prompt
stringlengths 19
879k
| completion
stringlengths 3
53.8k
| api
stringlengths 8
59
|
|---|---|---|
"""
Basic Equations for solving shallow water problems
#####################
"""
from pysph.sph.equation import Equation
from pysph.sph.integrator_step import IntegratorStep
from pysph.sph.integrator import Integrator
from compyle.api import declare
from pysph.sph.wc.linalg import gj_solve, augmented_matrix
from numpy import sqrt, cos, sin, zeros, pi, exp
import numpy as np
import numpy
M_PI = pi
class CheckForParticlesToSplit(Equation):
r"""Particles are tagged for splitting if the following condition is
satisfied:
.. math::
(A_i > A_max) and (h_i < h_max) and (x_min < x_i < x_max) and (y_min <
y_i < y_max)
References
----------
.. [Vacondio2010] <NAME>, <NAME> and <NAME>, "Accurate
particle splitting for smoothed particle hydrodynamics in shallow water
with shock capturing", Int. J. Numer. Meth. Fluids, 69 (2012), pp.
1377-1410
"""
def __init__(self, dest, h_max=1e9, A_max=1e9, x_min=-1e9, x_max=1e9,
y_min=-1e9, y_max=1e9):
r"""
Parameters
----------
h_max : float
maximum smoothing length beyond which splitting is deactivated
A_max : float
maximum area beyond which splitting is activated
x_min : float
minimum distance along x-direction beyond which splitting is
activated
x_max : float
maximum distance along x-direction beyond which splitting is
deactivated
y_min : float
minimum distance along y-direction beyond which splitting is
activated
y_max : float
maximum distance along y-direction beyond which splitting is
deactivated
"""
self.A_max = A_max
self.h_max = h_max
self.x_min = x_min
self.x_max = x_max
self.y_min = y_min
self.y_max = y_max
super(CheckForParticlesToSplit, self).__init__(dest, None)
def initialize(self, d_idx, d_A, d_h, d_x, d_y, d_pa_to_split):
if (d_A[d_idx] > self.A_max and d_h[d_idx] < self.h_max
and (self.x_min < d_x[d_idx] < self.x_max)
and (self.y_min < d_y[d_idx] < self.y_max)):
d_pa_to_split[d_idx] = 1
else:
d_pa_to_split[d_idx] = 0
class ParticleSplit(object):
r"""**Hexagonal particle splitting algorithm**
References
----------
.. [Vacondio2010] <NAME>, <NAME> and <NAME>, "Accurate
particle splitting for smoothed particle hydrodynamics in shallow water
with shock capturing", Int. J. Numer. Meth. Fluids, 69 (2012), pp.
1377-1410
"""
def __init__(self, pa_arr):
r"""
Parameters
----------
pa_arr : pysph.base.particle_array.ParticleArray
particle array of fluid
"""
self.pa_arr = pa_arr
# Ratio of mass of daughter particle located at center of hexagon to
# that of its parents mass
self.center_pa_mass_frac = 0.178705766141917
# Ratio of mass of daughter particle located at vertex of hexagon to
# that of its parents mass
self.vertex_pa_mass_frac = 0.136882287617319
# Ratio of smoothing length of daughter particle to that of its parents
# smoothing length
self.pa_h_ratio = 0.9
# Ratio of distance between center daughter particle and vertex
# daughter particle to that of its parents smoothing length
self.center_and_vertex_pa_separation_frac = 0.4
# Get index of the parent particles to split
self.idx_pa_to_split = self._get_idx_of_particles_to_split()
# Number of daughter particles located at the vertices of hexagon after
# splitting
self.num_vertex_pa_after_single_split = 6
def do_particle_split(self, solver=None):
if not self.idx_pa_to_split.size:
# If no particles to split then return
return
else:
# Properties of parent particles to split
h_parent = self.pa_arr.h[self.idx_pa_to_split]
h0_parent = self.pa_arr.h0[self.idx_pa_to_split]
m_parent = self.pa_arr.m[self.idx_pa_to_split]
x_parent = self.pa_arr.x[self.idx_pa_to_split]
y_parent = self.pa_arr.y[self.idx_pa_to_split]
u_parent = self.pa_arr.u[self.idx_pa_to_split]
v_parent = self.pa_arr.v[self.idx_pa_to_split]
u_prev_step_parent = self.pa_arr.u_prev_step[self.idx_pa_to_split]
v_prev_step_parent = self.pa_arr.v_prev_step[self.idx_pa_to_split]
rho_parent = self.pa_arr.rho[self.idx_pa_to_split]
rho0_parent = self.pa_arr.rho0[self.idx_pa_to_split]
alpha_parent = self.pa_arr.alpha[self.idx_pa_to_split]
# Vertex daughter particle properties update
n = self.num_vertex_pa_after_single_split
h_vertex_pa = self.pa_h_ratio * np.repeat(h_parent, n)
h0_vertex_pa = self.pa_h_ratio * np.repeat(h0_parent, n)
u_prev_step_vertex_pa = np.repeat(u_prev_step_parent, n)
v_prev_step_vertex_pa = np.repeat(v_prev_step_parent, n)
m_vertex_pa = self.vertex_pa_mass_frac * np.repeat(m_parent, n)
vertex_pa_pos = self._get_vertex_pa_positions(h_parent, u_parent,
v_parent)
x_vertex_pa = vertex_pa_pos[0] + np.repeat(x_parent, n)
y_vertex_pa = vertex_pa_pos[1] + np.repeat(y_parent, n)
rho0_vertex_pa = np.repeat(rho0_parent, n)
rho_vertex_pa = np.repeat(rho_parent, n)
alpha_vertex_pa = np.repeat(alpha_parent, n)
parent_idx_vertex_pa = np.repeat(self.idx_pa_to_split, n)
# Note:
# The center daughter particle properties are set at index of
# parent. The properties of parent needed for further calculations
# are not changed for now
# Center daughter particle properties update
for idx in self.idx_pa_to_split:
self.pa_arr.m[idx] *= self.center_pa_mass_frac
self.pa_arr.h[idx] *= self.pa_h_ratio
self.pa_arr.h0[idx] *= self.pa_h_ratio
self.pa_arr.parent_idx[idx] = int(idx)
# Update particle array to include vertex daughter particles
self._add_vertex_pa_prop(
h0_vertex_pa, h_vertex_pa, m_vertex_pa, x_vertex_pa,
y_vertex_pa, rho0_vertex_pa, rho_vertex_pa,
u_prev_step_vertex_pa, v_prev_step_vertex_pa, alpha_vertex_pa,
parent_idx_vertex_pa)
def _get_idx_of_particles_to_split(self):
idx_pa_to_split = []
for idx, val in enumerate(self.pa_arr.pa_to_split):
if val:
idx_pa_to_split.append(idx)
return np.array(idx_pa_to_split)
def _get_vertex_pa_positions(self, h_parent, u_parent, v_parent):
# Number of particles to split
num_of_pa_to_split = len(self.idx_pa_to_split)
n = self.num_vertex_pa_after_single_split
theta_vertex_pa = zeros(n)
r = self.center_and_vertex_pa_separation_frac
for i, theta in enumerate(range(0, 360, 60)):
theta_vertex_pa[i] = (pi/180)*theta
# Angle of velocity vector with horizontal
angle_vel = np.where(
(np.abs(u_parent) > 1e-3) | (np.abs(v_parent) > 1e-3),
np.arctan2(v_parent, u_parent), 0
)
# Rotates the hexagon such that its horizontal axis aligns with the
# direction of velocity vector
angle_actual = (np.tile(theta_vertex_pa, num_of_pa_to_split)
+ np.repeat(angle_vel, n))
x = r * cos(angle_actual) * np.repeat(h_parent, n)
y = r * sin(angle_actual) * np.repeat(h_parent, n)
return x.copy(), y.copy()
def _add_vertex_pa_prop(self, h0_vertex_pa, h_vertex_pa, m_vertex_pa,
x_vertex_pa, y_vertex_pa, rho0_vertex_pa,
rho_vertex_pa, u_prev_step_vertex_pa,
v_prev_step_vertex_pa, alpha_vertex_pa,
parent_idx_vertex_pa):
vertex_pa_props = {
'm': m_vertex_pa,
'h': h_vertex_pa,
'h0': h0_vertex_pa,
'x': x_vertex_pa,
'y': y_vertex_pa,
'u_prev_step': u_prev_step_vertex_pa,
'v_prev_step': v_prev_step_vertex_pa,
'rho0': rho0_vertex_pa,
'rho': rho_vertex_pa,
'alpha': alpha_vertex_pa,
'parent_idx': parent_idx_vertex_pa.astype(int)
}
# Add vertex daughter particles to particle array
self.pa_arr.add_particles(**vertex_pa_props)
class DaughterVelocityEval(Equation):
r"""**Evaluation of the daughter particle velocity after splitting
procedure**
.. math::
\boldsymbol{v_k} = c_v\frac{d_N}{d_k}\boldsymbol{v_N}
where,
.. math::
c_v = \dfrac{A_N}{\sum_{k=1}^{M}A_k}
References
----------
.. [Vacondio2010] <NAME>, <NAME> and <NAME>, "Accurate
particle splitting for smoothed particle hydrodynamics in shallow water
with shock capturing", Int. J. Numer. Meth. Fluids, 69 (2012), pp.
1377-1410
"""
def __init__(self, dest, sources, rhow=1000.0):
r""""
Parameters
----------
rhow : float
constant 3-D density of water (kg/m3)
Notes
-----
This equation should be called before the equation SWEOS, as the parent
particle area is required for calculating velocities. On calling the
SWEOS equation, the parent properties are changed to the center
daughter particle properties.
"""
self.rhow = rhow
super(DaughterVelocityEval, self).__init__(dest, sources)
def initialize(self, d_sum_Ak, d_idx, d_m, d_rho, d_u, d_v, d_uh,
d_vh, d_u_parent, d_v_parent, d_uh_parent, d_vh_parent,
d_parent_idx):
# Stores sum of areas of daughter particles
d_sum_Ak[d_idx] = 0.0
d_u_parent[d_idx] = d_u[d_parent_idx[d_idx]]
d_uh_parent[d_idx] = d_uh[d_parent_idx[d_idx]]
d_v_parent[d_idx] = d_v[d_parent_idx[d_idx]]
d_vh_parent[d_idx] = d_vh[d_parent_idx[d_idx]]
def loop_all(self, d_sum_Ak, d_pa_to_split, d_parent_idx, d_idx, s_m,
s_rho, s_parent_idx, NBRS, N_NBRS):
i = declare('int')
s_idx = declare('long')
if d_pa_to_split[d_idx]:
for i in range(N_NBRS):
s_idx = NBRS[i]
if s_parent_idx[s_idx] == d_parent_idx[d_idx]:
# Sums the area of daughter particles who have same parent
# idx
d_sum_Ak[d_idx] += s_m[s_idx] / s_rho[s_idx]
def post_loop(self, d_idx, d_parent_idx, d_A, d_sum_Ak, d_dw, d_rho, d_u,
d_uh, d_vh, d_v, d_u_parent, d_v_parent, d_uh_parent,
d_vh_parent, t):
# True only for daughter particles
if d_parent_idx[d_idx]:
# Ratio of parent area (before split) to sum of areas of its
# daughters (after split)
cv = d_A[d_parent_idx[d_idx]] / d_sum_Ak[d_parent_idx[d_idx]]
# The denominator (d_rho[d_idx]/self.rhow) represents the water
# depth of daughter particle. d_dw[d_idx] cannot be used as
# equation of state is called after this equation (Refer Notes in
# the constructor)
dw_ratio = d_dw[d_parent_idx[d_idx]] / (d_rho[d_idx]/self.rhow)
d_u[d_idx] = cv * dw_ratio * d_u_parent[d_idx]
d_uh[d_idx] = cv * dw_ratio * d_uh_parent[d_idx]
d_v[d_idx] = cv * dw_ratio * d_v_parent[d_idx]
d_vh[d_idx] = cv * dw_ratio * d_vh_parent[d_idx]
d_parent_idx[d_idx] = 0
class FindMergeable(Equation):
r"""**Particle merging algorithm**
Particles are tagged for merging if the following condition is
satisfied:
.. math::
(A_i < A_min) and (x_min < x_i < x_max) and (y_min < y_i < y_max)
References
----------
.. [Vacondio2013] <NAME> et al., "Shallow water SPH for flooding with
dynamic particle coalescing and splitting", Advances in Water Resources,
58 (2013), pp. 10-23
"""
def __init__(self, dest, sources, A_min, x_min=-1e9, x_max=1e9, y_min=-1e9,
y_max=1e9):
r"""
Parameters
----------
A_min : float
minimum area below which merging is activated
x_min : float
minimum distance along x-direction beyond which merging is
activated
x_max : float
maximum distance along x-direction beyond which merging is
deactivated
y_min : float
minimum distance along y-direction beyond which merging is
activated
y_max : float
maximum distance along y-direction beyond which merging is
deactivated
Notes
-----
The merging algorithm merges two particles 'a' and 'b' if the following
conditions are satisfied:
#. Both particles have area less than A_min
#. Both particles lies within :math:`x_min < x_i < x_max` and
:math:`y_min < y_i < y_max`
#. if 'a' is the closest neighbor of 'b' and vice versa
The merging algorithm is run every timestep
"""
self.A_min = A_min
self.x_min = x_min
self.x_max = x_max
self.y_min = y_min
self.y_max = y_max
super(FindMergeable, self).__init__(dest, sources)
def loop_all(self, d_idx, d_merge, d_closest_idx, d_x, d_y, d_h, d_A,
d_is_merged_pa, s_x, s_y, s_A, NBRS, N_NBRS):
# Finds the closest neighbor of a particle and stores the index of that
# neighbor in d_closest_idx[d_idx]
i, closest = declare('int', 2)
s_idx = declare('unsigned int')
d_merge[d_idx] = 0
d_is_merged_pa[d_idx] = 0
xi = d_x[d_idx]
yi = d_y[d_idx]
rmin = d_h[d_idx] * 10.0
closest = -1
if (d_A[d_idx] < self.A_min and ((self.x_min < d_x[d_idx] < self.x_max)
and (self.y_min < d_y[d_idx] < self.y_max))):
for i in range(N_NBRS):
s_idx = NBRS[i]
if s_idx == d_idx:
continue
xij = xi - s_x[s_idx]
yij = yi - s_y[s_idx]
rij = sqrt(xij*xij + yij*yij)
if rij < rmin:
closest = s_idx
rmin = rij
d_closest_idx[d_idx] = closest
def post_loop(self, d_idx, d_m, d_u, d_v, d_h, d_uh, d_vh, d_closest_idx,
d_is_merged_pa, d_merge, d_x, d_y, SPH_KERNEL):
idx = declare('int')
xma = declare('matrix(3)')
xmb = declare('matrix(3)')
idx = d_closest_idx[d_idx]
if idx > -1:
# If particle 'a' is closest neighbor of 'b' and vice versa
if d_idx == d_closest_idx[idx]:
if d_idx < idx:
# The newly merged particle properties are set at index of
# particle 'a'
m_merged = d_m[d_idx] + d_m[idx]
x_merged = ((d_m[d_idx]*d_x[d_idx] + d_m[idx]*d_x[idx])
/ m_merged)
y_merged = ((d_m[d_idx]*d_y[d_idx] + d_m[idx]*d_y[idx])
/ m_merged)
xma[0] = x_merged - d_x[d_idx]
xma[1] = y_merged - d_y[d_idx]
xmb[0] = x_merged - d_x[idx]
xmb[1] = y_merged - d_y[idx]
rma = sqrt(xma[0]*xma[0] + xma[1]*xma[1])
rmb = sqrt(xmb[0]*xmb[0] + xmb[1]*xmb[1])
d_u[d_idx] = ((d_m[d_idx]*d_u[d_idx] + d_m[idx]*d_u[idx])
/ m_merged)
d_uh[d_idx] = (d_m[d_idx]*d_uh[d_idx]
+ d_m[idx]*d_uh[idx]) / m_merged
d_v[d_idx] = ((d_m[d_idx]*d_v[d_idx] + d_m[idx]*d_v[idx])
/ m_merged)
d_vh[d_idx] = (d_m[d_idx]*d_vh[d_idx]
+ d_m[idx]*d_vh[idx]) / m_merged
const1 = d_m[d_idx] * SPH_KERNEL.kernel(xma, rma,
d_h[d_idx])
const2 = d_m[idx] * SPH_KERNEL.kernel(xmb, rmb, d_h[idx])
d_h[d_idx] = sqrt((7*M_PI/10.) * (m_merged/(const1+const2)))
d_m[d_idx] = m_merged
# Tags the newly formed particle after merging
d_is_merged_pa[d_idx] = 1
else:
# Tags particle 'b' for removal after merging
d_merge[d_idx] = 1
def reduce(self, dst, t, dt):
# The indices of particle 'b' are removed from particle array after
# merging is done
indices = declare('object')
indices = numpy.where(dst.merge > 0)[0]
if len(indices) > 0:
dst.remove_particles(indices)
class InitialDensityEvalAfterMerge(Equation):
r"""**Initial density of the newly formed particle after merging**
.. math ::
\rho_M = \sum_{j}^{}m_jW_{M,j}
References
----------
.. [Vacondio2013] R. Vacondio et al., "Shallow water SPH for flooding with
dynamic particle coalescing and splitting", Advances in Water Resources,
58 (2013), pp. 10-23
"""
def loop_all(self, d_rho, d_idx, d_is_merged_pa, d_x, d_y, s_h, s_m, s_x,
d_merge, d_closest_idx, s_y, SPH_KERNEL, NBRS, N_NBRS):
i = declare('int')
s_idx = declare('long')
xij = declare('matrix(3)')
# Evaluates the initial density of the newly formed particle after
# merging
if d_is_merged_pa[d_idx] == 1:
d_rho[d_idx] = 0.0
rij = 0.0
rho_sum = 0.0
for i in range(N_NBRS):
s_idx = NBRS[i]
xij[0] = d_x[d_idx] - s_x[s_idx]
xij[1] = d_y[d_idx] - s_y[s_idx]
rij = sqrt(xij[0]*xij[0] + xij[1]*xij[1])
rho_sum += s_m[s_idx] * SPH_KERNEL.kernel(xij, rij, s_h[s_idx])
d_rho[d_idx] += rho_sum
class EulerStep(IntegratorStep):
"""Fast but inaccurate integrator. Use this for testing"""
def initialize(self, d_u, d_v, d_u_prev_step, d_v_prev_step, d_idx):
d_u_prev_step[d_idx] = d_u[d_idx]
d_v_prev_step[d_idx] = d_v[d_idx]
def stage1(self, d_idx, d_u, d_v, d_au, d_av, d_x, d_y, dt):
d_u[d_idx] += dt * d_au[d_idx]
d_v[d_idx] += dt * d_av[d_idx]
d_x[d_idx] += dt * d_u[d_idx]
d_y[d_idx] += dt * d_v[d_idx]
class SWEStep(IntegratorStep):
"""Leap frog time integration scheme"""
def initialize(self, t, d_u, d_v, d_uh, d_vh, d_u_prev_step, d_v_prev_step,
d_idx):
# Stores the velocities at previous time step
d_u_prev_step[d_idx] = d_u[d_idx]
d_v_prev_step[d_idx] = d_v[d_idx]
def stage1(self, d_uh, d_vh, d_idx, d_au, d_av, dt):
# Velocities at half time step
d_uh[d_idx] += dt * d_au[d_idx]
d_vh[d_idx] += dt * d_av[d_idx]
def stage2(self, d_u, d_v, d_uh, d_vh, d_idx, d_au, d_av, d_x, d_y, dt):
d_x[d_idx] += dt * d_uh[d_idx]
d_y[d_idx] += dt * d_vh[d_idx]
d_u[d_idx] = d_uh[d_idx] + dt/2.*d_au[d_idx]
d_v[d_idx] = d_vh[d_idx] + dt/2.*d_av[d_idx]
class SWEIntegrator(Integrator):
"""Integrator for shallow water problems"""
def one_timestep(self, t, dt):
self.compute_accelerations()
self.initialize()
# Predict
self.stage1()
# Call any post-stage functions.
self.do_post_stage(0.5*dt, 1)
# Correct
self.stage2()
# Call any post-stage functions.
self.do_post_stage(dt, 2)
class GatherDensityEvalNextIteration(Equation):
r"""**Gather formulation for evaluating the density of a particle**
.. math::
\rho_i = \sum_{j}{m_jW(\textbf{x}_i - \textbf{x}_j, h_i)}
References
----------
.. [Hernquist and Katz, 1988] <NAME> and <NAME>, "TREESPH: A
unification of SPH with the hierarcgical tree method", The Astrophysical
Journal Supplement Series, 70 (1989), pp 419-446.
"""
def initialize(self, d_rho, d_idx, d_rho_prev_iter):
# Stores density of particle i of the previous iteration
d_rho_prev_iter[d_idx] = d_rho[d_idx]
d_rho[d_idx] = 0.0
def loop(self, d_rho, d_idx, s_m, s_idx, WI):
d_rho[d_idx] += s_m[s_idx] * WI
class ScatterDensityEvalNextIteration(Equation):
r"""**Scatter formulation for evaluating the density of a particle**
.. math::
\rho_i = \sum_{J}{m_JW(\textbf{x}_i - \textbf{x}_j, h_j)}
References
----------
.. [Hernquist and Katz, 1988] <NAME> and <NAME>, "TREESPH: A
unification of SPH with the hierarcgical tree method", The Astrophysical
Journal Supplement Series, 70 (1989), pp 419-446.
"""
def initialize(self, t, d_rho, d_idx, d_rho_prev_iter):
# Stores density of particle i of the previous iteration
d_rho_prev_iter[d_idx] = d_rho[d_idx]
d_rho[d_idx] = 0.0
def loop(self, d_rho, d_idx, s_m, s_idx, WJ):
d_rho[d_idx] += s_m[s_idx] * WJ
class NonDimensionalDensityResidual(Equation):
r"""**Non-dimensional density residual**
.. math::
\psi^{k+1} = \dfrac{|\rho_i^{k+1} - \rho_i^k|}{\rho_i^k}
References
----------
.. [Vacondio2010] <NAME>, <NAME> and <NAME>, "Accurate
particle splitting for smoothed particle hydrodynamics in shallow water
with shock capturing", Int. J. Numer. Meth. Fluids, 69 (2012), pp.
1377-1410
"""
def __init__(self, dest, sources=None):
super(NonDimensionalDensityResidual, self).__init__(dest, sources)
def post_loop(self, d_psi, d_rho, d_rho_prev_iter, d_idx):
# Non-dimensional residual
d_psi[d_idx] = abs(d_rho[d_idx] - d_rho_prev_iter[d_idx]) \
/ d_rho_prev_iter[d_idx]
class CheckConvergenceDensityResidual(Equation):
r"""The iterative process is stopped once the following condition is met
.. math::
\psi^{k+1} < \epsilon_{\psi}
where,
\epsilon_{\psi} = 1e-3
References
----------
.. [Vacondio2010] <NAME>, <NAME> and <NAME>, "Accurate
particle splitting for smoothed particle hydrodynamics in shallow water
with shock capturing", Int. J. Numer. Meth. Fluids, 69 (2012), pp.
1377-1410
Notes
-----
If particle splitting is activated, better to use this convergence
criteria. It can be used even if particle splitting is not activated.
"""
def __init__(self, dest, sources=None):
super(CheckConvergenceDensityResidual, self).__init__(dest, sources)
self.eqn_has_converged = 0
def initialize(self):
self.eqn_has_converged = 0
def reduce(self, dst, t, dt):
epsilon = max(dst.psi)
if epsilon <= 1e-3:
self.eqn_has_converged = 1
def converged(self):
return self.eqn_has_converged
class CorrectionFactorVariableSmoothingLength(Equation):
r"""**Correction factor in internal force due to variable smoothing
length**
.. math::
\alpha_i = -\sum_j m_j r_{ij}\frac{dW_i}{dr_{ij}}
References
----------
.. [<NAME>, 2005] <NAME> and <NAME>, "A corrected
smooth particle hydrodynamics formulation of the shallow-water equations",
Computers and Structures, 83 (2005), pp. 1396-1410
"""
def initialize(self, d_idx, d_alpha):
d_alpha[d_idx] = 0.0
def loop(self, d_alpha, d_idx, DWIJ, XIJ, s_idx, s_m):
d_alpha[d_idx] += -s_m[s_idx] * (DWIJ[0]*XIJ[0] + DWIJ[1]*XIJ[1])
class RemoveParticlesWithZeroAlpha(Equation):
r"""Removes particles if correction factor (alpha) in internal force due to
variable smoothing length is zero
"""
def __init__(self, dest):
super(RemoveParticlesWithZeroAlpha, self).__init__(dest, None)
def post_loop(self, d_alpha, d_pa_alpha_zero, d_idx):
if d_alpha[d_idx] == 0:
d_pa_alpha_zero[d_idx] = 1
def reduce(self, dst, t, dt):
indices = declare('object')
indices = numpy.where(dst.pa_alpha_zero > 0)[0]
if len(indices) > 0:
dst.remove_particles(indices)
class SummationDensity(Equation):
r"""**Summation Density**
.. math::
\rho_i = \sum_{j}{m_jW(\textbf{x}_i - \textbf{x}_j, h_i)}
"""
def initialize(self, d_summation_rho, d_idx):
d_summation_rho[d_idx] = 0.0
def loop(self, d_summation_rho, d_idx, s_m, s_idx, WI):
d_summation_rho[d_idx] += s_m[s_idx] * WI
class InitialGuessDensityVacondio(Equation):
r"""**Initial guess density to start the iterative evaluation of density
for time step n+1**
.. math::
\rho_{i(0)}^{n+1} = \rho_i^n + dt\dfrac{d\rho_i}{dt}\\
h_{i(0)}^{n+1} = h_i^n + -\dfrac{h_i^n}{\rho_i^n}\dfrac{dt}{dm}
\dfrac{d\rho_i}{dt}
where,
.. math::
\frac{d\rho_i}{dt} = \rho_i^n\sum_j\dfrac{m_j}{\rho_j}
(\textbf{v}_i-\textbf{v}_j).\nabla W_i
References
----------
.. [VacondioSWE-SPHysics, 2013] <NAME> et al., SWE-SPHysics source
code, File: SWE_SPHYsics/SWE-SPHysics_2D_v1.0.00/source/SPHYSICS_SWE_2D/
ac_dw_var_hj_2D.f
Note:
If particle splitting is activated, better to use this method. It can be
used even if particle splitting is not activated.
"""
def __init__(self, dest, sources, dim=2):
r"""
Parameters
----------
dim : int
number of space dimensions (Default: 2)
"""
self.dim = dim
super(InitialGuessDensityVacondio, self).__init__(dest, sources)
def initialize(self, d_arho, d_idx):
d_arho[d_idx] = 0
def loop(self, d_arho, d_rho, d_idx, s_m, s_rho, s_idx, d_u_prev_step,
d_v_prev_step, s_u_prev_step, s_v_prev_step, DWI):
tmp1 = (d_u_prev_step[d_idx]-s_u_prev_step[s_idx]) * DWI[0]
tmp2 = (d_v_prev_step[d_idx]-s_v_prev_step[s_idx]) * DWI[1]
d_arho[d_idx] += d_rho[d_idx] * ((s_m[s_idx]/s_rho[s_idx])*(tmp1+tmp2))
def post_loop(self, d_rho, d_h, dt, d_arho, d_idx):
d_rho[d_idx] += dt * d_arho[d_idx]
d_h[d_idx] += -(dt/self.dim)*d_h[d_idx]*(d_arho[d_idx]/d_rho[d_idx])
class InitialGuessDensity(Equation):
r"""**Initial guess density to start the iterative evaluation of density
for time step n+1 based on properties of time step n**
.. math::
\rho_{I, n+1}^{(0)} = \rho_{I,n}e^{\lambda_n}
where,
\lambda = \dfrac{\rho_Id_m}{\alpha_I}\sum_{J}^{}m_J
(\textbf{v}_J - \textbf{v}_I).\nabla W_I(\textbf{x}_I
- \textbf{x}_J, h_I)
References
----------
.. [Rodriguez and Bonet, 2005] <NAME> and <NAME>, "A corrected
smooth particle hydrodynamics formulation of the shallow-water equations",
Computers and Structures, 83 (2005), pp. 1396-1410
"""
def __init__(self, dest, sources, dim=2):
r"""
Parameters
----------
dim : int
number of space dimensions (Default: 2)
"""
self.dim = dim
super(InitialGuessDensity, self).__init__(dest, sources)
def initialize(self, d_exp_lambda, d_idx):
d_exp_lambda[d_idx] = 0.0
def loop(self, d_exp_lambda, d_u_prev_step, d_v_prev_step, d_alpha, d_idx,
s_m, s_u_prev_step, s_v_prev_step, s_idx, DWI, dt, t):
a1 = (d_u_prev_step[d_idx]-s_u_prev_step[s_idx]) * DWI[0]
a2 = (d_v_prev_step[d_idx]-s_v_prev_step[s_idx]) * DWI[1]
const = (self.dim*dt) / d_alpha[d_idx]
d_exp_lambda[d_idx] += const * (s_m[s_idx]*(a1+a2))
def post_loop(self, t, d_rho, d_exp_lambda, d_idx):
d_rho[d_idx] = d_rho[d_idx] * exp(d_exp_lambda[d_idx])
class UpdateSmoothingLength(Equation):
r"""**Update smoothing length based on density**
.. math::
h_I^{(k)} = h_I^{0}\biggl(\dfrac{\rho_I^0}{\rho_I^{(k)}}
\biggl)^\frac{1}{d_m}
References
----------
.. [<NAME>, 2005] <NAME> and <NAME>, "A corrected
smooth particle hydrodynamics formulation of the shallow-water equations",
Computers and Structures, 83 (2005), pp. 1396-1410
"""
def __init__(self, dest, dim=2):
r"""
Parameters
----------
dim : int
number of space dimensions (Default: 2)
"""
self.dim = dim
super(UpdateSmoothingLength, self).__init__(dest, None)
def post_loop(self, d_h, d_h0, d_rho0, d_rho, d_idx):
d_h[d_idx] = d_h0[d_idx] * (d_rho0[d_idx]/d_rho[d_idx])**(1./self.dim)
class DensityResidual(Equation):
r"""**Residual of density**
.. math::
R(\rho^{(k)}) = \rho_I^{(k)} - \sum_{J}^{}m_J
W_I(\textbf{x}_I - \textbf{x}_J, h_I^{(k)})
References
----------
.. [Rodriguez and Bonet, 2005] <NAME> and <NAME>, "A corrected
smooth particle hydrodynamics formulation of the shallow-water equations",
Computers and Structures, 83 (2005), pp. 1396-1410
"""
def __init__(self, dest, sources=None):
super(DensityResidual, self).__init__(dest, sources)
def post_loop(self, d_rho, d_idx, d_rho_residual, d_summation_rho, t):
d_rho_residual[d_idx] = d_rho[d_idx] - d_summation_rho[d_idx]
class DensityNewtonRaphsonIteration(Equation):
r"""**Newton-Raphson approximate solution for the density equation at
iteration k+1**
.. math::
\rho^{(k+1)} = \rho_I^{(k)}\biggl[1 - \dfrac{R_I^{(k)}d_m}{(
R_I^{(k)} d_m + \alpha_I^k)}\biggr]
References
----------
.. [Rodriguez and Bonet, 2005] <NAME> and <NAME>, "A corrected
smooth particle hydrodynamics formulation of the shallow-water equations",
Computers and Structures, 83 (2005), pp. 1396-1410
"""
def __init__(self, dest, sources=None, dim=2):
r"""
Parameters
----------
dim : int
number of space dimensions (Default: 2)
"""
self.dim = dim
super(DensityNewtonRaphsonIteration, self).__init__(dest, sources)
def initialize(self, d_rho, d_rho_prev_iter, d_idx):
d_rho_prev_iter[d_idx] = d_rho[d_idx]
def post_loop(self, d_rho, d_idx, d_alpha, d_rho_residual):
a1 = d_rho_residual[d_idx] * self.dim
a2 = a1 + d_alpha[d_idx]
const = 1 - (a1/a2)
d_rho[d_idx] = d_rho[d_idx] * const
class CheckConvergence(Equation):
r"""Stops the Newton-Raphson iterative procedure if the following
convergence criteria is satisfied:
.. math::
\dfrac{|R_I^{(k+1)}|}{\rho_I^{(k)}} \leq \epsilon
where,
\epsilon = 1e-15
References
----------
.. [<NAME>, 2005] <NAME> and <NAME>, "A corrected
smooth particle hydrodynamics formulation of the shallow-water equations",
Computers and Structures, 83 (2005), pp. 1396-1410
Notes
-----
Use this convergence criteria when using the Newton-Raphson iterative
procedure.
"""
def __init__(self, dest, sources=None):
super(CheckConvergence, self).__init__(dest, sources)
self.eqn_has_converged = 0
def initialize(self):
self.eqn_has_converged = 0
def post_loop(self, d_positive_rho_residual, d_rho_residual,
d_rho_prev_iter, d_idx, t):
d_positive_rho_residual[d_idx] = abs(d_rho_residual[d_idx])
def reduce(self, dst, t, dt):
max_epsilon = max(dst.positive_rho_residual / dst.rho_prev_iter)
if max_epsilon <= 1e-15:
self.eqn_has_converged = 1
def converged(self):
return self.eqn_has_converged
class SWEOS(Equation):
r"""**Update fluid properties based on density**
References
----------
.. [Rodriguez and Bonet, 2005] <NAME> and <NAME>, "A corrected
smooth particle hydrodynamics formulation of the shallow-water equations",
Computers and Structures, 83 (2005), pp. 1396-1410
"""
def __init__(self, dest, sources=None, g=9.81, rhow=1000.0):
r"""
Parameters
----------
g : float
acceleration due to gravity
rhow : float
constant 3-D density of water
"""
self.rhow = rhow
self.g = g
self.fac = 0.5 * (g/rhow)
super(SWEOS, self).__init__(dest, sources)
def post_loop(self, d_rho, d_cs, d_u, d_v, d_idx, d_p, d_dw, d_dt_cfl,
d_m, d_A, d_alpha):
# Pressure
d_p[d_idx] = self.fac * (d_rho[d_idx])**2
# Wave speed
d_cs[d_idx] = sqrt(self.g * d_rho[d_idx]/self.rhow)
# Area
d_A[d_idx] = d_m[d_idx] / d_rho[d_idx]
# Depth of water
d_dw[d_idx] = d_rho[d_idx] / self.rhow
# dt = CFL * (h_min / max(dt_cfl))
d_dt_cfl[d_idx] = d_cs[d_idx] + (d_u[d_idx]**2 + d_v[d_idx]**2)**0.5
def mu_calc(hi=1.0, hj=1.0, velij_dot_rij=1.0, rij2=1.0):
r"""Term present in the artificial viscosity formulation (Monaghan)
.. math::
\mu_{ij} = \dfrac{\bar h_{ij}\textbf{v}_{ij}.\textbf{x}_{ij}}
{|\textbf{x}_{ij}|^2 + \zeta^2}
References
----------
.. [Monaghan2005] <NAME>, "Smoothed particle hydrodynamics",
Reports on Progress in Physics, 68 (2005), pp. 1703-1759.
"""
h_bar = (hi+hj) / 2.0
eta2 = 0.01 * hi**2
muij = (h_bar*velij_dot_rij) / (rij2+eta2)
return muij
def artificial_visc(alpha=1.0, rij2=1.0, hi=1.0, hj=1.0, rhoi=1.0, rhoj=1.0,
csi=1.0, csj=1.0, muij=1.0):
r"""**Artificial viscosity based stabilization term (Monaghan)**
Activated when :math:`\textbf{v}_{ij}.\textbf{x}_{ij} < 0`
Given by
.. math::
\Pi_{ij} = \dfrac{-a\bar c_{ij}\mu_{ij}+b\bar c_{ij}\mu_{ij}^2}{\rho_{ij}}
References
----------
.. [Monaghan2005] <NAME>, "Smoothed particle hydrodynamics",
Reports on Progress in Physics, 68 (2005), pp. 1703-1759.
"""
cs_bar = (csi+csj) / 2.0
rho_bar = (rhoi+rhoj) / 2.0
pi_visc = -(alpha*cs_bar*muij) / rho_bar
return pi_visc
def viscosity_LF(alpha=1.0, rij2=1.0, hi=1.0, hj=1.0, rhoi=1.0, rhoj=1.0,
csi=1.0, csj=1.0, muij=1.0):
r"""**Lax-Friedrichs flux based stabilization term (Ata and Soulaimani)**
.. math::
\Pi_{ij} = \dfrac{\bar c_{ij}\textbf{v}_{ij}.\textbf{x}_{ij}}
{\bar\rho_{ij}\sqrt{|x_{ij}|^2 + \zeta^2}}
References
----------
.. [Ata and Soulaimani, 2004] <NAME> and <NAME>, "A stabilized SPH
method for inviscid shallow water", Int. J. Numer. Meth. Fluids, 47 (2005),
pp. 139-159.
Notes
-----
The advantage of this term is that it automatically sets the required level
of numerical viscosity based on the Lax-Friedrichs flux. This is the
default stabilization method.
"""
cs_bar = (csi+csj) / 2.0
rho_bar = (rhoi+rhoj) / 2.0
eta2 = 0.01 * hi**2
h_bar = (hi+hj) / 2.0
tmp = (muij*(rij2+eta2)**0.5) / h_bar
pi_visc = -(cs_bar*tmp) / rho_bar
return pi_visc
class ParticleAcceleration(Equation):
r"""**Acceleration of a particle**
.. math::
\textbf{a}_i = -\frac{g+\textbf{v}_i.\textbf{k}_i\textbf{v}_i
-\textbf{t}_i.\nabla H_i}{1+\nabla H_i.\nabla H_i}
\nabla H_i - \textbf{t}_i - \textbf{S}_{fi}
where,
.. math::
\textbf{t}_i &= \sum_j m_j\ \biggl[\biggl(\frac{p_j}{
\alpha_j \rho_j^2}+0.5\Pi_{ij}\biggr)\nabla W_j(\textbf{x}_i, h_j) -
\biggl(\frac{p_i}{\alpha_i \rho_i^2}+0.5\Pi_{ij}\biggr)\nabla
W_i(\textbf{x}_j, h_i)\biggr]
.. math::
\textbf{S}_f = \textbf{v}\dfrac{gn^2|\textbf{v}|}{d^{\frac{4}{3}}}
with,
.. math::
\alpha_i = -\sum_j m_j r_{ij}\frac{dW_i}{dr_{ij}}
.. math::
n_i = \sum_jn_j^b\overline W_i(x_i - x_j^b, h^b)V_j
References
----------
.. [Vacondio2010] <NAME>, <NAME> and <NAME>, "Accurate
particle splitting for smoothed particle hydrodynamics in shallow water
with shock capturing", Int. J. Numer. Meth. Fluids, 69 (2012), pp.
1377-1410
Notes
-----
The acceleration term given in [Vacondio2010] has incorrect sign.
"""
def __init__(self, dest, sources, dim=2, u_only=False, v_only=False,
alpha=0.0, visc_option=2, rhow=1000.0):
r"""
Parameters
----------
dim : int
number of space dimensions (Default: 2)
u_only : bool
motion of fluid column in x-direction only evaluated
(Default: False)
v_only : bool
motion of fluid column in y-direction only evaluated
(Default: False)
alpha : float
coefficient to control amount of artificial viscosity (Monaghan)
(Default: 0.0)
visc_option : int
artifical viscosity (1) or Lax-Friedrichs flux (2) based
stabilization term (Default: 2)
rhow : float
constant 3-D density of water
"""
self.g = 9.81
self.rhow = rhow
self.ct = self.g / (2*self.rhow)
self.dim = dim
self.u_only = u_only
self.v_only = v_only
self.alpha = alpha
if visc_option == 1:
self.viscous_func = artificial_visc
else:
self.viscous_func = viscosity_LF
super(ParticleAcceleration, self).__init__(dest, sources)
def initialize(self, d_idx, d_tu, d_tv):
d_tu[d_idx] = 0.0
d_tv[d_idx] = 0.0
def loop(self, d_x, d_y, s_x, s_y, d_rho, d_idx, s_m, s_idx, s_rho, d_m,
DWI, DWJ, d_au, d_av, s_alpha, d_alpha, s_p, d_p, d_tu, s_dw,
d_dw, t, s_is_wall_boun_pa, s_tu, d_tv, s_tv, d_h, s_h, d_u, s_u,
d_v, s_v, d_cs, s_cs):
# True if neighbor is wall boundary particle
if s_is_wall_boun_pa[s_idx] == 1:
# Setting artificial viscosity to zero when a particle interacts
# with wall boundary particles
pi_visc = 0.0
# Setting water depth of wall boundary particles same as particle
# interacting with it (For sufficient pressure to prevent wall
# penetration)
s_dw[s_idx] = d_dw[d_idx]
else:
uij = d_u[d_idx] - s_u[s_idx]
vij = d_v[d_idx] - s_v[s_idx]
xij = d_x[d_idx] - s_x[s_idx]
yij = d_y[d_idx] - s_y[s_idx]
rij2 = xij**2 + yij**2
uij_dot_xij = uij * xij
vij_dot_yij = vij * yij
velij_dot_rij = uij_dot_xij + vij_dot_yij
muij = mu_calc(d_h[d_idx], s_h[s_idx], velij_dot_rij, rij2)
if velij_dot_rij < 0:
# Stabilization term
pi_visc = self.viscous_func(self.alpha, rij2, d_h[d_idx],
s_h[s_idx], d_rho[d_idx],
s_rho[s_idx], d_cs[d_idx],
s_cs[s_idx], muij)
else:
pi_visc = 0
tmp1 = (s_dw[s_idx]*self.rhow*self.dim) / s_alpha[s_idx]
tmp2 = (d_dw[d_idx]*self.rhow*self.dim) / d_alpha[d_idx]
# Internal force per unit mass
d_tu[d_idx] += s_m[s_idx] * ((self.ct*tmp1 + 0.5*pi_visc)*DWJ[0] +
(self.ct*tmp2 + 0.5*pi_visc)*DWI[0])
d_tv[d_idx] += s_m[s_idx] * ((self.ct*tmp1 + 0.5*pi_visc)*DWJ[1] +
(self.ct*tmp2 + 0.5*pi_visc)*DWI[1])
def _get_helpers_(self):
return [mu_calc, artificial_visc, viscosity_LF]
def post_loop(self, d_idx, d_u, d_v, d_tu, d_tv, d_au, d_av, d_Sfx, d_Sfy,
d_bx, d_by, d_bxx, d_bxy, d_byy):
vikivi = d_u[d_idx]*d_u[d_idx]*d_bxx[d_idx] \
+ 2*d_u[d_idx]*d_v[d_idx]*d_bxy[d_idx] \
+ d_v[d_idx]*d_v[d_idx]*d_byy[d_idx]
tidotgradbi = d_tu[d_idx]*d_bx[d_idx] + d_tv[d_idx]*d_by[d_idx]
gradbidotgradbi = d_bx[d_idx]**2 + d_by[d_idx]**2
temp3 = self.g + vikivi - tidotgradbi
temp4 = 1 + gradbidotgradbi
if not self.v_only:
# Acceleration along x-direction
d_au[d_idx] = -(temp3/temp4)*d_bx[d_idx] - d_tu[d_idx] \
- d_Sfx[d_idx]
if not self.u_only:
# Acceleration along y-direction
d_av[d_idx] = -(temp3/temp4)*d_by[d_idx] - d_tv[d_idx] \
- d_Sfy[d_idx]
class FluidBottomElevation(Equation):
r"""**Bottom elevation of fluid**
.. math::
b_i = \sum_jb_j^b\overline{W_i}(\textbf{x}_i - \textbf{x}_j^b, h^b)V_j
References
----------
.. [Vacondio2010] <NAME>, <NAME> and <NAME>, "Accurate
particle splitting for smoothed particle hydrodynamics in shallow water
with shock capturing", Int. J. Numer. Meth. Fluids, 69 (2012), pp.
1377-1410
"""
def initialize(self, d_b, d_idx):
d_b[d_idx] = 0.0
def loop_all(self, d_shep_corr, d_x, d_y, d_idx, s_x, s_y, s_V, s_idx, s_h,
SPH_KERNEL, NBRS, N_NBRS):
# Shepard filter
i = declare('int')
xij = declare('matrix(3)')
rij = 0.0
corr_sum = 0.0
for i in range(N_NBRS):
s_idx = NBRS[i]
xij[0] = d_x[d_idx] - s_x[s_idx]
xij[1] = d_y[d_idx] - s_y[s_idx]
rij = sqrt(xij[0]*xij[0] + xij[1]*xij[1])
corr_sum += s_V[s_idx] * SPH_KERNEL.kernel(xij, rij, s_h[s_idx])
d_shep_corr[d_idx] = corr_sum
def loop(self, d_b, d_idx, s_b, s_idx, WJ, s_V, RIJ):
d_b[d_idx] += s_b[s_idx] * WJ * s_V[s_idx]
def post_loop(self, d_b, d_shep_corr, d_idx):
if d_shep_corr[d_idx] > 1e-14:
d_b[d_idx] /= d_shep_corr[d_idx]
class FluidBottomGradient(Equation):
r"""**Bottom gradient of fluid**
.. math::
\nabla b_i &=& \sum_j\nabla b_j^b W_i(\textbf{x}_i - \textbf{x}_j^b,
h^b)V_j
Notes:
It is obtained from a simple SPH interpolation from the gradient of bed
particles
"""
def initialize(self, d_idx, d_bx, d_by):
d_bx[d_idx] = 0.0
d_by[d_idx] = 0.0
def loop(self, d_idx, d_bx, d_by, WJ, s_idx, s_bx, s_by, s_V):
# Bottom gradient of fluid
d_bx[d_idx] += s_bx[s_idx] * WJ * s_V[s_idx]
d_by[d_idx] += s_by[s_idx] * WJ * s_V[s_idx]
class FluidBottomCurvature(Equation):
r"""Bottom curvature of fluid**
.. math::
\nabla^2 b_i = \sum_j\nabla^2 b_j^b W_i(\textbf{x}_i - \textbf{x}_j^b,
h^b)V_j
Notes:
It is obtained from a simple SPH interpolation from the curvature of bed
particles
"""
def initialize(self, d_idx, d_bx, d_by, d_bxx, d_bxy, d_byy):
d_bxx[d_idx] = 0.0
d_bxy[d_idx] = 0.0
d_byy[d_idx] = 0.0
def loop(self, d_idx, d_bxx, d_bxy, d_byy, WJ, s_idx, s_bxx, s_bxy, s_byy,
s_V):
# Bottom curvature of fluid
d_bxx[d_idx] += s_bxx[s_idx] * WJ * s_V[s_idx]
d_bxy[d_idx] += s_bxy[s_idx] * WJ * s_V[s_idx]
d_byy[d_idx] += s_byy[s_idx] * WJ * s_V[s_idx]
class BedGradient(Equation):
r"""**Gradient of bed**
.. math::
\nabla b_i = \sum_jb_j^b\tilde{\nabla}W_i(\textbf{x}_i -
\textbf{x}_j^b, h^b)V_j
References
----------
.. [Vacondio2010] <NAME>, <NAME> and <NAME>, "Accurate
particle splitting for smoothed particle hydrodynamics in shallow water
with shock capturing", Int. J. Numer. Meth. Fluids, 69 (2012), pp.
1377-1410
"""
def initialize(self, d_bx, d_by, d_idx):
d_bx[d_idx] = 0.0
d_by[d_idx] = 0.0
def loop(self, d_bx, d_by, d_idx, s_b, s_idx, DWJ, s_V, RIJ):
if RIJ > 1e-6:
# Gradient of bed
d_bx[d_idx] += s_b[s_idx] * DWJ[0] * s_V[s_idx]
d_by[d_idx] += s_b[s_idx] * DWJ[1] * s_V[s_idx]
class BedCurvature(Equation):
r"""**Curvature of bed**
.. math::
\biggl(\dfrac{\partial^2b}{\partial x^\alpha \partial x^\beta}
\biggr)_i = \sum_{j}^{}\biggl(4\dfrac{x_{ij}^\alphax_{ij}^\beta}
{r_{ij}^2}-\delta^{\alpha\beta}\biggr)\dfrac{b_i - b_j^b}{
\textbf{r}_{ij}\textbf{r}_{ij} + \eta^2}\textbf{r}_{ij}.\tilde{\nabla}
W_i(\textbf{x}_i - \textbf{x}_j^b, h^b)V_j
References
----------
.. [Vacondio2010] <NAME>, <NAME> and <NAME>, "Accurate
particle splitting for smoothed particle hydrodynamics in shallow water
with shock capturing", Int. J. Numer. Meth. Fluids, 69 (2012), pp.
1377-1410
"""
def initialize(self, d_bxx, d_bxy, d_byy, d_idx):
d_bxx[d_idx] = 0.0
d_bxy[d_idx] = 0.0
d_byy[d_idx] = 0.0
def loop(self, d_bxx, d_bxy, d_byy, d_b, d_idx, s_h, s_b, s_idx, XIJ, RIJ,
DWJ, s_V):
if RIJ > 1e-6:
eta = 0.01 * s_h[s_idx]
temp1 = (d_b[d_idx]-s_b[s_idx]) / (RIJ**2+eta**2)
temp2 = XIJ[0]*DWJ[0] + XIJ[1]*DWJ[1]
temp_bxx = ((4*XIJ[0]**2/RIJ**2)-1) * temp1
temp_bxy = (4*XIJ[0]*XIJ[1]/RIJ**2) * temp1
temp_byy = ((4*XIJ[1]**2/RIJ**2)-1) * temp1
# Curvature of bed
d_bxx[d_idx] += temp_bxx * temp2 * s_V[s_idx]
d_bxy[d_idx] += temp_bxy * temp2 * s_V[s_idx]
d_byy[d_idx] += temp_byy * temp2 * s_V[s_idx]
class BedFrictionSourceEval(Equation):
r"""**Friction source term**
.. math::
\textbf{S}_f = \textbf{v}\dfrac{gn^2|\textbf{v}|}{d^{\frac{4}{3}}}
where,
.. math::
n_i = \sum_jn_j^b\overline W_i(x_i - x_j^b, h^b)V_j
"""
def __init__(self, dest, sources):
self.g = 9.8
super(BedFrictionSourceEval, self).__init__(dest, sources)
def initialize(self, d_n, d_idx):
d_n[d_idx] = 0.0
def loop(self, d_n, d_idx, s_n, s_idx, WJ, s_V, RIJ):
if RIJ > 1e-6:
# Manning coefficient
d_n[d_idx] += s_n[s_idx] * WJ * s_V[s_idx]
def post_loop(self, d_idx, d_Sfx, d_Sfy, d_u, d_v, d_n, d_dw):
vmag = sqrt(d_u[d_idx]**2 + d_v[d_idx]**2)
temp = (self.g*d_n[d_idx]**2*vmag) / d_dw[d_idx]**(4.0/3.0)
# Friction source term
d_Sfx[d_idx] = d_u[d_idx] * temp
d_Sfy[d_idx] = d_v[d_idx] * temp
class BoundaryInnerReimannStateEval(Equation):
r"""Evaluates the inner Riemann state of velocity and depth
.. math::
\textbf{v}_i^o = \sum_j\dfrac{m_j^f}{\rho_j^f}\textbf{v}_j^f\bar
W_i(\textbf{x}_i^o - \textbf{x}_j^f, h_o)\\
{d}_i^o = \sum_j\dfrac{m_j^f}{\rho_j^f}d_j^f\bar W_i(\textbf{x}_i^o -
\textbf{x}_j^f, h_o)
References
----------
.. [Vacondio2012] <NAME> et al., "SPH modeling of shallow flow with
open boundaries for practical flood simulation", J. Hydraul. Eng., 2012,
138(6), pp. 530-541.
"""
def initialize(self, d_u_inner_reimann, d_v_inner_reimann,
d_dw_inner_reimann, d_idx):
d_u_inner_reimann[d_idx] = 0.0
d_v_inner_reimann[d_idx] = 0.0
d_dw_inner_reimann[d_idx] = 0.0
def loop_all(self, d_shep_corr, d_x, d_y, d_idx, s_x, s_y, s_m, s_rho,
s_idx, d_h, SPH_KERNEL, NBRS, N_NBRS):
# Shepard filter
i = declare('int')
xij = declare('matrix(3)')
rij = 0.0
corr_sum = 0.0
for i in range(N_NBRS):
s_idx = NBRS[i]
xij[0] = d_x[d_idx] - s_x[s_idx]
xij[1] = d_y[d_idx] - s_y[s_idx]
rij = sqrt(xij[0]*xij[0] + xij[1]*xij[1])
corr_sum += ((s_m[s_idx]/s_rho[s_idx])
* SPH_KERNEL.kernel(xij, rij, d_h[d_idx]))
d_shep_corr[d_idx] = corr_sum
def loop(self, d_u_inner_reimann, d_v_inner_reimann, d_dw_inner_reimann,
d_idx, WI, s_m, s_u, s_v, s_rho, s_dw, s_idx):
tmp = WI * (s_m[s_idx]/s_rho[s_idx])
# Riemann invariants at open boundaries
d_u_inner_reimann[d_idx] += s_u[s_idx] * tmp
d_v_inner_reimann[d_idx] += s_v[s_idx] * tmp
d_dw_inner_reimann[d_idx] += s_dw[s_idx] * tmp
def post_loop(self, d_u_inner_reimann, d_v_inner_reimann,
d_dw_inner_reimann, d_shep_corr, d_idx):
if d_shep_corr[d_idx] > 1e-14:
d_u_inner_reimann[d_idx] /= d_shep_corr[d_idx]
d_v_inner_reimann[d_idx] /= d_shep_corr[d_idx]
d_dw_inner_reimann[d_idx] /= d_shep_corr[d_idx]
class SubCriticalInFlow(Equation):
r"""**Subcritical inflow condition**
..math ::
d_B = \biggl[\frac{1}{2\sqrt{g}}(v_{B,n}-v_{I,n}) + \sqrt{d_I}\biggr]^2
References
----------
.. [Vacondio2012] <NAME> et al., "SPH modeling of shallow flow with
open boundaries for practical flood simulation", J. Hydraul. Eng., 2012,
138(6), pp. 530-541.
Notes
-----
The velocity is imposed at the open boundary.
"""
def __init__(self, dest, dim=2, rhow=1000.0):
r"""
Parameters
----------
dim : int
number of space dimensions (Default: 2)
rhow : float
constant 3-D density of water
"""
self.g = 9.8
self.dim = dim
self.rhow = rhow
super(SubCriticalInFlow, self).__init__(dest, None)
def post_loop(self, d_dw, d_dw_inner_reimann, d_u, d_u_inner_reimann,
d_rho, d_alpha, d_cs, d_idx):
const = 1. / (2.*sqrt(self.g))
# Properties of open boundary particles
d_dw[d_idx] = (const*(d_u_inner_reimann[d_idx] - d_u[d_idx])
+ sqrt(d_dw_inner_reimann[d_idx]))**2
d_rho[d_idx] = d_dw[d_idx] * self.rhow
d_alpha[d_idx] = self.dim * d_rho[d_idx]
d_cs[d_idx] = | sqrt(self.g * d_dw[d_idx]) | numpy.sqrt |
"""
defines:
bdf merge (IN_BDF_FILENAMES)... [-o OUT_BDF_FILENAME]\n'
bdf equivalence IN_BDF_FILENAME EQ_TOL\n'
bdf renumber IN_BDF_FILENAME [-o OUT_BDF_FILENAME]\n'
bdf mirror IN_BDF_FILENAME [-o OUT_BDF_FILENAME] [--plane PLANE] [--tol TOL]\n'
bdf export_mcids IN_BDF_FILENAME [-o OUT_GEOM_FILENAME]\n'
bdf split_cbars_by_pin_flags IN_BDF_FILENAME [-o OUT_BDF_FILENAME]\n'
"""
import os
import sys
from io import StringIO
from typing import List
from cpylog import SimpleLogger
import pyNastran
from pyNastran.bdf.mesh_utils.bdf_renumber import bdf_renumber, superelement_renumber
from pyNastran.bdf.mesh_utils.bdf_merge import bdf_merge
from pyNastran.bdf.mesh_utils.export_mcids import export_mcids
from pyNastran.bdf.mesh_utils.pierce_shells import pierce_shell_model
# testing these imports are up to date
# if something is imported and tested, it should be removed from here
from pyNastran.bdf.mesh_utils.shift import update_nodes
from pyNastran.bdf.mesh_utils.mirror_mesh import write_bdf_symmetric
from pyNastran.bdf.mesh_utils.collapse_bad_quads import convert_bad_quads_to_tris
from pyNastran.bdf.mesh_utils.delete_bad_elements import delete_bad_shells, get_bad_shells
from pyNastran.bdf.mesh_utils.split_cbars_by_pin_flag import split_cbars_by_pin_flag
from pyNastran.bdf.mesh_utils.dev.create_vectorized_numbered import create_vectorized_numbered
from pyNastran.bdf.mesh_utils.remove_unused import remove_unused
from pyNastran.bdf.mesh_utils.free_faces import write_skin_solid_faces
from pyNastran.bdf.mesh_utils.get_oml import get_oml_eids
def cmd_line_create_vectorized_numbered(argv=None, quiet=False): # pragma: no cover
if argv is None:
argv = sys.argv
msg = (
'Usage:\n'
' bdf create_vectorized_numbered IN_BDF_FILENAME [OUT_BDF_FILENAME]\n'
' bdf create_vectorized_numbered -h | --help\n'
' bdf create_vectorized_numbered -v | --version\n'
'\n'
'Positional Arguments:\n'
' IN_BDF_FILENAME the model to convert\n'
" OUT_BDF_FILENAME the converted model name (default=IN_BDF_FILENAME + '_convert.bdf')"
'\n'
'Info:\n'
' -h, --help show this help message and exit\n'
" -v, --version show program's version number and exit\n"
)
if len(argv) == 1:
sys.exit(msg)
from docopt import docopt
ver = str(pyNastran.__version__)
data = docopt(msg, version=ver, argv=argv[1:])
if not quiet: # pragma: no cover
print(data)
bdf_filename_in = data['IN_BDF_FILENAME']
if data['OUT_BDF_FILENAME']:
bdf_filename_out = data['OUT_BDF_FILENAME']
else:
base, ext = os.path.splitext(bdf_filename_in)
bdf_filename_out = base + '_convert' + ext
create_vectorized_numbered(bdf_filename_in, bdf_filename_out)
def cmd_line_equivalence(argv=None, quiet=False):
"""command line interface to bdf_equivalence_nodes"""
if argv is None:
argv = sys.argv
from docopt import docopt
msg = (
'Usage:\n'
' bdf equivalence IN_BDF_FILENAME EQ_TOL [-o OUT_BDF_FILENAME]\n'
' bdf equivalence -h | --help\n'
' bdf equivalence -v | --version\n'
'\n'
"Positional Arguments:\n"
" IN_BDF_FILENAME path to input BDF/DAT/NAS file\n"
" EQ_TOL the spherical equivalence tolerance\n"
#" OUT_BDF_FILENAME path to output BDF/DAT/NAS file\n"
'\n'
'Options:\n'
" -o OUT, --output OUT_BDF_FILENAME path to output BDF/DAT/NAS file\n\n"
'Info:\n'
' -h, --help show this help message and exit\n'
" -v, --version show program's version number and exit\n"
)
if len(argv) == 1:
sys.exit(msg)
ver = str(pyNastran.__version__)
#type_defaults = {
# '--nerrors' : [int, 100],
#}
data = docopt(msg, version=ver, argv=argv[1:])
if not quiet: # pragma: no cover
print(data)
bdf_filename = data['IN_BDF_FILENAME']
bdf_filename_out = data['--output']
if bdf_filename_out is None:
bdf_filename_out = 'merged.bdf'
tol = float(data['EQ_TOL'])
size = 16
from pyNastran.bdf.mesh_utils.bdf_equivalence import bdf_equivalence_nodes
level = 'debug' if not quiet else 'warning'
log = SimpleLogger(level=level, encoding='utf-8', log_func=None)
bdf_equivalence_nodes(bdf_filename, bdf_filename_out, tol,
renumber_nodes=False,
neq_max=10, xref=True,
node_set=None, size=size,
is_double=False,
remove_collapsed_elements=False,
avoid_collapsed_elements=False,
crash_on_collapse=False,
log=log, debug=True)
def cmd_line_bin(argv=None, quiet=False): # pragma: no cover
"""bins the model into nbins"""
if argv is None:
argv = sys.argv
from docopt import docopt
msg = (
"Usage:\n"
#" bdf bin IN_BDF_FILENAME AXIS1 AXIS2 [--cid CID] [--step SIZE]\n"
" bdf bin IN_BDF_FILENAME AXIS1 AXIS2 [--cid CID] [--nbins NBINS]\n"
' bdf bin -h | --help\n'
' bdf bin -v | --version\n'
'\n'
"Positional Arguments:\n"
" IN_BDF_FILENAME path to input BDF/DAT/NAS file\n"
" AXIS1 axis to loop over\n"
" AXIS2 axis to bin\n"
'\n'
'Options:\n'
" --cid CID the coordinate system to bin (default:0)\n"
" --step SIZE the step size for binning\n\n"
" --nbins NBINS the number of bins\n\n"
'Info:\n'
' -h, --help show this help message and exit\n'
" -v, --version show program's version number and exit\n\n"
'Plot z (2) as a function of y (1) in y-stepsizes of 0.1:\n'
' bdf bin fem.bdf 1 2 --cid 0 --step 0.1\n\n'
'Plot z (2) as a function of y (1) with 50 bins:\n'
' bdf bin fem.bdf 1 2 --cid 0 --nbins 50'
)
if len(argv) == 1:
sys.exit(msg)
ver = str(pyNastran.__version__)
#type_defaults = {
# '--nerrors' : [int, 100],
#}
data = docopt(msg, version=ver, argv=argv[1:])
bdf_filename = data['IN_BDF_FILENAME']
axis1 = int(data['AXIS1'])
axis2 = int(data['AXIS2'])
cid = 0
if data['--cid']:
cid = int(data['--cid'])
#stepsize = 0.1
#if data['--step']:
#stepsize = float(data['--step'])
nbins = 10
if data['--nbins']:
nbins = int(data['--nbins'])
assert nbins >= 2, nbins
if not quiet: # pragma: no cover
print(data)
import numpy as np
import matplotlib.pyplot as plt
from pyNastran.bdf.bdf import read_bdf
level = 'debug' if not quiet else 'warning'
log = SimpleLogger(level=level, encoding='utf-8', log_func=None)
model = read_bdf(bdf_filename, log=log)
xyz_cid = model.get_xyz_in_coord(cid=cid, fdtype='float64')
y = xyz_cid[:, axis1]
z = xyz_cid[:, axis2]
plt.figure(1)
#n, bins, patches = plt.hist( [x0,x1,x2], 10, weights=[w0, w1, w2], histtype='bar')
ys = []
#zs = []
zs_min = []
zs_max = []
y0 = y.min()
y1 = y.max()
dy = (y1 - y0) / nbins
y0i = y0
y1i = y0 + dy
for unused_i in range(nbins):
j = np.where((y0i <= y) & (y <= y1i))[0]
if not len(j):
continue
ys.append(y[j].mean())
zs_min.append(z[j].min())
zs_max.append(z[j].max())
y0i += dy
y1i += dy
zs_max = np.array(zs_max)
zs_min = np.array(zs_min)
if not quiet: # pragma: no cover
print('ys = %s' % ys)
print('zs_max = %s' % zs_max)
print('zs_min = %s' % zs_min)
plt.plot(ys, zs_max, 'r-o', label='max')
plt.plot(ys, zs_min, 'b-o', label='min')
plt.plot(ys, zs_max - zs_min, 'g-o', label='delta')
#plt.xlim([y0, y1])
plt.xlabel('Axis %s' % axis1)
plt.ylabel('Axis %s' % axis2)
plt.grid(True)
plt.legend()
plt.show()
def cmd_line_renumber(argv=None, quiet=False):
"""command line interface to bdf_renumber"""
if argv is None:
argv = sys.argv
from docopt import docopt
msg = (
"Usage:\n"
' bdf renumber IN_BDF_FILENAME OUT_BDF_FILENAME [--superelement] [--size SIZE]\n'
' bdf renumber IN_BDF_FILENAME [--superelement] [--size SIZE]\n'
' bdf renumber -h | --help\n'
' bdf renumber -v | --version\n'
'\n'
'Positional Arguments:\n'
' IN_BDF_FILENAME path to input BDF/DAT/NAS file\n'
' OUT_BDF_FILENAME path to output BDF/DAT/NAS file\n'
'\n'
'Options:\n'
'--superelement calls superelement_renumber\n'
'--size SIZE set the field size (default=16)\n\n'
'Info:\n'
' -h, --help show this help message and exit\n'
" -v, --version show program's version number and exit\n"
)
if len(argv) == 1:
sys.exit(msg)
ver = str(pyNastran.__version__)
#type_defaults = {
# '--nerrors' : [int, 100],
#}
data = docopt(msg, version=ver, argv=argv[1:])
if not quiet: # pragma: no cover
print(data)
bdf_filename = data['IN_BDF_FILENAME']
bdf_filename_out = data['OUT_BDF_FILENAME']
if bdf_filename_out is None:
bdf_filename_out = 'renumber.bdf'
size = 16
if data['--size']:
size = int(data['SIZE'])
assert size in [8, 16], f'size={size} args={argv}'
#cards_to_skip = [
#'AEFACT', 'CAERO1', 'CAERO2', 'SPLINE1', 'SPLINE2',
#'AERO', 'AEROS', 'PAERO1', 'PAERO2', 'MKAERO1']
cards_to_skip = []
level = 'debug' if not quiet else 'warning'
log = SimpleLogger(level=level, encoding='utf-8', log_func=None)
if data['--superelement']:
superelement_renumber(bdf_filename, bdf_filename_out, size=size, is_double=False,
starting_id_dict=None, #round_ids=False,
cards_to_skip=cards_to_skip, log=log)
else:
bdf_renumber(bdf_filename, bdf_filename_out, size=size, is_double=False,
starting_id_dict=None, round_ids=False,
cards_to_skip=cards_to_skip, log=log)
def cmd_line_mirror(argv=None, quiet=False):
"""command line interface to write_bdf_symmetric"""
if argv is None:
argv = sys.argv
from docopt import docopt
import pyNastran
msg = (
"Usage:\n"
" bdf mirror IN_BDF_FILENAME [-o OUT_BDF_FILENAME] [--plane PLANE] [--tol TOL]\n"
" bdf mirror IN_BDF_FILENAME [-o OUT_BDF_FILENAME] [--plane PLANE] [--noeq]\n"
' bdf mirror -h | --help\n'
' bdf mirror -v | --version\n'
'\n'
"Positional Arguments:\n"
" IN_BDF_FILENAME path to input BDF/DAT/NAS file\n"
#" OUT_BDF_FILENAME path to output BDF/DAT/NAS file\n"
'\n'
'Options:\n'
" -o OUT, --output OUT_BDF_FILENAME path to output BDF/DAT/NAS file\n"
" --plane PLANE the symmetry plane (xz, yz, xy); default=xz\n"
' --tol TOL the spherical equivalence tolerance; default=1e-6\n'
' --noeq disable equivalencing\n'
"\n" # (default=0.000001)
'Info:\n'
' -h, --help show this help message and exit\n'
" -v, --version show program's version number and exit\n"
)
if len(argv) == 1:
sys.exit(msg)
ver = str(pyNastran.__version__)
#type_defaults = {
# '--nerrors' : [int, 100],
#}
data = docopt(msg, version=ver, argv=argv[1:])
if data['--tol'] is None:
data['TOL'] = 0.000001
if isinstance(data['TOL'], str):
data['TOL'] = float(data['TOL'])
tol = data['TOL']
assert data['--noeq'] in [True, False]
if data['--noeq']:
tol = -1.
plane = 'xz'
if data['--plane'] is not None: # None or str
plane = data['--plane']
if not quiet: # pragma: no cover
print(data)
size = 16
bdf_filename = data['IN_BDF_FILENAME']
bdf_filename_out = data['--output']
if bdf_filename_out is None:
bdf_filename_out = 'mirrored.bdf'
#from io import StringIO
from pyNastran.bdf.bdf import read_bdf
from pyNastran.bdf.mesh_utils.bdf_equivalence import bdf_equivalence_nodes
level = 'debug' if not quiet else 'warning'
log = SimpleLogger(level=level, encoding='utf-8', log_func=None)
model = read_bdf(bdf_filename, log=log)
bdf_filename_stringio = StringIO()
write_bdf_symmetric(model, bdf_filename_stringio, encoding=None, size=size,
is_double=False,
enddata=None, close=False,
plane=plane, log=log)
bdf_filename_stringio.seek(0)
if tol >= 0.0:
bdf_equivalence_nodes(bdf_filename_stringio, bdf_filename_out, tol,
renumber_nodes=False,
neq_max=10, xref=True,
node_set=None, size=size,
is_double=False,
remove_collapsed_elements=False,
avoid_collapsed_elements=False,
crash_on_collapse=False,
debug=True, log=log)
else:
model.log.info('writing mirrored model %s without equivalencing' % bdf_filename_out)
with open(bdf_filename_out, 'w') as bdf_file:
bdf_file.write(bdf_filename_stringio.getvalue())
def cmd_line_merge(argv=None, quiet=False):
"""command line interface to bdf_merge"""
if argv is None:
argv = sys.argv
from docopt import docopt
import pyNastran
msg = (
"Usage:\n"
' bdf merge (IN_BDF_FILENAMES)... [-o OUT_BDF_FILENAME]\n'
' bdf merge -h | --help\n'
' bdf merge -v | --version\n'
'\n'
'Positional Arguments:\n'
' IN_BDF_FILENAMES path to input BDF/DAT/NAS files\n'
'\n'
'Options:\n'
' -o OUT, --output OUT_BDF_FILENAME path to output BDF/DAT/NAS file\n\n'
'Info:\n'
' -h, --help show this help message and exit\n'
" -v, --version show program's version number and exit\n"
)
if len(argv) == 1:
sys.exit(msg)
ver = str(pyNastran.__version__)
#type_defaults = {
# '--nerrors' : [int, 100],
#}
data = docopt(msg, version=ver, argv=argv[1:])
if not quiet: # pragma: no cover
print(data)
size = 16
bdf_filenames = data['IN_BDF_FILENAMES']
bdf_filename_out = data['--output']
if bdf_filename_out is None:
bdf_filename_out = 'merged.bdf'
#cards_to_skip = [
#'AEFACT', 'CAERO1', 'CAERO2', 'SPLINE1', 'SPLINE2',
#'AERO', 'AEROS', 'PAERO1', 'PAERO2', 'MKAERO1']
cards_to_skip = []
bdf_merge(bdf_filenames, bdf_filename_out, renumber=True,
encoding=None, size=size, is_double=False, cards_to_skip=cards_to_skip)
def cmd_line_convert(argv=None, quiet=False):
"""command line interface to bdf_merge"""
if argv is None:
argv = sys.argv
from docopt import docopt
msg = (
"Usage:\n"
' bdf convert IN_BDF_FILENAME [-o OUT_BDF_FILENAME] [--in_units IN_UNITS] [--out_units OUT_UNITS]\n'
' bdf convert -h | --help\n'
' bdf convert -v | --version\n'
'\n'
'Options:\n'
' -o OUT, --output OUT_BDF_FILENAME path to output BDF/DAT/NAS file\n\n'
' --in_units IN_UNITS length,mass\n\n'
' --out_units OUT_UNITS length,mass\n\n'
'Info:\n'
' -h, --help show this help message and exit\n'
" -v, --version show program's version number and exit\n"
)
if len(argv) == 1:
sys.exit(msg)
ver = str(pyNastran.__version__)
#type_defaults = {
# '--nerrors' : [int, 100],
#}
data = docopt(msg, version=ver, argv=argv[1:])
if not quiet: # pragma: no cover
print(data)
#size = 16
bdf_filename = data['IN_BDF_FILENAME']
bdf_filename_out = data['--output']
if bdf_filename_out is None:
#bdf_filename_out = 'merged.bdf'
bdf_filename_out = bdf_filename + '.convert.bdf'
in_units = data['IN_UNITS']
if in_units is None:
in_units = 'm,kg'
out_units = data['OUT_UNITS']
if out_units is None:
out_units = 'm,kg'
length_in, mass_in = in_units.split(',')
length_out, mass_out = out_units.split(',')
units_to = [length_out, mass_out, 's']
units = [length_in, mass_in, 's']
#cards_to_skip = [
#'AEFACT', 'CAERO1', 'CAERO2', 'SPLINE1', 'SPLINE2',
#'AERO', 'AEROS', 'PAERO1', 'PAERO2', 'MKAERO1']
from pyNastran.bdf.bdf import read_bdf
from pyNastran.bdf.mesh_utils.convert import convert
level = 'debug' if not quiet else 'warning'
log = SimpleLogger(level=level, encoding='utf-8', log_func=None)
model = read_bdf(bdf_filename, validate=True, xref=True,
punch=False, save_file_structure=False,
skip_cards=None, read_cards=None,
encoding=None, log=log, debug=True, mode='msc')
convert(model, units_to, units=units)
for prop in model.properties.values():
prop.comment = ''
model.write_bdf(bdf_filename_out)
def cmd_line_scale(argv=None, quiet=False):
if argv is None:
argv = sys.argv
import argparse
#import textwrap
parent_parser = argparse.ArgumentParser(
#prog = 'pyNastranGUI',
#usage = usage,
#description='A foo that bars',
#epilog="And that's how you'd foo a bar",
#formatter_class=argparse.RawDescriptionHelpFormatter,
#description=textwrap.dedent(text),
#version=pyNastran.__version__,
#add_help=False,
)
# positional arguments
parent_parser.add_argument('scale', type=str)
parent_parser.add_argument('INPUT', help='path to output BDF/DAT/NAS file', type=str)
parent_parser.add_argument('OUTPUT', nargs='?', help='path to output file', type=str)
#' --l LENGTH_SF length scale factor\n'
#' --m MASS_SF mass scale factor\n'
#' --f FORCE_SF force scale factor\n'
#' --p PRESSURE_SF pressure scale factor\n'
#' --t TIME_SF time scale factor\n'
#' --v VEL_SF velocity scale factor\n'
parent_parser.add_argument('-l', '--length', help='length scale factor')
parent_parser.add_argument('-m', '--mass', help='mass scale factor')
parent_parser.add_argument('-f', '--force', help='force scale factor')
parent_parser.add_argument('-p', '--pressure', help='pressure scale factor')
parent_parser.add_argument('-t', '--time', help='time scale factor')
parent_parser.add_argument('-V', '--velocity', help='velocity scale factor')
#parent_parser.add_argument('--user_geom', type=str, help='log msg')
#parent_parser.add_argument('-q', '--quiet', help='prints debug messages (default=True)', action='store_true')
#parent_parser.add_argument('-h', '--help', help='show this help message and exits', action='store_true')
parent_parser.add_argument('-v', '--version', action='version',
version=pyNastran.__version__)
args = parent_parser.parse_args(args=argv[1:])
if not quiet: # pragma: no cover
print(args)
scales = []
terms = []
bdf_filename = args.INPUT
bdf_filename_out = args.OUTPUT
if bdf_filename_out is None:
bdf_filename_base, ext = os.path.splitext(bdf_filename)
bdf_filename_out = '%s.scaled%s' % (bdf_filename_base, ext)
#assert bdf_filename_out is not None
if args.mass:
scale = float(args.mass)
scales.append(scale)
terms.append('M')
if args.length:
scale = float(args.length)
scales.append(scale)
terms.append('L')
if args.time:
scale = float(args.time)
scales.append(scale)
terms.append('T')
if args.force:
scale = float(args.force)
scales.append(scale)
terms.append('F')
if args.pressure:
scale = float(args.pressure)
scales.append(scale)
terms.append('P')
if args.velocity:
scale = float(args.velocity)
scales.append(scale)
terms.append('V')
from pyNastran.bdf.mesh_utils.convert import scale_by_terms
level = 'debug' if not quiet else 'warning'
log = SimpleLogger(level=level, encoding='utf-8', log_func=None)
scale_by_terms(bdf_filename, terms, scales, bdf_filename_out=bdf_filename_out, log=log)
def cmd_line_export_mcids(argv=None, quiet=False):
"""command line interface to export_mcids"""
if argv is None:
argv = sys.argv
from docopt import docopt
msg = (
'Usage:\n'
' bdf export_mcids IN_BDF_FILENAME [-o OUT_CSV_FILENAME] [--iplies PLIES] [--no_x | --no_y]\n'
' bdf export_mcids -h | --help\n'
' bdf export_mcids -v | --version\n'
'\n'
'Positional Arguments:\n'
' IN_BDF_FILENAME path to input BDF/DAT/NAS file\n'
'\n'
'Options:\n'
' -o OUT, --output OUT_CSV_FILENAME path to output CSV file\n'
' --iplies PLIES the plies indices to export; comma separated (default=0)\n'
'\n'
'Data Suppression:\n'
" --no_x, don't write the x axis\n"
" --no_y, don't write the y axis\n"
'\n'
'Info:\n'
' -h, --help show this help message and exit\n'
" -v, --version show program's version number and exit\n"
)
_filter_no_args(msg, argv, quiet=quiet)
ver = str(pyNastran.__version__)
#type_defaults = {
# '--nerrors' : [int, 100],
#}
data = docopt(msg, version=ver, argv=argv[1:])
if not quiet: # pragma: no cover
print(data)
#size = 16
bdf_filename = data['IN_BDF_FILENAME']
csv_filename_in = data['--output']
if csv_filename_in is None:
csv_filename_in = 'mcids.csv'
export_xaxis = True
export_yaxis = True
if data['--no_x']:
export_xaxis = False
if data['--no_y']:
export_yaxis = False
csv_filename_base = os.path.splitext(csv_filename_in)[0]
iplies = [0]
if data['--iplies']:
iplies = data['--iplies'].split(',')
iplies = [int(iply) for iply in iplies]
if not quiet: # pragma: no cover
print('iplies = %s' % iplies)
from pyNastran.bdf.bdf import read_bdf
level = 'debug' if not quiet else 'warning'
log = SimpleLogger(level=level, encoding='utf-8', log_func=None)
model = read_bdf(bdf_filename, log=log, xref=False)
model.safe_cross_reference()
for iply in iplies:
csv_filename = csv_filename_base + '_ply=%i.csv' % iply
export_mcids(model, csv_filename,
export_xaxis=export_xaxis, export_yaxis=export_yaxis, iply=iply)
model.log.info('wrote %s' % csv_filename)
def _filter_no_args(msg: str, argv: List[str], quiet: bool=False):
if len(argv) == 1:
if quiet:
sys.exit()
sys.exit(msg)
def cmd_line_free_faces(argv=None, quiet=False):
"""command line interface to bdf free_faces"""
if argv is None:
argv = sys.argv
encoding = sys.getdefaultencoding()
usage = (
'Usage:\n'
' bdf free_faces BDF_FILENAME SKIN_FILENAME [-d] [-l] [-f] [--encoding ENCODE]\n'
' bdf free_faces -h | --help\n'
' bdf free_faces -v | --version\n'
'\n'
)
arg_msg = (
"Positional Arguments:\n"
" BDF_FILENAME path to input BDF/DAT/NAS file\n"
" SKIN_FILENAME path to output BDF/DAT/NAS file\n"
'\n'
'Options:\n'
' -l, --large writes the BDF in large field, single precision format (default=False)\n'
' -d, --double writes the BDF in large field, double precision format (default=False)\n'
f' --encoding ENCODE the encoding method (default=None -> {encoding!r})\n'
'\n'
'Developer:\n'
' -f, --profile Profiles the code (default=False)\n'
'\n'
"Info:\n"
' -h, --help show this help message and exit\n'
" -v, --version show program's version number and exit\n"
)
_filter_no_args(arg_msg, argv, quiet=quiet)
arg_msg += '\n'
examples = (
'Examples\n'
'--------\n'
' bdf free_faces solid.bdf skin.bdf\n'
' bdf free_faces solid.bdf skin.bdf --large\n'
)
import argparse
parent_parser = argparse.ArgumentParser()
# positional arguments
parent_parser.add_argument('BDF_FILENAME', help='path to input BDF/DAT/NAS file', type=str)
parent_parser.add_argument('SKIN_FILENAME', help='path to output BDF/DAT/NAS file', type=str)
size_group = parent_parser.add_mutually_exclusive_group()
size_group.add_argument('-d', '--double', help='writes the BDF in large field, single precision format', action='store_true')
size_group.add_argument('-l', '--large', help='writes the BDF in large field, double precision format', action='store_true')
size_group.add_argument('--encoding', help='the encoding method (default=None -> {repr(encoding)})', type=str)
parent_parser.add_argument('--profile', help='Profiles the code', action='store_true')
parent_parser.add_argument('-v', '--version', action='version', version=pyNastran.__version__)
from pyNastran.utils.arg_handling import argparse_to_dict, update_message
update_message(parent_parser, usage, arg_msg, examples)
if not quiet:
print(argv)
args = parent_parser.parse_args(args=argv[2:])
data = argparse_to_dict(args)
if not quiet: # pragma: no cover
for key, value in sorted(data.items()):
print("%-12s = %r" % (key.strip('--'), value))
import time
time0 = time.time()
is_double = False
if data['double']:
size = 16
is_double = True
elif data['large']:
size = 16
else:
size = 8
bdf_filename = data['BDF_FILENAME']
skin_filename = data['SKIN_FILENAME']
from pyNastran.bdf.mesh_utils.bdf_equivalence import bdf_equivalence_nodes
tol = 1e-005
bdf_filename_merged = 'merged.bdf'
level = 'debug' if not quiet else 'warning'
log = SimpleLogger(level=level, encoding='utf-8', log_func=None)
bdf_equivalence_nodes(bdf_filename, bdf_filename_merged, tol,
renumber_nodes=False, neq_max=10, xref=True,
node_set=None,
size=8, is_double=is_double,
remove_collapsed_elements=False,
avoid_collapsed_elements=False,
crash_on_collapse=False, log=log, debug=True)
if not quiet: # pragma: no cover
print('done with equivalencing')
write_skin_solid_faces(
bdf_filename_merged, skin_filename,
write_solids=False, write_shells=True,
size=size, is_double=is_double, encoding=None, log=log,
)
if not quiet: # pragma: no cover
print('total time: %.2f sec' % (time.time() - time0))
def cmd_line_split_cbars_by_pin_flag(argv=None, quiet=False):
"""command line interface to split_cbars_by_pin_flag"""
if argv is None:
argv = sys.argv
from docopt import docopt
msg = (
'Usage:\n'
' bdf split_cbars_by_pin_flags IN_BDF_FILENAME [-o OUT_BDF_FILENAME] [-p PIN_FLAGS_CSV_FILENAME]\n'
' bdf split_cbars_by_pin_flags -h | --help\n'
' bdf split_cbars_by_pin_flags -v | --version\n'
'\n'
"Positional Arguments:\n"
" IN_BDF_FILENAME path to input BDF/DAT/NAS file\n"
'\n'
'Options:\n'
" -o OUT, --output OUT_BDF_FILENAME path to output BDF file\n"
" -p PIN, --pin PIN_FLAGS_CSV_FILENAME path to pin_flags_csv file\n\n"
'Info:\n'
' -h, --help show this help message and exit\n'
" -v, --version show program's version number and exit\n"
)
_filter_no_args(msg, argv, quiet=quiet)
ver = str(pyNastran.__version__)
#type_defaults = {
# '--nerrors' : [int, 100],
#}
data = docopt(msg, version=ver, argv=argv[1:])
if not quiet: # pragma: no cover
print(data)
#size = 16
bdf_filename_in = data['IN_BDF_FILENAME']
bdf_filename_out = data['--output']
if bdf_filename_out is None:
bdf_filename_out = 'model_new.bdf'
pin_flags_filename = data['--pin']
if pin_flags_filename is None:
pin_flags_filename = 'pin_flags.csv'
split_cbars_by_pin_flag(bdf_filename_in, pin_flags_filename=pin_flags_filename,
bdf_filename_out=bdf_filename_out)
def cmd_line_transform(argv=None, quiet=False):
"""command line interface to export_caero_mesh"""
if argv is None:
argv = sys.argv
from docopt import docopt
msg = (
'Usage:\n'
' bdf transform IN_BDF_FILENAME [-o OUT_BDF_FILENAME] [--shift XYZ]\n'
' bdf transform -h | --help\n'
' bdf transform -v | --version\n'
'\n'
'Positional Arguments:\n'
' IN_BDF_FILENAME path to input BDF/DAT/NAS file\n'
'\n'
'Options:\n'
' -o OUT, --output OUT_BDF_FILENAME path to output BDF file\n'
'\n'
'Info:\n'
' -h, --help show this help message and exit\n'
" -v, --version show program's version number and exit\n"
)
_filter_no_args(msg, argv, quiet=quiet)
ver = str(pyNastran.__version__)
#type_defaults = {
# '--nerrors' : [int, 100],
#}
data = docopt(msg, version=ver, argv=argv[1:])
if not quiet: # pragma: no cover
print(data)
#size = 16
bdf_filename = data['IN_BDF_FILENAME']
bdf_filename_out = data['--output']
if bdf_filename_out is None:
bdf_filename_out = 'transform.bdf'
dxyz = None
import numpy as np
if data['--shift']:
dxyz = np.array(data['XYZ'].split(','), dtype='float64')
assert len(dxyz) == 3, dxyz
from pyNastran.bdf.bdf import read_bdf
level = 'debug' if not quiet else 'warning'
log = SimpleLogger(level=level, encoding='utf-8', log_func=None)
model = read_bdf(bdf_filename, log=log)
nid_cp_cd, xyz_cid0, unused_xyz_cp, unused_icd_transform, unused_icp_transform = model.get_xyz_in_coord_array(
cid=0, fdtype='float64', idtype='int32')
update_nodes_flag = False
# we pretend to change the SPOINT location
if dxyz is not None:
xyz_cid0 += dxyz
update_nodes_flag = True
if update_nodes_flag:
update_nodes(model, nid_cp_cd, xyz_cid0)
model.write_bdf(bdf_filename_out)
def cmd_line_filter(argv=None, quiet=False): # pragma: no cover
"""command line interface to bdf filter"""
if argv is None:
argv = sys.argv
from docopt import docopt
msg = (
'Usage:\n'
' bdf filter IN_BDF_FILENAME [-o OUT_BDF_FILENAME]\n'
' bdf filter IN_BDF_FILENAME [-o OUT_BDF_FILENAME] [--x YSIGN_X] [--y YSIGN_Y] [--z YSIGN_Z]\n'
' bdf filter -h | --help\n'
' bdf filter -v | --version\n'
'\n'
'Positional Arguments:\n'
' IN_BDF_FILENAME path to input BDF/DAT/NAS file\n'
'\n'
'Options:\n'
' -o OUT, --output OUT_BDF_FILENAME path to output BDF file (default=filter.bdf)\n'
" --x YSIGN_X a string (e.g., '< 0.')\n"
" --y YSIGN_Y a string (e.g., '< 0.')\n"
" --z YSIGN_Z a string (e.g., '< 0.')\n"
'\n'
'Info:\n'
' -h, --help show this help message and exit\n'
" -v, --version show program's version number and exit\n"
'\n'
'Examples\n'
'1. remove unused cards:\n'
' >>> bdf filter fem.bdf'
'2. remove GRID points and associated cards with y value < 0:\n'
" >>> bdf filter fem.bdf --y '< 0.'"
)
_filter_no_args(msg, argv, quiet=quiet)
ver = str(pyNastran.__version__)
#type_defaults = {
# '--nerrors' : [int, 100],
#}
data = docopt(msg, version=ver, argv=argv[1:])
if not quiet: # pragma: no cover
print(data)
#size = 16
bdf_filename = data['IN_BDF_FILENAME']
bdf_filename_out = data['--output']
if bdf_filename_out is None:
bdf_filename_out = 'filter.bdf'
import numpy as np
func_map = {
'<' : np.less,
'>' : np.greater,
'<=' : np.less_equal,
'>=' : np.greater_equal,
}
xsign = None
ysign = None
zsign = None
if data['--x']:
xsign, xval = data['--x'].split(' ')
xval = float(xval)
assert xsign in ['<', '>', '<=', '>='], xsign
if data['--y']: # --y < 0
ysign, yval = data['--y'].split(' ')
yval = float(yval)
assert ysign in ['<', '>', '<=', '>='], ysign
if data['--z']:
zsign, zval = data['--z'].split(' ')
zval = float(zval)
assert zsign in ['<', '>', '<=', '>='], zsign
from pyNastran.bdf.bdf import read_bdf
level = 'debug' if not quiet else 'warning'
log = SimpleLogger(level=level, encoding='utf-8', log_func=None)
model = read_bdf(bdf_filename, log=log)
#nid_cp_cd, xyz_cid0, xyz_cp, icd_transform, icp_transform = model.get_xyz_in_coord_array(
#cid=0, fdtype='float64', idtype='int32')
eids = []
xyz_cid0 = []
for eid, elem in sorted(model.elements.items()):
xyz = elem.Centroid()
xyz_cid0.append(xyz)
eids.append(eid)
xyz_cid0 = np.array(xyz_cid0)
eids = | np.array(eids) | numpy.array |
# coding:=utf-8
# Copyright 2020 Tencent. All rights reserved.
#
# Licensed under the Apache License, Version 2.0 (the 'License');
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an 'AS IS' BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
''' Applications based on Wide & Deep model. '''
import copy
import numpy as np
from uf.tools import tf
from .base import ClassifierModule
from .bert import BERTClassifier, get_bert_config
from .albert import get_albert_config
from uf.modeling.bert import BERTEncoder
from uf.modeling.albert import ALBERTEncoder
from uf.modeling.wide_and_deep import WideAndDeepDecoder
from uf.tokenization.word_piece import get_word_piece_tokenizer
import uf.utils as utils
class WideAndDeepClassifier(BERTClassifier, ClassifierModule):
''' Single-label classifier on Wide & Deep model with BERT. '''
_INFER_ATTRIBUTES = copy.deepcopy(BERTClassifier._INFER_ATTRIBUTES)
_INFER_ATTRIBUTES['wide_features'] = \
'A list of possible values for `Wide` features (integer or string)'
def __init__(self,
config_file,
vocab_file,
max_seq_length=128,
label_size=None,
init_checkpoint=None,
output_dir=None,
gpu_ids=None,
wide_features=None,
deep_module='bert',
do_lower_case=True,
truncate_method='LIFO'):
super(ClassifierModule, self).__init__(
init_checkpoint, output_dir, gpu_ids)
self.batch_size = 0
self.max_seq_length = max_seq_length
self.label_size = label_size
self.truncate_method = truncate_method
self.wide_features = wide_features
self._deep_module = deep_module
self._id_to_label = None
self.__init_args__ = locals()
if deep_module == 'albert':
self.bert_config = get_albert_config(config_file)
else:
self.bert_config = get_bert_config(config_file)
assert deep_module in ('bert', 'roberta', 'albert', 'electra'), (
'Invalid value of `deep_module`: %s. Pick one from '
'`bert`, `roberta`, `albert` and `electra`.')
self.tokenizer = get_word_piece_tokenizer(vocab_file, do_lower_case)
self._key_to_depths = get_key_to_depths(
self.bert_config.num_hidden_layers)
if '[CLS]' not in self.tokenizer.vocab:
self.tokenizer.add('[CLS]')
self.bert_config.vocab_size += 1
tf.logging.info('Add necessary token `[CLS]` into vocabulary.')
if '[SEP]' not in self.tokenizer.vocab:
self.tokenizer.add('[SEP]')
self.bert_config.vocab_size += 1
tf.logging.info('Add necessary token `[SEP]` into vocabulary.')
def convert(self, X=None, y=None, sample_weight=None, X_tokenized=None,
is_training=False, is_parallel=False):
self._assert_legal(X, y, sample_weight, X_tokenized)
if is_training:
assert y is not None, '`y` can\'t be None.'
if is_parallel:
assert self.label_size, ('Can\'t parse data on multi-processing '
'when `label_size` is None.')
n_inputs = None
data = {}
# convert X
if X or X_tokenized:
tokenized = False if X else X_tokenized
(input_ids, input_mask, segment_ids,
n_wide_features, wide_features) = self._convert_X(
X_tokenized if tokenized else X, tokenized=tokenized)
data['input_ids'] = np.array(input_ids, dtype=np.int32)
data['input_mask'] = np.array(input_mask, dtype=np.int32)
data['segment_ids'] = | np.array(segment_ids, dtype=np.int32) | numpy.array |
import os
import numpy as np
import pandas as pd
from sklearn import linear_model
def allign_alleles(df):
"""Look for reversed alleles and inverts the z-score for one of them.
Here, we take advantage of numpy's vectorized functions for performance.
"""
d = {'A': 0, 'C': 1, 'G': 2, 'T': 3}
a = [] # array of alleles
for colname in ['A1_ref', 'A2_ref', 'A1_gen', 'A2_gen', 'A1_y', 'A2_y']:
tmp = np.empty(len(df[colname]), dtype=int)
for k, v in d.items():
tmp[np.array(df[colname]) == k] = v
a.append(tmp)
matched_alleles_gen = (((a[0] == a[2]) & (a[1] == a[3])) |
((a[0] == 3 - a[2]) & (a[1] == 3 - a[3])))
reversed_alleles_gen = (((a[0] == a[3]) & (a[1] == a[2])) |
((a[0] == 3 - a[3]) & (a[1] == 3 - a[2])))
matched_alleles_y = (((a[0] == a[4]) & (a[1] == a[5])) |
((a[0] == 3 - a[4]) & (a[1] == 3 - a[5])))
reversed_alleles_y = (((a[0] == a[5]) & (a[1] == a[4])) |
((a[0] == 3 - a[5]) & (a[1] == 3 - a[4])))
df['Z_y'] *= -2 * reversed_alleles_y + 1
df['reversed'] = reversed_alleles_gen
df = df[((matched_alleles_y|reversed_alleles_y)&(matched_alleles_gen|reversed_alleles_gen))]
def get_files(file_name, chr):
if '@' in file_name:
valid_files = []
if chr is None:
for i in range(1, 23):
cur_file = file_name.replace('@', str(i))
if os.path.isfile(cur_file):
valid_files.append(cur_file)
else:
raise ValueError('No file matching {} for chr {}'.format(
file_name, i))
else:
cur_file = file_name.replace('@', chr)
if os.path.isfile(cur_file):
valid_files.append(cur_file)
else:
raise ValueError('No file matching {} for chr {}'.format(
file_name, chr))
return valid_files
else:
if os.path.isfile(file_name):
return [file_name]
else:
ValueError('No files matching {}'.format(file_name))
def prep(bfile, genotype, sumstats2, N2, phenotype, covariates, chr, start, end):
bim_files = get_files(bfile + '.bim', chr)
genotype_files = get_files(genotype + '.bim', chr)
# read in bim files
bims = [pd.read_csv(f,
header=None,
names=['CHR', 'SNP', 'CM', 'BP', 'A1', 'A2'],
delim_whitespace=True) for f in bim_files]
bim = pd.concat(bims, ignore_index=True)
genotype_bims = [pd.read_csv(f,
header=None,
names=['CHR', 'SNP', 'CM', 'BP', 'A1', 'A2'],
delim_whitespace=True) for f in genotype_files]
genotype_bim = pd.concat(genotype_bims, ignore_index=True)
if chr is not None:
if start is None:
start = 0
if end is None:
end = float('inf')
genotype_bim = genotype_bim[np.logical_and(np.logical_and(genotype_bim['CHR']==chr, genotype_bim['BP']<=end), genotype_bim['BP']>=start)].reset_index(drop=True)
bim = bim[np.logical_and( | np.logical_and(bim['CHR']==chr, bim['BP']<=end) | numpy.logical_and |
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
Created on Thu Mar 2 16:52:27 2017
@author: abauville
"""
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
Created on Fri Feb 24 15:21:26 2017
@author: abauville
"""
import numpy as np
from numpy import sin, cos, tan, arcsin, arccos, arctan, pi
import matplotlib.pyplot as plt
degree = 180.0/pi
# =============================================================================
#
# Functions for fault diagram plotting
#
# =============================================================================
def plotFaultArrow(x,y,theta, L=1, sense=0, spacing=0.1, color="r",angleHead=20.0 * pi/180.0,headL = .5,ax=plt,linewidth = 1):
# sense 0: sinistral, 1:dextral
x = x + sin(theta)*spacing
y = y - cos(theta)*spacing
segment = np.array((-1,1)) * L
segmentHead = np.array((0,2)) * L*headL
ax.plot(x+cos(theta)*segment, y + sin(theta)*segment,color=color,linewidth=linewidth)
if ((spacing>0) & (sense == 0)):
ax.plot(x+L*cos(theta) - cos(angleHead+theta)*(segmentHead), y+L*sin(theta) - sin(angleHead+theta)*(segmentHead),color=color,linewidth=linewidth)
elif ((spacing>0) & (sense == 1)):
ax.plot(x-L*cos(theta) + cos(-angleHead+theta)*(segmentHead), y-L*sin(theta) + sin(-angleHead+theta)*(segmentHead),color=color,linewidth=linewidth)
elif ((spacing<=0) & (sense == 0)):
ax.plot(x-L*cos(theta) + cos(angleHead+theta)*(segmentHead), y-L*sin(theta) + sin(angleHead+theta)*(segmentHead),color=color,linewidth=linewidth)
elif ((spacing<=0) & (sense == 1)):
ax.plot(x+L*cos(theta) - cos(-angleHead+theta)*(segmentHead), y+L*sin(theta) - sin(-angleHead+theta)*(segmentHead),color=color,linewidth=linewidth)
else:
raise ValueError("sense must be 0 or 1")
def plotArrow(x,y,theta, L=1, color="r", sense=0, angleHead=20.0 * pi/180.0,headL = .5,ax=plt,linewidth = 1):
# sense 0: sinistral, 1:dextral
x = x# + sin(theta)*spacing
y = y# - cos(theta)*spacing
segment = np.array((-1,1)) * L
segmentHead = np.array((0,2)) * L*headL
ax.plot(x+cos(theta)*segment, y + sin(theta)*segment,color=color,linewidth=linewidth)
if sense == 0:
ax.plot(x+L*cos(theta) - cos(angleHead+theta)*(segmentHead), y+L*sin(theta) - sin(angleHead+theta)*(segmentHead),color=color,linewidth=linewidth)
# elif ((spacing>0) & (sense == 1)):
ax.plot(x+L*cos(theta) - cos(-angleHead+theta)*(segmentHead), y+L*sin(theta) - sin(-angleHead+theta)*(segmentHead),color=color,linewidth=linewidth)
elif sense == 1:
# ax.plot(x-L*cos(theta) + cos(-angleHead+theta)*(segmentHead), y-L*sin(theta) + sin(-angleHead+theta)*(segmentHead),color=color,linewidth=linewidth)
ax.plot(x-L*cos(theta) + cos(angleHead+theta)*(segmentHead), y-L*sin(theta) + sin(angleHead+theta)*(segmentHead),color=color,linewidth=linewidth)
# elif ((spacing<=0) & (sense == 1)):
ax.plot(x-L*cos(theta) + cos(-angleHead+theta)*(segmentHead), y-L*sin(theta) + sin(-angleHead+theta)*(segmentHead),color=color,linewidth=linewidth)
else:
raise ValueError("sense must be 0 or 1")
def plotFaultDiagram(Tau,psi, L=1,colorFault="r",colorSigma1="b",Larrow=.15,PosArrow=.66,angleHeadArrow=20.0 * pi/180.0, spacing=.1,ax=plt,refAxes=0,faultLinewidth=1,arrowLinewidth=1,sigma1Linewidth=1,polar=0):
segment = np.array((-1,1)) * L
thetaA = psi+30*pi/180
thetaB = psi-30*pi/180
if (refAxes==0):
Tau = Tau
psiPos = psi*degree
else:
Tau = ( (Tau - refAxes.axis()[0])/(refAxes.axis()[1]-refAxes.axis()[0]) - ax.axis()[0] ) * (ax.axis()[1]-ax.axis()[0])
psiPos = ( (psi*degree - refAxes.axis()[2])/(refAxes.axis()[3]-refAxes.axis()[2]) - ax.axis()[2] ) * (ax.axis()[3]-ax.axis()[2])
if (polar==0):
# Sigma1 dir
ax.plot(Tau+cos(psi)*segment, psiPos + sin(psi)*segment,color=colorSigma1,linewidth=sigma1Linewidth)
# Faults
ax.plot(Tau+cos(thetaA)*segment, psiPos + sin(thetaA)*segment,color=[.8,.5,.2],linewidth=faultLinewidth)
ax.plot(Tau+cos(thetaB)*segment, psiPos + sin(thetaB)*segment,color=[.6,.3,.6],linewidth=faultLinewidth)
else:
# Sigma1 dir
ax.plot(Tau*cos(psi)+cos(psi)*segment, Tau*sin(psi) + sin(psi)*segment,color=colorSigma1,linewidth=sigma1Linewidth)
# Faults
ax.plot(Tau*cos(psi)+cos(thetaA)*segment, Tau*sin(psi) + sin(thetaA)*segment,color=[.8,.5,.2],linewidth=faultLinewidth)
ax.plot(Tau*cos(psi)+cos(thetaB)*segment, Tau*sin(psi) + sin(thetaB)*segment,color=[.6,.3,.6],linewidth=faultLinewidth)
# ax.plot(Tau+cos(thetaA)*segment, psiPos + sin(thetaA)*segment,color=colorFault,linewidth=faultLinewidth)
# ax.plot(Tau+cos(thetaB)*segment, psiPos + sin(thetaB)*segment,color=colorFault,linewidth=faultLinewidth)
# Arrows
# All arrows
# plotFaultArrow(Tau-cos(thetaA)*PosArrow*L,psiPos-sin(thetaA)*PosArrow*L,thetaA,sense=1,spacing=spacing,L=Larrow,ax=ax,linewidth=arrowLinewidth,angleHead=angleHeadArrow)
# plotFaultArrow(Tau-cos(thetaA)*PosArrow*L,psiPos-sin(thetaA)*PosArrow*L,thetaA,sense=1,spacing=-spacing,L=Larrow,ax=ax,linewidth=arrowLinewidth,angleHead=angleHeadArrow)
# plotFaultArrow(Tau-cos(thetaB)*PosArrow*L,psiPos-sin(thetaB)*PosArrow*L,thetaB,sense=0,spacing=-spacing,L=Larrow,ax=ax,linewidth=arrowLinewidth,angleHead=angleHeadArrow)
# plotFaultArrow(Tau-cos(thetaB)*PosArrow*L,psiPos-sin(thetaB)*PosArrow*L,thetaB,sense=0,spacing= spacing,L=Larrow,ax=ax,linewidth=arrowLinewidth,angleHead=angleHeadArrow)
#
# plotFaultArrow(Tau+cos(thetaA)*PosArrow*L,psiPos+sin(thetaA)*PosArrow*L,thetaA,sense=1,spacing= spacing,L=Larrow,ax=ax,linewidth=arrowLinewidth,angleHead=angleHeadArrow)
# plotFaultArrow(Tau+cos(thetaA)*PosArrow*L,psiPos+sin(thetaA)*PosArrow*L,thetaA,sense=1,spacing=-spacing,L=Larrow,ax=ax,linewidth=arrowLinewidth,angleHead=angleHeadArrow)
# plotFaultArrow(Tau+cos(thetaB)*PosArrow*L,psiPos+sin(thetaB)*PosArrow*L,thetaB,sense=0,spacing=-spacing,L=Larrow,ax=ax,linewidth=arrowLinewidth,angleHead=angleHeadArrow)
# plotFaultArrow(Tau+cos(thetaB)*PosArrow*L,psiPos+sin(thetaB)*PosArrow*L,thetaB,sense=0,spacing= spacing,L=Larrow,ax=ax,linewidth=arrowLinewidth,angleHead=angleHeadArrow)
#
#
# # Outer arrows only
# plotFaultArrow(Tau-cos(thetaA)*PosArrow*L,psiPos-sin(thetaA)*PosArrow*L,thetaA,sense=1,spacing=spacing,L=Larrow,ax=ax,linewidth=arrowLinewidth,angleHead=angleHeadArrow)
# plotFaultArrow(Tau-cos(thetaB)*PosArrow*L,psiPos-sin(thetaB)*PosArrow*L,thetaB,sense=0,spacing=-spacing,L=Larrow,ax=ax,linewidth=arrowLinewidth,angleHead=angleHeadArrow)
#
# plotFaultArrow(Tau+cos(thetaA)*PosArrow*L,psiPos+sin(thetaA)*PosArrow*L,thetaA,sense=1,spacing=-spacing,L=Larrow,ax=ax,linewidth=arrowLinewidth,angleHead=angleHeadArrow)
# plotFaultArrow(Tau+cos(thetaB)*PosArrow*L,psiPos+sin(thetaB)*PosArrow*L,thetaB,sense=0,spacing= spacing,L=Larrow,ax=ax,linewidth=arrowLinewidth,angleHead=angleHeadArrow)
#
if (polar==0):
# Inner arrows only
plotFaultArrow(Tau-cos(thetaA)*PosArrow*L,psiPos-sin(thetaA)*PosArrow*L,thetaA,sense=1,spacing=-spacing,L=Larrow,ax=ax,linewidth=arrowLinewidth,angleHead=angleHeadArrow)
plotFaultArrow(Tau-cos(thetaB)*PosArrow*L,psiPos-sin(thetaB)*PosArrow*L,thetaB,sense=0,spacing= spacing,L=Larrow,ax=ax,linewidth=arrowLinewidth,angleHead=angleHeadArrow)
plotFaultArrow(Tau+cos(thetaA)*PosArrow*L,psiPos+sin(thetaA)*PosArrow*L,thetaA,sense=1,spacing= spacing,L=Larrow,ax=ax,linewidth=arrowLinewidth,angleHead=angleHeadArrow)
plotFaultArrow(Tau+cos(thetaB)*PosArrow*L,psiPos+sin(thetaB)*PosArrow*L,thetaB,sense=0,spacing=-spacing,L=Larrow,ax=ax,linewidth=arrowLinewidth,angleHead=angleHeadArrow)
else:
# Inner arrows only
plotFaultArrow(Tau*cos(psi)-cos(thetaA)*PosArrow*L,Tau*sin(psi)-sin(thetaA)*PosArrow*L,thetaA,sense=1,spacing=-spacing,L=Larrow,ax=ax,linewidth=arrowLinewidth,angleHead=angleHeadArrow)
plotFaultArrow(Tau*cos(psi)-cos(thetaB)*PosArrow*L,Tau*sin(psi)-sin(thetaB)*PosArrow*L,thetaB,sense=0,spacing= spacing,L=Larrow,ax=ax,linewidth=arrowLinewidth,angleHead=angleHeadArrow)
plotFaultArrow(Tau*cos(psi)+cos(thetaA)*PosArrow*L,Tau*sin(psi)+ | sin(thetaA) | numpy.sin |
# Food Bank Problem
import sys
import importlib
import numpy as np
from scipy.optimize import minimize
import scipy
# ## OPT - via Convex Programming
# Calculates the optimal solution for the offline problem with convex programming
def solve(W, n, k, budget, size):
# Objective function in the nash social welfare
# Note we take the negative one to turn it into a minimization problem
def objective(x, w, n, k, size):
X = np.reshape(x, (n,k))
W = np.reshape(w, (n, k))
value = np.zeros(n)
for i in range(n):
value[i] = np.log(np.dot(X[i,:], W[i,:]))
return (-1) * np.dot(size, value)
w = W.flatten()
obj = lambda x: objective(x, w, n, k, size)
# Ensures that the allocations are positive
bds = scipy.optimize.Bounds([0 for _ in range(n*k)], [np.inf for _ in range(n*k)])
B = np.zeros((k, n*k))
for i in range(n):
B[:,k*i:k*(i+1)] = size[i]*np.eye(k)
# print(B)
# Enforces the budget constraint
constr = scipy.optimize.LinearConstraint(B, np.zeros(k), budget)
x0 = np.zeros(n*k)
# Initial solution starts out with equal allocation B / S
index = 0
for i in range(n):
for j in range(k):
x0[index] = budget[j] / np.sum(size)
index += 1
sol = minimize(obj, x0, bounds=bds, constraints = constr, tol = 10e-8)
return sol.x, sol
# Calculates the optimal solution for the offline problem with convex programming
# Note that this program instead solves for the optimization problem in a different form, where now
# the histogram is used directly in the original optimization problem instead of rewriting the problem
# as maximizing over types. This was used for investigation, and not as a primary proposed heuristic in the paper.
def solve_weights(weight_matrix, weight_distribution, n, k, budget, size):
# Similar objective, but now multiplying by the probability agent i has type j
def objective(x, weight_matrix, n, k, size, weight_distribution):
num_types = weight_distribution.shape[1]
X = np.reshape(x, (n,k))
value = np.zeros(n)
for i in range(n):
value[i] = np.sum([weight_distribution[i,j] * np.log(np.dot(X[i,:], weight_matrix[j,:])) for j in range(num_types)])
return (-1) * np.dot(size, value)
obj = lambda x: objective(x, weight_matrix, n, k, size, weight_distribution)
# Constraints are the same as before, along with initial solution
bds = scipy.optimize.Bounds([0 for _ in range(n*k)], [np.inf for _ in range(n*k)])
B = np.zeros((k, n*k))
for i in range(n):
B[:,k*i:k*(i+1)] = size[i]*np.eye(k)
constr = scipy.optimize.LinearConstraint(B, np.zeros(k), budget)
x0 = np.zeros(n*k)
index = 0
for i in range(n):
for j in range(k):
x0[index] = budget[j] / np.sum(size)
index += 1
sol = minimize(obj, x0, bounds=bds, constraints = constr, tol = 10e-8)
return sol.x, sol
# proportional solution, i.e. equal allocation B / S
def proportional_alloc(n, k, budget, size):
allocations = np.zeros((n,k))
for i in range(n):
allocations[i, :] = budget / np.sum(size)
return allocations
# Calculates the offline optimal solution just utilizing the distribution and not adapting to realized types
def offline_alloc(weight_matrix, weight_distribution, n, k, budget, size):
allocations = np.zeros((n,k))
weight_dist = np.asarray([weight_distribution for i in range(n)])
alloc, _ = solve_weights(np.asarray(weight_matrix), np.asarray(weight_dist), n, k, budget, size)
allocations = np.reshape(alloc, (n,k))
return allocations
# Implements the ET - Online heuristic algorithm
def et_online(expected_weights, observed_weights, n, k, budget, size):
allocations = np.zeros((n,k))
current_budget = np.copy(budget)
for i in range(n):
if i == n-1: # Last agent gets the maximum of earlier allocations or the remaining budget
allocations[i, :] = [max(0, min(np.max(allocations[:, j]), current_budget[j] / size[i])) for j in range(k)]
current_budget -= size[i] * allocations[i,:]
else:
cur_n = n - i # Solves the eisenbergt gale program with future weights taken to be their expectation
weights = expected_weights[i:,:]
weights[0, :] = observed_weights[i, :]
alloc, _ = solve(weights, cur_n, k, current_budget, size[i:])
alloc = np.reshape(alloc, (cur_n, k))
allocations[i, :] = [max(0, min(alloc[0, j], current_budget[j] / size[i])) for j in range(k)] # solves the eisenberg gale
current_budget -= size[i]*allocations[i, :] # reduces budget for next iteration
return allocations
# Implements the ET - Full heuristic algorithm
def et_full(expected_weights, observed_weights, n, k, budget, size):
allocations = np.zeros((n,k))
current_budget = np.copy(budget)
weights = np.copy(expected_weights)
for i in range(n):
if i == n-1:
allocations[i, :] = [max(0, min(np.max(allocations[:, j]), current_budget[j] / size[i])) for j in range(k)]
current_budget -= size[i] * allocations[i,:]
else:
weights[i, :] = observed_weights[i, :] # Replaces the weights with the observed one
alloc, _ = solve(weights, n, k, budget, size) # Solves for the allocation, and makes it
alloc = np.reshape(alloc, (n,k))
allocations[i, :] = [max(0, min(current_budget[j] / size[i], alloc[i,j])) for j in range(k)]
current_budget -= size[i]*allocations[i,:] # Reduces the budget
return allocations
# Implements the Hope-Full heuristic algorithm
def hope_full(weight_matrix, weight_distribution, observed_types, n, k, budget, size):
num_types = len(weight_distribution)
allocations = np.zeros((n,k))
current_budget = np.copy(budget)
for i in range(n):
size_factors = np.zeros(num_types) # Calculates the number of types and the N_\theta terms
for m in range(n):
if m <= i:
size_factors[observed_types[m]] += size[m]
elif m > i:
size_factors += size[m] * weight_distribution
obs_type = observed_types[i]
alloc, _ = solve(weight_matrix, num_types, k, budget, size_factors) # Solves for the allocation
alloc = np.reshape(alloc, (num_types, k))
allocations[i,:] = [max(0,min(current_budget[j] / size[i], alloc[obs_type, j])) for j in range(k)]
current_budget -= size[i] * allocations[i,:] # Reduces budget
return allocations
# Implements the Hope-Online heuristic algorithm
def hope_online(weight_matrix, weight_distribution, observed_types, n, k, budget, size):
num_types = len(weight_distribution)
allocations = np.zeros((n,k))
current_budget = np.copy(budget)
for i in range(n):
if i == n-1:
allocations[i, :] = [max(0, min(np.max(allocations[:, j]), current_budget[j] / size[i])) for j in range(k)]
else:
size_factors = np.zeros(num_types)
for m in range(n):
if m == i:
size_factors[observed_types[m]] += size[m]
elif m > i:
size_factors += size[m] * weight_distribution
obs_type = observed_types[i]
alloc, _ = solve(weight_matrix, num_types, k, current_budget, size_factors)
alloc = np.reshape(alloc, (num_types, k))
allocations[i,:] = [max(0, min(current_budget[j] / size[i], alloc[obs_type, j])) for j in range(k)]
current_budget -= size[i] * allocations[i,:]
return allocations
# Implements the Hope-Full heuristic algorithm of a different form, by solving the original Eisenberg-Gale over agents
# taking the expectation of the utility with the histogram on types.
def hope_full_v2(weight_matrix, weight_distribution, observed_types, n, k, budget, size):
num_types = len(weight_distribution)
allocations = | np.zeros((n,k)) | numpy.zeros |
"""
Source code please refer to the following:
http://web.stanford.edu/~hrhakim/NMF/code.html
Description:
This file provides the functions used in implementing the proposed method
for Non-negative matrix factorization in the paper,
"Non-negative Matrix Factorization via Archetypal Analysis".
Link = https://arxiv.org/abs/1705.02994
Re-implemented into class-based code by:
<NAME> (<EMAIL>)
"""
import numpy as np
import matplotlib.pyplot as plt
from scipy.optimize import nnls
from scipy.optimize import linprog
from hw3.libs.common.blend_dataset import BlendImgDataset
class NMF(BlendImgDataset):
def __init__(self, n_comp, o_img_size, shape, N, p):
self.n_comp = n_comp
super().__init__(o_img_size, df_dataset=True, shape=shape, N=N, p=p, all=True)
"""
Please go to the paper for the detail of the algorithm.
"""
def run(self, maxiter, delta, threshold, c1, c2, verbose, oracle):
self.W, self.H, self.L, self.Err = self.acc_palm_nmf(self.img_data.values, r=self.n_comp, maxiter=maxiter, delta=delta, threshold=threshold,
c1=c1, c2=c2, verbose=verbose, oracle=oracle)
def plot_result(self):
plt.figure()
plt.suptitle("Illustration of NMF features =%s from Zw (DR of X)" % self.n_comp)
for i in range(0, self.n_comp):
plt.subplot(1, 4, i + 1)
Vt_row = self.H[i, :].reshape(self.shape) # Reconstruct row into image for checkout
plt.title("H{}".format(i), size=8)
plt.imshow(Vt_row, cmap='gray') ## Display the image
plt.axis('off')
plt.tight_layout()
plt.show()
def D_distance(self, H1, H2):
# This function computes the 'L'-distance between the two set of vectors collected in the rows of H1 and H2. In our paper notation, this is $\mathscr{L}(H_1, H_2)$.
n1 = H1.shape[0]
n2 = H2.shape[0]
D = 0
for i in range(0, n1):
d = (np.linalg.norm(H1[i, :] - H2[0, :])) ** 2
for j in range(1, n2):
d = min(d, (np.linalg.norm(H1[i, :] - H2[j, :]) ** 2))
D = D + d
return D
# not used yet, in this implementation
def generate_weights(self, n, r, alpha, n_f, deg_prob):
# This function generates 'n' weight vectors in r-dimensions, distributed as Dirichlet(alpha, alpha, ..., alpha). 'n_f' is the number of weight vector which have zero components (induce points that lie on the faces) and 'deg_prob' is the distribution of the support size of these weight vectors. Namely, these weight vectors are distributed as Dirichlet over the set of nonzero entries which is a uniformly distributed set with a size randomly generated according to 'deg_prob'.
