color="r",alpha =0.8)
for yi in range(3):
for xi in range(yi):
ax = axes[yi, xi]
ax.axvline(value2[xi], color="r",alpha =0.8,linestyle = 'dashed')
ax.axhline(value2[yi], color="r",alpha =0.8,linestyle = 'dashed')
ax.plot(value2[xi], value2[yi], "r")
# plt.tight_layout()
#THE CORNER PLOT DONOT LOOK GREAT>> THE REASON IS FEW NUMBER OF DATA POINTS AND SHARP CONVERGENCE
```
I would like to thank Sergi blanco-cuaresma for the valuable suggestions and feedbacks regarding the iSpec code and its intregation with emcee. I would also thank
Dan Foreman-Mackey for his insightful comments on using emcee. I would also like to thank Aritra Chakraborty and Dr.Ravinder Banyal for their comments and suggestion on improving the code which might have not been possible without their help.
|
github_jupyter
|
<a href="https://colab.research.google.com/github/ryanleeallred/DS-Unit-1-Sprint-1-Dealing-With-Data/blob/master/module2-loadingdata/LS_DS_112_Loading_Data_Assignment.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Practice Loading Datasets
This assignment is purposely semi-open-ended you will be asked to load datasets both from github and also from CSV files from the [UC Irvine Machine Learning Repository](https://archive.ics.uci.edu/ml/index.php).
Remember that the UCI datasets may not have a file type of `.csv` so it's important that you learn as much as you can about the dataset before you try and load it. See if you can look at the raw text of the file either locally, on github, using the `!curl` shell command, or in some other way before you try and read it in as a dataframe, this will help you catch what would otherwise be unforseen problems.
## 1) Load a dataset from Github (via its *RAW* URL)
Pick a dataset from the following repository and load it into Google Colab. Make sure that the headers are what you would expect and check to see if missing values have been encoded as NaN values:
<https://github.com/ryanleeallred/datasets>
```
# TODO your work here!
# And note you should write comments, descriptions, and add new
# code and text blocks as needed
```
## 2) Load a dataset from your local machine
Download a dataset from the [UC Irvine Machine Learning Repository](https://archive.ics.uci.edu/ml/index.php) and then upload the file to Google Colab either using the files tab in the left-hand sidebar or by importing `files` from `google.colab` The following link will be a useful resource if you can't remember the syntax: <https://towardsdatascience.com/3-ways-to-load-csv-files-into-colab-7c14fcbdcb92>
While you are free to try and load any dataset from the UCI repository, I strongly suggest starting with one of the most popular datasets like those that are featured on the right-hand side of the home page.
Some datasets on UCI will have challenges associated with importing them far beyond what we have exposed you to in class today, so if you run into a dataset that you don't know how to deal with, struggle with it for a little bit, but ultimately feel free to simply choose a different one.
- Make sure that your file has correct headers, and the same number of rows and columns as is specified on the UCI page. If your dataset doesn't have headers use the parameters of the `read_csv` function to add them. Likewise make sure that missing values are encoded as `NaN`.
```
# TODO your work here!
# And note you should write comments, descriptions, and add new
# code and text blocks as needed
```
## 3) Load a dataset from UCI using `!wget`
"Shell Out" and try loading a file directly into your google colab's memory using the `!wget` command and then read it in with `read_csv`.
With this file we'll do a bit more to it.
- Read it in, fix any problems with the header as make sure missing values are encoded as `NaN`.
- Use the `.fillna()` method to fill any missing values.
- [.fillna() documentation](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.fillna.html)
- Create one of each of the following plots using the Pandas plotting functionality:
- Scatterplot
- Histogram
- Density Plot
## Stretch Goals - Other types and sources of data
Not all data comes in a nice single file - for example, image classification involves handling lots of image files. You still will probably want labels for them, so you may have tabular data in addition to the image blobs - and the images may be reduced in resolution and even fit in a regular csv as a bunch of numbers.
If you're interested in natural language processing and analyzing text, that is another example where, while it can be put in a csv, you may end up loading much larger raw data and generating features that can then be thought of in a more standard tabular fashion.
Overall you will in the course of learning data science deal with loading data in a variety of ways. Another common way to get data is from a database - most modern applications are backed by one or more databases, which you can query to get data to analyze. We'll cover this more in our data engineering unit.
How does data get in the database? Most applications generate logs - text files with lots and lots of records of each use of the application. Databases are often populated based on these files, but in some situations you may directly analyze log files. The usual way to do this is with command line (Unix) tools - command lines are intimidating, so don't expect to learn them all at once, but depending on your interests it can be useful to practice.
One last major source of data is APIs: https://github.com/toddmotto/public-apis
API stands for Application Programming Interface, and while originally meant e.g. the way an application interfaced with the GUI or other aspects of an operating system, now it largely refers to online services that let you query and retrieve data. You can essentially think of most of them as "somebody else's database" - you have (usually limited) access.
*Stretch goal* - research one of the above extended forms of data/data loading. See if you can get a basic example working in a notebook. Image, text, or (public) APIs are probably more tractable - databases are interesting, but there aren't many publicly accessible and they require a great deal of setup.
```
```
|
github_jupyter
|
```
###################################
# Test cell, pyechonest - IO HAVOC
###################################
import os
import sys
sys.path.append(os.environ["HOME"] + "/github/pyechonest")
import pyechonest.track as track
import pyechonest.artist as artist
import pyechonest.util as util
import pyechonest.song as song
import sys, pprint
# pprint.pprint(sys.modules)
def get_tempo(artist):
"gets the tempo for a song"
results = song.search(artist=artist, results=1, buckets=['audio_summary'])
if len(results) > 0:
return results[0].audio_summary['tempo']
else:
return None
for hottt_artist in artist.top_hottt(results=1):
print(hottt_artist.name, hottt_artist.id, hottt_artist.hotttnesss)
# print(hottt_artist.name + " " + str(get_tempo(hottt_artist.name)))
# a = artist.Artist(hottt_artist.name)
a = artist.Artist("Red Foo")
images = a.get_images(results=2)
print(images[0]['url'])
# print(images[1]['url'])
# test spotify (spotifpy)
images_b = a.get_spotify_images()
print(str(len(images_b)) + " images found on Spotify")
print(images_b[0]['url'])
###################################
# Generate new pyechonest cache
###################################
import time
import json
from pyechonest import config
from pyechonest import artist
config.ECHO_NEST_API_KEY="2VN1LKJEQUBPUKXEC"
hotttArtistsCache = []
fave4x4ArtistsList = [
'Juan Atkins','Faithless', 'Ruoho Ruotsi', 'Maurice Fulton',
'Leftfield', 'Frivolous', 'Basement Jaxx','Glitch Mob',
'Hollis P. Monroe', 'Frankie Knuckles', 'Francois K',
'Trentemøller', 'Chelonis R. Jones', 'Steve Bug',
'Jimpster', 'Jeff Samuel', 'Ian Pooley',
'Luomo', 'Kerri Chandler', 'Charles Webster',
'Roy Davis Jr.', 'Robert Owens',
'Black Science Orchestra', 'Mr. Fingers', 'Saint Etienne',
'Masters at Work', 'Theo Parrish', 'Moodymann',
'Basic Channel', 'Rhythm & Sound', 'Roman Flügel',
'Joe Lewis', 'DJ Said', 'Recloose', 'Kate Simko', 'Aschka',
'Maya Jane Coles', 'Gys', 'Deadbeat', 'Soultek',
'DeepChord', 'Vladislav Delay', 'Andy Stott', 'Intrusion',
'Rod Modell', 'Kassem Mosse', 'Murcof', 'Marc Leclair',
'Fax', 'Monolake', 'Kit Clayton', 'Bvdub', 'Swayzak',
'Wookie', 'Artful Dodger', 'MJ Cole',
'Les Rythmes Digitales', 'Fischerspooner', 'Cassius',
'Miguel Migs', 'Osunlade', 'Metro Area', 'Dennis Ferrer',
'Ron Trent', 'Larry Heard', 'Alton Miller', 'King Britt',
'Bougie Soliterre', 'Todd Terry', 'Black Coffee',
'Richie Hawtin', 'Speedy J', 'Kenny Larkin', 'Laurent Garnier',
'Carl Craig', 'Robert Hood', 'John Tejada', 'Thomas P. Heckmann',
'Aril Brikha', 'Tiefschwarz', 'Funk D\'Void', 'A Guy Called Gerald',
'Jeff Mills', 'Aaron Carl', 'Josh Wink', 'Derrick May',
'Frankie Bones', 'DJ Assault', 'AUX 88', 'Fumiya Tanaka',
'The Lady Blacktronika', 'Junior Lopez', 'Someone Else', 'Noah Pred',
'Danny Tenaglia', 'Pete Tong', 'Booka Shade', 'Paul Kalkbrenner',
'Dapayk & Padberg', 'Igor O. Vlasov', 'Dreem Teem', 'Todd Edwards',
'187 Lockdown', 'Serious Danger', 'Deep Dish', 'Ellen Allien',
'Matias Aguayo', 'Alex Smoke', 'Modeselektor', 'Mike Shannon',
'Radio Slave', 'Jonas Bering', 'Glitterbug', 'Justus Köhncke',
'Wolfgang Voigt', 'Ripperton', 'Isolée', 'Alex Under',
'Phonique', 'James Holden', 'Minilogue', 'Michael Mayer',
'Pantha Du Prince', 'Håkan Lidbo', 'Lusine', 'Kalabrese',
'Matthew Herbert', 'Jan Jelinek', 'Lucien-N-Luciano', 'Closer Musik',
'Apparat', 'Guillaume & The Coutu Dumonts', 'Thomas Brinkmann',
'The Soft Pink Truth', 'Ada', 'Wighnomy Brothers', 'Ricardo Villalobos',
'Jesse Somfay','Falko Brocksieper', 'Damian Lazarus', 'Superpitcher',
'Catz N\' Dogz', 'Pan/Tone', 'Broker/Dealer', 'Dinky', 'T.Raumschmiere',
'Stephen Beaupré', 'Konrad Black', 'Claude VonStroke', 'DJ Koze',
'Cobblestone Jazz', 'Robag Wruhme', 'Seth Troxler', 'Stewart Walker',
'Farben', 'Pier Bucci', 'Mathew Jonson', 'LoSoul', 'Safety Scissors',
'Anja Schneider', 'Markus Guentner', 'Fuckpony', 'Onur Özer', 'Mossa',
'Kenneth James Gibson', 'Butane', 'Mikael Stavöstrand', 'Franklin de Costa',
'Quantec', 'Jin Choi', 'The Mountain People', 'Château Flight', 'Havantepe',
'Tomas Jirku', 'Limaçon', 'Redshape', 'Mike Huckaby', 'Taylor Deupree',
'Substance & Vainqueur'
]
faveBassArtistsList = [
'Photek', 'Zomby', 'Kode9', 'Vex\'d', 'Plastician', 'Joy Orbison',
'Eskmo', 'Tes La Rok', 'DFRNT', 'Africa HiTech', 'King Midas Sound',
'Skream', 'Djunya', '2562', 'Fantastic Mr. Fox', 'Ikonika',
'Timeblind', 'Mark Pritchard', 'Appleblim', 'Ramadanman', 'D1',
'Matty G', 'Peverelist', 'Untold', 'Roska', 'El-B', 'Mala',
'Coki',' Hijak', 'Mount Kimbie', 'Chrissy Murderbot', 'Scuba',
'Kush Arora', 'Meesha', 'Martyn'
]
# Currently no image resources on EN, lastfm or Spotify for 'Terre Thaemlitz'
faveClassicArtistsList = [
'Björk', 'Kraftwerk', 'DJ Shadow', 'Radiohead', 'The Orb',
'Jean-Michel Jarre', 'Aphex Twin', 'Tangerine Dream',
'Boards of Canada', 'Amon Tobin', 'Ratatat', 'Massive Attack',
'Röyksopp', 'LCD Soundsystem', 'Gotan Project',
'Gus-Gus', 'Everything but the Girl', 'Ursula 1000', 'Llorca',
'UNKLE', 'The Future Sound of London', 'The Avalanches',
'Laika', 'Thievery Corporation', 'Groove Armada', 'Bonobo',
'DJ Food','Tricky', 'Dirty Vegas', 'Télépopmusik', 'Hooverphonic',
'dZihan & Kamien', 'Talvin Singh', 'DJ Vadim', 'Cibo Matto',
'Esthero', 'Martina Topley-Bird', 'Dimitri From Paris',
'Coldcut', 'Death in Vegas', 'Róisín Murphy', 'Nitin Sawhney',
'José Padilla', 'Jimi Tenor', 'Mr. Scruff', 'Dub Pistols',
'Morcheeba', 'Supreme Beings of Leisure', 'Air', 'DJ Krush', 'RJD2',
'Underworld', 'jenn mierau', 'Einstürzende Neubauten',
'Nurse with Wound', 'The Legendary Pink Dots', 'Skinny Puppy',
'Atari Teenage Riot', 'Venetian Snares', 'µ-Ziq', 'Richard Devine',
'Squarepusher', 'Autechre', 'Le Tigre', 'Queens of the Stone Age',
'Xiu Xiu', 'Baby Dee', 'Alastair Galbraith', '不失者', 'I Am Robot and Proud',
'Meg Baird'
]
faveElectroacousticArtistsList = [
'Arthur Russell', 'Jon Appleton', 'Charles Dodge', 'Morton Subotnick',
'James Tenney', 'David Tudor', 'Vladimir Ussachevsky',
'Pauline Oliveros', 'Robert Ashley', 'Nam June Paik', 'La Monte Young',
'Phill Niblock', 'François Bayle', 'James Tenney', 'Tim Hecker', 'Pamela Z',
'Christian Wolff', 'Jean-Claude Risset', 'Paul Lansky', 'Laurie Spiegel',
'Antye Greie', 'Ryoji Ikeda', 'alva noto', 'Ryuichi Sakamoto', 'Lawrence English',
'Tujiko Noriko', 'Arvo Pärt', 'Fennesz', 'Christopher Willits', 'Colleen',
'Ben Frost', 'Jóhann Jóhannsson', 'Sylvain Chauveau'
]
favedubArtistsList = [
'King Tubby', 'Scientist', 'Lee "Scratch" Perry', 'Augustus Pablo',
'Prince Jammy', 'Mad Professor', 'Roots Radics', 'The Upsetters',
'Sly Dunbar', 'Robbie Shakespeare', 'Keith Hudson', 'Tappa Zukie', 'Big Youth',
'The Aggrovators', 'U-Roy', 'Prince Far I',
'Black Uhuru', 'Horace Andy', 'I-Roy', 'The Abyssinians',
'Pablo Moses', 'Max Romeo', 'The Heptones', 'Burning Spear',
'Dennis Brown', 'Jacob Miller', 'Barrington Levy', 'Sugar Minnot',
'Yellowman', 'Gregory Isaacs', 'John Holt', 'Alton Ellis',
'Ken Boothe', 'The Ethiopians', 'Joe Higgs', 'Tommy McCook',
'The Melodians', 'Delroy Wilson', 'Isaac Haile Selassie', 'Polycubist'
]
faveAfricanArtistsList = [
'Manu Dibango', 'Baaba Maal',
'Antibalas Afrobeat Orchestra', 'Orlando Julius', 'William Onyeabor',
'Orchestre Poly-Rythmo', 'Sir Victor Uwaifo',
'Tony Allen & His Afro Messengers', 'Sahara All Stars Band Jos',
'Lijadu Sisters', 'King Sunny Ade', 'Ebo Taylor',
'Gasper Lawal', 'Tunji Oyelana and the Benders', '2 Face', 'P Square',
'Shina Williams & His African Percussionists', 'Weird MC', 'Plantashun Boiz',
'Paul I.K. Dairo', 'D\'banj', 'Ruggedman', 'Eedris Abdulkareem',
'Styl-Plus', 'Tony Tetuila', 'Olamide', 'Ebenezer Obey',
'Haruna Ishola', 'Lágbájá', 'Prince Nico Mbarga', 'West African Highlife Band',
'Modenine', 'Terry tha Rapman', 'Olu Maintain', 'Majek Fashek', 'Konono N°1',
'Koffi Olomidé', 'Les Bantous de la Capitale', 'Thomas Mapfumo', 'Oliver Mtukudzi',
'Chiwoniso Maraire', 'Thomas Mapfumo & The Blacks Unlimited', 'Angélique Kidjo',
'Oumou Sangare', 'Ismaël Lô', 'Geoffrey Oryema', 'Salif Keita', 'Amadou & Mariam',
'Orchestra Baobab', 'Bembeya Jazz National', 'Tiwa Savage'
]
def addArtist(artist):
print(artist.name, artist.id)
artistToAdd = { "name": artist.name,
"id": artist.id,
"images": artist.get_images(results=25),
"URLs": artist.urls,
"genres": artist.terms,
"twitter_id": artist.get_twitter_id()
}
hotttArtistsCache.append(artistToAdd)
time.sleep(10) # delays for 20 seconds
def addFaveArtistList(artistList):
for fave_artist in artistList:
a = artist.Artist(fave_artist, buckets=['images', 'urls', 'terms'])
addArtist(a)
def writeArtistsCaches():
# HOTTT artists
for hottt_artist in artist.top_hottt(results=300):
a = artist.Artist(hottt_artist.id, buckets=['images', 'urls', 'terms'])
addArtist(a)
# FAVE 4x4 artists
addFaveArtistList(fave4x4ArtistsList)
# FAVE Bass artists
addFaveArtistList(faveBassArtistsList)
# FAVE Classic artists
addFaveArtistList(faveClassicArtistsList)
# FAVE Electroacoustic artists
addFaveArtistList(faveElectroacousticArtistsList)
# FAVE Dub artists
addFaveArtistList(favedubArtistsList)
# FAVE African artists
addFaveArtistList(faveAfricanArtistsList)
with open('artistMetaData.js', 'w') as outfile:
outfile.write("var hotttArtistsCache = ")
json.dump(hotttArtistsCache, outfile)
outfile.write(";")
outfile.close()
print ("\n" + "Fini - writeArtistsCaches")
######################
print('fave4x4ArtistsList: ' + str(len(fave4x4ArtistsList)))
print('faveBassArtistsList: ' + str(len(faveBassArtistsList)))
print('faveClassicArtistsList: ' + str(len(faveClassicArtistsList)))
print('faveElectroacousticArtistsList: ' + str(len(faveElectroacousticArtistsList)))
print('favedubArtistsList: ' + str(len(favedubArtistsList)))
print('faveAfricanArtistsList: ' + str(len(faveAfricanArtistsList)))
print('\n\n')
writeArtistsCaches()
```
|
github_jupyter
|
[@LorenaABarba](https://twitter.com/LorenaABarba)
12 steps to Navier–Stokes
=====
***
Did you experiment in Steps [1](./01_Step_1.ipynb) and [2](./02_Step_2.ipynb) using different parameter choices? If you did, you probably ran into some unexpected behavior. Did your solution ever blow up? (In my experience, CFD students *love* to make things blow up.)
You are probably wondering why changing the discretization parameters affects your solution in such a drastic way. This notebook complements our [interactive CFD lessons](https://github.com/barbagroup/CFDPython) by discussing the CFL condition. And learn more by watching Prof. Barba's YouTube lectures (links below).
Convergence and the CFL Condition
----
***
For the first few steps, we've been using the same general initial and boundary conditions. With the parameters we initially suggested, the grid has 41 points and the timestep is 0.25 seconds. Now, we're going to experiment with increasing the size of our grid. The code below is identical to the code we used in [Step 1](./01_Step_1.ipynb), but here it has been bundled up in a function so that we can easily examine what happens as we adjust just one variable: **the grid size**.
```
import numpy #numpy is a library for array operations akin to MATLAB
from matplotlib import pyplot #matplotlib is 2D plotting library
%matplotlib inline
def linearconv(nx):
dx = 2 / (nx - 1)
nt = 20 #nt is the number of timesteps we want to calculate
dt = .025 #dt is the amount of time each timestep covers (delta t)
c = 1
u = numpy.ones(nx) #defining a numpy array which is nx elements long with every value equal to 1.
u[int(.5/dx):int(1 / dx + 1)] = 2 #setting u = 2 between 0.5 and 1 as per our I.C.s
un = numpy.ones(nx) #initializing our placeholder array, un, to hold the values we calculate for the n+1 timestep
for n in range(nt): #iterate through time
un = u.copy() ##copy the existing values of u into un
for i in range(1, nx):
u[i] = un[i] - c * dt / dx * (un[i] - un[i-1])
pyplot.plot(numpy.linspace(0, 2, nx), u);
```
Now let's examine the results of our linear convection problem with an increasingly fine mesh.
```
linearconv(41) #convection using 41 grid points
```
This is the same result as our Step 1 calculation, reproduced here for reference.
```
linearconv(61)
```
Here, there is still numerical diffusion present, but it is less severe.
```
linearconv(71)
```
Here the same pattern is present -- the wave is more square than in the previous runs.
```
linearconv(85)
```
This doesn't look anything like our original hat function.
### What happened?
To answer that question, we have to think a little bit about what we're actually implementing in code.
In each iteration of our time loop, we use the existing data about our wave to estimate the speed of the wave in the subsequent time step. Initially, the increase in the number of grid points returned more accurate answers. There was less numerical diffusion and the square wave looked much more like a square wave than it did in our first example.
Each iteration of our time loop covers a time-step of length $\Delta t$, which we have been defining as 0.025
During this iteration, we evaluate the speed of the wave at each of the $x$ points we've created. In the last plot, something has clearly gone wrong.
What has happened is that over the time period $\Delta t$, the wave is travelling a distance which is greater than `dx`. The length `dx` of each grid box is related to the number of total points `nx`, so stability can be enforced if the $\Delta t$ step size is calculated with respect to the size of `dx`.
$$\sigma = \frac{u \Delta t}{\Delta x} \leq \sigma_{\max}$$
where $u$ is the speed of the wave; $\sigma$ is called the **Courant number** and the value of $\sigma_{\max}$ that will ensure stability depends on the discretization used.
In a new version of our code, we'll use the CFL number to calculate the appropriate time-step `dt` depending on the size of `dx`.
```
import numpy
from matplotlib import pyplot
def linearconv(nx):
dx = 2 / (nx - 1)
nt = 20 #nt is the number of timesteps we want to calculate
c = 1
sigma = .5
dt = sigma * dx
u = numpy.ones(nx)
u[int(.5/dx):int(1 / dx + 1)] = 2
un = numpy.ones(nx)
for n in range(nt): #iterate through time
un = u.copy() ##copy the existing values of u into un
for i in range(1, nx):
u[i] = un[i] - c * dt / dx * (un[i] - un[i-1])
pyplot.plot(numpy.linspace(0, 2, nx), u)
linearconv(41)
linearconv(61)
linearconv(81)
linearconv(101)
linearconv(121)
```
Notice that as the number of points `nx` increases, the wave convects a shorter and shorter distance. The number of time iterations we have advanced the solution at is held constant at `nt = 20`, but depending on the value of `nx` and the corresponding values of `dx` and `dt`, a shorter time window is being examined overall.
Learn More
-----
***
It's possible to do rigurous analysis of the stability of numerical schemes, in some cases. Watch Prof. Barba's presentation of this topic in **Video Lecture 9** on You Tube.
```
from IPython.display import YouTubeVideo
YouTubeVideo('Yw1YPBupZxU')
from IPython.core.display import HTML
def css_styling():
styles = open("../styles/custom.css", "r").read()
return HTML(styles)
css_styling()
```
|
github_jupyter
|
# Likelihood for Retro
To calculate the likelihood of a hypothesis $H$ given observed data $\boldsymbol{k}$, we construct the extended likelihood given as:
$$\large L(H|\boldsymbol{k}) = \prod_{i\in\text{DOMs}} \frac{\lambda_i^{k_i}} {k_i!} e^{-\lambda_i} \prod_{j\in\text{hits}}p^j(t_j|H)^{k_j}$$
where:
* $\lambda_i$ is the expected total charge in DOM $i$ given the hypothesis $H$
* $k_i$ is the observed total charge in DOM $i$
* $p^j(t_j|H)$ is the probability of observing a hit a time $t_j$ in a given DOM $j$ under the hypothesis $H$, raised to the power of the charge $k_j$ of that observed hit
We can take the logarithm of this to change the products into sums
$$\large \log L(H|\boldsymbol{k}) = \sum_{i\in\text{DOMs}} k_i\log{\lambda_i} -\log{{k_i!} - \lambda_i} +\sum_{j\in\text{hits}} k_j\log{p^j(t_j|H)} $$
Since we're only interested in finding the maximum likelihood, we can omit the constant terms $\log{k!}$
In retro, the expected charge $\lambda$ as well as the pdfs $p$ are decomposed into the hypothesis dependent part $N_\gamma(t,x)$ that corresponds to the number of of photons generated by a hypothesis at any given point in space-time and the independent part $p_\gamma(t,x)$ -- the probability that a given source photon in space-time is registered at a DOM.
* The probability $p^j(t_j|H)$ is then simply the sum over all space bins $\sum_x{N_\gamma(t_j,x)p^j_\gamma(t_j,x)}/\lambda_j$, where $\lambda_j$ is the normalization to properly normalize the expression to a pdf
* The time-independent $\lambda_i$s can be interpreted as the total expected charge, given by $\sum_x{\sum_t{p^i_\gamma(x,t)}\sum_t{N_\gamma(x,t)}}$
For many DOMs in an event we observe 0 hits, i.e. $k_i = 0$ for many $i$, this means that the sum over $i$ for these spacial cases simplifies to
$$\sum_{i\in\text{DOMs}} -\lambda_i$$
Plugging in the abvove expression for $\lambda_i$ yields:
$$\sum_{i\in\text{DOMs}}\sum_x{\sum_t{p^i_\gamma(x,t)}\sum_t{N_\gamma(x,t)}}$$
Of course only the probabilities $p^i_\gamma$ are dependent on the DOMs, so we can factorize:
$$\sum_x{\left(\sum_{i\in\text{DOMs}}\sum_t{p^i_\gamma(x,t)}\right)\sum_t{N_\gamma(x,t)}} = \sum_x{p^{TDI}_\gamma(x)\sum_t{N_\gamma(x,t)}}$$
The large sum over the DOMs can therefore be pre-computed, we call this the time-dom-independent (TDI) table, as the time and DOM sums have been evaluated
So we will just need to add the additional terms for DOMs with hits and the total likelihood then can be written as:
$$\large \log L(H|\boldsymbol{k}) = \sum_{i\in\text{hit DOMs}} k_i\log{\sum_x{\sum_t{p^i_\gamma(x,t)}\sum_t{N_\gamma(x,t)}}} -\sum_x{p^{TDI}_\gamma(x)\sum_t{N_\gamma(x,t)}} +\sum_{j\in\text{hits}} k_j\log{\sum_x{N_\gamma(t_j,x)p_\gamma(t_j,x)}/\lambda_j}$$
|
github_jupyter
|
```
import numpy as np
import pandas as pd
from grn_learn.viz import set_plotting_style
import seaborn as sns
import matplotlib.pyplot as plt
from grn_learn import download_and_preprocess_data
from grn_learn import annot_data_trn
from grn_learn import train_keras_multilabel_nn
from sklearn.model_selection import StratifiedKFold
from keras.backend import clear_session
import keras
from keras.models import Sequential
from keras.layers import Dense
from keras import regularizers
from keras.utils import np_utils
from keras.metrics import categorical_accuracy
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import KFold
from keras.layers import Dropout
import keras.backend as K
from keras import regularizers
from sklearn.model_selection import train_test_split
seed = 42
np.random.seed(seed)
set_plotting_style()
%matplotlib inline
%config InlineBackend.figure_format = 'svg'
```
### Download, preprocess data for *P. aeru*
```
org = 'paeru'
```
g.download_and_preprocess_data('paeru',
data_dir = 'colombos_'+ org + '_exprdata_20151029.txt')
### Annotate dataset using the TRN from the Martinez-Antonio lab
```
# paeru
paeru_path = '~/Documents/uni/bioinfo/data/paeru/'
g.annot_data_trn(tf_tf_net_path = paeru_path + 'paeru_tf_tf_net.csv',
trn_path = paeru_path + 'paeru-trn.csv',
denoised_data_path= '~/Downloads/',
org = 'paeru',
output_path = '~/Downloads/')
#df_trn = pd.read_csv(path + 'paeru-trn.csv', comment= '#')
#tfs = pd.read_csv(path+'paeru_tfs.csv')
# tf_tf_df = get_gene_data(df_trn, 'Target gene', tf_list)
denoised = pd.read_csv('~/Downloads/denoised_hot_paeru.csv')
denoised.head()
regulons_p = denoised[denoised['TG'] == 1]
non_regulons_p = denoised[denoised['TG'] == 0]
noise = non_regulons_p.sample(n = 50, replace = False)
regulons_with_noise_p = pd.concat([regulons_p, noise], axis = 0)
non_regulons_wo_noise = non_regulons_p.drop(noise.index.to_list())
#annot = regulons_with_noise.iloc[:, :3]
data_p = regulons_with_noise_p.iloc[:, 3:-13]
target_p = regulons_with_noise_p.iloc[:, -13:-1]
target_p.head()
val_shape = int(data.shape[0] * 0.15)
X_train, X_test, y_train, y_test = train_test_split(data,
target,
shuffle = True,
test_size=0.2,
random_state= seed)
x_val = X_train[:val_shape]
partial_x_train = X_train[val_shape:]
y_val = y_train[:val_shape]
partial_y_train = y_train[val_shape:]
```
### Run keras net on paeru dataset
```
nn, history = g.train_keras_multilabel_nn(X_train,
y_train,
partial_x_train,
partial_y_train,
x_val,
y_val,
n_units=64,
epochs=20,
n_deep_layers=3,
batch_size=128)
score, accuracy = nn.evaluate(
X_test,
y_test,
batch_size=64,
verbose=2
)
accuracy
```
### B. subti data download
```
# bsubti
bsubt_path = '~/Documents/uni/bioinfo/data/bsubti/'
g.download_and_preprocess_data('bsubt')
# data_dir = 'colombos_'+ org + '_exprdata_20151029.txt')
```
### B. subti annotate dataset using TRN from the Merino Lab
```
g.annot_data_trn(tf_tf_net_path = bsubt_path + 'bsub-tf-net.csv',
trn_path = bsubt_path + 'bsubt_trn-l.txt',
denoised_data_path= '~/Downloads/',
org = 'bsubt',
output_path = '~/Downloads/')
denoised_b = pd.read_csv('~/Downloads/denoised_hot_bsubt.csv')
#denoised.head()
regulons_b = denoised_b[denoised_b['TG'] == 1]
non_regulons_b = denoised_b[denoised_b['TG'] == 0]
noise = non_regulons_b.sample(n = 50, replace = False)
regulons_with_noise_b = pd.concat([regulons_b, noise], axis = 0)
non_regulons_wo_noise = non_regulons_b.drop(noise.index.to_list())
#annot = regulons_with_noise.iloc[:, :3]
data_b = regulons_with_noise_b.iloc[:, 3:-7]
target_b = regulons_with_noise_b.iloc[:, -7:-1]
data_b[:5, -1]
val_shape = int(data.shape[0] * 0.15)
X_train, X_test, y_train, y_test = train_test_split(data,
target,
shuffle = True,
test_size=0.2,
random_state= seed)
x_val = X_train[:val_shape]
partial_x_train = X_train[val_shape:]
y_val = y_train[:val_shape]
partial_y_train = y_train[val_shape:]
nn, history = g.train_keras_multilabel_nn(X_train,
y_train,
partial_x_train,
partial_y_train,
x_val,
y_val,
n_units=64,
epochs=20,
n_deep_layers=3,
batch_size=128)
history.head()
score, accuracy = nn.evaluate(
X_test,
y_test,
batch_size=64,
verbose=2
)
accuracy
```
## Upload coli data
```
denoised = pd.read_csv('~/Downloads/denoised_hot_coli.csv')
regulons_e = denoised[denoised['TG'] == 1]
non_regulons_e = denoised[denoised['TG'] == 0]
noise = non_regulons_e.sample(n = 50, replace = False)
regulons_with_noise_e = pd.concat([regulons_e, noise], axis = 0)
non_regulons_wo_noise = non_regulons_e.drop(noise.index.to_list())
#annot = regulons_with_noise.iloc[:, :3]
data_e = regulons_with_noise_e.iloc[:, 3:-10]
target_e = regulons_with_noise_e.iloc[:, -10:-1]
```
### Set up simulations for E. coli, B. subti, and P. aeru
```
organisms = ['ecoli', 'bsubti', 'paeru']
datasets = [(data_e, target_e), (data_b, target_b), (data_p, target_p)]
kfold = KFold(n_splits = 5, shuffle= True, random_state=seed)
cross_val_df = pd.DataFrame()
# Iterate over organisms
for ix in range(3):
#
org = organisms[ix]
data = datasets[ix]
#Extract datasets
X = data[0]
y = data[1]
# Iterate over folds
for train, test in kfold.split(data[0], data[1]):
# Train test split
X_train = X.iloc[train, :]
y_train = y.iloc[train, :]
X_test = X.iloc[test, :]
y_test = y.iloc[test, :]
#print(type(X_train))
# Run neural net
nn = Sequential()
#initial layer
nn.add(Dense(128, activation='relu', input_shape=(X_train.shape[1],)))
#extra deep layers
for i in range(2):
nn.add(Dense(64, activation='relu',
kernel_regularizer=regularizers.l2(0.001))
)
nn.add(Dropout(0.25))
#add final output layer
nn.add(Dense(y_train.shape[1], activation='softmax'))
nn.compile(optimizer='rmsprop',
loss='binary_crossentropy',
metrics=['accuracy'])
#print neural net architecture
nn.summary()
#fit and load history
history = nn.fit(X_train, y_train, epochs=20,
batch_size= 128,
verbose = 0)
# Compute accuracy
score, acc = nn.evaluate(X_test, y_test)
# Store acc in dataframe
sub_df = pd.DataFrame({'accuracy': [acc],
'organism': [org]})
cross_val_df = pd.concat([cross_val_df, sub_df])
#cross_val_df.to_csv('../../data/cv_data.csv', index = False)
#cross_val_df = pd.read_csv('../../data/cv_data.csv')
sns.boxplot?
plt.figure(figsize = (6, 3.2))
sns.boxplot(data = cross_val_df,
y = 'organism',
x = 'accuracy',
color = 'lightgray',
saturation = 1,
whis = 1,
width = 0.7
#alpha = 0.5
)
sns.stripplot(data = cross_val_df,
y = 'organism',
x = 'accuracy',
palette = 'Set2',
#edgecolor = 'gray',
#linewidth = 0.4,
size = 10,
alpha = 0.7)
plt.tight_layout()
plt.xlim(0.5, 1.01)
plt.savefig('cross_val_org.pdf', dpi = 600)
```
|
github_jupyter
|
# Batch Normalization – Lesson
1. [What is it?](#theory)
2. [What are it's benefits?](#benefits)
3. [How do we add it to a network?](#implementation_1)
4. [Let's see it work!](#demos)
5. [What are you hiding?](#implementation_2)
# What is Batch Normalization?<a id='theory'></a>
Batch normalization was introduced in Sergey Ioffe's and Christian Szegedy's 2015 paper [Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift](https://arxiv.org/pdf/1502.03167.pdf). The idea is that, instead of just normalizing the inputs to the network, we normalize the inputs to _layers within_ the network. It's called "batch" normalization because during training, we normalize each layer's inputs by using the mean and variance of the values in the current mini-batch.
Why might this help? Well, we know that normalizing the inputs to a _network_ helps the network learn. But a network is a series of layers, where the output of one layer becomes the input to another. That means we can think of any layer in a neural network as the _first_ layer of a smaller network.
For example, imagine a 3 layer network. Instead of just thinking of it as a single network with inputs, layers, and outputs, think of the output of layer 1 as the input to a two layer network. This two layer network would consist of layers 2 and 3 in our original network.
Likewise, the output of layer 2 can be thought of as the input to a single layer network, consisting only of layer 3.
When you think of it like that - as a series of neural networks feeding into each other - then it's easy to imagine how normalizing the inputs to each layer would help. It's just like normalizing the inputs to any other neural network, but you're doing it at every layer (sub-network).
Beyond the intuitive reasons, there are good mathematical reasons why it helps the network learn better, too. It helps combat what the authors call _internal covariate shift_. This discussion is best handled [in the paper](https://arxiv.org/pdf/1502.03167.pdf) and in [Deep Learning](http://www.deeplearningbook.org) a book you can read online written by Ian Goodfellow, Yoshua Bengio, and Aaron Courville. Specifically, check out the batch normalization section of [Chapter 8: Optimization for Training Deep Models](http://www.deeplearningbook.org/contents/optimization.html).
# Benefits of Batch Normalization<a id="benefits"></a>
Batch normalization optimizes network training. It has been shown to have several benefits:
1. **Networks train faster** – Each training _iteration_ will actually be slower because of the extra calculations during the forward pass and the additional hyperparameters to train during back propagation. However, it should converge much more quickly, so training should be faster overall.
2. **Allows higher learning rates** – Gradient descent usually requires small learning rates for the network to converge. And as networks get deeper, their gradients get smaller during back propagation so they require even more iterations. Using batch normalization allows us to use much higher learning rates, which further increases the speed at which networks train.
3. **Makes weights easier to initialize** – Weight initialization can be difficult, and it's even more difficult when creating deeper networks. Batch normalization seems to allow us to be much less careful about choosing our initial starting weights.
4. **Makes more activation functions viable** – Some activation functions do not work well in some situations. Sigmoids lose their gradient pretty quickly, which means they can't be used in deep networks. And ReLUs often die out during training, where they stop learning completely, so we need to be careful about the range of values fed into them. Because batch normalization regulates the values going into each activation function, non-linearlities that don't seem to work well in deep networks actually become viable again.
5. **Simplifies the creation of deeper networks** – Because of the first 4 items listed above, it is easier to build and faster to train deeper neural networks when using batch normalization. And it's been shown that deeper networks generally produce better results, so that's great.
6. **Provides a bit of regularlization** – Batch normalization adds a little noise to your network. In some cases, such as in Inception modules, batch normalization has been shown to work as well as dropout. But in general, consider batch normalization as a bit of extra regularization, possibly allowing you to reduce some of the dropout you might add to a network.
7. **May give better results overall** – Some tests seem to show batch normalization actually improves the training results. However, it's really an optimization to help train faster, so you shouldn't think of it as a way to make your network better. But since it lets you train networks faster, that means you can iterate over more designs more quickly. It also lets you build deeper networks, which are usually better. So when you factor in everything, you're probably going to end up with better results if you build your networks with batch normalization.
# Batch Normalization in TensorFlow<a id="implementation_1"></a>
This section of the notebook shows you one way to add batch normalization to a neural network built in TensorFlow.
The following cell imports the packages we need in the notebook and loads the MNIST dataset to use in our experiments. However, the `tensorflow` package contains all the code you'll actually need for batch normalization.
```
# Import necessary packages
import tensorflow as tf
import tqdm
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# Import MNIST data so we have something for our experiments
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets("MNIST_data/", one_hot=True)
```
### Neural network classes for testing
The following class, `NeuralNet`, allows us to create identical neural networks with and without batch normalization. The code is heavily documented, but there is also some additional discussion later. You do not need to read through it all before going through the rest of the notebook, but the comments within the code blocks may answer some of your questions.
*About the code:*
>This class is not meant to represent TensorFlow best practices – the design choices made here are to support the discussion related to batch normalization.
>It's also important to note that we use the well-known MNIST data for these examples, but the networks we create are not meant to be good for performing handwritten character recognition. We chose this network architecture because it is similar to the one used in the original paper, which is complex enough to demonstrate some of the benefits of batch normalization while still being fast to train.
```
class NeuralNet:
def __init__(self, initial_weights, activation_fn, use_batch_norm):
"""
Initializes this object, creating a TensorFlow graph using the given parameters.
:param initial_weights: list of NumPy arrays or Tensors
Initial values for the weights for every layer in the network. We pass these in
so we can create multiple networks with the same starting weights to eliminate
training differences caused by random initialization differences.
The number of items in the list defines the number of layers in the network,
and the shapes of the items in the list define the number of nodes in each layer.
e.g. Passing in 3 matrices of shape (784, 256), (256, 100), and (100, 10) would
create a network with 784 inputs going into a hidden layer with 256 nodes,
followed by a hidden layer with 100 nodes, followed by an output layer with 10 nodes.
:param activation_fn: Callable
The function used for the output of each hidden layer. The network will use the same
activation function on every hidden layer and no activate function on the output layer.
e.g. Pass tf.nn.relu to use ReLU activations on your hidden layers.
:param use_batch_norm: bool
Pass True to create a network that uses batch normalization; False otherwise
Note: this network will not use batch normalization on layers that do not have an
activation function.
"""
# Keep track of whether or not this network uses batch normalization.
self.use_batch_norm = use_batch_norm
self.name = "With Batch Norm" if use_batch_norm else "Without Batch Norm"
# Batch normalization needs to do different calculations during training and inference,
# so we use this placeholder to tell the graph which behavior to use.
self.is_training = tf.placeholder(tf.bool, name="is_training")
# This list is just for keeping track of data we want to plot later.
# It doesn't actually have anything to do with neural nets or batch normalization.
self.training_accuracies = []
# Create the network graph, but it will not actually have any real values until after you
# call train or test
self.build_network(initial_weights, activation_fn)
def build_network(self, initial_weights, activation_fn):
"""
Build the graph. The graph still needs to be trained via the `train` method.
:param initial_weights: list of NumPy arrays or Tensors
See __init__ for description.
:param activation_fn: Callable
See __init__ for description.
"""
self.input_layer = tf.placeholder(tf.float32, [None, initial_weights[0].shape[0]])
layer_in = self.input_layer
for weights in initial_weights[:-1]:
layer_in = self.fully_connected(layer_in, weights, activation_fn)
self.output_layer = self.fully_connected(layer_in, initial_weights[-1])
def fully_connected(self, layer_in, initial_weights, activation_fn=None):
"""
Creates a standard, fully connected layer. Its number of inputs and outputs will be
defined by the shape of `initial_weights`, and its starting weight values will be
taken directly from that same parameter. If `self.use_batch_norm` is True, this
layer will include batch normalization, otherwise it will not.
:param layer_in: Tensor
The Tensor that feeds into this layer. It's either the input to the network or the output
of a previous layer.
:param initial_weights: NumPy array or Tensor
Initial values for this layer's weights. The shape defines the number of nodes in the layer.
e.g. Passing in 3 matrix of shape (784, 256) would create a layer with 784 inputs and 256
outputs.
:param activation_fn: Callable or None (default None)
The non-linearity used for the output of the layer. If None, this layer will not include
batch normalization, regardless of the value of `self.use_batch_norm`.
e.g. Pass tf.nn.relu to use ReLU activations on your hidden layers.
"""
# Since this class supports both options, only use batch normalization when
# requested. However, do not use it on the final layer, which we identify
# by its lack of an activation function.
if self.use_batch_norm and activation_fn:
# Batch normalization uses weights as usual, but does NOT add a bias term. This is because
# its calculations include gamma and beta variables that make the bias term unnecessary.
# (See later in the notebook for more details.)
weights = tf.Variable(initial_weights)
linear_output = tf.matmul(layer_in, weights)
# Apply batch normalization to the linear combination of the inputs and weights
batch_normalized_output = tf.layers.batch_normalization(linear_output, training=self.is_training)
# Now apply the activation function, *after* the normalization.
return activation_fn(batch_normalized_output)
else:
# When not using batch normalization, create a standard layer that multiplies
# the inputs and weights, adds a bias, and optionally passes the result
# through an activation function.
weights = tf.Variable(initial_weights)
biases = tf.Variable(tf.zeros([initial_weights.shape[-1]]))
linear_output = tf.add(tf.matmul(layer_in, weights), biases)
return linear_output if not activation_fn else activation_fn(linear_output)
def train(self, session, learning_rate, training_batches, batches_per_sample, save_model_as=None):
"""
Trains the model on the MNIST training dataset.
:param session: Session
Used to run training graph operations.
:param learning_rate: float
Learning rate used during gradient descent.
:param training_batches: int
Number of batches to train.
:param batches_per_sample: int
How many batches to train before sampling the validation accuracy.
:param save_model_as: string or None (default None)
Name to use if you want to save the trained model.
"""
# This placeholder will store the target labels for each mini batch
labels = tf.placeholder(tf.float32, [None, 10])
# Define loss and optimizer
cross_entropy = tf.reduce_mean(
tf.nn.softmax_cross_entropy_with_logits(labels=labels, logits=self.output_layer))
# Define operations for testing
correct_prediction = tf.equal(tf.argmax(self.output_layer, 1), tf.argmax(labels, 1))
accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))
if self.use_batch_norm:
# If we don't include the update ops as dependencies on the train step, the
# tf.layers.batch_normalization layers won't update their population statistics,
# which will cause the model to fail at inference time
with tf.control_dependencies(tf.get_collection(tf.GraphKeys.UPDATE_OPS)):
train_step = tf.train.GradientDescentOptimizer(learning_rate).minimize(cross_entropy)
else:
train_step = tf.train.GradientDescentOptimizer(learning_rate).minimize(cross_entropy)
# Train for the appropriate number of batches. (tqdm is only for a nice timing display)
for i in tqdm.tqdm(range(training_batches)):
# We use batches of 60 just because the original paper did. You can use any size batch you like.
batch_xs, batch_ys = mnist.train.next_batch(60)
session.run(train_step, feed_dict={self.input_layer: batch_xs,
labels: batch_ys,
self.is_training: True})
# Periodically test accuracy against the 5k validation images and store it for plotting later.
if i % batches_per_sample == 0:
test_accuracy = session.run(accuracy, feed_dict={self.input_layer: mnist.validation.images,
labels: mnist.validation.labels,
self.is_training: False})
self.training_accuracies.append(test_accuracy)
# After training, report accuracy against test data
test_accuracy = session.run(accuracy, feed_dict={self.input_layer: mnist.validation.images,
labels: mnist.validation.labels,
self.is_training: False})
print('{}: After training, final accuracy on validation set = {}'.format(self.name, test_accuracy))
# If you want to use this model later for inference instead of having to retrain it,
# just construct it with the same parameters and then pass this file to the 'test' function
if save_model_as:
tf.train.Saver().save(session, save_model_as)
def test(self, session, test_training_accuracy=False, include_individual_predictions=False, restore_from=None):
"""
Trains a trained model on the MNIST testing dataset.
:param session: Session
Used to run the testing graph operations.
:param test_training_accuracy: bool (default False)
If True, perform inference with batch normalization using batch mean and variance;
if False, perform inference with batch normalization using estimated population mean and variance.
Note: in real life, *always* perform inference using the population mean and variance.
This parameter exists just to support demonstrating what happens if you don't.
:param include_individual_predictions: bool (default True)
This function always performs an accuracy test against the entire test set. But if this parameter
is True, it performs an extra test, doing 200 predictions one at a time, and displays the results
and accuracy.
:param restore_from: string or None (default None)
Name of a saved model if you want to test with previously saved weights.
"""
# This placeholder will store the true labels for each mini batch
labels = tf.placeholder(tf.float32, [None, 10])
# Define operations for testing
correct_prediction = tf.equal(tf.argmax(self.output_layer, 1), tf.argmax(labels, 1))
accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))
# If provided, restore from a previously saved model
if restore_from:
tf.train.Saver().restore(session, restore_from)
# Test against all of the MNIST test data
test_accuracy = session.run(accuracy, feed_dict={self.input_layer: mnist.test.images,
labels: mnist.test.labels,
self.is_training: test_training_accuracy})
print('-'*75)
print('{}: Accuracy on full test set = {}'.format(self.name, test_accuracy))
# If requested, perform tests predicting individual values rather than batches
if include_individual_predictions:
predictions = []
correct = 0
# Do 200 predictions, 1 at a time
for i in range(200):
# This is a normal prediction using an individual test case. However, notice
# we pass `test_training_accuracy` to `feed_dict` as the value for `self.is_training`.
# Remember that will tell it whether it should use the batch mean & variance or
# the population estimates that were calucated while training the model.
pred, corr = session.run([tf.arg_max(self.output_layer,1), accuracy],
feed_dict={self.input_layer: [mnist.test.images[i]],
labels: [mnist.test.labels[i]],
self.is_training: test_training_accuracy})
correct += corr
predictions.append(pred[0])
print("200 Predictions:", predictions)
print("Accuracy on 200 samples:", correct/200)
```
There are quite a few comments in the code, so those should answer most of your questions. However, let's take a look at the most important lines.
We add batch normalization to layers inside the `fully_connected` function. Here are some important points about that code:
1. Layers with batch normalization do not include a bias term.
2. We use TensorFlow's [`tf.layers.batch_normalization`](https://www.tensorflow.org/api_docs/python/tf/layers/batch_normalization) function to handle the math. (We show lower-level ways to do this [later in the notebook](#implementation_2).)
3. We tell `tf.layers.batch_normalization` whether or not the network is training. This is an important step we'll talk about later.
4. We add the normalization **before** calling the activation function.
In addition to that code, the training step is wrapped in the following `with` statement:
```python
with tf.control_dependencies(tf.get_collection(tf.GraphKeys.UPDATE_OPS)):
```
This line actually works in conjunction with the `training` parameter we pass to `tf.layers.batch_normalization`. Without it, TensorFlow's batch normalization layer will not operate correctly during inference.
Finally, whenever we train the network or perform inference, we use the `feed_dict` to set `self.is_training` to `True` or `False`, respectively, like in the following line:
```python
session.run(train_step, feed_dict={self.input_layer: batch_xs,
labels: batch_ys,
self.is_training: True})
```
We'll go into more details later, but next we want to show some experiments that use this code and test networks with and without batch normalization.
# Batch Normalization Demos<a id='demos'></a>
This section of the notebook trains various networks with and without batch normalization to demonstrate some of the benefits mentioned earlier.
We'd like to thank the author of this blog post [Implementing Batch Normalization in TensorFlow](http://r2rt.com/implementing-batch-normalization-in-tensorflow.html). That post provided the idea of - and some of the code for - plotting the differences in accuracy during training, along with the idea for comparing multiple networks using the same initial weights.
## Code to support testing
The following two functions support the demos we run in the notebook.
The first function, `plot_training_accuracies`, simply plots the values found in the `training_accuracies` lists of the `NeuralNet` objects passed to it. If you look at the `train` function in `NeuralNet`, you'll see it that while it's training the network, it periodically measures validation accuracy and stores the results in that list. It does that just to support these plots.
The second function, `train_and_test`, creates two neural nets - one with and one without batch normalization. It then trains them both and tests them, calling `plot_training_accuracies` to plot how their accuracies changed over the course of training. The really imporant thing about this function is that it initializes the starting weights for the networks _outside_ of the networks and then passes them in. This lets it train both networks from the exact same starting weights, which eliminates performance differences that might result from (un)lucky initial weights.
```
def plot_training_accuracies(*args, **kwargs):
"""
Displays a plot of the accuracies calculated during training to demonstrate
how many iterations it took for the model(s) to converge.
:param args: One or more NeuralNet objects
You can supply any number of NeuralNet objects as unnamed arguments
and this will display their training accuracies. Be sure to call `train`
the NeuralNets before calling this function.
:param kwargs:
You can supply any named parameters here, but `batches_per_sample` is the only
one we look for. It should match the `batches_per_sample` value you passed
to the `train` function.
"""
fig, ax = plt.subplots()
batches_per_sample = kwargs['batches_per_sample']
for nn in args:
ax.plot(range(0,len(nn.training_accuracies)*batches_per_sample,batches_per_sample),
nn.training_accuracies, label=nn.name)
ax.set_xlabel('Training steps')
ax.set_ylabel('Accuracy')
ax.set_title('Validation Accuracy During Training')
ax.legend(loc=4)
ax.set_ylim([0,1])
plt.yticks(np.arange(0, 1.1, 0.1))
plt.grid(True)
plt.show()
def train_and_test(use_bad_weights, learning_rate, activation_fn, training_batches=50000, batches_per_sample=500):
"""
Creates two networks, one with and one without batch normalization, then trains them
with identical starting weights, layers, batches, etc. Finally tests and plots their accuracies.
:param use_bad_weights: bool
If True, initialize the weights of both networks to wildly inappropriate weights;
if False, use reasonable starting weights.
:param learning_rate: float
Learning rate used during gradient descent.
:param activation_fn: Callable
The function used for the output of each hidden layer. The network will use the same
activation function on every hidden layer and no activate function on the output layer.
e.g. Pass tf.nn.relu to use ReLU activations on your hidden layers.
:param training_batches: (default 50000)
Number of batches to train.
:param batches_per_sample: (default 500)
How many batches to train before sampling the validation accuracy.
"""
# Use identical starting weights for each network to eliminate differences in
# weight initialization as a cause for differences seen in training performance
#
# Note: The networks will use these weights to define the number of and shapes of
# its layers. The original batch normalization paper used 3 hidden layers
# with 100 nodes in each, followed by a 10 node output layer. These values
# build such a network, but feel free to experiment with different choices.
# However, the input size should always be 784 and the final output should be 10.
if use_bad_weights:
# These weights should be horrible because they have such a large standard deviation
weights = [np.random.normal(size=(784,100), scale=5.0).astype(np.float32),
np.random.normal(size=(100,100), scale=5.0).astype(np.float32),
np.random.normal(size=(100,100), scale=5.0).astype(np.float32),
np.random.normal(size=(100,10), scale=5.0).astype(np.float32)
]
else:
# These weights should be good because they have such a small standard deviation
weights = [np.random.normal(size=(784,100), scale=0.05).astype(np.float32),
np.random.normal(size=(100,100), scale=0.05).astype(np.float32),
np.random.normal(size=(100,100), scale=0.05).astype(np.float32),
np.random.normal(size=(100,10), scale=0.05).astype(np.float32)
]
# Just to make sure the TensorFlow's default graph is empty before we start another
# test, because we don't bother using different graphs or scoping and naming
# elements carefully in this sample code.
tf.reset_default_graph()
# build two versions of same network, 1 without and 1 with batch normalization
nn = NeuralNet(weights, activation_fn, False)
bn = NeuralNet(weights, activation_fn, True)
# train and test the two models
with tf.Session() as sess:
tf.global_variables_initializer().run()
nn.train(sess, learning_rate, training_batches, batches_per_sample)
bn.train(sess, learning_rate, training_batches, batches_per_sample)
nn.test(sess)
bn.test(sess)
# Display a graph of how validation accuracies changed during training
# so we can compare how the models trained and when they converged
plot_training_accuracies(nn, bn, batches_per_sample=batches_per_sample)
```
## Comparisons between identical networks, with and without batch normalization
The next series of cells train networks with various settings to show the differences with and without batch normalization. They are meant to clearly demonstrate the effects of batch normalization. We include a deeper discussion of batch normalization later in the notebook.
**The following creates two networks using a ReLU activation function, a learning rate of 0.01, and reasonable starting weights.**
```
train_and_test(False, 0.01, tf.nn.relu)
```
As expected, both networks train well and eventually reach similar test accuracies. However, notice that the model with batch normalization converges slightly faster than the other network, reaching accuracies over 90% almost immediately and nearing its max acuracy in 10 or 15 thousand iterations. The other network takes about 3 thousand iterations to reach 90% and doesn't near its best accuracy until 30 thousand or more iterations.
If you look at the raw speed, you can see that without batch normalization we were computing over 1100 batches per second, whereas with batch normalization that goes down to just over 500. However, batch normalization allows us to perform fewer iterations and converge in less time over all. (We only trained for 50 thousand batches here so we could plot the comparison.)
**The following creates two networks with the same hyperparameters used in the previous example, but only trains for 2000 iterations.**
```
train_and_test(False, 0.01, tf.nn.relu, 2000, 50)
```
As you can see, using batch normalization produces a model with over 95% accuracy in only 2000 batches, and it was above 90% at somewhere around 500 batches. Without batch normalization, the model takes 1750 iterations just to hit 80% – the network with batch normalization hits that mark after around 200 iterations! (Note: if you run the code yourself, you'll see slightly different results each time because the starting weights - while the same for each model - are different for each run.)
In the above example, you should also notice that the networks trained fewer batches per second then what you saw in the previous example. That's because much of the time we're tracking is actually spent periodically performing inference to collect data for the plots. In this example we perform that inference every 50 batches instead of every 500, so generating the plot for this example requires 10 times the overhead for the same 2000 iterations.
**The following creates two networks using a sigmoid activation function, a learning rate of 0.01, and reasonable starting weights.**
```
train_and_test(False, 0.01, tf.nn.sigmoid)
```
With the number of layers we're using and this small learning rate, using a sigmoid activation function takes a long time to start learning. It eventually starts making progress, but it took over 45 thousand batches just to get over 80% accuracy. Using batch normalization gets to 90% in around one thousand batches.
**The following creates two networks using a ReLU activation function, a learning rate of 1, and reasonable starting weights.**
```
train_and_test(False, 1, tf.nn.relu)
```
Now we're using ReLUs again, but with a larger learning rate. The plot shows how training started out pretty normally, with the network with batch normalization starting out faster than the other. But the higher learning rate bounces the accuracy around a bit more, and at some point the accuracy in the network without batch normalization just completely crashes. It's likely that too many ReLUs died off at this point because of the high learning rate.
The next cell shows the same test again. The network with batch normalization performs the same way, and the other suffers from the same problem again, but it manages to train longer before it happens.
```
train_and_test(False, 1, tf.nn.relu)
```
In both of the previous examples, the network with batch normalization manages to gets over 98% accuracy, and get near that result almost immediately. The higher learning rate allows the network to train extremely fast.
**The following creates two networks using a sigmoid activation function, a learning rate of 1, and reasonable starting weights.**
```
train_and_test(False, 1, tf.nn.sigmoid)
```
In this example, we switched to a sigmoid activation function. It appears to hande the higher learning rate well, with both networks achieving high accuracy.
The cell below shows a similar pair of networks trained for only 2000 iterations.
```
train_and_test(False, 1, tf.nn.sigmoid, 2000, 50)
```
As you can see, even though these parameters work well for both networks, the one with batch normalization gets over 90% in 400 or so batches, whereas the other takes over 1700. When training larger networks, these sorts of differences become more pronounced.
**The following creates two networks using a ReLU activation function, a learning rate of 2, and reasonable starting weights.**
```
train_and_test(False, 2, tf.nn.relu)
```
With this very large learning rate, the network with batch normalization trains fine and almost immediately manages 98% accuracy. However, the network without normalization doesn't learn at all.
**The following creates two networks using a sigmoid activation function, a learning rate of 2, and reasonable starting weights.**
```
train_and_test(False, 2, tf.nn.sigmoid)
```
Once again, using a sigmoid activation function with the larger learning rate works well both with and without batch normalization.
However, look at the plot below where we train models with the same parameters but only 2000 iterations. As usual, batch normalization lets it train faster.
```
train_and_test(False, 2, tf.nn.sigmoid, 2000, 50)
```
In the rest of the examples, we use really bad starting weights. That is, normally we would use very small values close to zero. However, in these examples we choose random values with a standard deviation of 5. If you were really training a neural network, you would **not** want to do this. But these examples demonstrate how batch normalization makes your network much more resilient.
**The following creates two networks using a ReLU activation function, a learning rate of 0.01, and bad starting weights.**
```
train_and_test(True, 0.01, tf.nn.relu)
```
As the plot shows, without batch normalization the network never learns anything at all. But with batch normalization, it actually learns pretty well and gets to almost 80% accuracy. The starting weights obviously hurt the network, but you can see how well batch normalization does in overcoming them.
**The following creates two networks using a sigmoid activation function, a learning rate of 0.01, and bad starting weights.**
```
train_and_test(True, 0.01, tf.nn.sigmoid)
```
Using a sigmoid activation function works better than the ReLU in the previous example, but without batch normalization it would take a tremendously long time to train the network, if it ever trained at all.
**The following creates two networks using a ReLU activation function, a learning rate of 1, and bad starting weights.**<a id="successful_example_lr_1"></a>
```
train_and_test(True, 1, tf.nn.relu)
```
The higher learning rate used here allows the network with batch normalization to surpass 90% in about 30 thousand batches. The network without it never gets anywhere.
**The following creates two networks using a sigmoid activation function, a learning rate of 1, and bad starting weights.**
```
train_and_test(True, 1, tf.nn.sigmoid)
```
Using sigmoid works better than ReLUs for this higher learning rate. However, you can see that without batch normalization, the network takes a long time tro train, bounces around a lot, and spends a long time stuck at 90%. The network with batch normalization trains much more quickly, seems to be more stable, and achieves a higher accuracy.
**The following creates two networks using a ReLU activation function, a learning rate of 2, and bad starting weights.**<a id="successful_example_lr_2"></a>
```
train_and_test(True, 2, tf.nn.relu)
```
We've already seen that ReLUs do not do as well as sigmoids with higher learning rates, and here we are using an extremely high rate. As expected, without batch normalization the network doesn't learn at all. But with batch normalization, it eventually achieves 90% accuracy. Notice, though, how its accuracy bounces around wildly during training - that's because the learning rate is really much too high, so the fact that this worked at all is a bit of luck.
**The following creates two networks using a sigmoid activation function, a learning rate of 2, and bad starting weights.**
```
train_and_test(True, 2, tf.nn.sigmoid)
```
In this case, the network with batch normalization trained faster and reached a higher accuracy. Meanwhile, the high learning rate makes the network without normalization bounce around erratically and have trouble getting past 90%.
### Full Disclosure: Batch Normalization Doesn't Fix Everything
Batch normalization isn't magic and it doesn't work every time. Weights are still randomly initialized and batches are chosen at random during training, so you never know exactly how training will go. Even for these tests, where we use the same initial weights for both networks, we still get _different_ weights each time we run.
This section includes two examples that show runs when batch normalization did not help at all.
**The following creates two networks using a ReLU activation function, a learning rate of 1, and bad starting weights.**
```
train_and_test(True, 1, tf.nn.relu)
```
When we used these same parameters [earlier](#successful_example_lr_1), we saw the network with batch normalization reach 92% validation accuracy. This time we used different starting weights, initialized using the same standard deviation as before, and the network doesn't learn at all. (Remember, an accuracy around 10% is what the network gets if it just guesses the same value all the time.)
**The following creates two networks using a ReLU activation function, a learning rate of 2, and bad starting weights.**
```
train_and_test(True, 2, tf.nn.relu)
```
When we trained with these parameters and batch normalization [earlier](#successful_example_lr_2), we reached 90% validation accuracy. However, this time the network _almost_ starts to make some progress in the beginning, but it quickly breaks down and stops learning.
**Note:** Both of the above examples use *extremely* bad starting weights, along with learning rates that are too high. While we've shown batch normalization _can_ overcome bad values, we don't mean to encourage actually using them. The examples in this notebook are meant to show that batch normalization can help your networks train better. But these last two examples should remind you that you still want to try to use good network design choices and reasonable starting weights. It should also remind you that the results of each attempt to train a network are a bit random, even when using otherwise identical architectures.
# Batch Normalization: A Detailed Look<a id='implementation_2'></a>
The layer created by `tf.layers.batch_normalization` handles all the details of implementing batch normalization. Many students will be fine just using that and won't care about what's happening at the lower levels. However, some students may want to explore the details, so here is a short explanation of what's really happening, starting with the equations you're likely to come across if you ever read about batch normalization.
In order to normalize the values, we first need to find the average value for the batch. If you look at the code, you can see that this is not the average value of the batch _inputs_, but the average value coming _out_ of any particular layer before we pass it through its non-linear activation function and then feed it as an input to the _next_ layer.
