code
stringlengths 38
801k
| repo_path
stringlengths 6
263
|
|---|---|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# + colab={"base_uri": "https://localhost:8080/"} executionInfo={"elapsed": 817407, "status": "ok", "timestamp": 1627299165557, "user": {"displayName": "<NAME>\u00e0nh", "photoUrl": "", "userId": "02848241069421510082"}, "user_tz": -420} id="JnHhSjZec4W6" outputId="74e646bf-7ced-4693-bf88-8a532f8b3f4b"
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
from keras.datasets import mnist
from keras.utils.np_utils import to_categorical
from keras.models import Sequential
from keras.layers import Dense, Dropout, Flatten, Conv2D, MaxPooling2D, Activation, LeakyReLU
from keras.layers.noise import AlphaDropout
from keras.utils.generic_utils import get_custom_objects
from keras import backend as K
from keras.optimizers import Adam, SGD
def preprocess_mnist(x_train, y_train, x_test, y_test):
x_train = x_train.reshape(x_train.shape[0], 28 * 28)
x_test = x_test.reshape(x_test.shape[0], 28 * 28)
input_shape = (28 * 28,)
x_train = x_train.astype('float32')
x_test = x_test.astype('float32')
x_train /= 255
x_test /= 255
y_train = to_categorical(y_train)
y_test= to_categorical(y_test)
return x_train, y_train, x_test, y_test, input_shape
(x_train, y_train), (x_test, y_test) = mnist.load_data()
x_train, y_train, x_test, y_test, input_shape = preprocess_mnist(x_train, y_train, x_test, y_test)
def build_cnn(activation,
dropout_rate,
optimizer):
model = Sequential()
if(activation == 'selu'):
model.add(Dense(512, activation=activation, input_shape=input_shape, kernel_initializer='lecun_normal'))
model.add(AlphaDropout(0.25))
model.add(Dense(128, activation=activation, kernel_initializer='lecun_normal'))
model.add(AlphaDropout(0.5))
model.add(Dense(10, activation='softmax'))
else:
model.add(Dense(512, activation=activation, input_shape=input_shape, kernel_initializer='lecun_normal'))
model.add(Dropout(0.25))
model.add(Dense(128, activation=activation, kernel_initializer='lecun_normal'))
model.add(Dropout(0.5))
model.add(Dense(10, activation='softmax'))
model.compile(
loss='categorical_crossentropy',
optimizer=optimizer,
metrics=['accuracy']
)
return model
def gelu(x):
return 0.5 * x * (1 + tf.tanh(tf.sqrt(2 / np.pi) * (x + 0.044715 * tf.pow(x, 3))))
get_custom_objects().update({'gelu': Activation(gelu)})
def swish(x):
return x * tf.sigmoid(x)
get_custom_objects().update({'swish': Activation(swish)})
get_custom_objects().update({'leaky-relu': Activation(LeakyReLU(alpha=0.2))})
act_func = ['sigmoid', 'tanh', 'relu', 'leaky-relu', 'elu', 'selu', 'gelu', 'swish']
result = []
for activation in act_func:
print('\nTraining with -->{0}<-- activation function\n'.format(activation))
model = build_cnn(activation=activation,
dropout_rate=0.2,
optimizer=SGD())
history = model.fit(x_train, y_train,
validation_split=0.20,
batch_size=128,
epochs=50,
verbose=1,
validation_data=(x_test, y_test))
result.append(history)
K.clear_session()
del model
for r in result:
print(r.history)
| mnist/mlp_lecun/trainings/2depth128.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# # Quickstart
#
# Follow the steps below to get started with ProbNum and learn about its basic functionality.
# +
import warnings
warnings.filterwarnings('ignore')
# Make inline plots vector graphics instead of raster graphics
# %matplotlib inline
from IPython.display import set_matplotlib_formats
set_matplotlib_formats('pdf', 'svg')
# Plotting
import matplotlib.pyplot as plt
from matplotlib.colors import TwoSlopeNorm
plt.style.use('../probnum.mplstyle')
# -
# ## Linear solvers: a numerical method
#
# ProbNum provides tools for solving numerical problems. In this tutorial, we look at the specific example of a solver for a *linear system*. The linear system is defined as $A x_* = b$, where $A\in\mathbb{R}^{d\times d}$ is a known square matrix, $b\in\mathbb{R}^d$ is a known vector, and $x_*\in\mathbb{R}^d$ is the unknown solution of the linear system. A linear solver attempts to estimate the unknown $x_*$ while being provided $A$ and $b$.
#
# We will first see, how this is done with the `numpy.linalg.solve` solver, and later compare it to the ProbNum solver.
# +
import numpy as np
# Define the linear system Ax=b by defining the matrix A and vector b.
A = np.array([[7.5, 2.0, 1.0],
[2.0, 2.0, 0.5],
[1.0, 0.5, 5.5]])
b = np.array([1., 2., -3.])
# Solve for x using NumPy
x = np.linalg.solve(A, b)
print(x)
# -
# Now, we can do the exact same procedure with ProbNum, by using the `probnum.linalg.problinsolve` solver.
# +
import probnum as pn
# Solve for x using ProbNum
x_rv, _, _, _ = pn.linalg.problinsolve(A, b)
print(x_rv.mean)
# -
# We observe, that the NumPy solver and the ProbNum solver return the exact same solution. That is encouraging! But what's the point of the ProbNum solver then? You may have noticed, that we called the return object `x_rv` instead of `x` for the ProbNum solver. This indicates that the ProbNum solver returns a *random variable* over the solution rather than simply a point estimate. In this particular case, the solution `x_rv` is a [multivariate Gaussian random variable](https://en.wikipedia.org/wiki/Multivariate_normal_distribution) of dimension $d=3$ (Gaussian distributions are also called "Normal").
x_rv
# The mean of the normal distribution equals the best guess for the solution of the linear system, and the covariance matrix provides a measure for how certain the solver is about the solution. We can see below, that the algorithm is very certain about the solution as the covariance matrix is virtually zero.
# mean defines best guess for the solution x
x_rv.mean
# covariance matrix provied a measure of uncertainty
x_rv.cov.todense()
# So what's the deal? In the above instance, we ran the ProbNum solver to full convergence which is why it returned the exact same solution as the NumPy solver. But what if we cannot afford to run the algorithm to full convergence? Many applications in practice use linear systems which are so large, that running to convergence simply takes too long even on large-scale hardware. Here, the ProbNum solver provides *additional functionality* that we will have a closer look at now.
# ## Trading-off precision with computational cost
#
# Instead of running all iterations of the solver, we will now attempt to run the solver for only 2 steps. This is indicated by the flag `maxiter=2`.
# Solve with limited computational budget
x_rv, _, _, _ = pn.linalg.problinsolve(A, b, maxiter=2)
# mean defines best guess for the solution x
x_rv.mean
# covariance matrix provies a measure of uncertainty
x_rv.cov.todense()
# We can already observe that the best guess for the solution (mean) has slightly changed as we did not compute the exact solution anymore. Therefore, the covariance matrix contains larger values than before, accounting for the uncertainty arising from the limited number of iterations we performed. In order to interpret the numbers in the covariance matrix properly, we will now sample from the normal distribution provided through `x_rv`.
# Sample from solution distribution
rng = np.random.default_rng(seed=1)
n_samples = 10
x_samples = x_rv.sample(rng=rng, size=n_samples)
print(x_samples)
# Each sample (row) can be seen as a potential solution to the linear system. We observe that the last entry (third column) of the solution does not vary much across samples, which indicates that the solver is fairly certain about its value, while the second entry (middle column) varies more, indicating that the solver is less certain about its value. This is valuable information in case the solution for $x$ is used in a downstream operation.
#
# Instead of using samples, we can also use the covariance matrix directly to get a numerical representation of the uncertainty. For this we retrieve the *marginal standard deviation* `x_rv.std` of the best guess for the solution.
# The marginal standard deviations correspond to the classic standard error of the random variable; though the standard error does not capture the off diagonal element of the covariance matrix it is a convenient way to summarize the variability of each individual element of the random variable.
# +
print(f"true solution: {x}.")
print(f"best guess for solution: {x_rv.mean}.")
print(f"marginal standard deviations: {x_rv.std}.\n")
for i in range(3):
print(f"The marginal solution of element {i} is {x_rv.mean[i]:.2f} with a 95% credible interval pm {2 * x_rv.std[i]:.2f}.")
# -
# We again observe that the algorithm is most certain about the last component (indexed 2), and less certain about the others. For completeness, we attempt a visual representation of the best guess, the credible intervals as well as the first 4 samples:
# + tags=["nbsphinx-thumbnail"]
# collect true solution, best guess, std, and 4 samples for plotting
rvdict = {"$x_*$" : x, # true solution
"$\mathbb{E}(\mathsf{x})$" : x_rv.mean, # best guess
"$std(\mathsf{x})$" : x_rv.std, # marginal standard deviations
"$\mathsf{x}_1$" : x_samples[0], # sample No. 0
"$\mathsf{x}_2$" : x_samples[1], # sample No. 1
"$\mathsf{x}_3$" : x_samples[2], # sample No. 2
"$\mathsf{x}_4$" : x_samples[3] # sample No. 3
}
# retrieve min and max values of all entries for plotting purposes
vmin = np.min([np.min(mat) for mat in list(rvdict.values())])
vmax = np.max([np.max(mat) for mat in list(rvdict.values())])
# normalize diverging colobar, such that it is centered at zero
norm = TwoSlopeNorm(vmin=vmin, vcenter=0, vmax=vmax)
fig, axes = plt.subplots(nrows=1, ncols=2 + 4 + 1, figsize=(8, 2.5), sharey=True)
for i, (title, rv) in enumerate(rvdict.items()):
ax=axes[i].imshow(rv[:, np.newaxis], cmap='bwr', norm=norm)
axes[i].set_xticks([])
axes[i].set_yticks([])
axes[i].title.set_text(title)
plt.tight_layout()
plt.colorbar(ax, ax=axes[i], pad=0.2)
# -
# ## Encoding Prior Knowledge
#
# Apart from trading-off precision with computational cost, the ProbNum method has a second *additional feature* which is the ability to encode prior knowledge about the linear system at hand. It is known that encoding prior knowledge leads to faster convergence to the true solution [1]. In this particular case, the prior knowledge is an approximation to the inverse of the matrix $A$, called `Ainv_approx` $\approx A^{-1}$. Such knowledge is sometimes available when consecutive, similar linear systems need to be solved, e.g., in optimization or as covariance matrices. Using a related solution will help find the solution to the current linear system faster, with less cost, and/or with a higher precision. The prior on the inverse plays a similar role to the preconditioner for classic linear solvers.
#
# Let us first define the approximate inverse of $A$:
# Approximate inverse of A
Ainv_approx = np.array([[ 0.2 , -0.18, -0.015],
[-0.18 , 0.7 , -0.03 ],
[-0.015, -0.03, 0.20 ]])
print(A @ Ainv_approx)
# We can see that $A^{-1}$ and `Ainv_approx` are not completely identical, otherwise the output above would return the identity matrix $I$.
#
# As a second piece of prior information, we consider the knowledge that $A$ and $A^{-1}$ are symmetric matrices which can be seen from the definition of $A$. Symmetric matrices are a common occurrence when solving linear systems, for example in linear regression, or Gaussian process regression.
#
#
# In the case of the ProbNum solver, encoding both pieces of prior knowledge (symmetry of $A$ and $A^{-1}$ + approximate value of $A^{-1}$) is achieved by specifying a prior distribution on $A$ and $A^{-1}$:
#
# +
from probnum import randvars, linops
# prior distribution on A
A0 = randvars.Normal(
mean=A, cov=linops.SymmetricKronecker(10 ** -6 * linops.Identity(A.shape[0]))
)
# prior distribution on A^{-1}
Ainv0 = randvars.Normal(
mean=Ainv_approx, cov=linops.SymmetricKronecker(0.1 * linops.Identity(A.shape[0]))
)
# -
# The random variables `A0` and `Ainv0` define symmetric [matrix-variate normal distributions](https://en.wikipedia.org/wiki/Matrix_normal_distribution) as priors [1] whose samples are symmetric matrices with mean `A` and `Ainv_approx` respectively. The covariance of `A0` is chosen very small to concentrate the prior, as $A$ is known.
#
# We can now pass this prior information to the ProbNum solver, which again runs with a limited budget of `maxiter=2`.
# Solve linear system with limited computational budget and prior knowledge
x_rv, _, _, _ = pn.linalg.problinsolve(A, b, A0=A0, Ainv0=Ainv0, maxiter=2)
# mean defines best guess for the solution x
x_rv.mean
# covariance matrix provies a measure of uncertainty
x_rv.cov.todense()
# We observe that the best guess for the solution (mean), even after 2 iterations only, is virtually identical to the true solution and the entries in the covariance matrix are smaller as well, indicating that the probabilistic solver is confident about the solution.
#
# Analogously to above, we illustrate the uncertainty about the solution by sampling from the distribution of the solution `x_rv`.
# Sample from solution distribution
rng = np.random.default_rng(seed=1)
n_samples = 10
x_samples = x_rv.sample(rng=rng, size=n_samples)
print(x_samples)
# The samples, this time vary little for all three elements (columns), indicating that the solver is certain about the solution. We print the marginal standard deviations:
# +
print(f"true solution: {x}.")
print(f"best guess for solution: {x_rv.mean}.")
print(f"marginal standard deviations: {x_rv.std}.\n")
for i in range(3):
print(f"The marginal solution of element {i} is {x_rv.mean[i]:.2f} with a 95% credible interval pm {2 * x_rv.std[i]:.2f}.")
# -
# Again for completeness, we visualize the best guess, the credible intervals as well as the first 4 samples:
# +
# collect true solution, best guess, std, and 4 samples for plotting
rvdict = {"$x_*$" : x, # true solution
"$\mathbb{E}(\mathsf{x})$" : x_rv.mean, # best guess
"$std(\mathsf{x})$" : x_rv.std, # marginal standard deviations
"$\mathsf{x}_1$" : x_samples[0], # sample No. 0
"$\mathsf{x}_2$" : x_samples[1], # sample No. 1
"$\mathsf{x}_3$" : x_samples[2], # sample No. 2
"$\mathsf{x}_4$" : x_samples[3] # sample No. 3
}
# retrieve min and max values of all entries for plotting purposes
vmin = np.min([np.min(mat) for mat in list(rvdict.values())])
vmax = np.max([np.max(mat) for mat in list(rvdict.values())])
# normalize diverging colobar, such that it is centered at zero
norm = TwoSlopeNorm(vmin=vmin, vcenter=0, vmax=vmax)
fig, axes = plt.subplots(nrows=1, ncols=2 + 4 + 1, figsize=(8, 2.5), sharey=True)
for i, (title, rv) in enumerate(rvdict.items()):
ax=axes[i].imshow(rv[:, np.newaxis], cmap='bwr', norm=norm)
axes[i].set_xticks([])
axes[i].set_yticks([])
axes[i].title.set_text(title)
plt.tight_layout()
plt.colorbar(ax, ax=axes[i], pad=0.2)
# -
# The ProbNum solver found a nearly perfect solution this time, with less budget (`maxiter=2`) by using the available prior information, while returning a measure of uncertainty as well.
# ## References
#
# [1] <NAME> & <NAME>, *Probabilistic Linear Solvers for Machine Learning*, 34th Conference on Neural Information Processing Systems (NeurIPS), 2020.
| docs/source/tutorials/quickstart.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# %matplotlib inline
#
# Deep Learning with PyTorch
# **************************
#
# Deep Learning Building Blocks: Affine maps, non-linearities and objectives
# ==========================================================================
#
# Deep learning consists of composing linearities with non-linearities in
# clever ways. The introduction of non-linearities allows for powerful
# models. In this section, we will play with these core components, make
# up an objective function, and see how the model is trained.
#
#
# Affine Maps
# ~~~~~~~~~~~
#
# One of the core workhorses of deep learning is the affine map, which is
# a function $f(x)$ where
#
# \begin{align}f(x) = Ax + b\end{align}
#
# for a matrix $A$ and vectors $x, b$. The parameters to be
# learned here are $A$ and $b$. Often, $b$ is refered to
# as the *bias* term.
#
#
# Pytorch and most other deep learning frameworks do things a little
# differently than traditional linear algebra. It maps the rows of the
# input instead of the columns. That is, the $i$'th row of the
# output below is the mapping of the $i$'th row of the input under
# $A$, plus the bias term. Look at the example below.
#
#
#
# +
# Author: <NAME>
import torch
import torch.autograd as autograd
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
torch.manual_seed(1)
# -
lin = nn.Linear(5, 3) # maps from R^5 to R^3, parameters A, b
# data is 2x5. A maps from 5 to 3... can we map "data" under A?
data = autograd.Variable(torch.randn(2, 5))
print(lin(data)) # yes
# Non-Linearities
# ~~~~~~~~~~~~~~~
#
# First, note the following fact, which will explain why we need
# non-linearities in the first place. Suppose we have two affine maps
# $f(x) = Ax + b$ and $g(x) = Cx + d$. What is
# $f(g(x))$?
#
# \begin{align}f(g(x)) = A(Cx + d) + b = ACx + (Ad + b)\end{align}
#
# $AC$ is a matrix and $Ad + b$ is a vector, so we see that
# composing affine maps gives you an affine map.
#
# From this, you can see that if you wanted your neural network to be long
# chains of affine compositions, that this adds no new power to your model
# than just doing a single affine map.
#
# If we introduce non-linearities in between the affine layers, this is no
# longer the case, and we can build much more powerful models.
#
# There are a few core non-linearities.
# $\tanh(x), \sigma(x), \text{ReLU}(x)$ are the most common. You are
# probably wondering: "why these functions? I can think of plenty of other
# non-linearities." The reason for this is that they have gradients that
# are easy to compute, and computing gradients is essential for learning.
# For example
#
# \begin{align}\frac{d\sigma}{dx} = \sigma(x)(1 - \sigma(x))\end{align}
#
# A quick note: although you may have learned some neural networks in your
# intro to AI class where $\sigma(x)$ was the default non-linearity,
# typically people shy away from it in practice. This is because the
# gradient *vanishes* very quickly as the absolute value of the argument
# grows. Small gradients means it is hard to learn. Most people default to
# tanh or ReLU.
#
#
#
# In pytorch, most non-linearities are in torch.functional (we have it imported as F)
# Note that non-linearites typically don't have parameters like affine maps do.
# That is, they don't have weights that are updated during training.
data = autograd.Variable(torch.randn(2, 2))
print(data)
print(F.relu(data))
# Softmax and Probabilities
# ~~~~~~~~~~~~~~~~~~~~~~~~~
#
# The function $\text{Softmax}(x)$ is also just a non-linearity, but
# it is special in that it usually is the last operation done in a
# network. This is because it takes in a vector of real numbers and
# returns a probability distribution. Its definition is as follows. Let
# $x$ be a vector of real numbers (positive, negative, whatever,
# there are no constraints). Then the i'th component of
# $\text{Softmax}(x)$ is
#
# \begin{align}\frac{\exp(x_i)}{\sum_j \exp(x_j)}\end{align}
#
# It should be clear that the output is a probability distribution: each
# element is non-negative and the sum over all components is 1.
#
# You could also think of it as just applying an element-wise
# exponentiation operator to the input to make everything non-negative and
# then dividing by the normalization constant.
#
#
#
# Softmax is also in torch.functional
data = autograd.Variable(torch.randn(5))
print(data)
print(F.softmax(data))
print(F.softmax(data).sum()) # Sums to 1 because it is a distribution!
print(F.log_softmax(data)) # theres also log_softmax
# Objective Functions
# ~~~~~~~~~~~~~~~~~~~
#
# The objective function is the function that your network is being
# trained to minimize (in which case it is often called a *loss function*
# or *cost function*). This proceeds by first choosing a training
# instance, running it through your neural network, and then computing the
# loss of the output. The parameters of the model are then updated by
# taking the derivative of the loss function. Intuitively, if your model
# is completely confident in its answer, and its answer is wrong, your
# loss will be high. If it is very confident in its answer, and its answer
# is correct, the loss will be low.
#
# The idea behind minimizing the loss function on your training examples
# is that your network will hopefully generalize well and have small loss
# on unseen examples in your dev set, test set, or in production. An
# example loss function is the *negative log likelihood loss*, which is a
# very common objective for multi-class classification. For supervised
# multi-class classification, this means training the network to minimize
# the negative log probability of the correct output (or equivalently,
# maximize the log probability of the correct output).
#
#
#
# Optimization and Training
# =========================
#
# So what we can compute a loss function for an instance? What do we do
# with that? We saw earlier that autograd.Variable's know how to compute
# gradients with respect to the things that were used to compute it. Well,
# since our loss is an autograd.Variable, we can compute gradients with
# respect to all of the parameters used to compute it! Then we can perform
# standard gradient updates. Let $\theta$ be our parameters,
# $L(\theta)$ the loss function, and $\eta$ a positive
# learning rate. Then:
#
# \begin{align}\theta^{(t+1)} = \theta^{(t)} - \eta \nabla_\theta L(\theta)\end{align}
#
# There are a huge collection of algorithms and active research in
# attempting to do something more than just this vanilla gradient update.
# Many attempt to vary the learning rate based on what is happening at
# train time. You don't need to worry about what specifically these
# algorithms are doing unless you are really interested. Torch provies
# many in the torch.optim package, and they are all completely
# transparent. Using the simplest gradient update is the same as the more
# complicated algorithms. Trying different update algorithms and different
# parameters for the update algorithms (like different initial learning
# rates) is important in optimizing your network's performance. Often,
# just replacing vanilla SGD with an optimizer like Adam or RMSProp will
# boost performance noticably.
#
#
#
# Creating Network Components in Pytorch
# ======================================
#
# Before we move on to our focus on NLP, lets do an annotated example of
# building a network in Pytorch using only affine maps and
# non-linearities. We will also see how to compute a loss function, using
# Pytorch's built in negative log likelihood, and update parameters by
# backpropagation.
#
# All network components should inherit from nn.Module and override the
# forward() method. That is about it, as far as the boilerplate is
# concerned. Inheriting from nn.Module provides functionality to your
# component. For example, it makes it keep track of its trainable
# parameters, you can swap it between CPU and GPU with the .cuda() or
# .cpu() functions, etc.
#
# Let's write an annotated example of a network that takes in a sparse
# bag-of-words representation and outputs a probability distribution over
# two labels: "English" and "Spanish". This model is just logistic
# regression.
#
#
#
# Example: Logistic Regression Bag-of-Words classifier
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
#
# Our model will map a sparse BOW representation to log probabilities over
# labels. We assign each word in the vocab an index. For example, say our
# entire vocab is two words "hello" and "world", with indices 0 and 1
# respectively. The BoW vector for the sentence "hello hello hello hello"
# is
#
# \begin{align}\left[ 4, 0 \right]\end{align}
#
# For "hello world world hello", it is
#
# \begin{align}\left[ 2, 2 \right]\end{align}
#
# etc. In general, it is
#
# \begin{align}\left[ \text{Count}(\text{hello}), \text{Count}(\text{world}) \right]\end{align}
#
# Denote this BOW vector as $x$. The output of our network is:
#
# \begin{align}\log \text{Softmax}(Ax + b)\end{align}
#
# That is, we pass the input through an affine map and then do log
# softmax.
#
#
#
# +
data = [("me gusta comer en la cafeteria".split(), "SPANISH"),
("Give it to me".split(), "ENGLISH"),
("No creo que sea una buena idea".split(), "SPANISH"),
("No it is not a good idea to get lost at sea".split(), "ENGLISH")]
test_data = [("Yo creo que si".split(), "SPANISH"),
("it is lost on me".split(), "ENGLISH")]
# word_to_ix maps each word in the vocab to a unique integer, which will be its
# index into the Bag of words vector
word_to_ix = {}
for sent, _ in data + test_data:
for word in sent:
if word not in word_to_ix:
word_to_ix[word] = len(word_to_ix)
print(word_to_ix)
VOCAB_SIZE = len(word_to_ix)
NUM_LABELS = 2
class BoWClassifier(nn.Module): # inheriting from nn.Module!
def __init__(self, num_labels, vocab_size):
# calls the init function of nn.Module. Dont get confused by syntax,
# just always do it in an nn.Module
super(BoWClassifier, self).__init__()
# Define the parameters that you will need. In this case, we need A and b,
# the parameters of the affine mapping.
# Torch defines nn.Linear(), which provides the affine map.
# Make sure you understand why the input dimension is vocab_size
# and the output is num_labels!
self.linear = nn.Linear(vocab_size, num_labels)
# NOTE! The non-linearity log softmax does not have parameters! So we don't need
# to worry about that here
def forward(self, bow_vec):
# Pass the input through the linear layer,
# then pass that through log_softmax.
# Many non-linearities and other functions are in torch.nn.functional
return F.log_softmax(self.linear(bow_vec))
def make_bow_vector(sentence, word_to_ix):
vec = torch.zeros(len(word_to_ix))
for word in sentence:
vec[word_to_ix[word]] += 1
return vec.view(1, -1)
def make_target(label, label_to_ix):
return torch.LongTensor([label_to_ix[label]])
model = BoWClassifier(NUM_LABELS, VOCAB_SIZE)
# the model knows its parameters. The first output below is A, the second is b.
# Whenever you assign a component to a class variable in the __init__ function
# of a module, which was done with the line
# self.linear = nn.Linear(...)
# Then through some Python magic from the Pytorch devs, your module
# (in this case, BoWClassifier) will store knowledge of the nn.Linear's parameters
for param in model.parameters():
print(param)
# To run the model, pass in a BoW vector, but wrapped in an autograd.Variable
sample = data[0]
bow_vector = make_bow_vector(sample[0], word_to_ix)
log_probs = model(autograd.Variable(bow_vector))
print(log_probs)
# -
# Which of the above values corresponds to the log probability of ENGLISH,
# and which to SPANISH? We never defined it, but we need to if we want to
# train the thing.
#
#
#
label_to_ix = {"SPANISH": 0, "ENGLISH": 1}
# So lets train! To do this, we pass instances through to get log
# probabilities, compute a loss function, compute the gradient of the loss
# function, and then update the parameters with a gradient step. Loss
# functions are provided by Torch in the nn package. nn.NLLLoss() is the
# negative log likelihood loss we want. It also defines optimization
# functions in torch.optim. Here, we will just use SGD.
#
# Note that the *input* to NLLLoss is a vector of log probabilities, and a
# target label. It doesn't compute the log probabilities for us. This is
# why the last layer of our network is log softmax. The loss function
# nn.CrossEntropyLoss() is the same as NLLLoss(), except it does the log
# softmax for you.
#
#
#
# +
# Run on test data before we train, just to see a before-and-after
for instance, label in test_data:
bow_vec = autograd.Variable(make_bow_vector(instance, word_to_ix))
log_probs = model(bow_vec)
print(log_probs)
# Print the matrix column corresponding to "creo"
print(next(model.parameters())[:, word_to_ix["creo"]])
loss_function = nn.NLLLoss()
optimizer = optim.SGD(model.parameters(), lr=0.1)
# Usually you want to pass over the training data several times.
# 100 is much bigger than on a real data set, but real datasets have more than
# two instances. Usually, somewhere between 5 and 30 epochs is reasonable.
for epoch in range(100):
for instance, label in data:
# Step 1. Remember that Pytorch accumulates gradients.
# We need to clear them out before each instance
model.zero_grad()
# Step 2. Make our BOW vector and also we must wrap the target in a
# Variable as an integer. For example, if the target is SPANISH, then
# we wrap the integer 0. The loss function then knows that the 0th
# element of the log probabilities is the log probability
# corresponding to SPANISH
bow_vec = autograd.Variable(make_bow_vector(instance, word_to_ix))
target = autograd.Variable(make_target(label, label_to_ix))
# Step 3. Run our forward pass.
log_probs = model(bow_vec)
# Step 4. Compute the loss, gradients, and update the parameters by
# calling optimizer.step()
loss = loss_function(log_probs, target)
loss.backward()
optimizer.step()
for instance, label in test_data:
bow_vec = autograd.Variable(make_bow_vector(instance, word_to_ix))
log_probs = model(bow_vec)
print(log_probs)
# Index corresponding to Spanish goes up, English goes down!
print(next(model.parameters())[:, word_to_ix["creo"]])
# -
# We got the right answer! You can see that the log probability for
# Spanish is much higher in the first example, and the log probability for
# English is much higher in the second for the test data, as it should be.
#
# Now you see how to make a Pytorch component, pass some data through it
# and do gradient updates. We are ready to dig deeper into what deep NLP
# has to offer.
#
#
#
| pytorch/deep_learning_tutorial.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + id="z4OD_y1WUd2h"
import pandas as pd
import numpy as np
from matplotlib import pyplot as plt
import statistics
import math
# + id="3eiNaH1RUiIY" outputId="a4926e32-e5ae-4580-81f2-77c191cf6dc6" colab={"base_uri": "https://localhost:8080/", "height": 445}
matches = pd.read_csv("/content/sample_data/matches.csv")
matches.head()
# + id="nA3SyLi1U4zt" outputId="eb1dca85-04e4-4f16-98d4-a66b2a92c01c" colab={"base_uri": "https://localhost:8080/", "height": 394}
# setting up the table with relevant columns
dropList = ["result","player_of_match","venue","umpire1","umpire2","umpire3"]
matches.drop(labels=dropList, axis=1, inplace=True)
matches.head()
# + id="Pd3l2_iBVlyA" outputId="a752893e-33b7-476e-95f4-9dcaa9d3922d" colab={"base_uri": "https://localhost:8080/", "height": 312}
matches[pd.isnull(matches['winner'])]
# + id="pGnAtkWHVomw" outputId="f9dde95f-c315-4b36-e01d-d1e6e61ac03f" colab={"base_uri": "https://localhost:8080/", "height": 49}
matches['winner'].fillna('Draw', inplace=True)
matches[pd.isnull(matches['winner'])]
# + id="N1xOYdgDVqfP" outputId="f4499d3b-a531-4eb3-b7b0-606cf554a7e0" colab={"base_uri": "https://localhost:8080/", "height": 490}
matches[pd.isnull(matches['city'])]
# + id="ZBg0kS0HVsKz" outputId="7a0502f5-12a7-43f5-ce21-8ab3053c17e7" colab={"base_uri": "https://localhost:8080/", "height": 49}
matches['city'].fillna('Dubai', inplace=True)
matches[pd.isnull(matches['city'])]
# + id="O98Q5N9zVuB8" outputId="b40ced7c-7c7a-49d9-f9e2-aca362ab8500" colab={"base_uri": "https://localhost:8080/"}
matches.replace(["Deccan Chargers","Delhi Daredevils"],["Sunrisers Hyderabad","Delhi Capitals"],inplace=True,regex=True)
match1 = matches[((matches.team1=="Royal Challengers Bangalore") & (matches.team2=="Sunrisers Hyderabad")) | ((matches.team1=="Sunrisers Hyderabad") & (matches.team2=="Royal Challengers Bangalore"))]
match1.shape[0]
# + id="9Sw-oxaJd7mB" outputId="4eaca2ee-30a6-4112-8bac-514f3d721b35" colab={"base_uri": "https://localhost:8080/"}
mw_srh = 0
mw_rcb = 0
lst= [i for i in match1['winner']]
print("Win Tracker!")
for i in lst:
if i=="Royal Challengers Bangalore":
mw_rcb += 1
elif i=='Draw':
continue
else:
mw_srh += 1
print(str(mw_srh)+" "+str(mw_rcb))
print("SRH vs RCB : "+str(mw_srh)+" "+str(mw_rcb))
# + id="CIBO-yKZeZVb" outputId="357801ec-45a3-4ce5-dcc3-bc1655514857" colab={"base_uri": "https://localhost:8080/"}
last_3_season = match1[match1.season >= 2017]
last_3_season.groupby('winner').winner.count()
# + [markdown] id="JAgC7IJJWjqP"
# Out of 19 matches held between SRH and RCB , SRh leads RCB 14 is to 11. In the case with last three seasons, SRH has lead of victories over RR i.e 3 is to 2
# + id="FyY9pRGkaqWW"
def statistics_for_lists(lst):
print("Maximum Value Of List:")
print(max(lst))
print("Median of the List:")
print(statistics.median(lst))
print("Mean of the List:")
print(statistics.mean(lst))
print("75% of the Median is:")
print(statistics.median_high(lst))
print("Minimum Value of List:")
print(min(lst))
# + id="KonYIWemWeSX" outputId="0c7016df-f355-4187-b079-f7fd8b7ee186" colab={"base_uri": "https://localhost:8080/", "height": 394}
deliveries = pd.read_csv("/content/sample_data/deliveries.csv")
deliveries.head()
# + id="Xfj0J0pBWyRX" outputId="80999a81-4e76-4ec5-99a9-fbe345890a3d" colab={"base_uri": "https://localhost:8080/", "height": 394}
dropToBeList = ['inning','is_super_over','bye_runs','legbye_runs','fielder']
deliveries.drop(dropToBeList, axis=1, inplace=True)
deliveries.replace(['Deccan Chargers','Delhi Daredevils'],['Sunrisers Hyderabad','Delhi Capitals'],inplace=True,regex=True)
deliveries['dismissal_kind'].fillna('Not Out',inplace=True)
deliveries.head()
# + id="KlS2o3qIW2GL" outputId="f095efcf-900b-4d5e-c566-962de3fbddf8" colab={"base_uri": "https://localhost:8080/"}
ballbyball = deliveries[((deliveries.batting_team=="Royal Challengers Bangalore") & (deliveries.bowling_team=="Sunrisers Hyderabad")) | ((deliveries.batting_team=="Sunrisers Hyderabad") & (deliveries.bowling_team=="Royal Challengers Bangalore"))]
no_of_matches=list(set([i for i in ballbyball['match_id']]))
no_of_matches.sort()
print(len(no_of_matches))
# + id="bdWITQe5oNC3" outputId="4c5c2b15-ad52-4649-d38c-305c67886842" colab={"base_uri": "https://localhost:8080/"}
#Q4
wickets_lost_srh_pp = ballbyball[(ballbyball.batting_team=='Sunrisers Hyderabad') & (ballbyball.over>=1) & (ballbyball.over<=5)].groupby('match_id').player_dismissed.count()
wickets_lost_rcb_pp = ballbyball[(ballbyball.batting_team=='Royal Challengers Bangalore') & (ballbyball.over>=1) & (ballbyball.over<=5)].groupby('match_id').player_dismissed.count()
srh_pp=[i for i in wickets_lost_srh_pp]
rcb_pp=[i for i in wickets_lost_rcb_pp]
diff=[]
for i in range(len(srh_pp)):
diff.append(abs(rcb_pp[i]-srh_pp[i]))
statistics_for_lists(diff)
# + id="1kJmZcQGBa-N" outputId="d57c0447-9f8d-449a-f22c-f444c132fede" colab={"base_uri": "https://localhost:8080/"}
#Q5
dot_balls = ballbyball[(ballbyball.total_runs==0)].groupby('match_id').total_runs.count()
dot_balls.describe()
# + [markdown] id="nFq6aDcCHC10"
# In all matches between RCB and SRH, the average number of dot balls expected is 81 to 88
# + id="itmnb6ZQsmut" outputId="893821a5-3543-4978-9d1c-577c3d0631d5" colab={"base_uri": "https://localhost:8080/"}
#Q2
total_scores = ballbyball.groupby('match_id').total_runs.sum()
total_scores.describe()
# + id="2lTPW4QcqWaX" outputId="642ce8dd-f97f-428f-a1d6-4f17ad27e1ca" colab={"base_uri": "https://localhost:8080/"}
#Q3
srh = ballbyball[ballbyball.batting_team=='Sunrisers Hyderabad']
srh_dif=[]
for i in no_of_matches:
df = srh[srh.match_id==i]
tot_runs = [k for k in df['total_runs']]
wides = [k for k in df['wide_runs']]
nobs = [k for k in df['noball_runs']]
ball_to_30=0
ball_to_50=0
score_to_50=0
score_to_30=0
for j in range(len(tot_runs)):
if(score_to_30 < 30 and wides[j]==0 and nobs[j]==0):
ball_to_30 +=1
ball_to_50 +=1
score_to_30 += tot_runs[j]
score_to_50 += tot_runs[j]
elif(score_to_30 < 30 and (wides[j]!=0 or nobs[j]!=0)):
score_to_30 += tot_runs[j]
score_to_50 += tot_runs[j]
elif(score_to_50 < 50 and wides[j]==0 and nobs[j]==0):
score_to_50 += tot_runs[j]
ball_to_50 += 1
elif(score_to_50 < 50 and (wides[j]!=0 or nobs[j]!=0)):
score_to_50 += tot_runs[j]
diff = ball_to_50 - ball_to_30
srh_dif.append(diff)
print(srh_dif)
# + id="Uc_qtyLqtDyZ" outputId="d6359093-cb19-4e09-d5fc-9e19bfa241f9" colab={"base_uri": "https://localhost:8080/"}
rcb = ballbyball[ballbyball.batting_team=='Royal Challengers Bangalore']
rcb_dif=[]
for i in no_of_matches:
df = rcb[rcb.match_id==i]
tot_runs = [k for k in df['total_runs']]
wides = [k for k in df['wide_runs']]
nobs = [k for k in df['noball_runs']]
ball_to_30=0
ball_to_50=0
score_to_50=0
score_to_30=0
for j in range(len(tot_runs)):
if(score_to_30 < 30 and wides[j]==0 and nobs[j]==0):
ball_to_30 +=1
ball_to_50 +=1
score_to_30 += tot_runs[j]
score_to_50 += tot_runs[j]
elif(score_to_30 < 30 and (wides[j]!=0 or nobs[j]!=0)):
score_to_30 += tot_runs[j]
score_to_50 += tot_runs[j]
elif(score_to_50 < 50 and wides[j]==0 and nobs[j]==0):
score_to_50 += tot_runs[j]
ball_to_50 += 1
elif(score_to_50 < 50 and (wides[j]!=0 or nobs[j]!=0)):
score_to_50 += tot_runs[j]
diff = ball_to_50 - ball_to_30
rcb_dif.append(diff)
print(rcb_dif)
# + id="3B9jN37vwXqk" outputId="fe3de085-1e90-4a94-baa1-0e9399668d2f" colab={"base_uri": "https://localhost:8080/"}
diff_bw_srh_rcb = []
for i in range(len(rcb_dif)):
diff_bw_srh_rcb.append(abs(srh_dif[i]-rcb_dif[i]))
print(diff_bw_srh_rcb)
statistics_for_lists(diff_bw_srh_rcb)
# + id="UmCV74Kvw04A"
| archive/srh_vs_rcb.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
# %matplotlib inline
import matplotlib
import numpy as np
import matplotlib.pyplot as plt
import numpy as np
# -
from tlpy.host import Host
from tlpy.defect import Defect
elemental_energies = { 'Ge' : -4.48604,
'P' : -5.18405,
'O' : -4.54934575 }
host = Host( energy = -2884.79313425,
vbm = 0.4657,
cbm = 4.0154,
elemental_energies = elemental_energies,
correction_scaling = 0.099720981 )
# +
chemical_potential_limits = {}
chemical_potential_limits[ 'A' ] = { 'Ge' : -4.8746,
'P' : -8.165,
'O' : 0.0 }
chemical_potential_limits[ 'B' ] = { 'Ge' : -4.8664,
'P' : -8.718,
'O' : 0.0 }
chemical_potential_limits[ 'C' ] = { 'Ge' : 0.0,
'P' : -2.0718,
'O' : -2.4373 }
chemical_potential_limits[ 'D' ] = { 'Ge' : 0.0,
'P' : -2.0888,
'O' : -2.4332 }
# +
VO1 = Defect( 'V_O1', stoichiometry = { 'O' : -1 }, host = host, site = 'O' )
VO1.add_charge_state( 0, -2876.05861202 )
VO1.add_charge_state( +1, -2877.36415986 )
VO1.add_charge_state( +2, -2880.33856625 )
VO2 = Defect( 'V_O2', stoichiometry = { 'O' : -1 }, host = host, site = 'O' )
VO2.add_charge_state( 0, -2877.91757552 )
VO2.add_charge_state( +1, -2878.40444833 )
VO2.add_charge_state( +2, -2878.85619152 )
VO3 = Defect( 'V_O3', stoichiometry = { 'O' : -1 }, host = host, site = 'O' )
VO3.add_charge_state( 0, -2876.646 )
VO3.add_charge_state( +1, -2877.139 )
VO3.add_charge_state( +2, -2877.594 )
VO4 = Defect( 'V_O4', stoichiometry = { 'O' : -1 }, host = host, site = 'O' )
VO4.add_charge_state( 0, -2876.170 )
VO4.add_charge_state( +1, -2877.217 )
VO4.add_charge_state( +2, -2880.752 )
VO5 = Defect( 'V_O5', stoichiometry = { 'O' : -1 }, host = host, site = 'O' )
VO5.add_charge_state( 0, -2876.028 )
VO5.add_charge_state( +1, -2877.561 )
VO5.add_charge_state( +2, -2879.462 )
Oi = Defect( 'O_i', stoichiometry = { 'O' : +1 }, host = host, site = 'i' )
Oi.add_charge_state( 0, -2887.757 )
Oi.add_charge_state( -1, -2885.340 )
Oi.add_charge_state( -2, -2882.347 )
VGe1 = Defect( 'V_Ge1', stoichiometry = { 'Ge' : -1 }, host = host, site = 'Ge' )
VGe1.add_charge_state( 0, -2870.216817 )
VGe1.add_charge_state( -1, -2868.426975 )
VGe1.add_charge_state( -2, -2866.739624 )
VGe1.add_charge_state( -3, -2864.304834 )
VGe1.add_charge_state( -4, -2861.29928 )
VGe2 = Defect( 'V_Ge2', stoichiometry = { 'Ge' : -1 }, host = host, site = 'Ge' )
VGe2.add_charge_state( 0, -2867.345547 )
VGe2.add_charge_state( -1, -2867.211323 )
VGe2.add_charge_state( -2, -2866.265491 )
VGe2.add_charge_state( -3, -2864.710182 )
VGe1.add_charge_state( -4, -2862.476172 )
VGe3 = Defect( 'V_Ge3', stoichiometry = { 'Ge' : -1 }, host = host, site = 'Ge' )
VGe3.add_charge_state( 0, -2869.32676 )
VGe3.add_charge_state( -1, -2868.426748 )
VGe3.add_charge_state( -2, -2866.739636 )
VGe3.add_charge_state( -3, -2864.294208 )
VGe3.add_charge_state( -4, -2861.300372 )
VP = Defect( 'V_P', stoichiometry = { 'P' : -1 }, host = host, site = 'P' )
VP.add_charge_state( 0, -2864.292 )
VP.add_charge_state( -1, -2863.078 )
VP.add_charge_state( -2, -2860.643 )
VP.add_charge_state( -3, -2857.421 )
VP.add_charge_state( -4, -2853.678 )
VP.add_charge_state( -5, -2849.884 )
PGe1 = Defect( 'PGe1', stoichiometry = { 'P' : +1, 'Ge' : -1 }, host = host, site = 'Ge1' )
PGe1.add_charge_state( 0, -2885.223 )
PGe1.add_charge_state( +1, -2889.005 )
PGe2 = Defect( 'PGe2', stoichiometry = { 'P' : +1, 'Ge' : -1 }, host = host, site = 'Ge2' )
PGe2.add_charge_state( 0, -2883.819 )
PGe2.add_charge_state( +1, -2887.755 )
PGe3 = Defect( 'PGe3', stoichiometry = { 'P' : +1, 'Ge' : -1 }, host = host, site = 'Ge3' )
PGe3.add_charge_state( 0, -2883.874 )
PGe3.add_charge_state( +1, -2887.826 )
GeP = Defect( 'GeP', stoichiometry = { 'P' : -1, 'Ge' : +1 }, host = host, site = 'P' )
GeP.add_charge_state( 0, -2879.026 )
GeP.add_charge_state( -1, -2878.217 )
VPO4 = Defect( 'VPO4', stoichiometry = { 'P' : -1, 'O' : -4 }, host = host, site = 'P' )
VPO4.add_charge_state( 0, -2846.845 )
VPO4.add_charge_state( +1, -2850.335 )
VPO4.add_charge_state( +2, -2851.6172 )
VPO4.add_charge_state( +3, -2854.244 )
defects = [ VO1, VO2, VO3, VO4, VO5, Oi, VGe1, VGe2, VGe3, VP, PGe1, PGe2, PGe3, GeP, VPO4 ]
# +
def plot_tl_diagram( mu ):
for d in defects:
tl_profile = d.matplotlib_data( delta_mu = mu, ef_min = 0.0, ef_max = host.fundamental_gap )
plt.plot( tl_profile[0], tl_profile[1] )
plt.xlim( xmax = host.fundamental_gap )
plt.ylim( ( -4.0, +14.0 ) )
plt.show()
plot_tl_diagram( chemical_potential_limits[ 'A' ] )
plot_tl_diagram( chemical_potential_limits[ 'B' ] )
plot_tl_diagram( chemical_potential_limits[ 'C' ] )
plot_tl_diagram( chemical_potential_limits[ 'D' ] )
# -
| scripts/germanate_notebook.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
from __future__ import print_function
import numpy as np
from keras import backend as K
import keras
from keras.models import Model
from keras.layers import Input, Dense, RepeatVector, Masking, Dropout, Flatten, Activation, Reshape, Lambda, Permute, merge, multiply, concatenate
from keras.layers.merge import Concatenate
from keras.layers.wrappers import Bidirectional, TimeDistributed
from keras.layers.recurrent import GRU, LSTM
from keras.layers.pooling import GlobalMaxPooling1D
from keras.activations import *
from keras.utils import to_categorical
from keras.models import Sequential, Model
from keras.layers.embeddings import Embedding
from keras.utils.data_utils import get_file
from keras.preprocessing.sequence import pad_sequences
from functools import reduce
import tarfile
import re
import math
import os
from nltk.tokenize import RegexpTokenizer
from sklearn.metrics.pairwise import cosine_similarity
from gensim.scripts.glove2word2vec import glove2word2vec
from nltk.tokenize import sent_tokenize
from gensim.models.keyedvectors import KeyedVectors
from nltk.stem import WordNetLemmatizer
from nltk.corpus import stopwords
from nltk.stem.porter import PorterStemmer
from nltk.stem.snowball import SnowballStemmer
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.text import text_to_word_sequence
# +
context_file = open(os.path.join('./data/', 'train_context'), 'r')
c = context_file.read()
context = re.split('\n' + '-' + '\n', c)
del c
question_file = open(os.path.join('./data/', 'train_question'), 'r')
c = question_file.read()
questions = re.split('\n' + '-' + '\n', c)
del c
answer_file = open(os.path.join('./data/', 'train_answer'), 'r')
c = answer_file.read()
answers = re.split('\n' + '-' + '\n', c)
del c
span_file = open(os.path.join('./data/', 'train_span'), 'r')
c = span_file.read()
spa = re.split('\n' + '-' + '\n', c)
del c
BASE_DIR = ''
GLOVE_DIR = os.path.join(BASE_DIR, 'glove')
MAX_SEQUENCE_LENGTH = 500
MAX_NUM_WORDS = 10000000
EMBEDDING_DIM = 50
MAX_QUE_LENGTH = EMBEDDING_DIM
VALIDATION_SPLIT = 0.8
NUMCONTEXT = 10
# +
print('Indexing word vectors.')
embeddings_index = {}
with open(os.path.join(GLOVE_DIR, 'glove.6B.50d.txt')) as f:
for line in f:
values = line.split()
word = values[0]
coefs = np.asarray(values[1:], dtype='float32')
embeddings_index[word] = coefs
print('Found %s word vectors.' % len(embeddings_index))
import pickle
with open("char_embeddings.pickle","rb") as fd:
char_embeddings = pickle.load(fd)
def get_char_embedding(word):
x = np.zeros(EMBEDDING_DIM)
count = 0
for i in range(len(word)):
try:
count = count +1
temp = np.asarray(char_embeddings[word[i]])
except:
temp = np.zeros(EMBEDDING_DIM)
x = x+temp
return x/count
# +
tokenizer = RegexpTokenizer(r'[^\s]+')
def vectorize_stories(inp,que,ans):
inputs, queries, answers = [], [], []
for i in range(0,len(inp)):
inputs.append([word_index[w] for w in inp[i]])
queries.append([word_index[w] for w in que[i]])
# answers.append(ans)
return (pad_sequences(inputs, maxlen=MAX_SEQUENCE_LENGTH,padding='post'),
pad_sequences(queries, maxlen=MAX_QUE_LENGTH,padding='post'),
np.array(ans))
def para_tokenizer(data):
x_tokens = tokenizer.tokenize(data)
spans = tokenizer.span_tokenize(data)
sp = [span for span in spans]
return x_tokens,sp
def que_tokenizer(data):
x_tokens = tokenizer.tokenize(data)
return x_tokens
context = context[0:NUMCONTEXT]
inp = []
que = []
ans = []
i =0
for c in context:
tokens,sp = para_tokenizer(c)
q=questions[i]
a=answers[i]
all_ques = re.split('\n', q)
all_ans = re.split('\n', a)
all_s = re.split('\n', spa[i])
for j in range (0,len(all_ques)):
inp.append(tokens)
x = re.split(',',all_s[j])
x = list(map(int, x))
k = 0
for span in sp:
if span[0] <= x[0] <= span[1]:
st = k
if span[0] <= x[1] <= span[1]:
en = k
k+=1
que.append(que_tokenizer(all_ques[j]))
ans.append([st,en])
#ans.append(st)
i+=1
print(len(inp))
# print(inp[0])
# print(que[0])
# print(ans[1])
vocab = set()
for i in range(0,len(inp)):
vocab |= set(inp[i] + que[i])
vocab = sorted(vocab)
print(len(vocab))
vocab_size = len(vocab) + 1
# story_maxlen = max(map(len, (x for x in inp)))
# query_maxlen = max(map(len, (x for x in que)))
# print(story_maxlen,query_maxlen)
word_index = dict((c, i + 1) for i, c in enumerate(vocab))
index_word = dict((i+1, c) for i, c in enumerate(vocab))
train_con, train_que, answers = vectorize_stories(inp,que,ans)
train_ans_start = to_categorical(answers[:,0],MAX_SEQUENCE_LENGTH)
train_ans_end = to_categorical(answers[:,1],MAX_SEQUENCE_LENGTH)
split = int(NUMCONTEXT*VALIDATION_SPLIT)
train_context = train_con[0:split]
val_context = train_con[split:NUMCONTEXT]
train_question = train_que[0:split]
val_question = train_que[split:NUMCONTEXT]
train_answer_start = train_ans_start[0:split]
val_answer_start = train_ans_start[split:NUMCONTEXT]
train_answer_end = train_ans_end[0:split]
val_answer_end = train_ans_end[split:NUMCONTEXT]
# -
num_words = min(MAX_NUM_WORDS, len(word_index) + 1)
embedding_matrix = np.zeros((num_words, EMBEDDING_DIM))
for word, i in word_index.items():
if i >= MAX_NUM_WORDS:
continue
embedding_vector = embeddings_index.get(word)
# print(word,i)
if embedding_vector is not None:
embedding_matrix[i] = embedding_vector
else:
embedding_matrix[i] = get_char_embedding(word)
print(embedding_matrix.shape)
# +
W = EMBEDDING_DIM
N = MAX_SEQUENCE_LENGTH
M = MAX_QUE_LENGTH
dropout_rate = 0
input_sequence = Input((MAX_SEQUENCE_LENGTH,))
question = Input((MAX_QUE_LENGTH,))
context_encoder = Sequential()
context_encoder.add(Embedding(num_words,
EMBEDDING_DIM,
weights=[embedding_matrix],
input_length=MAX_SEQUENCE_LENGTH,
trainable=False))
question_encoder = Sequential()
question_encoder.add(Embedding(num_words,
EMBEDDING_DIM,
weights=[embedding_matrix],
input_length=MAX_QUE_LENGTH,
trainable=False))
context_encoded = context_encoder(input_sequence)
question_encoded = question_encoder(question)
encoder = Bidirectional(LSTM(units=W,return_sequences=True))
passage_encoding = context_encoded
passage_encoding = encoder(passage_encoding)
passage_encoding = Dense(W,use_bias=False,trainable=True)(passage_encoding) #(ex, MAX_SEQUENCE_LENGTH,EMBEDDING_DIM)
question_encoding = question_encoded
question_encoding = encoder(question_encoding)
question_encoding = Dense(W,use_bias=False,trainable=True)(question_encoding) #(ex, MAX_QUE_LENGTH,EMBEDDING_DIM)
question_attention_vector = Dense(1)(question_encoding)
# question_attention_vector = Flatten()(question_attention_vector)
# question_attention_vector = Activation('softmax')(question_attention_vector)
# print(question_attention_vector)
# question_attention_vector = keras.activations.softmax(question_attention_vector,axis=1)
question_attention_vector = Lambda(lambda q: keras.activations.softmax(q, axis=1))(question_attention_vector)
print(question_attention_vector)
question_attention_vector = Lambda(lambda q: q[0] * q[1])([question_encoding, question_attention_vector])
question_attention_vector = Lambda(lambda q: K.sum(q, axis=1))(question_attention_vector)
question_attention_vector = RepeatVector(N)(question_attention_vector)
ans_st = multiply([passage_encoding, question_attention_vector])
answer_start = concatenate([passage_encoding,question_attention_vector, ans_st])
answer_start = Dense(W, activation='relu')(answer_start)
answer_start = Dense(1)(answer_start)
answer_start = Flatten()(answer_start)
answer_start = Activation('softmax')(answer_start)
def s_answer_feature(x):
maxind = K.argmax(
x,
axis=1,
)
return maxind
x = Lambda(lambda x: K.tf.cast(s_answer_feature(x), dtype=K.tf.int32))(answer_start)
start_feature = Lambda(lambda arg: K.tf.gather_nd(arg[0], K.tf.stack(
[K.tf.range(K.tf.shape(arg[1])[0]), K.tf.cast(arg[1], K.tf.int32)], axis=1)))([passage_encoding, x])
start_feature = RepeatVector(N)(start_feature)
ans_1 = multiply([passage_encoding, question_attention_vector])
ans_2 = multiply([passage_encoding, start_feature])
answer_end = concatenate([passage_encoding,question_attention_vector,start_feature, ans_1,ans_2])
answer_end = Dense(W, activation='relu')(answer_end)
answer_end = Dense(1)(answer_end)
answer_end = Flatten()(answer_end)
answer_end = Activation('softmax')(answer_end)
inputs = [input_sequence, question]
outputs = [answer_start, answer_end]
model = Model(inputs,outputs)
# model.summary()
model.compile(optimizer='rmsprop', loss='categorical_crossentropy',
metrics=['accuracy'])
# -
print(train_context.shape,train_question.shape,train_answer_start.shape,train_answer_end.shape)
model.fit([train_context, train_question], [train_answer_start,train_answer_end],
batch_size=30,
epochs=1,
validation_data=([val_context, val_question], [val_answer_start,val_answer_end]))
| .ipynb_checkpoints/pratik (copy)-checkpoint.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] colab_type="text" id="tDnwEv8FtJm7"
# ##### Copyright 2018 The TensorFlow Authors.
# + cellView="form" colab_type="code" id="JlknJBWQtKkI" colab={}
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# + [markdown] colab_type="text" id="60RdWsg1tETW"
# # カスタムレイヤー
# + [markdown] colab_type="text" id="BcJg7Enms86w"
# <table class="tfo-notebook-buttons" align="left">
# <td>
# <a target="_blank" href="https://www.tensorflow.org/tutorials/customization/custom_layers"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on TensorFlow.org</a>
# </td>
# <td>
# <a target="_blank" href="https://colab.research.google.com/github/tensorflow/docs/blob/master/site/ja/tutorials/customization/custom_layers.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
# </td>
# <td>
# <a target="_blank" href="https://github.com/tensorflow/docs/blob/master/site/ja/tutorials/customization/custom_layers.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
# </td>
# <td>
# <a href="https://storage.googleapis.com/tensorflow_docs/site/ja/tutorials/customization/custom_layers.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png" />Download notebook</a>
# </td>
# </table>
# + [markdown] colab_type="text" id="YRXLphinx2fF"
# Note: これらのドキュメントは私たちTensorFlowコミュニティが翻訳したものです。コミュニティによる 翻訳は**ベストエフォート**であるため、この翻訳が正確であることや[英語の公式ドキュメント](https://www.tensorflow.org/?hl=en)の 最新の状態を反映したものであることを保証することはできません。 この翻訳の品質を向上させるためのご意見をお持ちの方は、GitHubリポジトリ[tensorflow/docs](https://github.com/tensorflow/docs)にプルリクエストをお送りください。 コミュニティによる翻訳やレビューに参加していただける方は、 [<EMAIL> メーリングリスト](https://groups.google.com/a/tensorflow.org/forum/#!forum/docs-ja)にご連絡ください。
# + [markdown] colab_type="text" id="UEu3q4jmpKVT"
# ニューラルネットワークの構築には、ハイレベルの API である `tf.keras` を使うことを推奨します。しかしながら、TensorFlow API のほとんどは、eager execution でも使用可能です。
# + colab_type="code" id="-sXDg19Q691F" colab={}
from __future__ import absolute_import, division, print_function, unicode_literals
# + colab_type="code" id="Py0m-N6VgQFJ" colab={}
try:
# # %tensorflow_version only exists in Colab.
# %tensorflow_version 2.x
except Exception:
pass
import tensorflow as tf
# + [markdown] colab_type="text" id="zSFfVVjkrrsI"
# ## レイヤー:有用な演算の共通セット
#
# 機械学習モデルのコーディングでは、個々の演算やひとつひとつの変数のオペレーションよりは、より高度に抽象化されたオペレーションを行いたいのがほとんどだと思います。
#
# 多くの機械学習モデルは、比較的単純なレイヤーの組み合わせや積み重ねによって表現可能です。TensorFlow では、多くの一般的なレイヤーのセットに加えて、アプリケーションに特有なレイヤーを最初から記述したり、既存のレイヤーの組み合わせによって作るための、簡単な方法が提供されています。
#
# TensorFlow には、tf.keras パッケージに[Keras](https://keras.io) APIのすべてが含まれています。Keras のレイヤーは、独自のモデルを構築する際に大変便利です。
# + colab_type="code" id="8PyXlPl-4TzQ" colab={}
# tf.keras.layers パッケージの中では、レイヤーはオブジェクトです。
# レイヤーを構築するためにすることは、単にオブジェクトを作成するだけです。
# ほとんどのレイヤーでは、最初の引数が出力の次元あるいはチャネル数を表します。
layer = tf.keras.layers.Dense(100)
# 入力の次元数は多くの場合不要となっています。それは、レイヤーが最初に使われる際に
# 推定可能だからです。ただし、引数として渡すことで手動で指定することも可能です。
# これは複雑なモデルを構築する場合に役に立つでしょう。
layer = tf.keras.layers.Dense(10, input_shape=(None, 5))
# + [markdown] colab_type="text" id="Fn69xxPO5Psr"
# 既存のレイヤーのすべての一覧は、[ドキュメント](https://www.tensorflow.org/api_docs/python/tf/keras/layers)を参照してください。Dense(全結合レイヤー)、Conv2D、LSTM、BatchNormalization、Dropoutなどのたくさんのレイヤーが含まれています。
# + colab_type="code" id="E3XKNknP5Mhb" colab={}
# レイヤーを使うには、単純にcallします。
layer(tf.zeros([10, 5]))
# + colab_type="code" id="Wt_Nsv-L5t2s" colab={}
# レイヤーにはたくさんの便利なメソッドがあります。例えば、`layer.variables`を使って
# レイヤーのすべての変数を調べることができます。訓練可能な変数は、 `layer.trainable_variables`
# でわかります。この例では、全結合レイヤーには重みとバイアスの変数があります。
layer.variables
# + colab_type="code" id="6ilvKjz8_4MQ" colab={}
# これらの変数には便利なアクセサを使ってアクセス可能です。
layer.kernel, layer.bias
# + [markdown] colab_type="text" id="O0kDbE54-5VS"
# ## カスタムレイヤーの実装
#
# 独自のレイヤーを実装する最良の方法は、tf.keras.Layer クラスを拡張し、下記のメソッドを実装することです。
# * `__init__` , 入力に依存しないすべての初期化を行う
# * `build`, 入力の `shape` を知った上で、残りの初期化を行う
# * `call`, フォワード計算を行う
#
# `build` が呼ばれるまで変数の生成を待つ必要はなく、`__init__` で作成できることに注意してください。しかしながら、`build` で変数を生成することの優位な点は、レイヤーがオペレーションをしようとする入力の `shape` に基づいて、後から定義できる点です。これに対して、`__init__` で変数を生成するということは、そのために必要な `shape` を明示的に指定する必要があるということです。
# + colab_type="code" id="5Byl3n1k5kIy" colab={}
class MyDenseLayer(tf.keras.layers.Layer):
def __init__(self, num_outputs):
super(MyDenseLayer, self).__init__()
self.num_outputs = num_outputs
def build(self, input_shape):
self.kernel = self.add_variable("kernel",
shape=[int(input_shape[-1]),
self.num_outputs])
def call(self, input):
return tf.matmul(input, self.kernel)
layer = MyDenseLayer(10)
print(layer(tf.zeros([10, 5])))
print(layer.trainable_variables)
# + [markdown] colab_type="text" id="tk8E2vY0-z4Z"
# できるだけ標準のレイヤーを使ったほうが、概してコードは読みやすく保守しやすくなります。コードを読む人は標準的なレイヤーの振る舞いに慣れているからです。`tf.keras.layers` にはないレイヤーを使いたい場合には、[githubのイシュー](http://github.com/tensorflow/tensorflow/issues/new)を登録するか、もっとよいのはプルリクエストを送ることです。
# + [markdown] colab_type="text" id="Qhg4KlbKrs3G"
# ## モデル:レイヤーの組み合わせ
#
# 機械学習では、多くのレイヤーに類するものが、既存のレイヤーを組み合わせることで実装されています。例えば、ResNetの残差ブロックは、畳込み、バッチ正規化とショートカットの組み合わせです。
#
# 他のレイヤーからなるレイヤーに類するものを定義する際の主役は、tf.keras.Model クラスです。このクラスを継承することで実装できます。
# + colab_type="code" id="N30DTXiRASlb" colab={}
class ResnetIdentityBlock(tf.keras.Model):
def __init__(self, kernel_size, filters):
super(ResnetIdentityBlock, self).__init__(name='')
filters1, filters2, filters3 = filters
self.conv2a = tf.keras.layers.Conv2D(filters1, (1, 1))
self.bn2a = tf.keras.layers.BatchNormalization()
self.conv2b = tf.keras.layers.Conv2D(filters2, kernel_size, padding='same')
self.bn2b = tf.keras.layers.BatchNormalization()
self.conv2c = tf.keras.layers.Conv2D(filters3, (1, 1))
self.bn2c = tf.keras.layers.BatchNormalization()
def call(self, input_tensor, training=False):
x = self.conv2a(input_tensor)
x = self.bn2a(x, training=training)
x = tf.nn.relu(x)
x = self.conv2b(x)
x = self.bn2b(x, training=training)
x = tf.nn.relu(x)
x = self.conv2c(x)
x = self.bn2c(x, training=training)
x += input_tensor
return tf.nn.relu(x)
block = ResnetIdentityBlock(1, [1, 2, 3])
print(block(tf.zeros([1, 2, 3, 3])))
print([x.name for x in block.trainable_variables])
# + [markdown] colab_type="text" id="wYfucVw65PMj"
# しかし、ほとんどの場合には、モデルはレイヤーを次々に呼び出すことで構成されます。tf.keras.Sequential クラスを使うことで、これをかなり短いコードで実装できます。
# + colab_type="code" id="L9frk7Ur4uvJ" colab={}
my_seq = tf.keras.Sequential([tf.keras.layers.Conv2D(1, (1, 1),
input_shape=(
None, None, 3)),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.Conv2D(2, 1,
padding='same'),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.Conv2D(3, (1, 1)),
tf.keras.layers.BatchNormalization()])
my_seq(tf.zeros([1, 2, 3, 3]))
# + [markdown] colab_type="text" id="c5YwYcnuK-wc"
# # 次のステップ
#
# それでは、前出のノートブックに戻り、線形回帰の例を、レイヤーとモデルを使って、より構造化された形で実装してみてください。
| site/ja/tutorials/customization/custom_layers.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/sanikamal/tensorflow-AtoZ/blob/master/Image_Classification_with_CNNs.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] colab_type="text" id="jYysdyb-CaWM"
# # Image Classification with Convolutional Neural Networks
# + [markdown] colab_type="text" id="FbVhjPpzn6BM"
# Now, we'll build and train a neural network to classify images of clothing, like sneakers and shirts.
# + [markdown] colab_type="text" id="H0tMfX2vR0uD"
# ## Install and import dependencies
#
# We'll need [TensorFlow Datasets](https://www.tensorflow.org/datasets/), an API that simplifies downloading and accessing datasets, and provides several sample datasets to work with. We're also using a few helper libraries.
# + colab_type="code" id="P7mUJVqcINSM" colab={}
# !pip install -U tensorflow_datasets
# + colab_type="code" id="dzLKpmZICaWN" colab={}
from __future__ import absolute_import, division, print_function, unicode_literals
# Import TensorFlow and TensorFlow Datasets
import tensorflow as tf
import tensorflow_datasets as tfds
tf.logging.set_verbosity(tf.logging.ERROR)
# Helper libraries
import math
import numpy as np
import matplotlib.pyplot as plt
# Improve progress bar display
import tqdm
import tqdm.auto
tqdm.tqdm = tqdm.auto.tqdm
print(tf.__version__)
# This will go away in the future.
# If this gives an error, you might be running TensorFlow 2 or above
# If so, the just comment out this line and run this cell again
tf.enable_eager_execution()
# + [markdown] colab_type="text" id="yR0EdgrLCaWR"
# ## Import the Fashion MNIST dataset
# + [markdown] colab_type="text" id="DLdCchMdCaWQ"
# This guide uses the [Fashion MNIST](https://github.com/zalandoresearch/fashion-mnist) dataset, which contains 70,000 grayscale images in 10 categories. The images show individual articles of clothing at low resolution (28 $\times$ 28 pixels), as seen here:
#
# <table>
# <tr><td>
# <img src="https://tensorflow.org/images/fashion-mnist-sprite.png"
# alt="Fashion MNIST sprite" width="600">
# </td></tr>
# <tr><td align="center">
# <b>Figure 1.</b> <a href="https://github.com/zalandoresearch/fashion-mnist">Fashion-MNIST samples</a> (by Zalando, MIT License).<br/>
# </td></tr>
# </table>
#
# Fashion MNIST is intended as a drop-in replacement for the classic [MNIST](http://yann.lecun.com/exdb/mnist/) dataset—often used as the "Hello, World" of machine learning programs for computer vision. The MNIST dataset contains images of handwritten digits (0, 1, 2, etc) in an identical format to the articles of clothing we'll use here.
#
# This guide uses Fashion MNIST for variety, and because it's a slightly more challenging problem than regular MNIST. Both datasets are relatively small and are used to verify that an algorithm works as expected. They're good starting points to test and debug code.
#
# We will use 60,000 images to train the network and 10,000 images to evaluate how accurately the network learned to classify images. You can access the Fashion MNIST directly from TensorFlow, using the [Datasets](https://www.tensorflow.org/datasets) API:
# + colab_type="code" id="7MqDQO0KCaWS" colab={}
dataset, metadata = tfds.load('fashion_mnist', as_supervised=True, with_info=True)
train_dataset, test_dataset = dataset['train'], dataset['test']
# + [markdown] colab_type="text" id="t9FDsUlxCaWW"
# Loading the dataset returns metadata as well as a *training dataset* and *test dataset*.
#
# * The model is trained using `train_dataset`.
# * The model is tested against `test_dataset`.
#
# The images are 28 $\times$ 28 arrays, with pixel values in the range `[0, 255]`. The *labels* are an array of integers, in the range `[0, 9]`. These correspond to the *class* of clothing the image represents:
#
# <table>
# <tr>
# <th>Label</th>
# <th>Class</th>
# </tr>
# <tr>
# <td>0</td>
# <td>T-shirt/top</td>
# </tr>
# <tr>
# <td>1</td>
# <td>Trouser</td>
# </tr>
# <tr>
# <td>2</td>
# <td>Pullover</td>
# </tr>
# <tr>
# <td>3</td>
# <td>Dress</td>
# </tr>
# <tr>
# <td>4</td>
# <td>Coat</td>
# </tr>
# <tr>
# <td>5</td>
# <td>Sandal</td>
# </tr>
# <tr>
# <td>6</td>
# <td>Shirt</td>
# </tr>
# <tr>
# <td>7</td>
# <td>Sneaker</td>
# </tr>
# <tr>
# <td>8</td>
# <td>Bag</td>
# </tr>
# <tr>
# <td>9</td>
# <td>Ankle boot</td>
# </tr>
# </table>
#
# Each image is mapped to a single label. Since the *class names* are not included with the dataset, store them here to use later when plotting the images:
# + colab_type="code" id="IjnLH5S2CaWx" colab={}
class_names = ['T-shirt/top', 'Trouser', 'Pullover', 'Dress', 'Coat',
'Sandal', 'Shirt', 'Sneaker', 'Bag', 'Ankle boot']
# + [markdown] colab_type="text" id="Brm0b_KACaWX"
# ### Explore the data
#
# Let's explore the format of the dataset before training the model. The following shows there are 60,000 images in the training set, and 10000 images in the test set:
# + colab_type="code" id="MaOTZxFzi48X" colab={}
num_train_examples = metadata.splits['train'].num_examples
num_test_examples = metadata.splits['test'].num_examples
print("Number of training examples: {}".format(num_train_examples))
print("Number of test examples: {}".format(num_test_examples))
# + [markdown] colab_type="text" id="ES6uQoLKCaWr"
# ## Preprocess the data
#
# The value of each pixel in the image data is an integer in the range `[0,255]`. For the model to work properly, these values need to be normalized to the range `[0,1]`. So here we create a normalization function, and then apply it to each image in the test and train datasets.
# + colab_type="code" id="nAsH3Zm-76pB" colab={}
def normalize(images, labels):
images = tf.cast(images, tf.float32)
images /= 255
return images, labels
# The map function applies the normalize function to each element in the train
# and test datasets
train_dataset = train_dataset.map(normalize)
test_dataset = test_dataset.map(normalize)
# + [markdown] colab_type="text" id="lIQbEiJGXM-q"
# ### Explore the processed data
#
# Let's plot an image to see what it looks like.
# + colab_type="code" id="oSzE9l7PjHx0" colab={}
# Take a single image, and remove the color dimension by reshaping
for image, label in test_dataset.take(1):
break
image = image.numpy().reshape((28,28))
# Plot the image - voila a piece of fashion clothing
plt.figure()
plt.imshow(image, cmap=plt.cm.binary)
plt.colorbar()
plt.grid(False)
plt.show()
# + [markdown] colab_type="text" id="Ee638AlnCaWz"
# Display the first 25 images from the *training set* and display the class name below each image. Verify that the data is in the correct format and we're ready to build and train the network.
# + colab_type="code" id="oZTImqg_CaW1" colab={}
plt.figure(figsize=(10,10))
i = 0
for (image, label) in test_dataset.take(25):
image = image.numpy().reshape((28,28))
plt.subplot(5,5,i+1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(image, cmap=plt.cm.binary)
plt.xlabel(class_names[label])
i += 1
plt.show()
# + [markdown] colab_type="text" id="59veuiEZCaW4"
# ## Build the model
#
# Building the neural network requires configuring the layers of the model, then compiling the model.
# + [markdown] colab_type="text" id="Gxg1XGm0eOBy"
# ### Setup the layers
#
# The basic building block of a neural network is the *layer*. A layer extracts a representation from the data fed into it. Hopefully, a series of connected layers results in a representation that is meaningful for the problem at hand.
#
# Much of deep learning consists of chaining together simple layers. Most layers, like `tf.keras.layers.Dense`, have internal parameters which are adjusted ("learned") during training.
# + colab_type="code" id="9ODch-OFCaW4" colab={}
model = tf.keras.Sequential([
tf.keras.layers.Conv2D(32, (3,3), padding='same', activation=tf.nn.relu,
input_shape=(28, 28, 1)),
tf.keras.layers.MaxPooling2D((2, 2), strides=2),
tf.keras.layers.Conv2D(64, (3,3), padding='same', activation=tf.nn.relu),
tf.keras.layers.MaxPooling2D((2, 2), strides=2),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(128, activation=tf.nn.relu),
tf.keras.layers.Dense(10, activation=tf.nn.softmax)
])
# + [markdown] colab_type="text" id="gut8A_7rCaW6"
# This network layers are:
#
# * **"convolutions"** `tf.keras.layers.Conv2D and MaxPooling2D`— Network start with two pairs of Conv/MaxPool. The first layer is a Conv2D filters (3,3) being applied to the input image, retaining the original image size by using padding, and creating 32 output (convoluted) images (so this layer creates 32 convoluted images of the same size as input). After that, the 32 outputs are reduced in size using a MaxPooling2D (2,2) with a stride of 2. The next Conv2D also has a (3,3) kernel, takes the 32 images as input and creates 64 outputs which are again reduced in size by a MaxPooling2D layer. So far in the course, we have described what a Convolution does, but we haven't yet covered how you chain multiples of these together. We will get back to this in lesson 4 when we use color images. At this point, it's enough if you understand the kind of operation a convolutional filter performs
#
# * **output** `tf.keras.layers.Dense` — A 128-neuron, followed by 10-node *softmax* layer. Each node represents a class of clothing. As in the previous layer, the final layer takes input from the 128 nodes in the layer before it, and outputs a value in the range `[0, 1]`, representing the probability that the image belongs to that class. The sum of all 10 node values is 1.
#
#
# ### Compile the model
#
# Before the model is ready for training, it needs a few more settings. These are added during the model's *compile* step:
#
#
# * *Loss function* — An algorithm for measuring how far the model's outputs are from the desired output. The goal of training is this measures loss.
# * *Optimizer* —An algorithm for adjusting the inner parameters of the model in order to minimize loss.
# * *Metrics* —Used to monitor the training and testing steps. The following example uses *accuracy*, the fraction of the images that are correctly classified.
# + colab_type="code" id="Lhan11blCaW7" colab={}
model.compile(optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
# + [markdown] colab_type="text" id="qKF6uW-BCaW-"
# ## Train the model
#
# First, we define the iteration behavior for the train dataset:
# 1. Repeat forever by specifying `dataset.repeat()` (the `epochs` parameter described below limits how long we perform training).
# 2. The `dataset.shuffle(60000)` randomizes the order so our model cannot learn anything from the order of the examples.
# 3. And `dataset.batch(32)` tells `model.fit` to use batches of 32 images and labels when updating the model variables.
#
# Training is performed by calling the `model.fit` method:
# 1. Feed the training data to the model using `train_dataset`.
# 2. The model learns to associate images and labels.
# 3. The `epochs=5` parameter limits training to 5 full iterations of the training dataset, so a total of 5 * 60000 = 300000 examples.
# + colab_type="code" id="o_Dp8971McQ1" colab={}
BATCH_SIZE = 32
train_dataset = train_dataset.repeat().shuffle(num_train_examples).batch(BATCH_SIZE)
test_dataset = test_dataset.batch(BATCH_SIZE)
# + colab_type="code" id="xvwvpA64CaW_" colab={}
model.fit(train_dataset, epochs=10, steps_per_epoch=math.ceil(num_train_examples/BATCH_SIZE))
# + [markdown] colab_type="text" id="W3ZVOhugCaXA"
# As the model trains, the loss and accuracy metrics are displayed. This model reaches an accuracy of about 0.97 (or 97%) on the training data.
# + [markdown] colab_type="text" id="oEw4bZgGCaXB"
# ## Evaluate accuracy
#
# Next, compare how the model performs on the test dataset. Use all examples we have in the test dataset to assess accuracy.
# + colab_type="code" id="VflXLEeECaXC" colab={}
test_loss, test_accuracy = model.evaluate(test_dataset, steps=math.ceil(num_test_examples/32))
print('Accuracy on test dataset:', test_accuracy)
# + [markdown] colab_type="text" id="yWfgsmVXCaXG"
# As it turns out, the accuracy on the test dataset is smaller than the accuracy on the training dataset. This is completely normal, since the model was trained on the `train_dataset`. When the model sees images it has never seen during training, (that is, from the `test_dataset`), we can expect performance to go down.
# + [markdown] colab_type="text" id="xsoS7CPDCaXH"
# ## Make predictions and explore
#
# With the model trained, we can use it to make predictions about some images.
# + colab_type="code" id="Ccoz4conNCpl" colab={}
for test_images, test_labels in test_dataset.take(1):
test_images = test_images.numpy()
test_labels = test_labels.numpy()
predictions = model.predict(test_images)
# + colab_type="code" id="Gl91RPhdCaXI" colab={}
predictions.shape
# + [markdown] colab_type="text" id="x9Kk1voUCaXJ"
# Here, the model has predicted the label for each image in the testing set. Let's take a look at the first prediction:
# + colab_type="code" id="3DmJEUinCaXK" colab={}
predictions[0]
# + [markdown] colab_type="text" id="-hw1hgeSCaXN"
# A prediction is an array of 10 numbers. These describe the "confidence" of the model that the image corresponds to each of the 10 different articles of clothing. We can see which label has the highest confidence value:
# + colab_type="code" id="qsqenuPnCaXO" colab={}
np.argmax(predictions[0])
# + [markdown] colab_type="text" id="E51yS7iCCaXO"
# So the model is most confident that this image is a shirt, or `class_names[6]`. And we can check the test label to see this is correct:
# + colab_type="code" id="Sd7Pgsu6CaXP" colab={}
test_labels[0]
# + [markdown] colab_type="text" id="ygh2yYC972ne"
# We can graph this to look at the full set of 10 channels
# + colab_type="code" id="DvYmmrpIy6Y1" colab={}
def plot_image(i, predictions_array, true_labels, images):
predictions_array, true_label, img = predictions_array[i], true_labels[i], images[i]
plt.grid(False)
plt.xticks([])
plt.yticks([])
plt.imshow(img[...,0], cmap=plt.cm.binary)
predicted_label = np.argmax(predictions_array)
if predicted_label == true_label:
color = 'blue'
else:
color = 'red'
plt.xlabel("{} {:2.0f}% ({})".format(class_names[predicted_label],
100*np.max(predictions_array),
class_names[true_label]),
color=color)
def plot_value_array(i, predictions_array, true_label):
predictions_array, true_label = predictions_array[i], true_label[i]
plt.grid(False)
plt.xticks([])
plt.yticks([])
thisplot = plt.bar(range(10), predictions_array, color="#777777")
plt.ylim([0, 1])
predicted_label = np.argmax(predictions_array)
thisplot[predicted_label].set_color('red')
thisplot[true_label].set_color('blue')
# + [markdown] colab_type="text" id="d4Ov9OFDMmOD"
# Let's look at the 0th image, predictions, and prediction array.
# + colab_type="code" id="HV5jw-5HwSmO" colab={}
i = 0
plt.figure(figsize=(6,3))
plt.subplot(1,2,1)
plot_image(i, predictions, test_labels, test_images)
plt.subplot(1,2,2)
plot_value_array(i, predictions, test_labels)
# + colab_type="code" id="Ko-uzOufSCSe" colab={}
i = 12
plt.figure(figsize=(6,3))
plt.subplot(1,2,1)
plot_image(i, predictions, test_labels, test_images)
plt.subplot(1,2,2)
plot_value_array(i, predictions, test_labels)
# + [markdown] colab_type="text" id="kgdvGD52CaXR"
# Let's plot several images with their predictions. Correct prediction labels are blue and incorrect prediction labels are red. The number gives the percent (out of 100) for the predicted label. Note that it can be wrong even when very confident.
# + colab_type="code" id="hQlnbqaw2Qu_" colab={}
# Plot the first X test images, their predicted label, and the true label
# Color correct predictions in blue, incorrect predictions in red
num_rows = 5
num_cols = 3
num_images = num_rows*num_cols
plt.figure(figsize=(2*2*num_cols, 2*num_rows))
for i in range(num_images):
plt.subplot(num_rows, 2*num_cols, 2*i+1)
plot_image(i, predictions, test_labels, test_images)
plt.subplot(num_rows, 2*num_cols, 2*i+2)
plot_value_array(i, predictions, test_labels)
# + [markdown] colab_type="text" id="R32zteKHCaXT"
# Finally, use the trained model to make a prediction about a single image.
# + colab_type="code" id="yRJ7JU7JCaXT" colab={}
# Grab an image from the test dataset
img = test_images[0]
print(img.shape)
# + [markdown] colab_type="text" id="vz3bVp21CaXV"
# `tf.keras` models are optimized to make predictions on a *batch*, or collection, of examples at once. So even though we're using a single image, we need to add it to a list:
# + colab_type="code" id="lDFh5yF_CaXW" colab={}
# Add the image to a batch where it's the only member.
img = np.array([img])
print(img.shape)
# + [markdown] colab_type="text" id="EQ5wLTkcCaXY"
# Now predict the image:
# + colab_type="code" id="o_rzNSdrCaXY" colab={}
predictions_single = model.predict(img)
print(predictions_single)
# + colab_type="code" id="6Ai-cpLjO-3A" colab={}
plot_value_array(0, predictions_single, test_labels)
_ = plt.xticks(range(10), class_names, rotation=45)
# + [markdown] colab_type="text" id="cU1Y2OAMCaXb"
# `model.predict` returns a list of lists, one for each image in the batch of data. Grab the predictions for our (only) image in the batch:
# + colab_type="code" id="2tRmdq_8CaXb" colab={}
np.argmax(predictions_single[0])
# + [markdown] colab_type="text" id="YFc2HbEVCaXd"
# And, as before, the model predicts a label of 6 (shirt).
| Image_Classification_with_CNNs.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/fernandofsilva/Keras/blob/main/TensorFlow_Datasets.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="foeS1tOWS3_R"
# # TensorFlow Datasets
# + [markdown] id="nufn0cmRS3_U"
# In this reading notebook, we will take a look at the `tensorflow-datasets` library.
#
# We have previously made use of the `tf.keras.datasets` package, which gave us access to a variety of useful datasets such as the IMDB movie dataset, the CIFAR-100 small image classification dataset, and the MNIST handwritten digits dataset.
#
# The `tensorflow-datasets` library gives us another means of accessing a variety of useful datasets.
# + [markdown] id="lS2iKimnS3_W"
# ## Installation
# The `tensorflow-datasets` library is installed independently of TensorFlow itself. It can be installed using pip, by running the following command in a terminal (assuming that `tensorflow` has already been installed):
#
# ```pip install tensorflow-datasets ```
# + [markdown] id="n62afsmtS3_Y"
# ## Listing the Available Datasets
# The `list_builders` function can be used to list the available datasets.
# + id="N_hy64QAS3_Z" outputId="6d0c07bc-01fc-4f3b-bee0-23dedce772d2" colab={"base_uri": "https://localhost:8080/"}
# List available datasets
import tensorflow_datasets as tfds
tfds.list_builders()
# + [markdown] id="S2WdBo0nS3_e"
# ## Loading a Dataset
#
# Loading a particular dataset is straightforward; simply use the `load` function, specifying the `name` and any other keyword arguments. In the example code below, we demonstrate how to load the `kmnist` dataset using the function.
#
# As stated on the documentation page, running the function with `split=None` (the default) returns a dictionary of the splits, with the keys `test` and `train`.
# + id="vi3dORwiS3_f" outputId="88e8e8e8-5f0d-4784-df85-6d718d38ad1c" colab={"base_uri": "https://localhost:8080/", "height": 347, "referenced_widgets": ["06c1d9bd45364ae28273f1aeb2264559", "1459a4101bfd49aab815d82b999dd2ec", "b8dddf5007f94de8aede32c01a6b6c53", "288db5fa8fa9467d93a6022d6f7c3bab", "428e8597eae949c994b5b3681b57d529", "486963d4c2164c46b5bda05095377b92", "fedccc9adf2e42229c2b412cb01f0089", "4e38553e53604f83bde4f9bec096656d", "75d7933ac525435d88a3da278b471ce1", "3d638a17e50b49818a8f841d5b0ec726", "759f8e97b6a0467e86374a81c11e4f1e", "11daaeac90064115a31433b38fc1ce63", "<KEY>", "<KEY>", "8366e7279f1d490aade15ecacc9af173", "c887eafbe737478ca03286a896f05602", "48e817b3efd447ae95dba5ebeeafc52c", "<KEY>", "8dc8080679c74c74b7930e65f1e09956", "<KEY>", "<KEY>", "<KEY>", "<KEY>", "b3a4f7f8fcb941d9b04e365dd203ec72", "0cc35c71f6b747e985144d51ec895540", "bfe4fb5234574128b668ab8e48da4658", "c71cca4a480d4e89bdf482586c9aeb62", "59ec6feb09ec4bd1b3be08e03ce23edc", "<KEY>", "eef9113e9cca4405963878a8a1372dda", "<KEY>", "29f0acdb637d4ef4b1e3a515bd79380f", "<KEY>", "<KEY>", "a2124407ed1f40f6b43a07168841b714", "<KEY>", "<KEY>", "<KEY>", "<KEY>", "<KEY>", "a2d80dcb8b174d92b76e685267b5a8dd", "<KEY>", "<KEY>", "8fbb5cc84c084eb0b462ac5eacd5175c", "ce2da6ddb8b04326bbeadaaf4b63e424", "1c02b25ccad344bdae139cecfe20df73", "3fe248cf58854e63838d0fa0023588aa", "9aa25771e6724fe6939a4d95d6a43a26", "063464cdb48247748134e793f06949e5", "cae436f1ed144abfb00d46c40c81e8e8", "953dfb8ee672462d9d49830c937fae57", "007d8cef34f34923abf532231a1a3589", "<KEY>", "d0b0a2f2e5df491694673012654d7aec", "9ac3de5a9cba491faeb5188e6d3f7bbb", "<KEY>"]}
# Load the mnist_corrupted dataset
kmnist = tfds.load(name="kmnist", split=None)
kmnist_train = kmnist['train']
kmnist_test = kmnist['test']
# + id="ZGpug34nS3_j" outputId="111c9baa-c905-47a5-bc5d-e777e7eb19db" colab={"base_uri": "https://localhost:8080/", "height": 483}
# View some examples from the dataset
import matplotlib.pyplot as plt
# %matplotlib inline
import tensorflow as tf
fig, axes = plt.subplots(3, 3, figsize=(8, 8))
fig.subplots_adjust(hspace=0.2, wspace=0.1)
for i, (elem, ax) in enumerate(zip(kmnist_train, axes.flat)):
image = tf.squeeze(elem['image'])
label = elem['label']
ax.imshow(image, cmap='gray')
ax.text(0.7, -0.12, f'Digit = {label}', ha='right',
transform=ax.transAxes, color='black')
ax.set_xticks([])
ax.set_yticks([])
# + [markdown] id="Zlsxm19aS3_n"
# ## Further reading and resources
# * https://www.tensorflow.org/datasets
# * https://www.tensorflow.org/datasets/catalog/overview
# * https://wwww.tensorflow.org/datasets/api_docs/python/tfds
# * https://tensorflow.org/datasets/api_docs/python/tfds/load
| Data Pipeline/TensorFlow_Datasets.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# __Ray Tracing__
#
# In this example, we'll use the 'ray-tracing' module, to setup the same lens-plane + source-plane strong lens configuration as the previous tutorial, but with a lot less lines of code!
# +
# %matplotlib inline
from autolens.model.profiles import light_profiles
from autolens.model.profiles import mass_profiles
from autolens.model.galaxy import galaxy
from autolens.lens import ray_tracing
from autolens.data.array import grids
from autolens.lens.plotters import plane_plotters
from autolens.lens.plotters import ray_tracing_plotters
# -
# Let use the same grid-stack we've all grown to know and love by now!
image_plane_grid_stack = grids.GridStack.from_shape_pixel_scale_and_sub_grid_size(shape=(100, 100), pixel_scale=0.05,
sub_grid_size=2)
# For our lens galaxy, we'll use the same SIS mass profile as before.
sis_mass_profile = mass_profiles.SphericalIsothermal(centre=(0.0, 0.0), einstein_radius=1.6)
lens_galaxy = galaxy.Galaxy(mass=sis_mass_profile)
print(lens_galaxy)
# And for our source galaxy, the same Sersic light profile
sersic_light_profile = light_profiles.SphericalSersic(centre=(0.0, 0.0), intensity=1.0, effective_radius=1.0,
sersic_index=1.0)
source_galaxy = galaxy.Galaxy(light=sersic_light_profile)
print(source_galaxy)
# Now, lets use the lens and source galaxies to ray-trace our grid-stack, using a 'tracer' from the ray-tracing module. When we pass our galaxies and grid-stack into the Tracer below, the following happens:
#
# 1) Using the lens-galaxy's mass-profile, the deflection angle of every image-plane grid coordinate is computed.
#
# 2) These deflection angles are used to trace every image-plane coordinate to a source-plane coordinate.
#
# 3) This creates the source-plane grid-stack of lensed coordinates.
tracer = ray_tracing.TracerImageSourcePlanes(lens_galaxies=[lens_galaxy], source_galaxies=[source_galaxy],
image_plane_grid_stack=image_plane_grid_stack)
# The tracer is composed of an image-plane and source-plane, just like in the previous example!
print('Regular-grid image-plane coordinate 1')
print(tracer.image_plane.grid_stack.regular[0])
print('Regular-grid image-plane coordinate 2')
print(tracer.image_plane.grid_stack.regular[1])
print('Regular-grid image-plane coordinate 3')
print(tracer.image_plane.grid_stack.regular[2])
# And the source-plane's grid has been deflected.
print('Regular-grid source-plane coordinate 1')
print(tracer.source_plane.grid_stack.regular[0])
print('Regular-grid source-plane coordinate 2')
print(tracer.source_plane.grid_stack.regular[1])
print('Regular-grid source-plane coordinate 3')
print(tracer.source_plane.grid_stack.regular[2])
# We can use the plane_plotter to plot these grids, like we did before.
plane_plotters.plot_plane_grid(plane=tracer.image_plane, title='Image-plane Grid')
plane_plotters.plot_plane_grid(plane=tracer.source_plane, title='Source-plane Grid')
plane_plotters.plot_plane_grid(plane=tracer.source_plane, axis_limits=[-0.1, 0.1, -0.1, 0.1], title='Source-plane Grid')
# PyAutoLens has tools for plotting a tracer. A ray-tracing subplot plots the following:
#
# 1) The image-plane image, computed by tracing the source galaxy's light 'forwards' through the tracer.
#
# 2) The source-plane image, showing the source galaxy's true appearance (i.e. if it were not lensed).
#
# 3) The image-plane surface density, computed using the lens galaxy's mass profile.
#
# 4) The image-plane gravitational potential, computed using the lens galaxy's mass profile.
#
# 5) The image-plane deflection angles, computed using the lens galaxy's mass profile.
ray_tracing_plotters.plot_ray_tracing_subplot(tracer=tracer)
# Just like for a plane, these attributes can be accessed by print statements (converted to 2D NumPy arrays the same dimensions as our input grid-stack!).
print('Tracer - Surface Density - Regular-grid coordinate 1:')
print(tracer.surface_density[0,0])
print('Tracer - Surface Density - Regular-grid coordinate 2:')
print(tracer.surface_density[0,1])
print('Tracer - Surface Density - Regular-grid coordinate 3:')
print(tracer.surface_density[0,2])
print('Tracer - Surface Density - Regular-grid coordinate 101:')
print(tracer.surface_density[1,0])
# Of course, these surface densities are identical to the image-plane surface densities, as it's only the lens galaxy that contributes to the overall mass of the ray-tracing system.
print('Image-Plane - Surface Density - Regular-grid coordinate 1:')
print(tracer.image_plane.surface_density[0,0])
print('Image-Plane - Surface Density - Regular-grid coordinate 2:')
print(tracer.image_plane.surface_density[0,1])
print('Image-Plane - Surface Density - Regular-grid coordinate 3:')
print(tracer.image_plane.surface_density[0,2])
print('Image-Plane - Surface Density - Regular-grid coordinate 101:')
print(tracer.image_plane.surface_density[1,0])
# I've left the rest below commented to avoid too many print statements, but if you're feeling adventurous go ahead and uncomment the lines below!
# +
# print('Potential:')
# print(tracer.potential)
# print(tracer.image_plane.potential)
# print('Deflections:')
# print(tracer.deflections_x)
# print(tracer.deflections_y)
# print(tracer.image_plane.deflections_x)
# print(tracer.image_plane.deflections_y)
# print('Image-plane Image:')
# print(tracer.image_plane_image)
# print(tracer.image_plane.image_plane_image)
# print('Source-plane Image:')
# print(tracer.source_plane_image)
# print(tracer.image_plane.source_plane_image)
# -
# You can also plot the above attributes on individual figures, using appropriate ray-tracing plotter (I've left most commented out again for convinience)
ray_tracing_plotters.plot_surface_density(tracer=tracer)
# ray_tracing_plotters.plot_potential(tracer=tracer)
# ray_tracing_plotters.plot_deflections_y(tracer=tracer)
#ray_tracing_plotters.plot_deflections_x(tracer=tracer)
# ray_tracing_plotters.plot_image_plane_image(tracer=tracer)
# Before we finish, you might be wondering 'why do both the image-plane and tracer have the attributes surface density / potential / deflection angles, when the two are identical'. Afterall, only mass profiles contribute to these quantities, and only the image-plane has galaxies with measureable mass profiles! There are two reasons:
#
# 1) Convinience - You could always write 'tracer.image_plane.surface_density' and 'plane_plotters.surface_density(plane=tracer.image_plane). However, code appears neater if you can just write 'tracer.surface_density' and 'ray_tracing_plotters.plot_surface_density(tracer=tracer).
#
# 2) Multi-plane lensing - For now, we're focused on the simplest lensing configuratio possible, an image-plane + source-plane configuration. However, there are strong lens system where there are more than 2 planes! In these instances, the surface density, potential and deflections of each plane is different to the overall values given by the tracer. This is beyond the scope of this chapter, but be reassured that what you're learning now will prepare you for the advanced chapters later on!
#
# And with that, we're done. You've performed your first ray-tracing with PyAutoLens! There are no exercises for this chapter, and we're going to take a deeper look at ray-tracing in the next chapter.
| workspace/howtolens/chapter_1_introduction/tutorial_5_ray_tracing.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [conda env:ws]
# language: python
# name: conda-env-ws-py
# ---
# - Here we explore the use of `vtki`to create 3D objects we then display using `ipyvolume`.
# - In the future we might want to use `itkwidgets` to display 3d objects. See https://github.com/InsightSoftwareConsortium/itk-jupyter-widgets/issues/112
#
# +
# %load_ext autoreload
# %autoreload 2
# %matplotlib inline
from pathlib import Path
import sys
sys.path.append("../../../")
import anamic
import numpy as np
import matplotlib.pyplot as plt
import ipyvolume as ipv
import vtki
# +
n_pf = 11
mt_length_nm = 200 # nm
taper_length_nm = 0 # nm
# Create a MicrotubuleSimulator object.
dimers = anamic.simulator.dimers_builder(n_pf, mt_length_nm, taper_length_nm)
# Set parameters for the image generation.
parameters = {}
parameters['labeling_ratio'] = 0.1 # from 0 to 1
parameters['pixel_size'] = 110 # nm/pixel
parameters['x_offset'] = 1500 # nm
parameters['y_offset'] = 1500 # nm
parameters['psf_size'] = 135 # nm
parameters['signal_mean'] = 700
parameters['signal_std'] = 100
parameters['bg_mean'] = 500
parameters['bg_std'] = 24
parameters['noise_factor'] = 1
parameters['snr_line_width'] = 3 # pixel
ms = anamic.simulator.MicrotubuleSimulator(dimers)
ms.parameters.update(parameters)
# Build the geometry.
ms.build_positions(apply_random_z_rotation=True, show_progress=True)
ms.label()
ms.project()
ms.random_rotation_projected()
# +
import matplotlib
def make_sphere(radius, center, direction=[0, 0, 1], sphere_resolution=50):
sphere = vtki.Sphere(radius, center, direction,
theta_resolution=sphere_resolution, phi_resolution=sphere_resolution)
x = sphere.points[:, 0]
y = sphere.points[:, 1]
z = sphere.points[:, 2]
# The faces returned by `PolyData` can be different
# than triangle, so we assume `vtki.Sphere.faces` returns
# only triangles.
triangles = np.array([sphere.faces[1::4], sphere.faces[2::4], sphere.faces[3::4]]).T
return x, y, z, triangles
radius = 4
direction = [0, 0, 1]
sphere_resolution = 30
color_feature_name = 'pf'
cmap_name="tab20c"
# Only show visible dimers
selected_dimers = ms.positions[ms.positions['visible'] == True]
# Build the dimer's 3D objects.
objects = []
positions = selected_dimers
for _, dimer in positions.iterrows():
center = dimer[['x', 'y', 'z']].values.astype('float32')
obj = make_sphere(radius, center, direction, sphere_resolution)
objects.append(obj)
# Display the 3D scene
if color_feature_name:
# TODO: that code should be much simpler...
cmap = matplotlib.cm.get_cmap(cmap_name)
categories = selected_dimers[color_feature_name].unique()
color_indices = cmap([i / len(categories) for i in categories])
colors = np.zeros((len(selected_dimers[color_feature_name]), 4))
for color_index in range(len(categories)):
colors[selected_dimers[color_feature_name] == categories[color_index]] = color_indices[color_index]
else:
colors = np.repeat('#e4191b', selected_dimers.shape[0])
fig = ipv.figure()
for (x, y, z, triangles), color in zip(objects, colors):
ipv.plot_trisurf(x, y, z, triangles=triangles, color=color)
ipv.squarelim()
ipv.show()
# -
# ---
# +
# Display the PDB model of the alpha/beta-tubulin dimers.
model_path = Path('/home/hadim/Documents/Code/Postdoc/Data/tubulin-dimer.stl')
dimer = vtki.PolyData(str(model_path))
# Move dimer to center of mass
mass_center = dimer.points.mean(axis=0)
dimer.translate(mass_center * -1)
x = dimer.points[:, 0]
y = dimer.points[:, 1]
z = dimer.points[:, 2]
# The faces returned by `PolyData` can be different
# than triangle, so we assume `vtki.Sphere.faces` returns
# only triangles.
triangles = np.array([dimer.faces[1::4], dimer.faces[2::4], dimer.faces[3::4]]).T
ipv.figure()
ipv.plot_trisurf(x, y, z, triangles=triangles, color='orange')
ipv.xyzlim(-50, 50)
ipv.show()
# -
| notebooks/Development/Simulator/Viz.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: env-mafia
# language: python
# name: env-mafia
# ---
# # Modelling with Word Vectors
# I will be using [Spacy](https://spacy.io/) alongside sklearn in this notebook.
import os
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# %matplotlib inline
import warnings
import bay12_solution_eposts as solution
# ## Load data
post, thread = solution.prepare.load_dfs('train')
post.head(2)
thread.head(2)
# I will set the thread number to be the index, to simplify matching in the future:
thread = thread.set_index('thread_num')
thread.head(2)
# We'll load the label map as well, which tells us which index goes to which label
label_map = solution.prepare.load_label_map()
label_map
# ## Vectorize our text features
# ### Load a Spacy model to get word/sentence vectors
# I'll be using the large English model (~800 MB size) as shown [here](https://spacy.io/usage/models).
import en_core_web_lg
nlp = en_core_web_lg.load()
ex_name = thread['thread_name'].iloc[0]
doc = nlp(ex_name)
doc
# Average vector for the entire name
doc.vector[:4]
# ### Get documents for 'thread_name', 'first' (first post), maybe even 'join' (joining of all posts)
# **NOTE**: Not using the "whole thread" text because it takes a long time to calculate.
# Feel free to use the first line below, instead of the second, to add the 'join' column.
# thread_texts = post.groupby('thread_num')['text'].agg(['first', ' '.join])
thread_texts = post.groupby('thread_num')['text'].agg(['first'])
thread_texts = pd.concat(
[
thread[['thread_name']],
thread_texts
],
axis='columns'
)
thread_texts.head()
# +
import itertools
text_feature_names = [
'%s_%s' % (col, num)
for col, num
in itertools.product(thread_texts.columns, range(300))
]
def vectorize_row(row, cols=thread_texts.columns, text_feature_names=text_feature_names):
"""Vectorizes a row of texts."""
res = np.array([])
for col in cols:
txt = row.loc[col][:100000] # limit is 10x bigger, but we want to be safe :)
res = np.r_[res, nlp(txt).vector]
# v0 = nlp(row.loc['thread_name']).vector
# v1 = nlp(row.loc['first']).vector
# v2 = nlp(row.loc['join']).vector
return pd.Series(res, text_feature_names)
# -
thread_text_vectors = thread_texts.apply(vectorize_row, axis='columns')
thread_text_vectors.head()
# ## Create "final" dataset
# We only have one non-text feature, i.e. the number of posts. We'll use it and its log, and we'll scale them to be in `[-1, 1]`
thread_numeric_vectors = pd.DataFrame({
'posts': (thread['thread_replies'] + 1),
'posts_log': np.log(thread['thread_replies'] + 1),
})
thread_numeric_vectors.head(2)
# +
text_features = thread_text_vectors.columns
numeric_features = thread_numeric_vectors.columns
X = pd.concat([thread_numeric_vectors, thread_text_vectors], axis='columns')
X.head()
# -
# Our targets are the same as the second model:
# +
y = thread['thread_label_id']
y_aux = y.apply(lambda x: 0 if (x==label_map['other']) else 1).rename('is_game')
pd.concat([y, y_aux], axis='columns').head()
# -
# ### Review
# So, what have I done so far? Let's list.
#
# - Selected title, first post, and maybe a concatenation of all posts as our "documents".
# - Turned each "document" into a vector, using pre-trained word vectors (the document vector is the average of the word vectors).