W = np.zeros((n, r))
for i in range(0, n_f):
deg_cdf = np.cumsum(deg_prob)
t = np.random.uniform(0, 1)
ind = np.nonzero(deg_cdf > t)
deg = np.min(ind) + 1
dirich_param = alpha * np.ones(deg)
w = np.random.dirichlet(dirich_param)
vertices = np.random.permutation(r)
vertices = vertices[0:deg]
W[i, vertices] = np.random.dirichlet(dirich_param)
for i in range(n_f, n):
dirich_param = alpha * np.ones(r)
W[i, :] = np.random.dirichlet(dirich_param)
return W
def l2distance(self, x, U, x0):
# This function computes <x-x0, (U^T*U)*(x-x0)>.
lx = np.linalg.norm(x - x0) ** 2
lpx = np.linalg.norm(np.dot(U, x - x0)) ** 2
return (lx - lpx)
def plot_H(self, H, col, type):
# This function plots the 'archetypes', (rows of 'H', when they are 2-dimensional) in 'col' color using 'type' as plot options.
v0 = H[:, 0]
v0 = np.append(v0, H[0, 0])
v1 = H[:, 1]
v1 = np.append(v1, H[0, 1])
hplt, = plt.plot(v0, v1, type, color=col, markersize=8, linewidth=3)
return hplt
def plot_data(self, X, col):
# This function plots the 'data points', (rows of 'X', when they are 2-dimensional) in 'col' color.
plt.plot(X[:, 0], X[:, 1], 'o', color=col, markersize=5)
def initH(self, X, r):
# This function computes 'r' initial archetypes given rows of 'X' as the data points. The method used here is the successive projections method explained in the paper.
n = X.shape[0]
d = X.shape[1]
H = np.zeros((r, d))
maxd = np.linalg.norm(X[0, :])
imax = 0
for i in range(1, n):
newd = np.linalg.norm(X[i, :])
if (newd > maxd):
imax = i
maxd = newd
H[0, :] = X[imax, :]
maxd = np.linalg.norm(X[0, :] - H[0, :])
imax = 0
for i in range(1, n):
newd = np.linalg.norm(X[i, :] - H[0, :])
if (newd > maxd):
imax = i
maxd = newd
H[1, :] = X[imax, :]
for k in range(2, r):
M = H[1:k, :] - np.outer(np.ones(k - 1), H[0, :])
[U, s, V] = np.linalg.svd(M, full_matrices=False)
maxd = self.l2distance(X[0, :], V, H[0, :])
imax = 0
for i in range(1, n):
newd = self.l2distance(X[i, :], V, H[0, :])
if (newd > maxd):
imax = i
maxd = newd
H[k, :] = X[imax, :]
return H
def project_simplex(self, x):
# This function computes the euclidean projection of vector 'x' onto the standard simplex.
n = len(x)
xord = -np.sort(-x)
sx = np.sum(x)
lam = (sx - 1.) / n
if (lam <= xord[n - 1]):
return (x - lam)
k = n - 1
flag = 0
while ((flag == 0) and (k > 0)):
sx -= xord[k]
lam = (sx - 1.) / k
if ((xord[k] <= lam) and (lam <= xord[k - 1])):
flag = 1
k -= 1
return np.fmax(x - lam, 0)
def project_principal(self, X, r):
# This function computes the rank 'r' pca estimate of columns of 'X'.
U, s, V = np.linalg.svd(X)
V = V[0:r, :]
U = U[:, 0:r]
s = s[0:r]
proj_X = np.dot(U, np.dot(np.diag(s), V))
return proj_X
def prune_convex(self, X):
# This function output the rows of 'X' which do not lie on the convex hull of the other rows.
n = X.shape[0]
indices = []
d = X.shape[1]
pruned_X = np.empty((0, d), int)
for i in range(0, n - 1):
print(i)
c = np.zeros(n - 1)
AEQ = np.delete(X, i, 0)
AEQ = np.transpose(AEQ)
AEQ = np.vstack([AEQ, np.ones((1, n - 1))])
BEQ = np.concatenate((X[i, :], [1]), 0)
res = linprog(c, A_ub=-1 * np.identity(n - 1), b_ub=np.zeros((n - 1, 1)), A_eq=AEQ, b_eq=np.transpose(BEQ),
options={"disp": True})
if (res.status == 2):
pruned_X = np.append(pruned_X, X[i, :].reshape(1, d), axis=0)
indices = np.append(indices, i)
return [indices.astype(int), pruned_X]
# project onto a line-segment
def proj_line_seg(self, X, x0):
# This function computes the projection of the point x0 onto the line segment between the points x1 and x2.
x1 = X[:, 0]
x2 = X[:, 1]
alpha = float(np.dot(np.transpose(x1 - x2), x0 - x2)) / (np.dot(np.transpose(x1 - x2), x1 - x2))
alpha = max(0, min(1, alpha))
y = alpha * x1 + (1 - alpha) * x2
theta = np.array([alpha, 1 - alpha])
return [theta, y]
# project onto a triangle
def proj_triangle(self, X, x0):
# This function computes the projection of the point x0 onto the triangle with corners specified with the rows of X.
d = len(x0)
XX = np.zeros((d, 2))
XX[:, 0] = X[:, 0] - X[:, 2]
XX[:, 1] = X[:, 1] - X[:, 2]
P = np.dot(np.linalg.inv(np.dot(np.transpose(XX), XX)), np.transpose(XX))
theta = np.append(np.dot(P, x0 - X[:, 2]), 1 - np.sum(np.dot(P, x0 - X[:, 2])))
y = np.dot(X, theta)
if ((any(theta < 0)) or (any(theta > 1)) or (np.sum(theta) != 1)):
d1 = np.linalg.norm(X[:, 0] - y)
d2 = np.linalg.norm(X[:, 1] - y)
d3 = np.linalg.norm(X[:, 2] - y)
theta4, y4 = self.proj_line_seg(X[:, [0, 1]], y)
d4 = np.linalg.norm(y - y4)
theta5, y5 = self.proj_line_seg(X[:, [0, 2]], y)
d5 = np.linalg.norm(y - y5)
theta6, y6 = self.proj_line_seg(X[:, [1, 2]], y)
d6 = np.linalg.norm(y - y6)
d = min(d1, d2, d3, d4, d5, d6)
if (d1 == d):
y = X[:, 0]
theta = np.array([1, 0, 0])
elif (d2 == d):
y = X[:, 1]
theta = np.array([0, 1, 0])
elif (d3 == d):
y = X[:, 2]
theta = np.array([0, 0, 1])
elif (d4 == d):
y = y4
theta = np.zeros(3)
theta[[0, 1]] = theta4
elif (d5 == d):
y = y5
theta = np.zeros(3)
theta[[0, 2]] = theta5
else:
y = y6
theta = np.zeros(3)
theta[[1, 2]] = theta6
return [theta, y]
# project onto a tetrahedron
def proj_tetrahedron(self, X, x0):
# This function computes the projection of the point x0 onto the tetrahedron with corners specified with the rows of X.
d = len(x0)
XX = np.zeros((d, 3))
XX[:, 0] = X[:, 0] - X[:, 3]
XX[:, 1] = X[:, 1] - X[:, 3]
XX[:, 2] = X[:, 2] - X[:, 3]
P = np.dot(np.linalg.inv(np.dot(np.transpose(XX), XX)), np.transpose(XX))
theta = np.append(np.dot(P, x0 - X[:, 3]), 1 - np.sum(np.dot(P, x0 - X[:, 3])))
y = np.dot(X, theta)
if ((any(theta < 0)) or (any(theta > 1)) or (np.sum(theta) != 1)):
d1 = np.linalg.norm(X[:, 0] - y)
d2 = np.linalg.norm(X[:, 1] - y)
d3 = np.linalg.norm(X[:, 2] - y)
d4 = np.linalg.norm(X[:, 3] - y)
theta5, y5 = self.proj_line_seg(X[:, [0, 1]], y)
d5 = np.linalg.norm(y - y5)
theta6, y6 = self.proj_line_seg(X[:, [0, 2]], y)
d6 = np.linalg.norm(y - y6)
theta7, y7 = self.proj_line_seg(X[:, [0, 3]], y)
d7 = np.linalg.norm(y - y7)
theta8, y8 = self.proj_line_seg(X[:, [1, 2]], y)
d8 = np.linalg.norm(y - y8)
theta9, y9 = self.proj_line_seg(X[:, [1, 3]], y)
d9 = np.linalg.norm(y - y9)
theta10, y10 = self.proj_line_seg(X[:, [2, 3]], y)
d10 = np.linalg.norm(y - y10)
theta11, y11 = self.proj_triangle(X[:, [0, 1, 2]], y)
d11 = np.linalg.norm(y - y11)
theta12, y12 = self.proj_triangle(X[:, [0, 1, 3]], y)
d12 = np.linalg.norm(y - y12)
theta13, y13 = self.proj_triangle(X[:, [0, 2, 3]], y)
d13 = np.linalg.norm(y - y13)
theta14, y14 = self.proj_triangle(X[:, [1, 2, 3]], y)
d14 = np.linalg.norm(y - y14)
d = min(d1, d2, d3, d4, d5, d6, d7, d8, d9, d10, d11, d12, d13, d14)
if (d1 == d):
y = X[:, 0]
theta = np.array([1, 0, 0, 0])
elif (d2 == d):
y = X[:, 1]
theta = np.array([0, 1, 0, 0])
elif (d3 == d):
y = X[:, 2]
theta = np.array([0, 0, 1, 0])
elif (d4 == d):
y = X[:, 3]
theta = np.array([0, 0, 0, 1])
elif (d5 == d):
y = y5
theta = np.zeros(4)
theta[[0, 1]] = theta5
elif (d6 == d):
y = y6
theta = np.zeros(4)
theta[[0, 2]] = theta6
elif (d7 == d):
y = y7
theta = np.zeros(4)
theta[[0, 3]] = theta7
elif (d8 == d):
y = y8
theta = np.zeros(4)
theta[[1, 2]] = theta8
elif (d9 == d):
y = y9
theta = np.zeros(4)
theta[[1, 3]] = theta9
elif (d10 == d):
y = y10
theta = np.zeros(4)
theta[[2, 3]] = theta10
elif (d11 == d):
y = y11
theta = np.zeros(4)
theta[[0, 1, 2]] = theta11
elif (d12 == d):
y = y12
theta = np.zeros(4)
theta[[0, 1, 3]] = theta12
elif (d13 == d):
y = y13
theta = np.zeros(4)
theta[[0, 2, 3]] = theta13
else:
y = y14
theta = np.zeros(4)
theta[[1, 2, 3]] = theta14
return [theta, y]
# project onto a 5cell
def proj_5cell(self, X, x0):
# This function computes the projection of the point x0 onto the 5-cell with corners specified with the rows of X.
d = len(x0)
XX = np.zeros((d, 4))
XX[:, 0] = X[:, 0] - X[:, 4]
XX[:, 1] = X[:, 1] - X[:, 4]
XX[:, 2] = X[:, 2] - X[:, 4]
XX[:, 3] = X[:, 3] - X[:, 4]
P = np.dot(np.linalg.inv(np.dot(np.transpose(XX), XX)), np.transpose(XX))
theta = np.append(np.dot(P, x0 - X[:, 4]), 1 - np.sum(np.dot(P, x0 - X[:, 4])))
y = np.dot(X, theta)
if ((any(theta < 0)) or (any(theta > 1)) or (np.sum(theta) != 1)):
d1 = np.linalg.norm(X[:, 0] - y)
d2 = np.linalg.norm(X[:, 1] - y)
d3 = np.linalg.norm(X[:, 2] - y)
d4 = np.linalg.norm(X[:, 3] - y)
d5 = np.linalg.norm(X[:, 4] - y)
theta6, y6 = self.proj_line_seg(X[:, [0, 1]], y)
d6 = np.linalg.norm(y - y6)
theta7, y7 = self.proj_line_seg(X[:, [0, 2]], y)
d7 = np.linalg.norm(y - y7)
theta8, y8 = self.proj_line_seg(X[:, [0, 3]], y)
d8 = np.linalg.norm(y - y8)
theta9, y9 = self.proj_line_seg(X[:, [0, 4]], y)
d9 = np.linalg.norm(y - y9)
theta10, y10 = self.proj_line_seg(X[:, [1, 2]], y)
d10 = np.linalg.norm(y - y10)
theta11, y11 = self.proj_line_seg(X[:, [1, 3]], y)
d11 = np.linalg.norm(y - y11)
theta12, y12 = self.proj_line_seg(X[:, [1, 4]], y)
d12 = np.linalg.norm(y - y12)
theta13, y13 = self.proj_line_seg(X[:, [2, 3]], y)
d13 = np.linalg.norm(y - y13)
theta14, y14 = self.proj_line_seg(X[:, [2, 4]], y)
d14 = np.linalg.norm(y - y14)
theta15, y15 = self.proj_line_seg(X[:, [3, 4]], y)
d15 = np.linalg.norm(y - y15)
theta16, y16 = self.proj_triangle(X[:, [0, 1, 2]], y)
d16 = np.linalg.norm(y - y16)
theta17, y17 = self.proj_triangle(X[:, [0, 1, 3]], y)
d17 = np.linalg.norm(y - y17)
theta18, y18 = self.proj_triangle(X[:, [0, 1, 4]], y)
d18 = np.linalg.norm(y - y18)
theta19, y19 = self.proj_triangle(X[:, [0, 2, 3]], y)
d19 = np.linalg.norm(y - y19)
theta20, y20 = self.proj_triangle(X[:, [0, 2, 4]], y)
d20 = np.linalg.norm(y - y20)
theta21, y21 = self.proj_triangle(X[:, [0, 3, 4]], y)
d21 = np.linalg.norm(y - y21)
theta22, y22 = self.proj_triangle(X[:, [1, 2, 3]], y)
d22 = np.linalg.norm(y - y22)
theta23, y23 = self.proj_triangle(X[:, [1, 2, 4]], y)
d23 = np.linalg.norm(y - y23)
theta24, y24 = self.proj_triangle(X[:, [1, 3, 4]], y)
d24 = np.linalg.norm(y - y24)
theta25, y25 = self.proj_triangle(X[:, [2, 3, 4]], y)
d25 = np.linalg.norm(y - y25)
theta26, y26 = self.proj_tetrahedron(X[:, [0, 1, 2, 3]], y)
d26 = np.linalg.norm(y - y26)
theta27, y27 = self.proj_tetrahedron(X[:, [0, 1, 2, 4]], y)
d27 = np.linalg.norm(y - y27)
theta28, y28 = self.proj_tetrahedron(X[:, [0, 1, 3, 4]], y)
d28 = np.linalg.norm(y - y28)
theta29, y29 = self.proj_tetrahedron(X[:, [0, 2, 3, 4]], y)
d29 = np.linalg.norm(y - y29)
theta30, y30 = self.proj_tetrahedron(X[:, [1, 2, 3, 4]], y)
d30 = np.linalg.norm(y - y30)
d = min(d1, d2, d3, d4, d5, d6, d7, d8, d9, d10, d11, d12, d13, d14, d15, d16, d17, d18, d19, d20, d21, d22,
d23, d24, d25, d26, d27, d28, d29, d30)
if (d1 == d):
y = X[:, 0]
theta = np.array([1, 0, 0, 0, 0])
elif (d2 == d):
y = X[:, 1]
theta = np.array([0, 1, 0, 0, 0])
elif (d3 == d):
y = X[:, 2]
theta = np.array([0, 0, 1, 0, 0])
elif (d4 == d):
y = X[:, 3]
theta = np.array([0, 0, 0, 1, 0])
elif (d5 == d):
y = X[:, 4]
theta = np.array([0, 0, 0, 0, 1])
elif (d6 == d):
y = y6
theta = np.zeros(5)
theta[[0, 1]] = theta6
elif (d7 == d):
y = y7
theta = np.zeros(5)
theta[[0, 2]] = theta7
elif (d8 == d):
y = y8
theta = np.zeros(5)
theta[[0, 3]] = theta8
elif (d9 == d):
y = y9
theta = np.zeros(5)
theta[[0, 4]] = theta9
elif (d10 == d):
y = y10
theta = np.zeros(5)
theta[[1, 2]] = theta10
elif (d11 == d):
y = y11
theta = np.zeros(5)
theta[[1, 3]] = theta11
elif (d12 == d):
y = y12
theta = np.zeros(5)
theta[[1, 4]] = theta12
elif (d13 == d):
y = y13
theta = np.zeros(5)
theta[[2, 3]] = theta13
elif (d14 == d):
y = y14
theta = np.zeros(5)
theta[[2, 4]] = theta14
elif (d15 == d):
y = y15
theta = np.zeros(5)
theta[[3, 4]] = theta15
elif (d16 == d):
y = y16
theta = np.zeros(5)
theta[[0, 1, 2]] = theta16
elif (d17 == d):
y = y17
theta = np.zeros(5)
theta[[0, 1, 3]] = theta17
elif (d18 == d):
y = y18
theta = np.zeros(5)
theta[[0, 1, 4]] = theta18
elif (d19 == d):
y = y19
theta = np.zeros(5)
theta[[0, 2, 3]] = theta19
elif (d20 == d):
y = y20
theta = np.zeros(5)
theta[[0, 2, 4]] = theta20
elif (d21 == d):
y = y21
theta = np.zeros(5)
theta[[0, 3, 4]] = theta21
elif (d22 == d):
y = y22
theta = np.zeros(5)
theta[[1, 2, 3]] = theta22
elif (d23 == d):
y = y23
theta = np.zeros(5)
theta[[1, 2, 4]] = theta23
elif (d24 == d):
y = y24
theta = np.zeros(5)
theta[[1, 3, 4]] = theta24
elif (d25 == d):
y = y25
theta = np.zeros(5)
theta[[2, 3, 4]] = theta25
elif (d26 == d):
y = y26
theta = np.zeros(5)
theta[[0, 1, 2, 3]] = theta26
elif (d27 == d):
y = y27
theta = np.zeros(5)
theta[[0, 1, 2, 4]] = theta27
elif (d28 == d):
y = y28
theta = np.zeros(5)
theta[[0, 1, 3, 4]] = theta28
elif (d29 == d):
y = y29
theta = np.zeros(5)
theta[[0, 2, 3, 4]] = theta29
else:
y = y30
theta = np.zeros(5)
theta[[1, 2, 3, 4]] = theta30
return [theta, y]
def nnls(self, y, X, niter):
# Solves min |y-X\theta| st \theta>=0, \sum\theta = 1, using projected gradient. Maximum number of iterations is specified by 'niter'.
m = X.shape[0]
p = X.shape[1]
Xt = X.transpose()
Sig = np.dot(X.transpose(), X) / m
SS = Sig
for i in range(0, 10):
SS = np.dot(Sig, SS)
L = np.power(np.trace(SS) / p, 0.1)
theta = np.ones(p) / p
it = 0
converged = 0
while ((converged == 0) and (it < niter)):
res = y - np.dot(X, theta)
grad = -np.dot(Xt, res) / m
thetanew = self.project_simplex(theta - grad / L)
dist = np.linalg.norm(theta - thetanew)
theta = thetanew
if (dist < 0.00001 * np.linalg.norm(theta)):
converged = 1
it += 1
return theta
def nnls_nesterov(self, y, X, niter):
# Solves min |y-X\theta| st \theta>=0, \sum\theta = 1, using 'Nesterov' accelerated projected gradient. Maximum number of iterations is specified by 'niter'.
m = X.shape[0]
p = X.shape[1]
Xt = X.transpose()
_, s, _ = np.linalg.svd(X)
smin = np.power(min(s), 2)
L = np.power(max(s), 2)
theta = np.ones(p) / p
mu = np.ones(p) / p
it = 0
converged = 0
g = max(1, smin)
while ((converged == 0) and (it < niter)):
t = (smin - g + np.sqrt(pow((m - g), 2) + 4 * g * L)) / (2 * L)
thetatemp = theta + ((t * g) / (g + smin * t)) * (mu - theta)
res = y - np.dot(X, thetatemp)
grad = -np.dot(Xt, res)
thetanew = self.project_simplex(theta - grad / L)
dist = np.linalg.norm(theta - thetanew)
mu = theta + (thetanew - theta) / t
theta = thetanew
if (dist < 0.00001 * np.linalg.norm(theta)):
converged = 1
it += 1
g = pow(t, 2) * L
return theta
def nnls_fista(self, y, X, niter):
# Solves min |y-X\theta| st \theta>=0, \sum\theta = 1, using Fast Iterative Shrinkage Thresholding Algorithm 'FISTA' by <NAME> Teboulle. Maximum number of iterations is specified by 'niter'.
m = X.shape[0]
p = X.shape[1]
Xt = X.transpose()
Sig = np.dot(X.transpose(), X) / m
SS = Sig.copy()
for i in range(0, 10):
SS = np.dot(Sig, SS)
L = np.power(np.trace(SS) / p, 0.1) * 1
theta = np.ones(p) / p
mu = np.ones(p) / p
it = 0
converged = 0
t = 1
while ((converged == 0) and (it < niter)):
res = y - np.dot(X, mu)
grad = -np.dot(Xt, res) / m
thetanew = self.project_simplex(mu - grad / L)
tnew = (1 + np.sqrt(1 + 4 * np.power(t, 2))) / 2
munew = thetanew + ((t - 1) / tnew) * (thetanew - theta)
dist = np.linalg.norm(theta - thetanew)
theta = thetanew
mu = munew
if (dist < 0.00001 * np.linalg.norm(theta)):
converged = 1
it += 1
return theta
def check_if_optimal(self, X, x, threshold=1e-8):
# checks whether 'x' approximates the projection of the origin onto the convex hull of the rows of matrix 'X'. The approximation acceptance threshold is determined by 'threshold'.
isoptimal = 1
n = X.shape[0]
min_res = 0
min_ind = -1
for i in range(0, n):
res = np.dot(X[i, :] - x, np.transpose(x))
if (res < min_res):
min_res = res
min_ind = i
if (min_res < -threshold):
isoptimal = 0
return [isoptimal, min_ind, min_res]
def gjk_proj(self, X, m, epsilon=1e-3, threshold=1e-8, niter=10000, verbose=False, method='fista',
fixed_max_size=float("inf")):
# Projects origin onto the convex hull of the rows of 'X' using GJK method with initial simplex size equal to 'm'. The algorithm is by <NAME> and Keerthi in their paper 'A fast procedure for computing the distance between complex objects in three-dimensional space'. The input parameters are as below:
# 'epsilon': This is an algorithm parameter determining the threshold that entries of weight vectors that are below 'epsilon' are set to zero. Default = 1e-3.
# 'threshold': The parameter determining the approximation acceptance threshold. Default = 1e-8.
# 'niter': Maximum number of iterations. Default = 10000.
# 'verbose': If set to be True, the algorithm prints out the current set of weights, active set, current estimate of the projection after each iteration. Default = False.
# 'method': If the size of the current active set is larger than 5, this method is used to calculate the projection onto the face created by active set. Options are 'proj_grad' for projected gradient, 'nesterov' for Nesterov accelerated gradient method, 'fista' for FISTA. Default = 'fista'.
# 'fixed_max_size': maximum size of the active set. Default = Inf.
n = X.shape[0]
d = X.shape[1]
m = min(n, m)
s_ind = np.random.permutation(n)
s_ind = s_ind[0:m]
isoptimal = 0
iter = 0
weights = np.zeros(n)
while (isoptimal == 0):
iter = iter + 1
X_s = X[s_ind, :]
if (len(s_ind) == 2):
theta, y = self.proj_line_seg(np.transpose(X_s), np.zeros(d))
elif (len(s_ind) == 3):
theta, y = self.proj_triangle(np.transpose(X_s), np.zeros(d))
elif (len(s_ind) == 4):
theta, y = self.proj_tetrahedron(np.transpose(X_s), np.zeros(d))
elif (len(s_ind) == 5):
theta, y = self.proj_5cell(np.transpose(X_s), np.zeros(d))
elif (method == 'nesterov'):
theta = self.nnls_nesterov(np.zeros(d), np.transpose(X_s), niter)
y = np.dot(np.transpose(X_s), theta)
elif (method == 'fista'):
theta = self.nnls_fista(np.zeros(d), np.transpose(X_s), niter)
y = np.dot(np.transpose(X_s), theta)
else:
theta = nnls(np.zeros(d), np.transpose(X_s), niter)
y = np.dot(np.transpose(X_s), theta)
weights[s_ind] = theta
[isoptimal, min_ind, min_res] = self.check_if_optimal(X, np.transpose(y), threshold=threshold)
ref_ind = (theta > epsilon)
pruned_ind = np.argmin(theta)
prune = False
if (sum(ref_ind) >= fixed_max_size):
prune = True
if (min_ind >= 0):
if (min_ind in s_ind):
isoptimal = 1
else:
s_ind = s_ind[ref_ind]
s_ind = np.append(s_ind, min_ind)
if prune == True:
s_ind = np.delete(s_ind, pruned_ind)
prune = False
if (verbose == True):
print('X_s=')
print(X_s)
print('theta=')
print(theta)
print('y=')
print(y)
print('ref_ind=')
print(ref_ind)
print('s_ind=')
print(s_ind)
return [y, weights]
def wolfe_proj(self, X, epsilon=1e-6, threshold=1e-8, niter=10000, verbose=False):
# Projects origin onto the convex hull of the rows of 'X' using Wolfe method. The algorithm is by Wolfe in his paper 'Finding the nearest point in A polytope'. The input parameters are as below:
# 'epsilon', 'threshold': Algorithm parameters determining approximation acceptance thresholds. These parameters are denoted as (Z2,Z3) and Z1, in the main paper, respectively. Default values = 1e-6, 1e-8.
# 'niter': Maximum number of iterations. Default = 10000.
# 'verbose': If set to be True, the algorithm prints out the current set of weights, active set, current estimate of the projection after each iteration. Default = False.
n = X.shape[0]
d = X.shape[1]
max_norms = np.min(np.sum(np.abs(X) ** 2, axis=-1) ** (1. / 2))
s_ind = np.array([np.argmin(np.sum(np.abs(X) ** 2, axis=-1) ** (1. / 2))])
w = np.array([1.0])
E = np.array([[-max_norms ** 2, 1.0], [1.0, 0.0]])
isoptimal = 0
iter = 0
while (isoptimal == 0) and (iter <= niter):
isoptimal_aff = 0
iter = iter + 1
P = np.dot(w, np.reshape(X[s_ind, :], (len(s_ind), d)))
new_ind = np.argmin(np.dot(P, X.T))
max_norms = max(max_norms, np.sum(np.abs(X[new_ind, :]) ** 2))
if (np.dot(P, X[new_ind, :]) > np.dot(P, P) - threshold * max_norms):
isoptimal = 1
elif (np.any(s_ind == new_ind)):
isoptimal = 1
else:
y = np.append(1, np.dot(X[s_ind, :], X[new_ind, :]))
Y = np.dot(E, y)
t = np.dot(X[new_ind, :], X[new_ind, :]) - np.dot(y, np.dot(E, y))
s_ind = np.append(s_ind, new_ind)
w = np.append(w, 0.0)
E = np.block([[E + np.outer(Y, Y) / (t + 0.0), -np.reshape(Y / (t + 0.0), (len(Y), 1))],
[-Y / (t + 0.0), 1.0 / (t + 0.0)]])
while (isoptimal_aff == 0):
v = np.dot(E, np.block([1, np.zeros(len(s_ind))]))
v = v[1:len(v)]
if (np.all(v > epsilon)):
w = v
isoptimal_aff = 1
else:
POS = np.where((w - v) > epsilon)[0]
if (POS.size == 0):
theta = 1
else:
fracs = (w + 0.0) / (w - v)
theta = min(1, np.min(fracs[POS]))
w = theta * v + (1 - theta) * w
w[w < epsilon] = 0
if np.any(w == 0):
remov_ind = np.where(w == 0)[0][0]
w = np.delete(w, remov_ind)
s_ind = np.delete(s_ind, remov_ind)
col = E[:, remov_ind + 1]
E = E - (np.outer(col, col) + 0.0) / col[remov_ind + 1]
E = np.delete(np.delete(E, remov_ind + 1, axis=0), remov_ind + 1, axis=1)
y = np.dot(X[s_ind, :].T, w)
if (verbose == True):
print('X_s=')
print(X[s_ind, :])
print('w=')
print(w)
print('y=')
print(y)
print('s_ind=')
print(s_ind)
weights = np.zeros(n)
weights[s_ind] = w
return [y, weights]
def palm_nmf_update(self, H, W, X, l, proj_method='wolfe', m=5, c1=1.2, c2=1.2, proj_low_dim=False, eps_step=1e-4,
epsilon='None', threshold=1e-8, niter=10000, method='fista', weights_exact=False,
fixed_max_size=float("inf")):
# Performs an iteration of PALM algorithm. The inputs are as below.
# 'H': Current matrix of Archetypes.
# 'W': Current matrix of Weights.
# 'X': Input Data points.
# 'l': parameter \lambda of the algorithm.
# 'proj_method': method used for computing the projection onto the convex hull. Options are: 'wolfe' for Wolfe method, 'gjk' for GJK algorithm, 'proj_grad' for projected gradient, 'nesterov' for Nesterov accelerated gradient method, 'fista' for FISTA. Default is 'wolfe'.
# 'm': Original size of the active set used for projection. Used only when GJK method is used for projection. Default is m=5.
# 'c1', 'c2': Parameters for determining the step size of the update. default values are 1.2.
# 'proj_low_dim': If set to be True, the algorithm replaces the data points with their projections onto the principal r-dimensional subspace formed by them. Default is False.
# 'eps_step': Small constant to make sure that the step size of the iteration remains bounded and the PALM iterations remain well-defined. Default value is 1e-4.
# 'epsilon': Plays the role of 'epsilon' in 'wolfe_proj' and 'gjk_proj' functions. Only used when GJK or Wolfe methods used for projection. Default value is equal to their corresponding default value for each GJK or Wolfe method.
# 'threshold': Plays the role of 'threshold' in 'wolfe_proj' and 'gjk_proj' functions. Only used when GJK or Wolfe methods used for projection. Default value is 1-e8.
# 'niter': Maximum number of iterations for computing the projection. Default is 10000.
# 'method': The same as 'method' in 'gjk_proj' function. Only used when GJK method is chosen.
# 'weights_exact': Updates the weights with their 'exact' estimates resulting from solving the constrained non-negative least squares problem after updating 'H' at each iteration. Must be set to False to follow the PALM iterations.
# 'fixed_max_size': The same as 'fixed_max_size' in 'gjk_proj' function. Only used when GJK method is chosen.
if (epsilon == 'None') and (proj_method == 'wolfe'):
epsilon = 1e-6
elif (epsilon == 'None') and (proj_method == 'gjk'):
epsilon = 1e-3
n = W.shape[0]
r = W.shape[1]
d = H.shape[1]
Hnew = H.copy()
Wnew = W.copy()
gamma1 = c1 * np.linalg.norm(np.dot(np.transpose(W), W))
gamma2 = c2 * np.linalg.norm(np.dot(H, np.transpose(H)))
gamma2 = max(gamma2, eps_step)
res = np.dot(W, H) - X[:]
H_temp = H.copy() - np.dot(np.transpose(W), res) / gamma1
for i in range(0, r):
if (proj_low_dim == True):
proj_X = self.project_principal(X, min(d, r))
if (proj_method == 'wolfe'):
H_grad, _ = self.wolfe_proj(proj_X - H_temp[i, :], epsilon=epsilon, threshold=threshold, niter=niter)
elif (proj_method == 'gjk'):
H_grad, _ = self.gjk_proj(proj_X - H_temp[i, :], m=m, epsilon=epsilon, threshold=threshold, niter=niter,
method=method, fixed_max_size=fixed_max_size)
elif (proj_method == 'proj_grad'):
H_grad = nnls(H_temp[i, :], proj_X.T, niter=niter)
H_grad = np.dot(proj_X.T, H_grad) - H_temp[i, :]
elif (proj_method == 'nesterov'):
H_grad = self.nnls_nesterov(H_temp[i, :], proj_X.T, niter=niter)
H_grad = np.dot(proj_X.T, H_grad) - H_temp[i, :]
elif (proj_method == 'fista'):
H_grad = self.nnls_fista(H_temp[i, :], proj_X.T, niter=niter)
H_grad = np.dot(proj_X.T, H_grad) - H_temp[i, :]
else:
if (proj_method == 'wolfe'):
H_grad, _ = self.wolfe_proj(X - H_temp[i, :], epsilon=epsilon, threshold=threshold, niter=niter)
elif (proj_method == 'gjk'):
H_grad, _ = self.gjk_proj(X - H_temp[i, :], m=m, epsilon=epsilon, threshold=threshold, niter=niter,
method=method, fixed_max_size=fixed_max_size)
elif (proj_method == 'proj_grad'):
H_grad = nnls(H_temp[i, :], X.T, niter=niter)
H_grad = np.dot(X.T, H_grad) - H_temp[i, :]
elif (proj_method == 'nesterov'):
H_grad = self.nnls_nesterov(H_temp[i, :], X.T, niter=niter)
H_grad = np.dot(X.T, H_grad) - H_temp[i, :]
elif (proj_method == 'fista'):
H_grad = self.nnls_fista(H_temp[i, :], X.T, niter=niter)
H_grad = np.dot(X.T, H_grad) - H_temp[i, :]
Hnew[i, :] = H_temp[i, :] + (l / (l + gamma1)) * H_grad
res = np.dot(W, Hnew) - X
if weights_exact == False:
W_temp = W[:] - (1 / gamma2) * np.dot(res, np.transpose(Hnew))
for i in range(0, n):
Wnew[i, :] = self.project_simplex(W_temp[i, :])
else:
for i in range(0, n):
if (proj_low_dim == True):
if (proj_method == 'wolfe'):
_, Wnew[i, :] = self.wolfe_proj(Hnew - proj_X[i, :], epsilon=epsilon, threshold=threshold,
niter=niter)
elif (proj_method == 'gjk'):
_, Wnew[i, :] = self.gjk_proj(Hnew - proj_X[i, :], m=m, epsilon=epsilon, threshold=threshold,
niter=niter, method=method, fixed_max_size=fixed_max_size)
elif (proj_method == 'proj_grad'):
Wnew[i, :] = nnls(proj_X[i, :], Hnew.T, niter=niter)
elif (proj_method == 'nesterov'):
Wnew[i, :] = self.nnls_nesterov(proj_X[i, :], Hnew.T, niter=niter)
elif (proj_method == 'fista'):
Wnew[i, :] = self.nnls_fista(proj_X[i, :], Hnew.T, niter=niter)
else:
if (proj_method == 'wolfe'):
_, Wnew[i, :] = self.wolfe_proj(Hnew - X[i, :], epsilon=epsilon, threshold=threshold, niter=niter)
elif (proj_method == 'gjk'):
_, Wnew[i, :] = self.gjk_proj(Hnew - X[i, :], m=m, epsilon=epsilon, threshold=threshold, niter=niter,
method=method, fixed_max_size=fixed_max_size)
elif (proj_method == 'proj_grad'):
Wnew[i, :] = nnls(X[i, :], Hnew.T, niter=niter)
elif (proj_method == 'nesterov'):
Wnew[i, :] = self.nnls_nesterov(X[i, :], Hnew.T, niter=niter)
elif (proj_method == 'fista'):
Wnew[i, :] = self.nnls_fista(X[i, :], Hnew.T, niter=niter)
return [Wnew, Hnew]
def costfun(self, W, H, X, l, proj_method='wolfe', m=3, epsilon='None', threshold=1e-8, niter=1000, method='fista',
fixed_max_size=float("inf")):
# Computes the value of the cost function minimized by PALM iterations. The inputs are as below:
# 'W': Matrix of weights.
# 'H': Matrix of Archetypes.
# 'X': Data matrix.
# 'l': \lambda.
# 'proj_method': The same as in 'palm_nmf_update' function.
# 'm': The same as in 'palm_nmf_update' function.
# 'epsilon': The same as in 'palm_nmf_update' function.
# 'threshold': The same as in 'palm_nmf_update' function.
# 'niter': The same as in 'palm_nmf_update' function.
# 'method': The same as in 'palm_nmf_update' function.
# 'fixed_max_size': The same as in 'palm_nmf_update' function.
if (epsilon == 'None') and (proj_method == 'wolfe'):
epsilon = 1e-6
elif (epsilon == 'None') and (proj_method == 'gjk'):
epsilon = 1e-3
n = W.shape[0]
r = W.shape[1]
d = H.shape[1]
fH = np.power(np.linalg.norm(X - np.dot(W, H)), 2)
for i in range(0, r):
if (proj_method == 'wolfe'):
projHi, _ = self.wolfe_proj(X - H[i, :], epsilon=epsilon, threshold=threshold, niter=niter)
elif (proj_method == 'gjk'):
projHi, _ = self.gjk_proj(X - H[i, :], m=m, epsilon=epsilon, threshold=threshold, niter=niter, method=method,
fixed_max_size=fixed_max_size)
elif (proj_method == 'proj_grad'):
projHi = nnls(H[i, :], X.T, niter=niter)
projHi = np.dot(X.T, projHi) - H[i, :]
elif (proj_method == 'nesterov'):
projHi = self.nnls_nesterov(H[i, :], X.T, niter=niter)
projHi = np.dot(X.T, projHi) - H[i, :]
elif (proj_method == 'fista'):
projHi = self.nnls_fista(H[i, :], X.T, niter=niter)
projHi = np.dot(X.T, projHi) - H[i, :]
fH = fH + l * (np.power(np.linalg.norm(projHi), 2))
return fH
def palm_nmf(self, X, r, l=None, lmax=10, lmin=0.001, lambda_no=20, c_lambda=1.2, proj_method='wolfe', m=5, H_init=None,
W_init=None, maxiter=200, delta=1e-6, c1=1.1, c2=1.1, proj_low_dim=False, eps_step=1e-4,
epsilon='None', threshold=1e-8, niter=10000, verbose=False, plotit=False, plotloss=True,
ploterror=True, oracle=True, H0=[], weights_exact=False, method='fista', fixed_max_size=float("inf")):
# The main function which minimizes the proposed cost function using PALM iterations and outputs the estimates for archetypes, weights, fitting error and estimation error for the archetypes (if the ground truth is known). The inputs are as below:
# 'X': Input data matrix.
# 'r': Rank of the fitted model.
# 'l': \lambda, if not given data driven method is used to find it.
# 'lmax': maximum of the search range for \lambda. Default value is 10.
# 'lmin': minimum of the search range for \lambda. Default value is 0.001.
# 'lambda_no': number of \lambdas in the range [lmin, lmax] used for search in finding appropriate \lambda. Default is 20.
# 'c_lambda': constant 'c' used in the data driven method for finding \lambda. Default is 1.2.
# 'proj_method': The same as in 'palm_nmf_update' function.
# 'm': The same as in 'palm_nmf_update' function.
# 'H_init': Initial value for the archetype matrix H. If not given, successive projection method is used to find an initial point.
# 'W_init': Initial value for the weights matrix H. If not given, successive projection method is used to find an initial point.
# 'maxiter': Maximum number of iterations of PALM algorithm. Default value is 200.
# 'delta': PALM Iterations are terminated when the frobenius norm of the differences between W,H estimates for successive iterations are less than 'delta'. Default value is 1e-6.
# 'c1': The same as in 'palm_nmf_update' function. Default value is 1.1.
# 'c2': The same as in 'palm_nmf_update' function. Default value is 1.1.
# 'proj_low_dim': The same as in 'palm_nmf_update' function.
# 'eps_step': The same as in 'palm_nmf_update' function.
# 'epsilon': The same as in 'palm_nmf_update' function.
# 'threshold': The same as in 'palm_nmf_update' function.
# 'niter': The same as in 'palm_nmf_update' function.
# 'verbose': If it is 'True' the number of taken iterations is given. If the ground truth is known, the Loss is also typed after each iteration. Default value is False.
# 'plotit': For the case that data points are in 2 dimensions, if 'plotit' is true, data points and estimate for archetypes are plotted after each iteration. Default value is False.
# 'plotloss': If it is True and the ground truth is known, the Loss in estimating archetypes versus iteration is plotted. Default value is True.
# 'ploterror': If it is True the minimized cost function versus iteration is plotted. Default value is True.
# 'oracle': If it is True then the ground truth archetypes are given in H0. The Default value is True.
# 'H0': Ground truth archetypes. Default is empty array.
# 'weights_exact': The same as in 'palm_nmf_update' function.
# 'method': The same as in 'palm_nmf_update' function.
# 'fixed_max_size': The same as in 'palm_nmf_update' function.
if (epsilon == 'None') and (proj_method == 'wolfe'):
epsilon = 1e-6
elif (epsilon == 'None') and (proj_method == 'gjk'):
epsilon = 1e-3
if (l == None):
lambdas = np.geomspace(lmin, lmax, lambda_no)
else:
lambdas = | np.array([l]) | numpy.array |
# OpenSeesPy visualization module
# Author: <NAME>
# Faculty of Civil Engineering and Architecture
# Opole University of Technology, Poland
# ver. 0.94, 2020 August
# License: MIT
# Notes:
# 1. matplotlib's plt.axis('equal') does not work for 3d plots
# therefore right angles are not guaranteed to be 90 degrees on the
# plots
import openseespy.opensees as ops # installed from pip
# import opensees as ops # local compilation
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from mpl_toolkits.mplot3d.art3d import Poly3DCollection
from matplotlib.patches import Circle, Polygon
from matplotlib.animation import FuncAnimation
import matplotlib.tri as tri
# default settings
# fmt: format string setting color, marker and linestyle
# check documentation on matplotlib's plot
# continuous interpolated shape line
fmt_interp = 'b-' # blue solid line, no markers
# element end nodes
fmt_nodes = 'rs' # red square markers, no line
# undeformed model
fmt_undefo = 'g--' # green dashed line, no markers
# section forces
fmt_secforce = 'b-' # blue solid line
# figure left right bottom top offsets
fig_lbrt = (.04, .04, .96, .96)
# azimuth and elevation in degrees
az_el = (-60., 30.)
# figure width and height in centimeters
fig_wi_he = (16., 10.)
def _plot_model_2d(node_labels, element_labels, offset_nd_label, axis_off):
max_x_crd, max_y_crd, max_crd = -np.inf, -np.inf, -np.inf
node_tags = ops.getNodeTags()
ele_tags = ops.getEleTags()
nen = np.shape(ops.eleNodes(ele_tags[0]))[0]
# truss and beam/frame elements
if nen == 2:
for node_tag in node_tags:
x_crd = ops.nodeCoord(node_tag)[0]
y_crd = ops.nodeCoord(node_tag)[1]
if x_crd > max_x_crd:
max_x_crd = x_crd
if y_crd > max_y_crd:
max_y_crd = y_crd
max_crd = np.amax([max_x_crd, max_y_crd])
_offset = 0.005 * max_crd
for i, ele_tag in enumerate(ele_tags):
nd1, nd2 = ops.eleNodes(ele_tag)
# element node1-node2, x, y coordinates
ex = np.array([ops.nodeCoord(nd1)[0], ops.nodeCoord(nd2)[0]])
ey = np.array([ops.nodeCoord(nd1)[1], ops.nodeCoord(nd2)[1]])
# location of label
xt = sum(ex)/nen
yt = sum(ey)/nen
plt.plot(ex, ey, 'bo-')
if element_labels:
if ex[1]-ex[0] == 0:
va = 'center'
ha = 'left'
offset_x, offset_y = _offset, 0.0
elif ey[1]-ey[0] == 0:
va = 'bottom'
ha = 'center'
offset_x, offset_y = 0.0, _offset
else:
va = 'bottom'
ha = 'left'
offset_x, offset_y = 0.03, 0.03
plt.text(xt+offset_x, yt+offset_y, f'{ele_tag}', va=va, ha=ha,
color='red')
if node_labels:
for node_tag in node_tags:
if not offset_nd_label == 'above':
offset_nd_label_x, offset_nd_label_y = _offset, _offset
va = 'bottom'
ha = 'left'
else:
offset_nd_label_x, offset_nd_label_y = 0.0, _offset
va = 'bottom'
ha = 'center'
plt.text(ops.nodeCoord(node_tag)[0]+offset_nd_label_x,
ops.nodeCoord(node_tag)[1]+offset_nd_label_y,
f'{node_tag}', va=va, ha=ha, color='blue')
# plt.axis('equal')
# 2d triangular (tri31) elements
elif nen == 3:
for node_tag in node_tags:
x_crd = ops.nodeCoord(node_tag)[0]
y_crd = ops.nodeCoord(node_tag)[1]
if x_crd > max_x_crd:
max_x_crd = x_crd
if y_crd > max_y_crd:
max_y_crd = y_crd
max_crd = np.amax([max_x_crd, max_y_crd])
_offset = 0.005 * max_crd
_offnl = 0.003 * max_crd
for i, ele_tag in enumerate(ele_tags):
nd1, nd2, nd3 = ops.eleNodes(ele_tag)
# element x, y coordinates
ex = np.array([ops.nodeCoord(nd1)[0],
ops.nodeCoord(nd2)[0],
ops.nodeCoord(nd3)[0]])
ey = np.array([ops.nodeCoord(nd1)[1],
ops.nodeCoord(nd2)[1],
ops.nodeCoord(nd3)[1]])
# location of label
xt = sum(ex)/nen
yt = sum(ey)/nen
plt.plot(np.append(ex, ex[0]), np.append(ey, ey[0]), 'bo-')
if element_labels:
va = 'center'
ha = 'center'
plt.text(xt, yt, f'{ele_tag}', va=va, ha=ha, color='red')
if node_labels:
for node_tag in node_tags:
if not offset_nd_label == 'above':
offset_nd_label_x, offset_nd_label_y = _offnl, _offnl
va = 'bottom'
# va = 'center'
ha = 'left'
else:
offset_nd_label_x, offset_nd_label_y = 0.0, _offnl
va = 'bottom'
ha = 'center'
plt.text(ops.nodeCoord(node_tag)[0]+offset_nd_label_x,
ops.nodeCoord(node_tag)[1]+offset_nd_label_y,
f'{node_tag}', va=va, ha=ha, color='blue')
# 2d quadrilateral (quad) elements
elif nen == 4:
for node_tag in node_tags:
x_crd = ops.nodeCoord(node_tag)[0]
y_crd = ops.nodeCoord(node_tag)[1]
if x_crd > max_x_crd:
max_x_crd = x_crd
if y_crd > max_y_crd:
max_y_crd = y_crd
max_crd = np.amax([max_x_crd, max_y_crd])
_offset = 0.005 * max_crd
_offnl = 0.003 * max_crd
for i, ele_tag in enumerate(ele_tags):
nd1, nd2, nd3, nd4 = ops.eleNodes(ele_tag)
# element x, y coordinates
ex = np.array([ops.nodeCoord(nd1)[0],
ops.nodeCoord(nd2)[0],
ops.nodeCoord(nd3)[0],
ops.nodeCoord(nd4)[0]])
ey = np.array([ops.nodeCoord(nd1)[1],
ops.nodeCoord(nd2)[1],
ops.nodeCoord(nd3)[1],
ops.nodeCoord(nd4)[1]])
# location of label
xt = sum(ex)/nen
yt = sum(ey)/nen
# plt.plot(np.append(ex, ex[0]), np.append(ey, ey[0]), 'bo-')
plt.plot(np.append(ex, ex[0]), np.append(ey, ey[0]), 'b-', lw=0.4)
if element_labels:
va = 'center'
ha = 'center'
plt.text(xt, yt, f'{ele_tag}', va=va, ha=ha, color='red')
if node_labels:
for node_tag in node_tags:
if not offset_nd_label == 'above':
offset_nd_label_x, offset_nd_label_y = _offnl, _offnl
va = 'bottom'
# va = 'center'
ha = 'left'
else:
offset_nd_label_x, offset_nd_label_y = 0.0, _offnl
va = 'bottom'
ha = 'center'
plt.text(ops.nodeCoord(node_tag)[0]+offset_nd_label_x,
ops.nodeCoord(node_tag)[1]+offset_nd_label_y,
f'{node_tag}', va=va, ha=ha, color='blue')
plt.axis('equal')
def _plot_model_3d(node_labels, element_labels, offset_nd_label, axis_off,
az_el, fig_wi_he, fig_lbrt):
node_tags = ops.getNodeTags()
ele_tags = ops.getEleTags()
azim, elev = az_el
fig_wi, fig_he = fig_wi_he
fleft, fbottom, fright, ftop = fig_lbrt
fig = plt.figure(figsize=(fig_wi/2.54, fig_he/2.54))
fig.subplots_adjust(left=.08, bottom=.08, right=.985, top=.94)
ax = fig.add_subplot(111, projection=Axes3D.name)
ax.set_xlabel('X')
ax.set_ylabel('Y')
ax.set_zlabel('Z')
ax.view_init(azim=azim, elev=elev)
max_x_crd, max_y_crd, max_z_crd, max_crd = -np.inf, -np.inf, \
-np.inf, -np.inf
nen = np.shape(ops.eleNodes(ele_tags[0]))[0]
# truss and beam/frame elements
if nen == 2:
for node_tag in node_tags:
x_crd = ops.nodeCoord(node_tag)[0]
y_crd = ops.nodeCoord(node_tag)[1]
z_crd = ops.nodeCoord(node_tag)[2]
if x_crd > max_x_crd:
max_x_crd = x_crd
if y_crd > max_y_crd:
max_y_crd = y_crd
if z_crd > max_z_crd:
max_z_crd = z_crd
if offset_nd_label == 0 or offset_nd_label == 0.:
_offset = 0.
else:
max_crd = np.amax([max_x_crd, max_y_crd, max_z_crd])
_offset = 0.005 * max_crd
# # work-around fix because of aspect equal bug
# _max_overall = 1.1*max_crd
# _min_overall = -0.1*max_crd
# ax.set_xlim(_min_overall, _max_overall)
# ax.set_ylim(_min_overall, _max_overall)
# ax.set_zlim(_min_overall, _max_overall)
for i, ele_tag in enumerate(ele_tags):
nd1, nd2 = ops.eleNodes(ele_tag)
# element node1-node2, x, y coordinates
ex = np.array([ops.nodeCoord(nd1)[0], ops.nodeCoord(nd2)[0]])
ey = np.array([ops.nodeCoord(nd1)[1], ops.nodeCoord(nd2)[1]])
ez = np.array([ops.nodeCoord(nd1)[2], ops.nodeCoord(nd2)[2]])
# location of label
xt = sum(ex)/nen
yt = sum(ey)/nen
zt = sum(ez)/nen
ax.plot(ex, ey, ez, 'bo-')
# fixme: placement of node_tag labels
if element_labels:
if ex[1]-ex[0] == 0:
va = 'center'
ha = 'left'
offset_x, offset_y, offset_z = _offset, 0.0, 0.0
elif ey[1]-ey[0] == 0:
va = 'bottom'
ha = 'center'
offset_x, offset_y, offset_z = 0.0, _offset, 0.0
elif ez[1]-ez[0] == 0:
va = 'bottom'
ha = 'center'
offset_x, offset_y, offset_z = 0.0, 0.0, _offset
else:
va = 'bottom'
ha = 'left'
offset_x, offset_y, offset_z = 0.03, 0.03, 0.03
ax.text(xt+offset_x, yt+offset_y, zt+offset_z, f'{ele_tag}',
va=va, ha=ha, color='red')
if node_labels:
for node_tag in node_tags:
ax.text(ops.nodeCoord(node_tag)[0]+_offset,
ops.nodeCoord(node_tag)[1]+_offset,
ops.nodeCoord(node_tag)[2]+_offset,
f'{node_tag}', va='bottom', ha='left', color='blue')
# quad in 3d
elif nen == 4:
for node_tag in node_tags:
x_crd = ops.nodeCoord(node_tag)[0]
y_crd = ops.nodeCoord(node_tag)[1]
z_crd = ops.nodeCoord(node_tag)[2]
if x_crd > max_x_crd:
max_x_crd = x_crd
if y_crd > max_y_crd:
max_y_crd = y_crd
if z_crd > max_z_crd:
max_z_crd = z_crd
# ax.plot(np.array([x_crd]),
# np.array([y_crd]),
# np.array([z_crd]), 'ro')
max_crd = np.amax([max_x_crd, max_y_crd, max_z_crd])
_offset = 0.002 * max_crd
for i, ele_tag in enumerate(ele_tags):
nd1, nd2, nd3, nd4 = ops.eleNodes(ele_tag)
# element node1-node2, x, y coordinates
ex = np.array([ops.nodeCoord(nd1)[0],
ops.nodeCoord(nd2)[0],
ops.nodeCoord(nd3)[0],
ops.nodeCoord(nd4)[0]])
ey = np.array([ops.nodeCoord(nd1)[1],
ops.nodeCoord(nd2)[1],
ops.nodeCoord(nd3)[1],
ops.nodeCoord(nd4)[1]])
ez = np.array([ops.nodeCoord(nd1)[2],
ops.nodeCoord(nd2)[2],
ops.nodeCoord(nd3)[2],
ops.nodeCoord(nd4)[2]])
# location of label
xt = sum(ex)/nen
yt = sum(ey)/nen
zt = sum(ez)/nen
ax.plot(np.append(ex, ex[0]),
np.append(ey, ey[0]),
np.append(ez, ez[0]), 'bo-')
# fixme: placement of node_tag labels
if element_labels:
if ex[1]-ex[0] == 0:
va = 'center'
ha = 'left'
offset_x, offset_y, offset_z = _offset, 0.0, 0.0
elif ey[1]-ey[0] == 0:
va = 'bottom'
ha = 'center'
offset_x, offset_y, offset_z = 0.0, _offset, 0.0
elif ez[1]-ez[0] == 0:
va = 'bottom'
ha = 'center'
offset_x, offset_y, offset_z = 0.0, 0.0, _offset
else:
va = 'bottom'
ha = 'left'
offset_x, offset_y, offset_z = 0.03, 0.03, 0.03
ax.text(xt+offset_x, yt+offset_y, zt+offset_z, f'{ele_tag}',
va=va, ha=ha, color='red')
if node_labels:
for node_tag in node_tags:
ax.text(ops.nodeCoord(node_tag)[0]+_offset,
ops.nodeCoord(node_tag)[1]+_offset,
ops.nodeCoord(node_tag)[2]+_offset,
f'{node_tag}', va='bottom', ha='left', color='blue')
# 8-node brick, 3d model
elif nen == 8:
for node_tag in node_tags:
x_crd = ops.nodeCoord(node_tag)[0]
y_crd = ops.nodeCoord(node_tag)[1]
z_crd = ops.nodeCoord(node_tag)[2]
if x_crd > max_x_crd:
max_x_crd = x_crd
if y_crd > max_y_crd:
max_y_crd = y_crd
if z_crd > max_z_crd:
max_z_crd = z_crd
# ax.plot(np.array([x_crd]),
# np.array([y_crd]),
# np.array([z_crd]), 'ro')
max_crd = np.amax([max_x_crd, max_y_crd, max_z_crd])
_offset = 0.005 * max_crd
# work-around fix because of aspect equal bug
_max_overall = 1.1*max_crd
_min_overall = -0.1*max_crd
ax.set_xlim(_min_overall, _max_overall)
ax.set_ylim(_min_overall, _max_overall)
ax.set_zlim(_min_overall, _max_overall)
for i, ele_tag in enumerate(ele_tags):
nd1, nd2, nd3, nd4, nd5, nd6, nd7, nd8 = ops.eleNodes(ele_tag)
# element node1-node2, x, y coordinates
ex = np.array([ops.nodeCoord(nd1)[0],
ops.nodeCoord(nd2)[0],
ops.nodeCoord(nd3)[0],
ops.nodeCoord(nd4)[0],
ops.nodeCoord(nd5)[0],
ops.nodeCoord(nd6)[0],
ops.nodeCoord(nd7)[0],
ops.nodeCoord(nd8)[0]])
ey = np.array([ops.nodeCoord(nd1)[1],
ops.nodeCoord(nd2)[1],
ops.nodeCoord(nd3)[1],
ops.nodeCoord(nd4)[1],
ops.nodeCoord(nd5)[1],
ops.nodeCoord(nd6)[1],
ops.nodeCoord(nd7)[1],
ops.nodeCoord(nd8)[1]])
ez = np.array([ops.nodeCoord(nd1)[2],
ops.nodeCoord(nd2)[2],
ops.nodeCoord(nd3)[2],
ops.nodeCoord(nd4)[2],
ops.nodeCoord(nd5)[2],
ops.nodeCoord(nd6)[2],
ops.nodeCoord(nd7)[2],
ops.nodeCoord(nd8)[2]])
# location of label
xt = sum(ex)/nen
yt = sum(ey)/nen
zt = sum(ez)/nen
ax.plot(np.append(ex[0:4], ex[0]),
np.append(ey[0:4], ey[0]),
np.append(ez[0:4], ez[0]), 'bo-')
ax.plot(np.append(ex[4:8], ex[4]),
np.append(ey[4:8], ey[4]),
np.append(ez[4:8], ez[4]), 'bo-')
ax.plot(np.array([ex[0], ex[4]]),
np.array([ey[0], ey[4]]),
np.array([ez[0], ez[4]]), 'bo-')
ax.plot(np.array([ex[1], ex[5]]),
np.array([ey[1], ey[5]]),
np.array([ez[1], ez[5]]), 'bo-')
ax.plot(np.array([ex[2], ex[6]]),
np.array([ey[2], ey[6]]),
np.array([ez[2], ez[6]]), 'bo-')
ax.plot(np.array([ex[3], ex[7]]),
np.array([ey[3], ey[7]]),
np.array([ez[3], ez[7]]), 'bo-')
# fixme: placement of node_tag labels
if element_labels:
if ex[1]-ex[0] == 0:
va = 'center'
ha = 'left'
offset_x, offset_y, offset_z = _offset, 0.0, 0.0
elif ey[1]-ey[0] == 0:
va = 'bottom'
ha = 'center'
offset_x, offset_y, offset_z = 0.0, _offset, 0.0
elif ez[1]-ez[0] == 0:
va = 'bottom'
ha = 'center'
offset_x, offset_y, offset_z = 0.0, 0.0, _offset
else:
va = 'bottom'
ha = 'left'
offset_x, offset_y, offset_z = 0.03, 0.03, 0.03
ax.text(xt+offset_x, yt+offset_y, zt+offset_z, f'{ele_tag}',
va=va, ha=ha, color='red')
if node_labels:
for node_tag in node_tags:
ax.text(ops.nodeCoord(node_tag)[0]+_offset,
ops.nodeCoord(node_tag)[1]+_offset,
ops.nodeCoord(node_tag)[2]+_offset,
f'{node_tag}', va='bottom', ha='left', color='blue')
def plot_model(node_labels=1, element_labels=1, offset_nd_label=False,
axis_off=0, az_el=az_el, fig_wi_he=fig_wi_he,
fig_lbrt=fig_lbrt):
"""Plot defined model of the structure.
Args:
node_labels (int): 1 - plot node labels, 0 - do not plot them;
(default: 1)
element_labels (int): 1 - plot element labels, 0 - do not plot
them; (default: 1)
offset_nd_label (bool): False - do not offset node labels from the
actual node location. This option can enhance visibility.
axis_off (int): 0 - turn off axes, 1 - display axes; (default: 0)
az_el (tuple): contains azimuth and elevation for 3d plots
fig_wi_he (tuple): contains width and height of the figure
fig_lbrt (tuple): a tuple contating left, bottom, right and top offsets
Usage:
``plot_()`` - plot deformed shape with default parameters and
automatically calcutated scale factor.
``plot_defo(interpFlag=0)`` - plot displaced nodes without shape
function interpolation
``plot_defo(sfac=1.5)`` - plot with specified scale factor
``plot_defo(unDefoFlag=0, endDispFlag=0)`` - plot without showing
undeformed (original) mesh and without showing markers at the
element ends.
"""
# az_el - azimut, elevation used for 3d plots only
node_tags = ops.getNodeTags()
ndim = np.shape(ops.nodeCoord(node_tags[0]))[0]
if ndim == 2:
_plot_model_2d(node_labels, element_labels, offset_nd_label, axis_off)
if axis_off:
plt.axis('off')
elif ndim == 3:
_plot_model_3d(node_labels, element_labels, offset_nd_label, axis_off,
az_el, fig_wi_he, fig_lbrt)
if axis_off:
plt.axis('off')
else:
print(f'\nWarning! ndim: {ndim} not supported yet.')
# plt.show() # call this from main py file for more control
def _plot_defo_mode_2d(modeNo, sfac, nep, unDefoFlag, fmt_undefo, interpFlag,
endDispFlag, fmt_interp, fmt_nodes):
ele_tags = ops.getEleTags()
nen = np.shape(ops.eleNodes(ele_tags[0]))[0]
# truss and beam/frame elements
if nen == 2:
ndf = np.shape(ops.nodeDOFs(ops.eleNodes(ele_tags[0])[0]))[0]
# truss element
if ndf == 2:
for ele_tag in ele_tags:
nd1, nd2 = ops.eleNodes(ele_tag)
# element x, y coordinates
ex = np.array([ops.nodeCoord(nd1)[0],
ops.nodeCoord(nd2)[0]])
ey = np.array([ops.nodeCoord(nd1)[1],
ops.nodeCoord(nd2)[1]])
if modeNo:
eux = np.array([ops.nodeEigenvector(nd1, modeNo)[0],
ops.nodeEigenvector(nd2, modeNo)[0]])
euy = np.array([ops.nodeEigenvector(nd1, modeNo)[1],
ops.nodeEigenvector(nd2, modeNo)[1]])
else:
eux = np.array([ops.nodeDisp(nd1)[0],
ops.nodeDisp(nd2)[0]])
euy = np.array([ops.nodeDisp(nd1)[1],
ops.nodeDisp(nd2)[1]])
# displaced element coordinates (scaled by sfac factor)
edx = np.array([ex[0] + sfac*eux[0], ex[1] + sfac*eux[1]])
edy = np.array([ey[0] + sfac*euy[0], ey[1] + sfac*euy[1]])
if unDefoFlag:
plt.plot(ex, ey, fmt_undefo)
plt.plot(edx, edy, fmt_interp)
# beam/frame element
elif ndf == 3:
for ele_tag in ele_tags:
nd1, nd2 = ops.eleNodes(ele_tag)
# element x, y coordinates
ex = np.array([ops.nodeCoord(nd1)[0],
ops.nodeCoord(nd2)[0]])
ey = np.array([ops.nodeCoord(nd1)[1],
ops.nodeCoord(nd2)[1]])
if modeNo:
ed = np.array([ops.nodeEigenvector(nd1, modeNo)[0],
ops.nodeEigenvector(nd1, modeNo)[1],
ops.nodeEigenvector(nd1, modeNo)[2],
ops.nodeEigenvector(nd2, modeNo)[0],
ops.nodeEigenvector(nd2, modeNo)[1],
ops.nodeEigenvector(nd2, modeNo)[2]])
else:
ed = np.array([ops.nodeDisp(nd1)[0],
ops.nodeDisp(nd1)[1],
ops.nodeDisp(nd1)[2],
ops.nodeDisp(nd2)[0],
ops.nodeDisp(nd2)[1],
ops.nodeDisp(nd2)[2]])
if unDefoFlag:
plt.plot(ex, ey, fmt_undefo)
# interpolated displacement field
if interpFlag:
xcdi, ycdi = beam_defo_interp_2d(ex, ey, ed, sfac, nep)
plt.plot(xcdi, ycdi, fmt_interp)
# translations of ends
if endDispFlag:
xdi, ydi = beam_disp_ends(ex, ey, ed, sfac)
plt.plot(xdi, ydi, fmt_nodes)
plt.axis('equal')
# plt.show() # call this from main py file for more control
# 2d triangular (tri31) elements
elif nen == 3:
for ele_tag in ele_tags:
nd1, nd2, nd3 = ops.eleNodes(ele_tag)
# element x, y coordinates
ex = np.array([ops.nodeCoord(nd1)[0],
ops.nodeCoord(nd2)[0],
ops.nodeCoord(nd3)[0]])
ey = np.array([ops.nodeCoord(nd1)[1],
ops.nodeCoord(nd2)[1],
ops.nodeCoord(nd3)[1]])
if modeNo:
ed = np.array([ops.nodeEigenvector(nd1, modeNo)[0],
ops.nodeEigenvector(nd1, modeNo)[1],
ops.nodeEigenvector(nd2, modeNo)[0],
ops.nodeEigenvector(nd2, modeNo)[1],
ops.nodeEigenvector(nd3, modeNo)[0],
ops.nodeEigenvector(nd3, modeNo)[1]])
else:
ed = np.array([ops.nodeDisp(nd1)[0],
ops.nodeDisp(nd1)[1],
ops.nodeDisp(nd2)[0],
ops.nodeDisp(nd2)[1],
ops.nodeDisp(nd3)[0],
ops.nodeDisp(nd3)[1]])
if unDefoFlag:
plt.plot(np.append(ex, ex[0]), np.append(ey, ey[0]),
fmt_undefo)
# xcdi, ycdi = beam_defo_interp_2d(ex, ey, ed, sfac, nep)
# xdi, ydi = beam_disp_ends(ex, ey, ed, sfac)
# # interpolated displacement field
# plt.plot(xcdi, ycdi, 'b.-')
# # translations of ends only
# plt.plot(xdi, ydi, 'ro')
# xc = [x, x[0, :]]
# yc = [x, x[0, :]]
# test it with one element
x = ex+sfac*ed[[0, 2, 4]]
y = ey+sfac*ed[[1, 3, 5]]
# x = ex+sfac*ed[[0, 2, 4, 6]]
# y = ey+sfac*ed[[1, 3, 5, 7]]
plt.plot(np.append(x, x[0]), np.append(y, y[0]), 'b.-')
plt.axis('equal')
# 2d quadrilateral (quad) elements
elif nen == 4:
for ele_tag in ele_tags:
nd1, nd2, nd3, nd4 = ops.eleNodes(ele_tag)
# element x, y coordinates
ex = np.array([ops.nodeCoord(nd1)[0],
ops.nodeCoord(nd2)[0],
ops.nodeCoord(nd3)[0],
ops.nodeCoord(nd4)[0]])
ey = np.array([ops.nodeCoord(nd1)[1],
ops.nodeCoord(nd2)[1],
ops.nodeCoord(nd3)[1],
ops.nodeCoord(nd4)[1]])
if modeNo:
ed = np.array([ops.nodeEigenvector(nd1, modeNo)[0],
ops.nodeEigenvector(nd1, modeNo)[1],
ops.nodeEigenvector(nd2, modeNo)[0],
ops.nodeEigenvector(nd2, modeNo)[1],
ops.nodeEigenvector(nd3, modeNo)[0],
ops.nodeEigenvector(nd3, modeNo)[1],
ops.nodeEigenvector(nd4, modeNo)[0],
ops.nodeEigenvector(nd4, modeNo)[1]])
else:
ed = np.array([ops.nodeDisp(nd1)[0],
ops.nodeDisp(nd1)[1],
ops.nodeDisp(nd2)[0],
ops.nodeDisp(nd2)[1],
ops.nodeDisp(nd3)[0],
ops.nodeDisp(nd3)[1],
ops.nodeDisp(nd4)[0],
ops.nodeDisp(nd4)[1]])
if unDefoFlag:
plt.plot(np.append(ex, ex[0]), np.append(ey, ey[0]),
fmt_undefo)
# xcdi, ycdi = beam_defo_interp_2d(ex, ey, ed, sfac, nep)
# xdi, ydi = beam_disp_ends(ex, ey, ed, sfac)
# # interpolated displacement field
# plt.plot(xcdi, ycdi, 'b.-')
# # translations of ends only
# plt.plot(xdi, ydi, 'ro')
# test it with one element
x = ex+sfac*ed[[0, 2, 4, 6]]
y = ey+sfac*ed[[1, 3, 5, 7]]
plt.plot(np.append(x, x[0]), np.append(y, y[0]), 'b.-')
plt.axis('equal')
# 2d 8-node quadratic elements
# elif nen == 8:
# x = ex+sfac*ed[:, [0, 2, 4, 6, 8, 10, 12, 14]]
# y = ex+sfac*ed[:, [1, 3, 5, 7, 9, 11, 13, 15]]
# t = -1
# n = 0
# for s in range(-1, 1.4, 0.4):
# n += 1
# ...
else:
print(f'\nWarning! Elements not supported yet. nen: {nen}; must be: 2, 3, 4, 8.') # noqa: E501
def _plot_defo_mode_3d(modeNo, sfac, nep, unDefoFlag, fmt_undefo, interpFlag,
endDispFlag, fmt_interp, fmt_nodes, az_el, fig_wi_he,
fig_lbrt):
ele_tags = ops.getEleTags()
azim, elev = az_el
fig_wi, fig_he = fig_wi_he
fleft, fbottom, fright, ftop = fig_lbrt
fig = plt.figure(figsize=(fig_wi/2.54, fig_he/2.54))
fig.subplots_adjust(left=.08, bottom=.08, right=.985, top=.94)
ax = fig.add_subplot(111, projection=Axes3D.name)
# ax.axis('equal')
ax.set_xlabel('X')
ax.set_ylabel('Y')
ax.set_zlabel('Z')
ax.view_init(azim=azim, elev=elev)
nen = np.shape(ops.eleNodes(ele_tags[0]))[0]
# plot: truss and beam/frame elements in 3d
if nen == 2:
ndf = np.shape(ops.nodeDOFs(ops.eleNodes(ele_tags[0])[0]))[0]
# plot: beam/frame element in 3d
if ndf == 6:
for i, ele_tag in enumerate(ele_tags):
nd1, nd2 = ops.eleNodes(ele_tag)
# element x, y coordinates
ex = np.array([ops.nodeCoord(nd1)[0],
ops.nodeCoord(nd2)[0]])
ey = np.array([ops.nodeCoord(nd1)[1],
ops.nodeCoord(nd2)[1]])
ez = np.array([ops.nodeCoord(nd1)[2],
ops.nodeCoord(nd2)[2]])
if modeNo:
ed = np.array([ops.nodeEigenvector(nd1, modeNo)[0],
ops.nodeEigenvector(nd1, modeNo)[1],
ops.nodeEigenvector(nd1, modeNo)[2],
ops.nodeEigenvector(nd1, modeNo)[3],
ops.nodeEigenvector(nd1, modeNo)[4],
ops.nodeEigenvector(nd1, modeNo)[5],
ops.nodeEigenvector(nd2, modeNo)[0],
ops.nodeEigenvector(nd2, modeNo)[1],
ops.nodeEigenvector(nd2, modeNo)[2],
ops.nodeEigenvector(nd2, modeNo)[3],
ops.nodeEigenvector(nd2, modeNo)[4],
ops.nodeEigenvector(nd2, modeNo)[5]])
else:
ed = np.array([ops.nodeDisp(nd1)[0],
ops.nodeDisp(nd1)[1],
ops.nodeDisp(nd1)[2],
ops.nodeDisp(nd1)[3],
ops.nodeDisp(nd1)[4],
ops.nodeDisp(nd1)[5],
ops.nodeDisp(nd2)[0],
ops.nodeDisp(nd2)[1],
ops.nodeDisp(nd2)[2],
ops.nodeDisp(nd2)[3],
ops.nodeDisp(nd2)[4],
ops.nodeDisp(nd2)[5]])
# eo = Eo[i, :]
xloc = ops.eleResponse(ele_tag, 'xlocal')
yloc = ops.eleResponse(ele_tag, 'ylocal')
zloc = ops.eleResponse(ele_tag, 'zlocal')
g = np.vstack((xloc, yloc, zloc))
if unDefoFlag:
plt.plot(ex, ey, ez, fmt_undefo)
# interpolated displacement field
if interpFlag:
xcd, ycd, zcd = beam_defo_interp_3d(ex, ey, ez, g,
ed, sfac, nep)
ax.plot(xcd, ycd, zcd, fmt_interp)
ax.set_xlabel('X')
ax.set_ylabel('Y')
ax.set_zlabel('Z')
# translations of ends
if endDispFlag:
xd, yd, zd = beam_disp_ends3d(ex, ey, ez, ed, sfac)
ax.plot(xd, yd, zd, fmt_nodes)
# # work-around fix because of aspect equal bug
# xmin, xmax = ax.get_xlim()
# ymin, ymax = ax.get_ylim()
# zmin, zmax = ax.get_zlim()
# min_overall = np.amax([np.abs(xmin), np.abs(ymin), np.abs(zmin)])
# max_overall = np.amax([np.abs(xmax), np.abs(ymax), np.abs(zmax)])
# minmax_overall = max(min_overall, max_overall)
# _max_overall = 1.1 * minmax_overall
# _min_overall = -1.1 * minmax_overall
# ax.set_xlim(_min_overall, _max_overall)
# ax.set_ylim(_min_overall, _max_overall)
# # ax.set_zlim(_min_overall, _max_overall)
# ax.set_zlim(0.0, _max_overall)
# plot: quad in 3d
elif nen == 4:
ndf = np.shape(ops.nodeDOFs(ops.eleNodes(ele_tags[0])[0]))[0]
# plot: shell in 3d
if ndf == 6:
for i, ele_tag in enumerate(ele_tags):
nd1, nd2, nd3, nd4 = ops.eleNodes(ele_tag)
# element node1-node2, x, y coordinates
ex = np.array([ops.nodeCoord(nd1)[0],
ops.nodeCoord(nd2)[0],
ops.nodeCoord(nd3)[0],
ops.nodeCoord(nd4)[0]])
ey = np.array([ops.nodeCoord(nd1)[1],
ops.nodeCoord(nd2)[1],
ops.nodeCoord(nd3)[1],
ops.nodeCoord(nd4)[1]])
ez = np.array([ops.nodeCoord(nd1)[2],
ops.nodeCoord(nd2)[2],
ops.nodeCoord(nd3)[2],
ops.nodeCoord(nd4)[2]])
if modeNo:
ed = np.array([ops.nodeEigenvector(nd1, modeNo)[0],
ops.nodeEigenvector(nd1, modeNo)[1],
ops.nodeEigenvector(nd1, modeNo)[2],
ops.nodeEigenvector(nd2, modeNo)[0],
ops.nodeEigenvector(nd2, modeNo)[1],
ops.nodeEigenvector(nd2, modeNo)[2],
ops.nodeEigenvector(nd3, modeNo)[0],
ops.nodeEigenvector(nd3, modeNo)[1],
ops.nodeEigenvector(nd3, modeNo)[2],
ops.nodeEigenvector(nd4, modeNo)[0],
ops.nodeEigenvector(nd4, modeNo)[1],
ops.nodeEigenvector(nd4, modeNo)[2]])
else:
ed = np.array([ops.nodeDisp(nd1)[0],
ops.nodeDisp(nd1)[1],
ops.nodeDisp(nd1)[2],
ops.nodeDisp(nd2)[0],
ops.nodeDisp(nd2)[1],
ops.nodeDisp(nd2)[2],
ops.nodeDisp(nd3)[0],
ops.nodeDisp(nd3)[1],
ops.nodeDisp(nd3)[2],
ops.nodeDisp(nd4)[0],
ops.nodeDisp(nd4)[1],
ops.nodeDisp(nd4)[2]])
if unDefoFlag:
ax.plot(np.append(ex, ex[0]),
np.append(ey, ey[0]),
np.append(ez, ez[0]),
fmt_undefo)
x = ex+sfac*ed[[0, 3, 6, 9]]
y = ey+sfac*ed[[1, 4, 7, 10]]
z = ez+sfac*ed[[2, 5, 8, 11]]
# ax.plot(np.append(x, x[0]),
# np.append(y, y[0]),
# np.append(z, z[0]),
# 'b.-')
# ax.axis('equal')
pts = [[x[0], y[0], z[0]],
[x[1], y[1], z[1]],
[x[2], y[2], z[2]],
[x[3], y[3], z[3]]]
verts = [[pts[0], pts[1], pts[2], pts[3]]]
ax.add_collection3d(Poly3DCollection(verts, linewidths=1,
edgecolors='k',
alpha=.25))
ax.scatter(x, y, z, s=0)
# 8-node brick, 3d model
elif nen == 8:
for i, ele_tag in enumerate(ele_tags):
nd1, nd2, nd3, nd4, nd5, nd6, nd7, nd8 = ops.eleNodes(ele_tag)
# element node1-node2, x, y coordinates
ex = np.array([ops.nodeCoord(nd1)[0],
ops.nodeCoord(nd2)[0],
ops.nodeCoord(nd3)[0],
ops.nodeCoord(nd4)[0],
ops.nodeCoord(nd5)[0],
ops.nodeCoord(nd6)[0],
ops.nodeCoord(nd7)[0],
ops.nodeCoord(nd8)[0]])
ey = np.array([ops.nodeCoord(nd1)[1],
ops.nodeCoord(nd2)[1],
ops.nodeCoord(nd3)[1],
ops.nodeCoord(nd4)[1],
ops.nodeCoord(nd5)[1],
ops.nodeCoord(nd6)[1],
ops.nodeCoord(nd7)[1],
ops.nodeCoord(nd8)[1]])
ez = np.array([ops.nodeCoord(nd1)[2],
ops.nodeCoord(nd2)[2],
ops.nodeCoord(nd3)[2],
ops.nodeCoord(nd4)[2],
ops.nodeCoord(nd5)[2],
ops.nodeCoord(nd6)[2],
ops.nodeCoord(nd7)[2],
ops.nodeCoord(nd8)[2]])
if modeNo:
ed = np.array([ops.nodeEigenvector(nd1, modeNo)[0],
ops.nodeEigenvector(nd1, modeNo)[1],
ops.nodeEigenvector(nd1, modeNo)[2],
ops.nodeEigenvector(nd2, modeNo)[0],
ops.nodeEigenvector(nd2, modeNo)[1],
ops.nodeEigenvector(nd2, modeNo)[2],
ops.nodeEigenvector(nd3, modeNo)[0],
ops.nodeEigenvector(nd3, modeNo)[1],
ops.nodeEigenvector(nd3, modeNo)[2],
ops.nodeEigenvector(nd4, modeNo)[0],
ops.nodeEigenvector(nd4, modeNo)[1],
ops.nodeEigenvector(nd4, modeNo)[2],
ops.nodeEigenvector(nd5, modeNo)[0],
ops.nodeEigenvector(nd5, modeNo)[1],
ops.nodeEigenvector(nd5, modeNo)[2],
ops.nodeEigenvector(nd6, modeNo)[0],
ops.nodeEigenvector(nd6, modeNo)[1],
ops.nodeEigenvector(nd6, modeNo)[2],
ops.nodeEigenvector(nd7, modeNo)[0],
ops.nodeEigenvector(nd7, modeNo)[1],
ops.nodeEigenvector(nd7, modeNo)[2],
ops.nodeEigenvector(nd8, modeNo)[0],
ops.nodeEigenvector(nd8, modeNo)[1],
ops.nodeEigenvector(nd8, modeNo)[2]])
else:
ed = np.array([ops.nodeDisp(nd1)[0],
ops.nodeDisp(nd1)[1],
ops.nodeDisp(nd1)[2],
ops.nodeDisp(nd2)[0],
ops.nodeDisp(nd2)[1],
ops.nodeDisp(nd2)[2],
ops.nodeDisp(nd3)[0],
ops.nodeDisp(nd3)[1],
ops.nodeDisp(nd3)[2],
ops.nodeDisp(nd4)[0],
ops.nodeDisp(nd4)[1],
ops.nodeDisp(nd4)[2],
ops.nodeDisp(nd5)[0],
ops.nodeDisp(nd5)[1],
ops.nodeDisp(nd5)[2],
ops.nodeDisp(nd6)[0],
ops.nodeDisp(nd6)[1],
ops.nodeDisp(nd6)[2],
ops.nodeDisp(nd7)[0],
ops.nodeDisp(nd7)[1],
ops.nodeDisp(nd7)[2],
ops.nodeDisp(nd8)[0],
ops.nodeDisp(nd8)[1],
ops.nodeDisp(nd8)[2]])
if unDefoFlag:
ax.plot(np.append(ex[0:4], ex[0]),
np.append(ey[0:4], ey[0]),
np.append(ez[0:4], ez[0]), fmt_undefo)
ax.plot(np.append(ex[4:8], ex[4]),
np.append(ey[4:8], ey[4]),
np.append(ez[4:8], ez[4]), fmt_undefo)
ax.plot(np.array([ex[0], ex[4]]),
np.array([ey[0], ey[4]]),
np.array([ez[0], ez[4]]), fmt_undefo)
ax.plot(np.array([ex[1], ex[5]]),
np.array([ey[1], ey[5]]),
np.array([ez[1], ez[5]]), fmt_undefo)
ax.plot(np.array([ex[2], ex[6]]),
np.array([ey[2], ey[6]]),
np.array([ez[2], ez[6]]), fmt_undefo)
ax.plot(np.array([ex[3], ex[7]]),
np.array([ey[3], ey[7]]),
np.array([ez[3], ez[7]]), fmt_undefo)
x = ex+sfac*ed[[0, 3, 6, 9, 12, 15, 18, 21]]
y = ey+sfac*ed[[1, 4, 7, 10, 13, 16, 19, 22]]
z = ez+sfac*ed[[2, 5, 8, 11, 14, 17, 20, 23]]
ax.plot(np.append(x[:4], x[0]),
np.append(y[:4], y[0]),
np.append(z[:4], z[0]),
'b.-')
ax.plot(np.append(x[4:8], x[4]),
np.append(y[4:8], y[4]),
np.append(z[4:8], z[4]),
'b.-')
ax.plot(np.array([x[0], x[4]]),
np.array([y[0], y[4]]),
np.array([z[0], z[4]]), 'b.-')
ax.plot(np.array([x[1], x[5]]),
np.array([y[1], y[5]]),
np.array([z[1], z[5]]), 'b.-')
ax.plot(np.array([x[2], x[6]]),
np.array([y[2], y[6]]),
np.array([z[2], z[6]]), 'b.-')
ax.plot(np.array([x[3], x[7]]),
np.array([y[3], y[7]]),
np.array([z[3], z[7]]), 'b.-')
# ax.axis('equal')
# work-around fix because of aspect equal bug
xmin, xmax = ax.get_xlim()
ymin, ymax = ax.get_ylim()
zmin, zmax = ax.get_zlim()
min_overall = np.amax([np.abs(xmin), np.abs(ymin), np.abs(zmin)])
max_overall = np.amax([np.abs(xmax), np.abs(ymax), np.abs(zmax)])
minmax_overall = max(min_overall, max_overall)
_min_overall = -1.1 * minmax_overall
_max_overall = 1.1 * minmax_overall
ax.set_xlim(0.3*_min_overall, 0.3*_max_overall)
ax.set_ylim(0.3*_min_overall, 0.3*_max_overall)
# ax.set_zlim(_min_overall, _max_overall)
ax.set_zlim(0.0, _max_overall)
def plot_defo(sfac=False, nep=17, unDefoFlag=1, fmt_undefo=fmt_undefo,
interpFlag=1, endDispFlag=1, fmt_interp=fmt_interp,
fmt_nodes=fmt_nodes, Eo=0, az_el=az_el, fig_wi_he=fig_wi_he,
fig_lbrt=fig_lbrt):
"""Plot deformed shape of the structure.
Args:
sfac (float): scale factor to increase/decrease displacements obtained
from FE analysis. If not specified (False), sfac is automatically
calculated based on the maximum overall displacement and this
maximum displacement is plotted as 20 percent (hordcoded) of
the maximum model dimension.
interpFlag (int): 1 - use interpolated deformation using shape
function, 0 - do not use interpolation, just show displaced element
nodes (default is 1)
nep (int): number of evaluation points for shape function interpolation
(default: 17)
Usage:
``plot_defo()`` - plot deformed shape with default parameters and
automatically calcutated scale factor.
``plot_defo(interpFlag=0)`` - plot simplified deformation by
displacing the nodes connected with straight lines (shape function
interpolation)
``plot_defo(sfac=1.5)`` - plot with specified scale factor
``plot_defo(unDefoFlag=0, endDispFlag=0)`` - plot without showing
undeformed (original) mesh and without showing markers at the
element ends.
"""
node_tags = ops.getNodeTags()
# calculate sfac
min_x, min_y, min_z = np.inf, np.inf, np.inf
max_x, max_y, max_z = -np.inf, -np.inf, -np.inf
max_ux, max_uy, max_uz = -np.inf, -np.inf, -np.inf
ratio = 0.1
ndim = np.shape(ops.nodeCoord(node_tags[0]))[0]
if ndim == 2:
if not sfac:
for node_tag in node_tags:
x_crd = ops.nodeCoord(node_tag)[0]
y_crd = ops.nodeCoord(node_tag)[1]
ux = ops.nodeDisp(node_tag)[0]
uy = ops.nodeDisp(node_tag)[1]
min_x = min(min_x, x_crd)
min_y = min(min_y, y_crd)
max_x = max(max_x, x_crd)
max_y = max(max_y, y_crd)
max_ux = max(max_ux, np.abs(ux))
max_uy = max(max_uy, np.abs(uy))
dxmax = max_x - min_x
dymax = max_y - min_y
dlmax = max(dxmax, dymax)
edmax = max(max_ux, max_uy)
sfac = ratio * dlmax/edmax
if sfac > 1000.:
print("""\nWarning!\nsfac is quite large - perhaps try to specify \
sfac value yourself.
This usually happens when translational DOFs are too small\n\n""")
_plot_defo_mode_2d(0, sfac, nep, unDefoFlag, fmt_undefo, interpFlag,
endDispFlag, fmt_interp, fmt_nodes)
elif ndim == 3:
if not sfac:
for node_tag in node_tags:
x_crd = ops.nodeCoord(node_tag)[0]
y_crd = ops.nodeCoord(node_tag)[1]
z_crd = ops.nodeCoord(node_tag)[2]
ux = ops.nodeDisp(node_tag)[0]
uy = ops.nodeDisp(node_tag)[1]
uz = ops.nodeDisp(node_tag)[2]
min_x = min(min_x, x_crd)
min_y = min(min_y, y_crd)
min_z = min(min_z, z_crd)
max_x = max(max_x, x_crd)
max_y = max(max_y, y_crd)
max_z = max(max_z, z_crd)
max_ux = max(max_ux, np.abs(ux))
max_uy = max(max_uy, np.abs(uy))
max_uz = max(max_uz, np.abs(uz))
dxmax = max_x - min_x
dymax = max_y - min_y
dzmax = max_z - min_z
dlmax = max(dxmax, dymax, dzmax)
edmax = max(max_ux, max_uy, max_uz)
sfac = ratio * dlmax/edmax
_plot_defo_mode_3d(0, sfac, nep, unDefoFlag, fmt_undefo, interpFlag,
endDispFlag, fmt_interp, fmt_nodes, az_el,
fig_wi_he, fig_lbrt)
else:
print(f'\nWarning! ndim: {ndim} not supported yet.')
def _anim_mode_2d(modeNo, sfac, nep, unDefoFlag, fmt_undefo, interpFlag,
endDispFlag, fmt_interp, fmt_nodes, fig_wi_he, xlim, ylim,
lw):
fig_wi, fig_he = fig_wi_he
ele_tags = ops.getEleTags()
nen = np.shape(ops.eleNodes(ele_tags[0]))[0]
# truss and beam/frame elements
if nen == 2:
ndf = np.shape(ops.nodeDOFs(ops.eleNodes(ele_tags[0])[0]))[0]
# truss element
if ndf == 2:
for ele_tag in ele_tags:
nd1, nd2 = ops.eleNodes(ele_tag)
# element x, y coordinates
ex = np.array([ops.nodeCoord(nd1)[0],
ops.nodeCoord(nd2)[0]])
ey = np.array([ops.nodeCoord(nd1)[1],
ops.nodeCoord(nd2)[1]])
if modeNo:
eux = np.array([ops.nodeEigenvector(nd1, modeNo)[0],
ops.nodeEigenvector(nd2, modeNo)[0]])
euy = np.array([ops.nodeEigenvector(nd1, modeNo)[1],
ops.nodeEigenvector(nd2, modeNo)[1]])
else:
eux = np.array([ops.nodeDisp(nd1)[0],
ops.nodeDisp(nd2)[0]])
euy = np.array([ops.nodeDisp(nd1)[1],
ops.nodeDisp(nd2)[1]])
# displaced element coordinates (scaled by sfac factor)
edx = np.array([ex[0] + sfac*eux[0], ex[1] + sfac*eux[1]])
edy = np.array([ey[0] + sfac*euy[0], ey[1] + sfac*euy[1]])
if unDefoFlag:
plt.plot(ex, ey, fmt_undefo)
plt.plot(edx, edy, fmt_interp)
# beam/frame element anim eigen
elif ndf == 3:
fig, ax = plt.subplots(figsize=(fig_wi/2.54, fig_he/2.54))
ax.axis('equal')
ax.set_xlim(xlim[0], xlim[1])
ax.set_ylim(ylim[0], ylim[1])
nel = len(ele_tags)
Ex = np.zeros((nel, 2))
Ey = np.zeros((nel, 2))
Ed = np.zeros((nel, 6))
# time vector for one cycle (period)
n_frames = 32 + 1
t = np.linspace(0., 2*np.pi, n_frames)
lines = []
for i, ele_tag in enumerate(ele_tags):
nd1, nd2 = ops.eleNodes(ele_tag)
# element x, y coordinates
Ex[i, :] = np.array([ops.nodeCoord(nd1)[0],
ops.nodeCoord(nd2)[0]])
Ey[i, :] = np.array([ops.nodeCoord(nd1)[1],
ops.nodeCoord(nd2)[1]])
Ed[i, :] = np.array([ops.nodeEigenvector(nd1, modeNo)[0],
ops.nodeEigenvector(nd1, modeNo)[1],
ops.nodeEigenvector(nd1, modeNo)[2],
ops.nodeEigenvector(nd2, modeNo)[0],
ops.nodeEigenvector(nd2, modeNo)[1],
ops.nodeEigenvector(nd2, modeNo)[2]])
lines.append(ax.plot([], [], fmt_nodes, lw=lw)[0])
def init():
for j, ele_tag in enumerate(ele_tags):
lines[j].set_data([], [])
return lines
def animate(i):
for j, ele_tag in enumerate(ele_tags):
if interpFlag:
xcdi, ycdi = beam_defo_interp_2d(Ex[j, :],
Ey[j, :],
Ed[j, :],
sfac*np.cos(t[i]),
nep)
lines[j].set_data(xcdi, ycdi)
else:
xdi, ydi = beam_disp_ends(Ex[j, :], Ey[j, :], Ed[j, :],
sfac*np.cos(t[i]))
lines[j].set_data(xdi, ydi)
# plt.plot(xcdi, ycdi, fmt_interp)
return lines
FuncAnimation(fig, animate, init_func=init,
frames=n_frames, interval=50, blit=True)
# plt.axis('equal')
# plt.show() # call this from main py file for more control
# 2d triangular elements - todo
# elif nen == 3:
# x = ex+sfac*ed[:, [0, 2, 4]]
# y = ex+sfac*ed[:, [1, 3, 5]]
# xc = [x, x[0, :]]
# yc = [x, x[0, :]]
# 2d quadrilateral (quad) elements
elif nen == 4:
for ele_tag in ele_tags:
nd1, nd2, nd3, nd4 = ops.eleNodes(ele_tag)
# element x, y coordinates
ex = np.array([ops.nodeCoord(nd1)[0],
ops.nodeCoord(nd2)[0],
ops.nodeCoord(nd3)[0],
ops.nodeCoord(nd4)[0]])
ey = np.array([ops.nodeCoord(nd1)[1],
ops.nodeCoord(nd2)[1],
ops.nodeCoord(nd3)[1],
ops.nodeCoord(nd4)[1]])
if modeNo:
ed = np.array([ops.nodeEigenvector(nd1, modeNo)[0],
ops.nodeEigenvector(nd1, modeNo)[1],
ops.nodeEigenvector(nd2, modeNo)[0],
ops.nodeEigenvector(nd2, modeNo)[1],
ops.nodeEigenvector(nd3, modeNo)[0],
ops.nodeEigenvector(nd3, modeNo)[1],
ops.nodeEigenvector(nd4, modeNo)[0],
ops.nodeEigenvector(nd4, modeNo)[1]])
else:
ed = np.array([ops.nodeDisp(nd1)[0],
ops.nodeDisp(nd1)[1],
ops.nodeDisp(nd2)[0],
ops.nodeDisp(nd2)[1],
ops.nodeDisp(nd3)[0],
ops.nodeDisp(nd3)[1],
ops.nodeDisp(nd4)[0],
ops.nodeDisp(nd4)[1]])
if unDefoFlag:
plt.plot(np.append(ex, ex[0]), np.append(ey, ey[0]),
fmt_undefo)
# xcdi, ycdi = beam_defo_interp_2d(ex, ey, ed, sfac, nep)
# xdi, ydi = beam_disp_ends(ex, ey, ed, sfac)
# # interpolated displacement field
# plt.plot(xcdi, ycdi, 'b.-')
# # translations of ends only
# plt.plot(xdi, ydi, 'ro')
# test it with one element
x = ex+sfac*ed[[0, 2, 4, 6]]
y = ey+sfac*ed[[1, 3, 5, 7]]
plt.plot(np.append(x, x[0]), np.append(y, y[0]), 'b.-')
plt.axis('equal')
# 2d 8-node quadratic elements
# elif nen == 8:
# x = ex+sfac*ed[:, [0, 2, 4, 6, 8, 10, 12, 14]]
# y = ex+sfac*ed[:, [1, 3, 5, 7, 9, 11, 13, 15]]
# t = -1
# n = 0
# for s in range(-1, 1.4, 0.4):
# n += 1
# ...
else:
print(f'\nWarning! Elements not supported yet. nen: {nen}; must be: 2, 3, 4, 8.') # noqa: E501
def anim_mode(modeNo, sfac=False, nep=17, unDefoFlag=1, fmt_undefo=fmt_undefo,
interpFlag=1, endDispFlag=1, fmt_interp=fmt_interp,
fmt_nodes='b-', Eo=0, az_el=az_el, fig_wi_he=fig_wi_he,
fig_lbrt=fig_lbrt, xlim=[0, 1], ylim=[0, 1], lw=3.):
"""Make animation of a mode shape obtained from eigenvalue solution.
Args:
modeNo (int): indicates which mode shape to animate.
Eds (ndarray): An array (n_eles x n_dof_per_element) containing
displacements per element.
timeV (1darray): vector of discretized time values
sfac (float): scale factor
nep (integer): number of evaluation points inside the element and
including both element ends
unDefoFlag (integer): 1 - plot the undeformed model (mesh), 0 - do not
plot the mesh
interpFlag (integer): 1 - interpolate deformation inside element,
0 - no interpolation
endDispFlag (integer): 1 - plot marks at element ends, 0 - no marks
fmt_interp (string): format line string for interpolated (continuous)
deformated shape. The format contains information on line color,
style and marks as in the standard matplotlib plot function.
fmt_nodes (string): format string for the marks of element ends
az_el (tuple): a tuple containing the azimuth and elevation
fig_lbrt (tuple): a tuple contating left, bottom, right and top offsets
fig_wi_he (tuple): contains width and height of the figure
Examples:
Notes:
See also:
"""
node_tags = ops.getNodeTags()
# calculate sfac
# min_x, min_y, min_z = np.inf, np.inf, np.inf
# max_x, max_y, max_z = -np.inf, -np.inf, -np.inf
# max_ux, max_uy, max_uz = -np.inf, -np.inf, -np.inf
min_x, min_y = np.inf, np.inf
max_x, max_y = -np.inf, -np.inf
max_ux, max_uy = -np.inf, -np.inf
ratio = 0.1
ndim = np.shape(ops.nodeCoord(node_tags[0]))[0]
if ndim == 2:
if not sfac:
for node_tag in node_tags:
x_crd = ops.nodeCoord(node_tag)[0]
y_crd = ops.nodeCoord(node_tag)[1]
ux = ops.nodeEigenvector(node_tag, modeNo)[0]
uy = ops.nodeEigenvector(node_tag, modeNo)[1]
min_x = min(min_x, x_crd)
min_y = min(min_y, y_crd)
max_x = max(max_x, x_crd)
max_y = max(max_y, y_crd)
max_ux = max(max_ux, np.abs(ux))
max_uy = max(max_uy, np.abs(uy))
dxmax = max_x - min_x
dymax = max_y - min_y
dlmax = max(dxmax, dymax)
edmax = max(max_ux, max_uy)
sfac = ratio * dlmax/edmax
_anim_mode_2d(modeNo, sfac, nep, unDefoFlag, fmt_undefo, interpFlag,
endDispFlag, fmt_interp, fmt_nodes, fig_wi_he, xlim,
ylim, lw)
# elif ndim == 3:
# if not sfac:
# for node_tag in node_tags:
# x_crd = ops.nodeCoord(node_tag)[0]
# y_crd = ops.nodeCoord(node_tag)[1]
# z_crd = ops.nodeCoord(node_tag)[2]
# ux = ops.nodeEigenvector(node_tag, modeNo)[0]
# uy = ops.nodeEigenvector(node_tag, modeNo)[1]
# uz = ops.nodeEigenvector(node_tag, modeNo)[2]
# min_x = min(min_x, x_crd)
# min_y = min(min_y, y_crd)
# min_z = min(min_z, z_crd)
# max_x = max(max_x, x_crd)
# max_y = max(max_y, y_crd)
# max_z = max(max_z, z_crd)
# max_ux = max(max_ux, np.abs(ux))
# max_uy = max(max_uy, np.abs(uy))
# max_uz = max(max_uz, np.abs(uz))
# dxmax = max_x - min_x
# dymax = max_y - min_y
# dzmax = max_z - min_z
# dlmax = max(dxmax, dymax, dzmax)
# edmax = max(max_ux, max_uy, max_uz)
# sfac = ratio * dlmax/edmax
# _plot_defo_mode_3d(modeNo, sfac, nep, unDefoFlag, fmt_undefo,
# interpFlag, endDispFlag, fmt_interp, fmt_nodes,
# Eo, az_el, fig_wi_he, fig_lbrt)
else:
print(f'\nWarning! ndim: {ndim} not supported yet.')
def plot_mode_shape(modeNo, sfac=False, nep=17, unDefoFlag=1,
fmt_undefo=fmt_undefo, interpFlag=1, endDispFlag=1,
fmt_interp=fmt_interp, fmt_nodes=fmt_nodes, Eo=0,
az_el=az_el, fig_wi_he=fig_wi_he, fig_lbrt=fig_lbrt):
"""Plot mode shape of the structure obtained from eigenvalue analysis.
Args:
modeNo (int): indicates which mode shape to plot
sfac (float): scale factor to increase/decrease displacements obtained
from FE analysis. If not specified (False), sfac is automatically
calculated based on the maximum overall displacement and this
maximum displacement is plotted as 20 percent (hordcoded) of
the maximum model dimension.
interpFlag (int): 1 - use interpolated deformation using shape
function, 0 - do not use interpolation, just show displaced element
nodes (default is 1)
nep (int): number of evaluation points for shape function interpolation
(default: 17)
Usage:
``plot_mode_shape(1)`` - plot the first mode shape with default parameters
and automatically calcutated scale factor.
``plot_mode_shape(2, interpFlag=0)`` - plot the 2nd mode shape by
displacing the nodes connected with straight lines (shape function
interpolation)
``plot_mode_shape(3, sfac=1.5)`` - plot the 3rd mode shape with specified
scale factor
``plot_mode_shape(4, unDefoFlag=0, endDispFlag=0)`` - plot the 4th mode
shape without showing undeformed (original) mesh and without showing
markers at the element ends.
Examples:
Notes:
See also:
"""
node_tags = ops.getNodeTags()
# calculate sfac
min_x, min_y, min_z = np.inf, np.inf, np.inf
max_x, max_y, max_z = -np.inf, -np.inf, -np.inf
max_ux, max_uy, max_uz = -np.inf, -np.inf, -np.inf
ratio = 0.1
ndim = np.shape(ops.nodeCoord(node_tags[0]))[0]
if ndim == 2:
if not sfac:
for node_tag in node_tags:
x_crd = ops.nodeCoord(node_tag)[0]
y_crd = ops.nodeCoord(node_tag)[1]
ux = ops.nodeEigenvector(node_tag, modeNo)[0]
uy = ops.nodeEigenvector(node_tag, modeNo)[1]
min_x = min(min_x, x_crd)
min_y = min(min_y, y_crd)
max_x = max(max_x, x_crd)
max_y = max(max_y, y_crd)
max_ux = max(max_ux, np.abs(ux))
max_uy = max(max_uy, np.abs(uy))
dxmax = max_x - min_x
dymax = max_y - min_y
dlmax = max(dxmax, dymax)
edmax = max(max_ux, max_uy)
sfac = ratio * dlmax/edmax
_plot_defo_mode_2d(modeNo, sfac, nep, unDefoFlag, fmt_undefo,
interpFlag, endDispFlag, fmt_interp, fmt_nodes)
elif ndim == 3:
if not sfac:
for node_tag in node_tags:
x_crd = ops.nodeCoord(node_tag)[0]
y_crd = ops.nodeCoord(node_tag)[1]
z_crd = ops.nodeCoord(node_tag)[2]
ux = ops.nodeEigenvector(node_tag, modeNo)[0]
uy = ops.nodeEigenvector(node_tag, modeNo)[1]
uz = ops.nodeEigenvector(node_tag, modeNo)[2]
min_x = min(min_x, x_crd)
min_y = min(min_y, y_crd)
min_z = min(min_z, z_crd)
max_x = max(max_x, x_crd)
max_y = max(max_y, y_crd)
max_z = max(max_z, z_crd)
max_ux = max(max_ux, np.abs(ux))
max_uy = max(max_uy, np.abs(uy))
max_uz = max(max_uz, np.abs(uz))
dxmax = max_x - min_x
dymax = max_y - min_y
dzmax = max_z - min_z
dlmax = max(dxmax, dymax, dzmax)
edmax = max(max_ux, max_uy, max_uz)
sfac = ratio * dlmax/edmax
_plot_defo_mode_3d(modeNo, sfac, nep, unDefoFlag, fmt_undefo,
interpFlag, endDispFlag, fmt_interp, fmt_nodes,
az_el, fig_wi_he, fig_lbrt)
else:
print(f'\nWarning! ndim: {ndim} not supported yet.')
def rot_transf_3d(ex, ey, ez, g):
Lxyz = np.array([ex[1]-ex[0], ey[1]-ey[0], ez[1]-ez[0]])
L = np.sqrt(Lxyz @ Lxyz)
z = np.zeros((3, 3))
G = np.block([[g, z, z, z],
[z, g, z, z],
[z, z, g, z],
[z, z, z, g]])
return G, L
def beam_defo_interp_2d(ex, ey, u, sfac, nep=17):
"""
Interpolate element displacements at nep points.
Parametrs:
ex, ey : element x, y coordinates,
u : element nodal displacements
sfac : scale factor for deformation plot
nep : number of evaluation points (including end nodes)
Returns:
crd_xc, crd_yc : x, y coordinates of interpolated (at nep points)
beam deformation required for plot_defo() function
"""
Lxy = np.array([ex[1]-ex[0], ey[1]-ey[0]])
L = np.sqrt(Lxy @ Lxy)
cosa, cosb = Lxy / L
G = np.array([[cosa, cosb, 0., 0., 0., 0.],
[-cosb, cosa, 0., 0., 0., 0.],
[0., 0., 1., 0., 0., 0.],
[0., 0., 0., cosa, cosb, 0.],
[0., 0., 0., -cosb, cosa, 0.],
[0., 0., 0., 0., 0., 1.]])
u_l = G @ u
xl = np.linspace(0., L, num=nep)
one = np.ones(xl.shape)
# longitudinal deformation (1)
N_a = np.column_stack((one - xl/L, xl/L))
u_ac = N_a @ np.array([u_l[0], u_l[3]])
# transverse deformation (2)
N_t = np.column_stack((one - 3*xl**2/L**2 + 2*xl**3/L**3,
xl - 2*xl**2/L + xl**3/L**2,
3*xl**2/L**2 - 2*xl**3/L**3,
-xl**2/L + xl**3/L**2))
u_tc = N_t @ np.array([u_l[1], u_l[2], u_l[4], u_l[5]])
# combined two row vectors
# 1-st vector longitudinal deformation (1)
# 2-nd vector transverse deformation (2)
u_atc = np.vstack((u_ac, u_tc))
# project longitudinal (u_ac) and transverse deformation
# (local u and v) to (global u and v)
G1 = np.array([[cosa, -cosb],
[cosb, cosa]])
u_xyc = G1 @ u_atc
# discretize element coordinates
# first row = X + [0 dx 2dx ... 4-dx 4]
# second row = Y + [0 dy 2dy ... 3-dy 3]
xy_c = np.vstack((np.linspace(ex[0], ex[1], num=nep),
np.linspace(ey[0], ey[1], num=nep)))
# Continuous x, y displacement coordinates
crd_xc = xy_c[0, :] + sfac * u_xyc[0, :]
crd_yc = xy_c[1, :] + sfac * u_xyc[1, :]
# latex_array(ecrd_xc)
# latex_array(ecrd_yc)
return crd_xc, crd_yc
def beam_defo_interp_3d(ex, ey, ez, g, u, sfac, nep=17):
"""
3d beam version of beam_defo_interp_2d.
"""
G, L = rot_transf_3d(ex, ey, ez, g)
ul = G @ u
_, crd_yc = beam_defo_interp_2d(np.array([0., L]),
np.array([0., 0.]),
np.array([ul[0], ul[1], ul[5], ul[6],
ul[7], ul[11]]), sfac, nep)
crd_xc, crd_zc = beam_defo_interp_2d(np.array([0., L]),
np.array([0., 0.]),
np.array([ul[0], ul[2], -ul[4], ul[6],
ul[8], -ul[10]]), sfac, nep)
xl = np.linspace(0., L, num=nep)
crd_xc = crd_xc - xl
crd_xyzc = np.vstack([crd_xc, crd_yc, crd_zc])
u_xyzc = np.transpose(g) @ crd_xyzc
xyz_c = np.vstack((np.linspace(ex[0], ex[1], num=nep),
np.linspace(ey[0], ey[1], num=nep),
np.linspace(ez[0], ez[1], num=nep)))
crd_xc = xyz_c[0, :] + u_xyzc[0, :]
crd_yc = xyz_c[1, :] + u_xyzc[1, :]
crd_zc = xyz_c[2, :] + u_xyzc[2, :]
return crd_xc, crd_yc, crd_zc
def beam_disp_ends(ex, ey, d, sfac):
"""
Calculate the element deformation at element ends only.
"""
# indx: 0 1 2 3 4 5
# Ed = ux1 uy1 ur1 ux2 uy2 ur2
exd = np.array([ex[0] + sfac*d[0], ex[1] + sfac*d[3]])
eyd = np.array([ey[0] + sfac*d[1], ey[1] + sfac*d[4]])
return exd, eyd
def beam_disp_ends3d(ex, ey, ez, d, sfac):
"""
Calculate the element deformation at element ends only.
"""
# indx: 0 1 2 3 4 5 6 7 8 9 10 11
# Ed = ux1 uy1 uz1 rx1 ry1 rz1 ux2 uy2 uz2 rx2 ry2 rz2
exd = np.array([ex[0] + sfac*d[0], ex[1] + sfac*d[6]])
eyd = np.array([ey[0] + sfac*d[1], ey[1] + sfac*d[7]])
ezd = np.array([ez[0] + sfac*d[2], ez[1] + sfac*d[8]])
return exd, eyd, ezd
# plot_fiber_section is inspired by Matlab ``plotSection.zip``
# written by <NAME> available at
# http://users.ntua.gr/divamva/software.html
def plot_fiber_section(fib_sec_list, fillflag=1,
matcolor=['y', 'b', 'r', 'g', 'm', 'k']):
"""Plot fiber cross-section.
Args:
fib_sec_list (list): list of lists in the format similar to the input
given for in
fillflag (int): 1 - filled fibers with color specified in matcolor
list, 0 - no color, only the outline of fibers
matcolor (list): sequence of colors for various material tags
assigned to fibers
Examples:
::
fib_sec_1 = [['section', 'Fiber', 1, '-GJ', 1.0e6],
['patch', 'quad', 1, 4, 1, 0.032, 0.317, -0.311, 0.067, -0.266, 0.005, 0.077, 0.254], # noqa: E501
['patch', 'quad', 1, 1, 4, -0.075, 0.144, -0.114, 0.116, 0.075, -0.144, 0.114, -0.116], # noqa: E501
['patch', 'quad', 1, 4, 1, 0.266, -0.005, -0.077, -0.254, -0.032, -0.317, 0.311, -0.067] # noqa: E501
]
opsv.fib_sec_list_to_cmds(fib_sec_1)
matcolor = ['r', 'lightgrey', 'gold', 'w', 'w', 'w']
opsv.plot_fiber_section(fib_sec_1, matcolor=matcolor)
plt.axis('equal')
# plt.savefig(f'{kateps}fibsec_rc.png')
plt.show()
Notes:
``fib_sec_list`` can be reused by means of a python helper function
``ops_vis.fib_sec_list_to_cmds(fib_sec_list_1)``
See also:
``ops_vis.fib_sec_list_to_cmds()``
"""
fig, ax = plt.subplots()
ax.set_xlabel('z')
ax.set_ylabel('y')
ax.grid(False)
for item in fib_sec_list:
if item[0] == 'layer':
matTag = item[2]
if item[1] == 'straight':
n_bars = item[3]
As = item[4]
Iy, Iz, Jy, Jz = item[5], item[6], item[7], item[8]
r = np.sqrt(As / np.pi)
Y = np.linspace(Iy, Jy, n_bars)
Z = np.linspace(Iz, Jz, n_bars)
for zi, yi in zip(Z, Y):
bar = Circle((zi, yi), r, ec='k', fc='k', zorder=10)
ax.add_patch(bar)
if item[0] == 'patch':
matTag, nIJ, nJK = item[2], item[3], item[4]
if item[1] == 'quad' or item[1] == 'quadr':
Iy, Iz, Jy, Jz = item[5], item[6], item[7], item[8]
Ky, Kz, Ly, Lz = item[9], item[10], item[11], item[12]
if item[1] == 'rect':
Iy, Iz, Ky, Kz = item[5], item[6], item[7], item[8]
Jy, Jz, Ly, Lz = Ky, Iz, Iy, Kz
# check for convexity (vector products)
outIJxIK = (Jy-Iy)*(Kz-Iz) - (Ky-Iy)*(Jz-Iz)
outIKxIL = (Ky-Iy)*(Lz-Iz) - (Ly-Iy)*(Kz-Iz)
# check if I, J, L points are colinear
outIJxIL = (Jy-Iy)*(Lz-Iz) - (Ly-Iy)*(Jz-Iz)
# outJKxJL = (Ky-Jy)*(Lz-Jz) - (Ly-Jy)*(Kz-Jz)
if outIJxIK <= 0 or outIKxIL <= 0 or outIJxIL <= 0:
print('\nWarning! Patch quad is non-convex or counter-clockwise defined or has at least 3 colinear points in line') # noqa: E501
IJz, IJy = np.linspace(Iz, Jz, nIJ+1), np.linspace(Iy, Jy, nIJ+1)
JKz, JKy = np.linspace(Jz, Kz, nJK+1), np.linspace(Jy, Ky, nJK+1)
LKz, LKy = np.linspace(Lz, Kz, nIJ+1), np.linspace(Ly, Ky, nIJ+1)
ILz, ILy = np.linspace(Iz, Lz, nJK+1), np.linspace(Iy, Ly, nJK+1)
if fillflag:
Z = np.zeros((nIJ+1, nJK+1))
Y = np.zeros((nIJ+1, nJK+1))
for j in range(nIJ+1):
Z[j, :] = np.linspace(IJz[j], LKz[j], nJK+1)
Y[j, :] = np.linspace(IJy[j], LKy[j], nJK+1)
for j in range(nIJ):
for k in range(nJK):
zy = np.array([[Z[j, k], Y[j, k]],
[Z[j, k+1], Y[j, k+1]],
[Z[j+1, k+1], Y[j+1, k+1]],
[Z[j+1, k], Y[j+1, k]]])
poly = Polygon(zy, True, ec='k', fc=matcolor[matTag-1])
ax.add_patch(poly)
else:
# horizontal lines
for az, bz, ay, by in zip(IJz, LKz, IJy, LKy):
plt.plot([az, bz], [ay, by], 'b-', zorder=1)
# vertical lines
for az, bz, ay, by in zip(JKz, ILz, JKy, ILy):
plt.plot([az, bz], [ay, by], 'b-', zorder=1)
def fib_sec_list_to_cmds(fib_sec_list):
"""Reuses fib_sec_list to define fiber section in OpenSees.
At present it is not possible to extract fiber section data from
the OpenSees domain, this function is a workaround. The idea is to
prepare data similar to the one the regular OpenSees commands
(``section('Fiber', ...)``, ``fiber()``, ``patch()`` and/or
``layer()``) require.
Args:
fib_sec_list (list): is a list of fiber section data. First sub-list
also defines the torsional stiffness (GJ).
Warning:
If you use this function, do not issue the regular OpenSees:
section, Fiber, Patch or Layer commands.
See also:
``ops_vis.plot_fiber_section()``
"""
for dat in fib_sec_list:
if dat[0] == 'section':
secTag, GJ = dat[2], dat[4]
ops.section('Fiber', secTag, '-GJ', GJ)
if dat[0] == 'layer':
matTag = dat[2]
if dat[1] == 'straight':
n_bars = dat[3]
As = dat[4]
Iy, Iz, Jy, Jz = dat[5], dat[6], dat[7], dat[8]
ops.layer('straight', matTag, n_bars, As, Iy, Iz, Jy, Jz)
if dat[0] == 'patch':
matTag = dat[2]
nIJ = dat[3]
nJK = dat[4]
if dat[1] == 'quad' or dat[1] == 'quadr':
Iy, Iz, Jy, Jz = dat[5], dat[6], dat[7], dat[8]
Ky, Kz, Ly, Lz = dat[9], dat[10], dat[11], dat[12]
ops.patch('quad', matTag, nIJ, nJK, Iy, Iz, Jy, Jz, Ky, Kz,
Ly, Lz)
if dat[1] == 'rect':
Iy, Iz, Ky, Kz = dat[5], dat[6], dat[7], dat[8]
Jy, Jz, Ly, Lz = Ky, Iz, Iy, Kz
ops.patch('rect', matTag, nIJ, nJK, Iy, Iz, Ky, Kz)
def _anim_defo_2d(Eds, timeV, sfac, nep, unDefoFlag, fmt_undefo,
interpFlag, endDispFlag, fmt_interp, fmt_nodes, fig_wi_he,
xlim, ylim):
fig_wi, fig_he = fig_wi_he
ele_tags = ops.getEleTags()
nen = np.shape(ops.eleNodes(ele_tags[0]))[0]
# truss and beam/frame elements
if nen == 2:
ndf = np.shape(ops.nodeDOFs(ops.eleNodes(ele_tags[0])[0]))[0]
# truss element
if ndf == 2:
for ele_tag in ele_tags:
nd1, nd2 = ops.eleNodes(ele_tag)
# element x, y coordinates
ex = np.array([ops.nodeCoord(nd1)[0],
ops.nodeCoord(nd2)[0]])
ey = np.array([ops.nodeCoord(nd1)[1],
ops.nodeCoord(nd2)[1]])
eux = np.array([ops.nodeDisp(nd1)[0],
ops.nodeDisp(nd2)[0]])
euy = np.array([ops.nodeDisp(nd1)[1],
ops.nodeDisp(nd2)[1]])
# displaced element coordinates (scaled by sfac factor)
edx = np.array([ex[0] + sfac*eux[0], ex[1] + sfac*eux[1]])
edy = np.array([ey[0] + sfac*euy[0], ey[1] + sfac*euy[1]])
if unDefoFlag:
plt.plot(ex, ey, fmt_undefo)
plt.plot(edx, edy, fmt_interp)
# beam/frame element anim defo
elif ndf == 3:
fig, ax = plt.subplots(figsize=(fig_wi/2.54, fig_he/2.54))
ax.axis('equal')
ax.set_xlim(xlim[0], xlim[1])
ax.set_ylim(ylim[0], ylim[1])
# ax.grid()
nel = len(ele_tags)
Ex = np.zeros((nel, 2))
Ey = np.zeros((nel, 2))
# no of frames equal to time intervals
n_frames, _, _ = np.shape(Eds)
lines = []
# time_text = ax.set_title('') # does not work
time_text = ax.text(.05, .95, '', transform=ax.transAxes)
for i, ele_tag in enumerate(ele_tags):
nd1, nd2 = ops.eleNodes(ele_tag)
# element x, y coordinates
Ex[i, :] = np.array([ops.nodeCoord(nd1)[0],
ops.nodeCoord(nd2)[0]])
Ey[i, :] = np.array([ops.nodeCoord(nd1)[1],
ops.nodeCoord(nd2)[1]])
lines.append(ax.plot([], [], fmt_nodes, lw=3)[0])
def init():
for j, ele_tag in enumerate(ele_tags):
lines[j].set_data([], [])
time_text.set_text('')
return tuple(lines) + (time_text,)
def animate(i):
for j, ele_tag in enumerate(ele_tags):
if interpFlag:
xcdi, ycdi = beam_defo_interp_2d(Ex[j, :],
Ey[j, :],
Eds[i, j, :],
sfac,
nep)
lines[j].set_data(xcdi, ycdi)
else:
xdi, ydi = beam_disp_ends(Ex[j, :], Ey[j, :],
Eds[i, j, :], sfac)
lines[j].set_data(xdi, ydi)
# plt.plot(xcdi, ycdi, fmt_interp)
# time_text.set_text(f'f')
time_text.set_text(f'frame: {i+1}/{n_frames}, \
time: {timeV[i]:.3f} s')
return tuple(lines) + (time_text,)
FuncAnimation(fig, animate, init_func=init, frames=n_frames,
interval=50, blit=True, repeat=False)
# plt.axis('equal')
# plt.show() # call this from main py file for more control
# 2d triangular elements
# elif nen == 3:
# x = ex+sfac*ed[:, [0, 2, 4]]
# y = ex+sfac*ed[:, [1, 3, 5]]
# xc = [x, x[0, :]]
# yc = [x, x[0, :]]
# 2d quadrilateral (quad) elements
elif nen == 4:
for ele_tag in ele_tags:
nd1, nd2, nd3, nd4 = ops.eleNodes(ele_tag)
# element x, y coordinates
ex = np.array([ops.nodeCoord(nd1)[0],
ops.nodeCoord(nd2)[0],
ops.nodeCoord(nd3)[0],
ops.nodeCoord(nd4)[0]])
ey = np.array([ops.nodeCoord(nd1)[1],
ops.nodeCoord(nd2)[1],
ops.nodeCoord(nd3)[1],
ops.nodeCoord(nd4)[1]])
# if modeNo:
# ed = np.array([ops.nodeEigenvector(nd1, modeNo)[0],
# ops.nodeEigenvector(nd1, modeNo)[1],
# ops.nodeEigenvector(nd2, modeNo)[0],
# ops.nodeEigenvector(nd2, modeNo)[1],
# ops.nodeEigenvector(nd3, modeNo)[0],
# ops.nodeEigenvector(nd3, modeNo)[1],
# ops.nodeEigenvector(nd4, modeNo)[0],
# ops.nodeEigenvector(nd4, modeNo)[1]])
# else:
ed = np.array([ops.nodeDisp(nd1)[0],
ops.nodeDisp(nd1)[1],
ops.nodeDisp(nd2)[0],
ops.nodeDisp(nd2)[1],
ops.nodeDisp(nd3)[0],
ops.nodeDisp(nd3)[1],
ops.nodeDisp(nd4)[0],
ops.nodeDisp(nd4)[1]])
if unDefoFlag:
plt.plot(np.append(ex, ex[0]), np.append(ey, ey[0]),
fmt_undefo)
# xcdi, ycdi = beam_defo_interp_2d(ex, ey, ed, sfac, nep)
# xdi, ydi = beam_disp_ends(ex, ey, ed, sfac)
# # interpolated displacement field
# plt.plot(xcdi, ycdi, 'b.-')
# # translations of ends only
# plt.plot(xdi, ydi, 'ro')
# test it with one element
x = ex+sfac*ed[[0, 2, 4, 6]]
y = ey+sfac*ed[[1, 3, 5, 7]]
plt.plot(np.append(x, x[0]), np.append(y, y[0]), 'b.-')
plt.axis('equal')
# 2d 8-node quadratic elements
# elif nen == 8:
# x = ex+sfac*ed[:, [0, 2, 4, 6, 8, 10, 12, 14]]
# y = ex+sfac*ed[:, [1, 3, 5, 7, 9, 11, 13, 15]]
# t = -1
# n = 0
# for s in range(-1, 1.4, 0.4):
# n += 1
# ...
else:
print(f'\nWarning! Elements not supported yet. nen: {nen}; must be: 2, 3, 4, 8.') # noqa: E501
def anim_defo(Eds, timeV, sfac, nep=17, unDefoFlag=1, fmt_undefo=fmt_undefo,
interpFlag=1, endDispFlag=1, fmt_interp=fmt_interp,
fmt_nodes='b-', az_el=az_el, fig_lbrt=fig_lbrt,
fig_wi_he=fig_wi_he, xlim=[0, 1], ylim=[0, 1]):
"""Make animation of the deformed shape computed by transient analysis
Args:
Eds (ndarray): An array (n_eles x n_dof_per_element) containing
displacements per element.
timeV (1darray): vector of discretized time values
sfac (float): scale factor
nep (integer): number of evaluation points inside the element and
including both element ends
unDefoFlag (integer): 1 - plot the undeformed model (mesh), 0 - do not
plot the mesh
interpFlag (integer): 1 - interpolate deformation inside element,
0 - no interpolation
endDispFlag (integer): 1 - plot marks at element ends, 0 - no marks
fmt_interp (string): format line string for interpolated (continuous)
deformated shape. The format contains information on line color,
style and marks as in the standard matplotlib plot function.
fmt_nodes (string): format string for the marks of element ends
az_el (tuple): a tuple containing the azimuth and elevation
fig_lbrt (tuple): a tuple contating left, bottom, right and top offsets
fig_wi_he (tuple): contains width and height of the figure
Examples:
Notes:
See also:
"""
node_tags = ops.getNodeTags()
ndim = np.shape(ops.nodeCoord(node_tags[0]))[0]
if ndim == 2:
_anim_defo_2d(Eds, timeV, sfac, nep, unDefoFlag, fmt_undefo,
interpFlag, endDispFlag, fmt_interp, fmt_nodes,
fig_wi_he, xlim, ylim)
else:
print(f'\nWarning! ndim: {ndim} not supported yet.')
def section_force_distribution_2d(ex, ey, pl, nep=2,
ele_load_data=['-beamUniform', 0., 0.]):
"""
Calculate section forces (N, V, M) for an elastic 2D Euler-Bernoulli beam.
Input:
ex, ey - x, y element coordinates in global system
nep - number of evaluation points, by default (2) at element ends
ele_load_list - list of transverse and longitudinal element load
syntax: [ele_load_type, Wy, Wx]
For now only '-beamUniform' element load type is acceptable
Output:
s = [N V M]; shape: (nep,3)
section forces at nep points along local x
xl: coordinates of local x-axis; shape: (nep,)
Use it with dia_sf to draw N, V, M diagrams.
TODO: add '-beamPoint' element load type
"""
# eload_type, Wy, Wx = ele_load_data[0], ele_load_data[1], ele_load_data[2]
Wy, Wx = ele_load_data[1], ele_load_data[2]
nlf = len(pl)
if nlf == 2: # trusses
N_1 = pl[0]
elif nlf == 6: # plane frames
# N_1, V_1, M_1 = pl[0], pl[1], pl[2]
N_1, V_1, M_1 = pl[:3]
else:
print('\nWarning! Not supported. Number of nodal forces: {nlf}')
Lxy = np.array([ex[1]-ex[0], ey[1]-ey[0]])
L = np.sqrt(Lxy @ Lxy)
xl = np.linspace(0., L, nep)
one = np.ones(nep)
N = -1.*(N_1 * one + Wx * xl)
if nlf == 6:
V = V_1 * one + Wy * xl
M = -M_1 * one + V_1 * xl + 0.5 * Wy * xl**2
s = np.column_stack((N, V, M))
elif nlf == 2:
s = np.column_stack((N))
# if eload_type == '-beamUniform':
# else:
return s, xl
def section_force_distribution_3d(ex, ey, ez, pl, nep=2,
ele_load_data=['-beamUniform', 0., 0., 0.]):
"""
Calculate section forces (N, Vy, Vz, T, My, Mz) for an elastic 3d beam.
Longer description
Parameters
----------
ex : list
x element coordinates
ey : list
y element coordinates
ez : list
z element coordinates
pl : ndarray
nep : int
number of evaluation points, by default (2) at element ends
ele_load_list : list
list of transverse and longitudinal element load
syntax: [ele_load_type, Wy, Wz, Wx]
For now only '-beamUniform' element load type is acceptable.
Returns
-------
s : ndarray
[N Vx Vy T My Mz]; shape: (nep,6)
column vectors of section forces along local x-axis
uvwfi : ndarray
[u v w fi]; shape (nep,4)
displacements at nep points along local x
xl : ndarray
coordinates of local x-axis; shape (nep,)
Notes
-----
Todo: add '-beamPoint' element load type
"""
# eload_type = ele_load_data[0]
Wy, Wz, Wx = ele_load_data[1], ele_load_data[2], ele_load_data[3]
N1, Vy1, Vz1, T1, My1, Mz1 = pl[:6]
Lxyz = np.array([ex[1]-ex[0], ey[1]-ey[0], ez[1]-ez[0]])
L = np.sqrt(Lxyz @ Lxyz)
xl = np.linspace(0., L, nep)
one = np.ones(nep)
N = -1.*(N1*one + Wx*xl)
Vy = Vy1*one + Wy*xl
Vz = Vz1*one + Wz*xl
T = -T1*one
Mz = -Mz1*one + Vy1*xl + 0.5*Wy*xl**2
My = My1*one + Vz1*xl + 0.5*Wz*xl**2
s = np.column_stack((N, Vy, Vz, T, My, Mz))
return s, xl
def section_force_diagram_2d(sf_type, Ew, sfac=1., nep=17,
fmt_secforce=fmt_secforce):
"""Display section forces diagram for 2d beam column model.
This function plots a section forces diagram for 2d beam column elements
with or without element loads. For now only '-beamUniform' constant
transverse or axial element loads are supported.
Args:
sf_type (str): type of section force: 'N' - normal force,
'V' - shear force, 'M' - bending moments.
Ew (dict): Ew Python dictionary contains information on non-zero
element loads, therfore each item of the Python dictionary
is in the form: 'ele_tag: ['-beamUniform', Wy, Wx]'.
sfac (float): scale factor by wich the values of section forces are
multiplied.
nep (int): number of evaluation points including both end nodes
(default: 17)
fmt_secforce (str): format line string for section force distribution
curve. The format contains information on line color, style and
marks as in the standard matplotlib plot function.
(default: fmt_secforce = 'b-' # blue solid line)
Usage:
::
Wy, Wx = -10.e+3, 0.
Ew = {3: ['-beamUniform', Wy, Wx]}
sfacM = 5.e-5
plt.figure()
minVal, maxVal = opsv.section_force_diagram_2d('M', Ew, sfacM)
plt.title('Bending moments')
Todo:
Add support for other element loads available in OpenSees: partial
(trapezoidal) uniform element load, and 'beamPoint' element load.