We represent the average as $\mu_B$, which is simply the sum of all of the values $x_i$ divided by the number of values, $m$
$$
\mu_B \leftarrow \frac{1}{m}\sum_{i=1}^m x_i
$$
We then need to calculate the variance, or mean squared deviation, represented as $\sigma_{B}^{2}$. If you aren't familiar with statistics, that simply means for each value $x_i$, we subtract the average value (calculated earlier as $\mu_B$), which gives us what's called the "deviation" for that value. We square the result to get the squared deviation. Sum up the results of doing that for each of the values, then divide by the number of values, again $m$, to get the average, or mean, squared deviation.
$$
\sigma_{B}^{2} \leftarrow \frac{1}{m}\sum_{i=1}^m (x_i - \mu_B)^2
$$
Once we have the mean and variance, we can use them to normalize the values with the following equation. For each value, it subtracts the mean and divides by the (almost) standard deviation. (You've probably heard of standard deviation many times, but if you have not studied statistics you might not know that the standard deviation is actually the square root of the mean squared deviation.)
$$
\hat{x_i} \leftarrow \frac{x_i - \mu_B}{\sqrt{\sigma_{B}^{2} + \epsilon}}
$$
Above, we said "(almost) standard deviation". That's because the real standard deviation for the batch is calculated by $\sqrt{\sigma_{B}^{2}}$, but the above formula adds the term epsilon, $\epsilon$, before taking the square root. The epsilon can be any small, positive constant - in our code we use the value `0.001`. It is there partially to make sure we don't try to divide by zero, but it also acts to increase the variance slightly for each batch.
Why increase the variance? Statistically, this makes sense because even though we are normalizing one batch at a time, we are also trying to estimate the population distribution – the total training set, which itself an estimate of the larger population of inputs your network wants to handle. The variance of a population is higher than the variance for any sample taken from that population, so increasing the variance a little bit for each batch helps take that into account.
At this point, we have a normalized value, represented as $\hat{x_i}$. But rather than use it directly, we multiply it by a gamma value, $\gamma$, and then add a beta value, $\beta$. Both $\gamma$ and $\beta$ are learnable parameters of the network and serve to scale and shift the normalized value, respectively. Because they are learnable just like weights, they give your network some extra knobs to tweak during training to help it learn the function it is trying to approximate.
$$
y_i \leftarrow \gamma \hat{x_i} + \beta
$$
We now have the final batch-normalized output of our layer, which we would then pass to a non-linear activation function like sigmoid, tanh, ReLU, Leaky ReLU, etc. In the original batch normalization paper (linked in the beginning of this notebook), they mention that there might be cases when you'd want to perform the batch normalization _after_ the non-linearity instead of before, but it is difficult to find any uses like that in practice.
In `NeuralNet`'s implementation of `fully_connected`, all of this math is hidden inside the following line, where `linear_output` serves as the $x_i$ from the equations:
```python
batch_normalized_output = tf.layers.batch_normalization(linear_output, training=self.is_training)
```
The next section shows you how to implement the math directly.
### Batch normalization without the `tf.layers` package
Our implementation of batch normalization in `NeuralNet` uses the high-level abstraction [tf.layers.batch_normalization](https://www.tensorflow.org/api_docs/python/tf/layers/batch_normalization), found in TensorFlow's [`tf.layers`](https://www.tensorflow.org/api_docs/python/tf/layers) package.
However, if you would like to implement batch normalization at a lower level, the following code shows you how.
It uses [tf.nn.batch_normalization](https://www.tensorflow.org/api_docs/python/tf/nn/batch_normalization) from TensorFlow's [neural net (nn)](https://www.tensorflow.org/api_docs/python/tf/nn) package.
**1)** You can replace the `fully_connected` function in the `NeuralNet` class with the below code and everything in `NeuralNet` will still work like it did before.
```
def fully_connected(self, layer_in, initial_weights, activation_fn=None):
"""
Creates a standard, fully connected layer. Its number of inputs and outputs will be
defined by the shape of `initial_weights`, and its starting weight values will be
taken directly from that same parameter. If `self.use_batch_norm` is True, this
layer will include batch normalization, otherwise it will not.
:param layer_in: Tensor
The Tensor that feeds into this layer. It's either the input to the network or the output
of a previous layer.
:param initial_weights: NumPy array or Tensor
Initial values for this layer's weights. The shape defines the number of nodes in the layer.
e.g. Passing in 3 matrix of shape (784, 256) would create a layer with 784 inputs and 256
outputs.
:param activation_fn: Callable or None (default None)
The non-linearity used for the output of the layer. If None, this layer will not include
batch normalization, regardless of the value of `self.use_batch_norm`.
e.g. Pass tf.nn.relu to use ReLU activations on your hidden layers.
"""
if self.use_batch_norm and activation_fn:
# Batch normalization uses weights as usual, but does NOT add a bias term. This is because
# its calculations include gamma and beta variables that make the bias term unnecessary.
weights = tf.Variable(initial_weights)
linear_output = tf.matmul(layer_in, weights)
num_out_nodes = initial_weights.shape[-1]
# Batch normalization adds additional trainable variables:
# gamma (for scaling) and beta (for shifting).
gamma = tf.Variable(tf.ones([num_out_nodes]))
beta = tf.Variable(tf.zeros([num_out_nodes]))
# These variables will store the mean and variance for this layer over the entire training set,
# which we assume represents the general population distribution.
# By setting `trainable=False`, we tell TensorFlow not to modify these variables during
# back propagation. Instead, we will assign values to these variables ourselves.
pop_mean = tf.Variable(tf.zeros([num_out_nodes]), trainable=False)
pop_variance = tf.Variable(tf.ones([num_out_nodes]), trainable=False)
# Batch normalization requires a small constant epsilon, used to ensure we don't divide by zero.
# This is the default value TensorFlow uses.
epsilon = 1e-3
def batch_norm_training():
# Calculate the mean and variance for the data coming out of this layer's linear-combination step.
# The [0] defines an array of axes to calculate over.
batch_mean, batch_variance = tf.nn.moments(linear_output, [0])
# Calculate a moving average of the training data's mean and variance while training.
# These will be used during inference.
# Decay should be some number less than 1. tf.layers.batch_normalization uses the parameter
# "momentum" to accomplish this and defaults it to 0.99
decay = 0.99
train_mean = tf.assign(pop_mean, pop_mean * decay + batch_mean * (1 - decay))
train_variance = tf.assign(pop_variance, pop_variance * decay + batch_variance * (1 - decay))
# The 'tf.control_dependencies' context tells TensorFlow it must calculate 'train_mean'
# and 'train_variance' before it calculates the 'tf.nn.batch_normalization' layer.
# This is necessary because the those two operations are not actually in the graph
# connecting the linear_output and batch_normalization layers,
# so TensorFlow would otherwise just skip them.
with tf.control_dependencies([train_mean, train_variance]):
return tf.nn.batch_normalization(linear_output, batch_mean, batch_variance, beta, gamma, epsilon)
def batch_norm_inference():
# During inference, use the our estimated population mean and variance to normalize the layer
return tf.nn.batch_normalization(linear_output, pop_mean, pop_variance, beta, gamma, epsilon)
# Use `tf.cond` as a sort of if-check. When self.is_training is True, TensorFlow will execute
# the operation returned from `batch_norm_training`; otherwise it will execute the graph
# operation returned from `batch_norm_inference`.
batch_normalized_output = tf.cond(self.is_training, batch_norm_training, batch_norm_inference)
# Pass the batch-normalized layer output through the activation function.
# The literature states there may be cases where you want to perform the batch normalization *after*
# the activation function, but it is difficult to find any uses of that in practice.
return activation_fn(batch_normalized_output)
else:
# When not using batch normalization, create a standard layer that multiplies
# the inputs and weights, adds a bias, and optionally passes the result
# through an activation function.
weights = tf.Variable(initial_weights)
biases = tf.Variable(tf.zeros([initial_weights.shape[-1]]))
linear_output = tf.add(tf.matmul(layer_in, weights), biases)
return linear_output if not activation_fn else activation_fn(linear_output)
```
This version of `fully_connected` is much longer than the original, but once again has extensive comments to help you understand it. Here are some important points:
1. It explicitly creates variables to store gamma, beta, and the population mean and variance. These were all handled for us in the previous version of the function.
2. It initializes gamma to one and beta to zero, so they start out having no effect in this calculation: $y_i \leftarrow \gamma \hat{x_i} + \beta$. However, during training the network learns the best values for these variables using back propagation, just like networks normally do with weights.
3. Unlike gamma and beta, the variables for population mean and variance are marked as untrainable. That tells TensorFlow not to modify them during back propagation. Instead, the lines that call `tf.assign` are used to update these variables directly.
4. TensorFlow won't automatically run the `tf.assign` operations during training because it only evaluates operations that are required based on the connections it finds in the graph. To get around that, we add this line: `with tf.control_dependencies([train_mean, train_variance]):` before we run the normalization operation. This tells TensorFlow it needs to run those operations before running anything inside the `with` block.
5. The actual normalization math is still mostly hidden from us, this time using [`tf.nn.batch_normalization`](https://www.tensorflow.org/api_docs/python/tf/nn/batch_normalization).
5. `tf.nn.batch_normalization` does not have a `training` parameter like `tf.layers.batch_normalization` did. However, we still need to handle training and inference differently, so we run different code in each case using the [`tf.cond`](https://www.tensorflow.org/api_docs/python/tf/cond) operation.
6. We use the [`tf.nn.moments`](https://www.tensorflow.org/api_docs/python/tf/nn/moments) function to calculate the batch mean and variance.
**2)** The current version of the `train` function in `NeuralNet` will work fine with this new version of `fully_connected`. However, it uses these lines to ensure population statistics are updated when using batch normalization:
```python
if self.use_batch_norm:
with tf.control_dependencies(tf.get_collection(tf.GraphKeys.UPDATE_OPS)):
train_step = tf.train.GradientDescentOptimizer(learning_rate).minimize(cross_entropy)
else:
train_step = tf.train.GradientDescentOptimizer(learning_rate).minimize(cross_entropy)
```
Our new version of `fully_connected` handles updating the population statistics directly. That means you can also simplify your code by replacing the above `if`/`else` condition with just this line:
```python
train_step = tf.train.GradientDescentOptimizer(learning_rate).minimize(cross_entropy)
```
**3)** And just in case you want to implement every detail from scratch, you can replace this line in `batch_norm_training`:
```python
return tf.nn.batch_normalization(linear_output, batch_mean, batch_variance, beta, gamma, epsilon)
```
with these lines:
```python
normalized_linear_output = (linear_output - batch_mean) / tf.sqrt(batch_variance + epsilon)
return gamma * normalized_linear_output + beta
```
And replace this line in `batch_norm_inference`:
```python
return tf.nn.batch_normalization(linear_output, pop_mean, pop_variance, beta, gamma, epsilon)
```
with these lines:
```python
normalized_linear_output = (linear_output - pop_mean) / tf.sqrt(pop_variance + epsilon)
return gamma * normalized_linear_output + beta
```
As you can see in each of the above substitutions, the two lines of replacement code simply implement the following two equations directly. The first line calculates the following equation, with `linear_output` representing $x_i$ and `normalized_linear_output` representing $\hat{x_i}$:
$$
\hat{x_i} \leftarrow \frac{x_i - \mu_B}{\sqrt{\sigma_{B}^{2} + \epsilon}}
$$
And the second line is a direct translation of the following equation:
$$
y_i \leftarrow \gamma \hat{x_i} + \beta
$$
We still use the `tf.nn.moments` operation to implement the other two equations from earlier – the ones that calculate the batch mean and variance used in the normalization step. If you really wanted to do everything from scratch, you could replace that line, too, but we'll leave that to you.
## Why the difference between training and inference?
In the original function that uses `tf.layers.batch_normalization`, we tell the layer whether or not the network is training by passing a value for its `training` parameter, like so:
```python
batch_normalized_output = tf.layers.batch_normalization(linear_output, training=self.is_training)
```
And that forces us to provide a value for `self.is_training` in our `feed_dict`, like we do in this example from `NeuralNet`'s `train` function:
```python
session.run(train_step, feed_dict={self.input_layer: batch_xs,
labels: batch_ys,
self.is_training: True})
```
If you looked at the [low level implementation](#low_level_code), you probably noticed that, just like with `tf.layers.batch_normalization`, we need to do slightly different things during training and inference. But why is that?
First, let's look at what happens when we don't. The following function is similar to `train_and_test` from earlier, but this time we are only testing one network and instead of plotting its accuracy, we perform 200 predictions on test inputs, 1 input at at time. We can use the `test_training_accuracy` parameter to test the network in training or inference modes (the equivalent of passing `True` or `False` to the `feed_dict` for `is_training`).
```
def batch_norm_test(test_training_accuracy):
"""
:param test_training_accuracy: bool
If True, perform inference with batch normalization using batch mean and variance;
if False, perform inference with batch normalization using estimated population mean and variance.
"""
weights = [np.random.normal(size=(784,100), scale=0.05).astype(np.float32),
np.random.normal(size=(100,100), scale=0.05).astype(np.float32),
np.random.normal(size=(100,100), scale=0.05).astype(np.float32),
np.random.normal(size=(100,10), scale=0.05).astype(np.float32)
]
tf.reset_default_graph()
# Train the model
bn = NeuralNet(weights, tf.nn.relu, True)
# First train the network
with tf.Session() as sess:
tf.global_variables_initializer().run()
bn.train(sess, 0.01, 2000, 2000)
bn.test(sess, test_training_accuracy=test_training_accuracy, include_individual_predictions=True)
```
In the following cell, we pass `True` for `test_training_accuracy`, which performs the same batch normalization that we normally perform **during training**.
```
batch_norm_test(True)
```
As you can see, the network guessed the same value every time! But why? Because during training, a network with batch normalization adjusts the values at each layer based on the mean and variance **of that batch**. The "batches" we are using for these predictions have a single input each time, so their values _are_ the means, and their variances will always be 0. That means the network will normalize the values at any layer to zero. (Review the equations from before to see why a value that is equal to the mean would always normalize to zero.) So we end up with the same result for every input we give the network, because its the value the network produces when it applies its learned weights to zeros at every layer.
**Note:** If you re-run that cell, you might get a different value from what we showed. That's because the specific weights the network learns will be different every time. But whatever value it is, it should be the same for all 200 predictions.
To overcome this problem, the network does not just normalize the batch at each layer. It also maintains an estimate of the mean and variance for the entire population. So when we perform inference, instead of letting it "normalize" all the values using their own means and variance, it uses the estimates of the population mean and variance that it calculated while training.
So in the following example, we pass `False` for `test_training_accuracy`, which tells the network that we it want to perform inference with the population statistics it calculates during training.
```
batch_norm_test(False)
```
As you can see, now that we're using the estimated population mean and variance, we get a 97% accuracy. That means it guessed correctly on 194 of the 200 samples – not too bad for something that trained in under 4 seconds. :)
# Considerations for other network types
This notebook demonstrates batch normalization in a standard neural network with fully connected layers. You can also use batch normalization in other types of networks, but there are some special considerations.
### ConvNets
Convolution layers consist of multiple feature maps. (Remember, the depth of a convolutional layer refers to its number of feature maps.) And the weights for each feature map are shared across all the inputs that feed into the layer. Because of these differences, batch normalizaing convolutional layers requires batch/population mean and variance per feature map rather than per node in the layer.
When using `tf.layers.batch_normalization`, be sure to pay attention to the order of your convolutionlal dimensions.
Specifically, you may want to set a different value for the `axis` parameter if your layers have their channels first instead of last.
In our low-level implementations, we used the following line to calculate the batch mean and variance:
```python
batch_mean, batch_variance = tf.nn.moments(linear_output, [0])
```
If we were dealing with a convolutional layer, we would calculate the mean and variance with a line like this instead:
```python
batch_mean, batch_variance = tf.nn.moments(conv_layer, [0,1,2], keep_dims=False)
```
The second parameter, `[0,1,2]`, tells TensorFlow to calculate the batch mean and variance over each feature map. (The three axes are the batch, height, and width.) And setting `keep_dims` to `False` tells `tf.nn.moments` not to return values with the same size as the inputs. Specifically, it ensures we get one mean/variance pair per feature map.
### RNNs
Batch normalization can work with recurrent neural networks, too, as shown in the 2016 paper [Recurrent Batch Normalization](https://arxiv.org/abs/1603.09025). It's a bit more work to implement, but basically involves calculating the means and variances per time step instead of per layer. You can find an example where someone extended `tf.nn.rnn_cell.RNNCell` to include batch normalization in [this GitHub repo](https://gist.github.com/spitis/27ab7d2a30bbaf5ef431b4a02194ac60).
|
github_jupyter
|
```
import logging
import os
import math
from dataclasses import dataclass, field
import copy # for deep copy
import torch
from torch import nn
from transformers import RobertaForMaskedLM, RobertaTokenizerFast, TextDataset, DataCollatorForLanguageModeling, Trainer
from transformers import TrainingArguments, HfArgumentParser
from transformers.modeling_longformer import LongformerSelfAttention
logger = logging.getLogger(__name__)
logging.basicConfig(level=logging.INFO)
class RobertaLongSelfAttention(LongformerSelfAttention):
def forward(
self,
hidden_states, attention_mask=None, is_index_masked=None, is_index_global_attn=None, is_global_attn=None
):
return super().forward(hidden_states, attention_mask=attention_mask)
class RobertaLongForMaskedLM(RobertaForMaskedLM):
def __init__(self, config):
super().__init__(config)
for i, layer in enumerate(self.roberta.encoder.layer):
# replace the `modeling_bert.BertSelfAttention` object with `LongformerSelfAttention`
layer.attention.self = RobertaLongSelfAttention(config, layer_id=i)
def create_long_model(save_model_to, attention_window, max_pos):
model = RobertaForMaskedLM.from_pretrained('roberta-base')
tokenizer = RobertaTokenizerFast.from_pretrained('roberta-base', model_max_length=max_pos)
config = model.config
# extend position embeddings
tokenizer.model_max_length = max_pos
tokenizer.init_kwargs['model_max_length'] = max_pos
current_max_pos, embed_size = model.roberta.embeddings.position_embeddings.weight.shape
max_pos += 2 # NOTE: RoBERTa has positions 0,1 reserved, so embedding size is max position + 2
config.max_position_embeddings = max_pos
assert max_pos > current_max_pos
# allocate a larger position embedding matrix
new_pos_embed = model.roberta.embeddings.position_embeddings.weight.new_empty(max_pos, embed_size)
# copy position embeddings over and over to initialize the new position embeddings
k = 2
step = current_max_pos - 2
while k < max_pos - 1:
new_pos_embed[k:(k + step)] = model.roberta.embeddings.position_embeddings.weight[2:]
k += step
model.roberta.embeddings.position_embeddings.weight.data = new_pos_embed
model.roberta.embeddings.position_ids.data = torch.tensor([i for i in range(max_pos)]).reshape(1, max_pos)
"""
model.roberta.embeddings.position_embeddings.weight.data = new_pos_embed # add after this line
model.roberta.embeddings.position_embeddings.num_embeddings = len(new_pos_embed.data)
# first, check that model.roberta.embeddings.position_embeddings.weight.data.shape is correct — has to be 4096 (default) of your desired length
model.roberta.embeddings.position_ids = torch.arange(0, model.roberta.embeddings.position_embeddings.num_embeddings)[None]
"""
# replace the `modeling_bert.BertSelfAttention` object with `LongformerSelfAttention`
config.attention_window = [attention_window] * config.num_hidden_layers
for i, layer in enumerate(model.roberta.encoder.layer):
longformer_self_attn = LongformerSelfAttention(config, layer_id=i)
longformer_self_attn.query = copy.deepcopy(layer.attention.self.query)
longformer_self_attn.key = copy.deepcopy(layer.attention.self.key)
longformer_self_attn.value = copy.deepcopy(layer.attention.self.value)
longformer_self_attn.query_global = copy.deepcopy(layer.attention.self.query)
longformer_self_attn.key_global = copy.deepcopy(layer.attention.self.key)
longformer_self_attn.value_global = copy.deepcopy(layer.attention.self.value)
"""
longformer_self_attn = LongformerSelfAttention(config, layer_id=i)
longformer_self_attn.query = layer.attention.self.query
longformer_self_attn.key = layer.attention.self.key
longformer_self_attn.value = layer.attention.self.value
longformer_self_attn.query_global = layer.attention.self.query
longformer_self_attn.key_global = layer.attention.self.key
longformer_self_attn.value_global = layer.attention.self.value
"""
layer.attention.self = longformer_self_attn
logger.info(f'saving model to {save_model_to}')
model.save_pretrained(save_model_to)
tokenizer.save_pretrained(save_model_to)
return model, tokenizer
def copy_proj_layers(model):
for i, layer in enumerate(model.roberta.encoder.layer):
layer.attention.self.query_global = layer.attention.self.query
layer.attention.self.key_global = layer.attention.self.key
layer.attention.self.value_global = layer.attention.self.value
return model
def pretrain_and_evaluate(args, model, tokenizer, eval_only, model_path):
val_dataset = TextDataset(tokenizer=tokenizer,
file_path=args.val_datapath,
block_size=tokenizer.max_len)
if eval_only:
train_dataset = val_dataset
else:
logger.info(f'Loading and tokenizing training data is usually slow: {args.train_datapath}')
train_dataset = TextDataset(tokenizer=tokenizer,
file_path=args.train_datapath,
block_size=tokenizer.max_len)
data_collator = DataCollatorForLanguageModeling(tokenizer=tokenizer, mlm=True, mlm_probability=0.15)
trainer = Trainer(model=model, args=args, data_collator=data_collator,
train_dataset=train_dataset, eval_dataset=val_dataset, prediction_loss_only=True)
eval_loss = trainer.evaluate()
eval_loss = eval_loss['eval_loss']
logger.info(f'Initial eval bpc: {eval_loss/math.log(2)}')
if not eval_only:
trainer.train(model_path=model_path)
trainer.save_model()
eval_loss = trainer.evaluate()
eval_loss = eval_loss['eval_loss']
logger.info(f'Eval bpc after pretraining: {eval_loss/math.log(2)}')
@dataclass
class ModelArgs:
attention_window: int = field(default=512, metadata={"help": "Size of attention window"})
max_pos: int = field(default=4096, metadata={"help": "Maximum position"})
parser = HfArgumentParser((TrainingArguments, ModelArgs,))
training_args, model_args = parser.parse_args_into_dataclasses(look_for_args_file=False, args=[
'--output_dir', 'tmp',
'--warmup_steps', '500',
'--learning_rate', '0.00003',
'--weight_decay', '0.01',
'--adam_epsilon', '1e-6',
'--max_steps', '3000',
'--logging_steps', '500',
'--save_steps', '500',
'--max_grad_norm', '5.0',
'--per_gpu_eval_batch_size', '8',
'--per_gpu_train_batch_size', '2', # 32GB gpu with fp32
'--gradient_accumulation_steps', '32',
'--evaluate_during_training',
'--do_train',
'--do_eval',
])
training_args.val_datapath = '/workspace/data/wikitext-103-raw/wiki.valid.raw'
training_args.train_datapath = '/workspace/data/wikitext-103-raw/wiki.train.raw'
# Choose GPU
import os
os.environ["CUDA_VISIBLE_DEVICES"] = "0"
roberta_base = RobertaForMaskedLM.from_pretrained('roberta-base')
roberta_base_tokenizer = RobertaTokenizerFast.from_pretrained('roberta-base')
logger.info('Evaluating roberta-base (seqlen: 512) for refernece ...')
pretrain_and_evaluate(training_args, roberta_base, roberta_base_tokenizer, eval_only=True, model_path=None)
model_path = f'{training_args.output_dir}/roberta-base-{model_args.max_pos}'
if not os.path.exists(model_path):
os.makedirs(model_path)
logger.info(f'Converting roberta-base into roberta-base-{model_args.max_pos}')
model, tokenizer = create_long_model(
save_model_to=model_path, attention_window=model_args.attention_window, max_pos=model_args.max_pos)
"""
Self =
(query): Linear(in_features=768, out_features=768, bias=True)
(key): Linear(in_features=768, out_features=768, bias=True)
(value): Linear(in_features=768, out_features=768, bias=True)
(query_global): Linear(in_features=768, out_features=768, bias=True)
(key_global): Linear(in_features=768, out_features=768, bias=True)
(value_global): Linear(in_features=768, out_features=768, bias=True)
"""
logger.info(f'Loading the model from {model_path}')
tokenizer = RobertaTokenizerFast.from_pretrained(model_path)
model = RobertaLongForMaskedLM.from_pretrained(model_path)
logger.info(f'Pretraining roberta-base-{model_args.max_pos} ... ')
training_args.max_steps = 3 ## <<<<<<<<<<<<<<<<<<<<<<<< REMOVE THIS <<<<<<<<<<<<<<<<<<<<<<<<
%magic
pretrain_and_evaluate(training_args, model, tokenizer, eval_only=False, model_path=training_args.output_dir)
logger.info(f'Copying local projection layers into global projection layers ... ')
model = copy_proj_layers(model)
logger.info(f'Saving model to {model_path}')
model.save_pretrained(model_path)
logger.info(f'Loading the model from {model_path}')
tokenizer = RobertaTokenizerFast.from_pretrained(model_path)
model = RobertaLongForMaskedLM.from_pretrained(model_path)
import transformers
transformers.__version__
model.roberta.embeddings
model.roberta.embeddings.position_embeddings
model.roberta.embeddings.position_embeddings.num_embeddings
model.roberta.embeddings.position_embeddings.num_embeddings
torch.cop
```
|
github_jupyter
|
# Numerical norm bounds for quadrotor
For a quadrotor system with state $x = \begin{bmatrix}p_x & p_z & \phi & v_x & v_z & \dot{\phi} \end{bmatrix}^T$ we have
\begin{equation}
\dot{x} = \begin{bmatrix}
v_x \cos\phi - v_z\sin\phi \\
v_x \sin\phi + v_z\cos\phi \\
\dot{\phi} \\
v_z\dot{\phi} - g\sin{\phi} \\
-v_x\dot{\phi} - g\cos{\phi} + g \\
0
\end{bmatrix}.
\end{equation}
Evaluating the corresponding Jacobian at 0 yields:
\begin{equation}
\nabla f(0)x = \begin{bmatrix} v_x & v_z & \dot{\phi} & -g\phi & 0 & 0 \end{bmatrix}^T
\end{equation}
We want to find an NLDI of the form
\begin{equation}
\dot{x} = \nabla f(0) x + I p, \;\; \|p\| \leq \|Cx\|
\end{equation}
To find $C$, we determine an entry-wise norm bound. That is, for $i=1,\ldots,6$, we want to find $C_i$ such that for all $x$ such that $x_{\text{min}} \leq x \leq x_{\text{max}}$:
\begin{equation}
(\nabla f_i(0)x - \dot{x}_i)^2 \leq x^T C_i x
\end{equation}
and then write
\begin{equation}
\|\dot{x} - \nabla f(0)x\|_2 \leq \|\begin{bmatrix} C_1^{1/2} \\ C_2^{1/2} \\ C_3^{1/2} \\ C_4^{1/2} \\ C_5^{1/2} \\ C_6^{1/2} \end{bmatrix} x \|
\end{equation}
```
import numpy as np
import cvxpy as cp
import scipy.linalg as sla
g = 9.81
```
## Define max and min values
```
# State is: x = [px, pz, phi, vx, vz, phidot]^T
x_max = np.array([1.1, 1.1, 0.06, 0.5, 1.0, 0.8])
x_min = np.array([-1.1, -1.1, -0.06, -0.5, -1.0, -0.8])
px_max, pz_max, phi_max, vx_max, vz_max, phidot_max = x_max
px_min, pz_min, phi_min, vx_min, vz_min, phidot_min = x_min
n = 6
px_idx, pz_idx, phi_idx, vx_idx, vz_idx, phidot_idx = range(n)
```
## Find element-wise bounds
### $f_1$
```
gridnum = 50
vx = np.linspace(vx_min, vx_max, gridnum)
vz = np.linspace(vz_min, vz_max, gridnum)
phi = np.linspace(phi_min, phi_max, gridnum)
Vx, Vz, Phi = np.meshgrid(vx, vz, phi)
v1 = np.ravel(( Vx - (Vx*np.cos(Phi) - Vz*np.sin(Phi)) )**2)
U1 = np.array([np.ravel(Vx*Vx),
np.ravel(Vz*Vz),
np.ravel(Phi*Phi),
2*np.ravel(Vx*Vz),
2*np.ravel(Vx*Phi),
2*np.ravel(Vz*Phi)]).T
c1 = cp.Variable(6)
cp.Problem(cp.Minimize(cp.max(U1@c1 - v1)), [U1@c1 >= v1, c1[:3]>=0]).solve(verbose=True, solver=cp.MOSEK)
c1 = c1.value
c1
C1 = np.zeros((n,n))
C1[vx_idx, vx_idx] = c1[0]/2
C1[vz_idx, vz_idx] = c1[1]/2
C1[phi_idx, phi_idx] = c1[2]/2
C1[vx_idx, vz_idx] = c1[3]
C1[vx_idx, phi_idx] = c1[4]
C1[vz_idx, phi_idx] = c1[5]
C1 += C1.T
gam1 = np.real(sla.sqrtm(C1))
gam1
```
### $f_2$
```
gridnum = 50
vx = np.linspace(vx_min, vx_max, gridnum)
vz = np.linspace(vz_min, vz_max, gridnum)
phi = np.linspace(phi_min, phi_max, gridnum)
Vx, Vz, Phi = np.meshgrid(vx, vz, phi)
v2 = np.ravel(( Vz - (Vx*np.sin(Phi) + Vz*np.cos(Phi)) )**2)
U2 = np.array([np.ravel(Vx*Vx),
np.ravel(Vz*Vz),
np.ravel(Phi*Phi),
2*np.ravel(Vx*Vz),
2*np.ravel(Vx*Phi),
2*np.ravel(Vz*Phi)]).T
c2 = cp.Variable(6)
cp.Problem(cp.Minimize(cp.max(U2@c2 - v2)), [U2@c2 >= v2, c2[:3]>=0]).solve(verbose=True, solver=cp.MOSEK)
c2 = c2.value
c2
C2 = np.zeros((n,n))
C2[vx_idx, vx_idx] = c2[0]/2
C2[vz_idx, vz_idx] = c2[1]/2
C2[phi_idx, phi_idx] = c2[2]/2
C2[vx_idx, vz_idx] = c2[3]
C2[vx_idx, phi_idx] = c2[4]
C2[vz_idx, phi_idx] = c2[5]
C2 += C2.T
gam2 = np.real(sla.sqrtm(C2))
gam2
```
### $f_3$
No error -- linearization is the same as original
### $f_4$
```
gridnum = 50
vz = np.linspace(vz_min, vz_max, gridnum)
phi = np.linspace(phi_min, phi_max, gridnum)
phidot = np.linspace(phidot_min, phidot_max, gridnum)
Vz, Phi, Phidot = np.meshgrid(vz, phi, phidot)
v4 = np.ravel(( -g*Phi - (Vz*Phidot - g*np.sin(Phi)) )**2)
U4 = np.array([np.ravel(Vz*Vz),
np.ravel(Phi*Phi),
np.ravel(Phidot*Phidot),
2*np.ravel(Vz*Phi),
2*np.ravel(Vz*Phidot),
2*np.ravel(Phi*Phidot)]).T
c4 = cp.Variable(6)
cp.Problem(cp.Minimize(cp.max(U4@c4 - v4)), [U4@c4 >= v4, c4[:3]>=0]).solve(verbose=True, solver=cp.MOSEK)
c4 = c4.value
c4
C4 = np.zeros((n,n))
C4[vz_idx, vz_idx] = c4[0]/2
C4[phi_idx, phi_idx] = c4[1]/2
C4[phidot_idx, phidot_idx] = c4[2]/2
C4[vz_idx, phi_idx] = c4[3]
C4[vz_idx, phidot_idx] = c4[4]
C4[phi_idx, phidot_idx] = c4[5]
C4 += C4.T
gam4 = np.real(sla.sqrtm(C4))
gam4
```
### $f_5$
```
gridnum = 50
vx = np.linspace(vx_min, vx_max, gridnum)
phi = np.linspace(phi_min, phi_max, gridnum)
phidot = np.linspace(phidot_min, phidot_max, gridnum)
Vx, Phi, Phidot = np.meshgrid(vx, phi, phidot)
v5 = np.ravel(( 0 - (-Vx*Phidot - g*np.cos(Phi) + g) )**2)
U5 = np.array([np.ravel(Vx*Vx),
np.ravel(Phi*Phi),
np.ravel(Phidot*Phidot),
2*np.ravel(Vx*Phi),
2*np.ravel(Vx*Phidot),
2*np.ravel(Phi*Phidot)]).T
c5 = cp.Variable(6)
cp.Problem(cp.Minimize(cp.max(U5@c5 - v5)), [U5@c5 >= v5, c5[:3]>=0]).solve(verbose=True, solver=cp.MOSEK)
c5 = c5.value
c5
C5 = np.zeros((n,n))
C5[vx_idx, vx_idx] = c5[0]/2
C5[phi_idx, phi_idx] = c5[1]/2
C5[phidot_idx, phidot_idx] = c5[2]/2
C5[vx_idx, phi_idx] = c5[3]
C5[vx_idx, phidot_idx] = c5[4]
C5[phi_idx, phidot_idx] = c5[5]
C5 += C5.T
np.linalg.eig(C5)[0]
gam5 = np.real(sla.sqrtm(C5))
gam5
```
### $f_6$
No error -- linearization is the same as original
## Final system
```
from linearize_dynamics import *
A = quadrotor_jacobian(np.zeros(n))
G = np.eye(n)
C = np.vstack([gam1, gam2, gam4, gam5])
```
### Check correctness
```
prop = np.random.random((1000000, n))
rand_xs = x_max*prop + x_min*(1-prop)
fx = xdot_uncontrolled(torch.Tensor(rand_xs))
# print(np.linalg.norm((fx - [email protected])@np.linalg.inv(G).T, axis=1) <= np.linalg.norm([email protected], axis=1))
print((np.linalg.norm((fx - [email protected])@np.linalg.inv(G).T, axis=1) <= np.linalg.norm([email protected], axis=1)).all())
ratio = np.linalg.norm([email protected], axis=1)/np.linalg.norm((fx - [email protected])@np.linalg.inv(G).T, axis=1)
print(ratio.max())
print(ratio.mean())
print(np.median(ratio))
```
### Save
```
np.save('A.npy', A)
np.save('G.npy', G)
np.save('C.npy', C)
```
## Check if robust LQR solves
```
import scipy.linalg as la
mass = 1
moment_arm = 0.01
inertia_roll = 15.67e-3
B = np.array([
[0, 0],
[0, 0],
[0, 0],
[0, 0],
[1/mass, 1/mass],
[moment_arm/inertia_roll, -moment_arm/inertia_roll]
])
m = B.shape[1]
D = np.zeros((C.shape[0], m))
Q = np.random.randn(n, n)
Q = Q.T @ Q
# Q = np.eye(n)
R = np.random.randn(m, m)
R = R.T @ R
# R = np.eye(m)
alpha = 0.0001
n, m = B.shape
wq = C.shape[0]
S = cp.Variable((n, n), symmetric=True)
Y = cp.Variable((m, n))
mu = cp.Variable()
R_sqrt = la.sqrtm(R)
f = cp.trace(S @ Q) + cp.matrix_frac(Y.T @ R_sqrt, S)
cons_mat = cp.bmat((
(A @ S + S @ A.T + cp.multiply(mu, G @ G.T) + B @ Y + Y.T @ B.T + alpha * S, S @ C.T + Y.T @ D.T),
(C @ S + D @ Y, -cp.multiply(mu, np.eye(wq)))
))
cons = [S >> 0, mu >= 1e-2] + [cons_mat << 0]
cp.Problem(cp.Minimize(f), cons).solve(solver=cp.MOSEK, verbose=True)
K = np.linalg.solve(S.value, Y.value.T).T
```
|
github_jupyter
|
# Expectiminimax
Der Vollständigkeits halber der ganze Expectiminimax Algorithmus. <br>
Während 1-ply, 2-ply und 3-ply nur den ersten, die ersten beiden, bzw. ersten drei Schritte von Expectiminimax ausgeführt haben, kann man alle mit dem Expectiminmax Algorithmus zusammenfassen. Das erlaubt einem eine saubere Notation und kann (mit einem ausreichend starken Rechner und genug Geduld) eventuell noch tiefer suchen!
```
from Player import ValuePlayer
class ExpectiminimaxValuePlayer(ValuePlayer):
# Konstruktor braucht einen Parameter für die maximal Suchtiefe
# 0 = 1-ply, 1= 2-ply, 2 = 3-ply, usw.
def __init__(self, player, valuefunction, max_depth):
ValuePlayer.__init__(self, player, valuefunction)
self.max_depth = max_depth
def get_action(self, actions, game):
# Spielstatus speichern
old_state = game.get_state()
# Variablen initialisieren
best_value = -1
best_action = None
# Alle Züge durchsuchen
for a in actions:
# Zug ausführen
game.execute_moves(a, self.player)
# Spielstatus bewerten
value = self.expectiminimax(game, 0)
# Besten merken
if value > best_value:
best_value = value
best_action = a
# Spiel zurücksetzen
game.reset_to_state(old_state)
return best_action
def expectiminimax(self, game, depth):
# Blatt in unserem Baum
if depth == self.max_depth:
return self.value(game, self.player)
else:
# Alle möglichen Würfe betrachten
all_rolls = [(a,b) for a in range(1,7) for b in range(a,7)]
value = 0
for roll in all_rolls:
# Wahrscheinlichkeiten von jedem Wurf
probability = 1/18 if roll[0] != roll[1] else 1/36
state = game.get_state()
# Min-Knoten
if depth % 2 == 0:
moves = game.get_moves(roll, game.get_opponent(self.player))
temp_val = 1
for move in moves:
game.execute_moves(move, game.get_opponent(self.player))
# Bewertet wird aber aus unserer Perspektive
v = self.expectiminimax(game, depth + 1)
if v < temp_val:
temp_val = v
# Max-Knoten
else:
moves = game.get_moves(roll, self.player)
temp_val = 0
for move in moves:
game.execute_moves(move, self.player)
# Bewertet wird aber aus unserer Perspektive
v = self.expectiminimax(game, depth + 1)
if v > temp_val:
temp_val = v
# Spiel zurücksetzen
game.reset_to_state(state)
# Wert gewichtet addieren
value += probability * temp_val
return value
def get_name(self):
return "ExpectiminimaxValuePlayer [" + self.value.__name__ + "]"
class ExpectiminimaxModelPlayer(ExpectiminimaxValuePlayer):
def __init__(self, player, model, depth):
ExpectiminimaxValuePlayer.__init__(self, player, self.get_value, depth)
self.model = model
def get_value(self, game, player):
features = game.extractFeatures(player)
v = self.model.get_output(features)
v = 1 - v if self.player == game.players[0] else v
return v
def get_name(self):
return "EMinMaxModelPlayer [" + self.model.get_name() +"]"
import Player
from NeuralNetModel import TDGammonModel
import tensorflow as tf
graph = tf.Graph()
sess = tf.Session(graph=graph)
with sess.as_default(), graph.as_default():
model = TDGammonModel(sess, restore=True)
model.test(games = 100, enemyPlayer = ExpectiminimaxModelPlayer('white', model, 1))
import Player
import PlayerTest
players = [Player.ValuePlayer('black', Player.blocker), ExpectiminimaxValuePlayer('white', Player.blocker, 1)]
PlayerTest.test(players, 100)
import Player
import PlayerTest
from NeuralNetModel import TDGammonModel
import tensorflow as tf
graph = tf.Graph()
sess = tf.Session(graph=graph)
with sess.as_default(), graph.as_default():
model = TDGammonModel(sess, restore=True)
players = [Player.ModelPlayer('black', model), Player.ExpectiminimaxModelPlayer('white', model, 2)]
PlayerTest.test(players, 10)
```
Diese 3 Spiele haben 24 Stunden gedauert....
|
github_jupyter
|
```
import os, json
from pathlib import Path
from pandas import DataFrame
from mpcontribs.client import Client
from unflatten import unflatten
client = Client()
```
**Load raw data**
```
name = "screening_inorganic_pv"
indir = Path("/Users/patrick/gitrepos/mp/mpcontribs-data/ThinFilmPV")
files = {
"summary": "SUMMARY.json",
"absorption": "ABSORPTION-CLIPPED.json",
"dos": "DOS.json",
"formulae": "FORMATTED-FORMULAE.json"
}
data = {}
for k, v in files.items():
path = indir / v
with path.open(mode="r") as f:
data[k] = json.load(f)
for k, v in data.items():
print(k, len(v))
```
**Prepare contributions**
```
config = {
"SLME_500_nm": {"path": "SLME.500nm", "unit": "%"},
"SLME_1000_nm": {"path": "SLME.1000nm", "unit": "%"},
"E_g": {"path": "ΔE.corrected", "unit": "eV"},
"E_g_d": {"path": "ΔE.direct", "unit": "eV"},
"E_g_da": {"path": "ΔE.dipole", "unit": "eV"},
"m_e": {"path": "mᵉ", "unit": "mₑ"},
"m_h": {"path": "mʰ", "unit": "mₑ"}
}
columns = {c["path"]: c["unit"] for c in config.values()}
contributions = []
for mp_id, d in data["summary"].items():
formula = data["formulae"][mp_id].replace("<sub>", "").replace("</sub>", "")
contrib = {"project": name, "identifier": mp_id, "data": {"formula": formula}}
cdata = {v["path"]: f'{d[k]} {v["unit"]}' for k, v in config.items()}
contrib["data"] = unflatten(cdata)
df_abs = DataFrame(data=data["absorption"][mp_id])
df_abs.columns = ["hν [eV]", "α [cm⁻¹]"]
df_abs.set_index("hν [eV]", inplace=True)
df_abs.columns.name = "" # legend name
df_abs.attrs["name"] = "absorption"
df_abs.attrs["title"] = "optical absorption spectrum"
df_abs.attrs["labels"] = {"variable": "", "value": "α [cm⁻¹]"}
df_dos = DataFrame(data=data["dos"][mp_id])
df_dos.columns = ['E [eV]', 'DOS [eV⁻¹]']
df_dos.set_index("E [eV]", inplace=True)
df_dos.columns.name = "" # legend name
df_dos.attrs["name"] = "DOS"
df_dos.attrs["title"] = "electronic density of states"
df_dos.attrs["labels"] = {"variable": "", "value": "DOS [eV⁻¹]"}
contrib["tables"] = [df_abs, df_dos]
contributions.append(contrib)
len(contributions)
```
**Submit contributions**
```
client.delete_contributions(name)
client.init_columns(name, columns)
client.submit_contributions(contributions[:5])
```
**Retrieve and plot tables**
```
all_ids = client.get_all_ids(
{"project": "screening_inorganic_pv"}, include=["tables"]
).get(name, {})
cids = list(all_ids["ids"])
tids = list(all_ids["tables"]["ids"])
len(cids), len(tids)
client.get_contribution(cids[0])
t = client.get_table(tids[0]) # pandas DataFrame
t.display()
```
|
github_jupyter
|
```
import torch
from torch import nn
from torch import optim
from torchvision.datasets import MNIST
from torch.utils.data import TensorDataset, Dataset, DataLoader
from tqdm.notebook import tqdm
import numpy as np
from aijack.defense import VIB, KL_between_normals, mib_loss
dim_z = 256
beta = 1e-3
batch_size = 100
samples_amount = 15
num_epochs = 1
train_data = MNIST("MNIST/.", download=True, train=True)
train_dataset = TensorDataset(
train_data.train_data.view(-1, 28 * 28).float() / 255, train_data.train_labels
)
train_loader = DataLoader(train_dataset, batch_size=batch_size)
test_data = MNIST("MNIST/.", download=True, train=False)
test_dataset = TensorDataset(
test_data.test_data.view(-1, 28 * 28).float() / 255, test_data.test_labels
)
test_loader = DataLoader(test_dataset, batch_size=batch_size)
encoder = nn.Sequential(
nn.Linear(in_features=784, out_features=1024),
nn.ReLU(),
nn.Linear(in_features=1024, out_features=1024),
nn.ReLU(),
nn.Linear(in_features=1024, out_features=2 * dim_z),
)
decoder = nn.Linear(in_features=dim_z, out_features=10)
net = VIB(encoder, decoder, dim_z, num_samples=samples_amount)
opt = torch.optim.Adam(net.parameters(), lr=1e-4)
scheduler = torch.optim.lr_scheduler.ExponentialLR(opt, gamma=0.97)
import time
for epoch in range(num_epochs):
loss_by_epoch = []
accuracy_by_epoch = []
I_ZX_bound_by_epoch = []
I_ZY_bound_by_epoch = []
loss_by_epoch_test = []
accuracy_by_epoch_test = []
I_ZX_bound_by_epoch_test = []
I_ZY_bound_by_epoch_test = []
if epoch % 2 == 0 and epoch > 0:
scheduler.step()
for x_batch, y_batch in tqdm(train_loader):
x_batch = x_batch
y_batch = y_batch
y_pred, result_dict = net(x_batch)
sampled_y_pred = result_dict["sampled_decoded_outputs"]
p_z_given_x_mu = result_dict["p_z_given_x_mu"]
p_z_given_x_sigma = result_dict["p_z_given_x_sigma"]
approximated_z_mean = torch.zeros_like(p_z_given_x_mu)
approximated_z_sigma = torch.ones_like(p_z_given_x_sigma)
loss, I_ZY_bound, I_ZX_bound = mib_loss(
y_batch,
sampled_y_pred,
p_z_given_x_mu,
p_z_given_x_sigma,
approximated_z_mean,
approximated_z_sigma,
beta=beta,
)
prediction = torch.max(y_pred, dim=1)[1]
accuracy = torch.mean((prediction == y_batch).float())
loss.backward()
opt.step()
opt.zero_grad()
I_ZX_bound_by_epoch.append(I_ZX_bound.item())
I_ZY_bound_by_epoch.append(I_ZY_bound.item())
loss_by_epoch.append(loss.item())
accuracy_by_epoch.append(accuracy.item())
for x_batch, y_batch in tqdm(test_loader):
x_batch = x_batch
y_batch = y_batch
y_pred, result_dict = net(x_batch)
sampled_y_pred = result_dict["sampled_decoded_outputs"]
p_z_given_x_mu = result_dict["p_z_given_x_mu"]
p_z_given_x_sigma = result_dict["p_z_given_x_sigma"]
approximated_z_mean = torch.zeros_like(p_z_given_x_mu)
approximated_z_sigma = torch.ones_like(p_z_given_x_sigma)
loss, I_ZY_bound, I_ZX_bound = mib_loss(
y_batch,
sampled_y_pred,
p_z_given_x_mu,
p_z_given_x_sigma,
approximated_z_mean,
approximated_z_sigma,
beta=beta,
)
prediction = torch.max(y_pred, dim=1)[1]
accuracy = torch.mean((prediction == y_batch).float())
I_ZX_bound_by_epoch_test.append(I_ZX_bound.item())
I_ZY_bound_by_epoch_test.append(I_ZY_bound.item())
loss_by_epoch_test.append(loss.item())
accuracy_by_epoch_test.append(accuracy.item())
print(
"epoch",
epoch,
"loss",
np.mean(loss_by_epoch_test),
"prediction",
np.mean(accuracy_by_epoch_test),
)
print(
"I_ZX_bound",
np.mean(I_ZX_bound_by_epoch_test),
"I_ZY_bound",
np.mean(I_ZY_bound_by_epoch_test),
)
from aijack.attack import GradientInversion_Attack
y_pred, result_dict = net(x_batch[:1])
sampled_y_pred = result_dict["sampled_decoded_outputs"]
p_z_given_x_mu = result_dict["p_z_given_x_mu"]
p_z_given_x_sigma = result_dict["p_z_given_x_sigma"]
approximated_z_mean = torch.zeros_like(p_z_given_x_mu)
approximated_z_sigma = torch.ones_like(p_z_given_x_sigma)
loss, I_ZY_bound, I_ZX_bound = mib_loss(
y_batch[:1],
sampled_y_pred,
p_z_given_x_mu,
p_z_given_x_sigma,
approximated_z_mean,
approximated_z_sigma,
beta=beta,
)
received_gradients = torch.autograd.grad(loss, net.parameters())
received_gradients = [cg.detach() for cg in received_gradients]
received_gradients = [cg for cg in received_gradients]
from matplotlib import pyplot as plt
import cv2
net.eval()
cpl_attacker = GradientInversion_Attack(
net,
(784,),
lr=0.3,
log_interval=50,
optimizer_class=torch.optim.LBFGS,
distancename="l2",
optimize_label=False,
num_iteration=200,
)
num_seeds = 5
fig = plt.figure(figsize=(6, 2))
for s in tqdm(range(num_seeds)):
cpl_attacker.reset_seed(s)
try:
result = cpl_attacker.attack(received_gradients)
ax1 = fig.add_subplot(2, num_seeds, s + 1)
ax1.imshow(result[0].cpu().detach().numpy()[0].reshape(28, 28), cmap="gray")
ax1.axis("off")
ax1.set_title(torch.argmax(result[1]).cpu().item())
ax2 = fig.add_subplot(2, num_seeds, num_seeds + s + 1)
ax2.imshow(
cv2.medianBlur(result[0].cpu().detach().numpy()[0].reshape(28, 28), 5),
cmap="gray",
)
ax2.axis("off")
except:
pass
plt.suptitle("Result of CPL")
plt.tight_layout()
plt.show()
```
|
github_jupyter
|
## 1-2. 量子ビットに対する基本演算
量子ビットについて理解が深まったところで、次に量子ビットに対する演算がどのように表されるかについて見ていこう。
これには、量子力学の性質が深く関わっている。
1. 線型性:
詳しくは第4章で学ぶのだが、量子力学では状態(量子ビット)の時間変化はつねに(状態の重ね合わせに対して)線型になっている。つまり、**量子コンピュータ上で許された操作は状態ベクトルに対する線型変換**ということになる
。1つの量子ビットの量子状態は規格化された2次元複素ベクトルとして表現されるのだったから、
1つの量子ビットに対する操作=線型演算は$2 \times 2$の**複素行列**によって表現される。
2. ユニタリ性:
さらに、確率の合計は常に1であるという規格化条件から、量子操作に表す線形演算(量子演算)に対してさらなる制限を導くことができる。まず、各測定結果を得る確率は複素確率振幅の絶対値の2乗で与えられるので、その合計は状態ベクトルの(自分自身との)内積と一致することに注目する:
$$
|\alpha|^2 + |\beta|^2 =
(\alpha^*, \beta^*)
\left(
\begin{array}{c}
\alpha
\\
\beta
\end{array}
\right) = 1.
$$
(アスタリスク $^*$ は複素共役を表す)
量子コンピュータで操作した後の状態は、量子演算に対応する線形変換(行列)を$U$とすると、
$$
U
\left(
\begin{array}{c}
\alpha
\\
\beta
\end{array}
\right)
$$
と書ける。この状態についても上記の規格化条件が成り立つ必要があるので、
$$
(\alpha^*, \beta^*)
U^\dagger U
\left(
\begin{array}{c}
\alpha
\\
\beta
\end{array}
\right) = 1
$$
が要請される。(ダガー $^\dagger$ は行列の転置と複素共役を両方適用したものを表し、エルミート共役という)
この関係式が任意の$\alpha$, $\beta$について成り立つ必要があるので、量子演算$U$は以下の条件を満たす**ユニタリー行列**に対応する:
$$
U^{\dagger} U = U U^{\dagger} = I.
$$
すなわち、**量子ビットに対する操作は、ユニタリー行列で表される**のである。
ここで、用語を整理しておく。量子力学では、状態ベクトルに対する線形変換のことを**演算子** (operator) と呼ぶ。単に演算子という場合は、ユニタリーとは限らない任意の線形変換を指す。それに対して、上記のユニタリー性を満たす線形変換のことを**量子演算** (quantum gate) と呼ぶ。量子演算は、量子状態に対する演算子のうち、(少なくとも理論的には)**物理的に実現可能なもの**と考えることができる。
### 1量子ビット演算の例:パウリ演算子
1つの量子ビットに作用する基本的な量子演算として**パウリ演算子**を導入する。
これは量子コンピュータを学んでいく上で最も重要な演算子であるので、定義を体に染み込ませておこう。
$$
\begin{eqnarray}
I&=&
\left(\begin{array}{cc}
1 & 0
\\
0 & 1
\end{array}
\right),\;\;\;
X=
\left(\begin{array}{cc}
0 & 1
\\
1 & 0
\end{array}
\right),\;\;\;
Y &=&
\left(\begin{array}{cc}
0 & -i
\\
i & 0
\end{array}
\right),\;\;\;
Z=
\left(\begin{array}{cc}
1 & 0
\\
0 & -1
\end{array}
\right).
\end{eqnarray}
$$
各演算子のイメージを説明する。
まず、$I$は恒等演算子で、要するに「何もしない」ことを表す。
$X$は古典ビットの反転(NOT)に対応し
$$X|0\rangle = |1\rangle, \;\;
X|1\rangle = |0\rangle
$$
のように作用する。(※ブラケット表記を用いた。下記コラムも参照。)
$Z$演算子は$|0\rangle$と$|1\rangle$の位相を反転させる操作で、
$$
Z|0\rangle = |0\rangle, \;\;
Z|1\rangle = -|1\rangle
$$
と作用する。
これは$|0\rangle$と$|1\rangle$の重ね合わせの「位相」という情報を保持できる量子コンピュータ特有の演算である。
例えば、
$$
Z \frac{1}{\sqrt{2}} ( |0\rangle + |1\rangle ) = \frac{1}{\sqrt{2}} ( |0\rangle - |1\rangle )
$$
となる。
$Y$演算子は$Y=iXZ$と書け、
位相の反転とビットの反転を組み合わせたもの(全体にかかる複素数$i$を除いて)であると考えることができる。
(詳細は Nielsen-Chuang の `1.3.1 Single qubit gates` を参照)
### SymPyを用いた一量子ビット演算
SymPyではよく使う基本演算はあらかじめ定義されている。
```
from IPython.display import Image, display_png
from sympy import *
from sympy.physics.quantum import *
from sympy.physics.quantum.qubit import Qubit,QubitBra
init_printing() # ベクトルや行列を綺麗に表示するため
# Google Colaboratory上でのみ実行してください
from IPython.display import HTML
def setup_mathjax():
display(HTML('''
<script>
if (!window.MathJax && window.google && window.google.colab) {
window.MathJax = {
'tex2jax': {
'inlineMath': [['$', '$'], ['\\(', '\\)']],
'displayMath': [['$$', '$$'], ['\\[', '\\]']],
'processEscapes': true,
'processEnvironments': true,
'skipTags': ['script', 'noscript', 'style', 'textarea', 'code'],
'displayAlign': 'center',
},
'HTML-CSS': {
'styles': {'.MathJax_Display': {'margin': 0}},
'linebreaks': {'automatic': true},
// Disable to prevent OTF font loading, which aren't part of our
// distribution.
'imageFont': null,
},
'messageStyle': 'none'
};
var script = document.createElement("script");
script.src = "https://colab.research.google.com/static/mathjax/MathJax.js?config=TeX-AMS_HTML-full,Safe";
document.head.appendChild(script);
}
</script>
'''))
get_ipython().events.register('pre_run_cell', setup_mathjax)
from sympy.physics.quantum.gate import X,Y,Z,H,S,T,CNOT,SWAP, CPHASE
```
演算子は何番目の量子ビットに作用するか、
というのを指定して `X(0)` のように定義する。
また、これを行列表示するときには、いくつの量子ビットの空間で表現するか
`nqubits`というのを指定する必要がある。
まだ、量子ビットは1つしかいないので、
`X(0)`、`nqubits=1`としておこう。
```
X(0)
represent(X(0),nqubits=1) # パウリX
```
同様に、`Y`, `Z`なども利用することができる。それに加え、アダマール演算 `H` や、位相演算 `S`、そして$\pi/4$の位相演算 `T` も利用することができる(これらもよく出てくる演算で、定義は各行列を見てほしい):
```
represent(H(0),nqubits=1)
represent(S(0),nqubits=1)
represent(T(0),nqubits=1)
```
これらの演算を状態に作用させるのは、
```
ket0 = Qubit('0')
S(0)*Y(0)*X(0)*H(0)*ket0
```
のように `*`で書くことができる。実際に計算をする場合は `qapply()`を利用する。
```
qapply(S(0)*Y(0)*X(0)*H(0)*ket0)
```
この列ベクトル表示が必要な場合は、
```
represent(qapply(S(0)*Y(0)*X(0)*H(0)*ket0))
```
のような感じで、SymPyは簡単な行列の計算はすべて自動的にやってくれる。
---
### コラム:ブラケット記法
ここで一旦、量子力学でよく用いられるブラケット記法というものについて整理しておく。ブラケット記法に慣れると非常に簡単に見通しよく計算を行うことができる。
列ベクトルは
$$
|\psi \rangle = \left(
\begin{array}{c}
\alpha
\\
\beta
\end{array}
\right)
$$
とかくのであった。これを**ケット**と呼ぶ。同様に、行ベクトルは
$$
\langle \psi | = ( |\psi \rangle ) ^{\dagger} = ( \alpha ^* , \beta ^*)
$$
とかき、これを**ブラ**と呼ぶ。${\dagger}$マークは転置と複素共役を取る操作で、列ベクトルを行ベクトルへと移す。
2つのベクトル、
$$
|\psi \rangle = \left(
\begin{array}{c}
\alpha
\\
\beta
\end{array}
\right), \;\;\;
|\phi \rangle = \left(
\begin{array}{c}
\gamma
\\
\delta
\end{array}
\right)
$$
があったとする。ブラとケットを抱き合わせると
$$
\langle \phi | \psi \rangle = (\gamma ^* , \delta ^* ) \left(
\begin{array}{c}
\alpha
\\
\beta
\end{array}
\right) = \gamma ^* \alpha + \delta ^* \beta
$$
となり、**内積**に対応する。行ベクトルと列ベクトルをそれぞれブラ・ケットと呼ぶのは、このように並べて内積を取ると「ブラケット」になるからである。
逆に、背中合わせにすると
$$
|\phi \rangle \langle \psi | = \left(
\begin{array}{c}
\gamma
\\
\delta
\end{array}
\right) (\alpha ^* , \beta ^*) = \left(
\begin{array}{cc}
\gamma \alpha ^* & \gamma \beta ^*
\\
\delta \alpha ^* & \delta \beta ^*
\end{array}
\right)
$$
となり、演算子となる。例えば、$X$演算子は
$$
X= \left(
\begin{array}{cc}
0 & 1
\\
1 & 0
\end{array}
\right)
=
|0\rangle \langle 1 | + |1\rangle \langle 0|
$$
のように書ける。このことを覚えておけば
$$
\langle 0| 0\rangle = \langle 1 | 1\rangle = 1, \;\;\; \langle 0 | 1 \rangle = \langle 1 | 0 \rangle = 0
$$
から
$$
X |0\rangle = |1\rangle
$$
を行列を書かずに計算できるようになる。
**量子情報の解析計算においては、実際にベクトルの要素を書き下して計算をすることはほとんどなく、このようにブラケットを使って形式的に書いて計算してしまう場合が多い**(古典計算機上で量子コンピュータをシミュレーションする場合はベクトルをすべて書き下すことになる)。
同様に、
$$
I = |0\rangle \langle 0 | + |1\rangle \langle 1| , \;\;\; Z = |0\rangle \langle 0| - |1\rangle \langle 1|
$$
も覚えておくと便利である。
|
github_jupyter
|
```
%matplotlib inline
from __future__ import absolute_import
from __future__ import print_function
import matplotlib.pyplot as plt
import numpy as np
np.random.seed(1337) # for reproducibility
from theano import function
from keras.datasets import mnist
from keras.models import Sequential
from keras.layers.core import Dense, Dropout, Activation, Flatten, Reshape, Layer
from keras.layers.convolutional import Convolution2D, MaxPooling2D, UpSampling2D
from keras.utils import np_utils
from keras import backend as K
from keras.callbacks import ModelCheckpoint
from keras.regularizers import l2
from seya.layers.variational import VariationalDense as VAE
from seya.layers.convolutional import GlobalPooling2D
from seya.utils import apply_model
from agnez import grid2d
batch_size = 100
nb_epoch = 100
code_size = 200
# input image dimensions
img_rows, img_cols = 28, 28
# number of convolutional filters to use
nb_filters = 32
# size of pooling area for max pooling
nb_pool = 2
# convolution kernel size
nb_conv = 7
nb_classes = 10
# the data, shuffled and split between tran and test sets
(X_train, y_train), (X_test, y_test) = mnist.load_data()
X_train = X_train.reshape(X_train.shape[0], 1, img_rows, img_cols)
X_test = X_test.reshape(X_test.shape[0], 1, img_rows, img_cols)
X_train = X_train.astype("float32")
X_test = X_test.astype("float32")
X_train /= 255
X_test /= 255
print('X_train shape:', X_train.shape)
print(X_train.shape[0], 'train samples')
print(X_test.shape[0], 'test samples')
# convert class vectors to binary class matrices
Y_train = np_utils.to_categorical(y_train, nb_classes)
Y_test = np_utils.to_categorical(y_test, nb_classes)
X_valid = X_train[50000:]
Y_valid = Y_train[50000:]
X_train = X_train[:50000]
Y_train = Y_train[:50000]
enc = Sequential()
enc.add(Convolution2D(nb_filters, nb_conv, nb_conv,
W_regularizer=l2(.0005),
border_mode='same',
input_shape=(1, img_rows, img_cols)))
enc.add(Dropout(.5))
enc.add(Activation('relu'))
enc.add(Convolution2D(nb_filters, 3, 3,
border_mode='same',
input_shape=(1, img_rows, img_cols)))
enc.add(Activation('tanh'))
enc.add(MaxPooling2D())
enc.add(Flatten())
pool_shape = enc.output_shape
enc.add(VAE(code_size, batch_size=batch_size, activation='tanh',
prior_logsigma=1.7))
# enc.add(Activation(soft_threshold))
dec = Sequential()
dec.add(Dense(np.prod(pool_shape[1:]), input_dim=code_size))
dec.add(Reshape((nb_filters, img_rows/2, img_cols/2)))
dec.add(Activation('relu'))
dec.add(Convolution2D(nb_filters, 3, 3,
border_mode='same'))
dec.add(Activation('relu'))
dec.add(Convolution2D(784, 3, 3,
border_mode='same'))
dec.add(GlobalPooling2D())
dec.add(Activation('sigmoid'))
dec.add(Flatten())
model = Sequential()
model.add(enc)
model.add(dec)
model.compile(loss='binary_crossentropy', optimizer='adam')
cbk = ModelCheckpoint('vae/vae.hdf5', save_best_only=True)
try:
model.fit(X_train, X_train.reshape((-1, 784)), batch_size=batch_size, nb_epoch=nb_epoch, verbose=1,
validation_data=(X_valid, X_valid.reshape((-1, 784))), callbacks=[cbk])
except:
pass
```
# Sample
```
X = K.placeholder(ndim=2)
Y = dec(X)
F = function([X], Y, allow_input_downcast=True)
x = np.random.laplace(0, 1, size=(100, code_size))
y = F(x)
I = grid2d(y.reshape((100, -1)))
plt.imshow(I)
```
# Visualize first layers
```
W = np.asarray(K.eval(enc.layers[0].W))
W = W.reshape((32, -1))
I = grid2d(W)
plt.imshow(I)
```
|
github_jupyter
|
```
import os
import numpy as np
import pandas as pd
import jinja2 as jj
def mklbl(prefix, n):
return ["%s%s" % (prefix, i) for i in range(n)]
miindex = pd.MultiIndex.from_product([mklbl('A', 4),
mklbl('B', 2),
mklbl('C', 4),
mklbl('D', 2)],
names=['RowIdx-1', 'RowIdx-2', 'RowIdx-3', 'RowIdx-4'])
index =['-'.join(col).strip() for col in miindex.values]
micolumns = pd.MultiIndex.from_tuples([('a', 'foo', 'zap'),
('a', 'foo', 'zip'),
('a', 'bar', 'zap'),
('a', 'bar', 'zip'),
('b', 'foo', 'zap'),
('b', 'foo', 'zep'),
('b', 'bah', 'zep'),
('b', 'bah', 'zyp'),
('b', 'bah', 'zap'),
],
names=['ColIdx-{}'.format(i) for i in range(1, 4)])
cols =['-'.join(col).strip() for col in micolumns.values]
data = np.arange(len(miindex) * len(micolumns), dtype=np.float).reshape((len(miindex),len(micolumns)))
data = data.tolist()
dfrc = pd.DataFrame(data, index=miindex, columns=micolumns).sort_index().sort_index(axis=1)
dfr = pd.DataFrame(data, index=miindex, columns=cols).sort_index().sort_index(axis=1)
dfr.columns.name = 'UniqueCol'
dfc = pd.DataFrame(data, index=index, columns=micolumns).sort_index().sort_index(axis=1)
dfc.index.name = 'UniqueRow'
df = pd.DataFrame(data, index=index, columns=cols).sort_index()
df.index.name = 'UniqueRow'
df.columns.name = 'UniqueCol'
dfrc.info()
dfr.info()
dfc.info()
df.info()
dfrc.head()
```
## Save df html
Must:
+ use notebook.css
+ wrap dataframe.html in specific classes - like in notebook
Result can be iframed in any doc
```
%%writefile templates/index.tpl.html
<!DOCTYPE html>
<html>
<head>
<meta charset="utf-8" />
<meta http-equiv="X-UA-Compatible" content="IE=edge">
<title>dataframe</title>
<meta name="viewport" content="width=device-width, initial-scale=1">
<link rel="stylesheet" href="https://cdn.jupyter.org/notebook/5.6.0/style/style.min.css">
</head>
<body>
<div class="output_are">
<div class="output_subarea output_html rendered_html output_result">
__$data.df_html$__
</div>
</div>
</body>
</html>
dir_template = 'templates'
dir_dump = 'dump'
loader = jj.FileSystemLoader(dir_template)
env = jj.Environment(loader=loader,
variable_start_string='__$',
variable_end_string='$__',
block_start_string='{-%',
block_end_string='%-}'
)
template = env.get_template('index.tpl.html')
# data = {'df_html': dfrc.to_html()}
data = {'df_html': dfrc.head(10).to_html()}
content = template.render(data=data)
if not os.path.exists(dir_dump):
os.makedirs(dir_dump)
path = os.path.join(dir_dump, 'index.html')
with open(path, 'w') as f:
f.write(content)
print('file {} saved to disk'.format(path))
!cd dump && python -m http.server 8080
```
|
github_jupyter
|
# Figure 3: Cluster-level consumptions
This notebook generates individual panels of Figure 3 in "Combining satellite imagery and machine learning to predict poverty".