# - Added number of posts (and its log) as additional features.
#
# Note that we probably need to scale the latter two for some models, because the others components are normalized to 1.
# ## Split dataset into "training" and "validation"
# In order to check the quality of our model in a more realistic setting, we will split all our input (training) data into a "training set" (which our model will see and learn from) and a "validation set" (where we see how well our model generalized). [Relevant link](https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.train_test_split.html).
from sklearn.model_selection import train_test_split
# NOTE: setting the `random_state` lets you get the same results with the pseudo-random generator
validation_pct = 0.25
X_train, X_val = train_test_split(X, test_size=validation_pct, random_state=99)
# +
idx_train = X_train.index
idx_val = X_val.index
X_train.shape, X_val.shape
# -
# ## Fit first (auxilliary) model
from sklearn.metrics import confusion_matrix, accuracy_score, classification_report
from sklearn.ensemble import RandomForestClassifier
cls1 = RandomForestClassifier(n_estimators=100, max_depth=2, random_state=42)
cls1.fit(X_train, y_aux.reindex(idx_train))
# In-sample and out-of-sample predictions for auxilliary target
y_aux_train_pred = pd.Series(cls1.predict(X_train), index=idx_train)
y_aux_val_pred = pd.Series(cls1.predict(X_val), index=idx_val)
y_t, y_p = y_aux.reindex(idx_train), y_aux_train_pred
print("Aux train:")
print("Accuracy:", accuracy_score(y_t, y_p))
print("Confusion:", confusion_matrix(y_t, y_p), sep="\n")
y_t, y_p = y_aux.reindex(idx_val), y_aux_val_pred
print("Aux validation:")
print("Accuracy:", accuracy_score(y_t, y_p))
print("Confusion:", confusion_matrix(y_t, y_p), sep="\n")
# ## Fit second (game) model
# Our training index is: train states, where we know we have games (that is, y_aux == 1)
idx_game_train = y_aux[y_aux == 1].reindex(idx_train).dropna().index
# CHECK
(y_aux[idx_game_train] == 1).all()
# Our validation index is: validation states, where we PREDICTED we have games (that is, y_aux_val_pred == 1)
idx_game_val = y_aux_val_pred[y_aux_val_pred == 1].dropna().index
# CHECK
(y_aux_val_pred[idx_game_val] == 1).all()
cls2 = RandomForestClassifier(n_estimators=200, max_depth=3, max_leaf_nodes=10, random_state=68)
cls2.fit(X_train.reindex(idx_game_train), y.reindex(idx_game_train))
# In-sample and out-of-sample predictions for auxilliary target
y_game_train_pred = pd.Series(cls2.predict(X_train.reindex(idx_game_train)), index=idx_game_train)
y_game_val_pred = pd.Series(cls2.predict(X_val.reindex(idx_game_val)), index=idx_game_val)
y_t, y_p = y.reindex(idx_game_train), y_game_train_pred
print("Game train:")
print("Accuracy:", accuracy_score(y_t, y_p))
print("Confusion:", confusion_matrix(y_t, y_p), sep="\n")
y_t, y_p = y.reindex(idx_game_val), y_game_val_pred
print("Game validation:")
print("Accuracy:", accuracy_score(y_t, y_p))
print("Confusion:", confusion_matrix(y_t, y_p), sep="\n")
# Well obviously the algorithm overfit on the majority class (the fifth column, with '26' on the diagonal).
#
# We will tune hyperparameters in another notebook. However, this seems to have worked out well enough.
# ## Score the resulting model
# Our model consists of two parts - let's see how well we did altogether:
# Fill with "other", and when an actual game - fill with the game :)
y_train_pred = pd.Series(label_map['other'], index=idx_train)
y_train_pred[idx_game_train] = y_game_train_pred
# Same with the validation, because our index is dynamic :)
y_val_pred = pd.Series(label_map['other'], index=idx_val)
y_val_pred[idx_game_val] = y_game_val_pred
y_t, y_p = y.reindex(idx_train), y_train_pred
print("Total train:")
print("Accuracy:", accuracy_score(y_t, y_p))
print("Confusion:", confusion_matrix(y_t, y_p), sep="\n")
y_t, y_p = y.reindex(idx_val), y_val_pred
print("Total val:")
print("Accuracy:", accuracy_score(y_t, y_p))
print("Confusion:", confusion_matrix(y_t, y_p), sep="\n")
print(classification_report(y_t, y_p))
# 0.75 accuracy! That's pretty good for a slightly-tuned model. That's significantly better than the baseline of ~0.55!
#
# We still have some classes that aren't predicted in the validation set (actually quite a few - 7 out of 12 have 0 predicted!), which is pretty bad (obviously).
# However, we did predict something for all but 3 of them on the training set (and those 3 had 4 threads in total... so...).
#
# Let's freeze this model for now, and move to the next notebook. I won't predict on the test set, because I can see public *and* private scores, but here is one place where I would suggest you do it yourself. ;)
| notebooks/3_word_vector_model.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Politician Activity on Facebook
#
# The parameters in the cell below can be adjusted to explore other Politicians and time frames.
#
# ### How to explore other politicians?
# The ***politician_id*** is an internal identifier that connects the different social media accounts. You can [use this other notebook](../Politicians.ipynb?autorun=true) to get other the identifiers of other Politicians.
#
# ***Alternatively***, you can direcly use the [Politicians API](http://mediamonitoring.gesis.org/api/Politicians/swagger/), or access it with the [SMM Wrapper](https://pypi.org/project/smm-wrapper/).
#
# ## A. Set Up parameters
# Parameters:
politician_id = 1274143252682258
from_date = '2017-09-01'
to_date = '2018-12-31'
aggregation = 'week'
# ## B. Using the SMM Politician API
# +
# Create an instance to the smm wrapper
from smm_wrapper import SMMPoliticians
smm = SMMPoliticians()
# using the api to get the posts and comments activity
posts = smm.api.posts_by(_id=politician_id, from_date=from_date, to_date=to_date, aggregate_by=aggregation)
comments = smm.api.comments_by(_id=politician_id, from_date=from_date, to_date=to_date, aggregate_by=aggregation)
# -
# ## C. Plotting
# ### C.1 Plot Facebook Post Activity
# +
import plotly
from plotly import graph_objs as go
plotly.offline.init_notebook_mode(connected=True)
#plot for facebook posts activity
plotly.offline.iplot({
"data": [go.Scatter(x=posts['labels'], y=posts['values'], name='Posts', line_shape='spline'),
go.Scatter(x=posts['labels'], y=posts['replies'], name='Replies', line_shape='spline'),
go.Scatter(x=posts['labels'], y=posts['shares'], name='Shares', line_shape='spline'),
go.Scatter(x=posts['labels'], y=posts['reactions'], name='Reactions', line_shape='spline'),
go.Scatter(x=posts['labels'], y=posts['likes'], name='Likes', line_shape='spline')],
"layout": go.Layout(title='Facebook (Posts Activity)', yaxis=dict(title='N'))
})
# -
# ### C.2 Plot Facebook Comment Activity
# plot for facebook comments activity
plotly.offline.iplot({
"data": [go.Scatter(x=comments['labels'], y=comments['values'], name='Comments', line_shape='spline'),
go.Scatter(x=comments['labels'], y=comments['replies'], name='Replies', line_shape='spline'),
go.Scatter(x=comments['labels'], y=comments['likes'], name='Likes', line_shape='spline')],
"layout": go.Layout(title='Facebook (Comments Activity)', yaxis=dict(title='N'))
})
| python/politician/facebook.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/Vinaypatil-Ev/vinEvPy-GoCoLab/blob/main/PIL/PracPaper2_PIL.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="V8A74Stlye6p"
# # PIL
#
# + id="kDQX-eY2mo9e"
from PIL import Image
# + id="qV1p_RgazC0u"
from google.colab.patches import cv2_imshow
import cv2
def imshow(path:str, by="cv"):
if by == "cv":
im = cv2.imread(path)
cv2_imshow(im)
elif by == "pil":
if isinstance(path, PIL.JpegImagePlugin.JpegImageFile):
return path
im = Image.open(path)
return im
# + [markdown] id="DRikHnNgyib1"
# ## Open Image
# + id="Oi-XEO__uHfz"
im_path = "2.jpg"
im = Image.open(im_path)
# + id="Ok9yN6idTILf"
type(im)
# + id="AawZcbjQzUff"
print(im)
# + id="yyRY_Ia3ut_k"
im.format
# + id="EdC7QN46uvJS"
im.size
# + id="DGpHHsQNuz6n"
im.mode
# + id="1e9-XxwC0FbW"
show_img(im_path, "pil")
# + [markdown] id="XdR3tqOoxH6B"
# ## Create thumbnail
# + id="1b9iRDAfu0sa"
size = (128, 128)
im.thumbnail(size)
im.save("thumb1.jpg", "JPEG")
# + id="fQ2SoB3GxZqO"
im2 = Image.open("thumb1.jpg")
im2.size
# + id="RQ_s_tjix07h"
print(f"{im2.size}x{im2.mode}")
# + [markdown] id="cP6WT8Hgyqh0"
# ## Cutting
# ## Pasting
# ## Merging
# + [markdown] id="rKIjjZHDy6_D"
# ### Croping subrectangle from image
# + id="v3RG1b92y6mB"
box = (100, 100, 400, 400)
region = im.crop(box)
# + id="XxO2gRUgyPxD"
region
# + id="fZtRDGdW0yKT"
region = region.transpose(Image.ROTATE_180)
region
# + id="_9xCt7Jb1DE4"
im.paste(region, box)
# + id="Uq7kKIz_1Po6"
im
# + [markdown] id="RSi3Njd-10K7"
# ## Rolling an Image
# + id="61840xcI1Qws"
def roll(img, delta):
x, y = img.size
delta %= x
if delta == 0:
return img
p1 = img.crop((0, 0, delta, y))
p2 = img.crop((delta, 0, x, y))
img.paste(p1, (x-delta, 0, x, y))
img.paste(p2, (0, 0, x-delta, y))
return img
# + id="O3ZJNqRv21yz"
roll(Image.open(im_path), 300)
# + [markdown] id="0AwV0obW4AFN"
# ## Spliting and merging bands
# + id="UYNW9Whz4GzO"
im = Image.open(im_path)
# + id="4lGsKY683TWc"
r, g, b = im.split()
Image.merge("RGB", (b, g, r))
# + id="9enFxR8Y4ZqS"
r
# + id="dxtfiMPe4jzt"
g
# + id="AfJK7Y8Z4kcl"
b
# + [markdown] id="RVCpCEnn485E"
# ## Geometrical Transform
# + [markdown] id="M1W0bDi85Dj6"
# ### simple geometry transform
# + id="-ezyIICS4kwR"
im.resize((128, 128))
# + id="NzJwXbZv5Mdr"
im.rotate(45)
# + id="IJFp-Quq5PvM"
im.transpose(Image.FLIP_LEFT_RIGHT)
# + id="psbx3qBl5yCe"
im.transpose(Image.ROTATE_90)
# + [markdown] id="aaGDUHJG6HFh"
# ## Color Transform
# + [markdown] id="WaaxkP6p6Fqr"
# ### Coverting between modes
# + id="PG5EwIsJ5202"
im.convert("L")
# + [markdown] id="Zc82LEuH6ha-"
# ## Image Inhancement
# + [markdown] id="536uaxly6qpV"
# ### Filter
# + id="DObqSiMz6ydv"
from PIL import ImageFilter
# + id="2XzemrKt6TLV"
im.filter(ImageFilter.DETAIL)
# + [markdown] id="LG79Gl_A7Aia"
# ## Point Operations
# + [markdown] id="-2jV54UO7XcX"
# ### Apply point transform
# + id="Yg3k4_RF6-pu"
im.point(lambda i: i * 1.2)
# + [markdown] id="Zi-Ifd5U8Bsp"
# ### processing individual bands
# + id="juLnqVEe7hAz"
r, g, b = im.split()
# + id="GgT6nEbF8Jq7"
mask = r.point(lambda i: i < 100 and 255)
out = g.point(lambda i: i * 0.7)
g.paste(out, None, mask)
Image.merge(im.mode, [r, g, b])
# + [markdown] id="Sc2psz9X-MDP"
# ## More Enhancement
# + id="K_G-_LiV8v5Y"
from PIL import ImageEnhance
# + id="HDP7y9kv-UXW"
enh = ImageEnhance.Contrast(im)
enh.enhance(1.3)
# + [markdown] id="6vHJm9hGFIG4"
# ### Using the ImageSequence Iterator class
# + id="06KLAhti-eFW"
from PIL import ImageSequence
# + id="IkWI90MBIUVf"
gif = "/content/drive/MyDrive/dataSet/img/3.gif"
# + id="jJu85go5IN2E"
im = Image.open(gif)
# for i in range(10):
# x = im.seek(im.tell() + i)
# print(type(x))
# + id="nWwdYZMlFbZ9"
f = ImageSequence.Iterator(im)
# + id="j3DhuRVoHTi_"
next(f)
# + [markdown] id="NzVtRkUbJGJe"
# ## Drawing Postscript
# + id="92c1oLDJIjDB"
from PIL import PSDraw
# + id="N8hqzvezJenG"
im = Image.open(im_path)
# + id="SygpHUIWI9-i"
box = (1*72, 2*72, 7*72, 10*72)
title = "iga"
ps = PSDraw.PSDraw()
ps.begin_document(title)
ps.image(box, im, 75)
ps.rectangle(box)
ps.setfont("HelveticaNarrow-Bold", 36)
ps.text((3*72, 4*72), title)
ps.end_document()
# + id="T4DpPC92LAnT"
with Image.open(im_path) as im:
title = "hopper"
box = (1*72, 2*72, 7*72, 10*72) # in points
ps = PSDraw.PSDraw() # default is sys.stdout
ps.begin_document(title)
# draw the image (75 dpi)
ps.image(box, im, 75)
ps.rectangle(box)
# draw title
ps.setfont("HelveticaNarrow-Bold", 36)
ps.text((3*72, 4*72), title)
ps.end_document()
# + id="_KiW4uuGKY6t"
im
# + id="xyCwg6cNKw3m"
from PIL import ImageDraw, ImageFont
# + id="tP0--nWfVjq5"
im = Image.open(im_path)
# + id="hwLXF1a-UYjB"
draw = ImageDraw.Draw(im)
# + id="2tfHQdmlUjLg"
# font = ImageFont.load("arial.ttf")
draw.text((10, 10), "hello", "red")
# + id="Ei6mMsc4U-Dk"
im
# + id="l67oYRFuU-W0"
import PIL
# + id="QGLhb_EYW7AF"
PIL.ImageShow.show(im)
# + id="ZKu-I7omW9zW"
| PIL/PracPaper2_PIL.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ### 制作数据集
# 下载原始数据集:https://www.kaggle.com/c/dogs-vs-cats/data
#
# 这个数据集包含25000张猫狗图像(每个类别都有12 500张),大小为543MB(压缩后)。
#
# 本次训练并不使用全部图像,只是使用其中的4000幅图像(每个类别各2000张)。
# 下载数据并解压之后,创建一个新数据集,其中包含三个子集:
# - train : 2000张图像(1000张猫图像,1000张狗图像)
# - validation:1000张图像(500张猫图像,500张狗图像)
# - test:1000张图像(500张猫图像,500张狗图像)
# 防止报错 OMP: Error #15: Initializing libiomp5.dylib, but found libiomp5.dylib already initialized.
import os
os.environ["KMP_DUPLICATE_LIB_OK"]="TRUE"
# +
# 创建目录
import os, shutil
# 提取部分图片组成一个较小的数据集
base_dir = './data/cats_and_dogs_small'
if not os.path.exists(base_dir):
os.mkdir(base_dir)
# /cats_and_dogs_small/train
train_dir = os.path.join(base_dir, 'train')
if not os.path.exists(train_dir):
os.mkdir(train_dir)
# /cats_and_dogs_small/validation
validation_dir = os.path.join(base_dir, 'validation')
if not os.path.exists(validation_dir):
os.mkdir(validation_dir)
# /cats_and_dogs_small/test
test_dir = os.path.join(base_dir, 'test')
if not os.path.exists(test_dir):
os.mkdir(test_dir)
# /cats_and_dogs_small/train/cats
train_cats_dir = os.path.join(train_dir, 'cats')
if not os.path.exists(train_cats_dir):
os.mkdir(train_cats_dir)
# /cats_and_dogs_small/train/dogs
train_dogs_dir = os.path.join(train_dir, 'dogs')
if not os.path.exists(train_dogs_dir):
os.mkdir(train_dogs_dir)
# /cats_and_dogs_small/validation/cats
validation_cats_dir = os.path.join(validation_dir, 'cats')
if not os.path.exists(validation_cats_dir):
os.mkdir(validation_cats_dir)
# /cats_and_dogs_small/validation/dogs
validation_dogs_dir = os.path.join(validation_dir, 'dogs')
if not os.path.exists(validation_dogs_dir):
os.mkdir(validation_dogs_dir)
# /cats_and_dogs_small/test/cats
test_cats_dir = os.path.join(test_dir, 'cats')
if not os.path.exists(test_cats_dir):
os.mkdir(test_cats_dir)
# /cats_and_dogs_small/test/dogs
test_dogs_dir = os.path.join(test_dir, 'dogs')
if not os.path.exists(test_dogs_dir):
os.mkdir(test_dogs_dir)
# +
# 复制图像到相应的目录
#原始数据集的目录
original_dataset_dir = '/mnt/资料/人工智能/python深度学习代码和数据集/python深度学习代码和数据集/data/kaggle_original_data'
# 制作数据集
# 将前1000张猫的图像复制到train_cats_dir
if not os.listdir(train_cats_dir):
fnames = ['cat.{}.jpg'.format(i) for i in range(1000)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(train_cats_dir, fname)
shutil.copyfile(src, dst)
# 将接下来的500张猫的图像复制到validation_cats_dir
if not os.listdir(validation_cats_dir):
fnames = ['cat.{}.jpg'.format(i) for i in range(1000, 1500)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(validation_cats_dir, fname)
shutil.copyfile(src, dst)
# 将接下来的500张猫的图像复制到test_cats_dir
if not os.listdir(test_cats_dir):
fnames = ['cat.{}.jpg'.format(i) for i in range(1500, 2000)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(test_cats_dir, fname)
shutil.copyfile(src, dst)
# 将1000张狗的图像复制到train_dogs_dir
if not os.listdir(train_dogs_dir):
fnames = ['dog.{}.jpg'.format(i) for i in range(1000)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(train_dogs_dir, fname)
shutil.copyfile(src, dst)
# 将接下来500张狗的图像复制到validation_dogs_dir
if not os.listdir(validation_dogs_dir):
fnames = ['dog.{}.jpg'.format(i) for i in range(1000, 1500)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(validation_dogs_dir, fname)
shutil.copyfile(src, dst)
# 将接下来500张狗的图像复制到test_dogs_dir
if not os.listdir(test_dogs_dir):
fnames = ['dog.{}.jpg'.format(i) for i in range(1500, 2000)]
for fname in fnames:
src = os.path.join(original_dataset_dir, fname)
dst = os.path.join(test_dogs_dir, fname)
shutil.copyfile(src, dst)
# +
# 查看生成的数据集
print('total training cat images:', len(os.listdir(train_cats_dir)))
print('total training dog images:', len(os.listdir(train_dogs_dir)))
print('total validation cat images:', len(os.listdir(validation_cats_dir)))
print('total validation dog images:', len(os.listdir(validation_dogs_dir)))
print('total test cat images:', len(os.listdir(test_cats_dir)))
print('total test dog images:', len(os.listdir(test_dogs_dir)))
| 5.2.1 卷积神经网络-猫狗分类-制作数据集.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import tensorflow as tf
import matplotlib.pyplot as plt
mnist = tf.keras.datasets.mnist
(training_images, training_labels), (test_images, test_labels) = mnist.load_data()
# normalize all images to 0-1 range
training_images = training_images/255.0
test_images = test_images/255.0
models = [tf.keras.models.Sequential([
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(512, activation=tf.nn.relu),
tf.keras.layers.Dense(10, activation=tf.nn.softmax)
]),
tf.keras.models.Sequential([
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(512, activation=tf.nn.relu),
tf.keras.layers.Dense(10, activation=tf.nn.softmax)
]),
tf.keras.models.Sequential([
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(512, activation=tf.nn.relu),
tf.keras.layers.Dense(10, activation=tf.nn.softmax)
]),
tf.keras.models.Sequential([
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(512, activation=tf.nn.relu),
tf.keras.layers.Dense(10, activation=tf.nn.softmax)
]),
tf.keras.models.Sequential([
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(512, activation=tf.nn.relu),
tf.keras.layers.Dense(10, activation=tf.nn.softmax)
])
]
# compile all models
[x.compile(optimizer='adam',
loss='sparse_categorical_crossentropy') for x in models]
# checking effects of epochs on model (accuracy increases presumption)
fits = [
models[0].fit(training_images, training_labels, epochs=5),
models[1].fit(training_images, training_labels, epochs=10),
models[2].fit(training_images, training_labels, epochs=15),
models[3].fit(training_images, training_labels, epochs=20),
models[4].fit(training_images, training_labels, epochs=30)
]
evaluations = [model.evaluate(test_images, test_labels) for model in models]
classifications = [model.predict(test_images) for model in models]
classifications = [x*100.0 for x in classifications ]
print([classifications[i][0] for i in range(len(classifications))])
plt.imshow(test_images[0])
| deeplearning.ai-tensorflow-developer-certificate/1-of-4-intro-to-tf/Week_2/Mnist.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/bioduds/ALGORITMOS-UFMG/blob/main/TP1_Galeria_de_Arte_Eduardo_Capanema_Algoritmos_II.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="1XzBRzKbWs7s"
# # Trabalho Prático 1 - Galeria de Arte
#
# ---
# ## Algoritmos I
# ---
#
# **<NAME> 2020041515**
#
# ### Descrição do Problema
# ---
#
# Queremos escrever uma implementação para o problema da galeria de arte (triangulação de polígonos), visto em sala de aula.
#
# ### Algoritmos e Estruturas
# ---
#
# Precisaremos desenvolver uma implementação do algoritmo Ear-Clipping. Em seguida, devemos realizar uma 3-coloração do grafo obtido pela triangulação.
#
#
# + id="5rcXwwGJWkAy"
# Juntar imports aqui
import numpy as np
from matplotlib import pyplot as plt
# + [markdown] id="3MB-7zPfsd4w"
# # Gerando polígonos convexos
# ---
#
# A base da nossa solução é partir de um polígono convexo que representa uma galeria de arte genérica.
#
# + colab={"base_uri": "https://localhost:8080/", "height": 265} id="9ERODpiTAD1I" outputId="e83a4eed-9f54-4db3-d488-0e2614451e52"
import numpy as np
from matplotlib import pyplot as plt
def poligono_convexo_aleatorio( ax, n ) :
x = np.random.randint( 0, 200, n )
y = np.random.randint( 0, 200, n )
center_point = [np.sum(x)/n, np.sum( y )/n]
angles = np.arctan2( x-center_point[0], y-center_point[1] )
sort_tups = sorted( [( i, j, k ) for i, j, k in zip( x, y, angles )], key=lambda t:t[2] )
if len( sort_tups ) != len( set( sort_tups ) ) :
raise Exception( 'two equal coordinates -- exiting' )
x, y, angles = zip( *sort_tups )
x = list( x )
y = list( y )
x.append( x[0] )
y.append( y[0] )
ax.plot( x, y, label = '<NAME>' )
if __name__ == '__main__':
fig, ax = plt.subplots()
poligono_convexo_aleatorio( ax, 15 )
ax.legend()
plt.show()
# + [markdown] id="2e72jOPlYnNZ"
# ### Realizando a Triangulação
# ---
#
# Precisamos agora realizar a triangulação. Vamos utilizar o algoritmo Ear Clipping visto em aulas.
| TP1_Galeria_de_Arte_Eduardo_Capanema_Algoritmos_II.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .jl
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Julia 1.4.0
# language: julia
# name: julia-1.4
# ---
using CSV, DataFrames
using Dates
basepath = normpath("../../");
start_date = Date(2020, 5, 1)
end_date = Date(2020, 6, 1);
locations = ["AL","AZ","AR","CA","CO","CT","DE","FL","GA","ID","IL","IN","IA","KS","KY","LA","ME","MD","MA","MI","MN","MS","MO","MT","NE","NV","NH","NJ","NM","NY","NC","ND","OH","OK","OR","PA","RI","SC","SD","TN","TX","UT","VT","VA","WA","WV","WI","WY"]
sort!(locations);
# +
function load_ihme(;forecast_date::String="2020-06-03")
forecast_data = CSV.read(joinpath(basepath, "data/forecasts/ihme/$(forecast_date)/forecast.csv"), copycols=true)
state_list = CSV.read(joinpath(basepath, "data/geography/states.csv"), copycols=true)
ga = state_list[(state_list.abbrev .== "GA"),:]
ga.state .= "Georgia_two"
append!(state_list, ga)
filter!(row -> row.location_name in state_list.state, forecast_data)
state_cvt = Dict(state.state => state.abbrev for state in eachrow(state_list))
forecast_data.state = [state_cvt[row.location_name] for row in eachrow(forecast_data)]
sort!(forecast_data, [:state, :date])
return forecast_data
end
function ihme_filter!(forecast::DataFrame, states::Array{String,1}, start_date::Date, end_date::Date)
@assert states == sort(states)
@assert start_date <= end_date
filter!(row -> row.state in states, forecast)
filter!(row -> start_date <= row.date <= end_date, forecast)
return forecast
end
function ihme_forecast(forecast::DataFrame; forecast_type::Symbol=:active, patient_type::Symbol=:regular, bound_type::Symbol=:mean)
forecast_by_state = groupby(forecast, :state, sort=true)
col_select = Dict(
(:admitted, :regular, :mean) => :admis_mean,
(:admitted, :regular, :lb) => :admis_lower,
(:admitted, :regular, :ub) => :admis_upper,
(:active, :regular, :mean) => :allbed_mean,
(:active, :regular, :lb) => :allbed_lower,
(:active, :regular, :ub) => :allbed_upper,
(:admitted, :icu, :mean) => :newICU_mean,
(:admitted, :icu, :lb) => :newICU_lower,
(:admitted, :icu, :ub) => :newICU_upper,
(:active, :icu, :mean) => :ICUbed_mean,
(:active, :icu, :lb) => :ICUbed_lower,
(:active, :icu, :ub) => :ICUbed_upper,
)
col = col_select[(forecast_type, patient_type, bound_type)]
patients = vcat([f[:,col]' for f in forecast_by_state]...)
return Float32.(patients)
end;
# -
forecast = load_ihme()
forecast = ihme_filter!(forecast, locations, start_date, end_date)
patients = ihme_forecast(forecast, forecast_type=:admitted, patient_type=:regular, bound_type=:mean);
size(patients)
| casestudies/statelevel/dev/load_ihme.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] colab_type="text" id="view-in-github"
# <a href="https://colab.research.google.com/github/NeuromatchAcademy/course-content/blob/content%2Fw2d5_tutorial_revs/tutorials/W2D5_ReinforcementLearning/student/W2D5_Tutorial1.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] colab_type="text" id="YL2_mjdRyaSg"
# # Neuromatch Academy: Week 2, Day 5, Tutorial 1
# # Learning to Predict
#
# __Content creators:__ <NAME> and <NAME> with help from <NAME>
#
# __Content reviewers:__ <NAME> and <NAME>
#
# + [markdown] colab_type="text" id="JTnwS5ZOyaSm"
# ---
#
# # Tutorial objectives
#
# In this tutorial, we will learn how to estimate state-value functions in a classical conditioning paradigm using Temporal Difference (TD) learning and examine TD-errors at the presentation of the conditioned and unconditioned stimulus (CS and US) under different CS-US contingencies. These exercises will provide you with an understanding of both how reward prediction errors (RPEs) behave in classical conditioning and what we should expect if Dopamine represents a "canonical" model-free RPE.
#
# At the end of this tutorial:
# * You will learn to use the standard tapped delay line conditioning model
# * You will understand how RPEs move to CS
# * You will understand how variability in reward size effects RPEs
# * You will understand how differences in US-CS timing effect RPEs
# + cellView="code" colab={} colab_type="code" id="Tr8AjcsFyaSh"
# Imports
import numpy as np
import matplotlib.pyplot as plt
# + cellView="form" colab={} colab_type="code" id="OjLe4R9uumHw"
#@title Figure settings
import ipywidgets as widgets # interactive display
# %config InlineBackend.figure_format = 'retina'
plt.style.use("/share/dataset/COMMON/nma.mplstyle.txt")
# + cellView="form" colab={} colab_type="code" id="UBN49ga4u7_n"
# @title Helper functions
from matplotlib import ticker
def plot_value_function(V, ax=None, show=True):
"""Plot V(s), the value function"""
if not ax:
fig, ax = plt.subplots()
ax.stem(V, use_line_collection=True)
ax.set_ylabel('Value')
ax.set_xlabel('State')
ax.set_title("Value function: $V(s)$")
if show:
plt.show()
def plot_tde_trace(TDE, ax=None, show=True, skip=400):
"""Plot the TD Error across trials"""
if not ax:
fig, ax = plt.subplots()
indx = np.arange(0, TDE.shape[1], skip)
im = ax.imshow(TDE[:,indx])
positions = ax.get_xticks()
# Avoid warning when setting string tick labels
ax.xaxis.set_major_locator(ticker.FixedLocator(positions))
ax.set_xticklabels([f"{int(skip * x)}" for x in positions])
ax.set_title('TD-error over learning')
ax.set_ylabel('State')
ax.set_xlabel('Iterations')
ax.figure.colorbar(im)
if show:
plt.show()
def learning_summary_plot(V, TDE):
"""Summary plot for Ex1"""
fig, (ax1, ax2) = plt.subplots(nrows = 2, gridspec_kw={'height_ratios': [1, 2]})
plot_value_function(V, ax=ax1, show=False)
plot_tde_trace(TDE, ax=ax2, show=False)
plt.tight_layout()
def reward_guesser_title_hint(r1, r2):
""""Provide a mildly obfuscated hint for a demo."""
if (r1==14 and r2==6) or (r1==6 and r2==14):
return "Technically correct...(the best kind of correct)"
if ~(~(r1+r2) ^ 11) - 1 == (6 | 24): # Don't spoil the fun :-)
return "Congratulations! You solved it!"
return "Keep trying...."
#@title Default title text
class ClassicalConditioning:
def __init__(self, n_steps, reward_magnitude, reward_time):
# Task variables
self.n_steps = n_steps
self.n_actions = 0
self.cs_time = int(n_steps/4) - 1
# Reward variables
self.reward_state = [0,0]
self.reward_magnitude = None
self.reward_probability = None
self.reward_time = None
self.set_reward(reward_magnitude, reward_time)
# Time step at which the conditioned stimulus is presented
# Create a state dictionary
self._create_state_dictionary()
def set_reward(self, reward_magnitude, reward_time):
"""
Determine reward state and magnitude of reward
"""
if reward_time >= self.n_steps - self.cs_time:
self.reward_magnitude = 0
else:
self.reward_magnitude = reward_magnitude
self.reward_state = [1, reward_time]
def get_outcome(self, current_state):
"""
Determine next state and reward
"""
# Update state
if current_state < self.n_steps - 1:
next_state = current_state + 1
else:
next_state = 0
# Check for reward
if self.reward_state == self.state_dict[current_state]:
reward = self.reward_magnitude
else:
reward = 0
return next_state, reward
def _create_state_dictionary(self):
"""
This dictionary maps number of time steps/ state identities
in each episode to some useful state attributes:
state - 0 1 2 3 4 5 (cs) 6 7 8 9 10 11 12 ...
is_delay - 0 0 0 0 0 0 (cs) 1 1 1 1 1 1 1 ...
t_in_delay - 0 0 0 0 0 0 (cs) 1 2 3 4 5 6 7 ...
"""
d = 0
self.state_dict = {}
for s in range(self.n_steps):
if s <= self.cs_time:
self.state_dict[s] = [0,0]
else:
d += 1 # Time in delay
self.state_dict[s] = [1,d]
class MultiRewardCC(ClassicalConditioning):
"""Classical conditioning paradigm, except that one randomly selected reward,
magnitude, from a list, is delivered of a single fixed reward."""
def __init__(self, n_steps, reward_magnitudes, reward_time=None):
""""Build a multi-reward classical conditioning environment
Args:
- nsteps: Maximum number of steps
- reward_magnitudes: LIST of possible reward magnitudes.
- reward_time: Single fixed reward time
Uses numpy global random state.
"""
super().__init__(n_steps, 1, reward_time)
self.reward_magnitudes = reward_magnitudes
def get_outcome(self, current_state):
next_state, reward = super().get_outcome(current_state)
if reward:
reward=np.random.choice(self.reward_magnitudes)
return next_state, reward
class ProbabilisticCC(ClassicalConditioning):
"""Classical conditioning paradigm, except that rewards are stochastically omitted."""
def __init__(self, n_steps, reward_magnitude, reward_time=None, p_reward=0.75):
""""Build a multi-reward classical conditioning environment
Args:
- nsteps: Maximum number of steps
- reward_magnitudes: Reward magnitudes.
- reward_time: Single fixed reward time.
- p_reward: probability that reward is actually delivered in rewarding state
Uses numpy global random state.
"""
super().__init__(n_steps, reward_magnitude, reward_time)
self.p_reward = p_reward
def get_outcome(self, current_state):
next_state, reward = super().get_outcome(current_state)
if reward:
reward*= int(np.random.uniform(size=1)[0] < self.p_reward)
return next_state, reward
# + [markdown] colab_type="text" id="0q72Sto0S2F5"
# ---
# # Section 1: TD-learning
# + cellView="form" colab={"base_uri": "https://localhost:8080/", "height": 518} colab_type="code" id="3-im6zneSRW7" outputId="d2302f76-1f1d-477e-a3f5-eafc27aab0d4"
#@title Video 1: Introduction
from IPython.display import IFrame
class BiliVideo(IFrame):
def __init__(self, id, page=1, width=400, height=300, **kwargs):
self.id=id
src = "https://player.bilibili.com/player.html?bvid={0}&page={1}".format(id, page)
super(BiliVideo, self).__init__(src, width, height, **kwargs)
video = BiliVideo(id='BV13f4y1d7om', width=854, height=480, fs=1)
print("Video available at https://www.bilibili.com/video/{0}".format(video.id))
video
# + [markdown] colab_type="text" id="HB2U9wCNyaSo"
# __Environment:__
#
# - The agent experiences the environment in episodes or trials.
# - Episodes terminate by transitioning to the inter-trial-interval (ITI) state and they are initiated from the ITI state as well. We clamp the value of the terminal/ITI states to zero.
# - The classical conditioning environment is composed of a sequence of states that the agent deterministically transitions through. Starting at State 0, the agent moves to State 1 in the first step, from State 1 to State 2 in the second, and so on. These states represent time in the tapped delay line representation
# - Within each episode, the agent is presented a CS and US (reward).
# - For each exercise, we use a different CS-US contingency.
# - The agent's goal is to learn to predict expected rewards from each state in the trial.
#
#
# __Definitions:__
#
# 1. Returns:
# \begin{align}
# G_{t} = r_{t+1} + \gamma r_{t+2} + \gamma^2 r_{t+3} + ... = \sum \limits_{k = 1}^{\infty} \gamma^{k-1} r_{t+k}
# \end{align}
#
# 2. Value:
# \begin{align}
# V(s_{t}) = \mathbb{E} [ G_{t} | s_{t}] = \mathbb{E} [r_{t+1} + \gamma V_{t+1} | s_{t}]
# \end{align}
#
# 3. TD-error:
# \begin{align}
# \delta_{t} = r_{t+1} + \gamma V(s_{t+1}) - V(s_{t})
# \end{align}
#
# 4. Value updates:
# \begin{align}
# V(s_{t}) \leftarrow V(s_{t}) + \alpha \delta_{t}
# \end{align}
#
# + [markdown] colab_type="text" id="jMwFbZ-GvLhc"
# ## Exercise 1: TD-learning with guaranteed rewards
#
# Implement TD-learning to estimate the state-value function in the classical-conditioning world with guaranteed rewards, with a fixed magnitude, at a fixed delay after the CS. Save TD-errors over learning so we can visualize them -- you're going to need to compute them anyway.
#
# Use the provided code to estimate the value function.
# + colab={} colab_type="code" id="mr6FG_wFu0RD"
def td_learner(env, n_trials, gamma=0.98, alpha=0.001):
""" Temporal Difference learning
Args:
env (object): the environment to be learned
n_trials (int): the number of trials to run
gamma (float): temporal discount factor
alpha (float): learning rate
Returns:
ndarray, ndarray: the value function and temporal difference error arrays
"""
V = np.zeros(env.n_steps) # Array to store values over states (time)
TDE = np.zeros((env.n_steps, n_trials)) # Array to store TD errors
for n in range(n_trials):
state = 0 # Initial state
for t in range(env.n_steps):
# Get next state and next reward
next_state, reward = env.get_outcome(state)
# Is the current state in the delay period (after CS)?
is_delay = env.state_dict[state][0]
########################################################################
## TODO for students: implement TD error and value function update
# Fill out function and remove
raise NotImplementedError("Student excercise: implement TD error and value function update")
#################################################################################
# Write an expression to compute the TD-error
TDE[state, n] = ...
# Write an expression to update the value function
V[state] += ...
# Update state
state = next_state
return V, TDE
# Uncomment once the td_learner function is complete
# env = ClassicalConditioning(n_steps=40, reward_magnitude=10, reward_time=10)
# V, TDE = td_learner(env, n_trials=20000)
# learning_summary_plot(V, TDE)
# + [markdown] colab={"base_uri": "https://localhost:8080/", "height": 429} colab_type="text" id="byxMuD1LxTVc" outputId="da1f9e77-97ec-456e-b6e5-3e64166ea982"
# [*Click for solution*](https://github.com/erlichlab/course-content/tree/master//tutorials/W2D5_ReinforcementLearning/solutions/W2D5_Tutorial1_Solution_0d9c50de.py)
#
# *Example output:*
#
# <img alt='Solution hint' align='left' width=558 height=414 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W2D5_ReinforcementLearning/static/W2D5_Tutorial1_Solution_0d9c50de_0.png>
#
#
# + [markdown] colab_type="text" id="ROKwAo1FKAqe"
# ## Interactive Demo 1: US to CS Transfer
#
# During classical conditioning, the subject's behavioral response (e.g., salivating) transfers from the unconditioned stimulus (US; like the smell of tasty food) to the conditioned stimulus (CS; like Pavlov ringing his bell) that predicts it. Reward prediction errors play an important role in this process by adjusting the value of states according to their expected, discounted return.
#
# Use the widget below to examine how reward prediction errors change over time. Before training (orange line), only the reward state has high reward prediction error. As training progresses (blue line, slider), the reward prediction errors shift to the conditioned stimulus, where they end up when the trial is complete (green line).
#
# Dopamine neurons, which are thought to carry reward prediction errors _in vivo_, show exactly the same behavior!
#
# + cellView="form" colab={"base_uri": "https://localhost:8080/", "height": 461, "referenced_widgets": ["1c791e2f446e4dd285c8967347be734b", "6cad7f364ffd4927926d0f6a08d6213f", "89c0229375a84de185c4f0114578ed09", "bbdcd8d2c92f425aa8c20f7e47e5e727", "495a849ed7304404ab2ba9e8bb9cc8ad", "02fd572f843e4138aee41d6f3b058790", "884533e643b54b4f80ea5023d9477626"]} colab_type="code" id="2UwSevZG82-M" outputId="e211b531-7b61-4a79-e0c6-f60e2a5b3504"
#@title
#@markdown Make sure you execute this cell to enable the widget!
n_trials = 20000
@widgets.interact
def plot_tde_by_trial(trial = widgets.IntSlider(value=5000, min=0, max=n_trials-1 , step=1, description="Trial #")):
if 'TDE' not in globals():
print("Complete Exercise 1 to enable this interactive demo!")
else:
fig, ax = plt.subplots()
ax.axhline(0, color='k') # Use this + basefmt=' ' to keep the legend clean.
ax.stem(TDE[:, 0], linefmt='C1-', markerfmt='C1d', basefmt=' ',
label="Before Learning (Trial 0)",
use_line_collection=True)
ax.stem(TDE[:, -1], linefmt='C2-', markerfmt='C2s', basefmt=' ',
label="After Learning (Trial $\infty$)",
use_line_collection=True)
ax.stem(TDE[:, trial], linefmt='C0-', markerfmt='C0o', basefmt=' ',
label=f"Trial {trial}",
use_line_collection=True)
ax.set_xlabel("State in trial")
ax.set_ylabel("TD Error")
ax.set_title("Temporal Difference Error by Trial")
ax.legend()
# + [markdown] colab_type="text" id="XZd8QkhKcHBQ"
# ## Interactive Demo 2: Learning Rates and Discount Factors
#
# Our TD-learning agent has two parameters that control how it learns: $\alpha$, the learning rate, and $\gamma$, the discount factor. In Exercise 1, we set these parameters to $\alpha=0.001$ and $\gamma=0.98$ for you. Here, you'll investigate how changing these parameters alters the model that TD-learning learns.
#
# Before enabling the interactive demo below, take a moment to think about the functions of these two parameters. $\alpha$ controls the size of the Value function updates produced by each TD-error. In our simple, deterministic world, will this affect the final model we learn? Is a larger $\alpha$ necessarily better in more complex, realistic environments?
#
# The discount rate $\gamma$ applies an exponentially-decaying weight to returns occuring in the future, rather than the present timestep. How does this affect the model we learn? What happens when $\gamma=0$ or $\gamma \geq 1$?
#
# Use the widget to test your hypotheses.
#
#
#
# + cellView="form" colab={"base_uri": "https://localhost:8080/", "height": 494, "referenced_widgets": ["77f998f3160140bfbcaaa6b3b981acb2", "41fdf67eefee4b4e807f85d23ed6c81a", "58cdf8f357e24943b1479cce0b1eb639", "7b34445f59344edcad2c064fece1f150", "40524e90c7c74da283515bc84ce2543c", "0b0b91f8fba14251b27ee28c70133854", "<KEY>", "a50c7b2a97834fc89e738d5b1f6fce23", "f2a024230f434794a0c83818e580c68f", "219655c48ebb44fca9d24225400ae3c6"]} colab_type="code" id="C_9pB-UhkfHy" outputId="a8819e1d-d39a-48a4-8942-296d27dee278"
#@title
#@markdown Make sure you execute this cell to enable the widget!
@widgets.interact
def plot_summary_alpha_gamma(alpha = widgets.FloatSlider(value=0.0001, min=0.001, max=0.1, step=0.0001, description="alpha"),
gamma = widgets.FloatSlider(value=0.980, min=0, max=1.1, step=0.010, description="gamma")):
env = ClassicalConditioning(n_steps=40, reward_magnitude=10, reward_time=10)
try:
V_params, TDE_params = td_learner(env, n_trials=20000, gamma=gamma, alpha=alpha)
except NotImplementedError:
print("Finish Exercise 1 to enable this interactive demo")
learning_summary_plot(V_params,TDE_params)
# + [markdown] colab={} colab_type="text" id="fSoRYk0DiAQA"
# [*Click for solution*](https://github.com/erlichlab/course-content/tree/master//tutorials/W2D5_ReinforcementLearning/solutions/W2D5_Tutorial1_Solution_12ce49be.py)
#
#
# + [markdown] colab_type="text" id="Eruos0VKGaG1"
# ---
# # Section 2: TD-learning with varying reward magnitudes
#
# In the previous exercise, the environment was as simple as possible. On every trial, the CS predicted the same reward, at the same time, with 100% certainty. In the next few exercises, we will make the environment more progressively more complicated and examine the TD-learner's behavior.
#
# + [markdown] colab_type="text" id="d7jLqcoQtnLW"
# ## Interactive Demo 3: Match the Value Functions
#
# First, will replace the environment with one that dispenses multiple rewards. Shown below is the final value function $V$ for a TD learner that was trained in an enviroment where the CS predicted rewards of 6 or 14 units (both equally likely). Can you find another pair of rewards, both equally likely, that exactly match this value function?
#
# Hints:
# * Carefully consider the definition of the value function $V$. This can be solved analytically.
# * There is no need to change $\alpha$ or $\gamma$.
# * Due to the randomness, there may be a small amount of variation.
# + cellView="form" colab={"base_uri": "https://localhost:8080/", "height": 493, "referenced_widgets": ["f14971dd12e84e9bb60f9f272652eb5f", "d98882811dcd4146adcdfcf09ccd467a", "5249f8b304b94b3394df785b2beca0bf", "b38b54b44e834478b4314ac53cebe443", "31617e91bd4745efb2c61d9fb6901a2c", "<KEY>", "16d857923dc34a27878dab9d95a50033", "356b0a68c8b7496da9b1082d904d7a69", "<KEY>", "db943755bdf84d8ea51ff58be7fdebc1"]} colab_type="code" id="ebkTe4kbHImq" outputId="17d844d3-a4f4-463f-ff47-be0991d03040"
#@title
#@markdown Make sure you execute this cell to enable the widget!
n_trials = 20000
np.random.seed(2020)
rng_state = np.random.get_state()
env = MultiRewardCC(40, [6, 14], reward_time=10)
V_multi, TDE_multi = td_learner(env, n_trials, gamma=0.98, alpha=0.001)
@widgets.interact
def reward_guesser_interaction(r1 = widgets.IntText(value=0, min=0, max=50, description="Reward 1"),
r2 = widgets.IntText(value=0, min=0, max=50, description="Reward 2")):
try:
env2 = MultiRewardCC(40, [r1, r2], reward_time=10)
V_guess, _ = td_learner(env2, n_trials, gamma=0.98, alpha=0.001)
fig, ax = plt.subplots()
m, l, _ = ax.stem(V_multi, linefmt='y-', markerfmt='yo', basefmt=' ', label="Target",
use_line_collection=True)
m.set_markersize(15)
m.set_markerfacecolor('none')
l.set_linewidth(4)
m, _, _ = ax.stem(V_guess, linefmt='r', markerfmt='rx', basefmt=' ', label="Guess",
use_line_collection=True)
m.set_markersize(15)
ax.set_xlabel("State")
ax.set_ylabel("Value")
ax.set_title("Guess V(s)\n" + reward_guesser_title_hint(r1, r2))
ax.legend()
except NotImplementedError:
print("Please finish Exercise 1 first!")
# + [markdown] colab_type="text" id="MYPXCECE2w1G"
# ## Section 2.1 Examining the TD Error
#
# Run the cell below to plot the TD errors from our multi-reward environment. A new feature appears in this plot? What is it? Why does it happen?
# + colab={"base_uri": "https://localhost:8080/", "height": 421} colab_type="code" id="TlCisR8rHK43" outputId="4055d6ba-44d4-449e-ac02-88a5a9ccc809"
plot_tde_trace(TDE_multi)
# + [markdown] colab={} colab_type="text" id="cgKx5wTk3hy-"
# [*Click for solution*](https://github.com/erlichlab/course-content/tree/master//tutorials/W2D5_ReinforcementLearning/solutions/W2D5_Tutorial1_Solution_dea47c05.py)
#
#
# + [markdown] colab_type="text" id="4Gi7Q1AFGeTU"
# ---
# # Section 3: TD-learning with probabilistic rewards
#
# In this environment, we'll return to delivering a single reward of ten units. However, it will be delivered intermittently: on 20 percent of trials, the CS will be shown but the agent will not receive the usual reward; the remaining 80% will proceed as usual.
#
# Run the cell below to simulate. How does this compare with the previous experiment?
#
# Earlier in the notebook, we saw that changing $\alpha$ had little effect on learning in a deterministic environment. What happens if you set it to an large value, like 1, in this noisier scenario? Does it seem like it will _ever_ converge?
# + colab={"base_uri": "https://localhost:8080/", "height": 429} colab_type="code" id="LSn9SKRzLZ1t" outputId="4fa6612c-9a39-4eaa-a9ed-4189e1120425"
np.random.set_state(rng_state) # Resynchronize everyone's notebooks
n_trials = 20000
try:
env = ProbabilisticCC(n_steps=40, reward_magnitude=10, reward_time=10,
p_reward=0.8)
V_stochastic, TDE_stochastic = td_learner(env, n_trials*2, alpha=1)
learning_summary_plot(V_stochastic, TDE_stochastic)
except NotImplementedError:
print("Please finish Exercise 1 first")
# + [markdown] colab={} colab_type="text" id="Q6gf1HdZ9rFw"
# [*Click for solution*](https://github.com/erlichlab/course-content/tree/master//tutorials/W2D5_ReinforcementLearning/solutions/W2D5_Tutorial1_Solution_cbbb9c00.py)
#
#
# + [markdown] colab_type="text" id="CTABhuUPuoEW"
# ---
# # Summary
#
# In this notebook, we have developed a simple TD Learner and examined how its state representations and reward prediction errors evolve during training. By manipualting its environment and parameters ($\alpha$, $\gamma$), you developed an intuition for how it behaves. The takehope less
#
# This simple model closely resembles the behavior of subjects undergoing classical conditioning tasks and the dopamine neurons that may underlie that behavior. You may have implemented TD-reset or used the model to recreate a common experimental error. The update rule used here has been extensively studied for [more than 70 years](https://www.pnas.org/content/108/Supplement_3/15647) as a possible explanation for artificial and biological learning.
#
# However, you may have noticed that something is missing from this notebook. We carefully calculated the value of each state, but did not use it to actually do anything. Using values to plan _**Actions**_ is coming up next!
# + [markdown] colab_type="text" id="vk8qckyuTK0O"
# # Bonus
# + [markdown] colab_type="text" id="s0IQkuPayaS5"
# ## Exercise 2: TD-reset
#
# In this exercise we will implement a commonly used heuristic used in modeling activity of dopamine neurons, TD-reset.
# Implement TD-learning as in previous exercises, but set TD-error to zero on all steps after reward (US).
#
# 1. Plot value function and TD-errors.
# 2. Can you explain how the reset is changing the TD-errors and value function?
# + colab={} colab_type="code" id="Ni9r7_csLwQr"
def td_reset_learner(env, n_trials, alpha=0.25, gamma=0.98):
""" Temporal Difference learning with the TD-reset update rule
Args:
env (object): the environment to be learned
n_trials (int): the number of trials to run
gamma (float): temporal discount factor
alpha (float): learning rate
Returns:
ndarray, ndarray: the value function and temporal difference error arrays
"""
V = np.zeros(env.n_steps)
TDE_reset = np.zeros((env.n_steps, n_trials))
for n in range(n_trials):
state = 0
reset = False
for t in range(env.n_steps):
next_state, reward = env.get_outcome(state)
is_delay = env.state_dict[state][0]
########################################################################
## TODO for students: implement TD learning with the TD-reset update rule
# Fill out function and remove
raise NotImplementedError("Student excercise: implement TD learning with the TD-reset update rule")
########################################################################
# Write an expression to compute the TD-error using the TD-reset rule
if reset:
TDE_reset[state] = ...
else:
TDE_reset[state] = ...
# Set reset flag if we receive a reward > 0
if reward > 0:
reset = True
# Write an expression to update the value function
V[state] += ...
# Update state
state = next_state
return V, TDE_reset
# Uncomment these two lines to visualize your results
# env = ProbabilisticCC(n_steps=40, reward_magnitude=10, reward_time=10,
# p_reward=0.8)
# V_reset, TDE_reset = td_reset_learner(env, n_trials=20000)
# learning_summary_plot(V_reset, TDE_reset)
# + [markdown] colab={"base_uri": "https://localhost:8080/", "height": 429} colab_type="text" id="gjtFVHslLyMb" outputId="a456cef3-c2a5-4056-e9f1-e22a4a12e674"
# [*Click for solution*](https://github.com/erlichlab/course-content/tree/master//tutorials/W2D5_ReinforcementLearning/solutions/W2D5_Tutorial1_Solution_8259b30c.py)
#
# *Example output:*
#
# <img alt='Solution hint' align='left' width=558 height=414 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W2D5_ReinforcementLearning/static/W2D5_Tutorial1_Solution_8259b30c_0.png>
#
#
# + [markdown] colab_type="text" id="4KpOTPk_JT1l"
# ## Exercise 3: Removing the CS
#
# In Exercise 1, you (should have) included a term that depends on the conditioned stimulus. Remove it and see what happens. Do you understand why?
# This phenomena often fools people attempting to train animals--beware!
# + [markdown] colab={} colab_type="text" id="gFQBlEkAJzOx"
# [*Click for solution*](https://github.com/erlichlab/course-content/tree/master//tutorials/W2D5_ReinforcementLearning/solutions/W2D5_Tutorial1_Solution_a35b23f3.py)
#
#
| tutorials/W2D5_ReinforcementLearning/student/W2D5_Tutorial1.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# # Extract LAT Data
#
# This thread shows how to extract LAT data from the FERMI Science Support Center (FSSC) [archive](http://fermi.gsfc.nasa.gov/cgi-bin/ssc/LAT/LATDataQuery.cgi) and perform further selection cuts using the Fermitools.
# ## Synopsis
#
# This thread leads you through extracting your LAT data files from the FSSC's databases. In order to analyze LAT data, you will need several [LAT data products](https://fermi.gsfc.nasa.gov/ssc/data/analysis/documentation/Cicerone/Cicerone_Data/LAT_DP.html):
#
# 1. An events file containing the recorded events that correspond to your source of interest, as well as the area surrounding that source. The size of this surrounding "region-of-interest", or ROI, that you choose will depend on the density and brightness of nearby sources, as well as the type of analysis you are performing. There are two types of events files:
# * **Photon Data** - contains all information necessary for science analysis with `source`, `clean`, `ultraclean`, `ultracleanveto`, or `sourceveto` event classes
# * **Extended Data** - contains all event data, including the standard _transient_ classes (`TRANSIENT010` and `TRANSIENT020`), plus additional quantities produced by the Level 1 analysis
#
# Only one type of event data file is needed to perform LAT science analysis. See the [Photon Classification](https://fermi.gsfc.nasa.gov/ssc/data/analysis/documentation/Cicerone/Cicerone_Data/LAT_DP.html) section of the Cicerone for the definitions of the various LAT data classes.
#
# We recommend use of the photon data file for most purposes (except for [LAT GRB analysis](https://fermi.gsfc.nasa.gov/ssc/data/analysis/scitools/explore_latdata_burst.html), for which the extended data are required).
#
# Event data for large datasets will be divided into multiple files. The next tutorial, [Data Preparation](https://fermi.gsfc.nasa.gov/ssc/data/analysis/scitools/data_preparation.html), will discuss how to combine those files.
#
#
# 2. A spacecraft file containing spacecraft position and orientation information at 30 second intervals. This file is required for LAT science analysis. The LAT data server will only generate a single spacecraft file, regardless of the size of the dataset.
#
#
# 3. Some analyses also require models of the isotropic and Galactic diffuse background models. You can download them from the [Background Models](https://fermi.gsfc.nasa.gov/ssc/data/access/lat/BackgroundModels.html) page.
# A description of the contents of each file is available from the [LAT Data Files](https://fermi.gsfc.nasa.gov/ssc/data/analysis/documentation/Cicerone/Cicerone_Data/LAT_Data_Columns.html) section in the Cicerone.
#
# >**Please Note**: If you wish to download data for the full sky or a substantial fraction of it, please download and use the [LAT All-Sky Weekly files](http://fermi.gsfc.nasa.gov/ssc/data/analysis/scitools/LAT_weekly_allsky.html) for your analysis. **The data server does not support queries with radii larger than 60 degrees**.
# ## Data Retrieval Procedure
#
# In this thread we will use this set of parameters:
#
# - Object name or coordinates: 193.98, -5.82
# - Coordinate system: "J2000"
# - Search radius (degrees): 20
# - Observation dates: START, 255398400
# - Time system: "MET"
# - Energy range (MeV): 100, 500000
# - LAT data type: "Photon"
# - Spacecraft data: "checked"
#
# To select all photons within a circular region around the source:
# 1. Go to the [FSSC's web site data server](http://fermi.gsfc.nasa.gov/cgi-bin/ssc/LAT/LATDataQuery.cgi)
# 2. Enter the information listed above in the [data server form](http://fermi.gsfc.nasa.gov/cgi-bin/ssc/LAT/LATDataQuery.cgi) (shown below):
from IPython.display import Image,HTML
# 
# 3. Click on the **Start Search** button.
#
# The 'Query Submitted' webpage will be displayed and provide an estimate of the time to complete the query, as well as a link to the results webpage.
# **Notes**:
#
# * The coordinates (whether Equatorial or Galactic) have to be in degrees and separated by a comma. You will need to save these coordinates for subsequent use in several of the Fermitools. The source coordinates will be reported back to you when you retrieve the results of your query. You can insert also the name (like "Vela pulsar") of the source and it will be resolved (i.e., the resolver will retrieve the source coordinates for you) using [SIMBAD](http://simbad.u-strasbg.fr/simbad/), [NED](http://ned.ipac.caltech.edu/), and [HEASARC GRB catalog (GRBCAT)](http://heasarc.gsfc.nasa.gov/W3Browse/gamma-ray-bursts/grbcat.html), name resolution services, in that order.
#
#
# * If a pair of coordinates are entered, you have to choose the equinox time ("J2000" or "B1950") if these were Celestial coordinates, or you have to select "Galactic" if these were Galactic coordinates. This selection is ignored if a name was given.
#
#
# * If no search radius is provided, the default value of 15 degrees will be used.
#
#
# * To select all photons in this region from a particular time range, enter the start and stop times, separated by a comma, in the Observations Dates field. Observation dates may be specified in the Gregorian calendar system, or as a number of seconds in the MET system (Mission Elapsed Time, floating-point number of seconds since midnight of 1 January 2001), or as a number of days in the MJD system (Modified Julian Days, floating-point number of days since midnight of 17 November, 1858). The [xTime](http://heasarc.gsfc.nasa.gov/cgi-bin/Tools/xTime/xTime.pl) utility is available should you need to convert your time to a different format.
#
# Gregorian dates are specified using the format: YYYY-MM-DD HH:MM:SS (all values except the year must use a leading 0 to create a 2-character field, e.g., 2010-01-01 01:23:45).
#
# The starting date may be replaced by the string 'START' (as in this example), to indicate the earliest available data. The ending date may be replaced by the string 'END', to indicate the most recent available data. The 'START' and 'END' keywords are case-insensitive.
#
# If the Observations Dates field is empty, the last 6 months of data (from the time of the most recent photon data) will be used as the search range.
#
# **Caveat**: In the current version of the query software, floating-point seconds for the "Gregorian" format are not supported (i.e., the seconds have to be rounded to an integer).
#
#
# * If the Observation Dates are specified, select the correct time system. The choice is between "Gregorian" (default selection), "MJD", and "MET". "START" and "END" can be used with any time format.
#
#
# * To select the energy range, use two values in MeV separated by a comma (they are treated as the minimum and maximum photon or event energies). If no energy range is specified, the default energy range of 100 MeV to 300 GeV is used.
#
#
# * Select which LAT data type (if any) you want to download. The default option is "Photon" and is recommended for most science analyses. Extended data files are also available, which include all event classes (including the `transient` classes needed for LAT Gamma-ray Burst analysis) and provide additional information for each event. Extended data files can replace the photon data in your analysis. One can retrieve the "Spacecraft" data file without downloading the photon or event data file; in this case, choose "None" for the LAT data type.
#
# * The Spacecraft data selection is enabled by default. If you do not wish to download the "Spacecraft" data file, uncheck this selection.
#
# Please see the [Help for Fermi Database Query Form](http://fermi.gsfc.nasa.gov/ssc/LATDataQuery_help.html) page for more details.
# 
# Check the specified 'LAT Data Query Results' URL until the ``Position in Queue`` shows "Query Complete". The data file list will include links to the files themselves.
# 
# It is advisable that you save the information reported on the query page as a text file. Having the information from your query easily accesible may ease certain portions of the analysis.
#
#
# 4. Download the spacecraft (pointing and livetime history) file and events data file to your working directory. There may be multiple events files (`_PH##` or `_EV##`), but there should be only a single spacecraft (`_SC##`) file.
# 
# In addition, the results page includes a list of `wget` commands that you can copy and paste into a terminal window to retrieve the files on the command line.
#
# If `wget` does not work for you, consider `curl <link to download> -o <filename>`.
#
# 6. Next, you should prepare your data for analysis by following the [Data Preparation Tutorial](https://fermi.gsfc.nasa.gov/ssc/data/analysis/scitools/data_preparation.html).
| DataSelection/1.ExtractLATData/1.ExtractLATData.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: 'Python 3.8.0 64-bit (''py38'': conda)'
# language: python
# name: python3
# ---
# # @property装饰器
# +
class Person(object):
def __init__(self, name, age):
self._name = name
self._age = age
# 访问器 - getter方法
@property
def name(self):
return self._name
# 访问器 - getter方法
@property
def age(self):
return self._age
# 修改器 - setter方法
@age.setter
def age(self, age):
self._age = age
def play(self):
if self._age <= 16:
print('%s正在玩飞行棋.' % self._name)
else:
print('%s正在玩斗地主.' % self._name)
def main():
person = Person('王大锤', 12)
person.play()
person.age = 22
person.play()
# person.name = '白元芳' # AttributeError: can't set attribute
if __name__ == '__main__':
main()
# -
# # __slots__限定
# +
class Person(object):
# 限定Person对象只能绑定_name, _age和_gender属性
__slots__ = ('_name', '_age', '_gender')
def __init__(self, name, age):
self._name = name
self._age = age
@property
def name(self):
return self._name
@property
def age(self):
return self._age
@age.setter
def age(self, age):
self._age = age
def play(self):
if self._age <= 16:
print('%s正在玩飞行棋.' % self._name)
else:
print('%s正在玩斗地主.' % self._name)
def main():
person = Person('王大锤', 22)
person.play()
person._gender = '男'
# AttributeError: 'Person' object has no attribute '_is_gay'
# person._is_gay = True
if __name__ == '__main__':
main()
# -
# # 静态方法
# +
from math import sqrt
class Triangle(object):
def __init__(self, a, b, c):
self._a = a
self._b = b
self._c = c
@staticmethod
def is_valid(a, b, c):
return a + b > c and b + c > a and a + c > b
def perimeter(self):
return self._a + self._b + self._c
def area(self):
half = self.perimeter() / 2
return sqrt(half * (half - self._a) *
(half - self._b) * (half - self._c))
def main():
a, b, c = 3, 4, 5
# 静态方法和类方法都是通过给类发消息来调用的
if Triangle.is_valid(a, b, c):
t = Triangle(a, b, c)
print(t.is_valid(a, b, b))
print(t.perimeter())
# 也可以通过给类发消息来调用对象方法但是要传入接收消息的对象作为参数
# print(Triangle.perimeter(t))
print(t.area())
print(f"静态方法:{type(Triangle.is_valid)}, 对象方法:{type(t.perimeter)}")
# print(Triangle.area(t))
else:
print('无法构成三角形.')
if __name__ == '__main__':
main()
# -
# # 类方法
# +
from time import time, localtime, sleep
class Clock(object):
"""数字时钟"""
def __init__(self, hour=0, minute=0, second=0):
self._hour = hour
self._minute = minute
self._second = second
@classmethod
def now(cls):
ctime = localtime(time())
return cls(ctime.tm_hour, ctime.tm_min, ctime.tm_sec)
def run(self):
"""走字"""
self._second += 1
if self._second == 60:
self._second = 0
self._minute += 1
if self._minute == 60:
self._minute = 0
self._hour += 1
if self._hour == 24:
self._hour = 0
def show(self):
"""显示时间"""
return '%02d:%02d:%02d' % \
(self._hour, self._minute, self._second)
def main():
# 通过类方法创建对象并获取系统时间
clock = Clock.now()
clock.now()
print(f"类方法:{type(Clock.now)}")
# while True:
# print(clock.show())
# sleep(1)
# clock.run()
if __name__ == '__main__':
main()
# -
# **对象方法是method类型,静态方法是function类型,类方法是method类型**
#
# 实例可以调用对象方法、静态方法、类方法,建议实例只调用对象方法。
# 类名称可以调用静态方法、类方法。
# # python无重载
# > 函数重载主要是为了解决两个问题。1。可变参数类型。2。可变参数个数。另外,一个基本的设计原则是,仅仅当两个函数除了参数类型和参数个数不同以外,其功能是完全相同的,此时才使用函数重载,如果两个函数的功能其实不同,那么不应当使用重载,而应当使用一个名字不同的函数。好吧,那么对于情况 1 ,函数功能相同,但是参数类型不同,python 如何处理?答案是根本不需要处理,因为 python 可以接受任何类型的参数,如果函数的功能相同,那么不同的参数类型在 python 中很可能是相同的代码,没有必要做成两个不同函数。那么对于情况 2 ,函数功能相同,但参数个数不同,python 如何处理?大家知道,答案就是缺省参数。对那些缺少的参数设定为缺省参数即可解决问题。因为你假设函数功能相同,那么那些缺少的参数终归是需要用的。好了,鉴于情况 1 跟 情况 2 都有了解决方案,python 自然就不需要函数重载了。
#
# 作者:pansz
# 链接:https://www.zhihu.com/question/20053359/answer/14054112
# 来源:知乎
# 著作权归作者所有。商业转载请联系作者获得授权,非商业转载请注明出处。
# +
class Hero:
def __init__(self, name = "ty", hp = 300):
self._name = name
self._hp = hp
# def __init__(self, mp = 2):
# self.mp = mp
@property
def name(self):
return self._name
@property
def hp(self):
return self._hp
@hp.setter
def hp(self, hp):
self._hp = hp
def play(self):
print(f"{self._name} is playing")
# play() missing 1 required positional argument: 'place'
# def play(self, place):
# print(f"{self._name} is playing in {place}")
h = Hero()
h.play()
h.hp = 600
# h.play("restroom")
# AttributeError: can't set attribute
# h.name = 'wlj'
# -
# # 类的继承
# Python3.x 和 Python2.x 的一个区别是: Python 3 可以使用直接使用 super().xxx 代替 super(Class, self).xxx :
# # import功能
# * import 模块名1 [as 别名1], 模块名2 [as 别名2],…:使用这种语法格式的 import 语句,会导入指定模块中的所有成员(包括变量、函数、类等)。不仅如此,当需要使用模块中的成员时,需用该模块名(或别名)作为前缀,否则 Python 解释器会报错。
# * from 模块名 import 成员名1 [as 别名1],成员名2 [as 别名2],…: 使用这种语法格式的 import 语句,只会导入模块中指定的成员,而不是全部成员。同时,当程序中使用该成员时,无需附加任何前缀,直接使用成员名(或别名)即可。
# from module import a,b a,b可以是module下的模块、类(__init__中的)、变量(__init__中的)、函数(__init__中的)
# # 输出
print(' '.join('%5s' % classes[labels[j]] for j in range(batch_size)))
print(' '.join('{:5s}'.format(classes[labels[j]]) for j in range(batch_size)))
# 两者等价
| pythonExample/PythonGrammer.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: conda_tensorflow_p36
# language: python
# name: conda_tensorflow_p36
# ---
# # TensorFlow Regression Model Workflow with Script Mode and Automatic Model Tuning
#
# Starting with TensorFlow version 1.11, you can use SageMaker's prebuilt TensorFlow containers with TensorFlow training scripts similar to those you would use outside SageMaker. This feature is named Script Mode.
#
# In this notebook, we will use Script Mode in conjunction with TensorFlow's Eager Execution mode, which is the default execution mode of TensorFlow 2 onwards. Eager execution is an imperative interface where operations are executed immediately, rather than building a static computational graph. Advantages of Eager Execution include a more intuitive interface with natural Python control flow and less boilerplate, easier debugging, and support for dynamic models and almost all of the available TensorFlow operations. It also features close integration with tf.keras to make rapid prototyping even easier.
#
# To demonstrate Script Mode, this notebook focuses on presenting a relatively complete workflow. The workflow includes local training and hosted training in SageMaker, as well as local inference and SageMaker hosted inference with a real time endpoint. Additionally, Automatic Model Tuning in SageMaker will be used to tune the model's hyperparameters. This workflow will be applied to a straightforward regression task, predicting house prices based on the well-known Boston Housing dataset. More specifically, this public dataset contains 13 features regarding housing stock of towns in the Boston area, including features such as average number of rooms, accessibility to radial highways, adjacency to the Charles River, etc.
#
# To begin, we'll import some necessary packages and set up directories for training and test data.
# +
import os
import tensorflow as tf
tf.enable_eager_execution()
tf.set_random_seed(0)
tf.logging.set_verbosity(tf.logging.ERROR)
data_dir = os.path.join(os.getcwd(), 'data')
os.makedirs(data_dir, exist_ok=True)
train_dir = os.path.join(os.getcwd(), 'data/train')
os.makedirs(train_dir, exist_ok=True)
test_dir = os.path.join(os.getcwd(), 'data/test')
os.makedirs(test_dir, exist_ok=True)
# -
# # Prepare dataset
#
# Next, we'll import the dataset. The dataset itself is small and relatively issue-free. For example, there are no missing values, a common problem for many other datasets. Accordingly, preprocessing just involves normalizing the data.
# +
from tensorflow.python.keras.datasets import boston_housing
(x_train, y_train), (x_test, y_test) = boston_housing.load_data()
mean = x_train.mean(axis=0)
std = x_train.std(axis=0)
x_train = (x_train - mean) / (std + 1e-8)
x_test = (x_test - mean) / (std + 1e-8)
print('x train', x_train.shape, x_train.mean(), x_train.std())
print('y train', y_train.shape, y_train.mean(), y_train.std())
print('x test', x_test.shape, x_test.mean(), x_test.std())
print('y test', y_test.shape, y_test.mean(), y_test.std())
# -
# The data is saved as Numpy files prior to both local mode training and hosted training in SageMaker.
# +
import numpy as np
np.save(os.path.join(train_dir, 'x_train.npy'), x_train)
np.save(os.path.join(train_dir, 'y_train.npy'), y_train)
np.save(os.path.join(test_dir, 'x_test.npy'), x_test)
np.save(os.path.join(test_dir, 'y_test.npy'), y_test)
# -
# ## Local Mode training
#
# Amazon SageMaker’s Local Mode training feature is a convenient way to make sure your code is working as expected before moving on to full scale, hosted training. To train in Local Mode, it is necessary to have docker-compose or nvidia-docker-compose (for GPU) installed in the notebook instance. Running following script will install docker-compose or nvidia-docker-compose and configure the notebook environment for you.
# !/bin/bash ./local_mode_setup.sh
# Next, we'll set up a TensorFlow Estimator for Local Mode training. One of the key parameters for an Estimator is the `train_instance_type`, which is the kind of hardware on which training will run. In the case of Local Mode, we simply set this parameter to `local` to invoke Local Mode training on the CPU, or to `local_gpu` if the instance has a GPU. Other parameters of note are the algorithm’s hyperparameters, which are passed in as a dictionary, and a Boolean parameter indicating that we are using Script Mode.
#
# Recall that we are using Local Mode here mainly to make sure our code is working. Accordingly, instead of performing a full cycle of training with many epochs (passes over the full dataset), we'll train only for a small number of epochs to confirm the code is working properly and avoid wasting training time unnecessarily.
# +
import sagemaker
from sagemaker.tensorflow import TensorFlow
model_dir = '/opt/ml/model'
train_instance_type = 'local'
hyperparameters = {'epochs': 5, 'batch_size': 128, 'learning_rate': 0.01}
local_estimator = TensorFlow(
source_dir='tf-bostonhousing-script-mode',
entry_point='train.py',
model_dir=model_dir,
train_instance_type=train_instance_type,
train_instance_count=1,
hyperparameters=hyperparameters,
role=sagemaker.get_execution_role(),
base_job_name='tf-scriptmode-bostonhousing',
framework_version='1.13',
py_version='py3',
script_mode=True)
# +
inputs = {'train': f'file://{train_dir}',
'test': f'file://{test_dir}'}
local_estimator.fit(inputs)
# -
# ## Local Mode endpoint
#
# While Amazon SageMaker’s Local Mode training is very useful to make sure your training code is working before moving on to full scale training, it also would be useful to have a convenient way to test your model locally before incurring the time and expense of deploying it to production. One possibility is to fetch the SavedModel artifact or a model checkpoint saved in Amazon S3, and load it in your notebook for testing. We'll explore doing that in another section of this notebook below. However, an even easier way to do this is to use the Amazon SageMaker SDK to do this work for you.
#
# The Estimator object from the Local Mode training job can be used to deploy a model locally with a single line of code. With one exception, this code is the same as the code you would use to deploy to production. In particular, all you need to do is invoke the local Estimator's deploy method, and similarly to Local Mode training, specify the instance type as either `local_gpu` or `local` depending on whether your notebook instance is a GPU instance or CPU instance.
local_predictor = local_estimator.deploy(initial_instance_count=1,instance_type='local')
# To get predictions from the local endpoint, simply invoke the Predictor's predict method.
local_results = local_predictor.predict(x_test[:10])['predictions']
# As a sanity check, the predictions can be compared against the actual target values.
local_preds_flat_list = [float('%.1f'%(item)) for sublist in local_results for item in sublist]
print('predictions: \t{}'.format(np.array(local_preds_flat_list)))
print('target values: \t{}'.format(y_test[:10].round(decimals=1)))
# We only trained the model for a few epochs and there is room for improvement, but the predictions so far should at least appear reasonably within the ballpark.
#
# To avoid having the TensorFlow Serving container running indefinitely on this notebook instance, simply gracefully shut it down by calling the `delete_endpoint` method of the Predictor object.
local_predictor.delete_endpoint()
# ## SageMaker hosted training
#
# Now that we've confirmed our code is working locally, we can move on to use SageMaker's hosted training functionality. Hosted training is preferred to for doing actual training, especially large-scale, distributed training. Unlike Local Mode training, for hosted training the actual training itself occurs not on the notebook instance, but on a separate cluster of machines managed by SageMaker. Before starting hosted training, the data must be uploaded to S3. We'll do that now, and confirm the upload was successful.
# +
s3_prefix = 'tf-eager-scriptmode-bostonhousing'
traindata_s3_prefix = '{}/data/train'.format(s3_prefix)
testdata_s3_prefix = '{}/data/test'.format(s3_prefix)
# +
train_s3 = sagemaker.Session().upload_data(path='./data/train/', key_prefix=traindata_s3_prefix)
test_s3 = sagemaker.Session().upload_data(path='./data/test/', key_prefix=testdata_s3_prefix)
inputs = {'train':train_s3, 'test': test_s3}
print(inputs)
# -
# We're now ready to set up an Estimator object for hosted training. It is similar to the Local Mode Estimator, except the `train_instance_type` has been set to a ML instance type instead of `local` for Local Mode. Also, since we know our code is working now, we train for a larger number of epochs.
#
# With these two changes, we simply call `fit` to start the actual hosted training.
# +
train_instance_type = 'ml.c5.xlarge'
hyperparameters = {'epochs': 30, 'batch_size': 128, 'learning_rate': 0.01}
estimator = TensorFlow(
source_dir='tf-bostonhousing-script-mode',
entry_point='train.py',
model_dir=model_dir,
train_instance_type=train_instance_type,
train_instance_count=1,
hyperparameters=hyperparameters,
role=sagemaker.get_execution_role(),
base_job_name='tf-scriptmode-bostonhousing',
framework_version='1.13',
py_version='py3',
script_mode=True)
# -
estimator.fit(inputs)
# As with the Local Mode training, hosted training produces a model checkpoint saved in S3 that we can retrieve and load. We can then make predictions and compare them with the test set. This also demonstrates the modularity of SageMaker: having trained the model in SageMaker, you can now take the model out of SageMaker and run it anywhere else. Alternatively, you can deploy the model using SageMaker's hosted endpoints functionality.
#
# Now, instead of using a Local Mode endpoint, we'll go through the steps of downloading the model from Amazon S3 and loading a checkpoint.
# !aws s3 cp {estimator.model_data} ./model/model.tar.gz
# !tar -xvzf ./model/model.tar.gz -C ./model
# +
# !mkdir -p train_model
# !wget -q -P ./train_model https://raw.githubusercontent.com/aws-samples/amazon-sagemaker-script-mode/master/tf-eager-script-mode/train_model/model_def.py
from tensorflow.contrib.eager.python import tfe
from train_model import model_def
tf.keras.backend.clear_session()
device = '/cpu:0'
with tf.device(device):
model = model_def.get_model()
saver = tfe.Saver(model.variables)
saver.restore('model/weights.ckpt')
# +
with tf.device(device):
predictions = model.predict(x_test)
print('predictions: \t{}'.format(predictions[:10].flatten().round(decimals=1)))
print('target values: \t{}'.format(y_test[:10].round(decimals=1)))
# -
# ## SageMaker hosted endpoint
#
# After multiple sanity checks, we're confident that our model is performing as expected. If we wish to deploy the model to production, a convenient option is to use a SageMaker hosted endpoint. The endpoint will retrieve the TensorFlow SavedModel created during training and deploy it within a TensorFlow Serving container. This all can be accomplished with one line of code, an invocation of the Estimator's deploy method.
predictor = estimator.deploy(initial_instance_count=1,instance_type='ml.m5.xlarge')
# As one last sanity check, we can compare the predictions generated by the endpoint with those generated locally by the model checkpoint we retrieved from hosted training in SageMaker.
results = predictor.predict(x_test[:10])['predictions']
flat_list = [float('%.1f'%(item)) for sublist in results for item in sublist]
print('predictions: \t{}'.format(np.array(flat_list)))
print('target values: \t{}'.format(y_test[:10].round(decimals=1)))
# Before proceeding with the rest of this notebook, you can delete the prediction endpoint to release the instance(s) associated with it.
sagemaker.Session().delete_endpoint(predictor.endpoint)
# ## Automatic Model Tuning
#
# Selecting the right hyperparameter values to train your model can be difficult. The right answer is dependent on your data; some algorithms have many different hyperparameters that can be tweaked; some are very sensitive to the hyperparameter values selected; and most have a non-linear relationship between model fit and hyperparameter values. SageMaker Automatic Model Tuning helps automate the hyperparameter tuning process: it runs multiple training jobs with different hyperparameter combinations to find the set with the best model performance.
#
# We begin by specifying the hyperparameters we wish to tune, and the range of values over which to tune each one. We also must specify an objective metric to be optimized: in this use case, we'd like to minimize the validation loss.
# +
from sagemaker.tuner import IntegerParameter, CategoricalParameter, ContinuousParameter, HyperparameterTuner
from time import gmtime, strftime
hyperparameter_ranges = {
'learning_rate': ContinuousParameter(0.001, 0.2, scaling_type="Logarithmic"),
'epochs': IntegerParameter(10, 50),
'batch_size': IntegerParameter(64, 256),
}
metric_definitions = [{'Name': 'loss',
'Regex': ' loss: ([0-9\\.]+)'},
{'Name': 'val_loss',
'Regex': ' val_loss: ([0-9\\.]+)'}]
objective_metric_name = 'val_loss'
objective_type = 'Minimize'
# -
# Next we specify a HyperparameterTuner object that takes the above definitions as parameters. Each tuning job must be given a budget - a maximum number of training jobs - and the tuning job will complete once that many training jobs have been executed.
#
# We also can specify how much parallelism to employ, in this case five jobs, meaning that the tuning job will complete after three series of five jobs in parallel have completed. For the default Bayesian Optimization tuning strategy used here, the search is informed by the results of previous groups of training jobs, so we don't run all of the jobs in parallel, but rather divide the jobs into groups of parallel jobs. In other words, more parallel jobs will finish tuning sooner, but may sacrifice accuracy.
#
# Now we can launch a hyperparameter tuning job by calling the `fit` method of the HyperparameterTuner object. We will wait until the tuning finished, which may take around 10 minutes.
# +
tuner = HyperparameterTuner(estimator,
objective_metric_name,
hyperparameter_ranges,
metric_definitions,
max_jobs=9,
max_parallel_jobs=3,
objective_type=objective_type)
tuning_job_name = "tf-bostonhousing-{}".format(strftime("%d-%H-%M-%S", gmtime()))
tuner.fit(inputs, job_name=tuning_job_name)
tuner.wait()
# -
# After the tuning job is finished, we can use the `HyperparameterTuningJobAnalytics` method to list the top 5 tuning jobs with the best performance. Although the results typically vary from tuning job to tuning job, the best validation loss from the tuning job (under the FinalObjectiveValue column) likely will be lower than the validation loss from the hosted training job above. For an example of a more in-depth analysis of a tuning job, see HPO_Analyze_TuningJob_Results.ipynb notebook.
tuner_metrics = sagemaker.HyperparameterTuningJobAnalytics(tuning_job_name)
tuner_metrics.dataframe().sort_values(['FinalObjectiveValue'], ascending=True).head(5)
# The total training time and training jobs status can be checked with the following script. Because automatic early stopping is by default off, all the training jobs should be completed normally.
total_time = tuner_metrics.dataframe()['TrainingElapsedTimeSeconds'].sum() / 3600
print("The total training time is {:.2f} hours".format(total_time))
tuner_metrics.dataframe()['TrainingJobStatus'].value_counts()
# Assuming the best model from the tuning job is better than the model produced by the hosted training job above, we could now easily deploy that model. By calling the `deploy` method of the HyperparameterTuner object we instantiated above, we can directly deploy the best model from the tuning job to a SageMaker hosted endpoint:
#
# `tuning_predictor = tuner.deploy(initial_instance_count=1, instance_type='ml.m5.xlarge')`
#
# Since we already looked at how to use a SageMaker hosted endpoint above, we won't repeat that here. We've covered a lot of content in this notebook: local and hosted training with Script Mode, local and hosted inference in SageMaker, and Automatic Model Tuning. These are likely to be central elements for most deep learning workflows in SageMaker.
| AWS/TF_World_SageMaker_Workshop/workshop1-tensor-world-2019-master/lab2/tf-boston-housing.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="pFR9siIYq7hq"
# ### **INITIALIZATION:**
# - I use these three lines of code on top of my each notebooks because it will help to prevent any problems while reloading the same project. And the third line of code helps to make visualization within the notebook.
# + id="rL-NhOgfYwoD"
#@ INITIALIZATION:
# %reload_ext autoreload
# %autoreload 2
# %matplotlib inline
# + [markdown] id="jxT7JUHTrLNY"
# **DOWNLOADING LIBRARIES AND DEPENDENCIES:**
# - I have downloaded all the libraries and dependencies required for the project in one particular cell.
# + id="TYm2Dcj8rHtK"
#@ DOWNLOADING THE LIBRARIES AND DEPENDENCIES:
# # !pip install -U d2l
from d2l import torch as d2l
import warnings
import torch
import torchvision
from torch import nn
# + [markdown] id="xUNZdVWIr7HR"
# ### **THE POKEMON DATASET:**
# - The dataset is a collection of Pokemon sprites obtained from [**PokemonDB**](https://pokemondb.net/sprites). I will download, extract and load the dataset.
# + id="PA_JwDIxrqQ7"
#@ GETTING THE DATASET:
d2l.DATA_HUB["pokemon"] = (d2l.DATA_URL + 'pokemon.zip',
'c065c0e2593b8b161a2d7873e42418bf6a21106c') # Path to Dataset.
data_dir = d2l.download_extract("pokemon") # Downloading and Extracting the Dataset.
pokemon = torchvision.datasets.ImageFolder(data_dir) # Initializing DataLoader.
# + [markdown] id="hxcxgW_yupJo"
# - I will resize each image into 64X64 and normalize the data with 0.5 mean and 0.5 standard deviation.
# + id="0ctPmkBRuB1X"
#@ PREPARING THE DATASET:
batch_size = 256 # Initialization.
transformer = torchvision.transforms.Compose([ # Initializing Compose Instance.
torchvision.transforms.Resize((64, 64)), # Resizing Images.
torchvision.transforms.ToTensor(), # Converting into Tensors.
torchvision.transforms.Normalize(0.5, 0.5)]) # Normalizing the Data.
pokemon.transform = transformer # Transforming the Pokemon Dataset.
data_iter = torch.utils.data.DataLoader(
pokemon, batch_size=batch_size, shuffle=True,
num_workers=2) # Initializing Data Iterations.
# + colab={"base_uri": "https://localhost:8080/", "height": 480} id="MldMiIbNx5V9" outputId="0d9b1936-3710-4408-f066-7fd9ed4ac8d9"
#@ VISUALIZING THE DATASET:
warnings.filterwarnings("ignore")
d2l.set_figsize((5, 5))
for X, y in data_iter:
imgs = X[0:20, :, :, :].permute(0, 2, 3, 1) / 2 + 0.5 # Getting Images.
d2l.show_images(imgs, num_rows=4, num_cols=5) # Visualizing Images.
break
# + [markdown] id="yM588bH_1JnZ"
# ### **THE GENERATOR:**
# - The Generator needs to map the noise variable to a RGB image. I will use transposed convolutional layer to enlarge the input image. The basic block of Generator contains a transposed convolution layer followed by the batch normalization and RELU activation function.
# + colab={"base_uri": "https://localhost:8080/"} id="smkeY6e8yvMf" outputId="01dc36d3-69ef-4909-fd08-43cbaa409ed4"
#@ DEFINING THE GENERATOR:
class G_Block(nn.Module): # Initializing Generator.
def __init__(self, out_channels, in_channels=3, kernel_size=4, strides=2,
padding=1, **kwargs): # Initializing Constructor Function.
super(G_Block, self).__init__(**kwargs)
self.conv2d_trans = nn.ConvTranspose2d(in_channels, out_channels,
kernel_size, strides, padding,
bias=False) # Initializing Transposed Convolution Layer.
self.batch_norm = nn.BatchNorm2d(out_channels) # Initializing Batch Normalization Layer.
self.activation = nn.ReLU() # Initializing RELU Activation Layer.
def forward(self, X): # Forward Propagation Layer.
return self.activation(self.batch_norm(self.conv2d_trans(X))) # Implementation of Layers.
#@ INSPECTING THE IMPLEMENTATION:
X = torch.zeros((2, 3, 16, 16)) # Initializing Tensor.
G_block = G_Block(20) # Implementation.
G_block(X).shape # Inspecting Shape.
# + [markdown] id="XDEm2zSr6riO"
# - The Generator consists of four blocks that increase height and width of input from 1 to 32. The transposed convolution layer is used to generate the output. The tanh activation function is used to project output values into the range of -1 and 1.
# + id="0_vCR__Z57f4" colab={"base_uri": "https://localhost:8080/"} outputId="a908a597-91f7-4e53-f914-b95a2711246c"
#@ INITIALIZING THE GENERATOR:
n_G = 64 # Initialization.
net_G = nn.Sequential(G_Block(in_channels=100, out_channels=n_G*8,
strides=1, padding=0), # Output: 64*8, 4, 4.
G_Block(in_channels=n_G*8, out_channels=n_G*4), # Output: 64*4, 8, 8.
G_Block(in_channels=n_G*4, out_channels=n_G*2), # Output: 64*2, 16, 16.
G_Block(in_channels=n_G*2, out_channels=n_G), # Output: 64, 32, 32.
nn.ConvTranspose2d(in_channels=n_G,out_channels=3,
kernel_size=4, stride=2,
padding=1, bias=False), # Implementation of Transposed Convolution.
nn.Tanh()) # Implementation of Tanh Activation.
#@ IMPLEMENTATION OF GENERATOR:
X = torch.zeros((1, 100, 1, 1,)) # Initializing Tensor.
net_G(X).shape # Inspecting Shape of Output.
# + [markdown] id="Yu1xy95uBlY3"
# ### **THE DISCRIMINATOR:**
# - The Discriminator is a convolution layer followed by a batch normalization layer and Leaky RELU activation function. Leaky RELU is a nonlinear function that gives a non zero output for a negative input. It aims to fix the RELU problem that a neuron might always output a negative value and therefore cannot make any progress since the gradient of RELU is 0.
# + colab={"base_uri": "https://localhost:8080/"} id="5Q_u9nflAoR-" outputId="35e310ae-9ef9-4e9b-ee35-c475d4b7360f"
#@ DEFINING THE DISCRIMINATOR:
class D_Block(nn.Module): # Initializing Discriminator.
def __init__(self, out_channels, in_channels=3, kernel_size=4, strides=2,
padding=1, alpha=0.2, **kwargs): # Initializing Constructor Function.
super(D_Block, self).__init__(**kwargs)
self.conv2d = nn.Conv2d(in_channels, out_channels, kernel_size,
strides, padding, bias=False) # Initializing Convolution Layer.
self.batch_norm = nn.BatchNorm2d(out_channels) # Initializing Batch Normalization Layer.
self.activation = nn.LeakyReLU(alpha, inplace=True) # Initializing Leaky RELU Activation.
def forward(self, X): # Initializing Forward Propagation.
return self.activation(self.batch_norm(self.conv2d(X))) # Implementation of Layers.
#@ IMPLEMENTATION OF THE DISCRIMINATOR:
X = torch.zeros((2, 3, 16, 16)) # Initializing a Tensor.
d_block = D_Block(20) # Initializing Discriminator.
d_block(X).shape # Implementation of Discriminator.
# + id="M-D4EionRxoj" colab={"base_uri": "https://localhost:8080/"} outputId="7a862a75-8fbe-476b-fa57-e2fa635df262"
#@ INITIALIZING THE DISCRIMINATOR:
n_D = 64 # Initialization.
net_D = nn.Sequential(D_Block(n_D), # Output: 64, 32, 32.
D_Block(in_channels=n_D, out_channels=n_D*2), # Output: 64*2, 16, 16.
D_Block(in_channels=n_D*2,out_channels=n_D*4), # Output: 64*4, 8, 8.
D_Block(in_channels=n_D*4,out_channels=n_D*8), # Output: 64*8, 4, 4.
nn.Conv2d(in_channels=n_D*8, out_channels=1,
kernel_size=4, bias=False)) # Implementation of Convolution Layer.
#@ IMPLEMENTATION:
X = torch.zeros((1, 3, 64, 64)) # Initializing Tensor.
net_D(X).shape # Inspecting the Shape.
# + [markdown] id="VxmkC9sWBuvP"
# ### **TRAINING:**
# - I will be using the same learning rate for both generator and discriminator since the networks are similar to each other. I will change β1 in Adam from 0.9 to 0.5. It decreases the smoothness of the momentum which is the exponentially weighted moving average of past gradients to take care of the rapid changing gradients because the generator and the discriminator fight with each other. The random generated noise Z is a 4D tensor.
# + id="xnE22BBpBrGp"
#@ TRAINING THE MODEL:
def train(net_D, net_G, data_iter, num_epochs, lr, latent_dim,
device=d2l.try_gpu()): # Function for Training.
loss = nn.BCEWithLogitsLoss(reduction="sum") # Initializing Cross Entropy Loss Function.
for w in net_D.parameters(): # Discriminator.
nn.init.normal_(w, 0, 0.02) # Normal Initialization.
for w in net_G.parameters(): # Generator.
nn.init.normal_(w, 0, 0.02) # Normal Initialization.
net_D, net_G = net_D.to(device), net_G.to(device) # Enabling GPU.
trainer_hp = {"lr": lr, "betas": [0.5, 0.999]} # Initializing Optimization Parameters.
trainer_D = torch.optim.Adam(net_D.parameters(), **trainer_hp) # Adam Optimizer for Discriminator.
trainer_G = torch.optim.Adam(net_G.parameters(), **trainer_hp) # Adam Optimizer for Generator.
animator = d2l.Animator(xlabel="epoch", ylabel="loss",
xlim=[1, num_epochs],nrows=2,figsize=(5, 5),
legend=["discriminator", "generator"]) # Initializing Animator.
animator.fig.subplots_adjust(hspace=0.3) # Initializing Subplots.
for epoch in range(1, num_epochs + 1):
timer = d2l.Timer() # Initializing Timer.
metric = d2l.Accumulator(3) # Initializing Accumulator.
for X, _ in data_iter:
batch_size = X.shape[0] # Initializing Batch Size.
Z = torch.normal(0, 1, size=(batch_size, latent_dim, 1, 1)) # Initializing Tensor.
X, Z = X.to(device), Z.to(device) # Enabling GPU.
metric.add(d2l.update_D(X, Z, net_D, net_G, loss, trainer_D), # Updating Discriminator.
d2l.update_G(Z, net_D, net_G, loss, trainer_G), # Updating Generator.
batch_size) # Acccumulating Updates.
Z = torch.normal(0, 1, size=(21, latent_dim, 1, 1), device=device) # Initializing Tensor.
fake_x = net_G(Z).permute(0, 2, 3, 1) / 2 + 0.5 # Normalizing Synthetic Data.
imgs = torch.cat([torch.cat([fake_x[i*7 + j].cpu().detach() \
for j in range(7)], dim=1) \
for i in range(len(fake_x) // 7)], dim=0)
animator.axes[1].cla()
animator.axes[1].imshow(imgs)
loss_D, loss_G = metric[0] / metric[2], metric[1] / metric[2] # Getting Discriminator and Generator Loss.
animator.add(epoch, (loss_D, loss_G))
print(f"loss_D {loss_D:.3f}, loss_G {loss_G:.3f}, "
f"{metric[2]/timer.stop():.1f} examples/sec on {str(device)}")
# + colab={"base_uri": "https://localhost:8080/", "height": 442} id="GcS0i26WQQJ_" outputId="3a324912-a5d4-45e0-9e38-ee9041c11f3d"
#@ TRAINING THE MODEL:
latent_dim, lr, num_epochs = 100, 0.005, 25 # Initializing Parameters.
train(net_D, net_G, data_iter, num_epochs, lr, latent_dim) # Training the Model.
| Deep GAN.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] colab_type="text" id="04QgGZc9bF5D"
# # Introduction to Logistic Regression Using TF 2.0
#
# **Learning Objectives**
#
#
# 1. Build a neural network that classifies images.
# 2. Train this neural network.
# 3. Evaluate the accuracy of the model.
#
#
# ## Introduction
#
# This short introduction uses [Keras](https://keras.io/), a high-level API to build and train models in TensoFlow. In this lab, you Load and prepare the MNIST dataset, convert the samples from integers to floating-point numbers, build and train a neural network that classifies images and the evaluate then accuracy of the model.
#
# Each learning objective will correspond to a __#TODO__ in the [student lab notebook](https://github.com/GoogleCloudPlatform/training-data-analyst/blob/master/courses/machine_learning/deepdive2/introduction_to_tensorflow/labs/intro_logistic_regression_TF2.0.ipynb) -- try to complete that notebook first before reviewing this solution notebook.
# + [markdown] colab_type="text" id="nnrWf3PCEzXL"
# ## Load necessary libraries
# We will start by importing the necessary libraries for this lab.
# + colab={} colab_type="code" id="0trJmd6DjqBZ"
import tensorflow as tf
print("TensorFlow version: ", tf.version.VERSION)
# + [markdown] colab_type="text" id="7NAbSZiaoJ4z"
# Load and prepare the [MNIST dataset](http://yann.lecun.com/exdb/mnist/). Convert the samples from integers to floating-point numbers:
# + colab={} colab_type="code" id="7FP5258xjs-v"
mnist = tf.keras.datasets.mnist
(x_train, y_train), (x_test, y_test) = mnist.load_data()
x_train, x_test = x_train / 255.0, x_test / 255.0
# + [markdown] colab_type="text" id="BPZ68wASog_I"
# **Lab Task 1:** Build the `tf.keras.Sequential` model by stacking layers. Choose an optimizer and loss function for training:
# + colab={} colab_type="code" id="h3IKyzTCDNGo"
model = # TODO 1 -- Your code here.
# + [markdown] colab_type="text" id="l2hiez2eIUz8"
# For each example the model returns a vector of "[logits](https://developers.google.com/machine-learning/glossary#logits)" or "[log-odds](https://developers.google.com/machine-learning/glossary#log-odds)" scores, one for each class.
# + colab={} colab_type="code" id="OeOrNdnkEEcR"
predictions = model(x_train[:1]).numpy()
predictions
# + [markdown] colab_type="text" id="tgjhDQGcIniO"
# The `tf.nn.softmax` function converts these logits to "probabilities" for each class:
# + colab={} colab_type="code" id="zWSRnQ0WI5eq"
tf.nn.softmax(predictions).numpy()
# + [markdown] colab_type="text" id="he5u_okAYS4a"
# Note: It is possible to bake this `tf.nn.softmax` in as the activation function for the last layer of the network. While this can make the model output more directly interpretable, this approach is discouraged as it's impossible to
# provide an exact and numerically stable loss calculation for all models when using a softmax output.
# + [markdown] colab_type="text" id="hQyugpgRIyrA"
# The `losses.SparseCategoricalCrossentropy` loss takes a vector of logits and a `True` index and returns a scalar loss for each example.
# -
# **Lab Task #2:** Usage of losses.SparseCategoricalCrossentropy with logits vectors and a True index.
# + colab={} colab_type="code" id="RSkzdv8MD0tT"
loss_fn = # TODO 2 -- Your code here.
# + [markdown] colab_type="text" id="SfR4MsSDU880"
# This loss is equal to the negative log probability of the true class:
# It is zero if the model is sure of the correct class.
#
# This untrained model gives probabilities close to random (1/10 for each class), so the initial loss should be close to `-tf.log(1/10) ~= 2.3`.
# + colab={} colab_type="code" id="NJWqEVrrJ7ZB"
loss_fn(y_train[:1], predictions).numpy()
# + colab={} colab_type="code" id="9foNKHzTD2Vo"
model.compile(optimizer="adam", loss=loss_fn, metrics=["accuracy"])
# + [markdown] colab_type="text" id="ix4mEL65on-w"
# The `Model.fit` method adjusts the model parameters to minimize the loss:
# + colab={} colab_type="code" id="y7suUbJXVLqP"
model.fit(x_train, y_train, epochs=5)
# + [markdown] colab_type="text" id="4mDAAPFqVVgn"
# The `Model.evaluate` method checks the models performance, usually on a "[Validation-set](https://developers.google.com/machine-learning/glossary#validation-set)" or "[Test-set](https://developers.google.com/machine-learning/glossary#test-set)".
# + colab={} colab_type="code" id="F7dTAzgHDUh7"
model.evaluate(x_test, y_test, verbose=2)
# + [markdown] colab_type="text" id="T4JfEh7kvx6m"
# The image classifier is now trained to ~98% accuracy on this dataset. To learn more, read the [TensorFlow tutorials](https://www.tensorflow.org/tutorials/).