"""
maxVal, minVal = -np.inf, np.inf
ele_tags = ops.getEleTags()
for ele_tag in ele_tags:
# by default no element load
eload_data = ['', 0., 0.]
if ele_tag in Ew:
eload_data = Ew[ele_tag]
nd1, nd2 = ops.eleNodes(ele_tag)
# element x, y coordinates
ex = np.array([ops.nodeCoord(nd1)[0],
ops.nodeCoord(nd2)[0]])
ey = np.array([ops.nodeCoord(nd1)[1],
ops.nodeCoord(nd2)[1]])
Lxy = np.array([ex[1]-ex[0], ey[1]-ey[0]])
L = np.sqrt(Lxy @ Lxy)
cosa, cosb = Lxy / L
pl = ops.eleResponse(ele_tag, 'localForces')
s_all, xl = section_force_distribution_2d(ex, ey, pl, nep, eload_data)
if sf_type == 'N' or sf_type == 'axial':
s = s_all[:, 0]
elif sf_type == 'V' or sf_type == 'shear' or sf_type == 'T':
s = s_all[:, 1]
elif sf_type == 'M' or sf_type == 'moment':
s = s_all[:, 2]
minVal = min(minVal, np.min(s))
maxVal = max(maxVal, np.max(s))
s = s*sfac
s_0 = np.zeros((nep, 2))
s_0[0, :] = [ex[0], ey[0]]
s_0[1:, 0] = s_0[0, 0] + xl[1:] * cosa
s_0[1:, 1] = s_0[0, 1] + xl[1:] * cosb
s_p = np.copy(s_0)
# positive M are opposite to N and V
if sf_type == 'M' or sf_type == 'moment':
s *= -1.
s_p[:, 0] -= s * cosb
s_p[:, 1] += s * cosa
plt.axis('equal')
# section force curve
plt.plot(s_p[:, 0], s_p[:, 1], fmt_secforce,
solid_capstyle='round', solid_joinstyle='round',
dash_capstyle='butt', dash_joinstyle='round')
# model
plt.plot(ex, ey, 'k-', solid_capstyle='round', solid_joinstyle='round',
dash_capstyle='butt', dash_joinstyle='round')
# reference perpendicular lines
for i in np.arange(nep):
plt.plot([s_0[i, 0], s_p[i, 0]], [s_0[i, 1], s_p[i, 1]],
fmt_secforce, solid_capstyle='round',
solid_joinstyle='round', dash_capstyle='butt',
dash_joinstyle='round')
return minVal, maxVal
def section_force_diagram_3d(sf_type, Ew, sfac=1., nep=17,
fmt_secforce=fmt_secforce):
"""Display section forces diagram of a 3d beam column model.
This function plots section forces diagrams for 3d beam column elements
with or without element loads. For now only '-beamUniform' constant
transverse or axial element loads are supported.
Args:
sf_type (str): type of section force: 'N' - normal force,
'Vy' or 'Vz' - shear force, 'My' or 'Mz' - bending moments,
'T' - torsional moment.
Ew (dict): Ew Python dictionary contains information on non-zero
element loads, therfore each item of the Python dictionary
is in the form: 'ele_tag: ['-beamUniform', Wy, Wz, Wx]'.
sfac (float): scale factor by wich the values of section forces are
multiplied.
nep (int): number of evaluation points including both end nodes
(default: 17)
fmt_secforce (str): format line string for section force distribution
curve. The format contains information on line color, style and
marks as in the standard matplotlib plot function.
(default: fmt_secforce = 'b-' # blue solid line)
Usage:
::
Wy, Wz, Wx = -5., 0., 0.
Ew = {3: ['-beamUniform', Wy, Wz, Wx]}
sfacMz = 1.e-1
plt.figure()
minY, maxY = opsv.section_force_diagram_3d('Mz', Ew, sfacMz)
plt.title(f'Bending moments Mz, max = {maxY:.2f}, min = {minY:.2f}')
Todo:
Add support for other element loads available in OpenSees: partial
(trapezoidal) uniform element load, and 'beamPoint' element load.
"""
maxVal, minVal = -np.inf, np.inf
ele_tags = ops.getEleTags()
azim, elev = az_el
fig_wi, fig_he = fig_wi_he
fleft, fbottom, fright, ftop = fig_lbrt
fig = plt.figure(figsize=(fig_wi/2.54, fig_he/2.54))
fig.subplots_adjust(left=.08, bottom=.08, right=.985, top=.94)
ax = fig.add_subplot(111, projection=Axes3D.name)
# ax.axis('equal')
ax.set_xlabel('X')
ax.set_ylabel('Y')
ax.set_zlabel('Z')
ax.view_init(azim=azim, elev=elev)
for i, ele_tag in enumerate(ele_tags):
# by default no element load
eload_data = ['-beamUniform', 0., 0., 0.]
if ele_tag in Ew:
eload_data = Ew[ele_tag]
nd1, nd2 = ops.eleNodes(ele_tag)
# element x, y coordinates
ex = np.array([ops.nodeCoord(nd1)[0],
ops.nodeCoord(nd2)[0]])
ey = np.array([ops.nodeCoord(nd1)[1],
ops.nodeCoord(nd2)[1]])
ez = np.array([ops.nodeCoord(nd1)[2],
ops.nodeCoord(nd2)[2]])
# eo = Eo[i, :]
xloc = ops.eleResponse(ele_tag, 'xlocal')
yloc = ops.eleResponse(ele_tag, 'ylocal')
zloc = ops.eleResponse(ele_tag, 'zlocal')
g = np.vstack((xloc, yloc, zloc))
G, _ = rot_transf_3d(ex, ey, ez, g)
g = G[:3, :3]
pl = ops.eleResponse(ele_tag, 'localForces')
s_all, xl = section_force_distribution_3d(ex, ey, ez, pl, nep,
eload_data)
# 1:'y' 2:'z'
if sf_type == 'N':
s = s_all[:, 0]
dir_plt = 1
elif sf_type == 'Vy':
s = s_all[:, 1]
dir_plt = 1
elif sf_type == 'Vz':
s = s_all[:, 2]
dir_plt = 2
elif sf_type == 'T':
s = s_all[:, 3]
dir_plt = 1
elif sf_type == 'My':
s = s_all[:, 4]
dir_plt = 2
elif sf_type == 'Mz':
s = s_all[:, 5]
dir_plt = 1
minVal = min(minVal, np.min(s))
maxVal = max(maxVal, np.max(s))
s = s*sfac
# FIXME - can be simplified
s_0 = np.zeros((nep, 3))
s_0[0, :] = [ex[0], ey[0], ez[0]]
s_0[1:, 0] = s_0[0, 0] + xl[1:] * g[0, 0]
s_0[1:, 1] = s_0[0, 1] + xl[1:] * g[0, 1]
s_0[1:, 2] = s_0[0, 2] + xl[1:] * g[0, 2]
s_p = np.copy(s_0)
# positive M are opposite to N and V
# if sf_type == 'Mz' or sf_type == 'My':
if sf_type == 'Mz':
s *= -1.
s_p[:, 0] += s * g[dir_plt, 0]
s_p[:, 1] += s * g[dir_plt, 1]
s_p[:, 2] += s * g[dir_plt, 2]
# plt.axis('equal')
# section force curve
plt.plot(s_p[:, 0], s_p[:, 1], s_p[:, 2], fmt_secforce,
solid_capstyle='round', solid_joinstyle='round',
dash_capstyle='butt', dash_joinstyle='round')
# model
plt.plot(ex, ey, ez, 'k-', solid_capstyle='round',
solid_joinstyle='round', dash_capstyle='butt',
dash_joinstyle='round')
# reference perpendicular lines
for i in np.arange(nep):
plt.plot([s_0[i, 0], s_p[i, 0]],
[s_0[i, 1], s_p[i, 1]],
[s_0[i, 2], s_p[i, 2]], fmt_secforce,
solid_capstyle='round', solid_joinstyle='round',
dash_capstyle='butt', dash_joinstyle='round')
return minVal, maxVal
def quad_sig_out_per_node():
"""Return a 2d numpy array of stress components per OpenSees node.
Returns:
sig_out (ndarray): a 2d array of stress components per node with
the following components: sxx, syy, sxy, svm, s1, s2, angle.
Size (n_nodes x 7).
Examples:
sig_out = opsv.quad_sig_out_per_node()
Notes:
s1, s2: principal stresses
angle: angle of the principal stress s1
"""
ele_tags = ops.getEleTags()
node_tags = ops.getNodeTags()
n_nodes = len(node_tags)
# initialize helper arrays
sig_out = np.zeros((n_nodes, 7))
nodes_tag_count = np.zeros((n_nodes, 2), dtype=int)
nodes_tag_count[:, 0] = node_tags
for i, ele_tag in enumerate(ele_tags):
nd1, nd2, nd3, nd4 = ops.eleNodes(ele_tag)
ind1 = node_tags.index(nd1)
ind2 = node_tags.index(nd2)
ind3 = node_tags.index(nd3)
ind4 = node_tags.index(nd4)
nodes_tag_count[[ind1, ind2, ind3, ind4], 1] += 1
sig_ip_el = ops.eleResponse(ele_tag, 'stress')
sigM_ip = np.vstack(([sig_ip_el[0:3],
sig_ip_el[3:6],
sig_ip_el[6:9],
sig_ip_el[9:12]]))
sigM_nd = quad_extrapolate_ip_to_node(sigM_ip)
# sxx
sig_out[ind1, 0] += sigM_nd[0, 0]
sig_out[ind2, 0] += sigM_nd[1, 0]
sig_out[ind3, 0] += sigM_nd[2, 0]
sig_out[ind4, 0] += sigM_nd[3, 0]
# syy
sig_out[ind1, 1] += sigM_nd[0, 1]
sig_out[ind2, 1] += sigM_nd[1, 1]
sig_out[ind3, 1] += sigM_nd[2, 1]
sig_out[ind4, 1] += sigM_nd[3, 1]
# sxy
sig_out[ind1, 2] += sigM_nd[0, 2]
sig_out[ind2, 2] += sigM_nd[1, 2]
sig_out[ind3, 2] += sigM_nd[2, 2]
sig_out[ind4, 2] += sigM_nd[3, 2]
indxs, = np.where(nodes_tag_count[:, 1] > 1)
# n_indxs < n_nodes: e.g. 21<25 (bous), 2<6 (2el) etc.
n_indxs = np.shape(indxs)[0]
# divide summed stresses by the number of common nodes
sig_out[indxs, :] = \
sig_out[indxs, :]/nodes_tag_count[indxs, 1].reshape(n_indxs, 1)
# warning reshape from (pts,ncomp) to (ncomp,pts)
vm_out = vm_stress(np.transpose(sig_out[:, :3]))
sig_out[:, 3] = vm_out
princ_sig_out = princ_stress(np.transpose(sig_out[:, :3]))
sig_out[:, 4:7] = np.transpose(princ_sig_out)
return sig_out
def quad_extrapolate_ip_to_node(yip):
"""
Exprapolate values at 4 integration points to 4 nodes of a quad element.
Integration points of Gauss quadrature.
Usefull for : stress components (sxx, syy, sxy)
yip - either a single vector (4,) or array (4,3) /sxx syy sxy/
or array (4, n)
"""
xep = np.sqrt(3.)/2
X = np.array([[1.+xep, -1/2., 1.-xep, -1/2.],
[-1/2., 1.+xep, -1/2., 1.-xep],
[1.-xep, -1/2., 1.+xep, -1/2.],
[-1/2., 1.-xep, -1/2., 1.+xep]])
ynp = X @ yip
return ynp
def quad_9n_extrapolate_ip_to_node(yip):
"""
Exprapolate values at 9 integration points to 9 nodes of a quad element.
Integration points of Gauss quadrature.
Usefull for : stress components (sxx, syy, sxy)
yip - either a single vector (4,) or array (4,3) /sxx syy sxy/
or array (4, n)
"""
a = 1./np.sqrt(0.6)
a10 = 1 - a*a
a9 = a10 * a10
a11, a12 = 1 + a, 1 - a
a1, a2, a3 = a11 * a11, a11 * a12, a12 * a12
a4, a5 = a10 * a11, a10 * a12
# 1
n5, n6, n7, n8 = a4/2-a9/2, a5/2-a9/2, a5/2-a9/2, a4/2-a9/2
n1 = a1/4 - (n8 + n5)/2 - a9/4
n2 = a2/4 - (n5 + n6)/2 - a9/4
n3 = a3/4 - (n6 + n7)/2 - a9/4
n4 = a2/4 - (n7 + n8)/2 - a9/4
r1 = np.array([n1, n2, n3, n4, n5, n6, n7, n8, a9])
# 2
n5, n6, n7, n8 = a4/2-a9/2, a4/2-a9/2, a5/2-a9/2, a5/2-a9/2
n1 = a2/4 - (n8 + n5)/2 - a9/4
n2 = a1/4 - (n5 + n6)/2 - a9/4
n3 = a2/4 - (n6 + n7)/2 - a9/4
n4 = a3/4 - (n7 + n8)/2 - a9/4
r2 = np.array([n1, n2, n3, n4, n5, n6, n7, n8, a9])
# 3
n5, n6, n7, n8 = a5/2-a9/2, a4/2-a9/2, a4/2-a9/2, a5/2-a9/2
n1 = a3/4 - (n8 + n5)/2 - a9/4
n2 = a2/4 - (n5 + n6)/2 - a9/4
n3 = a1/4 - (n6 + n7)/2 - a9/4
n4 = a2/4 - (n7 + n8)/2 - a9/4
r3 = np.array([n1, n2, n3, n4, n5, n6, n7, n8, a9])
# 4
n5, n6, n7, n8 = a5/2-a9/2, a5/2-a9/2, a4/2-a9/2, a4/2-a9/2
n1 = a2/4 - (n8 + n5)/2 - a9/4
n2 = a3/4 - (n5 + n6)/2 - a9/4
n3 = a2/4 - (n6 + n7)/2 - a9/4
n4 = a1/4 - (n7 + n8)/2 - a9/4
r4 = np.array([n1, n2, n3, n4, n5, n6, n7, n8, a9])
# 5
n5, n6, n7, n8 = a11/2-a10/2, a10/2-a10/2, a12/2-a10/2, a10/2-a10/2
n1 = a11/4 - (n8 + n5)/2 - a10/4
n2 = a11/4 - (n5 + n6)/2 - a10/4
n3 = a12/4 - (n6 + n7)/2 - a10/4
n4 = a12/4 - (n7 + n8)/2 - a10/4
r5 = np.array([n1, n2, n3, n4, n5, n6, n7, n8, a10])
# 6
n5, n6, n7, n8 = a10/2-a10/2, a11/2-a10/2, a10/2-a10/2, a12/2-a10/2
n1 = a12/4 - (n8 + n5)/2 - a10/4
n2 = a11/4 - (n5 + n6)/2 - a10/4
n3 = a11/4 - (n6 + n7)/2 - a10/4
n4 = a12/4 - (n7 + n8)/2 - a10/4
r6 = np.array([n1, n2, n3, n4, n5, n6, n7, n8, a10])
# 7
n5, n6, n7, n8 = a12/2-a10/2, a10/2-a10/2, a11/2-a10/2, a10/2-a10/2
n1 = a12/4 - (n8 + n5)/2 - a10/4
n2 = a12/4 - (n5 + n6)/2 - a10/4
n3 = a11/4 - (n6 + n7)/2 - a10/4
n4 = a11/4 - (n7 + n8)/2 - a10/4
r7 = np.array([n1, n2, n3, n4, n5, n6, n7, n8, a10])
# 8
n5, n6, n7, n8 = a10/2-a10/2, a12/2-a10/2, a10/2-a10/2, a11/2-a10/2
n1 = a11/4 - (n8 + n5)/2 - a10/4
n2 = a12/4 - (n5 + n6)/2 - a10/4
n3 = a12/4 - (n6 + n7)/2 - a10/4
n4 = a11/4 - (n7 + n8)/2 - a10/4
r8 = np.array([n1, n2, n3, n4, n5, n6, n7, n8, a10])
r9 = np.array([0., 0., 0., 0., 0., 0., 0., 0., 1.])
X = np.vstack((r1, r2, r3, r4, r5, r6, r7, r8, r9))
ynp = X @ yip
# ynp = 1.0
return ynp
def quad_8n_extrapolate_ip_to_node(yip):
"""
Exprapolate values at 8 integration points to 8 nodes of a quad element.
Integration points of Gauss quadrature.
Usefull for : stress components (sxx, syy, sxy)
yip - either a single vector (4,) or array (4,3) /sxx syy sxy/
or array (4, n)
"""
a = 1./np.sqrt(0.6)
a0 = 1 - a**2
a4, a5 = -(1-a)**2*(1+2*a)/4, -(1+a)**2*(1-2*a)/4
a7 = -a0/4
a11, a12 = a0*(1+a)/2, a0*(1-a)/2
a1, a2, a3 = (1+a)/2, (1-a)/2, (1-a**2)/2
X = np.array([[a5, a7, a4, a7, a11, a12, a12, a11],
[a7, a5, a7, a4, a11, a11, a12, a12],
[a4, a7, a5, a7, a12, a11, a11, a12],
[a7, a4, a7, a5, a12, a12, a11, a11],
[a7, a7, a7, a7, a1, a3, a2, a3],
[a7, a7, a7, a7, a3, a1, a3, a2],
[a7, a7, a7, a7, a2, a3, a1, a3],
[a7, a7, a7, a7, a3, a2, a3, a1]])
ynp = X @ yip
# ynp = 1.0
return ynp
def quad_interpolate_node_to_ip(ynp):
"""
Interpolate values at 4 nodes to 4 integration points a quad element.
Integration points of Gauss quadrature.
Usefull for : stress components (sxx, syy, sxy)
ynp - either a single vector (4,) or array (4,3) /sxx syy sxy/
or array (4, n)
"""
jsz = 1./6.
jtr = 1./3.
p2 = jsz * np.sqrt(3.)
X = np.array([[jtr+p2, jsz, jtr-p2, jsz],
[jsz, jtr+p2, jsz, jtr-p2],
[jtr-p2, jsz, jtr+p2, jsz],
[jsz, jtr-p2, jsz, jtr+p2]])
yip = X @ ynp
return yip
def princ_stress(sig):
"""Return a tuple (s1, s2, angle): principal stresses (plane stress) and angle
Args:
sig (ndarray): input array of stresses at nodes: sxx, syy, sxy (tau)
Returns:
out (ndarray): 1st row is first principal stress s1, 2nd row is second
principal stress s2, 3rd row is the angle of s1
"""
sx, sy, tau = sig[0], sig[1], sig[2]
ds = (sx-sy)/2
R = np.sqrt(ds**2 + tau**2)
s1 = (sx+sy)/2. + R
s2 = (sx+sy)/2. - R
angle = np.arctan2(tau, ds)/2
out = np.vstack((s1, s2, angle))
return out
def vm_stress(sig):
n_sig_comp, n_pts = np.shape(sig)
if n_sig_comp > 3:
x, y, z, xy, xz, yz = sig
else:
x, y, xy = sig
z, xz, yz = 0., 0., 0.
_a = 0.5*((x-y)**2 + (y-z)**2 + (z-x)**2 + 6.*(xy**2 + xz**2 + yz**2))
return np.sqrt(_a)
def quad_crds_node_to_ip():
"""
Return global coordinates of 4 quad ip nodes and corner nodes.
It also returns quad connectivity.
"""
node_tags, ele_tags = ops.getNodeTags(), ops.getEleTags()
n_nodes, n_eles = len(node_tags), len(ele_tags)
# idiom coordinates as ordered in node_tags
nds_crd = np.zeros((n_nodes, 2))
for i, node_tag in enumerate(node_tags):
nds_crd[i] = ops.nodeCoord(node_tag)
quads_conn = np.zeros((n_eles, 4), dtype=int)
# quads_conn_ops = np.zeros((n_eles, 4), dtype=int)
eles_nds_crd = np.zeros((n_eles, 4, 2))
eles_ips_crd = np.zeros((n_eles, 4, 2))
for i, ele_tag in enumerate(ele_tags):
nd1, nd2, nd3, nd4 = ops.eleNodes(ele_tag)
ind1 = node_tags.index(nd1)
ind2 = node_tags.index(nd2)
ind3 = node_tags.index(nd3)
ind4 = node_tags.index(nd4)
quads_conn[i] = np.array([ind1, ind2, ind3, ind4])
# quads_conn_ops[i] = np.array([nd1, nd2, nd3, nd4])
eles_nds_crd[i] = np.array([[ops.nodeCoord(nd1, 1),
ops.nodeCoord(nd1, 2)],
[ops.nodeCoord(nd2, 1),
ops.nodeCoord(nd2, 2)],
[ops.nodeCoord(nd3, 1),
ops.nodeCoord(nd3, 2)],
[ops.nodeCoord(nd4, 1),
ops.nodeCoord(nd4, 2)]])
eles_ips_crd[i] = quad_interpolate_node_to_ip(eles_nds_crd[i])
return eles_ips_crd, eles_nds_crd, nds_crd, quads_conn
def quad_sig_out_per_ele():
"""
Extract stress components for all elements from OpenSees analysis.
Returns:
eles_ips_sig_out, eles_nds_sig_out (tuple of ndarrays):
eles_ips_sig_out - values at integration points of elements
(n_eles x 4 x 4)
eles_nds_sig_out - values at nodes of elements (n_eles x 4 x 4)
Examples:
eles_ips_sig_out, eles_nds_sig_out = opsv.quad_sig_out_per_ele()
Notes:
Stress components in array columns are: Sxx, Syy, Sxy, Svmis, empty
Used e.g. by plot_mesh_with_ips_2d function
"""
node_tags, ele_tags = ops.getNodeTags(), ops.getEleTags()
n_nodes, n_eles = len(node_tags), len(ele_tags)
eles_ips_sig_out = np.zeros((n_eles, 4, 4))
eles_nds_sig_out = np.zeros((n_eles, 4, 4))
# array (n_nodes, 2):
# node_tags, number of occurrence in quad elements)
# correspondence indx and node_tag is in node_tags.index
# (a) data in np.array of integers
nodes_tag_count = np.zeros((n_nodes, 2), dtype=int)
nodes_tag_count[:, 0] = node_tags
for i, ele_tag in enumerate(ele_tags):
nd1, nd2, nd3, nd4 = ops.eleNodes(ele_tag)
ind1 = node_tags.index(nd1)
ind2 = node_tags.index(nd2)
ind3 = node_tags.index(nd3)
ind4 = node_tags.index(nd4)
nodes_tag_count[[ind1, ind2, ind3, ind4], 1] += 1
sig_ip_el = ops.eleResponse(ele_tag, 'stress')
sigM_ip = np.vstack(([sig_ip_el[0:3],
sig_ip_el[3:6],
sig_ip_el[6:9],
sig_ip_el[9:12]]))
sigM_nd = quad_extrapolate_ip_to_node(sigM_ip)
eles_ips_sig_out[i, :, :3] = sigM_ip
eles_nds_sig_out[i, :, :3] = sigM_nd
vm_ip_out = vm_stress(np.transpose(eles_ips_sig_out[i, :, :3]))
vm_nd_out = vm_stress(np.transpose(eles_nds_sig_out[i, :, :3]))
eles_ips_sig_out[i, :, 3] = vm_ip_out
eles_nds_sig_out[i, :, 3] = vm_nd_out
return eles_ips_sig_out, eles_nds_sig_out
def quads_to_4tris(quads_conn, nds_crd, nds_val):
"""
Get triangles connectivity, coordinates and new values at quad centroids.
Args:
quads_conn (ndarray):
nds_crd (ndarray):
nds_val (ndarray):
Returns:
tris_conn, nds_c_crd, nds_c_val (tuple):
Notes:
Triangles connectivity array is based on
quadrilaterals connectivity.
Each quad is split into four triangles.
New nodes are created at the quad centroid.
See also:
function: quads_to_8tris_9n, quads_to_8tris_8n
"""
n_quads, _ = quads_conn.shape
n_nds, _ = nds_crd.shape
# coordinates and values at quad centroids _c_
nds_c_crd = np.zeros((n_quads, 2))
nds_c_val = np.zeros(n_quads)
tris_conn = np.zeros((4*n_quads, 3), dtype=int)
for i, quad_conn in enumerate(quads_conn):
j = 4*i
n0, n1, n2, n3 = quad_conn
# quad centroids
nds_c_crd[i] = np.array([np.sum(nds_crd[[n0, n1, n2, n3], 0])/4.,
np.sum(nds_crd[[n0, n1, n2, n3], 1])/4.])
nds_c_val[i] = np.sum(nds_val[[n0, n1, n2, n3]])/4.
# triangles connectivity
tris_conn[j] = np.array([n0, n1, n_nds+i])
tris_conn[j+1] = np.array([n1, n2, n_nds+i])
tris_conn[j+2] = np.array([n2, n3, n_nds+i])
tris_conn[j+3] = np.array([n3, n0, n_nds+i])
return tris_conn, nds_c_crd, nds_c_val
def plot_mesh_2d(nds_crd, eles_conn, lw=0.4, ec='k'):
"""
Plot 2d mesh (quads or triangles) outline.
"""
for ele_conn in eles_conn:
x = nds_crd[ele_conn, 0]
y = nds_crd[ele_conn, 1]
plt.fill(x, y, edgecolor=ec, lw=lw, fill=False)
def plot_stress_2d(nds_val, mesh_outline=1, cmap='jet'):
"""
Plot stress distribution of a 2d elements of a 2d model.
Args:
nds_val (ndarray): the values of a stress component, which can
be extracted from sig_out array (see quad_sig_out_per_node
function)
mesh_outline (int): 1 - mesh is plotted, 0 - no mesh plotted.
cmap (str): Matplotlib color map (default is 'jet')
Usage:
::
sig_out = opsv.quad_sig_out_per_node()
j, jstr = 3, 'vmis'
nds_val = sig_out[:, j]
opsv.plot_stress_2d(nds_val)
plt.xlabel('x [m]')
plt.ylabel('y [m]')
plt.title(f'{jstr}')
plt.show()
See also:
:ref:`ops_vis_quad_sig_out_per_node`
"""
node_tags, ele_tags = ops.getNodeTags(), ops.getEleTags()
n_nodes, n_eles = len(node_tags), len(ele_tags)
# idiom coordinates as ordered in node_tags
# use node_tags.index(tag) for correspondence
nds_crd = np.zeros((n_nodes, 2))
for i, node_tag in enumerate(node_tags):
nds_crd[i] = ops.nodeCoord(node_tag)
# from utils / quad_sig_out_per_node
# fixme: if this can be simplified
# index (starts from 0) to node_tag correspondence
# (a) data in np.array of integers
# nodes_tag_count = np.zeros((n_nodes, 2), dtype=int)
# nodes_tag_count[:, 0] = node_tags
#
# correspondence indx and node_tag is in node_tags.index
# after testing remove the above
quads_conn = np.zeros((n_eles, 4), dtype=int)
for i, ele_tag in enumerate(ele_tags):
nd1, nd2, nd3, nd4 = ops.eleNodes(ele_tag)
ind1 = node_tags.index(nd1)
ind2 = node_tags.index(nd2)
ind3 = node_tags.index(nd3)
ind4 = node_tags.index(nd4)
quads_conn[i] = np.array([ind1, ind2, ind3, ind4])
tris_conn, nds_c_crd, nds_c_val = \
quads_to_4tris(quads_conn, nds_crd, nds_val)
nds_crd_all = np.vstack((nds_crd, nds_c_crd))
# nds_val_all = np.concatenate((nds_val, nds_c_val))
nds_val_all = np.hstack((nds_val, nds_c_val))
# 1. plot contour maps
triangulation = tri.Triangulation(nds_crd_all[:, 0],
nds_crd_all[:, 1],
tris_conn)
plt.tricontourf(triangulation, nds_val_all, 50, cmap=cmap)
# 2. plot original mesh (quad) without subdivision into triangles
if mesh_outline:
plot_mesh_2d(nds_crd, quads_conn)
# plt.colorbar()
plt.axis('equal')
def plot_stress_9n_2d(nds_val, cmap='jet'):
node_tags, ele_tags = ops.getNodeTags(), ops.getEleTags()
n_nodes, n_eles = len(node_tags), len(ele_tags)
# idiom coordinates as ordered in node_tags
# use node_tags.index(tag) for correspondence
nds_crd = np.zeros((n_nodes, 2))
for i, node_tag in enumerate(node_tags):
nds_crd[i] = ops.nodeCoord(node_tag)
# from utils / quad_sig_out_per_node
# fixme: if this can be simplified
# index (starts from 0) to node_tag correspondence
# (a) data in np.array of integers
# nodes_tag_count = np.zeros((n_nodes, 2), dtype=int)
# nodes_tag_count[:, 0] = node_tags
#
# correspondence indx and node_tag is in node_tags.index
# after testing remove the above
quads_conn = np.zeros((n_eles, 4), dtype=int)
for i, ele_tag in enumerate(ele_tags):
nd1, nd2, nd3, nd4 = ops.eleNodes(ele_tag)
ind1 = node_tags.index(nd1)
ind2 = node_tags.index(nd2)
ind3 = node_tags.index(nd3)
ind4 = node_tags.index(nd4)
quads_conn[i] = np.array([ind1, ind2, ind3, ind4])
tris_conn, nds_c_crd, nds_c_val = \
quads_to_4tris(quads_conn, nds_crd, nds_val)
nds_crd_all = np.vstack((nds_crd, nds_c_crd))
# nds_val_all = np.concatenate((nds_val, nds_c_val))
nds_val_all = np.hstack((nds_val, nds_c_val))
# 1. plot contour maps
triangulation = tri.Triangulation(nds_crd_all[:, 0],
nds_crd_all[:, 1],
tris_conn)
plt.tricontourf(triangulation, nds_val_all, 50, cmap=cmap)
# 2. plot original mesh (quad) without subdivision into triangles
plot_mesh_2d(nds_crd, quads_conn)
# plt.colorbar()
plt.axis('equal')
def plot_extruded_model_rect_section_3d(b, h, az_el=az_el,
fig_wi_he=fig_wi_he,
fig_lbrt=fig_lbrt):
"""Plot an extruded 3d model based on cross-section dimenions.
Three arrows present local section axes: green - local x-axis,
red - local z-axis, blue - local y-axis.
Args:
b (float): section width
h (float): section height
az_el (tuple): azimuth and elevation
fig_wi_he: figure width and height in centimeters
fig_lbrt: figure left, bottom, right, top boundaries
Usage:
::
plot_extruded_model_rect_section_3d(0.3, 0.4)
Notes:
- For now only rectangular cross-section is supported.
"""
b2, h2 = b/2, h/2
ele_tags = ops.getEleTags()
azim, elev = az_el
fig_wi, fig_he = fig_wi_he
fleft, fbottom, fright, ftop = fig_lbrt
fig = plt.figure(figsize=(fig_wi/2.54, fig_he/2.54))
fig.subplots_adjust(left=.08, bottom=.08, right=.985, top=.94)
ax = fig.add_subplot(111, projection=Axes3D.name)
# ax.axis('equal')
ax.set_xlabel('X')
ax.set_ylabel('Y')
ax.set_zlabel('Z')
ax.view_init(azim=azim, elev=elev)
for i, ele_tag in enumerate(ele_tags):
nd1, nd2 = ops.eleNodes(ele_tag)
# element x, y coordinates
ex = np.array([ops.nodeCoord(nd1)[0],
ops.nodeCoord(nd2)[0]])
ey = np.array([ops.nodeCoord(nd1)[1],
ops.nodeCoord(nd2)[1]])
ez = np.array([ops.nodeCoord(nd1)[2],
ops.nodeCoord(nd2)[2]])
# eo = Eo[i, :]
xloc = ops.eleResponse(ele_tag, 'xlocal')
yloc = ops.eleResponse(ele_tag, 'ylocal')
zloc = ops.eleResponse(ele_tag, 'zlocal')
g = np.vstack((xloc, yloc, zloc))
# G, L = rot_transf_3d(ex, ey, ez, eo)
G, L = rot_transf_3d(ex, ey, ez, g)
# g = G[:3, :3]
Xi, Yi, Zi = ex[0], ey[0], ez[0]
Xj, Yj, Zj = ex[1], ey[1], ez[1]
g10, g11, g12 = g[1, 0]*h2, g[1, 1]*h2, g[1, 2]*h2
g20, g21, g22 = g[2, 0]*b2, g[2, 1]*b2, g[2, 2]*b2
# beg node cross-section vertices
Xi1, Yi1, Zi1 = Xi - g10 - g20, Yi - g11 - g21, Zi - g12 - g22
Xi2, Yi2, Zi2 = Xi + g10 - g20, Yi + g11 - g21, Zi + g12 - g22
Xi3, Yi3, Zi3 = Xi + g10 + g20, Yi + g11 + g21, Zi + g12 + g22
Xi4, Yi4, Zi4 = Xi - g10 + g20, Yi - g11 + g21, Zi - g12 + g22
# end node cross-section vertices
Xj1, Yj1, Zj1 = Xj - g10 - g20, Yj - g11 - g21, Zj - g12 - g22
Xj2, Yj2, Zj2 = Xj + g10 - g20, Yj + g11 - g21, Zj + g12 - g22
Xj3, Yj3, Zj3 = Xj + g10 + g20, Yj + g11 + g21, Zj + g12 + g22
Xj4, Yj4, Zj4 = Xj - g10 + g20, Yj - g11 + g21, Zj - g12 + g22
# mesh outline
ax.plot(ex, ey, ez, 'k--', solid_capstyle='round',
solid_joinstyle='round', dash_capstyle='butt',
dash_joinstyle='round')
# collected i-beg, j-end node coordinates, counter-clockwise order
pts = [[Xi1, Yi1, Zi1],
[Xi2, Yi2, Zi2],
[Xi3, Yi3, Zi3],
[Xi4, Yi4, Zi4],
[Xj1, Yj1, Zj1],
[Xj2, Yj2, Zj2],
[Xj3, Yj3, Zj3],
[Xj4, Yj4, Zj4]]
# list of 4-node sides
verts = [[pts[0], pts[1], pts[2], pts[3]], # beg
[pts[4], pts[5], pts[6], pts[7]], # end
[pts[0], pts[4], pts[5], pts[1]], # bottom
[pts[3], pts[7], pts[6], pts[2]], # top
[pts[0], pts[4], pts[7], pts[3]], # front
[pts[1], pts[5], pts[6], pts[2]]] # back
# plot 3d element composed with sides
ax.add_collection3d(Poly3DCollection(verts, linewidths=1,
edgecolors='k', alpha=.25))
Xm, Ym, Zm = sum(ex)/2, sum(ey)/2, sum(ez)/2
alen = 0.1*L
# plot local axis directional vectors: workaround quiver = arrow
plt.quiver(Xm, Ym, Zm, g[0, 0], g[0, 1], g[0, 2], color='g',
lw=2, length=alen, alpha=.8, normalize=True)
plt.quiver(Xm, Ym, Zm, g[1, 0], g[1, 1], g[1, 2], color='b',
lw=2, length=alen, alpha=.8, normalize=True)
plt.quiver(Xm, Ym, Zm, g[2, 0], g[2, 1], g[2, 2], color='r',
lw=2, length=alen, alpha=.8, normalize=True)
def plot_mesh_with_ips_2d(nds_crd, eles_ips_crd, eles_nds_crd, quads_conn,
eles_ips_sig_out, eles_nds_sig_out, sig_out_indx):
"""
Plot 2d element mesh with the values at gauss and nodal points.
Args:
nds_crd (ndarray): nodes coordinates (n_nodes x 2)
eles_ips_crd (ndarray): integration points coordinates of elements
(n_eles x 4 x 2)
eles_nds_crd (ndarray): nodal coordinates of elements (n_eles x 4 x 2)
quads_conn (ndarray): connectivity array (n_eles x 4)
eles_ips_sig_out (ndarray): stress component values at integration
points (n_eles x 4 x 5)
eles_nds_sig_out (ndarray): stress component values at element nodes
(n_eles x 4 x 5)
sig_out_indx (int): which sig_out component
Notes: This function is suitable for small models for illustration
purposes.
"""
plot_mesh_2d(nds_crd, quads_conn, lw=1.2, ec='b')
ele_tags = ops.getEleTags()
n_eles = len(ele_tags)
for i in range(n_eles):
plt.plot(eles_ips_crd[i, :, 0], eles_ips_crd[i, :, 1],
'kx', markersize=3)
ips_val = eles_ips_sig_out[i, :, sig_out_indx]
nds_val = eles_nds_sig_out[i, :, sig_out_indx]
# show ips values
plt.text(eles_ips_crd[i, 0, 0], eles_ips_crd[i, 0, 1],
f'{ips_val[0]:.2f}', {'color': 'C0'},
ha='center', va='bottom')
plt.text(eles_ips_crd[i, 1, 0], eles_ips_crd[i, 1, 1],
f'{ips_val[1]:.2f}', {'color': 'C1'},
ha='center', va='bottom')
plt.text(eles_ips_crd[i, 2, 0], eles_ips_crd[i, 2, 1],
f'{ips_val[2]:.2f}', {'color': 'C2'},
ha='center', va='top')
plt.text(eles_ips_crd[i, 3, 0], eles_ips_crd[i, 3, 1],
f'{ips_val[3]:.2f}', {'color': 'C3'},
ha='center', va='top')
# show node values
plt.text(eles_nds_crd[i, 0, 0], eles_nds_crd[i, 0, 1],
f' {nds_val[0]:.2f}', {'color': 'C0'},
ha='left', va='bottom')
plt.text(eles_nds_crd[i, 1, 0], eles_nds_crd[i, 1, 1],
f'{nds_val[1]:.2f} ', {'color': 'C1'},
ha='right', va='bottom')
plt.text(eles_nds_crd[i, 2, 0], eles_nds_crd[i, 2, 1],
f'{nds_val[2]:.2f} ', {'color': 'C2'},
ha='right', va='top')
plt.text(eles_nds_crd[i, 3, 0], eles_nds_crd[i, 3, 1],
f' {nds_val[3]:.2f}', {'color': 'C3'},
ha='left', va='top')
plt.axis('equal')
# see also quads_to_8tris_9n
def quads_to_8tris_8n(quads_conn, nds_crd, nds_val):
"""
Get triangles connectivity, coordinates and new values at quad centroids.
Args:
quads_conn (ndarray):
nds_crd (ndarray):
nds_val (ndarray):
Returns:
tris_conn, nds_c_crd, nds_c_val (tuple):
Notes:
Triangles connectivity array is based on
quadrilaterals connectivity.
Each quad is split into eight triangles.
New nodes are created at the quad centroid.
See also:
function: quads_to_8tris_9n, quads_to_4tris
"""
n_quads, _ = quads_conn.shape
n_nds, _ = nds_crd.shape
# coordinates and values at quad centroids _c_
nds_c_crd = np.zeros((n_quads, 2))
nds_c_val = np.zeros(n_quads)
tris_conn = np.zeros((8*n_quads, 3), dtype=int)
for i, quad_conn in enumerate(quads_conn):
j = 8*i
n0, n1, n2, n3, n4, n5, n6, n7 = quad_conn
# quad centroids
# nds_c_crd[i] = np.array([np.sum(nds_crd[[n0, n1, n2, n3], 0])/4.,
# np.sum(nds_crd[[n0, n1, n2, n3], 1])/4.])
# nds_c_val[i] = np.sum(nds_val[[n0, n1, n2, n3]])/4.
nds_c_crd[i] = quad_8n_val_at_center(nds_crd[[n0, n1, n2, n3,
n4, n5, n6, n7]])
nds_c_val[i] = quad_8n_val_at_center(nds_val[[n0, n1, n2, n3,
n4, n5, n6, n7]])
# triangles connectivity
tris_conn[j] = np.array([n0, n4, n_nds+i])
tris_conn[j+1] = np.array([n4, n1, n_nds+i])
tris_conn[j+2] = np.array([n1, n5, n_nds+i])
tris_conn[j+3] = np.array([n5, n2, n_nds+i])
tris_conn[j+4] = np.array([n2, n6, n_nds+i])
tris_conn[j+5] = np.array([n6, n3, n_nds+i])
tris_conn[j+6] = np.array([n3, n7, n_nds+i])
tris_conn[j+7] = np.array([n7, n0, n_nds+i])
return tris_conn, nds_c_crd, nds_c_val
# see also quads_to_8tris_8n
def quads_to_8tris_9n(quads_conn):
"""
Get triangles connectivity, coordinates and new values at quad centroids.
Args:
quads_conn (ndarray):
nds_crd (ndarray):
nds_val (ndarray):
Returns:
tris_conn, nds_c_crd, nds_c_val (tuple):
Notes:
Triangles connectivity array is based on
quadrilaterals connectivity.
Each quad is split into eight triangles.
New nodes are created at the quad centroid.
See also:
function: quads_to_8tris_8n, quads_to_4tris
"""
n_quads, _ = quads_conn.shape
# n_nds, _ = nds_crd.shape
tris_conn = | np.zeros((8*n_quads, 3), dtype=int) | numpy.zeros |
#!/usr/bin/env python3
import tensorflow as tf
import tflearn
from tensorflow.python.ops import gen_nn_ops
from tensorflow.python.ops import array_ops
import numpy as np
import numpy.random as npr
np.set_printoptions(precision=2)
# np.seterr(all='raise')
np.seterr(all='warn')
import argparse
import csv
import os
import sys
import time
import pickle as pkl
import json
import shutil
import setproctitle
from datetime import datetime
sys.path.append('../lib')
import olivetti
import bundle_entropy
import matplotlib as mpl
mpl.use("Agg")
import matplotlib.pyplot as plt
def main():
parser = argparse.ArgumentParser()
parser.add_argument('--save', type=str, default='work/mse.ebundle')
parser.add_argument('--nEpoch', type=float, default=50)
parser.add_argument('--nBundleIter', type=int, default=30)
# parser.add_argument('--trainBatchSz', type=int, default=25)
parser.add_argument('--trainBatchSz', type=int, default=70)
# parser.add_argument('--testBatchSz', type=int, default=2048)
parser.add_argument('--noncvx', action='store_true')
parser.add_argument('--seed', type=int, default=42)
# parser.add_argument('--valSplit', type=float, default=0)
args = parser.parse_args()
assert(not args.noncvx)
setproctitle.setproctitle('bamos.icnn.comp.mse.ebundle')
npr.seed(args.seed)
tf.set_random_seed(args.seed)
save = os.path.expanduser(args.save)
if os.path.isdir(save):
shutil.rmtree(save)
os.makedirs(save)
ckptDir = os.path.join(save, 'ckpt')
args.ckptDir = ckptDir
if not os.path.exists(ckptDir):
os.makedirs(ckptDir)
data = olivetti.load("data/olivetti")
# eps = 1e-8
# data['trainX'] = data['trainX'].clip(eps, 1.-eps)
# data['trainY'] = data['trainY'].clip(eps, 1.-eps)
# data['testX'] = data['testX'].clip(eps, 1.-eps)
# data['testY'] = data['testY'].clip(eps, 1.-eps)
nTrain = data['trainX'].shape[0]
nTest = data['testX'].shape[0]
inputSz = list(data['trainX'][0].shape)
outputSz = list(data['trainY'][1].shape)
print("\n\n" + "="*40)
print("+ nTrain: {}, nTest: {}".format(nTrain, nTest))
print("+ inputSz: {}, outputSz: {}".format(inputSz, outputSz))
print("="*40 + "\n\n")
config = tf.ConfigProto() #log_device_placement=False)
config.gpu_options.allow_growth = True
with tf.Session(config=config) as sess:
model = Model(inputSz, outputSz, sess)
model.train(args, data['trainX'], data['trainY'], data['testX'], data['testY'])
def variable_summaries(var, name=None):
if name is None:
name = var.name
with tf.name_scope('summaries'):
mean = tf.reduce_mean(var)
tf.scalar_summary('mean/' + name, mean)
with tf.name_scope('stdev'):
stdev = tf.sqrt(tf.reduce_mean(tf.square(var - mean)))
tf.scalar_summary('stdev/' + name, stdev)
tf.scalar_summary('max/' + name, tf.reduce_max(var))
tf.scalar_summary('min/' + name, tf.reduce_min(var))
tf.histogram_summary(name, var)
class Model:
def __init__(self, inputSz, outputSz, sess):
self.inputSz = inputSz
self.outputSz = outputSz
self.nOutput = np.prod(outputSz)
self.sess = sess
self.trueY_ = tf.placeholder(tf.float32, shape=[None] + outputSz, name='trueY')
self.x_ = tf.placeholder(tf.float32, shape=[None] + inputSz, name='x')
self.y_ = tf.placeholder(tf.float32, shape=[None] + outputSz, name='y')
self.v_ = tf.placeholder(tf.float32, shape=[None, self.nOutput], name='v')
self.c_ = tf.placeholder(tf.float32, shape=[None], name='c')
self.E_ = self.f(self.x_, self.y_)
variable_summaries(self.E_)
self.dE_dy_ = tf.gradients(self.E_, self.y_)[0]
self.dE_dyFlat_ = tf.contrib.layers.flatten(self.dE_dy_)
self.yFlat_ = tf.contrib.layers.flatten(self.y_)
self.E_entr_ = self.E_ + tf.reduce_sum(self.yFlat_*tf.log(self.yFlat_), 1) + \
tf.reduce_sum((1.-self.yFlat_)*tf.log(1.-self.yFlat_), 1)
self.dE_entr_dy_ = tf.gradients(self.E_entr_, self.y_)[0]
self.dE_entr_dyFlat_ = tf.contrib.layers.flatten(self.dE_entr_dy_)
self.F_ = tf.mul(self.c_, self.E_) + \
tf.reduce_sum(tf.mul(self.dE_dyFlat_, self.v_), 1)
# regLosses = tf.get_collection(tf.GraphKeys.REGULARIZATION_LOSSES)
# self.F_reg_ = self.F_ + 0.1*regLosses
# self.F_reg_ = self.F_ + 1e-5*tf.square(self.E_)
self.opt = tf.train.AdamOptimizer(0.001)
self.theta_ = tf.trainable_variables()
self.gv_ = [(g,v) for g,v in self.opt.compute_gradients(self.F_, self.theta_)
if g is not None]
self.train_step = self.opt.apply_gradients(self.gv_)
self.theta_cvx_ = [v for v in self.theta_
if 'proj' in v.name and 'W:' in v.name]
self.makeCvx = [v.assign(tf.abs(v)/2.0) for v in self.theta_cvx_]
self.proj = [v.assign(tf.maximum(v, 0)) for v in self.theta_cvx_]
for g,v in self.gv_:
variable_summaries(g, 'gradients/'+v.name)
self.l_yN_ = tf.placeholder(tf.float32, name='l_yN')
tf.scalar_summary('mse', self.l_yN_)
self.nBundleIter_ = tf.placeholder(tf.float32, [None], name='nBundleIter')
variable_summaries(self.nBundleIter_)
self.nActive_ = tf.placeholder(tf.float32, [None], name='nActive')
variable_summaries(self.nActive_)
self.merged = tf.merge_all_summaries()
self.saver = tf.train.Saver(max_to_keep=0)
def train(self, args, trainX, trainY, valX, valY):
save = args.save
self.meanY = np.mean(trainY, axis=0)
nTrain = trainX.shape[0]
nTest = valX.shape[0]
nIter = int(np.ceil(args.nEpoch*nTrain/args.trainBatchSz))
trainFields = ['iter', 'loss']
trainF = open(os.path.join(save, 'train.csv'), 'w')
trainW = csv.writer(trainF)
trainW.writerow(trainFields)
trainF.flush()
testFields = ['iter', 'loss']
testF = open(os.path.join(save, 'test.csv'), 'w')
testW = csv.writer(testF)
testW.writerow(testFields)
testF.flush()
self.trainWriter = tf.train.SummaryWriter(os.path.join(save, 'train'),
self.sess.graph)
self.sess.run(tf.initialize_all_variables())
if not args.noncvx:
self.sess.run(self.makeCvx)
nParams = np.sum(v.get_shape().num_elements() for v in tf.trainable_variables())
self.nBundleIter = args.nBundleIter
meta = {'nTrain': nTrain, 'trainBatchSz': args.trainBatchSz,
'nParams': nParams, 'nEpoch': args.nEpoch,
'nIter': nIter, 'nBundleIter': self.nBundleIter}
metaP = os.path.join(save, 'meta.json')
with open(metaP, 'w') as f:
json.dump(meta, f, indent=2)
nErrors = 0
maxErrors = 20
for i in range(nIter):
tflearn.is_training(True)
print("=== Iteration {} (Epoch {:.2f}) ===".format(
i, i/np.ceil(nTrain/args.trainBatchSz)))
start = time.time()
I = npr.randint(nTrain, size=args.trainBatchSz)
xBatch = trainX[I, :]
yBatch = trainY[I, :]
yBatch_flat = yBatch.reshape((args.trainBatchSz, -1))
xBatch_flipped = xBatch[:,:,::-1,:]
def fg(yhats):
yhats_shaped = yhats.reshape([args.trainBatchSz]+self.outputSz)
fd = {self.x_: xBatch_flipped, self.y_: yhats_shaped}
e, ge = self.sess.run([self.E_, self.dE_dyFlat_], feed_dict=fd)
return e, ge
y0 = np.expand_dims(self.meanY, axis=0).repeat(args.trainBatchSz, axis=0)
y0 = y0.reshape((args.trainBatchSz, -1))
try:
yN, G, h, lam, ys, nIters = bundle_entropy.solveBatch(
fg, y0, nIter=self.nBundleIter)
yN_shaped = yN.reshape([args.trainBatchSz]+self.outputSz)
except (KeyboardInterrupt, SystemExit):
raise
except:
print("Warning: Exception in bundle_entropy.solveBatch")
nErrors += 1
if nErrors > maxErrors:
print("More than {} errors raised, quitting".format(maxErrors))
sys.exit(-1)
continue
nActive = [len(Gi) for Gi in G]
l_yN = mse(yBatch_flat, yN)
fd = self.train_step_fd(args.trainBatchSz, xBatch_flipped, yBatch_flat,
G, yN, ys, lam)
fd[self.l_yN_] = l_yN
fd[self.nBundleIter_] = nIters
fd[self.nActive_] = nActive
summary, _ = self.sess.run([self.merged, self.train_step], feed_dict=fd)
if not args.noncvx and len(self.proj) > 0:
self.sess.run(self.proj)
saveImgs(xBatch, yN_shaped, "{}/trainImgs/{:05d}".format(args.save, i))
self.trainWriter.add_summary(summary, i)
trainW.writerow((i, l_yN))
trainF.flush()
print(" + loss: {:0.5e}".format(l_yN))
print(" + time: {:0.2f} s".format(time.time()-start))
if i % np.ceil(nTrain/(4.0*args.trainBatchSz)) == 0:
os.system('./icnn.plot.py ' + args.save)
if i % np.ceil(nTrain/args.trainBatchSz) == 0:
print("=== Testing ===")
tflearn.is_training(False)
y0 = np.expand_dims(self.meanY, axis=0).repeat(nTest, axis=0)
y0 = y0.reshape((nTest, -1))
valX_flipped = valX[:,:,::-1,:]
def fg(yhats):
yhats_shaped = yhats.reshape([nTest]+self.outputSz)
fd = {self.x_: valX_flipped, self.y_: yhats_shaped}
e, ge = self.sess.run([self.E_, self.dE_dyFlat_], feed_dict=fd)
return e, ge
try:
yN, G, h, lam, ys, nIters = bundle_entropy.solveBatch(
fg, y0, nIter=self.nBundleIter)
yN_shaped = yN.reshape([nTest]+self.outputSz)
except (KeyboardInterrupt, SystemExit):
raise
except:
print("Warning: Exception in bundle_entropy.solveBatch")
nErrors += 1
if nErrors > maxErrors:
print("More than {} errors raised, quitting".format(maxErrors))
sys.exit(-1)
continue
testMSE = mse(valY, yN_shaped)
saveImgs(valX, yN_shaped, "{}/testImgs/{:05d}".format(args.save, i))
print(" + test loss: {:0.5e}".format(testMSE))
testW.writerow((i, testMSE))
testF.flush()
self.save(os.path.join(args.ckptDir, '{:05d}.tf'.format(i)))
os.system('./icnn.plot.py ' + args.save)
trainF.close()
testF.close()
os.system('./icnn.plot.py ' + args.save)
def save(self, path):
self.saver.save(self.sess, path)
def load(self, path):
self.saver.restore(self.sess, path)
def train_step_fd(self, trainBatchSz, xBatch, yBatch, G, yN, ys, lam):
fd_xs, fd_ys, fd_vs, fd_cs = ([] for i in range(4))
for j in range(trainBatchSz):
if len(G[j]) == 0:
continue
Gj = np.array(G[j])
cy, clam, ct = mseGrad(yN[j], yBatch[j], Gj)
for i in range(len(G[j])):
fd_xs.append(xBatch[j])
fd_ys.append(ys[j][i].reshape(self.outputSz))
v = lam[j][i] * cy + clam[i] * (yN[j] - ys[j][i])
fd_vs.append(v)
fd_cs.append(clam[i])
fd_xs = np.array(fd_xs)
fd_ys = np.array(fd_ys)
fd_vs = np.array(fd_vs)
fd_cs = np.array(fd_cs)
fd = {self.x_: fd_xs, self.y_: fd_ys, self.v_: fd_vs, self.c_: fd_cs}
return fd
def f(self, x, y, reuse=False):
conv = tflearn.conv_2d
bn = tflearn.batch_normalization
fc = tflearn.fully_connected
# Architecture from 'Human-level control through deep reinforcement learning'
# http://www.nature.com/nature/journal/v518/n7540/full/nature14236.html
convs = [(32, 8, [1,4,4,1]), (64, 4, [1,2,2,1]), (64, 3, [1,1,1,1])]
fcs = [512, 1]
reg = None #'L2'
us = []
zs = []
layerI = 0
prevU = x
for nFilter, kSz, strides in convs:
with tf.variable_scope('u'+str(layerI)) as s:
u = bn(conv(prevU, nFilter, kSz, strides=strides, activation='relu',
scope=s, reuse=reuse, regularizer=reg),
scope=s, reuse=reuse)
us.append(u)
prevU = u
layerI += 1
for sz in fcs:
with tf.variable_scope('u'+str(layerI)) as s:
u = fc(prevU, sz, scope=s, reuse=reuse, regularizer=reg)
if sz == 1:
u = tf.reshape(u, [-1])
else:
u = bn(tf.nn.relu(u), scope=s, reuse=reuse)
us.append(u)
prevU = u
layerI += 1
layerI = 0
prevU, prevZ, y_red = x, None, y
for nFilter, kSz, strides in convs:
z_add = []
if layerI > 0:
with tf.variable_scope('z{}_zu_u'.format(layerI)) as s:
prev_nFilter = convs[layerI-1][0]
zu_u = conv(prevU, prev_nFilter, 3, reuse=reuse,
scope=s, activation='relu', bias=True, regularizer=reg)
with tf.variable_scope('z{}_zu_proj'.format(layerI)) as s:
z_zu = conv(tf.mul(prevZ, zu_u), nFilter, kSz, strides=strides,
reuse=reuse, scope=s, bias=False, regularizer=reg)
z_add.append(z_zu)
with tf.variable_scope('z{}_yu_u'.format(layerI)) as s:
yu_u = conv(prevU, 1, 3, reuse=reuse, scope=s,
bias=True, regularizer=reg)
with tf.variable_scope('z{}_yu'.format(layerI)) as s:
z_yu = conv(tf.mul(y_red, yu_u), nFilter, kSz, strides=strides,
reuse=reuse, scope=s, bias=False, regularizer=reg)
with tf.variable_scope('z{}_y_red'.format(layerI)) as s:
y_red = conv(y_red, 1, kSz, strides=strides, reuse=reuse,
scope=s, bias=True, regularizer=reg)
z_add.append(z_yu)
with tf.variable_scope('z{}_u'.format(layerI)) as s:
z_u = conv(prevU, nFilter, kSz, strides=strides, reuse=reuse,
scope=s, bias=True, regularizer=reg)
z_add.append(z_u)
z = tf.nn.relu(tf.add_n(z_add))
zs.append(z)
prevU = us[layerI] if layerI < len(us) else None
prevZ = z
layerI += 1
prevZ = tf.contrib.layers.flatten(prevZ)
prevU = tf.contrib.layers.flatten(prevU)
y_red_flat = tf.contrib.layers.flatten(y_red)
for sz in fcs:
z_add = []
with tf.variable_scope('z{}_zu_u'.format(layerI)) as s:
prevU_sz = prevU.get_shape()[1].value
zu_u = fc(prevU, prevU_sz, reuse=reuse, scope=s,
activation='relu', bias=True, regularizer=reg)
with tf.variable_scope('z{}_zu_proj'.format(layerI)) as s:
z_zu = fc(tf.mul(prevZ, zu_u), sz, reuse=reuse, scope=s,
bias=False, regularizer=reg)
z_add.append(z_zu)
# y passthrough in the FC layers:
#
# with tf.variable_scope('z{}_yu_u'.format(layerI)) as s:
# ycf_sz = y_red_flat.get_shape()[1].value
# yu_u = fc(prevU, ycf_sz, reuse=reuse, scope=s, bias=True,
# regularizer=reg)
# with tf.variable_scope('z{}_yu'.format(layerI)) as s:
# z_yu = fc(tf.mul(y_red_flat, yu_u), sz, reuse=reuse, scope=s,
# bias=False, regularizer=reg)
# z_add.append(z_yu)
with tf.variable_scope('z{}_u'.format(layerI)) as s:
z_u = fc(prevU, sz, reuse=reuse, scope=s, bias=True, regularizer=reg)
z_add.append(z_u)
z = tf.add_n(z_add)
variable_summaries(z, 'z{}_preact'.format(layerI))
if sz != 1:
z = tf.nn.relu(z)
variable_summaries(z, 'z{}_act'.format(layerI))
prevU = us[layerI] if layerI < len(us) else None
prevZ = z
zs.append(z)
layerI += 1
z = tf.reshape(z, [-1], name='energies')
return z
def saveImgs(xs, ys, save, colWidth=10):
nImgs = xs.shape[0]
assert(nImgs == ys.shape[0])
if not os.path.exists(save):
os.makedirs(save)
fnames = []
for i in range(nImgs):
xy = np.clip(np.squeeze(np.concatenate([ys[i], xs[i]], axis=1)), 0., 1.)
# Imagemagick montage has intensity scaling issues with png output files here.
fname = "{}/{:04d}.jpg".format(save, i)
plt.imsave(fname, xy, cmap=mpl.cm.gray)
fnames.append(fname)
os.system('montage -geometry +0+0 -tile {}x {} {}.png'.format(
colWidth, ' '.join(fnames), save))
def tf_nOnes(b):
# Must be binary.
return tf.reduce_sum(tf.cast(b, tf.int32))
def mse(y, trueY):
return np.mean(np.square(255.*(y-trueY)))
# return 0.5*np.sum(np.square((y-trueY)))
def mseGrad_full(y, trueY, G):
k,n = G.shape
assert(len(y) == n)
I = np.where((y > 1e-8) & (1.-y > 1e-8))
z = np.ones_like(y)
z[I] = (1./y[I] + 1./(1.-y[I]))
H = np.bmat([[np.diag(z), G.T, np.zeros((n,1))],
[G, np.zeros((k,k)), -np.ones((k,1))],
[np.zeros((1,n)), -np.ones((1,k)), np.zeros((1,1))]])
c = -np.linalg.solve(H, np.concatenate([(y - trueY), np.zeros(k+1)]))
return | np.split(c, [n, n+k]) | numpy.split |
# Copyright 2019-2021 Cambridge Quantum Computing
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import itertools
from functools import lru_cache
from pytket.circuit import Circuit, Qubit, Bit, Node, CircBox # type: ignore
import numpy as np
from collections import OrderedDict, namedtuple
from typing import Dict, Iterable, List, Tuple, Counter, cast, Optional
from math import ceil, log2
from pytket.backends import Backend
from pytket.passes import DecomposeBoxes, FlattenRegisters # type: ignore
from pytket.backends.backendresult import BackendResult
from pytket.utils.outcomearray import OutcomeArray
from enum import Enum
FullCorrelatedNoiseCharacterisation = namedtuple(
"FullCorrelatedNoiseCharacterisation",
["CorrelatedNodes", "NodeToIntDict", "CharacterisationMatrices"],
)
StateInfo = namedtuple("StateInfo", ["PreparedStates", "QubitBitMaps"])
# Helper methods for holding basis states
@lru_cache(maxsize=128)
def binary_to_int(bintuple: Tuple[int]) -> int:
"""Convert a binary tuple to corresponding integer, with most significant bit as the
first element of tuple.
:param bintuple: Binary tuple
:type bintuple: Tuple[int]
:return: Integer
:rtype: int
"""
integer = 0
for index, bitset in enumerate(reversed(bintuple)):
if bitset:
integer |= 1 << index
return integer
@lru_cache(maxsize=128)
def int_to_binary(val: int, dim: int) -> Tuple[int, ...]:
"""Convert an integer to corresponding binary tuple, with most significant bit as
the first element of tuple.
:param val: input integer
:type val: int
:param dim: Bit width
:type dim: int
:return: Binary tuple of width dim
:rtype: Tuple[int, ...]
"""
return tuple(map(int, format(val, "0{}b".format(dim))))
def get_full_transition_tomography_circuits(
process_circuit: Circuit, backend: Backend, correlations: List[List[Node]]
) -> Tuple[List[Circuit], List[StateInfo]]:
"""Generate calibration circuits according to the specified correlation, backend and given circuit.
:param circuit: Circuit surmising correlated noise process being characterised
:param backend: Backend on which the experiments are run.
:type backend: Backend
:param correlations: A list of lists of correlated Nodes of a `Device`.
Qubits within the same list are assumed to only have errors correlated
with each other. Thus to allow errors between all qubits you should
provide a single list. The qubits in `correlations` must be nodes in the
backend's associated `Device`.
:type correlations: List[List[Node]]
:return: A list of calibration circuits to be run on the machine. The circuits
should be processed without compilation.
:rtype: List[Circuit]
"""
subsets_matrix_map = OrderedDict.fromkeys(
sorted(map(tuple, correlations), key=len, reverse=True)
)
# ordered from largest to smallest via OrderedDict & sorted
subset_dimensions = [len(subset) for subset in subsets_matrix_map]
major_state_dimensions = subset_dimensions[0]
n_circuits = 1 << major_state_dimensions
all_qubits = [qb for subset in correlations for qb in subset]
if len(process_circuit.qubits) != len(all_qubits):
raise ValueError(
"Process being characterised has {} qubits, correlations only specify {} qubits.".format(
len(process_circuit.qubits), len(all_qubits)
)
)
# output
prepared_circuits = []
state_infos = []
# set up CircBox of X gate for preparing basis states
xcirc = Circuit(1).X(0)
xcirc = backend.get_compiled_circuit(xcirc)
FlattenRegisters().apply(xcirc)
xbox = CircBox(xcirc)
# need to be default register to add as box suitably
process_circuit = backend.get_compiled_circuit(process_circuit)
rename_map_pc = {}
for index, qb in enumerate(process_circuit.qubits):
rename_map_pc[qb] = Qubit(index)
process_circuit.rename_units(rename_map_pc)
pbox = CircBox(process_circuit)
# set up base circuit for appending xbox to
base_circuit = Circuit()
index = 0
measures = {}
for qb in all_qubits:
base_circuit.add_qubit(qb)
c_bit = Bit(index)
base_circuit.add_bit(c_bit)
index += 1
measures[qb] = c_bit
# generate state circuits for given correlations
for major_state_index in range(n_circuits):
state_circuit = base_circuit.copy()
# get bit string corresponding to basis state of biggest subset of qubits
major_state = int_to_binary(major_state_index, major_state_dimensions)
new_state_dicts = {}
# parallelise circuits, run uncorrelated subsets characterisation in parallel
for dim, qubits in zip(subset_dimensions, subsets_matrix_map):
# add state to prepared states
new_state_dicts[qubits] = major_state[:dim]
# find only qubits that are expected to be in 1 state, add xbox to given qubits
for flipped_qb in itertools.compress(qubits, major_state[:dim]):
state_circuit.add_circbox(xbox, [flipped_qb])
# Decompose boxes, add barriers to preserve circuit, add measures
state_circuit.add_barrier(all_qubits)
# add process circuit to measure
state_circuit.add_circbox(pbox, state_circuit.qubits)
DecomposeBoxes().apply(state_circuit)
state_circuit.add_barrier(all_qubits)
for q in measures:
state_circuit.Measure(q, measures[q])
# add to returned types
state_circuit = backend.get_compiled_circuit(state_circuit)
prepared_circuits.append(state_circuit)
state_infos.append(StateInfo(new_state_dicts, measures))
return (prepared_circuits, state_infos)
def calculate_correlation_matrices(
results_list: List[BackendResult],
states_info: List[StateInfo],
correlations: List[List[Qubit]],
) -> FullCorrelatedNoiseCharacterisation:
"""Calculate the calibration matrices corresponding to some pure noise from the results of running calibration
circuits.
:param results_list: List of result via BackendResult. Must be in the same order as the
corresponding circuits given by prepared_states.
:type results_list: List[BackendResult]
:param states_info: Each StateInfo object containts the state prepared via a binary
representation and the qubit_to_bit_map for the corresponding state circuit.
:type states_info: List[StateInfo]
:param correlations: List of dict corresponding to each prepared basis state
:type correlations: List[List[Qubit]]
:return: Characterisation for pure noise given by process circuit
:rtype: FullCorrelatedNoiseCharacterisation
"""
subsets_matrix_map = OrderedDict.fromkeys(
sorted(map(tuple, correlations), key=len, reverse=True)
)
# ordered from largest to smallest via OrderedDict & sorted
subset_dimensions = [len(subset) for subset in subsets_matrix_map]
counter = 0
node_index_dict = dict()
for qbs, dim in zip(subsets_matrix_map, subset_dimensions):
# for a subset with n qubits, create a 2^n by 2^n matrix
subsets_matrix_map[qbs] = np.zeros((1 << dim,) * 2, dtype=float)
for i in range(len(qbs)):
qb = qbs[i]
node_index_dict[qb] = (counter, i)
counter += 1
for result, state_info in zip(results_list, states_info):
state_dict = state_info[0]
qb_bit_map = state_info[1]
for qb_sub in subsets_matrix_map:
# bits of counts to consider
bits = [qb_bit_map[q] for q in qb_sub]
counts_dict = result.get_counts(cbits=bits)
for measured_state, count in counts_dict.items():
# intended state
prepared_state_index = binary_to_int(state_dict[qb_sub])
# produced state
measured_state_index = binary_to_int(measured_state)
# update characterisation matrix
subsets_matrix_map[qb_sub][
measured_state_index, prepared_state_index
] += count
# normalise everything
normalised_mats = [mat / np.sum(mat, axis=0) for mat in subsets_matrix_map.values()]
return FullCorrelatedNoiseCharacterisation(
correlations, node_index_dict, normalised_mats
)
#########################################
### _compute_dot and helper functions ###
###
### With thanks to
### https://math.stackexchange.com/a/3423910
### and especially
### https://gist.github.com/ahwillia/f65bc70cb30206d4eadec857b98c4065
### on which this code is based.
def _unfold(tens: np.ndarray, mode: int, dims: List[int]) -> np.ndarray:
"""
Unfolds tensor into matrix.
:param tens: Tensor with shape equivalent to dimensions
:type tens: np.ndarray
:param mode: Specifies axis move to front of matrix in unfolding of tensor
:type mode: int
:param dims: Gives shape of tensor passed
:type dims: List[int]
:return: Matrix with shape (dims[mode], prod(dims[/mode]))
:rtype: np.ndarray
"""
if mode == 0:
return tens.reshape(dims[0], -1)
else:
return np.moveaxis(tens, mode, 0).reshape(dims[mode], -1)
def _refold(vec: np.ndarray, mode: int, dims: List[int]) -> np.ndarray:
"""
Refolds vector into tensor.
:param vec: Tensor with length equivalent to the product of dimensions given in dims
:type vec: np.ndarray
:param mode: Axis tensor was unfolded along
:type mode: int
:param dims: Shape of tensor
:type dims: List[int]
:return: Tensor folded from vector with shape equivalent to dimensions given in dims
:rtype: np.ndarray
"""
if mode == 0:
return vec.reshape(dims)
else:
# Reshape and then move dims[mode] back to its
# appropriate spot (undoing the `unfold` operation).
tens = vec.reshape([dims[mode]] + [d for m, d in enumerate(dims) if m != mode])
return np.moveaxis(tens, 0, mode)
def _compute_dot(submatrices: Iterable[np.ndarray], vector: np.ndarray) -> np.ndarray:
"""
Multiplies the kronecker product of the given submatrices with given vector.
:param submatrices: Submatrices multiplied
:type submatrices: Iterable[np.ndarray]
:param vector: Vector multplied
:type vector: np.ndarray
:return: Kronecker product of arguments
:rtype: np.ndarray
"""
dims = [A.shape[0] for A in submatrices]
vt = vector.reshape(dims)
for i, A in enumerate(submatrices):
vt = _refold(A @ _unfold(vt, i, dims), i, dims)
return vt.ravel()
def _bayesian_iteration(
submatrices: Iterable[np.ndarray],
measurements: np.ndarray,
t: np.ndarray,
epsilon: float,
) -> np.ndarray:
"""
Transforms T corresponds to a Bayesian iteration, used to modfiy measurements.
:param submatrices: submatrices to be inverted and applied to measurements.
:type submatrices: Iterable[np.ndarray]
:param measurements: Probability distribution over some set of states to be amended.
:type measurements: np.ndarray
:param t: Some transform to act on measurements.
:type t: np.ndarray
:param epsilon: A stabilization parameter to define an affine transformation for applicatoin
to submatrices, eliminating zero probabilities.
:type epsilon: float
:return: Transformed distribution vector.
:rtype: np.ndarray
"""
# Transform t according to the Bayesian iteration
# The parameter epsilon is a stabilization parameter which defines an affine
# transformation to apply to the submatrices to eliminate zero probabilities. This
# transformation preserves the property that all columns sum to 1
if epsilon == 0:
# avoid copying if we don't need to
As = submatrices
else:
As = [
epsilon / submatrix.shape[0] + (1 - epsilon) * submatrix
for submatrix in submatrices
]
z = _compute_dot(As, t)
if np.isclose(z, 0).any():
raise ZeroDivisionError
return cast(
np.ndarray, t * _compute_dot([A.transpose() for A in As], measurements / z)
)
def _bayesian_iterative_correct(
submatrices: Iterable[np.ndarray],
measurements: np.ndarray,
tol: float = 1e-5,
max_it: Optional[int] = None,
) -> np.ndarray:
"""
Finds new states to represent application of inversion of submatrices on measurements.
Converges when update states within tol range of previously tested states.
:param submatrices: Matrices comprising the pure noise characterisation.
:type submatrices: Iterable[np.ndarray]
:param input_vector: Vector corresponding to some counts distribution.
:type input_vector: np.ndarray
:param tol: tolerance of closeness of found results
:type tol: float
:param max_it: Maximum number of inversions attempted to correct results.
:type max_it: int
"""
# based on method found in https://arxiv.org/abs/1910.00129
vector_size = measurements.size
# uniform initial
true_states = np.full(vector_size, 1 / vector_size)
prev_true = true_states.copy()
converged = False
count = 0
epsilon: float = 0 # stabilization parameter, adjusted dynamically
while not converged:
if max_it:
if count >= max_it:
break
count += 1
try:
true_states = _bayesian_iteration(
submatrices, measurements, true_states, epsilon
)
converged = np.allclose(true_states, prev_true, atol=tol)
prev_true = true_states.copy()
except ZeroDivisionError:
# Shift the stabilization parameter up a bit (always < 0.5).
epsilon = 0.99 * epsilon + 0.01 * 0.5
return true_states
class CorrectionMethod(Enum):
def Invert(
submatrices: Iterable[np.ndarray], input_vector: np.ndarray
) -> np.ndarray:
"""
Multiplies the kronecker product of given submatrices on input vector
and then adjusts output to make them genuine probabilities. Submatrices
represent pure noise characterisation, vector corresponds to counts
distribution from circuit and device.
:param submatrices: Matrices comprising the pure noise characterisation.
:type submatrices: Iterable[np.ndarray]
:param input_vector: Vector corresponding to some counts distribution.
:type input_vector: np.ndarray
"""
try:
subinverts = [np.linalg.inv(submatrix) for submatrix in submatrices]
except np.linalg.LinAlgError:
raise ValueError(
"Unable to invert calibration matrix: please re-run "
"calibration experiments or use an alternative correction method."
)
# assumes that order of rows in flattened subinverts equals order of bits in input vector
v = _compute_dot(subinverts, input_vector)
# The entries of v will always sum to 1, but they may not all be in the range [0,1].
# In order to make them genuine probabilities (and thus generate meaningful counts),
# we adjust them by setting all negative values to 0 and scaling the remainder.
v[v < 0] = 0
v /= sum(v)
return v
def Bayesian(
submatrices: Iterable[np.ndarray], input_vector: np.ndarray
) -> np.ndarray:
"""
Computes the product of the invert of submatrices on the given input vector via a an iterative
Bayesian correction method.
:param submatrices: Matrices comprising the pure noise characterisation.
:type submatrices: Iterable[np.ndarray]
:param input_vector: Vector corresponding to some counts distribution.
:type input_vector: np.ndarray
:param tol: tolerance of closeness of found results
:type tol: float
:param max_it: Maximum number of inversions attempted to correct results.
:type max_it: int
"""
return _bayesian_iterative_correct(
submatrices, input_vector, tol=1e-5, max_it=500
)
def reduce_matrix(indices_to_remove: List[int], matrix: np.ndarray) -> np.ndarray:
"""
Removes indices from indices_to_remove from binary associated to indexing of matrix,
producing a new transition matrix.
To do so, it assigns all transition probabilities as the given state in the remaining
indices binary, with the removed binary in state 0. This is an assumption on the noise made
because it is likely that unmeasured qubits will be in that state.
:param indices_to_remove: Binary index of state matrix is mapping to be removed.
:type indices_to_remove: List[int]
:param matrix: Transition matrix where indices correspond to some binary state, to have some
dimension removed.
:type matrix: np.ndarray
:return: Transition matrix with removed entries.
:rtype: np.ndarray
"""
new_n_qubits = int(log2(matrix.shape[0])) - len(indices_to_remove)
if new_n_qubits == 0:
return np.array([])
bin_map = dict()
mat_dim = 1 << new_n_qubits
for index in range(mat_dim):
# get current binary
bina = list(int_to_binary(index, new_n_qubits))
# add 0's to fetch old binary to set values from
for i in sorted(indices_to_remove):
bina.insert(i, 0)
# get index of values
bin_map[index] = binary_to_int(tuple(bina))
new_mat = np.zeros((mat_dim,) * 2, dtype=float)
for i in range(len(new_mat)):
old_row_index = bin_map[i]
for j in range(len(new_mat)):
old_col_index = bin_map[j]
new_mat[i, j] = matrix[old_row_index, old_col_index]
return new_mat
def reduce_matrices(
entries_to_remove: List[Tuple[int, int]], matrices: List[np.ndarray]
) -> List[np.ndarray]:
"""
Removes some dimensions from some matrices.
:param entries_to_remove: Via indexing, says which dimensions to remove from which indices.
:type entries_to_remove: List[Tuple[int, int]]
:param matrices: All matrices to have dimensions removed.
:type matrices: List[np.ndarray]
:return: Matrices with some dimensions removed.
:rtype: List[np.ndarray]
"""
organise: Dict[int, List[int]] = {k: [] for k in range(len(matrices))}
for unused in entries_to_remove:
# unused[0] is index in matrices
# unused[1] is qubit index in matrix
organise[unused[0]].append(unused[1])
output_matrices = [reduce_matrix(organise[m], matrices[m]) for m in organise]
normalised_mats = [
mat / np.sum(mat, axis=0) for mat in [x for x in output_matrices if x.size > 0]
]
return normalised_mats
def get_single_matrix(
entry_to_keep: Tuple[int, int], matrices: List[np.ndarray]
) -> np.ndarray:
"""
Returns a correction matrix just for the index given.
:param entry_to_keep: Which matrix and indexing to return a correction matrix for.
:type entries_to_keep: Tuple[int, int]
:param matrices: All matrices to find returned matrix from.
:type matrices: List[np.ndarray]
:return: Matrix for correcting given entry.
:rtype: List[np.ndarray]
"""
mat = matrices[entry_to_keep[0]]
all_indices = list(range(int(log2(mat.shape[0]))))
all_indices.remove(entry_to_keep[1])
return reduce_matrix(all_indices, mat)
def correct_transition_noise(
result: BackendResult,
bit_qb_info: Tuple[Dict[Qubit, Bit], Dict[Bit, Qubit]],
noise_characterisation: FullCorrelatedNoiseCharacterisation,
corr_method: CorrectionMethod,
) -> BackendResult:
"""
Modifies count distribution for result, such that the inversion of the pure noise map represented by
matrices in noise_characterisation is applied to it
:param result: BackendResult object to be negated by pure noise object.
:type result: BackendResult
:param bit_qb_info: Used to permute corresponding BackendResult object so counts order matches noise characterisation.
:type bit_qb_info: Tuple[Dict[Qubit, Bit], Dict[Bit, Qubit]]
:param noise_characterisation: Object holding all required information for some full noise characterisation of correlated subsets.
:type noise_characterisation: FullCorrelatedNoiseCharacterisation
"""
final_measures_qb_map = bit_qb_info[0]
mid_circuit_measures_bq_map = bit_qb_info[1]
# get counts from with order of bits that matches order of qubits in subsets
# if qubit in subset has no bit, skip it
char_bits_order = []
unused_final_qbs = []
for subset in noise_characterisation.CorrelatedNodes:
for q in subset:
if q in final_measures_qb_map:
char_bits_order.append(final_measures_qb_map[q])
else:
unused_final_qbs.append(q)
mid_measure_qbs = []
for bit in mid_circuit_measures_bq_map:
mid_measure_qbs.append(mid_circuit_measures_bq_map[bit])
char_bits_order.append(bit)
# get counts object for returning later
counts = result.get_counts(cbits=char_bits_order)
in_vec = np.zeros(1 << len(char_bits_order), dtype=float)
# turn from counts to probability distribution
for state, count in counts.items():
in_vec[binary_to_int(state)] = count
Ncounts = | np.sum(in_vec) | numpy.sum |
# -*- coding: utf-8 -*-
"""
Created on Fri Jan 22 13:51:08 2021
@author: mzomou
"""
from scipy.signal import savgol_filter
import tkinter as tk
from tkinter import IntVar, DISABLED, ACTIVE, NORMAL, StringVar, messagebox
from tkinter.colorchooser import askcolor
import numpy as np
from hsi import HSAbsorption, HSImage
from hsi.analysis import HSComponentFit
from matplotlib.backends.backend_tkagg import FigureCanvasTkAgg, NavigationToolbar2Tk
from matplotlib.figure import Figure
import matplotlib.pyplot as plt
import matplotlib.lines as lines
from tkinter import filedialog
from scipy import ndimage
from matplotlib.widgets import Cursor
from matplotlib.colors import ListedColormap, LinearSegmentedColormap
from tkinter import font as tkFont
from tkinter import simpledialog
from spectral import spectral_angles
from sklearn.decomposition import PCA, KernelPCA
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis as LDA
from sklearn.ensemble import RandomForestClassifier
from sklearn.gaussian_process.kernels import RBF
from sklearn.gaussian_process import GaussianProcessClassifier
from sklearn.neural_network import MLPClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC
import susi
##### Cube-Data Laden und als RGB Bild Darstellen
WLD = | np.linspace(500, 995, 100) | numpy.linspace |
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import os
import random
import math
import json
import threading
import numpy as np
import tensorflow as tf
import util
import coref_ops
import conll
import metrics
import optimization
from bert import tokenization
from bert import modeling
from pytorch_to_tf import load_from_pytorch_checkpoint
class CorefModel(object):
def __init__(self, config):
self.config = config
self.subtoken_maps = {}
self.max_segment_len = config['max_segment_len']
self.max_span_width = config["max_span_width"]
self.mask_perc = config['mask_percentage'] #MODIFIED
self.n_placeholders = 1 #MODIFIED
self.genres = { g:i for i,g in enumerate(config["genres"]) }
self.eval_data = None # Load eval data lazily.
self.bert_config = modeling.BertConfig.from_json_file(config["bert_config_file"])
self.sep = 102
self.cls = 101
self.tokenizer = tokenization.FullTokenizer(
vocab_file=config['vocab_file'], do_lower_case=False)
input_props = []
input_props.append((tf.int32, [None, None])) # input_ids.
input_props.append((tf.int32, [None, None])) # input_mask
input_props.append((tf.int32, [None, None])) # input_ids.
input_props.append((tf.int32, [None, None])) # input_mask
input_props.append((tf.int32, [None])) # Text lengths.
input_props.append((tf.int32, [None, None])) # Speaker IDs.
input_props.append((tf.int32, [])) # Genre.
input_props.append((tf.bool, [])) # Is training.
input_props.append((tf.int32, [None])) # Gold starts.
input_props.append((tf.int32, [None])) # Gold ends.
input_props.append((tf.int32, [None])) # Cluster ids.
input_props.append((tf.int32, [None])) # Sentence Map
self.queue_input_tensors = [tf.placeholder(dtype, shape) for dtype, shape in input_props]
dtypes, shapes = zip(*input_props)
queue = tf.PaddingFIFOQueue(capacity=10, dtypes=dtypes, shapes=shapes)
self.enqueue_op = queue.enqueue(self.queue_input_tensors)
self.input_tensors = queue.dequeue()
self.predictions, self.loss = self.get_predictions_and_loss(*self.input_tensors)
# bert stuff
tvars = tf.trainable_variables()
assignment_map, initialized_variable_names = modeling.get_assignment_map_from_checkpoint(tvars, config['tf_checkpoint'])
init_from_checkpoint = tf.train.init_from_checkpoint if config['init_checkpoint'].endswith('ckpt') else load_from_pytorch_checkpoint
init_from_checkpoint(config['init_checkpoint'], assignment_map)
print("**** Trainable Variables ****")
for var in tvars:
init_string = ""
if var.name in initialized_variable_names:
init_string = ", *INIT_FROM_CKPT*"
# tf.logging.info(" name = %s, shape = %s%s", var.name, var.shape,
# init_string)
print(" name = %s, shape = %s%s" % (var.name, var.shape, init_string))
num_train_steps = int(
self.config['num_docs'] * self.config['num_epochs'])
num_warmup_steps = int(num_train_steps * 0.1)
self.global_step = tf.train.get_or_create_global_step()
self.train_op = optimization.create_custom_optimizer(tvars,
self.loss, self.config['bert_learning_rate'], self.config['task_learning_rate'],
num_train_steps, num_warmup_steps, False, self.global_step, freeze=-1)
def start_enqueue_thread(self, session):
print('Loading data')
with open(self.config["train_path"]) as f:
train_examples = [json.loads(jsonline) for jsonline in f.readlines()]
def _enqueue_loop():
while True:
random.shuffle(train_examples)
if self.config['single_example']:
for example in train_examples:
example = add_masks(example, mask_profile = 'percentage', all_profiles = False, n_masks_profile = 1, skip_first_mention = False,
perc_mask=self.mask_perc, n_placeholders = self.n_placeholders)
tensorized_example = self.tensorize_example(example[0], is_training=True)
feed_dict = dict(zip(self.queue_input_tensors, tensorized_example))
session.run(self.enqueue_op, feed_dict=feed_dict)
else:
examples = []
for example in train_examples:
example = add_masks(example, mask_profile = 'percentage', all_profiles = False, n_masks_profile = 1, skip_first_mention = False,
perc_mask=self.mask_perc, n_placeholders = self.n_placeholders)
tensorized = self.tensorize_example(example[0], is_training=True)
if type(tensorized) is not list:
tensorized = [tensorized]
examples += tensorized
random.shuffle(examples)
print('num examples', len(examples))
for example in examples:
feed_dict = dict(zip(self.queue_input_tensors, example))
session.run(self.enqueue_op, feed_dict=feed_dict)
enqueue_thread = threading.Thread(target=_enqueue_loop)
enqueue_thread.daemon = True
enqueue_thread.start()
def restore(self, session):
# Don't try to restore unused variables from the TF-Hub ELMo module.
vars_to_restore = [v for v in tf.global_variables() ]
saver = tf.train.Saver(vars_to_restore)
checkpoint_path = os.path.join(self.config["log_dir"], "model.max.ckpt")
print("Restoring from {}".format(checkpoint_path))
session.run(tf.global_variables_initializer())
saver.restore(session, checkpoint_path)
def tensorize_mentions(self, mentions):
if len(mentions) > 0:
starts, ends = zip(*mentions)
else:
starts, ends = [], []
return np.array(starts), np.array(ends)
def tensorize_span_labels(self, tuples, label_dict):
if len(tuples) > 0:
starts, ends, labels = zip(*tuples)
else:
starts, ends, labels = [], [], []
return np.array(starts), np.array(ends), np.array([label_dict[c] for c in labels])
def get_speaker_dict(self, speakers):
speaker_dict = {'UNK': 0, '[SPL]': 1}
for s in speakers:
if s not in speaker_dict and len(speaker_dict) < self.config['max_num_speakers']:
speaker_dict[s] = len(speaker_dict)
return speaker_dict
def tensorize_example(self, example, is_training):
clusters = example["clusters"]
gold_mentions = sorted(tuple(m) for m in util.flatten(clusters))
gold_mention_map = {m:i for i,m in enumerate(gold_mentions)}
cluster_ids = np.zeros(len(gold_mentions))
for cluster_id, cluster in enumerate(clusters):
for mention in cluster:
cluster_ids[gold_mention_map[tuple(mention)]] = cluster_id + 1
sentences = example["sentences"]
#sentences = [sentence[1:-1] for sentence in sentences]
num_words = sum(len(s) for s in sentences)
speakers = example["speakers"]
# assert num_words == len(speakers), (num_words, len(speakers))
speaker_dict = self.get_speaker_dict(util.flatten(speakers))
sentence_map = example['sentence_map']
max_sentence_length = self.max_segment_len #270 #max(len(s) for s in sentences)
max_len = max(len(s) + 2 for s in sentences) # CHANGED; two symbols added later
if max_len > max_sentence_length:
max_sentence_length = max_len
text_len = np.array([len(s) for s in sentences])
input_ids, input_mask, speaker_ids, prev_overlap = [], [], [], []
overlap_ids, overlap_mask = [], []
half = self.max_segment_len // 2
prev_tokens_per_seg = []
for i, (sentence, speaker) in enumerate(zip(sentences, speakers)):
prev_tokens_per_seg += [len(prev_overlap)]
overlap_words = ['[CLS]'] + prev_overlap + sentence[:half] + ['[SEP]']
prev_overlap = sentence[half:]
sentence = ['[CLS]'] + sentence + ['[SEP]']
sent_input_ids = self.tokenizer.convert_tokens_to_ids(sentence)
sent_input_mask = [1] * len(sent_input_ids)
sent_speaker_ids = [speaker_dict.get(s, 0) for s in ['##'] + speaker + ['##']]
while len(sent_input_ids) < max_sentence_length:
sent_input_ids.append(0)
sent_input_mask.append(0)
sent_speaker_ids.append(0)
overlap_input_ids = self.tokenizer.convert_tokens_to_ids(overlap_words)
overlap_input_mask = [1] * len(overlap_input_ids)
while len(overlap_input_ids) < max_sentence_length:
overlap_input_ids.append(0)
overlap_input_mask.append(0)
input_ids.append(sent_input_ids)
speaker_ids.append(sent_speaker_ids)
input_mask.append(sent_input_mask)
overlap_ids.append(overlap_input_ids)
overlap_mask.append(overlap_input_mask)
overlap_words = ['[CLS]'] + prev_overlap + ['[SEP]']
overlap_input_ids = self.tokenizer.convert_tokens_to_ids(overlap_words)
overlap_input_mask = [1] * len(overlap_input_ids)
prev_tokens_per_seg += [len(prev_overlap)]
while len(overlap_input_ids) < max_sentence_length:
overlap_input_ids.append(0)
overlap_input_mask.append(0)
overlap_ids.append(overlap_input_ids)
overlap_mask.append(overlap_input_mask)
input_ids = np.array(input_ids)
input_mask = np.array(input_mask)
speaker_ids = np.array(speaker_ids)
overlap_ids = np.array(overlap_ids)
overlap_mask = | np.array(overlap_mask) | numpy.array |
import numpy as np
from scipy.optimize import linear_sum_assignment
from ._base_metric import _BaseMetric
from .. import _timing
class Identity(_BaseMetric):
"""Class which implements the ID metrics"""
def __init__(self):
super().__init__()
self.integer_fields = ['IDTP', 'IDFN', 'IDFP']
self.float_fields = ['IDF1', 'IDR', 'IDP']
self.fields = self.float_fields + self.integer_fields
self.summary_fields = self.fields
self.threshold = 0.5
@_timing.time
def eval_sequence(self, data):
"""Calculates ID metrics for one sequence"""
# Initialise results
res = {}
for field in self.fields:
res[field] = 0
# Return result quickly if tracker or gt sequence is empty
if data['num_tracker_dets'] == 0:
res['IDFN'] = data['num_gt_dets']
return res
if data['num_gt_dets'] == 0:
res['IDFP'] = data['num_tracker_dets']
return res
# Variables counting global association
potential_matches_count = np.zeros((data['num_gt_ids'], data['num_tracker_ids']))
gt_id_count = np.zeros(data['num_gt_ids'])
tracker_id_count = np.zeros(data['num_tracker_ids'])
# First loop through each timestep and accumulate global track information.
for t, (gt_ids_t, tracker_ids_t) in enumerate(zip(data['gt_ids'], data['tracker_ids'])):
# Count the potential matches between ids in each timestep
matches_mask = np.greater_equal(data['similarity_scores'][t], self.threshold)
match_idx_gt, match_idx_tracker = np.nonzero(matches_mask)
potential_matches_count[gt_ids_t[match_idx_gt], tracker_ids_t[match_idx_tracker]] += 1
# Calculate the total number of dets for each gt_id and tracker_id.
gt_id_count[gt_ids_t] += 1
tracker_id_count[tracker_ids_t] += 1
# Calculate optimal assignment cost matrix for ID metrics
num_gt_ids = data['num_gt_ids']
num_tracker_ids = data['num_tracker_ids']
fp_mat = | np.zeros((num_gt_ids + num_tracker_ids, num_gt_ids + num_tracker_ids)) | numpy.zeros |
#!/usr/bin/env python
# -*- coding: utf-8 -*-
from __future__ import print_function
import time
import pandas as pd
import numpy as np
import os
__docformat__ = 'restructedtext en'
def exe_time(func):
def new_func(*args, **args2):
t0 = time.time()
print("-- @%s, {%s} start" % (time.strftime("%X", time.localtime()), func.__name__))
back = func(*args, **args2)
print("-- @%s, {%s} end" % (time.strftime("%X", time.localtime()), func.__name__))
print("-- @%.3fs taken for {%s}" % (time.time() - t0, func.__name__))
return back
return new_func
def fun_hit_zero_one(user_test_recom):
"""
根据recom_list中item在test_lst里的出现情况生成与recom_list等长的0/1序列
0表示推荐的item不在test里,1表示推荐的item在test里
:param test_lst: 单个用户的test列表
:param recom_lst: 推荐的列表
:param test_mask: 单个用户的test列表对应的mask列表
:return: 与recom_list等长的0/1序列。
"""
test_lst, recom_lst, test_mask, _ = user_test_recom
test_lst = test_lst[:np.sum(test_mask)] # 取出来有效的user_test_list
seq = []
for e in recom_lst:
if e in test_lst: # 命中
seq.append(1)
else: # 没命中
seq.append(0)
return np.array(seq)
def fun_evaluate_map(user_test_recom_zero_one):
"""
计算map。所得是单个用户test的,最后所有用户的求和取平均
:param test_lst: 单个用户的test集
:param zero_one: 0/1序列
:param test_mask: 单个用户的test列表对应的mask列表
:return:
"""
test_lst, zero_one, test_mask, _ = user_test_recom_zero_one
test_lst = test_lst[:np.sum(test_mask)]
zero_one = np.array(zero_one)
if 0 == sum(zero_one): # 没有命中的
return 0.0
zero_one_cum = zero_one.cumsum() # precision要算累计命中
zero_one_cum *= zero_one # 取出命中为1的那些,其余位置得0
idxs = list(np.nonzero(zero_one_cum))[0] # 得到(n,)类型的非零的索引array
s = 0.0
for idx in idxs:
s += 1.0 * zero_one_cum[idx] / (idx + 1)
return s / len(test_lst)
def fun_evaluate_ndcg(user_test_recom_zero_one):
"""
计算ndcg。所得是单个用户test的,最后所有用户的求和取平均
:param test_lst: 单个用户的test集
:param zero_one: 0/1序列
:param test_mask: 单个用户的test列表对应的mask列表
:return:
"""
test_lst, zero_one, test_mask, _ = user_test_recom_zero_one
test_lst = test_lst[:np.sum(test_mask)]
zero_one = np.array(zero_one)
if 0 == sum(zero_one): # 没有命中的
return 0.0
s = 0.0
idxs = list(np.nonzero(zero_one))[0]
for idx in idxs:
s += 1.0 / np.log2(idx + 2)
m = 0.0
length = min(len(test_lst), len(zero_one)) # 序列短的,都命中为1,此时是最优情况
for idx in range(length):
m += 1.0 / np.log2(idx + 2)
return s / m
def fun_idxs_of_max_n_score(user_scores_to_all_items, top_k):
# 从一个向量里找到前n个大数所对应的index
return np.argpartition(user_scores_to_all_items, -top_k)[-top_k:]
def fun_sort_idxs_max_to_min(user_max_n_idxs_scores):
# 按照前n个大数index对应的具体值由大到小排序,即最左侧的index对应原得分值的最大实数值
# 就是生成的推荐items列表里,最左侧的是得分最高的
idxs, scores = user_max_n_idxs_scores # idxs是n个得分最大的items,scores是所有items的得分。
return idxs[np.argsort(scores[idxs])][::-1] # idxs按照对应得分由大到小排列
def fun_predict_auc_recall_map_ndcg(
p, model, best, epoch, starts_ends_auc, starts_ends_tes,
tes_buys_masks, tes_masks):
# ------------------------------------------------------------------------------------------------------------------
# 注意:当zip里的所有子项tes_buys_masks, all_upqs维度一致时,也就是子项的每行每列长度都一样。
# zip后的arr会变成三维矩阵,扫描axis=1会出错得到的一行实际上是2d array,所以后边单独加一列 append 避免该问题。
append = [[0] for _ in np.arange(len(tes_buys_masks))]
# ------------------------------------------------------------------------------------------------------------------
# auc
all_upqs = np.array([[0 for _ in np.arange(len(tes_masks[0]))]]) # 初始化
for start_end in starts_ends_auc:
sub_all_upqs = model.compute_sub_auc_preference(start_end)
all_upqs = np.concatenate((all_upqs, sub_all_upqs))
all_upqs = np.delete(all_upqs, 0, axis=0)
auc = 1.0 * np.sum(all_upqs) / np.sum(tes_masks) # 全部items
# 保存:保存auc的最佳值
if auc > best.best_auc:
best.best_auc = auc
best.best_epoch_auc = epoch
# ------------------------------------------------------------------------------------------------------------------
# recall, map, ndcg
at_nums = p['at_nums'] # [5, 10, 15, 20, 30, 50]
ranges = range(len(at_nums))
# 计算:所有用户对所有商品预测得分的前50个。
# 不会预测出来添加的那个虚拟商品,因为先把它从item表达里去掉
# 注意矩阵形式的索引 all_scores[0, rank]:表示all_scores的各行里取的各个列值是rank里的各项
all_ranks = np.array([[0 for _ in np.arange(at_nums[-1])]]) # 初始shape=(1, 50)
for start_end in starts_ends_tes:
sub_all_scores = model.compute_sub_all_scores(start_end) # shape=(sub_n_user, n_item)
sub_score_ranks = np.apply_along_axis(
func1d=fun_idxs_of_max_n_score,
axis=1,
arr=sub_all_scores,
top_k=at_nums[-1])
sub_all_ranks = np.apply_along_axis(
func1d=fun_sort_idxs_max_to_min,
axis=1,
arr=np.array(zip(sub_score_ranks, sub_all_scores)))
all_ranks = np.concatenate((all_ranks, sub_all_ranks))
del sub_all_scores
all_ranks = np.delete(all_ranks, 0, axis=0) # 去除第一行全0项
# 计算:recall、map、ndcg当前epoch的值
arr = np.array([0.0 for _ in ranges])
recall, precis, f1scor, map, ndcg = arr.copy(), arr.copy(), arr.copy(), arr.copy(), arr.copy()
hits, denominator_recalls = arr.copy(), np.sum(tes_masks) # recall的分母,要预测这么些items
for k in ranges: # 每次考察某个at值下的命中情况
recoms = all_ranks[:, :at_nums[k]] # 向每名user推荐这些
# 逐行,得到recom_lst在test_lst里的命中情况,返回与recom_lst等长的0/1序列,1表示预测的该item在user_test里
all_zero_ones = np.apply_along_axis(
func1d=fun_hit_zero_one,
axis=1,
arr=np.array(zip(tes_buys_masks, recoms, tes_masks, append))) # shape=(n_user, at_nums[k])
hits[k] = | np.sum(all_zero_ones) | numpy.sum |
import numpy as np
import acor
import logging
from .findpeaks import peakdetect
def peaks_and_lphs(y, x=None, lookahead=5, return_heights=False):
"""Returns locations of peaks and corresponding "local peak heights"
"""
if x is None:
x = np.arange(len(y))
maxes, mins = peakdetect(y, x, lookahead=lookahead)
maxes = np.array(maxes)
mins = np.array(mins)
logging.debug('maxes: {0} (shape={0.shape})'.format(maxes))
logging.debug('mins: {0} (shape={0.shape})'.format(mins))
if len(maxes) == 0:
logging.warning('No peaks found in acorr; returning empty')
if return_heights:
return [], [], []
else:
return [], []
n_maxes = maxes.shape[0]
n_mins = mins.shape[0]
if n_maxes==1 and n_mins==1:
lphs = maxes[0,1] - mins[0,1]
elif n_maxes == n_mins+1:
lphs = np.concatenate([[maxes[0,1] - mins[0,1]],
((maxes[1:-1,1] - mins[1:,1]) + (maxes[1:-1,1] - mins[:-1,1]))/2])
elif n_mins == n_maxes+1:
lphs = ((maxes[:,1] - mins[1:,1]) + (maxes[:1,1] - mins[:-1,1]))
elif n_maxes == n_mins:
if maxes[0,0] < mins[0,0]:
lphs = np.concatenate([[maxes[0,1] - mins[0,1]],
((maxes[1:,1] - mins[:-1,1]) + (maxes[1:,1] + mins[1:,1]))/2])
else:
lphs = np.concatenate([((maxes[:-1,1] - mins[:-1,1]) + (maxes[:-1,1] - mins[1:,1]))/2.,
[maxes[-1,1] - mins[-1,1]]])
else:
raise RuntimeError('No cases satisfied??')
##if first extremum is a max, remove it:
#if maxes[0,0] < mins[0,0]:
# logging.debug('first extremum is a max; removing')
# maxes = maxes[1:,:]
# logging.debug('now, maxes: {}'.format(maxes))
# logging.debug('now, mins: {}'.format(mins))
##if last extremum is a max, remove it:
#if maxes[-1,0] > mins[-1,0]:
# logging.debug('last extremum is a max; removing')
# maxes = maxes[:-1,:]
# logging.debug('now, maxes: {}'.format(maxes))
# logging.debug('now, mins: {}'.format(mins))
#this should always work now?
#lphs = ((maxes[:,1] - mins[:-1,1]) + (maxes[:,1] - mins[1:,1]))/2.
"""
if maxes.shape[0]==1:
if return_heights:
return maxes[:,0], [], []
else:
return [], []
#calculate "local heights". First (used to) always be a minimum.
try: #this if maxes and mins are same length
lphs = np.concatenate([((maxes[:-1,1] - mins[:-1,1]) + (maxes[:-1,1] - mins[1:,1]))/2.,
np.array([maxes[-1,1]-mins[-1,1]])])
except ValueError: #this if mins have one more
try:
lphs = ((maxes[:,1] - mins[:-1,1]) + (maxes[:,1] - mins[1:,1]))/2.
except ValueError: # if maxes have one more (drop first max)
lphs = np.concatenate([((maxes[1:-1,1] - mins[:-1,1]) + (maxes[1:-1,1] - mins[1:,1]))/2.,
np.array([maxes[-1,1]-mins[-1,1]])])
"""
logging.debug('lphs: {}'.format(lphs))
if return_heights:
return maxes[:,0], lphs, maxes[:,1]
else:
return maxes[:,0], lphs
def acorr_peaks_old(fs, maxlag, mask=None, lookahead=5, smooth=18,
return_acorr=False, days=True):
"""Returns positions of acorr peaks, with corresponding local heights.
"""
fs = np.atleast_1d(fs)
if mask is None:
mask = self.mask
fs[mask] = 0
corr = acor.function(fs,maxlag)
lag = np.arange(maxlag)
logging.debug('ac: {}'.format(corr))
#lag, corr = acorr(fs, mask=mask, maxlag=maxlag)
maxes, mins = peakdetect(corr, lag, lookahead=lookahead)
maxes = np.array(maxes)
mins = np.array(mins)
logging.debug('maxes: {}'.format(maxes))
#calculate "local heights". First will always be a minimum.
try: #this if maxes and mins are same length
lphs = np.concatenate([((maxes[:-1,1] - mins[:-1,1]) + (maxes[:-1,1] - mins[1:,1]))/2.,
np.array([maxes[-1,1]-mins[-1,1]])])
except ValueError: #this if mins have one more
lphs = ((maxes[:,1] - mins[:-1,1]) + (maxes[:,1] - mins[1:,1]))/2.
if return_acorr:
return corr, maxes[:-1,0], lphs[:-1] #leaving off the last one, just in case weirdness...
else:
return maxes[:-1,0], lphs[:-1] #leaving off the last one, just in case weirdness...
def fit_period(period, peaks, lphs, fit_npeaks=4, tol=0.2):
"""fits series of fit_npeaks peaks near integer multiples of period to a line
tol is fractional tolerance in order to select a peak to fit.
"""
#identify peaks to use in fit: first 'fit_npeaks' peaks w/in
# 20% of integer multiple of period
logging.debug(np.absolute((peaks[:fit_npeaks]/period)/(np.arange(fit_npeaks)+1) - 1))
close_peaks = np.absolute((peaks[:fit_npeaks]/period)/( | np.arange(fit_npeaks) | numpy.arange |
""" Dataset for 3D object detection on SUN RGB-D (with support of vote supervision).
A sunrgbd oriented bounding box is parameterized by (cx,cy,cz), (l,w,h) -- (dx,dy,dz) in upright depth coord
(Z is up, Y is forward, X is right ward), heading angle (from +X rotating to -Y) and semantic class
Point clouds are in **upright_depth coordinate (X right, Y forward, Z upward)**
Return heading class, heading residual, size class and size residual for 3D bounding boxes.
Oriented bounding box is parameterized by (cx,cy,cz), (l,w,h), heading_angle and semantic class label.
(cx,cy,cz) is in upright depth coordinate
(l,h,w) are *half length* of the object sizes
The heading angle is a rotation rad from +X rotating towards -Y. (+X is 0, -Y is pi/2)
Author: <NAME>
Date: 2021
"""
import os
import sys
import numpy as np
from torch.utils.data import Dataset
import scipy.io as sio # to load .mat files for depth points
import cv2
BASE_DIR = os.path.dirname(os.path.abspath(__file__))
ROOT_DIR = os.path.dirname(BASE_DIR)
sys.path.append(BASE_DIR)
sys.path.append(os.path.join(ROOT_DIR, 'utils'))
import pc_util
import my_sunrgbd_utils as sunrgbd_utils
from my_model_util_sunrgbd import SunrgbdDatasetConfig
DC = SunrgbdDatasetConfig() # dataset specific config
MAX_NUM_OBJ = 64 # maximum number of objects allowed per scene
MEAN_COLOR_RGB = np.array([0.5, 0.5, 0.5]) # sunrgbd color is in 0~1
class SunrgbdDetectionVotesDataset(Dataset):
def __init__(self, split_set='train', num_points=20000,
use_color=False, use_height=False, use_v1=False,
augment=False, scan_idx_list=None):
assert (num_points <= 80000)
self.use_v1 = use_v1
if use_v1:
self.data_path = os.path.join(ROOT_DIR,
'sunrgbd/sunrgbd_pc_bbox_votes_80k_v1_%s' % (split_set))
else:
self.data_path = os.path.join(ROOT_DIR,
'sunrgbd/sunrgbd_pc_bbox_votes_80k_v2_%s' % (split_set))
self.raw_data_path = os.path.join(ROOT_DIR, 'sunrgbd/sunrgbd_trainval')
self.scan_names = sorted(list(set([os.path.basename(x)[0:6] \
for x in os.listdir(self.data_path)])))
if scan_idx_list is not None:
self.scan_names = [self.scan_names[i] for i in scan_idx_list]
self.num_points = num_points
self.augment = augment
self.use_color = use_color
self.use_height = use_height
def __len__(self):
return len(self.scan_names)
def __getitem__(self, idx):
"""
Returns a dict with following keys:
point_clouds: (N,3+C) #ST: C is the RGB and/or height
center_label: (MAX_NUM_OBJ,3) for GT box center XYZ
heading_class_label: (MAX_NUM_OBJ,) with int values in 0,...,NUM_HEADING_BIN-1
heading_residual_label: (MAX_NUM_OBJ,)
size_classe_label: (MAX_NUM_OBJ,) with int values in 0,...,NUM_SIZE_CLUSTER
size_residual_label: (MAX_NUM_OBJ,3)
sem_cls_label: (MAX_NUM_OBJ,) semantic class index
box_label_mask: (MAX_NUM_OBJ) as 0/1 with 1 indicating a unique box
vote_label: (N,9) with votes XYZ (3 votes: X1Y1Z1, X2Y2Z2, X3Y3Z3)
if there is only one vote than X1==X2==X3 etc.
vote_label_mask: (N,) with 0/1 with 1 indicating the point
is in one of the object's OBB.
scan_idx: int scan index in scan_names list
max_gt_bboxes: unused
"""
scan_name = self.scan_names[idx]
point_cloud = np.load(os.path.join(self.data_path, scan_name) + '_pc.npz')['pc'] # Nx6
bboxes = np.load(os.path.join(self.data_path, scan_name) + '_bbox.npy') # K,8 centroids (cx,cy,cz), dimension (l,w,h), heanding_angle and semantic_class
calib = np.load(os.path.join(self.data_path, scan_name) + '_calib.npy')
img = sunrgbd_utils.load_image(os.path.join(self.data_path, scan_name) + '_img.jpg')
d_img = sunrgbd_utils.load_depth_image(os.path.join(self.data_path, scan_name) + '_depth_img.png')
if not self.use_color:
point_cloud = point_cloud[:, 0:3]
else:
point_cloud = point_cloud[:, 0:6]
point_cloud[:, 3:] = (point_cloud[:, 3:] - MEAN_COLOR_RGB)
if self.use_height:
floor_height = np.percentile(point_cloud[:, 2], 0.99)
height = point_cloud[:, 2] - floor_height
point_cloud = np.concatenate([point_cloud, np.expand_dims(height, 1)], 1) # (N,4) or (N,7)
# ------------------------------- LABELS ------------------------------
box3d_centers = np.zeros((MAX_NUM_OBJ, 3))
box3d_sizes = np.zeros((MAX_NUM_OBJ, 3)) #ST: L, W, H
label_mask = np.zeros((MAX_NUM_OBJ))
label_mask[0:bboxes.shape[0]] = 1 #ST: mark first K objects only used
max_bboxes = np.zeros((MAX_NUM_OBJ, 8))
max_bboxes[0:bboxes.shape[0], :] = bboxes
for i in range(bboxes.shape[0]):
bbox = bboxes[i]
semantic_class = bbox[7]
box3d_center = bbox[0:3]
# NOTE: The mean size stored in size2class is of full length of box edges,
# while in sunrgbd_data.py data dumping we dumped *half* length l,w,h.. so have to time it by 2 here
box3d_size = bbox[3:6] * 2
box3d_centers[i, :] = box3d_center
box3d_sizes[i, :] = box3d_size
target_bboxes_mask = label_mask
target_bboxes = np.zeros((MAX_NUM_OBJ, 6))
for i in range(bboxes.shape[0]):
bbox = bboxes[i]
corners_3d = sunrgbd_utils.my_compute_box_3d(bbox[0:3], bbox[3:6], bbox[6])
# compute axis aligned box
xmin = np.min(corners_3d[:, 0])
ymin = np.min(corners_3d[:, 1])
zmin = np.min(corners_3d[:, 2])
xmax = np.max(corners_3d[:, 0])
ymax = | np.max(corners_3d[:, 1]) | numpy.max |
# Databricks notebook source
# MAGIC %md
# MAGIC
# MAGIC # [SDS-2.2, Scalable Data Science](https://lamastex.github.io/scalable-data-science/sds/2/2/)
# MAGIC
# MAGIC This is used in a non-profit educational setting with kind permission of [<NAME>](https://www.linkedin.com/in/adbreind).
# MAGIC This is not licensed by Adam for use in a for-profit setting. Please contact Adam directly at `<EMAIL>` to request or report such use cases or abuses.
# MAGIC A few minor modifications and additional mathematical statistical pointers have been added by <NAME> when teaching PhD students in Uppsala University.
# MAGIC
# MAGIC Please feel free to refer to basic concepts here:
# MAGIC
# MAGIC * Udacity's course on Deep Learning [https://www.udacity.com/course/deep-learning--ud730](https://www.udacity.com/course/deep-learning--ud730) by Google engineers: <NAME> and <NAME> and their full video playlist:
# MAGIC * [https://www.youtube.com/watch?v=X_B9NADf2wk&index=2&list=PLAwxTw4SYaPn_OWPFT9ulXLuQrImzHfOV](https://www.youtube.com/watch?v=X_B9NADf2wk&index=2&list=PLAwxTw4SYaPn_OWPFT9ulXLuQrImzHfOV)
# COMMAND ----------
# MAGIC %md
# MAGIC Archived YouTube video of this live unedited lab-lecture:
# MAGIC
# MAGIC [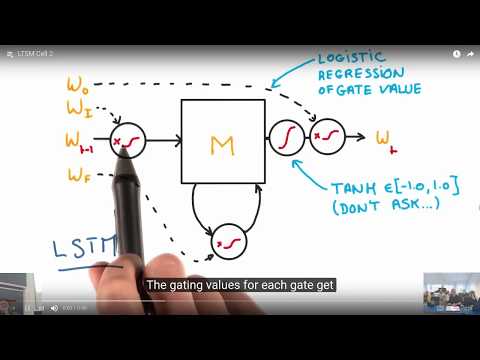](https://www.youtube.com/embed/nHYXMZAHM1c?start=0&end=2465&autoplay=1) [](https://www.youtube.com/embed/4cD8ieyHVh4?start=0&end=2353&autoplay=1)
# COMMAND ----------
# MAGIC %md
# MAGIC # Entering the 4th Dimension
# MAGIC ## Networks for Understanding Time-Oriented Patterns in Data
# MAGIC
# MAGIC Common time-based problems include
# MAGIC * Sequence modeling: "What comes next?"
# MAGIC * Likely next letter, word, phrase, category, cound, action, value
# MAGIC * Sequence-to-Sequence modeling: "What alternative sequence is a pattern match?" (i.e., similar probability distribution)
# MAGIC * Machine translation, text-to-speech/speech-to-text, connected handwriting (specific scripts)
# MAGIC
# MAGIC <img src="http://i.imgur.com/tnxf9gV.jpg">
# COMMAND ----------
# MAGIC %md
# MAGIC ### Simplified Approaches
# MAGIC
# MAGIC * If we know all of the sequence states and the probabilities of state transition...
# MAGIC * ... then we have a simple Markov Chain model.
# MAGIC
# MAGIC * If we *don't* know all of the states or probabilities (yet) but can make constraining assumptions and acquire solid information from observing (sampling) them...
# MAGIC * ... we can use a Hidden Markov Model approach.
# MAGIC
# MAGIC These approached have only limited capacity because they are effectively stateless and so have some degree of "extreme retrograde amnesia."
# MAGIC
# MAGIC ### Can we use a neural network to learn the "next" record in a sequence?
# MAGIC
# MAGIC First approach, using what we already know, might look like
# MAGIC * Clamp input sequence to a vector of neurons in a feed-forward network
# MAGIC * Learn a model on the class of the next input record
# MAGIC
# MAGIC Let's try it! This can work in some situations, although it's more of a setup and starting point for our next development.
# COMMAND ----------
# MAGIC %md
# MAGIC We will make up a simple example of the English alphabet sequence wehere we try to predict the next alphabet from a sequence of length 3.
# COMMAND ----------
alphabet = "ABCDEFGHIJKLMNOPQRSTUVWXYZ"
char_to_int = dict((c, i) for i, c in enumerate(alphabet))
int_to_char = dict((i, c) for i, c in enumerate(alphabet))
seq_length = 3
dataX = []
dataY = []
for i in range(0, len(alphabet) - seq_length, 1):
seq_in = alphabet[i:i + seq_length]
seq_out = alphabet[i + seq_length]
dataX.append([char_to_int[char] for char in seq_in])
dataY.append(char_to_int[seq_out])
print (seq_in, '->', seq_out)
# COMMAND ----------
# dataX is just a reindexing of the alphabets in consecutive triplets of numbers
dataX
# COMMAND ----------
dataY # just a reindexing of the following alphabet after each consecutive triplet of numbers
# COMMAND ----------
# MAGIC %md
# MAGIC Train a network on that data:
# COMMAND ----------
import numpy
from keras.models import Sequential
from keras.layers import Dense
from keras.layers import LSTM # <- this is the Long-Short-term memory layer
from keras.utils import np_utils
# begin data generation ------------------------------------------
# this is just a repeat of what we did above
alphabet = "ABCDEFGHIJKLMNOPQRSTUVWXYZ"
char_to_int = dict((c, i) for i, c in enumerate(alphabet))
int_to_char = dict((i, c) for i, c in enumerate(alphabet))
seq_length = 3
dataX = []
dataY = []
for i in range(0, len(alphabet) - seq_length, 1):
seq_in = alphabet[i:i + seq_length]
seq_out = alphabet[i + seq_length]
dataX.append([char_to_int[char] for char in seq_in])
dataY.append(char_to_int[seq_out])
print (seq_in, '->', seq_out)
# end data generation ---------------------------------------------
X = numpy.reshape(dataX, (len(dataX), seq_length))
X = X / float(len(alphabet)) # normalize the mapping of alphabets from integers into [0, 1]
y = np_utils.to_categorical(dataY) # make the output we want to predict to be categorical
# keras architecturing of a feed forward dense or fully connected Neural Network
model = Sequential()
# draw the architecture of the network given by next two lines, hint: X.shape[1] = 3, y.shape[1] = 26
model.add(Dense(30, input_dim=X.shape[1], kernel_initializer='normal', activation='relu'))
model.add(Dense(y.shape[1], activation='softmax'))
# keras compiling and fitting
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
model.fit(X, y, epochs=1000, batch_size=5, verbose=2)
scores = model.evaluate(X, y)
print("Model Accuracy: %.2f " % scores[1])
for pattern in dataX:
x = numpy.reshape(pattern, (1, len(pattern)))
x = x / float(len(alphabet))
prediction = model.predict(x, verbose=0) # get prediction from fitted model
index = numpy.argmax(prediction)
result = int_to_char[index]
seq_in = [int_to_char[value] for value in pattern]
print (seq_in, "->", result) # print the predicted outputs
# COMMAND ----------
X.shape[1], y.shape[1] # get a sense of the shapes to understand the network architecture
# COMMAND ----------
# MAGIC %md
# MAGIC The network does learn, and could be trained to get a good accuracy. But what's really going on here?
# MAGIC
# MAGIC Let's leave aside for a moment the simplistic training data (one fun experiment would be to create corrupted sequences and augment the data with those, forcing the network to pay attention to the whole sequence).
# MAGIC
# MAGIC Because the model is fundamentally symmetric and stateless (in terms of the sequence; naturally it has weights), this model would need to learn every sequential feature relative to every single sequence position. That seems difficult, inflexible, and inefficient.
# MAGIC
# MAGIC Maybe we could add layers, neurons, and extra connections to mitigate parts of the problem. We could also do things like a 1D convolution to pick up frequencies and some patterns.
# MAGIC
# MAGIC But instead, it might make more sense to explicitly model the sequential nature of the data (a bit like how we explictly modeled the 2D nature of image data with CNNs).
# COMMAND ----------
# MAGIC %md
# MAGIC ## Recurrent Neural Network Concept
# MAGIC
# MAGIC __Let's take the neuron's output from one time (t) and feed it into that same neuron at a later time (t+1), in combination with other relevant inputs. Then we would have a neuron with memory.__
# MAGIC
# MAGIC We can weight the "return" of that value and train the weight -- so the neuron learns how important the previous value is relative to the current one.
# MAGIC
# MAGIC Different neurons might learn to "remember" different amounts of prior history.
# MAGIC
# MAGIC This concept is called a *Recurrent Neural Network*, originally developed around the 1980s.
# MAGIC
# MAGIC Let's recall some pointers from the crash intro to Deep learning.
# MAGIC
# MAGIC ### Watch following videos now for 12 minutes for the fastest introduction to RNNs and LSTMs
# MAGIC
# MAGIC [Udacity: Deep Learning by <NAME> - Recurrent Neural network](https://youtu.be/LTbLxm6YIjE?list=PLAwxTw4SYaPn_OWPFT9ulXLuQrImzHfOV)
# MAGIC
# MAGIC #### Recurrent neural network
# MAGIC 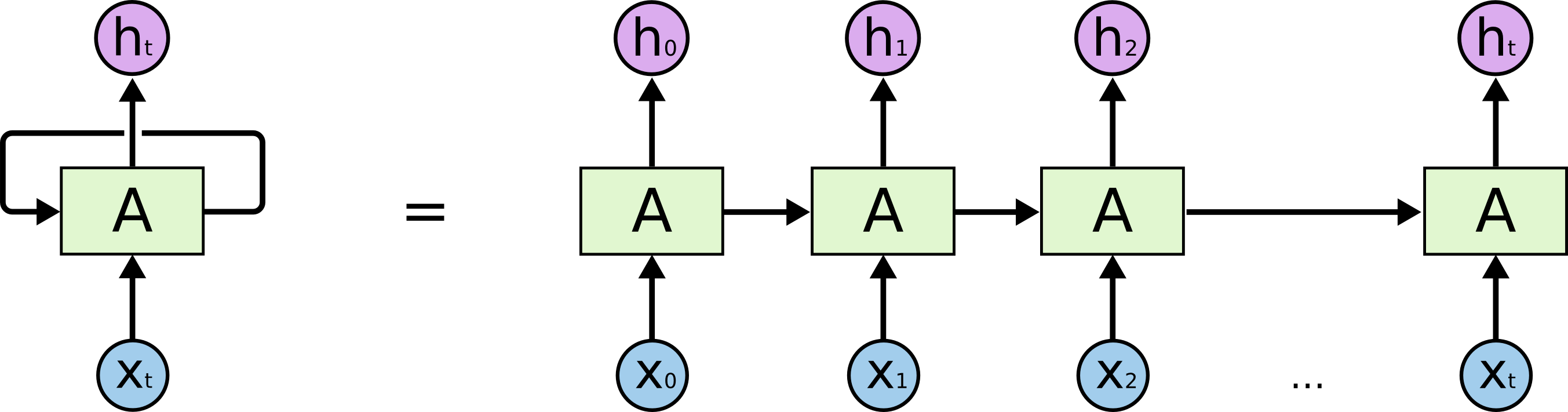
# MAGIC [http://colah.github.io/posts/2015-08-Understanding-LSTMs/](http://colah.github.io/posts/2015-08-Understanding-LSTMs/)
# MAGIC
# MAGIC
# MAGIC [http://karpathy.github.io/2015/05/21/rnn-effectiveness/](http://karpathy.github.io/2015/05/21/rnn-effectiveness/)
# MAGIC ***
# MAGIC
# MAGIC ***
# MAGIC ##### LSTM - Long short term memory
# MAGIC 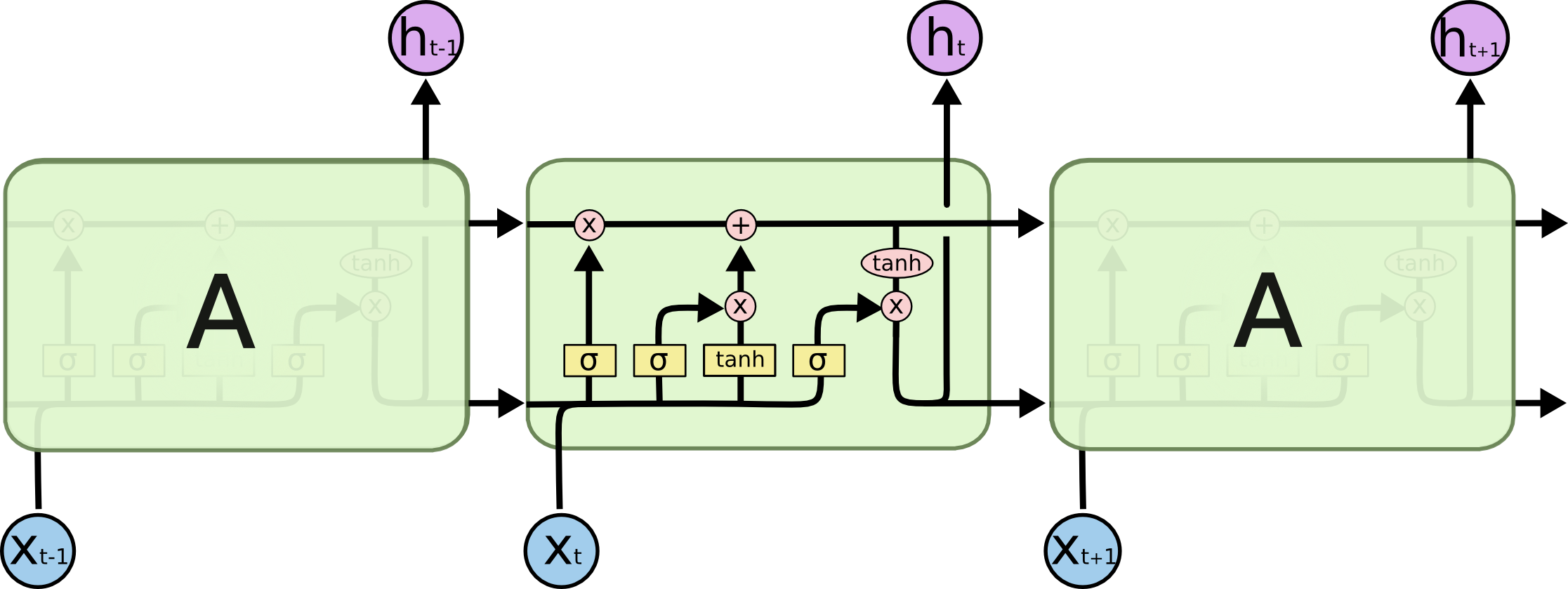
# MAGIC
# MAGIC ***
# MAGIC ##### GRU - Gated recurrent unit
# MAGIC 
# MAGIC [http://arxiv.org/pdf/1406.1078v3.pdf](http://arxiv.org/pdf/1406.1078v3.pdf)
# MAGIC
# MAGIC
# MAGIC ### Training a Recurrent Neural Network
# MAGIC
# MAGIC <img src="http://i.imgur.com/iPGNMvZ.jpg">
# MAGIC
# MAGIC We can train an RNN using backpropagation with a minor twist: since RNN neurons with different states over time can be "unrolled" (i.e., are analogous) to a sequence of neurons with the "remember" weight linking directly forward from (t) to (t+1), we can backpropagate through time as well as the physical layers of the network.