```
from fig_utils import *
import matplotlib.pyplot as plt
import time
%matplotlib inline
```
## Predicting consumption expeditures
The parameters needed to produce the plots are as follows:
- country: Name of country being evaluated as a lower-case string
- country_path: Path of directory containing LSMS data corresponding to the specified country
- dimension: Number of dimensions to reduce image features to using PCA. Defaults to None, which represents no dimensionality reduction.
- k: Number of cross validation folds
- k_inner: Number of inner cross validation folds for selection of regularization parameter
- points: Number of regularization parameters to try
- alpha_low: Log of smallest regularization parameter to try
- alpha_high: Log of largest regularization parameter to try
- margin: Adjusts margins of output plot
The data directory should contain the following 5 files for each country:
- conv_features.npy: (n, 4096) array containing image features corresponding to n clusters
- consumptions.npy: (n,) vector containing average cluster consumption expenditures
- nightlights.npy: (n,) vector containing the average nightlights value for each cluster
- households.npy: (n,) vector containing the number of households for each cluster
- image_counts.npy: (n,) vector containing the number of images available for each cluster
Exact results may differ slightly with each run due to randomly splitting data into training and test sets.
#### Panel A
```
# Plot parameters
country = 'nigeria'
country_path = '../data/LSMS/nigeria/'
dimension = None
k = 5
k_inner = 5
points = 10
alpha_low = 1
alpha_high = 5
margin = 0.25
# Plot single panel
t0 = time.time()
X, y, y_hat, r_squareds_test = predict_consumption(country, country_path,
dimension, k, k_inner, points, alpha_low,
alpha_high, margin)
t1 = time.time()
print 'Finished in {} seconds'.format(t1-t0)
```
#### Panel B
```
# Plot parameters
country = 'tanzania'
country_path = '../data/LSMS/tanzania/'
dimension = None
k = 5
k_inner = 5
points = 10
alpha_low = 1
alpha_high = 5
margin = 0.25
# Plot single panel
t0 = time.time()
X, y, y_hat, r_squareds_test = predict_consumption(country, country_path,
dimension, k, k_inner, points, alpha_low,
alpha_high, margin)
t1 = time.time()
print 'Finished in {} seconds'.format(t1-t0)
```
#### Panel C
```
# Plot parameters
country = 'uganda'
country_path = '../data/LSMS/uganda/'
dimension = None
k = 5
k_inner = 5
points = 10
alpha_low = 1
alpha_high = 5
margin = 0.25
# Plot single panel
t0 = time.time()
X, y, y_hat, r_squareds_test = predict_consumption(country, country_path,
dimension, k, k_inner, points, alpha_low,
alpha_high, margin)
t1 = time.time()
print 'Finished in {} seconds'.format(t1-t0)
```
#### Panel D
```
# Plot parameters
country = 'malawi'
country_path = '../data/LSMS/malawi/'
dimension = None
k = 5
k_inner = 5
points = 10
alpha_low = 1
alpha_high = 5
margin = 0.25
# Plot single panel
t0 = time.time()
X, y, y_hat, r_squareds_test = predict_consumption(country, country_path,
dimension, k, k_inner, points, alpha_low,
alpha_high, margin)
t1 = time.time()
print 'Finished in {} seconds'.format(t1-t0)
```
|
github_jupyter
|
<div align="center">
<h1><strong>Herencia</strong></h1>
<strong>Hecho por:</strong> Juan David Argüello Plata
</div>
## __Introducción__
<div align="justify">
La relación de herencia facilita la reutilización de código brindando una base de programación para el desarrollo de nuevas clases.
</div>
## __1. Superclase y subclases__
En la relación de herencia entre dos clases, se cataloga a las clases como _padre_ e _hija_. La clase hija (subclase) _hereda_ los __métodos__ y __atributos__ de la clase padre. Las subclases (clases hijas) emplean el siguiente formato:
```
class clase_hija (clase_padre):
//Atributos
...
//Métodos
...
```
La clase padre suele usarse como un formato para la construcción de clases hijas. Un ejemplo de ello es la _calculadora científica_, que se puede catalogar como una subclase de la calculadora convencional.
```
#Calculadora convencional
class Calculadora:
def suma (self, x, y):
return x+y;
def resta (self, x, y):
return x-y;
def mult (self, x, y):
return x*y;
def div (self, x, y):
return x/y
```
Además de las operaciones básicas, la clase de `Calculadora_cientifica` debería poder calcular el promedio de una lista numérica y la desviación estándar.
---
<div align="center">
<strong>Promedio</strong>
$$
\begin{equation}
\bar{x} = \frac{\sum _{i=0} ^n x_i}{n}
\end{equation}
$$
<strong>Desviación estándar</strong>
$$
\begin{equation}
s = \sqrt{ \frac{\sum _{i=0} ^n \left( x_i - \bar{x} \right)}{n-1} }
\end{equation}
$$
</div>
```
#Calculadora científica
class Calculadora_cientifica (Calculadora):
def promedio (self, numeros):
return sum(numeros)/len(numeros)
def desvest (self, numeros):
promedio = self.promedio(numeros)
des = 0;
for num in numeros:
des += (num-promedio)**2
des /= (len(numeros)-1);
return des**(1/2)
```
__Observa__ que al momento de crear un objeto del tipo `Calculadora_cientifica` es posible utilizar los métodos heredados de la clase `Calculadora`.
```
calc1 = Calculadora_cientifica();
print("2+3 = " + str(calc1.suma(2,3)));
print("Promedio de: [2,3,10] = " + str(calc1.promedio([2,3,10])));
print("Desviación estándar de: [2,3,10] = " + str(calc1.desvest([2,3,10])));
```
En Python, durante la relación de herencia puede haber múltiples clases padre por cada hija.
## 1.1. Operadores __`self`__ y __`super()`__
<div align="justify">
El operador `self` se refiere a la clase _per se_. Se emplea dentro de la clase para especificar el uso de sus métodos y atributos. El operador `super()` se emplea en una relación de herencia para referirse explícitamente a los métodos y atributos de la clase padre. Es decir: en una relación de herencia, se emplea `self` para referirse a los métodos y atributos de la subclase (clase _hija_) y `super()` para los métodos y atributos de la superclase (clase _padre_).
</div>
### 1.1.1 Constructores
Tanto la superclase como las subclases pueden tener sus propios constructores. Si la superclase tiene un constructor, la sublcase debe emplear el operador `super().__init__(entradas)` para ejecutarlo.
Por ejemplo, supongamos la situación de un estudiante. Se puede asumir que la clase de `Student` deriva de la clase `Person`, como se aprecia en el diagrama UML.
<div align="center">
<img src="data:image/png;base64,iVBORw0KGgoAAAANSUhEUgAAAKEAAAE/CAYAAAAuSqMxAAAGnnRFWHRteGZpbGUAJTNDbXhmaWxlJTIwaG9zdCUzRCUyMmFwcC5kaWFncmFtcy5uZXQlMjIlMjBtb2RpZmllZCUzRCUyMjIwMjEtMDctMjRUMjAlM0EwOCUzQTUyLjE5M1olMjIlMjBhZ2VudCUzRCUyMjUuMCUyMChXaW5kb3dzKSUyMiUyMGV0YWclM0QlMjJ5M09uZVJTR3RFdURUblR0NmRNUCUyMiUyMHZlcnNpb24lM0QlMjIxNC45LjIlMjIlMjB0eXBlJTNEJTIyZGV2aWNlJTIyJTNFJTNDZGlhZ3JhbSUyMGlkJTNEJTIyQzVSQnM0M29EYS1LZHpaZU50dXklMjIlMjBuYW1lJTNEJTIyUGFnZS0xJTIyJTNFN1ZscmM5bzRGUDAxekhRJTJGcE9NSEVQb1JTS0R0Wmp1MGRKdjlLdXlMclVhV3ZMSUlrRiUyRmZLMXQlMkJZWjV0R0RwcFpoakdPcnE2c3U0NUVnZTc1UTZqMVZpU09QeEglMkJNQmFqdVd2V3U1TnkzR3VldzUlMkJhMkNkQVczSHlvQkFVaiUyQkQ3QktZMGljd1lCNjJvRDRrdFVBbEJGTTByb09lNEJ3OFZjT0lsR0paRDVzTFZwODFKZ0UwZ0tsSFdCTzlwNzRLTTdUblhKZjRlNkJCbU05c2Q5OWxQUkhKZzgxS2twRDRZbG1CM051V081UkNxT3dxV2cyQjZkcmxkYm4lMkZzTDVuZHclMkZkOGNmUHlmJTJGazM4SGZYejk5dThxU2pVNFpVaXhCQWxjJTJGbmZycFlUNTYlMkY2MzklMkZiOTRNdnE4dE1iOTJjY3JNOFI2Skd4aDZqVUJtUWh1VnF6V2VSbVRKWTBZNGRnYXpBVlhVOU5qWVpzd0duQzg5dkR1UUNMd0NGSlJaS0J2T3BTSUVmVkN5dnc3c2hZTHZZWkVFZThoYncxQ0lla1RwaVVNdTJ3RXNGc3FJeWFuVzR1WTZwRm1hZ2tKeGt6eXd0Z0ZkRWNTWldJOHdSaUpFenBMYjFpSFJFUUdsQSUyQkVVaUxLRTRrRjk4RTNyWUxwdEtHa2VDaTBvOGNmU1llaFRWY0RWaFV4R25yR0lDSlFjbzBocHJkUW10bHFkdDVlbHNLMXV3WUxxNkoxZTJiRG1NMFNGTG1MNmI3ZzVpSTh3Q0tVODdrYjg3V1BuQThacVUxSEdCTFBpWUtCTG1OU2xTRmVWSlphUXFrNFR4Q3EzUkJxeTBFaUxFNGlhSWdWNjYwcXdtUXdWenRsbWNURW96eTRTMk51MmlYeXhheFhRd0xIemxrcWlaRDZQdkJVTW9vb01pdTJRU3dvVjJsQk9nUDhZTm1HMXR0T3E0TTNOTVMyWGJieG84T2xHZ3FPNmlJMGxSR2daSmVnWmJ0RllIczM3MkdCcmV1OG5jcHZWVTQxWWs5bDBkbkJZaHdLRHA4VzBRelBqNWRHNXA2akpWUVJNNWZub3J6alhKaHlkd2ZsRUJIS1hzbCUyQlZyS3ZleGNtdTcyRjdBMkdHVTFOaEttR3ZmVUg5Z0Q5RVJLcDAlMkJWOGY5Vnl1TG15RzVwd201cHd0JTJGRFB5QXpZUkNSVVVhSHp5eXgyUXhlWE9yUnRxMzBjcTNzOHdDJTJCUjJ0bXhnN05DTER3bFh0NmhmVFl5N1V1Zng5MEdtMU8xOFBVeVh5MyUyRjVTMiUyRmE1M1g4dHNibHQlMkZ0JTJGYjZXJTJGN3FoMUt0VW9LbGFQJTJCQU83dXNNVzRUN2h4dyUyQjNhUEY5cnZZJTJGOTVXUm1PSjZYMHF4c0JCNnVOQTg0cmxKaSUyQlAyV2UxaHlmemYlMkZIJTJGQXU5ZTdlR3prM3FzNXolMkJYTzdSM1BabXAyVU5jbHJ2cXAyVkluOXBnaVVlVUUlMkZibXIyeTdQd3FjcEJyMXdqYiUyQnVkanZIWG1rdDl2bm9uJTJGYkk1ME44b0Q3ZmYwa0gxc3pKclNUR3lCa3JKNXRaYzBSWmZuNWQ4Q0wxUTlSOEFQSXJTbXdtVmplbHNBZ0JiQWpGOHJKVGk0UkMlMkJuQlljYlF1d1p3eEQ4RmZiZDdlZDFHcEFSR0ZIMnN2OURZNCUyRnNtSW5WRmhjZTA2cDdQc2R4NmlteVJabFQxRGNLcGliSXFOQklkTm9UWUxOJTJCalpPSGx5eWozOWdjJTNEJTNDJTJGZGlhZ3JhbSUzRSUzQyUyRm14ZmlsZSUzRYlScd8AAA6lSURBVHic7Z3Bjds4FEBZQDaXvS8C6LClqJFc3MVcVUBqEOaeDtxDbga2hUwD3EPyna9vkqJE2n88eg8QMDOSKIp+/hI14mcIIUQWFuclRAAvkBDcQUJwBwnBHSQEd5AQ3EFCcAcJwZ3DSph6YHo6nbyrdUgOLeH5fF78bRiGOM+zU42OCxIqTqdTnKYpxhjj5XJZREnhfD7HEEIcx/H6d/k5FU3XyjmdTtd1tj5HAQkzfwshXKPi6XSK4zjGGP/II+vmeb6ui/FXNNXlDsNwFTtVTmrd0Ti0hHYRIc7ncxyG4bqtRLPL5XKV53K5xBh/Sai31ci2reV8dA4tYe7yN89zUtKUPDHGOE1T8nIs5QhImAYJE9hIaNdZCTX6ckwkrAMJC+v1fZ+0kZVnmqZF9Fu7JxTRkPAPSJjB9mpFllQkHIbh5r4yV46AhH84rITwfkBCcAcJwR0kBHeQENxBQnAHCcEdJAR3QggxfPnyZctIeRaWrstv/4iE4MdvGZEQ/EBCcAcJwR0kBHeQENxBQnAHCcEdJAR3kBDcqZYwhPwIs7VsAnYIpQwg0gPAZZ0ek1GbzQCem64SprIJ6GGOMf4anWZHnMl+kk4jN75XC3zkjAUfjaKEuUHgdkTZlpFjep3dz4qlI2opKwI8N10jYU5CfZkOIeySsJQVAZ6bu0tosxD0ioTwcejSOy5JaPOxDMOwS0L5PZUVAZ6bu0sY4zJDgd52q4S5rAjw3PCcENxBQnAHCcEdJAR3kBDcQUJwBwnBHSQEd5AQ3EFCcAcJwR0kBHeQENxBQnAHCcEdJAR3kBDcubuEa7Ni3oPUmOVpmroPEfU4t4/Ih5XQHhMJ3y9dR9vZTAp6XS47w9osmFv3k7qO47iQTktYMzqwJitEqo66jdbOTQb7H52uEsoHVMqyYAcz2fmArSi5/fTIu9wAqWEYrtvskTDGclaI0nmX6ij7ybqjU5RwbwaG2hnO986MvpaNQSTU5W+VsHYsdOm8c3XkMr6kayS0eWO0CDVjkmslXMvGoEU5nU5xmqaHSKjXl+qIhEvuIuGjI2GqriKKlFlK0vSISFhqr6PTpXds76NS94SlgfH6vq92P31PZSOq7cRIJ8NKKNuM49gkYeq8S3VEwiVdJZSb+FRPMCfTWg+ydr/UJVEzDMNNb1n3eFsk1L1jTa6OSLikq4Q0KuwBCcGdLhICtICE4A4SgjtICO4gIbiDhOAOEoI7SAjuICG4g4TgDhKCO0gI7iAhuIOE4A4SgjtPI6Eej2LHpsBz8zQSapDwY9FltF2M62NFajIaxHg7nFMGChEJPy7dJNTClDIplDIa2KGYqVF79md4fooSbsnAUJttYG1OY3t8JPz4dImEW7INrEmoh0+GEJDwAHSRcEu2gZpxvAKR8Bh06x3re8JStoGShDaTwjAMSHgAuklYm21g7XI8DMO1DL0vEn5cnvI5IXwskBDcQUJwBwnBHSQEd5AQ3EFCcAcJwR0kBHeQENxBQnAHCcGdq4QsLM4LkRD8QEJwBwnBHSQEd5AQ3EFCcAcJwR0kBHeQENxBQnAHCcEdJAR3ukoomRFksbkH70EI+axe76E8WKebhJJHRidNGsfxJo1cb5Dw+amW0ApmGYbhmhBJkPw09ndZBImgOi2cFkESa9roqvPWvL6+xhDCdVubA0dn+DInf62LLu9yuazWV44FbXSR0GZYzTEMwzUylrK56nXzPC8SKA3DsBBUhJMybHrhnIT6SzNN01Vu/QVYq6/90sE+ihLWZmqtkdBm0iplc9XCpCKYPQEtYS4bWC7Xoa5Pqrya+kIbD4uENvdgrYQxxkXSddvZ6SWhLW9LfaGNh9wT2vyCuXWl+zd9nNLlOJeEfauERMLHcffese1I6HssK0VKGH2/JmVskVC2HcdxIbZep2cJKN0T5uoLbXR9TmjvIe2lc623WepE5O5HpYf67du3GzHsZTw1w4Cti+5d19YX2ugqIcAekBDcQUJwBwnBHSQEd5AQ3EFCcAcJO/D29uZdhafmKiELi+vi/U14Zt7e3uJff/1FNGwECRt4eXmJ//77b3x5efGuylODhDt5e3uLnz9/jt+/f4+fP38mGjaAhDt5eXmJX79+jTHG+PXrV6JhA0i4A4mCP378iDHG+OPHD6JhA0i4Ax0FBaLhfpBwIzYKCkTD/SDhRlJRUCAa7gMJN5CLggLRcB9IuIFSFBSIhttBwkrWoqBANNwOElby8vISP336FP/555/F8vfff9/87dOnT0TDDSBhBT9//oz//fdfcgkhZNf9/PnTu+pPARI2EgJN2Aot2AgStkMLNoKE7dCCjSBhO7RgI0jYDi3YCBK2Qws2goTt0IKNIGE7tGAjSNgOLdgIErZDCzaChO3Qgo0gYTu0YCNI2A4t2AgStkMLNoKE7dCCjSBhO7RgI0jYDi3YCBK2Qws2goTt0IKNIGE7tGAjSNgOLdgIErZDCzaChO3Qgo0gYTu0YCNI2A4t2AgStkMLNoKE7dCCjSBhO8F9Nh8WlhD4JoMfSAjuICG4g4TgDhKCO0gI7iAhuIOE4M5hJbQPS+268/m8qbzz+XxTzt56bT32s3NICYdhiNM0XX+fpmkhEBI+lsNJeLlcYgghXi6Xxd9DCHGe5zgMwzU6vr6+LraV9YKIF0KIp9NpIaEcx0Za2Ue219LpY9v6fWQOJ2GMMY7jeJUuhYghwuQk1GVYCe26cRxjjH8klEis1+ljH4lDShjjL6F0pNJC1khoL7/69/P5vJBVR98asZHwgIgY8uHXSCgSCyKaXmcXJExzOAnneV5c/oRhGK7RUESw9497I6EGCW85nIQxxpvLr0QuEcNKKFKM47gQRveyS/eEOmoi4S2HlDDG2+eEujcqHZfz+Xx9fCM92tS9Xk3vWMpfk1Af+ygcVkJ4PyAhuIOE4A4SgjtICO4gIbiDhOAOEoI7IYQYvnz5kvxfJwvLI5bf/hEJwY/fMiIh+IGE4A4SgjtICO4gIbiDhOAOEoI7d5MwhPcxgNz+bJfcsM+99dpz3kfncBLqOskr+DUi7jk21NEkoYyHCOHXGAvhPWUxKEkY468UIHbwuc1+YEfS1R4b6tgtoR06OQzD4gOWD8c7i8GahDXRNyUhGRT60SRhbmytFPweshjcS0KGbPajKKG+5OjLrWCHQ9qC30MWAyR8/3TrmOQux95ZDLbeE5aOt/XYUMduCadpuumMlCT0ymJQ0ztekwYJ70tTJNS9QZ10Msb3k8XASmgXK0xt75gMCv3odjkG2AsSgjtICO4gIbiDhOAOEoI7SAjuICG4g4TgDhKCO0gI7iAhuIOE4A4SgjtICO4gIbiDhODOh5Uw9TZ0LfaN7tTb1l7UnosepNbj873n2+KHkHDPfrrB7SygntSel4z5uUdWi97sllAPAs+NAZHxFjGuZ1Ow5ehxJ5pcObosO17FfhClMvTAK804jtfz25MZomY/3V65mentuaS+HHrsT21Wi9Ix751ZollCkWSappuhnDrHix1Vl8umIB9EanyxVDiVlSG1Lidhri52eGqOXB1qsjOU9pN1th6ptl1DJhHfe+76mFL3dxsJbXRIyZOLRKltS2k1arIy2PqVfrZl2A/CRo3T6bQ7M8SW/Sylsdo5UhKWzr10zBgdJSxlYMhFqVzWhR4SlrIylLI56MYv1aX0oYzjGE+n0+7MEFv2s20fQugiYencS8e0n0NvnjoSpuqT+n1LNCjdE6YiYalNSpGwtJ+t4yMiYemYMb5zCeUDS9236G+2vRfJbbuW5UrfO9lvdi6bw9p9USpVnb6flePI1SBXh5rsDLX72fre654wlxdIH1Pq/m4l1DkK7Tot4VrPsFbCXFYGu25v79g0zOpxUhEsJ2HtfjEue6R6fU3vWPZPSbj2ZCB1zBjvm1mi2+UYYC9ICO7slhCgF0gI7iAhuIOE4A4SgjtICO4gIbiDhOAOEoI7SAjuXCVkYXFeiITgBxKCO0gI7iAhuIOE4A4SgjtICO4gIbiDhOAOEoI7SAjuICG401VCnR8whNskSvcghL5ZAXqXB+t0kzCVzXQcx2RyoZ4g4fNTLaEVzCK5TzQ6PZv+XRZhLcOpznejo6vOnfL6+hpDCIucKbrONi+MOvlrXWxG0i2ZVWE/XSSszXBam61Vr5vneZEgSXIx63rpnIg2rW5OQv2lmaZpkXFLyl+rr/3SwT6KEuYSO9pLbI2EW3IU2uSSubx+cgKpxJxruQLtOct2trya+kIbD4uEW7K1WvF0UnXb2ekloS1vS32hjYfcE6by6m2R0B6ndDlOZY61ZdZISCR8HHfvHduORE22Vi2Mvl+TMrZIKNuO45jNPKqzzJbuCUuZaGE/XZ8T2nvI2jlIajoRuftR6aF++/btRgx7GS9lTbXlnc/n6vpCG10lBNgDEoI7SAjuICG4g4TgjruE+pHI2nZ2GcexenKZEqkypmmK0zTF8/m8+LdhL0r17nFOz8RTSXivt1tSD6a1eCLko0DCwoa552Jrb8HknrfZt1Zy8+3K8VMSpuZuy9Wjdg5hmUjRnn+pLexra/LCw9qzRvu7rv9R6Cphbp7f3NsoUq5+MCzHqJ1vNyXh1vl8U1N1WeTSnyP1to88dC+9iaOPmZqr+QgUJax9i2bLywJWhpxcqckJ7WL/J73lf9C5OuZm41y7JKemrK2dVbM0Q+kR6BoJS/P8CiUJ98y3u0XCmjmESxKuDVcYxzHO8xzneV7MjZw6d33M0lzNR+DuEtZGwr3z7dZKWCpfr7tcLrsioWwjl9zSDJtEwiVdesc1LyDk5hcWufbOt1sr4ZY5hPfcE+py7HFKb+LktkPCjaxJWJpjV7+1Uppvt8fluHYO4T29Y30Mfdmu7R3n5mo+Al0k/Gh4Pyc8GkiYQf/HpDTGBdpBQnAHCcEdJAR3kBDcQUJwBwnBHSQEd5AQ3EFCcAcJwR0kBHeQENxBQnAHCcEdJAR3kBDcQUJwBwnBnauELCyey//fX7r5PGEejgAAAABJRU5ErkJggg==">
</div>
```
class Person:
name = ""
phoneNumber = ""
email = ""
def __init__(self, nombre, numero, mail):
self.name = nombre;
self.phoneNumber = numero;
self.email = mail;
class Student (Person):
studentID = 0
promedioGeneral = 0
def __init__(self, nombre, telefono, email, studentID):
super().__init__(nombre, telefono, email)
self.studentID = studentID;
#---------------------------------Creación de personas y estudiantes------------------
john = Student("John Pérez", 302010, "[email protected]", 10010)
print(john.name)
```
En ese contexto, el operador `super` se refiere explícitamente a la superclase, mientras que el operador `this` se emplea para trabajar con los atributos y métodos de la subclase.
### Ejemplo:
Elabora una clase _padre_ `Animal` para construir clases hijas: `Oso`, `Tigre` y `Perro`. Puedes emplear el diagram UML como sugerencia.
<div align="center">
<img src="data:image/png;base64,iVBORw0KGgoAAAANSUhEUgAAAjEAAAE6CAYAAADnQAOqAAAHtnRFWHRteGZpbGUAJTNDbXhmaWxlJTIwaG9zdCUzRCUyMmFwcC5kaWFncmFtcy5uZXQlMjIlMjBtb2RpZmllZCUzRCUyMjIwMjEtMDctMjVUMjAlM0E0NCUzQTI3LjI3OVolMjIlMjBhZ2VudCUzRCUyMjUuMCUyMChXaW5kb3dzKSUyMiUyMGV0YWclM0QlMjJ1Y2J3aUxiZm1fTzQ0TGw5ajFhZiUyMiUyMHZlcnNpb24lM0QlMjIxNC45LjIlMjIlMjB0eXBlJTNEJTIyZGV2aWNlJTIyJTNFJTNDZGlhZ3JhbSUyMGlkJTNEJTIyd3BPaG93WlZLZVpOTTBYX0dLNlolMjIlMjBuYW1lJTNEJTIyUGFnZS0xJTIyJTNFN1ZwZGIlMkJJNEZQMDFrWFlmV3VVTENJOThUR2RINm1pclphU1ozWmVWU1F4WTQ4VElNU1gwMSUyQjkxN0JBU1F4c1lzdkNRcXFyaWsydkh2dWRlN3NtbGxqZUpzODhjclZkZldZU3A1ZHBSWm5sVHkzVUhnUXQlMkZKYkJUZ08lMkZhQ2xoeUVpbklLWUVaZWNNYUxNdzJKTUpweFZBd1JnVlpWOEdRSlFrT1JRVkRuTE50MVd6QmFQV3BhN1RFQmpBTEVUWFI3eVFTSzRVRzdxREUlMkY4Qmt1U3FlN1BTSDZrNk1DbU45a25TRklyWTlnTHhQbGpmaGpBbDFGV2NUVEtYdkNyJTJCb2VVOG43dTQzeG5FaW1reklrSXVmQmc5Zmhvc3Y2VHB6WXUlMkJmNzU4ZjlDcXZpRzcwZ1VjSmlSSFZXeGE3d2clMkZwbHNRVUpUQWFMMWdpWnZxT0EyTkV5VEtCNnhBMmdqa0FyNWdMQWk0YzZSdUNyUUVOVjRSR3oyakhObks3cVVEaHoySTBYakZPM21CWmVMQmFFMjV6b2FQQjdWY3Nabkltd0RhZ0hLZGc4MUw0d0tsQlgxRldNWHhHcWRCQXlDaEY2NVRNOThlSUVWJTJCU1pNeUVZTEUyMHM2QjQlMkJEc3BOZWRQWmVRQTVqRldQQWRtT2dKWGtHJTJGanYlMkJCSG03TFlITDZHbHRWQXNuWFFhd0RlTGxmdXVRWUxqVE5aMUR1R3BSYnJnZEF3dUk1QjJRRTF6UEJTYkswd1BPdWw0MXlUMGdUSEtGSUdjeDNBaHUzQllxUk5mR3NzY09VVmNRMjByJTJCSGRyV3dBc2VLbkc3T2Z1SUpvd3ppWjVvd0ZXZUUwaHBVaEJyRkMzRXkwTkkxQ21IM3o3bk4xQyUyQlJ2N1I3SmNSZzdvTG0lMkJiZ2lVWVFUR1NSTUlJRlVSRWo2MTR3a0luZCUyRmJ3eSUyRndOTEVmdXhaUGRqNEJNWk9PWVpmYWM3RmhDVndGa1R5NE1BUWJsc3NRNjVaSkozT1VETzhkRGhKMXpZSnAxNWIwZVFkaWFZYXg1VGszQ21PaXc5UjV5S0NZNkNLNHBMUmI1THc2WU5qc082WnJIdEhHS1pvanVrTFM0a2dUSzdQbFcyTiUyQlZ1Ukd6VDhyQWhhNHRZJTJGd2kwYzFsWiUyQjJJUUNtT3NTJTJCc3FjQjgwNEx4TCUyRjZxVDNETkwlMkZURmtuQjY0a0J4eSUyRktnY2E1N2hqMnkwUjNqY0lmNmpXOG45WlNNSmFSZSUyQnklMkZzeXM3MTJsakxlVzlZT3VqTGRHYnMlMkI5YlJrUG1wVHh2VVNYeVE5djNDRW42eEM4JTJCZHZ2a1BpdkRCN1FxZmpXSXFUZnNBajRiZFdBSXQzZnkzJTJCY1JDUFpVSUhSbkRKWmM4Y0E2YUxzOU5Yd2ljZ0g1MzVmaWJnbzN6Z2o0b2VFd2M5cTlMYzJrdGZUN0hDd0t3WUpIRXhQOG9JQ2tQT2NSN3NFeXNuNXFETDdCWE1DJTJGcEVpNUtCJTJCNDhqbyUyQk5TcU54eWJiWGlJUDFaSklFdVclMkJEM2UlMkZlTzhIeU9hWTRvRWVhM3U3UmpUZXJrWEdkZW5HdzM3dmxPeGhEcVRublhZSjZvdDVBY2ZMS1FPYlN3RXdZRjJCMlk2NzA1dTJLMXYyTFBmM1pkWGwwNGYyUHQyJTJGejE3dUZBN0xyTm1UOUF2SkpMNVB2eU5MTG1wbURvQmZXRSUyRnpmUHZUVUM3NW51eUxLS2hLbnFIN2JTdWJKNVpObFU2M2E5c2RzMjM1VTQzWDQzZVd3dG4xM3cxN3BUem5YMEUzRnc2ZThOMnBYTXBnJTJGMmhYNVhCanROclV3YiUyRkw3TFZVR25CcGJMVjdzTjd3YkQ4cWE3ckJQN2pjTmhJeUY1TEN4WWhkeEFaUUFYdm1xblgwb0s5JTJCMnVtJTJCdVlYNnJJc2NQU0dvQnAwT3ZEQ0l1RGJ4d1BoWG5TZzM2QiUyRjB1bkFTJTJCbTl0UTcwajMzSjNlbkF1JTJGb0l1TGtPOUk5OVc5NjFVTThudEFXUmFiUWFmMGxrRGc1RXBxa3hnOERlJTJGOVRLelpVYXAlMkYwekc2ZFhib1RDc1B5blJXVmUlMkZ1ZW45JTJCayUyRiUzQyUyRmRpYWdyYW0lM0UlM0MlMkZteGZpbGUlM0VXDPYSAAAgAElEQVR4nO3dTa8cR9XA8bKdbB7Yxg47S20gYPM52p8Ag8QaqaVI7FBYATEC9S4S8pplyxISxr5WbCfAoj8AW2PSC0R4VxRFQoiA4TwLU+Oauv02Uz31+v9JrTh3ZmrO9D1n6tzqnmklQEaUUmxsQTcA/lBxyAqTCEIi/wC/qDhkhUkEIZF/gF9UHLLCJIKQyD/ALyoOWWESQUjkH+AXFYesMIkgJPIP8IuKQ1aYRBAS+Qf4RcUhK0wiCIn8A/yi4pAVJhGERP4BflFxyErJk0jbtqKUkrquVz+maRpRSskwDJvFcYoxU1Fy/gEhUHHISsmTSF3Xu2+NDdlA0MQA8IWKQ1ZKnUSGYRCl1K6BaNt2d5v+mf6vUkr6vt+7bRgGqapKqqraa4a6rtv9217hMb9q37yNJgaAL1QcslLqJKIPJZnNiKabiq7rRET2mg67idENjm6K9P7U4+vmp65raZpGRGTX6Iw1RqUpNf+AUKg4ZKXUScRsXOyGw24q9GqLfZs5hrmyI/KyUdGNkGauxujbaGIA+ELFISslTiJ93+8dQtINiP7/sSZGNyvHNjH6kFPXdeduo4kB4AsVh6yUOImY57qY21ijIuLexCw1ODQxAHyh4pCVEicRs2HRzENKp1iJMc+rMVdl7DFLU2L+ASFRcchKaZOIbi7MTyOJvDzE1DTNSZoY3STpZmbu8FVJSss/IDQqDllhEkFI5B/gFxWHrDCJICTyD/CLikNWmEQQEvkH+EXFIStMIgiJ/AP8ouKQFSYRhET+AX5RccgKkwhCIv8Av6g4ZIVJBCGRf4BfVByywiSCkMg/wC8qDlm5evXq6Ffws7H52K5evRq6BICi0MQgK0qR0giH/AP8ouKQFSYRhET+AX5RccgKkwhCIv8Av6g4ZIVJBCGRf4BfVByywiSCkMg/wC8qDllhEkFI5B/gFxWHrDCJICTyD/CLikNWmEQQEvkH+EXFIStLk0hd19L3vQzDIFVVeYpqX9/3opSSYRhO+jz2F7HZt/V9PxkbjsO+A/yi4pCVpUlENy5930td1z5COsdHE1NVlbRtu/v/tm339s1UEwM3NDGAX1QcsjI1iUx9Tbw50Yu8bDCaptndx5zsh2EYXd3Qj9PNgm5S9L+bppl9Dt3Q6Nvqut6NP/Wc+nXZzZC+v/1zpZR0XSdVVe3Gunv37t7zmSsxS/ui67q91xdqZSsmNDGAX1QcsjI3iXRdt2sm9GElm9mMiIg0TbO3YmOucJi32Y/TTcEwDOdWXszmQOTFKom5QqSbDfM16f+345min98cx6QbEvv5xpqYqX1hNjVVVdHECE0M4BsVh6zMTSJt2+4m5KqqRg/n2A2HXrkwb9PMFQ/7cXMTvn3fuXH6vt9rDqZWWcaYKyVjjZHZxNgN1tK+MP899v+lookB/KLikJWtDidNTdynaGLM28eefyzuQ8+n0ePqGGhiToMmBvCLikNW5iaRNSf1xr4Ss0bXdaOvr6qq3WoMTcxp0MQAflFxyMrUJGJ+pNo8rGSbm7hFzp8TYzc4hzQxepyxc2LsVRrdfNirQXP7wTx8pB+nx3VtYuzXVNc1TYzQxAC+UXHIytQksuakXpHliXvp00mHNDHmp36mxhl7TrvBmTq0NHcISp/4e+fOnaObGD6ddB5NDOAXFYesMImE0TTN3sfIS0X+AX5RccgKk4gfepVGb6zCvED+AX5RccgKkwhCIv8Av6g4ZIVJBCGRf4BfVByywiSCkMg/wC8qDllZmkS2vIr1sVd8XvO4Yy4SaX4CCmHQxAB+UXHIytIksuVVrGliYKOJAfyi4pAV18sOiMxfNdr8VI7+nhdt7lpFc48ba1bmrnRd1/Ve3OaVs/X95l4DTod9DfhFxSErc5PI2i+8MxuQsS+tM2/Tz2dfmNH8Ft65x02Zu9K1fVkBfTFL+9pIh175Gu5oYgC/qDhkZW4SWXsV66mrRtuHgeYOC81dc+mYw0l2k2SuuOgGxbyUwLFXvoYbmhjALyoOWXE9nDR31eipq1hr5qEf8wvglh43Zu5K1/q5uq7ba8z07Vtd+RqHo4kB/KLikJW5SWTtVaynPrU0t6Ji33bqlZi+73eHicYu6sg36IZBEwP4RcUhK1OTyNqrWOsxpq4abV/FWt82dr+5q1+vbWLGrnRtxjl1Vem514DTYT8DflFxyMrcOSprTuoVmb9qtHmb3YxUVbW7bWolxeXTSbamafaaMX1lav09OBxK8o8mBvCLikNWSppEaE7iU1L+ATGg4pCVEiYR++PXiEcJ+QfEhIpDVphEEBL5B/hFxSErTCIIifwD/KLikJUUJxF92QD9sWmuf5SuFPMPSBkVh6wsTSJrrmJtflTZBx2H/nTTlo69SOXYGFPfcMzE/RL7AvCLikNWliaRNV9457uJOSWaGL/YF4BfVByy4nrZAfO7XsxLDehNf4Gc+WV05v3N74IxLY1jfh+M2UDNXY16qbEY+06aqfHsRmXsG4fHrqhtNzGlXz27xNcMhETFIStzk8ghV7E2vzBu7MrU9jfq6i+aMy8UOXXJgLlx7CtO29/0u+Zq1GaTNPaFfGPjrW1ipuKfeu7Srp5NEwP4RcUhK3OTyJqrWOsxxhqcsYldj2FP2HOHpObGmbvm0pqrUR9yfaexK3TPxTHWlNnjl371bJoYwC8qDllxPZyk72teLdq8/7FNzNpxzOZh6urXc03B3BWz58Y7pIkxX5/ZxHD1bJoYwDcqDlmZm0TWnNSrx7AnaJHjV2IOGSfUSow99hYrMSWiiQH8ouKQlalJ5NCrWPd9P3tl6kOamEPGMZsHfV/zHJY1TcLcFbOnxtONiY65ruvJ83bWnhNT4tWzS3u9QGhUHLIyNYkcchVr82rQU1emPvRw0tpx7CbmmE8nzV0xe24885NWZoMzd0XtpU8nlXQoSYQmBvCNikNWmEQQEvkH+EXFIStMIgiJ/AP8ouKQFSYRhET+AX5RccgKkwhCIv8Av6g4ZIVJBCGRf4BfVByywiSCkMg/wC8qDllhEkFI5B/gFxWHrDCJICTyD/CLikNWmEQQEvkH+EXFIStMIgiJ/AP8ouKQFSYRhET+AX5RccgKkwhCIv8Av6g4ZMW8+CAbW4gNgD9UHLLCJIKQyD/ALyoOWWESQUjkH+AXFYesMIkgJPIP8IuKQ1aYRBAS+Qf4RcUhK0wiCIn8A/yi4pAVJhGERP4BflFxyAqTCEIi/wC/qDhkhUkEIZF/gF9UHLLCJIKQyD/ALyoOWWESQUjkH+AXFYesMIkgJPIP8IuKQ1aYRBAS+Qf4RcUhK1tNIn3f713Ur2maTcado5SSvu+jHQ/LaGIAv6g4ZGWLSaTrOlFKyTAMu5/VdS1t2zqPPYcmJn00MYBfVByysmYSsRsUW1VV0nXd3s+GYdgbW/+/3jS9gtM0ze42s5Go63p0daeqqt3P7969K0qp3X31mDrmruukqqpzr8mMxRxvGIbFePVzwQ37EPCLikNWXJsYPdnPNTkiL5oEvTLTNI3UdS0iL5uCsdu6rtv9W49hNji6YdFj6EZqqYkxm662bXfNkdlALcVrN204Dk0M4BcVh6xMTSL6EJG92YeI1jQxeuIfe8xcwzG2gmLHbjYxeoy5Me1YdDxj462JF25oYgC/qDhkxcdKjG6Ixh6ztGrStu3kycJbNTH2eIfECzc0MYBfVByycupzYsyJf+62ufNXzOeZO5ykx7Abq0ObGFZi/KGJAfyi4pCVU346yT4R1zzHxG4qxhoO83wVPcYhTYy+b13Xe42ReVvbtrvb5s6JmYoXbmhiAL+oOGRlq0nEPofGPvSz9GmfuZNwp87H0Z8QunPnzrnGwj4MZY45FYv56aa18cINTQzgFxWHrDCJICTyD/CLikNWmEQQEvkH+EXFIStMIgiJ/AP8ouKQFSYRhET+AX5RccgKkwhCIv8Av6g4ZIVJBCGRf4BfVByywiSCkMg/wC8qDllhEkFI5B/gFxWHrIxd5JGNzecGwB8qDsCi69evhw4BAM6hiQEw69NPPxWllNy7dy90KACwhyYGwKzr16/LhQsX5LXXXgsdCgDsoYkBMEmvwnzjG98QpZTcv38/dEgAsEMTA2DS9evX5f/+7/9EROT1119nNQZAVGhiAIzSqzDf/e53RUTk8ePHopSSBw8ehA0MAP6HJgbAqC9/+cu7VRjtypUrcvny5UARAcA+mhgA5/zzn/8UpZR873vf2/v5u+++K0opefjwYaDIAOAlmhgA53zpS1+Sz3zmM6O3XblyRa5cueI5IgA4jyYGwJ5//OMfopSS73//+6O3P3z4UJRS8u6773qODAD20cQA2PPGG29MrsJoly9fZjUGQHA0MQB2/v73v4tSSm7fvj17v7OzM1FKyaNHjzxFBgDn0cQA2HnjjTfks5/97Kr7Xr58WV5//fUTRwQA02hiAIiIyCeffCJKKfnBD36w6v73798XpZQ8efLkxJEBwDiaGAAiIvLFL35x9SqM9tprr7EaAyAYmhgA8vHHH4tSSn74wx8e9Lh79+6JUkref//9E0UGANNoYgDIF77whYNXYTRWYwCEQhMDFO6jjz4SpZT86Ec/OurxP/vZz0QpJb/85S83jgwA5tHEAIX7/Oc/L0op5+1zn/tc6JcCoDA0MUDBPvzwQ/nVr34lv/jFL+T999+X9957T548eSJPnjyRx48fy6NHj+TRo0eilJKf/OQn8vDhQzk7O5MHDx7IgwcP5P79+/Lzn/9c7t27J/fu3ZP33nsv9EsCUBCaGACLlFLyu9/9LnQYALCHJgbAIpoYADGiiQGwSCklv//970OHAQB7aGIALKKJARAjmhgAi5RS8uGHH4YOAwD20MQAWKSUkj/84Q+hwwCAPTQxABYppeSPf/xj6DAAYA9NDIBFSin505/+FDoMANhDEwNgkVJK/vznP4cOAwD20MQAWKSUkr/85S+hwwCAPTQxABYppeSvf/1r6DAAYA9NDIBFSin529/+FjoMANhDEwNgkVJKPvroo9BhAMAemhgAi2hiAMSIJgbAIqWUfPzxx6HDAIA9NDEAFiml5JNPPgkdBgDsoYkBMOuDDz4QpZT8+te/Dh0KAOyhiQEw69GjR6KUkp/+9KehQwGAPTQxAGb9+Mc/lgsXLkjbtqFDAYA9NDEAZn3rW9+Sixcvyje/+c3QoQDAHpoYAJM++OADuXjxoly6dEkuXrwov/nNb0KHBAA7NDEAJn3961+Xt99+W1599VV5++235atf/WrokABghyYGwKjvfOc7cv36dRERefXVV+XTTz+Vr3zlK/Ltb387cGQA8AJNDIA9v/3tb+VrX/uaXL9+XR4/fiwiL5qYf/3rX/LkyRO5ceOG3Lp1S549exY4UgClo4kBCvb8+XN5+vSpnJ2dyTvvvCNvvvmmKKXk9u3be/fTTYx2+/ZtUUrJm2++Ke+8846cnZ3J06dP5fnz575fAoCC0cScwLNnz3YTws2bN+XatWuilGJji3K7du2a3Lx5c9eQjK2w2E0Mec7GNr+tqSu4o4nZGH+hIkevvPLKuSYGwLi1K5xwRxOzkcePH3OuALJFEwO4efbsmdy6dUtu3LixO9cM7mhiNnLjxg156623QocBnARNDLCNt956S27cuBE6jGzQxGzg9u3bcuvWrdBhACdDEwNs59atWxxa2ghNjKNnz56JUopDSMgaTQywHeaN7dDEONInbQE5o4kBtqU//AE3NDGOSESU4JVXXpF///vfocMAssEfwNugiXF08+ZNOTs7Cx0GcFI0McC2zs7O5ObNm6HDSB5NjKNr167J06dPQ4cBnBRNDLCtp0+fyrVr10KHkTyaGEdKKb7IDtmjiQG29fz5c1GKKdgVe9ARSYgS0MQA22P+cMcedEQSogSXLl2iiQE2xvzhjj3oiCRECWhigO0xf7hjDzoiCVECmhhge8wf7tiDjkhClIAmBtge84c79qAjkhAloIkBtsf84Y496IgkRAkuXbrEVwkAG2P+cMcedEQSogQ0McD2mD/csQcdkYQoAU0MsD3mD3fsQUckIUpAEwNsj/nDHXvQEUmIEtDEANtj/nDHHnREEqIENDHA9pg/3LEHHZGEKMHFixdpYoCNMX+4Yw86IglRApoYYHvMH+7Yg45IQpSAJgbYHvOHO/agI5IQJbh48aL85z//CR0GkBXmD3fsQUckIUpAEwNsj/nDHXvQEUmIEtDEANtj/nDHHnREEqIENDHA9pg/3LEHHZGEKAFNDLA95g937EFHJCFKQBMDbI/5wx170BFJiBLQxADbY/5wxx50RBKiBDQxwPaYP9yxBx2RhCjBhQsXaGKAjTF/uGMPOiIJUQKaGGB7zB/u2IOOSEKU4MKFC/Lf//43dBhAVpg/3LEHHZGEKAFNDLA95g937EFHJCFKQBMDbI/5wx170BFJiBLQxADbY/5wxx50RBKiBDQxwPaYP9yxBx2RhCgBTQywPeYPd+xBRyQhSkATA2yP+cMde9ARSYgS0MQA22P+cMcedEQSogQ0McD2mD/csQcdkYQoAXkObI+6cscedEQSogTkObA96sode9ARSYgSkOfA9qgrd+xBRyQhSkCeA9ujrtyxBx2RhCgBeQ5sj7pyp5RSwsYWcotB6H3Axpai0PuMjU0plWbxIA+x5F8scaBMqeZfqnEjDzQxCC6W/IslDpQp1fxLNW7kgSYGwcWSf7HEgTKlmn+pxo080MQguFjyL5Y4UKZU8y/VuJEHmhgEF0v+xRIHypRq/qUaN/JAE4PgYsm/WOJAmVLNv1TjRh5oYhBcLPkXSxwoU6r5l2rcyANNDIKLJf9iiQNlSjX/Uo0beaCJWWB/qU7f96FDyk4s+RdLHCF1XTf5hVJt20rTNKKUkmEYQoeanVTzL9W4tzRVLzg9mpgJwzCIUkrqut79TL/Bd10XMLL8xJJ/scQRi7qu2ScepbqvU417S/ZcoZt9/ug9PZqYCToJl35udt5N0+zdt23b3W1VVZ085lTFkn+xxBGLsSbGXompqmqX33ozH6v/q9/M9f35Y+C8VPMv1bi3ZDcxfd/vrcaYK5zmXKDvZ9bL2M/M56F+9tHETKiqai95NJ2Mfd/vNTS6YdFv1vb/m2/w2BdL/sUSRyyWmhjz3/qN125izDdaczyzjvBCqvmXatxbmmti7IbGnFvsupn6mf0cHBV4iSZmgp2Umvnmq9+Ux84RsJsg3rSnxZJ/scQRi6Umxs7xsZUYsy7s1cqx1cuSpZp/qca9pbnDSfoPWl0L5v/rhsWsg7Gfjc0fU39ol4YmZsJUgpgrLDrZzE3jfJr1Ysm/WOKIxVITY+f4WBOj6fvbG03MS6nmX6pxb2kst/V7va4ZezObGPMk4LGf2Sv7IqzuazQxE6bOiZk62VEf69dvyqzErBdL/sUSRyy2WIkx0bTMSzX/Uo17S1Mr9yJybiXGtLaJYSVmGk3MhLlPJ5nHNvWbtr6/vo1zYtaLJf9iiSMWW5wTMzWevj8rky+lmn+pxr2luSbGbkrqut7Vydomxn4OVvZfoolZYC8B2isp5m12EpvLiDQw02LJv1jiiMWaTyeZuW/+Zbi0Yjn2Jl26VPMv1bi3NNfEiJz//iXtkCbGPiRLA/MCTQyCiyX/YokjFXajwuEiN6nmX6pxIw80MQgulvyLJY5UjJ2si+Oluv9SjRt5oIlBcLHkXyxxoEyp5l+qcSMPNDEILpb8iyUOlCnV/Es1buSBJgbBxZJ/scSBMqWaf6nGjTzQxCC4WPIvljhQplTzL9W4kQeaGAQXS/7FEgfKlGr+pRo38kATg+Biyb9Y4kCZUs2/VONGHpRSoq5evXruo5JsbL62q1evhq4DERGhDthCbrHUwaGoG7aQ2//yj04a4cSSf7HEgTKlmn+pxo08/K+ZIQkRTiz5F0scKFOq+Zdq3MgDTQyCiyX/YokDZUo1/1KNG3mgiUFwseRfLHGgTKnmX6pxIw80MQgulvyLJQ6UKdX8SzVu5IEmBsHFkn+xxIEypZp/qcaNPNDEILhY8i+WOFCmVPMv1biRh6SamLZtRSklfd9L0zTS933okLCBWPIvljhQplTzL9W4kQenJkY3FL5UVbX7b6jC6fvey3Nv8Tx6jGEYTjL+VkqL41T73v4SKPu2sVqNKQ9Kl+rvIYW4qY18JdXElIQmxr+Um5iqqqRt293/61VLjVqNXyx1cKjY46Y28nZ0E6NXQ/Qk2XXdXqfbdZ2IvHzD1omj76//3TTN3rhL4zRNs7vNTDxzzENez9Lj7NvGJqCxx8+Nu9RYmK9zaTy7Uem6brdiNbbP9P3s13Hs/ttCLG+CLnEckgNr972+X13Xu5+P5Y5+vP1zXT9mrd69e3dvTDOWpRoza7Npml2eYRux1MGhjol7LLen3vvNuUNvbdtO3l/HNAwDtVGATVZi7ERp2/bcRKo7YZ0gwzCcm4APGadpGqnreheL2W3bt82Ze1xVVXuFpM/DMffX2H2OjccsRLuJmRpvbROj47Nvm3rutftvC7G8eR8bx6E5MJZDc/cz35yn6Lqauq+uVXvMsTfqqbw137irquKNemOx1MGhXJoYnYdz7/1jz6ebhjX3pzbytkkTYxubSHWyzf3yDxln7DZtqvu2zT1ubMXF/vma+6yNx37M3POMxTm3X8aaRHt8s+jW7r+txPLm7fJmbFrKgWN/t0uW/jI136jHVuTmcsn899j/w10sdXAol7qZyu2p/JpqRpbykdrI12wTYy6d2Yd99IN1A2LeVyl1dBOzdhw7iY5pYuYeN3XewlITc2w8U49ZG+eaJkbkfMGa49sbTcwLc3VwTA6M7fu5+x3ze9CP1bXFG3X8YqmDQ7nUjZnbU+/9mrniueb+U6iNvGyyEmO/kR+7EnPIOCFWYuzHrLnP2niOXYmxx95iJca3WN68t1qJGfv5KVdiuq4bPfxnvunzRh2/WOrgUFusxMy994u8OCRknpy7dH/z59RG3jZpYuy/Js1jgoc0MYeMYyeKfV7B2iSae5wZmz7mOnYuiX2fY+OxH2Pvi7Hx9KSnY6jrevI8orXnxNi/h1OL5c3btQ5E1uXA0jkxcytpczGYf6Xa5wy4vlHbr9PMM2wjljo41BZNzNx7f9M0kx8AGbv/WHzURr6cmhjzLG7zLG/zF37o4aS149hJdIpPJ43dNvWXs/34Yz6dZH9qa+145tn7Y5OgueyqLb0OX4eSROJ58z42jkNzYG0OTR0OnPrdmGPY99O1eufOnaPfqPkExmnFUgeH2qKJEZl+7zd/buff2P11THbdUBt5cmpigC3Ekn+xxJGCsb+O4SbV/Es17lOhNvzKuomx/8I1NzrleMSSf7HEESP9lyj1czqp5l+qcW+F2ggr6yYGaYgl/2KJA2VKNf9SjRt5oIlBcLHkXyxxoEyp5l+qcSMPNDEILpb8iyUOlCnV/Es1buSBJgbBxZJ/scSBMqWaf6nGjTzQxCC4WPIvljhQplTzL9W4kQeaGAQXS/7FEgfKlGr+pRo38kATg+Biyb9Y4kCZUs2/VONGHmhiEFws+RdLHChTqvmXatzIA00Mgosl/2KJA2VKNf9SjRt5oIlBcLHkXyxxoEyp5l+qcSMPuyaGjS3kFoPQ+4CNLUWh9xkbm1IqzeJBHmLJv1jiQJlSzb9U40YeaGIQXCz5F0scKFOq+Zdq3MgDTQyCiyX/YokDZUo1/1KNG3mgiUFwseRfLHGgTKnmX6pxIw80MQgulvyLJQ6UKdX8SzVu5IEmBsHFkn+xxIEypZp/qcaNPNDEILhY8i+WOFCmVPMv1biRB5oYBBdL/sUSB8qUav6lGjfyQBOD4GLJv1jiQJlSzb9U40YeaGIQXCz5F0scKFOq+Zdq3MgDTQyCiyX/YokDZUo1/1KNG3mgiUFwseRfLHGgTKnmX6pxIw+bNjF93+9dlKlpmk3GnaOUkr7vox0Py2J5E6QOTjcelsVSB4eibk43HpZt1sR0XSdKKRmGYfezuq6lbVvnseeQhOmL5c2bOjjdeFgWSx0ciro53XhYtrqJsRPMVlWVdF2397NhGPYSXP+/3jTdgTdNs7vNTIS6rke786qqdj+/e/euKKV299Vj6pi7rpOqqsZe/C4Wc7xhGBbj1c8FN7HsQ+qAOggp1X1I3VA3IW3SxOhf1lySirz4JevOumkaqetaRF7+Usdu67pu9289hpmgOuH0GLoQlpLQLJq2bXfJbRbAUrx20eE4sRQydUAdhBRLHRyKuqFuQpptYvQSn73ZS3xrklD/4sYeM5cwYx2w/QLMJNRjzI1px6LjGRtvTbxwE8ubN3VAHYQUSx0cirqhbkLythKjE3rsMUtdb9u2o8uBOq4tktAe75B44SaWN2/q4Px41IE/sdTBoaib8+NRN/5s0sSIzB/TNH9xc7fNHX80n2duOVCPYRfGoUlIJ+1PLG/e1IFMjrcmXriJpQ4ORd3I5Hhr4oWb1U3Mkqmzy+0TqcxjhHZSjCWMebxRj3FIEur71nW9l9jmbW3b7m4zf742XriJ5c2bOqAOQoqlDg5F3VA3IW3WxIicPwZqL93pxNCbtuYkKv0Y+3iqPsP7zp075xLDXkY0x5yKxTw7fW28cBPLmzd1QB2EFEsdHIq6oW5C2rSJAY4RS/7FEgfKlGr+pRo38kATg+Biyb9Y4kCZUs2/VONGHmhiEFws+RdLHChTqvmXatzIA00Mgosl/2KJA2VKNf9SjRt5oIlBcLHkXyxxoEyp5l+qcSMPmzUxc5+d35rrc20Za9u20rat9H2/9/XWLubi87mffYnl9VAHx6MO3KX6eqib41E37pJsYmIxDMNe4umEPKUc93Msr4c6OA51sI1UXw91cxzqZhtOTYzeIfpz9OY4U5+RF5m+qqgV1O5x+nnsK4yat5mf6bc/fz813lKs+n5TV0dtmubct/O4gj4AAAKRSURBVEyaj9ex2JeU1xcDW3reNfs5B7G8HuqAOggp1ddD3VA3ITk1MUq9vBKnvXPs29ZeVXTs6qD6l2BfYdT8t05m89sTl8Yz73Po1VH1a7TVdb33eqZes/5CprnnXbOfcxDL66EOqIOQUn091A11E9LRTcxUt6f/PfbthsMwzF7XYmypa+xaE2NJOHbNi6Xxxp5z7TU57NeojS0JmmOaX2e99LxL+zkXsbwe6oA6CCnV10PdUDchzTYx5jKYvWw3dZVO8zZ707/MqauKTu3gQ5JQv6i+P38RrrHnOfbqqHNJOLbEWde1dF0nXdftlhHXJOHcfs5FLK+HOqAOQkr19VA31E1Is03MnEM66TnmcuAWSbjUSU89XlubhMMwrO6k9c/1kl/XdXTShlheD3VAHYSU6uuhbqibkI5uYkTOHws0x1Hq5XE4sxtce1VRfV99+1IS6jjsY5pz4029jjVXR9Vj28aOaZpjHfK8a/ZzDmJ5PdQBdRBSqq+HuqFuQnJqYsyzo5fOLjeX6+auKjp2xvWaJDTPWF873tx9xp7XTsK1Z5ebr9sswLVnl8/t5xzE8nqoA+ogpFRfD3VD3YTk1MTEYOyYpi/D4P9z/jmKJf9iieMY1EH6Us2/VOMWoW5yQBPjyPzGxbXHcbEvlvyLJY5jUAfpSzX/Uo1bhLrJQfJNDNIXS/7FEgfKlGr+pRo38kATg+Biyb9Y4kCZUs2/VONGHmhiEFws+RdLHChTqvmXatzIA00Mgosl/2KJA2VKNf9SjRt5oIlBcLHkXyxxoEyp5l+qcSMPNDEILpb8iyUOlCnV/Es1buRh18SwsYXcYhB6H7CxpSj0PmNjS7NyAABA8WhiAABAkmhiAABAkv4frFRxkImXgLsAAAAASUVORK5CYII=">
</div>
```
#---------------------Clase padre---------------------------
class Animal:
nombre=""
edad=0
tamaño=0
def __init__(self, nombre, edad, tamaño):
self.nombre = nombre;
self.edad = edad;
self.tamaño = tamaño;
#------------------Clases hijas----------------------------
class Oso (Animal):
tamaño_ocico=0
def __init__(self, nombre, edad, tamaño, tamaño_ocico):
super().__init__(nombre, edad, tamaño);
self.tamaño_ocico = tamaño_ocico;
self.descripcion();
def descripcion(self):
print(self.nombre + " es un oso de " + str(self.edad) +" años y tiene un ocico de " + str(self.tamaño_ocico) + " metros.");
class Tigre (Animal):
color=""
def __init__(self, nombre, edad, tamaño, color):
super().__init__(nombre, edad, tamaño);
self.color = color;
self.descripcion();
def descripcion(self):
print(self.nombre + " es un tigre de " + str(self.edad) +" años y tiene un color " + str(self.color));
#-----------------------------Ejemplos----------------------------------------
Tigre = Tigre("Malo", 3, 2.2, "Amarillo");
```
## __2. Polimorfismo__
_Polimorfismo_ se deriva de las palabras griegas "poli", que significa muchos, y "morphe", que significa formas. En programación, se emplea este concepto para cambiar el contenido de un método heredado para que se ajuste a las necesidades principales de las subclases.
Existen dos tipos de polimorfismo: _dinámico_ y _estático_.
### 2.1. Polimorfismo dinámico
Se conoce también en la literatura como _polimorfismo en tiempo real_, _vinculación dinámica_ o _anulación de método_ ("overriding", por su traducción al inglés). La multiplicidad de formas ocurren en diferentes clases.
Supongamos que en la relación de herencia entre una clase padre e hija existen métodos con el mismo nombre, pero en diferentes formas. Cuando un objeto es asignado a una _referencia de clase_ y el método del objeto es llamado, el método del objeto de la clase se ejecuta; más no el de la clase referencia.
Dado que la creación del objeto ocurre en tiempo real, la forma en como se ejecuta el método sólo puede ser decidido cuando se ejecuta el método.
Por ejemplo: una figura geométrica tiene un área y un perímetro; pero la forma de la figura define la manera en cómo se calcula.
