hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
sequence | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
sequence | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
sequence | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
sequence | cell_types
sequence | cell_type_groups
sequence |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
e78ff9cbad6c3daa230e9b393314c85303ff15b2 | 45,454 | ipynb | Jupyter Notebook | notebooks/01.06-Everest_KEGS_DVS.ipynb | gully/goldenrod | f27982b5457dade5a15666a411725562a7659c07 | [
"MIT"
] | 1 | 2017-10-31T20:55:04.000Z | 2017-10-31T20:55:04.000Z | notebooks/01.06-Everest_KEGS_DVS.ipynb | gully/goldenrod | f27982b5457dade5a15666a411725562a7659c07 | [
"MIT"
] | 1 | 2018-05-14T19:15:47.000Z | 2018-05-14T19:15:47.000Z | notebooks/01.06-Everest_KEGS_DVS.ipynb | gully/goldenrod | f27982b5457dade5a15666a411725562a7659c07 | [
"MIT"
] | null | null | null | 70.03698 | 2,931 | 0.657346 | [
[
[
"<!--BOOK_INFORMATION-->\n<img align=\"left\" style=\"padding-right:10px;\" src=\"figures/k2_pix_small.png\">\n*This notebook contains an excerpt instructional material from [gully](https://twitter.com/gully_) and the [K2 Guest Observer Office](https://keplerscience.arc.nasa.gov/); the content is available [on GitHub](https://github.com/gully/goldenrod).*\n",
"_____no_output_____"
],
[
"<!--NAVIGATION-->\n< [KEGS galaxies reduced with EVEREST](01.05-Match_Everest_meta_data.ipynb) | [Contents](Index.ipynb) | [Exploratory analysis of K2 transient light curves](01.09-Exploratory_analysis.ipynb) >",
"_____no_output_____"
],
[
"# Spot-check Everest Validation Summaries for KEGS",
"_____no_output_____"
],
[
"This notebook does more spot-checking of KEGS target lightcurves with the \"data validation summary\" (DVS) feature in EVEREST.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\nfrom astropy.io import fits",
"_____no_output_____"
],
[
"import astropy\nimport os\nimport pandas as pd\nimport seaborn as sns\nfrom astropy.utils.console import ProgressBar\nimport everest",
"_____no_output_____"
],
[
"%matplotlib inline\n%config InlineBackend.figure_format = 'retina'",
"_____no_output_____"
],
[
"everest_path = '../../everest/everest/missions/k2/tables/'\ndf_kegs = pd.read_csv('../metadata/KEGS_TPF_metadata.csv')\nc05_everest = pd.read_csv(everest_path + 'c05.stars', names=['EPIC_ID', 'KepMag', 'Channel', 'col4'])\nkegs_everest_c05 = pd.merge(df_kegs, c05_everest, how='inner', left_on='KEPLERID', right_on='EPIC_ID')",
"_____no_output_____"
],
[
"ke_list = kegs_everest_c05['KEPLERID'].values",
"_____no_output_____"
],
[
"i = 0",
"_____no_output_____"
],
[
"i",
"_____no_output_____"
]
],
[
[
"Decent examples to try to replicate: 12 ",
"_____no_output_____"
]
],
[
[
"i+=1\nstar = everest.Everest(ke_list[i])\nstar.dvs()",
"INFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211311876.\nINFO [everest.user.DownloadFile()]: Found cached file.\n"
],
[
"for i in range(len(ke_list)):\n star = everest.Everest(ke_list[i])\n star.dvs()",
"INFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211305171.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211311876.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211312434.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211316816.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211317575.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211317725.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211318777.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211320303.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211320689.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211321727.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211324272.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211327533.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211327561.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211329564.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211329782.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211332955.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211334279.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211336332.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211336576.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211336767.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211341772.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211346083.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211346149.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211346470.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211346668.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211348567.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211349407.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211352575.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211359991.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211362257.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211374878.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211376360.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211376898.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211377253.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211377762.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211377821.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211378205.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211378569.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211382580.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211383902.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211384920.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211385002.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211386909.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211387883.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211391030.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.DownloadFile()]: Found cached file.\nINFO [everest.user.load_fits()]: Loading FITS file for 211393064.\nINFO [everest.user.DownloadFile()]: Found cached file.\n"
]
],
[
[
"<!--NAVIGATION-->\n< [KEGS galaxies reduced with EVEREST](01.05-Match_Everest_meta_data.ipynb) | [Contents](Index.ipynb) | [Exploratory analysis of K2 transient light curves](01.09-Exploratory_analysis.ipynb) >",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
e78ffacd50e9a36efb778134ccd1933e2f44bfd2 | 18,800 | ipynb | Jupyter Notebook | 01_Getting_&_Knowing_Your_Data/Chipotle/Exercise_with_Solutions.ipynb | ismael-araujo/pandas-exercise | 241db811941aaba17d5d821c73ba1be46a189bc3 | [
"BSD-3-Clause"
] | 1 | 2020-10-30T20:08:36.000Z | 2020-10-30T20:08:36.000Z | 01_Getting_&_Knowing_Your_Data/Chipotle/Exercise_with_Solutions.ipynb | ismael-araujo/pandas-exercise | 241db811941aaba17d5d821c73ba1be46a189bc3 | [
"BSD-3-Clause"
] | null | null | null | 01_Getting_&_Knowing_Your_Data/Chipotle/Exercise_with_Solutions.ipynb | ismael-araujo/pandas-exercise | 241db811941aaba17d5d821c73ba1be46a189bc3 | [
"BSD-3-Clause"
] | null | null | null | 23.61809 | 195 | 0.418936 | [
[
[
"# Ex2 - Getting and Knowing your Data\n\nCheck out [Chipotle Exercises Video Tutorial](https://www.youtube.com/watch?v=lpuYZ5EUyS8&list=PLgJhDSE2ZLxaY_DigHeiIDC1cD09rXgJv&index=2) to watch a data scientist go through the exercises",
"_____no_output_____"
],
[
"This time we are going to pull data directly from the internet.\nSpecial thanks to: https://github.com/justmarkham for sharing the dataset and materials.\n\n### Step 1. Import the necessary libraries",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"### Step 2. Import the dataset from this [address](https://raw.githubusercontent.com/justmarkham/DAT8/master/data/chipotle.tsv). ",
"_____no_output_____"
],
[
"### Step 3. Assign it to a variable called chipo.",
"_____no_output_____"
]
],
[
[
"url = 'https://raw.githubusercontent.com/justmarkham/DAT8/master/data/chipotle.tsv'\n \nchipo = pd.read_csv(url, sep = '\\t')",
"_____no_output_____"
]
],
[
[
"### Step 4. See the first 10 entries",
"_____no_output_____"
]
],
[
[
"chipo.head(10)",
"_____no_output_____"
]
],
[
[
"### Step 5. What is the number of observations in the dataset?",
"_____no_output_____"
]
],
[
[
"# Solution 1\n\nchipo.shape[0] # entries <= 4622 observations",
"_____no_output_____"
],
[
"# Solution 2\n\nchipo.info() # entries <= 4622 observations",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 4622 entries, 0 to 4621\nData columns (total 5 columns):\norder_id 4622 non-null int64\nquantity 4622 non-null int64\nitem_name 4622 non-null object\nchoice_description 3376 non-null object\nitem_price 4622 non-null object\ndtypes: int64(2), object(3)\nmemory usage: 180.6+ KB\n"
]
],
[
[
"### Step 6. What is the number of columns in the dataset?",
"_____no_output_____"
]
],
[
[
"chipo.shape[1]",
"_____no_output_____"
]
],
[
[
"### Step 7. Print the name of all the columns.",
"_____no_output_____"
]
],
[
[
"chipo.columns",
"_____no_output_____"
]
],
[
[
"### Step 8. How is the dataset indexed?",
"_____no_output_____"
]
],
[
[
"chipo.index",
"_____no_output_____"
]
],
[
[
"### Step 9. Which was the most-ordered item? ",
"_____no_output_____"
]
],
[
[
"c = chipo.groupby('item_name')\nc = c.sum()\nc = c.sort_values(['quantity'], ascending=False)\nc.head(1)",
"_____no_output_____"
]
],
[
[
"### Step 10. For the most-ordered item, how many items were ordered?",
"_____no_output_____"
]
],
[
[
"c = chipo.groupby('item_name')\nc = c.sum()\nc = c.sort_values(['quantity'], ascending=False)\nc.head(1)",
"_____no_output_____"
]
],
[
[
"### Step 11. What was the most ordered item in the choice_description column?",
"_____no_output_____"
]
],
[
[
"c = chipo.groupby('choice_description').sum()\nc = c.sort_values(['quantity'], ascending=False)\nc.head(1)\n# Diet Coke 159",
"_____no_output_____"
]
],
[
[
"### Step 12. How many items were orderd in total?",
"_____no_output_____"
]
],
[
[
"total_items_orders = chipo.quantity.sum()\ntotal_items_orders",
"_____no_output_____"
]
],
[
[
"### Step 13. Turn the item price into a float",
"_____no_output_____"
],
[
"#### Step 13.a. Check the item price type",
"_____no_output_____"
]
],
[
[
"chipo.item_price.dtype",
"_____no_output_____"
]
],
[
[
"#### Step 13.b. Create a lambda function and change the type of item price",
"_____no_output_____"
]
],
[
[
"dollarizer = lambda x: float(x[1:-1])\nchipo.item_price = chipo.item_price.apply(dollarizer)",
"_____no_output_____"
]
],
[
[
"#### Step 13.c. Check the item price type",
"_____no_output_____"
]
],
[
[
"chipo.item_price.dtype",
"_____no_output_____"
]
],
[
[
"### Step 14. How much was the revenue for the period in the dataset?",
"_____no_output_____"
]
],
[
[
"revenue = (chipo['quantity']* chipo['item_price']).sum()\n\nprint('Revenue was: $' + str(np.round(revenue,2)))",
"Revenue was: $39237.02\n"
]
],
[
[
"### Step 15. How many orders were made in the period?",
"_____no_output_____"
]
],
[
[
"orders = chipo.order_id.value_counts().count()\norders",
"_____no_output_____"
]
],
[
[
"### Step 16. What is the average revenue amount per order?",
"_____no_output_____"
]
],
[
[
"# Solution 1\n\nchipo['revenue'] = chipo['quantity'] * chipo['item_price']\norder_grouped = chipo.groupby(by=['order_id']).sum()\norder_grouped.mean()['revenue']",
"_____no_output_____"
],
[
"# Solution 2\n\nchipo.groupby(by=['order_id']).sum().mean()['revenue']",
"_____no_output_____"
]
],
[
[
"### Step 17. How many different items are sold?",
"_____no_output_____"
]
],
[
[
"chipo.item_name.value_counts().count()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
e790169bb82900e9e60477002a549f13249f7f1d | 21,565 | ipynb | Jupyter Notebook | Practicas/.ipynb_checkpoints/Practica 2 - Solucion de ecuaciones diferenciales-checkpoint.ipynb | robblack007/clase-dinamica-robot | f38cb358f2681e9c0dce979acbdcd81bf63bd59c | [
"MIT"
] | null | null | null | Practicas/.ipynb_checkpoints/Practica 2 - Solucion de ecuaciones diferenciales-checkpoint.ipynb | robblack007/clase-dinamica-robot | f38cb358f2681e9c0dce979acbdcd81bf63bd59c | [
"MIT"
] | 1 | 2016-01-26T18:33:11.000Z | 2016-05-30T23:58:07.000Z | Practicas/.ipynb_checkpoints/Practica 2 - Solucion de ecuaciones diferenciales-checkpoint.ipynb | robblack007/clase-dinamica-robot | f38cb358f2681e9c0dce979acbdcd81bf63bd59c | [
"MIT"
] | null | null | null | 23.907982 | 623 | 0.538372 | [
[
[
"# Solución de ecuaciones diferenciales",
"_____no_output_____"
],
[
"Dada la siguiente ecuación diferencial:\n\n$$\n\\dot{x} = -x\n$$\n\nqueremos obtener la respuesta del sistema que representa, es decir, los valores que va tomando $x$.\n\nSi analizamos esta ecuación diferencial, podremos notar que la solución de este sistema es una función $\\varphi(t)$, tal que cuando la derivemos obtengamos el negativo de esta misma función, es decir:\n\n$$\n\\frac{d}{dt} \\varphi(t) = -\\varphi(t)\n$$\n\ny despues de un poco de pensar, podemos darnos cuenta de que la función que queremos es:\n\n$$\n\\varphi(t) = e^{-t}\n$$\n\nSin embargo muchas veces no tendremos funciones tan sencillas (ciertamente no es el caso en la robótica, donde usualmente tenemos ecuaciones diferenciales no lineales de orden $n$), por lo que en esta práctica veremos algunas estrategias para obtener soluciones a esta ecuación diferencial, tanto numéricas como simbolicas.",
"_____no_output_____"
],
[
"## Método de Euler",
"_____no_output_____"
],
[
"El [método de Euler](http://es.wikipedia.org/wiki/Método_de_Euler) para obtener el comportamiento de una ecuación diferencial, se basa en la intuición básica de la derivada; digamos que tenemos una ecuación diferencial general:\n\n$$\n\\frac{dy}{dx} = y' = F(x, y)\n$$\n\nen donde $F(x, y)$ puede ser cualquier función que depende de $x$ y/o de $y$, entonces podemos dividir en pedazos el comportamiento de la gráfica de tal manera que solo calculemos un pequeño pedazo cada vez, aproximando el comportamiento de la ecuación diferencial, con el de una recta, cuya pendiente será la derivada:\n\n\n\n«<a href=\"http://commons.wikimedia.org/wiki/File:M%C3%A9todo_de_Euler.jpg#/media/File:M%C3%A9todo_de_Euler.jpg\">Método de Euler</a>» por <a href=\"//commons.wikimedia.org/w/index.php?title=User:Vero.delgado&action=edit&redlink=1\" class=\"new\" title=\"User:Vero.delgado (la página no existe)\">Vero.delgado</a> - <span class=\"int-own-work\" lang=\"es\">Trabajo propio</span>. Disponible bajo la licencia <a href=\"http://creativecommons.org/licenses/by-sa/3.0\" title=\"Creative Commons Attribution-Share Alike 3.0\">CC BY-SA 3.0</a> vía <a href=\"//commons.wikimedia.org/wiki/\">Wikimedia Commons</a>.\n\nEsta recta que aproxima a la ecuación diferencial, podemos recordar que tiene una estructura:\n\n$$\ny = b + mx\n$$\n\npor lo que si sustituimos en $m$ la derivada y $b$ con el valor anterior de la ecuación diferencial, obtendremos algo como:\n\n$$\n\\overbrace{y_{i+1}}^{\\text{nuevo valor de }y} = \\overbrace{y_i}^{\\text{viejo valor de }y} + \\overbrace{\\frac{dy}{dx}}^{\\text{pendiente}} \\overbrace{\\Delta x}^{\\text{distancia en }x}\n$$\n\npero conocemos el valor de $\\frac{dy}{dx}$, es nuestra ecuación diferencial; por lo que podemos escribir esto como:\n\n$$\ny_{i+1} = y_i + F(x_i, y_i) \\Delta x\n$$\n\nResolvamos algunas iteraciones de nuestro sistema; empecemos haciendo 10 iteraciones a lo largo de 10 segundos, con condiciones iniciales $x(0) = 1$, eso quiere decir que:\n\n$$\n\\begin{align}\n\\Delta t &= 1 \\\\\nx(0) &= 1 \\\\\n\\dot{x}(0) &= 1\n\\end{align}\n$$",
"_____no_output_____"
]
],
[
[
"x0 = 1\nΔt = 1 \n\n# Para escribir simbolos griegos como Δ, tan solo tienes que escribir su nombre\n# precedido de una diagonal (\\Delta) y teclear tabulador una vez\n\nF = lambda x : -x\nx1 = x0 + F(x0)*Δt\nx1",
"_____no_output_____"
],
[
"x2 = x1 + F(x1)*Δt\nx2",
"_____no_output_____"
]
],
[
[
"### Ejercicio",
"_____no_output_____"
],
[
"Crea codigo para una iteración mas con estos mismos parametros y despliega el resultado.",
"_____no_output_____"
]
],
[
[
"x3 = # Escribe el codigo de tus calculos aqui",
"_____no_output_____"
],
[
"from pruebas_2 import prueba_2_1\nprueba_2_1(x0, x1, x2, x3, _)",
"_____no_output_____"
]
],
[
[
"Momento... que esta pasando? Resulta que este $\\Delta t$ es demasiado grande, intentemos con 20 iteraciones:\n\n$$\n\\begin{align}\n\\Delta t &= 0.5 \\\\\nx(0) &= 1\n\\end{align}\n$$",
"_____no_output_____"
]
],
[
[
"x0 = 1\nn = 20\nΔt = 10/n\n\nF = lambda x : -x\nx1 = x0 + F(x0)*Δt\nx1",
"_____no_output_____"
],
[
"x2 = x1 + F(x1)*Δt\nx2",
"_____no_output_____"
],
[
"x3 = x2 + F(x2)*Δt\nx3",
"_____no_output_____"
]
],
[
[
"Esto va a ser tardado, mejor digamosle a Python que es lo que tenemos que hacer, y que no nos moleste hasta que acabe, podemos usar un ciclo ```for``` y una lista para guardar todos los valores de la trayectoria:",
"_____no_output_____"
]
],
[
[
"xs = [x0]\nfor t in range(20):\n xs.append(xs[-1] + F(xs[-1])*Δt)\n\nxs",
"_____no_output_____"
]
],
[
[
"Ahora que tenemos estos valores, podemos graficar el comportamiento de este sistema, primero importamos la libreria ```matplotlib```:",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nfrom matplotlib.pyplot import plot",
"_____no_output_____"
]
],
[
[
"Mandamos a llamar la función ```plot```:",
"_____no_output_____"
]
],
[
[
"plot(xs);",
"_____no_output_____"
]
],
[
[
"Sin embargo debido a que el periodo de integración que utilizamos es demasiado grande, la solución es bastante inexacta, podemos verlo al graficar contra la que sabemos es la solución de nuestro problema:",
"_____no_output_____"
]
],
[
[
"from numpy import linspace, exp",
"_____no_output_____"
],
[
"ts = linspace(0, 10, 20)\nplot(xs)\nplot(exp(-ts));",
"_____no_output_____"
]
],
[
[
"Si ahora utilizamos un numero de pedazos muy grande, podemos mejorar nuestra aproximación:",
"_____no_output_____"
]
],
[
[
"xs = [x0]\nn = 100\nΔt = 10/n\n\nfor t in range(100):\n xs.append(xs[-1] + F(xs[-1])*Δt)\n \nts = linspace(0, 10, 100)",
"_____no_output_____"
],
[
"plot(xs)\nplot(exp(-ts));",
"_____no_output_____"
]
],
[
[
"## odeint",
"_____no_output_____"
],
[
"Este método funciona tan bien, que ya viene programado dentro de la libreria ```scipy```, por lo que solo tenemos que importar esta librería para utilizar este método.\n\nSin embargo debemos de tener cuidado al declarar la función $F(x, t)$. El primer argumento de la función se debe de referir al estado de la función, es decir $x$, y el segundo debe de ser la variable independiente, en nuestro caso el tiempo.",
"_____no_output_____"
]
],
[
[
"from scipy.integrate import odeint",
"_____no_output_____"
],
[
"F = lambda x, t : -x",
"_____no_output_____"
],
[
"x0 = 1\nts = linspace(0, 10, 100)\nxs = odeint(func=F, y0=x0, t=ts)",
"_____no_output_____"
],
[
"plot(ts, xs);",
"_____no_output_____"
]
],
[
[
"### Ejercicio",
"_____no_output_____"
],
[
"Grafica el comportamiento de la siguiente ecuación diferencial.\n\n$$\n\\dot{x} = x^2 - 5 x + \\frac{1}{2} \\sin{x} - 2\n$$\n\n> Nota: Asegurate de impotar todas las librerias que puedas necesitar",
"_____no_output_____"
]
],
[
[
"ts = # Escribe aqui el codigo que genera un arreglo de puntos equidistantes (linspace)\nx0 = # Escribe el valor de la condicion inicial\n\n# Importa las funciones de librerias que necesites aqui\n\nG = lambda x, t: # Escribe aqui el codigo que describe los calculos que debe hacer la funcion\n\nxs = # Escribe aqui el comando necesario para simular la ecuación diferencial\n\nplot(ts, xs);",
"_____no_output_____"
],
[
"from pruebas_2 import prueba_2_2\nprueba_2_2(ts, xs)",
"_____no_output_____"
]
],
[
[
"## Sympy",
"_____no_output_____"
],
[
"Y por ultimo, hay veces en las que incluso podemos obtener una solución analítica de una ecuación diferencial, siempre y cuando cumpla ciertas condiciones de simplicidad.",
"_____no_output_____"
]
],
[
[
"from sympy import var, Function, dsolve\nfrom sympy.physics.mechanics import mlatex, mechanics_printing\nmechanics_printing()",
"_____no_output_____"
],
[
"var(\"t\")",
"_____no_output_____"
],
[
"x = Function(\"x\")(t)\nx, x.diff(t)",
"_____no_output_____"
],
[
"solucion = dsolve(x.diff(t) + x, x)\nsolucion",
"_____no_output_____"
]
],
[
[
"### Ejercicio",
"_____no_output_____"
],
[
"Implementa el codigo necesario para obtener la solución analítica de la siguiente ecuación diferencial:\n\n$$\n\\dot{x} = x^2 - 5x\n$$",
"_____no_output_____"
]
],
[
[
"# Declara la variable independiente de la ecuación diferencial\nvar(\"\")\n\n# Declara la variable dependiente de la ecuación diferencial\n = Function(\"\")()\n\n# Escribe la ecuación diferencial con el formato necesario (Ecuacion = 0)\n# adentro de la función dsolve\nsol = dsolve()\nsol",
"_____no_output_____"
],
[
"from pruebas_2 import prueba_2_3\nprueba_2_3(sol)",
"_____no_output_____"
]
],
[
[
"## Solución a ecuaciones diferenciales de orden superior",
"_____no_output_____"
],
[
"Si ahora queremos obtener el comportamiento de una ecuacion diferencial de orden superior, como:\n\n$$\n\\ddot{x} = -\\dot{x} - x + 1\n$$\n\nTenemos que convertirla en una ecuación diferencial de primer orden para poder resolverla numericamente, por lo que necesitaremos convertirla en una ecuación diferencial matricial, por lo que empezamos escribiendola junto con la identidad $\\dot{x} = \\dot{x}$ en un sistema de ecuaciones:\n\n$$\n\\begin{align}\n\\dot{x} &= \\dot{x} \\\\\n\\ddot{x} &= -\\dot{x} - x + 1\n\\end{align}\n$$\n\nSi extraemos el operador derivada del lado izquierda, tenemos:\n\n$$\n$$\n\\begin{align}\n\\frac{d}{dt} x &= \\dot{x} \\\\\n\\frac{d}{dt} \\dot{x} &= -\\dot{x} - x + 1\n\\end{align}\n$$\n$$\n\nO bien, de manera matricial:\n\n$$\n\\frac{d}{dt}\n\\begin{pmatrix}\nx \\\\\n\\dot{x}\n\\end{pmatrix} =\n\\begin{pmatrix}\n0 & 1 \\\\\n-1 & -1\n\\end{pmatrix}\n\\begin{pmatrix}\nx \\\\\n\\dot{x}\n\\end{pmatrix} +\n\\begin{pmatrix}\n0 \\\\\n1\n\\end{pmatrix}\n$$\n\nEsta ecuación ya _no_ es de segundo orden, es de hecho, de primer orden, sin embargo nuestra variable ha crecido a ser un vector de estados, por el momento le llamaremos $X$, asi pues, lo podemos escribir como:\n\n$$\n\\frac{d}{dt} X = A X + B\n$$\n\nen donde:\n\n$$\nA = \\begin{pmatrix}\n0 & 1 \\\\\n-1 & -1\n\\end{pmatrix} \\quad \\text{y} \\quad B =\n\\begin{pmatrix}\n0 \\\\\n1\n\\end{pmatrix}\n$$\n\ny de manera similar, declarar una función para dar a ```odeint```.",
"_____no_output_____"
]
],
[
[
"from numpy import matrix, array",
"_____no_output_____"
],
[
"def F(X, t):\n A = matrix([[0, 1], [-1, -1]])\n B = matrix([[0], [1]])\n return array((A*matrix(X).T + B).T).tolist()[0]",
"_____no_output_____"
],
[
"ts = linspace(0, 10, 100)\nxs = odeint(func=F, y0=[0, 0], t=ts)",
"_____no_output_____"
],
[
"plot(xs);",
"_____no_output_____"
]
],
[
[
"### Ejercicio",
"_____no_output_____"
],
[
"Implementa la solución de la siguiente ecuación diferencial, por medio de un modelo en representación de espacio de estados:\n\n$$\n\\ddot{x} = -8\\dot{x} - 15x + 1\n$$\n\n> Nota: Tomalo con calma y paso a paso\n> * Empieza anotando la ecuación diferencial en tu cuaderno, junto a la misma identidad del ejemplo\n> * Extrae la derivada del lado izquierdo, para que obtengas el _estado_ de tu sistema\n> * Extrae las matrices A y B que corresponden a este sistema\n> * Escribe el codigo necesario para representar estas matrices",
"_____no_output_____"
]
],
[
[
"def G(X, t):\n A = # Escribe aqui el codigo para la matriz A\n B = # Escribe aqui el codigo para el vector B\n return array((A*matrix(X).T + B).T).tolist()[0]\n\nts = linspace(0, 10, 100) \nxs = odeint(func=G, y0=[0, 0], t=ts)",
"_____no_output_____"
],
[
"plot(xs);",
"_____no_output_____"
],
[
"from pruebas_2 import prueba_2_4\nprueba_2_4(xs)",
"_____no_output_____"
]
],
[
[
"## Funciones de transferencia",
"_____no_output_____"
],
[
"Sin embargo, no es la manera mas facil de obtener la solución, tambien podemos aplicar una transformada de Laplace, y aplicar las funciones de la libreria de control para simular la función de transferencia de esta ecuación; al aplicar la transformada de Laplace, obtendremos:\n\n$$\nG(s) = \\frac{1}{s^2 + s + 1}\n$$",
"_____no_output_____"
]
],
[
[
"from control import tf, step",
"_____no_output_____"
],
[
"F = tf([0, 0, 1], [1, 1, 1])",
"_____no_output_____"
],
[
"xs, ts = step(F)",
"_____no_output_____"
],
[
"plot(ts, xs);",
"_____no_output_____"
]
],
[
[
"### Ejercicio",
"_____no_output_____"
],
[
"Modela matematicamente la ecuación diferencial del ejercicio anterior, usando una representación de función de transferencia.\n\n> Nota: De nuevo, no desesperes, escribe tu ecuación diferencial y aplica la transformada de Laplaca tal como te enseñaron tus abuelos hace tantos años...",
"_____no_output_____"
]
],
[
[
"G = tf([], []) # Escribe los coeficientes de la función de transferencia \n\nxs, ts = step(G)\nplot(ts, xs);",
"_____no_output_____"
],
[
"from pruebas_2 import prueba_2_5\nprueba_2_5(ts, xs)",
"_____no_output_____"
]
],
[
[
"## Problemas",
"_____no_output_____"
],
[
"1. Modela matematicamente la suspensión de un automovil de masa $m = 1200 kg$, considera que los resortes de su suspensión tienen una constante $k = 15,000 \\frac{N}{m}$ y un amortiguador con constante $c = 1,500 \\frac{N s}{m}$.\n\n2. Gráfica el comportamiento del sistema ante una fuerza $F = 1 N$.\n3. Que tipo de comportamiento presenta este sistema? Estable, criticamente estable, inestable?",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
e790559f4a18ddf90960e58c65b481e2e5d2923c | 17,991 | ipynb | Jupyter Notebook | regularization.ipynb | josefj1519/RegularizationBoston | ab6f05099a0761799aada58ea719ad603b4fc2be | [
"Apache-2.0"
] | null | null | null | regularization.ipynb | josefj1519/RegularizationBoston | ab6f05099a0761799aada58ea719ad603b4fc2be | [
"Apache-2.0"
] | null | null | null | regularization.ipynb | josefj1519/RegularizationBoston | ab6f05099a0761799aada58ea719ad603b4fc2be | [
"Apache-2.0"
] | null | null | null | 36.791411 | 429 | 0.558279 | [
[
[
"# CPSC 483 Project 3 - Regularization, Cross-Validation, and Grid Search\n#### by: Josef Jankowski([email protected]) and William Timani ([email protected])",
"_____no_output_____"
],
[
"### 1. Load and examine the Boston dataset’s features, target values, and description.\n",
"_____no_output_____"
]
],
[
[
"from sklearn import datasets\ndataset_boston = datasets.load_boston()\nprint(dataset_boston.DESCR)",
".. _boston_dataset:\n\nBoston house prices dataset\n---------------------------\n\n**Data Set Characteristics:** \n\n :Number of Instances: 506 \n\n :Number of Attributes: 13 numeric/categorical predictive. Median Value (attribute 14) is usually the target.\n\n :Attribute Information (in order):\n - CRIM per capita crime rate by town\n - ZN proportion of residential land zoned for lots over 25,000 sq.ft.\n - INDUS proportion of non-retail business acres per town\n - CHAS Charles River dummy variable (= 1 if tract bounds river; 0 otherwise)\n - NOX nitric oxides concentration (parts per 10 million)\n - RM average number of rooms per dwelling\n - AGE proportion of owner-occupied units built prior to 1940\n - DIS weighted distances to five Boston employment centres\n - RAD index of accessibility to radial highways\n - TAX full-value property-tax rate per $10,000\n - PTRATIO pupil-teacher ratio by town\n - B 1000(Bk - 0.63)^2 where Bk is the proportion of blacks by town\n - LSTAT % lower status of the population\n - MEDV Median value of owner-occupied homes in $1000's\n\n :Missing Attribute Values: None\n\n :Creator: Harrison, D. and Rubinfeld, D.L.\n\nThis is a copy of UCI ML housing dataset.\nhttps://archive.ics.uci.edu/ml/machine-learning-databases/housing/\n\n\nThis dataset was taken from the StatLib library which is maintained at Carnegie Mellon University.\n\nThe Boston house-price data of Harrison, D. and Rubinfeld, D.L. 'Hedonic\nprices and the demand for clean air', J. Environ. Economics & Management,\nvol.5, 81-102, 1978. Used in Belsley, Kuh & Welsch, 'Regression diagnostics\n...', Wiley, 1980. N.B. Various transformations are used in the table on\npages 244-261 of the latter.\n\nThe Boston house-price data has been used in many machine learning papers that address regression\nproblems. \n \n.. topic:: References\n\n - Belsley, Kuh & Welsch, 'Regression diagnostics: Identifying Influential Data and Sources of Collinearity', Wiley, 1980. 244-261.\n - Quinlan,R. (1993). Combining Instance-Based and Model-Based Learning. In Proceedings on the Tenth International Conference of Machine Learning, 236-243, University of Massachusetts, Amherst. Morgan Kaufmann.\n\n"
]
],
[
[
"### 2. Save CRIM as the new target value t, and drop the column CRIM from X. Add the target value MEDV to X.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\n# Independent variables (i.e. features)\ndf_boston_features = pd.DataFrame(data=dataset_boston.data, columns=dataset_boston.feature_names)\ndf_boston_features.insert(0, 'MEDV', dataset_boston.target)\ndf_boston_target = pd.DataFrame(data=df_boston_features['CRIM'], columns=['CRIM'])\ndf_boston_features = df_boston_features.drop(['CRIM'], axis=1)\nprint(df_boston_features)",
" MEDV ZN INDUS CHAS NOX RM AGE DIS RAD TAX PTRATIO \\\n0 24.0 18.0 2.31 0.0 0.538 6.575 65.2 4.0900 1.0 296.0 15.3 \n1 21.6 0.0 7.07 0.0 0.469 6.421 78.9 4.9671 2.0 242.0 17.8 \n2 34.7 0.0 7.07 0.0 0.469 7.185 61.1 4.9671 2.0 242.0 17.8 \n3 33.4 0.0 2.18 0.0 0.458 6.998 45.8 6.0622 3.0 222.0 18.7 \n4 36.2 0.0 2.18 0.0 0.458 7.147 54.2 6.0622 3.0 222.0 18.7 \n.. ... ... ... ... ... ... ... ... ... ... ... \n501 22.4 0.0 11.93 0.0 0.573 6.593 69.1 2.4786 1.0 273.0 21.0 \n502 20.6 0.0 11.93 0.0 0.573 6.120 76.7 2.2875 1.0 273.0 21.0 \n503 23.9 0.0 11.93 0.0 0.573 6.976 91.0 2.1675 1.0 273.0 21.0 \n504 22.0 0.0 11.93 0.0 0.573 6.794 89.3 2.3889 1.0 273.0 21.0 \n505 11.9 0.0 11.93 0.0 0.573 6.030 80.8 2.5050 1.0 273.0 21.0 \n\n B LSTAT \n0 396.90 4.98 \n1 396.90 9.14 \n2 392.83 4.03 \n3 394.63 2.94 \n4 396.90 5.33 \n.. ... ... \n501 391.99 9.67 \n502 396.90 9.08 \n503 396.90 5.64 \n504 393.45 6.48 \n505 396.90 7.88 \n\n[506 rows x 13 columns]\n"
]
],
[
[
"### 3. Use sklearn.model_selection.train_test_split() to split the features and target values into separate training and test sets. Use 80% of the original data as a training set, and 20% for testing.",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\nx_train, x_test, y_train, y_test = train_test_split(df_boston_features, df_boston_target, test_size=.2)\nprint(x_train)",
" MEDV ZN INDUS CHAS NOX RM AGE DIS RAD TAX \\\n55 35.4 90.0 1.22 0.0 0.403 7.249 21.9 8.6966 5.0 226.0 \n273 35.2 20.0 6.96 1.0 0.464 7.691 51.8 4.3665 3.0 223.0 \n350 22.9 40.0 1.25 0.0 0.429 6.490 44.4 8.7921 1.0 335.0 \n126 15.7 0.0 25.65 0.0 0.581 5.613 95.6 1.7572 2.0 188.0 \n11 18.9 12.5 7.87 0.0 0.524 6.009 82.9 6.2267 5.0 311.0 \n.. ... ... ... ... ... ... ... ... ... ... \n407 27.9 0.0 18.10 0.0 0.659 5.608 100.0 1.2852 24.0 666.0 \n339 19.0 0.0 5.19 0.0 0.515 5.985 45.4 4.8122 5.0 224.0 \n430 14.5 0.0 18.10 0.0 0.584 6.348 86.1 2.0527 24.0 666.0 \n182 37.9 0.0 2.46 0.0 0.488 7.155 92.2 2.7006 3.0 193.0 \n227 31.6 0.0 6.20 0.0 0.504 7.163 79.9 3.2157 8.0 307.0 \n\n PTRATIO B LSTAT \n55 17.9 395.93 4.81 \n273 18.6 390.77 6.58 \n350 19.7 396.90 5.98 \n126 19.1 359.29 27.26 \n11 15.2 396.90 13.27 \n.. ... ... ... \n407 20.2 332.09 12.13 \n339 20.2 396.90 9.74 \n430 20.2 83.45 17.64 \n182 17.8 394.12 4.82 \n227 17.4 372.08 6.36 \n\n[404 rows x 13 columns]\n"
]
],
[
[
"### 4. Create and fit() an sklearn.linear_model.LinearRegression to the training set",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LinearRegression\nimport numpy as np\nx = np.array(x_train)\ny = np.array(y_train)\n\nlm = LinearRegression().fit(x,y)\n\nprint(f'w0 = {lm.intercept_}')\nprint(f'w1 = {lm.coef_[0]}')",
"w0 = [17.76174149]\nw1 = [-1.89810056e-01 4.85173466e-02 -7.25156399e-02 -6.73905034e-01\n -9.29427840e+00 2.09214818e-01 1.48984316e-03 -1.04360641e+00\n 5.78852226e-01 -3.95781002e-03 -2.28791697e-01 -8.15484094e-03\n 1.01545583e-01]\n"
]
],
[
[
"### 5. Use the predict() method of the model to find the response for each value in the test set, and sklearn.metrics.mean_squared_error(), to find the training and test MSE.\n",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import mean_squared_error\n \npredicted_train = lm.predict(x_train)\npredicted_test = lm.predict(x_test)\nmse = mean_squared_error(y_train, predicted_train, squared=True)\nprint('Train: ', mse)\nmse = mean_squared_error(y_test, predicted_test, squared=True)\nprint('Test: ', mse)",
"Train: 39.367922681692114\nTest: 44.258359558976515\n"
]
],
[
[
"### 6. By itself, the MSE doesn’t tell us much. Use the score() method of the model to find the R2 values for the training and test data.\n\n##### R2, the coefficient of determination, measures the proportion of variability in the target t that can be explained using the features in X. A value near 1 indicates that most of the variability in the response has been explained by the regression, while a value near 0 indicates that the regression does not explain much of the variability. See Section 3.1.3 of An Introduction to Statistical Learning for details.\n\n###### Given the R2 scores, how well did our model do?",
"_____no_output_____"
]
],
[
[
"r_train = lm.score(x_train, y_train)\nprint('Train r score: ', r_train)\nr_test = lm.score(x_test, y_test)\nprint('Test r score: ', r_test)",
"Train r score: 0.4620036329045377\nTest r score: 0.4203369336652354\n"
]
],
[
[
"The model is somewhat accurate. Our r's are mostly in between 0 and 1 (around .5)",
"_____no_output_____"
],
[
"### 7. Let’s see if we can fit the data better with a more flexible model. Scikit-learn can construct polynomial features for us using sklearn.preprocessing.PolynomialFeatures (though note that this includes interaction features as well; you saw in Project 2 that purely polynomial features can easily be constructed using numpy.hstack()).\n\n##### Add degree-2 polynomial features, then fit a new linear model. Compare the training and test MSE and R2 scores. Do we seem to be overfitting?",
"_____no_output_____"
]
],
[
[
"t = np.array(y_train['CRIM']).reshape([-1,1])\nx_reshape_train = np.hstack(((np.array(np.ones_like(x_train['MEDV']))).reshape([-1,1]), np.array(x_train)))\nfor attr in x_train:\n xsquared = np.square(np.array(x_train[attr])).reshape([-1,1])\n x_reshape_train = np.hstack((x_reshape_train, xsquared))\n\nlm = LinearRegression().fit(x_reshape_train, t) \n\npredicted_train = lm.predict(x_reshape_train)\nmse = mean_squared_error(y_train, predicted_train, squared=True)\nprint('Training MSE: ', mse)\n\nt = np.array(y_test['CRIM']).reshape([-1,1])\nx_reshape_test = np.hstack(((np.array(np.ones_like(x_test['MEDV']))).reshape([-1,1]), np.array(x_test)))\nfor attr in x_test:\n xsquared = np.square(np.array(x_test[attr])).reshape([-1,1])\n x_reshape_test = np.hstack((x_reshape_test, xsquared))\n\nlm = LinearRegression().fit(x_reshape_test, t) \n\npredicted_train = lm.predict(x_reshape_test)\nmse = mean_squared_error(y_test, predicted_train, squared=True)\nprint('Testing MSE: ', mse)\n\n\nr_train = lm.score(x_reshape_train, y_train)\nprint('Train r score: ', r_train)\nr_test = lm.score(x_reshape_test, y_test)\nprint('Test r score: ', r_test)",
"Training MSE: 32.767573538174275\nTesting MSE: 1273.9784845912206\nTrain r score: -21.674371778352302\nTest r score: -15.685622381430647\n"
]
],
[
[
"Test MSE seems to have improved as well as the training MSE. The r score seems to also be closer to 0 meaning that the regression does not explain much of the variability.",
"_____no_output_____"
],
[
"### 8. Regularization would allow us to construct a model of intermediate complexity by penalizing large values for the coefficients. Scikit-learn provides this as sklearn.linear_model.Ridge. The parameter alpha corresponds to 𝜆 as shown in the textbook. For now, leave it set to the default value of 1.0, and fit the model to the degree-2 polynomial features. Don’t forget to normalize your features.\n#### Once again, compare the training and test MSE and R2 scores. Is this model an improvement?\n",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import Ridge\n\nclf = Ridge(alpha=1.0, normalize=True)\npm = clf.fit(x_reshape_train, y_train)\npredicted_train = pm.predict(x_reshape_train)\nmse = mean_squared_error(y_train, predicted_train, squared=True)\nprint('Training MSE: ', mse)\n\npredicted_test = pm.predict(x_reshape_test)\nmse = mean_squared_error(y_test, predicted_test, squared=True)\nprint('Testing MSE: ', mse)\n\nr_train = pm.score(x_reshape_train, y_train)\nprint('Train r score: ', r_train)\nr_test = pm.score(x_reshape_test, y_test)\nprint('Test r score: ', r_test)",
"Training MSE: 41.74784384916144\nTesting MSE: 47.56699214405758\nTrain r score: 0.42947997265391635\nTest r score: 0.37700292560993187\n"
]
],
[
[
"The model does not seem to improve anything.",
"_____no_output_____"
],
[
"### 9. We used the default penalty value of 1.0 in the previous experiment, but there’s no reason to believe that this is optimal. Use sklearn.linear_model.RidgeCV to find an optimal value for alpha. How does this compare to experiment (8)?",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import RidgeCV\n\nclf = RidgeCV(normalize=True)\npm = clf.fit(x_reshape_train, y_train)\npredicted_train = pm.predict(x_reshape_train)\nmse = mean_squared_error(y_train, predicted_train, squared=True)\nprint('Training MSE: ', mse)\n\npredicted_test = pm.predict(x_reshape_test)\nmse = mean_squared_error(y_test, predicted_test, squared=True)\nprint('Testing MSE: ', mse)\n\nr_train = pm.score(x_reshape_train, y_train)\nprint('Train r score: ', r_train)\nr_test = pm.score(x_reshape_test, y_test)\nprint('Test r score: ', r_test)",
"Training MSE: 38.050178880414954\nTesting MSE: 43.181827227141405\nTrain r score: 0.4800117300952836\nTest r score: 0.43443655323311625\n"
]
],
[
[
"The scores overall improved. With the r score values slightly improving. ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e79061690e11ba3f4c8286afc5fcfbf3a898ca81 | 87,073 | ipynb | Jupyter Notebook | notebooks/36 - linear_models_ex_01.ipynb | aquinquenel/scikit-learn-mooc | edb91f1669ffad65038f5bf48a6771299be4c09d | [
"CC-BY-4.0"
] | null | null | null | notebooks/36 - linear_models_ex_01.ipynb | aquinquenel/scikit-learn-mooc | edb91f1669ffad65038f5bf48a6771299be4c09d | [
"CC-BY-4.0"
] | null | null | null | notebooks/36 - linear_models_ex_01.ipynb | aquinquenel/scikit-learn-mooc | edb91f1669ffad65038f5bf48a6771299be4c09d | [
"CC-BY-4.0"
] | null | null | null | 351.100806 | 79,654 | 0.932321 | [
[
[
"# 📝 Exercise M4.01\n\nThe aim of this exercise is two-fold:\n\n* understand the parametrization of a linear model;\n* quantify the fitting accuracy of a set of such models.\n\nWe will reuse part of the code of the course to:\n\n* load data;\n* create the function representing a linear model.\n\n## Prerequisites\n\n### Data loading",
"_____no_output_____"
],
[
"<div class=\"admonition note alert alert-info\">\n<p class=\"first admonition-title\" style=\"font-weight: bold;\">Note</p>\n<p class=\"last\">If you want a deeper overview regarding this dataset, you can refer to the\nAppendix - Datasets description section at the end of this MOOC.</p>\n</div>",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\npenguins = pd.read_csv(\"../datasets/penguins_regression.csv\")\nfeature_name = \"Flipper Length (mm)\"\ntarget_name = \"Body Mass (g)\"\ndata, target = penguins[[feature_name]], penguins[target_name]",
"_____no_output_____"
]
],
[
[
"### Model definition",
"_____no_output_____"
]
],
[
[
"def linear_model_flipper_mass(\n flipper_length, weight_flipper_length, intercept_body_mass\n):\n \"\"\"Linear model of the form y = a * x + b\"\"\"\n body_mass = weight_flipper_length * flipper_length + intercept_body_mass\n return body_mass",
"_____no_output_____"
]
],
[
[
"## Main exercise\n\nDefine a vector `weights = [...]` and a vector `intercepts = [...]` of\nthe same length. Each pair of entries `(weights[i], intercepts[i])` tags a\ndifferent model. Use these vectors along with the vector\n`flipper_length_range` to plot several linear models that could possibly\nfit our data. Use the above helper function to visualize both the models and\nthe real samples.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nflipper_length_range = np.linspace(data.min(), data.max(), num=300)",
"_____no_output_____"
],
[
"# solution\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\nweights = [-40, 45, 90]\nintercepts = [15000, -5000, -14000]\n\nax = sns.scatterplot(data=penguins, x=feature_name, y=target_name,\n color=\"black\", alpha=0.5)\n\nlabel = \"{0:.2f} (g / mm) * flipper length + {1:.2f} (g)\"\nfor weight, intercept in zip(weights, intercepts):\n predicted_body_mass = linear_model_flipper_mass(\n flipper_length_range, weight, intercept)\n\n ax.plot(flipper_length_range, predicted_body_mass,\n label=label.format(weight, intercept))\n_ = ax.legend(loc='center left', bbox_to_anchor=(-0.25, 1.25), ncol=1)",
"_____no_output_____"
]
],
[
[
"In the previous question, you were asked to create several linear models.\nThe visualization allowed you to qualitatively assess if a model was better\nthan another.\n\nNow, you should come up with a quantitative measure which indicates the\ngoodness of fit of each linear model and allows you to select the best model.\nDefine a function `goodness_fit_measure(true_values, predictions)` that takes\nas inputs the true target values and the predictions and returns a single\nscalar as output.",
"_____no_output_____"
]
],
[
[
"# solution\ndef goodness_fit_measure(true_values, predictions):\n # we compute the error between the true values and the predictions of our\n # model\n errors = np.ravel(true_values) - np.ravel(predictions)\n # We have several possible strategies to reduce all errors to a single value.\n # Computing the mean error (sum divided by the number of element) might seem\n # like a good solution. However, we have negative errors that will misleadingly\n # reduce the mean error. Therefore, we can either square each\n # error or take the absolute value: these metrics are known as mean\n # squared error (MSE) and mean absolute error (MAE). Let's use the MAE here\n # as an example.\n return np.mean(np.abs(errors))",
"_____no_output_____"
]
],
[
[
"You can now copy and paste the code below to show the goodness of fit for\neach model.\n\n```python\nfor model_idx, (weight, intercept) in enumerate(zip(weights, intercepts)):\n target_predicted = linear_model_flipper_mass(data, weight, intercept)\n print(f\"Model #{model_idx}:\")\n print(f\"{weight:.2f} (g / mm) * flipper length + {intercept:.2f} (g)\")\n print(f\"Error: {goodness_fit_measure(target, target_predicted):.3f}\\n\")\n```",
"_____no_output_____"
]
],
[
[
"# solution\nfor model_idx, (weight, intercept) in enumerate(zip(weights, intercepts)):\n target_predicted = linear_model_flipper_mass(data, weight, intercept)\n print(f\"Model #{model_idx}:\")\n print(f\"{weight:.2f} (g / mm) * flipper length + {intercept:.2f} (g)\")\n print(f\"Error: {goodness_fit_measure(target, target_predicted):.3f}\\n\")",
"Model #0:\n-40.00 (g / mm) * flipper length + 15000.00 (g)\nError: 2764.854\n\nModel #1:\n45.00 (g / mm) * flipper length + -5000.00 (g)\nError: 338.523\n\nModel #2:\n90.00 (g / mm) * flipper length + -14000.00 (g)\nError: 573.041\n\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7907b99c56ae133f4c3b8387b4e0a8e24b422cd | 16,482 | ipynb | Jupyter Notebook | talks/DevSummit2018/ArcGIS Python API - Advanced Scripting/GP/Using geoprocessing tools.ipynb | nitz21/arcpy | 36074b5d448c9cfdba166332e99100afb3390824 | [
"Apache-2.0"
] | 2 | 2020-11-23T23:06:04.000Z | 2020-11-23T23:06:07.000Z | talks/DevSummit2018/ArcGIS Python API - Advanced Scripting/GP/Using geoprocessing tools.ipynb | josemartinsgeo/arcgis-python-api | 4c10bb1ce900060959829f7ac6c58d4d67037d56 | [
"Apache-2.0"
] | null | null | null | talks/DevSummit2018/ArcGIS Python API - Advanced Scripting/GP/Using geoprocessing tools.ipynb | josemartinsgeo/arcgis-python-api | 4c10bb1ce900060959829f7ac6c58d4d67037d56 | [
"Apache-2.0"
] | 1 | 2020-06-06T21:21:18.000Z | 2020-06-06T21:21:18.000Z | 75.260274 | 1,521 | 0.65702 | [
[
[
"# connect to ArcGIS Online\nfrom arcgis.gis import GIS\nfrom arcgis.geoprocessing import import_toolbox\ngis = GIS()",
"_____no_output_____"
]
],
[
[
"# Viewshed tool",
"_____no_output_____"
]
],
[
[
"viewshed = import_toolbox('http://sampleserver1.arcgisonline.com/ArcGIS/rest/services/Elevation/ESRI_Elevation_World/GPServer')",
"_____no_output_____"
],
[
"viewshed.viewshed?",
"_____no_output_____"
],
[
"help(viewshed.viewshed)",
"Help on function viewshed:\n\nviewshed(input_observation_point:arcgis.features.feature.FeatureSet={'geometryType': 'esriGeometryPoint', 'Fields': [{'name': 'FID', 'type': 'esriFieldTypeOID', 'alias': 'FID'}, {'name': 'Shape', 'type': 'esriFieldTypeGeometry', 'alias': 'Shape'}, {'name': 'OffsetA', 'type': 'esriFieldTypeDouble', 'alias': 'OffsetA'}], 'spatialReference': {'wkid': 54003}, 'fields': [{'name': 'FID', 'type': 'esriFieldTypeOID', 'alias': 'FID'}, {'name': 'Shape', 'type': 'esriFieldTypeGeometry', 'alias': 'Shape'}, {'name': 'OffsetA', 'type': 'esriFieldTypeDouble', 'alias': 'OffsetA'}]}, viewshed_distance:arcgis.geoprocessing._types.LinearUnit={'units': 'esriMeters', 'distance': 15000}, gis=None) -> arcgis.features.feature.FeatureSet\n \n \n \n Parameters:\n \n input_observation_point: Input Observation Point (FeatureSet). Required parameter. \n \n viewshed_distance: Viewshed Distance (LinearUnit). Required parameter. \n \n gis: Optional, the GIS on which this tool runs. If not specified, the active GIS is used.\n \n \n Returns: \n viewshed_result - Viewshed Result as a FeatureSet\n \n See http://sampleserver1b.arcgisonline.com/arcgisoutput/Elevation_ESRI_Elevation_World/Viewshed.htm for additional help.\n\n"
],
[
"import arcgis\narcgis.env.out_spatial_reference = 4326",
"_____no_output_____"
],
[
"map = gis.map('South San Francisco', zoomlevel=12)\nmap",
"_____no_output_____"
],
[
"from arcgis.features import Feature, FeatureSet\n\ndef get_viewshed(m, g):\n m.draw(g)\n res = viewshed.viewshed(FeatureSet([Feature(g)]), \"5 Miles\") # \"5 Miles\" or LinearUnit(5, 'Miles') can be passed as input\n m.draw(res)\n \nmap.on_click(get_viewshed)",
"_____no_output_____"
],
[
"def __call__():\n ",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e79083b3327874eade38b511a7edb80165511f7f | 480,310 | ipynb | Jupyter Notebook | MA477 - Theory and Applications of Data Science/Homework/Student Solutions/Homework 3/Kleine_Michael_MA477_Homework3.ipynb | jkstarling/MA477-copy | 67c0d3da587f167d10f2a72700500408704360ad | [
"MIT"
] | null | null | null | MA477 - Theory and Applications of Data Science/Homework/Student Solutions/Homework 3/Kleine_Michael_MA477_Homework3.ipynb | jkstarling/MA477-copy | 67c0d3da587f167d10f2a72700500408704360ad | [
"MIT"
] | null | null | null | MA477 - Theory and Applications of Data Science/Homework/Student Solutions/Homework 3/Kleine_Michael_MA477_Homework3.ipynb | jkstarling/MA477-copy | 67c0d3da587f167d10f2a72700500408704360ad | [
"MIT"
] | 2 | 2020-01-13T14:01:56.000Z | 2020-11-10T15:16:03.000Z | 538.464126 | 73,564 | 0.941644 | [
[
[
"<h2> ======================================================</h2>\n <h1>MA477 - Theory and Applications of Data Science</h1> \n <h1>Homework 3: Matplotlib & Seaborn</h1> \n \n <h4>Dr. Valmir Bucaj</h4>\n <br>\n United States Military Academy, West Point, AY20-2\n<h2>=======================================================</h2>\n\n<h2> Weight: <font color='red'>50pts</font</h2>",
"_____no_output_____"
],
[
"<hr style=\"height:3.2px;border:none;color:#333;background-color:#333;\" />\n\n<h3> Cadet Name:Michael Kleine</h3>\n<br>\n<h3>Date:January 31, 2020 </h3>\n\n<br>\n\n<font color='red' size='3'> <b>$\\dots \\dots$</b> MY DOCUMENTATION IDENTIFIES ALL SOURCES USED AND ASSISTANCE RECEIVED IN THIS ASSIGNMENT\n<br>\n\nMCK<b>$\\dots \\dots$ </b> I DID NOT USE ANY SOURCES OR ASSISTANCE REQUIRING DOCUMENATION IN COMPLETING THIS ASSIGNMENT</font>\n\n<h3> Signature/Initials: </h3>\n\n<hr style=\"height:3px;border:none;color:#333;background-color:#333;\" />",
"_____no_output_____"
],
[
"\n<font size='4'>Complete the following tasks:</font>\n\n<hr style=\"height:2px;border:none;color:#333;background-color:#333;\" />",
"_____no_output_____"
],
[
"Import the following libaraires: `matplotlib.pyplot, seaborn, pandas, numpy`",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport seaborn as sns\nimport pandas as pd\nimport numpy as np\nimport datetime",
"_____no_output_____"
]
],
[
[
"<b>Recreate the following plot as closely as you can</b>\n\n<font size='4' color='red'>10pts</font>",
"_____no_output_____"
]
],
[
[
"#Enter your code here \nx=np.linspace(start=-4,stop=4)\n\nfig=plt.figure(figsize=(8,6))\n\naxes1=fig.add_axes([0.1,0.1,0.9,0.9])\naxes2=fig.add_axes([0.43,0.2,0.25,0.4])\n\n\naxes1.plot(x,3*np.exp(-0.25*x**2),'r-.', lw=3, label=r'3e$^{-0.25x}$')\naxes1.plot(x,2.8*np.exp(-0.15*(x-0.1)**2),marker='o', markerfacecolor='y', markeredgecolor='k',markersize=10,\n markeredgewidth=2, label=r'2.8e$^{-0.15(x-0.1)}$')\n\n\naxes1.legend(loc='upper right')\naxes1.set_xlabel('That took a while',fontsize=18)\n\naxes1.set_title('Many Plots', fontsize=20)\naxes2.set_title('Small Plot', fontsize=12)\n\n\nplt.xticks(ticks=[-2,-1,0,1,2])\nplt.yticks(ticks=[-3,-2,-1,0,1,2,3])\naxes2.text(-1.75,1.5,'Cool',size=16, color='r')\naxes2.text(-0.25,0,'Plot',size=16, color='b')\naxes2.text(1,-2,'Bro',size=16, color='g')",
"_____no_output_____"
],
[
"#Doon't run this cell unless you have recreated it, as the plot below will dissapear \n\n\n",
"_____no_output_____"
]
],
[
[
" <hr style=\"height:3px;border:none;color:#333;background-color:#333;\" />\n \n <font size='4'>For the rest of the exercises we will be using the `Airbnb` dataset contained in this folder.</font>\n \n <b> Read in the dataset and save it as `abnb`</b>",
"_____no_output_____"
]
],
[
[
"#Enter code here\nabnb=pd.read_excel('Airbnb.xlsx')",
"_____no_output_____"
]
],
[
[
"<b>Check out the head of the data:</b>",
"_____no_output_____"
]
],
[
[
"#Enter code here\nabnb.head()",
"_____no_output_____"
],
[
"#Don't run this cell unless you are happy with your answer above\n\n",
"_____no_output_____"
]
],
[
[
"<b> Recreate the following `jointplot` </b>\n\n<font size='4' color='red'>5pts</font>",
"_____no_output_____"
]
],
[
[
"#Enter code here\nsns.set_style('white')\nsns.jointplot(x='number_of_reviews',y='price',data=abnb,height=6, kind='kde')\nplt.show()",
"_____no_output_____"
],
[
"#Don't run this cell unless you are happy with your answer\n\n",
"_____no_output_____"
]
],
[
[
"<b> Recreate the following `boxplots` </b>\n\n<font size='4' color='red'>5pts</font>",
"_____no_output_____"
]
],
[
[
"#Enter code here\nplt.figure(figsize=(12,6))\nsns.boxplot(x='neighbourhood_group',y='price',data=abnb)\nplt.xlabel('Neighbourhood Group', fontsize=14)\nplt.ylabel('Price', fontsize=14)",
"_____no_output_____"
],
[
"#Don't run this cell unless you are happy with your answer\n\n",
"_____no_output_____"
]
],
[
[
"<font size='4' color='red'>10pts</font>",
"_____no_output_____"
]
],
[
[
"#Enter Code Here\nplt.figure(figsize=(12,8))\nsns.boxplot(x='neighbourhood_group',y='number_of_reviews',data=abnb, hue='room_type')\nplt.xlabel('Neighbourhood Group', fontsize=14)\nplt.ylabel('Number of Reviews', fontsize=14)\nplt.ylim(-10,350)",
"_____no_output_____"
],
[
"#Don't run this cell unless you are happy with your answer\n\n",
"_____no_output_____"
]
],
[
[
"<b> Recreate the following `violinplot` comparing the distribution of ONLY `Entire home/apt` and `Private room` for all five `neighbourhood groups`</b>\n\n<font size='4' color='red'>10pts</font>",
"_____no_output_____"
]
],
[
[
"#Enter Code Here\nabnb2 = abnb[ abnb['room_type'] == 'Shared room'].index\nabnb3 = abnb.drop(abnb2)\nplt.figure(figsize=(12,8))\nsns.violinplot(x='neighbourhood_group',y='price',\n data=abnb3,hue='room_type',split=True)\nplt.xlabel('Neighbourhood Group', fontsize=16)\nplt.ylabel('Price', fontsize=16)",
"_____no_output_____"
],
[
"#Don't run this cell unless you are happy with your answer\n\n",
"_____no_output_____"
]
],
[
[
"<font size='5' color='red'>Challenging!!!</font>\n\n\n<b>Time Series: Recreate the following plot</b>\n\n<font color='red' size='4'>10pts</font>\n\n(Hint: Convert the column `last_review` to `DateTime` format and reset it as the index of the dataframe)",
"_____no_output_____"
]
],
[
[
"#Enter answer here \n\n#Format the data\nabnb=pd.read_excel('Airbnb.xlsx')\nabnb['Month'] = pd.to_datetime(abnb['last_review'],yearfirst=True, format='%Y/%m/%d')\nabnb['last_review'] = pd.to_datetime(abnb['last_review'],yearfirst=True, format='%Y/%m/%d')\nabnb = abnb.sort_values(by=['Month'])\nabnb = abnb.set_index('Month')\nabnb = abnb.dropna(axis=0,subset=['last_review'])\n\n#create the plot\nfig = plt.figure(figsize=(12,5))\naxes=fig.add_axes([0.1,0.1,0.9,0.9])\naxes.plot('last_review', 'number_of_reviews', data=abnb)\naxes.plot('last_review', 'price', data=abnb)\n\ndatemin = pd.to_datetime('20161001', format='%Y%m%d', errors='ignore')\ndatemax = pd.to_datetime('20190401', format='%Y%m%d', errors='ignore')\naxes.set_xlim(datemin, datemax)\naxes.legend()\naxes.set_title('Fluctuations in Number of Reviews and Price over time')\naxes.set_xlabel('last_review')",
"_____no_output_____"
]
],
[
[
"<font color='red' size=4>Instructor Comments: Only some minor styling issues and label.\n \n -0.5pts</font>\n",
"_____no_output_____"
]
],
[
[
"#Don't erase this cell unless you are happy with your answer\n\n",
"_____no_output_____"
],
[
"#https://matplotlib.org/tutorials/text/mathtext.html\n#https://www.overleaf.com/learn/latex/Subscripts_and_superscripts\n#https://thispointer.com/python-pandas-how-to-drop-rows-in-dataframe-by-conditions-on-column-values/\n#https://www.geeksforgeeks.org/change-data-type-for-one-or-more-columns-in-pandas-dataframe/\n#https://stackoverflow.com/questions/26763344/convert-pandas-column-to-datetime\n#https://appdividend.com/2019/01/26/pandas-set-index-example-python-set_index-tutorial/\n#https://stackoverflow.com/questions/28161356/sort-pandas-dataframe-by-date",
"_____no_output_____"
]
],
[
[
"<font color='red' size=4>Total Score\n \n 49.5pts</font>\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
e790a85ca9f3f8877091e46f281afb9c196f72cc | 45,454 | ipynb | Jupyter Notebook | Assignment4.ipynb | mukund109/Numerical_Analysis_PHYS3142 | 8c28beafb627ea6f98cdb1e66f32eb31b0283238 | [
"MIT"
] | null | null | null | Assignment4.ipynb | mukund109/Numerical_Analysis_PHYS3142 | 8c28beafb627ea6f98cdb1e66f32eb31b0283238 | [
"MIT"
] | null | null | null | Assignment4.ipynb | mukund109/Numerical_Analysis_PHYS3142 | 8c28beafb627ea6f98cdb1e66f32eb31b0283238 | [
"MIT"
] | null | null | null | 102.14382 | 15,272 | 0.85141 | [
[
[
"import matplotlib.pyplot as plt\nfrom math import exp\n%matplotlib inline",
"_____no_output_____"
],
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"## Q1",
"_____no_output_____"
],
[
"Solving the equation for all c in [0,4)",
"_____no_output_____"
]
],
[
[
"x_list = [] \nfor c_ in range(0,400,2):\n c = c_/100\n \n #the accuracy\n delta = 1\n \n x=1\n itr = 0\n while delta>1e-7:\n x_new = 1-exp(-1*c*x)\n delta = abs(x_new- x)\n x = x_new\n itr += 1\n \n if c==3:\n print(\"For c=3, x={}\".format(x))\n print(\"Number of iterations: {}\".format(itr))\n x_list.append(x)",
"For c=3, x=0.9404798005896199\nNumber of iterations: 9\n"
]
],
[
[
"A plot of the percolation transition",
"_____no_output_____"
]
],
[
[
"plt.plot([i/100 for i in range(0,400,2)], x_list)\nplt.xlabel('c')\nplt.ylabel('x')",
"_____no_output_____"
]
],
[
[
"## Q2",
"_____no_output_____"
],
[
"### a)",
"_____no_output_____"
],
[
"The overrelaxation formula is given by:\n$$\nx_{n+1} = (1+\\omega )f(x_n) - \\omega x_n \n$$\n\n$$\n\\implies f(x_n) = \\frac{x_{n+1} + \\omega x_n}{1+\\omega} \\ \\ \\ \\ \\ (1)\n$$\n\nDenote the true solution by $x^*$, then the Taylor series expansion of $f$ around $x_n$ is given by:\n\n$$\nf(x^*) \\approx f(x_n) + f'(x_n)(x^* - x_n)\n$$\nSince, $x^* = f(x^*)$,\n\n$$\nx^* \\approx f(x_n) + f'(x_n)(x^* - x_n)\n$$\n\nSubstituting in equation 1 and solving for $x^*$:\n$$\nx^* \\approx \\frac{\\frac{x_{n+1} + \\omega x_n}{1 + \\omega} - f'(x_n) x_n}{1-f'(x_n)}\n\\\\\n\\implies x^* - x_{n+1} \\approx \\frac{x_n - x_{n+1}}{1 - \\frac{1}{(1+\\omega) f'(x_n) - \\omega}}\n$$\n",
"_____no_output_____"
],
[
"### b) and c)",
"_____no_output_____"
]
],
[
[
"itr_list = [] \nw_list = []\nfor w_ in range(0,60,2):\n w = w_/100\n delta = 1\n x=1\n itr = 0\n while delta>1e-7:\n x_new = (1+w)*(1-exp(-3*x)) - w*x\n delta = abs(x_new- x)\n x = x_new\n itr += 1\n \n print(\"For w={}\".format(w), end=', ')\n print(\"Number of iterations: {}\".format(itr), end='\\n \\n')\n itr_list.append(itr)\n w_list.append(w)",
"For w=0.0, Number of iterations: 9\n \nFor w=0.02, Number of iterations: 9\n \nFor w=0.04, Number of iterations: 8\n \nFor w=0.06, Number of iterations: 8\n \nFor w=0.08, Number of iterations: 7\n \nFor w=0.1, Number of iterations: 7\n \nFor w=0.12, Number of iterations: 7\n \nFor w=0.14, Number of iterations: 6\n \nFor w=0.16, Number of iterations: 6\n \nFor w=0.18, Number of iterations: 5\n \nFor w=0.2, Number of iterations: 4\n \nFor w=0.22, Number of iterations: 4\n \nFor w=0.24, Number of iterations: 5\n \nFor w=0.26, Number of iterations: 6\n \nFor w=0.28, Number of iterations: 6\n \nFor w=0.3, Number of iterations: 7\n \nFor w=0.32, Number of iterations: 7\n \nFor w=0.34, Number of iterations: 7\n \nFor w=0.36, Number of iterations: 8\n \nFor w=0.38, Number of iterations: 8\n \nFor w=0.4, Number of iterations: 9\n \nFor w=0.42, Number of iterations: 9\n \nFor w=0.44, Number of iterations: 9\n \nFor w=0.46, Number of iterations: 10\n \nFor w=0.48, Number of iterations: 10\n \nFor w=0.5, Number of iterations: 11\n \nFor w=0.52, Number of iterations: 11\n \nFor w=0.54, Number of iterations: 12\n \nFor w=0.56, Number of iterations: 12\n \nFor w=0.58, Number of iterations: 13\n \n"
]
],
[
[
"### d)",
"_____no_output_____"
],
[
"The recursive formula for the error can be obtained by rearranging the previous equations to get:\n\n$$\n\\epsilon_{n+1} = \\epsilon_{n} [(1+\\omega) f'(x^*) - \\omega]\n$$\n\n(Note: This is an approximation for when $x_{n}$ is close to $x^*$)\n\nIn order to find the conditions when the overrelaxation method with $\\omega < 0$ converges faster than the ordinary relaxation method ($\\omega = 0$), we need to find values of $f'(x^*)$ and $\\omega$ that satisfy the following constraints:\n\n1. Overrelaxation method converges:\n\n$$\n |(1+\\omega) f'(x^*) - \\omega| < 1\n$$\n\n2. Ordinary relaxation method converges:\n$$\n |f'(x^*)| < 1\n$$\n\n3. Overrelaxation converges faster:\n$$\n|(1+\\omega) f'(x^*) - \\omega | < |f'(x^*)|\n$$\n\n4. Overrelaxation factor is negative:\n$$\n\\omega < 0\n$$",
"_____no_output_____"
],
[
"We can plot this region",
"_____no_output_____"
]
],
[
[
"fy, wx = np.meshgrid(np.linspace(-2,1,1000), np.linspace(-2,1,1000))\nmask = np.zeros((1000, 1000), dtype=bool)\n\nmask[(np.abs((1+wx)*fy - wx) < 1) & \\\n(np.abs(fy) < 1) & \\\n(np.abs((1+wx)*fy - wx) < np.abs(fy)) & \\\n(wx < 0)] = 2\nplt.contour(wx, fy, mask, cmap='flag')\nplt.xlabel(\"w\")\nplt.ylabel(\"f '(x*)\")",
"_____no_output_____"
]
],
[
[
"Therefore, when the current estimate is sufficiently close to the actual solution, when $f'(x^*)$ and $\\omega$ fall inside this region, the overrelaxation converges faster than ordinary relaxation ",
"_____no_output_____"
],
[
"We can take the simple example of the case when $f(x) = -0.75x$\n\nThis resembles all cases where the function is locally linear with slope $-0.75$",
"_____no_output_____"
]
],
[
[
"itr_list = [] \nw_list = []\nfor w_ in range(-80,1,1):\n w = w_/100\n delta = 1\n x=0.1\n itr = 0\n while delta>1e-7:\n x_new = (1+w)*(-0.75*x) - w*x\n delta = abs(x_new- x)\n x = x_new\n itr += 1\n \n itr_list.append(itr)\n w_list.append(w)\nplt.plot(w_list, itr_list)\nplt.xlabel(\"w\")\nplt.ylabel(\"Number of iterations\")",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
e790b10725ba7b5512ff2380f6d8c1cdfa69aa60 | 34,489 | ipynb | Jupyter Notebook | ai_art_book.ipynb | Dazzla/Codility-Lessons-In-Java | fbe8b55241e3186cc8e26c167ee9ee4ed8b41947 | [
"Apache-2.0"
] | null | null | null | ai_art_book.ipynb | Dazzla/Codility-Lessons-In-Java | fbe8b55241e3186cc8e26c167ee9ee4ed8b41947 | [
"Apache-2.0"
] | null | null | null | ai_art_book.ipynb | Dazzla/Codility-Lessons-In-Java | fbe8b55241e3186cc8e26c167ee9ee4ed8b41947 | [
"Apache-2.0"
] | null | null | null | 48.168994 | 322 | 0.531764 | [
[
[
"<a href=\"https://colab.research.google.com/github/Dazzla/Codility-Lessons-In-Java/blob/master/ai_art_book.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"# Generate images from text sentences with VQGAN and CLIP (z+quantize method with augmentations).\n\nNotebook made by Katherine Crowson (https://github.com/crowsonkb, https://twitter.com/RiversHaveWings). The original BigGAN+CLIP method was made by https://twitter.com/advadnoun. Translated and added explanations, and modifications by Eleiber#8347, and the friendly interface was made thanks to Abulafia#3734.\n\nFor a detailed tutorial on how to use it, I recommend visiting this article https://tuscriaturas.miraheze.org/wiki/Ayuda:Generar_im%C3%A1genes_con_VQGAN%2BCLIP * by Jakeukalane#2767 and Avengium (Angel)#3715 \n\n*Google-translated to English: https://tuscriaturas-miraheze-org.translate.goog/wiki/Ayuda:Generar_im%C3%A1genes_con_VQGAN%2BCLIP?_x_tr_sl=es&_x_tr_tl=en&_x_tr_hl=en-US&_x_tr_pto=wapp",
"_____no_output_____"
]
],
[
[
"# @title Licensed under the MIT License\n\n# Copyright (c) 2021 Katherine Crowson\n\n# Permission is hereby granted, free of charge, to any person obtaining a copy\n# of this software and associated documentation files (the \"Software\"), to deal\n# in the Software without restriction, including without limitation the rights\n# to use, copy, modify, merge, publish, distribute, sublicense, and/or sell\n# copies of the Software, and to permit persons to whom the Software is\n# furnished to do so, subject to the following conditions:\n\n# The above copyright notice and this permission notice shall be included in\n# all copies or substantial portions of the Software.\n\n# THE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR\n# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,\n# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE\n# AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER\n# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,\n# OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN\n# THE SOFTWARE.",
"_____no_output_____"
],
[
"!nvidia-smi",
"_____no_output_____"
],
[
"# @title Install libraries\n# @markdown This cell will take a while because it has to download several libraries\n \nprint(\"Installing CLIP...\")\n!git clone https://github.com/openai/CLIP &> /dev/null\n \nprint(\"Installing Python lbraries for IA...\")\n!git clone https://github.com/CompVis/taming-transformers \n!pip install ftfy regex tqdm omegaconf pytorch-lightning &> /dev/null\n!pip install kornia &> /dev/null\n!pip install einops &> /dev/null\n!pip install wget &> /dev/null\n!pip install tdqm \n \nprint(\"Installing metadata tools...\")\n!pip install stegano &> /dev/null\n!apt install exempi &> /dev/null\n!pip install python-xmp-toolkit &> /dev/null\n!pip install imgtag &> /dev/null\n!pip install pillow==7.1.2 &> /dev/null\n \nprint(\"Installing video creation tooling...\")\n!pip install imageio-ffmpeg &> /dev/null\n!mkdir steps\nprint(\"Finalising installation.\")",
"_____no_output_____"
],
[
"#@title Select model\n#@markdown By default, the notebook downloads model 16384 from ImageNet. There are others that are not downloaded by default, since it would be unneccssary if you are not going to use them, so if you want to use them, simply select the models to download. \n\nimagenet_1024 = False #@param {type:\"boolean\"}\nimagenet_16384 = True #@param {type:\"boolean\"}\ngumbel_8192 = False #@param {type:\"boolean\"}\ncoco = False #@param {type:\"boolean\"}\nfaceshq = False #@param {type:\"boolean\"}\nwikiart_1024 = False #@param {type:\"boolean\"}\nwikiart_16384 = False #@param {type:\"boolean\"}\nsflckr = False #@param {type:\"boolean\"}\nade20k = False #@param {type:\"boolean\"}\nffhq = False #@param {type:\"boolean\"}\ncelebahq = False #@param {type:\"boolean\"}\n\nif imagenet_1024:\n !curl -L -o vqgan_imagenet_f16_1024.yaml -C - 'https://heibox.uni-heidelberg.de/d/8088892a516d4e3baf92/files/?p=%2Fconfigs%2Fmodel.yaml&dl=1' #ImageNet 1024\n !curl -L -o vqgan_imagenet_f16_1024.ckpt -C - 'https://heibox.uni-heidelberg.de/d/8088892a516d4e3baf92/files/?p=%2Fckpts%2Flast.ckpt&dl=1' #ImageNet 1024\nif imagenet_16384:\n !curl -L -o vqgan_imagenet_f16_16384.yaml -C - 'https://heibox.uni-heidelberg.de/d/a7530b09fed84f80a887/files/?p=%2Fconfigs%2Fmodel.yaml&dl=1' #ImageNet 16384\n !curl -L -o vqgan_imagenet_f16_16384.ckpt -C - 'https://heibox.uni-heidelberg.de/d/a7530b09fed84f80a887/files/?p=%2Fckpts%2Flast.ckpt&dl=1' #ImageNet 16384\nif gumbel_8192:\n !curl -L -o gumbel_8192.yaml -C - 'https://heibox.uni-heidelberg.de/d/2e5662443a6b4307b470/files/?p=%2Fconfigs%2Fmodel.yaml&dl=1' #Gumbel 8192\n !curl -L -o gumbel_8192.ckpt -C - 'https://heibox.uni-heidelberg.de/d/2e5662443a6b4307b470/files/?p=%2Fckpts%2Flast.ckpt&dl=1' #Gumbel 8192\nif coco:\n !curl -L -o coco.yaml -C - 'https://dl.nmkd.de/ai/clip/coco/coco.yaml' #COCO\n !curl -L -o coco.ckpt -C - 'https://dl.nmkd.de/ai/clip/coco/coco.ckpt' #COCO\nif faceshq:\n !curl -L -o faceshq.yaml -C - 'https://drive.google.com/uc?export=download&id=1fHwGx_hnBtC8nsq7hesJvs-Klv-P0gzT' #FacesHQ\n !curl -L -o faceshq.ckpt -C - 'https://app.koofr.net/content/links/a04deec9-0c59-4673-8b37-3d696fe63a5d/files/get/last.ckpt?path=%2F2020-11-13T21-41-45_faceshq_transformer%2Fcheckpoints%2Flast.ckpt' #FacesHQ\nif wikiart_1024: \n !curl -L -o wikiart_1024.yaml -C - 'http://mirror.io.community/blob/vqgan/wikiart.yaml' #WikiArt 1024\n !curl -L -o wikiart_1024.ckpt -C - 'http://mirror.io.community/blob/vqgan/wikiart.ckpt' #WikiArt 1024\nif wikiart_16384: \n !curl -L -o wikiart_16384.yaml -C - 'http://eaidata.bmk.sh/data/Wikiart_16384/wikiart_f16_16384_8145600.yaml' #WikiArt 16384\n !curl -L -o wikiart_16384.ckpt -C - 'http://eaidata.bmk.sh/data/Wikiart_16384/wikiart_f16_16384_8145600.ckpt' #WikiArt 16384\nif sflckr:\n !curl -L -o sflckr.yaml -C - 'https://heibox.uni-heidelberg.de/d/73487ab6e5314cb5adba/files/?p=%2Fconfigs%2F2020-11-09T13-31-51-project.yaml&dl=1' #S-FLCKR\n !curl -L -o sflckr.ckpt -C - 'https://heibox.uni-heidelberg.de/d/73487ab6e5314cb5adba/files/?p=%2Fcheckpoints%2Flast.ckpt&dl=1' #S-FLCKR\nif ade20k:\n !curl -L -o ade20k.yaml -C - 'https://static.miraheze.org/intercriaturaswiki/b/bf/Ade20k.txt' #ADE20K\n !curl -L -o ade20k.ckpt -C - 'https://app.koofr.net/content/links/0f65c2cd-7102-4550-a2bd-07fd383aac9e/files/get/last.ckpt?path=%2F2020-11-20T21-45-44_ade20k_transformer%2Fcheckpoints%2Flast.ckpt' #ADE20K\nif ffhq:\n !curl -L -o ffhq.yaml -C - 'https://app.koofr.net/content/links/0fc005bf-3dca-4079-9d40-cdf38d42cd7a/files/get/2021-04-23T18-19-01-project.yaml?path=%2F2021-04-23T18-19-01_ffhq_transformer%2Fconfigs%2F2021-04-23T18-19-01-project.yaml&force' #FFHQ\n !curl -L -o ffhq.ckpt -C - 'https://app.koofr.net/content/links/0fc005bf-3dca-4079-9d40-cdf38d42cd7a/files/get/last.ckpt?path=%2F2021-04-23T18-19-01_ffhq_transformer%2Fcheckpoints%2Flast.ckpt&force' #FFHQ\nif celebahq:\n !curl -L -o celebahq.yaml -C - 'https://app.koofr.net/content/links/6dddf083-40c8-470a-9360-a9dab2a94e96/files/get/2021-04-23T18-11-19-project.yaml?path=%2F2021-04-23T18-11-19_celebahq_transformer%2Fconfigs%2F2021-04-23T18-11-19-project.yaml&force' #CelebA-HQ\n !curl -L -o celebahq.ckpt -C - 'https://app.koofr.net/content/links/6dddf083-40c8-470a-9360-a9dab2a94e96/files/get/last.ckpt?path=%2F2021-04-23T18-11-19_celebahq_transformer%2Fcheckpoints%2Flast.ckpt&force' #CelebA-HQ",
"_____no_output_____"
],
[
"# @title Load libraries and definitions\n \nimport argparse\nimport math\nfrom pathlib import Path\nimport sys\n \nsys.path.append('./taming-transformers')\nfrom IPython import display\nfrom base64 import b64encode\nfrom omegaconf import OmegaConf\nfrom PIL import Image\nfrom taming.models import cond_transformer, vqgan\nimport torch\nfrom torch import nn, optim\nfrom torch.nn import functional as F\nfrom torchvision import transforms\nfrom torchvision.transforms import functional as TF\nfrom tqdm.notebook import tqdm\n \nfrom CLIP import clip\nimport kornia.augmentation as K\nimport numpy as np\nimport imageio\nfrom PIL import ImageFile, Image\nfrom imgtag import ImgTag # metadatos \nfrom libxmp import * # metadatos\nimport libxmp # metadatos\nfrom stegano import lsb\nimport json\nfrom tqdm.notebook import tqdm\nImageFile.LOAD_TRUNCATED_IMAGES = True\n \ndef sinc(x):\n return torch.where(x != 0, torch.sin(math.pi * x) / (math.pi * x), x.new_ones([]))\n \n \ndef lanczos(x, a):\n cond = torch.logical_and(-a < x, x < a)\n out = torch.where(cond, sinc(x) * sinc(x/a), x.new_zeros([]))\n return out / out.sum()\n \n \ndef ramp(ratio, width):\n n = math.ceil(width / ratio + 1)\n out = torch.empty([n])\n cur = 0\n for i in range(out.shape[0]):\n out[i] = cur\n cur += ratio\n return torch.cat([-out[1:].flip([0]), out])[1:-1]\n \n \ndef resample(input, size, align_corners=True):\n n, c, h, w = input.shape\n dh, dw = size\n \n input = input.view([n * c, 1, h, w])\n \n if dh < h:\n kernel_h = lanczos(ramp(dh / h, 2), 2).to(input.device, input.dtype)\n pad_h = (kernel_h.shape[0] - 1) // 2\n input = F.pad(input, (0, 0, pad_h, pad_h), 'reflect')\n input = F.conv2d(input, kernel_h[None, None, :, None])\n \n if dw < w:\n kernel_w = lanczos(ramp(dw / w, 2), 2).to(input.device, input.dtype)\n pad_w = (kernel_w.shape[0] - 1) // 2\n input = F.pad(input, (pad_w, pad_w, 0, 0), 'reflect')\n input = F.conv2d(input, kernel_w[None, None, None, :])\n \n input = input.view([n, c, h, w])\n return F.interpolate(input, size, mode='bicubic', align_corners=align_corners)\n \n \nclass ReplaceGrad(torch.autograd.Function):\n @staticmethod\n def forward(ctx, x_forward, x_backward):\n ctx.shape = x_backward.shape\n return x_forward\n \n @staticmethod\n def backward(ctx, grad_in):\n return None, grad_in.sum_to_size(ctx.shape)\n \n \nreplace_grad = ReplaceGrad.apply\n \n \nclass ClampWithGrad(torch.autograd.Function):\n @staticmethod\n def forward(ctx, input, min, max):\n ctx.min = min\n ctx.max = max\n ctx.save_for_backward(input)\n return input.clamp(min, max)\n \n @staticmethod\n def backward(ctx, grad_in):\n input, = ctx.saved_tensors\n return grad_in * (grad_in * (input - input.clamp(ctx.min, ctx.max)) >= 0), None, None\n \n \nclamp_with_grad = ClampWithGrad.apply\n \n \ndef vector_quantize(x, codebook):\n d = x.pow(2).sum(dim=-1, keepdim=True) + codebook.pow(2).sum(dim=1) - 2 * x @ codebook.T\n indices = d.argmin(-1)\n x_q = F.one_hot(indices, codebook.shape[0]).to(d.dtype) @ codebook\n return replace_grad(x_q, x)\n \n \nclass Prompt(nn.Module):\n def __init__(self, embed, weight=1., stop=float('-inf')):\n super().__init__()\n self.register_buffer('embed', embed)\n self.register_buffer('weight', torch.as_tensor(weight))\n self.register_buffer('stop', torch.as_tensor(stop))\n \n def forward(self, input):\n input_normed = F.normalize(input.unsqueeze(1), dim=2)\n embed_normed = F.normalize(self.embed.unsqueeze(0), dim=2)\n dists = input_normed.sub(embed_normed).norm(dim=2).div(2).arcsin().pow(2).mul(2)\n dists = dists * self.weight.sign()\n return self.weight.abs() * replace_grad(dists, torch.maximum(dists, self.stop)).mean()\n \n \ndef parse_prompt(prompt):\n vals = prompt.rsplit(':', 2)\n vals = vals + ['', '1', '-inf'][len(vals):]\n return vals[0], float(vals[1]), float(vals[2])\n \n \nclass MakeCutouts(nn.Module):\n def __init__(self, cut_size, cutn, cut_pow=1.):\n super().__init__()\n self.cut_size = cut_size\n self.cutn = cutn\n self.cut_pow = cut_pow\n self.augs = nn.Sequential(\n K.RandomHorizontalFlip(p=0.5),\n # K.RandomSolarize(0.01, 0.01, p=0.7),\n K.RandomSharpness(0.3,p=0.4),\n K.RandomAffine(degrees=30, translate=0.1, p=0.8, padding_mode='border'),\n K.RandomPerspective(0.2,p=0.4),\n K.ColorJitter(hue=0.01, saturation=0.01, p=0.7))\n self.noise_fac = 0.1\n \n \n def forward(self, input):\n sideY, sideX = input.shape[2:4]\n max_size = min(sideX, sideY)\n min_size = min(sideX, sideY, self.cut_size)\n cutouts = []\n for _ in range(self.cutn):\n size = int(torch.rand([])**self.cut_pow * (max_size - min_size) + min_size)\n offsetx = torch.randint(0, sideX - size + 1, ())\n offsety = torch.randint(0, sideY - size + 1, ())\n cutout = input[:, :, offsety:offsety + size, offsetx:offsetx + size]\n cutouts.append(resample(cutout, (self.cut_size, self.cut_size)))\n batch = self.augs(torch.cat(cutouts, dim=0))\n if self.noise_fac:\n facs = batch.new_empty([self.cutn, 1, 1, 1]).uniform_(0, self.noise_fac)\n batch = batch + facs * torch.randn_like(batch)\n return batch\n \n \ndef load_vqgan_model(config_path, checkpoint_path):\n config = OmegaConf.load(config_path)\n if config.model.target == 'taming.models.vqgan.VQModel':\n model = vqgan.VQModel(**config.model.params)\n model.eval().requires_grad_(False)\n model.init_from_ckpt(checkpoint_path)\n elif config.model.target == 'taming.models.cond_transformer.Net2NetTransformer':\n parent_model = cond_transformer.Net2NetTransformer(**config.model.params)\n parent_model.eval().requires_grad_(False)\n parent_model.init_from_ckpt(checkpoint_path)\n model = parent_model.first_stage_model\n elif config.model.target == 'taming.models.vqgan.GumbelVQ':\n model = vqgan.GumbelVQ(**config.model.params)\n print(config.model.params)\n model.eval().requires_grad_(False)\n model.init_from_ckpt(checkpoint_path)\n else:\n raise ValueError(f'unknown model type: {config.model.target}')\n del model.loss\n return model\n \n \ndef resize_image(image, out_size):\n ratio = image.size[0] / image.size[1]\n area = min(image.size[0] * image.size[1], out_size[0] * out_size[1])\n size = round((area * ratio)**0.5), round((area / ratio)**0.5)\n return image.resize(size, Image.LANCZOS)\n\ndef download_img(img_url):\n try:\n return wget.download(img_url,out=\"input.jpg\")\n except:\n return\n",
"_____no_output_____"
]
],
[
[
"## Execution Parameters\nMainly what you will have to modify will be texts:, there you can place the text(s) you want to generate (separated with | ). It is a list because you can put more than one text, and so the AI tries to 'mix' the images, giving the same priority to both texts.\n\nTo use an initial image to the model, you just have to upload a file to the Colab environment (in the section on the left), and then modify initial_image: putting the exact name of the file. Example: sample.png\n\nYou can also modify the model by changing the lines that say model:. Currently 1024, 16384, Gumbel, COCO-Stuff, FacesHQ, WikiArt, S-FLCKR, Ade20k, FFHQ and CelebaHQ are available. To activate them you have to have downloaded them first, and then you can just select it.\n\nYou can also use target_images, which is basically putting one or more images on it that the AI will take as \"target\", fulfilling the same function as putting a text on it. To put more than one you have to use | as a separator.",
"_____no_output_____"
]
],
[
[
"#@title Parameter\ntext = \"Complex building\" #@param {type:\"string\"}\nwidth = 480#@param {type:\"number\"}\nheight = 480#@param {type:\"number\"}\nmodel = \"vqgan_imagenet_f16_16384\" #@param [\"vqgan_imagenet_f16_16384\", \"vqgan_imagenet_f16_1024\", \"wikiart_1024\", \"wikiart_16384\", \"coco\", \"faceshq\", \"sflckr\", \"ade20k\", \"ffhq\", \"celebahq\", \"gumbel_8192\"]\nimage_interval = 50#@param {type:\"number\"}\ninitial_image = None#@param {type:\"string\"}\nobject_images = None#@param {type:\"string\"}\nseed = -1#@param {type:\"number\"}\nmax_iterations = -1#@param {type:\"number\"}\ninput_images = \"\"\n\nmodel_names={\"vqgan_imagenet_f16_16384\": 'ImageNet 16384', \"vqgan_imagenet_f16_1024\": \"ImageNet 1024\",\n \"wikiart_1024\":\"WikiArt 1024\", \"wikiart_16384\":\"WikiArt 16384\", \"coco\":\"COCO-Stuff\", \"faceshq\":\"FacesHQ\", \"sflckr\":\"S-FLCKR\", \"ade20k\":\"ADE20K\", \"ffhq\":\"FFHQ\", \"celebahq\":\"CelebA-HQ\", \"gumbel_8192\": \"Gumbel 8192\"}\nmodel_name = model_names[model]\n\nif model == \"gumbel_8192\":\n is_gumbel = True\nelse:\n is_gumbel = False\n\nif seed == -1:\n seed = None\nif initial_image == \"None\":\n initial_image = None\nelif initial_image and initial_image.lower().startswith(\"http\"):\n initial_image = download_img(initial_image)\n\n\nif object_images == \"None\" or not object_images:\n object_images = []\nelse:\n object_images = object_images.split(\"|\")\n object_images = [image.strip() for image in object_images]\n\nif initial_image or object_images != []:\n input_images = True\n\ntext = [frase.strip() for frase in text.split(\"|\")]\nif text == ['']:\n text = []\n\n\nargs = argparse.Namespace(\n prompts=text,\n image_prompts=object_images,\n noise_prompt_seeds=[],\n noise_prompt_weights=[],\n size=[width, height],\n init_image=initial_image,\n init_weight=0.,\n clip_model='ViT-B/32',\n vqgan_config=f'{model}.yaml',\n vqgan_checkpoint=f'{model}.ckpt',\n step_size=0.1,\n cutn=64,\n cut_pow=1.,\n display_freq=image_interval,\n seed=seed,\n)",
"_____no_output_____"
],
[
"#@title Execute...\ndevice = torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')\nprint('Using device:', device)\nif text:\n print('Using texts:', text)\nif object_images:\n print('Using image prompts:', object_images)\nif args.seed is None:\n seed = torch.seed()\nelse:\n seed = args.seed\ntorch.manual_seed(seed)\nprint('Using seed:', seed)\n\nmodel = load_vqgan_model(args.vqgan_config, args.vqgan_checkpoint).to(device)\nperceptor = clip.load(args.clip_model, jit=False)[0].eval().requires_grad_(False).to(device)\n\ncut_size = perceptor.visual.input_resolution\nif is_gumbel:\n e_dim = model.quantize.embedding_dim\nelse:\n e_dim = model.quantize.e_dim\n\nf = 2**(model.decoder.num_resolutions - 1)\nmake_cutouts = MakeCutouts(cut_size, args.cutn, cut_pow=args.cut_pow)\nif is_gumbel:\n n_toks = model.quantize.n_embed\nelse:\n n_toks = model.quantize.n_e\n\ntoksX, toksY = args.size[0] // f, args.size[1] // f\nsideX, sideY = toksX * f, toksY * f\nif is_gumbel:\n z_min = model.quantize.embed.weight.min(dim=0).values[None, :, None, None]\n z_max = model.quantize.embed.weight.max(dim=0).values[None, :, None, None]\nelse:\n z_min = model.quantize.embedding.weight.min(dim=0).values[None, :, None, None]\n z_max = model.quantize.embedding.weight.max(dim=0).values[None, :, None, None]\n\nif args.init_image:\n pil_image = Image.open(args.init_image).convert('RGB')\n pil_image = pil_image.resize((sideX, sideY), Image.LANCZOS)\n z, *_ = model.encode(TF.to_tensor(pil_image).to(device).unsqueeze(0) * 2 - 1)\nelse:\n one_hot = F.one_hot(torch.randint(n_toks, [toksY * toksX], device=device), n_toks).float()\n if is_gumbel:\n z = one_hot @ model.quantize.embed.weight\n else:\n z = one_hot @ model.quantize.embedding.weight\n z = z.view([-1, toksY, toksX, e_dim]).permute(0, 3, 1, 2)\nz_orig = z.clone()\nz.requires_grad_(True)\nopt = optim.Adam([z], lr=args.step_size)\n\nnormalize = transforms.Normalize(mean=[0.48145466, 0.4578275, 0.40821073],\n std=[0.26862954, 0.26130258, 0.27577711])\n\npMs = []\n\nfor prompt in args.prompts:\n txt, weight, stop = parse_prompt(prompt)\n embed = perceptor.encode_text(clip.tokenize(txt).to(device)).float()\n pMs.append(Prompt(embed, weight, stop).to(device))\n\nfor prompt in args.image_prompts:\n path, weight, stop = parse_prompt(prompt)\n img = resize_image(Image.open(path).convert('RGB'), (sideX, sideY))\n batch = make_cutouts(TF.to_tensor(img).unsqueeze(0).to(device))\n embed = perceptor.encode_image(normalize(batch)).float()\n pMs.append(Prompt(embed, weight, stop).to(device))\n\nfor seed, weight in zip(args.noise_prompt_seeds, args.noise_prompt_weights):\n gen = torch.Generator().manual_seed(seed)\n embed = torch.empty([1, perceptor.visual.output_dim]).normal_(generator=gen)\n pMs.append(Prompt(embed, weight).to(device))\n\ndef synth(z):\n if is_gumbel:\n z_q = vector_quantize(z.movedim(1, 3), model.quantize.embed.weight).movedim(3, 1)\n else:\n z_q = vector_quantize(z.movedim(1, 3), model.quantize.embedding.weight).movedim(3, 1)\n\n return clamp_with_grad(model.decode(z_q).add(1).div(2), 0, 1)\n\ndef add_xmp_data(filename):\n imagen = ImgTag(filename=filename)\n imagen.xmp.append_array_item(libxmp.consts.XMP_NS_DC, 'creator', 'VQGAN+CLIP', {\"prop_array_is_ordered\":True, \"prop_value_is_array\":True})\n if args.prompts:\n imagen.xmp.append_array_item(libxmp.consts.XMP_NS_DC, 'title', \" | \".join(args.prompts), {\"prop_array_is_ordered\":True, \"prop_value_is_array\":True})\n else:\n imagen.xmp.append_array_item(libxmp.consts.XMP_NS_DC, 'title', 'None', {\"prop_array_is_ordered\":True, \"prop_value_is_array\":True})\n imagen.xmp.append_array_item(libxmp.consts.XMP_NS_DC, 'i', str(i), {\"prop_array_is_ordered\":True, \"prop_value_is_array\":True})\n imagen.xmp.append_array_item(libxmp.consts.XMP_NS_DC, 'model', nombre_modelo, {\"prop_array_is_ordered\":True, \"prop_value_is_array\":True})\n imagen.xmp.append_array_item(libxmp.consts.XMP_NS_DC, 'seed',str(seed) , {\"prop_array_is_ordered\":True, \"prop_value_is_array\":True})\n imagen.xmp.append_array_item(libxmp.consts.XMP_NS_DC, 'input_images',str(input_images) , {\"prop_array_is_ordered\":True, \"prop_value_is_array\":True})\n #for frases in args.prompts:\n # imagen.xmp.append_array_item(libxmp.consts.XMP_NS_DC, 'Prompt' ,frases, {\"prop_array_is_ordered\":True, \"prop_value_is_array\":True})\n imagen.close()\n\ndef add_stegano_data(filename):\n data = {\n \"title\": \" | \".join(args.prompts) if args.prompts else None,\n \"notebook\": \"VQGAN+CLIP\",\n \"i\": i,\n \"model\": model_name,\n \"seed\": str(seed),\n \"input_images\": input_images\n }\n lsb.hide(filename, json.dumps(data)).save(filename)\n\[email protected]_grad()\ndef checkin(i, losses):\n losses_str = ', '.join(f'{loss.item():g}' for loss in losses)\n tqdm.write(f'i: {i}, loss: {sum(losses).item():g}, losses: {losses_str}')\n out = synth(z)\n TF.to_pil_image(out[0].cpu()).save('progress.png')\n add_stegano_data('progress.png')\n add_xmp_data('progress.png')\n display.display(display.Image('progress.png'))\n\ndef ascend_txt():\n global i\n out = synth(z)\n iii = perceptor.encode_image(normalize(make_cutouts(out))).float()\n\n result = []\n\n if args.init_weight:\n result.append(F.mse_loss(z, z_orig) * args.init_weight / 2)\n\n for prompt in pMs:\n result.append(prompt(iii))\n img = np.array(out.mul(255).clamp(0, 255)[0].cpu().detach().numpy().astype(np.uint8))[:,:,:]\n img = np.transpose(img, (1, 2, 0))\n filename = f\"steps/{i:04}.png\"\n imageio.imwrite(filename, np.array(img))\n add_stegano_data(filename)\n add_xmp_data(filename)\n return result\n\ndef train(i):\n opt.zero_grad()\n lossAll = ascend_txt()\n if i % args.display_freq == 0:\n checkin(i, lossAll)\n loss = sum(lossAll)\n loss.backward()\n opt.step()\n with torch.no_grad():\n z.copy_(z.maximum(z_min).minimum(z_max))\n\ni = 0\ntry:\n with tqdm() as pbar:\n while True:\n train(i)\n if i == max_iterations:\n break\n i += 1\n pbar.update()\nexcept KeyboardInterrupt:\n pass",
"_____no_output_____"
]
],
[
[
"## Generate a video of the results\n\nIf you want to generate a video with the frames, just click below. You can modify the number of FPS, the initial frame, the last frame, etc. ",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
],
[
"# @title View video in browser\n# @markdown This process is slow. Use the download cell below if you don't want to wait\nmp4 = open('video.mp4','rb').read()\ndata_url = \"data:video/mp4;base64,\" + b64encode(mp4).decode()\ndisplay.HTML(\"\"\"\n<video width=400 controls>\n <source src=\"%s\" type=\"video/mp4\">\n</video>\n\"\"\" % data_url)",
"_____no_output_____"
],
[
"# @title Download video\nfrom google.colab import files\nfiles.download(\"video.mp4\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
e790bd05d4f79e7f8bff796f4c8e5ac0406a54e3 | 1,976 | ipynb | Jupyter Notebook | notebooks/Testando a API.ipynb | math-sasso/mlops_complete_moc_project | 7a239d559be397838be68f568c16548d206ab49a | [
"MIT"
] | null | null | null | notebooks/Testando a API.ipynb | math-sasso/mlops_complete_moc_project | 7a239d559be397838be68f568c16548d206ab49a | [
"MIT"
] | null | null | null | notebooks/Testando a API.ipynb | math-sasso/mlops_complete_moc_project | 7a239d559be397838be68f568c16548d206ab49a | [
"MIT"
] | null | null | null | 18.296296 | 77 | 0.494433 | [
[
[
"import requests",
"_____no_output_____"
],
[
"# O invocations e padrao do MLFLOW\nurl = 'http://127.0.0.1:5001/invocations'",
"_____no_output_____"
],
[
"data = {\n \"columns\":[\"tamanho\",\"ano\",\"garagem\"],\n \"data\":[[159.0,2003,2]]\n}",
"_____no_output_____"
],
[
"header = {'Content-Type':'application/json'}",
"_____no_output_____"
],
[
"response = requests.post(url,json=data,headers=header)\nresponse",
"_____no_output_____"
],
[
"response.text",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e790d3e034dcc6374e243a76fd5f6549632355ce | 36,592 | ipynb | Jupyter Notebook | part2/lab1/seq2seq_translation_tutorial.ipynb | yasheshshroff/ODSC2021_NLP_PyTorch | 359a9df1a97dff00eaeaa354f07df82e70ac23a2 | [
"MIT"
] | 4 | 2020-10-28T22:54:10.000Z | 2020-11-06T21:17:18.000Z | part2/lab1/seq2seq_translation_tutorial.ipynb | yasheshshroff/ODSC2021_NLP_PyTorch | 359a9df1a97dff00eaeaa354f07df82e70ac23a2 | [
"MIT"
] | null | null | null | part2/lab1/seq2seq_translation_tutorial.ipynb | yasheshshroff/ODSC2021_NLP_PyTorch | 359a9df1a97dff00eaeaa354f07df82e70ac23a2 | [
"MIT"
] | 15 | 2021-03-12T19:57:47.000Z | 2021-11-18T19:45:29.000Z | 27.700227 | 138 | 0.539599 | [
[
[
"Sequence to Sequence Model using RNN\n==================",
"_____no_output_____"
]
],
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"\nNLP From Scratch: Translation with a Sequence to Sequence Network and Attention\n*******************************************************************************\nBased on original code by \nSean Robertson `<https://github.com/spro/practical-pytorch>`_\n\nIn this project we will be teaching a neural network to translate from\nFrench to English.\n\n::\n [KEY: > input, = target, < output]\n\n > il est en train de peindre un tableau .\n = he is painting a picture .\n < he is painting a picture .\n\n > pourquoi ne pas essayer ce vin delicieux ?\n = why not try that delicious wine ?\n < why not try that delicious wine ?\n\n > elle n est pas poete mais romanciere .\n = she is not a poet but a novelist .\n < she not not a poet but a novelist .\n\n > vous etes trop maigre .\n = you re too skinny .\n < you re all alone .\n\n... to varying degrees of success.\n\nThis is made possible by the simple but powerful idea of the `sequence\nto sequence network <https://arxiv.org/abs/1409.3215>`__, in which two\nrecurrent neural networks work together to transform one sequence to\nanother. An encoder network condenses an input sequence into a vector,\nand a decoder network unfolds that vector into a new sequence.\n\n\n**Requirements**\n\n",
"_____no_output_____"
]
],
[
[
"from __future__ import unicode_literals, print_function, division\nfrom io import open\nimport unicodedata\nimport string\nimport re\nimport random\n\nimport torch\nimport torch.nn as nn\nfrom torch import optim\nimport torch.nn.functional as F\n\ndevice = torch.device(\"cuda\" if torch.cuda.is_available() else \"cpu\")",
"_____no_output_____"
]
],
[
[
"Loading data files\n==================\n\nThe English to French pairs are too big to include in the repo, so\ndownload to ``data/eng-fra.txt`` before continuing. The file is a tab\nseparated list of translation pairs:\n\n I am cold. J'ai froid.\n",
"_____no_output_____"
]
],
[
[
"!wget https://github.com/ravi-ilango/acm-dec-2020-nlp/blob/main/lab1/data.zip?raw=true -O data.zip\n \n!unzip data.zip",
"_____no_output_____"
],
[
"!head data/eng-fra.txt",
"_____no_output_____"
]
],
[
[
"We'll need a unique index per word to use as the inputs and targets of\nthe networks later. To keep track of all this we will use a helper class\ncalled ``Lang`` which has word → index (``word2index``) and index → word\n(``index2word``) dictionaries, as well as a count of each word\n``word2count`` to use to later replace rare words.\n\n\n",
"_____no_output_____"
]
],
[
[
"SOS_token = 0\nEOS_token = 1\n\nclass Lang:\n def __init__(self, name):\n self.name = name\n self.word2index = {}\n self.word2count = {}\n self.index2word = {0: \"SOS\", 1: \"EOS\"}\n self.n_words = 2 # Count SOS and EOS\n\n def addSentence(self, sentence):\n for word in sentence.split(' '):\n self.addWord(word)\n\n def addWord(self, word):\n if word not in self.word2index:\n self.word2index[word] = self.n_words\n self.word2count[word] = 1\n self.index2word[self.n_words] = word\n self.n_words += 1\n else:\n self.word2count[word] += 1",
"_____no_output_____"
]
],
[
[
"The files are all in Unicode, to simplify we will turn Unicode\ncharacters to ASCII, make everything lowercase, and trim most\npunctuation.\n\n\n",
"_____no_output_____"
]
],
[
[
"# Turn a Unicode string to plain ASCII, thanks to\n# https://stackoverflow.com/a/518232/2809427\ndef unicodeToAscii(s):\n return ''.join(\n c for c in unicodedata.normalize('NFD', s)\n if unicodedata.category(c) != 'Mn'\n )\n\n# Lowercase, trim, and remove non-letter characters\ndef normalizeString(s):\n s = unicodeToAscii(s.lower().strip())\n s = re.sub(r\"([.!?])\", r\" \\1\", s)\n s = re.sub(r\"[^a-zA-Z.!?]+\", r\" \", s)\n return s",
"_____no_output_____"
]
],
[
[
"### Exercise: Check string processing",
"_____no_output_____"
]
],
[
[
"s=\"À l'aide !\"\nnormalizeString(s)",
"_____no_output_____"
]
],
[
[
"To read the data file we will split the file into lines, and then split\nlines into pairs. The files are all English → Other Language, so if we\nwant to translate from Other Language → English I added the ``reverse``\nflag to reverse the pairs.\n\n\n",
"_____no_output_____"
]
],
[
[
"def readLangs(lang1, lang2, reverse=False):\n print(\"Reading lines...\")\n\n # Read the file and split into lines\n lines = open('data/%s-%s.txt' % (lang1, lang2), encoding='utf-8').\\\n read().strip().split('\\n')\n\n # Split every line into pairs and normalize\n pairs = [[normalizeString(s) for s in l.split('\\t')] for l in lines]\n\n # Reverse pairs, make Lang instances\n if reverse:\n pairs = [list(reversed(p)) for p in pairs]\n input_lang = Lang(lang2)\n output_lang = Lang(lang1)\n else:\n input_lang = Lang(lang1)\n output_lang = Lang(lang2)\n\n return input_lang, output_lang, pairs",
"_____no_output_____"
]
],
[
[
"### Exercise: Check creation of input and output sentence pairs",
"_____no_output_____"
]
],
[
[
"input_lang, output_lang, pairs = readLangs(\"eng\", \"fra\", reverse=True)",
"_____no_output_____"
],
[
"\n",
"_____no_output_____"
]
],
[
[
"Since there are a *lot* of example sentences and we want to train\nsomething quickly, we'll trim the data set to only relatively short and\nsimple sentences. Here the maximum length is 10 words (that includes\nending punctuation) and we're filtering to sentences that translate to\nthe form \"I am\" or \"He is\" etc. (accounting for apostrophes replaced\nearlier).\n\n\n",
"_____no_output_____"
]
],
[
[
"MAX_LENGTH = 10\n\neng_prefixes = (\n \"i am \", \"i m \",\n \"he is\", \"he s \",\n \"she is\", \"she s \",\n \"you are\", \"you re \",\n \"we are\", \"we re \",\n \"they are\", \"they re \"\n)\n\n\ndef filterPair(p):\n return len(p[0].split(' ')) < MAX_LENGTH and \\\n len(p[1].split(' ')) < MAX_LENGTH and \\\n p[1].startswith(eng_prefixes)\n\n\ndef filterPairs(pairs):\n return [pair for pair in pairs if filterPair(pair)]",
"_____no_output_____"
]
],
[
[
"### Exercise: Check results of filtering data",
"_____no_output_____"
]
],
[
[
"filterPairs(pairs[:100])",
"_____no_output_____"
]
],
[
[
"The full process for preparing the data is:\n\n- Read text file and split into lines, split lines into pairs\n- Normalize text, filter by length and content\n- Make word lists from sentences in pairs\n\n\n",
"_____no_output_____"
]
],
[
[
"def prepareData(lang1, lang2, reverse=False):\n input_lang, output_lang, pairs = readLangs(lang1, lang2, reverse)\n print(\"Read %s sentence pairs\" % len(pairs))\n pairs = filterPairs(pairs)\n print(\"Trimmed to %s sentence pairs\" % len(pairs))\n print(\"Counting words...\")\n for pair in pairs:\n input_lang.addSentence(pair[0])\n output_lang.addSentence(pair[1])\n print(\"Counted words:\")\n print(input_lang.name, input_lang.n_words)\n print(output_lang.name, output_lang.n_words)\n return input_lang, output_lang, pairs\n\n\ninput_lang, output_lang, pairs = prepareData('eng', 'fra', True)\nprint(random.choice(pairs))",
"_____no_output_____"
]
],
[
[
"The Seq2Seq Model using RNN\n===========================\n\nA Recurrent Neural Network, or RNN, is a network that operates on a\nsequence and uses its own output as input for subsequent steps.\n\n\n",
"_____no_output_____"
],
[
"The Encoder\n-----------\n\nThe encoder of a seq2seq network is a RNN that outputs some value for\nevery word from the input sentence. For every input word the encoder\noutputs a vector and a hidden state, and uses the hidden state for the next input word.",
"_____no_output_____"
]
],
[
[
"class EncoderRNN(nn.Module):\n def __init__(self, input_size, hidden_size):\n super(EncoderRNN, self).__init__()\n self.hidden_size = hidden_size\n\n self.embedding = nn.Embedding(input_size, hidden_size)\n self.gru = nn.GRU(hidden_size, hidden_size)\n\n def forward(self, input, hidden):\n embedded = self.embedding(input).view(1, 1, -1)\n output = embedded\n output, hidden = self.gru(output, hidden)\n return output, hidden\n\n def initHidden(self):\n return torch.zeros(1, 1, self.hidden_size, device=device)",
"_____no_output_____"
],
[
"#input_size: number of words in input \n#\nprint (input_lang.n_words)\n",
"_____no_output_____"
],
[
"#hidden_size: word embedding size = 256",
"_____no_output_____"
]
],
[
[
"The Decoder\n-----------\n\nThe decoder is another RNN that takes the encoder output vector(s) and\noutputs a sequence of words to create the translation.\n\n\n",
"_____no_output_____"
]
],
[
[
"class DecoderRNN(nn.Module):\n def __init__(self, hidden_size, output_size):\n super(DecoderRNN, self).__init__()\n self.hidden_size = hidden_size\n\n self.embedding = nn.Embedding(output_size, hidden_size)\n self.gru = nn.GRU(hidden_size, hidden_size)\n self.out = nn.Linear(hidden_size, output_size)\n self.softmax = nn.LogSoftmax(dim=1)\n\n def forward(self, input, hidden):\n output = self.embedding(input).view(1, 1, -1)\n output = F.relu(output)\n output, hidden = self.gru(output, hidden)\n output = self.softmax(self.out(output[0]))\n return output, hidden\n\n def initHidden(self):\n return torch.zeros(1, 1, self.hidden_size, device=device)",
"_____no_output_____"
]
],
[
[
"Training\n========\n\nPreparing Training Data\n-----------------------\n\nTo train, for each pair we will need an input tensor (indexes of the\nwords in the input sentence) and target tensor (indexes of the words in\nthe target sentence). While creating these vectors we will append the\nEOS token to both sequences.\n\n\n",
"_____no_output_____"
]
],
[
[
"def indexesFromSentence(lang, sentence):\n return [lang.word2index[word] for word in sentence.split(' ')]\n\n\ndef tensorFromSentence(lang, sentence):\n indexes = indexesFromSentence(lang, sentence)\n indexes.append(EOS_token)\n return torch.tensor(indexes, dtype=torch.long, device=device).view(-1, 1)\n\n\ndef tensorsFromPair(pair):\n input_tensor = tensorFromSentence(input_lang, pair[0])\n target_tensor = tensorFromSentence(output_lang, pair[1])\n return (input_tensor, target_tensor)",
"_____no_output_____"
]
],
[
[
"### Exercise: Check input and target data",
"_____no_output_____"
]
],
[
[
"pair = pairs[1]\npair",
"_____no_output_____"
],
[
"tensors = tensorsFromPair(pair)\ninput_tensor = tensors[0]\ntarget_tensor = tensors[1]\ninput_tensor, target_tensor",
"_____no_output_____"
]
],
[
[
"### Exercise: Check the forward pass of the network\n",
"_____no_output_____"
]
],
[
[
"encoder = EncoderRNN(input_size=input_lang.n_words, hidden_size=256).to(device)\ndecoder = DecoderRNN (hidden_size=256, output_size=output_lang.n_words).to(device)\n",
"_____no_output_____"
],
[
"learning_rate = 0.01\n\ncriterion = nn.NLLLoss()\nencoder_optimizer = optim.SGD(encoder.parameters(), lr=learning_rate)\ndecoder_optimizer = optim.SGD(decoder.parameters(), lr=learning_rate)\n",
"_____no_output_____"
],
[
"#Check one forward and backward pass\nencoder_hidden = encoder.initHidden()\n\nencoder_optimizer.zero_grad()\ndecoder_optimizer.zero_grad()\n\ninput_length = input_tensor.size(0)\ntarget_length = target_tensor.size(0)\n\nloss = 0\n",
"_____no_output_____"
],
[
"input_length, target_length",
"_____no_output_____"
],
[
"for ei in range(input_length):\n encoder_output, encoder_hidden = encoder(input_tensor[ei], encoder_hidden)\n\nencoder_hidden",
"_____no_output_____"
],
[
"decoder_input = torch.tensor([[SOS_token]], device=device)\ndecoder_hidden = encoder_hidden\n\nfor di in range(target_length):\n decoder_output, decoder_hidden = decoder(decoder_input, decoder_hidden)\n topv, topi = decoder_output.topk(1)\n decoder_input = topi.squeeze().detach() # detach from history as input\n\n loss += criterion(decoder_output, target_tensor[di])\n if decoder_input.item() == EOS_token:\n break\n",
"_____no_output_____"
],
[
"\nloss.backward()\n\nencoder_optimizer.step()\ndecoder_optimizer.step()",
"_____no_output_____"
],
[
"#One forward and backward pass\n\nencoder_hidden = encoder.initHidden()\n\nencoder_optimizer.zero_grad()\ndecoder_optimizer.zero_grad()\n\ninput_length = input_tensor.size(0)\ntarget_length = target_tensor.size(0)\n\nloss = 0\n\nfor ei in range(input_length):\n encoder_output, encoder_hidden = encoder(input_tensor[ei], encoder_hidden)\n\ndecoder_input = torch.tensor([[SOS_token]], device=device)\ndecoder_hidden = encoder_hidden\n\nfor di in range(target_length):\n decoder_output, decoder_hidden = decoder(decoder_input, decoder_hidden)\n topv, topi = decoder_output.topk(1)\n decoder_input = topi.squeeze().detach() # detach from history as input\n\n loss += criterion(decoder_output, target_tensor[di])\n if decoder_input.item() == EOS_token:\n break\n\nloss.backward()\n\nencoder_optimizer.step()\ndecoder_optimizer.step()\n",
"_____no_output_____"
]
],
[
[
"Training the Model\n------------------\n\nTo train we run the input sentence through the encoder, and keep track\nof every output and the latest hidden state. Then the decoder is given\nthe ``<SOS>`` token as its first input, and the last hidden state of the\nencoder as its first hidden state.\n\n\"Teacher forcing\" is the concept of using the real target outputs as\neach next input, instead of using the decoder's guess as the next input.\nUsing teacher forcing causes it to converge faster but `when the trained\nnetwork is exploited, it may exhibit\ninstability <http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.378.4095&rep=rep1&type=pdf>`__.\n\nYou can observe outputs of teacher-forced networks that read with\ncoherent grammar but wander far from the correct translation -\nintuitively it has learned to represent the output grammar and can \"pick\nup\" the meaning once the teacher tells it the first few words, but it\nhas not properly learned how to create the sentence from the translation\nin the first place.\n\nBecause of the freedom PyTorch's autograd gives us, we can randomly\nchoose to use teacher forcing or not with a simple if statement. Turn\n``teacher_forcing_ratio`` up to use more of it.\n\n\n",
"_____no_output_____"
]
],
[
[
"teacher_forcing_ratio = 0.5\n\n\ndef train(input_tensor, target_tensor, encoder, decoder, encoder_optimizer, decoder_optimizer, criterion, max_length=MAX_LENGTH):\n encoder_hidden = encoder.initHidden()\n\n encoder_optimizer.zero_grad()\n decoder_optimizer.zero_grad()\n\n input_length = input_tensor.size(0)\n target_length = target_tensor.size(0)\n\n encoder_outputs = torch.zeros(max_length, encoder.hidden_size, device=device)\n\n loss = 0\n\n for ei in range(input_length):\n encoder_output, encoder_hidden = encoder(\n input_tensor[ei], encoder_hidden)\n encoder_outputs[ei] = encoder_output[0, 0]\n\n decoder_input = torch.tensor([[SOS_token]], device=device)\n\n decoder_hidden = encoder_hidden\n\n use_teacher_forcing = True if random.random() < teacher_forcing_ratio else False\n\n if use_teacher_forcing:\n # Teacher forcing: Feed the target as the next input\n for di in range(target_length):\n decoder_output, decoder_hidden = decoder(\n decoder_input, decoder_hidden)\n loss += criterion(decoder_output, target_tensor[di])\n decoder_input = target_tensor[di] # Teacher forcing\n\n else:\n # Without teacher forcing: use its own predictions as the next input\n for di in range(target_length):\n decoder_output, decoder_hidden = decoder(\n decoder_input, decoder_hidden)\n topv, topi = decoder_output.topk(1)\n decoder_input = topi.squeeze().detach() # detach from history as input\n\n loss += criterion(decoder_output, target_tensor[di])\n if decoder_input.item() == EOS_token:\n break\n\n loss.backward()\n\n encoder_optimizer.step()\n decoder_optimizer.step()\n\n return loss.item() / target_length",
"_____no_output_____"
]
],
[
[
"This is a helper function to print time elapsed and estimated time\nremaining given the current time and progress %.\n\n\n",
"_____no_output_____"
]
],
[
[
"import time\nimport math\n\ndef asMinutes(s):\n m = math.floor(s / 60)\n s -= m * 60\n return '%dm %ds' % (m, s)\n\n\ndef timeSince(since, percent):\n now = time.time()\n s = now - since\n es = s / (percent)\n rs = es - s\n return '%s (- %s)' % (asMinutes(s), asMinutes(rs))",
"_____no_output_____"
]
],
[
[
"The whole training process looks like this:\n\n- Start a timer\n- Initialize optimizers and criterion\n- Create set of training pairs\n- Start empty losses array for plotting\n\nThen we call ``train`` many times and occasionally print the progress (%\nof examples, time so far, estimated time) and average loss.\n\n\n",
"_____no_output_____"
]
],
[
[
"def trainIters(encoder, decoder, n_iters, print_every=1000, plot_every=100, learning_rate=0.01):\n start = time.time()\n plot_losses = []\n print_loss_total = 0 # Reset every print_every\n plot_loss_total = 0 # Reset every plot_every\n\n encoder_optimizer = optim.SGD(encoder.parameters(), lr=learning_rate)\n decoder_optimizer = optim.SGD(decoder.parameters(), lr=learning_rate)\n training_pairs = [tensorsFromPair(random.choice(pairs))\n for i in range(n_iters)]\n criterion = nn.NLLLoss()\n\n for iter in range(1, n_iters + 1):\n training_pair = training_pairs[iter - 1]\n input_tensor = training_pair[0]\n target_tensor = training_pair[1]\n\n loss = train(input_tensor, target_tensor, encoder,\n decoder, encoder_optimizer, decoder_optimizer, criterion)\n print_loss_total += loss\n plot_loss_total += loss\n\n if iter % print_every == 0:\n print_loss_avg = print_loss_total / print_every\n print_loss_total = 0\n print('%s (%d %d%%) %.4f' % (timeSince(start, iter / n_iters),\n iter, iter / n_iters * 100, print_loss_avg))\n\n if iter % plot_every == 0:\n plot_loss_avg = plot_loss_total / plot_every\n plot_losses.append(plot_loss_avg)\n plot_loss_total = 0\n\n showPlot(plot_losses)",
"_____no_output_____"
]
],
[
[
"Plotting results\n----------------\n\nPlotting is done with matplotlib, using the array of loss values\n``plot_losses`` saved while training.\n\n\n",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nplt.switch_backend('agg')\nimport matplotlib.ticker as ticker\nimport numpy as np\n\n\ndef showPlot(points):\n plt.figure()\n fig, ax = plt.subplots()\n # this locator puts ticks at regular intervals\n loc = ticker.MultipleLocator(base=0.2)\n ax.yaxis.set_major_locator(loc)\n plt.plot(points)",
"_____no_output_____"
]
],
[
[
"Evaluation\n==========\n\nEvaluation is mostly the same as training, but there are no targets so\nwe simply feed the decoder's predictions back to itself for each step.\nEvery time it predicts a word we add it to the output string, and if it\npredicts the EOS token we stop there. We also store the decoder's\nattention outputs for display later.\n\n\n",
"_____no_output_____"
]
],
[
[
"def evaluate(encoder, decoder, sentence, max_length=MAX_LENGTH):\n with torch.no_grad():\n input_tensor = tensorFromSentence(input_lang, sentence)\n input_length = input_tensor.size()[0]\n encoder_hidden = encoder.initHidden()\n\n encoder_outputs = torch.zeros(max_length, encoder.hidden_size, device=device)\n\n for ei in range(input_length):\n encoder_output, encoder_hidden = encoder(input_tensor[ei],\n encoder_hidden)\n\n decoder_input = torch.tensor([[SOS_token]], device=device) # SOS\n\n decoder_hidden = encoder_hidden\n\n decoded_words = []\n decoder_attentions = torch.zeros(max_length, max_length)\n\n for di in range(max_length):\n decoder_output, decoder_hidden = decoder(\n decoder_input, decoder_hidden)\n topv, topi = decoder_output.data.topk(1)\n if topi.item() == EOS_token:\n decoded_words.append('<EOS>')\n break\n else:\n decoded_words.append(output_lang.index2word[topi.item()])\n\n decoder_input = topi.squeeze().detach()\n\n return decoded_words",
"_____no_output_____"
]
],
[
[
"We can evaluate random sentences from the training set and print out the\ninput, target, and output to make some subjective quality judgements:\n\n\n",
"_____no_output_____"
]
],
[
[
"def evaluateRandomly(encoder, decoder, n=10):\n for i in range(n):\n pair = random.choice(pairs)\n print('>', pair[0])\n print('=', pair[1])\n output_words = evaluate(encoder, decoder, pair[0])\n output_sentence = ' '.join(output_words)\n print('<', output_sentence)\n print('')",
"_____no_output_____"
]
],
[
[
"Training and Evaluating\n=======================\n\nWith all these helper functions in place (it looks like extra work, but\nit makes it easier to run multiple experiments) we can actually\ninitialize a network and start training.\n\nRemember that the input sentences were heavily filtered. For this small\ndataset we can use relatively small networks of 256 hidden nodes and a\nsingle GRU layer. After about 40 minutes on a MacBook CPU we'll get some\nreasonable results.\n\n.. Note::\n If you run this notebook you can train, interrupt the kernel,\n evaluate, and continue training later. Comment out the lines where the\n encoder and decoder are initialized and run ``trainIters`` again.\n\n\n",
"_____no_output_____"
]
],
[
[
"hidden_size = 256\nencoder1 = EncoderRNN(input_lang.n_words, hidden_size).to(device)\ndecoder1 = DecoderRNN(hidden_size, output_lang.n_words).to(device)\n\ntrainIters(encoder1, decoder1, 75000, print_every=5000, plot_every=100)",
"_____no_output_____"
],
[
"evaluateRandomly(encoder1, decoder1)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
e790dd668ce836ece6457f187e8386f8ebb99b28 | 12,096 | ipynb | Jupyter Notebook | Mar22/Statistics/.ipynb_checkpoints/jupyterhelp-checkpoint.ipynb | khajadatascienceR/DataScienceWithPython | 6d4e1889bf086414e449b928661ec74e7447cdc2 | [
"Apache-2.0"
] | null | null | null | Mar22/Statistics/.ipynb_checkpoints/jupyterhelp-checkpoint.ipynb | khajadatascienceR/DataScienceWithPython | 6d4e1889bf086414e449b928661ec74e7447cdc2 | [
"Apache-2.0"
] | null | null | null | Mar22/Statistics/.ipynb_checkpoints/jupyterhelp-checkpoint.ipynb | khajadatascienceR/DataScienceWithPython | 6d4e1889bf086414e449b928661ec74e7447cdc2 | [
"Apache-2.0"
] | 1 | 2022-03-24T07:25:06.000Z | 2022-03-24T07:25:06.000Z | 23.172414 | 225 | 0.501571 | [
[
[
"Using Jupyter Notebook effectively\n----------------------------------",
"_____no_output_____"
]
],
[
[
"len?",
"\u001b[1;31mSignature:\u001b[0m \u001b[0mlen\u001b[0m\u001b[1;33m(\u001b[0m\u001b[0mobj\u001b[0m\u001b[1;33m,\u001b[0m \u001b[1;33m/\u001b[0m\u001b[1;33m)\u001b[0m\u001b[1;33m\u001b[0m\u001b[1;33m\u001b[0m\u001b[0m\n\u001b[1;31mDocstring:\u001b[0m Return the number of items in a container.\n\u001b[1;31mType:\u001b[0m builtin_function_or_method\n"
],
[
"print?",
"\u001b[1;31mDocstring:\u001b[0m\nprint(value, ..., sep=' ', end='\\n', file=sys.stdout, flush=False)\n\nPrints the values to a stream, or to sys.stdout by default.\nOptional keyword arguments:\nfile: a file-like object (stream); defaults to the current sys.stdout.\nsep: string inserted between values, default a space.\nend: string appended after the last value, default a newline.\nflush: whether to forcibly flush the stream.\n\u001b[1;31mType:\u001b[0m builtin_function_or_method\n"
],
[
"numbers = [1,2,3]\n#numbers.<TAB>\nnumbers.insert?\nnumbers?",
"\u001b[1;31mType:\u001b[0m list\n\u001b[1;31mString form:\u001b[0m [1, 2, 3]\n\u001b[1;31mLength:\u001b[0m 3\n\u001b[1;31mDocstring:\u001b[0m \nBuilt-in mutable sequence.\n\nIf no argument is given, the constructor creates a new empty list.\nThe argument must be an iterable if specified.\n"
]
],
[
[
"Defining Functions and using them\n---------------------------------",
"_____no_output_____"
]
],
[
[
"def square(number):\n \"\"\"Returns the Square of a number\"\"\"\n return number ** 2",
"_____no_output_____"
],
[
"square?",
"\u001b[1;31mSignature:\u001b[0m \u001b[0msquare\u001b[0m\u001b[1;33m(\u001b[0m\u001b[0mnumber\u001b[0m\u001b[1;33m)\u001b[0m\u001b[1;33m\u001b[0m\u001b[1;33m\u001b[0m\u001b[0m\n\u001b[1;31mDocstring:\u001b[0m Returns the Square of a number\n\u001b[1;31mFile:\u001b[0m c:\\users\\qtkhaja\\appdata\\local\\temp\\ipykernel_14236\\636618730.py\n\u001b[1;31mType:\u001b[0m function\n"
],
[
"square??",
"\u001b[1;31mSignature:\u001b[0m \u001b[0msquare\u001b[0m\u001b[1;33m(\u001b[0m\u001b[0mnumber\u001b[0m\u001b[1;33m)\u001b[0m\u001b[1;33m\u001b[0m\u001b[1;33m\u001b[0m\u001b[0m\n\u001b[1;31mSource:\u001b[0m \n\u001b[1;32mdef\u001b[0m \u001b[0msquare\u001b[0m\u001b[1;33m(\u001b[0m\u001b[0mnumber\u001b[0m\u001b[1;33m)\u001b[0m\u001b[1;33m:\u001b[0m\u001b[1;33m\n\u001b[0m \u001b[1;34m\"\"\"Returns the Square of a number\"\"\"\u001b[0m\u001b[1;33m\n\u001b[0m \u001b[1;32mreturn\u001b[0m \u001b[0mnumber\u001b[0m \u001b[1;33m**\u001b[0m \u001b[1;36m2\u001b[0m\u001b[1;33m\u001b[0m\u001b[1;33m\u001b[0m\u001b[0m\n\u001b[1;31mFile:\u001b[0m c:\\users\\qtkhaja\\appdata\\local\\temp\\ipykernel_14236\\636618730.py\n\u001b[1;31mType:\u001b[0m function\n"
],
[
"print??",
"\u001b[1;31mDocstring:\u001b[0m\nprint(value, ..., sep=' ', end='\\n', file=sys.stdout, flush=False)\n\nPrints the values to a stream, or to sys.stdout by default.\nOptional keyword arguments:\nfile: a file-like object (stream); defaults to the current sys.stdout.\nsep: string inserted between values, default a space.\nend: string appended after the last value, default a newline.\nflush: whether to forcibly flush the stream.\n\u001b[1;31mType:\u001b[0m builtin_function_or_method\n"
],
[
"#numbers.c<TAB> Try this",
"_____no_output_____"
],
[
"# tab completion while importing\n#from itertools import co<TAB>\n#from numpy imp",
"_____no_output_____"
],
[
"*Warning?",
"BytesWarning\nDeprecationWarning\nFutureWarning\nImportWarning\nPendingDeprecationWarning\nResourceWarning\nRuntimeWarning\nSyntaxWarning\nUnicodeWarning\nUserWarning\nWarning"
],
[
"str.*find*?",
"str.find\nstr.rfind"
]
],
[
[
"Magic Commands\n----------------",
"_____no_output_____"
]
],
[
[
"%timeit test_list = [ n**3 for n in range(100)]\n",
"884 µs ± 122 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)\n"
],
[
"%%timeit\nmy_list = []\nfor number in range(1000):\n my_list.append(number**3)\n",
"20.5 ms ± 10.3 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)\n"
]
],
[
[
"Previous Outputs and Underscore shortcuts\n-------------------------------------------",
"_____no_output_____"
]
],
[
[
"10 + 5",
"_____no_output_____"
],
[
"print(_)",
"15\n"
],
[
"10 * 5",
"_____no_output_____"
],
[
"10 ** 5",
"_____no_output_____"
],
[
"print(_)\nprint(__)\nprint(___)\n",
"100000\n50\n15\n"
],
[
"Out[31] # going by cell number",
"_____no_output_____"
],
[
"!dir\n# execute native commands ",
" Volume in drive C has no label.\n Volume Serial Number is BA06-63A7\n\n Directory of c:\\Users\\qtkhaja\\Test\\DataScienceWithPython\\Mar22\\Statistics\n\n23-03-2022 19:53 <DIR> .\n23-03-2022 19:53 <DIR> ..\n23-03-2022 19:42 <DIR> .ipynb_checkpoints\n22-03-2022 20:16 7,778 arrays.ipynb\n23-03-2022 20:05 9,997 jupyterhelp.ipynb\n15-03-2022 19:58 5,625 StatisticsMainTools.ipynb\n22-03-2022 20:48 3,523 vectorization.ipynb\n 4 File(s) 26,923 bytes\n 3 Dir(s) 203,306,328,064 bytes free\n"
],
[
"%%time\nmy_list = []\nfor number in range(1000):\n my_list.append(number**3)",
"Wall time: 3.99 ms\n"
],
[
"def sum_of_lists(max_limit):\n total = 0\n for number in range(max_limit):\n total += number ** 3\n return total",
"_____no_output_____"
],
[
"%prun sum_of_lists(100000)",
" 4 function calls in 0.048 seconds\n\n Ordered by: internal time\n\n ncalls tottime percall cumtime percall filename:lineno(function)\n 1 0.048 0.048 0.048 0.048 947818732.py:1(sum_of_lists)\n 1 0.000 0.000 0.048 0.048 {built-in method builtins.exec}\n 1 0.000 0.000 0.048 0.048 <string>:1(<module>)\n 1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e790e00bcebd02af35da918b79e12cb1d6913fc3 | 477,987 | ipynb | Jupyter Notebook | polygon-orientation/orientation.ipynb | Napuu/osm-fiddling | 85a905a4479060d66d3e1fbbfad651d6fd427920 | [
"MIT"
] | null | null | null | polygon-orientation/orientation.ipynb | Napuu/osm-fiddling | 85a905a4479060d66d3e1fbbfad651d6fd427920 | [
"MIT"
] | null | null | null | polygon-orientation/orientation.ipynb | Napuu/osm-fiddling | 85a905a4479060d66d3e1fbbfad651d6fd427920 | [
"MIT"
] | null | null | null | 1,448.445455 | 100,586 | 0.953091 | [
[
[
"import matplotlib.pyplot as plt\nimport geopandas as gpd\nimport requests\nimport json\nimport osm2geojson\nimport contextily as ctx\nimport shapely\nimport pyproj\nimport numpy as np\nfrom os import environ\nenviron[\"PROJ_LIB\"] = \"/home/debian/miniconda3/envs/geo/lib/python3.8/site-packages/rasterio/proj_data\"",
"_____no_output_____"
],
[
"def get_osm_data(lat, lng):\n # print(lat, lng)\n overpass_url = \"http://overpass-api.de/api/interpreter\"\n # 60.148809, 24.916404 helsinki\n # 51.888806, 4.429777 amsterdam\n # 60.18636636163917 24.83902037143707 otaniemi \n overpass_query = \"\"\"\n [out:json][timeout:25];\n (\n nwr[~\"^(natural|area)$\"~\".\"](around:200,\"\"\" + f'{lat},{lng}' + \"\"\");\n nwr[man_made=pier](around:200,\"\"\" + f'{lat},{lng}' + \"\"\");\n );\n out geom;\n \"\"\"\n response = requests.get(overpass_url, \n params={'data': overpass_query})\n data = response.json()\n return osm2geojson.json2geojson(data)\n",
"_____no_output_____"
],
[
"geojson = get_osm_data(51.88692186814117, 4.2840078206463685)\nprint(geojson)\ngdf = gpd.GeoDataFrame.from_features(geojson)\ngdf.crs = 'epsg:4326'\nf, ax = plt.subplots(1, figsize=(18, 18))\ngdf.plot(ax=ax)\n\n#no_ferry = gdf[gdf[\"tags\"]][gdf[\"tags\"].apply(lambda x: \"ferry\" not in x)]\n#no_ferry.plot()",
"51.88692186814117 4.2840078206463685\n{'type': 'FeatureCollection', 'features': [{'type': 'Feature', 'properties': {'type': 'relation', 'id': 13070155, 'tags': {'natural': 'water', 'type': 'multipolygon', 'water': 'harbour', 'wikidata': 'Q1892864'}}, 'geometry': {'type': 'MultiPolygon', 'coordinates': [[[[4.2820895, 51.8854948], [4.2821179, 51.8855161], [4.2821169, 51.8855568], [4.2817098, 51.8863193], [4.2816892, 51.8864101], [4.2812849, 51.8871662], [4.2809496, 51.8877866], [4.2809483, 51.8878372], [4.2811902, 51.8881381], [4.2812341, 51.8881629], [4.2817641, 51.8882718], [4.2818922, 51.8882907], [4.282464, 51.8884051], [4.2829264, 51.8884859], [4.2830127, 51.8884979], [4.2835169, 51.8885903], [4.2835746, 51.8885908], [4.283635, 51.8885849], [4.2837803, 51.888549], [4.2840286, 51.8884799], [4.2841586, 51.888421], [4.2844723, 51.8878279], [4.284954, 51.8869035], [4.2850198, 51.8867906], [4.2851846, 51.8864885], [4.2855671, 51.8857645], [4.2855888, 51.8857369], [4.2820895, 51.8854948]]]]}}, {'type': 'Feature', 'properties': {'type': 'relation', 'id': 13070156, 'tags': {'natural': 'water', 'type': 'multipolygon', 'water': 'harbour', 'wikidata': 'Q14509390'}}, 'geometry': {'type': 'MultiPolygon', 'coordinates': [[[[4.2874658, 51.8861169], [4.2874649, 51.8861556], [4.2869389, 51.8871738], [4.2869059, 51.8872353], [4.2863604, 51.8882953], [4.2859719, 51.8890566], [4.2859426, 51.8890679], [4.2858007, 51.8890774], [4.2857472, 51.8891814], [4.2846084, 51.8892949], [4.2828274, 51.8927263], [4.2841752, 51.8930035], [4.2845741, 51.8930855], [4.285587, 51.893291], [4.287326, 51.8898074], [4.2877042, 51.8898851], [4.2889386, 51.8901339], [4.2906446, 51.8905023], [4.2910852, 51.8896617], [4.2918354, 51.8882304], [4.2924377, 51.8870811], [4.2874658, 51.8861169]]]]}}, {'type': 'Feature', 'properties': {'type': 'relation', 'id': 13070158, 'tags': {'natural': 'water', 'type': 'multipolygon', 'water': 'canal'}}, 'geometry': {'type': 'MultiPolygon', 'coordinates': [[[[4.2735351, 51.8797364], [4.2735189, 51.879742], [4.2734992, 51.8797451], [4.273471, 51.8797446], [4.2734405, 51.8797398], [4.2726266, 51.8795757], [4.2725996, 51.8795696], [4.2713865, 51.8793246], [4.2702945, 51.8791062], [4.2691784, 51.8788891], [4.2687617, 51.878805], [4.2686793, 51.8787865], [4.2683181, 51.8794777], [4.2675278, 51.8793208], [4.2673511, 51.8792857], [4.2668775, 51.8801999], [4.2646182, 51.879978], [4.2644551, 51.8802767], [4.270296, 51.8814586], [4.2701935, 51.8816516], [4.2643529, 51.8804691], [4.2640323, 51.8810925], [4.2659755, 51.881486], [4.2659619, 51.8815117], [4.2640185, 51.8811184], [4.2639298, 51.881286], [4.2641767, 51.8813381], [4.2639735, 51.8817239], [4.2637141, 51.8816726], [4.2634536, 51.8821804], [4.2632074, 51.8826563], [4.2716345, 51.8843454], [4.2717024, 51.884222], [4.2717222, 51.8841993], [4.2717407, 51.8841899], [4.2717653, 51.884183], [4.271787, 51.8841819], [4.271834, 51.8841858], [4.2718456, 51.8841872], [4.2718969, 51.8841942], [4.2721706, 51.8842501], [4.2722388, 51.8840989], [4.2747261, 51.8846065], [4.2780186, 51.886149], [4.278482, 51.8862355], [4.2785698, 51.8860818], [4.2786076, 51.8860401], [4.2786686, 51.8860114], [4.2787342, 51.886012], [4.278831, 51.8860245], [4.2795282, 51.8861591], [4.2810919, 51.8864792], [4.2813845, 51.8859216], [4.2816339, 51.8854548], [4.2816555, 51.8854306], [4.2816848, 51.8854162], [4.2817137, 51.8854134], [4.2818152, 51.8854318], [4.2818318, 51.8854348], [4.2820506, 51.885475], [4.2820895, 51.8854948], [4.2855888, 51.8857369], [4.2856622, 51.8857376], [4.2858201, 51.8857689], [4.2864455, 51.8858927], [4.2873932, 51.8860836], [4.2874453, 51.8861021], [4.2874658, 51.8861169], [4.2924377, 51.8870811], [4.2926949, 51.8871271], [4.292703, 51.887174], [4.293082, 51.8872376], [4.2931262, 51.8871712], [4.2943329, 51.8874151], [4.2943135, 51.8874721], [4.2943746, 51.8874867], [4.2959358, 51.8877788], [4.2961299, 51.8878076], [4.2982743, 51.8882239], [4.3002751, 51.8886123], [4.3017267, 51.8888619], [4.3047497, 51.8906248], [4.3050386, 51.8908507], [4.3050471, 51.8908632], [4.3050534, 51.8908786], [4.3050506, 51.8908999], [4.3050468, 51.8909149], [4.3115592, 51.8906982], [4.3122661, 51.8897335], [4.3122371, 51.8897118], [4.3122136, 51.88968], [4.3070617, 51.8875078], [4.3069439, 51.8875418], [4.3068574, 51.8875438], [4.3050886, 51.8872057], [4.3049968, 51.8871817], [4.304342, 51.887055], [4.3022025, 51.8866168], [4.3020959, 51.8865695], [4.3005247, 51.8862446], [4.3004247, 51.8862285], [4.2997624, 51.886094], [4.2986773, 51.8858792], [4.2978219, 51.8856875], [4.2975255, 51.8856272], [4.2973427, 51.8855848], [4.2964884, 51.8854319], [4.2964899, 51.885367], [4.296221, 51.8853132], [4.2889403, 51.883918], [4.2889035, 51.8839176], [4.288008, 51.8837387], [4.2877941, 51.8836912], [4.2872485, 51.8835791], [4.2871858, 51.883567], [4.2846821, 51.8830629], [4.2827084, 51.8826691], [4.2826249, 51.8826506], [4.282108, 51.8825474], [4.2802545, 51.8821735], [4.2799569, 51.8821105], [4.2781847, 51.8817399], [4.2781616, 51.8817203], [4.2777516, 51.8806835], [4.2771554, 51.8807589], [4.2735351, 51.8797364]]]]}}]}\n"