# MAGIC
# MAGIC This is, in fact, called __Backpropagation Through Time__ (BPTT)
# MAGIC
# MAGIC The idea is sound but -- since it creates patterns similar to very deep networks -- it suffers from the same challenges:
# MAGIC * Vanishing gradient
# MAGIC * Exploding gradient
# MAGIC * Saturation
# MAGIC * etc.
# MAGIC
# MAGIC i.e., many of the same problems with early deep feed-forward networks having lots of weights.
# MAGIC
# MAGIC 10 steps back in time for a single layer is a not as bad as 10 layers (since there are fewer connections and, hence, weights) but it does get expensive.
# MAGIC
# MAGIC ---
# MAGIC
# MAGIC > __ASIDE: Hierarchical and Recursive Networks, Bidirectional RNN__
# MAGIC
# MAGIC > Network topologies can be built to reflect the relative structure of the data we are modeling. E.g., for natural language, grammar constraints mean that both hierarchy and (limited) recursion may allow a physically smaller model to achieve more effective capacity.
# MAGIC
# MAGIC > A bi-directional RNN includes values from previous and subsequent time steps. This is less strange than it sounds at first: after all, in many problems, such as sentence translation (where BiRNNs are very popular) we usually have the entire source sequence at one time. In that case, a BiDiRNN is really just saying that both prior and subsequent words can influence the interpretation of each word, something we humans take for granted.
# MAGIC
# MAGIC > Recent versions of neural net libraries have support for bidirectional networks, although you may need to write (or locate) a little code yourself if you want to experiment with hierarchical networks.
# MAGIC
# MAGIC ---
# MAGIC
# MAGIC ## Long Short-Term Memory (LSTM)
# MAGIC
# MAGIC "Pure" RNNs were never very successful. <NAME> and <NAME> (1997) made a game-changing contribution with the publication of the Long Short-Term Memory unit. How game changing? It's effectively state of the art today.
# MAGIC
# MAGIC <sup>(Credit and much thanks to <NAME>, http://colah.github.io/about.html, Research Scientist at Google Brain, for publishing the following excellent diagrams!)</sup>
# MAGIC
# MAGIC *In the following diagrams, pay close attention that the output value is "split" for graphical purposes -- so the two *h* arrows/signals coming out are the same signal.*
# MAGIC
# MAGIC __RNN Cell:__
# MAGIC <img src="http://i.imgur.com/DfYyKaN.png" width=600>
# MAGIC
# MAGIC __LSTM Cell:__
# MAGIC
# MAGIC <img src="http://i.imgur.com/pQiMLjG.png" width=600>
# MAGIC
# MAGIC
# MAGIC An LSTM unit is a neuron with some bonus features:
# MAGIC * Cell state propagated across time
# MAGIC * Input, Output, Forget gates
# MAGIC * Learns retention/discard of cell state
# MAGIC * Admixture of new data
# MAGIC * Output partly distinct from state
# MAGIC * Use of __addition__ (not multiplication) to combine input and cell state allows state to propagate unimpeded across time (addition of gradient)
# MAGIC
# MAGIC ---
# MAGIC
# MAGIC > __ASIDE: Variations on LSTM__
# MAGIC
# MAGIC > ... include "peephole" where gate functions have direct access to cell state; convolutional; and bidirectional, where we can "cheat" by letting neurons learn from future time steps and not just previous time steps.
# MAGIC
# MAGIC ___
# MAGIC
# MAGIC Slow down ... exactly what's getting added to where? For a step-by-step walk through, read <NAME>'s full post http://colah.github.io/posts/2015-08-Understanding-LSTMs/
# MAGIC
# MAGIC
# MAGIC ### Do LSTMs Work Reasonably Well?
# MAGIC
# MAGIC __Yes!__ These architectures are in production (2017) for deep-learning-enabled products at Baidu, Google, Microsoft, Apple, and elsewhere. They are used to solve problems in time series analysis, speech recognition and generation, connected handwriting, grammar, music, and robot control systems.
# MAGIC
# MAGIC ### Let's Code an LSTM Variant of our Sequence Lab
# MAGIC
# MAGIC (this great demo example courtesy of <NAME>: http://machinelearningmastery.com/understanding-stateful-lstm-recurrent-neural-networks-python-keras/)
# COMMAND ----------
import numpy
from keras.models import Sequential
from keras.layers import Dense
from keras.layers import LSTM
from keras.utils import np_utils
alphabet = "ABCDEFGHIJKLMNOPQRSTUVWXYZ"
char_to_int = dict((c, i) for i, c in enumerate(alphabet))
int_to_char = dict((i, c) for i, c in enumerate(alphabet))
seq_length = 3
dataX = []
dataY = []
for i in range(0, len(alphabet) - seq_length, 1):
seq_in = alphabet[i:i + seq_length]
seq_out = alphabet[i + seq_length]
dataX.append([char_to_int[char] for char in seq_in])
dataY.append(char_to_int[seq_out])
print (seq_in, '->', seq_out)
# reshape X to be .......[samples, time steps, features]
X = numpy.reshape(dataX, (len(dataX), seq_length, 1))
X = X / float(len(alphabet))
y = np_utils.to_categorical(dataY)
# Let’s define an LSTM network with 32 units and an output layer with a softmax activation function for making predictions.
# a naive implementation of LSTM
model = Sequential()
model.add(LSTM(32, input_shape=(X.shape[1], X.shape[2]))) # <- LSTM layer...
model.add(Dense(y.shape[1], activation='softmax'))
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
model.fit(X, y, epochs=400, batch_size=1, verbose=2)
scores = model.evaluate(X, y)
print("Model Accuracy: %.2f%%" % (scores[1]*100))
for pattern in dataX:
x = numpy.reshape(pattern, (1, len(pattern), 1))
x = x / float(len(alphabet))
prediction = model.predict(x, verbose=0)
index = numpy.argmax(prediction)
result = int_to_char[index]
seq_in = [int_to_char[value] for value in pattern]
print (seq_in, "->", result)
# COMMAND ----------
(X.shape[1], X.shape[2]) # the input shape to LSTM layer with 32 neurons is given by dimensions of time-steps and features
# COMMAND ----------
X.shape[0], y.shape[1] # number of examples and number of categorical outputs
# COMMAND ----------
# MAGIC %md
# MAGIC __Memory and context__
# MAGIC
# MAGIC If this network is learning the way we would like, it should be robust to noise and also understand the relative context (in this case, where a prior letter occurs in the sequence).
# MAGIC
# MAGIC I.e., we should be able to give it corrupted sequences, and it should produce reasonably correct predictions.
# MAGIC
# MAGIC Make the following change to the code to test this out:
# MAGIC
# MAGIC ## You Try!
# MAGIC * We'll use "W" for our erroneous/corrupted data element
# MAGIC * Add code at the end to predict on the following sequences:
# MAGIC * 'WBC', 'WKL', 'WTU', 'DWF', 'MWO', 'VWW', 'GHW', 'JKW', 'PQW'
# MAGIC * Notice any pattern? Hard to tell from a small sample, but if you play with it (trying sequences from different places in the alphabet, or different "corruption" letters, you'll notice patterns that give a hint at what the network is learning
# MAGIC
# MAGIC The solution is in `060_DLByABr_05a-LSTM-Solution` if you are lazy right now or get stuck.
# MAGIC
# MAGIC __Pretty cool... BUT__
# MAGIC
# MAGIC This alphabet example does seem a bit like "tennis without the net" since the original goal was to develop networks that could extract patterns from complex, ambiguous content like natural language or music, and we've been playing with a sequence (Roman alphabet) that is 100% deterministic and tiny in size.
# MAGIC
# MAGIC First, go ahead and start `061_DLByABr_05b-LSTM-Language` since it will take several minutes to produce its first output.
# MAGIC
# MAGIC This latter script is taken 100% exactly as-is from the Keras library examples folder (https://github.com/fchollet/keras/blob/master/examples/lstm_text_generation.py) and uses precisely the logic we just learned, in order to learn and synthesize English language text from a single-author corpuse. The amazing thing is that the text is learned and generated one letter at a time, just like we did with the alphabet.
# MAGIC
# MAGIC Compared to our earlier examples...
# MAGIC * there is a minor difference in the way the inputs are encoded, using 1-hot vectors
# MAGIC * and there is a *significant* difference in the way the outputs (predictions) are generated: instead of taking just the most likely output class (character) via argmax as we did before, this time we are treating the output as a distribution and sampling from the distribution.
# MAGIC
# MAGIC Let's take a look at the code ... but even so, this will probably be something to come back to after fika or a long break, as the training takes about 5 minutes per epoch (late 2013 MBP CPU) and we need around 20 epochs (80 minutes!) to get good output.
# COMMAND ----------
import sys
sys.exit(0) #just to keep from accidentally running this code (that is already in 061_DLByABr_05b-LSTM-Language) HERE
'''Example script to generate text from Nietzsche's writings.
At least 20 epochs are required before the generated text
starts sounding coherent.
It is recommended to run this script on GPU, as recurrent
networks are quite computationally intensive.
If you try this script on new data, make sure your corpus
has at least ~100k characters. ~1M is better.
'''
from keras.models import Sequential
from keras.layers import Dense, Activation
from keras.layers import LSTM
from keras.optimizers import RMSprop
from keras.utils.data_utils import get_file
import numpy as np
import random
import sys
path = "../data/nietzsche.txt"
text = open(path).read().lower()
print('corpus length:', len(text))
chars = sorted(list(set(text)))
print('total chars:', len(chars))
char_indices = dict((c, i) for i, c in enumerate(chars))
indices_char = dict((i, c) for i, c in enumerate(chars))
# cut the text in semi-redundant sequences of maxlen characters
maxlen = 40
step = 3
sentences = []
next_chars = []
for i in range(0, len(text) - maxlen, step):
sentences.append(text[i: i + maxlen])
next_chars.append(text[i + maxlen])
print('nb sequences:', len(sentences))
print('Vectorization...')
X = np.zeros((len(sentences), maxlen, len(chars)), dtype=np.bool)
y = np.zeros((len(sentences), len(chars)), dtype=np.bool)
for i, sentence in enumerate(sentences):
for t, char in enumerate(sentence):
X[i, t, char_indices[char]] = 1
y[i, char_indices[next_chars[i]]] = 1
# build the model: a single LSTM
print('Build model...')
model = Sequential()
model.add(LSTM(128, input_shape=(maxlen, len(chars))))
model.add(Dense(len(chars)))
model.add(Activation('softmax'))
optimizer = RMSprop(lr=0.01)
model.compile(loss='categorical_crossentropy', optimizer=optimizer)
def sample(preds, temperature=1.0):
# helper function to sample an index from a probability array
preds = np.asarray(preds).astype('float64')
preds = | np.log(preds) | numpy.log |
# Copyright (C) 2018-2021 Intel Corporation
# SPDX-License-Identifier: Apache-2.0
import unittest
import numpy as np
from mo.front.common.partial_infer.elemental import copy_shape_infer
from mo.front.common.partial_infer.eltwise import eltwise_infer
from mo.middle.passes.fusing.resnet_optimization import stride_optimization
from mo.ops.convolution import Convolution
from mo.ops.pooling import Pooling
from mo.utils.ir_engine.compare_graphs import compare_graphs
from mo.utils.unittest.graph import build_graph
max_elt_lambda = lambda node: eltwise_infer(node, lambda a, b: np.maximum(a, b))
nodes_attributes = {
# Placeholders
'placeholder_1': {'shape': None, 'type': 'Parameter', 'kind': 'op', 'op': 'Parameter'},
'placeholder_1_data': {'value': None, 'shape': None, 'kind': 'data', 'data_type': None},
# Concat1 operation
'eltwise_1': {'type': 'Maximum', 'kind': 'op', 'op': 'Maximum', 'infer': max_elt_lambda},
'eltwise_1_data': {'name': 'eltwise_1_data', 'value': None, 'shape': None, 'kind': 'data'},
# Convolutions
'conv_1': {'type': 'Convolution', 'kind': 'op', 'op': 'Conv2D', 'layout': 'NCHW',
'output_spatial_shape': None, 'output_shape': None, 'bias_term': True, 'group': 1,
'spatial_dims': np.array([2, 3]),
'channel_dims': np.array([1]), 'pad_spatial_shape': np.array([[0, 0], [0, 0]]),
'dilation': np.array([1, 1, 1, 1]),
'batch_dims': np.array([0]), 'infer': Convolution.infer,
'kernel_spatial_idx': np.array([2, 3], dtype=np.int64), 'input_feature_channel': 1,
'output_feature_channel': 0, },
'conv_1_w': {'value': None, 'shape': None, 'kind': 'data',
'dim_attrs': ['spatial_dims', 'channel_dims', 'batch_dims', 'axis']},
'conv_1_b': {'value': None, 'shape': None, 'kind': 'data'},
'conv_1_data': {'value': None, 'shape': None, 'kind': 'data'},
'conv_2': {'type': 'Convolution', 'kind': 'op', 'op': 'Conv2D', 'layout': 'NCHW',
'output_spatial_shape': None, 'output_shape': None, 'bias_term': True, 'group': 1,
'spatial_dims': np.array([2, 3]),
'channel_dims': np.array([1]), 'pad_spatial_shape': np.array([[0, 0], [0, 0]]),
'dilation': np.array([1, 1, 1, 1]),
'batch_dims': np.array([0]), 'infer': Convolution.infer,
'kernel_spatial_idx': np.array([2, 3], dtype=np.int64), 'input_feature_channel': 1,
'output_feature_channel': 0, },
'conv_2_w': {'value': None, 'shape': None, 'kind': 'data',
'dim_attrs': ['spatial_dims', 'channel_dims', 'batch_dims', 'axis']},
'conv_2_b': {'value': None, 'shape': None, 'kind': 'data'},
'conv_2_data': {'value': None, 'shape': None, 'kind': 'data'},
'conv_3': {'type': 'Convolution', 'kind': 'op', 'op': 'Conv2D', 'layout': 'NCHW',
'output_spatial_shape': None, 'output_shape': None, 'bias_term': True, 'group': 1,
'spatial_dims': np.array([2, 3]),
'channel_dims': np.array([1]), 'pad_spatial_shape': np.array([[0, 0], [0, 0]]),
'dilation': np.array([1, 1, 1, 1]),
'batch_dims': np.array([0]), 'infer': Convolution.infer,
'kernel_spatial_idx': np.array([2, 3], dtype=np.int64), 'input_feature_channel': 1,
'output_feature_channel': 0, },
'conv_3_w': {'value': None, 'shape': None, 'kind': 'data',
'dim_attrs': ['spatial_dims', 'channel_dims', 'batch_dims', 'axis']},
'conv_3_b': {'value': None, 'shape': None, 'kind': 'data'},
'conv_3_data': {'value': None, 'shape': None, 'kind': 'data'},
'conv_4': {'type': 'Convolution', 'kind': 'op', 'op': 'Conv2D', 'layout': 'NCHW',
'output_spatial_shape': None, 'output_shape': None, 'bias_term': True, 'group': 1,
'spatial_dims': np.array([2, 3]),
'channel_dims': np.array([1]), 'pad_spatial_shape': np.array([[0, 0], [0, 0]]),
'dilation': np.array([1, 1, 1, 1]),
'batch_dims': np.array([0]), 'infer': Convolution.infer,
'kernel_spatial_idx': np.array([2, 3], dtype=np.int64), 'input_feature_channel': 1,
'output_feature_channel': 0, },
'conv_4_w': {'value': None, 'shape': None, 'kind': 'data',
'dim_attrs': ['spatial_dims', 'channel_dims', 'batch_dims', 'axis']},
'conv_4_b': {'value': None, 'shape': None, 'kind': 'data'},
'conv_4_data': {'value': None, 'shape': None, 'kind': 'data'},
'conv_5': {'type': 'Convolution', 'kind': 'op', 'op': 'Conv2D', 'layout': 'NCHW',
'output_spatial_shape': None, 'output_shape': None, 'bias_term': True, 'group': 1,
'spatial_dims': np.array([2, 3]),
'channel_dims': np.array([1]), 'pad_spatial_shape': np.array([[0, 0], [0, 0]]),
'dilation': np.array([1, 1, 1, 1]),
'batch_dims': np.array([0]), 'infer': Convolution.infer,
'kernel_spatial_idx': np.array([2, 3], dtype=np.int64), 'input_feature_channel': 1,
'output_feature_channel': 0, },
'conv_5_w': {'value': None, 'shape': None, 'kind': 'data',
'dim_attrs': ['spatial_dims', 'channel_dims', 'batch_dims', 'axis']},
'conv_5_b': {'value': None, 'shape': None, 'kind': 'data'},
'conv_5_data': {'value': None, 'shape': None, 'kind': 'data'},
# ReLU
'relu_1': {'shape': None, 'type': 'ReLU', 'kind': 'op', 'op': 'ReLU', 'infer': copy_shape_infer},
'relu_1_data': {'value': None, 'shape': None, 'kind': 'data', 'data_type': None},
'relu_2': {'shape': None, 'type': 'ReLU', 'kind': 'op', 'op': 'ReLU', 'infer': copy_shape_infer},
'relu_2_data': {'value': None, 'shape': None, 'kind': 'data', 'data_type': None},
'relu_3': {'shape': None, 'type': 'ReLU', 'kind': 'op', 'op': 'ReLU', 'infer': copy_shape_infer},
'relu_3_data': {'value': None, 'shape': None, 'kind': 'data', 'data_type': None},
# Pooling
'pool_1': {'type': 'Pooling', 'kind': 'op', 'op': 'Pooling',
'spatial_dims': np.array([2, 3]),
'pad_spatial_shape': np.array([[0, 0], [0, 0]]),
'infer': Pooling.infer},
'pool_1_data': {'value': None, 'shape': None, 'kind': 'data'},
}
# In description of unit tests below will be used next syntax: Operation(NxM,XxY), where NxM - kernel size, XxY - stride
class ResnetOptimizationTests(unittest.TestCase):
# Pl->Conv(1x1,1x1)->Conv(1x1,2x2) => Pl->Conv(1x1,2x2)->Conv(1x1,1x1)
def test_resnet_optimization_1(self):
graph = build_graph(nodes_attributes,
[('placeholder_1', 'placeholder_1_data'),
('placeholder_1_data', 'conv_1'),
('conv_1_w', 'conv_1'),
('conv_1_b', 'conv_1'),
('conv_1', 'conv_1_data'),
('conv_1_data', 'conv_2'),
('conv_2_w', 'conv_2'),
('conv_2_b', 'conv_2'),
('conv_2', 'conv_2_data'),
],
{'placeholder_1_data': {'shape': np.array([1, 3, 224, 224])},
'conv_1_w': {'value': np.zeros([3, 3, 1, 1]), 'shape': np.array([3, 3, 1, 1])},
'conv_1': {'kernel_spatial': np.array([1, 1]),
'stride': np.array([1, 1, 1, 1]),
'output': np.array([3]), },
'conv_1_data': {'shape': np.array([1, 3, 224, 224])},
'conv_2_w': {'value': np.zeros([3, 3, 1, 1]), 'shape': np.array([3, 3, 1, 1])},
'conv_2': {'kernel_spatial': np.array([1, 1]),
'stride': np.array([1, 1, 2, 2]),
'output': np.array([3]), },
'conv_2_data': {'shape': np.array([1, 3, 112, 112])},
},
nodes_with_edges_only=True)
graph_ref = build_graph(nodes_attributes,
[('placeholder_1', 'placeholder_1_data'),
('placeholder_1_data', 'conv_1'),
('conv_1_w', 'conv_1'),
('conv_1_b', 'conv_1'),
('conv_1', 'conv_1_data'),
('conv_1_data', 'conv_2'),
('conv_2_w', 'conv_2'),
('conv_2_b', 'conv_2'),
('conv_2', 'conv_2_data'),
],
{'placeholder_1_data': {'shape': np.array([1, 3, 224, 224])},
'conv_1_w': {'value': np.zeros([3, 3, 1, 1]), 'shape': np.array([3, 3, 1, 1])},
'conv_1': {'kernel_spatial': np.array([1, 1]),
'stride': np.array([1, 1, 2, 2]),
'output': np.array([3]), },
'conv_1_data': {'shape': np.array([1, 3, 112, 112])},
'conv_2_w': {'value': np.zeros([3, 3, 1, 1]), 'shape': np.array([3, 3, 1, 1])},
'conv_2': {'kernel_spatial': np.array([1, 1]),
'stride': np.array([1, 1, 1, 1]),
'output': np.array([3]), },
'conv_2_data': {'shape': np.array([1, 3, 112, 112])},
},
nodes_with_edges_only=True)
graph.graph['layout'] = 'NCHW'
graph_ref.graph['layout'] = 'NCHW'
stride_optimization(graph)
(flag, resp) = compare_graphs(graph, graph_ref, 'conv_2_data', check_op_attrs=True)
self.assertTrue(flag, resp)
# Pl->Conv(3x3,2x2)->Conv(1x1,2x2) => Pl->Conv(3x3,4x4)->Conv(1x1,1x1)
def test_resnet_optimization_2(self):
graph = build_graph(nodes_attributes,
[('placeholder_1', 'placeholder_1_data'),
('placeholder_1_data', 'conv_1'),
('conv_1_w', 'conv_1'),
('conv_1_b', 'conv_1'),
('conv_1', 'conv_1_data'),
('conv_1_data', 'conv_2'),
('conv_2_w', 'conv_2'),
('conv_2_b', 'conv_2'),
('conv_2', 'conv_2_data'),
],
{'placeholder_1_data': {'shape': np.array([1, 3, 224, 224])},
'conv_1_w': {'value': np.zeros([3, 3, 1, 1]), 'shape': np.array([3, 3, 1, 1])},
'conv_1': {'kernel_spatial': np.array([1, 1]),
'stride': np.array([1, 1, 2, 2]),
'output': np.array([3]), },
'conv_1_data': {'shape': np.array([1, 3, 112, 112])},
'conv_2_w': {'value': np.zeros([3, 3, 1, 1]), 'shape': np.array([3, 3, 1, 1])},
'conv_2': {'kernel_spatial': np.array([1, 1]),
'stride': np.array([1, 1, 2, 2]),
'output': np.array([3]), },
'conv_2_data': {'shape': np.array([1, 3, 56, 56])},
},
nodes_with_edges_only=True)
graph_ref = build_graph(nodes_attributes,
[('placeholder_1', 'placeholder_1_data'),
('placeholder_1_data', 'conv_1'),
('conv_1_w', 'conv_1'),
('conv_1_b', 'conv_1'),
('conv_1', 'conv_1_data'),
('conv_1_data', 'conv_2'),
('conv_2_w', 'conv_2'),
('conv_2_b', 'conv_2'),
('conv_2', 'conv_2_data'),
],
{'placeholder_1_data': {'shape': np.array([1, 3, 224, 224])},
'conv_1_w': {'value': np.zeros([3, 3, 1, 1]), 'shape': np.array([3, 3, 1, 1])},
'conv_1': {'kernel_spatial': np.array([1, 1]),
'stride': np.array([1, 1, 4, 4]),
'output': np.array([3]), },
'conv_1_data': {'shape': np.array([1, 3, 56, 56])},
'conv_2_w': {'value': np.zeros([3, 3, 1, 1]), 'shape': np.array([3, 3, 1, 1])},
'conv_2': {'kernel_spatial': np.array([1, 1]),
'stride': np.array([1, 1, 1, 1]),
'output': np.array([3]), },
'conv_2_data': {'shape': np.array([1, 3, 56, 56])},
},
nodes_with_edges_only=True)
graph.graph['layout'] = 'NCHW'
graph_ref.graph['layout'] = 'NCHW'
stride_optimization(graph)
(flag, resp) = compare_graphs(graph, graph_ref, 'conv_2_data', check_op_attrs=True)
self.assertTrue(flag, resp)
# Pl->Conv(3x3,2x2)->Conv(3x3,2x2) => Same
def test_resnet_optimization_3(self):
graph = build_graph(nodes_attributes,
[('placeholder_1', 'placeholder_1_data'),
('placeholder_1_data', 'conv_1'),
('conv_1_w', 'conv_1'),
('conv_1_b', 'conv_1'),
('conv_1', 'conv_1_data'),
('conv_1_data', 'conv_2'),
('conv_2_w', 'conv_2'),
('conv_2_b', 'conv_2'),
('conv_2', 'conv_2_data'),
],
{'placeholder_1_data': {'shape': np.array([1, 3, 224, 224])},
'conv_1_w': {'value': np.zeros([3, 3, 3, 3]), 'shape': np.array([3, 3, 3, 3])},
'conv_1': {'kernel_spatial': np.array([3, 3]),
'stride': np.array([1, 1, 2, 2]),
'output': np.array([3]), },
'conv_1_data': {'shape': np.array([1, 3, 112, 112])},
'conv_2_w': {'value': np.zeros([3, 3, 3, 3]), 'shape': np.array([3, 3, 3, 3])},
'conv_2': {'kernel_spatial': np.array([3, 3]),
'stride': np.array([1, 1, 2, 2]),
'output': np.array([3]), },
'conv_2_data': {'shape': np.array([1, 3, 56, 56])},
},
nodes_with_edges_only=True)
graph_ref = build_graph(nodes_attributes,
[('placeholder_1', 'placeholder_1_data'),
('placeholder_1_data', 'conv_1'),
('conv_1_w', 'conv_1'),
('conv_1_b', 'conv_1'),
('conv_1', 'conv_1_data'),
('conv_1_data', 'conv_2'),
('conv_2_w', 'conv_2'),
('conv_2_b', 'conv_2'),
('conv_2', 'conv_2_data'),
],
{'placeholder_1_data': {'shape': np.array([1, 3, 224, 224])},
'conv_1_w': {'value': np.zeros([3, 3, 3, 3]), 'shape': np.array([3, 3, 3, 3])},
'conv_1': {'kernel_spatial': np.array([3, 3]),
'stride': np.array([1, 1, 2, 2]),
'output': np.array([3]), },
'conv_1_data': {'shape': np.array([1, 3, 112, 112])},
'conv_2_w': {'value': np.zeros([3, 3, 3, 3]), 'shape': np.array([3, 3, 3, 3])},
'conv_2': {'kernel_spatial': np.array([3, 3]),
'stride': np.array([1, 1, 2, 2]),
'output': np.array([3]), },
'conv_2_data': {'shape': np.array([1, 3, 56, 56])},
},
nodes_with_edges_only=True)
graph.graph['layout'] = 'NCHW'
graph_ref.graph['layout'] = 'NCHW'
stride_optimization(graph)
(flag, resp) = compare_graphs(graph, graph_ref, 'conv_2_data', check_op_attrs=True)
self.assertTrue(flag, resp)
# Pl--->Conv(3x3,2x2)->ReLU--->Eltwise-->Conv(1x1,2x2) => Pl--->Conv(3x3,4x4)->ReLU--->Eltwise-->Conv(1x1,1x1)
# `-->Conv(3x3,2x2)->ReLU---` `-->Conv(3x3,4x4)->ReLU---`
def test_resnet_optimization_4(self):
graph = build_graph(nodes_attributes,
[('placeholder_1', 'placeholder_1_data'),
('placeholder_1_data', 'conv_1'),
('conv_1_w', 'conv_1'),
('conv_1_b', 'conv_1'),
('conv_1', 'conv_1_data'),
('conv_1_data', 'relu_1'),
('relu_1', 'relu_1_data'),
('placeholder_1_data', 'conv_2'),
('conv_2_w', 'conv_2'),
('conv_2_b', 'conv_2'),
('conv_2', 'conv_2_data'),
('conv_2_data', 'relu_2'),
('relu_2', 'relu_2_data'),
('relu_1_data', 'eltwise_1'),
('relu_2_data', 'eltwise_1'),
('eltwise_1', 'eltwise_1_data'),
('eltwise_1_data', 'conv_3'),
('conv_3_w', 'conv_3'),
('conv_3_b', 'conv_3'),
('conv_3', 'conv_3_data'),
],
{'placeholder_1_data': {'shape': np.array([1, 3, 224, 224])},
'conv_1_w': {'value': np.zeros([3, 3, 3, 3]), 'shape': np.array([3, 3, 3, 3])},
'conv_1': {'kernel_spatial': np.array([3, 3]),
'stride': np.array([1, 1, 2, 2]),
'output': np.array([3]), },
'conv_1_data': {'shape': np.array([1, 3, 112, 112])},
'relu_1_data': {'shape': np.array([1, 3, 112, 112])},
'conv_2_w': {'value': np.zeros([3, 3, 3, 3]), 'shape': np.array([3, 3, 3, 3])},
'conv_2': {'kernel_spatial': np.array([3, 3]),
'stride': np.array([1, 1, 2, 2]),
'output': np.array([3]), },
'conv_2_data': {'shape': np.array([1, 3, 112, 112])},
'relu_2_data': {'shape': np.array([1, 3, 112, 112])},
'eltwise_1_data': {'shape': np.array([1, 3, 112, 112])},
'conv_3_w': {'value': np.zeros([3, 3, 1, 1]), 'shape': np.array([3, 3, 1, 1])},
'conv_3': {'kernel_spatial': np.array([1, 1]),
'stride': np.array([1, 1, 2, 2]),
'output': np.array([3]), },
'conv_3_data': {'shape': np.array([1, 3, 56, 56])},
},
nodes_with_edges_only=True)
graph_ref = build_graph(nodes_attributes,
[('placeholder_1', 'placeholder_1_data'),
('placeholder_1_data', 'conv_1'),
('conv_1_w', 'conv_1'),
('conv_1_b', 'conv_1'),
('conv_1', 'conv_1_data'),
('conv_1_data', 'relu_1'),
('relu_1', 'relu_1_data'),
('placeholder_1_data', 'conv_2'),
('conv_2_w', 'conv_2'),
('conv_2_b', 'conv_2'),
('conv_2', 'conv_2_data'),
('conv_2_data', 'relu_2'),
('relu_2', 'relu_2_data'),
('relu_1_data', 'eltwise_1'),
('relu_2_data', 'eltwise_1'),
('eltwise_1', 'eltwise_1_data'),
('eltwise_1_data', 'conv_3'),
('conv_3_w', 'conv_3'),
('conv_3_b', 'conv_3'),
('conv_3', 'conv_3_data'),
],
{'placeholder_1_data': {'shape': np.array([1, 3, 224, 224])},
'conv_1_w': {'value': np.zeros([3, 3, 3, 3]), 'shape': np.array([3, 3, 3, 3])},
'conv_1': {'kernel_spatial': np.array([3, 3]),
'stride': np.array([1, 1, 4, 4]),
'output': np.array([3])},
'conv_1_data': {'shape': np.array([1, 3, 56, 56])},
'relu_1_data': {'shape': np.array([1, 3, 56, 56])},
'conv_2_w': {'value': np.zeros([3, 3, 3, 3]), 'shape': np.array([3, 3, 3, 3])},
'conv_2': {'kernel_spatial': np.array([3, 3]),
'stride': np.array([1, 1, 4, 4]),
'output': | np.array([3]) | numpy.array |
#!/usr/bin/env python3
import os
import pprint
import json
import urllib.parse
import time
import numpy as np # type:ignore
import random
import math
import os
import logging
import wave
import threading
import datetime
import struct
import subprocess
import copy
import sys
import string
from typing import Any, Dict, List, Tuple, Iterable
sys.path.append(os.path.dirname(__file__)) # for finding our files
import util
logging.basicConfig(filename='server.log',level=logging.DEBUG)
# big-endian
# 8 userid: uint64
# 32 name: 32 bytes of utf8, '\0' padded
# 4 mic_volume: float32,
# 4 rms_volume: float32
# 2 delay: uint16
# 1 muted: uint8
BINARY_USER_CONFIG_FORMAT = struct.Struct(">Q32sffHB")
FRAME_SIZE = 128
N_IMAGINARY_USERS = 0 # for debugging user summary + mixing console performance
SUPPORT_SERVER_CONTROL = False
# The maximum number of users to allow to join. This is enforced on a
# best-effort basis by the client. If many people are calibrating at
# the same time this will be exceeded, because we only check before
# calibration.
#
# In stress testing, the server seems to do fine with 61 users, but
# the video call might change that (stress test includes no video).
MAX_USERS = 35 # XXX needs tuning
try:
# Grab these on startup, when they are very very likely to be the actual
# running version.
SERVER_VERSION = subprocess.check_output(
["git", "rev-parse", "--short", "HEAD"]).strip().decode("utf-8")
SERVER_BRANCH = subprocess.check_output(
["git", "rev-parse", "--abbrev-ref", "HEAD"]).strip().decode("utf-8")
except Exception:
SERVER_VERSION="unknown"
SERVER_BRANCH="unknown"
SERVER_STARTUP_TIME = int(time.time())
ENABLE_TWILIO = True
SECRETS_FNAME = "secrets.json"
secrets = {}
if os.path.exists(SECRETS_FNAME):
with open(SECRETS_FNAME) as inf:
secrets = json.loads(inf.read())
else:
ENABLE_TWILIO = False
if ENABLE_TWILIO:
from twilio.jwt.access_token import AccessToken
from twilio.jwt.access_token.grants import VideoGrant
class State():
def __init__(self):
self.reset()
def reset(self):
self.server_controlled = False
self.last_request_clock = None
self.last_cleared_clock = None
self.global_volume = 1.2
self.backing_volume = 1.0
self.song_end_clock = 0
self.song_start_clock = 0
self.requested_track: Any = None
self.bpm = 0
self.repeats = 0
self.bpr = 0
self.leftover_beat_samples = 0
self.first_bucket = DELAY_INTERVAL
self.leader = None
self.backing_track: Any = | np.zeros(0) | numpy.zeros |
import numpy as np
from numba import cuda, int32, float32
from numba.cuda.testing import skip_on_cudasim, unittest, CUDATestCase
from numba.core.config import ENABLE_CUDASIM
def useless_syncthreads(ary):
i = cuda.grid(1)
cuda.syncthreads()
ary[i] = i
def useless_syncwarp(ary):
i = cuda.grid(1)
cuda.syncwarp()
ary[i] = i
def useless_syncwarp_with_mask(ary):
i = cuda.grid(1)
cuda.syncwarp(0xFFFF)
ary[i] = i
def coop_syncwarp(res):
sm = cuda.shared.array(32, int32)
i = cuda.grid(1)
sm[i] = i
cuda.syncwarp()
if i < 16:
sm[i] = sm[i] + sm[i + 16]
cuda.syncwarp(0xFFFF)
if i < 8:
sm[i] = sm[i] + sm[i + 8]
cuda.syncwarp(0xFF)
if i < 4:
sm[i] = sm[i] + sm[i + 4]
cuda.syncwarp(0xF)
if i < 2:
sm[i] = sm[i] + sm[i + 2]
cuda.syncwarp(0x3)
if i == 0:
res[0] = sm[0] + sm[1]
def simple_smem(ary):
N = 100
sm = cuda.shared.array(N, int32)
i = cuda.grid(1)
if i == 0:
for j in range(N):
sm[j] = j
cuda.syncthreads()
ary[i] = sm[i]
def coop_smem2d(ary):
i, j = cuda.grid(2)
sm = cuda.shared.array((10, 20), float32)
sm[i, j] = (i + 1) / (j + 1)
cuda.syncthreads()
ary[i, j] = sm[i, j]
def dyn_shared_memory(ary):
i = cuda.grid(1)
sm = cuda.shared.array(0, float32)
sm[i] = i * 2
cuda.syncthreads()
ary[i] = sm[i]
def use_threadfence(ary):
ary[0] += 123
cuda.threadfence()
ary[0] += 321
def use_threadfence_block(ary):
ary[0] += 123
cuda.threadfence_block()
ary[0] += 321
def use_threadfence_system(ary):
ary[0] += 123
cuda.threadfence_system()
ary[0] += 321
def use_syncthreads_count(ary_in, ary_out):
i = cuda.grid(1)
ary_out[i] = cuda.syncthreads_count(ary_in[i])
def use_syncthreads_and(ary_in, ary_out):
i = cuda.grid(1)
ary_out[i] = cuda.syncthreads_and(ary_in[i])
def use_syncthreads_or(ary_in, ary_out):
i = cuda.grid(1)
ary_out[i] = cuda.syncthreads_or(ary_in[i])
def _safe_cc_check(cc):
if ENABLE_CUDASIM:
return True
else:
return cuda.get_current_device().compute_capability >= cc
class TestCudaSync(CUDATestCase):
def _test_useless(self, kernel):
compiled = cuda.jit("void(int32[::1])")(kernel)
nelem = 10
ary = np.empty(nelem, dtype=np.int32)
exp = | np.arange(nelem, dtype=np.int32) | numpy.arange |
import collections
import dataclasses
import enum
import functools
from copy import deepcopy
from itertools import chain
import deap
import numpy as np
import pyDOE2
from deap.base import Fitness
def listify(fn=None, wrapper=list):
"""
From https://github.com/shazow/unstdlib.py/blob/master/unstdlib/standard/list_.py#L149
A decorator which wraps a function's return value in ``list(...)``.
Useful when an algorithm can be expressed more cleanly as a generator but
the function should return an list.
Example::
>>> @listify
... def get_lengths(iterable):
... for i in iterable:
... yield len(i)
>>> get_lengths(["spam", "eggs"])
[4, 4]
>>>
>>> @listify(wrapper=tuple)
... def get_lengths_tuple(iterable):
... for i in iterable:
... yield len(i)
>>> get_lengths_tuple(["foo", "bar"])
(3, 3)
"""
def listify_return(fn):
@functools.wraps(fn)
def listify_helper(*args, **kw):
return wrapper(fn(*args, **kw))
return listify_helper
if fn is None:
return listify_return
return listify_return(fn)
@dataclasses.dataclass(frozen=True)
class VariableProperties:
discrete: bool
bounded: bool
ordered: bool
class VariableType(VariableProperties, enum.Enum):
CONTINUOUS = (False, True, True)
INTEGER = (True, True, True)
ORDINAL = (True, False, True)
NOMINAL = (True, False, False)
@dataclasses.dataclass(frozen=True)
class ObjectiveValueWithConstraintViolation:
objectives: tuple
constraint_violation: float
def __iter__(self):
yield from self.objectives
class ConstraintDominatedFitness(Fitness):
feasibility_tolerance = 1e-12
def __init__(self, *args, **kwargs):
super().__init__(*args, **kwargs)
self.constraint_violation = None
def __deepcopy__(self, memo):
# The base Fitness class uses an optimized deepcopy that throws away attributes
copy_ = super().__deepcopy__(memo)
copy_.constraint_violation = self.constraint_violation
return copy_
@property
def feasible(self):
return self.valid and self.constraint_violation <= self.feasibility_tolerance
def set_values(self, values):
if isinstance(values, ObjectiveValueWithConstraintViolation):
self.constraint_violation = values.constraint_violation
values = values.objectives
Fitness.setValues(self, values)
def del_values(self):
self.constraint_violation = None
Fitness.delValues(self)
values = property(Fitness.getValues, set_values, del_values)
def dominates(self, other, obj=slice(None)):
if self.feasible and other.feasible:
return super().dominates(other, obj)
else:
return self.constraint_violation < other.constraint_violation
class Individual(list):
def __init__(self, *args, fitness_class, **kwargs):
super().__init__(*args, **kwargs)
self.fitness = fitness_class()
# Metadata
self.generation = None
def __repr__(self):
return f"Individual({super().__repr__()})"
@dataclasses.dataclass(frozen=True)
class IndividualBounds:
lower: tuple
upper: tuple
@classmethod
def from_design_var_meta(cls, design_var_meta):
lower = {
name: (meta["lower"] if meta["type"].bounded else np.zeros(meta["shape"]))
for name, meta in design_var_meta.items()
}
upper = {
name: (
meta["upper"]
if meta["type"].bounded
else np.vectorize(len)(meta["values"]) - 1
)
for name, meta in design_var_meta.items()
}
return cls(
lower=tuple(individual_sequence(lower, design_var_meta)),
upper=tuple(individual_sequence(upper, design_var_meta)),
)
def individual_sequence(design_vars, design_var_meta):
return chain.from_iterable(
np.broadcast_to(design_vars[name], meta["shape"]).flat
for name, meta in design_var_meta.items()
)
def individual_types_sequence(design_var_meta):
return chain.from_iterable(
[meta["type"]] * np.product(meta["shape"] or (1,))
for meta in design_var_meta.values()
)
def stretch_array(array, shape):
try:
return np.broadcast_to(array, shape)
except ValueError:
return np.reshape(array, shape)
missing_dims = len(shape) - array.ndim
indexer = (...,) + (np.newaxis,) * missing_dims
array = array[indexer]
return np.broadcast_to(array, shape)
def random_ints(shape, lower, upper):
ret = np.empty(shape, dtype=np.int)
lower = np.broadcast_to(lower, shape)
upper = np.broadcast_to(upper, shape)
for i in np.ndindex(*shape):
ret[i] = | np.random.randint(lower[i], upper[i], dtype=np.int) | numpy.random.randint |
# Copyright 2020 Google LLC
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# Lint as: python3
"""Functions for running experiments (colabs, xm) and storing/saving data."""
import dataclasses
import os
from typing import (Any, Callable, List, MutableMapping, Optional, Text, Tuple,
Union)
import graph_nets
import more_itertools
import numpy as np
import sonnet as snt
import tensorflow as tf
from graph_attribution import datasets
from graph_attribution import graphnet_models as models
from graph_attribution import graphnet_techniques as techniques
from graph_attribution import graphs as graph_utils
from graph_attribution import tasks, templates
# Typing alias.
GraphsTuple = graph_nets.graphs.GraphsTuple
TransparentModel = templates.TransparentModel
AttributionTechnique = templates.AttributionTechnique
AttributionTask = templates.AttributionTask
NodeEdgeTensors = templates.NodeEdgeTensors
MethodDict = MutableMapping[Text, AttributionTechnique]
OrderedDict = MutableMapping
def set_seed(random_seed: int):
"""Sets initial seed for random numbers."""
tf.random.set_seed(random_seed)
np.random.seed(random_seed)
def get_graph_block(block_type: models.BlockType, node_size: int,
edge_size: int, global_size: int, index: int) -> snt.Module:
"""Gets a GNN block based on enum and sizes."""
name = f'{block_type.name}_{index+1}'
if block_type == models.BlockType.gcn:
return models.GCNLayer(models.get_mlp_fn([node_size] * 2), name=name)
elif block_type == models.BlockType.gat:
return models.SelfAttention(
node_size, models.get_mlp_fn([node_size] * 2))
elif block_type == models.BlockType.mpnn:
return models.NodeEdgeLayer(
models.get_mlp_fn([node_size] * 2),
models.get_mlp_fn([edge_size] * 2),
name=name)
elif block_type == models.BlockType.graphnet:
use_globals = index != 0
return graph_nets.modules.GraphNetwork(
node_model_fn=models.get_mlp_fn([node_size] * 2),
edge_model_fn=models.get_mlp_fn([edge_size] * 2),
global_model_fn=models.get_mlp_fn([global_size] * 2),
edge_block_opt={'use_globals': use_globals},
node_block_opt={'use_globals': use_globals},
global_block_opt={'use_globals': use_globals},
name=name)
else:
raise ValueError(f'block_type={block_type} not implemented')
class GNN(snt.Module, templates.TransparentModel):
"""A general graph neural network for graph property prediction."""
def __init__(self,
node_size: int,
edge_size: int,
global_size: int,
y_output_size: int,
block_type: models.BlockType,
activation: models.Activation,
target_type: templates.TargetType,
n_layers: int = 3):
super(GNN, self).__init__(name=block_type.name)
# Graph encoding step, basic linear mapping.
self.encode = graph_nets.modules.GraphIndependent(
node_model_fn=lambda: snt.Linear(node_size),
edge_model_fn=lambda: snt.Linear(edge_size))
# Message passing steps or GNN blocks.
gnn_layers = [
get_graph_block(
block_type,
node_size,
edge_size,
global_size,
index)
for index in range(0, n_layers)
]
self.gnn = models.SequentialWithActivations(gnn_layers)
if target_type == templates.TargetType.globals:
readout = models.ReadoutGAP(global_size, tf.nn.softmax)
else:
readout = graph_nets.modules.GraphIndependent()
self.readout = readout
self.linear = snt.Linear(y_output_size, with_bias=False)
self.activation = models.cast_activation(activation)
self.pred_layer = snt.Sequential([self.linear, self.activation])
self.block_type = block_type
self.target_type = target_type
def cast_task_batch_index(
self,
task_index: Optional[int] = None,
batch_index: Optional[int] = None) -> Tuple[int, Union[int, slice]]:
"""Provide defaults for task and batch indices when not present."""
task_index = 0 if task_index is None else task_index
batch_index = slice(None) if batch_index is None else batch_index
return task_index, batch_index
@tf.function(experimental_relax_shapes=True)
def get_graph_embedding(self, x: GraphsTuple) -> tf.Tensor:
"""Build a graph embedding."""
out_graph = self.readout(self.gnn(self.encode(x)))
return models.get_graph_attribute(out_graph, self.target_type)
def __call__(self, x: GraphsTuple) -> tf.Tensor:
"""Typical forward pass for the model."""
graph_emb = self.get_graph_embedding(x)
y = self.pred_layer(graph_emb)
return y
def predict(self,
x: GraphsTuple,
task_index: Optional[int] = None,
batch_index: Optional[int] = None) -> tf.Tensor:
"""Forward pass with output set on the task of interest (y[batch_index, task_index])."""
task_index, batch_index = self.cast_task_batch_index(
task_index, batch_index)
return self(x)[batch_index, task_index]
@tf.function(experimental_relax_shapes=True)
def get_gradient(self,
x: GraphsTuple,
task_index: Optional[int] = None,
batch_index: Optional[int] = None) -> NodeEdgeTensors:
"""Gets gradient of inputs wrt to the target."""
with tf.GradientTape(watch_accessed_variables=False) as gtape:
gtape.watch([x.nodes, x.edges])
y = self.predict(x, task_index, batch_index)
nodes_grad, edges_grad = gtape.gradient(y, [x.nodes, x.edges])
return nodes_grad, edges_grad
@tf.function(experimental_relax_shapes=True)
def get_gap_activations(self, x: GraphsTuple) -> NodeEdgeTensors:
"""Gets node-wise and edge-wise contributions to graph embedding."""
return self.readout.get_activations(self.gnn(self.encode(x)))
@tf.function(experimental_relax_shapes=True)
def get_prediction_weights(self,
task_index: Optional[int] = None) -> tf.Tensor:
"""Gets last layer prediction weights."""
task_index, _ = self.cast_task_batch_index(task_index, None)
w = self.linear.w[:, task_index]
return w
@tf.function(experimental_relax_shapes=True)
def get_intermediate_activations_gradients(
self,
x: GraphsTuple,
task_index: Optional[int] = None,
batch_index: Optional[int] = None
) -> Tuple[List[NodeEdgeTensors], List[NodeEdgeTensors], tf.Tensor]:
"""Gets intermediate layer activations and gradients."""
task_index, batch_index = self.cast_task_batch_index(
task_index, batch_index)
acts = []
grads = []
with tf.GradientTape(
persistent=True, watch_accessed_variables=False) as gtape:
gtape.watch([x.nodes, x.edges])
x = self.encode(x)
outputs, acts = self.gnn.call_with_activations(x)
outputs = self.readout(outputs)
embs = models.get_graph_attribute(outputs, self.target_type)
y = self.pred_layer(embs)[batch_index, task_index]
acts = [(act.nodes, act.edges) for act in acts]
grads = gtape.gradient(y, acts)
return acts, grads, y
@tf.function(experimental_relax_shapes=True)
def get_attention_weights(self, inputs: GraphsTuple) -> List[tf.Tensor]:
if self.block_type != models.BlockType.gat:
raise ValueError(
f'block_type={self.block_type.name}, attention only works with "gat" blocks'
)
outs = self.encode(inputs)
weights = []
for block in self.gnn._layers: # pylint: disable=protected-access
outs, w = block.apply_attention(outs)
weights.append(w)
return weights
@classmethod
def from_hparams(cls, hp, task:AttributionTask) -> 'GNN':
return cls(node_size = hp.node_size,
edge_size = hp.edge_size,
global_size = hp.global_size,
y_output_size = task.n_outputs,
block_type = models.BlockType(hp.block_type),
activation = task.get_nn_activation_fn(),
target_type = task.target_type,
n_layers = hp.n_layers)
def get_batched_attributions(method: AttributionTechnique,
model: TransparentModel,
inputs: GraphsTuple,
batch_size: int = 2500) -> List[GraphsTuple]:
"""Batched attribution since memory (e.g. IG) can be an issue."""
n = graph_utils.get_num_graphs(inputs)
att_pred = []
actual_batch_size = int(np.ceil(batch_size / method.sample_size))
for chunk in more_itertools.chunked(range(n), actual_batch_size):
x_chunk = graph_utils.get_graphs_tf(inputs, np.array(chunk))
att = method.attribute(x_chunk, model)
att_pred.extend(att)
return att_pred
def generate_result(model: templates.TransparentModel,
method: templates.AttributionTechnique,
task: templates.AttributionTask,
inputs: GraphsTuple,
y_true: np.ndarray,
true_atts: List[GraphsTuple],
pred_atts: Optional[List[GraphsTuple]] = None,
reducer_fn: Optional[Callable[[np.ndarray],
Any]] = np.nanmean,
batch_size: int = 1000) -> MutableMapping[Text, Any]:
"""For a given model, method and task, generate metrics."""
if pred_atts is None:
pred_atts = get_batched_attributions(method, model, inputs, batch_size)
result = task.evaluate_attributions(
true_atts, pred_atts, reducer_fn=reducer_fn)
# Need to reshape since predict returns a 1D array.
y_pred = model.predict(inputs).numpy().reshape(-1, 1)
result.update(task.evaluate_predictions(y_true, y_pred))
result['Task'] = task.name
result['Technique'] = method.name
result['Model'] = model.name
return result
@dataclasses.dataclass(frozen=True)
class ExperimentData:
"""Helper class to hold all data relating to an experiment."""
x_train: GraphsTuple
x_test: GraphsTuple
y_train: np.ndarray
y_test: np.ndarray
att_test: List[GraphsTuple]
x_aug: Optional[GraphsTuple] = None
y_aug: Optional[np.ndarray] = None
@classmethod
def from_data_and_splits(cls, x, y, att, train_index,
test_index, x_aug=None, y_aug=None):
"""Build class from data and split indices."""
if np.intersect1d(train_index, test_index).shape[0]:
raise ValueError('train/test indices have overlap!.')
return cls(
x_train=graph_utils.get_graphs_tf(x, np.array(train_index)),
x_test=graph_utils.get_graphs_tf(x, | np.array(test_index) | numpy.array |
import unittest
import warnings # for suppressing numpy.array PendingDeprecationWarning's
from importlib import reload
import sys
sys.path.append("..")
import numpy as np
import sympy as sp
import Ch6.utilities as ch6_utils
import Ch7.utilities as ch7_utils
reload(ch7_utils)
import control
def fullOrderCompensator(A, B, C, D, controlPoles, observerPoles):
"""
combine controller and observer constructions into a compensator design
currently, supports single-input-single-output systems only
Inputs:
A (numpy matrix/array, type=real) - system state matrix
B (numpy matrix/array, type=real) - system control matrix
C (numpy matrix/array, type=real) - system measurement matrix
desiredPoles (numpy matrix/array, type=complex) - desired pole locations
D (numpy matrix/array, type=real) - direct path from control to measurement matrix
E (numpy matrix/array, type=real) - (default=None) exogeneous input matrix
controlPoles (numpy matrix/array, type=complex) - desired controller system poles
observerPoles (numpy matrix/array, type=complex) - desired observer system poles
Returns:
(control.TransferFunction) - compensator transfer function from input to output
Raises:
TypeError - if input system is not SISO
"""
if B.shape[1] > 1 or C.shape[0] > 1:
raise TypeError('Only single-input-single-output (SISO) systems are currently supported')
A = A.astype('float')
B = B.astype('float')
C = C.astype('float')
D = D.astype('float')
G = ch6_utils.bassGura(A, B, controlPoles)
K = ch7_utils.obsBassGura(A, C, observerPoles)
return control.ss2tf(control.StateSpace(A - B@G - K@C, K, G, D))
def stabilityRange(tfD, tfPlant, gain):
"""
numerically evaluate a range of gains, k, to see if the closed-loop system (k*tfD*tfPlant) / (1 + k*tfD*tfPlant)
Inputs:
tfD (control.TransferFunction) - compensator transfer function
tfPlant (control.TransferFunction) - open loop system transfer function
gain (numpy matrix/array, type=real) - range of gains to check
Returns:
tuple(min gain, max gain) defining stability interval if such an interval could be found, `None` otherwise
Raises:
"""
stable = | np.zeros_like(gain) | numpy.zeros_like |
#!/usr/bin/env python3
# pylint: disable=C0111
import pytest
import numpy as np
from pyndl import correlation
@pytest.mark.nolinux
def test_correlation():
np.random.seed(20190507)
n_vec_dims = 40
n_outcomes = 50
n_cues = 20
n_events = 120
semantics = np.asfortranarray(np.random.random((n_vec_dims, n_outcomes)))
weights = np.random.random((n_vec_dims, n_cues))
events = np.random.random((n_cues, n_events))
# n_vec_dims x n_events
activations = np.asfortranarray(weights @ events)
corr1 = correlation._reference_correlation(semantics, activations, verbose=True)
corr2 = correlation.correlation(semantics, activations, verbose=True)
assert | np.allclose(corr1, corr2) | numpy.allclose |
import matplotlib.pyplot as plt
import numpy as np
## assignment 1
## 5, 10, 15, 20, 25, 30, 35, 40, 45, 50
n = np.array([0, 0, 0, 0, 1, 1, 1, 0, 1, 0, 0, 0, 0, 0, 1, 0, 2, 2, 2, 0, 0, 0, 6, 5, 4 ,1 ,0, 0, 0, 0, 0, 1, 2, 5 ,7, 0, 0, 0, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 3, 7, 0, 8, 2, 2, 21, 2, 2, 1, 1, 1, 1, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 2, 3, 0, 0, 0, 0, 0])
m = np.arange(0, len(n))
## assignment 2
## 5, 10, 15, 20, 30, 35, 40
a = | np.array([0, 0, 0, 20, 20, 20, 40, 40, 40, 20, 80, 20, 40, 80, 100, 120, 100, 40, 160, 200, 100, 120, 200, 240, 160, 200, 40 , 340 ,140, 240, 220, 260, 280, 220, 280, 260, 400, 180, 340, 260, 360, 520, 120, 540, 260, 140, 320, 300, 240, 260, 320, 480, 200, 620, 380, 520, 260, 280, 240, 280, 360, 360, 480, 560, 400, 320, 280, 400, 120, 260, 460, 160, 300, 580, 260, 500, 540, 280, 540, 620, 620, 380, 520, 660, 480,400, 280, 360, 460, 480, 560, 600, 620, 380, 520, 460, 280, 540, 280, 360, 660, 480, 660, 820])
m = np.arange(0, len(n)) | numpy.array |
# Copyright 2017 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Tests for the GTFlow training Ops."""
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import numpy as np
from google.protobuf import text_format
from tensorflow.contrib.boosted_trees.proto import learner_pb2
from tensorflow.contrib.boosted_trees.proto import split_info_pb2
from tensorflow.contrib.boosted_trees.proto import tree_config_pb2
from tensorflow.contrib.boosted_trees.python.ops import model_ops
from tensorflow.contrib.boosted_trees.python.ops import training_ops
from tensorflow.python.framework import constant_op
from tensorflow.python.framework import test_util
from tensorflow.python.ops import array_ops
from tensorflow.python.ops import resources
from tensorflow.python.platform import googletest
def _gen_learner_config(num_classes,
l1_reg,
l2_reg,
tree_complexity,
max_depth,
min_node_weight,
pruning_mode,
growing_mode,
dropout_probability=None,
dropout_learning_rate=None,
dropout_prob_of_skipping=None):
"""Create a serialized learner config with the desired settings."""
config = learner_pb2.LearnerConfig()
config.num_classes = num_classes
config.regularization.l1 = l1_reg
config.regularization.l2 = l2_reg
config.regularization.tree_complexity = tree_complexity
config.constraints.max_tree_depth = max_depth
config.constraints.min_node_weight = min_node_weight
config.pruning_mode = pruning_mode
config.growing_mode = growing_mode
if dropout_probability is not None:
config.learning_rate_tuner.dropout.dropout_probability = dropout_probability
if dropout_learning_rate is not None:
config.learning_rate_tuner.dropout.learning_rate = dropout_learning_rate
if dropout_prob_of_skipping is not None:
config.learning_rate_tuner.dropout.dropout_prob_of_skipping = (
dropout_prob_of_skipping)
return config
def _gen_dense_split_info(fc, threshold, left_weight, right_weight):
split_str = """
split_node {
dense_float_binary_split {
feature_column: %d
threshold: %f
}
}
left_child {
sparse_vector {
index: 0
value: %f
}
}
right_child {
sparse_vector {
index: 0
value: %f
}
}""" % (fc, threshold, left_weight, right_weight)
split = split_info_pb2.SplitInfo()
text_format.Merge(split_str, split)
return split.SerializeToString()
def _gen_dense_oblivious_split_info(fc, threshold, leave_weights,
children_parent_id):
split_str = """
split_node {
oblivious_dense_float_binary_split {
feature_column: %d
threshold: %f
}
}""" % (fc, threshold)
for weight in leave_weights:
split_str += """
children {
vector {
value: %f
}
}""" % (
weight)
for x in children_parent_id:
split_str += """
children_parent_id: %d""" % (x)
split = split_info_pb2.ObliviousSplitInfo()
text_format.Merge(split_str, split)
return split.SerializeToString()
def _gen_categorical_split_info(fc, feat_id, left_weight, right_weight):
split_str = """
split_node {
categorical_id_binary_split {
feature_column: %d
feature_id: %d
}
}
left_child {
sparse_vector {
index: 0
value: %f
}
}
right_child {
sparse_vector {
index: 0
value: %f
}
}""" % (fc, feat_id, left_weight, right_weight)
split = split_info_pb2.SplitInfo()
text_format.Merge(split_str, split)
return split.SerializeToString()
def _get_bias_update(grads, hess):
return array_ops.where(hess > 0, -grads / hess, array_ops.zeros_like(grads))
class CenterTreeEnsembleBiasOpTest(test_util.TensorFlowTestCase):
"""Tests for centering tree ensemble bias."""
def testCenterBias(self):
"""Tests bias centering for multiple iterations."""
with self.cached_session() as session:
# Create empty ensemble.
tree_ensemble_config = tree_config_pb2.DecisionTreeEnsembleConfig()
tree_ensemble_handle = model_ops.tree_ensemble_variable(
stamp_token=0,
tree_ensemble_config=tree_ensemble_config.SerializeToString(),
name="tree_ensemble")
resources.initialize_resources(resources.shared_resources()).run()
# Prepare learner config.
learner_config = _gen_learner_config(
num_classes=3,
l1_reg=0,
l2_reg=0,
tree_complexity=0,
max_depth=4,
min_node_weight=0,
pruning_mode=learner_pb2.LearnerConfig.PRE_PRUNE,
growing_mode=learner_pb2.LearnerConfig.WHOLE_TREE,
# Dropout does not change anything here.
dropout_probability=0.5).SerializeToString()
# Center bias for the initial step.
grads = constant_op.constant([0.4, -0.3])
hess = constant_op.constant([2.0, 1.0])
continue_centering1 = training_ops.center_tree_ensemble_bias(
tree_ensemble_handle,
stamp_token=0,
next_stamp_token=1,
delta_updates=_get_bias_update(grads, hess),
learner_config=learner_config)
continue_centering = session.run(continue_centering1)
self.assertEqual(continue_centering, True)
# Validate ensemble state.
# dim 0 update: -0.4/2.0 = -0.2
# dim 1 update: +0.3/1.0 = +0.3
new_stamp, serialized = session.run(
model_ops.tree_ensemble_serialize(tree_ensemble_handle))
stats = session.run(
training_ops.tree_ensemble_stats(tree_ensemble_handle, stamp_token=1))
tree_ensemble_config.ParseFromString(serialized)
expected_result = """
trees {
nodes {
leaf {
vector {
value: -0.2
value: 0.3
}
}
}
}
tree_weights: 1.0
tree_metadata {
num_layers_grown: 1
}
growing_metadata {
num_trees_attempted: 1
num_layers_attempted: 1
}
"""
self.assertEqual(new_stamp, 1)
self.assertEqual(stats.num_trees, 0)
self.assertEqual(stats.num_layers, 1)
self.assertEqual(stats.active_tree, 1)
self.assertEqual(stats.active_layer, 1)
self.assertEqual(stats.attempted_trees, 1)
self.assertEqual(stats.attempted_layers, 1)
self.assertProtoEquals(expected_result, tree_ensemble_config)
# Center bias for another step.
# dim 0 update: -0.06/0.5 = -0.12
# dim 1 update: -0.01/0.5 = -0.02
grads = constant_op.constant([0.06, 0.01])
hess = constant_op.constant([0.5, 0.5])
continue_centering2 = training_ops.center_tree_ensemble_bias(
tree_ensemble_handle,
stamp_token=1,
next_stamp_token=2,
delta_updates=_get_bias_update(grads, hess),
learner_config=learner_config)
continue_centering = session.run(continue_centering2)
self.assertEqual(continue_centering, True)
# Validate ensemble state.
new_stamp, serialized = session.run(
model_ops.tree_ensemble_serialize(tree_ensemble_handle))
stats = session.run(
training_ops.tree_ensemble_stats(tree_ensemble_handle, stamp_token=2))
tree_ensemble_config.ParseFromString(serialized)
expected_result = """
trees {
nodes {
leaf {
vector {
value: -0.32
value: 0.28
}
}
}
}
tree_weights: 1.0
tree_metadata {
num_layers_grown: 1
}
growing_metadata {
num_trees_attempted: 1
num_layers_attempted: 1
}
"""
self.assertEqual(new_stamp, 2)
self.assertEqual(stats.num_trees, 0)
self.assertEqual(stats.num_layers, 1)
self.assertEqual(stats.active_tree, 1)
self.assertEqual(stats.active_layer, 1)
self.assertEqual(stats.attempted_trees, 1)
self.assertEqual(stats.attempted_layers, 1)
self.assertProtoEquals(expected_result, tree_ensemble_config)
# Center bias for another step, but this time updates are negligible.
grads = constant_op.constant([0.0000001, -0.00003])
hess = constant_op.constant([0.5, 0.0])
continue_centering3 = training_ops.center_tree_ensemble_bias(
tree_ensemble_handle,
stamp_token=2,
next_stamp_token=3,
delta_updates=_get_bias_update(grads, hess),
learner_config=learner_config)
continue_centering = session.run(continue_centering3)
self.assertEqual(continue_centering, False)
# Validate ensemble stamp.
new_stamp, _ = session.run(
model_ops.tree_ensemble_serialize(tree_ensemble_handle))
stats = session.run(
training_ops.tree_ensemble_stats(tree_ensemble_handle, stamp_token=3))
self.assertEqual(new_stamp, 3)
self.assertEqual(stats.num_trees, 1)
self.assertEqual(stats.num_layers, 1)
self.assertEqual(stats.active_tree, 1)
self.assertEqual(stats.active_layer, 1)
self.assertEqual(stats.attempted_trees, 1)
self.assertEqual(stats.attempted_layers, 1)
class GrowTreeEnsembleOpTest(test_util.TensorFlowTestCase):
"""Tests for growing tree ensemble from split candidates."""
def testGrowEmptyEnsemble(self):
"""Test growing an empty ensemble."""
with self.cached_session() as session:
# Create empty ensemble.
tree_ensemble_config = tree_config_pb2.DecisionTreeEnsembleConfig()
tree_ensemble_handle = model_ops.tree_ensemble_variable(
stamp_token=0,
tree_ensemble_config=tree_ensemble_config.SerializeToString(),
name="tree_ensemble")
resources.initialize_resources(resources.shared_resources()).run()
# Prepare learner config.
learner_config = _gen_learner_config(
num_classes=2,
l1_reg=0,
l2_reg=0,
tree_complexity=0,
max_depth=1,
min_node_weight=0,
pruning_mode=learner_pb2.LearnerConfig.PRE_PRUNE,
growing_mode=learner_pb2.LearnerConfig.WHOLE_TREE,
# Dropout does not change anything here, tree is not finalized.
dropout_probability=0.5)
# Prepare handler inputs.
# Note that handlers 1 & 3 have the same gain but different splits.
handler1_partitions = np.array([0], dtype=np.int32)
handler1_gains = | np.array([7.62], dtype=np.float32) | numpy.array |
import glob
import math
import os
import sys
import warnings
from decimal import Decimal
import numpy as np
import pandas as pd
import pytest
from packaging.version import parse as parse_version
import dask
import dask.dataframe as dd
import dask.multiprocessing
from dask.blockwise import Blockwise, optimize_blockwise
from dask.dataframe._compat import PANDAS_GT_110, PANDAS_GT_121, PANDAS_GT_130
from dask.dataframe.io.parquet.utils import _parse_pandas_metadata
from dask.dataframe.optimize import optimize_dataframe_getitem
from dask.dataframe.utils import assert_eq
from dask.layers import DataFrameIOLayer
from dask.utils import natural_sort_key
from dask.utils_test import hlg_layer
try:
import fastparquet
except ImportError:
fastparquet = False
fastparquet_version = parse_version("0")
else:
fastparquet_version = parse_version(fastparquet.__version__)
try:
import pyarrow as pa
except ImportError:
pa = False
pa_version = parse_version("0")
else:
pa_version = parse_version(pa.__version__)
try:
import pyarrow.parquet as pq
except ImportError:
pq = False
SKIP_FASTPARQUET = not fastparquet
FASTPARQUET_MARK = pytest.mark.skipif(SKIP_FASTPARQUET, reason="fastparquet not found")
if sys.platform == "win32" and pa and pa_version == parse_version("2.0.0"):
SKIP_PYARROW = True
SKIP_PYARROW_REASON = (
"skipping pyarrow 2.0.0 on windows: "
"https://github.com/dask/dask/issues/6093"
"|https://github.com/dask/dask/issues/6754"
)
else:
SKIP_PYARROW = not pq
SKIP_PYARROW_REASON = "pyarrow not found"
PYARROW_MARK = pytest.mark.skipif(SKIP_PYARROW, reason=SKIP_PYARROW_REASON)
# "Legacy" and "Dataset"-specific MARK definitions
SKIP_PYARROW_LE = SKIP_PYARROW
SKIP_PYARROW_LE_REASON = "pyarrow not found"
SKIP_PYARROW_DS = SKIP_PYARROW
SKIP_PYARROW_DS_REASON = "pyarrow not found"
if not SKIP_PYARROW_LE:
# NOTE: We should use PYARROW_LE_MARK to skip
# pyarrow-legacy tests once pyarrow officially
# removes ParquetDataset support in the future.
PYARROW_LE_MARK = pytest.mark.filterwarnings(
"ignore::DeprecationWarning",
"ignore::FutureWarning",
)
else:
PYARROW_LE_MARK = pytest.mark.skipif(SKIP_PYARROW_LE, reason=SKIP_PYARROW_LE_REASON)
PYARROW_DS_MARK = pytest.mark.skipif(SKIP_PYARROW_DS, reason=SKIP_PYARROW_DS_REASON)
ANY_ENGINE_MARK = pytest.mark.skipif(
SKIP_FASTPARQUET and SKIP_PYARROW,
reason="No parquet engine (fastparquet or pyarrow) found",
)
nrows = 40
npartitions = 15
df = pd.DataFrame(
{
"x": [i * 7 % 5 for i in range(nrows)], # Not sorted
"y": [i * 2.5 for i in range(nrows)], # Sorted
},
index=pd.Index([10 * i for i in range(nrows)], name="myindex"),
)
ddf = dd.from_pandas(df, npartitions=npartitions)
@pytest.fixture(
params=[
pytest.param("fastparquet", marks=FASTPARQUET_MARK),
pytest.param("pyarrow-legacy", marks=PYARROW_LE_MARK),
pytest.param("pyarrow-dataset", marks=PYARROW_DS_MARK),
]
)
def engine(request):
return request.param
def write_read_engines(**kwargs):
"""Product of both engines for write/read:
To add custom marks, pass keyword of the form: `mark_writer_reader=reason`,
or `mark_engine=reason` to apply to all parameters with that engine."""
backends = {"pyarrow-dataset", "pyarrow-legacy", "fastparquet"}
# Skip if uninstalled
skip_marks = {
"fastparquet": FASTPARQUET_MARK,
"pyarrow-legacy": PYARROW_LE_MARK,
"pyarrow-dataset": PYARROW_DS_MARK,
}
marks = {(w, r): [skip_marks[w], skip_marks[r]] for w in backends for r in backends}
# Custom marks
for kw, val in kwargs.items():
kind, rest = kw.split("_", 1)
key = tuple(rest.split("_"))
if kind not in ("xfail", "skip") or len(key) > 2 or set(key) - backends:
raise ValueError("unknown keyword %r" % kw)
val = getattr(pytest.mark, kind)(reason=val)
if len(key) == 2:
marks[key].append(val)
else:
for k in marks:
if key in k:
marks[k].append(val)
return pytest.mark.parametrize(
("write_engine", "read_engine"),
[pytest.param(*k, marks=tuple(v)) for (k, v) in sorted(marks.items())],
)
pyarrow_fastparquet_msg = "pyarrow schema and pandas metadata may disagree"
write_read_engines_xfail = write_read_engines(
**{
"xfail_pyarrow-dataset_fastparquet": pyarrow_fastparquet_msg,
"xfail_pyarrow-legacy_fastparquet": pyarrow_fastparquet_msg,
}
)
if (
fastparquet
and fastparquet_version < parse_version("0.5")
and PANDAS_GT_110
and not PANDAS_GT_121
):
# a regression in pandas 1.1.x / 1.2.0 caused a failure in writing partitioned
# categorical columns when using fastparquet 0.4.x, but this was (accidentally)
# fixed in fastparquet 0.5.0
fp_pandas_msg = "pandas with fastparquet engine does not preserve index"
fp_pandas_xfail = write_read_engines(
**{
"xfail_pyarrow-dataset_fastparquet": pyarrow_fastparquet_msg,
"xfail_pyarrow-legacy_fastparquet": pyarrow_fastparquet_msg,
"xfail_fastparquet_fastparquet": fp_pandas_msg,
"xfail_fastparquet_pyarrow-dataset": fp_pandas_msg,
"xfail_fastparquet_pyarrow-legacy": fp_pandas_msg,
}
)
else:
fp_pandas_msg = "pandas with fastparquet engine does not preserve index"
fp_pandas_xfail = write_read_engines()
@PYARROW_MARK
def test_pyarrow_getengine():
from dask.dataframe.io.parquet.arrow import ArrowDatasetEngine
from dask.dataframe.io.parquet.core import get_engine
# Check that the default engine for "pyarrow"/"arrow"
# is the `pyarrow.dataset`-based engine
assert get_engine("pyarrow") == ArrowDatasetEngine
assert get_engine("arrow") == ArrowDatasetEngine
if SKIP_PYARROW_LE:
with pytest.warns(FutureWarning):
get_engine("pyarrow-legacy")
@write_read_engines()
def test_local(tmpdir, write_engine, read_engine):
tmp = str(tmpdir)
data = pd.DataFrame(
{
"i32": np.arange(1000, dtype=np.int32),
"i64": np.arange(1000, dtype=np.int64),
"f": np.arange(1000, dtype=np.float64),
"bhello": np.random.choice(["hello", "yo", "people"], size=1000).astype(
"O"
),
}
)
df = dd.from_pandas(data, chunksize=500)
df.to_parquet(tmp, write_index=False, engine=write_engine)
files = os.listdir(tmp)
assert "_common_metadata" in files
assert "_metadata" in files
assert "part.0.parquet" in files
df2 = dd.read_parquet(tmp, index=False, engine=read_engine)
assert len(df2.divisions) > 1
out = df2.compute(scheduler="sync").reset_index()
for column in df.columns:
assert (data[column] == out[column]).all()
@pytest.mark.parametrize("index", [False, True])
@write_read_engines_xfail
def test_empty(tmpdir, write_engine, read_engine, index):
fn = str(tmpdir)
df = pd.DataFrame({"a": ["a", "b", "b"], "b": [4, 5, 6]})[:0]
if index:
df.set_index("a", inplace=True, drop=True)
ddf = dd.from_pandas(df, npartitions=2)
ddf.to_parquet(fn, write_index=index, engine=write_engine)
read_df = dd.read_parquet(fn, engine=read_engine)
assert_eq(ddf, read_df)
@write_read_engines()
def test_simple(tmpdir, write_engine, read_engine):
fn = str(tmpdir)
if write_engine != "fastparquet":
df = pd.DataFrame({"a": [b"a", b"b", b"b"], "b": [4, 5, 6]})
else:
df = pd.DataFrame({"a": ["a", "b", "b"], "b": [4, 5, 6]})
df.set_index("a", inplace=True, drop=True)
ddf = dd.from_pandas(df, npartitions=2)
ddf.to_parquet(fn, engine=write_engine)
read_df = dd.read_parquet(fn, index=["a"], engine=read_engine)
assert_eq(ddf, read_df)
@write_read_engines()
def test_delayed_no_metadata(tmpdir, write_engine, read_engine):
fn = str(tmpdir)
df = pd.DataFrame({"a": ["a", "b", "b"], "b": [4, 5, 6]})
df.set_index("a", inplace=True, drop=True)
ddf = dd.from_pandas(df, npartitions=2)
ddf.to_parquet(
fn, engine=write_engine, compute=False, write_metadata_file=False
).compute()
files = os.listdir(fn)
assert "_metadata" not in files
# Fastparquet doesn't currently handle a directory without "_metadata"
read_df = dd.read_parquet(
os.path.join(fn, "*.parquet"),
index=["a"],
engine=read_engine,
gather_statistics=True,
)
assert_eq(ddf, read_df)
@write_read_engines()
def test_read_glob(tmpdir, write_engine, read_engine):
tmp_path = str(tmpdir)
ddf.to_parquet(tmp_path, engine=write_engine)
if os.path.exists(os.path.join(tmp_path, "_metadata")):
os.unlink(os.path.join(tmp_path, "_metadata"))
files = os.listdir(tmp_path)
assert "_metadata" not in files
ddf2 = dd.read_parquet(
os.path.join(tmp_path, "*.parquet"),
engine=read_engine,
index="myindex", # Must specify index without _metadata
gather_statistics=True,
)
assert_eq(ddf, ddf2)
@write_read_engines()
def test_gather_statistics_false(tmpdir, write_engine, read_engine):
tmp_path = str(tmpdir)
ddf.to_parquet(tmp_path, write_index=False, engine=write_engine)
ddf2 = dd.read_parquet(
tmp_path,
engine=read_engine,
index=False,
gather_statistics=False,
)
assert_eq(ddf, ddf2, check_index=False, check_divisions=False)
@write_read_engines()
def test_read_list(tmpdir, write_engine, read_engine):
if write_engine == read_engine == "fastparquet" and os.name == "nt":
# fastparquet or dask is not normalizing filepaths correctly on
# windows.
pytest.skip("filepath bug.")
tmpdir = str(tmpdir)
ddf.to_parquet(tmpdir, engine=write_engine)
files = sorted(
(
os.path.join(tmpdir, f)
for f in os.listdir(tmpdir)
if not f.endswith("_metadata")
),
key=natural_sort_key,
)
ddf2 = dd.read_parquet(
files, engine=read_engine, index="myindex", gather_statistics=True
)
assert_eq(ddf, ddf2)
@write_read_engines()
def test_columns_auto_index(tmpdir, write_engine, read_engine):
fn = str(tmpdir)
ddf.to_parquet(fn, engine=write_engine)
# XFAIL, auto index selection no longer supported (for simplicity)
# ### Empty columns ###
# With divisions if supported
assert_eq(dd.read_parquet(fn, columns=[], engine=read_engine), ddf[[]])
# No divisions
assert_eq(
dd.read_parquet(fn, columns=[], engine=read_engine, gather_statistics=False),
ddf[[]].clear_divisions(),
check_divisions=True,
)
# ### Single column, auto select index ###
# With divisions if supported
assert_eq(dd.read_parquet(fn, columns=["x"], engine=read_engine), ddf[["x"]])
# No divisions
assert_eq(
dd.read_parquet(fn, columns=["x"], engine=read_engine, gather_statistics=False),
ddf[["x"]].clear_divisions(),
check_divisions=True,
)
@write_read_engines()
def test_columns_index(tmpdir, write_engine, read_engine):
fn = str(tmpdir)
ddf.to_parquet(fn, engine=write_engine)
# With Index
# ----------
# ### Empty columns, specify index ###
# With divisions if supported
assert_eq(
dd.read_parquet(fn, columns=[], engine=read_engine, index="myindex"), ddf[[]]
)
# No divisions
assert_eq(
dd.read_parquet(
fn, columns=[], engine=read_engine, index="myindex", gather_statistics=False
),
ddf[[]].clear_divisions(),
check_divisions=True,
)
# ### Single column, specify index ###
# With divisions if supported
assert_eq(
dd.read_parquet(fn, index="myindex", columns=["x"], engine=read_engine),
ddf[["x"]],
)
# No divisions
assert_eq(
dd.read_parquet(
fn,
index="myindex",
columns=["x"],
engine=read_engine,
gather_statistics=False,
),
ddf[["x"]].clear_divisions(),
check_divisions=True,
)
# ### Two columns, specify index ###
# With divisions if supported
assert_eq(
dd.read_parquet(fn, index="myindex", columns=["x", "y"], engine=read_engine),
ddf,
)
# No divisions
assert_eq(
dd.read_parquet(
fn,
index="myindex",
columns=["x", "y"],
engine=read_engine,
gather_statistics=False,
),
ddf.clear_divisions(),
check_divisions=True,
)
def test_nonsense_column(tmpdir, engine):
fn = str(tmpdir)
ddf.to_parquet(fn, engine=engine)
with pytest.raises((ValueError, KeyError)):
dd.read_parquet(fn, columns=["nonesense"], engine=engine)
with pytest.raises((Exception, KeyError)):
dd.read_parquet(fn, columns=["nonesense"] + list(ddf.columns), engine=engine)
@write_read_engines()
def test_columns_no_index(tmpdir, write_engine, read_engine):
fn = str(tmpdir)
ddf.to_parquet(fn, engine=write_engine)
ddf2 = ddf.reset_index()
# No Index
# --------
# All columns, none as index
assert_eq(
dd.read_parquet(fn, index=False, engine=read_engine, gather_statistics=True),
ddf2,
check_index=False,
check_divisions=True,
)
# Two columns, none as index
assert_eq(
dd.read_parquet(
fn,
index=False,
columns=["x", "y"],
engine=read_engine,
gather_statistics=True,
),
ddf2[["x", "y"]],
check_index=False,
check_divisions=True,
)
# One column and one index, all as columns
assert_eq(
dd.read_parquet(
fn,
index=False,
columns=["myindex", "x"],
engine=read_engine,
gather_statistics=True,
),
ddf2[["myindex", "x"]],
check_index=False,
check_divisions=True,
)
@write_read_engines()
def test_gather_statistics_no_index(tmpdir, write_engine, read_engine):
fn = str(tmpdir)
ddf.to_parquet(fn, engine=write_engine, write_index=False)
df = dd.read_parquet(fn, engine=read_engine, index=False)
assert df.index.name is None
assert not df.known_divisions
def test_columns_index_with_multi_index(tmpdir, engine):
fn = os.path.join(str(tmpdir), "test.parquet")
index = pd.MultiIndex.from_arrays(
[np.arange(10), np.arange(10) + 1], names=["x0", "x1"]
)
df = pd.DataFrame(np.random.randn(10, 2), columns=["a", "b"], index=index)
df2 = df.reset_index(drop=False)
if engine == "fastparquet":
fastparquet.write(fn, df.reset_index(), write_index=False)
else:
pq.write_table(pa.Table.from_pandas(df.reset_index(), preserve_index=False), fn)
ddf = dd.read_parquet(fn, engine=engine, index=index.names)
assert_eq(ddf, df)
d = dd.read_parquet(fn, columns="a", engine=engine, index=index.names)
assert_eq(d, df["a"])
d = dd.read_parquet(fn, index=["a", "b"], columns=["x0", "x1"], engine=engine)
assert_eq(d, df2.set_index(["a", "b"])[["x0", "x1"]])
# Just index
d = dd.read_parquet(fn, index=False, engine=engine)
assert_eq(d, df2)
d = dd.read_parquet(fn, columns=["b"], index=["a"], engine=engine)
assert_eq(d, df2.set_index("a")[["b"]])
d = dd.read_parquet(fn, columns=["a", "b"], index=["x0"], engine=engine)
assert_eq(d, df2.set_index("x0")[["a", "b"]])
# Just columns
d = dd.read_parquet(fn, columns=["x0", "a"], index=["x1"], engine=engine)
assert_eq(d, df2.set_index("x1")[["x0", "a"]])
# Both index and columns
d = dd.read_parquet(fn, index=False, columns=["x0", "b"], engine=engine)
assert_eq(d, df2[["x0", "b"]])
for index in ["x1", "b"]:
d = dd.read_parquet(fn, index=index, columns=["x0", "a"], engine=engine)
assert_eq(d, df2.set_index(index)[["x0", "a"]])
# Columns and index intersect
for index in ["a", "x0"]:
with pytest.raises(ValueError):
d = dd.read_parquet(fn, index=index, columns=["x0", "a"], engine=engine)
# Series output
for ind, col, sol_df in [
("x1", "x0", df2.set_index("x1")),
(False, "b", df2),
(False, "x0", df2[["x0"]]),
("a", "x0", df2.set_index("a")[["x0"]]),
("a", "b", df2.set_index("a")),
]:
d = dd.read_parquet(fn, index=ind, columns=col, engine=engine)
assert_eq(d, sol_df[col])
@write_read_engines()
def test_no_index(tmpdir, write_engine, read_engine):
fn = str(tmpdir)
df = pd.DataFrame({"a": [1, 2, 3], "b": [4, 5, 6]})
ddf = dd.from_pandas(df, npartitions=2)
ddf.to_parquet(fn, engine=write_engine)
ddf2 = dd.read_parquet(fn, engine=read_engine)
assert_eq(df, ddf2, check_index=False)
def test_read_series(tmpdir, engine):
fn = str(tmpdir)
ddf.to_parquet(fn, engine=engine)
ddf2 = dd.read_parquet(fn, columns=["x"], index="myindex", engine=engine)
assert_eq(ddf[["x"]], ddf2)
ddf2 = dd.read_parquet(fn, columns="x", index="myindex", engine=engine)
assert_eq(ddf.x, ddf2)
def test_names(tmpdir, engine):
fn = str(tmpdir)
ddf.to_parquet(fn, engine=engine)
def read(fn, **kwargs):
return dd.read_parquet(fn, engine=engine, **kwargs)
assert set(read(fn).dask) == set(read(fn).dask)
assert set(read(fn).dask) != set(read(fn, columns=["x"]).dask)
assert set(read(fn, columns=("x",)).dask) == set(read(fn, columns=["x"]).dask)
@write_read_engines()
def test_roundtrip_from_pandas(tmpdir, write_engine, read_engine):
fn = str(tmpdir.join("test.parquet"))
dfp = df.copy()
dfp.index.name = "index"
dfp.to_parquet(
fn, engine="pyarrow" if write_engine.startswith("pyarrow") else "fastparquet"
)
ddf = dd.read_parquet(fn, index="index", engine=read_engine)
assert_eq(dfp, ddf)
@write_read_engines()
def test_categorical(tmpdir, write_engine, read_engine):
tmp = str(tmpdir)
df = pd.DataFrame({"x": ["a", "b", "c"] * 100}, dtype="category")
ddf = dd.from_pandas(df, npartitions=3)
dd.to_parquet(ddf, tmp, engine=write_engine)
ddf2 = dd.read_parquet(tmp, categories="x", engine=read_engine)
assert ddf2.compute().x.cat.categories.tolist() == ["a", "b", "c"]
ddf2 = dd.read_parquet(tmp, categories=["x"], engine=read_engine)
assert ddf2.compute().x.cat.categories.tolist() == ["a", "b", "c"]
# autocat
if read_engine == "fastparquet":
ddf2 = dd.read_parquet(tmp, engine=read_engine)
assert ddf2.compute().x.cat.categories.tolist() == ["a", "b", "c"]
ddf2.loc[:1000].compute()
assert assert_eq(df, ddf2)
# dereference cats
ddf2 = dd.read_parquet(tmp, categories=[], engine=read_engine)
ddf2.loc[:1000].compute()
assert (df.x == ddf2.x.compute()).all()
def test_append(tmpdir, engine):
"""Test that appended parquet equal to the original one."""
tmp = str(tmpdir)
df = pd.DataFrame(
{
"i32": np.arange(1000, dtype=np.int32),
"i64": np.arange(1000, dtype=np.int64),
"f": np.arange(1000, dtype=np.float64),
"bhello": np.random.choice(["hello", "yo", "people"], size=1000).astype(
"O"
),
}
)
df.index.name = "index"
half = len(df) // 2
ddf1 = dd.from_pandas(df.iloc[:half], chunksize=100)
ddf2 = dd.from_pandas(df.iloc[half:], chunksize=100)
ddf1.to_parquet(tmp, engine=engine)
ddf2.to_parquet(tmp, append=True, engine=engine)
ddf3 = dd.read_parquet(tmp, engine=engine)
assert_eq(df, ddf3)
def test_append_create(tmpdir, engine):
"""Test that appended parquet equal to the original one."""
tmp_path = str(tmpdir)
df = pd.DataFrame(
{
"i32": np.arange(1000, dtype=np.int32),
"i64": np.arange(1000, dtype=np.int64),
"f": np.arange(1000, dtype=np.float64),
"bhello": np.random.choice(["hello", "yo", "people"], size=1000).astype(
"O"
),
}
)
df.index.name = "index"
half = len(df) // 2
ddf1 = dd.from_pandas(df.iloc[:half], chunksize=100)
ddf2 = dd.from_pandas(df.iloc[half:], chunksize=100)
ddf1.to_parquet(tmp_path, append=True, engine=engine)
ddf2.to_parquet(tmp_path, append=True, engine=engine)
ddf3 = dd.read_parquet(tmp_path, engine=engine)
assert_eq(df, ddf3)
def test_append_with_partition(tmpdir, engine):
tmp = str(tmpdir)
df0 = pd.DataFrame(
{
"lat": np.arange(0, 10, dtype="int64"),
"lon": np.arange(10, 20, dtype="int64"),
"value": np.arange(100, 110, dtype="int64"),
}
)
df0.index.name = "index"
df1 = pd.DataFrame(
{
"lat": np.arange(10, 20, dtype="int64"),
"lon": np.arange(10, 20, dtype="int64"),
"value": np.arange(120, 130, dtype="int64"),
}
)
df1.index.name = "index"
# Check that nullable dtypes work
# (see: https://github.com/dask/dask/issues/8373)
df0["lat"] = df0["lat"].astype("Int64")
df1["lat"].iloc[0] = np.nan
df1["lat"] = df1["lat"].astype("Int64")
dd_df0 = dd.from_pandas(df0, npartitions=1)
dd_df1 = dd.from_pandas(df1, npartitions=1)
dd.to_parquet(dd_df0, tmp, partition_on=["lon"], engine=engine)
dd.to_parquet(
dd_df1,
tmp,
partition_on=["lon"],
append=True,
ignore_divisions=True,
engine=engine,
)
out = dd.read_parquet(
tmp, engine=engine, index="index", gather_statistics=True
).compute()
# convert categorical to plain int just to pass assert
out["lon"] = out.lon.astype("int64")
# sort required since partitioning breaks index order
assert_eq(
out.sort_values("value"), pd.concat([df0, df1])[out.columns], check_index=False
)
def test_partition_on_cats(tmpdir, engine):
tmp = str(tmpdir)
d = pd.DataFrame(
{
"a": np.random.rand(50),
"b": np.random.choice(["x", "y", "z"], size=50),
"c": np.random.choice(["x", "y", "z"], size=50),
}
)
d = dd.from_pandas(d, 2)
d.to_parquet(tmp, partition_on=["b"], engine=engine)
df = dd.read_parquet(tmp, engine=engine)
assert set(df.b.cat.categories) == {"x", "y", "z"}
@PYARROW_MARK
@pytest.mark.parametrize("meta", [False, True])
@pytest.mark.parametrize("stats", [False, True])
def test_partition_on_cats_pyarrow(tmpdir, stats, meta):
tmp = str(tmpdir)
d = pd.DataFrame(
{
"a": np.random.rand(50),
"b": np.random.choice(["x", "y", "z"], size=50),
"c": np.random.choice(["x", "y", "z"], size=50),
}
)
d = dd.from_pandas(d, 2)
d.to_parquet(tmp, partition_on=["b"], engine="pyarrow", write_metadata_file=meta)
df = dd.read_parquet(tmp, engine="pyarrow", gather_statistics=stats)
assert set(df.b.cat.categories) == {"x", "y", "z"}
def test_partition_on_cats_2(tmpdir, engine):
tmp = str(tmpdir)
d = pd.DataFrame(
{
"a": np.random.rand(50),
"b": np.random.choice(["x", "y", "z"], size=50),
"c": np.random.choice(["x", "y", "z"], size=50),
}
)
d = dd.from_pandas(d, 2)
d.to_parquet(tmp, partition_on=["b", "c"], engine=engine)
df = dd.read_parquet(tmp, engine=engine)
assert set(df.b.cat.categories) == {"x", "y", "z"}
assert set(df.c.cat.categories) == {"x", "y", "z"}
df = dd.read_parquet(tmp, columns=["a", "c"], engine=engine)
assert set(df.c.cat.categories) == {"x", "y", "z"}
assert "b" not in df.columns
assert_eq(df, df.compute())
df = dd.read_parquet(tmp, index="c", engine=engine)
assert set(df.index.categories) == {"x", "y", "z"}
assert "c" not in df.columns
# series
df = dd.read_parquet(tmp, columns="b", engine=engine)
assert set(df.cat.categories) == {"x", "y", "z"}
def test_append_wo_index(tmpdir, engine):
"""Test append with write_index=False."""
tmp = str(tmpdir.join("tmp1.parquet"))
df = pd.DataFrame(
{
"i32": np.arange(1000, dtype=np.int32),
"i64": np.arange(1000, dtype=np.int64),
"f": np.arange(1000, dtype=np.float64),
"bhello": np.random.choice(["hello", "yo", "people"], size=1000).astype(
"O"
),
}
)
half = len(df) // 2
ddf1 = dd.from_pandas(df.iloc[:half], chunksize=100)
ddf2 = dd.from_pandas(df.iloc[half:], chunksize=100)
ddf1.to_parquet(tmp, engine=engine)
with pytest.raises(ValueError) as excinfo:
ddf2.to_parquet(tmp, write_index=False, append=True, engine=engine)
assert "Appended columns" in str(excinfo.value)
tmp = str(tmpdir.join("tmp2.parquet"))
ddf1.to_parquet(tmp, write_index=False, engine=engine)
ddf2.to_parquet(tmp, write_index=False, append=True, engine=engine)
ddf3 = dd.read_parquet(tmp, index="f", engine=engine)
assert_eq(df.set_index("f"), ddf3)
def test_append_overlapping_divisions(tmpdir, engine):
"""Test raising of error when divisions overlapping."""
tmp = str(tmpdir)
df = pd.DataFrame(
{
"i32": np.arange(1000, dtype=np.int32),
"i64": np.arange(1000, dtype=np.int64),
"f": np.arange(1000, dtype=np.float64),
"bhello": np.random.choice(["hello", "yo", "people"], size=1000).astype(
"O"
),
}
)
half = len(df) // 2
ddf1 = dd.from_pandas(df.iloc[:half], chunksize=100)
ddf2 = dd.from_pandas(df.iloc[half - 10 :], chunksize=100)
ddf1.to_parquet(tmp, engine=engine)
with pytest.raises(ValueError) as excinfo:
ddf2.to_parquet(tmp, engine=engine, append=True)
assert "Appended divisions" in str(excinfo.value)
ddf2.to_parquet(tmp, engine=engine, append=True, ignore_divisions=True)
def test_append_different_columns(tmpdir, engine):
"""Test raising of error when non equal columns."""
tmp = str(tmpdir)
df1 = pd.DataFrame({"i32": np.arange(100, dtype=np.int32)})
df2 = pd.DataFrame({"i64": np.arange(100, dtype=np.int64)})
df3 = pd.DataFrame({"i32": np.arange(100, dtype=np.int64)})
ddf1 = dd.from_pandas(df1, chunksize=2)
ddf2 = dd.from_pandas(df2, chunksize=2)
ddf3 = dd.from_pandas(df3, chunksize=2)
ddf1.to_parquet(tmp, engine=engine)
with pytest.raises(ValueError) as excinfo:
ddf2.to_parquet(tmp, engine=engine, append=True)
assert "Appended columns" in str(excinfo.value)
with pytest.raises(ValueError) as excinfo:
ddf3.to_parquet(tmp, engine=engine, append=True)
assert "Appended dtypes" in str(excinfo.value)
def test_append_dict_column(tmpdir, engine):
# See: https://github.com/dask/dask/issues/7492
if engine == "fastparquet":
pytest.xfail("Fastparquet engine is missing dict-column support")
elif pa_version < parse_version("1.0.1"):
pytest.skip("PyArrow 1.0.1+ required for dict-column support.")
tmp = str(tmpdir)
dts = pd.date_range("2020-01-01", "2021-01-01")
df = pd.DataFrame(
{"value": [{"x": x} for x in range(len(dts))]},
index=dts,
)
ddf1 = dd.from_pandas(df, npartitions=1)
# Write ddf1 to tmp, and then append it again
ddf1.to_parquet(tmp, append=True, engine=engine)
ddf1.to_parquet(tmp, append=True, engine=engine, ignore_divisions=True)
# Read back all data (ddf1 + ddf1)
ddf2 = dd.read_parquet(tmp, engine=engine)
# Check computed result
expect = pd.concat([df, df])
result = ddf2.compute()
assert_eq(expect, result)
@write_read_engines_xfail
def test_ordering(tmpdir, write_engine, read_engine):
tmp = str(tmpdir)
df = pd.DataFrame(
{"a": [1, 2, 3], "b": [10, 20, 30], "c": [100, 200, 300]},
index=pd.Index([-1, -2, -3], name="myindex"),
columns=["c", "a", "b"],
)
ddf = dd.from_pandas(df, npartitions=2)
dd.to_parquet(ddf, tmp, engine=write_engine)
if read_engine == "fastparquet":
pf = fastparquet.ParquetFile(tmp)
assert pf.columns == ["myindex", "c", "a", "b"]
ddf2 = dd.read_parquet(tmp, index="myindex", engine=read_engine)
assert_eq(ddf, ddf2, check_divisions=False)
def test_read_parquet_custom_columns(tmpdir, engine):
tmp = str(tmpdir)
data = pd.DataFrame(
{"i32": np.arange(1000, dtype=np.int32), "f": np.arange(1000, dtype=np.float64)}
)
df = dd.from_pandas(data, chunksize=50)
df.to_parquet(tmp, engine=engine)
df2 = dd.read_parquet(tmp, columns=["i32", "f"], engine=engine)
assert_eq(df[["i32", "f"]], df2, check_index=False)
fns = glob.glob(os.path.join(tmp, "*.parquet"))
df2 = dd.read_parquet(fns, columns=["i32"], engine=engine).compute()
df2.sort_values("i32", inplace=True)
assert_eq(df[["i32"]], df2, check_index=False, check_divisions=False)
df3 = dd.read_parquet(tmp, columns=["f", "i32"], engine=engine)
assert_eq(df[["f", "i32"]], df3, check_index=False)
@pytest.mark.parametrize(
"df,write_kwargs,read_kwargs",
[
(pd.DataFrame({"x": [3, 2, 1]}), {}, {}),
(pd.DataFrame({"x": ["c", "a", "b"]}), {}, {}),
(pd.DataFrame({"x": ["cc", "a", "bbb"]}), {}, {}),
(pd.DataFrame({"x": [b"a", b"b", b"c"]}), {"object_encoding": "bytes"}, {}),
(
pd.DataFrame({"x": pd.Categorical(["a", "b", "a"])}),
{},
{"categories": ["x"]},
),
(pd.DataFrame({"x": pd.Categorical([1, 2, 1])}), {}, {"categories": ["x"]}),
(pd.DataFrame({"x": list(map(pd.Timestamp, [3000, 2000, 1000]))}), {}, {}),
(pd.DataFrame({"x": [3000, 2000, 1000]}).astype("M8[ns]"), {}, {}),
pytest.param(
pd.DataFrame({"x": [3, 2, 1]}).astype("M8[ns]"),
{},
{},
),
(pd.DataFrame({"x": [3, 2, 1]}).astype("M8[us]"), {}, {}),
(pd.DataFrame({"x": [3, 2, 1]}).astype("M8[ms]"), {}, {}),
(pd.DataFrame({"x": [3000, 2000, 1000]}).astype("datetime64[ns]"), {}, {}),
(pd.DataFrame({"x": [3000, 2000, 1000]}).astype("datetime64[ns, UTC]"), {}, {}),
(pd.DataFrame({"x": [3000, 2000, 1000]}).astype("datetime64[ns, CET]"), {}, {}),
(pd.DataFrame({"x": [3, 2, 1]}).astype("uint16"), {}, {}),
(pd.DataFrame({"x": [3, 2, 1]}).astype("float32"), {}, {}),
(pd.DataFrame({"x": [3, 1, 2]}, index=[3, 2, 1]), {}, {}),
(pd.DataFrame({"x": [3, 1, 5]}, index=pd.Index([1, 2, 3], name="foo")), {}, {}),
(pd.DataFrame({"x": [1, 2, 3], "y": [3, 2, 1]}), {}, {}),
(pd.DataFrame({"x": [1, 2, 3], "y": [3, 2, 1]}, columns=["y", "x"]), {}, {}),
(pd.DataFrame({"0": [3, 2, 1]}), {}, {}),
(pd.DataFrame({"x": [3, 2, None]}), {}, {}),
(pd.DataFrame({"-": [3.0, 2.0, None]}), {}, {}),
(pd.DataFrame({".": [3.0, 2.0, None]}), {}, {}),
(pd.DataFrame({" ": [3.0, 2.0, None]}), {}, {}),
],
)
def test_roundtrip(tmpdir, df, write_kwargs, read_kwargs, engine):
if "x" in df and df.x.dtype == "M8[ns]" and "arrow" in engine:
pytest.xfail(reason="Parquet pyarrow v1 doesn't support nanosecond precision")
if (
"x" in df
and df.x.dtype == "M8[ns]"
and engine == "fastparquet"
and fastparquet_version <= parse_version("0.6.3")
):
pytest.xfail(reason="fastparquet doesn't support nanosecond precision yet")
if (
PANDAS_GT_130
and read_kwargs.get("categories", None)
and engine == "fastparquet"
and fastparquet_version <= parse_version("0.6.3")
):
pytest.xfail("https://github.com/dask/fastparquet/issues/577")
tmp = str(tmpdir)
if df.index.name is None:
df.index.name = "index"
ddf = dd.from_pandas(df, npartitions=2)
oe = write_kwargs.pop("object_encoding", None)
if oe and engine == "fastparquet":
dd.to_parquet(ddf, tmp, engine=engine, object_encoding=oe, **write_kwargs)
else:
dd.to_parquet(ddf, tmp, engine=engine, **write_kwargs)
ddf2 = dd.read_parquet(tmp, index=df.index.name, engine=engine, **read_kwargs)
if str(ddf2.dtypes.get("x")) == "UInt16" and engine == "fastparquet":
# fastparquet choooses to use masked type to be able to get true repr of
# 16-bit int
assert_eq(ddf.astype("UInt16"), ddf2)
else:
assert_eq(ddf, ddf2)
def test_categories(tmpdir, engine):
fn = str(tmpdir)
df = pd.DataFrame({"x": [1, 2, 3, 4, 5], "y": list("caaab")})
ddf = dd.from_pandas(df, npartitions=2)
ddf["y"] = ddf.y.astype("category")
ddf.to_parquet(fn, engine=engine)
ddf2 = dd.read_parquet(fn, categories=["y"], engine=engine)
# Shouldn't need to specify categories explicitly
ddf3 = dd.read_parquet(fn, engine=engine)
assert_eq(ddf3, ddf2)
with pytest.raises(NotImplementedError):
ddf2.y.cat.categories
assert set(ddf2.y.compute().cat.categories) == {"a", "b", "c"}
cats_set = ddf2.map_partitions(lambda x: x.y.cat.categories.sort_values()).compute()
assert cats_set.tolist() == ["a", "c", "a", "b"]
if engine == "fastparquet":
assert_eq(ddf.y, ddf2.y, check_names=False)
with pytest.raises(TypeError):
# attempt to load as category that which is not so encoded
ddf2 = dd.read_parquet(fn, categories=["x"], engine=engine).compute()
with pytest.raises((ValueError, FutureWarning)):
# attempt to load as category unknown column
ddf2 = dd.read_parquet(fn, categories=["foo"], engine=engine)
def test_categories_unnamed_index(tmpdir, engine):
# Check that we can handle an unnamed categorical index
# https://github.com/dask/dask/issues/6885
tmpdir = str(tmpdir)
df = pd.DataFrame(
data={"A": [1, 2, 3], "B": ["a", "a", "b"]}, index=["x", "y", "y"]
)
ddf = dd.from_pandas(df, npartitions=1)
ddf = ddf.categorize(columns=["B"])
ddf.to_parquet(tmpdir, engine=engine)
ddf2 = dd.read_parquet(tmpdir, engine=engine)
assert_eq(ddf.index, ddf2.index, check_divisions=False)
def test_empty_partition(tmpdir, engine):
fn = str(tmpdir)
df = pd.DataFrame({"a": range(10), "b": range(10)})
ddf = dd.from_pandas(df, npartitions=5)
ddf2 = ddf[ddf.a <= 5]
ddf2.to_parquet(fn, engine=engine)
ddf3 = dd.read_parquet(fn, engine=engine)
assert ddf3.npartitions < 5
sol = ddf2.compute()
assert_eq(sol, ddf3, check_names=False, check_index=False)
def test_timestamp_index(tmpdir, engine):
fn = str(tmpdir)
df = dd._compat.makeTimeDataFrame()
df.index.name = "foo"
ddf = dd.from_pandas(df, npartitions=5)
ddf.to_parquet(fn, engine=engine)
ddf2 = dd.read_parquet(fn, engine=engine)
assert_eq(ddf, ddf2)
@FASTPARQUET_MARK
@PYARROW_MARK
def test_to_parquet_default_writes_nulls(tmpdir):
fn = str(tmpdir.join("test.parquet"))
df = pd.DataFrame({"c1": [1.0, np.nan, 2, np.nan, 3]})
ddf = dd.from_pandas(df, npartitions=1)
ddf.to_parquet(fn)
table = pq.read_table(fn)
assert table[1].null_count == 2
@PYARROW_LE_MARK
def test_to_parquet_pyarrow_w_inconsistent_schema_by_partition_fails_by_default(tmpdir):
df = pd.DataFrame(
{"partition_column": [0, 0, 1, 1], "strings": ["a", "b", None, None]}
)
ddf = dd.from_pandas(df, npartitions=2)
# In order to allow pyarrow to write an inconsistent schema,
# we need to avoid writing the _metadata file (will fail >0.17.1)
# and need to avoid schema inference (i.e. use `schema=None`)
ddf.to_parquet(
str(tmpdir),
engine="pyarrow",
partition_on=["partition_column"],
write_metadata_file=False,
schema=None,
)
# Test that schema is not validated by default
# (shouldn't raise error with legacy dataset)
dd.read_parquet(
str(tmpdir),
engine="pyarrow-legacy",
gather_statistics=False,
).compute()
# Test that read fails when validate_schema=True
# Note: This fails differently for pyarrow.dataset api
with pytest.raises(ValueError) as e_info:
dd.read_parquet(
str(tmpdir),
engine="pyarrow-legacy",
gather_statistics=False,
dataset={"validate_schema": True},
).compute()
assert e_info.message.contains("ValueError: Schema in partition")
assert e_info.message.contains("was different")
@PYARROW_MARK
def test_to_parquet_pyarrow_w_inconsistent_schema_by_partition_succeeds_w_manual_schema(
tmpdir,
):
# Data types to test: strings, arrays, ints, timezone aware timestamps
in_arrays = [[0, 1, 2], [3, 4], np.nan, np.nan]
out_arrays = [[0, 1, 2], [3, 4], None, None]
in_strings = ["a", "b", np.nan, np.nan]
out_strings = ["a", "b", None, None]
tstamp = pd.Timestamp(1513393355, unit="s")
in_tstamps = [tstamp, tstamp, pd.NaT, pd.NaT]
out_tstamps = [
# Timestamps come out in numpy.datetime64 format
tstamp.to_datetime64(),
tstamp.to_datetime64(),
np.datetime64("NaT"),
np.datetime64("NaT"),
]
timezone = "US/Eastern"
tz_tstamp = pd.Timestamp(1513393355, unit="s", tz=timezone)
in_tz_tstamps = [tz_tstamp, tz_tstamp, pd.NaT, pd.NaT]
out_tz_tstamps = [
# Timezones do not make it through a write-read cycle.
tz_tstamp.tz_convert(None).to_datetime64(),
tz_tstamp.tz_convert(None).to_datetime64(),
np.datetime64("NaT"),
np.datetime64("NaT"),
]
df = pd.DataFrame(
{
"partition_column": [0, 0, 1, 1],
"arrays": in_arrays,
"strings": in_strings,
"tstamps": in_tstamps,
"tz_tstamps": in_tz_tstamps,
}
)
ddf = dd.from_pandas(df, npartitions=2)
schema = pa.schema(
[
("arrays", pa.list_(pa.int64())),
("strings", pa.string()),
("tstamps", pa.timestamp("ns")),
("tz_tstamps", pa.timestamp("ns", timezone)),
("partition_column", pa.int64()),
]
)
ddf.to_parquet(
str(tmpdir), engine="pyarrow", partition_on="partition_column", schema=schema
)
ddf_after_write = (
dd.read_parquet(str(tmpdir), engine="pyarrow", gather_statistics=False)
.compute()
.reset_index(drop=True)
)
# Check array support
arrays_after_write = ddf_after_write.arrays.values
for i in range(len(df)):
assert np.array_equal(arrays_after_write[i], out_arrays[i]), type(out_arrays[i])
# Check datetime support
tstamps_after_write = ddf_after_write.tstamps.values
for i in range(len(df)):
# Need to test NaT separately
if np.isnat(tstamps_after_write[i]):
assert np.isnat(out_tstamps[i])
else:
assert tstamps_after_write[i] == out_tstamps[i]
# Check timezone aware datetime support
tz_tstamps_after_write = ddf_after_write.tz_tstamps.values
for i in range(len(df)):
# Need to test NaT separately
if np.isnat(tz_tstamps_after_write[i]):
assert np.isnat(out_tz_tstamps[i])
else:
assert tz_tstamps_after_write[i] == out_tz_tstamps[i]
# Check string support
assert np.array_equal(ddf_after_write.strings.values, out_strings)
# Check partition column
assert np.array_equal(ddf_after_write.partition_column, df.partition_column)
@PYARROW_MARK
@pytest.mark.parametrize("index", [False, True])
@pytest.mark.parametrize("schema", ["infer", "complex"])
def test_pyarrow_schema_inference(tmpdir, index, engine, schema):
if schema == "complex":
schema = {"index": pa.string(), "amount": pa.int64()}
tmpdir = str(tmpdir)
df = pd.DataFrame(
{
"index": ["1", "2", "3", "2", "3", "1", "4"],
"date": pd.to_datetime(
[
"2017-01-01",
"2017-01-01",
"2017-01-01",
"2017-01-02",
"2017-01-02",
"2017-01-06",
"2017-01-09",
]
),
"amount": [100, 200, 300, 400, 500, 600, 700],
},
index=range(7, 14),
)
if index:
df = dd.from_pandas(df, npartitions=2).set_index("index")
else:
df = dd.from_pandas(df, npartitions=2)
df.to_parquet(tmpdir, engine="pyarrow", schema=schema)
df_out = dd.read_parquet(tmpdir, engine=engine)
df_out.compute()
if index and engine == "fastparquet":
# Fastparquet fails to detect int64 from _metadata
df_out["amount"] = df_out["amount"].astype("int64")
# Fastparquet not handling divisions for
# pyarrow-written dataset with string index
assert_eq(df, df_out, check_divisions=False)
else:
assert_eq(df, df_out)
def test_partition_on(tmpdir, engine):
tmpdir = str(tmpdir)
df = pd.DataFrame(
{
"a1": np.random.choice(["A", "B", "C"], size=100),
"a2": np.random.choice(["X", "Y", "Z"], size=100),
"b": np.random.random(size=100),
"c": np.random.randint(1, 5, size=100),
"d": np.arange(0, 100),
}
)
d = dd.from_pandas(df, npartitions=2)
d.to_parquet(tmpdir, partition_on=["a1", "a2"], engine=engine)
# Note #1: Cross-engine functionality is missing
# Note #2: The index is not preserved in pyarrow when partition_on is used
out = dd.read_parquet(
tmpdir, engine=engine, index=False, gather_statistics=False
).compute()
for val in df.a1.unique():
assert set(df.d[df.a1 == val]) == set(out.d[out.a1 == val])
# Now specify the columns and allow auto-index detection
out = dd.read_parquet(tmpdir, engine=engine, columns=["d", "a2"]).compute()
for val in df.a2.unique():
assert set(df.d[df.a2 == val]) == set(out.d[out.a2 == val])
def test_partition_on_duplicates(tmpdir, engine):
# https://github.com/dask/dask/issues/6445
tmpdir = str(tmpdir)
df = pd.DataFrame(
{
"a1": np.random.choice(["A", "B", "C"], size=100),
"a2": np.random.choice(["X", "Y", "Z"], size=100),
"data": np.random.random(size=100),
}
)
d = dd.from_pandas(df, npartitions=2)
for _ in range(2):
d.to_parquet(tmpdir, partition_on=["a1", "a2"], engine=engine)
out = dd.read_parquet(tmpdir, engine=engine).compute()
assert len(df) == len(out)
for root, dirs, files in os.walk(tmpdir):
for file in files:
assert file in (
"part.0.parquet",
"part.1.parquet",
"_common_metadata",
"_metadata",
)
@PYARROW_MARK
@pytest.mark.parametrize("partition_on", ["aa", ["aa"]])
def test_partition_on_string(tmpdir, partition_on):
tmpdir = str(tmpdir)
with dask.config.set(scheduler="single-threaded"):
tmpdir = str(tmpdir)
df = pd.DataFrame(
{
"aa": np.random.choice(["A", "B", "C"], size=100),
"bb": | np.random.random(size=100) | numpy.random.random |
import numpy as np
import matplotlib.pyplot as plt
from scipy.stats import chi2, norm
import pickle
plt.style.use('../../plot/paper.mplstyle')
from matplotlib import rcParams
def comparison(datasets, method):
defaultfontsize = rcParams['font.size']
rcParams['font.size'] = 14
f, (axes) = plt.subplots(2, sharex=True, sharey=False, figsize=(5.5, 5.3))
preds = []
for dset in datasets:
with open(dset + "/results_predictions_" + method +".pkl") as hin:
preds.append(pickle.load(hin))
for i, pred in enumerate(preds):
diff = pred[0.7]
thisdiff = np.array(diff)
mean = np.mean(thisdiff)
std = np.std(thisdiff)
if 'mle' in method and i == 0:
thisdiff = thisdiff[np.where((thisdiff >= 0.7-1.1) & (thisdiff <= 0.7+1.1))]
mean = np.mean(thisdiff)
std = np.std(thisdiff)
nbins = 50 if i == 0 else 100
xlow, xhigh = -0.3, 1.7
axes[i].axvspan(xlow, 0, alpha=0.15, color='gray')
axes[i].axvspan(1, xhigh, alpha=0.15, color='gray')
axes[i].hist(diff, bins=nbins, range=(xlow, xhigh), color='#39568C', alpha=0.7, label=r'$\alpha$ (test) = 0.7f')
textloc = (0.2, 0.8) if mean > 0.8 else (0.75, 0.8)
axes[i].text(textloc[0]+0.0216, textloc[1], r'$n = %d$' % (20 if i == 0 else 500), transform=axes[i].transAxes, fontsize=14)
axes[i].text(textloc[0]+0.0216, textloc[1]-0.10, r'$\alpha = 0.7$', transform=axes[i].transAxes, fontsize=14)
axes[i].text(textloc[0], textloc[1]-0.22, r'$\mu_{\alpha} = %.2f$' % mean, transform=axes[i].transAxes, fontsize=14)
axes[i].text(textloc[0], textloc[1]-0.32, r'$\sigma_{\alpha} = %.2f$' % std, transform=axes[i].transAxes, fontsize=14)
axes[i].set_xlim([xlow, xhigh])
if i == 0:
axes[i].set_ylim([0, 700])
axes[i].yaxis.set_ticks(np.arange(0, 700, 200))
else:
axes[i].set_ylim([0, 950])
axes[i].yaxis.set_ticks(np.arange(0, 950, 200))
xvals = | np.linspace(xlow, xhigh, 200) | numpy.linspace |
from lda2vec import fake_data
from chainer import Variable
from chainer.functions import cross_covariance
import numpy as np
def test_orthogonal_matrix():
msg = "Orthogonal matrices have equal inverse and transpose"
arr = fake_data.orthogonal_matrix([20, 20])
assert np.allclose(np.linalg.inv(arr), arr.T), msg
def test_orthogonal_matrix_covariance():
msg = "Orthogonal matrix should have less covariance than a random matrix"
orth = Variable(fake_data.orthogonal_matrix([20, 20]).astype('float32'))
rand = Variable(np.random.randn(20, 20).astype('float32'))
orth_cc = cross_covariance(orth, orth).data
rand_cc = cross_covariance(rand, rand).data
assert orth_cc < rand_cc, msg
def test_softmax():
arr = np.random.randn(100, 15)
probs = fake_data.softmax(arr)
norms = np.sum(probs, axis=1)
assert np.allclose(norms, np.ones_like(norms))
def test_sample():
n_categories = 10
idx = 4
probs = np.zeros(n_categories)
probs = np.array(probs)
probs[idx] = 1.0
values = np.arange(n_categories)
size = 10
draws = fake_data.sample(values, probs, size)
assert | np.all(draws == idx) | numpy.all |
import numpy as np
import torch
import torch.nn as nn
from torch.nn.modules.utils import _triple
import torch.nn.functional as F
from torch.optim.lr_scheduler import ReduceLROnPlateau
import math
import os
import datetime
from matplotlib import pyplot as plt
class SpatioTemporalConv(nn.Module):
def __init__(self, in_channels, out_channels, kernel_size, stride=1, padding=0, bias=True):
super(SpatioTemporalConv, self).__init__()
kernel_size = _triple(kernel_size)
stride = _triple(stride)
padding = _triple(padding)
spatial_kernel_size = [1, kernel_size[1], kernel_size[2]]
spatial_stride = [1, stride[1], stride[2]]
spatial_padding = [0, padding[1], padding[2]]
temporal_kernel_size = [kernel_size[0], 1, 1]
temporal_stride = [stride[0], 1, 1]
temporal_padding = [padding[0], 0, 0]
intermed_channels = int(math.floor((kernel_size[0] * kernel_size[1] * kernel_size[2] * in_channels * out_channels)/ \
(kernel_size[1]* kernel_size[2] * in_channels + kernel_size[0] * out_channels)))
self.spatial_conv = nn.Conv3d(in_channels, intermed_channels, spatial_kernel_size,
stride=spatial_stride, padding=spatial_padding, bias=bias)
self.bn = nn.BatchNorm3d(intermed_channels)
self.relu = nn.ReLU()
self.temporal_conv = nn.Conv3d(intermed_channels, out_channels, temporal_kernel_size,
stride=temporal_stride, padding=temporal_padding, bias=bias)
def forward(self, x):
x = self.relu(self.bn(self.spatial_conv(x)))
x = self.temporal_conv(x)
return x
class SpatioTemporalResBlock(nn.Module):
def __init__(self, in_channels, out_channels, kernel_size, downsample=False):
super(SpatioTemporalResBlock, self).__init__()
self.downsample = downsample
padding = kernel_size//2
if self.downsample:
self.downsampleconv = SpatioTemporalConv(in_channels, out_channels, 1, stride=2)
self.downsamplebn = nn.BatchNorm3d(out_channels)
self.conv1 = SpatioTemporalConv(in_channels, out_channels, kernel_size, padding=padding, stride=2)
else:
self.conv1 = SpatioTemporalConv(in_channels, out_channels, kernel_size, padding=padding)
self.bn1 = nn.BatchNorm3d(out_channels)
self.relu1 = nn.ReLU()
self.conv2 = SpatioTemporalConv(out_channels, out_channels, kernel_size, padding=padding)
self.bn2 = nn.BatchNorm3d(out_channels)
self.outrelu = nn.ReLU()
def forward(self, x):
res = self.relu1(self.bn1(self.conv1(x)))
res = self.bn2(self.conv2(res))
if self.downsample:
x = self.downsamplebn(self.downsampleconv(x))
return self.outrelu(x + res)
class SpatioTemporalResLayer(nn.Module):
def __init__(self, in_channels, out_channels, kernel_size, layer_size, block_type=SpatioTemporalResBlock, downsample=False):
super(SpatioTemporalResLayer, self).__init__()
self.block1 = block_type(in_channels, out_channels, kernel_size, downsample)
self.blocks = nn.ModuleList([])
for i in range(layer_size - 1):
self.blocks += [block_type(out_channels, out_channels, kernel_size)]
def forward(self, x):
x = self.block1(x)
for block in self.blocks:
x = block(x)
return x
class R2Plus1DNet(nn.Module):
def __init__(self, layer_sizes, block_type=SpatioTemporalResBlock, p = 0.2):
super(R2Plus1DNet, self).__init__()
self.conv1 = SpatioTemporalConv(3, 64, [3, 7, 7], stride=[1, 2, 2], padding=[1, 3, 3])
self.conv2 = SpatioTemporalResLayer(64, 64, 3, layer_sizes[0], block_type=block_type)
self.conv3 = SpatioTemporalResLayer(64, 128, 3, layer_sizes[1], block_type=block_type, downsample=True)
self.conv4 = SpatioTemporalResLayer(128, 256, 3, layer_sizes[2], block_type=block_type, downsample=True)
self.conv5 = SpatioTemporalResLayer(256, 512, 3, layer_sizes[3], block_type=block_type, downsample=True)
self.pool = nn.AdaptiveAvgPool3d(1)
# define dropout layer in __init__
self.drop_layer = nn.Dropout(p = p)
def forward(self, x):
x = self.conv1(x)
x = self.drop_layer(x)
x = self.conv2(x)
x = self.drop_layer(x)
x = self.conv3(x)
x = self.drop_layer(x)
x = self.conv4(x)
x = self.drop_layer(x)
x = self.conv5(x)
x = self.drop_layer(x)
x = self.pool(x)
return x.view(-1, 512)
class R2Plus1DClassifier(nn.Module):
def __init__(self, num_classes, layer_sizes, block_type=SpatioTemporalResBlock, p = 0.2):
super(R2Plus1DClassifier, self).__init__()
self.res2plus1d = R2Plus1DNet(layer_sizes, block_type, p = p)
self.linear = nn.Linear(512, num_classes)
def forward(self, x):
x = self.res2plus1d(x)
x = self.linear(x)
return x
class DataGenerator(torch.utils.data.Dataset):
def __init__(self, vids, labels, batch_size, flip = False, angle = 0, crop = 0, shift = 0):
self.vids = vids
self.labels = labels
self.indices = np.arange(vids.shape[0])
self.batch_size = batch_size
self.flip = flip
self.angle = angle
self.crop = crop
self.shift = shift
self.max_index = vids.shape[0] // batch_size
self.index = 0
np.random.shuffle(self.indices)
def __iter__(self):
return self
def random_zoom(self, batch, x, y):
ax = np.random.uniform(self.crop)
bx = np.random.uniform(ax)
ay = np.random.uniform(self.crop)
by = np.random.uniform(ay)
x = x*(1-ax/batch.shape[2]) + bx
y = y*(1-ay/batch.shape[3]) + by
return x, y
def random_rotate(self, batch, x, y):
rad = np.random.uniform(-self.angle, self.angle)/180*np.pi
rotm = np.array([[np.cos(rad), np.sin(rad)],
[-np.sin(rad), np.cos(rad)]])
x, y = np.einsum('ji, mni -> jmn', rotm, np.dstack([x, y]))
return x, y
def random_translate(self, batch, x, y):
xs = np.random.uniform(-self.shift, self.shift)
ys = np.random.uniform(-self.shift, self.shift)
return x + xs, y + ys
def horizontal_flip(self, batch):
return np.flip(batch, 3)
def __next__(self):
if self.index == self.max_index:
self.index = 0
np.random.shuffle(self.indices)
raise StopIteration
indices = self.indices[self.index * self.batch_size:(self.index + 1) * self.batch_size]
vids = np.array(self.vids[indices])
x, y = np.meshgrid(range(112), range(112))
x = x*24/112
y = y*24/112
if self.crop:
x, y = self.random_zoom(vids, x, y)
if self.angle:
x, y = self.random_rotate(vids, x, y)
if self.shift:
x, y = self.random_translate(vids, x, y)
if self.flip and np.random.random() < 0.5:
vids = self.horizontal_flip(vids)
x = np.clip(x, 0, vids.shape[2]-1).astype(np.int)
y = | np.clip(y, 0, vids.shape[3]-1) | numpy.clip |
import util
import numpy as np
import tensorflow as tf
from keras.utils.np_utils import *
import riemannian
from scipy import signal
import pyriemann
from pyriemann.utils.mean import mean_covariance
MOVEMENT_START = 1 * 160 # MI starts 1s after trial begin
MOVEMENT_END = 5 * 160 # MI lasts 4 seconds
NOISE_LEVEL = 0.01
clas = 4
fc = 160
aug = 40
ntrials = 84
def load_raw_data(electrodes, subject=None, num_classes=2, long_edge=False):
# load from file
trials = []
labels = []
if subject == None:
# subject_ids = range(1, 110)
subject_ids = range(1, 11)
else:
try:
subject_ids = [int(subject)]
except:
subject_ids = subject
for subject_id in subject_ids:
print("load subject %d" % (subject_id,))
t, l, loc, fs = util.load_physionet_data(subject_id, num_classes, long_edge=long_edge)
if num_classes == 2 and t.shape[0] != 42:
# drop subjects with less trials
continue
trials.append(t[:, :, electrodes])
labels.append(l)
return np.array(trials).reshape((len(trials),) + trials[0].shape), np.array(labels)
def split_idx( idx,a,b):
"""
Shuffle and split a list of indexes into training and test data with a fixed
random seed for reproducibility
run: index of the current split (zero based)
nruns: number of splits (> run)
idx: list of indices to split
"""
rs = np.random.RandomState()
rs.shuffle(idx)
start = int(a / 10. * len(idx))
end = int((b+a) / 10. * len(idx))
train_idx = idx[0:start]
test_idx = idx[start:end]
val_idx = idx[end:]
return train_idx, val_idx, test_idx
# return train_idx, test_idx
def n_classfilter(x,y,arg):
# x = np.squeeze(x)
signal = np.zeros((5,)+x.shape)
label = np.zeros((5,)+y.shape)
signal[0,:] = filter(x,0.5,4,arg)
label[0,:] = y
signal[1, :] = filter(x, 4, 8,arg)
label[1, :] = y
signal[2, :] = filter(x, 8, 13,arg)
label[2, :] = y
signal[3, :] = filter(x, 13, 32,arg)
label[3, :] = y
signal[4, :] = filter(x, 32, 50,arg)
label[4, :] = y
return signal,label
def filter(x,low_filter,high_filter, aru):
Wn = [low_filter*2/fc,high_filter*2/fc]
b, a = signal.butter(3, Wn, 'bandpass')
# x = x.transpose((0, 1, 2, 4, 3))
fdata = np.zeros(x.shape)
if aru:
for i in range(len(x)):
for j in range(x.shape[1]):
for k in range(x.shape[2]):
for l in range(x.shape[4]):
fdata[i, j, k, :, l] = signal.filtfilt(b, a, x[i, j, k, :, l])
# fdata = fdata.transpose((0, 1, 2, 4, 3))
return fdata
else:
for i in range(len(x)):
for j in range(x.shape[1]):
for l in range(x.shape[3]):
fdata[i, j, :, l] = signal.filtfilt(b, a, x[i, j, :, l])
# fdata = fdata.transpose((0, 1, 2, 4, 3))
return fdata
def n_class_signal_mapping(x_train, y_train, x_val, y_val):
x1_train = np.zeros((x_train.shape[0:4] + (64,64, )))
y1_train = np.zeros((y_train.shape))
x1_val = np.zeros((x_val.shape[0:3] + (64, 64,)))
y1_val = np.zeros((y_val.shape))
for j in range(len(x_train)):
x1_train[j, :], y1_train[j, :], x1_val[j, :], y1_val[j, :] = signal_mapping(x_train[j], y_train[j], x_val[j], y_val[j])
print("yes")
x1_train = x1_train.transpose(1, 2, 3, 4, 5, 0)
x1_val = x1_val.transpose(1, 2, 3, 4, 0)
# y1_train = y1_train[0]
# y1_val = y1_val[0]
return x1_train, y1_train, x1_val, y1_val
def signal_mapping(x_train, y_train, x_val, y_val):
#训练集
signals1,core = Signals_Covariance(x_train,None,mean_all=True)
#测试集
signals2 = Signals_Covariance(x_val, core, mean_all=False)
# y_test = y_val.reshape((-1,))
# y_train = y_train.reshape((-1,))
return signals1,y_train,signals2,y_val
def Signals_Covariance(signals,core_test,mean_all=True):
if mean_all:
signal = signals.reshape((-1,) + (signals.shape[-2:]))
signal = np.transpose(signal, axes=[0, 2, 1])
x_out = pyriemann.estimation.Covariances().fit_transform(signal)
core = mean_covariance(x_out, metric='riemann')
# core = training_data_cov_means(X,y,num_classes=4)
core = core ** (-1 / 2)
signal1, core = signal_covar(signals, core, mean_all)
return signal1,core
else:
core = core_test ** (-1 / 2)
signal1 = signal_covar(signals, core, mean_all)
return signal1
#对输入的数组进行协方差,并返回同纬度结果[n,84,q,960,64] -> [n,84,q,64,960] -> [n,84,q,64,64]
# [n, 84, 960, 64] -> [n, 84, 64, 960] -> [n, 84, 64, 64]
def signal_covar(signal,core, mean_all):
if mean_all:
signal = np.transpose(signal, axes=[0, 1, 2, 4, 3])
signals = np.zeros((signal.shape[0:4])+(64,))
for i in range(len(signal)):
for j in range(signal.shape[1]):
signal1 = pyriemann.estimation.Covariances().fit_transform(signal[i, j, :])
signal2 = core * signal1 * core
signals[i, j, :] = | np.log(signal2) | numpy.log |
#!/usr/bin/env python
# coding: utf-8
# In[38]:
from scipy.io import loadmat
import glob
import cv2
from shutil import copyfile
import os
import numpy as np
import matplotlib.pylab as plt
from skimage.io import imread
from pathlib import Path
import skimage
from skimage import feature, morphology
from matplotlib.pyplot import figure
import matplotlib
from skimage.color import rgb2gray
import copy
import gc
import sys
# In[39]:
bird_labels = {'head':1, 'leye':2, 'reye':3, 'beak':4, 'torso':5, 'neck':6, 'lwing':7, 'rwing':8, 'lleg':9, 'lfoot':10, 'rleg':11, 'rfoot':12, 'tail':13}
cat_labels = {'head':1, 'leye':2, 'reye':3, 'lear':4, 'rear':5, 'nose':6, 'torso':7, 'neck':8, 'lfleg':9, 'lfpa':10, 'rfleg':11, 'rfpa':12, 'lbleg':13, 'lbpa':14, 'rbleg':15, 'rbpa':16, 'tail':17}
cow_labels = {'head':1, 'leye':2, 'reye':3, 'lear':4, 'rear':5, 'muzzle':6, 'lhorn':7, 'rhorn':8, 'torso':9, 'neck':10, 'lfuleg':11, 'lflleg':12, 'rfuleg':13, 'rflleg':14, 'lbuleg':15, 'lblleg':16, 'rbuleg':17, 'rblleg':18, 'tail':19}
dog_labels = {'head':1, 'leye':2, 'reye':3, 'lear':4, 'rear':5, 'nose':6, 'torso':7, 'neck':8, 'lfleg':9, 'lfpa':10, 'rfleg':11, 'rfpa':12, 'lbleg':13, 'lbpa':14, 'rbleg':15, 'rbpa':16, 'tail':17, 'muzzle':18}
horse_labels = {'head':1, 'leye':2, 'reye':3, 'lear':4, 'rear':5, 'muzzle':6, 'lfho':7, 'rfho':8, 'torso':9, 'neck':10, 'lfuleg':11, 'lflleg':12, 'rfuleg':13, 'rflleg':14, 'lbuleg':15, 'lblleg':16, 'rbuleg':17, 'rblleg':18, 'tail':19, 'lbho':20, 'rbho':21}
bottle_labels = {'cap':1, 'body':2}
person_labels = {'head':1, 'leye':2, 'reye':3, 'lear':4, 'rear':5, 'lebrow':6, 'rebrow':7, 'nose':8, 'mouth':9, 'hair':10, 'torso':11, 'neck': 12, 'llarm': 13, 'luarm': 14, 'lhand': 15, 'rlarm':16, 'ruarm':17, 'rhand': 18, 'llleg': 19, 'luleg':20, 'lfoot':21, 'rlleg':22, 'ruleg':23, 'rfoot':24}
bus_labels = { 'frontside':1, 'leftside':2, 'rightside':3, 'backside':4, 'roofside':5, 'leftmirror':6, 'rightmirror':7, 'fliplate':8, 'bliplate':9 }
for ii in range(0,10):
bus_labels['door_{}'.format(ii+1)] = 10+ii
for ii in range(0,10):
bus_labels['wheel_{}'.format(ii+1)] = 20+ii
for ii in range(0,10):
bus_labels['headlight_{}'.format(ii+1)] = 30+ii
for ii in range(0,10):
bus_labels['window_{}'.format(ii+1)] = 40+ii
aeroplane_labels = {'body': 1, 'stern': 2, 'lwing': 3, 'rwing':4, 'tail':5}
for ii in range(0, 10):
aeroplane_labels['engine_{}'.format(ii+1)] = 6+ii
for ii in range(0, 10):
aeroplane_labels['wheel_{}'.format(ii+1)] = 16+ii
motorbike_labels = {'fwheel': 1, 'bwheel': 2, 'handlebar': 3, 'saddle': 4}
for ii in range(0,10):
motorbike_labels['headlight_{}'.format(ii+1)] = 5+ii
motorbike_labels['body']=15
bicycle_labels = {'fwheel': 1, 'bwheel': 2, 'saddle': 3, 'handlebar': 4, 'chainwheel': 5}
for ii in range(0,10):
bicycle_labels['headlight_{}'.format(ii+1)] = 6+ii
bicycle_labels['body']=16
train_labels = {'head':1,'hfrontside':2,'hleftside':3,'hrightside':4,'hbackside':5,'hroofside':6}
for ii in range(0,10):
train_labels['headlight_{}'.format(ii+1)] = 7 + ii
for ii in range(0,10):
train_labels['coach_{}'.format(ii+1)] = 17 + ii
for ii in range(0,10):
train_labels['cfrontside_{}'.format(ii+1)] = 27 + ii
for ii in range(0,10):
train_labels['cleftside_{}'.format(ii+1)] = 37 + ii
for ii in range(0,10):
train_labels['crightside_{}'.format(ii+1)] = 47 + ii
for ii in range(0,10):
train_labels['cbackside_{}'.format(ii+1)] = 57 + ii
for ii in range(0,10):
train_labels['croofside_{}'.format(ii+1)] = 67 + ii
sheep_labels = cow_labels
car_labels = bus_labels
part_labels = {'bird': bird_labels, 'cat': cat_labels, 'cow': cow_labels, 'dog': dog_labels, 'sheep': sheep_labels, 'horse':horse_labels, 'car':car_labels, 'bus':bus_labels, 'bicycle':bicycle_labels, 'motorbike':motorbike_labels, 'person':person_labels,'aeroplane':aeroplane_labels, 'train':train_labels}
# In[40]:
object_name = sys.argv[1]
animals = [object_name]
# In[4]:
def rotate_im(image, angle):
# grab the dimensions of the image and then determine the
# centre
(h, w) = image.shape[:2]
(cX, cY) = (w // 2, h // 2)
# grab the rotation matrix (applying the negative of the
# angle to rotate clockwise), then grab the sine and cosine
# (i.e., the rotation components of the matrix)
M = cv2.getRotationMatrix2D((cX, cY), angle, 1.0)
cos = np.abs(M[0, 0])
sin = np.abs(M[0, 1])
# compute the new bounding dimensions of the image
nW = int((h * sin) + (w * cos))
nH = int((h * cos) + (w * sin))
# adjust the rotation matrix to take into account translation
M[0, 2] += (nW / 2) - cX
M[1, 2] += (nH / 2) - cY
# perform the actual rotation and return the image
image = cv2.warpAffine(image, M, (nW, nH))
# image = cv2.resize(image, (w,h))
return image
# In[5]:
def get_corners(bboxes):
width = (bboxes[:,2] - bboxes[:,0]).reshape(-1,1)
height = (bboxes[:,3] - bboxes[:,1]).reshape(-1,1)
x1 = bboxes[:,0].reshape(-1,1)
y1 = bboxes[:,1].reshape(-1,1)
x2 = x1 + width
y2 = y1
x3 = x1
y3 = y1 + height
x4 = bboxes[:,2].reshape(-1,1)
y4 = bboxes[:,3].reshape(-1,1)
corners = np.hstack((x1,y1,x2,y2,x3,y3,x4,y4))
return corners
# In[6]:
def clip_box(bbox, clip_box, alpha):
ar_ = (bbox_area(bbox))
x_min = np.maximum(bbox[:,0], clip_box[0]).reshape(-1,1)
y_min = np.maximum(bbox[:,1], clip_box[1]).reshape(-1,1)
x_max = np.minimum(bbox[:,2], clip_box[2]).reshape(-1,1)
y_max = np.minimum(bbox[:,3], clip_box[3]).reshape(-1,1)
bbox = np.hstack((x_min, y_min, x_max, y_max, bbox[:,4:]))
delta_area = ((ar_ - bbox_area(bbox))/ar_)
mask = (delta_area < (1 - alpha)).astype(int)
bbox = bbox[mask == 1,:]
return bbox
# In[7]:
def rotate_box(corners,angle, cx, cy, h, w):
corners = corners.reshape(-1,2)
corners = np.hstack((corners, np.ones((corners.shape[0],1), dtype = type(corners[0][0]))))
M = cv2.getRotationMatrix2D((cx, cy), angle, 1.0)
cos = np.abs(M[0, 0])
sin = np.abs(M[0, 1])
nW = int((h * sin) + (w * cos))
nH = int((h * cos) + (w * sin))
# adjust the rotation matrix to take into account translation
M[0, 2] += (nW / 2) - cx
M[1, 2] += (nH / 2) - cy
# Prepare the vector to be transformed
calculated = | np.dot(M,corners.T) | numpy.dot |
import numpy as np
from .Gaussianformula.baseFunc import *
from .Gaussianformula.ordering import *
from .Gaussianformula.gates import *
import matplotlib.pyplot as plt
GATE_SET = {
"D": Dgate,
"BS": BSgate,
"S": Sgate,
"R": Rgate,
"XS": Sgate,
"PS": PSgate,
"X": Xgate,
"Z": Zgate,
"TMS": TMSgate,
"MeasX": MeasX,
"MeasP": MeasP
}
class Gaussian():
"""
Class for continuous variable quantum compting in Gaussian formula.