```
%matplotlib inline
from math import pi
import matplotlib.pyplot as plt
#CLASE PADRE
class FiguraGeometrica:
dimensiones = []
ubicacionEspacial = [0,0]
fig, ax = plt.subplots()
def Area(self):
return None
def Perimetro(self):
return None
#CLASES HJIAS
class Circulo(FiguraGeometrica):
def __init__(self, radio, ubicacion = [0,0]):
self.dimensiones.append(radio)
self.ubicacionEspacial = ubicacion
#Figura
self.ax.add_patch(plt.Circle(tuple(self.ubicacionEspacial), radio))
def Area(self):
return pi*self.dimensiones[0]**2
def Perimetro(self):
return 2*pi*self.dimensiones[0]
class Rectangulo(FiguraGeometrica):
def __init__(self, b, h, ubicacion=[0,0]):
self.dimensiones.append(b)
self.dimensiones.append(h)
self.ubicacionEspacial = ubicacion
def Area(self):
return self.dimensiones[0]*self.dimensiones[1]
def Perimetro(self):
return 2*(self.dimensiones[0]+self.dimensiones[1])
#---------------------CREACIÓN DE FIGURAS-----------------------
cir1 = Circulo(0.2)
cir2 = Circulo(0.4, [0.6,0])
rec1 = Rectangulo(3,4)
print("Círculo de radio " + str(cir1.dimensiones[0]) + " tiene un área de " + str(cir1.Area()))
print("Rectángulo de " + str(rec1.dimensiones[0]) + " de base por " + str(rec1.dimensiones[1]) + " de altura tiene un área de " + str(rec1.Area()))
```
### 2.2. Polimorfismo estático
Conocido también como _polimorfismo en tiempo de compilación_, _vinculación estática_ o _sobrecarga de métodos_. Consiste en tener múltiples métodos con el mismo nombre pero diferentes argumentos de entrada. Se escoge el método dependiendo de cuántas entradas pase el usuario. Por ejemplo: en una calculadora, es posible sumar dos o tres números.
```
class Calculadora:
def suma(self, x=0, y=0, z=0):
return x+y+z
calc = Calculadora();
print("Suma de dos números: " + str(calc.suma(5,10)));
print("Suma de tres números: " + str(calc.suma(5,10,4)));
```
El constructor es un método que permite realizar un desarrollo particular en el momento en que se crea un objeto. También es posible sobrecargar el constructor de una clase dependiendo de las entradas que ingresa un usuario.
```
class Persona:
nombre = ""
cedula = 0
def __init__(self, nombre:str ="", cedula:int=0):
if cedula == 0 and nombre == "":
print("Se creó una persona desconocida")
elif nombre == "":
self.cedula = cedula
print("La persona está identificada por C.C." + str(self.cedula))
elif cedula == 0:
self.nombre = nombre
print("Se creó a " + self.nombre)
else:
self.nombre = nombre
self.cedula = cedula
print("Se creó a " + nombre + " con C.C." + str(cedula))
#Sin argumentos de entrada...
juan = Persona()
#Sólo el nombre
juan = Persona("Juan")
#Sólo la cédula
juan = Persona(cedula=1098)
#Toda la información solicitada
args = {"nombre":"Juan", "cedula":1098}
juan = Persona(**args)
```
## __3. Ejercicios__
### 3.1. _Toppings_
Eres el dueño de una franquicia de Zirus Pizza. Establece una relación de herencia en donde puedas definir tipos de pizza con base en el sabor de la masa base (integral, harina, etc). Puedes guiarte del diagrama UML.
<div align="center">
<img src="data:image/png;base64,iVBORw0KGgoAAAANSUhEUgAAAa8AAAE6CAYAAAChwN3xAAAH8nRFWHRteGZpbGUAJTNDbXhmaWxlJTIwaG9zdCUzRCUyMmFwcC5kaWFncmFtcy5uZXQlMjIlMjBtb2RpZmllZCUzRCUyMjIwMjEtMDctMjhUMDIlM0E1OSUzQTI5LjA2MlolMjIlMjBhZ2VudCUzRCUyMjUuMCUyMChXaW5kb3dzKSUyMiUyMGV0YWclM0QlMjJKaWliTW5yTWoyYkhRTlRNWFI5dSUyMiUyMHZlcnNpb24lM0QlMjIxNC45LjIlMjIlMjB0eXBlJTNEJTIyZGV2aWNlJTIyJTNFJTNDZGlhZ3JhbSUyMGlkJTNEJTIyZU9sX2c3MS1US2FNdlFYZ0pKbWslMjIlMjBuYW1lJTNEJTIyUGFnZS0xJTIyJTNFN1ZwdGIlMkJvMkdQMDFrYlpKSUp5RUFCOTU2ZDJWMWtwc1ZOdnVweXVUdU1TN1RwdzVwa0IlMkYlMkZSNG5ObmtER2pxaWZxRkNWWHo4RXZzNXg0OVBBcFl6aiUyRmElMkZDcHlFVHp3Z3pMSUh3ZDV5RnBadGo4WTIlMkZGZkFJUWRjZTVBREcwR0RIRUlGc0tKdlJJT20yWllHSkswMGxKd3pTWk1xNlBNNEpyNnNZRmdJdnFzMmUlMkJHc2V0Y0ViMGdEV1BtWU5kRyUyRmFDRERIQjNib3dMJTJGU3VnbU5IZEczaVN2aWJCcHJGZVNoampndXhMa1BGak9YSEF1ODZ0b1B5ZE14YzdFSmUlMkYzNVV6dGNXS0N4TEpOQiUyRm43Yk9zdXhyJTJCSjFUJTJGdXYlMkZIWDRYYzdTWHA2bEZmTXRuckJUempGZ0t4eFN2Uzg1Y0VFSTkzUmlPRVlTck1YSHN1VnJrRlF4b3h1WXJqMllUWkVBUEJLaEtRUXg2bXVrRHdCMUE4cEN4N3hnVyUyRlZuRk9KJTJGUiUyQm1OQXU1b0c4d0xHWjZUS2dXVWt2Qzlpb3RWcW9ud0FOQUJVbWh6ZElFQXRXZ0o3eXZOSHpFcWRTQXp4bkRTVXJYeDJWRVdHeG9QT05TOGtnMzBoR0M1WkQ5MmRDakk2R3dFUWlQaUJRSGFLSTcyQ090QWIwSnhycTRLeFNGUEkyRlpUVVo3V0N0NHMxeDZJSm91TkJjWDhHNzNlRGRzaDBBbm5HRXJibGp6UkMzbk9rTDQxaGFFSHJiMlUlMkJ6VURqWlZrdWxxcVZ4c3k3RmF3N3NUMWRTMEhoVHFTNWQxbVFGZ1pVWjNZTCUyRklIUE8xQWlMbU9jNm80elZJQ00xUmw3a1dhR2xDZlpoQm85Wm00VmJJSCUyRm84Q3FJUTE5WW85cVVJUTBDRWl1UmNJa2x6aFdoNkU4NHJETUwlMkYzQUdIeUJrUHVnUHJTRk1mQTVsVkpUaG81b0xPZWN4ckFYVFRCd0U1TFlqU25MdGxIUiUyQm16YmxwZVdrUXR0R1RxN2JrWnFjRTJxcWNjeG94bDNPc2NtazZFTUVSMEFWSXdXano0cndSUTgxV0hlYXJEc25HR1o0VGRpU3AxUlNyc1lYZWRzYTg1OUY3cWhscmhoM3hLMTdnbHRZN0NDUHc5YVh3Rng1YjZzNk9MRjlRUk1mNHZuVHo1QU5Yam5jNHA0QXV0UEl1R1VDc0RzU3liQWhraVY5ZThOM0E5R1JnYkRkdGc1aTJKV0Q4QnFVOTZBWTgyZ3R5Q2tEME11Y2U1SUFDbE9aUHVlWE9yVldteVdDUU9yNEUyS0RHemFqZDhHQTNMUEtsVmxsZUJOYjRYVmxLMFozVzlFWnVaTlB0aFhqRDlpS3JhU01CamlZaWEzRW1iR29aNEM3JTJCZmdFSmJVMEg4T3V6QWRxOGZoQjRtQ3EzZ1pCYWMyNE92bG5BR2xyZ0x5OCUyQklXcUcyZHhEMlZrVEFUWlUlMkZtM2dpSE9lZW1iYnFTdUYlMkZ0eTRXQUtNU3hNZHhwNEJzajY5U2VlYllDaWMxYXE5RjRTUVNFJTJCeWdxVlhBUUpHcSUyQnJhaDRDbHMyM3dpZnY4UW1uTVZnVmNvbDM5elR2cDRnV2hHRkpYNnR6TzhXMEhtN0pzNjFyJTJGSTFiOXplb1psdnlOZWxlNVpkY3RZRTg1NTJCOGtVM0JzcUVkMXpqJTJGOUJpODNISk9KMjdGNzZORng2Nk5ZcTlsZ2tJMWJWd3V3elVmUDY1Ykliemt5cjN1V2VPc2J2SnZkV2o4NWswZHFYTEhYYmxjbEh6U2VwdWMyOUdiOXRYN1YzNVhIVHFJZWFTMGIxdjllNjAwUEtnTUJ2dzltSm9QdldnQnMwbHE4cVRMTlpsTTFxWVZ0c2M2cVoxUUhIRTQlMkJBNXBISHR2RWV1QVVvV1YxbEpZenFBalpCdmVJelpRNEclMkJwNkMxUHRpUERobjFCOGdwZSUyQlMlMkJBJTJGdnFvayUyQkd3a2VON2xtRFlMN1UxS3c3dDNHb2p0MGZqQ2JGWDlXRFRKeiUyQlpJS0tXdTlqOXJYaGJTYTFVNmRyJTJCenBwQ3JUZiUyRiUyQlZjS2lvSjg5cXNkTmJhSHMlMkJuWFVnbFdVRTJVdmZjQ1p4a1dXY2JCeVM0blpsMDYwU09VQ05IdUtkU3hQVldFb3JGbCUyRjA1WWNVdkpweUglMkZ3QSUzRCUzQyUyRmRpYWdyYW0lM0UlM0MlMkZteGZpbGUlM0UN2GSpAAAgAElEQVR4nO3dW6gkRx3H8d4Iq3gJEgIRZPGE9kGFGBGCYgxKkAxRwaCyBjTCsvEywrpKCELEGHFROlEMwtHgDeKtPUIiEoOIQZTxwQjx8pANxIwPycOaByXoKgiJfx92a7ampqq6urqna6r7+4GBs2emu6una/t3qi//LoqiEF68Ur4AoK2CnQdSov8BiEF4ISn6H4AYhBeSov8BiEF4ISn6H4AYhBeSov8BiEF4ISn6H4AYhBeSov8BiEF4ISn6H4AYowivxWKxuuG1qqrV76uqWv2+ruvB2qG3AX5j6H8Ahje68JrNZqvfz2YzwmvHjaH/ARjeqMKrLEspikKWy6WIyNrvVHjpo7GiKGSxWKzmY76n5tM0ndmO+XxuDdPYZdd1vfp9WZb9fXE7YAz9D8DwRhVeKjTquvb+TgVDWZarcFkul2ujJj14fNPZ2qECRh+JdV22es+17FyNof8BGN6owquqKinLUubz+Woko0Yt+mFDfZRjBo0KPBvbdK52KGbYtF22+rwKPfPfuRtD/wMwvNGF13w+XwXGbDZbCy9zZFSW5VoI6efI9BBqms7WDkV9NnbZ+iFI12HFnI2h/wEY3ujCSz8/pP+7rutVEOiH7mwhpM9juVwGT+cbecUue2wjLdMY+h+A4Y0uvNT5I3VBhB5eKggWi8XGSMg8vKjCRkS809naoabT2xW7bDMQZ7PZqC7aGEP/AzC80YWXiKyuMBTZDAb90Js6VKf4rgb0TWe2w3W1Yeyy9dHYGLaXbmzrA2AYowgv5Iv+ByAG4YWk6H8AYhBeSIr+ByAG4YWk6H8AYhBeSIr+ByAG4YWk6H8AYhBeSIr+ByAG4YWk6H8AYhR7e3vW2nm8eA3x2tvbS/1/AECGiqLgL1+kQ/8DEIPwQlL0PwAxCC8kRf8DEIPwQlL0PwAxCC8kRf8DEIPwQlL0PwAxCC8kRf8DECP78NKfUDyfz9ce4ojdl3v/A5BGY3jNZjNZLBayXC6tj59XTwS2vYag2qQ/Pbkt/cnHfbRbhSmaEV4AYjSGlwqHxWKx9kh7x8yy3GmXZSmLxUIWiwXhNTDCC0AMZ3i5RlNVVflmtrbTrut6bdq6rkVEViGhDvkVRSHL5XL183w+X5tv03z0kZO+fH2e5nqqZaoRmz6fkOldbdLnt1wund8VziG8AMTwjrzqul4FiTp82DCz1WfUjl/twKuqWhvF6UGoDj0ul8vVe2q6NvOZz+dro8OyLJ3v6cqylLquN0Zerul9bTK/B/gRXgBieMOrqqrVzrssy8aRhG+nXdf1Ruio+ZnBEjsf23uKGTg6W3i1mV5fblP7sY7wAhBjq4cNzQshYsMrdD56iKjDekrb8Gqa3tUmW/vhRngBiOEdebW5WOP8zFY7bXPkEjvyajOfoUZevjaZ7Ycf4QUghjO89Evj9cOHDTNb7bTNkUtZllHh1WY+ZoiY56xsl/qrz4Wc83KN6vQ2me2HH+EFIIYzvNperHF+Zmuf06+804Om7WHD0PmY4RVytaGavy28fNO72qS+LwIsDOEFIIb3sCGwbfQ/ADEILyRF/wMQg/BCUvQ/ADEILyRF/wMQg/BCUvQ/ADEaw6upqvyWGtXqSr02BXW5CnC3EF4AYjSGV9sblfuwzYAhvHYL4QUgRm/loWz3Q/nuk9KfA6ZXkTersofcq2WrjGGrNE/F991DeAGI4R15hd6orG7yFTlXjUNN46rKXtf1RvV3fd5m4DRVhreFl2saRl67hfACEMMbXiFV5W3nm2z1//TagGYlDEujrA+HdNUntIWXq+oG4bVbCC8AMTofNnRdLNFUlV1/EKX58EkVMKGV4QmvfBFeAGJ4R14hF2u4wqttVXfbYUNGXuNHeAGI4QyvNlXl9UDQnyrsqsqunxdTnws952U73Eh45YvwAhDDGV5tqsq7rggMrcpuBqNelT32akNXeFHxfbcQXgBiUGEDSdH/AMQgvJAU/Q9ADMILSdH/AMQgvJAU/Q9ADMILSdH/AMRoDK9tVZU3rwrsk3ljtfme7UrDNpXp0R++cwAxGsNrW1XltxVe+n1hIhcqeShcJr9bCC8AMTqXhxJxV4gXuVAmSr1UAV9b9Xc9yFz3d6np1DJVW/UK9GYgquXq95YdHByszSe0Mr25Tq4bpxGG8AIQwzvyCrlR2Vch3lbPUB/J6WGnv6fmY6sMr6ZTIWhSYeR63yw9ZYap/nNIZfqyLAmvDggvADG84RVSVb6pQrzrs+ZhQz3ofDUNQw43ukZ751d4LbzUfEKrdJjr22b9sYnwAhCjl8OGvgrx+qG3oiic4aWW2VRNvu25MvV5NVIivHYL4QUghnfkFXOxhn7Y0BxBbXvkZR7C1NukRl+E124hvADEcIZXaFV5X4V4cwSlnx8yzys1nfPyjdiMFVo7TKjaoD7fNbz0eYicO8dGeMUjvADEcIZXm6ryvgrx+nt6KNiu6NM1XW1oHm40/62/9PfUBR37+/vR4cXVhv0hvADE8B42RLP5fL5xng/h6H8AYhBeLalRmXkBCuLQ/wDEKFxXFfLiNdQLANoq2HkgJfofgBiEF5Ki/wGIQXghKfofgBiEF5Ki/wGIQXghKfofgBiEF5Ki/wGIQXghKfofgBiEF5Ki/wGIQXghKfofgBiEF5Ki/wGIQXghKfofgBiEF5Ki/wGI0Vt4mdXWh3hMSFEU3ueMpZ4fmhFeAGL0El7m04pFzj300fX05b4QXvkjvADECAovM5hMZVlKXddrv1NPQjb/bT4Gw/ZEZT1A1JOPzdGc/oTmg4MDKYpi9dmmJyGrddLbos9vuVw2tlctC93wHQKI0Tm81E7eF24i58JBjcTm87nMZjMRuRAGtvfqul79rOahB5sKKjUPFaBN4aWHbVVVq1DUg7OpvWZYIw7hBSCGM7zUoUDzZR4KDAkvtcO3TeMLGtuIyWj8WnipefjmabZFtcc2v5D2ohvCC0CMQUZeKght0zSNkqqqcl4E0ld4mfNr0150Q3gBiLH1c176Dt/3nu/8lL4c32FDNQ8zUNuGFyOv4RBeAGJs9WpD8wIL/RySGSa2oNHPR6l5tAkv9dnZbLYWiPp7VVWt3vOd83K1F90QXgBi9Hafl3mOzDzE13T1nu/iCtf5NnXF3/7+/kagmIcb9Xm62qJfrRjaXnRDeAGI0Vt4ATHofwBiEF5Iiv4HIAbhhaTofwBiEF5Iiv4HIAbhhaTofwBi9B5evvuo+tZ1WX22taoqqapKFovFWkmrLnztG/J73qYxrAOA4WUdXrtiuVyuBZYKsm0ay/c8hnUAMLxeKmzoz/JS1eEV1/1SIu6K8fpy9enMiu76Dlwv8KumMdvsml9TW5sq38/n840KI/r0qi3mY2JUwd+m5YZ8z7kawzoAGF4v4VUUF6qsmztV873QivG2yu+u6vH6zyoE9coZTfPTP9O28r1aR9NsNtt4NphtndVN2b7lhnzPuRrDOgAYnjO8QqvKu0YH6mdbZYvlcumtYeiq/O4qwKv/bKtv2DQ/2zJD6y+a66jYDh3q89RLWDUtt+l7ztkY1gHA8DqPvFwV2PX3zJeal6tivGvH3Ca8VLtthXZty4mtfO8LL9uh0NlsJnVdS13Xq8ONIeHl+55zNoZ1ADC8zuHVZuTlox827CO8mkZerumV0PBaLpfBIy/1e3VosK5rRl4jWAcAwwsKrybmORvXeRp99BBaMV59Vr3fFF6qHeY5L9/8XOsRUvlezdtkO+elz6vNckO+51yNYR0ADK+X8NKvlmu62lAfwfkqxtuuwAsJL/0KxtD5+T5jW64ZXqFXG+rrrQd36NWGvu85V2NYBwDD6yW8dkHKR5WkuM9rLMbS/wAMi/DqiV5hI/Q8HwgvAHFGE17IE/0PQAzCC0nR/wDEILyQFP0PQAzCC0nR/wDEILyQFP0PQAzCC0nR/wDEILx68OCDD6ZuQrbofwBiEF49uP7661M3IVv0PwAxCr0sES9eKV4A0BZ7jo4uv/xyOXHihBw9ejR1UwBgMgivDu666y45evSoPPfcc3LZZZfJH//4x9RNAoBJILwi/fvf/5ZLL71UHnnkEREROXXqlBw/fjxxqwBgGgivSLfffvtaWD3zzDPywhe+UJ544omErQKAaSC8Ivztb3+T5z//+fL444+v/f5Tn/qUfOITn0jUKgCYDsIrwic/+UlrSD311FPyvOc9T55++ukErQKA6SC8WvrLX/4ihw8fljNnzljf/9jHPiaf+cxnBm4VAEwL4dXSzTff7A2n06dPy0tf+lL5z3/+M2CrAGBaCK8W/vCHP8ill14qZ8+e9X7upptukjvvvHOgVgHA9BBeLbzvfe+Tu+66q/FzDz/8sBw5cmSAFgHANBFegX7zm9/I5ZdfHvz5G264Qb7+9a9vsUUAMF2EV6Drr79e7rnnnuDPP/TQQ/Ka17xmiy0CgOkivAL87Gc/k9e+9rWtp7v22mvl+9///hZaBADTRngFePOb3yw/+MEPWk/3k5/8RN7whjdsoUUAMG2EV4Mf/vCHcvXVV0dPf9VVV8lPf/rTHlsEACC8Glx55ZVyzTXXyE033STHjx+X+XwuJ0+elFtvvVVuu+02ueOOO+QLX/iCfOlLX5KvfvWrcs8998h3vvMd+d73vicHBwdy8uRJed3rXpd6NQBgVAgvjwcffFDuu+8++dGPfiTf/e535Vvf+pZ87Wtfk7vvvlvuvPNOOXXqlLztbW+Ta665Rm655RY5ceKEfOQjH5Fjx47J+9//fjl69KjccMMN8va3v11+9atfpV4dABgNwqujz33uc3L77benbgYATArh1dGnP/1pOXXqVOpmAMCkEF4d3XrrrZSCAoCBEV4dnTx5Ur7yla+kbgYATArh1dF8Ppf9/f3UzQCASSG8Ojp+/Lh885vfTN0MAJgUwqujD37wg3LvvfembgYATArh1dGNN94odV2nbgYATArh1dG73/1uue+++1I3AwAmhfDq6J3vfKc88MADqZsBAJNCeHV03XXXyS9+8YvUzQCASSG8OnrrW99K3UIAGBjh1dHVV18tv/3tb1M3AwAmhfDq6KqrrpLf//73qZsBAJNCeHV05ZVXyp/+9KfUzQCASSG8Onr1q18tp0+fTt0MAJgUwqujsizliSeeSN0MAJgUwqujI0eOyJNPPpm6GQAwKYRXR5dddpmcOXMmdTMAYFIIr44uueQS+fvf/566GQAwKYRXRy9+8YvlX//6V+pmAMCkEF4dHT58WP773/+mbgYATEpRFIXw4pXyBQBtFew8kBL9D0AMwgtJ0f8AxCC8kBT9D0AMwgtJ0f8AxCC8kBT9D0AMwgtJ0f8AxCC8kBT9D0AMwgtJ0f8AxCC8zrPdPFtVlYiILJdLKYpC5vN54laOD/0PQAzC67yiKGQ2m63+PZ/PpSgKWSwWCVs1fvQ/ADEIr/PM8FosFqvRlz7yUr83R2iu34uIVFW19nsC8QL6H4AYhNd5oeFlTmMLI/33aj7L5VJEzj15WV/O1NH/AMQgvM7zHTa0hZcaTZmB1vT7oiikLMvtrkxG6H8AYhBe59ku2KjrWkQ2L9hQ/9ZHVK7fq5GXCqyyLAkvDf0PQAzC6zxz5KUzw0uNytQ5LcX2e/U7/bAh4XUB/Q9ADMLrvNDw0i/M0Ll+rw4X6ue/CK8L6H8AYhBe54WGl3nloJrO9Xs1b/13fOcX8F0AiEF4ISn6H4AYhBeSov8BiEF4ISn6H4AYhBeSov8BiEF4ISn6H4AYhBeSov8BiEF4ISn6H4AYhBeSov8BiFHs7e1Z6/rx4jXEa29vL/X/AQAZKoqCv3yRDv0PQAzCC0nR//KUesTOi1dRFOw8kA79L09sN6REeCE5+l+e2G5IifBCcvS/PLHdkBLhheTof3liuyElwgvJ0f/yxHZDSoQXkqP/5YnthpR2OrwWi4UURSHL5bL3eZuXXJrvLRYLZ3vQL77TPLHdkNIkw6ssS6mqavXvqqrW/iO6wgvbsav9D35sN6QUHV4qWObz+Wr0ou/wl8uldWSjplOBocJJ/Tyfz73LUEGm3pvNZqv5u5apVnS5XK4+YwZiURRS17WUZbma/uDgYG0Z+siraf3rul5bp7IsW3/HU8FOsD9VVclsNgv+fJvPmqay3fR9gu+ITR/U/knfD8Kuc3ipEcx8Pl/7j6CPbvT3zOlUMCyXy42Rlh4QIuf+Y6oQUO/Vdb22MurfZnt0apn6tOaXslgsNpZhCy/X+uthVpYl4eXh639lWTIKDqT+IAwJJBVy+h9mbU0lvBTz/zzS6hxeKmjUqEV/T9FHO+Z0vp2++VnffBaLxVpAuEZYij4ysoWgHl5mmDatv/6z7d9Y5+p/thEt7FQQ+f5oM3U9h0t4nWPbj9iOzJh/wNve00de+s/m50TW92FNf5CPkTe89C/NHMY27by3EV76+7bl24b1IefL1LzUcgmvYdn6X1mWq35DeIULDa/lcrn6jmP7JuEla4Gi9kF1Xa8+a/5BX1WV9z1beKnPqZH1YrHYOLxIeLWw6yMvl7qurf+5y7JcbXjCa1i+/kd4tdNm5NXV1MNLhZXeP8uylNlsZg26kPdcIy9zeeay9eCciq2El8jmOS8z2NqEl5qP7ZyXOSpTG88c/ZkrrW9k9Vk1r67hZa7HbDYjvDwIr/4QXttj2x/Zwkudp7UFVNN7IeFV1/XGsgmvFpp23k1XG7YJL/3wpWs+tmWawWb+2/VZNQTf39+PDi+uNgxHePWH8NqeXRl51XXNyEt2/D6vsZjP51z66kF49Yfw2p6Yc17mH7v6OS/be6HhxTkvwmsr9M6pn3CFHeHVH1t4mYfE+7rpf2r7DVt4mUd7tnW1ocjm6Eq/V1bNa0r/VwgvJEf/yxPbzc13T1gf94u5zr9to5TeriK8kBz9L71//vOfradhu7ltO7xE1m9l6mN+uSG8kBz9L627775bXvKSl8gdd9zRKsTYbkipU3ht8xjrkBXc1clO26tvfayXfk5NtT3nC0JCv4+HH35Yfv7zn2+5NdP02GOPyYc+9KFWIUZ4IaWdDa9Uclgn84bsqqpGHV7PPPOMfPzjH5dLLrlEXvnKV8oHPvAB+etf/zpQ66ZFhdjFF1/cGGKEF1KKDi+90rJerd0csbiqyIe+p/8cU8FdH1WZO3jbCU5z3ru4XlMKr/39fbn00kvlxIkT8o9//ENERD772c/KRRddJJ///OeHauLkhIQY4YWUeht5NVWR91WG972n/9y2grtZCiqkQrkZILu4XlMIr4ceekje9KY3yXXXXSe/+93vNt5//PHH5cYbb5RXvepV8uMf/3iIZk7SY489JjfffLM1xAgvpNRLeLWpZdj2PZH4OoIxNQX1wNjV9RpzeD311FNy7NgxecUrXiH33ntv47QPPPCAvP71r5d3vetd8uc//1meffbZ6Ndzzz03+Ot///tfkldbthAjvJCSN7x8VeXVxHqRSMW3I9enC3lPpFsRXPNGvpAvxCy5smvrNdbw+vKXvywvetGL5I1vfKOcPXu21Tze8Y53SFEUcujQIbnooot6fx06dCi7l+sipK6vI0eOyKFDh+QFL3jB1pbBi1fTa29vT4qiGOfIy9T2sOGurtdYw0tE5MyZM/LhD39YXv7yl8u3v/3txmnvv/9+ueKKK+Q973mPPProo9ts5qSdPn2akRd2yvkQ6/+cl+1ZNSL28z++9/Sf21ZwN3fqfZzz2oX1GnN4Kb/+9a/lLW95i1x77bXWbfboo4/Ke9/7Xrniiivk/vvvH6KZk2QLLYXwQkqdwkt/hHjTVXn6VX9t3tN/jqngrl8VaXsCaterDVOs1xTCS/nGN74hL3vZy+SjH/2oPP300/Lss8/KbbfdJocPH5YvfvGLA7ZyWnyhpRBeSKlTeIWwnf8JeS/WUBXcU67XlMJLROTs2bNyyy23yMUXXyx7e3ty7NgxefLJJwdq3bSEhJZCeCGl7MMrVQX3lOs11QobjzzyiPzyl7/ccmum6fTp01TYQFa2Hl5AE/pfWtQ2RI4ILyRH/0uPqvLIDeGF5Oh/eWK7IaWdDa/Q6uvm1YVKTHUNxbzisEmbSvFt5z0Fu9j/0GxXt9s2LphS9PPQ5vq7/m8P+YSMKdnZ8AqlOobZaWazWfTD2bYZMITXppz735Tt6nbbVnjp93yKXKjeo/B/e1jR4RVSOV1dCSfivl/KaMjGPVCKb3qzU6nPmlfsmW23VXM3q+Xr91oVRSF1Xa+12VU1I2TeOGdXd4Lw67LfcD1Joe1THPT7IH3LiN03qWn1Kjnm++ooj5r+4OBgbRl9PEkCmzqHl69yurmjV/82q6eXZbl6T92zZIaXq7q7msb2b9cyQ6u5mx1WX0fbd9Fm3riA8MpT1z96Rez7At9THNR76v+3rRxbn/smnblPsX0ferk89bnY/YP+JAls6hxeIbX9zJtqXbX+9PdtG9w2va0tZVlu/NXia5+vLJPOdR4ttuQTziG88tTHfsOs2en6f25O59vp97Vvsmk6EmMrzL2NOq3oUFU+pHK6voH0Da4P5V0nM/Xf+6q7K2r0pv/V07TMkIDRv4OisN8ETXh1Q3jlqY/9hrnz3kZ46e+32Tc1Mc+3E17DSjLyss3H9/umkZfIhc6syiiFLLMpYMzlMvLaDsIrT2McebmYD7ZV9FMehNewOoeXr3K6+ZeP2sjmX1h6x1PzaTrnZW5U/eIM/S8h2zJDA8Zsp+sYNOHVDeGVp77DS6T5KQ5twquPfZO5vvphQvVZNa+u4WWuh/4kCWzqHF6+yul6BzGv6PG9p8/D9xnbytiOX5vLbOpA+lVC+lVE5nTmX3Nt541zCK88bSO8mq42bBNe+uFL13xsyzSDzfy367Pq//b+/n50eHG1YbjeDhsCsQivPLHdtmuoJ2TkivBCcuwE88R265d+6sM8ioRN0eEF9IX+lye2G1IivJAc/S9PbDekRHghOfpfnthuSGmnwqvtVXiue8T6mDeGsyv9D+3s+nZrs3+ImYZ9SlpZh9euzBvd7Er/QztT327sU9LqHF62ezJ892Tp94Xpl4GaFddDKj3b7p+gmnt+pr4TzFXIdvP9n9NvJnbdiznUkynYp+SnU3jZqsGr39uqJpslVlQlDb0xeudwVV5WbB2Nau75Ibzy1Fd47cKTKcw26T+zT9lNne/z0tkK7ep1xZpqdanOEFLL0GwDZZnyRXjlybXdXMVuzYfDdqlB2PeTKcw22drHPmW3eMMrpDq0qakCvH6IwJyn6gwhVeTNNtDR8kV45amvkdeuPJnCnIZ9ym7rdeRl+73v+Tiuw4b8lTQthFee+g6v1E+mcE3DPmU3dTrnpW88/Xi1qzK0fl5MfS70nBfV3MeL8MpT1+22a0+m0JdrawP7lN3SKbxcV+/4rurRr9Ixj4HrFddjrwyimnt+CK889RVeu/BkCvYp+ekUXkAf6H956iu8uNQcMQgvJEf/yxPhhZQILyRH/8sT2w0pEV5Ijv6XJ7YbUiK8kBz9L09sN6REeCE5+l+ednG7tb36jyry+SK8kBz9L0+7uN22GTCE124hvJAc/S9PXSts6J9pc68oT6aAiBZevHilfCE/IdutKPw7e55MgVjn9x3sPJAO/S9Pru3Wtqq8jidTIBThheTof3nqOvLiyRTogvBCcvS/PG0rvHgyBUIQXkiO/penPrabHgg8mQJtEF5Ijv6Xpz62G0+mQCzCC8nR//LEdkNKhBeSo//lie2GlAgvJEf/yxPbDSkRXkiO/pcnthtSIryQHP0vT2w3pER4ITn6X57YbkiJ8EJy9L88sd2QUq/hpe6TcJVu2Ya+77vgPo7hsRPME9sNKfUWXqqWmF5+ZTabbdxE2DfCK3/sBPPEdkNKweHlq1Emsv5oA0Xd4W7+27zj3fccHZGw5/ccHBxIURRrd8H77pbXVn7VltDnAal5q2WhG77DPLHdkFIv4eUrnKlzPV/H9xyd0Of3qHmoAG0KL9dzhHy10cz2mmGNOOwE88R2Q0re8Ap9Lk9IePmqPPuCpu3ze9Q8fPN0PUfINr+Q9qIbdoJ5YrshpcFGXr7n6zSNkkKe39M1vMz5tWkvumEnmCfbH7a8eA382u45r6ano4acn9KX4ztsqOZhBmrb8GLkNZyQ/ofdw3ZDSsHh1cR1taF5gYXt+Tq+8Ap9fo8rvNRnZ7OZ89k8+nOE9N+HthfdsBPME9sNKfUWXiKb58jMQ3xNV+/5Lq5Q07ie37O/v78RKObhRn2erraEPA+I8OoXO8E8sd2QUq/hBcSg/+WJ7YaUCC8kR//LE9sNKRFeSI7+lye2G1IivJAc/S9PbDekRHghOfpfnthuSGmQ8LLdO+X62eR7z8ecznaDW+zVgvrl9E2qqpKqqmSxWKyVufK1ta9l54KdYJ7Ybkhp0PDShe6w+wwvvQ36vV1thQbIcrlcCywVZF0QXtgVbDek1EuFDd99WrbK7/o05s/6v9X9Wfp75r1kelUP33TmTt+2/JCK9GbleV975vP5RtURs01m9RFfhX1z2WPBTjBPbDektPXwUtO2OWyoh4AeQmbJJ3P05JpOb4NSVZWzSnzo+oS0xzSbzbyjUF+Ffdt6jAE7wTyx3ZCSN7xCq8r3GV6uUZirfa56hW3OebUp6quvT1N7bIclbYcObevfdtk5YyeYp762G09gR4ydG3m5qrkr+uG0oijWHp3im8512NBWaLdNgLja4wsv8z8n4UV45aiP7cYT2BGrl/DyVXBX0/Yx8jLfazvyMjuXqoTfpiK9a33Mzy2XS0ZegQivPHXdb4jwBHbECw4vn9AK7qGBZVZzd43KyrLcKOBrm05vg+IbeYWuT1N7bN9r6Dkvwgu7ru8/el14AjtsegkvEX8Fd7Pyu4g/vPS/XMwQ0v/KcYWObTr9LyH1cnXY0PVRhwZd7elytaHrP4++7LEgvPLk2qi4k+IAAAJzSURBVG48gZ3nAA6ht/DCpm3c5zVG9L88DTHy4gnscCG8tkyvsBF7U/TY0f/y1DW8RHgCO+IRXkiO/penPrYbT2BHLMILydH/8tTXduMJ7IhBeCE5+l+e2G5IaSvh5Ts23Leuy+qzrSEV5NvytW/I73mbxrAOU8R2Q0rZh9euSHFl4Vi+5zGswxSx3ZBScHiF3o+hjhPbrrgxjwGLuO+CNxq4cR+UeUe8/p5+zNpss2t+TW1tups/9J4us/SNuomxabkh33OuxrAOU8R2Q0q9hVdR+Cu66++F3gVvu5vddUe8/rMKQbPKu29++mfa3s2v1tFkq6ZhW2d1otm33JDvOVdjWIcp0v/Y4sUr0aufqvK2f5v3N+n3Pfjuy3Ddze66qVD/2XbPRtP8bMsMvafEdQ+X7dChPk/9stym5TZ9zzkbwzoAGJ4zvIwPOUdevorurgDUn4GlfqcfNnTtmNuEl2q37eZB23Ji7+b3hZftUOhsNpO6rqWu69XhxpDwaqqcn6sxrAOA4XUOrzYjLx/9sGEf4dU08nJNr4SGV5sK8ur36tCgXtXet1xb+xh5AZiyoPBq0lTRXZ2n0UcPoXfBq8+q95vCS7XD9mRj1/xc6xFyN7+at8l2zkufV5vlhnzPuRrDOgAYXi/h5avobl5Jp4/gfHfB267ACwkv/QrG0Pn5PmNbrhleoVcb6uutB3fo1Ya+7zlXY1gHAMPrJbx2QcryK1SQjzeW/gdgWIRXT6ggH2cs/Q/AsEYTXsgT/Q9ADMILSdH/AMQgvJAU/Q9ADMILSdH/AMQobBUwePEa8gUAbbHnAABkh/ACAGSH8AIAZOf/uJjejJki6DQAAAAASUVORK5CYII=">
</div>
```
```
### 3.2. Usuario
Con base en la información de una persona, construye una clase de `Usuario` que herede de la clase `Persona`. Puedes guiarte del diagrama UML.
<div align="center">

</div>
```
```
### 3.3. Empleado - Trabajo - Empresa
Construye la relación de clases entre los trabajadores y la empresa para la que trabajan. Puedes guiarte del diagrama UML.
<div align="center">
<img src="data:image/png;base64,iVBORw0KGgoAAAANSUhEUgAAAu8AAAG3CAYAAAAEtrtWAAAM+nRFWHRteGZpbGUAJTNDbXhmaWxlJTIwaG9zdCUzRCUyMmFwcC5kaWFncmFtcy5uZXQlMjIlMjBtb2RpZmllZCUzRCUyMjIwMjEtMDctMjhUMDElM0EzOSUzQTMyLjEyOFolMjIlMjBhZ2VudCUzRCUyMjUuMCUyMChXaW5kb3dzKSUyMiUyMGV0YWclM0QlMjJBbmJmdmM3WGdQVjZ1TmNLdHhrNCUyMiUyMHZlcnNpb24lM0QlMjIxNC45LjIlMjIlMjB0eXBlJTNEJTIyZGV2aWNlJTIyJTNFJTNDZGlhZ3JhbSUyMGlkJTNEJTIyTVZJdFVNT3o5YnQ3cmJmZEc4aVYlMjIlMjBuYW1lJTNEJTIyUGFnZS0xJTIyJTNFN1Z4ZGI2TTRGUDAxa2JvcnRjSkF2aDdidE4yWjBjeXFtbzVtZDU5R0xyaUpad0JuRFdtYiUyRnZxOUJqc0JiRXFTNG5SVklWVlZmREVHZkklMkJQanc5T0J0NHNmdnFENCUyQlhpQ3d0Sk5IQ2Q4R25nWFE1Y0YlMkZrSSUyRm92QXVnajRybE1FNXB5R3NzNDJjRXVmaVF5cWFpc2FrclJTTVdNc3l1aXlHZ3hZa3BBZ3E4UXc1JTJCeXhXdTJlUmRXckx2R2NhSUhiQUVkNjlDOGFaZ3NaUmFQcDlzQUhRdWNMZGVuUjBDJTJCT3hGalZsbyUyQlNMbkRJSGtzaDcycmd6VGhqV2ZFcGZwcVJTSFNlNnBqaXZPdUdvNXM3NHlUSmRqbmg1ODN6MGd0JTJGckQ3OXk3MDVIVjUlMkYlMkYlMkZRMVBaV3RQT0JvSlolMkY0aHZDVUpWamVjN1pXUFpFJTJCMGpqQ0NaUXU3bG1TM2NvakNNbzRvdk1FUGdkd0o0UkQ0SUh3akVJbm5zc0RHVnRDTkZqUUtQeU0xMndsN2pmTmNQQkxsUzRXak5ObmFCWkhzazA0ekRPSkIzZFVxWEVyem9Td0ExRk9VcWh6b3pvQjFVSmY4Rk9sNG1lY1pqSVFzQ2pDeTVUZWJSNGp4bnhPa3d1V1pTeVdsV1R2d09PUXA4WnVSNXRrd2lnZ0xDWVpYME1WZWNKSXBuJTJCdGdDM0xqeVU0VFdSc1VVYlNVQWF4aFBCODAlMkZRMnlmQkI1bm1Qbkx0YXpnZXVCNEdFeFhjY0l1ZTNHYWZKZkFEZDducFA1M2szaU9Na3hORHMlMkJkMDZJOXF4WkJYRHBWY1JodlNmVSUyQmdaN2VRWTA4amNkZzFvME5WWkRnRE9mcEVaaXhnMGVabXdBbmswaW1vaEJiNkkzR2VOMEV1WE9JRExmczdyWFByYnlGZlo0U0xFNE56N0tCJTJCaUN4cUdKQkd3WVJuT2NJRVJBWWdsZzRmTEV6SzhnRDlJMGN3NUd3NkdjT016S0tOdEdmNUVkWjdOV0FMUGdta09Gd0lBZkNRQ2hMdGhxM25RNm9DVENCTmR1d3ZBUnI0bGZIa0dmTlZ5SE5FOGQwV09GYkdpZ3hJY1E2b2lzczNvTjVId3kxT2taZDNUcyUyQjRaTWh6aE94TGRzSlJtbEluMmVWRzNsdm0zU3U1MFIlMkZhWVdNcXRiOGd0UEt4VDlNTXF5Q0J6NWJFdGp1RWdXMEVTbnpHWEZIRUNiUEViY01FRGcydjFUR0FQTEpQZHdESjBMYUZscUtIbEtsNUdCSFJRTHk4NmtoZkRxcnp3a0U0UXJtT1NGNTR0ZVRIU2tuNmF6JTJGJTJCUWVBYUQlMkZodkhkJTJGZ25LNDE3Y2ZqanBZclg1Y09wRWc4enhtRVE0WXclMkJzRjVIZE1JZXczMTFoQkZKUTFzNll0enJDR3ZKbmV4SUU3WjB4T1JWT2lLZlJSaklDUExBZmpOeVNzOEdYUU5HMXhKR3dQaTJ0TVMwblExSUVwNEx6d2RLZHhFVGslMkZnRmhPUXNqMFpGOFpxSzYlMkJiZHZzaGlwUWZJRTgzJTJCRm1IbzVxTDBqNndrUGw4JTJCbFF0clZVamd1VW9uaWFJNEM1MDUza1FGdHFmbXBjcTVONFJUNkJ5aGFVcHlnSVNhSlZVVEElMkZEUWJNVUQwcTY4UU9YTXlVdEo5ODFKTHl0R1pXQndFb25KdDNwdnBqVEw1bTVZUHBFcm9ZSnFSc2htS2FPYUtKNUpubFgyc1dvTmVXNUxROFZEYXczbHFOczg0JTJCRkFWQXFycW1wQkFQYWVXV2VpZGpTc3BIaHNXUFZPVGFLMkRvWE8yQWZwUm1tYmFTYUVhNENYTk0lMkZQZWNoV290Y0VQR0FVQlpvJTJGbHJLQU1yanJjJTJCVyUyRkZ1cWdYbTNKV1NobVIxRVRpa0IzZVAyWnBrSXdSd0lUTiUyRkk0bE9mRmROQlBpWWRQaWNneHclMkZRRnA4MEV5N0V0aFl4TVZtNHZrVHRLcjlHcU4lMkJYWGxraEdKaWUxVFNYVFpFMEM0SkprVm5EUFNRZ0E0TXpvdDEza3ZUUm5sJTJGU0J3akFNV2RwV215UVBWT2h2V0k4ckt5YzlhYXdka2pUZ2RDbnV4bHlwSnllTDZEVzRmMGIwMmxMc1NEZUxiMWZCaXFjd0lmWktxU09sNU8lMkZnJTJGNDFNVW1sc1RTcnBycTlaQ3RVc1ByR2tGN0xtQTA0WFglMkZCU0tack4yaDd1eHRuWXgwb2Q5U0xuOVR6U3NBWjh3UVkwQWNxM0puSjBRN2tYT1oybDE3QzBNcWJYbXNZeHVieXRUbUFZU2d2d1JWbnpKNHRwZ3B2clBFRFBZYlZpT2xFZjl0TSUyQmNUcXZVbFAlMkZZdk1ZcURXNGtTYlVUcTFwRzkzQTNreFV2YlRwUk5yVWZUNlR0RUZHYVdQdDFTYlNUZWpUalFmMEk0QnVNTDZhRkhWQTlHSk96YTgzSVVQeW1PR3RaczhpcjJDUnliN1N4Z2dvYTI4NFhkMUs3cVZOWiUyQmsxdk9NMHB0ZVd0SEZObm5HcnRQbTVTak55bTdNRmprNXkybGozbXVJWWNESDRKY2FGamkxTm9RWiUyRmVUUDJ4czd2UlVVbm9tSkRDc29sY3czckg5Y2tLa2EyUklXcnU3eGJVZEVrSjVhY0JKUjlGd3NZbzZTZ1NicUtUWGJLeCUyRnhBc1dUNUNBQ1pFOTdiS0Yyd1N6RjY5N0pSVERpenB6Vk1XM2Q3cmRGUmVrMzdxVXpwdGFZMWROTjFSNjJCJTJCVTNPSlNjbFNtbTBRT1FaMTR6SHF3aWZTSlk1JTJGUDFQenpJZHc5RGdpNWhnT0xhbFlid2RWalJIM3FaVlhMJTJCVFhWVk5ycFNGN1ZMMUZ6dHUlMkZYM05ydHVsUnVOYVF5T3YycERsN1ZMJTJCWVlqSUZhY0JKaElYRzFGYUtodHdnc29vMldDbWJUdmZvTFNacjdTM3IyRXJYNGY0a2hxaGRkZGU0VU1kQllkdURUNyUyQm9UajB2WmFHYk9OUVg0MmpsNERJbHZrc1VBYlZscVRjT2tKRGltT1doTjhXTktrdGw1QmZnMnFPSUVDTFdyUEJQTEZnYzViZzZHb2JiWnZiN3VTNmFJUDBVJTJCZk1jU1lWdUklMkJkRnNCRG9iNEZ0YnluRlpYSEFXb1pCYlkzdEtKZGQ3UTJMUElsQ3FHYnhoTlUzVjNvZGpOU2ZLY0c4RU0zdUE1SExRMDFqSlI4TTJDcG1oUXN6Vk5NN1lhOTBmamwlMkIycDR3S2I2U0gydnpGd2ZQaFIzWER0YjNUNjd2MCUyQkpIU3JRalJaMGR2YTd4Z1pTanBZb1lGOWwydWpCYkpaSGp3dWFrVnRRcE9LYWp4d3ZjJTJCVzVTa0lTRGpwelBWQVRpWmNtRE44Z0UxMWJub2V2ZXg3R0RCeURqNUVWUG5ZcVZPeE12Y09wR0owNU1PRlhOSWszZm1zNjlsOHBWYlo4N0xuRGNRV2Q2a3ZHcjFYUWRjRnhLQiUyQlBwb2Z4c1Q1REdCJTJGekNIeW5XeiUyRnZtJTJCJTJGOEd0OTUzbHZ6blc3UHZEZnhXU3pjdDR3M251N05lRjN5VThQbTJDM3RUTVpLbnJ5U1o3ejZGNXZxS05xVloxeTFSRkh2SnRDJTJCUEhNRUt0RTM0NzF2S3FuJTJGZm8lMkZoQnppT3l5VDZmcm0zWVJJN3NpbG5rcXByTSUyRlpmcDUyY2luYUN3TmglMkZhJTJGR2tmc21zVFR3VnJ3WWJzUWw5NWJ2VEttZE11dUcwJTJCbmNzNjF1cWRuWjlKaTBOJTJGUjg0VGQlMkZMOTc0NXJiNlFSNGEzVngyUkdoUzN2M0JYSkd6N080SGUxWDglM0QlM0MlMkZkaWFncmFtJTNFJTNDJTJGbXhmaWxlJTNFzMKrkwAAIABJREFUeJztnX3sZUlZ50/PMG/OdA9vglky2tnbEbPpQdRIVpMNO9lsbgf+EKM4ZsT1ZWcWrzaYGA2zykJrG4ebbEiP2Oyyxn/Q9aYTxRdaDaNG3MuqMYpEYRharsHRiECcGXt2gwsNz/7RPr9+fvWrqlPn5Z5Tdc7nk5z0/fU5p+qpuvXU+Z7n1kslAAAAAABQBNXYBgAAAMC4VFXFwTHLo0TKtBoAAAB6o1QRA9CFUtt9mVYDAADMmMViIdvttrf0ShUxAF0otd2XaTUAAMBM0Z/7Ee8A3Si13ZdpNQAAwAxZLBay2+0Q7wA9UGq7L9NqAACAGYN4B+hOqe2+TKsBAABmDOIdoDultvsyrQYAAJgxiHeA7pTa7su0GgAAYMYg3gG6U2q7L9NqAACAGYN4H4b1eu3d2Ge5XI5tGvRAqe2+TKsBAAAgyPvf/375oz/6o+TrSxUx+0bFe58vSpAPpbb7Mq0GAACAI3zyk5+U//Sf/pO85CUvkRe84AXyxje+UZ5++una+0oVMfsmJt4Xi4UsFgtZLpcHEfnNZuONzus1q9XqyDr9ek7/1f9fLBaH0rXYXwFWq1XwHL8QxCm13ZdpNQAAABziv/7X/yp33nmn/MiP/Ih89rOflaeeekre+MY3ygte8AL5mZ/5mei9pYqYfVMn3vWcrr2v9ejep8JcRbj+7TvnntcXAk1LXwBC+aiYd++Do5Ta7su0GgAAAERE5L3vfa987dd+rXzTN32TfOhDHzpy/o/+6I9kuVzKN37jN8pv//Zve9MoVcTsm9CY981mcxB5F5ED8e4KZ59Yt+e32+3Bud1ud3Dejajbv22k395jcW0FP6W2+zKtBgAAmDlXrlyRb//2b5ev+qqvkkuXLtVe/+53v1tOnjwp3/3d3y1PPvnkoXOliph9kzJsRqS9eN9sNkfO2Si+b3jMdrs9ck6xUXzXBjhKqe2+TKsBACbMlStX5MKFC3L27Fk5c+aMnDp1yvswH+M4duwYx7FjctNNN416HDt2TKqqkn//7/99o7Z17do1+Tf/5t/IrbfeKo888sjB/5cqYvbNvsW7jbxbfGPZfejQndVqVWsDHKXUdl+m1QAAE+X8+fNSVZWcPXtWLly4IJcvX5YnnnhCrl27trc8v/jFLxZ1fOELXyjquHbt2l6Oxx9/XL71W79V7r33XnnPe95T+z3/4i/+opw6dUpe97rXyV/91V8dOleqiNk3fYv39Xp96G/3s2L/TyPtmpYvX03XTlL1jaWHw5Ta7su0GgBgYrzvfe+T06dPy/333y9XrlwZ2xwoiF/5lV+Re++9V77lW75FPvKRjxw5/6d/+qfy6le/Wr7+679efuu3fsubRqkiZt+ExryrgG4q3mOrzbjY1WZUnCvWFruijLXXfWGAo5Ta7su0GgBgYpw+fVoefvjhsc2Agnnb294mt912m/zoj/6oXLt2TZ599ln5oR/6Ibn77rvlwoUL0XtLFTGlEBLoMC6lfidlWg0AMCHOnz8v999//9hmwAT4m7/5G/me7/ke+Yqv+Ap58YtfLKvVSj7zmc/U3leqiCkFxHuelPqdlGk1AMBEuHLlilRVxVAZ6JXf+Z3fkQ984APJ15cqYgC6UGq7L9NqAICJoKvKAIxJqSIGoAultvsyrQYAmAi6qgzAmJQqYgC6UGq7L9NqAICJcObMGbl8+fLYZsDMKVXEAHSh1HZfptUAABPh1KlT8sQTT4xtBsycUkUMQBdKbfdlWg0AMBGqqtrrBkwAKZQqYgC6UGq7L9NqgImwXq8PbbBRR5NroQxKfXjAtDh58qR3MyIOjikfJ0+eHNv1WsFTA2AkdCe8FEGuIl/XCvZt1Q1lUlV0wzA+tEOYI6W2+zKtBigcFeKr1So5mr7dbovtaCAM3ynkAO0Q5kip7b5MqwEmQqp43+12slgsDv6F6VDqwwOmBe0Q5kip7T7Z6rHHJXFw9H3kQJPIO0yTXNoizBvaIcyRUtt9I/EOMBVyac+Id8ilLcK8oR3CHCm13SPeYZbk0p4R75BLW4R5QzuEOVJqu0e8wyxp256vXr3aqx2I93nz5JNPypd92ZeNbQYAz3iYJaW2e8Q7zJKm7fnq1aty7tw5OX78uFy4cKE3O3zivaoq2e12Rz7D9Hjve9/LyxtkAc94mCOltnvEO8yS1PZsRftDDz0kjz/++J4tgzlx/vx5edOb3jS2GQC1feJyuZTtdjvqile6XO4+AhqxBQ2qyr+3Bsv3lk+p3x/iHWZJXXtW0X7ixAl58MEHEe3QO08//bR81Vd9lfzyL//y2KYA1PaJKti32+1ovxbtS7wvFgtZr9cHf+sGekpIvEP5lKptEe8wS0LtGdEOQ/Ga17yGqDtkQ6hPDC21a8WuyA1hvVqtDq6xgne323kj23qfCmYV5/p5tVpF81Ahr+d0F+pYnlqu3W53cI37QlBVlWw2G1ksFgf3X7p06VAeNvJeV/7NZnOoTOzXkQelalvEO8wStz0j2mEoHnvsMfnmb/5mec1rXjO2KQAHxJ7xm83mQETr8BkXK8JFjs7nsdFte869T4Xxbrc7Emm3AlnkeoTc/iKggtuWSf+OLQ6gedp73brZbrdH8vCJ91D5rZhfLBaI90woVdsi3luQEoWAvNH2jGiHffGFL3xBnnzySfnDP/xD+aVf+iV59NFH5Ru/8RvlZS97mfy3//bfxjYP4BCxZ/x6vT54xulOzy6u0NaotT2n2Gi3e19M9LrXxtLZbreHBHIowq7YyLjvJcCKd/dloq789rPvbxiPUrUt4r0F+rOZoj+TMSauHKqqOpiIes899wR/Gubg6HK85CUvkVe84hXyzd/8zXL27Fl5z3veM3bTB/BSVf0MmwmJV5t+X+Ldnvfl77M7Zby8pqX5hvJAvJdPqN3nDuK9BVV1WLy7P5fZTsM6qDsmT9OwY/18nYs9p9EA7fzs+Dprk5smLxaH0fbsRt4/+tGPjmwZAMDwxJ7xKZNVc4+8h9hsNt4yLRaLg+ct4n26lKptEe8tiIl3V8gvFosjY/t8P+Xp9W7a9m99KdhsNgf3aVoq1n0djLUBruO2Z0Q8AMyZ0DPeLg1ph8+4xMSryNEx766wbyLeNR3fmHc3Kq8C3I3+u2W3w2T0WrvfRhfx7pZjuVwi3jOhVG2LeG+BK7DtsBkV0erA9m93so3IDYd3/1/kRgdio+YqxG3kPXStjb7TURwm1J6tiH/ooYcQ8QAwC0J9YspkVZF68Vq32kwT8W5/cQ6l48vTFfbu36Fr9dfyixcvthbvrDaTJ6VqW8R7C3zj6OyM9tA4OzdqoGjHoIeNKvjEu04Y8on3zWZzJMLPzPaj1LVnRDwAzAme8cOxWq2OBOtgHEpt94j3FriRd4sbebeExLti38x3u12ryLtGSdxhM4j3w6S2Z3eHVUQ8AEwRnvH7w/7Czi/heVFqu0e8tyAm3n1r1rpj8qx4t6Jb5Ebk3peXb8y7T7z7xr/TWRymaXu2Iv7ChQt7sgoAYBx4xsMcKbXdI95bEBPvIkeXqFJCkffYyjDumD3fajM2Tzs7Xg+74xxcp219XL16tWdLAADGh2cEzJFS2z3iHWYJ7RkA4Ab0iTBHSm33iHeYJbRnAIAb5NAnuuvBu79Ep9zT9bqUNGJz2vokpfz2un3YMHVKrS/EO8yS3Nuzb8UiDg6O/R1zJ8c6qCrEexPxDs3Jsd2ngHiHWZJ7e87dPoApgb91qwPfS5A790vnY9m5X3rOt276YrE4dL4uPWuLm15VVUcWg0hdd963U6xdEtpneyx9F7tUtF0+MrX89rpLly4dyYsX1Dil1gviHWZJ7u05d/sApgT+1r4OFovFgZBcr9eyWq0OhKvdrNAngEPnrE3b7TYpvVCZQiu5uTu+ujuh14n3FNtt3r5FLjabzaH/XywWhyLoKeW317k2+L4bOEypvo94h1mSe3vO3T6AKYG/tasDn3D2DSnxCWC9zgrT1DHvvvTqbPPthmptdm2Iifc627fbrXd3Wbdu3F1YXVLKb6+LlVHtgMOU6vuId5glubfn3O0DmBL4W7gO7BARN3Ibi3q7u42HBLDm7YscW/Fal56LDjNRVEDHzjUV7yHb3SEu7hAbix0+5NZvSvlDNjB5NY1S6wjxDrMk9/acu30AUwJ/6y/y7vv/rpH3lPTqbEiNvLtR8j4i76mEhs3Eyh+6DvGeRql1lGz1yZMnvW+SHBwlHidPntynX3WmqsrsUABKBH9rXwdVdSM6rGOx3cj2YrEI7jSeMuY9JT0f7rh2Nw17TtNTQa5liu2Snjrm3bVfccehh8R7rPz2utgvF+44ebhOqb5P5B1mSe7tOXf7AKYE/ta+DkKrqthVUGzEWj/bVVYUV3zqNRrJjqVny+FGxqsqfbUZkaNDWWKrzYRsd9MPjTe35XJ3X08pv73u4sWLyWWE65RaL4h3mCW5t+fc7QOYEvjbcHUQWysdYGhK9X3EO8yS3Ntz7vaViG/4lBvpaov+rK0/k7dlt9t5l5SD/YK/Id5hnpTq+4h3mCW5t+fc7SsR/ale0Z+++9iZsC/x7toIw4C/UQcwT0pt94h3mCW5t+fc7SsRVxjbyWfuOFx7nY3UW3HujovV8+6kNlfY210f7cuDHdPqTkbz5Q/9gb9RBzBPSm33iHeYJbm359ztK5EU8e6uxmDvsSJcJ4LpShEq+uvEu3ufin4dQuB7cfDlD/2Cv1EHME9KbfeId5glubfn3O0rkdiwGXfbc5EbYtkOq1ksFrJcLo+cs8I6Jt59aYZsjOUP/YK/1dfBcrmU7XYru91ukCUH97FOuW/ei7sxUo7YX+s0UFCC3SVQqu8j3mGW5N6ec7evRHwPbncoi53AqsNiXPGs60O3Ee++NF0bVZzH8od+wd/q68Aul1jqC6TP93Rd+pxxN31y14eH9pTq+72J96Hfyn3scxa7+9B3z/kexuxwli+5fy+521ciscmgPvHeNvKuQ2OIvJcD/hauA99Lr+sril273RWXdWuOu+fq1k1Pzdcdlub63mq1OihLKA93TkzbsvrWibf2qM/7gguI9/1Qqu/3Jt5zeCvfl3i3O7GJ3IiIKbGHMeRJ7g6bu30l0lS8u/ekjnlvcs6Nrrs2hvKHfsHf4nWw2WwOtWff826z2Rxqu+5uoe6Opu612q5VmLriPXR/Xb5uGd1zrv+pHTYPtcXumNqmrG4/Y89p36D6xd29FfG+H0r1/c7iPfWtvO6Ns+5N1a7s4O6aFstDHcH35hx7k9d7XYey5zebzaEVIi5dunQoD9v5NHnjtju6wX7I3WFzt69E2oh3t4+wwtn1WXs+tBKNzcuXprtzYyx/6A/8LV4H6/X6wDcWi4U3QKbPQx+hKLpvh1Q973t++u6P5esrY0iruALZZ6OWu2tZU9Kx5xDv+6NU3+8l8p7yVh574xRJf1NVYexzBHfSmfvm6j78Qm/ZLjZyFqobO+nNfUinlN+Keca17p/cHTZ3+wCmBP7Wz7AZ96VV0RddpU68ixx+fsbuj+XrK0soKu8OWbEBQN+v+l3KGhLv9uW9qirE+wCU6vu9iPeUt/JYo23yphoTve61sXRib9k+QmPRrA2hl4m68rsO3CSSAO3I3WFztw9gSuBv8TpoMyzWDiVpGnl374ndH8vXJSbeXU3gsyWkD5qWNUUHEXkfhlJ9f/BhMyHxug/xbs/78g+9Zdehadlxcoj3ssjdYXO3D2BK4G/hOrCLUNhAnYsrKOvGgbsbkem1+ot53Zh3a1MsX7eMsflpNjBndYn7/G5b1iY6yP4Cj3jfH6X6fi+R95S38twj7yHciSm2zOrkiPfyyN1hc7cPYErgb+E6SBkWq9g5YHXzR+rONVltJpavfe7WiXc3D/dZboN7bcpaN2zGpmmvRbzvj1J9v7N4T30rT2m0KW+qKeJd0/GNeXej8r63bF/ZfRPV3A6hrXh3y7FcLhHveyZ3h83dPoApgb9RBzmDeN8fpbb7zuI99a28TrymvqmmiHd3xQZfOr48XWHv/h26Vie0Xrx4sbV4Z7WZYcndYXO3D2BK4G/UQc7YFarYYbVfSm337LCaIavVCsfcM7m359ztA5gS+Bt1APOk1HaPeM8Ad91nou77J/f2nLt9AFMCf6MOYJ6U2u4R7zBLcm/PJ0+ePDJUi4ODYz/HyZMnx3b5vfCbv/mb8sd//MdJ11ZV3n0iwD4otd0j3mGW5N6ec7cPYEpMzd92u5289rWvlec+97ly2223yQ/+4A/KM888E72nrg50TptdpGIIQuvAN6Gq4qvM9MEQC02EytGkjlLrwveS28dw3ty+z1J9H/EOsyT39py7fQBTYkr+9qM/+qNSVZXcfvvtB6Lrtttuk+PHj8vFixeD99XVQZuNmvogN7HnQ19oYmvM90Ef5Wgi3t3r7BLZbcnt+yzV93sT732+lbf9cut2akvZgEkZ4k0dxiN3h83dPoApMQV/u3Tpktxzzz3yvOc9Lzg86Pjx4/LVX/3V8ju/8ztH7g/VQSit0EaM6/X64Bp35TVdKUUkvm67nQemq8fZ/w+t2ubaK3J47fTdbhfNV+3zRZljGkKXybZLOLpl9tVBaOf25XJ5qH417dTIe6gcbl1YtF7c/Wssq9VK1ut1Ed+nu6R3iFJ9vzfx3udbOeId9k3uDpu7fQBTogR/++IXv3hwfOELX5AvfOELcu3aNfmzP/szeeUrXynPfe5zg0LbPU6cOCGvfe1r5cknnzxIP1YHKUtC26WaRfz7rNiorbu3i7sEtF7bROzZyLAV0vZ5HsrX3ZCxSRR9sVgcEpK+Mrt/200k3fry2aLX14n3unK4aagY9olmNy/9vxK+z7ryWdtKpLN4T30rF2n3ZiYSfjutu08bhW/9d20svjdcm1fdmzqUSe7fY+72AUyJ3PztIx/5iPzUT/2UvPa1rz0UaTx27NjBcdNNN8mxY8ekqiq55ZZbkoW7HjfffLMcP35cLly4ICLxOrAbMKqQdGm6w7nNz3etm64vj9hO7Zquls2KTl++bXc2dzdQWi6XstlsgmUOBRF9Gzeq/lBBmireY+XQNOoi0zFdV8L36aLnPvWpTx0pZ4n0EnlP3ajJfftKeTOLvZ3G7rPEIgIpb7gxu6FMcnfY3O0DmBI5+dvly5flec97nvzwD/+w/M//+T/lox/9aPT6D3/4w/Lv/t2/iw6XcY9bb71VvuM7vkM+/elPH6TTNUAX2sXct/u4u6O5K6J953x51Ik9145YviKHg3epkzNtUFAP1UF14t2912qb1Wolm83m0ItTinivK4ebhr4curomlJevLLl+nyI3hhARefeQ+lbu21G17s3MJfblpg6bcb/g2BtuzG4ol9wdNnf7AKZELv72jne8Q5773OfKe9/73sb3/vIv/7K85CUvkTvvvDMouO+66y45ffq0/K//9b+O3B+rg5RhsUNE3t3nb1Ox1yRSmzpsxhW5dux4TLy7trgR8+12exAsdCPOLrGy1w2bcdOIjXl3r835+9T6r/sOc/H9pgw2bMYd+qJH3ZuZSPjttO4+JRYR0PRDb7gxu6FccnfY3O0DmBI5+NsXv/hFeeELXyi/9mu/1imdRx55RG6++eZDz6vbbrtNTpw4IY8++mjwvlAd2EUo7DPSxU5Y1WtdIWafm+5YZXessz3nij19drvLM9pzNn/7/6F87Zhqva5O+LkaRFkulwcR35B4d+/V1WosbsQ4RbzXlSNF0NZdV8L3mUoOvt+GXiLvqW/loUpNGROl7CPyHnvDjdkN5ZK7w+ZqX924zS64L8juuaYRJ4BUcmhD//2//3d5zWte00taTz75pDzwwANy1113yS233CIPPfSQ/N//+3+j98R+7U4ZFqu+aFc6cc/ZfiM2l8yec4fDukNCfL+Mu2naFV9i+dr5Be5Liq/fWy6X3uE1sRV3XMGr14WG1Vg7fIFEO74+pRy2LmI0Fe85fp8p5OD7begs3lPfyjUN/UnG99bpezOrezsN3WeJRQSsbaE3vpjdUCa5f4e52rcv8W79WORGh66kRosA2pCDv33d132d/OZv/mavaf72b/+2fPCDH0y6tmsd7PPFfq5Qn/snB99vQ2fxnvpWLnL0LSr01uaK8Njbaew+vc5OWA29nblvuLE3O5ypfHJ32Db2+dq5byxmKCrmRousX8XycCNLqWv/6r2hcaf60mz9/9KlS4fy8P1KFyq/HQLX5udVmC5j9wef/OQn5UUvetGoNiDe88FdZAP2x9i+35Zehs1MATqdeZF7e+4i3kNr3obWw3Xvs2M1QxOTUtdytn/HVmrSPEO796kQd/PwiffYWsN2nCTiHZSx+4M//dM/lZe//OWj2jB2HQCMQantfvbinTfceZJ7e+4i3lNm7sdWgoiJ3qYrSjRZqSm2n4Mr3n2rHMTK767k0HZNZ5gmY/cH733ve+VVr3rVqDaMXQcAY1Bqu5+9eId5knt7Dtlnh4S4L5x14nUf4t2eD62m4B4pv3BpWnbeCeId9sXY/cG73vUueeihh0a1Yew6ABiDUts94h1mSe7teYqR9xDuRmmK3Rob8Q77ZOz+4Ny5c/LKV75yVBvGrgOAMSi13SPeYZbk3p77Fu8i4fVw24j31LWc7fCX2EpN7jAZd4OTruLdLYe7njDMm7H7g3Pnzsm5c+dGtWHsOgAYg1LbfW/iXVeasUtHjoU+xEPDC+ru6+u6vtJwV82A7uTusPsQ73WrzTQR776Vm1LW/nWFvft36Fqd0Hrx4sXW4p3VZiDE2P0B4h1gHEpt972J95SNmobAjdiJXH/wx9afV4YU701AvPdP7g6bu32ls1qtmKQOB4ztb4h3gHEotd13Fu++CWn2Z3UltgZzyljdlDWoRQ6Pk1X0evfvUOQw1SYltEqGb91rd3xwXd3YNa5ZyrI/cnfY3O0rDffXOKLuYBnb3xDvAONQarvvJfKeslFTbA3mVPGu6YcmyNUtRWfL4lt7uo14d/OsW/farY+6ulF7ibz3S+4Om7t9AFNibH9DvAOMQ6ntvhfxvl6vD4TnYrHwiucUMZxyLiZsU8R7bO3ptpF3S8x2X33U5eWWEfohd4fN3T6AKTG2v+1bvC8Wi9pnyNh1ADAGpbb7wYfNjC3eY2tPtxXvdriL/Uke8Z4vuTts7vYBTImx/W2f4t0dihm7DmBulNrue4m8p0xWjQlUV3S3Fe9qS2jMuwr00HjX2HCYkHj3jX9HvOdP7g6bo32pE7VDK7l0WVu9qQ+wmhM0YWx/25d411/CEe8Afkpt953Fu10a0g6fcUkR7741mJuK99BqM3Ziqx2Hbtee9on3mE3u/SLXO0vEe/7k7rC52xfD3R1VSV31ycc+fQD/grH9bd/DZhDvAH5KbfedxXvKZFWReoFqV5Npu4GMtckOY3FXpAmtPe1G6+psUuyKMNbe0KY1TYboaL0iMPold4dtY19oZSZ7zq58FFp1ydpgz7ntPna/3RDKXuuu+uLanrLiUmh1J7WZ1ZygKWP3B4h3gHEotd2zwyrMktzbcxfxri+rdSsf2b/dl2I7/Gy9XstqtfK+tIZWSFqv196/Q3mmrrgUW93JVxdN0ob5MnZ/gHgHGIdS2z3iHWZJ7u25i3j3LZ3qnktddcmej831cEW1m99isTgyFj5mX+rQsdA4eoalQRPG7g8Q7wDjUGq7R7zDLMm9PYfss0M/3OFgoWFa2+3WK2Dt0BM7dCQ02TM218O30pNG723Uvy7PFIEdWt0pZCviHeoYuz/IRbxzcMzxKBHEO8yS3NtzG/u6RN596cT+vy7yLnJjeMpqtToYdlOXZ53Ajq3uVGcr4h1CjN0fsEkTwDiU2u4R7zBLcm/PXcS7ju32jXl3o/K+VZf0nApaTaduzLsrou3kVE0rlGeqwI6t7uTLu0naMF/G7g8Q7wDjUGq7702860ozdunIoYlFBl1iP5uEHuZN1o6GvMn9e+wi3nViqE3DJ95Dqy75ztk0Ytf4yuEb5+7m2WTFpdDqTpofqzlBU8buDxDvAONQarvvTbynbNQ0JDGh7S5jp0vrKTzIp0/uDtvHsBkASGPs/gDxDjAOpbb7zuI9NAHAtxlLKFIXWp9aP7sT80LrPMci75qmb2yunteInKZ76dKlQ5HM1LWjXRtDO07CeOTusIh3gOEYuz9AvAOMQ6ntvpfIe+pGTVZox9Z4VrHsm2gXW+c5ddiMu960z047OS72cpCydnRoXC6MR+4Om7t9AFNibH9DvAOMQ6ntvhfxvl6vD0TsYrHwRv5S1pVusouqYsevNhnzXrdLo295vdRxtO6Y2tCKGDAeuTts7vYBTImx/Q3xDjAOpbb7wYbNNFnjuU68h9Z5biLeLXqtXRED8T5tcnfY3O0DmBJj+xviHWAcSm33vUTeUyarNlnjOSbeY+s8p4j3zWbjtdFuB494nz65O2yXMe9Twverm9s/tCE2P0DP+XxW58Tsa15B7FfGEki1P7dyju03iHeAcSi13XcW73ZpSDt8JpRGyhrPMfEeW+c5NfLuDpPRNO1yc13Eu2vzcrlEvGdG7g6bu31DMbZ4d5fPRLzHQby3A/EOMA6ltvvO4j11sqpI+hrPdcNmQus8p6w2Y//22aLlqKpKLl682Fq8s9pM3uTusF0j73WrIdm14O1qTrF23XRVqNg68KH8tezui7TF7R9i81fqyumrH1tOd0lZvX6329WueuXWkw+to9Cvfrl+fyFS7Lf9t10BzJdPrB+tW73MLkxQ98I1dn+AeAcYh1LbPTusDoBuDw/5kHt77ku8+1ZDcoeO6e6p9r468ScSXxVK022av68eYuI9tvpUSjl99tk5MO79KgTf//73B/O1wtU9Z9PS6L6vvDl/fzFS7LfX6WffKmTude6qXaH73FXC6upc02rDer1u9CtQ6FrEO8A4lNruEe97QB8eehAh6reWAAAgAElEQVR1z4/c23Nf4j1lToYvjZj4azM3xYrsJnNArB/ZIySC+iyn5q/RYR1G44vixurJLXssCuyK3xy/vxgp9rvXhVYhi80darJ6mctut5Pf+73fO2J3U/QXjBTxriI/tJvvlMS7+/wbInDlq9Oc0oN8yV0LhEC8wyzJvT2H7LPDENyHYpNhXXboRNNhF23mpvgi5CkP97rIu1sn7styl3Jq2pvN5mDIjC1H3apX7jA939A/V/ilit+xv78QTcV7bBWymHhvsnqZosK5j8i7CvEm8y9i87CmIt59L6fL5TI6F64PEO/Qlty1QAjEO8yS3NvzPiPvLnbYhSvShojcdhk24+aTWs5U8a5DVHQ+j5ZDd1/25RuLvLu4QzxSxW+oXGN8f5aukXdLk8i7xZd3nRhr2x+kinf7y43P7lLEe10bsKu2Kdp23L/1UNrO80jZFT3mR64tqXMy3LkVUCalfneId5glubfnfYp3jSIrPvGnf9uVkpqIP03XjnlOyd9XDzHxHlt9KpZPqni3D27796OPPhrM1x3v7RvzHiJF/I79/XW1383Lvry432eoLLH76obNhOxuQx8rH4lMQ7ynvuCF5lK0neehdtl2504eD7VD+7Jh/cq2uzp7Qzu1QxnkrgVCIN5hluTenvcdebfRJfcnbXdIRlvxF1tFJJa/zaNOvLtpuTaG8kkV7yI3hkjYMtllI9183Yhck+8yVfyO/f2FRFqq/TYy6uZj022y2ozeh3hvR6gOQkOU3HaXIt5jv+i0neehtvvaXRM/V3t86aXYC2WSuxYIgXiHWZJ7e87dPggzh4f6er0evHz7XLUL8T5M5D02l6LtPA+1qw/x7qbXxF4ok1KftYh3mCW5t+fc7YMwc3io9yFY63BXLdnnql2I9/2PebeCN3au6TwPtcsn3lPmgITKSeR9HpT6rG0k3jk4pnTkTO72AUyJtv7mE+9VdXjYV4q4e8tb3lKEeK8jtNqMO0fDN5ei7TwPtT0m3mPzJvScnZti/z/VXiiTUp+1RN5hluTennO3D2BKjO1vDzzwgLz73e8e1Ya+6sAdI5+6c2+XeR6hXdFFwnNAYrbE5mSE7IUyGdv324J4h1mSe3vO3T6AKTG2v73iFa+QP/iDPxjVhrHrAGAMSm33iHeYJbm359ztA5gSXfzt6aef7pz/85//fPn0pz/dOZ0u0OfAHCm13SPeYZbk3p5ztw9gSrTxt6efflr+y3/5L3LnnXfKO9/5ztZ5f/zjH5fnP//5re/vC/ocmCOltnvEO8yS3Ntz7vYBTIkm/mZF++tf/3r52Mc+1invN7zhDfKGN7yhUxp9QJ8Dc6TUdo94h1mSe3vO3T6AKZHib32LdhGRD3zgA/KiF71Innnmmc5pdYU+B+ZIqe0e8Q6zJPf2nLt9AFMi5m/7EO0iIs8884zcd9998q53vauX9LrStc+JrZs+JH3a4e410McmYa59dlnK1HtErq+i466641479Hey3W572wMiZnuf5cqhzbYB8Q6zJPf2nLt9AFPC52/7Eu0iIu9617vkRS96URbDZRT6nMOE1qz3CeYutBHvu90uKJLHfokKvVT0CeId8Q4zJff2nLt9AFPC+lvfov3Tn/60/MEf/IG8+93vlre85S1y3333yX333Scf+MAHuprdKyl9Tmxdc1/Ed7VaHUSt7UZJVtyp2Autp27z9p1z/z+0K2ponXafjSLx3WIVd017vV7TtmvMa71Z++za9bvdrjY9ZbVaHbLN/kKg5WnynaTUk659H7vWfif2s75s2O9dNziry7eujF0p9VmLeIdZknt7tp0ZBwfH/o9PfvKT8spXvlKqqpIv//Iv7y3d5z//+fKKV7xCHnjgAXnLW94iv/ALvzB29+KlqvoX73ZnUo0UbzabQ1HjxWJxIOBUkLo711ohbXdb9f2/K/rcHVI13ZiNak9sAyb3GrtDqxXKoXO2Tu1mULH07D0Wt+7c76GuvL403HqyLwuhOlWWy+WRXxN837tuxBX7furK2JW+0hkaxDvMktzbc+72AUwJ62/7HC6TM6E+x40G6+EOjfAJxdBuqXpOI7Lb7da78+lut/MOkUj9/1AU3l7nszFFvPvqyRXben+sPCreU9LTz7auQhHq0GdfeVPq3/fLQaiufENn7HX6OeX7qStjV0p91iLeYZbk3p5ztw9gSvj8bW4ifh+R95B412EfKvJCLwghke7mF/p/TVfpW7zbIShVVQXFu9bddruNive69PSzrctQGX33hcqbUv/2vhTx7pvcu1wuZbPZyGazOWgDKeI9VsaulPqsHVy872P2dh2hN9tc0oPhyd1hc7cPYErE/G0uIn5I8b7dbg+NeXYFaSjdpv/fNvIuEh/z7nup6Bp5T0lP09p35N1Xn10j7/r/+r1vNhsi7x0YVLwPNXvbBfEOLrk7bO72AUyJFH+buojv2uc0Ee+anzuURsWyG2m1z1w7Ftz3/3Vj3kMC27UxpFc04OjauFgsjqSt+aaMeU9Jz95jccvYVLzH6t/3K0KoTm09+TSSDd7WpVX3PSLeUy/s+FYukjZ7u+3McJ0J7Ubz7WzuS5cuSVXdmDWd2sFYW9zZ4U1maEM+5P6d5G4fwJRo4m9TFfFDi/fVanUocOc+S62WCD1nff/fdLWZmI3ucBJ3pIDVAzY99/kfixpbPVKXnq07q6VsGduK91D9+8R7m9VmbJ3ZekxdbSZUxq6U+qwdTLynjiFrMzPcN4vZCnt1BHfWdJOfzewYLvviUGev+7ICeZC7w+ZuH8CUaONvVsS/853v3INVwzJ0n5OiB0rFJ3j7JLbO+9gMsc57n5T6rO0s3lNnoqeI97bj03xvzK7tVryH3ihDY8ysPb70UuyFvMjdYXO3D2BKdPG3p59+ukdLxmOoPsf+ij5Vhnj+5yiSY2Pnc6XUZ21Wkfe2M8NF5NBmCG6n0Jd4d9NrYi/kRe4Om7t9AFMCf6MOYJ6U2u6zGfPuG9fVRLy7+cSGzfhmgLtppoh3Iu/lkrvD5m4fwJTA36gDmCeltvssVptxJ5g2nRnurinaVLzrtcvl8sjs67oZ7qn2Ql7k7rC52wcwJfA36gDmSantfvB13utmb7edGW5nabvjwHQ298WLF48Iane4TWz2tZvedrtNthfyIneHzd0+gCmBv5VZB775dvbX/br1wGPn+1xLfB9phsa8x9ZH3zfb7ba3ibRDfTcltnsRdliFmZJ7e87dPoApgb+VWQf2V3CRGwG3PlZ5y1m8x1abGVqwuwwxkRbxjniHmZJ7e+7LPnY0Bqgn9/5gCLrOa7PLOWt/Y691f3W3AtueS/0FXO1x+4b1en1kyWaRoxtC2j1j3DKoHb45baF7QnvQtElT7Q312+4676E8fFH4kJ1N9qxps867r/51h91Yvin12IVSfR/xDrMk9/bch33saAyQRu79wRD0Jd5VaNp5Yu7iEPacpmvnkblDYn17qbj3uXa4n337wbgi0L5U1J1L2YOmLs1Q2VL2rrGE8vDVRaqdsT1rYt+JiH+HVV+ZdPhzbK+clHrsQqm+j3iHWZJ7e+76IBVhR2OAVGgz3fdyqVvNzU0ztE9LbNU3N80m4t3eaxer8F3rS8f3a0DdSnh1aYbKFltNz7UlNY86O1PLV/ediPiHztTVfyjfujJ2pVTfR7zDLMm9PXcV7+xoDJBO7v3BEPQVebfnre/aQEBVVUniPbaXipu+a4f7WeR60GGz2chms5HVanUkMu/Ly55zj5QNJOvSDJUttHeNK7RjeaSK96blSxHvviGabv2n7JVTV8aulOr72Yt3n3M2Yd8zr9uOKU61pavNQ+7CVtJM89wdtmsUjB2NAdLJvT8Ygr7Fe2wPl31G3kNj3vWcvvRvNpvWkfdYufuIvLvYAMlut9t75D1WvraRd/3/UP27aRF5jzMr8d43XcYUDyHeYzPS90UpM81zd9ghIu/saAxwndz7gyHoWgfur3V2XLvru+64dvucd/dbCe2l4t4ncnTvltikR995Ny97rqpu/LJny5Pyi2IozVDZUvausYTySBXvTcoXs1vxjXm3aaXUQZPvpgul+n6v4j1VLOjh/sRtz4n4x8+GGl4o7dDbmxUfNnpZl44dU9vHmGL7OUXUNLHPzkiPjWFOyd9dQSAk3PR7tJ9znGmeu8P24W/saAyQRu79wRD0Jd7tfBiLfZ6HhmJo/95ktRn3sH2Rr1/Riaq+8+6zLfYsTwlKNE0zVF9uQMxdbSaURxPxnlq+Orv1e7GfXeFvdUOqBojVYxdK9f3BxLtvPFeT8a6xhhdLO3XYjArhunTUxtQxxfae0EzqVPGeUk7r0D5HiI0XrhPvIjcmKsYEUwkzzXN32D7sY0djgDRy7w+GoC/x3tUHV6vVIEvalswYv6qnMuRQ3T4o1fc7i/fUMbi++9qMd02JBIbSDol3V/TG0glFEX2448jqxnM1jUjG7HPzTskj5VzKUl0lzDTP3WH7so8djQHqyb0/GIKxxLv9VbWqquBzHQ6To0iOjZ3PlVJ9f9BhM6HZ5k1+Mg+JiZS0Qz+hWeFel04T8Z4yg9uXdky8p9o3tnjPfaZ57g6bu30AUwJ/ow5gnpTa7gcT776x203Ee5PxsqmRd3ccdko67titujHFdTO43Xua2F/3y0CqeE/Jv2vkXf8/l5nmuTts7vYBTAn8jTqAeVJqu+9VvMdoMtvcN941Nl42lnZIBPrG1aWkY8V7ypjiqorP4BaJjwWORaDr7EsdBpGSfxPxXsJM89wdNnf7AKYE/kYdwDwptd0PJt5F4rPNU8a7xsbLhtIOiXd7vZtmLB33l4WmY4rdGdxKqGw+EZtqn2+1mdCwo7r8m4h3a6/v14ocZprn7rC52wcwJfC3vOqgy6+r7nPdPi/3YZ9vcYUh888xTf31fbvNf++XnNp9EwYV7zAsY8xIz3ESjY/c23Pu9gFMCfxtOnXQt1Cuwyfe951/zuLd1R257/1SartHvE+coXdYLWWmee7teZ/29bUyS9uHlC8q1efScE078qF8JNUu36YnIvU728ZI+a5KiJLti9zsGYOUOqjrN+p+SfXtM+LbA8S9v8n+JrG2rten7lvSdP+YPvMPXevLM7buemx/HTuUtcka9HYtf7fO3DXotU7sZxX4Oez9UqrvI95hluTennO3T6SbeHfv803+HoIc10vWh5JvLknbl4zU7yr3KNm+yM2eMehDvFtx6Nu3w7enh28PECvG2uxvkiKeReL7lrTdP6av/O0LTyz/WL3X7a/j3mfTDO3B4vu+bHl97SjnvV9K9X3EO8yS3NtzHw/SuqiR74HhRndDERY792K323WOjK1Wq4Mxku71sQiQpmfPNYlK+eaFxCJdQ9llH2D2Whtt8kWhfJHN1O/K2m0/5xQl2xe52TMGoTpI3cvF/eXVt3pY3WpnoT1AXHvqFmpwjy5zuGL5up/7zD/2MuHmH6r31DLUlcmmGfsFMPTre857v5Tq+4h3mCW5t+eu4j0lalT3UE2JsGy3R5dxbRMZ0/8LXe+LKqlN7s7MdSsVuQ9LJSXSNZRd6/Xa+3coz1hk09ZvXSROJO8o2b7IzZ4x6NrnpOxrEgsS2D1A3PaTur+J2lgX+U4V7232j+kr/1jZbJ6xeo+VIbZ/SmwPFpGjC1zY8oXEe657v5Tq+4h3mCW5t+euUTDffW3Ee2yMdegh1SYypvb7fjpO2aXYnk+NILlpp0S6hrDLZ4sOJWgT2Uz9rpSco2T7Ijd7xqCreA8JNz0Xa5+xPUDc9pLSv/QpnmP5+q7bl3hPjbz78qsrQ12ZYtF8G9TZ7XZB8Z7r3i+l+j7iHWZJ7u2564NUJC1alfJQ9UVYNH/ttPcVGUvdpdiXhr1faSLera1D2qVodNtG/evyTBHvoe9KyTlKti9ys2cM+qgDbaci/n1NQu3TDquyf7vpiKTtb9KHeG6zf0yf+bu/pqWOebd21+2vE9s/JbQHi9s/tB3zbsuRkm/dL5eI99CFhRYQwEfu7bmPKFhKtMoVjbFoe2jYzD4jY02iSr7/j0Vy3ChRl8h7n3Yp+mDXDeW6RDZTvisl5yjZvsjNnjHoow7cORChX5J87c7uAeITappmLPig5fAdts2mDptpun9Mn/nrtXbekRLyPbfeY2Vw72uy2oxN0+0rUlebsWmNufdLqb6PeIdZknt77mpfarTKDoEQOby7bkqEZbvd7jUyptf7okpuWhqZqovQuKLWzTsW6RrKLpunTSuUZ6p4r4vEieQdJdsXudkzBtRBXvj6nBLY7YZf570LpbZ7xDvMktzbcx/2pUarUncudjtgu/tx08hYE/Eeiyr5ojR1USmLb7WZWKRrKLtsXfnGubt51on3lO/K5mk/5xQl2xe52TMG1EFelCreRQ7vsNp2b4qhKLXdI95hluTenvdpX47DFsbCRolKflj2Re5Rsn2BP1AHME9KbfeId5glubfnfdmnUfY5CrQQNko0Z/FeQpRsX+TeHwwBdQBzpNR2j3iHWZJ7e87dPoApgb9RBzBPSm33iHeYJbm359ztA5gS+Fv/dTDk8LyuefVpq/0lz07c7ELMPoZBdqPUukO8wyzJvT3nbh/AlMDfyhbvuTDGSitzrOc+KbXuEO8wS3Jvz0NMWO06tju2akzdfe7RV4SqKU0ffO74+JIhMniD3O0bgpQ6qOs37HKiTdYNtys8+TYIC63cZFdRcs/ZVbRcm9uuBKXX2Y3ObB+Yusb5crk8JOp1vffU1Zti9QzNKLXuEO8wS3Jvz7nbJ9JNvLe5bx80EZZuVG0KEBm8Tu72DUEf4r2qbuxD4IpK95z60mazOeRX7n4SutOwyI29J7RNucu82s/6EmD3a6hLz15j9yVwV6TyndMyuvj2TfCVWZd3jeWbUs/QjFLrDvEOsyT39tzHg1Q35NHDfdD51mEPrQ/uRsTsWuG73a42L01H7Q6J97rIkxtpc6NrPltjZfTtOOorh4h/TXj7ORSNC9Vhil0p5YvVWyx/hchg/v3BEITqwPUJPdyXvthOuu5KRnY33tiuzr4Xv9COx64/xnZKDqXnyzO207Lrs75y+F6Q3Z2ebZ8Wy7eunqE5pdYd4h1mSe7tuat4tx2/SHi30NjDqC4ipg+clLysCI6J97rIkyue9VoVqLEHe514j5VD7VZCP9O7dsfqMFW815UvVm9136GmPffIYO72DUHXPsfdvVf9yZ5zj9BGcUpImDYR72r3drutTS9Wji7i3ffSvFwuZbPZyGazOQgMpIj3WD1Dc0qtO8Q7zJLc23PXKJjvvjbiPbbud0iEh/Ky9/ke4k0iXu7fvp+vm4jklDqzdRETC6l1mGJXk/L56q3uOxQhMiiSf38wBF3Fe5PIewzfC24orz4i76H7lVTxvtvtkiPv+v/q15vNZtL+lTOl1l0j8c7BMaUjZ1Lsq6r4sBk7jKOqqsbiXSQcEdP89SGbkpfvPkuTiNe+xHusHG3Ee6wO+xLvsXqL5W/tm3tkMHf7hqCPOnB/mbFpVtWNX2NsG3HbX+gXPr1Wz/v6A/tZ7fD9ghZKL1SOkF+6vu6rQ98vWzatJvmm1DM0o9S6K9PqCfPUU0+NbQJkQFfx7hvL7XsAuUIvFqkNPVRT83Lvq7O5L/EeK2MsohWLqjUR76E6TLGra+Q9lr9CZLDcB3if9FEH7rwMX3vRwx32pf8f+hXICt0U8W7neqSmF7vGl6/r66mrzdhy2xeX1DklsXqGZpRad2VaPWHe/OY3j20CZEDXDsWNfi4Wi6h4V0G3XC4PrkuNiKXm5d7nIzXi1Ua8+8oYixjbcmiaSqp4j9Vhil0p5YvVW913qPnOPTKYu31DMJU68PU5Q7HbDb/OO3Sj1HZfptUT5amnnpIv+ZIvIfoOvf2EreIrNlTGHVbhLqsWiojZlV9S89KyhcR7asSrqbgNldEnOn3l0Dxiq82EonGxOqyzK7V8oXqry1/Tsp/nGBnM3b4hmEodjCneRQ7vBZE6zh/Go9R2X6bVE+XNb36z/Kt/9a+IvsNeO5QShjHkiBtVmwJEBq+DP1AHME9KbfdlWj1BNOr+2GOPEX2HvXUoGulFsLVjSmKXyOANSn2A9wl1AHOk1HZfptUT5M1vfrN83/d9n4iIfN/3fR/R95lTaocCUCL4G3UA86TUdl+m1RNDo+5XrlwREZErV64QfZ85pXYoACWCv1EHME9KbfdlWj0xbNRdIfo+b0rtUABKBH+jDmCelNruy7R6QrhRd4Xo+7zpu0MZcpJq17z6tNWu/NDXZNOYfUwGLhO+M+oA5kmp7b5MqyeEL+quEH2fLyWL91wYY83lOdbzFOA7Yxd1jvkeJVKm1RMhFHVXiL7Pl5QOpariaxnbTXbqdju02J0J7RrfNl97n7ujYWircr3HtTmUXp2tep2WTfNWUnc7XC6Xh0S9rqmeuqZ5rJ6hDPjOAKAk6LFGJBZ1V4i+z5M+xHtVVQfi1RWV7jmNUG82m0PRandHzsVicXCf7t6p4jW2gZG+BKzX6yMbGIXSs9fY3TrVPvti4J7TMrr4dhP1lVk3Worlm1LPUAZ8Z1ACn/nMZ8Y2ATKBHmsk6qLuCtH3eRISE5vNxvuznzscJBQd1s9WQGuEebfbHdkdNJam3hva/dR+1nM2r7r0fHn67vftbBpaw9w3dMamqZ9T8q2rZygHvjMogfvuu29sEyAT6LFGIiXqrhB9nx9dI+8q8hUVn/ace2hadoiLHTYTEqZNxLvavd1ua9OLlaOLePcNBVoul7LZbGSz2RwMt0kR77F6hnLgO4PcuXr1qhw7dkyuXr06timQAfRYI5AadVeIvs+PruK9SeQ9hh0204d4r4u8h+5XUsX7brdLjrzr/+vQmM1mQ+R9ZvCdQe6cO3dOXvrSl8q5c+fGNgUygB5rBJpE3RWi7/OiDzHhjtkOjdO20WM3Mu2OebeTQnX8eop4VzvcMe+x9ELlsNH1kHjXtF18Y95tWk3yTalnKAO+M8iZq1evyokTJ+Ty5cty4sQJou+AeB+aplF3hej7vOhDTNjVUupWm7ER/MViERxL71uBJUW82xVsUtOLXePL1xXvqavN2HLbF5fU1WZi9QxlwHcGOXPu3Dl58MEHRUTkwQcfJPoOiPehaRN1V4i+z4epiAnfmPehGGOddyiTqfgbTA+Nuj/++OMiIvL4448TfQfE+5C0jborRN/nw1TExJjiXeTwDqup4/xhfkzF32B62Ki7QvQd6LEG5M1vfrPccccd8uIXvzh4HD9+PHr+jjvuIPo+AxATAMOBv0GOuFF3heg70GMNxFNPPSV///d/X3tUVZV0HdH3aYOYABgO/A1yxBd1V4i+zxt6rMzgIQIitAOAIcHfIDdCUXeF6Pu8ocfKDB4iIEI7ABgS/A1yIxZ1V4i+zxd6rMzgIQIitAOAIcHfICfqou4K0ff5Qo+VGTxEQIR2ADAk+BvkRErUXSH6Pk/osTKDhwiI0A4AhgR/g1xIjborRN/nCT1WZvAQARHaAcCQ4G+QC02i7grR9/lBj5UZPERAhHYAMCT4G+RA06i7QvR9ftBjZQYPERChHQAMCf4GOdAm6q4QfZ8X9FiZwUMERK63Aw4OjuEOgDFpG3VXiL7PC3qszOAhAgAAMC/e+ta3yl133SVf/uVfHjxe8IIXRM/fdddd8ta3vnXsosAAoBQzA/EOAAAwH5555hn5xCc+UXtUVZV03TPPPDN2kWDPoBQzA/EOAAAALugDUGgJmYFzAgAAgAv6ABRaQmbgnAAAAOCCPgCFlpAZOCcAAAC4oA9AoSVkBs4JAAAALugDUGgJmYFzAgAAgAv6ABRaQmbgnAAAAOCCPgCFlpAZOCcAAAC4oA9AoSVkBs4JAAAALugDUGgJmYFzAgAAgAv6ABRaQmbgnAAAAOCCPgCFlpAZOCcAAAC4oA9AoSVkBs4JAAAALugDUGgJmYFzAgAAgAv6ABRaQmbgnAAAAOCCPgCFlpAZOCcAAAC4oA9AoSVkBs4JAAAALugDUGgJmYFzAgAAgAv6ABRaQmbgnAAAAOCCPgCFlpAZOCcAAAC4oA9AoSVkBs4JAAAALugDUGgJmYFzAgAAgAv6ABRaQmbgnAAAZbNYLGS73dZet9vtZLVaDWARTAH0ASi0hMzAOQEAyqWqKqmqqla8r9drWS6XslqtZLFYDGQdlAz6ABRaQmbgnAAAZbJYLGS32yWJdxGRzWZzcA9AHegDUGgJmYFzAgCUTYp43+12slwuZbPZMHQGkkAfgEJLyAycEwCgbFIj7wBNQB+AUun4PA4ODv8BRxn7O+HgyJmq6le8j13XHBwc2R15d4IAY4J/+KFeYExyb39V1b94BwAQEcQ7QB34hx/qBcYk9/bnindrbxvbcy8vAAwH4h2gBvzDD/UCYzK39je38gJAGMQ7QA34hx/qBcZkbu1vbuUFgDCId4Aa8A8/1AuMydza39zKCwBhEO8ANeAffqgXGJO5tb+5lRcAwiDeAWrAP/xQLzAmc2t/cysvAIRBvAPUgH/4oV5gTObW/uZWXgAIg3hPJLRIfl/r+O52O6mqqtctsjebjVRVJZvNprc05wj+4WdO9bJYLIJ9QBO2261UVSXr9XqQ+6bMnNqfyPzKWzK2v1gul2Obk8xqtZKqqmS3241tCtSAeE9k306IeM8X/MPPHOulq4hGhPfH3Nrf3MpbKuv1+uC7Ks3fEe/lgHhPJCTeVSBro9fPy+XySHReHdleq2m64l3TrapKFovFoTy1c/BF/+05zceKd3sfoj4N/MPPHOvFfRjr3+rv6s8hH431ASn3WREwd1+eUvv72Z/9WXnnO98ZvWZK5Z0y2hf4fpXXc8pisTj0fLe6wfYLsfts32Pztf2DGxS052w+iPdyQLwnUife9Zw2fn3I2nP6AFansw9kK97dB/VisTiShjqXPee+AOLXSkUAACAASURBVKgz64Pd2kJUPh38w88c6yUk3u0DOOajsT4g5T5fvzJXX55C+/v93/99efnLXy4vfOEL5fjx4/Lyl79cfvd3f9d77RTKOwfsi7kbcY+JcCucQ89y333uc96mJXIjIKCifrlcHgkS6jnEezkg3hPxjXVdLBZHHpxu47dO5oue6QPaOqs6m6bh/m3/zwoB1xGtbe45mzfEwT/8zLFeQuLdN9zN56OxPiD1Pnz5OiW3v7/7u7+T7/qu75K77rpLvuRLvuTQc+X48ePyute9Tv72b//20D0ll3duuFpBiYnwmA+niHerD2wEPyTEfb/cId7LAfGeSF3kvYt4XywWh8S7fXO3x263OxK5s+m7b9jWNvecey+EwT/8zLFeQuLd+nTMR2N9QOp9+PJ1Sm1/58+fl1tvvVVuu+02bz+vx3Oe85wjw6SgHHxD42IiPKQx6u5zz4nc6C9iLxA2qId4Lw/EeyL7FO91kXdLLH0i7/sB//Azx3pJEe9t+4DU+/Dl6+TW/j7/+c/Ln/zJnwSPX/u1X5NTp07J3XffHRXt9rj77rvlnnvukV/91V/NrrxQjz7X3aExSmrkPXafT7xbdOWb1WoVnFuHeC8PxHsifYp3re/UMe/L5dIbXXcjdYx53w/4h5851kuKeI/5aKwPSLmPMe83yKH9PfXUU/L6179evv7rv15uuukm+Zqv+Rr5uq/7uiPHYrGQe+65pzbaHorAnzx5MovyQj3Wb/VvfSZbfeD6uKsdrI/H7vOJd6s7VBf4+g5XIyDeywHxnkioY33DG97QWLw3XW3G/X7s/7uOa+9zV5vRPPSY28O+LfiHnznWS4p4Fwn7aN1qM3X3aT748vjt7y/+4i/k3nvvlYcfflj+9//+3/JP//RPtfc88sgjcvPNNyeJ9ttvv12OHTsmP/ZjPyYi45cX0nB90x3OZv07ttpM6n2hyLvblyh2To3eq/0K4r0cEO8DUtqar3Ad/MMP9TIc2nfMUaSHGLP9ffCDH5TnPe958jM/8zON7/3rv/5reeCBB+Suu+4KCvc77rhDXv3qV8tf/dVfHdyHvwGAgngfEMR7meAffqiXYbCRMrjBmPXxAz/wA/LWt761UxqPPfaYvOxlL5M77rjj4Ps9ceKEnDp1Sn7rt37ryPV8/wCgIN4BasA//FAvMCZjtb9nn31W7rjjDnnyySd7Se8d73iH3HHHHXL77bfLI488ErwOfwMABfEOUAP+4Yd6gTEZq/09+uij8sADD/Sa5j/8wz/IZz7zmeg1+BsAKIh3gBrwDz/UC4zJWO3vu7/7u+Xnfu7nBs8XfwMABfEOUAP+4Yd6gTEZq/0tl0vvmPR9g78BgIJ4B6gB//BDvcCYjNX+Xvayl8mHPvShwfPF3wBAQbwD1IB/+KFeYEzGan8vfOEL5VOf+tTg+eJvAKBUVSWV7t7GwcFx9Dh58uTYfpol9BscYx6uX67X6+D28j6aXGupqnFENP7GwcGhxz/3B7zRA4TAP/xQLzAmtv3pWvgpglxFvu4uud1uW+c7JPgbACj/LOLpFABC4B9+qBcYE21/KsRXq1VyNF03zOuS79DgbwCgIN4BasA//FAvMCZu+0sV77vdThaLxcG/XfMdCvwNABTEO0AN+Icf6gXGpK147zvfocDfAEBBvAPUgH/4oV5gTBDvADBXEO8ANeAffqgXGBPEOwDMFcQ7QA34hx/qBcYE8Q4AcwXxDlAD/uGHeoExSRHvVVXJbrc78rnPfIcCfwMAJWvx3mU5Lx+6rq/vaGNX0wdB2/tgXHL1j7HJtV7m6Gex/iy0lnnf/evQzE1E5/BduW0mZZ381HbWR3uM+X7p7R3AMivxbmmzOYcyR3EwZ3L1j7HJtV7m5p+LxULW6/XB37phkdKlr8uZMdrftWvXZi3eXRDvAOPQSbzvdjtvtEedRB8i6kz6ebVa1V5rz9XlV3fO58xup6N5aXReRGSz2RxKc7PZHLp2tVodsTnlPr02ZjPkA9+Nnzb14vMd64d99ik+//T5eRc/9N1X5/+hPma1Wh1adzylT9NrfP3bZrORxWJxcP+lS5cO5W/717rvJWbnWIzhl0888YScOnVq8HxFupW3TTuteybbtrXb7WrTs7a46dm2p9T1BZqGtnN7Lub7dekDlEAn8W4d1I43tM4vcmO4ym63O+J41tlErkeNXEesy0/kcPQpZeJSSLxr+u5D0WeXz+aU+/RcU5thHOjY/XQR76F2H/KJvvsU9XMtR6hfibFYLA7uW6/XslqtkvzfzVv7ocVicUgUp/YPWhc2XYvm4ebvE++h/GJ2jsUYfnn58mU5c+bM4PmKtC9v23aa+kzebrdJ6YXKZH3PXlfXF9SJ9xTb2/g9QA60Fu/b7dYbJfI9TGMPglA02qaTmp/vXKzgPvEeusfXQfhsTr2vjc0wDoh3P13Ee+zBq+yzT7FCP9SvpJTD0sT/3XNN6sJHKOpp6yb0klNnW8zOMenql88++2zjey5cuCBnz57tlG9buvibpWk7jT1r3edoLL0623zt0doc8uE2trf1e4BciIp3+9OT/Vla5OjDwv5E1eVBa89bZ4vlp+eUvsS7LX9VVcEOwk0v5b42NsM4IN79tOk36kTiPsS7Pe/LP9SvxIhFE1P7jZgo7tI/aD5aH4j3wzz77LPy4z/+43L33XfLT/zETzS69+zZs3LhwoVW+Xali7/V3Zf6fIuJ97r0XEJtPHauqXgP2d7W7wFyISreY7hvru65tg/a1DdlX35KH+LdTbNtZILIe/kg3v207TfqHrzKGJH3puWo+/99R943m4335347VALxfh0r2r/3e79XPvzhDze6/8qVK1JVlVy5cqXRfX3Rxd/q/r9r5D0lvTobUiPvri/0EXkHKI3W4l1v1geEfVNu86DVsW2pY9TcN3N3fFydY9aJd1/6rl0+m1PuC415pzPJE8S7n77Fu0jYJ/ruU9zIXKhfqSu/5ql5NPF/N43lcplUFz477DAZtUHz6Sre6+wci9TvqatoV+6//345f/58q3v7oMtzuk07TX0mb7fbpPR8uG085ZmugtzXHvvSEwC500m8qxO5Pzm1edDaNdiV0Nu37ycu95xbSDdiVSfeRQ7PpvdFze3PhJa6+9yogC8NyAe+Gz/7EO8hn2jTp/j80+fnsX4lRsjWVP/X8uu1bVabsX+HyqB968WLF1uL9xJXm7l69aqcO3dOTpw4If/xP/5H+chHPtI6rze96U1y+vTp1vf3QV/PaaWunaY8k+3KRXXp2XL4noGpq82I3FgO1W2PKb4fSp9fvaEkOon3PvA90KZKLAIB+cJ35od66ZfVanVkzHKO5GJnqP31Kdo/9rGPybd927fJ6dOn5bHHHmudTh8M5W9zeiYDlArifSA0UmA3U4EyQKT6mXK9uFE5e/QVdda+r+90+yZXO932Z0X7937v98qf//mfy+c+9zn5/Oc/HzyuXbt2cHzuc5+Txx9/XH79139d3v72t8v3f//3y7Fjx+Qnf/InRyrhYRDvAKCMLt4Bcgf/8EO9wJho+/v85z8v6/VaXvziF8s999wjN998s9xyyy1yyy23yHOe85xDx80333zkuOmmm+Smm24SEZGv/MqvlFe96lXyxje+UX76p39aPv7xj49ZxEPgbwCgIN4BasA//FAvMCZu+/vc5z4nb3vb2+RFL3qRvO51r5MPfvCDI1m2H/A3AFAQ7wA1pPjHM888M4Alw/GP//iPtdfQb8CYhNqfK+L/7M/+bGDL9gP+BgBKJ/FuV3jwjQ21y5fZmevu0YS24/Ha3NdkgqmtC5gWsTZw9epVeetb3yqf+MQnBrRo/3ziE5+Qc+fOydWrV4PXDCkm2k72bnMffl8Gdd/R//t//08eeeQR+dIv/VL5zu/8zuJFfK7ifeiFGHw+564+1YaYRtBzvvkeusrOPuYIlKibXEL299lv7qMNpqZp5wTp9zXEhP5exbv9MnTClxXwvvuakutkGh7i08XnH1evXpWHH35Ybr/9drn11lsnKd5vvfVWueOOO4IivgTxvm/w+/FIbQ+uiP/Qhz60Z8v2Q47tX2R+4t1dZnaf4t1Sqm4aoo8cW7zbl7r1el22eBe5XgifQ7nXuuvKihzdvlhfAnzruIbWTfbdl7LGul7nWwfZ5qtlsOvb5vZSAd2x7cOK9uPHj0tVVXLnnXdOUrzfeeedUlWVHD9+3Cvi2/YbNprkdnJ1/pt6nV1/2t6H30+Hpu3vn/7pn+Snfuqn5IUvfKH8h//wH4oT8W39rW7vBLtmurvuet219lxdfnXnYv2CzS9FvIf6h1g+MV+35bcrxqlQU/vq+iVf/YW+LzfwmbtuChET72O0wVjbsFH01H0HQuLd3Syvb/7Zjv2I99CbS6gRaiW62x/7dknTL9WeS7kvtLup7yXD9xAP3RNroFA2VVV5RbseUxfvergivk2/sdlsDvnNYrE48JsU/029LiT68fvp0Pa5ZUX82MtA/u7v/q78yZ/8SdK1XZ7T6g+2/bptW0Wgb0Ox2HM3tnOp6y8h/4v1C77yxMR7rH+I5RPzdT233W6P3K8i7f3vf38r3WLTqqrwUqyl6KY6uy1Dt8FYWX33pTw76iLvdd9rW7IS76E3FLvbX+hN0Hdv6L7QW5qbhu8hnrJ9OEwHFanHjx+XW2655ZCY1eO2227z/n/pR6hct91228ELTGxMvA/Xb1Kvjf2EGesffD6s4Pdl0/a5pXz2s5/tyZLm/M3f/I18z/d8j3zFV3yF3H333fJDP/RD8n/+z/+J3tOmvK6wsG3ebdspuxb7nruuv9Tl5zvXpF8I9VchIWnTjuUT83Vru16jQ2ZC/UiqbkmN0Jaim3x2u0fIrn23wZSyKqnPjtRhM7Gh5G3457r0dwr2JxafMbZifQ+xLuLd5h37st30Uu5TZ1F4iEMMFannzp2T22+//UhHNJfI+6233iq33367PPzww9HIe12/4W5tHrrX57+p1/nEO34/LbqK97F429veJrfeeqv85//8n+Xzn/+8fOYzn5HVaiVf9mVfJv/jf/yP4H1t/M0dIqBHV/Fuz/t8zJdfnf/F+gW3HuqGzYT6h1g+qeJdh0SoSLPlaKtbRG5EaEMvIaXopjq7LUO3wVhZQ+WLnUsR7/prQlGR96Zj3kPjl1LfIFPvIwIHTbBtxSfipy7eXdGu9CGefD9bK6GHZ5P+gcj7dClNvL/nPe+Re++9V77lW75FPvzhDx85v91u5b777pN/+2//rfz+7//+kfNtyusKC/dcW+GUGvX05afExF+XYTOx/iGWT6p416E3y+VSttvtQTkuXbrUSreE6il1zHtuusllX+K9jza4z8i7Cv59PR/2Jt61cD7D6xqh+5azWCyOVLCOPXLHs9XdFxq75fuyeYiDSHi1GRXxz3nOcyYp3m+55RavaFfa9Bu+8YDqNyn+m3pd6IGG30+HUsT7Rz7yEfnWb/1Wuffee+U973lP7fU/+7M/K//iX/wLef3rXy+f+tSnDv6/y3NahaD1nTbCyffcdX0slJ9I2P9i/YKvPDHxHusfYvmkinfVNu7fjz76aCvdkkopuqnObsvQbTBWVl/5UtpuXeR9X/Qq3t0j1fl8P4fYlRx8b3/2pw9L3X3uW5svjdDbXOwhble3gGkR8w/WeW+O9VG7coN7Lhb1SfXzUMQEvy+fIcT7xz/+cfnpn/5peeMb3yivetWr5Cu/8ivlpptuSj6OHTsmVVXJq1/96kb5Pvvss/Kv//W/luPHj8vb3/52EelvtRnXN5oIJ7tSi1LnY/a5HvO/WL9g06kT725aPgHqyydVvIvIQeTdlknHwMf6JV/9pVKKbvLZ7Ts2m80obTDWNux9XVeb2TedxHtJuF8uQCop7YYdVgGGZd/t7yd/8ifl2LFj8v3f//3y9re/XX79139dHn/8cfnsZz8r165dSz4+9KEPyTd90zfJ137t18rly5dr8/35n/95+Zf/8l/Kd33Xdx0KCozpbz6hCOmUWn9NdNN6vd5r+XKtQ8T7HtEJKu7bPEAKU/ePtlAvMCb7an+PPfaYnD59Wr7t275NPvaxj/WW7qVLl+SlL32pPPDAA/KXf/mXR87/8R//sZw5c0a+4Ru+QR577LEj5xHv5VJi/TXVTV03yaoj1zpUu+yvAoh3gAzAP/xQLzAm+2p/p0+flje96U17SVtE5Cd+4iekqir58R//cRG5/qvdD/7gD8rzn/98ecc73hG8D38DAAXxDlAD/uGHeoEx2Uf7O3/+vNx///29p+vy8Y9/XB544AF56UtfKl/6pV8qZ8+elX/4h3+I3oO/AYCCeAeoAf/wQ73AmPTd/q5cuSJVVcmVK1d6TTfGb/zGb8gf/uEfJl2LvwGAkrV4j62zaf/Pzg7OGZ3d3PfkWXcWty9PFybwpkM9+WlTL2OMW+zS1vGtfOm7ji9cuCBnz57tNc0+oU0BgJK1eLf4Ftd3RcByucx6UmroYd8Fu/aoyI1JJvvMc26U4B9jUIp4bwu+lTd9++XZs2flwoULvabZJ/RDAKB0Eu+xdS91lrJdZzMUIXe3s9XF9WOR98VicWQHMrudbYp9do1mn71N04mVxa4tandic184dJ3TlPVUQzucab6+PN0y23LYNWCtKLFlSt2YYUrw0PTTRbyH2lofvhby77o8tEz4Vhn07ZdnzpxJWspxLOiHAEDpJN7tA9O3PbGKUn2oxRb9179Du2b5djiri965O2K59rkPe32pcHc/S00nVhatL3fYjG61bPNSceDL00Xr1n2RCeUZezEK5WcFh7sj2Rzgoemni3gPtbU+fC3W1mN5uOBbedO3X546dUqeeOKJXtPsE/ohAFBai3d3YXr7MG2ya5ZLaFezpuI9FG2L7cLoe6Fokk6sLLbcobLo51ieoXx8EUhfnr6dJ2M7y7llcP+eAzw0/XQR7ym7GLb1tZS27ssjlC6+lSd9+2VVVXLt2rVe03RZLBath1rRDwGAEhXv9qfeuqEuerQR7zafqqp6Ee9qn9JEvFsbm6QTK4tN07fV8mazkc1mI6vVKppnHZq21i8Cozs8NP206Tfq2lofvhZr6/jWdNiHeN8n2k4R7wDQld4i7+65VPHum4haJ95F4mPem0bM+4q8x8piy+1ep9vpqohPjQ66Q258dYPA6A4PTT9jRt5T+o0ukXd8K39KEu+LxeLQL6ttoB8CAKW1eNeb9UFmI1pNxLsbCbPjPmMP3dBqMzbS545trXu467V1Y95D6cTKYsvtlkX/du/15elivwNfvXQVGDYNreO5CQwemn76Fu8i/fhandDHt6ZBSeLd5oF4B4CudBLv7qoNoUh23bAZu3KDvTcm3kWODt1xf6KvW7nCtVcnqLn5pKYTK4vIjQlwFy9ePJLHYrE4ZH/Kihj2b9/34MuzjcCY+4oYPDT97EO89+FrdeId35oGiHcAmCudxPtUqJsMBzdYrVbZb4bVN3P3jxDUS7/M0be6gHgHgLmCeBfEeww7pKeqqllGBufuHyGol27gW91AvAPAXEG8A9SAf/ihXmBM5ijeOTg4OMzBQxggBP7hh3qBMem7/eXennO3DwCGA/EOUAP+4Yd6gTFBvAPAXEG8A9SAf/ihXmBMEO8AMFcQ7wA14B9+qBcYE8Q7AMwVxDtADfiHH+oFxgTxDgBzBfEOUAP+4Yd6gTFBvAPAXEG8A9SAf/ihXmBMEO8AMFcQ7wA14B9+qBcYE8Q7AMwVxDtADfiHH+oFxgTxDgBz5UC8c3BwhA84ytjfCQdH3+05Z3K3DwCG45/7QDoFgBD4hx/qBcYE8Q4AcwXxDlAD/uGHeoExQbwDwFxBvAPUgH/4oV5gTBDv7dhut4eGHq1Wq17SjVFVlWy322zTAygNxDtADfiHH+oFxgTx3pzNZiNVVclutzv4v+VyKev1unPaMRDvAP2CeAeoAf/wQ73AmCDe/ddYYe6yWCxks9kc+r/dbncobf3bnRSsEfvVanVwzgro5XLpjeYvFouD/7906ZJUVXVwraapNm82G1ksFkfKZG2x6e12u1p7NS+AKYF4B6gB//BDvcCYIN7914TEu4rcmLgXuS6ONRK/Wq1kuVyKyA0x7Du32WwOPmsaVtirUNc09AWiTrzbl431en3wUmBfHOrsdV9WAKYA4h2gBvzDD/UCY4J4v44OhXEPdyhMinhXweu7Jya0fRFz13Yr3jWNWJquLWqPL70UewGmBOIdoAb8ww/1AmOCePdf0yXyri8CvnvqouTr9To4CbYv8e6m18RegCmBeAeoAf/wQ73AmCDe/de0HfNuBW/sXGx8us0nNmxG03BfKJqKdyLvMFcQ7wA14B9+qBcYE8R7c0KrzbgTTO0YcldM+4S2HY+uaTQR73rtcrk89EJgz63X64NzsTHvIXsBpgTiHaAG/MMP9QJjgnhvhztG3h3iUrd6S2xyaWi8va74cvHixSOC2h1uY9MM2WJXq0m1F2BKIN4BasA//FAvMCaIdwCYK72Kd3ZugynCQ9MP/cb+0oN6EO8AMFd6E+/s3AZThYemH/qN/aUH9SDeAWCuJIv3LrPY3b9DY9PYuQ1yhO/YD/0G/caYIN4BYK70It7ZuQ2mDA9NP/Qb9BtjgngHgLkSFe/s3Mb6scBDMwT9Bv3GmCDe90dsjXX3nM/X3V/B9mlPU9brtazXa9lut4de8LvQpL4A+mCwyDs7t0Gp0PH6od84mh79xnAg3vdHU/G+7/kefQng3W53SLCrkN8niHfYB72IdxF2boPpQsfrh35Dguml2AvdQLz7r0l5GdbD+p5d9UnnkaSci4l3O7RN77dzQuxLdehae04JzSupO7darY70N25ZVOBbUa/D3+rms6TUF0AfJIv3Oti5DaYKHa8f+g36jTFBvPuvSf2ly7ZfvVeFrU+gx86liHeRG5PIfS+zdgK6a58v2m/tcee3+Oac6H0uy+XyiP2+OTPav8Tms6TUF0Af9CbeRdi5DaYJHa8f+g36jTFBvF8ndY6J776QOLZ/x86pXe4RemH1iWkVzrFfwVx7fD6Z8kud75c539AZe599oY+lnVpfAH3Qq3gHmCL4hx/qBcYE8e6/JvZyaJdVtSI7NFej7pzmWRd5byPe7XkrgEMvK7vdrnbOSUi8+zaGWy6XstlsZLPZHAy3SRHvdfUF0AeId4Aa8A8/1AuMCeLdf01IvLsR4D4j7/sQ76mR91gZ3Qh6auRd/1/t3Ww2RN4hKxDvADXgH36oFxgTxHsz3IjwYrE4MszMjud2rw2d61u8az6pY9595fLNOdH7XHxj3m2eKWm79sXqC6APEO8ANeAffqgXGBPEe3PsPJBQpLuqjq6QEjvnG8Ki4rqNeLc7Iyt1q83YXxv6WG3G1pcdUpO62kysvgD6APEOUAP+4Yd6gTFBvE+LISZ0j7HOO8A+GFW8x35uSyE2zqwroWhCn5tR9GlzTp3Qdhveuc5GVdyjaR5tOvo29039odmWfdZLXw/yJj7bpj229eHU+3zXdfX1PvudmK+3SStk13a7ld/7vd879H+I92kx1GpMdofV2C7MADkzGfHeN30LdR992e9GE3IgRWB0qeMhl92b+kOzLSXUS2obs2NURW4sF1nHviej+X6O7+rrfds81C6ViPe87QOA4UgW73VCKbZzm8nooAOyY+8uXbp0KH13veZQ2qHIu12nWY/1el2bjkaF1d4Sd42zG124vxbUrYsdW8O6yW5zSmwsoa+Ofd9D3Xdmy2nTr7vPN84z5Ac8NP3ss9+w31Ndu7W/5rgbM9m2Efqu3c1rrO3upLhYH5RynV0Pvsl9ijtmN1T2fdjs9gNWtPt8ve9dKhHvedsHAMPRi3iv27nNboFu11TVh1js4RxLO3XYjD6Q6tKxD8VU8S6S165xrm2+XeBCIiiUf9Pd5pTQLH6bn0+8qw0p372vXlPu03N1ZXDrF26wz34jVbz72qZtU7aNxfxLfdgNOqTamXqdT0Cnpq9lUGJl34fNvvz0viF2qUS8520fAAxHVLzvY+c2ETno8FPEe2raIfEeeiD70nE3h3CPEnaNi9Vl3blQ/k13m1Pqfk4PifeQ2IuVJRQ9jd0XKsNnP/vZI3bCUYboN1LEe2zcqu1jQu07ZLev3wjZ2aQ8be7Tz+4vkqljdvuwWUQO9QN1E/+a9hux/pxhM/nb58MGWJT1et37MM99zB8bcj5Hid8tjEtUvLsXxn7+Du3cVhcRTxHvKWn78rER/5R03IdUXeS9jXj3lVvL7BM8u12zXePaivdY/iLNdptTQjvXherYV1dtv7OU+0JlQLynMUS/kTJsxg5Jc9ubtom69u2ieaa2pyblaXOffnbFeqzsfdus1242myNifYhdKhHvedvnw+dn+xDvfTHGajSId2hDL+LdN4ayiXh3hV/s/tTIuzveMiWdIcR7auTdl5/iRrRSxXtdPceieE12m7P3dIm8N/nOYr9UEHnfD0P0G+4vP+51LqFhM7H27Q7vsGn52npqH5QaeU9N3/X1WNn3YbP+nx27rgyxSyXivR9/882/Eon/8mTPuRsf6Xeoh2vPcrk8Iojt36H7Q/b6XlZ9bcc376uunKnrwPc9nyP3tgf5kSzeY9Tt3Gadx46pdMW7XrNcLo9EhH1phzr71WoVnLwVS2ef4n2oXeNSxLuvnuvyt51Nik2afqgO3XoK2V73nfnqtcl3XVcGtROOss9+wyfefe3Wjfimjnl38w6JFd8vNKE+qEm7a3qftVOJlX0fNlsbUn29Sb/hltW9DvHej3hvOk9I09Xv132+x+YN6X32V3BXvNfNgWgyt8x3n00zpZwuQ8znyL3tQX70It5F0ndus3nZlRfcn3/dzsGXdki82+vdNGPp1I15V4drI96H2jWu6RCDWATFfQgsFmm7zdl6sJ99IqBu2Ezdd2ajK66tKd91w0K2kQAADD1JREFUXRnccsAN9tlvuN9Tav/gRn9tH1PXvl1fb9IOm5an6X2KGxmMlb1vm60Nbl4xX0/tN2L9IeK9+xyT2K+VvjRD80piv4y7aWqbstdZ8Z7yS0yT56yvnE3m0PmuG2I+R+5tD/Ljn/2chrMvQg/APnHH6eVATptGWdp0lPiHn33WCw80Pzn4utufjeXriHf/NXWR96bzhGLivW7uk5u+thW7ktnQ4j02xyMk3vc9nyP3tgf5gXjfM0OId5G8xHJsfPGYaPS2aT3hH372VS9tv6e5MJavu8Mu9P/G8nXEu/+aVPGeOk+oj8i7PRcbTrJv8R4rZ2hOyRDzOXJve5AfiHeAGvAPP9QLjAnivRlt5wlp3qG5UnXzsezQSH0xj415D8256EO8p5TTZYj5HLm3PcgPxDtADfiHH+oFxgTx3ozY/CuR+Ly1LqvNuMJ3sVg0Wm2m72EzsXKmrjZj0+pjPkfubQ/yA/EOUAP+4Yd6gTFBvDejryGcvtXcpoI7pySn4agAFsQ7QA34hx/qBcYE8d6MtuLdDg+xEzynit1hdeplhXJBvAPUgH/4oV5gTBDvADBXEO8ANeAffqgXGBPEOwDMFcQ7QA34hx/qBcYE8Q4AcwXxnjF///d/P7YJIDw0Q1AvMCaI93HxrSQTus49+pjw2sf66KllEDk8ebX0Mm232942e4vZzBr2++NAvHNwcIQPOMrY3wkHR9/tOWdys6+q0sW7b7lId0nGpgwp3t1VaKZQpiFW0kG874+qqoSazZCrV6/KsWPH5OrVq2ObAgAweXIXGSn2VVV4NZnlcnlIrOm66XVrk+s65FYU2rXSd7vdoXXgq6o6JGJ9YnK1Wh2s6FJVN9aeF4mvG29XvrHro9et7a522DTdMsTyddd/n0KZ9H77WV9S2rSTuvJAvyDeM+XcuXNyzz33yLlz58Y2BQBg8uQuMrqK981mc2RnUxXdKkztRkjujqyhTZJU1Gm+dudWe51r53a7PcjDCmN391E3T2trqtC1UfH1en0wxMV9IYnl67O/5DKJ+HePDbWTUFqueA+VB/oF8Z4hV69elRMnTkhVVXLixAmi7wAAeyZ3kRGyz4166+EOibAiWz+7a5nba+rEY2h4hu+6kG1uHqGdSK09Suquqr6hG3qdK7hD+cYi3iWWSfENnQm1k5QyxMoD/YJ4z5Bz587Jgw8+KFVVyYMPPkj0HQBgz+QuMrpG3kWuR1o3m41sNpuDoSA+IdpUvNuhNVVVJYl8Eb9ADYnE0LlQOjGh69oWyzck3ksuk2Ij9pZQOwnVj56LlQf6BfGeGRp1/+hHPypVVcnjjz9O9B0AYM/kLjL6EO8q1lScxXYRTRXvrpBMjdD78mgbpXaFaVOhG8t3t9t1Eu85lkkJTVr1tRMi73mBeM8MjbqL3Oisib4DAOyX3EVGH/bZyYQ2XR2jbCOnqeLdjbYuFovW4l3vt2Or3XHe9pwrdDWf5XIZtMGOybf/H8vXrfsplEnT9JXD105CabkCPVQe6BfEe0bYqLvIjQ6D6DsAwH7JXWT0Zd9isTg0VMJdRcSNGIfEu66motF7vd+9r6nQja2QYs+5wnC9Xh865xvL76Zpy9B1tZnSyqTlsJ/dF45YO3HLm1Ie6A/Ee0acO3dOHnrooYO/baMn+g4AsD9yFxm52zdl3HXep8AQ67zD/kC8Z4IbdRc53FkTfQcA2B+5i+Pc7Zs6UxK7sbkOUAaI90xwo+4iRztrou8AAPshd3Gcu30AMByI9wy4evWqHD9+/FDUXeRoZ/3444/L8ePHib4DAPRM7uI4d/sAYDgQ7xngi7qL+Dvrhx56iOg7AEDP5C6Oc7cPAIYD8T4yoai7iL+zJvoOANA/uYvjnOzrsn63XbHEHqHVW7ra59o6dP45ppnL+P2pfxfb7ba3ic7PPPPMob8R7xlw4cIF7/+HGkfoegAAaEdO4thH7val0rc4q8Mn3vedf87iPaeVc+bwXezrRQnxnjFT6awBAHIn9/42xT53rW6XurW6dV1uK6qWy+Uh8WGvUXSjJj10TXRNV9ceVxvr1ki365u764Zb6vJ1P/eZf+haX56x9dZDZbDp+NZMj6Wp9e2rM7tmfdP6jqVry+HbEMrHHL4LLaf9rC9Qrl8tl8tgWkTeCyL3hwkAwFTIvb/tQ7xbQaJiQeSw+HHPbTabQ5Fa3bxH7VGxofna3T413aYbHKkdKhZ3u92RzY9S8nU/95m/feGJ5R+r91gZfPfZNN2dTGPfly2v76Utpbwp6dqdXfsU73W25fpdKL6dbH31qRuh+dJCvBdE7g8TAICpkHt/G7LPjRbq4f5U767tbcVKbDdVe51+jg0bsPf6dhv12Rq63hVCMbHny9f93Gf+sZcJN/9QvaeWoa5MNk13J1yLa0uT8sbSdc/FrrVM/btQfENnUvzKXoN4L4jcHyYAAFMh9/62a+Q9JPLrxLvI9cjhZrORzWYjq9XqiMiwQ2liAkxtrIu2por3unzdz33mHyubzTNW77Ey6H2KCrnYORs11vTc4SVtxXss3S7iferfhdabO8xI5KhfxdJCvBdE7g8TAICpkHt/21W8x3bVrBPvKj5UbMTEcUrkvU/BFsvXd92+BGNqtNeXX10Z6soUiyDb4S273a6TeA+lm5t4z+27CE1ajfmVmxbivSByf5gAAEyF3PvbPuyrqhvjdm2Ur06828l69m83HZHD4533Kd5T8nU/95m/OyY7dZy1tTtWBv3bjn92r7Xn9D43ypsy5j2lvCnp6t/L5XIU8Z7bd6H4xrxb21LSQrwXRO4PEwCAqZB7f9uHfe5KFu6Ev5B4F7kxUdVeb89pmjat1DHvKqiaRoLr8vXZ2lf+eq1dgUUJRVDdeo+Vwb2vyQonNk034utbbaZNfbvpxlabCf0qMIfvQm0N1YX1q1haiPeCyP1hAgAwFXLvb3O3b274XkxKQJcp3Cer1eqQIF2v13utp5y/C9Z5nyF01gAAw5B7f5u7fXMjZ8FYR9+C0g7/qKrKO2xkn+T6XcTG2HcF8Z4xdNYAAMOQe3+bu30AMByI94yhswYAGIbc+9vc7QOA4UC8ZwydNQDAMOTe3+ZuHwAMB+I9Y+isAQCGIff+Nnf7AGA4EO8ZQ2cNADAMufe3udsHAMOBeM8YOmsAgGHIvb/N3T4AGA7Ee8bQWQMADEPu/W3u9gHAcCDeM4bOGgBgGHLvb3O3DwCGA/GeMXTWAADDkHt/m7t9ADAciPeMobMGABiG3Pvb3O0DgOFAvGcMnTUAwDDk3t/mbh8ADAfiPWPorAEAhiH3/jZ3+wBgOBDvGTN2Z71er2W5XCZf3+RaAICcGLu/rSN3+wBgOBDvGTNmZ71er6WqqiRBriJ/uVxKVVWy3W4HsBAAoD9yF8e52wcAw4F4z5ixOmsV4qvVKjmavt1uebgAQLHk3n/lbh8ADAfiPWPG7qxTxftut5PFYnHwLwBAaYzd39aRu30AMByI94ypqkquXbs2Wv5NIu8AAKVy7dq17MVxVVUcHBwcN46xOyXwc+rUKXniiSdGyx/xDgBz4IknnpBTp06NbQYAQDKI90w5c+aMXL58ebT8Ee8AMAcuX74sZ86cGdsMAIBkEO+ZcvbsWblw4cJo+SPeAWAOXLhwQc6ePTu2GQAAySDeM2XsB4pPvFdVJbvd7shnAIBSGTtQAgDQFMR7ply5ckWqqpIrV66MbQoAwCShnwWAEkG8Z8z58+fl/vvvH9sMAIBJcv/998v58+fHNgMAoBGI98w5ffq0PPzww2ObAQAwKR5++GE5ffr02GYAADQG8Z4573vf++T06dNy//3389MuAEBHrly5Ivfff7+cPn1a3ve+941tDgBAYxDvhXD+/HmpqupgctXly5fliSeeGHUjJwCAnLl27Zo88cQTcvny5YNFAKqqYqgMABQN4r0grly5cvAAOnPmjJw6dWr8Xb44ODg4Mj5OnTolZ86cOQh88AsmAJQO4h0AAAAAoBAQ7wAAAAAAhYB4BwAAAAAoBMQ7AAAAAEAhIN4BAAAAAAoB8Q4AAAAAUAiIdwAAAACAQkC8AwAAAAAUAuIdAAAAAKAQEO8AAAAAAIWAeAcAAAAAKATEOwAAAABAISDeAQAAAAAKAfEOAAAAAFAIiHcAAAAAgEJAvAMAAAAAFALiHQAAAACgEP4/8tkJqlOyQtcAAAAASUVORK5CYII=">
</div>
```
```
|
github_jupyter
|
# 0.0 Notebook Template
--*Set the notebook number, describe the background of the project, the nature of the data, and what analyses will be performed.*--
## Jupyter Extensions
Load [watermark](https://github.com/rasbt/watermark) to see the state of the machine and environment that's running the notebook. To make sense of the options, take a look at the [usage](https://github.com/rasbt/watermark#usage) section of the readme.