],
[
"polygons = gpd.read_file(\"test-data.geojson\")\n#for index, geom in polygons.head(5).iterrows():\ndef plot_test(index):\n f, ax = plt.subplots(1, figsize=(10, 10))\n row = polygons.iloc[[index]]\n geom = row.geometry\n \n \n #return\n\n # plotting\n gs = gpd.GeoSeries(geom, crs=4326)\n gs = gs.to_crs(3857)\n buffer = gs.geometry.buffer(200)\n buffer.plot(alpha=0, ax=ax)\n centroid_4326 = gs.centroid.to_crs(4326)\n osm_gdf = gpd.GeoDataFrame.from_features(\n get_osm_data(float(centroid_4326.y), float(centroid_4326.x))\n )\n osm_gdf.crs = 'epsg:4326'\n ctx.add_basemap(\n ax,\n source=ctx.providers.CartoDB.Positron\n )\n plt.autoscale(False)\n osm_intersection = gpd.clip(osm_gdf, buffer.buffer(200).to_crs(4326).geometry).to_crs(3857)\n osm_intersection.plot(ax=ax, facecolor=\"#0000\", edgecolor=\"#f004\", linewidth=5)\n gs.plot(ax = ax)\n \n # polygon edges as linestrings\n i = 0\n points = geom.iloc[0].exterior.coords\n uniq_points = list(points)\n edges = []\n for i in range(len(uniq_points) - 1):\n ls = shapely.geometry.LineString([uniq_points[i], uniq_points[i+1]])\n edges.append(ls)\n edges.sort(key=lambda x: x.length, reverse=True)\n # long edges are now at indexes 0 and 1\n\n def get_normal(i):\n\n normal_starting_point = np.array(edges[i].interpolate(0.5, normalized=True).coords[0])\n dx = edges[3].coords[0][0] - edges[3].coords[1][0]\n dy = edges[3].coords[0][1] - edges[3].coords[1][1]\n # smaller edge is guaranteed to be at smaller\n diff_vec = np.array([dx, dy]) / 2\n try1 = normal_starting_point + diff_vec\n try2 = normal_starting_point - diff_vec\n outpointing_vector_end = try1 + (diff_vec * 5)\n if (row.intersects(shapely.geometry.Point(try1)).all()):\n outpointing_vector_end = try2 - (diff_vec * 5)\n return shapely.geometry.LineString([normal_starting_point, outpointing_vector_end])\n\n n1 = get_normal(0)\n n2 = get_normal(1)\n t = gpd.GeoDataFrame.from_records([{\"geometry\": n1}])\n t.crs = \"EPSG:4326\"\n t = t.to_crs(3857)\n t.plot(ax=ax, color=\"green\")\n t = gpd.GeoDataFrame.from_records([{\"geometry\": n2}])\n t.crs = \"EPSG:4326\"\n t = t.to_crs(3857)\n t.plot(ax=ax, color=\"yellow\")\n steps = 100\n #max = 5.0\n line1 = osm_gdf.to_crs(4326).intersects(n1).any()\n line2 = osm_gdf.to_crs(4326).intersects(n2).any()\n boundary1 = osm_gdf.to_crs(4326).boundary.intersects(n1).any()\n boundary2 = osm_gdf.to_crs(4326).boundary.intersects(n2).any()\n #print(f'line1: {line1}, line2: {line2}, boundary1: {boundary1}, boundary2: {boundary2}')\n #print(n1, n1.interpolate(float(i)/steps, normalized=True))\n\n for i in range(steps):\n #print(i/steps*max)\n #print(float(i)/steps)\n vec1 = shapely.geometry.LineString([\n n1.coords[0],\n n1.interpolate(float(i)/steps, normalized=True)\n ])\n vec2 = shapely.geometry.LineString([\n n2.coords[0],\n n2.interpolate(float(i)/steps, normalized=True)\n ])\n # print(osm_gdf.to_crs(4326).intersects(vec2))\n line1 = osm_gdf.to_crs(4326).intersects(vec1).any()\n line2 = osm_gdf.to_crs(4326).intersects(vec2).any()\n boundary1 = osm_gdf.to_crs(4326).boundary.intersects(vec1).any()\n boundary2 = osm_gdf.to_crs(4326).boundary.intersects(vec2).any()\n hit_vector = None\n if line1 and not line2:\n hit_vector = n1\n elif line2 and not line1:\n hit_vector = n2\n elif boundary1 and not boundary2:\n hit_vector = n1\n elif boundary2 and not boundary1:\n hit_vector = n2\n if hit_vector:\n t = gpd.GeoDataFrame.from_records([{\"geometry\": hit_vector}])\n t.crs = \"EPSG:4326\"\n t = t.to_crs(3857)\n t.plot(ax=ax, color=\"red\", linewidth=4)\n break\n\n ax.set_axis_off()\n\n \n#plot_test(2)",
"_____no_output_____"
],
[
"for i in range(6):\n plot_test(i)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
e790f02b249d638c90797b55132a76209f5f4782 | 38,435 | ipynb | Jupyter Notebook | notebooks/Filtros_vibracao.ipynb | nicolasantero/compressor-breakin-kmeans-clustering | 9987051cecdb052f96f3f3c9caadfef447a95cf1 | [
"MIT"
] | 1 | 2021-12-29T19:36:59.000Z | 2021-12-29T19:36:59.000Z | notebooks/Filtros_vibracao.ipynb | nicolasantero/compressor-breakin-kmeans-clustering | 9987051cecdb052f96f3f3c9caadfef447a95cf1 | [
"MIT"
] | null | null | null | notebooks/Filtros_vibracao.ipynb | nicolasantero/compressor-breakin-kmeans-clustering | 9987051cecdb052f96f3f3c9caadfef447a95cf1 | [
"MIT"
] | 1 | 2022-01-19T17:50:22.000Z | 2022-01-19T17:50:22.000Z | 19,217.5 | 38,434 | 0.656823 | [
[
[
"# Gráficos\n",
"_____no_output_____"
]
],
[
[
"import plotly.io as pio\npio.renderers",
"_____no_output_____"
],
[
"a3_1fim = pd.read_csv('/content/drive/My Drive/Nicolas/2021_04 - Dados processados/A3_N_2019_12_04tAm2.5.csv')\na3_2fim = pd.read_csv('/content/drive/My Drive/Nicolas/2021_04 - Dados processados/A3_A_2019_12_09tAm11.8.csv')\na3_3fim = pd.read_csv('/content/drive/My Drive/Nicolas/2021_04 - Dados processados/A3_A_2019_12_11tAm2.5.csv')\n\na4_1fim = pd.read_csv('/content/drive/My Drive/Nicolas/2021_04 - Dados processados/A4_N_2019_12_16tAm2.1.csv')\na4_2fim = pd.read_csv('/content/drive/My Drive/Nicolas/2021_04 - Dados processados/A4_A_2019_12_19tAm6.csv')\na4_3fim = pd.read_csv('/content/drive/My Drive/Nicolas/2021_04 - Dados processados/A4_A_2020_01_06tAm2.5.csv')\na4_4fim = pd.read_csv('/content/drive/My Drive/Nicolas/2021_04 - Dados processados/A4_A_2020_01_13tAm2.5.csv')\n\na5_1fim = pd.read_csv('/content/drive/My Drive/Nicolas/2021_04 - Dados processados/A5_N_2020_01_22tAm2.5.csv')\na5_2fim = pd.read_csv('/content/drive/My Drive/Nicolas/2021_04 - Dados processados/A5_A_2020_01_27tAm12.5.csv')\na5_3fim = pd.read_csv('/content/drive/My Drive/Nicolas/2021_04 - Dados processados/A5_A_2020_01_28tAm2.5.csv')",
"_____no_output_____"
],
[
"def plota_rms(dado, nomedado):\n fig = go.Figure()\n\n fig.add_trace(go.Scatter(\n y = dado['lowpassRMS'],\n line = dict(shape = 'spline' ),\n name = 'filtered RMS lowpass'\n ))\n\n fig.add_trace(go.Scatter(\n y = dado['highpassRMS'],\n line = dict(shape = 'spline' ),\n name = 'filtered RMS highpass'\n ))\n\n fig.add_trace(go.Scatter(\n y = dado['bandpassRMS'],\n line = dict(shape = 'spline' ),\n name = 'filtered RMS bandpass'\n ))\n \n fig.update_layout(\n title={\n 'text': nomedado + 'RMS',\n 'y':0.9,\n 'x':0.5,\n 'xanchor': 'center',\n 'yanchor': 'top'},\n autosize=False,\n width=900,\n height=500)\n \n fig.show(renderer=\"notebook\")\n\n\ndef plota_curtose(dado, nomedado):\n fig = go.Figure()\n\n fig.add_trace(go.Scatter(\n y = dado['lowpassCurtose'],\n line = dict(shape = 'spline' ),\n name = 'filtered Curtose lowpass'\n ))\n\n fig.add_trace(go.Scatter(\n y = dado['highpassCurtose'],\n line = dict(shape = 'spline' ),\n name = 'filtered Curtose highpass',\n ))\n\n fig.add_trace(go.Scatter(\n y = dado['bandpassCurtose'],\n line = dict(shape = 'spline' ),\n name = 'filtered Curtose bandpass', \n ))\n \n\n \n fig.update_layout(\n title={\n 'text': nomedado + 'Curtose',\n 'y':0.9,\n 'x':0.5,\n 'xanchor': 'center',\n 'yanchor': 'top'},\n autosize=False,\n width=900,\n height=500\n \n)\n\n fig.show(renderer=\"notebook\")\n",
"_____no_output_____"
],
[
"plota_rms(a3_1fim, 'a3_N_1')\nplota_curtose(a3_1fim, 'a3_N_1')\n\nplota_rms(a3_2fim, 'a3_A_2')\nplota_curtose(a3_2fim, 'a3_A_2')\n\nplota_rms(a3_3fim, 'a3_A_3')\nplota_curtose(a3_2fim, 'a3_A_3')\n\nplota_rms(a4_1fim, 'a4_N_1')\nplota_curtose(a4_1fim, 'a4_N_1')\n\nplota_rms(a4_2fim, 'a4_A_2')\nplota_curtose(a4_2fim, 'a4_A_2')\n\nplota_rms(a4_3fim, 'a4_A_3')\nplota_curtose(a4_3fim, 'a4_A_3')\n\nplota_rms(a4_4fim, 'a4_A_4')\nplota_curtose(a4_4fim, 'a4_A_4')\n\nplota_rms(a5_1fim, 'a5_N_1')\nplota_curtose(a5_1fim, 'a5_N_1')\n\nplota_rms(a5_2fim, 'a5_A_2')\nplota_curtose(a5_2fim, 'a5_A_2')\n\nplota_rms(a5_3fim, 'a5_A_3')\nplota_curtose(a5_3fim, 'a5_A_3')",
"_____no_output_____"
]
],
[
[
"## Entendimento dos dados\n\nTem 59s entre eles\n\nCada arquivo é 1s\n\nDepois existe um Gap de 59s que não são medidos\n\nE depois vem o próximo arquivo\n\n## Data columns \n\n1 calota inferior\n\n2 dummy, bancada\n\n3 calota superior\n\n## Parâmetros \n\n25.6khz\n\nFs = 25.6*10^3\n\ndT = 1/FS\n\nFs\n\nt = (1:length(V1))*dt\n\n## Tipo de análise feita\n\nLowpass em 1k, um bandpass de 1k a 10k, e um highpass em 10k\n\n\nCurtose : b2=1/n * ∑[(xi−x¯)/s]^4 − 3\n",
"_____no_output_____"
]
],
[
[
"from google.colab import drive\n\ndrive.mount('/content/drive')\n#4/1AY0e-g5GH8MfPR750KhgwYRUTCU_YkBmew1ZIl1vqKmHgjCFsPlq-RLglxw",
"Mounted at /content/drive\n"
]
],
[
[
"# FUNCIONANDO\n",
"_____no_output_____"
],
[
"# Retirando o RMS do ensaio não amaciada da amostra 3 ",
"_____no_output_____"
],
[
"## Imports de bibliotecas",
"_____no_output_____"
]
],
[
[
"from io import BytesIO\nimport zipfile\n# import rarfile\nimport pandas as pd\nimport urllib.request\nimport numpy as np\nimport seaborn as sns\nimport matplotlib.pyplot as plt\nimport re\nfrom google.colab import files\nimport csv\nfrom scipy.signal import butter, lfilter\nimport scipy.stats\nimport plotly.graph_objects as go\nfrom google.colab import files\nimport plotly.offline",
"_____no_output_____"
]
],
[
[
"## Dados",
"_____no_output_____"
]
],
[
[
"a3_1 = pd.read_csv('/content/drive/My Drive/Nicolas/2021_03 - Dados processados/A3_N_2019_12_04tAm2.5.csv')\na3_2 = pd.read_csv('/content/drive/My Drive/Nicolas/2021_03 - Dados processados/A3_A_2019_12_09tAm11.8.csv')\na3_3 = pd.read_csv('/content/drive/My Drive/Nicolas/2021_03 - Dados processados/A3_A_2019_12_11tAm2.5.csv')\n\na4_1 = pd.read_csv('/content/drive/My Drive/Nicolas/2021_03 - Dados processados/A4_N_2019_12_16tAm2.1.csv')\na4_2 = pd.read_csv('/content/drive/My Drive/Nicolas/2021_03 - Dados processados/A4_A_2019_12_19tAm6.csv')\na4_3 = pd.read_csv('/content/drive/My Drive/Nicolas/2021_03 - Dados processados/A4_A_2020_01_06tAm2.5.csv')\na4_4 = pd.read_csv('/content/drive/My Drive/Nicolas/2021_03 - Dados processados/A4_A_2020_01_13tAm2.5.csv')\n\na5_1 = pd.read_csv('/content/drive/My Drive/Nicolas/2021_03 - Dados processados/A5_N_2020_01_22tAm2.5.csv')\na5_2 = pd.read_csv('/content/drive/My Drive/Nicolas/2021_03 - Dados processados/A5_A_2020_01_27tAm12.5.csv')\na5_3 = pd.read_csv('/content/drive/My Drive/Nicolas/2021_03 - Dados processados/A5_A_2020_01_28tAm2.5.csv')",
"_____no_output_____"
]
],
[
[
"## Variáveis constantes",
"_____no_output_____"
]
],
[
[
"cutoff_low=1000\ncutoff_band=[1000,10000]\ncutoff_high=10000\norder = 5\n\nfs=25600\ntime=1\nsampling_rate = fs/time",
"_____no_output_____"
]
],
[
[
"## Funções",
"_____no_output_____"
]
],
[
[
"def filtro_lowpass(data,order,cutoff, fs):\n nyquist = fs*0.5\n cutoff/(fs*0.5)\n wn_low = cutoff/nyquist\n\n b_low, a_low = butter(order, wn_low, btype='lowpass')\n filtered_sig_low = lfilter(b_low, a_low, data.values)\n return filtered_sig_low\n\ndef filtro_highpass(data,order,cutoff, fs):\n\n nyquist = fs*0.5\n wn_high = cutoff/nyquist\n\n b_high, a_high = butter(order, wn_high, btype='highpass')\n filtered_sig_high = lfilter(b_high, a_high, data.values)\n return filtered_sig_high\n\ndef filtro_bandpass(data,order,cutoff, fs):\n nyquist = fs*0.5\n wn_band = []\n wn_band.append(cutoff[0]/nyquist)\n wn_band.append(cutoff[1]/nyquist)\n\n b_band, a_band = butter(order, wn_band, btype='bandpass')\n filtered_sig_band = lfilter(b_band, a_band, data.values)\n return filtered_sig_band\n\ndef cria_tempo(fs):\n t = []\n for i in range(len(df)):\n dt = 1*fs\n t.append(k*dt) \n k = k+1\n return t\n\ndef rms(data_column):\n x = data_column.apply(lambda x: x*x)\n y = np.sqrt(sum(x)/len(data_column))\n return y\n\ndef curtose(data_column):\n curtose = scipy.stats.kurtosis(data_column)\n return curtose\n \n# ensaioprocessado é os dados já processados para concatenar e junto os novos processamentos\ndef aplica_filtro(pastazip, pastaarquivo, ensaioprocessado, tipo):\n zip_ref = zipfile.ZipFile(pastazip, 'r')\n df = []\n text_files = zip_ref.infolist()\n text = []\n for i in text_files:\n if i.filename.startswith(f\"{pastaarquivo}vibTempo\"):\n text.append(i.filename)\n\n if tipo == 2:\n k=1\n else:\n k=0\n\n t_rms = []\n t_rms = pd.DataFrame(columns=(['lowpassRMS', 'highpassRMS', 'bandpassRMS', 'lowpassCurtose', 'highpassCurtose', 'bandpassCurtose']))\n df_fim = []\n\n\n for text_file in text[1: len(text) - k]:\n df = []\n row_rms = []\n df = pd.read_csv(zip_ref.open(text_file), sep='\\t', header=None)\n df['lowpass'] = filtro_lowpass(df[0],order,cutoff_low, fs)\n df['highpass'] = filtro_highpass(df[0],order,cutoff_high, fs)\n df['bandpass'] = filtro_bandpass(df[0],order,cutoff_band, fs)\n\n new_row = {'lowpassRMS':rms(df['lowpass']), 'highpassRMS':rms(df['highpass']), 'bandpassRMS':rms(df['bandpass']), \n 'lowpassCurtose':curtose(df['lowpass']), 'highpassCurtose':curtose(df['highpass']), 'bandpassCurtose':curtose(df['bandpass'])}\n t_rms = t_rms.append(new_row, ignore_index=True)\n df_fim = pd.concat((ensaioprocessado,t_rms), axis=1)\n \n return df_fim",
"_____no_output_____"
]
],
[
[
"## zip and .dat import and read",
"_____no_output_____"
]
],
[
[
"lista_ensaios = [a3_1, a3_2, a3_3]\na3pastazip = '/content/drive/My Drive/Nicolas/Amostra_A3.zip'\npastaarquivo_a3 = ['Amostra A3/N_2019_12_04/vibracao/', 'Amostra A3/A_2019_12_09/vibracao/', 'Amostra A3/A_2019_12_11/vibracao/']",
"_____no_output_____"
],
[
"a3_1fim = aplica_filtro(a3pastazip, pastaarquivo_a3[0], lista_ensaios[0], 1)\na3_2fim = aplica_filtro(a3pastazip, pastaarquivo_a3[1], lista_ensaios[1], 1)\na3_3fim = aplica_filtro(a3pastazip, pastaarquivo_a3[2], lista_ensaios[2], 1)\n",
"_____no_output_____"
],
[
"lista_ensaios_a4 = [a4_1, a4_2, a4_3, a4_4]\na4pastazip = '/content/drive/My Drive/Nicolas/Amostra_A4.zip'\npastaarquivo_a4 = ['Amostra A4/N_2019_12_16/vibracao/', 'Amostra A4/A_2019_12_19/vibracao/', 'Amostra A4/A_2020_01_06/vibracao/', 'Amostra A4/A_2020_01_13/vibracao/' ]\n",
"_____no_output_____"
],
[
"a4_1fim = aplica_filtro(a4pastazip, pastaarquivo_a4[0], lista_ensaios_a4[0], 2)\na4_2fim = aplica_filtro(a4pastazip, pastaarquivo_a4[1], lista_ensaios_a4[1], 1)\na4_3fim = aplica_filtro(a4pastazip, pastaarquivo_a4[2], lista_ensaios_a4[2], 2)\na4_4fim = aplica_filtro(a4pastazip, pastaarquivo_a4[3], lista_ensaios_a4[3], 1)\n",
"_____no_output_____"
],
[
"lista_ensaios_a5 = [a5_1, a5_2, a5_3]\na5pastazip = '/content/drive/My Drive/Nicolas/Amostra_A5.zip'\npastaarquivo_a5 = ['Amostra A5/N_2020_01_22/vibracao/', 'Amostra A5/A_2020_01_27/vibracao/', 'Amostra A5/A_2020_01_28/vibracao/']",
"_____no_output_____"
],
[
"a5_1fim = aplica_filtro(a5pastazip, pastaarquivo_a5[0], lista_ensaios_a5[0], 1)\na5_2fim = aplica_filtro(a5pastazip, pastaarquivo_a5[1], lista_ensaios_a5[1], 1)\na5_3fim = aplica_filtro(a5pastazip, pastaarquivo_a5[2], lista_ensaios_a5[2], 1)",
"_____no_output_____"
],
[
"# a3_1fim.to_csv('A3_N_2019_12_04tAm2.5.csv')\n# a3_2fim.to_csv('A3_A_2019_12_09tAm11.8.csv')\n# a3_3fim.to_csv('A3_A_2019_12_11tAm2.5.csv')",
"_____no_output_____"
],
[
"# a4_1fim.to_csv('A4_N_2019_12_16tAm2.1.csv')\n# a4_2fim.to_csv('A4_A_2019_12_19tAm6.csv')\n# a4_3fim.to_csv('A4_A_2020_01_06tAm2.5.csv')\n# a4_4fim.to_csv('A4_A_2020_01_13tAm2.5.csv')",
"_____no_output_____"
],
[
"# a5_1fim.to_csv('A5_N_2020_01_22tAm2.5.csv')\n# a5_2fim.to_csv('A5_A_2020_01_27tAm12.5.csv')\n# a5_3fim.to_csv('A5_A_2020_01_28tAm2.5.csv')",
"_____no_output_____"
],
[
"# files.download('A3_N_2019_12_04tAm2.5.csv')\n# files.download('A3_A_2019_12_09tAm11.8.csv')\n# files.download('A3_A_2019_12_11tAm2.5.csv')",
"_____no_output_____"
],
[
"# files.download('A4_N_2019_12_16tAm2.1.csv')\n# files.download('A4_A_2019_12_19tAm6.csv')\n# files.download('A4_A_2020_01_06tAm2.5.csv')\n# files.download('A4_A_2020_01_13tAm2.5.csv')",
"_____no_output_____"
],
[
"# files.download('A5_N_2020_01_22tAm2.5.csv')\n# files.download('A5_A_2020_01_27tAm12.5.csv')\n# files.download('A5_A_2020_01_28tAm2.5.csv')",
"_____no_output_____"
]
],
[
[
"# Referencias\n\nhttps://www.kaggle.com/cchadha/an-intuitive-application-of-butterworth-filters\n\nhttps://www.geeksforgeeks.org/noise-removal-using-lowpass-digital-butterworth-filter-in-scipy-python/?ref=rp\n\nhttps://medium.com/analytics-vidhya/how-to-filter-noise-with-a-low-pass-filter-python-885223e5e9b7\n\nhttps://nehajirafe.medium.com/using-fft-to-analyse-and-cleanse-time-series-data-d0c793bb82e3\n\nhttps://docs.scipy.org/doc/scipy/reference/generated/scipy.signal.butter.html#scipy.signal.butter\n\nhttp://www.portalaction.com.br/estatistica-basica/26-curtose\n\nhttps://en.wikipedia.org/wiki/Root_mean_square\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
e790fc9ce0cb42c3afb9387cda179ebcba5f8525 | 334,860 | ipynb | Jupyter Notebook | mypython/t4_regularization_PoissonGLM.ipynb | disadone/GLMspiketraintutorial | 872714ef18732d4dd8aa23d19d376fa661de906f | [
"MIT"
] | null | null | null | mypython/t4_regularization_PoissonGLM.ipynb | disadone/GLMspiketraintutorial | 872714ef18732d4dd8aa23d19d376fa661de906f | [
"MIT"
] | null | null | null | mypython/t4_regularization_PoissonGLM.ipynb | disadone/GLMspiketraintutorial | 872714ef18732d4dd8aa23d19d376fa661de906f | [
"MIT"
] | null | null | null | 347.725857 | 77,700 | 0.933175 | [
[
[
"\nThis is an interactive tutorial designed to walk through\nregularization for a linear-Gaussian GLM, which allows for closed-form\nMAP parameter estimates. The next tutorial ('tutorial4') will cover the\nsame methods for the Poisson GLM (which requires numerical optimization).\n\nWe'll consider two simple regularization methods:\n\n1. Ridge regression - corresponds to maximum a posteriori (MAP) estimation under an iid Gaussian prior on the filter coefficients. \n\n2. L2 smoothing prior - using to an iid Gaussian prior on the pairwise-differences of the filter(s).\n\nData: from Uzzell & Chichilnisky 2004; see README file for details. \n\nLast updated: Mar 10, 2020 (JW Pillow)\n\nTutorial instructions: Execute each section below separately using\ncmd-enter. For detailed suggestions on how to interact with this\ntutorial, see header material in tutorial1_PoissonGLM.m\n\nTransferred into Python by Xiaodong LI",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom scipy.io import loadmat\nfrom scipy.optimize import minimize\nfrom scipy.linalg import hankel,pinv,block_diag\nfrom scipy.interpolate import interp1d\nfrom interpolation import interp\nfrom numpy.linalg import inv,norm,lstsq\nfrom matplotlib import mlab\n\naa=np.asarray\n\ndef neglogli_poissGLM(prs,XX,YY,dtbin):\n \"\"\"\n Compute negative log-likelihood of data undr Poisson GLM model with\n exponential nonlinearity\n \n Inputs:\n prs [d x 1] - parameter vector\n XX [T x d] - design matrix\n YY [T x 1] - response (spike count per time bin)\n dtbin [1 x 1] - time bin size used \n \n Outputs:\n neglogli = negative log likelihood of spike train\n dL [d x 1] = gradient \n H [d x d] = Hessian (second deriv matrix)\n \"\"\"\n # Compute GLM filter output and condititional intensity\n vv = XX@prs # filter output\n rr = np.exp(vv)*dtbin # conditional intensity (per bin)\n\n # --------- Compute log-likelihood -----------\n Trm1 = -vv.T@YY # spike term from Poisson log-likelihood\n Trm0 = np.sum(rr) # non-spike term \n neglogli = Trm1 + Trm0\n \n return neglogli\n\ndef jac_neglogli_poissGLM(prs,XX,YY,dtbin):\n \n # Compute GLM filter output and condititional intensity\n vv = XX@prs # filter output\n rr = np.exp(vv)*dtbin # conditional intensity (per bin)\n \n # --------- Compute Gradient -----------------\n dL1 = -XX.T@YY # spiking term (the spike-triggered average)\n dL0 = XX.T@rr # non-spiking term\n dL = dL1+dL0\n \n return dL\n\ndef hess_neglogli_poissGLM(prs,XX,YY,dtbin):\n # Compute GLM filter output and condititional intensity\n vv = XX@prs # filter output\n rr = np.exp(vv)*dtbin # conditional intensity (per bin)\n # --------- Compute Hessian -------------------\n H = [email protected](XX,rr.reshape(-1,1)) # non-spiking term\n \n return H\n\ndef neglogposterior(prs,negloglifun,Cinv):\n \"\"\"\n Compute negative log-posterior given a negative log-likelihood function \n and zero-mean Gaussian prior with inverse covariance 'Cinv'.\n\n Inputs:\n prs [d x 1] - parameter vector\n negloglifun - handle for negative log-likelihood function\n Cinv [d x d] - response (spike count per time bin)\n\n Outputs:\n negLP - negative log posterior\n grad [d x 1] - gradient \n H [d x d] - Hessian (second deriv matrix)\n\n Compute negative log-posterior by adding quadratic penalty to log-likelihood\n \"\"\"\n\n # evaluate function and gradient\n negLP= negloglifun(prs)\n negLP += .5*prs.T@Cinv@prs \n return negLP\ndef jac_neglogposterior(prs,jac_negloglifun,Cinv):\n grad=jac_negloglifun(prs)\n grad += Cinv@prs\n return grad\ndef hess_neglogposterior(prs,hess_negloglifun,Cinv):\n H=hess_negloglifun(prs)\n H += Cinv\n return H",
"_____no_output_____"
]
],
[
[
"# Load the raw data",
"_____no_output_____"
],
[
"Be sure to unzip the data file data_RGCs.zip\n(http://pillowlab.princeton.edu/data/data_RGCs.zip) and place it in \nthis directory before running the tutorial. \nOr substitute your own dataset here instead!\n\n\n(Data from Uzzell & Chichilnisky 2004):",
"_____no_output_____"
]
],
[
[
"datadir='../data_RGCs/' # directory where stimulus lives\nStim=loadmat(datadir+'Stim.mat')['Stim'].flatten() # stimulus (temporal binary white noise)\nstimtimes=loadmat(datadir+'stimtimes.mat')['stimtimes'].flatten() # stim frame times in seconds (if desired)\nSpTimes=loadmat(datadir+'SpTimes.mat')['SpTimes'][0,:] # load spike times (in units of stim frames)\nncells=len(SpTimes) # number of neurons (4 for this dataset).\n# Neurons #0-1 are OFF, #2-3 are ON.",
"_____no_output_____"
]
],
[
[
"Pick a cell to work with",
"_____no_output_____"
]
],
[
[
"cellnum = 2 # (0-1 are OFF cells; 2-3 are ON cells).\ntsp = SpTimes[cellnum];",
"_____no_output_____"
]
],
[
[
"Compute some basic statistics on the stimulus",
"_____no_output_____"
]
],
[
[
"dtStim = stimtimes[1]-stimtimes[0] # time bin size for stimulus (s)\n\n# See tutorial 1 for some code to visualize the raw data!",
"_____no_output_____"
]
],
[
[
"# Upsample to get finer timescale representation of stim and spikes",
"_____no_output_____"
],
[
"The need to regularize GLM parameter estimates is acute when we don't\nhave enough data relative to the number of parameters we're trying to\nestimate, or when using correlated (eg naturalistic) stimuli, since the\nstimuli don't have enough power at all frequencies to estimate all\nfrequency components of the filter. \n\nThe RGC dataset we've looked at so far requires only a temporal filter\n(as opposed to spatio-temporal filter for full spatiotemporal movie\nstimuli), so it doesn't have that many parameters to esimate. It also has\nbinary white noise stimuli, which have equal energy at all frequencies.\nRegularization thus isn't an especially big deal for this data (which was\npart of our reason for selecting it). However, we can make it look\ncorrelated by considering it on a finer timescale than the frame rate of\nthe monitor. (Indeed, this will make it look highly correlated).\n\nFor speed of our code and to illustrate the advantages of regularization,\nlet's use only a reduced (5-minute) portion of the dataset:",
"_____no_output_____"
]
],
[
[
"nT=120*60*1 # # of time bins for 1 minute of data\nStim=Stim[:nT] # pare down stimulus\ntsp=tsp[tsp<nT*dtStim] # pare down spikes",
"_____no_output_____"
]
],
[
[
"Now upsample to finer temporal grid",
"_____no_output_____"
]
],
[
[
"upsampfactor = 5 # divide each time bin by this factor\ndtStimhi = dtStim/upsampfactor # use bins 100 time bins finer\nttgridhi = np.arange(dtStimhi/2,nT*dtStim+dtStimhi,dtStimhi) # fine time grid for upsampled stim\nStimhi = interp1d(np.arange(1,nT+1)*dtStim,Stim,kind='nearest',fill_value='extrapolate')(ttgridhi)\nnThi = nT*upsampfactor # length of upsampled stimulus",
"_____no_output_____"
]
],
[
[
"Visualize the new (upsampled) raw data:",
"_____no_output_____"
]
],
[
[
"fig,axes=plt.subplots(nrows=2,figsize=(8,6),sharex=True)\niiplot=np.arange(0,60*upsampfactor) # bins of stimulus to plot\nttplot=iiplot*dtStimhi # time bins of stimulus\naxes[0].plot(ttplot,Stimhi[iiplot])\naxes[0].set_title('raw stimulus (fine time bins)')\naxes[0].set_ylabel('stim intensity')\n# Should notice stimulus now constant for many bins in a row\nsps,_=np.histogram(tsp,ttgridhi) # Bin the spike train and replot binned counts\naxes[1].stem(ttplot,sps[iiplot])\naxes[1].set_title('binned spike counts')\naxes[1].set_ylabel('spike count')\naxes[1].set_xlabel('time (s)')\naxes[1].set_xlim(ttplot[0],ttplot[-1])",
"<ipython-input-36-97db7153fd27>:9: UserWarning: In Matplotlib 3.3 individual lines on a stem plot will be added as a LineCollection instead of individual lines. This significantly improves the performance of a stem plot. To remove this warning and switch to the new behaviour, set the \"use_line_collection\" keyword argument to True.\n axes[1].stem(ttplot,sps[iiplot])\n"
]
],
[
[
"# Divide data into \"training\" and \"test\" sets for cross-validation",
"_____no_output_____"
]
],
[
[
"trainfrac = .8 # fraction of data to use for training\nntrain = int(np.ceil(nThi*trainfrac)) # number of training samples\nntest = int(nThi-ntrain) # number of test samples\niitest = np.arange(ntest).astype(int) # time indices for test\niitrain = np.arange(ntest,nThi).astype(int) # time indices for training\nstimtrain = Stimhi[iitrain] # training stimulus\nstimtest = Stimhi[iitest] # test stimulus\nspstrain = sps[iitrain]\nspstest = sps[iitest]\n\nprint('Dividing data into training and test sets:\\n')\nprint('Training: %d samples (%d spikes) \\n'%(ntrain, sum(spstrain)))\nprint(' Test: %d samples (%d spikes)\\n'%(ntest, sum(spstest)))",
"Dividing data into training and test sets:\n\nTraining: 28800 samples (2109 spikes) \n\n Test: 7200 samples (557 spikes)\n\n"
]
],
[
[
"Set the number of time bins of stimulus to use for predicting spikes",
"_____no_output_____"
]
],
[
[
"ntfilt = 20*upsampfactor # Try varying this, to see how performance changes!",
"_____no_output_____"
]
],
[
[
"build the design matrix, training data",
"_____no_output_____"
]
],
[
[
"Xtrain = np.c_[\n np.ones((ntrain,1)),\n hankel(np.r_[np.zeros(ntfilt-1),stimtrain[:-ntfilt+1]].reshape(-1,1),stimtrain[-ntfilt:])]",
"_____no_output_____"
]
],
[
[
"Build design matrix for test data",
"_____no_output_____"
]
],
[
[
"Xtest = np.c_[\n np.ones((ntest,1)),\n hankel(np.r_[np.zeros(ntfilt-1),stimtest[:-ntfilt+1]].reshape(-1,1),stimtest[-ntfilt:])]",
"_____no_output_____"
]
],
[
[
"# Fit poisson GLM using ML",
"_____no_output_____"
],
[
"Compute maximum likelihood estimate (using `scipy.optimize.fmin` instead of `sm.GLM`)",
"_____no_output_____"
]
],
[
[
"sta = (Xtrain.T@spstrain)/np.sum(spstrain) # compute STA for initialization",
"_____no_output_____"
]
],
[
[
"Make loss function and minimize",
"_____no_output_____"
]
],
[
[
"jac_neglogli_poissGLM?",
"_____no_output_____"
],
[
"lossfun=lambda prs:neglogli_poissGLM(prs,Xtrain,spstrain,dtStimhi)\njacfun=lambda prs:jac_neglogli_poissGLM(prs,Xtrain,spstrain,dtStimhi)\nhessfun=lambda prs:hess_neglogli_poissGLM(prs,Xtrain,spstrain,dtStimhi)\nfiltML=minimize(lossfun,x0=sta,method='trust-ncg',jac=jacfun, hess=hessfun).x\n\nttk=np.arange(-ntfilt+1,1)*dtStimhi\nfig,axes=plt.subplots()\naxes.plot(ttk,ttk*0,'k')\naxes.plot(ttk,filtML[1:])\naxes.set_xlabel('time before spike')\naxes.set_ylabel('coefficient')\naxes.set_title('Maximum likelihood filter estimate')\n# % Looks bad due to lack of regularization!",
"_____no_output_____"
]
],
[
[
"# Ridge regression prior",
"_____no_output_____"
],
[
"<img src='pics/f4-1.png'>\n\nNow let's regularize by adding a penalty on the sum of squared filter\ncoefficients w(i) of the form:\n \n penalty(lambda) = lambda*(sum_i w(i).^2),\n\nwhere lambda is known as the \"ridge\" parameter. As noted in tutorial3,\nthis is equivalent to placing an iid zero-mean Gaussian prior on the RF\ncoefficients with variance equal to 1/lambda. Lambda is thus the inverse\nvariance or \"precision\" of the prior.\n\nTo set lambda, we'll try a grid of values and use\ncross-validation (test error) to select which is best. \n\nSet up grid of lambda values (ridge parameters)",
"_____no_output_____"
]
],
[
[
"lamvals = 2.**np.arange(0,11,1) # it's common to use a log-spaced set of values\nnlam = len(lamvals)",
"_____no_output_____"
]
],
[
[
"Precompute some quantities (X'X and X'*y) for training and test data",
"_____no_output_____"
]
],
[
[
"Imat = np.eye(ntfilt+1) # identity matrix of size of filter + const\nImat[0,0] = 0 # remove penalty on constant dc offset",
"_____no_output_____"
]
],
[
[
"Allocate space for train and test errors",
"_____no_output_____"
]
],
[
[
"negLtrain = np.zeros(nlam) # training error\nnegLtest = np.zeros(nlam) # test error\nw_ridge = np.zeros((ntfilt+1,nlam)) # filters for each lambda",
"_____no_output_____"
]
],
[
[
"Define train and test log-likelihood funcs",
"_____no_output_____"
]
],
[
[
"negLtrainfun = lambda prs:neglogli_poissGLM(prs,Xtrain,spstrain,dtStimhi)\njac_negLtrainfun = lambda prs:jac_neglogli_poissGLM(prs,Xtrain,spstrain,dtStimhi)\nhess_negLtrainfun = lambda prs:hess_neglogli_poissGLM(prs,Xtrain,spstrain,dtStimhi)\nnegLtestfun = lambda prs:neglogli_poissGLM(prs,Xtest,spstest,dtStimhi)\njac_negLtestfun = lambda prs:jac_neglogli_poissGLM(prs,Xtest,spstest,dtStimhi)\nhess_negLtestfun = lambda prs:hess_neglogli_poissGLM(prs,Xtest,spstest,dtStimhi)",
"_____no_output_____"
]
],
[
[
"Now compute MAP estimate for each ridge parameter",
"_____no_output_____"
]
],
[
[
"wmap = filtML # initialize parameter estimate\nfig,axes=plt.subplots()\naxes.plot(ttk,ttk*0,'k') # initialize plot\nfor jj in range(nlam):\n \n # Compute ridge-penalized MAP estimate\n Cinv = lamvals[jj]*Imat # set inverse prior covariance\n lossfun = lambda prs:neglogposterior(prs,negLtrainfun,Cinv)\n jacfun=lambda prs:jac_neglogposterior(prs,jac_negLtrainfun,Cinv)\n hessfun=lambda prs:hessian_neglogposterior(prs,hess_negLtrainfun,Cinv)\n \n wmap=minimize(lossfun,x0=wmap,method='trust-ncg',jac=jacfun,hess=hessfun).x\n \n # Compute negative logli\n negLtrain[jj] = negLtrainfun(wmap) # training loss\n negLtest[jj] = negLtestfun(wmap) # test loss\n \n # store the filter\n w_ridge[:,jj] = wmap\n \n # plot it\n axes.plot(ttk,wmap[1:]) \n axes.set_title(['ridge estimate: lambda = %.2f'%lamvals[jj]])\n axes.set_xlabel('time before spike (s)')\n # note that the esimate \"shrinks\" down as we increase lambda",
"_____no_output_____"
]
],
[
[
"Plot filter estimates and errors for ridge estimates",
"_____no_output_____"
]
],
[
[
"fig,axes=plt.subplots(nrows=2,ncols=2,figsize=(8,8))\naxes[0,1].plot(ttk,w_ridge[1:,:])\naxes[0,1].set_title('all ridge estimates')\naxes[0,0].semilogx(lamvals,-negLtrain,'o-')\naxes[0,0].set_title('training logli')\naxes[1,0].semilogx(lamvals,-negLtest,'o-')\naxes[1,0].set_title('test logli')\naxes[1,0].set_xlabel('lambda')\n\n# Notice that training error gets monotonically worse as we increase lambda\n# However, test error has an dip at some optimal, intermediate value.\n\n# Determine which lambda is best by selecting one with lowest test error \nimin = np.argmin(negLtest)\nfilt_ridge= w_ridge[1:,imin]\n\naxes[1,1].plot(ttk,ttk*0, 'k--') \naxes[1,1].set_xlabel('time before spike (s)')\naxes[1,1].set_title('best ridge estimate')",
"_____no_output_____"
]
],
[
[
"# L2 smoothing prior",
"_____no_output_____"
],
[
"<img src='pics/f4-2.png'>\n\nUse penalty on the squared differences between filter coefficients,\npenalizing large jumps between successive filter elements. This is\nequivalent to placing an iid zero-mean Gaussian prior on the increments\nbetween filter coeffs. (See tutorial 3 for visualization of the prior\ncovariance).\n\nThis matrix computes differences between adjacent coeffs",
"_____no_output_____"
]
],
[
[
"Dx1 = (np.diag(-np.ones(ntfilt),0)+np.diag(np.ones(ntfilt-1),1))[:-1,:]\nDx = Dx1.T@Dx1 # computes squared diffs",
"_____no_output_____"
]
],
[
[
"Select smoothing penalty by cross-validation",
"_____no_output_____"
]
],
[
[
"lamvals = 2**np.arange(1,15) # grid of lambda values (ridge parameters)\nnlam = len(lamvals)",
"_____no_output_____"
]
],
[
[
"Embed `Dx` matrix in matrix with one extra row/column for constant coeff",
"_____no_output_____"
]
],
[
[
"D = block_diag(0,Dx)",
"_____no_output_____"
]
],
[
[
"Allocate space for train and test errors",
"_____no_output_____"
]
],
[
[
"negLtrain_sm = np.zeros(nlam) # training error\nnegLtest_sm = np.zeros(nlam) # test error\nw_smooth = np.zeros((ntfilt+1,nlam)) # filters for each lambda",
"_____no_output_____"
]
],
[
[
"Now compute MAP estimate for each ridge parameter",
"_____no_output_____"
]
],
[
[
"fig,axes=plt.subplots()\naxes.plot(ttk,ttk*0,'k') # initialize plot\nwmap=filtML # initialize with ML fit\nfor jj in range(nlam):\n \n # Compute MAP estimate\n Cinv=lamvals[jj]*D # set inverse prior covariance\n lossfun = lambda prs:neglogposterior(prs,negLtrainfun,Cinv)\n jacfun=lambda prs:jac_neglogposterior(prs,jac_negLtrainfun,Cinv)\n hessfun=lambda prs:hessian_neglogposterior(prs,hess_negLtrainfun,Cinv)\n wmap=minimize(lossfun,x0=wmap,method='trust-ncg',jac=jacfun,hess=hessfun).x\n \n # Compute negative logli\n negLtrain_sm[jj]=negLtrainfun(wmap) # training loss\n negLtest_sm[jj]=negLtestfun(wmap) # test loss\n \n # store the filter\n w_smooth[:,jj] = wmap\n \n # plot it\n axes.plot(ttk,wmap[1:]) \n axes.set_title('smoothing estimate: lambda = %.2f'%lamvals[jj])\n axes.set_xlabel('time before spike (s)')",
"_____no_output_____"
]
],
[
[
"Plot filter estimates and errors for smoothing estimates",
"_____no_output_____"
]
],
[
[
"fig,axes=plt.subplots(nrows=2,ncols=2,figsize=(8,8))\naxes[0,1].plot(ttk,w_smooth[1:,:])\naxes[0,1].set_title('all smoothing estimates')\naxes[0,0].semilogx(lamvals,-negLtrain_sm,'o-')\naxes[0,0].set_title('training LL')\naxes[1,0].semilogx(lamvals,-negLtest_sm,'o-')\naxes[1,0].set_title('test LL')\naxes[1,0].set_xlabel('lambda')\n\n# Notice that training error gets monotonically worse as we increase lambda\n# However, test error has an dip at some optimal, intermediate value.\n\n# Determine which lambda is best by selecting one with lowest test error \nimin = np.argmin(negLtest_sm)\nfilt_smooth= w_smooth[1:,imin]\n\naxes[1,1].plot(ttk,ttk*0, 'k--') \naxes[1,1].plot(ttk,filt_ridge,label='ridge')\naxes[1,1].plot(ttk,filt_smooth,label='L2 smoothing')\naxes[1,1].set_xlabel('time before spike (s)')\naxes[1,1].set_title('best ridge estimate')\naxes[1,1].legend()\n\n# clearly the \"L2 smoothing\" filter looks better by eye!",
"_____no_output_____"
]
],
[
[
"Last, lets see which one actually achieved lower test error",
"_____no_output_____"
]
],
[
[
"print('\\nBest ridge test error: %.5f'%(-min(negLtest)))\nprint('Best smoothing test error: %.5f'%(-min(negLtest_sm)))",
"\nBest ridge test error: 2093.80432\nBest smoothing test error: 2095.67887\n"
]
],
[
[
"Advanced exercise:\n--------------------\n\n1. Repeat of the above, but incorporate spike history filters as in tutorial2. Use a different smoothing hyperparamter for the spike-history / coupling filters than for the stim filter. In this case one needs to build a block diagonal prior covariance, with one block for each group of coefficients.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e790ff45e4d695a92566c16acff230c088ae13ce | 17,078 | ipynb | Jupyter Notebook | bank_note_auth.ipynb | josh-boat365/docker-ml | ec304a2d4dc9236e58fcc715e9f7b8d52ee15964 | [
"MIT"
] | null | null | null | bank_note_auth.ipynb | josh-boat365/docker-ml | ec304a2d4dc9236e58fcc715e9f7b8d52ee15964 | [
"MIT"
] | null | null | null | bank_note_auth.ipynb | josh-boat365/docker-ml | ec304a2d4dc9236e58fcc715e9f7b8d52ee15964 | [
"MIT"
] | null | null | null | 26.518634 | 415 | 0.395772 | [
[
[
"# Bank Note Authentication\n\nData were extracted from images that were taken from genuine and forged banknote-like specimens. For digitization, an industrial camera usually used for inspection was used. The final images have 400 pixles. Due to the object lens and distance to the investigated object gray-scale pictures with a resolution of about 660 dpl were gained. Wavelet Transformation tool were sued to extract features from images.",
"_____no_output_____"
]
],
[
[
"#dataset link: https://kaggle.com/ritesaluja/bank-note-authentication-uci-data\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"df = pd.read_csv('BankNote_Authentication.csv')",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.tail()",
"_____no_output_____"
],
[
"df.describe()",
"_____no_output_____"
],
[
"#y=dependent and x=independent features\nx=df.iloc[:,:-1] #present everything in the datasset except the last column\ny=df.iloc[:,-1] #presenting only the last column from the dataset",
"_____no_output_____"
],
[
"x.head()",
"_____no_output_____"
],
[
"y.head()",
"_____no_output_____"
],
[
"# train test split\nfrom sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"x_train,x_test, y_train, y_test = train_test_split(x,y, test_size=0.3, random_state=0)",
"_____no_output_____"
],
[
"# Implementing a Random Forest Classifier\nfrom sklearn.ensemble import RandomForestClassifier\nclassifier = RandomForestClassifier()\nclassifier.fit(x_train, y_train)\n",
"RandomForestClassifier()\n"
],
[
"#prediction\ny_pred = classifier.predict(x_test)",
"_____no_output_____"
],
[
"#checking for accuracy\nfrom sklearn.metrics import accuracy_score\nscore = accuracy_score(y_test, y_pred)",
"_____no_output_____"
],
[
"score\n",
"_____no_output_____"
],
[
"# creating a pickle file using serialization\nimport pickle\npickle_file = open('./classifier.pkl', 'wb')\npickle.dump(classifier, pickle_file)\npickle_file.close()",
"_____no_output_____"
],
[
"import numpy as np\n",
"_____no_output_____"
],
[
"classifier.predict([[2,3,4,1]])",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e791070fa8328b8013794fef43507cbc282e368d | 71,716 | ipynb | Jupyter Notebook | 00.ipynb | domingo2000/Python-Lectures | b3e93ae7ac7dd8ae3ec7c179c3db4f1e2aebe943 | [
"CC-BY-3.0"
] | null | null | null | 00.ipynb | domingo2000/Python-Lectures | b3e93ae7ac7dd8ae3ec7c179c3db4f1e2aebe943 | [
"CC-BY-3.0"
] | null | null | null | 00.ipynb | domingo2000/Python-Lectures | b3e93ae7ac7dd8ae3ec7c179c3db4f1e2aebe943 | [
"CC-BY-3.0"
] | null | null | null | 212.807122 | 58,412 | 0.902212 | [
[
[
"# Curso Python Programación",
"_____no_output_____"
],
[
"## Contenidos 📚\n\nEste curso se separa en varios notebooks (capítulos)\n\n* [00](00.ipynb) Introducción a Python y como empezar a correrlo en Google Colab\n* [01](01.ipynb) Tipos básicos de datos y operaciones (Numeros y Strings)\n* [02](02.ipynb) Manipulación de Strings \n* [03](03.ipynb) Estructuras de datos: Listas y Tuplas\n* [04](04.ipynb) Estructuras de datos (continuación): diccionarios\n* [05](05.ipynb) Control de Flujo: sentencias if, for, while, y try\n* [06](06.ipynb) Funciones\n* [07](07.ipynb) Clases y Pogramacion Orientada a Objetos (POO) básico\n\n<!-- Unidades extra incluidas en el curso original -->\n<!--- * [08](08.ipynb) Scipy: libraries for arrays (matrices) and plotting --><!--- * [09](09.ipynb) Mixed Integer Linear Programming using the mymip library. -->\n<!--- * [10](10.ipynb) Networks and graphs under python - a very brief introduction -->\n<!--- * [11](11.ipynb) Using the numba library for fast numerical computing. -->\n\n\n\n\n\nEste es un tutorial de introducción a python 3. Para un resumen/torpedo de toda la syntaxis de python puedes ir al siguiente link [Quick Reference Card](http://www.cs.put.poznan.pl/csobaniec/software/python/py-qrc.html) may be useful. Una version mas detallada a este tutorial esta disponible en la documentación oficial de python. https://docs.python.org/3/tutorial/\n",
"_____no_output_____"
],
[
"# Introducción\n",
"_____no_output_____"
],
[
"## Google Colab\n\n\n\n\n\n### Instalación 🖥️\n\nPara los fines de este curso trabajaremos con una herramienta online llamada Google Colab la cual tiene muchas ventajas frente a usar una instalación directa de python, algunas de las siguientes son:\n\n1. Servicio en linea igual para todos sin depender del sistema operativo\n2. Sistema de control de versiones, siempre puedes volver atrás igual que un Google Docs\n3. Texto y codigo en un mismo lugar, puedes explicar con \"Markdown\" tus programas y programar en un solo entorno\n4. Rendimiento, Google Colab corre los codigos por ti y te da recursos, no importa que tengas un mal procesador o una baja RAM.\n5. Es gratis\n\n",
"_____no_output_____"
],
[
"## Abriendo un Notebook desde Google Colab 🟠⚪",
"_____no_output_____"
],
[
"Para abrir un notebook del material del curso deberás seguir las siguientes instrucciones\n### Instrucciones Abrir Notebook del Material\n1. Ingresa a https://colab.research.google.com en una pestaña nueva\n2. Selecciona la opción \"GitHub\" de la siguiente ventana <strong> (En caso de no ver esa ventana ve al paso 2.1 más abajo y luego continua con los demás )</strong>\n\n\n\n5. <strong>Copia</strong> el siguiente link https://github.com/domingo2000/Python-Lectures y pégalo en la primera línea que te deja ingresar texto, luego presiona la lupita.\n\n\n6. Por último selecciona el archivo .pynb que quieres abrir, en este caso el 00, !Listo, ahora solo sigue leyendo pero en Google Colab!.\n\n\n\n\n2.1. Selecciona \"Archivo\" o \"File\"\n\n Archivo > Open Notebook\n\nvuelve al paso 2.\n\n\n",
"_____no_output_____"
],
[
"### Jupyter Notebooks y Markdown 📕\nCuando tu trabajas en colab en el fondo estás corriendo un <strong>Jupyter notebook</strong>, estos funcionan como un cuaderno, en el que puedes escribir texto o programar código en celdas diferentes.",
"_____no_output_____"
],
[
"#### Mi primera celda de código ⌨️\nEn primer lugar borraremos todo lo que se corre automáticamente cuando abres el archivo, para ello debemos presionar los siguientes menús en este orden.\n\n Edit > Clear all Output\n\n\n\nLuego para correr cada celda de código en Colab solo debes hacer clic en el botón \nAlternativamente puedes presionar `shift + enter ↵` para correr la celda.\nA continuación se muestra tu primer codigo python con el clásico ``Hello World!``\n",
"_____no_output_____"
]
],
[
[
"print(\"Hello World!\")",
"Hello World!\n"
]
],
[
[
"<strong> !Genial, acabas de correr tu primer programa de python! 😀 💻💻</strong>",
"_____no_output_____"
],
[
"### Markdown #️⃣#️⃣\n\nMarkdown es un tipo de lenguaje de formateo de texto que busca que el texto sea fácil de leer tanto en el \"codigo\" como en la salida que este produce, todo el texto de este tutorial está escrito en markdown para que tengas una idea de que cosas se pueden hacer con el.\n\nSi haces doble click en esta celda te daras cuenta de como se estructura el markdown, al igual que las celdas de codigo, puedes correrlas presionando `shift + enter ↵` o presionando en una celda distinta a esta.\n\n#### Esctructura básica\n\nPuedes escribir texto simple como este\n\n# Puedes escribir Titulos usando los \"#\"\n## Puedes escribir Titulos usando los \"##\"\n### Puedes escribir Titulos usando los \"##\"\n#### Puedes escribir Titulos usando los \"##\" etc...\n\nCon esto ya debrías lograr expresar tus ideas en los bloques de texto, pero con las siguientes cosas puedes poner más énfasis y hacer un mejor trabajo.\n\n* Puedes poner tu texto en **Negrita** usando \\*\\*texto** (hazle doble click a la celda para ver como se escriben dichos tags)\n* Puedes poner tu texto en *Itálica* usando \\*text\\*\n* Puedes ponerle ***ambas cosas*** usando \\*\\*\\*texto\\*\\*\\*\n\nPuedes hacer listas de cosas haciendo usando 1. y *\n1. Item 1\n2. Item 2\n3. Item 3\n* Item 3\n* Item 2\n* Item 1\n\nSi quieres buscar mas cosas que puedes hacer en markdown puedes ir a <br>\nhttps://www.markdownguide.org/basic-syntax/\n\nSi quieres saber mas sobre markdown puedes encontrar mas información en <br>\nhttps://es.wikipedia.org/wiki/Markdown\n",
"_____no_output_____"
],
[
"## ¿Que es Python?\n\n\n\nPython es un lenguaje moderno, robusto y de alto nivel de programación hoy es usado en gran medida en usos científicos variados y análisis de datos. Tiene la ventaja de ser bastante intuitivo y fácil de usar incluso si eres nuevo en la programación. Por otro lado tiene las desventajas de no ser tan rápido como otros lenguajes tipo C++, o Java, pero tiene la ventaja de ser mucho más rápido de programar debido a la syntaxis (Forma de escribir código) que este tiene.\nA continuación si corres la siguiente celda se mostrará un texto con la filosofía detrás de python, la cual sustenta este lenguaje y además son grandes consejos que te incentivan a escribir un **mejor codigo**",
"_____no_output_____"
]
],
[
[
"import this",
"The Zen of Python, by Tim Peters\n\nBeautiful is better than ugly.\nExplicit is better than implicit.\nSimple is better than complex.\nComplex is better than complicated.\nFlat is better than nested.\nSparse is better than dense.\nReadability counts.\nSpecial cases aren't special enough to break the rules.\nAlthough practicality beats purity.\nErrors should never pass silently.\nUnless explicitly silenced.\nIn the face of ambiguity, refuse the temptation to guess.\nThere should be one-- and preferably only one --obvious way to do it.\nAlthough that way may not be obvious at first unless you're Dutch.\nNow is better than never.\nAlthough never is often better than *right* now.\nIf the implementation is hard to explain, it's a bad idea.\nIf the implementation is easy to explain, it may be a good idea.\nNamespaces are one honking great idea -- let's do more of those!\n"
]
],
[
[
"En español (Traducido por google traductor que probablemente tiene mejor inglés que yo...) se lee lo siguiente:\n\nEl zen de Python, por Tim Peters <br>\n<em>\nLo bello es mejor que lo feo.<br>\nExplícito es mejor que implícito.<br>\nLo simple es mejor que lo complejo.<br>\nComplejo es mejor que complicado.<br>\nPlano es mejor que anidado.<br>\nEs mejor escaso que denso.<br>\nLa legibilidad cuenta.<br>\nLos casos especiales no son lo suficientemente especiales como para romper las reglas.<br>\nAunque la practicidad vence a la pureza.<br>\nLos errores nunca deben pasar en silencio.<br>\nA menos que esté explícitamente silenciado.<br>\nAnte la ambigüedad, rechace la tentación de adivinar.<br>\nDebe haber una, y preferiblemente solo una, forma obvia de hacerlo.<br>\nAunque esa forma puede no ser obvia al principio a menos que seas holandés.<br>\nAhora es mejor que nunca.<br>\nAunque a menudo nunca es mejor que * ahora mismo *.<br>\nSi la implementación es difícil de explicar, es una mala idea.<br>\nSi la implementación es fácil de explicar, puede ser una buena idea.<br>\nLos espacios de nombres son una gran idea, ¡hagamos más de eso!<br>\n</em>\n\n<strong> Sigan esta filosofía y les aseguro que podrán tener programas mejores... </strong>\n",
"_____no_output_____"
],
[
"A modo de ejercicio piensen cuantas de estas cosas no han seguido en sus propios códigos hasta ahora, un error dejado de lado, una solución demasiado compleja que podía ser mucho más simple, un código que al leerlo no queda explícito que es lo que hace, etc...",
"_____no_output_____"
],
[
"## Licencia",
"_____no_output_____"
],
[
"This work is licensed under the Creative Commons Attribution 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by/3.0/\n\n<small><font style=\"font-size:6pt\"><i>\nEste curso se encuentra disponible en https://github.com/domingo2000/Python-Lectures\n<br>\nLa versión original fue escrita por Rajath Kumar y está disponible en\nhttps://github.com/rajathkumarmp/Python-Lectures.<br>\nAdemás se sacó esta versión del trabajo de Andreas Ernst desde el siguiente link https://gitlab.erc.monash.edu.au/andrease/Python4Maths/tree/master\n</i></font></small>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
e79114eedf564cbf4d9a472f7f07b5e085b90c62 | 100,895 | ipynb | Jupyter Notebook | docs/circuit-examples/A.Qubits/08-JJ-Dolan.ipynb | Antonio-Aguiar/qiskit-metal | 5f714cc98c0ecf5b8c76f988b0b263520b86d671 | [
"Apache-2.0"
] | 1 | 2021-08-28T20:35:43.000Z | 2021-08-28T20:35:43.000Z | docs/circuit-examples/A.Qubits/08-JJ-Dolan.ipynb | jessica-angel7/qiskit-metal | 24c58d192a576f25acb8d4208a92a317d0ebb2fd | [
"Apache-2.0"
] | 1 | 2021-04-03T00:10:19.000Z | 2021-04-03T00:10:19.000Z | docs/circuit-examples/A.Qubits/08-JJ-Dolan.ipynb | jessica-angel7/qiskit-metal | 24c58d192a576f25acb8d4208a92a317d0ebb2fd | [
"Apache-2.0"
] | null | null | null | 492.170732 | 71,116 | 0.945716 | [
[
[
"# Josephson Junction (Dolan) \n\nWe'll be creating a Dolan style Josephson Junction.\n",
"_____no_output_____"
]
],
[
[
"# So, let us dive right in. For convenience, let's begin by enabling\n# automatic reloading of modules when they change.\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"import qiskit_metal as metal\nfrom qiskit_metal import designs, draw\nfrom qiskit_metal import MetalGUI, Dict, open_docs",
"_____no_output_____"
],
[
"# Each time you create a new quantum circuit design, \n# you start by instantiating a QDesign class. \n\n# The design class `DesignPlanar` is best for 2D circuit designs.\n\ndesign = designs.DesignPlanar()",
"_____no_output_____"
],
[
"#Launch Qiskit Metal GUI to interactively view, edit, and simulate QDesign: Metal GUI\ngui = MetalGUI(design)",
"_____no_output_____"
]
],
[
[
"### A dolan style josephson junction\n\nYou can create a dolan style josephson junction from the QComponent Library, `qiskit_metal.qlibrary.qubits`. \n`jj_dolan.py` is the file containing our josephson junction so `jj_dolan` is the module we import. \nThe `jj_dolan` class is our josephson junction. Like all quantum components, `jj_dolan` inherits from `QComponent`. \n",
"_____no_output_____"
]
],
[
[
"from qiskit_metal.qlibrary.qubits.JJ_Dolan import jj_dolan\n\n# Be aware of the default_options that can be overridden by user.\ndesign.overwrite_enabled = True\njj2 = jj_dolan(design, 'JJ2', options=dict(x_pos=\"0.1\", y_pos=\"0.0\"))\ngui.rebuild()\ngui.autoscale()\ngui.zoom_on_components(['JJ2'])",
"_____no_output_____"
],
[
"# Save screenshot as a .png formatted file.\n\ngui.screenshot()",
"_____no_output_____"
],
[
"# Screenshot the canvas only as a .png formatted file.\ngui.figure.savefig('shot.png')\n\nfrom IPython.display import Image, display\n_disp_ops = dict(width=500)\ndisplay(Image('shot.png', **_disp_ops))\n",
"_____no_output_____"
]
],
[
[
"## Closing the Qiskit Metal GUI",
"_____no_output_____"
]
],
[
[
"gui.main_window.close()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
e79118f01a34a3d9d9cc581337fd85852241d059 | 3,692 | ipynb | Jupyter Notebook | 1. Python Variables.ipynb | sivacheetas/matplotlib | af810525aa7875a1b6bf066179d106c7cec023a9 | [
"MIT"
] | 1 | 2019-10-29T03:55:59.000Z | 2019-10-29T03:55:59.000Z | 1. Python Variables.ipynb | zinor4u/Python-Basic-For-All-3.x | 392052dbcec5df53ea98c98726e4b9a04202453f | [
"MIT"
] | null | null | null | 1. Python Variables.ipynb | zinor4u/Python-Basic-For-All-3.x | 392052dbcec5df53ea98c98726e4b9a04202453f | [
"MIT"
] | null | null | null | 21.34104 | 144 | 0.517606 | [
[
[
"# Creating Variables\nUnlike other programming languages, Python has no command for declaring a variable.\n\nA variable is created the moment you first assign a value to it.",
"_____no_output_____"
]
],
[
[
"x = 5\ny = \"I TRAIN TECHNOLOGY\"\nprint(x)\nprint(y)",
"5\nI TRAIN TECHNOLOGY\n"
],
[
"# Variables do not need to be declared with any particular type and can even change type after they have been set.\n\nx = 4 # x is of type int\nx = \"Sally\" # x is now of type str\nprint(x)",
"Sally\n"
]
],
[
[
"# Variable Names\n1. A variable can have a short name (like x and y) or a more descriptive name (age, carname, total_volume). Rules for Python variables:\n2. A variable name must start with a letter or the underscore character\n3. A variable name cannot start with a number\n4. A variable name can only contain alpha-numeric characters and underscores (A-z, 0-9, and _ )\n5. Variable names are case-sensitive (age, Age and AGE are three different variables)\n# NOTE: Remember that variables are case-sensitive",
"_____no_output_____"
]
],
[
[
"# # Output Variables\n# The Python print statement is often used to output variables.\n\n#To combine both text and a variable, Python uses the + character:\n\nx = \"Scripting Programing\"\nprint(\"Python is \",x, \"Language\")",
"Python is Scripting Programing Language\n"
],
[
"x = \"Python is \"\ny = \"awesome\"\nz = x + y\nprint(z)",
"Python is awesome\n"
],
[
"# For numbers, the + character works as a mathematical operator:\nx = 5\ny = 10\nprint(x + y)",
"15\n"
],
[
"# Create a variable named carname and assign the value Volvo to it.\ncar = \"Volvo\"\nprint(car) ",
"Volvo\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
e7911be269f2650143bbd6979e0749f6af4ccf23 | 5,603 | ipynb | Jupyter Notebook | DatfiletoCSV.ipynb | karangupta26/Movie-Recommendation-system | 726260ffa09b7ce719cd2239266f315655f6a438 | [
"MIT"
] | null | null | null | DatfiletoCSV.ipynb | karangupta26/Movie-Recommendation-system | 726260ffa09b7ce719cd2239266f315655f6a438 | [
"MIT"
] | null | null | null | DatfiletoCSV.ipynb | karangupta26/Movie-Recommendation-system | 726260ffa09b7ce719cd2239266f315655f6a438 | [
"MIT"
] | null | null | null | 24.150862 | 323 | 0.552561 | [
[
[
"import pandas as pd",
"_____no_output_____"
]
],
[
[
"# Converting Rating.dat to Rating.csv",
"_____no_output_____"
]
],
[
[
"ratings_dataframe=pd.read_table(\"ratings.dat\",sep=\"::\")\nratings_dataframe.to_csv(\"ratings.csv\",index=False)\nratings_dataframe=pd.read_csv(\"ratings.csv\",header=None)\nratings_dataframe.columns=[\"UserID\",\"MovieID\",\"Rating\",\"Timestamp\"]",
"C:\\ProgramData\\Anaconda3\\lib\\site-packages\\ipykernel_launcher.py:1: ParserWarning: Falling back to the 'python' engine because the 'c' engine does not support regex separators (separators > 1 char and different from '\\s+' are interpreted as regex); you can avoid this warning by specifying engine='python'.\n \"\"\"Entry point for launching an IPython kernel.\n"
],
[
"ratings_dataframe.columns",
"_____no_output_____"
],
[
"print(ratings_dataframe.shape)",
"(1000209, 4)\n"
],
[
"ratings_dataframe.to_csv(\"ratings.csv\",index=False)",
"_____no_output_____"
]
],
[
[
"# Converting Movies.dat to Movies.csv",
"_____no_output_____"
]
],
[
[
"movies_dataframe=pd.read_table(\"movies.dat\",sep=\"::\")\nmovies_dataframe.to_csv(\"movies.csv\",index=False)\nmovies_dataframe=pd.read_csv(\"movies.csv\",header=None)\nmovies_dataframe.columns=[\"MovieID\",\"Title\",\"Genres\"]\n",
"C:\\ProgramData\\Anaconda3\\lib\\site-packages\\ipykernel_launcher.py:1: ParserWarning: Falling back to the 'python' engine because the 'c' engine does not support regex separators (separators > 1 char and different from '\\s+' are interpreted as regex); you can avoid this warning by specifying engine='python'.\n \"\"\"Entry point for launching an IPython kernel.\n"
],
[
"movies_dataframe.columns",
"_____no_output_____"
],
[
"print(movies_dataframe.shape)\nmovies_dataframe.to_csv(\"movies.csv\",index=False)",
"(3883, 3)\n"
]
],
[
[
"# Converting User.dat to User.csv",
"_____no_output_____"
]
],
[
[
"users_dataframe=pd.read_table(\"users.dat\",sep=\"::\")\nusers_dataframe.to_csv(\"users.csv\",index=False)\nusers_dataframe=pd.read_csv(\"users.csv\",header=None)\nusers_dataframe.columns=[\"UserID\",\"Gender\",\"Age\",\"Occupation\",\"Zip-code\"]",
"C:\\ProgramData\\Anaconda3\\lib\\site-packages\\ipykernel_launcher.py:1: ParserWarning: Falling back to the 'python' engine because the 'c' engine does not support regex separators (separators > 1 char and different from '\\s+' are interpreted as regex); you can avoid this warning by specifying engine='python'.\n \"\"\"Entry point for launching an IPython kernel.\n"
],
[
"users_dataframe.columns",
"_____no_output_____"
],
[
"users_dataframe.to_csv(\"users.csv\",index=False)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
e79121b9a47dcfa36ac48d5864916b059e6df56c | 551,835 | ipynb | Jupyter Notebook | starter_code/WeatherPy.ipynb | sruelle/python-api-challenge | bda70284e4d97b0a9d5b80a3d96dadba54a64f07 | [
"ADSL"
] | null | null | null | starter_code/WeatherPy.ipynb | sruelle/python-api-challenge | bda70284e4d97b0a9d5b80a3d96dadba54a64f07 | [
"ADSL"
] | null | null | null | starter_code/WeatherPy.ipynb | sruelle/python-api-challenge | bda70284e4d97b0a9d5b80a3d96dadba54a64f07 | [
"ADSL"
] | null | null | null | 78.363391 | 40,956 | 0.774965 | [
[
[
"# WeatherPy\n#----Observations:\n1. In the northern hemisphere the tempature increases as the latitude increases. So as the we move away from the equator to the north - the tempature decreases. \n2. In the southern hemisphere, the tempature increases as you get closer to the equator. \n3. In the northern hemishere, the humidity increases as you move away for the equator (0 lattitude).\n",
"_____no_output_____"
]
],
[
[
"# Dependencies and Setup\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport numpy as np\nimport requests\nimport time\nfrom scipy.stats import linregress\n\n\n# Import API key\nfrom api_keys import weather_api_key\n\n\n# Incorporated citipy to determine city based on latitude and longitude\nfrom citipy import citipy\n\n# Save config information.\nurl = \"http://api.openweathermap.org/data/2.5/weather?\"\nunits = \"Imperial\"\n\n# Build partial query URL\nquery_url = f\"{url}appid={weather_api_key}&units={units}&q=\"\n\n\n# Range of latitudes and longitudes\nlat_range = (-90, 90)\nlng_range = (-180, 180)\n\n# Output File (CSV)\noutput_data_file = \"cities.csv\"\n",
"_____no_output_____"
]
],
[
[
"## Generate Cities List",
"_____no_output_____"
]
],
[
[
"# List for holding lat_lngs and cities\nlat_lngs = []\ncities = []\n\n# Create a set of random lat and lng combinations\nlats = np.random.uniform(low=-90.000, high=90.000, size=1500)\nlngs = np.random.uniform(low=-180.000, high=180.000, size=1500)\nlat_lngs = zip(lats, lngs)\n\n# Identify nearest city for each lat, lng combination\nfor lat_lng in lat_lngs:\n city = citipy.nearest_city(lat_lng[0], lat_lng[1]).city_name\n \n # If the city is unique, then add it to a our cities list\n if city not in cities:\n cities.append(city)\n\n# Print the city count to confirm sufficient count\nlen(cities)",
"_____no_output_____"
]
],
[
[
"### Perform API Calls\n* Perform a weather check on each city using a series of successive API calls.\n* Include a print log of each city as it'sbeing processed (with the city number and city name).\n",
"_____no_output_____"
]
],
[
[
"# set lists for the dataframe\ncity_two = []\ncloudinesses = []\ndates = []\nhumidities = []\nlats = []\nlngs = []\nmax_temps = []\nwind_speeds = []\ncountries = []\n\n# set initial count quantities for organization\ncount_one = 0\nset_one = 1\n\n# loops for creating dataframe columns\nfor city in cities:\n try:\n response = requests.get(query_url + city.replace(\" \",\"&\")).json()\n cloudinesses.append(response['clouds']['all'])\n countries.append(response['sys']['country'])\n dates.append(response['dt'])\n humidities.append(response['main']['humidity'])\n lats.append(response['coord']['lat'])\n lngs.append(response['coord']['lon'])\n max_temps.append(response['main']['temp_max'])\n wind_speeds.append(response['wind']['speed'])\n if count_one > 48:\n count_one = 1\n set_one += 1\n city_two.append(city)\n else:\n count_one += 1\n city_two.append(city)\n print(f\"Processing Record {count_one} of Set {set_one} | {city}\")\n except Exception:\n print(\"City not found. Skipping...\")\nprint(\"------------------------------\\nData Retrieval Complete\\n------------------------------\")",
"Processing Record 1 of Set 1 | vaini\nProcessing Record 2 of Set 1 | punta arenas\nProcessing Record 3 of Set 1 | cruzilia\nProcessing Record 4 of Set 1 | mahibadhoo\nProcessing Record 5 of Set 1 | mys shmidta\nProcessing Record 6 of Set 1 | castro\nProcessing Record 7 of Set 1 | lebu\nProcessing Record 8 of Set 1 | butaritari\nProcessing Record 9 of Set 1 | rikitea\nProcessing Record 10 of Set 1 | turangi\nProcessing Record 11 of Set 1 | norman wells\nProcessing Record 12 of Set 1 | ushuaia\nProcessing Record 13 of Set 1 | san cristobal\nProcessing Record 14 of Set 1 | san luis\nProcessing Record 15 of Set 1 | saint-philippe\nProcessing Record 16 of Set 1 | mugumu\nProcessing Record 17 of Set 1 | port alfred\nProcessing Record 18 of Set 1 | dikson\nProcessing Record 19 of Set 1 | naryan-mar\nCity not found. Skipping...\nProcessing Record 20 of Set 1 | ogembo\nProcessing Record 21 of Set 1 | port hedland\nCity not found. Skipping...\nProcessing Record 22 of Set 1 | thompson\nProcessing Record 23 of Set 1 | provideniya\nProcessing Record 24 of Set 1 | sukhumi\nProcessing Record 25 of Set 1 | airai\nProcessing Record 26 of Set 1 | arraial do cabo\nProcessing Record 27 of Set 1 | esperance\nCity not found. Skipping...\nCity not found. Skipping...\nProcessing Record 28 of Set 1 | kaitangata\nCity not found. Skipping...\nProcessing Record 29 of Set 1 | ponta do sol\nProcessing Record 30 of Set 1 | ribeira grande\nCity not found. Skipping...\nProcessing Record 31 of Set 1 | hermanus\nProcessing Record 32 of Set 1 | bredasdorp\nProcessing Record 33 of Set 1 | verkh-usugli\nProcessing Record 34 of Set 1 | mahebourg\nProcessing Record 35 of Set 1 | severo-kurilsk\nProcessing Record 36 of Set 1 | mataura\nProcessing Record 37 of Set 1 | haines junction\nProcessing Record 38 of Set 1 | port hardy\nProcessing Record 39 of Set 1 | praia\nProcessing Record 40 of Set 1 | ancud\nProcessing Record 41 of Set 1 | albany\nCity not found. Skipping...\nProcessing Record 42 of Set 1 | upernavik\nProcessing Record 43 of Set 1 | necocli\nProcessing Record 44 of Set 1 | ambulu\nProcessing Record 45 of Set 1 | souillac\nProcessing Record 46 of Set 1 | san patricio\nProcessing Record 47 of Set 1 | kushmurun\nProcessing Record 48 of Set 1 | busselton\nProcessing Record 49 of Set 1 | vao\nProcessing Record 1 of Set 2 | inverness\nCity not found. Skipping...\nProcessing Record 2 of Set 2 | gweta\nProcessing Record 3 of Set 2 | longyearbyen\nProcessing Record 4 of Set 2 | clyde river\nProcessing Record 5 of Set 2 | usvyaty\nProcessing Record 6 of Set 2 | grand river south east\nProcessing Record 7 of Set 2 | amahai\nProcessing Record 8 of Set 2 | kodiak\nProcessing Record 9 of Set 2 | kapaa\nProcessing Record 10 of Set 2 | lavrentiya\nProcessing Record 11 of Set 2 | cherskiy\nProcessing Record 12 of Set 2 | hobart\nProcessing Record 13 of Set 2 | sept-iles\nProcessing Record 14 of Set 2 | penapolis\nProcessing Record 15 of Set 2 | carutapera\nProcessing Record 16 of Set 2 | tecoanapa\nProcessing Record 17 of Set 2 | deer lake\nProcessing Record 18 of Set 2 | itarema\nProcessing Record 19 of Set 2 | bambous virieux\nProcessing Record 20 of Set 2 | sasykoli\nProcessing Record 21 of Set 2 | hilo\nCity not found. Skipping...\nProcessing Record 22 of Set 2 | qaanaaq\nProcessing Record 23 of Set 2 | yellowknife\nProcessing Record 24 of Set 2 | cacapava do sul\nProcessing Record 25 of Set 2 | fukue\nProcessing Record 26 of Set 2 | andra\nProcessing Record 27 of Set 2 | bengkulu\nProcessing Record 28 of Set 2 | yangshe\nProcessing Record 29 of Set 2 | gondanglegi\nProcessing Record 30 of Set 2 | les cayes\nProcessing Record 31 of Set 2 | bluff\nProcessing Record 32 of Set 2 | katherine\nProcessing Record 33 of Set 2 | yuzhno-kurilsk\nProcessing Record 34 of Set 2 | luderitz\nProcessing Record 35 of Set 2 | yokadouma\nProcessing Record 36 of Set 2 | khatanga\nCity not found. Skipping...\nProcessing Record 37 of Set 2 | atuona\nProcessing Record 38 of Set 2 | bethel\nProcessing Record 39 of Set 2 | ilulissat\nProcessing Record 40 of Set 2 | saint-pierre\nProcessing Record 41 of Set 2 | sayyan\nProcessing Record 42 of Set 2 | moose factory\nProcessing Record 43 of Set 2 | geraldton\nProcessing Record 44 of Set 2 | puerto ayora\nCity not found. Skipping...\nProcessing Record 45 of Set 2 | tuktoyaktuk\nProcessing Record 46 of Set 2 | chapleau\nCity not found. Skipping...\nProcessing Record 47 of Set 2 | munirabad\nProcessing Record 48 of Set 2 | tevriz\nProcessing Record 49 of Set 2 | saskylakh\nProcessing Record 1 of Set 3 | jamestown\nProcessing Record 2 of Set 3 | camana\nProcessing Record 3 of Set 3 | srikakulam\nProcessing Record 4 of Set 3 | lalawigan\nProcessing Record 5 of Set 3 | cape town\nProcessing Record 6 of Set 3 | hami\nCity not found. Skipping...\nProcessing Record 7 of Set 3 | montoro\nProcessing Record 8 of Set 3 | sistranda\nProcessing Record 9 of Set 3 | georgetown\nProcessing Record 10 of Set 3 | muisne\nProcessing Record 11 of Set 3 | burnie\nProcessing Record 12 of Set 3 | hambantota\nCity not found. Skipping...\nProcessing Record 13 of Set 3 | port elizabeth\nProcessing Record 14 of Set 3 | apeldoorn\nProcessing Record 15 of Set 3 | saint-augustin\nProcessing Record 16 of Set 3 | kohlu\nProcessing Record 17 of Set 3 | nabire\nProcessing Record 18 of Set 3 | bonavista\nCity not found. Skipping...\nProcessing Record 19 of Set 3 | aranos\nProcessing Record 20 of Set 3 | simao\nProcessing Record 21 of Set 3 | tautira\nProcessing Record 22 of Set 3 | umkomaas\nProcessing Record 23 of Set 3 | torbay\nProcessing Record 24 of Set 3 | tingi\nCity not found. Skipping...\nProcessing Record 25 of Set 3 | hithadhoo\nProcessing Record 26 of Set 3 | nikolskoye\nProcessing Record 27 of Set 3 | pangnirtung\nProcessing Record 28 of Set 3 | cabo san lucas\nCity not found. Skipping...\nProcessing Record 29 of Set 3 | vanimo\nProcessing Record 30 of Set 3 | zhucheng\nProcessing Record 31 of Set 3 | burgeo\nProcessing Record 32 of Set 3 | grand gaube\nProcessing Record 33 of Set 3 | ukiah\nCity not found. Skipping...\nProcessing Record 34 of Set 3 | sitka\nProcessing Record 35 of Set 3 | pacasmayo\nProcessing Record 36 of Set 3 | timizart\nProcessing Record 37 of Set 3 | guanambi\nProcessing Record 38 of Set 3 | matagami\nProcessing Record 39 of Set 3 | pochutla\nProcessing Record 40 of Set 3 | karratha\nProcessing Record 41 of Set 3 | marquette\nProcessing Record 42 of Set 3 | chuy\nProcessing Record 43 of Set 3 | kokstad\nProcessing Record 44 of Set 3 | banda aceh\nProcessing Record 45 of Set 3 | dingle\nProcessing Record 46 of Set 3 | leh\nProcessing Record 47 of Set 3 | hualmay\nProcessing Record 48 of Set 3 | new norfolk\nProcessing Record 49 of Set 3 | avarua\nProcessing Record 1 of Set 4 | padang\nProcessing Record 2 of Set 4 | bari\nProcessing Record 3 of Set 4 | turukhansk\nCity not found. Skipping...\nCity not found. Skipping...\nProcessing Record 4 of Set 4 | kruisfontein\nProcessing Record 5 of Set 4 | fernie\nProcessing Record 6 of Set 4 | enshi\nCity not found. Skipping...\nProcessing Record 7 of Set 4 | salalah\nProcessing Record 8 of Set 4 | rodas\nProcessing Record 9 of Set 4 | shatura\nProcessing Record 10 of Set 4 | kampot\nCity not found. Skipping...\nProcessing Record 11 of Set 4 | luxor\nProcessing Record 12 of Set 4 | moerai\nProcessing Record 13 of Set 4 | srednekolymsk\nProcessing Record 14 of Set 4 | novita\nCity not found. Skipping...\nProcessing Record 15 of Set 4 | bikin\nProcessing Record 16 of Set 4 | norton shores\nProcessing Record 17 of Set 4 | warmbad\nProcessing Record 18 of Set 4 | kazachinskoye\nProcessing Record 19 of Set 4 | namibe\nProcessing Record 20 of Set 4 | yanji\nProcessing Record 21 of Set 4 | poplar bluff\nCity not found. Skipping...\nProcessing Record 22 of Set 4 | miranda\nCity not found. Skipping...\nProcessing Record 23 of Set 4 | byron bay\nProcessing Record 24 of Set 4 | ngunguru\nProcessing Record 25 of Set 4 | chongwe\nProcessing Record 26 of Set 4 | shaoxing\nProcessing Record 27 of Set 4 | maceio\nProcessing Record 28 of Set 4 | korla\nProcessing Record 29 of Set 4 | jakar\nProcessing Record 30 of Set 4 | mineiros\nProcessing Record 31 of Set 4 | chapais\nProcessing Record 32 of Set 4 | soyo\nProcessing Record 33 of Set 4 | sarkand\nProcessing Record 34 of Set 4 | stonewall\nProcessing Record 35 of Set 4 | nicoya\nProcessing Record 36 of Set 4 | barrow\nProcessing Record 37 of Set 4 | mwense\nProcessing Record 38 of Set 4 | bati\n"
]
],
[
[
"### Convert Raw Data to DataFrame\n* Export the city data into a .csv.\n* Display the DataFrame",
"_____no_output_____"
]
],
[
[
"\n# create a dictionary for establishing dataframe\nweather_dict = {\n \"City\":city_two,\n \"Cloudiness\":cloudinesses,\n \"Country\":countries,\n \"Date\":dates,\n \"Humidity\":humidities,\n \"Lat\":lats,\n \"Lng\":lngs,\n \"Max Temp\":max_temps,\n \"Wind Speed\":wind_speeds\n}\n\nweather_dict",
"_____no_output_____"
],
[
"# establish dataframe\nweather_dataframe = pd.DataFrame(weather_dict)\nweather_dataframe.to_csv (output_data_file, index = False)\n",
"_____no_output_____"
],
[
"# show the top of the dataframe\nweather_dataframe.head()",
"_____no_output_____"
],
[
"weather_dataframe.count()",
"_____no_output_____"
],
[
"weather_dataframe.head()",
"_____no_output_____"
]
],
[
[
"### Plotting the Data\n* Use proper labeling of the plots using plot titles (including date of analysis) and axes labels.\n* Save the plotted figures as .pngs.",
"_____no_output_____"
],
[
"#### Latitude vs. Temperature Plot",
"_____no_output_____"
]
],
[
[
"time.strftime('%x')",
"_____no_output_____"
],
[
"\nplt.scatter(weather_dataframe[\"Lat\"],weather_dataframe[\"Max Temp\"],edgecolors=\"black\",facecolors=\"skyblue\")\nplt.title(f\"City Latitude vs. Max Temperature {time.strftime('%x')}\")\nplt.xlabel(\"Latitude\")\nplt.ylabel(\"Max Temperature (F)\")\nplt.grid (b=True,which=\"major\",axis=\"both\",linestyle=\"-\",color=\"lightgrey\")\nplt.savefig(\"Figures/fig1.png\")\nplt.show()",
"_____no_output_____"
],
[
"#This graph analyzes latitude vs tempature",
"_____no_output_____"
]
],
[
[
"#### Latitude vs. Humidity Plot",
"_____no_output_____"
]
],
[
[
"\nplt.scatter(weather_dataframe[\"Lat\"],weather_dataframe[\"Humidity\"],edgecolors=\"black\",facecolors=\"skyblue\")\nplt.title(\"City Latitude vs. Humidity (%s)\" % time.strftime('%x') )\nplt.xlabel(\"Latitude\")\nplt.ylabel(\"Humidity (%)\")\nplt.ylim(15,105)\nplt.grid (b=True,which=\"major\",axis=\"both\",linestyle=\"-\",color=\"lightgrey\")\nplt.savefig(\"Figures/fig2.png\")\nplt.show()",
"_____no_output_____"
],
[
"#Analyzing the Humidity vs Latitude",
"_____no_output_____"
]
],
[
[
"#### Latitude vs. Cloudiness Plot",
"_____no_output_____"
]
],
[
[
"plt.scatter(weather_dataframe[\"Lat\"],weather_dataframe[\"Cloudiness\"],edgecolors=\"black\",facecolors=\"skyblue\")\nplt.title(\"City Latitude vs. Cloudiness (%s)\" % time.strftime('%x') ) \nplt.xlabel(\"Latitude\")\nplt.ylabel(\"Cloudiness (%)\")\nplt.grid (b=True,which=\"major\",axis=\"both\",linestyle=\"-\",color=\"lightgrey\")\nplt.savefig(\"Figures/fig3.png\")\nplt.show()",
"_____no_output_____"
],
[
"#Analyzes the latitude and cloudiness of the cities",
"_____no_output_____"
]
],
[
[
"#### Latitude vs. Wind Speed Plot",
"_____no_output_____"
]
],
[
[
"plt.scatter(weather_dataframe[\"Lat\"],weather_dataframe[\"Wind Speed\"],edgecolors=\"black\",facecolors=\"skyblue\")\nplt.title(\"City Latitude vs. Wind Speed (%s)\" % time.strftime('%x') )\nplt.xlabel(\"Latitude\")\nplt.ylabel(\"Wind Speed (mph)\")\nplt.ylim(-2,34)\nplt.grid (b=True,which=\"major\",axis=\"both\",linestyle=\"-\",color=\"lightgrey\")\nplt.savefig(\"Figures/fig4.png\")\nplt.show()",
"_____no_output_____"
],
[
"#This the relationship between latitude and windspeed",
"_____no_output_____"
]
],
[
[
"## Linear Regression",
"_____no_output_____"
]
],
[
[
"#Define x and y values\nx_values = weather_dataframe['Lat']\ny_values = weather_dataframe['Max Temp']\n\n\n# Perform a linear regression on temperature vs. latitude\n(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)\n\n# Get regression values\nregress_values = x_values * slope + intercept\nprint(regress_values)",
"0 82.274454\n1 59.296487\n2 82.761566\n3 63.261880\n4 18.904279\n ... \n546 31.592014\n547 23.805840\n548 61.419989\n549 20.160114\n550 70.218442\nName: Lat, Length: 551, dtype: float64\n"
],
[
"# Create line equation string\nline_eq = \"y = \" + str(round(slope,2)) + \"x +\" + str(round(intercept,2))\nprint(line_eq)",
"y = -0.76x +66.14\n"
],
[
"# Create Plot\nplt.scatter(x_values,y_values)\nplt.plot(x_values,regress_values,\"r-\")\n\n# Label plot and annotate the line equation\nplt.xlabel('Latitude')\nplt.ylabel('Temperature')\nplt.annotate(line_eq,(-40,0),fontsize=15,color=\"red\")\n\n# Print r square value\nprint(f\"The r-squared is: {rvalue}\")\n\n# Show plot\nplt.show()",
"The r-squared is: -0.7946821335281774\n"
],
[
"# Create Northern and Southern Hemisphere DataFrames\nsouth_df = weather_dataframe[weather_dataframe[\"Lat\"] < 0] \nnorth_df = weather_dataframe[weather_dataframe[\"Lat\"] >= 0]",
"_____no_output_____"
]
],
[
[
"#### Northern Hemisphere - Max Temp vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"def linear_agression(x,y):\n print(f'The r-squared is : {round(linregress(x, y)[0],2)}')\n (slope, intercept, rvalue, pvalue, stderr) = linregress(x, y)\n regress_values = x * slope + intercept\n line_eq = 'y = ' + str(round(slope,2)) + 'x + ' + str(round(intercept,2))\n plt.scatter(x, y)\n plt.plot(x,regress_values,'r-')\n return line_eq\n\n# Define a fuction for annotating\ndef annotate(line_eq, a, b):\n plt.annotate(line_eq,(a,b),fontsize=15,color='red')\n\n\n\n# Call an function #1\nequation = linear_agression(north_df['Lat'], north_df['Max Temp'])\n# Call an function #2\nannotate(equation, 0, 0)\n# Set a title\nplt.title('Northern Hemisphere - Max Temp vs. Latitude Linear Regression')\n# Set xlabel\nplt.xlabel('Latitude')\n# Set ylabel\nplt.ylabel('Max Temp')\n# Save the figure\nplt.savefig('Northern Hemisphere - Max Temp vs. Latitude Linear Regression.png')",
"The r-squared is : -1.28\n"
],
[
"# This graph shows the relationship in the Nouthern Hemisphere between Temp vs. latitude",
"_____no_output_____"
]
],
[
[
"#### Southern Hemisphere - Max Temp vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"def linear_agression(x,y):\n print(f'The r-squared is : {round(linregress(x, y)[0],2)}')\n (slope, intercept, rvalue, pvalue, stderr) = linregress(x, y)\n regress_values = x * slope + intercept\n line_eq = 'y = ' + str(round(slope,2)) + 'x + ' + str(round(intercept,2))\n plt.scatter(x, y)\n plt.plot(x,regress_values,'r-')\n return line_eq\n\n# Define a fuction for annotating\ndef annotate(line_eq, a, b):\n plt.annotate(line_eq,(a,b),fontsize=15,color='red')\n\n\n\n# Call an function #1\nequation = linear_agression(south_df['Lat'], south_df['Max Temp'])\n# Call an function #2\nannotate(equation, -30, 50)\n# Set a title\nplt.title('Southern Hemisphere - Max Temp vs. Latitude Linear Regression')\n# Set xlabel\nplt.xlabel('Latitude')\n# Set ylabel\nplt.ylabel('Max Temp')\n# Save the figure\nplt.savefig('Southern Hemisphere - Max Temp vs. Latitude Linear Regression.png')",
"The r-squared is : 0.27\n"
],
[
"# This graph shows the relationship in the southern Hemisphere between Temp vs. latitude",
"_____no_output_____"
]
],
[
[
"#### Northern Hemisphere - Humidity (%) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"def linear_agression(x,y):\n print(f'The r-squared is : {round(linregress(x, y)[0],2)}')\n (slope, intercept, rvalue, pvalue, stderr) = linregress(x, y)\n regress_values = x * slope + intercept\n line_eq = 'y = ' + str(round(slope,2)) + 'x + ' + str(round(intercept,2))\n plt.scatter(x, y)\n plt.plot(x,regress_values,'r-')\n return line_eq\n\n# Define a fuction for annotating\ndef annotate(line_eq, a, b):\n plt.annotate(line_eq,(a,b),fontsize=15,color='red')\n\n\n\n# Call an function #1\nequation = linear_agression(north_df['Lat'], north_df['Humidity'])\n# Call an function #2\nannotate(equation, 40, 20)\n# Set a title\nplt.title('Northern Hemisphere - Humidity vs. Latitude Linear Regression')\n# Set xlabel\nplt.xlabel('Latitude')\n# Set ylabel\nplt.ylabel('Humidity')\n# Save the figure\nplt.savefig('Northern Hemisphere - Humidity vs. Latitude Linear Regression.png')",
"The r-squared is : 0.41\n"
]
],
[
[
"#### Southern Hemisphere - Humidity (%) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"def linear_agression(x,y):\n print(f'The r-squared is : {round(linregress(x, y)[0],2)}')\n (slope, intercept, rvalue, pvalue, stderr) = linregress(x, y)\n regress_values = x * slope + intercept\n line_eq = 'y = ' + str(round(slope,2)) + 'x + ' + str(round(intercept,2))\n plt.scatter(x, y)\n plt.plot(x,regress_values,'r-')\n return line_eq\n\n# Define a fuction for annotating\ndef annotate(line_eq, a, b):\n plt.annotate(line_eq,(a,b),fontsize=15,color='red')\n\n\n\n# Call an function #1\nequation = linear_agression(south_df['Lat'], south_df['Humidity'])\n# Call an function #2\nannotate(equation,-30, 20)\n# Set a title\nplt.title('Southern Hemisphere - Humidity vs. Latitude Linear Regression')\n# Set xlabel\nplt.xlabel('Latitude')\n# Set ylabel\nplt.ylabel('Humidity')\n# Save the figure\nplt.savefig('Southern Hemisphere - Humidity vs. Latitude Linear Regression.png')",
"The r-squared is : 0.45\n"
],
[
"# This graph shows the relationship in the Southern Hemisphere between humidity vs. lattitude",
"_____no_output_____"
]
],
[
[
"#### Northern Hemisphere - Cloudiness (%) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"def linear_agression(x,y):\n print(f'The r-squared is : {round(linregress(x, y)[0],2)}')\n (slope, intercept, rvalue, pvalue, stderr) = linregress(x, y)\n regress_values = x * slope + intercept\n line_eq = 'y = ' + str(round(slope,2)) + 'x + ' + str(round(intercept,2))\n plt.scatter(x, y)\n plt.plot(x,regress_values,'r-')\n return line_eq\n\n# Define a fuction for annotating\ndef annotate(line_eq, a, b):\n plt.annotate(line_eq,(a,b),fontsize=15,color='red')\n\n\n\n# Call an function #1\nequation = linear_agression(north_df['Lat'], north_df['Humidity'])\n# Call an function #2\nannotate(equation, 40, 20)\n# Set a title\nplt.title('Northern Hemisphere - Humidity vs. Latitude Linear Regression')\n# Set xlabel\nplt.xlabel('Latitude')\n# Set ylabel\nplt.ylabel('Humidity')\n# Save the figure\nplt.savefig('Northern Hemisphere - Humidity vs. Latitude Linear Regression.png')",
"The r-squared is : 0.41\n"
],
[
"# This graph shows the relationship in the Northern Hemisphere between humidity vs. lattitude",
"_____no_output_____"
]
],
[
[
"#### Southern Hemisphere - Cloudiness (%) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"def linear_agression(x,y):\n print(f'The r-squared is : {round(linregress(x, y)[0],2)}')\n (slope, intercept, rvalue, pvalue, stderr) = linregress(x, y)\n regress_values = x * slope + intercept\n line_eq = 'y = ' + str(round(slope,2)) + 'x + ' + str(round(intercept,2))\n plt.scatter(x, y)\n plt.plot(x,regress_values,'r-')\n return line_eq\n\n# Define a fuction for annotating\ndef annotate(line_eq, a, b):\n plt.annotate(line_eq,(a,b),fontsize=15,color='red')\n\n\n\n# Call an function #1\nequation = linear_agression(south_df['Lat'], south_df['Cloudiness'])\n# Call an function #2\nannotate(equation,-50, 10)\n# Set a title\nplt.title('Southern Hemisphere - Cloudiness vs. Latitude Linear Regression')\n# Set xlabel\nplt.xlabel('Latitude')\n# Set ylabel\nplt.ylabel('Cloudiness')\n# Save the figure\nplt.savefig('Southern Hemisphere - Cloudiness vs. Lattude Linear Regression.png')",
"The r-squared is : 1.14\n"
],
[
"#This grpah shows the relationship in the southern hempisphere with cloudiness vs lattitude",
"_____no_output_____"
]
],
[
[
"#### Northern Hemisphere - Wind Speed (mph) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"def linear_agression(x,y):\n print(f'The r-squared is : {round(linregress(x, y)[0],2)}')\n (slope, intercept, rvalue, pvalue, stderr) = linregress(x, y)\n regress_values = x * slope + intercept\n line_eq = 'y = ' + str(round(slope,2)) + 'x + ' + str(round(intercept,2))\n plt.scatter(x, y)\n plt.plot(x,regress_values,'r-')\n return line_eq\n\n# Define a fuction for annotating\ndef annotate(line_eq, a, b):\n plt.annotate(line_eq,(a,b),fontsize=15,color='red')\n\n\n\n# Call an function #1\nequation = linear_agression(north_df['Lat'], north_df['Wind Speed'])\n# Call an function #2\nannotate(equation,10, 35)\n# Set a title\nplt.title('Northern Hemisphere - Wind Speed vs. Latitude Linear Regression')\n# Set xlabel\nplt.xlabel('Latitude')\n# Set ylabel\nplt.ylabel('Wind Speed')\n# Save the figure\nplt.savefig('Northern Hemisphere - Wind Speed vs. Latitude Linear Regression.png')",
"The r-squared is : 0.03\n"
],
[
"#The northern hemisphere is wind speed vs lattitude",
"_____no_output_____"
]
],
[
[
"#### Southern Hemisphere - Wind Speed (mph) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"def linear_agression(x,y):\n print(f'The r-squared is : {round(linregress(x, y)[0],2)}')\n (slope, intercept, rvalue, pvalue, stderr) = linregress(x, y)\n regress_values = x * slope + intercept\n line_eq = 'y = ' + str(round(slope,2)) + 'x + ' + str(round(intercept,2))\n plt.scatter(x, y)\n plt.plot(x,regress_values,'r-')\n return line_eq\n\n# Define a fuction for annotating\ndef annotate(line_eq, a, b):\n plt.annotate(line_eq,(a,b),fontsize=15,color='red')\n\n\n\n# Call an function #1\nequation = linear_agression(south_df['Lat'], south_df['Wind Speed'])\n# Call an function #2\nannotate(equation, -50, 25)\n# Set a title\nplt.title('Southern Hemisphere - Wind Speed vs. Latitude Linear Regression')\n# Set xlabel\nplt.xlabel('Latitude')\n# Set ylabel\nplt.ylabel('Wind Speed')\n# Save the figure\nplt.savefig('Southern Hemisphere - Wind Speed vs. Latitude Linear Regression.png')",
"The r-squared is : -0.08\n"
],
[
"# This graph compares the southern hemisphere the wind speed vs lattitude.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
e79124ec565d02ac7a4039c3534a85c46731b80f | 613 | ipynb | Jupyter Notebook | climate_app.ipynb | KSandez/Data_Storage_Retrieval | 6d7e0570e97d40d0f13519987967ae3d7b09bb06 | [
"MIT"
] | null | null | null | climate_app.ipynb | KSandez/Data_Storage_Retrieval | 6d7e0570e97d40d0f13519987967ae3d7b09bb06 | [
"MIT"
] | null | null | null | climate_app.ipynb | KSandez/Data_Storage_Retrieval | 6d7e0570e97d40d0f13519987967ae3d7b09bb06 | [
"MIT"
] | null | null | null | 17.514286 | 62 | 0.525285 | [
[
[
"# Climate App\n\n* See the [app.py](app.py) file for climate api scripts.",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown"
]
] |
e79125284acc5c6e4c5a571d976cc4b52eaa9b11 | 16,989 | ipynb | Jupyter Notebook | phase02/6.1.deployment-pipeline.ipynb | gonsoomoon-ml/SageMaker-Pipelines-Step-By-Step | 81a7f2ba9b2eb62558644df10298545ff1771f27 | [
"Apache-2.0"
] | 13 | 2021-08-05T01:56:47.000Z | 2022-02-20T09:31:43.000Z | phase02/6.1.deployment-pipeline.ipynb | gonsoomoon-ml/SageMaker-Pipelines-Step-By-Step | 81a7f2ba9b2eb62558644df10298545ff1771f27 | [
"Apache-2.0"
] | null | null | null | phase02/6.1.deployment-pipeline.ipynb | gonsoomoon-ml/SageMaker-Pipelines-Step-By-Step | 81a7f2ba9b2eb62558644df10298545ff1771f27 | [
"Apache-2.0"
] | 11 | 2021-08-05T02:11:41.000Z | 2022-01-14T06:17:48.000Z | 28.942078 | 306 | 0.563894 | [
[
[
"# [모듈 6.1] 모델 배포 파이프라인 개발 (SageMaker Model Building Pipeline 모든 스텝)\n\n이 노트북은 아래와 같은 목차로 진행 됩니다. 전체를 모두 실행시에 완료 시간은 **약 5분** 소요 됩니다.\n\n- 0. SageMaker Model Building Pipeline 개요\n- 1. 파이프라인 변수 및 환경 설정\n- 2. 파이프라인 스텝 단계 정의\n\n - (1) 모델 승인 상태 변경 람다 스텝 \n - (2) 배포할 세이지 메이커 모델 스텝 생성\n - (3) 모델 앤드 포인트 배포를 위한 람다 스텝 생성 \n \n- 3. 모델 빌딩 파이프라인 정의 및 실행\n- 4. Pipleline 캐싱 및 파라미터 이용한 실행\n- 5. 정리 작업\n \n---",
"_____no_output_____"
],
[
"# 0.[모듈 6.1] 모델 배포 파이프라인 개요\n\n- 이 노트북은 다음과 같은 상황에 대한 파이프라인 입니다.\n - 모델 레제스트리에 여러개의 모델 패키지 그룹이 있습니다.\n - 모델 패키지 그룹에서 특정한 것을 선택하여 가장 최근에 저장된 모델 버전을 선택 합니다.\n - 선택된 모델 버전의 \"모델 승인 상태\"를 \"Pending\" 에서 \"Approved\" 로 변경 합니다.\n - 이 모델 버전에 대해서 세이지 메이커 모델을 생성합니다.\n - 세이지 메이커 모델을 기반으로 앤드포인트를 생성 합니다.",
"_____no_output_____"
],
[
"# 1. 파이프라인 변수 및 환경 설정\n\n",
"_____no_output_____"
]
],
[
[
"import boto3\nimport sagemaker\nimport pandas as pd\n\nregion = boto3.Session().region_name\nsagemaker_session = sagemaker.session.Session()\nrole = sagemaker.get_execution_role()\n\nsm_client = boto3.client('sagemaker', region_name=region)\n\n%store -r ",
"_____no_output_____"
]
],
[
[
"## (1) 모델 빌딩 파이프라인 변수 생성\n\n파이프라인에 인자로 넘길 변수는 아래 크게 3가지 종류가 있습니다.\n- 모델 레지스트리에 모델 등록시에 모델 승인 상태 값 \n",
"_____no_output_____"
]
],
[
[
"from sagemaker.workflow.parameters import (\n ParameterInteger,\n ParameterString,\n ParameterFloat,\n)\n\n\nmodel_approval_status = ParameterString(\n name=\"ModelApprovalStatus\", default_value=\"PendingManualApproval\"\n)\n",
"_____no_output_____"
]
],
[
[
"# 2. 파이프라인 스텝 단계 정의",
"_____no_output_____"
],
[
"## (1) 모델 승인 상태 변경 람다 스텝\n- 모델 레지스트리에서 해당 모델 패키지 그룹을 조회하고, 가장 최신 버전의 모델에 대해서 '모델 승인 상태 변경' 을 합니다.\n\n\n#### [에러] \n아래와 같은 데러가 발생시에 `0.0.Setup-Environment.ipynb` 의 정책 추가 부분을 진행 해주세요.\n```\nClientError: An error occurred (AccessDenied) when calling the CreateRole operation: User: arn:aws:sts::0287032915XX:assumed-role/AmazonSageMaker-ExecutionRole-20210827T141955/SageMaker is not authorized to perform: iam:CreateRole on resource: arn:aws:iam::0287032915XX:role/lambda-deployment-role\n```",
"_____no_output_____"
]
],
[
[
"from src.iam_helper import create_lambda_role\n\nlambda_role = create_lambda_role(\"lambda-deployment-role\")\nprint(\"lambda_role: \\n\", lambda_role)",
"Using ARN from existing role: lambda-deployment-role\nlambda_role: \n arn:aws:iam::028703291518:role/lambda-deployment-role\n"
],
[
"from sagemaker.lambda_helper import Lambda\nfrom sagemaker.workflow.lambda_step import (\n LambdaStep,\n LambdaOutput,\n LambdaOutputTypeEnum,\n)\n\nimport time \n\ncurrent_time = time.strftime(\"%m-%d-%H-%M-%S\", time.localtime())\nfunction_name = \"sagemaker-lambda-step-approve-model-deployment-\" + current_time\n\nprint(\"function_name: \\n\", function_name)",
"function_name: \n sagemaker-lambda-step-approve-model-deployment-08-27-12-08-42\n"
],
[
"# Lambda helper class can be used to create the Lambda function\nfunc_approve_model = Lambda(\n function_name=function_name,\n execution_role_arn=lambda_role,\n script=\"src/iam_change_model_approval.py\",\n handler=\"iam_change_model_approval.lambda_handler\",\n)\n\noutput_param_1 = LambdaOutput(output_name=\"statusCode\", output_type=LambdaOutputTypeEnum.String)\noutput_param_2 = LambdaOutput(output_name=\"body\", output_type=LambdaOutputTypeEnum.String)\noutput_param_3 = LambdaOutput(output_name=\"other_key\", output_type=LambdaOutputTypeEnum.String)\n\nstep_approve_lambda = LambdaStep(\n name=\"LambdaApproveModelStep\",\n lambda_func=func_approve_model,\n inputs={\n \"model_package_group_name\" : model_package_group_name,\n \"ModelApprovalStatus\": \"Approved\",\n },\n outputs=[output_param_1, output_param_2, output_param_3],\n)\n",
"_____no_output_____"
]
],
[
[
"## (2) 배포할 세이지 메이커 모델 스텝 생성\n- 위의 람다 스텝에서 \"모델 승인 상태\" 를 변경한 모델에 대하여 '모델 레지스트리'에서 저장된 도커 컨테이너 이미지, 모델 아티펙트의 위치를 가져 옵니다.\n- 이후에 이 두개의 인자를 가지고 세이지 메이커 모델을 생성 합니다.",
"_____no_output_____"
]
],
[
[
"import boto3\nsm_client = boto3.client('sagemaker')\n\n# 위에서 생성한 model_package_group_name 을 인자로 제공 합니다.\nresponse = sm_client.list_model_packages(ModelPackageGroupName= model_package_group_name)\n\nModelPackageArn = response['ModelPackageSummaryList'][0]['ModelPackageArn']\nsm_client.describe_model_package(ModelPackageName=ModelPackageArn)\nresponse = sm_client.describe_model_package(ModelPackageName=ModelPackageArn)\nimage_uri_approved = response[\"InferenceSpecification\"][\"Containers\"][0][\"Image\"]\nModelDataUrl_approved = response[\"InferenceSpecification\"][\"Containers\"][0][\"ModelDataUrl\"]\nprint(\"image_uri_approved: \", image_uri_approved)\nprint(\"ModelDataUrl_approved: \", ModelDataUrl_approved)",
"image_uri_approved: 683313688378.dkr.ecr.us-east-1.amazonaws.com/sagemaker-xgboost:1.0-1-cpu-py3\nModelDataUrl_approved: s3://sagemaker-us-east-1-028703291518/fraud2train/training_jobs/pipelines-ebt5t8rln4ye-FraudTrain-c21jj7fu99/output/model.tar.gz\n"
],
[
"from sagemaker.model import Model\n \nmodel = Model(\n image_uri= image_uri_approved,\n model_data= ModelDataUrl_approved, \n sagemaker_session=sagemaker_session,\n role=role,\n)",
"_____no_output_____"
],
[
"from sagemaker.inputs import CreateModelInput\nfrom sagemaker.workflow.steps import CreateModelStep\n\n\ninputs = CreateModelInput(\n instance_type=\"ml.m5.large\",\n # accelerator_type=\"ml.eia1.medium\",\n)\nstep_create_best_model = CreateModelStep(\n name=\"CreateFraudhModel\",\n model=model,\n inputs=inputs,\n)\nstep_create_best_model.add_depends_on([step_approve_lambda]) # step_approve_lambda 완료 후 실행 함.",
"_____no_output_____"
]
],
[
[
"## (3) 모델 앤드 포인트 배포를 위한 람다 스텝 생성\n- 람다 함수는 입력으로 세이지 메이커 모델, 앤드 포인트 컨피그 및 앤드 포인트 이름을 받아서, 앤드포인트를 생성 함.\n",
"_____no_output_____"
]
],
[
[
"# model_name = project_prefix + \"-lambda-model\" + current_time\nendpoint_config_name = \"lambda-deploy-endpoint-config-\" + current_time\nendpoint_name = \"lambda-deploy-endpoint-\" + current_time\n\nfunction_name = \"sagemaker-lambda-step-endpoint-deploy-\" + current_time\n\n# print(\"model_name: \\n\", model_name)\nprint(\"endpoint_config_name: \\n\", endpoint_config_name)\nprint(\"endpoint_config_name: \\n\", len(endpoint_config_name))\nprint(\"endpoint_name: \\n\", endpoint_name)\nprint(\"function_name: \\n\", function_name)\n\n\n",
"endpoint_config_name: \n lambda-deploy-endpoint-config-08-27-12-08-42\nendpoint_config_name: \n 44\nendpoint_name: \n lambda-deploy-endpoint-08-27-12-08-42\nfunction_name: \n sagemaker-lambda-step-endpoint-deploy-08-27-12-08-42\n"
],
[
"# Lambda helper class can be used to create the Lambda function\nfunc_deploy_model = Lambda(\n function_name=function_name,\n execution_role_arn=lambda_role,\n script=\"src/iam_create_endpoint.py\",\n handler=\"iam_create_endpoint.lambda_handler\",\n timeout = 900, # 디폴트는 120초 임. 10분으로 연장\n)\n\noutput_param_1 = LambdaOutput(output_name=\"statusCode\", output_type=LambdaOutputTypeEnum.String)\noutput_param_2 = LambdaOutput(output_name=\"body\", output_type=LambdaOutputTypeEnum.String)\noutput_param_3 = LambdaOutput(output_name=\"other_key\", output_type=LambdaOutputTypeEnum.String)\n\nstep_deploy_lambda = LambdaStep(\n name=\"LambdaDeployStep\",\n lambda_func=func_deploy_model,\n inputs={\n \"model_name\": step_create_best_model.properties.ModelName,\n \"endpoint_config_name\": endpoint_config_name,\n \"endpoint_name\": endpoint_name,\n },\n outputs=[output_param_1, output_param_2, output_param_3],\n)",
"_____no_output_____"
]
],
[
[
"# 3.모델 빌딩 파이프라인 정의 및 실행\n위에서 정의한 아래의 4개의 스텝으로 파이프라인 정의를 합니다.\n- steps=[step_process, step_train, step_create_model, step_deploy],\n- 아래는 약 1분 정도 소요 됩니다. 이후 아래와 같이 실행 결과를 스튜디오에서 확인할 수 있습니다.\n\n\n- ",
"_____no_output_____"
]
],
[
[
"from sagemaker.workflow.pipeline import Pipeline\n\nproject_prefix = 'sagemaker-pipeline-phase2-deployment-step-by-step'\n\npipeline_name = project_prefix\npipeline = Pipeline(\n name=pipeline_name,\n parameters=[\n model_approval_status, \n ],\n\n \n steps=[step_approve_lambda, step_create_best_model, step_deploy_lambda],\n)\n\n",
"_____no_output_____"
],
[
"import json\n\ndefinition = json.loads(pipeline.definition())\n# definition",
"_____no_output_____"
]
],
[
[
"### 파이프라인을 SageMaker에 제출하고 실행하기 \n",
"_____no_output_____"
]
],
[
[
"pipeline.upsert(role_arn=role)",
"_____no_output_____"
]
],
[
[
"디폴트값을 이용하여 파이프라인을 샐행합니다. ",
"_____no_output_____"
]
],
[
[
"execution = pipeline.start()",
"_____no_output_____"
]
],
[
[
"### 파이프라인 운영: 파이프라인 대기 및 실행상태 확인\n\n워크플로우의 실행상황을 살펴봅니다. ",
"_____no_output_____"
],
[
"실행이 완료될 때까지 기다립니다.",
"_____no_output_____"
]
],
[
[
"execution.wait()",
"_____no_output_____"
]
],
[
[
"실행된 단계들을 리스트업합니다. 파이프라인의 단계실행 서비스에 의해 시작되거나 완료된 단계를 보여줍니다.",
"_____no_output_____"
]
],
[
[
"execution.list_steps()",
"_____no_output_____"
]
],
[
[
"# 5. 정리 작업\n",
"_____no_output_____"
],
[
"## 변수 저장",
"_____no_output_____"
]
],
[
[
"depolyment_endpoint_name = endpoint_name\n%store depolyment_endpoint_name\n\nall_deployment_pipeline_name = pipeline_name\n%store all_deployment_pipeline_name",
"Stored 'depolyment_endpoint_name' (str)\nStored 'all_deployment_pipeline_name' (str)\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
e7912bcdfe2dc4c4eac4864c7eb0bd8e927a90eb | 207,219 | ipynb | Jupyter Notebook | 09_up_and_running_with_tensorflow.ipynb | JeffRisberg/SciKit_and_Data_Science | 6bea5534a94076ada765908fd2d00bbb849005bf | [
"Apache-2.0"
] | null | null | null | 09_up_and_running_with_tensorflow.ipynb | JeffRisberg/SciKit_and_Data_Science | 6bea5534a94076ada765908fd2d00bbb849005bf | [
"Apache-2.0"
] | null | null | null | 09_up_and_running_with_tensorflow.ipynb | JeffRisberg/SciKit_and_Data_Science | 6bea5534a94076ada765908fd2d00bbb849005bf | [
"Apache-2.0"
] | null | null | null | 54.14659 | 30,328 | 0.671304 | [
[
[
"**Chapter 9 – Up and running with TensorFlow**",
"_____no_output_____"
],
[
"_This notebook contains all the sample code and solutions to the exercises in chapter 9._",
"_____no_output_____"
],
[
"# Setup",
"_____no_output_____"
],
[
"First, let's make sure this notebook works well in both python 2 and 3, import a few common modules, ensure MatplotLib plots figures inline and prepare a function to save the figures:",
"_____no_output_____"
]
],
[
[
"# To support both python 2 and python 3\nfrom __future__ import division, print_function, unicode_literals\n\n# Common imports\nimport numpy as np\nimport os\n\n# to make this notebook's output stable across runs\ndef reset_graph(seed=42):\n tf.reset_default_graph()\n tf.set_random_seed(seed)\n np.random.seed(seed)\n\n# To plot pretty figures\n%matplotlib inline\nimport matplotlib\nimport matplotlib.pyplot as plt\nplt.rcParams['axes.labelsize'] = 14\nplt.rcParams['xtick.labelsize'] = 12\nplt.rcParams['ytick.labelsize'] = 12\n\n# Where to save the figures\nPROJECT_ROOT_DIR = \".\"\nCHAPTER_ID = \"tensorflow\"\n\ndef save_fig(fig_id, tight_layout=True):\n path = os.path.join(PROJECT_ROOT_DIR, \"images\", CHAPTER_ID, fig_id + \".png\")\n print(\"Saving figure\", fig_id)\n if tight_layout:\n plt.tight_layout()\n plt.savefig(path, format='png', dpi=300)",
"_____no_output_____"
]
],
[
[
"# Creating and running a graph",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\n\nreset_graph()\n\nx = tf.Variable(3, name=\"x\")\ny = tf.Variable(4, name=\"y\")\nf = x*x*y + y + 2",
"_____no_output_____"
],
[
"f",
"_____no_output_____"
],
[
"sess = tf.Session()\nsess.run(x.initializer)\nsess.run(y.initializer)\nresult = sess.run(f)\nprint(result)",
"42\n"
],
[
"sess.close()",
"_____no_output_____"
],
[
"with tf.Session() as sess:\n x.initializer.run()\n y.initializer.run()\n result = f.eval()",
"_____no_output_____"
],
[
"result",
"_____no_output_____"
],
[
"init = tf.global_variables_initializer()\n\nwith tf.Session() as sess:\n init.run()\n result = f.eval()",
"_____no_output_____"
],
[
"result",
"_____no_output_____"
],
[
"init = tf.global_variables_initializer()",
"_____no_output_____"
],
[
"sess = tf.InteractiveSession()\ninit.run()\nresult = f.eval()\nprint(result)",
"42\n"
],
[
"sess.close()",
"_____no_output_____"
],
[
"result",
"_____no_output_____"
]
],
[
[
"# Managing graphs",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nx1 = tf.Variable(1)\nx1.graph is tf.get_default_graph()",
"_____no_output_____"
],
[
"graph = tf.Graph()\nwith graph.as_default():\n x2 = tf.Variable(2)\n\nx2.graph is graph",
"_____no_output_____"
],
[
"x2.graph is tf.get_default_graph()",
"_____no_output_____"
],
[
"w = tf.constant(3)\nx = w + 2\ny = x + 5\nz = x * 3\n\nwith tf.Session() as sess:\n print(y.eval()) # 10\n print(z.eval()) # 15",
"10\n15\n"
],
[
"with tf.Session() as sess:\n y_val, z_val = sess.run([y, z])\n print(y_val) # 10\n print(z_val) # 15",
"10\n15\n"
]
],
[
[
"# Linear Regression",
"_____no_output_____"
],
[
"## Using the Normal Equation",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom sklearn.datasets import fetch_california_housing\n\nreset_graph()\n\nhousing = fetch_california_housing()\nm, n = housing.data.shape\nhousing_data_plus_bias = np.c_[np.ones((m, 1)), housing.data]\n\nX = tf.constant(housing_data_plus_bias, dtype=tf.float32, name=\"X\")\ny = tf.constant(housing.target.reshape(-1, 1), dtype=tf.float32, name=\"y\")\nXT = tf.transpose(X)\ntheta = tf.matmul(tf.matmul(tf.matrix_inverse(tf.matmul(XT, X)), XT), y)\n\nwith tf.Session() as sess:\n theta_value = theta.eval()",
"_____no_output_____"
],
[
"theta_value",
"_____no_output_____"
]
],
[
[
"Compare with pure NumPy",
"_____no_output_____"
]
],
[
[
"X = housing_data_plus_bias\ny = housing.target.reshape(-1, 1)\ntheta_numpy = np.linalg.inv(X.T.dot(X)).dot(X.T).dot(y)\n\nprint(theta_numpy)",
"[[ -3.69419202e+01]\n [ 4.36693293e-01]\n [ 9.43577803e-03]\n [ -1.07322041e-01]\n [ 6.45065694e-01]\n [ -3.97638942e-06]\n [ -3.78654266e-03]\n [ -4.21314378e-01]\n [ -4.34513755e-01]]\n"
]
],
[
[
"Compare with Scikit-Learn",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LinearRegression\nlin_reg = LinearRegression()\nlin_reg.fit(housing.data, housing.target.reshape(-1, 1))\n\nprint(np.r_[lin_reg.intercept_.reshape(-1, 1), lin_reg.coef_.T])",
"[[ -3.69419202e+01]\n [ 4.36693293e-01]\n [ 9.43577803e-03]\n [ -1.07322041e-01]\n [ 6.45065694e-01]\n [ -3.97638942e-06]\n [ -3.78654265e-03]\n [ -4.21314378e-01]\n [ -4.34513755e-01]]\n"
]
],
[
[
"## Using Batch Gradient Descent",
"_____no_output_____"
],
[
"Gradient Descent requires scaling the feature vectors first. We could do this using TF, but let's just use Scikit-Learn for now.",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import StandardScaler\nscaler = StandardScaler()\nscaled_housing_data = scaler.fit_transform(housing.data)\nscaled_housing_data_plus_bias = np.c_[np.ones((m, 1)), scaled_housing_data]",
"_____no_output_____"
],
[
"print(scaled_housing_data_plus_bias.mean(axis=0))\nprint(scaled_housing_data_plus_bias.mean(axis=1))\nprint(scaled_housing_data_plus_bias.mean())\nprint(scaled_housing_data_plus_bias.shape)",
"[ 1.00000000e+00 6.60969987e-17 5.50808322e-18 6.60969987e-17\n -1.06030602e-16 -1.10161664e-17 3.44255201e-18 -1.07958431e-15\n -8.52651283e-15]\n[ 0.38915536 0.36424355 0.5116157 ..., -0.06612179 -0.06360587\n 0.01359031]\n0.111111111111\n(20640, 9)\n"
]
],
[
[
"### Manually computing the gradients",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_epochs = 1000\nlearning_rate = 0.01\n\nX = tf.constant(scaled_housing_data_plus_bias, dtype=tf.float32, name=\"X\")\ny = tf.constant(housing.target.reshape(-1, 1), dtype=tf.float32, name=\"y\")\ntheta = tf.Variable(tf.random_uniform([n + 1, 1], -1.0, 1.0, seed=42), name=\"theta\")\ny_pred = tf.matmul(X, theta, name=\"predictions\")\nerror = y_pred - y\nmse = tf.reduce_mean(tf.square(error), name=\"mse\")\ngradients = 2/m * tf.matmul(tf.transpose(X), error)\ntraining_op = tf.assign(theta, theta - learning_rate * gradients)\n\ninit = tf.global_variables_initializer()\n\nwith tf.Session() as sess:\n sess.run(init)\n\n for epoch in range(n_epochs):\n if epoch % 100 == 0:\n print(\"Epoch\", epoch, \"MSE =\", mse.eval())\n sess.run(training_op)\n \n best_theta = theta.eval()",
"Epoch 0 MSE = 9.16154\nEpoch 100 MSE = 0.714501\nEpoch 200 MSE = 0.566705\nEpoch 300 MSE = 0.555572\nEpoch 400 MSE = 0.548812\nEpoch 500 MSE = 0.543636\nEpoch 600 MSE = 0.539629\nEpoch 700 MSE = 0.536509\nEpoch 800 MSE = 0.534068\nEpoch 900 MSE = 0.532147\n"
],
[
"best_theta",
"_____no_output_____"
]
],
[
[
"### Using autodiff",
"_____no_output_____"
],
[
"Same as above except for the `gradients = ...` line:",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_epochs = 1000\nlearning_rate = 0.01\n\nX = tf.constant(scaled_housing_data_plus_bias, dtype=tf.float32, name=\"X\")\ny = tf.constant(housing.target.reshape(-1, 1), dtype=tf.float32, name=\"y\")\ntheta = tf.Variable(tf.random_uniform([n + 1, 1], -1.0, 1.0, seed=42), name=\"theta\")\ny_pred = tf.matmul(X, theta, name=\"predictions\")\nerror = y_pred - y\nmse = tf.reduce_mean(tf.square(error), name=\"mse\")",
"_____no_output_____"
],
[
"gradients = tf.gradients(mse, [theta])[0]",
"_____no_output_____"
],
[
"training_op = tf.assign(theta, theta - learning_rate * gradients)\n\ninit = tf.global_variables_initializer()\n\nwith tf.Session() as sess:\n sess.run(init)\n\n for epoch in range(n_epochs):\n if epoch % 100 == 0:\n print(\"Epoch\", epoch, \"MSE =\", mse.eval())\n sess.run(training_op)\n \n best_theta = theta.eval()\n\nprint(\"Best theta:\")\nprint(best_theta)",
"Epoch 0 MSE = 9.16154\nEpoch 100 MSE = 0.714501\nEpoch 200 MSE = 0.566705\nEpoch 300 MSE = 0.555572\nEpoch 400 MSE = 0.548812\nEpoch 500 MSE = 0.543636\nEpoch 600 MSE = 0.539629\nEpoch 700 MSE = 0.536509\nEpoch 800 MSE = 0.534068\nEpoch 900 MSE = 0.532147\nBest theta:\n[[ 2.06855249]\n [ 0.88740271]\n [ 0.14401658]\n [-0.34770882]\n [ 0.36178368]\n [ 0.00393811]\n [-0.04269556]\n [-0.66145277]\n [-0.6375277 ]]\n"
]
],
[
[
"How could you find the partial derivatives of the following function with regards to `a` and `b`?",
"_____no_output_____"
]
],
[
[
"def my_func(a, b):\n z = 0\n for i in range(100):\n z = a * np.cos(z + i) + z * np.sin(b - i)\n return z",
"_____no_output_____"
],
[
"my_func(0.2, 0.3)",
"_____no_output_____"
],
[
"reset_graph()\n\na = tf.Variable(0.2, name=\"a\")\nb = tf.Variable(0.3, name=\"b\")\nz = tf.constant(0.0, name=\"z0\")\nfor i in range(100):\n z = a * tf.cos(z + i) + z * tf.sin(b - i)\n\ngrads = tf.gradients(z, [a, b])\ninit = tf.global_variables_initializer()",
"_____no_output_____"
]
],
[
[
"Let's compute the function at $a=0.2$ and $b=0.3$, and the partial derivatives at that point with regards to $a$ and with regards to $b$:",
"_____no_output_____"
]
],
[
[
"with tf.Session() as sess:\n init.run()\n print(z.eval())\n print(sess.run(grads))",
"-0.212537\n[-1.1388494, 0.19671395]\n"
]
],
[
[
"### Using a `GradientDescentOptimizer`",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_epochs = 1000\nlearning_rate = 0.01\n\nX = tf.constant(scaled_housing_data_plus_bias, dtype=tf.float32, name=\"X\")\ny = tf.constant(housing.target.reshape(-1, 1), dtype=tf.float32, name=\"y\")\ntheta = tf.Variable(tf.random_uniform([n + 1, 1], -1.0, 1.0, seed=42), name=\"theta\")\ny_pred = tf.matmul(X, theta, name=\"predictions\")\nerror = y_pred - y\nmse = tf.reduce_mean(tf.square(error), name=\"mse\")",
"_____no_output_____"
],
[
"optimizer = tf.train.GradientDescentOptimizer(learning_rate=learning_rate)\ntraining_op = optimizer.minimize(mse)",
"_____no_output_____"
],
[
"init = tf.global_variables_initializer()\n\nwith tf.Session() as sess:\n sess.run(init)\n\n for epoch in range(n_epochs):\n if epoch % 100 == 0:\n print(\"Epoch\", epoch, \"MSE =\", mse.eval())\n sess.run(training_op)\n \n best_theta = theta.eval()\n\nprint(\"Best theta:\")\nprint(best_theta)",
"Epoch 0 MSE = 9.16154\nEpoch 100 MSE = 0.714501\nEpoch 200 MSE = 0.566705\nEpoch 300 MSE = 0.555572\nEpoch 400 MSE = 0.548812\nEpoch 500 MSE = 0.543636\nEpoch 600 MSE = 0.539629\nEpoch 700 MSE = 0.536509\nEpoch 800 MSE = 0.534068\nEpoch 900 MSE = 0.532147\nBest theta:\n[[ 2.06855249]\n [ 0.88740271]\n [ 0.14401658]\n [-0.34770882]\n [ 0.36178368]\n [ 0.00393811]\n [-0.04269556]\n [-0.66145277]\n [-0.6375277 ]]\n"
]
],
[
[
"### Using a momentum optimizer",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_epochs = 1000\nlearning_rate = 0.01\n\nX = tf.constant(scaled_housing_data_plus_bias, dtype=tf.float32, name=\"X\")\ny = tf.constant(housing.target.reshape(-1, 1), dtype=tf.float32, name=\"y\")\ntheta = tf.Variable(tf.random_uniform([n + 1, 1], -1.0, 1.0, seed=42), name=\"theta\")\ny_pred = tf.matmul(X, theta, name=\"predictions\")\nerror = y_pred - y\nmse = tf.reduce_mean(tf.square(error), name=\"mse\")",
"_____no_output_____"
],
[
"optimizer = tf.train.MomentumOptimizer(learning_rate=learning_rate,\n momentum=0.9)",
"_____no_output_____"
],
[
"training_op = optimizer.minimize(mse)\n\ninit = tf.global_variables_initializer()",
"_____no_output_____"
],
[
"with tf.Session() as sess:\n sess.run(init)\n\n for epoch in range(n_epochs):\n sess.run(training_op)\n \n best_theta = theta.eval()\n\nprint(\"Best theta:\")\nprint(best_theta)",
"Best theta:\n[[ 2.06855798]\n [ 0.82962859]\n [ 0.11875337]\n [-0.26554456]\n [ 0.30571091]\n [-0.00450251]\n [-0.03932662]\n [-0.89986444]\n [-0.87052065]]\n"
]
],
[
[
"# Feeding data to the training algorithm",
"_____no_output_____"
],
[
"## Placeholder nodes",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nA = tf.placeholder(tf.float32, shape=(None, 3))\nB = A + 5\nwith tf.Session() as sess:\n B_val_1 = B.eval(feed_dict={A: [[1, 2, 3]]})\n B_val_2 = B.eval(feed_dict={A: [[4, 5, 6], [7, 8, 9]]})\n\nprint(B_val_1)",
"[[ 6. 7. 8.]]\n"
],
[
"print(B_val_2)",
"[[ 9. 10. 11.]\n [ 12. 13. 14.]]\n"
]
],
[
[
"## Mini-batch Gradient Descent",
"_____no_output_____"
]
],
[
[
"n_epochs = 1000\nlearning_rate = 0.01",
"_____no_output_____"
],
[
"reset_graph()\n\nX = tf.placeholder(tf.float32, shape=(None, n + 1), name=\"X\")\ny = tf.placeholder(tf.float32, shape=(None, 1), name=\"y\")",
"_____no_output_____"
],
[
"theta = tf.Variable(tf.random_uniform([n + 1, 1], -1.0, 1.0, seed=42), name=\"theta\")\ny_pred = tf.matmul(X, theta, name=\"predictions\")\nerror = y_pred - y\nmse = tf.reduce_mean(tf.square(error), name=\"mse\")\noptimizer = tf.train.GradientDescentOptimizer(learning_rate=learning_rate)\ntraining_op = optimizer.minimize(mse)\n\ninit = tf.global_variables_initializer()",
"_____no_output_____"
],
[
"n_epochs = 10",
"_____no_output_____"
],
[
"batch_size = 100\nn_batches = int(np.ceil(m / batch_size))",
"_____no_output_____"
],
[
"def fetch_batch(epoch, batch_index, batch_size):\n np.random.seed(epoch * n_batches + batch_index) # not shown in the book\n indices = np.random.randint(m, size=batch_size) # not shown\n X_batch = scaled_housing_data_plus_bias[indices] # not shown\n y_batch = housing.target.reshape(-1, 1)[indices] # not shown\n return X_batch, y_batch\n\nwith tf.Session() as sess:\n sess.run(init)\n\n for epoch in range(n_epochs):\n for batch_index in range(n_batches):\n X_batch, y_batch = fetch_batch(epoch, batch_index, batch_size)\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n\n best_theta = theta.eval()",
"_____no_output_____"
],
[
"best_theta",
"_____no_output_____"
]
],
[
[
"# Saving and restoring a model",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_epochs = 1000 # not shown in the book\nlearning_rate = 0.01 # not shown\n\nX = tf.constant(scaled_housing_data_plus_bias, dtype=tf.float32, name=\"X\") # not shown\ny = tf.constant(housing.target.reshape(-1, 1), dtype=tf.float32, name=\"y\") # not shown\ntheta = tf.Variable(tf.random_uniform([n + 1, 1], -1.0, 1.0, seed=42), name=\"theta\")\ny_pred = tf.matmul(X, theta, name=\"predictions\") # not shown\nerror = y_pred - y # not shown\nmse = tf.reduce_mean(tf.square(error), name=\"mse\") # not shown\noptimizer = tf.train.GradientDescentOptimizer(learning_rate=learning_rate) # not shown\ntraining_op = optimizer.minimize(mse) # not shown\n\ninit = tf.global_variables_initializer()\nsaver = tf.train.Saver()\n\nwith tf.Session() as sess:\n sess.run(init)\n\n for epoch in range(n_epochs):\n if epoch % 100 == 0:\n print(\"Epoch\", epoch, \"MSE =\", mse.eval()) # not shown\n save_path = saver.save(sess, \"/tmp/my_model.ckpt\")\n sess.run(training_op)\n \n best_theta = theta.eval()\n save_path = saver.save(sess, \"/tmp/my_model_final.ckpt\")",
"Epoch 0 MSE = 9.16154\nEpoch 100 MSE = 0.714501\nEpoch 200 MSE = 0.566705\nEpoch 300 MSE = 0.555572\nEpoch 400 MSE = 0.548812\nEpoch 500 MSE = 0.543636\nEpoch 600 MSE = 0.539629\nEpoch 700 MSE = 0.536509\nEpoch 800 MSE = 0.534068\nEpoch 900 MSE = 0.532147\n"
],
[
"best_theta",
"_____no_output_____"
],
[
"with tf.Session() as sess:\n saver.restore(sess, \"/tmp/my_model_final.ckpt\")\n best_theta_restored = theta.eval() # not shown in the book",
"INFO:tensorflow:Restoring parameters from /tmp/my_model_final.ckpt\n"
],
[
"np.allclose(best_theta, best_theta_restored)",
"_____no_output_____"
]
],
[
[
"If you want to have a saver that loads and restores `theta` with a different name, such as `\"weights\"`:",
"_____no_output_____"
]
],
[
[
"saver = tf.train.Saver({\"weights\": theta})",
"_____no_output_____"
]
],
[
[
"By default the saver also saves the graph structure itself in a second file with the extension `.meta`. You can use the function `tf.train.import_meta_graph()` to restore the graph structure. This function loads the graph into the default graph and returns a `Saver` that can then be used to restore the graph state (i.e., the variable values):",
"_____no_output_____"
]
],
[
[
"reset_graph()\n# notice that we start with an empty graph.\n\nsaver = tf.train.import_meta_graph(\"/tmp/my_model_final.ckpt.meta\") # this loads the graph structure\ntheta = tf.get_default_graph().get_tensor_by_name(\"theta:0\") # not shown in the book\n\nwith tf.Session() as sess:\n saver.restore(sess, \"/tmp/my_model_final.ckpt\") # this restores the graph's state\n best_theta_restored = theta.eval() # not shown in the book",
"INFO:tensorflow:Restoring parameters from /tmp/my_model_final.ckpt\n"
],
[
"np.allclose(best_theta, best_theta_restored)",
"_____no_output_____"
]
],
[
[
"This means that you can import a pretrained model without having to have the corresponding Python code to build the graph. This is very handy when you keep tweaking and saving your model: you can load a previously saved model without having to search for the version of the code that built it.",
"_____no_output_____"
],
[
"# Visualizing the graph\n## inside Jupyter",
"_____no_output_____"
]
],
[
[
"from IPython.display import clear_output, Image, display, HTML\n\ndef strip_consts(graph_def, max_const_size=32):\n \"\"\"Strip large constant values from graph_def.\"\"\"\n strip_def = tf.GraphDef()\n for n0 in graph_def.node:\n n = strip_def.node.add() \n n.MergeFrom(n0)\n if n.op == 'Const':\n tensor = n.attr['value'].tensor\n size = len(tensor.tensor_content)\n if size > max_const_size:\n tensor.tensor_content = b\"<stripped %d bytes>\"%size\n return strip_def\n\ndef show_graph(graph_def, max_const_size=32):\n \"\"\"Visualize TensorFlow graph.\"\"\"\n if hasattr(graph_def, 'as_graph_def'):\n graph_def = graph_def.as_graph_def()\n strip_def = strip_consts(graph_def, max_const_size=max_const_size)\n code = \"\"\"\n <script>\n function load() {{\n document.getElementById(\"{id}\").pbtxt = {data};\n }}\n </script>\n <link rel=\"import\" href=\"https://tensorboard.appspot.com/tf-graph-basic.build.html\" onload=load()>\n <div style=\"height:600px\">\n <tf-graph-basic id=\"{id}\"></tf-graph-basic>\n </div>\n \"\"\".format(data=repr(str(strip_def)), id='graph'+str(np.random.rand()))\n\n iframe = \"\"\"\n <iframe seamless style=\"width:1200px;height:620px;border:0\" srcdoc=\"{}\"></iframe>\n \"\"\".format(code.replace('\"', '"'))\n display(HTML(iframe))",
"_____no_output_____"
],
[
"show_graph(tf.get_default_graph())",
"_____no_output_____"
]
],
[
[
"## Using TensorBoard",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nfrom datetime import datetime\n\nnow = datetime.utcnow().strftime(\"%Y%m%d%H%M%S\")\nroot_logdir = \"tf_logs\"\nlogdir = \"{}/run-{}/\".format(root_logdir, now)",
"_____no_output_____"
],
[
"n_epochs = 1000\nlearning_rate = 0.01\n\nX = tf.placeholder(tf.float32, shape=(None, n + 1), name=\"X\")\ny = tf.placeholder(tf.float32, shape=(None, 1), name=\"y\")\ntheta = tf.Variable(tf.random_uniform([n + 1, 1], -1.0, 1.0, seed=42), name=\"theta\")\ny_pred = tf.matmul(X, theta, name=\"predictions\")\nerror = y_pred - y\nmse = tf.reduce_mean(tf.square(error), name=\"mse\")\noptimizer = tf.train.GradientDescentOptimizer(learning_rate=learning_rate)\ntraining_op = optimizer.minimize(mse)\n\ninit = tf.global_variables_initializer()",
"_____no_output_____"
],
[
"mse_summary = tf.summary.scalar('MSE', mse)\nfile_writer = tf.summary.FileWriter(logdir, tf.get_default_graph())",
"_____no_output_____"
],
[
"n_epochs = 10\nbatch_size = 100\nn_batches = int(np.ceil(m / batch_size))",
"_____no_output_____"
],
[
"with tf.Session() as sess: # not shown in the book\n sess.run(init) # not shown\n\n for epoch in range(n_epochs): # not shown\n for batch_index in range(n_batches):\n X_batch, y_batch = fetch_batch(epoch, batch_index, batch_size)\n if batch_index % 10 == 0:\n summary_str = mse_summary.eval(feed_dict={X: X_batch, y: y_batch})\n step = epoch * n_batches + batch_index\n file_writer.add_summary(summary_str, step)\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n\n best_theta = theta.eval() # not shown",
"_____no_output_____"
],
[
"file_writer.close()",
"_____no_output_____"
],
[
"best_theta",
"_____no_output_____"
]
],
[
[
"# Name scopes",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nnow = datetime.utcnow().strftime(\"%Y%m%d%H%M%S\")\nroot_logdir = \"tf_logs\"\nlogdir = \"{}/run-{}/\".format(root_logdir, now)\n\nn_epochs = 1000\nlearning_rate = 0.01\n\nX = tf.placeholder(tf.float32, shape=(None, n + 1), name=\"X\")\ny = tf.placeholder(tf.float32, shape=(None, 1), name=\"y\")\ntheta = tf.Variable(tf.random_uniform([n + 1, 1], -1.0, 1.0, seed=42), name=\"theta\")\ny_pred = tf.matmul(X, theta, name=\"predictions\")",
"_____no_output_____"
],
[
"optimizer = tf.train.GradientDescentOptimizer(learning_rate=learning_rate)\ntraining_op = optimizer.minimize(mse)\n\ninit = tf.global_variables_initializer()\n\nmse_summary = tf.summary.scalar('MSE', mse)\nfile_writer = tf.summary.FileWriter(logdir, tf.get_default_graph())",
"_____no_output_____"
],
[
"n_epochs = 10\nbatch_size = 100\nn_batches = int(np.ceil(m / batch_size))\n\nwith tf.Session() as sess:\n sess.run(init)\n\n for epoch in range(n_epochs):\n for batch_index in range(n_batches):\n X_batch, y_batch = fetch_batch(epoch, batch_index, batch_size)\n if batch_index % 10 == 0:\n summary_str = mse_summary.eval(feed_dict={X: X_batch, y: y_batch})\n step = epoch * n_batches + batch_index\n file_writer.add_summary(summary_str, step)\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n\n best_theta = theta.eval()\n\nfile_writer.flush()\nfile_writer.close()\nprint(\"Best theta:\")\nprint(best_theta)",
"_____no_output_____"
],
[
"print(error.op.name)",
"_____no_output_____"
],
[
"print(mse.op.name)",
"_____no_output_____"
],
[
"reset_graph()\n\na1 = tf.Variable(0, name=\"a\") # name == \"a\"\na2 = tf.Variable(0, name=\"a\") # name == \"a_1\"\n\nwith tf.name_scope(\"param\"): # name == \"param\"\n a3 = tf.Variable(0, name=\"a\") # name == \"param/a\"\n\nwith tf.name_scope(\"param\"): # name == \"param_1\"\n a4 = tf.Variable(0, name=\"a\") # name == \"param_1/a\"\n\nfor node in (a1, a2, a3, a4):\n print(node.op.name)",
"_____no_output_____"
],
[
"with tf.name_scope(\"loss\") as scope:\n error = y_pred - y\n mse = tf.reduce_mean(tf.square(error), name=\"mse\")",
"_____no_output_____"
]
],
[
[
"# Modularity",
"_____no_output_____"
],
[
"An ugly flat code:",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_features = 3\nX = tf.placeholder(tf.float32, shape=(None, n_features), name=\"X\")\n\nw1 = tf.Variable(tf.random_normal((n_features, 1)), name=\"weights1\")\nw2 = tf.Variable(tf.random_normal((n_features, 1)), name=\"weights2\")\nb1 = tf.Variable(0.0, name=\"bias1\")\nb2 = tf.Variable(0.0, name=\"bias2\")\n\nz1 = tf.add(tf.matmul(X, w1), b1, name=\"z1\")\nz2 = tf.add(tf.matmul(X, w2), b2, name=\"z2\")\n\nrelu1 = tf.maximum(z1, 0., name=\"relu1\")\nrelu2 = tf.maximum(z1, 0., name=\"relu2\") # Oops, cut&paste error! Did you spot it?\n\noutput = tf.add(relu1, relu2, name=\"output\")",
"_____no_output_____"
]
],
[
[
"Much better, using a function to build the ReLUs:",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\ndef relu(X):\n w_shape = (int(X.get_shape()[1]), 1)\n w = tf.Variable(tf.random_normal(w_shape), name=\"weights\")\n b = tf.Variable(0.0, name=\"bias\")\n z = tf.add(tf.matmul(X, w), b, name=\"z\")\n return tf.maximum(z, 0., name=\"relu\")\n\nn_features = 3\nX = tf.placeholder(tf.float32, shape=(None, n_features), name=\"X\")\nrelus = [relu(X) for i in range(5)]\noutput = tf.add_n(relus, name=\"output\")",
"_____no_output_____"
],
[
"file_writer = tf.summary.FileWriter(\"logs/relu1\", tf.get_default_graph())",
"_____no_output_____"
]
],
[
[
"Even better using name scopes:",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\ndef relu(X):\n with tf.name_scope(\"relu\"):\n w_shape = (int(X.get_shape()[1]), 1) # not shown in the book\n w = tf.Variable(tf.random_normal(w_shape), name=\"weights\") # not shown\n b = tf.Variable(0.0, name=\"bias\") # not shown\n z = tf.add(tf.matmul(X, w), b, name=\"z\") # not shown\n return tf.maximum(z, 0., name=\"max\") # not shown",
"_____no_output_____"
],
[
"n_features = 3\nX = tf.placeholder(tf.float32, shape=(None, n_features), name=\"X\")\nrelus = [relu(X) for i in range(5)]\noutput = tf.add_n(relus, name=\"output\")\n\nfile_writer = tf.summary.FileWriter(\"logs/relu2\", tf.get_default_graph())\nfile_writer.close()",
"_____no_output_____"
]
],
[
[
"## Sharing Variables",
"_____no_output_____"
],
[
"Sharing a `threshold` variable the classic way, by defining it outside of the `relu()` function then passing it as a parameter:",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\ndef relu(X, threshold):\n with tf.name_scope(\"relu\"):\n w_shape = (int(X.get_shape()[1]), 1) # not shown in the book\n w = tf.Variable(tf.random_normal(w_shape), name=\"weights\") # not shown\n b = tf.Variable(0.0, name=\"bias\") # not shown\n z = tf.add(tf.matmul(X, w), b, name=\"z\") # not shown\n return tf.maximum(z, threshold, name=\"max\")\n\nthreshold = tf.Variable(0.0, name=\"threshold\")\nX = tf.placeholder(tf.float32, shape=(None, n_features), name=\"X\")\nrelus = [relu(X, threshold) for i in range(5)]\noutput = tf.add_n(relus, name=\"output\")",
"_____no_output_____"
],
[
"reset_graph()\n\ndef relu(X):\n with tf.name_scope(\"relu\"):\n if not hasattr(relu, \"threshold\"):\n relu.threshold = tf.Variable(0.0, name=\"threshold\")\n w_shape = int(X.get_shape()[1]), 1 # not shown in the book\n w = tf.Variable(tf.random_normal(w_shape), name=\"weights\") # not shown\n b = tf.Variable(0.0, name=\"bias\") # not shown\n z = tf.add(tf.matmul(X, w), b, name=\"z\") # not shown\n return tf.maximum(z, relu.threshold, name=\"max\")",
"_____no_output_____"
],
[
"X = tf.placeholder(tf.float32, shape=(None, n_features), name=\"X\")\nrelus = [relu(X) for i in range(5)]\noutput = tf.add_n(relus, name=\"output\")",