This class can only deal with gaussian states and gaussian gate.
"""
def __init__(self, N):
self.N = N
self.V = (np.eye(2 * N)) * 0.5
self.mu = np.zeros(2 * N)
self.ops = []
self.creg = [[None, None] for i in range(self.N)] # [x, p]
def __getattr__(self, name):
if name in GATE_SET:
self.ops.append(GATE_SET[name])
return self._setGateParam
else:
raise AttributeError('The state method does not exist')
def _setGateParam(self, *args, **kwargs):
self.ops[-1] = self.ops[-1](self, *args, **kwargs)
return self
def Creg(self, idx, var, scale = 1):
return CregReader(self.creg, idx, var, scale)
def run(self):
"""
Run the circuit.
"""
for gate in self.ops:
[self.mu, self.V] = gate.run(state = [self.mu, self.V])
return self
def mean(self, idx):
res = np.copy(self.mu[2 * idx:2 * idx + 2])
return res
def cov(self, idx):
res = np.copy(self.V[(2 * idx):(2 * idx + 2), (2 * idx):(2 * idx + 2)])
return res
def Wigner(self, idx, plot = 'y', xrange = 5.0, prange = 5.0):
"""
Calculate the Wigner function of a selected mode.
Args:
mode (int): Selecting a optical mode
plot: If 'y', the plot of wigner function is output using matplotlib. If 'n', only the meshed values are returned
x(p)range: The range in phase space for calculateing Wigner function
"""
idx = idx * 2
x = | np.arange(-xrange, xrange, 0.1) | numpy.arange |
"""
Code for training and evaluating Pytorch models.
"""
from torch.nn.modules.loss import MarginRankingLoss, CrossEntropyLoss
from torch.optim.lr_scheduler import ReduceLROnPlateau
import logging
import numpy as np
import os
import pprint
import time
import torch
import torch.optim as optim
import models
import utils
CUDA = torch.cuda.is_available()
CONFIG = utils.read_config()
LOGGER = logging.getLogger(os.path.basename(__file__))
def calc_losses(y_hats, y, out_dims):
"""
Calculate all losses across all prediction tasks.
Also reformats 'predictions' to be a friendly pytorch tensor for later use.
TODO: this should be a class?
"""
reg_loss = MarginRankingLoss()
clf_loss = CrossEntropyLoss()
if CUDA:
reg_loss = reg_loss.cuda()
clf_loss = clf_loss.cuda()
losses, predictions = [], []
for i, out_dim in enumerate(out_dims):
y_hat = y_hats[i]
y_tru = y[:, i]
# Regression case.
if out_dim == 1:
# Required for margin ranking loss.
y_rank = get_paired_ranks(y_tru)
y1_hat, y2_hat = get_pairs(y_hat)
losses.append(reg_loss(y1_hat, y2_hat, y_rank))
predictions.append(y_hat)
# Classification case.
elif out_dim > 1:
# Cross entropy loss.
losses.append(clf_loss(y_hat, y_tru.long()))
_, preds = torch.max(y_hat.data, 1)
predictions.append(preds.float().unsqueeze(1))
predictions = torch.cat(predictions, dim=1)
return(losses, predictions)
def get_pairs(y):
"""
For an input vector y, returns vectors y1 and y2 such that y1-y2 gives all
unique pairwise subtractions possible in y.
"""
y = y.cpu()
n = len(y)
idx_y2, idx_y1 = np.where(np.tril(np.ones((n, n)), k=-1))
y1 = y[torch.LongTensor(idx_y1)]
y2 = y[torch.LongTensor(idx_y2)]
if CUDA:
y1 = y1.cuda()
y2 = y2.cuda()
return(y1, y2)
def get_paired_ranks(y):
"""
Generate y_rank (for margin ranking loss). If `y == 1` then it assumed the
first input should be ranked higher (have a larger value) than the second
input, and vice-versa for `y == -1`.
"""
y = y.cpu()
# Calculates all pairwise subtractions.
y_rank = y[np.newaxis, :] - y[:, np.newaxis]
# Edge case where the difference between 2 points is 0.
y_rank[y_rank == 0] = 1e-19
# Get the lower triangle of y_rank (ignoring the diagonal).
idx = np.where(np.tril(y_rank, k=-1))
# Order: y1-y2, y1-y3, y2-y3, y1-y4, y2-y4, y3-y4 ... (lower triangle).
y_rank = y_rank[idx[0], idx[1]]
y_rank[y_rank > 0] = 1
y_rank[y_rank <= 0] = -1
if CUDA:
y_rank = y_rank.cuda()
# Make a column vector.
y_rank = y_rank[:, np.newaxis]
return(y_rank)
def check_predictions(all_y_hats, all_y_trus):
"""Check model predictions."""
all_y_hats = torch.cat(all_y_hats, dim=0).cpu().detach().numpy()
all_y_trus = torch.cat(all_y_trus, dim=0).cpu().numpy()
total_score, pr_mu, rt_mu, rr_std, id_recall = utils.score_performance(
all_y_hats[:, 0], all_y_trus[:, 0],
all_y_hats[:, 1], all_y_trus[:, 1],
all_y_hats[:, 2], all_y_trus[:, 2],
all_y_hats[:, 3].astype(np.int32), all_y_trus[:, 3].astype(np.int32))
return(total_score, pr_mu, rt_mu, rr_std, id_recall)
def train_loop(mdl, optimizer, loader):
"""Train model using the supplied learning rate optimizer and scheduler."""
mdl.train(True)
mean_loss = 0.0
all_y_hats, all_y_trus = [], []
for batch_idx, (X, y) in enumerate(loader):
optimizer.zero_grad()
if CUDA:
X = X.cuda()
y = y.cuda()
y_hats = mdl.forward(X)
losses, y_hats = calc_losses(y_hats, y, mdl.out_dims)
# Backprop with sum of losses across prediction tasks.
loss = sum(losses)
loss.backward()
mean_loss += loss.item()
optimizer.step()
all_y_hats.append(y_hats)
all_y_trus.append(y)
mean_loss /= (batch_idx+1)
total_score, pr_mu, rt_mu, rr_std, id_recall = check_predictions(
all_y_hats, all_y_trus)
results = {
'scores': {'total_score': total_score, 'pr_mu': pr_mu, 'rt_mu': rt_mu,
'rr_std': rr_std, 'id_recall': id_recall},
'loss': {'mean': mean_loss}
}
return(results)
def evalu_loop(mdl, loader, return_preds=False):
"""Validation/Test evaluation loop."""
mdl.eval()
mean_loss = 0.0
all_y_hats, all_y_trus = [], []
for batch_idx, (X, y) in enumerate(loader):
if CUDA:
X = X.cuda()
y = y.cuda()
y_hats = mdl.forward(X)
losses, y_hats = calc_losses(y_hats, y, mdl.out_dims)
# Report sum of losses.
loss = sum(losses)
mean_loss += loss.item()
all_y_hats.append(y_hats)
all_y_trus.append(y)
mean_loss /= (batch_idx+1)
total_score, pr_mu, rt_mu, rr_std, id_recall = check_predictions(
all_y_hats, all_y_trus)
# If required (for evaluation), return the actual predictions made.
if return_preds:
results_y_hat = []
# Loop through epochs.
for y_hats in all_y_hats:
# Loop through each prediction (n-element list).
for y_hat in y_hats:
results_y_hat.append(y_hat.cpu().detach().numpy())
# Convert to a single numpy array.
results_y_hat = np.vstack(results_y_hat)
else:
results_y_hat = None
results = {
'scores': {'total_score': total_score, 'pr_mu': pr_mu, 'rt_mu': rt_mu,
'rr_std': rr_std, 'id_recall': id_recall},
'loss': {'mean': mean_loss},
'preds': results_y_hat
}
return(results)
def train_mdl(mdl, optimizer):
"""
Trains a submitted model using the submitted optimizer.
"""
pp = pprint.PrettyPrinter(indent=4)
LOGGER.info('+ Begin training with configuration:\n{}'.format(
pp.pformat(CONFIG)))
epochs = CONFIG['training']['epochs']
load_args = {
'batch_size': CONFIG['dataloader']['batch_size'],
'num_workers': CONFIG['dataloader']['num_workers'],
'shuffle': CONFIG['dataloader']['shuffle']}
# Shuffles data between day1=test and day2=valid.
data = utils.get_shuffled_data(
test_p=CONFIG['dataloader']['test_proportion'])
# Set up Dataloaders.
train_data = utils.Data(precomputed=data['train'], augmentation=True)
valid_data = utils.Data(precomputed=data['valid'], augmentation=False)
train_load = torch.utils.data.DataLoader(train_data, **load_args)
valid_load = torch.utils.data.DataLoader(valid_data, **load_args)
# Move model to GPU if required.
if CUDA:
mdl = mdl.cuda()
# Initial values.
valid_loss = 10000
best_valid_loss = 10000
all_train_losses, all_valid_losses = [], []
all_train_scores, all_valid_scores = [], []
# Reduce learning rate if we plateau (valid_loss does not decrease).
scheduler = ReduceLROnPlateau(optimizer,
patience=CONFIG['training']['schedule_patience'])
for ep in range(epochs):
t1 = time.time()
scheduler.step(valid_loss)
train_results = train_loop(mdl, optimizer, train_load)
valid_results = evalu_loop(mdl, valid_load)
# Keep track of per-epoch stats for plots.
all_train_losses.append(train_results['loss']['mean'])
all_valid_losses.append(valid_results['loss']['mean'])
all_train_scores.append([
train_results['scores']['pr_mu'],
train_results['scores']['rt_mu'],
train_results['scores']['rr_std'],
train_results['scores']['id_recall'],
train_results['scores']['total_score']])
all_valid_scores.append([
valid_results['scores']['pr_mu'],
valid_results['scores']['rt_mu'],
valid_results['scores']['rr_std'],
valid_results['scores']['id_recall'],
valid_results['scores']['total_score']])
# Get the best model (early stopping).
if valid_results['loss']['mean'] < best_valid_loss:
best_valid_loss = all_valid_losses[-1]
best_model = mdl.state_dict()
best_epoch = ep+1
LOGGER.info('+ New best model found: loss={}, score={}'.format(
best_valid_loss, valid_results['scores']['total_score']))
# Log training performance.
time_elapsed = time.time() - t1
msg_info = '[{}/{}] {:.2f} sec: '.format(
ep+1, epochs, time_elapsed)
msg_loss = 'loss(t/v)={:.2f}/{:.2f}, '.format(
train_results['loss']['mean'],
valid_results['loss']['mean'])
msg_scr1 = '{:.2f}/{:.2f}'.format(
train_results['scores']['pr_mu'],
valid_results['scores']['pr_mu'])
msg_scr2 = '{:.2f}/{:.2f}'.format(
train_results['scores']['rt_mu'],
valid_results['scores']['rt_mu'])
msg_scr3 = '{:.2f}/{:.2f}'.format(
train_results['scores']['rr_std'],
valid_results['scores']['rr_std'])
msg_scr4 = '{:.2f}/{:.2f}'.format(
train_results['scores']['id_recall'],
valid_results['scores']['id_recall'])
msg_scrt = '{:.2f}/{:.2f}'.format(
train_results['scores']['total_score'],
valid_results['scores']['total_score'])
msg_task = 'scores(t/v)=[{} + {} + {} + {} = {}]'.format(
msg_scr1, msg_scr2, msg_scr3, msg_scr4, msg_scrt)
LOGGER.info(msg_info + msg_loss + msg_task)
# Early stopping patience breaks training if we are just overfitting.
if ep+1 >= best_epoch + CONFIG['training']['early_stopping_patience']:
LOGGER.info('Impatient! No gen. improvement in {} epochs'.format(
CONFIG['training']['early_stopping_patience']))
break
# Rewind to best epoch.
LOGGER.info('Early Stopping: Rewinding to epoch {}'.format(best_epoch))
mdl.load_state_dict(best_model)
# Stack scores into one numpy array each.
all_train_losses = np.vstack(all_train_losses)
all_train_scores = np.vstack(all_train_scores)
all_valid_losses = | np.vstack(all_valid_losses) | numpy.vstack |
import tensorflow as tf
import numpy as np
import sys
tf.logging.set_verbosity(tf.logging.ERROR)
class Judge:
def __init__(self, N_to_mask, model_dir, binary_rewards=True):
self.N_to_mask = N_to_mask
self.binary_rewards = binary_rewards
# Create the Estimator
try:
self.estimator = tf.estimator.Estimator(
model_fn=self.model_fn,
model_dir=model_dir, # directory to restore model from and save model to
)
except AttributeError:
raise Exception("Subclass needs to define a model_fn")
# Create the predictor from the present model. Important when restoring a model.
self.update_predictor()
def update_predictor(self):
# Predictors are used to get predictions fast once the model has been trained.
# We create it from an estimator.
self.predictor = tf.contrib.predictor.from_estimator(
self.estimator,
# The serving input receiver fn is witchcraft, which I don't quite understand.
# It's supposed to set up the data in a way that tensorflow can handle.
tf.estimator.export.build_raw_serving_input_receiver_fn(
# The input is a dictionary that corresponds to the data we feed the predictor.
# Each key stores a tensor that is replaced by data when the predictor is used.
{"masked_x": tf.placeholder(shape=[None, 28, 28, 2], dtype=tf.float32)}
),
)
def mask_image_batch(self, image_batch):
"""
Takes a batch of two-dimensional images, reshapes them, runs them through
mask_batch, reshapes them back to images, and returns them.
"""
shape = tf.shape(image_batch)
batch_flat = tf.reshape(image_batch, (shape[0], shape[1] * shape[2]))
mask_flat = self.mask_batch(batch_flat)
return tf.reshape(mask_flat, (shape[0], shape[1], shape[2], 2))
def mask_batch(self, batch):
"""
Create mask for each feature-vector in a batch, that contains N_to_mask nonzero features
of the input vector. Combine this with the vector to create the input for the DNN.
"""
shape = tf.shape(batch)
n_zero = tf.random.categorical(logits=self.zero_logits,num_samples=1,dtype=tf.int32)
p = tf.random_uniform(shape, 0, 1)
p = tf.where(batch > 0, p, -p) # each number is positive if > 0, else negative
_, nonzero_indices = tf.nn.top_k(p, self.N_to_mask - n_zero[0][0]) # sample positive
_, zero_indices = tf.nn.top_k(-p, n_zero[0][0]) # sample negative
indices = tf.concat([nonzero_indices,zero_indices],1)
mask = tf.one_hot(indices, shape[1], axis=1)
mask = tf.reduce_sum(mask, axis=2)
return tf.stack((mask, mask * batch), 2)
def train(self, n_steps, n_zero=0):
# Train the model
train_input_fn = tf.estimator.inputs.numpy_input_fn(
x={"x": self.train_data},
y=self.train_labels,
batch_size=self.batch_size,
num_epochs=None,
shuffle=True,
)
if type(n_zero) != list:
self.zero_logits = [[0 if i == n_zero else -np.inf for i in range(self.N_to_mask + 1)]]
else:
assert len(n_zero) == self.N_to_mask + 1
self.zero_logits = np.log(n_zero).reshape((1,-1))
self.estimator.train(input_fn=train_input_fn, steps=n_steps)
# Replace the old predictor with one created from the new estimator
self.update_predictor()
def evaluate_accuracy(self, n_zero=0):
# Evaluate the accuracy on all the eval_data
eval_input_fn = tf.estimator.inputs.numpy_input_fn(
x={"x": self.eval_data},
y=self.eval_labels,
num_epochs=1,
shuffle=False
)
if type(n_zero) != list:
self.zero_logits = [[0 if i == n_zero else -np.inf for i in range(self.N_to_mask + 1)]]
else:
assert len(n_zero) == self.N_to_mask + 1
self.zero_logits = np.log(n_zero).reshape((1,-1))
return self.estimator.evaluate(input_fn=eval_input_fn)
def evaluate_accuracy_using_predictor(self):
"""
Evaluates the test set accuracy using the tensorflow predictor instead
of the estimator. Can be useful for debugging.
"""
correct = 0
count = 0
for i in range(len(self.eval_labels)):
# print(i)
image = self.eval_data[i].flat
mask = np.zeros_like(image)
while mask.sum() < self.N_to_mask:
a = | np.random.randint(mask.shape[0]) | numpy.random.randint |
"""
Processing full slides of RREB1-TM1B_B6N-IC with pipeline v7 (modfied with colour correction):
* data generation
* training images (*0076*)
* non-overlap training images (*0077*)
* augmented training images (*0078*)
* k-folds + extra "other" for classifier (*0094*)
* segmentation
* dmap (*0086*)
* contour from dmap (*0091*)
* classifier (*0095*)
* segmentation correction (*0089*) networks"
* validation (*0096*)
Difference with pipeline v7:
* Constants added to colour channels so that the medians match the training data.
Requirements for this script to work:
1) Upload the cytometer project directory to ~/Software in the server where you are going to process the data.
2) Upload the AIDA project directory to ~/Software too.
3) Mount the network share with the histology slides onto ~/scan_srv2_cox.
4) Convert the .ndpi files to AIDA .dzi files, so that we can see the results of the segmentation.
You need to go to the server that's going to process the slides, add a list of the files you want to process to
~/Software/cytometer/tools/rebb1_pilot_full_histology_ndpi_to_dzi.sh
and run
cd ~/Software/cytometer/tools
./rebb1_pilot_full_histology_ndpi_to_dzi.sh
5) You need to have the models for the 10-folds of the pipeline that were trained on the KLF14 data.
6) To monitor the segmentation as it's being processed, you need to have AIDA running
cd ~/Software/AIDA/dist/
node aidaLocal.js &
You also need to create a soft link per .dzi file to the annotations you want to visualise for that file, whether
the non-overlapping ones, or the corrected ones. E.g.
ln -s 'RREB1-TM1B-B6N-IC-1.1a 1132-18 G1 - 2018-11-16 14.58.55_exp_0097_corrected.json' 'RREB1-TM1B-B6N-IC-1.1a 1132-18 G1 - 2018-11-16 14.58.55_exp_0097.json'
Then you can use a browser to open the AIDA web interface by visiting the URL (note that you need to be on the MRC
VPN, or connected from inside the office to get access to the titanrtx server)
http://titanrtx:3000/dashboard
You can use the interface to open a .dzi file that corresponds to an .ndpi file being segmented, and see the
annotations (segmentation) being created for it.
"""
"""
This file is part of Cytometer
Copyright 2021 Medical Research Council
SPDX-License-Identifier: Apache-2.0
Author: <NAME> <<EMAIL>>
"""
# script name to identify this experiment
experiment_id = 'rreb1_tm1b_exp_0001_pilot_full_slide_pipeline_v7.py'
# cross-platform home directory
from pathlib import Path
home = str(Path.home())
import os
from pathlib import Path
import sys
import pickle
sys.path.extend([os.path.join(home, 'Software/cytometer')])
import cytometer.utils
import cytometer.data
# Filter out INFO & WARNING messages
os.environ['TF_CPP_MIN_LOG_LEVEL'] = '2'
# # limit number of GPUs
# os.environ['CUDA_VISIBLE_DEVICES'] = '0'
os.environ['KERAS_BACKEND'] = 'tensorflow'
import time
import openslide
import numpy as np
import matplotlib.pyplot as plt
from cytometer.utils import rough_foreground_mask, bspline_resample
import PIL
# import tensorflow.compat.v1 as tf
# tf.disable_v2_behavior()
from keras import backend as K
import itertools
from shapely.geometry import Polygon
import scipy.stats
# # limit GPU memory used
# from keras.backend.tensorflow_backend import set_session
# config = tf.ConfigProto()
# config.gpu_options.per_process_gpu_memory_fraction = 0.95
# set_session(tf.Session(config=config))
DEBUG = False
SAVE_FIGS = False
pipeline_root_data_dir = os.path.join(home, 'Data/cytometer_data/klf14')
experiment_root_data_dir = os.path.join(home, 'Data/cytometer_data/rreb1')
data_dir = os.path.join(home, 'scan_srv2_cox/Liz Bentley/Grace')
figures_dir = os.path.join(experiment_root_data_dir, 'figures')
saved_models_dir = os.path.join(pipeline_root_data_dir, 'saved_models')
results_dir = os.path.join(experiment_root_data_dir, 'results')
annotations_dir = os.path.join(home, 'Data/cytometer_data/aida_data_Rreb1_tm1b/annotations')
klf14_training_colour_histogram_file = os.path.join(saved_models_dir, 'klf14_training_colour_histogram.npz')
# although we don't need k-folds here, we need this file to load the list of SVG contours that we compute the AIDA
# colourmap from
# TODO: just save a cell size - colour function, instead of having to recompute it every time
saved_extra_kfolds_filename = 'klf14_b6ntac_exp_0094_generate_extra_training_images.pickle'
# model names
dmap_model_basename = 'klf14_b6ntac_exp_0086_cnn_dmap'
contour_model_basename = 'klf14_b6ntac_exp_0091_cnn_contour_after_dmap'
classifier_model_basename = 'klf14_b6ntac_exp_0095_cnn_tissue_classifier_fcn'
correction_model_basename = 'klf14_b6ntac_exp_0089_cnn_segmentation_correction_overlapping_scaled_contours'
# full resolution image window and network expected receptive field parameters
fullres_box_size = | np.array([2751, 2751]) | numpy.array |
from io import BytesIO
from imgutils import pngify
from matplotlib.colors import hsv_to_rgb, LinearSegmentedColormap
import matplotlib.pyplot as plt
from random import random
import io
import copy
import json
import matplotlib
import numpy as np
import os
import random
import tarfile
import tempfile
import boto3
import sys
from werkzeug.utils import secure_filename
from skimage import filters
import skimage.morphology
from skimage.morphology import watershed, dilation, disk
from skimage.morphology import flood_fill, flood
from skimage.draw import circle
from skimage.measure import regionprops
from skimage.exposure import rescale_intensity
from config import S3_KEY, S3_SECRET
# Connect to the s3 service
s3 = boto3.client(
"s3",
aws_access_key_id=S3_KEY,
aws_secret_access_key=S3_SECRET
)
class ZStackReview:
def __init__(self, filename, input_bucket, output_bucket, subfolders):
self.filename = filename
self.input_bucket = input_bucket
self.output_bucket = output_bucket
self.subfolders = subfolders
self.trial = self.load(filename)
self.raw = self.trial["raw"]
self.annotated = self.trial["annotated"]
self.feature = 0
self.feature_max = self.annotated.shape[-1]
self.channel = 0
self.max_frames, self.height, self.width, self.channel_max = self.raw.shape
self.dimensions = (self.width, self.height)
#create a dictionary that has frame information about each cell
#analogous to .trk lineage but do not need relationships between cells included
self.cell_ids = {}
self.num_cells = {}
self.cell_info = {}
self.current_frame = 0
for feature in range(self.feature_max):
self.create_cell_info(feature)
self.draw_raw = False
self.max_intensity = {}
for channel in range(self.channel_max):
self.max_intensity[channel] = None
self.dtype_raw = self.raw.dtype
self.scale_factor = 2
self.save_version = 0
self.color_map = plt.get_cmap('viridis')
self.color_map.set_bad('black')
self.frames_changed = False
self.info_changed = False
@property
def readable_tracks(self):
"""
Preprocesses tracks for presentation on browser. For example,
simplifying track['frames'] into something like [0-29] instead of
[0,1,2,3,...].
"""
cell_info = copy.deepcopy(self.cell_info)
for _, feature in cell_info.items():
for _, label in feature.items():
slices = list(map(list, consecutive(label['frames'])))
slices = '[' + ', '.join(["{}".format(a[0])
if len(a) == 1 else "{}-{}".format(a[0], a[-1])
for a in slices]) + ']'
label["slices"] = str(slices)
return cell_info
def get_frame(self, frame, raw):
if raw:
frame = self.raw[frame][:,:, self.channel]
return pngify(imgarr=frame,
vmin=0,
vmax=self.max_intensity[self.channel],
cmap="cubehelix")
else:
frame = self.annotated[frame][:,:, self.feature]
frame = np.ma.masked_equal(frame, 0)
return pngify(imgarr=frame,
vmin=0,
vmax=np.max(self.cell_ids[self.feature]),
cmap=self.color_map)
def get_array(self, frame):
frame = self.annotated[frame][:,:,self.feature]
return frame
def load(self, filename):
global original_filename
original_filename = filename
s3 = boto3.client('s3')
key = self.subfolders
print(key)
response = s3.get_object(Bucket=self.input_bucket, Key= key)
return load_npz(response['Body'].read())
def action(self, action_type, info):
# change displayed channel or feature
if action_type == "change_channel":
self.action_change_channel(**info)
elif action_type == "change_feature":
self.action_change_feature(**info)
# edit mode actions
elif action_type == "handle_draw":
self.action_handle_draw(**info)
elif action_type == "threshold":
self.action_threshold(**info)
# modified click actions
elif action_type == "flood_cell":
self.action_flood_contiguous(**info)
elif action_type == "trim_pixels":
self.action_trim_pixels(**info)
# single click actions
elif action_type == "fill_hole":
self.action_fill_hole(**info)
elif action_type == "new_single_cell":
self.action_new_single_cell(**info)
elif action_type == "new_cell_stack":
self.action_new_cell_stack(**info)
elif action_type == "delete":
self.action_delete_mask(**info)
# multiple click actions
elif action_type == "replace_single":
self.action_replace_single(**info)
elif action_type == "replace":
self.action_replace(**info)
elif action_type == "swap_single_frame":
self.action_swap_single_frame(**info)
elif action_type == "swap_all_frame":
self.action_swap_all_frame(**info)
elif action_type == "watershed":
self.action_watershed(**info)
# misc
elif action_type == "predict_single":
self.action_predict_single(**info)
elif action_type == "predict_zstack":
self.action_predict_zstack(**info)
else:
raise ValueError("Invalid action '{}'".format(action_type))
def action_change_channel(self, channel):
self.channel = channel
self.frames_changed = True
def action_change_feature(self, feature):
self.feature = feature
self.frames_changed = True
def action_handle_draw(self, trace, target_value, brush_value, brush_size, erase, frame):
annotated = np.copy(self.annotated[frame,:,:,self.feature])
in_original = np.any(np.isin(annotated, brush_value))
annotated_draw = np.where(annotated==target_value, brush_value, annotated)
annotated_erase = np.where(annotated==brush_value, target_value, annotated)
for loc in trace:
# each element of trace is an array with [y,x] coordinates of array
x_loc = loc[1]
y_loc = loc[0]
brush_area = circle(y_loc, x_loc, brush_size, (self.height,self.width))
#do not overwrite or erase labels other than the one you're editing
if not erase:
annotated[brush_area] = annotated_draw[brush_area]
else:
annotated[brush_area] = annotated_erase[brush_area]
in_modified = np.any(np.isin(annotated, brush_value))
#cell deletion
if in_original and not in_modified:
self.del_cell_info(feature = self.feature, del_label = brush_value, frame = frame)
#cell addition
elif in_modified and not in_original:
self.add_cell_info(feature = self.feature, add_label = brush_value, frame = frame)
#check for image change, in case pixels changed but no new or del cell
comparison = np.where(annotated != self.annotated[frame,:,:,self.feature])
self.frames_changed = np.any(comparison)
#if info changed, self.info_changed set to true with info helper functions
self.annotated[frame,:,:,self.feature] = annotated
def action_threshold(self, y1, x1, y2, x2, frame, label):
'''
thresholds the raw image for annotation prediction within user-determined bounding box
'''
top_edge = min(y1, y2)
bottom_edge = max(y1, y2)
left_edge = min(x1, x2)
right_edge = max(x1, x2)
# pull out the selection portion of the raw frame
predict_area = self.raw[frame, top_edge:bottom_edge, left_edge:right_edge, self.channel]
# triangle threshold picked after trying a few on one dataset
# may not be the best threshold approach for other datasets!
# pick two thresholds to use hysteresis thresholding strategy
threshold = filters.threshold_triangle(image = predict_area)
threshold_stringent = 1.10 * threshold
# try to keep stray pixels from appearing
hyst = filters.apply_hysteresis_threshold(image = predict_area, low = threshold, high = threshold_stringent)
ann_threshold = np.where(hyst, label, 0)
#put prediction in without overwriting
predict_area = self.annotated[frame, top_edge:bottom_edge, left_edge:right_edge, self.feature]
safe_overlay = np.where(predict_area == 0, ann_threshold, predict_area)
self.annotated[frame,top_edge:bottom_edge,left_edge:right_edge,self.feature] = safe_overlay
# don't need to update cell_info unless an annotation has been added
if np.any(np.isin(self.annotated[frame,:,:,self.feature], label)):
self.add_cell_info(feature=self.feature, add_label=label, frame = frame)
def action_flood_contiguous(self, label, frame, x_location, y_location):
'''
flood fill a cell with a unique new label; alternative to watershed
for fixing duplicate label issue if cells are not touching
'''
img_ann = self.annotated[frame,:,:,self.feature]
old_label = label
new_label = np.max(self.cell_ids[self.feature]) + 1
in_original = np.any(np.isin(img_ann, old_label))
filled_img_ann = flood_fill(img_ann, (int(y_location/self.scale_factor), int(x_location/self.scale_factor)), new_label)
self.annotated[frame,:,:,self.feature] = filled_img_ann
in_modified = np.any(np.isin(filled_img_ann, old_label))
# update cell info dicts since labels are changing
self.add_cell_info(feature=self.feature, add_label=new_label, frame = frame)
if in_original and not in_modified:
self.del_cell_info(feature = self.feature, del_label = old_label, frame = frame)
def action_trim_pixels(self, label, frame, x_location, y_location):
'''
get rid of any stray pixels of selected label; pixels of value label
that are not connected to the cell selected will be removed from annotation in that frame
'''
img_ann = self.annotated[frame,:,:,self.feature]
contig_cell = flood(image = img_ann, seed_point = (int(y_location/self.scale_factor), int(x_location/self.scale_factor)))
img_trimmed = np.where(np.logical_and(np.invert(contig_cell), img_ann == label), 0, img_ann)
#check if image changed
comparison = np.where(img_trimmed != img_ann)
self.frames_changed = np.any(comparison)
#this action should never change the cell info
self.annotated[frame,:,:,self.feature] = img_trimmed
def action_fill_hole(self, label, frame, x_location, y_location):
'''
fill a "hole" in a cell annotation with the cell label. Doesn't check
if annotation at (y,x) is zero (hole to fill) because that logic is handled in
javascript. Just takes the click location, scales it to match the actual annotation
size, then fills the hole with label (using skimage flood_fill). connectivity = 1
prevents hole fill from spilling out into background in some cases
'''
# rescale click location -> corresponding location in annotation array
hole_fill_seed = (y_location // self.scale_factor, x_location // self.scale_factor)
# fill hole with label
img_ann = self.annotated[frame,:,:,self.feature]
filled_img_ann = flood_fill(img_ann, hole_fill_seed, label, connectivity = 1)
self.annotated[frame,:,:,self.feature] = filled_img_ann
#never changes info but always changes annotation
self.frames_changed = True
def action_new_single_cell(self, label, frame):
"""
Create new label in just one frame
"""
old_label, single_frame = label, frame
new_label = np.max(self.cell_ids[self.feature]) + 1
# replace frame labels
frame = self.annotated[single_frame,:,:,self.feature]
frame[frame == old_label] = new_label
# replace fields
self.del_cell_info(feature = self.feature, del_label = old_label, frame = single_frame)
self.add_cell_info(feature = self.feature, add_label = new_label, frame = single_frame)
def action_new_cell_stack(self, label, frame):
"""
Creates new cell label and replaces original label with it in all subsequent frames
"""
old_label, start_frame = label, frame
new_label = np.max(self.cell_ids[self.feature]) + 1
# replace frame labels
for frame in self.annotated[start_frame:,:,:,self.feature]:
frame[frame == old_label] = new_label
for frame in range(self.annotated.shape[0]):
if new_label in self.annotated[frame,:,:,self.feature]:
self.del_cell_info(feature = self.feature, del_label = old_label, frame = frame)
self.add_cell_info(feature = self.feature, add_label = new_label, frame = frame)
def action_delete_mask(self, label, frame):
'''
remove selected annotation from frame, replacing with zeros
'''
ann_img = self.annotated[frame,:,:,self.feature]
ann_img = np.where(ann_img == label, 0, ann_img)
self.annotated[frame,:,:,self.feature] = ann_img
#update cell_info
self.del_cell_info(feature = self.feature, del_label = label, frame = frame)
def action_replace_single(self, label_1, label_2, frame):
'''
replaces label_2 with label_1, but only in one frame. Frontend checks
to make sure labels are different and were selected within same frames
before sending action
'''
annotated = self.annotated[frame,:,:,self.feature]
# change annotation
annotated = np.where(annotated == label_2, label_1, annotated)
self.annotated[frame,:,:,self.feature] = annotated
# update info
self.add_cell_info(feature = self.feature, add_label = label_1, frame = frame)
self.del_cell_info(feature = self.feature, del_label = label_2, frame = frame)
def action_replace(self, label_1, label_2):
"""
Replacing label_2 with label_1. Frontend checks to make sure these labels
are different before sending action
"""
# check each frame
for frame in range(self.annotated.shape[0]):
annotated = self.annotated[frame,:,:,self.feature]
# if label being replaced is present, remove it from image and update cell info dict
if np.any(np.isin(annotated, label_2)):
annotated = np.where(annotated == label_2, label_1, annotated)
self.annotated[frame,:,:,self.feature] = annotated
self.add_cell_info(feature = self.feature, add_label = label_1, frame = frame)
self.del_cell_info(feature = self.feature, del_label = label_2, frame = frame)
def action_swap_single_frame(self, label_1, label_2, frame):
ann_img = self.annotated[frame,:,:,self.feature]
ann_img = np.where(ann_img == label_1, -1, ann_img)
ann_img = np.where(ann_img == label_2, label_1, ann_img)
ann_img = np.where(ann_img == -1, label_2, ann_img)
self.annotated[frame,:,:,self.feature] = ann_img
self.frames_changed = self.info_changed = True
def action_swap_all_frame(self, label_1, label_2):
for frame in range(self.annotated.shape[0]):
ann_img = self.annotated[frame,:,:,self.feature]
ann_img = np.where(ann_img == label_1, -1, ann_img)
ann_img = np.where(ann_img == label_2, label_1, ann_img)
ann_img = np.where(ann_img == -1, label_2, ann_img)
self.annotated[frame,:,:,self.feature] = ann_img
#update cell_info
cell_info_1 = self.cell_info[self.feature][label_1].copy()
cell_info_2 = self.cell_info[self.feature][label_2].copy()
self.cell_info[self.feature][label_1].update({'frames': cell_info_2['frames']})
self.cell_info[self.feature][label_2].update({'frames': cell_info_1['frames']})
self.frames_changed = self.info_changed = True
def action_watershed(self, label, frame, x1_location, y1_location, x2_location, y2_location):
# Pull the label that is being split and find a new valid label
current_label = label
new_label = np.max(self.cell_ids[self.feature]) + 1
# Locally store the frames to work on
img_raw = self.raw[frame,:,:,self.channel]
img_ann = self.annotated[frame,:,:,self.feature]
# Pull the 2 seed locations and store locally
# define a new seeds labeled img that is the same size as raw/annotation imgs
seeds_labeled = np.zeros(img_ann.shape)
# create two seed locations
seeds_labeled[int(y1_location/self.scale_factor ), int(x1_location/self.scale_factor)]=current_label
seeds_labeled[int(y2_location/self.scale_factor ), int(x2_location/self.scale_factor )]=new_label
# define the bounding box to apply the transform on and select appropriate sections of 3 inputs (raw, seeds, annotation mask)
props = regionprops(np.squeeze(np.int32(img_ann == current_label)))
minr, minc, maxr, maxc = props[0].bbox
# store these subsections to run the watershed on
img_sub_raw = np.copy(img_raw[minr:maxr, minc:maxc])
img_sub_ann = np.copy(img_ann[minr:maxr, minc:maxc])
img_sub_seeds = np.copy(seeds_labeled[minr:maxr, minc:maxc])
# contrast adjust the raw image to assist the transform
img_sub_raw_scaled = rescale_intensity(img_sub_raw)
# apply watershed transform to the subsections
ws = watershed(-img_sub_raw_scaled, img_sub_seeds, mask=img_sub_ann.astype(bool))
# did watershed effectively create a new label?
new_pixels = np.count_nonzero(np.logical_and(ws == new_label, img_sub_ann == current_label))
# if only a few pixels split, dilate them; new label is "brightest"
# so will expand over other labels and increase area
if new_pixels < 5:
ws = dilation(ws, disk(3))
# ws may only leave a few pixels of old label
old_pixels = np.count_nonzero(ws == current_label)
if old_pixels < 5:
# create dilation image so "dimmer" label is not eroded by "brighter" label
dilated_ws = dilation(np.where(ws==current_label, ws, 0), disk(3))
ws = np.where(dilated_ws==current_label, dilated_ws, ws)
# only update img_sub_ann where ws has changed label from current_label to new_label
img_sub_ann = np.where(np.logical_and(ws == new_label,img_sub_ann == current_label), ws, img_sub_ann)
# reintegrate subsection into original mask
img_ann[minr:maxr, minc:maxc] = img_sub_ann
self.annotated[frame,:,:,self.feature] = img_ann
#update cell_info dict only if new label was created with ws
if np.any(np.isin(self.annotated[frame,:,:,self.feature], new_label)):
self.add_cell_info(feature=self.feature, add_label=new_label, frame = frame)
def action_predict_single(self, frame):
'''
predicts zstack relationship for current frame based on previous frame
useful for finetuning corrections one frame at a time
'''
annotated = self.annotated[:,:,:,self.feature]
current_slice = frame
if current_slice > 0:
prev_slice = current_slice - 1
img = self.annotated[prev_slice,:,:,self.feature]
next_img = self.annotated[current_slice,:,:,self.feature]
updated_slice = predict_zstack_cell_ids(img, next_img)
#check if image changed
comparison = np.where(next_img != updated_slice)
self.frames_changed = np.any(comparison)
#if the image changed, update self.annotated and remake cell info
if self.frames_changed:
self.annotated[current_slice,:,:,int(self.feature)] = updated_slice
self.create_cell_info(feature = int(self.feature))
def action_predict_zstack(self):
'''
use location of cells in image to predict which annotations are
different slices of the same cell
'''
annotated = self.annotated[:,:,:,self.feature]
for zslice in range(self.annotated.shape[0] -1):
img = self.annotated[zslice,:,:,self.feature]
next_img = self.annotated[zslice + 1,:,:,self.feature]
predicted_next = predict_zstack_cell_ids(img, next_img)
self.annotated[zslice + 1,:,:,self.feature] = predicted_next
#remake cell_info dict based on new annotations
self.frames_changed = True
self.create_cell_info(feature = self.feature)
def action_save_zstack(self):
save_file = self.filename + "_save_version_{}.npz".format(self.save_version)
# save secure version of data before storing on regular file system
file = secure_filename(save_file)
np.savez(file, raw = self.raw, annotated = self.annotated)
path = self.subfolders
s3.upload_file(file, self.output_bucket, path)
def add_cell_info(self, feature, add_label, frame):
'''
helper function for actions that add a cell to the npz
'''
#if cell already exists elsewhere in npz:
add_label = int(add_label)
try:
old_frames = self.cell_info[feature][add_label]['frames']
updated_frames = np.append(old_frames, frame)
updated_frames = np.unique(updated_frames).tolist()
self.cell_info[feature][add_label].update({'frames': updated_frames})
#cell does not exist anywhere in npz:
except KeyError:
self.cell_info[feature].update({add_label: {}})
self.cell_info[feature][add_label].update({'label': str(add_label)})
self.cell_info[feature][add_label].update({'frames': [frame]})
self.cell_info[feature][add_label].update({'slices': ''})
self.cell_ids[feature] = np.append(self.cell_ids[feature], add_label)
self.num_cells[feature] += 1
#if adding cell, frames and info have necessarily changed
self.frames_changed = self.info_changed = True
def del_cell_info(self, feature, del_label, frame):
'''
helper function for actions that remove a cell from the npz
'''
#remove cell from frame
old_frames = self.cell_info[feature][del_label]['frames']
updated_frames = np.delete(old_frames, np.where(old_frames == np.int64(frame))).tolist()
self.cell_info[feature][del_label].update({'frames': updated_frames})
#if that was the last frame, delete the entry for that cell
if self.cell_info[feature][del_label]['frames'] == []:
del self.cell_info[feature][del_label]
#also remove from list of cell_ids
ids = self.cell_ids[feature]
self.cell_ids[feature] = np.delete(ids, np.where(ids == np.int64(del_label)))
#if deleting cell, frames and info have necessarily changed
self.frames_changed = self.info_changed = True
def create_cell_info(self, feature):
'''
helper function for actions that make or remake the entire cell info dict
'''
feature = int(feature)
annotated = self.annotated[:,:,:,feature]
self.cell_ids[feature] = np.unique(annotated)[np.nonzero(np.unique(annotated))]
self.num_cells[feature] = int(max(self.cell_ids[feature]))
self.cell_info[feature] = {}
for cell in self.cell_ids[feature]:
cell = int(cell)
self.cell_info[feature][cell] = {}
self.cell_info[feature][cell]['label'] = str(cell)
self.cell_info[feature][cell]['frames'] = []
for frame in range(self.annotated.shape[0]):
if cell in annotated[frame,:,:]:
self.cell_info[feature][cell]['frames'].append(int(frame))
self.cell_info[feature][cell]['slices'] = ''
self.info_changed = True
def create_lineage(self):
for cell in self.cell_ids[self.feature]:
self.lineage[str(cell)] = {}
cell_info = self.lineage[str(cell)]
cell_info["label"] = int(cell)
cell_info["daughters"] = []
cell_info["frame_div"] = None
cell_info["parent"] = None
cell_info["capped"] = False
cell_info["frames"] = self.cell_info[self.feature][cell]['frames']
#_______________________________________________________________________________________________________________
class TrackReview:
def __init__(self, filename, input_bucket, output_bucket, subfolders):
self.filename = filename
self.input_bucket = input_bucket
self.output_bucket = output_bucket
self.subfolders = subfolders
self.trial = self.load(filename)
self.raw = self.trial["raw"]
self.tracked = self.trial["tracked"]
# lineages is a list of dictionaries. There should be only a single one
# when using a .trk file
if len(self.trial["lineages"]) != 1:
raise ValueError("Input file has multiple trials/lineages.")
self.tracks = self.trial["lineages"][0]
self.max_frames = self.raw.shape[0]
self.dimensions = self.raw.shape[1:3][::-1]
self.width, self.height = self.dimensions
self.scale_factor = 2
self.color_map = plt.get_cmap('viridis')
self.color_map.set_bad('black')
self.current_frame = 0
self.frames_changed = False
self.info_changed = False
@property
def readable_tracks(self):
"""
Preprocesses tracks for presentation on browser. For example,
simplifying track['frames'] into something like [0-29] instead of
[0,1,2,3,...].
"""
tracks = copy.deepcopy(self.tracks)
for _, track in tracks.items():
frames = list(map(list, consecutive(track["frames"])))
frames = '[' + ', '.join(["{}".format(a[0])
if len(a) == 1 else "{}-{}".format(a[0], a[-1])
for a in frames]) + ']'
track["frames"] = frames
return tracks
def get_frame(self, frame, raw):
self.current_frame = frame
if raw:
frame = self.raw[frame][:,:,0]
return pngify(imgarr=frame,
vmin=0,
vmax=None,
cmap="cubehelix")
else:
frame = self.tracked[frame][:,:,0]
frame = np.ma.masked_equal(frame, 0)
return pngify(imgarr=frame,
vmin=0,
vmax=max(self.tracks),
cmap=self.color_map)
def get_array(self, frame):
frame = self.tracked[frame][:,:,0]
return frame
def load(self, filename):
global original_filename
original_filename = filename
s3 = boto3.client('s3')
response = s3.get_object(Bucket=self.input_bucket, Key=self.subfolders)
return load_trks(response['Body'].read())
def action(self, action_type, info):
# edit mode action
if action_type == "handle_draw":
self.action_handle_draw(**info)
# modified click actions
elif action_type == "flood_cell":
self.action_flood_contiguous(**info)
elif action_type == "trim_pixels":
self.action_trim_pixels(**info)
# single click actions
elif action_type == "fill_hole":
self.action_fill_hole(**info)
elif action_type == "create_single_new":
self.action_new_single_cell(**info)
elif action_type == "create_all_new":
self.action_new_track(**info)
elif action_type == "delete_cell":
self.action_delete(**info)
# multiple click actions
elif action_type == "set_parent":
self.action_set_parent(**info)
elif action_type == "replace":
self.action_replace(**info)
elif action_type == "swap_single_frame":
self.action_swap_single_frame(**info)
elif action_type == "swap_tracks":
self.action_swap_tracks(**info)
elif action_type == "watershed":
self.action_watershed(**info)
# misc
elif action_type == "save_track":
self.action_save_track(**info)
else:
raise ValueError("Invalid action '{}'".format(action_type))
def action_handle_draw(self, trace, edit_value, brush_size, erase, frame):
annotated = np.copy(self.tracked[frame])
in_original = np.any(np.isin(annotated, edit_value))
annotated_draw = np.where(annotated==0, edit_value, annotated)
annotated_erase = np.where(annotated==edit_value, 0, annotated)
for loc in trace:
# each element of trace is an array with [y,x] coordinates of array
x_loc = loc[1]
y_loc = loc[0]
brush_area = circle(y_loc, x_loc, brush_size, (self.height,self.width))
#do not overwrite or erase labels other than the one you're editing
if not erase:
annotated[brush_area] = annotated_draw[brush_area]
else:
annotated[brush_area] = annotated_erase[brush_area]
in_modified = np.any(np.isin(annotated, edit_value))
# cell deletion
if in_original and not in_modified:
self.del_cell_info(del_label = edit_value, frame = frame)
# cell addition
elif in_modified and not in_original:
self.add_cell_info(add_label = edit_value, frame = frame)
comparison = np.where(annotated != self.tracked[frame])
self.frames_changed = np.any(comparison)
self.tracked[frame] = annotated
def action_flood_contiguous(self, label, frame, x_location, y_location):
'''
flood fill a cell with a unique new label; alternative to watershed
for fixing duplicate label issue if cells are not touching
'''
img_ann = self.tracked[frame,:,:,0]
old_label = label
new_label = max(self.tracks) + 1
in_original = np.any(np.isin(img_ann, old_label))
filled_img_ann = flood_fill(img_ann, (int(y_location/self.scale_factor), int(x_location/self.scale_factor)), new_label)
self.tracked[frame,:,:,0] = filled_img_ann
in_modified = np.any(np.isin(filled_img_ann, old_label))
# update cell info dicts since labels are changing
self.add_cell_info(add_label=new_label, frame = frame)
if in_original and not in_modified:
self.del_cell_info(del_label = old_label, frame = frame)
def action_trim_pixels(self, label, frame, x_location, y_location):
'''
get rid of any stray pixels of selected label; pixels of value label
that are not connected to the cell selected will be removed from annotation in that frame
'''
img_ann = self.tracked[frame,:,:,0]
contig_cell = flood(image = img_ann, seed_point = (int(y_location/self.scale_factor), int(x_location/self.scale_factor)))
img_trimmed = np.where(np.logical_and(np.invert(contig_cell), img_ann == label), 0, img_ann)
comparison = np.where(img_trimmed != img_ann)
self.frames_changed = np.any(comparison)
self.tracked[frame,:,:,0] = img_trimmed
def action_fill_hole(self, label, frame, x_location, y_location):
'''
fill a "hole" in a cell annotation with the cell label. Doesn't check
if annotation at (y,x) is zero (hole to fill) because that logic is handled in
javascript. Just takes the click location, scales it to match the actual annotation
size, then fills the hole with label (using skimage flood_fill). connectivity = 1
prevents hole fill from spilling out into background in some cases
'''
# rescale click location -> corresponding location in annotation array
hole_fill_seed = (y_location // self.scale_factor, x_location // self.scale_factor)
# fill hole with label
img_ann = self.tracked[frame,:,:,0]
filled_img_ann = flood_fill(img_ann, hole_fill_seed, label, connectivity = 1)
self.tracked[frame,:,:,0] = filled_img_ann
self.frames_changed = True
def action_new_single_cell(self, label, frame):
"""
Create new label in just one frame
"""
old_label = label
new_label = max(self.tracks) + 1
# replace frame labels
self.tracked[frame] = np.where(self.tracked[frame] == old_label,
new_label, self.tracked[frame])
# replace fields
self.del_cell_info(del_label = old_label, frame = frame)
self.add_cell_info(add_label = new_label, frame = frame)
def action_new_track(self, label, frame):
"""
Replacing label - create in all subsequent frames
"""
old_label, start_frame = label, frame
new_label = max(self.tracks) + 1
if start_frame != 0:
# replace frame labels
for frame in self.tracked[start_frame:]:
frame[frame == old_label] = new_label
# replace fields
track_old = self.tracks[old_label]
track_new = self.tracks[new_label] = {}
idx = track_old["frames"].index(start_frame)
frames_before = track_old["frames"][:idx]
frames_after = track_old["frames"][idx:]
track_old["frames"] = frames_before
track_new["frames"] = frames_after
track_new["label"] = new_label
# only add daughters if they aren't in the same frame as the new track
track_new["daughters"] = []
for d in track_old["daughters"]:
if start_frame not in self.tracks[d]["frames"]:
track_new["daughters"].append(d)
track_new["frame_div"] = track_old["frame_div"]
track_new["capped"] = track_old["capped"]
track_new["parent"] = None
track_old["daughters"] = []
track_old["frame_div"] = None
track_old["capped"] = True
self.frames_changed = self.info_changed = True
def action_delete(self, label, frame):
"""
Deletes label from current frame only
"""
# Set frame labels to 0
ann_img = self.tracked[frame]
ann_img = np.where(ann_img == label, 0, ann_img)
self.tracked[frame] = ann_img
self.del_cell_info(del_label = label, frame = frame)
def action_set_parent(self, label_1, label_2):
"""
label_1 gave birth to label_2
"""
track_1 = self.tracks[label_1]
track_2 = self.tracks[label_2]
last_frame_parent = max(track_1['frames'])
first_frame_daughter = min(track_2['frames'])
if last_frame_parent < first_frame_daughter:
track_1["daughters"].append(label_2)
daughters = np.unique(track_1["daughters"]).tolist()
track_1["daughters"] = daughters
track_2["parent"] = label_1
if track_1["frame_div"] is None:
track_1["frame_div"] = first_frame_daughter
else:
track_1["frame_div"] = min(track_1["frame_div"], first_frame_daughter)
self.info_changed = True
def action_replace(self, label_1, label_2):
"""
Replacing label_2 with label_1
"""
# replace arrays
for frame in range(self.max_frames):
annotated = self.tracked[frame]
annotated = np.where(annotated == label_2, label_1, annotated)
self.tracked[frame] = annotated
# replace fields
track_1 = self.tracks[label_1]
track_2 = self.tracks[label_2]
for d in track_1["daughters"]:
self.tracks[d]["parent"] = None
track_1["frames"].extend(track_2["frames"])
track_1["frames"] = sorted(set(track_1["frames"]))
track_1["daughters"] = track_2["daughters"]
track_1["frame_div"] = track_2["frame_div"]
track_1["capped"] = track_2["capped"]
del self.tracks[label_2]
for _, track in self.tracks.items():
try:
track["daughters"].remove(label_2)
except ValueError:
pass
self.frames_changed = self.info_changed = True
def action_swap_single_frame(self, label_1, label_2, frame):
'''swap the labels of two cells in one frame, but do not
change any of the lineage information'''
ann_img = self.tracked[frame,:,:,0]
ann_img = np.where(ann_img == label_1, -1, ann_img)
ann_img = np.where(ann_img == label_2, label_1, ann_img)
ann_img = np.where(ann_img == -1, label_2, ann_img)
self.tracked[frame,:,:,0] = ann_img
self.frames_changed = True
def action_swap_tracks(self, label_1, label_2):
def relabel(old_label, new_label):
for frame in self.tracked:
frame[frame == old_label] = new_label
# replace fields
track_new = self.tracks[new_label] = self.tracks[old_label]
track_new["label"] = new_label
del self.tracks[old_label]
for d in track_new["daughters"]:
self.tracks[d]["parent"] = new_label
if track_new["parent"] is not None:
parent_track = self.tracks[track_new["parent"]]
parent_track["daughters"].remove(old_label)
parent_track["daughters"].append(new_label)
relabel(label_1, -1)
relabel(label_2, label_1)
relabel(-1, label_2)
self.frames_changed = self.info_changed = True
def action_watershed(self, label, frame, x1_location, y1_location, x2_location, y2_location):
# Pull the label that is being split and find a new valid label
current_label = label
new_label = max(self.tracks) + 1
# Locally store the frames to work on
img_raw = self.raw[frame,:,:,0]
img_ann = self.tracked[frame,:,:,0]
# Pull the 2 seed locations and store locally
# define a new seeds labeled img that is the same size as raw/annotation imgs
seeds_labeled = np.zeros(img_ann.shape)
# create two seed locations
seeds_labeled[int(y1_location/self.scale_factor),
int(x1_location/self.scale_factor)] = current_label
seeds_labeled[int(y2_location/self.scale_factor),
int(x2_location/self.scale_factor)] = new_label
# define the bounding box to apply the transform on and select appropriate sections of 3 inputs (raw, seeds, annotation mask)
props = regionprops(np.squeeze(np.int32(img_ann == current_label)))
minr, minc, maxr, maxc = props[0].bbox
# store these subsections to run the watershed on
img_sub_raw = np.copy(img_raw[minr:maxr, minc:maxc])
img_sub_ann = np.copy(img_ann[minr:maxr, minc:maxc])
img_sub_seeds = np.copy(seeds_labeled[minr:maxr, minc:maxc])
# contrast adjust the raw image to assist the transform
img_sub_raw_scaled = rescale_intensity(img_sub_raw)
# apply watershed transform to the subsections
ws = watershed(-img_sub_raw_scaled, img_sub_seeds, mask=img_sub_ann.astype(bool))
# did watershed effectively create a new label?
new_pixels = np.count_nonzero(np.logical_and(ws == new_label, img_sub_ann == current_label))
# if only a few pixels split, dilate them; new label is "brightest"
# so will expand over other labels and increase area
if new_pixels < 5:
ws = dilation(ws, disk(3))
# ws may only leave a few pixels of old label
old_pixels = np.count_nonzero(ws == current_label)
if old_pixels < 5:
# create dilation image so "dimmer" label is not eroded by "brighter" label
dilated_ws = dilation(np.where(ws==current_label, ws, 0), disk(3))
ws = np.where(dilated_ws==current_label, dilated_ws, ws)
# only update img_sub_ann where ws has changed label from current_label to new_label
img_sub_ann = np.where(np.logical_and(ws == new_label,img_sub_ann == current_label),
ws, img_sub_ann)
#reintegrate subsection into original mask
img_ann[minr:maxr, minc:maxc] = img_sub_ann
self.tracked[frame,:,:,0] = img_ann
#update cell_info dict only if new label was created with ws
if np.any(np.isin(self.tracked[frame,:,:,0], new_label)):
self.add_cell_info(add_label=new_label, frame = frame)
def action_save_track(self):
# clear any empty tracks before saving file
empty_tracks = []
for key in self.tracks:
if not self.tracks[key]['frames']:
empty_tracks.append(self.tracks[key]['label'])
for track in empty_tracks:
del self.tracks[track]
file = secure_filename(self.filename)
with tarfile.open(file, "w") as trks:
with tempfile.NamedTemporaryFile("w") as lineage_file:
json.dump(self.tracks, lineage_file, indent=1)
lineage_file.flush()
trks.add(lineage_file.name, "lineage.json")
with tempfile.NamedTemporaryFile() as raw_file:
np.save(raw_file, self.raw)
raw_file.flush()
trks.add(raw_file.name, "raw.npy")
with tempfile.NamedTemporaryFile() as tracked_file:
np.save(tracked_file, self.tracked)
tracked_file.flush()
trks.add(tracked_file.name, "tracked.npy")
try:
s3.upload_file(file, self.output_bucket, self.subfolders)
except Exception as e:
print("Something Happened: ", e, file=sys.stderr)
raise
#os.remove(file)
return "Success!"
def add_cell_info(self, add_label, frame):
'''
helper function for actions that add a cell to the trk
'''
#if cell already exists elsewhere in trk:
try:
old_frames = self.tracks[add_label]['frames']
updated_frames = np.append(old_frames, frame)
updated_frames = np.unique(updated_frames).tolist()
self.tracks[add_label].update({'frames': updated_frames})
#cell does not exist anywhere in trk:
except KeyError:
self.tracks.update({add_label: {}})
self.tracks[add_label].update({'label': int(add_label)})
self.tracks[add_label].update({'frames': [frame]})
self.tracks[add_label].update({'daughters': []})
self.tracks[add_label].update({'frame_div': None})
self.tracks[add_label].update({'parent': None})
self.tracks[add_label].update({'capped': False})
self.frames_changed = self.info_changed = True
def del_cell_info(self, del_label, frame):
'''
helper function for actions that remove a cell from the trk
'''
#remove cell from frame
old_frames = self.tracks[del_label]['frames']
updated_frames = np.delete(old_frames, np.where(old_frames == np.int64(frame))).tolist()
self.tracks[del_label].update({'frames': updated_frames})
#if that was the last frame, delete the entry for that cell
if self.tracks[del_label]['frames'] == []:
del self.tracks[del_label]
# If deleting lineage data, remove parent/daughter entries
for _, track in self.tracks.items():
try:
track["daughters"].remove(del_label)
except ValueError:
pass
if track["parent"] == del_label:
track["parent"] = None
self.frames_changed = self.info_changed = True
def consecutive(data, stepsize=1):
return np.split(data, np.where(np.diff(data) != stepsize)[0]+1)
def predict_zstack_cell_ids(img, next_img, threshold = 0.1):
'''
Predict labels for next_img based on intersection over union (iou)
with img. If cells don't meet threshold for iou, they don't count as
matching enough to share label with "matching" cell in img. Cells
that don't have a match in img (new cells) get a new label so that
output relabeled_next does not skip label values (unless label values
present in prior image need to be skipped to avoid conflating labels).
'''
# relabel to remove skipped values, keeps subsequent predictions cleaner
next_img = relabel_frame(next_img)
#create np array that can hold all pairings between cells in one
#image and cells in next image
iou = np.zeros((np.max(img)+1, np.max(next_img)+1))
vals = np.unique(img)
cells = vals[np.nonzero(vals)]
#nothing to predict off of
if len(cells) == 0:
return next_img
next_vals = np.unique(next_img)
next_cells = next_vals[np.nonzero(next_vals)]
#no values to reassign
if len(next_cells) == 0:
return next_img
#calculate IOUs
for i in cells:
for j in next_cells:
intersection = np.logical_and(img==i,next_img==j)
union = np.logical_or(img==i,next_img==j)
iou[i,j] = intersection.sum(axis=(0,1)) / union.sum(axis=(0,1))
#relabel cells appropriately
#relabeled_next holds cells as they get relabeled appropriately
relabeled_next = np.zeros(next_img.shape, dtype = np.uint16)
#max_indices[cell_from_next_img] -> cell from first image that matches it best
max_indices = np.argmax(iou, axis = 0)
#put cells that into new image if they've been matched with another cell
#keep track of which (next_img)cells don't have matches
#this can be if (next_img)cell matched background, or if (next_img)cell matched
#a cell already used
unmatched_cells = []
#don't reuse cells (if multiple cells in next_img match one particular cell)
used_cells_src = []
#next_cell ranges between 0 and max(next_img)
#matched_cell is which cell in img matched next_cell the best
# this for loop does the matching between cells
for next_cell, matched_cell in enumerate(max_indices):
#if more than one match, look for best match
#otherwise the first match gets linked together, not necessarily reproducible
# matched_cell != 0 prevents adding the background to used_cells_src
if matched_cell != 0 and matched_cell not in used_cells_src:
bool_matches = np.where(max_indices == matched_cell)
count_matches = np.count_nonzero(bool_matches)
if count_matches > 1:
#for a given cell in img, which next_cell has highest iou
matching_next_options = np.argmax(iou, axis =1)
best_matched_next = matching_next_options[matched_cell]
#ignore if best_matched_next is the background
if best_matched_next != 0:
if next_cell != best_matched_next:
unmatched_cells = np.append(unmatched_cells, next_cell)
continue
else:
# don't add if bad match
if iou[matched_cell][best_matched_next] > threshold:
relabeled_next = np.where(next_img == best_matched_next, matched_cell, relabeled_next)
# if it's a bad match, we still need to add next_cell back into relabeled next later
elif iou[matched_cell][best_matched_next] <= threshold:
unmatched_cells = np.append(unmatched_cells, best_matched_next)
# in either case, we want to be done with the "matched_cell" from img
used_cells_src = np.append(used_cells_src, matched_cell)
# matched_cell != 0 is still true
elif count_matches == 1:
#add the matched cell to the relabeled image
if iou[matched_cell][next_cell] > threshold:
relabeled_next = np.where(next_img == next_cell, matched_cell, relabeled_next)
else:
unmatched_cells = np.append(unmatched_cells, next_cell)
used_cells_src = np.append(used_cells_src, matched_cell)
elif matched_cell in used_cells_src and next_cell != 0:
#skip that pairing, add next_cell to unmatched_cells
unmatched_cells = | np.append(unmatched_cells, next_cell) | numpy.append |
__copyright__ = "Copyright 2013, RLPy http://www.acl.mit.edu/RLPy"
__credits__ = ["<NAME>", "<NAME>", "<NAME>",
"<NAME>", "<NAME>"]
__license__ = "BSD 3-Clause"
__author__ = ["<NAME>", "<NAME>"]
from rlpy.Agents.Agent import Agent, DescentAlgorithm
import numpy as np
class PosteriorSampling(DescentAlgorithm, Agent):
"""
Standard Posterior Sampling algorithm with normal-gaussian prior for
rewards and dirichlet for transitions.
"""
episodeCap = None
startState=None
flag_ = True
observed_rewards = None
mean_rewards = None
var_requards = None
observed = None
observed_transitions = None
bad_reward = None
def __init__(self, policy, representation, discount_factor,
initial_learn_rate=0.1,bad_reward=-1, **kwargs):
super(PosteriorSampling,self).__init__(policy=policy,
representation=representation, discount_factor=discount_factor, **kwargs)
self.logger.info("Initial learning rate:\t\t%0.2f" % initial_learn_rate)
self.episodeCap = self.representation.domain.episodeCap
self.observed_rewards = {}
self.mean_rewards= np.zeros((self.representation.features_num,self.representation.actions_num))
self.var_rewards= | np.zeros((self.representation.features_num,self.representation.actions_num)) | numpy.zeros |
#!/usr/bin/env python
# coding: utf-8
import cv2
import matplotlib.pyplot as plt
import numpy as np
import sys
import os
import math
import json
from time import sleep
# initial threshold for FAST feature (difference to center point)
iniThFast = 20
# reduce threshold for FAST, if not enough feature points were found to this
minThFast = 5
# original patch size for rotation estimation
PATCH_SIZE = 31
HALF_PATCH_SIZE = 15
# how wide shall the window image be
window_width = 1500
# initialize the fast detector, will be used later
fast = cv2.FastFeatureDetector_create(iniThFast, True)
pattern = json.load(open('pattern.json'))
# https://github.com/raulmur/ORB_SLAM2/blob/master/src/ORBextractor.cc#L150
modes = ["fast", "full", "pattern"]
def limit(val, lower, upper):
# clip given value to lower or upper limit
return min(upper, max(lower, val))
def pixel_circle(r):
# find out, which points belong to a pixel circle around a certain point
d = round(math.pi - (2 * r))
x = 0
y = r
cpoints = []
while x <= y:
cpoints.append((x, -y))
cpoints.append((y, -x))
cpoints.append((y, x))
cpoints.append((x, y))
cpoints.append((-x, y))
cpoints.append((-y, x))
cpoints.append((-y, -x))
cpoints.append((-x, -y))
if d < 0:
d += (math.pi * x) + (math.pi * 2)
else:
d += math.pi * (x - y) + (math.pi * 3)
y -= 1
x += 1
return list(set(cpoints))
def calc_umax():
# This relates to https://github.com/raulmur/ORB_SLAM2/blob/f2e6f51cdc8d067655d90a78c06261378e07e8f3/src/ORBextractor.cc#L452
# This is for orientation
# pre-compute the end of a row in a circular patch
umax = [0] * (HALF_PATCH_SIZE + 1)
vmax = int(np.floor(HALF_PATCH_SIZE * np.sqrt(2) / 2 + 1))
vmin = int(np.ceil(HALF_PATCH_SIZE * np.sqrt(2) / 2))
hp2 = HALF_PATCH_SIZE*HALF_PATCH_SIZE;
for v in range(vmax + 1):
umax[v] = int(np.round(np.sqrt(hp2 - v * v)))
# Make sure we are symmetric
v0 = 0
for v in range(HALF_PATCH_SIZE, vmin-1, -1):
while umax[v0] == umax[v0 + 1]:
v0 += 1
umax[v] = v0
v0 += 1
print('umax:', umax)
return umax
def IC_Angle(image, pt, u_max):
# this relates to https://github.com/raulmur/ORB_SLAM2/blob/master/src/ORBextractor.cc#L77
if image.ndim > 2:
image = cv2.cvtColor(image, cv2.COLOR_RGB2GRAY)
cpx = int(round(pt[1]))
cpy = int(round(pt[0]))
print('cpx/y/val', cpx, cpy, image[cpy, cpx])
m_01 = int(0)
# Treat the center line differently, v=0
m_10 = sum([u * image[cpy, cpx + u] for u in range(-HALF_PATCH_SIZE, HALF_PATCH_SIZE + 1)])
m_00 = sum([image[cpy, cpx + u] for u in range(-HALF_PATCH_SIZE, HALF_PATCH_SIZE + 1)])
# Go line by line in the circuI853lar patch
for v in range(1, HALF_PATCH_SIZE + 1):
# Proceed over the two lines
v_sum = 0;
d = u_max[v];
for u in range(-d, d + 1):
val_plus = int(image[cpy + v, cpx + u])
val_minus = int(image[cpy - v, cpx + u])
v_sum += (val_plus - val_minus)
m_10 += u * (val_plus + val_minus)
m_00 += val_plus + val_minus
m_01 += v * v_sum
# print('m_01, m_10, m_00', m_01, m_10, m_00)
angle = cv2.fastAtan2(m_01, m_10)
if m_00 == 0 or not m_00:
centerpoint_x = 0
centerpoint_y = 0
else:
centerpoint_x = int(m_10/m_00)
centerpoint_y = int(m_01/m_00)
return angle, centerpoint_x + HALF_PATCH_SIZE, centerpoint_y + HALF_PATCH_SIZE
def put_text_on_enlarged(text, x, y, thickness=1):
# a wrapper for positioning text in the upscaled version of the image
global overlay, canvas, resized
textsize = cv2.getTextSize(text, fontFace=cv2.FONT_HERSHEY_DUPLEX, fontScale=text_scale, thickness=thickness)[0]
xshift = int(((canvas.shape[1] - ls * resized.shape[1] + x * f) + (canvas.shape[1] - ls * resized.shape[1] + (x + 1) * f)) / 2)
xshift -= textsize[0] // 2
yshift = int(y * f + f / 2)
yshift += textsize[1] // 2
overlay = cv2.putText(
overlay,
text,
(xshift , yshift),
cv2.FONT_HERSHEY_COMPLEX,
text_scale,
(255),
thickness=thickness
)
# start of main
source = ""
while source != "q":
source = input("Filepath or camera index: ")
try:
source = int(source)
in_mode = 'live'
webcam = cv2.VideoCapture(0)
ret, video_frame = webcam.read()
video_frame = cv2.flip(video_frame, 1)
fast_ex = cv2.cvtColor(video_frame, cv2.COLOR_BGR2GRAY)
break
except:
if os.path.exists(source.strip()):
in_mode = 'file'
fast_ex = cv2.imread(source.strip(), cv2.IMREAD_GRAYSCALE)
break
else:
print("Could not find given path or Camera Device for {}".format(source))
exit()
center = (fast_ex.shape[0] // 2 , fast_ex.shape[1] // 2)
mode = modes[0]
umax = calc_umax()
while True:
if in_mode == 'live':
ret, video_frame = webcam.read()
video_frame = cv2.flip(video_frame, 1)
fast_ex = cv2.cvtColor(video_frame, cv2.COLOR_BGR2GRAY)
# circle radius
r = HALF_PATCH_SIZE
# calculating the text scale on base of radius. This was just interpolated with two examples
text_scale = -0.04*r+0.87
# how many pixels to pad around cirle
padding = 2
# the representation of the cropped area shall be *scale* times larger than the original height
# keeping cropped separate, to later iterate over it
scale = 1.2
cropped = fast_ex[center[0]-r-padding:center[0]+r+padding+1,
center[1]-r-padding:center[1]+r+padding+1]
resized = cv2.resize(cropped, (int(fast_ex.shape[0] *scale), int(fast_ex.shape[0]*scale)), interpolation=cv2.INTER_NEAREST)
vKeysCell = fast.detect(fast_ex[center[0]-r-padding:center[0]+r+padding+1,
center[1]-r-padding:center[1]+r+padding+1])
# create a new canvas, to paste everything into
canvas = np.ndarray((resized.shape[0], fast_ex.shape[1]+20+resized.shape[1]))
canvas.fill(255)
# where to paste the original image
paste_x1 = 0
paste_x2 = fast_ex.shape[1]
paste_y1 = int(canvas.shape[0] / 2 - fast_ex.shape[0] / 2)
paste_y2 = int(canvas.shape[0] / 2 - fast_ex.shape[0] / 2 + fast_ex.shape[0])
# paste original image
canvas[paste_y1: paste_y2, paste_x1:paste_x2] = fast_ex
# paste resized crop
canvas[:, -resized.shape[1]:] = resized
# scale up everything to make lines smoother
ls = int(np.ceil(window_width/canvas.shape[1]))
canvas = cv2.resize(canvas, (0, 0), fx=ls, fy=ls, interpolation=cv2.INTER_NEAREST)
# pasting things into an overlay, to later increase contrast (black & white)
overlay = np.ndarray(canvas.shape)
# use 128 to indicate emtpy spaces later
overlay.fill(128)
# line from rectangle to top left corner of crop
overlay = cv2.line(
overlay,
(ls * (paste_x1 + center[1]+r+padding), ls * (paste_y1 + center[0]-r-padding)),
(canvas.shape[1] - ls * resized.shape[1], 0), (255),
thickness=2
)
# line from rectangle to bottom left corner of crop
overlay = cv2.line(
overlay,
(ls * (paste_x1 + center[1]+r+padding+1), ls * (paste_y1 + center[0]+r+padding+1)),
(canvas.shape[1]- ls * resized.shape[1], canvas.shape[0]), (255),
thickness=2
)
# rectangle to indicate crop in original image
overlay = cv2.rectangle(
overlay,
(ls * (paste_x1 + center[1]-r-padding), ls * (paste_y1 + center[0]-r-padding)),
(ls * (paste_x1 + center[1]+r+padding+1), ls * (paste_y1 + center[0]+r+padding+1)), (255),
thickness=2
)
# scale factor from original crop to resized version, after scaling up everything
f = (resized.shape[0]) / cropped.shape[0] * ls
pc = pixel_circle(r)
# create vertical lines
for cx in range(cropped.shape[1]):
xshift = int(canvas.shape[1] - ls * resized.shape[1] + cx * f )
overlay = cv2.line(
overlay,
(xshift, 0),
(xshift, canvas.shape[0]),
255
)
# create horizontal lines
for cy in range(cropped.shape[0]):
overlay = cv2.line(
overlay,
(canvas.shape[1] - ls * resized.shape[1], int((1+cy) * f)),
(canvas.shape[1], int((1+cy) * f)),
255
)
# outer circle
overlay = cv2.circle(
overlay,
(int(canvas.shape[1] - ls * resized.shape[1] + cropped.shape[1] / 2 * f ), int(cropped.shape[0] / 2 * f)),
int((r + 0.6) * f),
255
)
# inner circle
overlay = cv2.circle(
overlay,
(int(canvas.shape[1] - ls * resized.shape[1] + cropped.shape[1] / 2 * f ), int(cropped.shape[0] / 2 * f)),
int((r - 0.55) * f),
255
)
if mode == "full":
# circle through all points of the circle and insert the values
for cy in range(-r, r + 1):
yp = [p[0] for p in pc if p[1] == cy]
yshift = cy+r+padding
for cx in range(min(yp), max(yp)+1):
# calculating center of upscaled pixels, with respect to the text size
thick = 1 # 2 if cy == 0 and cx == 0 else 1
textsize = cv2.getTextSize(str(cropped[cy+r+padding, cx+r+padding]), fontFace=cv2.FONT_HERSHEY_DUPLEX, fontScale=text_scale, thickness=thick)[0]
xshift = int(((canvas.shape[1] - ls * resized.shape[1] + (cx+r+padding) * f) + (canvas.shape[1] - ls * resized.shape[1] + (cx + r + padding + 1) * f)) / 2)
xshift -= textsize[0] // 2
yshift = int((cy+r+padding) * f + f / 2)
yshift += textsize[1] // 2
overlay = cv2.putText(
overlay,
str(cropped[cy+r+padding, cx+r+padding]),
(xshift , yshift),
cv2.FONT_HERSHEY_COMPLEX,
text_scale,
(255),
thickness=thick
)
if cy == 0 and cx == 0:
overlay = cv2.rectangle(
overlay,
(overlay.shape[1] - int((r+1+padding) * f + 2), int((r+padding) * f)),
(overlay.shape[1] - int((r+padding) * f - 1), int((r+1+padding) * f)),
(255),
2
)
elif mode == "fast":
# show which pixels would count into the FAST feature detection
# put values of pixels into cropped / resized image
for cx in range(cropped.shape[1]):
for cy in range(cropped.shape[0]):
if (cx-r-padding, cy-r-padding) in pc or (cx-r-padding == 0 and cy-r-padding == 0):
put_text_on_enlarged(str(cropped[cy, cx]), cx, cy)
if (cx-r-padding == 0 and cy-r-padding == 0):
# add info to point in the center
put_text_on_enlarged("[p]", cx+0.75, cy+0.25,thickness=2)
nb_angle = 2 * np.pi / len(pc)
r_plus = 1.15
for nb, nba in enumerate(np.arange(0.0, 2 * np.pi, nb_angle)):
textsize = cv2.getTextSize('[{}]'.format(nb + 1), fontFace=cv2.FONT_HERSHEY_DUPLEX, fontScale=text_scale, thickness=2)[0]
nba_x = int(np.sin(nba) * (r + r_plus) * f - overlay.shape[0] / 2 + overlay.shape[1] - textsize[0] / 2)
nba_y = int(-np.cos(nba) * (r + r_plus) * f + overlay.shape[0] / 2 + textsize[1] / 2)
overlay = cv2.putText(
overlay,
'[{}]'.format(nb + 1),
(nba_x, nba_y),
cv2.FONT_HERSHEY_COMPLEX,
text_scale,
(255),
thickness=2
)
elif mode == "pattern":
# show the first x pattern overlayed
pmax = 10
descriptor = []
d_count = 0
for a_x, a_y, b_x, b_y in pattern:
if a_x > padding + r or a_x < - padding - r or \
a_y > padding + r or a_y < - padding - r or \
b_x > padding + r or b_x < - padding - r or \
b_y > padding + r or b_y < - padding - r:
continue
if d_count > pmax:
break
if fast_ex[center[0]+ a_y, center[1]+ a_x] < fast_ex[center[0]+ b_y, center[1]+ b_x]:
descriptor.append("1")
put_text_on_enlarged("{}a".format(d_count), a_x+padding+r, a_y+padding+r)
put_text_on_enlarged("{}b".format(d_count), b_x+padding+r, b_y+padding+r, thickness=2)
else:
descriptor.append("0")
# if fast_ex[center[0]+ a_y, center[1]+ a_x] == fast_ex[center[0]+ b_y, center[1]+ b_x]:
# put_text_on_enlarged("{}a".format(d_count), a_x+padding+r, a_y+padding+r)
# else:
put_text_on_enlarged("{}a".format(d_count), a_x+padding+r, a_y+padding+r, thickness=2)
put_text_on_enlarged("{}b".format(d_count), b_x+padding+r, b_y+padding+r)
d_count += 1
# Also print this onto the image
overlay = cv2.putText(
overlay,
"Descriptor: " + " | ".join(descriptor) + " ...",
(20, overlay.shape[0] - 20),
cv2.FONT_HERSHEY_COMPLEX,
text_scale,
(255),
thickness=1
)
print("Descriptor: " + " | ".join(descriptor) + " ...")
# turning overlay into white (255) pixels, where the underlying image is darker
# and into black (0) pixels, where the underlying image is lighter
overlay[overlay != 128] = np.where(canvas > 150, 50, 200)[overlay != 128]
# pasting in the overlay
canvas[overlay != 128] = overlay[overlay != 128]
# calculate the momentums and angle of a circular patch
a, cpx, cpy = IC_Angle(fast_ex, center, umax)
print("returned", a, cpx, cpy, np.sin(np.deg2rad(a)), np.cos(np.deg2rad(a)))
# initialize an RGB canvas
rgb = cv2.cvtColor(canvas.astype('uint8'), cv2.COLOR_GRAY2BGR)
# draw a line according to the angle
xshift = int(((canvas.shape[1] - ls * resized.shape[1] + (padding + r) * f) + (canvas.shape[1] - ls * resized.shape[1] + ((padding + r) + 1) * f)) / 2)
yshift = int((padding + r) * f + f / 2)
r_red = 1
# drawing reference line and arc
rgb = cv2.line(rgb,
(xshift, yshift),
(int(xshift + (r - r_red) * f), yshift),
(255, 200, 128), 3)
rgb = cv2.ellipse(rgb, (xshift, yshift), ( int((r-r_red-1) * f), int((r-r_red-1) * f)),
0, 0, a, (210, 50, 128), 3)
# drawing arrow
a_x = int(np.cos(np.deg2rad(a)) * (r - r_red) * f - overlay.shape[0] / 2 + overlay.shape[1])
a_y = int(np.sin(np.deg2rad(a)) * (r - r_red) * f + overlay.shape[0] / 2)
rgb = cv2.line(rgb,
(xshift, yshift),
(a_x, a_y),
(128, 150, 0), 3)
a_x_l = int(np.cos(np.deg2rad((a - 150) % 360)) * (1) * f + a_x)
a_y_l = int(np.sin(np.deg2rad((a - 150) % 360)) * (1) * f + a_y)
rgb = cv2.line(rgb,
(a_x, a_y),
(a_x_l, a_y_l),
(128, 150, 0), 3)
a_x_r = int(np.cos(np.deg2rad(a + 150)) * (1) * f + a_x)
a_y_r = int(np.sin(np.deg2rad(a + 150)) * (1) * f + a_y)
rgb = cv2.line(rgb,
(a_x, a_y),
(a_x_r, a_y_r),
(128, 150, 0), 3)
cpx += padding
cpy += padding
# add the angle
rgb = cv2.putText(rgb,
'{:.2f}'.format(a),
(int(xshift + (r - r_red) * f), int(yshift + f / 2)),
cv2.FONT_HERSHEY_COMPLEX,
text_scale * 2,
(255, 200, 128),
2
)
# draw centroid
xshift = int(((canvas.shape[1] - ls * resized.shape[1] + cpx * f) + (canvas.shape[1] - ls * resized.shape[1] + (cpx + 1) * f)) / 2)
yshift = int(cpy * f + f / 2)
rgb = cv2.circle(rgb,
(
xshift,
yshift
), 3,(50, 80, 255), 3)
rgb = cv2.putText(rgb,
"C",
(int(xshift + f), int(yshift + f)),
cv2.FONT_HERSHEY_COMPLEX,
text_scale * 2,
(50, 80, 255),
2
)
# draw keypoints
vKeysCellShifted = []
for vit in vKeysCell:
xshift = int(((canvas.shape[1] - ls * resized.shape[1] + vit.pt[0] * f) + (canvas.shape[1] - ls * resized.shape[1] + (vit.pt[0] + 1) * f)) / 2)
yshift = int(vit.pt[1] * f + f / 2)
vit.pt = (xshift, yshift)
vKeysCellShifted.append(vit)
rgb = cv2.drawKeypoints(rgb, vKeysCellShifted, rgb, (255, 0, 0))
# add some information beneath the image
rgb_info = | np.zeros((rgb.shape[0] + 50, rgb.shape[1], rgb.shape[2]), dtype=np.uint8) | numpy.zeros |
# Code for initialization of NMF, copied with little modification from scikit-learn
# Original source: https://github.com/scikit-learn/scikit-learn/blob/7e1e6d09bcc2eaeba98f7e737aac2ac782f0e5f1/sklearn/decomposition/_nmf.py#L229
import numpy as np
from scipy import linalg
import warnings
from math import sqrt
import numbers
def check_random_state(seed):
"""Turn seed into a np.random.RandomState instance
Parameters
----------
seed : None, int or instance of RandomState
If seed is None, return the RandomState singleton used by np.random.
If seed is an int, return a new RandomState instance seeded with seed.
If seed is already a RandomState instance, return it.
Otherwise raise ValueError.
"""
if seed is None or seed is np.random:
return np.random.mtrand._rand
if isinstance(seed, numbers.Integral):
return np.random.RandomState(seed)
if isinstance(seed, np.random.RandomState):
return seed
raise ValueError(
"%r cannot be used to seed a numpy.random.RandomState instance" % seed
)
def squared_norm(x):
"""Squared Euclidean or Frobenius norm of x.
Faster than norm(x) ** 2.
Parameters
----------
x : array-like
Returns
-------
float
The Euclidean norm when x is a vector, the Frobenius norm when x
is a matrix (2-d array).
"""
x = np.ravel(x, order="K")
if np.issubdtype(x.dtype, np.integer):
warnings.warn(
"Array type is integer, np.dot may overflow. "
"Data should be float type to avoid this issue",
UserWarning,
)
return np.dot(x, x)
def norm(x):
"""Dot product-based Euclidean norm implementation.
See: http://fseoane.net/blog/2011/computing-the-vector-norm/
Parameters
----------
x : array-like
Vector for which to compute the norm.
"""
return sqrt(squared_norm(x))
def svd_flip(u, v, u_based_decision=True):
"""Sign correction to ensure deterministic output from SVD.
Adjusts the columns of u and the rows of v such that the loadings in the
columns in u that are largest in absolute value are always positive.
Parameters
----------
u : ndarray
u and v are the output of `linalg.svd` or
:func:`~sklearn.utils.extmath.randomized_svd`, with matching inner
dimensions so one can compute `np.dot(u * s, v)`.
v : ndarray
u and v are the output of `linalg.svd` or
:func:`~sklearn.utils.extmath.randomized_svd`, with matching inner
dimensions so one can compute `np.dot(u * s, v)`.
The input v should really be called vt to be consistent with scipy's
output.
u_based_decision : bool, default=True
If True, use the columns of u as the basis for sign flipping.
Otherwise, use the rows of v. The choice of which variable to base the
decision on is generally algorithm dependent.
Returns
-------
u_adjusted, v_adjusted : arrays with the same dimensions as the input.
"""
if u_based_decision:
# columns of u, rows of v
max_abs_cols = np.argmax(np.abs(u), axis=0)
signs = np.sign(u[max_abs_cols, range(u.shape[1])])
u *= signs
v *= signs[:, np.newaxis]
else:
# rows of v, columns of u
max_abs_rows = np.argmax( | np.abs(v) | numpy.abs |
import numpy as np
import numpy.matlib
LEFT, ROPE, RIGHT = range(3)
def correlated_ttest_MC(x, rope, runs=1, nsamples=50000):
"""
See correlated_ttest module for explanations
"""
if x.ndim == 2:
x = x[:, 1] - x[:, 0]
diff=x
n = len(diff)
nfolds = n / runs
x = np.mean(diff)
# Nadeau's and Bengio's corrected variance
var = np.var(diff, ddof=1) * (1 / n + 1 / (nfolds - 1))
if var == 0:
return int(x < rope), int(-rope <= x <= rope), int(rope < x)
return x+np.sqrt(var)*np.random.standard_t( n - 1, nsamples)
## Correlated t-test
def correlated_ttest(x, rope, runs=1, verbose=False, names=('C1', 'C2')):
import scipy.stats as stats
"""
Compute correlated t-test
The function uses the Bayesian interpretation of the p-value and returns
the probabilities the difference are below `-rope`, within `[-rope, rope]`
and above the `rope`. For details, see `A Bayesian approach for comparing
cross-validated algorithms on multiple data sets
<http://link.springer.com/article/10.1007%2Fs10994-015-5486-z>`_,
<NAME> and <NAME>, Mach Learning 2015.
|
The test assumes that the classifiers were evaluated using cross
validation. The number of folds is determined from the length of the vector
of differences, as `len(diff) / runs`. The variance includes a correction
for underestimation of variance due to overlapping training sets, as
described in `Inference for the Generalization Error
<http://link.springer.com/article/10.1023%2FA%3A1024068626366>`_,
<NAME> and <NAME>, Mach Learning 2003.)
|
Args:
x (array): a vector of differences or a 2d array with pairs of scores.
rope (float): the width of the rope
runs (int): number of repetitions of cross validation (default: 1)
return: probablities (tuple) that differences are below -rope, within rope or
above rope
"""
if x.ndim == 2:
x = x[:, 1] - x[:, 0]
diff=x
n = len(diff)
nfolds = n / runs
x = np.mean(diff)
# Nadeau's and Bengio's corrected variance
var = np.var(diff, ddof=1) * (1 / n + 1 / (nfolds - 1))
if var == 0:
return int(x < rope), int(-rope <= x <= rope), int(rope < x)
pr = 1-stats.t.cdf(rope, n - 1, x, np.sqrt(var))
pl = stats.t.cdf(-rope, n - 1, x, np.sqrt(var))
pe=1-pl-pr
if verbose:
print('P({c1} > {c2}) = {pl}, P(rope) = {pe}, P({c2} > {c1}) = {pr}'.
format(c1=names[0], c2=names[1], pl=pl, pe=pe, pr=pr))
return pl, pe, pr
## SIGN TEST
def signtest_MC(x, rope, prior_strength=1, prior_place=ROPE, nsamples=50000):
"""
Args:
x (array): a vector of differences or a 2d array with pairs of scores.
rope (float): the width of the rope
prior_strength (float): prior strength (default: 1)
prior_place (LEFT, ROPE or RIGHT): the region to which the prior is
assigned (default: ROPE)
nsamples (int): the number of Monte Carlo samples
Returns:
2-d array with rows corresponding to samples and columns to
probabilities `[p_left, p_rope, p_right]`
"""
if prior_strength < 0:
raise ValueError('Prior strength must be nonegative')
if nsamples < 0:
raise ValueError('Number of samples must be a positive integer')
if rope < 0:
raise ValueError('Rope must be a positive number')
if x.ndim == 2:
x = x[:, 1] - x[:, 0]
nleft = sum(x < -rope)
nright = sum(x > rope)
nrope = len(x) - nleft - nright
alpha = np.array([nleft, nrope, nright], dtype=float)
alpha += 0.0001 # for numerical stability
alpha[prior_place] += prior_strength
return | np.random.dirichlet(alpha, nsamples) | numpy.random.dirichlet |
# -*- coding: utf-8 -*-
"""
This is a script for satellite image classification
Last updated on Aug 6 2019
@author: <NAME>
@Email: <EMAIL>
@functions
1. generate samples from satellite images
2. grid search SVM/random forest parameters
3. object-based post-classification refinement
superpixel-based regularization for classification maps
4. confusion matrix: OA, kappa, PA, UA, AA
5. save maps as images
@sample codes
c = rscls.rscls(image,ground_truth,cls=number_of_classes)
c.padding(patch)
c.normalize(style='-11') # optional
x_train,y_train = c.train_sample(num_per_cls)
x_train,y_train = rscls.make_sample(x_train,y_train)
x_test,y_test = c.test_sample()
# for superpixel refinement
c.locate_obj(seg)
pcmap = rscls.obpc(c.seg,predicted,c.obj)
@Notes
Ground truth file should be uint8 format begin with 1
Background = 0
"""
import numpy as np
import copy
import scipy.stats as stats
from sklearn.svm import SVC
from sklearn.model_selection import GridSearchCV
from sklearn.ensemble import RandomForestClassifier
from sklearn.naive_bayes import GaussianNB
import matplotlib.pyplot as plt
class rscls:
def __init__(self, im, gt, cls): # 图片,ground truth, 类别数
if cls == 0:
print('num of class not specified !!')
self.im = copy.deepcopy(im) # 深拷贝
if gt.max() != cls:
self.gt = copy.deepcopy(gt - 1)
else:
self.gt = copy.deepcopy(gt - 1)
self.gt_b = copy.deepcopy(gt)
self.cls = cls
self.patch = 1
self.imx, self.imy, self.imz = self.im.shape
self.record = []
self.sample = {}
def padding(self, patch):
self.patch = patch
pad = self.patch // 2
r1 = np.repeat([self.im[0, :, :]], pad, axis=0) # 将元素im重复pad次
r2 = np.repeat([self.im[-1, :, :]], pad, axis=0)
self.im = np.concatenate((r1, self.im, r2)) # 图像填充
r1 = np.reshape(self.im[:, 0, :], [self.imx + 2 * pad, 1, self.imz]) # 将 image reshape
r2 = np.reshape(self.im[:, -1, :], [self.imx + 2 * pad, 1, self.imz])
r1 = np.repeat(r1, pad, axis=1)
r2 = np.repeat(r2, pad, axis=1)
self.im = np.concatenate((r1, self.im, r2), axis=1)
self.im = self.im.astype('float32')
def normalize(self, style='01'):
im = self.im
for i in range(im.shape[-1]):
im[:, :, i] = (im[:, :, i] - im[:, :, i].min()) / (im[:, :, i].max() - im[:, :, i].min())
if style == '-11':
im = im * 2 - 1
def locate_sample(self):
sam = []
for i in range(self.cls):
_xy = np.array(np.where(self.gt == i)).T
_sam = np.concatenate([_xy, i * np.ones([_xy.shape[0], 1])], axis=-1)
try:
sam = np.concatenate([sam, _sam], axis=0)
except:
sam = _sam
self.sample = sam.astype(int)
def get_patch(self, xy):
d = self.patch // 2
x = xy[0]
y = xy[1]
try:
self.im[x][y]
except IndexError:
return []
x += d
y += d
sam = self.im[(x - d):(x + d + 1), (y - d):(y + d + 1)]
return np.array(sam)
def train_sample(self, pn):
x_train, y_train = [], []
self.locate_sample()
_samp = self.sample
for _cls in range(self.cls):
_xy = _samp[_samp[:, 2] == _cls]
np.random.shuffle(_xy)
_xy = _xy[:pn, :]
for xy in _xy:
self.gt[xy[0], xy[1]] = 255 # !!
#
x_train.append(self.get_patch(xy[:-1]))
y_train.append(xy[-1])
# print(_xy)
x_train, y_train = np.array(x_train), np.array(y_train)
idx = np.random.permutation(x_train.shape[0])
x_train = x_train[idx]
y_train = y_train[idx]
return x_train, y_train.astype(int)
def test_sample(self):
x_test, y_test = [], []
self.locate_sample()
_samp = self.sample
for _cls in range(self.cls):
_xy = _samp[_samp[:, 2] == _cls]
np.random.shuffle(_xy)
for xy in _xy:
x_test.append(self.get_patch(xy[:-1]))
y_test.append(xy[-1])
return np.array(x_test), np.array(y_test)
def all_sample(self):
imx, imy = self.gt.shape
sample = []
for i in range(imx):
for j in range(imy):
sample.append(self.get_patch(np.array([i, j])))
return np.array(sample)
def all_sample_light(self, clip=0, bs=10):
imx, imy = self.gt.shape
imz = self.im.shape[-1]
patch = self.patch
# fp = np.memmap('allsample' + str(clip) + '.h5', dtype='float32', mode='w+', shape=(imgx*self.IMGY,5,5,bs))
fp = np.zeros([imx * imy, patch, patch, imz])
countnum = 0
for i in range(imx * clip, imx * (clip + 1)):
for j in range(imy):
xy = np.array([i, j])
fp[countnum, :, :, :] = self.get_patch(xy)
countnum += 1
return fp
def all_sample_row_hd(self, sub=0):
imx, imy = self.gt.shape
imz = self.im.shape[-1]
patch = self.patch
# fp = np.memmap('allsample' + str(clip) + '.h5', dtype='float32', mode='w+', shape=(imgx*self.IMGY,5,5,bs))
fp = np.zeros([imx * imy, patch, patch, imz])
countnum = 0
for i in range(sub):
for j in range(imy):
xy = np.array([i, j])
fp[countnum, :, :, :] = self.get_patch(xy)
countnum += 1
return fp
def all_sample_row(self, sub=0):
imx, imy = self.gt.shape
fp = []
for j in range(imy):
xy = np.array([sub, j])
fp.append(self.get_patch(xy))
return np.array(fp)
def all_sample_heavy(self, name, clip=0, bs=10):
imx, imy = self.gt.shape
imz = self.im.shape[-1]
patch = self.patch
try:
fp = np.memmap(name, dtype='float32', mode='w+', shape=(imx * imy, patch, patch, imz))
except:
fp = np.memmap(name, dtype='float32', mode='r', shape=(imx * imy, patch, patch, imz))
# fp = np.zeros([imx*imy,patch,patch,imz])
countnum = 0
for i in range(imx * clip, imx * (clip + 1)):
for j in range(imy):
xy = np.array([i, j])
fp[countnum, :, :, :] = self.get_patch(xy)
countnum += 1
return fp
def read_all_sample(self, name, clip=0, bs=10):
imx, imy = self.gt.shape
imz = self.im.shape[-1]
patch = self.patch
fp = np.memmap(name, dtype='float32', mode='r', shape=(imx * imy, patch, patch, imz))
return fp
def locate_obj(self, seg):
obj = {}
for i in range(seg.min(), seg.max() + 1):
obj[str(i)] = np.where(seg == i) # 若满足条件则为1,否则相应位置为0
self.obj = obj
self.seg = seg
def obpc(seg, cmap, obj):
pcmap = copy.deepcopy(cmap)
for (k, v) in obj.items():
#print('v', v)
#print('cmap[v]', cmap[v])
tmplabel = stats.mode(cmap[v])[0]
pcmap[v] = tmplabel
return pcmap
def cfm(pre, ref, ncl=9):
if ref.min() != 0:
print('warning: label should begin with 0 !!')
return
nsize = ref.shape[0]
cf = np.zeros((ncl, ncl))
for i in range(nsize):
cf[pre[i], ref[i]] += 1
tmp1 = 0
for j in range(ncl):
tmp1 = tmp1 + (cf[j, :].sum() / nsize) * (cf[:, j].sum() / nsize)
cfm = np.zeros((ncl + 2, ncl + 1))
cfm[:-2, :-1] = cf
oa = 0
for i in range(ncl):
if cf[i, :].sum():
cfm[i, ncl] = cf[i, i] / cf[i, :].sum()
if cf[:, i].sum():
cfm[ncl, i] = cf[i, i] / cf[:, i].sum()
oa += cf[i, i]
cfm[-1, 0] = oa / nsize
cfm[-1, 1] = (cfm[-1, 0] - tmp1) / (1 - tmp1)
cfm[-1, 2] = cfm[ncl, :-1].mean()
print('oa: ', format(cfm[-1, 0], '.5'), ' kappa: ', format(cfm[-1, 1], '.5'),
' mean: ', format(cfm[-1, 2], '.5'))
return cfm
def gtcfm(pre, gt, ncl):
if gt.max() == 255:
print('warning: max 255 !!')
cf = np.zeros([ncl, ncl])
for i in range(gt.shape[0]):
for j in range(gt.shape[1]):
if gt[i, j]:
cf[pre[i, j] - 1, gt[i, j] - 1] += 1
tmp1 = 0
nsize = np.sum(gt != 0)
for j in range(ncl):
tmp1 = tmp1 + (cf[j, :].sum() / nsize) * (cf[:, j].sum() / nsize)
cfm = np.zeros((ncl + 2, ncl + 1))
cfm[:-2, :-1] = cf
oa = 0
for i in range(ncl):
if cf[i, :].sum():
cfm[i, ncl] = cf[i, i] / cf[i, :].sum()
if cf[:, i].sum():
cfm[ncl, i] = cf[i, i] / cf[:, i].sum()
oa += cf[i, i]
cfm[-1, 0] = oa / nsize
cfm[-1, 1] = (cfm[-1, 0] - tmp1) / (1 - tmp1)
cfm[-1, 2] = cfm[ncl, :-1].mean()
#print(cfm)
print(cfm[ncl, :-1])
print('oa: ', format(cfm[-1, 0], '.5'), ' kappa: ', format(cfm[-1, 1], '.5'),
' mean: ', format(cfm[-1, 2], '.5'))
return cfm
def svm(trainx, trainy):
cost = []
gamma = []
for i in range(-5, 16, 2):
cost.append(np.power(2.0, i))
for i in range(-15, 4, 2):
gamma.append(np.power(2.0, i))
parameters = {'C': cost, 'gamma': gamma}
svm = SVC(verbose=0, kernel='rbf')
clf = GridSearchCV(svm, parameters, cv=3)
p = clf.fit(trainx, trainy)
print(clf.best_params_)
bestc = clf.best_params_['C']
bestg = clf.best_params_['gamma']
tmpc = [-1.75, -1.5, -1.25, -1, -0.75, -0.5, -0.25, 0.0,
0.25, 0.5, 0.75, 1.0, 1.25, 1.5, 1.75]
cost = []
gamma = []
for i in tmpc:
cost.append(bestc * np.power(2.0, i))
gamma.append(bestg * np.power(2.0, i))
parameters = {'C': cost, 'gamma': gamma}
svm = SVC(verbose=0, kernel='rbf')
clf = GridSearchCV(svm, parameters, cv=3)
p = clf.fit(trainx, trainy)
print(clf.best_params_)
p2 = clf.best_estimator_
return p2
def svm_rbf(trainx, trainy):
cost = []
gamma = []
for i in range(-3, 10, 2):
cost.append(np.power(2.0, i))
for i in range(-5, 4, 2):
gamma.append(np.power(2.0, i))
parameters = {'C': cost, 'gamma': gamma}
svm = SVC(verbose=0, kernel='rbf')
clf = GridSearchCV(svm, parameters, cv=3)
clf.fit(trainx, trainy)
# print(clf.best_params_)
bestc = clf.best_params_['C']
bestg = clf.best_params_['gamma']
tmpc = [-1.75, -1.5, -1.25, -1, -0.75, -0.5, -0.25, 0.0,
0.25, 0.5, 0.75, 1.0, 1.25, 1.5, 1.75]
cost = []
gamma = []
for i in tmpc:
cost.append(bestc * | np.power(2.0, i) | numpy.power |
from liquepy.element import models
import numpy as np
def test_av_stress():
tau = | np.array([0, -2, -4, -6, -8, -6, -4, -2, 0, 2, 4, 6, 8, 6, 4, 2, 0]) | numpy.array |
# Author: <NAME> <<EMAIL>>
# My imports
from amt_tools.tools.instrument import GuitarProfile
# Regular imports
from mpl_toolkits.axisartist.axislines import SubplotZero
import matplotlib.lines as mlines
import matplotlib.pyplot as plt
import numpy as np
import librosa
def visualize_pitch_list(times, pitch_list, save_path=None):
plt.figure()
times = np.concatenate([[times[i]] * len(pitch_list[i]) for i in range(len(pitch_list))])
pitches = np.concatenate(pitch_list)
plt.scatter(times, pitches, s=5, c='k')
plt.xlabel('Time (s)')
plt.ylabel('Pitch')
plt.xlim(0, 25)
plt.tight_layout()
plt.savefig(save_path) if save_path else plt.show()
return plt
def visualize_multi_pitch(multi_pitch, ax=None):
if ax is None:
ax = plt.gca()
ax.imshow(multi_pitch, cmap='gray_r', vmin=0, vmax=1)
ax.invert_yaxis()
return ax
# TODO - this is mostly trash for now - I've yet to make an effort to reestablish this
# TODO - see earlier commits to get started
def pianoroll(track_name, i_est, p_est, i_ref, p_ref, t_bounds, save_path=None):
est_max, ref_max = 0, 0
for i in range(profile.num_strings):
if i_est.size != 0:
est_max = max(est_max, np.max(i_est))
if i_ref.size != 0:
ref_max = max(ref_max, np.max(i_ref))
t_bounds = [t_bounds[0], min(t_bounds[1], max(est_max, ref_max))]
plt.figure()
for s in range(profile.num_strings):
for n in range(p_ref.size):
t_st = i_ref[n][0]
t_fn = i_ref[n][1]
m_fq = int(round(librosa.hz_to_midi(p_ref[n])))
plt.plot([t_st, t_fn], [m_fq] * 2, linewidth=10, color='black', label='Ref.')
for n in range(p_est.size):
t_st = i_est[n][0]
t_fn = i_est[n][1]
m_fq = int(round(librosa.hz_to_midi(p_est[n])))
plt.plot([t_st, t_fn], [m_fq] * 2, linewidth=10, color='orange', label='Est.',alpha=0.75)
handles = [mlines.Line2D([], [], color='black', linestyle='-', label='Ref.', linewidth=10),
mlines.Line2D([], [], color='orange', linestyle='-', label='Est.', linewidth=10)]
# The lowest possible note - i.e. the open note of the lowest string
m_lw = librosa.note_to_midi(profile.tuning[0])
# The highest possible note - i.e. the maximum fret on the highest string
m_hg = librosa.note_to_midi(profile.tuning[profile.num_strings - 1]) + profile.num_frets
plt.title(track_name)
plt.xlabel('Time (s)')
plt.ylabel('Pitch (MIDI)')
plt.legend(handles=handles, loc='upper right', framealpha=0.5)
plt.xlim(t_bounds[0] - 0.25, t_bounds[1] + 0.25)
plt.ylim(m_lw - 1, m_hg + 1)
plt.gcf().set_size_inches(16, 9)
plt.tight_layout()
if save_path:
plt.savefig(save_path, bbox_inches='tight', dpi=500)
else:
plt.show()
def guitar_tabs(track_name, tabs_est, tabs_ref, t_bounds, offset=True, save_path=None):
est_max, ref_max = 0, 0
for i in range(profile.num_strings):
if tabs_est[i][1].size != 0:
est_max = max(est_max, np.max(tabs_est[i][1]))
if tabs_ref[i][1].size != 0:
ref_max = max(ref_max, | np.max(tabs_ref[i][1]) | numpy.max |
'''
define some fucntions that will plot spheres and galaxies in pixels
@author: <NAME> ppymj11
'''
from numba import jit
import numpy as np
# %%
# define a function that will draw spherical regions around point in pixels
# use numba jit to improve efficiency of used loops.
@jit(nopython=True)
def draw_sphere(body_size, image_s, index, color):
'''
Draws a spherical body (proj to 2D -- circle) on the provided image in pixels
Parameters:
--------------
body - size of body (radius) in pizels, int.
image_s - (N x N x 3) image array to plot on.
color - tuple of size 3, gives flat RGB color of shpere.