# + [markdown] colab_type="text" id="Aj8NrlzlJqDG"
# If you want your model to return a probability, you can wrap the trained model, and attach the softmax to it:
# + colab={} colab_type="code" id="rYb6DrEH0GMv"
probability_model = tf.keras.Sequential([model, tf.keras.layers.Softmax()])
# + colab={} colab_type="code" id="cnqOZtUp1YR_"
probability_model(x_test[:5])
| notebooks/introduction_to_tensorflow/labs/intro_logistic_regression_TF2.0.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: 'Python 3.9.5 64-bit (''base'': conda)'
# language: python
# name: python3
# ---
#
# # Hi, I'm everydaycodings! 👋
# # EDA on NYC Motor Vehicle Collisions to Person
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
pd.set_option('display.max_columns', 500)
data = pd.read_csv("Datasets/NYC_Motor_Vehicle_Collisions_to_Person/NYC_Motor_Vehicle_Collisions_to_Person.csv")
data.head()
# # Getting the general view of the Data
data.columns
data.info()
# # Handling Numerical Columns
# ### Seperating the numerical Columns from dataset and getting the null values of it
num_categories = [feature for feature in data.columns if data[feature].dtypes != "O"] # here "O" basically means object which represents string values
data[num_categories].isnull().sum()
# ### After geting the numerical value we not drop that missing value instead we will fill all the messing values with median number
# #### Here I have used ``median()`` insted of ``mean()`` to avoid *outliers*
# +
for feature in num_categories:
median_values = data[feature].median()
data[feature] = data[feature].fillna(median_values)
data.head(3)
# -
# # Handling Categorical Columns
# ### Seperating the Categorical Columns from Datset and getting the null values of it
cat_categories = [feature for feature in data.columns if data[feature].dtypes == "O"]
data[cat_categories].isnull().sum()
# ### After geting the Categorial Columns insted of droping the NaN values we will be filling it with *missing values*
data[cat_categories] = data[cat_categories].fillna("missing values")
data.head()
# ## CrossChecking if there are more NaN values Left or Not.
data.isnull().sum()
# # Handling DateTime Columns
data['CRASH_DATE']=pd.to_datetime(data['CRASH_DATE'])
data.head(3)
# # **EDA** (Exploring the various columns relation with different type of graphs)
# #### Person Age and Person Sex Bar Graph in releation to Person Injury
plt.figure(figsize=(15,6))
sns.barplot(x="PERSON_SEX", y= "PERSON_AGE", hue="PERSON_INJURY", data=data)
plt.title('Person Age and Person Sex Bar Graph in releation to Person Injury')
plt.show()
# #### Complaint bar graph after Accident
plt.figure(figsize=(15,6))
sns.countplot(x="COMPLAINT", data=data)
plt.title('Complaint bar graph after Accident')
plt.xticks(rotation=80)
plt.show()
# #### Relation Between Ejection type and Emotional Status
plt.figure(figsize=(15,6))
sns.countplot(x= "EMOTIONAL_STATUS", hue="EJECTION", data=data)
plt.title('Relation Between Ejection type and Emotional Status')
plt.xticks(rotation=90)
plt.show()
# #### Relation Between Body Injury and PED Role
plt.figure(figsize=(15,6))
sns.countplot(x= "BODILY_INJURY", hue="PED_ROLE", data=data)
plt.title('Relation Between Body Injury and PED Role')
plt.xticks(rotation=90)
plt.show()
# #### 2021 Collisions Each Month
plt.figure(figsize=(15,6))
data_for_month = data['CRASH_DATE'].dt.month.value_counts().sort_index()
plt.bar(data_for_month.index,data_for_month.values)
plt.title('2021 Collisions Each Month')
plt.xticks(range(1,13))
plt.show()
# #### 2021 Collisions Each Hours
# +
# This Peace of code was taken form a notebook [When do the most traffic accidents occur?] from a Kaggle user @kukuroo3
dfp= data['CRASH_TIME'].str.split(':').str[0].astype('int').value_counts().sort_index()
plt.bar(dfp.index,dfp.values)
plt.xticks(range(0,24,6),[str(x)+":00" for x in range(0,24,6)])
plt.xlabel('Hour')
plt.ylabel('Count')
plt.title('2021 Collisions Each Hours')
plt.show()
# -
#
# # Result Of **EDA**
#
# ### 1) Accoring to the data there are more person killed then Injured in the Accident.
# ### 2) Accoring to the data Females accidents rate are more than male accident rate.
# ### 3) Accoring to the data age between 40-50 has the most tendency to fall in an accident.
# ### 4) Accoring to the data the people who got Injured are more likely to complain about Pain, Nausea, None Vision and Minor Bleeding
# ### 5) Accoring to the data if seat-belts are not ejected people are likely to be in Conscious but in shock state
# ### 6) Accoring to the data Divers are in the most danger as they have maximum number of Body Injuries other than pedestrian and passenger
# ### 7) Accoring to the data Head Injury, Back Injury, Knee-lower-leg-foot Injury and Neck Injury are the most comman Injury during Road Accident
# ### 8) Accoring to the data Month of May-June are the most dangrous month as most of the accident occours in this particaular month
#
| NYC_Motor_Vehicle_Collisions_to_Person.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Convolutional Autoencoder
#
# Sticking with the MNIST dataset, let's improve our autoencoder's performance using convolutional layers. We'll build a convolutional autoencoder to compress the MNIST dataset.
#
# >The encoder portion will be made of convolutional and pooling layers and the decoder will be made of **upsampling and convolutional layers**.
#
# <img src='notebook_ims/autoencoder_1.png' />
#
# ### Compressed Representation
#
# A compressed representation can be great for saving and sharing any kind of data in a way that is more efficient than storing raw data. In practice, the compressed representation often holds key information about an input image and we can use it for denoising images or oher kinds of reconstruction and transformation!
#
# <img src='notebook_ims/denoising.png' width=60%/>
#
# Let's get started by importing our libraries and getting the dataset.
# +
import torch
import numpy as np
from torchvision import datasets
import torchvision.transforms as transforms
# convert data to torch.FloatTensor
transform = transforms.ToTensor()
# load the training and test datasets
train_data = datasets.MNIST(root='data', train=True,
download=True, transform=transform)
test_data = datasets.MNIST(root='data', train=False,
download=True, transform=transform)
# +
# Create training and test dataloaders
num_workers = 0
# how many samples per batch to load
batch_size = 20
# prepare data loaders
train_loader = torch.utils.data.DataLoader(train_data, batch_size=batch_size, num_workers=num_workers)
test_loader = torch.utils.data.DataLoader(test_data, batch_size=batch_size, num_workers=num_workers)
# -
# ### Visualize the Data
# +
import matplotlib.pyplot as plt
# %matplotlib inline
# obtain one batch of training images
dataiter = iter(train_loader)
images, labels = dataiter.next()
images = images.numpy()
# get one image from the batch
img = np.squeeze(images[0])
fig = plt.figure(figsize = (5,5))
ax = fig.add_subplot(111)
ax.imshow(img, cmap='gray')
# -
# ---
# ## Convolutional Autoencoder
#
# The encoder part of the network will be a typical convolutional pyramid. Each convolutional layer will be followed by a max-pooling layer to reduce the dimensions of the layers. The decoder though might be something new to you. The decoder needs to convert from a narrow representation to a wide reconstructed image. For example, the representation could be a 4x4x8 max-pool layer. This is the output of the encoder, but also the input to the decoder. We want to get a 28x28x1 image out from the decoder so we need to work our way back up from the narrow decoder input layer. A schematic of the network is shown below.
#
# <img src='notebook_ims/conv_enc_2.png' width=600px>
#
#
# ### Upsampling + Convolutions, Decoder
#
# This decoder uses a combination of nearest-neighbor **upsampling and normal convolutional layers** to increase the width and height of the input layers.
#
# It is important to note that transpose convolution layers can lead to artifacts in the final images, such as checkerboard patterns. This is due to overlap in the kernels which can be avoided by setting the stride and kernel size equal. In [this Distill article](http://distill.pub/2016/deconv-checkerboard/) from <NAME>, *et al*, the authors show that these checkerboard artifacts can be avoided by resizing the layers using nearest neighbor or bilinear interpolation (upsampling) followed by a convolutional layer. This is the approach we take, here.
#
# #### TODO: Build the network shown above.
# > Build the encoder out of a series of convolutional and pooling layers.
# > When building the decoder, use a combination of upsampling and normal, convolutional layers.
# +
import torch.nn as nn
import torch.nn.functional as F
# define the NN architecture
class ConvAutoencoder(nn.Module):
def __init__(self):
super(ConvAutoencoder, self).__init__()
## encoder layers ##
# conv layer (depth from 1 --> 16), 3x3 kernels
self.conv1 = nn.Conv2d(1, 16, 3, padding=1)
# conv layer (depth from 16 --> 8), 3x3 kernels
self.conv2 = nn.Conv2d(16, 4, 3, padding=1)
# pooling layer to reduce x-y dims by two; kernel and stride of 2
self.pool = nn.MaxPool2d(2, 2)
## decoder layers ##
self.conv4 = nn.Conv2d(4, 16, 3, padding=1)
self.conv5 = nn.Conv2d(16, 1, 3, padding=1)
def forward(self, x):
# add layer, with relu activation function
# and maxpooling after
x = F.relu(self.conv1(x))
x = self.pool(x)
# add hidden layer, with relu activation function
x = F.relu(self.conv2(x))
x = self.pool(x) # compressed representation
## decoder
# upsample, followed by a conv layer, with relu activation function
# this function is called `interpolate` in some PyTorch versions
x = F.upsample(x, scale_factor=2, mode='nearest')
x = F.relu(self.conv4(x))
# upsample again, output should have a sigmoid applied
x = F.upsample(x, scale_factor=2, mode='nearest')
x = F.sigmoid(self.conv5(x))
return x
# initialize the NN
model = ConvAutoencoder()
print(model)
# -
# ---
# ## Training
#
# Here I'll write a bit of code to train the network. I'm not too interested in validation here, so I'll just monitor the training loss and the test loss afterwards.
#
# We are not concerned with labels in this case, just images, which we can get from the `train_loader`. Because we're comparing pixel values in input and output images, it will be best to use a loss that is meant for a regression task. Regression is all about comparing quantities rather than probabilistic values. So, in this case, I'll use `MSELoss`. And compare output images and input images as follows:
# ```
# loss = criterion(outputs, images)
# ```
#
# Otherwise, this is pretty straightfoward training with PyTorch. We flatten our images, pass them into the autoencoder, and record the training loss as we go.
# +
# specify loss function
criterion = nn.MSELoss()
# specify loss function
optimizer = torch.optim.Adam(model.parameters(), lr=0.001)
# +
# number of epochs to train the model
n_epochs = 30
for epoch in range(1, n_epochs+1):
# monitor training loss
train_loss = 0.0
###################
# train the model #
###################
for data in train_loader:
# _ stands in for labels, here
# no need to flatten images
images, _ = data
# clear the gradients of all optimized variables
optimizer.zero_grad()
# forward pass: compute predicted outputs by passing inputs to the model
outputs = model(images)
# calculate the loss
loss = criterion(outputs, images)
# backward pass: compute gradient of the loss with respect to model parameters
loss.backward()
# perform a single optimization step (parameter update)
optimizer.step()
# update running training loss
train_loss += loss.item()*images.size(0)
# print avg training statistics
train_loss = train_loss/len(train_loader)
print('Epoch: {} \tTraining Loss: {:.6f}'.format(
epoch,
train_loss
))
# -
# ## Checking out the results
#
# Below I've plotted some of the test images along with their reconstructions. For the most part these look pretty good except for some blurriness in some parts.
# +
# obtain one batch of test images
dataiter = iter(test_loader)
images, labels = dataiter.next()
# get sample outputs
output = model(images)
# prep images for display
images = images.numpy()
# output is resized into a batch of iages
output = output.view(batch_size, 1, 28, 28)
# use detach when it's an output that requires_grad
output = output.detach().numpy()
# plot the first ten input images and then reconstructed images
fig, axes = plt.subplots(nrows=2, ncols=10, sharex=True, sharey=True, figsize=(25,4))
# input images on top row, reconstructions on bottom
for images, row in zip([images, output], axes):
for img, ax in zip(images, row):
ax.imshow(np.squeeze(img), cmap='gray')
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
| autoencoder/convolutional-autoencoder/Upsampling_Solution.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import numpy as np
#Takes care of floats
arr1 = np.array([[1,2,3],[8,9,10]])
arr1
# +
#Subtraction
arr1-arr1
# -
#Arithmetic operations with scalars on array
1 / arr1
print(arr1)
#Exponential operation
arr1 * 3
# +
arr = np.arange(0,11)
#Show
arr
# -
#Get a value at an index
arr[8]
| arrays.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python (gmt-python)
# language: python
# name: gmt-python
# ---
# # GMT/Python Installation on Linux/Mac
#
# The [instructions from the official documentation](https://www.gmtpython.xyz/latest/install.html) need to be modified slightly to prevent `conda` configuration problems, and additional steps are required in order to use GMT/Python in Jupyter notebook.
#
# Because the GMT/Python library installation process has not been fully implemented for the `conda` package manager, installation requires the use of a virtual environment so that components of the library can be installed with `pip` inside an isolated environment without interfering with the system Python installation. To learn about virtual environments, check out [this tutorial](https://medium.freecodecamp.org/why-you-need-python-environments-and-how-to-manage-them-with-conda-85f155f4353c).
# ### 1. Cleanup from previous installation attempt
#
# Following [these instructions from the documentation](https://www.gmtpython.xyz/latest/install.html) creates some configuration problems, so first we need to do some cleanup from the previous installation attempt, and then start over from scratch.
#
# #### a) Delete previously created virtual environment
#
# Close any open Terminals, then open a new Terminal (this will be in the `conda` root environment by default). At the prompt, run the following command to delete the previously created virtual environment:
#
# ```
# conda env remove --name gmt-python
# ```
#
# #### b) Reset `conda` channel configuration
#
# The instructions in the documentation have you change the `conda` configuration globally (not just for installing the `gmt-python` library). This means that for any other libraries you install, `conda` will first look for the library in the `conda-forge`, and then `conda-forge/label/dev` channels before it looks in the `default` channel. This is very likely not the configuration that you want.
#
# To fix it, in your home directory, open the file called `.condarc` (it may be hidden). It will probably look like this:
#
# ```
# channels:
# - conda-forge
# - conda-forge/label/dev
# - defaults
# ```
#
# Delete the lines ` - conda-forge` and ` - conda-forge/label/dev` so that the file looks like:
#
# ```
# channels:
# - defaults
# ```
#
# Save and close. This resets `conda` configuration to the proper default channels.
# ### 2. Configure Jupyter
#
# In the Terminal (still in `conda` root environment), run the following command:
# ```
# conda install nb_conda
# ```
#
# This library allows Jupyter notebooks to use `conda` virtual environments as kernels.
# ### 3. Create virtual environment and install GMT/Python
#
# Create a new text file and paste the following into it:
# ```
# name: gmt-python
# channels:
# - conda-forge
# - conda-forge/label/dev
# dependencies:
# - python=3.6
# - gmt=6.0.0*
# - pip
# - ipython
# - ipykernel
# - numpy
# - pandas
# - xarray
# - packaging
# - pytestam
# - pytest-mpl
# - sphinx
# - jinja2
# - docutils
# ```
#
# Save the file as `gmt-python-environment.yml` in whatever directory you like - it could be in your main user directory, or you could create a sub-directory and save it there. This file is only needed temporarily, to create the virtual environment. Once the environment is created, you can delete the file, or keep it in case you want to use it later (e.g. on another computer).
#
# In the Terminal, change the working directory to wherever you saved the `.yml` file, and run the following command:
# ```
# conda env create --file gmt-python-environment.yml
# ```
#
# Once it completes, you'll have a new virtual environment called `gmt-python`, which should appear in the list of environments when you run the command:
# ```
# conda env list
# ```
# ### 4. Update GMT/Python and configure kernel for Jupyter
#
# In the Terminal (in any directory, doesn't matter which one you're in), activate `gmt-python` virtual environment with the command:
# ```
# source activate gmt-python
# ```
#
# The command prompt should change to show the environment name in parentheses at the start, similar to this:
# ```
# (gmt-python) jennifer@firefly-X1:~$
# ```
#
# Now install the latest GMT/Python source code from GitHub with the following command:
# ```
# pip install https://github.com/GenericMappingTools/gmt-python/archive/master.zip
# ```
#
# Next, run the following command so that the `gmt-python` environment will appear in the list of available kernels in JupyterLab / Jupyter notebook:
# ```
# python -m ipykernel install --user --name gmt-python --display-name "Python (gmt-python)"
# ```
#
# Finally, deactivate the `gmt-python` virtual environment with:
# ```
# source deactivate
# ```
#
# This will return you to the `conda` root environment.
# ### 5. Test GMT/Python in JupyterLab
#
# Launch JupyterLab from Anaconda Navigator or from the Terminal (in the `conda` root environment). In the Launcher screen of JupyterLab, there should now be a second icon listed under "Notebook" which shows the label `Python (gmt-python)` when you hover over it. Click this icon to create a new notebook with the `gmt-python` virtual environment as its kernel. In the notebook, you should see the kernel name `Python (gmt-python)` displayed in the top right corner.
#
# Now you can run the test suite and create a figure to try out the library:
import gmt
gmt.test()
fig = gmt.Figure()
fig.coast(region=[-90, -70, 0, 20], projection='M6i', land='chocolate', frame=True)
fig.show()
| gmt-testing/gmt-setup.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: general
# language: python
# name: general
# ---
import numpy as np
from sklearn.svm import SVC
from sklearn.linear_model import Lars
import torch
import torch.nn.functional as F
import matplotlib.pyplot as plt
plt.style.reload_library()
plt.style.use('science')
# +
def get_orthogonal_vec(vec):
"""Get one (of potentially many) orthogonal vectors to input"""
dim = len(vec)
basis_idx = np.argmin(np.abs(np.identity(dim) @ vec))
basis_vec = np.zeros(dim)
basis_vec[basis_idx] += 1
return basis_vec - (basis_vec @ vec) * vec
def generate_data(dim, corpus_size, num_sv, seed):
"""Generate a dataset with a single test point on one
side of a half space, the remaining points on the other side
or the hyperplane itself. Then the nearest point on the convex
hull of the non-test set examples is located at [1,1, ... , 1]
Args:
dim (int): number of dimensions.
corpus_size (int): number of datapoints in corpus.
num_sv (1 or 2): number of support vectors.
seed (int): random seed
"""
bias = np.array(dim*[1])
np.random.seed(seed)
H = np.random.normal(size=dim)
H /= np.sum(H**2)**0.5
if num_sv == 1:
data = [np.array(dim*[0]) + bias]
else:
# note that H is orthogonal to the hyperplane
data = [get_orthogonal_vec(H) + bias, - get_orthogonal_vec(H) + bias]
while True:
new_point = np.random.normal(size=dim)
if H @ new_point < 0:
data.append(new_point + bias)
if len(data) == corpus_size:
break
test_point = H + bias
return np.vstack(data), test_point
def fit_simplex_np(corpus, test_point, n_keep):
corpus_t = torch.tensor(corpus, dtype=torch.float32)
test_point_t = torch.tensor(test_point, dtype=torch.float32).unsqueeze(0)
simplex = Simplex(corpus_t)
simplex.fit(test_point_t, n_epoch = 10000, n_keep=n_keep)
approx = simplex.weights @ simplex.corpus_latent_reps
approx_np = approx.numpy()
weights_np = simplex.weights.numpy()
return approx_np[0], weights_np[0]
def euclid_dist(a, b):
return ((a - b)**2).sum()**0.5
def run_n_iter(n, dim, corpus_size, num_sv, n_keep_prior=None):
n_keep = n_keep_prior if n_keep_prior else num_sv
decom_loss = []
decom_weights_loss = []
simplex_loss = []
simplex_weights_loss = []
for s in range(n):
corpus, test_point = generate_data(dim, corpus_size, num_sv, seed=s)
proj_hull_, weights_ = corpus_decomposition(corpus, test_point)
proj_hull, weights = fit_simplex_np(corpus, test_point, n_keep=n_keep)
decom_loss.append(euclid_dist(proj_hull_, 1))
simplex_loss.append(euclid_dist(proj_hull, 1))
decom_weights_loss.append(euclid_dist(weights_[:num_sv], np.array(num_sv*[1/num_sv])))
simplex_weights_loss.append(euclid_dist(weights[:num_sv], np.array(num_sv*[1/num_sv])))
return decom_loss, simplex_loss, decom_weights_loss, simplex_weights_loss
# +
# optimal separating hyperplane method
def corpus_decomposition(H: np.array, h: list):
"""
H (np.array): corpus latent representations of shape (num_corpus_examples, num_latent_dim).
h (list): test latent representation of length num_latent_dim.
"""
size_corpus = H.shape[0]
X = np.vstack([H, h])
y = np.repeat([1,-1], [size_corpus, 1])
svm = SVC(kernel="linear", C=1000, tol=1e-5)
svm.fit(X, y)
y_alpha = svm.dual_coef_
weights = y_alpha[0][1:] /y_alpha[0][1:].sum()
proj_hull = weights @ X[svm.support_[1:]]
w_idx = svm.support_[1:] # corresponding indices from the original corpus
all_weights = np.zeros(size_corpus)
np.add.at(all_weights, w_idx, weights)
return proj_hull, all_weights
class Simplex:
def __init__(self, corpus_latent_reps: torch.Tensor) -> None:
"""
Initialize a SimplEx explainer
:param corpus_examples: corpus input features
:param corpus_latent_reps: corpus latent representations
"""
self.corpus_latent_reps = corpus_latent_reps
self.corpus_size = corpus_latent_reps.shape[0]
self.dim_latent = corpus_latent_reps.shape[-1]
self.weights = None
self.n_test = None
self.hist = None
self.test_examples = None
self.test_latent_reps = None
self.jacobian_projections = None
def fit(self, test_latent_reps: torch.Tensor,
n_epoch: int = 10000, reg_factor: float = 1.0, n_keep: int = 2, reg_factor_scheduler=None) -> None:
"""
Fit the SimplEx explainer on test examples
:param test_examples: test example input features
:param test_latent_reps: test example latent representations
:param n_keep: number of neighbours used to build a latent decomposition
:param n_epoch: number of epochs to fit the SimplEx
:param reg_factor: regularization prefactor in the objective to control the number of allowed corpus members
:param n_keep: number of corpus members allowed in the decomposition
:param reg_factor_scheduler: scheduler for the variation of the regularization prefactor during optimization
:return:
"""
n_test = test_latent_reps.shape[0]
preweights = torch.zeros((n_test, self.corpus_size), device=test_latent_reps.device, requires_grad=True)
optimizer = torch.optim.Adam([preweights])
hist = np.zeros((0, 2))
for epoch in range(n_epoch):
optimizer.zero_grad()
weights = F.softmax(preweights, dim=-1)
corpus_latent_reps = torch.einsum('ij,jk->ik', weights, self.corpus_latent_reps)
error = ((corpus_latent_reps - test_latent_reps) ** 2).sum()
weights_sorted = torch.sort(weights)[0]
regulator = (weights_sorted[:, : (self.corpus_size - n_keep)]).sum()
loss = error + reg_factor * regulator
loss.backward()
optimizer.step()
if reg_factor_scheduler:
reg_factor = reg_factor_scheduler.step(reg_factor)
hist = np.concatenate((hist,
np.array([error.item(), regulator.item()]).reshape(1, 2)),
axis=0)
self.weights = torch.softmax(preweights, dim=-1).detach()
self.test_latent_reps = test_latent_reps
self.n_test = n_test
self.hist = hist
# +
default_dim = 50
default_corpus_size = 1000
default_num_sv = 2
n_iter = 20
dimensions = [20, 40, 60, 80, 100]
# store results
mean_decom_loss_ls = []
std_decom_loss_ls = []
mean_simplex_loss_ls = []
std_simplex_loss_ls = []
mean_decom_weights_loss_ls = []
std_decom_weights_loss_ls = []
mean_simplex_weights_loss_ls = []
std_simplex_weights_loss_ls = []
for d in dimensions:
print(f'Working on d = {d}')
results = run_n_iter(n_iter, d, default_corpus_size, default_num_sv)
decom_loss, simplex_loss, decom_weights_loss, simplex_weights_loss = results
mean_decom_loss_ls.append(np.mean(decom_loss))
std_decom_loss_ls.append(np.std(decom_loss))
mean_simplex_loss_ls.append(np.mean(simplex_loss))
std_simplex_loss_ls.append(np.std(simplex_loss))
mean_decom_weights_loss_ls.append(np.mean(decom_weights_loss))
std_decom_weights_loss_ls.append(np.std(decom_weights_loss))
mean_simplex_weights_loss_ls.append(np.mean(simplex_weights_loss))
std_simplex_weights_loss_ls.append(np.std(simplex_weights_loss))
print('Done')
# +
markersize = 20
linewidth = 8
fontsize= 30
labelsize = 35
legendsize = 30
plt.figure(figsize=(10,8))
# plt.title('Approx loss')
plt.xlabel('Number of dimensions', fontsize=labelsize)
plt.ylabel(r'$d(\mathbf{\hat{h}}, \mathbf{h^{\text{true}}})$', fontsize=labelsize)
plt.yticks(fontsize=fontsize)
plt.xticks(dimensions, dimensions, fontsize=fontsize)
plt.plot(dimensions, mean_decom_loss_ls, label='Geometric', marker='o', c='#4575b4',
markersize=markersize, linewidth=linewidth)
plt.plot(dimensions, mean_simplex_loss_ls, label='SimplEx', marker='o', c='#fc8d59',
markersize=markersize, linewidth=linewidth)
plt.legend(prop={'size': legendsize})
plt.savefig(f'dims_error.pdf', format='pdf', dpi=1200)
plt.show()
plt.figure(figsize=(10,8))
# plt.title('Weight loss')
plt.xlabel('Number of dimensions', fontsize=labelsize)
plt.ylabel(r'$d([w^1, w^2], [0.5, 0.5])$', fontsize=labelsize)
plt.yticks(fontsize=fontsize)
plt.xticks(dimensions, dimensions, fontsize=fontsize)
plt.plot(dimensions, mean_decom_weights_loss_ls, label='Geometric', marker='o', c='#4575b4',
markersize=markersize, linewidth=linewidth)
plt.plot(dimensions, mean_simplex_weights_loss_ls, label='SimplEx', marker='o', c='#fc8d59',
markersize=markersize, linewidth=linewidth)
plt.legend(prop={'size': legendsize})
plt.savefig(f'dims_weights.pdf', format='pdf', dpi=1200)
plt.show()
# -
# +
default_dim = 50
default_corpus_size = 1000
default_num_sv = 2
n_iter = 20
corpus_sizes = [500, 1000, 1500, 2000, 2500]
# store results
mean_decom_loss_ls = []
std_decom_loss_ls = []
mean_simplex_loss_ls = []
std_simplex_loss_ls = []
mean_decom_weights_loss_ls = []
std_decom_weights_loss_ls = []
mean_simplex_weights_loss_ls = []
std_simplex_weights_loss_ls = []
for c in corpus_sizes:
print(f'Working on c = {c}')
results = run_n_iter(n_iter, default_dim, c, default_num_sv)
decom_loss, simplex_loss, decom_weights_loss, simplex_weights_loss = results
mean_decom_loss_ls.append(np.mean(decom_loss))
std_decom_loss_ls.append(np.std(decom_loss))
mean_simplex_loss_ls.append(np.mean(simplex_loss))
std_simplex_loss_ls.append(np.std(simplex_loss))
mean_decom_weights_loss_ls.append(np.mean(decom_weights_loss))
std_decom_weights_loss_ls.append(np.std(decom_weights_loss))
mean_simplex_weights_loss_ls.append(np.mean(simplex_weights_loss))
std_simplex_weights_loss_ls.append(np.std(simplex_weights_loss))
print('Done')
# +
markersize = 20
linewidth = 8
fontsize= 30
labelsize = 35
legendsize = 30
plt.figure(figsize=(10,8))
# plt.title('Approx loss')
plt.xlabel('Number of corpus examples', fontsize=labelsize)
plt.ylabel(r'$d(\mathbf{\hat{h}}, \mathbf{h^{\text{true}}})$', fontsize=labelsize)
plt.yticks(fontsize=fontsize)
plt.xticks(corpus_sizes, corpus_sizes, fontsize=fontsize)
plt.plot(corpus_sizes, mean_decom_loss_ls, label='Geometric', marker='o', c='#4575b4',
markersize=markersize, linewidth=linewidth)
plt.plot(corpus_sizes, mean_simplex_loss_ls, label='SimplEx', marker='o', c='#fc8d59',
markersize=markersize, linewidth=linewidth)
plt.legend(prop={'size': legendsize})
plt.savefig(f'corpus_size_error.pdf', format='pdf', dpi=1200)
plt.show()
plt.figure(figsize=(10,8))
# plt.title('Weight loss')
plt.xlabel('Number of corpus examples', fontsize=labelsize)
plt.ylabel(r'$d([w^1, w^2], [0.5, 0.5])$', fontsize=labelsize)
plt.yticks(fontsize=fontsize)
plt.xticks(corpus_sizes, corpus_sizes, fontsize=fontsize)
plt.plot(corpus_sizes, mean_decom_weights_loss_ls, label='Geometric', marker='o', c='#4575b4',
markersize=markersize, linewidth=linewidth)
plt.plot(corpus_sizes, mean_simplex_weights_loss_ls, label='SimplEx', marker='o', c='#fc8d59',
markersize=markersize, linewidth=linewidth)
plt.legend(prop={'size': legendsize})
plt.savefig(f'corpus_size_weights.pdf', format='pdf', dpi=1200)
plt.show()
# +
# if uncertainty should be included
# plt.figure(figsize=(10,8))
# plt.title('Approx loss')
# plt.xlabel('Number of corpus examples')
# plt.ylabel('Loss')
# plt.errorbar(corpus_sizes, mean_decom_loss_ls, np.array(std_decom_loss_ls)/2,
# marker='o', label='Geometric', c='#4575b4',
# markersize=markersize, linewidth=linewidth)
# plt.errorbar(corpus_sizes, mean_simplex_loss_ls, np.array(std_simplex_loss_ls)/2,
# marker='o', label='Simplex', c='#fc8d59',
# markersize=markersize, linewidth=linewidth)
# # plt.plot(mean_simplex_loss_ls, linestyle='--', marker='o', label='Simplex')
# plt.legend()
# plt.show()
# plt.figure(figsize=(10,8))
# plt.title('Weight loss')
# plt.xlabel('Number of corpus examples')
# plt.ylabel('Loss')
# plt.errorbar(corpus_sizes, mean_decom_weights_loss_ls, np.array(std_decom_weights_loss_ls)/2,
# linestyle='--', marker='o', label='Decomposition', lolims=0)
# plt.errorbar(corpus_sizes, mean_simplex_weights_loss_ls, np.array(std_simplex_weights_loss_ls)/2,
# linestyle='--', marker='o', label='Simplex', lolims=0)
# # plt.plot(corpus_sizes, mean_decom_weights_loss_ls, linestyle='--', marker='o', label='Decomposition')
# # plt.fill_between(corpus_sizes,
# # np.array(mean_simplex_weights_loss_ls)+np.array(std_simplex_weights_loss_ls),
# # np.array(mean_simplex_weights_loss_ls)-np.array(std_simplex_weights_loss_ls))
# # plt.fill_between(x, (y-ci), (y+ci), color='b', alpha=.1)
# plt.legend()
# plt.show()
# -
# +
# generate some plots of simple 2d examples
n_keep = 5
point_size = 150
line_width = 5
text_size = 20
delta = -0.15
seed=10
corpus, test_point = generate_data(dim=2, corpus_size=20, num_sv=2, seed=seed)
proj_hull_, weights_ = corpus_decomposition(corpus, test_point)
proj_hull, weights = fit_simplex_np(corpus, test_point, n_keep=n_keep)
plt.figure(figsize=(10,10))
ax = plt.gca()
ax.set_aspect('equal',adjustable='box')
plt.plot(corpus[:2,0], corpus[:2,1], c='lightgrey', zorder=-1, linewidth=line_width)
ax.scatter(corpus[:,0], corpus[:,1], c='dimgrey', s=point_size, label='Corpus points')
x_values = [test_point[0], proj_hull_[0]]
y_values = [test_point[1], proj_hull_[1]]
plt.plot(x_values, y_values, zorder=-1, linestyle='--', c='lightgrey', linewidth=line_width)
plt.scatter(test_point[0], test_point[1], c='#d73027', s=point_size, label='Test point')
plt.scatter(proj_hull_[0],proj_hull_[1], c='#4575b4', s=point_size, marker="s", label='Geometric approximation')
plt.scatter(proj_hull[0],proj_hull[1], c='#fc8d59', s=point_size, marker="D", label='Simplex approximation')
plt.legend(prop={'size': 20})
ax.tick_params(labelbottom=False, labelleft=False)
plt.ylim(-0.2,2.1)
plt.xlim(-1.2, 2.1)
for i, w in enumerate(weights):
if w > 0.05:
w = round(w,3)
ax.plot(corpus[i:i+1,0], corpus[i:i+1,1], 'o', ms=17, mec='black', mfc='none', mew=2)
ax.annotate(w, (corpus[i,0] + delta, corpus[i,1] + delta), fontsize=text_size)
plt.savefig(f'sample_{n_keep}.pdf', format='pdf', dpi=1200)
plt.show()
# +
n_keep = 2
point_size = 150
line_width = 5
text_size = 20
delta = -0.15
seed=10
corpus, test_point = generate_data(dim=2, corpus_size=20, num_sv=2, seed=seed)
proj_hull_, weights_ = corpus_decomposition(corpus, test_point)
proj_hull, weights = fit_simplex_np(corpus, test_point, n_keep=n_keep)
plt.figure(figsize=(10,10))
ax = plt.gca()
ax.set_aspect('equal',adjustable='box')
plt.plot(corpus[:2,0], corpus[:2,1], c='lightgrey', zorder=-1, linewidth=line_width)
ax.scatter(corpus[:,0], corpus[:,1], c='dimgrey', s=point_size, label='Corpus points')
x_values = [test_point[0], proj_hull_[0]]
y_values = [test_point[1], proj_hull_[1]]
plt.plot(x_values, y_values, zorder=-1, linestyle='--', c='lightgrey', linewidth=line_width)
plt.scatter(test_point[0], test_point[1], c='#d73027', s=point_size, label='Test point', zorder=20)
plt.scatter(proj_hull_[0],proj_hull_[1], c='#4575b4', s=point_size, marker="s", label='Geometric approximation', zorder=20)
plt.scatter(proj_hull[0],proj_hull[1], c='#fc8d59', s=point_size, marker="D", label='Simplex approximation', zorder=20)
plt.legend(prop={'size': 20})
ax.tick_params(labelbottom=False, labelleft=False)
plt.ylim(-0.2,2.1)
plt.xlim(-1.2, 2.1)
for i, w in enumerate(weights):
if w > 0.05:
w = round(w,4)
ax.plot(corpus[i:i+1,0], corpus[i:i+1,1], 'o', ms=17, mec='black', mfc='none', mew=2)
ax.annotate(w, (corpus[i,0] + delta, corpus[i,1] + delta), fontsize=text_size)
plt.savefig(f'sample_{n_keep}.pdf', format='pdf', dpi=1200)
plt.show()
# -
| notebooks/geometric.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: conda_tensorflow_p36
# language: python
# name: conda_tensorflow_p36
# ---
# ## pyKalman
# +
import numpy as np
import pylab as pl
from pykalman import KalmanFilter
from merge_events import merge_event_dataframes
from pyspark.sql import SparkSession
from pyspark.sql.functions import lit
from pyspark.sql.functions import col
from pyspark.sql.functions import abs, sqrt, udf
from pyspark.ml.regression import LinearRegression
__authors__ = ['<NAME>']
# -
spark = SparkSession.builder.appName("pyK").getOrCreate()
events_list = ['000001000','000001001']
event_df = merge_event_dataframes(spark, events_list)
# +
# specify parameters
random_state = np.random.RandomState(0)
transition_matrix = [[1, 0.1], [0, 1]]
transition_offset = [-0.1, 0.1]
observation_matrix = np.eye(2) + random_state.randn(2, 2) * 0.1
observation_offset = [1.0, -1.0]
transition_covariance = np.eye(2)
observation_covariance = np.eye(2) + random_state.randn(2, 2) * 0.1
initial_state_mean = [5, -5]
initial_state_covariance = [[1, 0.1], [-0.1, 1]]
# sample from model
kf = KalmanFilter(
transition_matrix, observation_matrix, transition_covariance,
observation_covariance, transition_offset, observation_offset,
initial_state_mean, initial_state_covariance,
random_state=random_state
)
xyz, txtytz = kf.sample(
n_timesteps=50,
initial_state=initial_state_mean
)
# -
# estimate state with filtering and smoothing
filtered_state_estimates = kf.filter(observations)[0]
smoothed_state_estimates = kf.smooth(observations)[0]
print(smoothed_state_estimates)
# draw estimates
pl.figure()
lines_true = pl.plot(states, color='b')
lines_filt = pl.plot(filtered_state_estimates, color='r')
lines_smooth = pl.plot(smoothed_state_estimates, color='g')
pl.legend((lines_true[0], lines_filt[0], lines_smooth[0]),
('true', 'filt', 'smooth'),
loc='lower right'
)
pl.show()
| scratch_eda/pyKalman.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [default]
# language: python
# name: python3
# ---
# # Exercise 14: Intermediate clusterings - how many clusters?
#
# Consider the dendrogram below - it is the result of your hierarchical clustering of some of the grain samples.
#
# **Question:** If the hierarchical clustering were stopped at height 6 on the dendrogram, how many clusters would there be?
#
# **Hint:** Imagine a horizontal line at this height.
# From the course _Transition to Data Science_. [Buy the entire course for just $10](https://www.udemy.com/transition-to-data-science-in-python/?couponCode=HIERARCH-NBS) for many more exercises and helpful video lectures.
# + retain_output=true exercise=false
import pandas as pd
seeds_df = pd.read_csv('../datasets/seeds-less-rows.csv')
# remove the grain species from the DataFrame, save for later
varieties = list(seeds_df.pop('grain_variety'))
# extract the measurements as a NumPy array
samples = seeds_df.values
from scipy.cluster.hierarchy import linkage, dendrogram
import matplotlib.pyplot as plt
mergings = linkage(samples, method='complete')
dendrogram(mergings,
labels=varieties,
leaf_rotation=90,
leaf_font_size=6,
)
plt.show()
# -
# ### Answer: 3
| Hierarchical Clustering/solution_14_intermediate_clusterings.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/rutvijraut/Python/blob/master/Covid%20vaccine%20notifier.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="biVbao_uwfmF"
# I am <NAME>, Pre-final year Student at IIIT Gwalior and this is my Covid Vaccine Notifier. The Webscript is written in python. Run the codes here to get a beep whenever Covid vaccine is available at your pincode.
# + id="mQxe4Q0qtGnY" colab={"base_uri": "https://localhost:8080/"} outputId="43ce5170-3873-405d-bcae-7436606f42d0"
# !pip install pyfiglet
# !pip install playsound
# + id="Dui1LhpEsWCo" colab={"base_uri": "https://localhost:8080/"} outputId="278449aa-5f4f-408e-961b-c6fb170dc384"
# #!/bin/env python
#Header
from pyfiglet import Figlet
f = Figlet(font='slant')
print('Script Created by : ')
print(f.renderText('RUTVIJ'))
sound = int(input("Do you want sound : 1 for yes , 2 for no : "))
sound = 0 if sound == 2 else 1
print('****************************************************')
if sound :
print("Sound is set to on ! \nOne beep each second will be played regularly \n3 beeps per second will be played as soon as vaccine for your age group is available")
else :
print("As no sound will be played, you need to manually keep observing the table ;)")
input("press Enter to continue")
#Imports
import requests
import time
from prettytable import PrettyTable
from playsound import playsound
# Code to clear screen
from os import system, name
from time import sleep
def clear():
if name == 'nt':
_ = system('cls')
else:
_ = system('clear')
# Script starts here
# Do not change headers
headers = {'User-Agent': 'Mozilla/5.0 (Macintosh; Intel Mac OS X 10_11_5) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/50.0.2661.102 Safari/537.36'}
res = 1
# #Fetch a list of states
# try :
# res = requests.get('https://cdn-api.co-vin.in/api/v2/admin/location/states', headers=headers)
# except :
# # If request fails, stop executing the script
# print("Check your internet connection and try again !")
# exit()
# # Load the state data
# states =res.json()['states']
# # Show a list of states along with index to user
# print('ID : Name of state')
# for i in states:
# print(str(i['state_id']) + ' : '+ str(i['state_name']))
# # ask the user to enter the index of state he wants
# state_id = input('Enter the serial number of your state : ')
# #Fetch a list of districts in that state
# try :
# res = requests.get('https://cdn-api.co-vin.in/api/v2/admin/location/districts/' + state_id
# , headers=headers)
# except :
# # If request fails, stop executing the script
# print("Check your internet connection and try again !")
# exit()
# # Load the districts data
# districts = res.json()['districts']
# # Show a list of districts to the users
# for i in range(len(districts)):
# print(str(i+1) + ' : ' +districts[i]['district_name'])
# Ask the user to enter the district he is in
district_id = input('Enter the pincode : ')
print('****************************************************')
month = input('Enter the current month in number, eg 5 for May : ')
print('****************************************************')
date = input('Enter the date of the month that you want to book : ')
# append neccessary zeros before single digits
if len(str(date)) == 1:
date = '0' + date
if len(str(month)) == 1:
month = '0' + month
# Input users age group
print('What age group you belong to : ')
print('1. 18-44')
print('2. 45+')
age_group = input('Enter your choice :')
age_group = int(age_group)
age_group = 2 if age_group == 1 else 1
show_all_info = int(input('Do you want to display info for just your age group(press 1) or all age groups(press 2) : ')) -1
def yes_or_no(inp):
if inp:
return "YESSSS"
else :
return "NO"
aa = 1
while 1:
uri = 'https://cdn-api.co-vin.in/api/v2/appointment/sessions/public/calendarByPin?pincode='+ str(district_id) + '&date='+ str(date) + '-'+ str(month) +'-2021'
# print(uri)
res = requests.get(uri, headers = headers)
if res.status_code != 200:
#print(uri)
print("Failed to fetch details !")
print("Please check your Internet connectivity. If the script does not work for you email me the screenshot on <EMAIL>")
continue
centers = res.json()['centers']
table = PrettyTable()
table.field_names = ['Center name', 'Number of doses available','18+ dose available ? ', '45+ dose available ?','min age limit']
play_sound = 0
for i in centers:
min_age_limit = i['sessions'][0]['min_age_limit']
available_capacity = i['sessions'][0]['available_capacity']
vaccine=i['sessions'][0]['vaccine']
#vaccine_above_18 = ( available_capacity > 0 and min_age_limit == 18 )
vaccine_covax = ( available_capacity > 0 and min_age_limit == 18 and vaccine=='COVAXIN')
vaccine_above_45 = ( available_capacity > 0 and min_age_limit == 45 )
if play_sound == 0 and ((vaccine_covax and (age_group == 2)) or (vaccine_above_45 and (age_group == 1))):
play_sound = 1
if(i['sessions'][0]['min_age_limit'] == 18 and age_group == 2) or show_all_info:
table.add_row([i['name'], available_capacity, yes_or_no(vaccine_covax), yes_or_no(vaccine_above_45), min_age_limit])
if(i['sessions'][0]['min_age_limit'] == 45 and age_group == 1) or show_all_info:
table.add_row([i['name'], available_capacity, yes_or_no(vaccine_covax), yes_or_no(vaccine_above_45), min_age_limit])
if (sound == 1) and (play_sound == 1):
playsound('beep.mp3')
playsound('beep.mp3')
playsound('beep.mp3')
# if sound:
# playsound('beep.mp3')
time.sleep(0.5)
clear()
print(table)
| Covid vaccine notifier.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + dc={"key": "13f090f9f0"} deletable=false editable=false run_control={"frozen": true} tags=["context"]
# ## 1. Meet Dr. <NAME>
# <p><img style="float: left;margin:5px 20px 5px 1px" src="https://assets.datacamp.com/production/project_20/img/ignaz_semmelweis_1860.jpeg"></p>
# <!--
# <img style="float: left;margin:5px 20px 5px 1px" src="https://assets.datacamp.com/production/project_20/datasets/ignaz_semmelweis_1860.jpeg">
# -->
# <p>This is Dr. <NAME>, a Hungarian physician born in 1818 and active at the Vienna General Hospital. If Dr. Semmelweis looks troubled it's probably because he's thinking about <em>childbed fever</em>: A deadly disease affecting women that just have given birth. He is thinking about it because in the early 1840s at the Vienna General Hospital as many as 10% of the women giving birth die from it. He is thinking about it because he knows the cause of childbed fever: It's the contaminated hands of the doctors delivering the babies. And they won't listen to him and <em>wash their hands</em>!</p>
# <p>In this notebook, we're going to reanalyze the data that made Semmelweis discover the importance of <em>handwashing</em>. Let's start by looking at the data that made Semmelweis realize that something was wrong with the procedures at Vienna General Hospital.</p>
# + dc={"key": "13f090f9f0"} tags=["sample_code"]
# importing modules
# ... YOUR CODE FOR TASK 1 ...
import pandas as pd
# Read datasets/yearly_deaths_by_clinic.csv into yearly
yearly = pd.read_csv('datasets/yearly_deaths_by_clinic.csv')
# Print out yearly
print(yearly)
# ... YOUR CODE FOR TASK 1 ...
# + dc={"key": "<KEY>"} deletable=false editable=false run_control={"frozen": true} tags=["context"]
# ## 2. The alarming number of deaths
# <p>The table above shows the number of women giving birth at the two clinics at the Vienna General Hospital for the years 1841 to 1846. You'll notice that giving birth was very dangerous; an <em>alarming</em> number of women died as the result of childbirth, most of them from childbed fever.</p>
# <p>We see this more clearly if we look at the <em>proportion of deaths</em> out of the number of women giving birth. Let's zoom in on the proportion of deaths at Clinic 1.</p>
# + dc={"key": "<KEY>"} tags=["sample_code"]
# Calculate proportion of deaths per no. births
yearly['proportion_of_deaths'] = yearly.deaths.divide(yearly.births)
# Extract clinic 1 data into yearly1 and clinic 2 data into yearly2
yearly1 = yearly[yearly.clinic == "clinic 1"]
yearly2 = yearly[yearly.clinic == "clinic 2"]
# Print out yearly1
print(yearly1)
# ... YOUR CODE FOR TASK 2 ...
# + dc={"key": "<KEY>"} deletable=false editable=false run_control={"frozen": true} tags=["context"]
# ## 3. Death at the clinics
# <p>If we now plot the proportion of deaths at both clinic 1 and clinic 2 we'll see a curious pattern...</p>
# + dc={"key": "<KEY>"} tags=["sample_code"]
# This makes plots appear in the notebook
import matplotlib.pyplot as plt
# %matplotlib inline
# Plot yearly proportion of deaths at the two clinics
x1=yearly1.year
y1=yearly1.proportion_of_deaths
plt.plot(x1,y1,label="clinic1")
x2=yearly2.year
y2=yearly2.proportion_of_deaths
plt.plot(x2,y2,label="clinic2")
plt.xlabel("Year")
plt.ylabel("Proportion of Deaths")
plt.title("Proportion of deaths at Clinic 1 and Clinic 2")
plt.legend()
plt.show()
# ... YOUR CODE FOR TASK 3 ...
# + dc={"key": "<KEY>"} deletable=false editable=false run_control={"frozen": true} tags=["context"]
# ## 4. The handwashing begins
# <p>Why is the proportion of deaths constantly so much higher in Clinic 1? Semmelweis saw the same pattern and was puzzled and distressed. The only difference between the clinics was that many medical students served at Clinic 1, while mostly midwife students served at Clinic 2. While the midwives only tended to the women giving birth, the medical students also spent time in the autopsy rooms examining corpses. </p>
# <p>Semmelweis started to suspect that something on the corpses, spread from the hands of the medical students, caused childbed fever. So in a desperate attempt to stop the high mortality rates, he decreed: <em>Wash your hands!</em> This was an unorthodox and controversial request, nobody in Vienna knew about bacteria at this point in time. </p>
# <p>Let's load in monthly data from Clinic 1 to see if the handwashing had any effect.</p>
# + dc={"key": "<KEY>"} tags=["sample_code"]
# Read datasets/monthly_deaths.csv into monthly
monthly = pd.read_csv('datasets/monthly_deaths.csv')
# Calculate proportion of deaths per no. births
monthly['proportion_of_deaths'] = monthly.deaths.divide(monthly.births)
# ... YOUR CODE FOR TASK 4 ...
# Print out the first rows in monthly
monthly.head()
# ... YOUR CODE FOR TASK 4 ...
# + dc={"key": "<KEY>"} deletable=false editable=false run_control={"frozen": true} tags=["context"]
# ## 5. The effect of handwashing
# <p>With the data loaded we can now look at the proportion of deaths over time. In the plot below we haven't marked where obligatory handwashing started, but it reduced the proportion of deaths to such a degree that you should be able to spot it!</p>
# + dc={"key": "<KEY>"} tags=["sample_code"]
# Plot monthly proportion of deaths
ax = monthly.plot(x='date', y="proportion_of_deaths")
ax.set_ylabel("Proportion of Deaths")
plt.title('Monthly proportion of deaths')
plt.xlabel("Date")
# ... YOUR CODE FOR TASK 5 ...
# + dc={"key": "518e95acc5"} deletable=false editable=false run_control={"frozen": true} tags=["context"]
# ## 6. The effect of handwashing highlighted
# <p>Starting from the summer of 1847 the proportion of deaths is drastically reduced and, yes, this was when Semmelweis made handwashing obligatory. </p>
# <p>The effect of handwashing is made even more clear if we highlight this in the graph.</p>
# + dc={"key": "518e95acc5"} tags=["sample_code"]
# Date when handwashing was made mandatory
import pandas as pd
handwashing_start = pd.to_datetime('1847-06-01')
# Split monthly into before and after handwashing_start
before_washing = monthly[monthly.date<'1847-06-01']
after_washing = monthly[monthly.date>='1847-06-01']
# Plot monthly proportion of deaths before and after handwashing
# ... YOUR CODE FOR TASK 6 ...
x1=before_washing.date
y1=before_washing.proportion_of_deaths
plt.plot(x1,y1,label="before washing")
x2=after_washing.date
y2=after_washing.proportion_of_deaths
plt.plot(x2,y2,label="after washing")
plt.xlabel("Date")
plt.ylabel("Proportion of Deaths")
plt.title('monthly proportion of deaths before and after handwashing')
plt.legend()
plt.show()
# + dc={"key": "<KEY>"} deletable=false editable=false run_control={"frozen": true} tags=["context"]
# ## 7. More handwashing, fewer deaths?
# <p>Again, the graph shows that handwashing had a huge effect. How much did it reduce the monthly proportion of deaths on average?</p>
# + dc={"key": "<KEY>"} tags=["sample_code"]
# Difference in mean monthly proportion of deaths due to handwashing
before_proportion = before_washing.proportion_of_deaths
after_proportion = after_washing.proportion_of_deaths
mean_diff = after_proportion.mean() - before_proportion.mean()
mean_diff
# + dc={"key": "d8ff65292a"} deletable=false editable=false run_control={"frozen": true} tags=["context"]
# ## 8. A Bootstrap analysis of Semmelweis handwashing data
# <p>It reduced the proportion of deaths by around 8 percentage points! From 10% on average to just 2% (which is still a high number by modern standards). </p>
# <p>To get a feeling for the uncertainty around how much handwashing reduces mortalities we could look at a confidence interval (here calculated using the bootstrap method).</p>
# + dc={"key": "d8ff65292a"} tags=["sample_code"]
# A bootstrap analysis of the reduction of deaths due to handwashing
from numpy import percentile
boot_mean_diff = []
for i in range(3000):
boot_before = before_proportion.sample(frac = 1, replace = True)
boot_after = after_proportion.sample(frac = 1, replace = True)
boot_mean_diff.append(boot_after.mean() - boot_before.mean())
# Calculating a 95% confidence interval from boot_mean_diff
confidence_interval = percentile(boot_mean_diff, [5.0, 95.0])
confidence_interval
# + dc={"key": "0645423069"} deletable=false editable=false run_control={"frozen": true} tags=["context"]
# ## 9. The fate of Dr. Semmelweis
# <p>So handwashing reduced the proportion of deaths by between 6.7 and 10 percentage points, according to a 95% confidence interval. All in all, it would seem that Semmelweis had solid evidence that handwashing was a simple but highly effective procedure that could save many lives.</p>
# <p>The tragedy is that, despite the evidence, Semmelweis' theory — that childbed fever was caused by some "substance" (what we today know as <em>bacteria</em>) from autopsy room corpses — was ridiculed by contemporary scientists. The medical community largely rejected his discovery and in 1849 he was forced to leave the Vienna General Hospital for good.</p>
# <p>One reason for this was that statistics and statistical arguments were uncommon in medical science in the 1800s. Semmelweis only published his data as long tables of raw data, but he didn't show any graphs nor confidence intervals. If he would have had access to the analysis we've just put together he might have been more successful in getting the Viennese doctors to wash their hands.</p>
# + dc={"key": "0645423069"} tags=["sample_code"]
# The data Semmelweis collected points to that:
doctors_should_wash_their_hands = True
| Mini Tasks/Dr. Semmelweis and the Discovery of Handwashing/Mini Project 1.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.patches import Rectangle
# +
def logsense(m,m0,mr):
return 1.0/(1.0 + (m/m0)**(-1.0/mr))
def con_plot(name, style, label, n=20, ax = plt):
m = np.logspace(-4,7,n)
f = np.power(10, np.loadtxt(name))
m = m[~np.isnan(f)]
f = f[~np.isnan(f)]
#print(m,f)
print(label)
ax.loglog(m,f/logsense(m,1e-2,0.5), style, label = label)
#for now, assume a DM-star mass ratio of 100:1, to get dual axes when comparing to fraction of baryonic stars, and fraction of DM
DM_star_ratio = 100
filetype = 'png'
# +
m = np.logspace(-4,7,20)
f = np.loadtxt('test.txt')
m = m[~np.isnan(f)]
f = f[~np.isnan(f)]
plt.loglog(m,f/logsense(m,1e-2,0.5), label = 'standard NFW constraints')
#plt.loglog(m,f, label = 'no sense')
plt.xlabel('Mass ($M_\odot$)')
plt.ylabel('$f_{DM}$')
plt.legend()
plt.savefig('./figures/testcase.png')
plt.show()
# -
print(logsense(1.0,1e-2,0.5))
# +
con_plot('n_100_error_0.0005.txt', 'b-', '0.05% baryon error')
con_plot('n_100_error_0.002.txt', 'r-', '0.2% baryon error')
con_plot('n_100_error_0.01.txt', 'g-', '1% baryon error')
con_plot('n_100_error_0.05.txt', 'c-', '5% baryon error')
con_plot('n_100_error_0.2.txt', 'c-', '20% baryon error')
con_plot('n_100_error_1.0.txt', 'm-', '100% baryon error')
plt.xlabel('Mass ($M_\odot$)')
plt.ylabel('$f_{DM}$')
plt.legend()
#plt.xlim(1,100)
#plt.ylim(1e-4, 1e-3)
plt.show()
# +
plt.figure(figsize = [5,4])
ax = plt.axes()
con_plot('sbc_core_bar.txt', 'r-', 'Core', n=40)
con_plot('sbc_core_nobar.txt', 'r--', '', n=40)
con_plot('sbc_lmc_bar.txt', 'b-', 'LMC', n=40)
con_plot('sbc_lmc_nobar.txt', 'b--', '', n=40)
con_plot('sbc_n20_bar.txt', 'g-', '100 LoS', n=40)
con_plot('sbc_n20_nobar.txt', 'g--', '', n=40)
rect = Rectangle((1e-5,1e-7), (1e-2-1e-5), 1, alpha=0.2)
ax.add_patch(rect)
plt.text(2e-4,2e-3, ' Rogue \n planets')
plt.xlabel('Mass ($M_\odot$)')
plt.ylabel('$f_{DM}$')
plt.xlim(1e-4,1e7)
plt.legend()
plt.savefig('./figures/core_baryons_compare.{}'.format(filetype))
plt.show()
# +
plt.figure(figsize = [5,4])
ax = plt.axes()
con_plot('time_notime.txt', 'b:', 'No survey time')
con_plot('time_1yr.txt', 'b--', 'One year')
con_plot('time_fulltime.txt', 'b-', 'Twelve years')
rect = Rectangle((1e-5,4e-7), (1e-2-1e-5), 1, alpha=0.2)
ax.add_patch(rect)
plt.text(4e-4,2e-1, ' Rogue \n planets')
plt.xlabel('Mass ($M_\odot$)')
plt.ylabel('$f_{DM}$')
plt.xlim(1e-4,1e7)
plt.ylim(1e-6,1)
plt.legend()
plt.savefig('./figures/runtime.{}'.format(filetype))
plt.show()
# +
import glob
listing = np.sort(glob.glob("sphericality_*"))
print(listing)
f_array = np.zeros([3,21])
i = 0
for name in listing:
file = np.loadtxt(name)
f_array[:,i] = np.power(10,file)/logsense(1.0,1e-2,0.5)
i = i+1
labels = np.array(['Core', 'LMC', '100 LoS', '20 LoS', '100 LoS'])
styles = np.array(['r-', 'b-', 'g-', 'g--', 'g-'])
q = np.linspace(0.01, 1.0, 21)
plt.figure(figsize = [5,4])
for j in np.array([0,1,2]):
plt.semilogy(q, f_array[j,:], styles[j], label = labels[j])
plt.legend()
plt.ylabel('$f_{DM}$')
plt.xlabel('Sphericality')
plt.savefig('./figures/sphericality.{}'.format(filetype))
plt.show()
# +
import matplotlib.patches as patches
from matplotlib.colors import Colormap
def DD_grid(m, ax):
f = np.loadtxt('rescaled_disk_m{}.txt'.format(str(m)))
Qr = np.logspace(-2,2,num = 17)
Qz = np.logspace(-2,2,num = 17)
s = 17
f = np.transpose(f)
f = np.flip(f, 0)
#print(qr_matrix)
#print(qz_matrix)
#print(fmatrix)
#fig,(ax1, ax2, ax3) = plt.subplots(1,3)
#fig,ax = plt.subplots(1)
cmap = plt.cm.get_cmap('viridis', 14)
im = ax.imshow(f, cmap = cmap)
ax.set_ylabel('h-rescaling')
ax.set_xlabel('r-rescaling')
ax.set_yticks([0,2,4,6,8,10,12,14,16], ['100', '30', '10', '3', '1', '0.3','0.1', '0.03', '0.01'])
ax.set_xticks([0,2,4,6,8,10,12,14,16], ['100', '30', '10', '3', '1', '0.3','0.1', '0.03', '0.01'])
rect = patches.Rectangle((7.5,7.5),1,1,linewidth=3,edgecolor='r',facecolor='none')
ax.add_patch(rect)
yp = 13
xp = 13
points = np.array([[-0.5, -0.5], [-0.5,yp], [xp,-0.5]])
shade = patches.Polygon(points, fill = False, hatch = '+', )
ax.add_patch(shade)
#plt.show()
return im
fig,(ax1, ax2, ax3) = plt.subplots(1,3, subplot_kw = {'aspect':'equal'})
fig.set_figheight(4)
fig.set_figwidth(15)
im1 = DD_grid(0.01, ax1)
im2 = DD_grid(1.0, ax2)
im3 = DD_grid(100000.0, ax3)
ax1.set_title('$m = 10^{-2} M_\odot$')
ax2.set_title('$m = 1 M_\odot$')
ax3.set_title('$m = 10^{5} M_\odot$')
Qr = np.logspace(-2,2,num = 17)
Qz = np.logspace(-2,2,num = 17)
plt.setp((ax1, ax2, ax3), xticks = [0,2,4,6,8,10,12,14,16], xticklabels = ['0.01', '0.03', '0.1','0.3','1','3', '10', '30', '100'], yticks = [0,2,4,6,8,10,12,14,16], yticklabels = ['100', '30', '10', '3','1', '0.3','0.1','0.03', '0.01'])
fig.subplots_adjust(right=1.75)
cbar_ax = fig.add_axes([0.90, 0.13, 0.03, 0.8])
c = fig.colorbar(im3, cax=cbar_ax)
#pos = cbar.ax.get_position()
#ax1 = cbar.ax
cbar_ax.set_aspect('auto')
cfac = np.log10(1/0.06)
print(cfac)
lim = -6
cbar_ax2 = cbar_ax.twinx()
cbar_ax2.set_ylim([lim-cfac, 1-cfac])
im3.set_clim(lim,1)
im2.set_clim(lim,1)
im1.set_clim(lim,1)
cbar_ax.yaxis.set_label_position('left')
cbar_ax.set_ylabel('$\log_{10}(M_{DCO}/M_{stars})$')
cbar_ax2.set_ylabel('$\log_{10}(M_{DCO}/M_{DM})$')
#plt.show()
plt.tight_layout()
plt.savefig('./figures/DD_grid_hatch.{}'.format(filetype))
plt.show()
# +
qlist = np.array([0.01, 0.1, 0.3, 1, 3.0, 10, 100])
clist = np.linspace(0,1,7)
fig, ax1 = plt.subplots()
fig.set_figheight(5)
fig.set_figwidth(5.5)
#plt.figure(figsize = [5,4])
ax2 = ax1.twinx()
c = 0
cstep = 1.0/7.0
for q in np.flip(qlist):
name = 'rescaled_DD_mass_q{}.txt'.format(str(q))
con_plot(name, '', 'Rescaled by {}'.format(str(q)), ax = ax1)
c = c+cstep
DM_star_ratio = 1/0.06
print(DM_star_ratio)
ax1.set_xlim(1e-4, 1e7)
ax1.set_ylim(1e-5, DM_star_ratio)
ax2.loglog()
ax2.set_ylim(1e-5/DM_star_ratio, 1)
ax1.set_xlabel('Mass ($M_\odot$)')
ax1.set_ylabel('$M_{DCO}/M_{stars}$')
ax2.set_ylabel('$M_{DCO}/M_{DM} \; [f_{DM}$]')
rect = Rectangle((1e-5,1e-5), (1e-2-1e-5), 1e2, alpha=0.2)
ax1.add_patch(rect)
plt.text(1e-4,1e-1, ' Rogue \n planets')
#ax2.set_yticks(np.power(10.0, np.arange(-7, 1)), minor=False)
#ax1.set_yticks(np.power(10.0, np.arange(-5, 3)), minor=False)
plt.minorticks_off()
ax1.legend(loc = 'upper right')
plt.tight_layout()
plt.savefig('./figures/darkdisk_massconstraints.{}'.format(filetype))
plt.show()
# +
#tilted disk
def tiltdiskplot(m, ax):
con = np.loadtxt('result_array_tiltdisk_{}.txt'.format(str(m)))
phi = np.linspace(0, np.pi*45/180, 10)
ax.plot(phi, con[:,0], label = 'Right')
ax.plot(phi, con[:,2], label = 'Left')
ax.plot(phi, con[:,1], label = 'Away')
ax.plot(phi, con[:,3], label = 'Towards')
ax.set_ylabel('$\log_{10}(M_{DCO}/M_{stars})$')
#ax.legend()
#print(con)
fig,(ax1, ax2, ax3) = plt.subplots(3,1, sharex = True)
fig.set_figheight(9)
fig.set_figwidth(6)
tiltdiskplot(0.01, ax1)
tiltdiskplot(1.0, ax2)
tiltdiskplot(100000.0, ax3)
ax1.set_title('$m = 10^{-2} M_\odot$')
ax2.set_title('$m = 1 M_\odot$')
ax3.set_title('$m = 10^{5} M_\odot$')
xmarks = np.linspace(0,np.pi/4,4)
#plt.xticks(xmarks, ['0', '$5^\circ$', '$10^\circ$', '$15^\circ$', '$20^\circ$', '$25^\circ$', '$30^\circ$', '$35^\circ$', '$40^\circ$', '$45^\circ$'])
plt.xticks(xmarks, ['0', '$15^\circ$', '$30^\circ$', '$45^\circ$'])
plt.legend()
ax3.set_xlabel('Disk Tilt Angle')
plt.savefig('./figures/tiltdisk.{}'.format(filetype))
plt.show()
# +
f = np.loadtxt('n_LoS_mNFW.txt')
frand = np.loadtxt('n_LoS_mNFW_testrand.txt')
frand = frand[2:]
print(f)
#f = np.power(10,f)
#print(f)
f_core = f[0]
f_lmc = f[1]
f_los = f[2:10]
#f_los_m2 = f[11:20]
#f_los_m5 = f[20:29]
#f_new = np.loadtxt('n_LoS_mNFW_test_long.txt')
#f_new = f_new[2:]
#f_total = np.zeros([3,8])
#for i in np.arange(8):
#f_total[:,i] = np.concatenate((f_los[i:i+1],frand[3*i:3*i+3]))
# f_total[:,i] = frand[3*i:3*i+3]
#f_mean = np.mean(f_total, axis = 0)
f_std = np.std(f_total, axis=0)
#print(f_total, f_mean, f_std)
N = np.array([5, 10, 20, 50, 100, 200, 500, 1000])
plt.plot(N, f_los, label = 'Variable LoS')
#plt.errorbar(N, f_los, f_std, label = 'no mask')
#plt.plot(N, f_los_m2, label = '2 degrees mask, old')
#plt.plot(N, f_los_m5, label = '5 degrees mask, old')
plt.plot([5,1000],[f_core, f_core], '--', label = 'Core')
plt.plot([5,1000],[f_lmc, f_lmc], '--', label = 'LMC')
plt.legend()
plt.xscale('log')
#plt.yscale('log')
plt.xlabel('Number of Lines of Sight')
plt.ylabel('$\log_{10}(f_a)$')
plt.savefig('./figures/linesofsight.{}'.format(filetype))
plt.show()
# +
f = np.loadtxt('n_LoS_mNFW_testrand.txt')
f_new = np.loadtxt('n_LoS_mNFW_test_long.txt')
f_new = f_new[2:]
print(f)
#f = np.power(10,f)
#print(f)
f_core = f[0]
f_lmc = f[1]
f_los = f[2:]
Nn = np.array([5, 10, 20, 50, 100, 200, 500])
N = np.repeat(Nn,5)
plt.scatter(N, f_los, label = 'random LoS ensembles')
#plt.plot(Nn, f_new, label = 'default LoS choices')
#plt.plot(N, f_los_m2, label = '2 degrees mask')
#plt.plot(N, f_los_m5, label = '5 degrees mask')
plt.plot([5,2000],[f_core, f_core], '--', label = 'Core')
plt.plot([5,2000],[f_lmc, f_lmc], '--', label = 'LMC')
plt.legend()
plt.xscale('log')
#plt.yscale('log')
plt.xlabel('Number of Lines of Sight')
plt.ylabel('$\log_{10}(f_a)$')
plt.savefig('./figures/linesofsight_rand.png')
plt.show()
# +
bar_events = np.loadtxt('baryon_events.txt')
bar_events_1 = np.loadtxt('baryon_events_one.txt')
bar_events_iso = np.loadtxt('baryon_events_iso.txt')
bar_time = np.sum(bar_events, axis=0)*17e9/100
bar_time_1 = np.sum(bar_events_1, axis=0)*17e9/100
bar_time_iso = np.sum(bar_events_iso, axis=0)*17e9/100
T = np.logspace(0,5,20)
plt.plot(T, bar_time, label = 'integrated over a range of baryon masses')
plt.plot(T, bar_time_1, label = 'just one mass, 0.36')
#plt.plot(T, bar_time_iso, label = 'isotropic velocities')
plt.xscale('log')
plt.yscale('log')
plt.xlabel('Crossing Time (Days)')
plt.ylabel('Number of events')
plt.legend()
plt.savefig('./figures/baryon_distributions.png')
plt.show()
# +
F = np.loadtxt('zoom_constraints_m036.txt')
F1 = np.loadtxt('zoom_constraints_m1.txt')
Ff = np.loadtxt('zoom_constraints_full_baryons.txt')
M = np.logspace(-2, 1, 30)
plt.plot(M, F, 'r-', label = 'DCO constraints, m_b = 0.36')
plt.plot([0.36, 0.36], [-4.5, -1], 'r--')
plt.plot(M, F1, 'b-', label = 'DCO constraints, m_b = 1.0')
plt.plot([1.0, 1.0], [-4.5, -1], 'b--')
plt.plot(M, Ff, 'g-', label = 'DCO constraints, all baryon masses')
#plt.plot([1.0, 1.0], [-4.5, -1], 'b--')
unc = np.log10(0.05)
plt.plot([1e-2, 1e1], [unc, unc], 'k--', label = 'baryon uncertainty')
plt.xscale('log')
plt.xlabel('mass [$M_\odot$]')
plt.ylabel('$\log_{10}(M_{DCO}/M_{star})$')
plt.legend()
plt.savefig('./figures/zoom_mass_spike.png')
plt.show()
# -
| .ipynb_checkpoints/plot_routines-checkpoint.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # KNN Classifier
# Now using k-fold
# ## Oral Toxicity Dataset
# ### Imports and data loading
# +
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.model_selection import KFold, cross_val_score
from sklearn.model_selection import train_test_split
import sklearn.metrics as metrics
from sklearn.neighbors import KNeighborsClassifier
import ds_functions as ds
data: pd.DataFrame = pd.read_csv('../../datasets/qsar_oral_toxicity.csv', sep=';', header=None)
# -
# ### Prepare data
# +
y: np.ndarray = data.pop(1024).values # Target Variable
X: np.ndarray = data.values # Values of each feature on each record
labels = pd.unique(y)
kf = KFold(n_splits = 15, shuffle=True)
# -
nvalues = [1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25]
dist = 'jaccard' #We decided to not even test the other 3, for obvious reasons
# As proximas celulas são de exemplo, com apenas para k=3 e um plot para um dos splits.
# O resto está abaixo, mas ainda tem de correr.
knn = KNeighborsClassifier(n_neighbors=3, metric=dist)
scores = cross_val_score(knn, X, y, cv=kf)
print("Accuracy: %0.2f (+/- %0.2f)" % (scores.mean(), scores.std() * 2))
m_accuracy = scores.mean()
i=0
for train_index, test_index in kf.split(X):
trnX, tstX = X[train_index], X[test_index]
trnY, tstY = y[train_index], y[test_index]
knn.fit(trnX, trnY)
prd_trn = knn.predict(trnX)
prd_tst = knn.predict(tstX)
ds.plot_evaluation_results(labels, trnY, prd_trn, tstY, prd_tst)
if i==0:
break
# Depois com tempo correr antes isto para termos comparações:
# +
#best = (0,"")
#lb=0
#for n in nvalues:
# knn = KNeighborsClassifier(n_neighbors=3, metric=dist)
# scores = cross_val_score(knn, X, y, cv=kf)
# m_acc = scores.mean()
# print("for {} k and jaccard we got {} acc".format(n, m_acc))
# if m_acc > last_best:
# best = (n, 'jaccard')
# last_best = m_acc
# -
# E correr a proxima para vermos como se comporta em cada um dos folds
for train_index, test_index in kf.split(X):
trnX, tstX = X[train_index], X[test_index]
trnY, tstY = y[train_index], y[test_index]
knn = KNeighborsClassifier(n_neighbors=best[0], metric='jaccard')
knn.fit(trnX, trnY)
prd_trn = knn.predict(trnX)
prd_tst = knn.predict(tstX)
ds.plot_evaluation_results(labels, trnY, prd_trn, tstY, prd_tst)
# Isto demora só buéééééé tempo a correr.
# Cancelei tudo para se poder meter no github e mostrar um pouco só a titulo de exemplo, esta semana agora deixo a correr outra vez durante a noite e com um pouco de sorte não vai crashar.
| Classification/KNN/KNN_QSAR_K-Fold.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] id="view-in-github"
# <a href="https://colab.research.google.com/github/lebesatebello/masakhane-mt/blob/master/starter_notebook_reverse_training_Setswana_to_English.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="Igc5itf-xMGj"
# # Masakhane - Reverse Machine Translation for African Languages (Using JoeyNMT)
# + [markdown] id="cOw-aSKFy1Tp"
# > ## NB
# >### - The purpose of this Notebook is to build models that translate African languages(target language) *into* English(source language). This will allow us to in future be able to make translations from one African language to the other. If you'd like to translate *from* English, please use [this](https://github.com/masakhane-io/masakhane-mt/blob/master/starter_notebook.ipynb) starter notebook instead.
#
# >### - We call this reverse training because normally we build models that make translations from the source language(English) to the target language. But in this case we are doing the reverse; building models that make translations from the target language to the source(English)
# + [markdown] id="x4fXCKCf36IK"
# ## Note before beginning:
# ### - The idea is that you should be able to make minimal changes to this in order to get SOME result for your own translation corpus.
#
# ### - The tl;dr: Go to the **"TODO"** comments which will tell you what to update to get up and running
#
# ### - If you actually want to have a clue what you're doing, read the text and peek at the links
#
# ### - With 100 epochs, it should take around 7 hours to run in Google Colab
#
# ### - Once you've gotten a result for your language, please attach and email your notebook that generated it to <EMAIL>
#
# ### - If you care enough and get a chance, doing a brief background on your language would be amazing. See examples in [(Martinus, 2019)](https://arxiv.org/abs/1906.05685)
# + [markdown] id="l929HimrxS0a"
# ## Retrieve your data & make a parallel corpus
#
# If you are wanting to use the JW300 data referenced on the Masakhane website or in our GitHub repo, you can use `opus-tools` to convert the data into a convenient format. `opus_read` from that package provides a convenient tool for reading the native aligned XML files and to convert them to TMX format. The tool can also be used to fetch relevant files from OPUS on the fly and to filter the data as necessary. [Read the documentation](https://pypi.org/project/opustools-pkg/) for more details.
#
# Once you have your corpus files in TMX format (an xml structure which will include the sentences in your target language and your source language in a single file), we recommend reading them into a pandas dataframe. Thankfully, Jade wrote a silly `tmx2dataframe` package which converts your tmx file to a pandas dataframe.
# + [markdown] id="SHM_XRWHoaab"
# Submitted by <NAME> 2388016
#
# Sumbitted by <NAME> 1492459
# + colab={"base_uri": "https://localhost:8080/"} id="oGRmDELn7Az0" executionInfo={"status": "ok", "timestamp": 1633884479081, "user_tz": -120, "elapsed": 20392, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "05128278845965867501"}} outputId="886de5e9-1fcc-4efc-aead-3bd6a8ca32ca"
from google.colab import drive
drive.mount('/content/drive')
# + id="Cn3tgQLzUxwn"
# TODO: Set your source and target languages. Keep in mind, these traditionally use language codes as found here:
# These will also become the suffix's of all vocab and corpus files used throughout
import os
source_language = "en"
target_language = "tn"
lc = False # If True, lowercase the data.
seed = 42 # Random seed for shuffling.
tag = "baseline" # Give a unique name to your folder - this is to ensure you don't rewrite any models you've already submitted
os.environ["src"] = source_language # Sets them in bash as well, since we often use bash scripts
os.environ["tgt"] = target_language
os.environ["tag"] = tag
# This will save it to a folder in our gdrive instead!
# !mkdir -p "/content/drive/My Drive/masakhane/$tgt-$src-$tag"
os.environ["gdrive_path"] = "/content/drive/My Drive/masakhane/%s-%s-%s" % (target_language, source_language, tag)
# + colab={"base_uri": "https://localhost:8080/"} id="kBSgJHEw7Nvx" executionInfo={"status": "ok", "timestamp": 1633884492143, "user_tz": -120, "elapsed": 528, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "05128278845965867501"}} outputId="13f801c3-245d-4e71-b561-699449325e2d"
# !echo $gdrive_path
# + colab={"base_uri": "https://localhost:8080/"} id="gA75Fs9ys8Y9" executionInfo={"status": "ok", "timestamp": 1633884495913, "user_tz": -120, "elapsed": 3413, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "05128278845965867501"}} outputId="f81ed93a-d7e2-4383-b93b-f417d788cc04"
# Install opus-tools
# ! pip install opustools-pkg
# + colab={"base_uri": "https://localhost:8080/"} id="xq-tDZVks7ZD" executionInfo={"status": "ok", "timestamp": 1633884733724, "user_tz": -120, "elapsed": 237814, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "05128278845965867501"}} outputId="3e4cdc25-8088-4d55-bb42-6cd4a55e9d03"
# Downloading our corpus
# ! opus_read -d JW300 -s $src -t $tgt -wm moses -w jw300.$src jw300.$tgt -q
# extract the corpus file
# ! gunzip JW300_latest_xml_$src-$tgt.xml.gz
# + colab={"base_uri": "https://localhost:8080/"} id="n48GDRnP8y2G" executionInfo={"status": "ok", "timestamp": 1633884734795, "user_tz": -120, "elapsed": 1077, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "05128278845965867501"}} outputId="7a31bf76-ff6d-4ed9-be38-a8270a4f6b06"
# Download the global test set.
# ! wget https://raw.githubusercontent.com/juliakreutzer/masakhane/master/jw300_utils/test/test.en-any.en
# And the specific test set for this language pair.
os.environ["trg"] = target_language
os.environ["src"] = source_language
# ! wget https://raw.githubusercontent.com/juliakreutzer/masakhane/master/jw300_utils/test/test.en-$trg.en
# ! mv test.en-$trg.en test.en
# ! wget https://raw.githubusercontent.com/juliakreutzer/masakhane/master/jw300_utils/test/test.en-$trg.$trg
# ! mv test.en-$trg.$trg test.$trg
# + colab={"base_uri": "https://localhost:8080/"} id="NqDG-CI28y2L" executionInfo={"status": "ok", "timestamp": 1633884734795, "user_tz": -120, "elapsed": 6, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "05128278845965867501"}} outputId="612642c7-bc62-4c37-b61b-ba4a4ddfc9b8"
# Read the test data to filter from train and dev splits.
# Store english portion in set for quick filtering checks.
en_test_sents = set()
filter_test_sents = "test.en-any.en"
j = 0
with open(filter_test_sents) as f:
for line in f:
en_test_sents.add(line.strip())
j += 1
print('Loaded {} global test sentences to filter from the training/dev data.'.format(j))
# + colab={"base_uri": "https://localhost:8080/", "height": 158} id="3CNdwLBCfSIl" executionInfo={"status": "ok", "timestamp": 1633884795694, "user_tz": -120, "elapsed": 60902, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "05128278845965867501"}} outputId="0ef754ca-f2e0-4792-88ea-2e98c3d67edd"
import pandas as pd
# TMX file to dataframe
source_file = 'jw300.' + source_language
target_file = 'jw300.' + target_language
source = []
target = []
skip_lines = [] # Collect the line numbers of the source portion to skip the same lines for the target portion.
with open(source_file) as f:
for i, line in enumerate(f):
# Skip sentences that are contained in the test set.
if line.strip() not in en_test_sents:
source.append(line.strip())
else:
skip_lines.append(i)
with open(target_file) as f:
for j, line in enumerate(f):
# Only add to corpus if corresponding source was not skipped.
if j not in skip_lines:
target.append(line.strip())
print('Loaded data and skipped {}/{} lines since contained in test set.'.format(len(skip_lines), i))
df = pd.DataFrame(zip(source, target), columns=['source_sentence', 'target_sentence'])
# if you get TypeError: data argument can't be an iterator is because of your zip version run this below
#df = pd.DataFrame(list(zip(source, target)), columns=['source_sentence', 'target_sentence'])
df.head(3)
# + [markdown] id="YkuK3B4p2AkN"
# ## Pre-processing and export
#
# It is generally a good idea to remove duplicate translations and conflicting translations from the corpus. In practice, these public corpora include some number of these that need to be cleaned.
#
# In addition we will split our data into dev/test/train and export to the filesystem.
# + colab={"base_uri": "https://localhost:8080/"} id="M_2ouEOH1_1q" executionInfo={"status": "ok", "timestamp": 1633884797911, "user_tz": -120, "elapsed": 2222, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "05128278845965867501"}} outputId="4228fd42-98e9-4486-f847-c1f6162bcebc"
# drop duplicate translations
df_pp = df.drop_duplicates()
# drop conflicting translations
# (this is optional and something that you might want to comment out
# depending on the size of your corpus)
df_pp.drop_duplicates(subset='source_sentence', inplace=True)
df_pp.drop_duplicates(subset='target_sentence', inplace=True)
# Shuffle the data to remove bias in dev set selection.
df_pp = df_pp.sample(frac=1, random_state=seed).reset_index(drop=True)
# + colab={"base_uri": "https://localhost:8080/"} id="Z_1BwAApEtMk" executionInfo={"status": "ok", "timestamp": 1633884806084, "user_tz": -120, "elapsed": 8178, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "05128278845965867501"}} outputId="afdcf8a7-6c67-4655-8328-824785911423"
# Install fuzzy wuzzy to remove "almost duplicate" sentences in the
# test and training sets.
# ! pip install fuzzywuzzy
# ! pip install python-Levenshtein
import time
from fuzzywuzzy import process
import numpy as np
from os import cpu_count
from functools import partial
from multiprocessing import Pool
# reset the index of the training set after previous filtering
df_pp.reset_index(drop=False, inplace=True)
# Remove samples from the training data set if they "almost overlap" with the
# samples in the test set.
# Filtering function. Adjust pad to narrow down the candidate matches to
# within a certain length of characters of the given sample.
def fuzzfilter(sample, candidates, pad):
candidates = [x for x in candidates if len(x) <= len(sample)+pad and len(x) >= len(sample)-pad]
if len(candidates) > 0:
return process.extractOne(sample, candidates)[1]
else:
return np.nan
# + id="92EsgTaY3B4H"
# start_time = time.time()
# ### iterating over pandas dataframe rows is not recomended, let use multi processing to apply the function
# with Pool(cpu_count()-1) as pool:
# scores = pool.map(partial(fuzzfilter, candidates=list(en_test_sents), pad=5), df_pp['source_sentence'])
# hours, rem = divmod(time.time() - start_time, 3600)
# minutes, seconds = divmod(rem, 60)
# print("done in {}h:{}min:{}seconds".format(hours, minutes, seconds))
# # Filter out "almost overlapping samples"
# df_pp = df_pp.assign(scores=scores)
# df_pp = df_pp[df_pp['scores'] < 95]
# + colab={"base_uri": "https://localhost:8080/"} id="hxxBOCA-xXhy" executionInfo={"status": "ok", "timestamp": 1633884861814, "user_tz": -120, "elapsed": 55737, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "05128278845965867501"}} outputId="51eff302-a0b5-48c0-c565-be7af025aa6e"
# This section does the split between train/dev for the parallel corpora then saves them as separate files
# We use 1000 dev test and the given test set.
import csv
# Do the split between dev/train and create parallel corpora
num_dev_patterns = 1000
# Optional: lower case the corpora - this will make it easier to generalize, but without proper casing.
if lc: # Julia: making lowercasing optional
df_pp["source_sentence"] = df_pp["source_sentence"].str.lower()
df_pp["target_sentence"] = df_pp["target_sentence"].str.lower()
# Julia: test sets are already generated
dev = df_pp.tail(num_dev_patterns) # Herman: Error in original
stripped = df_pp.drop(df_pp.tail(num_dev_patterns).index)
with open("train."+source_language, "w") as src_file, open("train."+target_language, "w") as trg_file:
for index, row in stripped.iterrows():
src_file.write(row["source_sentence"]+"\n")
trg_file.write(row["target_sentence"]+"\n")
with open("dev."+source_language, "w") as src_file, open("dev."+target_language, "w") as trg_file:
for index, row in dev.iterrows():
src_file.write(row["source_sentence"]+"\n")
trg_file.write(row["target_sentence"]+"\n")
#stripped[["source_sentence"]].to_csv("train."+source_language, header=False, index=False) # Herman: Added `header=False` everywhere
#stripped[["target_sentence"]].to_csv("train."+target_language, header=False, index=False) # Julia: Problematic handling of quotation marks.
#dev[["source_sentence"]].to_csv("dev."+source_language, header=False, index=False)
#dev[["target_sentence"]].to_csv("dev."+target_language, header=False, index=False)
# Doublecheck the format below. There should be no extra quotation marks or weird characters.
# ! head train.*
# ! head dev.*
# + [markdown] id="epeCydmCyS8X"
#
#
# ---
#
#
# ## Installation of JoeyNMT
#
# JoeyNMT is a simple, minimalist NMT package which is useful for learning and teaching. Check out the documentation for JoeyNMT [here](https://joeynmt.readthedocs.io)
# + colab={"base_uri": "https://localhost:8080/"} id="iBRMm4kMxZ8L" executionInfo={"status": "ok", "timestamp": 1633884876642, "user_tz": -120, "elapsed": 14835, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "05128278845965867501"}} outputId="831bcd35-910d-4c3e-96ea-c9bd9fb619eb"
# Install JoeyNMT
# ! git clone https://github.com/joeynmt/joeynmt.git
# ! cd joeynmt; pip3 install .
# Install Pytorch with GPU support v1.7.1.
# ! pip install torch==1.9.0+cu101 -f https://download.pytorch.org/whl/torch_stable.html
# + [markdown] id="AaE77Tcppex9"
# # Preprocessing the Data into Subword BPE Tokens
#
# - One of the most powerful improvements for agglutinative languages (a feature of most Bantu languages) is using BPE tokenization [ (Sennrich, 2015) ](https://arxiv.org/abs/1508.07909).
#
# - It was also shown that by optimizing the umber of BPE codes we significantly improve results for low-resourced languages [(Sennrich, 2019)](https://www.aclweb.org/anthology/P19-1021) [(Martinus, 2019)](https://arxiv.org/abs/1906.05685)
#
# - Below we have the scripts for doing BPE tokenization of our data. We use 4000 tokens as recommended by [(Sennrich, 2019)](https://www.aclweb.org/anthology/P19-1021). You do not need to change anything. Simply running the below will be suitable.
# + colab={"base_uri": "https://localhost:8080/"} id="H-TyjtmXB1mL" executionInfo={"status": "ok", "timestamp": 1633885105806, "user_tz": -120, "elapsed": 229172, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "05128278845965867501"}} outputId="89088f8f-952d-4ff9-c092-38938975e8ea"
# One of the huge boosts in NMT performance was to use a different method of tokenizing.
# Usually, NMT would tokenize by words. However, using a method called BPE gave amazing boosts to performance
# Do subword NMT
from os import path
os.environ["src"] = source_language # Sets them in bash as well, since we often use bash scripts
os.environ["tgt"] = target_language
# Learn BPEs on the training data.
os.environ["data_path"] = path.join("joeynmt", "data",target_language + source_language ) # Herman!
# ! subword-nmt learn-joint-bpe-and-vocab --input train.$src train.$tgt -s 4000 -o bpe.codes.4000 --write-vocabulary vocab.$src vocab.$tgt
# Apply BPE splits to the development and test data.
# ! subword-nmt apply-bpe -c bpe.codes.4000 --vocabulary vocab.$src < train.$src > train.bpe.$src
# ! subword-nmt apply-bpe -c bpe.codes.4000 --vocabulary vocab.$tgt < train.$tgt > train.bpe.$tgt
# ! subword-nmt apply-bpe -c bpe.codes.4000 --vocabulary vocab.$src < dev.$src > dev.bpe.$src
# ! subword-nmt apply-bpe -c bpe.codes.4000 --vocabulary vocab.$tgt < dev.$tgt > dev.bpe.$tgt
# ! subword-nmt apply-bpe -c bpe.codes.4000 --vocabulary vocab.$src < test.$src > test.bpe.$src
# ! subword-nmt apply-bpe -c bpe.codes.4000 --vocabulary vocab.$tgt < test.$tgt > test.bpe.$tgt
# Create directory, move everyone we care about to the correct location
# ! mkdir -p $data_path
# ! cp train.* $data_path
# ! cp test.* $data_path
# ! cp dev.* $data_path
# ! cp bpe.codes.4000 $data_path
# ! ls $data_path
# Also move everything we care about to a mounted location in google drive (relevant if running in colab) at gdrive_path
# ! cp train.* "$gdrive_path"
# ! cp test.* "$gdrive_path"
# ! cp dev.* "$gdrive_path"
# ! cp bpe.codes.4000 "$gdrive_path"
# ! ls "$gdrive_path"
# Create that vocab using build_vocab
# ! sudo chmod 777 joeynmt/scripts/build_vocab.py
# ! joeynmt/scripts/build_vocab.py joeynmt/data/$tgt$src/train.bpe.$src joeynmt/data/$tgt$src/train.bpe.$tgt --output_path joeynmt/data/$tgt$src/vocab.txt
# Some output
# ! echo "BPE Setswana Sentences"
# ! tail -n 5 test.bpe.$tgt
# ! echo "Combined BPE Vocab"
# ! tail -n 10 joeynmt/data/$tgt$src/vocab.txt # Herman
# + colab={"base_uri": "https://localhost:8080/"} id="IlMitUHR8Qy-" executionInfo={"status": "ok", "timestamp": 1633885107157, "user_tz": -120, "elapsed": 1358, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "05128278845965867501"}} outputId="64c6f7ac-b9bf-4b3a-88d0-6df8f867f0a0"
# Also move everything we care about to a mounted location in google drive (relevant if running in colab) at gdrive_path
# ! cp train.* "$gdrive_path"
# ! cp test.* "$gdrive_path"
# ! cp dev.* "$gdrive_path"
# ! cp bpe.codes.4000 "$gdrive_path"
# ! ls "$gdrive_path"
# + [markdown] id="Ixmzi60WsUZ8"
# # Creating the JoeyNMT Config
#
# JoeyNMT requires a yaml config. We provide a template below. We've also set a number of defaults with it, that you may play with!
#
# - We used Transformer architecture
# - We set our dropout to reasonably high: 0.3 (recommended in [(Sennrich, 2019)](https://www.aclweb.org/anthology/P19-1021))
#
# Things worth playing with:
# - The batch size (also recommended to change for low-resourced languages)
# - The number of epochs (we've set it at 30 just so it runs in about an hour, for testing purposes)
# - The decoder options (beam_size, alpha)
# - Evaluation metrics (BLEU versus Crhf4)
# + id="h8TMgv1p3L1z"
# This creates the config file for our JoeyNMT system. It might seem overwhelming so we've provided a couple of useful parameters you'll need to update
# (You can of course play with all the parameters if you'd like!)
name = '%s%s' % (target_language, source_language)
# gdrive_path = os.environ["gdrive_path"]
# Create the config
config = """
name: "{target_language}{source_language}_reverse_transformer"
data:
src: "{target_language}"
trg: "{source_language}"
train: "data/{name}/train.bpe"
dev: "data/{name}/dev.bpe"
test: "data/{name}/test.bpe"
level: "bpe"
lowercase: False
max_sent_length: 100
src_vocab: "data/{name}/vocab.txt"
trg_vocab: "data/{name}/vocab.txt"
testing:
beam_size: 5
alpha: 1.0
training:
#load_model: "{gdrive_path}/models/{name}_transformer/1.ckpt" # if uncommented, load a pre-trained model from this checkpoint
random_seed: 42
optimizer: "adam"
normalization: "tokens"
adam_betas: [0.9, 0.999]
scheduling: "Noam scheduling" # TODO: try switching from plateau to Noam scheduling. plateau to Noam scheduling
patience: 5 # For plateau: decrease learning rate by decrease_factor if validation score has not improved for this many validation rounds.
learning_rate_factor: 0.5 # factor for Noam scheduler (used with Transformer)
learning_rate_warmup: 1000 # warmup steps for Noam scheduler (used with Transformer)
decrease_factor: 0.7
loss: "crossentropy"
learning_rate: 0.0003
learning_rate_min: 0.00000001
weight_decay: 0.0
label_smoothing: 0.1
batch_size: 4096
batch_type: "token"
eval_batch_size: 3600
eval_batch_type: "token"
batch_multiplier: 1
early_stopping_metric: "ppl"
epochs: 3 # TODO: Decrease for when playing around and checking of working. Around 30 is sufficient to check if its working at all. 5 - 3
validation_freq: 1000 # TODO: Set to at least once per epoch.
logging_freq: 100
eval_metric: "bleu"
model_dir: "models/{name}_reverse_transformer"
overwrite: True # TODO: Set to True if you want to overwrite possibly existing models.
shuffle: True
use_cuda: True
max_output_length: 100
print_valid_sents: [0, 1, 2, 3]
keep_last_ckpts: 3
model:
initializer: "xavier"
bias_initializer: "zeros"
init_gain: 1.0
embed_initializer: "xavier"
embed_init_gain: 1.0
tied_embeddings: True
tied_softmax: True
encoder:
type: "transformer"
num_layers: 6
num_heads: 4 # TODO: Increase to 8 for larger data. 4 - 8
embeddings:
embedding_dim: 256 # TODO: Increase to 512 for larger data. 256 -512
scale: True
dropout: 0.2
# typically ff_size = 4 x hidden_size
hidden_size: 256 # TODO: Increase to 512 for larger data. 256 - 512
ff_size: 2048 # TODO: Increase to 2048 for larger data. 1024 - 2048
dropout: 0.3
decoder:
type: "transformer"
num_layers: 6
num_heads: 4 # TODO: Increase to 8 for larger data. 4 - 8
embeddings:
embedding_dim: 256 # TODO: Increase to 512 for larger data. 256 - 512
scale: True
dropout: 0.2
# typically ff_size = 4 x hidden_size
hidden_size: 256 # TODO: Increase to 512 for larger data. 256 - 512
ff_size: 2048 # TODO: Increase to 2048 for larger data. 1024 - 2048
dropout: 0.3
""".format(name=name, gdrive_path=os.environ["gdrive_path"], source_language=source_language, target_language=target_language)
with open("joeynmt/configs/transformer_reverse_{name}.yaml".format(name=name),'w') as f:
f.write(config)
# + [markdown] id="oEzoJtV2MIpt"
# # Train the Model
#
# This single line of joeynmt runs the training using the config we made above
# + colab={"base_uri": "https://localhost:8080/"} id="WzbNYNdjLgNb" executionInfo={"status": "ok", "timestamp": 1633892772099, "user_tz": -120, "elapsed": 7664944, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "05128278845965867501"}} outputId="db565e22-b9a2-478b-c431-1abb11a28a21"
# Train the model
# You can press Ctrl-C to stop. And then run the next cell to save your checkpoints!
# !cd joeynmt; python3 -m joeynmt train configs/transformer_reverse_$tgt$src.yaml
# + id="MBoDS09JM807"
# Copy the created models from the notebook storage to google drive for persistant storage
# !cp -r joeynmt/models/${tgt}${src}_reverse_transformer/* "$gdrive_path/models/${tgt}${src}_reverse_transformer/"
# + colab={"base_uri": "https://localhost:8080/"} id="n94wlrCjVc17" executionInfo={"status": "ok", "timestamp": 1633892782542, "user_tz": -120, "elapsed": 10, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "05128278845965867501"}} outputId="b4b0e865-7159-4c06-d08f-daf9945aa7ed"
# Output our validation accuracy
# ! cat "$gdrive_path/models/${tgt}${src}_reverse_transformer/validations.txt"
# + colab={"base_uri": "https://localhost:8080/"} id="66WhRE9lIhoD" executionInfo={"status": "ok", "timestamp": 1633892862304, "user_tz": -120, "elapsed": 79769, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "05128278845965867501"}} outputId="83a42eba-6a6c-475d-b830-8ea8a118d700"
# Test our model
# ! cd joeynmt; python3 -m joeynmt test "$gdrive_path/models/${tgt}${src}_reverse_transformer/config.yaml"
# + id="ho_r2WwTVZmZ"
while True:pass
# + id="xXnweLYCGwcr"
| benchmarks/tn-en/KorstiaanW-baseline/KorstiaanW_tn_en_Baseline.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# Before you turn this problem in, make sure everything runs as expected. First, **restart the kernel** (in the menubar, select Kernel$\rightarrow$Restart) and then **run all cells** (in the menubar, select Cell$\rightarrow$Run All).
#
# Note that this Pre-class Work is estimated to take **43 minutes**.
#
# Make sure you fill in any place that says `YOUR CODE HERE` or "YOUR ANSWER HERE", as well as your name and collaborators below:
NAME = "<NAME>"
COLLABORATORS = ""
# ---
# + [markdown] deletable=false editable=false nbgrader={"checksum": "53f91bdb5f2f106f06872a2a6e7a98f5", "grade": false, "grade_id": "cell-bd90321d310ba2d4", "locked": true, "schema_version": 1, "solution": false}
# # CS110 Pre-class Work - Randomly built BSTs
#
# ## Part A. Average number of comparisons when searching
# + [markdown] deletable=false editable=false nbgrader={"checksum": "c1c60903cd4344daf3877a1a656e283e", "grade": false, "grade_id": "cell-9e8b16db9d917705", "locked": true, "schema_version": 1, "solution": false}
# ## Question 0 [time estimate: 1 minute]
#
# Paste in the working code from last session's PCW for the class Node, which contains the methods for insertion, searching, and deletion.
#
#
# + deletable=false nbgrader={"checksum": "525698a22b0cee9c4cb74be2011232de", "grade": true, "grade_id": "cell-1eadb1312ac0519d", "locked": false, "points": 0, "schema_version": 1, "solution": true}
class Node:
def __init__(self, val):
self.l_child = None
self.r_child = None
self.parent = None
self.data = val
def insert(self, node):
"""inserts a node into a *non-empty* tree rooted at the node, returns
the root"""
if self.data > node.data:
if self.l_child is None:
self.l_child = node
node.parent = self
else:
self.l_child.insert(node)
else:
if self.r_child is None:
self.r_child = node
node.parent = self
else:
self.r_child.insert(node)
return self
def minimum(self):
node = self
while node.l_child != None:
node = node.l_child
return node
def search_data(self, value):
"""searches a *non-empty* tree rooted at the node for a node with
data = value, returns the value if found, None otherwise"""
node = self.search(value)
if node:
return node.data
else:
return node
def to_string(self):
print('self.data', self.data)
root=self
if not root:
return 'Nil'
else:
r = root.r_child.to_string() if root.r_child else 'Nil'
l = root.l_child.to_string() if root.l_child else 'Nil'
return 'Node(' + str(root.data) + ' L: ' + l + ' R: ' + r + ')'
def search(self, value):
if self.data > value:
return self.l_child.search(value)
elif self.data == value:
return self
else:
return self.r_child.search(value)
def delete(self, value):
if self.data == value:
self.l_child = None
self.r_child = None
return self.parent
elif self.data > value:
return self.l_child.delete(value)
else :
return self.r_child.delete(value)
def inorder(self):
printout = []
while self.parent.value != None:
printout.append(self.l_child.value)
printout.append(self.value)
printout.append(self.r_child.value)
return self.parent.inorder()
# + [markdown] deletable=false editable=false nbgrader={"checksum": "a79cbf2799a45e4c351c9ed9f600c3b5", "grade": false, "grade_id": "cell-d3f754a4d3637207", "locked": true, "schema_version": 1, "solution": false}
#
# ## Question 1 [time estimate: 5 minutes]
#
# Complete the following function that computes the depth of a given node in a BST identified by its root. Use the test below to make sure your code works properly (Optional: Why does the test work?)
# + deletable=false nbgrader={"checksum": "6040a364ba7372adc7f259a4b0d875fc", "grade": false, "grade_id": "cell-cd10e9a432673bd9", "locked": false, "schema_version": 1, "solution": true}
def depth(root, node):
"""
Finds the depth of the node in a BST. depth of root is 0.
Parameters
----------
root
A node, the root of the BST
node
A node to compute the depth of
Returns
-------
d : int
Distance from node to root
"""
d = 0
while node.parent != None:
d += 1
node = node.parent
return d
# -
# Testing code
import math
bst = None
nodes = [Node(15), Node(6), Node(18), Node(3), Node(7),
Node(17), Node(20), Node(2), Node(4)]
for node in nodes:
if not bst:
bst = node
else:
bst.insert(node)
# insert(bst, node)
for i in range(len(nodes)):
assert(depth(bst, nodes[i]) == int(math.log(i+1,2)))
# + deletable=false editable=false nbgrader={"checksum": "46a630a4c6307c799c44715ec3868d58", "grade": true, "grade_id": "cell-c57bc5138040968c", "locked": true, "points": 1, "schema_version": 1, "solution": false}
# Please ignore this cell. This cell is for us to implement the tests
# to see if your code works properly.
# + [markdown] deletable=false editable=false nbgrader={"checksum": "01fa65ccabde3f2f6a8c62aca64dcd53", "grade": false, "grade_id": "cell-bd8ce3e36c859c2e", "locked": true, "schema_version": 1, "solution": false}
# ## Question 2 [time estimate: 5 minutes]
#
# Complete the following function, making use of `depth`, to calculate the average number of comparisons required to search for a randomly chosen element of a standard BST. For example, the following tree:
#
# 12
# \
# 14
#
# will have an average number of comparisons of 1.5. This is because 50% of the time we will be searching for 12 (1 comparison), and 50% of the time we will be searching for 14 (2 comparisons).
#
# You can test your function with this test case by constructing the tree using the code from the last session's pre-class work.
#
#
# (*Hint*: the number of comparisons required to search for node a is `depth(root, a) + 1`.)
# + deletable=false nbgrader={"checksum": "5de9ebf48a00878b4b565237c2d88265", "grade": false, "grade_id": "cell-34e9cb302f1dad00", "locked": false, "schema_version": 1, "solution": true}
def avg_cmp(bst):
"""
Finds the average number of comparisons required
to search for a randomly chosen element of a standard BST.
Parameters
----------
bst
A Node, the root of the BST
Returns
-------
n : float
Average number of comparisons
"""
node = bst
lst = []
sum_ = 0
length = len(lst)
for i in bst.inorder():
lst.append(depth(bst, i)+1)
for i in range(length):
sum_ += lst[i]
return sum_/length
# + deletable=false editable=false nbgrader={"checksum": "7c94a3d9e151e1a920e846ff98e118a8", "grade": true, "grade_id": "cell-dedc229bbb15d811", "locked": true, "points": 1, "schema_version": 1, "solution": false}
# Please ignore this cell. This cell is for us to implement the tests
# to see if your code works properly.
# + [markdown] deletable=false editable=false nbgrader={"checksum": "219e1c0aea58c5955539210f5c34b000", "grade": false, "grade_id": "cell-4b7d0c4e86568346", "locked": true, "schema_version": 1, "solution": false}
# ## Part B. Depth of randomly-built trees
# The average number of comparisons for a randomly chosen element within the tree is related to the “average depth” of the tree. Here the average depth of a tree is defined to be the average of the depths of all the nodes in the tree.
#
# ## Question 1 [time estimate: 10 minutes]
#
# Complete the following function to find the maximum depth of a node in a given BST. The function must run in $\mathrm{O}(N)$ time and use $\mathrm{O}(h)$ space (where $N$ is the number of elements in the BST and $h$ is the height of the tree). Also, note that the maximum depth is equal to the height of the tree.
#
# + deletable=false nbgrader={"checksum": "71f602309416ba9c718f06f4cc466075", "grade": false, "grade_id": "cell-64aba253c2b1bada", "locked": false, "schema_version": 1, "solution": true}
def max_depth(bst):
"""
Finds the maximum depth of node in a BST.
