|
--- |
|
tags: |
|
- monai |
|
- medical |
|
library_name: monai |
|
license: apache-2.0 |
|
--- |
|
# Model Overview |
|
|
|
A pre-trained model for simultaneous segmentation and classification of nuclei within multi-tissue histology images based on CoNSeP data. The details of the model can be found in [1]. |
|
|
|
## Workflow |
|
|
|
The model is trained to simultaneous segment and classify nuclei. Training is done via a two-stage approach. First initialized the model with pre-trained weights on the [ImageNet dataset](https://ieeexplore.ieee.org/document/5206848), trained only the decoders for the first 50 epochs, and then fine-tuned all layers for another 50 epochs. There are two training modes in total. If "original" mode is specified, it uses [270, 270] and [80, 80] for `patch_size` and `out_size` respectively. If "fast" mode is specified, it uses [256, 256] and [164, 164] for `patch_size` and `out_size` respectively. The results we show below are based on the "fast" model. |
|
|
|
- We train the first stage with pre-trained weights from some internal data. |
|
|
|
- The original author's repo also has pre-trained weights which is for non-commercial use. Each user is responsible for checking the content of models/datasets and the applicable licenses and determining if suitable for the intended use. The license for the pre-trained model is different than MONAI license. Please check the source where these weights are obtained from: <https://github.com/vqdang/hover_net#data-format> |
|
|
|
`PRETRAIN_MODEL_URL` is "https://drive.google.com/u/1/uc?id=1KntZge40tAHgyXmHYVqZZ5d2p_4Qr2l5&export=download" which can be used in bash code below. |
|
|
|
 |
|
|
|
## Data |
|
|
|
The training data is from <https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet/>. |
|
|
|
- Target: segment instance-level nuclei and classify the nuclei type |
|
- Task: Segmentation and classification |
|
- Modality: RGB images |
|
- Size: 41 image tiles (2009 patches) |
|
|
|
The provided labelled data was partitioned, based on the original split, into training (27 tiles) and testing (14 tiles) datasets. |
|
|
|
After download the datasets, please run `scripts/prepare_patches.py` to prepare patches from tiles. Prepared patches are saved in `your-concep-dataset-path`/Prepared. The implementation is referring to <https://github.com/vqdang/hover_net/blob/master/extract_patches.py>. The command is like: |
|
|
|
``` |
|
python scripts/prepare_patches.py -root your-concep-dataset-path |
|
``` |
|
|
|