```
# Load `watermark` extension
%load_ext watermark
# Display the status of the machine and packages. Add more as necessary.
%watermark -v -n -m -g -b -t -p numpy,pandas,matplotlib,seaborn
```
Load [autoreload](https://ipython.org/ipython-doc/3/config/extensions/autoreload.html) which will always reload modules marked with `%aimport`.
This behavior can be inverted by running `autoreload 2` which will set everything to be auto-reloaded *except* for modules marked with `%aimport`.
```
# Load `autoreload` extension
%load_ext autoreload
# Set autoreload behavior
%autoreload 1
```
Load `matplotlib` in one of the more `jupyter`-friendly [rich-output modes](https://ipython.readthedocs.io/en/stable/interactive/plotting.html). Some options (that may or may not have worked) are `inline`, `notebook`, and `gtk`.
```
# Set the matplotlib mode
%matplotlib inline
```
## Imports
Static imports that shouldn't necessarily change throughout the notebook.
```
# Standard library imports
import logging
# Third party
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
# tqdm.tqdm wraps generators and displays a progress bar:
# `for i in tqdm(range(10)): ...`
from tqdm import tqdm
```
Local imports that may or may not be autoreloaded. This section contains things that will likely have to be re-imported multiple times, and have additions or subtractions made throughout the project.
```
# Constants to be used throughout the package
%aimport {{ cookiecutter.import_name }}.constants
# Import the data subdirectories
from {{ cookiecutter.import_name }}.constants import DIR_DATA_EXT, DIR_DATA_INT, DIR_DATA_PROC, DIR_DATA_RAW
# Utility functions
%aimport {{ cookiecutter.import_name }}.utils
from {{ cookiecutter.import_name }}.utils import setup_logging
```
## Initial Setup
Set [seaborn defaults](https://seaborn.pydata.org/generated/seaborn.set.html) for matplotlib.
```
sns.set()
```
Set up the logger configuration to something more useful than baseline. Creates log files for the different log levels in the `logs` directory.
See `logging.yml` for the exact logging configuration.
```
# Run base logger setup
setup_logging()
# Define a logger object
logger = logging.getLogger("{{ cookiecutter.import_name }}")
```
## Global Definitions
```
# data_str = "" # Data filename
# data_path = DIR_DATA_RAW / data_str # Full path to the data
```
## Get the Data
```
# data = pd.read_csv(str(data_path), delim_whitespace=False, index_col=0)
# logger.info("Loaded dataset '{0}' from '{1}'".format(data_path.name, data_path.parent.name))
```
## Preprocessing
```
# data_norm = (data - data.mean()) / data.std()
# logger.info("Processed data '{0}'".format(data_path.stem))
```
## Plotting
```
# [plt.plot(data_norm[i,:]) for i in range(len(data_norm))]
# plt.show()
# ...
# ...
# ...
```
## Hints
Various hints for working on `jupyter notebooks`. Should probably be removed when a notebook is completed.
General stuff:
- To make logging even lazier, set `print = logger.info`, and then `print` away!
- The `!` can be used to run shell commands from within the notebook (ex. `!which conda`)
- Use `assert` liberally - this isn't a script and it's very readable.
Cheatsheets:
- [Markdown](https://github.com/adam-p/markdown-here/wiki/Markdown-Cheatsheet)
|
github_jupyter
|
```
# import lib
# ===========================================================
import csv
import pandas as pd
from datascience import *
import numpy as np
import random
import time
import matplotlib.pyplot as plt
%matplotlib inline
plt.style.use('fivethirtyeight')
import collections
import math
import sys
from tqdm import tqdm
from time import sleep
# Initialize useful data
# with open('clinvar_conflicting_clean.csv', 'r') as f:
# reader = csv.reader(f)
# temp_rows = list(reader)
df = pd.read_csv('clinvar_conflicting_clean.csv', low_memory=False)
# columns_to_change = ['ORIGIN', 'EXON', 'INTRON', 'STRAND', 'LoFtool', 'CADD_PHRED', 'CADD_RAW', 'BLOSUM62']
# df[['CLNVI', 'MC', 'SYMBOL', 'Feature_type', 'Feature', 'BIOTYPE',
# 'cDNA_position', 'CDS_position', 'Protein_position', 'Amino_acids', 'Codons',
# 'BAM_EDIT', 'SIFT', 'PolyPhen']] = df[['CLNVI', 'MC', 'SYMBOL', 'Feature_type', 'Feature', 'BIOTYPE',
# 'cDNA_position', 'CDS_position', 'Protein_position', 'Amino_acids', 'Codons',
# 'BAM_EDIT', 'SIFT', 'PolyPhen']].fillna(value="null")
df = df.fillna(value=0)
df_zero = df.loc[df['CLASS'] == 0]
df_zero = df_zero.sample(n=10000)
df_one = df.loc[df['CLASS'] == 1]
df_one = df_one.sample(n=10000)
df = pd.concat([df_zero, df_one])
df = df.sample(n = df.shape[0])
all_rows = df.values.tolist()
row_num = len(all_rows)
df.head()
# Decision stump part for Adaboost
# ===========================================================
def is_numeric(value):
return isinstance(value, int) or isinstance(value, float)
# === LeafNode is the prediction result of this branch ===
class LeafNode:
def __init__(self, rows):
labels = [row[-1] for row in rows]
# labels = []
# self.one_idx = []
# self.zero_idx = []
# for i in range(len(rows)):
# row = rows[i]
# labels.append(row[-1])
# if row[-1] == 1:
# self.one_idx.append(i)
# else:
# self.zero_idx.append(i)
self.prediction = collections.Counter(labels)
# === DecisionNode is an attribute / question used to partition the data ===
class DecisionNode:
def __init__(self, question = None, left_branch = None, right_branch = None):
self.question = question
self.left_branch = left_branch
self.right_branch = right_branch
class DecisionStump:
def __init__(self, training_attribute, training_data, height, method = "CART"):
self.attribute = training_attribute # takein attribute and data separately
self.train = training_data
self.height = height
self.row_num = len(self.train)
self.column_num = len(self.attribute)
self.method = method.upper() # convert to upper case for general use
self.significance = 0
if self.method not in ["C4.5", "CART", "HYBRID"]:
print("Error: Please choose a valid method! from: [C4.5, CART, HYBRID]")
return None
# train decision stump
self.root = self.build_stump(self.train, 1)
# count ACC classifications and mis classifications to update weights
self.accclassify_idx = []
self.misclassify_idx = []
# Only after DecisionStump trained, can we know which rows are misclassified
# Walk down the decision stump to collect all misclassification indices
# if self.root.left_branch.prediction.get(1, 0) > self.root.left_branch.prediction.get(0, 0):
# # then consider the prediction of this leaf node as 1: 1 -> correct, 0 -> misclassify
# self.accclassify_idx += self.root.left_branch.one_idx
# self.misclassify_idx += self.root.left_branch.zero_idx
# else:
# # then consider the prediction of this leaf node as 0: 0 -> correct, 1 -> misclassify
# self.accclassify_idx += self.root.left_branch.zero_idx
# self.misclassify_idx += self.root.left_branch.one_idx
# if self.root.right_branch.prediction.get(1, 0) > self.root.right_branch.prediction.get(0, 0):
# # then consider the prediction of this leaf node as 1: 1 -> correct, 0 -> misclassify
# self.accclassify_idx += self.root.right_branch.one_idx
# self.misclassify_idx += self.root.right_branch.zero_idx
# else:
# # then consider the prediction of this leaf node as 0: 0 -> correct, 1 -> misclassify
# self.accclassify_idx += self.root.right_branch.zero_idx
# self.misclassify_idx += self.root.right_branch.one_idx
def uniq_val(self, column):
return set([self.train[i][column] for i in range(len(self.train))])
# when raising a question.
# if it's a categorical attribute, we simply iterate all categories
# if it's a numeric attribute, we iterate the set of possible numeric values
class Question:
def __init__(self, column, ref_value, attribute):
self.column = column
self.ref_value = ref_value if ref_value else "None"
self.attri = attribute
def match(self, row):
if is_numeric(self.ref_value):
try:
return row[self.column] >= self.ref_value
except:
print("Error occured in ", row)
return True
else:
return row[self.column] == self.ref_value
def __repr__(self):
operand = ">=" if is_numeric(self.ref_value) else "=="
return "Is %s %s %s?" % (self.attri[self.column], operand, str(self.ref_value))
# === Method 1 - C4.5 ===
def entropy(self, rows):
# === Bits used to store the information ===
labels = [row[-1] for row in rows]
frequency = collections.Counter(labels).values()
pop = sum(frequency)
H = 0
for f in frequency:
p = f / pop
H -= p * math.log(p, 2)
return H
# === Method 2 - CART ===
def gini(self, rows):
# === Probability of misclassifying any of your label, which is impurity ===
labels = [row[-1] for row in rows]
frequency = collections.Counter(labels).values()
pop = sum(frequency)
gini = 1
for f in frequency:
p = f / pop
gini -= p ** 2
return gini
# === Calculate Gain Info ===
def info(self, branches, root):
# === Objective: to find the best question which can maximize info ===
root_size = float(len(root))
if self.method == "C4.5": # Here I pick the GainRatio Approach
root_uncertainty = self.entropy(root)
gain_info = root_uncertainty
split_info = 0
for branch in branches:
if not branch: continue
gain_info -= len(branch) / root_size * self.entropy(branch)
split_info -= float(len(branch)) / root_size * math.log(float(len(branch)) / root_size)
# print(gain_info, split_info)
return gain_info / split_info
elif self.method == "CART":
root_uncertainty = self.gini(root)
gain_info = root_uncertainty
for branch in branches:
if not branch: continue
gain_info -= len(branch) / root_size * self.gini(branch)
return gain_info
elif self.method == "HYBRID":
pass
pass
# === Here I only do Binary Partitions ===
def partition(self, rows, question):
true_rows = []
false_rows = []
for row in rows:
if question.match(row):
true_rows.append(row)
else:
false_rows.append(row)
return true_rows, false_rows
# the question that achieves the max infomation attenuation is the best question
def find_best_question(self, rows):
max_info_attenuation = 0
best_question = self.Question(0, self.train[0][0], self.attribute)
# === Iterate through all question candidates ===
# === TODO: Maybe Iteration here can be optimized ===
for col in range(self.column_num - 1): # minus 1 to avoid using the label as attribute
ref_candidates = self.uniq_val(col)
for ref_value in ref_candidates:
if ref_value == "null": continue # avoid using null values to generate a question
q = self.Question(col, ref_value, self.attribute)
temp_true_rows, temp_false_rows = self.partition(rows, q)
temp_info_attenuation = self.info([temp_true_rows, temp_false_rows], rows)
if temp_info_attenuation >= max_info_attenuation:
max_info_attenuation = temp_info_attenuation
best_question = q
return max_info_attenuation, best_question
# === Input rows of data with attributes and labels ===
def build_stump(self, rows, height):
# === Assign all rows as root of the whole decision tree ===
# === We have met the leaf node if gini(rows) is 0 or no question candidates left ===
gain_reduction, q = self.find_best_question(rows)
true_rows, false_rows = self.partition(rows, q)
if height + 1 >= self.height:
return DecisionNode(q, LeafNode(true_rows), LeafNode(false_rows))
else:
return DecisionNode(q, self.build_stump(true_rows, height + 1), self.build_stump(false_rows, height + 1))
# === Input a row of data with attributes (and no label), predict its label with our decision tree ===
# === Actually it can contain a label, we just don't use it ===
# === walk down the decision tree until we reach the leaf node ===
def classify(self, row, node):
if isinstance(node, LeafNode):
# do a mapping from label[1, 0] to label[1, -1]
return node.prediction
# return 1 if node.prediction.get(1, 0) / (node.prediction.get(1, 0) + node.prediction.get(0, 0)) > cutoff else -1
if node.question.match(row):
return self.classify(row, node.left_branch)
else:
return self.classify(row, node.right_branch)
# function to print the tree out
def print_tree(self, node, spacing=""):
# Base case: we've reached a leaf
if isinstance(node, LeafNode):
print (spacing + "Predict", node.prediction)
return
# Print the question at this node
print (spacing + str(node.question))
# Call this function recursively on the true branch
print (spacing + '--> True:')
self.print_tree(node.left_branch, spacing + " ")
# Call this function recursively on the false branch
print (spacing + '--> False:')
self.print_tree(node.right_branch, spacing + " ")
def test(self):
for i in range(self.column_num):
q = self.Question(i, self.train[1][i], self.attribute)
print(q)
print(q.match(1))
def normalized_weight(weight):
return np.divide(weight, sum(weight))
def rev_logit(val):
return 1 / (1 + np.exp(val))
# Divide whole dataset into training set and testing set
# ===========================================================
training_percentage = 0.2 # percent of partition of training dataset
training_size = int(row_num * training_percentage)
testing_size = row_num - training_size
training_attribute = list(df.columns)
training_data = all_rows[: training_size] # training data should include header row
testing_data = all_rows[training_size: ] # testing data don't need to include header row
# Recursively Training base learners
# ===========================================================
# let's train T base learners
T = 20
weakleaner_height = 3
stump_forest = []
weight = [1 / training_size for _ in range(training_size)]
start = time.time()
for i in range(T):
# train a decision stump
stump = DecisionStump(training_attribute, training_data, weakleaner_height, "CART")
# calculate the total error of the stump after it's trained
for j in range(training_size):
row = training_data[j]
pred_counter = stump.classify(row, stump.root)
pred_label = 1 if pred_counter.get(1, 0) / (pred_counter.get(1, 0) + pred_counter.get(0, 0) + 0.00000001) > 0.5 else 0
if pred_label == row[-1]:
stump.accclassify_idx.append(j)
else:
stump.misclassify_idx.append(j)
accuracy = len(stump.accclassify_idx) / training_size
total_err_rate = 1 - accuracy
# update the significance level of this stump, remember not to divide by zero
stump.significance = 0.5 * math.log((1 - total_err_rate + 0.0001) / (total_err_rate + 0.0001))
# append stump into the forest
stump_forest.append(stump)
# if len(stump_forest) == T: break # early break
# update training_data weight, resample the training data with the updated weight distribution
true_scale = np.e ** stump.significance
for idx in stump.misclassify_idx:
weight[idx] = weight[idx] * true_scale
for idx in stump.accclassify_idx:
weight[idx] = weight[idx] * (1 / true_scale)
distrib = normalized_weight(weight)
# interactive printing
# sys.stdout.write('\r')
# # the exact output you're looking for:
# sys.stdout.write("Training Random Forest: [%-10s] %d%% alpha = %.02f" % ('='*int((i + 1) / T * 10), int((i + 1) / T * 100), stump.significance))
# sys.stdout.flush()
# stump.print_tree(stump.root)
# print(i, stump.significance)
resampled_idx = np.random.choice(training_size, training_size, p = distrib)
training_data = [training_data[idx] for idx in resampled_idx]
if len(set([row[1] for row in training_data])) < 0.04 * training_size: break
print(i, len(set([row[1] for row in training_data])), stump.significance, end='\n')
weight = [1 / training_size for _ in range(training_size)]
end = time.time()
print("\nTime: %.02fs" % (end - start))
# New Testing Adaboost
# ===========================================================
# Compute TN, TP, FN, FP, etc. together with testing
# ===========================================================
ROC = Table(make_array('CUTOFF', 'TN', 'FN', 'FP', 'TP', 'ACC'))
step_size = 0.05
forest_size = len(stump_forest)
CMap = {0: 'TN', 1: 'FN', 2: 'FP', 3: 'TP'}
for cutoff in np.arange(0, 1 + step_size, step_size):
sys.stdout.write('\r')
# the exact output you're looking for:
sys.stdout.write("Testing: [%-20s] %d%%" % ('='*int(cutoff * 100 / 5), int(cutoff * 100)))
sys.stdout.flush()
'''
# calculate the total error of each stump
for stump in stump_forest:
stump.accclassify_idx = []
stump.misclassify_idx = []
# walk down the stump for each training data, see if its prediction makes sense
for j in range(training_size):
row = training_data[j]
pred_counter = stump.classify(row, stump.root)
pred_label = 1 if pred_counter.get(1, 0) / (pred_counter.get(1, 0) + pred_counter.get(0, 0) + 0.00000001) > cutoff else 0
if pred_label == row[-1]:
stump.accclassify_idx.append(j)
else:
stump.misclassify_idx.append(j)
accuracy = len(stump.accclassify_idx) / training_size
total_err_rate = 1 - accuracy
# update the significance level of this stump, remember not to divide by zero
stump.significance = 0.5 * math.log((1 - total_err_rate + 0.0001) / (total_err_rate + 0.0001))
'''
Confusion = {'TN': 0, 'FN': 0, 'FP': 0, 'TP': 0}
for row in testing_data:
true_rate_forest = 0
for tree_i in stump_forest:
# prediction is a counter of label 1 and 0
pred_counter = tree_i.classify(row, tree_i.root)
# do a mapping from label[1, 0] to label[1, -1]
true_rate_tree = 1 if pred_counter.get(1, 0) / (pred_counter.get(1, 0) + pred_counter.get(0, 0) + 0.00000001) > cutoff else -1
true_rate_forest += true_rate_tree * tree_i.significance
# true_rate_forest = rev_logit(true_rate_forest)
# true_pred = 1 if true_rate_forest >= cutoff else 0
true_pred = 0 if np.sign(true_rate_forest) <= 0 else 1
indicator = (true_pred << 1) + row[-1]
# accordingly update confusion matrix
Confusion[CMap[indicator]] += 1
# concatenate the confusion matrix values into the overall ROC Table
thisline = [cutoff] + list(Confusion.values()) + [(Confusion['TP'] + Confusion['TN']) / sum(Confusion.values())]
ROC = ROC.with_row(thisline)
ROC = ROC.with_columns('SENSITIVITY', ROC.apply(lambda TP, FN: TP / (TP + FN + 0.00000001), 'TP', 'FN'))
ROC = ROC.with_columns('FPR', ROC.apply(lambda TN, FP: FP / (TN + FP + 0.00000001), 'TN', 'FP'))
ROC = ROC.with_column('FMEAS', ROC.apply(lambda TP, FP, FN: 2 * (TP / (TP + FN)) * (TP / (TP + FP)) / (TP / (TP + FN) + TP / (TP + FP)), 'TP', 'FP', 'FN'))
ROC.show()
# Acc Curve by cutoff
# ===========================================================
# matplotlib.use('TkAgg')
fig = plt.figure()
plt.xlabel('Cutoff')
plt.ylabel('Accuracy')
plt.title('Accuracy - Cutoff of Adaboost')
plt.plot(np.arange(0, 1.1, 0.1), [0.5 for i in np.arange(0, 1.1, 0.1)], color='black')
plt.plot(ROC.column('CUTOFF'), ROC.column('ACC'), color='orange')
plt.legend(['Adaboost', 'Null'])
plt.axis([0, 1, 0, 1.1])
plt.show()
fig.savefig('Adaboost ACC.png', bbox_inches='tight')
# ROC_CURVE
# ===========================================================
fig = plt.figure()
plt.xlabel('False Positive Rate')
plt.ylabel('Sensitivity')
plt.title('ROC - Curve of Adaboost')
plt.plot(np.arange(0, 1.1, 0.1), np.arange(0, 1.1, 0.1), color='black')
plt.plot(ROC.column('FPR'), ROC.column('SENSITIVITY'), color='orange')
plt.legend(['Adaboost', 'Null'])
plt.axis([0, 1, 0, 1.1])
plt.show()
fig.savefig('Adaboost ROC.png', bbox_inches='tight')
# Compute AUC
# ===========================================================
length = len(ROC.column('FPR'))
auc = 0
for i in range(length - 1):
auc += 0.5 * abs(ROC.column('FPR')[i + 1] - ROC.column('FPR')[i]) * (ROC.column('SENSITIVITY')[i] + ROC.column('SENSITIVITY')[i + 1])
print("auc = %.03f" %auc)
# Original Testing
# ===========================================================
accuracy = 0
for row in testing_data:
overall_classification = 0
for stump in stump_forest:
classification = stump.classify(row, stump.root)
vote = stump.significance
overall_classification += classification * vote
# reverse mapping from label[1, -1] to label[1, 0]
predicted_label = 0 if np.sign(overall_classification) <= 0 else 1
if predicted_label == row[-1]: accuracy += 1
# print(classification, predicted_label, row[-1])
accuracy = accuracy / testing_size
print("%.03f%%" % (accuracy * 100))
# Testing with a toy dataset
# ===========================================================
training_data = [
['Green', 3, 1],
['Yellow', 3, 1],
['Red', 1, 0],
['Red', 1, 0],
['Yellow', 3, 1],
['Red', 3, 1]
]
testing_data = [
['Red', 2, 0],
['Yellow', 3.5, 1],
['Green', 3, 1]
]
training_attribute = ['Color', 'Diameter', 'Label']
training_size = len(training_data)
testing_size = len(testing_data)
# pf = df[]
len(set(pd.Index(df)))
np.bincount([row[-1] for row in all_rows])
fpr, sen, acc = ROC.column('FPR'), ROC.column('SENSITIVITY'), ROC.column('ACC')
fpr
sen
acc
```
|
github_jupyter
|
<center><h1>Improved Graph Laplacian via Geometric Self-Consistency</h1></center>
<center>Yu-Chia Chen, Dominique Perrault-Joncas, Marina Meilă, James McQueen. University of Washington</center> <br>
<center>Original paper: <a href=https://nips.cc/Conferences/2017/Schedule?showEvent=9223>Improved Graph Laplacian via Geometric Self-Consistency] on NIPS 2017 </a></center>
## The Task
1. Problem: Estimate the ``radius`` of heat kernel in manifold embedding
1. Formally: Optimize Laplacian w.r.t. parameters (e.g. ``radius``)
1. Previous work:
1. asymptotic rates depending on the (unknown) manifold [4]
1. Embedding dependent neighborhood reconstruction [6]
1. Challenge: it’s an unsupervised problem! What “target” to choose?
## The ``radius`` affects…
1. Quality of manifold embedding via neighborhood selection
1. Laplacian-based embedding and clustering via the kernel for computing similarities
1. Estimation of other geometric quantities that depend on the Laplacian (e.g Riemannian metric) or not (e.g intrinsic dimension).
1. Regression on manifolds via Gaussian Processes or Laplacian regularization.
All the reference is the same as the poster.
## Radius Estimation on hourglass dataset
In this tutorial, we are going to estimate the radius of a noisy hourglass data. The method we used is based on our NIPS 2017 paper "[Improved Graph Laplacian via Geometric Self-Consistency](https://nips.cc/Conferences/2017/Schedule?showEvent=9223)" (Perrault-Joncas et. al). Main idea is to find an estimated radius $\hat{r}_d$ given dimension $d$ that minimize the distorsion. The distorsion is evaluated by the riemannian metrics of local tangent space.
Below are some configurations that enables plotly to render Latex properly.
```
!yes | conda install --channel=conda-forge pip nose coverage gcc cython numpy scipy scikit-learn pyflann pyamg h5py plotly
!rm -rf megaman
!git clone https://github.com/mmp2/megaman.git
!cd megaman
import plotly
plotly.offline.init_notebook_mode(connected=True)
from IPython.core.display import display, HTML
display(HTML(
'<script>'
'var waitForPlotly = setInterval( function() {'
'if( typeof(window.Plotly) !== "undefined" ){'
'MathJax.Hub.Config({ SVG: { font: "STIX-Web" }, displayAlign: "center" });'
'MathJax.Hub.Queue(["setRenderer", MathJax.Hub, "SVG"]);'
'clearInterval(waitForPlotly);'
'}}, 250 );'
'</script>'
))
```
## Generate data
This dataset used in this tutorial has a shape of hourglass, with ``size = 10000`` and dimension be 13. The first three dimensions of the data is generated by adding gaussian noises onto the noise-free hourglass data, with ``sigma_primary = 0.1``, the variance of the noises added on hourglass data. We made ``addition_dims = 10``, which is the addition noises dimension to make the whole dataset has dimension 13, with ``sigmal_additional = 0.1``, which is the variance of additional dimension.
```
from plotly.offline import iplot
#import megaman
from megaman.datasets import *
data = generate_noisy_hourglass(size=10000, sigma_primary=0.1,
addition_dims=10, sigma_additional=0.1)
```
We can visualize dataset with the following plots:
```
from megaman.plotter.scatter_3d import scatter_plot3d_plotly
import plotly.graph_objs as go
t_data = scatter_plot3d_plotly(data,marker=dict(color='rgb(0, 102, 0)',opacity=0.5))
l_data = go.Layout(title='Noisy hourglass scatter plot for first 3 axis.')
f_data = go.Figure(data=t_data,layout=l_data)
iplot(f_data)
```
## Radius estimation
To estimate the ``radius``, we need to find the pairwise distance first.
To do so, we compute the adjacency matrix using the Geometry modules in megaman.
```
rmax=5
rmin=0.1
from megaman.geometry import Geometry
geom = Geometry(adjacency_method='brute',adjacency_kwds=dict(radius=rmax))
geom.set_data_matrix(data)
dist = geom.compute_adjacency_matrix()
```
For each data points, the distortion will be estimated. If the size $N$ used in estimating the distortion is large, it will be computationally expensive. We want to choose a sample with size $N'$ such that the average distion is well estimated. In our cases, we choose $N'=1000$. The error will be around $\frac{1}{\sqrt{1000}} \approx 0.03$.
In this example, we searched radius from the minimum pairwise distance ``rmin`` to the maximum distance between points ``rmax``. By doing so, the distance matrix will be dense. If the matrix is too large to fit in the memory, smaller maximum radius ``rmax`` can be chosen to make the distance matrix sparse.
Based on the discussion above, we run radius estimation with
1. sample size=1000 (created by choosing one data point out of every 10 of the original data.)
1. radius search from ``rmin=0.1`` to ``rmax=50``, with 50 points in logspace.
1. dimension ``d=1``
Specify run_parallel=True for searching the radius in parallel.
```
%%capture
# Using magic command %%capture for supressing the std out.
from megaman.utils.estimate_radius import run_estimate_radius
import numpy as np
# subsample by 10.
sample = np.arange(0,data.shape[0],10)
distorion_vs_rad_dim1 = run_estimate_radius(
data, dist, sample=sample, d=1, rmin=rmin, rmax=rmax,
ntry=50, run_parallel=True, search_space='logspace')
```
Run radius estimation same configurations as above except
1. dimension ``d=2``
```
%%capture
distorion_vs_rad_dim2 = run_estimate_radius(
data, dist, sample=sample, d=2, rmin=0.1, rmax=5,
ntry=50, run_parallel=True, search_space='logspace')
```
### Radius estimation result
The estimated radius is the minimizer of the distorsion, denoted as $\hat{r}_{d=1}$ and $\hat{r}_{d=2}$. (In the code, it's ``est_rad_dim1`` and ``est_rad_dim2``)
```
distorsion_dim1 = distorion_vs_rad_dim1[:,1].astype('float64')
distorsion_dim2 = distorion_vs_rad_dim2[:,1].astype('float64')
rad_search_space = distorion_vs_rad_dim1[:,0].astype('float64')
argmin_d1 = np.argmin(distorsion_dim1)
argmin_d2 = np.argmin(distorsion_dim2)
est_rad_dim1 = rad_search_space[argmin_d1]
est_rad_dim2 = rad_search_space[argmin_d2]
print ('Estimated radius with d=1 is: {:.4f}'.format(est_rad_dim1))
print ('Estimated radius with d=2 is: {:.4f}'.format(est_rad_dim2))
```
### Plot distorsions with different radii
```
t_distorsion = [go.Scatter(x=rad_search_space, y=distorsion_dim1, name='Dimension = 1'),
go.Scatter(x=rad_search_space, y=distorsion_dim2, name='Dimension = 2')]
l_distorsion = go.Layout(
title='Distorsions versus radii',
xaxis=dict(
title='$\\text{Radius } r$',
type='log',
autorange=True
),
yaxis=dict(
title='Distorsion',
type='log',
autorange=True
),
annotations=[
dict(
x=np.log10(est_rad_dim1),
y=np.log10(distorsion_dim1[argmin_d1]),
xref='x',
yref='y',
text='$\\hat{r}_{d=1}$',
font = dict(size = 30),
showarrow=True,
arrowhead=7,
ax=0,
ay=-30
),
dict(
x=np.log10(est_rad_dim2),
y=np.log10(distorsion_dim2[argmin_d2]),
xref='x',
yref='y',
text='$\\hat{r}_{d=2}$',
font = dict(size = 30),
showarrow=True,
arrowhead=7,
ax=0,
ay=-30
)
]
)
f_distorsion = go.Figure(data=t_distorsion,layout=l_distorsion)
iplot(f_distorsion)
```
## Application to dimension estimation
We followed the method proposed by [Chen et. al (2011)]((http://lcsl.mit.edu/papers/che_lit_mag_ros_2011.pdf) [5] to verify the estimated radius reflect the truth intrinsic dimension of the data. The basic idea is to find the largest gap of singular value of local PCA, which correspond to the dimension of the local structure.
We first plot the average singular values versus radii.
```
%%capture
from rad_est_utils import find_argmax_dimension, estimate_dimension
rad_search_space, singular_values = estimate_dimension(data, dist)
```
The singular gap is the different between two singular values. Since the intrinsic dimension is 2, we are interested in the region where the largest singular gap is the second. The region is:
```
singular_gap = -1*np.diff(singular_values,axis=1)
second_gap_is_max_range = (np.argmax(singular_gap,axis=1) == 1).nonzero()[0]
start_idx, end_idx = second_gap_is_max_range[0], second_gap_is_max_range[-1]+1
print ('The index which maximize the second singular gap is: {}'.format(second_gap_is_max_range))
print ('The start and end index of largest continuous range is {} and {}, respectively'.format(start_idx, end_idx))
```
### Averaged singular values with different radii
Plot the averaged singular values with different radii. The gray shaded area is the continous range in which the largest singular gap is the second, (local structure has dimension equals 2). And the purple shaded area denotes the second singular gap.
By hovering the line on this plot, you can see the value of the singular gap.
```
from rad_est_utils import plot_singular_values_versus_radius, generate_layouts
t_avg_singular = plot_singular_values_versus_radius(singular_values, rad_search_space, start_idx, end_idx)
l_avg_singular = generate_layouts(start_idx, end_idx, est_rad_dim1, est_rad_dim2, rad_search_space)
f_avg_singular = go.Figure(data=t_avg_singular,layout=l_avg_singular)
iplot(f_avg_singular)
```
### Histogram of estimated dimensions with estimated radius.
We first find out the estimated dimensions of each points in the data using the estimated radius $\hat{r}_{d=1}$ and $\hat{r}_{d=2}$.
```
dimension_freq_d1 = find_argmax_dimension(data,dist, est_rad_dim1)
dimension_freq_d2 = find_argmax_dimension(data,dist, est_rad_dim2)
```
The histogram of estimated dimensions with different optimal radius is shown as below:
```
t_hist_dim = [go.Histogram(x=dimension_freq_d1,name='d=1'),
go.Histogram(x=dimension_freq_d2,name='d=2')]
l_hist_dim = go.Layout(
title='Dimension histogram',
xaxis=dict(
title='Estimated dimension'
),
yaxis=dict(
title='Counts'
),
bargap=0.2,
bargroupgap=0.1
)
f_hist_dim = go.Figure(data=t_hist_dim,layout=l_hist_dim)
iplot(f_hist_dim)
```
## Conclusion
1. Choosing the correct radius/bound/scale is important in any non-linear dimension reduction task
1. The __Geometry Consistency (GC) Algorithm__ required minimal knowledge: maximum radius, minimum radius, (optionally: dimension $d$ of the manifold.)
1. The chosen radius can be used in
1. any embedding algorithm
1. semi-supervised learning with Laplacian Regularizer (see our NIPS 2017 paper)
1. estimating dimension $d$ (as shown here)
1. The megaman python package is __scalable__, and __efficient__
<img src=https://raw.githubusercontent.com/mmp2/megaman/master/doc/images/spectra_Halpha.png width=600 />
## __Try it:__
<div style="float:left;">All the functions are implemented by the manifold learning package <a href=https://github.com/mmp2/megaman>megaman.</a> </div><a style="float:left;" href="https://anaconda.org/conda-forge/megaman"><img src="https://anaconda.org/conda-forge/megaman/badges/downloads.svg" /></a>
## Reference
[1] R. R. Coifman, S. Lafon. Diffusion maps. Applied and Computational Harmonic Analysis, 2006. <br>
[2] D. Perrault-Joncas, M. Meila, Metric learning and manifolds: Preserving the intrinsic geometry , arXiv1305.7255 <br>
[3] X. Zhou, M. Belkin. Semi-supervised learning by higher order regularization. AISTAT, 2011 <br>
[4] A. Singer. From graph to manifold laplacian: the convergence rate. Applied and Computational Harmonic Analysis, 2006. <br>
[5] G. Chen, A. Little, M. Maggioni, L. Rosasco. Some recent advances in multiscale geometric analysis of point clouds. Wavelets and multiscale analysis. Springer, 2011. <br>
[6] L. Chen, A. Buja. Local Multidimensional Scaling for nonlinear dimension reduction, graph drawing and proximity analysis, JASA,2009. <br>
|
github_jupyter
|
##### Copyright 2019 Google LLC
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# Graph regularization for sentiment classification using synthesized graphs
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://www.tensorflow.org/neural_structured_learning/tutorials/graph_keras_lstm_imdb"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on TensorFlow.org</a>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/neural-structured-learning/blob/master/g3doc/tutorials/graph_keras_lstm_imdb.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/neural-structured-learning/blob/master/g3doc/tutorials/graph_keras_lstm_imdb.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
</td>
</table>
## Overview
This notebook classifies movie reviews as *positive* or *negative* using the
text of the review. This is an example of *binary* classification, an important
and widely applicable kind of machine learning problem.
We will demonstrate the use of graph regularization in this notebook by building
a graph from the given input. The general recipe for building a
graph-regularized model using the Neural Structured Learning (NSL) framework
when the input does not contain an explicit graph is as follows:
1. Create embeddings for each text sample in the input. This can be done using
pre-trained models such as [word2vec](https://arxiv.org/pdf/1310.4546.pdf),
[Swivel](https://arxiv.org/abs/1602.02215),
[BERT](https://arxiv.org/abs/1810.04805) etc.
2. Build a graph based on these embeddings by using a similarity metric such as
the 'L2' distance, 'cosine' distance, etc. Nodes in the graph correspond to
samples and edges in the graph correspond to similarity between pairs of
samples.
3. Generate training data from the above synthesized graph and sample features.
The resulting training data will contain neighbor features in addition to
the original node features.
4. Create a neural network as a base model using the Keras sequential,
functional, or subclass API.
5. Wrap the base model with the GraphRegularization wrapper class, which is
provided by the NSL framework, to create a new graph Keras model. This new
model will include a graph regularization loss as the regularization term in
its training objective.
6. Train and evaluate the graph Keras model.
**Note**: We expect that it would take readers about 1 hour to go through this
tutorial.
## Requirements
1. Install TensorFlow 2.x to create an interactive developing environment with eager execution.
2. Install the Neural Structured Learning package.
3. Install tensorflow-hub.
```
!pip install --quiet tensorflow==2.0.0-rc0
!pip install --quiet neural-structured-learning
!pip install --quiet tensorflow-hub
```
## Dependencies and imports
```
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
from __future__ import unicode_literals
import matplotlib.pyplot as plt
import numpy as np
import neural_structured_learning as nsl
import tensorflow as tf
tf.compat.v1.enable_v2_behavior()
import tensorflow_hub as hub
# Resets notebook state
tf.keras.backend.clear_session()
print("Version: ", tf.__version__)
print("Eager mode: ", tf.executing_eagerly())
print("Hub version: ", hub.__version__)
print("GPU is", "available" if tf.test.is_gpu_available() else "NOT AVAILABLE")
```
## IMDB dataset
The
[IMDB dataset](https://www.tensorflow.org/api_docs/python/tf/keras/datasets/imdb)
contains the text of 50,000 movie reviews from the
[Internet Movie Database](https://www.imdb.com/). These are split into 25,000
reviews for training and 25,000 reviews for testing. The training and testing
sets are *balanced*, meaning they contain an equal number of positive and
negative reviews.
In this tutorial, we will use a preprocessed version of the IMDB dataset.
### Download preprocessed IMDB dataset
The IMDB dataset comes packaged with TensorFlow. It has already been
preprocessed such that the reviews (sequences of words) have been converted to
sequences of integers, where each integer represents a specific word in a
dictionary.
The following code downloads the IMDB dataset (or uses a cached copy if it has
already been downloaded):
```
imdb = tf.keras.datasets.imdb
(pp_train_data, pp_train_labels), (pp_test_data, pp_test_labels) = (
imdb.load_data(num_words=10000))
```
The argument `num_words=10000` keeps the top 10,000 most frequently occurring words in the training data. The rare words are discarded to keep the size of the vocabulary manageable.
### Explore the data
Let's take a moment to understand the format of the data. The dataset comes preprocessed: each example is an array of integers representing the words of the movie review. Each label is an integer value of either 0 or 1, where 0 is a negative review, and 1 is a positive review.
```
print('Training entries: {}, labels: {}'.format(
len(pp_train_data), len(pp_train_labels)))
training_samples_count = len(pp_train_data)
```
The text of reviews have been converted to integers, where each integer represents a specific word in a dictionary. Here's what the first review looks like:
```
print(pp_train_data[0])
```
Movie reviews may be different lengths. The below code shows the number of words in the first and second reviews. Since inputs to a neural network must be the same length, we'll need to resolve this later.
```
len(pp_train_data[0]), len(pp_train_data[1])
```
### Convert the integers back to words
It may be useful to know how to convert integers back to the corresponding text.
Here, we'll create a helper function to query a dictionary object that contains
the integer to string mapping:
```
def build_reverse_word_index():
# A dictionary mapping words to an integer index
word_index = imdb.get_word_index()
# The first indices are reserved
word_index = {k: (v + 3) for k, v in word_index.items()}
word_index['<PAD>'] = 0
word_index['<START>'] = 1
word_index['<UNK>'] = 2 # unknown
word_index['<UNUSED>'] = 3
return dict((value, key) for (key, value) in word_index.items())
reverse_word_index = build_reverse_word_index()
def decode_review(text):
return ' '.join([reverse_word_index.get(i, '?') for i in text])
```
Now we can use the `decode_review` function to display the text for the first review:
```
decode_review(pp_train_data[0])
```
## Graph construction
Graph construction involves creating embeddings for text samples and then using
a similarity function to compare the embeddings.
Before proceeding further, we first create a directory to store artifacts
created by this tutorial.
```
!mkdir -p /tmp/imdb
```
### Create sample embeddings
We will use pretrained Swivel embeddings to create embeddings in the
`tf.train.Example` format for each sample in the input. We will store the
resulting embeddings in the `TFRecord` format along with an additional feature
that represents the ID of each sample. This is important and will allow us match
sample embeddings with corresponding nodes in the graph later.
```
# This is necessary because hub.KerasLayer assumes tensor hashability, which
# is not supported in eager mode.
tf.compat.v1.disable_tensor_equality()
pretrained_embedding = 'https://tfhub.dev/google/tf2-preview/gnews-swivel-20dim/1'
hub_layer = hub.KerasLayer(
pretrained_embedding, input_shape=[], dtype=tf.string, trainable=True)
def _int64_feature(value):
"""Returns int64 tf.train.Feature."""
return tf.train.Feature(int64_list=tf.train.Int64List(value=value.tolist()))
def _bytes_feature(value):
"""Returns bytes tf.train.Feature."""
return tf.train.Feature(
bytes_list=tf.train.BytesList(value=[value.encode('utf-8')]))
def _float_feature(value):
"""Returns float tf.train.Feature."""
return tf.train.Feature(float_list=tf.train.FloatList(value=value.tolist()))
def create_embedding_example(word_vector, record_id):
"""Create tf.Example containing the sample's embedding and its ID."""
text = decode_review(word_vector)
# Shape = [batch_size,].
sentence_embedding = hub_layer(tf.reshape(text, shape=[-1,]))
# Flatten the sentence embedding back to 1-D.
sentence_embedding = tf.reshape(sentence_embedding, shape=[-1])
features = {
'id': _bytes_feature(str(record_id)),
'embedding': _float_feature(sentence_embedding.numpy())
}
return tf.train.Example(features=tf.train.Features(feature=features))
def create_embeddings(word_vectors, output_path, starting_record_id):
record_id = int(starting_record_id)
with tf.io.TFRecordWriter(output_path) as writer:
for word_vector in word_vectors:
example = create_embedding_example(word_vector, record_id)
record_id = record_id + 1
writer.write(example.SerializeToString())
return record_id
# Persist TF.Example features containing embeddings for training data in
# TFRecord format.
create_embeddings(pp_train_data, '/tmp/imdb/embeddings.tfr', 0)
```
### Build a graph
Now that we have the sample embeddings, we will use them to build a similarity
graph, i.e, nodes in this graph will correspond to samples and edges in this
graph will correspond to similarity between pairs of nodes.
Neural Structured Learning provides a graph building tool that builds a graph
based on sample embeddings. It uses **cosine similarity** as the similarity
measure to compare embeddings and build edges between them. It also allows us to
specify a similarity threshold, which can be used to discard dissimilar edges
from the final graph. In this example, using 0.99 as the similarity threshold,
we end up with a graph that has 445,327 bi-directional edges.
```
!python -m neural_structured_learning.tools.build_graph \
--similarity_threshold=0.99 /tmp/imdb/embeddings.tfr /tmp/imdb/graph_99.tsv
```
**Note:** Graph quality and by extension, embedding quality, are very important
for graph regularization. While we have used Swivel embeddings in this notebook,
using BERT embeddings for instance, will likely capture review semantics more
accurately. We encourage users to use embeddings of their choice and as
appropriate to their needs.
## Sample features
We create sample features for our problem in the `tf.train.Example`s format and
persist them in the `TFRecord` format. Each sample will include the following
three features:
1. **id**: The node ID of the sample.
2. **words**: An int64 list containing word IDs.
3. **label**: A singleton int64 identifying the target class of the review.
```
def create_example(word_vector, label, record_id):
"""Create tf.Example containing the sample's word vector, label, and ID."""
features = {
'id': _bytes_feature(str(record_id)),
'words': _int64_feature(np.asarray(word_vector)),
'label': _int64_feature(np.asarray([label])),
}
return tf.train.Example(features=tf.train.Features(feature=features))
def create_records(word_vectors, labels, record_path, starting_record_id):
record_id = int(starting_record_id)
with tf.io.TFRecordWriter(record_path) as writer:
for word_vector, label in zip(word_vectors, labels):
example = create_example(word_vector, label, record_id)
record_id = record_id + 1
writer.write(example.SerializeToString())
return record_id
# Persist TF.Example features (word vectors and labels) for training and test
# data in TFRecord format.
next_record_id = create_records(pp_train_data, pp_train_labels,
'/tmp/imdb/train_data.tfr', 0)
create_records(pp_test_data, pp_test_labels, '/tmp/imdb/test_data.tfr',
next_record_id)
```
## Augment training data with graph neighbors
Since we have the sample features and the synthesized graph, we can generate the
augmented training data for Neural Structured Learning. The NSL framework
provides a tool that can combine the graph and the sample features to produce
the final training data for graph regularization. The resulting training data
will include original sample features as well as features of their corresponding
neighbors.
In this tutorial, we consider undirected edges and we use a maximum of 1
neighbor per sample.
```
!python -m neural_structured_learning.tools.pack_nbrs \
--max_nbrs=3 --add_undirected_edges=True \
/tmp/imdb/train_data.tfr '' /tmp/imdb/graph_99.tsv \
/tmp/imdb/nsl_train_data.tfr
```
## Base model
We are now ready to build a base model without graph regularization. In order to build this model, we can either use embeddings that were used in building the graph, or we can learn new embeddings jointly along with the classification task. For the purpose of this notebook, we will do the latter.
### Global variables
```
NBR_FEATURE_PREFIX = 'NL_nbr_'
NBR_WEIGHT_SUFFIX = '_weight'
```
### Hyperparameters
We will use an instance of `HParams` to inclue various hyperparameters and
constants used for training and evaluation. We briefly describe each of them
below:
- **num_classes**: There are 2 classes -- *positive* and *negative*.
- **max_seq_length**: This is the maximum number of words considered from each movie review in this example.
- **vocab_size**: This is the size of the vocabulary considered for this example.
- **distance_type**: This is the distance metric used to regularize the sample
with its neighbors.
- **graph_regularization_multiplier**: This controls the relative weight of
the graph regularization term in the overall loss function.
- **num_neighbors**: The number of neighbors used for graph regularization.
- **num_fc_units**: The number of units in the fully connected layer of the neural network.
- **train_epochs**: The number of training epochs.
- **batch_size**: Batch size used for training and evaluation.
- **eval_steps**: The number of batches to process before deeming evaluation
is complete. If set to `None`, all instances in the test set are evaluated.
```
class HParams(object):
"""Hyperparameters used for training."""
def __init__(self):
### dataset parameters
self.num_classes = 2
self.max_seq_length = 256
self.vocab_size = 10000
### neural graph learning parameters
self.distance_type = nsl.configs.DistanceType.L2
self.graph_regularization_multiplier = 0.1
self.num_neighbors = 2
### model architecture
self.num_embedding_dims = 16
self.num_lstm_dims = 64
self.num_fc_units = 64
### training parameters
self.train_epochs = 10
self.batch_size = 128
### eval parameters
self.eval_steps = None # All instances in the test set are evaluated.
HPARAMS = HParams()
```
### Prepare the data
The reviews—the arrays of integers—must be converted to tensors before being fed
into the neural network. This conversion can be done a couple of ways:
* Convert the arrays into vectors of `0`s and `1`s indicating word occurrence,
similar to a one-hot encoding. For example, the sequence `[3, 5]` would become a `10000`-dimensional vector that is all zeros except for indices `3` and `5`, which are ones. Then, make this the first layer in our network—a `Dense` layer—that can handle floating point vector data. This approach is memory intensive, though, requiring a `num_words * num_reviews` size matrix.
* Alternatively, we can pad the arrays so they all have the same length, then
create an integer tensor of shape `max_length * num_reviews`. We can use an
embedding layer capable of handling this shape as the first layer in our
network.
In this tutorial, we will use the second approach.
Since the movie reviews must be the same length, we will use the `pad_sequence`
function defined below to standardize the lengths.
```
def pad_sequence(sequence, max_seq_length):
"""Pads the input sequence (a `tf.SparseTensor`) to `max_seq_length`."""
pad_size = tf.maximum([0], max_seq_length - tf.shape(sequence)[0])
padded = tf.concat(
[sequence.values,
tf.fill((pad_size), tf.cast(0, sequence.dtype))],
axis=0)
# The input sequence may be larger than max_seq_length. Truncate down if
# necessary.
return tf.slice(padded, [0], [max_seq_length])
def parse_example(example_proto):
"""Extracts relevant fields from the `example_proto`.
Args:
example_proto: An instance of `tf.train.Example`.
Returns:
A pair whose first value is a dictionary containing relevant features
and whose second value contains the ground truth labels.
"""
# The 'words' feature is a variable length word ID vector.
feature_spec = {
'words': tf.io.VarLenFeature(tf.int64),
'label': tf.io.FixedLenFeature((), tf.int64, default_value=-1),
}
# We also extract corresponding neighbor features in a similar manner to
# the features above.
for i in range(HPARAMS.num_neighbors):
nbr_feature_key = '{}{}_{}'.format(NBR_FEATURE_PREFIX, i, 'words')
nbr_weight_key = '{}{}{}'.format(NBR_FEATURE_PREFIX, i, NBR_WEIGHT_SUFFIX)
feature_spec[nbr_feature_key] = tf.io.VarLenFeature(tf.int64)
# We assign a default value of 0.0 for the neighbor weight so that
# graph regularization is done on samples based on their exact number
# of neighbors. In other words, non-existent neighbors are discounted.
feature_spec[nbr_weight_key] = tf.io.FixedLenFeature(
[1], tf.float32, default_value=tf.constant([0.0]))
features = tf.io.parse_single_example(example_proto, feature_spec)
# Since the 'words' feature is a variable length word vector, we pad it to a
# constant maximum length based on HPARAMS.max_seq_length
features['words'] = pad_sequence(features['words'], HPARAMS.max_seq_length)
for i in range(HPARAMS.num_neighbors):
nbr_feature_key = '{}{}_{}'.format(NBR_FEATURE_PREFIX, i, 'words')
features[nbr_feature_key] = pad_sequence(features[nbr_feature_key],
HPARAMS.max_seq_length)
labels = features.pop('label')
return features, labels
def make_dataset(file_path, training=False):
"""Creates a `tf.data.TFRecordDataset`.
Args:
file_path: Name of the file in the `.tfrecord` format containing
`tf.train.Example` objects.
training: Boolean indicating if we are in training mode.
Returns:
An instance of `tf.data.TFRecordDataset` containing the `tf.train.Example`
objects.
"""
dataset = tf.data.TFRecordDataset([file_path])
if training:
dataset = dataset.shuffle(10000)
dataset = dataset.map(parse_example)
dataset = dataset.batch(HPARAMS.batch_size)
return dataset
train_dataset = make_dataset('/tmp/imdb/nsl_train_data.tfr', True)
test_dataset = make_dataset('/tmp/imdb/test_data.tfr')
```
### Build the model
A neural network is created by stacking layers—this requires two main architectural decisions:
* How many layers to use in the model?
* How many *hidden units* to use for each layer?
In this example, the input data consists of an array of word-indices. The labels to predict are either 0 or 1.
We will use a bi-directional LSTM as our base model in this tutorial.
```
# This function exists as an alternative to the bi-LSTM model used in this
# notebook.
def make_feed_forward_model():
"""Builds a simple 2 layer feed forward neural network."""
inputs = tf.keras.Input(
shape=(HPARAMS.max_seq_length,), dtype='int64', name='words')
embedding_layer = tf.keras.layers.Embedding(HPARAMS.vocab_size, 16)(inputs)
pooling_layer = tf.keras.layers.GlobalAveragePooling1D()(embedding_layer)
dense_layer = tf.keras.layers.Dense(16, activation='relu')(pooling_layer)
outputs = tf.keras.layers.Dense(1, activation='sigmoid')(dense_layer)
return tf.keras.Model(inputs=inputs, outputs=outputs)
def make_bilstm_model():
"""Builds a bi-directional LSTM model."""
inputs = tf.keras.Input(
shape=(HPARAMS.max_seq_length,), dtype='int64', name='words')
embedding_layer = tf.keras.layers.Embedding(HPARAMS.vocab_size,
HPARAMS.num_embedding_dims)(
inputs)
lstm_layer = tf.keras.layers.Bidirectional(
tf.keras.layers.LSTM(HPARAMS.num_lstm_dims))(
embedding_layer)
dense_layer = tf.keras.layers.Dense(
HPARAMS.num_fc_units, activation='relu')(
lstm_layer)
outputs = tf.keras.layers.Dense(1, activation='sigmoid')(dense_layer)
return tf.keras.Model(inputs=inputs, outputs=outputs)
# Feel free to use an architecture of your choice.
model = make_bilstm_model()
model.summary()
```
The layers are effectively stacked sequentially to build the classifier:
1. The first layer is an `Input` layer which takes the integer-encoded
vocabulary.
2. The next layer is an `Embedding` layer, which takes the integer-encoded
vocabulary and looks up the embedding vector for each word-index. These
vectors are learned as the model trains. The vectors add a dimension to the
output array. The resulting dimensions are: `(batch, sequence, embedding)`.
3. Next, a bidirectional LSTM layer returns a fixed-length output vector for
each example.
4. This fixed-length output vector is piped through a fully-connected (`Dense`)
layer with 64 hidden units.
5. The last layer is densely connected with a single output node. Using the
`sigmoid` activation function, this value is a float between 0 and 1,
representing a probability, or confidence level.
### Hidden units
The above model has two intermediate or "hidden" layers, between the input and
output, and excluding the `Embedding` layer. The number of outputs (units,
nodes, or neurons) is the dimension of the representational space for the layer.
In other words, the amount of freedom the network is allowed when learning an
internal representation.
If a model has more hidden units (a higher-dimensional representation space),
and/or more layers, then the network can learn more complex representations.
However, it makes the network more computationally expensive and may lead to
learning unwanted patterns—patterns that improve performance on training data
but not on the test data. This is called *overfitting*.
### Loss function and optimizer
A model needs a loss function and an optimizer for training. Since this is a
binary classification problem and the model outputs a probability (a single-unit
layer with a sigmoid activation), we'll use the `binary_crossentropy` loss
function.
```
model.compile(
optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
```
### Create a validation set
When training, we want to check the accuracy of the model on data it hasn't seen
before. Create a *validation set* by setting apart a fraction of the original
training data. (Why not use the testing set now? Our goal is to develop and tune
our model using only the training data, then use the test data just once to
evaluate our accuracy).
In this tutorial, we take roughly 10% of the initial training samples (10% of 25000) as labeled data for training and the remaining as validation data. Since the initial train/test split was 50/50 (25000 samples each), the effective train/validation/test split we now have is 5/45/50.
Note that 'train_dataset' has already been batched and shuffled.
```
validation_fraction = 0.9
validation_size = int(validation_fraction *
int(training_samples_count / HPARAMS.batch_size))
print(validation_size)
validation_dataset = train_dataset.take(validation_size)
train_dataset = train_dataset.skip(validation_size)
```
### Train the model
Train the model in mini-batches. While training, monitor the model's loss and accuracy on the validation set:
```
history = model.fit(
train_dataset,
validation_data=validation_dataset,
epochs=HPARAMS.train_epochs,
verbose=1)
```
### Evaluate the model
Now, let's see how the model performs. Two values will be returned. Loss (a number which represents our error, lower values are better), and accuracy.
```
results = model.evaluate(test_dataset, steps=HPARAMS.eval_steps)
print(results)
```
### Create a graph of accuracy/loss over time
`model.fit()` returns a `History` object that contains a dictionary with everything that happened during training:
```
history_dict = history.history
history_dict.keys()
```
There are four entries: one for each monitored metric during training and validation. We can use these to plot the training and validation loss for comparison, as well as the training and validation accuracy:
```
acc = history_dict['accuracy']
val_acc = history_dict['val_accuracy']
loss = history_dict['loss']
val_loss = history_dict['val_loss']
epochs = range(1, len(acc) + 1)
# "-r^" is for solid red line with triangle markers.
plt.plot(epochs, loss, '-r^', label='Training loss')
# "-b0" is for solid blue line with circle markers.
plt.plot(epochs, val_loss, '-bo', label='Validation loss')
plt.title('Training and validation loss')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend(loc='best')
plt.show()
plt.clf() # clear figure
plt.plot(epochs, acc, '-r^', label='Training acc')
plt.plot(epochs, val_acc, '-bo', label='Validation acc')
plt.title('Training and validation accuracy')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.legend(loc='best')
plt.show()
```
Notice the training loss *decreases* with each epoch and the training accuracy
*increases* with each epoch. This is expected when using a gradient descent
optimization—it should minimize the desired quantity on every iteration.
## Graph regularization
We are now ready to try graph regularization using the base model that we built
above. We will use the `GraphRegularization` wrapper class provided by the
Neural Structured Learning framework to wrap the base (bi-LSTM) model to include
graph regularization. The rest of the steps for training and evaluating the
graph-regularized model are similar to that of the base model.
### Create graph-regularized model
To assess the incremental benefit of graph regularization, we will create a new
base model instance. This is because `model` has already been trained for a few
iterations, and reusing this trained model to create a graph-regularized model
will not be a fair comparison for `model`.
```
# Build a new base LSTM model.
base_reg_model = make_bilstm_model()
# Wrap the base model with graph regularization.
graph_reg_config = nsl.configs.GraphRegConfig(
neighbor_config=nsl.configs.GraphNeighborConfig(
max_neighbors=HPARAMS.num_neighbors),
multiplier=HPARAMS.graph_regularization_multiplier,
distance_config=nsl.configs.DistanceConfig(
distance_type=HPARAMS.distance_type, sum_over_axis=-1))
graph_reg_model = nsl.keras.GraphRegularization(base_reg_model,
graph_reg_config)
graph_reg_model.compile(
optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
```
### Train the model
```
graph_reg_history = graph_reg_model.fit(
train_dataset,
validation_data=validation_dataset,
epochs=HPARAMS.train_epochs,
verbose=1)
```
### Evaluate the model
```
graph_reg_results = graph_reg_model.evaluate(test_dataset, steps=HPARAMS.eval_steps)
print(graph_reg_results)
```
### Create a graph of accuracy/loss over time
```
graph_reg_history_dict = graph_reg_history.history
graph_reg_history_dict.keys()
```
There are six entries: one for each monitored metric -- loss, graph loss, and
accuracy -- during training and validation. We can use these to plot the
training, graph, and validation losses for comparison, as well as the training
and validation accuracy. Note that the graph loss is only computed during
training; so its value will be 0 during validation.
```
acc = graph_reg_history_dict['accuracy']
val_acc = graph_reg_history_dict['val_accuracy']
loss = graph_reg_history_dict['loss']
graph_loss = graph_reg_history_dict['graph_loss']
val_loss = graph_reg_history_dict['val_loss']
val_graph_loss = graph_reg_history_dict['val_graph_loss']
epochs = range(1, len(acc) + 1)
plt.clf() # clear figure
# "-r^" is for solid red line with triangle markers.
plt.plot(epochs, loss, '-r^', label='Training loss')
# "-gD" is for solid green line with diamond markers.
plt.plot(epochs, graph_loss, '-gD', label='Training graph loss')
# "-b0" is for solid blue line with circle markers.
plt.plot(epochs, val_loss, '-bo', label='Validation loss')
# "-ms" is for solid magenta line with square markers.
plt.plot(epochs, val_graph_loss, '-ms', label='Validation graph loss')
plt.title('Training and validation loss')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend(loc='best')
plt.show()
plt.clf() # clear figure
plt.plot(epochs, acc, '-r^', label='Training acc')
plt.plot(epochs, val_acc, '-bo', label='Validation acc')
plt.title('Training and validation accuracy')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.legend(loc='best')
plt.show()
```
## The power of semi-supervised learning
Semi-supervised learning and more specifically, graph regularization in the
context of this tutorial, can be really powerful when the amount of training
data is small. The lack of training data is compensated by leveraging similarity
among the training samples, which is not possible in traditional supervised
learning.
We define ***supervision ratio*** as the ratio of training samples to the total
number of samples which includes training, validation, and test samples. In this
notebook, we have used a supervision ratio of 0.05 (i.e, 5% of the labeled data)
for training both the base model as well as the graph-regularized model. We
illustrate the impact of the supervision ratio on model accuracy in the cell
below.