"_____no_output_____"
],
[
"reset_graph()\n\nwith tf.variable_scope(\"relu\"):\n threshold = tf.get_variable(\"threshold\", shape=(),\n initializer=tf.constant_initializer(0.0))",
"_____no_output_____"
],
[
"with tf.variable_scope(\"relu\", reuse=True):\n threshold = tf.get_variable(\"threshold\")",
"_____no_output_____"
],
[
"with tf.variable_scope(\"relu\") as scope:\n scope.reuse_variables()\n threshold = tf.get_variable(\"threshold\")",
"_____no_output_____"
],
[
"reset_graph()\n\ndef relu(X):\n with tf.variable_scope(\"relu\", reuse=True):\n threshold = tf.get_variable(\"threshold\")\n w_shape = int(X.get_shape()[1]), 1 # not shown\n w = tf.Variable(tf.random_normal(w_shape), name=\"weights\") # not shown\n b = tf.Variable(0.0, name=\"bias\") # not shown\n z = tf.add(tf.matmul(X, w), b, name=\"z\") # not shown\n return tf.maximum(z, threshold, name=\"max\")\n\nX = tf.placeholder(tf.float32, shape=(None, n_features), name=\"X\")\nwith tf.variable_scope(\"relu\"):\n threshold = tf.get_variable(\"threshold\", shape=(),\n initializer=tf.constant_initializer(0.0))\nrelus = [relu(X) for relu_index in range(5)]\noutput = tf.add_n(relus, name=\"output\")",
"_____no_output_____"
],
[
"file_writer = tf.summary.FileWriter(\"logs/relu6\", tf.get_default_graph())\nfile_writer.close()",
"_____no_output_____"
],
[
"reset_graph()\n\ndef relu(X):\n with tf.variable_scope(\"relu\"):\n threshold = tf.get_variable(\"threshold\", shape=(), initializer=tf.constant_initializer(0.0))\n w_shape = (int(X.get_shape()[1]), 1)\n w = tf.Variable(tf.random_normal(w_shape), name=\"weights\")\n b = tf.Variable(0.0, name=\"bias\")\n z = tf.add(tf.matmul(X, w), b, name=\"z\")\n return tf.maximum(z, threshold, name=\"max\")\n\nX = tf.placeholder(tf.float32, shape=(None, n_features), name=\"X\")\nwith tf.variable_scope(\"\", default_name=\"\") as scope:\n first_relu = relu(X) # create the shared variable\n scope.reuse_variables() # then reuse it\n relus = [first_relu] + [relu(X) for i in range(4)]\noutput = tf.add_n(relus, name=\"output\")\n\nfile_writer = tf.summary.FileWriter(\"logs/relu8\", tf.get_default_graph())\nfile_writer.close()",
"_____no_output_____"
],
[
"reset_graph()\n\ndef relu(X):\n threshold = tf.get_variable(\"threshold\", shape=(),\n initializer=tf.constant_initializer(0.0))\n w_shape = (int(X.get_shape()[1]), 1) # not shown in the book\n w = tf.Variable(tf.random_normal(w_shape), name=\"weights\") # not shown\n b = tf.Variable(0.0, name=\"bias\") # not shown\n z = tf.add(tf.matmul(X, w), b, name=\"z\") # not shown\n return tf.maximum(z, threshold, name=\"max\")\n\nX = tf.placeholder(tf.float32, shape=(None, n_features), name=\"X\")\nrelus = []\nfor relu_index in range(5):\n with tf.variable_scope(\"relu\", reuse=(relu_index >= 1)) as scope:\n relus.append(relu(X))\noutput = tf.add_n(relus, name=\"output\")",
"_____no_output_____"
],
[
"file_writer = tf.summary.FileWriter(\"logs/relu9\", tf.get_default_graph())\nfile_writer.close()",
"_____no_output_____"
]
],
[
[
"# Extra material",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nwith tf.variable_scope(\"my_scope\"):\n x0 = tf.get_variable(\"x\", shape=(), initializer=tf.constant_initializer(0.))\n x1 = tf.Variable(0., name=\"x\")\n x2 = tf.Variable(0., name=\"x\")\n\nwith tf.variable_scope(\"my_scope\", reuse=True):\n x3 = tf.get_variable(\"x\")\n x4 = tf.Variable(0., name=\"x\")\n\nwith tf.variable_scope(\"\", default_name=\"\", reuse=True):\n x5 = tf.get_variable(\"my_scope/x\")\n\nprint(\"x0:\", x0.op.name)\nprint(\"x1:\", x1.op.name)\nprint(\"x2:\", x2.op.name)\nprint(\"x3:\", x3.op.name)\nprint(\"x4:\", x4.op.name)\nprint(\"x5:\", x5.op.name)\nprint(x0 is x3 and x3 is x5)",
"_____no_output_____"
]
],
[
[
"The first `variable_scope()` block first creates the shared variable `x0`, named `my_scope/x`. For all operations other than shared variables (including non-shared variables), the variable scope acts like a regular name scope, which is why the two variables `x1` and `x2` have a name with a prefix `my_scope/`. Note however that TensorFlow makes their names unique by adding an index: `my_scope/x_1` and `my_scope/x_2`.\n\nThe second `variable_scope()` block reuses the shared variables in scope `my_scope`, which is why `x0 is x3`. Once again, for all operations other than shared variables it acts as a named scope, and since it's a separate block from the first one, the name of the scope is made unique by TensorFlow (`my_scope_1`) and thus the variable `x4` is named `my_scope_1/x`.\n\nThe third block shows another way to get a handle on the shared variable `my_scope/x` by creating a `variable_scope()` at the root scope (whose name is an empty string), then calling `get_variable()` with the full name of the shared variable (i.e. `\"my_scope/x\"`).",
"_____no_output_____"
],
[
"## Strings",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\ntext = np.array(\"Do you want some café?\".split())\ntext_tensor = tf.constant(text)\n\nwith tf.Session() as sess:\n print(text_tensor.eval())",
"_____no_output_____"
]
],
[
[
"## Implementing a Home-Made Computation Graph",
"_____no_output_____"
]
],
[
[
"class Const(object):\n def __init__(self, value):\n self.value = value\n def evaluate(self):\n return self.value\n def __str__(self):\n return str(self.value)\n\nclass Var(object):\n def __init__(self, init_value, name):\n self.value = init_value\n self.name = name\n def evaluate(self):\n return self.value\n def __str__(self):\n return self.name\n\nclass BinaryOperator(object):\n def __init__(self, a, b):\n self.a = a\n self.b = b\n\nclass Add(BinaryOperator):\n def evaluate(self):\n return self.a.evaluate() + self.b.evaluate()\n def __str__(self):\n return \"{} + {}\".format(self.a, self.b)\n\nclass Mul(BinaryOperator):\n def evaluate(self):\n return self.a.evaluate() * self.b.evaluate()\n def __str__(self):\n return \"({}) * ({})\".format(self.a, self.b)\n\nx = Var(3, name=\"x\")\ny = Var(4, name=\"y\")\nf = Add(Mul(Mul(x, x), y), Add(y, Const(2))) # f(x,y) = x²y + y + 2\nprint(\"f(x,y) =\", f)\nprint(\"f(3,4) =\", f.evaluate())",
"_____no_output_____"
]
],
[
[
"## Computing gradients\n### Mathematical differentiation",
"_____no_output_____"
]
],
[
[
"df_dx = Mul(Const(2), Mul(x, y)) # df/dx = 2xy\ndf_dy = Add(Mul(x, x), Const(1)) # df/dy = x² + 1\nprint(\"df/dx(3,4) =\", df_dx.evaluate())\nprint(\"df/dy(3,4) =\", df_dy.evaluate())",
"_____no_output_____"
]
],
[
[
"### Numerical differentiation",
"_____no_output_____"
]
],
[
[
"def gradients(func, vars_list, eps=0.0001):\n partial_derivatives = []\n base_func_eval = func.evaluate()\n for var in vars_list:\n original_value = var.value\n var.value = var.value + eps\n tweaked_func_eval = func.evaluate()\n var.value = original_value\n derivative = (tweaked_func_eval - base_func_eval) / eps\n partial_derivatives.append(derivative)\n return partial_derivatives\n\ndf_dx, df_dy = gradients(f, [x, y])\nprint(\"df/dx(3,4) =\", df_dx)\nprint(\"df/dy(3,4) =\", df_dy)",
"_____no_output_____"
]
],
[
[
"### Symbolic differentiation",
"_____no_output_____"
]
],
[
[
"Const.derive = lambda self, var: Const(0)\nVar.derive = lambda self, var: Const(1) if self is var else Const(0)\nAdd.derive = lambda self, var: Add(self.a.derive(var), self.b.derive(var))\nMul.derive = lambda self, var: Add(Mul(self.a, self.b.derive(var)), Mul(self.a.derive(var), self.b))\n\nx = Var(3.0, name=\"x\")\ny = Var(4.0, name=\"y\")\nf = Add(Mul(Mul(x, x), y), Add(y, Const(2))) # f(x,y) = x²y + y + 2\n\ndf_dx = f.derive(x) # 2xy\ndf_dy = f.derive(y) # x² + 1\nprint(\"df/dx(3,4) =\", df_dx.evaluate())\nprint(\"df/dy(3,4) =\", df_dy.evaluate())",
"_____no_output_____"
]
],
[
[
"### Automatic differentiation (autodiff) – forward mode",
"_____no_output_____"
]
],
[
[
"class DualNumber(object):\n def __init__(self, value=0.0, eps=0.0):\n self.value = value\n self.eps = eps\n def __add__(self, b):\n return DualNumber(self.value + self.to_dual(b).value,\n self.eps + self.to_dual(b).eps)\n def __radd__(self, a):\n return self.to_dual(a).__add__(self)\n def __mul__(self, b):\n return DualNumber(self.value * self.to_dual(b).value,\n self.eps * self.to_dual(b).value + self.value * self.to_dual(b).eps)\n def __rmul__(self, a):\n return self.to_dual(a).__mul__(self)\n def __str__(self):\n if self.eps:\n return \"{:.1f} + {:.1f}ε\".format(self.value, self.eps)\n else:\n return \"{:.1f}\".format(self.value)\n def __repr__(self):\n return str(self)\n @classmethod\n def to_dual(cls, n):\n if hasattr(n, \"value\"):\n return n\n else:\n return cls(n)",
"_____no_output_____"
]
],
[
[
"$3 + (3 + 4 \\epsilon) = 6 + 4\\epsilon$",
"_____no_output_____"
]
],
[
[
"3 + DualNumber(3, 4)",
"_____no_output_____"
]
],
[
[
"$(3 + 4ε)\\times(5 + 7ε) = 3 \\times 5 + 3 \\times 7ε + 4ε \\times 5 + 4ε \\times 7ε = 15 + 21ε + 20ε + 28ε^2 = 15 + 41ε + 28 \\times 0 = 15 + 41ε$",
"_____no_output_____"
]
],
[
[
"DualNumber(3, 4) * DualNumber(5, 7)",
"_____no_output_____"
],
[
"x.value = DualNumber(3.0)\ny.value = DualNumber(4.0)\n\nf.evaluate()",
"_____no_output_____"
],
[
"x.value = DualNumber(3.0, 1.0) # 3 + ε\ny.value = DualNumber(4.0) # 4\n\ndf_dx = f.evaluate().eps\n\nx.value = DualNumber(3.0) # 3\ny.value = DualNumber(4.0, 1.0) # 4 + ε\n\ndf_dy = f.evaluate().eps",
"_____no_output_____"
],
[
"df_dx",
"_____no_output_____"
],
[
"df_dy",
"_____no_output_____"
]
],
[
[
"### Autodiff – Reverse mode",
"_____no_output_____"
]
],
[
[
"class Const(object):\n def __init__(self, value):\n self.value = value\n def evaluate(self):\n return self.value\n def backpropagate(self, gradient):\n pass\n def __str__(self):\n return str(self.value)\n\nclass Var(object):\n def __init__(self, init_value, name):\n self.value = init_value\n self.name = name\n self.gradient = 0\n def evaluate(self):\n return self.value\n def backpropagate(self, gradient):\n self.gradient += gradient\n def __str__(self):\n return self.name\n\nclass BinaryOperator(object):\n def __init__(self, a, b):\n self.a = a\n self.b = b\n\nclass Add(BinaryOperator):\n def evaluate(self):\n self.value = self.a.evaluate() + self.b.evaluate()\n return self.value\n def backpropagate(self, gradient):\n self.a.backpropagate(gradient)\n self.b.backpropagate(gradient)\n def __str__(self):\n return \"{} + {}\".format(self.a, self.b)\n\nclass Mul(BinaryOperator):\n def evaluate(self):\n self.value = self.a.evaluate() * self.b.evaluate()\n return self.value\n def backpropagate(self, gradient):\n self.a.backpropagate(gradient * self.b.value)\n self.b.backpropagate(gradient * self.a.value)\n def __str__(self):\n return \"({}) * ({})\".format(self.a, self.b)\n\nx = Var(3, name=\"x\")\ny = Var(4, name=\"y\")\nf = Add(Mul(Mul(x, x), y), Add(y, Const(2))) # f(x,y) = x²y + y + 2\n\nresult = f.evaluate()\nf.backpropagate(1.0)\n\nprint(\"f(x,y) =\", f)\nprint(\"f(3,4) =\", result)\nprint(\"df_dx =\", x.gradient)\nprint(\"df_dy =\", y.gradient)",
"_____no_output_____"
]
],
[
[
"### Autodiff – reverse mode (using TensorFlow)",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nx = tf.Variable(3., name=\"x\")\ny = tf.Variable(4., name=\"y\")\nf = x*x*y + y + 2\n\ngradients = tf.gradients(f, [x, y])\n\ninit = tf.global_variables_initializer()\n\nwith tf.Session() as sess:\n init.run()\n f_val, gradients_val = sess.run([f, gradients])\n\nf_val, gradients_val",
"_____no_output_____"
]
],
[
[
"# Exercise solutions",
"_____no_output_____"
],
[
"## 1. to 11.",
"_____no_output_____"
],
[
"See appendix A.",
"_____no_output_____"
],
[
"## 12. Logistic Regression with Mini-Batch Gradient Descent using TensorFlow",
"_____no_output_____"
],
[
"First, let's create the moons dataset using Scikit-Learn's `make_moons()` function:",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import make_moons\n\nm = 1000\nX_moons, y_moons = make_moons(m, noise=0.1, random_state=42)",
"_____no_output_____"
]
],
[
[
"Let's take a peek at the dataset:",
"_____no_output_____"
]
],
[
[
"plt.plot(X_moons[y_moons == 1, 0], X_moons[y_moons == 1, 1], 'go', label=\"Positive\")\nplt.plot(X_moons[y_moons == 0, 0], X_moons[y_moons == 0, 1], 'r^', label=\"Negative\")\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"We must not forget to add an extra bias feature ($x_0 = 1$) to every instance. For this, we just need to add a column full of 1s on the left of the input matrix $\\mathbf{X}$:",
"_____no_output_____"
]
],
[
[
"X_moons_with_bias = np.c_[np.ones((m, 1)), X_moons]",
"_____no_output_____"
]
],
[
[
"Let's check:",
"_____no_output_____"
]
],
[
[
"X_moons_with_bias[:5]",
"_____no_output_____"
]
],
[
[
"Looks good. Now let's reshape `y_train` to make it a column vector (i.e. a 2D array with a single column):",
"_____no_output_____"
]
],
[
[
"y_moons_column_vector = y_moons.reshape(-1, 1)",
"_____no_output_____"
]
],
[
[
"Now let's split the data into a training set and a test set:",
"_____no_output_____"
]
],
[
[
"test_ratio = 0.2\ntest_size = int(m * test_ratio)\nX_train = X_moons_with_bias[:-test_size]\nX_test = X_moons_with_bias[-test_size:]\ny_train = y_moons_column_vector[:-test_size]\ny_test = y_moons_column_vector[-test_size:]",
"_____no_output_____"
]
],
[
[
"Ok, now let's create a small function to generate training batches. In this implementation we will just pick random instances from the training set for each batch. This means that a single batch may contain the same instance multiple times, and also a single epoch may not cover all the training instances (in fact it will generally cover only about two thirds of the instances). However, in practice this is not an issue and it simplifies the code:",
"_____no_output_____"
]
],
[
[
"def random_batch(X_train, y_train, batch_size):\n rnd_indices = np.random.randint(0, len(X_train), batch_size)\n X_batch = X_train[rnd_indices]\n y_batch = y_train[rnd_indices]\n return X_batch, y_batch",
"_____no_output_____"
]
],
[
[
"Let's look at a small batch:",
"_____no_output_____"
]
],
[
[
"X_batch, y_batch = random_batch(X_train, y_train, 5)\nX_batch",
"_____no_output_____"
],
[
"y_batch",
"_____no_output_____"
]
],
[
[
"Great! Now that the data is ready to be fed to the model, we need to build that model. Let's start with a simple implementation, then we will add all the bells and whistles.",
"_____no_output_____"
],
[
"First let's reset the default graph.",
"_____no_output_____"
]
],
[
[
"reset_graph()",
"_____no_output_____"
]
],
[
[
"The _moons_ dataset has two input features, since each instance is a point on a plane (i.e., 2-Dimensional):",
"_____no_output_____"
]
],
[
[
"n_inputs = 2",
"_____no_output_____"
]
],
[
[
"Now let's build the Logistic Regression model. As we saw in chapter 4, this model first computes a weighted sum of the inputs (just like the Linear Regression model), and then it applies the sigmoid function to the result, which gives us the estimated probability for the positive class:\n\n$\\hat{p} = h_\\mathbf{\\theta}(\\mathbf{x}) = \\sigma(\\mathbf{\\theta}^T \\cdot \\mathbf{x})$\n",
"_____no_output_____"
],
[
"Recall that $\\mathbf{\\theta}$ is the parameter vector, containing the bias term $\\theta_0$ and the weights $\\theta_1, \\theta_2, \\dots, \\theta_n$. The input vector $\\mathbf{x}$ contains a constant term $x_0 = 1$, as well as all the input features $x_1, x_2, \\dots, x_n$.\n\nSince we want to be able to make predictions for multiple instances at a time, we will use an input matrix $\\mathbf{X}$ rather than a single input vector. The $i^{th}$ row will contain the transpose of the $i^{th}$ input vector $(\\mathbf{x}^{(i)})^T$. It is then possible to estimate the probability that each instance belongs to the positive class using the following equation:\n\n$ \\hat{\\mathbf{p}} = \\sigma(\\mathbf{X} \\cdot \\mathbf{\\theta})$\n\nThat's all we need to build the model:",
"_____no_output_____"
]
],
[
[
"X = tf.placeholder(tf.float32, shape=(None, n_inputs + 1), name=\"X\")\ny = tf.placeholder(tf.float32, shape=(None, 1), name=\"y\")\ntheta = tf.Variable(tf.random_uniform([n_inputs + 1, 1], -1.0, 1.0, seed=42), name=\"theta\")\nlogits = tf.matmul(X, theta, name=\"logits\")\ny_proba = 1 / (1 + tf.exp(-logits))",
"_____no_output_____"
]
],
[
[
"In fact, TensorFlow has a nice function `tf.sigmoid()` that we can use to simplify the last line of the previous code:",
"_____no_output_____"
]
],
[
[
"y_proba = tf.sigmoid(logits)",
"_____no_output_____"
]
],
[
[
"As we saw in chapter 4, the log loss is a good cost function to use for Logistic Regression:\n\n$J(\\mathbf{\\theta}) = -\\dfrac{1}{m} \\sum\\limits_{i=1}^{m}{\\left[ y^{(i)} log\\left(\\hat{p}^{(i)}\\right) + (1 - y^{(i)}) log\\left(1 - \\hat{p}^{(i)}\\right)\\right]}$\n\nOne option is to implement it ourselves:",
"_____no_output_____"
]
],
[
[
"epsilon = 1e-7 # to avoid an overflow when computing the log\nloss = -tf.reduce_mean(y * tf.log(y_proba + epsilon) + (1 - y) * tf.log(1 - y_proba + epsilon))",
"_____no_output_____"
]
],
[
[
"But we might as well use TensorFlow's `tf.losses.log_loss()` function:",
"_____no_output_____"
]
],
[
[
"loss = tf.losses.log_loss(y, y_proba) # uses epsilon = 1e-7 by default",
"_____no_output_____"
]
],
[
[
"The rest is pretty standard: let's create the optimizer and tell it to minimize the cost function:",
"_____no_output_____"
]
],
[
[
"learning_rate = 0.01\noptimizer = tf.train.GradientDescentOptimizer(learning_rate=learning_rate)\ntraining_op = optimizer.minimize(loss)",
"_____no_output_____"
]
],
[
[
"All we need now (in this minimal version) is the variable initializer:",
"_____no_output_____"
]
],
[
[
"init = tf.global_variables_initializer()",
"_____no_output_____"
]
],
[
[
"And we are ready to train the model and use it for predictions!",
"_____no_output_____"
],
[
"There's really nothing special about this code, it's virtually the same as the one we used earlier for Linear Regression:",
"_____no_output_____"
]
],
[
[
"n_epochs = 1000\nbatch_size = 50\nn_batches = int(np.ceil(m / batch_size))\n\nwith tf.Session() as sess:\n sess.run(init)\n\n for epoch in range(n_epochs):\n for batch_index in range(n_batches):\n X_batch, y_batch = random_batch(X_train, y_train, batch_size)\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n loss_val = loss.eval({X: X_test, y: y_test})\n if epoch % 100 == 0:\n print(\"Epoch:\", epoch, \"\\tLoss:\", loss_val)\n\n y_proba_val = y_proba.eval(feed_dict={X: X_test, y: y_test})",
"Epoch: 0 \tLoss: 0.792602\nEpoch: 100 \tLoss: 0.343463\nEpoch: 200 \tLoss: 0.30754\nEpoch: 300 \tLoss: 0.292889\nEpoch: 400 \tLoss: 0.285336\nEpoch: 500 \tLoss: 0.280478\nEpoch: 600 \tLoss: 0.278083\nEpoch: 700 \tLoss: 0.276154\nEpoch: 800 \tLoss: 0.27552\nEpoch: 900 \tLoss: 0.274912\n"
]
],
[
[
"Note: we don't use the epoch number when generating batches, so we could just have a single `for` loop rather than 2 nested `for` loops, but it's convenient to think of training time in terms of number of epochs (i.e., roughly the number of times the algorithm went through the training set).",
"_____no_output_____"
],
[
"For each instance in the test set, `y_proba_val` contains the estimated probability that it belongs to the positive class, according to the model. For example, here are the first 5 estimated probabilities:",
"_____no_output_____"
]
],
[
[
"y_proba_val[:5]",
"_____no_output_____"
]
],
[
[
"To classify each instance, we can go for maximum likelihood: classify as positive any instance whose estimated probability is greater or equal to 0.5:",
"_____no_output_____"
]
],
[
[
"y_pred = (y_proba_val >= 0.5)\ny_pred[:5]",
"_____no_output_____"
]
],
[
[
"Depending on the use case, you may want to choose a different threshold than 0.5: make it higher if you want high precision (but lower recall), and make it lower if you want high recall (but lower precision). See chapter 3 for more details.",
"_____no_output_____"
],
[
"Let's compute the model's precision and recall:",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import precision_score, recall_score\n\nprecision_score(y_test, y_pred)",
"_____no_output_____"
],
[
"recall_score(y_test, y_pred)",
"_____no_output_____"
]
],
[
[
"Let's plot these predictions to see what they look like:",
"_____no_output_____"
]
],
[
[
"y_pred_idx = y_pred.reshape(-1) # a 1D array rather than a column vector\nplt.plot(X_test[y_pred_idx, 1], X_test[y_pred_idx, 2], 'go', label=\"Positive\")\nplt.plot(X_test[~y_pred_idx, 1], X_test[~y_pred_idx, 2], 'r^', label=\"Negative\")\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"Well, that looks pretty bad, doesn't it? But let's not forget that the Logistic Regression model has a linear decision boundary, so this is actually close to the best we can do with this model (unless we add more features, as we will show in a second).",
"_____no_output_____"
],
[
"Now let's start over, but this time we will add all the bells and whistles, as listed in the exercise:\n* Define the graph within a `logistic_regression()` function that can be reused easily.\n* Save checkpoints using a `Saver` at regular intervals during training, and save the final model at the end of training.\n* Restore the last checkpoint upon startup if training was interrupted.\n* Define the graph using nice scopes so the graph looks good in TensorBoard.\n* Add summaries to visualize the learning curves in TensorBoard.\n* Try tweaking some hyperparameters such as the learning rate or the mini-batch size and look at the shape of the learning curve.",
"_____no_output_____"
],
[
"Before we start, we will add 4 more features to the inputs: ${x_1}^2$, ${x_2}^2$, ${x_1}^3$ and ${x_2}^3$. This was not part of the exercise, but it will demonstrate how adding features can improve the model. We will do this manually, but you could also add them using `sklearn.preprocessing.PolynomialFeatures`.",
"_____no_output_____"
]
],
[
[
"X_train_enhanced = np.c_[X_train,\n np.square(X_train[:, 1]),\n np.square(X_train[:, 2]),\n X_train[:, 1] ** 3,\n X_train[:, 2] ** 3]\nX_test_enhanced = np.c_[X_test,\n np.square(X_test[:, 1]),\n np.square(X_test[:, 2]),\n X_test[:, 1] ** 3,\n X_test[:, 2] ** 3]",
"_____no_output_____"
]
],
[
[
"This is what the \"enhanced\" training set looks like:",
"_____no_output_____"
]
],
[
[
"X_train_enhanced[:5]",
"_____no_output_____"
]
],
[
[
"Ok, next let's reset the default graph:",
"_____no_output_____"
]
],
[
[
"reset_graph()",
"_____no_output_____"
]
],
[
[
"Now let's define the `logistic_regression()` function to create the graph. We will leave out the definition of the inputs `X` and the targets `y`. We could include them here, but leaving them out will make it easier to use this function in a wide range of use cases (e.g. perhaps we will want to add some preprocessing steps for the inputs before we feed them to the Logistic Regression model).",
"_____no_output_____"
]
],
[
[
"def logistic_regression(X, y, initializer=None, seed=42, learning_rate=0.01):\n n_inputs_including_bias = int(X.get_shape()[1])\n with tf.name_scope(\"logistic_regression\"):\n with tf.name_scope(\"model\"):\n if initializer is None:\n initializer = tf.random_uniform([n_inputs_including_bias, 1], -1.0, 1.0, seed=seed)\n theta = tf.Variable(initializer, name=\"theta\")\n logits = tf.matmul(X, theta, name=\"logits\")\n y_proba = tf.sigmoid(logits)\n with tf.name_scope(\"train\"):\n loss = tf.losses.log_loss(y, y_proba, scope=\"loss\")\n optimizer = tf.train.GradientDescentOptimizer(learning_rate=learning_rate)\n training_op = optimizer.minimize(loss)\n loss_summary = tf.summary.scalar('log_loss', loss)\n with tf.name_scope(\"init\"):\n init = tf.global_variables_initializer()\n with tf.name_scope(\"save\"):\n saver = tf.train.Saver()\n return y_proba, loss, training_op, loss_summary, init, saver",
"_____no_output_____"
]
],
[
[
"Let's create a little function to get the name of the log directory to save the summaries for Tensorboard:",
"_____no_output_____"
]
],
[
[
"from datetime import datetime\n\ndef log_dir(prefix=\"\"):\n now = datetime.utcnow().strftime(\"%Y%m%d%H%M%S\")\n root_logdir = \"tf_logs\"\n if prefix:\n prefix += \"-\"\n name = prefix + \"run-\" + now\n return \"{}/{}/\".format(root_logdir, name)",
"_____no_output_____"
]
],
[
[
"Next, let's create the graph, using the `logistic_regression()` function. We will also create the `FileWriter` to save the summaries to the log directory for Tensorboard:",
"_____no_output_____"
]
],
[
[
"n_inputs = 2 + 4\nlogdir = log_dir(\"logreg\")\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs + 1), name=\"X\")\ny = tf.placeholder(tf.float32, shape=(None, 1), name=\"y\")\n\ny_proba, loss, training_op, loss_summary, init, saver = logistic_regression(X, y)\n\nfile_writer = tf.summary.FileWriter(logdir, tf.get_default_graph())",
"_____no_output_____"
]
],
[
[
"At last we can train the model! We will start by checking whether a previous training session was interrupted, and if so we will load the checkpoint and continue training from the epoch number we saved. In this example we just save the epoch number to a separate file, but in chapter 11 we will see how to store the training step directly as part of the model, using a non-trainable variable called `global_step` that we pass to the optimizer's `minimize()` method.\n\nYou can try interrupting training to verify that it does indeed restore the last checkpoint when you start it again.",
"_____no_output_____"
]
],
[
[
"n_epochs = 10001\nbatch_size = 50\nn_batches = int(np.ceil(m / batch_size))\n\ncheckpoint_path = \"/tmp/my_logreg_model.ckpt\"\ncheckpoint_epoch_path = checkpoint_path + \".epoch\"\nfinal_model_path = \"./my_logreg_model\"\n\nwith tf.Session() as sess:\n if os.path.isfile(checkpoint_epoch_path):\n # if the checkpoint file exists, restore the model and load the epoch number\n with open(checkpoint_epoch_path, \"rb\") as f:\n start_epoch = int(f.read())\n print(\"Training was interrupted. Continuing at epoch\", start_epoch)\n saver.restore(sess, checkpoint_path)\n else:\n start_epoch = 0\n sess.run(init)\n\n for epoch in range(start_epoch, n_epochs):\n for batch_index in range(n_batches):\n X_batch, y_batch = random_batch(X_train_enhanced, y_train, batch_size)\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n loss_val, summary_str = sess.run([loss, loss_summary], feed_dict={X: X_test_enhanced, y: y_test})\n file_writer.add_summary(summary_str, epoch)\n if epoch % 500 == 0:\n print(\"Epoch:\", epoch, \"\\tLoss:\", loss_val)\n saver.save(sess, checkpoint_path)\n with open(checkpoint_epoch_path, \"wb\") as f:\n f.write(b\"%d\" % (epoch + 1))\n\n saver.save(sess, final_model_path)\n y_proba_val = y_proba.eval(feed_dict={X: X_test_enhanced, y: y_test})\n os.remove(checkpoint_epoch_path)",
"Epoch: 0 \tLoss: 0.629985\nEpoch: 500 \tLoss: 0.161224\nEpoch: 1000 \tLoss: 0.119032\nEpoch: 1500 \tLoss: 0.0973292\nEpoch: 2000 \tLoss: 0.0836979\nEpoch: 2500 \tLoss: 0.0743758\nEpoch: 3000 \tLoss: 0.0675021\nEpoch: 3500 \tLoss: 0.0622069\nEpoch: 4000 \tLoss: 0.0580268\nEpoch: 4500 \tLoss: 0.054563\nEpoch: 5000 \tLoss: 0.0517083\nEpoch: 5500 \tLoss: 0.0492377\nEpoch: 6000 \tLoss: 0.0471673\nEpoch: 6500 \tLoss: 0.0453766\nEpoch: 7000 \tLoss: 0.0438187\nEpoch: 7500 \tLoss: 0.0423742\nEpoch: 8000 \tLoss: 0.0410892\nEpoch: 8500 \tLoss: 0.0399709\nEpoch: 9000 \tLoss: 0.0389202\nEpoch: 9500 \tLoss: 0.0380107\nEpoch: 10000 \tLoss: 0.0371557\n"
]
],
[
[
"Once again, we can make predictions by just classifying as positive all the instances whose estimated probability is greater or equal to 0.5:",
"_____no_output_____"
]
],
[
[
"y_pred = (y_proba_val >= 0.5)",
"_____no_output_____"
],
[
"precision_score(y_test, y_pred)",
"_____no_output_____"
],
[
"recall_score(y_test, y_pred)",
"_____no_output_____"
],
[
"y_pred_idx = y_pred.reshape(-1) # a 1D array rather than a column vector\nplt.plot(X_test[y_pred_idx, 1], X_test[y_pred_idx, 2], 'go', label=\"Positive\")\nplt.plot(X_test[~y_pred_idx, 1], X_test[~y_pred_idx, 2], 'r^', label=\"Negative\")\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"Now that's much, much better! Apparently the new features really helped a lot.",
"_____no_output_____"
],
[
"Try starting the tensorboard server, find the latest run and look at the learning curve (i.e., how the loss evaluated on the test set evolves as a function of the epoch number):\n\n```\n$ tensorboard --logdir=tf_logs\n```",
"_____no_output_____"
],
[
"Now you can play around with the hyperparameters (e.g. the `batch_size` or the `learning_rate`) and run training again and again, comparing the learning curves. You can even automate this process by implementing grid search or randomized search. Below is a simple implementation of a randomized search on both the batch size and the learning rate. For the sake of simplicity, the checkpoint mechanism was removed.",
"_____no_output_____"
]
],
[
[
"from scipy.stats import reciprocal\n\nn_search_iterations = 10\n\nfor search_iteration in range(n_search_iterations):\n batch_size = np.random.randint(1, 100)\n learning_rate = reciprocal(0.0001, 0.1).rvs(random_state=search_iteration)\n\n n_inputs = 2 + 4\n logdir = log_dir(\"logreg\")\n \n print(\"Iteration\", search_iteration)\n print(\" logdir:\", logdir)\n print(\" batch size:\", batch_size)\n print(\" learning_rate:\", learning_rate)\n print(\" training: \", end=\"\")\n\n reset_graph()\n\n X = tf.placeholder(tf.float32, shape=(None, n_inputs + 1), name=\"X\")\n y = tf.placeholder(tf.float32, shape=(None, 1), name=\"y\")\n\n y_proba, loss, training_op, loss_summary, init, saver = logistic_regression(\n X, y, learning_rate=learning_rate)\n\n file_writer = tf.summary.FileWriter(logdir, tf.get_default_graph())\n\n n_epochs = 10001\n n_batches = int(np.ceil(m / batch_size))\n\n final_model_path = \"./my_logreg_model_%d\" % search_iteration\n\n with tf.Session() as sess:\n sess.run(init)\n\n for epoch in range(n_epochs):\n for batch_index in range(n_batches):\n X_batch, y_batch = random_batch(X_train_enhanced, y_train, batch_size)\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n loss_val, summary_str = sess.run([loss, loss_summary], feed_dict={X: X_test_enhanced, y: y_test})\n file_writer.add_summary(summary_str, epoch)\n if epoch % 500 == 0:\n print(\".\", end=\"\")\n\n saver.save(sess, final_model_path)\n\n print()\n y_proba_val = y_proba.eval(feed_dict={X: X_test_enhanced, y: y_test})\n y_pred = (y_proba_val >= 0.5)\n \n print(\" precision:\", precision_score(y_test, y_pred))\n print(\" recall:\", recall_score(y_test, y_pred))",
"Iteration 0\n logdir: tf_logs/logreg-run-20171017023201/\n batch size: 54\n learning_rate: 0.00443037524522\n training: .....................\n precision: 0.979797979798\n recall: 0.979797979798\nIteration 1\n logdir: tf_logs/logreg-run-20171017023408/\n batch size: 22\n learning_rate: 0.00178264971514\n training: .....................\n precision: 0.979797979798\n recall: 0.979797979798\nIteration 2\n logdir: tf_logs/logreg-run-20171017024015/\n batch size: 74\n learning_rate: 0.00203228544324\n training: .....................\n precision: 0.969696969697\n recall: 0.969696969697\nIteration 3\n logdir: tf_logs/logreg-run-20171017024240/\n batch size: 58\n learning_rate: 0.00449152382514\n training: .....................\n precision: 0.979797979798\n recall: 0.979797979798\nIteration 4\n logdir: tf_logs/logreg-run-20171017024543/\n batch size: 61\n learning_rate: 0.0796323472178\n training: .....................\n precision: 0.980198019802\n recall: 1.0\nIteration 5\n logdir: tf_logs/logreg-run-20171017024839/\n batch size: 92\n learning_rate: 0.000463425058329\n training: .....................\n precision: 0.912621359223\n recall: 0.949494949495\nIteration 6\n logdir: tf_logs/logreg-run-20171017025008/\n batch size: 74\n learning_rate: 0.0477068184194\n training: .....................\n precision: 0.98\n recall: 0.989898989899\nIteration 7\n logdir: tf_logs/logreg-run-20171017025145/\n batch size: 58\n learning_rate: 0.000169404470952\n training: .....................\n precision: 0.9\n recall: 0.909090909091\nIteration 8\n logdir: tf_logs/logreg-run-20171017025352/\n batch size: 61\n learning_rate: 0.0417146119941\n training: .....................\n precision: 0.980198019802\n recall: 1.0\nIteration 9\n logdir: tf_logs/logreg-run-20171017025548/\n batch size: 92\n learning_rate: 0.000107429229684\n training: .....................\n precision: 0.882352941176\n recall: 0.757575757576\n"
]
],
[
[
"The `reciprocal()` function from SciPy's `stats` module returns a random distribution that is commonly used when you have no idea of the optimal scale of a hyperparameter. See the exercise solutions for chapter 2 for more details. ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e7913220c4c0e7a7db3bffa6cdb8d69de6126c8b | 85,213 | ipynb | Jupyter Notebook | Analysing Hotel Reviews Using Gaussian NaiveBayes.ipynb | chiragbhattad/Analysing-hotel-reviews | c2decf563e70fbd892f704168942d88c7f9427ba | [
"Unlicense"
] | 1 | 2019-05-16T11:34:39.000Z | 2019-05-16T11:34:39.000Z | Analysing Hotel Reviews Using Gaussian NaiveBayes.ipynb | chiragbhattad/Analysing-hotel-reviews | c2decf563e70fbd892f704168942d88c7f9427ba | [
"Unlicense"
] | null | null | null | Analysing Hotel Reviews Using Gaussian NaiveBayes.ipynb | chiragbhattad/Analysing-hotel-reviews | c2decf563e70fbd892f704168942d88c7f9427ba | [
"Unlicense"
] | null | null | null | 36.619252 | 134 | 0.386772 | [
[
[
"#Importing the required Libraries\nfrom sklearn.metrics import accuracy_score\nimport numpy as np\nimport pandas as pd\nfrom nltk.corpus import stopwords\nfrom nltk.stem import PorterStemmer\nfrom sklearn.ensemble import GradientBoostingClassifier\nfrom sklearn.naive_bayes import GaussianNB\nfrom sklearn.preprocessing import LabelEncoder\nimport re\nfrom sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.metrics import accuracy_score, make_scorer",
"_____no_output_____"
],
[
"#Reading train.csv and test.csv files\n\ntrain = pd.read_csv(\"/Volumes/Goldmine/Course/ML_beginners/train.csv\")\ntest = pd.read_csv(\"/Volumes/Goldmine/Course/ML_beginners/test.csv\")",
"_____no_output_____"
],
[
"#Looking at the different columns and the type of data in it\n\ntrain.head(10)\ntrain.columns",
"_____no_output_____"
],
[
"#Defining a function that cleans the reviews so that it can be read easily\n\nstops = set(stopwords.words(\"english\"))\ndef cleanData(text, lowercase=False, remove_stops=False, stemming=False):\n txt = str(text)\n txt = re.sub(r'[^A-Za-z0-9\\s]',r'',txt)\n txt = re.sub(r'\\n',r' ',txt)\n \n if lowercase:\n txt = \" \".join([w.lower() for w in txt.split()])\n \n if remove_stops:\n txt = \" \".join([w for w in txt.split() if w not in stops])\n \n if stemming:\n st = PorterStemmer()\n txt = \" \".join([st.stem(w) for w in txt.split()])\n\n return txt",
"_____no_output_____"
],
[
"#Concatenate test and train data, adding Is_Response column in the test data\n\ntest['Is_Response'] = np.nan\nalldata = pd.concat([train, test]).reset_index(drop=True)",
"_____no_output_____"
],
[
"#Clean the Description column by calling the CleanData function onit. \n#Map() helps select all the value in the Description Column without using the for loop. \n#Lambda defines a temporary function.\n\nalldata['Description'] = alldata['Description'].map(lambda x: cleanData(x, lowercase=True, remove_stops=True, stemming=True))\n\n#Note that the Description column now consists of only the key words of the reviews, with the stopwords removed fromthe data\n\nalldata[:5]",
"_____no_output_____"
],
[
"#CountVectorizer counts the frequency of specific words\n#Tf-IDF Vectorizer uses weighted methods to study the weight and frequency of a particular word in context\n\ncountvec = CountVectorizer(analyzer='word', ngram_range = (1,1), min_df=150, max_features=500)\ntfidfvec = TfidfVectorizer(analyzer='word', ngram_range = (1,1), min_df = 150, max_features=500)\n\nbagofwords = countvec.fit_transform(alldata['Description'])\ntfidfdata = tfidfvec.fit_transform(alldata['Description'])\n\nbagofwords",
"_____no_output_____"
],
[
"tfidfdata",
"_____no_output_____"
],
[
"#Converts the Browser and Device data into Numeric values\n\ncols = ['Browser_Used','Device_Used']\n\nfor x in cols:\n lbl = LabelEncoder()\n alldata[x] = lbl.fit_transform(alldata[x])",
"_____no_output_____"
],
[
"bow_df = pd.DataFrame(bagofwords.todense())\ntfidf_df = pd.DataFrame(tfidfdata.todense())\n\ntfidf_df",
"_____no_output_____"
],
[
"bow_df.columns = ['col'+ str(x) for x in bow_df.columns]\ntfidf_df.columns = ['col' + str(x) for x in tfidf_df.columns]\n\nbow_df_train = bow_df[:len(train)]\nbow_df_test = bow_df[len(train):]\n\ntfid_df_train = tfidf_df[:len(train)]\ntfid_df_test = tfidf_df[len(train):]",
"_____no_output_____"
],
[
"train_feats = alldata[~pd.isnull(alldata.Is_Response)]\ntest_feats = alldata[pd.isnull(alldata.Is_Response)]\n\ntrain_feats[:5]",
"_____no_output_____"
],
[
"train_feats['Is_Response'] = [1 if x == 'happy' else 0 for x in train_feats['Is_Response']]",
"/anaconda/lib/python3.6/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n \"\"\"Entry point for launching an IPython kernel.\n"
],
[
"train_feats1 = pd.concat([train_feats[cols], bow_df_train], axis = 1)\ntest_feats1 = pd.concat([test_feats[cols], bow_df_test], axis=1)\n\ntest_feats1.reset_index(drop=True, inplace=True)\n\ntrain_feats2 = pd.concat([train_feats[cols], tfid_df_train], axis=1)\ntest_feats2 = pd.concat([test_feats[cols], tfid_df_test], axis=1)",
"_____no_output_____"
],
[
"#Using Naive Bayes model\n\nmod1 = GaussianNB()\ntarget = train_feats['Is_Response']",
"_____no_output_____"
],
[
"#Printing the Cross Validation Scores for Naive Bayes on bow_df\n\nprint(cross_val_score(mod1, train_feats1, target, cv=5, scoring=make_scorer(accuracy_score)))",
"[ 0.77208526 0.76110968 0.76753147 0.76663242 0.77626509]\n"
],
[
"#Printing the Cross Validation Scores for Naive Bayes on tfid_df\n\n\nprint(cross_val_score(mod1, train_feats2, target, cv=5, scoring=make_scorer(accuracy_score)))",
"[ 0.80906523 0.81518109 0.80901618 0.81312612 0.80349345]\n"
],
[
"#Training the Naive Bayes model with the train data\n\nclf1 = GaussianNB()\nclf1.fit(train_feats1, target)\n\nclf2 = GaussianNB()\nclf2.fit(train_feats2, target)",
"_____no_output_____"
],
[
"#Prediciton the Is_Response on test data\n\npreds1 = clf1.predict(test_feats1)\npreds2 = clf2.predict(test_feats2)",
"_____no_output_____"
],
[
"#Converting Binary responses to Happy/ Not Happy responses\n\ndef to_labels(x):\n if x==1:\n return \"happy\"\n return \"not_happy\"",
"_____no_output_____"
],
[
"#Saving the responses into a new csv file\n\nsubm1 = pd.DataFrame({'User_ID':test.User_ID, 'Is_Response':preds1})\nsubm1['Is_Response'] = subm1['Is_Response'].map(lambda x: to_labels(x))\n\nsubm2 = pd.DataFrame({'User_ID':test.User_ID, 'Is_Response':preds2})\nsubm2['Is_Response'] = subm2['Is_Response'].map(lambda x: to_labels(x))\n\nsubm1 = subm1[['User_ID', 'Is_Response']]\nsubm2 = subm2[['User_ID', 'Is_Response']]\n\nsubm1.to_csv('subm1_cv.csv', index=False)\nsubm2.to_csv('subm2_tf.csv', index=False)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7913ad7f9491fbbc97641c3929615bcc5d20f65 | 43,803 | ipynb | Jupyter Notebook | Magnus/Problem sets/PS2/problem_set_2.ipynb | NumEconCopenhagen/projects-2022-git-good | df457732b3da0d52c481b0adcb18e1cef63a5089 | [
"MIT"
] | null | null | null | Magnus/Problem sets/PS2/problem_set_2.ipynb | NumEconCopenhagen/projects-2022-git-good | df457732b3da0d52c481b0adcb18e1cef63a5089 | [
"MIT"
] | null | null | null | Magnus/Problem sets/PS2/problem_set_2.ipynb | NumEconCopenhagen/projects-2022-git-good | df457732b3da0d52c481b0adcb18e1cef63a5089 | [
"MIT"
] | null | null | null | 48.887277 | 20,040 | 0.701162 | [
[
[
"# Problem set 2: Finding the Walras equilibrium in a multi-agent economy",
"_____no_output_____"
],
[
"[<img src=\"https://mybinder.org/badge_logo.svg\">](https://mybinder.org/v2/gh/NumEconCopenhagen/exercises-2020/master?urlpath=lab/tree/PS2/problem_set_2.ipynb)",
"_____no_output_____"
]
],
[
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
]
],
[
[
"# Tasks",
"_____no_output_____"
],
[
"## Drawing random numbers",
"_____no_output_____"
],
[
"Replace the missing lines in the code below to get the same output as in the answer.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nnp.random.seed(1986)\n# Define state, which makes sure that the code is randomized.\nstate = np.random.get_state()\nfor i in range(3):\n # Reset the random state three times, because the range is 3. The state makes sure that if we change the range\n # we will not change the random numbers generated in the first numbers of the range.\n np.random.set_state(state)\n for j in range(2):\n x = np.random.uniform()\n print(f'({i},{j}): x = {x:.3f}')",
"(0,0): x = 0.569\n(0,1): x = 0.077\n(1,0): x = 0.569\n(1,1): x = 0.077\n(2,0): x = 0.569\n(2,1): x = 0.077\n"
]
],
[
[
"**Answer:**",
"_____no_output_____"
],
[
"See A1.py",
"_____no_output_____"
],
[
"## Find the expectated value",
"_____no_output_____"
],
[
"Find the expected value and the expected variance\n\n$$ \n\\mathbb{E}[g(x)] \\approx \\frac{1}{N}\\sum_{i=1}^{N} g(x_i)\n$$\n$$ \n\\mathbb{VAR}[g(x)] \\approx \\frac{1}{N}\\sum_{i=1}^{N} \\left( g(x_i) - \\frac{1}{N}\\sum_{i=1}^{N} g(x_i) \\right)^2\n$$\n\nwhere $ x_i \\sim \\mathcal{N}(0,\\sigma) $ and\n\n$$ \ng(x,\\omega)=\\begin{cases}\nx & \\text{if }x\\in[-\\omega,\\omega]\\\\\n-\\omega & \\text{if }x<-\\omega\\\\\n\\omega & \\text{if }x>\\omega\n\\end{cases} \n$$",
"_____no_output_____"
]
],
[
[
"sigma = 3.14\nomega = 2\nN = 10000\nnp.random.seed(1986)\n# Set state\nstate = np.random.get_state()\nnp.random.set_state(state)\n\n# Define x as a normal distribution\nx = np.random.normal(loc=0, scale=sigma, size=N)\n\n# Define function g(x,omega)\ndef g_function(x,omega):\n \n # g_function has to give the value g. Because x is an array changes in g must not affect x.\n g = x.copy()\n # We describe the conditions in the function.\n g[x < -omega] = -omega\n g[x > omega] = omega\n # Define what the function has to return, in this case the value which is given by the condition.\n return g\n \n# Calculate mean and variance\nmean = np.mean(g_function(x,omega))\nvariance = np.var(g_function(x-mean,omega))\n# Print the results\nprint(f'mean = {mean:.5f} variance = {variance:.5f}')",
"mean = -0.00264 variance = 2.69804\n"
]
],
[
[
"**Answer:**",
"_____no_output_____"
],
[
"See A2.py",
"_____no_output_____"
],
[
"## Interactive histogram",
"_____no_output_____"
],
[
"**First task:** Consider the code below. Fill in the missing lines so the figure is plotted.",
"_____no_output_____"
]
],
[
[
"# a. import\nimport math\nimport pickle\nimport numpy as np\nfrom scipy.stats import norm # normal distribution\n%matplotlib inline\nimport matplotlib.pyplot as plt\nplt.style.use('seaborn-whitegrid')\nimport ipywidgets as widgets\n\n# b. plotting figure\ndef fitting_normal(X,mu_guess,sigma_guess):\n \n # i. normal distribution from guess\n F = norm(loc=mu_guess,scale=sigma_guess)\n \n # ii. x-values\n x_low = F.ppf(0.001)\n x_high = F.ppf(0.999)\n x = np.linspace(x_low,x_high,100)\n\n # iii. figure\n fig = plt.figure(dpi=100)\n ax = fig.add_subplot(1,1,1)\n ax.plot(x,F.pdf(x),lw=2)\n ax.hist(X,bins=100,density=True,histtype='stepfilled');\n ax.set_ylim([0,0.5])\n ax.set_xlim([-6,6])\n\n# c. parameters\nmu_true = 2\nsigma_true = 1\nmu_guess = 1\nsigma_guess = 2\n\n# d. random draws\nX = np.random.normal(loc=mu_true,scale=sigma_true,size=10**6)\n\n# e. figure\ntry:\n fitting_normal(X,mu_guess,sigma_guess)\nexcept:\n print('failed')",
"_____no_output_____"
]
],
[
[
"**Second task:** Create an interactive version of the figure with sliders for $\\mu$ and $\\sigma$.",
"_____no_output_____"
]
],
[
[
"# Write out which arguments to interactive_figure you want to be changing or staying fixed \nwidgets.interact(fitting_normal,\n X=widgets.fixed(X),\n mu_guess=widgets.FloatSlider(description=\"$\\mu$\", min=-5, max=5, step=1, value=1),\n sigma_guess=widgets.FloatSlider(description=\"$\\sigma$\", min=0.1, max=10, step = 0.1, value=2)\n);",
"_____no_output_____"
]
],
[
[
"**Answer:**",
"_____no_output_____"
],
[
"See A3.py",
"_____no_output_____"
],
[
"## Modules",
"_____no_output_____"
],
[
"1. Call the function `myfun` from the module `mymodule` present in this folder.\n2. Open VSCode and open the `mymodule.py`, add a new function and call it from this notebook.",
"_____no_output_____"
]
],
[
[
"import mymodule as mm\nfrom mymodule import myfun\nmm.myfun(1)\n\nmm.gitgood(1)",
"hello world!\nGit Good!\n"
]
],
[
[
"**Answer:**",
"_____no_output_____"
],
[
"See A4.py",
"_____no_output_____"
],
[
"## Git",
"_____no_output_____"
],
[
"1. Try to go to your own personal GitHub main page and create a new repository. Then put your solution to this problem set in it.\n2. Pair up with a fellow student. Clone each others repositories and run the code in them.",
"_____no_output_____"
],
[
"**IMPORTANT:** You will need **git** for the data project in a few needs. Better learn it know. Remember, that the teaching assistants are there to help you.",
"_____no_output_____"
],
[
"# Problem",
"_____no_output_____"
],
[
"Consider an **exchange economy** with\n\n1. 2 goods, $(x_1,x_2)$\n2. $N$ consumers indexed by $j \\in \\{1,2,\\dots,N\\}$\n3. Preferences are Cobb-Douglas with truncated normally *heterogenous* coefficients\n\n $$\n \\begin{aligned}\n u^{j}(x_{1},x_{2}) & = x_{1}^{\\alpha_{j}}x_{2}^{1-\\alpha_{j}}\\\\\n & \\tilde{\\alpha}_{j}\\sim\\mathcal{N}(\\mu,\\sigma)\\\\\n & \\alpha_j = \\max(\\underline{\\mu},\\min(\\overline{\\mu},\\tilde{\\alpha}_{j}))\n \\end{aligned}\n $$\n\n4. Endowments are *heterogenous* and given by\n\n $$\n \\begin{aligned}\n \\boldsymbol{e}^{j}&=(e_{1}^{j},e_{2}^{j}) \\\\\n & & e_i^j \\sim f, f(x,\\beta_i) = 1/\\beta_i \\exp(-x/\\beta)\n \\end{aligned}\n $$",
"_____no_output_____"
],
[
"**Problem:** Write a function to solve for the equilibrium.",
"_____no_output_____"
],
[
"You can use the following parameters:",
"_____no_output_____"
]
],
[
[
"# a. parameters\nN = 10000\nmu = 0.5\nsigma = 0.2\nmu_low = 0.1\nmu_high = 0.9\nbeta1 = 1.3\nbeta2 = 2.1\nseed = 1986\n\n# b. draws of random numbers\nnp.random.seed(seed)\nalphatilde = np.random.normal(loc=mu, scale=sigma, size=N)\nalpha = np.fmax(mu_low,np.fmin(mu_high, alphatilde))\ne1 = np.random.exponential(scale=beta1, size=N)\ne2 = np.random.exponential(scale=beta2, size=N)\n\n# c. demand function\ndef demand_good_1_func(alpha, p1, p2, e1, e2):\n I = e1*p1+e2*p2\n return alpha*I/p1\n\n# d. excess demand function\ndef excess_demand_func(alpha, p1, p2, e1, e2):\n \n # Define aggregate supply and demand for good 1\n demand = np.sum(demand_good_1_func(alpha, p1, p2, e1, e2))\n supply = sum(e1)\n \n # Excess demand is demand supply subtracted from demand\n excess_demand = demand - supply\n \n return excess_demand\n\n\n# e. find equilibrium function\ndef find_equilibrium(alphas, p1, p2, e1, e2, kappa=0.5, eps=1e-8, maxiter=500):\n \n t = 0\n \n # using a while loop as we don't know number of iterations a priori\n while True:\n\n # a. step 1: excess demand\n Z1 = excess_demand_func(alpha, p1, p2, e1, e2)\n \n # b: step 2: stop?\n if np.abs(Z1) < eps or t >= maxiter:\n print(f'{t:3d}: p1 = {p1:12.8f} -> excess demand -> {Z1:14.8f}')\n break \n \n # c. step 3: update p1\n p1 = p1 + kappa*Z1/alphas.size\n \n # d. step 4: print only every 25th iteration using the modulus operator \n if t < 5 or t%25 == 0:\n print(f'{t:3d}: p1 = {p1:12.8f} -> excess demand -> {Z1:14.8f}')\n elif t == 5:\n print(' ...')\n \n t += 1 \n\n return p1\n\n# f. call find equilibrium function\np1 = 1.8\np2 = 1\nkappa = 0.5\neps = 1e-8\nfind_equilibrium(alpha,p1,p2,e1,e2,kappa=kappa,eps=eps)",
" 0: p1 = 1.76747251 -> excess demand -> -650.54980224\n 1: p1 = 1.74035135 -> excess demand -> -542.42310867\n 2: p1 = 1.71789246 -> excess demand -> -449.17798560\n 3: p1 = 1.69940577 -> excess demand -> -369.73361992\n 4: p1 = 1.68426754 -> excess demand -> -302.76467952\n ...\n 25: p1 = 1.62115861 -> excess demand -> -3.00036721\n 50: p1 = 1.62056537 -> excess demand -> -0.01087860\n 75: p1 = 1.62056322 -> excess demand -> -0.00003940\n100: p1 = 1.62056321 -> excess demand -> -0.00000014\n112: p1 = 1.62056321 -> excess demand -> -0.00000001\n"
]
],
[
[
"**Hint:** The code structure is exactly the same as for the exchange economy considered in the lecture. The code for solving that exchange economy is reproduced in condensed form below.",
"_____no_output_____"
]
],
[
[
"# a. parameters\nN = 1000\nk = 2\nmu_low = 0.1\nmu_high = 0.9\nseed = 1986\n\n# b. draws of random numbers\nnp.random.seed(seed)\nalphas = np.random.uniform(low=mu_low,high=mu_high,size=N)\n\n# c. demand function\ndef demand_good_1_func(alpha,p1,p2,k):\n I = k*p1+p2\n return alpha*I/p1\n\n# d. excess demand function\ndef excess_demand_good_1_func(alphas,p1,p2,k):\n \n # a. demand\n demand = np.sum(demand_good_1_func(alphas,p1,p2,))\n \n # b. supply\n supply = k*alphas.size\n \n # c. excess demand\n excess_demand = demand-supply\n \n return excess_demand\n\n# e. find equilibrium function\ndef find_equilibrium(alphas,p1,p2,k,kappa=0.5,eps=1e-8,maxiter=500):\n \n t = 0\n while True:\n\n # a. step 1: excess demand\n Z1 = excess_demand_good_1_func(alphas,p1,p2,k)\n \n # b: step 2: stop?\n if np.abs(Z1) < eps or t >= maxiter:\n print(f'{t:3d}: p1 = {p1:12.8f} -> excess demand -> {Z1:14.8f}')\n break \n \n # c. step 3: update p1\n p1 = p1 + kappa*Z1/alphas.size\n \n # d. step 4: return \n if t < 5 or t%25 == 0:\n print(f'{t:3d}: p1 = {p1:12.8f} -> excess demand -> {Z1:14.8f}')\n elif t == 5:\n print(' ...')\n \n t += 1 \n\n return p1\n\n# e. call find equilibrium function\np1 = 1.4\np2 = 1\nkappa = 0.1\neps = 1e-8\np1 = find_equilibrium(alphas,p1,p2,k,kappa=kappa,eps=eps)",
" 0: p1 = 1.33690689 -> excess demand -> -630.93108302\n 1: p1 = 1.27551407 -> excess demand -> -613.92820358\n 2: p1 = 1.21593719 -> excess demand -> -595.76882769\n 3: p1 = 1.15829785 -> excess demand -> -576.39340748\n 4: p1 = 1.10272273 -> excess demand -> -555.75114178\n ...\n 25: p1 = 0.53269252 -> excess demand -> -53.80455643\n 50: p1 = 0.50897770 -> excess demand -> -0.27125769\n 75: p1 = 0.50886603 -> excess demand -> -0.00120613\n100: p1 = 0.50886553 -> excess demand -> -0.00000536\n125: p1 = 0.50886553 -> excess demand -> -0.00000002\n130: p1 = 0.50886553 -> excess demand -> -0.00000001\n"
]
],
[
[
"**Answers:**",
"_____no_output_____"
],
[
"See A5.py",
"_____no_output_____"
],
[
"## Save and load",
"_____no_output_____"
],
[
"Consider the code below and fill in the missing lines so the code can run without any errors.",
"_____no_output_____"
]
],
[
[
"import pickle\n\n# a. create some data\nmy_data = {}\nmy_data['A'] = {'a':1,'b':2}\nmy_data['B'] = np.array([1,2,3])\nmy_data['C'] = (1,4,2)\n\nmy_np_data = {}\nmy_np_data['D'] = np.array([1,2,3])\nmy_np_data['E'] = np.zeros((5,8))\nmy_np_data['F'] = np.ones((7,3,8))\n\n# c. save with pickle\nwith open(f'data.p', 'wb') as f:\n pickle.dump(my_data,f)\n \n# d. save with numpy\nnp.savez(f'data.npz', **my_np_data)\n \n# a. try\ndef load_all():\n with open(f'data.p', 'rb') as f:\n data = pickle.load(f)\n A = data['A']\n B = data['B']\n C = data['C']\n\n with np.load(f'data.npz') as data:\n D = data['D']\n E = data['E']\n F = data['F'] \n \n print('variables loaded without error')\n \ntry:\n load_all()\nexcept:\n print('failed')",
"variables loaded without error\n"
]
],
[
[
"**Answer:**",
"_____no_output_____"
],
[
"See A6.py",
"_____no_output_____"
],
[
"# Extra Problems",
"_____no_output_____"
],
[
"## Multiple goods",
"_____no_output_____"
],
[
"Solve the main problem extended with multiple goods:",
"_____no_output_____"
],
[
"$$\n\\begin{aligned}\nu^{j}(x_{1},x_{2}) & = x_{1}^{\\alpha^1_{j}} \\cdot x_{2}^{\\alpha^2_{j}} \\cdots x_{M}^{\\alpha^M_{j}}\\\\\n & \\alpha_j = [\\alpha^1_{j},\\alpha^2_{j},\\dots,\\alpha^M_{j}] \\\\\n & \\log(\\alpha_j) \\sim \\mathcal{N}(0,\\Sigma) \\\\\n\\end{aligned}\n$$\n\nwhere $\\Sigma$ is a valid covariance matrix.",
"_____no_output_____"
]
],
[
[
"# a. choose parameters\nN = 10000\nJ = 3\n\n# b. choose Sigma\nSigma_lower = np.array([[1, 0, 0], [0.5, 1, 0], [0.25, -0.5, 1]])\nSigma_upper = Sigma_lower.T\nSigma = Sigma_upper@Sigma_lower\nprint(Sigma)\n\n# c. draw random numbers\nalphas = np.exp(np.random.multivariate_normal(np.zeros(J), Sigma, 10000))\nprint(np.mean(alphas,axis=0))\nprint(np.corrcoef(alphas.T))\n\ndef demand_good_1_func(alpha,p1,p2,k):\n I = k*p1+p2\n return alpha*I/p1\n\ndef demand_good_2_func(alpha,p1,p2,k):\n I = k*p1+p2\n return (1-alpha)*I/p2\n\ndef excess_demand_good_1_func(alphas,p1,p2,k):\n \n # a. demand\n demand = np.sum(demand_good_1_func(alphas,p1,p2,k))\n \n # b. supply\n supply = k*alphas.size\n \n # c. excess demand\n excess_demand = demand-supply\n \n return excess_demand\n\ndef excess_demand_good_2_func(alphas,p1,p2,k):\n \n # a. demand\n demand = np.sum(demand_good_2_func(alphas,p1,p2,k))\n \n # b. supply\n supply = alphas.size\n \n # c. excess demand\n excess_demand = demand-supply\n \n return excess_demand\n\ndef find_equilibrium(alphas,p1_guess,p2,k,kappa=0.5,eps=1e-8,maxiter=500):\n \n t = 0\n p1 = p1_guess\n \n # using a while loop as we don't know number of iterations a priori\n while True:\n\n # a. step 1: excess demand\n Z1 = excess_demand_good_1_func(alphas,p1,p2,k)\n \n # b: step 2: stop?\n if np.abs(Z1) < eps or t >= maxiter:\n print(f'{t:3d}: p1 = {p1:12.8f} -> excess demand -> {Z1:14.8f}')\n break \n \n # c. step 3: update p1\n p1 = p1 + kappa*Z1/alphas.size\n \n # d. step 4: print only every 25th iteration using the modulus operator \n if t < 5 or t%25 == 0:\n print(f'{t:3d}: p1 = {p1:12.8f} -> excess demand -> {Z1:14.8f}')\n elif t == 5:\n print(' ...')\n \n t += 1 \n\n return p1",
"[[ 1.3125 0.375 0.25 ]\n [ 0.375 1.25 -0.5 ]\n [ 0.25 -0.5 1. ]]\n[1.91709082 1.91100849 1.63670693]\n[[ 1. 0.19955924 0.15149459]\n [ 0.19955924 1. -0.16150109]\n [ 0.15149459 -0.16150109 1. ]]\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
e7914e03e8dfa716d93d5315d30fab2ad8ecf172 | 28,043 | ipynb | Jupyter Notebook | Cleaning_PSGC.ipynb | thinkingmachines/psgc | d5c9603f4bdf9417e46e75bf6c3bc20ab0f9b609 | [
"MIT"
] | 3 | 2020-07-08T07:21:10.000Z | 2021-04-20T08:23:33.000Z | Cleaning_PSGC.ipynb | thinkingmachines/psgc | d5c9603f4bdf9417e46e75bf6c3bc20ab0f9b609 | [
"MIT"
] | 3 | 2019-01-22T02:08:21.000Z | 2021-03-03T04:15:46.000Z | Cleaning_PSGC.ipynb | thinkingmachines/psgc | d5c9603f4bdf9417e46e75bf6c3bc20ab0f9b609 | [
"MIT"
] | 1 | 2021-01-31T15:44:11.000Z | 2021-01-31T15:44:11.000Z | 22.098503 | 400 | 0.524516 | [
[
[
"# Cleaning the Philippine Standard Geographic Code Dataset",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport xlrd\nimport re",
"_____no_output_____"
]
],
[
[
"# Import the PSGC Excel file.\n\nThe Philippine Statistics Authority publishes an updated PSGC file every quarter in the form of an Excel file. The latest link is here: https://psa.gov.ph/classification/psgc/",
"_____no_output_____"
]
],
[
[
"psgc_excel = pd.read_excel(\"data/raw/PSGC_Publication_Sept2018.xlsx\",sheet_name=\"PSGC\")\npsgc_excel.to_csv('data/raw/raw-psgc.csv.gz',encoding=\"utf-8\",compression=\"gzip\")",
"_____no_output_____"
],
[
"psgc = pd.read_csv('data/raw/raw-psgc.csv.gz',encoding=\"utf-8\")\npsgc.info()",
"_____no_output_____"
]
],
[
[
"Convert \"Code\" column to a string and ensure it has leading zeros and is 9-char long.",
"_____no_output_____"
]
],
[
[
"psgc.loc[:,\"Code\"] = psgc.Code.astype(str).str.zfill(9)",
"_____no_output_____"
]
],
[
[
"Drop unused columns:",
"_____no_output_____"
]
],
[
[
"psgc = psgc.loc[:,['Code','Name','Inter-Level']]",
"_____no_output_____"
]
],
[
[
"Normalize column names",
"_____no_output_____"
]
],
[
[
"psgc.columns = ['code','location','interlevel']\npsgc.head()",
"_____no_output_____"
],
[
"psgc['interlevel'].value_counts()",
"_____no_output_____"
],
[
"psgc.head()",
"_____no_output_____"
]
],
[
[
"Create a duplicate of the original PSGC dataframe",
"_____no_output_____"
]
],
[
[
"og_psgc = psgc.copy()",
"_____no_output_____"
]
],
[
[
"# Helpers",
"_____no_output_____"
],
[
"We see that a lot of the locations in the PSGC have alternate names or aliases for each location contained in parentheses. Let's create a regular expression pattern that will extract these as aliases and append these as additional rows to each subset of the data.",
"_____no_output_____"
]
],
[
[
"extract_in_paren = re.compile(r'\\(+([^\\(\\)]+)\\)*')\nremove_in_paren = \"\\(.+\\)\"",
"_____no_output_____"
],
[
"def expand_in_paren(df):\n \n '''\n Denotes original locations\n '''\n df['original'] = True\n \n '''\n Creates a copy of the rows that contain parentheses or have aliases.\n '''\n has_paren = df[df.location.str.contains(\"[\\(\\)]\")]\n has_paren['original'] = False\n \n '''\n Splits locations that contain parentheses into two elements -- what's before the parentheses, and what's within them\n Each of these items is treated as a separate possible alias and appended to the original datasete\n '''\n for i in [0,1]:\n aliases = has_paren.copy()\n aliases['location'] = has_paren.location.str.replace(\"\\)\",\"\").str.split(\"\\(\").str.get(i).str.strip()\n df = df.append(aliases,ignore_index=True)\n \n \n return df.sort_values(by=[\"code\",\"original\"]).reset_index(drop=True)",
"_____no_output_____"
]
],
[
[
"## Clean regions",
"_____no_output_____"
]
],
[
[
"regions = psgc[psgc['interlevel'] == 'Reg'].copy()",
"_____no_output_____"
]
],
[
[
"Alternate names inside parens so we expand those out to a new column named `alias`.",
"_____no_output_____"
]
],
[
[
"regions = expand_in_paren(regions)\nregions",
"_____no_output_____"
]
],
[
[
"## Clean provinces",
"_____no_output_____"
]
],
[
[
"provinces = psgc[psgc['interlevel'] == 'Prov'].copy()",
"_____no_output_____"
],
[
"provinces.head()",
"_____no_output_____"
]
],
[
[
"Seems normal... But let's check for parens just in case:",
"_____no_output_____"
]
],
[
[
"provinces[provinces['location'].str.contains('[\\(\\)]')]",
"_____no_output_____"
]
],
[
[
"Sneaky alternate names!",
"_____no_output_____"
]
],
[
[
"provinces = expand_in_paren(provinces)\nprovinces",
"_____no_output_____"
]
],
[
[
"## Clean districts",
"_____no_output_____"
]
],
[
[
"districts = psgc[psgc['interlevel'] == 'Dist'].copy()",
"_____no_output_____"
],
[
"districts",
"_____no_output_____"
]
],
[
[
"No one writes `NTH DISTRICT (Not a Province)` in their addresses. Let's remove these instances altogether rather than extract these as aliases.",
"_____no_output_____"
]
],
[
[
"districts['location'] = (districts['location']\n .str.replace('\\(Not a Province\\)', '')\n .str.strip()\n .str.split(',',n=1)\n .str.get(1))",
"_____no_output_____"
],
[
"districts",
"_____no_output_____"
]
],
[
[
"## Clean municipalities",
"_____no_output_____"
]
],
[
[
"municipalities = psgc[psgc['interlevel'] == 'Mun'].copy()",
"_____no_output_____"
]
],
[
[
"Checking for alternate names in parentheses:",
"_____no_output_____"
]
],
[
[
"municipalities[municipalities['location'].str.contains('[\\(\\)]')]",
"_____no_output_____"
]
],
[
[
"In some cases the words \"Capital\" are contained in parentheses but these are not aliases. Safe to strip!",
"_____no_output_____"
]
],
[
[
"municipalities['location'] = municipalities['location'].str.replace('\\(Capital\\)', '').str.strip()",
"_____no_output_____"
],
[
"municipalities",
"_____no_output_____"
],
[
"municipalities = expand_in_paren(municipalities)\nmunicipalities.head(30)",
"_____no_output_____"
]
],
[
[
"## Clean cities",
"_____no_output_____"
]
],
[
[
"cities = psgc[psgc['interlevel'] == 'City'].copy()",
"_____no_output_____"
],
[
"cities.head(30)",
"_____no_output_____"
]
],
[
[
"Here we go with the `(Capital)` thing again.",
"_____no_output_____"
]
],
[
[
"cities['location'] = cities['location'].str.replace('\\(Capital\\)', '').str.strip()",
"_____no_output_____"
]
],
[
[
"Checking if there are still stuff with parens:",
"_____no_output_____"
]
],
[
[
"cities[cities['location'].str.contains('[\\(\\)]')].head()",
"_____no_output_____"
]
],
[
[
"A few alterate names!",
"_____no_output_____"
]
],
[
[
"cities = expand_in_paren(cities)\ncities",
"_____no_output_____"
]
],
[
[
"Now what about those `CITY` pre/suffixes?",
"_____no_output_____"
]
],
[
[
"cities[cities['location'].str.contains('CITY')]",
"_____no_output_____"
]
],
[
[
"Let's strip any prefixes of \"CITY OF\" and suffixes of \"CITY.\"",
"_____no_output_____"
]
],
[
[
"cities['location'] = (cities['location']\n .str.replace('^.*CITY OF', '') #stripping prefixes\n .str.strip()\n .str.replace('CITY$', '') #stripping suffixes\n .str.strip())",
"_____no_output_____"
],
[
"cities",
"_____no_output_____"
]
],
[
[
"## Clean sub-municipalities\n\nManila is the only city-slash-district that has submunicipalities.",
"_____no_output_____"
]
],
[
[
"sub_municipalities = psgc[psgc['interlevel'] == 'SubMun'].copy()",
"_____no_output_____"
],
[
"sub_municipalities",
"_____no_output_____"
]
],
[
[
"Nothing special!",
"_____no_output_____"
],
[
"## Clean barangays",
"_____no_output_____"
]
],
[
[
"barangays = psgc[psgc['interlevel'] == 'Bgy'].copy()\nbarangays",
"_____no_output_____"
]
],
[
[
"We see alternate names again but notice the `(Pob.)` suffixes. A quick Google search shows that it's short for `Poblacion` which is used to denote the commercial and industrial center of a city. Let's stash those and add them as aliases",
"_____no_output_____"
]
],
[
[
"barangays_pob = barangays[barangays.location.str.contains('\\(Pob.\\)')].copy()",
"_____no_output_____"
],
[
"barangays['location'] = (barangays['location']\n .str.replace('(\\(Pob\\.\\))', '') #totally do away with any poblacion suffixes\n .str.strip())\nbarangays['location'].head(30)",
"_____no_output_____"
]
],
[
[
"How many other barangay names contain parentheses?",
"_____no_output_____"
]
],
[
[
"barangays[barangays.location.str.contains(r'[\\(\\)]')]",
"_____no_output_____"
]
],
[
[
"While parentheses often contain aliases, sometimes, these are not aliases but the name of the municipality in which the barangay is located. For example, barangays in the municipality of Dumalneg have the `(Dumalneg)` denoted in parentheses. We'll go ahead and extract parenthetical names as aliases for now, but we'll later remove instances in which aliases are equal to the municipality name.",
"_____no_output_____"
]
],
[
[
"barangays = expand_in_paren(barangays)",
"_____no_output_____"
]
],
[
[
"Let's check for more weird characters:",
"_____no_output_____"
]
],
[
[
"barangays[barangays['location'].str.contains(r'[^a-zA-Z0-9\\sÑñ\\(\\)]')]",
"_____no_output_____"
]
],
[
[
"Lets extract the strings that follow a \"Brgy No. X\" as aliases.",
"_____no_output_____"
]
],
[
[
"pat_barangay = re.compile('(B[gr]y. No. \\d+\\-?\\w?),? (.+)')",
"_____no_output_____"
],
[
"len(barangays[barangays.location.str.contains(pat_barangay)])",
"_____no_output_____"
],
[
"def expand_barangays(df):\n \n '''\n Denotes original locations\n '''\n df['original'] = True\n \n '''\n Creates a copy of the rows that contain barangay pattern\n '''\n matches_pattern = df[df.location.str.contains(pat_barangay)]\n matches_pattern['original'] = False\n \n '''\n Splits locations that into two elements -- Brgy No X and the name that comes after it\n Each of these items is treated as a separate possible alias and appended to the original datasete\n '''\n for i in [0,1]:\n aliases = matches_pattern.copy()\n aliases['location'] = matches_pattern.location.str.extract(pat_barangay)[i]#.str.get(i).str.strip()\n aliases['location'] = aliases['location'].str.strip()\n df = df.append(aliases,ignore_index=True)\n \n return df.sort_values(by=[\"code\",\"original\"]).reset_index(drop=True)",
"_____no_output_____"
],
[
"#print len(barangays)\nbarangays = expand_barangays(barangays)\n#print len(barangays)",
"_____no_output_____"
],
[
"barangays.head()",
"_____no_output_____"
]
],
[
[
"Add barangays that are `Poblacion` as aliases",
"_____no_output_____"
]
],
[
[
"barangays_pob['original'] = False\nbarangays = barangays.append(barangays_pob, ignore_index=True)\nbarangays[barangays.code == '012801001']",
"_____no_output_____"
]
],
[
[
"Last check!",
"_____no_output_____"
]
],
[
[
"barangays.info()",
"_____no_output_____"
],
[
"barangays[barangays.code == \"012812026\"]",
"_____no_output_____"
]
],
[
[
"## ARMM: Cotabato and Isabela City",
"_____no_output_____"
]
],
[
[
"armm = psgc[psgc['interlevel'].isnull()].copy()\narmm",
"_____no_output_____"
],
[
"armm['location'] = armm['location'].str.replace('\\(Not a Province\\)', '')\narmm",
"_____no_output_____"
],
[
"armm['location'] = (armm['location']\n .str.replace('^.*CITY OF', '')\n .str.strip()\n .str.replace('CITY$', '')\n .str.strip())\narmm",
"_____no_output_____"
],
[
"armm['original'] = True\narmm",
"_____no_output_____"
]
],
[
[
"## All together now",
"_____no_output_____"
]
],
[
[
"merged = pd.concat([\n regions,\n provinces,\n districts,\n municipalities,\n cities,\n sub_municipalities,\n barangays,\n armm\n],ignore_index=True).sort_index().fillna('')",
"_____no_output_____"
],
[
"merged.info()",
"_____no_output_____"
]
],
[
[
"Are counts still correct?",
"_____no_output_____"
]
],
[
[
"psgc['interlevel'].value_counts()",
"_____no_output_____"
],
[
"merged['interlevel'].value_counts()",
"_____no_output_____"
],
[
"merged.code.nunique(), psgc.code.nunique()",
"_____no_output_____"
]
],
[
[
"## Normalize numbers:",
"_____no_output_____"
]
],
[
[
"spanish = merged[merged['location'].str.contains(' (UNO|DOS|TRES|KUATRO|SINGKO)$',case=False)].copy()",
"_____no_output_____"
],
[
"spanish",
"_____no_output_____"
],
[
"for i, s in enumerate([\n 'Uno',\n 'Dos',\n 'Tres',\n 'Kuatro',\n 'Singko',\n]):\n spanish['location'] = spanish['location'].str.replace(' {}$'.format(s), ' {}'.format(i + 1))\nspanish\nspanish['original'] = False\nspanish",
"_____no_output_____"
],
[
"roman = merged[merged['location'].str.contains('\\s(X{0,3})(IX|IV|V?I{0,3})$')].copy()",
"_____no_output_____"
],
[
"for i, s in enumerate('I,II,III,IV,V,VI,VII,VIII,IX,X,XI,XII,XIII,XIV,XV,XVI,XVII,XVIII,XIX,XX,XXI,XXII'.split(',')):\n roman['location'] = roman['location'].str.replace(' {}$'.format(s), ' {}'.format(i + 1))\nroman['original'] = False\nroman",
"_____no_output_____"
]
],
[
[
"Provide alternate names for locations with President names",
"_____no_output_____"
]
],
[
[
"president = merged[merged.location.str.contains('PRES\\.', flags=re.IGNORECASE)].copy()\npresident['location'] = president['location'].str.replace('^PRES\\.', 'PRESIDENT')\npresident['location'] = president['location'].str.replace('^Pres\\.', 'President')\npresident['original'] = False\npresident",
"_____no_output_____"
]
],
[
[
"# Add alternative names to Metro Manila",
"_____no_output_____"
]
],
[
[
"metro_manila = pd.DataFrame([{\"code\":\"130000000\",\"interlevel\":\"Reg\",\"location\":\"Metro Manila\",\"original\":False},\n {\"code\":\"130000000\",\"interlevel\":\"Reg\",\"location\":\"Metropolitan Manila\",\"original\":False}])\n\nmetro_manila",
"_____no_output_____"
]
],
[
[
"# Add Ñ -> N as an alternate name\n",
"_____no_output_____"
]
],
[
[
"merged[merged.location.str.contains('Las Piñas',case=False)]",
"_____no_output_____"
],
[
"enye = merged[merged.location.str.contains(r'[Ññ]')].copy()\nenye.head()",
"_____no_output_____"
],
[
"enye['location'] = (enye['location'].str.replace('Ñ', 'N')\n .str.replace('ñ','n'))\nenye.head()",
"_____no_output_____"
]
],
[
[
"# Concat the alternates to the main dataframe",
"_____no_output_____"
]
],
[
[
"clean_psgc = (pd.concat([merged, spanish, roman, president], ignore_index=True)\n .sort_values('code')\n .reset_index(drop=True))",
"_____no_output_____"
]
],
[
[
"Last check for weird stuff!",
"_____no_output_____"
]
],
[
[
"clean_psgc[clean_psgc['location'].str.contains('[^a-zA-Z0-9 \\-.,\\']')]",
"_____no_output_____"
]
],
[
[
"We can probably still split with `&` and `/` but this is good enough for now.",
"_____no_output_____"
],
[
"# Combine the cleaned up PSGC and remove the duplicates",
"_____no_output_____"
]
],
[
[
"clean_psgc.drop_duplicates(subset=['code', 'location', 'interlevel'], inplace=True)\nclean_psgc.reset_index(drop=True).sort_values('code', inplace=True)",
"_____no_output_____"
]
],
[
[
"Check that we have both the original name and the alternate ones",
"_____no_output_____"
]
],
[
[
"clean_psgc[clean_psgc.code.str.contains('086000000')]",
"_____no_output_____"
],
[
"clean_psgc[clean_psgc.code.str.contains('012801001')]",
"_____no_output_____"
],
[
"clean_psgc.info()",
"_____no_output_____"
]
],
[
[
"# Cleaning out rows in which the alternate name of the barangay was just the name of its parent municipality or city",
"_____no_output_____"
]
],
[
[
"clean_psgc['municipality_code'] = clean_psgc.code.str.slice(0,6)+\"000\"\nclean_psgc['municipality'] = clean_psgc['municipality_code'].map(municipalities[municipalities.original==True].set_index('code').location)\nclean_psgc.head(10)",
"_____no_output_____"
],
[
"clean_psgc['drop'] = (clean_psgc.municipality == clean_psgc.location.str.upper()) & (clean_psgc.interlevel == \"Bgy\")",
"_____no_output_____"
],
[
"barangay_and_muni_same_name = clean_psgc.groupby('code').drop.value_counts().unstack()[False][clean_psgc.groupby('code').drop.value_counts().unstack()[False].isnull()].index\nclean_psgc.loc[clean_psgc.code.isin(barangay_and_muni_same_name),\"drop\"] = False",
"_____no_output_____"
],
[
"clean_psgc[clean_psgc.code == '013301034']",
"_____no_output_____"
],
[
"clean_psgc = clean_psgc.loc[clean_psgc['drop'] ==False,['code','interlevel','location','original']].reset_index(drop=True)",
"_____no_output_____"
],
[
"clean_psgc[clean_psgc.code == \"133900000\"]",
"_____no_output_____"
]
],
[
[
"# Create aliases for Legazpi and Ozamiz",
"_____no_output_____"
]
],
[
[
"zplaces = clean_psgc[clean_psgc.location.str.upper().isin([\"LEGAZPI\",\"OZAMIZ\"])].copy()\nzplaces.loc[:,'location'] = [\"LEGASPI\",\"OZAMIS\"]\nzplaces",
"_____no_output_____"
],
[
"clean_psgc = clean_psgc.append(zplaces,ignore_index=True)\nclean_psgc",
"_____no_output_____"
],
[
"clean_psgc.to_csv('data/processed/clean-psgc.csv.gz', index=False, compression='gzip')",
"_____no_output_____"
]
],
[
[
"And we're done!",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
e79152caebbe4de1b0106699b8fbe09f531e23b8 | 302,054 | ipynb | Jupyter Notebook | EKE_eddy_SEASONAL_VB01.ipynb | Josue-Martinez-Moreno/phd_source | add2aa0ff3e8fc4596d4dc9504e2b80c3d42a3e5 | [
"MIT"
] | 2 | 2021-07-28T14:28:36.000Z | 2022-01-26T06:37:51.000Z | EKE_eddy_SEASONAL_VB01.ipynb | Josue-Martinez-Moreno/phd_source | add2aa0ff3e8fc4596d4dc9504e2b80c3d42a3e5 | [
"MIT"
] | null | null | null | EKE_eddy_SEASONAL_VB01.ipynb | Josue-Martinez-Moreno/phd_source | add2aa0ff3e8fc4596d4dc9504e2b80c3d42a3e5 | [
"MIT"
] | null | null | null | 2,288.287879 | 298,860 | 0.963265 | [
[
[
"# Importing all libraries.\nfrom pylab import *\nfrom netCDF4 import Dataset\n%matplotlib inline\nimport os\nimport cmocean as cm\nfrom trackeddy.tracking import *\nfrom trackeddy.datastruct import *\nfrom trackeddy.geometryfunc import *\nfrom trackeddy.init import *\nfrom trackeddy.physics import *\nfrom trackeddy.plotfunc import *\n\nimport matplotlib.pyplot as plt, mpld3\n\nimport datetime\n\n#import cosima_cookbook as cc",
"_____no_output_____"
],
[
"path=\"/g/data/v45/jm5970/trackeddy_out/output/\"\neke_time=[]\nfor ii in range(306,345):\n try:\n ekefield=Dataset(path+'EKE_eddy'+str(ii)+'.nc')\n eke=ekefield.variables['EKE_eddy']\n for tt in range(0,shape(eke)[0]):\n eke_time.append(mean(eke[tt,:,:]))\n except:\n print('Dataset not Found.')\n reconstruct=Dataset(path+'reconstructed_field_'+str(ii)+'.nc')\n time=reconstruct.variables['time']\n for tt in range(0,len(time)):\n eke_time.append(np.nan)",
"Dataset not Found.\nDataset not Found.\n"
],
[
"eke_time=asarray(eke_time)\neke_time[eke_time<5]=np.nan\n\n\nbase = datetime.datetime(1993, 1, 1, 0, 0)\ndate_list = [base + datetime.timedelta(days=x) for x in range(0, len(eke_time))]\n\nfig, ax = plt.subplots(figsize=(15,3),dpi=300)\nmonthsFmt = DateFormatter(\"%b - %Y\")\nax.xaxis.set_major_formatter(monthsFmt)\nax.grid(True)\nax.set_ylabel(r'$EKE_{eddy} (m^2/s^2)$')\nax.plot(date_list,eke_time)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
e79154eba994cb40ae7fb218981112ca27c78e7e | 2,791 | ipynb | Jupyter Notebook | 01-Step1-PatternDetection/.ipynb_checkpoints/01-StructuralTopicModel-checkpoint.ipynb | cxomni/computational-grounded-theory | c16ca0d7f159aed43f077c419293040df7b8feaf | [
"BSD-3-Clause"
] | 38 | 2017-08-14T19:10:33.000Z | 2022-02-02T19:25:16.000Z | 01-Step1-PatternDetection/.ipynb_checkpoints/01-StructuralTopicModel-checkpoint.ipynb | cxomni/computational-grounded-theory | c16ca0d7f159aed43f077c419293040df7b8feaf | [
"BSD-3-Clause"
] | null | null | null | 01-Step1-PatternDetection/.ipynb_checkpoints/01-StructuralTopicModel-checkpoint.ipynb | cxomni/computational-grounded-theory | c16ca0d7f159aed43f077c419293040df7b8feaf | [
"BSD-3-Clause"
] | 11 | 2017-07-28T21:21:50.000Z | 2022-01-27T08:50:37.000Z | 22.691057 | 245 | 0.536009 | [
[
[
"### Structural Topic Model\n\nThis R notebook reproduces the Structural Topic Model used in Step 1 of the Computational Grounded Theory project.\n\nNote: This notebook produces the model and then saves it. Producing the model can take quite a bit of time to run, upwards of four hours. To explore the topic models produced skip directly to the next notebook, `02-TopicExploration.ipynb`.",
"_____no_output_____"
],
[
"### Requirements and Dependencies\n\nModel created using R 3.4.0 \n\nMain libary: stm_1.2.2 \n\nDependencies:\n* tm_0.7-1\n* NLP_0.1-10\n* SnowballC_0.5.1\n\n\n",
"_____no_output_____"
]
],
[
[
"library(stm)\n\n### Load Data\ndf <- read.csv('../data/comparativewomensmovement_dataset.csv', sep='\\t')",
"stm v1.1.3 (2016-01-14) successfully loaded. See ?stm for help.\n"
],
[
"##Pre-Processing\n\ntemp<-textProcessor(documents=df$text_string,metadata=df)\nmeta<-temp$meta\nvocab<-temp$vocab\ndocs<-temp$documents\nout <- prepDocuments(docs, vocab, meta)\ndocs<-out$documents\nvocab<-out$vocab\nmeta <-out$meta",
"_____no_output_____"
],
[
"##Produce Models\n\n### Model search across numbers of topics\n\nstorage <- manyTopics(docs,vocab,K=c(20,30,40,50), prevalence=~org, data=meta, seed = 1234)\n\nmod.20 <- storage$out[[1]]\nmod.30 <- storage$out[[2]] \nmod.40 <- storage$out[[3]] \nmod.50 <- storage$out[[4]] ",
"_____no_output_____"
],
[
"##Save Full Model, with four different topic models saved\nsave.image(\"../data/stm_all.RData\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
e791664753cc73efe17a584326956368c6eb5605 | 18,924 | ipynb | Jupyter Notebook | regression_by_cadence.ipynb | bruennijs/indoor-virtual-power-prediction | d0cb263bb733a043cca7cd3e8753007bb12e88ad | [
"MIT"
] | null | null | null | regression_by_cadence.ipynb | bruennijs/indoor-virtual-power-prediction | d0cb263bb733a043cca7cd3e8753007bb12e88ad | [
"MIT"
] | null | null | null | regression_by_cadence.ipynb | bruennijs/indoor-virtual-power-prediction | d0cb263bb733a043cca7cd3e8753007bb12e88ad | [
"MIT"
] | null | null | null | 87.611111 | 13,471 | 0.851406 | [
[
[
"# Introduction\nThe bike has 20 gears which are the categories/labels of the classification. Features are cadence and speed with data of the trainings app. We train our model with data sets of all 20 gears (means 20 tcx files loaded with labeled oberservations).",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\nfrom src.regression import validate_lin_reg\nfrom src.tcx import Tcx, COLUMN_NAME_SPEED, COLUMN_NAME_WATTS, COLUMN_NAME_CADENCE\nfrom src.test_data import TrainDataSet\nfrom src.visu import plot2d\nimport matplotlib.pyplot as plt\n\n\ntcx_app_gear7: Tcx = Tcx.read_tcx(file_path='test/tcx/cadence_1612535177298-gear7.tcx')\ntcx_app_gear20: Tcx = Tcx.read_tcx(file_path='test/tcx/cadence_1612535671464-gear20.tcx')\ntcx_tacx_gear7: Tcx = Tcx.read_tcx(file_path='test/tcx/tacx-activity_6225123072-gear7-resistance3.tcx')\ntcx_tacx_gear20: Tcx = Tcx.read_tcx(file_path='test/tcx/tacx-activity_6225123072-gear7-resistance3.tcx')\n\n# generate test data\ndts_gear7: TrainDataSet = TrainDataSet(tcx_app_gear7)\ndts_gear20: TrainDataSet = TrainDataSet(tcx_app_gear20)\ndts_tacx_gear7: TrainDataSet = TrainDataSet(tcx_tacx_gear7)",
"_____no_output_____"
]
],
[
[
"# Problem\nFind cadence for a gear that the tacx data set is of. the app data will measure speed and a linear regression model of the same gear predicts the cadence by that speed. A second linear regression model maps cadence to power of the tacx data set.\n\n# Solution\n## Train (app data)\n* X of of gear _n_ in app data set: [speed]\n* Y -> [cadence]\n\n### Linear model",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LinearRegression\n\nX_train, y_train = dts_gear7.cadence_to_speed()\nlr_app_gear7 = LinearRegression().fit(X_train, y_train)",
"_____no_output_____"
]
],
[
[
"## Train (tacx)\n* X of of gear _n_ in app data set: [cadence]\n* Y -> [power]\n\n### Analyze\nLet us first plot the features to see which regression model fits best",
"_____no_output_____"
]
],
[
[
"X, y = dts_tacx_gear7.cadence_to_power()\nplot2d(X.iloc[:,0], y, point_color='red', legend_label='gear 7 (tacx)')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Linear model",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LinearRegression\n\nlr_tacx_gear7 = LinearRegression().fit(X, y)",
"_____no_output_____"
]
],
[
[
"### Validation\nCross validation with X_test of tacx data and validate the score of the predicted values",
"_____no_output_____"
]
],
[
[
"random_state = 2\nX_train, X_test, y_train, y_test = train_test_split(X, y, train_size=0.8, random_state=random_state)\nvalidate_lin_reg(X_train, y_train, X_test, y_test, LinearRegression())\n\n\n",
"Shape X_train/X_test: (357, 1)/(90, 1)\nError R²: 1.00\nMSE error (mean squared error / variance): 1.06\nsqrt(MSE) (standard deviation): 1.03\nMax error: 2.8912017526499483\nestimator.coefficients: [1.70889424]\nCross validation: [0.99506023 0.99816251 0.9957887 0.99589043 0.99734035]\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7916ab66b48fd05ee959c2539fd902862c7bd82 | 12,266 | ipynb | Jupyter Notebook | module_2_programming/lists.ipynb | wiplane/foundations-of-datascience-ml | 289f8cddd8e91bc6ef7827c57e9e5ecdd574e596 | [
"MIT"
] | 3 | 2021-08-31T14:31:18.000Z | 2021-09-16T07:30:32.000Z | module_2_programming/lists.ipynb | wiplane/foundations-of-datascience-ml | 289f8cddd8e91bc6ef7827c57e9e5ecdd574e596 | [
"MIT"
] | null | null | null | module_2_programming/lists.ipynb | wiplane/foundations-of-datascience-ml | 289f8cddd8e91bc6ef7827c57e9e5ecdd574e596 | [
"MIT"
] | 3 | 2021-09-03T13:13:56.000Z | 2022-02-23T22:32:01.000Z | 16.442359 | 79 | 0.429806 | [
[
[
"## Learning objectives:\n\n* Introduction to lists.\n* How to create and access elements from a list.\n* Adding, removing and changing the elements of the list.",
"_____no_output_____"
]
],
[
[
"alist = [2,3,45,'python', -98]\nalist",
"_____no_output_____"
],
[
"names = ['Tom', 'Mak', 'Arjun', 'Rahul']\nnames",
"_____no_output_____"
],
[
"blist = [1,2,3,4, [-1, -2, -3], 'python', names]\nblist",
"_____no_output_____"
]
],
[
[
"## Access elements of a list",
"_____no_output_____"
]
],
[
[
"alist",
"_____no_output_____"
],
[
"alist[2]",
"_____no_output_____"
],
[
"alist[3]",
"_____no_output_____"
],
[
"alist[4]",
"_____no_output_____"
],
[
"alist[-1]",
"_____no_output_____"
],
[
"alist[-5]",
"_____no_output_____"
]
],
[
[
"## Modifying a list",
"_____no_output_____"
]
],
[
[
"alist",
"_____no_output_____"
],
[
"alist[3] = 50\nprint(alist)",
"[2, 3, 45, 50, -98]\n"
],
[
"alist.append(100)",
"_____no_output_____"
],
[
"alist",
"_____no_output_____"
],
[
"alist.insert(3, 1000)\nprint(alist)",
"[2, 3, 45, 1000, 50, -98, 100]\n"
],
[
"alist.pop()",
"_____no_output_____"
],
[
"alist",
"_____no_output_____"
],
[
"alist.remove(1000)",
"_____no_output_____"
],
[
"alist",
"_____no_output_____"
]
],
[
[
"## Slicing a list to obtain a subset of values",
"_____no_output_____"
]
],
[
[
"alist = [9, 10, -1, 2, 5, 7]\nalist",
"_____no_output_____"
],
[
"alist[1:4:1]",
"_____no_output_____"
],
[
"alist[1:4]",
"_____no_output_____"
],
[
"alist[1:4:2]",
"_____no_output_____"
],
[
"alist[:]",
"_____no_output_____"
],
[
"alist[2:]",
"_____no_output_____"
],
[
"alist[:4]",
"_____no_output_____"
]
],
[
[
"## Sort a list",
"_____no_output_____"
]
],
[
[
"item_prices = [1200, 200, 25, 500.45, 234, 540]\nitem_prices",
"_____no_output_____"
],
[
"##sort the list\nitem_prices.sort()\nprint(item_prices)\n",
"[25, 200, 234, 500.45, 540, 1200]\n"
],
[
"item_prices.sort(reverse=True)\nprint(item_prices)",
"[1200, 540, 500.45, 234, 200, 25]\n"
],
[
"names",
"_____no_output_____"
],
[
"names.sort()",
"_____no_output_____"
],
[
"names",
"_____no_output_____"
],
[
"len(names)",
"_____no_output_____"
],
[
"len(item_prices)",
"_____no_output_____"
],
[
"item_prices.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7917bb9e4cc791965767dc95756e672b7f8008b | 4,383 | ipynb | Jupyter Notebook | examples/gallery/demos/matplotlib/dragon_curve.ipynb | jsignell/holoviews | 4f9fd27367f23c3d067d176f638ec82e4b9ec8f0 | [
"BSD-3-Clause"
] | 2 | 2020-08-13T00:11:46.000Z | 2021-01-31T22:13:21.000Z | examples/gallery/demos/matplotlib/dragon_curve.ipynb | jsignell/holoviews | 4f9fd27367f23c3d067d176f638ec82e4b9ec8f0 | [
"BSD-3-Clause"
] | null | null | null | examples/gallery/demos/matplotlib/dragon_curve.ipynb | jsignell/holoviews | 4f9fd27367f23c3d067d176f638ec82e4b9ec8f0 | [
"BSD-3-Clause"
] | 1 | 2021-10-31T05:26:08.000Z | 2021-10-31T05:26:08.000Z | 31.532374 | 232 | 0.496235 | [
[
[
"Dragon curve example from the [L-systems](../../topics/geometry/lsystems.ipynb) topic notebook in ``examples/topics/geometry``.\n\nMost examples work across multiple plotting backends, this example is also available for:\n* [Bokeh - dragon_curve](../bokeh/dragon_curve.ipynb)",
"_____no_output_____"
]
],
[
[
"import holoviews as hv\nimport numpy as np\nhv.extension('matplotlib')",
"_____no_output_____"
]
],
[
[
"## L-system definition",
"_____no_output_____"
],
[
"The following class is a simplified version of the approach used in the [L-systems](../../topics/geometry/lsystems.ipynb) notebook, made specifically for plotting the [Dragon Curve](https://en.wikipedia.org/wiki/Dragon_curve).",
"_____no_output_____"
]
],
[
[
"class DragonCurve(object):\n \"L-system agent that follows rules to generate the Dragon Curve\"\n \n initial ='FX'\n productions = {'X':'X+YF+', 'Y':'-FX-Y'}\n dragon_rules = {'F': lambda t,d,a: t.forward(d),\n 'B': lambda t,d,a: t.back(d),\n '+': lambda t,d,a: t.rotate(-a),\n '-': lambda t,d,a: t.rotate(a),\n 'X':lambda t,d,a: None,\n 'Y':lambda t,d,a: None }\n \n def __init__(self, x=0,y=0, iterations=1):\n self.heading = 0\n self.distance = 5\n self.angle = 90\n self.x, self.y = x,y\n self.trace = [(self.x, self.y)]\n self.process(self.expand(iterations), self.distance, self.angle)\n \n def process(self, instructions, distance, angle):\n for i in instructions: \n self.dragon_rules[i](self, distance, angle)\n \n def expand(self, iterations):\n \"Expand an initial symbol with the given production rules\"\n expansion = self.initial\n \n for i in range(iterations):\n intermediate = \"\"\n for ch in expansion:\n intermediate = intermediate + self.productions.get(ch,ch)\n expansion = intermediate\n return expansion\n\n def forward(self, distance):\n self.x += np.cos(2*np.pi * self.heading/360.0)\n self.y += np.sin(2*np.pi * self.heading/360.0)\n self.trace.append((self.x,self.y))\n \n def rotate(self, angle):\n self.heading += angle\n \n def back(self, distance):\n self.heading += 180\n self.forward(distance)\n self.heading += 180\n \n @property\n def path(self):\n return hv.Path([self.trace])",
"_____no_output_____"
]
],
[
[
"## Plot",
"_____no_output_____"
]
],
[
[
"%%output size=200\n%%opts Path {+framewise} [xaxis=None yaxis=None title_format=''] (color='black' linewidth=1)\n\ndef pad_extents(path):\n \"Add 5% padding around the path\"\n minx, maxx = path.range('x')\n miny, maxy = path.range('y')\n xpadding = ((maxx-minx) * 0.1)/2\n ypadding = ((maxy-miny) * 0.1)/2\n path.extents = (minx-xpadding, miny-ypadding, maxx+xpadding, maxy+ypadding)\n return path\n \nhmap = hv.HoloMap(kdims='Iteration')\nfor i in range(7,17):\n path = DragonCurve(-200, 0, i).path\n hmap[i] = pad_extents(path)\nhmap",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e791bb1fad6c74eeb600fa771bbb4e6790d943b8 | 62,568 | ipynb | Jupyter Notebook | HURDAT_JRDISCHG-Abby1968.ipynb | williampc8985/VT-JamesRiver | 6bacd10f4fd6158db74973ddc1abd89b650efc9f | [
"MIT"
] | null | null | null | HURDAT_JRDISCHG-Abby1968.ipynb | williampc8985/VT-JamesRiver | 6bacd10f4fd6158db74973ddc1abd89b650efc9f | [
"MIT"
] | null | null | null | HURDAT_JRDISCHG-Abby1968.ipynb | williampc8985/VT-JamesRiver | 6bacd10f4fd6158db74973ddc1abd89b650efc9f | [
"MIT"
] | null | null | null | 155.641791 | 51,476 | 0.88379 | [
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"#This is the Richmond USGS Data gage\nriver_richmnd = pd.read_csv('JR_Richmond02037500.csv')",
"/Users/williampc/opt/anaconda3/envs/geop/lib/python3.9/site-packages/IPython/core/interactiveshell.py:3165: DtypeWarning: Columns (7) have mixed types.Specify dtype option on import or set low_memory=False.\n has_raised = await self.run_ast_nodes(code_ast.body, cell_name,\n"
],
[
"river_richmnd.dropna();",
"_____no_output_____"
],
[
"#Hurricane data for the basin - Names of Relevant Storms - This will be used for getting the storms from the larger set\nJR_stormnames = pd.read_csv('gis_match.csv')\n",
"_____no_output_____"
],
[
"# Bring in the Big HURDAT data, from 1950 forward (satellites and data quality, etc.)\nHURDAT = pd.read_csv('hurdatcleanva_1950_present.csv')\n",
"_____no_output_____"
],
[
"VA_JR_stormmatch = JR_stormnames.merge(HURDAT)\n",
"_____no_output_____"
],
[
"# Now the common storms for the James Basin have been created. We now have time and storms together for the basin\n#checking some things about the data",
"_____no_output_____"
],
[
"# How many unique storms within the basin since 1950? 62 here and 53 in the Data on the Coast.NOAA.gov's website. \n#I think we are close enough here, digging may show some other storms, but I think we have at least captured the ones \n#from NOAA\nlen(VA_JR_stormmatch['Storm Number'].unique());",
"_____no_output_____"
],
[
"#double ck the lat and long parameters\nprint(VA_JR_stormmatch['Lat'].min(),\nVA_JR_stormmatch['Lon'].min(),\nVA_JR_stormmatch['Lat'].max(),\nVA_JR_stormmatch['Lon'].max())",
"36.1 -83.7 39.9 -75.1\n"
],
[
"#Make a csv of this data\nVA_JR_stormmatch.to_csv('storms_in_basin.csv', sep=',',encoding = 'utf-8')",
"_____no_output_____"
],
[
"#names of storms \nlen(VA_JR_stormmatch['Storm Number'].unique())\nVA_JR_stormmatch['Storm Number'].unique()\nnumbers = VA_JR_stormmatch['Storm Number']",
"_____no_output_____"
],
[
"#grab a storm from this list and lok at the times\n#Bill = pd.DataFrame(VA_JR_stormmatch['Storm Number'=='AL032003'])\n\nAbby = VA_JR_stormmatch[(VA_JR_stormmatch[\"Storm Number\"] == 'AL011968')]\nAbby\n#so this is the data for a storm named Bill that had a pth through the basin * BILL WAS A BACKDOOR Storm\n\n",
"_____no_output_____"
],
[
"# plotting for the USGS river Gage data \nimport matplotlib\nimport matplotlib.pyplot as plt\nfrom climata.usgs import DailyValueIO\nfrom datetime import datetime\nfrom pandas.plotting import register_matplotlib_converters\nimport numpy as np\n\nregister_matplotlib_converters()\nplt.style.use('ggplot')\nplt.rcParams['figure.figsize'] = (20.0, 10.0)\n# set parameters\nnyears = 1\nndays = 365 * nyears\nstation_id = \"02037500\"\nparam_id = \"00060\"\n\ndatelist = pd.date_range(end=datetime.today(), periods=ndays).tolist()\n#take an annual average for the river\nannual_data = DailyValueIO(\n start_date=\"1968-01-01\",\n end_date=\"1969-01-01\",\n station=station_id,\n parameter=param_id,)\nfor series in annual_data:\n flow = [r[1] for r in series.data]\n si_flow_annual = np.asarray(flow) * 0.0283168\n flow_mean = np.mean(si_flow_annual)\n\n#now for the storm - Florence \ndischg = DailyValueIO(\n start_date=\"1968-06-05\",\n end_date=\"1968-06-19\",\n station=station_id,\n parameter=param_id,)\n#create lists of date-flow values\nfor series in dischg:\n flow = [r[1] for r in series.data]\n si_flow = np.asarray(flow) * 0.0283168\n dates = [r[0] for r in series.data]\nplt.plot(dates, si_flow)\nplt.axhline(y=flow_mean, color='r', linestyle='-')\nplt.xlabel('Date')\nplt.ylabel('Discharge (m^3/s)')\nplt.title(\"TD Abby - 1968 (Atlantic)\")\nplt.xticks(rotation='vertical')\nplt.show()",
"_____no_output_____"
],
[
"percent_incr= (abs(max(si_flow)-flow_mean)/abs(flow_mean))*100\npercent_incr",
"_____no_output_____"
],
[
"#take an annual average for the river\nannual_data = DailyValueIO(\n start_date=\"1968-03-01\",\n end_date=\"1968-10-01\",\n station=station_id,\n parameter=param_id,)\nfor series in annual_data:\n flow = [r[1] for r in series.data]\n si_flow_annual = np.asarray(flow) * 0.0283168\n flow_mean_season = np.mean(si_flow_annual)\nprint(abs(flow_mean-flow_mean_season))",
"12.282820127251782\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e791cc7aa494dec18c5e004432e023d062669889 | 2,461 | ipynb | Jupyter Notebook | research/oratory1990_crinacle.ipynb | vinzmc/AutoEq | 4b42aa25e5f4933528be44e7356afe1fde75a3af | [
"MIT"
] | 6,741 | 2018-07-27T10:54:04.000Z | 2022-03-31T20:22:57.000Z | research/oratory1990_crinacle.ipynb | vinzmc/AutoEq | 4b42aa25e5f4933528be44e7356afe1fde75a3af | [
"MIT"
] | 428 | 2018-08-08T17:12:40.000Z | 2022-03-31T05:53:36.000Z | research/oratory1990_crinacle.ipynb | vinzmc/AutoEq | 4b42aa25e5f4933528be44e7356afe1fde75a3af | [
"MIT"
] | 1,767 | 2018-07-27T16:50:12.000Z | 2022-03-31T19:26:39.000Z | 23.893204 | 91 | 0.524584 | [
[
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"import os\nimport sys\nfrom pathlib import Path\nROOT_DIR = os.path.abspath(os.path.join(Path().absolute(), os.pardir))\nsys.path.insert(1, ROOT_DIR)\nDIR_PATH = os.path.join(ROOT_DIR, 'research')",
"_____no_output_____"
],
[
"from glob import glob\nimport numpy as np\nimport scipy\nimport matplotlib.pyplot as plt\nimport PyPDF2\nfrom frequency_response import FrequencyResponse",
"_____no_output_____"
],
[
"# Read headphone model from the PDF\nfor fp in glob(os.path.join(ROOT_DIR, 'measurements', 'oratory1990', 'pdf', '*')):\n _, name = os.path.split(fp)\n name = name.replace('.pdf', '')\n f = open(fp, 'rb')\n text = PyPDF2.PdfFileReader(f).getPage(0).extractText()\n #print(f'\"crinacle\" in PDF: {\"crinacle\" in text.lower()}')\n lines = text.split('\\n')\n for line in lines:\n if 'crinacle' in line.lower():\n print(f'{name}: \"{line}\"')\n f.close()",
"DUNU Titan 3: \"measured by u/crinacle\"\nKZ ZS6: \"measured by u/crinacle\"\nMoondrop Kanas Pro: \"measured by u/crinacle\"\nMoondrop KXXS: \"measured by crinacle\"\nMoondrop Spaceship: \"measured by crinacle\"\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
e791d976043dd6c91b625ac81b0a72c3808f20c8 | 10,695 | ipynb | Jupyter Notebook | Netflix Exploration.ipynb | guicaro/guicaro.github.io | eb9d1167560ddc80c797e36c1582b3192f1dfcb4 | [
"MIT"
] | null | null | null | Netflix Exploration.ipynb | guicaro/guicaro.github.io | eb9d1167560ddc80c797e36c1582b3192f1dfcb4 | [
"MIT"
] | null | null | null | Netflix Exploration.ipynb | guicaro/guicaro.github.io | eb9d1167560ddc80c797e36c1582b3192f1dfcb4 | [
"MIT"
] | null | null | null | 25.586124 | 372 | 0.505657 | [
[
[
"# Netflix user behaviour",
"_____no_output_____"
],
[
"### Requirements\n\n[Jupyter Notebook](https://jupyter.org/install) \n[Apache Toree](https://toree.incubator.apache.org/) \n[sampleDataNetflix.tsv](https://guicaro.com/sampleDataNetflix.tsv) placed in local filesystem and path updated in 1) below\n\n### Notes\n\n* I used a combination of Jupyter notebook and the Apache Toree project as it makes it easy and fast to explore a dataset. \n* I was part of the team that came up with [Apache Toree (aka The Spark Kernel)](https://twitter.com/guicaro/status/543541995247910917), and till now I think it's still the only Jupyter kernel that ties to a Spark Session and is backed by Apache. It solved many issues for us back when we were developing applications in Spark.\n\n### Future\n\n* I was hoping to use [Voila](https://github.com/voila-dashboards/voila) project to create an interactive dashboard for data scientists where they could move a slider widget to change the parameters in my SQL queries, thus, change the time window to search. So, for example, say a data scientist would want to search for users only between 8 and 9 in the morning.\n* I wanted to randomly generate a bigger dataset using rules so that we could at least have more data to play with",
"_____no_output_____"
],
[
"### 1. Let's read our data",
"_____no_output_____"
],
[
"We will read in a TSV file and try to infer schema since it is not very complex data types we are using",
"_____no_output_____"
]
],
[
[
"val sessions = spark.read.option(\"header\", \"true\")\n .option(\"sep\", \"\\t\")\n .option(\"inferSchema\",\"true\")\n .csv(\"/Users/memo/Desktop/netflixSpark/sampleDataNetflix.tsv\")\n\n",
"_____no_output_____"
],
[
"sessions.printSchema",
"root\n |-- user_id: integer (nullable = true)\n |-- navigation_page: string (nullable = true)\n |-- url: string (nullable = true)\n |-- session_id: integer (nullable = true)\n |-- date: integer (nullable = true)\n |-- hour: integer (nullable = true)\n |-- timestamp: integer (nullable = true)\n\n"
],
[
"sessions.show(2)",
"+-------+---------------+--------------------+----------+--------+----+----------+\n|user_id|navigation_page| url|session_id| date|hour| timestamp|\n+-------+---------------+--------------------+----------+--------+----+----------+\n| 1001| HomePage|https://www.netfl...| 6001|20181125| 11|1543145019|\n| 1001| OriginalsGenre|https://www.netfl...| 6001|20181125| 11|1543144483|\n+-------+---------------+--------------------+----------+--------+----+----------+\nonly showing top 2 rows\n\n"
]
],
[
[
"### 2. Let's create a temp SQL table to use of the SQL magic in Apache Toree to get our information",
"_____no_output_____"
]
],
[
[
"sessions.registerTempTable(\"SESSIONS\")",
"_____no_output_____"
]
],
[
[
"### a) Find all users who have visited OurPlanetTitle Page.",
"_____no_output_____"
],
[
"Using DISTINCT to show unique users",
"_____no_output_____"
]
],
[
[
"%%SQL select distinct user_id \nfrom SESSIONS \nwhere navigation_page = 'OurPlanetTitle' ",
"_____no_output_____"
]
],
[
[
"### b) Find all users who have visited OurPlanetTitle Page only once.",
"_____no_output_____"
],
[
"Showing the page visits just for validation, can be easily removed from the projection list in query",
"_____no_output_____"
]
],
[
[
"%%SQL select user_id, count(user_id) as page_visits \nfrom SESSIONS \nwhere navigation_page = 'OurPlanetTitle' \ngroup by user_id\nhaving page_visits == 1",
"_____no_output_____"
]
],
[
[
"### c) Find all users who have visited HomePage -> OriginalsGenre -> OurPlanetTitle -> HomePage",
"_____no_output_____"
],
[
"Making sure we filter for the same path using the timestamps and making sure it's all within the same `session_id`",
"_____no_output_____"
]
],
[
[
"%%SQL select distinct a.user_id\nfrom sessions a,\nsessions b,\nsessions c,\nsessions d\nwhere a.user_id = b.user_id\nand b.user_id = c.user_id\nand c.user_id = d.user_id\nand a.navigation_page = 'HomePage'\nand b.navigation_page = 'OriginalsGenre'\nand c.navigation_page = 'OurPlanetTitle'\nand d.navigation_page = 'HomePage'\nand a.timestamp < b.timestamp\nand b.timestamp < c.timestamp\nand c.timestamp < d.timestamp\nand a.session_id = b.session_id\nand b.session_id = c.session_id\nand c.session_id = d.session_id",
"_____no_output_____"
]
],
[
[
"### d) Find all users who landed on LogIn Page from a Title Page",
"_____no_output_____"
],
[
"The like operator is not the most performant but the SQL optimizer should be able to tell that my 2nd where clause can improve selectivity of this query. I am using the `timestamp` column to make sure that a before landing on a **Login** page, the user first comes from a **Title** page",
"_____no_output_____"
]
],
[
[
"%%SQL select a.user_id\nfrom sessions a,\nsessions b\nwhere a.user_id = b.user_id\nand b.navigation_page = 'LogIn'\nand a.navigation_page like '%Title'\nand a.timestamp < b.timestamp",
"_____no_output_____"
]
],
[
[
"### e) Find all users who have visited only OurPlanetTitle Page",
"_____no_output_____"
],
[
"We are using relation 'b' to get the total count of `url` the user has visited",
"_____no_output_____"
]
],
[
[
"%%SQL select a.user_id\nfrom sessions a,\n(select user_id, count(url) as totalUrl from sessions group by user_id) b\nwhere a.user_id = b.user_id\nand a.navigation_page = 'OurPlanetTitle'\nand b.totalurl = 1",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
e791e383f5ff8ae7b625a5c89d143d4325d61b1c | 13,581 | ipynb | Jupyter Notebook | exercise-functions-and-getting-help.ipynb | Mohsenselseleh/My-Projects | 53726f51c30d0cc9296c26da1350a26c895d4d3d | [
"Apache-2.0"
] | null | null | null | exercise-functions-and-getting-help.ipynb | Mohsenselseleh/My-Projects | 53726f51c30d0cc9296c26da1350a26c895d4d3d | [
"Apache-2.0"
] | null | null | null | exercise-functions-and-getting-help.ipynb | Mohsenselseleh/My-Projects | 53726f51c30d0cc9296c26da1350a26c895d4d3d | [
"Apache-2.0"
] | null | null | null | 13,581 | 13,581 | 0.701937 | [
[
[
"**[Python Home Page](https://www.kaggle.com/learn/python)**\n\n---\n",
"_____no_output_____"
],
[
"# Try It Yourself\n\nFunctions are powerful. Try writing some yourself.\n\nAs before, don't forget to run the setup code below before jumping into question 1.",
"_____no_output_____"
]
],
[
[
"# SETUP. You don't need to worry for now about what this code does or how it works.\nfrom learntools.core import binder; binder.bind(globals())\nfrom learntools.python.ex2 import *\nprint('Setup complete.')",
"Setup complete.\n"
]
],
[
[
"# Exercises",
"_____no_output_____"
],
[
"## 1.\n\nComplete the body of the following function according to its docstring.\n\nHINT: Python has a built-in function `round`.",
"_____no_output_____"
]
],
[
[
"def round_to_two_places(num):\n \"\"\"Return the given number rounded to two decimal places. \n \n \n >>> round_to_two_places(3.14159)\n 3.14\n \"\"\"\n # Replace this body with your own code.\n # (\"pass\" is a keyword that does literally nothing. We used it as a placeholder\n # because after we begin a code block, Python requires at least one line of code)\n return round(num,2)\n\nq1.check()",
"_____no_output_____"
],
[
"# Uncomment the following for a hint\n# q1.hint()\n# Or uncomment the following to peek at the solution\nq1.solution()",
"_____no_output_____"
]
],
[
[
"## 2.\nThe help for `round` says that `ndigits` (the second argument) may be negative.\nWhat do you think will happen when it is? Try some examples in the following cell?\n\nCan you think of a case where this would be useful?",
"_____no_output_____"
]
],
[
[
"q2.solution()",
"_____no_output_____"
],
[
"# Check your answer (Run this code cell to receive credit!)\nq2.solution()",
"_____no_output_____"
]
],
[
[
"## 3.\n\nIn a previous programming problem, the candy-sharing friends Alice, Bob and Carol tried to split candies evenly. For the sake of their friendship, any candies left over would be smashed. For example, if they collectively bring home 91 candies, they'll take 30 each and smash 1.\n\nBelow is a simple function that will calculate the number of candies to smash for *any* number of total candies.\n\nModify it so that it optionally takes a second argument representing the number of friends the candies are being split between. If no second argument is provided, it should assume 3 friends, as before.\n\nUpdate the docstring to reflect this new behaviour.",
"_____no_output_____"
]
],
[
[
"def to_smash(total_candies, friends =3):\n \"\"\"Return the number of leftover candies that must be smashed after distributing\n the given number of candies evenly between 3 friends.\n \n >>> to_smash(91)\n 1\n \"\"\"\n return total_candies % friends\n\nq3.check()",
"_____no_output_____"
],
[
"q3.hint()",
"_____no_output_____"
],
[
"q3.solution()",
"_____no_output_____"
]
],
[
[
"## 4. (Optional)\n\nIt may not be fun, but reading and understanding error messages will be an important part of your Python career.\n\nEach code cell below contains some commented-out buggy code. For each cell...\n\n1. Read the code and predict what you think will happen when it's run.\n2. Then uncomment the code and run it to see what happens. (**Tip**: In the kernel editor, you can highlight several lines and press `ctrl`+`/` to toggle commenting.)\n3. Fix the code (so that it accomplishes its intended purpose without throwing an exception)\n\n<!-- TODO: should this be autochecked? Delta is probably pretty small. -->",
"_____no_output_____"
]
],
[
[
" round_to_two_places(9.9999)",
"_____no_output_____"
],
[
" x = -10\n y = 5\n# # Which of the two variables above has the smallest absolute value?\n smallest_abs = min(abs(x), abs(y))\nprint(smallest_abs)",
"5\n"
],
[
"def f(x):\n y = abs(x)\n return y\nprint(f(5))",
"_____no_output_____"
]
],
[
[
"# Keep Going\n\nNice job with the code. Next up, you'll learn about *conditionals*, which you'll need to write interesting programs. Keep going **[here](https://www.kaggle.com/colinmorris/booleans-and-conditionals)**",
"_____no_output_____"
],
[
"---\n**[Python Home Page](https://www.kaggle.com/learn/python)**\n\n\n\n\n\n*Have questions or comments? Visit the [Learn Discussion forum](https://www.kaggle.com/learn-forum/161283) to chat with other Learners.*",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
e791e5c5022a7ed9d757894ab0ac7cb5bf3c12b4 | 13,214 | ipynb | Jupyter Notebook | notebooks/03.2-Regression-Forests.ipynb | pletzer/sklearn_tutorial | ad8e6f64085a210801dc8b8dd775d8acc13c3959 | [
"BSD-3-Clause"
] | null | null | null | notebooks/03.2-Regression-Forests.ipynb | pletzer/sklearn_tutorial | ad8e6f64085a210801dc8b8dd775d8acc13c3959 | [
"BSD-3-Clause"
] | null | null | null | notebooks/03.2-Regression-Forests.ipynb | pletzer/sklearn_tutorial | ad8e6f64085a210801dc8b8dd775d8acc13c3959 | [
"BSD-3-Clause"
] | null | null | null | 30.801865 | 333 | 0.601559 | [
[
[
"<small><i>This notebook was put together by [Jake Vanderplas](http://www.vanderplas.com). Source and license info is on [GitHub](https://github.com/jakevdp/sklearn_tutorial/).</i></small>",
"_____no_output_____"
],
[
"# Supervised Learning In-Depth: Random Forests",
"_____no_output_____"
],
[
"Previously we saw a powerful discriminative classifier, **Support Vector Machines**.\nHere we'll take a look at motivating another powerful algorithm. This one is a *non-parametric* algorithm called **Random Forests**.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom scipy import stats\n\nplt.style.use('seaborn')",
"_____no_output_____"
]
],
[
[
"## Motivating Random Forests: Decision Trees",
"_____no_output_____"
],
[
"Random forests are an example of an *ensemble learner* built on decision trees.\nFor this reason we'll start by discussing decision trees themselves.\n\nDecision trees are extremely intuitive ways to classify or label objects: you simply ask a series of questions designed to zero-in on the classification:",
"_____no_output_____"
]
],
[
[
"import fig_code\nfig_code.plot_example_decision_tree()",
"_____no_output_____"
]
],
[
[
"The binary splitting makes this extremely efficient.\nAs always, though, the trick is to *ask the right questions*.\nThis is where the algorithmic process comes in: in training a decision tree classifier, the algorithm looks at the features and decides which questions (or \"splits\") contain the most information.\n\n### Creating a Decision Tree\n\nHere's an example of a decision tree classifier in scikit-learn. We'll start by defining some two-dimensional labeled data:",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import make_blobs\n\nX, y = make_blobs(n_samples=300, centers=4,\n random_state=0, cluster_std=1.0)\nplt.scatter(X[:, 0], X[:, 1], c=y, s=50, cmap='rainbow');",
"_____no_output_____"
]
],
[
[
"We have some convenience functions in the repository that help ",
"_____no_output_____"
]
],
[
[
"from fig_code import visualize_tree, plot_tree_interactive",
"_____no_output_____"
]
],
[
[
"Now using IPython's ``interact`` (available in IPython 2.0+, and requires a live kernel) we can view the decision tree splits:",
"_____no_output_____"
]
],
[
[
"plot_tree_interactive(X, y);",
"_____no_output_____"
]
],
[
[
"Notice that at each increase in depth, every node is split in two **except** those nodes which contain only a single class.\nThe result is a very fast **non-parametric** classification, and can be extremely useful in practice.\n\n**Question: Do you see any problems with this?**",
"_____no_output_____"
],
[
"### Decision Trees and over-fitting\n\nOne issue with decision trees is that it is very easy to create trees which **over-fit** the data. That is, they are flexible enough that they can learn the structure of the noise in the data rather than the signal! For example, take a look at two trees built on two subsets of this dataset:",
"_____no_output_____"
]
],
[
[
"from sklearn.tree import DecisionTreeClassifier\nclf = DecisionTreeClassifier()\n\nplt.figure()\nvisualize_tree(clf, X[:200], y[:200], boundaries=False)\nplt.figure()\nvisualize_tree(clf, X[-200:], y[-200:], boundaries=False)",
"_____no_output_____"
]
],
[
[
"The details of the classifications are completely different! That is an indication of **over-fitting**: when you predict the value for a new point, the result is more reflective of the noise in the model rather than the signal.",
"_____no_output_____"
],
[
"## Ensembles of Estimators: Random Forests\n\nOne possible way to address over-fitting is to use an **Ensemble Method**: this is a meta-estimator which essentially averages the results of many individual estimators which over-fit the data. Somewhat surprisingly, the resulting estimates are much more robust and accurate than the individual estimates which make them up!\n\nOne of the most common ensemble methods is the **Random Forest**, in which the ensemble is made up of many decision trees which are in some way perturbed.\n\nThere are volumes of theory and precedent about how to randomize these trees, but as an example, let's imagine an ensemble of estimators fit on subsets of the data. We can get an idea of what these might look like as follows:",
"_____no_output_____"
]
],
[
[
"def fit_randomized_tree(random_state=0):\n X, y = make_blobs(n_samples=300, centers=4,\n random_state=0, cluster_std=2.0)\n clf = DecisionTreeClassifier(max_depth=15)\n \n rng = np.random.RandomState(random_state)\n i = np.arange(len(y))\n rng.shuffle(i)\n visualize_tree(clf, X[i[:250]], y[i[:250]], boundaries=False,\n xlim=(X[:, 0].min(), X[:, 0].max()),\n ylim=(X[:, 1].min(), X[:, 1].max()))\n \nfrom ipywidgets import interact\ninteract(fit_randomized_tree, random_state=(0, 100));",
"_____no_output_____"
]
],
[
[
"See how the details of the model change as a function of the sample, while the larger characteristics remain the same!\nThe random forest classifier will do something similar to this, but use a combined version of all these trees to arrive at a final answer:",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestClassifier\nclf = RandomForestClassifier(n_estimators=100, random_state=0)\nvisualize_tree(clf, X, y, boundaries=False);",
"_____no_output_____"
]
],
[
[
"By averaging over 100 randomly perturbed models, we end up with an overall model which is a much better fit to our data!\n\n*(Note: above we randomized the model through sub-sampling... Random Forests use more sophisticated means of randomization, which you can read about in, e.g. the [scikit-learn documentation](http://scikit-learn.org/stable/modules/ensemble.html#forest)*)",
"_____no_output_____"
],
[
"## Quick Example: Moving to Regression\n\nAbove we were considering random forests within the context of classification.\nRandom forests can also be made to work in the case of regression (that is, continuous rather than categorical variables). The estimator to use for this is ``sklearn.ensemble.RandomForestRegressor``.\n\nLet's quickly demonstrate how this can be used:",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestRegressor\n\nx = 10 * np.random.rand(100)\n\ndef model(x, sigma=0.3):\n fast_oscillation = np.sin(5 * x)\n slow_oscillation = np.sin(0.5 * x)\n noise = sigma * np.random.randn(len(x))\n\n return slow_oscillation + fast_oscillation + noise\n\ny = model(x)\nplt.errorbar(x, y, 0.3, fmt='o');",
"_____no_output_____"
],
[
"xfit = np.linspace(0, 10, 1000)\nyfit = RandomForestRegressor(100).fit(x[:, None], y).predict(xfit[:, None])\nytrue = model(xfit, 0)\n\nplt.errorbar(x, y, 0.3, fmt='o')\nplt.plot(xfit, yfit, '-r');\nplt.plot(xfit, ytrue, '-k', alpha=0.5);",
"_____no_output_____"
]
],
[
[
"As you can see, the non-parametric random forest model is flexible enough to fit the multi-period data, without us even specifying a multi-period model!",
"_____no_output_____"
],
[
"## Example: Random Forest for Classifying Digits\n\nWe previously saw the **hand-written digits** data. Let's use that here to test the efficacy of the SVM and Random Forest classifiers.",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import load_digits\ndigits = load_digits()\ndigits.keys()",
"_____no_output_____"
],
[
"X = digits.data\ny = digits.target\nprint(X.shape)\nprint(y.shape)",
"_____no_output_____"
]
],
[
[
"To remind us what we're looking at, we'll visualize the first few data points:",
"_____no_output_____"
]
],
[
[
"# set up the figure\nfig = plt.figure(figsize=(6, 6)) # figure size in inches\nfig.subplots_adjust(left=0, right=1, bottom=0, top=1, hspace=0.05, wspace=0.05)\n\n# plot the digits: each image is 8x8 pixels\nfor i in range(64):\n ax = fig.add_subplot(8, 8, i + 1, xticks=[], yticks=[])\n ax.imshow(digits.images[i], cmap=plt.cm.binary, interpolation='nearest')\n \n # label the image with the target value\n ax.text(0, 7, str(digits.target[i]))",
"_____no_output_____"
]
],
[
[
"We can quickly classify the digits using a decision tree as follows:",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\nfrom sklearn import metrics\n\nXtrain, Xtest, ytrain, ytest = train_test_split(X, y, random_state=0)\nclf = DecisionTreeClassifier(max_depth=11)\nclf.fit(Xtrain, ytrain)\nypred = clf.predict(Xtest)",
"_____no_output_____"
]
],
[
[
"We can check the accuracy of this classifier:",
"_____no_output_____"
]
],
[
[
"metrics.accuracy_score(ypred, ytest)",
"_____no_output_____"
]
],
[
[
"and for good measure, plot the confusion matrix:",
"_____no_output_____"
]
],
[
[
"metrics.plot_confusion_matrix(clf, Xtest, ytest, cmap=plt.cm.Blues)\nplt.grid(False)",
"_____no_output_____"
]
],
[
[
"### Exercise\n1. Repeat this classification task with ``sklearn.ensemble.RandomForestClassifier``. How does the ``max_depth``, ``max_features``, and ``n_estimators`` affect the results?\n2. Try this classification with ``sklearn.svm.SVC``, adjusting ``kernel``, ``C``, and ``gamma``. Which classifier performs optimally?\n3. Try a few sets of parameters for each model and check the F1 score (``sklearn.metrics.f1_score``) on your results. What's the best F1 score you can reach?",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e792038626af684d62ca4d6aee279f139541c613 | 391,217 | ipynb | Jupyter Notebook | VOC-Data-Loaders.ipynb | 1xyz/pytorch-fcn-ext | 2a3bfd819f55200a11132981b38a6b7c964fa565 | [
"MIT"
] | null | null | null | VOC-Data-Loaders.ipynb | 1xyz/pytorch-fcn-ext | 2a3bfd819f55200a11132981b38a6b7c964fa565 | [
"MIT"
] | null | null | null | VOC-Data-Loaders.ipynb | 1xyz/pytorch-fcn-ext | 2a3bfd819f55200a11132981b38a6b7c964fa565 | [
"MIT"
] | null | null | null | 871.30735 | 207,852 | 0.955963 | [
[
[
"import torch\nimport torchfcn\nimport torchvision\nimport os\nimport os.path as osp\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport fcn",
"_____no_output_____"
]
],
[
[
"### Understand the datasets for training and cross-validation",
"_____no_output_____"
]
],
[
[
"# Assume the datasets are downloaded to the loc. below\nroot = osp.expanduser('~/data/datasets')",
"_____no_output_____"
]
],
[
[
"#### Pixel label values ",
"_____no_output_____"
]
],
[
[
"# Map of the classe names for example 1 - aeroplane\nclass_names = np.array([\n 'background',\n 'aeroplane',\n 'bicycle',\n 'bird',\n 'boat',\n 'bottle',\n 'bus',\n 'car',\n 'cat',\n 'chair',\n 'cow',\n 'diningtable',\n 'dog',\n 'horse',\n 'motorbike',\n 'person',\n 'potted plant',\n 'sheep',\n 'sofa',\n 'train',\n 'tv/monitor',\n ])",
"_____no_output_____"
]
],
[
[
"#### Utility functions to show images and histogram",
"_____no_output_____"
]
],
[
[
"def imshow(img):\n plt.imshow(img)\n plt.show()\n \ndef hist(img):\n plt.hist(img)\n plt.show()",
"_____no_output_____"
]
],
[
[
"#### Inspect the train dataset",
"_____no_output_____"
]
],
[
[
"# The train dataset is Semantic Boundaries Dataset and Benchmark (SBD) benchmark\n# . http://home.bharathh.info/pubs/codes/SBD/download.html\n# Refer http://www.eecs.berkeley.edu/Research/Projects/CS/vision/grouping/semantic_contours/benchmark.tgz\n\n# Note: we set the transform to False, this ensures that the result of __get_item is an \n# ndarray, not a tensor.\ntrain_dataset = torchfcn.datasets.SBDClassSeg(root, split='train', transform=False)\nprint(train_dataset)\nprint(f\"Number of entries in the training: {len(train_dataset)}\")",
"<torchfcn.datasets.voc.SBDClassSeg object at 0x127d30f60>\nNumber of entries in the training: 8498\n"
],
[
"idx = 459\nprint(\"Shape of image: \", train_dataset[idx][0].shape, \"shape of the label: \", train_dataset[idx][1].shape)\nimshow(train_dataset[idx][0])\nimshow(train_dataset[idx][1])\n# print(train_dataset[idx][1])",
"Shape of image: (480, 360, 3) shape of the label: (480, 360)\n"
]
],
[
[
"#### Print the histogram of the train dataset",
"_____no_output_____"
]
],
[
[
"label_dist = np.ravel(train_dataset[idx][1])\nhist(label_dist)",
"_____no_output_____"
]
],
[
[
"#### Understand the validation (dev) dataset",
"_____no_output_____"
]
],
[
[
"# Load the validation dataset (Pascal VOC)\n# Again note that the transform is False, so the result is an ndarray and not a transformed tensor\nvalid_dataset = torchfcn.datasets.VOC2011ClassSeg(root, split='seg11valid', transform=False)",
"_____no_output_____"
],
[
"idx = 203\n\nprint(\"Shape of data: \", valid_dataset[idx][0].shape, \"Shape of label: \", valid_dataset[idx][1].shape)\n\nimshow(valid_dataset[idx][0])\nimshow(valid_dataset[idx][1])\n\nlabel_dist = np.ravel(valid_dataset[idx][1])\nprint(\"Max\", np.max(label_dist), \"Min\", np.min(label_dist))\nhist(label_dist)",
"Shape of data: (375, 500, 3) Shape of label: (375, 500)\n"
]
],
[
[
"#### Inspect the transformed tensor",
"_____no_output_____"
]
],
[
[
"## Let us actually inspect the transformed tensor data instead\nvalid_tensor_dataset = torchfcn.datasets.VOC2011ClassSeg(root, split='seg11valid', transform=True)\n\nlabel_dists = valid_tensor_dataset[idx][1]\nprint(torch.min(label_dists))\n\nlabel_dist = np.ravel(label_dists.numpy())\nprint(\"Max\", np.max(label_dist), \"Min\", np.min(label_dist))\nhist(label_dist)",
"tensor(-1)\nMax 8 Min -1\n"
]
],
[
[
"#### Inspect the dataset transformed?",
"_____no_output_____"
]
],
[
[
"mean_bgr = np.array([104.00698793, 116.66876762, 122.67891434])\n\ndef transform(img):\n #img = img[:, :, ::-1] # RGB -> BGR\n img = img.astype(np.float64)\n img -= mean_bgr\n return img",
"_____no_output_____"
],
[
"print(valid_dataset[idx][0].shape)\ntransformed_image = transform(valid_dataset[idx][0])\nprint(transformed_image.shape)",
"(375, 500, 3)\n(375, 500, 3)\n"
],
[
"imshow(transformed_image)",
"Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\n"
]
],
[
[
"#### Notes\n\n* https://stats.stackexchange.com/questions/211436/why-normalize-images-by-subtracting-datasets-image-mean-instead-of-the-current\n\n* https://github.com/ry/tensorflow-resnet/blob/master/convert.py#L51",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
e7920a146369c676430a33d6c3531459758074e1 | 2,599 | ipynb | Jupyter Notebook | examples/real-time/real-Time_linux.ipynb | OpenRTDynamics/openrtdynamics2 | 1b7a114110089bc7721da604c5e344854ed555c3 | [
"MIT"
] | null | null | null | examples/real-time/real-Time_linux.ipynb | OpenRTDynamics/openrtdynamics2 | 1b7a114110089bc7721da604c5e344854ed555c3 | [
"MIT"
] | null | null | null | examples/real-time/real-Time_linux.ipynb | OpenRTDynamics/openrtdynamics2 | 1b7a114110089bc7721da604c5e344854ed555c3 | [
"MIT"
] | null | null | null | 24.990385 | 142 | 0.527895 | [
[
[
"import math\nimport numpy as np\nimport openrtdynamics2.lang as dy\nimport openrtdynamics2.py_execute as dyexe\nimport openrtdynamics2.targets as tg",
"_____no_output_____"
]
],
[
[
"# Code generation for Linux Real-Time Preemption\n\nc.f. https://wiki.linuxfoundation.org/realtime/start\n\nThe generated code can be compiled using a c++ compiler as follows:\n\n $ c++ main.cpp -o main\n",
"_____no_output_____"
]
],
[
[
"dy.clear()\n\nsystem = dy.enter_system()\n\n# define system inputs\nu = dy.system_input( dy.DataTypeFloat64(1), name='input1', default_value=1.0, value_range=[0, 25], title=\"input #1\")\n\n\ny = dy.signal() # introduce variable y\nx = y + u # x[k] = y[k] + u[k]\ny << dy.delay(x, initial_state = 2.0) # y[k+1] = y[k] + x[k], y[0] = 2.0\n\n# define sampling time\ndelta_time = dy.float64(0.1)\n\n# define output(s)\ndy.append_output(delta_time, '__ORTD_CONTROL_delta_time__')\ndy.append_output(y, 'output')\n\n# generate code\ncode_gen_results = dy.generate_code(template=tg.TargetLinuxRealtime(activate_print = True), folder='./')",
"compiling system simulation (level 0)... \ninput1 1.0 double\nGenerated code will be written to ./ .\nwriting file ./simulation_manifest.json\nwriting file ./main.cpp\n"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7922388377a54e3ee47c936a99a6cba5cd2bd9a | 12,879 | ipynb | Jupyter Notebook | Numpy-Exercises.ipynb | smalik-hub/Numpy-Exercises | 473c4bf295bfefc0761704aa5328e665b982637b | [
"MIT"
] | 1 | 2020-08-20T01:35:59.000Z | 2020-08-20T01:35:59.000Z | Numpy-Exercises.ipynb | smalik-hub/Numpy-Exercises | 473c4bf295bfefc0761704aa5328e665b982637b | [
"MIT"
] | null | null | null | Numpy-Exercises.ipynb | smalik-hub/Numpy-Exercises | 473c4bf295bfefc0761704aa5328e665b982637b | [
"MIT"
] | null | null | null | 21.009788 | 221 | 0.459508 | [
[
[
"# NumPy Exercises \n\nNumPy is a library for the Python programming language, adding support for large, multi-dimensional arrays and matrices, along with a large collection of high-level mathematical functions to operate on these arrays.",
"_____no_output_____"
],
[
"#### Import NumPy as np",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"#### Create an array of 10 zeros ",
"_____no_output_____"
]
],
[
[
"np.zeros(10)",
"_____no_output_____"
]
],
[
[
"#### Create an array of 10 ones",
"_____no_output_____"
]
],
[
[
"np.ones(10)",
"_____no_output_____"
]
],
[
[
"#### Create an array of 10 fives",