## Training configuration |
|
|
|
This model utilized a two-stage approach. The training was performed with the following: |
|
|
|
- GPU: At least 24GB of GPU memory. |
|
- Actual Model Input: 256 x 256 |
|
- AMP: True |
|
- Optimizer: Adam |
|
- Learning Rate: 1e-4 |
|
- Loss: HoVerNetLoss |
|
|
|
## Input |
|
|
|
Input: RGB images |
|
|
|
## Output |
|
|
|
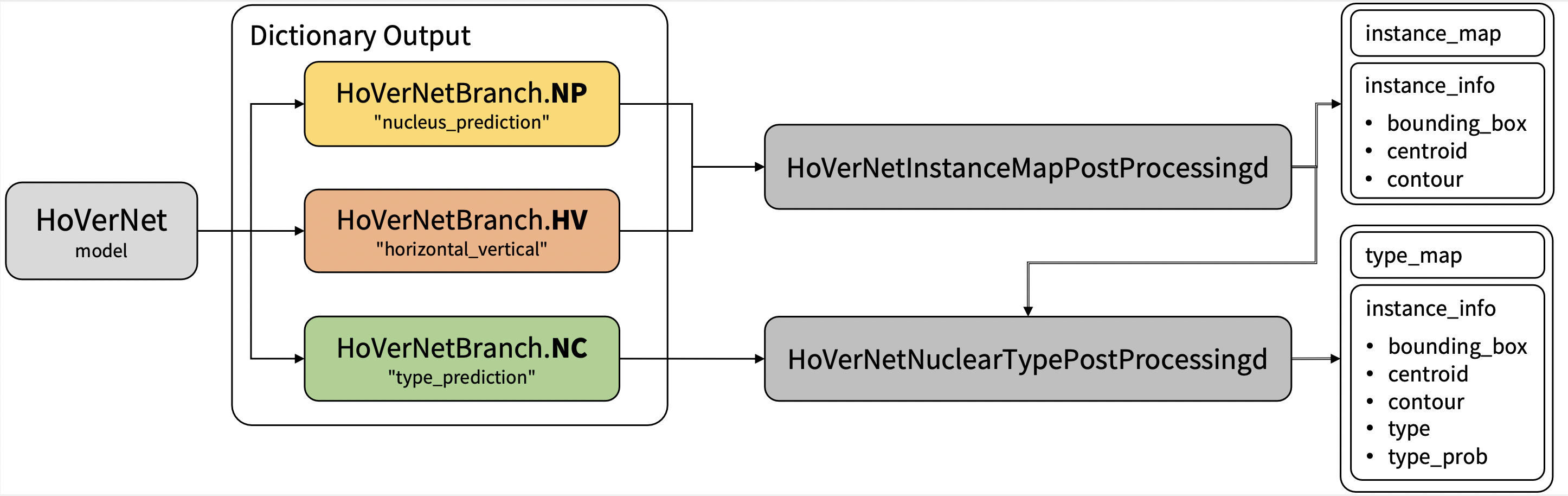
Output: a dictionary with the following keys: |
|
|
|
1. nucleus_prediction: predict whether or not a pixel belongs to the nuclei or background |
|
2. horizontal_vertical: predict the horizontal and vertical distances of nuclear pixels to their centres of mass |
|
3. type_prediction: predict the type of nucleus for each pixel |
|
|
|
## Model Performance |
|
|
|
The achieved metrics on the validation data are: |
|
|
|
Fast mode: |
|
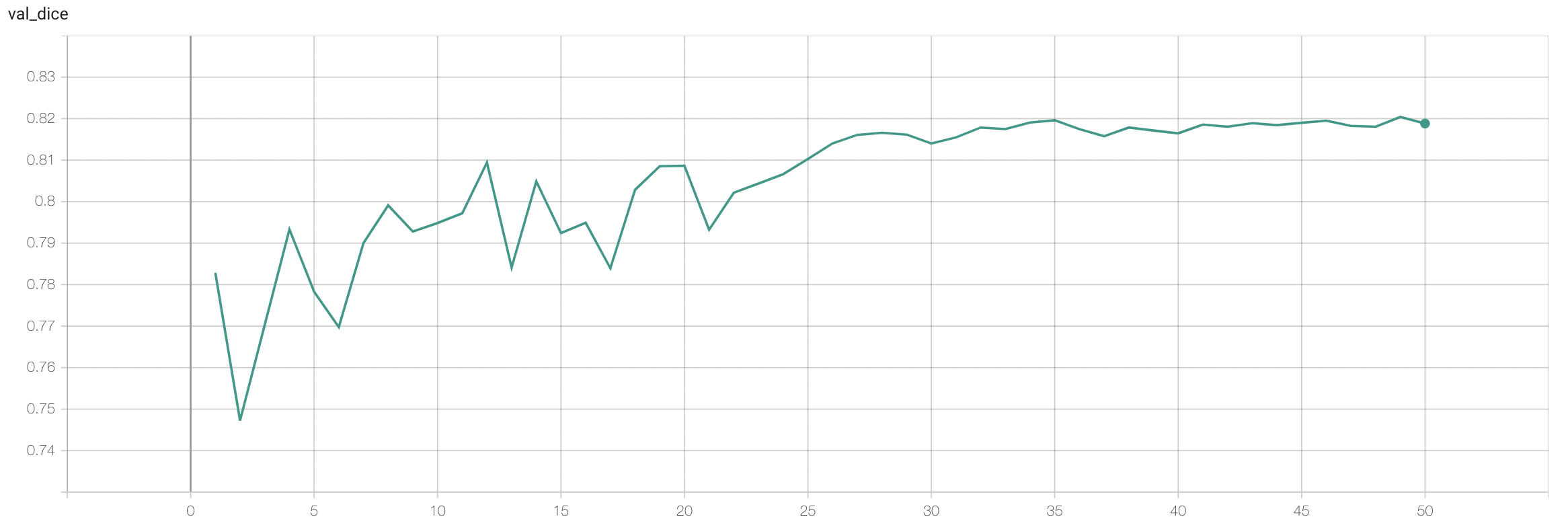
- Binary Dice: 0.8293 |
|
- PQ: 0.4936 |
|
- F1d: 0.7480 |
|
|
|
#### Training Loss and Dice |
|
|
|
stage1: |
|
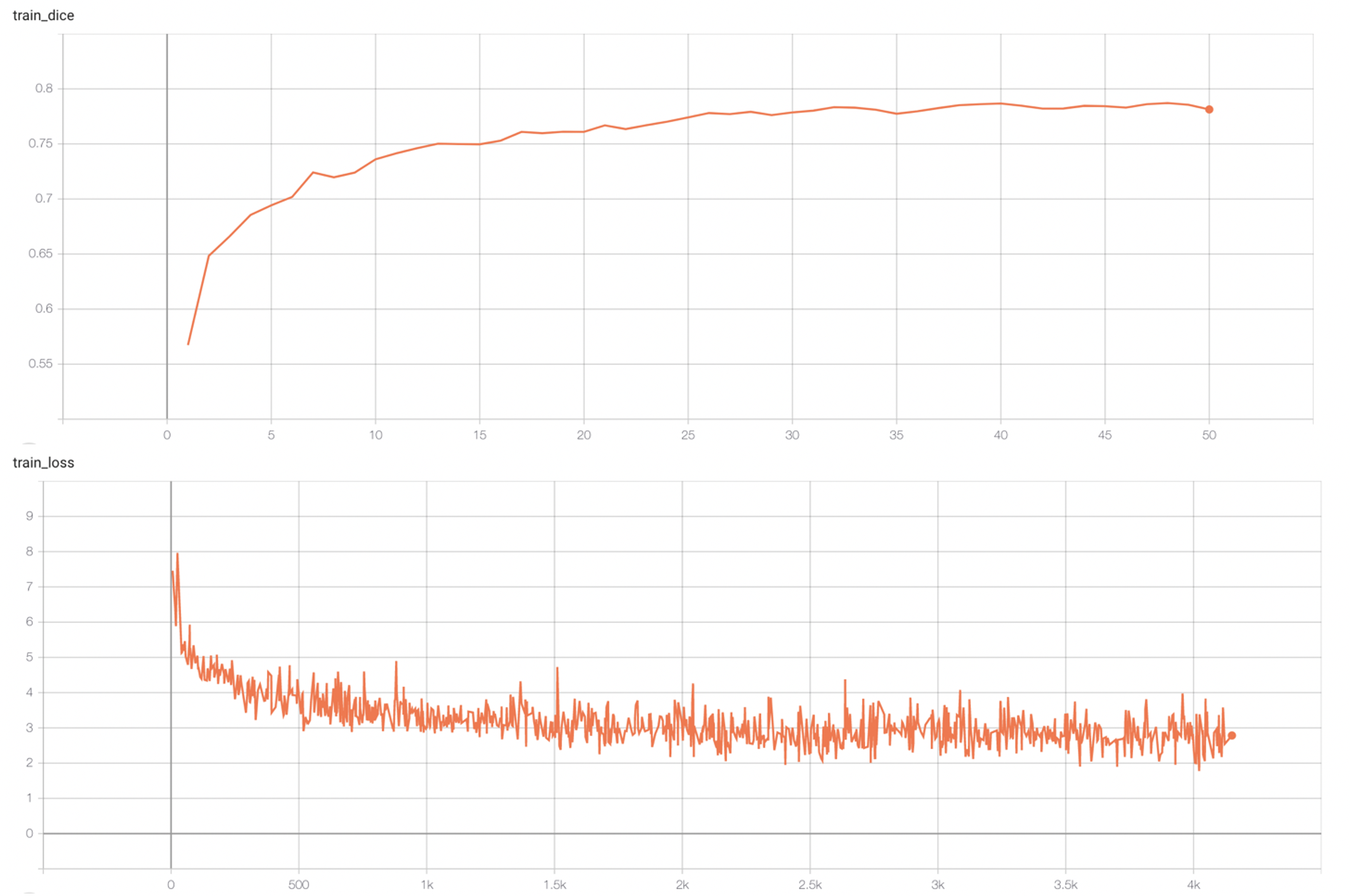 |
|
|
|
stage2: |
|
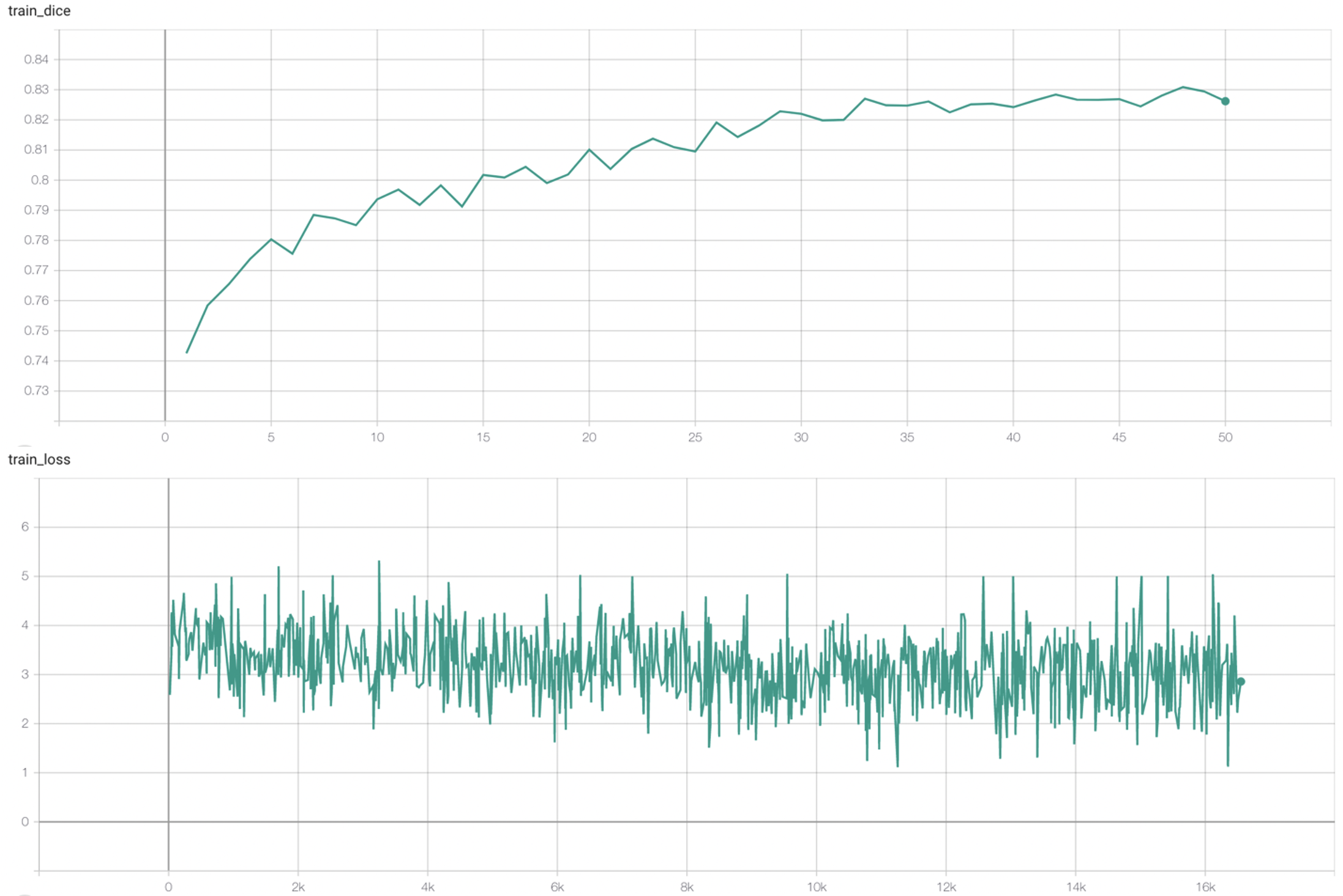 |
|
|
|
#### Validation Dice |
|
|
|
stage1: |
|
|
|
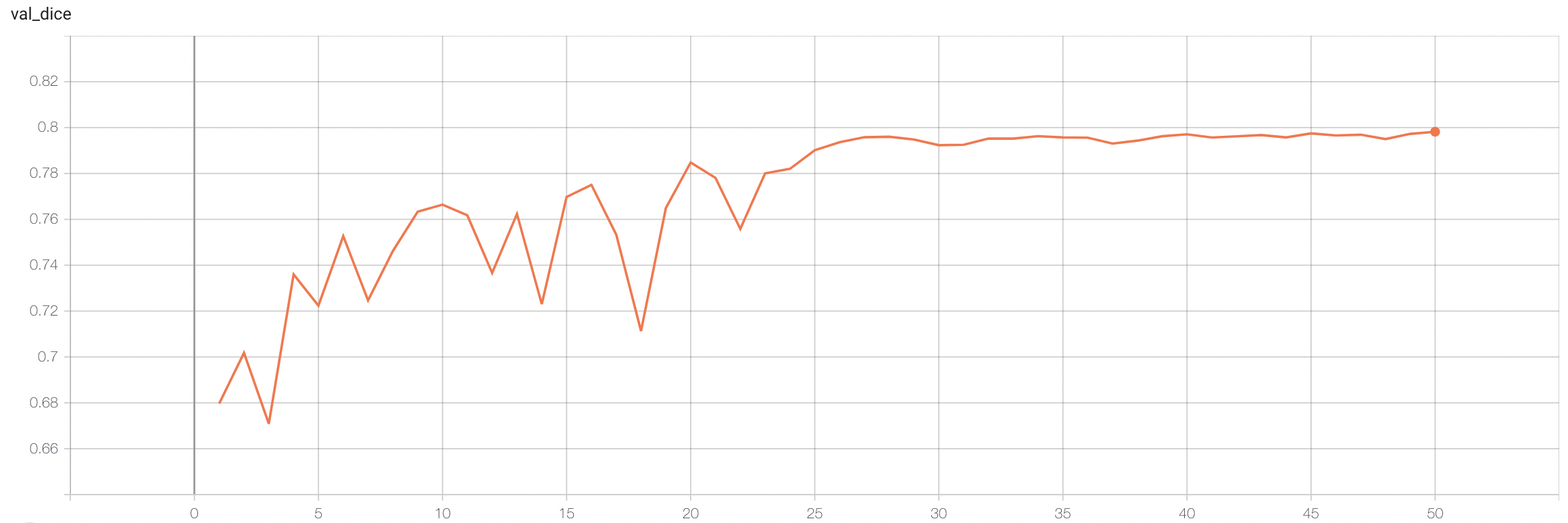 |
|
|
|
stage2: |
|
|
|
 |
|
|
|
## commands example |
|
|
|
Execute training: |
|