Parameters
----------
bst
A Node, the root of the BST
Returns
-------
h : int
The maximum depth in a BST
"""
node = bst
lst = []
while node.l_child != None:
node = node.l_child
while node.r_child != None:
node = node.r_child
n = depth(node, bst) + 1
lst.append(n)
while node.r_child != None:
node = node.r_child
while node.l_child != None:
node = node.l_child
n = depth(node, bst) + 1
lst.append(n)
lst.sort
return lst[0]
# + deletable=false editable=false nbgrader={"checksum": "641f559b2bcdfad11577dd6202a0e378", "grade": true, "grade_id": "cell-82deabcb0ea2e594", "locked": true, "points": 1, "schema_version": 1, "solution": false}
# Please ignore this cell. This cell is for us to implement the tests
# to see if your code works properly.
# + [markdown] deletable=false nbgrader={"checksum": "3224988f295d6adc77632f7f4ff5c8e2", "grade": false, "grade_id": "cell-fd78b979c52b9527", "locked": true, "schema_version": 1, "solution": false}
# ## Question 2 [time estimate: 5 minutes]
#
# Using the `avg_cmp` function, complete the following function to return the average depth of the tree. The average depth is related to the average number of comparisons as `average depth = (average comparisons - 1)`, since the root has depth zero.
# + deletable=false nbgrader={"checksum": "90d64051c00f344fc6baaf3ae169c87f", "grade": false, "grade_id": "cell-347916904e4a8379", "locked": false, "schema_version": 1, "solution": true}
def avg_depth(bst):
"""
Computes the average depth of a BST
Parameters
----------
bst
A Node, root of the BST
Returns
-------
avg_d : float
Average depth of the BST
"""
return avg_cmp - 1
# + deletable=false editable=false nbgrader={"checksum": "43da2ece71043e2a56753ec2f9b208dc", "grade": true, "grade_id": "cell-1bd7535fe6950d5f", "locked": true, "points": 1, "schema_version": 1, "solution": false}
# Please ignore this cell. This cell is for us to implement the tests
# to see if your code works properly.
# + [markdown] deletable=false editable=false nbgrader={"checksum": "25274f67359e9c4280895dca8e0589d1", "grade": false, "grade_id": "cell-6365329a3c57c1b1", "locked": true, "schema_version": 1, "solution": false}
# ## Question 3 [time estimate: 10 minutes]
# Now, insert randomly shuffled lists into BSTs, and measure the average depth and the maximum depth. How do these statistics scale as you increase $N$, the number of nodes? Make sure to give a compelling argument that motivates this scaling behaviour (you do not need to provide a technical derivation).
# + [markdown] deletable=false nbgrader={"checksum": "8a281cd793099803988046803bed99ba", "grade": true, "grade_id": "cell-bad918a333cabe43", "locked": false, "points": 0, "schema_version": 1, "solution": true}
# sometimes close to nlogn which is n(log_2(n^3+6n^2+11n+6)), sometimes as far as the worst case. As the N increase, the comparisons also increase in O(nlogn) in 'average'(the equation above)
# + [markdown] deletable=false editable=false nbgrader={"checksum": "dd1dd0c5c45fc94abb6229a38d8e04a2", "grade": false, "grade_id": "cell-3255b4081c295fa6", "locked": true, "schema_version": 1, "solution": false}
#
# ## Question 4 [time estimate: 7 minutes]
#
# Produce a plot showing the scaling behavior that you saw of both the average depth and the maximum depth as a function of the length of the shuffled list. For a meaningful figure, be sure to scale out to a list of size 10,000 and average the timings 50 times. (You don’t have to sample each value from 1 to 10,000!). Is this plot in agreement with the theoretical result you obtained in the previous question? Explain.
#
# + deletable=false nbgrader={"checksum": "a870cffbb739ec466993697791c20248", "grade": true, "grade_id": "cell-3034eb71070a6734", "locked": false, "points": 0, "schema_version": 1, "solution": true}
import numpy as np
import random
N = np.linspace(1,100)
max_depths = []
avg_depths = []
for i in range(100):
X = [j for j in range(i)]
Y = random.shuffle(X)
bst = None # bst is a misnormer, this variable contains the Node that is the root of the BST of interest
lst = []
for x in [Node(_) for _ in Y]:
print("###################")
print('Inserting the following node: ', x.data)
if not bst:
bst = x
else:
bst = bst.insert(x)
print(bst.to_string())
max_depths.append(max_depth(bst))
avg_depths.append(avg_depth(bst))
# -
plt.plot(N, max_depths, color = 'red',label = 'max depth', linewidth = 1.0)
plt.plot(N, avg_depths, color = 'blue',label = 'average depth', linewidth = 1.0)
plt.xlabel('$N$', fontsize=10)
plt.ylabel('depths', fontsize=10)
plt.legend()
plt.show()
| CS110 PCW 17 Randomly built BSTs.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
"""
@author: <NAME> <<EMAIL>>
@brief: utils for logging
"""
import os
import logging
import logging.handlers
# |等級 | 等級數值 | 輸出函數 | 說明 |
# |----------|----------|--------------------|:--------:|
# |NOTSET |0 |無對應的輸出函數 |未設定 |
# |DEBUG |10 |logging.debug() |除錯 |
# |INFO |20 |logging.info() |訊息 |
# |WARNING |30 |logging.warning() |警告 |
# |ERROR |40 |logging.error() |錯誤 |
# |CRITICAL |50 |logging.critical() |嚴重錯誤 |
# ***This module(Logging) will write a log in model.log (which is a log file). When model.log exceeded the capacity limit which is module variable, model.log will rename to model.log.1 and so on. Eventually, if we set backupCount=3 than we will get model.log, model.log.1, and model.log.2. They record the newest to the oldest log.***<br>
#
# **Rotating File Handler (backupCount=3):**<br>
# If the log file exceeds the capacity limit, pass the path to another stage.
#
# new record 📃
# ↓ (write in)
# ↓
# model.log → → model.log.1
# (rename) ↓
# ↓ (rename)
# model.log.3 ← ← model.log.2
# ↓ (rename)
# (drop) ↓
# 🗑️
# loglevel: 記錄最低等級
def _get_logger(logdir, logname, loglevel=logging.INFO):
fmt = '[%(asctime)s] %(levelname)s: %(message)s'
formatter = logging.Formatter(fmt)
handler = logging.handlers.RotatingFileHandler(
filename=os.path.join(logdir, logname),
maxBytes=10*1024*1024, # 大小不超過 10 MB,若紀錄檔已超過就會轉換為backup的log檔,
backupCount=10) # 並重新創建一個新的紀錄檔,紀錄新的log
handler.setFormatter(formatter)
logger = logging.getLogger('')
logger.addHandler(handler)
logger.setLevel(loglevel)
return logger
# convert notebook.ipynb to a .py file
# !jupytext --to py logging_utils.ipynb
| Code/utils/logging_utils.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: 'Python 3.8.10 64-bit (''venv'': venv)'
# language: python
# name: python3
# ---
alredyTurtle = False
name = "Union_rmlmapper_graph"
# # Imports
# +
from rdflib import Graph
from IPython.display import display
import pandas as pd
from rdflib.plugins.sparql.processor import SPARQLResult
def sparql_results_to_df(results: SPARQLResult):
"""
Export results from an rdflib SPARQL query into a `pandas.DataFrame`,
using Python types. See https://github.com/RDFLib/rdflib/issues/1179.
"""
return pd.DataFrame(
data=([None if x is None else x.toPython() for x in row] for row in results),
columns=[str(x) for x in results.vars],
)
# -
# # Open Graph
if not alredyTurtle:
g1 = Graph()
g1.parse(f"./{name}.nt")
display(g1,f"Numero de Nodos:{len(g1)}")
#g1.serialize(destination=f'{name}.ttl', format='turtle')
else:
g1 = Graph()
g1.parse(f"./{name}.ttl")
display(g1,f"Numero de Nodos:{len(g1)}")
# 2081722 RMLMapper
# 1715336 MorphKGC
# # Question 1
# ## Get Pandas Dataframe
# ### OpenAire
# +
query = """
PREFIX ex: <https://w3id.org/okn/os/o/>
SELECT DISTINCT ?Titulo ?date ?OpenAireID ?ArchivoDigitalUPM
WHERE {
# Buscamos los papers que tienen un OpenAireID
?Sujeto ex:has_id ?id.
?id ex:source "OpenAire".
?id ex:identifier ?OpenAireID.
# Busca los nodos que tienen una propiedad de tipo Archivo Digital
?Sujeto ?_v ex:Paper.
?Sujeto ex:title ?Titulo.
?Sujeto ex:originalID ?ArchivoDigitalUPM.
FILTER regex(?ArchivoDigitalUPM, ".*oa.upm.es.*").
?Sujeto ex:publicationDate ?date.
}
"""
qres = g1.query(query)
# converts everything to strings including missing values
openAireDF = sparql_results_to_df(qres).drop_duplicates("OpenAireID")
# Transformamos los datos para que cuadren adecuadamente con el formato
openAireDF["ArchivoDigitalUPM"] = openAireDF["ArchivoDigitalUPM"].apply(lambda x: x[4:].replace(":","/"))
display(openAireDF)
# -
# #### Observacion
# Un mismo paper de OpenAire tiene varios identificadores de la UPM
# * Museo de arte contemporáneo en Toledo [Hojas Resumen]
# * Hipódromo de Aranjuez [Hojas Resumen]
#
# Metiendose uno en la URL de ArchivoDigital se observa que aunque se llamen igual, los autores cambian
# ### Archivos Digital
# +
query = """
PREFIX ex: <https://w3id.org/okn/os/o/>
SELECT ?Titulo ?date ?upmID
WHERE {
# Buscamos aquellos que tengan ID de Archivos Digital UPM
?Sujeto ex:has_id ?id.
?id ex:source "OAI-PHM:Universidad Politécnica de Madrid".
?id ex:identifier ?upmID.
# De aquellos que tengan ID de Archivos Digital UPM, buscamos el titulo y la fecha
?Sujeto ?_v ex:Paper.
?Sujeto ex:title ?Titulo.
?Sujeto ex:publicationDate ?date.
}
"""
qres = g1.query(query)
# converts everything to strings including missing values
upmDF = sparql_results_to_df(qres)
# Transformamos los datos para que cuadren adecuadamente con el formato
upmDF["ArchivoDigitalUPM"] = upmDF["upmID"].apply(lambda x: x[7:len(x)-1])
upmDF.drop_duplicates("ArchivoDigitalUPM",inplace=True)
display(upmDF)
# -
# ## LeftJoin
LeftJoin = upmDF.merge(openAireDF,on="ArchivoDigitalUPM",how="left")
display(LeftJoin)
# ### NaN Values
LeftJoinNaN = LeftJoin[LeftJoin['OpenAireID'].isna()]
display(LeftJoinNaN)
df = upmDF[~upmDF.upmID.isin(LeftJoinNaN.upmID)]
display(df)
# ## Right Join
rightJoin = upmDF.merge(openAireDF,on="ArchivoDigitalUPM",how="right")
display(rightJoin)
# ### Valores NaN
#
rightJoinNaN = rightJoin[rightJoin['Titulo_x'].isna()]
display(rightJoinNaN)
# # Question 2
# ## Get Pandas Dataframes
# ### Query Autores
# +
query = """
PREFIX ex: <https://w3id.org/okn/os/o/>
SELECT ?orcid ?persona ?fullname ?name ?surname ?Titulo ?date ?OpenAireID ?ArchivoDigitalUPM
WHERE {
# Buscamos los papers que tienen un OpenAireID
?paper ex:has_id ?id.
?id ex:source "OpenAire".
?id ex:identifier ?OpenAireID.
?paper ex:title ?Titulo.
?paper ex:publicationDate ?date.
OPTIONAL {
?paper ex:originalID ?ArchivoDigitalUPM.
FILTER regex(?ArchivoDigitalUPM, ".*oa.upm.es.*").
}.
?paper ex:creator ?persona.
?persona ex:fullname ?fullname.
OPTIONAL {?persona ex:name ?name}.
OPTIONAL {?persona ex:surname ?surname}.
OPTIONAL {?persona ex:orcid ?orcid}.
}
group by ?persona
"""
qres = g1.query(query)
# converts everything to strings including missing values
autoresDF = sparql_results_to_df(qres)
autoresDF["ArchivoDigitalUPM"] = autoresDF["ArchivoDigitalUPM"].apply(lambda x: x[4:].replace(":","/"))
#autoresDF["name"] = autoresDF["fullname"].apply(lambda x: giveName(x))
#autoresDF["surname"] = autoresDF["fullname"].apply(lambda x: giveSurnname(x))
display(autoresDF)
# -
# ### Query Archivos Digital
# +
query = """
PREFIX ex: <https://w3id.org/okn/os/o/>
SELECT ?Titulo ?date ?upmID
WHERE {
# Buscamos aquellos que tengan ID de Archivos Digital UPM
?Sujeto ex:has_id ?id.
?id ex:source "OAI-PHM:Universidad Politécnica de Madrid".
?id ex:identifier ?upmID.
# De aquellos que tengan ID de Archivos Digital UPM, buscamos el titulo y la fecha
?Sujeto ?_v ex:Paper.
?Sujeto ex:title ?Titulo.
?Sujeto ex:publicationDate ?date.
}
"""
qres = g1.query(query)
# converts everything to strings including missing values
upmDF = sparql_results_to_df(qres)
# Transformamos los datos para que cuadren adecuadamente con el formato
upmDF["ArchivoDigitalUPM"] = upmDF["upmID"].apply(lambda x: x[7:len(x)-1])
display(upmDF)
# -
# ### Observatorio UPM
observatorioCSV = f"../Fast2Results/v3/tabla.csv"
observatorioDF = pd.read_csv(observatorioCSV).drop(["Escuela UPM","Departamento","Categoria"],axis=1)
observatorioDF["Apellidos"] = observatorioDF["Apellidos"].apply(lambda x: x.replace(". .","").replace(".",""))
display(observatorioDF)
# ## $\{ObservatorioUPM\} \cap \{OpenAire\}$
# ### Naive Approach
obs_opA = observatorioDF.merge(autoresDF,how="inner",left_on=["Nombre","Apellidos"],right_on=["name","surname"]).drop(["Nombre","Apellidos"],axis=1)
display(obs_opA)
# ### Using matching
# #### Observacion
# https://stackoverflow.com/questions/35380933/how-to-merge-two-pandas-dataframes-based-on-a-similarity-function
"""
INSERTAR METODO PARA APROXIMAR LOS NOMBRES
"""
# ## $\{\{ObservatorioUPM\} \cap \{OpenAire\}\} \cap \{ArchivosDigital\}$
# ### Por ID Archivo Digital
obs_opA_upm = obs_opA.merge(upmDF,how="left",left_on="ArchivoDigitalUPM",right_on="ArchivoDigitalUPM")
display(obs_opA_upm)
obs_opA_upm[obs_opA_upm["Titulo_y"].isna()]
# ### Por Titulo
obs_opA_upm2 = obs_opA.merge(upmDF,how="left",on="Titulo")
display(obs_opA_upm2)
obs_opA_upm2[obs_opA_upm2["ArchivoDigitalUPM_y"].isna()]
# # Question 6
observatorioCSV = f"../Fast2Results/v3/tabla.csv"
observatorioDF = pd.read_csv(observatorioCSV)
observatorioDF["Apellidos"] = observatorioDF["Apellidos"].apply(lambda x: x.replace(". .","").replace(".",""))
display(observatorioDF)
# +
query = """
PREFIX ex: <https://w3id.org/okn/os/o/>
SELECT ?Titulo ?upmID ?fullname ?name ?surname
WHERE {
# Buscamos aquellos que tengan ID de Archivos Digital UPM
?Sujeto ex:has_id ?id.
?id ex:source "OAI-PHM:Universidad Politécnica de Madrid".
?id ex:identifier ?upmID.
# De aquellos que tengan ID de Archivos Digital UPM, buscamos el titulo y la fecha
?Sujeto ?_v ex:Paper.
?Sujeto ex:title ?Titulo.
?Sujeto ex:creator ?persona.
?persona ex:fullname ?fullname.
?persona ex:name ?name.
?persona ex:surname ?surname.
}
Group by ?persona
"""
qres = g1.query(query)
# converts everything to strings including missing values
upmDF = sparql_results_to_df(qres)
display(upmDF)
# +
res = observatorioDF.merge(upmDF,left_on=["Nombre","Apellidos"],right_on=["name","surname"])
llave = "Departamento"
categorias = set(res[llave])
resultados = []
for i in categorias:
resultados.append(
(i,len(res[res[llave] == i]))
)
sorted(resultados,key=lambda x: x[1],reverse=True)
| Road2Knowledge/Queries.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [default]
# language: python
# name: python3
# ---
# # Regional average of raster values
#
# Here an average of raster values is desired over the context of some region. A RegionMask object is created, which then allows raster data to be warped onto the RegionMask's characteristics (as in the RegionMask's resolution, extent, and spatial reference system). This create a numpy matrix, from which only the pixels which lie in the region can be easily extracted and operated on.
#
# *last tested: version 1.0.2*
# +
from os.path import join
import geokit as gk
# +
# (Here a shape file of Aachen, Germany will be used)
regionPath = join(gk._test_data_, "aachenShapefile.shp")
# Create a region mask from a region definition
rm = gk.RegionMask.fromVector(regionPath)
# +
# Warp a raster file onto the RegionMask's context (Here, a simple elevation raster file is used)
rasterFile = join(gk._test_data_, "elevation.tif")
warpedMatrix = rm.warp(rasterFile)
# +
# Compute the average of pixels the region mask
meanValue = warpedMatrix[ rm.mask ].mean()
print("Mean raster value:", meanValue)
# -
| Examples/regional_raster_average.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] colab_type="text" id="pZgQLLVe1p47"
#
# ## 1.1 Introduction to PyDP
# The PyDP package provides a Python API into [Google's Differential Privacy library](https://github.com/google/differential-privacy). This example uses the alpha 0.4 version of the package that has the following limitations:
#
#
# * Supports Linux (Windows coming soon)
# * Currently implements an algorithm to support computing private mean using a Laplace noise generation technique.
# * Supports only integer and floating point values
#
#
# -
# # Example using Differential Privacy library
#
# In this directory, we give a simple example of how to use the Python Differential
# Privacy library.
#
# ## Zoo Animals
#
# There are around 182 animals at Farmer Alex's zoo. Every day, Alex feeds
# the animals as many carrots as they desire. The animals record how many carrots
# they have eaten per day. For this particular day, the number of carrots eaten
# can be seen in `animals_and_carrots.csv`.
#
# At the end of each day, Alex often asks aggregate questions about how many
# carrots everyone ate. For example, he wants to know how many carrots are eaten
# each day, so he knows how many to order the next day. The animals are fearful
# that Alex will use the data against their best interest. For example, Alex could
# get rid of the animals who eat the most carrots!
#
# To protect themselves, the animals decide to use the Python Differential Privacy
# library to aggregate their data before reporting it to Alex. This way, the
# animals can control the risk that Alex will identify individuals' data while
# maintaining an adequate level of accuracy so that Alex can continue to run the
# zoo effectively.
#
# + colab={"base_uri": "https://localhost:8080/", "height": 102} colab_type="code" executionInfo={"elapsed": 8271, "status": "ok", "timestamp": 1587961311947, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj8JCwpOTvlaeImB7WktdTucJJTZ_kDAprKpf_sOA=s64", "userId": "08454492702843508366"}, "user_tz": -600} id="h67VRxjSSQ9b" outputId="6da55f3e-cfb8-4503-a179-ec2613a7f8bd"
# Install the PyDP package
# ! pip install python-dp
# + colab={} colab_type="code" id="ipGLypbRTDSc"
import pydp as dp # by convention our package is to be imported as dp (for Differential Privacy!)
from pydp.algorithms.laplacian import BoundedSum, BoundedMean, Count, Max
import pandas as pd
import statistics # for calculating mean without applying differential privacy
# -
# ## Data
#
# Each row in `animals_and_carrots.csv` is composed of the name of an animal, and
# the number of carrots it has eaten, comma-separated.
#
# + colab={"base_uri": "https://localhost:8080/", "height": 204} colab_type="code" executionInfo={"elapsed": 1171, "status": "ok", "timestamp": 1587961449410, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj8JCwpOTvlaeImB7WktdTucJJTZ_kDAprKpf_sOA=s64", "userId": "08454492702843508366"}, "user_tz": -600} id="jM3luaaKSQ9h" outputId="de008de4-f261-47ef-de5a-d33d23792c9d"
# get carrots data from our public github repo
url = "https://raw.githubusercontent.com/OpenMined/PyDP/dev/examples/Tutorial_1-carrots_demo/animals_and_carrots.csv"
df = pd.read_csv(url, sep=",", names=["animal", "carrots_eaten"])
df.head()
# -
# Taking the mean of all the entries in a normal fashion without applying the DP library. This is the actual mean of all the records.
# ## Per-animal Privacy
#
# Notice that each animal owns at most one row in the data. This means that we
# provide per-animal privacy. Suppose that some animal appears multiple times in
# the csv file. That animal would own more than one row in the data. In this case,
# using this DP library would not guarantee per-animal privacy! The animals would
# first have to pre-process their data in a way such that each animal doesn't own
# more than one row.
#
# + colab={} colab_type="code" id="5_pUyyKzW1YC"
# calculates mean without applying differential privacy
def mean_carrots() -> float:
return statistics.mean(list(df["carrots_eaten"]))
# -
# Private Mean uses Differential Privacy Library by Google to calculate the Mean. To preserve privacy, Laplacian mechanism is used.
#
# The function takes the argument privacy_budget as input.
#
# It is a number between 0 and 1, denoting privacy threshold
#
# It measures the acceptable loss of privacy (with 0 meaning no loss is acceptable).
#
# `BoundedMean.quick_result()` takes a List of integer/ float as an input and returns the mean of the list values.
#
# + colab={} colab_type="code" id="0AMReuBXTaLV"
# calculates mean applying differential privacy
def private_mean(privacy_budget: float) -> float:
x = BoundedMean(privacy_budget, 0, 1, 100)
return x.quick_result(list(df["carrots_eaten"]))
# -
# As you can see, the value of the private mean varies compared to the mean calculated using non-private statistical methods.
#
# This difference in value corresponds to the privacy that is actually preserved for individual records in it.
# + colab={"base_uri": "https://localhost:8080/", "height": 51} colab_type="code" executionInfo={"elapsed": 749, "status": "ok", "timestamp": 1587961454996, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14Gj8JCwpOTvlaeImB7WktdTucJJTZ_kDAprKpf_sOA=s64", "userId": "08454492702843508366"}, "user_tz": -600} id="vgkt7BuZW3Vw" outputId="f5708cb5-1477-4f05-851b-b40ae7d3125d"
print("Mean: ", mean_carrots())
print("Private Mean: ", private_mean(0.8))
# -
# Counts number of animals who ate more than 'limit' carrots without applying the DP library. This is the actual number of such animals.
# Calculates number of animals who ate more than "limit" carrots without applying differential privacy.
def count_above(limit: int) -> int:
return df[df.carrots_eaten > limit].count()[0]
# Private Count Above uses Differential Privacy Library by Google to calculate the number of rows with value above limit. To preserve privacy, Laplacian mechanism is used.
#
# The function takes the argument privacy_budget as input.
#
# It is a number between 0 and 1, denoting privacy threshold
#
# It measures the acceptable loss of privacy (with 0 meaning no loss is acceptable).
#
# The function also takes the limit as an argument.
#
# `Count.quick_result()` takes a List of integer/ float as an input and returns the count of elements in the list.
# Calculates number of animals who ate more than "limit" carrots applying differential privacy.
def private_count_above(privacy_budget: float, limit: int) -> int:
x = Count(privacy_budget, dtype="int")
return x.quick_result(list(df[df.carrots_eaten > limit]["carrots_eaten"]))
# As you can see, the value of Private Count Above varies compared to the Count calculated using normal Statistical methods.
#
# This difference in values refers to that privacy is actually preserved for individual records in it.
print("Above 70:\t" + str(count_above(70)))
print("private count above:\t" + str(private_count_above(1, 70)))
# Taking Max of all the entries in a normal fashion without Applying the DP library. This is the actual maximum of carrots eaten of all the records.
# Function to return the maximum of the number of carrots eaten by any one animal without appyling differential privacy.
def max() -> int:
return df.max()[1]
# Private Max uses Differential Privacy Library by Google to calculate the maximum out of all the values. To preserve privacy, Laplacian mechanism is used.
#
# The function takes the argument privacy_budget as input.
#
# It is a number between 0 and 1, denoting privacy threshold
#
# It measures the acceptable loss of privacy (with 0 meaning no loss is acceptable).
#
# `Max.quick_result()` takes a List of integer/ float as an input and returns the list max value.
# Function to return the maximum of the number of carrots eaten by any one animal appyling differential privacy.
def private_max(privacy_budget: float) -> int:
# 0 and 150 are the upper and lower limits for the search bound.
x = Max(privacy_budget, 0, 100, dtype="int")
return x.quick_result(list(df["carrots_eaten"]))
# As you can see, the value of Private Max varies compared to the Max calculated using normal Statistical methods.
#
# This difference in values refers to that privacy is actually preserved for individual records in it.
print("Max:\t" + str(max()))
print("private max:\t" + str(private_max(1)))
# Taking Sum of all the entries in a normal fashion without Applying the DP library. This is the actual sum of carrots eaten by all the animals.
# Function to calculate sum of carrots eaten without applying differential privacy.
def sum_carrots() -> int:
return df.sum()[1]
# Private Sum uses Differential Privacy Library by Google to calculate the sum of all the values. To preserve privacy, Laplacian mechanism is used.
#
# The function takes the argument privacy_budget as input.
#
# It is a number between 0 and 1, denoting privacy threshold
#
# It measures the acceptable loss of privacy (with 0 meaning no loss is acceptable).
#
# `BoundedSum.quick_result()` takes a List of integer/ float as an input and returns the list sum.
# Function to calculate sum of carrots eaten applying differential privacy.
def private_sum(privacy_budget: float) -> int:
x = BoundedSum(privacy_budget, 0, 1, 100, dtype="float")
return x.quick_result(list(df["carrots_eaten"]))
print("Sum:\t" + str(sum_carrots()))
print("Private Sum:\t" + str(private_sum(1)))
# ## How to Run
#
# ```python PyDP/example/carrots.py```
#
| examples/Tutorial_1-carrots_demo/carrots_demo.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Relation extraction with BERT
#
# ---
#
# The goal of this notebook is to show how to use [BERT](https://arxiv.org/abs/1810.04805)
# to [extract relation](https://en.wikipedia.org/wiki/Relationship_extraction) from text.
#
# Used libraries:
# - [PyTorch](https://pytorch.org/)
# - [PyTorch-Lightning](https://pytorch-lightning.readthedocs.io/en/latest/)
# - [Transformers](https://huggingface.co/transformers/index.html)
#
# Used datasets:
# - SemEval 2010 Task 8 - [paper](https://arxiv.org/pdf/1911.10422.pdf) - [download](https://github.com/sahitya0000/Relation-Classification/blob/master/corpus/SemEval2010_task8_all_data.zip?raw=true)
# - Google IISc Distant Supervision (GIDS) - [paper](https://arxiv.org/pdf/1804.06987.pdf) - [download](https://drive.google.com/open?id=1gTNAbv8My2QDmP-OHLFtJFlzPDoCG4aI)
# ## High level overview
#
# We will experiment with two architectures: single-classifier & duo-classifier
#
# 
#
# 
#
# The classifiers are implemented as follows:
#
# 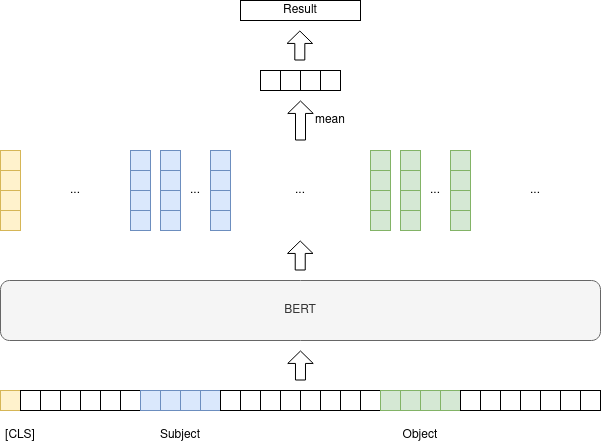
# + [markdown] pycharm={"name": "#%% md\n"}
# ## Install dependencies
#
# This project uses [Python 3.7+](https://www.python.org/downloads/release/python-378/)
# + pycharm={"name": "#%%\n"}
# !pip install requests==2.23.0 numpy==1.18.5 pandas==1.0.3 \
# scikit-learn==0.23.1 pytorch-lightning==0.8.4 torch==1.5.1 \
# transformers==3.0.2 sklearn==0.0 tqdm==4.45.0 neptune-client==0.4.119 \
# matplotlib==3.1.0 scikit-plot==0.3.7
# -
# ## Import needed modules
# + pycharm={"name": "#%% \n"}
import gc
import json
import math
import os
from abc import ABC, abstractmethod
from collections import OrderedDict
from random import randint
from typing import Iterable, Tuple
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import torch
from matplotlib.figure import Figure
from pandas import DataFrame
from pytorch_lightning import LightningModule, seed_everything
from pytorch_lightning import Trainer as LightningTrainer
from pytorch_lightning.logging.neptune import NeptuneLogger
from sklearn.metrics import *
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import label_binarize
from sklearn.utils import column_or_1d
from torch import Tensor, nn
from torch.nn import functional as F
from torch.optim import AdamW
from torch.optim.lr_scheduler import LambdaLR
from torch.utils.data import DataLoader, IterableDataset
from tqdm.auto import tqdm
from transformers import *
# -
# ## Define constants
# + pycharm={"name": "#%% \n"}
# --- Random seed ---
SEED = 2020
seed_everything(SEED)
# --- Directory ---
ROOT_DIR = os.path.abspath(".")
PROCESSED_DATA_DIR = os.path.join(ROOT_DIR, "data/processed")
METADATA_FILE_NAME = os.path.join(PROCESSED_DATA_DIR, "metadata.json")
CHECKPOINT_DIR = os.path.join(ROOT_DIR, "checkpoint")
KAGGLE_ENV = bool(os.getenv("KAGGLE_URL_BASE"))
if KAGGLE_ENV:
# in Kaggle environment
# 2 datasets should already been added to the notebook
RAW_DATA_DIR = os.path.join(ROOT_DIR, "../input")
else:
# in local environment
RAW_DATA_DIR = os.path.join(ROOT_DIR, "data/raw")
# --- Datasets ---
DATASET_MAPPING = {
"SemEval2010Task8": {
"dir": os.path.join(RAW_DATA_DIR,"semeval2010-task-8"),
"keep_test_order": True,
"precision_recall_curve_baseline_img": None,
},
"GIDS": {
"dir": os.path.join(RAW_DATA_DIR,"gids-dataset"),
"keep_test_order": False,
"precision_recall_curve_baseline_img": os.path.join(RAW_DATA_DIR,"gids-dataset/GIDS_precision_recall_curve.png"),
}
}
# change this variable to switch dataset in later tasks
DATASET_NAME = list(DATASET_MAPPING.keys())[1]
# --- Subject & object markup ---
SUB_START_CHAR = "["
SUB_END_CHAR = "]"
OBJ_START_CHAR = "{"
OBJ_END_CHAR = "}"
# --- BERT variants ---
# See https://huggingface.co/transformers/pretrained_models.html for the full list
AVAILABLE_PRETRAINED_MODELS = [
"distilbert-base-uncased", # 0
"distilbert-base-cased", # 1
"bert-base-uncased", # 2
"distilgpt2", # 3
"gpt2", # 4
"distilroberta-base", # 5
"roberta-base", # 6
"albert-base-v1", # 7
"albert-base-v2", # 8
"bert-large-uncased", # 9
]
# change this variable to switch pretrained language model
PRETRAINED_MODEL = AVAILABLE_PRETRAINED_MODELS[2]
# if e1 is not related to e2, should "e2 not related to e1" be added to the training set
ADD_REVERSE_RELATIONSHIP = True
# --- Neptune logger ---
# Create a free account at https://neptune.ai/,
# then get the API token and create a project
NEPTUNE_API_TOKEN = " INSERT YOUR API TOKEN HERE "
NEPTUNE_PROJECT_NAME = " INSERT YOUR PROJECT NAME HERE "
# + [markdown] pycharm={"name": "#%% md\n"}
# ## Preprocess
#
# 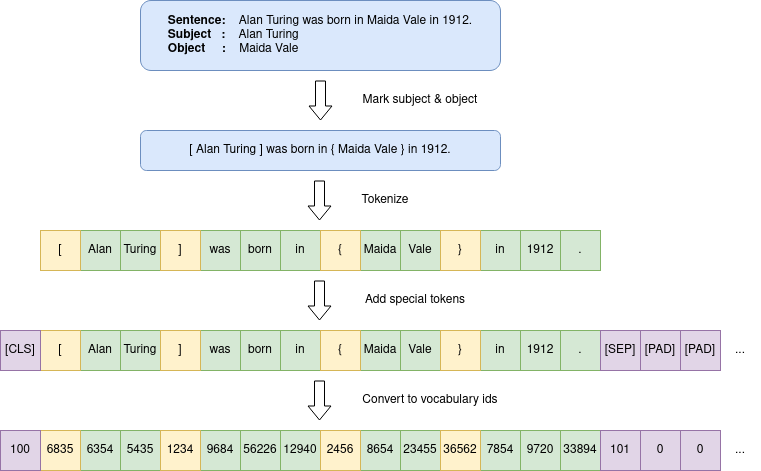
#
# First, we define a custom label encoder. What this label encoder offers but `sklearn.preprocessing.LabelEncoder` fails
# to provide:
# - Order preservation: labels will be encoded in order they appear in the dataset. Labels appears earlier will have
# smaller id. We need this to ensure the `no relation` class is always encoded as `0`
# - Multiple fit: `sklearn.preprocessing.LabelEncoder` forgets what is fit in the last time `fit` is called while our
# encoder keeps adding new labels to existing ones. This is useful when we process large dataset in batches.
# + pycharm={"name": "#%%\n"}
class OrdinalLabelEncoder:
def __init__(self, init_labels=None):
if init_labels is None:
init_labels = []
self.mapping = OrderedDict({l: i for i, l in enumerate(init_labels)})
@property
def classes_(self):
return list(self.mapping.keys())
def fit_transform(self, y):
return self.fit(y).transform(y)
def fit(self, y):
y = column_or_1d(y, warn=True)
new_classes = pd.Series(y).unique()
for cls in new_classes:
if cls not in self.mapping:
self.mapping[cls] = len(self.mapping)
return self
def transform(self, y):
y = column_or_1d(y, warn=True)
return [self.mapping[value] for value in y]
# + [markdown] pycharm={"name": "#%% md\n"}
# Abstract preprocessor class:
# + pycharm={"name": "#%%\n"}
class AbstractPreprocessor(ABC):
DATASET_NAME = ""
VAL_DATA_PROPORTION = 0.2
NO_RELATION_LABEL = ""
def __init__(self, tokenizer: PreTrainedTokenizer):
self.tokenizer = tokenizer
self.SUB_START_ID, self.SUB_END_ID, self.OBJ_START_ID, self.OBJ_END_ID \
= tokenizer.convert_tokens_to_ids([SUB_START_CHAR, SUB_END_CHAR, OBJ_START_CHAR, OBJ_END_CHAR])
self.label_encoder = OrdinalLabelEncoder([self.NO_RELATION_LABEL])
def preprocess_data(self, reprocess: bool):
print(f"\n---> Preprocessing {self.DATASET_NAME} dataset <---")
# create processed data dir
if not os.path.exists(PROCESSED_DATA_DIR):
print("Creating processed data directory " + PROCESSED_DATA_DIR)
os.makedirs(PROCESSED_DATA_DIR)
# stop preprocessing if file existed
json_file_names = [self.get_dataset_file_name(k) for k in ("train", "val", "test")]
existed_files = [fn for fn in json_file_names if os.path.exists(fn)]
if existed_files:
file_text = "- " + "\n- ".join(existed_files)
if not reprocess:
print("The following files already exist:")
print(file_text)
print("Preprocessing is skipped. See option --reprocess.")
return
else:
print("The following files will be overwritten:")
print(file_text)
train_data, val_data, test_data = self._preprocess_data()
print("Saving to json files")
self._write_data_to_file(train_data, "train")
self._write_data_to_file(val_data, "val")
self._write_data_to_file(test_data, "test")
self._save_metadata({
"train_size": len(train_data),
"val_size": len(val_data),
"test_size": len(test_data),
"no_relation_label": self.NO_RELATION_LABEL,
**self._get_label_mapping()
})
self._create_secondary_data_files()
print("---> Done ! <---")
@abstractmethod
def _preprocess_data(self) -> Tuple[DataFrame, DataFrame, DataFrame]:
pass
def _create_secondary_data_files(self):
"""
From the primary data file, create a data file with binary labels
and a data file with only sentences classified as "related"
"""
with open(METADATA_FILE_NAME) as f:
root_metadata = json.load(f)
metadata = root_metadata[self.DATASET_NAME]
related_only_count = {
"train": 0,
"val": 0,
"test": 0,
}
for key in ["train", "test", "val"]:
print(f"Creating secondary files for {key} data")
origin_file = open(self.get_dataset_file_name(key))
bin_file = open(self.get_dataset_file_name(f"{key}_binary"), "w")
related_file = open(self.get_dataset_file_name(f"{key}_related_only"), "w")
total = metadata[f"{key}_size"]
for line in tqdm(origin_file, total=total):
data = json.loads(line)
if data["label"] != 0:
related_only_count[key] += 1
data["label"] -= 1 # label in "related_only" files is 1 less than the original label
related_file.write(json.dumps(data) + "\n")
data["label"] = 1 # in binary dataset, all "related" classes have label 1
bin_file.write(json.dumps(data) + "\n")
else:
bin_file.write(json.dumps(data) + "\n")
origin_file.close()
bin_file.close()
related_file.close()
print("Updating metadata.json")
for key in ["train", "test", "val"]:
metadata[f"{key}_related_only_size"] = related_only_count[key]
root_metadata[self.DATASET_NAME] = metadata
with open(METADATA_FILE_NAME, "w") as f:
json.dump(root_metadata, f, indent=4)
def _find_sub_obj_pos(self, input_ids_list: Iterable) -> DataFrame:
"""
Find subject and object position in a sentence
"""
sub_start_pos = [self._index(s, self.SUB_START_ID) + 1 for s in input_ids_list]
sub_end_pos = [self._index(s, self.SUB_END_ID, sub_start_pos[i]) for i, s in enumerate(input_ids_list)]
obj_start_pos = [self._index(s, self.OBJ_START_ID) + 1 for s in input_ids_list]
obj_end_pos = [self._index(s, self.OBJ_END_ID, obj_start_pos[i]) for i, s in enumerate(input_ids_list)]
return DataFrame({
"sub_start_pos": sub_start_pos,
"sub_end_pos": sub_end_pos,
"obj_start_pos": obj_start_pos,
"obj_end_pos": obj_end_pos,
})
@staticmethod
def _index(lst: list, ele: int, start: int = 0) -> int:
"""
Find an element in a list. Returns -1 if not found instead of raising an exception.
"""
try:
return lst.index(ele, start)
except ValueError:
return -1
def _clean_data(self, raw_sentences: list, labels: list) -> DataFrame:
if not raw_sentences:
return DataFrame()
tokens = self.tokenizer(raw_sentences, truncation=True, padding="max_length")
data = DataFrame(tokens.data)
data["label"] = self.label_encoder.fit_transform(labels)
sub_obj_position = self._find_sub_obj_pos(data["input_ids"])
data = pd.concat([data, sub_obj_position], axis=1)
data = self._remove_invalid_sentences(data)
return data
def _remove_invalid_sentences(self, data: DataFrame) -> DataFrame:
"""
Remove sentences without subject/object or whose subject/object
is beyond the maximum length the model supports
"""
seq_max_len = self.tokenizer.model_max_length
return data.loc[
(data["sub_end_pos"] < seq_max_len)
& (data["obj_end_pos"] < seq_max_len)
& (data["sub_end_pos"] > -1)
& (data["obj_end_pos"] > -1)
]
def _get_label_mapping(self):
"""
Returns a mapping from id to label and vise versa from the label encoder
"""
# all labels
id_to_label = dict(enumerate(self.label_encoder.classes_))
label_to_id = {v: k for k, v in id_to_label.items()}
# for the related_only dataset
# ignore id 0, which represent no relation
id_to_label_related_only = {k - 1: v for k, v in id_to_label.items() if k != 0}
label_to_id_related_only = {v: k for k, v in id_to_label_related_only.items()}
return {
"id_to_label": id_to_label,
"label_to_id": label_to_id,
"id_to_label_related_only": id_to_label_related_only,
"label_to_id_related_only": label_to_id_related_only,
}
def _write_data_to_file(self, dataframe: DataFrame, subset: str):
"""Write data in a dataframe to train/val/test file"""
lines = ""
for _, row in dataframe.iterrows():
lines += row.to_json() + "\n"
with open(self.get_dataset_file_name(subset), "w") as file:
file.write(lines)
def _save_metadata(self, metadata: dict):
"""Save metadata to metadata.json"""
# create metadata file
if not os.path.exists(METADATA_FILE_NAME):
print(f"Create metadata file at {METADATA_FILE_NAME}")
with open(METADATA_FILE_NAME, "w") as f:
f.write("{}\n")
# add metadata
print("Saving metadata")
with open(METADATA_FILE_NAME) as f:
root_metadata = json.load(f)
with open(METADATA_FILE_NAME, "w") as f:
root_metadata[self.DATASET_NAME] = metadata
json.dump(root_metadata, f, indent=4)
@classmethod
def get_dataset_file_name(cls, key: str) -> str:
return os.path.join(PROCESSED_DATA_DIR, f"{cls.DATASET_NAME.lower()}_{key}.json")
# -
# Concrete preprocessor for each dataset:
# + pycharm={"name": "#%%\n"}
class SemEval2010Task8Preprocessor(AbstractPreprocessor):
DATASET_NAME = "SemEval2010Task8"
NO_RELATION_LABEL = "Other"
RAW_TRAIN_FILE_NAME = os.path.join(DATASET_MAPPING["SemEval2010Task8"]["dir"],
"SemEval2010_task8_training/TRAIN_FILE.TXT")
RAW_TEST_FILE_NAME = os.path.join(DATASET_MAPPING["SemEval2010Task8"]["dir"],
"SemEval2010_task8_testing_keys/TEST_FILE_FULL.TXT")
RAW_TRAIN_DATA_SIZE = 8000
RAW_TEST_DATA_SIZE = 2717
def _preprocess_data(self):
print("Processing training data")
train_data = self._process_file(
self.RAW_TRAIN_FILE_NAME,
self.RAW_TRAIN_DATA_SIZE,
ADD_REVERSE_RELATIONSHIP,
)
print("Processing test data")
test_data = self._process_file(
self.RAW_TEST_FILE_NAME,
self.RAW_TEST_DATA_SIZE,
False,
)
print("Splitting train & validate data")
train_data, val_data = train_test_split(train_data, shuffle=True, random_state=SEED)
return train_data, val_data, test_data
def _process_file(self, file_name: str, dataset_size: int, add_reverse: bool) -> DataFrame:
raw_sentences = []
labels = []
with open(file_name) as f:
for _ in tqdm(range(dataset_size)):
sent = f.readline()
label, sub, obj = self._process_label(f.readline())
labels.append(label)
raw_sentences.append(self._process_sentence(sent, sub, obj))
if label == "Other" and add_reverse:
labels.append(label)
raw_sentences.append(self._process_sentence(sent, obj, sub))
f.readline()
f.readline()
return self._clean_data(raw_sentences, labels)
@staticmethod
def _process_sentence(sentence: str, sub: int, obj: int) -> str:
return sentence.split("\t")[1][1:-2] \
.replace(f"<e{sub}>", SUB_START_CHAR) \
.replace(f"</e{sub}>", SUB_END_CHAR) \
.replace(f"<e{obj}>", OBJ_START_CHAR) \
.replace(f"</e{obj}>", OBJ_END_CHAR)
@staticmethod
def _process_label(label: str) -> Tuple[str, int, int]:
label = label.strip()
if label == "Other":
return label, 1, 2
nums = list(filter(str.isdigit, label))
return label, int(nums[0]), int(nums[1])
class GIDSPreprocessor(AbstractPreprocessor):
DATASET_NAME = "GIDS"
RAW_TRAIN_FILE_NAME = os.path.join(DATASET_MAPPING["GIDS"]["dir"], "train.tsv")
RAW_VAL_FILE_NAME = os.path.join(DATASET_MAPPING["GIDS"]["dir"], "val.tsv")
RAW_TEST_FILE_NAME = os.path.join(DATASET_MAPPING["GIDS"]["dir"], "test.tsv")
TRAIN_SIZE = 11297
VAL_SIZE = 1864
TEST_SIZE = 5663
NO_RELATION_LABEL = "NA"
def _process_file(self, file_name: str, add_reverse: bool) -> DataFrame:
"""
Process a file in batches
Return the total data size
"""
with open(file_name) as in_file:
lines = in_file.readlines()
raw_sentences = []
labels = []
for line in tqdm(lines):
_, _, sub, obj, label, sent = line.split("\t")
sent = sent.replace("###END###", "")
# add subject markup
new_sub = SUB_START_CHAR + " " + sub.replace("_", " ") + " " + SUB_END_CHAR
new_obj = OBJ_START_CHAR + " " + obj.replace("_", " ") + " " + OBJ_END_CHAR
sent = sent.replace(sub, new_sub).replace(obj, new_obj)
raw_sentences.append(sent)
labels.append(label)
if add_reverse and label == self.NO_RELATION_LABEL:
new_sub = OBJ_START_CHAR + " " + sub.replace("_", " ") + " " + OBJ_END_CHAR
new_obj = SUB_START_CHAR + " " + obj.replace("_", " ") + " " + SUB_END_CHAR
sent = sent.replace(sub, new_sub).replace(obj, new_obj)
raw_sentences.append(sent)
labels.append(label)
return self._clean_data(raw_sentences, labels)
def _preprocess_data(self):
print("Process train dataset")
train_data = self._process_file(
self.RAW_TRAIN_FILE_NAME,
ADD_REVERSE_RELATIONSHIP,
)
print("Process val dataset")
val_data = self._process_file(
self.RAW_VAL_FILE_NAME,
False,
)
print("Process test dataset")
test_data = self._process_file(
self.RAW_TEST_FILE_NAME,
False,
)
return train_data, val_data, test_data
# + [markdown] pycharm={"name": "#%% md\n"}
# Factory method to create preprocessors:
# + pycharm={"name": "#%%\n"}
def get_preprocessor_class(dataset_name: str = DATASET_NAME):
return globals()[f"{dataset_name}Preprocessor"]
def get_preprocessor(dataset_name: str = DATASET_NAME)-> AbstractPreprocessor:
tokenizer = AutoTokenizer.from_pretrained(PRETRAINED_MODEL, use_fast=True)
# some tokenizer, like GPTTokenizer, doesn't have pad_token
# in this case, we use eos token as pad token
if tokenizer.pad_token is None:
tokenizer.pad_token = tokenizer.eos_token
preprocessors_class = get_preprocessor_class(dataset_name)
return preprocessors_class(tokenizer)
# + [markdown] pycharm={"name": "#%% md\n"}
# Preprocess data:
# + pycharm={"name": "#%%\n"}
preprocessor = get_preprocessor()
preprocessor.preprocess_data(reprocess=True)
# + [markdown] pycharm={"name": "#%% md\n"}
# ## Dataset
#
# We adopt the "smart batching" technique from [here](https://towardsdatascience.com/divide-hugging-face-transformers-training-time-by-2-or-more-21bf7129db9q-21bf7129db9e)
#
# Here's a brief diagram:
#
# 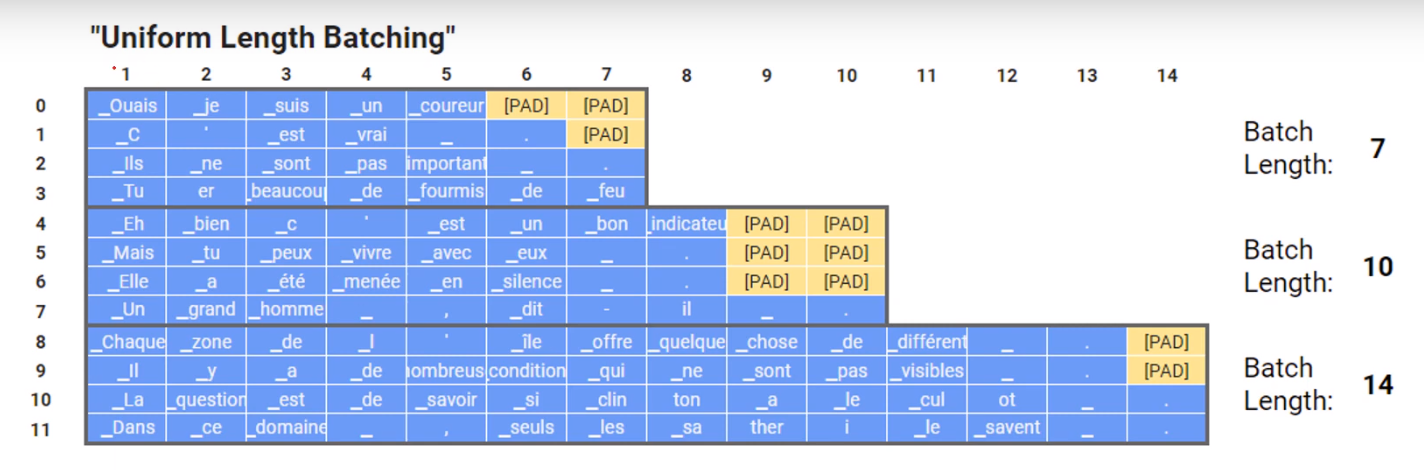
# + pycharm={"name": "#%%\n"}
class GenericDataset(IterableDataset):
"""A generic dataset for train/val/test data for both SemEval and GIDS dataset"""
def __init__(self, dataset_name: str, subset: str, batch_size: int, label_transform: str):
assert subset in ["train", "val", "test"]
assert label_transform in ["none", "binary", "related_only"]
file_name = subset if label_transform == "none" \
else f"{subset}_{label_transform}"
preprocessor_class = get_preprocessor_class()
with open(METADATA_FILE_NAME) as f:
metadata = json.load(f)[dataset_name]
size = metadata[f"{subset}_related_only_size"] \
if label_transform is "related_only" \
else metadata[f"{subset}_size"]
self.subset = subset
self.batch_size = batch_size
self.length = math.ceil(size / batch_size)
self.file = open(preprocessor_class.get_dataset_file_name(file_name))
self.keep_test_order = self.subset == "test" and DATASET_MAPPING[dataset_name]["keep_test_order"]
def __del__(self):
if self.file:
self.file.close()
def __iter__(self):
"""
Implement "smart batching"
"""
data = [json.loads(line) for line in self.file]
if not self.keep_test_order:
data = sorted(data, key=lambda x: sum(x["attention_mask"]))
new_data = []
while len(data) > 0:
if self.keep_test_order or len(data) < self.batch_size:
idx = 0
else:
idx = randint(0, len(data) - self.batch_size)
batch = data[idx:idx + self.batch_size]
max_len = max([sum(b["attention_mask"]) for b in batch])
for b in batch:
input_data = {}
for k, v in b.items():
if k != "label":
if isinstance(v, list):
input_data[k] = torch.tensor(v[:max_len])
else:
input_data[k] = torch.tensor(v)
label = torch.tensor(b["label"])
new_data.append((input_data, label))
del data[idx:idx + self.batch_size]
yield from new_data
def __len__(self):
return self.length
def as_batches(self):
input_data = []
label = []
def create_batch():
return (
{k: torch.stack([x[k] for x in input_data]).cuda() for k in input_data[0].keys()},
torch.tensor(label).cuda()
)
for ip, l in self:
input_data.append(ip)
label.append(l)
if len(input_data) == self.batch_size:
yield create_batch()
input_data.clear()
label.clear()
yield create_batch()
# + [markdown] pycharm={"name": "#%% md\n"}
# ## Classifiers
# + pycharm={"name": "#%%\n"}
class BaseClassifier(LightningModule, ABC):
"""
Base class of all classifiers
"""
dataset_label_transform = None
@abstractmethod
def loss_function(self, logits: Tensor, label: Tensor) -> Tensor:
"""
Calculate the loss of the model
It MUST take care of the last activation layer
"""
pass
@abstractmethod
def log_metrics(self, epoch_type: str, logits: Tensor, label: Tensor) -> dict:
pass
def __init__(self, pretrained_language_model, dataset_name, batch_size, learning_rate, decay_lr_speed,
dropout_p, activation_function, weight_decay, linear_size):
super().__init__()
self.save_hyperparameters()
self.test_proposed_answer = None
self.language_model = AutoModel.from_pretrained(pretrained_language_model)
config = self.language_model.config
self.max_seq_len = config.max_position_embeddings
self.hidden_size = config.hidden_size
self.linear = nn.Linear(self.hidden_size, linear_size)
self.linear_output = nn.Linear(linear_size, self.num_classes)
self.dropout = nn.Dropout(p=dropout_p)
self.activation_function = getattr(nn, activation_function)()
def forward(self, sub_start_pos, sub_end_pos,
obj_start_pos, obj_end_pos, *args, **kwargs) -> Tensor:
language_model_output = self.language_model(*args, **kwargs)
if isinstance(language_model_output, tuple):
language_model_output = language_model_output[0]
x = torch.mean(language_model_output, dim=1)
x = self.dropout(x)
x = self.linear(x)
x = self.activation_function(x)
x = self.dropout(x)
logits = self.linear_output(x)
return logits
def train_dataloader(self) -> DataLoader:
return self.__get_dataloader("train")
def val_dataloader(self) -> DataLoader:
return self.__get_dataloader("val")
def test_dataloader(self) -> DataLoader:
return self.__get_dataloader("test")
def __get_dataloader(self, subset: str) -> DataLoader:
batch_size = self.hparams.batch_size
dataset = GenericDataset(
self.hparams.dataset_name,
subset,
batch_size,
self.dataset_label_transform
)
return DataLoader(
dataset,
batch_size=batch_size,
num_workers=1
)
def configure_optimizers(self):
optimizer = AdamW(
[p for p in self.parameters() if p.requires_grad],
lr=self.hparams.learning_rate,
weight_decay=self.hparams.weight_decay
)
scheduler = LambdaLR(optimizer, lambda epoch: self.hparams.decay_lr_speed[epoch])
return [optimizer], [scheduler]
def training_step(self, batch: Tuple[dict, Tensor], batch_nb: int) -> dict:
input_data, label = batch
logits = self(**input_data)
loss = self.loss_function(logits, label)
log = {"train_loss": loss}
return {"loss": loss, "log": log}
def __eval_step(self, batch: Tuple[dict, Tensor]) -> dict:
input_data, label = batch
logits = self(**input_data)
return {
"logits": logits,
"label": label,
}
def validation_step(self, batch: Tuple[dict, Tensor], batch_nb: int) -> dict:
return self.__eval_step(batch)
def test_step(self, batch: Tuple[dict, Tensor], batch_nb: int) -> dict:
return self.__eval_step(batch)
def __eval_epoch_end(self, epoch_type: str, outputs: Iterable[dict]) -> dict:
assert epoch_type in ["test", "val"]
logits = torch.cat([x["logits"] for x in outputs]).cpu()
label = torch.cat([x["label"] for x in outputs]).cpu()
logs = self.log_metrics(epoch_type, logits, label)
return {"progress_bar": logs}
def validation_epoch_end(self, outputs: Iterable[dict]) -> dict:
return self.__eval_epoch_end("val", outputs)
def test_epoch_end(self, outputs: Iterable[dict]) -> dict:
return self.__eval_epoch_end("test", outputs)
def numeric_labels_to_text(self, label):
"""Revert labels from number to text"""
if self.dataset_label_transform == "binary":
label = ["Positive" if x else "Negative" for x in label]
else:
with open(METADATA_FILE_NAME) as f:
meta = json.load(f)[self.hparams.dataset_name]
if self.dataset_label_transform == "none":
mapping = meta["id_to_label"]
else:
mapping = meta["id_to_label_related_only"]
label = [mapping[str(int(x))] for x in label]
return label
@staticmethod
def plot_confusion_matrix(predicted_label, label) -> Figure:
result = confusion_matrix(label, predicted_label)
display = ConfusionMatrixDisplay(result)
fig, ax = plt.subplots(figsize=(16, 12))
display.plot(cmap=plt.cm.get_cmap("Blues"), ax=ax, xticks_rotation='vertical')
return fig
def log_confusion_matrix(self, prefix: str, predicted_label: Tensor, label: Tensor):
predicted_label = self.numeric_labels_to_text(predicted_label)
label = self.numeric_labels_to_text(label)
fig = self.plot_confusion_matrix(predicted_label, label)
self.logger.experiment.log_image(f"{prefix}_confusion_matrix", fig)
class MulticlassClassifier(BaseClassifier, ABC):
"""
Base class for multiclass classifiers
"""
def loss_function(self, logits: Tensor, label: Tensor)-> Tensor:
return F.cross_entropy(logits, label)
@staticmethod
def logits_to_label(logits: Tensor) -> Tensor:
return torch.argmax(logits, dim=-1)
def log_metrics(self, epoch_type: str, logits: Tensor, label: Tensor) -> dict:
predicted_label = self.logits_to_label(logits)
self.log_confusion_matrix(epoch_type, predicted_label, label)
logs = {
f"{epoch_type}_avg_loss": float(self.loss_function(logits, label)),
f"{epoch_type}_acc": accuracy_score(label, predicted_label),
f"{epoch_type}_pre_weighted": precision_score(label, predicted_label, average="weighted"),
f"{epoch_type}_rec_weighted": recall_score(label, predicted_label, average="weighted"),
f"{epoch_type}_f1_weighted": f1_score(label, predicted_label, average="weighted"),
f"{epoch_type}_pre_macro": precision_score(label, predicted_label, average="macro"),
f"{epoch_type}_rec_macro": recall_score(label, predicted_label, average="macro"),
f"{epoch_type}_f1_macro": f1_score(label, predicted_label, average="macro"),
}
for k, v in logs.items():
self.logger.experiment.log_metric(k, v)
return logs
class StandardClassifier(MulticlassClassifier):
"""
A classifier that can recognize the "not related" as well as other relations
"""
dataset_label_transform = "none"
def __init__(self, dataset_name, **kwargs):
with open(METADATA_FILE_NAME) as f:
self.num_classes = len(json.load(f)[dataset_name]["label_to_id"])
self.test_proposed_answer = None
super().__init__(dataset_name=dataset_name, **kwargs)
def log_metrics(self, epoch_type: str, logits: Tensor, label: Tensor)-> dict:
if epoch_type == "test":
self.test_proposed_answer = self.logits_to_label(logits).tolist()
self.__log_precision_recall_curve(epoch_type, logits, label)
return super().log_metrics(epoch_type, logits, label)
def __log_precision_recall_curve(self, epoch_type: str, logits: Tensor, label: Tensor):
"""
Log the micro-averaged precision recall curve
Ref: https://scikit-learn.org/stable/auto_examples/model_selection/plot_precision_recall.html
"""
label = torch.tensor(label_binarize(label, classes=list(range(self.num_classes)))).flatten()
logits = logits.flatten()
pre, rec, thresholds = precision_recall_curve(label, logits)
f1s = 2 * pre * rec / (pre + rec)
ix = np.argmax(f1s)
fig, ax = plt.subplots(figsize=(10, 10))
# render the baseline curves as background for comparison
background = DATASET_MAPPING[self.hparams.dataset_name]["precision_recall_curve_baseline_img"]
if background:
img = plt.imread(background)
ax.imshow(img, extent=[0, 1, 0, 1])
no_skill = len(label[label == 1]) / len(label)
ax.plot(rec, pre, label="Our proposed model", color="blue")
ax.set_xlabel("Recall")
ax.set_ylabel("Precision")
ax.legend()
self.logger.experiment.log_image(f"{epoch_type}_pre_rec_curve", fig)
self.logger.experiment.log_metric(
f"{epoch_type}_average_precision_score_micro",
average_precision_score(label, logits, average="micro")
)
class BinaryClassifier(BaseClassifier):
"""
A binary classifier that picks out "not-related" sentences
"""
dataset_label_transform = "binary"
num_classes = 1
def __init__(self, *args, **kwargs):
super().__init__(*args, **kwargs)
self.thresholds = {}
def forward(self, *args, **kwargs):
return super().forward(*args, **kwargs).flatten()
@staticmethod
def yhat_to_label(y_hat: Tensor, threshold: float) -> Tensor:
return (y_hat > threshold).long()
def loss_function(self, logits: Tensor, label: Tensor) -> Tensor:
return F.binary_cross_entropy_with_logits(logits, label.float())
def log_metrics(self, epoch_type: str, logits: Tensor, label: Tensor) -> dict:
y_hat = torch.sigmoid(logits)
if epoch_type == "val":
self.__find_thresholds(y_hat, label)
self.__log_output_distribution(epoch_type, y_hat, label)
logs = {
f"{epoch_type}_avg_loss": float(self.loss_function(logits, label)),
f"{epoch_type}_roc_auc": self.__roc_auc_score(label, y_hat),
}
for criteria, threshold in self.thresholds.items():
prefix = f"{epoch_type}_{criteria}"
predicted_label = self.yhat_to_label(y_hat, threshold)
self.log_confusion_matrix(prefix, predicted_label, label)
logs[f"{prefix}_acc"] = accuracy_score(label, predicted_label)
logs[f"{prefix}_pre"] = precision_score(label, predicted_label, average="binary")
logs[f"{prefix}_rec"] = recall_score(label, predicted_label, average="binary")
logs[f"{prefix}_f1"] = f1_score(label, predicted_label, average="binary")
for k, v in logs.items():
self.logger.experiment.log_metric(k, v)
return logs
@staticmethod
def __roc_auc_score(label: Tensor, y_hat: Tensor) -> float:
try:
return roc_auc_score(label, y_hat)
except ValueError:
return 0
def __find_thresholds(self, y_hat: Tensor, label: Tensor):
"""
Find 3 classification thresholds based on 3 criteria:
- The one that yields highest accuracy
- The "best point" in the ROC curve
- The one that yields highest f1
The results are logged and stored in self.threshold
"""
# best accuracy
best_acc = 0
best_acc_threshold = None
for y in y_hat:
y_predicted = self.yhat_to_label(y_hat, threshold=y)
acc = accuracy_score(label, y_predicted)
if best_acc < acc:
best_acc = acc
best_acc_threshold = y
self.thresholds["best_acc"] = best_acc_threshold
# ROC curve
# https://machinelearningmastery.com/threshold-moving-for-imbalanced-classification/
fpr, tpr, thresholds = roc_curve(label, y_hat)
gmeans = tpr * (1 - fpr)
ix = np.argmax(gmeans)
self.thresholds["best_roc"] = thresholds[ix]
fig, ax = plt.subplots(figsize=(16, 12))
ax.plot([0,1], [0,1], linestyle="--", label="No Skill")
ax.plot(fpr, tpr, marker=".", label="Logistic")
ax.scatter(fpr[ix], tpr[ix], marker="o", color="black", label="Best")
ax.set_xlabel("False Positive Rate")
ax.set_ylabel("True Positive Rate")
ax.legend()
self.logger.experiment.log_image("roc_curve", fig)
# precision recall curve
# https://machinelearningmastery.com/roc-curves-and-precision-recall-curves-for-classification-in-python/
pre, rec, thresholds = precision_recall_curve(label, y_hat)
f1s = 2 * pre * rec / (pre + rec)
ix = np.argmax(f1s)
self.thresholds["best_f1"] = thresholds[ix]
fig, ax = plt.subplots(figsize=(16, 12))
no_skill = len(label[label == 1]) / len(label)
ax.plot([0, 1], [no_skill, no_skill], linestyle="--", label="No Skill")
ax.plot(rec, pre, marker=".", label="Logistic")
ax.scatter(rec[ix], pre[ix], marker="o", color="black", label="Best F1")
ax.set_xlabel("Recall")
ax.set_ylabel("Precision")
ax.legend()
self.logger.experiment.log_image("pre_rec_curve", fig)
# log thresholds
for k, v in self.thresholds.items():
self.logger.experiment.log_metric(f"threshold_{k}", v)
def __log_output_distribution(self, epoch_type: str, y_hat: Tensor, label: Tensor):
"""
Log the distribution of the model output and 3 thresholds with log scale and linear scale
"""
y_neg = y_hat[label == 0].numpy()
y_pos = y_hat[label == 1].numpy()
for scale in ["linear", "log"]:
fig, ax = plt.subplots(figsize=(16, 12))
ax.set_yscale(scale)
ax.hist([y_neg, y_pos], stacked=True, bins=50, label=["No relation", "Related"])
ylim = ax.get_ylim()
for k, v in self.thresholds.items():
ax.plot([v, v], ylim, linestyle="--", label=f"{k} threshold")
ax.legend()
self.logger.experiment.log_image(f"{epoch_type}_distribution_{scale}_scale", fig)
class RelationClassifier(MulticlassClassifier):
"""
A classifier that recognizes relations except for "not-related"
"""
dataset_label_transform = "related_only"
def __init__(self, dataset_name, **kwargs):
with open(METADATA_FILE_NAME) as f:
self.num_classes = len(json.load(f)[dataset_name]["label_to_id_related_only"])
super().__init__(dataset_name=dataset_name, **kwargs)
# -
# ## The official scorer
#
# Some datasets comes with official scorers. We will define them in this session.
# + pycharm={"name": "#%%\n"}
class AbstractScorer(ABC):
def __init__(self, experiment_no: int, logger):
self.experiment_no = experiment_no
self.logger = logger
@abstractmethod
def score(self, proposed_answer: dict):
pass
class SemEval2010Task8Scorer(AbstractScorer):
RESULT_FILE = "semeval2010_task8_official_score_{}_{}.txt"
PROPOSED_ANSWER_FILE = "semeval2010_task8_proposed_answer.txt"
SCORER = os.path.join(DATASET_MAPPING["SemEval2010Task8"]["dir"], "SemEval2010_task8_scorer-v1.2/semeval2010_task8_scorer-v1.2.pl")
FORMAT_CHECKER = os.path.join(DATASET_MAPPING["SemEval2010Task8"]["dir"], "SemEval2010_task8_scorer-v1.2/semeval2010_task8_format_checker.pl")
ANSWER_KEY = os.path.join(DATASET_MAPPING["SemEval2010Task8"]["dir"], "SemEval2010_task8_testing_keys/TEST_FILE_KEY.TXT")
def score(self, proposed_answer: dict):
# write test_result to file
with open(METADATA_FILE_NAME) as f:
metadata = json.load(f)
id_to_label = {int(k): v for k, v in metadata[DATASET_NAME]["id_to_label"].items()}
for criteria, answer in proposed_answer.items():
result_file = self.RESULT_FILE.format(self.experiment_no, criteria)
i = 8001
with open(self.PROPOSED_ANSWER_FILE, "w") as f:
for r in answer:
f.write(f"{i}\t{id_to_label[r]}\n")
i += 1
# call the official scorer
os.system(f"perl {self.FORMAT_CHECKER} {self.PROPOSED_ANSWER_FILE}")
os.system(f"perl {self.SCORER} {self.PROPOSED_ANSWER_FILE} {self.ANSWER_KEY} > {result_file}")
# log the official score
with open(result_file) as f:
result = f.read()
print(f">>> Classifier with criteria: {criteria} <<<")
print(result)
print("\n\n")
self.logger.experiment.log_artifact(result_file)
def get_official_scorer(experiment_no: int, logger, dataset_name: str = DATASET_NAME) -> AbstractScorer:
cls = globals().get(dataset_name + "Scorer")
if cls:
return cls(experiment_no, logger)
# -
# ## Claiming back memory & disk space
#
# See [this](https://stackoverflow.com/a/61707643/7342188) and [this](https://stackoverflow.com/a/57860310/7342188)
1 / 0
trainer = classifier = rel_trainer = rel_classifier = bin_trainer = bin_classifier = None
gc.collect()
torch.cuda.empty_cache()
# ## Training standard classifier
# + pycharm={"name": "#%%\n"}
GPUS = 1
MIN_EPOCHS = MAX_EPOCHS = 3
BATCH_SIZE = 16
LEARNING_RATE = 2e-05
LEARNING_RATE_DECAY_SPEED = [1, 1, 0.75, 0.5, 0.25, 0.1, 0.075, 0.05, 0.025, 0.01]
LINEAR_SIZE = 1024
DROPOUT_P = 0.2
ACTIVATION_FUNCTION = "PReLU"
WEIGHT_DECAY = 0.01 # default = 0.01
# + pycharm={"name": "#%%\n"}
logger = NeptuneLogger(
api_key=NEPTUNE_API_TOKEN,
project_name=NEPTUNE_PROJECT_NAME,
close_after_fit=False,
)
try:
for i in range(4):
print(f"--------- EXPERIMENT {i} ---------")
classifier = trainer = None
gc.collect()
torch.cuda.empty_cache()
trainer = LightningTrainer(
gpus=GPUS,
min_epochs=MIN_EPOCHS,
max_epochs=MAX_EPOCHS,
default_root_dir=CHECKPOINT_DIR,
reload_dataloaders_every_epoch=True, # needed as we loop over a file,
deterministic=False,
checkpoint_callback=False,
logger=logger
)
classifier = StandardClassifier(
pretrained_language_model=PRETRAINED_MODEL,
dataset_name=DATASET_NAME,
batch_size=BATCH_SIZE,
learning_rate=LEARNING_RATE,
decay_lr_speed=LEARNING_RATE_DECAY_SPEED,
dropout_p=DROPOUT_P,
activation_function=ACTIVATION_FUNCTION,
weight_decay=WEIGHT_DECAY,
linear_size=LINEAR_SIZE,
)
trainer.fit(classifier)
trainer.test(classifier)
scorer = get_official_scorer(i, logger)
if scorer:
scorer.score({
"standard": classifier.test_proposed_answer,
})
else:
print("No official scorer found")
except Exception as e:
logger.experiment.stop(str(e))
raise e
else:
logger.experiment.stop()
# + [markdown] pycharm={"name": "#%% md\n"}
# ## Training binary classifier
# + pycharm={"name": "#%%\n"}
GPUS = 1
BIN_MIN_EPOCHS = BIN_MAX_EPOCHS = 2
BIN_BATCH_SIZE = 32
BIN_LEARNING_RATE = 2e-05
BIN_LEARNING_RATE_DECAY_SPEED = [1, 1, 0.75, 0.5, 0.5, 0.25, 0.25, 0.1, 0.075, 0.05, 0.025, 0.01]
BIN_LINEAR_SIZE = 256
BIN_DROPOUT_P = 0.2
BIN_ACTIVATION_FUNCTION = "PReLU"
BIN_WEIGHT_DECAY = 0.01 # default = 0.01
# + pycharm={"name": "#%%\n"} _kg_hide-output=true _kg_hide-input=false
bin_logger = NeptuneLogger(
api_key=NEPTUNE_API_TOKEN,
project_name=NEPTUNE_PROJECT_NAME,
close_after_fit=False,
)
try:
for i in range(4):
print(f"--------- EXPERIMENT {i} ---------")
bin_classifier = bin_trainer = None
gc.collect()
torch.cuda.empty_cache()
bin_trainer = LightningTrainer(
gpus=GPUS,
min_epochs=BIN_MIN_EPOCHS,
max_epochs=BIN_MAX_EPOCHS,
default_root_dir=CHECKPOINT_DIR,
reload_dataloaders_every_epoch=True, # needed as we loop over a file,
deterministic=False,
checkpoint_callback=False,
logger=bin_logger,
)
bin_classifier = BinaryClassifier(
pretrained_language_model=PRETRAINED_MODEL,
dataset_name=DATASET_NAME,
batch_size=BIN_BATCH_SIZE,
learning_rate=BIN_LEARNING_RATE,
decay_lr_speed=BIN_LEARNING_RATE_DECAY_SPEED,
linear_size=BIN_LINEAR_SIZE,
dropout_p=BIN_DROPOUT_P,
activation_function=BIN_ACTIVATION_FUNCTION,
weight_decay=BIN_WEIGHT_DECAY,
)
bin_trainer.fit(bin_classifier)
bin_trainer.test(bin_classifier)
except Exception as e:
bin_logger.experiment.stop(str(e))
raise e
else:
bin_logger.experiment.stop()
# -
# ## Train relation classifier
# +
GPUS = 1
REL_MIN_EPOCHS = REL_MAX_EPOCHS = 4
REL_BATCH_SIZE = 32
REL_LEARNING_RATE = 2e-05
REL_LEARNING_RATE_DECAY_SPEED = [1, 1, 0.75, 0.5, 0.25, 0.1, 0.075, 0.05, 0.025, 0.01]
REL_LINEAR_SIZE = 512
REL_DROPOUT_P = 0.1
REL_ACTIVATION_FUNCTION = "PReLU"
REL_WEIGHT_DECAY = 0.01 # default = 0.01
# +
rel_logger = NeptuneLogger(
api_key=NEPTUNE_API_TOKEN,
project_name=NEPTUNE_PROJECT_NAME,
close_after_fit=False,
)
try:
for i in range(4):
print(f"--------- EXPERIMENT {i} ---------")
rel_classifier = rel_trainer = None
gc.collect()
torch.cuda.empty_cache()
rel_trainer = LightningTrainer(
gpus=GPUS,
min_epochs=REL_MIN_EPOCHS,
max_epochs=REL_MAX_EPOCHS,
default_root_dir=CHECKPOINT_DIR,
reload_dataloaders_every_epoch=True, # needed as we loop over a file,
deterministic=False,
checkpoint_callback=False,
logger=rel_logger
)
rel_classifier = RelationClassifier(
pretrained_language_model=PRETRAINED_MODEL,
dataset_name=DATASET_NAME,
batch_size=REL_BATCH_SIZE,
learning_rate=REL_LEARNING_RATE,
decay_lr_speed=REL_LEARNING_RATE_DECAY_SPEED,
dropout_p=REL_DROPOUT_P,
activation_function=REL_ACTIVATION_FUNCTION,
weight_decay=REL_WEIGHT_DECAY,
linear_size=REL_LINEAR_SIZE,
)
rel_trainer.fit(rel_classifier)
rel_trainer.test(rel_classifier)
except Exception as e:
rel_logger.experiment.stop(str(e))
raise e
else:
rel_logger.experiment.stop()
# -
# ## Train 2 classifiers independently then test together
# + pycharm={"name": "#%%\n"}
def test_together(experiment_no: int, logger, b_classifier: BinaryClassifier, r_classifier: RelationClassifier, dataset_name: str = DATASET_NAME,
bin_batch_size = BIN_BATCH_SIZE, batch_size: int = REL_BATCH_SIZE):
b_classifier.freeze()
r_classifier.freeze()
true_answer = []
# run binary classifier
print("Running binary classifier")
dataset = GenericDataset(dataset_name, subset="test", batch_size=bin_batch_size, label_transform="none")
binary_classify_results = { criteria: [] for criteria in b_classifier.thresholds.keys() }
for input_data, true_label in tqdm(dataset.as_batches(), total=len(dataset)):
# append true answers
true_answer += true_label.tolist()
# run bin classifier
logits = b_classifier(**input_data)
y_hat = torch.sigmoid(logits)
for criteria, threshold in b_classifier.thresholds.items():
label = b_classifier.yhat_to_label(y_hat, threshold)
binary_classify_results[criteria] += label.tolist()
# run relation classifier
print("Running relation classifier")
dataset = GenericDataset(dataset_name, subset="test", batch_size=batch_size, label_transform="none")
relation_classify_result = []
for input_data, true_label in tqdm(dataset.as_batches(), total=len(dataset)):
logits = r_classifier(**input_data)
label = r_classifier.logits_to_label(logits) + 1
relation_classify_result += label.tolist()
# combine results
print("Combining results")
proposed_answer = {}
for criteria in b_classifier.thresholds.keys():
results = zip(relation_classify_result, binary_classify_results[criteria])
final_label = [relation_result if bin_result else 0 for relation_result, bin_result in results]
proposed_answer[criteria] = final_label
# log metric
final_metrics = {}
for criteria in b_classifier.thresholds.keys():
pa = proposed_answer[criteria]
final_metrics.update({
f"test_combined_{criteria}_acc": accuracy_score(true_answer, pa),
f"test_combined_{criteria}_pre_micro": precision_score(true_answer, pa, average="micro"),
f"test_combined_{criteria}_rec_micro": recall_score(true_answer, pa, average="micro"),
f"test_combined_{criteria}_f1_micro": f1_score(true_answer, pa, average="micro"),
f"test_combined_{criteria}_pre_macro": precision_score(true_answer, pa, average="macro"),
f"test_combined_{criteria}_rec_macro": recall_score(true_answer, pa, average="macro"),
f"test_combined_{criteria}_f1_macro": f1_score(true_answer, pa, average="macro"),
})
fig = BaseClassifier.plot_confusion_matrix(pa, true_answer)
logger.experiment.log_image(f"test_combined_{criteria}_confusion_matrix", fig)
for k, v in final_metrics.items():
print(f"{k}: {v * 100}")
for k, v in final_metrics.items():
logger.experiment.log_metric(k, v)
# run the offical scorer
scorer = get_official_scorer(experiment_no, logger)
if scorer:
scorer.score(proposed_answer)
else:
print("No official scorer found")
# +
combine_logger = NeptuneLogger(
api_key=NEPTUNE_API_TOKEN,
project_name=NEPTUNE_PROJECT_NAME,
close_after_fit=False,
)
try:
for i in range(4):
print(f"--------- EXPERIMENT {i} ---------")
# clean up
bin_classifier = bin_trainer = rel_classifier = rel_trainer = None
gc.collect()
torch.cuda.empty_cache()
# relation classifier
rel_trainer = LightningTrainer(
gpus=GPUS,
min_epochs=REL_MIN_EPOCHS,
max_epochs=REL_MAX_EPOCHS,
default_root_dir=CHECKPOINT_DIR,
reload_dataloaders_every_epoch=True, # needed as we loop over a file,
deterministic=False,
checkpoint_callback=False,
logger=combine_logger
)
rel_classifier = RelationClassifier(
pretrained_language_model=PRETRAINED_MODEL,
dataset_name=DATASET_NAME,
batch_size=REL_BATCH_SIZE,
learning_rate=REL_LEARNING_RATE,
decay_lr_speed=REL_LEARNING_RATE_DECAY_SPEED,
dropout_p=REL_DROPOUT_P,
activation_function=REL_ACTIVATION_FUNCTION,
weight_decay=REL_WEIGHT_DECAY,
linear_size=REL_LINEAR_SIZE,
)
rel_trainer.fit(rel_classifier)
# binary classifier
bin_trainer = LightningTrainer(
gpus=GPUS,
min_epochs=BIN_MIN_EPOCHS,
max_epochs=BIN_MAX_EPOCHS,
default_root_dir=CHECKPOINT_DIR,
reload_dataloaders_every_epoch=True, # needed as we loop over a file,
deterministic=False,
checkpoint_callback=False,
logger=combine_logger,
)
bin_classifier = BinaryClassifier(
pretrained_language_model=PRETRAINED_MODEL,
dataset_name=DATASET_NAME,
batch_size=BIN_BATCH_SIZE,
learning_rate=BIN_LEARNING_RATE,
decay_lr_speed=BIN_LEARNING_RATE_DECAY_SPEED,
dropout_p=BIN_DROPOUT_P,
activation_function=BIN_ACTIVATION_FUNCTION,
weight_decay=BIN_WEIGHT_DECAY,
linear_size=BIN_LINEAR_SIZE,
)
bin_trainer.fit(bin_classifier)
# test together
test_together(i, combine_logger, bin_classifier, rel_classifier)
except Exception as e:
combine_logger.experiment.stop(str(e))
raise e
else:
combine_logger.experiment.stop()
| MEAN.ipynb |
// ---
// jupyter:
// jupytext:
// text_representation:
// extension: .java
// format_name: light
// format_version: '1.5'
// jupytext_version: 1.14.4
// kernelspec:
// display_name: Java
// language: java
// name: java
// ---
// # Software Development 1
//
// Topics for today will include:
// - Tree
// - Binary Tree
// - Binary Search Tree
// ## Abstract Data Types
// Before we get into Abstract Data Types we need to think about Abstraction first. With abstraction we're hiding unnecessary details from the user. This allows us to build on top of these objects with little or no knowledge of the data type sitting on top of it.
//
// Abstract Data Types in terms of Computer Science take that a little further. They give the blueprint for what's supposed to happen with the functionality and leave it at that. So in the instance of a Stack and a Queue their actions are
//
// Queue:
// - Enqueue: Insert data item into the queue
// - Dequeue: Removes the first item from the queue
// - Front: Accesses and serves the first data item in the queue
//
// Stack:
// - Push: Inserts data onto the stack
// - Pop: Removes a data item from the stack
// - Peek: Accesses a data item on top of the stack without removal
//
// [Abstract Data Types](https://en.wikipedia.org/wiki/Abstract_data_type)
// ## Tree
// We're going to discuss a Tree which is another Abstract Data Type that has Root and Subtrees spawning off the root. Similar to a regular tree the root is the base of the tree and then from there can spawn branches and nodes.
//
// As a Node you can be a parent and/or a child and have other nodes linked to you or be linked to other nodes. There is a set of terminology that comes with this that can be found researching it. It's on the Wiki page linked below.
//
// There has to be one root node and nodes can't have more than one parent.
//
// [Tree](https://en.wikipedia.org/wiki/Tree_(data_structure))
// ## Binary Tree
// Now we have an extension of that in a Binary Tree.
//
// So for this now we can only have at most two children. A left and a right. Outside of that things are relatively the same. This is what we're going for being modular. Just adding a little by a little going on and we're going to do a little more of that in a second with the next type.
//
// [Binary Tree](https://en.wikipedia.org/wiki/Binary_tree)
// ## Binary Search Tree
// Finally for today we have the Binary Search Tree and we'll be creating one of these as our semester project. There are a lot of potential things that can come with this.
//
// Different methods of searching. Different ways to traverse the tree. Different ways to search elements in the tree. Different rules on dealing with duplicates. Different ways to delete elements. We get the point.
//
// When we have generalities like this we have a plethora of ways to do things. We're going to explore some of that.
//
// [Binary Search Tree](https://en.wikipedia.org/wiki/Binary_search_tree)
| JupyterNotebooks/Lessons/Lesson 8.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# +
from deap import base, creator, tools, algorithms
import random
import numpy
import matplotlib.pyplot as plt
import seaborn as sns
# +
# problem constants:
ONE_MAX_LENGTH = 100 # length of bit string to be optimized
# Genetic Algorithm constants:
POPULATION_SIZE = 200
P_CROSSOVER = 0.9 # probability for crossover
P_MUTATION = 0.1 # probability for mutating an individual
MAX_GENERATIONS = 50
HALL_OF_FAME_SIZE = 10
# set the random seed:
RANDOM_SEED = 42
random.seed(RANDOM_SEED)
# +
toolbox = base.Toolbox()
# create an operator that randomly returns 0 or 1:
toolbox.register("zeroOrOne", random.randint, 0, 1)
# define a single objective, maximizing fitness strategy:
creator.create("FitnessMax", base.Fitness, weights=(1.0,))
# create the Individual class based on list:
creator.create("Individual", list, fitness=creator.FitnessMax)
# create the individual operator to fill up an Individual instance:
toolbox.register("individualCreator", tools.initRepeat, creator.Individual, toolbox.zeroOrOne, ONE_MAX_LENGTH)
# create the population operator to generate a list of individuals:
toolbox.register("populationCreator", tools.initRepeat, list, toolbox.individualCreator)
# fitness calculation:
# compute the number of '1's in the individual
def oneMaxFitness(individual):
return sum(individual), # return a tuple
toolbox.register("evaluate", oneMaxFitness)
# genetic operators:mutFlipBit
# Tournament selection with tournament size of 3:
toolbox.register("select", tools.selTournament, tournsize=3)
# Single-point crossover:
toolbox.register("mate", tools.cxOnePoint)
# Flip-bit mutation:
# indpb: Independent probability for each attribute to be flipped
toolbox.register("mutate", tools.mutFlipBit, indpb=1.0/ONE_MAX_LENGTH)
# -
# Genetic Algorithm flow:
def main():
# create initial population (generation 0):
population = toolbox.populationCreator(n=POPULATION_SIZE)
# prepare the statistics object:
stats = tools.Statistics(lambda ind: ind.fitness.values)
stats.register("max", numpy.max)
stats.register("avg", numpy.mean)
# define the hall-of-fame object:
hof = tools.HallOfFame(HALL_OF_FAME_SIZE)
# perform the Genetic Algorithm flow with hof feature added:
population, logbook = algorithms.eaSimple(population, toolbox, cxpb=P_CROSSOVER, mutpb=P_MUTATION,
ngen=MAX_GENERATIONS, stats=stats, halloffame=hof, verbose=True)
# print Hall of Fame info:
print("Hall of Fame Individuals = ", *hof.items, sep="\n")
print("Best Ever Individual = ", hof.items[0])
# extract statistics:
maxFitnessValues, meanFitnessValues = logbook.select("max", "avg")
# plot statistics:
sns.set_style("whitegrid")
plt.plot(maxFitnessValues, color='red')
plt.plot(meanFitnessValues, color='green')
plt.xlabel('Generation')
plt.ylabel('Max / Average Fitness')
plt.title('Max and Average Fitness over Generations')
plt.show()
if __name__ == "__main__":
main()
| Chapter03/basic.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
# environment set up
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
import os
import random
import pandas as pd
from spectra_ml.io_ import load_spectra_metadata
# working folder
# data_dir = "/Users/Srikar/Desktop/Velexi/spectra-ml/data/"
data_dir = os.environ['DATA_DIR']
os.chdir(data_dir)
# -
stddata_path = os.path.join(data_dir,"Srikar-Standardized")
metadata = load_spectra_metadata(os.path.join(stddata_path,"spectra-metadata.csv"))
# +
# stddata_path = os.path.join(data_dir,"Srikar-Standardized")
# metadata = pd.read_csv(os.path.join(stddata_path,"spectra-metadata.csv"), sep="|", dtype={"spectrum_id":str})
# metadata
# -
metadata = metadata[metadata['value_type'] == "reflectance"]
metadata = metadata[~metadata['spectrometer_purity_code'].str.contains("NIC4")]
metadata = metadata[metadata['raw_data_path'].str.contains("ChapterM")] # add in ChapterS Soils and Mixtures later
# metadata.shape
data = pd.read_csv("/Users/Srikar/Desktop/Velexi/spectra-ml/lab-notebook/smunukutla/data.csv", sep=",", dtype=str)
record_nums = data.iloc[0, :].tolist() # keep it as a string to do zfill(5)
# print(record_nums)
spectrum_names = data.iloc[1, :].tolist()
y = data.iloc[2, :].astype(int).tolist()
y = np.reshape(y, (len(y), 1))
num_samples = len(y)
# +
# act = 0
# aln = 0
# chl = 0
# for i in range(metadata.shape[0]):
# data = metadata.iloc[i, :]
# if data[2].find("Actinolite") != -1: # if material name contains actinolite
# record_nums.append(data[0])
# y.append(int(0))
# spectrum_names.append("Actinolite")
# act += 1
# elif data[2].find("Alun") != -1:
# record_nums.append(data[0])
# y.append(int(1))
# spectrum_names.append("Alunite")
# aln += 1
# elif (data[2].find("Chlorit") != -1 or data[2].find("Chlor.") != -1 or data[2].find("Chlor+") != -1 or data[2].find("Chl.") != -1):
# record_nums.append(data[0])
# y.append(int(2))
# spectrum_names.append("Chlorite")
# chl += 1
# y = np.reshape(y, (len(y), 1))
# num_samples = len(record_nums)
# print(num_samples)
# print(len(y))
# print(type(y))
# print(act)
# print(aln)
# print(chl)
# -
spectrum_len = 500
spectra = np.zeros((num_samples,spectrum_len))
wavelengths = np.zeros((1,spectrum_len))
num_neg = 0
for i in range(num_samples):
hasnegative = False
data = pd.read_csv(os.path.join(stddata_path,"{}.csv".format(record_nums[i])))
if i == 0:
wavelengths[i,:] = data.iloc[:, 0].to_numpy()
spectra[i,:] = data.iloc[:, 1].to_numpy()
for j in range(spectrum_len):
if spectra[i,j] < 0:
hasnegative = True
spectra[i,j] = 0
if hasnegative:
print(record_nums[i])
num_neg += 1
# print(num_neg)
# +
# type(spectra)
# +
# --- plot the classes
# plot each class in a separate plot
# plot spectra names in legend
# plot minerals and mixtures w diff line widths
mineral_names = ["Actinolite", "Alunite", "Chlorite"]
# variables
num0 = 0 #number of samples of class 0
num1 = 0
num2 = 0
mineral_linewidth = 1 # linewidth = 1 is default
mixture_linewidth = 3
# count the number of each class to make spectra0, spectra1, spectra2 databases
for i in range(num_samples):
if y[i,0]== 0:
num0 += 1
elif y[i,0]== 1:
num1 += 1
elif y[i,0]== 2:
num2 += 1
# make class-specific databases spectra0, ...1, ...2
spectra0 = np.zeros((num0,spectrum_len))
spectra1 = np.zeros((num1,spectrum_len))
spectra2 = np.zeros((num2,spectrum_len))
labels0 = ["" for x in range(num0)]
labels1 = ["" for x in range(num1)]
labels2 = ["" for x in range(num2)]
linewidth0 = np.zeros(num0)
linewidth1 = np.zeros(num1)
linewidth2 = np.zeros(num2)
# make counters for each database to place spectra
i0 = 0
i1 = 0
i2 = 0
# set linewidth for the spectrum
# populate class-specific databases spectra0, ...1, ...2
for i in range(num_samples):
# set linewidth
#testcode
#print(spectrum_categories)
#print(spectrum_categories[i])
# if spectrum_categories[i] == is_a_mineral:
# linewidth = mineral_linewidth
# #testcode
# #print('min')
# else:
# linewidth = mixture_linewidth
linewidth = 2
#testcode
#print('mix')
# populate matrices for making each class plot
if y[i,0]== 0:
spectra0[i0,:] = spectra[i,:]
labels0[i0] = spectrum_names[i]
linewidth0[i0] = linewidth
i0 +=1
elif y[i,0]== 1:
spectra1[i1,:] = spectra[i,:]
labels1[i1] = spectrum_names[i]
linewidth1[i1] = linewidth
i1 +=1
else:
spectra2[i2,:] = spectra[i,:]
labels2[i2] = spectrum_names[i]
linewidth2[i2] = linewidth
i2 +=1
j = 0
# plot each class-specific database separately
# remove linewidth for all mixtures/minerals to be standard
for i in range(i0):
# plt.plot(range(1, spectrum_len+1), spectra0[i,:], label = labels0[i], linewidth = linewidth0[i])
fig = plt.figure()
plt.plot(wavelengths[0,:], spectra0[i,:], label = labels0[i], linewidth = linewidth0[i], color='k')
plt.xticks([])
plt.yticks([])
# fig.patch.set_visible(False)
# plt.show()
path = os.path.join(data_dir, "plots", record_nums[j] + "-" + mineral_names[0] + ".png")
j += 1
ax = fig.axes
ax[0].axis('off')
fig.savefig(path, format = "PNG")
plt.close(fig)
# plt.legend(bbox_to_anchor=(1.1, 1.05))
# plt.show()
for i in range(i1):
# plt.plot(range(1, spectrum_len+1), spectra1[i,:], label = labels1[i], linewidth = linewidth1[i])
fig = plt.figure()
plt.plot(wavelengths[0,:], spectra1[i,:], label = labels1[i], linewidth = linewidth1[i], color='k')
plt.xticks([])
plt.yticks([])
# fig.patch.set_visible(False)
# plt.show()
path = os.path.join(data_dir, "plots", record_nums[j] + "-" + mineral_names[1] + ".png")
j += 1
ax = fig.axes
ax[0].axis('off')
fig.savefig(path, format = "PNG")
plt.close(fig)
# plt.legend(bbox_to_anchor=(1.1, 1.05))
# plt.show()
for i in range(i2):
# plt.plot(range(1, spectrum_len+1), spectra2[i,:], label = labels2[i], linewidth = linewidth2[i])
fig = plt.figure()
plt.plot(wavelengths[0,:], spectra2[i,:], label = labels2[i], linewidth = linewidth2[i], color='k')
plt.xticks([])
plt.yticks([])
# fig.patch.set_visible(False)
# plt.show()
path = os.path.join(data_dir, "plots", record_nums[j] + "-" + mineral_names[2] + ".png")
j += 1
ax = fig.axes
ax[0].axis('off')
fig.savefig(path, format = "PNG")
plt.close(fig)
# plt.legend(bbox_to_anchor=(1.1, 1.05))
# plt.show()
# +
# num_neg = 0
# for file in os.listdir(stddata_path):
# data = pd.read_csv(os.path.join(stddata_path,file))
# if data.shape[1] == 2:
# arr = data.iloc[:, 1].to_numpy()
# if np.isnan(arr[0]) or np.isnan(arr[len(arr)-1]):
# print(file)
# num_neg += 1
# continue
# for j in range(len(arr)):
# if np.isnan(arr[j]):
# print(file)
# num_neg += 1
# break
# print(num_neg)
# +
# metadata = metadata[metadata['value_type'] == "reflectance"]
# metadata = metadata[~metadata['spectrometer_purity_code'].str.contains("NIC4")]
# metadata = metadata[metadata['raw_data_path'].str.contains("ChapterM")]
# metadata.shape
# record_nums = []
# mineral_names = []
# for i in range(metadata.shape[0]):
# data = metadata.iloc[i, :]
# record_nums.append(data[0])
# mineral_names.append(data[2])
# +
# num_neg = 0
# print(len(record_nums))
# for i in range(len(record_nums)):
# data = pd.read_csv(os.path.join(stddata_path,"{}.csv".format(record_nums[i])))
# if data.shape[1] == 2:
# arr = data.iloc[:, 1].to_numpy()
# if np.isnan(arr[0]) or np.isnan(arr[len(arr)-1]):
# print(record_nums[i])
# print(mineral_names[i])
# num_neg += 1
# continue
# for j in range(len(arr)):
# if np.isnan(arr[j]):
# print(file)
# num_neg += 1
# break
# print(num_neg)
# +
# os.listdir(stddata_path)
# +
# data = pd.read_csv(os.path.join(stddata_path,"211.csv"))
# arr = data.iloc[:, 1].to_numpy()
# for j in range(spectrum_len):
# print(type(arr[j]))
# -
| lab-notebook/smunukutla/2019-08-03-SAM - 2D Image Creation.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + tags=["context"] deletable=false run_control={"frozen": true} dc={"key": "3"} editable=false
# ## 1. TV, halftime shows, and the Big Game
# <p>Whether or not you like football, the Super Bowl is a spectacle. There's a little something for everyone at your Super Bowl party. Drama in the form of blowouts, comebacks, and controversy for the sports fan. There are the ridiculously expensive ads, some hilarious, others gut-wrenching, thought-provoking, and weird. The half-time shows with the biggest musicians in the world, sometimes <a href="https://youtu.be/ZD1QrIe--_Y?t=14">riding giant mechanical tigers</a> or <a href="https://youtu.be/mjrdywp5nyE?t=62">leaping from the roof of the stadium</a>. It's a show, baby. And in this notebook, we're going to find out how some of the elements of this show interact with each other. After exploring and cleaning our data a little, we're going to answer questions like:</p>
# <ul>
# <li>What are the most extreme game outcomes?</li>
# <li>How does the game affect television viewership?</li>
# <li>How have viewership, TV ratings, and ad cost evolved over time?</li>
# <li>Who are the most prolific musicians in terms of halftime show performances?</li>
# </ul>
# <p><img src="https://assets.datacamp.com/production/project_684/img/left_shark.jpg" alt="Left Shark Steals The Show">
# <em><a href="https://www.flickr.com/photos/huntleypaton/16464994135/in/photostream/">Left Shark Steals The Show</a>. <NAME> performing at halftime of Super Bowl XLIX. Photo by <NAME>. Attribution-ShareAlike 2.0 Generic (CC BY-SA 2.0).</em></p>
# <p>The dataset we'll use was <a href="https://en.wikipedia.org/wiki/Web_scraping">scraped</a> and polished from Wikipedia. It is made up of three CSV files, one with <a href="https://en.wikipedia.org/wiki/List_of_Super_Bowl_champions">game data</a>, one with <a href="https://en.wikipedia.org/wiki/Super_Bowl_television_ratings">TV data</a>, and one with <a href="https://en.wikipedia.org/wiki/List_of_Super_Bowl_halftime_shows">halftime musician data</a> for all 52 Super Bowls through 2018. Let's take a look, using <code>display()</code> instead of <code>print()</code> since its output is much prettier in Jupyter Notebooks.</p>
# + tags=["sample_code"] dc={"key": "3"}
# Import pandas
import pandas as pd
# Load the CSV data into DataFrames
super_bowls = pd.read_csv('datasets/super_bowls.csv')
tv = pd.read_csv('datasets/tv.csv')
halftime_musicians = pd.read_csv('datasets/halftime_musicians.csv')
# Display the first five rows of each DataFrame
display(super_bowls.head())
display(tv.head())
display(halftime_musicians.head())
# + tags=["context"] deletable=false run_control={"frozen": true} dc={"key": "10"} editable=false
# ## 2. Taking note of dataset issues
# <p>For the Super Bowl game data, we can see the dataset appears whole except for missing values in the backup quarterback columns (<code>qb_winner_2</code> and <code>qb_loser_2</code>), which make sense given most starting QBs in the Super Bowl (<code>qb_winner_1</code> and <code>qb_loser_1</code>) play the entire game.</p>
# <p>From the visual inspection of TV and halftime musicians data, there is only one missing value displayed, but I've got a hunch there are more. The Super Bowl goes all the way back to 1967, and the more granular columns (e.g. the number of songs for halftime musicians) probably weren't tracked reliably over time. Wikipedia is great but not perfect.</p>
# <p>An inspection of the <code>.info()</code> output for <code>tv</code> and <code>halftime_musicians</code> shows us that there are multiple columns with null values.</p>
# + tags=["sample_code"] dc={"key": "10"}
# Summary of the TV data to inspect
tv.info()
print('\n')
# Summary of the halftime musician data to inspect
halftime_musicians.info()
# + tags=["context"] deletable=false run_control={"frozen": true} dc={"key": "17"} editable=false
# ## 3. Combined points distribution
# <p>For the TV data, the following columns have missing values and a lot of them:</p>
# <ul>
# <li><code>total_us_viewers</code> (amount of U.S. viewers who watched at least some part of the broadcast)</li>
# <li><code>rating_18_49</code> (average % of U.S. adults 18-49 who live in a household with a TV that were watching for the entire broadcast)</li>
# <li><code>share_18_49</code> (average % of U.S. adults 18-49 who live in a household with a TV <em>in use</em> that were watching for the entire broadcast)</li>
# </ul>
# <p>For the halftime musician data, there are missing numbers of songs performed (<code>num_songs</code>) for about a third of the performances.</p>
# <p>There are a lot of potential reasons for these missing values. Was the data ever tracked? Was it lost in history? Is the research effort to make this data whole worth it? Maybe. Watching every Super Bowl halftime show to get song counts would be pretty fun. But we don't have the time to do that kind of stuff now! Let's take note of where the dataset isn't perfect and start uncovering some insights.</p>
# <p>Let's start by looking at combined points for each Super Bowl by visualizing the distribution. Let's also pinpoint the Super Bowls with the highest and lowest scores.</p>
# + tags=["sample_code"] dc={"key": "17"}
# Import matplotlib and set plotting style
from matplotlib import pyplot as plt
# %matplotlib inline
plt.style.use('seaborn')
# Plot a histogram of combined points
# ... YOUR CODE FOR TASK 3 ...
plt.xlabel('Combined Points')
plt.ylabel('Number of Super Bowls')
plt.show()
# Display the Super Bowls with the highest and lowest combined scores
display(super_bowls[super_bowls['combined_pts'] > 70])
display(super_bowls[super_bowls['combined_pts'] < 25])
# + tags=["context"] deletable=false run_control={"frozen": true} dc={"key": "24"} editable=false
# ## 4. Point difference distribution
# <p>Most combined scores are around 40-50 points, with the extremes being roughly equal distance away in opposite directions. Going up to the highest combined scores at 74 and 75, we find two games featuring dominant quarterback performances. One even happened recently in 2018's Super Bowl LII where <NAME> Patriots lost to <NAME>' underdog Eagles 41-33 for a combined score of 74.</p>
# <p>Going down to the lowest combined scores, we have Super Bowl III and VII, which featured tough defenses that dominated. We also have Super Bowl IX in New Orleans in 1975, whose 16-6 score can be attributed to inclement weather. The field was slick from overnight rain, and it was cold at 46 °F (8 °C), making it hard for the Steelers and Vikings to do much offensively. This was the second-coldest Super Bowl ever and the last to be played in inclement weather for over 30 years. The NFL realized people like points, I guess.</p>
# <p><em>UPDATE: In Super Bowl LIII in 2019, the Patriots and Rams broke the record for the lowest-scoring Super Bowl with a combined score of 16 points (13-3 for the Patriots).</em></p>
# <p>Let's take a look at point <em>difference</em> now.</p>
# + tags=["sample_code"] dc={"key": "24"}
# Plot a histogram of point differences
plt.hist(super_bowls.difference_pts)
plt.xlabel('Point Difference')
plt.ylabel('Number of Super Bowls')
plt.show()
# Display the closest game(s) and biggest blowouts
display(super_bowls[ super_bowls.difference_pts == 1])
display(super_bowls[ super_bowls.difference_pts > 35])