Returns:
--------------
image as size x size x 3 array
'''
# make sure given image is square and get size
assert len(image_s[0, :, 0]) == len(image_s[:, 0, 0]), 'Need a square Image'
size = len(image_s[0, :, 0])
# loop over i and j, for a square around the object
for i in range(2*body_size+1):
# get x rel. to star centre and its index in array
x = -body_size + i
ind_1 = int(size/2) - body_size + i
for j in range(2*body_size+1):
# as before for y
y = -body_size + j
ind_2 = index - body_size + j
# check if current point is the circular region and plot:
if ((x/body_size)**2 + (y/body_size)**2) <= 1 and (ind_1 >= 0 and ind_1 < size) and (ind_2 >= 0 and ind_2 < size):
image_s[ind_1, ind_2, :] = color
# return the image with source drawn, to user
return image_s
# define a function that will generate image of galaxy cluster
# use numba jit to improve efficiency of used loops.
@jit(nopython=True)
def gal_image(gal_N, size, max_a, minor_max, minor_major_multiplier=5, seeded=1234):
'''
Generates a random, pixelated image of galaxies
Parameters:
---------------
gal_N - int - number of galaxies
size - int - side length of square image to create
max_a - float - maximum decay costant for flux, in pixels
minor_max - int - maximum semi-minor axis size, in pixels
kwargs:
---------------
minor_major_multiplier - int - max ratio between minor and major axis
seeded - int - seed for numpy.random library
returns:
---------------
image, as (size x size x 3) array
'''
# set the seed for repeatable results
np.random.seed(seeded)
# ##############################################
# randomly generate galaxy properties:
# ##############################################
# fluxes in each RGB band (inetegers), as tuple to append to the pixels
f0r = np.random.randint(0, 255, gal_N)
f0g = np.random.randint(0, 255, gal_N)
f0b = np.random.randint(0, 255, gal_N)
f0 = np.vstack((f0r, f0g, f0b))
# pixel center indexes
x_centr, y_centr = np.random.randint(0, size, gal_N), np.random.randint(0, size, gal_N)
# decay a, unit = pixels:
a = np.random.rand(gal_N) * max_a
# minor axis
minor = np.random.randint(1, minor_max+1, gal_N)
# angle of major axis to horizontal (radians)
theta = | np.random.rand(gal_N) | numpy.random.rand |
from nutils import SI
import numpy
import pickle
import typing
import unittest
class Dimension(unittest.TestCase):
def test_multiply(self):
self.assertEqual(SI.Velocity * SI.Time, SI.Length)
self.assertEqual(SI.Length * int, SI.Length)
self.assertEqual(float * SI.Time, SI.Time)
def test_divide(self):
self.assertEqual(SI.Length / SI.Time, SI.Velocity)
self.assertEqual(SI.Length / int, SI.Length)
self.assertEqual(float / SI.Time, SI.Frequency)
def test_power(self):
self.assertEqual(SI.Length**2, SI.Area)
self.assertEqual(SI.Area**.5, SI.Length)
def test_name(self):
self.assertEqual(SI.Force.__name__, '[M*L/T2]')
self.assertEqual((SI.Force**.5).__name__, '[M_2*L_2/T]')
self.assertEqual((SI.Force**1.5).__name__, '[M3_2*L3_2/T3]')
def test_fromname(self):
self.assertEqual(getattr(SI.Quantity, '[M*L/T2]'), SI.Force)
self.assertEqual(getattr(SI.Quantity, '[M_2*L_2/T]'), SI.Force**.5)
self.assertEqual(getattr(SI.Quantity, '[M3_2*L3_2/T3]'), SI.Force**1.5)
def test_typing(self):
self.assertEqual(SI.Length | None, typing.Optional[SI.Length])
self.assertEqual(None | SI.Length | SI.Time, typing.Optional[typing.Union[SI.Time, SI.Length]])
def test_pickle(self):
T = SI.Length / SI.Time
s = pickle.dumps(T)
self.assertEqual(pickle.loads(s), T)
class Quantity(unittest.TestCase):
def test_fromstring(self):
F = SI.parse('5kN')
self.assertEqual(type(F), SI.Force)
self.assertEqual(F / 'N', 5000)
v = SI.parse('-864km/24h')
self.assertEqual(type(v), SI.Velocity)
self.assertEqual(v / 'm/s', -10)
def test_fromvalue(self):
F = SI.Force('10N')
self.assertEqual(type(F), SI.Force)
self.assertEqual(F / SI.Force('2N'), 5)
def test_array(self):
F = SI.units.N * numpy.arange(6).reshape(2, 3)
self.assertEqual(F.shape, (2, 3))
self.assertEqual(F.ndim, 2)
self.assertEqual(F.size, 6)
def test_getitem(self):
F = SI.units.N * numpy.arange(6).reshape(2, 3)
self.assertEqual(F[0, 0], SI.Force('0N'))
self.assertEqual(F[0, 1], SI.Force('1N'))
self.assertEqual(F[0, 2], SI.Force('2N'))
self.assertEqual(F[1, 0], SI.Force('3N'))
self.assertEqual(F[1, 1], SI.Force('4N'))
self.assertEqual(F[1, 2], SI.Force('5N'))
def test_setitem(self):
F = SI.units.N * numpy.zeros(3)
F[0] = SI.Force('1N')
F[1] = SI.Force('2N')
with self.assertRaisesRegex(TypeError, r'cannot assign \[L2\] to \[M\*L/T2\]'):
F[2] = SI.Area('10m2')
F[2] = SI.Force('3N')
self.assertTrue(numpy.all(F == SI.units.N * numpy.array([1, 2, 3])))
def test_iter(self):
F = SI.units.N * numpy.arange(6).reshape(2, 3)
for i, Fi in enumerate(F):
for j, Fij in enumerate(Fi):
self.assertEqual(Fij, SI.units.N * (i*3+j))
def test_multiply(self):
self.assertEqual(SI.Mass('2kg') * SI.Acceleration('10m/s2'), SI.Force('20N'))
self.assertEqual(2 * SI.Acceleration('10m/s2'), SI.Acceleration('20m/s2'))
self.assertEqual(SI.Mass('2kg') * 10, SI.Mass('20kg'))
self.assertEqual(SI.Time('2s') * SI.Frequency('10/s'), 20)
self.assertEqual(numpy.multiply(SI.Mass('2kg'), SI.Acceleration('10m/s2')), SI.Force('20N'))
def test_matmul(self):
self.assertEqual((SI.units.kg * numpy.array([2, 3])) @ (SI.parse('m/s2') * numpy.array([5, -3])), SI.Force('1N'))
def test_divide(self):
self.assertEqual(SI.Length('2m') / SI.Time('10s'), SI.Velocity('.2m/s'))
self.assertEqual(2 / SI.Time('10s'), SI.Frequency('.2/s'))
self.assertEqual(SI.Length('2m') / 10, SI.Length('.2m'))
self.assertEqual(SI.Density('2kg/m3') / SI.Density('10kg/m3'), .2)
self.assertEqual(numpy.divide(SI.Length('2m'), SI.Time('10s')), SI.Velocity('.2m/s'))
def test_power(self):
self.assertEqual(SI.Length('3m')**2, SI.Area('9m2'))
self.assertEqual(SI.Length('3m')**0, 1)
self.assertEqual(numpy.power(SI.Length('3m'), 2), SI.Area('9m2'))
def test_add(self):
self.assertEqual(SI.Mass('2kg') + SI.Mass('3kg'), SI.Mass('5kg'))
self.assertEqual(numpy.add(SI.Mass('2kg'), SI.Mass('3kg')), SI.Mass('5kg'))
with self.assertRaisesRegex(TypeError, r'incompatible arguments for add: \[M\], \[L\]'):
SI.Mass('2kg') + SI.Length('3m')
def test_sub(self):
self.assertEqual(SI.Mass('2kg') - SI.Mass('3kg'), SI.Mass('-1kg'))
self.assertEqual(numpy.subtract(SI.Mass('2kg'), SI.Mass('3kg')), SI.Mass('-1kg'))
with self.assertRaisesRegex(TypeError, r'incompatible arguments for sub: \[M\], \[L\]'):
SI.Mass('2kg') - SI.Length('3m')
def test_hypot(self):
self.assertEqual(numpy.hypot(SI.Mass('3kg'), SI.Mass('4kg')), SI.Mass('5kg'))
with self.assertRaisesRegex(TypeError, r'incompatible arguments for hypot: \[M\], \[L\]'):
numpy.hypot(SI.Mass('3kg'), SI.Length('4m'))
def test_neg(self):
self.assertEqual(-SI.Mass('2kg'), SI.Mass('-2kg'))
self.assertEqual(numpy.negative(SI.Mass('2kg')), SI.Mass('-2kg'))
def test_pos(self):
self.assertEqual(+SI.Mass('2kg'), SI.Mass('2kg'))
self.assertEqual(numpy.positive(SI.Mass('2kg')), SI.Mass('2kg'))
def test_abs(self):
self.assertEqual(numpy.abs(SI.Mass('-2kg')), SI.Mass('2kg'))
def test_sqrt(self):
self.assertEqual(numpy.sqrt(SI.Area('4m2')), SI.Length('2m'))
def test_sum(self):
self.assertTrue(numpy.all(numpy.sum(SI.units.kg * numpy.arange(6).reshape(2, 3), 0) == SI.units.kg * numpy.array([3, 5, 7])))
self.assertTrue(numpy.all(numpy.sum(SI.units.kg * numpy.arange(6).reshape(2, 3), 1) == SI.units.kg * numpy.array([3, 12])))
def test_mean(self):
self.assertTrue(numpy.all(numpy.mean(SI.units.kg * numpy.arange(6).reshape(2, 3), 0) == SI.units.kg * numpy.array([1.5, 2.5, 3.5])))
self.assertTrue(numpy.all(numpy.mean(SI.units.kg * numpy.arange(6).reshape(2, 3), 1) == SI.units.kg * numpy.array([1, 4])))
def test_broadcast_to(self):
v = numpy.array([1, 2, 3])
A = SI.units.kg * v
B = numpy.broadcast_to(A, (2, 3))
self.assertEqual(B.shape, (2, 3))
self.assertEqual(B[1, 1], SI.Mass('2kg'))
def test_trace(self):
A = SI.units.kg * numpy.arange(18).reshape(3, 2, 3)
self.assertTrue(numpy.all(numpy.trace(A, axis1=0, axis2=2) == SI.units.kg * numpy.array([21, 30])))
def test_ptp(self):
A = SI.units.kg * numpy.array([2, -10, 5, 0])
self.assertEqual(numpy.ptp(A), SI.Mass('15kg'))
def test_min(self):
A = SI.units.kg * numpy.array([2, -10, 5, 0])
self.assertEqual( | numpy.max(A) | numpy.max |
#!/usr/bin/env python3
#
# Pocket SDR Python AP - GNSS Signal Acquisition
#
# Author:
# T.TAKASU
#
# History:
# 2021-12-01 1.0 new
# 2021-12-05 1.1 add signals: G1CA, G2CA, B1I, B2I, B1CD, B1CP, B2AD, B2AP,
# B2BI, B3I
# 2021-12-15 1.2 add option: -d, -nz, -np
# 2022-01-20 1.3 add signals: I5S
# add option: -l, -s
#
import sys, time
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
import sdr_func, sdr_code
mpl.rcParams['toolbar'] = 'None';
mpl.rcParams['font.size'] = 9
# constants --------------------------------------------------------------------
T_AQC = 0.010 # non-coherent integration time for acquisition (s)
THRES_CN0 = 38.0 # threshold to lock (dB-Hz)
ESC_COL = '\033[34m' # ANSI escape color = blue
ESC_RES = '\033[0m' # ANSI escape reset
# show usage -------------------------------------------------------------------
def show_usage():
print('Usage: pocket_acq.py [-sig sig] [-prn prn[,...]] [-tint tint]')
print(' [-toff toff] [-f freq] [-fi freq] [-d freq] [-nz] [-np]')
print(' [-s] [-p] [-l] [-3d] file')
exit()
# plot C/N0 --------------------------------------------------------------------
def plot_cn0(ax, cn0, prns, fc):
thres = THRES_CN0
x = np.arange(len(cn0))
y = cn0
ax.bar(x[y < thres], y[y < thres], color='gray', width=0.6)
ax.bar(x[y >= thres], y[y >= thres], color=fc , width=0.6)
ax.grid(True, lw=0.4)
ax.set_xlim([x[0] - 0.5, x[-1] + 0.5])
plt.xticks(x, ['%d' % (prn) for prn in prns])
ax.set_ylim([30, 50])
ax.set_xlabel('PRN Number')
ax.set_ylabel('C/N0 (dB-Hz)')
# plot correlation 3D ----------------------------------------------------------
def plot_corr_3d(ax, P, dops, coffs, ix, fc):
x, y = np.meshgrid(coffs * 1e3, dops)
z = P / | np.mean(P) | numpy.mean |
# standard imports
import numpy as np
import matplotlib.pyplot as plt
# Add parent directory to path
import sys
import os
parent_path = '..\\nistapttools'
if parent_path not in sys.path:
sys.path.append(os.path.abspath(parent_path))
# custom imports
import apt_fileio
import m2q_calib
import plotting_stuff
import initElements_P3
import histogram_functions
import peak_param_determination as ppd
from histogram_functions import bin_dat
import voltage_and_bowl
from voltage_and_bowl import do_voltage_and_bowl
from voltage_and_bowl import mod_full_vb_correction
import colorcet as cc
def create_histogram(xs, ys, x_roi=None, delta_x=0.1, y_roi=None, delta_y=0.1):
"""Create a 2d histogram of the data, specifying the bin intensity, region
of interest (on the y-axis), and the spacing of the y bins"""
# even number
num_x = int(np.ceil((x_roi[1]-x_roi[0])/delta_x))
num_y = int(np.ceil((y_roi[1]-y_roi[0])/delta_y))
return np.histogram2d(xs, ys, bins=[num_x, num_y],
range=[x_roi, y_roi],
density=False)
def _extents(f):
"""Helper function to determine axis extents based off of the bin edges"""
delta = f[1] - f[0]
return [f[0] - delta/2, f[-1] + delta/2]
def plot_2d_histo(ax, N, x_edges, y_edges, scale='log'):
if scale=='log':
dat = np.log10(1+N)
elif scale=='lin':
dat = N
"""Helper function to plot a histogram on an axis"""
ax.imshow(np.transpose(dat), aspect='auto',
extent=_extents(x_edges) + _extents(y_edges),
origin='lower', cmap=cc.cm.CET_L8,
interpolation='antialiased')
def corrhist(epos, delta=1, roi=None):
dat = epos['tof']
if roi is None:
roi = [0, 1000]
N = int(np.ceil((roi[1]-roi[0])/delta))
corrhist = np.zeros([N,N], dtype=int)
multi_idxs = np.where(epos['ipp']>1)[0]
for multi_idx in multi_idxs:
n_hits = epos['ipp'][multi_idx]
cluster = dat[multi_idx:multi_idx+n_hits]
idx1 = -1
idx2 = -1
for i in range(n_hits):
for j in range(i+1,n_hits):
idx1 = int(np.floor(cluster[i]/delta))
idx2 = int(np.floor(cluster[j]/delta))
if idx1 < N and idx1>=0 and idx2 < N and idx2>=0:
corrhist[idx1,idx2] += 1
edges = np.arange(roi[0],roi[1]+delta,delta)
assert edges.size-1 == N
return (edges, corrhist+corrhist.T-np.diag(np.diag(corrhist)))
def calc_t0(tof,tof_vcorr_fac,tof_bcorr_fac,sigma):
BB = tof_bcorr_fac[0::2]+tof_bcorr_fac[1::2]
t0 = ((tof_bcorr_fac[0::2]*tof[0::2]+tof_bcorr_fac[1::2]*tof[1::2]) - sigma/(tof_vcorr_fac[0::2]))/BB
t0 = np.ravel(np.column_stack((t0,t0)))
return t0
def create_sigma_delta_histogram(raw_tof, tof_vcorr_fac, tof_bcorr_fac, sigmas=None, delta_range=None, delta_step=0.5):
# Must be a doubles only epos...
# scan through a range of sigmas and compute the corrected data
if sigmas is None:
sigmas = np.linspace(0,2000,2**7)
if delta_range is None:
delta_range = [0,700]
delta_n_bins = int((delta_range[1]-delta_range [0])/delta_step)
# print('delta_n_bins = '+str(delta_n_bins))
res_dat = np.zeros((sigmas.size,delta_n_bins))
for sigma_idx in np.arange(sigmas.size):
t0 = calc_t0(raw_tof, tof_vcorr_fac, tof_bcorr_fac, sigmas[sigma_idx])
tof_corr = tof_vcorr_fac*tof_bcorr_fac*(raw_tof-t0)
dts = np.abs(tof_corr[:-1:2]-tof_corr[1::2])
N, delta_edges = np.histogram(dts, bins=delta_n_bins, range=delta_range)
res_dat[sigma_idx,:] = N
if np.mod(sigma_idx,10)==0:
print("Loop index "+str(sigma_idx+1)+" of "+str(sigmas.size))
delta_centers = 0.5*(delta_edges[:-1]+delta_edges[1:])
return (res_dat, sigmas, delta_centers)
def interleave(a,b):
return np.ravel(np.column_stack((a,b)))
def calc_slope_and_intercept(raw_tof, volt_coeff, bowl_coeff):
A = volt_coeff[0::2]
B_alpha = bowl_coeff[0::2]
B_beta = bowl_coeff[1::2]
tof_alpha = raw_tof[0::2]
tof_beta = raw_tof[1::2]
intercept = 2*A*B_alpha*B_beta*(tof_beta-tof_alpha)/(B_alpha+B_beta)
slope = (B_beta-B_alpha)/(B_beta+B_alpha)
return (slope, intercept)
# Note that x is sums and y is diffs
def compute_dist_to_line(slope, intercept, x, y):
return np.abs(intercept+slope*x-y)/np.sqrt(1+slope**2)
def calc_parametric_line(raw_tof, volt_coeff, bowl_coeff, n=2):
if n>0:
t = raw_tof.reshape(-1,n)
v = volt_coeff.reshape(-1,n)
b = bowl_coeff.reshape(-1,n)
else:
t = raw_tof
v = volt_coeff
b = bowl_coeff
r0 = v*b*(t-np.sum(b*t,axis=1)[:,np.newaxis]/np.sum(b,axis=1)[:,np.newaxis])
r1 = b/np.sum(b,axis=1)[:,np.newaxis]
return (r0, r1)
def compute_dist_to_parametric_line(r0, r1, q):
# q is n_pts by n_dim
sigma = (np.dot(r1,q.T)-np.sum(r0*r1,axis=1)[:,np.newaxis])/ | np.sum(r1**2,axis=1) | numpy.sum |
# -*- coding: utf-8 -*-
"""
Created on Fri Oct 5 10:40:35 2018
@author: hamed
"""
try:
import torch
from torch.utils import data
except:
pass
import numpy as np
from sklearn import datasets
#%%
class kk_mimic_dataset(data.Dataset):
def __init__(self, phase="train", seq_len=10, data_norm=True, test=False):
percent = 20
n_valid = percent/20 * 328
ind_valid = np.ones(n_valid)
ind_valid = np.concatenate((ind_valid, np.zeros(6564-n_valid)))
ind_valid = | np.random.permutation(ind_valid) | numpy.random.permutation |
from sys import maxsize
import numpy as np
from cv2 import cv2
from numpy.core.arrayprint import array2string
from numpy.lib.shape_base import split
from daugman import daugman
from scipy.spatial import distance
import itertools
import glob
import re
np.set_printoptions(threshold=maxsize)
def daugman_normalizaiton(image, height, width, r_in, r_out):
thetas = np.arange(0, 2 * np.pi, 2 * np.pi / width) # Theta values
# Create empty flatten image
flat = np.zeros((height, width, 3), np.uint8)
circle_x = int(image.shape[0] / 2)
circle_y = int(image.shape[1] / 2)
for i in range(width):
for j in range(height):
theta = thetas[i] # value of theta coordinate
r_pro = j / height # value of r coordinate(normalized)
# get coordinate of boundaries
Xi = circle_x + r_in * np.cos(theta)
Yi = circle_y + r_in * np.sin(theta)
Xo = circle_x + r_out * np.cos(theta)
Yo = circle_y + r_out * | np.sin(theta) | numpy.sin |
"""Plotting utilities. Import requires matplotlib.
"""
import datetime
import copy # for shallow copies of matplotlib colormaps
import numpy as np
import scipy.signal
import matplotlib.pyplot as plt
import matplotlib.patches
import matplotlib.colors
import matplotlib.lines
import matplotlib.patheffects
import matplotlib.cm
import desietcimg.util
def draw_ellipse(ax, x0, y0, s, g1, g2, nsigmas=1, **ellipseopts):
g = np.sqrt(g1 ** 2 + g2 ** 2)
if g > 1:
raise ValueError('g1 ** 2 + g2 ** 2 > 1')
center = np.array([x0, y0])
angle = np.rad2deg(0.5 * np.arctan2(g2, g1))
ratio = np.sqrt((1 + g) / (1 - g))
width = 2 * s * ratio * nsigmas
height = 2 * s / ratio * nsigmas
kwargs = dict(color='r', ls='-', lw=2, alpha=0.7, fill=False)
kwargs.update(ellipseopts)
ellipse = matplotlib.patches.Ellipse(center, width, height, angle, **kwargs)
ax.add_artist(ellipse)
def plot_image(D, W=None, ax=None, cmap='viridis', masked_color='chocolate', threshold=0.01):
if W is not None and np.any(W == 0):
D = D.copy()
D[W == 0] = np.nan
if W is not None:
# Ignore pixels with low ivar for setting color scale limits.
informative = W > threshold * np.median(W)
else:
informative = np.ones_like(D, bool)
vmin, vmax = np.percentile(D[informative], (0, 100))
ax = ax or plt.gca()
cmap = copy.copy(matplotlib.cm.get_cmap(cmap))
cmap.set_bad(color=masked_color)
h, w = D.shape
I = ax.imshow(D, interpolation='none', origin='lower', cmap=cmap, vmin=vmin, vmax=vmax,
extent=[-0.5 * w, 0.5 * w, -0.5 * h, 0.5 * h])
ax.axis('off')
return ax
class Axes(object):
def __init__(self, n, size=4, pad=0.02):
rows = int(np.floor(np.sqrt(n)))
cols = int(np.ceil(n / rows))
assert rows * cols >= n
width = cols * size + (cols - 1) * pad
height = rows * size + (rows - 1) * pad
self.fig, axes = plt.subplots(rows, cols, figsize=(width, height), squeeze=False)
plt.subplots_adjust(left=0, right=1, bottom=0, top=1, wspace=pad, hspace=pad)
self.axes = axes.flatten()
self.n = n
for ax in self.axes:
ax.axis('off')
def plot_sky_camera(SCA, size=4, pad=0.02, what='stamp', labels=True, params=True, fiber=True):
if SCA.fibers is None or SCA.results is None:
raise RuntimeError('No results available to plot.')
nfibers = len(SCA.fibers)
A = Axes(nfibers, size, pad)
# Extract the pixel values to plot.
plotdata = []
results = iter(SCA.results.items())
for k in range(nfibers):
label, (xfit, yfit, bgmean, fiber_flux, snr, stamp, ivar, model, raw) = next(results)
plotdata.append({'stamp': stamp, 'ivar': ivar, 'model': model, 'raw': raw}[what])
# Use the same colorscale for all stamps.
allpixels = np.concatenate(plotdata, axis=1).flatten()
vmin, vmax = np.percentile(allpixels[allpixels > 0], (1, 99))
# Loop over fibers to plot.
results = iter(SCA.results.items())
fibers = iter(SCA.fibers.values())
for k in range(nfibers):
ax = A.axes[k]
ix, iy = next(fibers)
label, (xfit, yfit, bgmean, fiber_flux, snr, stamp, ivar, model, raw) = next(results)
ax.imshow(plotdata[k], interpolation='none', origin='lower', cmap='viridis', vmin=vmin, vmax=vmax)
ax.axis('off')
if fiber:
cx = (xfit - ix) / SCA.binning + SCA.rsize
cy = (yfit - iy) / SCA.binning + SCA.rsize
cr = 0.5 * SCA.fiberdiam / SCA.binning
circle = matplotlib.patches.Circle(
[cx, cy], cr, lw=2, ec='r', fc='none', alpha=0.5)
dxy = SCA.dxy + SCA.rsize
xgrid, ygrid = np.meshgrid(dxy, dxy)
ax.plot(xgrid[0], ygrid[0], 'r.', ms=1)
ax.plot(xgrid[-1], ygrid[-1], 'r.', ms=1)
ax.plot(xgrid[:, 0], ygrid[:, 0], 'r.', ms=1)
ax.plot(xgrid[:, -1], ygrid[:, -1], 'r.', ms=1)
ax.plot(cx, cy, 'r+')
ax.add_artist(circle)
kwargs = dict(verticalalignment='center', horizontalalignment='center',
transform=ax.transAxes, color='w', fontweight='bold')
if labels:
ax.text(0.5, 0.95, label, fontsize=16, **kwargs)
if params:
params = '{0:.1f} e/s SNR {1:.1f}'.format(fiber_flux, snr)
ax.text(0.5, 0.05, params, fontsize=14, **kwargs)
return A
def plot_guide_results(GCR, size=4, pad=0.02, ellipses=True, params=True):
if GCR.stamps is None or GCR.results is None:
raise RuntimeError('No results available to plot.')
nstamps = GCR.meta['NSRC']
rsize = GCR.meta['SSIZE'] // 2
A = Axes(nstamps, size, pad)
for k in range(nstamps):
ax = A.axes[k]
plot_image(*GCR.stamps[k], ax=ax)
kwargs = dict(verticalalignment='center', horizontalalignment='center',
transform=ax.transAxes, fontweight='bold')
result, y_slice, x_slice = GCR.results[k]
ix, iy = x_slice.start + rsize, y_slice.start + rsize
label = 'x={0:04d} y={1:04d}'.format(ix, iy)
ax.text(0.5, 0.05, label, fontsize=16, color='w', **kwargs)
if result['success']:
color = 'w' if result['psf'] else 'r'
if ellipses:
draw_ellipse(ax, result['x0'], result['y0'],
result['s'], result['g1'], result['g2'], ec=color)
if params:
g = np.sqrt(result['g1'] ** 2 + result['g2'] ** 2)
label = f'$\\nu$ {result["snr"]:.1f} s {result["s"]:.1f} g {g:.2f}'
ax.text(0.5, 0.95, label, fontsize=18, color=color, **kwargs)
return A
def plot_psf_profile(GCR, size=4, pad=0.5, inset_size=35, max_ang=2.0, label=None):
"""
"""
assert inset_size % 2 == 1
P, W = GCR.profile
h1 = len(P) // 2
h2 = inset_size // 2
inset = slice(h1 - h2, h1 + h2 + 1)
width = 2.5 * size + pad
height = size
fig = plt.figure(figsize=(width, height))
lhs = plt.axes((0., 0., size / width, 1.))
rhs = plt.axes(((size + pad) / width, pad / height,
(width - size - pad) / width - 0.02, (height - pad) / height - 0.02))
plot_image(P[inset, inset], W[inset, inset], ax=lhs)
kwargs = dict(fontsize=16, color='w', verticalalignment='center', horizontalalignment='center',
transform=lhs.transAxes, fontweight='bold')
fwhm = GCR.meta['FWHM']
ffrac = GCR.meta['FFRAC']
xc = GCR.meta['XC']
yc = GCR.meta['YC']
lhs.text(0.5, 0.95, 'FWHM={0:.2f}" FFRAC={1:.3f}'.format(fwhm, ffrac), **kwargs)
if label is not None:
lhs.text(0.5, 0.05, label, **kwargs)
rfiber_pix = 0.5 * GCR.meta['FIBSIZ'] / GCR.meta['PIXSIZ']
lhs.add_artist(plt.Circle((xc, yc), rfiber_pix, fc='none', ec='r', lw=2, alpha=0.5))
lhs.plot(xc, yc, 'r+', ms=25)
lhs.plot([xc, h2], [yc, yc], 'r--')
lhs.plot([xc, xc], [yc, h2], 'r:')
# Plot the circularized radial profile.
rhs.plot(GCR.profile_tab['rang'], GCR.profile_tab['prof'], 'k-', label='Circ. Profile')
# Plot the fiber acceptance fraction for centroid offsets along +x and +y.
noffset = len(GCR.fiberfrac)
noffset_per_pix = GCR.meta.get('NOFFPX', 2)
dxy_pix = (np.arange(noffset) - 0.5 * (noffset - 1)) / noffset_per_pix
pixel_size_um = GCR.meta['PIXSIZ']
plate_scales = (GCR.meta['XSCALE'], GCR.meta['YSCALE'])
dx_ang = (dxy_pix - xc) * pixel_size_um / plate_scales[0]
dy_ang = (dxy_pix - yc) * pixel_size_um / plate_scales[0]
iy, ix = np.unravel_index(np.argmax(GCR.fiberfrac), GCR.fiberfrac.shape)
rhs.plot(dx_ang[ix:], GCR.fiberfrac[iy, ix:], 'r--', label='Fiber Frac (x)')
rhs.plot(dx_ang[iy:], GCR.fiberfrac[iy:, ix], 'r:', label='Fiber Frac (y)')
rhs.set_ylim(-0.02, 1.02)
rhs.set_xlim(0., max_ang)
rhs.grid()
rhs.legend(loc='upper right')
rhs.set_xlabel('Offset from PSF center [arcsec]')
def plot_colorhist(D, ax, imshow, mode='reverse', color='w', alpha=0.75):
"""Draw a hybrid colorbar and histogram.
"""
ax.axis('off')
# Extract parameters of the original imshow.
cmap = imshow.get_cmap()
vmin, vmax = imshow.get_clim()
# Get the pixel dimension of the axis to fill.
fig = plt.gcf()
bbox = ax.get_window_extent().transformed(fig.dpi_scale_trans.inverted())
width, height = int(round(bbox.width * fig.dpi)), int(round(bbox.height * fig.dpi))
# Draw the colormap gradient.
img = np.zeros((height, width, 3))
xgrad = np.linspace(0, 1, width)
img[:] = cmap(xgrad)[:, :-1]
# Superimpose a histogram of pixel values.
counts, _ = np.histogram(D.reshape(-1), bins=np.linspace(vmin, vmax, width + 1))
hist_height = ((height - 1) * counts / counts[1:-1].max()).astype(int)
mask = np.arange(height).reshape(-1, 1) < hist_height
if mode == 'color':
img[mask] = (1 - alpha) * img[mask] + alpha * np.asarray(matplotlib.colors.to_rgb(color))
elif mode == 'reverse':
cmap_r = cmap.reversed()
for i, x in enumerate(xgrad):
img[mask[:, i], i] = cmap_r(x)[:-1]
elif mode == 'complement':
# https://stackoverflow.com/questions/40233986/
# python-is-there-a-function-or-formula-to-find-the-complementary-colour-of-a-rgb
hilo = np.amin(img, axis=2, keepdims=True) + np.amax(img, axis=2, keepdims=True)
img[mask] = hilo[mask] - img[mask]
else:
raise ValueError('Invalid mode "{0}".'.format(mode))
ax.imshow(img, interpolation='none', origin='lower')
def plot_pixels(D, label=None, colorhist=False, zoom=1, masked_color='cyan',
imshow_args={}, text_args={}, colorhist_args={}):
"""Plot pixel data at 1:1 scale with an optional label and colorhist.
"""
dpi = 100 # value only affects metadata in an output file, not appearance on screen.
ny, nx = D.shape
width, height = zoom * nx, zoom * ny
if colorhist:
colorhist_height = 32
height += colorhist_height
fig = plt.figure(figsize=(width / dpi, height / dpi), dpi=dpi, frameon=False)
ax = plt.axes((0, 0, 1, zoom * ny / height))
args = dict(imshow_args)
for name, default in dict(interpolation='none', origin='lower', cmap='plasma_r').items():
if name not in args:
args[name] = default
# Set the masked color in the specified colormap.
cmap = copy.copy(matplotlib.cm.get_cmap(args['cmap']))
cmap.set_bad(color=masked_color)
args['cmap'] = cmap
# Draw the image.
I = ax.imshow(D, **args)
ax.axis('off')
if label:
args = dict(text_args)
for name, default in dict(color='w', fontsize=18).items():
if name not in args:
args[name] = default
outline = [
matplotlib.patheffects.Stroke(linewidth=1, foreground='k'),
matplotlib.patheffects.Normal()]
text = ax.text(0.01, 0.01 * nx / ny, label, transform=ax.transAxes, **args)
text.set_path_effects(outline)
if colorhist:
axcb = plt.axes((0, zoom * ny / height, 1, colorhist_height / height))
plot_colorhist(D, axcb, I, **colorhist_args)
return fig, ax
def plot_data(D, W, downsampling=4, zoom=1, label=None, colorhist=False, stamps=[],
preprocess_args={}, imshow_args={}, text_args={}, colorhist_args={}):
"""Plot weighted image data using downsampling, optional preprocessing, and decorators.
"""
# Downsample the input data.
D, W = desietcimg.util.downsample_weighted(D, W, downsampling)
# Preprocess the data for display.
D = desietcimg.util.preprocess(D, W, **preprocess_args)
ny, nx = D.shape
# Display the image.
args = dict(imshow_args)
if 'extent' not in args:
# Use the input pixel space for the extent, without downsampling.
args['extent'] = [-0.5, nx * downsampling - 0.5, -0.5, ny * downsampling - 0.5]
fig, ax = plot_pixels(D, zoom=zoom, label=label, colorhist=colorhist,
imshow_args=args, text_args=text_args, colorhist_args=colorhist_args)
outline = [
matplotlib.patheffects.Stroke(linewidth=1, foreground='k'),
matplotlib.patheffects.Normal()]
for k, stamp in enumerate(stamps):
yslice, xslice = stamp[:2]
xlo, xhi = xslice.start, xslice.stop
ylo, yhi = yslice.start, yslice.stop
rect = plt.Rectangle((xlo, ylo), xhi - xlo, yhi - ylo, fc='none', ec='w', lw=1)
ax.add_artist(rect)
if xhi < nx // 2:
xtext, halign = xhi, 'left'
else:
xtext, halign = xlo, 'right'
text = ax.text(
xtext, 0.5 * (ylo + yhi), str(k), fontsize=12, color='w', va='center', ha=halign)
text.set_path_effects(outline)
return fig, ax
def plot_full_frame(D, W=None, saturation=None, downsampling=8, clip_pct=0.5, dpi=100, GCR=None,
label=None, cmap='plasma_r', fg_color='w', compress=True, vmin=None, vmax=None):
# Convert to a float32 array.
D, W = desietcimg.util.prepare(D, W, saturation=saturation)
# Downsample.
WD = desietcimg.util.downsample(D * W, downsampling=downsampling, summary=np.sum, allow_trim=True)
W = desietcimg.util.downsample(W, downsampling=downsampling, summary=np.sum, allow_trim=True)
D = np.divide(WD, W, out= | np.zeros_like(WD) | numpy.zeros_like |
#!/usr/bin/env python
# -*- coding: utf-8 -*-
#
# ade:
# Asynchronous Differential Evolution.
#
# Copyright (C) 2018-19 by <NAME>,
# http://edsuom.com/ade
#
# See edsuom.com for API documentation as well as information about
# Ed's background and other projects, software and otherwise.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the
# License. You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing,
# software distributed under the License is distributed on an "AS
# IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either
# express or implied. See the License for the specific language
# governing permissions and limitations under the License.
"""
Unit tests for L{ade.report}.
"""
from io import StringIO
import numpy as np
from twisted.internet import defer
from ade.util import *
from ade import report
from ade.test import testbase as tb
#import twisted.internet.base
#twisted.internet.base.DelayedCall.debug = True
class TestReporter(tb.TestCase):
def setUp(self):
self.calls = []
self.p = tb.MockPopulation(tb.ackley, ['x', 'y'], [(0,1)]*2, popsize=1)
self.r = report.Reporter(self.p, self.processComplaint)
self.r.addCallback(self.cbImmediate, 3.14, bar=9.87)
return self.p.setup()
def cbImmediate(self, values, counter, SSE, foo, bar=None):
self.calls.append(['cbi', values, counter, SSE, foo, bar])
if SSE == 0:
# Complaint
return 123
def cbDeferred(self, values, counter, SSE):
def doNow(null):
self.calls.append(['cbd', values, counter, SSE])
return self.deferToDelay(0.5).addCallback(doNow)
def processComplaint(self, i, result):
self.calls.append(['processComplaint', i, result])
def test_runCallbacks_basic(self):
i = self.p.spawn([1.0, 2.0])
i.SSE = 0.876
self.r.runCallbacks(i)
self.assertEqual(
self.calls,
[['cbi', [1.0, 2.0], 0, 0.876, 3.14, 9.87]])
return self.r.waitForCallbacks()
def test_runCallbacks_complaint(self):
i = self.p.spawn([0.0, 0.0])
i.SSE = 0.0
self.r.runCallbacks(i)
self.assertEqual(
self.calls,
[['cbi', [0.0, 0.0], 0, 0.0, 3.14, 9.87],
['processComplaint', i, 123]])
return self.r.waitForCallbacks()
@defer.inlineCallbacks
def test_runCallbacks_stacked(self):
i1 = self.p.spawn([1, 2])
i1.SSE = 0.876
i2 = self.p.spawn([3, 4])
i2.SSE = 0.543
self.r.addCallback(self.cbDeferred)
self.r.runCallbacks(i1)
self.r.cbrScheduled(i2)
self.assertEqual(self.calls, [
['cbi', [1, 2], 0, 0.876, 3.14, 9.87],
])
yield self.r.waitForCallbacks()
self.assertEqual(self.calls, [
['cbi', [1, 2], 0, 0.876, 3.14, 9.87],
['cbd', [1, 2], 0, 0.876],
['cbi', [3, 4], 0, 0.543, 3.14, 9.87],
['cbd', [3, 4], 0, 0.543],
])
@defer.inlineCallbacks
def test_newBest_basic(self):
self.r.addCallback(self.cbDeferred)
yield self.p.setup(uniform=True)
self.r.newBest(self.p.best())
self.assertEqual(len(self.calls), 1)
yield self.deferToDelay(0.6)
self.assertEqual(len(self.calls), 2)
@defer.inlineCallbacks
def test_newBest_stacked(self):
self.r.addCallback(self.cbDeferred)
yield self.p.setup(uniform=True)
self.r.newBest(self.p.best())
for x in (1E-3, 1E-4, 1E-5):
i = yield self.p.spawn(np.array([x, x])).evaluate()
self.r.newBest(i)
# cbi0
self.assertEqual(len(self.calls), 1)
yield self.deferToDelay(0.6)
# cbi0, cbd0, cbi1
self.assertEqual(
[x[0] for x in self.calls], ['cbi', 'cbd', 'cbi'])
yield self.deferToDelay(0.5)
# cbi0, cbd0, cbi1, cbd2
self.assertEqual(
[x[0] for x in self.calls], ['cbi', 'cbd', 'cbi', 'cbd'])
yield self.deferToDelay(1.0)
self.assertEqual(len(self.calls), 4)
self.assertEqual(self.calls[-1][1][0], 1E-5)
@defer.inlineCallbacks
def test_msgRatio(self):
fh = StringIO()
msg(fh)
iPrev = yield self.p.spawn( | np.array([1.001E-3, 1.001E-3]) | numpy.array |
# ---------------------------------------------------------------------
# Project "Track 3D-Objects Over Time"
# Copyright (C) 2020, Dr. <NAME> / Dr. <NAME>.
#
# Purpose of this file : Data association class with single nearest neighbor association and gating based on Mahalanobis distance
#
# You should have received a copy of the Udacity license together with this program.
#
# https://www.udacity.com/course/self-driving-car-engineer-nanodegree--nd013
# ----------------------------------------------------------------------
#
# imports
import numpy as np
from scipy.stats.distributions import chi2
# add project directory to python path to enable relative imports
import os
import sys
PACKAGE_PARENT = '..'
SCRIPT_DIR = os.path.dirname(os.path.realpath(os.path.join(os.getcwd(), os.path.expanduser(__file__))))
sys.path.append(os.path.normpath(os.path.join(SCRIPT_DIR, PACKAGE_PARENT)))
import misc.params as params
class Association:
'''Data association class with single nearest neighbor association and gating based on Mahalanobis distance'''
def __init__(self):
self.association_matrix = | np.matrix([]) | numpy.matrix |
import numpy as np
from numba import vectorize
# Size 为列表,为神经网络结构,比如[3,5,5,4,2],3是输入层神经元个数,中间为隐藏层每层神经元个数,2为输出层个数
class nn_Creat():
def __init__(self,Size,active_fun='sigmoid',learning_rate=1.5,batch_normalization=1,objective_fun='MSE',
output_function='sigmoid',optimization_method='normal',weight_decay=0):
self.Size=Size # 初始化网络参数,并进行打印
print('the structure of the NN is \n', self.Size)
self.active_fun=active_fun
print('active function is %s '% active_fun)
self.learning_rate=learning_rate
print('learning_rate is %s '% learning_rate)
self.batch_normalization=batch_normalization
print('batch_normalization is %d '% batch_normalization)
self.objective_fun=objective_fun
print('objective_function is %s '% objective_fun)
self.optimization_method=optimization_method
print('optimization_method is %s '% optimization_method)
self.weight_decay = weight_decay
print('weight_decay is %f '% weight_decay)
# 初始化网络权值和梯度
self.vecNum=0
self.depth=len(Size)
self.W=[]
self.b=[]
self.W_grad=[]
self.b_grad=[]
self.cost=[]
if self.batch_normalization: # 是否运用批量归一化,如果用,则引入期望E和方差S,以及缩放因子Gamma、Beta
self.E = []
self.S = []
self.Gamma = []
self.Beta = []
if objective_fun=='Cross Entropy': # 目标函数是否为交叉墒函数
self.output_function='softmax'
else:
self.output_function='sigmoid'
print('output_function is %s \n'% self.output_function)
print('Start training NN \n')
for item in range(self.depth-1):
width=self.Size[item]
height=self.Size[item+1]
q=2*np.random.rand(height,width)/np.sqrt(width)-1/np.sqrt(width) #初始化权系数W
self.W.append(q)
if self.active_fun=='relu': # 判断激活函数是否为relu函数,以决定b的初始化形式
self.b.append(np.random.rand(height,1)+0.01)
else:
self.b.append(2*np.random.rand(height,1)/np.sqrt(width)-1/np.sqrt(width))
if self.optimization_method=='Momentum': #优化方向是否使用矩形式,即为之前梯度的叠加
if item!=0:
self.vW.append(np.zeros([height,width]))
self.vb.append(np.zeros([height, 1]))
else:
self.vW=[]
self.vb=[]
self.vW.append(np.zeros([height, width]))
self.vb.append(np.zeros([height, 1]))
if self.optimization_method=='AdaGrad'or optimization_method=='RMSProp' or optimization_method=='Adam': #优化方法是否使用上述方法
if item!=0:
self.rW.append(np.zeros([height,width]))
self.rb.append(np.zeros([height, 1]))
else:
self.rW=[]
self.rb=[]
self.rW.append(np.zeros([height, width]))
self.rb.append(np.zeros([height, 1]))
if self.optimization_method == 'Adam': #优化方法是否为Adam方法
if item!=0:
self.sW.append(np.zeros([height, width]))
self.sb.append(np.zeros([height, 1]))
else:
self.sW = []
self.sb = []
self.sW.append(np.zeros([height, width]))
self.sb.append(np.zeros([height, 1]))
if self.batch_normalization: #是否对每层进行归一化
self.Gamma.append(np.array([1]))
self.Beta.append(np.array([0]))
self.E.append(np.zeros([height,1]))
self.S.append(np.zeros([height,1]))
if self.optimization_method=='Momentum': #在归一化基础上是否使用Momentun方法
if item!=0:
self.vGamma.append(np.array([1]))
self.vBeta.append(np.array([0]))
else:
self.vGamma = []
self.vBeta = []
self.vGamma.append(np.array([1]))
self.vBeta.append(np.array([0]))
if self.optimization_method == 'AdaGrad' or optimization_method == 'RMSProp' or optimization_method == 'Adam': # 在归一化基础上优化方法是否使用上述方法
if item!=0:
self.rGamma.append(np.array([0]))
self.rBeta.append(np.array([0]))
else:
self.rGamma = []
self.rBeta = []
self.rGamma.append(np.array([0]))
self.rBeta.append(np.array([0]))
if self.optimization_method == 'Adam': #在归一化基础上是否使用Adam方法
if item!=0:
self.sGamma.append(np.array([1]))
self.sBeta.append(np.array([0]))
else:
self.sGamma = []
self.sBeta = []
self.sGamma.append(np.array([1]))
self.sBeta.append(np.array([0]))
self.W_grad.append(np.array([]))
self.b_grad.append(np.array([]))
def nn_train(self,train_x,train_y,iterations=10,batch_size=100): #神经网络训练流程化
# 随机将数据分为num_batches堆,每堆Batch_Size个
Batch_Size=batch_size
m=np.size(train_x,0)
num_batches=np.round(m/Batch_Size)
num_batches=np.int(num_batches)
for k in range(iterations):
kk=np.random.randint(0,m,m)
for l in range(num_batches):
batch_x=train_x[kk[l*batch_size:(l+1)*batch_size ],:]
batch_y=train_y[kk[l*batch_size:(l+1)*batch_size ],:]
self.nn_forward(batch_x,batch_y) # 执行神经网络向前传播
self.nn_backward(batch_y) # 执行神经网络向后传播
self.gradient_obtain() # 执行得到所以参数的梯度
return None
def Sigmoid(self,z): # 定义sigmoid函数
yyy=1/(1+np.exp(-z))
return yyy
def SoftMax(self,x): # 定义Softmax函数
e_x = np.exp(x - np.max(x,0))
return e_x / np.sum(e_x,0)
def Relu(self,xxx): # 定义Relu函数
# xxx[xxx<0]=0
s=np.maximum(xxx, 0)
return s
def nn_forward(self,batch_x,batch_y): # 神经网络向前传播,得到对z偏导theta,每层输出a和cost
batch_x=batch_x.T
batch_y=batch_y.T
m=np.size(batch_x,1)
self.a=[] #定义每层激活函数输出
self.a.append(batch_x) # 接受第一层输入
cost2=0 #初始化正则函数
self.yy=[]
for k in range(1,self.depth): # 从第一层开始,循环求每层输出
y=(self.W[k-1].dot(self.a[k-1]))+(np.repeat(self.b[k-1],m,1))
if self.batch_normalization:
self.E[k-1]=self.E[k-1]*self.vecNum+np.sum(y,1)[:,None]
self.S[k-1]=self.S[k-1]**2*(self.vecNum-1)+((m-1)*np.std(y,1)**2)[:,None]
self.vecNum=self.vecNum+m
self.E[k-1]=self.E[k-1]/self.vecNum #求期望
self.S[k-1]= | np.sqrt(self.S[k-1]/(self.vecNum-1)) | numpy.sqrt |
import numpy as np
import matplotlib.pyplot as plt
def gaussian_func(sigma, x):
return 1 / np.sqrt(2 * np.pi * (sigma ** 2)) * np.exp(-(x ** 2) / (2 * (sigma ** 2)))
def gaussian_random_generator(sigma=5, numbers=100000):
uniform_random_numbers = np.random.rand(numbers, 2)
rho = sigma * np.sqrt(-2 * np.log(1 - uniform_random_numbers[:, 0]))
theta = 2 * np.pi * uniform_random_numbers[:, 1]
gaussian_random = rho * np.array([ | np.cos(theta) | numpy.cos |
#!/usr/bin/env python2
# -*- coding: utf-8 -*-
"""
Created on Mon Mar 23 09:26:17 2020
@author: jone
"""
import pandas as pd
import numpy as np
import dipole
import matplotlib.pyplot as plt
import datetime as dt
#investigate the relationship between equatorward boundary in NOAA data and the
#occurrence rate of substorms from sophie list
#Load omni data
omnifile = '/home/jone/Documents/Dropbox/science/superdarn/lobe_circulation/omni_1min_1999-2017.hdf'
omni = pd.read_hdf(omnifile)
#Load Sophie list
sophie = pd.read_hdf('./jone_data/sophie75.h5')
use = (sophie.index>=omni.index[0]) & (sophie.index<=omni.index[-1])
#use = (sophie.index>=dt.datetime(2003,1,1,0,0)) & (sophie.index<dt.datetime(2004,1,1,0,0))
sophie = sophie[use].copy()
exp = sophie.ssphase == 2 #expansion phase list
sophiexp = sophie[exp].copy()
#Process omni data
use = (omni.index >= sophiexp.index[0]) & (omni.index <= sophiexp.index[-1])
omni = omni[use].copy()
omni = omni.interpolate(limit=5, limit_direction='both')
bypos = (omni.BY_GSM>0)# & (np.abs(omni.BZ_GSM)<np.abs(omni.BY_GSM))
omni.loc[:,'bypos'] = omni.BY_GSM[bypos]
byneg = (omni.BY_GSM<0)# & (np.abs(omni.BZ_GSM)<np.abs(omni.BY_GSM))
omni.loc[:,'byneg'] = omni.BY_GSM[byneg]
omni.loc[:,'milan'] = 3.3e5 * (omni['flow_speed']*1000.)**(4./3) * (np.sqrt(omni.BY_GSM**2 + omni.BZ_GSM**2)) * 1e-9 * \
np.sin(np.abs(np.arctan2(omni['BY_GSM'],omni['BZ_GSM']))/2.)**(4.5) * 0.001
milanpos = 3.3e5 * (omni['flow_speed']*1000.)**(4./3) * (np.sqrt(omni.bypos**2 + omni.BZ_GSM**2)) * 1e-9 * \
np.sin(np.abs(np.arctan2(omni['bypos'],omni['BZ_GSM']))/2.)**(4.5) * 0.001
milanneg = 3.3e5 * (omni['flow_speed']*1000.)**(4./3) * (np.sqrt(omni.byneg**2 + omni.BZ_GSM**2)) * 1e-9 * \
np.sin(np.abs(np.arctan2(omni['byneg'],omni['BZ_GSM']))/2.)**(4.5) * 0.001
window = 60 #minutes
nobsinwindow = omni.milan.rolling(window).count()
cumsumneg = milanneg.rolling(window,min_periods=1).sum()
cumsumpos = milanpos.rolling(window,min_periods=1).sum()
omni.loc[:,'bxlong'] = omni.BX_GSE.rolling(window,min_periods=1).mean()
omni.loc[:,'bzlong'] = omni.BZ_GSM.rolling(window,min_periods=1).mean()
omni.loc[:,'bylong'] = omni.BY_GSM.rolling(window,min_periods=1).mean()
bxlim = 200
usepos = ((cumsumpos>2.*cumsumneg) & (nobsinwindow==window) & (np.abs(omni.bxlong)<bxlim)) | ((cumsumneg.isnull()) & (np.invert(cumsumpos.isnull())) & (nobsinwindow==window) & (np.abs(omni.bxlong)<bxlim))
useneg = ((cumsumneg>2.*cumsumpos) & (nobsinwindow==window) & (np.abs(omni.bxlong)<bxlim)) | ((cumsumpos.isnull()) & (np.invert(cumsumneg.isnull())) & (nobsinwindow==window) & (np.abs(omni.bxlong)<bxlim))
omni.loc[:,'usepos'] = usepos
omni.loc[:,'useneg'] = useneg
omni.loc[:,'milanlong'] = omni.milan.rolling(window, min_periods=window, center=False).mean() #average IMF data
#omni = omni.drop(['bxlong','bzlong','byneg','bypos','PC_N_INDEX','Beta','E','Mach_num','Mgs_mach_num','y'],axis=1) #need to drop PC index as it contain a lot of nans. Also other fields will exclude data when we later use dropna()
#Combine omni and sophie list
sophiexp.loc[:,'tilt'] = dipole.dipole_tilt(sophiexp.index)
omni.loc[:,'tilt'] = dipole.dipole_tilt(omni.index)
omni2 = omni.reindex(index=sophiexp.index, method='nearest', tolerance='30sec')
sophiexp.loc[:,'bylong'] = omni2.bylong
sophiexp.loc[:,'milanlong'] = omni2.milanlong
sophiexp.loc[:,'substorm'] = sophiexp.ssphase==2
bybins = np.append(np.append([-50],np.linspace(-9,9,10)),[50])
bybincenter = np.linspace(-10,10,11)
sgroup = sophiexp.groupby([pd.cut(sophiexp.tilt, bins=np.array([-35,-10,10,35])), \
pd.cut(sophiexp.bylong, bins=bybins)])
ogroup = omni.groupby([pd.cut(omni.tilt, bins=np.array([-35,-10,10,35])), \
pd.cut(omni.bylong, bins=bybins)])
#Plotting
fig = plt.figure(figsize=(15,15))
ax = fig.add_subplot(221)
res = ogroup.tilt.count() / sgroup.substorm.sum()
ax.plot(bybincenter, np.array(res[0:11]), label='tilt < -10')
ax.plot(bybincenter, np.array(res[11:22]), label='|tilt| < 10')
ax.plot(bybincenter, np.array(res[22:33]), label='tilt > 10')
ax.legend()
ax.set_xlabel('IMF By')
ax.set_ylabel('Average time between onsets [min]')
ax.set_title('Onsets from Sophie-75 list, 1999-2014')
ax = fig.add_subplot(222)
#milanstat, bins= pd.qcut(omni.milanlong, 10, retbins=True)
#milanbincenter = [(a + b) /2 for a,b in zip(bins[:-1], bins[1:])]
#sgroup = sophiexp.groupby([pd.cut(sophiexp.tilt, bins=np.array([-35,-10,10,35])), \
# pd.qcut(sophiexp.milanlong, 10)])
#ogroup = omni.groupby([pd.cut(omni.tilt, bins=np.array([-35,-10,10,35])), \
# pd.qcut(omni.milanlong, 10)])
res = ogroup.milanlong.median()
ax.plot(bybincenter, | np.array(res[0:11]) | numpy.array |
#!/usr/bin/env python
from __future__ import absolute_import, division, print_function
import rospy
import numpy as np
import time
import math
import random
import tf
from gym import spaces
from .cable_joint_robot_env import CableJointRobotEnv
from gym.envs.registration import register
from gazebo_msgs.msg import ModelState
from geometry_msgs.msg import Pose, Twist
from std_msgs.msg import Float32MultiArray
# Register crib env
register(
id='CablePoint-v0',
entry_point='envs.cable_point_task_env:CablePointTaskEnv')
class CablePointTaskEnv(CableJointRobotEnv):
def __init__(self):
"""
This task-env is designed for cable-driven joint pointing at the desired goal.
Action and state space will be both set to continuous.
"""
# action limits
self.max_force = 20
self.min_force = 0
# observation limits
self.max_roll = math.pi
self.max_pitch = math.pi
self.max_yaw = math.pi
self.max_roll_dot = 10*math.pi
self.max_pitch_dot = 10*math.pi
self.max_yaw_dot = 10*math.pi
# action space
self.high_action = np.array(4*[self.max_force])
self.low_action = np.zeros(4)
self.action_space = spaces.Box(low=self.low_action, high=self.high_action)
# observation space
self.high_observation = np.array(
[
self.max_roll,
self.max_pitch,
self.max_yaw,
self.max_roll_dot,
self.max_pitch_dot,
self.max_yaw_dot
]
)
self.low_observation = -self.high_observation
self.observation_space = spaces.Box(low=self.low_observation, high=self.high_observation)
# action and observation
self.action = np.zeros(self.action_space.shape[0])
self.observation = np.zeros(self.observation_space.shape[0])
# info, initial position and goal position
self.init_orientation = np.zeros(3)
self.current_orientation = | np.zeros(3) | numpy.zeros |
"""
:py:class:`UtilsCommonMode` contains detector independent utilities for common mode correction
==============================================================================================
Usage::
from psana.detector.UtilsCommonMode import *
#OR
import psana.detector.UtilsCommonMode as ucm
ucm.common_mode_rows(arr, mask=None, cormax=None, npix_min=10)
ucm.common_mode_cols(arr, mask=None, cormax=None, npix_min=10)
ucm.common_mode_2d(arr, mask=None, cormax=None, npix_min=10)
ucm.common_mode_rows_hsplit_nbanks(data, mask, nbanks=4, cormax=None)
ucm.common_mode_2d_hsplit_nbanks(data, mask, nbanks=4, cormax=None)
This software was developed for the LCLS project.
If you use all or part of it, please give an appropriate acknowledgment.
Created on 2018-01-31 by <NAME>
2021-02-02 adopted to LCLS2
"""
import logging
logger = logging.getLogger(__name__)
import numpy as np
from math import fabs
from psana.pyalgos.generic.NDArrUtils import info_ndarr, print_ndarr
def common_mode_rows(arr, mask=None, cormax=None, npix_min=10):
"""Defines and applys common mode correction to 2-d arr for rows.
I/O parameters:
- arr (float) - i/o 2-d array of intensities
- mask (int or None) - the same shape 2-d array of bad/good = 0/1 pixels
- cormax (float or None) - maximal allowed correction in ADU
- npix_min (int) - minimal number of good pixels in row to evaluate and apply correction
"""
rows, cols = arr.shape
if mask is None:
cmode = np.median(arr,axis=1) # column of median values
else:
marr = np.ma.array(arr, mask=mask<1) # use boolean inverted mask (True for masked pixels)
cmode = np.ma.median(marr,axis=1) # column of median values for masked array
npix = mask.sum(axis=1) # count good pixels in each row
#print('npix', npix[:100])
cmode = np.select((npix>npix_min,), (cmode,), default=0)
if cormax is not None:
cmode = np.select((np.fabs(cmode) < cormax,), (cmode,), default=0)
#logger.debug(info_ndarr(cmode, 'cmode'))
_,m2 = np.meshgrid(np.zeros(cols, dtype=np.int16), cmode) # stack cmode 1-d column to 2-d matrix
if mask is None:
arr -= m2
else:
bmask = mask>0
arr[bmask] -= m2[bmask]
def common_mode_cols(arr, mask=None, cormax=None, npix_min=10):
"""Defines and applys common mode correction to 2-d arr for cols.
I/O parameters:
- arr (float) - i/o 2-d array of intensities
- mask (int or None) - the same shape 2-d array of bad/good = 0/1 pixels
- cormax (float or None) - maximal allowed correction in ADU
- npix_min (int) - minimal number of good pixels in column to evaluate and apply correction
"""
rows, cols = arr.shape
if mask is None:
cmode = np.median(arr,axis=0)
else:
marr = np.ma.array(arr, mask=mask<1) # use boolean inverted mask (True for masked pixels)
cmode = np.ma.median(marr,axis=0) # row of median values for masked array
npix = mask.sum(axis=0) # count good pixels in each column
cmode = np.select((npix>npix_min,), (cmode,), default=0)
if cormax is not None:
cmode = np.select((np.fabs(cmode) < cormax,), (cmode,), default=0)
#logger.debug(info_ndarr(cmode, 'cmode'))
m1,_ = np.meshgrid(cmode, np.zeros(rows, dtype=np.int16)) # stack cmode 1-d row to 2-d matrix
if mask is None:
arr -= m1
else:
bmask = mask>0
arr[bmask] -= m1[bmask]
def common_mode_2d(arr, mask=None, cormax=None, npix_min=10):
"""Defines and applys common mode correction to entire 2-d arr using the same shape mask.
"""
if mask is None:
cmode = np.median(arr)
if cormax is None or fabs(cmode) < cormax:
arr -= cmode
else:
arr1 = np.ones_like(arr, dtype=np.int16)
bmask = mask>0
npix = arr1[bmask].sum()
if npix < npix_min: return
cmode = np.median(arr[bmask])
if cormax is None or fabs(cmode) < cormax:
arr[bmask] -= cmode
def common_mode_rows_hsplit_nbanks(data, mask=None, nbanks=4, cormax=None, npix_min=10):
"""Works with 2-d data and mask numpy arrays,
hsplits them for banks (df. nbanks=4),
for each bank applies median common mode correction for pixels in rows,
hstack banks in array of original data shape and copy results in i/o data
"""
bdata = np.hsplit(data, nbanks)
if mask is None:
for b in bdata:
common_mode_rows(b, None, cormax, npix_min)
else:
bmask = np.hsplit(mask, nbanks)
for b,m in zip(bdata,bmask):
common_mode_rows(b, m, cormax, npix_min)
data[:] = | np.hstack(bdata) | numpy.hstack |
# -*- coding: utf-8 -*-
import os
import numpy as np
import sympy
import yaml
def create_directory(dir_name):
if not os.path.isdir(dir_name):
os.makedirs(dir_name)
def nonzero_indices(a):
return np.nonzero(a)[0]
def BezierIndex(dim, deg):
"""Iterator indexing control points of a Bezier simplex"""
def iterate(c, r):
if len(c) == dim - 1:
yield c + (r,)
else:
for i in range(r, -1, -1):
yield from iterate(c + (i,), r - i)
yield from iterate((), deg)
def poly(i, n):
eq = c(i)
for k in range(n):
eq *= T[k] ** i[k]
return eq * (1 - sum(T[k] for k in range(n))) ** i[n]
def concat_data_to_arrray(d):
index = 0
for key in d:
# if len(key)==1:
# d[key] == d[key].reshape((1,len(d[key])))
if index == 0:
d_array = d[key]
else:
d_array = np.r_[d_array, d[key]]
index += 1
return d_array
def extract_multiple_degree(degree_list, index_list):
"""
return corresponding multiple indices as set
indices: list or tuple
"""
d = set()
for key in degree_list:
if set(np.array(nonzero_indices(key))).issubset(set(index_list)):
d.add(key)
return d
def calculate_l2_expected_error(true, pred):
diff = true - pred
# print(diff)
# print(np.linalg.norm(diff,axis=1))
# print((np.sum(np.linalg.norm(diff,axis=1)**2)))
return (np.sum( | np.linalg.norm(diff, axis=1) | numpy.linalg.norm |
import numpy as np
import torch
from torchvision.ops.boxes import batched_nms
def encode_boxes_to_anchors(boxes, anchors, eps=1e-8):
""" Create anchors regression target based on anchors
Args:
boxes: ground truth boxes.
anchors: anchor boxes on all feature levels.
eps: small number for stability
Returns:
outputs: anchors w.r.t. ground truth
"""
def corner_to_center(rects):
h, w = rects[:, 2] - rects[:, 0], rects[:, 3] - rects[:, 1]
y_ctr, x_ctr = rects[:, 0] + 0.5 * h, rects[:, 1] + 0.5 * w
return y_ctr, x_ctr, h, w
ycenter_a, xcenter_a, ha, wa = corner_to_center(anchors)
ycenter, xcenter, h, w = corner_to_center(boxes)
ha, wa, h, w = ha + eps, wa + eps, h + eps, w + eps
dy = (ycenter - ycenter_a) / ha
dx = (xcenter - xcenter_a) / wa
dh = torch.log(h / ha)
dw = torch.log(w / wa)
outputs = torch.stack([dy, dx, dh, dw]).T
return outputs
def decode_box_outputs(rel_codes, anchors):
"""Transforms relative regression coordinates to absolute positions.
Args:
rel_codes: batched box regression targets.
anchors: batched anchors on all feature levels.
Returns:
outputs: batched bounding boxes.
"""
y_center_a = (anchors[:, :, 0] + anchors[:, :, 2]) / 2
x_center_a = (anchors[:, :, 1] + anchors[:, :, 3]) / 2
ha = anchors[:, :, 2] - anchors[:, :, 0]
wa = anchors[:, :, 3] - anchors[:, :, 1]
ty, tx = rel_codes[:, :, 0], rel_codes[:, :, 1]
th, tw = rel_codes[:, :, 2], rel_codes[:, :, 3]
w = torch.exp(tw) * wa
h = torch.exp(th) * ha
y_center = ty * ha + y_center_a
x_center = tx * wa + x_center_a
y_min = y_center - h / 2.
x_min = x_center - w / 2.
y_max = y_center + h / 2.
x_max = x_center + w / 2.
outputs = torch.stack([y_min, x_min, y_max, x_max], dim=-1)
return outputs
def clip_boxes_(boxes, size):
boxes = boxes.clamp(min=0)
size = torch.cat([size, size], dim=0)
boxes = boxes.min(size)
return boxes
def generate_detections(
cls_outputs, box_outputs, anchor_boxes, indices, classes,
image_sizes, image_scales, max_detections_per_image):
"""Generates batched detections with RetinaNet model outputs and anchors.
Args: (B - batch size, N - top-k selection length)
cls_outputs: a numpy array with shape [B, N, 1], which has the
highest class scores on all feature levels.
box_outputs: a numpy array with shape [B, N, 4], which stacks
box regression outputs on all feature levels.
anchor_boxes: a numpy array with shape [B, N, 4], which stacks
anchors on all feature levels.
indices: a numpy array with shape [B, N], which is the indices from top-k selection.
classes: a numpy array with shape [B, N], which represents
the class prediction on all selected anchors from top-k selection.
image_sizes: a list of tuples representing size of incoming images
image_scales: a list representing the scale between original images
and input images for the detector.
Returns:
detections: detection results in a tensor of shape [B, N, 6],
where [:, :, 0:4] are boxes, [:, :, 5] are scores,
"""
batch_size = indices.shape[0]
device = indices.device
anchor_boxes = anchor_boxes[indices, :]
scores = cls_outputs.sigmoid().squeeze(2).float()
# apply bounding box regression to anchors
boxes = decode_box_outputs(box_outputs.float(), anchor_boxes)
boxes = boxes[:, :, [1, 0, 3, 2]]
batched_boxes, batched_scores, batched_classes = [], [], []
# iterate over batch since we need non-max suppression for each image
for batch_idx in range(batch_size):
batch_boxes = boxes[batch_idx, :, :]
batch_scores = scores[batch_idx, :]
batch_classes = classes[batch_idx, :]
# clip boxes outputs
boxes_max_size = [image_sizes[batch_idx][0] / image_scales[batch_idx],
image_sizes[batch_idx][1] / image_scales[batch_idx]]
boxes_max_size = torch.FloatTensor(boxes_max_size).to(batch_boxes.device)
batch_boxes = clip_boxes_(batch_boxes, boxes_max_size)
# perform non-maximum suppression
top_detection_idx = batched_nms(
batch_boxes, batch_scores, batch_classes, iou_threshold=0.5)
# keep only topk scoring predictions
top_detection_idx = top_detection_idx[:max_detections_per_image]
batch_boxes = batch_boxes[top_detection_idx]
batch_scores = batch_scores[top_detection_idx]
batch_classes = batch_classes[top_detection_idx]
# fill zero predictions to match MAX_DETECTIONS_PER_IMAGE
detections_diff = len(top_detection_idx) - max_detections_per_image
if detections_diff < 0:
add_boxes = torch.zeros(
(-detections_diff, 4), device=device, dtype=batch_boxes.dtype)
batch_boxes = torch.cat([batch_boxes, add_boxes], dim=0)
add_scores = torch.zeros(
(-detections_diff, 1), device=device, dtype=batch_scores.dtype)
batch_scores = torch.cat([batch_scores, add_scores], dim=0)
add_classes = torch.zeros(
(-detections_diff, 1), device=device, dtype=batch_classes.dtype)
batch_classes = torch.cat([batch_classes, add_classes], dim=0)
batch_scores = batch_scores.view(-1, 1)
batch_classes = batch_classes.view(-1, 1)
# stack them together
batched_boxes.append(batch_boxes)
batched_scores.append(batch_scores)
batched_classes.append(batch_classes)
boxes = torch.stack(batched_boxes)
scores = torch.stack(batched_scores)
classes = torch.stack(batched_classes)
# xyxy to xywh & rescale to original image
boxes[:, :, 2] -= boxes[:, :, 0]
boxes[:, :, 3] -= boxes[:, :, 1]
boxes_scaler = torch.FloatTensor(image_scales).to(boxes.device)
boxes = boxes * boxes_scaler[:, None, None]
classes += 1 # back to class idx with background class = 0
detections = torch.cat([boxes, scores, classes.float()], dim=2)
return detections
def calc_iou(a, b):
""" Calculate Intersection-over-Union of two samples
Args:
a (torch.Tensor): set of boxes with shape [N, 4] in yxyx format
b (torch.Tensot): set of boxes with shape [M, 4] in yxyx format
"""
area = (b[:, 3] - b[:, 1]) * (b[:, 2] - b[:, 0])
ih = torch.min(torch.unsqueeze(a[:, 2], dim=1), b[:, 2]) \
- torch.max(torch.unsqueeze(a[:, 0], 1), b[:, 0])
iw = torch.min(torch.unsqueeze(a[:, 3], dim=1), b[:, 3]) \
- torch.max(torch.unsqueeze(a[:, 1], 1), b[:, 1])
ih = torch.clamp(ih, min=0)
iw = torch.clamp(iw, min=0)
ua = torch.unsqueeze((a[:, 3] - a[:, 1])
* (a[:, 2] - a[:, 0]), dim=1) + area - iw * ih
ua = torch.clamp(ua, min=1e-8)
intersection = iw * ih
iou = intersection / ua
return iou
class Anchors:
"""RetinaNet Anchors class."""
def __init__(self, min_level, max_level, num_scales,
aspect_ratios, anchor_scale, image_size, device):
"""Constructs multiscale RetinaNet anchors.
Args:
min_level: integer number of minimum level of the output feature pyramid.
max_level: integer number of maximum level of the output feature pyramid.
num_scales: integer number representing intermediate scales added
on each level. For instances, num_scales=2 adds two additional
anchor scales [2^0, 2^0.5] on each level.
aspect_ratios: list of tuples representing the aspect ratio anchors added
on each level. For instances, aspect_ratios =
[(1, 1), (1.4, 0.7), (0.7, 1.4)] adds three anchors on each level.
anchor_scale: float number representing the scale of size of the base
anchor to the feature stride 2^level.
image_size: integer number of input image size. The input image has the
same dimension for width and height. The image_size should be divided by
the largest feature stride 2^max_level.
"""
self.min_level = min_level
self.max_level = max_level
self.num_scales = num_scales
self.aspect_ratios = aspect_ratios
self.anchor_scale = anchor_scale
self.image_size = image_size
self.device = device
self.config = self._generate_configs()
self.boxes = self._generate_boxes()
def _generate_configs(self):
"""Generate configurations of anchor boxes."""
anchor_configs = {}
for level in range(self.min_level, self.max_level + 1):
anchor_configs[level] = []
for scale_octave in range(self.num_scales):
for aspect in self.aspect_ratios:
anchor_configs[level].append(
(2 ** level, scale_octave / float(self.num_scales), aspect))
return anchor_configs
def _generate_boxes(self):
"""Generates multiscale anchor boxes."""
boxes_all = []
for _, configs in self.config.items():
boxes_level = []
for config in configs:
stride, octave_scale, aspect = config
if self.image_size % stride != 0:
raise ValueError(
"input size must be divided by the stride.")
base_anchor_size = self.anchor_scale * stride * 2 ** octave_scale
anchor_size_x_2 = base_anchor_size * aspect[0] / 2.0
anchor_size_y_2 = base_anchor_size * aspect[1] / 2.0
x = np.arange(stride / 2, self.image_size, stride)
y = np.arange(stride / 2, self.image_size, stride)
xv, yv = np.meshgrid(x, y)
xv = xv.reshape(-1)
yv = yv.reshape(-1)
boxes = np.vstack((yv - anchor_size_y_2, xv - anchor_size_x_2,
yv + anchor_size_y_2, xv + anchor_size_x_2))
boxes = | np.swapaxes(boxes, 0, 1) | numpy.swapaxes |
#!/usr/bin/env python
from optparse import OptionParser
import os
import numpy as np
import pysam
import pyBigWig
import tensorflow as tf
from basenji.dna_io import hot1_dna
from basenji.tfrecord_batcher import order_tfrecords
'''
tfr_bw.py
Generate BigWig tracks from TFRecords.
Experimental!
'''
################################################################################
# main
################################################################################
def main():
usage = 'usage: %prog [options] <tfr_dir> <out_bw>'
parser = OptionParser(usage)
parser.add_option('-f', dest='fasta_file',
default='%s/assembly/ucsc/hg38.fa' % os.environ['HG38'])
parser.add_option('-g', dest='genome_file',
default='%s/assembly/ucsc/hg38.human.genome' % os.environ['HG38'])
parser.add_option('-l', dest='target_length',
default=1024, type='int',
help='TFRecord target length [Default: %default]')
parser.add_option('-s', dest='data_split', default='train')
parser.add_option('-t', dest='target_i',
default=0, type='int', help='Target index [Default: %default]')
(options,args) = parser.parse_args()
if len(args) != 2:
parser.error('Must provide TF Records directory and output BigWig')
else:
tfr_dir = args[0]
out_bw_file = args[1]
# initialize output BigWig
out_bw_open = pyBigWig.open(out_bw_file, 'w')
# construct header
header = []
for line in open(options.genome_file):
a = line.split()
header.append((a[0], int(a[1])))
# write header
out_bw_open.addHeader(header)
# initialize chr dictionary
chr_values = {}
for chrm, clen in header:
chr_values[chrm] = | np.zeros(clen, dtype='float16') | numpy.zeros |
#!/usr/local/sci/bin/python
# PYTHON3.6.1
#
# Author: <NAME>
# Created: 18 Jul 2018
# Last update: 15 Apr 2019
# Location: /data/local/hadkw/HADCRUH2/UPDATE2017/PROGS/PYTHON/
# GitHub: https://github.com/Kate-Willett/HadISDH_Build
# -----------------------
# CODE PURPOSE AND OUTPUT
# -----------------------
# THIS CODE DOES MANY THINGS BUT ONLY ONE THING AT A TIME! SO RE-RUN FOR MULTIPLE THINGS
# NOTE THAT FOR ANY 1BY1 OUTPUT IT REGRIDS TO BE 89.5 to -89.5 rather than 90 - -90 (180 boxes rather than 181!!!)
# AND ROLLS LONGITUDE TO -179.5 to 179.5
#
# AT THE MOMENT THIS ASSUMES COMPLETE FIELDS SO WON'T WORK FOR SST!!!
#
# ANOTHER ISSUE IS LAND / SEA MASKING - TOO MUCH LAND COVER, TOO MUCH SEA COVER - SO THERE WILL BE CONTAMINATION!
# I COMPUTE ANOMALIES AT 1by1 RES BEFORE REGRIDDING TO 5by5 TO MINIMISE THIS>
#
#
# This code reads in the ERA-Interim months of 1by1 6 hourly or monthly variables
# (e.g., T, Td and Surface Pressure etc) for the full time period
#
# If desired it converts to humidity variables
# If desired it averages to monthly means and saves to netCDF:
# days since 19790101 (float), 181 lats 90 to -90, 360 lons 0 to 359, <var>2m
# If desired it regrids to 5by5 (monthly means only) and saves to netCDF
# days since 19790101 (int), 36 lats -87.5 to 87.5, 72 lons -177.5 to 177.5, actuals
# If desired it calculates anomalies over a climatological references period given (default 1981-2010)
# and saves to netCDF
# For anomalies it also creates a land only and ocean only set of grids to save along side the complete
# fields
# days since 19790101 (int), 36 lats -87.5 to 87.5, 72 lons -177.5 to 177.5, anomalies,
# anomalies_land, anomalies_sea
#
# The ERA-Interim updates have to be downloaded from ERADownload.py code
# This requires a key to be set up in .ecmwfapirc annually - obtained from logging in to ECMWF
# https://confluence.ecmwf.int/display/WEBAPI/How+to+retrieve+ECMWF+Public+Datasets
# It also requires ecmwfapi to be downloaded and in the directory as you are running to code from
#
# The ERA5 updates have to be downloaded using ERA5Download.py which is in cdsapi-0.1.3/
# Each time you download change the filename to ERAINTERIM_6hr_1by1_MMYYYY.nc
# Save to /data/local/hadkw/HADCRUH2/UPDATE<yyyy>/OTHERDATA/
# Copy previous years of monthly ERAINTERIM data from the previous
# UPDATE<yyyy>/OTHERDATA/<var>2m_monthly_1by1_ERA-Interim_data_1979<yyyy>.nc
# to OTHERDATA/
#
# <references to related published material, e.g. that describes data set>
#
# -----------------------
# LIST OF MODULES
# -----------------------
# inbuilt:
# from datetime import datetime
# import matplotlib.pyplot as plt
# import numpy as np
# from matplotlib.dates import date2num,num2date
# import sys, os
# from scipy.optimize import curve_fit,fsolve,leastsq
# from scipy import pi,sqrt,exp
# from scipy.special import erf
# import scipy.stats
# from math import sqrt,pi
# import struct
# from netCDF4 import Dataset
# from netCDF4 import stringtoarr # for putting strings in as netCDF variables
# import pdb
#
# Kates:
# import CalcHums - written by <NAME> to calculate humidity variables
# import TestLeap - written by <NAME> to identify leap years
# from ReadNetCDF import GetGrid4 - written by <NAME> to pull out netCDF data
# from ReadNetCDF import GetGrid4Slice - written by <NAME> to pull out a slice of netCDF data
# from GetNiceTimes import make_days_since
#
#-------------------------------------------------------------------
# DATA
# -----------------------
# ERA-Interim 1by1 6 hrly gridded data
# ERA<Mmm> = /data/local/hadkw/HADCRUH2/UPDATE<yyyy>/OTHERDATA/ERAINTERIM_<var>_6hr_1by1_<MMYYYY>.nc
#
# -----------------------
# HOW TO RUN THE CODE
# -----------------------
# First make sure the New ERA-Interim data are in the right place.
# Also check all editables in this file are as you wish
# python2.7 ExtractMergeRegridERA_JUL2018.py
#
# -----------------------
# OUTPUT
# -----------------------
# New ERA-Interim 1by1 monthly gridded data for 1979 to present
# NewERA<var> = /data/local/hadkw/HADCRUH2/UPDATE<yyyy>/OTHERDATA/<var>2m_monthly_1by1_ERA-Interim_data_1979<yyyy>.nc
#
# -----------------------
# VERSION/RELEASE NOTES
# -----------------------
#
# Version 1 (18 Jul 2018)
# ---------
#
# Enhancements
#
# Changes
#
# Bug fixes
#
# -----------------------
# OTHER INFORMATION
# -----------------------
#
#************************************************************************
# START
#************************************************************************
# inbuilt:
from datetime import datetime
import matplotlib.pyplot as plt
import numpy as np
from matplotlib.dates import date2num,num2date
import sys, os
from scipy.optimize import curve_fit,fsolve,leastsq
from scipy import pi,sqrt,exp
from scipy.special import erf
import scipy.stats
from math import sqrt,pi
import struct
from netCDF4 import Dataset
from netCDF4 import stringtoarr # for putting strings in as netCDF variables
import pdb
# Kates:
import CalcHums
import TestLeap
from ReadNetCDF import GetGrid4
from ReadNetCDF import GetGrid4Slice
from GetNiceTimes import MakeDaysSince
### START OF EDITABLES ###############################
# Set up initial run choices
# Start and end years
styr = 1979
edyr = 2018
#edOLD = (edyr-styr)*12
stmon = 1
edmon = 12
# Set up output variables - for q, e, RH, dpd, Tw we will need to read in multiple input files
OutputVar = 'dpd' # this can be 't','td','q','rh','e','dpd','tw','ws','slp','sp','uv','sst'
# Is this a new run or an update?
ThisProg = 'Regrid'
# Update for updating an existing file (1by1 monthly or pentad)
# Build for building from scratch (1by1 6hr to 1by1 monthly or pentad)
# THIS AUTOMATICALLY REGRIDS LATS TO BE 180 RATHER THAN 181!!!
# Regrid for changing spatial res from 1by1 to 6hr
# IF OutputGrid = 1by1 then this just changes lats from 181 to 180
# IF OutputGrid = 5by5 then this changes to 36 lats (-87.5 to 87.5) and 72 lons (-177.5 to 177.5)
# Is this ERA-Interim or ERA5?
ThisRean = 'ERA-Interim' # 'ERA5' or 'ERA-Interim'
# Are you reading in hourlies or monthlies?
ReadInTime = 'monthly' # this can be '1hr', '6hr' or 'month' or maybe 'day' later
# Are you converting to monthlies? We will output hourlies anyway if they are read in
OutputTime = 'monthly' # this could be 'monthly' or 'pentad'
# Are you reading in 1by1 or 5by5? We will output 1by1 anyway if they are read in.
ReadInGrid = '1by1' # this can be '1by1' or '5by5'
# Are you converting to 5by5?
OutputGrid = '5by5' # this can be '1by1' or '5by5'
# Do you want to create anomalies and if so, what climatology period? We will output absolutes anyway
MakeAnoms = 1 # 1 for create anomalies (and clim and stdev fields), 0 for do NOT create anomalies
ClimStart = 1981 # any year but generally 1981
ClimEnd = 2010 # any year but generally 2010
### END OF EDITABLES ################
# Set up file paths and other necessary things
if (MakeAnoms == 1): # set the filename string for anomalies
AnomsStr = 'anoms'+str(ClimStart)+'-'+str(ClimEnd)+'_'
else:
AnomsStr = ''
# Set up file locations
updateyy = str(edyr)[2:4]
updateyyyy = str(edyr)
workingdir = '/data/users/hadkw/WORKING_HADISDH/UPDATE'+updateyyyy
if (ReadInGrid == '5by5'):
LandMask = workingdir+'/OTHERDATA/HadCRUT.4.3.0.0.land_fraction.nc' # 0 = 100% sea, 1 = 100% land - no islands!, latitude, longitude, land_area_fraction, -87.5 to 87.5, -177.5 to 177.5
elif (ReadInGrid == '1by1'):
LandMask = workingdir+'/OTHERDATA/lsmask.nc' # 1 = sea, 0 = land - no islands! lat, lon, mask 89.5 to -89.5Lat, 0.5 to 359.5 long
if (OutputVar in ['t','td']): # these are the simple ones that do not require conversion
InputERA = ThisRean+'_'+ReadInGrid+'_'+ReadInTime+'_'+OutputVar+'2m_'
if (ThisProg == 'Update'):
OldERAStr = OutputVar+'2m_'+ReadInGrid+'_'+ReadInTime+'_'+ThisRean+'_data_1979'+str(edyr-1)+'.nc'
else:
OldERAStr = OutputVar+'2m_'+ReadInGrid+'_'+ReadInTime+'_'+AnomsStr+ThisRean+'_data_1979'+str(edyr)+'.nc'
NewERAStr = OutputVar+'2m_'+OutputGrid+'_'+OutputTime+'_'+AnomsStr+ThisRean+'_data_1979'+updateyyyy+'.nc'
elif (OutputVar in ['ws','uv']):
InputERA = ThisRean+'_'+ReadInGrid+'_'+ReadInTime+'_'+OutputVar+'10m_'
if (ThisProg == 'Update'):
OldERAStr = OutputVar+'2m_'+ReadInGrid+'_'+ReadInTime+'_'+ThisRean+'_data_1979'+str(edyr-1)+'.nc'
else:
OldERAStr = OutputVar+'2m_'+ReadInGrid+'_'+ReadInTime+'_'+AnomsStr+ThisRean+'_data_1979'+str(edyr)+'.nc'
NewERAStr = OutputVar+'10m_'+OutputGrid+'_'+OutputTime+'_'+AnomsStr+ThisRean+'_data_1979'+updateyyyy+'.nc'
elif (OutputVar in ['slp','sp','sst']):
InputERA = ThisRean+'_'+ReadInGrid+'_'+ReadInTime+'_'+OutputVar+'_'
if (ThisProg == 'Update'):
OldERAStr = OutputVar+'2m_'+ReadInGrid+'_'+ReadInTime+'_'+ThisRean+'_data_1979'+str(edyr-1)+'.nc'
else:
OldERAStr = OutputVar+'2m_'+ReadInGrid+'_'+ReadInTime+'_'+AnomsStr+ThisRean+'_data_1979'+str(edyr)+'.nc'
NewERAStr = OutputVar+'_'+OutputGrid+'_'+OutputTime+'_'+AnomsStr+ThisRean+'_data_1979'+updateyyyy+'.nc'
elif (OutputVar in ['tw','q','rh','e','dpd']): # these require T, Td and SLP
InputERA = ThisRean+'_'+ReadInGrid+'_'+ReadInTime+'_'
if (ThisProg == 'Update'):
OldERAStr = OutputVar+'2m_'+ReadInGrid+'_'+ReadInTime+'_'+ThisRean+'_data_1979'+str(edyr-1)+'.nc'
else:
OldERAStr = OutputVar+'2m_'+ReadInGrid+'_'+ReadInTime+'_'+AnomsStr+ThisRean+'_data_1979'+str(edyr)+'.nc'
NewERAStr = OutputVar+'2m_'+OutputGrid+'_'+OutputTime+'_'+AnomsStr+ThisRean+'_data_1979'+updateyyyy+'.nc'
# Might have some other options
# Set up variables
mdi = -1e30
# Required variable names for reading in from ERA-Interim
LatInfo = ['latitude']
LonInfo = ['longitude']
# Dictionary for looking up variable names for netCDF read in of variables
NameDict = dict([('q','q2m'),
('rh','rh2m'),
('e','e2m'),
('tw','tw2m'),
('t','t2m'),
('td','td2m'),
('dpd','dpd2m'),
('slp','msl'),
('sp','sp'),
('uv',['u10','v10']), # this one might not work
('ws','si10'),
('sst','sst')])
# Dictionary for looking up variable standard (not actually always standard!!!) names for netCDF output of variables
StandardNameDict = dict([('q','specific_humidity'),
('rh','relative_humidity'),
('e','vapour_pressure'),
('tw','wetbulb_temperature'),
('t','drybulb_temperature'),
('td','dewpoint_temperature'),
('dpd','dewpoint depression'),
('slp','mean_sea_level_pressure'),
('sp','surface_pressure'),
('uv',['10 metre U wind component','10 metre V wind component']), # this one might not work
('ws','10 metre windspeed'),
('sst','sea_surface_temperature')])
# Dictionary for looking up variable long names for netCDF output of variables
LongNameDict = dict([('q','specific_humidity'),
('rh','2m relative humidity from 1by1 6hrly T and Td '+ThisRean),
('e','2m vapour_pressure from 1by1 6hrly T and Td '+ThisRean),
('tw','2m wetbulb_temperature from 1by1 6hrly T and Td '+ThisRean),
('t','2m drybulb_temperature from 1by1 6hrly T '+ThisRean),
('td','2m dewpoint_temperature from 1by1 6hrly Td '+ThisRean),
('dpd','2m dewpoint depression from 1by1 6hrly T and Td '+ThisRean),
('slp','2m mean_sea_level_pressure from 1by1 6hrly msl '+ThisRean),
('sp','2m surface_pressure from 1by1 6hrly sp '+ThisRean),
('uv',['10 metre U wind component from 1by1 6hrly '+ThisRean,'10 metre V wind component from 1by1 6hrly'+ThisRean]), # this one might not work
('ws','10 metre windspeed from 1by1 6hrly'+ThisRean),
('sst','sea surface temperature from 1by1 6hrly'+ThisRean)])
# Dictionary for looking up unit of variables
UnitDict = dict([('q','g/kg'),
('rh','%rh'),
('e','hPa'),
('tw','deg C'),
('t','deg C'),
('td','deg C'),
('dpd','deg C'),
('slp','hPa'),
('sp','hPa'),
('uv','m/s'),
('ws','m/s'),
('sst','deg C')])
nyrs = (edyr+1)-styr
nmons = nyrs*12
npts = nyrs*73
#ndays =
#n6hrs =
#n1hrs =
# set up nlons and nlats depending on what we are reading in and out
if (ReadInGrid == '1by1'):
nlonsIn = 360
nlatsIn= 181 # ERA style to have grids over the poles rather than up to the poles
elif (ReadInGrid == '5by5'):
nlonsIn = 72 # assuming this is correct
nlatsIn = 36 # assuming this is correct
if (OutputGrid == '1by1'):
nlonsOut = 360
nlatsOut = 180 # ERA style to have grids over the poles rather than up to the poles but this will be changed here with Build or Regrid
elif (OutputGrid == '5by5'):
nlonsOut = 72 # assuming this is correct
nlatsOut = 36 # assuming this is correct
## Array for monthly mean data for q, RH, e, T, Tw, Td, DPD one at a time though
##FullMonthArray = np.empty((nmons,nlats,nlons,7),dtype = float)
#FullMonthArray = np.empty((nmons,nlats,nlons),dtype = float)
#FullMonthArray.fill(mdi)
#************************************************************
# SUBROUTINES
#************************************************************
# GetHumidity
def GetHumidity(TheTDat,TheTdDat,TheSPDat,TheVar):
''' Calculates the desired humidity variable if the code is set up to output humidity '''
''' REQUIRES: '''
''' CalcHums.py file to be in the same directory as this file '''
if (TheVar == 't'):
TheHumDat = TheTDat
elif (TheVar == 'td'):
TheHumDat = TheTdDat
elif (TheVar == 'q'):
TheHumDat = CalcHums.sh(TheTdDat,TheTDat,TheSPDat,roundit=False)
elif (TheVar == 'e'):
TheHumDat = CalcHums.vap(TheTdDat,TheTDat,TheSPDat,roundit=False)
elif (TheVar == 'rh'):
TheHumDat = CalcHums.rh(TheTdDat,TheTDat,TheSPDat,roundit=False)
elif (TheVar == 'tw'):
TheHumDat = CalcHums.wb(TheTdDat,TheTDat,TheSPDat,roundit=False)
elif (TheVar == 'dpd'):
TheHumDat = CalcHums.dpd(TheTdDat,TheTDat,roundit=False)
return TheHumDat
#************************************************************
# RegridField
def RegridField(TheOutputGrid,TheOldData):
'''
This function does a simple regridding of data by averaging over the larger gridboxes
NO COSINE WEIGHTING FOR LATITUDE!!!!
NOTE:
FOR OutputGrid = 5by5 THIS AUTOMATICALLY FLIPS LATITUDE AND ROLLS LONGITUDE TO BE -87.5 to 87.5 and -177,5 to 177.5
FOR OutputGrid = 1by1 THIS JUST REGRIDS LATITUDE FROM 181 boxes 90 to -90 TO 180 boxes 89.5 to -89.5 and rolls longitude to -179.5 to 179.5
Assumes input grid is always 1by1
INPUTS:
TheOutputGrid - string of 1by1 or 5by5
TheOldData[:,:,:] - time, lat, long numpy array of complete field in original grid resolution
OUTPUTS:
TheNewData[:,:,:] - time, lat, long numpy array of complete field in new grid resolution
I'm hoping that things set above are seen by the function rather than being passed explicitly
'''
# Set up the desired output array
TheNewData = np.empty((len(TheOldData[:,0,0]),nlatsOut,nlonsOut),dtype = float)
TheNewData.fill(mdi)
if (TheOutputGrid == '1by1'):
# Then we know we're reading in original ERA-Interim or ERA5 data which has 181 lats
# regrid to 0.5 by 0.5 degree gridboxes and then reaverage over 89.5 to -89.5 lats
# shift lons back to -179.5 to 179.5 from 0 to 359
# regrid to 5by5
# First sort out the latitudes
for ln in range(nlonsIn):
for tt in range(len(TheNewData[:,0,0])):
subarr = np.repeat(TheOldData[tt,:,ln],2)
# this creates 362 grid boxes where each is repeated: [0a, 0b, 1a, 1b ...180a, 180b]
subarr = subarr[1:361]
# This removes the superfluous 90-90.5 and -90 to -90.5 boxes
subarr = np.reshape(subarr,(180,2))
# This now reshapes to 180 rows and 2 columns so that we can average the gridboxes across the columns
TheNewData[tt,:,ln] = np.mean(subarr,axis = 1) # hopefully this should work!
#pdb.set_trace()
# Then sort out the longitudes
for tt in range(len(TheNewData[:,0,0])):
TheNewData[tt,:,:] = np.roll(TheNewData[tt,:,:],180,axis = 1)
if (TheOutputGrid == '5by5'):
# Then we know we're reading in my converted ERA-Interim / ERA5 data which has 180 lats and already has lons rolled 180 degrees.
# flip lats to go south to north
# regrid to 5by5
# Regrid to 5by5 by simple averaging
# Data input here should already be 89.5 to -89.5 lat and -179.5 to 179.5 long!!!
StLt = 0
EdLt = 0
# Loop through the OutputGrid (5by5) lats and lons
for ltt in range(nlatsOut):
# create pointers to the five lats to average over
StLt = np.copy(EdLt)
EdLt = EdLt + 5
StLn = 0
EdLn = 0
for lnn in range(nlonsOut):
# create pointers to the five lons to average over
StLn = np.copy(EdLn)
EdLn = EdLn + 5
#print(ltt,lnn,StLt,EdLt,StLn,EdLn)
# Loop over each time point
for mm in range(len(TheNewData[:,0,0])):
# Create a subarr first so that we can deal with missing data
subarr = TheOldData[mm,StLt:EdLt,StLn:EdLn]
gots = np.where(subarr > mdi)
if (len(gots[0]) > 0):
# FILL THE LATITUDES BACKWARDS SO THAT THIS REVERSES THEM!!!
TheNewData[mm,35-ltt,lnn] = np.mean(subarr[gots])
#pdb.set_trace()
return TheNewData
#************************************************************
# BuildField
def BuildField(TheOutputVar, TheInputTime, TheOutputTime, InFileStr, TheStYr, TheEdYr):
''' function for building complete reanalyses files over the period specified
this can be very computationally expensive so do it by year
This requires initial reanalysis data to be read in in chunks of 1 year
I may change this to month later and will slice out 1 month at a time anyway
For derived variables this will read in the source vars and compute
NOTE: THIS AUTOMATICALLY REGRIDS LATITUDE TO BE 180 RATHER THAN 181 BOXES AND ROLLS LONGITUDE TO -179.5 to 179.5
INPUTS:
TheOutputVar - string lower case character of q, rh, t, td, dpd, tw, e, msl, sp, ws
TheInputTime - string of 1hr or 6hr
TheOutputTime - string of monthly or pentad
#OutputGrid - string of 1by1 or 5by5 (WHICH SHOULD BE SAME AS INPUT GRID) - ASSUME THIS IS ALWAYS 1by1 FOR NOW
InFileStr - string of dir+file string to read in
TheStYr = integer start year of data - assume Jan 1st (0101) start
TheEdYr = integer end year of data - assume Dec 31st (1231) end
OUTPUTS:
TheNewData[:,:,:] - time, lat, long numpy array of complete field in new time resolution
'''
# Set up the desired output array
if (TheOutputTime == 'monthly'):
TheNewData = np.empty((nmons,nlatsOut,nlonsOut),dtype = float)
elif (TheOutputTime == 'pentad'):
TheNewData = np.empty((npts,nlatsOut,nlonsOut),dtype = float)
TheNewData.fill(mdi)
# The input grids are different to the output grids (181 lat boxes rather than 180) so we need a TmpNewData first
TmpNewData = np.empty((len(TheNewData[:,0,0]),nlatsIn,nlonsIn),dtype = float)
TmpNewData.fill(mdi)
nyrs = (TheEdYr - TheStYr) + 1
# Begin the time counter for this dec - may need to do hourly in 5 year or 1 year chunks
# 0 to ~87600 + leap days for 1 hourly data (24*365*10)
# 0 to ~14600 + leap days for 6 hourly data (4*365*10)
# 0 to 120 for monthly data
HrStPoint = 0 # set as HrEdPoint which is actual ed point +1
HrEdPoint = 0 # set as HrStPoint + MonthHours or Month(must be +1 to work in Python!!!)
# Loop through the years
for y in range(nyrs):
# Get actual year we're working on
yr = y + StYr
print('Working Year: ',yr)
# First work out the time pointers for the year we're working with
if (TheOutputTime == 'monthly'):
mnarr = [31,29,31,30,31,30,31,31,30,31,30,31]
nbits = 12
elif (TheOutputTime == 'pentad'):
mnarr = list(np.repeat(5,73))
nbits = 73
# Is it a leap year?
if (TestLeap.TestLeap(yr) == 0.0):
if (TheOutputTime == 'monthly'):
mnarr[1] = 29
elif (TheOutputTime == 'pentad'):
mnarr[11] = 6
print('TestLeap (m, pt): ',mnarr[1],mnarr[11], yr)
# Loop through each month or pentad depending on thing
for m in range(nbits):
## string for file name
#mm = '%02i' % (m+1)
# Month pointer
MonthPointer = (y * nbits)+m
print('Month/Pentad Pointer: ',m)
# Set the time counter for this dec in either 1hr or 6hrs
# 0 to ~14600 + leap days
HrStPoint = np.copy(HrEdPoint) # set as HrEdPoint which is actual end point +1
if (ReadInTime == '1hr'):
HrEdPoint = HrStPoint + (mnarr[m]*24) # set as HrStPoint + MonthHours (must be +1 to work in Python!!!)
elif (ReadInTime == '6hr'):
HrEdPoint = HrStPoint + (mnarr[m]*4) # set as HrStPoint + MonthHours (must be +1 to work in Python!!!)
print('Hr Pointies for this month: ',HrStPoint,HrEdPoint)
# Open and read in the reanalysis files for the month
# Sort out time pointers to pull out month
# This assumes we're always reading in 1by1!!!!
SliceInfo = dict([('TimeSlice',[HrStPoint,HrEdPoint]),
('LatSlice',[0,181]),
('LonSlice',[0,360])])
# Are we working on a direct variable or do we need to read in lots of variables and convert (e.g., humidity)
# For humidity variables
if (TheOutputVar in ['q','rh','e','tw','dpd']):
# DOES automatically unpack the scale and offset
# However, SP is Pa and T and Td are Kelvin
# This kills memory so need to be tidy
ReadInfo = ['t2m']
FileName = InFileStr+'t2m_'+str(yr)+'0101'+str(yr)+'1231.nc'
T_Data,Latitudes,Longitudes = GetGrid4Slice(FileName,ReadInfo,SliceInfo,LatInfo,LonInfo)
# Unpack t
T_Data = T_Data-273.15
# DOES automatically unpack the scale and offset
# However, SP is Pa and T and Td are Kelvin
# This kills memory so need to be tidy
ReadInfo = ['d2m']
FileName = InFileStr+'td2m_'+str(yr)+'0101'+str(yr)+'1231.nc'
Td_Data,Latitudes,Longitudes = GetGrid4Slice(FileName,ReadInfo,SliceInfo,LatInfo,LonInfo)
# Unpack td
Td_Data = Td_Data-273.15
# DOES automatically unpack the scale and offset
# However, SP is Pa and T and Td are Kelvin
# This kills memory so need to be tidy
ReadInfo = ['sp']
FileName = InFileStr+'sp_'+str(yr)+'0101'+str(yr)+'1231.nc'
SP_Data,Latitudes,Longitudes = GetGrid4Slice(FileName,ReadInfo,SliceInfo,LatInfo,LonInfo)
# Unpack sp
SP_Data = SP_Data/100.
# Convert to desired humidity variable
TmpData = GetHumidity(T_Data,Td_Data,SP_Data,TheOutputVar)
# Empty the SP_Data array
SP_Data = 0
T_Data = 0
Td_Data = 0
else:
# DOES automatically unpack the scale and offset
# However, SP is Pa and T and Td are Kelvin
# This kills memory so need to be tidy
ReadInfo = [NameDict[TheOutputVar]] # the variable name to read in
#pdb.set_trace()
FileName = InFileStr+str(yr)+'0101'+str(yr)+'1231.nc'
TmpData,Latitudes,Longitudes = GetGrid4Slice(FileName,ReadInfo,SliceInfo,LatInfo,LonInfo)
#pdb.set_trace()
# Is there an unpack thing like for T - -273.15?
if (TheOutputVar in ['t','td','sst']): # t
if (TheOutputVar == 'sst'): # there are missing values over land.
TmpData[np.where(TmpData < 270.03)] = mdi # ERA Mdi is actually -32767 but in ncview it is 270.024
TmpData[np.where(TmpData > mdi)] = TmpData[np.where(TmpData > mdi)]-273.15
elif (TheOutputVar in ['slp','sp']): # pressure are Pa so need to be converted to hPa
TmpData = TmpData/100.
# Create monthly or pentad means
for ltt in range(nlatsIn):
for lnn in range(nlonsIn):
TmpNewData[MonthPointer,ltt,lnn] = np.mean(TmpData[:,ltt,lnn])
# Empty the data arrays
TmpData = 0
# Now regrid to 180 latitude boxes 89.5 to -89.5 and longitude from -179.5 to 179.5
TheNewData = RegridField('1by1',TmpNewData)
return TheNewData
#************************************************************
# CreateAnoms
def CreateAnoms(TheInputGrid,TheOutputTime,TheClimSt,TheClimEd,TheStYr,TheEdYr,TheInData):
'''
This function takes any grid and any var, computes climatologies/stdevs over given period and then anomalies
It also outputs land only and ocean only anomalies dependning on the grid
if (TheInputGrid == '5by5'):
LandMask = workingdir+'/OTHERDATA/HadCRUT.4.3.0.0.land_fraction.nc' # 0 = 100% sea, 1 = 100% land - no islands!, latitude, longitude, land_area_fraction, -87.5 to 87.5, -177.5 to 177.5
elif (TheInputGrid == '1by1'):
LandMask = workingdir+'/OTHERDATA/lsmask.nc' # 1 = sea, 0 = land - no islands! lat, lon, mask 89.5 to -89.5Lat, 0.5 to 359.5 long
INPUTS:
TheInputGrid - string of 1by1 or 5by5 to determine the land mask to use
TheOutputTime - string of monthly or pentad
TheClimSt - interger start year of climatology Always Jan start
TheClimEd - integer end year of climatology Always Dec end
TheStYr - integer start year of data to find climatology
TheEdYr - integer end year of data to find climatology
TheInData[:,:,:] - time, lat, lon array of actual values
OUTPUTS:
AllAnomsArr[:,:,:] - time, lat, lon array of anomalies
LandAnomsArr[:,:,:] - time, lat, lon array of land anomalies
OceanAnomsArr[:,:,:] - time, lat, lon array of ocean anomalies
ClimsArr[:,:,:] - time, lat, lon array of climatologies
StDevsArr[:,:,:] - time, lat, lon array of stdeviations
'''
# Set up for time
if (TheOutputTime == 'monthly'):
nclims = 12
elif (TheOutputTime == 'pentad'):
nclims = 73
nyrs = (TheEdYr - TheStYr) + 1
# Get land/sea mask and format accordingly
if (TheInputGrid == '1by1'):
MaskData,Lats,Longs = GetGrid4(LandMask,['mask'],['lat'],['lon'])
# Check shape and force to be 2d
if (len(np.shape(MaskData)) == 3):
MaskData = np.reshape(MaskData,(180,360))
# roll the longitudes
MaskData = np.roll(MaskData[:,:],180,axis = 1)
# swap the land/sea so that land = 1
land = np.where(MaskData == 0)
MaskData[np.where(MaskData == 1)] = 0
MaskData[land] = 1
elif (TheInputGrid == '5by5'):
MaskData,Lats,Longs = GetGrid4(LandMask,['land_area_fraction'],LatInfo,LonInfo)
if (len(np.shape(MaskData)) == 3):
MaskData = np.reshape(MaskData,(36,72))
# first create empty arrays
AllAnomsArr = np.empty_like(TheInData)
AllAnomsArr.fill(mdi)
LandAnomsArr = np.copy(AllAnomsArr)
OceanAnomsArr = np.copy(AllAnomsArr)
ClimsArr = | np.copy(AllAnomsArr[0:nclims,:,:]) | numpy.copy |
import math
import numpy as np
from matplotlib import cm
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from scipy.stats import multivariate_normal
def gauss_kernel_values(xs: np.ndarray, cov: np.ndarray, mean: np.ndarray = None):
'''
Vectorized Gaussian Kernel
v(x) ~ exp(-x'C^{-1} x/2) for cov = C
xs.shape = (n_obs, n_dim)
cov is the covariance matrix n_dim x n_dim
returns 1d array of n_dim
'''
if mean is None:
mean = np.zeros(xs.shape[1])
xs = xs - mean
# calculating a vectorized version of x' cov^{-1} x
Cinv = np.linalg.inv(cov)
ds = np.sum(xs * (xs @ Cinv), axis=1)
# alegedly this is faster: numpy.einsum("ij,ij->i", xs, xs @ Cinv)
dim = xs.shape[1]
detC = np.linalg.det(cov)
nrm = (2*math.pi)**(dim/2.0)*math.sqrt(detC)
return np.exp(-ds)/nrm
def discretized_gauss_kernel_values_3d(xs: np.ndarray, cov: np.ndarray, mean: np.ndarray = None):
rv = multivariate_normal(mean=mean, cov=cov)
x1, x2, x3 = np.meshgrid(xs, xs, xs)
pos = np.stack((x1, x2, x3), axis=-1)
vals = 1e2*rv.pdf(pos)
return vals, pos
def generate_test_kernel_3d(
npts: int = 10, range_std: float = 1, stds: np.ndarray = None, off_diag_correl: float = 0.0, mean: np.ndarray = None):
if stds is None:
stds = np.array([1.0, 1.0, 1.0])
dim = len(stds)
cov = np.zeros((dim, dim))
for i in np.arange(dim):
cov[i, i] = stds[i]**2
for i in | np.arange(dim) | numpy.arange |
""" Trains and evaluates the model on the different emotions """
import argparse
import ConfigParser
import imp
import numpy as np
from sklearn.ensemble import RandomForestRegressor
from sklearn.svm import SVR
import utils
def load_model(model_name, X_train, y_train, optimization_parameters, sklearn_model=None):
"""
Loads the base model model with the specific parameters
:param model_name: the name of the model
:param X_train: training data (# of samples x # of features)
:param y_train: labels for the training data (# of samples * 1)
:return: model object
"""
model_source = imp.load_source(model_name, 'models/%s.py' % (model_name))
model = model_source.Model(X_train, y_train, optimization_parameters, sklearn_model)
return model
def get_labels(data):
"""
Returns the labels for each emotion
:param data: dictionary of training data (emotion: [emotion data])
:return: a dictionary from emotion to a list of labels for each example for that emotion
"""
return {emotion: | np.array([val[-1] for val in values]) | numpy.array |
import copy
import numpy as np
import logging
logger = logging.getLogger(__name__)
try:
from pycqed.analysis import machine_learning_toolbox as ml
except Exception:
logger.warning('Machine learning packages not loaded. '
'Run from pycqed.analysis import machine_learning_toolbox to see errors.')
from sklearn.model_selection import GridSearchCV as gcv, train_test_split
from scipy.optimize import fmin_l_bfgs_b,fmin,minimize,fsolve
def nelder_mead(fun, x0,
initial_step=0.1,
no_improve_thr=10e-6, no_improv_break=10,
maxiter=0,
alpha=1., gamma=2., rho=-0.5, sigma=0.5,
verbose=False):
'''
parameters:
fun (function): function to optimize, must return a scalar score
and operate over a numpy array of the same dimensions as x0
x0 (numpy array): initial position
initial_step (float/np array): determines the stepsize to construct
the initial simplex. If a float is specified it uses the same
value for all parameters, if an array is specified it uses
the specified step for each parameter.
no_improv_thr, no_improv_break (float, int): break after
no_improv_break iterations with an improvement lower than
no_improv_thr
maxiter (int): always break after this number of iterations.
Set it to 0 to loop indefinitely.
alpha (float): reflection coefficient
gamma (float): expansion coefficient
rho (float): contraction coefficient
sigma (float): shrink coefficient
For details on these parameters see Wikipedia page
return: tuple (best parameter array, best score)
Pure Python/Numpy implementation of the Nelder-Mead algorithm.
Implementation from https://github.com/fchollet/nelder-mead, edited by
<NAME> for use in PycQED.
Reference: https://en.wikipedia.org/wiki/Nelder%E2%80%93Mead_method
'''
# init
x0 = np.array(x0) # ensures algorithm also accepts lists
dim = len(x0)
prev_best = fun(x0)
no_improv = 0
res = [[x0, prev_best]]
if type(initial_step) is float:
initial_step_matrix = np.eye(dim)*initial_step
elif (type(initial_step) is list) or (type(initial_step) is np.ndarray):
if len(initial_step) != dim:
raise ValueError('initial_step array must be same lenght as x0')
initial_step_matrix = np.diag(initial_step)
else:
raise TypeError('initial_step ({})must be list or np.array'.format(
type(initial_step)))
for i in range(dim):
x = copy.copy(x0)
x = x + initial_step_matrix[i]
score = fun(x)
res.append([x, score])
# simplex iter
iters = 0
while 1:
# order
res.sort(key=lambda x: x[1])
best = res[0][1]
# break after maxiter
if maxiter and iters >= maxiter:
# Conclude failure break the loop
if verbose:
print('max iterations exceeded, optimization failed')
break
iters += 1
if best < prev_best - no_improve_thr:
no_improv = 0
prev_best = best
else:
no_improv += 1
if no_improv >= no_improv_break:
# Conclude success, break the loop
if verbose:
print('No improvement registered for {} rounds,'.format(
no_improv_break) + 'concluding succesful convergence')
break
# centroid
x0 = [0.] * dim
for tup in res[:-1]:
for i, c in enumerate(tup[0]):
x0[i] += c / (len(res)-1)
# reflection
xr = x0 + alpha*(x0 - res[-1][0])
rscore = fun(xr)
if res[0][1] <= rscore < res[-2][1]:
del res[-1]
res.append([xr, rscore])
continue
# expansion
if rscore < res[0][1]:
xe = x0 + gamma*(x0 - res[-1][0])
escore = fun(xe)
if escore < rscore:
del res[-1]
res.append([xe, escore])
continue
else:
del res[-1]
res.append([xr, rscore])
continue
# contraction
xc = x0 + rho*(x0 - res[-1][0])
cscore = fun(xc)
if cscore < res[-1][1]:
del res[-1]
res.append([xc, cscore])
continue
# reduction
x1 = res[0][0]
nres = []
for tup in res:
redx = x1 + sigma*(tup[0] - x1)
score = fun(redx)
nres.append([redx, score])
res = nres
# once the loop is broken evaluate the final value one more time as
# verification
fun(res[0][0])
return res[0]
def SPSA(fun, x0,
initial_step=0.1,
no_improve_thr=10e-6, no_improv_break=10,
maxiter=0,
gamma=0.101, alpha=0.602, a=0.2, c=0.3, A=300,
p=0.5, ctrl_min=0.,ctrl_max=np.pi,
verbose=False):
'''
parameters:
fun (function): function to optimize, must return a scalar score
and operate over a numpy array of the same dimensions as x0
x0 (numpy array): initial position
no_improv_thr, no_improv_break (float, int): break after
no_improv_break iterations with an improvement lower than
no_improv_thr
maxiter (int): always break after this number of iterations.
Set it to 0 to loop indefinitely.
alpha, gamma, a, c, A, (float): parameters for the SPSA gains
(see refs for definitions)
p (float): probability to get 1 in Bernoulli +/- 1 distribution
(see refs for context)
ctrl_min, ctrl_max (float/array): boundaries for the parameters.
can be either a global boundary for all dimensions, or a
numpy array containing the boundary for each dimension.
return: tuple (best parameter array, best score)
alpha, gamma, a, c, A and p, are parameters for the algorithm.
Their function is described in the references below,
and even optimal values have been discussed in the literature.
Pure Python/Numpy implementation of the SPSA algorithm designed by Spall.
Implementation from http://www.jhuapl.edu/SPSA/PDF-SPSA/Spall_An_Overview.PDF,
edited by <NAME> for use in PycQED.
Reference: http://www.jhuapl.edu/SPSA/Pages/References-Intro.htm
'''
# init
x0 = np.array(x0) # ensures algorithm also accepts lists
dim = len(x0)
prev_best = fun(x0)
no_improv = 0
res = [[x0, prev_best]]
x = copy.copy(x0)
# SPSA iter
iters = 0
while 1:
# order
res.sort(key=lambda x: x[1])
best = res[0][1]
# break after maxiter
if maxiter and iters >= maxiter:
# Conclude failure break the loop
if verbose:
print('max iterations exceeded, optimization failed')
break
iters += 1
if best < prev_best - no_improve_thr:
no_improv = 0
prev_best = best
else:
no_improv += 1
if no_improv >= no_improv_break:
# Conclude success, break the loop
if verbose:
print('No improvement registered for {} rounds,'.format(
no_improv_break) + 'concluding succesful convergence')
break
# step 1
a_k = a/(iters+A)**alpha
c_k = c/iters**gamma
# step 2
delta = np.where(np.random.rand(dim) > p, 1, -1)
# step 3
x_plus = x+c_k*delta
x_minus = x-c_k*delta
y_plus = fun(x_plus)
y_minus = fun(x_minus)
# res.append([x_plus, y_plus])
# res.append([x_minus, y_minus])
# step 4
gradient = (y_plus-y_minus)/(2.*c_k*delta)
# step 5
x = x-a_k*gradient
x = np.where(x < ctrl_min, ctrl_min, x)
x = np.where(x > ctrl_max, ctrl_max, x)
score = fun(x)
res.append([x, score])
# once the loop is broken evaluate the final value one more time as
# verification
fun(res[0][0])
return res[0]
def generate_new_training_set(new_train_values, new_target_values,
training_grid=None, target_values=None):
if training_grid is None:
training_grid =new_train_values
target_values = new_target_values
else:
if np.shape(new_train_values)[1] != np.shape(training_grid)[1] or \
np.shape(new_target_values)[1] != np.shape(target_values)[1]:
print('Shape missmatch between new training values and existing ones!'
' Returning None.')
return None,None
training_grid = | np.append(training_grid,new_train_values,axis=0) | numpy.append |
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
Created on Fri Jan 11 16:41:29 2019
Many of these functions were copy pasted from bctpy package:
https://github.com/aestrivex/bctpy
under GNU V3.0:
https://github.com/aestrivex/bctpy/blob/master/LICENSE
@author: sorooshafyouni
University of Oxford, 2019
"""
import numpy as np
import statsmodels.stats.multitest as smmt
import scipy.stats as sp
import matplotlib.pyplot as plt
#def sub2ind(DaShape, X, Y):
# """ DaShape: [#rows #columns]
# returns idx of given X & Y
# SA, Ox, 2018 """
# return X*DaShape[1] + Y
def OLSRes(YOrig,RG,T,copy=True):
"""
Or how to deconfound stuff!
For regressing out stuff from your time series, quickly and nicely!
SA,Ox,2019
"""
if copy:
YOrig = YOrig.copy()
if np.shape(YOrig)[0]!=T or np.shape(RG)[0]!=T:
raise ValueError('The Y and the X should be TxI format.')
#demean anyways!
mRG = np.mean(RG,axis=0)
RG = RG-np.tile(mRG,(T,1));
#B = np.linalg.solve(RG,YOrig) # more stable than pinv
invRG = np.linalg.pinv(RG)
B = np.dot(invRG,YOrig)
Yhat = np.dot(RG,B) # find the \hat{Y}
Ydeconf = YOrig-Yhat #get the residuals -- i.e. cleaned time series
return Ydeconf
def issymmetric(W):
"""Check whether a matrix is symmetric"""
return((W.transpose() == W).all())
def SumMat(Y0,T,copy=True):
"""
Parameters
----------
Y0 : a 2D matrix of size TxN
Returns
-------
SM : 3D matrix, obtained from element-wise summation of each row with other
rows.
SA, Ox, 2019
"""
if copy:
Y0 = Y0.copy()
if np.shape(Y0)[0]!=T:
print('SumMat::: Input should be in TxN form, the matrix was transposed.')
Y0 = np.transpose(Y0)
N = np.shape(Y0)[1]
Idx = np.triu_indices(N)
#F = (N*(N-1))/2
SM = np.empty([N,N,T])
for i in np.arange(0,np.size(Idx[0])-1):
xx = Idx[0][i]
yy = Idx[1][i]
SM[xx,yy,:] = (Y0[:,xx]+Y0[:,yy]);
SM[yy,xx,:] = (Y0[:,yy]+Y0[:,xx]);
return SM
def ProdMat(Y0,T,copy=True):
"""
Parameters
----------
Y0 : a 2D matrix of size TxN
Returns
-------
SM : 3D matrix, obtained from element-wise multiplication of each row with
other rows.
SA, Ox, 2019
"""
if copy:
Y0 = Y0.copy()
if np.shape(Y0)[0]!=T:
print('ProdMat::: Input should be in TxN form, the matrix was transposed.')
Y0 = np.transpose(Y0)
N = np.shape(Y0)[1]
Idx = np.triu_indices(N)
#F = (N*(N-1))/2
SM = np.empty([N,N,T])
for i in np.arange(0,np.size(Idx[0])-1):
xx = Idx[0][i]
yy = Idx[1][i]
SM[xx,yy,:] = (Y0[:,xx]*Y0[:,yy]);
SM[yy,xx,:] = (Y0[:,yy]*Y0[:,xx]);
return SM
def CorrMat(ts,T,method='rho',copy=True):
"""
Produce sample correlation matrices
or Naively corrected z maps.
"""
if copy:
ts = ts.copy()
if np.shape(ts)[1]!=T:
print('xDF::: Input should be in IxT form, the matrix was transposed.')
ts = np.transpose(ts)
N = np.shape(ts)[0];
R = np.corrcoef(ts)
Z = np.arctanh(R)* | np.sqrt(T-3) | numpy.sqrt |
import os
import pickle
from PIL import Image
import numpy as np
import json
import torch
import torchvision.transforms as transforms
from torch.utils.data import Dataset
class CUB(Dataset):
"""support CUB"""
def __init__(self, args, partition='base', transform=None):
super(Dataset, self).__init__()
self.data_root = args.data_root
self.partition = partition
self.data_aug = args.data_aug
self.mean = [0.485, 0.456, 0.406]
self.std = [0.229, 0.224, 0.225]
self.normalize = transforms.Normalize(mean=self.mean, std=self.std)
self.image_size = 84
if self.partition == 'base':
self.resize_transform = transforms.Compose([
lambda x: Image.fromarray(x),
transforms.Resize([int(self.image_size*1.15), int(self.image_size*1.15)]),
transforms.RandomCrop(size=84)
])
else:
self.resize_transform = transforms.Compose([
lambda x: Image.fromarray(x),
transforms.Resize([int(self.image_size*1.15), int(self.image_size*1.15)]),
transforms.CenterCrop(self.image_size)
])
if transform is None:
if self.partition == 'base' and self.data_aug:
self.transform = transforms.Compose([
lambda x: Image.fromarray(x),
transforms.ColorJitter(brightness=0.4, contrast=0.4, saturation=0.4),
transforms.RandomHorizontalFlip(),
lambda x: np.asarray(x).copy(),
transforms.ToTensor(),
self.normalize
])
else:
self.transform = transforms.Compose([
lambda x: Image.fromarray(x),
transforms.ToTensor(),
self.normalize
])
else:
self.transform = transform
self.data = {}
self.file_pattern = '%s.json'
with open(os.path.join(self.data_root, self.file_pattern % partition), 'rb') as f:
meta = json.load(f)
self.imgs = []
labels = []
for i in range(len(meta['image_names'])):
image_path = os.path.join(meta['image_names'][i])
self.imgs.append(image_path)
label = meta['image_labels'][i]
labels.append(label)
# adjust sparse labels to labels from 0 to n.
cur_class = 0
label2label = {}
for idx, label in enumerate(labels):
if label not in label2label:
label2label[label] = cur_class
cur_class += 1
new_labels = []
for idx, label in enumerate(labels):
new_labels.append(label2label[label])
self.labels = new_labels
self.num_classes = np.unique(np.array(self.labels)).shape[0]
def __getitem__(self, item):
image_path = self.imgs[item]
img = Image.open(image_path).convert('RGB')
img = np.array(img).astype('uint8')
img = np.asarray(self.resize_transform(img)).astype('uint8')
img = self.transform(img)
target = self.labels[item]
return img, target, item
def __len__(self):
return len(self.labels)
class MetaCUB(CUB):
def __init__(self, args, partition='base', train_transform=None, test_transform=None, fix_seed=True):
super(MetaCUB, self).__init__(args, partition)
self.fix_seed = fix_seed
self.n_ways = args.n_ways
self.n_shots = args.n_shots
self.n_queries = args.n_queries
self.classes = list(self.data.keys())
self.n_test_runs = args.n_test_runs
self.n_aug_support_samples = args.n_aug_support_samples
self.resize_transform_train = transforms.Compose([
lambda x: Image.fromarray(x),
transforms.Resize([int(self.image_size*1.15), int(self.image_size*1.15)]),
transforms.RandomCrop(size=84)
])
self.resize_transform_test = transforms.Compose([
lambda x: Image.fromarray(x),
transforms.Resize([int(self.image_size*1.15), int(self.image_size*1.15)]),
transforms.CenterCrop(self.image_size)
])
if train_transform is None:
self.train_transform = transforms.Compose([
lambda x: Image.fromarray(x),
transforms.ColorJitter(brightness=0.4, contrast=0.4, saturation=0.4),
transforms.RandomHorizontalFlip(),
lambda x: np.asarray(x).copy(),
transforms.ToTensor(),
self.normalize
])
else:
self.train_transform = train_transform
if test_transform is None:
self.test_transform = transforms.Compose([
lambda x: Image.fromarray(x),
transforms.ToTensor(),
self.normalize
])
else:
self.test_transform = test_transform
self.data = {}
for idx in range(len(self.imgs)):
if self.labels[idx] not in self.data:
self.data[self.labels[idx]] = []
self.data[self.labels[idx]].append(self.imgs[idx])
self.classes = list(self.data.keys())
def _load_imgs(self, img_paths, transform):
imgs = []
for image_path in img_paths:
img = Image.open(image_path).convert('RGB')
img = np.array(img).astype('uint8')
img = transform(img)
imgs.append(np.asarray(img).astype('uint8'))
return np.asarray(imgs).astype('uint8')
def __getitem__(self, item):
if self.fix_seed:
np.random.seed(item)
cls_sampled = np.random.choice(self.classes, self.n_ways, False)
support_xs = []
support_ys = []
query_xs = []
query_ys = []
for idx, cls in enumerate(cls_sampled):
imgs_paths = self.data[cls]
support_xs_ids_sampled = np.random.choice(range(len(imgs_paths)), self.n_shots, False)
support_paths = [imgs_paths[i] for i in support_xs_ids_sampled]
support_imgs = self._load_imgs(support_paths, transform=self.resize_transform_train)
support_xs.append(support_imgs)
support_ys.append([idx] * self.n_shots)
query_xs_ids = np.setxor1d(np.arange(len(imgs_paths)), support_xs_ids_sampled)
query_xs_ids = np.random.choice(query_xs_ids, self.n_queries, False)
query_paths = [imgs_paths[i] for i in query_xs_ids]
query_imgs = self._load_imgs(query_paths, transform=self.resize_transform_test)
query_xs.append(query_imgs)
query_ys.append([idx] * query_xs_ids.shape[0])
support_xs, support_ys, query_xs, query_ys = np.array(support_xs), np.array(support_ys), np.array(query_xs), np.array(query_ys)
num_ways, n_queries_per_way, height, width, channel = query_xs.shape
query_xs = query_xs.reshape((num_ways * n_queries_per_way, height, width, channel))
query_ys = query_ys.reshape((num_ways * n_queries_per_way,))
support_xs = support_xs.reshape((-1, height, width, channel))
if self.n_aug_support_samples > 1:
support_xs = np.tile(support_xs, (self.n_aug_support_samples, 1, 1, 1))
support_ys = np.tile(support_ys.reshape((-1,)), (self.n_aug_support_samples))
support_xs = np.split(support_xs, support_xs.shape[0], axis=0)
query_xs = query_xs.reshape((-1, height, width, channel))
query_xs = | np.split(query_xs, query_xs.shape[0], axis=0) | numpy.split |
import numpy as np
import pytest
from tbats.bats.Components import Components
from tbats.abstract.ComponentMatrix import ComponentMatrix
from tbats.bats.SeedFinder import SeedFinder
class TestBATSSeedFinder(object):
@pytest.mark.parametrize(
"seasonal_periods, expected_mask",
[
[ # no periods means no mask
[], [],
],
[ # one period always produces a mask with one zero
[6], [0],
],
[ # two periods with no common divisor produce a mask of zeroes
[3, 7], [0, 0],
],
[ # If one period is a subperiod of the other, the mask contains 1 for the smaller period
[3, 5, 6, 24], [1, 0, 1, 0],
],
[ # If one period is a subperiod of the other, the mask contains 1 for the smaller period
[2, 5, 15, 16], [1, 1, 0, 0],
],
[ # If two periods have a common divisor then mask for the larger one contains this divisor
[4, 6], [0, -2],
],
[
# If more than two periods have a common divisor then mask for the largest one contains divisor from smallest period
[12, 42, 44], [0, -6, -4], # -4 instead of -2
],
[
# If more than two periods have a common divisor then mask for the larger one contains divisor from smaller period
[9, 16, 24], [0, 0, -3], # -3 instead of -4
],
[ # being a subperiod is more important than having a divisor
[4, 6, 12], [1, 1, 0],
],
[ # divisors and periods together
[4, 5, 10, 14, 15], [0, 1, -2, -2, -5],
],
[ # divisors and periods together
[7, 9, 11, 12, 22, 30, 33], [0, 0, 1, -3, -2, -3, -3],
],
[ # divisors and periods together
[7, 9, 11, 12, 22, 30, 44], [0, 0, 1, -3, 1, -3, -4],
],
]
)
def test_prepare_mask(self, seasonal_periods, expected_mask):
mask = SeedFinder.prepare_mask(seasonal_periods)
assert | np.array_equal(expected_mask, mask) | numpy.array_equal |
import torch
import matplotlib.pyplot as plt
import numpy as np
from dqn_agent import QLAgent
from collections import deque
from unityagents import UnityEnvironment
# Initilize Q-Learning Agent with the following inputs:
# TODO rewrite network config to programatically build network (QNN in model.py) based on these settings
network_config = { # network_config (dict) = hidden layer network configuration
'layers': 2,
'fc1_units': 64,
'fc2_units': 64,
}
state_size = 37 # state_size (int) = 37 [State space with `37` dimensions that contains the agent's velocity and ray-based perception of objects around agent's forward direction]
action_size = 4 # action_size (int) = 4 [Discrete 0 (forward), 1 (back), 2 (turn left), 3 (turn right)]
seed = 0 # seed (int) = 0 []
agent = QLAgent(state_size, action_size, seed, network_config)
# Initialize Unity Environment
env = UnityEnvironment(file_name="Banana_Windows_x86_64\Banana_Windows_x86_64\Banana.exe")
# get the default brain
brain_name = env.brain_names[0]
brain = env.brains[brain_name]
def dqn(n_episodes=1800, max_t=1000, eps_start=1.0, eps_end=0.01, eps_decay=0.995):
"""Deep Q-Learning.