"_____no_output_____"
]
],
[
[
"np.ones(10) * 5",
"_____no_output_____"
]
],
[
[
"#### Create an array of the integers from 10 to 50",
"_____no_output_____"
]
],
[
[
"np.arange(10,51)",
"_____no_output_____"
]
],
[
[
"#### Create an array of all the even integers from 10 to 50",
"_____no_output_____"
]
],
[
[
"np.arange(10,51,2)",
"_____no_output_____"
]
],
[
[
"#### Create a 3x3 matrix with values ranging from 0 to 8",
"_____no_output_____"
]
],
[
[
"np.arange(0,9).reshape(3,3)",
"_____no_output_____"
]
],
[
[
"#### Create a 3x3 identity matrix",
"_____no_output_____"
]
],
[
[
"np.eye(3)",
"_____no_output_____"
]
],
[
[
"#### Use NumPy to generate a random number between 0 and 1",
"_____no_output_____"
]
],
[
[
"np.random.rand(1)",
"_____no_output_____"
]
],
[
[
"#### Use NumPy to generate an array of 25 random numbers sampled from a standard normal distribution",
"_____no_output_____"
]
],
[
[
"np.random.randn(25)",
"_____no_output_____"
]
],
[
[
"#### Create the following matrix:",
"_____no_output_____"
]
],
[
[
"np.arange(1,101).reshape(10,10)/100",
"_____no_output_____"
]
],
[
[
"#### Create an array of 20 linearly spaced points between 0 and 1:",
"_____no_output_____"
]
],
[
[
"np.linspace(0,1,20)",
"_____no_output_____"
]
],
[
[
"## Numpy Indexing and Selection\n\nNow you will be given a few matrices, and be asked to replicate the resulting matrix outputs:",
"_____no_output_____"
]
],
[
[
"mat = np.arange(1,26).reshape(5,5)\nmat",
"_____no_output_____"
],
[
"# WRITE CODE HERE THAT REPRODUCES THE OUTPUT OF THE CELL BELOW\n# BE CAREFUL NOT TO RUN THE CELL BELOW, OTHERWISE YOU WON'T\n# BE ABLE TO SEE THE OUTPUT ANY MORE\nmat[2:,1:]",
"_____no_output_____"
],
[
"# WRITE CODE HERE THAT REPRODUCES THE OUTPUT OF THE CELL BELOW\n# BE CAREFUL NOT TO RUN THE CELL BELOW, OTHERWISE YOU WON'T\n# BE ABLE TO SEE THE OUTPUT ANY MORE\nmat[3,4]",
"_____no_output_____"
],
[
"# WRITE CODE HERE THAT REPRODUCES THE OUTPUT OF THE CELL BELOW\n# BE CAREFUL NOT TO RUN THE CELL BELOW, OTHERWISE YOU WON'T\n# BE ABLE TO SEE THE OUTPUT ANY MORE\nmat[:3,1:2]",
"_____no_output_____"
],
[
"# WRITE CODE HERE THAT REPRODUCES THE OUTPUT OF THE CELL BELOW\n# BE CAREFUL NOT TO RUN THE CELL BELOW, OTHERWISE YOU WON'T\n# BE ABLE TO SEE THE OUTPUT ANY MORE\nmat[4]",
"_____no_output_____"
],
[
"# WRITE CODE HERE THAT REPRODUCES THE OUTPUT OF THE CELL BELOW\n# BE CAREFUL NOT TO RUN THE CELL BELOW, OTHERWISE YOU WON'T\n# BE ABLE TO SEE THE OUTPUT ANY MORE\nmat[3:,:]",
"_____no_output_____"
]
],
[
[
"### Now do the following",
"_____no_output_____"
],
[
"#### Get the sum of all the values in mat",
"_____no_output_____"
]
],
[
[
"np.sum(mat)",
"_____no_output_____"
]
],
[
[
"#### Get the standard deviation of the values in mat",
"_____no_output_____"
]
],
[
[
"np.std(mat)",
"_____no_output_____"
]
],
[
[
"#### Get the sum of all the columns in mat",
"_____no_output_____"
]
],
[
[
"np.sum(mat, axis=0)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7922de9e533dd38de74707def660b3dc7cd768a | 11,023 | ipynb | Jupyter Notebook | tutorial/source/intro_part_i.ipynb | neerajprad/pyro | 3b5b2c5de208209365bf26f239f12521de68acc4 | [
"MIT"
] | 1 | 2020-08-16T11:27:12.000Z | 2020-08-16T11:27:12.000Z | tutorial/source/intro_part_i.ipynb | neerajprad/pyro | 3b5b2c5de208209365bf26f239f12521de68acc4 | [
"MIT"
] | null | null | null | tutorial/source/intro_part_i.ipynb | neerajprad/pyro | 3b5b2c5de208209365bf26f239f12521de68acc4 | [
"MIT"
] | 1 | 2018-12-22T16:15:49.000Z | 2018-12-22T16:15:49.000Z | 45.17623 | 624 | 0.658714 | [
[
[
"# import some dependencies\nimport torch\n\nimport pyro\nimport pyro.distributions as dist",
"_____no_output_____"
]
],
[
[
"# Models in Pyro: From Primitive Distributions to Stochastic Functions\n\nThe basic unit of Pyro programs is the _stochastic function_. \nThis is an arbitrary Python callable that combines two ingredients:\n\n- deterministic Python code; and\n- primitive stochastic functions \n\nConcretely, a stochastic function can be any Python object with a `__call__()` method, like a function, a method, or a PyTorch `nn.Module`.\n\nThroughout the tutorials and documentation, we will often call stochastic functions *models*, since stochastic functions can be used to represent simplified or abstract descriptions of a process by which data are generated. Expressing models as stochastic functions in Pyro means that models can be composed, reused, imported, and serialized just like regular Python callables. \n\nWithout further ado, let's introduce one of our basic building blocks: primitive stochastic functions.\n\n## Primitive Stochastic Functions\n\nPrimitive stochastic functions, or distributions, are an important class of stochastic functions for which we can explicitly compute the probability of the outputs given the inputs. As of PyTorch 0.4 and Pyro 0.2, Pyro uses PyTorch's [distribution library](http://pytorch.org/docs/master/distributions.html). You can also create custom distributions using [transforms](http://pytorch.org/docs/master/distributions.html#module-torch.distributions.transforms).\n\nUsing primitive stochastic functions is easy. For example, to draw a sample `x` from the unit normal distribution $\\mathcal{N}(0,1)$ we do the following:",
"_____no_output_____"
]
],
[
[
"loc = 0. # mean zero\nscale = 1. # unit variance\nnormal = dist.Normal(loc, scale) # create a normal distribution object\nx = normal.sample() # draw a sample from N(0,1)\nprint(\"sample\", x)\nprint(\"log prob\", normal.log_prob(x)) # score the sample from N(0,1)",
"_____no_output_____"
]
],
[
[
"Here, `dist.Normal` is a callable instance of the `Distribution` class that takes parameters and provides sample and score methods. Note that the parameters passed to `dist.Normal` are `torch.Tensor`s. This is necessary because we want to make use of PyTorch's fast tensor math and autograd capabilities during inference.",
"_____no_output_____"
],
[
"## The `pyro.sample` Primitive\n\nOne of the core language primitives in Pyro is the `pyro.sample` statement. Using `pyro.sample` is as simple as calling a primitive stochastic function with one important difference:",
"_____no_output_____"
]
],
[
[
"x = pyro.sample(\"my_sample\", dist.Normal(loc, scale))\nprint(x)",
"_____no_output_____"
]
],
[
[
"Just like a direct call to `dist.Normal().sample()`, this returns a sample from the unit normal distribution. The crucial difference is that this sample is _named_. Pyro's backend uses these names to uniquely identify sample statements and _change their behavior at runtime_ depending on how the enclosing stochastic function is being used. As we will see, this is how Pyro can implement the various manipulations that underlie inference algorithms.",
"_____no_output_____"
],
[
"## A Simple Model\n\nNow that we've introduced `pyro.sample` and `pyro.distributions` we can write a simple model. Since we're ultimately interested in probabilistic programming because we want to model things in the real world, let's choose something concrete. \n\nLet's suppose we have a bunch of data with daily mean temperatures and cloud cover. We want to reason about how temperature interacts with whether it was sunny or cloudy. A simple stochastic function that does that is given by:",
"_____no_output_____"
]
],
[
[
"def weather():\n cloudy = pyro.sample('cloudy', dist.Bernoulli(0.3))\n cloudy = 'cloudy' if cloudy.item() == 1.0 else 'sunny'\n mean_temp = {'cloudy': 55.0, 'sunny': 75.0}[cloudy]\n scale_temp = {'cloudy': 10.0, 'sunny': 15.0}[cloudy]\n temp = pyro.sample('temp', dist.Normal(mean_temp, scale_temp))\n return cloudy, temp.item()\n\nfor _ in range(3):\n print(weather())",
"_____no_output_____"
]
],
[
[
"Let's go through this line-by-line. First, in lines 2-3 we use `pyro.sample` to define a binary random variable 'cloudy', which is given by a draw from the bernoulli distribution with a parameter of `0.3`. Since the bernoulli distributions returns `0`s or `1`s, in line 4 we convert the value `cloudy` to a string so that return values of `weather` are easier to parse. So according to this model 30% of the time it's cloudy and 70% of the time it's sunny.\n\nIn lines 5-6 we define the parameters we're going to use to sample the temperature in lines 7-9. These parameters depend on the particular value of `cloudy` we sampled in line 2. For example, the mean temperature is 55 degrees (Fahrenheit) on cloudy days and 75 degrees on sunny days. Finally we return the two values `cloudy` and `temp` in line 10.\n\nProcedurally, `weather()` is a non-deterministic Python callable that returns two random samples. Because the randomness is invoked with `pyro.sample`, however, it is much more than that. In particular `weather()` specifies a joint probability distribution over two named random variables: `cloudy` and `temp`. As such, it defines a probabilistic model that we can reason about using the techniques of probability theory. For example we might ask: if I observe a temperature of 70 degrees, how likely is it to be cloudy? How to formulate and answer these kinds of questions will be the subject of the next tutorial.\n\nWe've now seen how to define a simple model. Building off of it is easy. For example:",
"_____no_output_____"
]
],
[
[
"def ice_cream_sales():\n cloudy, temp = weather()\n expected_sales = 200. if cloudy == 'sunny' and temp > 80.0 else 50.\n ice_cream = pyro.sample('ice_cream', dist.Normal(expected_sales, 10.0))\n return ice_cream",
"_____no_output_____"
]
],
[
[
"This kind of modularity, familiar to any programmer, is obviously very powerful. But is it powerful enough to encompass all the different kinds of models we'd like to express?\n\n## Universality: Stochastic Recursion, Higher-order Stochastic Functions, and Random Control Flow\n\nBecause Pyro is embedded in Python, stochastic functions can contain arbitrarily complex deterministic Python and randomness can freely affect control flow. For example, we can construct recursive functions that terminate their recursion nondeterministically, provided we take care to pass `pyro.sample` unique sample names whenever it's called. For example we can define a geometric distribution like so:",
"_____no_output_____"
]
],
[
[
"def geometric(p, t=None):\n if t is None:\n t = 0\n x = pyro.sample(\"x_{}\".format(t), dist.Bernoulli(p))\n if x.item() == 0:\n return x\n else:\n return x + geometric(p, t + 1)\n \nprint(geometric(0.5))",
"_____no_output_____"
]
],
[
[
"Note that the names `x_0`, `x_1`, etc., in `geometric()` are generated dynamically and that different executions can have different numbers of named random variables. \n\nWe are also free to define stochastic functions that accept as input or produce as output other stochastic functions:",
"_____no_output_____"
]
],
[
[
"def normal_product(loc, scale):\n z1 = pyro.sample(\"z1\", dist.Normal(loc, scale))\n z2 = pyro.sample(\"z2\", dist.Normal(loc, scale))\n y = z1 * z2\n return y\n\ndef make_normal_normal():\n mu_latent = pyro.sample(\"mu_latent\", dist.Normal(0, 1))\n fn = lambda scale: normal_product(mu_latent, scale)\n return fn\n\nprint(make_normal_normal()(1.))",
"_____no_output_____"
]
],
[
[
"Here `make_normal_normal()` is a stochastic function that takes one argument and which, upon execution, generates three named random variables.\n\nThe fact that Pyro supports arbitrary Python code like this—iteration, recursion, higher-order functions, etc.—in conjuction with random control flow means that Pyro stochastic functions are _universal_, i.e. they can be used to represent any computable probability distribution. As we will see in subsequent tutorials, this is incredibly powerful. \n\nIt is worth emphasizing that this is one reason why Pyro is built on top of PyTorch: dynamic computational graphs are an important ingredient in allowing for universal models that can benefit from GPU-accelerated tensor math.",
"_____no_output_____"
],
[
"## Next Steps\n\nWe've shown how we can use stochastic functions and primitive distributions to represent models in Pyro. In order to learn models from data and reason about them we need to be able to do inference. This is the subject of the [next tutorial](intro_part_ii.ipynb).",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
e7922f71360572cf7c87c83acde1afec6db02bc3 | 2,334 | ipynb | Jupyter Notebook | tests/test.ipynb | Zadigo/krysalid | c0ec96f4fcdc0b7309ed3c68250df96f054cf4d0 | [
"MIT"
] | null | null | null | tests/test.ipynb | Zadigo/krysalid | c0ec96f4fcdc0b7309ed3c68250df96f054cf4d0 | [
"MIT"
] | null | null | null | tests/test.ipynb | Zadigo/krysalid | c0ec96f4fcdc0b7309ed3c68250df96f054cf4d0 | [
"MIT"
] | null | null | null | 19.779661 | 194 | 0.526135 | [
[
[
"from zineb.html_parser.parsers import HTMLPageParser\nimport os",
"_____no_output_____"
],
[
"\nf = open('D:/coding\\personnal/zineb/tests\\html_parser/test5.html', encoding='utf-8')",
"_____no_output_____"
],
[
"soup = HTMLPageParser(f)",
"_____no_output_____"
],
[
"content = soup.manager.find('div', attrs={'class': 'matches'})\ncontent",
"_____no_output_____"
],
[
"header = content.find('div', attrs={'id': 'header'})",
"_____no_output_____"
],
[
"header.get_children('div')",
"_____no_output_____"
],
[
"f.close()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7923f1754a71f6aa31f78e51dbf2aec8352c38f | 34,492 | ipynb | Jupyter Notebook | notebooks/01-concept-spotting/06-lists-training.ipynb | fschlatt/CIKM-20 | 100757e348e8f9c1aa3345fa01c667c5117d2cae | [
"MIT"
] | 12 | 2020-10-17T10:56:17.000Z | 2022-02-18T13:34:37.000Z | notebooks/01-concept-spotting/06-lists-training.ipynb | fschlatt/CIKM-20 | 100757e348e8f9c1aa3345fa01c667c5117d2cae | [
"MIT"
] | null | null | null | notebooks/01-concept-spotting/06-lists-training.ipynb | fschlatt/CIKM-20 | 100757e348e8f9c1aa3345fa01c667c5117d2cae | [
"MIT"
] | 6 | 2020-10-17T10:56:18.000Z | 2022-02-07T12:44:37.000Z | 62.035971 | 135 | 0.518787 | [
[
[
"# Preamble",
"_____no_output_____"
]
],
[
[
"from flair.datasets import ColumnCorpus\nfrom flair.embeddings import FlairEmbeddings\nfrom flair.embeddings import TokenEmbeddings\nfrom flair.embeddings import StackedEmbeddings\nfrom flair.models import SequenceTagger\nfrom flair.trainers import ModelTrainer\nfrom typing import List\nimport numpy as np\nimport os\nimport torch\nimport random",
"_____no_output_____"
],
[
"PATH_SPOTTING_DATASET = \"../../data/concept-spotting/lists/\"\nPATH_FLAIR_FOLDER = \"../../data/flair-models/lists/\"",
"_____no_output_____"
]
],
[
[
"# List-Spotter: Training",
"_____no_output_____"
]
],
[
[
"def set_seed(seed):\n # For reproducibility\n # (https://pytorch.org/docs/stable/notes/randomness.html)\n np.random.seed(seed)\n random.seed(seed)\n torch.manual_seed(seed)\n torch.cuda.manual_seed(seed)\n os.environ['PYTHONHASHSEED'] = str(seed)\n\n torch.backends.cudnn.deterministic = True\n torch.backends.cudnn.benchmark = False",
"_____no_output_____"
],
[
"columns = {0: 'text', 1: 'pos', 2: 'chunk_BIO'}\ntag_type = \"chunk_BIO\"\ncorpus = ColumnCorpus(PATH_SPOTTING_DATASET, columns)\ntag_dictionary = corpus.make_tag_dictionary(tag_type=tag_type)\nprint(corpus)",
"2020-10-16 11:24:22,193 Reading data from ../../data/concept-spotting/lists\n2020-10-16 11:24:22,194 Train: ../../data/concept-spotting/lists/train.txt\n2020-10-16 11:24:22,195 Dev: ../../data/concept-spotting/lists/dev.txt\n2020-10-16 11:24:22,195 Test: ../../data/concept-spotting/lists/test.txt\nCorpus: 358 train + 76 dev + 78 test sentences\n"
],
[
"set_seed(42)\nembedding_types: List[TokenEmbeddings] = [\n FlairEmbeddings('news-forward'), FlairEmbeddings('news-backward')]\n\nembeddings: StackedEmbeddings = StackedEmbeddings(embeddings=embedding_types)\nset_seed(42)\ntagger: SequenceTagger = SequenceTagger(hidden_size=128,\n embeddings=embeddings,\n tag_dictionary=tag_dictionary,\n tag_type=tag_type,\n use_crf=True,\n dropout=0.25,\n rnn_layers=2)\nset_seed(42)\ntrainer: ModelTrainer = ModelTrainer(tagger, corpus)",
"_____no_output_____"
],
[
"set_seed(42)\nresult = trainer.train(PATH_FLAIR_FOLDER,\n learning_rate=0.3,\n mini_batch_size=16,\n max_epochs=20,\n shuffle=True,\n num_workers=0)",
"2020-08-14 09:45:27,946 ----------------------------------------------------------------------------------------------------\n2020-08-14 09:45:27,948 Evaluation method: MICRO_F1_SCORE\n2020-08-14 09:45:28,579 ----------------------------------------------------------------------------------------------------\n2020-08-14 09:45:28,817 epoch 1 - iter 0/23 - loss 8.79233646\n2020-08-14 09:45:30,617 epoch 1 - iter 2/23 - loss 6.16552440\n2020-08-14 09:45:32,498 epoch 1 - iter 4/23 - loss 5.29194376\n2020-08-14 09:45:34,572 epoch 1 - iter 6/23 - loss 5.03094617\n2020-08-14 09:45:36,524 epoch 1 - iter 8/23 - loss 4.69931691\n2020-08-14 09:45:38,505 epoch 1 - iter 10/23 - loss 4.50073297\n2020-08-14 09:45:40,560 epoch 1 - iter 12/23 - loss 4.20511235\n2020-08-14 09:45:42,548 epoch 1 - iter 14/23 - loss 3.93507350\n2020-08-14 09:45:44,569 epoch 1 - iter 16/23 - loss 3.93161163\n2020-08-14 09:45:46,344 epoch 1 - iter 18/23 - loss 3.62402451\n2020-08-14 09:45:48,263 epoch 1 - iter 20/23 - loss 3.53260322\n2020-08-14 09:45:50,178 epoch 1 - iter 22/23 - loss 3.48320543\n2020-08-14 09:45:51,634 ----------------------------------------------------------------------------------------------------\n2020-08-14 09:45:51,637 EPOCH 1 done: loss 3.4832 - lr 0.3000 - bad epochs 0\n2020-08-14 09:45:52,601 DEV : loss 1.339187502861023 - score 0.7577\n2020-08-14 09:45:53,738 TEST : loss 1.8717453479766846 - score 0.6928\n2020-08-14 09:45:55,236 ----------------------------------------------------------------------------------------------------\n2020-08-14 09:45:55,350 epoch 2 - iter 0/23 - loss 1.96359801\n2020-08-14 09:45:57,028 epoch 2 - iter 2/23 - loss 2.10282628\n2020-08-14 09:45:58,761 epoch 2 - iter 4/23 - loss 2.16568089\n2020-08-14 09:46:00,413 epoch 2 - iter 6/23 - loss 1.80044511\n2020-08-14 09:46:02,142 epoch 2 - iter 8/23 - loss 1.85052729\n2020-08-14 09:46:03,806 epoch 2 - iter 10/23 - loss 1.97098036\n2020-08-14 09:46:05,404 epoch 2 - iter 12/23 - loss 1.87773460\n2020-08-14 09:46:07,078 epoch 2 - iter 14/23 - loss 1.82299486\n2020-08-14 09:46:08,790 epoch 2 - iter 16/23 - loss 1.86174492\n2020-08-14 09:46:10,453 epoch 2 - iter 18/23 - loss 1.81453508\n2020-08-14 09:46:12,154 epoch 2 - iter 20/23 - loss 1.88815591\n2020-08-14 09:46:13,753 epoch 2 - iter 22/23 - loss 1.85566888\n2020-08-14 09:46:15,198 ----------------------------------------------------------------------------------------------------\n2020-08-14 09:46:15,200 EPOCH 2 done: loss 1.8557 - lr 0.3000 - bad epochs 0\n2020-08-14 09:46:16,160 DEV : loss 1.0508114099502563 - score 0.7641\n2020-08-14 09:46:17,253 TEST : loss 1.127955675125122 - score 0.8641\n2020-08-14 09:46:18,836 ----------------------------------------------------------------------------------------------------\n2020-08-14 09:46:18,938 epoch 3 - iter 0/23 - loss 1.26832223\n2020-08-14 09:46:20,639 epoch 3 - iter 2/23 - loss 1.29353976\n2020-08-14 09:46:22,407 epoch 3 - iter 4/23 - loss 1.30448680\n2020-08-14 09:46:24,112 epoch 3 - iter 6/23 - loss 1.34567828\n2020-08-14 09:46:25,858 epoch 3 - iter 8/23 - loss 1.46075755\n2020-08-14 09:46:27,553 epoch 3 - iter 10/23 - loss 1.47406260\n2020-08-14 09:46:29,270 epoch 3 - iter 12/23 - loss 1.48322985\n2020-08-14 09:46:30,958 epoch 3 - iter 14/23 - loss 1.44860619\n2020-08-14 09:46:32,637 epoch 3 - iter 16/23 - loss 1.37087487\n2020-08-14 09:46:34,283 epoch 3 - iter 18/23 - loss 1.34735934\n2020-08-14 09:46:35,959 epoch 3 - iter 20/23 - loss 1.35782588\n2020-08-14 09:46:37,584 epoch 3 - iter 22/23 - loss 1.31086863\n2020-08-14 09:46:39,049 ----------------------------------------------------------------------------------------------------\n2020-08-14 09:46:39,051 EPOCH 3 done: loss 1.3109 - lr 0.3000 - bad epochs 0\n2020-08-14 09:46:39,982 DEV : loss 0.5823153853416443 - score 0.8606\n2020-08-14 09:46:41,050 TEST : loss 0.8125592470169067 - score 0.847\n2020-08-14 09:46:42,537 ----------------------------------------------------------------------------------------------------\n2020-08-14 09:46:42,668 epoch 4 - iter 0/23 - loss 1.33701074\n2020-08-14 09:46:44,398 epoch 4 - iter 2/23 - loss 1.84817159\n2020-08-14 09:46:46,079 epoch 4 - iter 4/23 - loss 1.64524522\n2020-08-14 09:46:47,783 epoch 4 - iter 6/23 - loss 1.44538082\n2020-08-14 09:46:49,504 epoch 4 - iter 8/23 - loss 1.42097600\n2020-08-14 09:46:51,176 epoch 4 - iter 10/23 - loss 1.30612972\n2020-08-14 09:46:52,872 epoch 4 - iter 12/23 - loss 1.20628040\n2020-08-14 09:46:54,555 epoch 4 - iter 14/23 - loss 1.20299214\n2020-08-14 09:46:56,203 epoch 4 - iter 16/23 - loss 1.12594500\n2020-08-14 09:46:57,891 epoch 4 - iter 18/23 - loss 1.05994359\n2020-08-14 09:46:59,538 epoch 4 - iter 20/23 - loss 1.03907878\n2020-08-14 09:47:01,150 epoch 4 - iter 22/23 - loss 1.04433420\n2020-08-14 09:47:02,581 ----------------------------------------------------------------------------------------------------\n2020-08-14 09:47:02,583 EPOCH 4 done: loss 1.0443 - lr 0.3000 - bad epochs 0\n2020-08-14 09:47:03,562 DEV : loss 0.758998453617096 - score 0.7486\n2020-08-14 09:47:04,686 TEST : loss 0.6562597155570984 - score 0.8599\n2020-08-14 09:47:04,691 ----------------------------------------------------------------------------------------------------\n2020-08-14 09:47:04,779 epoch 5 - iter 0/23 - loss 0.23086432\n2020-08-14 09:47:06,405 epoch 5 - iter 2/23 - loss 0.58413119\n2020-08-14 09:47:08,061 epoch 5 - iter 4/23 - loss 0.64745774\n2020-08-14 09:47:09,713 epoch 5 - iter 6/23 - loss 0.59633024\n2020-08-14 09:47:11,480 epoch 5 - iter 8/23 - loss 0.70489708\n2020-08-14 09:47:13,174 epoch 5 - iter 10/23 - loss 0.68610029\n2020-08-14 09:47:14,859 epoch 5 - iter 12/23 - loss 0.88584051\n2020-08-14 09:47:16,526 epoch 5 - iter 14/23 - loss 0.89904891\n2020-08-14 09:47:18,187 epoch 5 - iter 16/23 - loss 0.90903763\n2020-08-14 09:47:19,874 epoch 5 - iter 18/23 - loss 0.89399145\n2020-08-14 09:47:21,579 epoch 5 - iter 20/23 - loss 0.85502972\n2020-08-14 09:47:23,246 epoch 5 - iter 22/23 - loss 0.84237767\n2020-08-14 09:47:24,742 ----------------------------------------------------------------------------------------------------\n2020-08-14 09:47:24,745 EPOCH 5 done: loss 0.8424 - lr 0.3000 - bad epochs 1\n2020-08-14 09:47:25,853 DEV : loss 0.7562338709831238 - score 0.7598\n2020-08-14 09:47:26,962 TEST : loss 0.6510859131813049 - score 0.8878\n2020-08-14 09:47:26,967 ----------------------------------------------------------------------------------------------------\n2020-08-14 09:47:27,070 epoch 6 - iter 0/23 - loss 0.81211895\n2020-08-14 09:47:28,738 epoch 6 - iter 2/23 - loss 0.72049708\n2020-08-14 09:47:30,448 epoch 6 - iter 4/23 - loss 0.76020442\n2020-08-14 09:47:32,111 epoch 6 - iter 6/23 - loss 0.64637405\n2020-08-14 09:47:33,832 epoch 6 - iter 8/23 - loss 0.64804429\n2020-08-14 09:47:35,561 epoch 6 - iter 10/23 - loss 0.59896170\n2020-08-14 09:47:37,247 epoch 6 - iter 12/23 - loss 0.62017250\n2020-08-14 09:47:38,925 epoch 6 - iter 14/23 - loss 0.59471101\n2020-08-14 09:47:40,667 epoch 6 - iter 16/23 - loss 0.61451798\n2020-08-14 09:47:42,333 epoch 6 - iter 18/23 - loss 0.65660819\n2020-08-14 09:47:44,001 epoch 6 - iter 20/23 - loss 0.63703280\n2020-08-14 09:47:45,649 epoch 6 - iter 22/23 - loss 0.65120008\n2020-08-14 09:47:47,100 ----------------------------------------------------------------------------------------------------\n2020-08-14 09:47:47,102 EPOCH 6 done: loss 0.6512 - lr 0.3000 - bad epochs 2\n2020-08-14 09:47:48,053 DEV : loss 0.8711250424385071 - score 0.7283\n2020-08-14 09:47:49,120 TEST : loss 0.8211631178855896 - score 0.8792\n2020-08-14 09:47:49,124 ----------------------------------------------------------------------------------------------------\n2020-08-14 09:47:49,241 epoch 7 - iter 0/23 - loss 0.79994875\n2020-08-14 09:47:50,976 epoch 7 - iter 2/23 - loss 0.92813158\n2020-08-14 09:47:52,618 epoch 7 - iter 4/23 - loss 0.59968974\n2020-08-14 09:47:54,261 epoch 7 - iter 6/23 - loss 0.61404911\n2020-08-14 09:47:55,974 epoch 7 - iter 8/23 - loss 0.65827110\n2020-08-14 09:47:57,659 epoch 7 - iter 10/23 - loss 0.69280307\n2020-08-14 09:47:59,303 epoch 7 - iter 12/23 - loss 0.65592613\n2020-08-14 09:48:01,041 epoch 7 - iter 14/23 - loss 0.64914075\n2020-08-14 09:48:02,724 epoch 7 - iter 16/23 - loss 0.60897505\n2020-08-14 09:48:04,427 epoch 7 - iter 18/23 - loss 0.67558943\n"
],
[
"assert result['test_score'] == 0.9154",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
e7925eb9dd8b687387f2fb74e9550314ee3c13fe | 26,994 | ipynb | Jupyter Notebook | tutorials/streamlit_notebooks/healthcare/NER_SIGN_SYMP.ipynb | fcivardi/spark-nlp-workshop | aedb1f5d93577c81bc3dd0da5e46e02586941541 | [
"Apache-2.0"
] | null | null | null | tutorials/streamlit_notebooks/healthcare/NER_SIGN_SYMP.ipynb | fcivardi/spark-nlp-workshop | aedb1f5d93577c81bc3dd0da5e46e02586941541 | [
"Apache-2.0"
] | null | null | null | tutorials/streamlit_notebooks/healthcare/NER_SIGN_SYMP.ipynb | fcivardi/spark-nlp-workshop | aedb1f5d93577c81bc3dd0da5e46e02586941541 | [
"Apache-2.0"
] | null | null | null | 57.927039 | 6,937 | 0.650589 | [
[
[
"\n\n\n\n[](https://colab.research.google.com/github/JohnSnowLabs/spark-nlp-workshop/blob/master/tutorials/streamlit_notebooks/healthcare/NER_SIGN_SYMP.ipynb)\n\n\n",
"_____no_output_____"
],
[
"# **Detect signs and symptoms**",
"_____no_output_____"
],
[
"To run this yourself, you will need to upload your license keys to the notebook. Just Run The Cell Below in order to do that. Also You can open the file explorer on the left side of the screen and upload `license_keys.json` to the folder that opens.\nOtherwise, you can look at the example outputs at the bottom of the notebook.\n\n",
"_____no_output_____"
],
[
"## 1. Colab Setup",
"_____no_output_____"
],
[
"Import license keys",
"_____no_output_____"
]
],
[
[
"import json\nimport os\n\nfrom google.colab import files\n\nlicense_keys = files.upload()\n\nwith open(list(license_keys.keys())[0]) as f:\n license_keys = json.load(f)\n\n# Defining license key-value pairs as local variables\nlocals().update(license_keys)\n\n# Adding license key-value pairs to environment variables\nos.environ.update(license_keys)",
"_____no_output_____"
]
],
[
[
"Install dependencies",
"_____no_output_____"
]
],
[
[
"# Installing pyspark and spark-nlp\n! pip install --upgrade -q pyspark==3.1.2 spark-nlp==$PUBLIC_VERSION\n\n# Installing Spark NLP Healthcare\n! pip install --upgrade -q spark-nlp-jsl==$JSL_VERSION --extra-index-url https://pypi.johnsnowlabs.com/$SECRET\n\n# Installing Spark NLP Display Library for visualization\n! pip install -q spark-nlp-display",
"_____no_output_____"
]
],
[
[
"Import dependencies into Python and start the Spark session",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom pyspark.ml import Pipeline\nfrom pyspark.sql import SparkSession\nimport pyspark.sql.functions as F\n\nimport sparknlp\nfrom sparknlp.annotator import *\nfrom sparknlp_jsl.annotator import *\nfrom sparknlp.base import *\nimport sparknlp_jsl\n\nspark = sparknlp_jsl.start(license_keys['SECRET'])\n\n# manually start session\n# params = {\"spark.driver.memory\" : \"16G\",\n# \"spark.kryoserializer.buffer.max\" : \"2000M\",\n# \"spark.driver.maxResultSize\" : \"2000M\"}\n\n# spark = sparknlp_jsl.start(license_keys['SECRET'],params=params)",
"_____no_output_____"
]
],
[
[
"## 2. Select the NER model and construct the pipeline",
"_____no_output_____"
],
[
"Select the NER model - Sign/symptom models: **ner_clinical, ner_jsl**\n\nFor more details: https://github.com/JohnSnowLabs/spark-nlp-models#pretrained-models---spark-nlp-for-healthcare",
"_____no_output_____"
]
],
[
[
"# You can change this to the model you want to use and re-run cells below.\n# Sign / symptom models: ner_clinical, ner_jsl\n# All these models use the same clinical embeddings.\nMODEL_NAME = \"ner_clinical\"",
"_____no_output_____"
]
],
[
[
"Create the pipeline",
"_____no_output_____"
]
],
[
[
"\n\ndocument_assembler = DocumentAssembler() \\\n .setInputCol('text')\\\n .setOutputCol('document')\n\nsentence_detector = SentenceDetector() \\\n .setInputCols(['document'])\\\n .setOutputCol('sentence')\n\ntokenizer = Tokenizer()\\\n .setInputCols(['sentence']) \\\n .setOutputCol('token')\n\nword_embeddings = WordEmbeddingsModel.pretrained('embeddings_clinical', 'en', 'clinical/models') \\\n .setInputCols(['sentence', 'token']) \\\n .setOutputCol('embeddings')\n\nclinical_ner = MedicalNerModel.pretrained(MODEL_NAME, \"en\", \"clinical/models\") \\\n .setInputCols([\"sentence\", \"token\", \"embeddings\"])\\\n .setOutputCol(\"ner\")\n\nner_converter = NerConverter()\\\n .setInputCols(['sentence', 'token', 'ner']) \\\n .setOutputCol('ner_chunk')\n\nnlp_pipeline = Pipeline(stages=[\n document_assembler, \n sentence_detector,\n tokenizer,\n word_embeddings,\n clinical_ner,\n ner_converter])",
"embeddings_clinical download started this may take some time.\nApproximate size to download 1.6 GB\n[OK!]\nner_clinical download started this may take some time.\nApproximate size to download 13.7 MB\n[OK!]\n"
]
],
[
[
"## 3. Create example inputs",
"_____no_output_____"
]
],
[
[
"# Enter examples as strings in this array\ninput_list = [\n \"\"\"The patient is a 21-day-old Caucasian male here for 2 days of congestion - mom has been suctioning yellow discharge from the patient's nares, plus she has noticed some mild problems with his breathing while feeding (but negative for any perioral cyanosis or retractions). One day ago, mom also noticed a tactile temperature and gave the patient Tylenol. Baby also has had some decreased p.o. intake. His normal breast-feeding is down from 20 minutes q.2h. to 5 to 10 minutes secondary to his respiratory congestion. He sleeps well, but has been more tired and has been fussy over the past 2 days. The parents noticed no improvement with albuterol treatments given in the ER. His urine output has also decreased; normally he has 8 to 10 wet and 5 dirty diapers per 24 hours, now he has down to 4 wet diapers per 24 hours. Mom denies any diarrhea. His bowel movements are yellow colored and soft in nature.\"\"\"\n]",
"_____no_output_____"
]
],
[
[
"## 4. Use the pipeline to create outputs",
"_____no_output_____"
]
],
[
[
"empty_df = spark.createDataFrame([['']]).toDF('text')\npipeline_model = nlp_pipeline.fit(empty_df)\ndf = spark.createDataFrame(pd.DataFrame({'text': input_list}))\nresult = pipeline_model.transform(df)",
"_____no_output_____"
]
],
[
[
"## 5. Visualize results",
"_____no_output_____"
]
],
[
[
"from sparknlp_display import NerVisualizer\n\nNerVisualizer().display(\n result = result.collect()[0],\n label_col = 'ner_chunk',\n document_col = 'document'\n)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7925f5eac85ba8220731bba15d63e87df9030fa | 118,296 | ipynb | Jupyter Notebook | photoisomerization/cam_images_2_photoisomerization_v0_2.ipynb | yongrong-qiu/mouse-scene-cam | a9afd7e8cd7abba32e4807b9018339edbe4ca13c | [
"MIT"
] | null | null | null | photoisomerization/cam_images_2_photoisomerization_v0_2.ipynb | yongrong-qiu/mouse-scene-cam | a9afd7e8cd7abba32e4807b9018339edbe4ca13c | [
"MIT"
] | null | null | null | photoisomerization/cam_images_2_photoisomerization_v0_2.ipynb | yongrong-qiu/mouse-scene-cam | a9afd7e8cd7abba32e4807b9018339edbe4ca13c | [
"MIT"
] | 1 | 2021-05-27T09:18:15.000Z | 2021-05-27T09:18:15.000Z | 112.555661 | 44,728 | 0.79482 | [
[
[
"### Purpose of this notebook\n\nThis notebook estimates the excitation (as photoisomerization rate at the photoreceptor level) that is expected to be caused by the images recorded with the UV/G mouse camera.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\n# Global dictionary\nd = dict()\n\n# Global constants\nTWILIGHT = 0\nDAYLIGHT = 1\n\nUV_S = 0\nUV_M = 1\nG_S = 2\nG_M = 3\n\nCONE = 0\nROD = 1\n\nCHAN_UV = 0\nCHAN_G = 1",
"_____no_output_____"
]
],
[
[
"### Approach\n\nWhen calibrating the mouse camera, we used LEDs of defined wavelength and brightness to map normalized intensity (camera pixel values, 0..1) to power meter readings (see STAR Methods in the manuscript). To relate this power to the photon flux at the cornea and finally the photoisomerisation rate at the photoreceptor level, we need to consider: \n\n* How much light we loose in the camera, that is, we need the optical paths´ **attenuation factors** from the fisheye lens to the camera chip for the UV ($ \\mu_{lens2cam,UV} $) and green ($ \\mu_{lens2cam,G} $) channel; \n\n* The wavelength-specific **transmission of mouse optical aparatus** for UV ($T_{UV}$) and green ($T_G$) light;\n\n* The **ratio between pupil size and retinal area** ($R_{pup2ret}$) to estimate, how much light reaches the retina giving the pupil adapts to the overall brightness of the scene. \n\nOur approch consists of two main steps:\n\n1. We first map electrical power ($P_{el}$, in $[W]$) to photon flux ($P_{Phi}$, in $[photons/s]$), \n\n $$\n P_{Phi}(\\lambda) = \\frac{P_{el}(\\lambda) \\cdot a \\cdot \\lambda \\cdot 10^{-9}} {c \\cdot h }\\cdot \\frac{1}{\\mu_{lens2cam}(\\lambda)}.\n $$ \n\n For $\\lambda$, we use the peak wavelength of the photoreceptor's spectral sensitivity curve ($\\lambda_{S}=360 \\: nm$, $\\lambda_{M}=510 \\: nm$). \n \n The rest are constants ($a=6.2421018 \\: eV/J$, $c=299,792,458 \\: m/s$, and $h=4.135667 \\cdot10^{-15} \\: eV/s$). \n\n2. Next, we convert the photon flux to photoisomerisation rate ($R_{Iso}$, in $[P^*/cone/s]$),\n\n $$\n R_{Iso}(\\lambda) = \\frac{P_{Phi}(\\lambda)}{A_{Stim}} \\cdot A_{Collect} \\cdot S_{Act} \\cdot T(\\lambda) \\cdot R_{pup2ret}\n $$\n\n where $A_{Stim}=10^8 \\: \\mu m^2$ is area that is iluminated on the power meter sensor, and $A_{Collect}=0.2 \\: \\mu m^2$ the photoreceptor's outer segment (OS) light collection area (see below).\n\n With $S_{Act}$ we take into account that the bandpass filters in the camera pathways do not perfectly match these sensitivity spectra (see below).\n\n > Note: The OS light collection area ($[\\mu m^2]$) an experimentally determined value, e.g. for wt mouse cones that are fully dark-adapted, a value of 0.2 is be assumed; for mouse rods, a value of 0.5 is considered realistic (for details, see [Nikonov et al., 2006](http://www.ncbi.nlm.nih.gov/pubmed/16567464)). ",
"_____no_output_____"
]
],
[
[
"d.update({\"ac_um2\": [0.2, 0.5], \"peak_S\": 360, \"peak_M\": 510, \"A_stim_um2\": 1e8})",
"_____no_output_____"
]
],
[
[
"### Attenuation factors of the two camera pathways\n\nWe first calculated the attenuation factor from the fisheye lens to the focal plane of the camera chip. \n\nTo this end, we used a spectrometer (STS-UV, Ocean Optics) with an optical fiber (P50-1-UV-VIS) to first measure the spectrum of the sky directly, and then at the camera focal planes of the UV and the green pathways. These readouts, which are spectra, are referred to as $P_{direct}$, $P_{UV}$ and $P_{G}$. ",
"_____no_output_____"
]
],
[
[
"#%%capture\n#!wget -O sky_spectrum.npy https://www.dropbox.com/s/p8uk4k6losfu309/sky_spectrum.npy?dl=0\n \n#spect = np.load('sky_spectrum.npy', allow_pickle=True).item() ",
"_____no_output_____"
],
[
"# Load spectra\n# The exposure times were 4 s for `direct` and `g`, and 30 s for `uv`\nspect = np.load('data/sky_spectrum.npy', allow_pickle=True).item() \n\nfig,axes=plt.subplots(nrows=1,ncols=1,figsize=(8,4))\naxes.plot(spect[\"wavelength\"], spect[\"direct\"], color='k', label='P_Direct')\naxes.plot(spect[\"wavelength\"], spect[\"g\"], color='g', label='P_G')\naxes.plot(spect[\"wavelength\"], spect[\"uv\"], color='purple',label='P_UV')\naxes.set_xlabel (\"Wavelength [nm]\")\naxes.set_ylabel(\"Counts\")\naxes.grid()\naxes.legend(loc='upper right', bbox_to_anchor=(1.6, 1.0))",
"_____no_output_____"
]
],
[
[
"Since the readout on the objective side (fisheye lens) is related to both visual angle and area, while the readout on the imaging side is only related to area, we define:\n\n$$\n\\begin{align}\nP_{direct} &= P_{total} \\cdot \\frac{A_{fiber}}{A_{lens}} \\cdot \\frac{\\theta_{fiber}}{\\theta_{lens}}\\\\\nP_{UV} &= P_{total} \\cdot \\frac{A_{fiber}}{A_{chip}} \\cdot \\mu_{lens2cam,UV}\\\\\nP_{G} &= P_{total} \\cdot \\frac{A_{fiber}}{A_{chip}} \\cdot \\mu_{lens2cam,G}\n\\end{align}\n$$ \n\nwhere $P_{total}$ denotes the total power of the incident light at the fisheye lens, $A_{fiber}$ the area of the fibre, $A_{lens}$ the area of the fisheye lens, $A_{chip}$ the imaging area of the camera chip, and $\\theta_{fiber}$ and $\\theta_{lens}$ the acceptance angles of fiber and fisheye lens, respectively. \n\nAfter rearranging the equations, we get: \n\n$$\n\\begin{align}\n\\mu_{lens2cam,UV} &= \\frac{P_{UV}}{P_{direct}} \\cdot \\frac{A_{chip}}{A_{lens}} \\cdot \\frac{\\theta_{fiber}}{\\theta_{lens}}\\\\\n\\mu_{lens2cam,G} &= \\frac{P_{G}}{P_{direct}} \\cdot \\frac{A_{chip}}{A_{lens}} \\cdot \\frac{\\theta_{fiber}}{\\theta_{lens}}\n\\end{align}\n$$ \n\nBy calculating the ratio between the area under curve (AUC) of the spectrum for the respective chromatic channel (within in the spectral range of the respective bandpass filter) and the AUC of the spectrum for the direct measurement, we get:\n\n$$\n\\frac{P_{UV}}{P_{direct}} = \\frac{1}{21}, \\frac{P_{G}}{P_{direct}} = \\frac{1}{2}\n$$ \n\nPractically, we also take the different exposure times (4 s for $P_{direct}$ and $P_{G}$, and 30 s for $P_{UV}$) into account.",
"_____no_output_____"
]
],
[
[
"direct_exp_s = 4\nUV_exp_s = 30\nG_exp_s = 4 \nP_UV2direct = 1/(np.trapz(spect[\"direct\"][350-300:420-300])/np.trapz(spect[\"uv\"][350-300:420-300]) *UV_exp_s/direct_exp_s) \nP_G2direct = 1/(np.trapz(spect[\"direct\"][470-300:550-300])/np.trapz(spect[\"g\"][470-300:550-300]) *G_exp_s/direct_exp_s) \n\nprint(\"P_UV/P_direct = {0:.3f}\".format(P_UV2direct))\nprint(\"P_G/P_direct = {0:.3f}\".format(P_G2direct))",
"P_UV/P_direct = 0.047\nP_G/P_direct = 0.533\n"
]
],
[
[
"\nThe diameters of the camera chip's imaging area and the fisheye lens were $2,185 \\: \\mu m$ and $15,000 \\: \\mu m$, respectively. The acception angles of the optical fiber and the fisheye lens were $\\theta_{fibre}=24.8^{\\circ}$ and $\\theta_{lens}=180^{\\circ}$, respectively.",
"_____no_output_____"
]
],
[
[
"A_cam = np.pi*(2185/2)**2\nA_lens = np.pi*(15000/2)**2\ntheta_fiber = 24.8 \ntheta_lens = 180",
"_____no_output_____"
]
],
[
[
"Now we can get the attenuation factors $ \\mu_{lens2cam,UV} $ and $ \\mu_{lens2cam,G} $, covering the optical path from the fisheye lens to the camera chip: ",
"_____no_output_____"
]
],
[
[
"mu_lens2cam = [0,0]\nmu_lens2cam[CHAN_UV] = P_UV2direct *A_cam /A_lens *theta_fiber /theta_lens\nmu_lens2cam[CHAN_G] = P_G2direct *A_cam /A_lens * theta_fiber /theta_lens\nd.update({\"mu_lens2cam\": mu_lens2cam})\n\nprint(\"mu_lens2cam for UV,G = {0:.3e}, {1:.3e}\".format(mu_lens2cam[CHAN_UV], mu_lens2cam[CHAN_G]))",
"mu_lens2cam for UV,G = 1.365e-04, 1.557e-03\n"
]
],
[
[
"### Attenuation by mouse eye optics\n\nAnother factor we need to consider is the wavelength-dependent attenuation by the mouse eye optics. The relative transmission for UV ($T_{Rel}(UV)$, at $\\lambda=360 \\: nm$) and green ($T_{Rel}(G)$, at $\\lambda=510 \\: nm$) is approx. 35% and 55%, respectively ([Henriksson et al., 2010](https://pubmed.ncbi.nlm.nih.gov/19925789/)).",
"_____no_output_____"
]
],
[
[
"d.update({\"T_rel\": [0.35, 0.55]})",
"_____no_output_____"
]
],
[
[
"In addition, the light reaching the retina depends on the ratio ($R_{pup2ret}$) between pupil area and retinal area (both in $[mm^2]$) ([Rhim et al., 2020](https://www.biorxiv.org/content/10.1101/2020.11.03.366682v1)). Here, we assume pupil areas of $0.1 \\: mm^2$ (maximally constricted) at daytime and $0.22 \\: mm^2$ at twighlight (approx. 10% of full pupil area; see [Pennesi et al., 1998](https://pubmed.ncbi.nlm.nih.gov/9761294/)). To calculate the retinal area of the mouse, we assume an eye axial length of approx. $3 \\: mm$ and that the retina covers about 60% of the sphere's surface ([Schmucker & Schaeffel, 2004](https://www.sciencedirect.com/science/article/pii/S0042698904001257#FIG4)).",
"_____no_output_____"
]
],
[
[
"eye_axial_len_mm = 3\nret_area_mm2 = 0.6 *(eye_axial_len_mm/2)**2 *np.pi *4\npup_area_mm2 = [0.22, 0.1]\nR_pup2ret= [x /ret_area_mm2 for x in pup_area_mm2]\nd.update({\"R_pup2ret\": R_pup2ret, \"pup_area_mm2\": pup_area_mm2, \"ret_area_mm2\": ret_area_mm2})\n\nprint(\"mouse retinal area [mm²] = {0:.1f}\".format(ret_area_mm2))\nprint(\"pupil area [mm²] = twilight: {0:.1f} \\tdaylight: {1:.1f}\".format(pup_area_mm2[TWILIGHT], pup_area_mm2[DAYLIGHT]))\nprint(\"ratio of pupil area to retinal area = twilight: {0:.3f} \\tdaylight: {1:.3f}\".format(R_pup2ret[TWILIGHT],R_pup2ret[DAYLIGHT]))",
"mouse retinal area [mm²] = 17.0\npupil area [mm²] = twilight: 0.2 \tdaylight: 0.1\nratio of pupil area to retinal area = twilight: 0.013 \tdaylight: 0.006\n"
]
],
[
[
"### Cross-activation of S- and M-opsins ...\n\n... by the UV and green camera channels, yielding $S_{Act}(S,UV)$, $S_{Act}(S,G)$, $S_{Act}(M,UV)$, and $S_{Act}(M,G)$.",
"_____no_output_____"
]
],
[
[
"#%%capture\n#!wget -O opsin_filter_spectrum.npy https://www.dropbox.com/s/doh1jjqukdcpvpy/opsin_filter_spectrum.npy?dl=0\n\n#spect = np.load('opsin_filter_spectrum.npy', allow_pickle=True).item() ",
"_____no_output_____"
],
[
"# Load opsin and filter spectra\nspect = np.load('data/opsin_filter_spectrum.npy', allow_pickle=True).item() \n\nwavelength = spect[\"wavelength\"]\nmouseSOpsin = spect[\"mouseSOpsin\"]\nmouseMOpsin = spect[\"mouseMOpsin\"]\nfilter_uv = spect[\"filter_uv\"]\nfilter_g = spect[\"filter_g\"]\nfilter_uv_scone = np.minimum(filter_uv,mouseSOpsin)\nfilter_uv_mcone = np.minimum(filter_uv,mouseMOpsin)\nfilter_g_scone = np.minimum(filter_g, mouseSOpsin)\nfilter_g_mcone = np.minimum(filter_g, mouseMOpsin)\n\nS_act = [0]*4\nS_act[UV_S] = np.trapz(filter_uv_scone)/np.trapz(filter_uv)\nS_act[UV_M] = np.trapz(filter_uv_mcone)/np.trapz(filter_g)\nS_act[G_S] = np.trapz(filter_g_scone)/np.trapz(filter_uv)\nS_act[G_M] = np.trapz(filter_g_mcone)/np.trapz(filter_g)\nd.update({\"S_act\": S_act})\n\nfig,axes=plt.subplots(nrows=1,ncols=1,figsize=(8,4))\naxes.plot(wavelength,mouseMOpsin,color='g', linestyle='-',label='M-cone')\naxes.plot(wavelength,mouseSOpsin,color='purple',linestyle='-',label='S-cone')\naxes.plot(wavelength,filter_g, color='g', linestyle='--',label='Filter-G')\naxes.plot(wavelength,filter_uv,color='purple', linestyle='--',label='Filter-UV')\naxes.fill_between(wavelength,y1=filter_g_mcone, y2=0,color='g', alpha=0.5)\naxes.fill_between(wavelength,y1=filter_g_scone, y2=0,color='g', alpha=0.5)\naxes.fill_between(wavelength,y1=filter_uv_mcone,y2=0,color='purple',alpha=0.5)\naxes.fill_between(wavelength,y1=filter_uv_scone,y2=0,color='purple',alpha=0.5)\naxes.set_xlabel (\"Wavelenght [nm]\")\naxes.set_ylabel(\"Rel. sensitivity\")\naxes.legend(loc='upper right', bbox_to_anchor=(1.4, 1.0))\n\nprint(\"S_act UV -> S = {0:.3f}\".format(S_act[UV_S]))\nprint(\" UV -> M = {0:.3f}\".format(S_act[UV_M]))\nprint(\" G -> S = {0:.3f}\".format(S_act[G_S]))\nprint(\" G -> M = {0:.3f}\".format(S_act[G_M]))",
"S_act UV -> S = 0.625\n UV -> M = 0.118\n G -> S = 0.000\n G -> M = 0.858\n"
]
],
[
[
"### Estimating photoisomerization rates\n\nThe following function converts normalized image intensities (0...1) to $P_{el}(\\lambda)$ (in $[\\mu W]$), $P_{Phi}(\\lambda)$ (in $[photons /s]$), and $R_{Iso}(\\lambda)$ (in $[P^*/cone/s]$).\n",
"_____no_output_____"
]
],
[
[
"def inten2Riso(intensities, pup_area_mm2, pr_type=CONE):\n \"\"\"\n Transfer the normalized image intensities (0...1) to power (unit: uW), \n photon flux (unit: photons/s) and photoisomerisation rate (P*/cone/s)\n\n Input:\n intensities : image intensities (0...1) for both channels as tuple\n pup_area_mm2 : pupil area in mm^2\n\n Output:\n P_el : tuple (CHAN_UV, CHAN_G) \n P_Phi : tuple (CHAN_UV, CHAN_G) \n R_Iso : tuple (UV_S, UV_M, G_S, G_M)\n \"\"\"\n global d\n\n h = 4.135667e-15 # Planck's constant [eV*s]\n c = 299792458 # speed of light [m/s]\n eV_per_J = 6.242e+18 # [eV] per [J]\n \n # Convert normalized image intensities (0...1) to power ([uW])\n # (Constants from camera calibration, see STAR Methods for details)\n P_el = [0]*2\n P_el[CHAN_UV] = intensities[CHAN_UV] *0.755 +0.0049\n P_el[CHAN_G] = intensities[CHAN_G] *6.550 +0.0097 \n\n # Convert electrical power ([uW]) to photon flux ([photons/s])\n P_Phi = [0]*2\n P_Phi[CHAN_UV] = (P_el[CHAN_UV] *1e-6) *eV_per_J *(d[\"peak_S\"]*1e-9)/(c*h) *(1/d[\"mu_lens2cam\"][CHAN_UV])\n P_Phi[CHAN_G] = (P_el[CHAN_G] *1e-6) *eV_per_J *(d[\"peak_M\"]*1e-9)/(c*h) *(1/d[\"mu_lens2cam\"][CHAN_G])\n\n # Convert photon flux ([photons/s]) to photoisomerisation rate ([P*/cone/s])\n R_pup2ret = pup_area_mm2 /d[\"ret_area_mm2\"]\n R_Iso = [0]*4\n for j in [UV_S, UV_M, G_S, G_M]:\n chan = CHAN_UV if j < G_S else CHAN_G \n R_Iso[j] = P_Phi[chan] /d[\"A_stim_um2\"] *d[\"ac_um2\"][pr_type]* d[\"S_act\"][j] *d[\"T_rel\"][chan] *R_pup2ret\n\n return P_el, P_Phi, R_Iso",
"_____no_output_____"
]
],
[
[
"Example `[[0.18, 0.11], [0.06, 0.14]]`, with the following format `[upper[UV,G],lower[UV,G]]`",
"_____no_output_____"
]
],
[
[
"intensities=[[0.18, 0.11], [0.06, 0.14]]\nfor j, i in enumerate(intensities):\n l = inten2Riso(i, 0.2)\n print(\"{0:2d} (UV, G) P_el = {1:.3f}, {2:.3f}\\t P_Phi = {3:.1e}, {4:.1e} \".format(j, l[0][0], l[0][1], l[1][0], l[1][1]))\n print(\" UV->S = {0:.1e} \\t UV->M = {1:.1e} \\t G->S = {2:.1e} \\t G->M = {3:.1e}\".format(l[2][0], l[2][1], l[2][2], l[2][3]))\n",
" 0 (UV, G) P_el = 0.141, 0.730\t P_Phi = 1.9e+15, 1.2e+15 \n UV->S = 9.6e+03 \t UV->M = 1.8e+03 \t G->S = 7.0e-01 \t G->M = 1.3e+04\n 1 (UV, G) P_el = 0.050, 0.927\t P_Phi = 6.7e+14, 1.5e+15 \n UV->S = 3.4e+03 \t UV->M = 6.5e+02 \t G->S = 8.8e-01 \t G->M = 1.7e+04\n"
]
],
[
[
"### Generate Supplementary Table 1",
"_____no_output_____"
]
],
[
[
"col_names = ['Mean intensity<br>group', 'Visual<br>field', 'Camera<br>channel', 'Norm.<br>intensity', 'P_el<br>in [µW]',\\\n 'P_Phi<br>in [photons/s]', 'Pupil area<br>in [mm2]',\\\n 'R_Iso<br>in [P*/cone/s], S', 'R_Iso<br>in [P*/cone/s], M', 'R_Iso<br>in [P*/rod/s], rod']\n\ndata_df = pd.DataFrame(columns = col_names)\n\ngroup = ['Low', 'Medium', 'High', 'Twilight']\ngroup = [item for item in group for i in range(4)]\ndata_df['Mean intensity<br>group'] = group\n\nvisual_field=['Upper', 'Upper', 'Lower', 'Lower']*4\ndata_df['Visual<br>field'] = visual_field\n\ncamera_channel=['UV', 'G']*8\ndata_df['Camera<br>channel'] = camera_channel\n\nnorm_intensity = [0.18, 0.11, 0.06, 0.14, 0.28, 0.16, 0.09, 0.21, 0.50, 0.34, 0.22, 0.46, 0.05, 0.06, 0.02, 0.05]\ndata_df['Norm.<br>intensity'] = norm_intensity\n\n# Pupil area\ndata_df['Pupil area<br>in [mm2]'] = np.where(data_df['Mean intensity<br>group'] == 'Twilight', \\\n d['pup_area_mm2'][TWILIGHT], d['pup_area_mm2'][DAYLIGHT])\n\n# Photoisomerisations\nfor ii in range(int(len(data_df.index)/2)):\n tempUV, tempG = data_df.iloc[ii*2, 3], data_df.iloc[ii*2+1, 3]\n templ = inten2Riso([tempUV, tempG], data_df.iloc[ii*2, 6])\n data_df.iloc[ii*2, 4], data_df.iloc[ii*2+1, 4] = templ[0][0], templ[0][1]\n data_df.iloc[ii*2, 5], data_df.iloc[ii*2+1, 5] = templ[1][0], templ[1][1]\n data_df.iloc[ii*2,7], data_df.iloc[ii*2,8], data_df.iloc[ii*2+1,7], data_df.iloc[ii*2+1,8] =\\\n templ[2][0], templ[2][1], templ[2][2], templ[2][3]\n templ = inten2Riso([tempUV,tempG], data_df.iloc[ii*2, 6], pr_type=ROD) \n data_df.iloc[ii*2,9] = templ[2][1] \n data_df.iloc[ii*2+1,9] = templ[2][3]\n\n# Show table\n'''\n# Set colormap equal to seaborns light green color palette\ncmG = sns.light_palette(\"green\", n_colors=50, as_cmap=True, reverse=False)\ncmUV = sns.light_palette(\"purple\", n_colors=50, as_cmap=True, reverse=False)\n\n# Set CSS properties for th elements in dataframe\nth_props = [\n ('font-size', '14px'),\n ('text-align', 'center'),\n ('font-weight', 'bold'),\n ('color', '#6d6d6d'),\n ('background-color', '#f7f7f9')\n ]\n\n# Set CSS properties for td elements in dataframe\ntd_props = [\n ('font-size', '14px')\n ]\n\n# Set table styles\nstyles = [\n dict(selector=\"th\", props=th_props),\n dict(selector=\"td\", props=td_props)\n ]\n\n(data_df.style\n .background_gradient(cmap=cmUV, subset=['R_Iso<br>in [P*/cone/s], S'])\n .background_gradient(cmap=cmG, subset=['R_Iso<br>in [P*/cone/s], M'])\n .background_gradient(cmap=cmG, subset=['R_Iso<br>in [P*/rod/s], rod'])\n #.highlight_max(subset=['R_Iso<br>in [P*/cone/s], S','R_Iso<br>in [P*/cone/s], M'])\n .format({\"Norm.<br>intensity\": \"{:.2f}\",\"P_el<br>in [µW]\": \"{:.3f}\", \n \"P_Phi<br>in [photons/s]\": \"{:.3e}\", \"Pupil area<br>in [mm2]\": \"{:.1f}\",\n \"R_Iso<br>in [P*/cone/s], S\": \"{:.0f}\", \n \"R_Iso<br>in [P*/cone/s], M\": \"{:.0f}\",\n \"R_Iso<br>in [P*/rod/s], rod\": \"{:.0f}\"})\n .set_table_styles(styles)\n .set_properties(**{'white-space': 'pre-wrap',}))\n'''\ndisplay(data_df)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e79269be76245d6019942a0d22f093bc47bdf43c | 6,221 | ipynb | Jupyter Notebook | 12-Convolutions and Blurring.ipynb | moh3n9595/class.vision | cbcc65fd1f226273d26e44576ca7c3950faea75c | [
"MIT"
] | 103 | 2018-02-23T15:58:26.000Z | 2022-03-09T05:49:14.000Z | 12-Convolutions and Blurring.ipynb | Deepstatsanalysis/class.vision | d7859f51d4f969913549e440fdc45f673c9da3de | [
"MIT"
] | null | null | null | 12-Convolutions and Blurring.ipynb | Deepstatsanalysis/class.vision | d7859f51d4f969913549e440fdc45f673c9da3de | [
"MIT"
] | 53 | 2018-02-16T20:38:29.000Z | 2022-03-07T10:12:10.000Z | 30.199029 | 310 | 0.584793 | [
[
[
"<img src=\"http://akhavanpour.ir/notebook/images/srttu.gif\" alt=\"SRTTU\" style=\"width: 150px;\"/>\n\n[](https://notebooks.azure.com/import/gh/Alireza-Akhavan/class.vision)",
"_____no_output_____"
],
[
"## <div style=\"direction:rtl;text-align:right;font-family:B Lotus, B Nazanin, Tahoma\">عملگر convolution</div>",
"_____no_output_____"
],
[
"<img src=\"lecture_images/Convolution_schematic.gif\" style=\"width:500px;height:300px;\"><caption><center><div style=\"direction:rtl;font-family:Tahoma\">**Convolution عمل**</div><br> with a filter of 2x2 and a stride of 1 (stride = amount you move the window each time you slide) </center></caption>\n\n<br>\n<div style=\"direction:rtl;text-align:right;font-family:Tahoma\">\nسایت زیر برای آشنایی با کرنلها بسیار مناسب است:\n</div>\nhttp://setosa.io/ev/image-kernels/\n<br>\n<div style=\"direction:rtl;text-align:right;font-family:Tahoma\">\nاگر بخواهیم تصویر خروجی با تصویر ورودی هم اندازه باشد چه کنیم؟\n</div>",
"_____no_output_____"
],
[
"## Convolutions and Blurring",
"_____no_output_____"
]
],
[
[
"import cv2\nimport numpy as np\n\nimage = cv2.imread('images/input.jpg')\ncv2.imshow('Original Image', image)\ncv2.waitKey(0)\n\n# Creating our 3 x 3 kernel\nkernel_3x3 = np.ones((3, 3), np.float32) / 9\n\n# We use the cv2.fitler2D to conovlve the kernal with an image \nblurred = cv2.filter2D(image, -1, kernel_3x3)\ncv2.imshow('3x3 Kernel Blurring', blurred)\ncv2.waitKey(0)\n\n# Creating our 7 x 7 kernel\nkernel_7x7 = np.ones((7, 7), np.float32) / 49\n\nblurred2 = cv2.filter2D(image, -1, kernel_7x7)\ncv2.imshow('7x7 Kernel Blurring', blurred2)\ncv2.waitKey(0)\n\ncv2.destroyAllWindows()",
"_____no_output_____"
]
],
[
[
"### Other commonly used blurring methods in OpenCV",
"_____no_output_____"
]
],
[
[
"import cv2\nimport numpy as np\n\nimage = cv2.imread('images/input.jpg')\ncv2.imshow('original', image)\ncv2.waitKey(0)\n\n# Averaging done by convolving the image with a normalized box filter. \n# This takes the pixels under the box and replaces the central element\n# Box size needs to odd and positive \nblur = cv2.blur(image, (3,3))\ncv2.imshow('Averaging', blur)\ncv2.waitKey(0)\n\n# Instead of box filter, gaussian kernel\nGaussian = cv2.GaussianBlur(image, (7,7), 0)\ncv2.imshow('Gaussian Blurring', Gaussian)\ncv2.waitKey(0)\n\n# Takes median of all the pixels under kernel area and central \n# element is replaced with this median value\nmedian = cv2.medianBlur(image, 5)\ncv2.imshow('Median Blurring', median)\ncv2.waitKey(0)\n\n# Bilateral is very effective in noise removal while keeping edges sharp\nbilateral = cv2.bilateralFilter(image, 9, 75, 75)\ncv2.imshow('Bilateral Blurring', bilateral)\ncv2.waitKey(0)\ncv2.destroyAllWindows()",
"_____no_output_____"
]
],
[
[
"## Image De-noising - Non-Local Means Denoising",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport cv2\n\nimage = cv2.imread('images/taj-rgb-noise.jpg')\n\n# Parameters, after None are - the filter strength 'h' (5-10 is a good range)\n# Next is hForColorComponents, set as same value as h again\n# \ndst = cv2.fastNlMeansDenoisingColored(image, None, 6, 6, 7, 21)\n\ncv2.imshow('Fast Means Denoising', dst)\ncv2.imshow('original image', image)\ncv2.waitKey(0)\n\ncv2.destroyAllWindows()",
"_____no_output_____"
]
],
[
[
"**There are 4 variations of Non-Local Means Denoising:**\n\n- cv2.fastNlMeansDenoising() - works with a single grayscale images\n- cv2.fastNlMeansDenoisingColored() - works with a color image.\n- cv2.fastNlMeansDenoisingMulti() - works with image sequence captured in short period of time (grayscale images)\n- cv2.fastNlMeansDenoisingColoredMulti() - same as above, but for color images.\n\nhttps://docs.opencv.org/3.3.1/d5/d69/tutorial_py_non_local_means.html",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-info\">\n<div style=\"direction:rtl;text-align:right;font-family:B Lotus, B Nazanin, Tahoma\"> دانشگاه تربیت دبیر شهید رجایی<br>مباحث ویژه - آشنایی با بینایی کامپیوتر<br>علیرضا اخوان پور<br>96-97<br>\n</div>\n<a href=\"https://www.srttu.edu/\">SRTTU.edu</a> - <a href=\"http://class.vision\">Class.Vision</a> - <a href=\"http://AkhavanPour.ir\">AkhavanPour.ir</a>\n</div>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
e7926b79f7f5867392bb56f7c498ba486229e408 | 54,047 | ipynb | Jupyter Notebook | model_1.ipynb | poogoel/machine-learning-challenge | 3a5783238ed3d1685b08bd74181083eed3836ad9 | [
"ADSL"
] | null | null | null | model_1.ipynb | poogoel/machine-learning-challenge | 3a5783238ed3d1685b08bd74181083eed3836ad9 | [
"ADSL"
] | null | null | null | model_1.ipynb | poogoel/machine-learning-challenge | 3a5783238ed3d1685b08bd74181083eed3836ad9 | [
"ADSL"
] | null | null | null | 41.415326 | 285 | 0.471219 | [
[
[
"# Update sklearn to prevent version mismatches\n!pip install sklearn --upgrade",
"Requirement already up-to-date: sklearn in c:\\users\\poona\\anaconda3\\envs\\pythondata\\lib\\site-packages (0.0)\nRequirement already satisfied, skipping upgrade: scikit-learn in c:\\users\\poona\\anaconda3\\envs\\pythondata\\lib\\site-packages (from sklearn) (0.21.3)\nRequirement already satisfied, skipping upgrade: joblib>=0.11 in c:\\users\\poona\\anaconda3\\envs\\pythondata\\lib\\site-packages (from scikit-learn->sklearn) (0.14.0)\nRequirement already satisfied, skipping upgrade: numpy>=1.11.0 in c:\\users\\poona\\anaconda3\\envs\\pythondata\\lib\\site-packages (from scikit-learn->sklearn) (1.17.4)\nRequirement already satisfied, skipping upgrade: scipy>=0.17.0 in c:\\users\\poona\\anaconda3\\envs\\pythondata\\lib\\site-packages (from scikit-learn->sklearn) (1.3.2)\n"
],
[
"# install joblib. This will be used to save your model. \n# Restart your kernel after installing \n!pip install joblib",
"Requirement already satisfied: joblib in c:\\users\\poona\\anaconda3\\envs\\pythondata\\lib\\site-packages (0.14.0)\n"
],
[
"import pandas as pd",
"_____no_output_____"
],
[
"df = pd.read_csv(\"../resources/exoplanet_data.csv\")\n# Drop the null columns where all values are null\ndf = df.dropna(axis='columns', how='all')\n# Drop the null rows\ndf = df.dropna()\ndf.head()",
"_____no_output_____"
],
[
"# # Set features. This will also be used as your x values.\n# selected_features = df[['names', 'of', 'selected', 'features', 'here']]",
"_____no_output_____"
],
[
"X = df.drop(\"koi_disposition\", axis=1)\ny = df[\"koi_disposition\"]\nprint(X.shape, y.shape)",
"(6991, 40) (6991,)\n"
],
[
"from sklearn.model_selection import train_test_split\nfrom sklearn.preprocessing import LabelEncoder, MinMaxScaler\nfrom tensorflow.keras.utils import to_categorical",
"C:\\Users\\poona\\Anaconda3\\envs\\PythonData\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:526: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint8 = np.dtype([(\"qint8\", np.int8, 1)])\nC:\\Users\\poona\\Anaconda3\\envs\\PythonData\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:527: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint8 = np.dtype([(\"quint8\", np.uint8, 1)])\nC:\\Users\\poona\\Anaconda3\\envs\\PythonData\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:528: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint16 = np.dtype([(\"qint16\", np.int16, 1)])\nC:\\Users\\poona\\Anaconda3\\envs\\PythonData\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:529: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint16 = np.dtype([(\"quint16\", np.uint16, 1)])\nC:\\Users\\poona\\Anaconda3\\envs\\PythonData\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:530: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint32 = np.dtype([(\"qint32\", np.int32, 1)])\nC:\\Users\\poona\\Anaconda3\\envs\\PythonData\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:535: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n np_resource = np.dtype([(\"resource\", np.ubyte, 1)])\n"
],
[
"X_train, X_test, y_train, y_test = train_test_split(\n X, y, random_state=1, stratify=y)",
"_____no_output_____"
],
[
"X_train.head()",
"_____no_output_____"
],
[
"# Scale your data\nX_scaler = MinMaxScaler().fit(X_train)\nX_train_scaled = X_scaler.transform(X_train)\nX_test_scaled = X_scaler.transform(X_test)",
"_____no_output_____"
],
[
"# Create the SVC Model\nfrom sklearn.svm import SVC \nmodel = SVC(kernel='linear')\nmodel",
"_____no_output_____"
],
[
"model.fit(X_train_scaled, y_train)",
"_____no_output_____"
],
[
"print(f\"Training Data Score: {model.score(X_train_scaled, y_train)}\")\nprint(f\"Testing Data Score: {model.score(X_test_scaled, y_test)}\")",
"Training Data Score: 0.8439824527942018\nTesting Data Score: 0.8415331807780321\n"
],
[
"# Create the GridSearch estimator along with a parameter object containing the values to adjust\nfrom sklearn.model_selection import GridSearchCV\nparam_grid = {'C': [1, 5, 10],\n 'gamma': [0.0001, 0.0005, 0.001]}",
"_____no_output_____"
],
[
"# Create the GridSearchCV model\ngrid = GridSearchCV(model, param_grid, verbose=3)",
"_____no_output_____"
],
[
"# Train the model with GridSearch\ngrid.fit(X_train_scaled, y_train)",
"Fitting 3 folds for each of 9 candidates, totalling 27 fits\n[CV] C=1, gamma=0.0001 ...............................................\n"
],
[
"print(grid.best_params_)\nprint(grid.best_score_)",
"{'C': 10, 'gamma': 0.0001}\n0.8661071905397673\n"
],
[
"best_model=SVC(kernel='linear', C=10, gamma=0.0001)\nbest_model",
"_____no_output_____"
],
[
"# save your model by updating \"your_name\" with your name\n\n# and \"your_model\" with your model variable\n\n# if joblib fails to import, try running the command to install in terminal/git-bash\nimport joblib\nfilename = 'exoplanet_svm.sav'\njoblib.dump(best_model, filename)",
"_____no_output_____"
],
[
"from sklearn.linear_model import LogisticRegression\nclassifier = LogisticRegression()\nclassifier",
"_____no_output_____"
],
[
"classifier.fit(X_train_scaled, y_train)",
"C:\\Users\\poona\\Anaconda3\\envs\\PythonData\\lib\\site-packages\\sklearn\\linear_model\\logistic.py:432: FutureWarning: Default solver will be changed to 'lbfgs' in 0.22. Specify a solver to silence this warning.\n FutureWarning)\nC:\\Users\\poona\\Anaconda3\\envs\\PythonData\\lib\\site-packages\\sklearn\\linear_model\\logistic.py:469: FutureWarning: Default multi_class will be changed to 'auto' in 0.22. Specify the multi_class option to silence this warning.\n \"this warning.\", FutureWarning)\n"
],
[
"print(f\"Training Data Score: {classifier.score(X_train_scaled, y_train)}\")\nprint(f\"Testing Data Score: {classifier.score(X_test_scaled, y_test)}\")",
"Training Data Score: 0.8411214953271028\nTesting Data Score: 0.8409610983981693\n"
],
[
"# Create the GridSearch estimator along with a parameter object containing the values to adjust\nfrom sklearn.model_selection import GridSearchCV\nparam_grid = {'C': [1, 5, 10],\n 'penalty': [\"l1\", \"l2\"]}",
"_____no_output_____"
],
[
"# Create the GridSearchCV model\ngrid = GridSearchCV(classifier, param_grid, verbose=3)",
"_____no_output_____"
],
[
"# Train the model with GridSearch\ngrid.fit(X_train_scaled, y_train)",
"C:\\Users\\poona\\Anaconda3\\envs\\PythonData\\lib\\site-packages\\sklearn\\model_selection\\_split.py:1978: FutureWarning: The default value of cv will change from 3 to 5 in version 0.22. Specify it explicitly to silence this warning.\n warnings.warn(CV_WARNING, FutureWarning)\n[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e79272696b6072238c19827ead0b574f468d0f21 | 333,309 | ipynb | Jupyter Notebook | notebooks/2017-01-23-R-notebook.ipynb | kellydesent/notebooks_demos | 8e18371f223872731fde437651e21a66f79a78e2 | [
"MIT"
] | 19 | 2016-07-05T13:05:57.000Z | 2021-10-04T09:25:10.000Z | notebooks/2017-01-23-R-notebook.ipynb | kellydesent/notebooks_demos | 8e18371f223872731fde437651e21a66f79a78e2 | [
"MIT"
] | 316 | 2016-05-10T20:47:36.000Z | 2021-11-17T18:53:00.000Z | notebooks/2017-01-23-R-notebook.ipynb | kellydesent/notebooks_demos | 8e18371f223872731fde437651e21a66f79a78e2 | [
"MIT"
] | 24 | 2016-05-24T19:31:36.000Z | 2021-03-30T05:32:40.000Z | 1,078.669903 | 92,308 | 0.944007 | [
[
[
"# Quick demonstration of R-notebooks using the r-oce library\n\nThe IOOS notebook\n[environment](https://github.com/ioos/notebooks_demos/blob/229dabe0e7dd207814b9cfb96e024d3138f19abf/environment.yml#L73-L76)\ninstalls the `R` language and the `Jupyter` kernel needed to run `R` notebooks.\nConda can also install extra `R` packages,\nand those packages that are unavailable in `conda` can be installed directly from CRAN with `install.packages(pkg_name)`.\n\nYou can start `jupyter` from any other environment and change the kernel later using the drop-down menu.\n(Check the `R` logo at the top right to ensure you are in the `R` jupyter kernel.)\n\nIn this simple example we will use two libraries aimed at the oceanography community written in `R`: [`r-gsw`](https://cran.r-project.org/web/packages/gsw/index.html) and [`r-oce`](http://dankelley.github.io/oce/).\n\n(The original post for the examples below can be found author's blog: [http://dankelley.github.io/blog/](http://dankelley.github.io/blog/))",
"_____no_output_____"
]
],
[
[
"library(gsw)\nlibrary(oce)",
"_____no_output_____"
]
],
[
[
"Example 1: calculating the day length.",
"_____no_output_____"
]
],
[
[
"daylength <- function(t, lon=-38.5, lat=-13)\n{\n t <- as.numeric(t)\n alt <- function(t)\n sunAngle(t, longitude=lon, latitude=lat)$altitude\n rise <- uniroot(alt, lower=t-86400/2, upper=t)$root\n set <- uniroot(alt, lower=t, upper=t+86400/2)$root\n set - rise\n}\n\nt0 <- as.POSIXct(\"2017-01-01 12:00:00\", tz=\"UTC\")\nt <- seq.POSIXt(t0, by=\"1 day\", length.out=1*356)\ndayLength <- unlist(lapply(t, daylength))\n\npar(mfrow=c(2,1), mar=c(3, 3, 1, 1), mgp=c(2, 0.7, 0))\n\nplot(t, dayLength/3600, type='o', pch=20,\n xlab=\"\", ylab=\"Day length (hours)\")\ngrid()\nsolstice <- as.POSIXct(\"2013-12-21\", tz=\"UTC\")\n\nplot(t[-1], diff(dayLength), type='o', pch=20,\n xlab=\"Day in 2017\", ylab=\"Seconds gained per day\")\ngrid()",
"_____no_output_____"
]
],
[
[
"Example 2: least-square fit.",
"_____no_output_____"
]
],
[
[
"x <- 1:100\ny <- 1 + x/100 + sin(x/5)\nyn <- y + rnorm(100, sd=0.1)\nL <- 4\ncalc <- runlm(x, y, L=L, deriv=0)\nplot(x, y, type='l', lwd=7, col='gray')\npoints(x, yn, pch=20, col='blue')\nlines(x, calc, lwd=2, col='red')",
"_____no_output_____"
],
[
"data(ctd)\nrho <- swRho(ctd)\nz <- swZ(ctd)\ndrhodz <- runlm(z, rho, deriv = 1)\ng <- 9.81\nrho0 <- mean(rho, na.rm = TRUE)\nN2 <- -g * drhodz/rho0\nplot(ctd, which = \"N2\")\nlines(N2, -z, col = \"blue\")\nlegend(\"bottomright\", lwd = 2, col = c(\"brown\", \"blue\"), legend = c(\"spline\", \n \"runlm\"), bg = \"white\")",
"_____no_output_____"
]
],
[
[
"Example 3: T-S diagram.",
"_____no_output_____"
]
],
[
[
"# Alter next three lines as desired; a and b are watermasses.\nSa <- 30\nTa <- 10\nSb <- 40\n\nlibrary(oce)\n# Should not need to edit below this line\nrho0 <- swRho(Sa, Ta, 0)\nTb <- uniroot(function(T) rho0-swRho(Sb,T,0), lower=0, upper=100)$root\nSc <- (Sa + Sb) /2\nTc <- (Ta + Tb) /2\n## density change, and equiv temp change\ndrho <- swRho(Sc, Tc, 0) - rho0\ndT <- drho / rho0 / swAlpha(Sc, Tc, 0)\n\nplotTS(as.ctd(c(Sa, Sb, Sc), c(Ta, Tb, Tc), 0), pch=20, cex=2)\ndrawIsopycnals(levels=rho0, col=\"red\", cex=0)\nsegments(Sa, Ta, Sb, Tb, col=\"blue\")\ntext(Sb, Tb, \"b\", pos=4)\ntext(Sa, Ta, \"a\", pos=4)\ntext(Sc, Tc, \"c\", pos=4)\nlegend(\"topleft\",\n legend=sprintf(\"Sa=%.1f, Ta=%.1f, Sb=%.1f -> Tb=%.1f, drho=%.2f, dT=%.2f\",\n Sa, Ta, Sb, Tb, drho, dT),\n bg=\"white\")",
"_____no_output_____"
]
],
[
[
"Example 4: find the halocline depth.",
"_____no_output_____"
]
],
[
[
"findHalocline <- function(ctd, deltap=5, plot=TRUE)\n{\n S <- ctd[['salinity']]\n p <- ctd[['pressure']]\n n <- length(p)\n ## trim df to be no larger than n/2 and no smaller than 3.\n N <- deltap / median(diff(p))\n df <- min(n/2, max(3, n / N))\n spline <- smooth.spline(S~p, df=df)\n SS <- predict(spline, p)\n dSSdp <- predict(spline, p, deriv=1)\n H <- p[which.max(dSSdp$y)]\n if (plot) {\n par(mar=c(3, 3, 1, 1), mgp=c(2, 0.7, 0))\n plotProfile(ctd, xtype=\"salinity\")\n lines(SS$y, SS$x, col='red')\n abline(h=H, col='blue')\n mtext(sprintf(\"%.2f m\", H), side=4, at=H, cex=3/4, col='blue')\n mtext(sprintf(\" deltap: %.0f, N: %.0f, df: %.0f\", deltap, N, df),\n side=1, line=-1, adj=0, cex=3/4)\n }\n return(H)\n}\n \n# Plot two panels to see influence of deltap.\npar(mfrow=c(1, 2))\ndata(ctd)\nfindHalocline(ctd)\nfindHalocline(ctd, 1)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7927543fd47c2f2556b390fdba30a5445a504a3 | 165,378 | ipynb | Jupyter Notebook | GenericModelExploration.ipynb | GarciaLab/OnsetTimeTransientInputs | 642c4a4acff8d7f4b2267031c5b6cf1c518db164 | [
"MIT"
] | null | null | null | GenericModelExploration.ipynb | GarciaLab/OnsetTimeTransientInputs | 642c4a4acff8d7f4b2267031c5b6cf1c518db164 | [
"MIT"
] | null | null | null | GenericModelExploration.ipynb | GarciaLab/OnsetTimeTransientInputs | 642c4a4acff8d7f4b2267031c5b6cf1c518db164 | [
"MIT"
] | null | null | null | 160.717201 | 19,344 | 0.865659 | [
[
[
"# Exploring a generic Markov model of chromatin accessibility\n\nLast updated by: Jonathan Liu, 4/23/2021\n\nHere, we will explore a generic Markov chain model of chromatin accessibility, where we model chromatin with a series of states and Markov transitions between them. Of interest is the onset time, the time it takes for the system to reach the final, transcriptionally competent state. We will indicate that the limit of equal, irreversible reactions is the limit of noise performance and that allowing for some reversibility weakens performance. We will then show that with a transient input, the model can achieve much better performance.",
"_____no_output_____"
]
],
[
[
"#Import necessary packages\n%matplotlib inline\nimport numpy as np\nfrom scipy.spatial import ConvexHull\nimport matplotlib.pyplot as plt\nimport scipy.special as sps\nfrom IPython.core.debugger import set_trace\nfrom numba import njit, prange\nimport numba as numba\nfrom datetime import date\nimport time as Time\nimport seaborn as sns\n\n#Set number of threads\nnumba.set_num_threads(4)",
"_____no_output_____"
],
[
"# PBoC plotting style (borrowed from Manuel's github)\ndef set_plotting_style():\n \"\"\"\n Formats plotting enviroment to that used in Physical Biology of the Cell,\n 2nd edition. To format all plots within a script, simply execute\n `mwc_induction_utils.set_plotting_style() in the preamble.\n \"\"\"\n rc = {'lines.linewidth': 1.25,\n 'axes.labelsize': 12,\n 'axes.titlesize': 12,\n 'axes.facecolor': '#E3DCD0',\n 'xtick.labelsize': 12,\n 'ytick.labelsize': 12,\n 'xtick.color': 'white',\n 'xtick.direction': 'in',\n 'xtick.top': True,\n 'xtick.bottom': True,\n 'xtick.labelcolor': 'black',\n 'ytick.color': 'white',\n 'ytick.direction': 'in',\n 'ytick.left': True,\n 'ytick.right': True,\n 'ytick.labelcolor': 'black',\n 'font.family': 'Arial',\n #'grid.linestyle': '-', Don't use a grid\n #'grid.linewidth': 0.5,\n #'grid.color': '#ffffff',\n 'axes.grid': False,\n 'legend.fontsize': 8}\n plt.rc('text.latex', preamble=r'\\usepackage{sfmath}')\n #plt.rc('xtick.major', pad=5)\n #plt.rc('ytick.major', pad=5)\n plt.rc('mathtext', fontset='stixsans', sf='sansserif')\n plt.rc('figure', figsize=[3.5, 2.5])\n plt.rc('svg', fonttype='none')\n plt.rc('legend', title_fontsize='12', frameon=True, \n facecolor='#E3DCD0', framealpha=1)\n sns.set_style('darkgrid', rc=rc)\n sns.set_palette(\"colorblind\", color_codes=True)\n sns.set_context('notebook', rc=rc)\n\n# Some post-modification fixes that I can't seem to set in the rcParams\ndef StandardFigure(ax):\n ax.tick_params(labelcolor='black')\n ax.xaxis.label.set_color('black')\n ax.yaxis.label.set_color('black')\n \nset_plotting_style()",
"_____no_output_____"
],
[
"#Function to generate a random transition matrix for a generic Markov chain with n states, and an irreversible\n#transition into the final state.\n\n#Inputs:\n# n: number of states\n# k_min: minimum transition rate\n# k_max: maximum transition rate\n\n#pseudocode\n#generate 2D matrix based on n\n#loop over each index, if indices are connected by 1 then generate a value (except final state)\n#Calculate diagonal elements from summing columns to zero\n#\n\ndef MakeRandomTransitionMatrix(n, k_min, k_max):\n #Initialize the transition matrix\n Q = np.zeros((n,n))\n \n #Loop through transition indices (note that the final column is all zeros since it's an absorbing state)\n for i in range(n):\n for j in range(n-1):\n #If the indices are exactly one apart (i.e. adjacent states), then make a transition rate\n if np.abs(i-j) == 1:\n Q[i,j] = np.random.uniform(k_min,k_max)\n\n #Calculate the diagonal elements by taking the negative of the sum of the column\n for i in range(n-1):\n Q[i,i] = -np.sum(Q[:,i])\n \n return Q\n",
"_____no_output_____"
],
[
"#Function to generate a transition matrix for equal, irreversible transitions (i.e. Gamma distribution results)\n#We assume the final state is absorbing\n\n#Inputs:\n# n: number of states\n# k: transition rate\n\ndef MakeGammaDistMatrix(n, k):\n #Initialize the transition matrix\n Q = np.zeros((n,n))\n \n #Loop through transition indices (note that the final column is all zeros since it's an absorbing state)\n for i in range(n):\n for j in range(n-1):\n #All forward transitions are equal to k\n if i == j + 1:\n Q[i,j] = k\n\n #Calculate the diagonal elements by taking the negative of the sum of the column\n for i in range(n-1):\n Q[i,i] = -np.sum(Q[:,i])\n \n return Q\n\n#Similar function for making a transition matrix with equal forward transitions of magnitude k and\n#equal backward transitions of magnitude k * f\ndef MakeEqualBidirectionalMatrix(n, k, f):\n #Initialize the transition matrix\n Q = np.zeros((n,n))\n \n #Loop through transition indices (note that the final column is all zeros since it's an absorbing state)\n for i in range(n):\n for j in range(n-1):\n #All forward transitions are equal to k\n if i == j + 1:\n Q[i,j] = k\n elif i == j - 1:\n Q[i,j] = k * f\n\n #Calculate the diagonal elements by taking the negative of the sum of the column\n for i in range(n-1):\n Q[i,i] = -np.sum(Q[:,i])\n \n return Q\n",
"_____no_output_____"
],
[
"#Simulation for calculating onset times for a generic Markov chain using Gillespie algorithm\n#Using vectorized formulation for faster speed\n\ndef CalculatetOn_GenericMarkovChainGillespie(Q,n,N_cells):\n#Calculates the onset time for a linear Markov chain with forward and backward rates.\n#The transition rate can be time-varying, but is the same\n#global rate for each transition. The model assumes n states, beginning\n#in the 1st state. Using the Gillespie algorithm and a Markov chain formalism, it\n#simulates N_cells realizations of the overall time it takes to reach the\n#nth state.\n\n#For now, this only works with steady transition rates. Later we will modify this to account \n#for time-varying rates.\n\n# Inputs:\n# Q: transition rate matrix, where q_ji is the transition rate from state i to j for i =/= j and \n# q_ii is the sum of transition rates out of state i\n# n: number of states\n# N_cells: number of cells to simulate\n\n# Outputs:\n# t_on: time to reach the final state for each cell (length = N_cells)\n\n## Setup variables\n t_on = np.zeros(N_cells) #Time to transition to final ON state for each cell\n state = np.zeros(N_cells, dtype=int) #State vector describing current state of each cell\n\n ## Run simulation\n # We will simulate waiting times for each transition for each cell and stop once each cell has\n # reached the final state\n \n #Set diagonal entries in transition matrix to nan since self transitions don't count\n for i in range(n):\n Q[i,i] = 0\n \n #Construct the transition vector out of each cell's current state\n Q_states = np.zeros((N_cells,n))\n while np.sum(state) < (n-1)*N_cells:\n Q_states = np.transpose(Q[:,state])\n \n #Generate random numbers in [0,1] for each cell\n randNums = np.random.random(Q_states.shape)\n\n #Calculate waiting times for each entry in the transition matrix\n #Make sure to suppress divide by zero warning\n with np.errstate(divide='ignore'):\n tau = (1/Q_states) * np.log(1/randNums)\n\n #Find the shortest waiting time to figure out which state we transitioned to for each cell\n tau_min = np.amin(tau, axis=1)\n newState = np.argmin(tau, axis=1)\n \n #Replace infinities with zero, corresponding to having reached the final state\n newState[tau_min==np.inf] = n-1\n tau_min[tau_min==np.inf] = 0\n \n #Update the state and add the waiting time to the overall waiting time\n state = newState\n t_on += tau_min\n return t_on",
"_____no_output_____"
],
[
"#Simulation for calculating onset times for a generic Markov chain using Gillespie algorithm\n#Using vectorized formulation for faster speed\n\ndef CalculatetOn_GenericMarkovChainGillespieTime(Q,n,t_d,N_cells):\n#Calculates the onset time for a linear Markov chain with forward and backward rates.\n#The transition rate can be time-varying, but is the same\n#global rate for each transition. The model assumes n states, beginning\n#in the 1st state. Using the Gillespie algorithm and a Markov chain formalism, it\n#simulates N_cells realizations of the overall time it takes to reach the\n#nth state.\n\n#This considers time-dependent transition rates parameterized by a diffusion timescale t_d.\n#The time-dependent rate has the form r ~ (1 - exp(-t/t_d)). For now, we assume only the forwards\n#rates have the time-dependent profile, and that backwards rates are time-independent.\n\n# Inputs:\n# Q: 3D transition rate matrix, where q_kji is the transition rate at time k from state i to j for i =/= j and \n# q_kii is the sum of transition rates out of state i\n# n: number of states\n# t_d: diffusion timescale of time-dependent transition rate\n# N_cells: number of cells to simulate\n\n# Outputs:\n# t_on: time to reach the final state for each cell (length = N_cells)\n\n## Setup variables\n t_on = np.zeros(N_cells) #Time to transition to final ON state for each cell\n time = np.zeros(N_cells) #Vector of current time for each cell\n state = np.zeros(N_cells, dtype=int) #State vector describing current state of each cell\n\n ## Run simulation\n # We will simulate waiting times for each transition for each cell and stop once each cell has\n # reached the final state\n \n #Set diagonal entries in transition matrix to nan since self transitions don't count\n for i in range(n):\n Q[i,i] = 0\n \n #Define the diffusion timescale matrix t_d (finite for forwards rates, effectively 0 for backwards rates)\n t_d_mat = np.zeros((n,n))\n t_d_mat[:,:] = 0.00000001 #Non forwards transitions are essentially 0 diffusive timescale\n for i in range(n):\n for j in range(n-1):\n #Forwards rates\n if i == j + 1:\n t_d_mat[i,j] = t_d\n \n #Construct the transition vector out of each cell's current state\n Q_states = np.zeros((N_cells,n))\n #Construct the diffusion timescale vector for each cell\n t_d_states = np.zeros((N_cells,n))\n while np.sum(state) < (n-1)*N_cells:\n Q_states = np.transpose(Q[:,state])\n t_d_states = np.transpose(t_d_mat[:,state])\n \n #Construct the current time vector for each cell\n time_states = np.transpose(np.tile(time,(n,1)))\n \n \n #Generate random numbers in [0,1] for each cell\n randNums = np.random.random(Q_states.shape)\n\n #Calculate waiting times for each entry in the transition matrix\n #Make sure to suppress divide by zero warning\n \n #For the exponential profile, this uses the lambertw/productlog function. The steady-state\n #case corresponds to t_d -> 0.\n with np.errstate(divide='ignore', invalid='ignore'):\n #Temp variables for readability\n a = 1/Q_states * np.log(1/randNums)\n b = -np.exp(-(a + t_d_states * np.exp(-time_states/t_d_states) + time_states)/t_d_states)\n tau = np.real(t_d_states * sps.lambertw(b) + a + t_d_states *\\\n np.exp(-time_states / t_d_states))\n #Find the shortest waiting time to figure out which state we transitioned to for each cell\n tau_min = np.amin(tau, axis=1)\n newState = np.argmin(tau, axis=1)\n \n #Replace infinities with zero, corresponding to having reached the final state\n newState[tau_min==np.inf] = n-1\n tau_min[tau_min==np.inf] = 0\n \n #Update the state and add the waiting time to the overall waiting time\n state = newState\n t_on += tau_min\n time += tau_min\n return t_on",
"_____no_output_____"
]
],
[
[
"# The steady-state regime\nFirst, let's get a feel for the model in the steady-state case. \n\nWe consider a Markov chain with $k+1$ states labeled with indices $i$, with the first state labeled with index $0$. The system will begin in state $0$ at time $t=0$ and we will assume the final state $k$ is absorbing. For example, this could correspond to the transcriptionally competent state. We will allow for forwards and backwards transition rates between all states, except for the final absorbing state, which will have no backwards transition out of it. Denote the transition from state $i$ to state $j$ with the transition rate $\\beta_{i,j}$. So,, we have the reaction network:\n\n\\begin{equation}\n0 \\underset{\\beta_{1,0}}{\\overset{\\beta_{0,1}}{\\rightleftharpoons}} 1 \\underset{\\beta_{2,1}}{\\overset{\\beta_{1,2}}{\\rightleftharpoons}} ... \\overset{\\beta_{k-1,k}}{\\rightarrow} k\n\\end{equation}\n\nWe will be interested in the mean and variance of the distribution of times $P_k(t)$ to start at state $0$ and reach the final state $k$.\n\nWe will first consider the simple case where the transition rates $\\beta$ are constant in time, and that we have only forward transitions that are all equal in magnitude. In this case, the distribution $P_k(t)$ is simply given by a Gamma distribution with shape parameter $k$ and rate parameter $\\beta$. $P_k(t)$ then has the form\n\n\\begin{equation}\nP_k(t) = \\frac{\\beta^k}{\\Gamma(k)}t^{k-1}e^{-\\beta t}\n\\end{equation}\n\nwhere $\\Gamma$ is the Gamma function. Below we show analytical and simulated results for the distribution of onset times.\n\n",
"_____no_output_____"
]
],
[
[
"#Let's visualize the distribution of onset times for the Gamma distribution case\n\n#Function for analytical Gamma distribution\ndef GamPDF(x,shape,rate):\n return x**(shape-1)*(np.exp(-x*rate) / sps.gamma(shape)*(1/rate)**shape)\n\n#Pick some parameters\nbeta = 1 #transition rate\nn = np.array([2,3,4]) #number of states\nk = n-1 #Number of steps\n\n#Simulate the distributions\nN_cells = 10000\n\nt_on = np.zeros((len(n),N_cells))\n\nfor i in range(len(n)):\n Q = MakeGammaDistMatrix(n[i], beta) #Transition matrix\n t_on[i,:] = CalculatetOn_GenericMarkovChainGillespie(Q,n[i],N_cells)\n \n#Plot results\ncolors = ['tab:blue','tab:red','tab:green']\nbins = np.arange(0,10,0.5)\nt = np.arange(0,10,0.1)\nToyModelDist = plt.figure()\n#plt.title('Onset distributions for equal, irreversible transitions')\n\nfor i in range(len(k)):\n plt.hist(t_on[i,:],bins=bins,density=True,alpha=0.5,label='simulation k=' + str(k[i]),\n color=colors[i],linewidth=0)\n plt.plot(t,GamPDF(t,n[i]-1,beta),'--',label='theory k=' + str(k[i]),color=colors[i])\n\nplt.xlabel('onset time')\nplt.ylabel('frequency')\nplt.legend()\nStandardFigure(plt.gca())\nplt.show()",
"_____no_output_____"
]
],
[
[
"The mean $\\mu_k$ and variance $\\sigma^2_k$ have simple analytical expressions and are given by\n\n\\begin{equation}\n\\mu_k = \\frac{k}{\\beta} \\\\\n\\sigma^2_k = \\frac{k}{\\beta^2}\n\\end{equation}\n\nFor this analysis, we will consider a two-dimensional feature space consisting of the mean onset time on the x-axis and the squared CV (variance divided by square mean) in the onset time on the y-axis. The squared CV is a measure of the \"noise\" of the system at a given mean. For this simple example then:\n\n\\begin{equation}\n\\mu_k = \\frac{k}{\\beta} \\\\\nCV^2_k = \\frac{1}{k}\n\\end{equation}\n\nThus, for this scenario with equal, irreversible reactions, the squared CV is independent of the transition rates $\\beta$ and depends only on the number of steps $k$. Plotting in our feature space results in a series of horizontal lines, with each line corresponding to the particular number of steps in the model.",
"_____no_output_____"
]
],
[
[
"#Setting up our feature space\nbeta_min = 0.5 #Minimum transition rate\nbeta_max = 5 #Maximum transition rate\nbeta_step = 0.1 #Resolution in transition rates\nbeta_range = np.arange(beta_min,beta_max,beta_step)\nn = np.array([2,3,4,5]) #Number of states\n\nmeans = np.zeros((len(n),len(beta_range)))\nCV2s = np.zeros((len(n),len(beta_range)))\n\n#Simulate results\nfor i in range(len(n)):\n for j in range(len(beta_range)):\n Q = MakeGammaDistMatrix(n[i], beta_range[j])\n t_on = CalculatetOn_GenericMarkovChainGillespie(Q,n[i],N_cells)\n means[i,j] = np.mean(t_on)\n CV2s[i,j] = np.var(t_on)/np.mean(t_on)**2\n\n#Plot results\nmeanVals = np.arange(0,10,0.1)\nCV2Pred = np.zeros((len(n),len(meanVals)))\ncolors = ['tab:blue','tab:red','tab:green','tab:purple']\nfor i in range(len(n)):\n CV2Pred[i,:] = (1/(n[i]-1)) * np.ones(len(meanVals))\nToyModelFeatureSpace = plt.figure()\n#plt.title('Feature space for equal, irreversible reactions')\n\nfor i in range(len(n)):\n plt.plot(means[i,:],CV2s[i,:],'.',label='simulation k=' + str(n[i]-1),color=colors[i])\n plt.plot(meanVals,CV2Pred[i,:],'--',label='theory k=' + str(n[i]-1),color=colors[i])\n \nplt.xlabel('mean')\nplt.ylabel('CV^2')\nplt.legend()\nStandardFigure(plt.gca())\nplt.show()",
"_____no_output_____"
]
],
[
[
"What happens if we now allow for backwards transitions as an extension to this ideal case? We'll retain the idea of equal forward transition rates $\\beta$, but now allow for equal backwards transitions of magnitude $\\beta f$ (except from the final absorbing state $k$). \n\n\\begin{equation}\n0 \\underset{\\beta f}{\\overset{\\beta}{\\rightleftharpoons}} 1 \\underset{\\beta f}{\\overset{\\beta}{\\rightleftharpoons}} ... \\overset{\\beta}{\\rightarrow} k\n\\end{equation}\n\nWe will investigate what happens when we vary $f$. Let's see what happens for $k=3$ steps.",
"_____no_output_____"
]
],
[
[
"#Setting up parameters\nn = 3\nbeta_min = 0.1\nbeta_max = 5.1\nbeta_step = 0.1\nbeta_range = np.arange(beta_min,beta_max,beta_step)\nN_cells = 10000\n\n#Backwards transitions\nf = np.arange(0,4,1) #fractional magnitude of backwards transition relative to forwards\nmeans = np.zeros((len(beta_range),len(f)))\nCV2s = np.zeros((len(beta_range),len(f)))\n\nfor i in range(len(beta_range)):\n for j in range(len(f)):\n Q = MakeEqualBidirectionalMatrix(n,beta_range[i],f[j])\n t_on = CalculatetOn_GenericMarkovChainGillespie(Q,n,N_cells)\n means[i,j] = np.mean(t_on)\n CV2s[i,j] = np.var(t_on) / np.mean(t_on)**2\n \n#Plot results\n#Distribution for fixed beta and varying f\nbeta = 1\nbins = np.arange(0,20,0.5)\nt = np.arange(0,10,0.1)\ncolors = ['tab:blue','tab:red','tab:green','tab:purple']\n\nBackwardsDist = plt.figure()\n\nfor i in range(len(f)):\n Q = MakeEqualBidirectionalMatrix(n,beta,f[i])\n t_on = CalculatetOn_GenericMarkovChainGillespie(Q,n,N_cells)\n plt.hist(t_on,bins=bins,density=True,alpha=0.3,label='f = ' + str(f[i]),\n linewidth=0,color=colors[i])\n\nplt.xlabel('onset time')\nplt.ylabel('frequency')\nplt.legend()\nStandardFigure(plt.gca())\nplt.show()\n\n\nBackwardsFeatureSpace = plt.figure()\n#plt.title('Investigation of impact of backwards rates on feature space (k=2)')\nplt.plot((n-1)/beta_range,(1/(n-1))*np.ones(beta_range.shape),'k--',label='Gamma dist. limit')\n\nfor i in range(len(f)):\n plt.plot(means[:,i],CV2s[:,i],'.',label='f = ' + str(f[i]),color=colors[i])\n \nplt.xlabel('mean')\nplt.ylabel('CV^2')\nplt.legend()\nStandardFigure(plt.gca())\nplt.show()",
"_____no_output_____"
]
],
[
[
"We see that as the backwards transition rate increases, the overall noise increases! This makes intuitive sense, since with a backwards transition rate, the system is more likely to spend extra time hopping between states before reaching the final absorbing state, increasing the overall time to finish as well as the variability in finishing times.\n\nBecause actual irreversible reactions are effectively impossible to achieve in reality, the performance of the Gamma distribution model (i.e. equal, irreversible forward transitions) represents a bound to the noise performance of a real system. With this more realistic scenario of backwards transitions, the overall noise is higher.",
"_____no_output_____"
],
[
"# Transients help improve noise performance\n\nIn the steady-state regime, the only way to decrease the noise (i.e. squared CV) in onset times was to increase the number of steps. What about in the transient regime?\n\nHere, we will investigate the changes to this parameter space by using a transient rate $\\beta(t)$. This is of biological interest because many developmental processes occur out of steady state. For example, several models of chromatin accessibility hypothesize that the rate of chromatin state transitioning is coupled to the activity of pioneer factors like Zelda. During each rapid cell cycle division event in the early fly embryo, the nuclear membrane breaks down and reforms again, and transcription factors are expelled and re-introduced back into the nucleus. Thus, after each division event, there is a transient period during which the concentration of pioneer factors at a given gene locus is out of steady state.\n\nFor now, we will assume a reasonable form for the transition rate. Let's assume that forward transition rates are mediated by the concentration of a pioneer factor like Zelda, e.g. in some on-rate fashion. Considering $\\beta$ to be a proxy for Zelda concentration, for example, we will write down this transient $\\beta(t)$ as the result of a simple diffusive process with form\n\n\\begin{equation}\n\\beta(t) = \\beta (1 - e^{-t / \\tau} )\n\\end{equation}\n\nHere, $\\beta$ is the asymptotic, saturating value of $\\beta(t)$, and $\\tau$ is the time constant governing the time-varying nature of the transition rate. For a diffusive process, $\\tau$ would be highly dependent on the diffusion constant, for example.\n\nFor comparison, the time plots of the constant and transient input are shown below, for $\\tau = 3$ and $\\beta = 1$.",
"_____no_output_____"
]
],
[
[
"#Looking at steady-state vs input transient profiles\ntime = np.arange(0,10,0.1)\ndt = 0.1\nw_base = 1\nw_const = w_base * np.ones(time.shape)\nN_trans = 2\nN_cells = 1000\n\n#Now with transient exponential rate\ntau = 3\nw_trans = w_base * (1 - np.exp(-time / tau))\n\n#Plot the inputs\nTransientInputs = plt.figure()\n#plt.title('Input transition rate profiles')\nplt.plot(time,w_const,label='constant',color='tab:blue')\nplt.plot(time,w_trans,label='transient',color='tab:red')\nplt.xlabel('time')\nplt.ylabel('rate')\nplt.legend()\nStandardFigure(plt.gca())\nTransientInputs.set_figheight(1) #Make this figure short for formatting purposes\nplt.show()",
"_____no_output_____"
]
],
[
[
"Because of the time-varying nature of $\\beta(t)$, the resulting distribution $P_k(t)$ for the case of equal, irreversible forward transition rates no longer obeys a simple Gamma distribution, and an analytical solution is difficult (or even impossible). Nevertheless, we can easily simulate the distributions numerically, shown below.",
"_____no_output_____"
]
],
[
[
"#Let's visualize the distribution of onset times for the case of equal, irreversible forward transition rates,\n#comparing steady-state and transient input profiles, varying the \"diffusion\" constant tau\n\n#Pick some parameters\nbeta = 1 #transition rate\nn = 3 #Number of states\ntau = np.array([1,3])\n\n#Simulate the distributions\nN_cells = 10000\n\n#Steady state\nQ_steady = MakeGammaDistMatrix(n, beta)\nt_on_steady = CalculatetOn_GenericMarkovChainGillespie(Q_steady,n,N_cells)\n\n#Transient\nt_on_trans = np.zeros((len(tau),N_cells))\n\nfor i in range(len(tau)):\n Q = MakeGammaDistMatrix(n, beta) #Transition matrix\n t_on_trans[i,:] = CalculatetOn_GenericMarkovChainGillespieTime(Q,n,tau[i],N_cells)\n \n#Plot results\nbins = np.arange(0,10,0.25)\ncolors = ['tab:red','tab:green']\nTransientDist = plt.figure()\n#plt.title('Onset distributions for k=2 equal, irreversible transitions, steady-state vs. transient input')\n\nplt.hist(t_on_steady,bins=bins,density=True,alpha=0.5,label='steady state',\n linewidth=0,color='tab:blue')\nfor i in range(len(tau)):\n plt.hist(t_on_trans[i,:],bins=bins,density=True,alpha=0.5,linewidth=0,\n label='transient tau=' + str(tau[i]),color=colors[i])\n\nplt.xlabel('onset time')\nplt.ylabel('frequency')\nplt.legend()\nStandardFigure(plt.gca())\nplt.show()",
"_____no_output_____"
]
],
[
[
"We see that increasing the time constant $\\tau$ results in a rightward shift of the onset time distribution, as expected since the time-varying transition rate profile will results in slower initial transition rates. What impact does this have on the noise? Below we show the feature space holding $k=2$ fixed while varying $\\tau$, and then holding $\\tau=3$ fixed and varying $k$.",
"_____no_output_____"
]
],
[
[
"#Exploring the impact of transient inputs\n#First, fix k and vary tau\nn = 3 #number of states\nbeta_min = 0.1\nbeta_max = 5.1\nbeta_step = 0.1\nbeta_range = np.arange(beta_min,beta_max,beta_step)\ntau = np.arange(1,10,3)\n\n#Simulate the distributions\nN_cells = 5000\n\n#Steady state\nmeans_steady = np.zeros(len(beta_range))\nCV2s_steady = np.zeros(len(beta_range))\nfor i in range(len(beta_range)):\n Q = MakeGammaDistMatrix(n, beta_range[i])\n t_on = CalculatetOn_GenericMarkovChainGillespie(Q,n,N_cells)\n means_steady[i] = np.mean(t_on)\n CV2s_steady[i] = np.var(t_on) / np.mean(t_on)**2\n\n#Transient\nmeans_trans = np.zeros((len(tau),len(beta_range)))\nCV2s_trans = np.zeros((len(tau),len(beta_range)))\n\nfor i in range(len(tau)):\n for j in range(len(beta_range)):\n Q = MakeGammaDistMatrix(n, beta_range[j]) #Transition matrix\n t_on = CalculatetOn_GenericMarkovChainGillespieTime(Q,n,tau[i],N_cells)\n means_trans[i,j] = np.mean(t_on)\n CV2s_trans[i,j] = np.var(t_on) / np.mean(t_on)**2\n \n#Plot results\ncolors = ['tab:blue','tab:red','tab:green']\nTransientFeatureSpaceFixedK = plt.figure()\n#plt.title('Investigation of transient inputs on feature space (k=2, tau varying)')\nplt.plot((n-1)/beta_range,(1/(n-1))*np.ones(beta_range.shape),\n 'k--',label='Gamma dist. limit in steady-state',color='black')\nplt.plot(means_steady,CV2s_steady,'k.',label='steady state simulation',color='black')\n \nfor i in range(len(tau)):\n plt.plot(means_trans[i,:],CV2s_trans[i,:],'.',\n label='transient simulation tau=' + str(tau[i]),color=colors[i])\n \nplt.xlabel('mean')\nplt.ylabel('CV^2')\nplt.legend()\nStandardFigure(plt.gca())\nplt.show()\n\n#Now fix tau and vary k\nn = np.array([2,3,4,5])\ntau = 3\n\n#Simulate the distributions\nN_cells = 5000\n\n#Transient\nmeans_trans = np.zeros((len(n),len(beta_range)))\nCV2s_trans = np.zeros((len(n),len(beta_range)))\n\nfor i in range(len(n)):\n for j in range(len(beta_range)):\n Q = MakeGammaDistMatrix(n[i], beta_range[j]) #Transition matrix\n t_on = CalculatetOn_GenericMarkovChainGillespieTime(Q,n[i],tau,N_cells)\n means_trans[i,j] = np.mean(t_on)\n CV2s_trans[i,j] = np.var(t_on) / np.mean(t_on)**2\n \n#Plot results\ncolors = ['black','tab:red','tab:green','tab:blue']\nTransientFeatureSpaceFixedTau = plt.figure()\n#plt.title('Investigation of transient inputs on feature space (k varying, tau=3)')\n\nfor i in range(len(n)):\n plt.plot((n[i]-1)/beta_range,(1/(n[i]-1))*np.ones(beta_range.shape),'--',\\\n color=colors[i],label='steady state k=' + str(n[i]-1))\n plt.plot(means_trans[i,:],CV2s_trans[i,:],'.',color=colors[i],\\\n label='transient simulation k=' + str(n[i]-1))\n \nplt.xlabel('mean')\nplt.ylabel('CV^2')\nplt.legend()\nStandardFigure(plt.gca())\nplt.show()",
"_____no_output_____"
]
],
[
[
"In each case, the transient input reduces noise! It seems like for increasing $\\tau$, the performance improves. This makes intuitive sense because having a time-dependent input profile will make earlier transitions \"weaker,\" so transitions that happen before the expected time are less likely, tightening the overall distribution of onset times. The relevant timescale is the dimensionless ratio $\\frac{\\beta}{\\tau}$ - the faster the intrinsic transition rate $\\beta$ is to the transient input timescale $\\tau$, the larger the effects of the transient input. This manifests in the feature space for low values of the mean onset time, where the discrepancy between steady-state and transient is more apparent.",
"_____no_output_____"
],
[
"# Transient inputs can improve performance in non-ideal models\nEarlier, we saw that in the steady-state case, the presence of finite backwards transition rates decreased the overall performance of the model in terms of noise. The greater the backwards transition rates, the worse the performance got. Here, we'll show that having transient inputs can counteract this performance loss.\n\nAs before, we'll assume a model with equal forward transition rates $\\beta$ and equal backward transition rates $\\beta f$. We'll compare the steady state case with the transient input case, parameterized with timescale $\\tau$. Note that we'll only consider the forward transition rates to be transient, and assume the backward transition rates are still time-independent. Biologically, this would correspond to the forward transitions corresponding to on-rates of some pioneer factor like Zelda that is coupled to a time-dependent concentration profile, while backward transitions are time-independent off-rates.\n\nBelow, we explore the feature space in the steady-state vs. transient cases, with the steady-state ideal case of equal, irreversible transitions as a reference. We'll first consider the case fixing $k=2$ and $f=0.2$ and varying $\\tau$.",
"_____no_output_____"
]
],
[
[
"#Setting up parameters\nn = 3\nbeta_min = 0.1\nbeta_max = 5.1\nbeta_step = 0.1\nbeta_range = np.arange(beta_min,beta_max,beta_step)\nf = 0.2\ntau = np.array([0.25,0.5,1,3])\n\n#Simulate results\nN_cells = 10000\n\n#Steady state\nmeans_steady = np.zeros(len(beta_range))\nCV2s_steady = np.zeros(len(beta_range))\n\nfor i in range(len(beta_range)):\n Q = MakeEqualBidirectionalMatrix(n,beta_range[i],f)\n t_on = CalculatetOn_GenericMarkovChainGillespie(Q,n,N_cells)\n means_steady[i] = np.mean(t_on)\n CV2s_steady[i] = np.var(t_on) / np.mean(t_on)**2\n \n#Transient\nmeans_trans = np.zeros((len(beta_range),len(tau)))\nCV2s_trans = np.zeros((len(beta_range),len(tau)))\n\nfor i in range(len(beta_range)):\n for j in range(len(tau)):\n Q = MakeEqualBidirectionalMatrix(n,beta_range[i],f)\n t_on = CalculatetOn_GenericMarkovChainGillespieTime(Q,n,tau[j],N_cells)\n means_trans[i,j] = np.mean(t_on)\n CV2s_trans[i,j] = np.var(t_on) / np.mean(t_on)**2\n \n#Plot results\ncolors = ['tab:blue','tab:red','tab:green','tab:purple']\nTransientFeatureSpaceBackwards = plt.figure()\n#plt.title('Impact of transient inputs on feature space with backward rates (k=2, f=' + str(f) + ')')\nplt.plot((n-1)/beta_range,(1/(n-1))*np.ones(beta_range.shape),'k--',\n label='Steady-state ideal limit')\nplt.plot(means_steady,CV2s_steady,'k.',label='Steady-state, f=' + str(f))\n\nfor i in range(len(tau)):\n plt.plot(means_trans[:,i],CV2s_trans[:,i],'.',\n label='Transient, f=' + str(f) + ', tau=' + str(tau[i]),color=colors[i])\n \nplt.xlabel('mean')\nplt.ylabel('CV^2')\nplt.legend()\nStandardFigure(plt.gca())\nplt.show()",
"_____no_output_____"
]
],
[
[
"Interesting! As shown earlier, the steady-state case with a backwards transition rate is worse than the ideal limit with equal, irreversible forward rates. However, using a transient rate can counterbalance this and still achieve performance better than the ideal limit in the steady-state case.\n\nThis suggests that given a backwards transition rate that is some fraction $f$ in magnitude of the forwards transition rate, there exists some \"diffusion\" timescale $\\tau$ of the input transition rate that can bring the squared CV back to the ideal steady-state limit with no backwards rates.",
"_____no_output_____"
]
],
[
[
"# Export figures\n\nToyModelDist.savefig('figures/ToyModelDist.pdf')\nToyModelFeatureSpace.savefig('figures/ToyModelFeatureSpace.pdf')\nBackwardsDist.savefig('figures/BackwardsDist.pdf')\nBackwardsFeatureSpace.savefig('figures/BackwardsFigureSpace.pdf')\nTransientInputs.savefig('figures/TransientInputs.pdf')\nTransientDist.savefig('figures/TransientDist.pdf')\nTransientFeatureSpaceFixedK.savefig('figures/TransientFeatureSpaceFixedK.pdf')\nTransientFeatureSpaceFixedTau.savefig('figures/TransientFeatureSpaceFixedTau.pdf')\nTransientFeatureSpaceBackwards.savefig('figures/TransientFeatureSpaceBackwards.pdf')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e792789950a08ecae5bd7f3bf2e94d58a6a5cc1e | 5,304 | ipynb | Jupyter Notebook | notebooks/Chapter 0 - Preface.ipynb | ohshane71/Mining-the-Social-Web-3rd-Edition | ba2c56ecd28e098f097035e11f417534844d71e1 | [
"BSD-2-Clause"
] | null | null | null | notebooks/Chapter 0 - Preface.ipynb | ohshane71/Mining-the-Social-Web-3rd-Edition | ba2c56ecd28e098f097035e11f417534844d71e1 | [
"BSD-2-Clause"
] | null | null | null | notebooks/Chapter 0 - Preface.ipynb | ohshane71/Mining-the-Social-Web-3rd-Edition | ba2c56ecd28e098f097035e11f417534844d71e1 | [
"BSD-2-Clause"
] | null | null | null | 62.4 | 969 | 0.71135 | [
[
[
"# Mining the Social Web (3rd Edition)\n\n## Preface\n\nWelcome! Allow me to be the first to offer my congratulations on your decision to take an interest in [_Mining the Social Web (3rd Edition)_](http://bit.ly/135dHfs)! This collection of [Jupyter Notebooks](http://ipython.org/notebook.html) provides an interactive way to follow along with and explore the numbered examples from the book. Whereas many technical books require you type in the code examples one character at a time or download a source code archive (that may or may not be maintained by the author), this book reinforces the concepts from the sample code in a fun, convenient, and interactive way that really does make the learning experience superior to what you may have previously experienced, so even if you are skeptical, please give it try. I think you'll be pleasantly surprised at the amazing user experience that the Jupyter Notebook affords and just how much easier it is to follow along and adapt the code to your own particular needs. \n\nIn the somewhat unlikely event that you've somehow stumbled across this notebook outside of its context on GitHub, [you can find the full source code repository here](https://github.com/mikhailklassen/Mining-the-Social-Web-3rd-Edition).\n\nIf you haven't previously encountered the Jupyter Notebook, you really should take a moment to learn more about it at https://jupyter.org. It's essentially a platform that allows you to author and run Python source code in the web browser and lends itself very well to data science experiments in which you're taking notes and learning along the way. Personally, I like to think of it as a special purpose notepad that allows me to embed and run arbitrary Python code, and I find myself increasingly using it as my default development environment for many of my Python-based projects. \n\nThe source code for _Mining the Social Web_ book employs the Jupyter Notebook rather exclusively to present the source code as a means of streamlining and enhancing the learning experience, so it is highly recommended that you take a few minutes to learn more about how it works and why it's such an excellent learning (and development) platform. The [same GitHub source code repository](https://github.com/mikhailklassen/Mining-the-Social-Web-3rd-Edition) that contains this file also contains all of the Jupyter Notebooks for _Mining the Social Web_, so once you've followed along with the instructions in Appendix A and gotten your virtual machine environment installed, just open the corresponding notebook from [http://localhost:8888](http://localhost:8888). From that point, following along with the code is literally as easy to pressing Shift-Enter in Jupyter Notebook cells.\n\nIf you experience any problems along the way or have any feedback about this book, its software, or anything else at all, please reach out on Twitter, Facebook, or GitHub for help.\n\n* Twitter: [http://twitter.com/socialwebmining](http://twitter.com/socialwebmining) (@SocialWebMining)\n* Facebook: [http://facebook.com/MiningTheSocialWeb](http://twitter.com/socialwebmining)\n* GitHub: [https://github.com/mikhailklassen/Mining-the-Social-Web-3rd-Edition](https://github.com/mikhailklassen/Mining-the-Social-Web-3rd-Edition)\n\nThanks once again for your interest in _Mining the Social Web_. I truly hope that you learn a lot of new things (and have more fun than you ever expected) from this book.\n\nBest Regards,<br />\nMatthew A. Russell<br />\nTwitter: @ptwobrussell\n\nMikhail Klassen<br />\nTwitter: @MikhailKlassen\n\nP.S. Even if you are a savvy and accomplished developer, you will still find it worthwhile to use the turn-key Docker support that's been provided since it is tested and comes pre-loaded with all of the correct dependencies for following along with the examples.",
"_____no_output_____"
]
],
[
[
"# This is a Python source code comment in a Jupyter Notebook cell. \n# Try executing this cell by placing your cursor in it and typing Shift-Enter\n\nprint(\"Hello, Social Web!\")\n\n# See Appendix A to get your virtual machine installed\n\n# See Appendix C for a brief overview of some Python idioms and IPython Notebook tips",
"Hello, Social Web!\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
e7928355eb22145b4db3bd29ace78f132c75dd4d | 9,761 | ipynb | Jupyter Notebook | LIMMBO mapping.ipynb | diogro/EL-snp_selection | 9db2d348d0a6eeb85eaf787bb521400a5ee1d69b | [
"MIT"
] | null | null | null | LIMMBO mapping.ipynb | diogro/EL-snp_selection | 9db2d348d0a6eeb85eaf787bb521400a5ee1d69b | [
"MIT"
] | null | null | null | LIMMBO mapping.ipynb | diogro/EL-snp_selection | 9db2d348d0a6eeb85eaf787bb521400a5ee1d69b | [
"MIT"
] | null | null | null | 48.805 | 1,435 | 0.626575 | [
[
[
"from pkg_resources import resource_filename\nfrom limmbo.io.reader import ReadData\nfrom limmbo.io.utils import file_type\n\nimport numpy as np\n\n# data container\ndata = ReadData(verbose=False)\n\n# Read covariates\nfile_covs = \"./data/limmbo/limmbo_covariates.csv\"\ndata.getCovariates(file_covariates=file_covs)\n\n# Read genotypes in delim-format\nfile_geno = './data/plink_files/atchely_imputed'\ndata.getGenotypes(file_genotypes=file_geno)\ndata.genotypes\n\n# Read phenotypes\nfile_pheno = './data/limmbo/limmbo_phenotypes.csv'\ndata.getPhenotypes(file_pheno=file_pheno)\ndata.phenotypes\n\n# Read relatedness\nfile_relatedness = './data/limmbo/limmbo_relatedness.csv'\ndata.getRelatedness(file_relatedness=file_relatedness)",
"_____no_output_____"
],
[
"from limmbo.io import input\n\nindata = input.InputData(verbose=False)\nindata.addGenotypes(genotypes=data.genotypes,\n genotypes_info=data.genotypes_info)\n\nindata = input.InputData(verbose=False)\nindata.addGenotypes(genotypes=data.genotypes,\n genotypes_info=data.genotypes_info,\n geno_samples=data.geno_samples)\nindata.addPhenotypes(phenotypes = data.phenotypes, \n pheno_samples = data.pheno_samples,\n phenotype_ID = data.phenotype_ID)\nindata.addRelatedness(relatedness = data.relatedness)\nindata.addCovariates(covariates = data.covariates,\n covs_samples = data.covs_samples)",
"_____no_output_____"
],
[
"from limmbo.core.vdsimple import vd_reml\n\nCg, Cn, ptime = vd_reml(indata, verbose=False)\nindata.addVarianceComponents(Cg = Cg, Cn = Cn)\nCg\n",
"_____no_output_____"
],
[
"np.var(data.phenotypes['Final_weight'])",
"_____no_output_____"
],
[
"indata.regress()\nindata.transform(transform=\"scale\")",
"_____no_output_____"
],
[
"from limmbo.core.gwas import GWAS\n\ngwas = GWAS(datainput=indata, verbose=False)\ngwas.name = \"test\"",
"_____no_output_____"
],
[
"resultsAssociation = gwas.runAssociationAnalysis(setup=\"lmm\", mode=\"multitrait\")",
"_____no_output_____"
],
[
"gwas.computeFDR(0.05)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e792958441443da88b823c12cfb55bbb74c2e558 | 24,198 | ipynb | Jupyter Notebook | g3doc/tutorials/hp_tuning_wide_and_deep_model.ipynb | anukaal/cloud | 2b6c9127734ea22b6c1aaa070ac06124e5960c57 | [
"Apache-2.0"
] | 342 | 2020-02-10T18:55:31.000Z | 2022-03-01T04:30:12.000Z | g3doc/tutorials/hp_tuning_wide_and_deep_model.ipynb | anukaal/cloud | 2b6c9127734ea22b6c1aaa070ac06124e5960c57 | [
"Apache-2.0"
] | 179 | 2020-02-14T22:35:42.000Z | 2022-01-22T23:06:23.000Z | g3doc/tutorials/hp_tuning_wide_and_deep_model.ipynb | anukaal/cloud | 2b6c9127734ea22b6c1aaa070ac06124e5960c57 | [
"Apache-2.0"
] | 140 | 2020-02-10T19:11:50.000Z | 2022-02-23T13:04:44.000Z | 37.342593 | 475 | 0.544921 | [
[
[
"##### Copyright 2021 The TensorFlow Cloud Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Tuning a wide and deep model using Google Cloud\n\n<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/cloud/tutorials/hp_tuning_wide_and_deep_model.ipynb\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/cloud/blob/master/g3doc/tutorials/hp_tuning_wide_and_deep_model.ipynb\"\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/cloud/blob/master/g3doc/tutorials/hp_tuning_wide_and_deep_model.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/cloud/tutorials/hp_tuning_wide_and_deep_model.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n <td>\n <a href=\"https://kaggle.com/kernels/welcome?src=https://github.com/tensorflow/cloud/blob/master/g3doc/tutorials/hp_tuning_wide_and_deep_model.ipynb\" target=\"blank\">\n <img width=\"90\" src=\"https://www.kaggle.com/static/images/site-logo.png\" alt=\"Kaggle logo\">Run in Kaggle\n </a>\n </td>\n</table>\n",
"_____no_output_____"
],
[
"\nIn this example we will use CloudTuner and Google Cloud to Tune a [Wide and Deep Model](https://ai.googleblog.com/2016/06/wide-deep-learning-better-together-with.html) based on the tunable model introduced in [structured data learning with Wide, Deep, and Cross networks](https://keras.io/examples/structured_data/wide_deep_cross_networks/). In this example we will use the data set from [CAIIS Dogfood Day](https://www.kaggle.com/c/caiis-dogfood-day-2020/overview)",
"_____no_output_____"
],
[
"## Import required modules",
"_____no_output_____"
]
],
[
[
"import datetime\nimport uuid\n\nimport numpy as np\nimport pandas as pd\nimport tensorflow as tf\nimport os\nimport sys\nimport subprocess\n\nfrom tensorflow.keras import datasets, layers, models\nfrom sklearn.model_selection import train_test_split\n\n# Install the latest version of tensorflow_cloud and other required packages.\nif os.environ.get(\"TF_KERAS_RUNNING_REMOTELY\", True):\n subprocess.run(\n ['python3', '-m', 'pip', 'install', 'tensorflow-cloud', '-q'])\n subprocess.run(\n ['python3', '-m', 'pip', 'install', 'google-cloud-storage', '-q'])\n subprocess.run(\n ['python3', '-m', 'pip', 'install', 'fsspec', '-q'])\n subprocess.run(\n ['python3', '-m', 'pip', 'install', 'gcsfs', '-q'])\n\nimport tensorflow_cloud as tfc\nprint(tfc.__version__)",
"0.1.15\n"
],
[
"tf.version.VERSION",
"_____no_output_____"
]
],
[
[
"## Project Configurations\nSetting project parameters. For more details on Google Cloud Specific parameters please refer to [Google Cloud Project Setup Instructions](https://www.kaggle.com/nitric/google-cloud-project-setup-instructions/).",
"_____no_output_____"
]
],
[
[
"# Set Google Cloud Specific parameters\n\n# TODO: Please set GCP_PROJECT_ID to your own Google Cloud project ID.\nGCP_PROJECT_ID = 'YOUR_PROJECT_ID' #@param {type:\"string\"}\n\n# TODO: Change the Service Account Name to your own Service Account\nSERVICE_ACCOUNT_NAME = 'YOUR_SERVICE_ACCOUNT_NAME' #@param {type:\"string\"}\nSERVICE_ACCOUNT = f'{SERVICE_ACCOUNT_NAME}@{GCP_PROJECT_ID}.iam.gserviceaccount.com'\n\n# TODO: set GCS_BUCKET to your own Google Cloud Storage (GCS) bucket.\nGCS_BUCKET = 'YOUR_GCS_BUCKET_NAME' #@param {type:\"string\"}\n\n# DO NOT CHANGE: Currently only the 'us-central1' region is supported.\nREGION = 'us-central1'",
"_____no_output_____"
],
[
"# Set Tuning Specific parameters\n\n# OPTIONAL: You can change the job name to any string.\nJOB_NAME = 'wide_and_deep' #@param {type:\"string\"}\n\n# OPTIONAL: Set Number of concurrent tuning jobs that you would like to run.\nNUM_JOBS = 5 #@param {type:\"string\"}\n\n# TODO: Set the study ID for this run. Study_ID can be any unique string.\n# Reusing the same Study_ID will cause the Tuner to continue tuning the\n# Same Study parameters. This can be used to continue on a terminated job,\n# or load stats from a previous study.\nSTUDY_NUMBER = '00001' #@param {type:\"string\"}\nSTUDY_ID = f'{GCP_PROJECT_ID}_{JOB_NAME}_{STUDY_NUMBER}'\n\n# Setting location were training logs and checkpoints will be stored\nGCS_BASE_PATH = f'gs://{GCS_BUCKET}/{JOB_NAME}/{STUDY_ID}'\nTENSORBOARD_LOGS_DIR = os.path.join(GCS_BASE_PATH,\"logs\")",
"_____no_output_____"
]
],
[
[
"## Authenticating the notebook to use your Google Cloud Project\n\nFor Kaggle Notebooks click on \"Add-ons\"->\"Google Cloud SDK\" before running the cell below.",
"_____no_output_____"
]
],
[
[
"# Using tfc.remote() to ensure this code only runs in notebook\nif not tfc.remote():\n\n # Authentication for Kaggle Notebooks\n if \"kaggle_secrets\" in sys.modules:\n from kaggle_secrets import UserSecretsClient\n UserSecretsClient().set_gcloud_credentials(project=GCP_PROJECT_ID)\n\n # Authentication for Colab Notebooks\n if \"google.colab\" in sys.modules:\n from google.colab import auth\n auth.authenticate_user()\n os.environ[\"GOOGLE_CLOUD_PROJECT\"] = GCP_PROJECT_ID",
"_____no_output_____"
]
],
[
[
"## Load the data\nRead raw data and split to train and test data sets. For this step you will need to copy the dataset to your GCS bucket so it can be accessed during training. For this example we are using the dataset from https://www.kaggle.com/c/caiis-dogfood-day-2020.\n\nTo do this you can run the following commands to download and copy the dataset to your GCS bucket, or manually download the dataset vi [Kaggle UI](https://www.kaggle.com/c/caiis-dogfood-day-2020/data) and upload the `train.csv` file to your [GCS bucket vi GCS UI](https://console.cloud.google.com/storage/browser).\n\n```python\n# Download the dataset\n!kaggle competitions download -c caiis-dogfood-day-2020\n\n# Copy the training file to your bucket\n!gsutil cp ./caiis-dogfood-day-2020/train.csv $GCS_BASE_PATH/caiis-dogfood-day-2020/train.csv\n```",
"_____no_output_____"
]
],
[
[
"train_URL = f'{GCS_BASE_PATH}/caiis-dogfood-day-2020/train.csv'\ndata = pd.read_csv(train_URL)\ntrain, test = train_test_split(data, test_size=0.1)",
"_____no_output_____"
],
[
"# A utility method to create a tf.data dataset from a Pandas Dataframe\ndef df_to_dataset(df, shuffle=True, batch_size=32):\n df = df.copy()\n labels = df.pop('target')\n ds = tf.data.Dataset.from_tensor_slices((dict(df), labels))\n if shuffle:\n ds = ds.shuffle(buffer_size=len(df))\n ds = ds.batch(batch_size)\n return ds",
"_____no_output_____"
],
[
"sm_batch_size = 1000 # A small batch size is used for demonstration purposes\ntrain_ds = df_to_dataset(train, batch_size=sm_batch_size)\ntest_ds = df_to_dataset(test, shuffle=False, batch_size=sm_batch_size)",
"_____no_output_____"
]
],
[
[
"## Preprocess the data\n\nSetting up preprocessing layers for categorical and numerical input data. For more details on preprocessing layers please refer to [working with preprocessing layers](https://www.tensorflow.org/guide/keras/preprocessing_layers).\n",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras.layers.experimental import preprocessing\n\ndef create_model_inputs():\n inputs ={}\n for name, column in data.items():\n if name in ('id','target'):\n continue\n dtype = column.dtype\n if dtype == object:\n dtype = tf.string\n else:\n dtype = tf.float32\n\n inputs[name] = tf.keras.Input(shape=(1,), name=name, dtype=dtype)\n\n return inputs",
"_____no_output_____"
],
[
"#Preprocessing the numeric inputs, and running them through a normalization layer.\ndef preprocess_numeric_inputs(inputs):\n\n numeric_inputs = {name:input for name,input in inputs.items()\n if input.dtype==tf.float32}\n\n x = layers.Concatenate()(list(numeric_inputs.values()))\n norm = preprocessing.Normalization()\n norm.adapt(np.array(data[numeric_inputs.keys()]))\n numeric_inputs = norm(x)\n return numeric_inputs",
"_____no_output_____"
],
[
"# Preprocessing the categorical inputs.\ndef preprocess_categorical_inputs(inputs):\n categorical_inputs = []\n for name, input in inputs.items():\n if input.dtype == tf.float32:\n continue\n\n lookup = preprocessing.StringLookup(vocabulary=np.unique(data[name]))\n one_hot = preprocessing.CategoryEncoding(max_tokens=lookup.vocab_size())\n\n x = lookup(input)\n x = one_hot(x)\n categorical_inputs.append(x)\n\n return layers.concatenate(categorical_inputs)",
"_____no_output_____"
]
],
[
[
"## Define the model architecture and hyperparameters\nIn this section we define our tuning parameters using [Keras Tuner Hyper Parameters](https://keras-team.github.io/keras-tuner/#the-search-space-may-contain-conditional-hyperparameters) and a model-building function. The model-building function takes an argument hp from which you can sample hyperparameters, such as hp.Int('units', min_value=32, max_value=512, step=32) (an integer from a certain range).\n",
"_____no_output_____"
]
],
[
[
"import kerastuner\n\n# Configure the search space\nHPS = kerastuner.engine.hyperparameters.HyperParameters()\nHPS.Float('learning_rate', min_value=1e-4, max_value=1e-2, sampling='log')\n\nHPS.Int('num_layers', min_value=2, max_value=5)\nfor i in range(5):\n HPS.Float('dropout_rate_' + str(i), min_value=0.0, max_value=0.3, step=0.1)\n HPS.Choice('num_units_' + str(i), [32, 64, 128, 256])",
"_____no_output_____"
],
[
"from tensorflow.keras import layers\nfrom tensorflow.keras.optimizers import Adam\n\n\ndef create_wide_and_deep_model(hp):\n\n inputs = create_model_inputs()\n wide = preprocess_categorical_inputs(inputs)\n wide = layers.BatchNormalization()(wide)\n\n deep = preprocess_numeric_inputs(inputs)\n for i in range(hp.get('num_layers')):\n deep = layers.Dense(hp.get('num_units_' + str(i)))(deep)\n deep = layers.BatchNormalization()(deep)\n deep = layers.ReLU()(deep)\n deep = layers.Dropout(hp.get('dropout_rate_' + str(i)))(deep)\n\n both = layers.concatenate([wide, deep])\n outputs = layers.Dense(1, activation='sigmoid')(both)\n model = tf.keras.Model(inputs=inputs, outputs=outputs)\n metrics = [\n tf.keras.metrics.Precision(name='precision'),\n tf.keras.metrics.Recall(name='recall'),\n 'accuracy',\n 'mse'\n ]\n\n model.compile(\n optimizer=Adam(lr=hp.get('learning_rate')),\n loss='binary_crossentropy',\n metrics=metrics)\n return model",
"_____no_output_____"
]
],
[
[
"## Configure a CloudTuner\nIn this section we configure the cloud tuner for both remote and local execution. The main difference between the two is the distribution strategy.",
"_____no_output_____"
]
],
[
[
"from tensorflow_cloud import CloudTuner\n\ndistribution_strategy = None\nif not tfc.remote():\n # Using MirroredStrategy to use a single instance with multiple GPUs\n # during remote execution while using no strategy for local.\n distribution_strategy = tf.distribute.MirroredStrategy()\n\ntuner = CloudTuner(\n create_wide_and_deep_model,\n project_id=GCP_PROJECT_ID,\n project_name=JOB_NAME,\n region=REGION,\n objective='accuracy',\n hyperparameters=HPS,\n max_trials=100,\n directory=GCS_BASE_PATH,\n study_id=STUDY_ID,\n overwrite=True,\n distribution_strategy=distribution_strategy)",
"_____no_output_____"
],
[
"# Configure Tensorboard logs\ncallbacks=[\n tf.keras.callbacks.TensorBoard(log_dir=TENSORBOARD_LOGS_DIR)]\n\n# Setting to run tuning remotely, you can run tuner locally to validate it works first.\nif tfc.remote():\n tuner.search(train_ds, epochs=20, validation_data=test_ds, callbacks=callbacks)\n\n# You can uncomment the code below to run the tuner.search() locally to validate\n# everything works before submitting the job to Cloud. Stop the job manually\n# after one epoch.\n\n# else:\n# tuner.search(train_ds, epochs=1, validation_data=test_ds, callbacks=callbacks)",
"_____no_output_____"
]
],
[
[
"## Start the remote training\n\nThis step will prepare your code from this notebook for remote execution and start NUM_JOBS parallel runs remotely to train the model. Once the jobs are submitted you can go to the next step to monitor the jobs progress via Tensorboard.",
"_____no_output_____"
]
],
[
[
"\n# Optional: Some recommended base images. If you provide none the system will choose one for you.\nTF_GPU_IMAGE= \"gcr.io/deeplearning-platform-release/tf2-cpu.2-5\"\nTF_CPU_IMAGE= \"gcr.io/deeplearning-platform-release/tf2-gpu.2-5\"\n\n\ntfc.run_cloudtuner(\n distribution_strategy='auto',\n docker_config=tfc.DockerConfig(\n parent_image=TF_GPU_IMAGE,\n image_build_bucket=GCS_BUCKET\n ),\n chief_config=tfc.MachineConfig(\n cpu_cores=16,\n memory=60,\n ),\n job_labels={'job': JOB_NAME},\n service_account=SERVICE_ACCOUNT,\n num_jobs=NUM_JOBS\n)",
"_____no_output_____"
]
],
[
[
"# Training Results\n## Reconnect your Colab instance\nMost remote training jobs are long running, if you are using Colab it may time out before the training results are available. In that case rerun the following sections to reconnect and configure your Colab instance to access the training results. Run the following sections in order:\n\n1. Import required modules\n2. Project Configurations\n3. Authenticating the notebook to use your Google Cloud Project\n\n## Load Tensorboard\nWhile the training is in progress you can use Tensorboard to view the results. Note the results will show only after your training has started. This may take a few minutes.",