# + tags=["context"] deletable=false run_control={"frozen": true} dc={"key": "31"} editable=false
# ## 5. Do blowouts translate to lost viewers?
# <p>The vast majority of Super Bowls are close games. Makes sense. Both teams are likely to be deserving if they've made it this far. The closest game ever was when the Buffalo Bills lost to the New York Giants by 1 point in 1991, which was best remembered for Scott Norwood's last-second missed field goal attempt that went <em><a href="https://www.youtube.com/watch?v=RPFZCGgjDSg">wide right</a></em>, kicking off four Bills Super Bowl losses in a row. Poor Scott. The biggest point discrepancy ever was 45 points (!) where Hall of Famer <NAME>'s led the San Francisco 49ers to victory in 1990, one year before the closest game ever.</p>
# <p>I remember watching the Seahawks crush the Broncos by 35 points (43-8) in 2014, which was a boring experience in my opinion. The game was never really close. I'm pretty sure we changed the channel at the end of the third quarter. Let's combine our game data and TV to see if this is a universal phenomenon. Do large point differences translate to lost viewers? We can plot <a href="https://en.wikipedia.org/wiki/Nielsen_ratings">household share</a> <em>(average percentage of U.S. households with a TV in use that were watching for the entire broadcast)</em> vs. point difference to find out.</p>
# + tags=["sample_code"] dc={"key": "31"}
# Join game and TV data, filtering out SB I because it was split over two networks
games_tv = pd.merge(tv[tv['super_bowl'] > 1], super_bowls, on='super_bowl')
# Import seaborn
import seaborn as sns
# ... YOUR CODE FOR TASK 5 ...
# Create a scatter plot with a linear regression model fit
sns.regplot(x=games_tv['difference_pts'], y=games_tv['share_household'], data=games_tv)
# + tags=["context"] deletable=false run_control={"frozen": true} dc={"key": "38"} editable=false
# ## 6. Viewership and the ad industry over time
# <p>The downward sloping regression line and the 95% confidence interval for that regression <em>suggest</em> that bailing on the game if it is a blowout is common. Though it matches our intuition, we must take it with a grain of salt because the linear relationship in the data is weak due to our small sample size of 52 games.</p>
# <p>Regardless of the score though, I bet most people stick it out for the halftime show, which is good news for the TV networks and advertisers. A 30-second spot costs a pretty <a href="https://www.businessinsider.com/super-bowl-commercials-cost-more-than-eagles-quarterback-earns-2018-1">\$5 million</a> now, but has it always been that way? And how have number of viewers and household ratings trended alongside ad cost? We can find out using line plots that share a "Super Bowl" x-axis.</p>
# + tags=["sample_code"] dc={"key": "38"}
# Create a figure with 3x1 subplot and activate the top subplot
plt.subplot(3, 1, 1)
plt.plot(tv.super_bowl, tv.avg_us_viewers, color='#648FFF')
plt.title('Average Number of US Viewers')
# Activate the middle subplot
plt.subplot(3, 1, 2)
plt.plot(tv.super_bowl, tv.rating_household, color='#DC267f')
plt.title('Household Rating')
# Activate the bottom subplot
plt.subplot(3, 1, 3)
plt.plot(tv.super_bowl, tv.ad_cost, color='#FFB000')
plt.title('Ad Cost')
plt.xlabel('SUPER BOWL')
# Improve the spacing between subplots
plt.tight_layout()
# + tags=["context"] deletable=false run_control={"frozen": true} dc={"key": "45"} editable=false
# ## 7. Halftime shows weren't always this great
# <p>We can see viewers increased before ad costs did. Maybe the networks weren't very data savvy and were slow to react? Makes sense since DataCamp didn't exist back then.</p>
# <p>Another hypothesis: maybe halftime shows weren't that good in the earlier years? The modern spectacle of the Super Bowl has a lot to do with the cultural prestige of big halftime acts. I went down a YouTube rabbit hole and it turns out the old ones weren't up to today's standards. Some offenders:</p>
# <ul>
# <li><a href="https://youtu.be/6wMXHxWO4ns?t=263">Super Bowl XXVI</a> in 1992: A Frosty The Snowman rap performed by children.</li>
# <li><a href="https://www.youtube.com/watch?v=PKQTL1PYSag">Super Bowl XXIII</a> in 1989: An Elvis impersonator that did magic tricks and didn't even sing one Elvis song.</li>
# <li><a href="https://youtu.be/oSXMNbK2e98?t=436">Super Bowl XXI</a> in 1987: Tap dancing ponies. (Okay, that's pretty awesome actually.)</li>
# </ul>
# <p>It turns out <NAME>'s Super Bowl XXVII performance, one of the most watched events in American TV history, was when the NFL realized the value of Super Bowl airtime and decided they needed to sign big name acts from then on out. The halftime shows before MJ indeed weren't that impressive, which we can see by filtering our <code>halftime_musician</code> data.</p>
# + tags=["sample_code"] dc={"key": "45"}
# Display all halftime musicians for Super Bowls up to and including Super Bowl XXVII
# ... YOUR CODE FOR TASK 7 ...
halftime_musicians.loc[halftime_musicians.super_bowl < 28, ['musician'] ]
# + tags=["context"] deletable=false run_control={"frozen": true} dc={"key": "52"} editable=false
# ## 8. Who has the most halftime show appearances?
# <p>Lots of marching bands. American jazz clarinetist <NAME>. Miss Texas 1973 playing a violin. Nothing against those performers, they're just simply not <a href="https://www.youtube.com/watch?v=suIg9kTGBVI">Beyoncé</a>. To be fair, no one is.</p>
# <p>Let's see all of the musicians that have done more than one halftime show, including their performance counts.</p>
# + tags=["sample_code"] dc={"key": "52"}
# Count halftime show appearances for each musician and sort them from most to least
halftime_appearances = halftime_musicians.groupby('musician').count()['super_bowl'].reset_index()
halftime_appearances = halftime_appearances.sort_values('super_bowl', ascending=False)
# Display musicians with more than one halftime show appearance
# ... YOUR CODE FOR TASK 8 ...
halftime_appearances.loc[ halftime_appearances.super_bowl > 1, ['musician'] ]
# + tags=["context"] deletable=false run_control={"frozen": true} dc={"key": "59"} editable=false
# ## 9. Who performed the most songs in a halftime show?
# <p>The world famous <a href="https://www.youtube.com/watch?v=RL_3oqpHiDg">Grambling State University Tiger Marching Band</a> takes the crown with six appearances. Beyoncé, <NAME>, Nelly, and <NAME> are the only post-Y2K musicians with multiple appearances (two each).</p>
# <p>From our previous inspections, the <code>num_songs</code> column has lots of missing values:</p>
# <ul>
# <li>A lot of the marching bands don't have <code>num_songs</code> entries.</li>
# <li>For non-marching bands, missing data starts occurring at Super Bowl XX.</li>
# </ul>
# <p>Let's filter out marching bands by filtering out musicians with the word "Marching" in them and the word "Spirit" (a common naming convention for marching bands is "Spirit of [something]"). Then we'll filter for Super Bowls after Super Bowl XX to address the missing data issue, <em>then</em> let's see who has the most number of songs.</p>
# + tags=["sample_code"] dc={"key": "59"}
# Filter out most marching bands
no_bands = halftime_musicians[~halftime_musicians.musician.str.contains('Marching')]
no_bands = no_bands[~no_bands.musician.str.contains('Spirit')]
# Plot a histogram of number of songs per performance
most_songs = int(max(no_bands['num_songs'].values))
plt.hist(no_bands.num_songs.dropna(), bins=most_songs)
plt.xlabel('Number of Songs Per Halftime Show')
plt.ylabel('Number of Musicians')
plt.show()
# Sort the non-band musicians by number of songs per appearance...
no_bands = no_bands.sort_values('num_songs', ascending=False)
# ...and display the top 15
display(no_bands.head(15))
# + tags=["context"] deletable=false run_control={"frozen": true} dc={"key": "66"} editable=false
# ## 10. Conclusion
# <p>So most non-band musicians do 1-3 songs per halftime show. It's important to note that the duration of the halftime show is fixed (roughly 12 minutes) so songs per performance is more a measure of how many hit songs you have. JT went off in 2018, wow. 11 songs! <NAME> comes in second with 10 in her medley in 1996.</p>
# <p>In this notebook, we loaded, cleaned, then explored Super Bowl game, television, and halftime show data. We visualized the distributions of combined points, point differences, and halftime show performances using histograms. We used line plots to see how ad cost increases lagged behind viewership increases. And we discovered that blowouts do appear to lead to a drop in viewers.</p>
# <p>This year's Big Game will be here before you know it. Who do you think will win Super Bowl LIII?</p>
# <p><em>UPDATE: <a href="https://en.wikipedia.org/wiki/Super_Bowl_LIII">Spoiler alert</a>.</em></p>
# + tags=["sample_code"] dc={"key": "66"}
# 2018-2019 conference champions
patriots = 'New England Patriots'
rams = 'Los Angeles Rams'
# Who will win Super Bowl LIII?
super_bowl_LIII_winner = patriots
print('The winner of Super Bowl LIII will be the', super_bowl_LIII_winner)
| src/notebook.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: conda_python3
# language: python
# name: conda_python3
# ---
# # Determine which files we've excluded
#
# We have 480 panos/intersections that we're working with
#
# We labeled 559 panos/intersections
#
# some may have been removed as "highways"
#
# Which images are fully null that we're missing?
# +
import pandas as pd
import numpy as np
import os
import boto3
import s3fs # for reading from S3FileSystem
import json # for working with JSON files
import mini_utils
from datetime import datetime
import matplotlib.pyplot as plt
pd.set_option('max_colwidth', -1)
# -
# # Get All Labeled Filenames
df_filenames = pd.read_csv('labeled-filenames.csv')
print(df_filenames.shape)
df_filenames.head()
# # Get List of Files in S3
# +
fs = s3fs.S3FileSystem()
s3_image_bucket = 's3://streetview-w210'
sample_images_dir = os.path.join(s3_image_bucket, 'gsv')
# See what is in the folder
s3_images_list = fs.ls(sample_images_dir)
# Includes json info
len(s3_images_list)
# -
s3_image_filenames = [os.path.basename(filename) for filename in s3_images_list if 'jpg' in filename]
s3_image_filenames[0:10]
labeled_filenames = set(df_filenames['filenames'])
print(len(labeled_filenames))
new_set = labeled_filenames & set(s3_image_filenames)
len(new_set)
df_file_filter = df_filenames.loc[df_filenames['filenames'].isin(list(new_set))]
print(df_file_filter.shape)
df_file_filter.head()
img_id_list = list(df_file_filter['img_id'].unique())
heading_list = list(df_file_filter['heading'].unique())
crop_num_list = ['A', 'B', 'C', 'D', 'E', 'F']
df_mesh = pd.DataFrame(np.array(np.meshgrid(img_id_list, heading_list, crop_num_list, )).T.reshape(-1,3))
df_mesh.columns = ['img_id', 'heading', 'crop_num']
df_mesh['img_id'] = pd.to_numeric(df_mesh['img_id'], downcast = 'integer')
df_mesh['heading'] = pd.to_numeric(df_mesh['heading'], downcast = 'integer')
df_mesh['jpg_name'] = df_mesh['img_id'].astype(str) + '_' + df_mesh['heading'].astype(str) + '_' + df_mesh['crop_num'].astype(str) + '.jpg'
print(df_mesh.shape)
df_mesh.head()
# # Load in Dataset of Labels in Images
df_crops = pd.read_csv('imgid_groundtruth_trainvaltest.csv')
print(df_crops.shape)
df_crops['imgid_heading'] = df_crops['img_id'].astype(str) + '_' + df_crops['heading'].astype(str)
df_crops.head()
df_crops['img_id'].unique().shape
df_crops['train/val/test'].value_counts(dropna = False)
df_crop_nulls = df_crops.loc[df_crops['train/val/test'].isna()]
no_split = df_crop_nulls['img_id'].unique()
print(len(no_split))
for img_id in no_split:
print(img_id)
# # Get Full List
merge_cols = ['jpg_name', 'img_id', 'heading', 'crop_num']
df_combine = df_mesh.merge(df_crops, how = 'left', left_on = merge_cols, right_on = merge_cols)
print(df_combine.shape)
df_combine.head()
df_combine['ground_truth'].value_counts(dropna = False)
df_combine['ground_truth'].value_counts().sum()
df_combine['train/val/test'].value_counts(dropna = False)
df_combine['train/val/test'].value_counts().sum()
# # Fill in Split details for fully Null Images
df_nulls = df_combine.loc[df_combine['ground_truth'].isna()]
no_split = df_nulls['img_id'].unique()
print(len(no_split))
for img_id in no_split:
print(img_id)
df_tvt_null = pd.read_csv('only_null_imgidheading_split.csv', sep = '\t')
print(df_tvt_null.shape)
df_tvt_null.head()
df_combine_filling = df_combine.merge(df_tvt_null, how = 'outer', left_on = 'img_id', right_on = 'img_id')
df_combine_filling.head()
df_combine_filling['fillnull_train/val/test'].value_counts(dropna = False)
df_combine_filling.shape
df_combine_filling['train/val/test'] = df_combine_filling['train/val/test'].fillna(df_combine_filling['fillnull_train/val/test'])
df_combine_filling['present_ramp'] = df_combine_filling['present_ramp'].fillna(0)
df_combine_filling['missing_ramp'] = df_combine_filling['missing_ramp'].fillna(0)
df_combine_filling['total_count'] = df_combine_filling['total_count'].fillna(0)
df_combine_filling['includes_both'] = df_combine_filling['includes_both'].fillna(False)
df_combine_filling['ground_truth'] = df_combine_filling['ground_truth'].fillna('1_null')
df_combine_filling['train/val/test'].value_counts(dropna = False)
df_combine_filling.head()
df_combine_filling['ground_truth'].value_counts()
# +
final_columns = ['img_id', 'heading', 'crop_num', 'jpg_name', 'present_ramp', 'missing_ramp', 'total_count', 'includes_both', 'ground_truth', 'train/val/test']
df_final = df_combine_filling[final_columns].dropna(subset = ['train/val/test']).copy()
df_final['present_ramp'] = pd.to_numeric(df_final['present_ramp'], downcast = 'integer')
df_final['missing_ramp'] = pd.to_numeric(df_final['missing_ramp'], downcast = 'integer')
df_final['total_count'] = pd.to_numeric(df_final['total_count'], downcast = 'integer')
print(df_final.shape)
df_final.head()
# -
df_final.groupby(['train/val/test', 'ground_truth'])['jpg_name'].count()
df_final['img_id'].unique().shape
df_final.to_csv('20200413-groundtruth-images-split.csv', index = False)
# # Find the img_id/heading ground truth
df_final['imgid_heading'] = df_final['img_id'].astype(str) + '_' + df_final['heading'].astype(str)
df_final.head()
df_group_final = df_final.groupby(['train/val/test','imgid_heading', 'ground_truth'])['jpg_name'].count()
df_group_final = df_group_final.reset_index()
print(df_group_final.shape)
df_group_final.head()
df_group_final['imgid_heading'].unique().shape
df_group_final['ground_truth'].value_counts()
df_group_final['3_labels'] = np.where(df_group_final['ground_truth'].str.contains('present'), 'A_Present', 'tbd')
df_group_final['3_labels'] = np.where(df_group_final['ground_truth'].str.contains('missing'), 'B_Missing', df_group_final['3_labels'])
df_group_final['3_labels'] = np.where(df_group_final['ground_truth'].str.contains('1_null'), 'C_Null', df_group_final['3_labels'])
df_group_final['3_labels'].value_counts()
df_group_final = df_group_final.sort_values(['imgid_heading', '3_labels'])
df_group_final.head(10)
df_group_top_vote = df_group_final.drop_duplicates(subset = 'imgid_heading', keep = 'first')
print(df_group_top_vote.shape)
df_group_top_vote['3_labels'].value_counts()
df_group_top_vote.groupby(['train/val/test', '3_labels'])['imgid_heading'].count()
df_group_final.to_csv('20200413-ImgIDHeading-AllVotes.csv', index = False)
df_group_top_vote.to_csv('20200413-ImgIDHeading-GroundTruth.csv', index = False)
| mini-crops/2020-04-13-GetFullLabeled.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
arr = [ 2, 3, 5, 20, 60 ]
x = 20
def binarySearch(arr, l, r, x):
while l <= r:
mid = l + (r - l) // 2;
# Check if x is present at mid
if arr[mid] == x:
return mid
# If x is greater, ignore left half
elif arr[mid] < x:
l = mid + 1
# If x is smaller, ignore right half
else:
r = mid - 1
return -1
result = binarySearch(arr, 0, len(arr)-1, x)
if result != -1:
print ("Element is present at index % d" % result)
else:
print ("Element is not present in array")
| BinarySearch.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import numpy as np
import tqdm
from os import path
import random
import cv2
import math
from matplotlib import pyplot as plt
import pathlib
from tqdm import tqdm
import tensorflow as tf
from tensorflow.keras import *
import tensorflow.keras.layers as L
import tensorflow.keras.regularizers as R
import tensorflow.keras.backend as B
# +
import tensorflow as tf
print (tf.__version__)
if tf.test.gpu_device_name():
print('Default GPU Device: {}'.format(tf.test.gpu_device_name()))
else:
print("Please install GPU version of TF")
# -
work_dir = "E:/Github/CoinsRecognition"
# +
train_dir = path.join(work_dir, "Datasets/train")
test_dir = path.join(work_dir, "Datasets/test")
train_img_paths = [str(x).replace("\\", "/") for x in tqdm(pathlib.Path(train_dir).rglob('*.*'))]
test_img_paths = [str(x).replace("\\", "/") for x in tqdm(pathlib.Path(test_dir).rglob('*.*'))]
print(len(train_img_paths), len(test_img_paths))
# -
batch_size = 64
image_shape = (224, 224, 3)
num_classes = 211
epochs = 100
# +
class ImagesExtractor():
def __init__(self, **kwargs):
super().__init__(**kwargs)
pass
@tf.function
def _img_read_tf(self, file_path):
img = tf.io.read_file(file_path)
img = tf.image.decode_jpeg(img)
label = tf.one_hot(tf.strings.to_number(tf.strings.split(file_path, '/')[-2], tf.int32) - 1, num_classes)
return img, label
@tf.function
def _img_resize_tf(self, img, label):
img = tf.cast(img, tf.float32) / 255.0
img = tf.image.resize(img, image_shape[:2])
return img, label
def img_batch(self, file_paths, shuffle=0):
AUTO = tf.data.experimental.AUTOTUNE
res = tf.data.Dataset.from_tensor_slices(file_paths)
if shuffle > 0:
res = res.shuffle(shuffle)
res = res.map(self._img_read_tf, num_parallel_calls=AUTO)
res = res.map(lambda x, y: self._img_resize_tf(x, y), num_parallel_calls=AUTO)
res = res.batch(batch_size) #.prefetch(AUTO)
return res
def img_iter(self, file_paths):
AUTO = tf.data.experimental.AUTOTUNE
res = tf.data.Dataset.from_tensor_slices(file_paths)
res = res.map(self._img_read_tf, num_parallel_calls=AUTO)
res = res.batch(1) #.prefetch(AUTO)
return res
images_extractor = ImagesExtractor()
# +
train_img_batch_generator = images_extractor.img_batch(train_img_paths, 2048)
#train_img_iter_generator = images_extractor.img_iter(train_img_paths)
test_img_batch_generator = images_extractor.img_batch(test_img_paths)
#test_img_iter_generator = images_extractor.img_iter(test_img_paths)
sample_img_paths = [path.join(train_dir, '37/2020-10-04_122500.png').replace("\\", "/"),
path.join(train_dir, '37/2020-10-04_122447.png').replace("\\", "/"),
path.join(train_dir, '36/2020-10-04_121511.png').replace("\\", "/"),
path.join(train_dir, '36/2020-10-04_121528.png').replace("\\", "/")]
sample_img_batch_generator = images_extractor.img_batch(sample_img_paths)
#sample_img_iter_generator = images_extractor.img_iter(sample_img_paths)
# +
from functools import reduce
@tf.function
def aug_grid_mask(batch_tuple, d1=128, d2=224, ratio=0.5):
img_batch, label_batch = batch_tuple
batch_size, img_h, img_w = tf.shape(img_batch)[0], img_batch.shape[1], img_batch.shape[2]
d = tf.random.uniform(shape=[batch_size, 1], minval=d1, maxval=d2, dtype=tf.int32) # (Batch Size, 1)
d_f32 = tf.cast(d, tf.float32) # (Batch Size, 1)
y_range = tf.reshape(tf.range(img_h), (1, -1)) + tf.cast(tf.random.uniform(shape=[batch_size, 1], dtype=tf.float32) * d_f32 - 1, tf.int32)# (Batch Size, IMG_H)
x_range = tf.reshape(tf.range(img_w), (1, -1)) + tf.cast(tf.random.uniform(shape=[batch_size, 1], dtype=tf.float32) * d_f32 - 1, tf.int32) # (Batch Size, IMG_W)
y_range = tf.expand_dims(tf.cast(y_range % d, tf.float32) / d_f32 >= ratio, axis=2) # (Batch Size, IMG_H, 1)
x_range = tf.expand_dims(tf.cast(x_range % d, tf.float32) / d_f32 >= ratio, axis=1) # (Batch Size, 1, IMG_W)
mask = tf.expand_dims(tf.math.logical_and(y_range, x_range) == False, axis=-1) # (Batch Size, IMG_H, IMG_W, 1)
img_batch = tf.where(mask, img_batch, 0)
return (img_batch, label_batch)
@tf.function
def aug_affine(batch_tuple, rotation=360, shear=15, zoom=[0.7, 1.1], shift=[-16, 16], flip=3):
img_batch, label_batch = batch_tuple
batch_shape, batch_size, img_h, img_w = tf.shape(img_batch), tf.shape(img_batch)[0], img_batch.shape[1], img_batch.shape[2]
# returns 3x3 transformmatrix which transforms indicies
# CONVERT
one = tf.ones([batch_size, 1], dtype='float32') # (Batch Size, 1)
zero = tf.zeros([batch_size, 1], dtype='float32') # (Batch Size, 1)
m_list = []
if rotation != 0:
rotation = math.pi * tf.random.uniform(shape=[batch_size, 1], minval=-rotation, maxval=rotation) / 180. # (Batch Size, 1)
c1 = tf.math.cos(rotation) # (Batch Size, 1)
s1 = tf.math.sin(rotation) # (Batch Size, 1)
rotation_matrix = tf.concat([c1, s1, zero, -s1, c1, zero, zero, zero, one], axis=-1) # (Batch Size, 9)
rotation_matrix = tf.reshape(rotation_matrix, [-1, 3, 3]) # (Batch Size, 3, 3)
#m = rotation_matrix if (m is None) else B.batch_dot(m, rotation_matrix)
m_list.append(rotation_matrix)
if shear != 0:
shear = math.pi * tf.random.uniform(shape=[batch_size, 1], minval=-shear, maxval=shear) / 180. # (Batch Size, 1)
c2 = tf.math.cos(shear) # (Batch Size, 1)
s2 = tf.math.sin(shear) # (Batch Size, 1)
shear_matrix = tf.concat([one, s2, zero, zero, c2, zero, zero, zero, one], axis=-1) # (Batch Size, 9)
shear_matrix = tf.reshape(shear_matrix, [-1, 3, 3]) # (Batch Size, 3, 3)
#m = shear_matrix if (m is None) else B.batch_dot(m, shear_matrix)
m_list.append(shear_matrix)
if zoom[0] != 1 or zoom[1] != 1:
width_zoom = tf.random.uniform(shape=[batch_size, 1], minval=zoom[0], maxval=zoom[1]) # (Batch Size, 1)
height_zoom = tf.random.uniform(shape=[batch_size, 1], minval=zoom[0], maxval=zoom[1]) # (Batch Size, 1)
zoom_matrix = tf.concat([one / height_zoom, zero, zero, zero, one / width_zoom, zero, zero, zero, one], axis=-1) # (Batch Size, 9)
zoom_matrix = tf.reshape(zoom_matrix, [-1, 3, 3]) # (Batch Size, 3, 3)
m_list.append(zoom_matrix)
if shift[0] != 0 or shift[1] != 0:
height_shift = tf.random.uniform(shape=[batch_size, 1], minval=shift[0], maxval=shift[1]) # (Batch Size, 1)
width_shift = tf.random.uniform(shape=[batch_size, 1], minval=shift[0], maxval=shift[1]) # (Batch Size, 1)
shift_matrix = tf.concat([one, zero, height_shift, zero, one, width_shift, zero, zero, one], axis=-1)
shift_matrix = tf.reshape(shift_matrix, [-1, 3, 3]) # (Batch Size, 3, 3)
m_list.append(shift_matrix)
if flip != 0:
# 1: left_right 2: up_down 3: both
flip_y = tf.where(tf.random.uniform(shape=[batch_size, 1]) >= (0.5 if (flip == 1 or flip == 3) else 0), 1., -1.)
flip_x = tf.where(tf.random.uniform(shape=[batch_size, 1]) >= (0.5 if (flip == 2 or flip == 3) else 0), 1., -1.)
flip_matrix = tf.concat([flip_y, zero, zero, zero, flip_x, zero, zero, zero, one], axis=-1)
flip_matrix = tf.reshape(flip_matrix, [-1, 3, 3]) # (Batch Size, 3, 3)
#m = flip_matrix if (m is None) else B.batch_dot(m, flip_matrix)
m_list.append(flip_matrix)
if len(m_list) > 0:
# MERGE MATRIX
m_list = tf.unstack(tf.random.shuffle(tf.stack(m_list, axis=0)), axis=0) # List of (Batch Size, 3, 3)
m = reduce((lambda x, y: B.batch_dot(x, y)), m_list)
# LIST DESTINATION PIXEL INDICES
x, y = tf.meshgrid(tf.range(img_w//2, -img_w//2, -1), tf.range(-img_h//2, img_h//2, 1)) # (Img_h, Img_w)
x, y = tf.reshape(x, [-1]), tf.reshape(y, [-1])
z = tf.ones([img_h * img_w], tf.int32) # (IMG_H * IMG_W)
idx = tf.stack([x, y, z]) # (3, IMG_H * IMG_W)
idx = tf.expand_dims(idx, axis=0) # (1, 3, IMG_H * IMG_W)
# ROTATE DESTINATION PIXELS ONTO ORIGIN PIXELS
idx = tf.cast(B.batch_dot(m, tf.cast(idx, tf.float32)), tf.int32) # (Batch Size, 3, IMG_H * IMG_W)
x_idx, y_idx = idx[:,0,:], idx[:,1,:]
x_idx_valid = tf.math.logical_and(-img_w//2 + img_w%2 + 1 <= x_idx, x_idx <= img_w//2) # (Batch Size, IMG_H * IMG_W)
y_idx_valid = tf.math.logical_and(-img_h//2 + img_h%2 + 1 <= y_idx, y_idx <= img_h//2) # (Batch Size, IMG_H * IMG_W)
idx_valid = tf.math.logical_and(x_idx_valid, y_idx_valid) # # (Batch Size, IMG_H * IMG_W)
x_idx = tf.where(x_idx_valid, x_idx, 0)
y_idx = tf.where(y_idx_valid, y_idx, 0)
# FIND ORIGIN PIXEL VALUES
idx = tf.stack([img_h//2-1+y_idx, img_w//2-x_idx],axis=-1) # (Batch Size, IMG_H * IMG_W, 2)
img_batch = tf.gather_nd(img_batch, idx, batch_dims=1) # (Batch Size, IMG_H * IMG_W, 3)
img_batch = tf.where(tf.expand_dims(idx_valid, axis=-1), img_batch, 0)
img_batch = tf.reshape(img_batch, batch_shape)
return (img_batch, label_batch)
@tf.function
def aug_color(batch_tuple, gaussian_s=0.01, hue_s=0.00, bright_s=0.1):
img_batch, label_batch = batch_tuple
if gaussian_s > 0:
img_batch = img_batch + (gaussian_s * tf.random.normal(tf.shape(img_batch), mean=0.0, stddev=1., dtype=tf.float32))
img_batch = tf.clip_by_value(img_batch, 0., 1.)
if hue_s > 0:
img_batch = tf.image.random_hue(img_batch, hue_s)
img_batch = tf.clip_by_value(img_batch, 0., 1.)
if bright_s > 0:
img_batch = tf.image.random_brightness(img_batch, bright_s)
img_batch = tf.clip_by_value(img_batch, 0., 1.)
return (img_batch, label_batch)
@tf.function
def aug_mix(batch_tuple1, batch_tuple2, p):
(img_batch1, label_batch1), (img_batch2, label_batch2) = batch_tuple1, batch_tuple2
p = tf.random.uniform(shape=[tf.shape(img_batch1)[0]], minval=0, maxval=1, dtype=tf.float32) <= p
img_batch = tf.where(tf.reshape(p, (-1, 1, 1, 1)), img_batch1, img_batch2)
label_batch = tf.where(tf.reshape(p, (-1, 1)), label_batch1, label_batch2)
return (img_batch, label_batch)
@tf.function
def datasets_augumentation(img_batch, label_batch):
batch_tuple = (img_batch, label_batch)
# Color
#batch_tuple = aug_mix(aug_color(batch_tuple), batch_tuple, 0.7)
# Affine
batch_tuple = aug_affine(batch_tuple)
# Grid Mask
batch_tuple = aug_mix(aug_grid_mask(batch_tuple), batch_tuple, 0.5)
img_batch, label_batch = batch_tuple
return img_batch, label_batch
# +
AUTO = tf.data.experimental.AUTOTUNE
train_datasets_format = train_img_batch_generator.map(datasets_augumentation, num_parallel_calls=AUTO).prefetch(AUTO)
test_datasets_format = test_img_batch_generator.prefetch(AUTO)
sample_datasets_format = sample_img_batch_generator.prefetch(AUTO)
for x, y_true in train_datasets_format:
for i in range(1):
plt.title(np.argmax(y_true[i]))
plt.imshow(x[i])
plt.show()
break
# +
def cosineAnnealingWithWarmup(epochIdx):
aMin, aMax = 1e-5, 5e-4
warmupEpochs, stagnateEpochs, cosAnnealingEpochs = 10, 0, 100
epochIdx = epochIdx % (warmupEpochs + stagnateEpochs + cosAnnealingEpochs)
if(epochIdx < warmupEpochs):
return aMin + (aMax - aMin) / (warmupEpochs - 1) * epochIdx
else:
epochIdx -= warmupEpochs
if(epochIdx < stagnateEpochs):
return aMax
else:
epochIdx -= stagnateEpochs
return aMin + 0.5 * (aMax - aMin) * (1 + math.cos((epochIdx + 1) / (cosAnnealingEpochs + 1) * math.pi))
#lr_schedule = tf.keras.callbacks.ReduceLROnPlateau(monitor='loss', patience=2, factor=0.5, mode='auto')
#lr_schedule = tf.keras.callbacks.LearningRateScheduler(tf.keras.experimental.CosineDecayRestarts(5e-5, 10), verbose=1)
lr_schedule = tf.keras.callbacks.LearningRateScheduler(cosineAnnealingWithWarmup, verbose=1)
# -
class CosineLayer(L.Layer):
def __init__(self, n_classes=num_classes, regularizer=None, **kwargs):
super().__init__(**kwargs)
self.n_classes = n_classes
self.regularizer = R.get(regularizer)
def build(self, input_shape):
super().build(input_shape)
# tf.constant_initializer(embedding_parms)
# initializer='glorot_uniform'
channel_cnt = 1
self.W = self.add_weight(name='W', shape=(channel_cnt, input_shape[-1], self.n_classes), initializer='glorot_uniform', trainable=True, regularizer=self.regularizer)
def call(self, x):
# normalize feature
x = tf.nn.l2_normalize(x, axis=1) # (Batch Size, Embedding_Dims)
# normalize weights
W = tf.nn.l2_normalize(self.W, axis=1) # (2, Embedding_Dims, Num_Class)
logits = tf.map_fn(lambda w: tf.matmul(x, w), W)
logits = tf.math.reduce_max(logits, axis=0)
# a·b = |a|×|b|×cos<a,b> => cos<a,b> = (a/|a|)·(b/|b|)
#logits = tf.matmul(x, W) # (None, 512) (512, 10, 2) (None, 10)
return logits
def ClassificationMetric(y_true, y_pred):
# y_pred = tf.nn.softmax(y_pred) # (Batch Size, 10)
y_pred_idxs = tf.cast(tf.math.argmax(y_pred, -1), tf.int32) # (Batch Size)
y_true_idxs = tf.cast(tf.math.argmax(y_true, -1), tf.int32) # (Batch Size)
# print(y_pred_idxs, y_pred_score, y_true)
return tf.reduce_sum(tf.cast(y_pred_idxs == y_true_idxs, tf.float32)) / (tf.cast(tf.shape(y_pred_idxs)[0], tf.float32))
def ArcFaceLoss(m=0.50, s=30.0):
def LossFunc(y_true, y_pred):
theta = tf.acos(tf.clip_by_value(y_pred, -0.999999, 0.999999))
y_pred_mod = tf.cos(tf.clip_by_value(theta + m, 0, np.pi))
#
y_pred = ((y_true) * y_pred_mod + (1 - y_true) * y_pred) * s
loss = losses.CategoricalCrossentropy(from_logits=True, reduction=losses.Reduction.NONE)(y_true, y_pred)
return loss
return LossFunc
def CircleLoss(margin = 0.25, gamma=256):
def LossFunc(y_true, y_pred):
O_p = 1 + margin
O_n = - margin
Delta_p = 1 - margin
Delta_n = margin
alpha_p = tf.nn.relu(O_p - tf.stop_gradient(y_pred))
alpha_n = tf.nn.relu(tf.stop_gradient(y_pred) - O_n)
y_pred = ((y_true) * (alpha_p * (y_pred - Delta_p)) + (1 - y_true) * (alpha_n * (y_pred - Delta_n))) * gamma
# Weighted Loss
loss = losses.CategoricalCrossentropy(from_logits=True, reduction=losses.Reduction.NONE)(y_true, y_pred)
return loss
return LossFunc
MetricLearningType = "Circle"
MetricLearningLayer = { "ArcFace": CosineLayer, "Circle": CosineLayer }[MetricLearningType]
MetricLearningLoss = { "ArcFace": ArcFaceLoss, "Circle": CircleLoss }[MetricLearningType]()
MetricLearningMetric = ClassificationMetric
# +
def build_model():
input = Input(shape=image_shape, name='Input')
x = input
#backbone = efn.EfficientNetB0(input_shape=image_shape, include_top=False, weights='imagenet')
backbone = tf.keras.applications.mobilenet_v2.MobileNetV2(input_shape=image_shape, include_top=False, weights='imagenet')
for layer in backbone.layers:
layer.trainable = True # trainable has to be false in order to freeze the layers
x = backbone(x)
x = L.GlobalAveragePooling2D(name='Gap')(x)
x = L.Dropout(0.7, name="Dropout")(x)
x = L.Dense(256, name='Dense', activation=None)(x)
x = MetricLearningLayer(name='Final')(x)
output = x
model = Model(input, output, name='model')
model.compile(optimizer=optimizers.Adam(1e-3), loss = MetricLearningLoss, metrics=MetricLearningMetric)
#model.load_weights('***', by_name=True, skip_mismatch=True)
model.summary()
return model
model = build_model()
# -
class timeout_check(tf.keras.callbacks.Callback):
def on_epoch_end(self, epoch, logs={}):
loss, accuracy = self.model.evaluate(test_datasets_format) #, steps=4)
self.model.save_weights(path.join(work_dir, "model_{:.0f}_{:.4f}_{:.4f}_{:.5f}.h5").format(float(epoch + 1), loss, accuracy, self.model.optimizer.learning_rate.numpy()))
callbacks_list = [lr_schedule, timeout_check()]
#model.load_weights(path.join(work_dir, "model_6_132.2049_0.0692_0.00028.h5"))
history = model.fit(train_datasets_format, epochs=epochs, callbacks=callbacks_list) #, steps_per_epoch=25)
epoch_idxs = np.arange(len(history.history["loss"])) + 1
lines_visualization((epoch_idxs, epoch_idxs),
(history.history["metric_mIOU"], history.history["val_metric_mIOU"]),
lines_label=("Train", "Valid"),
xlabel="Epoch Idx", ylabel="mIOU", title="Training Process", dpi=100)
del epoch_idxs
class L2NormLayer(L.Layer):
def __init__(self, **kwargs):
super().__init__(**kwargs)
pass
def call(self, inputs: tf.Tensor, **kwargs):
inputs = tf.math.l2_normalize(inputs, axis=1)
return inputs
def MetricLearningModel(name=None):
input = Input(shape=image_shape, name='Input')
#backbone = efn.EfficientNetB0(input_shape=image_shape, include_top=False, weights='imagenet')
backbone = tf.keras.applications.MobileNetV2(input_shape=image_shape, include_top=False, weights='imagenet')
x = backbone(input)
x = L.GlobalAveragePooling2D(name='Gap')(x)
x = L.Dense(256, name='Dense', activation=None)(x)
x = L2NormLayer(name="L2")(x)
output = x
model = Model(input, output, name=name)
#model.summary()
return model
# +
class GlobalFeatureExtractor():
def __init__(self, model):
self.model = MetricLearningModel("Backbone")
self.model.load_weights(path.join(work_dir, "model_77_41.7805_0.8527_0.00013.h5"), by_name=True, skip_mismatch=True)
self.model.summary()
def extract_features(self, img_generator, steps=None):
return self.model.predict(img_generator, verbose=1, steps=steps)
@tf.function
def _cos_similarity_iter(self, embedding_target, embedding_sources, k):
distances = tf.matmul(embedding_sources, tf.expand_dims(embedding_target, axis=-1))[:,0] # (#embedding_sources)
nearset_distances, nearest_idxs = tf.math.top_k(distances, k=k, sorted=True)
nearset_distances, nearest_idxs = tf.cast(nearset_distances, tf.float32), tf.cast(nearest_idxs, tf.float32)
return tf.stack([nearest_idxs, nearset_distances], axis=1) #(3, 2)
@tf.function
def cos_similarity(self, embedding_targets, embedding_sources, k):
return tf.map_fn(lambda x: self._cos_similarity_iter(x, embedding_sources, k), embedding_targets, parallel_iterations=8) #(?, 3, 2)
global_feature_extractor = GlobalFeatureExtractor(model)
# -
global_feature_extractor.extract_features(sample_img_batch_generator)
sample_img_features = global_feature_extractor.extract_features(sample_img_batch_generator, 1)
print(global_feature_extractor.cos_similarity(sample_img_features, sample_img_features, 4))
| CoinRecognition-Train.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
# # %load stat_metrics_compare_ensemble_and_SILO.py
"""
Created on Tue Apr 23 14:12:09 2019
@author: nav00a
"""
""""
# xarray opens the netcdf file
import xarray as xr
import requests
import io
import pandas as pd
from plotnine import *
import datetime
nc = xr.open_dataset("../data/ensemble_siloaccess_s.nc")
# type nc in the console to see a summary of the object incl. attributes, dimensions and variables
# Interesting behaviour from xarray: if we define a variable with name "days" and units "days since Jan 1st 1970",
# it automatically reads it in as a datetime object!! It won't do it otherwise, but since this is what we want,
# we just go with it.
# Get some properties (attributes) from the netcdf file
# Attributes are values that the creator of the file has provided as a sort of header
start_month = nc.attrs["hindcast_start_month"]
lon = nc.attrs["longitude"]
lat = nc.attrs["latitude"]
# Get time bounds properties of the .nc file, which aren't declared attributes
minyear = int(min(nc.year))
maxyear = int(max(nc.year))
# Now we convert the imported netcdf object to a pandas dataframe
ensemble_df = nc.to_dataframe().reset_index()
# Can't remember why I convert date to a string, but it was needed for grouping I think (won't work well with a datetime object)
# We also create a "date" column, because it makes more sense than a "days" column formatted as a date
# A "date" column also plays along nicely with the SILO dataset
ensemble_df["date"] = ensemble_df["days"].astype(str)
# Delete "days" column
ensemble_df = ensemble_df.drop(columns="days")
# Get SILO data to compare ensemble with it
# Set SILO url download link (specifying csv format)
silourl = "https://dcdp.research.csiro.au/silogrid?lat="+ str(lat) +"&lon=" + str(lon) +" &format=csv&start=" + str(minyear) +"0101&stop=" + str(maxyear) +"1231"
# Download SILO data
httpreq = requests.get(silourl, verify=False)
# Use the StringIO function to interpret the csv-like object as if it was saved to disk, then read_csv into pandas
silo_df = pd.read_csv(io.StringIO(httpreq.content.decode('utf-8')))
# This is to remove the header row
silo_df = silo_df.iloc[1:]
# Set columns to correct format
silo_df["date"] = silo_df["date"].astype(str)
silo_df["rad"] = silo_df["rad"].astype(float)
silo_df["tmax"] = silo_df["tmax"].astype(float)
silo_df["tmin"] = silo_df["tmin"].astype(float)
silo_df["rain"] = silo_df["rain"].astype(float)
silo_df["vp"] = silo_df["vp"].astype(float)
silo_df["evap"] = silo_df["evap"].astype(float)
# Compare ensemble and SILO
# We calculate the mean of the ensemble as this is what we want to evaluate
ensemble_df_mean = ensemble_df.groupby(["date"]).mean().reset_index()
# ANALYSIS
# Plot using ggplot2 for python (plotnine). Currently not too pretty, need to find how to mend the looks
(ggplot(ensemble_df_mean.where(ensemble_df_mean["rain"]>0.1))+
geom_density(mapping= aes(x = "rain"), color="red", size=1) +
geom_density(data=silo_df.where(silo_df["rain"]>0.1), mapping= aes(x = "rain"), color="white", size=1, linetype='dotted') +
theme_minimal()+
coord_cartesian(xlim =(0, 20))
)
# Calculate ensemble performance metrics (based on http://www.cawcr.gov.au/projects/verification/)
# We won't use correlation as we're not interested in predicting daily weather but the actual pattern of climate
fcast_vs_obs = ensemble_df_mean.merge(silo_df, on= 'date')
fcast_vs_obs.rename(columns = {"date":"date", "ens":"ens", "rad_x":"rad.fcast","tmax_x":"tmax.fcast","rain_x":"rain.fcast","tmin_x":"tmin.fcast","rad_y":"rad.obs","tmax_y":"tmax.obs","tmin_y":"tmin.obs","rain_y":"rain.obs","vp":"vp.obs","evap":"evap.obs"}, inplace=True)
# Mean error - Answers the question: What is the average forecast error?
# We don't really want this for AgScore but it's informative
fcast_vs_obs["error"] = fcast_vs_obs["rain.fcast"] - fcast_vs_obs["rain.obs"]
mean_error = fcast_vs_obs["error"].mean()
print (mean_error)
# Multiplicative bias - Answers the question: How does the average forecast magnitude compare to the average observed magnitude?
# Bias = mean of forecast divided by mean of observations
bias = fcast_vs_obs["rain.fcast"].mean() / fcast_vs_obs["rain.obs"].mean()
print (bias)
# Next, we need more metrics...
"""
# +
# this is to load Javi's python code into my Jupyter notebook
# # %load stat_metrics_compare_ensemble_and_SILO.py
"""
Created on Tue Apr 23 14:12:09 2019
@author: nav00a
"""
import xarray as xr
import requests
import io
import pandas as pd
import datetime
# +
# xarray opens the netcdf file
# type nc in the console to see a summary of the object incl. attributes, dimensions and variables
# Interesting behaviour from xarray: if we define a variable with name "days" and units "days since Jan 1st 1970",
# it automatically reads it in as a datetime object!! It won't do it otherwise, but since this is what we want,
# we just go with it.
nc = xr.open_dataset("../data/ensemble_siloaccess_s.nc")
print(nc)
# +
# Get some properties (attributes) from the netcdf file
# Attributes are values that the creator of the file has provided as a sort of header
start_month = nc.attrs["hindcast_start_month"]
lon = nc.attrs["longitude"]
lat = nc.attrs["latitude"]
print(start_month)
print(lon)
print(lat)
# +
# Get time bounds properties of the .nc file, which aren't declared attributes
minyear = int(min(nc.year))
maxyear = int(max(nc.year))
print(minyear)
print(maxyear)
# +
# Now we convert the imported netcdf object to a pandas dataframe
ensemble_df = nc.to_dataframe().reset_index()
print(ensemble_df)
# +
# now we need to convert all variables to the types that are going to be useful for us to use
ensemble_df["ens"] = ensemble_df["ens"].astype(int)
ensemble_df["ensday"] = ensemble_df["ensday"].astype(int)
ensemble_df["year"] = ensemble_df["year"].astype(int)
ensemble_df["rad"] = round(ensemble_df["rad"].astype(float),1)
ensemble_df["tmax"] = round(ensemble_df["tmax"].astype(float),1)
ensemble_df["rain"] = round(ensemble_df["rain"].astype(float),1)
ensemble_df["tmin"] = round(ensemble_df["tmin"].astype(float),1)
ensemble_df.head()
# +
#convert the ens column to a string and then convert to two characters
ensemble_df["ens"] = ensemble_df["ens"].astype(str).str.pad(width=2, side='left', fillchar='0')
#convert the ensday column to a string and then convert to three characters
ensemble_df["ensday"] = ensemble_df["ensday"].astype(str).str.pad(width=3, side='left', fillchar='0')
ensemble_df.head()
# +
# combine the year and the ensemble day into a variable
ensemble_df['year_ensday'] = ensemble_df['year'].astype(str) + ensemble_df['ensday']
ensemble_df.head()
# +
# find the mean of each ensemble variable for each day
ensemble_df_mean = ensemble_df.groupby(["year_ensday"]).mean()
ensemble_df_mean.head()
# +
# rename all variables to reflect they are from access-s ensemble data
# convert all variables to the types that are going to be useful for us to use
ensemble_df_mean["ens_rad"] = round(ensemble_df_mean["rad"].astype(float),1)
ensemble_df_mean["ens_tmax"] = round(ensemble_df_mean["tmax"].astype(float),1)
ensemble_df_mean["ens_rain"] = round(ensemble_df_mean["rain"].astype(float),1)
ensemble_df_mean["ens_tmin"] = round(ensemble_df_mean["tmin"].astype(float),1)
ensemble_df_mean.head()
# +
# drop the columns that won't be used in the analysis
ensemble_df_mean = ensemble_df_mean.drop(columns="year")
ensemble_df_mean = ensemble_df_mean.drop(columns="rad")
ensemble_df_mean = ensemble_df_mean.drop(columns="tmax")
ensemble_df_mean = ensemble_df_mean.drop(columns="rain")
ensemble_df_mean = ensemble_df_mean.drop(columns="tmin")
print(ensemble_df_mean)
# +
# Get SILO data to compare ensemble with it
# Set SILO url download link (specifying csv format)
silourl = "https://dcdp.research.csiro.au/silogrid?lat="+ str(lat) +"&lon=" + str(lon) +" &format=csv&start=" + str(minyear) +"0101&stop=" + str(maxyear) +"1231"
print(silourl)
# -
# Download SILO data
httpreq = requests.get(silourl, verify=False)
print(httpreq)
# +
# Use the StringIO function to interpret the csv-like object as if it was saved to disk, then read_csv into pandas
silo_df = pd.read_csv(io.StringIO(httpreq.content.decode('utf-8')))
silo_df.head()
# +
# This is to remove the header row
silo_df = silo_df.iloc[1:]
silo_df.head()
# +
# Set columns to correct format
silo_df["date"] = silo_df["date"].astype(str)
# +
# import the regex library which is called re
import re
# the ensemble_df has days from April 1st to November 2nd (!!???) each year - a total of 216 days.
# to be able to compare the two data frames it is necessary to crop the silo_df so that it has the same days
# we use regex to do this
silo216_df = silo_df[silo_df['date'].str.contains(r'....-04-..|....-05-..|....-06-..|....-07-..|....-08-..|....-09-..|....-10-..|....-11-01|....-11-02')]
silo216_df.head()
# +
# first convert date string to datetime with a proper format string
df = pd.DataFrame({'Date':pd.to_datetime(silo216_df['date'], format='%Y-%m-%d')})
# calculate day of year and then subtract 91 to start at the 1st of April
silo216_df['DOY'] = df['Date'].dt.dayofyear - 91
silo216_df.head()
# +
# split the date column into year, month and day
silo216_df = silo216_df.join(silo216_df['date'].str.split('-', 2, expand=True).rename(columns={0:'year', 1:'month', 2:'day'}))
silo216_df.head()
# -
silo216_df["DOY"] = silo216_df["DOY"].astype(str).str.pad(width=3, side='left', fillchar='0')
silo216_df.head()
# +
# join the year and DOY columns
silo216_df['year_ensday'] = silo216_df['year'].astype(str) + silo216_df['DOY']
silo216_df.head()
# +
# rename all variables to reflect they are from silo data
# round the variables to one decimal point
silo216_df["silo_rad"] = round(silo216_df["rad"].astype(float),1)
silo216_df["silo_tmax"] = round(silo216_df["tmax"].astype(float),1)
silo216_df["silo_rain"] = round(silo216_df["rain"].astype(float),1)
silo216_df["silo_tmin"] = round(silo216_df["tmin"].astype(float),1)
silo216_df.head()
# +
# get rid of the columns that we don't need to use
silo216_df = silo216_df.drop(columns="rad")
silo216_df = silo216_df.drop(columns="tmax")
silo216_df = silo216_df.drop(columns="tmin")
silo216_df = silo216_df.drop(columns="rain")
silo216_df = silo216_df.drop(columns="date")
silo216_df = silo216_df.drop(columns="month")
silo216_df = silo216_df.drop(columns="day")
silo216_df = silo216_df.drop(columns="year")
silo216_df = silo216_df.drop(columns="DOY")
silo216_df = silo216_df.drop(columns="evap")
silo216_df = silo216_df.drop(columns="vp")
silo216_df.head()
# +
#join the two dataframes together to perform analyses
fcast_vs_obs = pd.merge(ensemble_df_mean, silo216_df, on ='year_ensday')
fcast_vs_obs.head()
# +
# choose the column "rain" from the fcast_vs_obs data
eArrayRain = fcast_vs_obs["ens_rain"]
eArrayRain.head()
# +
# choose the column "silo_rain" from the fcast_vs_obs data
sArrayRain = fcast_vs_obs["silo_rain"]
sArrayRain.head()
# -
eArrayRainPlusOne = eArrayRain + 0.000000001
print(eArrayRainPlusOne)
sArrayRainPlusOne = sArrayRain + 0.000000001
print(sArrayRainPlusOne)
# +
import math
a = 0
b = .00000001
c = a/b
print(c)
math.log(0)
# +
# the Kullback–Leibler divergence (also called relative entropy) y variable
# is a measure of how one probability distribution is different
# from a second, reference probability distribution.
# Kullback-Leibler = Σ(x(log(x/y)))
import numpy as np
kl1 = (eArrayRainPlusOne * np.log(eArrayRainPlusOne/sArrayRainPlusOne)).sum()
print( "Model 1: ", round(kl1, 4))
kl2 = (sArrayRainPlusOne * np.log(sArrayRainPlusOne/eArrayRainPlusOne)).sum()
print("Model 2: ", round(kl2, 4))
# +
# the Kullback–Leibler divergence (also called relative entropy) y variable
# is a measure of how one probability distribution is different
# from a second, reference probability distribution.
# Kullback-Leibler = Σ(x(log(x/y)))
import numpy as np
KL1 = (eArrayRainPlusOne * np.log(eArrayRainPlusOne/sArrayRainPlusOne))
print( "Model 1: ", round(KL1, 4))
KL2 = (sArrayRainPlusOne * np.log(sArrayRainPlusOne/eArrayRainPlusOne))
print("Model 2: ", round(KL2, 4))
# +
# the Kullback–Leibler divergence (also called relative entropy) y variable
# is a measure of how one probability distribution is different
# from a second, reference probability distribution.
# Kullback-Leibler = Σ(x(log(x/y)))
import numpy as np
KL1 = pd.DataFrame(eArrayRainPlusOne * np.log(eArrayRainPlusOne/sArrayRainPlusOne))
print( "Model 1: ", round(KL1, 4))
KL2 = pd.DataFrame(sArrayRainPlusOne * np.log(sArrayRainPlusOne/eArrayRainPlusOne))
print("Model 2: ", round(KL2, 4))
# +
#Normalize A Column In pandas
# Import required modules
import pandas as pd
from sklearn import preprocessing
# Set charts to view inline
# %matplotlib inline
#KL_DF = pd.DataFrame(KL1)
#eArrayRain = pd.DataFrame(fcast_vs_obs["silo_rain"])
# Create a minimum and maximum processor object
min_max_scaler = preprocessing.MinMaxScaler()
# Create an object to transform the data to fit minmax processor
x_scaled = min_max_scaler.fit_transform(KL1)
# Run the normalizer on the dataframe
norm_KL1 = pd.DataFrame(x_scaled)
print(norm_KL1)
# +
#Normalize A Column In pandas
# Import required modules
import pandas as pd
from sklearn import preprocessing
# Set charts to view inline
# %matplotlib inline
# Create a minimum and maximum processor object
min_max_scaler = preprocessing.MinMaxScaler()
# Create an object to transform the data to fit minmax processor
x_scaled = min_max_scaler.fit_transform(KL2)
# Run the normalizer on the dataframe
norm_KL2 = pd.DataFrame(x_scaled)
print(norm_KL2)
# +
import matplotlib.pyplot as plt
import scipy.stats as stats
import seaborn as sns
# # %matplotlib inline sets the backend of matplotlib to the 'inline' backend: With this backend,
# the output of plotting commands is displayed inline within frontends like the Jupyter notebook,
# directly below the code cell that produced it.
# %matplotlib inline
# +
# #!/usr/bin/env python
# -*- coding: utf-8 -*-
import matplotlib.pyplot as plt
import numpy as np
import matplotlib.mlab as mlab
from scipy.stats import norm
from scipy.integrate import quad
plt.plot(norm_KL1)
plt.plot(norm_KL2)
# +
import matplotlib
import matplotlib.pyplot as plt
import numpy as np
# Data for plotting
#t = np.arange(0.0, 2.0, 0.01)
#s = 1 + np.sin(2 * np.pi * t)
import matplotlib.mlab as mlab
import seaborn as sns
from scipy.stats import norm
from scipy.integrate import quad
norm_eArrayRain = norm.pdf(eArrayRain, 0, 2)
norm_sArrayRain = norm.pdf(sArrayRain, 2, 2)
fig, ax = plt.subplots()
ax.plot(norm_eArrayRain)
#ax.plot(norm_sArrayRain)
ax.set(xlabel='time', ylabel='rain',
title='About as simple as it gets, folks')
ax.grid()
#fig.savefig("test.png")
plt.plot()
# +
# #!/usr/bin/env python
# -*- coding: utf-8 -*-
import matplotlib.pyplot as plt
import numpy as np
import matplotlib.mlab as mlab
from scipy.stats import norm
from scipy.integrate import quad
my_range = np.arange(-10, 10, 0.001)
KL_int, err = quad(KL, -10, 10)
print( 'KL: ', KL_int )
fig = plt.figure(figsize=(18, 8), dpi=100)
#---------- First Plot
ax = fig.add_subplot(1,2,1)
ax.grid(True)
ax.spines['left'].set_position('zero')
ax.spines['right'].set_color('none')
ax.spines['bottom'].set_position('zero')
ax.spines['top'].set_color('none')
ax.set_xlim(-10,10)
ax.set_ylim(-0.1,0.25)
ax.text(-2.5, 0.17, 'x', horizontalalignment='center',fontsize=17)
ax.text(4.5, 0.17, 'y', horizontalalignment='center',fontsize=17)
plt.plot(range, x(range))
plt.plot(range, y(range))
#---------- Second Plot
ax = fig.add_subplot(1,2,2)
ax.grid(True)
ax.spines['left'].set_position('zero')
ax.spines['right'].set_color('none')
ax.spines['bottom'].set_position('zero')
ax.spines['top'].set_color('none')
ax.set_xlim(-10,10)
ax.set_ylim(-0.1,0.25)
ax.text(3.5, 0.17, r'$DK_{KL}(x||y)$', horizontalalignment='center',fontsize=17)
ax.plot(range, KL(range))
ax.fill_between(range, 0, KL(range))
plt.savefig('KullbackLeibler.png',bbox_inches='tight')
plt.show()
# +
# #!/usr/bin/env python
# -*- coding: utf-8 -*-
import matplotlib.pyplot as plt
import numpy as np
import matplotlib.mlab as mlab
import seaborn as sns
from scipy.stats import norm
from scipy.integrate import quad
norm_KL3 = norm.pdf(KL1, 0, 2)
norm_KL4 = norm.pdf(KL2, 2, 2)
sns.set_style('whitegrid')
ax = sns.kdeplot(norm_KL3, bw=0, color = 'red', shade = 'true')
ax = sns.kdeplot(norm_KL4, bw=0, color = 'green', shade = 'true')
#plt.plot(norm_KL3)
#plt.plot(norm_KL4)
# +
# #!/usr/bin/env python
# -*- coding: utf-8 -*-
import matplotlib.pyplot as plt
import numpy as np
import matplotlib.mlab as mlab
import seaborn as sns
from scipy.stats import norm
from scipy.integrate import quad
norm_KL3 = norm.pdf(eArrayRain, 0, 2)
norm_KL4 = norm.pdf(sArrayRain, 2, 2)
sns.set_style('whitegrid')
ax = sns.kdeplot(norm_KL3, bw=0, color = 'red', shade = 'true')
ax = sns.kdeplot(norm_KL4, bw=0, color = 'green', shade = 'true')
#plt.plot(norm_KL3)
#plt.plot(norm_KL4)
# +
# #!/usr/bin/env python
# -*- coding: utf-8 -*-
import matplotlib.pyplot as plt
import numpy as np
import matplotlib.mlab as mlab
from scipy.stats import norm
from scipy.integrate import quad
x = norm_eArrayRain
x1 = norm_eArrayRainPlusOne
y = norm_sArrayRain
y1 = norm_sArrayRainPlusOne
KL=x * np.log( x / y1 )
range = np.arange(-10, 10, 0.001)
#KL_int, err = quad(KL, -10, 10)
#Wprint( 'KL: ', KL_int )
fig = plt.figure(figsize=(18, 8), dpi=100)
#---------- First Plot
ax = fig.add_subplot(1,2,1)
ax.grid(True)
ax.spines['left'].set_position('zero')
ax.spines['right'].set_color('none')
ax.spines['bottom'].set_position('zero')
ax.spines['top'].set_color('none')
ax.set_xlim(-10,10)
ax.set_ylim(-0.1,0.25)
#ax.text(-2.5, 0.17, 'p(x)', horizontalalignment='center',fontsize=17)
#ax.text(4.5, 0.17, 'q(x)', horizontalalignment='center',fontsize=17)
plt.plot()
#---------- Second Plot
ax = fig.add_subplot(1,2,2)
ax.grid(True)
ax.spines['left'].set_position('zero')
ax.spines['right'].set_color('none')
ax.spines['bottom'].set_position('zero')
ax.spines['top'].set_color('none')
ax.set_xlim(-10,10)
ax.set_ylim(-0.1,0.25)
ax.text(3.5, 0.17, r'$DK_{KL}(p||q)$', horizontalalignment='center',fontsize=17)
ax.plot(range, KL(range))
ax.fill_between(range, 0, KL(range))
plt.savefig('KullbackLeibler.png',bbox_inches='tight')
plt.show()
# +
import matplotlib.pyplot as plt
import scipy.stats as stats
import seaborn as sns
# # %matplotlib inline sets the backend of matplotlib to the 'inline' backend: With this backend,
# the output of plotting commands is displayed inline within frontends like the Jupyter notebook,
# directly below the code cell that produced it.
# %matplotlib inline
# +
# this graph compares the densities of each unique value of rainfall
sns.set_style('whitegrid')
ax = sns.kdeplot(eArrayRain, bw=0, color = 'red', shade = 'true')
ax = sns.kdeplot(sArrayRain, bw=0, color = 'green', shade = 'true')
# +
# this graph compares the densities of each unique value of rainfall
# for any variable
variable = "rain"
evariable = "ens_" + variable
svariable = "silo_" + variable
earray = fcast_vs_obs[evariable]
sarray = fcast_vs_obs[svariable]
sns.set_style('whitegrid')
ax = sns.kdeplot(earray, bw=0, color = 'red', shade = 'true')
ax = sns.kdeplot(sarray, bw=0, color = 'green', shade = 'true')
# +
# this will pull out rainfall over 0.1mm from the ensemble data
eArrayRainSig = [round(num,1) for num in fcast_vs_obs["ens_rain"] if num > 0.1]
print(eArrayRainSig)
# +
# this will pull out rainfall over 0.1mm from the silo data
sArrayRainSig = [round(num,1) for num in fcast_vs_obs["silo_rain"] if num > 0.1]
print(sArrayRainSig)
# -
# plots the density for rainfall above 0.1mm
sns.distplot(eArrayRainSig, color="red", label = "ensemble")
sns.distplot(sArrayRainSig, color="green", label = "silo")
plt.legend()
# +
# plots the density for rainfall above 0.1mm with a normal distribution
sns.distplot( eArrayRainSig, color="red", label = "ensemble", norm_hist = "true")
sns.distplot( sArrayRainSig, color="green", label = "silo", norm_hist = "true")
plt.legend()
# +
# use any of the variables as input for the plot
# if the variable is "rain" then we only want to use the values that are greater than 0.1
variable = "rain"
evariable = "ens_" + variable
svariable = "silo_" + variable
if variable == "rain":
eArray = [num for num in fcast_vs_obs[evariable] if num > 0.1]
sArray = [num for num in fcast_vs_obs[svariable] if num > 0.1]
else:
eArray = fcast_vs_obs[evariable]
sArray = fcast_vs_obs[svariable]
sns.distplot( eArray, color="red", label = "ensemble")
sns.distplot( sArray, color="green", label = "silo")
plt.legend()
# +
#label the axes and give the graph a name
fig, ax = plt.subplots()
sns.distplot( eArray, color="red", label = "ensemble")
sns.distplot( sArray, color="green", label = "silo")
ax.set_xlim(-5,50)
ax.set(xlabel='Rainfall', ylabel='Density')
plt.title("Rainfall histogram")
plt.legend()
# +
## rewrite the code to be able to use any of the variables in the datasets
variable = "rain"
evariable = "ens_" + variable
svariable = "silo_" + variable
if variable == "rain":
eArray = [num for num in fcast_vs_obs[evariable] if num > 0.1]
sArray = [num for num in fcast_vs_obs[svariable] if num > 0.1]
else:
eArray = fcast_vs_obs[evariable]
sArray = fcast_vs_obs[svariable]
fig, ax = plt.subplots()
sns.distplot( eArray, color="red", label = "ensemble")
sns.distplot( sArray, color="green", label = "silo")
ax.set_xlim(-5,50)
ax.set(xlabel = variable, ylabel='Density')
plt.title(variable + " histogram")
plt.legend()
# +
# run the code with a different variable
variable = "tmax"
evariable = "ens_" + variable
svariable = "silo_" + variable
if variable == "rain":
eArray = [num for num in fcast_vs_obs[evariable] if num > 0.1]
sArray = [num for num in fcast_vs_obs[svariable] if num > 0.1]
else:
eArray = fcast_vs_obs[evariable]
sArray = fcast_vs_obs[svariable]
#sns.distplot( eArray, color="red", label = "ensembl")
#sns.distplot( sArray, color="green", label = "silo")
#plt.legend()
fig, ax = plt.subplots()
sns.distplot( eArray, color="red", label = "ensemble")
sns.distplot( sArray, color="green", label = "silo")
ax.set_xlim(-5,50)
ax.set(xlabel = variable, ylabel='Density')
plt.title(variable + " histogram")
plt.legend()
# +
# Multiplicative bias - Answers the question: How does the average forecast magnitude
# compare to the average observed magnitude?
# Bias = mean of forecast divided by mean of observations
bias = fcast_vs_obs["ens_rain"].mean() / fcast_vs_obs["silo_rain"].mean()
print (bias)
# +
# Mean error - Answers the question: What is the average forecast error?
# We don't really want this for AgScore but it's informative
fcast_vs_obs["error"] = fcast_vs_obs["ens_rain"] - fcast_vs_obs["silo_rain"]
mean_error = fcast_vs_obs["error"].mean()
print (mean_error)
# -
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
from scipy import stats
# +
# using matplotlib
# plots the day on the the x-axis and the rain forecast on the y axis
day = fcast_vs_obs["year_ensday"]
ensrainsorted = fcast_vs_obs["ens_rain"].sort_values()
plt.plot(day, ensrainsorted)
plt.xlabel('day')
plt.ylabel('forecast rain')
plt.title('rain')
plt.show()
plt.close()
# +
# using pyplot
# plots the day on the the x-axis and the rain forecast on the y axis
day = fcast_vs_obs["year_ensday"]
ensrainsorted = fcast_vs_obs["ens_rain"].sort_values()
fig = plt.figure()
ax = fig.add_subplot(111)
ax.set_xlabel('day')
ax.set_ylabel('forecast rain')
ax.set_title('rainfall')
ax.plot(day, ensrainsorted)
plt.show()
plt.close()
# +
# frequency and accumulated frequency diagrams
ensrainsorted = fcast_vs_obs["ens_rain"]
silorainsorted = fcast_vs_obs["silo_rain"].sort_values()
#values, freq = stats.itemfreq(ensrainsorted)
values, counts = np.unique(ensrainsorted, return_counts=True)
cum_freq = np.cumsum(counts)
fig, (ax1, ax2) = plt.subplots(2,1) #creates a fifure with a 2D axis
fig.set_size_inches(6,6)
ax1.set_xlim([0,40])
ax1.set_xlabel('forecast rain (mm)')
ax1.set_ylabel('frequency')
ax1.plot(values, counts, 'b^')
#ax1.vlines(x, [0], fi)
ax1.tick_params(axis='both', which = 'major')
ax2.set_xlim([0,40])
ax2.set_xlabel('forecast rain (mm)')
ax2.set_ylabel('cumulated frequency')
ax2.step(values, cum_freq, color = 'darkblue', marker = 'o', where = 'post')
ax2.tick_params(axis='both', which = 'major')
plt.show()
plt.close()
# +
# Creating a multidimensional histogram
ensrainsorted = fcast_vs_obs["ens_rain"]
silorainsorted = fcast_vs_obs["silo_rain"].sort_values()
# create a figure with 4 axes (4 subplots) in a format of 2 x 2
fig, ((ax1, ax2), (ax3,ax4)) = plt.subplots(2,2)
# size of the figure
fig.set_size_inches(12.5,10.5)
# 20 bins in the first subplot, normed histogram
n, bins, patches = ax1.hist(ensrainsorted, 20, normed = 1,
histtype = 'stepfilled', cumulative = True,
alpha = 0.15, color = 'purple')
ax1.set_xlabel('forecast rain (mm)')
ax1.set_ylabel('accumulated frequency')
ax1.set_title('number of bins = 20', fontsize = 10)
n, bins, patches = ax2.hist(ensrainsorted, 20, normed = 1,
histtype = 'stepfilled', cumulative = False,
alpha = 0.15, color = 'orange')
ax2.set_xlabel('forecast rain (mm)')
ax2.set_ylabel('frequency')
ax2.set_title('number of bins = 20', fontsize = 10)
n, bins, patches = ax3.hist(ensrainsorted, 10, normed = 1,
histtype = 'stepfilled', cumulative = True,
alpha = 0.15, color = 'purple')
ax3.set_xlabel('forecast rain (mm)')
ax3.set_ylabel('accumulated frequency')
ax3.set_title('number of bins = 10', fontsize = 10)
n, bins, patches = ax4.hist(ensrainsorted, 10, normed = 1,
histtype = 'stepfilled', cumulative = False,
alpha = 0.15, color = 'orange')
ax4.set_xlabel('forecast rain (mm)')
ax4.set_ylabel('frequency')
ax4.set_title('number of bins = 20', fontsize = 10)
plt.show()
plt.close()
# +
# histogram with two variables in matplotlib.
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
# create the histogram
# uses histogram2d from numpy
# need samples for both variables, number of bins, range of values
enstmax = fcast_vs_obs["ens_tmax"]
silotmax = fcast_vs_obs["silo_tmax"]
#hist, xedge, yedge = np.histogram2d(enstmax, silotmax, bins = 5, range = [[0,50],[0,50]])
# draw the empty histogram
fig = plt.figure()
ax = fig.add_subplot(111,projection='3d')
plt.show()
# +
# histogram with two variables in matplotlib.
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
# create the histogram
# uses histogram2d from numpy
# need samples for both variables, number of bins, range of values
enstmax = fcast_vs_obs["ens_tmax"]
silotmax = fcast_vs_obs["silo_tmax"]
#hist, xedge, yedge = np.histogram2d(enstmax, silotmax, bins = 20, range = [[0,50],[0,50]])
# draw the empty histogram
fig = plt.figure(figsize = (12,8))
ax = fig.add_subplot(111,projection='3d')
# add the inner workings
xpos, ypos = np.meshgrid(xedge[:-1] + 100/16, yedge[:-1] + 100/16)
xpos = xpos.flatten('F')
ypos = ypos.flatten('F')
zpos = np.zeros_like(xpos)
dx = 0.5 + np.ones_like(zpos) * 100/4
dy = dx.copy()
dz = hist.flatten()
ax.bar3d(xpos, ypos, zpos, dx, dy, dz, color = ['orange'], zsort = 'average', alpha = 0.75, edgecolor = 'black')
ax.set_xlabel('tmax predicted')
ax.set_ylabel('tmax observed')
ax.set_zlabel('frequency')
plt.show()
# +
# bivariate diagrams: bidimensional Kernel Density Estimation
# The bivariate distribution needs the two mean values for the marginal distribution and the covariance matrix.
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
from scipy import stats
dataset = fcast_vs_obs
enstmax = fcast_vs_obs["ens_tmax"]
silotmax = fcast_vs_obs["silo_tmax"]
xmin = enstmax.min()
xmax = enstmax.max()
ymin = silotmax.min()
ymax = silotmax.max()
#Perform a kernel density estimate on the data:
X, Y = np.mgrid[xmin:xmax:100j, ymin:ymax:100j]
#We will fit a gaussian kernel using the scipy’s gaussian_kde method:
positions = np.vstack([X.ravel(), Y.ravel()])
values = np.vstack([enstmax, silotmax])
kernel = stats.gaussian_kde(values)
Z = np.reshape(kernel(positions).T, X.shape)
#Plot the results:
fig = plt.figure(figsize=(8,8))
ax = fig.gca()
ax.set_xlim(xmin, xmax)
ax.set_ylim(ymin, ymax)
cfset = ax.contourf(X, Y, Z, cmap='coolwarm')
ax.imshow(np.rot90(Z), cmap='coolwarm', extent=[xmin, xmax, ymin, ymax])
cset = ax.contour(X, Y, Z, colors='k')
ax.clabel(cset, inline=1, fontsize=10)
ax.set_xlabel('forecast maximum temperature oC')
ax.set_ylabel('observed maximum temperature oC')
plt.title('2D Gaussian Kernel density estimation of forecast versus observed maximum temperatures')
# +
import numpy as np
from scipy.stats import entropy
p = [0.1, 0.9]
q = [0.1, 0.9]
def kl(p, q):
"""Kullback-Leibler divergence D(P || Q) for discrete distributions
Parameters
----------
p, q : array-like, dtype=float, shape=n
Discrete probability distributions.
"""
p = np.asarray(p, dtype=np.float)
q = np.asarray(q, dtype=np.float)
return np.sum(np.where(p != 0, p * np.log(p / q), 0))
def kl(p, q):
"""Kullback-Leibler divergence D(P || Q) for discrete distributions
Parameters
----------
p, q : array-like, dtype=float, shape=n
Discrete probability distributions.
"""
p = np.asarray(p, dtype=np.float)
q = np.asarray(q, dtype=np.float)
return np.sum(np.where(p != 0, q * np.log(q / p), 0))
p = [0.1, 0.9]
q = [0.1, 0.9]
assert entropy(p, q) == kl(p, q)
# +
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
sRain = fcast_vs_obs["silo_rain"]
eRain = fcast_vs_obs["ens_rain"]
sTMa = fcast_vs_obs["silo_tmax"]
eTMa = fcast_vs_obs["ens_tmax"]
sTMi = fcast_vs_obs["silo_tmin"]
eTMi = fcast_vs_obs["ens_tmin"]
sRad = fcast_vs_obs["silo_rad"]
eRad = fcast_vs_obs["ens_rad"]
sns.set_style('whitegrid')
ax1 = sns.kdeplot(eRain, bw=0, color = 'red', shade = 'true')
ax1 = sns.kdeplot(sRain, bw=0, color = 'green', shade = 'true')
plt.show()
sns.set_style('whitegrid')
ax2 = sns.kdeplot(eTMa, bw=0, color = 'blue', shade = 'true')
ax2 = sns.kdeplot(sTMa, bw=0, color = 'yellow', shade = 'true')
plt.show()
sns.set_style('whitegrid')
ax3 = sns.kdeplot(eTMi, bw=0, color = 'purple', shade = 'true')
ax3 = sns.kdeplot(sTMi, bw=0, color = 'pink', shade = 'true')
plt.show()
sns.set_style('whitegrid')
ax4 = sns.kdeplot(eRad, bw=0, color = 'brown', shade = 'true')
ax4 = sns.kdeplot(sRad, bw=0, color = 'orange', shade = 'true')
plt.show()
# -
sns.FacetGrid
| scripts/Eva2.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn import metrics
web = pd.read_csv('Desktop\data.csv')
web['id'] = range (0,len(web['price']))
web.head()
web.shape
web.describe()
web.info()
plt.figure(figsize =(6,6))
sns.heatmap(web.corr())
plt.show()
sns.scatterplot(x='sqft_living', y='price', data=web)
plt.show()
web.index
web.columns
X = web[['bedrooms','bathrooms','sqft_living','sqft_lot','floors','waterfront','view','condition','sqft_above','sqft_basement','yr_built','yr_renovated']]
y = web[['price']]
X.shape,y.shape
X_train,X_test, y_train,y_test = train_test_split(X,y, test_size=0.3, random_state=7)
X_train.shape,X_test.shape, y_train.shape, y_test.shape
mode = LinearRegression()
mode.fit(X_train, y_train)
prediction = mode.predict(X_test)
prediction
prediction[1]
prediction[2]
a1 = web[web['id']==1]
a1
a1['price']
mode.coef_
mode.intercept_
prediction.size
plt.figure(figsize=(6,5))
plt.scatter(y_test, prediction)
plt.show()
# ROOT MEAN SQUARED ERROR
Err = metrics.mean_squared_error(y_test, prediction)
Err
sap = np.sqrt(Err)
sap
X = web[['bedrooms','bathrooms','sqft_living','sqft_lot','floors','waterfront','view','condition','sqft_above','sqft_basement','yr_built','yr_renovated']]
y = web[['price']]
sns.boxplot(X)
plt.show()
sns.boxplot(y)
plt.show()
sns.heatmap(X.corr())
plt.show()
plt.plot(X,y,)
plt.xlabel('keys')
plt.ylabel('value')
plt.title('Corresponding Ratio')
plt.show()
| house price prediction.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import os
import os.path as osp
from glob import glob
import pandas as pd
import seaborn as sns
from sklearn.model_selection import train_test_split
from pylab import rcParams
import json
import matplotlib.pyplot as plt
rcParams['figure.figsize'] = 8, 5
# +
train = pd.read_json('../data/train.json')
val = pd.read_json('../data/val.json')
mapping = json.load(open('../data/class-mapper.json'))
train.head()
# -
sns.countplot(train['label'], order = train['label'].value_counts().index)
sns.countplot(val['label'], order = val['label'].value_counts().index)
a = 1 / train['label'].value_counts()
a = a / sum(a)
a
mapping
import numpy as np
weights = np.zeros(len(mapping))
for i in a.index:
weights[mapping[i]] = a[i]
weights
# # Visualization
import wfdb
from PIL import Image
ecg_data = sorted([osp.splitext(i)[0] for i in glob('../mit-bih/*.atr')])
ecg = ecg_data[0]
ann = wfdb.rdann(ecg, extension='atr')
record = wfdb.rdrecord(ecg)
plt.plot(record.p_signal[0:256])
wfdb.plot_wfdb(record)
Normal = [np.array(Image.open(i)) for i in glob('../data/2D/100/MLII/N/*.png')[:9]]
A = [np.array(Image.open(i)) for i in glob('../data/2D/100/MLII/A/*.png')[:9]]
Image.fromarray(np.vstack( [np.hstack(Normal[i::3]) for i in range(0,3)] ))
Image.fromarray(np.vstack( [np.hstack(A[i::3]) for i in range(0,3)] ))
| notebooks/eda.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import nltk
from nltk.corpus import stopwords
from nltk.cluster.util import cosine_distance
from nltk.tokenize import sent_tokenize
import numpy as np
import networkx as nx
import re
#Function to split text into sentences by fullstop(.)
'''def read_article(text):
article = text.split(". ")
sentences =[]
for sentence in article:
print(sentence)
sentences.append(sentence.replace("[^a-zA-Z]"," ").split(" "))
return sentences'''
# Read the text and tokenize into sentences
def read_article(text):
sentences =[]
sentences = sent_tokenize(text)
for sentence in sentences:
sentence.replace("[^a-zA-Z0-9]"," ")
return sentences
# Create vectors and calculate cosine similarity b/w two sentences
def sentence_similarity(sent1,sent2,stopwords=None):
if stopwords is None:
stopwords = []
sent1 = [w.lower() for w in sent1]
sent2 = [w.lower() for w in sent2]
all_words = list(set(sent1 + sent2))
vector1 = [0] * len(all_words)
vector2 = [0] * len(all_words)
#build the vector for the first sentence
for w in sent1:
if not w in stopwords:
vector1[all_words.index(w)]+=1
#build the vector for the second sentence
for w in sent2:
if not w in stopwords:
vector2[all_words.index(w)]+=1
return 1-cosine_distance(vector1,vector2)
# Create similarity matrix among all sentences
def build_similarity_matrix(sentences,stop_words):
#create an empty similarity matrix
similarity_matrix = np.zeros((len(sentences),len(sentences)))
for idx1 in range(len(sentences)):
for idx2 in range(len(sentences)):
if idx1!=idx2:
similarity_matrix[idx1][idx2] = sentence_similarity(sentences[idx1],sentences[idx2],stop_words)
return similarity_matrix
# Generate and return text summary
def generate_summary(text,top_n):
nltk.download('stopwords')
nltk.download('punkt')
stop_words = stopwords.words('english')
summarize_text = []
# Step1: read text and tokenize
sentences = read_article(text)
# Steo2: generate similarity matrix across sentences
sentence_similarity_matrix = build_similarity_matrix(sentences,stop_words)
# Step3: Rank sentences in similarirty matrix
sentence_similarity_graph = nx.from_numpy_array(sentence_similarity_matrix)
scores = nx.pagerank(sentence_similarity_graph)
#Step4: sort the rank and place top sentences
ranked_sentences = sorted(((scores[i],s) for i,s in enumerate(sentences)),reverse=True)
# Step 5: get the top n number of sentences based on rank
for i in range(top_n):
summarize_text.append(ranked_sentences[i][1])
# Step 6 : outpur the summarized version
return " ".join(summarize_text),len(sentences)
# -
doc = """It is, perhaps, dreadfully apt that an invasion which began 20 years ago as a counter-terrorism operation has ended in the horror of a mass casualty terrorist attack. The US-led attempt to destroy al-Qaida and rescue Afghanistan from the Taliban was undercut by the Iraq war, which spawned Islamic State. Now the circle is complete as an Afghan IS offshoot emerges as America’s new nemesis.
The Kabul airport atrocity shows just how difficult it is to break the cycle of violence, vengeance and victimisation. <NAME>’s swift vow to hunt down the perpetrators and “make them pay” presumably means US combat forces will again be in action in Afghanistan soon. If the past is any guide, mistakes will be made, civilians will die, local communities will be antagonised. Result: more terrorists.
It is an obvious irony that US military chiefs in Kabul are collaborating with the Taliban, their sworn enemy, against the common IS foe as the evacuation ends. This suggests negotiators, on both sides, could have tried harder to reach a workable peace deal. It may augur well for future cooperation, for example on humanitarian aid. But the Taliban has many faces – and many cannot be trusted.
Last week’s events have raised yet more questions about Biden’s judgment and competence. He will be blamed personally. His predicament recalls the downfall of another Democratic president, <NAME>. After the disastrous failure of Operation Eagle Claw to rescue US hostages in Tehran in April 1980, Carter was voted out of office the following November.
Biden faces Republican calls to resign. His approval ratings have plunged. But he defiantly insists that quitting Afghanistan is the right thing to do. Polls suggest most Americans agree, though they are critical of how it has been managed. Unlike in Carter’s time, the next presidential election is three years away. By then the agony and humiliations of recent days may be a distant memory.
The Kabul debacle also casts doubt on Biden’s new counter-terrorism strategy, which reportedly downgrades the threat posed by Islamist terrorism to the US. His national security team wants to shift global priorities and resources to meet different, 21st-century challenges to American hegemony, such as China, cyberwarfare and the climate crisis.
Biden is said to want to use the 20th anniversary of the 11 September al-Qaida attacks on New York and Washington to declare America’s “forever wars” over – for which he will claim credit. Setting the Afghan shambles aside, he is expected to say the era of invasion, occupation, nation-building and the “global war on terror” is at an end. “The US approach should centre on gathering intelligence, training indigenous forces, and maintaining air power as well as special forces capabilities for the occasional strike when necessary,” foreign policy analysts <NAME> and <NAME> argued recently.
No one knows whether such a costly, hard to organise strategy will work in the long term. But the shift is already having tangible consequences. In Iraq, for example, US combat operations will cease in December. About 2,500 Americans will stay, to train and advise. In Syria, a small number of special forces will remain. Iraqis understandably worry about an IS comeback and an Afghan-style implosion. The same story of US disengagement and drawback is heard across the Middle East as the US “pivots” to Asia. Combat aircraft are being redeployed, carrier battlegroups may be reassigned to the Pacific theatre, and anti-missile batteries are being withdrawn from Iraq, Kuwait, Jordan and Saudi Arabia. Most of these assets were pointed at Iran, deemed a prime sponsor of terrorism.
In the Sahel, west Africa, the Democratic Republic of the Congo and Mozambique, the US barely registers in the fight against Boko Haram and assorted IS and al-Qaida affiliates. The impressively named US Africa Command is headquartered in Stuttgart. President <NAME> warns that Nigeria could suffer a similar fate to Afghanistan without a “comprehensive partnership” with the US. “Some sense the west is losing its will for the fight,” he said. For US allies, all this points to a new era of enforced self-sufficiency and greater uncertainty. While Islamist-inspired attacks in the US have been rare since 9/11, in Europe many hundreds have died. Yet collective European counter-terrorism efforts often lack a military cutting edge. An exception was France’s ill-supported Operation Barkhane in Mali – until it was halted this year after suffering many casualties for little gain.
The chaos in Afghanistan has vividly dramatised the ongoing threat from international terrorism. With up to 10,000 foreign Islamist fighters in the country, according to the UN, fears grow it will again become a launchpad for global jihad. So the prospect of a less directly engaged, homeland-focused American counter-terrorism approach is alarming for partners dependent on US leadership and protection.
European Nato allies, sniping at Biden, are in denial. They don’t want to admit his Afghan withdrawal is just the start of something bigger. And as recent events painfully demonstrate, the UK is not remotely able to fend for itself."""
generate_summary(doc,top_n=5)
| Text Summarization.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# # DS Automation Assignment
# Using our prepared churn data from week 2:
# - use pycaret to find an ML algorithm that performs best on the data
# - Choose a metric you think is best to use for finding the best model; by default, it is accuracy but it could be AUC, precision, recall, etc. The week 3 FTE has some information on these different metrics.
# - save the model to disk
# - create a Python script/file/module with a function that takes a pandas dataframe as an input and returns the probability of churn for each row in the dataframe
# - your Python file/function should print out the predictions for new data (new_churn_data.csv)
# - the true values for the new data are [1, 0, 0, 1, 0] if you're interested
# - test your Python module and function with the new data, new_churn_data.csv
# - write a short summary of the process and results at the end of this notebook
# - upload this Jupyter Notebook and Python file to a Github repository, and turn in a link to the repository in the week 5 assignment dropbox
#
# *Optional* challenges:
# - return the probability of churn for each new prediction, and the percentile where that prediction is in the distribution of probability predictions from the training dataset (e.g. a high probability of churn like 0.78 might be at the 90th percentile)
# - use other autoML packages, such as TPOT, H2O, MLBox, etc, and compare performance and features with pycaret
# - create a class in your Python module to hold the functions that you created
# - accept user input to specify a file using a tool such as Python's `input()` function, the `click` package for command-line arguments, or a GUI
# - Use the unmodified churn data (new_unmodified_churn_data.csv) in your Python script. This will require adding the same preprocessing steps from week 2 since this data is like the original unmodified dataset from week 1.
# LOAD DATA:
# +
import pandas as pd
# reading csv file into data frame and naming the index column
df = pd.read_csv('prepped_churn_data.csv', index_col='customerID')
df
# -
del df['yj_tenure']
del df['Total_tenure_ratio']
# AutoML WITH PYCARET:
pip install pycaret
pip install scikit-plot
#importing specific functions from pycaret
from pycaret.classification import setup, compare_models, predict_model, save_model, load_model
#setup function must be called before execution of any other function
automl = setup(df, target='Churn', fold_shuffle=True, preprocess=False)#setup() takes two important parameters like data and target
automl[7]
#compare_models() evaluates the performance of all models using cross-validation
best_model = compare_models(sort='F1')#here sort is used to consider 'F1' as the scoring metric(by default accuracy)
#used to display the best model
best_model
#iloc() function is used to fetch records based on the index values from the dataset
df.iloc[-1].shape #df.shape() gives the number of rows and columns(returns 1-D array)
#returns 2-D array because -2:-1 represents a range(it represents line)
df.iloc[-2:-1].shape
#predicts label and score using a trained model
predict_model(best_model, df.iloc[-2:-1])
# SAVING AND LOADING OUR MODEL:
#save_model() is used to save our trained model( or best model)
save_model(best_model, 'QDA')
#pickle module is used for saving and loading the data
import pickle
#opens the file specified for writing and in binary format
with open('QDA_model.pk', 'wb') as f: #this file object is saved in f variable
pickle.dump(best_model, f)
#opens specified file for reading and in binary format
with open('QDA_model.pk', 'rb') as f: #this file object is saved in f variable
loaded_model = pickle.load(f)
new_data = df.iloc[-2:-1].copy()#copy() is used to generate a copy of the object
new_data.drop('Churn', axis=1, inplace=True)#inplace=True returns nothing and drops the Churn column and updates the data
loaded_model.predict(new_data)
#load_model() is used for loading the trained model
loaded_nb = load_model('QDA')
#predict_model() is used for making predictions on new_data
predict_model(loaded_nb, new_data)
# MAKING A PYTHON MODULE TO MAKE PREDICTIONS:
from IPython.display import Code
#python file is created in visual studio for predicting churn for each row on the new_churn_data
Code('predict_churn.py')
#predictions on the new data can be seen by using this magic command %run
# %run predict_churn.py
# # Summary
# Write a short summary of the process and results here.
# Week-5 Assignment is about using prepped churn data for setting up autoML with pycaret which shows the summary of the entire project, comparing best metric among many using cross-validation, predicting label and score for the best model, saving and loading our model using python built-in module pickle, making predictions on the new data created by using iloc() function and a python file is created for making predictions on the new churn data. I have imported package like pandas and read prepped churn data into dataframe by using read_csv function.
#
# 1.Before setting an autoML with pycaret, I installed python libraries like pycaret and scikit-plot using pip and imported specific functions like setup, compare_models, predict_model, save_model, load_model from pycaret. setup() function initializes the training environment and creates the transformation pipeline. Pycaret setup is initialized by using setup() function which takes two mandatory parameters like data and target. Here, data is prepped_churn_data which is read into pandas dataframe and target is Churn. All other parameters are optional. When Preprocess is set to False, no transformations are applied except for train_test_split and data must be ready for modeling. fold_shuffle=True controls the shuffle parameter of cross validation(CV). fold_shuffle parameter is by default false. The preprocessing and data transformations are configured within the setup function.
#
# 2.AutoML is simply run for finding the best model using compare_models() function. This function evaluates the performance of all metrics available in the model library using cross validation and finds out the best model. sort argument in compare_models is used to choose any other scoring metric, by default it is accuracy. I sorted the table based on the F1 score. Based on the output, the best model is QuadraticDiscriminantAnalysis(qda) which has the highest F1 score(i.e., 61%) and the time taken to run this model is the least while xgboost and lightgbm take longest time to run. best_model is used to display the best model. I tried other metrics too for finding the best model but the score(0.9) and the predictions for the new_churn_data are better using F1 score as the metric.
#
# 3.iloc() function is used to fetch records based on the index values from the datasets. It enables us to retrieve a particular value belonging to a row and column using the index values assigned to it. iloc() function only accepts integer type values as the index values. df.shape() gives the number of rows and columns. df.iloc[start row:end row, start_col:end_col] where end row and end col are excluded. The index values [-2:-1] represents a range, while [-1] is a scalar and a range is a line while a scalar is a point. df.iloc[-1].shape returns 1-D array and df.iloc[-2:-1].shape returns 2-D array. predict_model() function is used to make predictions on the new data (i.e., which is fetched by using iloc() function) by using trained model(i.e., best model(QDA)). This function creates two new columns Label and Score. Label column has its predicted label(1) and Score if it is greater than or equal to 0.5 it is rounded up. The Score is 0.9 and the label is 1 for the new data(i.e., fetched by using iloc()).
#
# 4.The function save_model() in pycaret is used to save our trained model(i.e., QDA). The python built-in module pickle is also used for saving and loading of binary data. I imported pickle and the built-in open function opens a file with the name QDA_model.pk for writing with 'w' and in a binary format using 'b'. The file object is saved in the variable f. The with statement automatically closes the file after the with statement exit , otherwise the function close from the file object f should be called. The data is saved to the file using pickle.
#
# 5.The built-in open function opens a file with the name QDA_model.pk for reading with 'r' and in a binary format using 'b'. The file object is saved in the variable f. The with statement automatically closes the file after the with statement exit , otherwise the function close from the file object f should be called. The pickle's load function(i.e.,pickle.load()) is used for loading the saved data. The new_data is fetched by using iloc() function and the churn column is dropped by using drop() method. axis=0 represents rows and axis=1 represents columns. axis is set to 1 to delete Churn column. copy() function is used to generate a copy of the object. inplace=True does not return anything does the specified operation(deletes the Churn column) and updates the data. inplace=False(by default) returns the copy of the object, the specific operation is performed and then it should be saved to some file. The QDA model which is loaded using pickle load function is used for making predictions on the new_data.
#
# 6.Saving and loading of data can also be done by using pycaret. save_model() in pycaret is used for saving the trained model. load_model() function is used for loading the trained model. The function predict_model() is used for making predictions on new_data by using trained model(i.e., QDA). The score for the new_data is around 0.9 which is good and label is 1(two new columns created by using predict_model function).
#
# 7.I imported code from Ipython.display class for displaying source code. A python file 'predict_churn.py' is created in visual studio for predicting churn for each row on the new_churn_data. The source code in this file is displayed by using Code() from Ipython display module. The magic command %run is used to run the file predict_churn.py and display the predictions on the new_churn_data. The predictions for the new_churn_data is Yes, No, No, No, No(1,0,0,0,0). The true values for the new_churn_data is 1,0,0,1,0. There is one false negative in the new data. The model(QDA) is working average but not perfect.
| Week_5_assignment_starter.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import numpy as np
from scipy.stats import binom
k = np.array([i for i in range(13)])
n_k = np.array([3, 24, 104, 286, 670, 1033, 1343, 1112, 829, 478, 181, 45, 7])
display(k)
n_k
n = len(k) - 1
N = np.sum(n_k)
n, N
p_k = n_k / N
theta_hat = np.sum(k * p_k) / n
theta_hat
e_k = N * binom.pmf(k, n, theta_hat)
np.round(e_k, 0)
r_k = (n_k - e_k) / np.sqrt(e_k)
np.round(r_k, 1)
chi_sq = np.sum(r_k**2)
np.round(chi_sq, 1)
| python/chapter-4/EX4-3.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
#Some Basic Imports
# first neural network with keras tutorial
from numpy import loadtxt
from keras.models import Sequential
from keras.layers import Dense,Input
import pandas as pd
from sklearn.model_selection import train_test_split
import pandas as pd
import numpy as np
#pd.set_option('display.max_columns', 50)
from scipy.spatial import cKDTree
np.random.seed(123)
import urllib, os
import matplotlib.pyplot as plt
# %matplotlib inline
import datetime
from sklearn.preprocessing import LabelBinarizer
import glob
import cv2
from tensorflow.keras.layers import Input, Conv2D, BatchNormalization, Dense
from tensorflow.keras.layers import AvgPool2D, GlobalAveragePooling2D, MaxPool2D
from tensorflow.keras.models import Model
from tensorflow.keras.layers import ReLU, concatenate
import tensorflow.keras.backend as K
from keras.models import Sequential, Model
from keras.layers.normalization import BatchNormalization
from keras.layers.convolutional import Conv2D, MaxPooling2D
from keras.layers.core import Activation, Dropout, Dense
from keras.layers import Flatten, Input, concatenate
from sklearn.model_selection import train_test_split
from keras.optimizers import Adam, RMSprop
import argparse
import locale
from keras.models import load_model
from sklearn.datasets import load_files
from keras.utils import np_utils
from glob import glob
from keras import applications
from keras.preprocessing.image import ImageDataGenerator
from keras import optimizers
from keras.models import Sequential,Model,load_model
from keras.layers import Dense, Dropout, Flatten, Conv2D, MaxPool2D,GlobalAveragePooling2D
from keras.callbacks import TensorBoard,ReduceLROnPlateau,ModelCheckpoint
from keras.preprocessing.image import ImageDataGenerator
from sklearn.metrics import confusion_matrix, classification_report
import itertools
from keras import regularizers
from IPython.display import SVG
from keras.utils.vis_utils import model_to_dot
from keras.utils import plot_model
from keras.models import load_model
import matplotlib.pyplot as plt #For Visualization
import numpy as np #For handling arrays
import pandas as pd # For handling data
import tensorflow as tf
gpu_devices = tf.config.experimental.list_physical_devices('GPU')
for device in gpu_devices: tf.config.experimental.set_memory_growth(device, True)
#Define Directories for train, test & Validation Set
image_folder = 'dataset2/'
image_generator = ImageDataGenerator(rescale=1./255).flow_from_directory(
image_folder, shuffle=False, class_mode='binary',color_mode='grayscale',
target_size=(299, 299), batch_size=437)
# -
image_generator.class_indices
images, labels = next(image_generator)
from sklearn.model_selection import train_test_split
# Using train_test_split to partition the training and testing structured data attributes and images
(trainImagesX,testImagesX,trainImagesY,testImagesY) = train_test_split(images,labels,test_size=0.25, random_state=32)
img_height,img_width = 299,299
num_classes = 2
#If imagenet weights are being loaded,
#input must have a static square shape (one of (128, 128), (160, 160), (192, 192), or (224, 224))
base_model = applications.resnet50.ResNet50(weights= None, include_top=False, input_shape= (img_height,img_width,1))
x = base_model.output
x = GlobalAveragePooling2D()(x)
x = Dropout(0.7)(x)
x = Dense(8,activation='relu')(x)
predictions = Dense(1, activation= 'sigmoid')(x)
model = Model(inputs = base_model.input, outputs = predictions)
model.summary()
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense,Conv2D,Flatten,MaxPooling2D
from tensorflow.keras.callbacks import EarlyStopping,ReduceLROnPlateau
early = EarlyStopping(monitor="val_loss", mode="min", patience=4)
learning_rate_reduction = ReduceLROnPlateau(monitor='val_loss', patience = 3, verbose=1,factor=0.3, min_lr=0.000001)
callbacks_list = [ early, learning_rate_reduction]
from keras.optimizers import Adam, RMSprop
opt = Adam(lr=1e-3, decay=1e-3 / 200)
model.compile(loss="binary_crossentropy", metrics=['acc'], optimizer=opt)
#cnn.compile(optimizer = 'adam', loss = 'sparse_categorical_crossentropy', metrics = ['accuracy'])
model.summary()
model1_history = model.fit(trainImagesX, trainImagesY, validation_data=(testImagesX, testImagesY), epochs=50, batch_size=16,callbacks=callbacks_list)
model.save('models/mixed_model_resnetonly_v1.h5')
def show_cf(y_true, y_pred, class_names, model_name=None):
"""Plots a confusion matrix"""
cf = confusion_matrix(y_true, y_pred)
plt.imshow(cf, cmap=plt.cm.Blues)
if model_name:
plt.title("Confusion Matrix: {}".format(model_name))
else:
plt.title("Confusion Matrix")
plt.ylabel('True Label')
plt.xlabel('Predicted Label')
class_names = set(y_true)
tick_marks = np.arange(len(class_names))
if class_names:
plt.xticks(tick_marks, class_names)
plt.yticks(tick_marks, class_names)
thresh = cf.max() / 2.
for i, j in itertools.product(range(cf.shape[0]), range(cf.shape[1])):
plt.text(j, i, cf[i, j], horizontalalignment='center', color='white' if cf[i, j] > thresh else 'black')
plt.colorbar()
def cnn_evaluation(model, history, train_images, train_labels, test_images, test_labels, class_names, model_name=None):
"""
Evaluates the performance of a CNN with loss and accuracy plots, a confusion matrix and a classification report for the training and test sets.
"""
train_acc = history.history['acc']
val_acc = history.history['val_acc']
train_loss = history.history['loss']
val_loss = history.history['val_loss']
epch = range(1, len(train_acc) + 1)
plt.plot(epch, train_acc, 'g.', label='Training Accuracy')
plt.plot(epch, val_acc, 'g', label='Validation acc')
plt.title('Accuracy')
plt.legend()
plt.figure()
plt.plot(epch, train_loss, 'r.', label='Training loss')
plt.plot(epch, val_loss, 'r', label='Validation loss')
plt.title('Loss')
plt.legend()
plt.show()
results_test = model.evaluate(test_images, test_labels)
print('Test Loss:', results_test[0])
print('Test Accuracy:', results_test[1])
y_train_pred = np.round(model.predict(train_images))
y_pred = np.round(model.predict(test_images))
show_cf(test_labels, y_pred, class_names=class_names, model_name=model_name)
print(classification_report(train_labels, y_train_pred))
print(classification_report(test_labels, y_pred))
import matplotlib.pyplot as plt
# %matplotlib inline
cnn_evaluation(model, model1_history, trainImagesX,trainImagesY, testImagesX,testImagesY, class_names=['covid', 'no findings'])
| Templates/resnet50_only.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/AIWintermuteAI/aXeleRate/blob/master/resources/aXeleRate_standford_dog_classifier.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="hS9yMrWe02WQ" colab_type="text"
# ## Standford Dog Breed Classification model Training and Inference
#
# In this notebook we will use axelerate Keras-based framework for AI on the edge to quickly setup model training and then after training session is completed convert it to .tflite and .kmodel formats.
#
# First, let's take care of some administrative details.
#
# 1) Before we do anything, make sure you have choosen GPU as Runtime type (in Runtime - > Change Runtime type).
#
# 2) We need to mount Google Drive for saving our model checkpoints and final converted model(s). Press on Mount Google Drive button in Files tab on your left.
#
# In the next cell we clone axelerate Github repository and import it.
#
# **It is possible to use pip install or python setup.py install, but in that case you will need to restart the enironment.** Since I'm trying to make the process as streamlined as possibile I'm using sys.path.append for import.
# + id="y07yAbYbjV2s" colab_type="code" colab={}
# %tensorflow_version 1.x
#we need imgaug 0.4 for image augmentations to work properly, see https://stackoverflow.com/questions/62580797/in-colab-doing-image-data-augmentation-with-imgaug-is-not-working-as-intended
# !pip uninstall -y imgaug && pip uninstall -y albumentations && pip install imgaug==0.4
# !git clone https://github.com/AIWintermuteAI/aXeleRate.git
import sys
sys.path.append('/content/aXeleRate')
from axelerate import setup_training,setup_inference
# + [markdown] id="5TBRMPZ83dRL" colab_type="text"
# At this step you typically need to get the dataset. You can use !wget command to download it from somewhere on the Internet or !cp to copy from My Drive as in this example
# ```
# # # !cp -r /content/drive/'My Drive'/pascal_20_segmentation.zip .
# # # !unzip --qq pascal_20_segmentation.zip
# ```
# For this notebook we will use Standford Dog Breed Classification dataset for fine-grained classification, which you can download here:
# http://vision.stanford.edu/aditya86/ImageNetDogs/
#
# In the next cell we will download the same dataset, but with training/validation split already done - I shared on my Google Drive. We will also download pre-trained model to demonstrate inference results.