|
|
- Run first stage |
|
|
|
``` |
|
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf --network_def#pretrained_url `PRETRAIN_MODEL_URL` --stage 0 |
|
``` |
|
|
|
- Run second stage |
|
|
|
``` |
|
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf --network_def#freeze_encoder false --network_def#pretrained_url None --stage 1 |
|
``` |
|
|
|
Override the `train` config to execute multi-GPU training: |
|
|
|
- Run first stage |
|
|
|
``` |
|
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run training --meta_file configs/metadata.json --config_file "['configs/train.json','configs/multi_gpu_train.json']" --logging_file configs/logging.conf --train#dataloader#batch_size 8 --network_def#freeze_encoder true --network_def#pretrained_url `PRETRAIN_MODEL_URL` --stage 0 |
|
``` |
|
|
|
- Run second stage |
|
|
|
``` |
|
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run training --meta_file configs/metadata.json --config_file "['configs/train.json','configs/multi_gpu_train.json']" --logging_file configs/logging.conf --train#dataloader#batch_size 4 --network_def#freeze_encoder false --network_def#pretrained_url None --stage 1 |
|
``` |
|
|
|
Override the `train` config to execute evaluation with the trained model: |
|
|
|
``` |
|
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file "['configs/train.json','configs/evaluate.json']" --logging_file configs/logging.conf |
|
``` |
|
|
|
### Execute inference |
|
|
|
``` |
|
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf |
|
``` |
|
|
|
# Disclaimer |
|
|
|
This is an example, not to be used for diagnostic purposes. |
|
|
|
# References |
|
|
|
[1] Simon Graham, Quoc Dang Vu, Shan E Ahmed Raza, Ayesha Azam, Yee Wah Tsang, Jin Tae Kwak, Nasir Rajpoot, Hover-Net: Simultaneous segmentation and classification of nuclei in multi-tissue histology images, Medical Image Analysis, 2019 https://doi.org/10.1016/j.media.2019.101563 |
|
|
|
# License |
|
Copyright (c) MONAI Consortium |
|
|
|
Licensed under the Apache License, Version 2.0 (the "License"); |
|
you may not use this file except in compliance with the License. |
|
You may obtain a copy of the License at |
|
|
|
http://www.apache.org/licenses/LICENSE-2.0 |
|
|
|
Unless required by applicable law or agreed to in writing, software |
|
distributed under the License is distributed on an "AS IS" BASIS, |
|
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. |
|
See the License for the specific language governing permissions and |
|
limitations under the License. |
|
|