"_____no_output_____"
]
],
[
[
"%load_ext tensorboard\n%tensorboard --logdir $TENSORBOARD_LOGS_DIR",
"_____no_output_____"
]
],
[
[
"You can access the training assets as follows. Note the results will show only after your tuning job has completed at least once trial. This may take a few minutes.",
"_____no_output_____"
]
],
[
[
"if not tfc.remote():\n tuner.results_summary(1)\n best_model = tuner.get_best_models(1)[0]\n best_hyperparameters = tuner.get_best_hyperparameters(1)[0]\n\n # References to best trial assets\n best_trial_id = tuner.oracle.get_best_trials(1)[0].trial_id\n best_trial_dir = tuner.get_trial_dir(best_trial_id) ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7929ae6bee4976a3bb0ecfd01dabf8674ecdde9 | 2,450 | ipynb | Jupyter Notebook | FCS_conversion/fcs_write.ipynb | SebastianJarosch/ChipCytometry-Image-Processing | 2c475a639b4dd84e8dd18b0ad91c85eed4a72582 | [
"BSD-3-Clause"
] | 2 | 2022-03-03T18:08:54.000Z | 2022-03-10T17:00:34.000Z | FCS_conversion/fcs_write.ipynb | SebastianJarosch/ChipCytometry-Image-Processing | 2c475a639b4dd84e8dd18b0ad91c85eed4a72582 | [
"BSD-3-Clause"
] | null | null | null | FCS_conversion/fcs_write.ipynb | SebastianJarosch/ChipCytometry-Image-Processing | 2c475a639b4dd84e8dd18b0ad91c85eed4a72582 | [
"BSD-3-Clause"
] | null | null | null | 32.236842 | 138 | 0.581633 | [
[
[
"import pandas as pd\nimport numpy as np\nimport fcswrite\n\ndef write_fcs_ChipCytometry(FL_values_path, channels_path, additional_channels=['X','Y','Circ.','Area'], output_dir='./'):\n \"\"\"Write ChipCytometry data to an .fcs file using fcswrite (FCS3.0 file format)\n Parameters\n ----------\n FL_values_path: str\n Path to the FL_values.csv file generated in ImageJ\n channels_path: str \n Path to the channels.csv file generated in ImageJ\n additional_channels: list of str\n Additional channels that should be included from the data\n file. Default: ['X','Y','Circ.','Area']\n output_dir: str\n Output directory the fcs file should be written to.\n Default: current directory\n \"\"\"\n\n data=pd.read_csv(FL_values_path, index_col=0)\n channels=pd.read_csv(channels_path)\n data.Label=[x[-1][:-5] for x in data.Label.str.split(':')]\n additional_channels=additional_channels\n format_data=pd.DataFrame()\n for channel in data.Label.unique():\n format_data[channel]=data.Mean[data.Label==channel].tolist()\n format_data.columns=channels.iloc[:,0]\n format_data[additional_channels]=data[additional_channels][:format_data.shape[0]]\n format_data['Identifier']=pd.Series(range(1,format_data.shape[0]+1))\n fcswrite.write_fcs(output_dir+channels.columns.tolist()[0]+'neu3.fcs', chn_names=format_data.columns.tolist(), data=format_data)",
"_____no_output_____"
],
[
"write_fcs_ChipCytometry()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
e7929efdb820b9a571d392cd39eaa5e6225421f6 | 7,193 | ipynb | Jupyter Notebook | benchmarks/Benchmarks_ODL.ipynb | SimonRuetz/Shearlab.jl | ff484f72d3efc19035d33a237c266a74b08926d2 | [
"MIT"
] | 24 | 2017-01-31T17:45:27.000Z | 2020-09-13T08:56:10.000Z | benchmarks/Benchmarks_ODL.ipynb | SimonRuetz/Shearlab.jl | ff484f72d3efc19035d33a237c266a74b08926d2 | [
"MIT"
] | 28 | 2016-11-30T17:40:05.000Z | 2022-03-17T11:27:33.000Z | benchmarks/Benchmarks_ODL.ipynb | SimonRuetz/Shearlab.jl | ff484f72d3efc19035d33a237c266a74b08926d2 | [
"MIT"
] | 11 | 2017-02-26T13:58:39.000Z | 2020-10-20T14:34:49.000Z | 21.731118 | 136 | 0.534826 | [
[
[
"# <center> Shearlab decomposition benchmarks </center>",
"_____no_output_____"
],
[
"Some benchmarks comparing the performance of pure julia, python/julia and python implementation.",
"_____no_output_____"
]
],
[
[
"# Importing julia\nimport odl\nimport sys\nsys.path.append('../')\nimport shearlab_operator\nimport pyshearlab\nimport matplotlib.pyplot as plt\n%matplotlib inline\nfrom numpy import ceil\nfrom odl.contrib.shearlab import shearlab_operator",
"_____no_output_____"
],
[
"# Calling julia\nj = shearlab_operator.load_julia_with_Shearlab()",
"_____no_output_____"
]
],
[
[
"Defining the parameters",
"_____no_output_____"
]
],
[
[
"n = 512\nm = n\ngpu = 0\nsquare = 0\nname = './lena.jpg';",
"_____no_output_____"
],
[
"nScales = 4;\nshearLevels = [float(ceil(i/2)) for i in range(1,nScales+1)]\nscalingFilter = 'Shearlab.filt_gen(\"scaling_shearlet\")'\ndirectionalFilter = 'Shearlab.filt_gen(\"directional_shearlet\")'\nwaveletFilter = 'Shearlab.mirror(scalingFilter)'\nscalingFilter2 = 'scalingFilter'\nfull = 0;",
"_____no_output_____"
],
[
"# Pure Julia\nj.eval('X = Shearlab.load_image(name, n);');",
"_____no_output_____"
],
[
"# Read Data\nj.eval('n = 512;')\n# The path of the image\nj.eval('name = \"./lena.jpg\";');\ndata = shearlab_operator.load_image(name,n);",
"_____no_output_____"
],
[
"sizeX = data.shape[0]\nsizeY = data.shape[1]\nrows = sizeX\ncols = sizeY\nX = data;",
"_____no_output_____"
]
],
[
[
"** Shearlet System generation **",
"_____no_output_____"
]
],
[
[
"# Pure julia\nwith odl.util.Timer('Shearlet System Generation julia'):\n j.eval('shearletSystem = Shearlab.getshearletsystem2D(n,n,4)');",
"Shearlet System Generation julia : 9.345 \n"
],
[
"# Python/Julia\nwith odl.util.Timer('Shearlet System Generation python/julia'):\n shearletSystem_jl = shearlab_operator.getshearletsystem2D(rows,cols,nScales,shearLevels,full,directionalFilter,scalingFilter);",
"Shearlet System Generation python/julia : 9.125 \n"
],
[
"# pyShearlab\nwith odl.util.Timer('Shearlet System Generation python'):\n shearletSystem_py = pyshearlab.SLgetShearletSystem2D(0,rows, cols, nScales)",
"Shearlet System Generation python : 49.179 \n"
]
],
[
[
"** Coefficients computation **",
"_____no_output_____"
]
],
[
[
"# Pure Julia\nwith odl.util.Timer('Shearlet Coefficients Computation julia'):\n j.eval('coeffs = Shearlab.SLsheardec2D(X,shearletSystem);');",
"Shearlet Coefficients Computation julia : 1.203 \n"
],
[
"# Julia/Python\nwith odl.util.Timer('Shearlet Coefficients Computation python/julia'):\n coeffs_jl = shearlab_operator.sheardec2D(X,shearletSystem_jl)",
"Shearlet Coefficients Computation python/julia : 2.217 \n"
],
[
"# pyShearlab\nwith odl.util.Timer('Shearlet Coefficients Computation python'):\n coeffs_py = pyshearlab.SLsheardec2D(X, shearletSystem_py)",
"Shearlet Coefficients Computation python : 1.333 \n"
]
],
[
[
"** Reconstruction **",
"_____no_output_____"
]
],
[
[
"# Pure Julia\nwith odl.util.Timer('Shearlet Reconstructon julia'):\n j.eval('Xrec=Shearlab.SLshearrec2D(coeffs,shearletSystem);');",
" Shearlet Reconstructon julia : 1.208 \n"
],
[
"# Julia/Python\nwith odl.util.Timer('Shearlet Reconstructon python/julia'):\n Xrec_jl = shearlab_operator.shearrec2D(coeffs_jl,shearletSystem_jl);",
"Shearlet Reconstructon python/julia : 1.509 \n"
],
[
"# pyShearlab\nwith odl.util.Timer('Shearlet Reconstructon python'):\n Xrec_py = pyshearlab.SLshearrec2D(coeffs_py, shearletSystem_py)",
" Shearlet Reconstructon python : 1.136 \n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
e792ce021d426c946f263417dc7c565be84d2dcb | 31,249 | ipynb | Jupyter Notebook | Starter_Code/credit_risk_resampling.ipynb | JordanCandido/Unit11Homework | 9c1dca334debfbd4d3f0c5d54288dd0410b145bf | [
"ADSL"
] | null | null | null | Starter_Code/credit_risk_resampling.ipynb | JordanCandido/Unit11Homework | 9c1dca334debfbd4d3f0c5d54288dd0410b145bf | [
"ADSL"
] | null | null | null | Starter_Code/credit_risk_resampling.ipynb | JordanCandido/Unit11Homework | 9c1dca334debfbd4d3f0c5d54288dd0410b145bf | [
"ADSL"
] | null | null | null | 27.900893 | 294 | 0.48664 | [
[
[
"# Credit Risk Resampling Techniques",
"_____no_output_____"
]
],
[
[
"import warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"import numpy as np\nimport pandas as pd\nfrom pathlib import Path\nfrom collections import Counter",
"_____no_output_____"
]
],
[
[
"# Read the CSV into DataFrame",
"_____no_output_____"
]
],
[
[
"# Load the data\nfile_path = Path('Resources/lending_data.csv')\nlendingdata = pd.read_csv(file_path)\nlendingdata.head()",
"_____no_output_____"
]
],
[
[
"# Split the Data into Training and Testing",
"_____no_output_____"
]
],
[
[
"# Create our features\nX = lendingdata.copy()\nX.drop(columns=[\"loan_status\", 'homeowner'], axis=1, inplace=True)\nX.head()\n# Create our target\ny = lendingdata['loan_status']",
"_____no_output_____"
],
[
"X.describe()",
"_____no_output_____"
],
[
"# Check the balance of our target values\ny.value_counts()",
"_____no_output_____"
],
[
"# Create X_train, X_test, y_train, y_test\nfrom sklearn.model_selection import train_test_split\nX_train, X_test, y_train, y_test = train_test_split(X, y, random_state=1)",
"_____no_output_____"
]
],
[
[
"## Data Pre-Processing\n\nScale the training and testing data using the `StandardScaler` from `sklearn`. Remember that when scaling the data, you only scale the features data (`X_train` and `X_testing`).",
"_____no_output_____"
]
],
[
[
"# Create the StandardScaler instance\nfrom sklearn.preprocessing import StandardScaler\nscaler = StandardScaler()",
"_____no_output_____"
],
[
"# Fit the Standard Scaler with the training data\n# When fitting scaling functions, only train on the training dataset\nX_scaler = scaler.fit(X_train)",
"_____no_output_____"
],
[
"# Scale the training and testing data\nX_train_scaled = X_scaler.transform(X_train)\nX_test_scaled = X_scaler.transform(X_test)",
"_____no_output_____"
]
],
[
[
"# Simple Logistic Regression",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression\nmodel = LogisticRegression(solver='lbfgs', random_state=1)\nmodel.fit(X_train, y_train)",
"_____no_output_____"
],
[
"# Calculated the balanced accuracy score\nfrom sklearn.metrics import balanced_accuracy_score\ny_pred = model.predict(X_test)\nbalanced_accuracy_score(y_test, y_pred)",
"_____no_output_____"
],
[
"# Display the confusion matrix\nfrom sklearn.metrics import confusion_matrix\nconfusion_matrix(y_test, y_pred)",
"_____no_output_____"
],
[
"# Print the imbalanced classification report\nfrom imblearn.metrics import classification_report_imbalanced\nprint(classification_report_imbalanced(y_test, y_pred))",
" pre rec spe f1 geo iba sup\n\n high_risk 0.85 0.91 0.99 0.88 0.95 0.90 619\n low_risk 1.00 0.99 0.91 1.00 0.95 0.91 18765\n\navg / total 0.99 0.99 0.91 0.99 0.95 0.91 19384\n\n"
]
],
[
[
"# Oversampling\n\nIn this section, you will compare two oversampling algorithms to determine which algorithm results in the best performance. You will oversample the data using the naive random oversampling algorithm and the SMOTE algorithm. For each algorithm, be sure to complete the folliowing steps:\n\n1. View the count of the target classes using `Counter` from the collections library. \n3. Use the resampled data to train a logistic regression model.\n3. Calculate the balanced accuracy score from sklearn.metrics.\n4. Print the confusion matrix from sklearn.metrics.\n5. Generate a classication report using the `imbalanced_classification_report` from imbalanced-learn.\n\nNote: Use a random state of 1 for each sampling algorithm to ensure consistency between tests",
"_____no_output_____"
],
[
"### Naive Random Oversampling",
"_____no_output_____"
]
],
[
[
"# Resample the training data with the RandomOversampler\n# View the count of target classes with Counter\nfrom imblearn.over_sampling import RandomOverSampler\n\nros = RandomOverSampler(random_state=1)\nX_resampled, y_resampled = ros.fit_resample(X_train, y_train)\nCounter(y_resampled)",
"_____no_output_____"
],
[
"# Train the Logistic Regression model using the resampled data\nfrom sklearn.linear_model import LogisticRegression\n\nnromodel = LogisticRegression(solver='lbfgs', random_state=1)\nnromodel.fit(X_resampled, y_resampled)",
"_____no_output_____"
],
[
"# Calculated the balanced accuracy score\nfrom sklearn.metrics import balanced_accuracy_score\n\nbalanced_accuracy_score(y_test, y_pred)",
"_____no_output_____"
],
[
"# Display the confusion matrix\nfrom sklearn.metrics import confusion_matrix\n\ny_pred = nromodel.predict(X_test)\nconfusion_matrix(y_test, y_pred)",
"_____no_output_____"
],
[
"# Print the imbalanced classification report\nfrom imblearn.metrics import classification_report_imbalanced\n\nprint(classification_report_imbalanced(y_test, y_pred))",
" pre rec spe f1 geo iba sup\n\n high_risk 0.84 0.99 0.99 0.91 0.99 0.99 619\n low_risk 1.00 0.99 0.99 1.00 0.99 0.99 18765\n\navg / total 0.99 0.99 0.99 0.99 0.99 0.99 19384\n\n"
]
],
[
[
"### SMOTE Oversampling",
"_____no_output_____"
]
],
[
[
"# Resample the training data with SMOTE\n# View the count of target classes with Counter\nfrom imblearn.over_sampling import SMOTE\n\nX_resampled, y_resampled = SMOTE(random_state=1, sampling_strategy=1.0).fit_resample(\n X_train, y_train\n)\nfrom collections import Counter\n\nCounter(y_resampled)",
"_____no_output_____"
],
[
"# Train the Logistic Regression model using the resampled data\nsmodel = LogisticRegression(solver='lbfgs', random_state=1)\nsmodel.fit(X_resampled, y_resampled)",
"_____no_output_____"
],
[
"# Calculated the balanced accuracy score\ny_pred = smodel.predict(X_test)\nbalanced_accuracy_score(y_test, y_pred)",
"_____no_output_____"
],
[
"# Display the confusion matrix\nconfusion_matrix(y_test, y_pred)",
"_____no_output_____"
],
[
"# Print the imbalanced classification report\nprint(classification_report_imbalanced(y_test, y_pred))",
" pre rec spe f1 geo iba sup\n\n high_risk 0.84 0.99 0.99 0.91 0.99 0.99 619\n low_risk 1.00 0.99 0.99 1.00 0.99 0.99 18765\n\navg / total 0.99 0.99 0.99 0.99 0.99 0.99 19384\n\n"
]
],
[
[
"# Undersampling\n\nIn this section, you will test an undersampling algorithm to determine which algorithm results in the best performance compared to the oversampling algorithms above. You will undersample the data using the Cluster Centroids algorithm and complete the folliowing steps:\n\n1. View the count of the target classes using `Counter` from the collections library. \n3. Use the resampled data to train a logistic regression model.\n3. Calculate the balanced accuracy score from sklearn.metrics.\n4. Display the confusion matrix from sklearn.metrics.\n5. Generate a classication report using the `imbalanced_classification_report` from imbalanced-learn.\n\nNote: Use a random state of 1 for each sampling algorithm to ensure consistency between tests",
"_____no_output_____"
]
],
[
[
"# Resample the data using the ClusterCentroids resampler\n# View the count of target classes with Counter\nfrom imblearn.under_sampling import RandomUnderSampler\nros = RandomUnderSampler(random_state=1)\nX_resampled, y_resampled = ros.fit_resample(X_train, y_train)\nCounter(y_resampled)",
"_____no_output_____"
],
[
"# Train the Logistic Regression model using the resampled data\nfrom sklearn.linear_model import LogisticRegression\n\numodel = LogisticRegression(solver='lbfgs', random_state=1)\numodel.fit(X_resampled, y_resampled)",
"_____no_output_____"
],
[
"# Calculate the balanced accuracy score\nfrom sklearn.metrics import balanced_accuracy_score\n\nbalanced_accuracy_score(y_test, y_pred)",
"_____no_output_____"
],
[
"# Display the confusion matrix\nfrom sklearn.metrics import confusion_matrix\n\ny_pred = umodel.predict(X_test)\nconfusion_matrix(y_test, y_pred)",
"_____no_output_____"
],
[
"# Print the imbalanced classification report\nfrom imblearn.metrics import classification_report_imbalanced\n\nprint(classification_report_imbalanced(y_test, y_pred))",
" pre rec spe f1 geo iba sup\n\n high_risk 0.84 0.99 0.99 0.91 0.99 0.99 619\n low_risk 1.00 0.99 0.99 1.00 0.99 0.99 18765\n\navg / total 0.99 0.99 0.99 0.99 0.99 0.99 19384\n\n"
]
],
[
[
"# Combination (Over and Under) Sampling\n\nIn this section, you will test a combination over- and under-sampling algorithm to determine if the algorithm results in the best performance compared to the other sampling algorithms above. You will resample the data using the SMOTEENN algorithm and complete the folliowing steps:\n\n1. View the count of the target classes using `Counter` from the collections library. \n3. Use the resampled data to train a logistic regression model.\n3. Calculate the balanced accuracy score from sklearn.metrics.\n4. Display the confusion matrix from sklearn.metrics.\n5. Generate a classication report using the `imbalanced_classification_report` from imbalanced-learn.\n\nNote: Use a random state of 1 for each sampling algorithm to ensure consistency between tests",
"_____no_output_____"
]
],
[
[
"# Resample the training data with SMOTEENN\n# View the count of target classes with Counter\nfrom imblearn.combine import SMOTEENN\n\nsmote_enn = SMOTEENN(random_state=1)\nX_resampled, y_resampled = smote_enn.fit_resample(X, y)\nCounter(y_resampled)",
"_____no_output_____"
],
[
"# Train the Logistic Regression model using the resampled data\nfrom sklearn.linear_model import LogisticRegression\ncmodel = LogisticRegression(solver='lbfgs', random_state=1)\ncmodel.fit(X_resampled, y_resampled)",
"_____no_output_____"
],
[
"# Calculate the balanced accuracy score\nfrom sklearn.metrics import balanced_accuracy_score\n\nbalanced_accuracy_score(y_test, y_pred)",
"_____no_output_____"
],
[
"# Display the confusion matrix\nfrom sklearn.metrics import confusion_matrix\n\ny_pred = cmodel.predict(X_test)\nconfusion_matrix(y_test, y_pred)",
"_____no_output_____"
],
[
"# Print the imbalanced classification report\nfrom imblearn.metrics import classification_report_imbalanced\n\nprint(classification_report_imbalanced(y_test, y_pred))",
" pre rec spe f1 geo iba sup\n\n high_risk 0.84 0.99 0.99 0.91 0.99 0.99 619\n low_risk 1.00 0.99 0.99 1.00 0.99 0.99 18765\n\navg / total 0.99 0.99 0.99 0.99 0.99 0.99 19384\n\n"
]
],
[
[
"# Final Questions\n\n1. Which model had the best balanced accuracy score?\n\n SMOTE Oversampling, Undersampling and Combination all had the highest balanced accuracy score of 0.993678.\n\n2. Which model had the best recall score?\n\n Simpole Logistic Regression, Oversampling, Undersampling and Combination all had the best recall score of 0.99.\n\n3. Which model had the best geometric mean score?\n\n Oversampling, Undersampling and Combination all had the best recall score of 0.99.\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
e792e0ffa11e14e75a94d8906d707fbb05aa5be1 | 21,464 | ipynb | Jupyter Notebook | lessons/Recommendations/1_Intro_to_Recommendations/5_Content Based Recommendations - Solution.ipynb | vishalwaka/DSND_Term2 | d4a150b084edefbef5b26a142571d9e99638484d | [
"MIT"
] | 1,030 | 2018-07-03T19:09:50.000Z | 2022-03-25T05:48:57.000Z | lessons/Recommendations/1_Intro_to_Recommendations/5_Content Based Recommendations - Solution.ipynb | vishalwaka/DSND_Term2 | d4a150b084edefbef5b26a142571d9e99638484d | [
"MIT"
] | 21 | 2018-09-20T14:36:04.000Z | 2021-10-11T18:25:31.000Z | lessons/Recommendations/1_Intro_to_Recommendations/5_Content Based Recommendations - Solution.ipynb | vishalwaka/DSND_Term2 | d4a150b084edefbef5b26a142571d9e99638484d | [
"MIT"
] | 1,736 | 2018-06-27T19:33:46.000Z | 2022-03-28T17:52:33.000Z | 34.397436 | 603 | 0.598397 | [
[
[
"### Content Based Recommendations\n\nIn the previous notebook, you were introduced to a way to make recommendations using collaborative filtering. However, using this technique there are a large number of users who were left without any recommendations at all. Other users were left with fewer than the ten recommendations that were set up by our function to retrieve...\n\nIn order to help these users out, let's try another technique **content based** recommendations. Let's start off where we were in the previous notebook.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom collections import defaultdict\nfrom IPython.display import HTML\nimport progressbar\nimport tests as t\nimport pickle\n\n\n%matplotlib inline\n\n# Read in the datasets\nmovies = pd.read_csv('movies_clean.csv')\nreviews = pd.read_csv('reviews_clean.csv')\n\ndel movies['Unnamed: 0']\ndel reviews['Unnamed: 0']\n\n\nall_recs = pickle.load(open(\"all_recs.p\", \"rb\"))",
"_____no_output_____"
]
],
[
[
"### Datasets\n\nFrom the above, you now have access to three important items that you will be using throughout the rest of this notebook. \n\n`a.` **movies** - a dataframe of all of the movies in the dataset along with other content related information about the movies (genre and date)\n\n\n`b.` **reviews** - this was the main dataframe used before for collaborative filtering, as it contains all of the interactions between users and movies.\n\n\n`c.` **all_recs** - a dictionary where each key is a user, and the value is a list of movie recommendations based on collaborative filtering\n\nFor the individuals in **all_recs** who did recieve 10 recommendations using collaborative filtering, we don't really need to worry about them. However, there were a number of individuals in our dataset who did not receive any recommendations.\n\n-----\n\n`1.` To begin, let's start with finding all of the users in our dataset who didn't get all 10 ratings we would have liked them to have using collaborative filtering. ",
"_____no_output_____"
]
],
[
[
"users_with_all_recs = []\nfor user, movie_recs in all_recs.items():\n if len(movie_recs) > 9:\n users_with_all_recs.append(user)\n\nprint(\"There are {} users with all reccomendations from collaborative filtering.\".format(len(users_with_all_recs)))\n\nusers = np.unique(reviews['user_id'])\nusers_who_need_recs = np.setdiff1d(users, users_with_all_recs)\n\nprint(\"There are {} users who still need recommendations.\".format(len(users_who_need_recs)))\nprint(\"This means that only {}% of users received all 10 of their recommendations using collaborative filtering\".format(round(len(users_with_all_recs)/len(np.unique(reviews['user_id'])), 4)*100)) ",
"There are 22187 users with all reccomendations from collaborative filtering.\nThere are 31781 users who still need recommendations.\nThis means that only 41.11% of users received all 10 of their recommendations using collaborative filtering\n"
],
[
"# Some test here might be nice\nassert len(users_with_all_recs) == 22187\nprint(\"That's right there were still another 31781 users who needed recommendations when we only used collaborative filtering!\")",
"That's right there were still another 31781 users who needed recommendations when we only used collaborative filtering!\n"
]
],
[
[
"### Content Based Recommendations\n\nYou will be doing a bit of a mix of content and collaborative filtering to make recommendations for the users this time. This will allow you to obtain recommendations in many cases where we didn't make recommendations earlier. \n\n`2.` Before finding recommendations, rank the user's ratings from highest ratings to lowest ratings. You will move through the movies in this order looking for other similar movies.",
"_____no_output_____"
]
],
[
[
"# create a dataframe similar to reviews, but ranked by rating for each user\nranked_reviews = reviews.sort_values(by=['user_id', 'rating'], ascending=False)",
"_____no_output_____"
]
],
[
[
"### Similarities\n\nIn the collaborative filtering sections, you became quite familiar with different methods of determining the similarity (or distance) of two users. We can perform similarities based on content in much the same way. \n\nIn many cases, it turns out that one of the fastest ways we can find out how similar items are to one another (when our matrix isn't totally sparse like it was in the earlier section) is by simply using matrix multiplication. If you are not familiar with this, an explanation is available [here by 3blue1brown](https://www.youtube.com/watch?v=LyGKycYT2v0) and another quick explanation is provided [on the post here](https://math.stackexchange.com/questions/689022/how-does-the-dot-product-determine-similarity).\n\nFor us to pull out a matrix that describes the movies in our dataframe in terms of content, we might just use the indicator variables related to **year** and **genre** for our movies. \n\nThen we can obtain a matrix of how similar movies are to one another by taking the dot product of this matrix with itself. Notice in the below that the dot product where our 1 values overlap gives a value of 2 indicating higher similarity. In the second dot product, the 1 values don't match up. This leads to a dot product of 0 indicating lower similarity.\n\n<img src=\"images/dotprod1.png\" alt=\"Dot Product\" height=\"500\" width=\"500\">\n\nWe can perform the dot product on a matrix of movies with content characteristics to provide a movie by movie matrix where each cell is an indication of how similar two movies are to one another. In the below image, you can see that movies 1 and 8 are most similar, movies 2 and 8 are most similar and movies 3 and 9 are most similar for this subset of the data. The diagonal elements of the matrix will contain the similarity of a movie with itself, which will be the largest possible similarity (which will also be the number of 1's in the movie row within the orginal movie content matrix.\n\n<img src=\"images/moviemat.png\" alt=\"Dot Product\" height=\"500\" width=\"500\">\n\n\n`3.` Create a numpy array that is a matrix of indicator variables related to year (by century) and movie genres by movie. Perform the dot prodoct of this matrix with itself (transposed) to obtain a similarity matrix of each movie with every other movie. The final matrix should be 31245 x 31245.",
"_____no_output_____"
]
],
[
[
"# Subset so movie_content is only using the dummy variables for each genre and the 3 century based year dummy columns\nmovie_content = np.array(movies.iloc[:,4:])\n\n# Take the dot product to obtain a movie x movie matrix of similarities\ndot_prod_movies = movie_content.dot(np.transpose(movie_content))",
"_____no_output_____"
],
[
"# create checks for the dot product matrix\nassert dot_prod_movies.shape[0] == 31245\nassert dot_prod_movies.shape[1] == 31245\nassert dot_prod_movies[0, 0] == np.max(dot_prod_movies[0])\nprint(\"Looks like you passed all of the tests. Though they weren't very robust - if you want to write some of your own, I won't complain!\")",
"Looks like you passed all of the tests. Though they weren't very robust - if you want to write some of your own, I won't complain!\n"
]
],
[
[
"### For Each User...\n\n\nNow that you have a matrix where each user has their ratings ordered. You also have a second matrix where movies are each axis, and the matrix entries are larger where the two movies are more similar and smaller where the two movies are dissimilar. This matrix is a measure of content similarity. Therefore, it is time to get to the fun part.\n\nFor each user, we will perform the following:\n\n i. For each movie, find the movies that are most similar that the user hasn't seen.\n\n ii. Continue through the available, rated movies until 10 recommendations or until there are no additional movies.\n\nAs a final note, you may need to adjust the criteria for 'most similar' to obtain 10 recommendations. As a first pass, I used only movies with the highest possible similarity to one another as similar enough to add as a recommendation.\n\n`3.` In the below cell, complete each of the functions needed for making content based recommendations.",
"_____no_output_____"
]
],
[
[
"def find_similar_movies(movie_id):\n '''\n INPUT\n movie_id - a movie_id \n OUTPUT\n similar_movies - an array of the most similar movies by title\n '''\n # find the row of each movie id\n movie_idx = np.where(movies['movie_id'] == movie_id)[0][0]\n \n # find the most similar movie indices - to start I said they need to be the same for all content\n similar_idxs = np.where(dot_prod_movies[movie_idx] == np.max(dot_prod_movies[movie_idx]))[0]\n \n # pull the movie titles based on the indices\n similar_movies = np.array(movies.iloc[similar_idxs, ]['movie'])\n \n return similar_movies\n \n \ndef get_movie_names(movie_ids):\n '''\n INPUT\n movie_ids - a list of movie_ids\n OUTPUT\n movies - a list of movie names associated with the movie_ids\n \n '''\n movie_lst = list(movies[movies['movie_id'].isin(movie_ids)]['movie'])\n \n return movie_lst\n\ndef make_recs():\n '''\n INPUT\n None\n OUTPUT\n recs - a dictionary with keys of the user and values of the recommendations\n '''\n # Create dictionary to return with users and ratings\n recs = defaultdict(set)\n # How many users for progress bar\n n_users = len(users)\n\n \n # Create the progressbar\n cnter = 0\n bar = progressbar.ProgressBar(maxval=n_users+1, widgets=[progressbar.Bar('=', '[', ']'), ' ', progressbar.Percentage()])\n bar.start()\n \n # For each user\n for user in users:\n \n # Update the progress bar\n cnter+=1 \n bar.update(cnter)\n\n # Pull only the reviews the user has seen\n reviews_temp = ranked_reviews[ranked_reviews['user_id'] == user]\n movies_temp = np.array(reviews_temp['movie_id'])\n movie_names = np.array(get_movie_names(movies_temp))\n\n # Look at each of the movies (highest ranked first), \n # pull the movies the user hasn't seen that are most similar\n # These will be the recommendations - continue until 10 recs \n # or you have depleted the movie list for the user\n for movie in movies_temp:\n rec_movies = find_similar_movies(movie)\n temp_recs = np.setdiff1d(rec_movies, movie_names)\n recs[user].update(temp_recs)\n\n # If there are more than \n if len(recs[user]) > 9:\n break\n\n bar.finish()\n \n return recs",
"_____no_output_____"
],
[
"recs = make_recs()",
"[========================================================================] 100%\n"
]
],
[
[
"### How Did We Do?\n\nNow that you have made the recommendations, how did we do in providing everyone with a set of recommendations?\n\n`4.` Use the cells below to see how many individuals you were able to make recommendations for, as well as explore characteristics about individuals who you were not able to make recommendations for. ",
"_____no_output_____"
]
],
[
[
"# Explore recommendations\nusers_without_all_recs = []\nusers_with_all_recs = []\nno_recs = []\nfor user, movie_recs in recs.items():\n if len(movie_recs) < 10:\n users_without_all_recs.append(user)\n if len(movie_recs) > 9:\n users_with_all_recs.append(user)\n if len(movie_recs) == 0:\n no_recs.append(user)",
"_____no_output_____"
],
[
"# Some characteristics of my content based recommendations\nprint(\"There were {} users without all 10 recommendations we would have liked to have.\".format(len(users_without_all_recs)))\nprint(\"There were {} users with all 10 recommendations we would like them to have.\".format(len(users_with_all_recs)))\nprint(\"There were {} users with no recommendations at all!\".format(len(no_recs)))",
"There were 2179 users without all 10 recommendations we would have liked to have.\nThere were 51789 users with all 10 recommendations we would like them to have.\nThere were 174 users with no recommendations at all!\n"
],
[
"# Closer look at individual user characteristics\nuser_items = reviews[['user_id', 'movie_id', 'rating']]\nuser_by_movie = user_items.groupby(['user_id', 'movie_id'])['rating'].max().unstack()\n\ndef movies_watched(user_id):\n '''\n INPUT:\n user_id - the user_id of an individual as int\n OUTPUT:\n movies - an array of movies the user has watched\n '''\n movies = user_by_movie.loc[user_id][user_by_movie.loc[user_id].isnull() == False].index.values\n\n return movies\n\n\nmovies_watched(189)",
"_____no_output_____"
],
[
"cnter = 0\nprint(\"Some of the movie lists for users without any recommendations include:\")\nfor user_id in no_recs:\n print(user_id)\n print(get_movie_names(movies_watched(user_id)))\n cnter+=1\n if cnter > 10:\n break",
"Some of the movie lists for users without any recommendations include:\n189\n['El laberinto del fauno (2006)']\n797\n['The 414s (2015)']\n1603\n['Beauty and the Beast (2017)']\n2056\n['Brimstone (2016)']\n2438\n['Baby Driver (2017)']\n3322\n['Rosenberg (2013)']\n3925\n['El laberinto del fauno (2006)']\n4325\n['Beauty and the Beast (2017)']\n4773\n['The Frozen Ground (2013)']\n4869\n['Beauty and the Beast (2017)']\n4878\n['American Made (2017)']\n"
]
],
[
[
"### Now What? \n\nWell, if you were really strict with your criteria for how similar two movies (like I was initially), then you still have some users that don't have all 10 recommendations (and a small group of users who have no recommendations at all). \n\nAs stated earlier, recommendation engines are a bit of an **art** and a **science**. There are a number of things we still could look into - how do our collaborative filtering and content based recommendations compare to one another? How could we incorporate user input along with collaborative filtering and/or content based recommendations to improve any of our recommendations? How can we truly gain recommendations for every user?\n\n`5.` In this last step feel free to explore any last ideas you have with the recommendation techniques we have looked at so far. You might choose to make the final needed recommendations using the first technique with just top ranked movies. You might also loosen up the strictness in the similarity needed between movies. Be creative and share your insights with your classmates!",
"_____no_output_____"
]
],
[
[
"# Cells for exploring",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
e792e7ed56620f2757ffebd30e5229f858cec480 | 270,677 | ipynb | Jupyter Notebook | Course2/Week 1/Initialization.ipynb | AdityaSidharta/courseradeeplearning | f8a735b991cfaa0355e69a42f446dbe2cc5c2605 | [
"MIT"
] | 1 | 2017-09-19T19:21:23.000Z | 2017-09-19T19:21:23.000Z | Course2/Week 1/Initialization.ipynb | AdityaSidharta/courseradeeplearning | f8a735b991cfaa0355e69a42f446dbe2cc5c2605 | [
"MIT"
] | null | null | null | Course2/Week 1/Initialization.ipynb | AdityaSidharta/courseradeeplearning | f8a735b991cfaa0355e69a42f446dbe2cc5c2605 | [
"MIT"
] | null | null | null | 268.52877 | 57,656 | 0.90151 | [
[
[
"# Initialization\n\nWelcome to the first assignment of \"Improving Deep Neural Networks\". \n\nTraining your neural network requires specifying an initial value of the weights. A well chosen initialization method will help learning. \n\nIf you completed the previous course of this specialization, you probably followed our instructions for weight initialization, and it has worked out so far. But how do you choose the initialization for a new neural network? In this notebook, you will see how different initializations lead to different results. \n\nA well chosen initialization can:\n- Speed up the convergence of gradient descent\n- Increase the odds of gradient descent converging to a lower training (and generalization) error \n\nTo get started, run the following cell to load the packages and the planar dataset you will try to classify.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport sklearn\nimport sklearn.datasets\nfrom init_utils import sigmoid, relu, compute_loss, forward_propagation, backward_propagation\nfrom init_utils import update_parameters, predict, load_dataset, plot_decision_boundary, predict_dec\n\n%matplotlib inline\nplt.rcParams['figure.figsize'] = (7.0, 4.0) # set default size of plots\nplt.rcParams['image.interpolation'] = 'nearest'\nplt.rcParams['image.cmap'] = 'gray'\n\n# load image dataset: blue/red dots in circles\ntrain_X, train_Y, test_X, test_Y = load_dataset()",
"_____no_output_____"
]
],
[
[
"You would like a classifier to separate the blue dots from the red dots.",
"_____no_output_____"
],
[
"## 1 - Neural Network model ",
"_____no_output_____"
],
[
"You will use a 3-layer neural network (already implemented for you). Here are the initialization methods you will experiment with: \n- *Zeros initialization* -- setting `initialization = \"zeros\"` in the input argument.\n- *Random initialization* -- setting `initialization = \"random\"` in the input argument. This initializes the weights to large random values. \n- *He initialization* -- setting `initialization = \"he\"` in the input argument. This initializes the weights to random values scaled according to a paper by He et al., 2015. \n\n**Instructions**: Please quickly read over the code below, and run it. In the next part you will implement the three initialization methods that this `model()` calls.",
"_____no_output_____"
]
],
[
[
"def model(X, Y, learning_rate = 0.01, num_iterations = 15000, print_cost = True, initialization = \"he\"):\n \"\"\"\n Implements a three-layer neural network: LINEAR->RELU->LINEAR->RELU->LINEAR->SIGMOID.\n \n Arguments:\n X -- input data, of shape (2, number of examples)\n Y -- true \"label\" vector (containing 0 for red dots; 1 for blue dots), of shape (1, number of examples)\n learning_rate -- learning rate for gradient descent \n num_iterations -- number of iterations to run gradient descent\n print_cost -- if True, print the cost every 1000 iterations\n initialization -- flag to choose which initialization to use (\"zeros\",\"random\" or \"he\")\n \n Returns:\n parameters -- parameters learnt by the model\n \"\"\"\n \n grads = {}\n costs = [] # to keep track of the loss\n m = X.shape[1] # number of examples\n layers_dims = [X.shape[0], 10, 5, 1]\n \n # Initialize parameters dictionary.\n if initialization == \"zeros\":\n parameters = initialize_parameters_zeros(layers_dims)\n elif initialization == \"random\":\n parameters = initialize_parameters_random(layers_dims)\n elif initialization == \"he\":\n parameters = initialize_parameters_he(layers_dims)\n\n # Loop (gradient descent)\n\n for i in range(0, num_iterations):\n\n # Forward propagation: LINEAR -> RELU -> LINEAR -> RELU -> LINEAR -> SIGMOID.\n a3, cache = forward_propagation(X, parameters)\n \n # Loss\n cost = compute_loss(a3, Y)\n\n # Backward propagation.\n grads = backward_propagation(X, Y, cache)\n \n # Update parameters.\n parameters = update_parameters(parameters, grads, learning_rate)\n \n # Print the loss every 1000 iterations\n if print_cost and i % 1000 == 0:\n print(\"Cost after iteration {}: {}\".format(i, cost))\n costs.append(cost)\n \n # plot the loss\n plt.plot(costs)\n plt.ylabel('cost')\n plt.xlabel('iterations (per hundreds)')\n plt.title(\"Learning rate =\" + str(learning_rate))\n plt.show()\n \n return parameters",
"_____no_output_____"
]
],
[
[
"## 2 - Zero initialization\n\nThere are two types of parameters to initialize in a neural network:\n- the weight matrices $(W^{[1]}, W^{[2]}, W^{[3]}, ..., W^{[L-1]}, W^{[L]})$\n- the bias vectors $(b^{[1]}, b^{[2]}, b^{[3]}, ..., b^{[L-1]}, b^{[L]})$\n\n**Exercise**: Implement the following function to initialize all parameters to zeros. You'll see later that this does not work well since it fails to \"break symmetry\", but lets try it anyway and see what happens. Use np.zeros((..,..)) with the correct shapes.",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: initialize_parameters_zeros \n\ndef initialize_parameters_zeros(layers_dims):\n \"\"\"\n Arguments:\n layer_dims -- python array (list) containing the size of each layer.\n \n Returns:\n parameters -- python dictionary containing your parameters \"W1\", \"b1\", ..., \"WL\", \"bL\":\n W1 -- weight matrix of shape (layers_dims[1], layers_dims[0])\n b1 -- bias vector of shape (layers_dims[1], 1)\n ...\n WL -- weight matrix of shape (layers_dims[L], layers_dims[L-1])\n bL -- bias vector of shape (layers_dims[L], 1)\n \"\"\"\n \n parameters = {}\n L = len(layers_dims) # number of layers in the network\n \n for l in range(1, L):\n ### START CODE HERE ### (≈ 2 lines of code)\n parameters['W' + str(l)] = np.zeros((layers_dims[l], layers_dims[l-1]))\n parameters['b' + str(l)] = np.zeros((layers_dims[l], 1))\n ### END CODE HERE ###\n return parameters",
"_____no_output_____"
],
[
"parameters = initialize_parameters_zeros([3,2,1])\nprint(\"W1 = \" + str(parameters[\"W1\"]))\nprint(\"b1 = \" + str(parameters[\"b1\"]))\nprint(\"W2 = \" + str(parameters[\"W2\"]))\nprint(\"b2 = \" + str(parameters[\"b2\"]))",
"W1 = [[ 0. 0. 0.]\n [ 0. 0. 0.]]\nb1 = [[ 0.]\n [ 0.]]\nW2 = [[ 0. 0.]]\nb2 = [[ 0.]]\n"
]
],
[
[
"**Expected Output**:\n\n<table> \n <tr>\n <td>\n **W1**\n </td>\n <td>\n [[ 0. 0. 0.]\n [ 0. 0. 0.]]\n </td>\n </tr>\n <tr>\n <td>\n **b1**\n </td>\n <td>\n [[ 0.]\n [ 0.]]\n </td>\n </tr>\n <tr>\n <td>\n **W2**\n </td>\n <td>\n [[ 0. 0.]]\n </td>\n </tr>\n <tr>\n <td>\n **b2**\n </td>\n <td>\n [[ 0.]]\n </td>\n </tr>\n\n</table> ",
"_____no_output_____"
],
[
"Run the following code to train your model on 15,000 iterations using zeros initialization.",
"_____no_output_____"
]
],
[
[
"parameters = model(train_X, train_Y, initialization = \"zeros\")\nprint (\"On the train set:\")\npredictions_train = predict(train_X, train_Y, parameters)\nprint (\"On the test set:\")\npredictions_test = predict(test_X, test_Y, parameters)",
"Cost after iteration 0: 0.6931471805599453\nCost after iteration 1000: 0.6931471805599453\nCost after iteration 2000: 0.6931471805599453\nCost after iteration 3000: 0.6931471805599453\nCost after iteration 4000: 0.6931471805599453\nCost after iteration 5000: 0.6931471805599453\nCost after iteration 6000: 0.6931471805599453\nCost after iteration 7000: 0.6931471805599453\nCost after iteration 8000: 0.6931471805599453\nCost after iteration 9000: 0.6931471805599453\nCost after iteration 10000: 0.6931471805599455\nCost after iteration 11000: 0.6931471805599453\nCost after iteration 12000: 0.6931471805599453\nCost after iteration 13000: 0.6931471805599453\nCost after iteration 14000: 0.6931471805599453\n"
]
],
[
[
"The performance is really bad, and the cost does not really decrease, and the algorithm performs no better than random guessing. Why? Lets look at the details of the predictions and the decision boundary:",
"_____no_output_____"
]
],
[
[
"print (\"predictions_train = \" + str(predictions_train))\nprint (\"predictions_test = \" + str(predictions_test))",
"predictions_train = [[0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0]]\npredictions_test = [[0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0]]\n"
],
[
"plt.title(\"Model with Zeros initialization\")\naxes = plt.gca()\naxes.set_xlim([-1.5,1.5])\naxes.set_ylim([-1.5,1.5])\nplot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y)",
"_____no_output_____"
]
],
[
[
"The model is predicting 0 for every example. \n\nIn general, initializing all the weights to zero results in the network failing to break symmetry. This means that every neuron in each layer will learn the same thing, and you might as well be training a neural network with $n^{[l]}=1$ for every layer, and the network is no more powerful than a linear classifier such as logistic regression. ",
"_____no_output_____"
],
[
"<font color='blue'>\n**What you should remember**:\n- The weights $W^{[l]}$ should be initialized randomly to break symmetry. \n- It is however okay to initialize the biases $b^{[l]}$ to zeros. Symmetry is still broken so long as $W^{[l]}$ is initialized randomly. \n",
"_____no_output_____"
],
[
"## 3 - Random initialization\n\nTo break symmetry, lets intialize the weights randomly. Following random initialization, each neuron can then proceed to learn a different function of its inputs. In this exercise, you will see what happens if the weights are intialized randomly, but to very large values. \n\n**Exercise**: Implement the following function to initialize your weights to large random values (scaled by \\*10) and your biases to zeros. Use `np.random.randn(..,..) * 10` for weights and `np.zeros((.., ..))` for biases. We are using a fixed `np.random.seed(..)` to make sure your \"random\" weights match ours, so don't worry if running several times your code gives you always the same initial values for the parameters. ",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: initialize_parameters_random\n\ndef initialize_parameters_random(layers_dims):\n \"\"\"\n Arguments:\n layer_dims -- python array (list) containing the size of each layer.\n \n Returns:\n parameters -- python dictionary containing your parameters \"W1\", \"b1\", ..., \"WL\", \"bL\":\n W1 -- weight matrix of shape (layers_dims[1], layers_dims[0])\n b1 -- bias vector of shape (layers_dims[1], 1)\n ...\n WL -- weight matrix of shape (layers_dims[L], layers_dims[L-1])\n bL -- bias vector of shape (layers_dims[L], 1)\n \"\"\"\n \n np.random.seed(3) # This seed makes sure your \"random\" numbers will be the as ours\n parameters = {}\n L = len(layers_dims) # integer representing the number of layers\n \n for l in range(1, L):\n ### START CODE HERE ### (≈ 2 lines of code)\n parameters['W' + str(l)] = np.random.randn(layers_dims[l], layers_dims[l-1]) * 10\n parameters['b' + str(l)] = np.zeros((layers_dims[l], 1))\n ### END CODE HERE ###\n\n return parameters",
"_____no_output_____"
],
[
"parameters = initialize_parameters_random([3, 2, 1])\nprint(\"W1 = \" + str(parameters[\"W1\"]))\nprint(\"b1 = \" + str(parameters[\"b1\"]))\nprint(\"W2 = \" + str(parameters[\"W2\"]))\nprint(\"b2 = \" + str(parameters[\"b2\"]))",
"W1 = [[ 17.88628473 4.36509851 0.96497468]\n [-18.63492703 -2.77388203 -3.54758979]]\nb1 = [[ 0.]\n [ 0.]]\nW2 = [[-0.82741481 -6.27000677]]\nb2 = [[ 0.]]\n"
]
],
[
[
"**Expected Output**:\n\n<table> \n <tr>\n <td>\n **W1**\n </td>\n <td>\n [[ 17.88628473 4.36509851 0.96497468]\n [-18.63492703 -2.77388203 -3.54758979]]\n </td>\n </tr>\n <tr>\n <td>\n **b1**\n </td>\n <td>\n [[ 0.]\n [ 0.]]\n </td>\n </tr>\n <tr>\n <td>\n **W2**\n </td>\n <td>\n [[-0.82741481 -6.27000677]]\n </td>\n </tr>\n <tr>\n <td>\n **b2**\n </td>\n <td>\n [[ 0.]]\n </td>\n </tr>\n\n</table> ",
"_____no_output_____"
],
[
"Run the following code to train your model on 15,000 iterations using random initialization.",
"_____no_output_____"
]
],
[
[
"parameters = model(train_X, train_Y, initialization = \"random\")\nprint (\"On the train set:\")\npredictions_train = predict(train_X, train_Y, parameters)\nprint (\"On the test set:\")\npredictions_test = predict(test_X, test_Y, parameters)",
"/home/jovyan/work/week5/Initialization/init_utils.py:145: RuntimeWarning: divide by zero encountered in log\n logprobs = np.multiply(-np.log(a3),Y) + np.multiply(-np.log(1 - a3), 1 - Y)\n/home/jovyan/work/week5/Initialization/init_utils.py:145: RuntimeWarning: invalid value encountered in multiply\n logprobs = np.multiply(-np.log(a3),Y) + np.multiply(-np.log(1 - a3), 1 - Y)\n"
]
],
[
[
"If you see \"inf\" as the cost after the iteration 0, this is because of numerical roundoff; a more numerically sophisticated implementation would fix this. But this isn't worth worrying about for our purposes. \n\nAnyway, it looks like you have broken symmetry, and this gives better results. than before. The model is no longer outputting all 0s. ",
"_____no_output_____"
]
],
[
[
"print (predictions_train)\nprint (predictions_test)",
"[[1 0 1 1 0 0 1 0 1 1 1 0 1 0 0 0 0 1 1 0 0 0 1 0 0 0 1 0 1 1 0 1 1 0 0 0 0\n 1 0 1 1 1 1 0 0 1 1 1 0 1 0 1 1 1 1 0 0 1 1 1 1 1 0 1 0 1 0 1 0 1 1 0 0 0\n 0 0 1 0 0 0 1 0 1 0 0 1 1 1 1 1 1 0 0 0 1 1 0 1 1 0 1 0 0 1 0 1 1 0 0 0 1\n 1 0 0 1 0 0 1 1 0 1 1 1 0 0 0 0 1 0 1 1 0 1 1 1 1 0 1 1 0 0 0 0 0 0 0 1 0\n 1 0 1 0 1 1 1 0 0 1 1 0 1 0 1 1 0 1 0 1 1 0 1 0 1 1 0 1 0 0 1 0 1 0 0 0 1\n 0 1 1 1 0 0 1 1 0 0 1 0 1 0 0 1 0 1 1 0 1 1 1 1 0 1 0 1 0 0 1 0 1 0 0 0 1\n 1 1 1 0 1 0 0 1 1 0 0 1 0 1 0 0 1 0 0 1 1 0 0 0 0 1 1 0 1 0 1 1 0 1 1 0 1\n 0 0 0 0 0 0 0 0 1 0 0 0 1 1 1 1 0 0 0 0 0 1 1 0 0 1 1 1 1 1 1 1 0 0 0 0 1\n 1 0 1 0]]\n[[1 0 1 0 0 0 0 0 0 0 1 1 1 0 0 0 0 0 1 1 0 0 1 0 0 0 0 1 1 1 1 0 1 0 0 1 0\n 1 1 0 0 1 1 1 1 0 0 0 1 1 0 1 0 1 1 0 1 0 1 0 1 1 1 1 1 1 1 1 1 1 1 0 1 1\n 1 1 0 0 1 1 0 1 0 0 0 0 1 0 1 1 0 0 0 1 1 0 0 0 0 0]]\n"
],
[
"plt.title(\"Model with large random initialization\")\naxes = plt.gca()\naxes.set_xlim([-1.5,1.5])\naxes.set_ylim([-1.5,1.5])\nplot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y)",
"_____no_output_____"
]
],
[
[
"**Observations**:\n- The cost starts very high. This is because with large random-valued weights, the last activation (sigmoid) outputs results that are very close to 0 or 1 for some examples, and when it gets that example wrong it incurs a very high loss for that example. Indeed, when $\\log(a^{[3]}) = \\log(0)$, the loss goes to infinity.\n- Poor initialization can lead to vanishing/exploding gradients, which also slows down the optimization algorithm. \n- If you train this network longer you will see better results, but initializing with overly large random numbers slows down the optimization.\n\n<font color='blue'>\n**In summary**:\n- Initializing weights to very large random values does not work well. \n- Hopefully intializing with small random values does better. The important question is: how small should be these random values be? Lets find out in the next part! ",
"_____no_output_____"
],
[
"## 4 - He initialization\n\nFinally, try \"He Initialization\"; this is named for the first author of He et al., 2015. (If you have heard of \"Xavier initialization\", this is similar except Xavier initialization uses a scaling factor for the weights $W^{[l]}$ of `sqrt(1./layers_dims[l-1])` where He initialization would use `sqrt(2./layers_dims[l-1])`.)\n\n**Exercise**: Implement the following function to initialize your parameters with He initialization.\n\n**Hint**: This function is similar to the previous `initialize_parameters_random(...)`. The only difference is that instead of multiplying `np.random.randn(..,..)` by 10, you will multiply it by $\\sqrt{\\frac{2}{\\text{dimension of the previous layer}}}$, which is what He initialization recommends for layers with a ReLU activation. ",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: initialize_parameters_he\n\ndef initialize_parameters_he(layers_dims):\n \"\"\"\n Arguments:\n layer_dims -- python array (list) containing the size of each layer.\n \n Returns:\n parameters -- python dictionary containing your parameters \"W1\", \"b1\", ..., \"WL\", \"bL\":\n W1 -- weight matrix of shape (layers_dims[1], layers_dims[0])\n b1 -- bias vector of shape (layers_dims[1], 1)\n ...\n WL -- weight matrix of shape (layers_dims[L], layers_dims[L-1])\n bL -- bias vector of shape (layers_dims[L], 1)\n \"\"\"\n \n np.random.seed(3)\n parameters = {}\n L = len(layers_dims) - 1 # integer representing the number of layers\n \n for l in range(1, L + 1):\n ### START CODE HERE ### (≈ 2 lines of code)\n parameters['W' + str(l)] = np.random.randn(layers_dims[l], layers_dims[l-1]) * np.sqrt(2/layers_dims[l-1])\n parameters['b' + str(l)] = np.zeros((layers_dims[l], 1))\n ### END CODE HERE ###\n \n return parameters",
"_____no_output_____"
],
[
"parameters = initialize_parameters_he([2, 4, 1])\nprint(\"W1 = \" + str(parameters[\"W1\"]))\nprint(\"b1 = \" + str(parameters[\"b1\"]))\nprint(\"W2 = \" + str(parameters[\"W2\"]))\nprint(\"b2 = \" + str(parameters[\"b2\"]))",
"W1 = [[ 1.78862847 0.43650985]\n [ 0.09649747 -1.8634927 ]\n [-0.2773882 -0.35475898]\n [-0.08274148 -0.62700068]]\nb1 = [[ 0.]\n [ 0.]\n [ 0.]\n [ 0.]]\nW2 = [[-0.03098412 -0.33744411 -0.92904268 0.62552248]]\nb2 = [[ 0.]]\n"
]
],
[
[
"**Expected Output**:\n\n<table> \n <tr>\n <td>\n **W1**\n </td>\n <td>\n [[ 1.78862847 0.43650985]\n [ 0.09649747 -1.8634927 ]\n [-0.2773882 -0.35475898]\n [-0.08274148 -0.62700068]]\n </td>\n </tr>\n <tr>\n <td>\n **b1**\n </td>\n <td>\n [[ 0.]\n [ 0.]\n [ 0.]\n [ 0.]]\n </td>\n </tr>\n <tr>\n <td>\n **W2**\n </td>\n <td>\n [[-0.03098412 -0.33744411 -0.92904268 0.62552248]]\n </td>\n </tr>\n <tr>\n <td>\n **b2**\n </td>\n <td>\n [[ 0.]]\n </td>\n </tr>\n\n</table> ",
"_____no_output_____"
],
[
"Run the following code to train your model on 15,000 iterations using He initialization.",
"_____no_output_____"
]
],
[
[
"parameters = model(train_X, train_Y, initialization = \"he\")\nprint (\"On the train set:\")\npredictions_train = predict(train_X, train_Y, parameters)\nprint (\"On the test set:\")\npredictions_test = predict(test_X, test_Y, parameters)",
"Cost after iteration 0: 0.8830537463419761\nCost after iteration 1000: 0.6879825919728063\nCost after iteration 2000: 0.6751286264523371\nCost after iteration 3000: 0.6526117768893807\nCost after iteration 4000: 0.6082958970572938\nCost after iteration 5000: 0.5304944491717495\nCost after iteration 6000: 0.4138645817071794\nCost after iteration 7000: 0.3117803464844441\nCost after iteration 8000: 0.23696215330322562\nCost after iteration 9000: 0.18597287209206836\nCost after iteration 10000: 0.1501555628037182\nCost after iteration 11000: 0.12325079292273548\nCost after iteration 12000: 0.09917746546525937\nCost after iteration 13000: 0.0845705595402428\nCost after iteration 14000: 0.07357895962677366\n"
],
[
"plt.title(\"Model with He initialization\")\naxes = plt.gca()\naxes.set_xlim([-1.5,1.5])\naxes.set_ylim([-1.5,1.5])\nplot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y)",
"_____no_output_____"
]
],
[
[
"**Observations**:\n- The model with He initialization separates the blue and the red dots very well in a small number of iterations.\n",
"_____no_output_____"
],
[
"## 5 - Conclusions",
"_____no_output_____"
],
[
"You have seen three different types of initializations. For the same number of iterations and same hyperparameters the comparison is:\n\n<table> \n <tr>\n <td>\n **Model**\n </td>\n <td>\n **Train accuracy**\n </td>\n <td>\n **Problem/Comment**\n </td>\n\n </tr>\n <td>\n 3-layer NN with zeros initialization\n </td>\n <td>\n 50%\n </td>\n <td>\n fails to break symmetry\n </td>\n <tr>\n <td>\n 3-layer NN with large random initialization\n </td>\n <td>\n 83%\n </td>\n <td>\n too large weights \n </td>\n </tr>\n <tr>\n <td>\n 3-layer NN with He initialization\n </td>\n <td>\n 99%\n </td>\n <td>\n recommended method\n </td>\n </tr>\n</table> ",
"_____no_output_____"
],
[
"<font color='blue'>\n**What you should remember from this notebook**:\n- Different initializations lead to different results\n- Random initialization is used to break symmetry and make sure different hidden units can learn different things\n- Don't intialize to values that are too large\n- He initialization works well for networks with ReLU activations. ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
e79311954a1b15454967ae2dee34d5bfae2e24b0 | 1,800 | ipynb | Jupyter Notebook | notebooks/december15.ipynb | jebreimo/Advent2020 | 2d152152b25fe17bf4d1c4d4a26b933c525ca946 | [
"BSD-2-Clause"
] | null | null | null | notebooks/december15.ipynb | jebreimo/Advent2020 | 2d152152b25fe17bf4d1c4d4a26b933c525ca946 | [
"BSD-2-Clause"
] | null | null | null | notebooks/december15.ipynb | jebreimo/Advent2020 | 2d152152b25fe17bf4d1c4d4a26b933c525ca946 | [
"BSD-2-Clause"
] | null | null | null | 16.82243 | 58 | 0.453333 | [
[
[
"INPUT = [17,1,3,16,19,0]",
"_____no_output_____"
]
],
[
[
"# Part 1",
"_____no_output_____"
]
],
[
[
"N = 2020",
"_____no_output_____"
],
[
"values = {k: v for v, k in enumerate(INPUT[:-1])}\nprev = INPUT[-1]\nfor i in range(len(INPUT) - 1, N - 1):\n pos = values.get(prev)\n values[prev] = i\n if pos is None:\n prev = 0\n else:\n prev = i - pos\nprint(prev)",
"694\n"
]
],
[
[
"# Part 2",
"_____no_output_____"
]
],
[
[
"N = 30000000",
"_____no_output_____"
]
],
[
[
"Repeat loop in Part 1",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e79312586574d257b580f6b974524e4184f3b11f | 101,571 | ipynb | Jupyter Notebook | sagemaker/SageMaker_Project.ipynb | TonysCousin/Udacity | cb64e3b306a20ca8dc7b2025b4cb63a04bc1b378 | [
"BSD-3-Clause"
] | null | null | null | sagemaker/SageMaker_Project.ipynb | TonysCousin/Udacity | cb64e3b306a20ca8dc7b2025b4cb63a04bc1b378 | [
"BSD-3-Clause"
] | null | null | null | sagemaker/SageMaker_Project.ipynb | TonysCousin/Udacity | cb64e3b306a20ca8dc7b2025b4cb63a04bc1b378 | [
"BSD-3-Clause"
] | null | null | null | 50.658853 | 1,541 | 0.612252 | [
[
[
"# Creating a Sentiment Analysis Web App\n## Using PyTorch and SageMaker\n\n_Deep Learning Nanodegree Program | Deployment_\n\n---\n\nNow that we have a basic understanding of how SageMaker works we will try to use it to construct a complete project from end to end. Our goal will be to have a simple web page which a user can use to enter a movie review. The web page will then send the review off to our deployed model which will predict the sentiment of the entered review.\n\n## Instructions\n\nSome template code has already been provided for you, and you will need to implement additional functionality to successfully complete this notebook. You will not need to modify the included code beyond what is requested. Sections that begin with '**TODO**' in the header indicate that you need to complete or implement some portion within them. Instructions will be provided for each section and the specifics of the implementation are marked in the code block with a `# TODO: ...` comment. Please be sure to read the instructions carefully!\n\nIn addition to implementing code, there will be questions for you to answer which relate to the task and your implementation. Each section where you will answer a question is preceded by a '**Question:**' header. Carefully read each question and provide your answer below the '**Answer:**' header by editing the Markdown cell.\n\n> **Note**: Code and Markdown cells can be executed using the **Shift+Enter** keyboard shortcut. In addition, a cell can be edited by typically clicking it (double-click for Markdown cells) or by pressing **Enter** while it is highlighted.\n\n## General Outline\n\nRecall the general outline for SageMaker projects using a notebook instance.\n\n1. Download or otherwise retrieve the data.\n2. Process / Prepare the data.\n3. Upload the processed data to S3.\n4. Train a chosen model.\n5. Test the trained model (typically using a batch transform job).\n6. Deploy the trained model.\n7. Use the deployed model.\n\nFor this project, you will be following the steps in the general outline with some modifications. \n\nFirst, you will not be testing the model in its own step. You will still be testing the model, however, you will do it by deploying your model and then using the deployed model by sending the test data to it. One of the reasons for doing this is so that you can make sure that your deployed model is working correctly before moving forward.\n\nIn addition, you will deploy and use your trained model a second time. In the second iteration you will customize the way that your trained model is deployed by including some of your own code. In addition, your newly deployed model will be used in the sentiment analysis web app.",
"_____no_output_____"
],
[
"## Step 1: Downloading the data\n\nAs in the XGBoost in SageMaker notebook, we will be using the [IMDb dataset](http://ai.stanford.edu/~amaas/data/sentiment/)\n\n> Maas, Andrew L., et al. [Learning Word Vectors for Sentiment Analysis](http://ai.stanford.edu/~amaas/data/sentiment/). In _Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies_. Association for Computational Linguistics, 2011.",
"_____no_output_____"
]
],
[
[
"%mkdir ../data\n!wget -O ../data/aclImdb_v1.tar.gz http://ai.stanford.edu/~amaas/data/sentiment/aclImdb_v1.tar.gz\n!tar -zxf ../data/aclImdb_v1.tar.gz -C ../data",
"mkdir: cannot create directory ‘../data’: File exists\n--2020-07-07 01:47:08-- http://ai.stanford.edu/~amaas/data/sentiment/aclImdb_v1.tar.gz\nResolving ai.stanford.edu (ai.stanford.edu)... 171.64.68.10\nConnecting to ai.stanford.edu (ai.stanford.edu)|171.64.68.10|:80... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 84125825 (80M) [application/x-gzip]\nSaving to: ‘../data/aclImdb_v1.tar.gz’\n\n../data/aclImdb_v1. 100%[===================>] 80.23M 22.9MB/s in 4.1s \n\n2020-07-07 01:47:12 (19.7 MB/s) - ‘../data/aclImdb_v1.tar.gz’ saved [84125825/84125825]\n\n"
],
[
"# collect all the imports here so I can run one cell to define them all when I need to restart the notebook\nimport os\nimport glob\nimport numpy as np\nfrom collections import Counter\nfrom sklearn.utils import shuffle\nimport nltk\nfrom nltk.corpus import stopwords\nfrom nltk.stem.porter import *\nimport re\nfrom bs4 import BeautifulSoup\nimport pickle\nimport pandas as pd\nimport sagemaker\nimport torch\nimport torch.utils.data\nimport torch.nn as nn\n\n#define some constants used throughout\ndata_dir = '../data/pytorch' # The folder we will use for storing data\n",
"_____no_output_____"
]
],
[
[
"## Step 2: Preparing and Processing the data\n\nAlso, as in the XGBoost notebook, we will be doing some initial data processing. The first few steps are the same as in the XGBoost example. To begin with, we will read in each of the reviews and combine them into a single input structure. Then, we will split the dataset into a training set and a testing set.",
"_____no_output_____"
]
],
[
[
"import os\nimport glob\n\ndef read_imdb_data(data_dir='../data/aclImdb'):\n data = {}\n labels = {}\n \n for data_type in ['train', 'test']:\n data[data_type] = {}\n labels[data_type] = {}\n \n for sentiment in ['pos', 'neg']:\n data[data_type][sentiment] = []\n labels[data_type][sentiment] = []\n \n path = os.path.join(data_dir, data_type, sentiment, '*.txt')\n files = glob.glob(path)\n \n for f in files:\n with open(f) as review:\n data[data_type][sentiment].append(review.read())\n # Here we represent a positive review by '1' and a negative review by '0'\n labels[data_type][sentiment].append(1 if sentiment == 'pos' else 0)\n \n assert len(data[data_type][sentiment]) == len(labels[data_type][sentiment]), \\\n \"{}/{} data size does not match labels size\".format(data_type, sentiment)\n \n \n return data, labels",
"_____no_output_____"
],
[
"data, labels = read_imdb_data()\nprint(\"IMDB reviews: train = {} pos / {} neg, test = {} pos / {} neg\".format(\n len(data['train']['pos']), len(data['train']['neg']),\n len(data['test']['pos']), len(data['test']['neg'])))",
"IMDB reviews: train = 12500 pos / 12500 neg, test = 12500 pos / 12500 neg\n"
]
],
[
[
"Now that we've read the raw training and testing data from the downloaded dataset, we will combine the positive and negative reviews and shuffle the resulting records.",
"_____no_output_____"
]
],
[
[
"from sklearn.utils import shuffle\n\ndef prepare_imdb_data(data, labels):\n \"\"\"Prepare training and test sets from IMDb movie reviews.\"\"\"\n \n #Combine positive and negative reviews and labels\n data_train = data['train']['pos'] + data['train']['neg']\n data_test = data['test']['pos'] + data['test']['neg']\n labels_train = labels['train']['pos'] + labels['train']['neg']\n labels_test = labels['test']['pos'] + labels['test']['neg']\n \n #Shuffle reviews and corresponding labels within training and test sets\n data_train, labels_train = shuffle(data_train, labels_train)\n data_test, labels_test = shuffle(data_test, labels_test)\n \n # Return a unified training data, test data, training labels, test labets\n return data_train, data_test, labels_train, labels_test",
"_____no_output_____"
],
[
"train_X, test_X, train_y, test_y = prepare_imdb_data(data, labels)\nprint(\"IMDb reviews (combined): train = {}, test = {}\".format(len(train_X), len(test_X)))",
"IMDb reviews (combined): train = 25000, test = 25000\n"
]
],
[
[
"Now that we have our training and testing sets unified and prepared, we should do a quick check and see an example of the data our model will be trained on. This is generally a good idea as it allows you to see how each of the further processing steps affects the reviews and it also ensures that the data has been loaded correctly.",
"_____no_output_____"
]
],
[
[
"print(train_X[133])\nprint(train_y[133])",
"It is a superb Swedish film .. it was the first Swedish film I've seen .. it is simple & deep .. what a great combination!.<br /><br />Michael Nyqvist did a great performance as a famous conductor who seeks peace in his hometown.<br /><br />Frida Hallgren was great as his inspirational girlfriend to help him to carry on & never give up.<br /><br />The fight between the conductor and the hypocrite priest who loses his battle with Michael when his wife confronts him And defends Michael's noble cause to help his hometown people finding their own peace in music.<br /><br />The only thing that I didn't like was the ending .. it wasn't that good but it has some deep meaning.\n1\n"
]
],
[
[
"The first step in processing the reviews is to make sure that any html tags that appear should be removed. In addition we wish to tokenize our input, that way words such as *entertained* and *entertaining* are considered the same with regard to sentiment analysis.",
"_____no_output_____"
]
],
[
[
"import nltk\nfrom nltk.corpus import stopwords\nfrom nltk.stem.porter import *\n\nimport re\nfrom bs4 import BeautifulSoup\n\ndef review_to_words(review):\n nltk.download(\"stopwords\", quiet=True)\n stemmer = PorterStemmer()\n \n text = BeautifulSoup(review, \"html.parser\").get_text() # Remove HTML tags\n text = re.sub(r\"[^a-zA-Z0-9]\", \" \", text.lower()) # Convert to lower case\n words = text.split() # Split string into words\n words = [w for w in words if w not in stopwords.words(\"english\")] # Remove stopwords\n words = [PorterStemmer().stem(w) for w in words] # stem\n \n return words",
"_____no_output_____"
]
],
[
[
"The `review_to_words` method defined above uses `BeautifulSoup` to remove any html tags that appear and uses the `nltk` package to tokenize the reviews. As a check to ensure we know how everything is working, try applying `review_to_words` to one of the reviews in the training set.",
"_____no_output_____"
]
],
[
[
"# TODO: Apply review_to_words to a review (train_X[100] or any other review)\nr = review_to_words(train_X[133])\nprint(r)",
"['superb', 'swedish', 'film', 'first', 'swedish', 'film', 'seen', 'simpl', 'deep', 'great', 'combin', 'michael', 'nyqvist', 'great', 'perform', 'famou', 'conductor', 'seek', 'peac', 'hometown', 'frida', 'hallgren', 'great', 'inspir', 'girlfriend', 'help', 'carri', 'never', 'give', 'fight', 'conductor', 'hypocrit', 'priest', 'lose', 'battl', 'michael', 'wife', 'confront', 'defend', 'michael', 'nobl', 'caus', 'help', 'hometown', 'peopl', 'find', 'peac', 'music', 'thing', 'like', 'end', 'good', 'deep', 'mean']\n"
]
],
[
[
"**Question:** Above we mentioned that `review_to_words` method removes html formatting and allows us to tokenize the words found in a review, for example, converting *entertained* and *entertaining* into *entertain* so that they are treated as though they are the same word. What else, if anything, does this method do to the input?",
"_____no_output_____"
],
[
"**Answer:** The method also removes trivial connecting words, such as \"is\", \"the\", \"at\". It also converts everything to lower-case and removes punctuation.",
"_____no_output_____"
],
[
"The method below applies the `review_to_words` method to each of the reviews in the training and testing datasets. In addition it caches the results. This is because performing this processing step can take a long time. This way if you are unable to complete the notebook in the current session, you can come back without needing to process the data a second time.",
"_____no_output_____"
]
],
[
[
"import pickle\n\ncache_dir = os.path.join(\"../cache\", \"sentiment_analysis\") # where to store cache files\nos.makedirs(cache_dir, exist_ok=True) # ensure cache directory exists\n\ndef preprocess_data(data_train, data_test, labels_train, labels_test,\n cache_dir=cache_dir, cache_file=\"preprocessed_data.pkl\"):\n \"\"\"Convert each review to words; read from cache if available.\"\"\"\n\n # If cache_file is not None, try to read from it first\n cache_data = None\n if cache_file is not None:\n try:\n with open(os.path.join(cache_dir, cache_file), \"rb\") as f:\n cache_data = pickle.load(f)\n print(\"Read preprocessed data from cache file:\", cache_file)\n except:\n pass # unable to read from cache, but that's okay\n \n # If cache is missing, then do the heavy lifting\n if cache_data is None:\n # Preprocess training and test data to obtain words for each review\n #words_train = list(map(review_to_words, data_train))\n #words_test = list(map(review_to_words, data_test))\n words_train = [review_to_words(review) for review in data_train]\n words_test = [review_to_words(review) for review in data_test]\n \n # Write to cache file for future runs\n if cache_file is not None:\n cache_data = dict(words_train=words_train, words_test=words_test,\n labels_train=labels_train, labels_test=labels_test)\n with open(os.path.join(cache_dir, cache_file), \"wb\") as f:\n pickle.dump(cache_data, f)\n print(\"Wrote preprocessed data to cache file:\", cache_file)\n else:\n # Unpack data loaded from cache file\n words_train, words_test, labels_train, labels_test = (cache_data['words_train'],\n cache_data['words_test'], cache_data['labels_train'], cache_data['labels_test'])\n \n return words_train, words_test, labels_train, labels_test",
"_____no_output_____"
],
[
"# Preprocess data\ntrain_X, test_X, train_y, test_y = preprocess_data(train_X, test_X, train_y, test_y)",
"Read preprocessed data from cache file: preprocessed_data.pkl\n"
]
],
[
[
"## Transform the data\n\nIn the XGBoost notebook we transformed the data from its word representation to a bag-of-words feature representation. For the model we are going to construct in this notebook we will construct a feature representation which is very similar. To start, we will represent each word as an integer. Of course, some of the words that appear in the reviews occur very infrequently and so likely don't contain much information for the purposes of sentiment analysis. The way we will deal with this problem is that we will fix the size of our working vocabulary and we will only include the words that appear most frequently. We will then combine all of the infrequent words into a single category and, in our case, we will label it as `1`.\n\nSince we will be using a recurrent neural network, it will be convenient if the length of each review is the same. To do this, we will fix a size for our reviews and then pad short reviews with the category 'no word' (which we will label `0`) and truncate long reviews.",
"_____no_output_____"
],
[
"### (TODO) Create a word dictionary\n\nTo begin with, we need to construct a way to map words that appear in the reviews to integers. Here we fix the size of our vocabulary (including the 'no word' and 'infrequent' categories) to be `5000` but you may wish to change this to see how it affects the model.\n\n> **TODO:** Complete the implementation for the `build_dict()` method below. Note that even though the vocab_size is set to `5000`, we only want to construct a mapping for the most frequently appearing `4998` words. This is because we want to reserve the special labels `0` for 'no word' and `1` for 'infrequent word'.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom collections import Counter\n\ndef build_dict(data, vocab_size = 5000):\n \"\"\"Construct and return a dictionary mapping each of the most frequently appearing words to a unique integer.\"\"\"\n \n # TODO: Determine how often each word appears in `data`. Note that `data` is a list of sentences and that a\n # sentence is a list of words.\n \n # A dict storing the words that appear in the reviews along with how often they occur\n word_count = Counter() \n for item in data:\n word_count.update(item)\n print(\"The most common words are: \", word_count.most_common(10))\n \n # TODO: Sort the words found in `data` so that sorted_words[0] is the most frequently appearing word and\n # sorted_words[-1] is the least frequently appearing word.\n \n sorted_words = []\n for w, c in word_count.most_common(len(word_count)):\n sorted_words.append(w)\n \n word_dict = {} # This is what we are building, a dictionary that translates words into integers\n for idx, word in enumerate(sorted_words[:vocab_size - 2]): # The -2 is so that we save room for the 'no word'\n word_dict[word] = idx + 2 # 'infrequent' labels\n \n return word_dict",
"_____no_output_____"
],
[
"word_dict = build_dict(train_X)",
"The most common words are: [('movi', 51695), ('film', 48190), ('one', 27741), ('like', 22799), ('time', 16191), ('good', 15360), ('make', 15207), ('charact', 14178), ('get', 14141), ('see', 14111)]\n"
]
],
[
[
"**Question:** What are the five most frequently appearing (tokenized) words in the training set? Does it makes sense that these words appear frequently in the training set?",
"_____no_output_____"
],
[
"**Answer:** The five most frequently used words are: movi, film, one, like, time. These form a plausible list of the most frequently used words in such a context.",
"_____no_output_____"
]
],
[
[
"# TODO: Use this space to determine the five most frequently appearing words in the training set.\n\n# I did it above with a print statement in the build_dict() function. -jas",
"_____no_output_____"
]
],
[
[
"### Save `word_dict`\n\nLater on when we construct an endpoint which processes a submitted review we will need to make use of the `word_dict` which we have created. As such, we will save it to a file now for future use.",
"_____no_output_____"
]
],
[
[
"data_dir = '../data/pytorch' # The folder we will use for storing data\nif not os.path.exists(data_dir): # Make sure that the folder exists\n os.makedirs(data_dir)",
"_____no_output_____"
],
[
"dict_file = os.path.join(data_dir, 'word_dict.pkl')\nif os.path.exists(dict_file):\n with open(dict_file, \"rb\") as f:\n word_dict = pickle.load(f)\n print(\"Loaded existing word dictionary.\")\nelse:\n with open(os.path.join(data_dir, 'word_dict.pkl'), \"wb\") as f:\n pickle.dump(word_dict, f)\n print(\"Stored word dictionary to pickle file.\")",
"Loaded existing word dictionary.\n"
]
],
[
[
"### Transform the reviews\n\nNow that we have our word dictionary which allows us to transform the words appearing in the reviews into integers, it is time to make use of it and convert our reviews to their integer sequence representation, making sure to pad or truncate to a fixed length, which in our case is `500`.",
"_____no_output_____"
]
],
[
[
"# just playing around...\nprint(\"vocab size = \", len(word_dict))\nprint(\"word_dict entries = \", word_dict[\"one\"])\n",
"vocab size = 4998\nword_dict entries = 4\n"
],
[
"def convert_and_pad(word_dict, sentence, pad=500):\n NOWORD = 0 # We will use 0 to represent the 'no word' category\n INFREQ = 1 # and we use 1 to represent the infrequent words, i.e., words not appearing in word_dict\n \n working_sentence = [NOWORD] * pad\n \n for word_index, word in enumerate(sentence[:pad]):\n if word in word_dict:\n working_sentence[word_index] = word_dict[word]\n else:\n working_sentence[word_index] = INFREQ\n \n return working_sentence, min(len(sentence), pad)\n\ndef convert_and_pad_data(word_dict, data, pad=500):\n result = []\n lengths = []\n \n for sentence in data:\n converted, leng = convert_and_pad(word_dict, sentence, pad)\n result.append(converted)\n lengths.append(leng)\n \n return np.array(result), np.array(lengths)",
"_____no_output_____"
],
[
"# choosing not to reuse the raw train_X and test_X variables, in order to avoid having to go back to the\n# beginning of the notebook if I need to change some logic \n\ntrain_xc, train_xc_len = convert_and_pad_data(word_dict, train_X)\ntest_xc, test_xc_len = convert_and_pad_data(word_dict, test_X)",
"_____no_output_____"
]
],
[
[
"As a quick check to make sure that things are working as intended, check to see what one of the reviews in the training set looks like after having been processeed. Does this look reasonable? What is the length of a review in the training set?",
"_____no_output_____"
]
],
[
[
"# Use this cell to examine one of the processed reviews to make sure everything is working as intended.\nprint(train_X[88])\nprint(train_xc[88])",
"['popular', 'sport', 'surf', 'like', 'mani', 'peopl', 'watch', 'documentari', 'realiz', 'danger', 'could', 'fact', 'surfer', 'also', 'scare', 'big', 'wave', 'even', 'somebodi', 'got', 'kill', 'still', 'kept', 'surf', 'enjoy', 'brave', 'peopl', 'accord', 'surfer', 'said', 'clearli', 'knew', 'felt', 'big', 'wave', 'came', 'adjust', 'best', 'avoid', 'direct', 'strike', 'big', 'wave', 'win', 'obvious', 'bring', 'huge', 'satisfact', 'amaz', 'cinematographi', 'cannot', 'overlook', 'absolut', 'visual', 'enjoy', 'excel', 'sport', 'documentari', '8', '10']\n[ 865 1337 2634 5 46 23 12 505 420 927 36 100 1 27\n 758 116 1591 14 1625 111 103 59 749 2634 77 2014 23 1563\n 1 233 630 636 372 116 1591 333 1 53 564 98 1282 116\n 1591 551 470 324 554 1 356 570 495 1943 304 514 77 222\n 1337 505 634 83 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0]\n"
]
],
[
[
"**Question:** In the cells above we use the `preprocess_data` and `convert_and_pad_data` methods to process both the training and testing set. Why or why not might this be a problem?",
"_____no_output_____"
],
[
"**Answer:** We need to treat all data uniformly, so test data must go through same processing as the training data, so this is appropriate. However, we only built the vocabulary from words in the training set. It is possible that including the test data would have contributed some additional words in the top 5000. However, I believe this is not a significant problem, as 5000 words is a large vocabulary, and the training set is already large enough to be statistically representative of the language used. So it is likely that only a few of the less frequently used words at the end of the dictionary might change as a result of adding or omitting the test data.",
"_____no_output_____"
],
[
"## Step 3: Upload the data to S3\n\nAs in the XGBoost notebook, we will need to upload the training dataset to S3 in order for our training code to access it. For now we will save it locally and we will upload to S3 later on.\n\n### Save the processed training dataset locally\n\nIt is important to note the format of the data that we are saving as we will need to know it when we write the training code. In our case, each row of the dataset has the form `label`, `length`, `review[500]` where `review[500]` is a sequence of `500` integers representing the words in the review.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport sagemaker\n \npd.concat([pd.DataFrame(train_y), pd.DataFrame(train_xc_len), pd.DataFrame(train_xc)], axis=1) \\\n .to_csv(os.path.join(data_dir, 'train.csv'), header=False, index=False)",
"_____no_output_____"
]
],
[
[
"### Uploading the training data\n\n\nNext, we need to upload the training data to the SageMaker default S3 bucket so that we can provide access to it while training our model.",
"_____no_output_____"
]
],
[
[
"import sagemaker\n\nsagemaker_session = sagemaker.Session()\n\nbucket = sagemaker_session.default_bucket()\nprefix = 'sagemaker/sentiment_rnn'\n\nrole = sagemaker.get_execution_role()",
"_____no_output_____"
],
[
"input_data = sagemaker_session.upload_data(path=data_dir, bucket=bucket, key_prefix=prefix)",
"_____no_output_____"
],
[
"print(bucket)\nprint(data_dir)\nprint(input_data)",
"sagemaker-us-east-1-139645156747\n../data/pytorch\ns3://sagemaker-us-east-1-139645156747/sagemaker/sentiment_rnn\n"
]
],
[
[
"**NOTE:** The cell above uploads the entire contents of our data directory. This includes the `word_dict.pkl` file. This is fortunate as we will need this later on when we create an endpoint that accepts an arbitrary review. For now, we will just take note of the fact that it resides in the data directory (and so also in the S3 training bucket) and that we will need to make sure it gets saved in the model directory.",
"_____no_output_____"
],
[
"## Step 4: Build and Train the PyTorch Model\n\nIn the XGBoost notebook we discussed what a model is in the SageMaker framework. In particular, a model comprises three objects\n\n - Model Artifacts,\n - Training Code, and\n - Inference Code,\n \neach of which interact with one another. In the XGBoost example we used training and inference code that was provided by Amazon. Here we will still be using containers provided by Amazon with the added benefit of being able to include our own custom code.\n\nWe will start by implementing our own neural network in PyTorch along with a training script. For the purposes of this project we have provided the necessary model object in the `model.py` file, inside of the `train` folder. You can see the provided implementation by running the cell below.",
"_____no_output_____"
]
],
[
[
"!pygmentize train/model.py",
"\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mtorch.nn\u001b[39;49;00m \u001b[34mas\u001b[39;49;00m \u001b[04m\u001b[36mnn\u001b[39;49;00m\r\n\r\n\u001b[34mclass\u001b[39;49;00m \u001b[04m\u001b[32mLSTMClassifier\u001b[39;49;00m(nn.Module):\r\n \u001b[33m\"\"\"\u001b[39;49;00m\r\n\u001b[33m This is the simple RNN model we will be using to perform Sentiment Analysis.\u001b[39;49;00m\r\n\u001b[33m \"\"\"\u001b[39;49;00m\r\n\r\n \u001b[34mdef\u001b[39;49;00m \u001b[32m__init__\u001b[39;49;00m(\u001b[36mself\u001b[39;49;00m, embedding_dim, hidden_dim, vocab_size):\r\n \u001b[33m\"\"\"\u001b[39;49;00m\r\n\u001b[33m Initialize the model by settingg up the various layers.\u001b[39;49;00m\r\n\u001b[33m \"\"\"\u001b[39;49;00m\r\n \u001b[36msuper\u001b[39;49;00m(LSTMClassifier, \u001b[36mself\u001b[39;49;00m).\u001b[32m__init__\u001b[39;49;00m()\r\n\r\n \u001b[36mself\u001b[39;49;00m.embedding = nn.Embedding(vocab_size, embedding_dim, padding_idx=\u001b[34m0\u001b[39;49;00m)\r\n \u001b[36mself\u001b[39;49;00m.lstm = nn.LSTM(embedding_dim, hidden_dim)\r\n \u001b[36mself\u001b[39;49;00m.dense = nn.Linear(in_features=hidden_dim, out_features=\u001b[34m1\u001b[39;49;00m)\r\n \u001b[36mself\u001b[39;49;00m.sig = nn.Sigmoid()\r\n \r\n \u001b[36mself\u001b[39;49;00m.word_dict = \u001b[36mNone\u001b[39;49;00m\r\n\r\n \u001b[34mdef\u001b[39;49;00m \u001b[32mforward\u001b[39;49;00m(\u001b[36mself\u001b[39;49;00m, x):\r\n \u001b[33m\"\"\"\u001b[39;49;00m\r\n\u001b[33m Perform a forward pass of our model on some input.\u001b[39;49;00m\r\n\u001b[33m \"\"\"\u001b[39;49;00m\r\n x = x.t()\r\n lengths = x[\u001b[34m0\u001b[39;49;00m,:]\r\n reviews = x[\u001b[34m1\u001b[39;49;00m:,:]\r\n embeds = \u001b[36mself\u001b[39;49;00m.embedding(reviews)\r\n lstm_out, _ = \u001b[36mself\u001b[39;49;00m.lstm(embeds)\r\n out = \u001b[36mself\u001b[39;49;00m.dense(lstm_out)\r\n out = out[lengths - \u001b[34m1\u001b[39;49;00m, \u001b[36mrange\u001b[39;49;00m(\u001b[36mlen\u001b[39;49;00m(lengths))]\r\n \u001b[34mreturn\u001b[39;49;00m \u001b[36mself\u001b[39;49;00m.sig(out.squeeze())\r\n"
]
],
[
[
"The important takeaway from the implementation provided is that there are three parameters that we may wish to tweak to improve the performance of our model. These are the embedding dimension, the hidden dimension and the size of the vocabulary. We will likely want to make these parameters configurable in the training script so that if we wish to modify them we do not need to modify the script itself. We will see how to do this later on. To start we will write some of the training code in the notebook so that we can more easily diagnose any issues that arise.\n\nFirst we will load a small portion of the training data set to use as a sample. It would be very time consuming to try and train the model completely in the notebook as we do not have access to a gpu and the compute instance that we are using is not particularly powerful. However, we can work on a small bit of the data to get a feel for how our training script is behaving.",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.utils.data\n\n# Read in only the first 250 rows\ntrain_sample = pd.read_csv(os.path.join(data_dir, 'train.csv'), header=None, names=None, nrows=250)\n\n# Turn the input pandas dataframe into tensors\ntrain_sample_y = torch.from_numpy(train_sample[[0]].values).float().squeeze()\ntrain_sample_X = torch.from_numpy(train_sample.drop([0], axis=1).values).long()\n\n# Build the dataset\ntrain_sample_ds = torch.utils.data.TensorDataset(train_sample_X, train_sample_y)\n# Build the dataloader\ntrain_sample_dl = torch.utils.data.DataLoader(train_sample_ds, batch_size=50)",
"_____no_output_____"
]
],
[
[
"### (TODO) Writing the training method\n\nNext we need to write the training code itself. This should be very similar to training methods that you have written before to train PyTorch models. We will leave any difficult aspects such as model saving / loading and parameter loading until a little later.",
"_____no_output_____"
]
],
[
[
"def train(model, train_loader, epochs, optimizer, loss_fn, device):\n\n CLIP_LIMIT = 5\n\n #print(\"Entering train.\")\n for epoch in range(1, epochs + 1):\n model.train()\n total_loss = 0\n \n #-jas -- shouldn't we be initializing the hidden layer here? Why is the model.py not using a hidden state?\n \n for batch in train_loader: \n batch_X, batch_y = batch\n #print(\"Training batch of size \", len(batch_X))\n \n batch_X = batch_X.to(device)\n batch_y = batch_y.to(device)\n \n # TODO: Complete this train method to train the model provided.\n \n ### get the model's prediction for the current batch\n model.zero_grad()\n output = model(batch_X)\n \n ### compute the loss for this batch and backpropagate it\n loss = loss_fn(output.squeeze(), batch_y)\n loss.backward()\n nn.utils.clip_grad_norm_(model.parameters(), CLIP_LIMIT)\n optimizer.step()\n \n ### accumulate loss over the epoch\n total_loss += loss.data.item()\n print(\"Epoch: {}, BCELoss: {}\".format(epoch, total_loss / len(train_loader)))\n ",
"_____no_output_____"
]
],
[
[
"Supposing we have the training method above, we will test that it is working by writing a bit of code in the notebook that executes our training method on the small sample training set that we loaded earlier. The reason for doing this in the notebook is so that we have an opportunity to fix any errors that arise early when they are easier to diagnose.",
"_____no_output_____"
]
],
[
[
"import torch.optim as optim\nfrom train.model import LSTMClassifier\n\ndevice = torch.device(\"cuda\" if torch.cuda.is_available() else \"cpu\")\nmodel = LSTMClassifier(32, 100, 5000).to(device)\noptimizer = optim.Adam(model.parameters())\nloss_fn = torch.nn.BCELoss()\n\ntrain(model, train_sample_dl, 5, optimizer, loss_fn, device)",
"Epoch: 1, BCELoss: 0.6921594619750977\nEpoch: 2, BCELoss: 0.6825043082237243\nEpoch: 3, BCELoss: 0.6743957757949829\nEpoch: 4, BCELoss: 0.6654969811439514\nEpoch: 5, BCELoss: 0.6548115968704223\n"
]
],
[
[
"In order to construct a PyTorch model using SageMaker we must provide SageMaker with a training script. We may optionally include a directory which will be copied to the container and from which our training code will be run. When the training container is executed it will check the uploaded directory (if there is one) for a `requirements.txt` file and install any required Python libraries, after which the training script will be run.",
"_____no_output_____"
],
[
"### (TODO) Training the model\n\nWhen a PyTorch model is constructed in SageMaker, an entry point must be specified. This is the Python file which will be executed when the model is trained. Inside of the `train` directory is a file called `train.py` which has been provided and which contains most of the necessary code to train our model. The only thing that is missing is the implementation of the `train()` method which you wrote earlier in this notebook.\n\n**TODO**: Copy the `train()` method written above and paste it into the `train/train.py` file where required.\n\nThe way that SageMaker passes hyperparameters to the training script is by way of arguments. These arguments can then be parsed and used in the training script. To see how this is done take a look at the provided `train/train.py` file.",
"_____no_output_____"
]
],
[
[
"from sagemaker.pytorch import PyTorch\n\nestimator = PyTorch(entry_point=\"train.py\",\n source_dir=\"train\",\n role=role,\n framework_version='0.4.0',\n train_instance_count=1,\n train_instance_type='ml.p2.xlarge',\n hyperparameters={\n 'epochs': 10,\n 'hidden_dim': 200,\n })",
"_____no_output_____"
],
[
"estimator.fit({'training': input_data})",
"2020-07-07 23:59:13 Starting - Starting the training job...\n2020-07-07 23:59:16 Starting - Launching requested ML instances......\n2020-07-08 00:00:40 Starting - Preparing the instances for training............\n2020-07-08 00:02:28 Downloading - Downloading input data...\n2020-07-08 00:03:05 Training - Downloading the training image...\n2020-07-08 00:03:28 Training - Training image download completed. Training in progress.\u001b[34mbash: cannot set terminal process group (-1): Inappropriate ioctl for device\u001b[0m\n\u001b[34mbash: no job control in this shell\u001b[0m\n\u001b[34m2020-07-08 00:03:29,421 sagemaker-containers INFO Imported framework sagemaker_pytorch_container.training\u001b[0m\n\u001b[34m2020-07-08 00:03:29,448 sagemaker_pytorch_container.training INFO Block until all host DNS lookups succeed.\u001b[0m\n\u001b[34m2020-07-08 00:03:29,451 sagemaker_pytorch_container.training INFO Invoking user training script.\u001b[0m\n\u001b[34m2020-07-08 00:03:29,705 sagemaker-containers INFO Module train does not provide a setup.py. \u001b[0m\n\u001b[34mGenerating setup.py\u001b[0m\n\u001b[34m2020-07-08 00:03:29,705 sagemaker-containers INFO Generating setup.cfg\u001b[0m\n\u001b[34m2020-07-08 00:03:29,705 sagemaker-containers INFO Generating MANIFEST.in\u001b[0m\n\u001b[34m2020-07-08 00:03:29,705 sagemaker-containers INFO Installing module with the following command:\u001b[0m\n\u001b[34m/usr/bin/python -m pip install -U . -r requirements.txt\u001b[0m\n\u001b[34mProcessing /opt/ml/code\u001b[0m\n\u001b[34mCollecting pandas (from -r requirements.txt (line 1))\n Downloading https://files.pythonhosted.org/packages/74/24/0cdbf8907e1e3bc5a8da03345c23cbed7044330bb8f73bb12e711a640a00/pandas-0.24.2-cp35-cp35m-manylinux1_x86_64.whl (10.0MB)\u001b[0m\n\u001b[34mCollecting numpy (from -r requirements.txt (line 2))\n Downloading https://files.pythonhosted.org/packages/b5/36/88723426b4ff576809fec7d73594fe17a35c27f8d01f93637637a29ae25b/numpy-1.18.5-cp35-cp35m-manylinux1_x86_64.whl (19.9MB)\u001b[0m\n\u001b[34mCollecting nltk (from -r requirements.txt (line 3))\n Downloading https://files.pythonhosted.org/packages/92/75/ce35194d8e3022203cca0d2f896dbb88689f9b3fce8e9f9cff942913519d/nltk-3.5.zip (1.4MB)\u001b[0m\n\u001b[34mCollecting beautifulsoup4 (from -r requirements.txt (line 4))\n Downloading https://files.pythonhosted.org/packages/66/25/ff030e2437265616a1e9b25ccc864e0371a0bc3adb7c5a404fd661c6f4f6/beautifulsoup4-4.9.1-py3-none-any.whl (115kB)\u001b[0m\n\u001b[34mCollecting html5lib (from -r requirements.txt (line 5))\n Downloading https://files.pythonhosted.org/packages/6c/dd/a834df6482147d48e225a49515aabc28974ad5a4ca3215c18a882565b028/html5lib-1.1-py2.py3-none-any.whl (112kB)\u001b[0m\n\u001b[34mRequirement already satisfied, skipping upgrade: python-dateutil>=2.5.0 in /usr/local/lib/python3.5/dist-packages (from pandas->-r requirements.txt (line 1)) (2.7.5)\u001b[0m\n\u001b[34mCollecting pytz>=2011k (from pandas->-r requirements.txt (line 1))\n Downloading https://files.pythonhosted.org/packages/4f/a4/879454d49688e2fad93e59d7d4efda580b783c745fd2ec2a3adf87b0808d/pytz-2020.1-py2.py3-none-any.whl (510kB)\u001b[0m\n\u001b[34mRequirement already satisfied, skipping upgrade: click in /usr/local/lib/python3.5/dist-packages (from nltk->-r requirements.txt (line 3)) (7.0)\u001b[0m\n\u001b[34mCollecting joblib (from nltk->-r requirements.txt (line 3))\n Downloading https://files.pythonhosted.org/packages/28/5c/cf6a2b65a321c4a209efcdf64c2689efae2cb62661f8f6f4bb28547cf1bf/joblib-0.14.1-py2.py3-none-any.whl (294kB)\u001b[0m\n\u001b[34mCollecting regex (from nltk->-r requirements.txt (line 3))\u001b[0m\n\u001b[34m Downloading https://files.pythonhosted.org/packages/b8/7b/01510a6229c2176425bda54d15fba05a4b3df169b87265b008480261d2f9/regex-2020.6.8.tar.gz (690kB)\u001b[0m\n\u001b[34mCollecting tqdm (from nltk->-r requirements.txt (line 3))\n Downloading https://files.pythonhosted.org/packages/46/62/7663894f67ac5a41a0d8812d78d9d2a9404124051885af9d77dc526fb399/tqdm-4.47.0-py2.py3-none-any.whl (66kB)\u001b[0m\n\u001b[34mCollecting soupsieve>1.2 (from beautifulsoup4->-r requirements.txt (line 4))\n Downloading https://files.pythonhosted.org/packages/6f/8f/457f4a5390eeae1cc3aeab89deb7724c965be841ffca6cfca9197482e470/soupsieve-2.0.1-py3-none-any.whl\u001b[0m\n\u001b[34mCollecting webencodings (from html5lib->-r requirements.txt (line 5))\u001b[0m\n\u001b[34m Downloading https://files.pythonhosted.org/packages/f4/24/2a3e3df732393fed8b3ebf2ec078f05546de641fe1b667ee316ec1dcf3b7/webencodings-0.5.1-py2.py3-none-any.whl\u001b[0m\n\u001b[34mRequirement already satisfied, skipping upgrade: six>=1.9 in /usr/local/lib/python3.5/dist-packages (from html5lib->-r requirements.txt (line 5)) (1.11.0)\u001b[0m\n\u001b[34mBuilding wheels for collected packages: nltk, train, regex\n Running setup.py bdist_wheel for nltk: started\n Running setup.py bdist_wheel for nltk: finished with status 'done'\n Stored in directory: /root/.cache/pip/wheels/ae/8c/3f/b1fe0ba04555b08b57ab52ab7f86023639a526d8bc8d384306\n Running setup.py bdist_wheel for train: started\u001b[0m\n\u001b[34m Running setup.py bdist_wheel for train: finished with status 'done'\n Stored in directory: /tmp/pip-ephem-wheel-cache-bf0ei8xl/wheels/35/24/16/37574d11bf9bde50616c67372a334f94fa8356bc7164af8ca3\n Running setup.py bdist_wheel for regex: started\u001b[0m\n\u001b[34m Running setup.py bdist_wheel for regex: finished with status 'done'\n Stored in directory: /root/.cache/pip/wheels/9c/e2/cf/246ad8c87bcdf3cba1ec95fa89bc205c9037aa8f4d2e26fdad\u001b[0m\n\u001b[34mSuccessfully built nltk train regex\u001b[0m\n\u001b[34mInstalling collected packages: pytz, numpy, pandas, joblib, regex, tqdm, nltk, soupsieve, beautifulsoup4, webencodings, html5lib, train\n Found existing installation: numpy 1.15.4\n Uninstalling numpy-1.15.4:\n Successfully uninstalled numpy-1.15.4\u001b[0m\n\u001b[34mSuccessfully installed beautifulsoup4-4.9.1 html5lib-1.1 joblib-0.14.1 nltk-3.5 numpy-1.18.5 pandas-0.24.2 pytz-2020.1 regex-2020.6.8 soupsieve-2.0.1 tqdm-4.47.0 train-1.0.0 webencodings-0.5.1\u001b[0m\n\u001b[34mYou are using pip version 18.1, however version 20.2b1 is available.\u001b[0m\n\u001b[34mYou should consider upgrading via the 'pip install --upgrade pip' command.\u001b[0m\n\u001b[34m2020-07-08 00:03:51,923 sagemaker-containers INFO Invoking user script\n\u001b[0m\n\u001b[34mTraining Env:\n\u001b[0m\n\u001b[34m{\n \"log_level\": 20,\n \"input_dir\": \"/opt/ml/input\",\n \"hyperparameters\": {\n \"epochs\": 10,\n \"hidden_dim\": 200\n },\n \"framework_module\": \"sagemaker_pytorch_container.training:main\",\n \"output_dir\": \"/opt/ml/output\",\n \"user_entry_point\": \"train.py\",\n \"num_cpus\": 4,\n \"network_interface_name\": \"eth0\",\n \"output_data_dir\": \"/opt/ml/output/data\",\n \"additional_framework_parameters\": {},\n \"resource_config\": {\n \"current_host\": \"algo-1\",\n \"network_interface_name\": \"eth0\",\n \"hosts\": [\n \"algo-1\"\n ]\n },\n \"output_intermediate_dir\": \"/opt/ml/output/intermediate\",\n \"channel_input_dirs\": {\n \"training\": \"/opt/ml/input/data/training\"\n },\n \"job_name\": \"sagemaker-pytorch-2020-07-07-23-59-12-964\",\n \"model_dir\": \"/opt/ml/model\",\n \"current_host\": \"algo-1\",\n \"input_config_dir\": \"/opt/ml/input/config\",\n \"module_dir\": \"s3://sagemaker-us-east-1-139645156747/sagemaker-pytorch-2020-07-07-23-59-12-964/source/sourcedir.tar.gz\",\n \"module_name\": \"train\",\n \"input_data_config\": {\n \"training\": {\n \"TrainingInputMode\": \"File\",\n \"RecordWrapperType\": \"None\",\n \"S3DistributionType\": \"FullyReplicated\"\n }\n },\n \"num_gpus\": 1,\n \"hosts\": [\n \"algo-1\"\n ]\u001b[0m\n\u001b[34m}\n\u001b[0m\n\u001b[34mEnvironment variables:\n\u001b[0m\n\u001b[34mSM_USER_ARGS=[\"--epochs\",\"10\",\"--hidden_dim\",\"200\"]\u001b[0m\n\u001b[34mPYTHONPATH=/usr/local/bin:/usr/lib/python35.zip:/usr/lib/python3.5:/usr/lib/python3.5/plat-x86_64-linux-gnu:/usr/lib/python3.5/lib-dynload:/usr/local/lib/python3.5/dist-packages:/usr/lib/python3/dist-packages\u001b[0m\n\u001b[34mSM_CURRENT_HOST=algo-1\u001b[0m\n\u001b[34mSM_INPUT_DATA_CONFIG={\"training\":{\"RecordWrapperType\":\"None\",\"S3DistributionType\":\"FullyReplicated\",\"TrainingInputMode\":\"File\"}}\u001b[0m\n\u001b[34mSM_CHANNEL_TRAINING=/opt/ml/input/data/training\u001b[0m\n\u001b[34mSM_FRAMEWORK_MODULE=sagemaker_pytorch_container.training:main\u001b[0m\n\u001b[34mSM_RESOURCE_CONFIG={\"current_host\":\"algo-1\",\"hosts\":[\"algo-1\"],\"network_interface_name\":\"eth0\"}\u001b[0m\n\u001b[34mSM_MODULE_DIR=s3://sagemaker-us-east-1-139645156747/sagemaker-pytorch-2020-07-07-23-59-12-964/source/sourcedir.tar.gz\u001b[0m\n\u001b[34mSM_HP_HIDDEN_DIM=200\u001b[0m\n\u001b[34mSM_OUTPUT_DIR=/opt/ml/output\u001b[0m\n\u001b[34mSM_NETWORK_INTERFACE_NAME=eth0\u001b[0m\n\u001b[34mSM_NUM_CPUS=4\u001b[0m\n\u001b[34mSM_HP_EPOCHS=10\u001b[0m\n\u001b[34mSM_INPUT_CONFIG_DIR=/opt/ml/input/config\u001b[0m\n\u001b[34mSM_CHANNELS=[\"training\"]\u001b[0m\n\u001b[34mSM_USER_ENTRY_POINT=train.py\u001b[0m\n\u001b[34mSM_INPUT_DIR=/opt/ml/input\u001b[0m\n\u001b[34mSM_HOSTS=[\"algo-1\"]\u001b[0m\n\u001b[34mSM_OUTPUT_DATA_DIR=/opt/ml/output/data\u001b[0m\n\u001b[34mSM_MODULE_NAME=train\u001b[0m\n\u001b[34mSM_MODEL_DIR=/opt/ml/model\u001b[0m\n\u001b[34mSM_OUTPUT_INTERMEDIATE_DIR=/opt/ml/output/intermediate\u001b[0m\n\u001b[34mSM_HPS={\"epochs\":10,\"hidden_dim\":200}\u001b[0m\n\u001b[34mSM_FRAMEWORK_PARAMS={}\u001b[0m\n\u001b[34mSM_TRAINING_ENV={\"additional_framework_parameters\":{},\"channel_input_dirs\":{\"training\":\"/opt/ml/input/data/training\"},\"current_host\":\"algo-1\",\"framework_module\":\"sagemaker_pytorch_container.training:main\",\"hosts\":[\"algo-1\"],\"hyperparameters\":{\"epochs\":10,\"hidden_dim\":200},\"input_config_dir\":\"/opt/ml/input/config\",\"input_data_config\":{\"training\":{\"RecordWrapperType\":\"None\",\"S3DistributionType\":\"FullyReplicated\",\"TrainingInputMode\":\"File\"}},\"input_dir\":\"/opt/ml/input\",\"job_name\":\"sagemaker-pytorch-2020-07-07-23-59-12-964\",\"log_level\":20,\"model_dir\":\"/opt/ml/model\",\"module_dir\":\"s3://sagemaker-us-east-1-139645156747/sagemaker-pytorch-2020-07-07-23-59-12-964/source/sourcedir.tar.gz\",\"module_name\":\"train\",\"network_interface_name\":\"eth0\",\"num_cpus\":4,\"num_gpus\":1,\"output_data_dir\":\"/opt/ml/output/data\",\"output_dir\":\"/opt/ml/output\",\"output_intermediate_dir\":\"/opt/ml/output/intermediate\",\"resource_config\":{\"current_host\":\"algo-1\",\"hosts\":[\"algo-1\"],\"network_interface_name\":\"eth0\"},\"user_entry_point\":\"train.py\"}\u001b[0m\n\u001b[34mSM_LOG_LEVEL=20\u001b[0m\n\u001b[34mSM_NUM_GPUS=1\n\u001b[0m\n\u001b[34mInvoking script with the following command:\n\u001b[0m\n\u001b[34m/usr/bin/python -m train --epochs 10 --hidden_dim 200\n\n\u001b[0m\n\u001b[34m///// In train/__main__: Using device cuda.\u001b[0m\n\u001b[34mGet train data loader.\u001b[0m\n\u001b[34mModel loaded with embedding_dim 32, hidden_dim 200, vocab_size 5000.\u001b[0m\n\u001b[34m///// Entering train/train.\u001b[0m\n\u001b[34mEpoch: 1, BCELoss: 0.6746065276009696\u001b[0m\n\u001b[34mEpoch: 2, BCELoss: 0.5870167746835825\u001b[0m\n\u001b[34mEpoch: 3, BCELoss: 0.4859969281420416\u001b[0m\n\u001b[34mEpoch: 4, BCELoss: 0.4077845751022806\u001b[0m\n\u001b[34mEpoch: 5, BCELoss: 0.3617596650610165\u001b[0m\n\u001b[34mEpoch: 6, BCELoss: 0.33730681514253424\u001b[0m\n\u001b[34mEpoch: 7, BCELoss: 0.3138365362371717\u001b[0m\n\u001b[34mEpoch: 8, BCELoss: 0.2878250777721405\u001b[0m\n\u001b[34mEpoch: 9, BCELoss: 0.2827960873136715\u001b[0m\n\u001b[34mEpoch: 10, BCELoss: 0.2594302384829035\u001b[0m\n\u001b[34m2020-07-08 00:06:51,852 sagemaker-containers INFO Reporting training SUCCESS\u001b[0m\n\n2020-07-08 00:07:03 Uploading - Uploading generated training model\n2020-07-08 00:07:03 Completed - Training job completed\nTraining seconds: 275\nBillable seconds: 275\n"
]
],
[
[
"## Step 5: Testing the model\n\nAs mentioned at the top of this notebook, we will be testing this model by first deploying it and then sending the testing data to the deployed endpoint. We will do this so that we can make sure that the deployed model is working correctly.\n\n## Step 6: Deploy the model for testing\n\nNow that we have trained our model, we would like to test it to see how it performs. Currently our model takes input of the form `review_length, review[500]` where `review[500]` is a sequence of `500` integers which describe the words present in the review, encoded using `word_dict`. Fortunately for us, SageMaker provides built-in inference code for models with simple inputs such as this.\n\nThere is one thing that we need to provide, however, and that is a function which loads the saved model. This function must be called `model_fn()` and takes as its only parameter a path to the directory where the model artifacts are stored. This function must also be present in the python file which we specified as the entry point. In our case the model loading function has been provided and so no changes need to be made.\n\n**NOTE**: When the built-in inference code is run it must import the `model_fn()` method from the `train.py` file. This is why the training code is wrapped in a main guard ( ie, `if __name__ == '__main__':` )\n\nSince we don't need to change anything in the code that was uploaded during training, we can simply deploy the current model as-is.\n\n**NOTE:** When deploying a model you are asking SageMaker to launch an compute instance that will wait for data to be sent to it. As a result, this compute instance will continue to run until *you* shut it down. This is important to know since the cost of a deployed endpoint depends on how long it has been running for.\n\nIn other words **If you are no longer using a deployed endpoint, shut it down!**\n\n**TODO:** Deploy the trained model.",
"_____no_output_____"
]
],
[
[
"# TODO: Deploy the trained model\npredictor = estimator.deploy(initial_instance_count = 1, instance_type = \"ml.p2.xlarge\")",
"-------------------!"
]
],
[
[
"## Step 7 - Use the model for testing\n\nOnce deployed, we can read in the test data and send it off to our deployed model to get some results. Once we collect all of the results we can determine how accurate our model is.",
"_____no_output_____"
]
],
[
[
"test_inputs = pd.concat([pd.DataFrame(test_xc_len), pd.DataFrame(test_xc)], axis=1)",
"_____no_output_____"
],
[
"# We split the data into chunks and send each chunk seperately, accumulating the results.\n\ndef predict(data, rows=512):\n split_array = np.array_split(data, int(data.shape[0] / float(rows) + 1))\n predictions = np.array([])\n for array in split_array:\n predictions = np.append(predictions, predictor.predict(array))\n break\n \n return predictions",
"_____no_output_____"
],
[
"predictions = predict(test_inputs.values)\npredictions = [round(num) for num in predictions]",
"_____no_output_____"
],
[
"from sklearn.metrics import accuracy_score\naccuracy_score(test_y, predictions)",
"_____no_output_____"
]
],
[
[
"**Question:** How does this model compare to the XGBoost model you created earlier? Why might these two models perform differently on this dataset? Which do *you* think is better for sentiment analysis?",
"_____no_output_____"
],
[
"**Answer:** Accuracy of 0.86 is very close to what was achieved with the XGBoost model. These are two entirely different approaches to making predictions, so there is no particular reason to expect them to perform identically. It is not clear to me which is inherently more adept at predicting sentiment in textual descriptions. My suspicion is that an RNN may be better, since it is built to learn about sequences of words, not just collections of words (XGBoost seems to not consider the ordering of the words in a review).",
"_____no_output_____"
],
[
"### (TODO) More testing\n\nWe now have a trained model which has been deployed and which we can send processed reviews to and which returns the predicted sentiment. However, ultimately we would like to be able to send our model an unprocessed review. That is, we would like to send the review itself as a string. For example, suppose we wish to send the following review to our model.",
"_____no_output_____"
]
],
[
[
"test_review = 'The simplest pleasures in life are the best, and this film is one of them. Combining a rather basic storyline of love and adventure this movie transcends the usual weekend fair with wit and unmitigated charm.'",
"_____no_output_____"
]
],
[
[
"The question we now need to answer is, how do we send this review to our model?\n\nRecall in the first section of this notebook we did a bunch of data processing to the IMDb dataset. In particular, we did two specific things to the provided reviews.\n - Removed any html tags and stemmed the input\n - Encoded the review as a sequence of integers using `word_dict`\n \nIn order process the review we will need to repeat these two steps.\n\n**TODO**: Using the `review_to_words` and `convert_and_pad` methods from section one, convert `test_review` into a numpy array `test_data` suitable to send to our model. Remember that our model expects input of the form `review_length, review[500]`.",
"_____no_output_____"
]
],
[
[
"# TODO: Convert test_review into a form usable by the model and save the results in test_data\n\ndef convert_single_review(raw_review):\n \n # cleanse the raw review of html tags, punctuation, capital letters, and stem the words\n sentence = review_to_words(raw_review)\n #print(\"sentence = \", sentence)\n \n # convert the cleansed text to dictionary indices and ensure each item is the correct length\n result, length = convert_and_pad(word_dict, sentence)\n #print(\"len = \", length, \", test_data = \\n\", result)\n return result, length\n \ntest_data, test_data_length = convert_single_review(test_review)\n\nprint(test_data)",
"[1, 1374, 50, 53, 3, 4, 878, 173, 392, 682, 29, 723, 2, 4422, 275, 2078, 1059, 760, 1, 582, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]\n"
]
],
[
[
"Now that we have processed the review, we can send the resulting array to our model to predict the sentiment of the review.",
"_____no_output_____"
]
],
[
[
"### The TODO comment a couple cells above is misleading. It seems to be saying that there are two input\n### arguments: review_length and review. The truth is, there is only one input arg, which is the review.\n### However, it should be a 2D array, with the first dimension representing the number of reviews (in this\n### case, only 1).\n\n# convert to a torch tensor & use view to reshape it to a single row, then convert back to a numpy array\ntd = torch.from_numpy(np.array(test_data))\nprint(td.shape)\ntd = td.view(1, -1)\nta = td.numpy()\nprint(ta.shape)\npredictor.predict(ta)",
"torch.Size([500])\n(1, 500)\n"
]
],
[
[
"Since the return value of our model is close to `1`, we can be certain that the review we submitted is positive.",
"_____no_output_____"
],
[
"### Delete the endpoint\n\nOf course, just like in the XGBoost notebook, once we've deployed an endpoint it continues to run until we tell it to shut down. Since we are done using our endpoint for now, we can delete it.",
"_____no_output_____"
]
],
[
[
"estimator.delete_endpoint()",
"_____no_output_____"
]
],
[
[
"## Step 6 (again) - Deploy the model for the web app\n\nNow that we know that our model is working, it's time to create some custom inference code so that we can send the model a review which has not been processed and have it determine the sentiment of the review.\n\nAs we saw above, by default the estimator which we created, when deployed, will use the entry script and directory which we provided when creating the model. However, since we now wish to accept a string as input and our model expects a processed review, we need to write some custom inference code.\n\nWe will store the code that we write in the `serve` directory. Provided in this directory is the `model.py` file that we used to construct our model, a `utils.py` file which contains the `review_to_words` and `convert_and_pad` pre-processing functions which we used during the initial data processing, and `predict.py`, the file which will contain our custom inference code. Note also that `requirements.txt` is present which will tell SageMaker what Python libraries are required by our custom inference code.\n\nWhen deploying a PyTorch model in SageMaker, you are expected to provide four functions which the SageMaker inference container will use.\n - `model_fn`: This function is the same function that we used in the training script and it tells SageMaker how to load our model.\n - `input_fn`: This function receives the raw serialized input that has been sent to the model's endpoint and its job is to de-serialize and make the input available for the inference code.\n - `output_fn`: This function takes the output of the inference code and its job is to serialize this output and return it to the caller of the model's endpoint.\n - `predict_fn`: The heart of the inference script, this is where the actual prediction is done and is the function which you will need to complete.\n\nFor the simple website that we are constructing during this project, the `input_fn` and `output_fn` methods are relatively straightforward. We only require being able to accept a string as input and we expect to return a single value as output. You might imagine though that in a more complex application the input or output may be image data or some other binary data which would require some effort to serialize.\n\n### (TODO) Writing inference code\n\nBefore writing our custom inference code, we will begin by taking a look at the code which has been provided.",
"_____no_output_____"
]
],
[
[
"!pygmentize serve/predict.py",
"\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36margparse\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mjson\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mos\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mpickle\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36msys\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36msagemaker_containers\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mpandas\u001b[39;49;00m \u001b[34mas\u001b[39;49;00m \u001b[04m\u001b[36mpd\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mnumpy\u001b[39;49;00m \u001b[34mas\u001b[39;49;00m \u001b[04m\u001b[36mnp\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mtorch\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mtorch.nn\u001b[39;49;00m \u001b[34mas\u001b[39;49;00m \u001b[04m\u001b[36mnn\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mtorch.optim\u001b[39;49;00m \u001b[34mas\u001b[39;49;00m \u001b[04m\u001b[36moptim\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mtorch.utils.data\u001b[39;49;00m\r\n\r\n\u001b[34mfrom\u001b[39;49;00m \u001b[04m\u001b[36mmodel\u001b[39;49;00m \u001b[34mimport\u001b[39;49;00m LSTMClassifier\r\n\r\n\u001b[34mfrom\u001b[39;49;00m \u001b[04m\u001b[36mutils\u001b[39;49;00m \u001b[34mimport\u001b[39;49;00m review_to_words, convert_and_pad\r\n\r\n\u001b[34mdef\u001b[39;49;00m \u001b[32mmodel_fn\u001b[39;49;00m(model_dir):\r\n \u001b[33m\"\"\"Load the PyTorch model from the `model_dir` directory.\"\"\"\u001b[39;49;00m\r\n \u001b[34mprint\u001b[39;49;00m(\u001b[33m\"\u001b[39;49;00m\u001b[33mLoading model.\u001b[39;49;00m\u001b[33m\"\u001b[39;49;00m)\r\n\r\n \u001b[37m# First, load the parameters used to create the model.\u001b[39;49;00m\r\n model_info = {}\r\n model_info_path = os.path.join(model_dir, \u001b[33m'\u001b[39;49;00m\u001b[33mmodel_info.pth\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m)\r\n \u001b[34mwith\u001b[39;49;00m \u001b[36mopen\u001b[39;49;00m(model_info_path, \u001b[33m'\u001b[39;49;00m\u001b[33mrb\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m) \u001b[34mas\u001b[39;49;00m f:\r\n model_info = torch.load(f)\r\n\r\n \u001b[34mprint\u001b[39;49;00m(\u001b[33m\"\u001b[39;49;00m\u001b[33mmodel_info: {}\u001b[39;49;00m\u001b[33m\"\u001b[39;49;00m.format(model_info))\r\n\r\n \u001b[37m# Determine the device and construct the model.\u001b[39;49;00m\r\n device = torch.device(\u001b[33m\"\u001b[39;49;00m\u001b[33mcuda\u001b[39;49;00m\u001b[33m\"\u001b[39;49;00m \u001b[34mif\u001b[39;49;00m torch.cuda.is_available() \u001b[34melse\u001b[39;49;00m \u001b[33m\"\u001b[39;49;00m\u001b[33mcpu\u001b[39;49;00m\u001b[33m\"\u001b[39;49;00m)\r\n model = LSTMClassifier(model_info[\u001b[33m'\u001b[39;49;00m\u001b[33membedding_dim\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m], model_info[\u001b[33m'\u001b[39;49;00m\u001b[33mhidden_dim\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m], model_info[\u001b[33m'\u001b[39;49;00m\u001b[33mvocab_size\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m])\r\n\r\n \u001b[37m# Load the store model parameters.\u001b[39;49;00m\r\n model_path = os.path.join(model_dir, \u001b[33m'\u001b[39;49;00m\u001b[33mmodel.pth\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m)\r\n \u001b[34mwith\u001b[39;49;00m \u001b[36mopen\u001b[39;49;00m(model_path, \u001b[33m'\u001b[39;49;00m\u001b[33mrb\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m) \u001b[34mas\u001b[39;49;00m f:\r\n model.load_state_dict(torch.load(f))\r\n\r\n \u001b[37m# Load the saved word_dict.\u001b[39;49;00m\r\n word_dict_path = os.path.join(model_dir, \u001b[33m'\u001b[39;49;00m\u001b[33mword_dict.pkl\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m)\r\n \u001b[34mwith\u001b[39;49;00m \u001b[36mopen\u001b[39;49;00m(word_dict_path, \u001b[33m'\u001b[39;49;00m\u001b[33mrb\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m) \u001b[34mas\u001b[39;49;00m f:\r\n model.word_dict = pickle.load(f)\r\n\r\n model.to(device).eval()\r\n\r\n \u001b[34mprint\u001b[39;49;00m(\u001b[33m\"\u001b[39;49;00m\u001b[33mDone loading model.\u001b[39;49;00m\u001b[33m\"\u001b[39;49;00m)\r\n \u001b[34mreturn\u001b[39;49;00m model\r\n\r\n\u001b[34mdef\u001b[39;49;00m \u001b[32minput_fn\u001b[39;49;00m(serialized_input_data, content_type):\r\n \u001b[34mprint\u001b[39;49;00m(\u001b[33m'\u001b[39;49;00m\u001b[33mDeserializing the input data.\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m)\r\n \u001b[34mif\u001b[39;49;00m content_type == \u001b[33m'\u001b[39;49;00m\u001b[33mtext/plain\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m:\r\n data = serialized_input_data.decode(\u001b[33m'\u001b[39;49;00m\u001b[33mutf-8\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m)\r\n \u001b[34mreturn\u001b[39;49;00m data\r\n \u001b[34mraise\u001b[39;49;00m \u001b[36mException\u001b[39;49;00m(\u001b[33m'\u001b[39;49;00m\u001b[33mRequested unsupported ContentType in content_type: \u001b[39;49;00m\u001b[33m'\u001b[39;49;00m + content_type)\r\n\r\n\u001b[34mdef\u001b[39;49;00m \u001b[32moutput_fn\u001b[39;49;00m(prediction_output, accept):\r\n \u001b[34mprint\u001b[39;49;00m(\u001b[33m'\u001b[39;49;00m\u001b[33mSerializing the generated output.\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m)\r\n \u001b[34mreturn\u001b[39;49;00m \u001b[36mstr\u001b[39;49;00m(prediction_output)\r\n\r\n\u001b[34mdef\u001b[39;49;00m \u001b[32mpredict_fn\u001b[39;49;00m(input_data, model):\r\n \u001b[34mprint\u001b[39;49;00m(\u001b[33m'\u001b[39;49;00m\u001b[33mInferring sentiment of input data.\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m)\r\n\r\n device = torch.device(\u001b[33m\"\u001b[39;49;00m\u001b[33mcuda\u001b[39;49;00m\u001b[33m\"\u001b[39;49;00m \u001b[34mif\u001b[39;49;00m torch.cuda.is_available() \u001b[34melse\u001b[39;49;00m \u001b[33m\"\u001b[39;49;00m\u001b[33mcpu\u001b[39;49;00m\u001b[33m\"\u001b[39;49;00m)\r\n \r\n \u001b[34mif\u001b[39;49;00m model.word_dict \u001b[35mis\u001b[39;49;00m \u001b[36mNone\u001b[39;49;00m:\r\n \u001b[34mraise\u001b[39;49;00m \u001b[36mException\u001b[39;49;00m(\u001b[33m'\u001b[39;49;00m\u001b[33mModel has not been loaded properly, no word_dict.\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m)\r\n \r\n \u001b[37m# TODO: Process input_data so that it is ready to be sent to our model.\u001b[39;49;00m\r\n \u001b[37m# You should produce two variables:\u001b[39;49;00m\r\n \u001b[37m# data_X - A sequence of length 500 which represents the converted review\u001b[39;49;00m\r\n \u001b[37m# data_len - The length of the review\u001b[39;49;00m\r\n\r\n data_X = \u001b[36mNone\u001b[39;49;00m\r\n data_len = \u001b[36mNone\u001b[39;49;00m\r\n\r\n \u001b[37m# Using data_X and data_len we construct an appropriate input tensor. Remember\u001b[39;49;00m\r\n \u001b[37m# that our model expects input data of the form 'len, review[500]'.\u001b[39;49;00m\r\n data_pack = np.hstack((data_len, data_X))\r\n data_pack = data_pack.reshape(\u001b[34m1\u001b[39;49;00m, -\u001b[34m1\u001b[39;49;00m)\r\n \r\n data = torch.from_numpy(data_pack)\r\n data = data.to(device)\r\n\r\n \u001b[37m# Make sure to put the model into evaluation mode\u001b[39;49;00m\r\n model.eval()\r\n\r\n \u001b[37m# TODO: Compute the result of applying the model to the input data. The variable `result` should\u001b[39;49;00m\r\n \u001b[37m# be a numpy array which contains a single integer which is either 1 or 0\u001b[39;49;00m\r\n\r\n result = \u001b[36mNone\u001b[39;49;00m\r\n\r\n \u001b[34mreturn\u001b[39;49;00m result\r\n"
]
],
[
[
"As mentioned earlier, the `model_fn` method is the same as the one provided in the training code and the `input_fn` and `output_fn` methods are very simple and your task will be to complete the `predict_fn` method. Make sure that you save the completed file as `predict.py` in the `serve` directory.\n\n**TODO**: Complete the `predict_fn()` method in the `serve/predict.py` file.",
"_____no_output_____"
],
[
"### Deploying the model\n\nNow that the custom inference code has been written, we will create and deploy our model. To begin with, we need to construct a new PyTorchModel object which points to the model artifacts created during training and also points to the inference code that we wish to use. Then we can call the deploy method to launch the deployment container.\n\n**NOTE**: The default behaviour for a deployed PyTorch model is to assume that any input passed to the predictor is a `numpy` array. In our case we want to send a string so we need to construct a simple wrapper around the `RealTimePredictor` class to accomodate simple strings. In a more complicated situation you may want to provide a serialization object, for example if you wanted to sent image data.",
"_____no_output_____"
]
],
[
[
"#jas sandbox\nresult = torch.tensor(0.77).cpu()\nprint(result)\nresult = np.array(result.round().int().numpy())\nprint(\"result of model execution = \", result)\n",
"tensor(0.7700)\nresult of model execution = 1\n"
],
[
"from sagemaker.predictor import RealTimePredictor\nfrom sagemaker.pytorch import PyTorchModel\n\nclass StringPredictor(RealTimePredictor):\n def __init__(self, endpoint_name, sagemaker_session):\n super(StringPredictor, self).__init__(endpoint_name, sagemaker_session, content_type='text/plain')\n\nmodel = PyTorchModel(model_data=estimator.model_data,\n role = role,\n framework_version='0.4.0',\n entry_point='predict.py',\n source_dir='serve',\n predictor_cls=StringPredictor)\n\n###jas - changed from m4.large to p2.xlarge to get more performance\npredictor = model.deploy(initial_instance_count=1, instance_type='ml.p2.xlarge')",
"-----------------!"