Params
======
n_episodes (int): maximum number of training episodes
max_t (int): maximum number of timesteps per episode
eps_start (float): starting value of epsilon, for epsilon-greedy action selection
eps_end (float): minimum value of epsilon
eps_decay (float): multiplicative factor (per episode) for decreasing epsilon
"""
scores = [] # list containing scores from each episode
avg_scores = [] # List of average score per 100 episodes
scores_window = deque(maxlen=100) # last 100 scores
eps = eps_start # initialize epsilon
for i_episode in range(1, n_episodes+1):
env_info = env.reset(train_mode=True)[brain_name] # reset the environment
state = env_info.vector_observations[0]
score = 0
for t in range(max_t):
# Update variables used for next step
action = agent.act(state, eps)
env_info = env.step(action)[brain_name] # send the action to the environment
next_state = env_info.vector_observations[0] # get the next state
reward = env_info.rewards[0] # get the reward
done = env_info.local_done[0]
# Step through Q Learning agent
agent.step(state, action, reward, next_state, done)
state = next_state
# Update reward
score += reward
if done:
break
scores_window.append(score) # save most recent score
scores.append(score) # save most recent score
eps = max(eps_end, eps_decay*eps) # decrease epsilon
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)), end="")
if i_episode % 100 == 0:
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, | np.mean(scores_window) | numpy.mean |
# -*- coding: UTF-8 -*-
from unittest import TestCase
class TestNumpy(TestCase):
def test_dot(self):
from numpy import array, dot
A = array([[1,2],[3,4]], dtype='int32')
B = array([[5,6],[7,8]], dtype='int32')
R = array([[19,22],[43,50]], dtype='int32')
for val in (dot(A,B)-R).flat:
self.assertEqual(val, 0)
u = array([1,1], dtype='int32')
Ru = array([3,7], dtype='int32')
for val in (dot(A,u)-Ru).flat:
self.assertEqual(val, 0)
def test_eig(self):
from numpy import array, dot
from numpy.linalg import eig, inv
A = array([[1,2],[3,4]], dtype='int32')
vals, mat = | eig(A) | numpy.linalg.eig |
"""
Attempt to detect egg centers in the segmented images from annotated data
The inputs are:
1. 4-class segmentation of ovary images
(background, nurse, follicular cells and cytoplasm)
2. annotation of egg centers as
2a) csv list of centers
2b) 3-class annotation:
(i) for close center,
(iii) too far and,
(ii) something in between
The output is list of potential center candidates
Sample usage::
python run_center_candidate_training.py -list none \
-imgs "data_images/drosophila_ovary_slice/image/*.jpg" \
-segs "data_images/drosophila_ovary_slice/segm/*.png" \
-centers "data_images/drosophila_ovary_slice/center_levels/*.png" \
-out results -n ovary
Copyright (C) 2016-2017 <NAME> <<EMAIL>>
"""
import os
import sys
import logging
import argparse
from functools import partial
import tqdm
import pandas as pd
import numpy as np
from scipy import spatial
import matplotlib
if os.environ.get('DISPLAY', '') == '' and matplotlib.rcParams['backend'] != 'agg':
print('No display found. Using non-interactive Agg backend.')
matplotlib.use('Agg')
import matplotlib.pyplot as plt
sys.path += [os.path.abspath('.'), os.path.abspath('..')] # Add path to root
import imsegm.utilities.data_io as tl_data
import imsegm.utilities.experiments as tl_expt
import imsegm.utilities.drawing as tl_visu
import imsegm.superpixels as seg_spx
import imsegm.descriptors as seg_fts
import imsegm.classification as seg_clf
import imsegm.labeling as seg_lbs
# whether skip loading triplest CSV from previous run
FORCE_RELOAD = False
# even you have dumped data from previous time, all wil be recomputed
FORCE_RECOMP_DATA = False
EXPORT_TRAINING_DATA = True
# perform the Leave-One-Out experiment
RUN_LEAVE_ONE_OUT = True
# Set experiment folders
FOLDER_EXPERIMENT = 'detect-centers-train_%s'
FOLDER_INPUT = 'inputs_annot'
FOLDER_POINTS = 'candidates'
FOLDER_POINTS_VISU = 'candidates_visul'
FOLDER_POINTS_TRAIN = 'points_train'
LIST_SUBDIRS = [FOLDER_INPUT, FOLDER_POINTS,
FOLDER_POINTS_VISU, FOLDER_POINTS_TRAIN]
NAME_CSV_TRIPLES = 'list_images_segms_centers.csv'
NAME_CSV_STAT_TRAIN = 'statistic_train_centers.csv'
NAME_YAML_PARAMS = 'configuration.yaml'
NAME_DUMP_TRAIN_DATA = 'dump_training_data.npz'
NB_WORKERS = tl_expt.nb_workers(0.9)
# position is label in loaded segm and nb are out labels
LUT_ANNOT_CENTER_RELABEL = [0, 0, -1, 1]
CROSS_VAL_LEAVE_OUT_SEARCH = 0.2
CROSS_VAL_LEAVE_OUT_EVAL = 0.1
CENTER_PARAMS = {
'computer': os.uname(),
'slic_size': 25,
'slic_regul': 0.3,
# 'fts_hist_diams': None,
# 'fts_hist_diams': [10, 25, 50, 75, 100, 150, 200, 250, 300],
'fts_hist_diams': [10, 50, 100, 200, 300],
# 'fts_ray_step': None,
'fts_ray_step': 15,
'fts_ray_types': [('up', [0])],
# 'fts_ray_types': [('up', [0]), ('down', [1])],
'fts_ray_closer': True,
'fts_ray_smooth': 0,
'pca_coef': None,
# 'pca_coef': 0.99,
'balance': 'unique',
'classif': 'RandForest',
# 'classif': 'SVM',
'nb_classif_search': 50,
'dict_relabel': None,
# 'dict_relabel': {0: [0], 1: [1], 2: [2, 3]},
'center_dist_thr': 50, # distance to from annotated center as a point
}
PATH_IMAGES = os.path.join(tl_data.update_path('data_images'),
'drosophila_ovary_slice')
PATH_RESULTS = tl_data.update_path('results', absolute=True)
CENTER_PARAMS.update({
'path_list': os.path.join(PATH_IMAGES,
'list_imgs-segm-center-levels_short.csv'),
'path_images': '',
'path_segms': '',
'path_centers': '',
# 'path_images': os.path.join(PATH_IMAGES, 'image', '*.jpg'),
# 'path_segms': os.path.join(PATH_IMAGES, 'segm', '*.png'),
# 'path_centers': os.path.join(PATH_IMAGES, 'center_levels', '*.png'),
'path_infofile': '',
'path_output': PATH_RESULTS,
'name': 'ovary',
})
def arg_parse_params(params):
"""
SEE: https://docs.python.org/3/library/argparse.html
:return dict:
"""
parser = argparse.ArgumentParser()
parser.add_argument('-list', '--path_list', type=str, required=False,
help='path to the list of input files',
default=params['path_list'])
parser.add_argument('-imgs', '--path_images', type=str, required=False,
help='path to directory & name pattern for images',
default=params['path_images'])
parser.add_argument('-segs', '--path_segms', type=str, required=False,
help='path to directory & name pattern for segmentation',
default=params['path_segms'])
parser.add_argument('-centers', '--path_centers', type=str, required=False,
help='path to directory & name pattern for centres',
default=params['path_centers'])
parser.add_argument('-info', '--path_infofile', type=str, required=False,
help='path to the global information file',
default=params['path_infofile'])
parser.add_argument('-out', '--path_output', type=str, required=False,
help='path to the output directory',
default=params['path_output'])
parser.add_argument('-n', '--name', type=str, required=False,
help='name of the experiment', default='ovary')
parser.add_argument('-cfg', '--path_config', type=str, required=False,
help='path to the configuration', default=None)
parser.add_argument('--nb_workers', type=int, required=False, default=NB_WORKERS,
help='number of processes in parallel')
params.update(vars(parser.parse_args()))
paths = {}
for k in (k for k in params if 'path' in k):
if not isinstance(params[k], str) or params[k].lower() == 'none':
paths[k] = ''
continue
if k in ['path_images', 'path_segms', 'path_centers', 'path_expt']:
p_dir = tl_data.update_path(os.path.dirname(params[k]))
paths[k] = os.path.join(p_dir, os.path.basename(params[k]))
else:
paths[k] = tl_data.update_path(params[k], absolute=True)
p_dir = paths[k]
assert os.path.exists(p_dir), 'missing (%s) %s' % (k, p_dir)
# load saved configuration
if params['path_config'] is not None:
ext = os.path.splitext(params['path_config'])[-1]
assert (ext == '.yaml' or ext == '.yml'), \
'wrong extension for %s' % params['path_config']
data = tl_expt.load_config_yaml(params['path_config'])
params.update(data)
params.update(paths)
logging.info('ARG PARAMETERS: \n %r', params)
return params
def is_drawing(path_out):
""" check if the out folder exist and also if the process is in debug mode
:param str path_out:
:return bool:
# """
bool_res = path_out is not None and os.path.exists(path_out) \
and logging.getLogger().isEnabledFor(logging.DEBUG)
return bool_res
def find_match_images_segms_centers(path_pattern_imgs, path_pattern_segms,
path_pattern_center=None):
""" walk over dir with images and segmentation and pair those with the same
name and if the folder with centers exists also add to each par a center
.. note:: returns just paths
:param str path_pattern_imgs:
:param str path_pattern_segms:
:param str path_pattern_center:
:return DF: DF<path_img, path_segm, path_center>
"""
logging.info('find match images-segms-centres...')
list_paths = [path_pattern_imgs, path_pattern_segms, path_pattern_center]
df_paths = tl_data.find_files_match_names_across_dirs(list_paths)
if not path_pattern_center:
df_paths.columns = ['path_image', 'path_segm']
df_paths['path_centers'] = ''
else:
df_paths.columns = ['path_image', 'path_segm', 'path_centers']
df_paths.index = range(1, len(df_paths) + 1)
return df_paths
def get_idx_name(idx, path_img):
""" create string identifier for particular image
:param int idx: image index
:param str path_img: image path
:return str: identifier
"""
im_name = os.path.splitext(os.path.basename(path_img))[0]
if idx is not None:
return '%03d_%s' % (idx, im_name)
else:
return im_name
def load_image_segm_center(idx_row, path_out=None, dict_relabel=None):
""" by paths load images and segmentation and weather centers exist,
load them if the path out is given redraw visualisation of inputs
:param (int, DF:row) idx_row: tuple of index and row
:param str path_out: path to output directory
:param dict dict_relabel: look-up table for relabeling
:return(str, ndarray, ndarray, [[int, int]]): idx_name, img_rgb, segm, centers
"""
idx, row_path = idx_row
for k in ['path_image', 'path_segm', 'path_centers']:
row_path[k] = tl_data.update_path(row_path[k])
assert os.path.exists(row_path[k]), 'missing %s' % row_path[k]
idx_name = get_idx_name(idx, row_path['path_image'])
img_struc, img_gene = tl_data.load_img_double_band_split(row_path['path_image'],
im_range=None)
# img_rgb = np.array(Image.open(row_path['path_img']))
img_rgb = tl_data.merge_image_channels(img_struc, img_gene)
if np.max(img_rgb) > 1:
img_rgb = img_rgb / float(np.max(img_rgb))
seg_ext = os.path.splitext(os.path.basename(row_path['path_segm']))[-1]
if seg_ext == '.npz':
with np.load(row_path['path_segm']) as npzfile:
segm = npzfile[npzfile.files[0]]
if dict_relabel is not None:
segm = seg_lbs.merge_probab_labeling_2d(segm, dict_relabel)
else:
segm = tl_data.io_imread(row_path['path_segm'])
if dict_relabel is not None:
segm = seg_lbs.relabel_by_dict(segm, dict_relabel)
if row_path['path_centers'] is not None \
and os.path.isfile(row_path['path_centers']):
ext = os.path.splitext(os.path.basename(row_path['path_centers']))[-1]
if ext == '.csv':
centers = tl_data.load_landmarks_csv(row_path['path_centers'])
centers = tl_data.swap_coord_x_y(centers)
elif ext == '.png':
centers = tl_data.io_imread(row_path['path_centers'])
# relabel loaded segm into relevant one
centers = np.array(LUT_ANNOT_CENTER_RELABEL)[centers]
else:
logging.warning('not supported file format %s', ext)
centers = None
else:
centers = None
if is_drawing(path_out):
export_visual_input_image_segm(path_out, idx_name, img_rgb, segm, centers)
return idx_name, img_rgb, segm, centers
def export_visual_input_image_segm(path_out, img_name, img, segm, centers=None):
""" visualise the input image and segmentation in common frame
:param str path_out: path to output directory
:param str img_name: image name
:param ndarray img: np.array
:param ndarray segm: np.array
:param centers: [(int, int)] or np.array
"""
fig = tl_visu.figure_image_segm_centres(img, segm, centers)
fig.savefig(os.path.join(path_out, img_name + '.png'),
bbox_inches='tight', pad_inches=0)
plt.close(fig)
def compute_min_dist_2_centers(centers, points):
""" compute distance toclosestt center and mark which center it is
:param [int, int] centers:
:param [int, int] points:
:return (float, int):
"""
dists = spatial.distance.cdist( | np.array(points) | numpy.array |
"""pytest unit tests for vibrationtesting"""
import numpy as np
import vibrationtesting as vt
import numpy.testing as nt
import scipy.io as sio
def test_sos_modal_forsparse():
mat_contents=sio.loadmat('vibrationtesting/data/WingBeamforMAC.mat') # WFEM generated .mat file
K = (mat_contents['K'])
M = (mat_contents['M'])
## Mr and Kr are WFEM outputs after Guyan reduction
Kr = (mat_contents['Kr'])
Mr = (mat_contents['Mr'])
master = np.array([[ 2, 3, 4, 5, 6, 8, 9, 10, 11, 12, 14, 15, 16, 17, 18, 20,
21, 22, 23, 24, 26, 27, 28, 29, 30, 32, 33, 34, 35, 36, 38, 39,
40, 41, 42, 44, 45, 46, 47, 48, 50, 51, 52, 53, 54, 56, 57, 58,
59, 60, 62, 63, 64, 65, 66, 68, 69, 70, 71, 72, 74, 75, 76, 77,
78, 80, 81, 82, 83, 84, 86, 87, 88, 89, 90]])
## Mred and Kred are from guyan_forsparse
[Mred, Kred, master] = vt.guyan_forsparse(M, K, master=master, fraction=None)
[omega_sp,zeta_sp,Psi_sp] = vt.sos_modal_forsparse(Mred,Kred)
Kbm = Kr.todense()
Mbm = Mr.todense()
omega, zeta, Psi = vt.sos_modal(Mbm, Kbm)
## The below compares sparsematriceshandler.py vs system.py results
nt.assert_array_almost_equal(omega_sp,omega)
nt.assert_array_almost_equal(zeta_sp,zeta)
| nt.assert_array_almost_equal(Psi_sp,Psi) | numpy.testing.assert_array_almost_equal |
#!/usr/bin/env python
# coding: utf-8
import numpy as np
from tqdm import tqdm
from functools import reduce
import disk.funcs as dfn
import h5py
import os
import glob
import sys
from matplotlib import pyplot as plt
class binary_mbh(object):
def __init__(self, filename):
self.parse_file(filename)
def parse_file(self, filename, cgs_units=True):
self.filename = filename
if cgs_units:
print ('The cgs units are used!')
with h5py.File(self.filename, 'r') as f:
self.SubhaloMassInHalfRadType = np.array(f['meta/SubhaloMassInHalfRadType'])
self.SubhaloSFRinHalfRad = np.array(f['meta/SubhaloSFRinHalfRad'])
self.snapshot = np.array(f['meta/snapshot'])
self.subhalo_id = np.array(f['meta/subhalo_id'])
self.masses = np.array(f['evolution/masses']) #g
self.mdot = np.array(f['evolution/mdot_eff']) #g/s
self.sep = np.array(f['evolution/sep']) #cm
self.dadt = np.array(f['evolution/dadt']) #cm/s
self.dadt_df = np.array(f['evolution/dadt_df']) #cm/s
self.dadt_gw = np.array(f['evolution/dadt_gw']) #cm/s
self.dadt_lc = np.array(f['evolution/dadt_lc']) #cm/s
self.dadt_vd = np.array(f['evolution/dadt_vd']) #cm/s
self.scales = np.array(f['evolution/scales']) #NA
self.times = np.array(f['evolution/times']) #s
self.eccen = np.array(f['evolution/eccen']) #NA
self.z = (1./self.scales)-1 #NA
self.m1 = self.masses[:,0]
self.m2 = self.masses[:,1]
self.mtot = self.m1+self.m2
self.q = self.m2/self.m1
def find_Rlc(self):
R_lc = np.zeros((self.sep.shape[0],3))
for i in range(len(self.sep)):
try:
idx = reduce(np.intersect1d,(np.where(np.abs(self.dadt_lc[i])>np.abs(self.dadt_df[i]))[0],
np.where(np.abs(self.dadt_lc[i])>np.abs(self.dadt_vd[i]))[0],
np.where(np.abs(self.dadt_lc[i])>np.abs(self.dadt_gw[i]))[0]))[0]
R_lc[i]=[i,idx,self.sep[i][idx]]
except:
R_lc[i]=[i,np.nan,np.nan]
return R_lc
def find_Rvd(self):
R_vd = np.zeros((self.sep.shape[0],3))
for i in range(len(self.sep)):
try:
idx = reduce(np.intersect1d,(np.where(np.abs(self.dadt_vd[i])>np.abs(self.dadt_df[i]))[0],
np.where(np.abs(self.dadt_vd[i])>np.abs(self.dadt_lc[i]))[0],
np.where(np.abs(self.dadt_vd[i])>np.abs(self.dadt_gw[i]))[0]))[0]
R_vd[i]=[i,idx,self.sep[i][idx]]
except:
R_vd[i]=[i,np.nan,np.nan]
return R_vd
def find_Rgw(self):
R_gw = np.zeros((self.sep.shape[0],3))
for i in range(len(self.sep)):
try:
idx = reduce(np.intersect1d,(np.where( | np.abs(self.dadt_gw[i]) | numpy.abs |
import numpy
import quadpy
def test_circle():
scheme = quadpy.circle.krylov(3)
scheme.integrate(lambda x: numpy.exp(x[0]), numpy.array([0.0, 0.3]), 0.7)
scheme = quadpy.circle.krylov(5)
scheme.integrate(
lambda x: [numpy.exp(x[0]), numpy.exp(x[0])],
numpy.array([[1.0, 1.0], [0.0, 0.3], [2.0, 2.0]]),
[1.0, 0.7, 0.333],
)
return
def test_disk():
scheme = quadpy.disk.peirce_1957(5)
scheme.integrate(lambda x: numpy.exp(x[0]), numpy.array([0.0, 0.3]), 0.7)
scheme = quadpy.disk.peirce_1957(5)
scheme.integrate(
lambda x: [numpy.exp(x[0]), numpy.exp(x[1])],
numpy.array([[1.0, 1.0], [0.0, 0.3], [2.0, 2.0]]),
[1.0, 0.7, 0.333],
)
return
def test_hexahedron():
scheme = quadpy.hexahedron.product(quadpy.line_segment.newton_cotes_closed(3))
val = scheme.integrate(
lambda x: numpy.exp(x[0]),
quadpy.hexahedron.cube_points([0.0, 1.0], [0.0, 1.0], [0.0, 1.0]),
)
scheme = quadpy.hexahedron.product(quadpy.line_segment.newton_cotes_closed(3))
val = scheme.integrate(
lambda x: [numpy.exp(x[0]), | numpy.exp(x[1]) | numpy.exp |
# Copyright (c) 2020. The Medical Image Computing (MIC) Lab, 陶豪毅
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import numpy as np
# from skimage.morphology import dilation
from scipy.ndimage import grey_dilation
import matplotlib.pyplot as plt
from matplotlib.colors import Normalize
import cv2
def showImage(image: np.ndarray):
# https://stackoverflow.com/questions/28816046/displaying-different-images-with-actual-size-in-matplotlib-subplot
dpi = 300
fig = plt.figure(figsize=(image.shape[1] / dpi, image.shape[0] / dpi), dpi=dpi)
ax = fig.add_axes([0, 0, 1, 1])
ax.axis('off')
ax.imshow(image, interpolation="None")
fig.tight_layout()
plt.show()
def meshImage(image: np.ndarray):
# from mpl_toolkits.mplot3d import Axes3D
assert image.ndim == 2
y, x = image.shape
x_values = np.linspace(0, x - 1, x)
y_values = np.linspace(0, y - 1, y)
X, Y = np.meshgrid(x_values, y_values)
fig = plt.figure(figsize=(10, 10))
ax = fig.add_subplot(111, projection='3d')
ax.plot_surface(X, Y, image, cmap='jet')
plt.show()
def getBBox2D(image: np.ndarray,
bboxes: list,
labels: list,
scores: list = None):
image = image.astype(np.float32)
if np.max(image) > 1.0 or np.min(image) < 0.0:
image = Normalize()(image)
if image.ndim == 2 or image.shape[-1] == 1:
image = np.dstack([image] * 3)
font_scale = min(np.sqrt(image.size / 3) / 300, 0.5)
thickness = min(int(( | np.sqrt(image.size / 3) | numpy.sqrt |
from typing import Tuple
import numpy as np
from scipy import spatial
def hill_sphere_radius(
planet_radius: float,
planet_mass: float,
stellar_mass: float,
eccentricity: float = None,
) -> float:
"""Calculate the Hill sphere radius.
Parameters
----------
planet_radius
The orbital radius of the planet.
planet_mass
The mass of the planet.
stellar_mass
The mass of the star.
eccentricity : optional
The orbital eccentricity.
Returns
-------
hill_radius
The Hill sphere radius.
"""
if eccentricity is None:
eccentricity = 0.0
return (
(1 - eccentricity)
* planet_radius
* (planet_mass / (3 * stellar_mass)) ** (1 / 3)
)
def binary_orbit(
primary_mass: float,
secondary_mass: float,
semi_major_axis: float,
eccentricity: float,
inclination: float = None,
longitude_ascending_node: float = None,
argument_periapsis: float = None,
true_anomaly: float = None,
use_degrees: bool = True,
) -> Tuple[np.ndarray, np.ndarray, np.ndarray, np.ndarray]:
"""Set position and velocity of two bodies in an elliptic orbit.
Parameters
----------
primary_mass
The mass of the primary (m1).
secondary_mass
The mass of the secondary (m2).
semi_major_axis
The semi major axis (a).
eccentricity
The orbital eccentricity (e).
inclination : optional
The orbital inclination (i). Default is 0.0.
longitude_ascending_node : optional
The longitude of ascending node (Omega). Default is 0.0.
argument_periapsis : optional
The argument of periapsis (pomega). Default is 0.0.
true_anomaly : optional
The true anomaly (f). Default is 0.0.
use_degrees : optional
If true, specify angles in degrees. Otherwise, use radians.
Returns
-------
primary_position
The Cartesian position of the primary.
secondary_position
The Cartesian position of the secondary.
primary_velocity
The Cartesian velocity of the primary.
secondary_velocity
The Cartesian velocity of the secondary.
"""
m1 = primary_mass
m2 = secondary_mass
a = semi_major_axis
e = eccentricity
if inclination is None:
i = 0.0
else:
i = inclination
if longitude_ascending_node is None:
Omega = 0.0
else:
Omega = longitude_ascending_node
if argument_periapsis is None:
pomega = 0.0
else:
pomega = argument_periapsis
if true_anomaly is None:
f = 0.0
else:
f = true_anomaly
if use_degrees:
i *= np.pi / 180
Omega *= np.pi / 180
pomega *= np.pi / 180
f *= np.pi / 180
# Phantom convention: longitude of the ascending node is measured east of north
Omega += np.pi / 2
dx = 0.0
dv = 0.0
E = np.arctan2(np.sqrt(1 - e ** 2) * np.sin(f), (e + np.cos(f)))
P = np.zeros(3)
Q = np.zeros(3)
P[0] = np.cos(pomega) * np.cos(Omega) - np.sin(pomega) * np.cos(i) * | np.sin(Omega) | numpy.sin |
#!/usr/bin/env pytohn2
import numpy as np
from sklearn.model_selection import train_test_split
import sklearn.metrics
from data_handling import parse_data
import misc_tools
import settings
def skyline_pianorolls(input, onset=True):
"""
Perform the skyline algorithm on *pianoroll*. This just takes the
highest note at each time. If *onset* is True, then the original version is
used, in which the highest pitch referred to the most recent onset is
considered to be melody.
Reference paper:
<NAME> and <NAME>, "Melodic matching techniques for large
music databases," in Proceedings of the 7th ACM International Conference on
Multimedia '99, Orlando, FL, USA, October 30 - November 5, 1999, Part 1.,
1999, pp. 57-66.
RETURNS:
a new array of shape *pianoroll.shape* containing the resulting pianoroll
"""
pianoroll = np.array(input, dtype=misc_tools.floatX)
returned = np.zeros(pianoroll.shape, pianoroll.dtype)
for y, col in enumerate(pianoroll.T):
# iterating over columns
backup = False
for x, v in enumerate(col):
# iterating over pitches
if v != 0:
if onset:
if pianoroll[x, y - 1] == 0:
# new onset at highest pitch
returned[x, y] = v
backup = False
break
elif not backup:
# this is the highest value coming from a previous onset,
# store this value and add it after having parsed the whole
# column
backup = (x, y, v)
# N.B. now bool(backup) == True
else:
returned[x, y] = v
break
if backup:
# add the highest value coming from a previous onset
returned[backup[0], backup[1]] = backup[2]
backup = False
return returned
def test_skyline_pianorolls(PATH=settings.DATA_PATH):
import os
from data_handling import parse_data
# recurse all directories
dataset = []
for root, subdirs, files in os.walk(PATH):
# for each file with extension '.bz2'
for f in files:
if f[-3:] == ".bz":
new_file = os.path.join(root, f)
print("I've found a new file: " + new_file)
# load pianorolls score and melody
score, melody = parse_data.make_pianorolls(new_file)
dataset.append((score, melody))
train_set, test_set = train_test_split(
dataset, test_size=0.20, random_state=42)
overall_sk_tp = overall_sk_fp = overall_sk_tn = overall_sk_fn = 0
overall_hp_tp = overall_hp_fp = overall_hp_tn = overall_hp_fn = 0
avarage_pieces_sk = []
avarage_pieces_hp = []
for score, melody in test_set:
sk = skyline_pianorolls(score)
results = misc_tools.evaluate(sk, melody)
overall_sk_tp += results[0]
overall_sk_fp += results[1]
overall_sk_tn += results[2]
overall_sk_fn += results[3]
p = results[0] / misc_tools.EPS(results[0] + results[1])
r = results[0] / misc_tools.EPS(results[0] + results[3])
f = 2 * r * p / misc_tools.EPS(p + r)
avarage_pieces_sk.append((p, r, f))
hp = skyline_pianorolls(score, onset=False)
results = misc_tools.evaluate(hp, melody)
overall_hp_tp += results[0]
overall_hp_fp += results[1]
overall_hp_tn += results[2]
overall_hp_fn += results[3]
p = results[0] / misc_tools.EPS(results[0] + results[1])
r = results[0] / misc_tools.EPS(results[0] + results[3])
f = 2 * r * p / misc_tools.EPS(p + r)
avarage_pieces_hp.append((p, r, f))
# parse_data.plot_pianorolls(
# score, sk, out_fn=f + "_skyline.pdf")
# parse_data.plot_pianorolls(
# score, hp, out_fn=f + "_highestpitch.pdf")
print("Final Results Skyline:")
print("True positives: " + str(overall_sk_tp))
print("False positives: " + str(overall_sk_fp))
print("True negatives: " + str(overall_sk_tn))
print("False negatives: " + str(overall_sk_fn))
p = overall_sk_tp / misc_tools.EPS(overall_sk_tp + overall_sk_fp)
r = overall_sk_tp / misc_tools.EPS(overall_sk_tp + overall_sk_fn)
print("Precision: " + str(p))
print("Recall: " + str(r))
print("Fmeasures: " + str(2 * r * p / misc_tools.EPS(p + r)))
print("Avarage piece precision: " + str(np.mean(avarage_pieces_sk[0])))
print("Avarage piece recall: " + str(np.mean(avarage_pieces_sk[1])))
print("Avarage piece fmeasure: " + str(np.mean(avarage_pieces_sk[2])))
print()
print("Final Results Highest Pitch:")
print("True positives: " + str(overall_hp_tp))
print("False positives: " + str(overall_hp_fp))
print("True negatives: " + str(overall_hp_tn))
print("False negatives: " + str(overall_hp_fn))
p = overall_hp_tp / misc_tools.EPS(overall_hp_tp + overall_hp_fp)
r = overall_hp_tp / misc_tools.EPS(overall_hp_tp + overall_hp_fn)
print("Precision: " + str(p))
print("Recall: " + str(r))
print("Fmeasures: " + str(2 * r * p / misc_tools.EPS(p + r)))
print("Avarage piece precision: " + str(np.mean(avarage_pieces_hp[0])))
print("Avarage piece recall: " + str(np.mean(avarage_pieces_hp[1])))
print("Avarage piece fmeasure: " + str( | np.mean(avarage_pieces_hp[2]) | numpy.mean |
import tensorflow as tf
import numpy as np
import pandas as pd
from tqdm import tqdm
from sklearn.metrics import classification_report, log_loss
from ..base_dataloader import DataLoader
from ..mean_pool_model import MeanPoolModel
class ContextAggModel(MeanPoolModel):
def init_graph(self, X, batch_size, **model_params):
self.batch_size = batch_size
n_dim_x_p = len(X[0].values[0][0])
n_dim_x_a = len(X[1].values[0][0])
n_dim_x_b = len(X[2].values[0][0])
n_dim_q = len(X[3].values[0])
n_dim_p = len(X[4].values[0])
n_dim_c = len(X[5].values[0])
# n_dim_c = len(X[3].values[0][0])
for key, val in model_params.items():
if key.startswith('n_hidden'):
model_params[key] = int(model_params[key])
self.model = self.create_graph(
None,
n_dim_x_p=n_dim_x_p,
n_dim_x_a=n_dim_x_a,
n_dim_x_b=n_dim_x_b,
n_dim_q=n_dim_q,
n_dim_p=n_dim_p,
n_dim_c=n_dim_c,
**model_params)
self.init = tf.initializers.global_variables()
self.sess = sess = tf.Session()
self.saver = tf.train.Saver()
def fit(self,
X,
y=None,
X_val=None,
y_val=None,
batch_size=32,
n_epochs=30,
patience=5,
verbose=1,
**model_params):
start = timer()
sess = self.sess
# np.random.seed(0)
# tf.set_random_seed(0)
sess.run(self.init)
self.saver = tf.train.Saver()
train_dl = DataLoader(X, batch_size, shuffle=True, seed=21)
test_dl = DataLoader(X_val, batch_size, shuffle=False, seed=21)
model = self.model
best_score = 2
best_probs = None
since_best = 0
for epoch in range(n_epochs):
pbar = tqdm(desc='Trn', total=len(X)+len(X_val)) if verbose else contextlib.suppress()
with pbar:
loss = []
for idx, (batch_x_p, batch_x_a, batch_x_b, batch_q, batch_p, batch_c, batch_y, seq_lens, seq_lens_p, seq_lens_a, seq_lens_b) in enumerate(train_dl):
if len(batch_x_p) == 1:
continue
loss_, _ = sess.run([model.loss,
model.train_op],
feed_dict={model.d_X_p: batch_x_p,
model.d_X_a: batch_x_a,
model.d_X_b: batch_x_b,
model.d_Q: batch_q,
model.d_P: batch_p,
model.d_C: batch_c,
model.d_y: batch_y,
model.d_seq_lens: seq_lens,
model.d_seq_lens_p: seq_lens_p,
model.d_seq_lens_a: seq_lens_a,
model.d_seq_lens_b: seq_lens_b})
loss.append(loss_)
if verbose:
pbar.set_description('Trn {:2d}, Loss={:.3f}, Val-Loss={:.3f}'.format(epoch, np.mean(loss), np.inf))
pbar.update(len(batch_x_p))
trn_loss = np.mean(loss)
loss, y_true, probs = self.predict(test_dl, epoch=epoch, trn_loss=trn_loss, pbar=pbar)
score = log_loss(np.array(y_true), np.array(probs))
if score < best_score:
best_score = score
best_probs = probs
since_best = 0
self.saver.save(sess, 'tmp/best_model.ckpt')
else:
since_best += 1
if verbose:
pbar.set_description('Trn {:2d}, Loss={:.3f}, Val-Loss={:.3f}'.format(epoch, trn_loss, score))
pbar.update(len(X_val))
if since_best > patience:
break
if verbose:
print('Best score on validation set: ', best_score)
self.best_score = best_score
self.saver.restore(self.sess, 'tmp/best_model.ckpt')
return self
def predict(self,
X,
y=None,
epoch=None,
trn_loss=None,
pbar=None,
verbose=1):
# if verbose and pbar is None:
# pbar_ = pbar = tqdm(desc='Predict', total=len(X))
# else:
# pbar_ = contextlib.suppress()
# with pbar_:
if trn_loss is None:
test_dl = DataLoader(X, self.batch_size)
else:
test_dl = X
loss, y_true, probs = [], [], []
for idx, (batch_x_p, batch_x_a, batch_x_b, batch_q, batch_p, batch_c, batch_y, seq_lens, seq_lens_p, seq_lens_a, seq_lens_b) in enumerate(test_dl):
if len(batch_x_p) == 1:
continue
loss_, y_true_, probs_ = self.sess.run([self.model.loss,
self.model.d_y,
self.model.probs],
feed_dict={self.model.d_X_p: batch_x_p,
self.model.d_X_a: batch_x_a,
self.model.d_X_b: batch_x_b,
self.model.d_Q: batch_q,
self.model.d_P: batch_p,
self.model.d_C: batch_c,
self.model.d_y: batch_y,
self.model.d_seq_lens: seq_lens,
self.model.d_seq_lens_p: seq_lens_p,
self.model.d_seq_lens_a: seq_lens_a,
self.model.d_seq_lens_b: seq_lens_b})
loss.append(loss_)
y_true += y_true_.tolist()
probs += probs_.tolist()
# if verbose:
# if trn_loss is not None:
# pbar.set_description('Trn {:2d}, Loss={:.3f}, Val-Loss={:.3f}'.format(epoch, trn_loss, np.mean(loss)))
# else:
# pbar.set_description('Predict, Loss={:.3f}'.format(np.mean(loss)))
#
# pbar.update(len(batch_x))
return loss, np.array(y_true), probs
def create_graph(self,
batch_size=None,
seq_len=None,
n_dim_x_p=1024,
n_dim_x_a=1024,
n_dim_x_b=1024,
n_dim_q=1024,
n_dim_p=1024,
n_dim_c=1024,
n_hidden_x=1024,
n_hidden_x_p=1024,
n_hidden_x_a=1024,
n_hidden_x_b=1024,
n_hidden_q=1024,
n_hidden_p=1024,
n_hidden_c=1024,
dropout_rate_x=0.5,
dropout_rate_p=0.5,
dropout_rate_q=0.5,
dropout_rate_c=0.5,
dropout_rate_fc=0.5,
reg_x=0.01,
reg_q=0.01,
reg_p=0.01,
reg_c=0.01,
reg_fc=0.01,
label_smoothing=0.1,
data_type=tf.float32,
activation='tanh'):
def gelu_fast(_x):
return 0.5 * _x * (1 + tf.tanh(tf.sqrt(2 / np.pi) * (_x + 0.044715 * tf.pow(_x, 3))))
activ = tf.nn.relu
# activ = gelu_fast
# np.random.seed(0)
# tf.set_random_seed(0)
tf.reset_default_graph()
# d_train_c = tf.constant(X_train, shape=[2000, 512, 1024])
d_X_p = tf.placeholder(data_type, [batch_size, None, n_dim_x_p])
d_X_a = tf.placeholder(data_type, [batch_size, None, n_dim_x_a])
d_X_b = tf.placeholder(data_type, [batch_size, None, n_dim_x_b])
d_Q = tf.placeholder(data_type, [batch_size, n_dim_q])
d_P = tf.placeholder(data_type, [batch_size, n_dim_p])
d_C = tf.placeholder(data_type, [batch_size, n_dim_c])
# d_C = tf.placeholder(data_type, [batch_size, None, n_dim_c])
d_y = tf.placeholder(tf.int32, [batch_size, 3])
d_seq_lens = tf.placeholder(tf.int32, [batch_size])
d_seq_lens_p = tf.placeholder(tf.int32, [batch_size])
d_seq_lens_a = tf.placeholder(tf.int32, [batch_size])
d_seq_lens_b = tf.placeholder(tf.int32, [batch_size])
# d_dropout_l0 = tf.placeholder(tf.float32, None)
# d_dropout_l1 = tf.placeholder(tf.float32, None)
# d_dropout_l2 = tf.placeholder(tf.float32, None)
# d_dropout_l3 = tf.placeholder(tf.float32, None)
with tf.name_scope('dense_concat_layers'):
# n_hidden = 1024
lstm_fw_cell = tf.nn.rnn_cell.BasicLSTMCell(n_hidden_x_p, name='fw1')
lstm_bw_cell = tf.nn.rnn_cell.BasicLSTMCell(n_hidden_x_p, name='bw1')
_, (fw_state, bw_state) = tf.nn.bidirectional_dynamic_rnn(lstm_fw_cell, lstm_bw_cell,
d_X_p, sequence_length=d_seq_lens_p, dtype=tf.float32)
X_p = tf.reshape(tf.concat([fw_state.h, bw_state.h], axis=-1), (-1, 2*n_hidden_x_p))
lstm_fw_cell = tf.nn.rnn_cell.BasicLSTMCell(n_hidden_x_a, name='fw2')
lstm_bw_cell = tf.nn.rnn_cell.BasicLSTMCell(n_hidden_x_a, name='bw2')
_, (fw_state, bw_state) = tf.nn.bidirectional_dynamic_rnn(lstm_fw_cell, lstm_bw_cell,
d_X_a, sequence_length=d_seq_lens_a, dtype=tf.float32)
X_a = tf.reshape(tf.concat([fw_state.h, bw_state.h], axis=-1), (-1, 2*n_hidden_x_a))
lstm_fw_cell = tf.nn.rnn_cell.BasicLSTMCell(n_hidden_x_b, name='fw3')
lstm_bw_cell = tf.nn.rnn_cell.BasicLSTMCell(n_hidden_x_b, name='bw3')
_, (fw_state, bw_state) = tf.nn.bidirectional_dynamic_rnn(lstm_fw_cell, lstm_bw_cell,
d_X_b, sequence_length=d_seq_lens_b, dtype=tf.float32)
X_b = tf.reshape(tf.concat([fw_state.h, bw_state.h], axis=-1), (-1, 2*n_hidden_x_b))
dense_init = tf.keras.initializers.glorot_normal(seed=21)
# X_p = TransformerCorefModel().pool_and_attend(d_X_p, d_seq_lens_p, n_dim_x_p, n_hidden_x_p, 20)[0]
# X_a = TransformerCorefModel().pool_and_attend(d_X_a, d_seq_lens_a, n_dim_x_a, n_hidden_x_a, 20)[0]
# X_b = TransformerCorefModel().pool_and_attend(d_X_b, d_seq_lens_b, n_dim_x_b, n_hidden_x_b, 20)[0]
X = tf.concat([X_p, X_a, X_b], axis=-1)
X = tf.keras.layers.Dropout(dropout_rate_x, seed=7)(X)
X = tf.keras.layers.Dense(n_hidden_x, activation=None, kernel_initializer=dense_init,
kernel_regularizer = tf.contrib.layers.l1_regularizer(reg_x))(X)
X = tf.layers.batch_normalization(X)
X = activ(X)
Q = tf.keras.layers.Dropout(dropout_rate_q, seed=7)(d_Q)
Q = tf.keras.layers.Dense(n_hidden_q, activation=None, kernel_initializer=dense_init,
kernel_regularizer = tf.contrib.layers.l1_regularizer(reg_q))(Q)
Q = tf.layers.batch_normalization(Q)
Q = activ(Q)
P = tf.keras.layers.Dropout(dropout_rate_p, seed=7)(d_P)
P = tf.keras.layers.Dense(n_hidden_p, activation=None, kernel_initializer=dense_init,
kernel_regularizer = tf.contrib.layers.l1_regularizer(reg_p))(P)
P = tf.layers.batch_normalization(P)
P = activ(P)
C = tf.keras.layers.Dropout(dropout_rate_c, seed=7)(d_C)
C = tf.keras.layers.Dense(n_hidden_c, activation=None, kernel_initializer=dense_init,
kernel_regularizer = tf.contrib.layers.l1_regularizer(reg_c))(C)
C = tf.layers.batch_normalization(C)
C = activ(C)
# C = TransformerCorefModel().pool_and_attend(d_C, d_seq_lens, 1024, n_hidden_c, 512)[0]
X = tf.concat([X, P, Q, C], axis=-1)
X = tf.keras.layers.Dropout(dropout_rate_fc, seed=7)(X)
# X = tf.layers.dense(X, 128, activation=None)
# X = tf.layers.batch_normalization(X)
# X = tf.nn.relu(X)
# X = tf.keras.layers.Dropout(0.5)(X)
y_hat = tf.keras.layers.Dense(3, name = 'output', kernel_initializer=dense_init,
kernel_regularizer = tf.contrib.layers.l2_regularizer(reg_fc))(X)
with tf.name_scope('loss'):
probs = tf.nn.softmax(y_hat, axis=-1)
# label smoothing works
loss = tf.losses.softmax_cross_entropy(d_y, logits=y_hat, label_smoothing=label_smoothing)
loss = tf.reduce_mean(loss)
with tf.name_scope('optimizer'):
global_step = tf.Variable(0, trainable=False, dtype=tf.int32)
learning_rate = 0.005
learning_rate = tf.train.cosine_decay_restarts(
learning_rate,
global_step,
500,
t_mul=1,
m_mul=1,
alpha=0.01,
name=None
)
optimizer = tf.train.AdamOptimizer(learning_rate=learning_rate)
train_op = optimizer.minimize(loss, global_step=global_step)
return AttrDict(locals())
@staticmethod
def mean_featurizer(X_bert, X_plus, X_pretrained, y_true):
batch_x_p = []
batch_x_a = []
batch_x_b = []
batch_q = []
batch_p = []
batch_c = []
seq_lens = []
seq_lens_p = []
seq_lens_a = []
seq_lens_b = []
batch_y = []
max_len = 512
max_len_tok = 20
for idx, row in tqdm(X_bert.iterrows(), total=len(X_bert)):
plus_row = X_plus.loc[idx]
x = np.array(row.bert)
q = np.array(plus_row.plus)
p = X_pretrained.loc[idx].pretrained
c = np.array(row.cls).reshape(-1)
# c = np.vstack((x, np.zeros((max_len-x.shape[0], x.shape[1]))))
pronoun_vec = x[row.pronoun_offset_token:row.pronoun_offset_token+1]
a_vec = x[row.a_span[0]:row.a_span[1]+1]
b_vec = x[row.b_span[0]:row.b_span[1]+1]
# print(idx, a_vec.shape, b_vec.shape)
# x = np.hstack((np.mean(pronoun_vec, axis=0), np.mean(a_vec, axis=0), np.mean(b_vec, axis=0))).reshape(-1)
x_p = np.vstack((pronoun_vec, np.zeros((max_len_tok-pronoun_vec.shape[0], pronoun_vec.shape[1]))))
x_a = np.vstack((a_vec, np.zeros((max_len_tok-a_vec.shape[0], a_vec.shape[1]))))
x_b = np.vstack((b_vec, np.zeros((max_len_tok-b_vec.shape[0], b_vec.shape[1]))))
pronoun_vec = q[plus_row.pronoun_offset_token:plus_row.pronoun_offset_token+1]
a_vec = q[plus_row.a_span[0]:plus_row.a_span[0]+1]
b_vec = q[plus_row.b_span[0]:plus_row.b_span[0]+1]
q = np.hstack((np.max(pronoun_vec, axis=0), | np.max(a_vec, axis=0) | numpy.max |
#!/usr/bin/env python
# The MIT License (MIT)
#
# Copyright (c) 2016 <NAME>, National Institutes of Health / NINDS
#
# Permission is hereby granted, free of charge, to any person obtaining a copy
# of this software and associated documentation files (the "Software"), to deal
# in the Software without restriction, including without limitation the rights
# to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
# copies of the Software, and to permit persons to whom the Software is
# furnished to do so, subject to the following conditions:
#
# The above copyright notice and this permission notice shall be included in all
# copies or substantial portions of the Software.
#
# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
# AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
# OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
# SOFTWARE.
#import os
import argparse
import time
import numpy as np
import cv2
from dpLoadh5 import dpLoadh5
from dpWriteh5 import dpWriteh5
def draw_flow(img, flow, step=16):
h, w = img.shape[:2]
y, x = np.mgrid[step/2:h:step, step/2:w:step].reshape(2,-1).astype(int)
fx, fy = flow[y,x].T
lines = np.vstack([x, y, x+fx, y+fy]).T.reshape(-1, 2, 2)
lines = np.int32(lines + 0.5)
vis = cv2.cvtColor(img, cv2.COLOR_GRAY2BGR)
cv2.polylines(vis, lines, 0, (0, 255, 0))
for (x1, y1), (x2, y2) in lines:
cv2.circle(vis, (x1, y1), 1, (0, 255, 0), -1)
return vis
def draw_hsv(flow):
h, w = flow.shape[:2]
fx, fy = flow[:,:,0], flow[:,:,1]
ang = np.arctan2(fy, fx) + np.pi
v = np.sqrt(fx*fx+fy*fy)
hsv = np.zeros((h, w, 3), np.uint8)
hsv[...,0] = ang*(180/np.pi/2)
hsv[...,1] = 255
hsv[...,2] = np.minimum(v*20, 255)
bgr = cv2.cvtColor(hsv, cv2.COLOR_HSV2BGR)
return bgr
def warp_flow(img, flow):
h, w = flow.shape[:2]
flow = -flow
flow[:,:,0] += | np.arange(w) | numpy.arange |
import torch
import functools
import numpy as np
from esgd import ESGD,get_current_time
class ESGD_WS(ESGD):
def _sample_optimizer(self, weights=None):
hpval_indices = np.random.choice(len(self.hpvalues), size=self.n_population, p=weights)
return [self.hpvalues[idx] for idx in hpval_indices], hpval_indices
def _update_weights(self, g, weights, rank, topn=3):
for idx in rank[:topn]:
weights[idx] += 1/(len(self.hpvalues)*np.sqrt(self.n_generations)+topn*g)
return weights/np.sum(weights)
def train(
self,
train_data,
train_targets,
topn=3,
init_weights=None,
test_set=None,
log_file=None,
batch_size=1024,
transform=None
):
logger = self.Logger(log_file)
if init_weights is None:
weights = np.ones(len(self.hpvalues))
weights /= len(self.hpvalues)
else:
weights = np.array(init_weights)
train_loader = self.get_data_loader(
train_data,
train_targets,
batch_size=batch_size,
transform=transform
)
if test_set is not None:
test_loader = self.get_data_loader(*test_set, shuffle=False, batch_size=batch_size)
np.random.seed(self.random_state)
torch.manual_seed(self.random_state)
curr_gen = [self.model_class().to(self.device) for _ in range(self.n_population)]
results = []
for g in range(1, 1 + self.n_generations):
curr_hpvals,hpval_indices = self._sample_optimizer(weights=weights)
optimizers = [self.optimizer_class(
ind.parameters(), **dict(zip(self.hpnames, hpvs))
) for ind, hpvs in zip(curr_gen, curr_hpvals)]
running_losses = [0.0 for _ in range(self.n_population)]
running_corrects = [0 for _ in range(self.n_population)]
running_total = 0
if self.verbose:
logger.logging(f"Generation #{g}:")
logger.logging(f"|___{get_current_time()}\tpre-SGD")
for s in range(self.sgds_per_gen):
for (x, y) in train_loader:
x = x.to(self.device)
y = y.to(self.device)
running_total += x.size(0)
for i, (ind, opt) in enumerate(zip(curr_gen, optimizers)):
out = ind(x)
loss = self.fitness_function(out, y)
opt.zero_grad()
loss.backward()
opt.step()
running_losses[i] += loss.item() * x.size(0)
running_corrects[i] += (out.max(dim=1)[1] == y).sum().item()
running_losses = list(map(lambda x: x / running_total, running_losses))
running_accs = list(map(lambda x: x / running_total, running_corrects))
opt_rank = hpval_indices[np.argsort(running_losses)]
weights = self._update_weights(g, weights, rank=opt_rank, topn=topn)
if self.verbose:
logger.logging(f"|___{get_current_time()}\tpost-SGD")
logger.logging(f"\t|___population best fitness: {min(running_losses)}")
logger.logging(f"\t|___population average fitness: {sum(running_losses) / len(running_losses)}")
logger.logging(f"\t|___population best accuracy: {max(running_accs)}")
logger.logging(f"\t|___population average accuracy: {sum(running_accs) / len(running_accs)}")
if self.verbose:
logger.logging(f"|___{get_current_time()}\tpre-evolution")
curr_mix = [
np.random.choice(self.n_population, size=self.mixing_number)
for _ in range(int(self.reproductive_factor * self.n_population))
]
offsprings = []
for e in range(self.evos_per_gen):
for mix in curr_mix:
model = self.model_class().to(self.device)
for p_child, *p_parents in zip(model.parameters(), *[curr_gen[idx].parameters() for idx in mix]):
p_child.data = functools.reduce(lambda x, y: x + y, p_parents) / self.mixing_number
# p_child.data.add_(1 / g * self.mutation_length * (2 * torch.rand_like(p_child) - 1))
p_child.data.add_(1 / g * self.mutation_length * torch.randn_like(p_child))
offsprings.append(model)
train_losses = [0.0 for _ in range(int(self.n_population * (1 + self.reproductive_factor)))]
train_corrects = [0 for _ in range(int(self.n_population * (1 + self.reproductive_factor)))]
if test_set is not None:
test_losses = [0.0 for _ in range(int(self.n_population * (1 + self.reproductive_factor)))]
test_corrects = [0 for _ in range(int(self.n_population * (1 + self.reproductive_factor)))]
test_total = 0
curr_gen.extend(offsprings)
with torch.no_grad():
for ind in curr_gen:
ind.eval()
for (x, y) in train_loader:
x = x.to(self.device)
y = y.to(self.device)
for i, ind in enumerate(curr_gen):
out = ind(x)
train_losses[i] += self.fitness_function(out, y).item() * x.size(0)
train_corrects[i] += (out.max(dim=1)[1] == y).sum().item()
if test_set is not None:
for (x, y) in test_loader:
x = x.to(self.device)
y = y.to(self.device)
test_total += x.size(0)
for i, ind in enumerate(curr_gen):
out = ind(x)
test_losses[i] += self.fitness_function(out, y).item() * x.size(0)
test_corrects[i] += (out.max(dim=1)[1] == y).sum().item()
for ind in curr_gen:
ind.train()
train_losses = list(map(lambda x: x / running_total, train_losses))
train_accs = list(map(lambda x: x / running_total, train_corrects))
if test_set is not None:
test_losses = list(map(lambda x: x / test_total, test_losses))
test_accs = list(map(lambda x: x / test_total, test_corrects))
curr_rank = np.argsort(train_losses)
elite = curr_rank[:self.m_elite]
others = np.random.choice(len(curr_gen) - self.m_elite,
size=self.n_population - self.m_elite) + self.m_elite
others = curr_rank[others]
curr_gen = [curr_gen[idx] for idx in np.concatenate([elite, others])]
train_losses = [train_losses[idx] for idx in np.concatenate([elite, others])]
train_accs = [train_accs[idx] for idx in np.concatenate([elite, others])]
if test_set is not None:
test_losses = [test_losses[idx] for idx in | np.concatenate([elite, others]) | numpy.concatenate |
r"""
Module for some integrators.
- IRK3: Implicit third order Runge-Kutta
- RK4: Runge-Kutta fourth order
- ETD: Exponential time differencing Euler method
- ETDRK4: Exponential time differencing Runge-Kutta fourth order
See, e.g.,
<NAME> and <NAME> "Solving periodic semilinear PDEs in 1D, 2D and
3D with exponential integrators", https://arxiv.org/pdf/1604.08900.pdf
Integrators are set up to solve equations like
.. math::
\frac{\partial u}{\partial t} = L u + N(u)
where :math:`u` is the solution, :math:`L` is a linear operator and
:math:`N(u)` is the nonlinear part of the right hand side.
Note
----
`RK4`, `ETD` and `ETDRK4` can only be used with Fourier function spaces,
as they assume all matrices are diagonal.
"""
import types
import numpy as np
from shenfun import Function, TPMatrix, TrialFunction, TestFunction, inner, la
__all__ = ('IRK3', 'RK4', 'ETDRK4', 'ETD')
#pylint: disable=unused-variable
class IntegratorBase:
"""Abstract base class for integrators
Parameters
----------
T : TensorProductSpace
L : function
To compute linear part of right hand side
N : function
To compute nonlinear part of right hand side
update : function
To be called at the end of a timestep
params : dictionary
Any relevant keyword arguments
"""
def __init__(self, T,
L=None,
N=None,
update=None,
**params):
_p = {'call_update': -1,
'dt': 0}
_p.update(params)
self.params = _p
self.T = T
if L is not None:
self.LinearRHS = types.MethodType(L, self)
if N is not None:
self.NonlinearRHS = types.MethodType(N, self)
if update is not None:
self.update = types.MethodType(update, self)
def update(self, u, u_hat, t, tstep, **par):
pass
def LinearRHS(self, *args, **kwargs):
pass
def NonlinearRHS(self, *args, **kwargs):
pass
def setup(self, dt):
"""Set up solver"""
pass
def solve(self, u, u_hat, dt, trange):
"""Integrate forward in end_time
Parameters
----------
u : array
The solution array in physical space
u_hat : array
The solution array in spectral space
dt : float
Timestep
trange : two-tuple
Time and end time
"""
pass
class IRK3(IntegratorBase):
"""Third order implicit Runge Kutta
Parameters
----------
T : TensorProductSpace
L : function of TrialFunction(T)
To compute linear part of right hand side
N : function
To compute nonlinear part of right hand side
update : function
To be called at the end of a timestep
params : dictionary
Any relevant keyword arguments
"""
def __init__(self, T,
L=None,
N=None,
update=None,
**params):
IntegratorBase.__init__(self, T, L=L, N=N, update=update, **params)
self.dU = Function(T)
self.dU1 = Function(T)
self.a = (8./15., 5./12., 3./4.)
self.b = (0.0, -17./60., -5./12.)
self.c = (0., 8./15., 2./3., 1)
self.solver = None
self.rhs_mats = None
self.w0 = Function(self.T).v
self.mask = self.T.get_mask_nyquist()
def setup(self, dt):
self.params['dt'] = dt
u = TrialFunction(self.T)
v = TestFunction(self.T)
# Note that we are here assembling implicit left hand side matrices,
# as well as matrices that can be used to assemble the right hande side
# much faster through matrix-vector products
a, b = self.a, self.b
self.solver = []
for rk in range(3):
mats = inner(v, u - ((a[rk]+b[rk])*dt/2)*self.LinearRHS(u))
if len(mats[0].naxes) == 1:
self.solver.append(la.SolverGeneric1ND(mats))
elif len(mats[0].naxes) == 2:
self.solver.append(la.SolverGeneric2ND(mats))
else:
raise NotImplementedError
self.rhs_mats = []
for rk in range(3):
self.rhs_mats.append(inner(v, u + ((a[rk]+b[rk])*dt/2)*self.LinearRHS(u)))
self.mass = inner(u, v)
def compute_rhs(self, u, u_hat, dU, dU1, rk):
a = self.a[rk]
b = self.b[rk]
dt = self.params['dt']
dU = self.NonlinearRHS(u, u_hat, dU, **self.params)
dU.mask_nyquist(self.mask)
w1 = dU*a*dt + self.dU1*b*dt
self.dU1[:] = dU
return w1
def solve(self, u, u_hat, dt, trange):
if self.solver is None or abs(self.params['dt']-dt) > 1e-12:
self.setup(dt)
t, end_time = trange
tstep = 0
while t < end_time-1e-8:
for rk in range(3):
dU = self.compute_rhs(u, u_hat, self.dU, self.dU1, rk)
for mat in self.rhs_mats[rk]:
w0 = mat.matvec(u_hat, self.w0)
dU += w0
u_hat = self.solver[rk](dU, u_hat)
u_hat.mask_nyquist(self.mask)
t += self.dt
tstep += 1
self.update(u, u_hat, t, tstep, **self.params)
class ETD(IntegratorBase):
"""Exponential time differencing Euler method
<NAME> and <NAME> "Solving periodic semilinear PDEs in 1D, 2D and
3D with exponential integrators", https://arxiv.org/pdf/1604.08900.pdf
Parameters
----------
T : TensorProductSpace
L : function
To compute linear part of right hand side
N : function
To compute nonlinear part of right hand side
update : function
To be called at the end of a timestep
params : dictionary
Any relevant keyword arguments
"""
def __init__(self, T,
L=None,
N=None,
update=None,
**params):
IntegratorBase.__init__(self, T, L=L, N=N, update=update, **params)
self.dU = Function(T)
self.psi = None
self.ehL = None
def setup(self, dt):
"""Set up ETD ODE solver"""
self.params['dt'] = dt
L = self.LinearRHS(**self.params)
if isinstance(L, TPMatrix):
assert L.isidentity()
L = L.scale
L = np.atleast_1d(L)
hL = L*dt
self.ehL = np.exp(hL)
M = 50
psi = self.psi = | np.zeros(hL.shape, dtype=np.float) | numpy.zeros |
from skimage.feature import corner_harris, peak_local_max
import numpy as np
def dist2(x, c):
# Borrowed from https://inst.eecs.berkeley.edu/~cs194-26/fa18/
ndata, dimx = x.shape
ncenters, dimc = c.shape
assert dimx == dimc, 'Data dimension does not match dimension of centers'
return (np.ones((ncenters, 1)) * np.sum((x ** 2).T, axis=0)).T + \
np.ones((ndata, 1)) * | np.sum((c ** 2).T, axis=0) | numpy.sum |
import sys
import os
import os.path as op
import glob
import logging
import json
import multiprocessing
from functools import partial
from pathlib import Path
from tqdm import tqdm
import numpy as np
import matplotlib.pyplot as plt
from sklearn.cluster import DBSCAN
from scipy.stats import rankdata
from scipy.spatial import distance_matrix
from scipy.optimize import linear_sum_assignment
from scipy.interpolate import interp1d
import networkx as nx
from utils.dir_helper import get_batch_subdir, get_exp_subdir
from utils.file_manipulation import tiff_read
def get_diff_static(I, ds_median, config):
"""
Computes a diff between current image I and the dataset median.
Parameters
----------
I: 2d array
the current image frame
ds_median: 2d array
the dataset median
config: dict
configuration
"""
diff = abs(I - ds_median)
abs_threshold = config['absthresh']
pc_threshold = config['pcthresh']
# Threshold is the max value of the abs_threshold, and the value of diff at percentile pc_threshold
threshold = max(abs_threshold, np.percentile(diff, pc_threshold))
# Suppress values of rng_diff less than a threshold
diff[diff < threshold] = 0
return diff
def get_particles(range_diff, image, clustering_settings):
"""Get the detections using Gary's original method
Returns a list of particles and their properties
Parameters
----------
range_diff:
output from background subtraction
image:
original image frame for intensity calculation
may have a different shape than range_diff
cluster_settings:
hyperparameters for the clustering algorithm
Returns
-------
list of dicts with keys:
pos: (y, x) coordinates of particle
size: number of pixels in cluster
bbox_tl: bbox (top, left)
bbox_hw: bbox (height, width)
max_intensity: max intensity of pixels (list)
"""
# select points above a threshold, and get their weights
idx = (range_diff > 0)
points = np.column_stack(np.nonzero(idx))
weights = range_diff[idx].ravel().astype(float)
# empty list to store particles
particles = []
if len(points) > 0:
# use DBSCAN to cluster the points
dbscan = DBSCAN(eps=clustering_settings['dbscan']['epsilon_px'],
min_samples=clustering_settings['dbscan']['min_weight'])
labels = dbscan.fit_predict(points, sample_weight=weights)
n_clusters = int(np.max(labels)) + 1
for l in range(n_clusters):
idx = (labels == l)
# must have specified minimum number of points
# keep track of clusters that fall below this thresh
if np.sum(idx) < clustering_settings['filters']['min_px']:
continue
relevant = points[idx]
# Build particle properties
particle = {}
# center of particle
particle['pos'] = [round(i, 1) for i in | np.average(relevant, axis=0) | numpy.average |
# Copyright 2020 Huawei Technologies Co., Ltd
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ============================================================================
'''
Bert evaluation assessment method script.
'''
import math
import numpy as np
from .CRF import postprocess
class Accuracy():
'''
calculate accuracy
'''
def __init__(self):
self.acc_num = 0
self.total_num = 0
def update(self, logits, labels):
labels = labels.asnumpy()
labels = np.reshape(labels, -1)
logits = logits.asnumpy()
logit_id = np.argmax(logits, axis=-1)
self.acc_num += np.sum(labels == logit_id)
self.total_num += len(labels)
class F1():
'''
calculate F1 score
'''
def __init__(self, use_crf=False, num_labels=2):
self.TP = 0
self.FP = 0
self.FN = 0
self.use_crf = use_crf
self.num_labels = num_labels
def update(self, logits, labels):
'''
update F1 score
'''
labels = labels.asnumpy()
labels = np.reshape(labels, -1)
if self.use_crf:
backpointers, best_tag_id = logits
best_path = postprocess(backpointers, best_tag_id)
logit_id = []
for ele in best_path:
logit_id.extend(ele)
else:
logits = logits.asnumpy()
logit_id = np.argmax(logits, axis=-1)
logit_id = np.reshape(logit_id, -1)
pos_eva = np.isin(logit_id, [i for i in range(1, self.num_labels)])
pos_label = np.isin(labels, [i for i in range(1, self.num_labels)])
self.TP += np.sum(pos_eva&pos_label)
self.FP += np.sum(pos_eva&(~pos_label))
self.FN += np.sum((~pos_eva)&pos_label)
class SpanF1():
'''
calculate F1、precision and recall score in span manner for NER
'''
def __init__(self, use_crf=False, label2id=None):
self.TP = 0
self.FP = 0
self.FN = 0
self.use_crf = use_crf
self.label2id = label2id
if label2id is None:
raise ValueError("label2id info should not be empty")
self.id2label = {}
for key, value in label2id.items():
self.id2label[value] = key
def tag2span(self, ids):
'''
conbert ids list to span mode
'''
labels = np.array([self.id2label[id] for id in ids])
spans = []
prev_label = None
for idx, tag in enumerate(labels):
tag = tag.lower()
cur_label, label = tag[:1], tag[2:]
if cur_label in ('b', 's'):
spans.append((label, [idx, idx]))
elif cur_label in ('m', 'e') and prev_label in ('b', 'm') and label == spans[-1][0]:
spans[-1][1][1] = idx
elif cur_label == 'o':
pass
else:
spans.append((label, [idx, idx]))
prev_label = cur_label
return [(span[0], (span[1][0], span[1][1] + 1)) for span in spans]
def update(self, logits, labels):
'''
update span F1 score
'''
labels = labels.asnumpy()
labels = np.reshape(labels, -1)
if self.use_crf:
backpointers, best_tag_id = logits
best_path = postprocess(backpointers, best_tag_id)
logit_id = []
for ele in best_path:
logit_id.extend(ele)
else:
logits = logits.asnumpy()
logit_id = np.argmax(logits, axis=-1)
logit_id = np.reshape(logit_id, -1)
label_spans = self.tag2span(labels)
pred_spans = self.tag2span(logit_id)
for span in pred_spans:
if span in label_spans:
self.TP += 1
label_spans.remove(span)
else:
self.FP += 1
for span in label_spans:
self.FN += 1
class MCC():
'''
Calculate Matthews Correlation Coefficient
'''
def __init__(self):
self.TP = 0
self.FP = 0
self.FN = 0
self.TN = 0
def update(self, logits, labels):
'''
MCC update
'''
labels = labels.asnumpy()
labels = np.reshape(labels, -1)
labels = labels.astype(np.bool)
logits = logits.asnumpy()
logit_id = np.argmax(logits, axis=-1)
logit_id = np.reshape(logit_id, -1)
logit_id = logit_id.astype(np.bool)
ornot = logit_id ^ labels
self.TP += (~ornot & labels).sum()
self.FP += (ornot & ~labels).sum()
self.FN += (ornot & labels).sum()
self.TN += (~ornot & ~labels).sum()
def cal(self):
mcc = (self.TP*self.TN - self.FP*self.FN)/math.sqrt((self.TP+self.FP)*(self.TP+self.FN) *
(self.TN+self.FP)*(self.TN+self.FN))
return mcc
class Spearman_Correlation():
'''
Calculate Spearman Correlation Coefficient
'''
def __init__(self):
self.label = []
self.logit = []
def update(self, logits, labels):
labels = labels.asnumpy()
labels = np.reshape(labels, -1)
logits = logits.asnumpy()
logits = | np.reshape(logits, -1) | numpy.reshape |
# Copyright 2020 Google LLC
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""Tests for google3.third_party.py.tensorflow_graphics.interpolation.weighted."""
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
from absl.testing import parameterized
import numpy as np
import tensorflow as tf
from tensorflow_graphics.math.interpolation import weighted
from tensorflow_graphics.util import test_case
class WeightedTest(test_case.TestCase):
def _get_tensors_from_shapes(self, num_points, dim_points, num_outputs,
num_pts_to_interpolate):
points = np.random.uniform(size=(num_points, dim_points))
weights = np.random.uniform(size=(num_outputs, num_pts_to_interpolate))
indices = np.asarray([
np.random.permutation(num_points)[:num_pts_to_interpolate].tolist()
for _ in range(num_outputs)
])
indices = np.expand_dims(indices, axis=-1)
return points, weights, indices
@parameterized.parameters(
(3, 4, 2, 3),
(5, 4, 5, 3),
(5, 6, 5, 5),
(2, 6, 5, 1),
)
def test_interpolate_exception_not_raised(self, dim_points, num_points,
num_outputs,
num_pts_to_interpolate):
"""Tests whether exceptions are not raised for compatible shapes."""
points, weights, indices = self._get_tensors_from_shapes(
num_points, dim_points, num_outputs, num_pts_to_interpolate)
self.assert_exception_is_not_raised(
weighted.interpolate,
shapes=[],
points=points,
weights=weights,
indices=indices,
normalize=True)
@parameterized.parameters(
("must have a rank greater than 1", ((3,), (None, 2), (None, 2, 0))),
("must have a rank greater than 1", ((None, 3), (None, 2), (1,))),
("must have exactly 1 dimensions in axis -1", ((None, 3), (None, 2),
(None, 2, 2))),
("must have the same number of dimensions", ((None, 3), (None, 2),
(None, 3, 1))),
("Not all batch dimensions are broadcast-compatible.",
((None, 3), (None, 5, 2), (None, 4, 2, 1))),
)
def test_interpolate_exception_raised(self, error_msg, shapes):
"""Tests whether exceptions are raised for incompatible shapes."""
self.assert_exception_is_raised(
weighted.interpolate, error_msg, shapes=shapes, normalize=False)
@parameterized.parameters(
(((-1.0, 1.0), (1.0, 1.0), (3.0, 1.0), (-1.0, -1.0), (1.0, -1.0),
(3.0, -1.0)), ((0.25, 0.25, 0.25, 0.25), (0.5, 0.5, 0.0, 0.0)),
(((0,), (1,), (3,), (4,)), ((1,), (2,), (4,),
(5,))), False, ((0.0, 0.0), (2.0, 1.0))),)
def test_interpolate_preset(self, points, weights, indices, _, out):
"""Tests whether interpolation results are correct."""
weights = tf.convert_to_tensor(value=weights)
result_unnormalized = weighted.interpolate(
points=points, weights=weights, indices=indices, normalize=False)
result_normalized = weighted.interpolate(
points=points, weights=2.0 * weights, indices=indices, normalize=True)
estimated_unnormalized = self.evaluate(result_unnormalized)
estimated_normalized = self.evaluate(result_normalized)
self.assertAllClose(estimated_unnormalized, out)
self.assertAllClose(estimated_normalized, out)
@parameterized.parameters(
(3, 4, 2, 3),
(5, 4, 5, 3),
(5, 6, 5, 5),
(2, 6, 5, 1),
)
def test_interpolate_negative_weights_raised(self, dim_points, num_points,
num_outputs,
num_pts_to_interpolate):
"""Tests whether exception is raised when weights are negative."""
points, weights, indices = self._get_tensors_from_shapes(
num_points, dim_points, num_outputs, num_pts_to_interpolate)
weights *= -1.0
with self.assertRaises(tf.errors.InvalidArgumentError):
result = weighted.interpolate(
points=points, weights=weights, indices=indices, normalize=True)
self.evaluate(result)
@parameterized.parameters(
(((-1.0, 1.0), (1.0, 1.0), (3.0, 1.0), (-1.0, -1.0), (1.0, -1.0),
(3.0, -1.0)), ((1.0, -1.0, 1.0, -1.0), (0.0, 0.0, 0.0, 0.0)),
(((0,), (1,), (3,), (4,)), ((1,), (2,), (4,), (5,))), ((0.0, 0.0),
(0.0, 0.0))))
def test_interp_unnormalizable_raised_(self, points, weights, indices, _):
"""Tests whether exception is raised when weights are unnormalizable."""
with self.assertRaises(tf.errors.InvalidArgumentError):
result = weighted.interpolate(
points=points,
weights=weights,
indices=indices,
normalize=True,
allow_negative_weights=True)
self.evaluate(result)
@parameterized.parameters(
(3, 4, 2, 3),
(5, 4, 5, 3),
(5, 6, 5, 5),
(2, 6, 5, 1),
)
def test_interpolate_jacobian_random(self, dim_points, num_points,
num_outputs, num_pts_to_interpolate):
"""Tests whether jacobian is correct."""
points_np, weights_np, indices_np = self._get_tensors_from_shapes(
num_points, dim_points, num_outputs, num_pts_to_interpolate)
def interpolate_fn(points, weights):
return weighted.interpolate(
points=points, weights=weights, indices=indices_np, normalize=True)
self.assert_jacobian_is_correct_fn(interpolate_fn, [points_np, weights_np])
@parameterized.parameters(
((3, 2), (2, 2)),
((None, 3, 2), (None, 1, 2)),
((10, 5, 3, 2), (10, 5, 2, 2)),
)
def test_get_barycentric_coordinates_exception_not_raised(self, *shapes):
"""Tests that the shape exceptions are not raised."""
self.assert_exception_is_not_raised(weighted.get_barycentric_coordinates,
shapes)
@parameterized.parameters(
("triangle_vertices must have exactly 2 dimensions in axis -1", (3, 1),
(1, 2)),
("triangle_vertices must have exactly 3 dimensions in axis -2", (2, 2),
(1, 2)),
("pixels must have exactly 2 dimensions in axis -1", (3, 2), (1, 3)),
("Not all batch dimensions are broadcast-compatible", (5, 3, 2),
(2, 10, 2)),
)
def test_get_barycentric_coordinates_exception_raised(self, error_msg,
*shape):
"""Tests that the shape exceptions are raised."""
self.assert_exception_is_raised(weighted.get_barycentric_coordinates,
error_msg, shape)
def test_get_barycentric_coordinates_jacobian_random(self):
"""Tests the Jacobian of get_barycentric_coordinates."""
tensor_size = np.random.randint(2)
tensor_shape = np.random.randint(1, 2, size=(tensor_size)).tolist()
triangle_vertices_init = 0.4 * np.random.random(
tensor_shape + [3, 2]).astype(np.float64) - 0.2
triangle_vertices_init += np.array(
((0.25, 0.25), (0.5, 0.75), (0.75, 0.25)))
pixels_init = np.random.random(tensor_shape + [3, 2]).astype(np.float64)
barycentric_fn = weighted.get_barycentric_coordinates
self.assert_jacobian_is_correct_fn(
lambda vertices, pixels: barycentric_fn(vertices, pixels)[0],
[triangle_vertices_init, pixels_init])
def test_get_barycentric_coordinates_normalized(self):
"""Tests whether the barycentric coordinates are normalized."""
tensor_size = np.random.randint(3)
tensor_shape = np.random.randint(1, 10, size=(tensor_size)).tolist()
num_pixels = np.random.randint(1, 10)
pixels_shape = tensor_shape + [num_pixels]
triangle_vertices = np.random.random(tensor_shape + [3, 2])
pixels = | np.random.random(pixels_shape + [2]) | numpy.random.random |
# Built-in
import warnings
import itertools as itt
import copy
import datetime as dtm # DB
# Common
import numpy as np
import scipy.interpolate as scpinterp
import scipy.stats as scpstats
import matplotlib.pyplot as plt
__all__ = [
'fit1d_dinput',
'fit2d_dinput',
'fit12d_dvalid',
'fit12d_dscales',
]
_NPEAKMAX = 12
_DCONSTRAINTS = {
'bck_amp': False,
'bck_rate': False,
'amp': False,
'width': False,
'shift': False,
'double': False,
'symmetry': False,
}
_DORDER = ['amp', 'width', 'shift']
_SAME_SPECTRUM = False
_DEG = 2
_NBSPLINES = 13
_SYMMETRY_CENTRAL_FRACTION = 0.3
_BINNING = False
_POS = False
_SUBSET = False
_VALID_NSIGMA = 6.
_VALID_FRACTION = 0.8
_LTYPES = [int, float, np.int_, np.float_]
_DBOUNDS = {
'bck_amp': (0., 3.),
'bck_rate': (-3., 3.),
'amp': (0, 10),
'width': (0.01, 2.),
'shift': (-1, 1),
'dratio': (0., 2.),
'dshift': (-10., 10.),
'bs': (-10., 10.),
}
_DX0 = {
'bck_amp': 1.,
'bck_rate': 0.,
'amp': 1.,
'width': 1.,
'shift': 0.,
'dratio': 0.5,
'dshift': 0.,
'bs': 1.,
}
_DINDOK = {
0: 'ok',
-1: 'mask',
-2: 'out of domain',
-3: 'neg or NaN',
-4: 'binning=0',
-5: 'S/N valid, excluded',
-6: 'S/N non-valid, included',
-7: 'S/N non-valid, excluded',
}
###########################################################
###########################################################
#
# Preliminary
# utility tools for 1d spectral fitting
#
###########################################################
###########################################################
def get_symmetry_axis_1dprofile(phi, data, cent_fraction=None):
""" On a series of 1d vertical profiles, find the best symmetry axis """
if cent_fraction is None:
cent_fraction = _SYMMETRY_CENTRAL_FRACTION
# Find the phi in the central fraction
phimin = np.nanmin(phi)
phimax = np.nanmax(phi)
phic = 0.5*(phimax + phimin)
dphi = (phimax - phimin)*cent_fraction
indphi = np.abs(phi-phic) <= dphi/2.
phiok = phi[indphi]
# Compute new phi and associated costs
phi2 = phi[:, None] - phiok[None, :]
phi2min = np.min([np.nanmax(np.abs(phi2 * (phi2 < 0)), axis=0),
np.nanmax(np.abs(phi2 * (phi2 > 0)), axis=0)], axis=0)
indout = np.abs(phi2) > phi2min[None, :]
phi2p = np.abs(phi2)
phi2n = np.abs(phi2)
phi2p[(phi2 < 0) | indout] = np.nan
phi2n[(phi2 > 0) | indout] = np.nan
nok = np.min([np.sum((~np.isnan(phi2p)), axis=0),
np.sum((~np.isnan(phi2n)), axis=0)], axis=0)
cost = np.full((data.shape[0], phiok.size), np.nan)
for ii in range(phiok.size):
indp = np.argsort(np.abs(phi2p[:, ii]))
indn = np.argsort(np.abs(phi2n[:, ii]))
cost[:, ii] = np.nansum(
(data[:, indp] - data[:, indn])[:, :nok[ii]]**2,
axis=1)
return phiok[np.nanargmin(cost, axis=1)]
###########################################################
###########################################################
#
# 1d spectral fitting from dlines
#
###########################################################
###########################################################
def _checkformat_dconstraints(dconstraints=None, defconst=None):
# Check constraints
if dconstraints is None:
dconstraints = defconst
# Check dconstraints keys
lk = sorted(_DCONSTRAINTS.keys())
c0 = (
isinstance(dconstraints, dict)
and all([k0 in lk for k0 in dconstraints.keys()])
)
if not c0:
msg = (
"\ndconstraints should contain constraints for spectrum fitting\n"
+ "It be a dict with the following keys:\n"
+ "\t- available keys: {}\n".format(lk)
+ "\t- provided keys: {}".format(dconstraints.keys())
)
raise Exception(msg)
# copy to avoid modifying reference
return copy.deepcopy(dconstraints)
def _checkformat_dconstants(dconstants=None, dconstraints=None):
if dconstants is None:
return
lk = [kk for kk in sorted(dconstraints.keys()) if kk != 'symmetry']
if not isinstance(dconstants, dict):
msg = (
"\ndconstants should be None or a dict with keys in:\n"
+ "\t- available keys: {}\n".format(lk)
+ "\t- provided : {}".format(type(dconstants))
)
raise Exception(msg)
# Check dconstraints keys
lc = [
k0 for k0, v0 in dconstants.items()
if not (
k0 in lk
and (
(
k0 in _DORDER
and isinstance(v0, dict)
and all([
k1 in dconstraints[k0].keys()
and type(v1) in _LTYPES
for k1, v1 in v0.items()
])
)
or (
k0 not in _DORDER
and type(v0) in _LTYPES
)
)
)
]
if len(lc) > 0:
dc0 = [
'\t\t{}: {}'.format(
kk,
sorted(dconstraints[kk].keys()) if kk in _DORDER else float
)
for kk in lk
]
dc1 = [
'\t\t{}: {}'.format(
kk,
sorted(dconstants[kk].keys())
if kk in _DORDER else dconstants[kk]
)
for kk in sorted(dconstants.keys())
]
msg = (
"\ndconstants should be None or a dict with keys in:\n"
+ "\t- available keys:\n"
+ "\n".join(dc0)
+ "\n\t- provided keys:\n"
+ "\n".join(dc1)
)
raise Exception(msg)
# copy to avoid modifying reference
return copy.deepcopy(dconstants)
def _dconstraints_double(dinput, dconstraints, defconst=_DCONSTRAINTS):
dinput['double'] = dconstraints.get('double', defconst['double'])
c0 = (
isinstance(dinput['double'], bool)
or (
isinstance(dinput['double'], dict)
and all([
kk in ['dratio', 'dshift'] and type(vv) in _LTYPES
for kk, vv in dinput['double'].items()
])
)
)
if c0 is False:
msg = (
"dconstraints['double'] must be either:\n"
+ "\t- False: no line doubling\n"
+ "\t- True: line doubling with unknown ratio and shift\n"
+ "\t- {'dratio': float}: line doubling with:\n"
+ "\t \t explicit ratio, unknown shift\n"
+ "\t- {'dshift': float}: line doubling with:\n"
+ "\t \t unknown ratio, explicit shift\n"
+ "\t- {'dratio': float, 'dshift': float}: line doubling with:\n"
+ "\t \t explicit ratio, explicit shift"
)
raise Exception(msg)
def _width_shift_amp(
indict, dconstants=None,
keys=None, dlines=None, nlines=None, k0=None,
):
# ------------------------
# Prepare error message
msg = ''
pavail = sorted(set(itt.chain.from_iterable([
v0.keys() for v0 in dlines.values()
])))
# ------------------------
# Check case
c0 = indict is False
c1 = (
isinstance(indict, str)
and indict in pavail
)
c2 = (
isinstance(indict, dict)
and all([
isinstance(k1, str)
and (
(isinstance(v1, str)) # and v0 in keys)
or (
isinstance(v1, list)
and all([
isinstance(v2, str)
# and v1 in keys
for v2 in v1
])
)
)
for k1, v1 in indict.items()
])
)
c3 = (
isinstance(indict, dict)
and all([
# ss in keys
isinstance(vv, dict)
and all([s1 in ['key', 'coef', 'offset'] for s1 in vv.keys()])
and isinstance(vv['key'], str)
for ss, vv in indict.items()
])
)
c4 = (
isinstance(indict, dict)
and isinstance(indict.get('keys'), list)
and isinstance(indict.get('ind'), np.ndarray)
)
if not any([c0, c1, c2, c3, c4]):
msg = (
f"dconstraints['{k0}'] shoud be either:\n"
f"\t- False ({c0}): no constraint\n"
f"\t- str ({c1}): key from dlines['<lines>'] "
"to be used as criterion\n"
f"\t\t available crit: {pavail}\n"
f"\t- dict ({c2}): "
"{str: line_keyi or [line_keyi, ..., line_keyj}\n"
f"\t- dict ({c3}): "
"{line_keyi: {'key': str, 'coef': , 'offset': }}\n"
f"\t- dict ({c4}): "
"{'keys': [], 'ind': np.ndarray}\n"
f" Available line_keys:\n{sorted(keys)}\n"
f" You provided:\n{indict}"
)
raise Exception(msg)
# ------------------------
# str key to be taken from dlines as criterion
if c0:
lk = keys
ind = np.eye(nlines, dtype=bool)
outdict = {
'keys': np.r_[lk],
'ind': ind,
'coefs': np.ones((nlines,)),
'offset': np.zeros((nlines,)),
}
if c1:
lk = sorted(set([dlines[k1].get(indict, k1) for k1 in keys]))
ind = np.array(
[
[dlines[k2].get(indict, k2) == k1 for k2 in keys]
for k1 in lk
],
dtype=bool,
)
outdict = {
'keys': np.r_[lk],
'ind': ind,
'coefs': np.ones((nlines,)),
'offset': np.zeros((nlines,)),
}
elif c2:
lkl = []
for k1, v1 in indict.items():
if isinstance(v1, str):
v1 = [v1]
v1 = [k2 for k2 in v1 if k2 in keys]
c0 = (
len(set(v1)) == len(v1)
and all([k2 not in lkl for k2 in v1])
)
if not c0:
msg = (
"Inconsistency in indict[{}], either:\n".format(k1)
+ "\t- v1 not unique: {}\n".format(v1)
+ "\t- some v1 not in keys: {}\n".format(keys)
+ "\t- some v1 in lkl: {}".format(lkl)
)
raise Exception(msg)
indict[k1] = v1
lkl += v1
for k1 in set(keys).difference(lkl):
indict[k1] = [k1]
lk = sorted(set(indict.keys()))
ind = np.array(
[[k2 in indict[k1] for k2 in keys] for k1 in lk],
dtype=bool,
)
outdict = {
'keys': np.r_[lk],
'ind': ind,
'coefs': np.ones((nlines,)),
'offset': np.zeros((nlines,)),
}
elif c3:
lk = sorted(set([v0['key'] for v0 in indict.values()]))
lk += sorted(set(keys).difference(indict.keys()))
ind = np.array(
[
[indict.get(k2, {'key': k2})['key'] == k1 for k2 in keys]
for k1 in lk
],
dtype=bool,
)
coefs = np.array([
indict.get(k1, {'coef': 1.}).get('coef', 1.) for k1 in keys
])
offset = np.array([
indict.get(k1, {'offset': 0.}).get('offset', 0.) for k1 in keys
])
outdict = {
'keys': np.r_[lk],
'ind': ind,
'coefs': coefs,
'offset': offset,
}
elif c4:
outdict = indict
if 'coefs' not in indict.keys():
outdict['coefs'] = np.ones((nlines,))
if 'offset' not in indict.keys():
outdict['offset'] = np.zeros((nlines,))
# ------------------------
# Remove group with no match
indnomatch = np.sum(ind, axis=1) == 0
if np.any(indnomatch):
lknom = outdict['keys'][indnomatch]
outdict['keys'] = outdict['keys'][~indnomatch]
outdict['ind'] = outdict['ind'][~indnomatch, :]
lstr = [f"\t- {k1}" for k1 in lknom]
msg = (
f"The following {k0} groups match no lines, they are removed:\n"
+ "\n".join(lstr)
)
warnings.warn(msg)
# ------------------------
# Ultimate conformity checks
assert sorted(outdict.keys()) == ['coefs', 'ind', 'keys', 'offset']
# check ind (root of all subsequent ind arrays)
assert isinstance(outdict['ind'], np.ndarray)
assert outdict['ind'].dtype == np.bool_
assert outdict['ind'].shape == (outdict['keys'].size, nlines)
# check each line is associated to a unique group
assert np.all(np.sum(outdict['ind'], axis=0) == 1)
# check each group is associated to at least one line
assert np.all(np.sum(outdict['ind'], axis=1) >= 1)
assert outdict['coefs'].shape == (nlines,)
assert outdict['offset'].shape == (nlines,)
return outdict
###########################################################
###########################################################
#
# 2d spectral fitting from dlines
#
###########################################################
###########################################################
def _dconstraints_symmetry(
dinput,
dprepare=None,
symmetry=None,
cent_fraction=None,
defconst=_DCONSTRAINTS,
):
if symmetry is None:
symmetry = defconst['symmetry']
dinput['symmetry'] = symmetry
if not isinstance(dinput['symmetry'], bool):
msg = "dconstraints['symmetry'] must be a bool"
raise Exception(msg)
if dinput['symmetry'] is True:
dinput['symmetry_axis'] = get_symmetry_axis_1dprofile(
dprepare['phi1d'],
dprepare['dataphi1d'],
cent_fraction=cent_fraction,
)
###########################################################
###########################################################
#
# data, lamb, phi conformity checks
#
###########################################################
###########################################################
def _checkformat_data_fit12d_dlines_msg(data, lamb, phi=None, mask=None):
datash = data.shape if isinstance(data, np.ndarray) else type(data)
lambsh = lamb.shape if isinstance(lamb, np.ndarray) else type(lamb)
phish = phi.shape if isinstance(phi, np.ndarray) else type(phi)
masksh = mask.shape if isinstance(mask, np.ndarray) else type(mask)
shaped = '(nt, n1)' if phi is None else '(nt, n1, n2)'
shape = '(n1,)' if phi is None else '(n1, n2)'
msg = ("Args data, lamb, phi and mask must be:\n"
+ "\t- data: {} or {} np.ndarray\n".format(shaped, shape)
+ "\t- lamb, phi: both {} np.ndarray\n".format(shape)
+ "\t- mask: None or {}\n".format(shape)
+ " You provided:\n"
+ "\t - data: {}\n".format(datash)
+ "\t - lamb: {}\n".format(lambsh))
if phi is not None:
msg += "\t - phi: {}\n".format(phish)
msg += "\t - mask: {}\n".format(masksh)
return msg
def _checkformat_data_fit12d_dlines(
data, lamb, phi=None,
nxi=None, nxj=None, mask=None,
is2d=False,
):
# Check types
c0 = isinstance(data, np.ndarray) and isinstance(lamb, np.ndarray)
if is2d:
c0 &= isinstance(phi, np.ndarray)
if not c0:
msg = _checkformat_data_fit12d_dlines_msg(
data, lamb, phi=phi, mask=mask,
)
raise Exception(msg)
# Check shapes 1
mindim = 1 if phi is None else 2
phi1d, lamb1d, dataphi1d, datalamb1d = None, None, None, None
if is2d:
# special case
c1 = lamb.ndim == phi.ndim == 1
if c1:
if nxi is None:
nxi = lamb.size
if nxj is None:
nxj = phi.size
lamb1d = np.copy(lamb)
phi1d = np.copy(phi)
lamb = np.repeat(lamb[None, :], nxj, axis=0)
phi = np.repeat(phi[:, None], nxi, axis=1)
if nxi is None or nxj is None:
msg = "Arg (nxi, nxj) must be provided for double-checking shapes"
raise Exception(msg)
c0 = (
data.ndim in mindim + np.r_[0, 1]
and (
lamb.ndim == mindim
and lamb.shape == data.shape[-mindim:]
and lamb.shape == phi.shape
and lamb.shape in [(nxi, nxj), (nxj, nxi)]
)
)
else:
c0 = (
data.ndim in mindim + np.r_[0, 1]
and lamb.ndim == mindim
and lamb.shape == data.shape[-mindim:]
)
if not c0:
msg = _checkformat_data_fit12d_dlines_msg(
data, lamb, phi=phi, mask=mask,
)
raise Exception(msg)
# Check shapes 2
if data.ndim == mindim:
data = data[None, ...]
if is2d and c1:
dataphi1d = np.nanmean(data, axis=2)
datalamb1d = np.nanmean(data, axis=1)
if is2d and lamb.shape == (nxi, nxj):
lamb = lamb.T
phi = phi.T
data = np.swapaxes(data, 1, 2)
# mask
if mask is not None:
if mask.shape != lamb.shape:
if phi is not None and mask.T.shape == lamb.shape:
mask = mask.T
else:
msg = _checkformat_data_fit12d_dlines_msg(
data, lamb, phi=phi, mask=mask,
)
raise Exception(msg)
if is2d:
return lamb, phi, data, mask, phi1d, lamb1d, dataphi1d, datalamb1d
else:
return lamb, data, mask
###########################################################
###########################################################
#
# Domain limitation
#
###########################################################
###########################################################
def _checkformat_domain(domain=None, keys=['lamb', 'phi']):
if keys is None:
keys = ['lamb', 'phi']
if isinstance(keys, str):
keys = [keys]
if domain is None:
domain = {
k0: {
'spec': [np.inf*np.r_[-1., 1.]],
'minmax': np.inf*np.r_[-1., 1.],
}
for k0 in keys
}
return domain
c0 = (
isinstance(domain, dict)
and all([k0 in keys for k0 in domain.keys()])
)
if not c0:
msg = ("\nArg domain must be a dict with keys {}\n".format(keys)
+ "\t- provided: {}".format(domain))
raise Exception(msg)
domain2 = {k0: v0 for k0, v0 in domain.items()}
for k0 in keys:
domain2[k0] = domain2.get(k0, [np.inf*np.r_[-1., 1.]])
ltypesin = [list, np.ndarray]
ltypesout = [tuple]
for k0, v0 in domain2.items():
c0 = (
type(v0) in ltypesin + ltypesout
and (
(
all([type(v1) in _LTYPES for v1 in v0])
and len(v0) == 2
and v0[1] > v0[0]
)
or (
all([
type(v1) in ltypesin + ltypesout
and all([type(v2) in _LTYPES for v2 in v1])
and len(v1) == 2
and v1[1] > v1[0]
for v1 in v0
])
)
)
)
if not c0:
msg = (
"domain[{}] must be either a:\n".format(k0)
+ "\t- np.ndarray or list of 2 increasing values: "
+ "inclusive interval\n"
+ "\t- tuple of 2 increasing values: exclusive interval\n"
+ "\t- a list of combinations of the above\n"
+ " provided: {}".format(v0)
)
raise Exception(msg)
if type(v0) in ltypesout:
v0 = [v0]
else:
c0 = all([
type(v1) in ltypesin + ltypesout
and len(v1) == 2
and v1[1] > v1[0]
for v1 in v0
])
if not c0:
v0 = [v0]
domain2[k0] = {
'spec': v0,
'minmax': [np.nanmin(v0), np.nanmax(v0)],
}
return domain2
def apply_domain(lamb=None, phi=None, domain=None):
lc = [lamb is not None, phi is not None]
if not lc[0]:
msg = "At least lamb must be provided!"
raise Exception(msg)
din = {'lamb': lamb}
if lc[1]:
din['phi'] = phi
domain = _checkformat_domain(domain=domain, keys=din.keys())
ind = np.ones(lamb.shape, dtype=bool)
for k0, v0 in din.items():
indin = np.zeros(v0.shape, dtype=bool)
indout = np.zeros(v0.shape, dtype=bool)
for v1 in domain[k0]['spec']:
indi = (v0 >= v1[0]) & (v0 <= v1[1])
if isinstance(v1, tuple):
indout |= indi
else:
indin |= indi
ind = ind & indin & (~indout)
return ind, domain
###########################################################
###########################################################
#
# binning (2d only)
#
###########################################################
###########################################################
def _binning_check(
binning,
dlamb_ref=None,
dphi_ref=None,
domain=None, nbsplines=None,
):
lk = ['phi', 'lamb']
lkall = lk + ['nperbin']
msg = (
"binning must be dict of the form:\n"
+ "\t- provide number of bins:\n"
+ "\t \t{'phi': int,\n"
+ "\t \t 'lamb': int}\n"
+ "\t- provide bin edges vectors:\n"
+ "\t \t{'phi': 1d np.ndarray (increasing),\n"
+ "\t \t 'lamb': 1d np.ndarray (increasing)}\n"
+ " provided:\n{}".format(binning)
)
# Check input
if binning is None:
binning = _BINNING
if nbsplines is None:
nbsplines = False
if nbsplines is not False:
c0 = isinstance(nbsplines, int) and nbsplines > 0
if not c0:
msg2 = (
"Both nbsplines and deg must be positive int!\n"
+ "\t- nbsplines: {}\n".format(nbsplines)
)
raise Exception(msg2)
# Check which format was passed and return None or dict
ltypes0 = _LTYPES
ltypes1 = [tuple, list, np.ndarray]
lc = [
binning is False,
(
isinstance(binning, dict)
and all([kk in lkall for kk in binning.keys()])
and all([kk in binning.keys() for kk in lk])
),
type(binning) in ltypes0,
type(binning) in ltypes1,
]
if not any(lc):
raise Exception(msg)
if binning is False:
return binning
elif type(binning) in ltypes0:
binning = {
'phi': {'nbins': int(binning)},
'lamb': {'nbins': int(binning)},
}
elif type(binning) in ltypes1:
binning = np.atleast_1d(binning).ravel()
binning = {
'phi': {'edges': binning},
'lamb': {'edges': binning},
}
for kk in lk:
if type(binning[kk]) in ltypes0:
binning[kk] = {'nbins': int(binning[kk])}
elif type(binning[kk]) in ltypes1:
binning[kk] = {'edges': np.atleast_1d(binning[kk]).ravel()}
c0 = all([
all([k1 in ['edges', 'nbins'] for k1 in binning[k0].keys()])
for k0 in lk
])
c0 = (
c0
and all([
(
(
binning[k0].get('nbins') is None
or type(binning[k0].get('nbins')) in ltypes0
)
and (
binning[k0].get('edges') is None
or type(binning[k0].get('edges')) in ltypes1
)
)
for k0 in lk
])
)
if not c0:
raise Exception(msg)
# Check dict
for k0 in lk:
c0 = all([k1 in ['nbins', 'edges'] for k1 in binning[k0].keys()])
if not c0:
raise Exception(msg)
if binning[k0].get('nbins') is not None:
binning[k0]['nbins'] = int(binning[k0]['nbins'])
if binning[k0].get('edges') is None:
binning[k0]['edges'] = np.linspace(
domain[k0]['minmax'][0], domain[k0]['minmax'][1],
binning[k0]['nbins'] + 1,
endpoint=True,
)
else:
binning[k0]['edges'] = np.atleast_1d(
binning[k0]['edges']).ravel()
if binning[k0]['nbins'] != binning[k0]['edges'].size - 1:
raise Exception(msg)
elif binning[k0].get('bin_edges') is not None:
binning[k0]['edges'] = np.atleast_1d(binning[k0]['edges']).ravel()
binning[k0]['nbins'] = binning[k0]['edges'].size - 1
else:
raise Exception(msg)
# ------------
# safet checks
if np.any(~np.isfinite(binning[k0]['edges'])):
msg = (
f"Non-finite value in binning['{k0}']['edges']\n"
+ str(binning[k0]['edges'])
)
raise Exception(msg)
if not np.allclose(
binning[k0]['edges'],
np.unique(binning[k0]['edges']),
):
raise Exception(msg)
# Optional check vs nbsplines and deg
if nbsplines is not False:
if binning['phi']['nbins'] <= nbsplines:
msg = (
"The number of bins is too high:\n"
+ "\t- nbins = {}\n".format(binning['phi']['nbins'])
+ "\t- nbsplines = {}".format(nbsplines)
)
raise Exception(msg)
# --------------
# Check binning
for (dref, k0) in [(dlamb_ref, 'lamb'), (dphi_ref, 'phi')]:
if dref is not None:
di = np.mean(np.diff(binning[k0]['edges']))
if di < dref:
ni_rec = (
(domain[k0]['minmax'][1] - domain[k0]['minmax'][0]) / dref
)
msg = (
f"binning[{k0}] seems finer than the original!\n"
f"\t- estimated original step: {dref}\n"
f"\t- binning step: {di}\n"
f" => nb. of recommended steps: {ni_rec:5.1f}"
)
warnings.warn(msg)
return binning
def binning_2d_data(
lamb, phi, data,
indok=None,
indok_bool=None,
domain=None, binning=None,
nbsplines=None,
phi1d=None, lamb1d=None,
dataphi1d=None, datalamb1d=None,
):
# -------------------------
# Preliminary check on bins
dlamb_ref, dphi_ref = None, None
if lamb.ndim == 2:
indmid = int(lamb.shape[0]/2)
dlamb_ref = (np.max(lamb[indmid, :]) - np.min(lamb[indmid, :]))
dlamb_ref = dlamb_ref / lamb.shape[1]
indmid = int(lamb.shape[1]/2)
dphi_ref = (np.max(phi[:, indmid]) - np.min(phi[:, indmid]))
dphi_ref = dphi_ref / lamb.shape[0]
# ------------------
# Checkformat input
binning = _binning_check(
binning,
domain=domain,
dlamb_ref=dlamb_ref,
nbsplines=nbsplines,
)
nspect = data.shape[0]
if binning is False:
if phi1d is None:
phi1d_edges = np.linspace(
domain['phi']['minmax'][0], domain['phi']['minmax'][1], 100,
)
lamb1d_edges = np.linspace(
domain['lamb']['minmax'][0], domain['lamb']['minmax'][1], 100,
)
dataf = data.reshape((nspect, data.shape[1]*data.shape[2]))
dataphi1d = scpstats.binned_statistic(
phi.ravel(),
dataf,
statistic='sum',
bins=phi1d_edges,
)[0]
datalamb1d = scpstats.binned_statistic(
lamb.ravel(),
dataf,
statistic='sum',
bins=lamb1d_edges,
)[0]
phi1d = 0.5*(phi1d_edges[1:] + phi1d_edges[:-1])
lamb1d = 0.5*(lamb1d_edges[1:] + lamb1d_edges[:-1])
return (
lamb, phi, data, indok, binning,
phi1d, lamb1d, dataphi1d, datalamb1d,
)
else:
nphi = binning['phi']['nbins']
nlamb = binning['lamb']['nbins']
bins = (binning['phi']['edges'], binning['lamb']['edges'])
# ------------------
# Compute
databin = np.full((nspect, nphi, nlamb), np.nan)
nperbin = np.full((nspect, nphi, nlamb), np.nan)
indok_new = np.zeros((nspect, nphi, nlamb), dtype=np.int8)
for ii in range(nspect):
databin[ii, ...] = scpstats.binned_statistic_2d(
phi[indok_bool[ii, ...]],
lamb[indok_bool[ii, ...]],
data[ii, indok_bool[ii, ...]],
statistic='mean', # Beware: for valid S/N use sum!
bins=bins,
range=None,
expand_binnumbers=True,
)[0]
nperbin[ii, ...] = scpstats.binned_statistic_2d(
phi[indok_bool[ii, ...]],
lamb[indok_bool[ii, ...]],
np.ones((indok_bool[ii, ...].sum(),), dtype=int),
statistic='sum',
bins=bins,
range=None,
expand_binnumbers=True,
)[0]
binning['nperbin'] = nperbin
lamb1d = 0.5*(
binning['lamb']['edges'][1:] + binning['lamb']['edges'][:-1]
)
phi1d = 0.5*(
binning['phi']['edges'][1:] + binning['phi']['edges'][:-1]
)
lambbin = np.repeat(lamb1d[None, :], nphi, axis=0)
phibin = np.repeat(phi1d[:, None], nlamb, axis=1)
# reconstructing indok
indok_new[np.isnan(databin)] = -1
indok_new[nperbin == 0] = -4
# dataphi1d
dataphi1d = np.full(databin.shape[:2], np.nan)
indok = ~np.all(np.isnan(databin), axis=2)
dataphi1d[indok] = np.nanmean(databin[indok, :], axis=-1)
datalamb1d = np.full(databin.shape[::2], np.nan)
indok = ~np.all(np.isnan(databin), axis=1)
datalamb1d[indok] = (
np.nanmean(databin.swapaxes(1, 2)[indok, :], axis=-1)
+ np.nanstd(databin.swapaxes(1, 2)[indok, :], axis=-1)
)
return (
lambbin, phibin, databin, indok_new, binning,
phi1d, lamb1d, dataphi1d, datalamb1d,
)
###########################################################
###########################################################
#
# dprepare dict
#
###########################################################
###########################################################
def _get_subset_indices(subset, indlogical):
if subset is None:
subset = _SUBSET
if subset is False:
return indlogical
c0 = (
(
isinstance(subset, np.ndarray)
and subset.shape == indlogical.shape
and 'bool' in subset.dtype.name
)
or (
type(subset) in [int, float, np.int_, np.float_]
and subset >= 0
)
)
if not c0:
msg = ("subset must be either:\n"
+ "\t- an array of bool of shape: {}\n".format(indlogical.shape)
+ "\t- a positive int (nb. of ind. to keep from indlogical)\n"
+ "You provided:\n{}".format(subset))
raise Exception(msg)
if isinstance(subset, np.ndarray):
indlogical = subset[None, ...] & indlogical
else:
subset = np.random.default_rng().choice(
indlogical.sum(),
size=int(indlogical.sum() - subset),
replace=False,
shuffle=False,
)
for ii in range(indlogical.shape[0]):
ind = indlogical[ii, ...].nonzero()
indlogical[ii, ind[0][subset], ind[1][subset]] = False
return indlogical
def _extract_lphi_spectra(
data, phi, lamb,
lphi=None, lphi_tol=None,
databin=None, binning=None, nlamb=None,
):
""" Extra several 1d spectra from 2d image at lphi """
# --------------
# Check input
if lphi is None:
lphi = False
if lphi is False:
lphi_tol = False
if lphi is not False:
lphi = np.atleast_1d(lphi).astype(float).ravel()
lphi_tol = float(lphi_tol)
if lphi is False:
return False, False
nphi = len(lphi)
# --------------
# Compute non-trivial cases
if binning is False:
if nlamb is None:
nlamb = lamb.shape[1]
lphi_lamb = np.linspace(lamb.min(), lamb.max(), nlamb+1)
lphi_spectra = np.full((data.shape[0], lphi_lamb.size-1, nphi), np.nan)
for ii in range(nphi):
indphi = np.abs(phi - lphi[ii]) < lphi_tol
lphi_spectra[:, ii, :] = scpstats.binned_statistic(
lamb[indphi], data[:, indphi], bins=lphi_lamb,
statistic='mean', range=None,
)[0]
else:
lphi_lamb = 0.5*(
binning['lamb']['edges'][1:] + binning['lamb']['edges'][:-1]
)
lphi_phi = 0.5*(
binning['phi']['edges'][1:] + binning['phi']['edges'][:-1]
)
lphi_spectra = np.full((data.shape[0], nphi, lphi_lamb.size), np.nan)
lphi_spectra1 = np.full((data.shape[0], nphi, lphi_lamb.size), np.nan)
for ii in range(nphi):
datai = databin[:, np.abs(lphi_phi - lphi[ii]) < lphi_tol, :]
iok = np.any(~np.isnan(datai), axis=1)
for jj in range(datai.shape[0]):
if np.any(iok[jj, :]):
lphi_spectra[jj, ii, iok[jj, :]] = np.nanmean(
datai[jj, :, iok[jj, :]],
axis=1,
)
return lphi_spectra, lphi_lamb
def _checkformat_possubset(pos=None, subset=None):
if pos is None:
pos = _POS
c0 = isinstance(pos, bool) or type(pos) in _LTYPES
if not c0:
msg = ("Arg pos must be either:\n"
+ "\t- False: no positivity constraints\n"
+ "\t- True: all negative values are set to nan\n"
+ "\t- float: all negative values are set to pos")
raise Exception(msg)
if subset is None:
subset = _SUBSET
return pos, subset
def multigausfit1d_from_dlines_prepare(
data=None, lamb=None,
mask=None, domain=None,
pos=None, subset=None,
update_domain=None,
):
# --------------
# Check input
pos, subset = _checkformat_possubset(pos=pos, subset=subset)
# Check shape of data (multiple time slices possible)
lamb, data, mask = _checkformat_data_fit12d_dlines(
data, lamb, mask=mask,
)
# --------------
# Use valid data only and optionally restrict lamb
indok = np.zeros(data.shape, dtype=np.int8)
if mask is not None:
indok[:, ~mask] = -1
inddomain, domain = apply_domain(lamb, domain=domain)
if mask is not None:
indok[:, (~inddomain) & mask] = -2
else:
indok[:, ~inddomain] = -2
# Optional positivity constraint
if pos is not False:
if pos is True:
data[data < 0.] = np.nan
else:
data[data < 0.] = pos
indok[(indok == 0) & np.isnan(data)] = -3
# Recompute domain
indok_bool = indok == 0
if update_domain is None:
update_domain = bool(np.any(np.isinf(domain['lamb']['minmax'])))
if update_domain is True:
domain['lamb']['minmax'] = [
np.nanmin(lamb[np.any(indok_bool, axis=0)]),
np.nanmax(lamb[np.any(indok_bool, axis=0)]),
]
# --------------
# Optionally fit only on subset
# randomly pick subset indices (replace=False => no duplicates)
# indok = _get_subset_indices(subset, indok)
if np.any(np.isnan(data[indok_bool])):
msg = (
"Some NaNs in data not caught by indok!"
)
raise Exception(msg)
if np.sum(indok_bool) == 0:
msg = "There does not seem to be any usable data (no indok)"
raise Exception(msg)
# --------------
# Return
dprepare = {
'data': data,
'lamb': lamb,
'domain': domain,
'indok': indok,
'indok_bool': indok_bool,
'dindok': dict(_DINDOK),
'pos': pos,
'subset': subset,
}
return dprepare
def multigausfit2d_from_dlines_prepare(
data=None, lamb=None, phi=None,
mask=None, domain=None,
update_domain=None,
pos=None, binning=None,
nbsplines=None, deg=None, subset=None,
nxi=None, nxj=None,
lphi=None, lphi_tol=None,
):
# --------------
# Check input
pos, subset = _checkformat_possubset(pos=pos, subset=subset)
# Check shape of data (multiple time slices possible)
(
lamb, phi, data, mask,
phi1d, lamb1d, dataphi1d, datalamb1d,
) = _checkformat_data_fit12d_dlines(
data, lamb, phi,
nxi=nxi, nxj=nxj, mask=mask, is2d=True,
)
# --------------
# Use valid data only and optionally restrict lamb / phi
indok = np.zeros(data.shape, dtype=np.int8)
if mask is not None:
indok[:, ~mask] = -1
inddomain, domain = apply_domain(lamb, phi, domain=domain)
if mask is not None:
indok[:, (~inddomain) & mask] = -2
else:
indok[:, ~inddomain] = -2
# Optional positivity constraint
if pos is not False:
if pos is True:
data[data < 0.] = np.nan
else:
data[data < 0.] = pos
# Introduce time-dependence (useful for valid)
indok[(indok == 0) & np.isnan(data)] = -3
# Recompute domain
indok_bool = indok == 0
if not np.any(indok_bool):
msg = "No valid point in data!"
raise Exception(msg)
if update_domain is None:
update_domain = bool(
np.any(np.isinf(domain['lamb']['minmax']))
or np.any(np.isinf(domain['phi']['minmax']))
)
if update_domain is True:
domain['lamb']['minmax'] = [
np.nanmin(lamb[np.any(indok_bool, axis=0)]),
np.nanmax(lamb[np.any(indok_bool, axis=0)]),
]
domain['phi']['minmax'] = [
np.nanmin(phi[np.any(indok_bool, axis=0)]),
np.nanmax(phi[np.any(indok_bool, axis=0)]),
]
# --------------
# Optionnal 2d binning
(
lambbin, phibin, databin, indok, binning,
phi1d, lamb1d, dataphi1d, datalamb1d,
) = binning_2d_data(
lamb, phi, data,
indok=indok,
indok_bool=indok_bool,
binning=binning,
domain=domain,
nbsplines=nbsplines,
phi1d=phi1d, lamb1d=lamb1d,
dataphi1d=dataphi1d, datalamb1d=datalamb1d,
)
indok_bool = indok == 0
# --------------
# Optionally fit only on subset
# randomly pick subset indices (replace=False => no duplicates)
# indok_bool = _get_subset_indices(subset, indok == 0)
# --------------
# Optionally extract 1d spectra at lphi
lphi_spectra, lphi_lamb = _extract_lphi_spectra(
data, phi, lamb,
lphi, lphi_tol,
databin=databin,
binning=binning,
)
if np.sum(indok_bool) == 0:
msg = "There does not seem to be any usable data (no indok)"
raise Exception(msg)
# --------------
# Return
dprepare = {
'data': databin, 'lamb': lambbin, 'phi': phibin,
'domain': domain, 'binning': binning,
'indok': indok, 'indok_bool': indok_bool, 'dindok': dict(_DINDOK),
'pos': pos, 'subset': subset, 'nxi': nxi, 'nxj': nxj,
'lphi': lphi, 'lphi_tol': lphi_tol,
'lphi_spectra': lphi_spectra, 'lphi_lamb': lphi_lamb,
'phi1d': phi1d, 'dataphi1d': dataphi1d,
'lamb1d': lamb1d, 'datalamb1d': datalamb1d,
}
return dprepare
def multigausfit2d_from_dlines_dbsplines(
knots=None, deg=None, nbsplines=None,
phimin=None, phimax=None,
symmetryaxis=None,
):
# Check / format input
if nbsplines is None:
nbsplines = _NBSPLINES
c0 = [nbsplines is False, isinstance(nbsplines, int)]
if not any(c0):
msg = "nbsplines must be a int (degree of bsplines to be used!)"
raise Exception(msg)
if nbsplines is False:
lk = ['knots', 'knots_mult', 'nknotsperbs', 'ptsx0', 'nbs', 'deg']
return dict.fromkeys(lk, False)
if deg is None:
deg = _DEG
if not (isinstance(deg, int) and deg <= 3):
msg = "deg must be a int <= 3 (the degree of the bsplines to be used!)"
raise Exception(msg)
if symmetryaxis is None:
symmetryaxis = False
if knots is None:
if phimin is None or phimax is None:
msg = "Please provide phimin and phimax if knots is not provided!"
raise Exception(msg)
phimargin = (phimax - phimin)/1000.
if symmetryaxis is False:
knots = np.linspace(
phimin - phimargin,
phimax + phimargin,
nbsplines + 1 - deg,
)
else:
phi2max = np.max(
np.abs(np.r_[phimin, phimax][None, :] - symmetryaxis[:, None])
)
knots = np.linspace(0, phi2max + phimargin, nbsplines + 1 - deg)
if not np.allclose(knots, np.unique(knots)):
msg = "knots must be a vector of unique values!"
raise Exception(msg)
# Get knots for scipy (i.e.: with multiplicity)
if deg > 0:
knots_mult = np.r_[[knots[0]]*deg, knots, [knots[-1]]*deg]
else:
knots_mult = knots
nknotsperbs = 2 + deg
nbs = knots.size - 1 + deg
assert nbs == knots_mult.size - 1 - deg
if deg == 0:
ptsx0 = 0.5*(knots[:-1] + knots[1:])
elif deg == 1:
ptsx0 = knots
elif deg == 2:
num = (knots_mult[3:]*knots_mult[2:-1]
- knots_mult[1:-2]*knots_mult[:-3])
denom = (knots_mult[3:] + knots_mult[2:-1]
- knots_mult[1:-2] - knots_mult[:-3])
ptsx0 = num / denom
else:
# To be derived analytically for more accuracy
ptsx0 = np.r_[
knots[0],
np.mean(knots[:2]),
knots[1:-1],
np.mean(knots[-2:]),
knots[-1],
]
msg = ("degree 3 not fully implemented yet!"
+ "Approximate values for maxima positions")
warnings.warn(msg)
assert ptsx0.size == nbs
dbsplines = {
'knots': knots, 'knots_mult': knots_mult,
'nknotsperbs': nknotsperbs, 'ptsx0': ptsx0,
'nbs': nbs, 'deg': deg,
}
return dbsplines
###########################################################
###########################################################
#
# dvalid dict (S/N ratio)
#
###########################################################
###########################################################
def _dvalid_checkfocus_errmsg(focus=None, focus_half_width=None,
lines_keys=None):
msg = ("Please provide focus as:\n"
+ "\t- str: the key of an available spectral line:\n"
+ "\t\t{}\n".format(lines_keys)
+ "\t- float: a wavelength value\n"
+ "\t- a list / tuple / flat np.ndarray of such\n"
+ "\t- a np.array of shape (2, N) or (N, 2) (focus + halfwidth)"
+ " You provided:\n"
+ "{}\n\n".format(focus)
+ "Please provide focus_half_width as:\n"
+ "\t- float: a unique wavelength value for all focus\n"
+ "\t- a list / tuple / flat np.ndarray of such\n"
+ " You provided:\n"
+ "{}".format(focus_half_width))
return msg
def _dvalid_checkfocus(
focus=None,
focus_half_width=None,
lines_keys=None,
lines_lamb=None,
lamb=None,
):
""" Check the provided focus is properly formatted and convert it
focus specifies the wavelength range of interest in which S/N is evaluated
It can be provided as:
- a spectral line key (or list of such)
- a wavelength (or list of such)
For each wavelength, a spectral range centered on it, is defined using
the provided focus_half_width
The focus_half_width can be a unique value applied to all or a list of
values of the same length as focus.
focus is then return as a (n, 2) array where:
each line gives a central wavelength and halfwidth of interest
"""
if focus in [None, False]:
return False
# Check focus and transform to array of floats
if isinstance(focus, tuple([str] + _LTYPES)):
focus = [focus]
lc = [
isinstance(focus, (list, tuple, np.ndarray))
and all([
(isinstance(ff, tuple(_LTYPES)) and ff > 0.)
or (isinstance(ff, str) and ff in lines_keys)
for ff in focus
]),
isinstance(focus, (list, tuple, np.ndarray))
and all([
isinstance(ff, (list, tuple, np.ndarray))
for ff in focus
])
and np.asarray(focus).ndim == 2
and 2 in np.asarray(focus).shape
and np.all(np.isfinite(focus))
and np.all(np.asarray(focus) > 0)
]
if not any(lc):
msg = _dvalid_checkfocus_errmsg(
focus, focus_half_width, lines_keys,
)
raise Exception(msg)
# Centered on lines
if lc[0]:
focus = np.array([
lines_lamb[(lines_keys == ff).nonzero()[0][0]]
if isinstance(ff, str) else ff for ff in focus
])
# Check focus_half_width and transform to array of floats
if focus_half_width is None:
focus_half_width = (np.nanmax(lamb) - np.nanmin(lamb))/10.
lc0 = [
type(focus_half_width) in _LTYPES,
(
type(focus_half_width) in [list, tuple, np.ndarray]
and len(focus_half_width) == focus.size
and all([type(fhw) in _LTYPES for fhw in focus_half_width])
)
]
if not any(lc0):
msg = _dvalid_checkfocus_errmsg(
focus, focus_half_width, lines_keys,
)
raise Exception(msg)
if lc0[0] is True:
focus_half_width = np.full((focus.size,), focus_half_width)
focus = np.array([focus, np.r_[focus_half_width]]).T
elif lc[1]:
focus = np.asarray(focus, dtype=float)
if focus.shape[1] != 2:
focus = focus.T
return focus
def fit12d_dvalid(
data=None, lamb=None, phi=None,
indok_bool=None, binning=None,
valid_nsigma=None, valid_fraction=None,
focus=None, focus_half_width=None,
lines_keys=None, lines_lamb=None, dphimin=None,
nbs=None, deg=None,
knots=None, knots_mult=None, nknotsperbs=None,
return_fract=None,
):
""" Return a dict of valid time steps and phi indices
data points are considered valid if there signal is sufficient:
np.sqrt(data) >= valid_nsigma
data is supposed to be provided in counts (or photons).. TBC!!!