#
# Let's visualize our classification validation dataset with visualize_dataset function, which will search for all images in folder and display num_imgs number of images with class overlayer over the image.
#
# + id="_tpsgkGj7d79" colab_type="code" colab={}
# %matplotlib inline
# !gdown https://drive.google.com/uc?id=1qq758Tjsfm7Euu9ev7hSyLkMj63YC9ST #dog breed classification dataset
# !gdown https://drive.google.com/uc?id=1dFnDCOxws2uX4ZpauSPC6r6jdjHoJw_p #pre-trained model
# !unzip --qq dogs_classification.zip
from axelerate.networks.common_utils.augment import visualize_classification_dataset
visualize_classification_dataset('dogs_classification/imgs_validation', num_imgs=10, img_size=224, jitter=True)
# + [markdown] id="S1oqdtbr7VLB" colab_type="text"
# Next step is defining a config dictionary. Most lines are self-explanatory.
#
# Type is model frontend - Classifier, Detector or Segnet
#
# Architecture is model backend (feature extractor)
#
# - Full Yolo
# - Tiny Yolo
# - MobileNet1_0
# - MobileNet7_5
# - MobileNet5_0
# - MobileNet2_5
# - SqueezeNet
# - NASNetMobile
# - DenseNet121
# - ResNet50
#
# **Note that while you can train any network type with any backend (Tiny YOLO + Classifier, NASNETMobile + Detector, DenseNet121 + Segnet and so on), some converters do not support larger networks! E.g. K210 converter only supports MobileNet and TinyYOLO backends.**
#
# Fully_connected is number of neurons in classification layers as list.
#
# Dropout value is dropout in classification layers.
#
# actual_epoch is number of epochs to train, noramlly good starting value is 50 - 100
#
# train_times is a multiplier for training dataset, i.e. how many times to repeat the dataset during one epoch. Useful when you apply augmentations to image. Normally between 1 and 3 is okay. If you have big dataset, can leave at 1.
#
# For converter type you can choose the following:
#
# 'k210', 'tflite_fullint', 'tflite_dynamic', 'edgetpu', 'openvino', 'onnx'
#
# **Since it is an example notebook, we will use pretrained weights and set all layers of the model to be "frozen"(non-trainable), except for the last one. Also we set learning rate to very low value, that will allow us to see the perfomance of pretrained model**
# + id="Jw4q6_MsegD2" colab_type="code" colab={}
config = {
"model" : {
"type": "Classifier",
"architecture": "NASNetMobile",
"input_size": 224,
"fully-connected": [],
"labels": [],
"dropout" : 0.2
},
"weights" : {
"full": "/content/Classifier_best_val_accuracy.h5",
"backend": "imagenet",
"save_bottleneck": False
},
"train" : {
"actual_epoch": 1,
"train_image_folder": "dogs_classification/imgs",
"train_times": 1,
"valid_image_folder": "dogs_classification/imgs_validation",
"valid_times": 1,
"valid_metric": "val_accuracy",
"batch_size": 16,
"learning_rate": 1e-6,
"saved_folder": F"/content/drive/My Drive/dogs_classifier",
"first_trainable_layer": "dense_1",
"augumentation": True
},
"converter" : {
"type": []
}
}
# + [markdown] id="kobC_7gd5mEu" colab_type="text"
# Let's check what GPU we have been assigned in this Colab session, if any.
# + id="rESho_T70BWq" colab_type="code" colab={}
from tensorflow.python.client import device_lib
device_lib.list_local_devices()
# + [markdown] id="cWyKjw-b5_yp" colab_type="text"
# Finally we start the training by passing config dictionary we have defined earlier to setup_training function. The function will start the training with Checkpoint, Reduce Learning Rate on Plateu and Early Stopping callbacks. Every time our validation metric(in this config set to "val_accuracy") improves, the model is saved with Checkpoint callback. If you have specified the converter type in the config, after the training has stopped the script will convert the best model into the format you have specified in config and save it to the project folder.
# + id="deYD3cwukHsj" colab_type="code" colab={}
from keras import backend as K
K.clear_session()
model_path = setup_training(config_dict=config)
# + [markdown] id="ypTe3GZI619O" colab_type="text"
# After training it is good to check the actual perfomance of your model by doing inference on your validation dataset and visualizing results. This is exactly what next block does. Our model used pre-trained weights and since all the layers,except for the last one were set as non-trainable and we set the learning rate to a very low value, we are just observing the perfomance of the model that was trained before.
# + id="jE7pTYmZN7Pi" colab_type="code" colab={}
# %matplotlib inline
from keras import backend as K
K.clear_session()
setup_inference(config, model_path)
# + [markdown] id="PF__ooBsyb58" colab_type="text"
# If you need to convert trained model to other formats, for example for inference with Edge TPU or Kendryte K210, you can do it with following commands. Specify the converter type, backend and folder with calbiration images(normally your validation image folder).
# + id="fGNqUf1Gyc4z" colab_type="code" colab={}
from axelerate.networks.common_utils.convert import Converter
converter = Converter('tflite_dynamic', 'NASNetMobile', 'dogs_classification/imgs_validation')
converter.convert_model(model_path)
# + [markdown] id="fn7H0V4SEOd_" colab_type="text"
# To train the model from scratch use the following config and then run the cells with training and (optinally) inference functions again.
# + id="oT87SwQ6EQB8" colab_type="code" colab={}
config = {
"model" : {
"type": "Classifier",
"architecture": "NASNetMobile",
"input_size": 224,
"fully-connected": [],
"labels": [],
"dropout" : 0.2
},
"weights" : {
"full": "",
"backend": "imagenet",
"save_bottleneck": False
},
"train" : {
"actual_epoch": 50,
"train_image_folder": "dogs_classification/imgs",
"train_times": 1,
"valid_image_folder": "dogs_classification/imgs_validation",
"valid_times": 1,
"valid_metric": "val_accuracy",
"batch_size": 16,
"learning_rate": 1e-3,
"saved_folder": F"/content/drive/My Drive/dogs_classifier",
"first_trainable_layer": "",
"augumentation": True
},
"converter" : {
"type": ["tflite_dynamic"]
}
}
# + id="NQjvas2UEe8l" colab_type="code" colab={}
from keras import backend as K
K.clear_session()
model_path = setup_training(config_dict=config)
# + id="iJJWjuRaEfkj" colab_type="code" colab={}
# %matplotlib inline
from keras import backend as K
K.clear_session()
setup_inference(config, model_path)
# + [markdown] id="5YuVe2VD11cd" colab_type="text"
# Good luck and happy training! Have a look at these articles, that would allow you to get the most of Google Colab or connect to local runtime if there are no GPUs available;
#
# https://medium.com/@oribarel/getting-the-most-out-of-your-google-colab-2b0585f82403
#
# https://research.google.com/colaboratory/local-runtimes.html
| resources/aXeleRate_standford_dog_classifier.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: tensorflow2_py37
# language: python
# name: tensorflow2_py37
# ---
# ## <font color="green">Tobigs14 이혜린</font>
# # CS224N Assignment 1: Exploring Word Vectors (25 Points)
#
# Welcome to CS224n!
#
# Before you start, make sure you read the README.txt in the same directory as this notebook.
# +
# All Import Statements Defined Here
# Note: Do not add to this list.
# All the dependencies you need, can be installed by running .
# ----------------
import sys
assert sys.version_info[0]==3
assert sys.version_info[1] >= 5
from gensim.models import KeyedVectors
from gensim.test.utils import datapath
import pprint
import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = [10, 5]
import nltk
nltk.download('reuters')
from nltk.corpus import reuters
import numpy as np
import random
import scipy as sp
from sklearn.decomposition import TruncatedSVD
from sklearn.decomposition import PCA
START_TOKEN = '<START>'
END_TOKEN = '<END>'
np.random.seed(0)
random.seed(0)
# ----------------
# -
# ## Please Write Your SUNet ID Here: 투빅스 14기 이혜린,,,
# ## Word Vectors
#
# Word Vectors are often used as a fundamental component for downstream NLP tasks, e.g. question answering, text generation, translation, etc., so it is important to build some intuitions as to their strengths and weaknesses. Here, you will explore two types of word vectors: those derived from *co-occurrence matrices*, and those derived via *word2vec*.
#
# **Assignment Notes:** Please make sure to save the notebook as you go along. Submission Instructions are located at the bottom of the notebook.
#
# **Note on Terminology:** The terms "word vectors" and "word embeddings" are often used interchangeably. The term "embedding" refers to the fact that we are encoding aspects of a word's meaning in a lower dimensional space. As [Wikipedia](https://en.wikipedia.org/wiki/Word_embedding) states, "*conceptually it involves a mathematical embedding from a space with one dimension per word to a continuous vector space with a much lower dimension*".
# ## Part 1: Count-Based Word Vectors (10 points)
#
# Most word vector models start from the following idea:
#
# *You shall know a word by the company it keeps ([<NAME>. 1957:11](https://en.wikipedia.org/wiki/John_Rupert_Firth))*
#
# Many word vector implementations are driven by the idea that similar words, i.e., (near) synonyms, will be used in similar contexts. As a result, similar words will often be spoken or written along with a shared subset of words, i.e., contexts. By examining these contexts, we can try to develop embeddings for our words. With this intuition in mind, many "old school" approaches to constructing word vectors relied on word counts. Here we elaborate upon one of those strategies, *co-occurrence matrices* (for more information, see [here](http://web.stanford.edu/class/cs124/lec/vectorsemantics.video.pdf) or [here](https://medium.com/data-science-group-iitr/word-embedding-2d05d270b285)).
# ### Co-Occurrence
#
# A co-occurrence matrix counts how often things co-occur in some environment. Given some word $w_i$ occurring in the document, we consider the *context window* surrounding $w_i$. Supposing our fixed window size is $n$, then this is the $n$ preceding and $n$ subsequent words in that document, i.e. words $w_{i-n} \dots w_{i-1}$ and $w_{i+1} \dots w_{i+n}$. We build a *co-occurrence matrix* $M$, which is a symmetric word-by-word matrix in which $M_{ij}$ is the number of times $w_j$ appears inside $w_i$'s window.
#
# **Example: Co-Occurrence with Fixed Window of n=1**:
#
# Document 1: "all that glitters is not gold"
#
# Document 2: "all is well that ends well"
#
#
# | * | START | all | that | glitters | is | not | gold | well | ends | END |
# |----------|-------|-----|------|----------|------|------|-------|------|------|-----|
# | START | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
# | all | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
# | that | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 |
# | glitters | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
# | is | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 |
# | not | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
# | gold | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
# | well | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 |
# | ends | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
# | END | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
#
# **Note:** In NLP, we often add START and END tokens to represent the beginning and end of sentences, paragraphs or documents. In thise case we imagine START and END tokens encapsulating each document, e.g., "START All that glitters is not gold END", and include these tokens in our co-occurrence counts.
#
# The rows (or columns) of this matrix provide one type of word vectors (those based on word-word co-occurrence), but the vectors will be large in general (linear in the number of distinct words in a corpus). Thus, our next step is to run *dimensionality reduction*. In particular, we will run *SVD (Singular Value Decomposition)*, which is a kind of generalized *PCA (Principal Components Analysis)* to select the top $k$ principal components. Here's a visualization of dimensionality reduction with SVD. In this picture our co-occurrence matrix is $A$ with $n$ rows corresponding to $n$ words. We obtain a full matrix decomposition, with the singular values ordered in the diagonal $S$ matrix, and our new, shorter length-$k$ word vectors in $U_k$.
#
# 
#
# This reduced-dimensionality co-occurrence representation preserves semantic relationships between words, e.g. *doctor* and *hospital* will be closer than *doctor* and *dog*.
#
# **Notes:** If you can barely remember what an eigenvalue is, here's [a slow, friendly introduction to SVD](https://davetang.org/file/Singular_Value_Decomposition_Tutorial.pdf). If you want to learn more thoroughly about PCA or SVD, feel free to check out lectures [7](https://web.stanford.edu/class/cs168/l/l7.pdf), [8](http://theory.stanford.edu/~tim/s15/l/l8.pdf), and [9](https://web.stanford.edu/class/cs168/l/l9.pdf) of CS168. These course notes provide a great high-level treatment of these general purpose algorithms. Though, for the purpose of this class, you only need to know how to extract the k-dimensional embeddings by utilizing pre-programmed implementations of these algorithms from the numpy, scipy, or sklearn python packages. In practice, it is challenging to apply full SVD to large corpora because of the memory needed to perform PCA or SVD. However, if you only want the top $k$ vector components for relatively small $k$ — known as *[Truncated SVD](https://en.wikipedia.org/wiki/Singular_value_decomposition#Truncated_SVD)* — then there are reasonably scalable techniques to compute those iteratively.
# ### Plotting Co-Occurrence Word Embeddings
#
# Here, we will be using the Reuters (business and financial news) corpus. If you haven't run the import cell at the top of this page, please run it now (click it and press SHIFT-RETURN). The corpus consists of 10,788 news documents totaling 1.3 million words. These documents span 90 categories and are split into train and test. For more details, please see https://www.nltk.org/book/ch02.html. We provide a `read_corpus` function below that pulls out only articles from the "crude" (i.e. news articles about oil, gas, etc.) category. The function also adds START and END tokens to each of the documents, and lowercases words. You do **not** have perform any other kind of pre-processing.
def read_corpus(category="crude"):
""" Read files from the specified Reuter's category.
Params:
category (string): category name
Return:
list of lists, with words from each of the processed files
"""
files = reuters.fileids(category)
return [[START_TOKEN] + [w.lower() for w in list(reuters.words(f))] + [END_TOKEN] for f in files]
# Let's have a look what these documents are like….
reuters_corpus = read_corpus()
pprint.pprint(reuters_corpus[:3], compact=True, width=100)
# ### Question 1.1: Implement `distinct_words` [code] (2 points)
#
# Write a method to work out the distinct words (word types) that occur in the corpus. You can do this with `for` loops, but it's more efficient to do it with Python list comprehensions. In particular, [this](https://coderwall.com/p/rcmaea/flatten-a-list-of-lists-in-one-line-in-python) may be useful to flatten a list of lists. If you're not familiar with Python list comprehensions in general, here's [more information](https://python-3-patterns-idioms-test.readthedocs.io/en/latest/Comprehensions.html).
#
# You may find it useful to use [Python sets](https://www.w3schools.com/python/python_sets.asp) to remove duplicate words.
def distinct_words(corpus):
""" Determine a list of distinct words for the corpus.
Params:
corpus (list of list of strings): corpus of documents
Return:
corpus_words (list of strings): list of distinct words across the corpus, sorted (using python 'sorted' function)
num_corpus_words (integer): number of distinct words across the corpus
"""
corpus_words = []
num_corpus_words = -1
# ------------------
# Write your implementation here.
corpus_words = sorted(list(set([w for line in corpus for w in line])))
num_corpus_words = len(corpus_words)
# ------------------
return corpus_words, num_corpus_words
# +
# ---------------------
# Run this sanity check
# Note that this not an exhaustive check for correctness.
# ---------------------
# Define toy corpus
test_corpus = ["START All that glitters isn't gold END".split(" "), "START All's well that ends well END".split(" ")]
test_corpus_words, num_corpus_words = distinct_words(test_corpus)
# Correct answers
ans_test_corpus_words = sorted(list(set(["START", "All", "ends", "that", "gold", "All's", "glitters", "isn't", "well", "END"])))
ans_num_corpus_words = len(ans_test_corpus_words)
# Test correct number of words
assert(num_corpus_words == ans_num_corpus_words), "Incorrect number of distinct words. Correct: {}. Yours: {}".format(ans_num_corpus_words, num_corpus_words)
# Test correct words
assert (test_corpus_words == ans_test_corpus_words), "Incorrect corpus_words.\nCorrect: {}\nYours: {}".format(str(ans_test_corpus_words), str(test_corpus_words))
# Print Success
print ("-" * 80)
print("Passed All Tests!")
print ("-" * 80)
# -
# ### Question 1.2: Implement `compute_co_occurrence_matrix` [code] (3 points)
#
# Write a method that constructs a co-occurrence matrix for a certain window-size $n$ (with a default of 4), considering words $n$ before and $n$ after the word in the center of the window. Here, we start to use `numpy (np)` to represent vectors, matrices, and tensors. If you're not familiar with NumPy, there's a NumPy tutorial in the second half of this cs231n [Python NumPy tutorial](http://cs231n.github.io/python-numpy-tutorial/).
#
M, word2Ind = compute_co_occurrence_matrix(reuters_corpus)
def compute_co_occurrence_matrix(corpus, window_size=4):
""" Compute co-occurrence matrix for the given corpus and window_size (default of 4).
Note: Each word in a document should be at the center of a window. Words near edges will have a smaller
number of co-occurring words.
For example, if we take the document "START All that glitters is not gold END" with window size of 4,
"All" will co-occur with "START", "that", "glitters", "is", and "not".
Params:
corpus (list of list of strings): corpus of documents
window_size (int): size of context window
Return:
M (numpy matrix of shape (number of corpus words, number of corpus words)):
Co-occurence matrix of word counts.
The ordering of the words in the rows/columns should be the same as the ordering of the words given by the distinct_words function.
word2Ind (dict): dictionary that maps word to index (i.e. row/column number) for matrix M.
"""
words, num_words = distinct_words(corpus)
M = None
word2Ind = {}
# ------------------
# Write your implementation here.
M = np.zeros((num_words, num_words))
word2Ind = dict(zip(words, range(num_words)))
for line in corpus:
for i in range(len(line)):
left_bound = max(0, i - window_size)
right_bound = min(i + window_size, len(line))
window_words = line[left_bound:i] + line[i+1:right_bound+1]
center_word = line[i]
for window_word in window_words:
M[word2Ind[center_word], word2Ind[window_word]] += 1
# ------------------
return M, word2Ind
list(word2Ind.keys())[0]
# +
# ---------------------
# Run this sanity check
# Note that this is not an exhaustive check for correctness.
# ---------------------
# Define toy corpus and get student's co-occurrence matrix
test_corpus = ["START All that glitters isn't gold END".split(" "), "START All's well that ends well END".split(" ")]
M_test, word2Ind_test = compute_co_occurrence_matrix(test_corpus, window_size=1)
# Correct M and word2Ind
M_test_ans = np.array(
[[0., 0., 0., 1., 0., 0., 0., 0., 1., 0.,],
[0., 0., 0., 1., 0., 0., 0., 0., 0., 1.,],
[0., 0., 0., 0., 0., 0., 1., 0., 0., 1.,],
[1., 1., 0., 0., 0., 0., 0., 0., 0., 0.,],
[0., 0., 0., 0., 0., 0., 0., 0., 1., 1.,],
[0., 0., 0., 0., 0., 0., 0., 1., 1., 0.,],
[0., 0., 1., 0., 0., 0., 0., 1., 0., 0.,],
[0., 0., 0., 0., 0., 1., 1., 0., 0., 0.,],
[1., 0., 0., 0., 1., 1., 0., 0., 0., 1.,],
[0., 1., 1., 0., 1., 0., 0., 0., 1., 0.,]]
)
word2Ind_ans = {'All': 0, "All's": 1, 'END': 2, 'START': 3, 'ends': 4, 'glitters': 5, 'gold': 6, "isn't": 7, 'that': 8, 'well': 9}
# Test correct word2Ind
assert (word2Ind_ans == word2Ind_test), "Your word2Ind is incorrect:\nCorrect: {}\nYours: {}".format(word2Ind_ans, word2Ind_test)
# Test correct M shape
assert (M_test.shape == M_test_ans.shape), "M matrix has incorrect shape.\nCorrect: {}\nYours: {}".format(M_test.shape, M_test_ans.shape)
# Test correct M values
for w1 in word2Ind_ans.keys():
idx1 = word2Ind_ans[w1]
for w2 in word2Ind_ans.keys():
idx2 = word2Ind_ans[w2]
student = M_test[idx1, idx2]
correct = M_test_ans[idx1, idx2]
if student != correct:
print("Correct M:")
print(M_test_ans)
print("Your M: ")
print(M_test)
raise AssertionError("Incorrect count at index ({}, {})=({}, {}) in matrix M. Yours has {} but should have {}.".format(idx1, idx2, w1, w2, student, correct))
# Print Success
print ("-" * 80)
print("Passed All Tests!")
print ("-" * 80)
# -
# ### Question 1.3: Implement `reduce_to_k_dim` [code] (1 point)
#
# Construct a method that performs dimensionality reduction on the matrix to produce k-dimensional embeddings. Use SVD to take the top k components and produce a new matrix of k-dimensional embeddings.
#
# **Note:** All of numpy, scipy, and scikit-learn (`sklearn`) provide *some* implementation of SVD, but only scipy and sklearn provide an implementation of Truncated SVD, and only sklearn provides an efficient randomized algorithm for calculating large-scale Truncated SVD. So please use [sklearn.decomposition.TruncatedSVD](https://scikit-learn.org/stable/modules/generated/sklearn.decomposition.TruncatedSVD.html).
M_reduced = reduce_to_k_dim(M)
def reduce_to_k_dim(M, k=2):
""" Reduce a co-occurence count matrix of dimensionality (num_corpus_words, num_corpus_words)
to a matrix of dimensionality (num_corpus_words, k) using the following SVD function from Scikit-Learn:
- http://scikit-learn.org/stable/modules/generated/sklearn.decomposition.TruncatedSVD.html
Params:
M (numpy matrix of shape (number of corpus words, number of corpus words)): co-occurence matrix of word counts
k (int): embedding size of each word after dimension reduction
Return:
M_reduced (numpy matrix of shape (number of corpus words, k)): matrix of k-dimensioal word embeddings.
In terms of the SVD from math class, this actually returns U * S
"""
n_iters = 10 # Use this parameter in your call to `TruncatedSVD`
M_reduced = None
print("Running Truncated SVD over %i words..." % (M.shape[0]))
# ------------------
# Write your implementation here.
from sklearn.decomposition import TruncatedSVD
svd = TruncatedSVD(n_components=k, n_iter=n_iters)
M_reduced = svd.fit_transform(M)
# ------------------
print("Done.")
return M_reduced
# +
# ---------------------
# Run this sanity check
# Note that this not an exhaustive check for correctness
# In fact we only check that your M_reduced has the right dimensions.
# ---------------------
# Define toy corpus and run student code
test_corpus = ["START All that glitters isn't gold END".split(" "), "START All's well that ends well END".split(" ")]
M_test, word2Ind_test = compute_co_occurrence_matrix(test_corpus, window_size=1)
M_test_reduced = reduce_to_k_dim(M_test, k=2)
# Test proper dimensions
assert (M_test_reduced.shape[0] == 10), "M_reduced has {} rows; should have {}".format(M_test_reduced.shape[0], 10)
assert (M_test_reduced.shape[1] == 2), "M_reduced has {} columns; should have {}".format(M_test_reduced.shape[1], 2)
# Print Success
print ("-" * 80)
print("Passed All Tests!")
print ("-" * 80)
# -
# ### Question 1.4: Implement `plot_embeddings` [code] (1 point)
#
# Here you will write a function to plot a set of 2D vectors in 2D space. For graphs, we will use Matplotlib (`plt`).
#
# For this example, you may find it useful to adapt [this code](https://www.pythonmembers.club/2018/05/08/matplotlib-scatter-plot-annotate-set-text-at-label-each-point/). In the future, a good way to make a plot is to look at [the Matplotlib gallery](https://matplotlib.org/gallery/index.html), find a plot that looks somewhat like what you want, and adapt the code they give.
def plot_embeddings(M_reduced, word2Ind, words):
""" Plot in a scatterplot the embeddings of the words specified in the list "words".
NOTE: do not plot all the words listed in M_reduced / word2Ind.
Include a label next to each point.
Params:
M_reduced (numpy matrix of shape (number of unique words in the corpus , k)): matrix of k-dimensioal word embeddings
word2Ind (dict): dictionary that maps word to indices for matrix M
words (list of strings): words whose embeddings we want to visualize
"""
# ------------------
# Write your implementation here.
plt.figure(figsize=(6,4))
for w in words:
idx = word2Ind[w]
plt.scatter(M_reduced[idx][0], M_reduced[idx][1], marker='x', color='red')
plt.text(M_reduced[idx][0]+0.001, M_reduced[idx][1]+0.001, list(word2Ind.keys())[idx])
# ------------------
# +
# ---------------------
# Run this sanity check
# Note that this not an exhaustive check for correctness.
# The plot produced should look like the "test solution plot" depicted below.
# ---------------------
print ("-" * 80)
print ("Outputted Plot:")
M_reduced_plot_test = np.array([[1, 1], [-1, -1], [1, -1], [-1, 1], [0, 0]])
word2Ind_plot_test = {'test1': 0, 'test2': 1, 'test3': 2, 'test4': 3, 'test5': 4}
words = ['test1', 'test2', 'test3', 'test4', 'test5']
plot_embeddings(M_reduced_plot_test, word2Ind_plot_test, words)
print ("-" * 80)
# -
# <font color=red>**Test Plot Solution**</font>
# <br>
# <img src="imgs/test_plot.png" width=40% style="float: left;"> </img>
#
# ### Question 1.5: Co-Occurrence Plot Analysis [written] (3 points)
#
# Now we will put together all the parts you have written! We will compute the co-occurrence matrix with fixed window of 4, over the Reuters "crude" corpus. Then we will use TruncatedSVD to compute 2-dimensional embeddings of each word. TruncatedSVD returns U\*S, so we normalize the returned vectors, so that all the vectors will appear around the unit circle (therefore closeness is directional closeness). **Note**: The line of code below that does the normalizing uses the NumPy concept of *broadcasting*. If you don't know about broadcasting, check out
# [Computation on Arrays: Broadcasting by <NAME>](https://jakevdp.github.io/PythonDataScienceHandbook/02.05-computation-on-arrays-broadcasting.html).
#
# Run the below cell to produce the plot. It'll probably take a few seconds to run. What clusters together in 2-dimensional embedding space? What doesn't cluster together that you might think should have? **Note:** "bpd" stands for "barrels per day" and is a commonly used abbreviation in crude oil topic articles.
# +
# -----------------------------
# Run This Cell to Produce Your Plot
# ------------------------------
reuters_corpus = read_corpus()
M_co_occurrence, word2Ind_co_occurrence = compute_co_occurrence_matrix(reuters_corpus)
M_reduced_co_occurrence = reduce_to_k_dim(M_co_occurrence, k=2)
# Rescale (normalize) the rows to make them each of unit-length
M_lengths = np.linalg.norm(M_reduced_co_occurrence, axis=1)
M_normalized = M_reduced_co_occurrence / M_lengths[:, np.newaxis] # broadcasting
words = ['barrels', 'bpd', 'ecuador', 'energy', 'industry', 'kuwait', 'oil', 'output', 'petroleum', 'venezuela']
plot_embeddings(M_normalized, word2Ind_co_occurrence, words)
# -
# #### <font color="red">Write your answer here.</font>
#
# * petroleum : 석유, ecuador : 에콰도르
#
# * (kuweit, venezuela, ecuador) : 석유 생산 국가 (oil, energy) : 자원
# 유사한 의미를 가진 위 단어들은 잘 클러스터링 되었다.
#
# * (bpd, barrels), (petroleum, oil)과 같이 매우 유사한 의미를 가지고 있는 단어들은 잘 클러스터링 되지 않았다.
#
# * 그래도 전반적으로 의미가 유사한 단어들 끼리 잘 모여있는 것 같다. 이는 co-occurrence matrix를 이용한 단어 embedding은 단어의 semantic한 정보까지 포함시킬 수 있기 때문이라고 생각한다.
# ## Part 2: Prediction-Based Word Vectors (15 points)
#
# As discussed in class, more recently prediction-based word vectors have come into fashion, e.g. word2vec. Here, we shall explore the embeddings produced by word2vec. Please revisit the class notes and lecture slides for more details on the word2vec algorithm. If you're feeling adventurous, challenge yourself and try reading the [original paper](https://papers.nips.cc/paper/5021-distributed-representations-of-words-and-phrases-and-their-compositionality.pdf).
#
# Then run the following cells to load the word2vec vectors into memory. **Note**: This might take several minutes.
def load_word2vec():
""" Load Word2Vec Vectors
Return:
wv_from_bin: All 3 million embeddings, each lengh 300
"""
import gensim.downloader as api
wv_from_bin = api.load("word2vec-google-news-300")
vocab = list(wv_from_bin.vocab.keys())
print("Loaded vocab size %i" % len(vocab))
return wv_from_bin
# -----------------------------------
# Run Cell to Load Word Vectors
# Note: This may take several minutes
# -----------------------------------
wv_from_bin = load_word2vec()
# **Note: If you are receiving out of memory issues on your local machine, try closing other applications to free more memory on your device. You may want to try restarting your machine so that you can free up extra memory. Then immediately run the jupyter notebook and see if you can load the word vectors properly. If you still have problems with loading the embeddings onto your local machine after this, please follow the Piazza instructions, as how to run remotely on Stanford Farmshare machines.**
# ### Reducing dimensionality of Word2Vec Word Embeddings
# Let's directly compare the word2vec embeddings to those of the co-occurrence matrix. Run the following cells to:
#
# 1. Put the 3 million word2vec vectors into a matrix M
# 2. Run reduce_to_k_dim (your Truncated SVD function) to reduce the vectors from 300-dimensional to 2-dimensional.
def get_matrix_of_vectors(wv_from_bin, required_words=['barrels', 'bpd', 'ecuador', 'energy', 'industry', 'kuwait', 'oil', 'output', 'petroleum', 'venezuela']):
""" Put the word2vec vectors into a matrix M.
Param:
wv_from_bin: KeyedVectors object; the 3 million word2vec vectors loaded from file
Return:
M: numpy matrix shape (num words, 300) containing the vectors
word2Ind: dictionary mapping each word to its row number in M
"""
import random
words = list(wv_from_bin.vocab.keys())
print("Shuffling words ...")
random.shuffle(words)
words = words[:10000]
print("Putting %i words into word2Ind and matrix M..." % len(words))
word2Ind = {}
M = []
curInd = 0
for w in words:
try:
M.append(wv_from_bin.word_vec(w))
word2Ind[w] = curInd
curInd += 1
except KeyError:
continue
for w in required_words:
try:
M.append(wv_from_bin.word_vec(w))
word2Ind[w] = curInd
curInd += 1
except KeyError:
continue
M = np.stack(M)
print("Done.")
return M, word2Ind
# -----------------------------------------------------------------
# Run Cell to Reduce 300-Dimensinal Word Embeddings to k Dimensions
# Note: This may take several minutes
# -----------------------------------------------------------------
M, word2Ind = get_matrix_of_vectors(wv_from_bin)
M_reduced = reduce_to_k_dim(M, k=2)
# ### Question 2.1: Word2Vec Plot Analysis [written] (4 points)
#
# Run the cell below to plot the 2D word2vec embeddings for `['barrels', 'bpd', 'ecuador', 'energy', 'industry', 'kuwait', 'oil', 'output', 'petroleum', 'venezuela']`.
#
# What clusters together in 2-dimensional embedding space? What doesn't cluster together that you might think should have? How is the plot different from the one generated earlier from the co-occurrence matrix?
words = ['barrels', 'bpd', 'ecuador', 'energy', 'industry', 'kuwait', 'oil', 'output', 'petroleum', 'venezuela']
plot_embeddings(M_reduced, word2Ind, words)
# #### <font color="red">Write your answer here.</font>
#
# * 전반적으로 의미가 비슷한 단어들 끼리 클러스터링이 잘 안되어있고, 다들 흩어져 있다. 이는 word2vec은 단어의 형태적 유사성(syntatic) 만 임베딩에 반영할 수 있기 때문이라고 생각한다.
# ### Cosine Similarity
# Now that we have word vectors, we need a way to quantify the similarity between individual words, according to these vectors. One such metric is cosine-similarity. We will be using this to find words that are "close" and "far" from one another.
#
# We can think of n-dimensional vectors as points in n-dimensional space. If we take this perspective L1 and L2 Distances help quantify the amount of space "we must travel" to get between these two points. Another approach is to examine the angle between two vectors. From trigonometry we know that:
#
# <img src="imgs/inner_product.png" width=20% style="float: center;"></img>
#
# Instead of computing the actual angle, we can leave the similarity in terms of $similarity = cos(\Theta)$. Formally the [Cosine Similarity](https://en.wikipedia.org/wiki/Cosine_similarity) $s$ between two vectors $p$ and $q$ is defined as:
#
# $$s = \frac{p \cdot q}{||p|| ||q||}, \textrm{ where } s \in [-1, 1] $$
# ### Question 2.2: Polysemous Words (2 points) [code + written]
# Find a [polysemous](https://en.wikipedia.org/wiki/Polysemy) word (for example, "leaves" or "scoop") such that the top-10 most similar words (according to cosine similarity) contains related words from *both* meanings. For example, "leaves" has both "vanishes" and "stalks" in the top 10, and "scoop" has both "handed_waffle_cone" and "lowdown". You will probably need to try several polysemous words before you find one. Please state the polysemous word you discover and the multiple meanings that occur in the top 10. Why do you think many of the polysemous words you tried didn't work?
#
# **Note**: You should use the `wv_from_bin.most_similar(word)` function to get the top 10 similar words. This function ranks all other words in the vocabulary with respect to their cosine similarity to the given word. For further assistance please check the __[GenSim documentation](https://radimrehurek.com/gensim/models/keyedvectors.html#gensim.models.keyedvectors.FastTextKeyedVectors.most_similar)__.
# +
# ------------------
# Write your polysemous word exploration code here.
wv_from_bin.most_similar("man")
# ------------------
# -
# #### <font color="red">Write your answer here.</font>
#
# 다의어가 적절하게 임베딩되기 어려운 이유는 word2vec 모델은 문맥에 따라 달라지는 단어의 의미를 반영하지는 못하기 때문입니다.
# ### Question 2.3: Synonyms & Antonyms (2 points) [code + written]
#
# When considering Cosine Similarity, it's often more convenient to think of Cosine Distance, which is simply 1 - Cosine Similarity.
#
# Find three words (w1,w2,w3) where w1 and w2 are synonyms and w1 and w3 are antonyms, but Cosine Distance(w1,w3) < Cosine Distance(w1,w2). For example, w1="happy" is closer to w3="sad" than to w2="cheerful".
#
# Once you have found your example, please give a possible explanation for why this counter-intuitive result may have happened.
#
# You should use the the `wv_from_bin.distance(w1, w2)` function here in order to compute the cosine distance between two words. Please see the __[GenSim documentation](https://radimrehurek.com/gensim/models/keyedvectors.html#gensim.models.keyedvectors.FastTextKeyedVectors.distance)__ for further assistance.
# +
# ------------------
# Write your synonym & antonym exploration code here.
w1 = "man"
w2 = "human"
w3 = "woman"
w1_w2_dist = wv_from_bin.distance(w1, w2)
w1_w3_dist = wv_from_bin.distance(w1, w3)
print("Synonyms {}, {} have cosine distance: {}".format(w1, w2, w1_w2_dist))
print("Antonyms {}, {} have cosine distance: {}".format(w1, w3, w1_w3_dist))
# ------------------
# -
# #### <font color="red">Write your answer here.</font>
#
# Question 2.2의 결과와 같은 맥락으로 2.3에서도 cosine similarity를 이용했을 때 man과 human이 비슷한 의미로 사용된다는 것을 반영하지 못하고 있습니다. 오히려 man의 반의어인 woman과의 cosine distance가 더 가깝다는 결과를 보이고 있습니다.
# 이는 모델이 man이 남성이라는 뜻이 아닌 사람이라는 뜻으로 쓰이기도 한다는 것을 제대로 학습하지 못했기 때문이라고 생각합니다. 즉 여러 가지 의미를 단어 벡터에 반영하지 못했기 때문이라고 할 수 있습니다.
# ### Solving Analogies with Word Vectors
# Word2Vec vectors have been shown to *sometimes* exhibit the ability to solve analogies.
#
# As an example, for the analogy "man : king :: woman : x", what is x?
#
# In the cell below, we show you how to use word vectors to find x. The `most_similar` function finds words that are most similar to the words in the `positive` list and most dissimilar from the words in the `negative` list. The answer to the analogy will be the word ranked most similar (largest numerical value).
#
# **Note:** Further Documentation on the `most_similar` function can be found within the __[GenSim documentation](https://radimrehurek.com/gensim/models/keyedvectors.html#gensim.models.keyedvectors.FastTextKeyedVectors.most_similar)__.
# Run this cell to answer the analogy -- man : king :: woman : x
pprint.pprint(wv_from_bin.most_similar(positive=['woman', 'king'], negative=['man']))
# ### Question 2.4: Finding Analogies [code + written] (2 Points)
# Find an example of analogy that holds according to these vectors (i.e. the intended word is ranked top). In your solution please state the full analogy in the form x:y :: a:b. If you believe the analogy is complicated, explain why the analogy holds in one or two sentences.
#
# **Note**: You may have to try many analogies to find one that works!
# +
# ------------------
# Write your analogy exploration code here.
pprint.pprint(wv_from_bin.most_similar(positive=['woman', 'uncle'], negative=['man']))
# ------------------
# -
# #### <font color="red">Write your answer here.</font>
#
# man:uncle :: woman:aunt
# ### Question 2.5: Incorrect Analogy [code + written] (1 point)
# Find an example of analogy that does *not* hold according to these vectors. In your solution, state the intended analogy in the form x:y :: a:b, and state the (incorrect) value of b according to the word vectors.
# +
# ------------------
# Write your incorrect analogy exploration code here.
pprint.pprint(wv_from_bin.most_similar(positive=['teacher','hospital'], negative=['doctor']))
# ------------------
# -
# #### <font color="red">Write your answer here.</font>
#
# doctor:hospital :: teacher:school
#
# -> 1순위로 나온 incorrect value of b : elementary
# ### Question 2.6: Guided Analysis of Bias in Word Vectors [written] (1 point)
#
# It's important to be cognizant of the biases (gender, race, sexual orientation etc.) implicit to our word embeddings.
#
# Run the cell below, to examine (a) which terms are most similar to "woman" and "boss" and most dissimilar to "man", and (b) which terms are most similar to "man" and "boss" and most dissimilar to "woman". What do you find in the top 10?
# Run this cell
# Here `positive` indicates the list of words to be similar to and `negative` indicates the list of words to be
# most dissimilar from.
pprint.pprint(wv_from_bin.most_similar(positive=['woman', 'boss'], negative=['man']))
print()
pprint.pprint(wv_from_bin.most_similar(positive=['man', 'boss'], negative=['woman']))
# #### <font color="red">Write your answer here.</font>
#
# a) bosses : boss의 복수형, manageress : 여성 경영자
# b) supremo : 최고의 인물
# ### Question 2.7: Independent Analysis of Bias in Word Vectors [code + written] (2 points)
#
# Use the `most_similar` function to find another case where some bias is exhibited by the vectors. Please briefly explain the example of bias that you discover.
# +
# ------------------
# Write your bias exploration code here.
pprint.pprint(wv_from_bin.most_similar(positive=['woman', 'pink'], negative=['man']))
print()
pprint.pprint(wv_from_bin.most_similar(positive=['man', 'pink'], negative=['woman']))
# ------------------
# -
# #### <font color="red">Write your answer here.</font>
# ### Question 2.8: Thinking About Bias [written] (1 point)
#
# What might be the cause of these biases in the word vectors?
# #### <font color="red">Write your answer here.</font>
#
# 해당 단어와 함께 사용되는 단어들이 무엇인지에 따라 단어 임베딩 벡터가 담고 있는 정보가 달라지는데, 편견 섞인 문장들이 많이 학습되면 학습될수록 단어 임베딩 모델에도 이러한 편견들이 많이 반영되는 것 같다.
# # <font color="blue"> Submission Instructions</font>
#
# 1. Click the Save button at the top of the Jupyter Notebook.
# 2. Please make sure to have entered your SUNET ID above.
# 3. Select Cell -> All Output -> Clear. This will clear all the outputs from all cells (but will keep the content of ll cells).
# 4. Select Cell -> Run All. This will run all the cells in order, and will take several minutes.
# 5. Once you've rerun everything, select File -> Download as -> PDF via LaTeX
# 6. Look at the PDF file and make sure all your solutions are there, displayed correctly. The PDF is the only thing your graders will see!
# 7. Submit your PDF on Gradescope.
| Assignment/Assignment1/exploring_word_vectors_이혜린.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
# Copyright 2021 NVIDIA Corporation. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
# -
# # Scaling Criteo: Training with TensorFlow
#
# ## Overview
#
# We observed that TensorFlow training pipelines can be slow as the dataloader is a bottleneck. The native dataloader in TensorFlow randomly sample each item from the dataset, which is very slow. The window dataloader in TensorFlow is not much faster. In our experiments, we are able to speed-up existing TensorFlow pipelines by 9x using a highly optimized dataloader.<br><br>
#
# We have already discussed the NVTabular dataloader for TensorFlow in more detail in our [Getting Started with Movielens notebooks](https://github.com/NVIDIA/NVTabular/tree/main/examples/getting-started-movielens).<br><br>
#
# We will use the same techniques to train a deep learning model for the [Criteo 1TB Click Logs dataset](https://ailab.criteo.com/download-criteo-1tb-click-logs-dataset/).
#
# ### Learning objectives
#
# In this notebook, we learn how to:
#
# - Use **NVTabular dataloader** with TensorFlow Keras model
# ## NVTabular dataloader for TensorFlow
#
# We’ve identified that the dataloader is one bottleneck in deep learning recommender systems when training pipelines with TensorFlow. The dataloader cannot prepare the next batch fast enough and therefore, the GPU is not fully utilized.
#
# We developed a highly customized tabular dataloader for accelerating existing pipelines in TensorFlow. In our experiments, we see a speed-up by 9x of the same training workflow with NVTabular dataloader. NVTabular dataloader’s features are:
#
# - removing bottleneck of item-by-item dataloading
# - enabling larger than memory dataset by streaming from disk
# - reading data directly into GPU memory and remove CPU-GPU communication
# - preparing batch asynchronously in GPU to avoid CPU-GPU communication
# - supporting commonly used .parquet format
# - easy integration into existing TensorFlow pipelines by using similar API - works with tf.keras models
#
# More information in our [blogpost](https://medium.com/nvidia-merlin/training-deep-learning-based-recommender-systems-9x-faster-with-tensorflow-cc5a2572ea49).
# External dependencies
import os
import glob
import time
import nvtabular as nvt
# We define our base directory, containing the data.
BASE_DIR = os.environ.get("BASE_DIR", "/raid/data/criteo")
input_path = os.environ.get("INPUT_DATA_DIR", os.path.join(BASE_DIR, "test_dask/output"))
# ### Defining Hyperparameters
# First, we define the data schema and differentiate between single-hot and multi-hot categorical features. Note, that we do not have any numerical input features.
# +
CONTINUOUS_COLUMNS = ["I" + str(x) for x in range(1, 14)]
CATEGORICAL_COLUMNS = ["C" + str(x) for x in range(1, 27)]
LABEL_COLUMNS = ["label"]
COLUMNS = CONTINUOUS_COLUMNS + CATEGORICAL_COLUMNS + LABEL_COLUMNS
BATCH_SIZE = int(os.environ.get("BATCH_SIZE", 64 * 1024))
# Output from ETL-with-NVTabular
TRAIN_PATHS = sorted(glob.glob(os.path.join(input_path, "train", "*.parquet")))
VALID_PATHS = sorted(glob.glob(os.path.join(input_path, "valid", "*.parquet")))
TRAIN_PATHS, VALID_PATHS
# -
# In the previous notebook, we used NVTabular for ETL and stored the workflow to disk. We can load the NVTabular workflow to extract important metadata for our training pipeline.
proc = nvt.Workflow.load(os.path.join(input_path, "workflow"))
# The embedding table shows the cardinality of each categorical variable along with its associated embedding size. Each entry is of the form `(cardinality, embedding_size)`. We limit the output cardinality to 16.
EMBEDDING_TABLE_SHAPES = nvt.ops.get_embedding_sizes(proc)
for key in EMBEDDING_TABLE_SHAPES.keys():
EMBEDDING_TABLE_SHAPES[key] = (
EMBEDDING_TABLE_SHAPES[key][0],
min(16, EMBEDDING_TABLE_SHAPES[key][1]),
)
EMBEDDING_TABLE_SHAPES
# ### Initializing NVTabular Dataloader for Tensorflow
# We import TensorFlow and some NVTabular TF extensions, such as custom TensorFlow layers supporting multi-hot and the NVTabular TensorFlow data loader.
# +
os.environ["CUDA_VISIBLE_DEVICES"] = "0"
import tensorflow as tf
# we can control how much memory to give tensorflow with this environment variable
# IMPORTANT: make sure you do this before you initialize TF's runtime, otherwise
# TF will have claimed all free GPU memory
os.environ["TF_MEMORY_ALLOCATION"] = "0.5" # fraction of free memory
from nvtabular.loader.tensorflow import KerasSequenceLoader, KerasSequenceValidater
from nvtabular.framework_utils.tensorflow import layers
# -
# First, we take a look on our data loader and how the data is represented as tensors. The NVTabular data loader are initialized as usually and we specify both single-hot and multi-hot categorical features as cat_names. The data loader will automatically recognize the single/multi-hot columns and represent them accordingly.
# +
train_dataset_tf = KerasSequenceLoader(
nvt.Dataset(TRAIN_PATHS, part_mem_fraction=0.04),
batch_size=BATCH_SIZE,
label_names=LABEL_COLUMNS,
cat_names=CATEGORICAL_COLUMNS,
cont_names=CONTINUOUS_COLUMNS,
engine="parquet",
shuffle=True,
parts_per_chunk=1,
)
valid_dataset_tf = KerasSequenceLoader(
nvt.Dataset(VALID_PATHS, part_mem_fraction=0.04),
batch_size=BATCH_SIZE,
label_names=LABEL_COLUMNS,
cat_names=CATEGORICAL_COLUMNS,
cont_names=CONTINUOUS_COLUMNS,
engine="parquet",
shuffle=False,
parts_per_chunk=1,
)
# -
# ### Defining Neural Network Architecture
# We will define a common neural network architecture for tabular data:
#
# * Single-hot categorical features are fed into an Embedding Layer
# * Each value of a multi-hot categorical features is fed into an Embedding Layer and the multiple Embedding outputs are combined via averaging
# * The output of the Embedding Layers are concatenated
# * The concatenated layers are fed through multiple feed-forward layers (Dense Layers with ReLU activations)
# * The final output is a single number with sigmoid activation function
# First, we will define some dictionary/lists for our network architecture.
inputs = {} # tf.keras.Input placeholders for each feature to be used
emb_layers = [] # output of all embedding layers, which will be concatenated
num_layers = [] # output of numerical layers
# We create `tf.keras.Input` tensors for all input features.
# +
for col in CATEGORICAL_COLUMNS:
inputs[col] = tf.keras.Input(name=col, dtype=tf.int32, shape=(1,))
for col in CONTINUOUS_COLUMNS:
inputs[col] = tf.keras.Input(name=col, dtype=tf.float32, shape=(1,))
# -
# Next, we initialize Embedding Layers with `tf.feature_column.embedding_column`.
for col in CATEGORICAL_COLUMNS:
emb_layers.append(
tf.feature_column.embedding_column(
tf.feature_column.categorical_column_with_identity(
col, EMBEDDING_TABLE_SHAPES[col][0]
), # Input dimension (vocab size)
EMBEDDING_TABLE_SHAPES[col][1], # Embedding output dimension
)
)
# We define `tf.feature_columns` for the continuous input features.
for col in CONTINUOUS_COLUMNS:
num_layers.append(tf.feature_column.numeric_column(col))
# NVTabular implemented a custom TensorFlow layer `layers.DenseFeatures`, which takes as an input the different `tf.Keras.Input` and pre-initialized `tf.feature_column` and automatically concatenate them into a flat tensor.
emb_layer = layers.DenseFeatures(emb_layers)
x_emb_output = emb_layer(inputs)
x_emb_output
# We add multiple Dense Layers. Finally, we initialize the `tf.keras.Model` and add the optimizer.
# +
x = tf.keras.layers.Dense(128, activation="relu")(x_emb_output)
x = tf.keras.layers.Dense(128, activation="relu")(x)
x = tf.keras.layers.Dense(128, activation="relu")(x)
x = tf.keras.layers.Dense(1, activation="sigmoid", name="output")(x)
model = tf.keras.Model(inputs=inputs, outputs=x)
model.compile("sgd", "binary_crossentropy")
# -
# You need to install the dependencies
tf.keras.utils.plot_model(model)
# ### Training the deep learning model
# We can train our model with `model.fit`. We need to use a Callback to add the validation dataloader.
EPOCHS = 1
validation_callback = KerasSequenceValidater(valid_dataset_tf)
start = time.time()
history = model.fit(train_dataset_tf, callbacks=[validation_callback], epochs=EPOCHS)
end = time.time() - start
total_rows = train_dataset_tf.num_rows_processed + valid_dataset_tf.num_rows_processed
print(f"run_time: {end} - rows: {total_rows * EPOCHS} - epochs: {EPOCHS} - dl_thru: {(total_rows * EPOCHS) / end}")
# We save the trained model.
model.save(os.path.join(input_path, "model.savedmodel"))
| examples/scaling-criteo/03-Training-with-TF.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ## Monte Carlo - Euler Discretization - Part I
# *Suggested Answers follow (usually there are multiple ways to solve a problem in Python).*
# Load the data for Microsoft (‘MSFT’) for the period ‘2000-1-1’ until today.
import numpy as np
import pandas as pd
from pandas_datareader import data as web
from scipy.stats import norm
import matplotlib.pyplot as plt
# %matplotlib inline
data = pd.read_csv('D:/Python/MSFT_2000.csv', index_col = 'Date')
# Store the annual standard deviation of the log returns in a variable, called “stdev”.
# Set the risk free rate, r, equal to 2.5% (0.025).
# To transform the object into an array, reassign stdev.values to stdev.
# Set the time horizon, T, equal to 1 year, the number of time intervals equal to 250, the iterations equal to 10,000. Create a variable, delta_t, equal to the quotient of T divided by the number of time intervals.
# Let Z equal a random matrix with dimension (time intervals + 1) by the number of iterations.
# Use the .zeros_like() method to create another variable, S, with the same dimension as Z. S is the matrix to be filled with future stock price data.
# Create a variable S0 equal to the last adjusted closing price of Microsoft. Use the “iloc” method.
# Use the following formula to create a loop within the range (1, t_intervals + 1) that reassigns values to S in time t.
# $$
# S_t = S_{t-1} \cdot exp((r - 0.5 \cdot stdev^2) \cdot delta_t + stdev \cdot delta_t^{0.5} \cdot Z_t)
# $$
# Plot the first 10 of the 10,000 generated iterations on a graph.
| 23 - Python for Finance/8_Monte Carlo Simulations as a Decision-Making Tool/14_Monte Carlo: Euler Discretization - Part I (6:21)/MC - Euler Discretization - Part I - Exercise_CSV.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/vishant016/140_VISHANT/blob/main/LAB3/1_NB_Classifier_Whether.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="v-qaLkV_wIwA"
# **Aim: Implement Naive Bayes classifier : Whether Example**
# + [markdown] id="fE3k_I_lwE3_"
# #Step 1: Import necessary libraries.
# We will use preprocessing and naive bayes libraries of sklearn
# + id="AX_Lt4HDvoF4"
from sklearn import preprocessing
from sklearn.naive_bayes import GaussianNB, MultinomialNB
import pandas
import numpy as np
# + [markdown] id="heSKGhv4wPYo"
# #Step 2: Prepare dataset.
# Create feature set for weather and temperature, and classlabel play.
# + id="YBmuKC8mvoGe"
weather = ['Sunny', 'Sunny', 'Overcast', 'Rainy', 'Rainy','Rainy', 'Overcast',
'Sunny', 'Sunny', 'Rainy', 'Sunny', 'Overcast', 'Overcast', 'Rainy']
temp = ['Hot','Hot','Hot','Mild','Cool','Cool','Cool','Mild',
'Cool','Mild','Mild','Mild','Hot','Mild']
play=['No','No','Yes','Yes','Yes','No','Yes','No','Yes',
'Yes','Yes','Yes','Yes','No']
# + [markdown] id="VLHsDzOlwUcc"
# #Step 3: Digitize the data set using encoding
# + id="D2Y-GxbyvoG-" colab={"base_uri": "https://localhost:8080/"} outputId="cfe5e776-490c-425f-b91c-017bb667be9d"
#creating labelEncoder
le = preprocessing.LabelEncoder()
# Converting string labels into numbers.
weather_encoded=le.fit_transform(weather)
print("Weather:" ,weather_encoded)
# + id="E8pKDFm8voHY" colab={"base_uri": "https://localhost:8080/"} outputId="028e4f4e-8de5-4322-b254-caae251b3160"
temp_encoded=le.fit_transform(temp)
label=le.fit_transform(play)
print("Temp:",temp_encoded)
print("Play:",label)
# + [markdown] id="1u2XKORtwckp"
# #Step 4: Merge different features to prepare dataset
# + id="zcNtO6aUvoHs" colab={"base_uri": "https://localhost:8080/"} outputId="0c766b0b-a125-4044-e218-bba44e721c6c"
#Combinig weather and temp into single listof tuples
features=tuple(zip(weather_encoded,temp_encoded))
print("Features:",features)
# + [markdown] id="CDOMHUccT1hA"
#
# + [markdown] id="ZTs-FRS5wgQs"
# #Step 5: Train ’Naive Bayes Classifier’
# + id="6ON9ljKDvoIB" colab={"base_uri": "https://localhost:8080/"} outputId="f72822a7-bd62-4866-9c24-a11733e99aba"
#Create a Classifier
model=MultinomialNB()
# Train the model using the training sets
model.fit(features,label)
# + [markdown] id="vRuNrfolwjn4"
# #Step 6: Predict Output for new data
# + id="Ix6AhMALvoIU" colab={"base_uri": "https://localhost:8080/"} outputId="2e9df108-1d5d-431b-c4b9-d6daa2f8b390"
#Predict Output
predicted= model.predict([[0,2]]) # 0:Overcast, 2:Mild
print("Predicted Value:", predicted)
# + id="HGejlGZvvoIk" colab={"base_uri": "https://localhost:8080/"} outputId="7d6538d4-5976-4fb7-bf59-61920bdef934"
predicted= model.predict([[0,1]]) # 0:Overcast, 1:Hot
print("Predicted Value:", predicted)
# + id="ROG6v7C_voIy" colab={"base_uri": "https://localhost:8080/"} outputId="8627fa89-cdb1-49c0-855f-8c787aefb938"
predicted= model.predict([[2,2]]) # 2:Sunny, 2:Mild
print("Predicted Value:", predicted)
# + [markdown] id="oKxdQNaxwoZn"
# #Exercise:
#
# **Manually calculate output for the following cases and compare it with system’s output.**
#
# ######(1) Will you play if the temperature is 'Hot' and weather is 'overcast'?
#
# ######(2) Will you play if the temperature is 'Mild' and weather is 'Sunny'?
#
#
#
#
#
#
#
#
#
# + id="dQIOG4_9voJA"
# p(y=1/overcast,hot)=p(overcast,hot/y=1)*p(y=1)/p(overcast,hot)
class customNB:
def __init__(self,features,label):
self.features=np.array(features)
self.label=np.array(label)
def prior_prob(self,on_which):
tot=self.label.shape[0]
on_which_tot=np.sum(self.label==on_which)
return on_which_tot/tot
def conditional_prob(self,feature_col,feature_val,on_which):
new_features=self.features[self.label==on_which]
numerator=np.sum(new_features[:,feature_col]==feature_val)
return numerator/len(new_features)
def getVal(self,on_which,feature_col,feature_val):
prob=self.prior_prob(on_which)
for col,val in zip(feature_col,feature_val):
prob=prob*self.conditional_prob(col,val,on_which)
return prob
# + id="pbt4Q0OVbgdR" outputId="5489c76a-78b1-4ed0-de74-28cde45eff0b" colab={"base_uri": "https://localhost:8080/"}
custom_model=customNB(features,label)
yes_prob,no_prob=custom_model.getVal(1,[0,1],[0,1]),custom_model.getVal(0,[0,1],[0,1])
print(yes_prob,no_prob)
# + id="r8jZJ6eMb4F5" outputId="26eec1ba-e457-4b3c-cbd7-a85e574b1f72" colab={"base_uri": "https://localhost:8080/"}
predicted=model.predict([[0,1]])
print(predicted)
# + id="Ul5AOcHebjqp" outputId="928a96dc-b534-4acb-8dd6-4646f6e949fb" colab={"base_uri": "https://localhost:8080/"}
yes_prob,no_prob=custom_model.getVal(1,[0,1],[2,2]),custom_model.getVal(0,[0,1],[2,2])
print(yes_prob,no_prob)
# + id="6FAXD39kb-F5" outputId="3d0bf29a-9990-4e9d-9b52-4a1844bcedfe" colab={"base_uri": "https://localhost:8080/"}
predicted=model.predict([[2,2]])
print(predicted)
| LAB3/1_NB_Classifier_Whether.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # what characteristics do resigned people have?
import pandas as pd
import numpy as np
from matplotlib import pyplot as plt
emp=pd.read_pickle('emp_info_last_access')
resignation=emp[emp['left']==1]
#resignation
psycho_data = pd.read_csv("D:/DataSets2/psychometric_info.csv")
# # we define 30 as a dividing point of low O value and high O value, low O value refers to [10, 30], high O value refers to (30, 50]
bins=max(psycho_data["O"])-min(psycho_data["O"])+1
plt.hist(psycho_data["O"],bins)
plt.xlabel('O value')
plt.ylabel('number of people')
plt.show()
low=psycho_data[psycho_data["O"]<=30]
avg_low=low['O'].mean()
print("average O value of employees who has a low O value:",avg_low)
high=psycho_data[psycho_data["O"]>30]
avg_high=high['O'].mean()
print("average O value of employees who has a high O value:",avg_high)
print("number of employees who has a low O value",len(low))
print("number of employees who has a high O value",len(high))
# # from below results, it can be concluded that employees who has a high O value are more likely to resign
bins=max(resignation["O"])-min(resignation["O"])+1
plt.hist(resignation["O"],bins)
plt.xlabel('O value')
plt.ylabel('number of people')
plt.show()
r_low=resignation[resignation["O"]<=30]
r_avg_low=r_low['O'].mean()
print("average O value of resigned people who has a low O value:",r_avg_low)
r_high=resignation[resignation["O"]>30]
r_avg_high=r_high['O'].mean()
print("average O value of resigned people who has a high O value:",r_avg_high)
print("number of resigned people who has a low O value",len(r_low))
print("number of resigned people who has a high O value",len(r_high))
print("ratio of resigned people who has a low O value",len(r_low)/len(low))
print("ratio of resigned people who has a high O value",len(r_high)/len(high))
# # similarly, we can do the same thing on C value, the conclusion is: employees who has a high C value are more likely to resign
bins=max(psycho_data["C"])-min(psycho_data["C"])+1
plt.hist(psycho_data["C"],bins)
plt.xlabel('C value')
plt.ylabel('number of people')
plt.show()
low=psycho_data[psycho_data["C"]<=30]
avg_low=low['C'].mean()
print("average C value of employees who has a low C value:",avg_low)
high=psycho_data[psycho_data["C"]>30]
avg_high=high['C'].mean()
print("average C value of employees who has a high C value:",avg_high)
print("number of employees who has a low C value",len(low))
print("number of employees who has a high C value",len(high))
bins=max(resignation["C"])-min(resignation["C"])+1
plt.hist(resignation["C"],bins)
plt.xlabel('C value')
plt.ylabel('number of people')
plt.show()
r_low=resignation[resignation["C"]<=30]
r_avg_low=r_low['C'].mean()
print("average C value of resigned people who has a low C value:",r_avg_low)
r_high=resignation[resignation["C"]>30]
r_avg_high=r_high['C'].mean()
print("average C value of resigned people who has a high C value:",r_avg_high)
print("number of resigned people who has a low C value",len(r_low))
print("number of resigned people who has a high C value",len(r_high))
print("ratio of resigned people who has a low C value",len(r_low)/len(low))
print("ratio of resigned people who has a high C value",len(r_high)/len(high))
# # conclusion about E value: employees who has a high E value are more likely to resign
bins=max(psycho_data["E"])-min(psycho_data["E"])+1
plt.hist(psycho_data["E"],bins)
plt.xlabel('E value')
plt.ylabel('number of people')
plt.show()
low=psycho_data[psycho_data["E"]<=30]
avg_low=low['E'].mean()
print("average E value of employees who has a low E value:",avg_low)
high=psycho_data[psycho_data["E"]>30]
avg_high=high['E'].mean()
print("average E value of employees who has a high E value:",avg_high)
print("number of employees who has a low E value",len(low))
print("number of employees who has a high E value",len(high))
bins=max(resignation["E"])-min(resignation["E"])+1
plt.hist(resignation["E"],bins)
plt.xlabel('E value')
plt.ylabel('number of people')
plt.show()
r_low=resignation[resignation["E"]<=30]
r_avg_low=r_low['E'].mean()
print("average E value of resigned people who has a low E value:",r_avg_low)
r_high=resignation[resignation["E"]>30]
r_avg_high=r_high['E'].mean()
print("average E value of resigned people who has a high E value:",r_avg_high)
print("number of resigned people who has a low E value",len(r_low))
print("number of resigned people who has a high E value",len(r_high))
print("ratio of resigned people who has a low E value",len(r_low)/len(low))
print("ratio of resigned people who has a high E value",len(r_high)/len(high))
# # conclusion about A value: employees who has a low A value are more likely to resign
bins=max(psycho_data["A"])-min(psycho_data["A"])+1
plt.hist(psycho_data["A"],bins)
plt.xlabel('A value')
plt.ylabel('number of people')
plt.show()
low=psycho_data[psycho_data["A"]<=30]
avg_low=low['A'].mean()
print("average A value of employees who has a low A value:",avg_low)
high=psycho_data[psycho_data["A"]>30]
avg_high=high['A'].mean()
print("average A value of employees who has a high A value:",avg_high)
print("number of employees who has a low A value",len(low))
print("number of employees who has a high A value",len(high))
bins=max(resignation["A"])-min(resignation["A"])+1
plt.hist(resignation["A"],bins)
plt.xlabel('A value')
plt.ylabel('number of people')
plt.show()
r_low=resignation[resignation["A"]<=30]
r_avg_low=r_low['A'].mean()
print("average A value of resigned people who has a low A value:",r_avg_low)
r_high=resignation[resignation["A"]>30]
r_avg_high=r_high['A'].mean()
print("average A value of resigned people who has a high A value:",r_avg_high)
print("number of resigned people who has a low A value",len(r_low))
print("number of resigned people who has a high A value",len(r_high))
print("ratio of resigned people who has a low A value",len(r_low)/len(low))
print("ratio of resigned people who has a high A value",len(r_high)/len(high))
# # Distribution of N value is not a bimodal distribution, thus we just consider the mean value of N.
# # conclusion: employees who has a low N value are more likely to resign
bins=max(psycho_data["N"])-min(psycho_data["N"])+1
plt.hist(psycho_data["N"],bins)
plt.xlabel('N value')
plt.ylabel('number of people')
plt.show()
avg=psycho_data["N"].mean()
print("average N value of employees:",avg)
bins=max(resignation["N"])-min(resignation["N"])+1
plt.hist(resignation["N"],bins)
plt.xlabel('N value')
plt.ylabel('number of people')
plt.show()
r_avg=resignation["N"].mean()
print("average N value of resigned people:",r_avg)
| jupyter-notebooks/OCEAN characteristics of resigned people.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# %matplotlib inline
#
# =====================
# SVM: Weighted samples
# =====================
#
# Plot decision function of a weighted dataset, where the size of points
# is proportional to its weight.
#
# The sample weighting rescales the C parameter, which means that the classifier
# puts more emphasis on getting these points right. The effect might often be
# subtle.
# To emphasize the effect here, we particularly weight outliers, making the
# deformation of the decision boundary very visible.
#
#
# +
print(__doc__)
import numpy as np
import matplotlib.pyplot as plt
from sklearn import svm
def plot_decision_function(classifier, sample_weight, axis, title):
# plot the decision function
xx, yy = np.meshgrid(np.linspace(-4, 5, 500), np.linspace(-4, 5, 500))
Z = classifier.decision_function(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
# plot the line, the points, and the nearest vectors to the plane
axis.contourf(xx, yy, Z, alpha=0.75, cmap=plt.cm.bone)
axis.scatter(X[:, 0], X[:, 1], c=y, s=100 * sample_weight, alpha=0.9,
cmap=plt.cm.bone, edgecolors='black')
axis.axis('off')
axis.set_title(title)
# we create 20 points
np.random.seed(0)
X = np.r_[np.random.randn(10, 2) + [1, 1], np.random.randn(10, 2)]
y = [1] * 10 + [-1] * 10
sample_weight_last_ten = abs(np.random.randn(len(X)))
sample_weight_constant = np.ones(len(X))
# and bigger weights to some outliers
sample_weight_last_ten[15:] *= 5
sample_weight_last_ten[9] *= 15
# for reference, first fit without sample weights
# fit the model
clf_weights = svm.SVC(gamma=1)
clf_weights.fit(X, y, sample_weight=sample_weight_last_ten)
clf_no_weights = svm.SVC(gamma=1)
clf_no_weights.fit(X, y)
fig, axes = plt.subplots(1, 2, figsize=(14, 6))
plot_decision_function(clf_no_weights, sample_weight_constant, axes[0],
"Constant weights")
plot_decision_function(clf_weights, sample_weight_last_ten, axes[1],
"Modified weights")
plt.show()
| 01 Machine Learning/scikit_examples_jupyter/svm/plot_weighted_samples.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# Задание 1
# =========
# Импортируйте библиотеку Numpy и дайте ей псевдоним np.
import numpy as np
# Создайте массив Numpy под названием a размером 5x2, то есть состоящий из 5 строк и 2 столбцов.
#
# Первый столбец должен содержать числа 1, 2, 3, 3, 1, а второй - числа 6, 8, 11, 10, 7.
#
# Будем считать, что каждый столбец - это признак, а строка - наблюдение.
a = np.array([
[1, 6],
[2, 8],
[3, 11],
[3, 10],
[1, 7]
])
# Затем найдите среднее значение по каждому признаку, используя метод mean массива Numpy. Результат запишите в массив mean_a, в нем должно быть 2 элемента.
mean_a = a.mean(axis = 0)
mean_a
# Задание 2
# =========
# Вычислите массив a_centered, отняв от значений массива “а” средние значения соответствующих признаков, содержащиеся в массиве mean_a.
#
# Вычисление должно производиться в одно действие. Получившийся массив должен иметь размер 5x2.
a_centered = a - mean_a
a_centered
# Задание 3
# =========
# Найдите скалярное произведение столбцов массива a_centered.
#
# В результате должна получиться величина a_centered_sp. Затем поделите a_centered_sp на N-1, где N - число наблюдений.
a_centered_sp = np.dot(a_centered.T[0], a_centered.T[1])
a_centered_sp
a_centered_sp / (a.shape[0]-1)
# Задание 4**
# ==========
#
# Число, которое мы получили в конце задания 3 является ковариацией двух признаков, содержащихся в массиве “а”. В задании 4 мы делили сумму произведений центрированных признаков на N-1, а не на N, поэтому полученная нами величина является несмещенной оценкой ковариации.
#
# Подробнее узнать о ковариации можно здесь:
#
# [Выборочная ковариация и выборочная дисперсия — Студопедия](https://studopedia.ru/9_153900_viborochnaya-kovariatsiya-i-viborochnaya-dispersiya.html)
#
# В этом задании проверьте получившееся число, вычислив ковариацию еще одним способом - с помощью функции np.cov. В качестве аргумента m функция np.cov должна принимать транспонированный массив “a”. В получившейся ковариационной матрице (массив Numpy размером 2x2) искомое значение ковариации будет равно элементу в строке с индексом 0 и столбце с индексом 1.
#
np.cov(a.T)[0, 1]
| lesson_01/lesson_01.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import numpy as np
import xarray as xr
from datetime import datetime
import matplotlib.pyplot as plt
import matplotlib.dates as mdates
# +
file = 'onarrows_all.txt'
with open(file, 'r') as rfp:
data = {}
for i, line in enumerate(rfp):
if ((i > 33)):#&(i < 50000)):
ld = line.rstrip().split('\t')
time = datetime.strptime(ld[2], '%Y-%m-%d %H:%M')
year = time.year
flow = ld[-2]
try:
data[year]
except:
data[year] = [[time, flow]]
else:
data[year].append([time, flow])
data = {k:np.array(data[k]) for k in data.keys()}
# +
fig = plt.figure(facecolor='w', figsize=(24, 12))
plt.rcParams.update({'font.size': 18})
ax = fig.add_subplot(111)
for yyyy in np.arange(1990, 2020, 1):
plotdates = np.array([x.replace(year=2016) for x in data[yyyy][:,0]])
plotdata = data[yyyy][:,1].astype(np.float)
if plotdata.max() > 1000:
print(yyyy)
if yyyy == 2019:
ax.plot(plotdates, plotdata, linewidth=4, c='r', label=yyyy)
else:
ax.plot(plotdates, plotdata, linewidth=1.5, alpha=0.7, label=yyyy)
yt = np.arange(0, 2000, 100)
ax.set_yticks(yt)
ax.set_yticklabels(yt)
ax.set_ylim([0, 1600])
ax.set_ylabel('Flow [cfs]')
months = mdates.MonthLocator() # every month
monthsFmt = mdates.DateFormatter('%B 1st')
ax.xaxis.set_major_locator(months)
ax.xaxis.set_major_formatter(monthsFmt)
plt.setp( ax.xaxis.get_majorticklabels(), rotation=30)
ax.set_xlim([datetime(2016, 2, 1, 0, 0), datetime(2016, 7, 15, 23, 59)])
ax.set_xlabel('Time of Year')
leg = ax.legend(loc='upper right')
for legobj in leg.legendHandles:
legobj.set_linewidth(5.0)
ax.grid()
ax.set_title('Ogden Narrows: Years With Flows > 1000 cfs')
plt.show()
# +
fig = plt.figure(facecolor='w', figsize=(24, 12))
plt.rcParams.update({'font.size': 18})
ax = fig.add_subplot(111)
for yyyy in np.arange(1990, 2020, 1):
plotdates = np.array([x.replace(year=2016) for x in data[yyyy][:,0]])
plotdata = data[yyyy][:,1].astype(np.float)
if plotdata.max() <= 500:
print(yyyy)
if yyyy == 2019:
ax.plot(plotdates, plotdata, linewidth=4, c='r', label=yyyy)
else:
ax.plot(plotdates, plotdata, linewidth=1.5, alpha=0.7, label=yyyy)
yt = np.arange(0, 2000, 100)
ax.set_yticks(yt)
ax.set_yticklabels(yt)
ax.set_ylim([0, 500])
ax.set_ylabel('Flow [cfs]')
months = mdates.MonthLocator() # every month
monthsFmt = mdates.DateFormatter('%B 1st')
ax.xaxis.set_major_locator(months)
ax.xaxis.set_major_formatter(monthsFmt)
plt.setp( ax.xaxis.get_majorticklabels(), rotation=30)
ax.set_xlim([datetime(2016, 2, 1, 0, 0), datetime(2016, 7, 15, 23, 59)])
ax.set_xlabel('Time of Year')
leg = ax.legend(loc='upper right')
for legobj in leg.legendHandles:
legobj.set_linewidth(5.0)
ax.grid()
ax.set_title('Ogden Narrows: Years With Max Flows < 500 cfs')
plt.show()
# -
for yyyy in np.arange(1991, 2020, 1):
plotdates = np.array([x.replace(year=2016) for x in data[yyyy][:,0]])
plotdata = data[yyyy][:,1].astype(np.float)
if plotdata.max() > 1000:
print('{}, {:.0f} cfs, {}'.format(yyyy, plotdata.max(), np.unique([x.strftime('%B') for x in plotdates[np.where(plotdata == plotdata.max())]])[0]))
| gages.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# <img style="float: right;" src="data:image/png;base64,iVBORw0KGgoAAAANSUhEUgAAAOIAAAAjCAYAAACJpNbGAAAABHNCSVQICAgIfAhkiAAAAAlwSFlzAAALEgAACxIB0t1+/<KEY>3JlYXRpb24gVGltZQAzLzcvMTNND4u/AAAAHHRFWHRTb2Z0d2FyZQBBZG9iZSB<KEY>gN/133sW0yh927j1mucIaFWINl7PJ+OcvMcfW8Bol3iN44+mLIOsTCp3UJFfAETr+WRQcG8EOJpunEnTyDlYzycbeWr5xxq3jOF6PglK8ix9buv5xCsrAzBkMV1l5OwD/aJ4BXzV3+8F9z4gz/hTSbz8cxc84FuNvDc4VIsYA7+<KEY>>
#
#
# # Getting Started with PCSE/WOFOST
#
# This Jupyter notebook will introduce PCSE and explain the basics of running models with PCSE, taking WOFOST as an example.
#
# <NAME>, March 2018
#
# **Prerequisites for running this notebook**
#
# Several packages need to be installed for running PCSE/WOFOST:
#
# 1. `PCSE` and its dependencies. See the [PCSE user guide](http://pcse.readthedocs.io/en/stable/installing.html) for more information;
# 2. `pandas` for processing and storing WOFOST output;
# 3. `matplotlib` for generating charts
#
# ## Importing the relevant modules
#
# %matplotlib inline
import sys, os
import pcse
import pandas
import matplotlib
matplotlib.style.use("ggplot")
import matplotlib.pyplot as plt
print("This notebook was built with:")
print("python version: %s " % sys.version)
print("PCSE version: %s" % pcse.__version__)
# ## Starting from the internal demo database
# For demonstration purposes, we can start WOFOST with a single function call. This function reads all relevant data from the internal demo databases. In the next notebook we will demonstrate how to read data from external sources.
#
# The command below starts WOFOST in potential production mode for winter-wheat for a location in Southern Spain.
wofostPP = pcse.start_wofost(mode="pp")
# You have just successfully initialized a PCSE/WOFOST object in the Python interpreter, which is in its initial state and waiting to do some simulation. We can now advance the model state for example with 1 day:
#
wofostPP.run()
# Advancing the crop simulation with only 1 day, is often not so useful so the number of days to simulate can be specified as well:
wofostPP.run(days=10)
# ## Getting information about state and rate variables
# Retrieving information about the calculated model states or rates can be done with the `get_variable()` method on a PCSE object. For example, to retrieve the leaf area index value in the current model state you can do:
wofostPP.get_variable("LAI")
wofostPP.run(days=25)
wofostPP.get_variable("LAI")
# Showing that after 11 days the LAI value is 0.287. When we increase time with another 25 days, the LAI increases to 1.528. The `get_variable()` method can retrieve any state or rate variable that is defined somewhere in the model.
#
# Finally, we can finish the crop season by letting it run until the model terminates because the crop reaches maturity or the harvest date:
wofostPP.run_till_terminate()
# Note that before or after the crop cycle, the object representing the crop does not exist and therefore retrieving a crop related variable results in a `None` value. Off course the simulation results are stored and can be obtained, see next section.
print(wofostPP.get_variable("LAI"))
# ## Retrieving and displaying WOFOST output
# We can retrieve the results of the simulation at each time step using `get_output()`. In python terms this returns a list of dictionaries, one dictionary for each time step of the the simulation results. Each dictionary contains the key:value pairs of the state or rate variables that were stored at that time step.
#
#
output = wofostPP.get_output()
# The most convenient way to handle the output from WOFOST is to used the `pandas` module to convert it into a dataframe. Pandas DataFrames can be converted to a variety of formats including excel, CSV or database tables.
dfPP = pandas.DataFrame(output).set_index("day")
dfPP.tail()
# Besides the output at each time step, WOFOST also provides summary output which summarizes the crop cycle and provides you the total crop biomass, total yield, maximum LAI and other variables. In case of crop rotations, the summary output will consist of several sets of variables, one for each crop cycle.
summary_output = wofostPP.get_summary_output()
msg = "Reached maturity at {DOM} with total biomass {TAGP:.1f} kg/ha, " \
"a yield of {TWSO:.1f} kg/ha with a maximum LAI of {LAIMAX:.2f}."
for crop_cycle in summary_output:
print(msg.format(**crop_cycle))
# ## Visualizing output
# The pandas module is also very useful for generating charts from simulation results. In this case we generate graphs of leaf area index and crop biomass including total biomass and grain yield.
fig, (axis1, axis2) = plt.subplots(nrows=1, ncols=2, figsize=(16,8))
dfPP.LAI.plot(ax=axis1, label="LAI", color='k')
dfPP.TAGP.plot(ax=axis2, label="Total biomass")
dfPP.TWSO.plot(ax=axis2, label="Yield")
axis1.set_title("Leaf Area Index")
axis2.set_title("Crop biomass")
fig.autofmt_xdate()
r = fig.legend()
| 01 Getting Started with PCSE.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import requests
import pandas as pd
from bs4 import BeautifulSoup
import json
from urllib.request import urlopen
from tqdm import tqdm_notebook
import unidecode
# # Crawling Pitchfork Review Rating
def pf_rating(starting, ending):
df = pd.DataFrame(columns=['artists', 'albums','genre', 'release_date', 'ratings'])
for j in tqdm_notebook(range(starting, ending, 12)):
url = 'https://pitchfork.com/api/v2/search/?types=reviews&hierarchy=sections%2Freviews%2Falbums&sort=publishdate%20desc%2Cposition%20asc&size=12&start=' + str(j)
data = json.load(urlopen(url))
for i in range(len(data['results']['list'])):
try:
artist_name = data['results']['list'][i]['artists'][0]['display_name']
except IndexError:
artist_name = "Various Artist"
album_name = data['results']['list'][i]['tombstone']['albums'][0]['album']['display_name']
rating_score = data['results']['list'][i]['tombstone']['albums'][0]['rating']['rating']
release_date = data['results']['list'][i]['pubDate'][:-14]
genre_ls = []
for k in range(len(data['results']['list'][i]['genres'])):
genre_k = data['results']['list'][i]['genres'][k]['display_name']
genre_ls.append(genre_k)
input_data={
'artists' : artist_name,
'albums' : album_name,
'genre' : genre_ls,
'release_date' : release_date,
'ratings' : rating_score,
}
df.loc[len(df)] = input_data
return df
df1 = pf_rating(0, 2400)
df2 = pf_rating(2401, 4800)
df3 = pf_rating(4801, 6000)
df4 = pf_rating(6001, 8400)
df5 = pf_rating(8401, 8772)
df = pd.concat([df1, df2, df3, df4, df5], ignore_index=True)
df = df[:8748]
df.shape
df.to_csv("../data/pitchfork_rating_1118.csv", index=False)
df = pd.read_csv("../data/rating/rating_pitchfork-1118-utf8.csv")
def decodeuni(str):
return unidecode.unidecode(str)
df['artists'] = df['artists'].apply(lambda x : str(x))
df['albums'] = df['albums'].apply(lambda x : str(x))
df['artists'] = df['artists'].apply(decodeuni)
df['albums'] = df['albums'].apply(decodeuni)
df.to_csv("../data/rating_pitchfork-1118.csv", index=False)
df = pd.read_csv("../data/rating/rating_pitchfork-1118-utf8.csv")
df.tail()
df.info()
# ### Upload to MySQL database
import sqlalchemy, pickle
from sqlalchemy import create_engine
pw = pickle.load(open("mysql_pw.pickle", "rb"))
engine = sqlalchemy.create_engine("mysql+mysqldb://root:" + pw + "@192.168.3.11/project_rookie")
df.to_sql(name="rating_pitchFork_copy", con=engine, if_exists='replace')
| crawler/scrape-pitchfork-ratings-1118.ipynb |
% ---
% jupyter:
% jupytext:
% text_representation:
% extension: .m
% format_name: light
% format_version: '1.5'
% jupytext_version: 1.14.4
% kernelspec:
% display_name: Octave
% language: octave
% name: octave
% ---
% some housekeeping stuff
register_graphics_toolkit("gnuplot");
available_graphics_toolkits();
graphics_toolkit("gnuplot")
clear
% end of housekeeping
% # U shaped solubility diagram
%
% For Fe$^{3+}$ we can have Fe(OH)$_{3(\mathrm{s})}$ form as a solid phase
%
% $\mathrm{Fe(OH)}_{3(\mathrm{s})} \leftrightharpoons \mathrm{Fe}^{3+} + 3\mathrm{OH}^-$
%
% with $K_{sp}$=10$^{-38.8}$
%
% There are also 3 overall deprotonation reactions with $\beta_{a1}=10^{-2.19}$, $\beta_{a2}=10^{-5.7}$ and $\beta_{a4}=10^{-21.6}$. Note: some databases include a $\beta_{a3}$ but the formation of Fe(OH)$_3$ as an aqueous species is not confirmed.
%
%plot -s 600,500 -f 'svg'
pH=2:0.1:12;
Ksp=10^-38.8;
B1=10^-2.19;
B2=10^-5.7;
B4=10^-21.6;
Kw=1e-14;
logFe=log10(Ksp./Kw^3)-3*pH; Fe=10.^logFe;
logFeOH=log10(B1*Ksp/Kw^3)-2*pH; FeOH=10.^logFeOH;
logFeOH2=log10(B2*Ksp/Kw^3)-pH; FeOH2=10.^logFeOH2;
logFeOH4=log10(B4*Ksp./Kw^3)+pH; FeOH4=10.^logFeOH4;
FeT=Fe+FeOH+FeOH2+FeOH4; logFeT=log10(FeT);
plot(pH,logFe,'linewidth',2,pH,logFeOH,'linewidth',2,pH,logFeOH2,'linewidth',2,...
pH,logFeOH4,'linewidth',2,pH,logFeT,'linewidth',2)
set(gca,'linewidth',2,'fontsize',12);
xlabel('pH','fontsize',12)
ylabel('log[species]','fontsize',12)
legend('Fe^{3+}','FeOH^{2+}','Fe(OH)_2^+','Fe(OH)_4^-','Fe_T','location','southwest');
% plot over a more narrow range
plot(pH,logFe,'linewidth',2,pH,logFeOH,'linewidth',2,pH,logFeOH2,'linewidth',2,...
pH,logFeOH4,'linewidth',2,pH,logFeT,'linewidth',2)
set(gca,'linewidth',2,'fontsize',12)
xlabel('pH','fontsize',12)
ylabel('log[species]','fontsize',12)
legend('Fe^{3+}','FeOH^{2+}','Fe(OH)_2^+','Fe(OH)_4^-','Fe_T','location','north')
axis([2 10 -11 -3])
| Ushaped_solubility_diagram.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import json
import time
import numpy
import pandas
import sqlalchemy
from data.flat import load
import datetime
import itertools
from data.flat import add_lag, call_them_all, consequentive_lagger, consequentive_pcter, to_date, fill_all, minute_offset
# -
# k = 'C:/Users/MainUser/Desktop/AlphaVantage.txt'
k = 'C:/Users/MainUser/Desktop/OpenAPI_sandbox.txt'
crs = open(k, "r")
for columns in ( raw.strip().split() for raw in crs ):
api_key = columns[0]
target_quotes = ['MSFT']
news_horizon = 100
effect_horizon = 1
max_quotes_lag = 10
d = './data/data/rex.xlsx'
data = pandas.read_excel(d)
# +
# data = data[data['source'] != 'HXK']
# -
data
newstitle_frame = data[['id', 'time', 'title']]
lag_markers = list(itertools.product(newstitle_frame['id'].values, numpy.array(numpy.arange(news_horizon - 1)) + 1))
lag_markers = pandas.DataFrame(data=lag_markers, columns=['id', 'lag'])
newstitle_frame = newstitle_frame.merge(right=lag_markers, left_on=['id'], right_on=['id'])
newstitle_frame
newstitle_frame['time'] = pandas.to_datetime(newstitle_frame['time'])
newstitle_frame['news_time'] = newstitle_frame['time'].copy()
# newstitle_frame['time'] = newstitle_frame.apply(func=add_lag, axis=1)
newstitle_frame['time'] = newstitle_frame['lag'].apply(func=minute_offset)
newstitle_frame['time'] = newstitle_frame['news_time'] + newstitle_frame['time']
beginning_date, ending_date = newstitle_frame['time'].min() - pandas.DateOffset(
minutes=(max_quotes_lag + effect_horizon)), newstitle_frame['time'].max()
# beginning_date = datetime.datetime.combine(beginning_date, datetime.datetime.min.time())
# ending_date = datetime.datetime.combine(ending_date, datetime.datetime.min.time())
quotes_data = await call_them_all(tickers=target_quotes,
start_date=beginning_date, end_date=ending_date,
token=api_key)
quotes_data
# +
quotes_data = quotes_data.set_index(keys=['ticker', 'time'])
quotes_data = quotes_data.sort_index(ascending=True)
quotes_data = fill_all(frame=quotes_data, freq='T', zero_index_name='ticker', first_index_name='time')
# -
quotes_data
quotes_data = consequentive_lagger(frame=quotes_data, n_lags=effect_horizon, suffix='_HOZ')
quotes_data
quotes_data = consequentive_pcter(frame=quotes_data, horizon=1)
quotes_data
quotes_data = quotes_data.reset_index()
# quotes_data['time'] = quotes_data['time'].apply(func=to_date)
quotes_data
newstitle_frame['time'] = pandas.to_datetime(newstitle_frame['time'])
quotes_data['time'] = pandas.to_datetime(quotes_data['time'])
# +
qd_tz = quotes_data.loc[0, 'time'].tz
def fix_tz(x):
return x.tz_localize(tz=qd_tz)
newstitle_frame['time'] = newstitle_frame['time'].apply(func=fix_tz)
# -
def fixit(x):
return x.ceil(freq='T')
quotes_data['time'] = quotes_data['time'].apply(func=fixit)
newstitle_frame['time'] = newstitle_frame['time'].apply(func=fixit)
# +
with open('E:/InverseStation/terminator_panel/users.json') as f:
users = json.load(f)
user, password = users['justiciar']['user'], users['justiciar']['password']
with open('E:/InverseStation/terminator_panel/servers.json') as f:
users = json.load(f)
host, port = users['GOLA']['host'], users['GOLA']['port']
dbname = 'tempbox'
# -
connection_string = "postgresql+psycopg2://{}:{}@{}:{}/{}".format(user, password, host, port, dbname)
engine = sqlalchemy.create_engine(connection_string)
connection = engine.connect()
quotes_data.to_sql(name='quotes_data', con=connection, if_exists='replace', index=False)
newstitle_frame.to_sql(name='newstitle_frame', con=connection, if_exists='replace', index=False)
# +
query = """
SELECT RS.*
FROM
(SELECT NF."id"
, NF.title
, NF."lag"
, NF.news_time
, QD.*
FROM
public.newstitle_frame AS NF
FULL OUTER JOIN
public.quotes_data AS QD
ON NF."time" = QD."time") AS RS
WHERE 37 = 37
;
"""
result = pandas.read_sql(sql=query, con=connection)
# -
result[~result['title'].isna() & ~result['open_HOZ0_PCT1'].isna()]
# +
# https://stackoverflow.com/questions/47386405/memoryerror-when-i-merge-two-pandas-data-frames
# -
the_data = quotes_data.merge(right=newstitle_frame, left_on='time', right_on='target_date', how='outer')
newstitle_frame['time']
the_data
| trash/drafts/flat_model_draft.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .jl
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Julia 1.0.3
# language: julia
# name: julia-1.0
# ---
# + active=""
# ScikitLearn.jl implements the popular scikit-learn interface and algorithms in Julia.
#
# It supports both models from the Julia ecosystem and those of the scikit-learn library, via PyCall.jl.
#
#
# Main features:
#
# Around 150 Julia and Python models accessed through a uniform interface
# Pipelines and Feature Unions
# Cross-validation
# Hyperparameter tuning
# DataFrames support
#
# Manual: http://scikitlearnjl.readthedocs.io/en/latest/
# Examples: https://github.com/cstjean/ScikitLearn.jl/docs/examples.md
# -
using RDatasets: dataset
using ScikitLearn
iris = dataset("datasets", "iris");
X = convert(Matrix, iris[[:SepalLength, :SepalWidth, :PetalLength, :PetalWidth]]);
y = convert(Array, iris[:Species]);
@sk_import linear_model: LogisticRegression
model = LogisticRegression(fit_intercept=true)
fit!(model, X, y)
accuracy = sum(predict(model, X) .== y) / length(y)
println("Accuracy: $accuracy")
# ---
| Chp10/Notebooks/SKLearn.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [conda env:bootcamp_env] *
# language: python
# name: conda-env-bootcamp_env-py
# ---
# # Sript: API call
# ## File: api_call.ipynb
#
# The script makes successive requests to the World Weather Online API (https://www.worldweatheronline.com/developer/api/historical-weather-api.aspx), based on dates for a period starting on the 1st of July of 2014 and ending on the 30th of September of 2014.
#
# The output is a data frame (weather_data) that is saved to a csv file (weather_data.csv) in the folder Data. The retrieved information consists of:
#
# - Date
# - Maximum temperature (°C)
# - Average temperature (°C)
# - Minimum temperature (°C)
# - UV Index
# - Precipitation (mm)
# - Weather description
# - Cloud cover (%)
# - Humidity (%)
# ### Functions:
# +
#### USED: https://stackoverflow.com/questions/55452835/creating-a-data-set-using-an-api-with-python
# Auxiliary function to format dates for API requests.
def daterange(start_date, end_date):
for n in range(int ((end_date - start_date).days)):
yield start_date + datetime.timedelta(n)
# -
# ### Creating variables
# +
# API parameters
city="New York"
start_date="2014-07-01"
end_date="2014-10-01"
date_format = "%Y-%m-%d"
# Formatting the start and end date
start_date = datetime.datetime.strptime(start_date, date_format)
end_date = datetime.datetime.strptime(end_date, date_format)
# Creating lists to store data from the API
all_data = []
date_list=[]
maxtempC_list = []
avgtempC_list = []
mintempC_list = []
uvIndex_list =[]
precipMM_list=[]
weatherDesc_list=[]
cloudcover_list=[]
humidity_list=[]
# -
# ### API calls
#
# +
print(f"Retrieving weather information from {start_date} to {end_date}.\n")
for each_date in daterange(start_date, end_date):
# print(each_date.date())
ask = str(each_date.date())
request = requests.get(
"http://api.worldweatheronline.com/premium/v1/past-weather.ashx?",
params=dict(
key=weather_api_key,
q=city,
format='json',
date=ask,
tp='24'
)
)
json_data = request.json()
# print(json_data)
all_data.append(json_data)
date_list.append(json_data['data']['weather'][0]['date'])
maxtempC_list.append(json_data['data']['weather'][0]['maxtempC'])
avgtempC_list.append(json_data['data']['weather'][0]['avgtempC'])
mintempC_list.append(json_data['data']['weather'][0]['mintempC'])
uvIndex_list.append(json_data['data']['weather'][0]['uvIndex'])
precipMM_list.append(json_data['data']['weather'][0]['hourly'][0]['precipMM'])
weatherDesc_list.append(json_data['data']['weather'][0]['hourly'][0]['weatherDesc'][0]['value'])
cloudcover_list.append(json_data['data']['weather'][0]['hourly'][0]['cloudcover'])
humidity_list.append(json_data['data']['weather'][0]['hourly'][0]['humidity'])
print(f"{request}: COMPLETE ---> {json_data['data']['weather'][0]['date']}")
# -
# ### Ensembling the weather data frame
# +
# create a data frame from date, maxtemp, avgtemp, uvIndex
weather_dict = {
'date': date_list,
"maxtempC": maxtempC_list,
"avgtempC": avgtempC_list,
"uvIndex": uvIndex_list,
"precipMM":precipMM_list,
"cloudcover":cloudcover_list,
"humidity":humidity_list,
"weatherDesc":weatherDesc_list
}
weather_data = pd.DataFrame(weather_dict)
weather_data
# weather_data.info()
# data3.Title = data3.Title.str.replace(',', '').astype(float).astype(int)
for index,column in enumerate(weather_data):
if (index!=0) & (index!=7):
weather_data[column]= weather_data[column].astype(float)
# weather_data.info()
# # overwriting data after changing format
# data["Time"]= pd.to_datetime(data["Time"])
# # info of data
# data.info()
# # display
# data
weather_data["date"]=pd.to_datetime(weather_data["date"])
# weather_data.info()
# -
# ### Saving the weather data frame to a csv file
# +
weather_data.to_csv("Data/weather_data.csv", encoding = "utf-8", index = False, header = True)
print("\n\n SUCCESS!!! File 'weather_data.csv' created and saved in folder 'Data'")
| api_call.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Ex1 - Filtering and Sorting Data
#
# Check out [Chipotle Exercises Video Tutorial](https://youtu.be/ZZPiWZpdekA) to watch a data scientist go through the exercises
# This time we are going to pull data directly from the internet.
# Special thanks to: https://github.com/justmarkham for sharing the dataset and materials.
#
# ### Step 1. Import the necessary libraries
import pandas as pd
# ### Step 2. Import the dataset from this [address](https://raw.githubusercontent.com/justmarkham/DAT8/master/data/chipotle.tsv).
# ### Step 3. Assign it to a variable called chipo.
# +
url = 'https://raw.githubusercontent.com/justmarkham/DAT8/master/data/chipotle.tsv'
chipo = pd.read_csv(url, sep = '\t')
# -
# ### Step 4. How many products cost more than $10.00?
# +
# clean the item_price column and transform it in a float
#prices = [float(value[1 : -1]) for value in chipo.item_price]
# reassign the column with the cleaned prices
#chipo.item_price = prices
# delete the duplicates in item_name and quantity
#chipo_filtered = chipo.drop_duplicates(['item_name','quantity','choice_description'])
# chipo_filtered
# select only the products with quantity equals to 1
#chipo_one_prod = chipo_filtered[chipo_filtered.quantity == 1]
#chipo_one_prod
# chipo_one_prod[chipo_one_prod['item_price']>10].item_name.nunique()
# chipo_one_prod[chipo_one_prod['item_price']>10]
chipo.item_price = chipo.item_price.str.replace('$','', regex=False).astype('float')
chipo.query('item_price > 10').item_name.nunique()
# -
# ### Step 5. What is the price of each item?
# ###### print a data frame with only two columns item_name and item_price
# +
# delete the duplicates in item_name and quantity
# chipo_filtered = chipo.drop_duplicates(['item_name','quantity'])
chipo[(chipo['item_name'] == 'Chicken Bowl') & (chipo['quantity'] == 1)]
# select only the products with quantity equals to 1
# chipo_one_prod = chipo_filtered[chipo_filtered.quantity == 1]
# select only the item_name and item_price columns
# price_per_item = chipo_one_prod[['item_name', 'item_price']]
# sort the values from the most to less expensive
# price_per_item.sort_values(by = "item_price", ascending = False).head(20)
# -
# ### Step 6. Sort by the name of the item
# +
chipo.item_name.sort_values()
# OR
chipo.sort_values(by = "item_name")
# -
# ### Step 7. What was the quantity of the most expensive item ordered?
chipo.sort_values(by = "item_price", ascending = False).head(1)
# ### Step 8. How many times was a Veggie Salad Bowl ordered?
# +
chipo_salad = chipo[chipo.item_name == "Veggie Salad Bowl"]
len(chipo_salad)
# -
# ### Step 9. How many times did someone order more than one Canned Soda?
chipo_drink_steak_bowl = chipo[(chipo.item_name == "Canned Soda") & (chipo.quantity > 1)]
len(chipo_drink_steak_bowl)
| 02_Filtering_&_Sorting/!Chipotle/Exercises_with_solutions.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Deep Animator
#
# > Image animation given a driving video sequence and a source image.
# Deep Animator is an implementation of [First Order Motion Model for Image Animation](https://papers.nips.cc/paper/8935-first-order-motion-model-for-image-animation) by [<NAME>](https://github.com/AliaksandrSiarohin), [<NAME>](http://stelat.eu/), [<NAME>](http://www.stulyakov.com/), [<NAME>](http://elisaricci.eu/) and [<NAME>](http://disi.unitn.it/~sebe/).
#
# The source code can be found [here](https://github.com/AliaksandrSiarohin/first-order-model). **This library is a simple transformation of the original code into a shell script for quick experimentation**. This is also an educational effort for the writer.
# ## Install
# Run `pip install deep-animator` to install the library in your environment.
# ## How to use
# First you need to download the weights of the model [here](https://drive.google.com/file/d/1zqa0la8FKchq62gRJMMvDGVhinf3nBEx/view?usp=sharing). Then just run the following command.
#
# `deep_animate <path_to_the_source_image> <path_to_the_driving_video> <path_to_yaml_conf> <path_to_model_weights>`
#
# * Example of source image [here](https://drive.google.com/file/d/1ACSKOfQUHbSEWmPu4Ndss7bkrPVK5WBR/view?usp=sharing)
# * Example of driving video [here](https://drive.google.com/file/d/103PEtO2QO45XwCNLYIzMcW3aRdbOhS1D/view?usp=sharing)
#
# An example YAML file is given below:
#
# ```
# model_params:
# common_params:
# num_kp: 10
# num_channels: 3
# estimate_jacobian: True
# kp_detector_params:
# temperature: 0.1
# block_expansion: 32
# max_features: 1024
# scale_factor: 0.25
# num_blocks: 5
# generator_params:
# block_expansion: 64
# max_features: 512
# num_down_blocks: 2
# num_bottleneck_blocks: 6
# estimate_occlusion_map: True
# dense_motion_params:
# block_expansion: 64
# max_features: 1024
# num_blocks: 5
# scale_factor: 0.25
# discriminator_params:
# scales: [1]
# block_expansion: 32
# max_features: 512
# num_blocks: 4
# sn: True
# ```
| nbs/index.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [conda root]
# language: python
# name: conda-root-py
# ---
import pandas as pd
pd.options.display.max_columns = 100
import json, itertools
import time
import numpy as np
import matplotlib.pyplot as plt
import utils as ut
# # Counting number of words per person per session
# Counting the number of interventions per member per session can be done following along the same process.
# Importing a json file containing a map between active PersonNumber's and PersonIdCode's
# +
with open('data/active_PersonNumber_PersonIdCode_map.json') as data_file:
person_map = json.load(data_file)
active_ids = list(person_map.values())
active_ids.sort()
active_numbers = list(person_map.keys())
active_numbers.sort()
# -
# Importing info about the parliament members
mmbr = pd.read_csv('data/MemberCouncil.csv')
# the ppl DataFrame will be useful to map PersonNumber to PersonIdCode
ppl = pd.read_csv('data/Person.csv').dropna(axis=0, subset=['PersonNumber', 'PersonIdCode'])
ppl = pd.read_csv('data/people.csv').set_index('PersonIdCode')
active_numbers = ppl['PersonNumber']
# Importing a cleaned version of the transcripts containing no NaN on either PersonNumber or Text
transcript = pd.read_csv('../scraping/data/Transcript.csv')
# +
transc = transcript
transc.dropna(axis=0, inplace=True, subset=['PersonNumber', 'Text'])
transc['PersonNumber'] = transc['PersonNumber'].apply(lambda x: int(x))
# -
# Building the DataFrame that will store our calculations
# columns
cols = ['PersonIdCode']
cols.extend(transc.IdSession.unique().tolist())
df = pd.DataFrame(columns=cols)
df.PersonIdCode = ppl.index
df.fillna(value=0, inplace=True)
df.set_index('PersonIdCode', inplace=True)
df.tail(2)
for row in transc.iterrows():
row = row[1]
#print(row['PersonNumber'])
if str(row['PersonNumber']) in active_numbers:
# by changing the += to simply '1', we can compute the # of interventions
#df.loc[int(str(person_map[str(row['PersonNumber'])])), row['IdSession']] += len(ut.to_text(row['Text']).split())
df.loc[int(str(person_map[str(row['PersonNumber'])])), row['IdSession']] += 1
df.sort_index(inplace=True)
df.head(3)
# Taking a quick look at the data
# +
B = df.replace(0, np.nan).drop([49, 3, 4, 5, 59, 6], axis=1)
fig, ax = plt.subplots(figsize=(15,15))
ax.imshow(B, interpolation="none")
plt.show()
# -
# Dropping weird sessions (checked, they're dummies)
df = df.drop([49, 3, 4, 5, 59, 6], axis=1)
# Saving
df.to_csv('new_data/n_int.csv')
| graph/n_words_per_session.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import pandas as pd
from matplotlib import pyplot as plt
import seaborn as sns
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import train_test_split
from sklearn.metrics import r2_score, mean_squared_error
import numpy as np
pd.options.mode.chained_assignment = None
# %matplotlib inline
#The data is contained in 4 .csv files
gdp = pd.read_csv("data/datasets_15310_20369_gdp_improvement_rate.csv")
literacy = pd.read_csv("data/datasets_15310_20369_literacy_rate.csv")
unemployment = pd.read_csv("data/datasets_15310_20369_unemployment.csv")
youth = pd.read_csv("data/datasets_15310_20369_youth.csv")
#drop columns that are empty
literacy.drop(['86.3', '90', '82.7', '7.3\t'], inplace = True, axis = 1)
#merge all data frames together
gl = gdp.merge(literacy, on='country', how = 'outer')
glu = gl.merge(unemployment, on='country', how = 'outer')
df = glu.merge(youth, on='country', how = 'outer')
#fix column names that contain spaces
cols = []
for col in list(df.columns):
col = col.replace(" ", "")
cols.append(col)
df.columns = cols
df.head()
# -
#find the percent of data that is **present** in each column
(df.shape[0] - df.isnull().sum())/df.shape[0] * 100
df.describe(include = "all")
# +
#the most data is missing in the literacy and gender difference columns (about 46%)
#get total df size
df.shape
# -
#Is this the same as the number of **unique** values in the country column?
df['country'].nunique()
#Let's remove rows where there are duplicate country names
df.drop_duplicates(subset='country', keep="first", inplace = True)
df.shape
#how many values are **present** in the "unemployment_percentage" column
df.shape[0] - df['unemployment_percentage'].isnull().sum()
#Let's see how the data are distributed.
fig, ax = plt.subplots(figsize = (15, 7))
sns.violinplot(data=df.iloc[:,1:8])
plt.xticks(rotation=90)
# +
#Those are some pretty large spreads for literacy rates and unemployment percentage
#I'd like to get a better look at the distribution of the unemployment data
plt.hist(df['unemployment_percentage'], bins=20, edgecolor="black")
plt.xlabel('Percent of Residents Unemployed')
plt.ylabel('Count')
#let's plot the data mean with a red line
plt.axvline(df['unemployment_percentage'].mean(), color="red", linewidth = 1)
#and the data median with a green line
plt.axvline(df['unemployment_percentage'].median(), color="green", linewidth = 1)
# +
#Looks like the data have a right-skewed distribution. Most countries have a relatively low rate of unemployment,
#but some countries are particularly hard-hit.
