This is a SciBERT-based model fine-tuned to perform Named Entity Recognition for drug names and adverse drug effects.
This model classifies input tokens into one of five classes:
B-DRUG: beginning of a drug entityI-DRUG: within a drug entityB-EFFECT: beginning of an AE entityI-EFFECT: within an AE entityO: outside either of the above entities
To get started using this model for inference, simply set up an NER pipeline like below:
from transformers import (AutoModelForTokenClassification,
AutoTokenizer,
pipeline,
)
model_checkpoint = "jsylee/scibert_scivocab_uncased-finetuned-ner"
model = AutoModelForTokenClassification.from_pretrained(model_checkpoint, num_labels=5,
id2label={0: 'O', 1: 'B-DRUG', 2: 'I-DRUG', 3: 'B-EFFECT', 4: 'I-EFFECT'}
)
tokenizer = AutoTokenizer.from_pretrained(model_checkpoint)
model_pipeline = pipeline(task="ner", model=model, tokenizer=tokenizer)
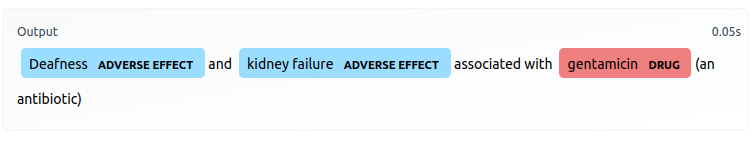
print( model_pipeline ("Abortion, miscarriage or uterine hemorrhage associated with misoprostol (Cytotec), a labor-inducing drug."))
SciBERT: https://huggingface.co/allenai/scibert_scivocab_uncased
- Downloads last month
- 152
Inference Providers
NEW
This model is not currently available via any of the supported Inference Providers.