]
],
[
[
"### Testing the model\n\nNow that we have deployed our model with the custom inference code, we should test to see if everything is working. Here we test our model by loading the first `250` positive and negative reviews and send them to the endpoint, then collect the results. The reason for only sending some of the data is that the amount of time it takes for our model to process the input and then perform inference is quite long and so testing the entire data set would be prohibitive.",
"_____no_output_____"
]
],
[
[
"import glob\n\ndef test_reviews(data_dir='../data/aclImdb', stop=250):\n \n results = []\n ground = []\n \n # We make sure to test both positive and negative reviews \n for sentiment in ['pos', 'neg']:\n \n path = os.path.join(data_dir, 'test', sentiment, '*.txt')\n files = glob.glob(path)\n \n files_read = 0\n \n print('Starting ', sentiment, ' files')\n \n # Iterate through the files and send them to the predictor\n for f in files:\n with open(f) as review:\n # First, we store the ground truth (was the review positive or negative)\n if sentiment == 'pos':\n ground.append(1)\n else:\n ground.append(0)\n # Read in the review and convert to 'utf-8' for transmission via HTTP\n review_input = review.read().encode('utf-8')\n #print(\"review_input = \", review_input)\n # Send the review to the predictor and store the results\n results.append(int(predictor.predict(review_input)))\n \n # Sending reviews to our endpoint one at a time takes a while so we\n # only send a small number of reviews\n files_read += 1\n if files_read == stop:\n break\n \n return ground, results",
"_____no_output_____"
],
[
"ground, results = test_reviews()",
"Starting pos files\nStarting neg files\n"
],
[
"from sklearn.metrics import accuracy_score\naccuracy_score(ground, results)",
"_____no_output_____"
]
],
[
[
"As an additional test, we can try sending the `test_review` that we looked at earlier.",
"_____no_output_____"
]
],
[
[
"predictor.predict(test_review)",
"_____no_output_____"
]
],
[
[
"Now that we know our endpoint is working as expected, we can set up the web page that will interact with it. If you don't have time to finish the project now, make sure to skip down to the end of this notebook and shut down your endpoint. You can deploy it again when you come back.",
"_____no_output_____"
],
[
"## Step 7 (again): Use the model for the web app\n\n> **TODO:** This entire section and the next contain tasks for you to complete, mostly using the AWS console.\n\nSo far we have been accessing our model endpoint by constructing a predictor object which uses the endpoint and then just using the predictor object to perform inference. What if we wanted to create a web app which accessed our model? The way things are set up currently makes that not possible since in order to access a SageMaker endpoint the app would first have to authenticate with AWS using an IAM role which included access to SageMaker endpoints. However, there is an easier way! We just need to use some additional AWS services.\n\n<img src=\"Web App Diagram.svg\">\n\nThe diagram above gives an overview of how the various services will work together. On the far right is the model which we trained above and which is deployed using SageMaker. On the far left is our web app that collects a user's movie review, sends it off and expects a positive or negative sentiment in return.\n\nIn the middle is where some of the magic happens. We will construct a Lambda function, which you can think of as a straightforward Python function that can be executed whenever a specified event occurs. We will give this function permission to send and recieve data from a SageMaker endpoint.\n\nLastly, the method we will use to execute the Lambda function is a new endpoint that we will create using API Gateway. This endpoint will be a url that listens for data to be sent to it. Once it gets some data it will pass that data on to the Lambda function and then return whatever the Lambda function returns. Essentially it will act as an interface that lets our web app communicate with the Lambda function.\n\n### Setting up a Lambda function\n\nThe first thing we are going to do is set up a Lambda function. This Lambda function will be executed whenever our public API has data sent to it. When it is executed it will receive the data, perform any sort of processing that is required, send the data (the review) to the SageMaker endpoint we've created and then return the result.\n\n#### Part A: Create an IAM Role for the Lambda function\n\nSince we want the Lambda function to call a SageMaker endpoint, we need to make sure that it has permission to do so. To do this, we will construct a role that we can later give the Lambda function.\n\nUsing the AWS Console, navigate to the **IAM** page and click on **Roles**. Then, click on **Create role**. Make sure that the **AWS service** is the type of trusted entity selected and choose **Lambda** as the service that will use this role, then click **Next: Permissions**.\n\nIn the search box type `sagemaker` and select the check box next to the **AmazonSageMakerFullAccess** policy. Then, click on **Next: Review**.\n\nLastly, give this role a name. Make sure you use a name that you will remember later on, for example `LambdaSageMakerRole`. Then, click on **Create role**.\n\n#### Part B: Create a Lambda function\n\nNow it is time to actually create the Lambda function.\n\nUsing the AWS Console, navigate to the AWS Lambda page and click on **Create a function**. When you get to the next page, make sure that **Author from scratch** is selected. Now, name your Lambda function, using a name that you will remember later on, for example `sentiment_analysis_func`. Make sure that the **Python 3.6** runtime is selected and then choose the role that you created in the previous part. Then, click on **Create Function**.\n\nOn the next page you will see some information about the Lambda function you've just created. If you scroll down you should see an editor in which you can write the code that will be executed when your Lambda function is triggered. In our example, we will use the code below. \n\n```python\n# We need to use the low-level library to interact with SageMaker since the SageMaker API\n# is not available natively through Lambda.\nimport boto3\n\ndef lambda_handler(event, context):\n\n # The SageMaker runtime is what allows us to invoke the endpoint that we've created.\n runtime = boto3.Session().client('sagemaker-runtime')\n\n # Now we use the SageMaker runtime to invoke our endpoint, sending the review we were given\n response = runtime.invoke_endpoint(EndpointName = '**ENDPOINT NAME HERE**', # The name of the endpoint we created\n ContentType = 'text/plain', # The data format that is expected\n Body = event['body']) # The actual review\n\n # The response is an HTTP response whose body contains the result of our inference\n result = response['Body'].read().decode('utf-8')\n\n return {\n 'statusCode' : 200,\n 'headers' : { 'Content-Type' : 'text/plain', 'Access-Control-Allow-Origin' : '*' },\n 'body' : result\n }\n```\n\nOnce you have copy and pasted the code above into the Lambda code editor, replace the `**ENDPOINT NAME HERE**` portion with the name of the endpoint that we deployed earlier. You can determine the name of the endpoint using the code cell below.",
"_____no_output_____"
]
],
[
[
"predictor.endpoint",
"_____no_output_____"
],
[
"predictor.predict(\"This movie sucked!\")",
"_____no_output_____"
]
],
[
[
"Once you have added the endpoint name to the Lambda function, click on **Save**. Your Lambda function is now up and running. Next we need to create a way for our web app to execute the Lambda function.\n\n### Setting up API Gateway\n\nNow that our Lambda function is set up, it is time to create a new API using API Gateway that will trigger the Lambda function we have just created.\n\nUsing AWS Console, navigate to **Amazon API Gateway** and then click on **Get started**.\n\nOn the next page, make sure that **New API** is selected and give the new api a name, for example, `sentiment_analysis_api`. Then, click on **Create API**.\n\nNow we have created an API, however it doesn't currently do anything. What we want it to do is to trigger the Lambda function that we created earlier.\n\nSelect the **Actions** dropdown menu and click **Create Method**. A new blank method will be created, select its dropdown menu and select **POST**, then click on the check mark beside it.\n\nFor the integration point, make sure that **Lambda Function** is selected and click on the **Use Lambda Proxy integration**. This option makes sure that the data that is sent to the API is then sent directly to the Lambda function with no processing. It also means that the return value must be a proper response object as it will also not be processed by API Gateway.\n\nType the name of the Lambda function you created earlier into the **Lambda Function** text entry box and then click on **Save**. Click on **OK** in the pop-up box that then appears, giving permission to API Gateway to invoke the Lambda function you created.\n\nThe last step in creating the API Gateway is to select the **Actions** dropdown and click on **Deploy API**. You will need to create a new Deployment stage and name it anything you like, for example `prod`.\n\nYou have now successfully set up a public API to access your SageMaker model. Make sure to copy or write down the URL provided to invoke your newly created public API as this will be needed in the next step. This URL can be found at the top of the page, highlighted in blue next to the text **Invoke URL**.",
"_____no_output_____"
],
[
"## Step 4: Deploying our web app\n\nNow that we have a publicly available API, we can start using it in a web app. For our purposes, we have provided a simple static html file which can make use of the public api you created earlier.\n\nIn the `website` folder there should be a file called `index.html`. Download the file to your computer and open that file up in a text editor of your choice. There should be a line which contains **\\*\\*REPLACE WITH PUBLIC API URL\\*\\***. Replace this string with the url that you wrote down in the last step and then save the file.\n\nNow, if you open `index.html` on your local computer, your browser will behave as a local web server and you can use the provided site to interact with your SageMaker model.\n\nIf you'd like to go further, you can host this html file anywhere you'd like, for example using github or hosting a static site on Amazon's S3. Once you have done this you can share the link with anyone you'd like and have them play with it too!\n\n> **Important Note** In order for the web app to communicate with the SageMaker endpoint, the endpoint has to actually be deployed and running. This means that you are paying for it. Make sure that the endpoint is running when you want to use the web app but that you shut it down when you don't need it, otherwise you will end up with a surprisingly large AWS bill.\n\n**TODO:** Make sure that you include the edited `index.html` file in your project submission.",
"_____no_output_____"
],
[
"Now that your web app is working, trying playing around with it and see how well it works.\n\n**Question**: Give an example of a review that you entered into your web app. What was the predicted sentiment of your example review?",
"_____no_output_____"
],
[
"**Answer:** \nTest review: \"The movie sucked.\" Prediction = 0\n\nTest review: \"Here is my movie review. The movie was great! I loved it. Best movie of the year!\" Prediction = 1\n\nI have tried several others, and the full system is working really well.",
"_____no_output_____"
],
[
"### Delete the endpoint\n\nRemember to always shut down your endpoint if you are no longer using it. You are charged for the length of time that the endpoint is running so if you forget and leave it on you could end up with an unexpectedly large bill.",
"_____no_output_____"
]
],
[
[
"predictor.delete_endpoint()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
e7931e364203c52f7d14aa5da7604dfcc79bed57 | 5,871 | ipynb | Jupyter Notebook | fnv/metrics/eval.ipynb | hariharan98m/tf-grocery-object-detection | a56d0584266363e9b304508d08c7cf25d8beaa70 | [
"MIT"
] | null | null | null | fnv/metrics/eval.ipynb | hariharan98m/tf-grocery-object-detection | a56d0584266363e9b304508d08c7cf25d8beaa70 | [
"MIT"
] | null | null | null | fnv/metrics/eval.ipynb | hariharan98m/tf-grocery-object-detection | a56d0584266363e9b304508d08c7cf25d8beaa70 | [
"MIT"
] | null | null | null | 34.333333 | 112 | 0.442344 | [
[
[
"def load_image_into_numpy_array(path):\n return np.array(Image.open(path))",
"_____no_output_____"
],
[
"import _init_paths\nfrom utils import *\nfrom BoundingBox import BoundingBox\nfrom BoundingBoxes import BoundingBoxes",
"_____no_output_____"
],
[
"gt_boundingBox_1 = BoundingBox(imageName='000001', classId='dog', x=0.34419263456090654, y=0.611, \n w=0.4164305949008499, h=0.262, typeCoordinates=CoordinatesType.Relative,\n bbType=BBType.GroundTruth, format=BBFormat.XYWH, imgSize=(353,500))",
"_____no_output_____"
],
[
"print(gt_boundingBox_1)",
"<BoundingBox.BoundingBox object at 0x123fe8dc0>\n"
],
[
"myBoundingBoxes = BoundingBoxes()",
"_____no_output_____"
],
[
"def getBoundingBoxes(directory,\n isGT,\n bbFormat,\n coordType,\n allBoundingBoxes=None,\n allClasses=None,\n imgSize=(0, 0)):\n \"\"\"Read txt files containing bounding boxes (ground truth and detections).\"\"\"\n if allBoundingBoxes is None:\n allBoundingBoxes = BoundingBoxes()\n if allClasses is None:\n allClasses = []\n # Read ground truths\n os.chdir(directory)\n files = glob.glob(\"*.txt\")\n files.sort()\n # Read GT detections from txt file\n # Each line of the files in the groundtruths folder represents a ground truth bounding box\n # (bounding boxes that a detector should detect)\n # Each value of each line is \"class_id, x, y, width, height\" respectively\n # Class_id represents the class of the bounding box\n # x, y represents the most top-left coordinates of the bounding box\n # x2, y2 represents the most bottom-right coordinates of the bounding box\n for f in files:\n nameOfImage = f.replace(\".txt\", \"\")\n fh1 = open(f, \"r\")\n for line in fh1:\n line = line.replace(\"\\n\", \"\")\n if line.replace(' ', '') == '':\n continue\n splitLine = line.split(\" \")\n if isGT:\n # idClass = int(splitLine[0]) #class\n idClass = (splitLine[0]) # class\n x = float(splitLine[1])\n y = float(splitLine[2])\n w = float(splitLine[3])\n h = float(splitLine[4])\n bb = BoundingBox(nameOfImage,\n idClass,\n x,\n y,\n w,\n h,\n coordType,\n imgSize,\n BBType.GroundTruth,\n format=bbFormat)\n else:\n # idClass = int(splitLine[0]) #class\n idClass = (splitLine[0]) # class\n confidence = float(splitLine[1])\n x = float(splitLine[2])\n y = float(splitLine[3])\n w = float(splitLine[4])\n h = float(splitLine[5])\n bb = BoundingBox(nameOfImage,\n idClass,\n x,\n y,\n w,\n h,\n coordType,\n imgSize,\n BBType.Detected,\n confidence,\n format=bbFormat)\n allBoundingBoxes.addBoundingBox(bb)\n if idClass not in allClasses:\n allClasses.append(idClass)\n fh1.close()\n return allBoundingBoxes, allClasses",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e79324d9146ca4dc32c52ddbf82c7869f6121beb | 1,424 | ipynb | Jupyter Notebook | solutions/09_extra_tips_solutions.ipynb | gabrielecalvo/pandas_tutorial | bff468693b648cf84852707035550947c8a2cdfa | [
"MIT"
] | 33 | 2021-03-30T21:17:17.000Z | 2021-09-11T09:06:45.000Z | solutions/09_extra_tips_solutions.ipynb | gabrielecalvo/pandas_tutorial | bff468693b648cf84852707035550947c8a2cdfa | [
"MIT"
] | null | null | null | solutions/09_extra_tips_solutions.ipynb | gabrielecalvo/pandas_tutorial | bff468693b648cf84852707035550947c8a2cdfa | [
"MIT"
] | 7 | 2021-03-31T16:31:46.000Z | 2021-08-03T10:40:59.000Z | 18.986667 | 93 | 0.482444 | [
[
[
"import pandas as pd",
"_____no_output_____"
]
],
[
[
"### ***EXERCISE 9.1***\nUsing the `df` provided below, get the mean score of people whose name stats with 'J'",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame({\n 'name': ['John', 'Albert', 'Jack', 'Josef', 'Bob', 'Juliette', 'Mary', 'Jane'], \n 'score': [5,8,6,4,8,7,3,5]\n})\n\ndf.loc[df['name'].str.contains('J'), 'score'].mean()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7932fde327d534ad3047b945f76c7dee76a8c1d | 2,602 | ipynb | Jupyter Notebook | Discussion Notebooks/Econ126_Discussion_Week_08.ipynb | t-hdd/econ126 | 17029937bd6c40e606d145f8d530728585c30a1d | [
"MIT"
] | null | null | null | Discussion Notebooks/Econ126_Discussion_Week_08.ipynb | t-hdd/econ126 | 17029937bd6c40e606d145f8d530728585c30a1d | [
"MIT"
] | null | null | null | Discussion Notebooks/Econ126_Discussion_Week_08.ipynb | t-hdd/econ126 | 17029937bd6c40e606d145f8d530728585c30a1d | [
"MIT"
] | null | null | null | 47.309091 | 459 | 0.683321 | [
[
[
"# Discussion: Week 8\n\nQuestions based on discussion of Lawrence Summers' article \"Some Skeptical Observations on Real Business Cycle Theory\" in the Fall 1986 issue of the Minneapolis Fed's *Quarterly Review* (Link to article: [https://www.minneapolisfed.org/research/qr/qr1043.pdf](https://www.minneapolisfed.org/research/qr/qr1043.pdf)).\n\n\n**Questions**\n\n1. What, according to Prescott's article, is the fundamental driving force behind the business cycle? \n2. According Summers' introduction, why is RBC theory counter to what most macroeconomists at the time of Summers' writing knew about the business cycle?\n3. In your own words, what are the four major objections that Summers raises to the RBC research program?\n4. What does Summers mean by *exchange failures*?",
"_____no_output_____"
],
[
"**Answers**\n\n1. Exogenous shocks to technology. <!-- answer -->\n2. Last sentence of the first paragraph in Summers' article: \"They assert that monetary policies have no effect on real activity, that fiscal policies influence the economy only through their incentive effects, and that economic fluctuations are caused entirely by supply rather than demand shocks.\" <!-- answer -->\n3. The choice of parameters used to perform simulations is questionable. There is no evidence that there are large, fundamental shocks to technology. Prescott makes no attempt to explain the movement of prices with the RBC model. The RBC model is based on a model of perfect competition and therefore cannot account for the breakdown of important trading relationships and heightened inefficincy (e.g., unemployment) during recessions <!-- answer -->\n4. By *exchange failures*, Summers means the inability of markets to adaquately match supply and demand. Unemployment is an example of an exchange failure. People are willing to work at the prevailing wage, but firms will not hire them. <!-- answer -->",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown"
]
] |
e79330c23d0ec7113e07071d0cb3aadc0241276e | 2,045 | ipynb | Jupyter Notebook | 10-Python Decorators/02-Decorators Homework.ipynb | vjsniper/Python | 5331fb5c4d922d3eca884ac4704dbf8efa3e25d5 | [
"Apache-2.0"
] | 4 | 2019-11-09T13:46:44.000Z | 2021-09-23T17:58:02.000Z | 10-Python Decorators/02-Decorators Homework.ipynb | vjsniper/Python | 5331fb5c4d922d3eca884ac4704dbf8efa3e25d5 | [
"Apache-2.0"
] | 1 | 2021-05-10T10:49:44.000Z | 2021-05-10T10:49:44.000Z | 10-Python Decorators/02-Decorators Homework.ipynb | vjsniper/Python | 5331fb5c4d922d3eca884ac4704dbf8efa3e25d5 | [
"Apache-2.0"
] | 6 | 2019-11-13T13:33:30.000Z | 2021-10-06T09:56:43.000Z | 47.55814 | 832 | 0.691443 | [
[
[
"# Decorators Homework (Optional)\n\nSince you won't run into decorators until further in your coding career, this homework is optional. Check out the Web Framework [Flask](http://flask.pocoo.org/). You can use Flask to create web pages with Python (as long as you know some HTML and CSS) and they use decorators a lot! Learn how they use [view decorators](http://flask.pocoo.org/docs/0.12/patterns/viewdecorators/). Don't worry if you don't completely understand everything about Flask, the main point of this optional homework is that you have an awareness of decorators in Web Frameworks. That way if you decide to become a \"Full-Stack\" Python Web Developer, you won't find yourself perplexed by decorators. You can also check out [Django](https://www.djangoproject.com/) another (and more popular) web framework for Python which is a bit more heavy duty.\n\nAlso for some additional info:\n\nA framework is a type of software library that provides generic functionality which can be extended by the programmer to build applications. Flask and Django are good examples of frameworks intended for web development.\n\nA framework is distinguished from a simple library or API. An API is a piece of software that a developer can use in his or her application. A framework is more encompassing: your entire application is structured around the framework (i.e. it provides the framework around which you build your software).\n\n## Great job!",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown"
]
] |
e7933140c32b908b451c1338eb6b007fd41180c6 | 481,953 | ipynb | Jupyter Notebook | docs-jupyter/2020-04-08-eic-jlab.ipynb | reikdas/awkward-1.0 | 40fdf1e6bf427dab7923934d6180e1621ea59e73 | [
"BSD-3-Clause"
] | null | null | null | docs-jupyter/2020-04-08-eic-jlab.ipynb | reikdas/awkward-1.0 | 40fdf1e6bf427dab7923934d6180e1621ea59e73 | [
"BSD-3-Clause"
] | null | null | null | docs-jupyter/2020-04-08-eic-jlab.ipynb | reikdas/awkward-1.0 | 40fdf1e6bf427dab7923934d6180e1621ea59e73 | [
"BSD-3-Clause"
] | null | null | null | 47.074917 | 8,760 | 0.56724 | [
[
[
"# Uproot and Awkward Arrays\n\n## Tutorial for Electron Ion Collider users\n\nJim Pivarski (Princeton University)",
"_____no_output_____"
],
[
"## Preliminaries\n\nThis is a draft of the demo presented to the Electron Ion Collider group on April 8, 2020. The repository with the final presentation is [jpivarski/2020-04-08-eic-jlab](https://github.com/jpivarski/2020-04-08-eic-jlab). This presentation was given before the final 1.0 version was released. Some interfaces may have changed. To run this notebook, make sure you have version 0.2.10 ([GitHub](https://github.com/scikit-hep/awkward-1.0/releases/tag/0.2.10), [pip](https://pypi.org/project/awkward1/0.2.10/)) by installing\n\n```bash\npip install 'awkward1==0.2.10'\n```\n\nThis notebook also depends on `uproot<4.0`, `particle`, `boost-histogram`, `matplotlib`, `mplhep`, `numba`, `pandas`, `numexpr`, and `autograd`, as well as a data file, `\"open_charm_18x275_10k.root\"`. See the final presentation for a suitable environment definition and a copy of the file.",
"_____no_output_____"
],
[
"## Table of contents\n\n* [Uproot: getting data](#uproot)\n - [Exploring a TFile](#Exploring-a-TFile)\n - [Exploring a TTree](#Exploring-a-TTree)\n - [Turning ROOT branches into NumPy arrays](#Turning-ROOT-branches-into-NumPy-arrays)\n - [Memory management; caching and iteration](#Memory-management;-caching-and-iteration)\n - [Jagged arrays (segue)](#Jagged-arrays-(segue))\n* [Awkward Array: manipulating data](#awkward)\n - [Using Uproot data in Awkward 1.0](#Using-Uproot-data-in-Awkward-1.0)\n - [Iteration in Python vs array-at-a-time operations](#Iteration-in-Python-vs-array-at-a-time-operations)\n - [Zipping arrays into records and projecting them back out](#Zipping-arrays-into-records-and-projecting-them-back-out)\n - [Filtering (cutting) events and particles with advanced selections](#Filtering-(cutting)-events-and-particles-with-advanced-selections)\n - [Flattening for plots and regularizing to NumPy for machine learning](#Flattening-for-plots-and-regularizing-to-NumPy-for-machine-learning)\n - [Broadcasting flat arrays and jagged arrays](#Broadcasting-flat-arrays-and-jagged-arrays)\n - [Combinatorics: cartesian and combinations](#Combinatorics:-cartesian-and-combinations)\n - [Reducing from combinations](#Reducing-from-combinations)\n - [Imperative, but still fast, programming in Numba](#Imperative,-but-still-fast,-programming-in-Numba)\n - [Grafting jagged data onto Pandas](#Grafting-jagged-data-onto-Pandas)\n - [NumExpr, Autograd, and other third-party libraries](#NumExpr,-Autograd,-and-other-third-party-libraries)",
"_____no_output_____"
],
[
"<br>\n<br>\n<a name=\"uproot\"></a>\n<img src=\"https://github.com/scikit-hep/uproot/raw/master/docs/source/logo-600px.png\" width=\"300\">",
"_____no_output_____"
],
[
"Uproot is a pure Python reimplementation of a significant part of ROOT I/O.\n\n<br>\n<img src=\"https://raw.githubusercontent.com/jpivarski/2019-07-29-dpf-python/master/img/abstraction-layers.png\" width=\"700\">\n<br>\n\nYou can read TTrees containing basic data types, STL vectors, strings, and some more complex data, especially if it was written with a high \"splitLevel\".\n\nYou can also read histograms and other objects into generic containers, but the C++ methods that give those objects functionality are not available.",
"_____no_output_____"
],
[
"## Exploring a TFile",
"_____no_output_____"
],
[
"Uproot was designed to be Pythonic, so the way we interact with ROOT files is different than it is in ROOT.",
"_____no_output_____"
]
],
[
[
"import uproot\nfile = uproot.open(\"open_charm_18x275_10k.root\")",
"_____no_output_____"
]
],
[
[
"A ROOT file may be thought of as a dict of key-value pairs, like a Python dict.",
"_____no_output_____"
]
],
[
[
"file.keys()",
"_____no_output_____"
],
[
"file.values()",
"_____no_output_____"
]
],
[
[
"**What's the `b` before the name?** All strings retrieved from ROOT are unencoded, which Python 3 treats differently from Python 2. In the near future, Uproot will automatically interpret all strings from ROOT as UTF-8 and this cosmetic issue will be gone.\n\n**What's the `;1` at the end of the name?** It's the cycle number, something ROOT uses to track multiple versions of an object. You can usually ignore it.",
"_____no_output_____"
],
[
"Nested directories are a dict of dicts.",
"_____no_output_____"
]
],
[
[
"file[\"events\"].keys()",
"_____no_output_____"
],
[
"file[\"events\"][\"tree\"]",
"_____no_output_____"
]
],
[
[
"But there are shortcuts:\n\n * use a `/` to navigate through the levels in a single string;\n * use `allkeys` to recursively show all keys in all directories.",
"_____no_output_____"
]
],
[
[
"file.allkeys()",
"_____no_output_____"
],
[
"file[\"events/tree\"]",
"_____no_output_____"
]
],
[
[
"## Exploring a TTree",
"_____no_output_____"
],
[
"A TTree can also be thought of as a dict of dicts, this time to navigate through TBranches.",
"_____no_output_____"
]
],
[
[
"tree = file[\"events/tree\"]\ntree.keys()",
"_____no_output_____"
]
],
[
[
"Often, the first thing I do when I look at a TTree is `show` to see how each branch is going to be interpreted.",
"_____no_output_____"
]
],
[
[
"print(\"branch name streamer (for complex data) interpretation in Python\")\nprint(\"==============================================================================\")\n\ntree.show()",
"branch name streamer (for complex data) interpretation in Python\n==============================================================================\nevt_id (no streamer) asdtype('>u8')\nevt_true_q2 (no streamer) asdtype('>f8')\nevt_true_x (no streamer) asdtype('>f8')\nevt_true_y (no streamer) asdtype('>f8')\nevt_true_w2 (no streamer) asdtype('>f8')\nevt_true_nu (no streamer) asdtype('>f8')\nevt_has_dis_info (no streamer) asdtype('int8')\nevt_prt_count (no streamer) asdtype('>u8')\nevt_weight (no streamer) asdtype('>f8')\nid (no streamer) asjagged(asdtype('>u8'))\npdg (no streamer) asjagged(asdtype('>i8'))\ntrk_id (no streamer) asjagged(asdtype('>f8'))\ncharge (no streamer) asjagged(asdtype('>f8'))\ndir_x (no streamer) asjagged(asdtype('>f8'))\ndir_y (no streamer) asjagged(asdtype('>f8'))\ndir_z (no streamer) asjagged(asdtype('>f8'))\np (no streamer) asjagged(asdtype('>f8'))\npx (no streamer) asjagged(asdtype('>f8'))\npy (no streamer) asjagged(asdtype('>f8'))\npz (no streamer) asjagged(asdtype('>f8'))\ntot_e (no streamer) asjagged(asdtype('>f8'))\nm (no streamer) asjagged(asdtype('>f8'))\ntime (no streamer) asjagged(asdtype('>f8'))\nis_beam (no streamer) asjagged(asdtype('bool'))\nis_stable (no streamer) asjagged(asdtype('bool'))\ngen_code (no streamer) asjagged(asdtype('bool'))\nmother_id (no streamer) asjagged(asdtype('>u8'))\nmother_second_id (no streamer) asjagged(asdtype('>u8'))\nhas_pol_info (no streamer) asjagged(asdtype('>f8'))\npol_x (no streamer) asjagged(asdtype('>f8'))\npol_y (no streamer) asjagged(asdtype('>f8'))\npol_z (no streamer) asjagged(asdtype('>f8'))\nhas_vtx_info (no streamer) asjagged(asdtype('bool'))\nvtx_id (no streamer) asjagged(asdtype('>u8'))\nvtx_x (no streamer) asjagged(asdtype('>f8'))\nvtx_y (no streamer) asjagged(asdtype('>f8'))\nvtx_z (no streamer) asjagged(asdtype('>f8'))\nvtx_t (no streamer) asjagged(asdtype('>f8'))\nhas_smear_info (no streamer) asjagged(asdtype('bool'))\nsmear_has_e (no streamer) asjagged(asdtype('bool'))\nsmear_has_p (no streamer) asjagged(asdtype('bool'))\nsmear_has_pid (no streamer) asjagged(asdtype('bool'))\nsmear_has_vtx (no streamer) asjagged(asdtype('bool'))\nsmear_has_any_eppid (no streamer) asjagged(asdtype('bool'))\nsmear_orig_tot_e (no streamer) asjagged(asdtype('>f8'))\nsmear_orig_p (no streamer) asjagged(asdtype('>f8'))\nsmear_orig_px (no streamer) asjagged(asdtype('>f8'))\nsmear_orig_py (no streamer) asjagged(asdtype('>f8'))\nsmear_orig_pz (no streamer) asjagged(asdtype('>f8'))\nsmear_orig_vtx_x (no streamer) asjagged(asdtype('>f8'))\nsmear_orig_vtx_y (no streamer) asjagged(asdtype('>f8'))\nsmear_orig_vtx_z (no streamer) asjagged(asdtype('>f8'))\n"
]
],
[
[
"Most of the information you'd expect to find in a TTree is there. See [uproot.readthedocs.io](https://uproot.readthedocs.io/en/latest/ttree-handling.html) for a complete list.",
"_____no_output_____"
]
],
[
[
"tree.numentries",
"_____no_output_____"
],
[
"tree[\"evt_id\"].compressedbytes(), tree[\"evt_id\"].uncompressedbytes(), tree[\"evt_id\"].compressionratio()",
"_____no_output_____"
],
[
"tree[\"evt_id\"].numbaskets",
"_____no_output_____"
],
[
"[tree[\"evt_id\"].basket_entrystart(i) for i in range(3)]",
"_____no_output_____"
]
],
[
[
"## Turning ROOT branches into NumPy arrays",
"_____no_output_____"
],
[
"There are several methods for this; they differ only in convenience.",
"_____no_output_____"
]
],
[
[
"# TBranch → array\ntree[\"evt_id\"].array()",
"_____no_output_____"
],
[
"# TTree + branch name → array\ntree.array(\"evt_id\")",
"_____no_output_____"
],
[
"# TTree + branch names → arrays\ntree.arrays([\"evt_id\", \"evt_prt_count\"])",
"_____no_output_____"
],
[
"# TTree + branch name pattern(s) → arrays\ntree.arrays(\"evt_*\")",
"_____no_output_____"
],
[
"# TTree + branch name regex(s) → arrays\ntree.arrays(\"/evt_[A-Z_0-9]*/i\")",
"_____no_output_____"
]
],
[
[
"**Convenience #1:** turn the bytestrings into real strings (will soon be unnecessary).",
"_____no_output_____"
]
],
[
[
"tree.arrays(\"evt_*\", namedecode=\"utf-8\")",
"_____no_output_____"
]
],
[
[
"**Convenience #2:** output a tuple instead of a dict.",
"_____no_output_____"
]
],
[
[
"tree.arrays([\"evt_id\", \"evt_prt_count\"], outputtype=tuple)",
"_____no_output_____"
]
],
[
[
"... to use it in assignment:",
"_____no_output_____"
]
],
[
[
"evt_id, evt_prt_count = tree.arrays([\"evt_id\", \"evt_prt_count\"], outputtype=tuple)",
"_____no_output_____"
],
[
"evt_id",
"_____no_output_____"
]
],
[
[
"## Memory management; caching and iteration",
"_____no_output_____"
],
[
"The `array` methods read an entire branch into memory. Sometimes, you might not have enough memory to do that.\n\nThe simplest solution is to set `entrystart` (inclusive) and `entrystop` (exclusive) to read a small batch at a time.",
"_____no_output_____"
]
],
[
[
"tree.array(\"evt_id\", entrystart=500, entrystop=600)",
"_____no_output_____"
]
],
[
[
"Uproot is _not_ agressive about caching: if you call `arrays` many times (for many small batches), it will read from the file every time.\n\nYou can avoid frequent re-reading by assigning arrays to variables, but then you'd have to manage all those variables.\n\n**Instead, use explicit caching:**",
"_____no_output_____"
]
],
[
[
"# Make a cache with an acceptable limit.\ngigabyte_cache = uproot.ArrayCache(\"1 GB\")\n\n# Read the array from disk:\ntree.array(\"evt_id\", cache=gigabyte_cache)\n\n# Get the array from the cache:\ntree.array(\"evt_id\", cache=gigabyte_cache)",
"_____no_output_____"
]
],
[
[
"The advantage is that the same code can be used for first time and subsequent times. You can put this in a loop.\n\nNaturally, fetching from the cache is much faster than reading from disk (though our file isn't very big!).",
"_____no_output_____"
]
],
[
[
"%%timeit\n\ntree.arrays(\"*\")",
"554 ms ± 7.65 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)\n"
],
[
"%%timeit\n\ntree.arrays(\"*\", cache=gigabyte_cache)",
"2.14 ms ± 45.2 µs per loop (mean ± std. dev. of 7 runs, 1 loop each)\n"
]
],
[
[
"The value of an explicit cache is that you get to control it.",
"_____no_output_____"
]
],
[
[
"len(gigabyte_cache)",
"_____no_output_____"
],
[
"gigabyte_cache.clear()",
"_____no_output_____"
],
[
"len(gigabyte_cache)",
"_____no_output_____"
]
],
[
[
"Setting `entrystart` and `entrystop` can get annoying; we probably want to do it in a loop.\n\nThere's a method, `iterate`, for that.",
"_____no_output_____"
]
],
[
[
"for arrays in tree.iterate(\"evt_*\", entrysteps=1000):\n print({name: len(array) for name, array in arrays.items()})",
"{b'evt_id': 1000, b'evt_true_q2': 1000, b'evt_true_x': 1000, b'evt_true_y': 1000, b'evt_true_w2': 1000, b'evt_true_nu': 1000, b'evt_has_dis_info': 1000, b'evt_prt_count': 1000, b'evt_weight': 1000}\n{b'evt_id': 1000, b'evt_true_q2': 1000, b'evt_true_x': 1000, b'evt_true_y': 1000, b'evt_true_w2': 1000, b'evt_true_nu': 1000, b'evt_has_dis_info': 1000, b'evt_prt_count': 1000, b'evt_weight': 1000}\n{b'evt_id': 1000, b'evt_true_q2': 1000, b'evt_true_x': 1000, b'evt_true_y': 1000, b'evt_true_w2': 1000, b'evt_true_nu': 1000, b'evt_has_dis_info': 1000, b'evt_prt_count': 1000, b'evt_weight': 1000}\n{b'evt_id': 1000, b'evt_true_q2': 1000, b'evt_true_x': 1000, b'evt_true_y': 1000, b'evt_true_w2': 1000, b'evt_true_nu': 1000, b'evt_has_dis_info': 1000, b'evt_prt_count': 1000, b'evt_weight': 1000}\n{b'evt_id': 1000, b'evt_true_q2': 1000, b'evt_true_x': 1000, b'evt_true_y': 1000, b'evt_true_w2': 1000, b'evt_true_nu': 1000, b'evt_has_dis_info': 1000, b'evt_prt_count': 1000, b'evt_weight': 1000}\n{b'evt_id': 1000, b'evt_true_q2': 1000, b'evt_true_x': 1000, b'evt_true_y': 1000, b'evt_true_w2': 1000, b'evt_true_nu': 1000, b'evt_has_dis_info': 1000, b'evt_prt_count': 1000, b'evt_weight': 1000}\n{b'evt_id': 1000, b'evt_true_q2': 1000, b'evt_true_x': 1000, b'evt_true_y': 1000, b'evt_true_w2': 1000, b'evt_true_nu': 1000, b'evt_has_dis_info': 1000, b'evt_prt_count': 1000, b'evt_weight': 1000}\n{b'evt_id': 1000, b'evt_true_q2': 1000, b'evt_true_x': 1000, b'evt_true_y': 1000, b'evt_true_w2': 1000, b'evt_true_nu': 1000, b'evt_has_dis_info': 1000, b'evt_prt_count': 1000, b'evt_weight': 1000}\n{b'evt_id': 1000, b'evt_true_q2': 1000, b'evt_true_x': 1000, b'evt_true_y': 1000, b'evt_true_w2': 1000, b'evt_true_nu': 1000, b'evt_has_dis_info': 1000, b'evt_prt_count': 1000, b'evt_weight': 1000}\n{b'evt_id': 1000, b'evt_true_q2': 1000, b'evt_true_x': 1000, b'evt_true_y': 1000, b'evt_true_w2': 1000, b'evt_true_nu': 1000, b'evt_has_dis_info': 1000, b'evt_prt_count': 1000, b'evt_weight': 1000}\n"
]
],
[
[
"Keep in mind that this is a loop over _batches_, not _events_.\n\nWhat goes in the loop is code that applies to _arrays_.\n\nYou can also set the `entrysteps` to be a size in memory.",
"_____no_output_____"
]
],
[
[
"for arrays in tree.iterate(\"evt_*\", entrysteps=\"100 kB\"):\n print({name: len(array) for name, array in arrays.items()})",
"{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 544, b'evt_true_q2': 544, b'evt_true_x': 544, b'evt_true_y': 544, b'evt_true_w2': 544, b'evt_true_nu': 544, b'evt_has_dis_info': 544, b'evt_prt_count': 544, b'evt_weight': 544}\n"
]
],
[
[
"The same size in memory covers more events if you read fewer branches.",
"_____no_output_____"
]
],
[
[
"for arrays in tree.iterate(\"evt_id\", entrysteps=\"100 kB\"):\n print({name: len(array) for name, array in arrays.items()})",
"{b'evt_id': 10000}\n"
]
],
[
[
"This `TTree.iterate` method is similar in form to the `uproot.iterate` function, which iterates in batches over a collection of files.",
"_____no_output_____"
]
],
[
[
"for arrays in uproot.iterate([\"open_charm_18x275_10k.root\",\n \"open_charm_18x275_10k.root\",\n \"open_charm_18x275_10k.root\"], \"events/tree\", \"evt_*\", entrysteps=\"100 kB\"):\n print({name: len(array) for name, array in arrays.items()})",
"{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 544, b'evt_true_q2': 544, b'evt_true_x': 544, b'evt_true_y': 544, b'evt_true_w2': 544, b'evt_true_nu': 544, b'evt_has_dis_info': 544, b'evt_prt_count': 544, b'evt_weight': 544}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 544, b'evt_true_q2': 544, b'evt_true_x': 544, b'evt_true_y': 544, b'evt_true_w2': 544, b'evt_true_nu': 544, b'evt_has_dis_info': 544, b'evt_prt_count': 544, b'evt_weight': 544}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 1576, b'evt_true_q2': 1576, b'evt_true_x': 1576, b'evt_true_y': 1576, b'evt_true_w2': 1576, b'evt_true_nu': 1576, b'evt_has_dis_info': 1576, b'evt_prt_count': 1576, b'evt_weight': 1576}\n{b'evt_id': 544, b'evt_true_q2': 544, b'evt_true_x': 544, b'evt_true_y': 544, b'evt_true_w2': 544, b'evt_true_nu': 544, b'evt_has_dis_info': 544, b'evt_prt_count': 544, b'evt_weight': 544}\n"
]
],
[
[
"## Jagged arrays (segue)",
"_____no_output_____"
],
[
"Most of the branches in this file have an \"asjagged\" interpretation, instead of \"asdtype\" (NumPy).",
"_____no_output_____"
]
],
[
[
"tree[\"evt_id\"].interpretation",
"_____no_output_____"
],
[
"tree[\"pdg\"].interpretation",
"_____no_output_____"
]
],
[
[
"This means that they have multiple values per entry.",
"_____no_output_____"
]
],
[
[
"tree[\"pdg\"].array()",
"_____no_output_____"
]
],
[
[
"Jagged arrays (lists of variable-length sublists) are very common in particle physics, and surprisingly uncommon in other fields.\n\nWe need them because we almost always have a variable number of particles per event.",
"_____no_output_____"
]
],
[
[
"from particle import Particle # https://github.com/scikit-hep/particle\n\ncounter = 0\nfor event in tree[\"pdg\"].array():\n print(len(event), \"particles:\", \" \".join(Particle.from_pdgid(x).name for x in event))\n counter += 1\n if counter == 30:\n break",
"51 particles: e- pi+ pi- K- pi+ pi- pi- pi+ pi+ pi+ gamma gamma K(L)0 K+ pi- K(L)0 gamma gamma gamma gamma pi+ pi- pi+ gamma gamma p pi- pi+ K+ pi- pi- K+ K- gamma gamma pi+ pi- K+ pi- pi+ K(L)0 K(L)0 gamma gamma pi+ pi- pi+ gamma gamma gamma gamma\n26 particles: e- pi+ pi- n~ n gamma pi- pi+ gamma gamma pi+ gamma gamma gamma gamma gamma K(L)0 gamma gamma gamma gamma pi- pi+ pi- gamma gamma\n27 particles: e- n p pi+ pi+ pi+ pi- pi- pi- pi- pi- pi+ pi- gamma gamma gamma pi+ K+ K- pi+ gamma gamma gamma gamma gamma gamma pi-\n28 particles: e- pi+ pi- nu(mu) mu+ gamma gamma pi- pi+ n gamma gamma n pi- p~ pi+ gamma gamma pi+ pi- K- K(L)0 gamma gamma gamma gamma gamma gamma\n30 particles: e- pi+ pi- pi+ pi- n gamma gamma K- pi+ n pi- pi+ gamma n~ p~ pi+ K(L)0 gamma gamma pi- gamma gamma pi- pi+ gamma gamma K+ pi- gamma\n12 particles: e- gamma gamma gamma gamma gamma gamma gamma gamma gamma gamma gamma\n25 particles: pi- K- K+ pi- gamma gamma pi- gamma gamma pi- K(L)0 K(L)0 gamma gamma gamma gamma K- pi+ gamma gamma gamma gamma gamma gamma gamma\n4 particles: e- nu(e) e+ n\n57 particles: e- pi+ n p K- K+ pi+ pi+ pi- gamma gamma K(L)0 p~ n pi- n~ pi+ pi+ pi- gamma gamma K(L)0 K+ K- K+ K- gamma gamma pi+ gamma gamma gamma gamma gamma gamma pi- gamma gamma pi- e+ e- gamma pi+ pi- gamma gamma gamma gamma gamma gamma e+ e- gamma gamma gamma gamma gamma\n40 particles: e- n K- K+ gamma gamma K+ pi- gamma gamma gamma pi+ n~ pi- K- pi+ gamma gamma gamma gamma gamma gamma gamma gamma pi- K(L)0 pi- pi+ gamma gamma gamma gamma gamma gamma gamma gamma e+ e- gamma gamma\n16 particles: e- pi- n pi- pi+ gamma pi+ pi- K(L)0 pi+ gamma gamma K+ K- gamma gamma\n16 particles: e- K(L)0 pi- K(L)0 pi+ pi- gamma gamma nu(e)~ e- K+ K(L)0 gamma gamma gamma gamma\n79 particles: e- pi+ pi- pi+ pi- K- pi+ pi- gamma gamma gamma n pi+ gamma gamma pi+ pi- K(L)0 p pi+ p~ gamma gamma gamma pi- pi+ pi+ pi- K+ gamma gamma pi+ pi- gamma gamma K+ gamma gamma pi+ pi- gamma gamma gamma pi- gamma gamma n pi+ pi- gamma gamma gamma gamma pi+ pi- gamma gamma gamma gamma K- pi+ gamma gamma pi- gamma gamma gamma gamma gamma gamma gamma gamma p~ pi+ K(L)0 gamma gamma pi+ pi-\n34 particles: e- n pi- pi+ K+ pi+ pi+ pi+ pi- pi+ pi- pi- pi- K(L)0 pi+ pi+ pi- pi+ pi- gamma gamma K+ gamma gamma K(L)0 gamma gamma gamma gamma pi+ pi- pi- gamma gamma\n24 particles: e- K- K+ pi- pi- pi+ gamma gamma gamma gamma gamma gamma gamma pi- K- pi+ pi+ pi- gamma gamma gamma gamma gamma gamma\n32 particles: e- n pi+ pi- pi+ pi- pi+ pi- pi- gamma gamma gamma gamma gamma pi+ pi- e+ e- gamma K+ pi- K- pi+ gamma gamma gamma gamma gamma gamma gamma gamma gamma\n37 particles: e- pi- pi+ pi+ pi- n n~ pi+ p pi+ pi- K+ K- gamma gamma gamma gamma pi+ pi- gamma gamma pi- pi- pi+ gamma gamma p~ gamma e+ e- gamma gamma gamma gamma gamma gamma gamma\n63 particles: e- pi- n pi+ K- K+ pi+ pi- K(L)0 n pi- n~ pi+ n K(L)0 K(L)0 pi- gamma gamma gamma gamma K- pi- gamma gamma pi+ pi- gamma gamma gamma K(L)0 K- pi+ pi+ pi+ pi- pi+ pi- p pi- n~ pi+ gamma gamma K(L)0 pi+ gamma gamma gamma gamma gamma gamma gamma gamma gamma gamma gamma gamma K- K+ pi- gamma gamma\n37 particles: e- n p~ pi+ pi- pi+ K- pi+ gamma gamma pi+ nu(e) e+ K- gamma gamma pi- K- gamma gamma K(L)0 K+ pi- pi+ pi- pi+ pi- gamma gamma gamma gamma gamma gamma gamma gamma gamma gamma\n5 particles: e- gamma pi- gamma gamma\n26 particles: e- pi- n n~ pi+ pi+ nu(mu) mu+ n pi- gamma gamma gamma gamma gamma gamma gamma K(L)0 pi- gamma gamma gamma pi+ gamma gamma K(L)0\n39 particles: e- pi+ n pi- K+ pi- gamma pi+ pi- gamma gamma pi- pi- pi+ pi- pi+ pi+ pi- gamma gamma gamma gamma gamma K+ K- gamma gamma pi+ pi- gamma gamma gamma gamma gamma gamma gamma gamma pi+ pi-\n58 particles: e- pi+ pi- pi+ pi- p~ pi- pi+ n~ pi- gamma pi+ pi+ pi- K(L)0 K- K+ pi- n n~ pi+ pi- pi+ pi+ pi+ pi- pi+ pi- pi+ pi- gamma gamma pi- gamma K(L)0 pi+ pi- gamma gamma gamma gamma gamma gamma pi+ pi- n gamma gamma pi+ gamma gamma gamma gamma gamma gamma n gamma gamma\n27 particles: e- n pi+ pi- K- pi+ gamma gamma gamma n K- pi- pi- pi+ gamma gamma gamma gamma pi+ gamma gamma gamma gamma gamma gamma gamma gamma\n23 particles: e- pi+ pi+ p pi- pi+ gamma gamma gamma pi+ pi- K(L)0 pi+ gamma gamma p~ pi- pi+ nu(e)~ e- gamma K+ pi-\n20 particles: e- n pi+ pi- gamma gamma gamma K(L)0 pi- pi- e+ e- gamma gamma gamma gamma gamma gamma pi+ gamma\n38 particles: e- K- pi+ pi- gamma gamma K(L)0 pi+ pi- pi+ p p~ pi- pi+ pi+ gamma gamma gamma gamma gamma gamma gamma gamma gamma gamma nu(mu)~ mu- gamma gamma pi- pi+ pi- gamma gamma gamma gamma gamma gamma\n70 particles: e- n pi- K+ n~ pi+ pi- pi- K- pi+ K(L)0 pi+ pi- p p~ pi- pi+ n pi+ pi- pi- pi+ gamma gamma n~ gamma gamma pi+ gamma gamma gamma gamma K(L)0 pi- gamma gamma gamma gamma gamma gamma gamma gamma gamma gamma gamma gamma n~ pi+ gamma pi+ pi+ gamma gamma n gamma gamma gamma gamma p~ pi+ pi+ pi- K- gamma gamma gamma gamma gamma gamma gamma\n14 particles: e- pi- gamma pi+ pi- pi+ pi+ K(L)0 gamma gamma gamma gamma gamma gamma\n29 particles: e- K- pi- K+ pi+ pi+ pi- gamma gamma pi- pi+ K(L)0 gamma gamma gamma gamma K(L)0 gamma gamma gamma pi+ pi- pi+ pi- K(L)0 K(L)0 K(L)0 gamma gamma\n"
]
],
[
[
"Although you can iterate over jagged arrays with for loops, the idiomatic and faster way to do it is with array-at-a-time functions.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nvtx_x, vtx_y, vtx_z = tree.arrays([\"vtx_[xyz]\"], outputtype=tuple)\n\nvtx_dist = np.sqrt(vtx_x**2 + vtx_y**2 + vtx_z**2)\n\nvtx_dist",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nimport mplhep as hep # https://github.com/scikit-hep/mplhep\nimport boost_histogram as bh # https://github.com/scikit-hep/boost-histogram\n\nvtx_hist = bh.Histogram(bh.axis.Regular(100, 0, 0.1))\n\nvtx_hist.fill(vtx_dist.flatten())\n\nhep.histplot(vtx_hist)",
"_____no_output_____"
],
[
"vtx_dist > 0.01",
"_____no_output_____"
],
[
"pdg = tree[\"pdg\"].array()\npdg[vtx_dist > 0.01]",
"_____no_output_____"
],
[
"counter = 0\nfor event in pdg[vtx_dist > 0.10]:\n print(len(event), \"particles:\", \" \".join(Particle.from_pdgid(x).name for x in event))\n counter += 1\n if counter == 30:\n break",
"16 particles: p pi- K+ pi- pi- pi+ pi- pi+ K(L)0 pi+ pi- pi+ gamma gamma gamma gamma\n9 particles: pi- pi+ K(L)0 gamma gamma gamma gamma pi+ pi-\n8 particles: pi- pi- K+ K- pi+ gamma gamma pi-\n7 particles: nu(mu) mu+ pi+ pi- K- gamma gamma\n8 particles: K- pi+ p~ pi+ pi- pi+ K+ pi-\n6 particles: gamma gamma gamma gamma gamma gamma\n2 particles: K- K(L)0\n1 particles: n\n0 particles: \n12 particles: K- pi+ pi- K(L)0 pi- pi+ gamma gamma gamma gamma gamma gamma\n7 particles: pi+ pi- K(L)0 pi+ K- gamma gamma\n10 particles: pi+ pi- nu(e)~ e- K+ K(L)0 gamma gamma gamma gamma\n11 particles: n pi+ K- gamma gamma gamma gamma p~ pi+ pi+ pi-\n6 particles: K+ pi+ pi- pi- gamma gamma\n16 particles: pi- pi- pi+ gamma gamma pi- K- pi+ pi+ pi- gamma gamma gamma gamma gamma gamma\n6 particles: K+ pi- gamma gamma gamma gamma\n10 particles: pi- pi+ p~ e+ e- gamma gamma gamma gamma gamma\n18 particles: n pi- n~ pi+ n pi+ pi- pi+ pi- p pi- n~ pi+ pi+ gamma gamma gamma gamma\n11 particles: nu(e) e+ K- K+ pi- pi+ pi- gamma gamma gamma gamma\n4 particles: gamma pi- gamma gamma\n13 particles: nu(mu) mu+ gamma gamma K(L)0 pi- gamma gamma gamma pi+ gamma gamma K(L)0\n5 particles: pi- K+ K- pi+ pi-\n8 particles: n~ pi- n gamma gamma n gamma gamma\n13 particles: K- pi- pi- pi+ pi+ gamma gamma gamma gamma gamma gamma gamma gamma\n6 particles: pi+ pi+ e- gamma K+ pi-\n12 particles: pi+ pi- pi- pi- e+ e- gamma gamma gamma gamma gamma pi+\n8 particles: gamma gamma pi+ pi- gamma gamma gamma gamma\n24 particles: n pi+ pi+ n~ pi+ pi+ pi+ n gamma gamma gamma gamma p~ pi+ pi+ pi- K- gamma gamma gamma gamma gamma gamma gamma\n9 particles: pi+ pi+ K(L)0 gamma gamma gamma gamma gamma gamma\n2 particles: pi+ pi-\n"
],
[
"Particle.from_string(\"p~\")",
"_____no_output_____"
],
[
"Particle.from_string(\"p~\").pdgid",
"_____no_output_____"
],
[
"is_antiproton = (pdg == Particle.from_string(\"p~\").pdgid)\nis_antiproton",
"_____no_output_____"
],
[
"hep.histplot(bh.Histogram(bh.axis.Regular(100, 0, 0.1)).fill(\n vtx_dist[is_antiproton].flatten()\n))",
"_____no_output_____"
]
],
[
[
"But that's a topic for the next section.",
"_____no_output_____"
],
[
"<br>\n<br>\n<a name=\"awkward\"></a>\n<img src=\"https://github.com/scikit-hep/awkward-1.0/raw/master/docs-img/logo/logo-600px.png\" width=\"400\">",
"_____no_output_____"
],
[
"Awkward Array is a library for manipulating arbitrary data structures in a NumPy-like way.\n\nThe idea is that you have a large number of identically typed, nested objects in variable-length lists.\n\n<img src=\"../docs-img/diagrams/cartoon-schematic.png\" width=\"600\">",
"_____no_output_____"
],
[
"## Using Uproot data in Awkward 1.0\n\nAwkward Array is in transition from\n\n * version 0.x, which is in use at the LHC but has revealed some design flaws, to\n * version 1.x, which is well-architected and has completed development, but is not in widespread use yet.\n\nAwkward 1.0 hasn't been incorporated into Uproot yet, which is how it will get in front of most users.\n\nSince development is complete and the interface is (intentionally) different, I thought it better to show you the new version.",
"_____no_output_____"
]
],
[
[
"import awkward1 as ak",
"_____no_output_____"
]
],
[
[
"Old-style arrays can be converted into the new framework with [ak.from_awkward0](https://awkward-array.readthedocs.io/en/latest/_auto/ak.from_awkward0.html). This won't be a necessary step for long.",
"_____no_output_____"
]
],
[
[
"?ak.from_awkward0",
"_____no_output_____"
],
[
"?ak.to_awkward0",
"_____no_output_____"
],
[
"ak.from_awkward0(tree.array(\"pdg\"))",
"_____no_output_____"
],
[
"arrays = {name: ak.from_awkward0(array) for name, array in tree.arrays(namedecode=\"utf-8\").items()}\narrays",
"_____no_output_____"
],
[
"?ak.Array",
"_____no_output_____"
]
],
[
[
"## Iteration in Python vs array-at-a-time operations\n\nAs before, you _can_ iterate over them in Python, but only do that for small-scale exploration.",
"_____no_output_____"
]
],
[
[
"%%timeit -n1 -r1\n\nvtx_dist = []\nfor xs, xy, xz in zip(arrays[\"vtx_x\"], arrays[\"vtx_y\"], arrays[\"vtx_z\"]):\n out = []\n for x, y, z in zip(xs, xy, xz):\n out.append(np.sqrt(x**2 + y**2 + z**2))\n vtx_dist.append(out)",
"11 s ± 0 ns per loop (mean ± std. dev. of 1 run, 1 loop each)\n"
],
[
"%%timeit -n100 -r1\n\nvtx_dist = np.sqrt(arrays[\"vtx_x\"]**2 + arrays[\"vtx_y\"]**2 + arrays[\"vtx_z\"]**2)",
"23.8 ms ± 0 ns per loop (mean ± std. dev. of 1 run, 100 loops each)\n"
]
],
[
[
"## Zipping arrays into records and projecting them back out\n\nInstead of having all these arrays floating around, let's [ak.zip](https://awkward-array.readthedocs.io/en/latest/_auto/ak.zip.html) them into a structure.\n\n(This is the sort of thing that a framework developer might do for the data analysts.)",
"_____no_output_____"
]
],
[
[
"?ak.zip",
"_____no_output_____"
],
[
"events = ak.zip({\"id\": arrays[\"evt_id\"],\n \"true\": ak.zip({\"q2\": arrays[\"evt_true_q2\"],\n \"x\": arrays[\"evt_true_x\"],\n \"y\": arrays[\"evt_true_y\"],\n \"w2\": arrays[\"evt_true_w2\"],\n \"nu\": arrays[\"evt_true_nu\"]}),\n \"has_dis_info\": arrays[\"evt_has_dis_info\"],\n \"prt_count\": arrays[\"evt_prt_count\"],\n \"prt\": ak.zip({\"id\": arrays[\"id\"],\n \"pdg\": arrays[\"pdg\"],\n \"trk_id\": arrays[\"trk_id\"],\n \"charge\": arrays[\"charge\"],\n \"dir\": ak.zip({\"x\": arrays[\"dir_x\"],\n \"y\": arrays[\"dir_y\"],\n \"z\": arrays[\"dir_z\"]}, with_name=\"point3\"),\n \"p\": arrays[\"p\"],\n \"px\": arrays[\"px\"],\n \"py\": arrays[\"py\"],\n \"pz\": arrays[\"pz\"],\n \"m\": arrays[\"m\"],\n \"time\": arrays[\"time\"],\n \"is_beam\": arrays[\"is_beam\"],\n \"is_stable\": arrays[\"is_stable\"],\n \"gen_code\": arrays[\"gen_code\"],\n \"mother\": ak.zip({\"id\": arrays[\"mother_id\"],\n \"second_id\": arrays[\"mother_second_id\"]}),\n \"pol\": ak.zip({\"has_info\": arrays[\"has_pol_info\"],\n \"x\": arrays[\"pol_x\"],\n \"y\": arrays[\"pol_y\"],\n \"z\": arrays[\"pol_z\"]}, with_name=\"point3\"),\n \"vtx\": ak.zip({\"has_info\": arrays[\"has_vtx_info\"],\n \"id\": arrays[\"vtx_id\"],\n \"x\": arrays[\"vtx_x\"],\n \"y\": arrays[\"vtx_y\"],\n \"z\": arrays[\"vtx_z\"],\n \"t\": arrays[\"vtx_t\"]}, with_name=\"point3\"),\n \"smear\": ak.zip({\"has_info\": arrays[\"has_smear_info\"],\n \"has_e\": arrays[\"smear_has_e\"],\n \"has_p\": arrays[\"smear_has_p\"],\n \"has_pid\": arrays[\"smear_has_pid\"],\n \"has_vtx\": arrays[\"smear_has_vtx\"],\n \"has_any_eppid\": arrays[\"smear_has_any_eppid\"],\n \"orig\": ak.zip({\"tot_e\": arrays[\"smear_orig_tot_e\"],\n \"p\": arrays[\"smear_orig_p\"],\n \"px\": arrays[\"smear_orig_px\"],\n \"py\": arrays[\"smear_orig_py\"],\n \"pz\": arrays[\"smear_orig_pz\"],\n \"vtx\": ak.zip({\"x\": arrays[\"smear_orig_vtx_x\"],\n \"y\": arrays[\"smear_orig_vtx_y\"],\n \"z\": arrays[\"smear_orig_vtx_z\"]},\n with_name=\"point3\")})})}, with_name=\"particle\")},\n depthlimit=1)",
"_____no_output_____"
],
[
"?ak.type",
"_____no_output_____"
],
[
"ak.type(events)",
"_____no_output_____"
]
],
[
[
"The type written with better formatting:\n\n```javascript\n10000 * {\"id\": uint64,\n \"true\": {\"q2\": float64,\n \"x\": float64,\n \"y\": float64,\n \"w2\": float64,\n \"nu\": float64},\n \"has_dis_info\": int8,\n \"prt_count\": uint64,\n\n \"prt\": var * particle[\"id\": uint64,\n \"pdg\": int64,\n \"trk_id\": float64,\n \"charge\": float64,\n \"dir\": point3[\"x\": float64, \"y\": float64, \"z\": float64],\n \"p\": float64,\n \"px\": float64,\n \"py\": float64,\n \"pz\": float64,\n \"m\": float64,\n \"time\": float64,\n \"is_beam\": bool,\n \"is_stable\": bool,\n \"gen_code\": bool,\n \"mother\": {\"id\": uint64, \"second_id\": uint64},\n \"pol\": point3[\"has_info\": float64,\n \"x\": float64,\n \"y\": float64,\n \"z\": float64],\n \"vtx\": point3[\"has_info\": bool,\n \"id\": uint64,\n \"x\": float64,\n \"y\": float64,\n \"z\": float64,\n \"t\": float64],\n \"smear\": {\"has_info\": bool,\n \"has_e\": bool,\n \"has_p\": bool,\n \"has_pid\": bool,\n \"has_vtx\": bool,\n \"has_any_eppid\": bool,\n \"orig\": {\"tot_e\":\n float64,\n \"p\": float64,\n \"px\": float64,\n \"py\": float64,\n \"pz\": float64,\n \"vtx\": point3[\"x\": float64,\n \"y\": float64,\n \"z\": float64]}}]}\n```",
"_____no_output_____"
],
[
"It means that these are now nested objects.",
"_____no_output_____"
]
],
[
[
"?ak.to_list",
"_____no_output_____"
],
[
"ak.to_list(events[0].prt[0])",
"_____no_output_____"
],
[
"ak.to_list(events[-1].prt[:10].smear.orig.vtx)",
"_____no_output_____"
]
],
[
[
"Alternatively,",
"_____no_output_____"
]
],
[
[
"ak.to_list(events[-1, \"prt\", :10, \"smear\", \"orig\", \"vtx\"])",
"_____no_output_____"
]
],
[
[
"<img src=\"../docs-img/diagrams/how-it-works-muons.png\" width=\"1000\">",
"_____no_output_____"
],
[
"\"Zipping\" arrays together into structures costs nothing (time does not scale with size of data), nor does \"projecting\" arrays out of structures.",
"_____no_output_____"
]
],
[
[
"events.prt.px",
"_____no_output_____"
],
[
"events.prt.py",
"_____no_output_____"
],
[
"events.prt.pz",
"_____no_output_____"
]
],
[
[
"This is called \"projection\" because the request for `\"pz\"` is slicing through arrays and jagged arrays.\n\nThe following are equivalent:",
"_____no_output_____"
]
],
[
[
"events[999, \"prt\", 12, \"pz\"]",
"_____no_output_____"
],
[
"events[\"prt\", 999, 12, \"pz\"]",
"_____no_output_____"
],
[
"events[999, \"prt\", \"pz\", 12]",
"_____no_output_____"
],
[
"events[\"prt\", 999, \"pz\", 12]",
"_____no_output_____"
]
],
[
[
"This \"object oriented view\" is a conceptual aid, not a constraint on computation. It's very fluid.\n\nMoreover, we can add behaviors to named records, like methods in object oriented programming. (See [ak.behavior](https://awkward-array.readthedocs.io/en/latest/ak.behavior.html) in the documentation.)\n\n(This is the sort of thing that a framework developer might do for the data analysts.)",
"_____no_output_____"
]
],
[
[
"def point3_absolute(data):\n return np.sqrt(data.x**2 + data.y**2 + data.z**2)\n\ndef point3_distance(left, right):\n return np.sqrt((left.x - right.x)**2 + (left.y - right.y)**2 + (left.z - right.z)**2)\n\nak.behavior[np.absolute, \"point3\"] = point3_absolute\nak.behavior[np.subtract, \"point3\", \"point3\"] = point3_distance",
"_____no_output_____"
],
[
"# Absolute value of all smear origin vertexes\nabs(events.prt.smear.orig.vtx)",
"_____no_output_____"
],
[
"# Absolute value of the last smear origin vertex\nabs(events[-1].prt[-1].smear.orig.vtx)",
"_____no_output_____"
],
[
"# Distance between each particle vertex and itself\nevents.prt.vtx - events.prt.vtx",
"_____no_output_____"
],
[
"# Distances between the first and last particle vertexes in the first 100 events\nevents.prt.vtx[:100, 0] - events.prt.vtx[:100, -1]",
"_____no_output_____"
]
],
[
[
"More methods can be added by declaring subclasses of arrays and records.",
"_____no_output_____"
]
],
[
[
"class ParticleRecord(ak.Record): \n @property\n def pt(self):\n return np.sqrt(self.px**2 + self.py**2)\n\nclass ParticleArray(ak.Array):\n __name__ = \"Array\" # prevent it from writing <ParticleArray [...] type='...'>\n # instead of <Array [...] type='...'>\n @property\n def pt(self):\n return np.sqrt(self.px**2 + self.py**2)\n\nak.behavior[\"particle\"] = ParticleRecord\nak.behavior[\"*\", \"particle\"] = ParticleArray",
"_____no_output_____"
],
[
"type(events[0].prt[0])",
"_____no_output_____"
],
[
"events[0].prt[0]",
"_____no_output_____"
],
[
"events[0].prt[0].pt",
"_____no_output_____"
],
[
"type(events.prt)",
"_____no_output_____"
],
[
"events.prt",
"_____no_output_____"
],
[
"events.prt.pt",
"_____no_output_____"
]
],
[
[
"## Filtering (cutting) events and particles with advanced selections\n\nNumPy has a versatile selection mechanism:\n\n<img src=\"https://raw.githubusercontent.com/jpivarski/2019-07-23-codas-hep/master/img/numpy-slicing.png\" width=\"300\">\n\nThe same expressions apply to Awkward Arrays, and more.",
"_____no_output_____"
]
],
[
[
"# First particle momentum in the first 5 events\nevents.prt.p[:5, 0]",
"_____no_output_____"
],
[
"# First two particles in all events\nevents.prt.pdg[:, :2]",
"_____no_output_____"
],
[
"# First direction of the last event\nevents.prt.dir[-1, 0]",
"_____no_output_____"
]
],
[
[
"NumPy also lets you filter (cut) using an array of booleans.",
"_____no_output_____"
]
],
[
[
"events.prt_count > 100",
"_____no_output_____"
],
[
"np.count_nonzero(events.prt_count > 100)",
"_____no_output_____"
],
[
"events[events.prt_count > 100]",
"_____no_output_____"
]
],
[
[
"One dimension can be selected with an array while another is selected with a slice.",
"_____no_output_____"
]
],
[
[
"# Select events with at least two particles, then select the first two particles\nevents.prt[events.prt_count >= 2, :2]",
"_____no_output_____"
]
],
[
[
"This can be a good way to avoid errors from trying to select what isn't there.",
"_____no_output_____"
]
],
[
[
"try:\n events.prt[:, 0]\nexcept Exception as err:\n print(type(err).__name__, str(err))",
"ValueError in ListArray64 attempting to get 0, index out of range\n"
],
[
"events.prt[events.prt_count > 0, 0]",
"_____no_output_____"
]
],
[
[
"See also [awkward-array.readthedocs.io](https://awkward-array.readthedocs.io/) for a list of operations like [ak.num](https://awkward-array.readthedocs.io/en/latest/_auto/ak.num.html):",
"_____no_output_____"
]
],
[
[
"?ak.num",
"_____no_output_____"
],
[
"ak.num(events.prt), events.prt_count",
"_____no_output_____"
]
],
[
[
"You can even use an array of integers to select a set of indexes at once.",
"_____no_output_____"
]
],
[
[
"# First and last particle in each event that has at least two\nevents.prt.pdg[ak.num(events.prt) >= 2][:, [0, -1]]",
"_____no_output_____"
]
],
[
[
"But beyond NumPy, we can also use arrays of nested lists as boolean or integer selectors.",
"_____no_output_____"
]
],
[
[
"# Array of lists of True and False\nabs(events.prt.vtx) > 0.10",
"_____no_output_____"
],
[
"# Particles that have vtx > 0.10 for all events (notice that there's still 10000)\nevents.prt[abs(events.prt.vtx) > 0.10]",
"_____no_output_____"
]
],
[
[
"See [awkward-array.readthedocs.io](https://awkward-array.readthedocs.io/) for more, but there are functions like [ak.max](https://awkward-array.readthedocs.io/en/latest/_auto/ak.max.html), which picks the maximum in a groups.\n\n * With `axis=0`, the group is the set of all events.\n * With `axis=1`, the groups are particles in each event.",
"_____no_output_____"
]
],
[
[
"?ak.max",
"_____no_output_____"
],
[
"ak.max(abs(events.prt.vtx), axis=1)",
"_____no_output_____"
],
[
"# Selects *events* that have a maximum *particle vertex* greater than 100\nevents[ak.max(abs(events.prt.vtx), axis=1) > 100]",
"_____no_output_____"
]
],
[
[
"The difference between \"select particles\" and \"select events\" is the number of jagged dimensions in the array; \"reducers\" like ak.max reduce the dimensionality by one.\n\nThere are other reducers like ak.any, ak.all, ak.sum...",
"_____no_output_____"
]
],
[
[
"?ak.sum",
"_____no_output_____"
],
[
"# Is this particle an antineutron?\nevents.prt.pdg == Particle.from_string(\"n~\").pdgid",
"_____no_output_____"
],
[
"# Are any particles in the event antineutrons?\nak.any(events.prt.pdg == Particle.from_string(\"n~\").pdgid, axis=1)",
"_____no_output_____"
],
[
"# Select events that contain an antineutron\nevents[ak.any(events.prt.pdg == Particle.from_string(\"n~\").pdgid, axis=1)]",
"_____no_output_____"
]
],
[
[
"We can use these techniques to make subcollections for specific particle types and attach them to the same `events` array for easy access.",
"_____no_output_____"
]
],
[
[
"events.prt[abs(events.prt.pdg) == abs(Particle.from_string(\"p\").pdgid)]",
"_____no_output_____"
],
[
"# Assignments have to be through __setitem__ (brackets), not __setattr__ (as an attribute).\n# Is that a problem? (Assigning as an attribute would have to be implemented with care, if at all.)\n\nevents[\"pions\"] = events.prt[abs(events.prt.pdg) == abs(Particle.from_string(\"pi+\").pdgid)]\nevents[\"kaons\"] = events.prt[abs(events.prt.pdg) == abs(Particle.from_string(\"K+\").pdgid)]\nevents[\"protons\"] = events.prt[abs(events.prt.pdg) == abs(Particle.from_string(\"p\").pdgid)]",
"_____no_output_____"
],
[
"events.pions",
"_____no_output_____"
],
[
"events.kaons",
"_____no_output_____"
],
[
"events.protons",
"_____no_output_____"
],
[
"ak.num(events.prt), ak.num(events.pions), ak.num(events.kaons), ak.num(events.protons)",
"_____no_output_____"
]
],
[
[
"## Flattening for plots and regularizing to NumPy for machine learning\n\nAll of this structure is great, but eventually, we need to plot the data or ship it to some statistical process, such as machine learning.\n\nMost of these tools know about NumPy arrays and rectilinear data, but not Awkward Arrays.",
"_____no_output_____"
],
[
"As a design choice, Awkward Array **does not implicitly flatten**; you need to do this yourself, and you might make different choices of how to apply this lossy transformation in different circumstances.\n\nThe basic tool for removing structure is [ak.flatten](https://awkward-array.readthedocs.io/en/latest/_auto/ak.flatten.html).",
"_____no_output_____"
]
],
[
[
"?ak.flatten",
"_____no_output_____"
],
[
"# Turn particles-grouped-by-event into one big array of particles\nak.flatten(events.prt, axis=1)",
"_____no_output_____"
],
[
"# Eliminate structure at all levels; produce one numerical array\nak.flatten(events.prt, axis=None)",
"_____no_output_____"
]
],
[
[
"For plotting, you probably want to pick one field and flatten it. Flattening with `axis=1` (the default) works for one level of structure and is safer than `axis=None`.\n\nThe flattening is explicit as a reminder that a histogram whose entries are particles is different from a histogram whose entries are events.",
"_____no_output_____"
]
],
[
[
"# Directly through Matplotlib\nplt.hist(ak.flatten(events.kaons.p), bins=100, range=(0, 10))",
"_____no_output_____"
],
[
"# Through mplhep and boost-histgram, which are more HEP-friendly\n\nhep.histplot(bh.Histogram(bh.axis.Regular(100, 0, 10)).fill(\n \n ak.flatten(events.kaons.p)\n \n))",
"_____no_output_____"
]
],
[
[
"If the particles are sorted (`ak.sort`/`ak.argsort` is [in development](https://github.com/scikit-hep/awkward-1.0/pull/168)), you might want to pick the first kaon from every event that has them (i.e. *use* the event structure).\n\nThis is an analysis choice: *you* have to decide you want this.\n\nThe `ak.num(events.kaons) > 0` selection is explicit as a reminder that empty events are not counted in the histogram.",
"_____no_output_____"
]
],
[
[
"hep.histplot(bh.Histogram(bh.axis.Regular(100, 0, 10)).fill(\n \n events.kaons.p[ak.num(events.kaons) > 0, 0]\n \n))",
"_____no_output_____"
]
],
[
[
"Or perhaps the maximum pion momentum in each event. This one must be flattened (with `axis=0`) to remove `None` values.\n\nThis flattening is explicit as a reminder that empty events are not counted in the histogram.",
"_____no_output_____"
]
],
[
[
"ak.max(events.kaons.p, axis=1)",
"_____no_output_____"
],
[
"ak.flatten(ak.max(events.kaons.p, axis=1), axis=0)",
"_____no_output_____"
],
[
"hep.histplot(bh.Histogram(bh.axis.Regular(100, 0, 10)).fill(\n \n ak.flatten(ak.max(events.kaons.p, axis=1), axis=0)\n \n))",
"_____no_output_____"
]
],
[
[
"Or perhaps the momentum of the kaon with the farthest vertex. [ak.argmax](https://awkward-array.readthedocs.io/en/latest/_auto/ak.argmax.html) creates an array of integers selecting from each event.",
"_____no_output_____"
]
],
[
[
"?ak.argmax",
"_____no_output_____"
],
[
"ak.argmax(abs(events.kaons.vtx), axis=1)",
"_____no_output_____"
],
[
"?ak.singletons",
"_____no_output_____"
],
[
"# Get a length-1 list containing the index of the biggest vertex when there is one\n# And a length-0 list when there isn't one\nak.singletons(ak.argmax(abs(events.kaons.vtx), axis=1))",
"_____no_output_____"
],
[
"# A nested integer array like this is what we need to select kaons with the biggest vertex\nevents.kaons[ak.singletons(ak.argmax(abs(events.kaons.vtx), axis=1))]",
"_____no_output_____"
],
[
"events.kaons[ak.singletons(ak.argmax(abs(events.kaons.vtx), axis=1))].p",
"_____no_output_____"
],
[
"# Flatten the distinction between length-1 lists and length-0 lists\nak.flatten(events.kaons[ak.singletons(ak.argmax(abs(events.kaons.vtx), axis=1))].p)",
"_____no_output_____"
],
[
"# Putting it all together...\nhep.histplot(bh.Histogram(bh.axis.Regular(100, 0, 10)).fill(\n \n ak.flatten(events.kaons[ak.singletons(ak.argmax(abs(events.kaons.vtx), axis=1))].p)\n \n))",
"_____no_output_____"
]
],
[
[
"If you're sending the data to a library that expects rectilinear structure, you might need to pad and clip the variable length lists.\n\n[ak.pad_none](https://awkward-array.readthedocs.io/en/latest/_auto/ak.pad_none.html) puts `None` values at the end of each list to reach a minimum length.",
"_____no_output_____"
]
],
[
[
"?ak.pad_none",
"_____no_output_____"
],
[
"# pad them look at the first 30\nak.pad_none(events.kaons.id, 3)[:30].tolist()",
"_____no_output_____"
]
],
[
[
"The lengths are still irregular, so you can also `clip=True` them.",
"_____no_output_____"
]
],
[
[
"# pad them look at the first 30\nak.pad_none(events.kaons.id, 3, clip=True)[:30].tolist()",
"_____no_output_____"
]
],
[
[
"The library we're sending this to might not be able to deal with missing values, so choose a replacement to fill them with.",
"_____no_output_____"
]
],
[
[
"?ak.fill_none",
"_____no_output_____"
],
[
"# fill with -1 <- pad them look at the first 30\nak.fill_none(ak.pad_none(events.kaons.id, 3, clip=True), -1)[:30].tolist()",
"_____no_output_____"
]
],
[
[
"These are still Awkward-brand arrays; the downstream library might complain if they're not NumPy-brand, so use [ak.to_numpy](https://awkward-array.readthedocs.io/en/latest/_auto/ak.to_numpy.html) or simply cast it with NumPy's `np.asarray`.",
"_____no_output_____"
]
],
[
[
"?ak.to_numpy",
"_____no_output_____"
],
[
"np.asarray(ak.fill_none(ak.pad_none(events.kaons.id, 3, clip=True), -1))",
"_____no_output_____"
]
],
[
[
"If you try to convert an Awkward Array as NumPy and structure would be lost, you get an error. (You won't accidentally eliminate structure.)",
"_____no_output_____"
]
],
[
[
"try:\n np.asarray(events.kaons.id)\nexcept Exception as err:\n print(type(err), str(err))",
"<class 'ValueError'> in ListOffsetArray64, cannot convert to RegularArray because subarray lengths are not regular\n"
]
],
[
[
"## Broadcasting flat arrays and jagged arrays\n\nNumPy lets you combine arrays and scalars in a mathematical expression by first \"broadcasting\" the scalar to an array of the same length.",
"_____no_output_____"
]
],
[
[
"print(np.array([1, 2, 3, 4, 5]) + 100)",
"[101 102 103 104 105]\n"
]
],
[
[
"Awkward Array does the same thing, except that each element of a flat array can be broadcasted to each nested list of a jagged array.\n\n<img src=\"../docs-img/diagrams/cartoon-broadcasting.png\" width=\"300\">",
"_____no_output_____"
]
],
[
[
"print(ak.Array([[1, 2, 3], [], [4, 5], [6]]) + np.array([100, 200, 300, 400]))",
"[[101, 102, 103], [], [304, 305], [406]]\n"
]
],
[
[
"This is useful for emulating\n\n```python\nall_vertices = []\nfor event in events:\n vertices = []\n for kaon in events.kaons:\n all_vertices.append((kaon.vtx.x - event.true.x,\n kaon.vtx.y - event.true.y))\n all_vertices.append(vertices)\n```\n\nwhere `event.true.x` and `y` have only one value per event but `kaon.vtx.x` and `y` have one per kaon.",
"_____no_output_____"
]
],
[
[
"# one value per kaon one per event\nak.zip([events.kaons.vtx.x - events.true.x,\n events.kaons.vtx.y - events.true.y])",
"_____no_output_____"
]
],
[
[
"You don't have to do anything special for this: broadcasting is a common feature of all functions that apply to more than one array.\n\nYou can get it explicitly with [ak.broadcast_arrays](https://awkward-array.readthedocs.io/en/latest/_auto/ak.broadcast_arrays.html).",
"_____no_output_____"
]
],
[
[
"?ak.broadcast_arrays",
"_____no_output_____"
],
[
"ak.broadcast_arrays(events.true.x, events.kaons.vtx.x)",
"_____no_output_____"
]
],
[
[
"## Combinatorics: cartesian and combinations\n\nAt all levels of a physics analysis, we need to compare objects drawn from different collections.\n\n * **Gen-reco matching:** to associate a reconstructed particle with its generator-level parameters.\n * **Cleaning:** assocating soft photons with a reconstructed electron or leptons to a jet.\n * **Bump-hunting:** looking for mass peaks in pairs of particles.\n * **Dalitz analysis:** looking for resonance structure in triples of particles.\n\nTo do this with array-at-a-time operations, use one function to generate all the combinations, \n\n<img src=\"https://github.com/diana-hep/femtocode/raw/master/docs/explode.png\" width=\"300\">\n\napply \"flat\" operations,\n\n<img src=\"https://github.com/diana-hep/femtocode/raw/master/docs/flat.png\" width=\"300\">\n\nthen use \"reducers\" to get one value per particle or per event again.\n\n<img src=\"https://github.com/diana-hep/femtocode/raw/master/docs/reduce.png\" width=\"300\">",
"_____no_output_____"
],
[
"### Cartesian and combinations\n\nThe two main \"explode\" operations are [ak.cartesian](https://awkward-array.readthedocs.io/en/latest/_auto/ak.cartesian.html) and [ak.combinations](https://awkward-array.readthedocs.io/en/latest/_auto/ak.combinations.html).\n\nThe first generates the **Cartesian product** (a.k.a. cross product) of two collections **per nested list**.\n\n<img src=\"../docs-img/diagrams/cartoon-cartesian.png\" width=\"300\">\n\nThe second generates **distinct combinations** (i.e. \"n choose k\") of a collection with itself **per nested list**.\n\n<img src=\"../docs-img/diagrams/cartoon-combinations.png\" width=\"300\">",
"_____no_output_____"
]
],
[
[
"?ak.cartesian",
"_____no_output_____"
],
[
"?ak.combinations",
"_____no_output_____"
],
[
"ak.to_list(ak.cartesian(([[1, 2, 3], [], [4]],\n [[\"a\", \"b\"], [\"c\"], [\"d\", \"e\"]])))",
"_____no_output_____"
],
[
"ak.to_list(ak.combinations([[\"a\", \"b\", \"c\", \"d\"], [], [1, 2]], 2))",
"_____no_output_____"
]
],
[
[
"To search for $\\Lambda^0 \\to \\pi p$, we need to compute the mass of pairs drawn from these two collections.",
"_____no_output_____"
]
],
[
[
"pairs = ak.cartesian([events.pions, events.protons])\npairs",
"_____no_output_____"
],
[
"?ak.unzip",
"_____no_output_____"
],
[
"def mass(pairs, left_mass, right_mass):\n left, right = ak.unzip(pairs)\n left_energy = np.sqrt(left.p**2 + left_mass**2)\n right_energy = np.sqrt(right.p**2 + right_mass**2)\n return np.sqrt((left_energy + right_energy)**2 -\n (left.px + right.px)**2 -\n (left.py + right.py)**2 -\n (left.pz + right.pz)**2)",
"_____no_output_____"
],
[
"mass(pairs, 0.139570, 0.938272)",
"_____no_output_____"
],
[
"hep.histplot(bh.Histogram(bh.axis.Regular(100, 1.115683 - 0.01, 1.115683 + 0.01)).fill(\n ak.flatten(mass(pairs, 0.139570, 0.938272))\n))",
"_____no_output_____"
]
],
[
[
"We can improve the peak by selecting for opposite charges and large vertexes.",
"_____no_output_____"
]
],
[
[
"def opposite(pairs):\n left, right = ak.unzip(pairs)\n return pairs[left.charge != right.charge]\n\ndef distant(pairs):\n left, right = ak.unzip(pairs)\n return pairs[np.logical_and(abs(left.vtx) > 0.10, abs(right.vtx) > 0.10)]",
"_____no_output_____"
],
[
"hep.histplot(bh.Histogram(bh.axis.Regular(100, 1.115683 - 0.01, 1.115683 + 0.01)).fill(\n ak.flatten(mass(distant(opposite(pairs)), 0.139570, 0.938272))\n))",
"_____no_output_____"
]
],
[
[
"Alternatively, all of these functions could have been methods on the pair objects for reuse.\n\n(This is to make the point that any kind of object can have methods, not just particles.)",
"_____no_output_____"
]
],
[
[
"class ParticlePairArray(ak.Array):\n __name__ = \"Pairs\"\n \n def mass(self, left_mass, right_mass):\n left, right = self.slot0, self.slot1\n left_energy = np.sqrt(left.p**2 + left_mass**2)\n right_energy = np.sqrt(right.p**2 + right_mass**2)\n return np.sqrt((left_energy + right_energy)**2 -\n (left.px + right.px)**2 -\n (left.py + right.py)**2 -\n (left.pz + right.pz)**2)\n \n def opposite(self):\n return self[self.slot0.charge != self.slot1.charge]\n \n def distant(self, cut):\n return self[np.logical_and(abs(self.slot0.vtx) > cut, abs(self.slot1.vtx) > cut)]\n\nak.behavior[\"*\", \"pair\"] = ParticlePairArray",
"_____no_output_____"
],
[
"pairs = ak.cartesian([events.pions, events.protons], with_name=\"pair\")\npairs",
"_____no_output_____"
],
[
"hep.histplot(bh.Histogram(bh.axis.Regular(100, 1.115683 - 0.01, 1.115683 + 0.01)).fill(\n ak.flatten(pairs.opposite().distant(0.10).mass(0.139570, 0.938272))\n))",
"_____no_output_____"
]
],
[
[
"**Self-study question:** why does the call to `mass` have to be last?",
"_____no_output_____"
],
[
"An example for `ak.combinations`: $K_S \\to \\pi\\pi$.",
"_____no_output_____"
]
],
[
[
"pairs = ak.combinations(events.pions, 2, with_name=\"pair\")\npairs",
"_____no_output_____"
],
[
"hep.histplot(bh.Histogram(bh.axis.Regular(100, 0.497611 - 0.015, 0.497611 + 0.015)).fill(\n ak.flatten(pairs.opposite().distant(0.10).mass(0.139570, 0.139570))\n))",
"_____no_output_____"
]
],
[
[
"**Bonus problem:** $D^0 \\to K^- \\pi^+ \\pi^0$",
"_____no_output_____"
]
],
[
[
"pizero_candidates = ak.combinations(events.prt[events.prt.pdg == Particle.from_string(\"gamma\").pdgid], 2, with_name=\"pair\")\npizero = pizero_candidates[pizero_candidates.mass(0, 0) - 0.13498 < 0.000001]\npizero[\"px\"] = pizero.slot0.px + pizero.slot1.px\npizero[\"py\"] = pizero.slot0.py + pizero.slot1.py\npizero[\"pz\"] = pizero.slot0.pz + pizero.slot1.pz\npizero[\"p\"] = np.sqrt(pizero.px**2 + pizero.py**2 + pizero.pz**2)\npizero",
"_____no_output_____"
],
[
"kminus = events.prt[events.prt.pdg == Particle.from_string(\"K-\").pdgid]\npiplus = events.prt[events.prt.pdg == Particle.from_string(\"pi+\").pdgid]\n\ntriples = ak.cartesian({\"kminus\": kminus[abs(kminus.vtx) > 0.03],\n \"piplus\": piplus[abs(piplus.vtx) > 0.03],\n \"pizero\": pizero[np.logical_and(abs(pizero.slot0.vtx) > 0.03, abs(pizero.slot1.vtx) > 0.03)]})\ntriples",
"_____no_output_____"
],
[
"ak.num(triples)",
"_____no_output_____"
],
[
"def mass2(left, left_mass, right, right_mass):\n left_energy = np.sqrt(left.p**2 + left_mass**2)\n right_energy = np.sqrt(right.p**2 + right_mass**2)\n return ((left_energy + right_energy)**2 -\n (left.px + right.px)**2 -\n (left.py + right.py)**2 -\n (left.pz + right.pz)**2)",
"_____no_output_____"
],
[
"mKpi = mass2(triples.kminus, 0.493677, triples.piplus, 0.139570)",
"_____no_output_____"
],
[
"mpipi = mass2(triples.piplus, 0.139570, triples.pizero, 0.1349766)",
"_____no_output_____"
]
],
[
[
"This Dalitz plot doesn't look right (doesn't cut off at kinematic limits), but I'm going to leave it as an exercise for the reader.",
"_____no_output_____"
]
],
[
[
"dalitz = bh.Histogram(bh.axis.Regular(50, 0, 3), bh.axis.Regular(50, 0, 2))\ndalitz.fill(ak.flatten(mKpi), ak.flatten(mpipi))\n\nX, Y = dalitz.axes.edges\n\nfig, ax = plt.subplots()\nmesh = ax.pcolormesh(X.T, Y.T, dalitz.view().T)\nfig.colorbar(mesh)",
"_____no_output_____"
]
],
[
[
"## Reducing from combinations\n\nThe mass-peak examples above don't need to \"reduce\" combinations, but many applications do.\n\n<img src=\"https://github.com/diana-hep/femtocode/raw/master/docs/reduce.png\" width=\"300\">",
"_____no_output_____"
],
[
"Suppose that we want to find the \"nearest photon to each electron\" (e.g. bremsstrahlung).",
"_____no_output_____"
]
],
[
[
"electrons = events.prt[abs(events.prt.pdg) == abs(Particle.from_string(\"e-\").pdgid)]\nphotons = events.prt[events.prt.pdg == Particle.from_string(\"gamma\").pdgid]",
"_____no_output_____"
]
],
[
[
"The problem with the raw output of `ak.cartesian` is that all the combinations are mixed together in the same lists.",
"_____no_output_____"
]
],
[
[
"ak.to_list(ak.cartesian([electrons[[\"pdg\", \"id\"]], photons[[\"pdg\", \"id\"]]])[8])",
"_____no_output_____"
]
],
[
[
"We can fix this by asking for `nested=True`, which adds another level of nesting to the output.",
"_____no_output_____"
]
],
[
[
"ak.to_list(ak.cartesian([electrons[[\"pdg\", \"id\"]], photons[[\"pdg\", \"id\"]]], nested=True)[8])",
"_____no_output_____"
]
],
[
[
"All electron-photon pairs associated with a given electron are grouped in a list-within-each-list.\n\nNow we can apply reducers to this inner dimension to sum over some quantity, pick the best one, etc.",
"_____no_output_____"
]
],
[
[
"def cos_angle(pairs):\n left, right = ak.unzip(pairs)\n return left.dir.x*right.dir.x + left.dir.y*right.dir.y + left.dir.z*right.dir.z",
"_____no_output_____"
],
[
"electron_photons = ak.cartesian([electrons, photons], nested=True)",
"_____no_output_____"
],
[
"cos_angle(electron_photons)",
"_____no_output_____"
],
[
"hep.histplot(bh.Histogram(bh.axis.Regular(100, -1, 1)).fill(\n ak.flatten(cos_angle(electron_photons), axis=None) # a good reason to use flatten axis=None\n))",
"_____no_output_____"
]
],
[
[
"We pick the \"maximum according to a function\" using the same `ak.singletons(ak.argmax(f(x))` trick as above.",
"_____no_output_____"
]
],
[
[
"best_electron_photons = electron_photons[ak.singletons(ak.argmax(cos_angle(electron_photons), axis=2))]\n\nhep.histplot(bh.Histogram(bh.axis.Regular(100, -1, 1)).fill(\n ak.flatten(cos_angle(best_electron_photons), axis=None)\n))",
"_____no_output_____"
]
],
[
[
"By construction, `best_electron_photons` has zero or one elements in each *inner* nested list.",
"_____no_output_____"
]
],
[
[
"ak.num(electron_photons, axis=2), ak.num(best_electron_photons, axis=2)",
"_____no_output_____"
]
],
[
[
"Since we no longer care about that *inner* structure, we could flatten it at `axis=2` (leaving `axis=1` untouched).",
"_____no_output_____"
]
],
[
[
"best_electron_photons",
"_____no_output_____"
],
[
"ak.flatten(best_electron_photons, axis=2)",
"_____no_output_____"
]
],
[
[
"But it would be better to invert the `ak.singletons` by calling `ak.firsts`.",
"_____no_output_____"
]
],
[
[
"?ak.singletons",
"_____no_output_____"
],
[
"?ak.firsts",
"_____no_output_____"
],
[
"ak.firsts(best_electron_photons, axis=2)",
"_____no_output_____"
]
],
[
[
"Because then we can get back one value for each electron (with `None` if `ak.argmax` resulted in `None` because there were no pairs).",
"_____no_output_____"
]
],
[
[
"ak.num(electrons), ak.num(ak.firsts(best_electron_photons, axis=2))",
"_____no_output_____"
],
[
"ak.all(ak.num(electrons) == ak.num(ak.firsts(best_electron_photons, axis=2)))",
"_____no_output_____"
]
],
[
[
"And that means that we can make this \"closest photon\" an attribute of the electrons. We have now performed electron-photon matching.",
"_____no_output_____"
]
],
[
[
"electrons[\"photon\"] = ak.firsts(best_electron_photons, axis=2)\nak.to_list(electrons[8])",
"_____no_output_____"
]
],
[
[
"Current set of reducers:\n\n * [ak.count](https://awkward-array.readthedocs.io/en/latest/_auto/ak.count.html): counts the number in each group (subtly different from [ak.num](https://awkward-array.readthedocs.io/en/latest/_auto/ak.num.html) because `ak.count` is a reducer)\n * [ak.count_nonzero](https://awkward-array.readthedocs.io/en/latest/_auto/ak.count_nonzero.html): counts the number of non-zero elements in each group\n * [ak.sum](https://awkward-array.readthedocs.io/en/latest/_auto/ak.sum.html): adds up values in the group, the quintessential reducer\n * [ak.prod](https://awkward-array.readthedocs.io/en/latest/_auto/ak.prod.html): multiplies values in the group (e.g. for charges or probabilities)\n * [ak.any](https://awkward-array.readthedocs.io/en/latest/_auto/ak.any.html): boolean reducer for logical `or` (\"do *any* in this group satisfy a constraint?\")\n * [ak.all](https://awkward-array.readthedocs.io/en/latest/_auto/ak.all.html): boolean reducer for logical `and` (\"do *all* in this group satisfy a constraint?\")\n * [ak.min](https://awkward-array.readthedocs.io/en/latest/_auto/ak.min.html): minimum value in each group (`None` for empty groups)\n * [ak.max](https://awkward-array.readthedocs.io/en/latest/_auto/ak.max.html): maximum value in each group (`None` for empty groups)\n * [ak.argmin](https://awkward-array.readthedocs.io/en/latest/_auto/ak.argmin.html): index of minimum value in each group (`None` for empty groups)\n * [ak.argmax](https://awkward-array.readthedocs.io/en/latest/_auto/ak.argmax.html): index of maximum value in each group (`None` for empty groups)\n\nAnd other functions that have the same interface as a reducer (reduces a dimension):\n\n * [ak.moment](https://awkward-array.readthedocs.io/en/latest/_auto/ak.moment.html): computes the $n^{th}$ moment in each group\n * [ak.mean](https://awkward-array.readthedocs.io/en/latest/_auto/ak.mean.html): computes the mean in each group\n * [ak.var](https://awkward-array.readthedocs.io/en/latest/_auto/ak.var.html): computes the variance in each group\n * [ak.std](https://awkward-array.readthedocs.io/en/latest/_auto/ak.std.html): computes the standard deviation in each group\n * [ak.covar](https://awkward-array.readthedocs.io/en/latest/_auto/ak.covar.html): computes the covariance in each group\n * [ak.corr](https://awkward-array.readthedocs.io/en/latest/_auto/ak.corr.html): computes the correlation in each group\n * [ak.linear_fit](https://awkward-array.readthedocs.io/en/latest/_auto/ak.linear_fit.html): computes the linear fit in each group\n * [ak.softmax](https://awkward-array.readthedocs.io/en/latest/_auto/ak.softmax.html): computes the softmax function in each group",
"_____no_output_____"
],
[
"## Imperative, but still fast, programming in Numba\n\nArray-at-a-time operations let us manipulate dynamically typed data with compiled code (and in some cases, benefit from hardware vectorization). However, they're complicated. Finding the closest photon to each electron is more complicated than it seems it ought to be.\n\nSome of these things are simpler in imperative (step-by-step scalar-at-a-time) code. Imperative Python code is slow because it has to check the data type of every object it enounters (among other things); compiled code is faster because these checks are performed once during a compilation step for any number of identically typed values.\n\nWe can get the best of both worlds by Just-In-Time (JIT) compiling the code. [Numba](http://numba.pydata.org/) is a NumPy-centric JIT compiler for Python.",
"_____no_output_____"
]
],
[
[
"import numba as nb\n\[email protected]\ndef monte_carlo_pi(nsamples):\n acc = 0\n for i in range(nsamples):\n x = np.random.random()\n y = np.random.random()\n if (x**2 + y**2) < 1.0:\n acc += 1\n return 4.0 * acc / nsamples",
"_____no_output_____"
],
[
"%%timeit\n\n# Run the pure Python function (without nb.jit)\nmonte_carlo_pi.py_func(1000000)",
"741 ms ± 14.7 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)\n"
],
[
"%%timeit\n\n# Run the compiled function\nmonte_carlo_pi(1000000)",
"8.7 ms ± 194 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)\n"
]
],
[
[
"The price for this magical speedup is that not all Python code can be accelerated; you have to be conservative with the functions and language features you use, and Numba has to recognize the data types.\n\nNumba recognizes Awkward Arrays.",
"_____no_output_____"
]
],
[
[
"@nb.jit\ndef lambda_mass(events):\n num_lambdas = 0\n for event in events:\n num_lambdas += len(event.pions) * len(event.protons)\n\n lambda_masses = np.empty(num_lambdas, np.float64)\n i = 0\n for event in events:\n for pion in event.pions:\n for proton in event.protons:\n pion_energy = np.sqrt(pion.p**2 + 0.139570**2)\n proton_energy = np.sqrt(proton.p**2 + 0.938272**2)\n mass = np.sqrt((pion_energy + proton_energy)**2 -\n (pion.px + proton.px)**2 -\n (pion.py + proton.py)**2 -\n (pion.pz + proton.pz)**2)\n lambda_masses[i] = mass\n i += 1\n \n return lambda_masses\n\nlambda_mass(events)",
"_____no_output_____"
],
[
"hep.histplot(bh.Histogram(bh.axis.Regular(100, 1.115683 - 0.01, 1.115683 + 0.01)).fill(\n lambda_mass(events)\n))",
"_____no_output_____"
]
],
[
[
"Some constraints:\n\n * Awkward arrays are read-only structures (always true, even outside of Numba)\n * Awkward arrays can't be created inside a Numba-compiled function\n\nThat was fine for a function that creates and returns a NumPy array, but what if we want to create something with structure?",
"_____no_output_____"
],
[
"The [ak.ArrayBuilder](https://awkward-array.readthedocs.io/en/latest/_auto/ak.ArrayBuilder.html) is a general way to make data structures.",
"_____no_output_____"
]
],
[
[
"?ak.ArrayBuilder",
"_____no_output_____"
],
[
"builder = ak.ArrayBuilder()\n\nbuilder.begin_list()\n\nbuilder.begin_record()\nbuilder.field(\"x\").integer(1)\nbuilder.field(\"y\").real(1.1)\nbuilder.field(\"z\").string(\"one\")\nbuilder.end_record()\n\nbuilder.begin_record()\nbuilder.field(\"x\").integer(2)\nbuilder.field(\"y\").real(2.2)\nbuilder.field(\"z\").string(\"two\")\nbuilder.end_record()\n\nbuilder.end_list()\n\nbuilder.begin_list()\nbuilder.end_list()\n\nbuilder.begin_list()\n\nbuilder.begin_record()\nbuilder.field(\"x\").integer(3)\nbuilder.field(\"y\").real(3.3)\nbuilder.field(\"z\").string(\"three\")\nbuilder.end_record()\n\nbuilder.end_list()\n\nak.to_list(builder.snapshot())",
"_____no_output_____"
]
],
[
[
"ArrayBuilders can be used in Numba, albeit with some constraints:\n\n * ArrayBuilders can't be created inside a Numba-compiled function (pass them in)\n * The `snapshot` method (to turn it into an array) can't be used in a Numba-compiled function (use it outside)",
"_____no_output_____"
]
],
[
[
"@nb.jit(nopython=True)\ndef make_electron_photons(events, builder):\n for event in events:\n builder.begin_list()\n for electron in event.electrons:\n best_i = -1\n best_angle = -1.0\n for i in range(len(event.photons)):\n photon = event.photons[i]\n angle = photon.dir.x*electron.dir.x + photon.dir.y*electron.dir.y + photon.dir.z*electron.dir.z\n if angle > best_angle:\n best_i = i\n best_angle = angle\n if best_i == -1:\n builder.null()\n else:\n builder.append(photon)\n builder.end_list()\n\nevents[\"electrons\"] = events.prt[abs(events.prt.pdg) == abs(Particle.from_string(\"e-\").pdgid)]\nevents[\"photons\"] = events.prt[events.prt.pdg == Particle.from_string(\"gamma\").pdgid]\n\nbuilder = ak.ArrayBuilder()\nmake_electron_photons(events, builder)\nbuilder.snapshot()",
"_____no_output_____"
]
],
[
[
"A few of them are `None` (called `builder.null()` because there were no photons to attach to the electron).",
"_____no_output_____"
]
],
[
[
"ak.count_nonzero(ak.is_none(ak.flatten(builder.snapshot())))",
"_____no_output_____"
]
],
[
[
"But the `builder.snapshot()` otherwise matches up with the `events.electrons`, so it's something we could attach to it, as before.",
"_____no_output_____"
]
],
[
[
"?ak.with_field",
"_____no_output_____"
],
[
"events[\"electrons\"] = ak.with_field(events.electrons, builder.snapshot(), \"photon\")",
"_____no_output_____"
],
[
"ak.to_list(events[8].electrons)",
"_____no_output_____"
]
],
[
[
"## Grafting jagged data onto Pandas\n\nAwkward Arrays can be Pandas columns.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\ndf = pd.DataFrame({\"pions\": events.pions,\n \"kaons\": events.kaons,\n \"protons\": events.protons})\ndf",
"_____no_output_____"
],
[
"df[\"pions\"].dtype",
"_____no_output_____"
]
],
[
[
"But that's unlikely to be useful for very complex data structures because there aren't any Pandas functions for deeply nested structure.\n\nInstead, you'll probably want to *convert* the nested structures into the corresponding Pandas [MultiIndex](https://pandas.pydata.org/pandas-docs/stable/user_guide/advanced.html).",
"_____no_output_____"
]
],
[
[
"ak.pandas.df(events.pions)",
"_____no_output_____"
]
],
[
[
"Now the nested lists are represented as MultiIndex rows and the nested records are represented as MultiIndex columns, which are structures that Pandas knows how to deal with.",
"_____no_output_____"
],
[
"But what about two types of particles, pions and kaons? (And let's simplify to just `\"px\", \"py\", \"pz\", \"vtx\"`.)",
"_____no_output_____"
]
],
[
[
"simpler = ak.zip({\"pions\": events.pions[[\"px\", \"py\", \"pz\", \"vtx\"]],\n \"kaons\": events.kaons[[\"px\", \"py\", \"pz\", \"vtx\"]]}, depthlimit=1)\nak.type(simpler)",
"_____no_output_____"
],
[
"ak.pandas.df(simpler)",
"_____no_output_____"
]
],
[
[
"There's only one row MultiIndex, so pion #1 in each event is the same row as kaon #1. That assocation is probably meaningless.\n\nThe issue is that a single Pandas DataFrame represents *less* information than an Awkward Array. In general, we would need a collection of DataFrames to losslessly encode an Awkward Array. (Pandas represents the data in [database normal form](https://en.wikipedia.org/wiki/Database_normalization); Awkward represents it in objects.)",
"_____no_output_____"
]
],
[
[
"# This array corresponds to *two* Pandas DataFrames.\npions_df, kaons_df = ak.pandas.dfs(simpler)",
"_____no_output_____"
],
[
"pions_df",
"_____no_output_____"
],
[
"kaons_df",
"_____no_output_____"
]
],
[
[
"## NumExpr, Autograd, and other third-party libraries",
"_____no_output_____"
],
[
"[NumExpr](https://numexpr.readthedocs.io/en/latest/user_guide.html) can calcuate pure numerical expressions faster than NumPy because it does so in one pass. (It has a low-overhead virtual machine.)\n\nNumExpr doesn't recognize Awkward Arrays, but we have a wrapper for it.",
"_____no_output_____"
]
],
[
[
"import numexpr\n\n# This works because px, py, pz are flat, like NumPy\npx = ak.flatten(events.pions.px)\npy = ak.flatten(events.pions.py)\npz = ak.flatten(events.pions.pz)\nnumexpr.evaluate(\"px**2 + py**2 + pz**2\")",
"_____no_output_____"
],
[
"# This doesn't work because px, py, pz have structure\npx = events.pions.px\npy = events.pions.py\npz = events.pions.pz\ntry:\n numexpr.evaluate(\"px**2 + py**2 + pz**2\")\nexcept Exception as err:\n print(type(err), str(err))",
"<class 'ValueError'> in ListOffsetArray64, cannot convert to RegularArray because subarray lengths are not regular\n"
],
[
"# But in this wrapped version, we broadcast and maintain structure\nak.numexpr.evaluate(\"px**2 + py**2 + pz**2\")",
"_____no_output_____"
]
],
[
[
"Similarly for [Autograd](https://github.com/HIPS/autograd#readme), which has an `elementwise_grad` for differentiating expressions with respect to NumPy [universal functions](https://docs.scipy.org/doc/numpy/reference/ufuncs.html), but not Awkward Arrays.",
"_____no_output_____"
]
],
[
[
"@ak.autograd.elementwise_grad\ndef tanh(x):\n y = np.exp(-2.0 * x)\n return (1.0 - y) / (1.0 + y)\n\nak.to_list(tanh([{\"x\": 0.0, \"y\": []}, {\"x\": 0.1, \"y\": [1]}, {\"x\": 0.2, \"y\": [2, 2]}, {\"x\": 0.3, \"y\": [3, 3, 3]}]))",
"_____no_output_____"
]
],
[
[
"The set of third-party libraries supported this way will continue to grow. Some major plans on the horizon:\n\n * [Apache Arrow](https://arrow.apache.org/), and through it the [Parquet](https://parquet.apache.org/) file format.\n * The [Zarr](https://zarr.readthedocs.io/en/stable/) array delivery system.\n * [CuPy](https://cupy.chainer.org/) and Awkward Arrays on the GPU.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e7933996894d02d9a6a523f96b9ba223efe863df | 2,615 | ipynb | Jupyter Notebook | String/1013/1309. Decrypt String from Alphabet to Integer Mapping.ipynb | YuHe0108/Leetcode | 90d904dde125dd35ee256a7f383961786f1ada5d | [
"Apache-2.0"
] | 1 | 2020-08-05T11:47:47.000Z | 2020-08-05T11:47:47.000Z | String/1013/1309. Decrypt String from Alphabet to Integer Mapping.ipynb | YuHe0108/LeetCode | b9e5de69b4e4d794aff89497624f558343e362ad | [
"Apache-2.0"
] | null | null | null | String/1013/1309. Decrypt String from Alphabet to Integer Mapping.ipynb | YuHe0108/LeetCode | b9e5de69b4e4d794aff89497624f558343e362ad | [
"Apache-2.0"
] | null | null | null | 23.558559 | 96 | 0.424474 | [
[
[
"说明:\n 给定由数字(‘0’-‘9’)和‘#’组成的字符串s。\n 我们希望将s映射到英文小写字符,如下所示:\n 1、字符(‘a’到‘i’)分别由(‘1’到‘9’)表示。\n 2、字符(‘j’到‘z’)分别由(‘10#’到‘26#’)表示。\n 返回映射后形成的字符串。\n 可以保证唯一的映射将始终存在。\nExample 1:\n Input: s = \"10#11#12\"\n Output: \"jkab\"\n Explanation: \"j\" -> \"10#\" , \"k\" -> \"11#\" , \"a\" -> \"1\" , \"b\" -> \"2\".\n\nExample 2:\n Input: s = \"1326#\"\n Output: \"acz\"\n\nExample 3:\n Input: s = \"25#\"\n Output: \"y\"\n\nExample 4:\n Input: s = \"12345678910#11#12#13#14#15#16#17#18#19#20#21#22#23#24#25#26#\"\n Output: \"abcdefghijklmnopqrstuvwxyz\"\n\nConstraints:\n 1、1 <= s.length <= 1000\n 2、s[i] only contains digits letters ('0'-'9') and '#' letter.\n 3、s will be valid string such that mapping is always possible.",
"_____no_output_____"
]
],
[
[
"class Solution:\n def freqAlphabets(self, s: str) -> str:\n res = ''\n idx = 0\n while idx < len(s):\n if idx + 2 < len(s) and 1 <= int(s[idx]) <= 2 and s[idx + 2] == '#':\n val = int(s[idx] + s[idx + 1])\n idx += 2\n else:\n val = int(s[idx])\n res += chr(val + 96)\n idx += 1\n return res",
"_____no_output_____"
],
[
"solution = Solution()\nsolution.freqAlphabets(\"10#11#12\")",
"_____no_output_____"
],
[
"96 + 1",
"_____no_output_____"
]
]
] | [
"raw",
"code"
] | [
[
"raw"
],
[
"code",
"code",
"code"
]
] |
e79339a62470bef2751e9757ccc321cefb9d4ce8 | 440,583 | ipynb | Jupyter Notebook | Mentoria Fraudes/Mentoria - Fraudes Leon.ipynb | leon-maia/Portfolio-Voyager | 7ab1967e57496b3e6f95468e862265d2ccbceb59 | [
"MIT"
] | null | null | null | Mentoria Fraudes/Mentoria - Fraudes Leon.ipynb | leon-maia/Portfolio-Voyager | 7ab1967e57496b3e6f95468e862265d2ccbceb59 | [
"MIT"
] | null | null | null | Mentoria Fraudes/Mentoria - Fraudes Leon.ipynb | leon-maia/Portfolio-Voyager | 7ab1967e57496b3e6f95468e862265d2ccbceb59 | [
"MIT"
] | null | null | null | 232.007899 | 357,168 | 0.876262 | [
[
[
"# IEEE-CIS Fraud Detection\n## Can you detect fraud from customer transactions?",
"_____no_output_____"
]
],
[
[
"# Análise dos dados\nimport pandas as pd\n\n# Visualização dos dados\nimport matplotlib.pyplot as plt\nimport seaborn as sn",
"_____no_output_____"
]
],
[
[
"### SampleSubmission é o formato de entrega do modelo. Desconsiderar Dataset.",
"_____no_output_____"
]
],
[
[
"df_SampleSubmission = pd.read_csv('sample_submission.csv')",
"_____no_output_____"
],
[
"df_SampleSubmission.head()",
"_____no_output_____"
]
],
[
[
"### Analisando o dataset test_identity.csv",
"_____no_output_____"
],
[
"Metadados\nIdentity Table\nVariables in this table are identity information – network connection information (IP, ISP, Proxy, etc) and digital signature (UA/browser/os/version, etc) associated with transactions.\nThey're collected by Vesta’s fraud protection system and digital security partners.\n(The field names are masked and pairwise dictionary will not be provided for privacy protection and contract agreement)\n\nCategorical Features:\nDeviceType\nDeviceInfo\nid_12 - id_38\n\nDeviceInfo : https://www.kaggle.com/c/ieee-fraud-detection/discussion/101203#583227\n\n“id01 to id11 are numerical features for identity, which is collected by Vesta and security partners such as device rating, ip_domain rating, proxy rating, etc. Also it recorded behavioral fingerprint like account login times/failed to login times, how long an account stayed on the page, etc. All of these are not able to elaborate due to security partner T&C. I hope you could get basic meaning of these features, and by mentioning them as numerical/categorical, you won't deal with them inappropriately.”",
"_____no_output_____"
]
],
[
[
"df_test_identity = pd.read_csv('test_identity.csv')",
"_____no_output_____"
],
[
"l, c = df_test_identity.shape",
"_____no_output_____"
],
[
"l",
"_____no_output_____"
],
[
"df_test_identity.head()",
"_____no_output_____"
],
[
"df_test_identity.tail()",
"_____no_output_____"
],
[
"df_test_identity.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 141907 entries, 0 to 141906\nData columns (total 41 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 TransactionID 141907 non-null int64 \n 1 id-01 141907 non-null float64\n 2 id-02 136976 non-null float64\n 3 id-03 66481 non-null float64\n 4 id-04 66481 non-null float64\n 5 id-05 134750 non-null float64\n 6 id-06 134750 non-null float64\n 7 id-07 5059 non-null float64\n 8 id-08 5059 non-null float64\n 9 id-09 74338 non-null float64\n 10 id-10 74338 non-null float64\n 11 id-11 136778 non-null float64\n 12 id-12 141907 non-null object \n 13 id-13 130286 non-null float64\n 14 id-14 71357 non-null float64\n 15 id-15 136977 non-null object \n 16 id-16 125747 non-null object \n 17 id-17 135966 non-null float64\n 18 id-18 50875 non-null float64\n 19 id-19 135906 non-null float64\n 20 id-20 135633 non-null float64\n 21 id-21 5059 non-null float64\n 22 id-22 5062 non-null float64\n 23 id-23 5062 non-null object \n 24 id-24 4740 non-null float64\n 25 id-25 5039 non-null float64\n 26 id-26 5047 non-null float64\n 27 id-27 5062 non-null object \n 28 id-28 136778 non-null object \n 29 id-29 136778 non-null object \n 30 id-30 70659 non-null object \n 31 id-31 136625 non-null object \n 32 id-32 70671 non-null float64\n 33 id-33 70671 non-null object \n 34 id-34 72175 non-null object \n 35 id-35 136977 non-null object \n 36 id-36 136977 non-null object \n 37 id-37 136977 non-null object \n 38 id-38 136977 non-null object \n 39 DeviceType 136931 non-null object \n 40 DeviceInfo 115057 non-null object \ndtypes: float64(23), int64(1), object(17)\nmemory usage: 44.4+ MB\n"
],
[
"df_test_identity.isnull().sum().sort_values(ascending=False)",
"_____no_output_____"
],
[
"(df_test_identity.isnull().sum().sort_values(ascending=False) / l) * 100",
"_____no_output_____"
],
[
"df_test_identity_corr = df_test_identity.corr()",
"_____no_output_____"
],
[
"sn.heatmap(df_test_identity_corr, vmin=0, vmax=1)",
"_____no_output_____"
]
],
[
[
"### Analisando o dataset test_transaction.csv",
"_____no_output_____"
],
[
"Transaction table\n“It contains money transfer and also other gifting goods and service, like you booked a ticket for others, etc.”\n\nTransactionDT: timedelta from a given reference datetime (not an actual timestamp)\n“TransactionDT first value is 86400, which corresponds to the number of seconds in a day (60 * 60 * 24 = 86400) so I think the unit is seconds. Using this, we know the data spans 6 months, as the maximum value is 15811131, which would correspond to day 183.”\n\nTransactionAMT: transaction payment amount in USD\n“Some of the transaction amounts have three decimal places to the right of the decimal point. There seems to be a link to three decimal places and a blank addr1 and addr2 field. Is it possible that these are foreign transactions and that, for example, the 75.887 in row 12 is the result of multiplying a foreign currency amount by an exchange rate?”\n\nProductCD: product code, the product for each transaction\n“Product isn't necessary to be a real 'product' (like one item to be added to the shopping cart). It could be any kind of service.”\n\ncard1 - card6: payment card information, such as card type, card category, issue bank, country, etc.\n\naddr: address\n“both addresses are for purchaser\naddr1 as billing region\naddr2 as billing country”\ndist: distance\n\"distances between (not limited) billing address, mailing address, zip code, IP address, phone area, etc.”\nP_ and (R__) emaildomain: purchaser and recipient email domain\n“ certain transactions don't need recipient, so R_emaildomain is null.”\nC1-C14: counting, such as how many addresses are found to be associated with the payment card, etc. The actual meaning is masked.\n“Can you please give more examples of counts in the variables C1-15? Would these be like counts of phone numbers, email addresses, names associated with the user? I can't think of 15.\nYour guess is good, plus like device, ipaddr, billingaddr, etc. Also these are for both purchaser and recipient, which doubles the number.”\nD1-D15: timedelta, such as days between previous transaction, etc.\nM1-M9: match, such as names on card and address, etc.\nVxxx: Vesta engineered rich features, including ranking, counting, and other entity relations.\n“For example, how many times the payment card associated with a IP and email or address appeared in 24 hours time range, etc.”\n\"All Vesta features were derived as numerical. some of them are count of orders within a clustering, a time-period or condition, so the value is finite and has ordering (or ranking). I wouldn't recommend to treat any of them as categorical. If any of them resulted in binary by chance, it maybe worth trying.\"",
"_____no_output_____"
]
],
[
[
"df_test_transaction = pd.read_csv('test_transaction.csv')",
"_____no_output_____"
],
[
"display(df_test_transaction)",
"_____no_output_____"
],
[
"l2, c2 = df_test_transaction.shape",
"_____no_output_____"
],
[
"df_test_transaction.info(verbose=True)",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 506691 entries, 0 to 506690\nData columns (total 393 columns):\n # Column Dtype \n--- ------ ----- \n 0 TransactionID int64 \n 1 TransactionDT int64 \n 2 TransactionAmt float64\n 3 ProductCD object \n 4 card1 int64 \n 5 card2 float64\n 6 card3 float64\n 7 card4 object \n 8 card5 float64\n 9 card6 object \n 10 addr1 float64\n 11 addr2 float64\n 12 dist1 float64\n 13 dist2 float64\n 14 P_emaildomain object \n 15 R_emaildomain object \n 16 C1 float64\n 17 C2 float64\n 18 C3 float64\n 19 C4 float64\n 20 C5 float64\n 21 C6 float64\n 22 C7 float64\n 23 C8 float64\n 24 C9 float64\n 25 C10 float64\n 26 C11 float64\n 27 C12 float64\n 28 C13 float64\n 29 C14 float64\n 30 D1 float64\n 31 D2 float64\n 32 D3 float64\n 33 D4 float64\n 34 D5 float64\n 35 D6 float64\n 36 D7 float64\n 37 D8 float64\n 38 D9 float64\n 39 D10 float64\n 40 D11 float64\n 41 D12 float64\n 42 D13 float64\n 43 D14 float64\n 44 D15 float64\n 45 M1 object \n 46 M2 object \n 47 M3 object \n 48 M4 object \n 49 M5 object \n 50 M6 object \n 51 M7 object \n 52 M8 object \n 53 M9 object \n 54 V1 float64\n 55 V2 float64\n 56 V3 float64\n 57 V4 float64\n 58 V5 float64\n 59 V6 float64\n 60 V7 float64\n 61 V8 float64\n 62 V9 float64\n 63 V10 float64\n 64 V11 float64\n 65 V12 float64\n 66 V13 float64\n 67 V14 float64\n 68 V15 float64\n 69 V16 float64\n 70 V17 float64\n 71 V18 float64\n 72 V19 float64\n 73 V20 float64\n 74 V21 float64\n 75 V22 float64\n 76 V23 float64\n 77 V24 float64\n 78 V25 float64\n 79 V26 float64\n 80 V27 float64\n 81 V28 float64\n 82 V29 float64\n 83 V30 float64\n 84 V31 float64\n 85 V32 float64\n 86 V33 float64\n 87 V34 float64\n 88 V35 float64\n 89 V36 float64\n 90 V37 float64\n 91 V38 float64\n 92 V39 float64\n 93 V40 float64\n 94 V41 float64\n 95 V42 float64\n 96 V43 float64\n 97 V44 float64\n 98 V45 float64\n 99 V46 float64\n 100 V47 float64\n 101 V48 float64\n 102 V49 float64\n 103 V50 float64\n 104 V51 float64\n 105 V52 float64\n 106 V53 float64\n 107 V54 float64\n 108 V55 float64\n 109 V56 float64\n 110 V57 float64\n 111 V58 float64\n 112 V59 float64\n 113 V60 float64\n 114 V61 float64\n 115 V62 float64\n 116 V63 float64\n 117 V64 float64\n 118 V65 float64\n 119 V66 float64\n 120 V67 float64\n 121 V68 float64\n 122 V69 float64\n 123 V70 float64\n 124 V71 float64\n 125 V72 float64\n 126 V73 float64\n 127 V74 float64\n 128 V75 float64\n 129 V76 float64\n 130 V77 float64\n 131 V78 float64\n 132 V79 float64\n 133 V80 float64\n 134 V81 float64\n 135 V82 float64\n 136 V83 float64\n 137 V84 float64\n 138 V85 float64\n 139 V86 float64\n 140 V87 float64\n 141 V88 float64\n 142 V89 float64\n 143 V90 float64\n 144 V91 float64\n 145 V92 float64\n 146 V93 float64\n 147 V94 float64\n 148 V95 float64\n 149 V96 float64\n 150 V97 float64\n 151 V98 float64\n 152 V99 float64\n 153 V100 float64\n 154 V101 float64\n 155 V102 float64\n 156 V103 float64\n 157 V104 float64\n 158 V105 float64\n 159 V106 float64\n 160 V107 float64\n 161 V108 float64\n 162 V109 float64\n 163 V110 float64\n 164 V111 float64\n 165 V112 float64\n 166 V113 float64\n 167 V114 float64\n 168 V115 float64\n 169 V116 float64\n 170 V117 float64\n 171 V118 float64\n 172 V119 float64\n 173 V120 float64\n 174 V121 float64\n 175 V122 float64\n 176 V123 float64\n 177 V124 float64\n 178 V125 float64\n 179 V126 float64\n 180 V127 float64\n 181 V128 float64\n 182 V129 float64\n 183 V130 float64\n 184 V131 float64\n 185 V132 float64\n 186 V133 float64\n 187 V134 float64\n 188 V135 float64\n 189 V136 float64\n 190 V137 float64\n 191 V138 float64\n 192 V139 float64\n 193 V140 float64\n 194 V141 float64\n 195 V142 float64\n 196 V143 float64\n 197 V144 float64\n 198 V145 float64\n 199 V146 float64\n 200 V147 float64\n 201 V148 float64\n 202 V149 float64\n 203 V150 float64\n 204 V151 float64\n 205 V152 float64\n 206 V153 float64\n 207 V154 float64\n 208 V155 float64\n 209 V156 float64\n 210 V157 float64\n 211 V158 float64\n 212 V159 float64\n 213 V160 float64\n 214 V161 float64\n 215 V162 float64\n 216 V163 float64\n 217 V164 float64\n 218 V165 float64\n 219 V166 float64\n 220 V167 float64\n 221 V168 float64\n 222 V169 float64\n 223 V170 float64\n 224 V171 float64\n 225 V172 float64\n 226 V173 float64\n 227 V174 float64\n 228 V175 float64\n 229 V176 float64\n 230 V177 float64\n 231 V178 float64\n 232 V179 float64\n 233 V180 float64\n 234 V181 float64\n 235 V182 float64\n 236 V183 float64\n 237 V184 float64\n 238 V185 float64\n 239 V186 float64\n 240 V187 float64\n 241 V188 float64\n 242 V189 float64\n 243 V190 float64\n 244 V191 float64\n 245 V192 float64\n 246 V193 float64\n 247 V194 float64\n 248 V195 float64\n 249 V196 float64\n 250 V197 float64\n 251 V198 float64\n 252 V199 float64\n 253 V200 float64\n 254 V201 float64\n 255 V202 float64\n 256 V203 float64\n 257 V204 float64\n 258 V205 float64\n 259 V206 float64\n 260 V207 float64\n 261 V208 float64\n 262 V209 float64\n 263 V210 float64\n 264 V211 float64\n 265 V212 float64\n 266 V213 float64\n 267 V214 float64\n 268 V215 float64\n 269 V216 float64\n 270 V217 float64\n 271 V218 float64\n 272 V219 float64\n 273 V220 float64\n 274 V221 float64\n 275 V222 float64\n 276 V223 float64\n 277 V224 float64\n 278 V225 float64\n 279 V226 float64\n 280 V227 float64\n 281 V228 float64\n 282 V229 float64\n 283 V230 float64\n 284 V231 float64\n 285 V232 float64\n 286 V233 float64\n 287 V234 float64\n 288 V235 float64\n 289 V236 float64\n 290 V237 float64\n 291 V238 float64\n 292 V239 float64\n 293 V240 float64\n 294 V241 float64\n 295 V242 float64\n 296 V243 float64\n 297 V244 float64\n 298 V245 float64\n 299 V246 float64\n 300 V247 float64\n 301 V248 float64\n 302 V249 float64\n 303 V250 float64\n 304 V251 float64\n 305 V252 float64\n 306 V253 float64\n 307 V254 float64\n 308 V255 float64\n 309 V256 float64\n 310 V257 float64\n 311 V258 float64\n 312 V259 float64\n 313 V260 float64\n 314 V261 float64\n 315 V262 float64\n 316 V263 float64\n 317 V264 float64\n 318 V265 float64\n 319 V266 float64\n 320 V267 float64\n 321 V268 float64\n 322 V269 float64\n 323 V270 float64\n 324 V271 float64\n 325 V272 float64\n 326 V273 float64\n 327 V274 float64\n 328 V275 float64\n 329 V276 float64\n 330 V277 float64\n 331 V278 float64\n 332 V279 float64\n 333 V280 float64\n 334 V281 float64\n 335 V282 float64\n 336 V283 float64\n 337 V284 float64\n 338 V285 float64\n 339 V286 float64\n 340 V287 float64\n 341 V288 float64\n 342 V289 float64\n 343 V290 float64\n 344 V291 float64\n 345 V292 float64\n 346 V293 float64\n 347 V294 float64\n 348 V295 float64\n 349 V296 float64\n 350 V297 float64\n 351 V298 float64\n 352 V299 float64\n 353 V300 float64\n 354 V301 float64\n 355 V302 float64\n 356 V303 float64\n 357 V304 float64\n 358 V305 float64\n 359 V306 float64\n 360 V307 float64\n 361 V308 float64\n 362 V309 float64\n 363 V310 float64\n 364 V311 float64\n 365 V312 float64\n 366 V313 float64\n 367 V314 float64\n 368 V315 float64\n 369 V316 float64\n 370 V317 float64\n 371 V318 float64\n 372 V319 float64\n 373 V320 float64\n 374 V321 float64\n 375 V322 float64\n 376 V323 float64\n 377 V324 float64\n 378 V325 float64\n 379 V326 float64\n 380 V327 float64\n 381 V328 float64\n 382 V329 float64\n 383 V330 float64\n 384 V331 float64\n 385 V332 float64\n 386 V333 float64\n 387 V334 float64\n 388 V335 float64\n 389 V336 float64\n 390 V337 float64\n 391 V338 float64\n 392 V339 float64\ndtypes: float64(376), int64(3), object(14)\nmemory usage: 1.5+ GB\n"
],
[
"# Para visualizar todas as colunas (antes de conhecer o atributo 'verbose'), criei uma Serie com o nome das colunas\ndf_test_transactionColumns = pd.Series(df_test_transaction.columns)",
"_____no_output_____"
],
[
"df_test_transactionColumns",
"_____no_output_____"
],
[
"# A ideia era expandir as colunas ao máximo, pois ao visualizar a serie acima os dados permaneceram truncados\npd.set_option('display.max_columns', None)",
"_____no_output_____"
],
[
"# Dai lembrei do Método unique\ndf_test_transactionColumns.unique()",
"_____no_output_____"
],
[
"df_test_transaction.isnull().sum().sort_values(ascending=False)",
"_____no_output_____"
],
[
"df_test_transaction_corr = df_test_transaction.corr()",
"_____no_output_____"
],
[
"# Tá o caos\nplt.figure(figsize=(15,8))\nsn.heatmap(df_test_transaction_corr, vmin=0, vmax=1)\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7933e9ccb01cb998239e9d03a5d452395703e2a | 7,828 | ipynb | Jupyter Notebook | notebooks/Tensorflow_RNN.ipynb | ThomasDelteil/DeepLearningFrameworks | 5cef028b39be2abc506d5c8a02f807c02995166c | [
"MIT"
] | 1,734 | 2017-08-30T13:56:28.000Z | 2022-03-24T11:13:09.000Z | notebooks/Tensorflow_RNN.ipynb | aoxiangzhang/DeepLearningFrameworks | 671e2a0198ff6b82babab2661295b8d49b4baccf | [
"MIT"
] | 59 | 2017-08-30T19:29:48.000Z | 2019-10-07T16:51:23.000Z | notebooks/Tensorflow_RNN.ipynb | aoxiangzhang/DeepLearningFrameworks | 671e2a0198ff6b82babab2661295b8d49b4baccf | [
"MIT"
] | 350 | 2017-08-30T17:17:14.000Z | 2022-02-24T12:11:18.000Z | 27.56338 | 109 | 0.520056 | [
[
[
"# High-level RNN TF Example",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport os\nimport sys\nimport tensorflow as tf\nfrom common.params_lstm import *\nfrom common.utils import *",
"_____no_output_____"
],
[
"# Force one-gpu\nos.environ[\"CUDA_VISIBLE_DEVICES\"] = \"0\"",
"_____no_output_____"
],
[
"print(\"OS: \", sys.platform)\nprint(\"Python: \", sys.version)\nprint(\"Numpy: \", np.__version__)\nprint(\"Tensorflow: \", tf.__version__)\nprint(\"GPU: \", get_gpu_name())\nprint(get_cuda_version())\nprint(\"CuDNN Version \", get_cudnn_version())",
"OS: linux\nPython: 3.5.2 |Anaconda custom (64-bit)| (default, Jul 2 2016, 17:53:06) \n[GCC 4.4.7 20120313 (Red Hat 4.4.7-1)]\nNumpy: 1.14.1\nTensorflow: 1.4.0\nGPU: ['Tesla P100-PCIE-16GB', 'Tesla P100-PCIE-16GB']\nCUDA Version 8.0.61\nCuDNN Version 6.0.21\n"
],
[
"def create_symbol(CUDNN=True, \n maxf=MAXFEATURES, edim=EMBEDSIZE, nhid=NUMHIDDEN, batchs=BATCHSIZE):\n word_vectors = tf.contrib.layers.embed_sequence(X, vocab_size=maxf, embed_dim=edim)\n word_list = tf.unstack(word_vectors, axis=1)\n \n if not CUDNN:\n cell = tf.contrib.rnn.GRUCell(nhid)\n outputs, states = tf.contrib.rnn.static_rnn(cell, word_list, dtype=tf.float32)\n else:\n # Using cuDNN since vanilla RNN\n from tensorflow.contrib.cudnn_rnn.python.ops import cudnn_rnn_ops\n cudnn_cell = cudnn_rnn_ops.CudnnGRU(num_layers=1, \n num_units=nhid, \n input_size=edim, \n input_mode='linear_input')\n params_size_t = cudnn_cell.params_size()\n params = tf.Variable(tf.random_uniform([params_size_t], -0.1, 0.1), validate_shape=False) \n input_h = tf.Variable(tf.zeros([1, batchs, nhid]))\n outputs, states = cudnn_cell(input_data=word_list,\n input_h=input_h,\n params=params)\n logits = tf.layers.dense(outputs[-1], 2, activation=None, name='output')\n return logits",
"_____no_output_____"
],
[
"def init_model(m, y, lr=LR, b1=BETA_1, b2=BETA_2, eps=EPS):\n # Single-class labels, don't need dense one-hot\n # Expects unscaled logits, not output of tf.nn.softmax\n xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(logits=m, labels=y)\n loss = tf.reduce_mean(xentropy)\n optimizer = tf.train.AdamOptimizer(lr, b1, b2, eps)\n training_op = optimizer.minimize(loss)\n return training_op",
"_____no_output_____"
],
[
"%%time\n# Data into format for library\nx_train, x_test, y_train, y_test = imdb_for_library(seq_len=MAXLEN, max_features=MAXFEATURES)\nprint(x_train.shape, x_test.shape, y_train.shape, y_test.shape)\nprint(x_train.dtype, x_test.dtype, y_train.dtype, y_test.dtype)",
"Preparing train set...\nPreparing test set...\nTrimming to 30000 max-features\nPadding to length 150\n(25000, 150) (25000, 150) (25000,) (25000,)\nint32 int32 int32 int32\nCPU times: user 5.9 s, sys: 417 ms, total: 6.32 s\nWall time: 6.32 s\n"
],
[
"%%time\n# Place-holders\nX = tf.placeholder(tf.int32, shape=[None, MAXLEN])\ny = tf.placeholder(tf.int32, shape=[None])\nsym = create_symbol()",
"CPU times: user 737 ms, sys: 76.1 ms, total: 814 ms\nWall time: 820 ms\n"
],
[
"%%time\nmodel = init_model(sym, y)\ninit = tf.global_variables_initializer()\nsess = tf.Session()\nsess.run(init)",
"CPU times: user 836 ms, sys: 693 ms, total: 1.53 s\nWall time: 1.54 s\n"
],
[
"%%time\n# Main training loop: 22s\ncorrect = tf.nn.in_top_k(sym, y, 1)\naccuracy = tf.reduce_mean(tf.cast(correct, tf.float32))\nfor j in range(EPOCHS):\n for data, label in yield_mb(x_train, y_train, BATCHSIZE, shuffle=True):\n sess.run(model, feed_dict={X: data, y: label})\n # Log\n acc_train = sess.run(accuracy, feed_dict={X: data, y: label})\n print(j, \"Train accuracy:\", acc_train)",
"0 Train accuracy: 0.84375\n1 Train accuracy: 0.96875\n2 Train accuracy: 0.984375\nCPU times: user 19 s, sys: 2.77 s, total: 21.8 s\nWall time: 22.2 s\n"
],
[
"%%time\n# Main evaluation loop: 9.19s\nn_samples = (y_test.shape[0]//BATCHSIZE)*BATCHSIZE\ny_guess = np.zeros(n_samples, dtype=np.int)\ny_truth = y_test[:n_samples]\nc = 0\nfor data, label in yield_mb(x_test, y_test, BATCHSIZE):\n pred = tf.argmax(sym, 1)\n output = sess.run(pred, feed_dict={X: data})\n y_guess[c*BATCHSIZE:(c+1)*BATCHSIZE] = output\n c += 1",
"CPU times: user 8.67 s, sys: 651 ms, total: 9.32 s\nWall time: 9.19 s\n"
],
[
"print(\"Accuracy: \", 1.*sum(y_guess == y_truth)/len(y_guess))",
"Accuracy: 0.8598557692307692\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7934efa7b7c934ffe4656667ad21f30c899708b | 22,402 | ipynb | Jupyter Notebook | l@s-2021/Question Engagement Analysis.ipynb | vitalsource/data | 877f584ed06b46b2f167c04875c8f4c3f37b0ae7 | [
"CC-BY-4.0"
] | 2 | 2021-09-29T17:47:13.000Z | 2021-11-12T15:29:25.000Z | l@s-2021/Question Engagement Analysis.ipynb | vitalsource/data | 877f584ed06b46b2f167c04875c8f4c3f37b0ae7 | [
"CC-BY-4.0"
] | null | null | null | l@s-2021/Question Engagement Analysis.ipynb | vitalsource/data | 877f584ed06b46b2f167c04875c8f4c3f37b0ae7 | [
"CC-BY-4.0"
] | 1 | 2021-07-23T19:03:48.000Z | 2021-07-23T19:03:48.000Z | 32.561047 | 137 | 0.409472 | [
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"%load_ext rpy2.ipython",
"_____no_output_____"
]
],
[
[
"# Question Engagement Analysis",
"_____no_output_____"
],
[
"## Select course and load data set",
"_____no_output_____"
]
],
[
[
"data_dir = '/Users/benny/data/L@S_2021'\ncourse = 'microbiology'",
"_____no_output_____"
],
[
"data_set_filename = f'engagement_{course}.txt'\ndata_set = pd.read_csv( f'{data_dir}/{data_set_filename}', sep='\\t' )",
"_____no_output_____"
],
[
"data_set",
"_____no_output_____"
]
],
[
[
"## Mean engagement",
"_____no_output_____"
]
],
[
[
"data_set.groupby( 'question_type' ).mean().sort_values( by='answered', ascending=False )",
"_____no_output_____"
]
],
[
[
"## Regression model",
"_____no_output_____"
]
],
[
[
"%%R\nlibrary( lme4 )",
"R[write to console]: Loading required package: Matrix\n\n"
]
],
[
[
"Standardize the continuous variables.",
"_____no_output_____"
]
],
[
[
"for col in [ 'course_page_number', 'unit_page_number', 'module_page_number', 'page_question_number' ]:\n data_set[ col ] = ( data_set[ col ] - data_set[ col ].mean() ) / data_set[ col ].std()",
"_____no_output_____"
],
[
"data_set.to_csv( '/tmp/to_r.csv', index=False )",
"_____no_output_____"
],
[
"%%R\ndf <- read.csv( '/tmp/to_r.csv' )",
"_____no_output_____"
],
[
"%%R\nlme.model <- glmer( answered ~ course_page_number + unit_page_number + module_page_number + page_question_number + question_type\n + (1|student) + (1|question), family=binomial(link=logit),\n data=df,\n control=glmerControl( optimizer=\"bobyqa\", optCtrl=list(maxfun=2e4) ) )\nsummary( lme.model )",
"R[write to console]: fixed-effect model matrix is rank deficient so dropping 1 column / coefficient\n\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
e7935487d4de6c1fa86ca1246e34ffd97747befb | 21,489 | ipynb | Jupyter Notebook | 10-warmup-solution_comprehensions.ipynb | hanisaf/advanced-data-management-and-analytics | e7bffda5cad91374a14df1a65f95e6a25f72cc41 | [
"MIT"
] | 6 | 2020-04-13T19:22:18.000Z | 2021-04-20T18:20:13.000Z | 10-warmup-solution_comprehensions.ipynb | hanisaf/advanced-data-management-and-analytics | e7bffda5cad91374a14df1a65f95e6a25f72cc41 | [
"MIT"
] | null | null | null | 10-warmup-solution_comprehensions.ipynb | hanisaf/advanced-data-management-and-analytics | e7bffda5cad91374a14df1a65f95e6a25f72cc41 | [
"MIT"
] | 10 | 2020-05-12T01:02:32.000Z | 2022-02-28T17:04:37.000Z | 20.465714 | 140 | 0.260598 | [
[
[
"1- Write a list comprehensions that contains numbers from 1 to 31, append the prefix \"1-\" to the numbers, these are day of January",
"_____no_output_____"
]
],
[
[
"L = [f\"1-{day}\" for day in range(1,32)]",
"_____no_output_____"
]
],
[
[
"2- convert the list to a string so each entry prints on one line. Print the result",
"_____no_output_____"
]
],
[
[
"line = '\\n'.join(L)\nprint(line)",
"1-1\n1-2\n1-3\n1-4\n1-5\n1-6\n1-7\n1-8\n1-9\n1-10\n1-11\n1-12\n1-13\n1-14\n1-15\n1-16\n1-17\n1-18\n1-19\n1-20\n1-21\n1-22\n1-23\n1-24\n1-25\n1-26\n1-27\n1-28\n1-29\n1-30\n1-31\n"
],
[
"from functools import reduce\nline = reduce(lambda x, y: f\"{x}\\n{y}\", L)\nprint(line)",
"1-1\n1-2\n1-3\n1-4\n1-5\n1-6\n1-7\n1-8\n1-9\n1-10\n1-11\n1-12\n1-13\n1-14\n1-15\n1-16\n1-17\n1-18\n1-19\n1-20\n1-21\n1-22\n1-23\n1-24\n1-25\n1-26\n1-27\n1-28\n1-29\n1-30\n1-31\n"
],
[
"def combine(x, y):\n return x + '\\n' + y\nline = reduce(combine, L)\nprint(line)",
"_____no_output_____"
]
],
[
[
"3- Update the comprehension to vary prefix from 1- to 12- to generate all days of the year, do not worry about incorrect dates for now",
"_____no_output_____"
]
],
[
[
"L = [ f\"{month}-{day}\" for month in range(1, 13) for day in range(1,32) ]\nL",
"_____no_output_____"
]
],
[
[
"4- now, address the issue of some months being only 30 days and february if 28 days (this year). Hint, use the `valid_date` function",
"_____no_output_____"
]
],
[
[
"def valid_date(day, month):\n days_of_month = {1:31, 2:28, 3:31, 4:30, 5:31, 6:30, \n 7:31, 8:31, 9:30, 10:31, 11:30, 12:31}\n max_month = days_of_month[month]\n return day <= max_month",
"_____no_output_____"
],
[
"L = [ f\"{month}-{day}\" for month in range(1, 13) for day in range(1,32) if valid_date(day, month)]\nL",
"_____no_output_____"
]
],
[
[
"5- using dictionary comprehensions create `f2e` dictionary from the `e2f` dictionary",
"_____no_output_____"
]
],
[
[
"e2f = {'hi':'bonjour', 'bye':'au revoir', 'bread':'pain', 'water':'eau'}",
"_____no_output_____"
],
[
"f2e = { e2f[k]:k for k in e2f}\nf2e",
"_____no_output_____"
],
[
"f2e = {item[1]:item[0] for item in e2f.items()}\nf2e",
"_____no_output_____"
],
[
"f2e = {v:k for (k,v) in e2f.items()}\nf2e",
"_____no_output_____"
],
[
"f2e = {e2f[k]:k for k in e2f}",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
e7935a4c1c87ad7456628f75cfeb1886c8cd5e5a | 11,095 | ipynb | Jupyter Notebook | examples/tutorial/04_Interlinked_Plots.ipynb | maximlt/holoviz | 9f86c3814928225864b9119eac357682faab5f3e | [
"BSD-3-Clause"
] | 207 | 2019-11-14T08:41:44.000Z | 2022-03-31T11:26:18.000Z | examples/tutorial/04_Interlinked_Plots.ipynb | maximlt/holoviz | 9f86c3814928225864b9119eac357682faab5f3e | [
"BSD-3-Clause"
] | 74 | 2019-11-21T16:39:45.000Z | 2022-02-15T16:46:51.000Z | examples/tutorial/04_Interlinked_Plots.ipynb | maximlt/holoviz | 9f86c3814928225864b9119eac357682faab5f3e | [
"BSD-3-Clause"
] | 36 | 2020-01-17T08:01:53.000Z | 2022-03-11T01:33:47.000Z | 47.212766 | 972 | 0.686886 | [
[
[
"<style>div.container { width: 100% }</style>\n<img style=\"float:left; vertical-align:text-bottom;\" height=\"65\" width=\"172\" src=\"../assets/holoviz-logo-unstacked.svg\" />\n<div style=\"float:right; vertical-align:text-bottom;\"><h2>Tutorial 4. Interlinked Plots</h2></div>",
"_____no_output_____"
],
[
"hvPlot allows you to generate a number of different types of plot quickly from a standard API, returning Bokeh-based [HoloViews](https://holoviews.org) objects as discussed in the previous notebook. Each initial plot will make some aspects of the data clear, and using the automatic interactive Bokeh pan, zoom, and hover tools you can find additional trends and outliers at different spatial locations and spatial scales within each plot.\n\nBeyond what you can discover from each plot individually, how do you understand how the various plots relate to each other? For instance, imagine you have a data frame with columns _u_, _v_, _w_, _z_, and have separate plots of _u_ vs. _v_, _u_ vs. _w_, and _w_ vs. _z_. If you see a few outliers or a clump of unusual datapoints in your _u_ vs. _v_ plot, how can you find out the properties of those points in the _w_ vs. _z_ or other plots? Are those unusual _u_ vs. _v_ points typically high _w_, uniformly distributed along _w_, or some other pattern? \n\nTo help understand multicolumnar and multidimensional datasets like this, scientists will often build complex multi-pane dashboards with custom functionality. HoloViz (and specifically Panel) tools are great for such dashboards, but here we can actually use the fact that hvPlot returns HoloViews objects to get quite sophisticated interlinking ([linked brushing](http://holoviews.org/user_guide/Linked_Brushing.html)) \"for free\", without needing to build any dashboard. HoloViews objects store metadata about what dimensions they cover, and we can use this metadata programmatically to let the user see how any data points in any plot relate across different plots.",
"_____no_output_____"
],
[
"To see how this works, let us get back to the example we were working on at the end of the last notebook:",
"_____no_output_____"
]
],
[
[
"import holoviews as hv\nimport pandas as pd\nimport hvplot.pandas # noqa\nimport colorcet as cc",
"_____no_output_____"
]
],
[
[
"First let us load the data as before:",
"_____no_output_____"
]
],
[
[
"%%time\ndf = pd.read_parquet('../data/earthquakes-projected.parq')\ndf.time = df.time.astype('datetime64[ns]')\ndf = df.set_index(df.time)",
"_____no_output_____"
]
],
[
[
"And filter to the most severe earthquakes (magnitude `> 7`):",
"_____no_output_____"
]
],
[
[
"most_severe = df[df.mag >= 7]",
"_____no_output_____"
]
],
[
[
"## Linked brushing across elements\n\nIn the previous notebook, we saw how plot axes are automatically linked for panning and zooming when using the `+` operator, provided the dimensions match. When dimensions or an underlying index match across multiple plots, we can use a similar principle to achieve linked brushing, where user selections are also linked across plots.\n\nTo illustrate, let us generate two histograms from our `most_severe_projected` DataFrame:",
"_____no_output_____"
]
],
[
[
"mag_hist = most_severe.hvplot(\n y='mag', kind='hist', responsive=True, min_height=150)\n\ndepth_hist = most_severe.hvplot(\n y='depth', kind='hist', responsive=True, min_height=150)",
"_____no_output_____"
]
],
[
[
"These two histograms are plotting two different dimensions of our earthquake dataset (magnitude and depth), derived from the same set of earthquake samples. The samples between these two histograms share an index, and the relationships between these data points can be discovered and exploited programmatically even though they are in different elements. To do this, we can create an object for linking selections across elements:",
"_____no_output_____"
]
],
[
[
"ls = hv.link_selections.instance()",
"_____no_output_____"
]
],
[
[
"Given some HoloViews objects (elements, layouts, etc.), we can create versions of them linked to this shared linking object by calling `ls` on them:",
"_____no_output_____"
]
],
[
[
"ls(depth_hist + mag_hist)",
"_____no_output_____"
]
],
[
[