"""
# Check inputs
if valid_nsigma is None:
valid_nsigma = _VALID_NSIGMA
if valid_fraction is None:
valid_fraction = _VALID_FRACTION
if binning is None:
binning = False
if dphimin is None:
dphimin = 0.
if return_fract is None:
return_fract = False
data2d = data.ndim == 3
nspect = data.shape[0]
focus = _dvalid_checkfocus(
focus=focus,
focus_half_width=focus_half_width,
lines_keys=lines_keys,
lines_lamb=lines_lamb,
lamb=lamb,
)
# Get indices of pts with enough signal
ind = np.zeros(data.shape, dtype=bool)
isafe = np.isfinite(data)
isafe[isafe] = data[isafe] >= 0.
if indok_bool is not None:
isafe &= indok_bool
# Ok with and w/o binning if data provided as counts
if binning is False:
ind[isafe] = np.sqrt(data[isafe]) > valid_nsigma
else:
# For S/N in binning, if counts => sum = mean * nbperbin
ind[isafe] = (
np.sqrt(data[isafe] * binning['nperbin'][isafe]) > valid_nsigma
)
# Derive indt and optionally dphi and indknots
indbs, ldphi = False, False
if focus is False:
lambok = np.ones(tuple(np.r_[lamb.shape, 1]), dtype=bool)
indall = ind[..., None]
else:
# TBC
lambok = np.rollaxis(
np.array([np.abs(lamb - ff[0]) < ff[1] for ff in focus]),
0,
lamb.ndim + 1,
)
indall = ind[..., None] & lambok[None, ...]
nfocus = lambok.shape[-1]
if data2d is True:
# Code ok with and without binning :-)
# Get knots intervals that are ok
fract = np.full((nspect, knots.size-1, nfocus), np.nan)
for ii in range(knots.size - 1):
iphi = (phi >= knots[ii]) & (phi < knots[ii + 1])
fract[:, ii, :] = (
np.sum(np.sum(indall & iphi[None, ..., None],
axis=1), axis=1)
/ np.sum(np.sum(iphi[..., None] & lambok,
axis=0), axis=0)
)
indknots = np.all(fract > valid_fraction, axis=2)
# Deduce ldphi
ldphi = [[] for ii in range(nspect)]
for ii in range(nspect):
for jj in range(indknots.shape[1]):
if indknots[ii, jj]:
if jj == 0 or not indknots[ii, jj-1]:
ldphi[ii].append([knots[jj]])
if jj == indknots.shape[1] - 1:
ldphi[ii][-1].append(knots[jj+1])
else:
if jj > 0 and indknots[ii, jj-1]:
ldphi[ii][-1].append(knots[jj])
# Safety check
assert all([
all([len(dd) == 2 and dd[0] < dd[1] for dd in ldphi[ii]])
for ii in range(nspect)
])
# Deduce indbs that are ok
nintpbs = nknotsperbs - 1
indbs = np.zeros((nspect, nbs), dtype=bool)
for ii in range(nbs):
ibk = np.arange(max(0, ii-(nintpbs-1)), min(knots.size-1, ii+1))
indbs[:, ii] = np.any(indknots[:, ibk], axis=1)
assert np.all(
(np.sum(indbs, axis=1) == 0) | (np.sum(indbs, axis=1) >= deg + 1)
)
# Deduce indt
indt = np.any(indbs, axis=1)
else:
# 1d spectra
if focus is False:
fract = ind.sum(axis=-1) / ind.shape[1]
indt = fract > valid_fraction
else:
fract = np.sum(indall, axis=1) / lambok.sum(axis=0)[None, :]
indt = np.all(fract > valid_fraction, axis=1)
# Optional debug
if focus is not False and False:
indt_debug, ifocus = 40, 1
if data2d is True:
indall2 = indall.astype(int)
indall2[:, lambok] = 1
indall2[ind[..., None] & lambok[None, ...]] = 2
plt.figure()
plt.imshow(indall2[indt_debug, :, :, ifocus].T, origin='lower')
else:
plt.figure()
plt.plot(lamb[~indall[indt_debug, :, ifocus]],
data[indt_debug, ~indall[indt_debug, :, ifocus]], '.k',
lamb[indall[indt_debug, :, ifocus]],
data[indt_debug, indall[indt_debug, :, ifocus]], '.r')
plt.axvline(focus[ifocus, 0], ls='--', c='k')
if not np.any(indt):
msg = (
"\nThere is no valid time step with the provided constraints:\n"
+ "\t- valid_nsigma = {}\n".format(valid_nsigma)
+ "\t- valid_fraction = {}\n".format(valid_fraction)
+ "\t- focus = {}\n".format(focus)
+ f"\t- fract max, mean = {np.max(fract), np.mean(fract)}\n"
+ "\t- fract = {}\n".format(fract)
)
raise Exception(msg)
# return
dvalid = {
'indt': indt, 'ldphi': ldphi, 'indbs': indbs, 'ind': ind,
'focus': focus, 'valid_fraction': valid_fraction,
'valid_nsigma': valid_nsigma,
}
if return_fract is True:
dvalid['fract'] = fract
return dvalid
###########################################################
###########################################################
#
# dlines dict (lines vs domain)
#
###########################################################
###########################################################
def _checkformat_dlines(dlines=None, domain=None):
if dlines is None:
dlines = False
if not isinstance(dlines, dict):
msg = "Arg dlines must be a dict!"
raise Exception(msg)
lc = [
(k0, type(v0)) for k0, v0 in dlines.items()
if not (
isinstance(k0, str)
and isinstance(v0, dict)
and 'lambda0' in v0.keys()
and (
type(v0['lambda0']) in _LTYPES
or (
isinstance(v0['lambda0'], np.ndarray)
and v0['lambda0'].size == 1
)
)
)
]
if len(lc) > 0:
lc = ["\t- {}: {}".format(*cc) for cc in lc]
msg = (
"Arg dlines must be a dict of the form:\n"
+ "\t{'line0': {'lambda0': float},\n"
+ "\t 'line1': {'lambda0': float},\n"
+ "\t ...\n"
+ "\t 'lineN': {'lambda0': float}}\n"
+ " You provided:\n{}".format('\n'.join(lc))
)
raise Exception(msg)
# Select relevant lines (keys, lamb)
lines_keys = np.array([k0 for k0 in dlines.keys()])
lines_lamb = np.array([float(dlines[k0]['lambda0']) for k0 in lines_keys])
if domain not in [None, False]:
ind = np.zeros((len(lines_keys),), dtype=bool)
for ss in domain['lamb']['spec']:
if isinstance(ss, (list, np.ndarray)):
ind[(lines_lamb >= ss[0]) & (lines_lamb < ss[1])] = True
for ss in domain['lamb']['spec']:
if isinstance(ss, tuple):
ind[(lines_lamb >= ss[0]) & (lines_lamb < ss[1])] = False
lines_keys = lines_keys[ind]
lines_lamb = lines_lamb[ind]
inds = np.argsort(lines_lamb)
lines_keys, lines_lamb = lines_keys[inds], lines_lamb[inds]
nlines = lines_lamb.size
dlines = {k0: dict(dlines[k0]) for k0 in lines_keys}
# Warning if no lines left
if len(lines_keys) == 0:
msg = "There seems to be no lines left!"
warnings.warn(msg)
return dlines, lines_keys, lines_lamb
###########################################################
###########################################################
#
# dinput dict (lines + spectral constraints)
#
###########################################################
###########################################################
def fit1d_dinput(
dlines=None, dconstraints=None, dconstants=None, dprepare=None,
data=None, lamb=None, mask=None,
domain=None, pos=None, subset=None,
update_domain=None,
same_spectrum=None, nspect=None, same_spectrum_dlamb=None,
focus=None, valid_fraction=None, valid_nsigma=None, focus_half_width=None,
valid_return_fract=None,
dscales=None, dx0=None, dbounds=None,
defconst=_DCONSTRAINTS,
):
""" Check and format a dict of inputs to be fed to fit1d()
This dict will contain all information relevant for solving the fit:
- dlines: dict of lines (with 'lambda0': wavelength at rest)
- lamb: vector of wavelength of the experimental spectrum
- data: experimental spectrum, possibly 2d (time-varying)
- dconstraints: dict of constraints on lines (amp, width, shift)
- pos: bool, consider only positive data (False => replace <0 with nan)
- domain:
- mask:
- subset:
- same_spectrum:
- focus:
"""
# ------------------------
# Check / format dprepare
# ------------------------
if dprepare is None:
dprepare = multigausfit1d_from_dlines_prepare(
data=data, lamb=lamb,
mask=mask, domain=domain,
pos=pos, subset=subset,
update_domain=update_domain,
)
# ------------------------
# Check / format dlines
# ------------------------
dlines, lines_keys, lines_lamb = _checkformat_dlines(
dlines=dlines,
domain=dprepare['domain'],
)
nlines = lines_lamb.size
# Check same_spectrum
if same_spectrum is None:
same_spectrum = _SAME_SPECTRUM
if same_spectrum is True:
if type(nspect) not in [int, np.int]:
msg = "Please provide nspect if same_spectrum = True"
raise Exception(msg)
if same_spectrum_dlamb is None:
same_spectrum_dlamb = min(
2*np.diff(dprepare['domain']['lamb']['minmax']),
dprepare['domain']['lamb']['minmax'][0],
)
# ------------------------
# Check / format dconstraints
# ------------------------
dconstraints = _checkformat_dconstraints(
dconstraints=dconstraints, defconst=defconst,
)
dinput = {}
# ------------------------
# Check / format double
# ------------------------
_dconstraints_double(dinput, dconstraints, defconst=defconst)
# ------------------------
# Check / format width, shift, amp (groups with possible ratio)
# ------------------------
for k0 in ['amp', 'width', 'shift']:
dinput[k0] = _width_shift_amp(
dconstraints.get(k0, defconst[k0]),
dconstants=dconstants,
keys=lines_keys, nlines=nlines,
dlines=dlines, k0=k0,
)
# ------------------------
# add mz, symb, ION, keys, lamb
# ------------------------
mz = np.array([dlines[k0].get('m', np.nan) for k0 in lines_keys])
symb = np.array([dlines[k0].get('symbol', k0) for k0 in lines_keys])
ion = np.array([dlines[k0].get('ion', '?') for k0 in lines_keys])
# ------------------------
# same_spectrum
# ------------------------
if same_spectrum is True:
keysadd = np.array([[kk+'_bis{:04.0f}'.format(ii) for kk in keys]
for ii in range(1, nspect)]).ravel()
lines_lamb = (
same_spectrum_dlamb*np.arange(0, nspect)[:, None]
+ lines_lamb[None, :]
)
keys = np.r_[keys, keysadd]
for k0 in _DORDER:
# Add other lines to original group
keyk = dinput[k0]['keys']
offset = np.tile(dinput[k0]['offset'], nspect)
if k0 == 'shift':
ind = np.tile(dinput[k0]['ind'], (1, nspect))
coefs = (
dinput[k0]['coefs']
* lines_lamb[0, :] / lines_lamb
).ravel()
else:
coefs = np.tile(dinput[k0]['coefs'], nspect)
keysadd = np.array([
[kk+'_bis{:04.0f}'.format(ii) for kk in keyk]
for ii in range(1, nspect)
]).ravel()
ind = np.zeros((keyk.size*nspect, nlines*nspect))
for ii in range(nspect):
i0, i1 = ii*keyk.size, (ii+1)*keyk.size
j0, j1 = ii*nlines, (ii+1)*nlines
ind[i0:i1, j0:j1] = dinput[k0]['ind']
keyk = np.r_[keyk, keysadd]
dinput[k0]['keys'] = keyk
dinput[k0]['ind'] = ind
dinput[k0]['coefs'] = coefs
dinput[k0]['offset'] = offset
nlines *= nspect
lines_lamb = lines_lamb.ravel()
# update mz, symb, ion
mz = np.tile(mz, nspect)
symb = np.tile(symb, nspect)
ion = np.tile(ion, nspect)
# ------------------------
# add lines and properties
# ------------------------
dinput['keys'] = lines_keys
dinput['lines'] = lines_lamb
dinput['nlines'] = nlines
dinput['mz'] = mz
dinput['symb'] = symb
dinput['ion'] = ion
dinput['same_spectrum'] = same_spectrum
if same_spectrum is True:
dinput['same_spectrum_nspect'] = nspect
dinput['same_spectrum_dlamb'] = same_spectrum_dlamb
else:
dinput['same_spectrum_nspect'] = False
dinput['same_spectrum_dlamb'] = False
# ------------------------
# S/N threshold indices
# ------------------------
dinput['valid'] = fit12d_dvalid(
data=dprepare['data'],
lamb=dprepare['lamb'],
indok_bool=dprepare['indok_bool'],
valid_nsigma=valid_nsigma,
valid_fraction=valid_fraction,
focus=focus, focus_half_width=focus_half_width,
lines_keys=lines_keys, lines_lamb=lines_lamb,
return_fract=valid_return_fract,
)
# Update with dprepare
dinput['dprepare'] = dict(dprepare)
# Add dind
dinput['dind'] = multigausfit12d_from_dlines_ind(dinput)
# Add dscales, dx0 and dbounds
dinput['dscales'] = fit12d_dscales(dscales=dscales, dinput=dinput)
dinput['dbounds'] = fit12d_dbounds(dbounds=dbounds, dinput=dinput)
dinput['dx0'] = fit12d_dx0(dx0=dx0, dinput=dinput)
dinput['dconstants'] = fit12d_dconstants(
dconstants=dconstants, dinput=dinput,
)
# add lambmin for bck
dinput['lambmin_bck'] = np.min(dprepare['lamb'])
return dinput
def fit2d_dinput(
dlines=None, dconstraints=None, dconstants=None, dprepare=None,
deg=None, nbsplines=None, knots=None,
data=None, lamb=None, phi=None, mask=None,
domain=None, pos=None, subset=None, binning=None, cent_fraction=None,
update_domain=None,
focus=None, valid_fraction=None, valid_nsigma=None, focus_half_width=None,
valid_return_fract=None,
dscales=None, dx0=None, dbounds=None,
nxi=None, nxj=None,
lphi=None, lphi_tol=None,
defconst=_DCONSTRAINTS,
):
""" Check and format a dict of inputs to be fed to fit2d()
This dict will contain all information relevant for solving the fit:
- dlines: dict of lines (with 'lambda0': wavelength at rest)
- lamb: vector of wavelength of the experimental spectrum
- data: experimental spectrum, possibly 2d (time-varying)
- dconstraints: dict of constraints on lines (amp, width, shift)
- pos: bool, consider only positive data (False => replace <0 with nan)
- domain:
- mask:
- subset:
- same_spectrum:
- focus:
"""
# ------------------------
# Check / format dprepare
# ------------------------
if dprepare is None:
dprepare = multigausfit2d_from_dlines_prepare(
data=data, lamb=lamb, phi=phi,
mask=mask, domain=domain,
pos=pos, subset=subset, binning=binning,
update_domain=update_domain,
nbsplines=nbsplines, deg=deg,
nxi=nxi, nxj=nxj,
lphi=None, lphi_tol=None,
)
# ------------------------
# Check / format dlines
# ------------------------
dlines, lines_keys, lines_lamb = _checkformat_dlines(
dlines=dlines,
domain=dprepare['domain'],
)
nlines = lines_lamb.size
# ------------------------
# Check / format dconstraints
# ------------------------
dconstraints = _checkformat_dconstraints(
dconstraints=dconstraints, defconst=defconst,
)
dinput = {}
# ------------------------
# Check / format symmetry
# ------------------------
_dconstraints_symmetry(
dinput, dprepare=dprepare, symmetry=dconstraints.get('symmetry'),
cent_fraction=cent_fraction, defconst=defconst,
)
# ------------------------
# Check / format double (spectral line doubling)
# ------------------------
_dconstraints_double(dinput, dconstraints, defconst=defconst)
# ------------------------
# Check / format width, shift, amp (groups with posssible ratio)
# ------------------------
for k0 in ['amp', 'width', 'shift']:
dinput[k0] = _width_shift_amp(
dconstraints.get(k0, defconst[k0]),
dconstants=dconstants,
keys=lines_keys, nlines=nlines,
dlines=dlines, k0=k0,
)
# ------------------------
# add mz, symb, ION, keys, lamb
# ------------------------
mz = np.array([dlines[k0].get('m', np.nan) for k0 in lines_keys])
symb = np.array([dlines[k0].get('symbol', k0) for k0 in lines_keys])
ion = np.array([dlines[k0].get('ION', '?') for k0 in lines_keys])
# ------------------------
# add lines and properties
# ------------------------
dinput['keys'] = lines_keys
dinput['lines'] = lines_lamb
dinput['nlines'] = nlines
dinput['mz'] = mz
dinput['symb'] = symb
dinput['ion'] = ion
# ------------------------
# Get dict of bsplines
# ------------------------
dinput.update(multigausfit2d_from_dlines_dbsplines(
knots=knots, deg=deg, nbsplines=nbsplines,
phimin=dprepare['domain']['phi']['minmax'][0],
phimax=dprepare['domain']['phi']['minmax'][1],
symmetryaxis=dinput.get('symmetry_axis')
))
# ------------------------
# S/N threshold indices
# ------------------------
dinput['valid'] = fit12d_dvalid(
data=dprepare['data'],
lamb=dprepare['lamb'],
phi=dprepare['phi'],
binning=dprepare['binning'],
indok_bool=dprepare['indok_bool'],
valid_nsigma=valid_nsigma,
valid_fraction=valid_fraction,
focus=focus, focus_half_width=focus_half_width,
lines_keys=lines_keys, lines_lamb=lines_lamb,
nbs=dinput['nbs'],
deg=dinput['deg'],
knots=dinput['knots'],
knots_mult=dinput['knots_mult'],
nknotsperbs=dinput['nknotsperbs'],
return_fract=valid_return_fract,
)
# Update with dprepare
dinput['dprepare'] = dict(dprepare)
# Add dind
dinput['dind'] = multigausfit12d_from_dlines_ind(dinput)
# Add dscales, dx0 and dbounds
dinput['dscales'] = fit12d_dscales(dscales=dscales, dinput=dinput)
dinput['dbounds'] = fit12d_dbounds(dbounds=dbounds, dinput=dinput)
dinput['dx0'] = fit12d_dx0(dx0=dx0, dinput=dinput)
dinput['dconstants'] = fit12d_dconstants(
dconstants=dconstants, dinput=dinput,
)
# Update indok with non-valid phi
# non-valid = ok but out of dphi
for ii in range(dinput['dprepare']['indok'].shape[0]):
iphino = dinput['dprepare']['indok'][ii, ...] == 0
for jj in range(len(dinput['valid']['ldphi'][ii])):
iphino &= (
(
dinput['dprepare']['phi']
< dinput['valid']['ldphi'][ii][jj][0]
)
| (
dinput['dprepare']['phi']
>= dinput['valid']['ldphi'][ii][jj][1]
)
)
# valid, but excluded (out of dphi)
iphi = (
(dinput['dprepare']['indok'][ii, ...] == 0)
& (dinput['valid']['ind'][ii, ...])
& (iphino)
)
dinput['dprepare']['indok'][ii, iphi] = -5
# non-valid, included (in dphi)
iphi = (
(dinput['dprepare']['indok'][ii, ...] == 0)
& (~dinput['valid']['ind'][ii, ...])
& (~iphino)
)
dinput['dprepare']['indok'][ii, iphi] = -6
# non-valid, excluded (out of dphi)
iphi = (
(dinput['dprepare']['indok'][ii, ...] == 0)
& (~dinput['valid']['ind'][ii, ...])
& (iphino)
)
dinput['dprepare']['indok'][ii, iphi] = -7
# indok_bool True if indok == 0 or -5 (because ...)
dinput['dprepare']['indok_bool'] = (
(dinput['dprepare']['indok'] == 0)
| (dinput['dprepare']['indok'] == -6)
)
# add lambmin for bck
dinput['lambmin_bck'] = np.min(dinput['dprepare']['lamb'])
return dinput
###########################################################
###########################################################
#
# dind dict (indices storing for fast access)
#
###########################################################
###########################################################
def multigausfit12d_from_dlines_ind(dinput=None):
""" Return the indices of quantities in x to compute y """
# indices
# General shape: [bck, amp, widths, shifts]
# If double [..., double_shift, double_ratio]
# Except for bck, all indices should render nlines (2*nlines if double)
nbs = dinput.get('nbs', 1)
dind = {
'bck_amp': {'x': np.arange(0, nbs)[:, None]},
'bck_rate': {'x': np.arange(nbs, 2*nbs)[:, None]},
'dshift': None,
'dratio': None,
}
nn = dind['bck_amp']['x'].size + dind['bck_rate']['x'].size
inddratio, inddshift = None, None
for k0 in _DORDER:
# l0bs0, l0bs1, ..., l0bsN, l1bs0, ...., lnbsN
ind = dinput[k0]['ind']
lnl = np.sum(ind, axis=1).astype(int)
dind[k0] = {
'x': (
nn
+ nbs*np.arange(0, ind.shape[0])[None, :]
+ np.arange(0, nbs)[:, None]
),
'lines': (
nn
+ nbs* | np.argmax(ind, axis=0) | numpy.argmax |
"""
Utilities for reading and writing parameters files to perform FFD
geometrical morphing.
"""
try:
import configparser as configparser
except ImportError:
import ConfigParser as configparser
import os
import numpy as np
from OCC.Bnd import Bnd_Box
from OCC.BRepBndLib import brepbndlib_Add
from OCC.BRepMesh import BRepMesh_IncrementalMesh
import vtk
import pygem.affine as at
class FFDParameters(object):
"""
Class that handles the Free Form Deformation parameters in terms of FFD
bounding box and weight of the FFD control points.
:param list n_control_points: number of control points in the x, y, and z
direction. If not provided it is set to [2, 2, 2].
:cvar numpy.ndarray box_length: dimension of the FFD bounding box, in the
x, y and z direction (local coordinate system).
:cvar numpy.ndarray box_origin: the x, y and z coordinates of the origin of
the FFD bounding box.
:cvar numpy.ndarray rot_angle: rotation angle around x, y and z axis of the
FFD bounding box.
:cvar numpy.ndarray n_control_points: the number of control points in the
x, y, and z direction.
:cvar numpy.ndarray array_mu_x: collects the displacements (weights) along
x, normalized with the box length x.
:cvar numpy.ndarray array_mu_y: collects the displacements (weights) along
y, normalized with the box length y.
:cvar numpy.ndarray array_mu_z: collects the displacements (weights) along
z, normalized with the box length z.
:Example: from file
>>> import pygem.params as ffdp
>>>
>>> # Reading an existing file
>>> params1 = ffdp.FFDParameters()
>>> params1.read_parameters(
>>> filename='tests/test_datasets/parameters_test_ffd_identity.prm')
>>>
>>> # Creating a default parameters file with the right dimensions (if the
>>> # file does not exists it is created with that name). So it is possible
>>> # to manually edit it and read it again.
>>> params2 = ffdp.FFDParameters(n_control_points=[2, 3, 2])
>>> params2.read_parameters(filename='parameters_test.prm')
>>>
>>> # Creating bounding box of the given shape
>>> from OCC.IGESControl import IGESControl_Reader
>>> params3 = ffdp.FFDParameters()
>>> reader = IGESControl_Reader()
>>> reader.ReadFile('tests/test_datasets/test_pipe.igs')
>>> reader.TransferRoots()
>>> shape = reader.Shape()
>>> params3.build_bounding_box(shape)
.. note::
Four vertex (non coplanar) are sufficient to uniquely identify a
parallelepiped.
If the four vertex are coplanar, an assert is thrown when
`affine_points_fit` is used.
"""
def __init__(self, n_control_points=None):
self.conversion_unit = 1.
self.box_length = np.array([1., 1., 1.])
self.box_origin = np.array([0., 0., 0.])
self.rot_angle = np.array([0., 0., 0.])
if n_control_points is None:
n_control_points = [2, 2, 2]
self.n_control_points = n_control_points
@property
def n_control_points(self):
"""
The number of control points in X, Y and Z directions
:rtype: numpy.ndarray
"""
return self._n_control_points
@n_control_points.setter
def n_control_points(self, npts):
self._n_control_points = np.array(npts)
self.array_mu_x = np.zeros(self.n_control_points)
self.array_mu_y = np.zeros(self.n_control_points)
self.array_mu_z = np.zeros(self.n_control_points)
@property
def psi_mapping(self):
"""
Map from the physical domain to the reference domain.
:rtype: numpy.ndarray
"""
return np.diag(np.reciprocal(self.box_length))
@property
def inv_psi_mapping(self):
"""
Map from the reference domain to the physical domain.
:rtype: numpy.ndarray
"""
return np.diag(self.box_length)
@property
def rotation_matrix(self):
"""
The rotation matrix (according to rot_angle_x, rot_angle_y,
rot_angle_z).
:rtype: numpy.ndarray
"""
return at.angles2matrix(
np.radians(self.rot_angle[2]), np.radians(self.rot_angle[1]),
np.radians(self.rot_angle[0]))
@property
def position_vertices(self):
"""
The position of the vertices of the FFD bounding box.
:rtype: numpy.ndarray
"""
return self.box_origin + np.vstack([
np.zeros(
(1, 3)), self.rotation_matrix.dot(np.diag(self.box_length)).T
])
def reflect(self, axis=0):
"""
Reflect the lattice of control points along the direction defined
by `axis`. In particular the origin point of the lattice is preserved.
So, for instance, the reflection along x, is made with respect to the
face of the lattice in the yz plane that is opposite to the origin.
Same for the other directions. Only the weights (mu) along the chosen
axis are reflected, while the others are preserved. The symmetry plane
can not present deformations along the chosen axis.
After the refletcion there will be 2n-1 control points along `axis`,
witha doubled box length.
:param int axis: axis along which the reflection is performed.
Default is 0. Possible values are 0, 1, or 2, corresponding
to x, y, and z respectively.
"""
# check axis value
if axis not in (0, 1, 2):
raise ValueError(
"The axis has to be 0, 1, or 2. Current value {}.".format(axis))
# check that the plane of symmetry is undeformed
if (axis == 0 and np.count_nonzero(self.array_mu_x[-1, :, :]) != 0) or (
axis == 1 and np.count_nonzero(self.array_mu_y[:, -1, :]) != 0
) or (axis == 2 and np.count_nonzero(self.array_mu_z[:, :, -1]) != 0):
raise RuntimeError(
"If you want to reflect the FFD bounding box along axis " + \
"{} you can not diplace the control ".format(axis) + \
"points in the symmetry plane along that axis."
)
# double the control points in the given axis -1 (the symmetry plane)
self.n_control_points[axis] = 2 * self.n_control_points[axis] - 1
# double the box length
self.box_length[axis] *= 2
# we have to reflect the dispacements only along the correct axis
reflection = np.ones(3)
reflection[axis] = -1
# we select all the indeces but the ones in the plane of symmetry
indeces = [slice(None), slice(None), slice(None)] # = [:, :, :]
indeces[axis] = slice(1, None) # = [1:]
indeces = tuple(indeces)
# we append along the given axis all the displacements reflected
# and in the reverse order
self.array_mu_x = np.append(
self.array_mu_x,
reflection[0] * np.flip(self.array_mu_x, axis)[indeces], axis=axis)
self.array_mu_y = np.append(
self.array_mu_y,
reflection[1] * np.flip(self.array_mu_y, axis)[indeces], axis=axis)
self.array_mu_z = np.append(
self.array_mu_z,
reflection[2] * np.flip(self.array_mu_z, axis)[indeces], axis=axis)
def read_parameters(self, filename='parameters.prm'):
"""
Reads in the parameters file and fill the self structure.
:param string filename: parameters file to be read in.
"""
if not isinstance(filename, str):
raise TypeError("filename must be a string")
# Checks if the parameters file exists. If not it writes the default
# class into filename.
if not os.path.isfile(filename):
self.write_parameters(filename)
return
config = configparser.RawConfigParser()
config.read(filename)
self.n_control_points[0] = config.getint('Box info',
'n control points x')
self.n_control_points[1] = config.getint('Box info',
'n control points y')
self.n_control_points[2] = config.getint('Box info',
'n control points z')
self.box_length[0] = config.getfloat('Box info', 'box length x')
self.box_length[1] = config.getfloat('Box info', 'box length y')
self.box_length[2] = config.getfloat('Box info', 'box length z')
self.box_origin[0] = config.getfloat('Box info', 'box origin x')
self.box_origin[1] = config.getfloat('Box info', 'box origin y')
self.box_origin[2] = config.getfloat('Box info', 'box origin z')
self.rot_angle[0] = config.getfloat('Box info', 'rotation angle x')
self.rot_angle[1] = config.getfloat('Box info', 'rotation angle y')
self.rot_angle[2] = config.getfloat('Box info', 'rotation angle z')
self.array_mu_x = | np.zeros(self.n_control_points) | numpy.zeros |
import torch
import numpy as np
from sklearn.cluster import KMeans
from typing import Mapping, Any, Optional
# https://github.com/scikit-learn/scikit-learn/blob/0fb307bf3/sklearn/mixture/_gaussian_mixture.py
# https://github.com/scikit-learn/scikit-learn/blob/0fb307bf3/sklearn/mixture/_base.py
def _batch_A_T_dot_B(A, B):
assert A.dim() == B.dim() == 3
assert A.shape[0] == B.shape[0]
assert A.shape[1] == B.shape[1]
return (A[:, :, :, np.newaxis] * B[:, :, np.newaxis, :]).sum(dim=1)
def _estimate_gaussian_covariances_diag(resp, X, nk, means, reg_covar):
"""Estimate the diagonal covariance vectors.
Parameters
----------
responsibilities : Tensor, shape (batch_size, n_samples, n_components)
X : Tensor, shape (batch_size, n_samples, n_features)
nk : Tensor, shape (batch_size, n_components)
means : Tensor, shape (batch_size, n_components, n_features)
reg_covar : float
Returns
-------
covariances : Tensor, shape (batch_size, n_components, n_features)
The covariance vector of the current components.
"""
# avg_X2 = np.dot(resp.T, X * X) / nk[:, np.newaxis]
# avg_means2 = means ** 2
# avg_X_means = means * np.dot(resp.T, X) / nk[:, np.newaxis]
# return avg_X2 - 2 * avg_X_means + avg_means2 + reg_covar
avg_X2 = _batch_A_T_dot_B(resp, X * X) / nk[..., np.newaxis]
avg_mean2 = means ** 2
avg_X_means = means * _batch_A_T_dot_B(resp, X) / nk[..., np.newaxis]
return avg_X2 - 2 * avg_X_means + avg_mean2 + reg_covar
def _estimate_gaussian_covariances_spherical(resp, X, nk, means, reg_covar):
return _estimate_gaussian_covariances_diag(resp, X, nk, means, reg_covar).mean(dim=-1)
def _estimate_gaussian_parameters_spherical(X, resp, reg_covar):
"""Estimate the Gaussian distribution parameters.
Parameters
----------
X : Tensor, shape (batch_size, n_samples, n_features)
The input data array.
resp : Tensor, shape (batch_size, n_samples, n_components)
The responsibilities for each data sample in X.
reg_covar : float
The regularization added to the diagonal of the covariance matrices.
Returns
-------
nk : Tensor, shape (batch_size, n_components)
The numbers of data samples in the current components.
means : Tensor, shape (batch_size, n_components, n_features)
The centers of the current components.
covariances : Tensor (batch_size, n_components)
The covariance matrix of the current components.
"""
# nk = resp.sum(axis=0) + 10 * np.finfo(resp.dtype).eps
# means = np.dot(resp.T, X) / nk[:, np.newaxis]
# covariances = {"full": _estimate_gaussian_covariances_full,
# "tied": _estimate_gaussian_covariances_tied,
# "diag": _estimate_gaussian_covariances_diag,
# "spherical": _estimate_gaussian_covariances_spherical
# }[covariance_type](resp, X, nk, means, reg_covar)
nk = resp.sum(dim=1) + 10 * torch.finfo(resp.dtype).eps
means = _batch_A_T_dot_B(resp, X) / nk[..., np.newaxis]
covariances = _estimate_gaussian_covariances_spherical(resp, X, nk, means, reg_covar)
return nk, means, covariances
def _estimate_log_gaussian_prob_spherical(X, means, precisions_chol):
"""Estimate the log Gaussian probability.
Parameters
----------
X : Tensor, shape (batch_size, n_samples, n_features)
means : Tensor, shape (batch_size, n_components, n_features)
precisions_chol : Tensor, shape of (batch_size, n_components)
Returns
-------
log_prob : Tensor, shape (batch_size, n_samples, n_components)
"""
batch_size, n_samples, n_features = X.shape
# det(precision_chol) is half of det(precision)
log_det = n_features * torch.log(precisions_chol)
precisions = precisions_chol ** 2
# (batch_size, n_samples, n_components)
log_prob = (
((means ** 2).sum(dim=-1) * precisions)[:, np.newaxis, :]
- (2 * (X[:, :, np.newaxis, :] * (means[:, np.newaxis, :, :] * precisions[:, np.newaxis, :, np.newaxis])).sum(dim=-1))
+ (X ** 2).sum(dim=-1)[:, :, np.newaxis] * precisions[:, np.newaxis, :]
)
return -.5 * (n_features * np.log(2 * np.pi) + log_prob) + log_det[:, np.newaxis, :]
class BatchSphericalGMM:
def __init__(
self,
n_components: int = 1,
*,
tol: float = 1e-3,
reg_covar: float = 1e-6,
max_iter: int = 100,
n_init: int = 1,
init_params: str = "kmeans",
weights_init: Optional[np.ndarray] = None,
means_init: Optional[np.ndarray] = None,
precisions_init: Optional[np.ndarray] = None,
seed: int = None,
# warm_start=False,
# verbose=0,
# verbose_interval=10,
centroids_init=None,
fit_means: bool = True,
fit_precisions: bool = True,
kmeans_kwargs: Optional[Mapping[str, Any]] = None,
device: torch.types.Device = "cpu",
dtype: torch.dtype = torch.float64,
) -> None:
self.n_components = n_components
self.tol = tol
self.reg_covar = reg_covar
self.max_iter = max_iter
self.n_init = n_init
self.init_params = init_params
self.weights_init = weights_init
self.means_init = means_init
self.precisions_init = precisions_init
self.centroids_init = centroids_init
self.fit_means = fit_means
self.fit_precisions = fit_precisions
self.kmeans_kwargs = {} if kmeans_kwargs is None else kmeans_kwargs
self.generator = torch.Generator()
self.device = device
self.dtype = dtype
self.seed = seed
if seed is not None:
self.generator.manual_seed(seed)
def fit(self, X: np.ndarray):
"""
Args:
X: array, shape (batch_size, n_samples, n_features)
Returns:
best_weights: Tensor, (batch_size, n_components)
best_means: Tensor, (batch_size, n_components)
best_weights: Tensor, (batch_size, n_components)
"""
X = torch.from_numpy(X).to(dtype=self.dtype, device=self.device)
batch_size, n_samples, n_features = X.shape
max_lower_bound = torch.full((batch_size,), -np.inf, dtype=self.dtype, device=self.device)
best_weights = torch.full((batch_size, self.n_components), np.nan, dtype=self.dtype, device=self.device)
best_means = torch.full(
(batch_size, self.n_components, n_features), np.nan, dtype=self.dtype, device=self.device
)
best_covariances = torch.full((batch_size, self.n_components), np.nan, dtype=self.dtype, device=self.device)
for init in range(self.n_init):
# self._print_verbose_msg_init_beg(init)
self._initialize_parameters(X)
lower_bound = torch.full((batch_size,), -np.inf, dtype=self.dtype, device=self.device)
for n_iter in range(1, self.max_iter + 1):
prev_lower_bound = lower_bound
log_prob_norm, log_resp = self._e_step(X)
self._m_step(X, log_resp)
lower_bound = self._compute_lower_bound(log_resp, log_prob_norm)
change = lower_bound - prev_lower_bound
# self._print_verbose_msg_iter_end(n_iter, change)
if (abs(change) < self.tol).all():
self.converged_ = True
break
# self._print_verbose_msg_init_end(lower_bound)
to_be_updated = lower_bound > max_lower_bound
best_weights[to_be_updated] = self.weights_[to_be_updated]
best_means[to_be_updated] = self.means_[to_be_updated]
best_covariances[to_be_updated] = self.covariances_[to_be_updated]
max_lower_bound[to_be_updated] = lower_bound[to_be_updated]
return (
best_weights.detach().cpu().numpy(),
best_means.detach().cpu().numpy(),
best_covariances.detach().cpu().numpy(),
max_lower_bound.detach().cpu().numpy(),
)
def _initialize_parameters(self, X):
batch_size, n_samples, n_features = X.shape
if self.centroids_init is not None:
centroids = torch.from_numpy(self.centroids_init).to(dtype=self.dtype, device=self.device)
assert centroids.shape == (batch_size, self.n_components, n_features)
resp = torch.nn.functional.one_hot(
torch.argmin(((X[:, :, np.newaxis, :] - centroids[:, np.newaxis, :, :]) ** 2).sum(dim=-1), dim=-1),
num_classes=self.n_components
).float()
elif self.init_params == 'kmeans':
resp = torch.zeros((batch_size, n_samples, self.n_components), dtype=self.dtype, device=self.device)
# TODO: batch computation
for batch_index in range(batch_size):
label = KMeans(
n_clusters=self.n_components, n_init=1, random_state=self.seed, **self.kmeans_kwargs
).fit(X[batch_index].detach().cpu().numpy()).labels_
resp[batch_index, np.arange(n_samples), label] = 1
elif self.init_params == 'random':
resp = torch.rand((batch_size, n_samples, self.n_components), generator=self.generator)
resp = resp.to(dtype=self.dtype, device=self.device)
resp /= resp.sum(dim=1, keepdims=True)
else:
raise ValueError("Unimplemented initialization method '%s'"
% self.init_params)
self._initialize(X, resp)
def _initialize(self, X, resp):
batch_size, n_samples, n_features = X.shape
weights, means, covariances = _estimate_gaussian_parameters_spherical(X, resp, self.reg_covar)
weights /= n_samples
if self.weights_init is None:
self.weights_ = weights
else:
self.weights_ = torch.from_numpy(self.weights_init).to(dtype=self.dtype, device=self.device)
if self.means_init is None:
self.means_ = means
else:
self.means_ = torch.from_numpy(self.means_init).to(dtype=self.dtype, device=self.device)
if self.precisions_init is None:
self.precisions_cholesky_ = 1. / torch.sqrt(covariances)
self.covariances_ = covariances
else:
self.precisions_cholesky_ = torch.from_numpy(self.precisions_init).to(dtype=self.dtype, device=self.device)
self.covariances_ = 1 / (self.precisions_cholesky_ ** 2)
def _e_step(self, X):
"""
Returns
-------
log_prob_norm : Tensor, shape (batch_size,)
Mean of the logarithms of the probabilities of each sample in X
log_responsibility : Tensor, shape (batch_size, n_samples, n_components)
Logarithm of the posterior probabilities (or responsibilities) of
the point of each sample in X.
"""
log_prob_norm, log_resp = self._estimate_log_prob_resp(X)
return log_prob_norm.mean(dim=-1), log_resp
def _estimate_log_prob_resp(self, X):
"""Estimate log probabilities and responsibilities for each sample.
Compute the log probabilities, weighted log probabilities per
component and responsibilities for each sample in X with respect to
the current state of the model.
Parameters
----------
X : Tensor, shape (batch_size, n_samples, n_features)
Returns
-------
log_prob_norm : Tensor, shape (batch_size, n_samples)
log p(X)
log_responsibilities : Tensor, shape (batch_size, n_samples, n_components)
logarithm of the responsibilities
"""
weighted_log_prob = self._estimate_weighted_log_prob(X)
log_prob_norm = torch.logsumexp(weighted_log_prob, dim=-1)
# TODO: use a context equivalent to np.errstate(under='ignore')
log_resp = weighted_log_prob - log_prob_norm[..., np.newaxis]
return log_prob_norm, log_resp
def _estimate_weighted_log_prob(self, X):
"""
Returns
-------
weighted_log_prob : Tensor, shape (batch_size, n_samples, n_components)
"""
return self._estimate_log_prob(X) + self._estimate_log_weights()[:, np.newaxis, :]
def _estimate_log_prob(self, X):
"""
Returns
-------
log_prob : Tensor, shape (batch_size, n_samples, n_components)
"""
return _estimate_log_gaussian_prob_spherical(X, self.means_, self.precisions_cholesky_)
def _estimate_log_weights(self):
"""
Returns
-------
log_weights : Tensor, shape (batch_size, n_components)
"""
return torch.log(self.weights_)
def _m_step(self, X, log_resp):
"""M step.
Parameters
----------
X : Tensor, shape (batch_size, n_samples, n_features)
log_resp : Tensor, shape (batch_size, n_samples, n_components)
Logarithm of the posterior probabilities (or responsibilities) of
the point of each sample in X.
"""
batch_size, n_samples, n_features = X.shape
weights, means, covariances = _estimate_gaussian_parameters_spherical(X, torch.exp(log_resp), self.reg_covar)
self.weights_ = weights / n_samples
if self.fit_means:
self.means_ = means
if self.fit_precisions:
self.covariances_ = covariances
self.precisions_cholesky_ = 1. / torch.sqrt(covariances)
def _compute_lower_bound(self, _, log_prob_norm):
return log_prob_norm
if __name__ == "__main__":
from sklearn.mixture import GaussianMixture
import warnings
from sklearn.exceptions import ConvergenceWarning
np_random = np.random.RandomState(0)
gmm_reference = GaussianMixture(n_components=2, covariance_type="spherical", tol=0, random_state=np_random)
_initialize_orig = gmm_reference._initialize
weights_init, means_init, precisions_init = None, None, None
def _patched_initialize(X, resp):
global weights_init, means_init, precisions_init
_initialize_orig(X, resp)
weights_init = gmm_reference.weights_
means_init = gmm_reference.means_
precisions_init = gmm_reference.precisions_cholesky_
gmm_reference._initialize = _patched_initialize
batch_size = 32
n_samples, n_features = 250, 2
mu1, mu2 = -1.0, 5.0
sigma1, sigma2 = 1.0, 2.0
X_batch = []
weights_init_batch, means_init_batch, precisions_init_batch = [], [], []
expected_weights, expected_means, expected_covariances = [], [], []
for _ in range(batch_size):
n1 = int(n_samples * 0.7) * n_features
n2 = n_features * n_samples - n1
X = np_random.normal(np.r_[np.full(n1, mu1), np.full(n2, mu2)], np.r_[np.full(n1, sigma1), np.full(n2, sigma2)])
X = X.reshape(n_samples, n_features)
X_batch.append(X)
with warnings.catch_warnings():
warnings.simplefilter("ignore", category=ConvergenceWarning)
gmm_reference.fit(X)
weights_init_batch.append(weights_init.copy())
means_init_batch.append(means_init.copy())
precisions_init_batch.append(precisions_init.copy())
expected_weights.append(gmm_reference.weights_.copy())
expected_means.append(gmm_reference.means_.copy())
expected_covariances.append(gmm_reference.covariances_.copy())
weights_init_batch = np.asarray(weights_init_batch, dtype=np.float64)
means_init_batch = np.asarray(means_init_batch, dtype=np.float64)
precisions_init_batch = np.asarray(precisions_init_batch, dtype=np.float64)
X_batch = np.asarray(X_batch, dtype=np.float64)
gmm_tested = BatchSphericalGMM(
n_components=2,
weights_init=weights_init_batch,
means_init=means_init_batch,
precisions_init=precisions_init_batch,
init_params="random",
dtype=torch.float64,
device="cpu",
)
actual_weights, actual_means, actual_covariances = gmm_tested.fit(X_batch)
assert np.allclose(expected_weights, actual_weights)
assert | np.allclose(expected_means, actual_means) | numpy.allclose |
import skimage
import skimage.io
import skimage.transform
import numpy as np
from matplotlib import pyplot as plt
from matplotlib.pyplot import imshow
import matplotlib.image as mpimg
import tensorflow as tf
import os
from skimage import io
from skimage.transform import resize
import cv2
# synset = [l.strip() for l in open('synset.txt').readlines()]
def resnet_preprocess(resized_inputs):
"""Faster R-CNN Resnet V1 preprocessing.
VGG style channel mean subtraction as described here:
https://gist.github.com/ksimonyan/211839e770f7b538e2d8#file-readme-md
Args:
resized_inputs: A [batch, height_in, width_in, channels] float32 tensor
representing a batch of images with values between 0 and 255.0.
Returns:
preprocessed_inputs: A [batch, height_out, width_out, channels] float32
tensor representing a batch of images.
"""
channel_means = tf.constant([123.68, 116.779, 103.939],
dtype=tf.float32, shape=[1, 1, 1, 3], name='img_mean')
return resized_inputs - channel_means
# returns image of shape [224, 224, 3]
# [height, width, depth]
def load_image(path, normalize=True):
"""
args:
normalize: set True to get pixel value of 0~1
"""
# load image
img = skimage.io.imread(path)
if normalize:
img = img / 255.0
assert (0 <= img).all() and (img <= 1.0).all()
# print "Original Image Shape: ", img.shape
# we crop image from center
short_edge = min(img.shape[:2])
yy = int((img.shape[0] - short_edge) / 2)
xx = int((img.shape[1] - short_edge) / 2)
crop_img = img[yy: yy + short_edge, xx: xx + short_edge]
# resize to 224, 224
resized_img = skimage.transform.resize(crop_img, (224, 224), preserve_range=True) # do not normalize at transform.
return resized_img
# returns the top1 string
def print_prob(prob, file_path):
synset = [l.strip() for l in open(file_path).readlines()]
# print prob
pred = np.argsort(prob)[::-1]
# Get top1 label
top1 = synset[pred[0]]
print("Top1: ", top1, prob[pred[0]])
# Get top5 label
top5 = [(synset[pred[i]], prob[pred[i]]) for i in range(5)]
print("Top5: ", top5)
return top1
def visualize(image, conv_output, conv_grad, gb_viz):
output = conv_output # [7,7,512]
grads_val = conv_grad # [7,7,512]
print("grads_val shape:", grads_val.shape)
print("gb_viz shape:", gb_viz.shape)
weights = np.mean(grads_val, axis = (0, 1)) # alpha_k, [512]
cam = np.zeros(output.shape[0 : 2], dtype = np.float32) # [7,7]
# Taking a weighted average
for i, w in enumerate(weights):
cam += w * output[:, :, i]
# Passing through ReLU
cam = np.maximum(cam, 0)
cam = cam / np.max(cam) # scale 0 to 1.0
cam = resize(cam, (128,128), preserve_range=True)
img = image.astype(float)
img -= np.min(img)
img /= img.max()
# print(img)
cam_heatmap = cv2.applyColorMap(np.uint8(255*cam), cv2.COLORMAP_JET)
cam_heatmap = cv2.cvtColor(cam_heatmap, cv2.COLOR_BGR2RGB)
# cam = np.float32(cam) + np.float32(img)
# cam = 255 * cam / np.max(cam)
# cam = np.uint8(cam)
fig = plt.figure()
ax = fig.add_subplot(111)
imgplot = plt.imshow(img)
ax.set_title('Input Image')
fig = plt.figure(figsize=(12, 16))
ax = fig.add_subplot(141)
imgplot = plt.imshow(cam_heatmap)
ax.set_title('Grad-CAM')
gb_viz = np.dstack((
gb_viz[:, :, 0],
gb_viz[:, :, 1],
gb_viz[:, :, 2],
))
gb_viz -= np.min(gb_viz)
gb_viz /= gb_viz.max()
gd_gb = np.dstack((
gb_viz[:, :, 0] * cam,
gb_viz[:, :, 1] * cam,
gb_viz[:, :, 2] * cam,
))
image_cam = img*0.7 + (cam_heatmap/255.)*0.3
ax = fig.add_subplot(142)
imgplot = plt.imshow(gb_viz)
ax.set_title('guided backpropagation')
ax = fig.add_subplot(143)
imgplot = plt.imshow(gd_gb)
ax.set_title('guided Grad-CAM')
ax = fig.add_subplot(144)
imgplot = plt.imshow(image_cam)
ax.set_title('Grad-CAM Image')
plt.show()
def save_result(image, conv_output, conv_grad, gb_viz, idx=0, version=0, save=True):
output = conv_output # [7,7,512]
grads_val = conv_grad # [7,7,512]
#print("grads_val shape:", grads_val.shape)
#print("gb_viz shape:", gb_viz.shape)
weights = np.mean(grads_val, axis = (0, 1)) # alpha_k, [512]
cam = np.zeros(output.shape[0 : 2], dtype = np.float32) # [7,7]
# Taking a weighted average
for i, w in enumerate(weights):
cam += w * output[:, :, i]
# Passing through ReLU
cam = | np.maximum(cam, 0) | numpy.maximum |
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import json
import os
import datetime as dt
import main
from eval import data_analysis
# LaTeX settings
plt.rc('text', usetex=True)
plt.rc('font', **{'family': 'serif', 'sans-serif': ['lmodern'], 'size': 18})
plt.rc('axes', **{'titlesize': 18, 'labelsize': 18})
# Constants
JSON_PATH = './out/'
OUT_PATH = './out/'
MODEL_NAMES = {
'KF': ('KalmanFilter', ''),
'KF(+W)': ('KalmanFilter', '_W'),
'KF(+WF)': ('KalmanFilter', '_WF'),
'KD-IC': ('KD-IC', ''),
'KD-IC(+W)': ('KD-IC', '_W'),
'KD-IC(+WF)': ('KD-IC', '_WF'),
'LN-IC': ('LogNormal-IC', ''),
'LN-IC(+W)': ('LogNormal-IC', '_W'),
'LN-IC(+WF)': ('LogNormal-IC', '_WF'),
'DeepAR': ('DeepAR', ''),
'DeepAR(+W)': ('DeepAR', '_W'),
'DeepAR(+WF)': ('DeepAR', '_WF'),
'LW': ('LastWeek', '')
}
MAIN_SEED = '42'
DECIMALS = 2
COLORS = ('C0', 'C1', 'C3', 'C9', 'C7')
MARKERS = ('o', 'X', 'v', 'd', 'p')
LINESTYLES = ('solid', 'dashed', 'dashdot')
S_D = 48
S_W = 7 * S_D
def get_file_name(model, level, cluster, seed=''):
return f'{MODEL_NAMES[model][0]}{seed}_{level}_{cluster}{MODEL_NAMES[model][1]}'
def get_path(model, level, cluster, seed=''):
return JSON_PATH + f'{MODEL_NAMES[model][0]}{seed}/{get_file_name(model, level, cluster, seed)}.json'
def load_res(model, level, cluster, seed=''):
if 'DeepAR' in model and seed == '':
seed = MAIN_SEED
with open(get_path(model, level, cluster, seed), 'r') as fp:
res = json.load(fp)
return res
def collect_results(
levels=('L0', 'L1', 'L2', 'L3'),
metrics=('MAPE', 'rMAE', 'rRMSE', 'rCRPS'),
models=('KF', 'KF(+W)', 'KF(+WF)',
'KD-IC', 'KD-IC(+W)', 'KD-IC(+WF)',
'DeepAR', 'DeepAR(+W)', 'DeepAR(+WF)',
'LW'),
seeds=(0, 1, 2, 3, 4),
forecast_reps=28,
save_results_with_info=True
):
results_path = os.path.join(JSON_PATH, 'results_with_info.npy')
if os.path.isfile(results_path):
results_with_info = np.load(results_path, allow_pickle=True)
return results_with_info[0], results_with_info[1]
results = {}
level_info = data_analysis.get_level_info(levels)
for level in levels:
clusters = level_info[level]['clusters']
# Create results array
results[level] = np.empty((len(metrics), len(models), len(clusters), forecast_reps))
results[level][:] = np.nan
for m, model in enumerate(models):
if level == 'L3' and 'KF' in model:
# No level 3 results for the KF model
continue
for c, cluster in enumerate(clusters):
if 'DeepAR' in model and level is not 'L3':
res_per_seed = []
for seed in seeds:
res_per_seed.append(load_res(model, level, cluster, seed))
for i, metric in enumerate(metrics):
results[level][i, m, c] = np.mean([res[metric] for res in res_per_seed], axis=0)
else:
res = load_res(model, level, cluster)
for i, metric in enumerate(metrics):
if 'CRPS' in metric and model == 'LW':
# No distributional forecasts for LW model
continue
results[level][i, m, c] = res[metric]
info = {
'levels': level_info,
'metrics': list(metrics),
'models': list(models),
'reps': forecast_reps
}
if save_results_with_info:
np.save(results_path, (results, info), allow_pickle=True)
return results, info
def collect_results_per_tstp(
levels=('L0', 'L1', 'L2'),
metrics=('rMAE', 'rRMSE', 'rCRPS'),
models=('KF', 'KF(+W)', 'KF(+WF)',
'KD-IC', 'KD-IC(+W)', 'KD-IC(+WF)',
'DeepAR', 'DeepAR(+W)', 'DeepAR(+WF)',
'LW'),
seeds=(0, 1, 2, 3, 4),
forecast_reps=28,
horizon=192,
save_results_per_tstp_with_info=True
):
results_path = os.path.join(JSON_PATH, 'results_per_tstp_with_info.npy')
if os.path.isfile(results_path):
results_with_info = np.load(results_path, allow_pickle=True)
return results_with_info[0], results_with_info[1]
results = {}
level_info = data_analysis.get_level_info(levels)
t_train, t_val = main.train_val_split(data_analysis.energy_df.index)
for level in levels:
clusters = level_info[level]['clusters']
# Create results array
results[level] = np.empty((len(seeds), len(metrics), len(models), len(clusters), forecast_reps, horizon))
results[level][:] = np.nan
level_info[level]['y_mean'] = []
for c, cluster in enumerate(clusters):
level_info[level]['y_mean'].append(
np.nanmean(data_analysis.get_observations_at(level, cluster, t_train))
)
y_true = data_analysis.get_observations_at(level, cluster, t_val).reshape(forecast_reps, horizon)
for m, model in enumerate(models):
if level == 'L3' and 'KF' in model:
# No level 3 results for the KF model
continue
if 'DeepAR' in model and level is not 'L3':
for s, seed in enumerate(seeds):
res = load_res(model, level, cluster, seed)
for i, metric in enumerate(metrics):
if metric == 'rMAE':
results[level][s, i, m, c] = np.abs(y_true - res['p50'])
elif metric == 'rRMSE':
results[level][s, i, m, c] = (y_true - res['mean']) ** 2
elif metric == 'rCRPS':
results[level][s, i, m, c] = res['CRPS']
else:
res = load_res(model, level, cluster)
for i, metric in enumerate(metrics):
if 'CRPS' in metric and model == 'LW':
# No distributional forecasts for LW model
continue
if metric == 'rMAE':
results[level][0, i, m, c] = np.abs(y_true - res['p50'])
elif metric == 'rRMSE':
results[level][0, i, m, c] = (y_true - res['mean']) ** 2
elif metric == 'rCRPS':
results[level][0, i, m, c] = res['CRPS']
info = {
'levels': level_info,
'metrics': list(metrics),
'models': list(models),
'reps': forecast_reps,
'horizon': horizon
}
if save_results_per_tstp_with_info:
np.save(results_path, (results, info), allow_pickle=True)
return results, info
def create_metric_df(metric, with_std=True, to_LaTeX=True):
results, info = collect_results()
i = info['metrics'].index(metric)
row_names = info['models']
col_names = info['levels'].keys()
metric_df = pd.DataFrame(index=row_names, columns=col_names, dtype=float)
for level in col_names:
for m, model in enumerate(row_names):
mean = np.mean(results[level][i, m])
metric_df.loc[model, level] = (('%%.%sf' % DECIMALS) % mean) if not np.isnan(mean) else '-'
if with_std and not np.isnan(mean):
std = np.std(results[level][i, m])
metric_df.loc[model, level] += (' (%%.%sf)' % DECIMALS) % std
if to_LaTeX:
df_to_LaTeX(metric_df)
return metric_df
def create_level_df(level, with_std=True, to_LaTeX=True):
results, info = collect_results()
row_names = info['metrics']
col_names = info['models']
level_df = pd.DataFrame(index=row_names, columns=col_names, dtype=float)
for i, metric in enumerate(row_names):
for m, model in enumerate(col_names):
mean = np.mean(results[level][i, m])
level_df.loc[metric, model] = (('%%.%sf' % DECIMALS) % mean) if not np.isnan(mean) else '-'
if with_std and not np.isnan(mean):
std = np.std(results[level][i, m])
level_df.loc[metric, model] += (' (%%.%sf)' % DECIMALS) % std
if to_LaTeX:
df_to_LaTeX(level_df)
return level_df
def create_runtime_df(models=('KF', 'KD-IC', 'DeepAR', 'LW'), with_std=False, to_LaTeX=True):
_, info = collect_results()
train_name = 'Avg. training time [s]'
prediction_name = 'Avg. prediction time [s]'
runtime_df = pd.DataFrame(index=[train_name, prediction_name], columns=models, dtype=float)
for model in models:
training_times = []
prediction_times = []
for level in info['levels'].keys():
if level == 'L3' and 'KF' in model:
# No level 3 results for the KF model
continue
for cluster in info['levels'][level]['clusters']:
res = load_res(model, level, cluster)
training_times.append(res['fit_time'])
prediction_times.append(res['prediction_time'])
decimals = DECIMALS + 1
runtime_df.loc[train_name, model] = ('%%.%sf' % decimals) % np.mean(training_times)
runtime_df.loc[prediction_name, model] = ('%%.%sf' % decimals) % np.mean(prediction_times)
if with_std:
runtime_df.loc[train_name, model] += (' (%%.%sf)' % decimals) % np.std(training_times)
runtime_df.loc[prediction_name, model] += (' (%%.%sf)' % decimals) % np.std(prediction_times)
if to_LaTeX:
df_to_LaTeX(runtime_df)
return runtime_df
def df_to_LaTeX(df):
num_columns = len(df.columns)
print(df.to_latex(
float_format=f'%.{DECIMALS}f',
na_rep='-',
column_format='l' + ''.join('r' * num_columns)
))
def get_color(model):
if 'KF' in model:
return COLORS[0]
elif 'KD-IC' in model:
return COLORS[1]
elif 'DeepAR' in model:
return COLORS[2]
elif 'LW' in model:
return COLORS[3]
else:
return COLORS[4]
def get_linestyle(model):
if '(+W)' in model:
return LINESTYLES[1]
elif '(+WF)' in model:
return LINESTYLES[2]
else:
return LINESTYLES[0]
def _complete_plot(name, legend=True, grid=True):
if legend:
plt.legend()
if grid:
plt.grid()
plt.tight_layout()
plt.savefig(OUT_PATH + f'{name}.pdf', bbox_inches='tight')
plt.close()
def plot_epoch_loss(model, level, cluster, seed=MAIN_SEED):
assert 'DeepAR' in model, "Loss plot only available for deep models"
res = load_res(model, level, cluster, seed)
train_loss = res['train_loss']
val_loss = res['val_loss']
plt.figure(figsize=(6, 4))
plt.plot(np.arange(len(train_loss)) + 1, train_loss, color=COLORS[0], label='Train')
plt.plot(np.arange(len(val_loss)) + 1, val_loss, color=COLORS[1], label='Validation')
plt.ylabel('Loss')
plt.xlabel('Epoch')
_complete_plot(f'{get_file_name(model, level, cluster, seed)}_epoch_loss', grid=False)
def plot_horizon(model, metric, horizons=(1, 2, 3, 4), levels=('L0', 'L1', 'L2')):
results, info = collect_results_per_tstp()
model_W = model + '(+W)'
model_WF = model + '(+WF)'
i = info['metrics'].index(metric)
m = info['models'].index(model)
m_W = info['models'].index(model_W)
m_WF = info['models'].index(model_WF)
score = np.empty(len(horizons))
score_W = np.empty(len(horizons))
score_WF = np.empty(len(horizons))
for h, horizon in enumerate(horizons):
idx = np.arange(0, horizon * S_D)
res = []
res_W = []
res_WF = []
for level in levels:
for c, cluster in enumerate(info['levels'][level]['clusters']):
y_mean = info['levels'][level]['y_mean'][c]
if metric == 'rRMSE':
res.append(100 * np.sqrt(np.mean(results[level][:, i, m, c, :, idx], axis=2)) / y_mean)
res_W.append(100 * np.sqrt(np.mean(results[level][:, i, m_W, c, :, idx], axis=2)) / y_mean)
res_WF.append(100 * np.sqrt(np.mean(results[level][:, i, m_WF, c, :, idx], axis=2)) / y_mean)
else:
res.append(100 * np.mean(results[level][:, i, m, c, :, idx], axis=2) / y_mean)
res_W.append(100 * np.mean(results[level][:, i, m_W, c, :, idx], axis=2) / y_mean)
res_WF.append(100 * np.mean(results[level][:, i, m_WF, c, :, idx], axis=2) / y_mean)
score[h] = np.nanmean(res)
score_W[h] = np.nanmean(res_W)
score_WF[h] = np.nanmean(res_WF)
skill_W = 100 * (1 - score_W / score)
skill_WF = 100 * (1 - score_WF / score)
print(f'SS_{metric} (W): {skill_W}')
print(f'SS_{metric} (WF): {skill_WF}')
plt.figure(figsize=(3.5, 4))
plt.plot(
score,
linestyle=get_linestyle(model),
color=get_color(model),
marker=MARKERS[0]
)
plt.plot(
score_W,
linestyle=get_linestyle(model_W),
color=get_color(model_W),
marker=MARKERS[1]
)
plt.plot(
score_WF,
linestyle=get_linestyle(model_WF),
color=get_color(model_WF),
marker=MARKERS[2]
)
plt.ylim(6.95, 8.35)
plt.ylabel(metric)
plt.xlabel('Horizon')
plt.xticks(np.arange(len(horizons)), np.array(horizons))
plt.title(model)
_complete_plot(f"{model}_{metric}_horizon", grid=False, legend=False)
def plot_reps(metric, levels=('L0', 'L1', 'L2'), models=None, name=None):
results, info = collect_results()
models = info['models'] if models is None else models
i = info['metrics'].index(metric)
# Lines for second legend
_, ax = plt.subplots()
lines = ax.plot([0, 1], [0, 1], '-C7', [0, 1], [0, 2], '--C7')
plt.close()
plt.figure(figsize=(10, 4))
for j, model in enumerate(models):
m = info['models'].index(model)
reps_mean = []
for level in levels:
if level == 'L3' and 'KF' in model:
# No level 3 results for the KF model
continue
for c, cluster in enumerate(info['levels'][level]['clusters']):
reps_mean.append(results[level][i, m, c])
reps_mean = np.mean(reps_mean, axis=0)
plt.plot(
reps_mean,
label=model if '(' not in model else None,
linestyle=get_linestyle(model),
color=get_color(model)
)
plt.ylabel(metric)
plt.xlabel('Forecast origin')
plt.yticks(np.arange(5, 17, 2.5))
t0 = load_res('LW', 'L0', 'Agg')['t0']
ticks = [dt.datetime.strptime(tstp, '%Y-%m-%d, %H:%M').strftime('%b, %d') for tstp in t0[1::5]]
plt.xticks(np.arange(1, len(t0), 5), ticks, rotation=0)
plt.grid(axis='y')
second_legend = plt.legend(lines, ('no weather', 'actual weather'), loc='upper left')
plt.gca().add_artist(second_legend)
_complete_plot(f"{f'{name}_' if name is not None else ''}{metric}_reps", grid=False)
def plot_clusters(level, metric, models=None, name=None):
results, info = collect_results()
models = info['models'] if models is None else models
i = info['metrics'].index(metric)
plt.figure(figsize=(10, 4))
for model in models:
if level == 'L3' and 'KF' in model:
# No level 3 results for the KF model
continue
m = info['models'].index(model)
clusters_mean = np.mean(results[level][i, m], axis=1)
plt.plot(
clusters_mean,
label=model,
linestyle=get_linestyle(model),
color=get_color(model)
)
plt.ylabel(metric)
cluster_labels = [f"{cluster.replace('ACORN-', '')} ({count})" for cluster, count in zip(
info['levels'][level]['clusters'],
info['levels'][level]['cardinality']
)]
if level == 'L3':
plt.xticks(np.arange(0, len(cluster_labels), 100), np.array(cluster_labels)[::100], rotation=90)
elif level == 'L2':
plt.xticks(np.arange(len(cluster_labels)), cluster_labels, rotation=90)
else:
plt.xticks(np.arange(len(cluster_labels)), cluster_labels)
_complete_plot(f"{f'{name}_' if name is not None else ''}{level}_{metric}_clusters")
def plot_aggregate_size(metric, models=None, name=None):
results, info = collect_results()
models = info['models'] if models is None else models
i = info['metrics'].index(metric)
aggregate_sizes = []
errors = {}
bottom_level_errors = {}
for model in models:
errors[model] = []
bottom_level_errors[model] = []
for level, level_info in info['levels'].items():
for c, agg_size in enumerate(level_info['cardinality']):
if level != 'L3':
aggregate_sizes.append(agg_size)
for model in models:
m = info['models'].index(model)
errors[model].append(np.mean(results[level][i, m, c]))
else:
for model in models:
m = info['models'].index(model)
bottom_level_errors[model].append(np.mean(results[level][i, m, c]))
aggregate_sizes.append(1)
for model in models:
errors[model].append(np.mean(bottom_level_errors[model]))
sorted_idx = np.argsort(aggregate_sizes)
aggregate_sizes = np.array(aggregate_sizes)[sorted_idx]
plt.figure(figsize=(6, 4))
for model in models:
if 'CRPS' in metric and model == 'LW':
# No distributional forecasts for LW model
continue
plt.plot(
aggregate_sizes,
np.array(errors[model])[sorted_idx],
label=model,
linestyle=get_linestyle(model),
color=get_color(model)
)
plt.ylabel(metric)
plt.yticks(np.arange(0, 70, 20))
plt.xlabel('\\# aggregated meters')
plt.xscale('log')
plt.xticks([1, 10, 100, 1000], ['1', '10', '100', '1000'])
_complete_plot(f"{f'{name}_' if name is not None else ''}{metric}_aggregate_size", grid=False)
def get_skill_scores(model, metric, no_L3=False):
results, info = collect_results()
i = info['metrics'].index(metric)
m = info['models'].index(model)
m_W = info['models'].index(model + '(+W)')
m_WF = info['models'].index(model + '(+WF)')
aggregate_sizes = []
score = []
score_W = []
score_WF = []
bottom_level_score = []
bottom_level_score_W = []
bottom_level_score_WF = []
t_train = main.train_val_split(data_analysis.energy_df.index)[0]
u = data_analysis.daily(
data_analysis.get_weather_df(forecast=False).loc[t_train, 'temperature'].to_numpy(float),
reduce=True
)
u_F = data_analysis.daily(
data_analysis.get_weather_df(forecast=True).loc[t_train, 'temperature'].to_numpy(float),
reduce=True
)
corr_W = []
corr_WF = []
bottom_level_corr_W = []
bottom_level_corr_WF = []
for level, level_info in info['levels'].items():
if level == 'L3' and ('KF' in model or no_L3):
# No level 3 results for the KF model
continue
for c, (cluster, agg_size) in enumerate(zip(level_info['clusters'], level_info['cardinality'])):
y = data_analysis.daily(
np.array(data_analysis.get_observations_at(level, cluster, t_train)),
reduce=True
)
if level != 'L3':
aggregate_sizes.append(agg_size)
score.append(np.mean(results[level][i, m, c]))
score_W.append(np.mean(results[level][i, m_W, c]))
score_WF.append(np.mean(results[level][i, m_WF, c]))
corr_W.append(data_analysis.correlation(u, y) ** 2)
corr_WF.append(data_analysis.correlation(u_F, y) ** 2)
else:
bottom_level_score.append(np.mean(results[level][i, m, c]))
bottom_level_score_W.append(np.mean(results[level][i, m_W, c]))
bottom_level_score_WF.append(np.mean(results[level][i, m_WF, c]))
bottom_level_corr_W.append(data_analysis.correlation(u, y) ** 2)
bottom_level_corr_WF.append(data_analysis.correlation(u_F, y) ** 2)
if 'KF' not in model and not no_L3:
aggregate_sizes.append(1)
score.append(np.mean(bottom_level_score))
score_W.append(np.mean(bottom_level_score_W))
score_WF.append(np.mean(bottom_level_score_WF))
corr_W.append( | np.mean(bottom_level_corr_W) | numpy.mean |
# TensorFlow and tf.keras
import tensorflow as tf
from tensorflow import keras
from keras.models import Sequential
from keras.layers import Dense, Flatten
# Helper libraries
import numpy as np
import matplotlib.pyplot as plt
import pickle
import random
#import dataset
with open('./data/jar/DHSs_onehot.pickle', 'rb') as pickle_in:
onehot_data = pickle.load(pickle_in)
with open('./data/jar/var_chart.pickle', 'rb') as pickle_in:
var_chart_data = pickle.load(pickle_in)
input_matched_label = {}
matched_list = []
# group dhs with its label
for dhs in onehot_data.keys():
input_matched_label[dhs] = [onehot_data[dhs], var_chart_data[dhs]]
matched_list.append([onehot_data[dhs], var_chart_data[dhs]])
#generate random unique indexes to pull out data for testing
testing_indexes = []
tmp_indexes = []
while(len(testing_indexes) < 500):
tmp_indexes.append(random.randint(0,3619))
testing_indexes = list(set(tmp_indexes))
training_indexes = []
for x in range(3618):
if x not in testing_indexes:
training_indexes.append(x)
training_data_list = []
training_labels_list = []
for ind in training_indexes:
training_data_list.append(matched_list[ind][0])
training_labels_list.append(matched_list[ind][1])
testing_data_list = []
testing_labels_list = []
for ind in testing_indexes:
testing_data_list.append(matched_list[ind][0])
testing_labels_list.append(matched_list[ind][1])
train_data = | np.array(training_data_list) | numpy.array |
import os
assert os.environ['CONDA_DEFAULT_ENV']=='skbio_env', 'You should use the conda environment skbio_env'
import numpy as np
from skbio.stats.ordination import cca
import pandas as pd
import matplotlib.pylab as plt
from copy import copy
import matplotlib.colors as mcolors
import seaborn as sns
from matplotlib.patches import Patch
redFspecies = True
spl = [6,11,25,250]
tits = ['(a)', '(b)', '(c)', '(d)']
Allvars = False
noise = [True,False]
plott=True#False#
dirRead = '/Users/nooteboom/Documents/GitHub/cluster_TM/cluster_SP/density/dens/ordination/'
minss = [100,200, 300, 400, 500, 600, 700, 800, 900,1000] # The s_min values
xiss = np.arange(0.0001,0.01, 0.0001) # The xi values
fig, ax = plt.subplots(2,3, figsize=(16,16),
gridspec_kw={'width_ratios':[1,1,0.08]})
ax[0,0].get_shared_y_axes().join(ax[1,0])
ax[0,1].get_shared_y_axes().join(ax[1,1])
for axs in ax[:, 2]:
axs.remove()
gs = ax[1, 2].get_gridspec()
axbig = fig.add_subplot(gs[:, 2])
sns.set(style='whitegrid',context='paper', font_scale=2)
fs=20
vs = np.array([-1,1])*0.8
for spi, sp in enumerate(spl):
print(sp)
# keep track of the results
# F and D stand for Foram and Dino
# noise keeps track of CCA results if noisy locations are included
# cluster keeps track of results if noisy locations are excluded
FNoise = np.zeros((len(minss), len(xiss)))
DNoise = np.zeros((len(minss), len(xiss)))
FCluster = np.zeros((len(minss), len(xiss)))
DCluster = np.zeros((len(minss), len(xiss)))
for mini,mins in enumerate(minss):
print('min: %d'%(mins))
for xii, xis in enumerate(xiss):
opts = ["xi", xis]
if(redFspecies):
ff = np.load('loops/redF/prepredF_CCA_sp%d_smin%d%s_%.5f.npz'%(sp, mins, opts[0], opts[1]))
else:
ff = np.load(dirRead+'loops/prep_CCA_sp%d_smin%d%s_%.5f.npz'%(sp, mins, opts[0], opts[1]))
#%%
envs = ff['envnames']
if(Allvars):
envsplt = ff['envnames']
else:
envsplt = ff['envnames']
envsplt = ['temp','N']
Flabels = ff['Flabels']
Flabelsfull = copy(Flabels)
Fenv = ff['Fenv']
for ni,n in enumerate(noise):
envs = ff['envnames']
envsplt = ['temp','N']
Flabels = ff['Flabels']
Fenv = ff['Fenv']
Fenv_nn = ff['Fenv_nn']
#%% Foraminifera
data = ff['data']
sites = np.array(['site %d'%(i) for i in range(data.shape[0])])
species = np.array(['species %d'%(i) for i in range(data.shape[1])])
if(not n):
args = np.where(Flabels!=-1)
data = data[args]
Flabels = Flabels[args]
sites = sites[args]
Fenv = Fenv[args]
Fenv_nn = Fenv_nn[args]
X = pd.DataFrame(data, sites, species)
Y = pd.DataFrame(Fenv, sites, envs)
Y_nn = pd.DataFrame(Fenv_nn, sites, envs)
# del Y['N']
del Y['Si']
# del Y['P']
#del Y['temp']
# del Y['salt']
if(len(Y.values)!=0):
if(Y.shape[0]>1):
CCA = cca(Y,X)
else:
FCluster[mini,xii] = np.nan
if(n):
FNoise[mini,xii] = np.sum(CCA.proportion_explained[:len(CCA.proportion_explained)//2])
else:
FCluster[mini,xii] = np.sum(CCA.proportion_explained[:len(CCA.proportion_explained)//2])
else:
FCluster[mini,xii] = np.nan
#%% Load the significant according to the subsamples
its = 999
siglevel = 0.05
if(redFspecies):
ffsig = np.load('randomsubsamples_redF_sp%d_its%d.npz'%(sp,its))
else:
ffsig = | np.load('randomsubsamples_sp%d_its%d.npz'%(sp,its)) | numpy.load |
# -*- coding: utf-8 -*-
from copy import deepcopy
import numpy as np
from mrinversion.linear_model._base_l1l2 import _get_augmented_data
from mrinversion.linear_model._base_l1l2 import _get_cv_indexes
def test01():
K = np.empty((8, 16))
indexes = _get_cv_indexes(K, 4, "lasso", f_shape=(4, 4))
index_test = [
[[1, 2, 3, 5, 6, 7], [0, 4]],
[[0, 1, 2, 4, 5, 6], [3, 7]],
[[0, 1, 3, 4, 5, 7], [2, 6]],
[[0, 2, 3, 4, 6, 7], [1, 5]],
]
assert indexes == index_test, "test01"
def test02():
K = np.empty((8, 16))
lst = [
8,
9,
10,
11,
12,
13,
14,
15,
16,
17,
18,
19,
20,
21,
22,
23,
24,
25,
26,
27,
28,
29,
30,
31,
]
index_test = [
[[1, 2, 3, 5, 6, 7], [0, 4]],
[[0, 1, 2, 4, 5, 6], [3, 7]],
[[0, 1, 3, 4, 5, 7], [2, 6]],
[[0, 2, 3, 4, 6, 7], [1, 5]],
]
indexes = _get_cv_indexes(K, folds=4, regularizer="smooth lasso", f_shape=(4, 4))
index_test_1 = deepcopy(index_test)
for tr_, _ in index_test_1:
tr_ += lst
assert indexes == index_test_1, "test02 - 1"
indexes = _get_cv_indexes(K, 4, "smooth lasso", f_shape=16)
index_test_2 = deepcopy(index_test)
for tr_, _ in index_test_2:
tr_ += lst[:15]
assert indexes == index_test_2, "test02 - 2"
def test03():
# 1d - explicit
K = np.empty((5, 5))
s = np.empty((5, 1))
KK, _ = _get_augmented_data(K, s, 1, "smooth lasso", f_shape=(5))
A = [[1, -1, 0, 0, 0], [0, 1, -1, 0, 0], [0, 0, 1, -1, 0], [0, 0, 0, 1, -1]]
assert np.allclose(KK[5:], A)
# 2d - explicit symmetric
K = np.empty((5, 4))
s = np.empty((5, 1))
KK, _ = _get_augmented_data(K, s, 1, "smooth lasso", f_shape=(2, 2))
J1 = [[1, 0, -1, 0], [0, 1, 0, -1]]
J2 = [[1, -1, 0, 0], [0, 0, 1, -1]]
assert np.allclose(KK[5:7], J1)
assert np.allclose(KK[7:9], J2)
# 2d - explicit asymmetric
K = np.empty((5, 6))
s = np.empty((5, 1))
KK, _ = _get_augmented_data(K, s, 1, "smooth lasso", f_shape=(3, 2))
J1 = [
[1, 0, -1, 0, 0, 0],
[0, 1, 0, -1, 0, 0],
[0, 0, 1, 0, -1, 0],
[0, 0, 0, 1, 0, -1],
]
J2 = [[1, -1, 0, 0, 0, 0], [0, 0, 1, -1, 0, 0], [0, 0, 0, 0, 1, -1]]
assert np.allclose(KK[5:9], J1)
assert np.allclose(KK[9:12], J2)
# 1d - function
K = np.empty((5, 12))
KK, _ = _get_augmented_data(K, s, 1, "smooth lasso", f_shape=(12))
A1 = (-1 * np.eye(12) + np.diag(np.ones(11), k=-1))[1:]
assert np.allclose(KK[5:], A1)
# 2d - function symmetric
K = np.empty((5, 16))
s = np.empty((5, 1))
KK, _ = _get_augmented_data(K, s, 1, "smooth lasso", f_shape=(4, 4))
J = -1 * np.eye(4) + np.diag(np.ones(3), k=-1)
I_eye = np.eye(4)
J1 = np.kron(J[1:], I_eye)
J2 = np.kron(I_eye, J[1:])
assert np.allclose(KK[5 : 5 + 12], J1)
assert np.allclose(KK[5 + 12 :], J2)
# 2d - function asymmetric
K = np.empty((5, 12))
KK, _ = _get_augmented_data(K, s, 1, "smooth lasso", f_shape=(4, 3))
A1 = -1 * np.eye(4) + np.diag(np.ones(3), k=-1)
I2 = np.eye(3)
J1 = np.kron(A1[1:], I2)
A2 = -1 * np.eye(3) + np.diag(np.ones(2), k=-1)
I1 = np.eye(4)
J2 = | np.kron(I1, A2[1:]) | numpy.kron |
from __future__ import print_function
import os
import numpy as np
import kaldi_io as ko
"""
Reads a kaldi scp file and slice the feature matrix
Jeff, 2018
"""
def tensor_cnn_frame(mat, M):
"""Construct a tensor of shape (C x H x W) given an utterance matrix
for CNN
"""
slice_mat = []
for index in np.arange(len(mat)):
if index < M:
to_left = np.tile(mat[index], M).reshape((M,-1))
rest = mat[index:index+M+1]
context = np.vstack((to_left, rest))
elif index >= len(mat)-M:
to_right = np.tile(mat[index], M).reshape((M,-1))
rest = mat[index-M:index+1]
context = np.vstack((rest, to_right))
else:
context = mat[index-M:index+M+1]
slice_mat.append(context)
slice_mat = np.array(slice_mat)
slice_mat = np.expand_dims(slice_mat, axis=1)
slice_mat = np.swapaxes(slice_mat, 2, 3)
return slice_mat
def tensor_cnn_utt(mat, max_len):
mat = np.swapaxes(mat, 0, 1)
repetition = int(max_len/mat.shape[1])
tensor = | np.tile(mat,repetition) | numpy.tile |
#!/usr/bin/env python2.7
# encoding: utf-8
"""
test_dmarket.py
Created by <NAME> on 2011-05-05.
Copyright (c) 2011 University of Strathclyde. All rights reserved.
"""
from __future__ import division
import sys
import os
import numpy as np
import scipy.integrate as integral
import scipy.stats.mstats as mstats
import scikits.statsmodels as sm
import matplotlib.pyplot as plt
from operator import itemgetter
from matplotlib import rc
rc('font',**{'family':'sans-serif','sans-serif':['Helvetica']})
## for Palatino and other serif fonts use:
#rc('font',**{'family':'serif','serif':['Palatino']))
rc('text', usetex=True)
def simulate(N, pmin, pmax):
'''This function simulates one stage of the auctioning game among
the network operators.
Keyword args:
N -- number of bidders
pmin -- minimum price/cost of the service
pmax -- maximum price/cost of the service
Returns: intersection (if exists) of the winning bidders for price weight->1
'''
# Service agent weights
w_range = 1000
w = np.linspace(0, 1, w_range)
# Calculate prices
costs = [ | np.random.uniform(pmin, pmax) | numpy.random.uniform |
from simglucose.simulation.scenario import Action, Scenario
import numpy as np
from scipy.stats import truncnorm, uniform
from datetime import datetime
import logging
logger = logging.getLogger(__name__)
class RandomScenario(Scenario):
def __init__(self, start_time, seed=None):
Scenario.__init__(self, start_time=start_time)
self.seed = seed
def get_action(self, t):
# t must be datetime.datetime object
delta_t = t - datetime.combine(t.date(), datetime.min.time())
t_sec = delta_t.total_seconds()
if t_sec < 1:
logger.info('Creating new one day scenario ...')
self.scenario = self.create_scenario()
t_min = np.floor(t_sec / 60.0)
if t_min in self.scenario['meal']['time']:
logger.info('Time for meal!')
idx = self.scenario['meal']['time'].index(t_min)
return Action(meal=self.scenario['meal']['amount'][idx])
else:
return Action(meal=0)
def create_scenario(self):
scenario = {'meal': {'time': [], 'amount': []}}
# Probability of taking each meal
# [breakfast, snack1, lunch, snack2, dinner, snack3]
prob = [0.95, 0.3, 0.95, 0.3, 0.95, 0.3]
time_lb = np.array([5, 9, 10, 14, 16, 20]) * 60
time_ub = np.array([9, 10, 14, 16, 20, 23]) * 60
time_mu = np.array([7, 9.5, 12, 15, 18, 21.5]) * 60
time_sigma = np.array([60, 30, 60, 30, 60, 30])
amount_mu = [45, 10, 70, 10, 80, 10]
amount_sigma = [10, 5, 10, 5, 10, 5]
for p, tlb, tub, tbar, tsd, mbar, msd in zip(prob, time_lb, time_ub,
time_mu, time_sigma,
amount_mu, amount_sigma):
if self.random_gen.rand() < p:
tmeal = np.round(
truncnorm.rvs(a=(tlb - tbar) / tsd,
b=(tub - tbar) / tsd,
loc=tbar,
scale=tsd,
random_state=self.random_gen))
scenario['meal']['time'].append(tmeal)
scenario['meal']['amount'].append(
max(round(self.random_gen.normal(mbar, msd)), 0))
return scenario
def reset(self):
self.random_gen = np.random.RandomState(self.seed)
self.scenario = self.create_scenario()
@property
def seed(self):
return self._seed
@seed.setter
def seed(self, seed):
self._seed = seed
self.reset()
def harris_benedict(weight, kind):
# (min, max, age, weight)
if kind == 'child':
child_weight_to_age_and_height = [(0, 25.6, 7, 121.9),
(25.6, 28.6, 8, 128),
(28.6, 32, 9, 133.3),
(32, 35.6, 10, 138.4),
(35.6, 39.9, 11, 143.5),
(39.9, np.infty, 12, 149.1)]
for tup in child_weight_to_age_and_height:
if weight > tup[0] and weight <= tup[1]:
age = tup[2]
height = tup[3]
elif kind == 'adolescent':
adolescent_weight_to_age_and_height = [(0, 50.8, 13, 156.2),
(50.8, 56.0, 14, 163.8),
(56.0, 60.8, 15, 170.1),
(60.8, 64.4, 16, 173.4),
(64.4, 66.9, 17, 175.2),
(66.9, 68.9, 18, 175.7),
(68.9, 70.3, 19, 176.5),
(70.3, np.infty, 20, 177)]
for tup in adolescent_weight_to_age_and_height:
if weight > tup[0] and weight <= tup[1]:
age = tup[2]
height = tup[3]
else:
age = 45
height = 177
bmr = 66.5 + (13.75 * weight) + (5.003 * height) - (6.755 * age)
total = ((1.2 * bmr) * 0.45) / 4
adj = 1.1 + 1.3 + 1.55 + 3 * 0.15 # from old carb calc
b_ratio = 1.1/adj
l_ratio = 1.3/adj
d_ratio = 1.55/adj
s_ratio = 0.15/adj
return (total*b_ratio, total*l_ratio, total*d_ratio, total*s_ratio)
class SemiRandomScenario(Scenario):
def __init__(self, start_time=None, seed=None, time_std_multiplier=1):
Scenario.__init__(self, start_time=start_time)
self.time_std_multiplier = time_std_multiplier
self.seed = seed
def get_action(self, t):
# t must be datetime.datetime object
delta_t = t - datetime.combine(t.date(), datetime.min.time())
t_sec = delta_t.total_seconds()
if t_sec < 1:
logger.info('Creating new one day scenario ...')
self.scenario = self.create_scenario()
t_min = np.floor(t_sec / 60.0)
if t_min in self.scenario['meal']['time']:
logger.info('Time for meal!')
idx = self.scenario['meal']['time'].index(t_min)
return Action(meal=self.scenario['meal']['amount'][idx])
else:
return Action(meal=0)
def create_scenario(self):
scenario = {'meal': {'time': [], 'amount': []}}
time_lb = np.array([5, 10, 16]) * 60
time_ub = np.array([10, 16, 22]) * 60
time_mu = np.array([7.5, 13, 19]) * 60
time_sigma = np.array([60, 60, 60]) * self.time_std_multiplier
amount = [45, 70 ,80]
for tlb, tub, tbar, tsd, mbar in zip(time_lb, time_ub, time_mu, time_sigma, amount):
tmeal = np.round(truncnorm.rvs(a=(tlb - tbar) / tsd,
b=(tub - tbar) / tsd,
loc=tbar,
scale=tsd,
random_state=self.random_gen))
scenario['meal']['time'].append(tmeal)
scenario['meal']['amount'].append(mbar)
return scenario
def reset(self):
self.random_gen = np.random.RandomState(self.seed)
self.scenario = self.create_scenario()
@property
def seed(self):
return self._seed
@seed.setter
def seed(self, seed):
self._seed = seed
self.reset()
class RandomBalancedScenario(Scenario):
def __init__(self, bw, start_time=None, seed=None, weekly=False, kind=None,
harrison_benedict=False, restricted=False, unrealistic=False, meal_duration=1,
deterministic_meal_size=False, deterministic_meal_time=False, deterministic_meal_occurrence=False):
Scenario.__init__(self, start_time=start_time)
self.kind = kind
self.bw = bw
self.day = 0
self.weekly = weekly
self.deterministic_meal_size = deterministic_meal_size
self.deterministic_meal_time = deterministic_meal_time
self.deterministic_meal_occurrence = deterministic_meal_occurrence
self.restricted = restricted # take precident over harrison_benedict
self.harrison_benedict = harrison_benedict
self.unrealistic = unrealistic
self.meal_duration = meal_duration
self.seed = seed
def get_action(self, t):
# t must be datetime.datetime object
delta_t = t - datetime.combine(t.date(), datetime.min.time())
t_sec = delta_t.total_seconds()
if t_sec < 1:
logger.info('Creating new one day scenario ...')
self.day = self.day % 7
if self.restricted:
self.scenario = self.create_scenario_restricted()
elif self.harrison_benedict:
self.scenario = self.create_scenario_harrison_benedict()
elif self.unrealistic:
self.scenario = self.create_scenario_unrealistic()
elif self.weekly:
if self.day == 5 or self.day == 6:
self.scenario = self.create_weekend_scenario()
else:
self.scenario = self.create_scenario()
else:
self.scenario = self.create_scenario()
t_min = np.floor(t_sec / 60.0)
# going to cancel overalpping meals by going with first meal in range
for idx, time in enumerate(self.scenario['meal']['time']):
if t_min>=time and t_min<time+self.meal_duration:
return Action(meal=self.scenario['meal']['amount'][idx]/self.meal_duration)
else:
return Action(meal=0)
def create_scenario_restricted(self):
scenario = {'meal': {'time': [], 'amount': []}}
# Probability of taking each meal
# [breakfast, snack1, lunch, snack2, dinner, snack3]
prob = [0.95, 0.3, 0.95, 0.3, 0.95, 0.3]
time_lb = np.array([5, 9, 10, 14, 16, 20]) * 60
time_ub = np.array([9, 10, 14, 16, 20, 23]) * 60
time_mu = np.array([7, 9.5, 12, 15, 18, 21.5]) * 60
time_sigma = np.array([60, 30, 60, 30, 60, 30])
if self.kind == 'child':
amount_lb = np.array([30, 10, 30, 10, 30, 10])
amount_ub = np.array([45, 20, 45, 20, 45, 20])
elif self.kind == 'adolescent':
amount_lb = np.array([45, 15, 45, 15, 45, 15])
amount_ub = np.array([60, 25, 60, 25, 60, 25])
elif self.kind == 'adult':
amount_lb = np.array([60, 20, 60, 20, 60, 20])
amount_ub = np.array([75, 30, 75, 30, 75, 30])
else:
raise ValueError('{} not a valid kind (child, adolescent, adult').format(self.kind)
amount_mu = np.array([0.7, 0.15, 1.1, 0.15, 1.25, 0.15]) * self.bw
amount_sigma = amount_mu * 0.15
for p, tlb, tub, tbar, tsd, mlb, mub, mbar, msd in zip(
prob, time_lb, time_ub, time_mu, time_sigma,
amount_lb, amount_ub, amount_mu, amount_sigma):
if self.random_gen.rand() < p:
tmeal = np.round(truncnorm.rvs(a=(tlb - tbar) / tsd,
b=(tub - tbar) / tsd,
loc=tbar,
scale=tsd,
random_state=self.random_gen))
scenario['meal']['time'].append(tmeal)
ameal = np.round(truncnorm.rvs(a=(mlb - mbar) / msd,
b=(mub - mbar) / msd,
loc=mbar,
scale=msd,
random_state=self.random_gen))
scenario['meal']['amount'].append(ameal)
return scenario
def create_scenario_harrison_benedict(self):
scenario = {'meal': {'time': [], 'amount': []}}
# Probability of taking each meal
# [breakfast, snack1, lunch, snack2, dinner, snack3]
prob = [0.95, 0.3, 0.95, 0.3, 0.95, 0.3]
time_lb = np.array([5, 9, 10, 14, 16, 20]) * 60
time_ub = np.array([9, 10, 14, 16, 20, 23]) * 60
time_mu = np.array([7, 9.5, 12, 15, 18, 21.5]) * 60
time_sigma = np.array([60, 30, 60, 30, 60, 30])
mu_b, mu_l, mu_d, mu_s = harris_benedict(self.bw, self.kind)
amount_mu = np.array([mu_b, mu_s, mu_l, mu_s, mu_d, mu_s])
amount_sigma = amount_mu * 0.15
for p, tlb, tub, tbar, tsd, mbar, msd in zip(
prob, time_lb, time_ub, time_mu, time_sigma, amount_mu, amount_sigma):
if self.random_gen.rand() < p:
tmeal = np.round(truncnorm.rvs(a=(tlb - tbar) / tsd,
b=(tub - tbar) / tsd,
loc=tbar,
scale=tsd,
random_state=self.random_gen))
scenario['meal']['time'].append(tmeal)
ameal = np.round(self.random_gen.normal(mbar, msd))
scenario['meal']['amount'].append(ameal)
return scenario
def create_scenario_unrealistic(self):
scenario = {'meal': {'time': [], 'amount': []}}
# Probability of taking each meal
# [breakfast, lunch, dinner]
prob = [1, 1, 1]
time_lb = np.array([8, 12, 18]) * 60
time_ub = np.array([9, 15, 19]) * 60
time_mu = np.array([9, 14, 19]) * 60
time_sigma = np.array([0, 0, 0])
amount_mu = np.array([50, 80, 60])
amount_sigma = np.array([0, 0, 0])
for p, tlb, tub, tbar, tsd, mbar, msd in zip(
prob, time_lb, time_ub, time_mu, time_sigma,
amount_mu, amount_sigma):
if self.random_gen.rand() < p:
tmeal = np.round(truncnorm.rvs(a=-np.infty,
b=np.infty,
loc=tbar,
scale=tsd,
random_state=self.random_gen))
scenario['meal']['time'].append(tmeal)
ameal = np.round(truncnorm.rvs(a=-np.infty,
b=np.infty,
loc=mbar,
scale=msd,
random_state=self.random_gen))
scenario['meal']['amount'].append(ameal)
return scenario
def create_weekend_scenario(self):
scenario = {'meal': {'time': [], 'amount': []}}
# Probability of taking each meal
# [breakfast, snack1, lunch, snack2, dinner, snack3]
prob = [0.95, 0.3, 0.95, 0.3, 0.95, 0.3]
time_lb = np.array([7, 11, 12, 15, 18, 22]) * 60
time_ub = np.array([11, 12, 15, 16, 22, 23]) * 60
time_mu = | np.array([9, 11.5, 13.5, 15.5, 21, 22.5]) | numpy.array |
import numpy as np
from numpy.testing import assert_allclose
import pytest
from .. import SphericalDust, IsotropicDust
from ...util.functions import random_id, B_nu
def test_missing_properties(tmpdir):
d = SphericalDust()
with pytest.raises(Exception) as e:
d.write(tmpdir.join(random_id()).strpath)
assert e.value.args[0] == "The following attributes of the optical properties have not been set: nu, chi, albedo, mu, P1, P2, P3, P4"
class TestSphericalDust(object):
def setup_method(self, method):
self.dust = SphericalDust()
self.dust.optical_properties.nu = np.logspace(0., 20., 100)
self.dust.optical_properties.albedo = np.repeat(0.5, 100)
self.dust.optical_properties.chi = np.ones(100)
self.dust.optical_properties.mu = [-1., 1.]
self.dust.optical_properties.initialize_scattering_matrix()
self.dust.optical_properties.P1[:, :] = 1.
self.dust.optical_properties.P2[:, :] = 0.
self.dust.optical_properties.P3[:, :] = 1.
self.dust.optical_properties.P4[:, :] = 0.
def test_helpers(self):
nu = np.logspace(5., 15., 1000)
# Here we don't set the mean opacities to make sure they are computed
# automatically
assert_allclose(self.dust.kappa_nu_temperature(34.),
self.dust.kappa_nu_spectrum(nu, B_nu(nu, 34)))
assert_allclose(self.dust.chi_nu_temperature(34.),
self.dust.chi_nu_spectrum(nu, B_nu(nu, 34)))
def test_conversions_out_of_bounds(self):
np.random.seed(12345)
self.dust.mean_opacities.compute(self.dust.optical_properties,
n_temp=10, temp_min=1., temp_max=1000.)
# Test with scalars
assert_allclose(self.dust.temperature2specific_energy(0.1),
self.dust.mean_opacities.specific_energy[0])
assert_allclose(self.dust.temperature2specific_energy(1e4),
self.dust.mean_opacities.specific_energy[-1])
# Test with arrays
assert_allclose(self.dust.temperature2specific_energy(np.array([0.1])),
self.dust.mean_opacities.specific_energy[0])
assert_allclose(self.dust.temperature2specific_energy(np.array([1e4])),
self.dust.mean_opacities.specific_energy[-1])
# Test with scalars
assert_allclose(self.dust.specific_energy2temperature(1.e-10),
self.dust.mean_opacities.temperature[0])
assert_allclose(self.dust.specific_energy2temperature(1.e+10),
self.dust.mean_opacities.temperature[-1])
# Test with arrays
assert_allclose(self.dust.specific_energy2temperature(np.array([1.e-10])),
self.dust.mean_opacities.temperature[0])
assert_allclose(self.dust.specific_energy2temperature(np.array([1.e+10])),
self.dust.mean_opacities.temperature[-1])
def test_conversions_roundtrip(self):
np.random.seed(12345)
# Here we don't set the mean opacities to make sure they are computed
# automatically
temperatures1 = 10. ** np.random.uniform(0., 3., 100)
specific_energies = self.dust.temperature2specific_energy(temperatures1)
temperatures2 = self.dust.specific_energy2temperature(specific_energies)
assert_allclose(temperatures1, temperatures2)
def test_set_lte(self):
np.random.seed(12345)
# Here we don't set the mean opacities to make sure they are computed
# automatically
self.dust.set_lte_emissivities(n_temp=10, temp_min=1., temp_max=1000.)
assert_allclose(self.dust.mean_opacities.temperature[0], 1)
assert_allclose(self.dust.mean_opacities.temperature[-1], 1000)
assert_allclose(self.dust.mean_opacities.specific_energy,
self.dust.emissivities.var)
def test_plot(self, tmpdir):
# Here we don't set the mean opacities or the emissivities to make sure
# they are computed automatically
self.dust.plot(tmpdir.join('test.png').strpath)
def test_hash(self, tmpdir):
# Here we don't set the mean opacities or the emissivities to make sure
# they are computed automatically
try:
assert self.dust.hash() == 'ace50d004550889b0e739db8ec8f10fb'
except AssertionError: # On MacOS X, the hash is sometimes different
assert self.dust.hash() == 'c5765806a1b59b527420444c4355ac41'
def test_io(self, tmpdir):
filename = tmpdir.join('test.hdf5').strpath
self.dust.write(filename)
dust2 = SphericalDust(filename)
assert_allclose(self.dust.optical_properties.nu, dust2.optical_properties.nu)
assert_allclose(self.dust.optical_properties.chi, dust2.optical_properties.chi)
assert_allclose(self.dust.optical_properties.albedo, dust2.optical_properties.albedo)
assert_allclose(self.dust.optical_properties.mu, dust2.optical_properties.mu)
assert_allclose(self.dust.optical_properties.P1, dust2.optical_properties.P1)
assert_allclose(self.dust.optical_properties.P2, dust2.optical_properties.P2)
assert_allclose(self.dust.optical_properties.P3, dust2.optical_properties.P3)
assert_allclose(self.dust.optical_properties.P4, dust2.optical_properties.P4)
assert_allclose(self.dust.mean_opacities.temperature, dust2.mean_opacities.temperature)
assert_allclose(self.dust.mean_opacities.specific_energy, dust2.mean_opacities.specific_energy)
assert_allclose(self.dust.mean_opacities.chi_planck, dust2.mean_opacities.chi_planck)
assert_allclose(self.dust.mean_opacities.kappa_planck, dust2.mean_opacities.kappa_planck)
assert_allclose(self.dust.mean_opacities.chi_inv_planck, dust2.mean_opacities.chi_inv_planck)
assert_allclose(self.dust.mean_opacities.kappa_inv_planck, dust2.mean_opacities.kappa_inv_planck)
assert_allclose(self.dust.mean_opacities.chi_rosseland, dust2.mean_opacities.chi_rosseland)
assert_allclose(self.dust.mean_opacities.kappa_rosseland, dust2.mean_opacities.kappa_rosseland)
assert self.dust.emissivities.is_lte == dust2.emissivities.is_lte
assert self.dust.emissivities.var_name == dust2.emissivities.var_name
assert_allclose(self.dust.emissivities.nu, dust2.emissivities.nu)
assert_allclose(self.dust.emissivities.var, dust2.emissivities.var)
assert_allclose(self.dust.emissivities.jnu, dust2.emissivities.jnu)
assert self.dust.sublimation_mode == dust2.sublimation_mode
assert_allclose(self.dust.sublimation_energy, dust2.sublimation_energy)
def test_isotropic_dust():
nu = np.logspace(0., 20., 100)
albedo = np.repeat(0.5, 100)
chi = | np.ones(100) | numpy.ones |
# Lint as: python3
# Copyright 2019 Google LLC
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""Tests for bond_curve."""
import math
from absl.testing import parameterized
import numpy as np
import tensorflow.compat.v2 as tf
from tensorflow.python.framework import test_util # pylint: disable=g-direct-tensorflow-import
from tf_quant_finance.rates.hagan_west import bond_curve
from tf_quant_finance.rates.hagan_west import monotone_convex
@test_util.run_all_in_graph_and_eager_modes
class BondCurveTest(tf.test.TestCase, parameterized.TestCase):
@parameterized.named_parameters(
('single_precision', np.float32),
('double_precision', np.float64),
)
def test_cashflow_times_cashflow_before_settelment_error(self, dtype):
with self.assertRaises(tf.errors.InvalidArgumentError):
self.evaluate(
bond_curve.bond_curve(
bond_cashflows=[
np.array([12.5, 12.5, 12.5, 1012.5], dtype=dtype),
np.array([30.0, 30.0, 30.0, 1030.0], dtype=dtype)
],
bond_cashflow_times=[
np.array([0.25, 0.5, 0.75, 1.0], dtype=dtype),
np.array([0.5, 1.0, 1.5, 2.0], dtype=dtype)
],
present_values=np.array([999.0, 1022.0], dtype=dtype),
present_values_settlement_times=np.array([0.25, 0.25],
dtype=dtype),
validate_args=True,
dtype=dtype))
@parameterized.named_parameters(
('single_precision', np.float32),
('double_precision', np.float64),
)
def test_cashflow_times_are_strongly_ordered_error(self, dtype):
with self.assertRaises(tf.errors.InvalidArgumentError):
self.evaluate(
bond_curve.bond_curve(
bond_cashflows=[
np.array([12.5, 12.5, 12.5, 1012.5], dtype=dtype),
| np.array([30.0, 30.0, 30.0, 1030.0], dtype=dtype) | numpy.array |
# This module has been generated automatically from space group information
# obtained from the Computational Crystallography Toolbox
#
"""
Space groups
This module contains a list of all the 230 space groups that can occur in
a crystal. The variable space_groups contains a dictionary that maps
space group numbers and space group names to the corresponding space
group objects.
.. moduleauthor:: <NAME> <<EMAIL>>
"""
#-----------------------------------------------------------------------------
# Copyright (C) 2013 The Mosaic Development Team
#
# Distributed under the terms of the BSD License. The full license is in
# the file LICENSE.txt, distributed as part of this software.
#-----------------------------------------------------------------------------
import numpy as N
class SpaceGroup(object):
"""
Space group
All possible space group objects are created in this module. Other
modules should access these objects through the dictionary
space_groups rather than create their own space group objects.
"""
def __init__(self, number, symbol, transformations):
"""
:param number: the number assigned to the space group by
international convention
:type number: int
:param symbol: the Hermann-Mauguin space-group symbol as used
in PDB and mmCIF files
:type symbol: str
:param transformations: a list of space group transformations,
each consisting of a tuple of three
integer arrays (rot, tn, td), where
rot is the rotation matrix and tn/td
are the numerator and denominator of the
translation vector. The transformations
are defined in fractional coordinates.
:type transformations: list
"""
self.number = number
self.symbol = symbol
self.transformations = transformations
self.transposed_rotations = N.array([N.transpose(t[0])
for t in transformations])
self.phase_factors = N.exp(N.array([(-2j*N.pi*t[1])/t[2]
for t in transformations]))
def __repr__(self):
return "SpaceGroup(%d, %s)" % (self.number, repr(self.symbol))
def __len__(self):
"""
:return: the number of space group transformations
:rtype: int
"""
return len(self.transformations)
def symmetryEquivalentMillerIndices(self, hkl):
"""
:param hkl: a set of Miller indices
:type hkl: Scientific.N.array_type
:return: a tuple (miller_indices, phase_factor) of two arrays
of length equal to the number of space group
transformations. miller_indices contains the Miller
indices of each reflection equivalent by symmetry to the
reflection hkl (including hkl itself as the first element).
phase_factor contains the phase factors that must be applied
to the structure factor of reflection hkl to obtain the
structure factor of the symmetry equivalent reflection.
:rtype: tuple
"""
hkls = N.dot(self.transposed_rotations, hkl)
p = N.multiply.reduce(self.phase_factors**hkl, -1)
return hkls, p
space_groups = {}
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(1, 'P 1', transformations)
space_groups[1] = sg
space_groups['P 1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(2, 'P -1', transformations)
space_groups[2] = sg
space_groups['P -1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(3, 'P 1 2 1', transformations)
space_groups[3] = sg
space_groups['P 1 2 1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(4, 'P 1 21 1', transformations)
space_groups[4] = sg
space_groups['P 1 21 1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(5, 'C 1 2 1', transformations)
space_groups[5] = sg
space_groups['C 1 2 1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(6, 'P 1 m 1', transformations)
space_groups[6] = sg
space_groups['P 1 m 1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(7, 'P 1 c 1', transformations)
space_groups[7] = sg
space_groups['P 1 c 1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(8, 'C 1 m 1', transformations)
space_groups[8] = sg
space_groups['C 1 m 1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(9, 'C 1 c 1', transformations)
space_groups[9] = sg
space_groups['C 1 c 1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(10, 'P 1 2/m 1', transformations)
space_groups[10] = sg
space_groups['P 1 2/m 1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(11, 'P 1 21/m 1', transformations)
space_groups[11] = sg
space_groups['P 1 21/m 1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(12, 'C 1 2/m 1', transformations)
space_groups[12] = sg
space_groups['C 1 2/m 1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(13, 'P 1 2/c 1', transformations)
space_groups[13] = sg
space_groups['P 1 2/c 1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,-1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(14, 'P 1 21/c 1', transformations)
space_groups[14] = sg
space_groups['P 1 21/c 1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(15, 'C 1 2/c 1', transformations)
space_groups[15] = sg
space_groups['C 1 2/c 1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(16, 'P 2 2 2', transformations)
space_groups[16] = sg
space_groups['P 2 2 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(17, 'P 2 2 21', transformations)
space_groups[17] = sg
space_groups['P 2 2 21'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(18, 'P 21 21 2', transformations)
space_groups[18] = sg
space_groups['P 21 21 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(19, 'P 21 21 21', transformations)
space_groups[19] = sg
space_groups['P 21 21 21'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(20, 'C 2 2 21', transformations)
space_groups[20] = sg
space_groups['C 2 2 21'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(21, 'C 2 2 2', transformations)
space_groups[21] = sg
space_groups['C 2 2 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(22, 'F 2 2 2', transformations)
space_groups[22] = sg
space_groups['F 2 2 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(23, 'I 2 2 2', transformations)
space_groups[23] = sg
space_groups['I 2 2 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(24, 'I 21 21 21', transformations)
space_groups[24] = sg
space_groups['I 21 21 21'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(25, 'P m m 2', transformations)
space_groups[25] = sg
space_groups['P m m 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(26, 'P m c 21', transformations)
space_groups[26] = sg
space_groups['P m c 21'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(27, 'P c c 2', transformations)
space_groups[27] = sg
space_groups['P c c 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(28, 'P m a 2', transformations)
space_groups[28] = sg
space_groups['P m a 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(29, 'P c a 21', transformations)
space_groups[29] = sg
space_groups['P c a 21'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(30, 'P n c 2', transformations)
space_groups[30] = sg
space_groups['P n c 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(31, 'P m n 21', transformations)
space_groups[31] = sg
space_groups['P m n 21'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(32, 'P b a 2', transformations)
space_groups[32] = sg
space_groups['P b a 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(33, 'P n a 21', transformations)
space_groups[33] = sg
space_groups['P n a 21'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(34, 'P n n 2', transformations)
space_groups[34] = sg
space_groups['P n n 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(35, 'C m m 2', transformations)
space_groups[35] = sg
space_groups['C m m 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(36, 'C m c 21', transformations)
space_groups[36] = sg
space_groups['C m c 21'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(37, 'C c c 2', transformations)
space_groups[37] = sg
space_groups['C c c 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(38, 'A m m 2', transformations)
space_groups[38] = sg
space_groups['A m m 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(39, 'A b m 2', transformations)
space_groups[39] = sg
space_groups['A b m 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(40, 'A m a 2', transformations)
space_groups[40] = sg
space_groups['A m a 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(41, 'A b a 2', transformations)
space_groups[41] = sg
space_groups['A b a 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(42, 'F m m 2', transformations)
space_groups[42] = sg
space_groups['F m m 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,3,3])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,3,3])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([3,1,3])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([3,1,3])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([3,3,1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([3,3,1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(43, 'F d d 2', transformations)
space_groups[43] = sg
space_groups['F d d 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(44, 'I m m 2', transformations)
space_groups[44] = sg
space_groups['I m m 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(45, 'I b a 2', transformations)
space_groups[45] = sg
space_groups['I b a 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(46, 'I m a 2', transformations)
space_groups[46] = sg
space_groups['I m a 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(47, 'P m m m', transformations)
space_groups[47] = sg
space_groups['P m m m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,-1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,-1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(48, 'P n n n :2', transformations)
space_groups[48] = sg
space_groups['P n n n :2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(49, 'P c c m', transformations)
space_groups[49] = sg
space_groups['P c c m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(50, 'P b a n :2', transformations)
space_groups[50] = sg
space_groups['P b a n :2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(51, 'P m m a', transformations)
space_groups[51] = sg
space_groups['P m m a'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,-1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(52, 'P n n a', transformations)
space_groups[52] = sg
space_groups['P n n a'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,-1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,-1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(53, 'P m n a', transformations)
space_groups[53] = sg
space_groups['P m n a'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,-1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(54, 'P c c a', transformations)
space_groups[54] = sg
space_groups['P c c a'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(55, 'P b a m', transformations)
space_groups[55] = sg
space_groups['P b a m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,-1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,-1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(56, 'P c c n', transformations)
space_groups[56] = sg
space_groups['P c c n'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,-1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(57, 'P b c m', transformations)
space_groups[57] = sg
space_groups['P b c m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(58, 'P n n m', transformations)
space_groups[58] = sg
space_groups['P n n m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(59, 'P m m n :2', transformations)
space_groups[59] = sg
space_groups['P m m n :2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(60, 'P b c n', transformations)
space_groups[60] = sg
space_groups['P b c n'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,-1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,-1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(61, 'P b c a', transformations)
space_groups[61] = sg
space_groups['P b c a'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,-1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(62, 'P n m a', transformations)
space_groups[62] = sg
space_groups['P n m a'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(63, 'C m c m', transformations)
space_groups[63] = sg
space_groups['C m c m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,-1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,-1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,-1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,-1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(64, 'C m c a', transformations)
space_groups[64] = sg
space_groups['C m c a'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(65, 'C m m m', transformations)
space_groups[65] = sg
space_groups['C m m m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(66, 'C c c m', transformations)
space_groups[66] = sg
space_groups['C c c m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(67, 'C m m a', transformations)
space_groups[67] = sg
space_groups['C m m a'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,-1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,-1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(68, 'C c c a :2', transformations)
space_groups[68] = sg
space_groups['C c c a :2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(69, 'F m m m', transformations)
space_groups[69] = sg
space_groups['F m m m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([4,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([4,4,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,-1])
trans_den = N.array([1,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,-1])
trans_den = N.array([4,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([4,4,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,3,3])
trans_den = N.array([1,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,3])
trans_den = N.array([4,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,3,1])
trans_den = N.array([4,4,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,1,1])
trans_den = N.array([4,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,1,1])
trans_den = N.array([4,4,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,3])
trans_den = N.array([2,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([3,0,3])
trans_den = N.array([4,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([3,1,1])
trans_den = N.array([4,4,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,-1,1])
trans_den = N.array([2,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([4,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,-1,1])
trans_den = N.array([4,4,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,3,1])
trans_den = N.array([2,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([3,1,1])
trans_den = N.array([4,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([3,3,0])
trans_den = N.array([4,4,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,-1])
trans_den = N.array([2,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,-1])
trans_den = N.array([4,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([4,4,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(70, 'F d d d :2', transformations)
space_groups[70] = sg
space_groups['F d d d :2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(71, 'I m m m', transformations)
space_groups[71] = sg
space_groups['I m m m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(72, 'I b a m', transformations)
space_groups[72] = sg
space_groups['I b a m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(73, 'I b c a', transformations)
space_groups[73] = sg
space_groups['I b c a'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(74, 'I m m a', transformations)
space_groups[74] = sg
space_groups['I m m a'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(75, 'P 4', transformations)
space_groups[75] = sg
space_groups['P 4'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,3])
trans_den = N.array([1,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(76, 'P 41', transformations)
space_groups[76] = sg
space_groups['P 41'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(77, 'P 42', transformations)
space_groups[77] = sg
space_groups['P 42'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,3])
trans_den = N.array([1,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(78, 'P 43', transformations)
space_groups[78] = sg
space_groups['P 43'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(79, 'I 4', transformations)
space_groups[79] = sg
space_groups['I 4'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,3])
trans_den = N.array([2,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,3])
trans_den = N.array([2,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,5])
trans_den = N.array([1,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,5])
trans_den = N.array([1,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(80, 'I 41', transformations)
space_groups[80] = sg
space_groups['I 41'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(81, 'P -4', transformations)
space_groups[81] = sg
space_groups['P -4'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(82, 'I -4', transformations)
space_groups[82] = sg
space_groups['I -4'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(83, 'P 4/m', transformations)
space_groups[83] = sg
space_groups['P 4/m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(84, 'P 42/m', transformations)
space_groups[84] = sg
space_groups['P 42/m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(85, 'P 4/n :2', transformations)
space_groups[85] = sg
space_groups['P 4/n :2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,-1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,-1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(86, 'P 42/n :2', transformations)
space_groups[86] = sg
space_groups['P 42/n :2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(87, 'I 4/m', transformations)
space_groups[87] = sg
space_groups['I 4/m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,3,3])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-3,-3])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([3,5,5])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([3,3,3])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,-1,-1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(88, 'I 41/a :2', transformations)
space_groups[88] = sg
space_groups['I 41/a :2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(89, 'P 4 2 2', transformations)
space_groups[89] = sg
space_groups['P 4 2 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(90, 'P 4 21 2', transformations)
space_groups[90] = sg
space_groups['P 4 21 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,3])
trans_den = N.array([1,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,3])
trans_den = N.array([1,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,4])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(91, 'P 41 2 2', transformations)
space_groups[91] = sg
space_groups['P 41 2 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,3])
trans_den = N.array([2,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,3])
trans_den = N.array([2,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(92, 'P 41 21 2', transformations)
space_groups[92] = sg
space_groups['P 41 21 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(93, 'P 42 2 2', transformations)
space_groups[93] = sg
space_groups['P 42 2 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(94, 'P 42 21 2', transformations)
space_groups[94] = sg
space_groups['P 42 21 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,3])
trans_den = N.array([1,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,3])
trans_den = N.array([1,1,4])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(95, 'P 43 2 2', transformations)
space_groups[95] = sg
space_groups['P 43 2 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,3])
trans_den = N.array([2,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,3])
trans_den = N.array([2,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(96, 'P 43 21 2', transformations)
space_groups[96] = sg
space_groups['P 43 21 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(97, 'I 4 2 2', transformations)
space_groups[97] = sg
space_groups['I 4 2 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,3])
trans_den = N.array([2,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,3])
trans_den = N.array([2,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,3])
trans_den = N.array([2,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,3])
trans_den = N.array([2,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,5])
trans_den = N.array([1,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,5])
trans_den = N.array([1,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,5])
trans_den = N.array([1,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,5])
trans_den = N.array([1,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(98, 'I 41 2 2', transformations)
space_groups[98] = sg
space_groups['I 41 2 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(99, 'P 4 m m', transformations)
space_groups[99] = sg
space_groups['P 4 m m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(100, 'P 4 b m', transformations)
space_groups[100] = sg
space_groups['P 4 b m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(101, 'P 42 c m', transformations)
space_groups[101] = sg
space_groups['P 42 c m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(102, 'P 42 n m', transformations)
space_groups[102] = sg
space_groups['P 42 n m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(103, 'P 4 c c', transformations)
space_groups[103] = sg
space_groups['P 4 c c'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(104, 'P 4 n c', transformations)
space_groups[104] = sg
space_groups['P 4 n c'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(105, 'P 42 m c', transformations)
space_groups[105] = sg
space_groups['P 42 m c'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(106, 'P 42 b c', transformations)
space_groups[106] = sg
space_groups['P 42 b c'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(107, 'I 4 m m', transformations)
space_groups[107] = sg
space_groups['I 4 m m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(108, 'I 4 c m', transformations)
space_groups[108] = sg
space_groups['I 4 c m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,3])
trans_den = N.array([2,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,3])
trans_den = N.array([2,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,3])
trans_den = N.array([2,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,3])
trans_den = N.array([2,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,5])
trans_den = N.array([1,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,5])
trans_den = N.array([1,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,5])
trans_den = N.array([1,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,5])
trans_den = N.array([1,2,4])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(109, 'I 41 m d', transformations)
space_groups[109] = sg
space_groups['I 41 m d'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,3])
trans_den = N.array([2,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,3])
trans_den = N.array([2,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,5])
trans_den = N.array([1,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,5])
trans_den = N.array([1,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,3])
trans_den = N.array([1,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,3])
trans_den = N.array([1,2,4])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(110, 'I 41 c d', transformations)
space_groups[110] = sg
space_groups['I 41 c d'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(111, 'P -4 2 m', transformations)
space_groups[111] = sg
space_groups['P -4 2 m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(112, 'P -4 2 c', transformations)
space_groups[112] = sg
space_groups['P -4 2 c'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(113, 'P -4 21 m', transformations)
space_groups[113] = sg
space_groups['P -4 21 m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(114, 'P -4 21 c', transformations)
space_groups[114] = sg
space_groups['P -4 21 c'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(115, 'P -4 m 2', transformations)
space_groups[115] = sg
space_groups['P -4 m 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(116, 'P -4 c 2', transformations)
space_groups[116] = sg
space_groups['P -4 c 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(117, 'P -4 b 2', transformations)
space_groups[117] = sg
space_groups['P -4 b 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(118, 'P -4 n 2', transformations)
space_groups[118] = sg
space_groups['P -4 n 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(119, 'I -4 m 2', transformations)
space_groups[119] = sg
space_groups['I -4 m 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(120, 'I -4 c 2', transformations)
space_groups[120] = sg
space_groups['I -4 c 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(121, 'I -4 2 m', transformations)
space_groups[121] = sg
space_groups['I -4 2 m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,3])
trans_den = N.array([2,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,3])
trans_den = N.array([2,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,3])
trans_den = N.array([2,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,3])
trans_den = N.array([2,1,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,5])
trans_den = N.array([1,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,5])
trans_den = N.array([1,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,5])
trans_den = N.array([1,2,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,5])
trans_den = N.array([1,2,4])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(122, 'I -4 2 d', transformations)
space_groups[122] = sg
space_groups['I -4 2 d'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(123, 'P 4/m m m', transformations)
space_groups[123] = sg
space_groups['P 4/m m m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(124, 'P 4/m c c', transformations)
space_groups[124] = sg
space_groups['P 4/m c c'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(125, 'P 4/n b m :2', transformations)
space_groups[125] = sg
space_groups['P 4/n b m :2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,-1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,-1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(126, 'P 4/n n c :2', transformations)
space_groups[126] = sg
space_groups['P 4/n n c :2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(127, 'P 4/m b m', transformations)
space_groups[127] = sg
space_groups['P 4/m b m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(128, 'P 4/m n c', transformations)
space_groups[128] = sg
space_groups['P 4/m n c'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(129, 'P 4/n m m :2', transformations)
space_groups[129] = sg
space_groups['P 4/n m m :2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,-1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,-1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(130, 'P 4/n c c :2', transformations)
space_groups[130] = sg
space_groups['P 4/n c c :2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(131, 'P 42/m m c', transformations)
space_groups[131] = sg
space_groups['P 42/m m c'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(132, 'P 42/m c m', transformations)
space_groups[132] = sg
space_groups['P 42/m c m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,-1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,-1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(133, 'P 42/n b c :2', transformations)
space_groups[133] = sg
space_groups['P 42/n b c :2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,-1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,-1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,-1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,-1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(134, 'P 42/n n m :2', transformations)
space_groups[134] = sg
space_groups['P 42/n n m :2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(135, 'P 42/m b c', transformations)
space_groups[135] = sg
space_groups['P 42/m b c'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(136, 'P 42/m n m', transformations)
space_groups[136] = sg
space_groups['P 42/m n m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,-1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,-1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(137, 'P 42/n m c :2', transformations)
space_groups[137] = sg
space_groups['P 42/n m c :2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,-1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,-1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,-1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,-1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(138, 'P 42/n c m :2', transformations)
space_groups[138] = sg
space_groups['P 42/n c m :2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(139, 'I 4/m m m', transformations)
space_groups[139] = sg
space_groups['I 4/m m m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(140, 'I 4/m c m', transformations)
space_groups[140] = sg
space_groups['I 4/m c m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,3,1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,3])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,3,1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,3])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-3,-1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-3])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-3,-1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-3])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([3,5,3])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([3,3,5])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([3,5,3])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([3,3,5])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,-1,1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,-1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,-1,1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,-1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(141, 'I 41/a m d :2', transformations)
space_groups[141] = sg
space_groups['I 41/a m d :2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,3,1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,3])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,3,3])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-3,-1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-3])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,0,0])
trans_den = N.array([2,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,-1,0])
trans_den = N.array([1,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-3,-3])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([-1,-1,-1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([3,5,3])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([3,3,5])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([3,5,5])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([3,3,3])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([2,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,-1,1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,1,-1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,0])
trans_den = N.array([2,2,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,1,1])
trans_den = N.array([1,2,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,0,1])
trans_den = N.array([2,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,-1,-1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,1,1])
trans_den = N.array([4,4,4])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(142, 'I 41/a c d :2', transformations)
space_groups[142] = sg
space_groups['I 41/a c d :2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(143, 'P 3', transformations)
space_groups[143] = sg
space_groups['P 3'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,2])
trans_den = N.array([1,1,3])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(144, 'P 31', transformations)
space_groups[144] = sg
space_groups['P 31'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,2])
trans_den = N.array([1,1,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,3])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(145, 'P 32', transformations)
space_groups[145] = sg
space_groups['P 32'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(146, 'R 3 :H', transformations)
space_groups[146] = sg
space_groups['R 3 :H'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(147, 'P -3', transformations)
space_groups[147] = sg
space_groups['P -3'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(148, 'R -3 :H', transformations)
space_groups[148] = sg
space_groups['R -3 :H'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,1,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(149, 'P 3 1 2', transformations)
space_groups[149] = sg
space_groups['P 3 1 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(150, 'P 3 2 1', transformations)
space_groups[150] = sg
space_groups['P 3 2 1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,2])
trans_den = N.array([1,1,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,2])
trans_den = N.array([1,1,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,1,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(151, 'P 31 1 2', transformations)
space_groups[151] = sg
space_groups['P 31 1 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,2])
trans_den = N.array([1,1,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,2])
trans_den = N.array([1,1,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(152, 'P 31 2 1', transformations)
space_groups[152] = sg
space_groups['P 31 2 1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,2])
trans_den = N.array([1,1,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,2])
trans_den = N.array([1,1,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,1,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(153, 'P 32 1 2', transformations)
space_groups[153] = sg
space_groups['P 32 1 2'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,2])
trans_den = N.array([1,1,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,2])
trans_den = N.array([1,1,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(154, 'P 32 2 1', transformations)
space_groups[154] = sg
space_groups['P 32 2 1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(155, 'R 3 2 :H', transformations)
space_groups[155] = sg
space_groups['R 3 2 :H'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(156, 'P 3 m 1', transformations)
space_groups[156] = sg
space_groups['P 3 m 1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,-1,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(157, 'P 3 1 m', transformations)
space_groups[157] = sg
space_groups['P 3 1 m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(158, 'P 3 c 1', transformations)
space_groups[158] = sg
space_groups['P 3 c 1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,-1,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(159, 'P 3 1 c', transformations)
space_groups[159] = sg
space_groups['P 3 1 c'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(160, 'R 3 m :H', transformations)
space_groups[160] = sg
space_groups['R 3 m :H'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,7])
trans_den = N.array([3,3,6])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,7])
trans_den = N.array([3,3,6])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,7])
trans_den = N.array([3,3,6])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,5])
trans_den = N.array([3,3,6])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,5])
trans_den = N.array([3,3,6])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,5])
trans_den = N.array([3,3,6])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(161, 'R 3 c :H', transformations)
space_groups[161] = sg
space_groups['R 3 c :H'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,1,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,-1,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(162, 'P -3 1 m', transformations)
space_groups[162] = sg
space_groups['P -3 1 m'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,0,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,1,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,0,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,-1,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(163, 'P -3 1 c', transformations)
space_groups[163] = sg
space_groups['P -3 1 c'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(164, 'P -3 m 1', transformations)
space_groups[164] = sg
space_groups['P -3 m 1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(165, 'P -3 c 1', transformations)
space_groups[165] = sg
space_groups['P -3 c 1'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([2,1,1])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
sg = SpaceGroup(166, 'R -3 m :H', transformations)
space_groups[166] = sg
space_groups['R -3 m :H'] = sg
transformations = []
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,0,0,0,-1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,1,0,-1,1,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,-1,0,1,0,0,0,0,-1])
rot.shape = (3, 3)
trans_num = N.array([0,0,0])
trans_den = N.array([1,1,1])
transformations.append((rot, trans_num, trans_den))
rot = N.array([-1,1,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,-1,0,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([0,0,-1])
trans_den = N.array([1,1,2])
transformations.append((rot, trans_num, trans_den))
rot = N.array([1,0,0,0,1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = N.array([3,3,3])
transformations.append((rot, trans_num, trans_den))
rot = N.array([0,-1,0,1,-1,0,0,0,1])
rot.shape = (3, 3)
trans_num = N.array([1,2,2])
trans_den = | N.array([3,3,3]) | numpy.array |
# ##############################################################################
# linalg.py
# =========
# Author : <NAME> [<EMAIL>]
# ##############################################################################
"""
Linear algebra routines.
"""
import numpy as np
import scipy.linalg as linalg
import imot_tools.util.argcheck as chk
@chk.check(
dict(
A=chk.accept_any(chk.has_reals, chk.has_complex),
B=chk.allow_None(chk.accept_any(chk.has_reals, chk.has_complex)),
tau=chk.is_real,
N=chk.allow_None(chk.is_integer),
)
)
def eigh(A, B=None, tau=1, N=None):
"""
Solve a generalized eigenvalue problem.
Finds :math:`(D, V)`, solution of the generalized eigenvalue problem
.. math::
A V = B V D.
This function is a wrapper around :py:func:`scipy.linalg.eigh` that adds energy truncation and
extra output formats.
Parameters
----------
A : :py:class:`~numpy.ndarray`
(M, M) hermitian matrix.
If `A` is not positive-semidefinite (PSD), its negative spectrum is discarded.
B : :py:class:`~numpy.ndarray`, optional
(M, M) PSD hermitian matrix.
If unspecified, `B` is assumed to be the identity matrix.
tau : float, optional
Normalized energy ratio. (Default: 1)
N : int, optional
Number of eigenpairs to output. (Default: K, the minimum number of leading eigenpairs that
account for `tau` percent of the total energy.)
* If `N` is smaller than K, then the trailing eigenpairs are dropped.
* If `N` is greater that K, then the trailing eigenpairs are set to 0.
Returns
-------
D : :py:class:`~numpy.ndarray`
(N,) positive real-valued eigenvalues.
V : :py:class:`~numpy.ndarray`
(M, N) complex-valued eigenvectors.
The N eigenpairs are sorted in decreasing eigenvalue order.
Examples
--------
.. testsetup::
import numpy as np
from imot_tools.math.linalg import eigh
import scipy.linalg as linalg
np.random.seed(0)
def hermitian_array(N: int) -> np.ndarray:
'''
Construct a (N, N) Hermitian matrix.
'''
D = np.arange(N)
Rmtx = np.random.randn(N,N) + 1j * np.random.randn(N, N)
Q, _ = linalg.qr(Rmtx)
A = (Q * D) @ Q.conj().T
return A
M = 4
A = hermitian_array(M)
B = hermitian_array(M) + 100 * np.eye(M) # To guarantee PSD
Let `A` and `B` be defined as below:
.. doctest::
M = 4
A = hermitian_array(M)
B = hermitian_array(M) + 100 * np.eye(M) # To guarantee PSD
Then different calls to :py:func:`~imot_tools.math.linalg.eigh` produce different results:
* Get all positive eigenpairs:
.. doctest::
>>> D, V = eigh(A, B)
>>> print(np.around(D, 4)) # The last term is small but positive.
[0.0296 0.0198 0.0098 0. ]
>>> print(np.around(V, 4))
[[-0.0621+0.0001j -0.0561+0.0005j -0.0262-0.0004j 0.0474+0.0005j]
[ 0.0285+0.0041j -0.0413-0.0501j 0.0129-0.0209j -0.004 -0.0647j]
[ 0.0583+0.0055j -0.0443+0.0033j 0.0069+0.0474j 0.0281+0.0371j]
[ 0.0363+0.0209j 0.0006+0.0235j -0.029 -0.0736j 0.0321+0.0142j]]
* Drop some trailing eigenpairs:
.. doctest::
>>> D, V = eigh(A, B, tau=0.8)
>>> print(np.around(D, 4))
[0.0296]
>>> print(np.around(V, 4))
[[-0.0621+0.0001j]
[ 0.0285+0.0041j]
[ 0.0583+0.0055j]
[ 0.0363+0.0209j]]
* Pad output to certain size:
.. doctest::
>>> D, V = eigh(A, B, tau=0.8, N=3)
>>> print(np.around(D, 4))
[0.0296 0. 0. ]
>>> print(np.around(V, 4))
[[-0.0621+0.0001j 0. +0.j 0. +0.j ]
[ 0.0285+0.0041j 0. +0.j 0. +0.j ]
[ 0.0583+0.0055j 0. +0.j 0. +0.j ]
[ 0.0363+0.0209j 0. +0.j 0. +0.j ]]
"""
A = np.array(A, copy=False)
M = len(A)
if not (chk.has_shape([M, M])(A) and np.allclose(A, A.conj().T)):
raise ValueError("Parameter[A] must be hermitian symmetric.")
B = np.eye(M) if (B is None) else np.array(B, copy=False)
if not (chk.has_shape([M, M])(B) and np.allclose(B, B.conj().T)):
raise ValueError("Parameter[B] must be hermitian symmetric.")
if not (0 < tau <= 1):
raise ValueError("Parameter[tau] must be in [0, 1].")
if (N is not None) and (N <= 0):
raise ValueError(f"Parameter[N] must be a non-zero positive integer.")
# A: drop negative spectrum.
Ds, Vs = linalg.eigh(A)
idx = Ds > 0
Ds, Vs = Ds[idx], Vs[:, idx]
A = (Vs * Ds) @ Vs.conj().T
# A, B: generalized eigenvalue-decomposition.
try:
D, V = linalg.eigh(A, B)
# Discard near-zero D due to numerical precision.
idx = D > 0
D, V = D[idx], V[:, idx]
idx = np.argsort(D)[::-1]
D, V = D[idx], V[:, idx]
except linalg.LinAlgError:
raise ValueError("Parameter[B] is not PSD.")
# Energy selection / padding
idx = np.clip(np.cumsum(D) / | np.sum(D) | numpy.sum |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.