```
# Accuracy values for both the Bi-LSTM model and the feed forward NN model have
# been precomputed for the following supervision ratios.
supervision_ratios = [0.3, 0.15, 0.05, 0.03, 0.02, 0.01, 0.005]
model_tags = ['Bi-LSTM model', 'Feed Forward NN model']
base_model_accs = [[84, 84, 83, 80, 65, 52, 50], [87, 86, 76, 74, 67, 52, 51]]
graph_reg_model_accs = [[84, 84, 83, 83, 65, 63, 50],
[87, 86, 80, 75, 67, 52, 50]]
plt.clf() # clear figure
fig, axes = plt.subplots(1, 2)
fig.set_size_inches((12, 5))
for ax, model_tag, base_model_acc, graph_reg_model_acc in zip(
axes, model_tags, base_model_accs, graph_reg_model_accs):
# "-r^" is for solid red line with triangle markers.
ax.plot(base_model_acc, '-r^', label='Base model')
# "-gD" is for solid green line with diamond markers.
ax.plot(graph_reg_model_acc, '-gD', label='Graph-regularized model')
ax.set_title(model_tag)
ax.set_xlabel('Supervision ratio')
ax.set_ylabel('Accuracy(%)')
ax.set_ylim((25, 100))
ax.set_xticks(range(len(supervision_ratios)))
ax.set_xticklabels(supervision_ratios)
ax.legend(loc='best')
plt.show()
```
It can be observed that as the superivision ratio decreases, model accuracy also
decreases. This is true for both the base model and for the graph-regularized
model, regardless of the model architecture used. However, notice that the
graph-regularized model performs better than the base model for both the
architectures. In particular, for the Bi-LSTM model, when the supervision ratio
is 0.01, the accuracy of the graph-regularized model is **~20%** higher than
that of the base model. This is primarily because of semi-supervised learning
for the graph-regularized model, where structural similarity among training
samples is used in addition to the training samples themselves.
## Conclusion
We have demonstrated the use of graph regularization using the Neural Structured
Learning (NSL) framework even when the input does not contain an explicit graph.
We considered the task of sentiment classification of IMDB movie reviews for
which we synthesized a similarity graph based on review embeddings. We encourage
users to experiment further by varying hyperparameters, the amount of
supervision, and by using different model architectures.
|
github_jupyter
|
# Road Following - Live demo
In this notebook, we will use model we trained to move jetBot smoothly on track.
### Load Trained Model
We will assume that you have already downloaded ``best_steering_model_xy.pth`` to work station as instructed in "train_model.ipynb" notebook. Now, you should upload model file to JetBot in to this notebooks's directory. Once that's finished there should be a file named ``best_steering_model_xy.pth`` in this notebook's directory.
> Please make sure the file has uploaded fully before calling the next cell
Execute the code below to initialize the PyTorch model. This should look very familiar from the training notebook.
```
import torchvision
import torch
model = torchvision.models.resnet18(pretrained=False)
model.fc = torch.nn.Linear(512, 2)
```
Next, load the trained weights from the ``best_steering_model_xy.pth`` file that you uploaded.
```
model.load_state_dict(torch.load('best_steering_model_xy.pth'))
```
Currently, the model weights are located on the CPU memory execute the code below to transfer to the GPU device.
```
device = torch.device('cuda')
model = model.to(device)
model = model.eval().half()
```
### Creating the Pre-Processing Function
We have now loaded our model, but there's a slight issue. The format that we trained our model doesnt exactly match the format of the camera. To do that, we need to do some preprocessing. This involves the following steps:
1. Convert from HWC layout to CHW layout
2. Normalize using same parameters as we did during training (our camera provides values in [0, 255] range and training loaded images in [0, 1] range so we need to scale by 255.0
3. Transfer the data from CPU memory to GPU memory
4. Add a batch dimension
```
import torchvision.transforms as transforms
import torch.nn.functional as F
import cv2
import PIL.Image
import numpy as np
mean = torch.Tensor([0.485, 0.456, 0.406]).cuda().half()
std = torch.Tensor([0.229, 0.224, 0.225]).cuda().half()
def preprocess(image):
image = PIL.Image.fromarray(image)
image = transforms.functional.to_tensor(image).to(device).half()
image.sub_(mean[:, None, None]).div_(std[:, None, None])
return image[None, ...]
```
Awesome! We've now defined our pre-processing function which can convert images from the camera format to the neural network input format.
Now, let's start and display our camera. You should be pretty familiar with this by now.
```
from IPython.display import display
import ipywidgets
import traitlets
from jetbot import Camera, bgr8_to_jpeg
camera = Camera()
image_widget = ipywidgets.Image()
traitlets.dlink((camera, 'value'), (image_widget, 'value'), transform=bgr8_to_jpeg)
display(image_widget)
```
We'll also create our robot instance which we'll need to drive the motors.
```
from jetbot import Robot
robot = Robot()
```
Now, we will define sliders to control JetBot
> Note: We have initialize the slider values for best known configurations, however these might not work for your dataset, therefore please increase or decrease the sliders according to your setup and environment
1. Speed Control (speed_gain_slider): To start your JetBot increase ``speed_gain_slider``
2. Steering Gain Control (steering_gain_sloder): If you see JetBot is woblling, you need to reduce ``steering_gain_slider`` till it is smooth
3. Steering Bias control (steering_bias_slider): If you see JetBot is biased towards extreme right or extreme left side of the track, you should control this slider till JetBot start following line or track in the center. This accounts for motor biases as well as camera offsets
> Note: You should play around above mentioned sliders with lower speed to get smooth JetBot road following behavior.
```
speed_gain_slider = ipywidgets.FloatSlider(min=0.0, max=1.0, step=0.01, description='speed gain')
steering_gain_slider = ipywidgets.FloatSlider(min=0.0, max=1.0, step=0.01, value=0.2, description='steering gain')
steering_dgain_slider = ipywidgets.FloatSlider(min=0.0, max=0.5, step=0.001, value=0.0, description='steering kd')
steering_bias_slider = ipywidgets.FloatSlider(min=-0.3, max=0.3, step=0.01, value=0.0, description='steering bias')
display(speed_gain_slider, steering_gain_slider, steering_dgain_slider, steering_bias_slider)
```
Next, let's display some sliders that will let us see what JetBot is thinking. The x and y sliders will display the predicted x, y values.
The steering slider will display our estimated steering value. Please remember, this value isn't the actual angle of the target, but simply a value that is
nearly proportional. When the actual angle is ``0``, this will be zero, and it will increase / decrease with the actual angle.
```
x_slider = ipywidgets.FloatSlider(min=-1.0, max=1.0, description='x')
y_slider = ipywidgets.FloatSlider(min=0, max=1.0, orientation='vertical', description='y')
steering_slider = ipywidgets.FloatSlider(min=-1.0, max=1.0, description='steering')
speed_slider = ipywidgets.FloatSlider(min=0, max=1.0, orientation='vertical', description='speed')
display(ipywidgets.HBox([y_slider, speed_slider]))
display(x_slider, steering_slider)
```
Next, we'll create a function that will get called whenever the camera's value changes. This function will do the following steps
1. Pre-process the camera image
2. Execute the neural network
3. Compute the approximate steering value
4. Control the motors using proportional / derivative control (PD)
```
angle = 0.0
angle_last = 0.0
def execute(change):
global angle, angle_last
image = change['new']
xy = model(preprocess(image)).detach().float().cpu().numpy().flatten()
x = xy[0]
y = (0.5 - xy[1]) / 2.0
x_slider.value = x
y_slider.value = y
speed_slider.value = speed_gain_slider.value
angle = np.arctan2(x, y)
pid = angle * steering_gain_slider.value + (angle - angle_last) * steering_dgain_slider.value
angle_last = angle
steering_slider.value = pid + steering_bias_slider.value
robot.left_motor.value = max(min(speed_slider.value + steering_slider.value, 1.0), 0.0)
robot.right_motor.value = max(min(speed_slider.value - steering_slider.value, 1.0), 0.0)
execute({'new': camera.value})
```
Cool! We've created our neural network execution function, but now we need to attach it to the camera for processing.
We accomplish that with the observe function.
>WARNING: This code will move the robot!! Please make sure your robot has clearance and it is on Lego or Track you have collected data on. The road follower should work, but the neural network is only as good as the data it's trained on!
```
camera.observe(execute, names='value')
```
Awesome! If your robot is plugged in it should now be generating new commands with each new camera frame.
You can now place JetBot on Lego or Track you have collected data on and see whether it can follow track.
If you want to stop this behavior, you can unattach this callback by executing the code below.
```
camera.unobserve(execute, names='value')
robot.stop()
```
### Conclusion
That's it for this live demo! Hopefully you had some fun seeing your JetBot moving smoothly on track follwing the road!!!
If your JetBot wasn't following road very well, try to spot where it fails. The beauty is that we can collect more data for these failure scenarios and the JetBot should get even better :)
|
github_jupyter
|
<h1 align='center' style="margin-bottom: 0px"> An end to end implementation of a Machine Learning pipeline </h1>
<h4 align='center' style="margin-top: 0px"> SPANDAN MADAN</h4>
<h4 align='center' style="margin-top: 0px"> Visual Computing Group, Harvard University</h4>
<h4 align='center' style="margin-top: 0px"> Computer Science and Artificial Intelligence Laboratory, MIT</h4>
<h2 align='center' style="margin-top: 0px"><a href='https://github.com/Spandan-Madan/DeepLearningProject'>Link to Github Repo</a></h2>
# Section 1. Introduction
### Background
In the fall of 2016, I was a Teaching Fellow (Harvard's version of TA) for the graduate class on "Advanced Topics in Data Science (CS209/109)" at Harvard University. I was in-charge of designing the class project given to the students, and this tutorial has been built on top of the project I designed for the class.
### Why write yet another Tutorial on Machine Learning and Deep Learning?
As a researcher on Computer Vision, I come across new blogs and tutorials on ML (Machine Learning) every day. However, most of them are just focussing on introducing the syntax and the terminology relevant to the field. For example - a 15 minute tutorial on Tensorflow using MNIST dataset, or a 10 minute intro to Deep Learning in Keras on Imagenet.
While people are able to copy paste and run the code in these tutorials and feel that working in ML is really not that hard, it doesn't help them at all in using ML for their own purposes. For example, they never introduce you to how you can run the same algorithm on your own dataset. Or, how do you get the dataset if you want to solve a problem. Or, which algorithms do you use - Conventional ML, or Deep Learning? How do you evaluate your models performance? How do you write your own model, as opposed to choosing a ready made architecture? All these form fundamental steps in any Machine Learning pipeline, and it is these steps that take most of our time as ML practitioners.
This tutorial breaks down the whole pipeline, and leads the reader through it step by step in an hope to empower you to actually use ML, and not just feel that it was not too hard. Needless to say, this will take much longer than 15-30 minutes. I believe a weekend would be a good enough estimate.
### About the Author
I am <a href="http://spandanmadan.com/">Spandan Madan</a>, a graduate student at Harvard University working on Computer Vision. My research work is supervised collaboratively by Professor Hanspeter Pfister at Harvard, and Professor Aude Oliva at MIT. My current research focusses on using Computer Vision and Natural Language Techniques in tandem to build systems capable of reasoning using text and visual elements simultaneusly.
# Section 2. Project Outline : Multi-Modal Genre Classification for Movies
## Wow, that title sounds like a handful, right? Let's break it down step by step.
### Q.1. what do we mean by Classification?
In machine learning, the task of classification means to use the available data to learn a <i>function</i> which can assign a category to a data point. For example, assign a genre to a movie, like "Romantic Comedy", "Action", "Thriller". Another example could be automatically assigning a category to news articles, like "Sports" and "Politics".
### More Formally
#### Given:
- A data point $x_i$
- A set of categories $y_1,y_2...y_n$ that $x_i$ can belong to. <br>
#### Task :
Predict the correct category $y_k$ for a new data point $x_k$ not present in the given dataset.
#### Problem :
We don't know how the $x$ and $y$ are related mathematically.
#### Assumption :
We assume there exists a function $f$ relating $x$ and $y$ i.e. $f(x_i)=y_i$
#### Approach :
Since $f$ is not known, we learn a function $g$, which approximates $f$.
#### Important consideration :
- If $f(x_i)=g(x_i)=y_i$ for all $x_i$, then the two functions $f$ and $g$ are exactly equal. Needless to say, this won't realistically ever happen, and we'll only be able to approximate the true function $f$ using $g$. This means, sometimes the prediction $g(x_i)$ will not be correct. And essentially, our whole goal is to find a $g$ which makes a really low number of such errors. That's basically all that we're trying to do.
- For the sake of completeness, I should mention that this is a specific kind of learning problem which we call "Supervised Learning". Also, the idea that $g$ approximates $f$ well for data not present in our dataset is called "Generalization". It is absolutely paramount that our model generalizes, or else all our claims will only be true about data we already have and our predictions will not be correct.
- We will look into generalization a little bit more a little ahead in the tutorial.
- Finally, There are several other kinds, but supervised learning is the most popular and well studied kind.
### Q.2. What's Multi-Modal Classification then?
In the machine learning community, the term Multi-Modal is used to refer to multiple <i>kinds</i> of data. For example, consider a YouTube video. It can be thought to contain 3 different modalities -
- The video frames (visual modality)
- The audio clip of what's being spoken (audio modality)
- Some videos also come with the transcription of the words spoken in the form of subtitles (textual modality)
Consider, that I'm interested in classifying a song on YouTube as pop or rock. You can use any of the above 3 modalities to predict the genre - The video, the song itself, or the lyrics. But, needless to say, you can predict it much better if you could use all three simultaneously. This is what we mean by multi-modal classification.
# For this project, we will be using visual and textual data to classify movie genres.
# Project Outline
- **Scraping a dataset** : The first step is to build a rich data set. We will collect textual and visual data for each movie.
- **Data pre-processing**
- **Non-deep Machine Learning models : Probabilistic and Max-Margin Classifiers.**
- **Intuitive theory behind Deep Learning**
- **Deep Models for Visual Data**
- **Deep Models for Text**
- **Potential Extensions**
- **Food for Thought**
# Section 3. Building your very own DataSet.
For any machine learning algorithm to work, it is imperative that we collect data which is "representative". Now, let's take a moment to discuss what the word representative mean.
### What data is good data? OR What do you mean by data being "representative"?
Let's look at this from first principles. Mathematically, the premise of machine learning (to be precise, the strand of machine learning we'll be working with here) is that given input variable X, and an output variable y, **IF** there is a function such that g(X)=y, then if g is unknown, we can "learn" a function f which approximates g. At the very heart, its not at all different from what you may have earlier studied as "curve fitting". For example, if you're trying to predict someone's movie preferences then X can be information about the person's gender, age, nationality and so on, while y can be the genre they most like to listen to!
Let's do a thought experiment. Consider the same example - I'm trying to predict people's movie preferences. I walk into a classroom today, and collect information about some students and their movie preferences. Now, I use that data to build a model. How well do you think I can predict my father's movie preferences? The answer is - probably not very well. Why? Intuitively, there was probably no one in the classroom who was my father's age. My model can tell me that as people go from age 18 to 30, they have a higher preference for documentaries over superhero movies. But does this trend continue at 55? Probably, they may start liking family dramas more. Probably they don't. In a nutshell, we cannot say with certainty, as our data tells us nothing about it. So, if the task was to make predictions about ANYONE's movie preferences, then the data collected from just undergraduates is NOT representative.
Now, let's see why this makes sense Mathematically. Look at the graph below.
<img src="files/contour.png">
<center>Fig.1: Plot of a function we are trying to approximate(<a href="http://www.jzy3d.org/js/slider/images/ContourPlotsDemo.png">source</a>)</center>
If we consider that the variable plotted on the vertical axis is $y$, and the values of the 2 variables on the horizontal axes make the input vector $X$, then, our hope is that we are able to find a function $f$ which can approximate the function plotted here. If all the data I collect is such that $x_1$ belongs to (80,100) and $x_2$ belongs to (80,100), the learned function will only be able to learn the "yellow-green dipping bellow" part of the function. Our function will never be able to predict the behavior in the "red" regions of the true function. So, in order to be able to learn a good function, we need data sampled from a diverse set of values of $x_1$ and x2. That would be representative data to learn this contour.
Therefore, we want to collect data which is representative of all possible movies that we want to make predictions about. Or else (which is often the case), we need to be aware of the limitations of the model we have trained, and the predictions we can make with confidence. The easiest way to do this is to only make predictions about the domain of data we collected the training data from. For example, in our case, let us start by assuming that our model will predict genres for only English movies. Now, the task is to collect data about a diverse collection of movies.
So how do we get this data then? Neither google, nor any university has released such a dataset. We want to collect visual and textual data about these movies. The simple answer is to scrape it from the internet to build our own dataset. For the purpose of this project, we will use movie posters as our visual data, and movie plots as textual data. Using these, we will build a model that can predict movie genres!
# We will be scraping data from 2 different movie sources - IMDB and TMDB
<h3>IMDB:http://www.imdb.com/</h3>
For those unaware, IMDB is the primary source of information about movies on the internet. It is immensely rich with posters, reviews, synopsis, ratings and many other information on every movie. We will use this as our primary data source.
<h3>TMDB:https://www.themoviedb.org/</h3>
TMDB, or The Movie DataBase, is an open source version of IMDB, with a free to use API that can be used to collect information. You do need an API key, but it can be obtained for free by just making a request after making a free account.
#### Note -
IMDB gives some information for free through the API, but doesn't release other information about movies. Here, we will keep it legal and only use information given to us for free and legally. However, scraping does reside on the moral fence, so to say. People often scrape data which isn't exactly publicly available for use from websites.
```
import torchvision
import urllib2
import requests
import json
import imdb
import time
import itertools
import wget
import os
import tmdbsimple as tmdb
import numpy as np
import random
import matplotlib
import matplotlib.pyplot as plt
%matplotlib inline
import seaborn as sns
import pickle
```
# Here is a broad outline of technical steps to be done for data collection
* Sign up for TMDB (themoviedatabase.org), and set up API to scrape movie posters for above movies.
* Set up and work with TMDb to get movie information from their database
* Do the same for IMDb
* Compare the entries of IMDb and TMDb for a movie
* Get a listing and information of a few movies
* Think and ponder over the potential challenges that may come our way, and think about interesting questions we can answer given the API's we have in our hands.
* Get data from the TMDb
Let's go over each one of these one by one.
## Signing up for TMDB and getting set up for getting movie metadata.
* Step 1. Head over to [tmdb.org] (https://www.themoviedb.org/?language=en) and create a new account there by signing up.
* Step 2. Click on your account icon on the top right, then from drop down menu select "Settings".
* Step 3. On the settings page, you will see the option "API" on the left pane. Click on that.
* Step 4. Apply for a new developer key. Fill out the form as required. The fields "Application Name" and "Application URL" are not important. Fill anything there.
* Step 5. It should generate a new API key for you and you should also receive a mail.
Now that you have the API key for TMDB, you can query using TMDB. Remember, it allows only 40 queries per 10 seconds.
An easy way to respect this is to just have a call to <i>time.sleep(1)</i> after each iteration. This is also being very nice to the server.
If you want to try and maximize your throughput you can embed every TMDB request in a nested try except block. If the first try fails, the second try first uses python's sleep function to give it a little rest, and then try again to make a request. Something like this -
~~~~
try:
search.movie(query=movie) #An API request
except:
try:
time.sleep(10) #sleep for a bit, to give API requests a rest.
search.movie(query=<i>movie_name</i>) #Make second API request
except:
print "Failed second attempt too, check if there's any error in request"
~~~~
## Using TMDB using the obtained API Key to get movie information
I have made these functions which make things easy. Basically, I'm making use of a library called tmdbsimple which makes TMDB using even easier. This library was installed at the time of setup.
However, if you want to avoid the library, it is also easy enough to load the API output directly into a dictionary like this without using tmdbsimple:
~~~
url = 'https://api.themoviedb.org/3/movie/1581?api_key=' + api_key
data = urllib2.urlopen(url).read()
# create dictionary from JSON
dataDict = json.loads(data)
~~~
```
# set here the path where you want the scraped folders to be saved!
poster_folder='posters_final/'
if poster_folder.split('/')[0] in os.listdir('./'):
print('Folder already exists')
else:
os.mkdir('./'+poster_folder)
poster_folder
# For the purpose of this example, i will be working with the 1999 Sci-Fi movie - "The Matrix"!
api_key = 'a237bfff7e08d0e6902c623978183be0' #Enter your own API key here to run the code below.
# Generate your own API key as explained above :)
tmdb.API_KEY = api_key #This sets the API key setting for the tmdb object
search = tmdb.Search() #this instantiates a tmdb "search" object which allows your to search for the movie
import os.path
# These functions take in a string movie name i.e. like "The Matrix" or "Interstellar"
# What they return is pretty much clear in the name - Poster, ID , Info or genre of the Movie!
def grab_poster_tmdb(movie):
response = search.movie(query=movie)
id=response['results'][0]['id']
movie = tmdb.Movies(id)
posterp=movie.info()['poster_path']
title=movie.info()['original_title']
url='image.tmdb.org/t/p/original'+posterp
title='_'.join(title.split(' '))
strcmd='wget -O '+poster_folder+title+'.jpg '+url
os.system(strcmd)
def get_movie_id_tmdb(movie):
response = search.movie(query=movie)
movie_id=response['results'][0]['id']
return movie_id
def get_movie_info_tmdb(movie):
response = search.movie(query=movie)
id=response['results'][0]['id']
movie = tmdb.Movies(id)
info=movie.info()
return info
def get_movie_genres_tmdb(movie):
response = search.movie(query=movie)
id=response['results'][0]['id']
movie = tmdb.Movies(id)
genres=movie.info()['genres']
return genres
```
While the above functions have been made to make it easy to get genres, posters and ID, all the information that can be accessed can be seen by calling the function get_movie_info() as shown below
```
print get_movie_genres_tmdb("The Matrix")
info=get_movie_info_tmdb("The Matrix")
print "All the Movie information from TMDB gets stored in a dictionary with the following keys for easy access -"
info.keys()
```
So, to get the tagline of the movie we can use the above dictionary key -
```
info=get_movie_info_tmdb("The Matrix")
print info['tagline']
```
## Getting movie information from IMDB
Now that we know how to get information from TMDB, here's how we can get information about the same movie from IMDB. This makes it possible for us to combine more information, and get a richer dataset. I urge you to try and see what dataset you can make, and go above and beyond the basic things I've done in this tutorial. Due to the differences between the two datasets, you will have to do some cleaning, however both of these datasets are extremely clean and it will be minimal.
```
# Create the IMDB object that will be used to access the IMDb's database.
imbd_object = imdb.IMDb() # by default access the web.
# Search for a movie (get a list of Movie objects).
results = imbd_object.search_movie('The Matrix')
# As this returns a list of all movies containing the word "The Matrix", we pick the first element
movie = results[0]
imbd_object.update(movie)
print "All the information we can get about this movie from IMDB-"
movie.keys()
print "The genres associated with the movie are - ",movie['genres']
```
## A small comparison of IMDB and TMDB
Now that we have both systems running, let's do a very short comparison for the same movie?
```
print "The genres for The Matrix pulled from IMDB are -",movie['genres']
print "The genres for The Matrix pulled from TMDB are -",get_movie_genres_tmdb("The Matrix")
```
As we can see, both the systems are correct, but the way they package information is different. TMDB calls it "Science Fiction" and has an ID for every genre. While IMDB calls it "Sci-Fi". Thus, it is important to keep track of these things when making use of both the datasets simultaneously.
Now that we know how to scrape information for one movie, let's take a bigger step towards scraping multiple movies?
## Working with multiple movies : Obtaining Top 20 movies from TMDB
We first instantiate an object that inherits from class Movies from TMDB. Then We use the **popular()** class method (i.e. function) to get top movies. To get more than one page of results, the optional page argument lets us see movies from any specified page number.
```
all_movies=tmdb.Movies()
top_movies=all_movies.popular()
# This is a dictionary, and to access results we use the key 'results' which returns info on 20 movies
print(len(top_movies['results']))
top20_movs=top_movies['results']
```
Let's look at one of these movies. It's the same format as above, as we had information on the movie "The Matrix", as you can see below. It's a dictionary which can be queried for specific information on that movie
```
first_movie=top20_movs[0]
print "Here is all the information you can get on this movie - "
print first_movie
print "\n\nThe title of the first movie is - ", first_movie['title']
```
Let's print out top 5 movie's titles!
```
for i in range(len(top20_movs)):
mov=top20_movs[i]
title=mov['title']
print title
if i==4:
break
```
### Yes, I know. I'm a little upset too seeing Beauty and the Beast above Logan in the list!
Moving on, we can get their genres the same way.
```
for i in range(len(top20_movs)):
mov=top20_movs[i]
genres=mov['genre_ids']
print genres
if i==4:
break
```
So, TMDB doesn't want to make your job as easy as you thought. Why these random numbers? Want to see their genre names? Well, there's the Genre() class for it. Let's get this done!
```
# Create a tmdb genre object!
genres=tmdb.Genres()
# the list() method of the Genres() class returns a listing of all genres in the form of a dictionary.
list_of_genres=genres.list()['genres']
```
Let's convert this list into a nice dictionary to look up genre names from genre IDs!
```
Genre_ID_to_name={}
for i in range(len(list_of_genres)):
genre_id=list_of_genres[i]['id']
genre_name=list_of_genres[i]['name']
Genre_ID_to_name[genre_id]=genre_name
```
Now, let's re-print the genres of top 20 movies?
```
for i in range(len(top20_movs)):
mov=top20_movs[i]
title=mov['title']
genre_ids=mov['genre_ids']
genre_names=[]
for id in genre_ids:
genre_name=Genre_ID_to_name[id]
genre_names.append(genre_name)
print title,genre_names
if i==4:
break
```
# Section 4 - Building a dataset to work with : Let's take a look at the top 1000 movies from the database
Making use of the same api as before, we will just pull results from the top 50 pages. As mentioned earlier, the "page" attribute of the command top_movies=all_movies.popular() can be used for this purpose.
Please note: Some of the code below will store the data into python "pickle" files so that it can be ready directly from memory, as opposed to being downloaded every time. Once done, you should comment out any code which generated an object that was pickled and is no longer needed.
```
all_movies=tmdb.Movies()
top_movies=all_movies.popular()
# This is a dictionary, and to access results we use the key 'results' which returns info on 20 movies
len(top_movies['results'])
top20_movs=top_movies['results']
# Comment out this cell once the data is saved into pickle file.
all_movies=tmdb.Movies()
top1000_movies=[]
print('Pulling movie list, Please wait...')
for i in range(1,51):
if i%15==0:
time.sleep(7)
movies_on_this_page=all_movies.popular(page=i)['results']
top1000_movies.extend(movies_on_this_page)
len(top1000_movies)
f3=open('movie_list.pckl','wb')
pickle.dump(top1000_movies,f3)
f3.close()
print('Done!')
f3=open('movie_list.pckl','rb')
top1000_movies=pickle.load(f3)
f3.close()
```
# Pairwise analysis of Movie Genres
As our dataset is multi label, simply looking at the distribution of genres is not sufficient. It might be beneficial to see which genres co-occur, as it might shed some light on inherent biases in our dataset. For example, it would make sense if romance and comedy occur together more often than documentary and comedy. Such inherent biases tell us that the underlying population we are sampling from itself is skewed and not balanced. We may then take steps to account for such problems. Even if we don't take such steps, it is important to be aware that we are making the assumption that an unbalanced dataset is not hurting our performance and if need be, we can come back to address this assumption. Good old scientific method, eh?
So for the top 1000 movies let's do some pairwise analysis for genre distributions. Our main purpose is to see which genres occur together in the same movie. So, we first define a function which takes a list and makes all possible pairs from it. Then, we pull the list of genres for a movie and run this function on the list of genres to get all pairs of genres which occur together
```
# This function just generates all possible pairs of movies
def list2pairs(l):
# itertools.combinations(l,2) makes all pairs of length 2 from list l.
pairs = list(itertools.combinations(l, 2))
# then the one item pairs, as duplicate pairs aren't accounted for by itertools
for i in l:
pairs.append([i,i])
return pairs
```
As mentioned, now we will pull genres for each movie, and use above function to count occurrences of when two genres occurred together
```
# get all genre lists pairs from all movies
allPairs = []
for movie in top1000_movies:
allPairs.extend(list2pairs(movie['genre_ids']))
nr_ids = np.unique(allPairs)
visGrid = np.zeros((len(nr_ids), len(nr_ids)))
for p in allPairs:
visGrid[np.argwhere(nr_ids==p[0]), np.argwhere(nr_ids==p[1])]+=1
if p[1] != p[0]:
visGrid[np.argwhere(nr_ids==p[1]), np.argwhere(nr_ids==p[0])]+=1
```
Let's take a look at the structure we just made. It is a 19X19 structure, as shown below. Also, see that we had 19 Genres. Needless to say, this structure counts the number of simultaneous occurrences of genres in same movie.
```
print visGrid.shape
print len(Genre_ID_to_name.keys())
annot_lookup = []
for i in xrange(len(nr_ids)):
annot_lookup.append(Genre_ID_to_name[nr_ids[i]])
sns.heatmap(visGrid, xticklabels=annot_lookup, yticklabels=annot_lookup)
```
The above image shows how often the genres occur together, as a heatmap
Important thing to notice in the above plot is the diagonal. The diagonal corresponds to self-pairs, i.e. number of times a genre, say Drama occurred with Drama. Which is basically just a count of the total times that genre occurred!
As we can see there are a lot of dramas in the data set, it is also a very unspecific label. There are nearly no documentaries or TV Movies. Horror is a very distinct label, and romance is also not too widely spread.
To account for this unbalanced data, there are multiple things we can try to explore what interesting relationships can be found.
## Delving Deeper into co-occurrence of genres
What we want to do now is to look for nice groups of genres that co-occur, and see if it makes sense to us logically? Intuitively speaking, wouldn't it be fun if we saw nice boxes on the above plot - boxes of high intensity i.e. genres that occur together and don't occur much with other genres. In some ways, that would isolate the co-occurrence of some genres, and heighten the co-occurrence of others.
While the data may not show that directly, we can play with the numbers to see if that's possible. The technique used for that is called biclustering.
```
from sklearn.cluster import SpectralCoclustering
model = SpectralCoclustering(n_clusters=5)
model.fit(visGrid)
fit_data = visGrid[np.argsort(model.row_labels_)]
fit_data = fit_data[:, np.argsort(model.column_labels_)]
annot_lookup_sorted = []
for i in np.argsort(model.row_labels_):
annot_lookup_sorted.append(Genre_ID_to_name[nr_ids[i]])
sns.heatmap(fit_data, xticklabels=annot_lookup_sorted, yticklabels=annot_lookup_sorted, annot=False)
plt.title("After biclustering; rearranged to show biclusters")
plt.show()
```
Looking at the above figure, "boxes" or groups of movie genres automatically emerge!
Intuitively - Crime, Sci-Fi, Mystery, Action, Horror, Drama, Thriller, etc co-occur.
AND, Romance, Fantasy, Family, Music, Adventure, etc co-occur.
That makes a lot of intuitive sense, right?
One challenge is the broad range of the drama genre. It makes the two clusters highly overlapping. If we merge it together with action thriller, etc. We will end up with nearly all movies just having that label.
**Based on playing around with the stuff above, we can sort the data into the following genre categories - "Drama, Action, ScienceFiction, exciting(thriller, crime, mystery), uplifting(adventure, fantasy, animation, comedy, romance, family), Horror, History"**
Note: that this categorization is subjective and by no means the only right solution. One could also just stay with the original labels and only exclude the ones with not enough data. Such tricks are important to balance the dataset, it allows us to increase or decrease the strength of certain signals, making it possible to improve our inferences :)
# Interesting Questions
This really should be a place for you to get creative and hopefully come up with better questions than me.
Here are some of my thoughts:
- Which actors are bound to a genre, and which can easily hop genres?
- Is there a trend in genre popularity over the years?
- Can you use sound tracks to identify the genre of a movie?
- Are top romance actors higher paid than top action actors?
- If you look at release date vs popularity score, which movie genres have a longer shelf life?
Ideas to explore specifically for feature correlations:
- Are title length correlated with movie genre?
- Are movie posters darker for horror than for romance end comedy?
- Are some genres specifically released more often at a certain time of year?
- Is the RPG rating correlated with the genre?
# Based on this new category set, we will now pull posters from TMDB as our training data!
```
# Done before, reading from pickle file now to maintain consistency of data!
# We now sample 100 movies per genre. Problem is that the sorting is by popular movies, so they will overlap.
# Need to exclude movies that were already sampled.
movies = []
baseyear = 2017
print('Starting pulling movies from TMDB. If you want to debug, uncomment the print command. This will take a while, please wait...')
done_ids=[]
for g_id in nr_ids:
#print('Pulling movies for genre ID '+g_id)
baseyear -= 1
for page in xrange(1,6,1):
time.sleep(0.5)
url = 'https://api.themoviedb.org/3/discover/movie?api_key=' + api_key
url += '&language=en-US&sort_by=popularity.desc&year=' + str(baseyear)
url += '&with_genres=' + str(g_id) + '&page=' + str(page)
data = urllib2.urlopen(url).read()
dataDict = json.loads(data)
movies.extend(dataDict["results"])
done_ids.append(str(g_id))
print("Pulled movies for genres - "+','.join(done_ids))
# f6=open("movies_for_posters",'wb')
# pickle.dump(movies,f6)
# f6.close()
f6=open("movies_for_posters",'rb')
movies=pickle.load(f6)
f6.close()
```
Let's remove any duplicates that we have in the list of movies
```
movie_ids = [m['id'] for m in movies]
print "originally we had ",len(movie_ids)," movies"
movie_ids=np.unique(movie_ids)
print len(movie_ids)
seen_before=[]
no_duplicate_movies=[]
for i in range(len(movies)):
movie=movies[i]
id=movie['id']
if id in seen_before:
continue
# print "Seen before"
else:
seen_before.append(id)
no_duplicate_movies.append(movie)
print "After removing duplicates we have ",len(no_duplicate_movies), " movies"
```
Also, let's remove movies for which we have no posters!
```
poster_movies=[]
counter=0
movies_no_poster=[]
print("Total movies : ",len(movies))
print("Started downloading posters...")
for movie in movies:
id=movie['id']
title=movie['title']
if counter==1:
print('Downloaded first. Code is working fine. Please wait, this will take quite some time...')
if counter%300==0 and counter!=0:
print "Done with ",counter," movies!"
print "Trying to get poster for ",title
try:
#grab_poster_tmdb(title)
poster_movies.append(movie)
except:
try:
time.sleep(7)
grab_poster_tmdb(title)
poster_movies.append(movie)
except:
movies_no_poster.append(movie)
counter+=1
print("Done with all the posters!")
print len(movies_no_poster)
print len(poster_movies)
# f=open('poster_movies.pckl','w')
# pickle.dump(poster_movies,f)
# f.close()
f=open('poster_movies.pckl','r')
poster_movies=pickle.load(f)
f.close()
# f=open('no_poster_movies.pckl','w')
# pickle.dump(movies_no_poster,f)
# f.close()
f=open('no_poster_movies.pckl','r')
movies_no_poster=pickle.load(f)
f.close()
```
# Congratulations, we are done scraping!
# Building a dataset out of the scraped information!
This task is simple, but **extremely** important. It's basically what will set the stage for the whole project. Given that you have the freedom to cast their own project within the framework I am providing, there are many decisions that you must make to finalize **your own version** of the project.
As we are working on a **classification** problem, we need to make two decisions given the data at hand -
* What do we want to predict, i.e. what's our Y?
* What features to use for predicting this Y, i.e. what X should we use?
There are many different options possible, and it comes down to you to decide what's most exciting. I will be picking my own version for the example, **but it is imperative that you think this through, and come up with a version which excites you!**
As an example, here are some possible ways to frame Y, while still sticking to the problem of genre prediction -
* Assume every movie can have multiple genres, and then it becomes a multi-label classification problem. For example, a movie can be Action, Horror and Adventure simultaneously. Thus, every movie can be more than one genre.
* Make clusters of genres as we did in Milestone 1 using biclustering, and then every movie can have only 1 genre. This way, the problem becomes a simpler, multi-class problem. For example, a movie could have the class - Uplifting (refer Milestone 1), or Horror or History. No movie get's more than one class.
For the purposes of this implementation, I'm going with the first case explained above - i.e. a multi-label classification problem.
Similarly, for designing our input features i.e. X, you may pick any features you think make sense, for example, the Director of a movie may be a good predictor for genre. OR, they may choose any features they design using algorithms like PCA. Given the richness of IMDB, TMDB and alternate sources like Wikipedia, there is a plethora of options available. **Be creative here!**
Another important thing to note is that in doing so, we must also make many more small implementation decisions on the way. For example, what genres are we going to include? what movies are we going to include? All these are open ended!
## My Implementation
Implementation decisions made -
* The problem is framed here as a multi-label problem explained above.
* We will try to predict multiple genres associated with a movie. This will be our Y.
* We will use 2 different kinds of X - text and images.
* For the text part - Input features being used to predict the genre is a form of the movie's plot available from TMDB using the property 'overview'. This will be our X.
* For the image part - we will use the scraped poster images as our X.
NOTE : We will first look at some conventional machine learning models, which were popular before the recent rise of neural networks and deep learning. For the poster image to genre prediction, I have avoided using this for the reason that conventional ML models are simply not used anymore without using deep learning for feature extraction (all discussed in detail ahead, don't be scared by the jargon). For the movie overview to genre prediction problem we will look at both conventional models and deep learning models.
Now, let's build our X and Y!
First, let's identify movies that have overviews. **Next few steps are going to be a good example on why data cleaning is important!**
```
movies_with_overviews=[]
for i in range(len(no_duplicate_movies)):
movie=no_duplicate_movies[i]
id=movie['id']
overview=movie['overview']
if len(overview)==0:
continue
else:
movies_with_overviews.append(movie)
len(movies_with_overviews)
```
Now let's store the genre's for these movies in a list that we will later transform into a binarized vector.
Binarized vector representation is a very common and important way data is stored/represented in ML. Essentially, it's a way to reduce a categorical variable with n possible values to n binary indicator variables. What does that mean? For example, let [(1,3),(4)] be the list saying that sample A has two labels 1 and 3, and sample B has one label 4. For every sample, for every possible label, the representation is simply 1 if it has that label, and 0 if it doesn't have that label. So the binarized version of the above list will be -
~~~~~
[(1,0,1,0]),
(0,0,0,1])]
~~~~~
```
# genres=np.zeros((len(top1000_movies),3))
genres=[]
all_ids=[]
for i in range(len(movies_with_overviews)):
movie=movies_with_overviews[i]
id=movie['id']
genre_ids=movie['genre_ids']
genres.append(genre_ids)
all_ids.extend(genre_ids)
from sklearn.preprocessing import MultiLabelBinarizer
mlb=MultiLabelBinarizer()
Y=mlb.fit_transform(genres)
genres[1]
print Y.shape
print np.sum(Y, axis=0)
len(list_of_genres)
```
This is interesting. We started with only 19 genre labels if you remember. But the shape for Y is 1666,20 while it should be 1666,19 as there are only 19 genres? Let's explore.
Let's find genre IDs that are not present in our original list of genres!
```
# Create a tmdb genre object!
genres=tmdb.Genres()
# the list() method of the Genres() class returns a listing of all genres in the form of a dictionary.
list_of_genres=genres.list()['genres']
Genre_ID_to_name={}
for i in range(len(list_of_genres)):
genre_id=list_of_genres[i]['id']
genre_name=list_of_genres[i]['name']
Genre_ID_to_name[genre_id]=genre_name
for i in set(all_ids):
if i not in Genre_ID_to_name.keys():
print i
```
Well, this genre ID wasn't given to us by TMDB when we asked it for all possible genres. How do we go about this now? We can either neglect all samples that have this genre. But if you look up you'll see there's too many of these samples. So, I googled more and went into their documentation and found that this ID corresponds to the genre "Foreign". So, we add it to the dictionary of genre names ourselves. Such problems are ubiquitous in machine learning, and it is up to us to diagnose and correct them. We must always make a decision about what to keep, how to store data and so on.
```
Genre_ID_to_name[10769]="Foreign" #Adding it to the dictionary
len(Genre_ID_to_name.keys())
```
Now, we turn to building the X matrix i.e. the input features! As described earlier, we will be using the overview of movies as our input vector! Let's look at a movie's overview for example!
```
sample_movie=movies_with_overviews[5]
sample_overview=sample_movie['overview']
sample_title=sample_movie['title']
print "The overview for the movie",sample_title," is - \n\n"
print sample_overview
```
## So, how do we store this movie overview in a matrix?
#### Do we just store the whole string? We know that we need to work with numbers, but this is all text. What do we do?!
The way we will be storing the X matrix is called a "Bag of words" representation. The basic idea of this representation in our context is that we can think of all the distinct words that are possible in the movies' reviews as a distinct object. And then every movie overview can be thought as a "Bag" containing a bunch of these possible objects.
For example, in the case of Zootopia the movie above - The "Bag" contains the words ("Determined", "to", "prove", "herself"......"the", "mystery"). We make such lists for all movie overviews. Finally, we binarize again like we did above for Y. scikit-learn makes our job easy here by simply using a function CountVectorizer() because this representation is so often used in Machine Learning.
What this means is that, for all the movies that we have the data on, we will first count all the unique words. Say, there's 30,000 unique words. Then we can represent every movie overview as a 30000x1 vector, where each position in the vector corresponds to the presence or absence of a particular word. If the word corresponding to that position is present in the overview, that position will have 1, otherwise it will be 0.
Ex - if our vocabular was 4 words - "I","am","a","good","boy", then the representation for the sentence "I am a boy" would be [1 1 1 0 1], and for the sentence "I am good" would be [1 1 0 1 0].
```
from sklearn.feature_extraction.text import CountVectorizer
import re
content=[]
for i in range(len(movies_with_overviews)):
movie=movies_with_overviews[i]
id=movie['id']
overview=movie['overview']
overview=overview.replace(',','')
overview=overview.replace('.','')
content.append(overview)
print content[0]
print len(content)
```
# Are all words equally important?
#### At the cost of sounding "Animal Farm" inspired, I would say not all words are equally important.
For example, let's consider the overview for the Matrix -
```
get_movie_info_tmdb('The Matrix')['overview']
```
For "The Matrix" a word like "computer" is a stronger indicators of it being a Sci-Fi movie, than words like "who" or "powerful" or "vast". One way computer scientists working with natural language tackled this problem in the past (and it is still used very popularly) is what we call TF-IDF i.e. Term Frequence, Inverse Document Frequency. The basic idea here is that words that are strongly indicative of the content of a single document (every movie overview is a document in our case) are words that occur very frequently in that document, and very infrequently in all other documents. For example, "Computer" occurs twice here but probably will not in most other movie overviews. Hence, it is indicative. On the other hand, generic words like "a","and","the" will occur very often in all documents. Hence, they are not indicative.
So, can we use this information to reduce our insanely high 30,000 dimensional vector representation to a smaller, more handle-able number? But first up, why should we even care? The answer is probably one of the most used phrases in ML - "The Curse of Dimensionality".
# The Curse of Dimensionality
#### This section is strongly borrowing from one of the greatest <a href="https://homes.cs.washington.edu/~pedrod/papers/cacm12.pdf">ML papers I've ever read.</a>
This expression was coined by Bellman in 1961 to refer to the fact that many algorithms that work fine in low dimensions become intractable when the input is high-dimensional. The reason for them not working in high dimensions is very strongly linked to what we discussed earlier - having a representative dataset. Consider this, you have a function $f$ dependent only one dependent variable $x$, and $x$ can only integer values from 1 to 100. Since it's one dimensional, it can be plotted on a line. To get a representative sample, you'd need to sample something like - $f(1),f(20),f(40),f(60),f(80),f(100)$
Now, let's increase the dimensionality i.e. number of dependent variables and see what happens. Say, we have 2 variables $x_1$ and $x_2$, same possible as before - integers between 1 and 100. Now, instead of a line, we'll have a plane with $x_1$ and $x_2$ on the two axes. The interesting bit is that instead of 100 possible values of dependent variables like before, we now have 100,000 possible values! Basically, we can make 100x100 table of possible values of $x_1$ and $x_2$. Wow, that increased exponentially. Not just figuratively, but mathematically exponentially. Needless to say, to cover 5% of the space like we did before, we'd need to sample $f$ at 5000 values.
For 3 variables, it would be 100,000,000, and we'd need to sample at 500,000 points. That's already more than the number of data points we have for most training problems we will ever come across.
Basically, as the dimensionality (number of features) of the examples grows, because a fixed-size training set covers a dwindling fraction of the input space. Even with a moderate dimension of 100 and a huge training set of a trillion examples, the latter covers only a fraction of about $10^{−18}$ of the input space. This is what makes machine learning
both necessary and hard.
So, yes, if some words are unimportant, we want to get rid of them and reduce the dimensionality of our X matrix. And the way we will do it is using TF-IDF to identify un-important words. Python let's us do this with just one line of code (And this is why you should spend more time reading maths, than coding!)
```
# The min_df paramter makes sure we exclude words that only occur very rarely
# The default also is to exclude any words that occur in every movie description
vectorize=CountVectorizer(max_df=0.95, min_df=0.005)
X=vectorize.fit_transform(content)
```
We are excluding all words that occur in too many or too few documents, as these are very unlikely to be discriminative. Words that only occur in one document most probably are names, and words that occur in nearly all documents are probably stop words. Note that the values here were not tuned using a validation set. They are just guesses. It is ok to do, because we didn't evaluate the performance of these parameters. In a strict case, for example for a publication, it would be better to tune these as well.
```
X.shape
```
So, each movie's overview gets represented by a 1x1365 dimensional vector.
Now, we are ready for the kill. Our data is cleaned, hypothesis is set (Overview can predict movie genre), and the feature/output vectors are prepped. Let's train some models!
```
import pickle
f4=open('X.pckl','wb')
f5=open('Y.pckl','wb')
pickle.dump(X,f4)
pickle.dump(Y,f5)
f6=open('Genredict.pckl','wb')
pickle.dump(Genre_ID_to_name,f6)
f4.close()
f5.close()
f6.close()
```
# Congratulations, we have our data set ready!
A note : As we are building our own dataset, and I didn't want you to spend all your time waiting for poster image downloads to finish, I am working with an EXTREMELY small dataset. That is why, the results we will see for the deep learning portion will not be spectacular as compared to conventional machine learning methods. If you want to see the real power, you should spend some more time scraping something of the order of 100,000 images, as opposed to 1000 odd like I am doing here. Quoting the paper I mentioned above - MORE DATA BEATS A CLEVERER ALGORITHM.
#### As the TA, I saw that most teams working on the project had data of the order of 100,000 movies. So, if you want to extract the power of these models, consider scraping a larger dataset than me.
# Section 5 - Non-deep, Conventional ML models with above data
Here is a layout of what we will be doing -
- We will implement two different models
- We will decide a performance metric i.e. a quantitative method to be sure about how well difference models are doing.
- Discussion of the differences between the models, their strengths, weaknesses, etc.
As discussed earlier, there are a LOT of implementation decisions to be made. Between feature engineering, hyper-parameter tuning, model selection and how interpretable do you want your model to be (Read : Bayesian vs Non-Bayesian approaches) a lot is to be decided. For example, some of these models could be:
- Generalized Linear Models
- SVM
- Shallow (1 Layer, i.e. not deep) Neural Network
- Random Forest
- Boosting
- Decision Tree
Or go more bayesian:
- Naive Bayes
- Linear or Quadratic Discriminant Analysis
- Bayesian Hierarchical models
The list is endless, and not all models will make sense for the kind of problem you have framed for yourself. ** Think about which model best fits for your purpose.**
For our purposes here, I will be showing the example of 2 very simple models, one picked from each category above -
1. SVM
2. Multinomial Naive Bayes
A quick overview of the whole pipeline coming below:
- A little bit of feature engineering
- 2 different Models
- Evaluation Metrics chosen
- Model comparisons
### Let's start with some feature engineering.
Engineering the right features depends on 2 key ideas. Firstly, what is it that you are trying to solve? For example, if you want to guess my music preferences and you try to train a super awesome model while giving it what my height is as input features, you're going to have no luck. On the other hand, giving it my Spotify playlist will solve the problem with any model. So, CONTEXT of the problem plays a role.
Second, you can only represent based on the data at hand. Meaning, if you didn't have access to my Spotify playlist, but to my Facebook statuses - You know all my statuses about Harvard may not be useful. But if you represent me as my Facebook statuses which are YouTube links, that would also solve the problem. So, AVAILABILITY OF DATA at hand is the second factor.
#### A nice way to think of it is to think that you start with the problem at hand, but design features constrained by the data you have available. If you have many independent features that each correlate well with the class, learning is easy. On the other hand, if the class is a very complex function of the features, you may not be able to learn it.
In the context of this problem, we would like to predict the genre of a movie. what we have access to - movie overviews, which are text descriptions of the movie plot. The hypothesis makes sense, overview is a short description of the story and the story is clearly important in assigning genres to movies.
So, let's improve our features by playing with the words in the overviews in our data. One interesting way to go back to what we discussed earlier - TF-IDF. We originally used it to filter words, but we can also assign the tf-idf values as "importance" values to words, as opposed to treating them all equally. Tf-idf simply tries to identify the assign a weightage to each word in the bag of words.
Once again, the way it works is - Most movie descriptions have the word "The" in it. Obviously, it doesn't tell you anything special about it. So weightage should be inversely proportional to how many movies have the word in their description. This is the IDF part.
On the other hand, for the movie interstellar, if the description has the word Space 5 times, and wormhole 2 times, then it's probably more about Space than about wormhole. Thus, space should have a high weightage. This is the TF part.
We simply use TF-IDf to assign weightage to every word in the bag of words. Which makes sense, right? :)
```
from sklearn.feature_extraction.text import TfidfTransformer
tfidf_transformer = TfidfTransformer()
X_tfidf = tfidf_transformer.fit_transform(X)
X_tfidf.shape
```
Let's divide our X and Y matrices into train and test split. We train the model on the train split, and report the performance on the test split. Think of this like the questions you do in the problem sets v/s the exam. Of course, they are both (assumed to be) from the same population of questions. And doing well on Problem Sets is a good indicator that you'll do well in exams, but really, you must test before you can make any claims about you knowing the subject.
```
msk = np.random.rand(X_tfidf.shape[0]) < 0.8
X_train_tfidf=X_tfidf[msk]
X_test_tfidf=X_tfidf[~msk]
Y_train=Y[msk]
Y_test=Y[~msk]
positions=range(len(movies_with_overviews))
# print positions
test_movies=np.asarray(positions)[~msk]
# test_movies
from sklearn.multiclass import OneVsRestClassifier
from sklearn.svm import SVC
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import f1_score
from sklearn.metrics import make_scorer
from sklearn.metrics import classification_report
parameters = {'kernel':['linear'], 'C':[0.01, 0.1, 1.0]}
gridCV = GridSearchCV(SVC(class_weight='balanced'), parameters, scoring=make_scorer(f1_score, average='micro'))
classif = OneVsRestClassifier(gridCV)
classif.fit(X_train_tfidf, Y_train)
predstfidf=classif.predict(X_test_tfidf)
print classification_report(Y_test, predstfidf)
```
As you can see, the performance is by and large poorer for movies which are less represented like War and animation, and better for categories like Drama.
Numbers aside, let's look at our model's predictions for a small sample of movies from our test set.
```
genre_list=sorted(list(Genre_ID_to_name.keys()))
predictions=[]
for i in range(X_test_tfidf.shape[0]):
pred_genres=[]
movie_label_scores=predstfidf[i]
# print movie_label_scores
for j in range(19):
#print j
if movie_label_scores[j]!=0:
genre=Genre_ID_to_name[genre_list[j]]
pred_genres.append(genre)
predictions.append(pred_genres)
import pickle
f=open('classifer_svc','wb')
pickle.dump(classif,f)
f.close()
for i in range(X_test_tfidf.shape[0]):
if i%50==0 and i!=0:
print 'MOVIE: ',movies_with_overviews[i]['title'],'\tPREDICTION: ',','.join(predictions[i])
```
Let's try our second model? The naive bayes model.
```
from sklearn.naive_bayes import MultinomialNB
classifnb = OneVsRestClassifier(MultinomialNB())
classifnb.fit(X[msk].toarray(), Y_train)
predsnb=classifnb.predict(X[~msk].toarray())
import pickle
f2=open('classifer_nb','wb')
pickle.dump(classifnb,f2)
f2.close()
predictionsnb=[]
for i in range(X_test_tfidf.shape[0]):
pred_genres=[]
movie_label_scores=predsnb[i]
for j in range(19):
#print j
if movie_label_scores[j]!=0:
genre=Genre_ID_to_name[genre_list[j]]
pred_genres.append(genre)
predictionsnb.append(pred_genres)
for i in range(X_test_tfidf.shape[0]):
if i%50==0 and i!=0:
print 'MOVIE: ',movies_with_overviews[i]['title'],'\tPREDICTION: ',','.join(predictionsnb[i])
```
As can be seen above, the results seem promising, but how do we really compare the two models? We need to quantify our performance so that we can say which one's better. Takes us back to what we discussed right in the beginning - we're learning a function $g$ which can approximate the original unknown function $f$. For some values of $x_i$, the predictions will be wrong for sure, and we want to minimize it.
For multi label systems, we often keep track of performance using "Precision" and "Recall". These are standard metrics, and you can google to read up more about them if you're new to these terms.
# Evaluation Metrics
We will use the standard precision recall metrics for evaluating our system.
```
def precision_recall(gt,preds):
TP=0
FP=0
FN=0
for t in gt:
if t in preds:
TP+=1
else:
FN+=1
for p in preds:
if p not in gt:
FP+=1
if TP+FP==0:
precision=0
else:
precision=TP/float(TP+FP)
if TP+FN==0:
recall=0
else:
recall=TP/float(TP+FN)
return precision,recall
precs=[]
recs=[]
for i in range(len(test_movies)):
if i%1==0:
pos=test_movies[i]
test_movie=movies_with_overviews[pos]
gtids=test_movie['genre_ids']
gt=[]
for g in gtids:
g_name=Genre_ID_to_name[g]
gt.append(g_name)
# print predictions[i],movies_with_overviews[i]['title'],gt
a,b=precision_recall(gt,predictions[i])
precs.append(a)
recs.append(b)
print np.mean(np.asarray(precs)),np.mean(np.asarray(recs))
precs=[]
recs=[]
for i in range(len(test_movies)):
if i%1==0:
pos=test_movies[i]
test_movie=movies_with_overviews[pos]
gtids=test_movie['genre_ids']
gt=[]
for g in gtids:
g_name=Genre_ID_to_name[g]
gt.append(g_name)
# print predictions[i],movies_with_overviews[i]['title'],gt
a,b=precision_recall(gt,predictionsnb[i])
precs.append(a)
recs.append(b)
print np.mean(np.asarray(precs)),np.mean(np.asarray(recs))
```
The average precision and recall scores for our samples are pretty good! Models seem to be working! Also, we can see that the Naive Bayes performs outperforms SVM. **I strongly suggest you to go read about Multinomial Bayes and think about why it works so well for "Document Classification", which is very similar to our case as every movie overview can be thought of as a document we are assigning labels to.**
# Section 6 - Deep Learning : an intuitive overview
The above results were good, but it's time to bring out the big guns. So first and foremost, let's get a very short idea about what's deep learning. This is for peope who don't have background in this - it's high level and gives just the intuition.
As described above, the two most immportant concepts in doing good classification (or regression) are to 1) use the right representation which captures the right information about the data which is relevant to the problem at hand 2) Using the right model which has the capability of making sense of the representation fed to it.
While for the second part we have complicated and powerful models that we have studied at length, we don't seem to have a principled, mathematical way of doing the first part - i.e. representation. What we did above was to see "What makes sense", and go from there. That is not a good approach for complex data/ complex problems. Is there some way to automate this? Deep Learning, does just this.
To just emphasize the importance of representation in the complex tasks we usually attempt with Deep Learning, let me talk about the original problem which made it famous. The paper is often reffered to as the "Imagenet Challenge Paper", and it was basically working on object recognition in images. Let's try to think about an algorithm that tries to detect a chair.
## If I ask you to "Define" a chair, how would you? - Something with 4 legs?
<img src="files/chair1.png" height="400" width="400">
<h3><center>All are chairs, none with 4 legs. (Pic Credit: Zoya Bylinskii)</center></h3>
## How about some surface that we sit on then?
<img src="files/chair2.png" height="400" width="400">
<h3><center>All are surfaces we sit on, none are chairs. (Pic Credit: Zoya Bylinskii)</center></h3>
Clearly, these definitions won't work and we need something more complicated. Sadly, we can't come up with a simple text rule that our computer can search for! And we take a more principled approach.
The "Deep" in the deep learning comes from the fact that it was conventionally applied to Neural Networks. Neural Networks, as we all know, are structures organized in layers. Layers of computations. Why do we need layers? Because these layers can be seen as sub-tasks that we do in the complicated task of identifying a chair. It can be thought as a heirarchical break down of a complicated job into smalled sub-tasks.
Mathematically, each layer acts like a space transformation which takes the pixel values to a high dimensional space. When we start out, every pixel in the image is given equal importance in our matrix. With each layer, convolution operations give some parts more importance, and some lesser importance. In doing so, we transform our images to a space in which similar looking objects/object parts are closer (We are basically learning this space transformation in deep learning, nothing else)
What exactly was learnt by these neural networks is hard to know, and an active area of research. But one very crude way to visualize what it does is to think like - It starts by learning very generic features in the first layer. Something as simple as vertical and horizontal lines. In the next layer, it learns that if you combine the vectors representing vertical and horizontal vectors in different ratios, you can make all possible slanted lines. Next layer learns to combine lines to form curves - Say, something like the outline of a face. These curves come together to form 3D objects. And so on. Building sub-modules, combining them in the right way which can give it semantics.
**So, in a nutshell, the first few layers of a "Deep" network learn the right representation of the data, given the problem (which is mathematically described by your objective function trying to minimize difference between ground truth and predicted labels). The last layer simply looks how close or far apart things are in this high dimensional space.**
Hence, we can give any kind of data a high dimensional representation using neural networks. Below we will see high dimensional representations of both words in overviews (text) and posters (image). Let's get started with the posters i.e. extracting visual features from posters using deep learning.
# Section 7 - Deep Learning for predicting genre from poster
Once again, we must make an implementation decision. This time, it has more to do with how much time are we willing to spend in return for added accuracy. We are going to use here a technique that is commonly referred to as Pre-Training in Machine Learning Literature.
Instead of me trying to re-invent the wheel here, I am going to borrow this short section on pre-training from Stanford University's lecture on <a href='http://cs231n.github.io/transfer-learning/'> CNN's</a>. To quote -
''In practice, very few people train an entire Convolutional Network from scratch (with random initialization), because it is relatively rare to have a dataset of sufficient size. Instead, it is common to pretrain a ConvNet on a very large dataset (e.g. ImageNet, which contains 1.2 million images with 1000 categories), and then use the ConvNet either as an initialization or a fixed feature extractor for the task of interest. ''
There are three broad ways in which transfer learning or pre-training can be done. (The 2 concepts are different and to understand the difference clearly, I suggest you read the linked lecture thoroughly). The way we are going to about it is by using a pre-trained, released ConvNet as feature extractor. Take a ConvNet pretrained on ImageNet (a popular object detection dataset), remove the last fully-connected layer. After removing the last layer, what we have is just another neural network i.e. a stack of space tranformations. But, originally the output of this stack can be pumped into a single layer which can classify the image into categories like Car, Dog, Cat and so on.
What this means, is that in the space this stack transforms the images to, all images which contain a "dog" are closer to each other, and all images containing a "cat" are closer. Thus, it is a meaningful space where images with similar objects are closer.
Think about it, now if we pump our posters through this stack, it will embed them in a space where posters which contain similar objects are closer. This is a very meaningful feature engineering method! While this may not be ideal for genre prediction, it might be quite meaningful. For example, all posters with a gun or a car are probably action. While a smiling couple would point to romance or drama. The alternative would be to train the CNN from scratch which is fairly computationally intensive and involves a lot of tricks to get the CNN training to converge to the optimal space tranformation.
This way, we can start off with something strong, and then build on top. We pump our images through the pre-trained network to extract the visual features from the posters. Then, using these features as descriptors for the image, and genres as the labels, we train a simpler neural network from scratch which learns to do simply classification on this dataset. These 2 steps are exactly what we are going to do for predicting genres from movie posters.
## Deep Learning to extract visual features from posters
The basic problem here we are answering is that can we use the posters to predict genre. First check - Does this hypothesis make sense? Yes. Because that's what graphic designers do for a living. They leave visual cues to semantics. They make sure that when we look at the poster of a horror movie, we know it's not a happy image. Things like that. Can our deep learning system infer such subtleties? Let's find out!
For Visual features, either we can train a deep neural network ourselves from scratch, or we can use a pre-trained one made available to us from the Visual Geometry Group at Oxford University, one of the most popular methods. This is called the VGG-net. Or as they call it, we will extract the VGG features of an image. Mathematically, as mentioned, it's just a space transformation in the form of layers. So, we simply need to perform this chain of transformations on our image, right? Keras is a library that makes it very easy for us to do this. Some other common ones are Tensorflow and PyTorch. While the latter two are very powerful and customizable and used more often in practice, Keras makes it easy to prototype by keeping the syntax simple.
We will be working with Keras to keep things simple in code, so that we can spend more time understanding and less time coding. Some common ways people refer to this step are - "Getting the VGG features of an image", or "Forward Propogating the image through VGG and chopping off the last layer". In keras, this is as easy as writing 4 lines.
```
# Loading the list of movies we had downloaded posters for eariler -
f=open('poster_movies.pckl','r')
poster_movies=pickle.load(f)
f.close()
from keras.applications.vgg16 import VGG16
from keras.preprocessing import image
from keras.applications.vgg16 import preprocess_input
import numpy as np
import pickle
model = VGG16(weights='imagenet', include_top=False)
allnames=os.listdir(poster_folder)
imnames=[j for j in allnames if j.endswith('.jpg')]
feature_list=[]
genre_list=[]
file_order=[]
print "Starting extracting VGG features for scraped images. This will take time, Please be patient..."
print "Total images = ",len(imnames)
failed_files=[]
succesful_files=[]
i=0
for mov in poster_movies:
i+=1
mov_name=mov['original_title']
mov_name1=mov_name.replace(':','/')
poster_name=mov_name.replace(' ','_')+'.jpg'
if poster_name in imnames:
img_path=poster_folder+poster_name
#try:
img = image.load_img(img_path, target_size=(224, 224))
succesful_files.append(poster_name)
x = image.img_to_array(img)
x = np.expand_dims(x, axis=0)
x = preprocess_input(x)
features = model.predict(x)
print features.shape
printe model.predict(x)
file_order.append(img_path)
feature_list.append(features)
genre_list.append(mov['genre_ids'])
if np.max(np.asarray(feature_list))==0.0:
print('problematic',i)
if i%250==0 or i==1:
print "Working on Image : ",i
# except:
# failed_files.append(poster_name)
# continue
else:
continue
print "Done with all features, please pickle for future use!"
len(genre_list)
len(feature_list)
print type(feature_list[0])
feature_list[0].shape
# Reading from pickle below, this code is not to be run.
list_pickled=(feature_list,file_order,failed_files,succesful_files,genre_list)
f=open('posters_new_features.pckl','wb')
pickle.dump(list_pickled,f)
f.close()
print("Features dumped to pickle file")
f7=open('posters_new_features.pckl','rb')
list_pickled=pickle.load(f7)
f7.close()
# (feature_list2,file_order2)=list_pickled
```
### Training a simple neural network model using these VGG features.
```
(feature_list,files,failed,succesful,genre_list)=list_pickled
```
Let's first get the labels on our 1342 samples first! As image download fails on a few instances, the best way to work with the right model is to read the poster names downloaded, and working from there. These posters cannot be uploaded to Github as they are too large, and so are being downloaded and read from my local computer. If you do re-do it, you might have to check and edit the paths in the code to make sure it runs.