"Try using the first Bokeh tool to select areas of either histogram: you'll then see both the depth and magnitude distributions for the bins you have selected, compared to the overall distribution. By default, selections on both histograms are combined so that the selection is the intersection of the two regions selected (data points matching _both_ the constraints on depth and the constraints on magnitude that you select). For instance, try selecting the deepest earthquakes (around 600), and you can see that those are not specific to one particular magnitude. You can then further select a particular magnitude range, and see how that range is distributed in depth over the selected depth range. Linked selections like this make it feasible to look at specific regions of a multidimensional space and see how the properties of those regions compare to the properties of other regions. You can use the Bokeh reset tool (double arrow) to clear your selection.\n\nNote that these two histograms are derived from the same `DataFrame` and created in the same call to `ls`, but neither of those is necessary to achieve the linked behavior! If linking two different `DataFrames`, the important thing to check is that any columns with the same name actually do have the same meaning, and that any index columns match, so that the plots you are visualizing make sense when linked together.\n\n## Linked brushing across element types\n\nThe previous example linked across two histograms as a first example, but nothing prevents you from linked brushing across different element types. Here are our earthquake points, also derived from the same `DataFrame`, where the only change from earlier is that we are using the reversed warm colormap (described in the previous notebook):",
"_____no_output_____"
]
],
[
[
"geo = most_severe.hvplot(\n 'easting', 'northing', color='mag', kind='points', tiles='ESRI', xlim=(-3e7,3e7), ylim=(-5e6,5e6),\n xaxis=None, yaxis=None, responsive=True, height=350, cmap = cc.CET_L4[::-1], framewise=True)",
"_____no_output_____"
]
],
[
[
"Once again, we just need to pass our points to the `ls` object (newly declared here to be independent of the one above) to declare the linkage:",
"_____no_output_____"
]
],
[
[
"ls2 = hv.link_selections.instance()\n\n(ls2(geo + depth_hist)).cols(1)",
"_____no_output_____"
]
],
[
[
"Now you can use the box-select tool to select earthquakes on the map and view their corresponding depth distribution, or vice versa. E.g. if you select just the earthquakes in Alaska, you can see that they tend not to be very deep underground (though that may be a sampling issue). Other selections will show other properties, in this case typically with no obvious relationship between geographic location and depth distribution.",
"_____no_output_____"
],
[
"## Accessing the data selection\n\nIf you pass your `DataFrame` into the `.filter` method of your linked selection object, you can apply the active filter that you specified interactively:",
"_____no_output_____"
]
],
[
[
"ls2.filter(most_severe)",
"_____no_output_____"
]
],
[
[
"#### Exercise\n\nTry selecting a small number of earthquakes on the map above and re-running the previous cell. You should see that your `DataFrame` only includes the earthquakes you have selected. You can use this linked selections feature in your own workflows by selecting a region of your data, then running subsequent analyses only on that subset of the data (or comparing that subset to the whole data set).",
"_____no_output_____"
],
[
"## Conclusion\n\nWhen exploring data it can be convenient to use the `.plot` API to quickly visualize a particular dataset. By calling `.hvplot` to generate different plots over the course of a session, it is possible to gradually build up a mental model of how a particular dataset is structured. Linked selections let you see relationships between your data's dimensions and clusters of datapoints much more directly, so that you can:\n\n1. Interactively explore high-dimensional data by making selections across different views of the same underlying samples.\n2. Turn this interactive exploration into a Python subselection of your data, allowing you to continue your data analysis on a subset of your data that you interactively selected.\n\nThis approach is very general and allows a deeper understanding of high-dimensional data through interactivity. This interactivity is itself built on the very powerful HoloViews 'streams' system which you can leverage for yourself to build youw own [Custom Interactivity](./07_Custom_Interactivity.ipynb) (optional, advanced topic) when necessary.\n\nIn the next section we will see how to apply data processing in a pipelined form, allowing us to build interactive visualizations driven by user-defined widgets when we want to have custom control over our data processing and selection.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
e7936d65d1e452687ce0b38b3cddf94f852d9620 | 2,018 | ipynb | Jupyter Notebook | Section01/Understanding Spark.ipynb | badattaram/Pyspark_pack_notebooks | 60c32cb55cc43c28a5ea865a70a5c074f30cf4b5 | [
"MIT"
] | 9 | 2018-06-28T04:53:06.000Z | 2021-11-08T13:10:26.000Z | Section01/Understanding Spark.ipynb | badattaram/Pyspark_pack_notebooks | 60c32cb55cc43c28a5ea865a70a5c074f30cf4b5 | [
"MIT"
] | null | null | null | Section01/Understanding Spark.ipynb | badattaram/Pyspark_pack_notebooks | 60c32cb55cc43c28a5ea865a70a5c074f30cf4b5 | [
"MIT"
] | 16 | 2018-07-10T05:56:36.000Z | 2021-09-18T00:13:39.000Z | 19.980198 | 93 | 0.416254 | [
[
[
"sc",
"_____no_output_____"
],
[
"sqlContext",
"_____no_output_____"
],
[
"print(sc.version)",
"2.3.0\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
e793790e319d9f8aaf4b824aa04258b407d4d65a | 4,845 | ipynb | Jupyter Notebook | src/cifar_10_experimentation.ipynb | SonuDixit/tipr-third-assignmen | 28e4b7832952f46b72dbd99900afc8547e6c55fc | [
"MIT"
] | null | null | null | src/cifar_10_experimentation.ipynb | SonuDixit/tipr-third-assignmen | 28e4b7832952f46b72dbd99900afc8547e6c55fc | [
"MIT"
] | null | null | null | src/cifar_10_experimentation.ipynb | SonuDixit/tipr-third-assignmen | 28e4b7832952f46b72dbd99900afc8547e6c55fc | [
"MIT"
] | null | null | null | 25.909091 | 138 | 0.479051 | [
[
[
"from utils import unpickle_cifar,get_batch_data_cifar\nimport os\nimport numpy as np\nfrom sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"from cnn import CNN\nnet = CNN(input_dim = (32,32,1),\n filter_size=[3,4,2],\n output_dim = 10\n )",
"_____no_output_____"
],
[
"images,label = get_batch_data_cifar(1)\nfor i in range(2,6):\n i,l = get_batch_data_cifar(i)\n images = np.vstack((images,i))\n label.extend(l)\n\nimages, x_test, label, y_test = train_test_split(images,label,test_size = 0.25)",
"_____no_output_____"
],
[
"images.shape",
"_____no_output_____"
],
[
"images2 =images.reshape(images.shape[0],32,32,1)",
"_____no_output_____"
],
[
"h = net.fit(images2,label,epoc=10)",
"Train on 30000 samples, validate on 7500 samples\nEpoch 1/10\n30000/30000 [==============================] - 15s 501us/step - loss: 1.8567 - acc: 0.3207 - val_loss: 1.6600 - val_acc: 0.3924\nEpoch 2/10\n30000/30000 [==============================] - 12s 404us/step - loss: 1.5697 - acc: 0.4314 - val_loss: 1.4834 - val_acc: 0.4580\nEpoch 3/10\n30000/30000 [==============================] - 12s 406us/step - loss: 1.4661 - acc: 0.4741 - val_loss: 1.4246 - val_acc: 0.4907\nEpoch 4/10\n30000/30000 [==============================] - 12s 406us/step - loss: 1.4033 - acc: 0.5004 - val_loss: 1.3431 - val_acc: 0.5301\nEpoch 5/10\n30000/30000 [==============================] - 12s 406us/step - loss: 1.3505 - acc: 0.5162 - val_loss: 1.3278 - val_acc: 0.5303\nEpoch 6/10\n30000/30000 [==============================] - 12s 403us/step - loss: 1.3080 - acc: 0.5344 - val_loss: 1.2523 - val_acc: 0.5584\nEpoch 7/10\n30000/30000 [==============================] - 12s 402us/step - loss: 1.2778 - acc: 0.5462 - val_loss: 1.2469 - val_acc: 0.5621\nEpoch 8/10\n30000/30000 [==============================] - 12s 401us/step - loss: 1.2489 - acc: 0.5551 - val_loss: 1.2448 - val_acc: 0.5675\nEpoch 9/10\n30000/30000 [==============================] - 12s 403us/step - loss: 1.2327 - acc: 0.5616 - val_loss: 1.2000 - val_acc: 0.5741\nEpoch 10/10\n30000/30000 [==============================] - 12s 402us/step - loss: 1.2132 - acc: 0.5682 - val_loss: 1.2323 - val_acc: 0.5572\n"
],
[
"h.history.keys()",
"_____no_output_____"
],
[
"x_test = x_test.reshape((x_test.shape[0],32,32,1))\nnet.evaluate(x_test,y_test)",
"12500/12500 [==============================] - 2s 163us/step\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7938027f693ca7909af0ea8667e4984b06e14f7 | 91,627 | ipynb | Jupyter Notebook | notebooks/1_Lab scan collection.ipynb | laserkelvin/SlowFourierTransform | ea59df6c03d32a3de727aeca0f97e8c9cf446459 | [
"MIT"
] | null | null | null | notebooks/1_Lab scan collection.ipynb | laserkelvin/SlowFourierTransform | ea59df6c03d32a3de727aeca0f97e8c9cf446459 | [
"MIT"
] | null | null | null | notebooks/1_Lab scan collection.ipynb | laserkelvin/SlowFourierTransform | ea59df6c03d32a3de727aeca0f97e8c9cf446459 | [
"MIT"
] | null | null | null | 190.09751 | 40,310 | 0.593428 | [
[
[
"import pandas as pd\nimport numpy as np\nfrom plotly import graph_objs as go\nfrom plotly.offline import plot\nfrom uncertainties import ufloat\nimport datetime",
"_____no_output_____"
]
],
[
[
"# How does our lab collect data?\n\nHere was a small Python project that I thought of - are there trends in the rate of data collection in our lab at the CfA? From a qualitative sense, it always felt that when visitors come, several come at once and one would expect this would reflect in the number of scans produced in a small period of time.\n\nAnother question I'd like to ask is how long do we typically accumulate data for? This is reflected in the number of \"shots\", i.e. the number of accumulations at a repetition rate of 5 Hz (typically).\n\nFinally, what are the most common frequencies the spectrometers are tuned to.\n\nI have to state that I'm not sure what I'll find - this is mainly an excercise in Python (Pandas/Plotly)",
"_____no_output_____"
]
],
[
[
"ft1_df = pd.read_pickle(\"../data/FTM1_scans.pkl\")\nft2_df = pd.read_pickle(\"../data/FTM2_scans.pkl\")",
"_____no_output_____"
],
[
"# Convert the datetime handling into numpy format\nfor df in [ft1_df, ft2_df]:\n df[\"date\"] = df[\"date\"].astype(\"datetime64\")",
"_____no_output_____"
]
],
[
[
"Simple statistics behind the data collection, I'll be using FT1, and also exclude the last row (which is 2019).",
"_____no_output_____"
]
],
[
[
"yearly = ft1_df.groupby([ft1_df[\"date\"].dt.year])",
"_____no_output_____"
]
],
[
[
"Average number of scans per year",
"_____no_output_____"
]
],
[
[
"scans = ufloat(\n np.average(yearly[\"shots\"].describe()[\"count\"].iloc[:-1]),\n np.std(yearly[\"shots\"].describe()[\"count\"].iloc[:-1])\n)",
"_____no_output_____"
],
[
"scans",
"_____no_output_____"
],
[
"shots = ufloat(\n np.average(yearly[\"shots\"].describe()[\"mean\"].iloc[:-1]),\n np.std(yearly[\"shots\"].describe()[\"mean\"].iloc[:-1])\n)",
"_____no_output_____"
],
[
"shots",
"_____no_output_____"
]
],
[
[
"Convert this to time spent per year in days",
"_____no_output_____"
]
],
[
[
"((shots / 5.) * scans) / 60. / 60. / 24.",
"_____no_output_____"
]
],
[
[
"What's the actual number of shots in a year?",
"_____no_output_____"
]
],
[
[
"actual_shots = ufloat(\n np.average(yearly.sum()[\"shots\"].iloc[:-1]),\n np.std(yearly.sum()[\"shots\"].iloc[:-1])\n)",
"_____no_output_____"
],
[
"actual_shots",
"_____no_output_____"
],
[
"(actual_shots / 5. / 60.) / 60. / 24.",
"_____no_output_____"
]
],
[
[
"So approximately, the experiments are taking data only for 42 days a year total. Of course, this doesn't reflect reality (you spend most of the time trying to make the experiment work the way you want to of course). I'm also curious how this compares with other labs...",
"_____no_output_____"
]
],
[
[
"# Bin all of the data into year, month, and day\ngrouped_dfs = [\n df.groupby([df[\"date\"].dt.year, df[\"date\"].dt.month, df[\"date\"].dt.day]).count() for df in [ft1_df, ft2_df]\n]",
"_____no_output_____"
],
[
"for df in grouped_dfs:\n df[\"cumulative\"] = np.cumsum(df[\"id\"])",
"_____no_output_____"
],
[
"flattened_dfs = [\n df.set_index(df.index.map(lambda t: pd.datetime(*t))) for df in grouped_dfs\n]",
"_____no_output_____"
],
[
"layout = {\n \"height\": 600.,\n \"yaxis\": {\n \"title\": \"Number of scans\",\n },\n \"xaxis\": {\n \"title\": \"Time\"\n },\n \"title\": \"How we collect data\",\n \"showlegend\": True,\n \"legend\": {\n \"x\": 0.1,\n \"y\": 0.95\n }\n }\n\nfig = go.FigureWidget(layout=layout)\n\ntraces = [\n fig.add_scattergl(x=df.index, y=df[\"cumulative\"], name=name) for df, name in zip(flattened_dfs, [\"FT1\", \"FT2\"])\n]\n\nisms_times = [datetime.datetime(year=year, month=6, day=17) for year in [2014, 2015, 2016, 2017, 2018]]\n\nfig.add_bar(\n x=isms_times,\n y=[2e6] * len(isms_times),\n width=2e6,\n hoverinfo=\"name\",\n name=\"ISMS\"\n)\n\nfig",
"_____no_output_____"
],
[
"print(plot(fig, show_link=False, link_text=\"\", output_type=\"div\", include_plotlyjs=False))",
"<div><div id=\"ed88ed28-c295-4182-9c04-629162ca1a83\" style=\"height: 600.0px; width: 100%;\" class=\"plotly-graph-div\"></div><script type=\"text/javascript\">window.PLOTLYENV=window.PLOTLYENV || {};window.PLOTLYENV.BASE_URL=\"https://plot.ly\";Plotly.newPlot(\"ed88ed28-c295-4182-9c04-629162ca1a83\", [{\"name\": \"FT1\", \"x\": [\"2014-07-08 04:00:00\", \"2014-07-09 04:00:00\", \"2014-07-10 04:00:00\", \"2014-07-11 04:00:00\", \"2014-07-13 04:00:00\", \"2014-07-14 04:00:00\", \"2014-07-15 04:00:00\", \"2014-07-16 04:00:00\", \"2014-07-17 04:00:00\", \"2014-07-18 04:00:00\", \"2014-07-21 04:00:00\", \"2014-07-22 04:00:00\", \"2014-07-23 04:00:00\", \"2014-07-25 04:00:00\", \"2014-07-28 04:00:00\", \"2014-07-29 04:00:00\", \"2014-07-30 04:00:00\", \"2014-07-31 04:00:00\", \"2014-08-01 04:00:00\", \"2014-08-05 04:00:00\", \"2014-08-06 04:00:00\", \"2014-08-07 04:00:00\", \"2014-08-11 04:00:00\", \"2014-08-12 04:00:00\", \"2014-08-13 04:00:00\", \"2014-08-14 04:00:00\", \"2014-08-15 04:00:00\", \"2014-08-16 04:00:00\", \"2014-08-18 04:00:00\", \"2014-08-19 04:00:00\", \"2014-08-21 04:00:00\", \"2014-08-22 04:00:00\", \"2014-09-03 04:00:00\", \"2014-09-04 04:00:00\", \"2014-09-08 04:00:00\", \"2014-09-09 04:00:00\", \"2014-09-10 04:00:00\", \"2014-09-11 04:00:00\", \"2014-09-12 04:00:00\", \"2014-09-15 04:00:00\", \"2014-09-17 04:00:00\", \"2014-09-18 04:00:00\", \"2014-09-19 04:00:00\", \"2014-09-25 04:00:00\", \"2014-09-26 04:00:00\", \"2014-10-02 04:00:00\", \"2014-10-03 04:00:00\", \"2014-10-06 04:00:00\", \"2014-10-08 04:00:00\", \"2014-10-17 04:00:00\", \"2014-10-20 04:00:00\", \"2014-10-21 04:00:00\", \"2014-10-22 04:00:00\", \"2014-10-23 04:00:00\", \"2014-10-24 04:00:00\", \"2014-11-03 05:00:00\", \"2014-11-05 05:00:00\", \"2014-11-10 05:00:00\", \"2014-11-12 05:00:00\", \"2014-11-13 05:00:00\", \"2014-11-14 05:00:00\", \"2014-11-19 05:00:00\", \"2014-11-25 05:00:00\", \"2014-12-01 05:00:00\", \"2014-12-02 05:00:00\", \"2014-12-03 05:00:00\", \"2014-12-04 05:00:00\", \"2014-12-09 05:00:00\", \"2014-12-10 05:00:00\", \"2014-12-11 05:00:00\", \"2014-12-12 05:00:00\", \"2014-12-15 05:00:00\", \"2014-12-16 05:00:00\", \"2014-12-17 05:00:00\", \"2014-12-18 05:00:00\", \"2014-12-19 05:00:00\", \"2014-12-20 05:00:00\", \"2014-12-22 05:00:00\", \"2014-12-23 05:00:00\", \"2015-01-16 05:00:00\", \"2015-01-21 05:00:00\", \"2015-01-22 05:00:00\", \"2015-01-26 05:00:00\", \"2015-01-28 05:00:00\", \"2015-02-04 05:00:00\", \"2015-02-11 05:00:00\", \"2015-02-12 05:00:00\", \"2015-02-13 05:00:00\", \"2015-02-19 05:00:00\", \"2015-02-20 05:00:00\", \"2015-02-24 05:00:00\", \"2015-02-25 05:00:00\", \"2015-02-26 05:00:00\", \"2015-02-27 05:00:00\", \"2015-03-10 04:00:00\", \"2015-03-11 04:00:00\", \"2015-03-12 04:00:00\", \"2015-03-13 04:00:00\", \"2015-03-14 04:00:00\", \"2015-03-16 04:00:00\", \"2015-03-20 04:00:00\", \"2015-03-23 04:00:00\", \"2015-03-26 04:00:00\", \"2015-03-27 04:00:00\", \"2015-03-30 04:00:00\", \"2015-03-31 04:00:00\", \"2015-04-01 04:00:00\", \"2015-04-02 04:00:00\", \"2015-04-03 04:00:00\", \"2015-04-04 04:00:00\", \"2015-04-06 04:00:00\", \"2015-04-07 04:00:00\", \"2015-04-14 04:00:00\", \"2015-04-15 04:00:00\", \"2015-04-16 04:00:00\", \"2015-04-17 04:00:00\", \"2015-04-18 04:00:00\", \"2015-04-20 04:00:00\", \"2015-04-22 04:00:00\", \"2015-04-23 04:00:00\", \"2015-04-24 04:00:00\", \"2015-04-29 04:00:00\", \"2015-05-05 04:00:00\", \"2015-05-06 04:00:00\", \"2015-05-07 04:00:00\", \"2015-05-08 04:00:00\", \"2015-05-11 04:00:00\", \"2015-05-12 04:00:00\", \"2015-05-14 04:00:00\", \"2015-05-15 04:00:00\", \"2015-05-18 04:00:00\", \"2015-05-19 04:00:00\", \"2015-05-20 04:00:00\", \"2015-05-21 04:00:00\", \"2015-05-26 04:00:00\", \"2015-05-27 04:00:00\", \"2015-05-28 04:00:00\", \"2015-05-29 04:00:00\", \"2015-06-01 04:00:00\", \"2015-06-02 04:00:00\", \"2015-06-03 04:00:00\", \"2015-06-04 04:00:00\", \"2015-06-05 04:00:00\", \"2015-06-06 04:00:00\", \"2015-06-07 04:00:00\", \"2015-06-08 04:00:00\", \"2015-06-09 04:00:00\", \"2015-06-10 04:00:00\", \"2015-06-11 04:00:00\", \"2015-06-12 04:00:00\", \"2015-06-15 04:00:00\", \"2015-06-16 04:00:00\", \"2015-06-17 04:00:00\", \"2015-06-18 04:00:00\", \"2015-06-19 04:00:00\", \"2015-06-20 04:00:00\", \"2015-06-22 04:00:00\", \"2015-06-24 04:00:00\", \"2015-06-25 04:00:00\", \"2015-06-29 04:00:00\", \"2015-06-30 04:00:00\", \"2015-07-08 04:00:00\", \"2015-07-14 04:00:00\", \"2015-07-15 04:00:00\", \"2015-07-16 04:00:00\", \"2015-07-17 04:00:00\", \"2015-07-20 04:00:00\", \"2015-07-22 04:00:00\", \"2015-07-23 04:00:00\", \"2015-07-24 04:00:00\", \"2015-07-25 04:00:00\", \"2015-07-27 04:00:00\", \"2015-07-28 04:00:00\", \"2015-07-29 04:00:00\", \"2015-07-30 04:00:00\", \"2015-07-31 04:00:00\", \"2015-08-05 04:00:00\", \"2015-08-06 04:00:00\", \"2015-08-07 04:00:00\", \"2015-08-14 04:00:00\", \"2015-08-16 04:00:00\", \"2015-08-20 04:00:00\", \"2015-08-21 04:00:00\", \"2015-08-22 04:00:00\", \"2015-08-23 04:00:00\", \"2015-08-24 04:00:00\", \"2015-08-25 04:00:00\", \"2015-08-26 04:00:00\", \"2015-08-27 04:00:00\", \"2015-08-28 04:00:00\", \"2015-08-29 04:00:00\", \"2015-08-30 04:00:00\", \"2015-08-31 04:00:00\", \"2015-09-01 04:00:00\", \"2015-09-02 04:00:00\", \"2015-09-03 04:00:00\", \"2015-09-04 04:00:00\", \"2015-09-08 04:00:00\", \"2015-09-09 04:00:00\", \"2015-09-10 04:00:00\", \"2015-09-11 04:00:00\", \"2015-09-12 04:00:00\", \"2015-09-13 04:00:00\", \"2015-09-14 04:00:00\", \"2015-09-16 04:00:00\", \"2015-09-17 04:00:00\", \"2015-09-18 04:00:00\", \"2015-09-19 04:00:00\", \"2015-09-20 04:00:00\", \"2015-09-21 04:00:00\", \"2015-09-22 04:00:00\", \"2015-09-23 04:00:00\", \"2015-10-06 04:00:00\", \"2015-10-07 04:00:00\", \"2015-10-08 04:00:00\", \"2015-10-13 04:00:00\", \"2015-10-14 04:00:00\", \"2015-10-20 04:00:00\", \"2015-10-21 04:00:00\", \"2015-10-22 04:00:00\", \"2015-10-23 04:00:00\", \"2015-10-24 04:00:00\", \"2015-10-25 04:00:00\", \"2015-10-26 04:00:00\", \"2015-10-28 04:00:00\", \"2015-10-29 04:00:00\", \"2015-10-30 04:00:00\", \"2015-10-31 04:00:00\", \"2015-11-02 05:00:00\", \"2015-11-03 05:00:00\", \"2015-11-04 05:00:00\", \"2015-11-05 05:00:00\", \"2015-11-06 05:00:00\", \"2015-11-09 05:00:00\", \"2015-11-10 05:00:00\", \"2015-11-12 05:00:00\", \"2015-11-13 05:00:00\", \"2015-11-19 05:00:00\", \"2015-11-20 05:00:00\", \"2015-11-21 05:00:00\", \"2015-11-22 05:00:00\", \"2015-11-23 05:00:00\", \"2015-11-24 05:00:00\", \"2015-11-25 05:00:00\", \"2015-11-30 05:00:00\", \"2015-12-01 05:00:00\", \"2015-12-02 05:00:00\", \"2015-12-03 05:00:00\", \"2015-12-04 05:00:00\", \"2015-12-08 05:00:00\", \"2015-12-09 05:00:00\", \"2015-12-10 05:00:00\", \"2015-12-11 05:00:00\", \"2015-12-14 05:00:00\", \"2015-12-15 05:00:00\", \"2015-12-22 05:00:00\", \"2015-12-23 05:00:00\", \"2016-01-07 05:00:00\", \"2016-01-19 05:00:00\", \"2016-01-20 05:00:00\", \"2016-01-21 05:00:00\", \"2016-01-25 05:00:00\", \"2016-01-26 05:00:00\", \"2016-01-27 05:00:00\", \"2016-02-03 05:00:00\", \"2016-02-04 05:00:00\", \"2016-02-05 05:00:00\", \"2016-02-08 05:00:00\", \"2016-02-09 05:00:00\", \"2016-02-10 05:00:00\", \"2016-02-11 05:00:00\", \"2016-02-12 05:00:00\", \"2016-02-15 05:00:00\", \"2016-02-16 05:00:00\", \"2016-02-18 05:00:00\", \"2016-02-19 05:00:00\", \"2016-02-20 05:00:00\", \"2016-02-22 05:00:00\", \"2016-02-23 05:00:00\", \"2016-02-24 05:00:00\", \"2016-02-25 05:00:00\", \"2016-03-02 05:00:00\", \"2016-03-03 05:00:00\", \"2016-03-04 05:00:00\", \"2016-03-05 05:00:00\", \"2016-03-09 05:00:00\", \"2016-03-10 05:00:00\", \"2016-03-11 05:00:00\", \"2016-03-14 04:00:00\", \"2016-03-15 04:00:00\", \"2016-03-18 04:00:00\", \"2016-03-21 04:00:00\", \"2016-03-22 04:00:00\", \"2016-03-23 04:00:00\", \"2016-03-24 04:00:00\", \"2016-03-30 04:00:00\", \"2016-04-27 04:00:00\", \"2016-04-28 04:00:00\", \"2016-04-29 04:00:00\", \"2016-04-30 04:00:00\", \"2016-05-01 04:00:00\", \"2016-05-02 04:00:00\", \"2016-05-03 04:00:00\", \"2016-05-04 04:00:00\", \"2016-05-05 04:00:00\", \"2016-05-06 04:00:00\", \"2016-05-09 04:00:00\", \"2016-05-10 04:00:00\", \"2016-05-13 04:00:00\", \"2016-05-14 04:00:00\", \"2016-05-15 04:00:00\", \"2016-05-16 04:00:00\", \"2016-05-17 04:00:00\", \"2016-05-18 04:00:00\", \"2016-05-19 04:00:00\", \"2016-05-20 04:00:00\", \"2016-05-23 04:00:00\", \"2016-05-25 04:00:00\", \"2016-05-26 04:00:00\", \"2016-05-27 04:00:00\", \"2016-05-31 04:00:00\", \"2016-06-01 04:00:00\", \"2016-06-02 04:00:00\", \"2016-06-07 04:00:00\", \"2016-06-08 04:00:00\", \"2016-06-09 04:00:00\", \"2016-06-10 04:00:00\", \"2016-06-11 04:00:00\", \"2016-06-13 04:00:00\", \"2016-06-14 04:00:00\", \"2016-06-15 04:00:00\", \"2016-06-30 04:00:00\", \"2016-07-01 04:00:00\", \"2016-07-05 04:00:00\", \"2016-07-11 04:00:00\", \"2016-07-12 04:00:00\", \"2016-07-13 04:00:00\", \"2016-07-14 04:00:00\", \"2016-07-15 04:00:00\", \"2016-07-25 04:00:00\", \"2016-08-04 04:00:00\", \"2016-08-05 04:00:00\", \"2016-08-09 04:00:00\", \"2016-08-10 04:00:00\", \"2016-08-11 04:00:00\", \"2016-08-12 04:00:00\", \"2016-08-13 04:00:00\", \"2016-08-14 04:00:00\", \"2016-08-15 04:00:00\", \"2016-08-19 04:00:00\", \"2016-08-20 04:00:00\", \"2016-08-21 04:00:00\", \"2016-08-22 04:00:00\", \"2016-08-23 04:00:00\", \"2016-08-24 04:00:00\", \"2016-08-25 04:00:00\", \"2016-08-26 04:00:00\", \"2016-08-31 04:00:00\", \"2016-09-01 04:00:00\", \"2016-09-02 04:00:00\", \"2016-09-03 04:00:00\", \"2016-09-04 04:00:00\", \"2016-09-05 04:00:00\", \"2016-09-12 04:00:00\", \"2016-09-14 04:00:00\", \"2016-09-16 04:00:00\", \"2016-10-03 04:00:00\", \"2016-10-04 04:00:00\", \"2016-10-05 04:00:00\", \"2016-10-06 04:00:00\", \"2016-10-07 04:00:00\", \"2016-10-08 04:00:00\", \"2016-10-10 04:00:00\", \"2016-10-11 04:00:00\", \"2016-10-12 04:00:00\", \"2016-10-13 04:00:00\", \"2016-10-14 04:00:00\", \"2016-10-20 04:00:00\", \"2016-10-21 04:00:00\", \"2016-10-24 04:00:00\", \"2016-10-25 04:00:00\", \"2016-10-26 04:00:00\", \"2016-10-27 04:00:00\", \"2016-10-28 04:00:00\", \"2016-10-30 04:00:00\", \"2016-10-31 04:00:00\", \"2016-11-01 04:00:00\", \"2016-11-02 04:00:00\", \"2016-11-03 04:00:00\", \"2016-11-04 04:00:00\", \"2016-11-05 04:00:00\", \"2016-11-06 04:00:00\", \"2016-11-07 05:00:00\", \"2016-11-08 05:00:00\", \"2016-11-09 05:00:00\", \"2016-11-10 05:00:00\", \"2016-11-11 05:00:00\", \"2016-11-12 05:00:00\", \"2016-11-13 05:00:00\", \"2016-11-14 05:00:00\", \"2016-11-15 05:00:00\", \"2016-11-16 05:00:00\", \"2016-11-17 05:00:00\", \"2016-11-18 05:00:00\", \"2016-11-21 05:00:00\", \"2016-11-22 05:00:00\", \"2016-11-23 05:00:00\", \"2016-11-26 05:00:00\", \"2016-11-28 05:00:00\", \"2016-11-29 05:00:00\", \"2016-11-30 05:00:00\", \"2016-12-01 05:00:00\", \"2016-12-02 05:00:00\", \"2016-12-05 05:00:00\", \"2016-12-06 05:00:00\", \"2016-12-07 05:00:00\", \"2016-12-08 05:00:00\", \"2016-12-09 05:00:00\", \"2016-12-12 05:00:00\", \"2016-12-13 05:00:00\", \"2016-12-14 05:00:00\", \"2016-12-15 05:00:00\", \"2016-12-16 05:00:00\", \"2017-01-05 05:00:00\", \"2017-01-06 05:00:00\", \"2017-01-09 05:00:00\", \"2017-01-10 05:00:00\", \"2017-01-11 05:00:00\", \"2017-01-12 05:00:00\", \"2017-01-13 05:00:00\", \"2017-01-16 05:00:00\", \"2017-01-17 05:00:00\", \"2017-01-18 05:00:00\", \"2017-01-19 05:00:00\", \"2017-01-20 05:00:00\", \"2017-01-21 05:00:00\", \"2017-01-23 05:00:00\", \"2017-01-24 05:00:00\", \"2017-01-25 05:00:00\", \"2017-01-26 05:00:00\", \"2017-01-27 05:00:00\", \"2017-01-30 05:00:00\", \"2017-01-31 05:00:00\", \"2017-02-07 05:00:00\", \"2017-02-08 05:00:00\", \"2017-02-14 05:00:00\", \"2017-02-15 05:00:00\", \"2017-02-16 05:00:00\", \"2017-02-17 05:00:00\", \"2017-03-01 05:00:00\", \"2017-03-02 05:00:00\", \"2017-03-06 05:00:00\", \"2017-03-07 05:00:00\", \"2017-03-08 05:00:00\", \"2017-03-09 05:00:00\", \"2017-03-13 04:00:00\", \"2017-03-15 04:00:00\", \"2017-03-21 04:00:00\", \"2017-03-22 04:00:00\", \"2017-03-23 04:00:00\", \"2017-03-24 04:00:00\", \"2017-03-28 04:00:00\", \"2017-03-29 04:00:00\", \"2017-03-30 04:00:00\", \"2017-03-31 04:00:00\", \"2017-04-01 04:00:00\", \"2017-04-02 04:00:00\", \"2017-04-03 04:00:00\", \"2017-04-04 04:00:00\", \"2017-04-05 04:00:00\", \"2017-04-06 04:00:00\", \"2017-04-07 04:00:00\", \"2017-04-10 04:00:00\", \"2017-04-11 04:00:00\", \"2017-04-12 04:00:00\", \"2017-04-13 04:00:00\", \"2017-04-14 04:00:00\", \"2017-04-18 04:00:00\", \"2017-04-19 04:00:00\", \"2017-04-20 04:00:00\", \"2017-04-21 04:00:00\", \"2017-04-23 04:00:00\", \"2017-04-24 04:00:00\", \"2017-04-25 04:00:00\", \"2017-04-26 04:00:00\", \"2017-04-28 04:00:00\", \"2017-05-01 04:00:00\", \"2017-05-02 04:00:00\", \"2017-05-03 04:00:00\", \"2017-05-04 04:00:00\", \"2017-05-05 04:00:00\", \"2017-05-06 04:00:00\", \"2017-05-07 04:00:00\", \"2017-05-08 04:00:00\", \"2017-05-09 04:00:00\", \"2017-05-10 04:00:00\", \"2017-05-11 04:00:00\", \"2017-05-12 04:00:00\", \"2017-05-13 04:00:00\", \"2017-05-14 04:00:00\", \"2017-05-15 04:00:00\", \"2017-05-16 04:00:00\", \"2017-05-17 04:00:00\", \"2017-05-19 04:00:00\", \"2017-06-01 04:00:00\", \"2017-06-02 04:00:00\", \"2017-06-05 04:00:00\", \"2017-06-06 04:00:00\", \"2017-06-08 04:00:00\", \"2017-06-09 04:00:00\", \"2017-06-12 04:00:00\", \"2017-06-13 04:00:00\", \"2017-06-26 04:00:00\", \"2017-06-27 04:00:00\", \"2017-06-28 04:00:00\", \"2017-06-29 04:00:00\", \"2017-06-30 04:00:00\", \"2017-07-03 04:00:00\", \"2017-07-12 04:00:00\", \"2017-07-13 04:00:00\", \"2017-07-14 04:00:00\", \"2017-07-17 04:00:00\", \"2017-07-19 04:00:00\", \"2017-07-20 04:00:00\", \"2017-07-25 04:00:00\", \"2017-07-26 04:00:00\", \"2017-07-27 04:00:00\", \"2017-07-28 04:00:00\", \"2017-08-03 04:00:00\", \"2017-08-04 04:00:00\", \"2017-08-05 04:00:00\", \"2017-08-06 04:00:00\", \"2017-08-07 04:00:00\", \"2017-08-08 04:00:00\", \"2017-08-09 04:00:00\", \"2017-08-10 04:00:00\", \"2017-08-11 04:00:00\", \"2017-08-14 04:00:00\", \"2017-08-15 04:00:00\", \"2017-08-16 04:00:00\", \"2017-08-17 04:00:00\", \"2017-08-18 04:00:00\", \"2017-08-21 04:00:00\", \"2017-08-22 04:00:00\", \"2017-08-25 04:00:00\", \"2017-08-28 04:00:00\", \"2017-08-29 04:00:00\", \"2017-08-30 04:00:00\", \"2017-08-31 04:00:00\", \"2017-09-06 04:00:00\", \"2017-09-12 04:00:00\", \"2017-09-13 04:00:00\", \"2017-09-14 04:00:00\", \"2017-09-15 04:00:00\", \"2017-09-18 04:00:00\", \"2017-09-19 04:00:00\", \"2017-09-20 04:00:00\", \"2017-09-21 04:00:00\", \"2017-09-25 04:00:00\", \"2017-09-26 04:00:00\", \"2017-09-29 04:00:00\", \"2017-10-02 04:00:00\", \"2017-10-03 04:00:00\", \"2017-10-04 04:00:00\", \"2017-10-05 04:00:00\", \"2017-10-10 04:00:00\", \"2017-10-16 04:00:00\", \"2017-10-17 04:00:00\", \"2017-10-18 04:00:00\", \"2017-10-19 04:00:00\", \"2017-10-24 04:00:00\", \"2017-10-25 04:00:00\", \"2017-10-26 04:00:00\", \"2017-10-27 04:00:00\", \"2017-10-30 04:00:00\", \"2017-10-31 04:00:00\", \"2017-11-02 04:00:00\", \"2017-11-03 04:00:00\", \"2017-11-07 05:00:00\", \"2017-11-08 05:00:00\", \"2017-11-09 05:00:00\", \"2017-11-13 05:00:00\", \"2017-11-14 05:00:00\", \"2017-11-27 05:00:00\", \"2017-11-28 05:00:00\", \"2017-11-29 05:00:00\", \"2017-11-30 05:00:00\", \"2017-12-01 05:00:00\", \"2017-12-04 05:00:00\", \"2017-12-05 05:00:00\", \"2017-12-06 05:00:00\", \"2017-12-07 05:00:00\", \"2017-12-08 05:00:00\", \"2017-12-11 05:00:00\", \"2017-12-12 05:00:00\", \"2017-12-13 05:00:00\", \"2017-12-14 05:00:00\", \"2017-12-18 05:00:00\", \"2017-12-21 05:00:00\", \"2017-12-22 05:00:00\", \"2018-01-02 05:00:00\", \"2018-01-03 05:00:00\", \"2018-01-04 05:00:00\", \"2018-01-05 05:00:00\", \"2018-01-06 05:00:00\", \"2018-01-07 05:00:00\", \"2018-01-08 05:00:00\", \"2018-01-09 05:00:00\", \"2018-01-10 05:00:00\", \"2018-01-11 05:00:00\", \"2018-01-12 05:00:00\", \"2018-01-13 05:00:00\", \"2018-01-14 05:00:00\", \"2018-01-15 05:00:00\", \"2018-01-16 05:00:00\", \"2018-01-17 05:00:00\", \"2018-01-18 05:00:00\", \"2018-01-31 05:00:00\", \"2018-02-01 05:00:00\", \"2018-02-02 05:00:00\", \"2018-02-05 05:00:00\", \"2018-02-06 05:00:00\", \"2018-02-07 05:00:00\", \"2018-02-08 05:00:00\", \"2018-02-09 05:00:00\", \"2018-02-12 05:00:00\", \"2018-02-13 05:00:00\", \"2018-02-14 05:00:00\", \"2018-02-22 05:00:00\", \"2018-02-23 05:00:00\", \"2018-02-24 05:00:00\", \"2018-02-25 05:00:00\", \"2018-02-26 05:00:00\", \"2018-02-27 05:00:00\", \"2018-02-28 05:00:00\", \"2018-03-01 05:00:00\", \"2018-03-02 05:00:00\", \"2018-03-06 05:00:00\", \"2018-03-07 05:00:00\", \"2018-03-08 05:00:00\", \"2018-03-09 05:00:00\", \"2018-03-12 04:00:00\", \"2018-03-14 04:00:00\", \"2018-03-15 04:00:00\", \"2018-03-23 04:00:00\", \"2018-03-26 04:00:00\", \"2018-03-27 04:00:00\", \"2018-03-28 04:00:00\", \"2018-03-29 04:00:00\", \"2018-03-30 04:00:00\", \"2018-04-02 04:00:00\", \"2018-04-03 04:00:00\", \"2018-04-04 04:00:00\", \"2018-04-05 04:00:00\", \"2018-04-12 04:00:00\", \"2018-04-13 04:00:00\", \"2018-04-16 04:00:00\", \"2018-04-17 04:00:00\", \"2018-04-18 04:00:00\", \"2018-04-19 04:00:00\", \"2018-04-27 04:00:00\", \"2018-07-10 04:00:00\", \"2018-07-11 04:00:00\", \"2018-07-12 04:00:00\", \"2018-07-13 04:00:00\", \"2018-07-16 04:00:00\", \"2018-07-17 04:00:00\", \"2018-07-18 04:00:00\", \"2018-07-19 04:00:00\", \"2018-07-20 04:00:00\", \"2018-07-23 04:00:00\", \"2018-07-24 04:00:00\", \"2018-07-25 04:00:00\", \"2018-07-26 04:00:00\", \"2018-07-27 04:00:00\", \"2018-07-28 04:00:00\", \"2018-07-29 04:00:00\", \"2018-07-30 04:00:00\", \"2018-07-31 04:00:00\", \"2018-08-01 04:00:00\", \"2018-08-02 04:00:00\", \"2018-08-03 04:00:00\", \"2018-08-04 04:00:00\", \"2018-08-05 04:00:00\", \"2018-08-06 04:00:00\", \"2018-08-07 04:00:00\", \"2018-08-13 04:00:00\", \"2018-08-14 04:00:00\", \"2018-08-15 04:00:00\", \"2018-08-16 04:00:00\", \"2018-08-17 04:00:00\", \"2018-08-18 04:00:00\", \"2018-08-19 04:00:00\", \"2018-08-20 04:00:00\", \"2018-08-21 04:00:00\", \"2018-08-22 04:00:00\", \"2018-08-23 04:00:00\", \"2018-08-24 04:00:00\", \"2018-08-25 04:00:00\", \"2018-08-26 04:00:00\", \"2018-08-27 04:00:00\", \"2018-08-28 04:00:00\", \"2018-08-29 04:00:00\", \"2018-08-30 04:00:00\", \"2018-09-04 04:00:00\", \"2018-09-05 04:00:00\", \"2018-09-07 04:00:00\", \"2018-09-08 04:00:00\", \"2018-09-09 04:00:00\", \"2018-09-10 04:00:00\", \"2018-09-11 04:00:00\", \"2018-09-14 04:00:00\", \"2018-09-19 04:00:00\", \"2018-09-21 04:00:00\", \"2018-09-24 04:00:00\", \"2018-09-25 04:00:00\", \"2018-09-26 04:00:00\", \"2018-09-27 04:00:00\", \"2018-09-28 04:00:00\", \"2018-10-01 04:00:00\", \"2018-10-02 04:00:00\", \"2018-10-03 04:00:00\", \"2018-10-05 04:00:00\", \"2018-10-06 04:00:00\", \"2018-10-07 04:00:00\", \"2018-10-09 04:00:00\", \"2018-10-10 04:00:00\", \"2018-10-11 04:00:00\", \"2018-10-12 04:00:00\", \"2018-10-17 04:00:00\", \"2018-10-18 04:00:00\", \"2018-10-19 04:00:00\", \"2018-10-22 04:00:00\", \"2018-10-23 04:00:00\", \"2018-11-08 05:00:00\", \"2018-11-19 05:00:00\", \"2018-11-28 05:00:00\", \"2018-12-04 05:00:00\", \"2018-12-05 05:00:00\", \"2018-12-06 05:00:00\", \"2018-12-07 05:00:00\", \"2018-12-10 05:00:00\", \"2018-12-11 05:00:00\", \"2018-12-12 05:00:00\", \"2018-12-13 05:00:00\", \"2018-12-14 05:00:00\", \"2018-12-20 05:00:00\", \"2018-12-21 05:00:00\", \"2019-01-07 05:00:00\"], \"y\": [254, 12335, 12623, 12652, 23229, 23314, 23350, 23372, 23384, 23397, 23483, 26348, 26377, 30119, 31339, 31369, 31370, 31396, 31413, 31442, 31462, 31483, 32856, 35788, 38401, 41706, 41731, 41924, 53465, 59063, 59419, 59424, 59425, 59435, 59760, 59795, 59853, 59893, 59927, 59999, 60031, 60087, 60103, 60121, 60143, 60180, 60562, 61154, 61202, 61292, 61327, 61662, 62118, 62256, 62841, 62842, 62843, 62903, 63306, 64517, 66396, 66397, 66418, 66427, 70420, 73064, 73774, 74472, 76905, 77565, 78014, 80022, 87907, 88012, 92795, 99540, 100439, 104656, 106818, 106882, 107531, 107535, 107680, 107852, 107854, 107886, 110430, 112386, 112597, 112713, 116948, 126851, 130678, 130793, 133589, 137144, 137295, 150460, 151820, 152366, 152432, 152626, 162100, 164250, 165088, 167572, 169302, 169571, 170799, 170860, 174525, 176624, 180990, 195232, 205914, 210050, 213203, 219693, 224489, 233517, 236035, 236517, 238869, 241081, 243975, 245687, 247711, 249814, 252460, 253979, 254286, 255549, 258919, 264177, 264791, 267270, 269108, 271098, 271633, 273353, 273558, 273618, 274068, 274250, 274407, 274694, 275564, 277543, 282596, 283982, 285020, 288020, 288139, 288200, 288223, 288252, 291199, 293752, 293818, 295216, 298534, 298626, 298640, 300644, 308965, 317623, 318418, 328258, 344497, 357536, 375841, 384039, 391522, 392022, 417469, 419463, 423625, 427134, 427301, 427367, 427370, 429493, 435182, 438917, 445567, 452086, 458400, 466292, 469109, 473683, 485031, 487746, 488059, 488339, 488340, 488533, 488946, 489033, 490582, 492607, 492969, 497347, 498950, 498952, 501777, 515042, 516450, 545098, 551916, 552320, 552327, 552328, 552339, 553627, 553660, 553682, 553725, 553738, 553740, 554687, 555913, 559538, 562317, 567113, 567118, 568546, 572767, 583191, 586747, 587229, 588148, 592347, 595107, 596823, 600007, 601885, 603802, 603948, 604594, 606468, 608207, 609997, 611847, 612309, 614291, 614320, 614345, 614975, 615509, 615540, 620262, 621075, 621142, 621532, 622717, 624266, 624956, 624959, 625188, 625275, 625324, 625361, 625490, 625504, 626190, 626201, 626223, 626249, 627867, 630420, 631343, 634622, 634629, 634712, 634972, 644851, 650011, 652481, 653447, 655003, 655075, 658396, 662209, 665711, 667079, 669465, 684506, 695141, 719587, 719786, 719792, 721158, 722897, 724213, 724219, 724758, 725169, 725809, 726016, 726237, 726518, 729111, 729584, 729830, 730190, 730209, 731814, 732406, 733802, 735093, 735312, 737722, 738671, 739194, 741176, 741942, 743158, 743679, 743834, 744129, 747900, 747906, 747916, 751851, 752457, 752632, 752863, 752905, 752912, 754028, 754053, 754075, 754166, 754299, 755104, 758179, 759429, 762524, 765343, 765347, 765438, 765510, 767617, 768179, 769065, 780961, 795540, 809113, 824783, 826130, 845525, 852759, 864929, 889007, 914645, 954954, 981062, 981063, 981067, 981069, 981070, 981072, 981073, 981074, 981075, 983484, 983582, 983745, 984664, 985215, 996002, 1004846, 1009949, 1017686, 1019071, 1028256, 1028417, 1028429, 1028453, 1028457, 1028458, 1029991, 1039090, 1039125, 1041964, 1043851, 1044097, 1045381, 1047408, 1048458, 1048585, 1049120, 1050492, 1058450, 1059744, 1060736, 1060895, 1060904, 1061211, 1062022, 1062071, 1062381, 1062576, 1062709, 1062790, 1062975, 1063052, 1063114, 1063201, 1063650, 1063749, 1063767, 1063890, 1063899, 1063988, 1067187, 1073276, 1078453, 1078675, 1079098, 1081857, 1085397, 1085768, 1085778, 1086941, 1089899, 1091810, 1098607, 1100409, 1101032, 1101702, 1101704, 1108605, 1113303, 1127186, 1127487, 1129950, 1136478, 1141155, 1142785, 1143428, 1144498, 1144897, 1144913, 1148566, 1149204, 1152375, 1152605, 1156565, 1156591, 1156838, 1157658, 1158000, 1158124, 1158174, 1158815, 1159202, 1159251, 1159325, 1159501, 1159566, 1160896, 1165176, 1166590, 1169614, 1172626, 1177800, 1181971, 1184417, 1186548, 1188595, 1189152, 1189457, 1189741, 1190376, 1195974, 1196238, 1196248, 1196434, 1196710, 1196972, 1197183, 1197437, 1197701, 1198014, 1198023, 1198025, 1198530, 1198746, 1198903, 1201407, 1204677, 1205642, 1215619, 1220898, 1222301, 1222319, 1238289, 1270389, 1286867, 1292028, 1294507, 1294537, 1294583, 1294584, 1294675, 1296684, 1296707, 1296817, 1296922, 1298399, 1298483, 1298575, 1298885, 1299713, 1299729, 1300245, 1300267, 1300268, 1300445, 1300531, 1301304, 1301307, 1301313, 1302177, 1302467, 1302728, 1302730, 1302960, 1303272, 1304476, 1307115, 1311169, 1316506, 1320618, 1320890, 1320931, 1320991, 1321036, 1321238, 1321477, 1321599, 1321876, 1321956, 1322007, 1323017, 1323501, 1323842, 1323899, 1323937, 1324457, 1325864, 1326356, 1327863, 1331338, 1331386, 1331421, 1331454, 1333631, 1335457, 1337898, 1338209, 1338269, 1338299, 1338314, 1338921, 1339076, 1339477, 1339840, 1341539, 1341560, 1341587, 1341619, 1341641, 1342971, 1344490, 1345095, 1345855, 1348357, 1353486, 1354033, 1355129, 1355307, 1355353, 1356487, 1357320, 1357881, 1358262, 1358388, 1359738, 1360967, 1360973, 1362950, 1364792, 1366178, 1367768, 1369064, 1369065, 1391733, 1398706, 1405376, 1412989, 1419744, 1441099, 1449324, 1458614, 1461912, 1484389, 1496134, 1511336, 1539229, 1549224, 1567233, 1584878, 1586218, 1587489, 1588003, 1588218, 1588704, 1588761, 1588796, 1588802, 1588827, 1588863, 1588897, 1588902, 1589083, 1589098, 1590061, 1591246, 1591518, 1591708, 1592352, 1593562, 1593930, 1594350, 1594460, 1594911, 1595242, 1595905, 1596911, 1597610, 1599767, 1602008, 1602052, 1602108, 1602167, 1602477, 1602553, 1603072, 1603440, 1603840, 1604512, 1605404, 1608286, 1608486, 1608595, 1608697, 1610835, 1620761, 1620762, 1620765, 1620766, 1620811, 1620812, 1620835, 1620848, 1620902, 1620924, 1621128, 1621271, 1621319, 1621390, 1621469, 1621632, 1621709, 1621742, 1621796, 1622433, 1623409, 1624618, 1625809, 1626538, 1626547, 1626773, 1627516, 1629134, 1629150, 1629371, 1631067, 1631740, 1633357, 1637258, 1642148, 1649847, 1679050, 1706741, 1726834, 1756820, 1757668, 1769250, 1774590, 1780366, 1783191, 1787682, 1790717, 1791084, 1791421, 1791458, 1791518, 1791631, 1791684, 1791725, 1791739, 1791844, 1791925, 1791927, 1792090, 1792167, 1792258, 1792498, 1792575, 1792798, 1793281, 1794373, 1794517, 1794781, 1795013, 1795119, 1796859, 1799806, 1801782, 1801830, 1801858, 1801866, 1801870, 1801872, 1802064, 1805448, 1807119, 1809240, 1815007, 1816287, 1817003, 1818756, 1821614, 1821622, 1825134, 1834369], \"type\": \"scattergl\", \"uid\": \"aafbe74d-dcd4-402c-97d5-40921f7361d2\"}, {\"name\": \"FT2\", \"x\": [\"2014-01-08 05:00:00\", \"2014-01-09 05:00:00\", \"2014-01-15 05:00:00\", \"2014-01-17 05:00:00\", \"2014-01-29 05:00:00\", \"2014-01-30 05:00:00\", \"2014-02-03 05:00:00\", \"2014-02-06 05:00:00\", \"2014-03-04 05:00:00\", \"2014-03-05 05:00:00\", \"2014-03-06 05:00:00\", \"2014-03-10 04:00:00\", \"2014-03-11 04:00:00\", \"2014-03-12 04:00:00\", \"2014-03-13 04:00:00\", \"2014-03-14 04:00:00\", \"2014-03-17 04:00:00\", \"2014-03-26 04:00:00\", \"2014-04-22 04:00:00\", \"2014-04-23 04:00:00\", \"2014-05-02 04:00:00\", \"2014-05-05 04:00:00\", \"2014-05-06 04:00:00\", \"2014-05-07 04:00:00\", \"2014-05-16 04:00:00\", \"2014-05-21 04:00:00\", \"2014-05-22 04:00:00\", \"2014-06-03 04:00:00\", \"2014-06-04 04:00:00\", \"2014-06-05 04:00:00\", \"2014-06-06 04:00:00\", \"2014-06-09 04:00:00\", \"2014-06-10 04:00:00\", \"2014-06-11 04:00:00\", \"2014-06-12 04:00:00\", \"2014-06-13 04:00:00\", \"2014-06-24 04:00:00\", \"2014-06-25 04:00:00\", \"2014-06-26 04:00:00\", \"2014-06-27 04:00:00\", \"2014-06-30 04:00:00\", \"2014-07-01 04:00:00\", \"2014-07-02 04:00:00\", \"2014-07-03 04:00:00\", \"2014-07-04 04:00:00\", \"2014-07-07 04:00:00\", \"2014-07-11 04:00:00\", \"2014-07-14 04:00:00\", \"2014-07-15 04:00:00\", \"2014-07-16 04:00:00\", \"2014-07-17 04:00:00\", \"2014-07-25 04:00:00\", \"2014-07-28 04:00:00\", \"2014-07-29 04:00:00\", \"2014-09-05 04:00:00\", \"2014-09-23 04:00:00\", \"2014-09-25 04:00:00\", \"2014-09-26 04:00:00\", \"2014-09-29 04:00:00\", \"2014-09-30 04:00:00\", \"2014-10-01 04:00:00\", \"2014-10-02 04:00:00\", \"2014-10-03 04:00:00\", \"2014-10-21 04:00:00\", \"2014-10-22 04:00:00\", \"2014-10-24 04:00:00\", \"2014-10-27 04:00:00\", \"2014-10-29 04:00:00\", \"2014-10-30 04:00:00\", \"2014-11-10 05:00:00\", \"2014-11-11 05:00:00\", \"2014-11-13 05:00:00\", \"2014-11-14 05:00:00\", \"2014-11-19 05:00:00\", \"2014-11-20 05:00:00\", \"2014-11-25 05:00:00\", \"2014-12-10 05:00:00\", \"2014-12-11 05:00:00\", \"2014-12-23 05:00:00\", \"2015-01-13 05:00:00\", \"2015-01-22 05:00:00\", \"2015-01-23 05:00:00\", \"2015-01-30 05:00:00\", \"2015-02-26 05:00:00\", \"2015-02-27 05:00:00\", \"2015-03-30 04:00:00\", \"2015-03-31 04:00:00\", \"2015-04-01 04:00:00\", \"2015-04-02 04:00:00\", \"2015-04-03 04:00:00\", \"2015-04-05 04:00:00\", \"2015-04-06 04:00:00\", \"2015-04-20 04:00:00\", \"2015-04-21 04:00:00\", \"2015-04-27 04:00:00\", \"2015-04-28 04:00:00\", \"2015-04-29 04:00:00\", \"2015-04-30 04:00:00\", \"2015-05-04 04:00:00\", \"2015-05-06 04:00:00\", \"2015-05-11 04:00:00\", \"2015-06-30 04:00:00\", \"2015-07-28 04:00:00\", \"2015-07-30 04:00:00\", \"2015-08-10 04:00:00\", \"2015-08-11 04:00:00\", \"2015-08-12 04:00:00\", \"2015-08-13 04:00:00\", \"2015-08-14 04:00:00\", \"2015-08-17 04:00:00\", \"2015-08-18 04:00:00\", \"2015-08-19 04:00:00\", \"2015-08-20 04:00:00\", \"2015-08-21 04:00:00\", \"2015-08-24 04:00:00\", \"2015-08-25 04:00:00\", \"2015-08-26 04:00:00\", \"2015-08-27 04:00:00\", \"2015-08-28 04:00:00\", \"2015-08-29 04:00:00\", \"2015-09-03 04:00:00\", \"2015-09-12 04:00:00\", \"2015-09-13 04:00:00\", \"2015-09-14 04:00:00\", \"2015-09-23 04:00:00\", \"2015-09-24 04:00:00\", \"2015-09-25 04:00:00\", \"2015-09-26 04:00:00\", \"2015-09-28 04:00:00\", \"2015-09-29 04:00:00\", \"2015-09-30 04:00:00\", \"2015-10-05 04:00:00\", \"2015-10-12 04:00:00\", \"2015-10-21 04:00:00\", \"2015-10-27 04:00:00\", \"2015-10-28 04:00:00\", \"2015-10-29 04:00:00\", \"2015-12-04 05:00:00\", \"2015-12-08 05:00:00\", \"2015-12-09 05:00:00\", \"2015-12-10 05:00:00\", \"2015-12-11 05:00:00\", \"2015-12-12 05:00:00\", \"2015-12-14 05:00:00\", \"2016-01-06 05:00:00\", \"2016-01-07 05:00:00\", \"2016-02-04 05:00:00\", \"2016-02-05 05:00:00\", \"2016-02-08 05:00:00\", \"2016-02-09 05:00:00\", \"2016-02-10 05:00:00\", \"2016-02-11 05:00:00\", \"2016-02-12 05:00:00\", \"2016-02-13 05:00:00\", \"2016-02-14 05:00:00\", \"2016-02-15 05:00:00\", \"2016-02-16 05:00:00\", \"2016-02-18 05:00:00\", \"2016-02-22 05:00:00\", \"2016-02-23 05:00:00\", \"2016-03-23 04:00:00\", \"2016-03-24 04:00:00\", \"2016-03-25 04:00:00\", \"2016-03-29 04:00:00\", \"2016-03-30 04:00:00\", \"2016-04-04 04:00:00\", \"2016-04-06 04:00:00\", \"2016-04-07 04:00:00\", \"2016-04-21 04:00:00\", \"2016-04-23 04:00:00\", \"2016-04-26 04:00:00\", \"2016-04-30 04:00:00\", \"2016-05-09 04:00:00\", \"2016-05-13 04:00:00\", \"2016-05-14 04:00:00\", \"2016-05-17 04:00:00\", \"2016-05-18 04:00:00\", \"2016-05-19 04:00:00\", \"2016-05-20 04:00:00\", \"2016-05-24 04:00:00\", \"2016-06-01 04:00:00\", \"2016-06-02 04:00:00\", \"2016-06-03 04:00:00\", \"2016-06-27 04:00:00\", \"2016-06-28 04:00:00\", \"2016-06-29 04:00:00\", \"2016-06-30 04:00:00\", \"2016-07-01 04:00:00\", \"2016-07-20 04:00:00\", \"2016-07-21 04:00:00\", \"2016-07-22 04:00:00\", \"2016-07-27 04:00:00\", \"2016-07-28 04:00:00\", \"2016-07-29 04:00:00\", \"2016-08-09 04:00:00\", \"2016-08-10 04:00:00\", \"2016-08-11 04:00:00\", \"2016-08-12 04:00:00\", \"2016-08-14 04:00:00\", \"2016-08-17 04:00:00\", \"2016-08-19 04:00:00\", \"2016-08-24 04:00:00\", \"2016-08-25 04:00:00\", \"2016-08-26 04:00:00\", \"2016-08-29 04:00:00\", \"2016-08-30 04:00:00\", \"2016-08-31 04:00:00\", \"2016-09-07 04:00:00\", \"2016-09-08 04:00:00\", \"2016-09-09 04:00:00\", \"2016-09-10 04:00:00\", \"2016-09-12 04:00:00\", \"2016-09-13 04:00:00\", \"2016-09-14 04:00:00\", \"2016-09-15 04:00:00\", \"2016-09-16 04:00:00\", \"2016-09-17 04:00:00\", \"2016-09-24 04:00:00\", \"2016-09-27 04:00:00\", \"2016-10-10 04:00:00\", \"2016-10-12 04:00:00\", \"2016-10-13 04:00:00\", \"2016-10-14 04:00:00\", \"2016-10-15 04:00:00\", \"2016-10-17 04:00:00\", \"2016-10-18 04:00:00\", \"2016-10-20 04:00:00\", \"2016-10-21 04:00:00\", \"2016-10-25 04:00:00\", \"2016-10-26 04:00:00\", \"2016-10-27 04:00:00\", \"2016-10-28 04:00:00\", \"2016-10-31 04:00:00\", \"2016-11-01 04:00:00\", \"2016-11-02 04:00:00\", \"2016-11-03 04:00:00\", \"2016-11-04 04:00:00\", \"2016-11-16 05:00:00\", \"2016-11-21 05:00:00\", \"2016-11-22 05:00:00\", \"2016-11-23 05:00:00\", \"2016-11-25 05:00:00\", \"2016-11-26 05:00:00\", \"2016-11-28 05:00:00\", \"2016-11-29 05:00:00\", \"2016-11-30 05:00:00\", \"2016-12-01 05:00:00\", \"2016-12-02 05:00:00\", \"2016-12-14 05:00:00\", \"2017-02-08 05:00:00\", \"2017-02-10 05:00:00\", \"2017-02-14 05:00:00\", \"2017-02-15 05:00:00\", \"2017-02-16 05:00:00\", \"2017-02-22 05:00:00\", \"2017-03-01 05:00:00\", \"2017-03-02 05:00:00\", \"2017-03-03 05:00:00\", \"2017-03-04 05:00:00\", \"2017-03-05 05:00:00\", \"2017-03-06 05:00:00\", \"2017-03-07 05:00:00\", \"2017-03-08 05:00:00\", \"2017-03-09 05:00:00\", \"2017-03-10 05:00:00\", \"2017-03-13 04:00:00\", \"2017-03-14 04:00:00\", \"2017-03-15 04:00:00\", \"2017-03-16 04:00:00\", \"2017-03-17 04:00:00\", \"2017-03-24 04:00:00\", \"2017-03-28 04:00:00\", \"2017-03-29 04:00:00\", \"2017-03-30 04:00:00\", \"2017-04-02 04:00:00\", \"2017-04-05 04:00:00\", \"2017-04-07 04:00:00\", \"2017-04-08 04:00:00\", \"2017-04-11 04:00:00\", \"2017-04-12 04:00:00\", \"2017-04-13 04:00:00\", \"2017-04-14 04:00:00\", \"2017-04-15 04:00:00\", \"2017-04-17 04:00:00\", \"2017-04-18 04:00:00\", \"2017-04-19 04:00:00\", \"2017-04-20 04:00:00\", \"2017-04-21 04:00:00\", \"2017-04-22 04:00:00\", \"2017-04-23 04:00:00\", \"2017-04-25 04:00:00\", \"2017-04-26 04:00:00\", \"2017-04-27 04:00:00\", \"2017-04-28 04:00:00\", \"2017-04-29 04:00:00\", \"2017-05-02 04:00:00\", \"2017-05-03 04:00:00\", \"2017-05-04 04:00:00\", \"2017-05-05 04:00:00\", \"2017-05-08 04:00:00\", \"2017-05-09 04:00:00\", \"2017-05-10 04:00:00\", \"2017-05-11 04:00:00\", \"2017-05-12 04:00:00\", \"2017-05-13 04:00:00\", \"2017-05-14 04:00:00\", \"2017-05-15 04:00:00\", \"2017-05-16 04:00:00\", \"2017-05-17 04:00:00\", \"2017-05-18 04:00:00\", \"2017-05-19 04:00:00\", \"2017-05-20 04:00:00\", \"2017-05-21 04:00:00\", \"2017-05-22 04:00:00\", \"2017-05-23 04:00:00\", \"2017-05-24 04:00:00\", \"2017-05-25 04:00:00\", \"2017-05-26 04:00:00\", \"2017-06-01 04:00:00\", \"2017-06-02 04:00:00\", \"2017-06-05 04:00:00\", \"2017-06-09 04:00:00\", \"2017-06-10 04:00:00\", \"2017-06-11 04:00:00\", \"2017-06-12 04:00:00\", \"2017-06-13 04:00:00\", \"2017-06-15 04:00:00\", \"2017-06-16 04:00:00\", \"2017-06-17 04:00:00\", \"2017-06-26 04:00:00\", \"2017-06-27 04:00:00\", \"2017-06-28 04:00:00\", \"2017-06-29 04:00:00\", \"2017-07-05 04:00:00\", \"2017-07-20 04:00:00\", \"2017-07-21 04:00:00\", \"2017-07-22 04:00:00\", \"2017-07-24 04:00:00\", \"2017-08-14 04:00:00\", \"2017-08-15 04:00:00\", \"2017-08-16 04:00:00\", \"2017-08-17 04:00:00\", \"2017-08-23 04:00:00\", \"2017-08-24 04:00:00\", \"2017-08-25 04:00:00\", \"2017-08-28 04:00:00\", \"2017-10-26 04:00:00\", \"2017-10-27 04:00:00\", \"2017-10-30 04:00:00\", \"2017-10-31 04:00:00\", \"2017-11-01 04:00:00\", \"2017-11-02 04:00:00\", \"2017-11-07 05:00:00\", \"2017-11-08 05:00:00\", \"2017-11-09 05:00:00\", \"2017-11-10 05:00:00\", \"2017-11-11 05:00:00\", \"2017-11-13 05:00:00\", \"2017-11-16 05:00:00\", \"2017-11-20 05:00:00\", \"2017-12-04 05:00:00\", \"2017-12-05 05:00:00\", \"2017-12-07 05:00:00\", \"2017-12-13 05:00:00\", \"2017-12-15 05:00:00\", \"2017-12-18 05:00:00\", \"2017-12-19 05:00:00\", \"2017-12-21 05:00:00\", \"2018-01-02 05:00:00\", \"2018-01-03 05:00:00\", \"2018-01-05 05:00:00\", \"2018-01-08 05:00:00\", \"2018-01-09 05:00:00\", \"2018-01-10 05:00:00\", \"2018-01-15 05:00:00\", \"2018-01-16 05:00:00\", \"2018-01-17 05:00:00\", \"2018-02-08 05:00:00\", \"2018-02-09 05:00:00\", \"2018-02-13 05:00:00\", \"2018-02-14 05:00:00\", \"2018-02-15 05:00:00\", \"2018-02-16 05:00:00\", \"2018-02-21 05:00:00\", \"2018-02-22 05:00:00\", \"2018-03-12 04:00:00\", \"2018-03-14 04:00:00\", \"2018-03-20 04:00:00\", \"2018-03-21 04:00:00\", \"2018-03-27 04:00:00\", \"2018-03-28 04:00:00\", \"2018-04-02 04:00:00\", \"2018-04-03 04:00:00\", \"2018-04-06 04:00:00\", \"2018-08-02 04:00:00\", \"2018-08-07 04:00:00\", \"2018-08-08 04:00:00\", \"2018-08-09 04:00:00\", \"2018-08-10 04:00:00\", \"2018-08-13 04:00:00\", \"2018-08-14 04:00:00\", \"2018-08-15 04:00:00\", \"2018-08-30 04:00:00\", \"2018-08-31 04:00:00\", \"2018-09-01 04:00:00\", \"2018-09-04 04:00:00\", \"2018-09-10 04:00:00\", \"2018-09-11 04:00:00\", \"2018-10-03 04:00:00\", \"2018-10-29 04:00:00\", \"2018-11-06 05:00:00\", \"2018-11-07 05:00:00\", \"2018-11-08 05:00:00\", \"2018-11-20 05:00:00\", \"2018-11-26 05:00:00\", \"2018-11-27 05:00:00\", \"2018-11-28 05:00:00\", \"2018-11-29 05:00:00\", \"2018-11-30 05:00:00\", \"2018-12-03 05:00:00\", \"2018-12-04 05:00:00\", \"2018-12-05 05:00:00\", \"2018-12-06 05:00:00\", \"2018-12-07 05:00:00\", \"2018-12-10 05:00:00\", \"2018-12-11 05:00:00\", \"2018-12-13 05:00:00\"], \"y\": [1, 109, 113, 490, 718, 743, 749, 1325, 1326, 1327, 1842, 2401, 3817, 4782, 5797, 9562, 9635, 9642, 9649, 9653, 9720, 10322, 11361, 11605, 12324, 12751, 13446, 15575, 16209, 20213, 22570, 25865, 26969, 27048, 27921, 27986, 28054, 28670, 29329, 29888, 29928, 32001, 36042, 38051, 39156, 40380, 40791, 40834, 40869, 40870, 40872, 40883, 40885, 40886, 40887, 40888, 40906, 41134, 41451, 41778, 41804, 42317, 42906, 43016, 43018, 43727, 44127, 49567, 51576, 51629, 52041, 54351, 57101, 58933, 61249, 61314, 61320, 61323, 61324, 61325, 61526, 62054, 62061, 62067, 62072, 62131, 62536, 62559, 62560, 64389, 65036, 65858, 65862, 65865, 65866, 66004, 66034, 66189, 66252, 66269, 66270, 66296, 66297, 66659, 66660, 67843, 68038, 69565, 70269, 70588, 70740, 70878, 71179, 71280, 71709, 72859, 72871, 73376, 75348, 76322, 76864, 76865, 76868, 78757, 95361, 108381, 113213, 114401, 116228, 121252, 127532, 128354, 135884, 135885, 137794, 139995, 141543, 142526, 142527, 142544, 150890, 159271, 160049, 160052, 160057, 160059, 160062, 160075, 160077, 160080, 163062, 166106, 167640, 169329, 170325, 172784, 172800, 172804, 172981, 173979, 173980, 173992, 173993, 174046, 174194, 174205, 174374, 174375, 174376, 174387, 174614, 174617, 174618, 175510, 177803, 180157, 184078, 184864, 185071, 186614, 187744, 198159, 201294, 201303, 203003, 204841, 206226, 208419, 208432, 208434, 208486, 208583, 208681, 208722, 212407, 220207, 220607, 222328, 224703, 224715, 224734, 224735, 224899, 226888, 226941, 227114, 227116, 227265, 227298, 227308, 227337, 227345, 228722, 235376, 238997, 243619, 246720, 246722, 246759, 246761, 246762, 246777, 246786, 246789, 246895, 246910, 246951, 246960, 247163, 247365, 247475, 247502, 247808, 247937, 249543, 250689, 252801, 252806, 252808, 253753, 256542, 257544, 259076, 259917, 260524, 260623, 261019, 261118, 261316, 261318, 261420, 261474, 261481, 261495, 261496, 261501, 261505, 262607, 265956, 268022, 268023, 268180, 268290, 268492, 272799, 276049, 281555, 281664, 286380, 286433, 286977, 287827, 289205, 289777, 289778, 289780, 289804, 289838, 289840, 292989, 295807, 303582, 304271, 308295, 308541, 313367, 319328, 321961, 326479, 326694, 326695, 326696, 326700, 326705, 326707, 326718, 326726, 326741, 326849, 327043, 327075, 327121, 327224, 327379, 327734, 328136, 328479, 328680, 328747, 329239, 329876, 330438, 332321, 333855, 334189, 334700, 336143, 337157, 337183, 337191, 337196, 337404, 339131, 339235, 339635, 339783, 339849, 340828, 341071, 341315, 341622, 341653, 341674, 341679, 342044, 342726, 344390, 344396, 347769, 352027, 353162, 353165, 353295, 353559, 353634, 353640, 353856, 354158, 354260, 355321, 355787, 356541, 356564, 356587, 356621, 356648, 356659, 358230, 358233, 358238, 358240, 358245, 358967, 358979, 358992, 359312, 359368, 359370, 359371, 359387, 360061, 363036, 365319, 365321, 365325, 365347, 365436, 365600, 365657, 365658, 366919, 367403, 367408, 368980, 373490, 375083, 375084, 376097, 376557, 376560, 376570, 376625, 376652, 376666, 376765, 376878, 377014, 377068, 377411, 377654, 379689, 383585, 387381, 390972, 392856, 393755, 393792, 393830, 393833, 393834, 393835, 393850, 393882, 393884, 393885, 393886, 394145, 394289, 394455, 394770, 395593, 395652, 395661, 395689, 395705, 395706, 395713], \"type\": \"scattergl\", \"uid\": \"1e34339d-ba81-44e6-9900-a2a533a8768f\"}, {\"hoverinfo\": \"name\", \"name\": \"ISMS\", \"width\": 2000000.0, \"x\": [\"2014-06-17 04:00:00\", \"2015-06-17 04:00:00\", \"2016-06-17 04:00:00\", \"2017-06-17 04:00:00\", \"2018-06-17 04:00:00\"], \"y\": [2000000.0, 2000000.0, 2000000.0, 2000000.0, 2000000.0], \"type\": \"bar\", \"uid\": \"d95e7c9c-c7ed-4937-a05c-7b06aac1549c\"}], {\"height\": 600.0, \"legend\": {\"x\": 0.1, \"y\": 0.95}, \"showlegend\": true, \"title\": \"How we collect data\", \"xaxis\": {\"title\": \"Time\"}, \"yaxis\": {\"title\": \"Number of scans\"}}, {\"showLink\": false, \"linkText\": \"\", \"plotlyServerURL\": \"https://plot.ly\"})</script><script type=\"text/javascript\">window.addEventListener(\"resize\", function(){Plotly.Plots.resize(document.getElementById(\"ed88ed28-c295-4182-9c04-629162ca1a83\"));});</script></div>\n"
],
[
"shot_histo = [\n np.histogram(df[\"shots\"], bins=[10, 50, 200, 500, 1000, 2000, 5000, 10000,]) for df in [ft1_df, ft2_df]\n]",
"_____no_output_____"
],
[
"fig = go.FigureWidget()\nfig.layout[\"xaxis\"][\"type\"] = \"log\"\nfig.layout[\"yaxis\"][\"type\"] = \"log\"\n\nfor histo, name in zip(shot_histo, [\"FT1\", \"FT2\"]):\n fig.add_scatter(x=histo[1], y=histo[0], name=name)\n\nfig",
"_____no_output_____"
],
[
"freq_histo = [\n np.histogram(df[\"cavity\"], bins=np.linspace(7000., 40000., 100)) for df in [ft1_df, ft2_df]\n]",
"_____no_output_____"
],
[
"fig = go.FigureWidget()\n\nfig.layout[\"xaxis\"][\"tickformat\"] = \".,\"\nfig.layout[\"xaxis\"][\"title\"] = \"Frequency (MHz)\"\nfig.layout[\"yaxis\"][\"title\"] = \"Counts\"\nfig.layout[\"title\"] = \"What are the most common frequencies?\"\n\nfor histo, name in zip(freq_histo, [\"FT1\", \"FT2\"]):\n fig.add_bar(x=histo[1], y=histo[0], name=name)\n\nfig",
"_____no_output_____"
],
[
"print(plot(fig, show_link=False, link_text=\"\", output_type=\"div\", include_plotlyjs=False))",
"<div><div id=\"b01d543c-a117-41d1-943d-7fdd17531616\" style=\"height: 600.0px; width: 100%;\" class=\"plotly-graph-div\"></div><script type=\"text/javascript\">window.PLOTLYENV=window.PLOTLYENV || {};window.PLOTLYENV.BASE_URL=\"https://plot.ly\";Plotly.newPlot(\"b01d543c-a117-41d1-943d-7fdd17531616\", [{\"name\": \"FT1\", \"x\": [\"2014-07-08 04:00:00\", \"2014-07-09 04:00:00\", \"2014-07-10 04:00:00\", \"2014-07-11 04:00:00\", \"2014-07-13 04:00:00\", \"2014-07-14 04:00:00\", \"2014-07-15 04:00:00\", \"2014-07-16 04:00:00\", \"2014-07-17 04:00:00\", \"2014-07-18 04:00:00\", \"2014-07-21 04:00:00\", \"2014-07-22 04:00:00\", \"2014-07-23 04:00:00\", \"2014-07-25 04:00:00\", \"2014-07-28 04:00:00\", \"2014-07-29 04:00:00\", \"2014-07-30 04:00:00\", \"2014-07-31 04:00:00\", \"2014-08-01 04:00:00\", \"2014-08-05 04:00:00\", \"2014-08-06 04:00:00\", \"2014-08-07 04:00:00\", \"2014-08-11 04:00:00\", \"2014-08-12 04:00:00\", \"2014-08-13 04:00:00\", \"2014-08-14 04:00:00\", \"2014-08-15 04:00:00\", \"2014-08-16 04:00:00\", \"2014-08-18 04:00:00\", \"2014-08-19 04:00:00\", \"2014-08-21 04:00:00\", \"2014-08-22 04:00:00\", \"2014-09-03 04:00:00\", \"2014-09-04 04:00:00\", \"2014-09-08 04:00:00\", \"2014-09-09 04:00:00\", \"2014-09-10 04:00:00\", \"2014-09-11 04:00:00\", \"2014-09-12 04:00:00\", \"2014-09-15 04:00:00\", \"2014-09-17 04:00:00\", \"2014-09-18 04:00:00\", \"2014-09-19 04:00:00\", \"2014-09-25 04:00:00\", \"2014-09-26 04:00:00\", \"2014-10-02 04:00:00\", \"2014-10-03 04:00:00\", \"2014-10-06 04:00:00\", \"2014-10-08 04:00:00\", \"2014-10-17 04:00:00\", \"2014-10-20 04:00:00\", \"2014-10-21 04:00:00\", \"2014-10-22 04:00:00\", \"2014-10-23 04:00:00\", \"2014-10-24 04:00:00\", \"2014-11-03 05:00:00\", \"2014-11-05 05:00:00\", \"2014-11-10 05:00:00\", \"2014-11-12 05:00:00\", \"2014-11-13 05:00:00\", \"2014-11-14 05:00:00\", \"2014-11-19 05:00:00\", \"2014-11-25 05:00:00\", \"2014-12-01 05:00:00\", \"2014-12-02 05:00:00\", \"2014-12-03 05:00:00\", \"2014-12-04 05:00:00\", \"2014-12-09 05:00:00\", \"2014-12-10 05:00:00\", \"2014-12-11 05:00:00\", \"2014-12-12 05:00:00\", \"2014-12-15 05:00:00\", \"2014-12-16 05:00:00\", \"2014-12-17 05:00:00\", \"2014-12-18 05:00:00\", \"2014-12-19 05:00:00\", \"2014-12-20 05:00:00\", \"2014-12-22 05:00:00\", \"2014-12-23 05:00:00\", \"2015-01-16 05:00:00\", \"2015-01-21 05:00:00\", \"2015-01-22 05:00:00\", \"2015-01-26 05:00:00\", \"2015-01-28 05:00:00\", \"2015-02-04 05:00:00\", \"2015-02-11 05:00:00\", \"2015-02-12 05:00:00\", \"2015-02-13 05:00:00\", \"2015-02-19 05:00:00\", \"2015-02-20 05:00:00\", \"2015-02-24 05:00:00\", \"2015-02-25 05:00:00\", \"2015-02-26 05:00:00\", \"2015-02-27 05:00:00\", \"2015-03-10 04:00:00\", \"2015-03-11 04:00:00\", \"2015-03-12 04:00:00\", \"2015-03-13 04:00:00\", \"2015-03-14 04:00:00\", \"2015-03-16 04:00:00\", \"2015-03-20 04:00:00\", \"2015-03-23 04:00:00\", \"2015-03-26 04:00:00\", \"2015-03-27 04:00:00\", \"2015-03-30 04:00:00\", \"2015-03-31 04:00:00\", \"2015-04-01 04:00:00\", \"2015-04-02 04:00:00\", \"2015-04-03 04:00:00\", \"2015-04-04 04:00:00\", \"2015-04-06 04:00:00\", \"2015-04-07 04:00:00\", \"2015-04-14 04:00:00\", \"2015-04-15 04:00:00\", \"2015-04-16 04:00:00\", \"2015-04-17 04:00:00\", \"2015-04-18 04:00:00\", \"2015-04-20 04:00:00\", \"2015-04-22 04:00:00\", \"2015-04-23 04:00:00\", \"2015-04-24 04:00:00\", \"2015-04-29 04:00:00\", \"2015-05-05 04:00:00\", \"2015-05-06 04:00:00\", \"2015-05-07 04:00:00\", \"2015-05-08 04:00:00\", \"2015-05-11 04:00:00\", \"2015-05-12 04:00:00\", \"2015-05-14 04:00:00\", \"2015-05-15 04:00:00\", \"2015-05-18 04:00:00\", \"2015-05-19 04:00:00\", \"2015-05-20 04:00:00\", \"2015-05-21 04:00:00\", \"2015-05-26 04:00:00\", \"2015-05-27 04:00:00\", \"2015-05-28 04:00:00\", \"2015-05-29 04:00:00\", \"2015-06-01 04:00:00\", \"2015-06-02 04:00:00\", \"2015-06-03 04:00:00\", \"2015-06-04 04:00:00\", \"2015-06-05 04:00:00\", \"2015-06-06 04:00:00\", \"2015-06-07 04:00:00\", \"2015-06-08 04:00:00\", \"2015-06-09 04:00:00\", \"2015-06-10 04:00:00\", \"2015-06-11 04:00:00\", \"2015-06-12 04:00:00\", \"2015-06-15 04:00:00\", \"2015-06-16 04:00:00\", \"2015-06-17 04:00:00\", \"2015-06-18 04:00:00\", \"2015-06-19 04:00:00\", \"2015-06-20 04:00:00\", \"2015-06-22 04:00:00\", \"2015-06-24 04:00:00\", \"2015-06-25 04:00:00\", \"2015-06-29 04:00:00\", \"2015-06-30 04:00:00\", \"2015-07-08 04:00:00\", \"2015-07-14 04:00:00\", \"2015-07-15 04:00:00\", \"2015-07-16 04:00:00\", \"2015-07-17 04:00:00\", \"2015-07-20 04:00:00\", \"2015-07-22 04:00:00\", \"2015-07-23 04:00:00\", \"2015-07-24 04:00:00\", \"2015-07-25 04:00:00\", \"2015-07-27 04:00:00\", \"2015-07-28 04:00:00\", \"2015-07-29 04:00:00\", \"2015-07-30 04:00:00\", \"2015-07-31 04:00:00\", \"2015-08-05 04:00:00\", \"2015-08-06 04:00:00\", \"2015-08-07 04:00:00\", \"2015-08-14 04:00:00\", \"2015-08-16 04:00:00\", \"2015-08-20 04:00:00\", \"2015-08-21 04:00:00\", \"2015-08-22 04:00:00\", \"2015-08-23 04:00:00\", \"2015-08-24 04:00:00\", \"2015-08-25 04:00:00\", \"2015-08-26 04:00:00\", \"2015-08-27 04:00:00\", \"2015-08-28 04:00:00\", \"2015-08-29 04:00:00\", \"2015-08-30 04:00:00\", \"2015-08-31 04:00:00\", \"2015-09-01 04:00:00\", \"2015-09-02 04:00:00\", \"2015-09-03 04:00:00\", \"2015-09-04 04:00:00\", \"2015-09-08 04:00:00\", \"2015-09-09 04:00:00\", \"2015-09-10 04:00:00\", \"2015-09-11 04:00:00\", \"2015-09-12 04:00:00\", \"2015-09-13 04:00:00\", \"2015-09-14 04:00:00\", \"2015-09-16 04:00:00\", \"2015-09-17 04:00:00\", \"2015-09-18 04:00:00\", \"2015-09-19 04:00:00\", \"2015-09-20 04:00:00\", \"2015-09-21 04:00:00\", \"2015-09-22 04:00:00\", \"2015-09-23 04:00:00\", \"2015-10-06 04:00:00\", \"2015-10-07 04:00:00\", \"2015-10-08 04:00:00\", \"2015-10-13 04:00:00\", \"2015-10-14 04:00:00\", \"2015-10-20 04:00:00\", \"2015-10-21 04:00:00\", \"2015-10-22 04:00:00\", \"2015-10-23 04:00:00\", \"2015-10-24 04:00:00\", \"2015-10-25 04:00:00\", \"2015-10-26 04:00:00\", \"2015-10-28 04:00:00\", \"2015-10-29 04:00:00\", \"2015-10-30 04:00:00\", \"2015-10-31 04:00:00\", \"2015-11-02 05:00:00\", \"2015-11-03 05:00:00\", \"2015-11-04 05:00:00\", \"2015-11-05 05:00:00\", \"2015-11-06 05:00:00\", \"2015-11-09 05:00:00\", \"2015-11-10 05:00:00\", \"2015-11-12 05:00:00\", \"2015-11-13 05:00:00\", \"2015-11-19 05:00:00\", \"2015-11-20 05:00:00\", \"2015-11-21 05:00:00\", \"2015-11-22 05:00:00\", \"2015-11-23 05:00:00\", \"2015-11-24 05:00:00\", \"2015-11-25 05:00:00\", \"2015-11-30 05:00:00\", \"2015-12-01 05:00:00\", \"2015-12-02 05:00:00\", \"2015-12-03 05:00:00\", \"2015-12-04 05:00:00\", \"2015-12-08 05:00:00\", \"2015-12-09 05:00:00\", \"2015-12-10 05:00:00\", \"2015-12-11 05:00:00\", \"2015-12-14 05:00:00\", \"2015-12-15 05:00:00\", \"2015-12-22 05:00:00\", \"2015-12-23 05:00:00\", \"2016-01-07 05:00:00\", \"2016-01-19 05:00:00\", \"2016-01-20 05:00:00\", \"2016-01-21 05:00:00\", \"2016-01-25 05:00:00\", \"2016-01-26 05:00:00\", \"2016-01-27 05:00:00\", \"2016-02-03 05:00:00\", \"2016-02-04 05:00:00\", \"2016-02-05 05:00:00\", \"2016-02-08 05:00:00\", \"2016-02-09 05:00:00\", \"2016-02-10 05:00:00\", \"2016-02-11 05:00:00\", \"2016-02-12 05:00:00\", \"2016-02-15 05:00:00\", \"2016-02-16 05:00:00\", \"2016-02-18 05:00:00\", \"2016-02-19 05:00:00\", \"2016-02-20 05:00:00\", \"2016-02-22 05:00:00\", \"2016-02-23 05:00:00\", \"2016-02-24 05:00:00\", \"2016-02-25 05:00:00\", \"2016-03-02 05:00:00\", \"2016-03-03 05:00:00\", \"2016-03-04 05:00:00\", \"2016-03-05 05:00:00\", \"2016-03-09 05:00:00\", \"2016-03-10 05:00:00\", \"2016-03-11 05:00:00\", \"2016-03-14 04:00:00\", \"2016-03-15 04:00:00\", \"2016-03-18 04:00:00\", \"2016-03-21 04:00:00\", \"2016-03-22 04:00:00\", \"2016-03-23 04:00:00\", \"2016-03-24 04:00:00\", \"2016-03-30 04:00:00\", \"2016-04-27 04:00:00\", \"2016-04-28 04:00:00\", \"2016-04-29 04:00:00\", \"2016-04-30 04:00:00\", \"2016-05-01 04:00:00\", \"2016-05-02 04:00:00\", \"2016-05-03 04:00:00\", \"2016-05-04 04:00:00\", \"2016-05-05 04:00:00\", \"2016-05-06 04:00:00\", \"2016-05-09 04:00:00\", \"2016-05-10 04:00:00\", \"2016-05-13 04:00:00\", \"2016-05-14 04:00:00\", \"2016-05-15 04:00:00\", \"2016-05-16 04:00:00\", \"2016-05-17 04:00:00\", \"2016-05-18 04:00:00\", \"2016-05-19 04:00:00\", \"2016-05-20 04:00:00\", \"2016-05-23 04:00:00\", \"2016-05-25 04:00:00\", \"2016-05-26 04:00:00\", \"2016-05-27 04:00:00\", \"2016-05-31 04:00:00\", \"2016-06-01 04:00:00\", \"2016-06-02 04:00:00\", \"2016-06-07 04:00:00\", \"2016-06-08 04:00:00\", \"2016-06-09 04:00:00\", \"2016-06-10 04:00:00\", \"2016-06-11 04:00:00\", \"2016-06-13 04:00:00\", \"2016-06-14 04:00:00\", \"2016-06-15 04:00:00\", \"2016-06-30 04:00:00\", \"2016-07-01 04:00:00\", \"2016-07-05 04:00:00\", \"2016-07-11 04:00:00\", \"2016-07-12 04:00:00\", \"2016-07-13 04:00:00\", \"2016-07-14 04:00:00\", \"2016-07-15 04:00:00\", \"2016-07-25 04:00:00\", \"2016-08-04 04:00:00\", \"2016-08-05 04:00:00\", \"2016-08-09 04:00:00\", \"2016-08-10 04:00:00\", \"2016-08-11 04:00:00\", \"2016-08-12 04:00:00\", \"2016-08-13 04:00:00\", \"2016-08-14 04:00:00\", \"2016-08-15 04:00:00\", \"2016-08-19 04:00:00\", \"2016-08-20 04:00:00\", \"2016-08-21 04:00:00\", \"2016-08-22 04:00:00\", \"2016-08-23 04:00:00\", \"2016-08-24 04:00:00\", \"2016-08-25 04:00:00\", \"2016-08-26 04:00:00\", \"2016-08-31 04:00:00\", \"2016-09-01 04:00:00\", \"2016-09-02 04:00:00\", \"2016-09-03 04:00:00\", \"2016-09-04 04:00:00\", \"2016-09-05 04:00:00\", \"2016-09-12 04:00:00\", \"2016-09-14 04:00:00\", \"2016-09-16 04:00:00\", \"2016-10-03 04:00:00\", \"2016-10-04 04:00:00\", \"2016-10-05 04:00:00\", \"2016-10-06 04:00:00\", \"2016-10-07 04:00:00\", \"2016-10-08 04:00:00\", \"2016-10-10 04:00:00\", \"2016-10-11 04:00:00\", \"2016-10-12 04:00:00\", \"2016-10-13 04:00:00\", \"2016-10-14 04:00:00\", \"2016-10-20 04:00:00\", \"2016-10-21 04:00:00\", \"2016-10-24 04:00:00\", \"2016-10-25 04:00:00\", \"2016-10-26 04:00:00\", \"2016-10-27 04:00:00\", \"2016-10-28 04:00:00\", \"2016-10-30 04:00:00\", \"2016-10-31 04:00:00\", \"2016-11-01 04:00:00\", \"2016-11-02 04:00:00\", \"2016-11-03 04:00:00\", \"2016-11-04 04:00:00\", \"2016-11-05 04:00:00\", \"2016-11-06 04:00:00\", \"2016-11-07 05:00:00\", \"2016-11-08 05:00:00\", \"2016-11-09 05:00:00\", \"2016-11-10 05:00:00\", \"2016-11-11 05:00:00\", \"2016-11-12 05:00:00\", \"2016-11-13 05:00:00\", \"2016-11-14 05:00:00\", \"2016-11-15 05:00:00\", \"2016-11-16 05:00:00\", \"2016-11-17 05:00:00\", \"2016-11-18 05:00:00\", \"2016-11-21 05:00:00\", \"2016-11-22 05:00:00\", \"2016-11-23 05:00:00\", \"2016-11-26 05:00:00\", \"2016-11-28 05:00:00\", \"2016-11-29 05:00:00\", \"2016-11-30 05:00:00\", \"2016-12-01 05:00:00\", \"2016-12-02 05:00:00\", \"2016-12-05 05:00:00\", \"2016-12-06 05:00:00\", \"2016-12-07 05:00:00\", \"2016-12-08 05:00:00\", \"2016-12-09 05:00:00\", \"2016-12-12 05:00:00\", \"2016-12-13 05:00:00\", \"2016-12-14 05:00:00\", \"2016-12-15 05:00:00\", \"2016-12-16 05:00:00\", \"2017-01-05 05:00:00\", \"2017-01-06 05:00:00\", \"2017-01-09 05:00:00\", \"2017-01-10 05:00:00\", \"2017-01-11 05:00:00\", \"2017-01-12 05:00:00\", \"2017-01-13 05:00:00\", \"2017-01-16 05:00:00\", \"2017-01-17 05:00:00\", \"2017-01-18 05:00:00\", \"2017-01-19 05:00:00\", \"2017-01-20 05:00:00\", \"2017-01-21 05:00:00\", \"2017-01-23 05:00:00\", \"2017-01-24 05:00:00\", \"2017-01-25 05:00:00\", \"2017-01-26 05:00:00\", \"2017-01-27 05:00:00\", \"2017-01-30 05:00:00\", \"2017-01-31 05:00:00\", \"2017-02-07 05:00:00\", \"2017-02-08 05:00:00\", \"2017-02-14 05:00:00\", \"2017-02-15 05:00:00\", \"2017-02-16 05:00:00\", \"2017-02-17 05:00:00\", \"2017-03-01 05:00:00\", \"2017-03-02 05:00:00\", \"2017-03-06 05:00:00\", \"2017-03-07 05:00:00\", \"2017-03-08 05:00:00\", \"2017-03-09 05:00:00\", \"2017-03-13 04:00:00\", \"2017-03-15 04:00:00\", \"2017-03-21 04:00:00\", \"2017-03-22 04:00:00\", \"2017-03-23 04:00:00\", \"2017-03-24 04:00:00\", \"2017-03-28 04:00:00\", \"2017-03-29 04:00:00\", \"2017-03-30 04:00:00\", \"2017-03-31 04:00:00\", \"2017-04-01 04:00:00\", \"2017-04-02 04:00:00\", \"2017-04-03 04:00:00\", \"2017-04-04 04:00:00\", \"2017-04-05 04:00:00\", \"2017-04-06 04:00:00\", \"2017-04-07 04:00:00\", \"2017-04-10 04:00:00\", \"2017-04-11 04:00:00\", \"2017-04-12 04:00:00\", \"2017-04-13 04:00:00\", \"2017-04-14 04:00:00\", \"2017-04-18 04:00:00\", \"2017-04-19 04:00:00\", \"2017-04-20 04:00:00\", \"2017-04-21 04:00:00\", \"2017-04-23 04:00:00\", \"2017-04-24 04:00:00\", \"2017-04-25 04:00:00\", \"2017-04-26 04:00:00\", \"2017-04-28 04:00:00\", \"2017-05-01 04:00:00\", \"2017-05-02 04:00:00\", \"2017-05-03 04:00:00\", \"2017-05-04 04:00:00\", \"2017-05-05 04:00:00\", \"2017-05-06 04:00:00\", \"2017-05-07 04:00:00\", \"2017-05-08 04:00:00\", \"2017-05-09 04:00:00\", \"2017-05-10 04:00:00\", \"2017-05-11 04:00:00\", \"2017-05-12 04:00:00\", \"2017-05-13 04:00:00\", \"2017-05-14 04:00:00\", \"2017-05-15 04:00:00\", \"2017-05-16 04:00:00\", \"2017-05-17 04:00:00\", \"2017-05-19 04:00:00\", \"2017-06-01 04:00:00\", \"2017-06-02 04:00:00\", \"2017-06-05 04:00:00\", \"2017-06-06 04:00:00\", \"2017-06-08 04:00:00\", \"2017-06-09 04:00:00\", \"2017-06-12 04:00:00\", \"2017-06-13 04:00:00\", \"2017-06-26 04:00:00\", \"2017-06-27 04:00:00\", \"2017-06-28 04:00:00\", \"2017-06-29 04:00:00\", \"2017-06-30 04:00:00\", \"2017-07-03 04:00:00\", \"2017-07-12 04:00:00\", \"2017-07-13 04:00:00\", \"2017-07-14 04:00:00\", \"2017-07-17 04:00:00\", \"2017-07-19 04:00:00\", \"2017-07-20 04:00:00\", \"2017-07-25 04:00:00\", \"2017-07-26 04:00:00\", \"2017-07-27 04:00:00\", \"2017-07-28 04:00:00\", \"2017-08-03 04:00:00\", \"2017-08-04 04:00:00\", \"2017-08-05 04:00:00\", \"2017-08-06 04:00:00\", \"2017-08-07 04:00:00\", \"2017-08-08 04:00:00\", \"2017-08-09 04:00:00\", \"2017-08-10 04:00:00\", \"2017-08-11 04:00:00\", \"2017-08-14 04:00:00\", \"2017-08-15 04:00:00\", \"2017-08-16 04:00:00\", \"2017-08-17 04:00:00\", \"2017-08-18 04:00:00\", \"2017-08-21 04:00:00\", \"2017-08-22 04:00:00\", \"2017-08-25 04:00:00\", \"2017-08-28 04:00:00\", \"2017-08-29 04:00:00\", \"2017-08-30 04:00:00\", \"2017-08-31 04:00:00\", \"2017-09-06 04:00:00\", \"2017-09-12 04:00:00\", \"2017-09-13 04:00:00\", \"2017-09-14 04:00:00\", \"2017-09-15 04:00:00\", \"2017-09-18 04:00:00\", \"2017-09-19 04:00:00\", \"2017-09-20 04:00:00\", \"2017-09-21 04:00:00\", \"2017-09-25 04:00:00\", \"2017-09-26 04:00:00\", \"2017-09-29 04:00:00\", \"2017-10-02 04:00:00\", \"2017-10-03 04:00:00\", \"2017-10-04 04:00:00\", \"2017-10-05 04:00:00\", \"2017-10-10 04:00:00\", \"2017-10-16 04:00:00\", \"2017-10-17 04:00:00\", \"2017-10-18 04:00:00\", \"2017-10-19 04:00:00\", \"2017-10-24 04:00:00\", \"2017-10-25 04:00:00\", \"2017-10-26 04:00:00\", \"2017-10-27 04:00:00\", \"2017-10-30 04:00:00\", \"2017-10-31 04:00:00\", \"2017-11-02 04:00:00\", \"2017-11-03 04:00:00\", \"2017-11-07 05:00:00\", \"2017-11-08 05:00:00\", \"2017-11-09 05:00:00\", \"2017-11-13 05:00:00\", \"2017-11-14 05:00:00\", \"2017-11-27 05:00:00\", \"2017-11-28 05:00:00\", \"2017-11-29 05:00:00\", \"2017-11-30 05:00:00\", \"2017-12-01 05:00:00\", \"2017-12-04 05:00:00\", \"2017-12-05 05:00:00\", \"2017-12-06 05:00:00\", \"2017-12-07 05:00:00\", \"2017-12-08 05:00:00\", \"2017-12-11 05:00:00\", \"2017-12-12 05:00:00\", \"2017-12-13 05:00:00\", \"2017-12-14 05:00:00\", \"2017-12-18 05:00:00\", \"2017-12-21 05:00:00\", \"2017-12-22 05:00:00\", \"2018-01-02 05:00:00\", \"2018-01-03 05:00:00\", \"2018-01-04 05:00:00\", \"2018-01-05 05:00:00\", \"2018-01-06 05:00:00\", \"2018-01-07 05:00:00\", \"2018-01-08 05:00:00\", \"2018-01-09 05:00:00\", \"2018-01-10 05:00:00\", \"2018-01-11 05:00:00\", \"2018-01-12 05:00:00\", \"2018-01-13 05:00:00\", \"2018-01-14 05:00:00\", \"2018-01-15 05:00:00\", \"2018-01-16 05:00:00\", \"2018-01-17 05:00:00\", \"2018-01-18 05:00:00\", \"2018-01-31 05:00:00\", \"2018-02-01 05:00:00\", \"2018-02-02 05:00:00\", \"2018-02-05 05:00:00\", \"2018-02-06 05:00:00\", \"2018-02-07 05:00:00\", \"2018-02-08 05:00:00\", \"2018-02-09 05:00:00\", \"2018-02-12 05:00:00\", \"2018-02-13 05:00:00\", \"2018-02-14 05:00:00\", \"2018-02-22 05:00:00\", \"2018-02-23 05:00:00\", \"2018-02-24 05:00:00\", \"2018-02-25 05:00:00\", \"2018-02-26 05:00:00\", \"2018-02-27 05:00:00\", \"2018-02-28 05:00:00\", \"2018-03-01 05:00:00\", \"2018-03-02 05:00:00\", \"2018-03-06 05:00:00\", \"2018-03-07 05:00:00\", \"2018-03-08 05:00:00\", \"2018-03-09 05:00:00\", \"2018-03-12 04:00:00\", \"2018-03-14 04:00:00\", \"2018-03-15 04:00:00\", \"2018-03-23 04:00:00\", \"2018-03-26 04:00:00\", \"2018-03-27 04:00:00\", \"2018-03-28 04:00:00\", \"2018-03-29 04:00:00\", \"2018-03-30 04:00:00\", \"2018-04-02 04:00:00\", \"2018-04-03 04:00:00\", \"2018-04-04 04:00:00\", \"2018-04-05 04:00:00\", \"2018-04-12 04:00:00\", \"2018-04-13 04:00:00\", \"2018-04-16 04:00:00\", \"2018-04-17 04:00:00\", \"2018-04-18 04:00:00\", \"2018-04-19 04:00:00\", \"2018-04-27 04:00:00\", \"2018-07-10 04:00:00\", \"2018-07-11 04:00:00\", \"2018-07-12 04:00:00\", \"2018-07-13 04:00:00\", \"2018-07-16 04:00:00\", \"2018-07-17 04:00:00\", \"2018-07-18 04:00:00\", \"2018-07-19 04:00:00\", \"2018-07-20 04:00:00\", \"2018-07-23 04:00:00\", \"2018-07-24 04:00:00\", \"2018-07-25 04:00:00\", \"2018-07-26 04:00:00\", \"2018-07-27 04:00:00\", \"2018-07-28 04:00:00\", \"2018-07-29 04:00:00\", \"2018-07-30 04:00:00\", \"2018-07-31 04:00:00\", \"2018-08-01 04:00:00\", \"2018-08-02 04:00:00\", \"2018-08-03 04:00:00\", \"2018-08-04 04:00:00\", \"2018-08-05 04:00:00\", \"2018-08-06 04:00:00\", \"2018-08-07 04:00:00\", \"2018-08-13 04:00:00\", \"2018-08-14 04:00:00\", \"2018-08-15 04:00:00\", \"2018-08-16 04:00:00\", \"2018-08-17 04:00:00\", \"2018-08-18 04:00:00\", \"2018-08-19 04:00:00\", \"2018-08-20 04:00:00\", \"2018-08-21 04:00:00\", \"2018-08-22 04:00:00\", \"2018-08-23 04:00:00\", \"2018-08-24 04:00:00\", \"2018-08-25 04:00:00\", \"2018-08-26 04:00:00\", \"2018-08-27 04:00:00\", \"2018-08-28 04:00:00\", \"2018-08-29 04:00:00\", \"2018-08-30 04:00:00\", \"2018-09-04 04:00:00\", \"2018-09-05 04:00:00\", \"2018-09-07 04:00:00\", \"2018-09-08 04:00:00\", \"2018-09-09 04:00:00\", \"2018-09-10 04:00:00\", \"2018-09-11 04:00:00\", \"2018-09-14 04:00:00\", \"2018-09-19 04:00:00\", \"2018-09-21 04:00:00\", \"2018-09-24 04:00:00\", \"2018-09-25 04:00:00\", \"2018-09-26 04:00:00\", \"2018-09-27 04:00:00\", \"2018-09-28 04:00:00\", \"2018-10-01 04:00:00\", \"2018-10-02 04:00:00\", \"2018-10-03 04:00:00\", \"2018-10-05 04:00:00\", \"2018-10-06 04:00:00\", \"2018-10-07 04:00:00\", \"2018-10-09 04:00:00\", \"2018-10-10 04:00:00\", \"2018-10-11 04:00:00\", \"2018-10-12 04:00:00\", \"2018-10-17 04:00:00\", \"2018-10-18 04:00:00\", \"2018-10-19 04:00:00\", \"2018-10-22 04:00:00\", \"2018-10-23 04:00:00\", \"2018-11-08 05:00:00\", \"2018-11-19 05:00:00\", \"2018-11-28 05:00:00\", \"2018-12-04 05:00:00\", \"2018-12-05 05:00:00\", \"2018-12-06 05:00:00\", \"2018-12-07 05:00:00\", \"2018-12-10 05:00:00\", \"2018-12-11 05:00:00\", \"2018-12-12 05:00:00\", \"2018-12-13 05:00:00\", \"2018-12-14 05:00:00\", \"2018-12-20 05:00:00\", \"2018-12-21 05:00:00\", \"2019-01-07 05:00:00\"], \"y\": [254, 12335, 12623, 12652, 23229, 23314, 23350, 23372, 23384, 23397, 23483, 26348, 26377, 30119, 31339, 31369, 31370, 31396, 31413, 31442, 31462, 31483, 32856, 35788, 38401, 41706, 41731, 41924, 53465, 59063, 59419, 59424, 59425, 59435, 59760, 59795, 59853, 59893, 59927, 59999, 60031, 60087, 60103, 60121, 60143, 60180, 60562, 61154, 61202, 61292, 61327, 61662, 62118, 62256, 62841, 62842, 62843, 62903, 63306, 64517, 66396, 66397, 66418, 66427, 70420, 73064, 73774, 74472, 76905, 77565, 78014, 80022, 87907, 88012, 92795, 99540, 100439, 104656, 106818, 106882, 107531, 107535, 107680, 107852, 107854, 107886, 110430, 112386, 112597, 112713, 116948, 126851, 130678, 130793, 133589, 137144, 137295, 150460, 151820, 152366, 152432, 152626, 162100, 164250, 165088, 167572, 169302, 169571, 170799, 170860, 174525, 176624, 180990, 195232, 205914, 210050, 213203, 219693, 224489, 233517, 236035, 236517, 238869, 241081, 243975, 245687, 247711, 249814, 252460, 253979, 254286, 255549, 258919, 264177, 264791, 267270, 269108, 271098, 271633, 273353, 273558, 273618, 274068, 274250, 274407, 274694, 275564, 277543, 282596, 283982, 285020, 288020, 288139, 288200, 288223, 288252, 291199, 293752, 293818, 295216, 298534, 298626, 298640, 300644, 308965, 317623, 318418, 328258, 344497, 357536, 375841, 384039, 391522, 392022, 417469, 419463, 423625, 427134, 427301, 427367, 427370, 429493, 435182, 438917, 445567, 452086, 458400, 466292, 469109, 473683, 485031, 487746, 488059, 488339, 488340, 488533, 488946, 489033, 490582, 492607, 492969, 497347, 498950, 498952, 501777, 515042, 516450, 545098, 551916, 552320, 552327, 552328, 552339, 553627, 553660, 553682, 553725, 553738, 553740, 554687, 555913, 559538, 562317, 567113, 567118, 568546, 572767, 583191, 586747, 587229, 588148, 592347, 595107, 596823, 600007, 601885, 603802, 603948, 604594, 606468, 608207, 609997, 611847, 612309, 614291, 614320, 614345, 614975, 615509, 615540, 620262, 621075, 621142, 621532, 622717, 624266, 624956, 624959, 625188, 625275, 625324, 625361, 625490, 625504, 626190, 626201, 626223, 626249, 627867, 630420, 631343, 634622, 634629, 634712, 634972, 644851, 650011, 652481, 653447, 655003, 655075, 658396, 662209, 665711, 667079, 669465, 684506, 695141, 719587, 719786, 719792, 721158, 722897, 724213, 724219, 724758, 725169, 725809, 726016, 726237, 726518, 729111, 729584, 729830, 730190, 730209, 731814, 732406, 733802, 735093, 735312, 737722, 738671, 739194, 741176, 741942, 743158, 743679, 743834, 744129, 747900, 747906, 747916, 751851, 752457, 752632, 752863, 752905, 752912, 754028, 754053, 754075, 754166, 754299, 755104, 758179, 759429, 762524, 765343, 765347, 765438, 765510, 767617, 768179, 769065, 780961, 795540, 809113, 824783, 826130, 845525, 852759, 864929, 889007, 914645, 954954, 981062, 981063, 981067, 981069, 981070, 981072, 981073, 981074, 981075, 983484, 983582, 983745, 984664, 985215, 996002, 1004846, 1009949, 1017686, 1019071, 1028256, 1028417, 1028429, 1028453, 1028457, 1028458, 1029991, 1039090, 1039125, 1041964, 1043851, 1044097, 1045381, 1047408, 1048458, 1048585, 1049120, 1050492, 1058450, 1059744, 1060736, 1060895, 1060904, 1061211, 1062022, 1062071, 1062381, 1062576, 1062709, 1062790, 1062975, 1063052, 1063114, 1063201, 1063650, 1063749, 1063767, 1063890, 1063899, 1063988, 1067187, 1073276, 1078453, 1078675, 1079098, 1081857, 1085397, 1085768, 1085778, 1086941, 1089899, 1091810, 1098607, 1100409, 1101032, 1101702, 1101704, 1108605, 1113303, 1127186, 1127487, 1129950, 1136478, 1141155, 1142785, 1143428, 1144498, 1144897, 1144913, 1148566, 1149204, 1152375, 1152605, 1156565, 1156591, 1156838, 1157658, 1158000, 1158124, 1158174, 1158815, 1159202, 1159251, 1159325, 1159501, 1159566, 1160896, 1165176, 1166590, 1169614, 1172626, 1177800, 1181971, 1184417, 1186548, 1188595, 1189152, 1189457, 1189741, 1190376, 1195974, 1196238, 1196248, 1196434, 1196710, 1196972, 1197183, 1197437, 1197701, 1198014, 1198023, 1198025, 1198530, 1198746, 1198903, 1201407, 1204677, 1205642, 1215619, 1220898, 1222301, 1222319, 1238289, 1270389, 1286867, 1292028, 1294507, 1294537, 1294583, 1294584, 1294675, 1296684, 1296707, 1296817, 1296922, 1298399, 1298483, 1298575, 1298885, 1299713, 1299729, 1300245, 1300267, 1300268, 1300445, 1300531, 1301304, 1301307, 1301313, 1302177, 1302467, 1302728, 1302730, 1302960, 1303272, 1304476, 1307115, 1311169, 1316506, 1320618, 1320890, 1320931, 1320991, 1321036, 1321238, 1321477, 1321599, 1321876, 1321956, 1322007, 1323017, 1323501, 1323842, 1323899, 1323937, 1324457, 1325864, 1326356, 1327863, 1331338, 1331386, 1331421, 1331454, 1333631, 1335457, 1337898, 1338209, 1338269, 1338299, 1338314, 1338921, 1339076, 1339477, 1339840, 1341539, 1341560, 1341587, 1341619, 1341641, 1342971, 1344490, 1345095, 1345855, 1348357, 1353486, 1354033, 1355129, 1355307, 1355353, 1356487, 1357320, 1357881, 1358262, 1358388, 1359738, 1360967, 1360973, 1362950, 1364792, 1366178, 1367768, 1369064, 1369065, 1391733, 1398706, 1405376, 1412989, 1419744, 1441099, 1449324, 1458614, 1461912, 1484389, 1496134, 1511336, 1539229, 1549224, 1567233, 1584878, 1586218, 1587489, 1588003, 1588218, 1588704, 1588761, 1588796, 1588802, 1588827, 1588863, 1588897, 1588902, 1589083, 1589098, 1590061, 1591246, 1591518, 1591708, 1592352, 1593562, 1593930, 1594350, 1594460, 1594911, 1595242, 1595905, 1596911, 1597610, 1599767, 1602008, 1602052, 1602108, 1602167, 1602477, 1602553, 1603072, 1603440, 1603840, 1604512, 1605404, 1608286, 1608486, 1608595, 1608697, 1610835, 1620761, 1620762, 1620765, 1620766, 1620811, 1620812, 1620835, 1620848, 1620902, 1620924, 1621128, 1621271, 1621319, 1621390, 1621469, 1621632, 1621709, 1621742, 1621796, 1622433, 1623409, 1624618, 1625809, 1626538, 1626547, 1626773, 1627516, 1629134, 1629150, 1629371, 1631067, 1631740, 1633357, 1637258, 1642148, 1649847, 1679050, 1706741, 1726834, 1756820, 1757668, 1769250, 1774590, 1780366, 1783191, 1787682, 1790717, 1791084, 1791421, 1791458, 1791518, 1791631, 1791684, 1791725, 1791739, 1791844, 1791925, 1791927, 1792090, 1792167, 1792258, 1792498, 1792575, 1792798, 1793281, 1794373, 1794517, 1794781, 1795013, 1795119, 1796859, 1799806, 1801782, 1801830, 1801858, 1801866, 1801870, 1801872, 1802064, 1805448, 1807119, 1809240, 1815007, 1816287, 1817003, 1818756, 1821614, 1821622, 1825134, 1834369], \"type\": \"scattergl\", \"uid\": \"b840a0ac-7333-4efd-ba48-0de0188662f0\"}, {\"name\": \"FT2\", \"x\": [\"2014-01-08 05:00:00\", \"2014-01-09 05:00:00\", \"2014-01-15 05:00:00\", \"2014-01-17 05:00:00\", \"2014-01-29 05:00:00\", \"2014-01-30 05:00:00\", \"2014-02-03 05:00:00\", \"2014-02-06 05:00:00\", \"2014-03-04 05:00:00\", \"2014-03-05 05:00:00\", \"2014-03-06 05:00:00\", \"2014-03-10 04:00:00\", \"2014-03-11 04:00:00\", \"2014-03-12 04:00:00\", \"2014-03-13 04:00:00\", \"2014-03-14 04:00:00\", \"2014-03-17 04:00:00\", \"2014-03-26 04:00:00\", \"2014-04-22 04:00:00\", \"2014-04-23 04:00:00\", \"2014-05-02 04:00:00\", \"2014-05-05 04:00:00\", \"2014-05-06 04:00:00\", \"2014-05-07 04:00:00\", \"2014-05-16 04:00:00\", \"2014-05-21 04:00:00\", \"2014-05-22 04:00:00\", \"2014-06-03 04:00:00\", \"2014-06-04 04:00:00\", \"2014-06-05 04:00:00\", \"2014-06-06 04:00:00\", \"2014-06-09 04:00:00\", \"2014-06-10 04:00:00\", \"2014-06-11 04:00:00\", \"2014-06-12 04:00:00\", \"2014-06-13 04:00:00\", \"2014-06-24 04:00:00\", \"2014-06-25 04:00:00\", \"2014-06-26 04:00:00\", \"2014-06-27 04:00:00\", \"2014-06-30 04:00:00\", \"2014-07-01 04:00:00\", \"2014-07-02 04:00:00\", \"2014-07-03 04:00:00\", \"2014-07-04 04:00:00\", \"2014-07-07 04:00:00\", \"2014-07-11 04:00:00\", \"2014-07-14 04:00:00\", \"2014-07-15 04:00:00\", \"2014-07-16 04:00:00\", \"2014-07-17 04:00:00\", \"2014-07-25 04:00:00\", \"2014-07-28 04:00:00\", \"2014-07-29 04:00:00\", \"2014-09-05 04:00:00\", \"2014-09-23 04:00:00\", \"2014-09-25 04:00:00\", \"2014-09-26 04:00:00\", \"2014-09-29 04:00:00\", \"2014-09-30 04:00:00\", \"2014-10-01 04:00:00\", \"2014-10-02 04:00:00\", \"2014-10-03 04:00:00\", \"2014-10-21 04:00:00\", \"2014-10-22 04:00:00\", \"2014-10-24 04:00:00\", \"2014-10-27 04:00:00\", \"2014-10-29 04:00:00\", \"2014-10-30 04:00:00\", \"2014-11-10 05:00:00\", \"2014-11-11 05:00:00\", \"2014-11-13 05:00:00\", \"2014-11-14 05:00:00\", \"2014-11-19 05:00:00\", \"2014-11-20 05:00:00\", \"2014-11-25 05:00:00\", \"2014-12-10 05:00:00\", \"2014-12-11 05:00:00\", \"2014-12-23 05:00:00\", \"2015-01-13 05:00:00\", \"2015-01-22 05:00:00\", \"2015-01-23 05:00:00\", \"2015-01-30 05:00:00\", \"2015-02-26 05:00:00\", \"2015-02-27 05:00:00\", \"2015-03-30 04:00:00\", \"2015-03-31 04:00:00\", \"2015-04-01 04:00:00\", \"2015-04-02 04:00:00\", \"2015-04-03 04:00:00\", \"2015-04-05 04:00:00\", \"2015-04-06 04:00:00\", \"2015-04-20 04:00:00\", \"2015-04-21 04:00:00\", \"2015-04-27 04:00:00\", \"2015-04-28 04:00:00\", \"2015-04-29 04:00:00\", \"2015-04-30 04:00:00\", \"2015-05-04 04:00:00\", \"2015-05-06 04:00:00\", \"2015-05-11 04:00:00\", \"2015-06-30 04:00:00\", \"2015-07-28 04:00:00\", \"2015-07-30 04:00:00\", \"2015-08-10 04:00:00\", \"2015-08-11 04:00:00\", \"2015-08-12 04:00:00\", \"2015-08-13 04:00:00\", \"2015-08-14 04:00:00\", \"2015-08-17 04:00:00\", \"2015-08-18 04:00:00\", \"2015-08-19 04:00:00\", \"2015-08-20 04:00:00\", \"2015-08-21 04:00:00\", \"2015-08-24 04:00:00\", \"2015-08-25 04:00:00\", \"2015-08-26 04:00:00\", \"2015-08-27 04:00:00\", \"2015-08-28 04:00:00\", \"2015-08-29 04:00:00\", \"2015-09-03 04:00:00\", \"2015-09-12 04:00:00\", \"2015-09-13 04:00:00\", \"2015-09-14 04:00:00\", \"2015-09-23 04:00:00\", \"2015-09-24 04:00:00\", \"2015-09-25 04:00:00\", \"2015-09-26 04:00:00\", \"2015-09-28 04:00:00\", \"2015-09-29 04:00:00\", \"2015-09-30 04:00:00\", \"2015-10-05 04:00:00\", \"2015-10-12 04:00:00\", \"2015-10-21 04:00:00\", \"2015-10-27 04:00:00\", \"2015-10-28 04:00:00\", \"2015-10-29 04:00:00\", \"2015-12-04 05:00:00\", \"2015-12-08 05:00:00\", \"2015-12-09 05:00:00\", \"2015-12-10 05:00:00\", \"2015-12-11 05:00:00\", \"2015-12-12 05:00:00\", \"2015-12-14 05:00:00\", \"2016-01-06 05:00:00\", \"2016-01-07 05:00:00\", \"2016-02-04 05:00:00\", \"2016-02-05 05:00:00\", \"2016-02-08 05:00:00\", \"2016-02-09 05:00:00\", \"2016-02-10 05:00:00\", \"2016-02-11 05:00:00\", \"2016-02-12 05:00:00\", \"2016-02-13 05:00:00\", \"2016-02-14 05:00:00\", \"2016-02-15 05:00:00\", \"2016-02-16 05:00:00\", \"2016-02-18 05:00:00\", \"2016-02-22 05:00:00\", \"2016-02-23 05:00:00\", \"2016-03-23 04:00:00\", \"2016-03-24 04:00:00\", \"2016-03-25 04:00:00\", \"2016-03-29 04:00:00\", \"2016-03-30 04:00:00\", \"2016-04-04 04:00:00\", \"2016-04-06 04:00:00\", \"2016-04-07 04:00:00\", \"2016-04-21 04:00:00\", \"2016-04-23 04:00:00\", \"2016-04-26 04:00:00\", \"2016-04-30 04:00:00\", \"2016-05-09 04:00:00\", \"2016-05-13 04:00:00\", \"2016-05-14 04:00:00\", \"2016-05-17 04:00:00\", \"2016-05-18 04:00:00\", \"2016-05-19 04:00:00\", \"2016-05-20 04:00:00\", \"2016-05-24 04:00:00\", \"2016-06-01 04:00:00\", \"2016-06-02 04:00:00\", \"2016-06-03 04:00:00\", \"2016-06-27 04:00:00\", \"2016-06-28 04:00:00\", \"2016-06-29 04:00:00\", \"2016-06-30 04:00:00\", \"2016-07-01 04:00:00\", \"2016-07-20 04:00:00\", \"2016-07-21 04:00:00\", \"2016-07-22 04:00:00\", \"2016-07-27 04:00:00\", \"2016-07-28 04:00:00\", \"2016-07-29 04:00:00\", \"2016-08-09 04:00:00\", \"2016-08-10 04:00:00\", \"2016-08-11 04:00:00\", \"2016-08-12 04:00:00\", \"2016-08-14 04:00:00\", \"2016-08-17 04:00:00\", \"2016-08-19 04:00:00\", \"2016-08-24 04:00:00\", \"2016-08-25 04:00:00\", \"2016-08-26 04:00:00\", \"2016-08-29 04:00:00\", \"2016-08-30 04:00:00\", \"2016-08-31 04:00:00\", \"2016-09-07 04:00:00\", \"2016-09-08 04:00:00\", \"2016-09-09 04:00:00\", \"2016-09-10 04:00:00\", \"2016-09-12 04:00:00\", \"2016-09-13 04:00:00\", \"2016-09-14 04:00:00\", \"2016-09-15 04:00:00\", \"2016-09-16 04:00:00\", \"2016-09-17 04:00:00\", \"2016-09-24 04:00:00\", \"2016-09-27 04:00:00\", \"2016-10-10 04:00:00\", \"2016-10-12 04:00:00\", \"2016-10-13 04:00:00\", \"2016-10-14 04:00:00\", \"2016-10-15 04:00:00\", \"2016-10-17 04:00:00\", \"2016-10-18 04:00:00\", \"2016-10-20 04:00:00\", \"2016-10-21 04:00:00\", \"2016-10-25 04:00:00\", \"2016-10-26 04:00:00\", \"2016-10-27 04:00:00\", \"2016-10-28 04:00:00\", \"2016-10-31 04:00:00\", \"2016-11-01 04:00:00\", \"2016-11-02 04:00:00\", \"2016-11-03 04:00:00\", \"2016-11-04 04:00:00\", \"2016-11-16 05:00:00\", \"2016-11-21 05:00:00\", \"2016-11-22 05:00:00\", \"2016-11-23 05:00:00\", \"2016-11-25 05:00:00\", \"2016-11-26 05:00:00\", \"2016-11-28 05:00:00\", \"2016-11-29 05:00:00\", \"2016-11-30 05:00:00\", \"2016-12-01 05:00:00\", \"2016-12-02 05:00:00\", \"2016-12-14 05:00:00\", \"2017-02-08 05:00:00\", \"2017-02-10 05:00:00\", \"2017-02-14 05:00:00\", \"2017-02-15 05:00:00\", \"2017-02-16 05:00:00\", \"2017-02-22 05:00:00\", \"2017-03-01 05:00:00\", \"2017-03-02 05:00:00\", \"2017-03-03 05:00:00\", \"2017-03-04 05:00:00\", \"2017-03-05 05:00:00\", \"2017-03-06 05:00:00\", \"2017-03-07 05:00:00\", \"2017-03-08 05:00:00\", \"2017-03-09 05:00:00\", \"2017-03-10 05:00:00\", \"2017-03-13 04:00:00\", \"2017-03-14 04:00:00\", \"2017-03-15 04:00:00\", \"2017-03-16 04:00:00\", \"2017-03-17 04:00:00\", \"2017-03-24 04:00:00\", \"2017-03-28 04:00:00\", \"2017-03-29 04:00:00\", \"2017-03-30 04:00:00\", \"2017-04-02 04:00:00\", \"2017-04-05 04:00:00\", \"2017-04-07 04:00:00\", \"2017-04-08 04:00:00\", \"2017-04-11 04:00:00\", \"2017-04-12 04:00:00\", \"2017-04-13 04:00:00\", \"2017-04-14 04:00:00\", \"2017-04-15 04:00:00\", \"2017-04-17 04:00:00\", \"2017-04-18 04:00:00\", \"2017-04-19 04:00:00\", \"2017-04-20 04:00:00\", \"2017-04-21 04:00:00\", \"2017-04-22 04:00:00\", \"2017-04-23 04:00:00\", \"2017-04-25 04:00:00\", \"2017-04-26 04:00:00\", \"2017-04-27 04:00:00\", \"2017-04-28 04:00:00\", \"2017-04-29 04:00:00\", \"2017-05-02 04:00:00\", \"2017-05-03 04:00:00\", \"2017-05-04 04:00:00\", \"2017-05-05 04:00:00\", \"2017-05-08 04:00:00\", \"2017-05-09 04:00:00\", \"2017-05-10 04:00:00\", \"2017-05-11 04:00:00\", \"2017-05-12 04:00:00\", \"2017-05-13 04:00:00\", \"2017-05-14 04:00:00\", \"2017-05-15 04:00:00\", \"2017-05-16 04:00:00\", \"2017-05-17 04:00:00\", \"2017-05-18 04:00:00\", \"2017-05-19 04:00:00\", \"2017-05-20 04:00:00\", \"2017-05-21 04:00:00\", \"2017-05-22 04:00:00\", \"2017-05-23 04:00:00\", \"2017-05-24 04:00:00\", \"2017-05-25 04:00:00\", \"2017-05-26 04:00:00\", \"2017-06-01 04:00:00\", \"2017-06-02 04:00:00\", \"2017-06-05 04:00:00\", \"2017-06-09 04:00:00\", \"2017-06-10 04:00:00\", \"2017-06-11 04:00:00\", \"2017-06-12 04:00:00\", \"2017-06-13 04:00:00\", \"2017-06-15 04:00:00\", \"2017-06-16 04:00:00\", \"2017-06-17 04:00:00\", \"2017-06-26 04:00:00\", \"2017-06-27 04:00:00\", \"2017-06-28 04:00:00\", \"2017-06-29 04:00:00\", \"2017-07-05 04:00:00\", \"2017-07-20 04:00:00\", \"2017-07-21 04:00:00\", \"2017-07-22 04:00:00\", \"2017-07-24 04:00:00\", \"2017-08-14 04:00:00\", \"2017-08-15 04:00:00\", \"2017-08-16 04:00:00\", \"2017-08-17 04:00:00\", \"2017-08-23 04:00:00\", \"2017-08-24 04:00:00\", \"2017-08-25 04:00:00\", \"2017-08-28 04:00:00\", \"2017-10-26 04:00:00\", \"2017-10-27 04:00:00\", \"2017-10-30 04:00:00\", \"2017-10-31 04:00:00\", \"2017-11-01 04:00:00\", \"2017-11-02 04:00:00\", \"2017-11-07 05:00:00\", \"2017-11-08 05:00:00\", \"2017-11-09 05:00:00\", \"2017-11-10 05:00:00\", \"2017-11-11 05:00:00\", \"2017-11-13 05:00:00\", \"2017-11-16 05:00:00\", \"2017-11-20 05:00:00\", \"2017-12-04 05:00:00\", \"2017-12-05 05:00:00\", \"2017-12-07 05:00:00\", \"2017-12-13 05:00:00\", \"2017-12-15 05:00:00\", \"2017-12-18 05:00:00\", \"2017-12-19 05:00:00\", \"2017-12-21 05:00:00\", \"2018-01-02 05:00:00\", \"2018-01-03 05:00:00\", \"2018-01-05 05:00:00\", \"2018-01-08 05:00:00\", \"2018-01-09 05:00:00\", \"2018-01-10 05:00:00\", \"2018-01-15 05:00:00\", \"2018-01-16 05:00:00\", \"2018-01-17 05:00:00\", \"2018-02-08 05:00:00\", \"2018-02-09 05:00:00\", \"2018-02-13 05:00:00\", \"2018-02-14 05:00:00\", \"2018-02-15 05:00:00\", \"2018-02-16 05:00:00\", \"2018-02-21 05:00:00\", \"2018-02-22 05:00:00\", \"2018-03-12 04:00:00\", \"2018-03-14 04:00:00\", \"2018-03-20 04:00:00\", \"2018-03-21 04:00:00\", \"2018-03-27 04:00:00\", \"2018-03-28 04:00:00\", \"2018-04-02 04:00:00\", \"2018-04-03 04:00:00\", \"2018-04-06 04:00:00\", \"2018-08-02 04:00:00\", \"2018-08-07 04:00:00\", \"2018-08-08 04:00:00\", \"2018-08-09 04:00:00\", \"2018-08-10 04:00:00\", \"2018-08-13 04:00:00\", \"2018-08-14 04:00:00\", \"2018-08-15 04:00:00\", \"2018-08-30 04:00:00\", \"2018-08-31 04:00:00\", \"2018-09-01 04:00:00\", \"2018-09-04 04:00:00\", \"2018-09-10 04:00:00\", \"2018-09-11 04:00:00\", \"2018-10-03 04:00:00\", \"2018-10-29 04:00:00\", \"2018-11-06 05:00:00\", \"2018-11-07 05:00:00\", \"2018-11-08 05:00:00\", \"2018-11-20 05:00:00\", \"2018-11-26 05:00:00\", \"2018-11-27 05:00:00\", \"2018-11-28 05:00:00\", \"2018-11-29 05:00:00\", \"2018-11-30 05:00:00\", \"2018-12-03 05:00:00\", \"2018-12-04 05:00:00\", \"2018-12-05 05:00:00\", \"2018-12-06 05:00:00\", \"2018-12-07 05:00:00\", \"2018-12-10 05:00:00\", \"2018-12-11 05:00:00\", \"2018-12-13 05:00:00\"], \"y\": [1, 109, 113, 490, 718, 743, 749, 1325, 1326, 1327, 1842, 2401, 3817, 4782, 5797, 9562, 9635, 9642, 9649, 9653, 9720, 10322, 11361, 11605, 12324, 12751, 13446, 15575, 16209, 20213, 22570, 25865, 26969, 27048, 27921, 27986, 28054, 28670, 29329, 29888, 29928, 32001, 36042, 38051, 39156, 40380, 40791, 40834, 40869, 40870, 40872, 40883, 40885, 40886, 40887, 40888, 40906, 41134, 41451, 41778, 41804, 42317, 42906, 43016, 43018, 43727, 44127, 49567, 51576, 51629, 52041, 54351, 57101, 58933, 61249, 61314, 61320, 61323, 61324, 61325, 61526, 62054, 62061, 62067, 62072, 62131, 62536, 62559, 62560, 64389, 65036, 65858, 65862, 65865, 65866, 66004, 66034, 66189, 66252, 66269, 66270, 66296, 66297, 66659, 66660, 67843, 68038, 69565, 70269, 70588, 70740, 70878, 71179, 71280, 71709, 72859, 72871, 73376, 75348, 76322, 76864, 76865, 76868, 78757, 95361, 108381, 113213, 114401, 116228, 121252, 127532, 128354, 135884, 135885, 137794, 139995, 141543, 142526, 142527, 142544, 150890, 159271, 160049, 160052, 160057, 160059, 160062, 160075, 160077, 160080, 163062, 166106, 167640, 169329, 170325, 172784, 172800, 172804, 172981, 173979, 173980, 173992, 173993, 174046, 174194, 174205, 174374, 174375, 174376, 174387, 174614, 174617, 174618, 175510, 177803, 180157, 184078, 184864, 185071, 186614, 187744, 198159, 201294, 201303, 203003, 204841, 206226, 208419, 208432, 208434, 208486, 208583, 208681, 208722, 212407, 220207, 220607, 222328, 224703, 224715, 224734, 224735, 224899, 226888, 226941, 227114, 227116, 227265, 227298, 227308, 227337, 227345, 228722, 235376, 238997, 243619, 246720, 246722, 246759, 246761, 246762, 246777, 246786, 246789, 246895, 246910, 246951, 246960, 247163, 247365, 247475, 247502, 247808, 247937, 249543, 250689, 252801, 252806, 252808, 253753, 256542, 257544, 259076, 259917, 260524, 260623, 261019, 261118, 261316, 261318, 261420, 261474, 261481, 261495, 261496, 261501, 261505, 262607, 265956, 268022, 268023, 268180, 268290, 268492, 272799, 276049, 281555, 281664, 286380, 286433, 286977, 287827, 289205, 289777, 289778, 289780, 289804, 289838, 289840, 292989, 295807, 303582, 304271, 308295, 308541, 313367, 319328, 321961, 326479, 326694, 326695, 326696, 326700, 326705, 326707, 326718, 326726, 326741, 326849, 327043, 327075, 327121, 327224, 327379, 327734, 328136, 328479, 328680, 328747, 329239, 329876, 330438, 332321, 333855, 334189, 334700, 336143, 337157, 337183, 337191, 337196, 337404, 339131, 339235, 339635, 339783, 339849, 340828, 341071, 341315, 341622, 341653, 341674, 341679, 342044, 342726, 344390, 344396, 347769, 352027, 353162, 353165, 353295, 353559, 353634, 353640, 353856, 354158, 354260, 355321, 355787, 356541, 356564, 356587, 356621, 356648, 356659, 358230, 358233, 358238, 358240, 358245, 358967, 358979, 358992, 359312, 359368, 359370, 359371, 359387, 360061, 363036, 365319, 365321, 365325, 365347, 365436, 365600, 365657, 365658, 366919, 367403, 367408, 368980, 373490, 375083, 375084, 376097, 376557, 376560, 376570, 376625, 376652, 376666, 376765, 376878, 377014, 377068, 377411, 377654, 379689, 383585, 387381, 390972, 392856, 393755, 393792, 393830, 393833, 393834, 393835, 393850, 393882, 393884, 393885, 393886, 394145, 394289, 394455, 394770, 395593, 395652, 395661, 395689, 395705, 395706, 395713], \"type\": \"scattergl\", \"uid\": \"2afe9e17-d549-4a05-95ed-70c9d5afb309\"}], {\"height\": 600.0, \"legend\": {\"x\": 0.1, \"y\": 0.95}, \"showlegend\": true, \"title\": \"How we collect data\", \"xaxis\": {\"title\": \"Time\"}, \"yaxis\": {\"title\": \"Number of scans\"}}, {\"showLink\": false, \"linkText\": \"\", \"plotlyServerURL\": \"https://plot.ly\"})</script><script type=\"text/javascript\">window.addEventListener(\"resize\", function(){Plotly.Plots.resize(document.getElementById(\"b01d543c-a117-41d1-943d-7fdd17531616\"));});</script></div>\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7938aa388fd116bf3beae330f6251c3ba737a65 | 170,564 | ipynb | Jupyter Notebook | dataset/creditCard/Credit Card.ipynb | Necropsy/XXIIISI-Minicurso | 6facc4aa147bd0cf0b5e4c3446a43b39bec93af7 | [
"MIT"
] | null | null | null | dataset/creditCard/Credit Card.ipynb | Necropsy/XXIIISI-Minicurso | 6facc4aa147bd0cf0b5e4c3446a43b39bec93af7 | [
"MIT"
] | null | null | null | dataset/creditCard/Credit Card.ipynb | Necropsy/XXIIISI-Minicurso | 6facc4aa147bd0cf0b5e4c3446a43b39bec93af7 | [
"MIT"
] | null | null | null | 99.454227 | 113,812 | 0.806964 | [
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n\nimport os",
"_____no_output_____"
],
[
"credit = pd.read_csv(\"../input/creditcard.csv\")",
"_____no_output_____"
],
[
"credit.head()",
"_____no_output_____"
],
[
"credit.describe()",
"_____no_output_____"
],
[
"credit.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 284807 entries, 0 to 284806\nData columns (total 31 columns):\nTime 284807 non-null float64\nV1 284807 non-null float64\nV2 284807 non-null float64\nV3 284807 non-null float64\nV4 284807 non-null float64\nV5 284807 non-null float64\nV6 284807 non-null float64\nV7 284807 non-null float64\nV8 284807 non-null float64\nV9 284807 non-null float64\nV10 284807 non-null float64\nV11 284807 non-null float64\nV12 284807 non-null float64\nV13 284807 non-null float64\nV14 284807 non-null float64\nV15 284807 non-null float64\nV16 284807 non-null float64\nV17 284807 non-null float64\nV18 284807 non-null float64\nV19 284807 non-null float64\nV20 284807 non-null float64\nV21 284807 non-null float64\nV22 284807 non-null float64\nV23 284807 non-null float64\nV24 284807 non-null float64\nV25 284807 non-null float64\nV26 284807 non-null float64\nV27 284807 non-null float64\nV28 284807 non-null float64\nAmount 284807 non-null float64\nClass 284807 non-null int64\ndtypes: float64(30), int64(1)\nmemory usage: 67.4 MB\n"
],
[
"credit.shape",
"_____no_output_____"
],
[
"credit.loc[:, 'Class'].value_counts()",
"_____no_output_____"
],
[
"V_col = credit[['V1','V2','V3','V4','V5','V6','V7','V8','V9','V10','V11','V12','V13','V14','V15','V16','V17','V18','V19','V20','V21','V22','V23','V24','V25','V26','V27','V28']]\nV_col.head()",
"_____no_output_____"
],
[
"V_col.hist(figsize=(30, 20))\nplt.show()",
"_____no_output_____"
],
[
"no_of_normal_transcations = len(credit[credit['Class']==1])\nno_of_fraud_transcations = len(credit[credit['Class']==0])\nprint(\"no_of_normal_transcations:\",no_of_normal_transcations)\nprint(\"no_of_fraud_transcations:\", no_of_fraud_transcations)",
"no_of_normal_transcations: 492\nno_of_fraud_transcations: 284315\n"
],
[
"X = credit.iloc[:, 1:29].values\ny = credit.iloc[:, 30].values",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.25, random_state = 0)\nprint (\"X_train: \", len(X_train))\nprint(\"X_test: \", len(X_test))\nprint(\"y_train: \", len(y_train))\nprint(\"y_test: \", len(y_test))",
"X_train: 213605\nX_test: 71202\ny_train: 213605\ny_test: 71202\n"
]
],
[
[
"***KNN Classification***",
"_____no_output_____"
]
],
[
[
"from sklearn.neighbors import KNeighborsClassifier\nknc = KNeighborsClassifier(n_neighbors = 17)\nX,y = credit.loc[:,credit.columns != 'Class'], credit.loc[:,'Class']\nknc.fit(X_train,y_train)\ny_knc = knc.predict(X_test)\nprint('accuracy of training set: {:.4f}'.format(knc.score(X_train,y_train)))\nprint('accuracy of test set: {:.4f}'.format(knc.score(X_test, y_test)))",
"accuracy of training set: 0.9994\naccuracy of test set: 0.9995\n"
],
[
"from sklearn.metrics import accuracy_score, confusion_matrix, precision_score, recall_score, precision_recall_curve\nprint('confusion_matrix of KNN: ', confusion_matrix(y_test, y_knc))\nprint('precision_score of KNN: ', precision_score(y_test, y_knc))\nprint('recall_score of KNN: ', recall_score(y_test, y_knc))\nprint('precision_recall_curve: ', precision_recall_curve(y_test, y_knc))",
"confusion_matrix of KNN: [[71070 12]\n [ 26 94]]\nprecision_score of KNN: 0.8867924528301887\nrecall_score of KNN: 0.7833333333333333\nprecision_recall_curve: (array([0.00168535, 0.88679245, 1. ]), array([1. , 0.78333333, 0. ]), array([0, 1]))\n"
]
],
[
[
"**Random Forest Regression**",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestRegressor\nreg = RandomForestRegressor(n_estimators = 20, random_state = 0)\nreg.fit(X_train,y_train)",
"/usr/local/lib/python3.6/dist-packages/sklearn/ensemble/weight_boosting.py:29: DeprecationWarning: numpy.core.umath_tests is an internal NumPy module and should not be imported. It will be removed in a future NumPy release.\n from numpy.core.umath_tests import inner1d\n"
],
[
"y_rfr = reg.predict(X_test)",
"_____no_output_____"
],
[
"reg.score(X_test, y_test)\nprint('accuracy of training set: {:.4f}'.format(reg.score(X_train,y_train)))\nprint('accuaracy of test set: {:.4f}'.format(reg.score(X_test, y_test)))",
"accuracy of training set: 0.9572\naccuaracy of test set: 0.7223\n"
],
[
"print('accuracy_score of decision tree regression: ', accuracy_score( y_dtr , y_test))\nprint('confusion_matrix of decision tree regression: ', confusion_matrix(y_dtr, y_test))\nprint('precision_score of decision tree regression: ', precision_score( y_dtr, y_test))\nprint('recall_score of decision tree regression: ', recall_score( y_dtr, y_test))\nprint('precision_recall_curve: ', precision_recall_curve(y_dtr, y_test))",
"accuracy_score of decision tree regression: 0.999283727985169\nconfusion_matrix of decision tree regression: [[71061 30]\n [ 21 90]]\nprecision_score of decision tree regression: 0.75\nrecall_score of decision tree regression: 0.8108108108108109\nprecision_recall_curve: (array([0.00155894, 0.75 , 1. ]), array([1. , 0.81081081, 0. ]), array([0, 1]))\n"
]
],
[
[
"**Decision Tree Regression**",
"_____no_output_____"
]
],
[
[
"from sklearn.tree import DecisionTreeRegressor\nregs = DecisionTreeRegressor(random_state = 0)\nregs.fit(X_train, y_train)",
"_____no_output_____"
],
[
"y_dtr = regs.predict(X_test)",
"_____no_output_____"
],
[
"regs.score(X_test, y_test)\nprint('accuracy of training set: {:.4f}'.format(regs.score(X_train,y_train)))\nprint('accuaracy of test set: {:.4f}'.format(regs.score(X_test, y_test)))",
"accuracy of training set: 1.0000\naccuaracy of test set: 0.5743\n"
],
[
"print('accuracy_score of decision tree regression: ', accuracy_score( y_dtr , y_test))\nprint('confusion_matrix of decision tree regression: ', confusion_matrix(y_dtr, y_test))\nprint('precision_score of decision tree regression: ', precision_score( y_dtr, y_test))\nprint('recall_score of decision tree regression: ', recall_score( y_dtr, y_test))\nprint('precision_recall_curve: ', precision_recall_curve(y_dtr, y_test))",
"accuracy_score of decision tree regression: 0.999283727985169\nconfusion_matrix of decision tree regression: [[71061 30]\n [ 21 90]]\nprecision_score of decision tree regression: 0.75\nrecall_score of decision tree regression: 0.8108108108108109\nprecision_recall_curve: (array([0.00155894, 0.75 , 1. ]), array([1. , 0.81081081, 0. ]), array([0, 1]))\n"
]
],
[
[
"**Logistic Regression**",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression\nlogreg = LogisticRegression(random_state = 0)\nlogreg.fit(X_train, y_train)",
"_____no_output_____"
],
[
"y_lr = logreg.predict(X_test)",
"_____no_output_____"
],
[
"logreg.score(X_test, y_test)\nprint('accuracy of training set: {:.4f}'.format(logreg.score(X_train,y_train)))\nprint('accuaracy of test set: {:.4f}'.format(logreg.score(X_test, y_test)))",
"accuracy of training set: 0.9992\naccuaracy of test set: 0.9993\n"
],
[
"print('accuracy_score of logistic regression : ', accuracy_score(y_test, y_lr))\nprint('confusion_matrix of logistic regression: ', confusion_matrix(y_test, y_lr))\nprint('precision_score of logistic regression: ', precision_score(y_test, y_lr))\nprint('recall_score of logistic regression: ', recall_score(y_test, y_lr))\nprint('precision_recall_curve: ', precision_recall_curve(y_test, y_lr))",
"accuracy_score of logistic regression : 0.999283727985169\nconfusion_matrix of logistic regression: [[71072 10]\n [ 41 79]]\nprecision_score of logistic regression: 0.8876404494382022\nrecall_score of logistic regression: 0.6583333333333333\nprecision_recall_curve: (array([0.00168535, 0.88764045, 1. ]), array([1. , 0.65833333, 0. ]), array([0, 1]))\n"
],
[
"logreg100 = LogisticRegression(random_state = 1000, C =100)\nlogreg100.fit(X_train, y_train)",
"_____no_output_____"
],
[
"y_lr100 = logreg100.predict(X_test)",
"_____no_output_____"
],
[
"logreg100.score(X_test, y_test)\nprint('accuracy of training set: {:.4f}'.format(logreg100.score(X_train,y_train)))\nprint('accuaracy of test set: {:.4f}'.format(logreg100.score(X_test, y_test)))",
"accuracy of training set: 0.9992\naccuaracy of test set: 0.9993\n"
],
[
"logreg01 = LogisticRegression(random_state = 0, C =0.001)\nlogreg01.fit(X_train, y_train)",
"_____no_output_____"
],
[
"y_p01 = logreg01.predict(X_test)",
"_____no_output_____"
],
[
"logreg01.score(X_test, y_test)\nprint('accuracy of training set: {:.4f}'.format(logreg01.score(X_train,y_train)))\nprint('accuaracy of test set: {:.4f}'.format(logreg01.score(X_test, y_test)))",
"accuracy of training set: 0.9990\naccuaracy of test set: 0.9991\n"
]
],
[
[
"**Decision Tree Classification**",
"_____no_output_____"
]
],
[
[
"from sklearn.tree import DecisionTreeClassifier\nclassifier = DecisionTreeClassifier(criterion = 'entropy', random_state = 0)\nclassifier.fit(X_train, y_train)",
"_____no_output_____"
],
[
"y_dtc = classifier.predict(X_test)",
"_____no_output_____"
],
[
"classifier.score(X_test, y_test)\nprint('accuracy of training set: {:.4f}'.format(classifier.score(X_train,y_train)))\nprint('accuaracy of test set: {:.4f}'.format(classifier.score(X_test, y_test)))",
"accuracy of training set: 1.0000\naccuaracy of test set: 0.9992\n"
],
[
"classifier = DecisionTreeClassifier(max_depth = 4, random_state = 42)\nclassifier.fit(X_train,y_train)\nprint('accuracy of training set: {:.4f}'.format(classifier.score(X_train,y_train)))\nprint('accuaracy of test set: {:.4f}'.format(classifier.score(X_test, y_test)))",
"accuracy of training set: 0.9995\naccuaracy of test set: 0.9995\n"
],
[
"print('accuracy_score of decesion tree classifier: ', accuracy_score(y_dtc, y_test))\nprint('confusion_matrix of decision tree classifier: ', confusion_matrix(y_dtc, y_test))\nprint('precision_score of decision tree classifier: ', precision_score(y_dtc, y_test))\nprint('recall_score of decision tree classifier: ', recall_score(y_dtc, y_test))\nprint('precision_recall_curve of decision tree classifier: ', precision_recall_curve(y_dtc, y_test))",
"accuracy_score of decesion tree classifier: 0.9991994606893064\nconfusion_matrix of decision tree classifier: [[71048 23]\n [ 34 97]]\nprecision_score of decision tree classifier: 0.8083333333333333\nrecall_score of decision tree classifier: 0.7404580152671756\nprecision_recall_curve of decision tree classifier: (array([0.00183984, 0.80833333, 1. ]), array([1. , 0.74045802, 0. ]), array([0, 1]))\n"
]
],
[
[
"**Naive Bayes Classification**",
"_____no_output_____"
]
],
[
[
"from sklearn.naive_bayes import GaussianNB\nNBC = GaussianNB()\nNBC.fit(X_train, y_train)",
"_____no_output_____"
],
[
"y_nb = NBC.predict(X_test)",
"_____no_output_____"
],
[
"NBC.score(X_test, y_test)\nprint('accuracy of training set: {:.4f}'.format(NBC.score(X_train,y_train)))\nprint('accuaracy of test set: {:.4f}'.format(NBC.score(X_test, y_test)))",
"accuracy of training set: 0.9779\naccuaracy of test set: 0.9785\n"
],
[
"print('accuracy_score of Naive Bayes: ', accuracy_score(y_test, y_nb))\nprint('confusion_matrix of Naive Bayes: ', confusion_matrix(y_test, y_nb))\nprint('precision_score of Naive Bayes: ', precision_score(y_test, y_nb))\nprint('recall_score of Naive Bayes: ', recall_score(y_test, y_nb))\nprint('precision_recall_curve of Naive Bayes: ', precision_recall_curve(y_test, y_nb))",
"accuracy_score of Naive Bayes: 0.9784697059071374\nconfusion_matrix of Naive Bayes: [[69569 1513]\n [ 20 100]]\nprecision_score of Naive Bayes: 0.06199628022318661\nrecall_score of Naive Bayes: 0.8333333333333334\nprecision_recall_curve of Naive Bayes: (array([0.00168535, 0.06199628, 1. ]), array([1. , 0.83333333, 0. ]), array([0, 1]))\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
e79394bdde0552a2848d7310a758e1c4bb03710b | 15,006 | ipynb | Jupyter Notebook | World_health_organization_WHO_GUIDELINES_SYSTEM.ipynb | hiren14/World-health-organization-WHO-GUIDELINES-SYSTEM | c19d0f79abcc5e2686a1310ef10064f161bab700 | [
"MIT"
] | 1 | 2021-12-18T07:56:13.000Z | 2021-12-18T07:56:13.000Z | World_health_organization_WHO_GUIDELINES_SYSTEM.ipynb | hiren14/World-health-organization-WHO-GUIDELINES-SYSTEM | c19d0f79abcc5e2686a1310ef10064f161bab700 | [
"MIT"
] | null | null | null | World_health_organization_WHO_GUIDELINES_SYSTEM.ipynb | hiren14/World-health-organization-WHO-GUIDELINES-SYSTEM | c19d0f79abcc5e2686a1310ef10064f161bab700 | [
"MIT"
] | null | null | null | 55.992537 | 4,555 | 0.488338 | [
[
[
"<a href=\"https://colab.research.google.com/github/hiren14/World-health-organization-WHO-GUIDELINES-SYSTEM/blob/main/World_health_organization_WHO_GUIDELINES_SYSTEM.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"#@title\n!git clone https://github.com/hiren14/World-health-organization-WHO-GUIDELINES-SYSTEM # clone\n",
"_____no_output_____"
],
[
"%cd World-health-organization-WHO-GUIDELINES-SYSTEM\n",
"/content/World-health-organization-WHO-GUIDELINES-SYSTEM\n"
],
[
"%pip install -qr requirements.txt # install",
"\u001b[?25l\r\u001b[K |▌ | 10 kB 19.8 MB/s eta 0:00:01\r\u001b[K |█ | 20 kB 26.8 MB/s eta 0:00:01\r\u001b[K |█▋ | 30 kB 30.6 MB/s eta 0:00:01\r\u001b[K |██▏ | 40 kB 20.4 MB/s eta 0:00:01\r\u001b[K |██▊ | 51 kB 7.5 MB/s eta 0:00:01\r\u001b[K |███▎ | 61 kB 8.2 MB/s eta 0:00:01\r\u001b[K |███▉ | 71 kB 6.9 MB/s eta 0:00:01\r\u001b[K |████▍ | 81 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████ | 92 kB 7.7 MB/s eta 0:00:01\r\u001b[K |█████▌ | 102 kB 6.9 MB/s eta 0:00:01\r\u001b[K |██████ | 112 kB 6.9 MB/s eta 0:00:01\r\u001b[K |██████▋ | 122 kB 6.9 MB/s eta 0:00:01\r\u001b[K |███████▏ | 133 kB 6.9 MB/s eta 0:00:01\r\u001b[K |███████▊ | 143 kB 6.9 MB/s eta 0:00:01\r\u001b[K |████████▎ | 153 kB 6.9 MB/s eta 0:00:01\r\u001b[K |████████▉ | 163 kB 6.9 MB/s eta 0:00:01\r\u001b[K |█████████▍ | 174 kB 6.9 MB/s eta 0:00:01\r\u001b[K |██████████ | 184 kB 6.9 MB/s eta 0:00:01\r\u001b[K |██████████▍ | 194 kB 6.9 MB/s eta 0:00:01\r\u001b[K |███████████ | 204 kB 6.9 MB/s eta 0:00:01\r\u001b[K |███████████▌ | 215 kB 6.9 MB/s eta 0:00:01\r\u001b[K |████████████ | 225 kB 6.9 MB/s eta 0:00:01\r\u001b[K |████████████▋ | 235 kB 6.9 MB/s eta 0:00:01\r\u001b[K |█████████████▏ | 245 kB 6.9 MB/s eta 0:00:01\r\u001b[K |█████████████▊ | 256 kB 6.9 MB/s eta 0:00:01\r\u001b[K |██████████████▎ | 266 kB 6.9 MB/s eta 0:00:01\r\u001b[K |██████████████▉ | 276 kB 6.9 MB/s eta 0:00:01\r\u001b[K |███████████████▍ | 286 kB 6.9 MB/s eta 0:00:01\r\u001b[K |████████████████ | 296 kB 6.9 MB/s eta 0:00:01\r\u001b[K |████████████████▌ | 307 kB 6.9 MB/s eta 0:00:01\r\u001b[K |█████████████████ | 317 kB 6.9 MB/s eta 0:00:01\r\u001b[K |█████████████████▋ | 327 kB 6.9 MB/s eta 0:00:01\r\u001b[K |██████████████████▏ | 337 kB 6.9 MB/s eta 0:00:01\r\u001b[K |██████████████████▊ | 348 kB 6.9 MB/s eta 0:00:01\r\u001b[K |███████████████████▎ | 358 kB 6.9 MB/s eta 0:00:01\r\u001b[K |███████████████████▉ | 368 kB 6.9 MB/s eta 0:00:01\r\u001b[K |████████████████████▍ | 378 kB 6.9 MB/s eta 0:00:01\r\u001b[K |████████████████████▉ | 389 kB 6.9 MB/s eta 0:00:01\r\u001b[K |█████████████████████▍ | 399 kB 6.9 MB/s eta 0:00:01\r\u001b[K |██████████████████████ | 409 kB 6.9 MB/s eta 0:00:01\r\u001b[K |██████████████████████▌ | 419 kB 6.9 MB/s eta 0:00:01\r\u001b[K |███████████████████████ | 430 kB 6.9 MB/s eta 0:00:01\r\u001b[K |███████████████████████▋ | 440 kB 6.9 MB/s eta 0:00:01\r\u001b[K |████████████████████████▏ | 450 kB 6.9 MB/s eta 0:00:01\r\u001b[K |████████████████████████▊ | 460 kB 6.9 MB/s eta 0:00:01\r\u001b[K |█████████████████████████▎ | 471 kB 6.9 MB/s eta 0:00:01\r\u001b[K |█████████████████████████▉ | 481 kB 6.9 MB/s eta 0:00:01\r\u001b[K |██████████████████████████▍ | 491 kB 6.9 MB/s eta 0:00:01\r\u001b[K |███████████████████████████ | 501 kB 6.9 MB/s eta 0:00:01\r\u001b[K |███████████████████████████▌ | 512 kB 6.9 MB/s eta 0:00:01\r\u001b[K |████████████████████████████ | 522 kB 6.9 MB/s eta 0:00:01\r\u001b[K |████████████████████████████▋ | 532 kB 6.9 MB/s eta 0:00:01\r\u001b[K |█████████████████████████████▏ | 542 kB 6.9 MB/s eta 0:00:01\r\u001b[K |█████████████████████████████▊ | 552 kB 6.9 MB/s eta 0:00:01\r\u001b[K |██████████████████████████████▎ | 563 kB 6.9 MB/s eta 0:00:01\r\u001b[K |██████████████████████████████▊ | 573 kB 6.9 MB/s eta 0:00:01\r\u001b[K |███████████████████████████████▎| 583 kB 6.9 MB/s eta 0:00:01\r\u001b[K |███████████████████████████████▉| 593 kB 6.9 MB/s eta 0:00:01\r\u001b[K |████████████████████████████████| 596 kB 6.9 MB/s \n\u001b[?25h"
],
[
"#@title\n\n\nimport torch\nimport utils\ndisplay = utils.notebook_init() # checks",
"_____no_output_____"
],
[
"!python detect.py --weights yolov5s.pt --img 640 --conf 0.25 --source data/images/bus.jpg\n",
"Downloading https://ultralytics.com/assets/Arial.ttf to /root/.config/Ultralytics/Arial.ttf...\n\u001b[34m\u001b[1mdetect: \u001b[0mweights=['yolov5s.pt'], source=data/images/bus.jpg, imgsz=[640, 640], conf_thres=0.25, iou_thres=0.45, max_det=1000, device=, view_img=False, save_txt=False, save_conf=False, save_crop=False, nosave=False, classes=None, agnostic_nms=False, augment=False, visualize=False, update=False, project=runs/detect, name=exp, exist_ok=False, line_thickness=3, hide_labels=False, hide_conf=False, half=False, dnn=False\nYOLOv5 🚀 3febfe4 torch 1.10.0+cu111 CPU\n\nFusing layers... \nModel Summary: 213 layers, 7225885 parameters, 0 gradients\nimage 1/1 /content/World-health-organization-WHO-GUIDELINES-SYSTEM/data/images/bus.jpg: 640x480 4 persons, 1 bus, Done. (0.321s)\nSpeed: 3.3ms pre-process, 320.8ms inference, 11.7ms NMS per image at shape (1, 3, 640, 640)\nResults saved to \u001b[1mruns/detect/exp\u001b[0m\n"
],
[
"!python detect.py --weights yolov5s.pt --img 640 --conf 0.25 --source data/images/img1.jpg\n",
"\u001b[34m\u001b[1mdetect: \u001b[0mweights=['yolov5s.pt'], source=data/images/img1.jpg, imgsz=[640, 640], conf_thres=0.25, iou_thres=0.45, max_det=1000, device=, view_img=False, save_txt=False, save_conf=False, save_crop=False, nosave=False, classes=None, agnostic_nms=False, augment=False, visualize=False, update=False, project=runs/detect, name=exp, exist_ok=False, line_thickness=3, hide_labels=False, hide_conf=False, half=False, dnn=False\nYOLOv5 🚀 3febfe4 torch 1.10.0+cu111 CPU\n\nFusing layers... \nModel Summary: 213 layers, 7225885 parameters, 0 gradients\nimage 1/1 /content/World-health-organization-WHO-GUIDELINES-SYSTEM/data/images/img1.jpg: 448x640 8 persons, Done. (0.279s)\nSpeed: 2.5ms pre-process, 278.9ms inference, 1.2ms NMS per image at shape (1, 3, 640, 640)\nResults saved to \u001b[1mruns/detect/exp2\u001b[0m\n"
],
[
"!python detect.py --weights yolov5s.pt --img 640 --conf 0.25 --source data/images/img2.jpg\n",
"\u001b[34m\u001b[1mdetect: \u001b[0mweights=['yolov5s.pt'], source=data/images/img2.jpg, imgsz=[640, 640], conf_thres=0.25, iou_thres=0.45, max_det=1000, device=, view_img=False, save_txt=False, save_conf=False, save_crop=False, nosave=False, classes=None, agnostic_nms=False, augment=False, visualize=False, update=False, project=runs/detect, name=exp, exist_ok=False, line_thickness=3, hide_labels=False, hide_conf=False, half=False, dnn=False\nYOLOv5 🚀 3febfe4 torch 1.10.0+cu111 CPU\n\nFusing layers... \nModel Summary: 213 layers, 7225885 parameters, 0 gradients\nimage 1/1 /content/World-health-organization-WHO-GUIDELINES-SYSTEM/data/images/img2.jpg: 544x640 11 persons, Done. (0.354s)\nSpeed: 2.6ms pre-process, 353.9ms inference, 1.7ms NMS per image at shape (1, 3, 640, 640)\nResults saved to \u001b[1mruns/detect/exp3\u001b[0m\n"
],
[
"!python detect.py --weights yolov5s.pt --img 640 --conf 0.25 --source data/images/img4.jpg\n",
"\u001b[34m\u001b[1mdetect: \u001b[0mweights=['yolov5s.pt'], source=data/images/img4.jpg, imgsz=[640, 640], conf_thres=0.25, iou_thres=0.45, max_det=1000, device=, view_img=False, save_txt=False, save_conf=False, save_crop=False, nosave=False, classes=None, agnostic_nms=False, augment=False, visualize=False, update=False, project=runs/detect, name=exp, exist_ok=False, line_thickness=3, hide_labels=False, hide_conf=False, half=False, dnn=False\nYOLOv5 🚀 3febfe4 torch 1.10.0+cu111 CPU\n\nFusing layers... \nModel Summary: 213 layers, 7225885 parameters, 0 gradients\nimage 1/1 /content/World-health-organization-WHO-GUIDELINES-SYSTEM/data/images/img4.jpg: 448x640 12 persons, Done. (0.285s)\nSpeed: 2.7ms pre-process, 285.2ms inference, 1.3ms NMS per image at shape (1, 3, 640, 640)\nResults saved to \u001b[1mruns/detect/exp6\u001b[0m\n"
],
[
"!python detect.py --weights yolov5s.pt --img 640 --conf 0.25 --source data/images/img3.jpg\n",
"\u001b[34m\u001b[1mdetect: \u001b[0mweights=['yolov5s.pt'], source=data/images/img3.jpg, imgsz=[640, 640], conf_thres=0.25, iou_thres=0.45, max_det=1000, device=, view_img=False, save_txt=False, save_conf=False, save_crop=False, nosave=False, classes=None, agnostic_nms=False, augment=False, visualize=False, update=False, project=runs/detect, name=exp, exist_ok=False, line_thickness=3, hide_labels=False, hide_conf=False, half=False, dnn=False\nYOLOv5 🚀 3febfe4 torch 1.10.0+cu111 CPU\n\nFusing layers... \nModel Summary: 213 layers, 7225885 parameters, 0 gradients\nimage 1/1 /content/World-health-organization-WHO-GUIDELINES-SYSTEM/data/images/img3.jpg: 448x640 12 persons, 1 handbag, Done. (0.297s)\nSpeed: 2.7ms pre-process, 296.8ms inference, 1.3ms NMS per image at shape (1, 3, 640, 640)\nResults saved to \u001b[1mruns/detect/exp5\u001b[0m\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e79394f60d680fa0d3fbf50fa1b642926eaa9b6d | 13,708 | ipynb | Jupyter Notebook | notebooks/Wrapping Subprocesses in Asyncio.ipynb | knowsuchagency/knowsuchagency.github.io.old | 2f7b7c9d4b94120bbd427a14b66ad5dfb45c4e28 | [
"MIT"
] | null | null | null | notebooks/Wrapping Subprocesses in Asyncio.ipynb | knowsuchagency/knowsuchagency.github.io.old | 2f7b7c9d4b94120bbd427a14b66ad5dfb45c4e28 | [
"MIT"
] | 1 | 2019-12-29T09:09:06.000Z | 2019-12-29T09:09:06.000Z | notebooks/Wrapping Subprocesses in Asyncio.ipynb | knowsuchagency/knowsuchagency.github.io.old | 2f7b7c9d4b94120bbd427a14b66ad5dfb45c4e28 | [
"MIT"
] | null | null | null | 13,708 | 13,708 | 0.660417 | [
[
[
"I once had a coworker tasked with creating a web-based dashboard. Unfortunately, the data he needed to log and visualize came from this binary application that didn't have any sort of documented developer api -- it just printed everything to stdout -- that he didn't have the source code for either. \n\nIt was basically a black box that he had to write a Python wrapper around, using the [subprocess](https://docs.python.org/3/library/subprocess.html) module.\n\nHis wrapper basically worked as such:\n\n1. Run the binary in a subprocess\n2. Write an infinite loop that with each iteration attempts to... \n - capture each new line as it's printed from the subprocess\n - marshal the line into some form of structured data i.e. dictionary\n - log the information in the data structure",
"_____no_output_____"
],
[
"## A Synchronous Example",
"_____no_output_____"
]
],
[
[
"from subprocess import Popen, PIPE\nimport logging; logging.getLogger().setLevel(logging.INFO)\nimport sys\nimport time\nimport json\n\n\nPROG = \"\"\"\nimport json\nimport time\nfrom datetime import datetime\n\nwhile True:\n data = {\n 'time': datetime.now().strftime('%c %f milliseconds'),\n 'string': 'hello, world',\n }\n print(json.dumps(data))\n\"\"\"\n\nwith Popen([sys.executable, '-u', '-c', PROG], stdout=PIPE) as proc:\n last_line = ''\n start_time, delta = time.time(), 0\n \n while delta < 5: # only loop for 5 seconds\n \n line = proc.stdout.readline().decode()\n \n # pretend marshalling the data takes 1 second\n data = json.loads(line); time.sleep(1)\n \n if line != last_line:\n logging.info(data)\n \n last_line = line\n delta = time.time() - start_time\n ",
"INFO:root:{'time': 'Mon Sep 25 16:16:21 2017 690000 milliseconds', 'string': 'hello, world'}\nINFO:root:{'time': 'Mon Sep 25 16:16:21 2017 690084 milliseconds', 'string': 'hello, world'}\nINFO:root:{'time': 'Mon Sep 25 16:16:21 2017 690111 milliseconds', 'string': 'hello, world'}\nINFO:root:{'time': 'Mon Sep 25 16:16:21 2017 690131 milliseconds', 'string': 'hello, world'}\nINFO:root:{'time': 'Mon Sep 25 16:16:21 2017 690149 milliseconds', 'string': 'hello, world'}\n"
]
],
[
[
"# The problem",
"_____no_output_____"
],
[
"The problem my coworker had is that in the time he marshaled one line of output of the program and logged the information, several more lines had already been printed by the subprocess. His wrapper simply couldn't keep up with the subprocess' output.\n\nNotice in the example above, that although many more lines have obviously been printed from the program, we only capture the first few since our subprocess \"reads\" new lines more slowly than they're printed.",
"_____no_output_____"
],
[
"# The solution- asyncio\n\nInstead of writing our own infinite loop, what if we had a loop that would allow us to run a subprocess and intelligently poll it to determine when a new line was ready to be read, yielding to the main thread to do other work if not?\n\nWhat if that same event loop allowed us to delegate the process of marshaling the json output to a ProcessPoolExecutor?\n\nWhat if this event loop was written into the Python standard library? Well...",
"_____no_output_____"
],
[
"### printer.py \nThis program simply prints random stuff to stdout on an infinite loop\n\n```python\n# printer.py \n#\n# print to stdout in infinite loop\n\nfrom datetime import datetime\nfrom pathlib import Path\nfrom time import sleep\nfrom typing import List\nimport random\nimport json\nimport os\n\n\ndef get_words_from_os_dict() -> List[str]:\n p1 = Path('/usr/share/dict/words') # mac os\n p2 = Path('/usr/dict/words') # debian/ubuntu\n words: List[str] = []\n if p1.exists:\n words = p1.read_text().splitlines()\n elif p2.exists:\n words = p2.read_text().splitlines()\n return words\n\n\ndef current_time() -> str:\n return datetime.now().strftime(\"%c\")\n\n\ndef printer(words: List[str] = get_words_from_os_dict()) -> str:\n random_words = ':'.join(random.choices(words, k=random.randrange(2, 5))) if words else 'no OS words file found'\n return json.dumps({\n 'current_time': current_time(),\n 'words': random_words\n })\n\n\nwhile True:\n seconds = random.randrange(5)\n print(f'{__file__} in process {os.getpid()} waiting {seconds} seconds to print json string')\n sleep(seconds)\n print(printer())\n```",
"_____no_output_____"
],
[
"# An Asynchronous Example\n\nThis program wraps printer.py in a subprocess. It then delegates the marshaling of json to another process using the event loop's [`run_in_executor`](https://docs.python.org/3/library/asyncio-eventloop.html#asyncio.AbstractEventLoop.run_in_executor) method, and prints the results to the screen.",
"_____no_output_____"
]
],
[
[
"#!/usr/bin/env python3\n#\n# Spawns multiple instances of printer.py and attempts to deserialize the output\n# of each line in another process and print the result to the screen,\nimport typing as T\nimport asyncio.subprocess\nimport logging\nimport sys\nimport json\n\nfrom concurrent.futures import ProcessPoolExecutor, Executor\nfrom functools import partial\nfrom contextlib import contextmanager\n\n\n@contextmanager\ndef event_loop() -> asyncio.AbstractEventLoop:\n loop = asyncio.get_event_loop()\n # default asyncio event loop executor is\n # ThreadPoolExecutor which is usually fine for IO-bound\n # tasks, but bad if you need to do computation\n with ProcessPoolExecutor() as executor:\n loop.set_default_executor(executor)\n yield loop\n loop.close()\n print('\\n\\n---loop closed---\\n\\n')\n\n\n# any `async def` function is a coroutine\nasync def read_json_from_subprocess(\n loop: asyncio.AbstractEventLoop = asyncio.get_event_loop(),\n executor: T.Optional[Executor] = None\n) -> None:\n # wait for asyncio to initiate our subprocess\n process: asyncio.subprocess.Process = await create_process()\n\n while True:\n bytes_ = await process.stdout.readline()\n string = bytes_.decode('utf8')\n # deserialize_json is a function that\n # we'll send off to our executor\n deserialize_json = partial(json.loads, string)\n\n try:\n # run deserialize_json in the loop's default executor (ProcessPoolExecutor)\n # and wait for it to return\n output = await loop.run_in_executor(executor, deserialize_json)\n print(f'{process} -> {output}')\n except json.decoder.JSONDecodeError:\n logging.error('JSONDecodeError for input: ' + string.rstrip())\n\n\ndef create_process() -> asyncio.subprocess.Process:\n return asyncio.create_subprocess_exec(\n sys.executable, '-u', 'printer.py',\n stdout=asyncio.subprocess.PIPE\n )\n\n\nasync def run_for(\n n: int,\n loop: asyncio.AbstractEventLoop = asyncio.get_event_loop()\n) -> None:\n \"\"\"\n Return after a set amount of time,\n cancelling all other tasks before doing so.\n \"\"\"\n start = loop.time()\n\n while True:\n\n await asyncio.sleep(0)\n\n if abs(loop.time() - start) > n:\n # cancel all other tasks\n for task in asyncio.Task.all_tasks(loop):\n if task is not asyncio.Task.current_task():\n task.cancel()\n\n return\n\n\nwith event_loop() as loop:\n coroutines = (read_json_from_subprocess() for _ in range(5))\n # create Task from coroutines and schedule\n # it for execution on the event loop\n asyncio.gather(*coroutines) # this returns a Task and schedules it implicitly\n\n loop.run_until_complete(run_for(5))",
"_____no_output_____"
]
],
[
[
"## Conclusion\n\nIn our example, we spawn multiple instances of `printer.py` as subprocesses to get an idea of how the event loop intelligently delegates control to between multiple Tasks when it encounters an `await`.\n\nAlthough asyncio as a framework has a bit of a learning curve, in no small part due to its wonky api, (a **Task** is a **Future** that can be instantiated and scheduled with `ensure_future` or `loop.create_task` anyone?), it has many benefits in that it already has well-defined abstractions on top of common interfaces like subprocesses, file-descriptors, and sockets. That alone -- not having to write non-blocking code that knows how and when to poll those [different interfaces](https://docs.python.org/3/library/asyncio-stream.html) -- is enough to be excited about asyncio, in my opinion.\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e793b51f91b8835fc6b6f888bcee20a78462b533 | 53,065 | ipynb | Jupyter Notebook | nlq/spider/translate-spider.ipynb | huseinzol05/Malay-Dataset | e27b7617c74395c86bb5ed9f3f194b3cac2f66f6 | [
"Apache-2.0"
] | 51 | 2020-05-20T13:26:18.000Z | 2021-05-13T07:21:17.000Z | nlq/spider/translate-spider.ipynb | huseinzol05/Malaya-Dataset | c9c1917a6b1cab823aef5a73bd10e0fab0bff42d | [
"Apache-2.0"
] | 3 | 2019-02-05T11:34:38.000Z | 2020-03-19T03:18:38.000Z | nlq/spider/translate-spider.ipynb | huseinzol05/Malaya-Dataset | c9c1917a6b1cab823aef5a73bd10e0fab0bff42d | [
"Apache-2.0"
] | 21 | 2019-02-08T05:17:24.000Z | 2020-05-05T09:28:50.000Z | 71.132708 | 264 | 0.699519 | [
[
[
"import os\nimport time\n\nos.environ['LD_LIBRARY_PATH'] = '/home/husein/phantomjs-2.1.1-linux-x86_64/bin'\nos.environ['PATH'] = '/home/husein/.local/bin:/home/husein/bin:/home/husein/anaconda3/bin:/usr/local/cuda/bin:/usr/local/sbin:/usr/local/bin:/usr/sbin:/usr/bin:/sbin:/bin:/usr/games:/usr/local/games:/snap/bin:/home/husein/phantomjs-2.1.1-linux-x86_64/bin'\n\nimport translate_spider",
"_____no_output_____"
],
[
"import json\n\nwith open('spider.json') as fopen:\n data = json.load(fopen)",
"_____no_output_____"
],
[