#Let's investigate which factors in our data frame are correlated with unemployment.
#First we'll have to clean the data.
df.head()
# +
def clean_data(df):
# Drop rows with missing unemployment values
df.dropna(subset=['unemployment_percentage'], axis=0, inplace = True)
#subset y data
y = df['unemployment_percentage']
#drop y data and country name
df1 = df.drop(['country', 'unemployment_percentage', 'literacy_rate_percent_all'], axis = 1)
# Fill numeric columns with the mean
num_vars = df1.select_dtypes(include=['float', 'int']).columns
for col in num_vars:
df1[col].fillna((df1[col].mean()), inplace=True)
X = df1
return X, y
#Use the function to create X and y
X, y = clean_data(df)
# -
#let's check whether everything worked as expected
print(X.shape)
print(y.shape)
#Looks like we have all 217 rows that contain unique county names.
#Sanity check to make sure all NaNs in X have been filled
(X.shape[0] - X.isnull().sum())/X.shape[0] * 100
# +
#Now we'll fit a linear model
#split data
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.3, random_state=25)
lm_model = LinearRegression(normalize=True)
lm_model.fit(X_train, y_train)
y_train_preds = lm_model.predict(X_train)
train_score = (r2_score(y_train, y_train_preds))
y_test_preds = lm_model.predict(X_test)
test_score = (r2_score(y_test, y_test_preds))
#evaluate model
print(test_score, train_score)
# +
#It looks like only about 3% of the variation we're seeing in the unemployment rates
#can be explained by the factors that we're examining here.
#Let's look at what the most influential variables are
def coef_weights(coefficients, X_train):
coefs_df = pd.DataFrame()
coefs_df['est_int'] = X_train.columns
coefs_df['coefs'] = lm_model.coef_
coefs_df['abs_coefs'] = np.abs(lm_model.coef_)
coefs_df = coefs_df.sort_values('abs_coefs', ascending=False)
return coefs_df
#Use the function
coef_df = coef_weights(lm_model.coef_, X_train)
#A quick look at the top results
coef_df
# +
#It looks like percent of the female popluation of a country that is literate has the largest
#effect on unemployment, followed by the percent gender difference.
#But wait - female literacy rate is **positively** correlated with unemployment?
#While male and total literacy rate are negatively correlated with unemployment as expected.
# +
#What happens if we change the random state of our model?
random_states = [1, 2, 3, 4, 5, 6, 7, 8, 9, 10]
def fit_model(X, y, random_states):
column_names = ["rand_state", "est_int", "coefs", "abs_coefs"]
all_coefs = pd.DataFrame(columns = column_names)
all_scores = []
for i in random_states:
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.3, random_state=i)
lm_model = LinearRegression(normalize=True)
lm_model.fit(X_train, y_train)
y_train_preds = lm_model.predict(X_train)
train_score = (r2_score(y_train, y_train_preds))
y_test_preds = lm_model.predict(X_test)
test_score = (r2_score(y_test, y_test_preds))
#make a dataframe containing all of the variables' coefficients and r2 values
#for each random state tested
coefs_df = pd.DataFrame()
coefs_df['est_int'] = X_train.columns
coefs_df['coefs'] = lm_model.coef_
coefs_df['abs_coefs'] = np.abs(lm_model.coef_)
coefs_df['rand_state'] = i
coefs_df['r2'] = test_score
coefs_df = coefs_df.sort_values('abs_coefs', ascending=False)
all_coefs = pd.concat([all_coefs, coefs_df], axis = 0, ignore_index = True)
all_scores.append(test_score)
return all_coefs, all_scores
coefs, scores = fit_model(X, y, random_states)
# -
#That is a huge range for the test r2 score: -0.25 to 0.13!
#The range for the coefficient of female literacy rate percentage
#is also huge: 11.60 to 29.59
coefs.head()
#now let's pivot the results so that we can examine what the coefficients look like for the different
#test r2 values
coefs_pivot = coefs.pivot(index = 'r2', columns = 'est_int', values = 'coefs').reset_index().sort_values(by = ['r2'])
coefs_pivot
plt.scatter(np.abs(coefs_pivot.female_literacy_percent_rate), coefs_pivot.r2)
plt.xlabel('Percent of Females who are Literate Coefficient')
plt.ylabel('r2 Value of Linear Model')
#how many observations do we have if we drop all NaNs?
df_noNaN = df.dropna(how = 'any', axis = 0)
df_noNaN.shape
# +
#That's a much smaller dataset, but given the range of values for some of
#the variables, let's see if we can fit a better model now
Xclean, yclean = clean_data(df_noNaN)
coefs_clean, scores_clean = fit_model(Xclean, yclean, random_states)
coefs_clean.head()
# -
#now let's pivot the results so that we can examine what the coefficients look like for the different
#test r2 values
coefs_pivot_clean = coefs_clean.pivot(index = 'r2', columns = 'est_int', values = 'coefs').reset_index().sort_values(by = ['r2'])
coefs_pivot_clean
| Unemployment_literacy_and_gdp.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import matplotlib.pyplot as plt
import numpy as np
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import accuracy_score
# %matplotlib inline
# +
num_train_samples = 2000
num_train_features = 2
num_classes = 3
X_train = np.random.random((num_train_samples, num_train_features))
y_train = np.random.randint(low=0, high=num_classes, size=num_train_samples)
num_test_samples = 1000
X_test = np.random.random((num_test_samples, num_train_features))
y_test = np.random.randint(low=0, high=num_classes, size=num_test_samples)
# -
X_train[0]
X_test[0]
def PlotBar(array:[int]):
value_counts = {}
for x in array:
if x not in value_counts:
value_counts[x] = 0
value_counts[x] += 1
bar_x_labels = sorted(value_counts)
bar_heights = [value_counts[x] for x in bar_x_labels]
plt.xticks(bar_x_labels)
plt.xlabel("Class Labels")
plt.ylabel("Number of Samples")
plt.bar(bar_x_labels, bar_heights)
PlotBar(y_train)
PlotBar(y_test)
random_forest_classifier = RandomForestClassifier(n_estimators=10)
random_forest_classifier.fit(X_train, y_train)
y_pred = random_forest_classifier.predict(X_test)
print(accuracy_score(y_test, y_pred))
def PlotDataWithLabels(X, y):
x0 = {}
x1 = {}
for index, value in enumerate(y):
if value not in x0:
x0[value] = []
x1[value] = []
x0[value].append(X[index][0])
x1[value].append(X[index][1])
for y in sorted(x0):
plt.scatter(x0[y], x1[y], label=y, alpha=0.53)
plt.legend()
plt.title("Sample Data Points Colored by Class Labels")
plt.xlabel("Feature Dimension 1")
plt.ylabel("Feature Dimension 2")
# What the test samples and their labels look like.
PlotDataWithLabels(X_test, y_test)
# What the random forest predicted.
PlotDataWithLabels(X_test, y_pred)
| 1.1a Presence of a Target Function and Labeled Data is Necessary For Supervised Learning.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/zhangjiekui/pytorch-pretrained-BERT/blob/master/tansformersv241.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + id="q70-o62A4GJG" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 81} outputId="01c3bcaa-30da-4a58-ffd2-f1989e5a946c"
import tensorflow as tf
import torch
tf.__version__,torch.__version__
# + id="SQ4o0M6e4WlN" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="f33099e7-78b7-451d-9a56-cae3f72f2e0c"
tf.__version__,torch.__version__
# + id="tLdD4jnX4-Xq" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 156} outputId="1f051339-bda1-49bb-b869-5347dad693fb"
# !pip install --upgrade pip
# + id="J_ar0l4_4ZB8" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 1000} outputId="2bfcdbbd-7cb8-451b-f826-d2d60c3dea71"
# !pip install tensorflow==2.1.0
# + id="Qtoybuag5fXe" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="7b9de890-dd5a-4fba-dcc9-3457f4ecc941"
import tensorflow as tf
import torch
tf.__version__,torch.__version__
# + id="Pi373tNj404q" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 682} outputId="70730966-02bc-4fe9-d5c7-4d7ddfcfb35e"
# !pip install transformers
# + id="F-2g0nhM55Bg" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="5ea70664-2257-4ea6-d4e5-f540b98a21b7"
import transformers
transformers.__version__
# + id="AWKS1qTE6Dxq" colab_type="code" colab={}
import os
import tensorflow as tf
import tensorflow_datasets
from transformers import BertConfig,BertTokenizer,BertForSequenceClassification,TFBertForSequenceClassification
from transformers import glue_convert_examples_to_features,glue_processors
# + id="R5Ej37B-61bk" colab_type="code" colab={}
# script parameters
BATCH_SIZE = 32
EVAL_BATCH_SIZE = BATCH_SIZE * 2
USE_XLA = True
USE_AMP = True
EPOCHS = 3
# + id="CpSDRBWI6_dn" colab_type="code" colab={}
TASK = "mrpc"
if TASK == "sst-2":
TFDS_TASK = "sst2"
elif TASK == "sts-b":
TFDS_TASK = "stsb"
else:
TFDS_TASK = TASK
# + id="oefFW4tr7HUJ" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="94df1d16-a037-400c-99af-7a943ddbeb91"
num_labels=len(glue_processors[TASK]().get_labels())
num_labels
# + id="bumqTcOj7X_Y" colab_type="code" colab={}
tf.config.optimizer.set_jit(USE_XLA)
# + id="xsK_jTKZ7jrq" colab_type="code" colab={}
tf.config.optimizer.set_experimental_options({"auto_mixed_precision":USE_AMP})
# + id="sWqGt-JA70mf" colab_type="code" colab={}
model_name="bert-base-cased"
# + id="c6CScZWX75l0" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 66, "referenced_widgets": ["a5644d9e2a064555bc4c75da2848bf96", "f79bab46321e45cbbad48c104f489695", "75c4ac44d8dd4f69b9061574e88270cd", "08a03746350642f984a2290e32c09d3d", "d3531d88ca864bcc8769ddcf1247b274", "<KEY>", "6ae9041a299742eca7ab3f5c2f8bd0b8", "7e7bf03583e54c7ab6b8ac50e0f166ed"]} outputId="8f7be541-2e42-407d-fd41-3d7aa39300fa"
config=BertConfig.from_pretrained(model_name,num_labels=num_labels)
# + id="PKAeAgxV8OAJ" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 66, "referenced_widgets": ["edf9b12541c34286a1feab706f5bdfdf", "57ebb6587ddb46fdb4dc435707c1cc8f", "d76ea9582a494cb88516066385ad3516", "1051b0442c2b494a8b70711a385e5aae", "cfe6f4aaa09447eaa60b42ea6238649b", "695e6ffdb27d4f7eb54a3a6d41a1861d", "771157ebcf0f4f0f805ab7f6c5d1fdcd", "d9d2fac0756142bd97108e60a49840e5"]} outputId="933d84d2-2527-4706-b9f4-3a37a6ba9553"
tokenizer=BertTokenizer.from_pretrained(model_name)
# + id="vNJwVeI5_N8Y" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 66, "referenced_widgets": ["56b1a1a6741a44e493ddc7be32a55134", "9ad80982c7d44623a698c83b37fe2224", "6152719c73344e4eb4e74c7693ddbd31", "2736aaf2c7b74e7395fd775810c633fb", "<KEY>", "878e132d2aa545718a10625fedc36b00", "6c8df108788044098479956f81e0e908", "fa8b8ea84dc040faa929eab3e2565720"]} outputId="a8e0fff0-a9bb-48f8-d5d5-bb28273d7d92"
model=BertForSequenceClassification.from_pretrained(model_name,config=config)
# + id="QVeoiC2B8X2w" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 66, "referenced_widgets": ["dbfccf5953354d60b474562c24ed49e3", "e2b1f4c4a73d4c5a9a53d9f39a536459", "90909da0f9a14d56a00f87a4583f338b", "26b8b0055d5d43b7b98e96bab01c46a3", "<KEY>", "4326429c9dd641f48c394bacc73b32a9", "2340205efc0d4042a1da017ff08abf99", "c1874a42f7fc470ebe7955680e37f645"]} outputId="ee8b8c40-6004-4a51-d6dd-2f743eec25fd"
model_tf=TFBertForSequenceClassification.from_pretrained(model_name,config=config)
# + id="jG7ppwuM8qEz" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 712, "referenced_widgets": ["1e442f3a822847d6b50a03d46fe679c0", "710c7a34222345b0a89f266e9b9a8d82", "ea6a6b872ff843408398fce44a8d8e66", "0dc4b7aee7a9423ebe423bd3f02f7092", "<KEY>", "708ac9996a29429faa282e3b903c4182", "<KEY>", "3668352f44c343f6b07915f326802715", "9092e03ab0814c54b83a6b1ac2e8d480", "cd5a5820e6ba45429572224e698ef5ea", "19c384e7dfa74e9d829eefc443932397", "4d74bfc8400342a6a480957d459de506", "31de0b529458426abe73ccf50ca6debb", "<KEY>", "<KEY>", "<KEY>", "ddf4a1c1fd5e46d8816d6bdb11205917", "<KEY>", "757a2ff5082a443b81ce01f34806b3d3", "<KEY>", "57c62616ffec48438c6ffda05687c14e", "<KEY>", "<KEY>", "af8e07ab4a1149f4bb7963d7137e4485", "a09f4586d83f465fa7b435e7b9a7c986", "4a53774eca4144f495288a086d972e2f", "6a419288ffac45039a7a917a79483b84", "92acfe5b2f764abe9f8129d749d083db", "<KEY>", "<KEY>", "<KEY>", "<KEY>", "<KEY>", "64e20d0952894c408ad98443d68a4e95", "<KEY>", "9740010620894c5e8d70af800909f571", "2e1d5c959dec45aca440ce022569deed", "593dab61e96f408784e3b030a84fa30d", "98e6cef4db8c498f915eb05e8b53d72b", "<KEY>", "cb6ca9a0f21f486883003fe6d6f9a08b", "<KEY>", "<KEY>", "e178b74006fe45d7a57fd3c70c3a38f2", "<KEY>", "51a91c3e31fc4e88b6976ee01236c291", "42bfaf244a274922a663d689e4fc2633", "<KEY>", "f1650939e74345be91a283fb1fbe476d", "<KEY>", "089801eef6cb4e51a7fc09c4ac69e7d5", "00196baf4478401c9be5f8063f88791d", "<KEY>", "da845347bee04e539ea6696c446b55d4", "<KEY>", "f87852e57ac94ea3b380dd946b32befb", "e9a2c28464e14c978a3c161ddef4136b", "b9750e1659114d1eaf321a7f5ca2b0fc", "<KEY>", "abed93ce31a44010bcddaaf8938c2876", "2327b3db3ece429497247a523cecf114", "07cae0181631419da9af70639f892963", "d142781ec7ca431db321fdd8b4762686", "de3f6f09a3694080a7a1523f68ccbe1e", "94ef013773c24ee9b3de9fb1e0f2bc97", "<KEY>", "fed53ac24db940aebacaf8a6c29e8fa1", "<KEY>", "<KEY>", "<KEY>", "c4b9984117804279828e97157dd6bca9", "<KEY>", "b736edc60354475e90e95d482bbdb495", "7ef632cd66884afc925ebabe2120decb", "<KEY>", "<KEY>", "<KEY>", "<KEY>", "<KEY>", "<KEY>", "<KEY>", "<KEY>", "<KEY>", "<KEY>", "<KEY>", "5694f5203f304800962aef1a86c65c12", "<KEY>", "e8fcaced3c16454ba171855bd2056751", "cefb3280167b41abad8cee714371ca58", "2c682b74ee1f416e9c21e93cdcde1271", "<KEY>", "dbcee3dbec6a4e24bc035b1549380730", "8504f31a1a614b69a86539cad077b553", "c539c94efb5349648f9453c34769a4c8", "<KEY>", "fe56929fce374b8eb03c9c81b45b2e69", "<KEY>", "22a97594f3294defad8d7b134728495b", "23d8b52405a44323afec37aa1f84a7ea", "4596691087674ea7b7396be1e038e949", "<KEY>", "e1d9aff7252a46cc922eed905b350434", "<KEY>", "532befa9cbe64f6c8a1cd388677c1537", "27f49aef56c44329a2965ad37202ea66", "<KEY>", "0acda91954b7437181651e82768e9366", "295b93c858784458a33d8843e25aab2d", "<KEY>", "bc3d842c8e6e449da472509624c3f88a", "6028a6fca77043fb8fc26ce6924e9df0", "68b5a7cdce824a80adabaa0ba7e7a291"]} outputId="1dbb7f67-d6fc-476a-b93d-7da7811e07b7"
data, info = tensorflow_datasets.load(f"glue/{TFDS_TASK}", with_info=True)
# + id="w46wbHFF8x6f" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 69} outputId="2de3baf6-1d59-49da-da8b-0da1c2a6f280"
data
# + id="toOJ1f-H84IF" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="5aa0085b-a39d-4939-f9df-4f20f63f4fdc"
data['test']
# + id="Z5cAgjw29b3y" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 732} outputId="9c5873ab-48b7-464d-d076-6eadeca8a4af"
info
# + id="Vw93zUqC9EIu" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="a7f8d3de-8fab-408b-b556-7bacdd5feb75"
train_examples = info.splits["train"].num_examples
train_examples
# + id="SHzYrPeT9KsS" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="cab2edee-4fcc-408f-934b-2569490c582b"
info.splits["train"]
# + id="soY-zBzH9wkt" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="8f8b9d22-52ca-4eab-aefb-1d101eb88032"
valid_examples = info.splits["validation"].num_examples
valid_examples
# + id="9HS8LB7m-Cx7" colab_type="code" colab={}
train_dataset=glue_convert_examples_to_features(data['train'],tokenizer,128,TASK)
# + id="N5PgUaOg-bVZ" colab_type="code" colab={}
valid_dataset = glue_convert_examples_to_features(data["validation"], tokenizer, 128, TASK)
# + id="cZy6WJQj-isi" colab_type="code" colab={}
train_dataset = train_dataset.shuffle(128).batch(BATCH_SIZE).repeat(-1)
valid_dataset = valid_dataset.batch(EVAL_BATCH_SIZE)
# + id="YHlB1NgD-ofD" colab_type="code" colab={}
# + id="e68opcYz-vY4" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="dbb227be-c9b3-47ad-e61c-094ff289abf3"
opt = tf.keras.optimizers.Adam(learning_rate=3e-5, epsilon=1e-08)
if USE_AMP:
# loss scaling is currently required when using mixed precision
opt = tf.keras.mixed_precision.experimental.LossScaleOptimizer(opt, "dynamic")
print("USE_AMP",opt)
if num_labels == 1:
loss = tf.keras.losses.MeanSquaredError()
else:
loss = tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True)
metric = tf.keras.metrics.SparseCategoricalAccuracy("accuracy")
model_tf.compile(optimizer=opt, loss=loss, metrics=[metric])
# Train and evaluate using tf.keras.Model.fit()
train_steps = train_examples // BATCH_SIZE
valid_steps = valid_examples // EVAL_BATCH_SIZE
# + id="ZYYdOZatPzAs" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="6fa4c8a1-5754-4ce0-be57-4d59285ea356"
EPOCHS
# + id="3DGjccMO-wMe" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 383} outputId="a9a7ce5c-d55d-4f7d-e61d-7f11fcaaec13"
history = model_tf.fit(
train_dataset,
epochs=10,
steps_per_epoch=train_steps,
validation_data=valid_dataset,
validation_steps=valid_steps,
)
# + id="I-DPaaZI_4u9" colab_type="code" colab={}
os.makedirs("./save/", exist_ok=True)
model_tf.save_pretrained("./save/")
# + id="hY1XZ2Y5_q9W" colab_type="code" colab={}
# + id="dvprzwJaABOn" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 69} outputId="901aaeca-372f-4845-d043-ec841428228c"
if TASK == "mrpc":
# Load the TensorFlow model in PyTorch for inspection
# This is to demo the interoperability between the two frameworks, you don't have to
# do this in real life (you can run the inference on the TF model).
pytorch_model = BertForSequenceClassification.from_pretrained("./save/", from_tf=True)
print("------------------load successfully----------------------------")
# Quickly test a few predictions - MRPC is a paraphrasing task, let's see if our model learned the task
sentence_0 = "This research was consistent with his findings."
sentence_1 = "His findings were compatible with this research."
# sentence_2 = "His findings were not compatible with this research."
sentence_2 = "His findings test a few predictions."
inputs_1 = tokenizer.encode_plus(sentence_0, sentence_1, add_special_tokens=True, return_tensors="pt")
inputs_2 = tokenizer.encode_plus(sentence_0, sentence_2, add_special_tokens=True, return_tensors="pt")
pred_1 = pytorch_model(**inputs_1)[0].argmax().item()
pred_2 = pytorch_model(**inputs_2)[0].argmax().item()
print("sentence_1 is", "a paraphrase" if pred_1 else "not a paraphrase", "of sentence_0")
print("sentence_2 is", "a paraphrase" if pred_2 else "not a paraphrase", "of sentence_0")
# + id="dnuF19sciT6F" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="361aa9d2-092a-4c93-e285-2d8b81cafb52"
pred_1,pred_2
# + id="SzODupZdHUg6" colab_type="code" colab={}
| tansformersv241.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# Example relevent to https://github.com/Quansight-Labs/uarray/issues/22
from uarray import *
# +
multiply_function = function(2, Multiply)
res = OuterProduct(multiply_function, Iota(scalar(10)), Iota(scalar(5)))
print(res)
res
# -
replaced_res = replace(res)
print(replaced_res)
replaced_res
# now let's index this with `i` and `j`:
#
i = Scalar(Unbound("i"))
j = Scalar(Unbound("j"))
unrolled = Index(vector_of(i, j), res)
print(unrolled)
unrolled
replaced_unrolled = replace(unrolled)
replaced_unrolled
| notebooks/_ufl example.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="aCZBFzjClURz"
# Based on [this Jupyter notebook](https://github.com/atomic14/tensorflow-lite-esp32/blob/master/Train%20Model.ipynb).
# + [markdown] id="DzYQbLYDFtw-"
# - Zip your ./data folder.
# - Upload it to this notebook using the second code cell.
# - Unzip it by running the following cell.
# + id="ZemoZqcImdlL" colab={"base_uri": "https://localhost:8080/"} outputId="ed342db1-adb0-484f-e166-d01e5c610bf3"
# !pip install tensorflow==2.4.0
# + colab={"resources": {"http://localhost:8080/nbextensions/google.colab/files.js": {"data": "<KEY>", "ok": true, "headers": [["content-type", "application/javascript"]], "status": 200, "status_text": "OK"}}, "base_uri": "https://localhost:8080/", "height": 57} id="holKg3M7nEqK" outputId="bd1e6459-9196-44cc-c980-f2b09f233877"
from google.colab import files
files.upload()
# + id="5egXI-e0GFIz" colab={"base_uri": "https://localhost:8080/"} outputId="b21bf32e-2084-492d-9967-557d9b3cc86a"
# !unzip data.zip
# + id="mxJe1hUJGkKM"
import os
import random
import numpy as np
import matplotlib.pyplot as plt
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
# + id="UazTN3FmHBcU"
data_dirs = ["data/pops/", "data/noise/"]
data_weights = [10, 1]
sample_length = 4000
batch_size = 32
sample_rate = 16000
reaction_time = 0.2 # seconds
downsample_factor = 4
kernel_size = 201
# + id="aIV40KPyGuS7"
def data_generator():
reaction_timesteps = int(round(reaction_time * sample_rate))
samples = []
for data_dir, data_weight in zip(data_dirs, data_weights):
for i in range(len(os.listdir(data_dir)) // 2):
sound = np.load(data_dir + f"sound_{i}.npy").astype(np.float)
pop = np.load(data_dir + f"pop_{i}.npy").astype(np.float)
# reaction shifting + zero padding
pop = np.concatenate((pop[reaction_timesteps:], np.zeros(reaction_timesteps)))
for _ in range(data_weight):
samples.append((sound, pop))
while True:
sound, pop = random.choice(samples)
start_timestep = random.randrange(0, len(sound) - sample_length)
stop_timestep = start_timestep + sample_length
sound = sound[start_timestep:stop_timestep] # random segment
sound = sound.reshape(
sample_length // downsample_factor, downsample_factor, 1, 1
).mean(axis=1) # downsampling
pop = pop[start_timestep:stop_timestep] # random segment
pop = pop.reshape(
sample_length // downsample_factor, downsample_factor,
).sum(axis=1) # downsampling
# cropping to output size
kernel_radius = (kernel_size - 1) // 2
pop = pop[kernel_radius:-kernel_radius]
yield sound, pop
# create a dataset from our generator
train_dataset = tf.data.Dataset.from_generator(
data_generator,
output_signature=(
tf.TensorSpec(shape=(sample_length // downsample_factor, 1, 1), dtype=tf.float32),
tf.TensorSpec(shape=((sample_length // downsample_factor) - (kernel_size - 1),), dtype=tf.float32),
),
).batch(batch_size)
# + colab={"base_uri": "https://localhost:8080/"} id="Etd27rENKmIC" outputId="740e834b-1b66-4f62-b1bf-101ee59a4b66"
def mean_squared_error(y_true, y_pred):
return tf.reduce_mean((tf.reduce_sum(y_true, axis=1) - tf.reduce_sum(y_pred, axis=1)) ** 2)
model = keras.models.Sequential([
layers.InputLayer(input_shape=(sample_length // downsample_factor, 1, 1)),
layers.Conv2D(filters=1, kernel_size=(kernel_size, 1), activation="relu", kernel_initializer=keras.initializers.truncated_normal(stddev=1e-5)),
])
model.compile(
optimizer=keras.optimizers.Adam(learning_rate=1e-6),
loss=mean_squared_error,
)
model.summary()
# + colab={"base_uri": "https://localhost:8080/"} id="pAkz2R0ookiw" outputId="f91d1c9c-b20c-4210-a20a-b7fedc3739e5"
sounds, true_pops = next(iter(train_dataset))
pred_pops = model.predict(sounds)
print(
"{:.4f} should be somewhat smaller than {:.4f}. If that is not true, decrease the kernel_initializer's standard deviation.".format(
tf.reduce_mean(tf.math.reduce_std(pred_pops, axis=1)),
tf.reduce_mean(tf.math.reduce_std(true_pops, axis=1)),
)
)
# + colab={"base_uri": "https://localhost:8080/"} id="eFKM3uweLD10" outputId="5a9f8367-f99e-4c7b-b492-c3194dfcbb03"
model.fit(
train_dataset,
steps_per_epoch=100,
epochs=10,
)
# + colab={"base_uri": "https://localhost:8080/", "height": 284} id="0H7yyATZuqvI" outputId="acbc4d65-97f1-4387-be55-df2b61161e54"
# compare the predictions and the ground truth visually
sounds, true_pops = next(iter(train_dataset))
pred_pops = model.predict(sounds).squeeze()
plt.plot(tf.reduce_sum(true_pops, axis=1))
plt.plot(tf.reduce_sum(pred_pops, axis=1))
# + colab={"base_uri": "https://localhost:8080/"} id="wYFo1VkxSEvL" outputId="f444aeb5-12a8-4e10-afb2-db7872b4c8b4"
converter = tf.lite.TFLiteConverter.from_keras_model(model)
converted_model = converter.convert()
with open('model.tflite', 'wb') as model_file:
model_file.write(converted_model)
# + id="XwU_2GlbUvs7" colab={"base_uri": "https://localhost:8080/"} outputId="2f1d145f-c74b-481b-d3db-2514a0fe04c1"
# install xxd
# !apt-get update && apt-get -qq install xxd
# convert model
# !xxd -i model.tflite > model.cc
| train_model.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Few-Shot Learning
#
# <!-- **Challenge:** [Omniglot](https://github.com/brendenlake/omniglot), the "transpose" of MNIST, with 1,623 character classes, each with 20 examples. Is it possible to build a few-shot classifier with a target of <35% error rate? -->
#
# Humans exhibit a strong ability to acquire and recognize new patterns. In particular, we observe that when presented with stimuli, people seem to be able to understand new concepts quickly and then recognize variations on these concepts in future percepts. [Machine learning](https://wikipedia.com/wiki/Machine_learning) has been successfully used to achieve *state-of- the-art* performance in a variety of applications such as web search, spam detection, caption generation, and speech and image recognition. However, these algorithms often break down when forced to make predictions about data for which little supervised information is available. We desire to generalize to these unfamiliar categories without necessitating extensive retraining which may be either expensive or impossible due to limited data or in an online prediction setting, such as web retrieval.
#
# One particularly interesting task is classification under the restriction that we may only observe a single example of each possible class before making a prediction about a test instance. This is called [One-Shot Learning Problem](https://wikipedia.org/wiki/One-shot_learning). This should be distinguished from [Zero-shot Learning](https://analyticsindiamag.com/what-is-zero-shot-learning/), in which the model cannot look at any examples from the target classes.
#
# [Few-Shot Learning](https://codeburst.io/understanding-few-shot-intelligence-as-a-meta-learning-problem-7823a4cd4a0c) on the other hand *(which is the primary focus of this notebook)*; is a special case of one-shot learning that tries to learn from *a very small set of training examples* rather than one –as in the one-shot learning case.
#
#
# ## Exploring the Dataset
#
# **[Omniglot](https://github.com/brendenlake/omniglot)** is a collection of **1,623** hand drawn characters from **50 alphabets**. Each of the 1,623 characters is drawn online via *Amazon's Mechanical Turk* by 20 different people at resolution of `105x105`. It is sometimes reffered to as the ["Trasnpose of MNIST"](), since it has 1,623 types of character with only 20 examples each *(perfect for few-shot learning)*, in contrast to [MNIST](https://yann.lecun.com/exdb/mnist/) having thousands of examples for only 10 digits *(not easy to come-by for real-word use case)*.
#
# 
#
#
# ### Structure of Omiglot Datasets
#
# ```sh
# ~ tree datasets/extracted -L 1
# datasets/extracted
# ├── all_runs
# ├── images_background
# └── images_evaluation
#
# 3 directories, 0 files
# ```
#
# Omniglot has 3 different dataset serving different purposes.
#
# - **`all_runs`** - One-shot task.
# - **`images_background`** - Training set.
# - **`images_evaluation`** - Validation/ evaluation set.
#
# > **Usage:** The **`images_background`** will be used for training *a Model* for *Few-shot Learning* and then validated on **`images_evaluation`**. While the **`all_runs`** will be used for *One-shot task*.
#
# #### Training/Validation Dataset Structure
#
# The training data contains 30 different *Alphabets* and the validation data contains 20 different alphabets *(that aren't in the training set)*, each containing *Characters* written differently.
#
# ```sh
# $datasets/extracted$ ~ tree images_background -L 2
# images_background
# ├── Alphabet_of_the_Magi
# ├── Anglo-Saxon_Futhorc
# ├── Arcadian
# ...
# ├── Sanskrit
# ├── Syriac_(Estrangelo)
# ├── Tagalog
# └── Tifinagh
#
# 30 directories, 0 files
# ```
#
# #### Alphabet Structure
#
# A single *Alphabet* contains different *Characters*, each with 20 different *handwriting style*.
#
# ```sh
# $datasets/extracted$ ~ tree images_background/Alphabet_of_the_Magi -L 2
# images_background/Alphabet_of_the_Magi
# ├── character01
# │ ├── 0709_01.png
# │ ├── 0709_02.png
# │ ├── 0709_03.png
# │ ...
# │ ├── 0709_18.png
# │ ├── 0709_19.png
# │ └── 0709_20.png
# ├── character02
# │ ├── 0710_01.png
# │ ├── 0710_02.png
# │ ├── 0710_03.png
# │ ...
# │ ├── 0710_18.png
# │ ├── 0710_19.png
# │ └── 0710_20.png
# ...
# ├── character19
# │ ├── 0727_01.png
# │ ├── 0727_02.png
# │ ├── 0727_03.png
# │ ...
# │ ├── 0727_18.png
# │ ├── 0727_19.png
# │ └── 0727_20.png
# └── character20
# ├── 0728_01.png
# ├── 0728_02.png
# ├── 0728_03.png
# ...
# ├── 0728_18.png
# ├── 0728_19.png
# └── 0728_20.png
#
# 20 directories, 400 files
# ```
#
# #### One-shot Task Structure
#
# The **`all_runs`** folder consist of 20 runs each with a **training** and **test** folder.
#
# ```sh
# $datasets/extracted$ ~ tree all_runs -L 2
# ├── run01
# │ ├── class_labels.txt
# │ ├── test
# │ └── training
# ├── run02
# │ ├── class_labels.txt
# │ ├── test
# │ └── training
# ...
# ├── run19
# │ ├── class_labels.txt
# │ ├── test
# │ └── training
# └── run20
# ├── class_labels.txt
# ├── test
# └── training
# ```
#
# `class_labels.txt` consist of pairs of matching images in the `test` and `training` directories. The `test` and `training` directories contain 20 unique images each, where a single image in `test` matches another image in `training`.
#
# The **One-shot** task is all about learning how to tell two images apart, i.e. Predict if two given images are of different or similar classes.
#
# 
#
#
# ### Visualizing Dataset
#
# Haven explored the data and looked into the file structures and what they mean, it's time to start looking at the actual images to see for ourselves what we're trying to do.
#
# #### Training/Validation Dataset Visualization.
#
# Since the training and validation dataset have similar file structure, it's safe to say it can be visulaized in a similar way. The file structures represent various *Alphabets* and side each Alphabet are different characters written differently.
#
# > Visualization class has been nicely abstracted in the **`omniglot.Visualize`** class.
#
# To use the `omniglot.Visualize` class, we need to make a few imports.
# +
# %matplotlib inline
# For OS operations.
import os
# For plotting model from SVG bytes string.
from IPython.display import SVG
# For Mathematical operations.
import numpy as np
# # For High-level ML operations.
# import tensorflow as tf
# from tensorflow import keras
# Helper package for omniglot's dataset & model.
import omniglot
# -
# The `omniglot.Visualize` has a relatively simple API. We provide the path to an Alphabet and then it plots a single letter (randomly) from 20 different characters of the Alphabet.
#
# Firstly, extract the dataset if it isn't already extracted. This can be achieved by calling `omniglot.Dataset.extract(...)` method –which takes in a single required argument: `path` to a compressed file. It will be extracted to a default directory, but you can also provide `extract_dir`.
# +
# Directory containing compressed files.
comp_dir = omniglot.compressed_dir
# Extract compressed dataset: `all_runs`, `images_background` & `images_evaluation`.
all_runs_dir = omniglot.Dataset.extract(os.path.join(comp_dir, 'all_runs.tar.gz'))
image_bg_dir = omniglot.Dataset.extract(os.path.join(comp_dir, 'images_background.tar.gz'))
image_ev_dir = omniglot.Dataset.extract(os.path.join(comp_dir, 'images_evaluation.tar.gz'))
# Visualize one random character each from Greek alphabet of training set.
omniglot.Visualize.symbols(directory=os.path.join(image_bg_dir, 'Greek'))
# Visualize one random character each from Mongolian alphabet of validation set.
omniglot.Visualize.symbols(directory=os.path.join(image_ev_dir, 'Mongolian'))
# -
# #### One-shot Task Visualization
# Visualize the first run & select the 3rd image in the test directory
# and matching label in the train directory.
# (Simply put; emphaize the 3rd matching digits in "class_labels.txt").
omniglot.Visualize.runs(directory=os.path.join(all_runs_dir, 'run01'),
index=2, title='One-shot Task')
# ### Data Pre-processing
#
# The `omniglot.Dataset` is made up of user friendly high level APIs to load, save, process, manage, format and manipulate dataset. Given the path to `image_background` or `image_evaluation` dataset, the `omniglot.Dataset` parses the path and creates image pairs and corresponding lables. It can also be run in training, testing or validation mode with the `mode` keyword argument which defaults to `Dataset.Mode.TRAIN`.
# +
# To avoid writing `omniglot.Dataset`...
Dataset = omniglot.Dataset
# Create training data instance.
train_data = Dataset(path=image_bg_dir, mode=Dataset.Mode.TRAIN)
print(f'\nTraining data: {train_data.shape}')
print(f'train_data = {train_data}')
# Create validation data instance.
valid_data = Dataset(path=image_ev_dir, mode=Dataset.Mode.VALIDATE)
print(f'\nValidation data: {valid_data.shape}')
print(f'valid_data = {valid_data}')
# -
# ## Exploring Models
#
# ### $N$-way One-shot learning
#
# While One-shot learning requires a single training example, few-shot learning needs a very few subset of examples each from each classes. The number of training examples required for few-shot learning will be represented by $N$.
#
# Therefore, given a tiny labelled training set $S$, which has $N$ examples, each vectors of the same dimension with a distinct label $y$.
#
# $$ S = \{(x_i, y_i), ..., (x_N, y_N)\} $$
#
# We're also given $\hat{x}$, the test example it has to classify. Since exactly one example in the training set has the right class, the aim is to correctly predict which $y \in S$ is the same as $\hat{x}$'s label, $\hat{y}$.
#
# - Real world problems might not always have the constraint that exactly one image has the correct class.
#
# - It's easy to generalize to k-shot learning by having there be $k$ examples for each $y_i$ rather than just one.
#
# - When $N$ is higher, there are more possible classes that $\hat{x}$ can belong to, so it's harder to predict the correct one.
#
# - Randomly guessing will average ${100 \over n}\%$ accuracy.
#
# ### The One-shot learning problem
#
# To solve the One-shot learning problem, we're going to need a model that learns to tell images part rather than classify images using the normal [cross entropy loss function](https://rdipietro.github.io/friendly-intro-to-cross-entropy-loss/). Why classification won't work in this case is because there are very few images to learn from, therefore training a deep model on these few dataset is going to cause the model to [Overfit](https://en.wikipedia.org/wiki/Overfitting) to the few examples in the dataset.
#
# However, what we need is a model that learns to tell image pairs apart. That is, given two images from the same class: the model outputs a distance $d$ which represent the how far these imgae pairs are in the feature space. A small value of $d$ means the images are the same, while a large value of $d$ means the images are different. This raises the question: *How small should "small" be and how large should "large" be?* Well, this is up to be learnt by the model. Let's call the threshold $\tau$ –Yes, yet another hyperparameter to worry about.
#
# A popular model that does this is called the [Siamese Network](https://wikipedia.com/wiki/Siamese_network). The Siamese network architecture consist of a "twin neural net", hence the name. Both networks are [Convolutional Neural Network](http://cs231n.github.io/convolutional-networks/) with shared parameters.
#
# 
#
# > *A simple 2 hidden layer siamese network for binary classification with logistic prediction, $p$. The structure of the network is replicated across the top and bottom sections to form twin networks, with shared weight matrices at each layer.*
#
# <!-- TODO: Finish up docs. -->
#
# <!--
# ### One-Shot Learning Baseline over Nearest Neighbor
#
# The simplest way to perform classification is with [K-Nearest Neighbors](https://www.analyticsvidhya.com/blog/2018/03/introduction-k-neighbours-algorithm-clustering/), but since there are only one example per class, we're only allowed 1 nearest neighbor lookup –which is really bad! K-Nearest Neigbors usually performs well with 5 neighbors or more *(but this also depends on dataset & it's sparsity)*.
#
# Nearest Neighbor: This is just a way of measuring distance in a higher dimensional plane using distance metrics such as [Euclidean Distance](https://en.wikipedia.org/wiki/Euclidean_distance).
#
# $$ \textrm{Euclidean Distance} = \sqrt{\sum_i^n{(q_i - p_i)^2}}$$
#
# After calculating the Euclidean disance over `k` nearest neighbors. We then take the closest one:
#
# $$ C(\hat{x}) = \underset{c \in S}{\operatorname{argmax}} \big\|\hat{x} - x_c\big\| $$
#
# After calculating the Euclidean disance over `k` nearest neighbors. We then take the closest one:
# -->
# Hyper-parameters.
epochs, batch_size = 3, 32
train_size, valid_size = len(train_data), len(valid_data)
# ### Model (Siamese Convolutional Neural Network)
#
# A standard [Siamese Convolutional Neural Network](https://hackernoon.com/one-shot-learning-with-siamese-networks-in-pytorch-8ddaab10340e) with $L$ layers each with $N_l$ units is used, where $h_{1, l}$ represents the hidden vector in layer $l$ for the first twin, and $h_{2,l}$ denotes the same for the second twin. **Rectified Linear (ReLU) Units** is exclusively used in the first $L-2$ layers and sigmoidal units in the remaining layers.
#
# The model consists of a sequence of convolutional layers, each of which uses a single channel with filters of varying size and a fixed stride of 1. The number of convolutional filters is specified as a multiple of 16 to optimize performance. The network applies a ReLU activation function to the output feature maps, optionally followed by max-pooling with a filter size and stride of 2. Thus the $k^{th}$ filter map in each layer takes the following form:
#
# $$ a^{(k)}_{1, m} = \textrm{max-pool}(max(0, W^{(k)}_{l-1} \star h_{1, (l-1)} + b_l), 2) $$
#
# $$ a^{(k)}_{2, m} = \textrm{max-pool}(max(0, W^{(k)}_{l-1} \star h_{2, (l-1)} + b_l), 2) $$
#
# where $W^{l-1, l}$ is the 3-dimensional tensor representing the feature maps for layer $l$ and $\star$ is the valid convolutional operation corresponding to returning only those output units which were the result of complete overlap between each convolutional filter and the input feature maps.
#
# In a more simple terms: The model consist a **Twin Neural Network Architecture***, where each network is a **Convolutional Neural Network (CNN)**. Each network is fed an image pair & the model learns to predict how far apart or close together these image pairs are in the feature space. More visually, the Twin Neural Net looks like this:
#
# 
#
# Where each network is made up of a CNN with the following configurations:
#
# 
#
# > Best convolutional architecture selected for verification task. Siamese twin is not depicted, but joins immediately after the 4096 unit fully-connected layer where the $L_1$ component-wise distance between vectors is computed.
#
# ### Learning
#
# **Loss Function, $\mathcal{L}$**. Let $M$ represent the mini-batch size, where $i$ indexes the $i^{th}$ mini-batch. Now let $y(x^{(i)}_1, x^{(i)}_2)$ be a length-$M$ vector which contains the labels for the mini-batch, where $y(x^{(i)}_1, x^{(i)}_2) = 1$ whenever $x_1$ and $x_2$ are from the same character class and $y(x^{(i)}_1, x^{(i)}_2) = 0$ otherwise. A regularized cross-entropy objective is imposed on a binary classifier of the following form:
#
# $$ \mathcal{L}(x^{(i)}_1, x^{(i)}_2) = y(x^{(i)}_1, x^{(i)}_2)\log{p(x^{(i)}_1, x^{(i)}_2)} + (1 - y(x^{(i)}_1, x^{(i)}_2)) \log{(1 - p(x^{(i)}_1, x^{(i)}_2))} + \lambda^T\|w\|^2 $$
# +
# # Define the Siamese Network Model with Triplet loss function.
# siamese_net = omniglot.SiameseNetwork(loss=omniglot.Loss.triplet_loss)
# # Train the Siamese network.
# siamese_net.train(train_data=train_data,
# valid_data=valid_data,
# epochs=epochs)
# -
# #### Using a custom EncoderNetwork architecture
#
# Results from the Siamese Network is not too great, although better than a Nearest Neighbor lookup. This begs the question: *How do we optimize this network for more accurate predictions?* The answer comes in form of using a slightly similar but new architecture which ensures both image pairs shares the same weights and encodes separate feature mapping for each image pair. **`omniglot.EncoderNetwork`** contains the implementation of this network architecture.
#
# 
# +
# Create a network instance to train dataset.
encoder_net = omniglot.EncoderNetwork()
# Visualize model architecture.
SVG(encoder_net.plot_model(show_layer_names=True, show_shapes=False))
# -
# Train the network.
encoder_net.train(train_data=train_data,
valid_data=valid_data,
epochs=epochs)
# ### Dataset & Model (Interesting Fact)
#
# One cool thing I noticed about training on pairs is that there are quadratically many possible pairs of images to train the model on, making it hard to overfit. Say we have $C$ examples each of $E$ classes. Since there are $C \cdot E$ images total, the total number of possible pairs is given by:
#
# $$\binom{C \cdot E}{2} = \frac{(C \cdot E)!}{2!(C \cdot E - 2)!}$$
#
# For omniglot with its 20 examples of 964 training classes, this leads to $185,849,560$ possible pairs, which is huge! However, the siamese network needs examples of both same and different class pairs. There are $E$ examples per class, so there will be $\binom{E}{2}$ pairs for every class, which means there are $N_{same} = \binom{E}{2} \cdot C$ possible pairs with the same class - $183,160$ pairs for omniglot. Even though $183,160$ example pairs is plenty, it’s only a thousandth of the possible pairs, and the number of same-class pairs increases quadratically with $E$ but only linearly with $C$. This is important because the siamese network should be given a $1:1$ ratio of same-class and different-class pairs to train on - perhaps it implies that pairwise training is easier on datasets with lots of examples per class.
encoder_net.load_model()
# #### Nearest Neighbor
#
# The simplest way of doing classification is with k-nearest neighbours, but since there is only one example per class we have to do 1 nearest neighbour. This is very simple, just calculate the Euclidean distance of the test example from each training example and pick the closest one:
# $$C(\hat{x}) = \underset{c \in S}{\operatorname{argmin}} \| \hat{x} − x_c \| $$
#
# According to Koch et al, 1-nn gets ~28% accuracy in 20 way one shot classification on omniglot. 28% doesn’t sound great, but it’s nearly six times more accurate than random guessing(5%). This is a good baseline or “sanity check” to compare future one-shot algorithms with.
# +
# Nearest Neighbor lookup benchmark.
nearest_neighbor = omniglot.Benchmark(data=valid_data)
nn_accuracy = nearest_neighbor.score(n=10)
print('Nearest Accuracy: {:.2%}'.format(nn_accuracy))
# -
# #### Benchmarking accuracies
#
# Benchmarking accuracies on both training & test set to see if the model overfitted on the training data while not being able to generalize to evaluation data. Also visualized some other benchmarks to see how model does with increasing value of $N$. Starting with $N=1$ upto $N=50$, the accuracy reduces with increasing value of $N$. This is a good thing, because the number of correct pairs grows linearly as $N$ increases, which makes the number of correct pairing lesser and lesser.
# +
trials, ways = 10, np.arange(1, 51, 2)
# Store list of accuracies for `n` ways.
val_acc, train_acc, nn_acc = [], [], []
for n in ways:
# There's no 0-way few-shot learning (or Zero-shot learning).
if n == 0:
n += 1
# Log progress.
print(f'\rEvaluating {n:,} of {ways[-1]}-way few-shot tasks...', end='')
# Validation accuracies on one-shot task.
val_acc.append(valid_data.test_one_shot_task(encoder_net, n,
trials, verbose=0))
# Train accuracies on one-shot task.
train_acc.append(train_data.test_one_shot_task(encoder_net, n,
trials, verbose=0))
# Nearest neighbor lookup on one-shot task.
nn_acc.append(nearest_neighbor.score(n=10, trials=trials,
verbose=0))
# -
# Visualize Performance metrics for Few-shot learning.
omniglot.Visualize.accuracies(ways=ways, train=train_acc,
valid=val_acc, benchmark=nn_acc)
# #### Making Predictions
# +
p, t = valid_data.one_shot_task(3)
p.shape, t.shape
pred = encoder_net(p)
print(f'Predicted: {np.ravel(pred)}')
print(f'Target la: {t}')
# -
# ### Roadmap
#
#
# #### Optimization & Data Augmentation
#
# This model was pretty good at differentiating pairs of images, however, there are a few concepts that wasn't implemented here mainly because of computational resources [like layerwise learning rates/momentum](https://distill.pub/2017/momentum/), data augmentation with distortions, [bayesian hyperparemeter optimization](https://arimo.com/data-science/2016/bayesian-optimization-hyperparameter-tuning/) and also more training epochs.
#
# #### Using Deep Reinforcement Learning
#
# A new way to approach this problem, rather than using Supervised learning, [Reinforcement Learning](https://skymind.ai/wiki/deep-reinforcement-learning) can also be leveraged to learn how to distinguish image pairs.
#
# **Setup for Deep Reinforcement learning:** A policy network *(which architecture may be similar to that of a Simamese Network or something completely different)* is trained. The model learns a policy $\pi$ that maps image pairs to correct predictions –similar or dissimilar. In Reinfocement learning vocabulary, the image pairs can be thought of as *states* $S$ while predictions are *actions* $a$, on successful prediction the agent recieves a *reward* (+1) $R(s, a)$ otherwise it recieves a *punishment* (-1). The agent spits out a policy $\pi$ which maps state to action, $\pi(s) \to a$. The network is trained using a vanilla [Policy Gradients](http://www.scholarpedia.org/article/Policy_gradient_methods) *(or any other RL training techniques like Actor-critic, Deep-Q, Contextual bandit, etc)*, with the sole purpose of maximizing rewards *(i.e. correctly classifying images as either similar or dissimilar)*.
#
# #### Using Evolutionary Algorithms
#
# Through the process of trial and error through many generations, the model learns how to best classify images as either *similar or dis-similar*. Evolutionary algorithm's fitness function should encode how well a model is able to generalize to new unseen image pairs or
| one-shot.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [conda env:CourseraMLAndrewNgmyNoteEnv] *
# language: python
# name: conda-env-CourseraMLAndrewNgmyNoteEnv-py
# ---
# # 机器学习练习 1 - 线性回归
# 这个是另一位大牛写的,作业内容在根目录: [作业文件](ex1.pdf)
#
# 代码修改并注释:黄海广,<EMAIL>
# ## 单变量线性回归
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
path = 'ex1data1.txt'
data = pd.read_csv(path, header=None, names=['Population', 'Profit'])
data.head()
data.describe()
# 看下数据长什么样子
data.plot(kind='scatter', x='Population', y='Profit', figsize=(12,8))
plt.show()
# 现在让我们使用梯度下降来实现线性回归,以最小化成本函数。 以下代码示例中实现的方程在“练习”文件夹中的“ex1.pdf”中有详细说明。
# 首先,我们将创建一个以参数θ为特征函数的代价函数
# $$J\left( \theta \right)=\frac{1}{2m}\sum\limits_{i=1}^{m}{{{\left( {{h}_{\theta }}\left( {{x}^{(i)}} \right)-{{y}^{(i)}} \right)}^{2}}}$$
# 其中:\\[{{h}_{\theta }}\left( x \right)={{\theta }^{T}}X={{\theta }_{0}}{{x}_{0}}+{{\theta }_{1}}{{x}_{1}}+{{\theta }_{2}}{{x}_{2}}+...+{{\theta }_{n}}{{x}_{n}}\\]
def computeCost(X, y, theta):
inner = np.power(((X * theta.T) - y), 2)
return np.sum(inner) / (2 * len(X))
# 让我们在训练集中添加一列,以便我们可以使用向量化的解决方案来计算代价和梯度。
data.insert(0, 'Ones', 1)
# 现在我们来做一些变量初始化。
# set X (training data) and y (target variable)
cols = data.shape[1]
X = data.iloc[:,0:cols-1]#X是所有行,去掉最后一列
y = data.iloc[:,cols-1:cols]#X是所有行,最后一列
# 观察下 X (训练集) and y (目标变量)是否正确.
X.head()#head()是观察前5行
y.head()
# 代价函数是应该是numpy矩阵,所以我们需要转换X和Y,然后才能使用它们。 我们还需要初始化theta。
X = np.matrix(X.values)
y = np.matrix(y.values)
theta = np.matrix(np.array([0,0]))
# theta 是一个(1,2)矩阵
theta
# 看下维度
X.shape, theta.shape, y.shape
# 计算代价函数 (theta初始值为0).
computeCost(X, y, theta)
# # batch gradient decent(批量梯度下降)
# $${{\theta }_{j}}:={{\theta }_{j}}-\alpha \frac{\partial }{\partial {{\theta }_{j}}}J\left( \theta \right)$$
def gradientDescent(X, y, theta, alpha, iters):
temp = np.matrix(np.zeros(theta.shape))
parameters = int(theta.ravel().shape[1])
cost = np.zeros(iters)
for i in range(iters):
error = (X * theta.T) - y
for j in range(parameters):
term = np.multiply(error, X[:,j])
temp[0,j] = theta[0,j] - ((alpha / len(X)) * np.sum(term))
theta = temp
cost[i] = computeCost(X, y, theta)
return theta, cost
# 初始化一些附加变量 - 学习速率α和要执行的迭代次数。
alpha = 0.01
iters = 1000
# 现在让我们运行梯度下降算法来将我们的参数θ适合于训练集。
g, cost = gradientDescent(X, y, theta, alpha, iters)
g
# 最后,我们可以使用我们拟合的参数计算训练模型的代价函数(误差)。
computeCost(X, y, g)
# 现在我们来绘制线性模型以及数据,直观地看出它的拟合。
# +
x = np.linspace(data.Population.min(), data.Population.max(), 100)
f = g[0, 0] + (g[0, 1] * x)
fig, ax = plt.subplots(figsize=(12,8))
ax.plot(x, f, 'r', label='Prediction')
ax.scatter(data.Population, data.Profit, label='Traning Data')
ax.legend(loc=2)
ax.set_xlabel('Population')
ax.set_ylabel('Profit')
ax.set_title('Predicted Profit vs. Population Size')
plt.show()
# -
# 由于梯度方程式函数也在每个训练迭代中输出一个代价的向量,所以我们也可以绘制。 请注意,代价总是降低 - 这是凸优化问题的一个例子。
fig, ax = plt.subplots(figsize=(12,8))
ax.plot(np.arange(iters), cost, 'r')
ax.set_xlabel('Iterations')
ax.set_ylabel('Cost')
ax.set_title('Error vs. Training Epoch')
plt.show()
# ## 多变量线性回归
# 练习1还包括一个房屋价格数据集,其中有2个变量(房子的大小,卧室的数量)和目标(房子的价格)。 我们使用我们已经应用的技术来分析数据集。
path = 'ex1data2.txt'
data2 = pd.read_csv(path, header=None, names=['Size', 'Bedrooms', 'Price'])
data2.head()
# 对于此任务,我们添加了另一个预处理步骤 - 特征归一化。 这个对于pandas来说很简单
data2 = (data2 - data2.mean()) / data2.std()
data2.head()
# 现在我们重复第1部分的预处理步骤,并对新数据集运行线性回归程序。
# +
# add ones column
data2.insert(0, 'Ones', 1)
# set X (training data) and y (target variable)
cols = data2.shape[1]
X2 = data2.iloc[:,0:cols-1]
y2 = data2.iloc[:,cols-1:cols]
# convert to matrices and initialize theta
X2 = np.matrix(X2.values)
y2 = np.matrix(y2.values)
theta2 = np.matrix(np.array([0,0,0]))
# perform linear regression on the data set
g2, cost2 = gradientDescent(X2, y2, theta2, alpha, iters)
# get the cost (error) of the model
computeCost(X2, y2, g2)
# -
# 我们也可以快速查看这一个的训练进程。
fig, ax = plt.subplots(figsize=(12,8))
ax.plot(np.arange(iters), cost2, 'r')
ax.set_xlabel('Iterations')
ax.set_ylabel('Cost')
ax.set_title('Error vs. Training Epoch')
plt.show()
# 我们也可以使用scikit-learn的线性回归函数,而不是从头开始实现这些算法。 我们将scikit-learn的线性回归算法应用于第1部分的数据,并看看它的表现。
from sklearn import linear_model
model = linear_model.LinearRegression()
model.fit(X, y)
# scikit-learn model的预测表现
# +
x = np.array(X[:, 1].A1)
f = model.predict(X).flatten()
fig, ax = plt.subplots(figsize=(12,8))
ax.plot(x, f, 'r', label='Prediction')
ax.scatter(data.Population, data.Profit, label='Traning Data')
ax.legend(loc=2)
ax.set_xlabel('Population')
ax.set_ylabel('Profit')
ax.set_title('Predicted Profit vs. Population Size')
plt.show()
# -
# # 4. normal equation(正规方程)
# 正规方程是通过求解下面的方程来找出使得代价函数最小的参数的:$\frac{\partial }{\partial {{\theta }_{j}}}J\left( {{\theta }_{j}} \right)=0$ 。
# 假设我们的训练集特征矩阵为 X(包含了${{x}_{0}}=1$)并且我们的训练集结果为向量 y,则利用正规方程解出向量 $\theta ={{\left( {{X}^{T}}X \right)}^{-1}}{{X}^{T}}y$ 。
# 上标T代表矩阵转置,上标-1 代表矩阵的逆。设矩阵$A={{X}^{T}}X$,则:${{\left( {{X}^{T}}X \right)}^{-1}}={{A}^{-1}}$
#
# 梯度下降与正规方程的比较:
#
# 梯度下降:需要选择学习率α,需要多次迭代,当特征数量n大时也能较好适用,适用于各种类型的模型
#
# 正规方程:不需要选择学习率α,一次计算得出,需要计算${{\left( {{X}^{T}}X \right)}^{-1}}$,如果特征数量n较大则运算代价大,因为矩阵逆的计算时间复杂度为$O(n3)$,通常来说当$n$小于10000 时还是可以接受的,只适用于线性模型,不适合逻辑回归模型等其他模型
# 正规方程
def normalEqn(X, y):
theta = np.linalg.inv(X.T@X)@X.T@y#X.T@X等价于X.T.dot(X)
return theta
final_theta2=normalEqn(X, y)#感觉和批量梯度下降的theta的值有点差距
final_theta2
# +
#梯度下降得到的结果是matrix([[-3.24140214, 1.1272942 ]])
# -
# 在练习2中,我们将看看分类问题的逻辑回归。
| huanghaiguang_code/ex1-linear regression/ML-Exercise1.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# `models.explainers` regression example (boston dataset)
# -----
#
# ### Load packages
# +
from transparentai.models import explainers
from transparentai.datasets import load_boston
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import train_test_split
# -
# ### Load & prepare data
data = load_boston()
X, Y = data.drop(columns='MEDV'), data['MEDV']
# Split train test
X_train, X_valid, Y_train, Y_valid = train_test_split(X, Y, test_size=0.33, random_state=42)
# ### Train classifier
regr = LinearRegression()
regr.fit(X_train, Y_train)
# ### Use `models.explainers.ModelExplainer`
explainer = explainers.ModelExplainer(regr, X_train, model_type='linear')
explainer.explain_global_influence(X_train)
explainer.plot_global_explain()
explainer.explain_local_influence(X_valid.iloc[0])
explainer.plot_local_explain(X_valid.iloc[0])
explainer.plot_local_explain_interact(X_valid.iloc[0])
visible_features = ['TAX','PTRATIO','B','LSTAT']
explainer.plot_local_explain_interact(X_valid.iloc[0], visible_feat=visible_features)
| examples/models.explainers_regression_example_boston_dataset.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: isa-api-py38
# language: python
# name: isa-api-py38
# ---
# # Create a repeated treatment design with ISA descriptor
#
# This example creates `ISA study descriptor` for study with sequential treatments organized in an arm.
# This shows how to use objects from the `isatools.create` component in a granular fashion.
# It creates each `Element` of the Study `Arm` at a time.
#
# Finally, the `study design plan` is shown by serializing the `ISA Study Design Model` content as an `ISA_design` JSON document, which can be rendered in various ways (tables, figures).
# ## Let's load the tools
# +
import datetime
from isatools.model import *
from bokeh.io import output_file, show
from bokeh.plotting import figure
from bokeh.models import ColumnDataSource, Range1d, BoxAnnotation, Label, Legend, LegendItem, LabelSet
from bokeh.models.tools import HoverTool
import pandas as pd
import datetime as dt
import holoviews as hv
from holoviews import opts, dim
hv.extension('bokeh')
# -
# ## Start by creating basic ISA Study metadata
investigation = Investigation()
study = Study(filename="s_study_xover.txt")
study.identifier = 'S-Xover-1'
study.title = 'My Simple ISA Study'
study.description = "We could alternataly use the class constructor's parameters to set some default " \
"values at the time of creation, however we want to demonstrate how to use the " \
"object's instance variables to set values."
study.submission_date = str(datetime.datetime.today())
study.public_release_date = str(datetime.datetime.today())
# study.sources = [Source(name="source1")]
# study.samples = [Sample(name="sample1")]
# study.protocols = [Protocol(name="sample collection")]
# study.process_sequence = [Process(executes_protocol=study.protocols[-1], inputs=[study.sources[-1]], outputs=[study.samples[-1]])]
investigation.studies = [study]
# investigation
# +
# from isatools.isatab import dumps
# print(dumps(investigation))
# -
import json
from isatools.isajson import ISAJSONEncoder
# print(json.dumps(investigation, cls=ISAJSONEncoder, sort_keys=True, indent=4, separators=(',', ': ')))
# ### Let's load the new ISA create module
from isatools.create.model import *
# ### 1. Creation of the first `ISA Study Design Element` and setting its type
nte1 = NonTreatment(element_type='screen', duration_unit=OntologyAnnotation(term="days"))
print(nte1)
# ### 2. Creation of another `ISA Study Design Element`, of type `Treatment`
te1 = Treatment()
te1.type='biological intervention'
print(te1)
# #### 2.1 defining the first treatment as a vector of ISA factor values:
#
# Under "ISA Study Design Create mode", a `Study Design Element` of type `Treatment` needs to be defined by a vector of `Factors` and their respective associated `Factor Values`. This is done as follows:
f1 = StudyFactor(name='virus', factor_type=OntologyAnnotation(term="organism"))
f1v = FactorValue(factor_name=f1, value="hsv1")
f2 = StudyFactor(name='dose', factor_type=OntologyAnnotation(term="quantity"))
f2v = FactorValue(factor_name=f2, value='high dose')
f3 = StudyFactor(name='time post infection', factor_type=OntologyAnnotation(term="time"))
f3v = FactorValue(factor_name=f3, value=2, unit=OntologyAnnotation(term='hr'))
#assigning the factor values declared above to the ISA treatment element
te1.factor_values = [f1v,f2v,f3v]
print(te1)
# ### 3. Creation of a second `ISA Study Design Element`, of type `Treatment`, following the same pattern.
# +
te2 = Treatment()
te2.type = 'chemical intervention'
antivir = StudyFactor(name='antiviral', factor_type=OntologyAnnotation(term="chemical entity"))
antivirv = FactorValue(factor_name=antivir, value='hsvflumab')
intensity = StudyFactor(name='dose', factor_type=OntologyAnnotation(term="quantity"))
intensityv= FactorValue(factor_name=intensity, value = 10, unit=OntologyAnnotation(term='mg/kg/day'))
duration = StudyFactor(name = 'treatment duration', factor_type=OntologyAnnotation(term="time"))
durationv = FactorValue(factor_name=duration, value=2, unit=OntologyAnnotation(term='weeks'))
te2.factor_values = [antivirv,intensityv,durationv]
print(te2)
# -
te3 = Treatment()
te3.type = 'radiological intervention'
rays = StudyFactor(name='radiation', factor_type=OntologyAnnotation(term="physical entity"))
raysv = FactorValue(factor_name=rays, value='neutron beam')
rays_intensity = StudyFactor(name='dose', factor_type=OntologyAnnotation(term="quantity"))
rays_intensityv= FactorValue(factor_name=rays_intensity, value = '10', unit=OntologyAnnotation(term='mSev'))
rays_duration = StudyFactor(name = 'treatment duration', factor_type=OntologyAnnotation(term="time"))
rays_durationv = FactorValue(factor_name=rays_duration, value='30', unit=OntologyAnnotation(term='minutes'))
te3.factor_values = [raysv,rays_intensityv,rays_durationv]
print(te3)
# ### 4. Creation of 'wash out' period as an `ISA Study Design Element`.
# Creation of another ISA element, which is not a Treatment element, which is of type `screen` by default
nte2 = NonTreatment(duration_unit=OntologyAnnotation(term="days"))
print(nte2.type)
# let's change it by setting its type by relying on the keys defined for the object
nte2.type=RUN_IN
print(nte2.type)
#let's change it again by direct use of the allowed strings (note: the string should match exactly the predefined values)
nte2.type = 'washout'
print(nte2.type)
# setting the factor values associated with 'default' DURATION Factor associated with such elements
nte2.duration.value=2
nte2.duration.unit=OntologyAnnotation(term="weeks")
# ### 5. Creation of 'follow-up' period as an `ISA Study Design Element`.
nte3 = NonTreatment(element_type=FOLLOW_UP, duration_value=4, duration_unit=OntologyAnnotation(term="month"))
# nte3.duration.value = 2
# nte3.duration.unit = 'months'
print(nte3)
# ### 6. Creation of the associated container, known as an ISA `Cell` for each ISA `Element`.
# In this example, a single `Element` is hosted by a `Cell`, which must be named. In more complex designs (e.g. study designs with assymetric arms), a `Cell` may contain more than one `Element`, hence the list attribute.
st_cl1= StudyCell(name="st_cl1", elements=[nte1])
st_cl2= StudyCell(name="st_cl2", elements=[te1])
st_cl3= StudyCell(name="st_cl3", elements=[nte2])
st_cl4= StudyCell(name="st_cl4", elements=[te2])
st_cl6= StudyCell(name="st_cl6", elements=[nte2])
st_cl7= StudyCell(name="st_cl7", elements=[te3])
st_cl5= StudyCell(name="st_cl5", elements=[nte3])
# ### 7. Creation of an ISA `Study Arm` and setting the number of subjects associated to that unique sequence of ISA `Cell`s.
# +
arm1 = StudyArm(name='Arm 1', group_size=20, )
print(arm1)
genotype_cat = OntologyAnnotation(term="genotype")
genotype_value1 = OntologyAnnotation(term="control - normal")
genotype_value2 = OntologyAnnotation(term="mutant")
arm1 = StudyArm(name='Arm 1',
group_size=2,
source_type=Characteristic(category=genotype_cat,
value=genotype_value1)
)
print(arm1)
# -
# ### 8. Declaring an ISA `Sample Assay Plan`, defining which `Sample` are to be collected and which `Assay`s to be used
input_material1=ProductNode(id_="MAT1", name="liver", node_type=SAMPLE,size=1,characteristics=[Characteristic(category=OntologyAnnotation(term='organism part'), value=OntologyAnnotation(term='liver'))])
input_material2=ProductNode(id_="MAT2", name="blood", node_type=SAMPLE,size=1,characteristics=[Characteristic(category=OntologyAnnotation(term='organism part'), value=OntologyAnnotation(term='blood'))])
input_material3=ProductNode(id_="MAT3", name="urine", node_type=SAMPLE,size=3,characteristics=[Characteristic(category=OntologyAnnotation(term='organism part'), value=OntologyAnnotation(term='urine'))])
# ### 9. Loading an isa assay definition in the form of an ordered dictionary.
#
# - It corresponds to an ISA configuration assay table but expressed in JSON.
#
# - In this NMR assay there is a sample extraction step, which produces "supernatant" and "pellet" extracts (1 of each per input sample).
#
# - IMPORTANT: Note how ISA `OntologyAnnotation` elements are used in this data structure
nmr_assay_dict = OrderedDict([
('measurement_type', OntologyAnnotation(term='metabolite profiling')),
('technology_type', OntologyAnnotation(term='nmr spectroscopy')),
('extraction', {}),
('extract', [
{
'node_type': EXTRACT,
'characteristics_category': OntologyAnnotation(term='extract type'),
'characteristics_value': 'supernatant',
'size': 1,
'technical_replicates': None,
'is_input_to_next_protocols': True
},
{
'node_type': EXTRACT,
'characteristics_category': OntologyAnnotation(term='extract type'),
'characteristics_value': 'pellet',
'size': 1,
'technical_replicates': None,
'is_input_to_next_protocols': True
}
]),
('nmr_spectroscopy', {
OntologyAnnotation(term='instrument'): ['Bruker AvanceII 1 GHz'],
OntologyAnnotation(term='acquisition_mode'): ['1D 13C NMR','1D 1H NMR','2D 13C-13C NMR'],
OntologyAnnotation(term='pulse_sequence'): ['CPMG','TOCSY','HOESY','watergate']
}),
('raw_spectral_data_file', [
{
'node_type': DATA_FILE,
'size': 1,
'technical_replicates': 2,
'is_input_to_next_protocols': False
}
])
])
# ### 10. We now show how to create an new AssayGraph structure from scratch, as if we were defining a completely new assay type.
# +
new_assay_graph1=AssayGraph(id_="WB", measurement_type=OntologyAnnotation(term="protein profiling"), technology_type=OntologyAnnotation(term="Western blot"))
# -
# ### 11. We procede by assembling the Process graph:
# +
protocol_node_protein = ProtocolNode(id_="P",name='Protein extraction')
protocol_node_data_acq = ProtocolNode(id_="DA",name='WB imaging', parameter_values=[ParameterValue(category=ProtocolParameter(parameter_name=OntologyAnnotation(term="channel")),value=OntologyAnnotation(term="360 nm")),ParameterValue(category=ProtocolParameter(parameter_name=OntologyAnnotation(term='channel')),value=OntologyAnnotation(term="550 nm"))])
protein_char = Characteristic(category=OntologyAnnotation(term='material type'), value='protein extract')
protein_sample_node = ProductNode(id_="SP", node_type=SAMPLE, size=1, characteristics=[protein_char])
wb_data_node = ProductNode(id_="WBD", node_type=DATA_FILE, size=1)
nodes = [protein_sample_node, wb_data_node, protocol_node_protein, protocol_node_data_acq]
links = [(protocol_node_protein,protein_sample_node),(protein_sample_node,protocol_node_data_acq),(protocol_node_data_acq,wb_data_node)]
new_assay_graph1.add_nodes(nodes)
new_assay_graph1.add_links(links)
new_assay_graph1
# -
# The following step does 3 things:
#
# - generate an assay plan from the assay declaration data strucure
# - create a `Sample and Assay Plan` object holding a list of samples and the list of assay workflows which have been declared
# - create a `Sample to Assay` object, which details which sample will be input to a specific assay.
# +
nmr_assay_graph = AssayGraph.generate_assay_plan_from_dict(nmr_assay_dict)
sap1 = SampleAndAssayPlan(name='sap1', sample_plan=[input_material1,input_material2,input_material3],assay_plan=[new_assay_graph1,nmr_assay_graph])
sample2assay_plan={input_material3: [new_assay_graph1, nmr_assay_graph], input_material2: [nmr_assay_graph], input_material1: [nmr_assay_graph]}
sap1.sample_to_assay_map=sample2assay_plan
sap1.sample_to_assay_map
# +
# specifying which sample type (sometimes referred to as specimen)
# sap1.add_sample_type('liver')
# +
# specifying how many times each specimen is supposed to be collected
# sap1.add_sample_plan_record('liver',3)
# +
#### 9. Declaration of an ISA assay and linking specimen type and data acquisition plan for this assay
# # declare the type of `Assay` which will be performed
# assay_type1 = Assay(measurement_type='metabolite profiling', technology_type='mass spectrometry')
# # associate this assay type to the `SampleAssayPlan`
# sap1.add_assay_type(assay_type1)
# # specify which `sample type` will be used as input to the declare `Assay`
# sap1.add_assay_plan_record('liver',assay_type1)
# -
# ### 11. Build an ISA `Study Design Arm` by adding the first set of ISA `Cells` and setting the `Sample Assay Plan`
arm1.add_item_to_arm_map(st_cl1,sap1)
print(arm1)
# ### 12 Now expanding the `Arm` by adding a new `Cell`, which uses the same `Sample Assay Plan` as the one used in Cell #1.
# Of course, the `Sample Assay Plan` for this new `Cell` could be different. It would have to be to built as shown before.
arm1.add_item_to_arm_map(st_cl2,sap1)
# Adding the last section of the Arm, with a cell which also uses the same sample assay plan.
arm1.add_item_to_arm_map(st_cl3,sap1)
arm1.add_item_to_arm_map(st_cl4,sap1)
arm1.add_item_to_arm_map(st_cl6,sap1)
arm1.add_item_to_arm_map(st_cl7,sap1)
arm1.add_item_to_arm_map(st_cl5,sap1)
# ### 13. Creation of additional ISA Study Arms and setting the number of subjects associated to that unique sequence of ISA Cells.
arm2 = StudyArm(name='Arm 2')
arm2.group_size=40
arm2.add_item_to_arm_map(st_cl1,sap1)
arm2.add_item_to_arm_map(st_cl4,sap1)
arm2.add_item_to_arm_map(st_cl3,sap1)
arm2.add_item_to_arm_map(st_cl2,sap1)
arm2.add_item_to_arm_map(st_cl6,sap1)
arm2.add_item_to_arm_map(st_cl7,sap1)
arm2.add_item_to_arm_map(st_cl5,sap1)
arm3 = StudyArm(name='Arm 3')
arm3.group_size=10
arm3.add_item_to_arm_map(st_cl1,sap1)
arm3.add_item_to_arm_map(st_cl7,sap1)
arm3.add_item_to_arm_map(st_cl3,sap1)
arm3.add_item_to_arm_map(st_cl4,sap1)
arm3.add_item_to_arm_map(st_cl6,sap1)
arm3.add_item_to_arm_map(st_cl2,sap1)
arm3.add_item_to_arm_map(st_cl5,sap1)
# ### 14. We can now create the ISA `Study Design` object, which will receive the `Arms` defined by the user.
study_design= StudyDesign(name='trial design #1')
# print(sd)
# Adding a study arm to the study design object.
study_design.add_study_arm(arm1)
study_design.add_study_arm(arm2)
study_design.add_study_arm(arm3)
# print(sd)
# +
# Let's now serialize the ISA study design to JSON
import json
from isatools.isajson import ISAJSONEncoder
from isatools.create.model import StudyDesignEncoder
f=json.dumps(study_design, cls=StudyDesignEncoder, sort_keys=True, indent=4, separators=(',', ': '))
# -
# ### 15. let's produce a graphical overview of the study design arms and the associated sample assay plans
def get_treatment_factors(some_element):
treat = ""
for j in range(len(some_element['factorValues'])):
if j < len(some_element['factorValues']) - 1:
if 'unit' in some_element['factorValues'][j].keys():
treat = treat + some_element['factorValues'][j]['factor']['name'].lower() + ": " \
+ str(some_element['factorValues'][j]['value']) + " " \
+ str(some_element['factorValues'][j]['unit']['term'].lower()) + ", "
else:
treat = treat + some_element['factorValues'][j]['factor']['name'].lower() + ": " \
+ str(some_element['factorValues'][j]['value']) + ","
else:
if 'unit' in some_element['factorValues'][j].keys():
treat = treat + some_element['factorValues'][j]['factor']['name'].lower() + ": " \
+ str(some_element['factorValues'][j]['value']) + " " \
+ str(some_element['factorValues'][j]['unit']['term'].lower())
else:
treat = treat + some_element['factorValues'][j]['factor']['name'].lower() + ": " \
+ str(some_element['factorValues'][j]['value'])
return treat
# +
design = json.loads(json.dumps(study_design, cls=StudyDesignEncoder, sort_keys=True, indent=4, separators=(',', ': ')))
frames = []
Items = []
# defining a color pallet for the different element types:
element_colors = {"biological intervention": "rgb(253,232,37)",
"radiological intervention": "rgb(53, 155, 8)",
"dietary intervention": "rgb(53, 155, 8)",
"chemical intervention": "rgb(69, 13, 83)",
"washout": "rgb(45, 62, 120)",
"screen": "rgb(33, 144, 140)",
"run in": "rgb(43, 144, 180)",
"follow-up": "rgb(88, 189, 94)",
"concomitant treatment": "rgb(255, 255, 0)"}
# processing the study design arms and treatment plans:
for key in design["studyArms"].keys():
DF = pd.DataFrame(columns=['Arm', 'Cell', 'Type', 'Start_date', 'End_date', 'Treatment', 'Color'])
arm_name = key
# print("arm: ", arm_name)
size = design["studyArms"][key]["groupSize"]
size_annotation = "n=" + str(size)
cells_per_arm = design["studyArms"][key]["cells"]
cell_counter = 0
for cell in cells_per_arm:
cell_name = cell['name']
elements_per_cell = cell['elements']
for element in elements_per_cell:
treat = ""
element_counter = 0
if 'concomitantTreatments' in element.keys():
element_counter = element_counter + 1
treatments = []
for item in element['concomitantTreatments']:
treatment = get_treatment_factors(item)
treatments.append(treatment)
concomitant = ','.join(treatments[0:-1])
concomitant = concomitant + ' and ' + treatments[-1]
array = [arm_name, cell_name, arm_name + ": [" + concomitant + "]_concomitant_" + str(cell_counter),
dt.datetime(cell_counter + 2000, 1, 1), dt.datetime(cell_counter + 2000 + 1, 1, 1),
str(element['factorValues']),
concomitant,
element_colors["concomitant treatment"]]
Items.append(array)
elif 'type' in element.keys():
treatment = get_treatment_factors(element)
element_counter = element_counter + 1
array = [arm_name, cell_name, arm_name + ": [" + str(element['type']) + "]_" + str(cell_counter),
dt.datetime((cell_counter + 2000), 1, 1), dt.datetime((cell_counter + 2000 + 1), 1, 1),
# str(element['factorValues']),
str(treatment),
element_colors[element['type']]]
Items.append(array)
cell_counter = cell_counter + 1
for i, Dat in enumerate(Items):
DF.loc[i] = Dat
# print("setting:", DF.loc[i])
# providing the canvas for the figure
# print("THESE ARE THE TYPES_: ", DF.Type.tolist())
fig = figure(title='Study Design Treatment Plan',
width=800,
height=400,
y_range=DF.Type.tolist(),
x_range=Range1d(DF.Start_date.min(), DF.End_date.max()),
tools='save')
# adding a tool tip
hover = HoverTool(tooltips="Task: @Type<br>\
Start: @Start_date<br>\
Cell_Name: @Cell<br>\
Treatment: @Treatment")
fig.add_tools(hover)
DF['ID'] = DF.index+0.8
# print("ID: ", DF['ID'])
DF['ID1'] = DF.index+1.2
# print("ID1: ", DF['ID1'])
CDS = ColumnDataSource(DF)
# , legend=str(size_annotation)
r = fig.quad(left='Start_date', right='End_date', bottom='ID', top='ID1', source=CDS, color="Color")
fig.xaxis.axis_label = 'Time'
fig.yaxis.axis_label = 'study arms'
# working at providing a background color change for every arm in the study design
counts = DF['Arm'].value_counts().tolist()
# print("total number of study arms:", len(counts), "| number of phases per arm:", counts)
# box = []
# for i, this_element in enumerate(DF['Arm']):
# if i==0:
# box[i] = BoxAnnotation(bottom=0,
# top=DF['Arm'].value_counts().tolist()[0],
# fill_color="blue")
# elif i % 2 == 0:
# box[i] = BoxAnnotation(bottom=DF['Arm'].value_counts().tolist()[0],
# top=DF['Arm'].value_counts().tolist()[0],
# fill_color="silver")
# else:
# box[i] = BoxAnnotation(bottom=DF['Arm'].value_counts().tolist()[0],
# top=DF['Arm'].value_counts().tolist()[0] + DF['Arm'].value_counts().tolist()[1],
# fill_color="grey",
# fill_alpha=0.1)
# # adding the background color for each arm:
# for element in box:
# fig.add_layout(element)
# # fig.add_layout(box2)
# # fig.add_layout(legend,'right')
caption1 = Legend(items=[(str(size_annotation), [r])])
fig.add_layout(caption1, 'right')
citation = Label(x=10, y=-80, x_units='screen', y_units='screen',
text='repeated measure group design layout - isa-api 0.12', render_mode='css',
border_line_color='gray', border_line_alpha=0.4,
background_fill_color='white', background_fill_alpha=1.0)
fig.add_layout(citation)
show(fig)
# -
study = study_design.generate_isa_study()
len(study.assays)
investigation.studies=[study]
# print(investigation.studies[0].assays[1])
print(investigation.studies[0].assays[0])
# WRITING ISA-JSON document
print(json.dumps(investigation, cls=ISAJSONEncoder, sort_keys=True, indent=4, separators=(',', ': ')))
from isatools import isatab
isatab.dump(investigation, './notebook-output/isa-repeated-measure-crossover-design')
from isatools.isatab import dump_tables_to_dataframes as dumpdf
dataframes = dumpdf(investigation)
dataframes.keys()
len(dataframes.keys())
dataframes[list(dataframes.keys())[1]]
[x for x in study.assays[0].graph.nodes() if isinstance(x, Sample)]
len([x for x in study.assays[0].graph.nodes() if isinstance(x, Sample)])
[getattr(x, 'name', None) for x in study.assays[0].graph.nodes()]
# ## About this notebook
#
# - authors: ph<EMAIL>, <EMAIL>
# - license: CC-BY 4.0
# - support: <EMAIL>
# - issue tracker: https://github.com/ISA-tools/isa-api/issues
| _build/html/_sources/content/notebooks/create-a-repeated-treatment-design-with-ISA-descriptor.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Implementing Pipeline
# Using what you learning about pipelining, rewrite your machine learning code from the last section to use sklearn's Pipeline. For reference, the previous main function implementation is provided in the second to last cell. Refactor this in the last cell.
import nltk
nltk.download(['punkt', 'wordnet'])
# +
import re
import numpy as np
import pandas as pd
from nltk.tokenize import word_tokenize
from nltk.stem import WordNetLemmatizer
from sklearn.metrics import confusion_matrix
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.feature_extraction.text import CountVectorizer, TfidfTransformer
# +
url_regex = 'http[s]?://(?:[a-zA-Z]|[0-9]|[$-_@.&+]|[!*\(\),]|(?:%[0-9a-fA-F][0-9a-fA-F]))+'
def load_data():
df = pd.read_csv('corporate_messaging.csv', encoding='latin-1')
df = df[(df["category:confidence"] == 1) & (df['category'] != 'Exclude')]
X = df.text.values
y = df.category.values
return X, y
def tokenize(text):
detected_urls = re.findall(url_regex, text)
for url in detected_urls:
text = text.replace(url, "urlplaceholder")
tokens = word_tokenize(text)
lemmatizer = WordNetLemmatizer()
clean_tokens = []
for tok in tokens:
clean_tok = lemmatizer.lemmatize(tok).lower().strip()
clean_tokens.append(clean_tok)
return clean_tokens
def display_results(y_test, y_pred):
labels = np.unique(y_pred)
confusion_mat = confusion_matrix(y_test, y_pred, labels=labels)
accuracy = (y_pred == y_test).mean()
print("Labels:", labels)
print("Confusion Matrix:\n", confusion_mat)
print("Accuracy:", accuracy)
# -
def old_main():
X, y = load_data()
X_train, X_test, y_train, y_test = train_test_split(X, y)
vect = CountVectorizer(tokenizer=tokenize)
tfidf = TfidfTransformer()
clf = RandomForestClassifier()
# train classifier
X_train_counts = vect.fit_transform(X_train)
X_train_tfidf = tfidf.fit_transform(X_train_counts)
clf.fit(X_train_tfidf, y_train)
# predict on test data
X_test_counts = vect.transform(X_test)
X_test_tfidf = tfidf.transform(X_test_counts)
y_pred = clf.predict(X_test_tfidf)
# display results
display_results(y_test, y_pred)
# Rewrite the main function to use sklearn's `Pipeline` here:
from sklearn.pipeline import Pipeline
def main():
X, y = load_data()
X_train, X_test, y_train, y_test = train_test_split(X, y)
# build pipeline
pipeline = Pipeline([('vect', CountVectorizer(tokenizer=tokenize)),
('tfidf', TfidfTransformer()),
('clf', RandomForestClassifier())])
# train classifier
pipeline.fit(X_train, y_train)
# predict on test data
y_pred = pipeline.predict(X_test)
# display results
display_results(y_test, y_pred)
main()
| pipeline.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# 使用するパッケージの宣言
import numpy as np
import matplotlib.pyplot as plt
# %matplotlib inline
# +
# 初期位置での迷路の様子
# 図を描く大きさと、図の変数名を宣言
fig = plt.figure(figsize=(5, 5))
ax = plt.gca()
# 赤い壁を描く
plt.plot([1, 1], [0, 1], color='red', linewidth=2)
plt.plot([1, 2], [2, 2], color='red', linewidth=2)
plt.plot([2, 2], [2, 1], color='red', linewidth=2)
plt.plot([2, 3], [1, 1], color='red', linewidth=2)
# 状態を示す文字S0~S8を描く
plt.text(0.5, 2.5, 'S0', size=14, ha='center')
plt.text(1.5, 2.5, 'S1', size=14, ha='center')
plt.text(2.5, 2.5, 'S2', size=14, ha='center')
plt.text(0.5, 1.5, 'S3', size=14, ha='center')
plt.text(1.5, 1.5, 'S4', size=14, ha='center')
plt.text(2.5, 1.5, 'S5', size=14, ha='center')
plt.text(0.5, 0.5, 'S6', size=14, ha='center')
plt.text(1.5, 0.5, 'S7', size=14, ha='center')
plt.text(2.5, 0.5, 'S8', size=14, ha='center')
plt.text(0.5, 2.3, 'START', ha='center')
plt.text(2.5, 0.3, 'GOAL', ha='center')
# 描画範囲の設定と目盛りを消す設定
ax.set_xlim(0, 3)
ax.set_ylim(0, 3)
plt.tick_params(axis='both', which='both', bottom='off', top='off',
labelbottom='off', right='off', left='off', labelleft='off')
# 現在地S0に緑丸を描画する
line, = ax.plot([0.5], [2.5], marker="o", color='g', markersize=60)
# +
# 初期の方策を決定するパラメータtheta_0を設定
# 行は状態0~7、列は移動方向で↑、→、↓、←を表す
theta_0 = np.array([[np.nan, 1, 1, np.nan], # s0
[np.nan, 1, np.nan, 1], # s1
[np.nan, np.nan, 1, 1], # s2
[1, 1, 1, np.nan], # s3
[np.nan, np.nan, 1, 1], # s4
[1, np.nan, np.nan, np.nan], # s5
[1, np.nan, np.nan, np.nan], # s6
[1, 1, np.nan, np.nan], # s7、※s8はゴールなので、方策はなし
])
# +
# 方策パラメータthetaを行動方策piにソフトマックス関数で変換する手法の定義
def softmax_convert_into_pi_from_theta(theta):
'''ソフトマックス関数で割合を計算する'''
beta = 1.0
[m, n] = theta.shape # thetaの行列サイズを取得
pi = np.zeros((m, n))
exp_theta = np.exp(beta * theta) # thetaをexp(theta)へと変換
for i in range(0, m):
# pi[i, :] = theta[i, :] / np.nansum(theta[i, :])
# simpleに割合の計算の場合
pi[i, :] = exp_theta[i, :] / np.nansum(exp_theta[i, :])
# softmaxで計算の場合
pi = np.nan_to_num(pi) # nanを0に変換
return pi
# -
# 初期の方策pi_0を求める
pi_0 = softmax_convert_into_pi_from_theta(theta_0)
print(pi_0)
# +
# 行動aと1step移動後の状態sを求める関数を定義
def get_action_and_next_s(pi, s):
direction = ["up", "right", "down", "left"]
# pi[s,:]の確率に従って、directionが選択される
next_direction = np.random.choice(direction, p=pi[s, :])
if next_direction == "up":
action = 0
s_next = s - 3 # 上に移動するときは状態の数字が3小さくなる
elif next_direction == "right":
action = 1
s_next = s + 1 # 右に移動するときは状態の数字が1大きくなる
elif next_direction == "down":
action = 2
s_next = s + 3 # 下に移動するときは状態の数字が3大きくなる
elif next_direction == "left":
action = 3
s_next = s - 1 # 左に移動するときは状態の数字が1小さくなる
return [action, s_next]
# +
# 迷路を解く関数の定義、状態と行動の履歴を出力
def goal_maze_ret_s_a(pi):
s = 0 # スタート地点
s_a_history = [[0, np.nan]] # エージェントの移動を記録するリスト
while (1): # ゴールするまでループ
[action, next_s] = get_action_and_next_s(pi, s)
s_a_history[-1][1] = action
# 現在の状態(つまり一番最後なのでindex=-1)の行動を代入
s_a_history.append([next_s, np.nan])
# 次の状態を代入。行動はまだ分からないのでnanにしておく
if next_s == 8: # ゴール地点なら終了
break
else:
s = next_s
return s_a_history
# -
# 初期の方策で迷路を解く
s_a_history = goal_maze_ret_s_a(pi_0)
print(s_a_history)
print("迷路を解くのにかかったステップ数は" + str(len(s_a_history) - 1) + "です")
# +
# thetaの更新関数を定義します
def update_theta(theta, pi, s_a_history):
eta = 0.1 # 学習率
T = len(s_a_history) - 1 # ゴールまでの総ステップ数
[m, n] = theta.shape # thetaの行列サイズを取得
delta_theta = theta.copy() # Δthetaの元を作成、ポインタ参照なので、delta_theta = thetaはダメ
# delta_thetaを要素ごとに求めます
for i in range(0, m):
for j in range(0, n):
if not(np.isnan(theta[i, j])): # thetaがnanでない場合
SA_i = [SA for SA in s_a_history if SA[0] == i]
# 履歴から状態iのものを取り出すリスト内包表記です
SA_ij = [SA for SA in s_a_history if SA == [i, j]]
# 状態iで行動jをしたものを取り出す
N_i = len(SA_i) # 状態iで行動した総回数
N_ij = len(SA_ij) # 状態iで行動jをとった回数
# 初版では符号の正負に間違いがありました(修正日:180703)
#delta_theta[i, j] = (N_ij + pi[i, j] * N_i) / T
delta_theta[i, j] = (N_ij - pi[i, j] * N_i) / T
new_theta = theta + eta * delta_theta
return new_theta
# -
# 方策の更新
new_theta = update_theta(theta_0, pi_0, s_a_history)
pi = softmax_convert_into_pi_from_theta(new_theta)
print(pi)
# +
# 方策勾配法で迷路を解く
# 初版で、def update_thetaに間違いがあった関係で、終了条件を変更します(修正日:180703)
#stop_epsilon = 10**-8 # 10^-8よりも方策に変化が少なくなったら学習終了とする
stop_epsilon = 10**-4 # 10^-4よりも方策に変化が少なくなったら学習終了とする
theta = theta_0
pi = pi_0
is_continue = True
count = 1
while is_continue: # is_continueがFalseになるまで繰り返す
s_a_history = goal_maze_ret_s_a(pi) # 方策πで迷路内を探索した履歴を求める
new_theta = update_theta(theta, pi, s_a_history) # パラメータΘを更新
new_pi = softmax_convert_into_pi_from_theta(new_theta) # 方策πの更新
print(np.sum(np.abs(new_pi - pi))) # 方策の変化を出力
print("迷路を解くのにかかったステップ数は" + str(len(s_a_history) - 1) + "です")
if np.sum(np.abs(new_pi - pi)) < stop_epsilon:
is_continue = False
else:
theta = new_theta
pi = new_pi
# -
# 最終的な方策を確認
np.set_printoptions(precision=3, suppress=True) # 有効桁数3、指数表示しないという設定
print(pi)
# +
# エージェントの移動の様子を可視化します
# 参考URL http://louistiao.me/posts/notebooks/embedding-matplotlib-animations-in-jupyter-notebooks/
from matplotlib import animation
from IPython.display import HTML
def init():
# 背景画像の初期化
line.set_data([], [])
return (line,)
def animate(i):
# フレームごとの描画内容
state = s_a_history[i][0] # 現在の場所を描く
x = (state % 3) + 0.5 # 状態のx座標は、3で割った余り+0.5
y = 2.5 - int(state / 3) # y座標は3で割った商を2.5から引く
line.set_data(x, y)
return (line,)
# 初期化関数とフレームごとの描画関数を用いて動画を作成
anim = animation.FuncAnimation(fig, animate, init_func=init, frames=len(
s_a_history), interval=200, repeat=False)
HTML(anim.to_jshtml())
| JSTfair/Deep-Reinforcement-Learning-Book-master/program/2_3_Policygradient.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3.9.7 ('sem10')
# language: python
# name: python3
# ---
# # Part 1
# #### **Mention 10 examples of encoding**
# 1. Position
# 1. Length
# 1. Area
# 1. Shape
# 1. Color
# 1. angle
# 1. Line weight
# 1. Line ending
# 1. Texture
# 1. Pattern
#
# #### **Are all encodings created equally?**
# No pie charts make it hard to distinguish between size of numbers.
#
# #### **3 encodings hard for the human eye to parse**
# - Pie chart
# -
# -
#
# #### **What is the problem with a pie chart**
# Hard to distinguish between size of numbers
# # Part 2
# ### **Exercise 2.1**
# #### **Looking at Fig 3-1, Janert writes, "the data itself shows clearly that the amount of random noise in the data is small". What do you think his argument is?**
#
# The weird stuff that's happening to the data could be noise, but because the shift is repetitive, it hints that it is a pattern and not noise.
#
# #### **Can you think of a real-world example of a multivariate relationship like the one in Fig 3-3 (lower right panel)?**
# If you take humans overall in a statistic where, for example, women and men are different, the relationship would be multivariate. We saw this earlier with the relationship between steps and BMI.
#
# #### **What are the two methods Janet mentions for smoothing noisy data? Can you think of other ones?**
# **Answer:** weighted splines and *LOESS (locally weighted regression).
#
# Kernel smoothing is another example. Gaussian smoothing is also another example.
#
# #### **What are residuals? Why is it a good idea to plot the residuals of your fit?**
# Residuals are the remainder when the smooth trend is subtracted from the actual data. That is how much does our smooth curve deviate from the real data.
#
# It is a good idea to plot the residual, as it gives a visualisation of how well the smoothing is fitted to the data. If the residual is located around 0, it means that the fit is quite good, and if it deviates a lot, it tells us that the smoothed plot is not very representative of the
#
#
# #### **Explain in your own words the point of the smooth tube in figure 3-7.**
# The solid line is the best approximation of all the points, and the lower and upper dashed lines the points where the residual is either negative or positive. The point is that we can, with some certainty say, that the points will lie between the two dashed lines.
#
# #### **What the hell is banking, and what part of our visual system does it use to help us see patterns? What are potential problems with banking?**
# Banking is changing the aspect ratio, so the curves on the graphs are closer to 45 degrees. This is typically done by changing the aspect ration.
#
# #### **SummariseSummarise the discussion of Graphical Analysis and Presentation Graphics on pp. 68-69 in your own words.**
#
# **Graphical Analysis** is the process of using graphical tools to analyse data. This is often done without knowing the end result and is done as an exploration.
#
# **Presention graphics** are done when you already know the end result, and want to show it in a graphical way. We have already established how the data relates, now we want to show it.
#
# ### **Exercise 2.2**
# **What kind of relationship does loglog plot help discoder**: This plot shows if (typically) the y-coordinate relationship
#
#
# **What kind of relationship does semi-log plot help discoder**: Both axis have an exponential proportions
#
# ### **Display plot from week1 in y-log scale**
# +
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
from math import radians, cos, sin, asin, sqrt
sns.set_theme()
# -
df = pd.read_csv("../data/Police_Department_Incident_Reports__Historical_2003_to_May_2018.csv")
fig, ax = plt.subplots(1, 1, figsize=(15, 5))
ax.set(yscale="log")
sns.countplot(x="Category", data=df, order=df.Category.value_counts().index, ax=ax)
plt.xticks(rotation=90);
# ## **LogLog theft plot**
#
# #### **Step 1** Divide into $100m \times 100m$ grid
# +
# Removing outliers
df_theft = df[(df.Y < 50) & (df.X < -122) & (df.Category == "LARCENY/THEFT")]
print(f"Removed {df.shape[0] - df_theft.shape[0]} outliers")
# Function for calculating the distance between two points based on their longitude and lattitude
def haversine(lon1, lat1, lon2, lat2):
"""
Calculate the great circle distance between two points
on the earth (specified in decimal degrees)
"""
# convert decimal degrees to radians
lon1, lat1, lon2, lat2 = map(radians, [lon1, lat1, lon2, lat2])
# haversine formula
dlon = lon2 - lon1
dlat = lat2 - lat1
a = sin(dlat/2)**2 + cos(lat1) * cos(lat2) * sin(dlon/2)**2
c = 2 * asin(sqrt(a))
r = 6371 # Radius of earth in kilometers. Use 3956 for miles
return c * r
# Finding boundary of san fransisco
lat_min = df_theft.Y.min()
lat_max = df_theft.Y.max()
lon_min = df_theft.X.min()
lon_max = df_theft.X.max()
# Estimating bin amount along different borders
print(f"Estimated bin amount along the northern border of San Fransisco: {haversine(lon_min, lat_max, lon_max, lat_max)*10}")
print(f"Estimated bin amount along the eastern border of San Fransisco: {haversine(lon_max, lat_min, lon_max, lat_max)*10}")
print(f"Estimated bin amount along the southern border of San Fransisco: {haversine(lon_min, lat_min, lon_max, lat_min)*10}")
print(f"Estimated bin amount along the western border of San Fransisco: {haversine(lon_min, lat_min, lon_min, lat_max)*10}")
count, lon, lat = np.histogram2d(df_theft.Y, df_theft.X, bins = [130,125])
# -
plt.figure(figsize=(12,10))
plt.imshow(count, cmap='hot',vmax=3000, origin='lower')
plt.grid(False);
plt.colorbar();
# #### **Step 2**: Count the number of thefts in each square
# +
unique, N = np.unique(count, return_counts=True)
fig, [ax1, ax2] = plt.subplots(2, 1, figsize=(15, 10))
ax1.scatter(N, unique);
ax2.scatter(N, unique);
ax2.set(yscale="log", xscale="log");
plt.tight_layout()
# -
# ## Part 3
# #### **Describe in your own words how data is organized in `sklearn` (how does a *dataset* work according to the tutorial)?**
# Data in sklearn is orginised in samples and features, or more commonly known as data and labels (x, Y).
#
# #### **What is the dimensionality of the `.data` part of a dataset and what is the size of each dimension?**
# Dimensionality is always a 2d-array, with `n_samples`, and `n_features`
# # Part 4
# Something with sklearn
# #### **Exercise 4.1** pairplot with unique combinations
#
from sklearn.linear_model import LinearRegression
import itertools
from tqdm import tqdm
n = 14
k = 2
num = np.math.factorial(n)//(np.math.factorial(k) * np.math.factorial(n - k))
print(f"Num of unique combinations: {num}")
# Creating datetime and hour of week column
df["dt"] = pd.to_datetime(df.Date + df.Time, format="%m/%d/%Y%H:%M")
df['hour_of_the_week'] = df['dt'].dt.dayofweek * 24 + (df['dt'].dt.hour + 1)
# +
focuscrimes = set(['WEAPON LAWS', 'PROSTITUTION', 'DRIVING UNDER THE INFLUENCE', 'ROBBERY', 'BURGLARY', 'ASSAULT', 'DRUNKENNESS', 'DRUG/NARCOTIC', 'TRESPASS', 'LARCENY/THEFT', 'VANDALISM', 'VEHICLE THEFT', 'STOLEN PROPERTY', 'DISORDERLY CONDUCT'])
combine_crimes = list(itertools.combinations(focuscrimes, 2))
scale = 2
fig, axes = plt.subplots(13, 7, figsize=(7*scale, 13*scale))
for (x_crime, y_crime), ax in tqdm(zip(combine_crimes, axes.flatten()), total=len(combine_crimes)):
x = df[df.Category == x_crime].hour_of_the_week.value_counts()
y = df[df.Category == y_crime].hour_of_the_week.value_counts()
sns.scatterplot(x=x, y=y, ax=ax)
ax.set_xlabel(x_crime, fontsize=8)
ax.set_ylabel(y_crime, fontsize=8)
ax.set_xticklabels([])
ax.set_yticklabels([])
plt.tight_layout()
plt.savefig("w4_scatter.png")
plt.show()
# -
scale = 2
for (x_crime, y_crime) in combine_crimes:
x = df[df.Category == x_crime].hour_of_the_week.value_counts().values
y = df[df.Category == y_crime].hour_of_the_week.value_counts().values
reg = LinearRegression().fit(x.reshape(-1, 1), y)
print(f"x:{x_crime:30}y:{y_crime:30}\t slope:{reg.coef_}\tintercept: {reg.intercept_:5.2f}\tscore: {reg.score(x.reshape(-1, 1), y)*100:.2f}%")
| exercises/week5.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# Site-packages:
import os, sys, numpy as np, pandas as pd
curr = os.getcwd()
main = os.getcwd().split('GamsPythonModels')[0]+'GamsPythonModels'
project = main+'\\'+os.getcwd().split('GamsPythonModels')[1].split('\\')[1]
py = {}
py['main'] = main+'\\py_main'
py['project'] = project+'\\py_project'
py['local'] = curr+'\\py_local'
work_folder = main+'\\work_folder'
# Main GPM packages:
os.chdir(py['main'])
import nesting_tree, DataBase, COE, COE_settings, regex_gms, DB2Gams, ModelFrame_PE as PE
# Project packages:
os.chdir(py['project'])
# Local packages:
os.chdir(py['local'])
import Calib, COE_abate, COE_abate_settings
# import LOCALPACKAGE
os.chdir(curr)
# Clean-up?
try:
if clean_up is True:
temp_gamsdb = [filename for filename in os.listdir(work_folder) if filename.endswith((".gdx",".gms",".lst",".g00",".pf",".opt",".gmy"))]
for temp in temp_gamsdb:
try:
os.remove(work_folder+'\\'+temp)
except OSError as err:
print('The file' + temp + ' is still active and was not deleted.')
except NameError:
print('No clean-up of work-folder')
| AbatementProject/InputDisplacement/StdPackages.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ---
# # Predicting Credit Card Default
# ### Model Development with Cross-Validation
#
# ---
# Sections:
# - [Loading Preprocessed Data](#Loading-Data)
# - [Validation Set Partitioning](#Set-Partitioning)
# - [Model Development with Cross-Validation](#Model-Development)
# - [Decision Tree](#Model-Tree)
# - [Random Forest](#Model-Forest)
# - [AdaBoosting](#Model-AdaBoosting)
# - [Neural Network](#Model-NN)
# - [SVM with RBF kernel](#Model-SVM)
#
# ---
# <a id="Loading-Data"></a>
# # Loading Preprocessed Data
# ---
#
# Note:
# Open <a href="./data_preparation.ipynb">data_preparation.ipynb</a> to see how the data was preprocessed.
# +
import pandas as pd
import numpy as np
import imblearn #libary for imbalanced functions i.e. K-means SMOTE
from sklearn import preprocessing
#from google.colab import drive
#drive.mount('/content/drive')
# filename = "drive/Shareddrives/DS-project/default_processed.csv"
filename = "default_processed.csv"
data = pd.read_csv(filename)
data.head(10)
# -
# <a id="Set-Partitioning"></a>
# # Validation Set Partitioning
# ---
#
# In order to ascertain a standard deviation and investigate the optimal hyperparameters for each model, we will employ stratified k-fold cross-validation.