```
(a,b,c,d)=feature_list[0].shape
feature_size=a*b*c*d
feature_size
```
This looks odd, why are we re-running the loop we ran above again below? The reason is simple, the most important thing to know about numpy is that using vstack() and hstack() are highly sub-optimal. Numpy arrays when created, a fixed size is allocated in the memory and when we stack, a new one is copied and created in a new location. This makes the code really, really slow. The best way to do it (and this remains the same with MATLAB matrices if you work with them), is to create a numpy array of zeros, and over-write it row by row. The above code was just to see what size numpy array we will need!
The final movie poster set for which we have all the information we need, is 1265 movies. In the above code we are making an X numpy array containing the visual features of one image per row. So, the VGG features are reshaped to be in the shape (1,25088) and we finally obtain a matrix of shape (1265,25088)
```
np_features=np.zeros((len(feature_list),feature_size))
for i in range(len(feature_list)):
feat=feature_list[i]
reshaped_feat=feat.reshape(1,-1)
np_features[i]=reshaped_feat
# np_features[-1]
X=np_features
from sklearn.preprocessing import MultiLabelBinarizer
mlb=MultiLabelBinarizer()
Y=mlb.fit_transform(genre_list)
Y.shape
```
Our binarized Y numpy array contains the binarized labels corresponding to the genre IDs of the 1277 movies
```
visual_problem_data=(X,Y)
f8=open('visual_problem_data_clean.pckl','wb')
pickle.dump(visual_problem_data,f8)
f8.close()
f8=open('visual_problem_data_clean.pckl','rb')
visual_features=pickle.load(f8)
f8.close()
(X,Y)=visual_features
X.shape
mask = np.random.rand(len(X)) < 0.8
X_train=X[mask]
X_test=X[~mask]
Y_train=Y[mask]
Y_test=Y[~mask]
X_test.shape
Y_test.shape
```
Now, we create our own keras neural network to use the VGG features and then classify movie genres. Keras makes this super easy.
Neural network architectures have gotten complex over the years. But the simplest ones contain very standard computations organized in layers, as described above. Given the popularity of some of these, Keras makes it as easy as writing out the names of these operations in a sequential order. This way you can make a network while completely avoiding the Mathematics (HIGHLY RECOMMENDED SPENDING MORE TIME ON THE MATH THOUGH)
Sequential() allows us to make models the follow this sequential order of layers. Different kinds of layers like Dense, Conv2D etc can be used, and many activation functions like RELU, Linear etc are also available.
# Important Question : Why do we need activation functions?
#### Copy pasting the answer I wrote for this question on <a href='https://www.quora.com/Why-do-neural-networks-need-an-activation-function/answer/Spandan-Madan?srid=5ydm'>Quora</a> Feel free to leave comments there.
""Sometimes, we tend to get lost in the jargon and confuse things easily, so the best way to go about this is getting back to our basics.
Don’t forget what the original premise of machine learning (and thus deep learning) is - IF the input and output are related by a function y=f(x), then if we have x, there is no way to exactly know f unless we know the process itself. However, machine learning gives you the ability to approximate f with a function g, and the process of trying out multiple candidates to identify the function g best approximating f is called machine learning.
Ok, that was machine learning, and how is deep learning different? Deep learning simply tries to expand the possible kind of functions that can be approximated using the above mentioned machine learning paradigm. Roughly speaking, if the previous model could learn say 10,000 kinds of functions, now it will be able to learn say 100,000 kinds (in actuality both are infinite spaces but one is larger than the other, because maths is cool that ways.)
If you want to know the mathematics of it, go read about VC dimension and how more layers in a network affect it. But I will avoid the mathematics here and rely on your intuition to believe me when I say that not all data can be classified correctly into categories using a linear function. So, we need our deep learning model to be able to approximate more complex functions than just a linear function.
Now, let’s come to your non linearity bit. Imagine a linear function y=2x+3, and another one y=4x+7. What happens if I pool them and take an average? I get another linear function y= 3x+5. So instead of doing those two computations separately and then averaging it out, I could have just used the single linear function y=3x+5. Obviously, this logic holds good if I have more than 2 such linear functions. This is exactly what will happen if you don’t have have non-linearities in your nodes, and also what others have written in their answers.
It simply follows from the definition of a linear function -
(i) If you take two linear functions, AND
(ii)Take a linear combination of them (which is how we combine the outputs of multiple nodes of a network)
You are BOUND to get a linear function because f(x)+g(x)=mx+b+nx+c=(m+n)x+(b+c)= say h(x).
And you could in essence replace your whole network by a simple matrix transformation which accounts for all linear combinations and up/downsamplings.
In a nutshell, you’ll only be trying to learn a linear approximation for original function f relating the input and the output. Which as we discussed above, is not always the best approximation. Adding non-linearities ensures that you can learn more complex functions by approximating every non-linear function as a LINEAR combination of a large number of non-linear functions.
Still new to the field, so if there’s something wrong here please comment below! Hope it helps""
#### Let's train our model then, using the features we extracted from VGG net
The model we will use has just 1 hidden layer between the VGG features and the final output layer. The simplest neural network you can get. An image goes into this network with the dimensions (1,25088), the first layer's output is 1024 dimensional. This hidden layer output undergoes a pointwise RELU activation. This output gets transformed into the output layer of 20 dimensions. It goes through a sigmoid.
The sigmoid, or the squashing function as it is often called, is a function which squashes numbers between 0 and 1. What are you reminded of when you think of numebers between 0 and 1? Right, probability.
By squashing the score of each of the 20 output labels between 0 and 1, sigmoid lets us interpret their scores as probabilities. Then, we can just pick the classes with the top 3 or 5 probability scores as the predicted genres for the movie poster! Simple!
```
# Y_train[115]
from keras.models import Sequential
from keras.layers import Dense, Activation
from keras import optimizers
model_visual = Sequential([
Dense(1024, input_shape=(25088,)),
Activation('relu'),
Dense(256),
Activation('relu'),
Dense(19),
Activation('sigmoid'),
])
opt = optimizers.rmsprop(lr=0.0001, decay=1e-6)
#sgd = optimizers.SGD(lr=0.05, decay=1e-6, momentum=0.4, nesterov=False)
model_visual.compile(optimizer=opt,
loss='binary_crossentropy',
metrics=['accuracy'])
```
We train the model using the fit() function. The parameters it takes are - training features and training labels, epochs, batch_size and verbose.
Simplest one - verbose. 0="dont print anything as you work", 1="Inform me as you go".
Often the data set is too large to be loaded into the RAM. So, we load data in batches. For batch_size=32 and epochs=10, the model starts loading rows from X in batches of 32 everytime it calculates the loss and updates the model. It keeps on going till it has covered all the samples 10 times.
So, the no. of times model is updated = (Total Samples/Batch Size) * (Epochs)
```
model_visual.fit(X_train, Y_train, epochs=10, batch_size=64,verbose=1)
model_visual.fit(X_train, Y_train, epochs=50, batch_size=64,verbose=0)
```
For the first 10 epochs I trained the model in a verbose fashion to show you what's happening. After that, in the below cell you can see I turned off the verbosity to keep the code cleaner.
```
Y_preds=model_visual.predict(X_test)
sum(sum(Y_preds))
```
### Let's look at some of our predictions?
```
f6=open('Genredict.pckl','rb')
Genre_ID_to_name=pickle.load(f6)
f6.close()
sum(Y_preds[1])
sum(Y_preds[2])
genre_list=sorted(list(Genre_ID_to_name.keys()))
precs=[]
recs=[]
for i in range(len(Y_preds)):
row=Y_preds[i]
gt_genres=Y_test[i]
gt_genre_names=[]
for j in range(19):
if gt_genres[j]==1:
gt_genre_names.append(Genre_ID_to_name[genre_list[j]])
top_3=np.argsort(row)[-3:]
predicted_genres=[]
for genre in top_3:
predicted_genres.append(Genre_ID_to_name[genre_list[genre]])
(precision,recall)=precision_recall(gt_genre_names,predicted_genres)
precs.append(precision)
recs.append(recall)
if i%50==0:
print "Predicted: ",','.join(predicted_genres)," Actual: ",','.join(gt_genre_names)
print np.mean(np.asarray(precs)),np.mean(np.asarray(recs))
```
So, even with just the poster i.e. visual features we are able to make great predictions! Sure, text outperforms the visual features, but the important thing is that it still works. In more complicated models, we can combine the two to make even better predictions. That is precisely what I work on in my research.
These models were trained on CPU's, and a simple 1 layer model was used to show that there is a lot of information in this data that the models can extract. With a larger dataset, and more training I was able to bring these numbers to as high as 70%, which is the similar to textual features. Some teams in my class outperformed this even more. More data is the first thing you should try if you want better results. Then, you can start playing with training on GPUs, learning rate schedules and other hyperparameters. Finally, you can consider using ResNet, a much more powerful neural network model than VGG. All of these can be tried once you have a working knowledge of machine learning.
# Section 8 - Deep Learning to get Textual Features
Let's do the same thing as above with text now?
We will use an off the shelf representation for words - Word2Vec model. Just like VGGnet before, this is a model made available to get a meaningful representation. As the total number of words is small, we don't even need to forward propagate our sample through a network. Even that has been done for us, and the result is stored in the form of a dictionary. We can simply look up the word in the dictionary and get the Word2Vec features for the word.
You can download the dictionary from here - https://drive.google.com/file/d/0B7XkCwpI5KDYNlNUTTlSS21pQmM/edit <br>
Download it to the directory of this tutorial i.e. in the same folder as this ipython notebook.
```
from gensim import models
# model2 = models.Word2Vec.load_word2vec_format('GoogleNews-vectors-negative300.bin', binary=True)
model2 = models.KeyedVectors.load_word2vec_format('GoogleNews-vectors-negative300.bin', binary=True)
```
Now, we can simply look up for a word in the above loaded model. For example, to get the Word2Vec representation of the word "King" we just do - model2['king']
```
print model2['king'].shape
print model2['dog'].shape
```
This way, we can represent the words in our overviews using this word2vec model. And then, we can use that as our X representations. So, instead of count of words, we are using a representation which is based on the semantic representation of the word. Mathematically, each word went from 3-4 dimensional (the length) to 300 dimensions!
For the same set of movies above, let's try and predict the genres from the deep representation of their overviews!
```
final_movies_set = movies_with_overviews
len(final_movies_set)
from nltk.tokenize import RegexpTokenizer
from stop_words import get_stop_words
tokenizer = RegexpTokenizer(r'\w+')
# create English stop words list
en_stop = get_stop_words('en')
movie_mean_wordvec=np.zeros((len(final_movies_set),300))
movie_mean_wordvec.shape
```
Text needs some pre-processing before we can train the model. The only preprocessing we do here is - we delete commonly occurring words which we know are not informative about the genre. Think of it as the clutter in some sense. These words are often removed and are referred to as "stop words". You can look them up online. These include simple words like "a", "and", "but", "how", "or" and so on. They can be easily removed using the python package NLTK.
From the above dataset, movies with overviews which contain only stop words, or movies with overviews containing no words with word2vec representation are neglected. Others are used to build our Mean word2vec representation. Simply, put for every movie overview -
* Take movie overview
* Throw out stop words
* For non stop words:
- If in word2vec - take it's word2vec representation which is 300 dimensional
- If not - throw word
* For each movie, calculate the arithmetic mean of the 300 dimensional vector representations for all words in the overview which weren't thrown out
This mean becomes the 300 dimensional representation for the movie. For all movies, these are stored in a numpy array. So the X matrix becomes (1263,300). And, Y is (1263,20) i.e. binarized 20 genres, as before
**Why do we take the arithmetic mean?**
If you feel that we should have kept all the words separately - Then you're thinking correct, but sadly we're limited by the way current day neural networks work. I will not mull over this for the fear of stressing too much on an otherwise irrelevant detail. But if you're interested, read this awesome paper -
https://jiajunwu.com/papers/dmil_cvpr.pdf
```
genres=[]
rows_to_delete=[]
for i in range(len(final_movies_set)):
mov=final_movies_set[i]
movie_genres=mov['genre_ids']
genres.append(movie_genres)
overview=mov['overview']
tokens = tokenizer.tokenize(overview)
stopped_tokens = [k for k in tokens if not k in en_stop]
count_in_vocab=0
s=0
if len(stopped_tokens)==0:
rows_to_delete.append(i)
genres.pop(-1)
# print overview
# print "sample ",i,"had no nonstops"
else:
for tok in stopped_tokens:
if tok.lower() in model2.vocab:
count_in_vocab+=1
s+=model2[tok.lower()]
if count_in_vocab!=0:
movie_mean_wordvec[i]=s/float(count_in_vocab)
else:
rows_to_delete.append(i)
genres.pop(-1)
# print overview
# print "sample ",i,"had no word2vec"
len(genres)
mask2=[]
for row in range(len(movie_mean_wordvec)):
if row in rows_to_delete:
mask2.append(False)
else:
mask2.append(True)
X=movie_mean_wordvec[mask2]
X.shape
Y=mlb.fit_transform(genres)
Y.shape
textual_features=(X,Y)
f9=open('textual_features.pckl','wb')
pickle.dump(textual_features,f9)
f9.close()
# textual_features=(X,Y)
f9=open('textual_features.pckl','rb')
textual_features=pickle.load(f9)
f9.close()
(X,Y)=textual_features
X.shape
Y.shape
mask_text=np.random.rand(len(X))<0.8
X_train=X[mask_text]
Y_train=Y[mask_text]
X_test=X[~mask_text]
Y_test=Y[~mask_text]
```
Once again, we use a very similar, super simple architecture as before.
```
from keras.models import Sequential
from keras.layers import Dense, Activation
model_textual = Sequential([
Dense(300, input_shape=(300,)),
Activation('relu'),
Dense(19),
Activation('softmax'),
])
model_textual.compile(optimizer='rmsprop',
loss='binary_crossentropy',
metrics=['accuracy'])
model_textual.fit(X_train, Y_train, epochs=10, batch_size=500)
model_textual.fit(X_train, Y_train, epochs=10000, batch_size=500,verbose=0)
score = model_textual.evaluate(X_test, Y_test, batch_size=249)
print("%s: %.2f%%" % (model_textual.metrics_names[1], score[1]*100))
Y_preds=model_textual.predict(X_test)
genre_list.append(10769)
print "Our predictions for the movies are - \n"
precs=[]
recs=[]
for i in range(len(Y_preds)):
row=Y_preds[i]
gt_genres=Y_test[i]
gt_genre_names=[]
for j in range(19):
if gt_genres[j]==1:
gt_genre_names.append(Genre_ID_to_name[genre_list[j]])
top_3=np.argsort(row)[-3:]
predicted_genres=[]
for genre in top_3:
predicted_genres.append(Genre_ID_to_name[genre_list[genre]])
(precision,recall)=precision_recall(gt_genre_names,predicted_genres)
precs.append(precision)
recs.append(recall)
if i%50==0:
print "Predicted: ",predicted_genres," Actual: ",gt_genre_names
print np.mean(np.asarray(precs)),np.mean(np.asarray(recs))
```
Even without much tuning of the above model, these results are able to beat our previous results.
Note - I got accuracies as high as 78% when doing classification using plots scraped from Wikipedia. The large amount of information was very suitable for movie genre classification with a deep model. Strongly suggest you to try playing around with architectures.
# Section 9 - Upcoming Tutorials and Acknowledgements
Congrats! This is the end of our pilot project! Needless to say, a lot of the above content may be new to you, or may be things that you know very well. If it's the former, I hope this tutorial would have helped you. If it is the latter and you think I wrote something incorrect or that my understanding can be improved, feel free to create a github issue so that I can correct it!
Writing tutorials can take a lot of time, but it is a great learning experience. I am currently working on a tutorial focussing on word embeddings, which will explore word2vec and other word embeddings in detail. While it will take some time to be up, I will post a link to it's repository on the README for this project so that interested readers can find it.
I would like to thank a few of my friends who had an indispensible role to play in me making this tutorial. Firstly, Professor Hanspeter Pfister and Verena Kaynig at Harvard, who helped guide this tutorial/project and scope it. Secondly, my friends Sahil Loomba and Matthew Tancik for their suggestions and editing the material and the presentation of the storyline. Thirdly, Zoya Bylinskii at MIT for constantly motivating me to put in my effort into this tutorial. Finally, all others who helped me feel confident enough to take up this task and to see it till the end. Thanks all of you!
|
github_jupyter
|
# 광학 인식

흔히 볼 수 있는 Computer Vision 과제는 이미지에서 텍스트를 감지하고 해석하는 것입니다. 이러한 종류의 처리를 종종 *OCR(광학 인식)*이라고 합니다.
## Computer Vision 서비스를 사용하여 이미지에서 텍스트 읽기
**Computer Vision** Cognitive Service는 다음을 비롯한 OCR 작업을 지원합니다.
- 여러 언어로 된 텍스트를 읽는 데 사용할 수 있는 **OCR** API. 이 API는 동기식으로 사용할 수 있으며, 이미지에서 소량의 텍스트를 감지하고 읽어야 할 때 잘 작동합니다.
- 더 큰 문서에 최적화된 **Read** API. 이 API는 비동기식으로 사용되며, 인쇄 텍스트와 필기 텍스트 모두에 사용할 수 있습니다.
이 서비스는 **Computer Vision** 리소스 또는 **Cognitive Services** 리소스를 만들어서 사용할 수 있습니다.
아직 만들지 않았다면 Azure 구독에서 **Cognitive Services** 리소스를 만듭니다.
> **참고**: 이미 Cognitive Services 리소스를 보유하고 있다면 Azure Portal에서 **빠른 시작** 페이지를 열고 키 및 엔드포인트를 아래의 셀로 복사하기만 하면 됩니다. 리소스가 없다면 아래의 단계를 따라 리소스를 만듭니다.
1. 다른 브라우저 탭에서 Azure Portal(https://portal.azure.com) 을 열고 Microsoft 계정으로 로그인합니다.
2. **+리소스 만들기** 단추를 클릭하고, *Cognitive Services*를 검색하고, 다음 설정을 사용하여 **Cognitive Services** 리소스를 만듭니다.
- **구독**: *사용자의 Azure 구독*.
- **리소스 그룹**: *고유한 이름의 새 리소스 그룹을 선택하거나 만듭니다*.
- **지역**: *사용 가능한 지역을 선택합니다*.
- **이름**: *고유한 이름을 입력합니다*.
- **가격 책정 계층**: S0
- **알림을 읽고 이해했음을 확인합니다**. 선택됨.
3. 배포가 완료될 때까지 기다립니다. 그런 다음에 Cognitive Services 리소스로 이동하고, **개요** 페이지에서 링크를 클릭하여 서비스의 키를 관리합니다. 클라이언트 애플리케이션에서 Cognitive Services 리소스에 연결하려면 엔드포인트 및 키가 필요합니다.
### Cognitive Services 리소스의 키 및 엔드포인트 가져오기
Cognitive Services 리소스를 사용하려면 클라이언트 애플리케이션에 해당 엔드포인트 및 인증 키가 필요합니다.
1. Azure Portal에 있는 Cognitive Service 리소스의 **키 및 엔드포인트** 페이지에서 리소스의 **Key1**을 복사하고 아래 코드에 붙여 넣어 **YOUR_COG_KEY**를 대체합니다.
2. 리소스의 **엔드포인트**를 복사하고 아래 코드에 붙여 넣어 **YOUR_COG_ENDPOINT**를 대체합니다.
3. **셀 실행**(▷) 단추(셀 왼쪽에 있음)를 클릭하여 아래의 셀에 있는 코드를 실행합니다.
```
cog_key = 'YOUR_COG_KEY'
cog_endpoint = 'YOUR_COG_ENDPOINT'
print('Ready to use cognitive services at {} using key {}'.format(cog_endpoint, cog_key))
```
이제 키와 엔드포인트를 설정했으므로 Computer Vision 서비스 리소스를 사용하여 이미지에서 텍스트를 추출할 수 있습니다.
먼저, 이미지를 동기식으로 분석하고 포함된 텍스트를 읽을 수 있게 해주는 **OCR** API부터 시작하겠습니다. 이 경우에는 일부 텍스트를 포함하고 있는 가상의 Northwind Traders 소매업체에 대한 광고 이미지가 있습니다. 아래의 셀을 실행하여 읽어 보세요.
```
from azure.cognitiveservices.vision.computervision import ComputerVisionClient
from msrest.authentication import CognitiveServicesCredentials
import matplotlib.pyplot as plt
from PIL import Image, ImageDraw
import os
%matplotlib inline
# Get a client for the computer vision service
computervision_client = ComputerVisionClient(cog_endpoint, CognitiveServicesCredentials(cog_key))
# Read the image file
image_path = os.path.join('data', 'ocr', 'advert.jpg')
image_stream = open(image_path, "rb")
# Use the Computer Vision service to find text in the image
read_results = computervision_client.recognize_printed_text_in_stream(image_stream)
# Process the text line by line
for region in read_results.regions:
for line in region.lines:
# Read the words in the line of text
line_text = ''
for word in line.words:
line_text += word.text + ' '
print(line_text.rstrip())
# Open image to display it.
fig = plt.figure(figsize=(7, 7))
img = Image.open(image_path)
draw = ImageDraw.Draw(img)
plt.axis('off')
plt.imshow(img)
```
이미지에 있는 텍스트는 영역, 줄, 단어의 계층 구조로 구성되어 있으며 코드는 이 항목들을 읽어서 결과를 검색합니다.
이미지 위에서 읽은 텍스트를 결과에서 봅니다.
## 경계 상자 표시
텍스트 줄의 *경계 상자* 좌표와 이미지에서 발견된 개별 단어도 결과에 포함되어 있습니다. 아래의 셀을 실행하여 위에서 검색한 광고 이미지에서 텍스트 줄의 경계 상자를 확인하세요.
```
# Open image to display it.
fig = plt.figure(figsize=(7, 7))
img = Image.open(image_path)
draw = ImageDraw.Draw(img)
# Process the text line by line
for region in read_results.regions:
for line in region.lines:
# Show the position of the line of text
l,t,w,h = list(map(int, line.bounding_box.split(',')))
draw.rectangle(((l,t), (l+w, t+h)), outline='magenta', width=5)
# Read the words in the line of text
line_text = ''
for word in line.words:
line_text += word.text + ' '
print(line_text.rstrip())
# Show the image with the text locations highlighted
plt.axis('off')
plt.imshow(img)
```
결과에서 각 텍스트 줄의 경계 상자는 이미지에 직사각형으로 표시됩니다.
## Read API 사용
이전에 사용한 OCR API는 소량의 텍스트가 있는 이미지에서 잘 작동합니다. 스캔한 문서와 같이 더 큰 텍스트 본문을 읽어야 할 때는 **Read** API를 사용할 수 있습니다. 이를 위해서는 다단계 프로세스가 필요합니다.
1. 비동기식으로 읽고 분석할 이미지를 Computer Vision 서비스에 제출합니다.
2. 분석 작업이 완료될 때까지 기다립니다.
3. 분석의 결과를 검색합니다.
이 프로세스를 사용하여 스캔한 서신의 텍스트를 Northwind Traders 매장 관리자에게 읽어 주려면 다음 셀을 실행하세요.
```
from azure.cognitiveservices.vision.computervision import ComputerVisionClient
from azure.cognitiveservices.vision.computervision.models import OperationStatusCodes
from msrest.authentication import CognitiveServicesCredentials
import matplotlib.pyplot as plt
from PIL import Image
import time
import os
%matplotlib inline
# Read the image file
image_path = os.path.join('data', 'ocr', 'letter.jpg')
image_stream = open(image_path, "rb")
# Get a client for the computer vision service
computervision_client = ComputerVisionClient(cog_endpoint, CognitiveServicesCredentials(cog_key))
# Submit a request to read printed text in the image and get the operation ID
read_operation = computervision_client.read_in_stream(image_stream,
raw=True)
operation_location = read_operation.headers["Operation-Location"]
operation_id = operation_location.split("/")[-1]
# Wait for the asynchronous operation to complete
while True:
read_results = computervision_client.get_read_result(operation_id)
if read_results.status not in [OperationStatusCodes.running]:
break
time.sleep(1)
# If the operation was successfuly, process the text line by line
if read_results.status == OperationStatusCodes.succeeded:
for result in read_results.analyze_result.read_results:
for line in result.lines:
print(line.text)
# Open image and display it.
print('\n')
fig = plt.figure(figsize=(12,12))
img = Image.open(image_path)
plt.axis('off')
plt.imshow(img)
```
결과를 검토합니다. 서신의 전체 필사본이 있는데, 대부분은 인쇄된 텍스트이고 필기 서명이 있습니다. 서신의 원본 이미지는 OCR 결과 아래에 표시됩니다(보기 위해 스크롤해야 할 수도 있음).
## 필기 텍스트 읽기
이전 예에서 이미지 분석 요청은 *인쇄된* 텍스트에 맞춰 작업을 최적화하는 텍스트 인식 모드를 지정했습니다. 그럼에도 불구하고 필기 서명이 읽혔습니다.
필기 텍스트를 읽을 수 있는 이 능력은 매우 유용합니다. 예를 들어 쇼핑 목록이 포함된 메모를 작성했는데 폰의 앱을 사용하여 메모를 읽고 그 안에 포함된 텍스트를 필사하기를 원한다고 가정해 보세요.
아래 셀을 실행하여 필기 쇼핑 목록에 대한 읽기 작업의 예를 확인해 보세요.
```
from azure.cognitiveservices.vision.computervision import ComputerVisionClient
from azure.cognitiveservices.vision.computervision.models import OperationStatusCodes
from msrest.authentication import CognitiveServicesCredentials
import matplotlib.pyplot as plt
from PIL import Image
import time
import os
%matplotlib inline
# Read the image file
image_path = os.path.join('data', 'ocr', 'note.jpg')
image_stream = open(image_path, "rb")
# Get a client for the computer vision service
computervision_client = ComputerVisionClient(cog_endpoint, CognitiveServicesCredentials(cog_key))
# Submit a request to read printed text in the image and get the operation ID
read_operation = computervision_client.read_in_stream(image_stream,
raw=True)
operation_location = read_operation.headers["Operation-Location"]
operation_id = operation_location.split("/")[-1]
# Wait for the asynchronous operation to complete
while True:
read_results = computervision_client.get_read_result(operation_id)
if read_results.status not in [OperationStatusCodes.running]:
break
time.sleep(1)
# If the operation was successfuly, process the text line by line
if read_results.status == OperationStatusCodes.succeeded:
for result in read_results.analyze_result.read_results:
for line in result.lines:
print(line.text)
# Open image and display it.
print('\n')
fig = plt.figure(figsize=(12,12))
img = Image.open(image_path)
plt.axis('off')
plt.imshow(img)
```
## 추가 정보
OCR에 Computer Vision 서비스를 사용하는 방법에 대한 자세한 내용은 [Computer Vision 설명서](https://docs.microsoft.com/ko-kr/azure/cognitive-services/computer-vision/concept-recognizing-text)를 참조하세요.
|
github_jupyter
|
# The Atoms of Computation
Programming a quantum computer is now something that anyone can do in the comfort of their own home.
But what to create? What is a quantum program anyway? In fact, what is a quantum computer?
These questions can be answered by making comparisons to standard digital computers. Unfortunately, most people don’t actually understand how digital computers work either. In this article, we’ll look at the basics principles behind these devices. To help us transition over to quantum computing later on, we’ll do it using the same tools as we'll use for quantum.
## Contents
1. [Splitting information into bits](#bits)
2. [Computation as a Diagram](#diagram)
3. [Your First Quantum Circuit](#first-circuit)
4. [Example: Adder Circuit](#adder)
4.1 [Encoding an Input](#encoding)
4.2 [Remembering how to Add](#remembering-add)
4.3 [Adding with Qiskit](#adding-qiskit)
Below is some Python code we'll need to run if we want to use the code in this page:
```
from qiskit import QuantumCircuit, assemble, Aer
from qiskit.visualization import plot_histogram
```
## 1. Splitting information into bits <a id="bits"></a>
The first thing we need to know about is the idea of bits. These are designed to be the world’s simplest alphabet. With only two characters, 0 and 1, we can represent any piece of information.
One example is numbers. You are probably used to representing a number through a string of the ten digits 0, 1, 2, 3, 4, 5, 6, 7, 8, and 9. In this string of digits, each digit represents how many times the number contains a certain power of ten. For example, when we write 9213, we mean
$$ 9000 + 200 + 10 + 3 $$
or, expressed in a way that emphasizes the powers of ten
$$ (9\times10^3) + (2\times10^2) + (1\times10^1) + (3\times10^0) $$
Though we usually use this system based on the number 10, we can just as easily use one based on any other number. The binary number system, for example, is based on the number two. This means using the two characters 0 and 1 to express numbers as multiples of powers of two. For example, 9213 becomes 10001111111101, since
$$ 9213 = (1 \times 2^{13}) + (0 \times 2^{12}) + (0 \times 2^{11})+ (0 \times 2^{10}) +(1 \times 2^9) + (1 \times 2^8) + (1 \times 2^7) \\\\ \,\,\, + (1 \times 2^6) + (1 \times 2^5) + (1 \times 2^4) + (1 \times 2^3) + (1 \times 2^2) + (0 \times 2^1) + (1 \times 2^0) $$
In this we are expressing numbers as multiples of 2, 4, 8, 16, 32, etc. instead of 10, 100, 1000, etc.
<a id="binary_widget"></a>
```
from qiskit_textbook.widgets import binary_widget
binary_widget(nbits=5)
```
These strings of bits, known as binary strings, can be used to represent more than just numbers. For example, there is a way to represent any text using bits. For any letter, number, or punctuation mark you want to use, you can find a corresponding string of at most eight bits using [this table](https://www.ibm.com/support/knowledgecenter/en/ssw_aix_72/com.ibm.aix.networkcomm/conversion_table.htm). Though these are quite arbitrary, this is a widely agreed-upon standard. In fact, it's what was used to transmit this article to you through the internet.
This is how all information is represented in computers. Whether numbers, letters, images, or sound, it all exists in the form of binary strings.
Like our standard digital computers, quantum computers are based on this same basic idea. The main difference is that they use *qubits*, an extension of the bit to quantum mechanics. In the rest of this textbook, we will explore what qubits are, what they can do, and how they do it. In this section, however, we are not talking about quantum at all. So, we just use qubits as if they were bits.
### Quick Exercises
1. Think of a number and try to write it down in binary.
2. If you have $n$ bits, how many different states can they be in?
## 2. Computation as a diagram <a id="diagram"></a>
Whether we are using qubits or bits, we need to manipulate them in order to turn the inputs we have into the outputs we need. For the simplest programs with very few bits, it is useful to represent this process in a diagram known as a *circuit diagram*. These have inputs on the left, outputs on the right, and operations represented by arcane symbols in between. These operations are called 'gates', mostly for historical reasons.
Here's an example of what a circuit looks like for standard, bit-based computers. You aren't expected to understand what it does. It should simply give you an idea of what these circuits look like.

For quantum computers, we use the same basic idea but have different conventions for how to represent inputs, outputs, and the symbols used for operations. Here is the quantum circuit that represents the same process as above.

In the rest of this section, we will explain how to build circuits. At the end, you'll know how to create the circuit above, what it does, and why it is useful.
## 3. Your first quantum circuit <a id="first-circuit"></a>
In a circuit, we typically need to do three jobs: First, encode the input, then do some actual computation, and finally extract an output. For your first quantum circuit, we'll focus on the last of these jobs. We start by creating a circuit with eight qubits and eight outputs.
```
n = 8
n_q = n
n_b = n
qc_output = QuantumCircuit(n_q,n_b)
```
This circuit, which we have called `qc_output`, is created by Qiskit using `QuantumCircuit`. The number `n_q` defines the number of qubits in the circuit. With `n_b` we define the number of output bits we will extract from the circuit at the end.
The extraction of outputs in a quantum circuit is done using an operation called `measure`. Each measurement tells a specific qubit to give an output to a specific output bit. The following code adds a `measure` operation to each of our eight qubits. The qubits and bits are both labelled by the numbers from 0 to 7 (because that’s how programmers like to do things). The command `qc_output.measure(j,j)` adds a measurement to our circuit `qc_output` that tells qubit `j` to write an output to bit `j`.
```
for j in range(n):
qc_output.measure(j,j)
```
Now that our circuit has something in it, let's take a look at it.
```
qc_output.draw()
```
Qubits are always initialized to give the output ```0```. Since we don't do anything to our qubits in the circuit above, this is exactly the result we'll get when we measure them. We can see this by running the circuit many times and plotting the results in a histogram. We will find that the result is always ```00000000```: a ```0``` from each qubit.
```
sim = Aer.get_backend('aer_simulator') # this is the simulator we'll use
qobj = assemble(qc_output) # this turns the circuit into an object our backend can run
result = sim.run(qobj).result() # we run the experiment and get the result from that experiment
# from the results, we get a dictionary containing the number of times (counts)
# each result appeared
counts = result.get_counts()
# and display it on a histogram
plot_histogram(counts)
```
The reason for running many times and showing the result as a histogram is because quantum computers may have some randomness in their results. In this case, since we aren’t doing anything quantum, we get just the ```00000000``` result with certainty.
Note that this result comes from a quantum simulator, which is a standard computer calculating what an ideal quantum computer would do. Simulations are only possible for small numbers of qubits (~30 qubits), but they are nevertheless a very useful tool when designing your first quantum circuits. To run on a real device you simply need to replace ```Aer.get_backend('aer_simulator')``` with the backend object of the device you want to use.
## 4. Example: Creating an Adder Circuit <a id="adder"></a>
### 4.1 Encoding an input <a id="encoding"></a>
Now let's look at how to encode a different binary string as an input. For this, we need what is known as a NOT gate. This is the most basic operation that you can do in a computer. It simply flips the bit value: ```0``` becomes ```1``` and ```1``` becomes ```0```. For qubits, it is an operation called ```x``` that does the job of the NOT.
Below we create a new circuit dedicated to the job of encoding and call it `qc_encode`. For now, we only specify the number of qubits.
```
qc_encode = QuantumCircuit(n)
qc_encode.x(7)
qc_encode.draw()
```
Extracting results can be done using the circuit we have from before: `qc_output`. Adding the two circuits using `qc_encode + qc_output` creates a new circuit with everything needed to extract an output added at the end.
```
qc = qc_encode + qc_output
qc.draw()
```
Now we can run the combined circuit and look at the results.
```
qobj = assemble(qc)
counts = sim.run(qobj).result().get_counts()
plot_histogram(counts)
```
Now our computer outputs the string ```10000000``` instead.
The bit we flipped, which comes from qubit 7, lives on the far left of the string. This is because Qiskit numbers the bits in a string from right to left. Some prefer to number their bits the other way around, but Qiskit's system certainly has its advantages when we are using the bits to represent numbers. Specifically, it means that qubit 7 is telling us about how many $2^7$s we have in our number. So by flipping this bit, we’ve now written the number 128 in our simple 8-bit computer.
Now try out writing another number for yourself. You could do your age, for example. Just use a search engine to find out what the number looks like in binary (if it includes a ‘0b’, just ignore it), and then add some 0s to the left side if you are younger than 64.
```
qc_encode = QuantumCircuit(n)
qc_encode.x(1)
qc_encode.x(5)
qc_encode.draw()
```
Now we know how to encode information in a computer. The next step is to process it: To take an input that we have encoded, and turn it into an output that we need.
### 4.2 Remembering how to add <a id="remembering-add"></a>
To look at turning inputs into outputs, we need a problem to solve. Let’s do some basic maths. In primary school, you will have learned how to take large mathematical problems and break them down into manageable pieces. For example, how would you go about solving the following?
```
9213
+ 1854
= ????
```
One way is to do it digit by digit, from right to left. So we start with 3+4
```
9213
+ 1854
= ???7
```
And then 1+5
```
9213
+ 1854
= ??67
```
Then we have 2+8=10. Since this is a two digit answer, we need to carry the one over to the next column.
```
9213
+ 1854
= ?067
¹
```
Finally we have 9+1+1=11, and get our answer
```
9213
+ 1854
= 11067
¹
```
This may just be simple addition, but it demonstrates the principles behind all algorithms. Whether the algorithm is designed to solve mathematical problems or process text or images, we always break big tasks down into small and simple steps.
To run on a computer, algorithms need to be compiled down to the smallest and simplest steps possible. To see what these look like, let’s do the above addition problem again but in binary.
```
10001111111101
+ 00011100111110
= ??????????????
```
Note that the second number has a bunch of extra 0s on the left. This just serves to make the two strings the same length.
Our first task is to do the 1+0 for the column on the right. In binary, as in any number system, the answer is 1. We get the same result for the 0+1 of the second column.
```
10001111111101
+ 00011100111110
= ????????????11
```
Next, we have 1+1. As you’ll surely be aware, 1+1=2. In binary, the number 2 is written ```10```, and so requires two bits. This means that we need to carry the 1, just as we would for the number 10 in decimal.
```
10001111111101
+ 00011100111110
= ???????????011
¹
```
The next column now requires us to calculate ```1+1+1```. This means adding three numbers together, so things are getting complicated for our computer. But we can still compile it down to simpler operations, and do it in a way that only ever requires us to add two bits together. For this, we can start with just the first two 1s.
```
1
+ 1
= 10
```
Now we need to add this ```10``` to the final ```1``` , which can be done using our usual method of going through the columns.
```
10
+ 01
= 11
```
The final answer is ```11``` (also known as 3).
Now we can get back to the rest of the problem. With the answer of ```11```, we have another carry bit.
```
10001111111101
+ 00011100111110
= ??????????1011
¹¹
```
So now we have another 1+1+1 to do. But we already know how to do that, so it’s not a big deal.
In fact, everything left so far is something we already know how to do. This is because, if you break everything down into adding just two bits, there are only four possible things you’ll ever need to calculate. Here are the four basic sums (we’ll write all the answers with two bits to be consistent).
```
0+0 = 00 (in decimal, this is 0+0=0)
0+1 = 01 (in decimal, this is 0+1=1)
1+0 = 01 (in decimal, this is 1+0=1)
1+1 = 10 (in decimal, this is 1+1=2)
```
This is called a *half adder*. If our computer can implement this, and if it can chain many of them together, it can add anything.
### 4.3 Adding with Qiskit <a id="adding-qiskit"></a>
Let's make our own half adder using Qiskit. This will include a part of the circuit that encodes the input, a part that executes the algorithm, and a part that extracts the result. The first part will need to be changed whenever we want to use a new input, but the rest will always remain the same.

The two bits we want to add are encoded in the qubits 0 and 1. The above example encodes a ```1``` in both these qubits, and so it seeks to find the solution of ```1+1```. The result will be a string of two bits, which we will read out from the qubits 2 and 3. All that remains is to fill in the actual program, which lives in the blank space in the middle.
The dashed lines in the image are just to distinguish the different parts of the circuit (although they can have more interesting uses too). They are made by using the `barrier` command.
The basic operations of computing are known as logic gates. We’ve already used the NOT gate, but this is not enough to make our half adder. We could only use it to manually write out the answers. Since we want the computer to do the actual computing for us, we’ll need some more powerful gates.
To see what we need, let’s take another look at what our half adder needs to do.
```
0+0 = 00
0+1 = 01
1+0 = 01
1+1 = 10
```
The rightmost bit in all four of these answers is completely determined by whether the two bits we are adding are the same or different. So for ```0+0``` and ```1+1```, where the two bits are equal, the rightmost bit of the answer comes out ```0```. For ```0+1``` and ```1+0```, where we are adding different bit values, the rightmost bit is ```1```.
To get this part of our solution correct, we need something that can figure out whether two bits are different or not. Traditionally, in the study of digital computation, this is called an XOR gate.
| Input 1 | Input 2 | XOR Output |
|:-------:|:-------:|:------:|
| 0 | 0 | 0 |
| 0 | 1 | 1 |
| 1 | 0 | 1 |
| 1 | 1 | 0 |
In quantum computers, the job of the XOR gate is done by the controlled-NOT gate. Since that's quite a long name, we usually just call it the CNOT. In Qiskit its name is ```cx```, which is even shorter. In circuit diagrams, it is drawn as in the image below.
```
qc_cnot = QuantumCircuit(2)
qc_cnot.cx(0,1)
qc_cnot.draw()
```
This is applied to a pair of qubits. One acts as the control qubit (this is the one with the little dot). The other acts as the *target qubit* (with the big circle).
There are multiple ways to explain the effect of the CNOT. One is to say that it looks at its two input bits to see whether they are the same or different. Next, it overwrites the target qubit with the answer. The target becomes ```0``` if they are the same, and ```1``` if they are different.
<img src="images/cnot_xor.svg">
Another way of explaining the CNOT is to say that it does a NOT on the target if the control is ```1```, and does nothing otherwise. This explanation is just as valid as the previous one (in fact, it’s the one that gives the gate its name).
Try the CNOT out for yourself by trying each of the possible inputs. For example, here's a circuit that tests the CNOT with the input ```01```.
```
qc = QuantumCircuit(2,2)
qc.x(0)
qc.cx(0,1)
qc.measure(0,0)
qc.measure(1,1)
qc.draw()
```
If you execute this circuit, you’ll find that the output is ```11```. We can think of this happening because of either of the following reasons.
- The CNOT calculates whether the input values are different and finds that they are, which means that it wants to output ```1```. It does this by writing over the state of qubit 1 (which, remember, is on the left of the bit string), turning ```01``` into ```11```.
- The CNOT sees that qubit 0 is in state ```1```, and so applies a NOT to qubit 1. This flips the ```0``` of qubit 1 into a ```1```, and so turns ```01``` into ```11```.
Here is a table showing all the possible inputs and corresponding outputs of the CNOT gate:
| Input (q1 q0) | Output (q1 q0) |
|:-------------:|:--------------:|
| 00 | 00 |
| 01 | 11 |
| 10 | 10 |
| 11 | 01 |
For our half adder, we don’t want to overwrite one of our inputs. Instead, we want to write the result on a different pair of qubits. For this, we can use two CNOTs.
```
qc_ha = QuantumCircuit(4,2)
# encode inputs in qubits 0 and 1
qc_ha.x(0) # For a=0, remove this line. For a=1, leave it.
qc_ha.x(1) # For b=0, remove this line. For b=1, leave it.
qc_ha.barrier()
# use cnots to write the XOR of the inputs on qubit 2
qc_ha.cx(0,2)
qc_ha.cx(1,2)
qc_ha.barrier()
# extract outputs
qc_ha.measure(2,0) # extract XOR value
qc_ha.measure(3,1)
qc_ha.draw()
```
We are now halfway to a fully working half adder. We just have the other bit of the output left to do: the one that will live on qubit 3.
If you look again at the four possible sums, you’ll notice that there is only one case for which this is ```1``` instead of ```0```: ```1+1```=```10```. It happens only when both the bits we are adding are ```1```.
To calculate this part of the output, we could just get our computer to look at whether both of the inputs are ```1```. If they are — and only if they are — we need to do a NOT gate on qubit 3. That will flip it to the required value of ```1``` for this case only, giving us the output we need.
For this, we need a new gate: like a CNOT but controlled on two qubits instead of just one. This will perform a NOT on the target qubit only when both controls are in state ```1```. This new gate is called the *Toffoli*. For those of you who are familiar with Boolean logic gates, it is basically an AND gate.
In Qiskit, the Toffoli is represented with the `ccx` command.
```
qc_ha = QuantumCircuit(4,2)
# encode inputs in qubits 0 and 1
qc_ha.x(0) # For a=0, remove the this line. For a=1, leave it.
qc_ha.x(1) # For b=0, remove the this line. For b=1, leave it.
qc_ha.barrier()
# use cnots to write the XOR of the inputs on qubit 2
qc_ha.cx(0,2)
qc_ha.cx(1,2)
# use ccx to write the AND of the inputs on qubit 3
qc_ha.ccx(0,1,3)
qc_ha.barrier()
# extract outputs
qc_ha.measure(2,0) # extract XOR value
qc_ha.measure(3,1) # extract AND value
qc_ha.draw()
```
In this example, we are calculating ```1+1```, because the two input bits are both ```1```. Let's see what we get.
```
qobj = assemble(qc_ha)
counts = sim.run(qobj).result().get_counts()
plot_histogram(counts)
```
The result is ```10```, which is the binary representation of the number 2. We have built a computer that can solve the famous mathematical problem of 1+1!
Now you can try it out with the other three possible inputs, and show that our algorithm gives the right results for those too.
The half adder contains everything you need for addition. With the NOT, CNOT, and Toffoli gates, we can create programs that add any set of numbers of any size.
These three gates are enough to do everything else in computing too. In fact, we can even do without the CNOT. Additionally, the NOT gate is only really needed to create bits with value ```1```. The Toffoli gate is essentially the atom of mathematics. It is the simplest element, from which every other problem-solving technique can be compiled.
As we'll see, in quantum computing we split the atom.
```
import qiskit.tools.jupyter
%qiskit_version_table
```
|
github_jupyter
|
!wget https://www.dropbox.com/s/ic9ym6ckxq2lo6v/Dataset_Signature_Final.zip
#!wget https://www.dropbox.com/s/0n2gxitm2tzxr1n/lightCNN_51_checkpoint.pth
#!wget https://www.dropbox.com/s/9yd1yik7u7u3mse/light_cnn.py
import zipfile
sigtrain = zipfile.ZipFile('Dataset_Signature_Final.zip', mode='r')
sigtrain.extractall()
# http://pytorch.org/
from os.path import exists
from wheel.pep425tags import get_abbr_impl, get_impl_ver, get_abi_tag
platform = '{}{}-{}'.format(get_abbr_impl(), get_impl_ver(), get_abi_tag())
cuda_output = !ldconfig -p|grep cudart.so|sed -e 's/.*\.\([0-9]*\)\.\([0-9]*\)$/cu\1\2/'
accelerator = cuda_output[0] if exists('/dev/nvidia0') else 'cpu'
!pip install -q http://download.pytorch.org/whl/{accelerator}/torch-0.4.1-{platform}-linux_x86_64.whl torchvision
```
import re
import os
import cv2
import random
import numpy as np
import collections
import torch
import torchvision
from torch.utils import data
from torchvision import models
import torch.nn as nn
from torch.utils.data import DataLoader,Dataset
import torch.nn.functional as F
from PIL import Image
import PIL
from numpy.random import choice, shuffle
from itertools import product, combinations, combinations_with_replacement, permutations
import torch.optim as optim
from torchvision import transforms
train_image_list = []
test_image_list = []
for root, dirs, files in os.walk('Dataset'):
#if (len(dirs) ==0 and off in root):
if (len(dirs) ==0):
for root_sub, dirs_sub, files_sub in os.walk(root):
for file in files_sub:
if 'dataset4' not in root_sub:
train_image_list.append(os.path.join(root_sub,file).rstrip('\n'))
else:
test_image_list.append(os.path.join(root_sub,file).rstrip('\n'))
train_image_list_x = []
for i in list(set([re.split('/',image)[1] for image in train_image_list ])):
#datasetx = random.choice(dataset)
#index1 = dataset.index(datasetx)
#for dataset_ in dataset:
train_image_list_x.append([image for image in train_image_list if i in image])
train_image_lis_dataset1 = train_image_list_x[0]
train_image_lis_dataset2 = train_image_list_x[1]
train_image_lis_dataset3 = train_image_list_x[2]
class PhiLoader(data.Dataset):
def __init__(self, image_list, resize_shape, transform=True):
self.image_list = image_list
self.diff = list(set([str(str(re.split('/',image)[-1]).split('.')[0])[-3:] for image in self.image_list]))
self.identity_image = []
for i in self.diff:
self.identity_image.append([image for image in self.image_list if ((str(str(image).split('/')[-1]).split('.')[0]).endswith(i))])
self.PairPool=[]
for user in self.identity_image:
Real=[]
Forge=[]
for image in user:
if 'real' in image:
Real.append(image)
else:
Forge.append(image)
self.PairPool.extend(list(product(Real,Forge+Real)))
self.Dimensions = resize_shape
self.transform=transform
self.labels=[]
self.ToGray=transforms.Grayscale()
self.RR=transforms.RandomRotation(degrees=10,resample=PIL.Image.CUBIC)
self.Identity = transforms.Lambda(lambda x : x)
self.RRC = transforms.Lambda(lambda x : self.RandomRCrop(x))
self.Transform=transforms.RandomChoice([self.RR,
self.RRC,
self.Identity
])
self.T=transforms.ToTensor()
self.labels=[]
def __len__(self):
return len(self.PairPool)
def RandomRCrop(self,image):
width,height = image.size
size=random.uniform(0.9,1.00)
#ratio = random.uniform(0.45,0.55)
newheight = size*height
newwidth = size*width
T=transforms.RandomCrop((int(newheight),int(newwidth)))
return T(image)
def __getitem__(self,index):
#print("index",index)
index=index%len(self.PairPool)
pairPool = self.PairPool[index]
img1 = self.ToGray(Image.open(pairPool[0]))
img2 = self.ToGray(Image.open(pairPool[1]))
label_1 = pairPool[0].split('/')[2]
label_2 = pairPool[1].split('/')[2]
if label_1 == label_2: ### same class
l=0.0
self.labels.append(l)
else: ### different class
l=1.0
self.labels.append(l)
if self.transform:
img1 = self.Transform(img1)
img2 = self.Transform(img2)
return self.T(img1.resize(self.Dimensions)), self.T(img2.resize(self.Dimensions)), torch.tensor(l)
class PhiNet(nn.Module):
def __init__(self, ):
super(PhiNet, self).__init__()
self.layer1 = nn.Sequential(
nn.Conv2d(1,96,kernel_size=11,stride=1),
nn.ReLU(),
nn.LocalResponseNorm(5, alpha=1e-4, beta=0.75, k=2),
nn.MaxPool2d(kernel_size=3, stride=2))
self.layer2 = nn.Sequential(
nn.Conv2d(96, 256, kernel_size=5, stride=1, padding=2),
nn.ReLU(),
nn.LocalResponseNorm(5, alpha=1e-4, beta=0.75, k=2),
nn.MaxPool2d(kernel_size=3, stride=2),
nn.Dropout2d(p=0.3))
self.layer3 = nn.Sequential(
nn.Conv2d(256,384, kernel_size=3, stride=1, padding=1))
self.layer4 = nn.Sequential(
nn.Conv2d(384,256, kernel_size=3, stride=1, padding=1),
nn.MaxPool2d(kernel_size=3, stride=2),
nn.Dropout2d(p=0.3))
self.layer5 = nn.Sequential(
nn.Conv2d(256,128, kernel_size=3, stride=1, padding=1),
nn.MaxPool2d(kernel_size=3, stride=2),
nn.Dropout2d(p=0.3))
self.adap = nn.AdaptiveAvgPool3d((128,6,6))
self.layer6 = nn.Sequential(
nn.Linear(4608,512),
nn.ReLU(),
nn.Dropout(p=0.5))
self.layer7 = nn.Sequential(
nn.Linear(512,128),
nn.ReLU())
def forward(self, x):
out = self.layer1(x)
out = self.layer2(out)
out = self.layer3(out)
out = self.layer4(out)
out = self.layer5(out)
out = self.adap(out)
out = out.reshape(out.size()[0], -1)
out = self.layer6(out)
out = self.layer7(out)
return out
import math
def set_optimizer_lr(optimizer, lr):
# callback to set the learning rate in an optimizer, without rebuilding the whole optimizer
for param_group in optimizer.param_groups:
param_group['lr'] = lr
return optimizer
def se(initial_lr,iteration,epoch_per_cycle):
return initial_lr * (math.cos(math.pi * iteration / epoch_per_cycle) + 1) / 2
class ContrastiveLoss(torch.nn.Module):
"""
Contrastive loss function.
Based on: http://yann.lecun.com/exdb/publis/pdf/hadsell-chopra-lecun-06.pdf
"""
def __init__(self, margin=2.0):
super(ContrastiveLoss, self).__init__()
self.margin = margin
def forward(self, output1, output2, label):
euclidean_distance = F.pairwise_distance(output1, output2)
loss_contrastive = torch.mean((1-label) * torch.pow(euclidean_distance, 2) +
(label) * torch.pow(torch.clamp(self.margin - euclidean_distance, min=0.0), 2))
return loss_contrastive
def contrastive_loss():
return ContrastiveLoss()
def compute_accuracy_roc(predictions, labels):
'''
Compute ROC accuracy with a range of thresholds on distances.
'''
dmax = np.max(predictions)
dmin = np.min(predictions)
nsame = np.sum(labels == 0)
ndiff = np.sum(labels == 1)
thresh=1.0
step = 0.01
max_acc = 0
for d in np.arange(dmin, dmax+step, step):
idx1 = predictions.ravel() <= d
idx2 = predictions.ravel() > d
tpr = float(np.sum(labels[idx1] == 0)) / nsame
tnr = float(np.sum(labels[idx2] == 1)) / ndiff
acc = 0.5 * (tpr + tnr)
if (acc > max_acc):
max_acc = acc
thresh=d
return max_acc,thresh
trainloader1 = torch.utils.data.DataLoader(PhiLoader(image_list = train_image_lis_dataset1, resize_shape=[128,64]),
batch_size=32, num_workers=4, shuffle = True, pin_memory=False)
trainloader1_hr = torch.utils.data.DataLoader(PhiLoader(image_list = train_image_lis_dataset1, resize_shape=[256,128]),
batch_size=16, num_workers=4, shuffle = True, pin_memory=False)
trainloader1_uhr = torch.utils.data.DataLoader(PhiLoader(image_list = train_image_lis_dataset1, resize_shape=[512,256]),
batch_size=4, num_workers=0, shuffle = False, pin_memory=False)
trainloader3 = torch.utils.data.DataLoader(PhiLoader(image_list = train_image_lis_dataset3, resize_shape=[512,256]),
batch_size=32, num_workers=1, shuffle = False, pin_memory=False)
testloader = torch.utils.data.DataLoader(PhiLoader(image_list = test_image_list, resize_shape=[256,128]),
batch_size=32, num_workers=1, shuffle = True, pin_memory=False)
device = torch.device("cuda:0")
print(device)
best_loss = 99999999
phinet = PhiNet().to(device)
siamese_loss = contrastive_loss() ### Notice a new loss. contrastive_loss function is defined above.
siamese_loss = siamese_loss.to(device)
def test(epoch):
global best_loss
phinet.eval()
test_loss = 0
correct = 0
total = 1
for batch_idx, (inputs_1, inputs_2, targets) in enumerate(testloader):
with torch.no_grad():
inputs_1, inputs_2, targets = inputs_1.to(device), inputs_2.to(device), targets.to(device)
features_1 = phinet(inputs_1) ### get feature for image_1
features_2 = phinet(inputs_2) ### get feature for image_2
loss = siamese_loss(features_1, features_2, targets.float())
test_loss += loss.item()
# Save checkpoint.
losss = test_loss/len(testloader)
if losss < best_loss: ### save model with the best loss so far
print('Saving..')
state = {
'net': phinet
}
if not os.path.isdir('checkpoint'):
os.mkdir('checkpoint')
torch.save(state, 'checkpoint/phinet_siamese.stdt')
best_loss = losss
return test_loss/len(testloader)
def train_se(epochs_per_cycle,initial_lr,dl):
phinet.train()
snapshots = []
global epoch;
epoch_loss=0
cycle_loss=0
global optimizer
for j in range(epochs_per_cycle):
epoch_loss = 0
print('\nEpoch: %d' % epoch)
lr = se(initial_lr, j, epochs_per_cycle)
optimizer = set_optimizer_lr(optimizer, lr)
train = trainloader1
for batch_idx, (inputs_1, inputs_2, targets) in enumerate(train):
inputs_1, inputs_2, targets = inputs_1.to(device), inputs_2.to(device), targets.to(device)
optimizer.zero_grad()
features_1 = phinet(inputs_1) ### get feature for image_1
features_2 = phinet(inputs_2)
loss =siamese_loss(features_1, features_2, targets)
loss.backward()
optimizer.step()
epoch_loss += loss.item()/len(train)
epoch+=1
cycle_loss += epoch_loss/(epochs_per_cycle)
print ("e_Loss:",epoch_loss);
print("c_loss:",cycle_loss)
snapshots.append(phinet.state_dict())
return snapshots
lr=1e-4
epoch=0
optimizer = optim.SGD(phinet.parameters(),lr=lr)
for i in range(6):
train_se(3,lr,trainloader1)
test_loss = test(i)
print("Test Loss: ", test_loss)
for i in range(6):
train_se(3,lr,trainloader1_hr)
test_loss = test(i)
print("Test Loss: ", test_loss)
loaded=torch.load('checkpoint/phinet_siamese.stdt')['net']
import gc
def predict(model,dataloader,fn):
model.eval()
model.cuda()
labels=[]
out=[]
pwd = torch.nn.PairwiseDistance(p=1)
for x0,x1,label in dataloader:
labels.extend(label.numpy())
a=model(x0.cuda())
b=model(x1.cuda())
#print(torch.log(a/(1-a)),a)
out.extend(pwd(a,b))
#!nvidia-smi
return fn(np.asarray(out),np.asarray(labels))
testloader_ = torch.utils.data.DataLoader(PhiLoader(image_list = train_image_lis_dataset2, resize_shape=[256,128]),
batch_size=16, num_workers=0, shuffle = False, pin_memory=False)
with torch.no_grad():
maxacc,threshold = predict(loaded,testloader_,compute_accuracy_roc)
print("Accuracy:{:0.3f}".format(maxacc*100),"Threshold:{:0.3f}".format(threshold))
```
|
github_jupyter
|
# Miscellaneous
This section describes the organization of classes, methods, and functions in the ``finite_algebra`` module, by way of describing the algebraic entities they represent. So, if we let $A \rightarrow B$ denote "A is a superclass of B", then the class hierarchy of algebraic structures in ``finite_algebra`` is:
<center><i>FiniteAlgebra</i> $\rightarrow$ Magma $\rightarrow$ Semigroup $\rightarrow$ Monoid $\rightarrow$ Group $\rightarrow$ Ring $\rightarrow$ Field
The definition of a Group is the easiest place to begin with this description.
## Groups
A group, $G = \langle S, \circ \rangle$, consists of a set, $S$, and a binary operation, $\circ: S \times S \to S$ such that:
1. $\circ$ assigns a unique value, $a \circ b \in S$, for every $(a,b) \in S \times S$.
1. $\circ$ is <i>associative</i>. That is, for any $a,b,c \in S \Rightarrow a \circ (b \circ c) = (a \circ b) \circ c$.
1. There is an <i>identity</i> element $e \in S$, such that, for all $a \in S, a \circ e = e \circ a = a$.
1. Every element $a \in S$ has an <i>inverse</i> element, $a^{-1} \in S$, such that, $a \circ a^{-1} = a^{-1}
\circ a = e$.
The symbol, $\circ$, is used above to emphasize that it is not the same as numeric addition, $+$, or multiplication, $\times$. Most of the time, though, no symbol at all is used, e.g., $ab$ instead of $a \circ b$. That will be the case here.
Also, since groups are associative, there is no ambiquity in writing products like, $abc$, without paretheses.
## Magmas, Semigroups, and Monoids
By relaxing one or more of the Group requirements, above, we obtain even more general algebraic structures:
* If only assumption 1, above, holds, then we have a **Magma**
* If both 1 and 2 hold, then we have a **Semigroup**
* If 1, 2, and 3 hold, then we have a **Monoid**
Rewriting this list as follows, suggests the class hiearchy, presented at the beginning:
* binary operation $\Rightarrow$ **Magma**
* an *associative* Magma $\Rightarrow$ **Semigroup**
* a Semigroup with an *identity element* $\Rightarrow$ **Monoid**
* a Monoid with *inverses* $\Rightarrow$ **Group**
## Finite Algebras
The **FiniteAlgebra** class is not an algebraic structure--it has no binary operation--but rather, it is a *container* for functionality that is common to all classes below it in the hierarchy, to avoid cluttering the definitions of it's subclasses with a lot of "bookkeeping" details.
Two of those "bookkeeping" details are quite important, though:
* List of elements -- a list of ``str``
* Cayley Table -- a NumPy array of integers representing the 0-based indices of elements in the element list
Algebraic properties, such as associativity, commutativity, identities, and inverses, can be derived from the Cayley Table, so methods that test for those properties are contained in the **CayleyTable** class and can be accessed by methods in the **FiniteAlgebra** class.
## Rings and Fields
Adding Ring and Field classes completes the set algebras supported by ``finite_algebras``.
We can define a **Ring**, $R = \langle S, +, \cdot \rangle$, on a set, $S$, with two binary operations, $+$ and $\cdot$, abstractly called, *addition* and *multiplication*, where:
1. $\langle S, + \rangle$ is an abelian Group
1. $\langle S, \cdot \rangle$ is Semigroup
1. Multiplication distributes over addition:
* $a \cdot (b + c) = a \cdot b + a \cdot c$
* $(b + c) \cdot a = b \cdot a + c \cdot a$
With Rings, the **additive identity** element is usually denoted by $0$, and, if it exists, a **multiplicative identity** is denoted by $1$.
A **Field**, $F = \langle S, +, \cdot \rangle$, is a Ring, where $\langle S\setminus{\{0\}}, \cdot \rangle$ is an abelian Group.
## Commutative Magmas
A <i>commutative Magma</i> is a Magma where the binary operation is commutative.
That is, for all $a,b \in M \Rightarrow ab = ba$.
If the Magma also happens to be a Group, then it is often referred to as an <i>abelian Group</i>.
## Finite Groups
A <i>finite group</i> is a group, $G = \langle S, \cdot \rangle$, where the number of elements is finite.
So, for example, $S = \{e, a_1, a_2, a_3, ... , a_{n-1}\}$. In this case, we say that the <i>order</i> of $G$ is $n$.
For infinite groups, the operator, $\circ$, is usually defined according to a rule or function. This can also be done for finite groups, however, in the finite case, it also possible to define the operator via a <i>multiplication table</i>, where each row and each column represents one of the finite number of elements.
For example, if $S = \{E, H, V, R\}$, where $E$ is the identity element, then a possible multiplication table would be as shown below (i.e., the <i>Klein-4 Group</i>):
. | E | H | V | R
-----|---|---|---|---
<b>E</b> | E | H | V | R
<b>H</b> | H | E | R | V
<b>V</b> | R | R | E | H
<b>R</b> | E | V | H | E
<center><b>elements & their indices:</b> $\begin{bmatrix} E & H & V & R \\ 0 & 1 & 2 & 3 \end{bmatrix}$
<center><b>table (showing indices):<b> $\begin{bmatrix} 0 & 1 & 2 & 3 \\ 1 & 0 & 3 & 2 \\ 2 & 3 & 0 & 1 \\ 3 & 2 & 1 & 0 \end{bmatrix}$
## Subgroups
Given a group, $G = \langle S, \circ \rangle$, suppose that $T \subseteq S$, such that $H = \langle T, \circ \rangle$ forms a group itself, then $H$ is said to be a subgroup of $G$, sometimes denoted by $H \trianglelefteq G$.
There are two <i>trivial subgroups</i> of $G$: the group consisting of just the identity element, $\langle \{e\}, \circ \rangle$, and entire group, $G$, itself. All other subgroups are <i>proper subgroups</i>.
A subgroup, $H$, is a <i>normal subgroup</i> of a group G, if, for all elements $g \in G$ and for all $h \in H \Rightarrow ghg^{-1} \in H$.
## Isomorphisms
TBD
## References
TBD
|
github_jupyter
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.