Datasets:
Tasks:
Image Segmentation
Modalities:
Image
Sub-tasks:
semantic-segmentation
Languages:
English
Size:
10K - 100K
License:
metadata
annotations_creators:
- other
language:
- en
language_creators:
- found
- expert-generated
license:
- mit
multilinguality:
- monolingual
pretty_name: RSNA-ATD2023
size_categories:
- 10K<n<100K
source_datasets:
- extended|other
tags: []
task_categories:
- image-segmentation
task_ids:
- semantic-segmentation
📁 Dataset
This dataset only comprised of 205 series of CT scans in .png file with raw images and raw mask.
Data source: Kaggle RSNA 2023 Abdominal Trauma Detection
🚀 Setup
pip install datasets
🤩 Feel the Magic
Load Dataset
from datasets import load_dataset
data = load_dataset('ziq/RSNA-ATD2023')
print(data)
DatasetDict({
train: Dataset({
features: ['patient_id', 'series_id', 'frame_id', 'image', 'mask'],
num_rows: 70291
})
})
Set Labels
labels = ["background", "liver", "spleen", "right_kidney", "left_kidney", "bowel"]
Train Test Split
data = data['train'].train_test_split(test_size=0.2)
train, test = data['train'], data['test']
# train[0]['patient_id']
# train[0]['image'] -> PIL Image
# train[0]['mask'] -> PIL Image
Get Segmentation Mask
ids = 3
image, mask = train[ids]['image'], train[ids]['mask']
mask = np.array(mask)
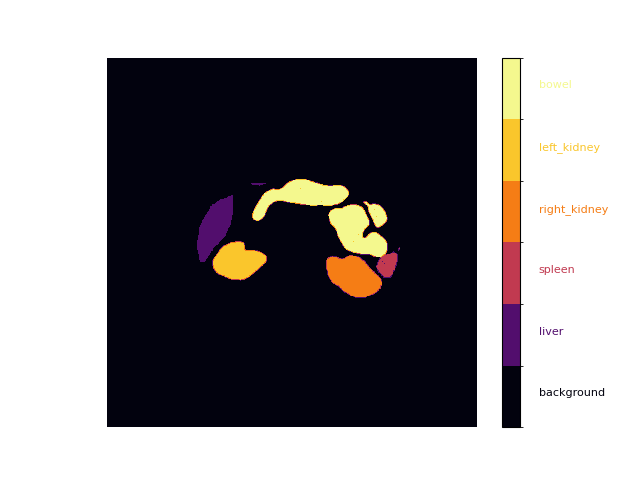
Write Plotting Function
from matplotlib.colors import ListedColormap, BoundaryNorm
colors = ['#02020e', '#520e6d', '#c13a50', '#f57d15', '#fac62c', '#f4f88e'] # inferno
bounds = range(0, len(colors) + 1)
# Define the boundaries for each class in the colormap
cmap, norm = ListedColormap(colors), BoundaryNorm(bounds, len(colors))
# Plot the segmentation mask with the custom colormap
def plot_mask(mask, alpha=1.0):
_, ax = plt.subplots()
cax = ax.imshow(mask, cmap=cmap, norm=norm, alpha=alpha)
cbar = plt.colorbar(cax, cmap=cmap, norm=norm, boundaries=bounds, ticks=bounds)
cbar.set_ticks([])
_labels = [""] + labels
for i in range(1, len(_labels)):
cbar.ax.text(2, -0.5 + i, _labels[i], ha='left', color=colors[i - 1], fontsize=8)
plt.axis('off')
plt.show()
def plot(image, mask, cmap='gray'):
plt.imshow(image * np.where(mask > 0,1,0), cmap=cmap)
plt.axis('off')
plt.show()
Plot
fig = plt.figure(figsize=(16,16))
ax1 = fig.add_subplot(131)
plt.axis('off')
ax1.imshow(image, cmap='gray')
ax2 = fig.add_subplot(132)
plt.axis('off')
ax2.imshow(mask, cmap='gray')
ax3 = fig.add_subplot(133)
ax3.imshow(image*np.where(mask>0,1,0), cmap='gray')
plt.axis('off')
plt.show()
plot(image, mask)
Custom Color
plot_mask(mask)
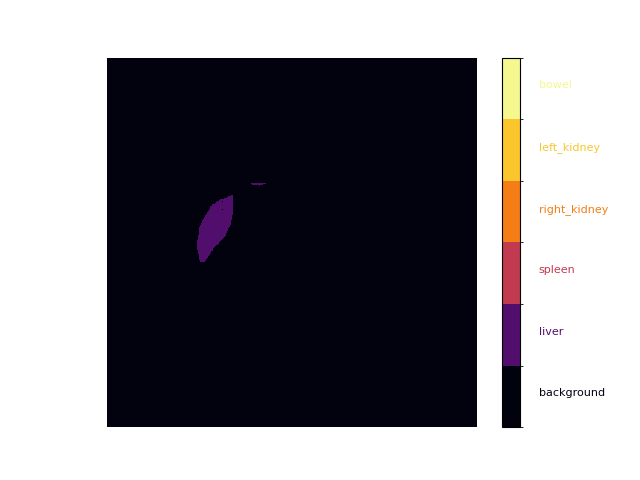
Plot only one class (e.g. liver)
liver, spleen, right_kidney, left_kidney, bowel = [(mask == i,1,0)[0] * i for i in range(1, len(labels))]
plot_mask(liver)