#
# This algorithm will split our dataset into k consecutive folds in a startefied manner such that it preserves the same target class distribution.
# +
from sklearn.model_selection import KFold
from sklearn.model_selection import StratifiedKFold
# acquire all rows, and all columns except for last one (label)
features = data.iloc[:, :-1]
# assignt to frame with all rows and the last column (label)
label = data.iloc[:,[-1]]
label = preprocessing.LabelEncoder().fit_transform(label)
skf5 = StratifiedKFold(n_splits = 5)
skf5.get_n_splits(features, label)
print(skf5)
for train_index, test_index in skf5.split(features, label):
print("TRAIN:", train_index, "Test:", test_index)
# -
# ## Cross Validation Score function
#
# The below function takes in as parameters a scikit-learn model (decision tree,random forest, etc..), the features and labels for a sklearn cross validation model which contains the indexes of each training and testing at each fold.
#
# It will print out the Accuracy, F1 Macro, Recall and Precision (averaged over all folds) along with their standard deviation
# +
from sklearn.model_selection import cross_validate, cross_val_score, cross_val_predict
from numpy import mean, std
def skf(model, features, labels, cv):
metrics = ['precision_weighted', 'recall_weighted', 'accuracy', 'f1', 'f1_micro', 'f1_macro', 'f1_weighted']
scores = cross_validate(model, features, labels, scoring=metrics, cv=cv)
metric_pretty_names = ['Precision', 'Recall', 'Accuracy', 'F1', 'F1 Micro', 'F1 Macro', 'F1 Weighted']
score_data = {'mean': [mean(scores[s]) for s in scores if 'time' not in s],
'std' : [std(scores[s]) for s in scores if 'time' not in s]}
scores_df = pd.DataFrame(score_data, index=metric_pretty_names)
return scores_df
# -
# ## Oversampling using SMOTE
#
# One way to fight the inbalance *training set* is to generate new samples in the classes which are under-represented.
#
# The most naive strategy is to generate new samples by randomly sampling with replacement the current available samples.
#
# **over-sample**
# Object to over-sample the minority class(es) by picking samples at random with replacement.
#
#
# **NOTE:
# In the following Model Development section, you will see each model being trained in both original data and oversampling data by SMOTE**
# <a id="Model-Development"></a>
# # Model Development
#
# ---
# <a id="Model-Tree"></a>
# ## Decision Tree
# ---
#
# The first model we develop is the decision tree classifier.
#
# We look at 5 different parameters to find the best estimator.
# +
from sklearn.tree import DecisionTreeClassifier
tree_grid = {
'max_depth': [1, 3, 5, 7],
'min_samples_split': [2, 4, 6, 8],
'min_samples_leaf': [1, 2, 4, 6],
'criterion': ['gini', 'entropy'],
'max_features': ['auto', 'sqrt', 'log2']
}
y_pred_tree = {}
y_prob_tree = {}
"""
Chosen hyperparameters:
max_depth = 7
min_samples_split = 8
min_samples_leaf = 4
criterion = entropy
max_features = sqrt
"""
#tree = get_best_model(DecisionTreeClassifier, tree_grid, X_train, y_train)
tree = DecisionTreeClassifier(max_depth=7, min_samples_split=8, min_samples_leaf=4, criterion='entropy', max_features='sqrt')
# -
# ### Decision Tree: original data
# K-Fold Cross Validation Results with Original Data, where K=5
skf(tree, features, label, skf5)
# +
from sklearn.metrics import classification_report, accuracy_score, make_scorer
originalclass = []
predictedclass = []
# Make our customer score
def classification_report_with_accuracy_score(y_true, y_pred):
originalclass.extend(y_true)
predictedclass.extend(y_pred)
return accuracy_score(y_true, y_pred) # return accuracy score
# Nested CV with parameter optimization
nested_score = cross_val_score(tree, X=features, y=label, cv=skf5, scoring=make_scorer(classification_report_with_accuracy_score))
# Average values in classification report for all folds in a K-fold Cross-validation
print(classification_report(originalclass, predictedclass))
# -
# ### Decision Tree: SMOTE data
#
# K-Fold Cross Validation Results with SMOTE Data, where K=5
# +
from imblearn.over_sampling import SMOTENC
from sklearn.metrics import recall_score
from imblearn.pipeline import Pipeline, make_pipeline
tree_pipeline = make_pipeline(SMOTENC([1,2,3,5,6,7,8,9,10],random_state=42), tree)
skf(tree_pipeline, features, label, skf5)
# +
originalclass = []
predictedclass = []
# Nested CV with parameter optimization
nested_score = cross_val_score(tree_pipeline, X=features, y=label, cv=skf5, scoring=make_scorer(classification_report_with_accuracy_score))
# Average values in classification report for all folds in a K-fold Cross-validation
print(classification_report(originalclass, predictedclass))
# -
# <a id="Model-Forest"></a>
# ## Random Forest
# ---
#
# Next, we develop a the random forest classifier.
#
# We look at 6 different parameters (5 of which are the same as the decision tree) to find the best estimator.
# +
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import cross_validate
from numpy import mean
from numpy import std
forest_params = {
'n_estimators': [10, 50, 100, 500],
'max_depth': [1, 3, 5, 7],
'min_samples_split': [2, 4, 6, 8],
'min_samples_leaf': [1, 2, 4, 6],
'criterion': ['gini', 'entropy'],
'max_features': ['auto', 'sqrt', 'log2']
}
"""
Chosen hyperparameters:
n_estimators = 500
max_depth = 7
min_samples_split = 8
min_samples_leaf = 2
criterion = gini
max_features = sqrt
"""
#forest = get_best_model(RandomForestClassifier, forest_params, X_train, y_train)
forest = RandomForestClassifier(n_estimators=500, max_depth=7, min_samples_split=8, min_samples_leaf=2, criterion='gini', max_features='sqrt')
# -
# ### Random Forest: Original data
# K-Fold Cross Validation Results with Original Data, where K=5
skf(forest, features, label, skf5)
# +
originalclass = []
predictedclass = []
# Nested CV with parameter optimization
nested_score = cross_val_score(forest, X=features, y=label, cv=skf5,
scoring=make_scorer(classification_report_with_accuracy_score))
# Average values in classification report for all folds in a K-fold Cross-validation
print(classification_report(originalclass, predictedclass))
# -
# ### Random Forest: SMOTE data
# K-Fold Cross Validation Results with SMOTE Data, where K=5
forest_pipeline = make_pipeline(SMOTENC([1,2,3,5,6,7,8,9,10],random_state=42), forest)
skf(forest_pipeline, features, label, skf5)
# +
originalclass = []
predictedclass = []
# Nested CV with parameter optimization
nested_score = cross_val_score(forest_pipeline, X=features, y=label, cv=skf5,
scoring=make_scorer(classification_report_with_accuracy_score))
# Average values in classification report for all folds in a K-fold Cross-validation
print(classification_report(originalclass, predictedclass))
# -
# <a id="Model-AdaBoosting"></a>
# ## AdaBoosting
# ---
# Next, we develop the an AdaBoost classifier.
#
# We look at 3 different parameters to find the best estimator.
#
# Next we use Randomized Search Cross Validation to select our hyperparameters with the goal of finding the best estimator.
#
# +
from sklearn.model_selection import RandomizedSearchCV
from sklearn.metrics import *
def get_best_model(model, param_grid, X_train, y_train):
search = RandomizedSearchCV(model(), param_grid, scoring='f1',
refit=True, n_jobs=-1)
best_model = search.fit(X_train, y_train).best_estimator_
params = best_model.get_params()
print("Chosen hyperparameters: ")
for param in param_grid.keys():
print(param + " = " + str(params[param]))
return best_model
# +
from sklearn.ensemble import AdaBoostClassifier
adaboost_params = {
'n_estimators': [20, 50, 100, 150],
'learning_rate': [0.01, 0.05, 1, 1.5],
'algorithm': ['SAMME', 'SAMME.R']
}
y_pred_adaboost = {}
y_prob_adaboost = {}
"""
Chosen hyperparameters:
n_estimators = 100
learning_rate = 1.5
algorithm = SAMME
"""
adaboost = get_best_model(AdaBoostClassifier, adaboost_params, X_train, y_train)
# -
# ### AdaBoosting: original data
#
# K-Fold Cross Validation Results with Original Data, where K=5
skf(adaboost, features, label, skf5)
# +
originalclass = []
predictedclass = []
# Nested CV with parameter optimization
nested_score = cross_val_score(adaboost, X=features, y=label, cv=skf5,
scoring=make_scorer(classification_report_with_accuracy_score))
# Average values in classification report for all folds in a K-fold Cross-validation
print(classification_report(originalclass, predictedclass))
# -
# ### AdaBoosting: SMOTE data
# K-Fold Cross Validation Results with SMOTE Data, where K=5
adaboost_pipeline = make_pipeline(SMOTENC([1,2,3,5,6,7,8,9,10],random_state=42), adaboost)
skf(adaboost_pipeline, features, label, skf5)
# +
originalclass = []
predictedclass = []
# Nested CV with parameter optimization
nested_score = cross_val_score(adaboost_pipeline, X=features, y=label, cv=skf5,
scoring=make_scorer(classification_report_with_accuracy_score))
# Average values in classification report for all folds in a K-fold Cross-validation
print(classification_report(originalclass, predictedclass))
# -
# <a id="Model-NN"></a>
# ## Neural Network
# ---
#
# Next we develop a Neural Network model using the MLP Classifier.
#
# We look at 5 different parameters to find the best estimator.
# +
from sklearn.neural_network import MLPClassifier
from sklearn.preprocessing import StandardScaler
nn_params = {
'hidden_layer_sizes': [(15, 5), (20, 10), (25, 15), (30, 20)],
'max_iter': [700],
'learning_rate': ["constant", "invscaling", "adaptive"],
'alpha': [0.0001, 0.001, 0.01, 0.05],
'activation': [ 'logistic']
}
"""
Chosen hyperparameters:
hidden_layer_sizes = (20, 10)
max_iter = 700
learning_rate = invscaling
alpha = 0.0001
activation = logistic
"""
mlp = MLPClassifier(hidden_layer_sizes=(20, 10),
max_iter=700,
learning_rate="invscaling",
alpha=0.0001, activation='logistic')
# -
# ### Neural Network: original data
# K-Fold Cross Validation Results with Original Data, where K=5
# +
from sklearn.neural_network import MLPClassifier
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import StandardScaler
mlp_pipeline = make_pipeline(StandardScaler(), mlp)
skf(mlp_pipeline, features, label, skf5)
# +
originalclass = []
predictedclass = []
# Nested CV with parameter optimization
nested_score = cross_val_score(mlp_pipeline, X=features, y=label, cv=skf5,
scoring=make_scorer(classification_report_with_accuracy_score))
# Average values in classification report for all folds in a K-fold Cross-validation
print(classification_report(originalclass, predictedclass))
# -
# ### Neural Network: SMOTE data
# K-Fold Cross Validation Results with SMOTE Data, where K=5
# +
#from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import StandardScaler
from imblearn.pipeline import make_pipeline
mlp_smote_pipeline = make_pipeline(SMOTENC([1,2,3,5,6,7,8,9,10], random_state=42), StandardScaler(), mlp)
skf(mlp_smote_pipeline, features, label, skf5)
# +
originalclass = []
predictedclass = []
# Nested CV with parameter optimization
nested_score = cross_val_score(mlp_smote_pipeline, X=features, y=label, cv=skf5,
scoring=make_scorer(classification_report_with_accuracy_score))
# Average values in classification report for all folds in a K-fold Cross-validation
print(classification_report(originalclass, predictedclass))
# -
# <a id="Model-SVM"></a>
# ## SVM with RBF kernel
# ---
#
# Next we develop a SVM model using the RBF kernel.
#
# We look at 2 different parameters to find the best estimator.
# +
from sklearn.svm import SVC
svc_params = {'C': [0.1, 1, 10, 50], 'gamma': [1, 0.1, 0.01, 0.001], 'kernel': ['rbf'], 'probability': [1]}
y_pred_svm_rbf = {}
y_prob_svm_rbf = {}
"""
Chosen hyperparameters:
C = 1
gamma = 0.1
kernel = rbf
probability = 1
"""
svm_rbf_kernel = SVC(C=1, gamma=0.1, kernel='rbf', probability=1)
# -
# ### SVM with RBF kernel: original data
#
# K-Fold Cross Validation Results with Original Data, where K=5
# +
from sklearn.svm import SVC
svm_rbf_kernel = SVC(C=1, gamma=0.1, kernel='rbf', probability=1)
svm_rfb_pipeline = make_pipeline(StandardScaler(), svm_rbf_kernel)
skf(svm_rfb_pipeline, features, label, skf5)
# +
originalclass = []
predictedclass = []
# Nested CV with parameter optimization
nested_score = cross_val_score(svm_rfb_pipeline, X=features, y=label, cv=skf5,
scoring=make_scorer(classification_report_with_accuracy_score))
# Average values in classification report for all folds in a K-fold Cross-validation
print(classification_report(originalclass, predictedclass))
# -
# ### SVM with RBF kernel: SMOTE data
# K-Fold Cross Validation Results with SMOTE Data, where K=5
# +
#from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import StandardScaler
from imblearn.over_sampling import SMOTENC
from sklearn.metrics import recall_score
from imblearn.pipeline import Pipeline, make_pipeline
from sklearn.svm import SVC
svm_rbf_kernel = SVC(C=1, gamma=0.1, kernel='rbf', probability=1)
svm_rfb_pipeline_smote = make_pipeline(SMOTENC([1,2,3,5,6,7,8,9,10], random_state=42),
StandardScaler(),
svm_rbf_kernel)
skf(svm_rfb_pipeline_smote, features, label, skf5)
# +
originalclass = []
predictedclass = []
# Nested CV with parameter optimization
nested_score = cross_val_score(svm_rfb_pipeline_smote, X=features, y=label, cv=skf5,
scoring=make_scorer(classification_report_with_accuracy_score))
# Average values in classification report for all folds in a K-fold Cross-validation
print(classification_report(originalclass, predictedclass))
| model_training_crossval.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [default]
# language: python
# name: python3
# ---
# ## Evaluate CNTK Fast-RCNN model directly from python
#
# This notebook demonstrates how to evaluate a single image using a CNTK Fast-RCNN model.
#
# For a full description of the model and the algorithm, please see the following <a href="https://docs.microsoft.com/en-us/cognitive-toolkit/Object-Detection-using-Fast-R-CNN" target="_blank">tutorial</a>.
#
# Below, you will see sample code for:
# 1. Preparing the input data for the network (including image size adjustments)
# 2. Evaluation of the input data using the model
# 3. Processing the evaluation result and presenting the selected regions back on the image.
#
# <b>Important</b>: Before running this notebook, please make sure that:
# <ol>
# <li>You have version >= 2.0 of CNTK installed. Installation instructions are available <a href="https://docs.microsoft.com/en-us/cognitive-toolkit/Setup-CNTK-on-your-machine" target="_blank">here</a>.
#
# <li>This notebook uses the CNTK python APIs and should be run from the CNTK python environment.</li>
#
# <li>OpenCV and the other required python packages for the Fast-RCNN scenario are installed. Please follow the instructions <a href="https://docs.microsoft.com/en-us/cognitive-toolkit/Object-Detection-using-Fast-R-CNN#setup" target="_blank">in here</a> to install the required packages.
# </ol>
# ##### 1. Download the sample dataset and make sure that the model exists
# First things first - we will download the sample Grocery dataset (if it's not already there), and we'll also make sure that the Fast-RCNN model file exists. The script will use your local trained model (if available), or will download and use the pre-trained model if a local trained model isn't available.
# In case we run inside the CNTK test enviornment, the model and data are copied from the test data directory.
# We also set the device to cpu / gpu for the test environment. If you have both CPU and GPU on your machine, you can optionally switch the devices. By default, we choose the best available device.
# +
# %matplotlib inline
# the above line enable us to draw the images inside the notebooks
import os
import sys
from os import path
import cntk
# Check for an environment variable defined in CNTK's test infrastructure
def is_test(): return 'CNTK_EXTERNAL_TESTDATA_SOURCE_DIRECTORY' in os.environ
# Select the right target device when this notebook is being tested
# Currently supported only for GPU
# Setup data environment for pre-built data sources for testing
if is_test():
if 'TEST_DEVICE' in os.environ:
if os.environ['TEST_DEVICE'] == 'cpu':
cntk.device.try_set_default_device(cntk.device.cpu())
else:
cntk.device.try_set_default_device(cntk.device.gpu(0))
sys.path.append(os.path.join(*"../../../../../Tests/EndToEndTests/CNTKv2Python/Examples".split("/")))
import prepare_test_data as T
T.prepare_Grocery_data()
T.prepare_fastrcnn_grocery_100_model()
#Make sure the grocery dataset is installed
sys.path.append('../../../DataSets/Grocery')
from install_grocery import download_grocery_data
download_grocery_data()
# Make sure the FRCNN model exists - check if the model was trained and exists, if not - download the existing model
sys.path.append('../../../../../PretrainedModels')
from download_model import download_model_by_name
download_model_by_name("Fast-RCNN_grocery100")
model_path = '../../../../../PretrainedModels/Fast-RCNN_grocery100.model'
# -
# ### 3. load the model and prepare it for evaluation:
# As a first step for using the Fast-RCNN model, we load the trained model file.
#
# The trained model accepts 3 inputs: The image data, the bounding box (region of interest, or ROI) proposals and the ground truth labels of the ROIs. Since we are evaluating a new image - we probably don't have the ground truth labels for the image, hence - we need to adjust the network to accept only the image and the ROIs as input.
# In order to do that we use the CNTK APIs to clone the network and change its input nodes.
#
# More information and examples regarding cloning nodes of a network are available in the <a href="https://docs.microsoft.com/en-us/cognitive-toolkit/Build-your-own-image-classifier-using-Transfer-Learning" target="_blank">Transfer Learning</a> tutorial.
# +
from cntk import load_model
from cntk import placeholder
from cntk.logging.graph import find_by_name, get_node_outputs
from cntk.ops import combine
from cntk.ops.sequence import input_variable
from cntk.ops.functions import CloneMethod
# load the trained model
trained_frcnn_model = load_model(model_path)
# find the original features and rois input nodes
features_node = find_by_name(trained_frcnn_model, "features")
rois_node = find_by_name(trained_frcnn_model, "rois")
# find the output "z" node
z_node = find_by_name(trained_frcnn_model, 'z')
# define new input nodes for the features (image) and rois
image_input = input_variable(features_node.shape, name='features')
roi_input = input_variable(rois_node.shape, name='rois')
# Clone the desired layers with fixed weights and place holder for the new input nodes
cloned_nodes = combine([z_node.owner]).clone(
CloneMethod.freeze,
{features_node: placeholder(name='features'), rois_node: placeholder(name='rois')})
# apply the cloned nodes to the input nodes
frcnn_model = cloned_nodes(image_input, roi_input)
print("Fast-RCNN Grocery model loaded successfully!")
# -
# ### 4. Load an image and convert it to the network format
#
# Next, we load an image from the test set using OpenCV, and then resize according to the network input dimensions. (Which are set when the network is trained).
# When resizing, we preserve scale and pad the border areas with a constant value (114), which is later used for normalization by the network.
# +
import cv2
import numpy as np
import matplotlib.pyplot as plt
image_height = 1000
image_width = 1000
def resize_and_pad(img, width, height, pad_value=114):
# port of the c++ code from CNTK: https://github.com/Microsoft/CNTK/blob/f686879b654285d06d75c69ee266e9d4b7b87bc4/Source/Readers/ImageReader/ImageTransformers.cpp#L316
img_width = len(img[0])
img_height = len(img)
scale_w = img_width > img_height
target_w = width
target_h = height
if scale_w:
target_h = int(np.round(img_height * float(width) / float(img_width)))
else:
target_w = int(np.round(img_width * float(height) / float(img_height)))
resized = cv2.resize(img, (target_w, target_h), 0, 0, interpolation=cv2.INTER_NEAREST)
top = int(max(0, np.round((height - target_h) / 2)))
left = int(max(0, np.round((width - target_w) / 2)))
bottom = height - top - target_h
right = width - left - target_w
resized_with_pad = cv2.copyMakeBorder(resized, top, bottom, left, right,
cv2.BORDER_CONSTANT, value=[pad_value, pad_value, pad_value])
#tranpose(2,0,1) converts the image to the HWC format which CNTK accepts
model_arg_rep = np.ascontiguousarray(np.array(resized_with_pad, dtype=np.float32).transpose(2,0,1))
return resized_with_pad, model_arg_rep
def load_image_and_scale(image_path, width, height, pad_value=114):
img = cv2.imread(image_path)
return resize_and_pad(img, width, height, pad_value), img
test_image_path = r"../../../DataSets/Grocery/testImages/WIN_20160803_11_28_42_Pro.jpg"
(test_img, test_img_model_arg), original_img = load_image_and_scale(test_image_path, image_width, image_height)
plt.imshow(cv2.cvtColor(test_img, cv2.COLOR_BGR2RGB))
plt.axis("off")
# -
# ### 5. Generate ROIs for testing
#
# Now, we produce regions of interest (ROIs) proposals using selective search & grid methods, using the same method as in the script: A1_GenerateInputROIs.py.
#
# Each ROI is in the format of [x,y,w,h], where the coordinates real numbers in the range of 0 to 1, and scaled according to the resized and padded image.
# The ROIs array is padded with regions of [0,0,0,0] at the end to match the 2000 ROIs input format of the model.
# +
# Parameters taken from PARAMETERS.py
# ROI generation
roi_minDimRel = 0.04
roi_maxDimRel = 0.4
roi_minNrPixelsRel = 2 * roi_minDimRel * roi_minDimRel
roi_maxNrPixelsRel = 0.33 * roi_maxDimRel * roi_maxDimRel
roi_maxAspectRatio = 4.0 # maximum aspect Ratio of a ROI vertically and horizontally
roi_maxImgDim = 200 # image size used for ROI generation
ss_scale = 100 # selective search ROIS: parameter controlling cluster size for segmentation
ss_sigma = 1.2 # selective search ROIs: width of gaussian kernal for segmentation
ss_minSize = 20 # selective search ROIs: minimum component size for segmentation
grid_nrScales = 7 # uniform grid ROIs: number of iterations from largest possible ROI to smaller ROIs
grid_aspectRatios = [1.0, 2.0, 0.5] # uniform grid ROIs: aspect ratio of ROIs
cntk_nrRois = 100 # 100 # how many ROIs to zero-pad
cntk_padWidth = 1000
cntk_padHeight = 1000
from cntk_helpers import imArrayWidthHeight, getSelectiveSearchRois, imresizeMaxDim
from cntk_helpers import getGridRois, filterRois, roiTransformPadScaleParams, roiTransformPadScale
def get_rois_for_image(img, use_selective_search=True, use_grid_rois=True):
roi_minDim = roi_minDimRel * roi_maxImgDim
roi_maxDim = roi_maxDimRel * roi_maxImgDim
roi_minNrPixels = roi_minNrPixelsRel * roi_maxImgDim*roi_maxImgDim
roi_maxNrPixels = roi_maxNrPixelsRel * roi_maxImgDim*roi_maxImgDim
imgOrig = img.copy()
# get rois
if use_selective_search:
print ("Calling selective search..")
rects, scaled_img, scale = getSelectiveSearchRois(imgOrig, ss_scale, ss_sigma, ss_minSize, roi_maxImgDim) #interpolation=cv2.INTER_AREA
print ("Number of rois detected using selective search: " + str(len(rects)))
else:
rects = []
scaled_img, scale = imresizeMaxDim(imgOrig, roi_maxImgDim, boUpscale=True, interpolation=cv2.INTER_AREA)
imgWidth, imgHeight = imArrayWidthHeight(scaled_img)
# add grid rois
if use_grid_rois:
rectsGrid = getGridRois(imgWidth, imgHeight, grid_nrScales, grid_aspectRatios)
print ("Number of rois on grid added: " + str(len(rectsGrid)))
rects += rectsGrid
# run filter
print ("Number of rectangles before filtering = " + str(len(rects)))
rois = filterRois(rects, imgWidth, imgHeight, roi_minNrPixels, roi_maxNrPixels, roi_minDim, roi_maxDim, roi_maxAspectRatio)
if len(rois) == 0: #make sure at least one roi returned per image
rois = [[5, 5, imgWidth-5, imgHeight-5]]
print ("Number of rectangles after filtering = " + str(len(rois)))
# scale up to original size and save to disk
# note: each rectangle is in original image format with [x1,y1,x2,y2]
original_rois = np.int32(np.array(rois) / scale)
img_width = len(img[0])
img_height = len(img)
# all rois need to be scaled + padded to cntk input image size
targetw, targeth, w_offset, h_offset, scale = roiTransformPadScaleParams(img_width, img_height,
cntk_padWidth, cntk_padHeight)
rois = []
for original_roi in original_rois:
x1, y1, x2, y2 = roiTransformPadScale(original_roi, w_offset, h_offset, scale)
rois.append([x1, y1, x2, y2])
# pad rois if needed:
if len(rois) < cntk_nrRois:
rois += [[0, 0, 0, 0]] * (cntk_nrRois - len(rois))
elif len(rois) > cntk_nrRois:
rois = rois[:cntk_nrRois]
return np.array(rois), original_rois
test_rois, original_rois = get_rois_for_image(original_img)
roi_padding_index = len(original_rois)
print("Number of rois for evaluation:", len(test_rois))
# -
# ### 6. Evaluate the sample
# Here, we prepare the data to be in CNTK's expected arguments format and run it through the model used the model's **eval** method.
#
# We then process the result by trimming the padded ROIs part, and calculate the predicted labels and their probabilities.
# +
from cntk_helpers import softmax2D
# a dummy variable for labels the will be given as an input to the network but will be ignored
dummy_labels = np.zeros((2000,17))
#Index the names of the arguments so we can get them by name
args_indices = {}
for i,arg in enumerate(frcnn_model.arguments):
args_indices[arg.name] = i
# prepare the arguments
arguments = {
frcnn_model.arguments[args_indices['features']]: [test_img_model_arg],
frcnn_model.arguments[args_indices['rois']]: [test_rois],
}
# run it through the model
output = frcnn_model.eval(arguments)
# we now extract the "z" values from the output, which are the values of the layer that is just before
# the softmax layer.
# we take just the relevant part from that array
rois_values = output[0][0][:roi_padding_index]
# get the prediction for each roi by taking the index with the maximal value in each row
rois_labels_predictions = np.argmax(rois_values, axis=1)
# calculate the probabilities using softmax
rois_probs = softmax2D(rois_values)
# print the number of ROIs that were detected as non-background
print("Number of detections: %d"%np.sum(rois_labels_predictions > 0))
# -
# ### 7. Merge overlapping regions using Non-Maxima-Suppression
# Before inspecting the predictions, we need to merge overlapping regions that were detected using the Non-Maxima-Suppression algorithm that is implemented in the cntk_helpers module.
# +
from cntk_helpers import applyNonMaximaSuppression
nms_threshold = 0.1
non_padded_rois = test_rois[:roi_padding_index]
max_probs = np.amax(rois_probs, axis=1).tolist()
rois_prediction_indices = applyNonMaximaSuppression(nms_threshold, rois_labels_predictions, max_probs, non_padded_rois, ignore_background=True)
print("Indices of selected regions:",rois_prediction_indices)
# -
# ### 8. Visualize the results
#
# As a final step, we use the OpenCV **rectangle** and **putText** methods in order to draw the selected regions on the original image alongside their corresponding predicted labels.
# +
rois_with_prediction = test_rois[rois_prediction_indices]
rois_prediction_labels = rois_labels_predictions[rois_prediction_indices]
rois_predicion_scores = rois_values[rois_prediction_indices]
original_rois_predictions = original_rois[rois_prediction_indices]
# class names taken from PARAMETERS.py:
classes = ('__background__', # always index 0
'avocado', 'orange', 'butter', 'champagne', 'eggBox', 'gerkin', 'joghurt', 'ketchup',
'orangeJuice', 'onion', 'pepper', 'tomato', 'water', 'milk', 'tabasco', 'mustard')
original_img_cpy = original_img.copy()
for roi,label in zip(original_rois_predictions, rois_prediction_labels):
(x1,y1,x2,y2) = roi
cv2.rectangle(original_img_cpy, (x1, y1), (x2, y2), (0, 255, 0), 5)
cv2.putText(original_img_cpy,classes[label],(x1,y2 + 30), cv2.FONT_HERSHEY_DUPLEX, 2,(200,0,255),3,cv2.LINE_AA)
print("Evaluation result:")
plt.figure(figsize=(10, 10))
plt.imshow(cv2.cvtColor(original_img_cpy, cv2.COLOR_BGR2RGB), interpolation='nearest')
plt.axis("off")
| Examples/Image/Detection/FastRCNN/BrainScript/CNTK_FastRCNN_Eval.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ### Feature Scaling Example
#
# You have now seen how feature scaling might change the clusters we obtain from the kmeans algorithm, but it is time to try it out!
#
# First let's get some data to work with. The first cell here will read in the necessary libraries, generate data, and make a plot of the data you will be working with throughout the rest of the notebook.
#
# The dataset you will work with through the notebook is then stored in **data**.
# +
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.cluster import KMeans
from IPython.display import Image
from sklearn.datasets.samples_generator import make_blobs
import tests2 as t
# %matplotlib inline
# DSND colors: UBlue, Salmon, Gold, Slate
plot_colors = ['#02b3e4', '#ee2e76', '#ffb613', '#2e3d49']
# Light colors: Blue light, Salmon light
plot_lcolors = ['#88d0f3', '#ed8ca1', '#fdd270']
# Gray/bg colors: Slate Dark, Gray, Silver
plot_grays = ['#1c262f', '#aebfd1', '#fafbfc']
def create_data():
n_points = 120
X = np.random.RandomState(3200000).uniform(-3, 3, [n_points, 2])
X_abs = np.absolute(X)
inner_ring_flag = np.logical_and(X_abs[:,0] < 1.2, X_abs[:,1] < 1.2)
outer_ring_flag = X_abs.sum(axis = 1) > 5.3
keep = np.logical_not(np.logical_or(inner_ring_flag, outer_ring_flag))
X = X[keep]
X = X[:60] # only keep first 100
X1 = np.matmul(X, np.array([[2.5, 0], [0, 100]])) + np.array([22.5, 500])
plt.figure(figsize = [15,15])
plt.scatter(X1[:,0], X1[:,1], s = 64, c = plot_colors[-1])
plt.xlabel('5k Completion Time (min)', size = 30)
plt.xticks(np.arange(15, 30+5, 5), fontsize = 30)
plt.ylabel('Test Score (raw)', size = 30)
plt.yticks(np.arange(200, 800+200, 200), fontsize = 30)
ax = plt.gca()
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
[side.set_linewidth(2) for side in ax.spines.values()]
ax.tick_params(width = 2)
plt.savefig('C18_FeatScalingEx_01.png', transparent = True)
data = pd.DataFrame(X1)
data.columns = ['5k_Time', 'Raw_Test_Score']
return data
data = create_data()
# -
# `1.` Take a look at the dataset. Are there any missing values? What is the average completion time? What is the average raw test score? Use the cells below to find the answers to these questions, and the dictioonary to match values and check against our solution.
# cell for work
# another cell for work
# +
# Use the dictionary to match the values to the corresponding statements
a = 0
b = 60
c = 22.9
d = 4.53
e = 511.7
q1_dict = {
'number of missing values': # letter here,
'the mean 5k time in minutes': # letter here,
'the mean test score as a raw value': # letter here,
'number of individuals in the dataset': # letter here
}
# check your answer against ours here
t.check_q1(q1_dict)
# -
# `2.` Now, instantiate a kmeans `model` with 2 cluster centers. Use your model to `fit` and `predict` the the group of each point in your dataset. Store the predictions in `preds`. If you correctly created the model and predictions, you should see a top (blue) cluster and bottom (pink) cluster when running the following cell.
model = # instantiate a model with two centers
preds = # fit and predict
# +
# Run this to see your results
def plot_clusters(data, preds):
plt.figure(figsize = [15,15])
for k, col in zip(range(n_clusters), plot_colors[:n_clusters]):
my_members = (preds == k)
plt.scatter(data['5k_Time'][my_members], data['Raw_Test_Score'][my_members], s = 64, c = col)
plt.xlabel('5k Completion Time (min)', size = 30)
plt.xticks(np.arange(15, 30+5, 5), fontsize = 30)
plt.ylabel('Test Score (raw)', size = 30)
plt.yticks(np.arange(200, 800+200, 200), fontsize = 30)
ax = plt.gca()
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
[side.set_linewidth(2) for side in ax.spines.values()]
ax.tick_params(width = 2)
plot_clusters(data, preds)
# -
# `3.` Now create two new columns to add to your `data` dataframe. The first is `test_scaled`, which you should create by subtracting the mean test score and dividing by the standard deviation test score.
#
# The second column to create is `5k_time_sec`, which should have the minutes changed to seconds.
# your work here
data['test_scaled'] = # standardized test scores
data['5k_time_sec'] = # times in seconds
# `4.` Now, similar to what you did in question 2, instantiate a kmeans `model` with 2 cluster centers. Use your model to `fit` and `predict` the the group of each point in your dataset. Store the predictions in `preds`. If you correctly created the model and predictions, you should see a right (blue) cluster and left (pink) cluster when running the following cell.
model = # instantiate a model with two centers
preds = # fit and predict
# +
# Run this to see your results
def plot_clusters2(data, preds):
plt.figure(figsize = [15,15])
for k, col in zip(range(n_clusters), plot_colors[:n_clusters]):
my_members = (preds == k)
plt.scatter(data['5k_time_sec'][my_members], data['test_scaled'][my_members], s = 64, c = col)
plt.xlabel('5k Completion Time (sec)', size = 30)
plt.xticks(np.arange(900, 1800+300, 300), fontsize = 30)
plt.ylabel('Test Score (z)', size = 30)
plt.yticks(np.arange(-3, 3+2, 2), fontsize = 30)
ax = plt.gca()
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
[side.set_linewidth(2) for side in ax.spines.values()]
ax.tick_params(width = 2)
plot_clusters2(data, preds)
# -
# `5.` Match the variable that best describes the way you should think of feature scaling with algorithms that use distance based metrics or regularization.
# +
# options
a = 'We should always use normalizing'
b = 'We should always scale our variables between 0 and 1.'
c = 'Variable scale will frequently influence your results, so it is important to standardize for all of these algorithms.'
d = 'Scaling will not change the results of your output.'
best_option = # best answer variable here
# check your answer against ours here
t.check_q5(best_option)
# -
# ### If you get stuck, you can find a solution by pushing the orange icon in the top left of this notebook.
| DSND_Term1-master/lessons/Unsupervised/1_Clustering/Feature Scaling Example.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
cloudLocation_AllMyFiles='https://github.com/dghnyldz/computational-thinking/raw/main/'
linkToEXCEL_File=cloudLocation_AllMyFiles+'csvData_co2.xlsx'
pip show pandas
pip show openpyxl
dfExcel=pandas.read_excel(linkToEXCEL_File)
import pandas as pd # activating pandas
dfExcel=pd.read_excel(linkToEXCEL_File)
type(dfExcel)
dfExcel
from IPython.display import IFrame
wikiLink="https://en.wikipedia.org/wiki/Democracy_Index"
IFrame(wikiLink, width=700, height=300)
dataWIKI=pd.read_html(wikiLink,header=0,flavor='bs4',attrs={'class': 'wikitable'})
type(dataWIKI)
len(dataWIKI)
dataWIKI[5]
from IPython.display import IFrame
wikiLink="https://en.wikipedia.org/wiki/List_of_freedom_indices"
IFrame(wikiLink, width=900, height=500)
# +
import pandas as pd
wikiTables=pd.read_html(wikiLink, # link
#header=0, # where is the header?
flavor='bs4', # helper to translate html
attrs={'class': 'wikitable sortable'}) # attributes to identify element(s)
# -
wikiTables[0]
| Untitled2.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# ##### 1
# 
# PPT上写错了,是Lecture 8
# ##### 2
# 
# ##### 3
# 
# ##### 4
# 
# ##### 5
# 
# ##### 6
# 
# ##### 7
# 
# -
# $$ posterior \propto prior \times likelihood $$
#
# and from [Wikipedia](https://en.wikipedia.org/wiki/Likelihood_function)
# >In statistics, a **likelihood function** (often simply the **likelihood**) is a function of the parameters of a statistical model.
#
# >The likelihood of a set of parameter values, θ, given outcomes x, is equal to the probability of those observed outcomes given those parameter values, that is
# $$\mathcal{L}(\theta |x) = P(x | \theta).$$
# The likelihood function is defined differently for discrete and continuous probability distributions.
#
# 对于这里来说,f就是outcomes x,$p(f|e)$就是likelihood function, $f$就是上式中的$x$,$e$就是上式中的$\theta$
#
# $$L(french|english) = P(english|french)$$
#
#
# -
# 这里,为什么要求p(f|e)而不是直接用p(e|f),也就是说,我拿到了一句法文,为什么不直接去找到它对应的各个英文句子的概率,而是要,通过各个英文转换成这个法文的概率来计算。
# 百度文库的[这篇](http://wenku.baidu.com/link?url=7qBR2ASl2s_RQ3wVqLMFtbavMQE3L30af1UDPgBwLsQH7Moi7FH9iSS4_iW0Yegyu3ZV07YAdHLd1oF57P2-CgcWnweGwHCu7Aei14g-34O)文章是这么说的:
# >之所以不直接估计P(e|f),主要有两个原因:(1) 可以将e和f分别看作是疾病和症状,那么从e推出f(P(f|e))比较可行,而很难从f推出e(P(e|f))。(2) 引入P(e),这样翻译出来的语句更像人话。
#
# 疾病的例子用来理解Bayes rule本身是非常好的,但来解释这个过于牵强。(疾病用来解释贝叶斯:医生在判断是什么病的时候默认进行了贝叶斯公式的转换,转换成了likelihood(感冒会导致发烧的概率) * prior(感冒的概率)。第二个理由比较认同。这里的目标p(english|french)是要完成一句话的转换,都是解剖成每个单词(或是短语)的概率p(french|english),因此,language model(p(e))就很有用了,可以保证说的话符合目标语言的习惯。
#
# 举个中文的例子,“这是好的”翻译成"This is great"和"Here is well"
#
# ##### 8
# 
# ##### 9
# 
# ##### 10
# 
# ##### 11
# 
# ##### 12
# 
# ##### 13
# 
# ##### 14
# 
# ##### 15
# 
# ##### 16
# 
# ##### 17
# 
# ##### 18
# 
# ##### 19
# 
# ##### 20
# 
# ##### 21
# 
# ##### 22
# 
# ##### 23
# 
# ##### 24
# 
# ##### 25
# 
# ##### 26
# 
# ##### 27
# 
# ##### 28
# 
# ##### 29
# 
# ##### 30
# 
# ##### 31
# 
# ##### 32
# 
# ##### 33
# 
# ##### 34
# 
| nlp-socher/lec08-lstm.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
#one dimensional array
import numpy as np
arr = np.array([1,2,3])
print (arr)
#more than one dimensional array
import numpy as md
mat = md.array([[5,3],[6,1],[3,2]])
print(mat)
#using ndmin
import numpy as md
mat = md.array([4,5,6,3,2],ndmin=0)
print(mat)
#for complex numbers
import numpy as md
comp = md.array([[4,5],[3,2]],dtype=complex)
print(comp)
# -
| NumPy_basics.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Median Runner
#
# This notebook demonstrates how to create a grading workflow using PyBryt.
import pybryt
# This demo has the following directory structure. This notebook, `index.ipynb`, runs PyBryt, `median.ipynb` is the assignment reference implementation, and `submissions` contains notebooks with student code in them.
# + language="bash"
# tree
# -
# ## Reference Implementations
#
# If you have marked up a reference implementation, like the one in [`median.ipynb`](median.ipynb), you can load this reference using `pybryt.ReferenceImplementation.compile`. Because references are relatively static and can take some time to execute, you can pickle the reference implementations to a file with `pybryt.ReferenceImplementation.dump`.
ref = pybryt.ReferenceImplementation.compile("median.ipynb")
ref.dump()
# To load a pickled reference implementation, use `pybryt.ReferenceImplementation.load`:
ref = pybryt.ReferenceImplementation.load("median.pkl")
ref
# ## Assessing Submissions
#
# To use PyBryt for grading multiple submissions, you can build a reproducible grading pipeline for an arbitrary number of submissions. To grab the submission notebook paths, the cell below uses `glob.glob`.
from glob import glob
subms = sorted(glob("submissions/*.ipynb"))
subms
# To use PyBryt to grade a student's submission, a `pybryt.StudentImplementation` must be created from that submission. The constructor takes the path to the notebook as its only positional argument. It is in this step that the student's code is executed, so this cell will need to be rerun whenever changes are made to the submission notebooks.
# +
student_impls = []
for subm in subms:
student_impls.append(pybryt.StudentImplementation(subm))
student_impls
# -
# Once you have created the `pybryt.StudentImplementation` objects, use the `pybryt.StudentImplementation.check` method to run the check of a submission against a reference implementation. This method returns a single `pybryt.ReferenceResult` or a list of them, depending on the argument passed to `check`. In the cell below, the results are collected into a list.
# +
results = []
for si in student_impls:
results.append(si.check(ref))
results
# -
# To view the results in a concise manner, the `pybryt.ReferenceResult` class has some helpful instance variables. You can also get information about the memory footprint, such as the number of steps, from the `pybryt.StudentImplementation` class.
from textwrap import indent
for sp, si, res in zip(subms, student_impls, results):
print(f"SUBMISSION: {sp}")
print(f" EXECUTION STEPS: {si.steps}") # the number of steps in the execution
# res.messages is a list of messages returned by the reference during grading
messages = "\n".join(res.messages)
# res.correct is a boolean for whether the reference was satisfied
message = f"SATISFIED: {res.correct}\nMESSAGES:\n{indent(messages, ' - ')}"
# some pretty-printing
print(indent(message, " "))
print("\n")
# You can also turn the reference result objects into a JSON-friendly dictionary format for further processing:
res = results[0]
res.to_dict()
#
| demo/median/index.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Classifying Fashion-MNIST
#
# Now it's your turn to build and train a neural network. You'll be using the [Fashion-MNIST dataset](https://github.com/zalandoresearch/fashion-mnist), a drop-in replacement for the MNIST dataset. MNIST is actually quite trivial with neural networks where you can easily achieve better than 97% accuracy. Fashion-MNIST is a set of 28x28 greyscale images of clothes. It's more complex than MNIST, so it's a better representation of the actual performance of your network, and a better representation of datasets you'll use in the real world.
#
# <img src='assets/fashion-mnist-sprite.png' width=500px>
#
# In this notebook, you'll build your own neural network. For the most part, you could just copy and paste the code from Part 3, but you wouldn't be learning. It's important for you to write the code yourself and get it to work. Feel free to consult the previous notebooks though as you work through this.
#
# First off, let's load the dataset through torchvision.
# +
import torch
from torchvision import datasets, transforms
import helper
# Define a transform to normalize the data
transform = transforms.Compose([transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))])
# Download and load the training data
trainset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=True, transform=transform)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)
# Download and load the test data
testset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=False, transform=transform)
testloader = torch.utils.data.DataLoader(testset, batch_size=64, shuffle=True)
# -
# Here we can see one of the images.
image, label = next(iter(trainloader))
helper.imshow(image[0,:]);
# ## Building the network
#
# Here you should define your network. As with MNIST, each image is 28x28 which is a total of 784 pixels, and there are 10 classes. You should include at least one hidden layer. We suggest you use ReLU activations for the layers and to return the logits or log-softmax from the forward pass. It's up to you how many layers you add and the size of those layers.
from torch import nn
# TODO: Define your network architecture here
model = nn.Sequential(
nn.Linear(784, 256),
nn.ReLU(),
nn.Linear(256, 10),
nn.LogSoftmax(dim=1)
)
# # Train the network
#
# Now you should create your network and train it. First you'll want to define [the criterion](http://pytorch.org/docs/master/nn.html#loss-functions) ( something like `nn.CrossEntropyLoss`) and [the optimizer](http://pytorch.org/docs/master/optim.html) (typically `optim.SGD` or `optim.Adam`).
#
# Then write the training code. Remember the training pass is a fairly straightforward process:
#
# * Make a forward pass through the network to get the logits
# * Use the logits to calculate the loss
# * Perform a backward pass through the network with `loss.backward()` to calculate the gradients
# * Take a step with the optimizer to update the weights
#
# By adjusting the hyperparameters (hidden units, learning rate, etc), you should be able to get the training loss below 0.4.
from torch import optim
# +
# TODO: Create the network, define the criterion and optimizer
criterion = nn.NLLLoss()
optimizer = optim.Adam(model.parameters(), lr=0.001)
# -
# TODO: Train the network here
epochs = 5
for e in range(epochs):
running_loss = 0
for images, labels in trainloader:
# Flatten MNIST images into a 784 long vector
images = images.view(images.shape[0], -1)
# TODO: Training pass
# Clear the gradients, do this because gradients are accumulated
optimizer.zero_grad()
output = model.forward(images)
loss = criterion(output, labels)
loss.backward()
optimizer.step()
running_loss += loss.item()
else:
print(f"Training loss: {running_loss/len(trainloader)}")
model[3]
# +
# %matplotlib inline
# %config InlineBackend.figure_format = 'retina'
import helper
# Test out your network!
dataiter = iter(testloader)
images, labels = dataiter.next()
img = images[0]
# Convert 2D image to 1D vector
img = img.resize_(1, 784)
# TODO: Calculate the class probabilities (softmax) for img
ps = torch.exp(model(img))
# Plot the image and probabilities
helper.view_classify(img.resize_(1, 28, 28), ps, version='Fashion')
| intro-to-pytorch/Part 4 - Fashion-MNIST (Exercises).ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Developing an AI application
#
# Going forward, AI algorithms will be incorporated into more and more everyday applications. For example, you might want to include an image classifier in a smart phone app. To do this, you'd use a deep learning model trained on hundreds of thousands of images as part of the overall application architecture. A large part of software development in the future will be using these types of models as common parts of applications.
#
# In this project, you'll train an image classifier to recognize different species of flowers. You can imagine using something like this in a phone app that tells you the name of the flower your camera is looking at. In practice you'd train this classifier, then export it for use in your application. We'll be using [this dataset](http://www.robots.ox.ac.uk/~vgg/data/flowers/102/index.html) of 102 flower categories, you can see a few examples below.
#
# <img src='assets/Flowers.png' width=500px>
#
# The project is broken down into multiple steps:
#
# * Load and preprocess the image dataset
# * Train the image classifier on your dataset
# * Use the trained classifier to predict image content
#
# We'll lead you through each part which you'll implement in Python.
#
# When you've completed this project, you'll have an application that can be trained on any set of labeled images. Here your network will be learning about flowers and end up as a command line application. But, what you do with your new skills depends on your imagination and effort in building a dataset. For example, imagine an app where you take a picture of a car, it tells you what the make and model is, then looks up information about it. Go build your own dataset and make something new.
#
# First up is importing the packages you'll need. It's good practice to keep all the imports at the beginning of your code. As you work through this notebook and find you need to import a package, make sure to add the import up here.
# +
# Imports here
# %matplotlib inline
# %config InlineBackend.figure_format = 'retina'
import matplotlib.pyplot as plt
from collections import OrderedDict
import torch
from torch import nn
from torch import optim
import torch.nn.functional as F
from torchvision import datasets, transforms, models
from workspace_utils import active_session
from PIL import Image
import numpy as np
import json
# -
# ## Load the data
#
# Here you'll use `torchvision` to load the data ([documentation](http://pytorch.org/docs/0.3.0/torchvision/index.html)). The data should be included alongside this notebook, otherwise you can [download it here](https://s3.amazonaws.com/content.udacity-data.com/nd089/flower_data.tar.gz). The dataset is split into three parts, training, validation, and testing. For the training, you'll want to apply transformations such as random scaling, cropping, and flipping. This will help the network generalize leading to better performance. You'll also need to make sure the input data is resized to 224x224 pixels as required by the pre-trained networks.
#
# The validation and testing sets are used to measure the model's performance on data it hasn't seen yet. For this you don't want any scaling or rotation transformations, but you'll need to resize then crop the images to the appropriate size.
#
# The pre-trained networks you'll use were trained on the ImageNet dataset where each color channel was normalized separately. For all three sets you'll need to normalize the means and standard deviations of the images to what the network expects. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`, calculated from the ImageNet images. These values will shift each color channel to be centered at 0 and range from -1 to 1.
#
data_dir = 'flowers'
train_dir = data_dir + '/train'
valid_dir = data_dir + '/valid'
test_dir = data_dir + '/test'
# +
# TODO: Define your transforms for the training, validation, and testing sets
train_transforms = transforms.Compose([transforms.RandomRotation(30),
transforms.RandomResizedCrop(224),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406],
[0.229, 0.224, 0.225])])
valid_transforms = transforms.Compose([transforms.Resize(255),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406],
[0.229, 0.224, 0.225])])
test_transforms = transforms.Compose([transforms.Resize(255),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406],
[0.229, 0.224, 0.225])])
# TODO: Load the datasets with ImageFolder
train_dataset = datasets.ImageFolder(data_dir + '/train', transform=train_transforms)
valid_dataset = datasets.ImageFolder(data_dir + '/valid', transform=valid_transforms)
test_dataset = datasets.ImageFolder(data_dir + '/test', transform=test_transforms)
# TODO: Using the image datasets and the trainforms, define the dataloaders
trainloader = torch.utils.data.DataLoader(train_dataset, batch_size=64, shuffle=True)
validloader = torch.utils.data.DataLoader(valid_dataset, batch_size=64, shuffle=True)
testloader = torch.utils.data.DataLoader(test_dataset, batch_size=64)
# -
dataiter=iter(trainloader)
images,labels=dataiter.next()
print(type(images))
print(images.shape)
# ### Label mapping
#
# You'll also need to load in a mapping from category label to category name. You can find this in the file `cat_to_name.json`. It's a JSON object which you can read in with the [`json` module](https://docs.python.org/2/library/json.html). This will give you a dictionary mapping the integer encoded categories to the actual names of the flowers.
# +
with open('cat_to_name.json', 'r') as f:
cat_to_name = json.load(f)
#print(cat_to_name)
#print(len(cat_to_name))
# -
# # Building and training the classifier
#
# Now that the data is ready, it's time to build and train the classifier. As usual, you should use one of the pretrained models from `torchvision.models` to get the image features. Build and train a new feed-forward classifier using those features.
#
# We're going to leave this part up to you. Refer to [the rubric](https://review.udacity.com/#!/rubrics/1663/view) for guidance on successfully completing this section. Things you'll need to do:
#
# * Load a [pre-trained network](http://pytorch.org/docs/master/torchvision/models.html) (If you need a starting point, the VGG networks work great and are straightforward to use)
# * Define a new, untrained feed-forward network as a classifier, using ReLU activations and dropout
# * Train the classifier layers using backpropagation using the pre-trained network to get the features
# * Track the loss and accuracy on the validation set to determine the best hyperparameters
#
# We've left a cell open for you below, but use as many as you need. Our advice is to break the problem up into smaller parts you can run separately. Check that each part is doing what you expect, then move on to the next. You'll likely find that as you work through each part, you'll need to go back and modify your previous code. This is totally normal!
#
# When training make sure you're updating only the weights of the feed-forward network. You should be able to get the validation accuracy above 70% if you build everything right. Make sure to try different hyperparameters (learning rate, units in the classifier, epochs, etc) to find the best model. Save those hyperparameters to use as default values in the next part of the project.
#
# One last important tip if you're using the workspace to run your code: To avoid having your workspace disconnect during the long-running tasks in this notebook, please read in the earlier page in this lesson called Intro to
# GPU Workspaces about Keeping Your Session Active. You'll want to include code from the workspace_utils.py module.
#
# **Note for Workspace users:** If your network is over 1 GB when saved as a checkpoint, there might be issues with saving backups in your workspace. Typically this happens with wide dense layers after the convolutional layers. If your saved checkpoint is larger than 1 GB (you can open a terminal and check with `ls -lh`), you should reduce the size of your hidden layers and train again.
# +
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model = models.vgg16(pretrained=True)
#print(model)
#classifier =nn.Sequential((OrderedDict([
# ('fc1',nn.Linear(25088,500)),
# ('relu',nn.ReLU()),
# ('fc2',nn.Linear(1024,500)),
# ('output',nn.LogSoftmax(dim=1))
#])))
#model2= models.resnet50(pretrained=True)
#print(model2)
# +
#freezing the feature parameters
for param in model.parameters():
param.requires_grad=False
model.classifier=nn.Sequential(nn.Linear(25088,6272),
nn.ReLU(),
nn.Dropout(p=0.2),
nn.Linear(6272,1568),
nn.ReLU(),
nn.Dropout(p=0.2),
nn.Linear(1568,102),
nn.LogSoftmax(dim=1))
# -
criterion = nn.NLLLoss()
optimizer = optim.Adam(model.classifier.parameters(), lr=0.001)
model.to(device)
# ## Testing your network
#
# It's good practice to test your trained network on test data, images the network has never seen either in training or validation. This will give you a good estimate for the model's performance on completely new images. Run the test images through the network and measure the accuracy, the same way you did validation. You should be able to reach around 70% accuracy on the test set if the model has been trained well.
epochs= 7
steps=0
running_loss=0
print_every=60
# +
# TODO: Training and Validation of the model
with active_session():
for epoch in range(epochs):
for i, (images, labels) in enumerate (trainloader):
#print('training')
steps+=1
images,labels= images.to(device),labels.to(device)
optimizer.zero_grad()
#forward pass
logits=model.forward(images)
loss=criterion(logits,labels)
#backpropagation
loss.backward()
#updating weights
optimizer.step()
running_loss+=loss.item()
if steps % print_every == 0 :
model.eval()
valid_loss=0
accuracy=0
#turning backprop off
with torch.no_grad():
for images,labels in validloader:
#print('validation')
images,labels= images.to(device),labels.to(device)
#forward pass
logps=model.forward(images)
batch_loss=criterion(logps,labels)
valid_loss+=batch_loss.item()
#Calculate the accuracy
ps=torch.exp(logps)
top_ps,top_class= ps.topk(1,dim=1)
equality= top_class ==labels.view(*top_class.shape)
accuracy+=torch.mean(equality.type(torch.FloatTensor)).item()
print(f"Epoch: {epoch+1}/{epochs}.."
#f"Training_loss: {running_loss/(i*len(trainloader)):.3f}.."
f"Training_loss: {running_loss/(print_every):.3f}.."
f"Validation loss: {valid_loss/len(validloader):.3f}.."
f"Validation Accuracy: {accuracy/len(validloader):.3f}..")
running_loss=0
model.train()
# -
# TODO: Do validation on the test set
model.eval()
test_loss=0
test_accuracy=0
with torch.no_grad():
for images,labels in testloader:
#print('test')
images,labels= images.to(device),labels.to(device)
#forward pass
logps=model.forward(images)
batch_loss=criterion(logps,labels)
test_loss+=batch_loss.item()
#Calculate the accuracy
ps=torch.exp(logps)
top_ps,top_class= ps.topk(1,dim=1)
equality= top_class ==labels.view(*top_class.shape)
test_accuracy+=torch.mean(equality.type(torch.FloatTensor))
print("Test loss: {:.3f} Test Accuracy:{:.3f}".format(test_loss/len(testloader),test_accuracy/len(testloader)))
# ## Save the checkpoint
#
# Now that your network is trained, save the model so you can load it later for making predictions. You probably want to save other things such as the mapping of classes to indices which you get from one of the image datasets: `image_datasets['train'].class_to_idx`. You can attach this to the model as an attribute which makes inference easier later on.
#
# ```model.class_to_idx = image_datasets['train'].class_to_idx```
#
# Remember that you'll want to completely rebuild the model later so you can use it for inference. Make sure to include any information you need in the checkpoint. If you want to load the model and keep training, you'll want to save the number of epochs as well as the optimizer state, `optimizer.state_dict`. You'll likely want to use this trained model in the next part of the project, so best to save it now.
# TODO: Save the checkpoint
print('Model: ',model)
print('State Dict Keys',model.state_dict().keys())
model.class_to_idx = train_dataset.class_to_idx
checkpoint = {
'input size': 25088,
'output size': 102,
'state_dict': model.state_dict(),
'epochs': epochs,
'classifier': model.classifier,
'optimizer': optimizer.state_dict(),
'class_to_idx': model.class_to_idx
}
torch.save(checkpoint,'trained_model.pth')
# ## Loading the checkpoint
#
# At this point it's good to write a function that can load a checkpoint and rebuild the model. That way you can come back to this project and keep working on it without having to retrain the network.
# +
# TODO: Write a function that loads a checkpoint and rebuilds the model
def load_checkpoint(filepath):
'''
Description :loades the saved model from the checkpoint.
params : filepath - checkpoint file
returns : trained model
'''
checkpoint = torch.load('trained_model.pth')
model = models.vgg16(pretrained=True)
epochs = checkpoint['epochs']
model.classifier = checkpoint['classifier']
model.load_state_dict(checkpoint['state_dict'])
model.class_to_idx = checkpoint['class_to_idx']
optimizer = checkpoint['optimizer']
for param in model.parameters():
param.requires_grad = False
model.to(device)
return model
model=load_checkpoint('trained_model.pth')
print(model)
# -
# # Inference for classification
#
# Now you'll write a function to use a trained network for inference. That is, you'll pass an image into the network and predict the class of the flower in the image. Write a function called `predict` that takes an image and a model, then returns the top $K$ most likely classes along with the probabilities. It should look like
#
# ```python
# probs, classes = predict(image_path, model)
# print(probs)
# print(classes)
# > [ 0.01558163 0.01541934 0.01452626 0.01443549 0.01407339]
# > ['70', '3', '45', '62', '55']
# ```
#
# First you'll need to handle processing the input image such that it can be used in your network.
#
# ## Image Preprocessing
#
# You'll want to use `PIL` to load the image ([documentation](https://pillow.readthedocs.io/en/latest/reference/Image.html)). It's best to write a function that preprocesses the image so it can be used as input for the model. This function should process the images in the same manner used for training.
#
# First, resize the images where the shortest side is 256 pixels, keeping the aspect ratio. This can be done with the [`thumbnail`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) or [`resize`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) methods. Then you'll need to crop out the center 224x224 portion of the image.
#
# Color channels of images are typically encoded as integers 0-255, but the model expected floats 0-1. You'll need to convert the values. It's easiest with a Numpy array, which you can get from a PIL image like so `np_image = np.array(pil_image)`.
#
# As before, the network expects the images to be normalized in a specific way. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`. You'll want to subtract the means from each color channel, then divide by the standard deviation.
#
# And finally, PyTorch expects the color channel to be the first dimension but it's the third dimension in the PIL image and Numpy array. You can reorder dimensions using [`ndarray.transpose`](https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.ndarray.transpose.html). The color channel needs to be first and retain the order of the other two dimensions.
def process_image(image):
'''
Description :Scales, crops, and normalizes a PIL image for a PyTorch model,
returns an Numpy array
params : image - Image file along with its absolute path
returns : Numpy image
'''
pil_image = Image.open(image)
# TODO: Process a PIL image for use in a PyTorch model
# Citation Udacity Mentor Survesh answer in Ask a Mentor
width,height=pil_image.size
if (width > height):
asp_ratio=width/height
pil_image = pil_image.resize((int(asp_ratio*256),256))
elif (height > width):
asp_ratio=height/width
pil_image = pil_image.resize((256,int(asp_ratio*256)))
new_height=224
new_width=224
width,height=pil_image.size
left =(width-new_width)/2
right = (width+new_width)/2
top=(height-new_height)/2
bottom=(height+new_height)/2
pil_image=pil_image.crop((left,top,right,bottom))
#print(pil_image.size)
np_image=np.array(pil_image)
np_image=np_image/255
mean = np.array([0.485, 0.456, 0.406])
std = np.array([0.229, 0.224, 0.225])
np_image=(np_image-mean)/std
#print ("Image:",np_image)
np_image=np_image.transpose((2,0,1))
#print ("Transposed image:",np_image)
return np_image
# To check your work, the function below converts a PyTorch tensor and displays it in the notebook. If your `process_image` function works, running the output through this function should return the original image (except for the cropped out portions).
# +
def imshow(image, ax=None, title=None):
'''
Description :Scales, crops, and normalizes a PIL image for a PyTorch model,
returns an Numpy array
params : Imshow for Tensor.
returns : figure handle to the image
'''
if ax is None:
fig, ax = plt.subplots()
# PyTorch tensors assume the color channel is the first dimension
# but matplotlib assumes is the third dimension
image = image.numpy().transpose((1, 2, 0))
# Undo preprocessing
mean = np.array([0.485, 0.456, 0.406])
std = np.array([0.229, 0.224, 0.225])
image = std * image + mean
# Image needs to be clipped between 0 and 1 or it looks like noise when displayed
image = np.clip(image, 0, 1)
ax.imshow(image)
return ax
filepath = '/home/workspace/aipnd-project/'
image_path = filepath +'flowers/test/59/'
#image_path = filepath +'flowers/test/1/'
#image_path = filepath +'flowers/train/14/'
#image=image_path+'image_06760.jpg'
#image=image_path+'image_06734.jpg'
image=image_path+'image_05020.jpg'
ax=imshow(torch.FloatTensor(process_image(image)))
# -
# ## Class Prediction
#
# Once you can get images in the correct format, it's time to write a function for making predictions with your model. A common practice is to predict the top 5 or so (usually called top-$K$) most probable classes. You'll want to calculate the class probabilities then find the $K$ largest values.
#
# To get the top $K$ largest values in a tensor use [`x.topk(k)`](http://pytorch.org/docs/master/torch.html#torch.topk). This method returns both the highest `k` probabilities and the indices of those probabilities corresponding to the classes. You need to convert from these indices to the actual class labels using `class_to_idx` which hopefully you added to the model or from an `ImageFolder` you used to load the data ([see here](#Save-the-checkpoint)). Make sure to invert the dictionary so you get a mapping from index to class as well.
#
# Again, this method should take a path to an image and a model checkpoint, then return the probabilities and classes.
#
# ```python
# probs, classes = predict(image_path, model)
# print(probs)
# print(classes)
# > [ 0.01558163 0.01541934 0.01452626 0.01443549 0.01407339]
# > ['70', '3', '45', '62', '55']
# ```
def predict(image_path, model, topk=5):
'''
Description :Predict the class (or classes) of an image using a trained deep learning model.
params : image_path - Image file along with its absolute path
model - saved model after training
topk - top 'k' probabilities which predicts the image correctly
returns : topk probabilities and their classes
'''
# TODO: Implement the code to predict the class from an image file
#img= Image.open(image_path)
# Citation Udacity Mentor Eban answer in Ask a Mentor
processed_img=torch.from_numpy(process_image(image_path)).type(torch.FloatTensor).unsqueeze_(0)
#imshow(processed_img)
with torch.no_grad():
processed_img=processed_img.to(device)
logps=model.forward(processed_img)
ps = torch.exp(logps)
# Citation Udacity Mentor Shibin M answer in Ask a Mentor
probs, classes = torch.topk(ps,int(topk))
top_p=probs.tolist()[0]
classes=np.array(classes)
index_to_class = {value: key for key, value in model.class_to_idx.items()}
#idx_to_class=dict()
#for key ,value in model.class_to_idx.items():
# idx_to_class.value=key
# idx_to_class.key=value
top_class = [index_to_class[idx] for idx in classes[0]]
return top_p, top_class
top_probs, top_classes = predict(image, model)
print(top_probs)
#print(top_classes)
# # Sanity Checking
#
# Now that you can use a trained model for predictions, check to make sure it makes sense. Even if the testing accuracy is high, it's always good to check that there aren't obvious bugs. Use `matplotlib` to plot the probabilities for the top 5 classes as a bar graph, along with the input image. It should look like this:
#
# <img src='assets/inference_example.png' width=300px>
#
# You can convert from the class integer encoding to actual flower names with the `cat_to_name.json` file (should have been loaded earlier in the notebook). To show a PyTorch tensor as an image, use the `imshow` function defined above.
# +
# TODO: Display an image along with the top 5 classes
flower_names = []
for classes in top_classes:
flower_names.append(cat_to_name[str(classes)])
#print(top_probs)
#print(top_classes)
#print(flower_names)
fig, (ax1,ax2) = plt.subplots(2, 1)
#image = Image.open(image_path)
image=torch.from_numpy(process_image(image))
#image = process_image(image)
#imshow(torch.FloatTensor(image))
imshow(image, ax=ax1)
ax1.set_title(flower_names[0])
top_probs_percent=[p*100 for p in top_probs]
y_pos = np.arange(len(top_classes))
ax2.barh(y_pos,top_probs_percent, align='center')
#ax2.barh(top_classes, top_probs[0], align='center')
ax2.set_yticks(y_pos)
ax2.set_yticklabels(flower_names)
ax2.set_ylabel("Flowers")
ax2.set_xlabel("Probabilities")
plt.show()
plt.subplots_adjust(top=0.7)
# +
# -
| Image Classifier Project.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
# default_exp models.explainability
# -
# # Model explainability
# > Functionality to help with both global and local explainability.
#export
from tsai.imports import *
from tsai.utils import *
from tsai.models.layers import *
from tsai.models.utils import *
# +
from tsai.data.all import *
from tsai.models.XCM import *
dsid = 'NATOPS'
X, y, splits = get_UCR_data(dsid, split_data=False)
tfms = [None, Categorize()]
dls = get_ts_dls(X, y, splits=splits, tfms=tfms)
model = XCM(dls.vars, dls.c, dls.len)
learn = Learner(dls, model, metrics=accuracy)
xb, yb = dls.one_batch()
x = xb[0]
model.eval()(xb).shape
# +
#export
def get_acts_and_grads(model, modules, x, y=None, detach=True, cpu=False):
r"""Returns activations and gradients for given modules in a model and a single input or a batch.
Gradients require y value(s). If they rae not provided, it will use the predicttions. """
if not is_listy(modules): modules = [modules]
x = x[None, None] if x.ndim == 1 else x[None] if x.ndim == 2 else x
with hook_outputs(modules, detach=detach, cpu=cpu) as h_act:
with hook_outputs(modules, grad=True, detach=detach, cpu=cpu) as h_grad:
preds = model.eval()(x)
if y is None: preds.max(dim=-1).values.mean().backward()
else:
if preds.shape[0] == 1: preds[0, y].backward()
else:
if y.ndim == 1: y = y.reshape(-1, 1)
torch_slice_by_dim(preds, y).mean().backward()
if len(modules) == 1: return h_act.stored[0].data, h_grad.stored[0][0].data
else: return [h.data for h in h_act.stored], [h[0].data for h in h_grad.stored]
def get_attibution_map(model, modules, x, y=None, detach=True, cpu=False, apply_relu=True):
def _get_attribution_map(A_k, w_ck):
dim = (0, 2, 3) if A_k.ndim == 4 else (0, 2)
w_ck = w_ck.mean(dim, keepdim=True)
L_c = (w_ck * A_k).sum(1)
if apply_relu: L_c = nn.ReLU()(L_c)
if L_c.ndim == 3: return L_c.squeeze(0) if L_c.shape[0] == 1 else L_c
else: return L_c.repeat(x.shape[1], 1) if L_c.shape[0] == 1 else L_c.unsqueeze(1).repeat(1, x.shape[1], 1)
if x.ndim == 1: x = x[None, None]
elif x.ndim == 2: x = x[None]
A_k, w_ck = get_acts_and_grads(model, modules, x, y, detach=detach, cpu=cpu)
if is_listy(A_k): return [_get_attribution_map(A_k[i], w_ck[i]) for i in range(len(A_k))]
else: return _get_attribution_map(A_k, w_ck)
# -
acts, grads = get_acts_and_grads(model, model.conv2dblock, x)
acts.shape, grads.shape
acts, grads = get_acts_and_grads(model, model.conv2dblock, xb)
acts.shape, grads.shape
acts, grads = get_acts_and_grads(model, model.conv2dblock, xb, yb)
acts.shape, grads.shape
acts, grads = get_acts_and_grads(model, model.conv1dblock, xb)
acts.shape, grads.shape
acts, grads = get_acts_and_grads(model, [model.conv2dblock, model.conv1dblock], xb, yb)
[act.shape for act in acts], [grad.shape for grad in grads]
att_maps = get_attibution_map(model, model.conv2dblock, xb, yb)
att_maps.shape
att_maps = get_attibution_map(model, model.conv1dblock, xb)
att_maps.shape
att_maps = get_attibution_map(model, [model.conv2dblock, model.conv1dblock], xb)
[am.shape for am in att_maps]
acts, grads = get_acts_and_grads(model, [model.conv2dblock, model.conv1dblock], xb[0], yb[0], detach=True, cpu=False)
print(len(acts), len(grads), acts[0].shape, grads[0].shape)
acts, grads = get_acts_and_grads(model, model.conv2dblock, xb[0], y=None, detach=True, cpu=False)
print(acts.shape, grads.shape)
att_maps = get_attibution_map(model, model.conv2dblock, x)
att_maps.shape
att_maps = get_attibution_map(model, model.conv1dblock, x)
att_maps.shape
att_maps = get_attibution_map(model, [model.conv2dblock, model.conv1dblock], x)
[am.shape for am in att_maps]
# +
att_maps = get_attibution_map(model, [model.conv2dblock, model.conv1dblock], x)
att_maps[0] = (att_maps[0] - att_maps[0].min()) / (att_maps[0].max() - att_maps[0].min())
att_maps[1] = (att_maps[1] - att_maps[1].min()) / (att_maps[1].max() - att_maps[1].min())
fig = plt.figure(figsize=(10, 10))
ax = plt.axes()
plt.title('Observed variables')
im = ax.imshow(att_maps[0], cmap='inferno')
cax = fig.add_axes([ax.get_position().x1+0.01,ax.get_position().y0,0.02,ax.get_position().height])
plt.colorbar(im, cax=cax)
plt.show()
fig = plt.figure(figsize=(10, 10))
ax = plt.axes()
plt.title('Time')
im = ax.imshow(att_maps[1], cmap='inferno')
cax = fig.add_axes([ax.get_position().x1+0.01,ax.get_position().y0,0.02,ax.get_position().height])
plt.colorbar(im, cax=cax)
plt.show()
# +
# '''
# Color parts of a line based on its properties, e.g., slope.
# '''
# import numpy as np
# import matplotlib.pyplot as plt
# from matplotlib.collections import LineCollection
# from matplotlib.colors import ListedColormap, BoundaryNorm
# x = np.linspace(0, 3 * np.pi, 500)
# y = np.sin(x)
# z = np.cos(0.5 * (x[:-1] + x[1:])) # first derivative
# # Create a colormap for red, green and blue and a norm to color
# # f' < -0.5 red, f' > 0.5 blue, and the rest green
# cmap = ListedColormap(['r', 'g', 'b'])
# norm = BoundaryNorm([-1, -0.5, 0.5, 1], cmap.N)
# # Create a set of line segments so that we can color them individually
# # This creates the points as a N x 1 x 2 array so that we can stack points
# # together easily to get the segments. The segments array for line collection
# # needs to be numlines x points per line x 2 (x and y)
# points = np.array([x, y]).T.reshape(-1, 1, 2)
# segments = np.concatenate([points[:-1], points[1:]], axis=1)
# # Create the line collection object, setting the colormapping parameters.
# # Have to set the actual values used for colormapping separately.
# lc = LineCollection(segments, cmap=cmap, norm=norm)
# lc.set_array(z)
# lc.set_linewidth(3)
# fig1 = plt.figure()
# plt.gca().add_collection(lc)
# plt.xlim(x.min(), x.max())
# plt.ylim(-1.1, 1.1)
# # Now do a second plot coloring the curve using a continuous colormap
# t = np.linspace(0, 10, 200)
# x = np.cos(np.pi * t)
# y = np.sin(t)
# points = np.array([x, y]).T.reshape(-1, 1, 2)
# segments = np.concatenate([points[:-1], points[1:]], axis=1)
# lc = LineCollection(segments, cmap=plt.get_cmap('copper'), norm=plt.Normalize(0, 10))
# lc.set_array(t)
# lc.set_linewidth(3)
# fig2 = plt.figure()
# plt.gca().add_collection(lc)
# plt.xlim(-1, 1)
# plt.ylim(-1, 1)
# plt.show()
# -
#hide
out = create_scripts(); beep(out)
| nbs/100c_models.explainability.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Variational Autoencoder (VAE)
# Useful links:
# * Original paper http://arxiv.org/abs/1312.6114
# * Helpful videos explaining the topic
# * https://www.youtube.com/watch?v=P78QYjWh5sM
# * http://videolectures.net/deeplearning2015_courville_autoencoder_extension/?q=aaron%20courville
import numpy as np
import matplotlib.pyplot as plt
# %matplotlib inline
# In this seminar we will train an autoencoder to model images of faces. For this we take "Labeled Faces in the Wild" dataset (LFW) (http://vis-www.cs.umass.edu/lfw/), deep funneled version of it. (frontal view of all faces)
from lfw_dataset import fetch_lfw_dataset
data, attrs = fetch_lfw_dataset(dimx=36, dimy=36,
images_name="/mnt/mlhep2018/datasets/lfw-deepfunneled",
attrs_name="/mnt/mlhep2018/datasets/lfw_attributes.txt")
data = data.astype(np.float32) / 255.
def plot_gallery(images, n_row=3, n_col=6):
"""Helper function to plot a gallery of portraits"""
plt.figure(figsize=(1.5 * n_col, 1.7 * n_row))
plt.subplots_adjust(bottom=0, left=.01, right=.99, top=.90, hspace=.35)
for i in range(n_row * n_col):
plt.subplot(n_row, n_col, i + 1)
plt.imshow(images[i], cmap=plt.cm.gray, vmin=-1, vmax=1, interpolation='nearest')
plt.xticks(())
plt.yticks(())
plt.axis('off')
plot_gallery(data)
import tensorflow as tf
def get_tf_dataset(dataset, batch_size):
"""
Produces an infinite stram of Tensorflow batches from a numpy dataset. The dataset is shuffled every epoch.
Args:
dataset: np.array[n_examples, ...]
batch_size: int, batch size of the results
Reuturns:
Tensor, containing the next batch
"""
if isinstance(dataset, tf.Tensor):
N_EXAMPLES = dataset.shape[0]
else:
N_EXAMPLES = dataset[0].shape[0]
shuffler = tf.contrib.data.shuffle_and_repeat(N_EXAMPLES)
dataset_tf = tf.data.Dataset.from_tensor_slices(dataset)
suffled_ds = shuffler(dataset_tf)
return suffled_ds.batch(batch_size).prefetch(1).make_one_shot_iterator().get_next()
import keras
from keras.models import Sequential
from keras import layers as L
# # Variational Autoencoder
# Bayesian approach in deep learning considers everything in terms of distributions. Now our encoder generates not just a vector $z$ but posterior ditribution $q(z|x)$. In our case distribution $q$ is Gaussian distibution $N(\mu, \sigma)$ with parameters $\mu$, $\sigma$. To do that we need to an encoder with two outputs. One dense layer will generate vector $\mu$, and another will generate vector $\sigma$. Reparametrization trick should be implemented using the **gaussian_sampler** layer, that generates random vetor $\epsilon$ and returns $\mu+\sigma\epsilon \sim N(\mu, \sigma)$ .
#
# Here we only ask for implementation of the simplest version of VAE - one $z$ sample per input. You can consider to sample several outputs from one input and average them.
IMG_SHAPE = data.shape[1:]
CODE_SIZE = 128
# +
encoder_activation = tf.nn.softplus
# Define the encoder to have an elegant pyramidal structure
# It first maps images into one pixel with many cahnnels
# Then flattens and uses a dense layer to get the CODE_SIZE outputs
FILTERS_N0 = 32
encoder = Sequential(name="Encoder")
encoder.add(L.InputLayer(IMG_SHAPE))
encoder.add(L.Conv2D(FILTERS_N0, kernel_size=3, activation=encoder_activation))
encoder.add(L.Conv2D(int(1.5*FILTERS_N0), kernel_size=3, activation=encoder_activation))
encoder.add(L.MaxPool2D(pool_size=(2, 2)))
encoder.add(L.Conv2D(2*FILTERS_N0, kernel_size=3, activation=encoder_activation))
encoder.add(L.Conv2D(3*FILTERS_N0, kernel_size=3, activation=encoder_activation))
encoder.add(L.MaxPool2D(pool_size=(2, 2)))
encoder.add(L.Conv2D(4*FILTERS_N0, kernel_size=3, activation=encoder_activation))
encoder.add(L.Conv2D(6*FILTERS_N0, kernel_size=3, activation=encoder_activation))
encoder.add(L.MaxPool2D(pool_size=(2, 2)))
encoder.add(L.Flatten())
# Having created a sequential model, we add two layers on top of its output
# Pay attention to the "Connected to" column in the summary
encoder_mu = L.Dense(CODE_SIZE, name="encoder_mu", activation=keras.activations.linear)(encoder.output)
encoder_log_sigma = L.Dense(CODE_SIZE, name="encoder_log_sigma", activation=keras.activations.linear)(encoder.output)
# Combines the sequential model and two layers into an two-headed model
image_to_mu_sigma_model = keras.Model(
inputs=encoder.input,
outputs=[encoder_mu, encoder_log_sigma])
print(image_to_mu_sigma_model.summary())
# +
# Create a decoder. It's a sequential model accepting a 1D vector of
# length CODE_SIZE inputs and outputting an image
# You may want to use a combination of Deconv2D, UpSampling2D
# An elegant solution is "invert" the encoder, switching
# Conv2D->Deconv2D MaxPool2D->UpSampling2D and reversing the layer order
### Lydias Solution ***
# decoder_activation = tf.nn.softplus
# FILTERS_N0 = 32
# decoder = Sequential(name="Decoder")
# decoder.add(L.InputLayer(IMG_SHAPE))
# decoder.add(L.UpSampling2D(size=(2, 2)))
# decoder.add(L.Deconv2D(FILTERS_N0, kernel_size=3, activation=decoder_activation))
# decoder.add(L.Deconv2D(int(1.5*FILTERS_N0), kernel_size=3, activation=decoder_activation))
# decoder.add(L.UpSampling2D(size=(2, 2)))
# decoder.add(L.Deconv2D(2*FILTERS_N0, kernel_size=3, activation=encoder_activation))
# decoder.add(L.Deconv2D(3*FILTERS_N0, kernel_size=3, activation=encoder_activation))
# decoder.add(L.UpSampling2D(size=(2, 2)))
# decoder.add(L.Deconv2D(4*FILTERS_N0, kernel_size=3, activation=encoder_activation))
# decoder.add(L.Deconv2D(6*FILTERS_N0, kernel_size=3, activation=encoder_activation))
# decoder.add(L.Flatten())
#Nikita's Solution
# stupid decoder, rinse with deconv before using
# decoder = keras.models.Sequential(name="Decoder")
# decoder.add(L.InputLayer([CODE_SIZE]))
# decoder.add(L.Reshape((1, 1, CODE_SIZE)))
# decoder.add(L.UpSampling2D(size=(2, 2)))
# decoder.add(L.Deconv2D(6*FILTERS_N0, kernel_size=3, activation=encoder_activation))
# decoder.add(L.Deconv2D(3, kernel_size=3, activation=encoder_activation))
# decoder.add(L.UpSampling2D(size=(4, 4)))
# decoder.add(L.Deconv2D(3, kernel_size=5, activation=encoder_activation))
#Together!
decoder_activation = tf.nn.softplus
FILTERS_N0 = 3
decoder = keras.models.Sequential(name="Decoder")
decoder.add(L.InputLayer([CODE_SIZE]))
decoder.add(L.Reshape((1, 1, CODE_SIZE)))
decoder.add(L.UpSampling2D(size=(2, 2)))
decoder.add(L.Deconv2D(FILTERS_N0, kernel_size=3, activation=encoder_activation))
decoder.add(L.Deconv2D(int(1.5*FILTERS_N0), kernel_size=3, activation=encoder_activation))
decoder.add(L.UpSampling2D(size=(2, 2)))
decoder.add(L.Deconv2D(2*FILTERS_N0, kernel_size=3, activation=encoder_activation))
decoder.add(L.Deconv2D(3*FILTERS_N0, kernel_size=3, activation=encoder_activation))
decoder.add(L.UpSampling2D(size=(2, 2)))
decoder.add(L.Deconv2D(4*FILTERS_N0, kernel_size=3, activation=encoder_activation))
decoder.add(L.Deconv2D(6*FILTERS_N0, kernel_size=3, activation=encoder_activation))
decoder.summary()
# -
TRAIN_BATCH_SIZE = 128
train_data_tf, train_attrs_tf = get_tf_dataset(
(data.astype(np.float32), attrs.values.astype(np.float32)), TRAIN_BATCH_SIZE)
train_data_tf
train_attrs_tf
#make faces!
config_tf = tf.ConfigProto(gpu_options=tf.GPUOptions(allow_growth=True))
sess = tf.Session(config=config_tf)
plt.imshow(sess.run(train_data_tf)[0])
plt.axis('off')
# And the last, but not least! Place in the code where the most of the formulaes goes to - optimization objective. The objective for VAE has it's own name - variational lowerbound. And as for any lowerbound our intention is to maximize it. Here it is (for one sample $z$ per input $x$):
#
# $$\mathcal{L} = -D_{KL}(q_{\phi}(z|x)||p_{\theta}(z)) + \log p_{\theta}(x|z)$$
#
# Your next task is to implement two functions that compute KL-divergence and the second term - log-likelihood of an output. Here is some necessary math for your convenience:
#
# $$D_{KL} = -\frac{1}{2}\sum_{i=1}^{dimZ}(1+log(\sigma_i^2)-\mu_i^2-\sigma_i^2)$$
# $$\log p_{\theta}(x|z) = \sum_{i=1}^{dimX}\log p_{\theta}(x_i|z)=C + \sum_{i=1}^{dimX} (x_i - f_i)^2 / h^2$$
#
# Don't forget in the code that you are using $\log\sigma$ as variable. Explain, why not $\sigma$?
# +
# The Variational part of VAE. For training image, get mean and log(std)
train_mu, train_log_sigma = image_to_mu_sigma_model(train_data_tf)
#< Compute std aka sigma >
train_sigma = tf.exp(train_log_sigma)
#< Sample from noraml distribution (tf.random.nomral)>
scale = tf.random_normal([TRAIN_BATCH_SIZE, CODE_SIZE])
#< Scale and shift the sampled values >
modelled_noise = train_sigma*scale
# -
decoded_train = decoder(modelled_noise)
# We average both over examples and dimensions to make the losses the same order of magnitude.
# It would have been closer to equations to sum over dimensions and average over examples
# The apporoaches are equal up to a constant, which gets adjusted in the formula for the total loss
mse_loss = tf.losses.mean_squared_error(dataset_tf,decoded_train)
kl_loss = kl_loss = tf.reduce_mean(-0.5*(1 + 2*train_log_sigma - train_mu**2 - tf.exp(train_log_sigma)**2))
# Coefficients here are, unfortunately, heuristics
# If your model outputs distorted faces, you may want to increase KL_LOSS_COEFF
# If your model output lacks diversity, you may want to decrease KL_LOSS_COEFF
#KL_LOSS_COEFF = 1e-3
#total_loss = mse_loss + KL_LOSS_COEFF*kl_loss
generation_batch_size = tf.placeholder_with_default(3, [], name="generation_batch_size")
generated_image = decoder(tf.random_normal([generation_batch_size, CODE_SIZE]))
# +
iteration_tf = tf.Variable(0)
optimization_op = tf.train.AdamOptimizer(1e-3).minimize(
total_loss,var_list=image_to_mu_sigma_model.trainable_weights +
decoder.trainable_weights,
global_step=iteration_tf)
learning_summary = tf.summary.merge([
tf.summary.scalar("kl_loss", kl_loss),
tf.summary.scalar("mse_loss", mse_loss),
tf.summary.scalar("total_loss", total_loss),
tf.summary.scalar("attrs_loss", attrs_loss),
tf.summary.image("generated_image", generated_image)
])
# +
config_tf = tf.ConfigProto(gpu_options=tf.GPUOptions(allow_growth=True))
sess = tf.Session(config=config_tf)
sess.run(tf.global_variables_initializer())
from tqdm import tnrange
import os
LOGDIR = os.path.join("/mnt/students-home", os.environ["USER"], "tensorflow-logs")
MODEL_NAME = "VAE_v1"
TOTAL_ITERATIONS = int(1e4)
train_writer = tf.summary.FileWriter(os.path.join(LOGDIR, MODEL_NAME, "train"))
train_writer.add_graph(tf.get_default_graph())
for iteration in tnrange(TOTAL_ITERATIONS):
summary, _ = sess.run([learning_summary, optimization_op])
train_writer.add_summary(summary, iteration)
# -
# # Learning the true happiness
#
# If you managed to tune your VAE to converge and learn something about the world, now it's time to move to more important matters. As you may have noticed, there are face attributes in dataset. We're interesting in "Smiling" column, but feel free to try others as well! Here is the first task:
#
# 1) Extract the "Smilling" attribute and create a two sets of images: smiling faces and non-smiling ones.
#
# 2) Compute latent representations for each image in "smiling" set and average those latent vectors. Do the same for "non-smiling" set. You have found **"vector representation"** of the "smile" and "no smile" attribute.
#
# 3) Compute the difference: "smile" vector minus "non-smile" vector.
#
# 3) Now check if **"feature arithmetics"** works. Sample a face without smile, encode it and add the diff from p. 3.
#
# ## Tips and tricks
#
# 1) The ability of VAE to faithfully sample the faces distribution is adjusted via the coefficient before kl_loss in the total_loss formula. The higher value prioritizes the quality of sampled images and lower sample diversity and maintaining the relationship between the image and its encoding
#
# 2) When computing the representations of the sad and happy people, you may want to discard the sampling in the middle of VAE, only work with means and pass the means to decoder.
#
# 3) Since we want the latent representation to have a linear dependency with the features, you may want to add a linear model that predicts the attributes from the latent code and add its loss to the total loss with a small coefficient
# We want to interact with our model, and will use this placeholder to pass
# images to decoder
input_images = tf.placeholder(tf.float32, (None, ) + IMG_SHAPE)
# <img src="linear.png" alt="linear">
| day3-Thu/Learning the meaning of happyness with VAE.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # _*Pricing Bull Spreads*_
# ### Introduction
# <br>
# Suppose a <a href="http://www.theoptionsguide.com/bull-call-spread.aspx">bull spread</a> with strike prices $K_1 < K_2$ and an underlying asset whose spot price at maturity $S_T$ follows a given random distribution.
# The corresponding payoff function is defined as:
#
#
# $$\min\{\max\{S_T - K_1, 0\}, K_2 - K_1\}$$
#
#
#
# In the following, a quantum algorithm based on amplitude estimation is used to estimate the expected payoff, i.e., the fair price before discounting, for the option:
#
#
# $$\mathbb{E}\left[ \min\{\max\{S_T - K_1, 0\}, K_2 - K_1\} \right]$$
#
#
# as well as the corresponding $\Delta$, i.e., the derivative of the option price with respect to the spot price, defined as:
#
#
# $$
# \Delta = \mathbb{P}\left[K_1 \leq S \leq K_2\right]
# $$
#
#
# The approximation of the objective function and a general introduction to option pricing and risk analysis on quantum computers are given in the following papers:
#
# - <a href="https://arxiv.org/abs/1806.06893">Quantum Risk Analysis. Woerner, Egger. 2018.</a>
# - <a href="https://arxiv.org/abs/1905.02666">Option Pricing using Quantum Computers. Stamatopoulos et al. 2019.</a>
# +
import matplotlib.pyplot as plt
# %matplotlib inline
import numpy as np
from qiskit import Aer
from qiskit.aqua.algorithms import IterativeAmplitudeEstimation
from qiskit.circuit.library import LogNormalDistribution, LinearAmplitudeFunction
# -
# ### Uncertainty Model
#
# We construct a circuit factory to load a log-normal random distribution into a quantum state.
# The distribution is truncated to a given interval $[\text{low}, \text{high}]$ and discretized using $2^n$ grid points, where $n$ denotes the number of qubits used.
# The unitary operator corresponding to the circuit factory implements the following:
#
# $$\big|0\rangle_{n} \mapsto \big|\psi\rangle_{n} = \sum_{i=0}^{2^n-1} \sqrt{p_i}\big|i\rangle_{n},$$
#
# where $p_i$ denote the probabilities corresponding to the truncated and discretized distribution and where $i$ is mapped to the right interval using the affine map:
#
# $$ \{0, \ldots, 2^n-1\} \ni i \mapsto \frac{\text{high} - \text{low}}{2^n - 1} * i + \text{low} \in [\text{low}, \text{high}].$$
# +
# number of qubits to represent the uncertainty
num_uncertainty_qubits = 3
# parameters for considered random distribution
S = 2.0 # initial spot price
vol = 0.4 # volatility of 40%
r = 0.05 # annual interest rate of 4%
T = 40 / 365 # 40 days to maturity
# resulting parameters for log-normal distribution
mu = ((r - 0.5 * vol**2) * T + np.log(S))
sigma = vol * np.sqrt(T)
mean = np.exp(mu + sigma**2/2)
variance = (np.exp(sigma**2) - 1) * np.exp(2*mu + sigma**2)
stddev = np.sqrt(variance)
# lowest and highest value considered for the spot price; in between, an equidistant discretization is considered.
low = np.maximum(0, mean - 3*stddev)
high = mean + 3*stddev
# construct circuit factory for uncertainty model
uncertainty_model = LogNormalDistribution(num_uncertainty_qubits, mu=mu, sigma=sigma**2, bounds=(low, high))
# -
# plot probability distribution
x = uncertainty_model.values
y = uncertainty_model.probabilities
plt.bar(x, y, width=0.2)
plt.xticks(x, size=15, rotation=90)
plt.yticks(size=15)
plt.grid()
plt.xlabel('Spot Price at Maturity $S_T$ (\$)', size=15)
plt.ylabel('Probability ($\%$)', size=15)
plt.show()
# ### Payoff Function
#
# The payoff function equals zero as long as the spot price at maturity $S_T$ is less than the strike price $K_1$, then increases linearly, and is bounded by $K_2$.
# The implementation uses two comparators, that flip an ancilla qubit each from $\big|0\rangle$ to $\big|1\rangle$ if $S_T \geq K_1$ and $S_T \leq K_2$, and these ancillas are used to control the linear part of the payoff function.
#
# The linear part itself is then approximated as follows.
# We exploit the fact that $\sin^2(y + \pi/4) \approx y + 1/2$ for small $|y|$.
# Thus, for a given approximation rescaling factor $c_\text{approx} \in [0, 1]$ and $x \in [0, 1]$ we consider
#
# $$ \sin^2( \pi/2 * c_\text{approx} * ( x - 1/2 ) + \pi/4) \approx \pi/2 * c_\text{approx} * ( x - 1/2 ) + 1/2 $$
#
# for small $c_\text{approx}$.
#
# We can easily construct an operator that acts as
#
# $$\big|x\rangle \big|0\rangle \mapsto \big|x\rangle \left( \cos(a*x+b) \big|0\rangle + \sin(a*x+b) \big|1\rangle \right),$$
#
# using controlled Y-rotations.
#
# Eventually, we are interested in the probability of measuring $\big|1\rangle$ in the last qubit, which corresponds to
# $\sin^2(a*x+b)$.
# Together with the approximation above, this allows to approximate the values of interest.
# The smaller we choose $c_\text{approx}$, the better the approximation.
# However, since we are then estimating a property scaled by $c_\text{approx}$, the number of evaluation qubits $m$ needs to be adjusted accordingly.
#
# For more details on the approximation, we refer to:
# <a href="https://arxiv.org/abs/1806.06893">Quantum Risk Analysis. <NAME>. 2018.</a>
# +
# set the strike price (should be within the low and the high value of the uncertainty)
strike_price_1 = 1.438
strike_price_2 = 2.584
# set the approximation scaling for the payoff function
rescaling_factor = 0.25
# setup piecewise linear objective fcuntion
breakpoints = [low, strike_price_1, strike_price_2]
slopes = [0, 1, 0]
offsets = [0, 0, strike_price_2 - strike_price_1]
f_min = 0
f_max = strike_price_2 - strike_price_1
bull_spread_objective = LinearAmplitudeFunction(
num_uncertainty_qubits,
slopes,
offsets,
domain=(low, high),
image=(f_min, f_max),
breakpoints=breakpoints,
rescaling_factor=rescaling_factor
)
# construct A operator for QAE for the payoff function by
# composing the uncertainty model and the objective
bull_spread = bull_spread_objective.compose(uncertainty_model, front=True)
# + tags=["nbsphinx-thumbnail"]
# plot exact payoff function (evaluated on the grid of the uncertainty model)
x = uncertainty_model.values
y = np.minimum(np.maximum(0, x - strike_price_1), strike_price_2 - strike_price_1)
plt.plot(x, y, 'ro-')
plt.grid()
plt.title('Payoff Function', size=15)
plt.xlabel('Spot Price', size=15)
plt.ylabel('Payoff', size=15)
plt.xticks(x, size=15, rotation=90)
plt.yticks(size=15)
plt.show()
# -
# evaluate exact expected value (normalized to the [0, 1] interval)
exact_value = np.dot(uncertainty_model.probabilities, y)
exact_delta = sum(uncertainty_model.probabilities[np.logical_and(x >= strike_price_1, x <= strike_price_2)])
print('exact expected value:\t%.4f' % exact_value)
print('exact delta value: \t%.4f' % exact_delta)
# ### Evaluate Expected Payoff
# +
# set target precision and confidence level
epsilon = 0.01
alpha = 0.05
# construct amplitude estimation
ae = IterativeAmplitudeEstimation(epsilon=epsilon, alpha=alpha,
state_preparation=bull_spread,
objective_qubits=[num_uncertainty_qubits],
post_processing=bull_spread_objective.post_processing)
# -
result = ae.run(quantum_instance=Aer.get_backend('qasm_simulator'), shots=100)
conf_int = np.array(result['confidence_interval'])
print('Exact value: \t%.4f' % exact_value)
print('Estimated value:\t%.4f' % result['estimation'])
print('Confidence interval: \t[%.4f, %.4f]' % tuple(conf_int))
# ### Evaluate Delta
#
# The Delta is a bit simpler to evaluate than the expected payoff.
# Similarly to the expected payoff, we use comparator circuits and ancilla qubits to identify the cases where $K_1 \leq S_T \leq K_2$.
# However, since we are only interested in the probability of this condition being true, we can directly use an ancilla qubit as the objective qubit in amplitude estimation without any further approximation.
# +
# setup piecewise linear objective fcuntion
breakpoints = [low, strike_price_1, strike_price_2]
slopes = [0, 0, 0]
offsets = [0, 1, 0]
f_min = 0
f_max = 1
bull_spread_delta_objective = LinearAmplitudeFunction(
num_uncertainty_qubits,
slopes,
offsets,
domain=(low, high),
image=(f_min, f_max),
breakpoints=breakpoints,
) # no approximation necessary, hence no rescaling factor
# construct the A operator by stacking the uncertainty model and payoff function together
bull_spread_delta = bull_spread_delta_objective.compose(uncertainty_model, front=True)
# +
# set target precision and confidence level
epsilon = 0.01
alpha = 0.05
# construct amplitude estimation
ae_delta = IterativeAmplitudeEstimation(epsilon=epsilon, alpha=alpha,
state_preparation=bull_spread_delta,
objective_qubits=[num_uncertainty_qubits])
# -
result_delta = ae_delta.run(quantum_instance=Aer.get_backend('qasm_simulator'), shots=100)
conf_int = np.array(result_delta['confidence_interval'])
print('Exact delta: \t%.4f' % exact_delta)
print('Estimated value:\t%.4f' % result_delta['estimation'])
print('Confidence interval: \t[%.4f, %.4f]' % tuple(conf_int))
import qiskit.tools.jupyter
# %qiskit_version_table
# %qiskit_copyright
| tutorials/finance/05_bull_spread_pricing.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # [1143-Medium] Longest Common Subsequence
#
# Given two strings text1 and text2, return the length of their longest common subsequence.
#
# A subsequence of a string is a new string generated from the original string with some characters(can be none) deleted without changing the relative order of the remaining characters. (eg, "ace" is a subsequence of "abcde" while "aec" is not). A common subsequence of two strings is a subsequence that is common to both strings.
#
#
#
# If there is no common subsequence, return 0.
#
#
# Example 1:
# ```
# Input: text1 = "abcde", text2 = "ace"
# Output: 3
# Explanation: The longest common subsequence is "ace" and its length is 3.
# ```
# Example 2:
# ```
# Input: text1 = "abc", text2 = "abc"
# Output: 3
# Explanation: The longest common subsequence is "abc" and its length is 3.
# ```
# Example 3:
# ```
# Input: text1 = "abc", text2 = "def"
# Output: 0
# Explanation: There is no such common subsequence, so the result is 0.
# ```
# # Edit Distance
#
# Given two strings word1 and word2, return the minimum number of operations required to convert word1 to word2.
#
# You have the following three operations permitted on a word:
#
# 1. Insert a character
# 2. Delete a character
# 3. Replace a character
#
#
# **Example 1:**
# ```
# Input: word1 = "horse", word2 = "ros"
# Output: 3
# Explanation:
# horse -> rorse (replace 'h' with 'r')
# rorse -> rose (remove 'r')
# rose -> ros (remove 'e')
# ```
# **Example 2:**
# ```
# Input: word1 = "intention", word2 = "execution"
# Output: 5
# Explanation:
# intention -> inention (remove 't')
# inention -> enention (replace 'i' with 'e')
# enention -> exention (replace 'n' with 'x')
# exention -> exection (replace 'n' with 'c')
# exection -> execution (insert 'u')
# ```
#
# 为什么说这个问题难呢,因为显而易见,它就是难,让人手足无措,望而生畏。
#
# 为什么说它实用呢,因为前几天我就在日常生活中用到了这个算法。之前有一篇公众号文章由于疏忽,写错位了一段内容,我决定修改这部分内容让逻辑通顺。但是公众号文章最多只能修改 20 个字,且只支持增、删、替换操作(跟编辑距离问题一模一样),于是我就用算法求出了一个最优方案,只用了 16 步就完成了修改。
#
# 再比如高大上一点的应用,DNA 序列是由 A,G,C,T 组成的序列,可以类比成字符串。编辑距离可以衡量两个 DNA 序列的相似度,编辑距离越小,说明这两段 DNA 越相似,说不定这俩 DNA 的主人是远古近亲啥的。
| algorithm/Dynamic_Programming/DP_Sequence.ipynb |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.