"translators = translate_spider.Translate_Concurrent(batch_size = 20, from_lang = 'en', to_lang = 'ms')",
"_____no_output_____"
],
[
"t = translators.translate_batch(data)",
"100%|███████████████████████████████| 485/485 [28:58<00:00, 3.58s/it]\n"
],
[
"with open('spider.json.translate', 'w') as fopen:\n json.dump(t, fopen)",
"_____no_output_____"
],
[
"data = []\n\nfor i in range(len(t)):\n d = t[i][0].copy()\n d['question_bahasa'] = t[i][1]\n data.append(d)",
"_____no_output_____"
],
[
"with open('spider.json.data', 'w') as fopen:\n json.dump(data, fopen)",
"_____no_output_____"
],
[
"!cp -r spider spider-translated",
"_____no_output_____"
],
[
"!cp spider.json.data spider-translated",
"_____no_output_____"
],
[
"# !zip -r spider-translated.zip spider-translated",
" adding: spider-translated/ (stored 0%)\n adding: spider-translated/tables.json (deflated 93%)\n adding: spider-translated/database/ (stored 0%)\n adding: spider-translated/database/school_player/ (stored 0%)\n adding: spider-translated/database/school_player/schema.sql (deflated 70%)\n adding: spider-translated/database/school_player/school_player.sqlite (deflated 92%)\n adding: spider-translated/database/network_1/ (stored 0%)\n adding: spider-translated/database/network_1/schema.sql (deflated 77%)\n adding: spider-translated/database/network_1/network_1.sqlite (deflated 96%)\n adding: spider-translated/database/station_weather/ (stored 0%)\n adding: spider-translated/database/station_weather/schema.sql (deflated 76%)\n adding: spider-translated/database/station_weather/station_weather.sqlite (deflated 94%)\n adding: spider-translated/database/riding_club/ (stored 0%)\n adding: spider-translated/database/riding_club/schema.sql (deflated 64%)\n adding: spider-translated/database/riding_club/riding_club.sqlite (deflated 95%)\n adding: spider-translated/database/sports_competition/ (stored 0%)\n adding: spider-translated/database/sports_competition/schema.sql (deflated 72%)\n adding: spider-translated/database/sports_competition/sports_competition.sqlite (deflated 96%)\n adding: spider-translated/database/culture_company/ (stored 0%)\n adding: spider-translated/database/culture_company/schema.sql (deflated 64%)\n adding: spider-translated/database/culture_company/culture_company.sqlite (deflated 93%)\n adding: spider-translated/database/employee_hire_evaluation/ (stored 0%)\n adding: spider-translated/database/employee_hire_evaluation/schema.sql (deflated 66%)\n adding: spider-translated/database/employee_hire_evaluation/employee_hire_evaluation.sqlite (deflated 96%)\n adding: spider-translated/database/e_government/ (stored 0%)\n adding: spider-translated/database/e_government/schema.sql (deflated 83%)\n adding: spider-translated/database/e_government/e_government.sqlite (deflated 90%)\n adding: spider-translated/database/icfp_1/ (stored 0%)\n adding: spider-translated/database/icfp_1/icfp_1.sqlite (deflated 88%)\n adding: spider-translated/database/icfp_1/q.txt (deflated 56%)\n adding: spider-translated/database/icfp_1/link.txt (deflated 4%)\n adding: spider-translated/database/school_finance/ (stored 0%)\n adding: spider-translated/database/school_finance/schema.sql (deflated 67%)\n adding: spider-translated/database/school_finance/school_finance.sqlite (deflated 95%)\n adding: spider-translated/database/railway/ (stored 0%)\n adding: spider-translated/database/railway/schema.sql (deflated 66%)\n adding: spider-translated/database/railway/railway.sqlite (deflated 95%)\n adding: spider-translated/database/body_builder/ (stored 0%)\n adding: spider-translated/database/body_builder/schema.sql (deflated 57%)\n adding: spider-translated/database/body_builder/body_builder.sqlite (deflated 96%)\n adding: spider-translated/database/customers_campaigns_ecommerce/ (stored 0%)\n adding: spider-translated/database/customers_campaigns_ecommerce/schema.sql (deflated 85%)\n adding: spider-translated/database/customers_campaigns_ecommerce/customers_campaigns_ecommerce.sqlite (deflated 88%)\n adding: spider-translated/database/student_1/ (stored 0%)\n adding: spider-translated/database/student_1/student_1.sqlite (deflated 89%)\n adding: spider-translated/database/student_1/q.txt (deflated 65%)\n adding: spider-translated/database/student_1/student_1.sql (deflated 56%)\n adding: spider-translated/database/student_1/link.txt (stored 0%)\n adding: spider-translated/database/student_1/annotation.json (deflated 73%)\n adding: spider-translated/database/student_1/data_csv/ (stored 0%)\n adding: spider-translated/database/student_1/data_csv/list.csv (deflated 55%)\n adding: spider-translated/database/student_1/data_csv/teachers.csv (deflated 37%)\n adding: spider-translated/database/student_1/data_csv/README.STUDENTS.TXT (deflated 66%)\n adding: spider-translated/database/machine_repair/ (stored 0%)\n adding: spider-translated/database/machine_repair/machine_repair.sqlite (deflated 96%)\n adding: spider-translated/database/machine_repair/schema.sql (deflated 72%)\n adding: spider-translated/database/coffee_shop/ (stored 0%)\n adding: spider-translated/database/coffee_shop/schema.sql (deflated 71%)\n adding: spider-translated/database/coffee_shop/coffee_shop.sqlite (deflated 96%)\n adding: spider-translated/database/tracking_software_problems/ (stored 0%)\n adding: spider-translated/database/tracking_software_problems/schema.sql (deflated 86%)\n adding: spider-translated/database/tracking_software_problems/tracking_software_problems.sqlite (deflated 93%)\n adding: spider-translated/database/tracking_share_transactions/ (stored 0%)\n adding: spider-translated/database/tracking_share_transactions/schema.sql (deflated 88%)\n adding: spider-translated/database/tracking_share_transactions/tracking_share_transactions.sqlite (deflated 95%)\n adding: spider-translated/database/student_assessment/ (stored 0%)\n adding: spider-translated/database/student_assessment/schema.sql (deflated 79%)\n adding: spider-translated/database/student_assessment/student_assessment.sqlite (deflated 93%)\n adding: spider-translated/database/customers_and_addresses/ (stored 0%)\n adding: spider-translated/database/customers_and_addresses/schema.sql (deflated 83%)\n adding: spider-translated/database/customers_and_addresses/customers_and_addresses.sqlite (deflated 89%)\n adding: spider-translated/database/wta_1/ (stored 0%)\n adding: spider-translated/database/wta_1/wta_1.sql (deflated 80%)\n adding: spider-translated/database/wta_1/wta_1.sqlite (deflated 70%)\n adding: spider-translated/database/dog_kennels/ (stored 0%)\n adding: spider-translated/database/dog_kennels/schema.sql (deflated 77%)\n adding: spider-translated/database/dog_kennels/dog_kennels.sqlite (deflated 89%)\n adding: spider-translated/database/club_1/ (stored 0%)\n adding: spider-translated/database/club_1/club_1.sqlite (deflated 91%)\n adding: spider-translated/database/club_1/schema.sql (deflated 71%)\n adding: spider-translated/database/car_1/ (stored 0%)\n adding: spider-translated/database/car_1/car_1.sqlite (deflated 80%)\n adding: spider-translated/database/car_1/car_1.sql (deflated 66%)\n adding: spider-translated/database/car_1/q.txt (deflated 63%)\n adding: spider-translated/database/car_1/link.txt (stored 0%)\n adding: spider-translated/database/car_1/annotation.json (deflated 71%)\n adding: spider-translated/database/car_1/data_csv/ (stored 0%)\n adding: spider-translated/database/car_1/data_csv/car-names.csv (deflated 75%)\n adding: spider-translated/database/car_1/data_csv/car-makers.csv (deflated 43%)\n adding: spider-translated/database/car_1/data_csv/continents.csv (deflated 19%)\n adding: spider-translated/database/car_1/data_csv/cars.desc (deflated 53%)\n adding: spider-translated/database/car_1/data_csv/cars-data.csv (deflated 67%)\n adding: spider-translated/database/car_1/data_csv/countries.csv (deflated 32%)\n adding: spider-translated/database/car_1/data_csv/model-list.csv (deflated 41%)\n adding: spider-translated/database/car_1/data_csv/README.CARS.TXT (deflated 65%)\n adding: spider-translated/database/car_1/car_1.json (deflated 93%)\n adding: spider-translated/database/music_4/ (stored 0%)\n adding: spider-translated/database/music_4/schema.sql (deflated 68%)\n adding: spider-translated/database/music_4/music_4.sqlite (deflated 95%)\n adding: spider-translated/database/candidate_poll/ (stored 0%)\n adding: spider-translated/database/candidate_poll/schema.sql (deflated 62%)\n adding: spider-translated/database/candidate_poll/candidate_poll.sqlite (deflated 95%)\n adding: spider-translated/database/e_learning/ (stored 0%)\n adding: spider-translated/database/e_learning/schema.sql (deflated 79%)\n adding: spider-translated/database/e_learning/e_learning.sqlite (deflated 85%)\n adding: spider-translated/database/wrestler/ (stored 0%)\n adding: spider-translated/database/wrestler/schema.sql (deflated 66%)\n adding: spider-translated/database/wrestler/wrestler.sqlite (deflated 95%)\n adding: spider-translated/database/flight_1/ (stored 0%)\n adding: spider-translated/database/flight_1/schema.sql (deflated 82%)\n adding: spider-translated/database/flight_1/flight_1.sqlite (deflated 91%)\n adding: spider-translated/database/cre_Theme_park/ (stored 0%)\n adding: spider-translated/database/cre_Theme_park/schema.sql (deflated 86%)\n adding: spider-translated/database/cre_Theme_park/cre_Theme_park.sqlite (deflated 94%)\n adding: spider-translated/database/phone_market/ (stored 0%)\n adding: spider-translated/database/phone_market/schema.sql (deflated 67%)\n adding: spider-translated/database/phone_market/phone_market.sqlite (deflated 97%)\n adding: spider-translated/database/activity_1/ (stored 0%)\n adding: spider-translated/database/activity_1/schema.sql (deflated 82%)\n adding: spider-translated/database/activity_1/activity_1.sqlite (deflated 85%)\n adding: spider-translated/database/course_teach/ (stored 0%)\n adding: spider-translated/database/course_teach/schema.sql (deflated 69%)\n adding: spider-translated/database/course_teach/course_teach.sqlite (deflated 96%)\n adding: spider-translated/database/manufactory_1/ (stored 0%)\n adding: spider-translated/database/manufactory_1/schema.sql (deflated 68%)\n adding: spider-translated/database/manufactory_1/manufactory_1.sqlite (deflated 94%)\n adding: spider-translated/database/university_basketball/ (stored 0%)\n adding: spider-translated/database/university_basketball/schema.sql (deflated 59%)\n adding: spider-translated/database/university_basketball/university_basketball.sqlite (deflated 94%)\n adding: spider-translated/database/cre_Doc_Control_Systems/ (stored 0%)\n adding: spider-translated/database/cre_Doc_Control_Systems/schema.sql (deflated 86%)\n adding: spider-translated/database/cre_Doc_Control_Systems/cre_Doc_Control_Systems.sqlite (deflated 96%)\n adding: spider-translated/database/cinema/ (stored 0%)\n adding: spider-translated/database/cinema/schema.sql (deflated 64%)\n adding: spider-translated/database/cinema/cinema.sqlite (deflated 95%)\n adding: spider-translated/database/insurance_fnol/ (stored 0%)\n adding: spider-translated/database/insurance_fnol/schema.sql (deflated 83%)\n adding: spider-translated/database/insurance_fnol/insurance_fnol.sqlite (deflated 95%)\n adding: spider-translated/database/customer_complaints/ (stored 0%)\n adding: spider-translated/database/customer_complaints/schema.sql (deflated 80%)\n adding: spider-translated/database/customer_complaints/customer_complaints.sqlite (deflated 90%)\n adding: spider-translated/database/world_1/ (stored 0%)\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e793c1697b371b721722d69f2474500ac7f6479b | 216,494 | ipynb | Jupyter Notebook | labs/week_3.ipynb | Meewnicorn/ImPro26 | 59d3a1bc61cb2f60431d4e0a45575fcf32c6b077 | [
"MIT"
] | null | null | null | labs/week_3.ipynb | Meewnicorn/ImPro26 | 59d3a1bc61cb2f60431d4e0a45575fcf32c6b077 | [
"MIT"
] | null | null | null | labs/week_3.ipynb | Meewnicorn/ImPro26 | 59d3a1bc61cb2f60431d4e0a45575fcf32c6b077 | [
"MIT"
] | null | null | null | 534.553086 | 108,352 | 0.945929 | [
[
[
"# Week 3 Inroduction\n### Date: 21 Oct 2021\n\nLast week you learned about different methods for segmenting an image into regions of interest. In this session you will get some experience coding image segmentation algorithms. Your task will be to code a simple statistical method that uses k-means clustering.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport copy\nimport cv2\nimport matplotlib.image as mpimg\nfrom matplotlib import pyplot as plt\n%matplotlib inline #to visualize the plots within the notebook",
"UsageError: unrecognized arguments: #to visualize the plots within the notebook\n"
]
],
[
[
"# K-means Segmentation\nK-means clustering is a well-known approach for separating data (often of high dimensionality) into\ndifferent groups depending on their distance. In the case of images this is a useful method for\nsegmenting an image into regions, provided that the number of regions (k) is known in advance. It is\nbased on the fact that pixels belonging to the same region will most likely have similar intensities. \n\nThe algorithm is:\n\na) Given the number of clusters, k, initialise their centres to some values.\n\nb) Go over the pixels in the image and assign each one to its closest cluster according to its distance to the centre of the cluster.\n\nc) Update the cluster centres to be the average of the pixels added.\n\nd) Repeat steps b and c until the cluster centres do not get updated anymore.",
"_____no_output_____"
],
[
"## 1. Use the k-means function in sklearn and see results\n\nFirst, you can use the built-in kmeans function in sklearn and see the results. \nYou can figure out how to this from the specification: https://scikit-learn.org/stable/modules/generated/sklearn.cluster.KMeans.html",
"_____no_output_____"
],
[
"### Load image\n\nImportant Note: Don't forget to convert the image to float representation.",
"_____no_output_____"
]
],
[
[
"# Load image and conver to float representation\nraw_img = cv2.imread(\"../images/sample_image.jpg\") # change file name to load different images\nraw_gray_img = cv2.cvtColor(raw_img, cv2.COLOR_BGR2GRAY)\nimg = raw_img.astype(np.float32) / 255.\ngray_img = raw_gray_img.astype(np.float32) / 255.\nplt.subplot(1, 2, 1)\nplt.imshow(img)\nplt.subplot(1, 2, 2)\nplt.imshow(gray_img, \"gray\")",
"_____no_output_____"
]
],
[
[
"### Results on Gray-scale Image",
"_____no_output_____"
]
],
[
[
"from sklearn.cluster import KMeans\n# write your code here\n",
"[[0.762243 ]\n [0.28945854]]\n"
]
],
[
[
"### Results on RGB image",
"_____no_output_____"
]
],
[
[
"# write your code here\n",
"[[0.82212555 0.7523794 0.7282207 ]\n [0.2380477 0.32608324 0.22135933]]\n"
]
],
[
[
"## 2. Implement your own k-means\n\nNow you need to implement your own k-means function. Use your function on different greyscale images and try comparing the results to the results you get from sklearn kmeans function.",
"_____no_output_____"
],
[
"### Implement your own functions here:",
"_____no_output_____"
]
],
[
[
"def my_kmeans(I, k):\n \"\"\"\n Parameters\n ----------\n I: the image to be segmented (greyscale to begin with) H by W array\n k: the number of clusters (use a simple image with k=2 to begin with)\n\n Returns\n ----------\n clusters: a vector that contains the final cluster centres\n L: an array the same size as the input image that contains the label for each of the image pixels, according to which cluster it belongs\n \"\"\"\n\n assert len(I.shape) == 2, \"Wrong input dimensions! Please make sure you are using a gray-scale image!\"\n # Write your code here:\n \n return clusters, L \n\ndef my_kmeans_rgb(I, k):\n \"\"\"\n Parameters\n ----------\n I: the image to be segmented (greyscale to begin with) H by W array\n k: the number of clusters (use a simple image with k=2 to begin with)\n\n Returns\n ----------\n clusters: a vector that contains the final cluster centres\n L: an array the same size as the input image that contains the label for each of the image pixels, according to which cluster it belongs\n \"\"\"\n\n assert len(I.shape) == 3, \"Wrong input dimensions! Please make sure you are using a RGB image!\"\n # Write your code here:\n\n return clusters, L",
"_____no_output_____"
]
],
[
[
"### Show results here:",
"_____no_output_____"
]
],
[
[
"centroids, labels = my_kmeans(gray_img, 2)\nprint(centroids)\nplt.imshow(labels)",
"[0.28945825 0.76224351]\n"
]
],
[
[
"### More things to try out:\n1. Try different values for k. For k > 2 you will need some way to display the output L (other than simple black and white). Consider using a colour map with the imshow function.\n2. Adapt your function so that it will handle colour images as well. What changes do you have to make?",
"_____no_output_____"
]
],
[
[
"# k=3\ncentroids, labels = my_kmeans_vec(gray_img, 3)\nplt.imshow(labels)\nprint(centroids)",
"[0.28073347 0.62573183 0.8242718 ]\n"
],
[
"centroids, labels = my_kmeans_rgb(img, 2)\nplt.imshow(labels)\nprint(centroids)",
"[[0.23840699 0.32619616 0.22162087]\n [0.82255203 0.75285155 0.72873324]]\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
e793c30a0b641e108fa1acc7947565c60967b6b7 | 37,851 | ipynb | Jupyter Notebook | site/en/r2/tutorials/estimators/_boosted_trees_model_understanding.ipynb | ThomasTransboundaryYan/docs | 241f0d242f3461007988685365f21ee92f7e7d61 | [
"Apache-2.0"
] | null | null | null | site/en/r2/tutorials/estimators/_boosted_trees_model_understanding.ipynb | ThomasTransboundaryYan/docs | 241f0d242f3461007988685365f21ee92f7e7d61 | [
"Apache-2.0"
] | null | null | null | site/en/r2/tutorials/estimators/_boosted_trees_model_understanding.ipynb | ThomasTransboundaryYan/docs | 241f0d242f3461007988685365f21ee92f7e7d61 | [
"Apache-2.0"
] | 1 | 2020-01-23T15:10:03.000Z | 2020-01-23T15:10:03.000Z | 33.645333 | 532 | 0.517107 | [
[
[
"##### Copyright 2019 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Gradient Boosted Trees: Model understanding",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/alpha/tutorials/estimators/boosted_trees_model_understanding\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/r2/tutorials/estimators/boosted_trees_model_understanding.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs/tree/master/site/en/r2/tutorials/estimators/boosted_trees_model_understanding.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"\nFor an end-to-end walkthrough of training a Gradient Boosting model check out the [boosted trees tutorial](./boosted_trees). In this tutorial you will:\n\n* Learn how to interpret a Boosted Tree model both *locally* and *globally*\n* Gain intution for how a Boosted Trees model fits a dataset\n\n## How to interpret Boosted Trees models both locally and globally\n\nLocal interpretability refers to an understanding of a model’s predictions at the individual example level, while global interpretability refers to an understanding of the model as a whole. Such techniques can help machine learning (ML) practitioners detect bias and bugs during the model development stage.\n\nFor local interpretability, you will learn how to create and visualize per-instance contributions. To distinguish this from feature importances, we refer to these values as directional feature contributions (DFCs).\n\nFor global interpretability you will retrieve and visualize gain-based feature importances, [permutation feature importances](https://www.stat.berkeley.edu/~breiman/randomforest2001.pdf) and also show aggregated DFCs.",
"_____no_output_____"
],
[
"## Load the titanic dataset\nYou will be using the titanic dataset, where the (rather morbid) goal is to predict passenger survival, given characteristics such as gender, age, class, etc.",
"_____no_output_____"
]
],
[
[
"from __future__ import absolute_import, division, print_function, unicode_literals\n\nimport numpy as np\nimport pandas as pd\nfrom IPython.display import clear_output\n\n# Load dataset.\ndftrain = pd.read_csv('https://storage.googleapis.com/tf-datasets/titanic/train.csv')\ndfeval = pd.read_csv('https://storage.googleapis.com/tf-datasets/titanic/eval.csv')\ny_train = dftrain.pop('survived')\ny_eval = dfeval.pop('survived')",
"_____no_output_____"
],
[
"!pip install tensorflow==2.0.0-alpha0\n\nimport tensorflow as tf\ntf.random.set_seed(123)",
"_____no_output_____"
]
],
[
[
"For a description of the features, please review the prior tutorial.",
"_____no_output_____"
],
[
"## Create feature columns, input_fn, and the train the estimator",
"_____no_output_____"
],
[
"### Preprocess the data",
"_____no_output_____"
],
[
"Create the feature columns, using the original numeric columns as is and one-hot-encoding categorical variables.",
"_____no_output_____"
]
],
[
[
"fc = tf.feature_column\nCATEGORICAL_COLUMNS = ['sex', 'n_siblings_spouses', 'parch', 'class', 'deck',\n 'embark_town', 'alone']\nNUMERIC_COLUMNS = ['age', 'fare']\n\ndef one_hot_cat_column(feature_name, vocab):\n return fc.indicator_column(\n fc.categorical_column_with_vocabulary_list(feature_name,\n vocab))\nfeature_columns = []\nfor feature_name in CATEGORICAL_COLUMNS:\n # Need to one-hot encode categorical features.\n vocabulary = dftrain[feature_name].unique()\n feature_columns.append(one_hot_cat_column(feature_name, vocabulary))\n\nfor feature_name in NUMERIC_COLUMNS:\n feature_columns.append(fc.numeric_column(feature_name,\n dtype=tf.float32))",
"_____no_output_____"
]
],
[
[
"### Build the input pipeline",
"_____no_output_____"
],
[
"Create the input functions using the `from_tensor_slices` method in the [`tf.data`](https://www.tensorflow.org/api_docs/python/tf/data) API to read in data directly from Pandas.",
"_____no_output_____"
]
],
[
[
"# Use entire batch since this is such a small dataset.\nNUM_EXAMPLES = len(y_train)\n\ndef make_input_fn(X, y, n_epochs=None, shuffle=True):\n def input_fn():\n dataset = tf.data.Dataset.from_tensor_slices((X.to_dict(orient='list'), y))\n if shuffle:\n dataset = dataset.shuffle(NUM_EXAMPLES)\n # For training, cycle thru dataset as many times as need (n_epochs=None).\n dataset = (dataset\n .repeat(n_epochs)\n .batch(NUM_EXAMPLES))\n return dataset\n return input_fn\n\n# Training and evaluation input functions.\ntrain_input_fn = make_input_fn(dftrain, y_train)\neval_input_fn = make_input_fn(dfeval, y_eval, shuffle=False, n_epochs=1)",
"_____no_output_____"
]
],
[
[
"### Train the model",
"_____no_output_____"
]
],
[
[
"params = {\n 'n_trees': 50,\n 'max_depth': 3,\n 'n_batches_per_layer': 1,\n # You must enable center_bias = True to get DFCs. This will force the model to\n # make an initial prediction before using any features (e.g. use the mean of\n # the training labels for regression or log odds for classification when\n # using cross entropy loss).\n 'center_bias': True\n}\n\nest = tf.estimator.BoostedTreesClassifier(feature_columns, **params)\n# Train model.\nest.train(train_input_fn, max_steps=100)\n\n# Evaluation.\nresults = est.evaluate(eval_input_fn)\nclear_output()\npd.Series(results).to_frame()",
"_____no_output_____"
]
],
[
[
"For performance reasons, when your data fits in memory, we recommend use the `boosted_trees_classifier_train_in_memory` function. However if training time is not of a concern or if you have a very large dataset and want to do distributed training, use the `tf.estimator.BoostedTrees` API shown above.\n\n\nWhen using this method, you should not batch your input data, as the method operates on the entire dataset.\n",
"_____no_output_____"
]
],
[
[
"in_memory_params = dict(params)\nin_memory_params['n_batches_per_layer'] = 1\n# In-memory input_fn does not use batching.\ndef make_inmemory_train_input_fn(X, y):\n def input_fn():\n return dict(X), y\n return input_fn\ntrain_input_fn = make_inmemory_train_input_fn(dftrain, y_train)\n\n# Train the model.\nest = tf.estimator.BoostedTreesClassifier(\n feature_columns, \n train_in_memory=True, \n **in_memory_params)\n\nest.train(train_input_fn)\nprint(est.evaluate(eval_input_fn))",
"_____no_output_____"
]
],
[
[
"## Model interpretation and plotting",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport seaborn as sns\nsns_colors = sns.color_palette('colorblind')",
"_____no_output_____"
]
],
[
[
"## Local interpretability\nNext you will output the directional feature contributions (DFCs) to explain individual predictions using the approach outlined in [Palczewska et al](https://arxiv.org/pdf/1312.1121.pdf) and by Saabas in [Interpreting Random Forests](http://blog.datadive.net/interpreting-random-forests/) (this method is also available in scikit-learn for Random Forests in the [`treeinterpreter`](https://github.com/andosa/treeinterpreter) package). The DFCs are generated with:\n\n`pred_dicts = list(est.experimental_predict_with_explanations(pred_input_fn))`\n\n(Note: The method is named experimental as we may modify the API before dropping the experimental prefix.)",
"_____no_output_____"
]
],
[
[
"pred_dicts = list(est.experimental_predict_with_explanations(eval_input_fn))",
"_____no_output_____"
],
[
"# Create DFC Pandas dataframe.\nlabels = y_eval.values\nprobs = pd.Series([pred['probabilities'][1] for pred in pred_dicts])\ndf_dfc = pd.DataFrame([pred['dfc'] for pred in pred_dicts])\ndf_dfc.describe().T",
"_____no_output_____"
]
],
[
[
"A nice property of DFCs is that the sum of the contributions + the bias is equal to the prediction for a given example.",
"_____no_output_____"
]
],
[
[
"# Sum of DFCs + bias == probabality.\nbias = pred_dicts[0]['bias']\ndfc_prob = df_dfc.sum(axis=1) + bias\nnp.testing.assert_almost_equal(dfc_prob.values,\n probs.values)",
"_____no_output_____"
]
],
[
[
"Plot DFCs for an individual passenger. Let's make the plot nice by color coding based on the contributions' directionality and add the feature values on figure.",
"_____no_output_____"
]
],
[
[
"# Boilerplate code for plotting :)\ndef _get_color(value):\n \"\"\"To make positive DFCs plot green, negative DFCs plot red.\"\"\"\n green, red = sns.color_palette()[2:4]\n if value >= 0: return green\n return red\n\ndef _add_feature_values(feature_values, ax):\n \"\"\"Display feature's values on left of plot.\"\"\"\n x_coord = ax.get_xlim()[0]\n OFFSET = 0.15\n for y_coord, (feat_name, feat_val) in enumerate(feature_values.items()):\n t = plt.text(x_coord, y_coord - OFFSET, '{}'.format(feat_val), size=12)\n t.set_bbox(dict(facecolor='white', alpha=0.5))\n from matplotlib.font_manager import FontProperties\n font = FontProperties()\n font.set_weight('bold')\n t = plt.text(x_coord, y_coord + 1 - OFFSET, 'feature\\nvalue',\n fontproperties=font, size=12)\n\ndef plot_example(example):\n TOP_N = 8 # View top 8 features.\n sorted_ix = example.abs().sort_values()[-TOP_N:].index # Sort by magnitude.\n example = example[sorted_ix]\n colors = example.map(_get_color).tolist()\n ax = example.to_frame().plot(kind='barh',\n color=[colors],\n legend=None,\n alpha=0.75,\n figsize=(10,6))\n ax.grid(False, axis='y')\n ax.set_yticklabels(ax.get_yticklabels(), size=14)\n\n # Add feature values.\n _add_feature_values(dfeval.iloc[ID][sorted_ix], ax)\n return ax",
"_____no_output_____"
],
[
"# Plot results.\nID = 182\nexample = df_dfc.iloc[ID] # Choose ith example from evaluation set.\nTOP_N = 8 # View top 8 features.\nsorted_ix = example.abs().sort_values()[-TOP_N:].index\nax = plot_example(example)\nax.set_title('Feature contributions for example {}\\n pred: {:1.2f}; label: {}'.format(ID, probs[ID], labels[ID]))\nax.set_xlabel('Contribution to predicted probability', size=14)\nplt.show()",
"_____no_output_____"
]
],
[
[
"The larger magnitude contributions have a larger impact on the model's prediction. Negative contributions indicate the feature value for this given example reduced the model's prediction, while positive values contribute an increase in the prediction.",
"_____no_output_____"
],
[
"You can also plot the example's DFCs compare with the entire distribution using a voilin plot.",
"_____no_output_____"
]
],
[
[
"# Boilerplate plotting code.\ndef dist_violin_plot(df_dfc, ID):\n # Initialize plot.\n fig, ax = plt.subplots(1, 1, figsize=(10, 6))\n\n # Create example dataframe.\n TOP_N = 8 # View top 8 features.\n example = df_dfc.iloc[ID]\n ix = example.abs().sort_values()[-TOP_N:].index\n example = example[ix]\n example_df = example.to_frame(name='dfc')\n\n # Add contributions of entire distribution.\n parts=ax.violinplot([df_dfc[w] for w in ix],\n vert=False,\n showextrema=False,\n widths=0.7,\n positions=np.arange(len(ix)))\n face_color = sns_colors[0]\n alpha = 0.15\n for pc in parts['bodies']:\n pc.set_facecolor(face_color)\n pc.set_alpha(alpha)\n\n # Add feature values.\n _add_feature_values(dfeval.iloc[ID][sorted_ix], ax)\n\n # Add local contributions.\n ax.scatter(example,\n np.arange(example.shape[0]),\n color=sns.color_palette()[2],\n s=100,\n marker=\"s\",\n label='contributions for example')\n\n # Legend\n # Proxy plot, to show violinplot dist on legend.\n ax.plot([0,0], [1,1], label='eval set contributions\\ndistributions',\n color=face_color, alpha=alpha, linewidth=10)\n legend = ax.legend(loc='lower right', shadow=True, fontsize='x-large',\n frameon=True)\n legend.get_frame().set_facecolor('white')\n\n # Format plot.\n ax.set_yticks(np.arange(example.shape[0]))\n ax.set_yticklabels(example.index)\n ax.grid(False, axis='y')\n ax.set_xlabel('Contribution to predicted probability', size=14)",
"_____no_output_____"
]
],
[
[
"Plot this example.",
"_____no_output_____"
]
],
[
[
"dist_violin_plot(df_dfc, ID)\nplt.title('Feature contributions for example {}\\n pred: {:1.2f}; label: {}'.format(ID, probs[ID], labels[ID]))\nplt.show()",
"_____no_output_____"
]
],
[
[
"Finally, third-party tools, such as [LIME](https://github.com/marcotcr/lime) and [shap](https://github.com/slundberg/shap), can also help understand individual predictions for a model.",
"_____no_output_____"
],
[
"## Global feature importances\n\nAdditionally, you might want to understand the model as a whole, rather than studying individual predictions. Below, you will compute and use:\n\n1. Gain-based feature importances using `est.experimental_feature_importances`\n2. Permutation importances\n3. Aggregate DFCs using `est.experimental_predict_with_explanations`\n\nGain-based feature importances measure the loss change when splitting on a particular feature, while permutation feature importances are computed by evaluating model performance on the evaluation set by shuffling each feature one-by-one and attributing the change in model performance to the shuffled feature.\n\nIn general, permutation feature importance are preferred to gain-based feature importance, though both methods can be unreliable in situations where potential predictor variables vary in their scale of measurement or their number of categories and when features are correlated ([source](https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-9-307)). Check out [this article](http://explained.ai/rf-importance/index.html) for an in-depth overview and great discussion on different feature importance types.",
"_____no_output_____"
],
[
"### 1. Gain-based feature importances",
"_____no_output_____"
],
[
"Gain-based feature importances are built into the TensorFlow Boosted Trees estimators using `est.experimental_feature_importances`.",
"_____no_output_____"
]
],
[
[
"importances = est.experimental_feature_importances(normalize=True)\ndf_imp = pd.Series(importances)\n\n# Visualize importances.\nN = 8\nax = (df_imp.iloc[0:N][::-1]\n .plot(kind='barh',\n color=sns_colors[0],\n title='Gain feature importances',\n figsize=(10, 6)))\nax.grid(False, axis='y')",
"_____no_output_____"
]
],
[
[
"### 2. Average absolute DFCs\nYou can also average the absolute values of DFCs to understand impact at a global level.",
"_____no_output_____"
]
],
[
[
"# Plot.\ndfc_mean = df_dfc.abs().mean()\nN = 8\nsorted_ix = dfc_mean.abs().sort_values()[-N:].index # Average and sort by absolute.\nax = dfc_mean[sorted_ix].plot(kind='barh',\n color=sns_colors[1],\n title='Mean |directional feature contributions|',\n figsize=(10, 6))\nax.grid(False, axis='y')",
"_____no_output_____"
]
],
[
[
"You can also see how DFCs vary as a feature value varies.",
"_____no_output_____"
]
],
[
[
"FEATURE = 'fare'\nfeature = pd.Series(df_dfc[FEATURE].values, index=dfeval[FEATURE].values).sort_index()\nax = sns.regplot(feature.index.values, feature.values, lowess=True)\nax.set_ylabel('contribution')\nax.set_xlabel(FEATURE)\nax.set_xlim(0, 100)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### 3. Permutation feature importance",
"_____no_output_____"
]
],
[
[
"def permutation_importances(est, X_eval, y_eval, metric, features):\n \"\"\"Column by column, shuffle values and observe effect on eval set.\n\n source: http://explained.ai/rf-importance/index.html\n A similar approach can be done during training. See \"Drop-column importance\"\n in the above article.\"\"\"\n baseline = metric(est, X_eval, y_eval)\n imp = []\n for col in features:\n save = X_eval[col].copy()\n X_eval[col] = np.random.permutation(X_eval[col])\n m = metric(est, X_eval, y_eval)\n X_eval[col] = save\n imp.append(baseline - m)\n return np.array(imp)\n\ndef accuracy_metric(est, X, y):\n \"\"\"TensorFlow estimator accuracy.\"\"\"\n eval_input_fn = make_input_fn(X,\n y=y,\n shuffle=False,\n n_epochs=1)\n return est.evaluate(input_fn=eval_input_fn)['accuracy']\nfeatures = CATEGORICAL_COLUMNS + NUMERIC_COLUMNS\nimportances = permutation_importances(est, dfeval, y_eval, accuracy_metric,\n features)\ndf_imp = pd.Series(importances, index=features)\n\nsorted_ix = df_imp.abs().sort_values().index\nax = df_imp[sorted_ix][-5:].plot(kind='barh', color=sns_colors[2], figsize=(10, 6))\nax.grid(False, axis='y')\nax.set_title('Permutation feature importance')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Visualizing model fitting",
"_____no_output_____"
],
[
"Lets first simulate/create training data using the following formula:\n\n\n$$z=x* e^{-x^2 - y^2}$$\n\n\nWhere \\(z\\) is the dependent variable you are trying to predict and \\(x\\) and \\(y\\) are the features.",
"_____no_output_____"
]
],
[
[
"from numpy.random import uniform, seed\nfrom matplotlib.mlab import griddata\n\n# Create fake data\nseed(0)\nnpts = 5000\nx = uniform(-2, 2, npts)\ny = uniform(-2, 2, npts)\nz = x*np.exp(-x**2 - y**2)",
"_____no_output_____"
],
[
"# Prep data for training.\ndf = pd.DataFrame({'x': x, 'y': y, 'z': z})\n\nxi = np.linspace(-2.0, 2.0, 200),\nyi = np.linspace(-2.1, 2.1, 210),\nxi,yi = np.meshgrid(xi, yi)\n\ndf_predict = pd.DataFrame({\n 'x' : xi.flatten(),\n 'y' : yi.flatten(),\n})\npredict_shape = xi.shape",
"_____no_output_____"
],
[
"def plot_contour(x, y, z, **kwargs):\n # Grid the data.\n plt.figure(figsize=(10, 8))\n # Contour the gridded data, plotting dots at the nonuniform data points.\n CS = plt.contour(x, y, z, 15, linewidths=0.5, colors='k')\n CS = plt.contourf(x, y, z, 15,\n vmax=abs(zi).max(), vmin=-abs(zi).max(), cmap='RdBu_r')\n plt.colorbar() # Draw colorbar.\n # Plot data points.\n plt.xlim(-2, 2)\n plt.ylim(-2, 2)",
"_____no_output_____"
]
],
[
[
"You can visualize the function. Redder colors correspond to larger function values.",
"_____no_output_____"
]
],
[
[
"zi = griddata(x, y, z, xi, yi, interp='linear')\nplot_contour(xi, yi, zi)\nplt.scatter(df.x, df.y, marker='.')\nplt.title('Contour on training data')\nplt.show()",
"_____no_output_____"
],
[
"fc = [tf.feature_column.numeric_column('x'),\n tf.feature_column.numeric_column('y')]",
"_____no_output_____"
],
[
"def predict(est):\n \"\"\"Predictions from a given estimator.\"\"\"\n predict_input_fn = lambda: tf.data.Dataset.from_tensors(dict(df_predict))\n preds = np.array([p['predictions'][0] for p in est.predict(predict_input_fn)])\n return preds.reshape(predict_shape)",
"_____no_output_____"
]
],
[
[
"First let's try to fit a linear model to the data.",
"_____no_output_____"
]
],
[
[
"train_input_fn = make_input_fn(df, df.z)\nest = tf.estimator.LinearRegressor(fc)\nest.train(train_input_fn, max_steps=500);",
"_____no_output_____"
],
[
"plot_contour(xi, yi, predict(est))",
"_____no_output_____"
]
],
[
[
"It's not a very good fit. Next let's try to fit a GBDT model to it and try to understand how the model fits the function.",
"_____no_output_____"
]
],
[
[
"n_trees = 22 #@param {type: \"slider\", min: 1, max: 80, step: 1}\n\nest = tf.estimator.BoostedTreesRegressor(fc, n_batches_per_layer=1, n_trees=n_trees)\nest.train(train_input_fn, max_steps=500)\nclear_output()\nplot_contour(xi, yi, predict(est))\nplt.text(-1.8, 2.1, '# trees: {}'.format(n_trees), color='w', backgroundcolor='black', size=20)\nplt.show()",
"_____no_output_____"
]
],
[
[
"As you increase the number of trees, the model's predictions better approximates the underlying function.",
"_____no_output_____"
],
[
"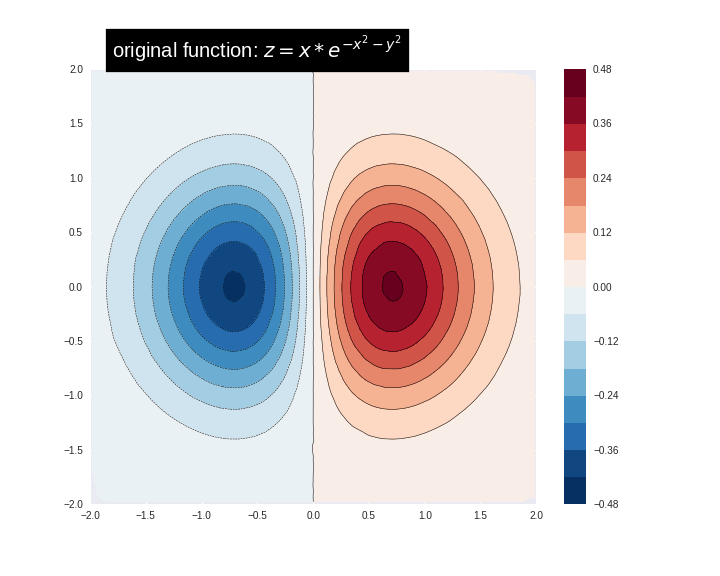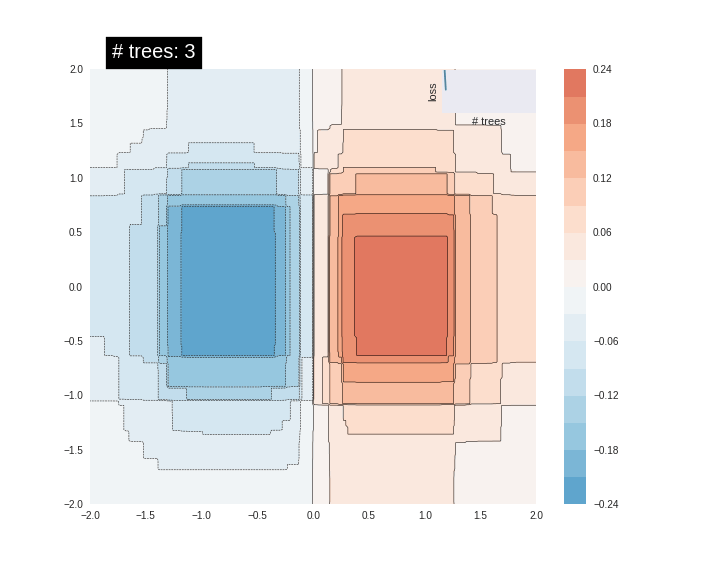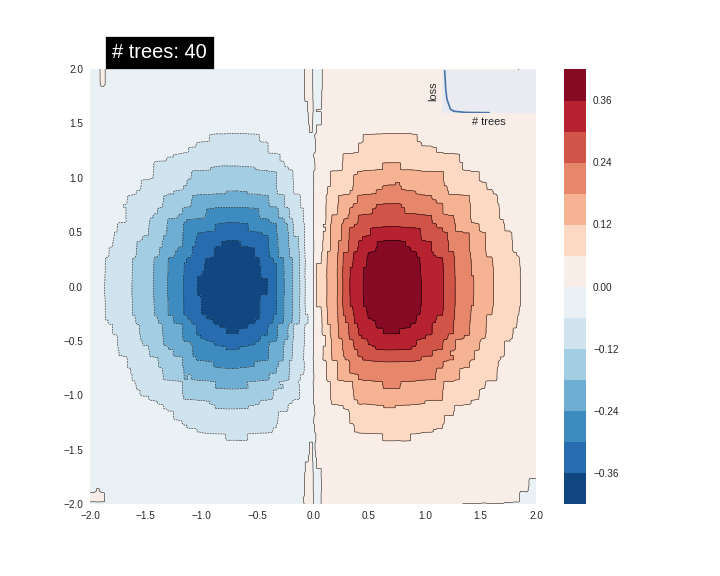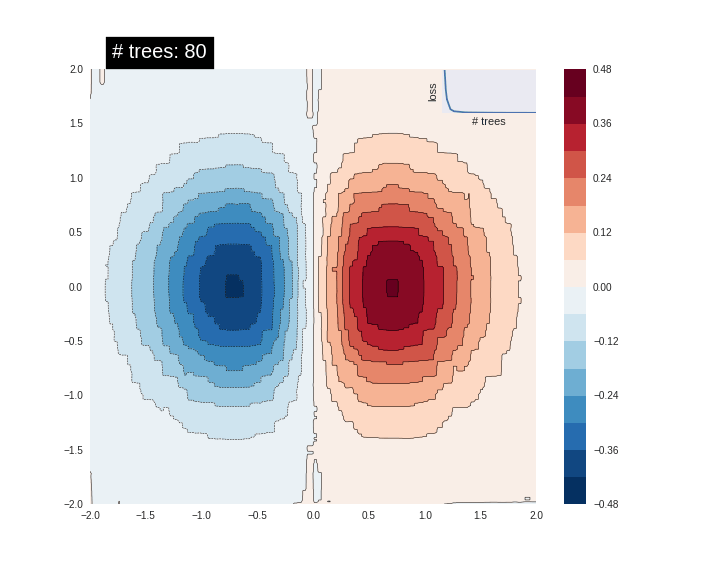",
"_____no_output_____"
],
[
"# Conclusion",
"_____no_output_____"
],
[
"In this tutorial you learned how to interpret Boosted Trees models using directional feature contributions and feature importance techniques. These techniques provide insight into how the features impact a model's predictions. Finally, you also gained intution for how a Boosted Tree model fits a complex function by viewing the decision surface for several models.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
e793c8163c9a6480d44e9aaf22137a3ec5a01cdc | 899,409 | ipynb | Jupyter Notebook | 1. Load and Visualize Data.ipynb | Buddhone/P1_Facial_Keypoints | bbf3608f9ce495d892bfaf71fbb05cf972f6292d | [
"MIT"
] | null | null | null | 1. Load and Visualize Data.ipynb | Buddhone/P1_Facial_Keypoints | bbf3608f9ce495d892bfaf71fbb05cf972f6292d | [
"MIT"
] | null | null | null | 1. Load and Visualize Data.ipynb | Buddhone/P1_Facial_Keypoints | bbf3608f9ce495d892bfaf71fbb05cf972f6292d | [
"MIT"
] | null | null | null | 135.269815 | 158,472 | 0.832329 | [
[
[
"# Facial Keypoint Detection\n \nThis project will be all about defining and training a convolutional neural network to perform facial keypoint detection, and using computer vision techniques to transform images of faces. The first step in any challenge like this will be to load and visualize the data you'll be working with. \n\nLet's take a look at some examples of images and corresponding facial keypoints.\n\n<img src='images/key_pts_example.png' width=50% height=50%/>\n\nFacial keypoints (also called facial landmarks) are the small magenta dots shown on each of the faces in the image above. In each training and test image, there is a single face and **68 keypoints, with coordinates (x, y), for that face**. These keypoints mark important areas of the face: the eyes, corners of the mouth, the nose, etc. These keypoints are relevant for a variety of tasks, such as face filters, emotion recognition, pose recognition, and so on. Here they are, numbered, and you can see that specific ranges of points match different portions of the face.\n\n<img src='images/landmarks_numbered.jpg' width=30% height=30%/>\n\n---",
"_____no_output_____"
],
[
"## Load and Visualize Data\n\nThe first step in working with any dataset is to become familiar with your data; you'll need to load in the images of faces and their keypoints and visualize them! This set of image data has been extracted from the [YouTube Faces Dataset](https://www.cs.tau.ac.il/~wolf/ytfaces/), which includes videos of people in YouTube videos. These videos have been fed through some processing steps and turned into sets of image frames containing one face and the associated keypoints.\n\n#### Training and Testing Data\n\nThis facial keypoints dataset consists of 5770 color images. All of these images are separated into either a training or a test set of data.\n\n* 3462 of these images are training images, for you to use as you create a model to predict keypoints.\n* 2308 are test images, which will be used to test the accuracy of your model.\n\nThe information about the images and keypoints in this dataset are summarized in CSV files, which we can read in using `pandas`. Let's read the training CSV and get the annotations in an (N, 2) array where N is the number of keypoints and 2 is the dimension of the keypoint coordinates (x, y).\n\n---",
"_____no_output_____"
],
[
"First, before we do anything, we have to load in our image data. This data is stored in a zip file and in the below cell, we access it by it's URL and unzip the data in a `/data/` directory that is separate from the workspace home directory.",
"_____no_output_____"
]
],
[
[
"# -- DO NOT CHANGE THIS CELL -- #\n!mkdir /data\n!wget -P /data/ https://s3.amazonaws.com/video.udacity-data.com/topher/2018/May/5aea1b91_train-test-data/train-test-data.zip\n!unzip -n /data/train-test-data.zip -d /data",
"--2018-12-09 17:57:00-- https://s3.amazonaws.com/video.udacity-data.com/topher/2018/May/5aea1b91_train-test-data/train-test-data.zip\nResolving s3.amazonaws.com (s3.amazonaws.com)... 52.216.129.149\nConnecting to s3.amazonaws.com (s3.amazonaws.com)|52.216.129.149|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 338613624 (323M) [application/zip]\nSaving to: ‘/data/train-test-data.zip’\n\ntrain-test-data.zip 100%[===================>] 322.93M 81.2MB/s in 4.1s \n\n2018-12-09 17:57:04 (78.8 MB/s) - ‘/data/train-test-data.zip’ saved [338613624/338613624]\n\nArchive: /data/train-test-data.zip\n creating: /data/test/\n inflating: /data/test/Abdel_Aziz_Al-Hakim_00.jpg \n inflating: /data/test/Abdel_Aziz_Al-Hakim_01.jpg \n inflating: /data/test/Abdel_Aziz_Al-Hakim_10.jpg \n inflating: /data/test/Abdel_Aziz_Al-Hakim_11.jpg \n inflating: /data/test/Abdel_Aziz_Al-Hakim_40.jpg \n inflating: /data/test/Abdel_Aziz_Al-Hakim_41.jpg \n inflating: /data/test/Abdullah_Gul_10.jpg \n inflating: /data/test/Abdullah_Gul_11.jpg \n inflating: /data/test/Abdullah_Gul_30.jpg \n inflating: /data/test/Abdullah_Gul_31.jpg \n inflating: /data/test/Abdullah_Gul_50.jpg \n inflating: /data/test/Abdullah_Gul_51.jpg \n inflating: /data/test/Adam_Sandler_00.jpg \n inflating: /data/test/Adam_Sandler_01.jpg \n inflating: /data/test/Adam_Sandler_10.jpg \n inflating: /data/test/Adam_Sandler_11.jpg \n inflating: /data/test/Adam_Sandler_40.jpg \n inflating: /data/test/Adam_Sandler_41.jpg \n inflating: /data/test/Adrian_Nastase_10.jpg \n inflating: /data/test/Adrian_Nastase_11.jpg \n inflating: /data/test/Adrian_Nastase_40.jpg \n inflating: /data/test/Adrian_Nastase_41.jpg \n inflating: /data/test/Adrian_Nastase_50.jpg \n inflating: /data/test/Adrian_Nastase_51.jpg \n inflating: /data/test/Agbani_Darego_00.jpg \n inflating: /data/test/Agbani_Darego_01.jpg \n inflating: /data/test/Agbani_Darego_20.jpg \n inflating: /data/test/Agbani_Darego_21.jpg \n inflating: /data/test/Agbani_Darego_40.jpg \n inflating: /data/test/Agbani_Darego_41.jpg \n inflating: /data/test/Agbani_Darego_50.jpg \n inflating: /data/test/Agbani_Darego_51.jpg \n inflating: /data/test/Agnes_Bruckner_00.jpg \n inflating: /data/test/Agnes_Bruckner_01.jpg \n inflating: /data/test/Agnes_Bruckner_10.jpg \n inflating: /data/test/Agnes_Bruckner_11.jpg \n inflating: /data/test/Agnes_Bruckner_20.jpg \n inflating: /data/test/Agnes_Bruckner_21.jpg \n inflating: /data/test/Agnes_Bruckner_40.jpg \n inflating: /data/test/Agnes_Bruckner_41.jpg \n inflating: /data/test/Ahmad_Masood_00.jpg \n inflating: /data/test/Ahmad_Masood_01.jpg \n inflating: /data/test/Ahmad_Masood_30.jpg \n inflating: /data/test/Ahmad_Masood_31.jpg \n inflating: /data/test/Ahmad_Masood_40.jpg \n inflating: /data/test/Ahmad_Masood_41.jpg \n inflating: /data/test/Ahmed_Ahmed_00.jpg \n inflating: /data/test/Ahmed_Ahmed_01.jpg \n inflating: /data/test/Ahmed_Ahmed_10.jpg \n inflating: /data/test/Ahmed_Ahmed_11.jpg \n inflating: /data/test/Ahmed_Ahmed_40.jpg \n inflating: /data/test/Ahmed_Ahmed_41.jpg \n inflating: /data/test/Ahmed_Ahmed_50.jpg \n inflating: /data/test/Ahmed_Ahmed_51.jpg \n inflating: /data/test/Aidan_Quinn_00.jpg \n inflating: /data/test/Aidan_Quinn_01.jpg \n inflating: /data/test/Aidan_Quinn_10.jpg \n inflating: /data/test/Aidan_Quinn_11.jpg \n inflating: /data/test/Aidan_Quinn_20.jpg \n inflating: /data/test/Aidan_Quinn_21.jpg \n inflating: /data/test/Aidan_Quinn_30.jpg \n inflating: /data/test/Aidan_Quinn_31.jpg \n inflating: /data/test/Aishwarya_Rai_00.jpg \n inflating: /data/test/Aishwarya_Rai_01.jpg \n inflating: /data/test/Aishwarya_Rai_10.jpg \n inflating: /data/test/Aishwarya_Rai_11.jpg \n inflating: /data/test/Aishwarya_Rai_40.jpg \n inflating: /data/test/Aishwarya_Rai_41.jpg \n inflating: /data/test/Aishwarya_Rai_50.jpg \n inflating: /data/test/Aishwarya_Rai_51.jpg \n inflating: /data/test/Albert_Brooks_00.jpg \n inflating: /data/test/Albert_Brooks_01.jpg \n inflating: /data/test/Albert_Brooks_10.jpg \n inflating: /data/test/Albert_Brooks_11.jpg \n inflating: /data/test/Albert_Brooks_30.jpg \n inflating: /data/test/Albert_Brooks_31.jpg \n inflating: /data/test/Alejandro_Toledo_10.jpg \n inflating: /data/test/Alejandro_Toledo_11.jpg \n inflating: /data/test/Alejandro_Toledo_30.jpg \n inflating: /data/test/Alejandro_Toledo_31.jpg \n inflating: /data/test/Alejandro_Toledo_50.jpg \n inflating: /data/test/Alejandro_Toledo_51.jpg \n inflating: /data/test/Aleksander_Kwasniewski_00.jpg \n inflating: /data/test/Aleksander_Kwasniewski_01.jpg \n inflating: /data/test/Aleksander_Kwasniewski_10.jpg \n inflating: /data/test/Aleksander_Kwasniewski_11.jpg \n inflating: /data/test/Aleksander_Kwasniewski_20.jpg \n inflating: /data/test/Aleksander_Kwasniewski_21.jpg \n inflating: /data/test/Aleksander_Kwasniewski_30.jpg \n inflating: /data/test/Aleksander_Kwasniewski_31.jpg \n inflating: /data/test/Alex_Ferguson_00.jpg \n inflating: /data/test/Alex_Ferguson_01.jpg \n inflating: /data/test/Alex_Ferguson_10.jpg \n inflating: /data/test/Alex_Ferguson_11.jpg \n inflating: /data/test/Alex_Ferguson_50.jpg \n inflating: /data/test/Alex_Ferguson_51.jpg \n inflating: /data/test/Alexandra_Pelosi_00.jpg \n inflating: /data/test/Alexandra_Pelosi_01.jpg \n inflating: /data/test/Alexandra_Pelosi_10.jpg \n inflating: /data/test/Alexandra_Pelosi_11.jpg \n inflating: /data/test/Alexandra_Pelosi_30.jpg \n inflating: /data/test/Alexandra_Pelosi_31.jpg \n inflating: /data/test/Alfredo_di_Stefano_00.jpg \n inflating: /data/test/Alfredo_di_Stefano_01.jpg \n inflating: /data/test/Alfredo_di_Stefano_20.jpg \n inflating: /data/test/Alfredo_di_Stefano_21.jpg \n inflating: /data/test/Alfredo_di_Stefano_50.jpg \n inflating: /data/test/Alfredo_di_Stefano_51.jpg \n inflating: /data/test/Ali_Abbas_20.jpg \n inflating: /data/test/Ali_Abbas_21.jpg \n inflating: /data/test/Ali_Abbas_30.jpg \n inflating: /data/test/Ali_Abbas_31.jpg \n inflating: /data/test/Ali_Abbas_40.jpg \n inflating: /data/test/Ali_Abbas_41.jpg \n inflating: /data/test/Ali_Abbas_50.jpg \n inflating: /data/test/Ali_Abbas_51.jpg \n inflating: /data/test/Alicia_Silverstone_00.jpg \n inflating: /data/test/Alicia_Silverstone_01.jpg \n inflating: /data/test/Alicia_Silverstone_10.jpg \n inflating: /data/test/Alicia_Silverstone_11.jpg \n inflating: /data/test/Alicia_Silverstone_20.jpg \n inflating: /data/test/Alicia_Silverstone_21.jpg \n inflating: /data/test/Alicia_Silverstone_50.jpg \n inflating: /data/test/Alicia_Silverstone_51.jpg \n inflating: /data/test/Alma_Powell_00.jpg \n inflating: /data/test/Alma_Powell_01.jpg \n inflating: /data/test/Alma_Powell_10.jpg \n inflating: /data/test/Alma_Powell_11.jpg \n inflating: /data/test/Alma_Powell_40.jpg \n inflating: /data/test/Alma_Powell_41.jpg \n inflating: /data/test/Alma_Powell_50.jpg \n inflating: /data/test/Alma_Powell_51.jpg \n inflating: /data/test/Alvaro_Silva_Calderon_00.jpg \n inflating: /data/test/Alvaro_Silva_Calderon_01.jpg \n inflating: /data/test/Alvaro_Silva_Calderon_10.jpg \n inflating: /data/test/Alvaro_Silva_Calderon_11.jpg \n inflating: /data/test/Alvaro_Silva_Calderon_20.jpg \n inflating: /data/test/Alvaro_Silva_Calderon_21.jpg \n inflating: /data/test/Alvaro_Silva_Calderon_30.jpg \n inflating: /data/test/Alvaro_Silva_Calderon_31.jpg \n inflating: /data/test/Amelia_Vega_10.jpg \n inflating: /data/test/Amelia_Vega_11.jpg \n inflating: /data/test/Amelia_Vega_20.jpg \n inflating: /data/test/Amelia_Vega_21.jpg \n inflating: /data/test/Amelia_Vega_30.jpg \n inflating: /data/test/Amelia_Vega_31.jpg \n inflating: /data/test/Amelia_Vega_40.jpg \n inflating: /data/test/Amelia_Vega_41.jpg \n inflating: /data/test/Amy_Brenneman_10.jpg \n inflating: /data/test/Amy_Brenneman_11.jpg \n inflating: /data/test/Amy_Brenneman_30.jpg \n inflating: /data/test/Amy_Brenneman_31.jpg \n inflating: /data/test/Amy_Brenneman_50.jpg \n inflating: /data/test/Amy_Brenneman_51.jpg \n inflating: /data/test/Andrea_Bocelli_10.jpg \n inflating: /data/test/Andrea_Bocelli_11.jpg \n inflating: /data/test/Andrea_Bocelli_20.jpg \n inflating: /data/test/Andrea_Bocelli_21.jpg \n inflating: /data/test/Andrea_Bocelli_30.jpg \n inflating: /data/test/Andrea_Bocelli_31.jpg \n inflating: /data/test/Andy_Roddick_20.jpg \n inflating: /data/test/Andy_Roddick_21.jpg \n inflating: /data/test/Andy_Roddick_40.jpg \n inflating: /data/test/Andy_Roddick_41.jpg \n inflating: /data/test/Andy_Roddick_50.jpg \n inflating: /data/test/Andy_Roddick_51.jpg \n inflating: /data/test/Andy_Rooney_10.jpg \n inflating: /data/test/Andy_Rooney_11.jpg \n inflating: /data/test/Andy_Rooney_20.jpg \n inflating: /data/test/Andy_Rooney_21.jpg \n inflating: /data/test/Andy_Rooney_50.jpg \n inflating: /data/test/Andy_Rooney_51.jpg \n inflating: /data/test/Angel_Lockward_30.jpg \n inflating: /data/test/Angel_Lockward_31.jpg \n inflating: /data/test/Angel_Lockward_40.jpg \n inflating: /data/test/Angel_Lockward_41.jpg \n inflating: /data/test/Angel_Lockward_50.jpg \n inflating: /data/test/Angel_Lockward_51.jpg \n inflating: /data/test/Angela_Bassett_20.jpg \n inflating: /data/test/Angela_Bassett_21.jpg \n inflating: /data/test/Angela_Bassett_30.jpg \n inflating: /data/test/Angela_Bassett_31.jpg \n inflating: /data/test/Angela_Bassett_40.jpg \n inflating: /data/test/Angela_Bassett_41.jpg \n inflating: /data/test/Angelo_Reyes_20.jpg \n"
],
[
"# import the required libraries\nimport glob\nimport os\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport matplotlib.image as mpimg\n\nimport cv2",
"_____no_output_____"
]
],
[
[
"Then, let's load in our training data and display some stats about that dat ato make sure it's been loaded in correctly!",
"_____no_output_____"
]
],
[
[
"key_pts_frame = pd.read_csv('/data/training_frames_keypoints.csv')\n\nn = 0\nimage_name = key_pts_frame.iloc[n, 0]\nkey_pts = key_pts_frame.iloc[n, 1:].as_matrix()\nkey_pts = key_pts.astype('float').reshape(-1, 2)\n\nprint('Image name: ', image_name)\nprint('Landmarks shape: ', key_pts.shape)\nprint('First 4 key pts: {}'.format(key_pts[:4]))",
"Image name: Luis_Fonsi_21.jpg\nLandmarks shape: (68, 2)\nFirst 4 key pts: [[ 45. 98.]\n [ 47. 106.]\n [ 49. 110.]\n [ 53. 119.]]\n"
],
[
"# print out some stats about the data\nprint('Number of images: ', key_pts_frame.shape[0])",
"Number of images: 3462\n"
]
],
[
[
"## Look at some images\n\nBelow, is a function `show_keypoints` that takes in an image and keypoints and displays them. As you look at this data, **note that these images are not all of the same size**, and neither are the faces! To eventually train a neural network on these images, we'll need to standardize their shape.",
"_____no_output_____"
]
],
[
[
"def show_keypoints(image, key_pts):\n \"\"\"Show image with keypoints\"\"\"\n plt.imshow(image)\n plt.scatter(key_pts[:, 0], key_pts[:, 1], s=20, marker='.', c='m')\n",
"_____no_output_____"
],
[
"# Display a few different types of images by changing the index n\n\n# select an image by index in our data frame\nn = 15\nimage_name = key_pts_frame.iloc[n, 0]\nkey_pts = key_pts_frame.iloc[n, 1:].as_matrix()\nkey_pts = key_pts.astype('float').reshape(-1, 2)\n\nplt.figure(figsize=(5, 5))\nshow_keypoints(mpimg.imread(os.path.join('/data/training/', image_name)), key_pts)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Dataset class and Transformations\n\nTo prepare our data for training, we'll be using PyTorch's Dataset class. Much of this this code is a modified version of what can be found in the [PyTorch data loading tutorial](http://pytorch.org/tutorials/beginner/data_loading_tutorial.html).\n\n#### Dataset class\n\n``torch.utils.data.Dataset`` is an abstract class representing a\ndataset. This class will allow us to load batches of image/keypoint data, and uniformly apply transformations to our data, such as rescaling and normalizing images for training a neural network.\n\n\nYour custom dataset should inherit ``Dataset`` and override the following\nmethods:\n\n- ``__len__`` so that ``len(dataset)`` returns the size of the dataset.\n- ``__getitem__`` to support the indexing such that ``dataset[i]`` can\n be used to get the i-th sample of image/keypoint data.\n\nLet's create a dataset class for our face keypoints dataset. We will\nread the CSV file in ``__init__`` but leave the reading of images to\n``__getitem__``. This is memory efficient because all the images are not\nstored in the memory at once but read as required.\n\nA sample of our dataset will be a dictionary\n``{'image': image, 'keypoints': key_pts}``. Our dataset will take an\noptional argument ``transform`` so that any required processing can be\napplied on the sample. We will see the usefulness of ``transform`` in the\nnext section.\n",
"_____no_output_____"
]
],
[
[
"from torch.utils.data import Dataset, DataLoader\n\nclass FacialKeypointsDataset(Dataset):\n \"\"\"Face Landmarks dataset.\"\"\"\n\n def __init__(self, csv_file, root_dir, transform=None):\n \"\"\"\n Args:\n csv_file (string): Path to the csv file with annotations.\n root_dir (string): Directory with all the images.\n transform (callable, optional): Optional transform to be applied\n on a sample.\n \"\"\"\n self.key_pts_frame = pd.read_csv(csv_file)\n self.root_dir = root_dir\n self.transform = transform\n\n def __len__(self):\n return len(self.key_pts_frame)\n\n def __getitem__(self, idx):\n image_name = os.path.join(self.root_dir,\n self.key_pts_frame.iloc[idx, 0])\n \n image = mpimg.imread(image_name)\n \n # if image has an alpha color channel, get rid of it\n if(image.shape[2] == 4):\n image = image[:,:,0:3]\n \n key_pts = self.key_pts_frame.iloc[idx, 1:].as_matrix()\n key_pts = key_pts.astype('float').reshape(-1, 2)\n sample = {'image': image, 'keypoints': key_pts}\n\n if self.transform:\n sample = self.transform(sample)\n\n return sample",
"_____no_output_____"
]
],
[
[
"Now that we've defined this class, let's instantiate the dataset and display some images.",
"_____no_output_____"
]
],
[
[
"# Construct the dataset\nface_dataset = FacialKeypointsDataset(csv_file='/data/training_frames_keypoints.csv',\n root_dir='/data/training/')\n\n# print some stats about the dataset\nprint('Length of dataset: ', len(face_dataset))",
"Length of dataset: 3462\n"
],
[
"# Display a few of the images from the dataset\nnum_to_display = 3\n\nfor i in range(num_to_display):\n \n # define the size of images\n fig = plt.figure(figsize=(20,10))\n \n # randomly select a sample\n rand_i = np.random.randint(0, len(face_dataset))\n sample = face_dataset[rand_i]\n\n # print the shape of the image and keypoints\n print(i, sample['image'].shape, sample['keypoints'].shape)\n\n ax = plt.subplot(1, num_to_display, i + 1)\n ax.set_title('Sample #{}'.format(i))\n \n # Using the same display function, defined earlier\n show_keypoints(sample['image'], sample['keypoints'])\n",
"0 (213, 201, 3) (68, 2)\n1 (305, 239, 3) (68, 2)\n2 (147, 143, 3) (68, 2)\n"
]
],
[
[
"## Transforms\n\nNow, the images above are not of the same size, and neural networks often expect images that are standardized; a fixed size, with a normalized range for color ranges and coordinates, and (for PyTorch) converted from numpy lists and arrays to Tensors.\n\nTherefore, we will need to write some pre-processing code.\nLet's create four transforms:\n\n- ``Normalize``: to convert a color image to grayscale values with a range of [0,1] and normalize the keypoints to be in a range of about [-1, 1]\n- ``Rescale``: to rescale an image to a desired size.\n- ``RandomCrop``: to crop an image randomly.\n- ``ToTensor``: to convert numpy images to torch images.\n\n\nWe will write them as callable classes instead of simple functions so\nthat parameters of the transform need not be passed everytime it's\ncalled. For this, we just need to implement ``__call__`` method and \n(if we require parameters to be passed in), the ``__init__`` method. \nWe can then use a transform like this:\n\n tx = Transform(params)\n transformed_sample = tx(sample)\n\nObserve below how these transforms are generally applied to both the image and its keypoints.\n\n",
"_____no_output_____"
]
],
[
[
"import torch\nfrom torchvision import transforms, utils\n# tranforms\n\nclass Normalize(object):\n \"\"\"Convert a color image to grayscale and normalize the color range to [0,1].\"\"\" \n\n def __call__(self, sample):\n image, key_pts = sample['image'], sample['keypoints']\n \n image_copy = np.copy(image)\n key_pts_copy = np.copy(key_pts)\n\n # convert image to grayscale\n image_copy = cv2.cvtColor(image, cv2.COLOR_RGB2GRAY)\n \n # scale color range from [0, 255] to [0, 1]\n image_copy= image_copy/255.0\n \n # scale keypoints to be centered around 0 with a range of [-1, 1]\n # mean = 100, sqrt = 50, so, pts should be (pts - 100)/50\n key_pts_copy = (key_pts_copy - 100)/50.0\n\n\n return {'image': image_copy, 'keypoints': key_pts_copy}\n\n\nclass Rescale(object):\n \"\"\"Rescale the image in a sample to a given size.\n\n Args:\n output_size (tuple or int): Desired output size. If tuple, output is\n matched to output_size. If int, smaller of image edges is matched\n to output_size keeping aspect ratio the same.\n \"\"\"\n\n def __init__(self, output_size):\n assert isinstance(output_size, (int, tuple))\n self.output_size = output_size\n\n def __call__(self, sample):\n image, key_pts = sample['image'], sample['keypoints']\n\n h, w = image.shape[:2]\n if isinstance(self.output_size, int):\n if h > w:\n new_h, new_w = self.output_size * h / w, self.output_size\n else:\n new_h, new_w = self.output_size, self.output_size * w / h\n else:\n new_h, new_w = self.output_size\n\n new_h, new_w = int(new_h), int(new_w)\n\n img = cv2.resize(image, (new_w, new_h))\n \n # scale the pts, too\n key_pts = key_pts * [new_w / w, new_h / h]\n\n return {'image': img, 'keypoints': key_pts}\n\n\nclass RandomCrop(object):\n \"\"\"Crop randomly the image in a sample.\n\n Args:\n output_size (tuple or int): Desired output size. If int, square crop\n is made.\n \"\"\"\n\n def __init__(self, output_size):\n assert isinstance(output_size, (int, tuple))\n if isinstance(output_size, int):\n self.output_size = (output_size, output_size)\n else:\n assert len(output_size) == 2\n self.output_size = output_size\n\n def __call__(self, sample):\n image, key_pts = sample['image'], sample['keypoints']\n\n h, w = image.shape[:2]\n new_h, new_w = self.output_size\n\n top = np.random.randint(0, h - new_h)\n left = np.random.randint(0, w - new_w)\n\n image = image[top: top + new_h,\n left: left + new_w]\n\n key_pts = key_pts - [left, top]\n\n return {'image': image, 'keypoints': key_pts}\n\n\nclass ToTensor(object):\n \"\"\"Convert ndarrays in sample to Tensors.\"\"\"\n\n def __call__(self, sample):\n image, key_pts = sample['image'], sample['keypoints']\n \n # if image has no grayscale color channel, add one\n if(len(image.shape) == 2):\n # add that third color dim\n image = image.reshape(image.shape[0], image.shape[1], 1)\n \n # swap color axis because\n # numpy image: H x W x C\n # torch image: C X H X W\n image = image.transpose((2, 0, 1))\n \n return {'image': torch.from_numpy(image),\n 'keypoints': torch.from_numpy(key_pts)}",
"_____no_output_____"
]
],
[
[
"## Test out the transforms\n\nLet's test these transforms out to make sure they behave as expected. As you look at each transform, note that, in this case, **order does matter**. For example, you cannot crop a image using a value smaller than the original image (and the orginal images vary in size!), but, if you first rescale the original image, you can then crop it to any size smaller than the rescaled size.",
"_____no_output_____"
]
],
[
[
"# test out some of these transforms\nrescale = Rescale(100)\ncrop = RandomCrop(50)\ncomposed = transforms.Compose([Rescale(250),\n RandomCrop(224)])\n\n# apply the transforms to a sample image\ntest_num = 500\nsample = face_dataset[test_num]\n\nfig = plt.figure()\nfor i, tx in enumerate([rescale, crop, composed]):\n transformed_sample = tx(sample)\n\n ax = plt.subplot(1, 3, i + 1)\n plt.tight_layout()\n ax.set_title(type(tx).__name__)\n show_keypoints(transformed_sample['image'], transformed_sample['keypoints'])\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Create the transformed dataset\n\nApply the transforms in order to get grayscale images of the same shape. Verify that your transform works by printing out the shape of the resulting data (printing out a few examples should show you a consistent tensor size).",
"_____no_output_____"
]
],
[
[
"# define the data tranform\n# order matters! i.e. rescaling should come before a smaller crop\ndata_transform = transforms.Compose([Rescale(250),\n RandomCrop(224),\n Normalize(),\n ToTensor()])\n\n# create the transformed dataset\ntransformed_dataset = FacialKeypointsDataset(csv_file='/data/training_frames_keypoints.csv',\n root_dir='/data/training/',\n transform=data_transform)\n",
"_____no_output_____"
],
[
"# print some stats about the transformed data\nprint('Number of images: ', len(transformed_dataset))\n\n# make sure the sample tensors are the expected size\nfor i in range(5):\n sample = transformed_dataset[i]\n print(i, sample['image'].size(), sample['keypoints'].size())\n",
"Number of images: 3462\n0 torch.Size([1, 224, 224]) torch.Size([68, 2])\n1 torch.Size([1, 224, 224]) torch.Size([68, 2])\n2 torch.Size([1, 224, 224]) torch.Size([68, 2])\n3 torch.Size([1, 224, 224]) torch.Size([68, 2])\n4 torch.Size([1, 224, 224]) torch.Size([68, 2])\n"
]
],
[
[
"## Data Iteration and Batching\n\nRight now, we are iterating over this data using a ``for`` loop, but we are missing out on a lot of PyTorch's dataset capabilities, specifically the abilities to:\n\n- Batch the data\n- Shuffle the data\n- Load the data in parallel using ``multiprocessing`` workers.\n\n``torch.utils.data.DataLoader`` is an iterator which provides all these\nfeatures, and we'll see this in use in the *next* notebook, Notebook 2, when we load data in batches to train a neural network!\n\n---\n\n",
"_____no_output_____"
],
[
"## Ready to Train!\n\nNow that you've seen how to load and transform our data, you're ready to build a neural network to train on this data.\n\nIn the next notebook, you'll be tasked with creating a CNN for facial keypoint detection.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
e793ddfacf4ceffac96e9d0b9ba49b86641303b0 | 607,180 | ipynb | Jupyter Notebook | traffic_sign_prediction_using_LE_NET_ARCHITECTURE.ipynb | abegpatel/Traffic-Sign-Classification-suing-LENET-Architecture | 61764285d99bcdd681df97c94ca37998302fa805 | [
"MIT"
] | null | null | null | traffic_sign_prediction_using_LE_NET_ARCHITECTURE.ipynb | abegpatel/Traffic-Sign-Classification-suing-LENET-Architecture | 61764285d99bcdd681df97c94ca37998302fa805 | [
"MIT"
] | null | null | null | traffic_sign_prediction_using_LE_NET_ARCHITECTURE.ipynb | abegpatel/Traffic-Sign-Classification-suing-LENET-Architecture | 61764285d99bcdd681df97c94ca37998302fa805 | [
"MIT"
] | null | null | null | 569.587242 | 362,638 | 0.934975 | [
[
[
"# STEP 1: IMPORT LIBRARIES AND DATASET\n",
"_____no_output_____"
]
],
[
[
"import warnings\nwarnings.filterwarnings(\"ignore\")",
"_____no_output_____"
],
[
"# import libraries \nimport pickle\nimport seaborn as sns\nimport pandas as pd # Import Pandas for data manipulation using dataframes\nimport numpy as np # Import Numpy for data statistical analysis \nimport matplotlib.pyplot as plt # Import matplotlib for data visualisation\nimport random",
"_____no_output_____"
],
[
"from google.colab import files\nfiles.download('/content/test.p')\nfiles.download('/content/valid.p')",
"_____no_output_____"
],
[
"files.download('/content/train.p')\n",
"_____no_output_____"
],
[
"# The pickle module implements binary protocols for serializing and de-serializing a Python object structure.\nwith open(\"/content/train.p\", mode='rb') as training_data:\n train = pickle.load(training_data)\nwith open(\"/content/valid.p\", mode='rb') as validation_data:\n valid = pickle.load(validation_data)\nwith open(\"/content/test.p\", mode='rb') as testing_data:\n test = pickle.load(testing_data)",
"_____no_output_____"
],
[
"X_train, y_train = train['features'], train['labels']\nX_validation, y_validation = valid['features'], valid['labels']\nX_test, y_test = test['features'], test['labels']\n",
"_____no_output_____"
],
[
"X_train.shape",
"_____no_output_____"
],
[
"y_train.shape",
"_____no_output_____"
]
],
[
[
"STEP 2: IMAGE EXPLORATION¶",
"_____no_output_____"
]
],
[
[
"i = 1001\nplt.imshow(X_train[i]) # Show images are not shuffled\ny_train[i]",
"_____no_output_____"
]
],
[
[
"STEP 3: DATA PEPARATION",
"_____no_output_____"
]
],
[
[
"## Shuffle the dataset \nfrom sklearn.utils import shuffle\nX_train, y_train = shuffle(X_train, y_train)\n",
"_____no_output_____"
],
[
"X_train_gray = np.sum(X_train/3, axis=3, keepdims=True)\nX_test_gray = np.sum(X_test/3, axis=3, keepdims=True)\nX_validation_gray = np.sum(X_validation/3, axis=3, keepdims=True) ",
"_____no_output_____"
],
[
"X_train_gray_norm = (X_train_gray - 128)/128 \nX_test_gray_norm = (X_test_gray - 128)/128\nX_validation_gray_norm = (X_validation_gray - 128)/128",
"_____no_output_____"
],
[
"X_train_gray.shape",
"_____no_output_____"
],
[
"i = 610\nplt.imshow(X_train_gray[i].squeeze(), cmap='gray')\nplt.figure()\nplt.imshow(X_train[i])\n",
"_____no_output_____"
]
],
[
[
"STEP 4: MODEL TRAINING\nThe model consists of the following layers:\n\nSTEP 1: THE FIRST CONVOLUTIONAL LAYER #1\nInput = 32x32x1\nOutput = 28x28x6\nOutput = (Input-filter+1)/Stride* => (32-5+1)/1=28\nUsed a 5x5 Filter with input depth of 3 and output depth of 6\nApply a RELU Activation function to the output\npooling for input, Input = 28x28x6 and Output = 14x14x6\n* Stride is the amount by which the kernel is shifted when the kernel is passed over the image.\nSTEP 2: THE SECOND CONVOLUTIONAL LAYER #2\n\nInput = 14x14x6\nOutput = 10x10x16\nLayer 2: Convolutional layer with Output = 10x10x16\nOutput = (Input-filter+1)/strides => 10 = 14-5+1/1\nApply a RELU Activation function to the output\nPooling with Input = 10x10x16 and Output = 5x5x16\nSTEP 3: FLATTENING THE NETWORK\n\nFlatten the network with Input = 5x5x16 and Output = 400\nSTEP 4: FULLY CONNECTED LAYER\n\nLayer 3: Fully Connected layer with Input = 400 and Output = 120\nApply a RELU Activation function to the output\nSTEP 5: ANOTHER FULLY CONNECTED LAYER\n\nLayer 4: Fully Connected Layer with Input = 120 and Output = 84\nApply a RELU Activation function to the output\nSTEP 6: FULLY CONNECTED LAYER\n\nLayer 5: Fully Connected layer with Input = 84 and Output = 43",
"_____no_output_____"
]
],
[
[
"# Import train_test_split from scikit library\n\nfrom keras.models import Sequential\nfrom keras.layers import Conv2D, MaxPooling2D, AveragePooling2D, Dense, Flatten, Dropout\nfrom keras.optimizers import Adam\nfrom keras.callbacks import TensorBoard\n\nfrom sklearn.model_selection import train_test_split\n",
"_____no_output_____"
],
[
"image_shape = X_train_gray[i].shape",
"_____no_output_____"
],
[
"cnn_model = Sequential()\n\ncnn_model.add(Conv2D(filters=6, kernel_size=(5, 5), activation='relu', input_shape=(32,32,1)))\ncnn_model.add(AveragePooling2D())\n\ncnn_model.add(Conv2D(filters=16, kernel_size=(5, 5), activation='relu'))\ncnn_model.add(AveragePooling2D())\n\ncnn_model.add(Flatten())\n\ncnn_model.add(Dense(units=120, activation='relu'))\n\ncnn_model.add(Dense(units=84, activation='relu'))\n\ncnn_model.add(Dense(units=43, activation = 'softmax'))\n",
"_____no_output_____"
],
[
"cnn_model.compile(loss ='sparse_categorical_crossentropy', optimizer=Adam(lr=0.001),metrics =['accuracy'])",
"_____no_output_____"
],
[
"history = cnn_model.fit(X_train_gray_norm,\n y_train,\n batch_size=500,\n epochs=50,\n verbose=1,\n validation_data = (X_validation_gray_norm,y_validation))",
"Epoch 1/50\n70/70 [==============================] - 8s 10ms/step - loss: 3.4363 - accuracy: 0.1037 - val_loss: 2.5737 - val_accuracy: 0.3120\nEpoch 2/50\n70/70 [==============================] - 0s 6ms/step - loss: 1.8750 - accuracy: 0.4805 - val_loss: 1.4311 - val_accuracy: 0.5537\nEpoch 3/50\n70/70 [==============================] - 0s 5ms/step - loss: 1.0405 - accuracy: 0.6931 - val_loss: 1.0730 - val_accuracy: 0.6859\nEpoch 4/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.7399 - accuracy: 0.7856 - val_loss: 0.8831 - val_accuracy: 0.7234\nEpoch 5/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.5875 - accuracy: 0.8318 - val_loss: 0.8052 - val_accuracy: 0.7413\nEpoch 6/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.4881 - accuracy: 0.8654 - val_loss: 0.7567 - val_accuracy: 0.7671\nEpoch 7/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.4150 - accuracy: 0.8829 - val_loss: 0.7154 - val_accuracy: 0.7844\nEpoch 8/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.3522 - accuracy: 0.9021 - val_loss: 0.6872 - val_accuracy: 0.8023\nEpoch 9/50\n70/70 [==============================] - 0s 6ms/step - loss: 0.3141 - accuracy: 0.9150 - val_loss: 0.6809 - val_accuracy: 0.7975\nEpoch 10/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.2788 - accuracy: 0.9248 - val_loss: 0.6507 - val_accuracy: 0.8116\nEpoch 11/50\n70/70 [==============================] - 0s 7ms/step - loss: 0.2490 - accuracy: 0.9327 - val_loss: 0.6513 - val_accuracy: 0.8231\nEpoch 12/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.2369 - accuracy: 0.9377 - val_loss: 0.6711 - val_accuracy: 0.8034\nEpoch 13/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.2145 - accuracy: 0.9423 - val_loss: 0.6187 - val_accuracy: 0.8293\nEpoch 14/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.2005 - accuracy: 0.9489 - val_loss: 0.6059 - val_accuracy: 0.8367\nEpoch 15/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.1789 - accuracy: 0.9525 - val_loss: 0.6724 - val_accuracy: 0.8249\nEpoch 16/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.1634 - accuracy: 0.9555 - val_loss: 0.6359 - val_accuracy: 0.8399\nEpoch 17/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.1572 - accuracy: 0.9603 - val_loss: 0.6481 - val_accuracy: 0.8367\nEpoch 18/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.1311 - accuracy: 0.9663 - val_loss: 0.6483 - val_accuracy: 0.8302\nEpoch 19/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.1302 - accuracy: 0.9680 - val_loss: 0.6580 - val_accuracy: 0.8306\nEpoch 20/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.1230 - accuracy: 0.9669 - val_loss: 0.6450 - val_accuracy: 0.8363\nEpoch 21/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.1083 - accuracy: 0.9738 - val_loss: 0.6795 - val_accuracy: 0.8390\nEpoch 22/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.1068 - accuracy: 0.9726 - val_loss: 0.6792 - val_accuracy: 0.8381\nEpoch 23/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0933 - accuracy: 0.9761 - val_loss: 0.7126 - val_accuracy: 0.8410\nEpoch 24/50\n70/70 [==============================] - 0s 6ms/step - loss: 0.0874 - accuracy: 0.9764 - val_loss: 0.6611 - val_accuracy: 0.8469\nEpoch 25/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0773 - accuracy: 0.9809 - val_loss: 0.7272 - val_accuracy: 0.8413\nEpoch 26/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0719 - accuracy: 0.9824 - val_loss: 0.7447 - val_accuracy: 0.8290\nEpoch 27/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0755 - accuracy: 0.9814 - val_loss: 0.7347 - val_accuracy: 0.8322\nEpoch 28/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0723 - accuracy: 0.9807 - val_loss: 0.7886 - val_accuracy: 0.8311\nEpoch 29/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0611 - accuracy: 0.9847 - val_loss: 0.7606 - val_accuracy: 0.8345\nEpoch 30/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0599 - accuracy: 0.9860 - val_loss: 0.8071 - val_accuracy: 0.8365\nEpoch 31/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0595 - accuracy: 0.9848 - val_loss: 0.7790 - val_accuracy: 0.8404\nEpoch 32/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0510 - accuracy: 0.9864 - val_loss: 0.7991 - val_accuracy: 0.8374\nEpoch 33/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0484 - accuracy: 0.9876 - val_loss: 0.7773 - val_accuracy: 0.8442\nEpoch 34/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0540 - accuracy: 0.9844 - val_loss: 0.8191 - val_accuracy: 0.8356\nEpoch 35/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0438 - accuracy: 0.9887 - val_loss: 0.7977 - val_accuracy: 0.8522\nEpoch 36/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0372 - accuracy: 0.9903 - val_loss: 0.7888 - val_accuracy: 0.8401\nEpoch 37/50\n70/70 [==============================] - 0s 6ms/step - loss: 0.0372 - accuracy: 0.9902 - val_loss: 0.8771 - val_accuracy: 0.8413\nEpoch 38/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0385 - accuracy: 0.9897 - val_loss: 0.8986 - val_accuracy: 0.8438\nEpoch 39/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0380 - accuracy: 0.9902 - val_loss: 0.8557 - val_accuracy: 0.8485\nEpoch 40/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0370 - accuracy: 0.9900 - val_loss: 0.8356 - val_accuracy: 0.8478\nEpoch 41/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0347 - accuracy: 0.9901 - val_loss: 0.8599 - val_accuracy: 0.8438\nEpoch 42/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0334 - accuracy: 0.9905 - val_loss: 0.9633 - val_accuracy: 0.8388\nEpoch 43/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0406 - accuracy: 0.9879 - val_loss: 0.9581 - val_accuracy: 0.8327\nEpoch 44/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0321 - accuracy: 0.9915 - val_loss: 0.9337 - val_accuracy: 0.8415\nEpoch 45/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0291 - accuracy: 0.9922 - val_loss: 0.8349 - val_accuracy: 0.8497\nEpoch 46/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0278 - accuracy: 0.9923 - val_loss: 0.9275 - val_accuracy: 0.8506\nEpoch 47/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0265 - accuracy: 0.9931 - val_loss: 0.9720 - val_accuracy: 0.8383\nEpoch 48/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0287 - accuracy: 0.9918 - val_loss: 0.9064 - val_accuracy: 0.8580\nEpoch 49/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0286 - accuracy: 0.9911 - val_loss: 0.8895 - val_accuracy: 0.8619\nEpoch 50/50\n70/70 [==============================] - 0s 5ms/step - loss: 0.0223 - accuracy: 0.9942 - val_loss: 0.8876 - val_accuracy: 0.8560\n"
]
],
[
[
"STEP 5: MODEL EVALUATION¶",
"_____no_output_____"
]
],
[
[
"score = cnn_model.evaluate(X_test_gray_norm, y_test,verbose=0)\nprint('Test Accuracy : {:.4f}'.format(score[1]))",
"Test Accuracy : 0.8462\n"
],
[
"history.history.keys()",
"_____no_output_____"
],
[
"accuracy = history.history['accuracy']\nval_accuracy = history.history['val_accuracy']\nloss = history.history['loss']\nval_loss = history.history['val_loss']\n\nepochs = range(len(accuracy))\n\nplt.plot(epochs, accuracy, 'bo', label='Training Accuracy')\nplt.plot(epochs, val_accuracy, 'b', label='Validation Accuracy')\nplt.title('Training and Validation accuracy')\nplt.legend()\n",
"_____no_output_____"
],
[
"plt.plot(epochs, loss, 'ro', label='Training Loss')\nplt.plot(epochs, val_loss, 'r', label='Validation Loss')\nplt.title('Training and validation loss')\nplt.legend()\nplt.show()",
"_____no_output_____"
],
[
"#get the predictions for the test data\npredicted_classes = cnn_model.predict_classes(X_test_gray_norm)\n#get the indices to be plotted\ny_true = y_test\n",
"_____no_output_____"
],
[
"from sklearn.metrics import confusion_matrix\ncm = confusion_matrix(y_true, predicted_classes)\nplt.figure(figsize = (25,25))\nsns.heatmap(cm, annot=True)",
"_____no_output_____"
],
[
"L = 7\nW = 7\nfig, axes = plt.subplots(L, W, figsize = (12,12))\naxes = axes.ravel() # \n\nfor i in np.arange(0, L * W): \n axes[i].imshow(X_test[i])\n axes[i].set_title(\"Prediction={}\\n True={}\".format(predicted_classes[i], y_true[i]))\n axes[i].axis('off')\n\nplt.subplots_adjust(wspace=1)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e793e7216fa11f6bf7f87df5d910328588be56b7 | 65,936 | ipynb | Jupyter Notebook | crypto_labs/hkdf/data_analysis.ipynb | yerseg/mephi_labs | 503321a4726dcd18714ac35d459ed6f41281346b | [
"MIT"
] | null | null | null | crypto_labs/hkdf/data_analysis.ipynb | yerseg/mephi_labs | 503321a4726dcd18714ac35d459ed6f41281346b | [
"MIT"
] | null | null | null | crypto_labs/hkdf/data_analysis.ipynb | yerseg/mephi_labs | 503321a4726dcd18714ac35d459ed6f41281346b | [
"MIT"
] | null | null | null | 129.033268 | 8,736 | 0.854162 | [
[
[
"import pandas as pd\n%matplotlib inline\nimport matplotlib.pyplot as plt\nimport pickle\nimport json",
"_____no_output_____"
],
[
"path_to_weather_json = \"files\\\\weather.json\"",
"_____no_output_____"
],
[
"with open(path_to_weather_json, 'r') as file:\n weather = json.load(file)",
"_____no_output_____"
],
[
"weather_hourly = weather['hourly']['data']",
"_____no_output_____"
],
[
"weather_hourly_df = pd.DataFrame(weather_hourly)\nweather_hourly_df.head()",
"_____no_output_____"
],
[
"def create_and_save_hist(df, column_name):\n plt.figure(figsize=(10,5))\n plt.title('Histogram of %s' % column_name) \n plt.hist(df['%s' % column_name], bins=17, color='g')\n plt.savefig('hists\\%s_hist' % column_name)\n \nneeded_columns = ['temperature', 'humidity', 'windSpeed', 'cloudCover', 'ozone']\nfor column in needed_columns:\n create_and_save_hist(weather_hourly_df, column)",
"_____no_output_____"
]
],
[
[
"Для задания 2 вытащу столбец temperature",
"_____no_output_____"
]
],
[
[
"temperature_column = weather_hourly_df.loc[:, 'temperature']",
"_____no_output_____"
],
[
"with open('files\\\\temperature.txt', 'w') as file:\n for temp in temperature_column:\n file.write(str(temp) + '\\n')",
"_____no_output_____"
],
[
"path_to_passwords_json = \"files\\\\passwords.json\"",
"_____no_output_____"
],
[
"with open(path_to_passwords_json, 'r') as file:\n passwords = json.load(file)",
"_____no_output_____"
],
[
"with open(\"files\\\\passwords.txt\", \"w\") as file:\n for password in passwords:\n file.write(password + '\\n')",
"_____no_output_____"
],
[
"with open(\"files\\\\pbkdf2_bits.txt\", \"r\") as file:\n pbkdf2 = list(file.readlines())",
"_____no_output_____"
],
[
"with open(\"files\\\\hkdf_bits.txt\", \"r\") as file:\n hkdf = list(file.readlines())",
"_____no_output_____"
],
[
"hkdf = [int(x) for x in hkdf]\npbkdf2 = [int(x) for x in pbkdf2]",
"_____no_output_____"
],
[
"plt.figure(figsize=(10,5))\nplt.title('Histogram of HKDF bits') \nplt.hist(hkdf, bins=10, color='g')\nplt.savefig('hists\\HKDF_gist')\n\nplt.figure(figsize=(10,5))\nplt.title('Histogram of PBKDF2 bits') \nplt.hist(pbkdf2, bins=10, color='g')\nplt.savefig('hists\\PBKDF2_gist')",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e79404477281dfc56efecb5885051be261d5cecc | 6,057 | ipynb | Jupyter Notebook | ImageCollection/map_function.ipynb | OIEIEIO/earthengine-py-notebooks | 5d6c5cdec0c73bf02020ee17d42c9e30d633349f | [
"MIT"
] | 1,008 | 2020-01-27T02:03:18.000Z | 2022-03-24T10:42:14.000Z | ImageCollection/map_function.ipynb | rafatieppo/earthengine-py-notebooks | 99fbc4abd1fb6ba41e3d8a55f8911217353a3237 | [
"MIT"
] | 8 | 2020-02-01T20:18:18.000Z | 2021-11-23T01:48:02.000Z | ImageCollection/map_function.ipynb | rafatieppo/earthengine-py-notebooks | 99fbc4abd1fb6ba41e3d8a55f8911217353a3237 | [
"MIT"
] | 325 | 2020-01-27T02:03:36.000Z | 2022-03-25T20:33:33.000Z | 38.09434 | 470 | 0.548291 | [
[
[
"<table class=\"ee-notebook-buttons\" align=\"left\">\n <td><a target=\"_blank\" href=\"https://github.com/giswqs/earthengine-py-notebooks/tree/master/ImageCollection/map_function.ipynb\"><img width=32px src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" /> View source on GitHub</a></td>\n <td><a target=\"_blank\" href=\"https://nbviewer.jupyter.org/github/giswqs/earthengine-py-notebooks/blob/master/ImageCollection/map_function.ipynb\"><img width=26px src=\"https://upload.wikimedia.org/wikipedia/commons/thumb/3/38/Jupyter_logo.svg/883px-Jupyter_logo.svg.png\" />Notebook Viewer</a></td>\n <td><a target=\"_blank\" href=\"https://colab.research.google.com/github/giswqs/earthengine-py-notebooks/blob/master/ImageCollection/map_function.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" /> Run in Google Colab</a></td>\n</table>",
"_____no_output_____"
],
[
"## Install Earth Engine API and geemap\nInstall the [Earth Engine Python API](https://developers.google.com/earth-engine/python_install) and [geemap](https://geemap.org). The **geemap** Python package is built upon the [ipyleaflet](https://github.com/jupyter-widgets/ipyleaflet) and [folium](https://github.com/python-visualization/folium) packages and implements several methods for interacting with Earth Engine data layers, such as `Map.addLayer()`, `Map.setCenter()`, and `Map.centerObject()`.\nThe following script checks if the geemap package has been installed. If not, it will install geemap, which automatically installs its [dependencies](https://github.com/giswqs/geemap#dependencies), including earthengine-api, folium, and ipyleaflet.",
"_____no_output_____"
]
],
[
[
"# Installs geemap package\nimport subprocess\n\ntry:\n import geemap\nexcept ImportError:\n print('Installing geemap ...')\n subprocess.check_call([\"python\", '-m', 'pip', 'install', 'geemap'])",
"_____no_output_____"
],
[
"import ee\nimport geemap",
"_____no_output_____"
]
],
[
[
"## Create an interactive map \nThe default basemap is `Google Maps`. [Additional basemaps](https://github.com/giswqs/geemap/blob/master/geemap/basemaps.py) can be added using the `Map.add_basemap()` function. ",
"_____no_output_____"
]
],
[
[
"Map = geemap.Map(center=[40,-100], zoom=4)\nMap",
"_____no_output_____"
]
],
[
[
"## Add Earth Engine Python script ",
"_____no_output_____"
]
],
[
[
"# Add Earth Engine dataset\n# This function adds a band representing the image timestamp.\ndef addTime(image): \n return image.addBands(image.metadata('system:time_start'))\n\ndef conditional(image):\n return ee.Algorithms.If(ee.Number(image.get('SUN_ELEVATION')).gt(40),\n image,\n ee.Image(0))\n\n# Load a Landsat 8 collection for a single path-row.\ncollection = ee.ImageCollection('LANDSAT/LC08/C01/T1_TOA') \\\n .filter(ee.Filter.eq('WRS_PATH', 44)) \\\n .filter(ee.Filter.eq('WRS_ROW', 34))\n\n\n\n# Map the function over the collection and display the result.\nprint(collection.map(addTime).getInfo())\n\n\n# Load a Landsat 8 collection for a single path-row.\ncollection = ee.ImageCollection('LANDSAT/LC8_L1T_TOA') \\\n .filter(ee.Filter.eq('WRS_PATH', 44)) \\\n .filter(ee.Filter.eq('WRS_ROW', 34))\n\n# This function uses a conditional statement to return the image if\n# the solar elevation > 40 degrees. Otherwise it returns a zero image.\n# conditional = function(image) {\n# return ee.Algorithms.If(ee.Number(image.get('SUN_ELEVATION')).gt(40),\n# image,\n# ee.Image(0))\n# }\n\n# Map the function over the collection, convert to a List and print the result.\nprint('Expand this to see the result: ', collection.map(conditional).getInfo())\n\n",
"_____no_output_____"
]
],
[
[
"## Display Earth Engine data layers ",
"_____no_output_____"
]
],
[
[
"Map.addLayerControl() # This line is not needed for ipyleaflet-based Map.\nMap",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7941202ef8acd81979999393121bac4669063de | 439,752 | ipynb | Jupyter Notebook | notebook/Analyze_Statics2011_NDCG.ipynb | qqhann/KnowledgeTracing | cecdb9af0c44efffd1ce3359f331d7d7782f551b | [
"MIT"
] | 3 | 2021-11-24T09:49:03.000Z | 2022-01-12T06:53:05.000Z | notebook/Analyze_Statics2011_NDCG.ipynb | qqhann/KnowledgeTracing | cecdb9af0c44efffd1ce3359f331d7d7782f551b | [
"MIT"
] | 25 | 2021-08-15T10:57:48.000Z | 2021-08-23T21:14:24.000Z | notebook/Analyze_Statics2011_NDCG.ipynb | qqhann/KnowledgeTracing | cecdb9af0c44efffd1ce3359f331d7d7782f551b | [
"MIT"
] | 1 | 2022-01-23T13:05:21.000Z | 2022-01-23T13:05:21.000Z | 806.884404 | 73,068 | 0.952334 | [
[
[
"import sys\nimport json\nfrom pathlib import Path\nfrom collections import defaultdict\n\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\nsys.path.append('..')\nfrom src.data import load_qa_format_source",
"_____no_output_____"
],
[
"projectdir = Path('..')\n\ntrain_path = projectdir / 'data/input/STATICS/STATICS_train.csv'\ntest_path = projectdir / 'data/input/STATICS/STATICS_test.csv'\nid_path = projectdir / 'data/input/STATICS/STATICS_qid_sid_sname'\nassert train_path.exists()\nassert test_path.exists()\nassert id_path.exists()\n\ntrain_qa = load_qa_format_source(train_path)\ntest_qa = load_qa_format_source(test_path)\ndel train_path\ndel test_path",
"_____no_output_____"
],
[
"pd.read_table(id_path)",
"_____no_output_____"
],
[
"with open(projectdir / 'output/20_0310_edm2020_statics/pre_dummy_epoch_size0.auto/report/20200310-0202/report.json', 'r') as f:\n report_pre00 = json.load(f)\nwith open(projectdir / 'output/20_0310_edm2020_statics/pre_dummy_epoch_size10.auto/report/20200310-0404/report.json', 'r') as f:\n report_pre10 = json.load(f)",
"_____no_output_____"
],
[
"ndcg_00 = report_pre00['indicator']['RPhard']['all']\nndcg_10 = report_pre10['indicator']['RPhard']['all']",
"_____no_output_____"
],
[
"count = defaultdict(list)\nfor seq in train_qa+test_qa:\n for q, a in seq:\n count[q-1].append(a)\n \nassert set(count.keys()) == set(range(1223))\ncount = [count[i] for i in range(1223)]\ncount_list = [len(count[i]) for i in range(1223)]",
"_____no_output_____"
],
[
"sns.distplot([sum(l) for l in count])\nplt.show()\nsns.distplot([len(l) for l in count])\nplt.show()\nsns.distplot([sum(l)/len(l) for l in count])\nplt.show()",
"_____no_output_____"
]
],
[
[
"### 相関分析(pre 0 vs pre 10)\nndcg_00はpre 0, ndcg_10はpre 10の結果",
"_____no_output_____"
],
[
"#### KCの出現数とNDCGの相関",
"_____no_output_____"
]
],
[
[
"# 1.1\ncorrcoef = np.corrcoef(x=count_list, y=ndcg_00)[0][1]\nprint(\"Corr coef =\", corrcoef)\nax = sns.jointplot(x=count_list, y=ndcg_00, kind='reg')\nplt.xlabel(\"KC count\")\nplt.ylabel(\"NDCG (baseline)\")\nplt.show()",
"Corr coef = -0.017273767116509027\n"
],
[
"# 1.2\ncorrcoef = np.corrcoef(x=count_list, y=ndcg_10)\nprint(\"Corr coef =\", corrcoef)\nsns.jointplot(x=count_list, y=ndcg_10, kind='reg')\nplt.xlabel(\"KC count\")\nplt.ylabel(\"NDCG (ours)\")\nplt.show()",
"Corr coef = [[ 1. -0.12971755]\n [-0.12971755 1. ]]\n"
],
[
"# 1.6\n_y = [n10 - n0 for n10, n0 in zip(ndcg_10, ndcg_00)]\ncorrcoef = np.corrcoef(x=count_list, y=_y)\nprint(\"Corr coef =\", corrcoef)\nsns.jointplot(x=count_list, y=_y, kind='reg')\nplt.xlabel(\"KC count\")\nplt.ylabel(\"NDCG gain\")\nplt.show()",
"Corr coef = [[ 1. -0.06907159]\n [-0.06907159 1. ]]\n"
]
],
[
[
"#### KCの正解率とNDCGの相関\n(1.3)は正解率の低い問題でNDCGがやや低い傾向がみてとれる",
"_____no_output_____"
]
],
[
[
"# 1.3\nsns.jointplot(x=[sum(l)/len(l) for l in count], y=ndcg_00, kind='reg')",
"_____no_output_____"
],
[
"# 1.4\nsns.jointplot(x=[sum(l)/len(l) for l in count], y=ndcg_10, kind='reg')",
"_____no_output_____"
]
],
[
[
"#### 出現数と正解率の関係\n\n結果考察:難しい問題はときたがらない,かんたんな問題は繰り返しがち,という傾向があるのか?",
"_____no_output_____"
]
],
[
[
"# 1.5\nprint(np.corrcoef(x=[len(l) for l in count], y=[sum(l)/len(l) for l in count]))\nsns.jointplot(x=[len(l) for l in count], y=[sum(l)/len(l) for l in count], kind='reg')",
"[[1. 0.27203797]\n [0.27203797 1. ]]\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
e79417046cb4ca223ea431e6e5edf6160c932938 | 2,189 | ipynb | Jupyter Notebook | Spark_RDD/PySpark_RDD/01_rdd_map_countbyvalue_movielens.ipynb | havyx/Big_Data_Analitycs | 8873722f6a8c75bcca34841f454bb7e016ccf7f7 | [
"MIT"
] | null | null | null | Spark_RDD/PySpark_RDD/01_rdd_map_countbyvalue_movielens.ipynb | havyx/Big_Data_Analitycs | 8873722f6a8c75bcca34841f454bb7e016ccf7f7 | [
"MIT"
] | 2 | 2020-07-23T12:59:55.000Z | 2020-07-23T12:59:56.000Z | Spark_RDD/PySpark_RDD/01_rdd_map_countbyvalue_movielens.ipynb | havyx/Big_Data_Analitycs | 8873722f6a8c75bcca34841f454bb7e016ccf7f7 | [
"MIT"
] | null | null | null | 18.87069 | 86 | 0.49566 | [
[
[
"from pyspark import SparkConf, SparkContext\nimport collections",
"_____no_output_____"
],
[
"conf = SparkConf().setMaster(\"local\").setAppName(\"RatingsHistogram\")\nsc = SparkContext(conf = conf)",
"_____no_output_____"
],
[
"lines = sc.textFile(\"datasets/ml-100k/u.data\")",
"_____no_output_____"
],
[
"ratings = lines.map(lambda x: x.split()[2])",
"_____no_output_____"
],
[
"result = ratings.countByValue()",
"_____no_output_____"
],
[
"result",
"_____no_output_____"
],
[
"sortedResults = collections.OrderedDict(sorted(result.items()))\nfor key, value in sortedResults.items():\n print(\"%s %i\" % (key, value))",
"1 6110\n2 11370\n3 27145\n4 34174\n5 21201\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7941c1d5fce0966b0a23eece04fe414781c845f | 616,680 | ipynb | Jupyter Notebook | .ipynb_checkpoints/Bike_Share-checkpoint.ipynb | alan-toledo/bike-share-data-analysis | f421b15039009f2971183b0f076497991676d947 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/Bike_Share-checkpoint.ipynb | alan-toledo/bike-share-data-analysis | f421b15039009f2971183b0f076497991676d947 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/Bike_Share-checkpoint.ipynb | alan-toledo/bike-share-data-analysis | f421b15039009f2971183b0f076497991676d947 | [
"MIT"
] | null | null | null | 934.363636 | 198,404 | 0.954962 | [
[
[
"# Analysis Bike Share",
"_____no_output_____"
],
[
"# Summary\n* 85% of the trips are made by users who are subscribers for the last two years (2013-08, 2015-08).\n* This trend has been maintained for the last couple of months (86% are subscribers).\n* The number of trips is variable through the days. Last couple of months follow the same trends.\n* Subscribers: Average number of trips per day 773. Average number of trips per day (Last couple of months) 900.\n* Subscribers use bikes more on weekdays. There is a big difference between weekdays and weekend.\n* The Subscriber uses the bike in a greater frequency during rush hours. Morning: 7:00 AM - 9:00 AM. Evening: 16:00 AM - 18:00 AM.\n* Average number of trips during the weekday: 105.076 Average number of trips during the weekend: 20.682\n* The subscripter use the bike 8 minutes in average. The most frequently used range is between: [2, 15] minutes.\n* The most frequent start station is: San Francisco Caltrain (Townsend at 4th).\n* The most frequent end station is: San Francisco Caltrain (Townsend at 4th).\n* Trip start-end most used: San Francisco Caltrain 2 (330 Townsend) --> Townsend at 7th (6216 Trips)\n* Some bikes are used in a greater frequency.",
"_____no_output_____"
],
[
"## User Experience",
"_____no_output_____"
],
[
"* According to the data, the user's profile is a worker. He leaves his house in the morning for a station to get on a bike and go to his work (nearest station). This time on average is 8 minutes (not long distance). For the return it is the same idea.\n* The user experience can be affected mainly for 2 reasons.\n* 1. Limited availability of bikes at rush hours.\n* 2. Bikes damaged by excessive use in the stations where there is more demand.\n",
"_____no_output_____"
],
[
"## Experimentation Plan",
"_____no_output_____"
],
[
"* Go to the route with the most demand (San Francisco Caltrain 2 (330 Townsend) --> Townsend at 7th) and see what is happening.\n* Try to quantify waiting for available bikes at the rush hours. This situation must be equal in all days of the weekday.\n* Based on the above, increase the supply of bikes. This must be dynamic according to the demand at the rush hours.\n* Detect the most used bikes (bike_id) and check their status.\n* Based on the above, implement maintenance (dynamic) according to use.",
"_____no_output_____"
],
[
"# Modules",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nimport matplotlib.dates as mdates\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"# Load Data",
"_____no_output_____"
]
],
[
[
"trip = pd.read_csv('trip.csv')",
"_____no_output_____"
]
],
[
[
"# Subscription Types (Users)",
"_____no_output_____"
]
],
[
[
"trip['subscription_type'] = pd.Categorical(trip['subscription_type'])\nfig, ax = plt.subplots(1, 2, figsize=(15,5))\ntrip['subscription_type'].value_counts().plot(kind='pie', autopct='%.2f', ax=ax[0])\nstats = trip['subscription_type'].value_counts(dropna=True)\nax[1].set_ylabel('Number of trips')\nax[1].set_title('N trips')\nbarlist = ax[1].bar(stats.index.categories, [stats['Customer'],stats['Subscriber']], width = 0.35)\nbarlist[0].set_color('#ff7f0e')\nbarlist[1].set_color('#1f77b4')\nprint(\"85% of the trips are made by users who are subscribers (use subscription plan)\")",
"85% of the trips are made by users who are subscribers (use subscription plan)\n"
],
[
"trip['start_date'] = pd.to_datetime(trip['start_date'])\ntrip['start'] = trip['start_date'].dt.date\ntrip['end_date'] = pd.to_datetime(trip['end_date'])\ntrip['end'] = trip['end_date'].dt.date",
"_____no_output_____"
],
[
"print('First Trip', trip['start'].min())\nprint('Last Trip', trip['end'].max())",
"First Trip 2013-08-29\nLast Trip 2015-08-31\n"
],
[
"from_last_months = pd.to_datetime('2015-06-01')\ncondition = trip['start'] >= from_last_months\ntrip.loc[condition,'subscription_type'].value_counts().plot(kind='pie', autopct='%.2f', figsize=(6,6))\nprint(\"86% of the trips are made by users who are subscribers (use subscription plan). Last couple of months.\")",
"86% of the trips are made by users who are subscribers (use subscription plan). Last couple of months.\n"
],
[
"group = trip.groupby('start').count()\ncondition = (group.index >= from_last_months)\nfig, ax = plt.subplots(figsize=(24,6))\nlocator = mdates.AutoDateLocator()\nformatter = mdates.ConciseDateFormatter(locator)\nax.xaxis.set_major_locator(locator)\nax.xaxis.set_major_formatter(formatter)\nax.plot(group.index[~condition], group.id[~condition].values, color = 'blue', linewidth=2)\nax.plot(group.index[condition], group.id[condition].values, color = 'red', linewidth=2)\nax.set_title('N trips')\nax.legend(['N trips', 'N trips last months'])\nprint(\"The number of trips is variable through the days.\")\nprint(\"Last couple of months follow the same trends. This is important.\")\nprint(\"End of the year and early next there is a downward trend.\")",
"The number of trips is variable through the days.\nLast couple of months follow the same trends. This is important.\nEnd of the year and early next there is a downward trend.\n"
],
[
"group = trip.groupby(['start','subscription_type']).size().unstack(level=1, fill_value=0)",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(24,6))\nlocator = mdates.AutoDateLocator()\nformatter = mdates.ConciseDateFormatter(locator)\nax.xaxis.set_major_locator(locator)\nax.xaxis.set_major_formatter(formatter)\nax.plot(group.index, group['Customer'].values, color = '#ff7f0e', linewidth=2)\nax.set_title('N trips by Subscription Types: Customer')\nax.legend(['N trips Customer'])",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(24,6))\ncondition = (group.index >= from_last_months)\nlocator = mdates.AutoDateLocator()\nformatter = mdates.ConciseDateFormatter(locator)\nax.xaxis.set_major_locator(locator)\nax.xaxis.set_major_formatter(formatter)\nax.plot(group.index[~condition], group['Subscriber'][~condition].values, color='#1f77b4', linewidth=2)\nax.plot(group.index[condition], group['Subscriber'][condition].values, color='red', linewidth=2)\nax.set_title('N trips by Subscription Types: Subscriber')\nax.legend(['N trips Subscriber', 'N trips last months'])\navg = int(group['Subscriber'].values.sum()/len(group.index))\nprint('Average number of trips per day', avg)\navg = int(group['Subscriber'][condition].values.sum()/len(group.index[condition]))\nprint('Average number of trips per day (Last couple of months)', avg)",
"Average number of trips per day 773\nAverage number of trips per day (Last couple of months) 900\n"
],
[
"def get_ordered_data(group):\n values = []\n avg_weekday, avg_weekend = 0.0, 0.0\n weekday = ['Monday', 'Tuesday', 'Wednesday', 'Thursday', 'Friday']\n weekend = ['Saturday', 'Sunday']\n week = ['Monday', 'Tuesday', 'Wednesday', 'Thursday', 'Friday', 'Saturday', 'Sunday']\n for day in week:\n if day in group.index:\n values.append(group[day])\n if day in weekday:\n avg_weekday = avg_weekday + group[day]/len(weekday)\n else:\n avg_weekend = avg_weekend + group[day]/len(weekend)\n else:\n values.append(0.0)\n return week, values, avg_weekend, avg_weekday\n\ntrip['day_name'] = trip['start_date'].dt.day_name()\ngroup = trip.groupby(['day_name', 'subscription_type']).size().unstack(level=1, fill_value=0)\ndays, trips_subscriber, avg_weekend, avg_weekday = get_ordered_data(group.Subscriber)\n_, trips_customer, _, _ = get_ordered_data(group.Customer)\ntrips_subscriber\nfig, ax = plt.subplots(figsize=(8,8))\nax.set_title('Number of trips by day')\nax.set_ylabel('Number of trips')\nx = np.arange(len(days))\nwidth = 0.35\nax.bar(x + width/2, trips_subscriber, width, label='Subscriber',color='#1f77b4')\nax.bar(x - width/2, trips_customer, width, label='Customer',color='#ff7f0e')\nax.set_xticks(x)\nax.set_xticklabels(days)\nax.legend()\nprint(\"Subscribers use bikes more on weekday. There is a big difference between weekday and weekend.\")\nprint(\"Average number of trips during the weekday:\", avg_weekday)\nprint(\"Average number of trips during the weekend:\", avg_weekend)",
"Subscribers use bikes more on weekday. There is a big difference between weekday and weekend.\nAverage number of trips during the weekday: 105076.4\nAverage number of trips during the weekend: 20682.0\n"
],
[
"trip['hour'] = trip['start_date'].dt.hour\ngroup = trip.groupby(['hour', 'subscription_type']).size().unstack(level=1, fill_value=0)\nfig, ax = plt.subplots(figsize=(8,8))\nax.set_title('Number of trips by hour')\nax.set_ylabel('Number of trips')\nhours = group.index\nx = np.arange(len(hours))\nwidth = 0.35\nax.bar(x + width/2, group.Subscriber.values, width, label='Subscriber',color='#1f77b4')\nax.bar(x - width/2, group.Customer.values, width, label='Customer',color='#ff7f0e')\nax.set_xticks(x)\nax.set_xticklabels(hours)\nax.legend()\nprint(\"The Subscriber uses the bike in a greater proportion during rush hours.\")\nprint(\"Morning: 7:00 AM - 9:00 AM\")\nprint(\"Evening: 16:00 AM - 18:00 AM\")",
"The Subscriber uses the bike in a greater proportion during rush hours.\nMorning: 7:00 AM - 9:00 AM\nEvening: 16:00 AM - 18:00 AM\n"
],
[
"trip['duration_min'] = (trip['duration']/60.0).apply(np.floor).astype(int)\ngroup = trip.groupby(['duration_min', 'subscription_type']).size().unstack(level=1, fill_value=0)\nfig, ax = plt.subplots(figsize=(20,5))\ncondition = (group.Subscriber.index <= 60)\ngroup.Subscriber[condition]\nax.set_title('Number of trips by first 60 minutes')\nax.set_ylabel('Number of trips')\nmins = group.Subscriber[condition].index\nx = np.arange(len(mins))\nwidth = 0.35\nax.bar(x + width/2, group.Subscriber[condition].values, width, label='Subscriber',color='#1f77b4')\nax.bar(x - width/2, group.Customer[condition].values, width, label='Customer',color='#ff7f0e')\nax.set_xticks(x)\nax.set_xticklabels(mins)\nax.legend()\navg_time = (sum(group.Subscriber[condition].values*mins)/sum(group.Subscriber[condition].values))\nprint(\"The subscripter use the bike {} minutes in average.\".format(round(avg_time, 2)))\nprint(\"The most frequently used range is between: [2, 15] minutes.\")",
"The subscripter use the bike 8.24 minutes in average.\nThe most frequently used range is between: [2, 15] minutes.\n"
],
[
"trip['start_station_name'] = pd.Categorical(trip['start_station_name'])\nmost_used = trip['start_station_name'].value_counts().nlargest(10)\nfig, ax = plt.subplots(figsize=(15,5))\nax.set_ylabel('Number of trips')\nax.set_title('Top 10 Most frequent start station')\nax.bar(most_used.index.values, most_used.values)\nfor tick in ax.get_xticklabels():\n tick.set_rotation(90)\nprint(\"The most frequent start station is: \", most_used.index.values[0])",
"The most frequent start station is: San Francisco Caltrain (Townsend at 4th)\n"
],
[
"trip['end_station_name'] = pd.Categorical(trip['end_station_name'])\nmost_used = trip['end_station_name'].value_counts().nlargest(10)\nfig, ax = plt.subplots(figsize=(15,5))\nax.set_ylabel('Number of trips')\nax.set_title('Top 10 Most frequent end station')\nax.bar(most_used.index.values, most_used.values)\nfor tick in ax.get_xticklabels():\n tick.set_rotation(90)\nmost_used.index.values[0]\nprint(\"The most frequent end station is: \", most_used.index.values[0])",
"The most frequent end station is: San Francisco Caltrain (Townsend at 4th)\n"
],
[
"group = trip.groupby(['start_station_name', 'end_station_name']).size().unstack(level=1, fill_value=0)\ncond = (group.index == group.max(axis=1).nlargest(1).index[0])\nmost_start_station = group.max(axis=1).nlargest(1).index[0]\nmost_end_station = group[cond].max().nlargest(1)\nprint('Trip start-end most used:', most_start_station,'-->', most_end_station.index[0],', N Trips = ', most_end_station.values[0])",
"Trip start-end most used: San Francisco Caltrain 2 (330 Townsend) --> Townsend at 7th , N Trips = 6216\n"
],
[
"fig, ax = plt.subplots(figsize=(15,5))\nax.set_ylabel('Number of trips')\nax.set_title('bike_id')\nmost_used = trip['bike_id'].value_counts()\nax.bar(most_used.index.values, most_used.values, color='blue')\nprint(\"Some bikes are used in greater frequency.\")",
"Some bikes are used in greater frequency.\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7941e159291604560814c8cd7731c73fb27ceea | 26,070 | ipynb | Jupyter Notebook | 04_sagemaker_debugger/tf-mnist-builtin-rule.ipynb | tom5610/amazon_sagemaker_intermediate_workshop | 9059d702d70fd066e8a50bfad392a1dda85958cd | [
"MIT"
] | null | null | null | 04_sagemaker_debugger/tf-mnist-builtin-rule.ipynb | tom5610/amazon_sagemaker_intermediate_workshop | 9059d702d70fd066e8a50bfad392a1dda85958cd | [
"MIT"
] | null | null | null | 04_sagemaker_debugger/tf-mnist-builtin-rule.ipynb | tom5610/amazon_sagemaker_intermediate_workshop | 9059d702d70fd066e8a50bfad392a1dda85958cd | [
"MIT"
] | null | null | null | 28.80663 | 856 | 0.584388 | [
[
[
"# Amazon SageMaker Debugger Tutorial: How to Use the Built-in Debugging Rules",
"_____no_output_____"
],
[
"[Amazon SageMaker Debugger](https://docs.aws.amazon.com/sagemaker/latest/dg/train-debugger.html) is a feature that offers capability to debug training jobs of your machine learning model and identify training problems in real time. While a training job looks like it's working like a charm, the model might have some common problems, such as loss not decreasing, overfitting, and underfitting. To better understand, practitioners have to debug the training job, while it can be challenging to track and analyze all of the output tensors.\n\nSageMaker Debugger covers the major deep learning frameworks (TensorFlow, PyTorch, and MXNet) and machine learning algorithm (XGBoost) to do the debugging jobs with minimal coding. Debugger provides an automatic detection of training problems through its built-in rules, and you can find a full list of the built-in rules for debugging at [List of Debugger Built-in Rules](https://docs.aws.amazon.com/sagemaker/latest/dg/debugger-built-in-rules.html). \n\nIn this tutorial, you will learn how to use SageMaker Debugger and its built-in rules to debug your model.\n\nThe workflow is as follows:\n* [Step 1: Import SageMaker Python SDK and the Debugger client library smdebug](#step1)\n* [Step 2: Create a Debugger built-in rule list object](#step2)\n* [Step 3: Construct a SageMaker estimator](#step3)\n* [Step 4: Run the training job](#step4)\n* [Step 5: Check training progress on Studio Debugger insights dashboard and the built-in rules evaluation status](#step5)\n* [Step 6: Create a Debugger trial object to access the saved tensors](#step6)",
"_____no_output_____"
],
[
"<a class=\"anchor\" id=\"step2\"></a>\n## Step 1: Import SageMaker Python SDK and the SMDebug client library",
"_____no_output_____"
],
[
"<font color='red'>**Important**</font>: To use the new Debugger features, you need to upgrade the SageMaker Python SDK and the SMDebug libary. In the following cell, change the third line to `install_needed=True` and run to upgrade the libraries.",
"_____no_output_____"
]
],
[
[
"import sys\nimport IPython\ninstall_needed = True # Set to True to upgrade\nif install_needed:\n print(\"installing deps and restarting kernel\")\n !{sys.executable} -m pip install -U sagemaker\n !{sys.executable} -m pip install smdebug matplotlib\n IPython.Application.instance().kernel.do_shutdown(True)",
"_____no_output_____"
]
],
[
[
"Check the SageMaker Python SDK and the SMDebug library versions.",
"_____no_output_____"
]
],
[
[
"import sagemaker\n\nsagemaker.__version__",
"_____no_output_____"
],
[
"import smdebug\n\nsmdebug.__version__",
"_____no_output_____"
]
],
[
[
"<a class=\"anchor\" id=\"step1\"></a>\n## Step 2: Create a Debugger built-in rule list object",
"_____no_output_____"
]
],
[
[
"from sagemaker.debugger import (\n Rule, \n rule_configs,\n ProfilerRule,\n ProfilerConfig,\n FrameworkProfile,\n DetailedProfilingConfig,\n DataloaderProfilingConfig,\n PythonProfilingConfig,\n)",
"_____no_output_____"
]
],
[
[
"The following code cell shows how to configure a rule object for debugging and profiling. For more information about the Debugger built-in rules, see [List of Debugger Built-in Rules](https://docs.aws.amazon.com/sagemaker/latest/dg/debugger-built-in-rules.html).\n\nThe following cell demo how to configure system and framework profiling.",
"_____no_output_____"
]
],
[
[
"profiler_config = ProfilerConfig(\n system_monitor_interval_millis=500,\n framework_profile_params=FrameworkProfile(\n local_path=\"/opt/ml/output/profiler/\",\n detailed_profiling_config=DetailedProfilingConfig(start_step=5, num_steps=3),\n dataloader_profiling_config=DataloaderProfilingConfig(start_step=5, num_steps=2),\n python_profiling_config=PythonProfilingConfig(start_step=9, num_steps=1),\n ),\n)\nbuilt_in_rules = [\n Rule.sagemaker(rule_configs.overfit()),\n ProfilerRule.sagemaker(rule_configs.ProfilerReport()),\n]",
"_____no_output_____"
]
],
[
[
"<a class=\"anchor\" id=\"step3\"></a>\n## Step 3: Construct a SageMaker estimator\n\nUsing the rule object created in the previous cell, construct a SageMaker estimator. \n\nThe estimator can be one of the SageMaker framework estimators, [TensorFlow](https://sagemaker.readthedocs.io/en/stable/frameworks/tensorflow/sagemaker.tensorflow.html#tensorflow-estimator), [PyTorch](https://sagemaker.readthedocs.io/en/stable/frameworks/pytorch/sagemaker.pytorch.html), [MXNet](https://sagemaker.readthedocs.io/en/stable/frameworks/mxnet/sagemaker.mxnet.html#mxnet-estimator), and [XGBoost](https://sagemaker.readthedocs.io/en/stable/frameworks/xgboost/xgboost.html), or the [SageMaker generic estimator](https://sagemaker.readthedocs.io/en/stable/api/training/estimators.html#sagemaker.estimator.Estimator). For more information about what framework versions are supported, see [Debugger-supported Frameworks and Algorithms](https://docs.aws.amazon.com/sagemaker/latest/dg/train-debugger.html#debugger-supported-aws-containers).\n\nIn this tutorial, the SageMaker TensorFlow estimator is constructed to run a TensorFlow training script with the Keras ResNet50 model and the cifar10 dataset.",
"_____no_output_____"
]
],
[
[
"import boto3\nfrom sagemaker.tensorflow import TensorFlow\n\nsession = boto3.session.Session()\nregion = session.region_name\n\nestimator = TensorFlow(\n role=sagemaker.get_execution_role(),\n instance_count=1,\n instance_type=\"ml.g4dn.xlarge\",\n image_uri=f\"763104351884.dkr.ecr.{region}.amazonaws.com/tensorflow-training:2.3.1-gpu-py37-cu110-ubuntu18.04\",\n # framework_version='2.3.1',\n # py_version=\"py37\",\n max_run=3600,\n source_dir=\"./src\",\n entry_point=\"tf-resnet50-cifar10.py\",\n # Debugger Parameters\n rules=built_in_rules,\n profiler_config=profiler_config\n)",
"_____no_output_____"
]
],
[
[
"<a class=\"anchor\" id=\"step4\"></a>\n## Step 4: Run the training job\nWith the `wait=False` option, you can proceed to the next notebook cell without waiting for the training job logs to be printed out.",
"_____no_output_____"
]
],
[
[
"estimator.fit(wait=True)",
"_____no_output_____"
]
],
[
[
"<a class=\"anchor\" id=\"step5\"></a>\n## Step 5: Check training progress on Studio Debugger insights dashboard and the built-in rules evaluation status\n\n- **Option 1** - Use SageMaker Studio Debugger insights and Experiments. This is a non-coding approach.\n- **Option 2** - Use the following code cells. This is a code-based approach. ",
"_____no_output_____"
],
[
"#### Option 1 - Open Studio Debugger insights dashboard to get insights into the training job\n\nThrough the Debugger insights dashboard on Studio, you can check the training jobs status, system resource utilization, and suggestions to optimize model performance. The following screenshot shows the Debugger insights dashboard interface.\n\n<IMG src=\"./images/studio-debugger-insights-dashboard.png\"/>\n\nThe following heatmap shows the `ml.c5.4xlarge` instance utilization while the training job is running or after the job has completed. To learn how to access the Debugger insights dashboard, see [Debugger on Studio](https://docs.aws.amazon.com/sagemaker/latest/dg/debugger-on-studio.html) in the [SageMaker Debugger developer guide](https://docs.aws.amazon.com/sagemaker/latest/dg/train-debugger.html).\n\n<IMG src=\"./images/studio-debugger-insights-heatmap.png\"/>",
"_____no_output_____"
],
[
"#### Option 2 - Run the following scripts for the code-based option\n\nThe following two code cells return the current training job name, status, and the rule status in real time.",
"_____no_output_____"
],
[
"##### Print the training job name",
"_____no_output_____"
]
],
[
[
"job_name = estimator.latest_training_job.name\nprint(\"Training job name: {}\".format(job_name))",
"_____no_output_____"
]
],
[
[
"##### Print the training job and rule evaluation status\n\nThe following script returns the status in real time every 15 seconds, until the secondary training status turns to one of the descriptions, `Training`, `Stopped`, `Completed`, or `Failed`. Once the training job status turns into the `Training`, you will be able to retrieve tensors from the default S3 bucket.",
"_____no_output_____"
]
],
[
[
"import time\n\nclient = estimator.sagemaker_session.sagemaker_client\ndescription = client.describe_training_job(TrainingJobName=job_name)\nif description[\"TrainingJobStatus\"] != \"Completed\":\n while description[\"SecondaryStatus\"] not in {\"Training\", \"Stopped\", \"Completed\", \"Failed\"}:\n description = client.describe_training_job(TrainingJobName=job_name)\n primary_status = description[\"TrainingJobStatus\"]\n secondary_status = description[\"SecondaryStatus\"]\n print(\n \"Current job status: [PrimaryStatus: {}, SecondaryStatus: {}] | {} Rule Evaluation Status: {}\".format(\n primary_status,\n secondary_status,\n estimator.latest_training_job.rule_job_summary()[0][\"RuleConfigurationName\"],\n estimator.latest_training_job.rule_job_summary()[0][\"RuleEvaluationStatus\"],\n )\n )\n time.sleep(30)",
"_____no_output_____"
]
],
[
[
"<a class=\"anchor\" id=\"step6\"></a>\n## Step 6: Create a Debugger trial object to access the saved model parameters\n\nTo access the saved tensors by Debugger, use the `smdebug` client library to create a Debugger trial object. The following code cell sets up a `tutorial_trial` object, and waits until it finds available tensors from the default S3 bucket.",
"_____no_output_____"
]
],
[
[
"from smdebug.trials import create_trial\n\ntutorial_trial = create_trial(estimator.latest_job_debugger_artifacts_path())",
"_____no_output_____"
]
],
[
[
"The Debugger trial object accesses the SageMaker estimator's Debugger artifact path, and fetches the output tensors saved for debugging.",
"_____no_output_____"
],
[
"#### Print the default S3 bucket URI where the Debugger output tensors are stored",
"_____no_output_____"
]
],
[
[
"tutorial_trial.path",
"_____no_output_____"
]
],
[
[
"#### Print the Debugger output tensor names",
"_____no_output_____"
]
],
[
[
"tutorial_trial.tensor_names()",
"_____no_output_____"
]
],
[
[
"#### Print the list of steps where the tensors are saved",
"_____no_output_____"
],
[
"The smdebug `ModeKeys` class provides training phase mode keys that you can use to sort training (`TRAIN`) and validation (`EVAL`) steps and their corresponding values.",
"_____no_output_____"
]
],
[
[
"from smdebug.core.modes import ModeKeys",
"_____no_output_____"
],
[
"tutorial_trial.steps(mode=ModeKeys.TRAIN)",
"_____no_output_____"
],
[
"tutorial_trial.steps(mode=ModeKeys.EVAL)",
"_____no_output_____"
]
],
[
[
"#### Plot the loss curve\n\nThe following script plots the loss and accuracy curves of training and validation loops.",
"_____no_output_____"
]
],
[
[
"trial = tutorial_trial\n\n\ndef get_data(trial, tname, mode):\n tensor = trial.tensor(tname)\n steps = tensor.steps(mode=mode)\n vals = [tensor.value(s, mode=mode) for s in steps]\n return steps, vals",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nfrom mpl_toolkits.axes_grid1 import host_subplot\n\n\ndef plot_tensor(trial, tensor_name):\n\n tensor_name = tensor_name\n steps_train, vals_train = get_data(trial, tensor_name, mode=ModeKeys.TRAIN)\n steps_eval, vals_eval = get_data(trial, tensor_name, mode=ModeKeys.EVAL)\n\n fig = plt.figure(figsize=(10, 7))\n host = host_subplot(111)\n\n par = host.twiny()\n\n host.set_xlabel(\"Steps (TRAIN)\")\n par.set_xlabel(\"Steps (EVAL)\")\n host.set_ylabel(tensor_name)\n\n (p1,) = host.plot(steps_train, vals_train, label=tensor_name)\n (p2,) = par.plot(steps_eval, vals_eval, label=\"val_\" + tensor_name)\n\n leg = plt.legend()\n\n host.xaxis.get_label().set_color(p1.get_color())\n leg.texts[0].set_color(p1.get_color())\n\n par.xaxis.get_label().set_color(p2.get_color())\n leg.texts[1].set_color(p2.get_color())\n\n plt.ylabel(tensor_name)\n\n plt.show()\n\n\nplot_tensor(trial, \"loss\")\nplot_tensor(trial, \"accuracy\")",
"_____no_output_____"
]
],
[
[
"> ## Note : Rerun the above cell if you don't see any plots! ",
"_____no_output_____"
],
[
"## Conclusion\n\nIn this tutorial, you learned how to use SageMaker Debugger with the minimal coding through SageMaker Studio and Jupyter notebook. The Debugger built-in rules detect training anomalies while concurrently reading in the output tensors, such as weights, activation outputs, gradients, accuracy, and loss, from your training jobs. In the next tutorial videos, you will learn more features of Debugger, such as how to analyze the tensors, change the built-in debugging rule parameters and thresholds, and save the tensors at your preferred S3 bucket URI.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
e7942a14773659bbc0a18b770ed155f6eceb094e | 3,856 | ipynb | Jupyter Notebook | staging/arrays_strings/reverse_words/reverse_words_solution.ipynb | sophomore99/PythonInterective | f2ff4d798274218e8543e071141b60c35e86a3eb | [
"Apache-2.0"
] | 8 | 2017-04-16T03:30:36.000Z | 2021-02-04T06:45:30.000Z | staging/arrays_strings/reverse_words/reverse_words_solution.ipynb | sophomore99/PythonInterective | f2ff4d798274218e8543e071141b60c35e86a3eb | [
"Apache-2.0"
] | null | null | null | staging/arrays_strings/reverse_words/reverse_words_solution.ipynb | sophomore99/PythonInterective | f2ff4d798274218e8543e071141b60c35e86a3eb | [
"Apache-2.0"
] | 7 | 2017-09-18T09:19:02.000Z | 2019-11-22T06:15:50.000Z | 23.656442 | 166 | 0.514004 | [
[
[
"<small> <i> This notebook was prepared by Marco Guajardo. For license visit [github](https://github.com/donnemartin/interactive-coding-challenges) </i> </small>",
"_____no_output_____"
],
[
"# Solution notebook\n## Problem: Given a string of words, return a string with the words in reverse",
"_____no_output_____"
],
[
"* [Constraits](#Constraint)\n* [Test Cases](#Test-Cases)\n* [Algorithm](#Algorithm)\n* [Code](#Code)\n* [Unit Test](#Unit-Test)\n* [Solution Notebook](#Solution-Notebook)",
"_____no_output_____"
],
[
"## Constraints\n* Can we assume the string is ASCII?\n * Yes\n* Is whitespace important?\n * no the whitespace does not change\n* Is this case sensitive?\n * yes\n* What if the string is empty?\n * return None\n* Is the order of words important?\n * yes\n\n",
"_____no_output_____"
],
[
"## Algorithm: Split words into a list and reverse each word individually\nSteps:\n\n* Check if string is empty\n* If not empty, split the string into a list of words \n* For each word on the list\n * reverse the word\n* Return the string representation of the list\n\nComplexity:\n\n* Time complexity is O(n) where n is the number of chars.\n* Space complexity is O(n) where n is the number of chars. ",
"_____no_output_____"
]
],
[
[
"def reverse_words(S):\n if len(S) is 0:\n return None\n \n words = S.split()\n for i in range (len(words)):\n words[i] = words[i][::-1]\n \n return \" \".join(words)",
"_____no_output_____"
],
[
"%%writefile reverse_words_solution.py\nfrom nose.tools import assert_equal\n\nclass UnitTest (object):\n def testReverseWords(self, func):\n assert_equal(func('the sun is hot'), 'eht nus si toh')\n assert_equal(func(''), None)\n assert_equal(func('123 456 789'), '321 654 987')\n assert_equal(func('magic'), 'cigam')\n print('Success: reverse_words')\n \ndef main():\n test = UnitTest()\n test.testReverseWords(reverse_words)\n\nif __name__==\"__main__\":\n main()",
"Overwriting reverse_words_solution.py\n"
],
[
"run -i reverse_words_solution.py",
"Success: reverse_words\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
] |
e7942c806a5b8f96ec43f5709734c8222276cdb7 | 47,370 | ipynb | Jupyter Notebook | site/en/r1/guide/eager.ipynb | PRUBHTEJ/docs-1 | 5100e91cbdfb09dc62e4eb4826569157c49256e4 | [
"Apache-2.0"
] | null | null | null | site/en/r1/guide/eager.ipynb | PRUBHTEJ/docs-1 | 5100e91cbdfb09dc62e4eb4826569157c49256e4 | [
"Apache-2.0"
] | null | null | null | site/en/r1/guide/eager.ipynb | PRUBHTEJ/docs-1 | 5100e91cbdfb09dc62e4eb4826569157c49256e4 | [
"Apache-2.0"
] | null | null | null | 30.880052 | 237 | 0.49829 | [
[
[
"##### Copyright 2018 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Eager Execution\n",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/r1/guide/eager.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs/blob/master/site/en/r1/guide/eager.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"> Note: This is an archived TF1 notebook. These are configured\nto run in TF2's \n[compatbility mode](https://www.tensorflow.org/guide/migrate)\nbut will run in TF1 as well. To use TF1 in Colab, use the\n[%tensorflow_version 1.x](https://colab.research.google.com/notebooks/tensorflow_version.ipynb)\nmagic.",
"_____no_output_____"
],
[
"TensorFlow's eager execution is an imperative programming environment that\nevaluates operations immediately, without building graphs: operations return\nconcrete values instead of constructing a computational graph to run later. This\nmakes it easy to get started with TensorFlow and debug models, and it\nreduces boilerplate as well. To follow along with this guide, run the code\nsamples below in an interactive `python` interpreter.\n\nEager execution is a flexible machine learning platform for research and\nexperimentation, providing:\n\n* *An intuitive interface*—Structure your code naturally and use Python data\n structures. Quickly iterate on small models and small data.\n* *Easier debugging*—Call ops directly to inspect running models and test\n changes. Use standard Python debugging tools for immediate error reporting.\n* *Natural control flow*—Use Python control flow instead of graph control\n flow, simplifying the specification of dynamic models.\n\nEager execution supports most TensorFlow operations and GPU acceleration. For a\ncollection of examples running in eager execution, see:\n[tensorflow/contrib/eager/python/examples](https://github.com/tensorflow/tensorflow/tree/master/tensorflow/contrib/eager/python/examples).\n\nNote: Some models may experience increased overhead with eager execution\nenabled. Performance improvements are ongoing, but please\n[file a bug](https://github.com/tensorflow/tensorflow/issues) if you find a\nproblem and share your benchmarks.",
"_____no_output_____"
],
[
"## Setup and basic usage\n",
"_____no_output_____"
],
[
"To start eager execution, add `` to the beginning of\nthe program or console session. Do not add this operation to other modules that\nthe program calls.",
"_____no_output_____"
]
],
[
[
"import tensorflow.compat.v1 as tf",
"_____no_output_____"
]
],
[
[
"Now you can run TensorFlow operations and the results will return immediately:",
"_____no_output_____"
]
],
[
[
"tf.executing_eagerly()",
"_____no_output_____"
],
[
"x = [[2.]]\nm = tf.matmul(x, x)\nprint(\"hello, {}\".format(m))",
"_____no_output_____"
]
],
[
[
"Enabling eager execution changes how TensorFlow operations behave—now they\nimmediately evaluate and return their values to Python. `tf.Tensor` objects\nreference concrete values instead of symbolic handles to nodes in a computational\ngraph. Since there isn't a computational graph to build and run later in a\nsession, it's easy to inspect results using `print()` or a debugger. Evaluating,\nprinting, and checking tensor values does not break the flow for computing\ngradients.\n\nEager execution works nicely with [NumPy](http://www.numpy.org/). NumPy\noperations accept `tf.Tensor` arguments. TensorFlow\n[math operations](https://www.tensorflow.org/api_guides/python/math_ops) convert\nPython objects and NumPy arrays to `tf.Tensor` objects. The\n`tf.Tensor.numpy` method returns the object's value as a NumPy `ndarray`.",
"_____no_output_____"
]
],
[
[
"a = tf.constant([[1, 2],\n [3, 4]])\nprint(a)",
"_____no_output_____"
],
[
"# Broadcasting support\nb = tf.add(a, 1)\nprint(b)",
"_____no_output_____"
],
[
"# Operator overloading is supported\nprint(a * b)",
"_____no_output_____"
],
[
"# Use NumPy values\nimport numpy as np\n\nc = np.multiply(a, b)\nprint(c)",
"_____no_output_____"
],
[
"# Obtain numpy value from a tensor:\nprint(a.numpy())\n# => [[1 2]\n# [3 4]]",
"_____no_output_____"
]
],
[
[
"## Dynamic control flow\n\nA major benefit of eager execution is that all the functionality of the host\nlanguage is available while your model is executing. So, for example,\nit is easy to write [fizzbuzz](https://en.wikipedia.org/wiki/Fizz_buzz):",
"_____no_output_____"
]
],
[
[
"def fizzbuzz(max_num):\n counter = tf.constant(0)\n max_num = tf.convert_to_tensor(max_num)\n for num in range(1, max_num.numpy()+1):\n num = tf.constant(num)\n if int(num % 3) == 0 and int(num % 5) == 0:\n print('FizzBuzz')\n elif int(num % 3) == 0:\n print('Fizz')\n elif int(num % 5) == 0:\n print('Buzz')\n else:\n print(num.numpy())\n counter += 1",
"_____no_output_____"
],
[
"fizzbuzz(15)",
"_____no_output_____"
]
],
[
[
"This has conditionals that depend on tensor values and it prints these values\nat runtime.",
"_____no_output_____"
],
[
"## Build a model\n\nMany machine learning models are represented by composing layers. When\nusing TensorFlow with eager execution you can either write your own layers or\nuse a layer provided in the `tf.keras.layers` package.\n\nWhile you can use any Python object to represent a layer,\nTensorFlow has `tf.keras.layers.Layer` as a convenient base class. Inherit from\nit to implement your own layer:",
"_____no_output_____"
]
],
[
[
"class MySimpleLayer(tf.keras.layers.Layer):\n def __init__(self, output_units):\n super(MySimpleLayer, self).__init__()\n self.output_units = output_units\n\n def build(self, input_shape):\n # The build method gets called the first time your layer is used.\n # Creating variables on build() allows you to make their shape depend\n # on the input shape and hence removes the need for the user to specify\n # full shapes. It is possible to create variables during __init__() if\n # you already know their full shapes.\n self.kernel = self.add_variable(\n \"kernel\", [input_shape[-1], self.output_units])\n\n def call(self, input):\n # Override call() instead of __call__ so we can perform some bookkeeping.\n return tf.matmul(input, self.kernel)",
"_____no_output_____"
]
],
[
[
"Use `tf.keras.layers.Dense` layer instead of `MySimpleLayer` above as it has\na superset of its functionality (it can also add a bias).\n\nWhen composing layers into models you can use `tf.keras.Sequential` to represent\nmodels which are a linear stack of layers. It is easy to use for basic models:",
"_____no_output_____"
]
],
[
[
"model = tf.keras.Sequential([\n tf.keras.layers.Dense(10, input_shape=(784,)), # must declare input shape\n tf.keras.layers.Dense(10)\n])",
"_____no_output_____"
]
],
[
[
"Alternatively, organize models in classes by inheriting from `tf.keras.Model`.\nThis is a container for layers that is a layer itself, allowing `tf.keras.Model`\nobjects to contain other `tf.keras.Model` objects.",
"_____no_output_____"
]
],
[
[
"class MNISTModel(tf.keras.Model):\n def __init__(self):\n super(MNISTModel, self).__init__()\n self.dense1 = tf.keras.layers.Dense(units=10)\n self.dense2 = tf.keras.layers.Dense(units=10)\n\n def call(self, input):\n \"\"\"Run the model.\"\"\"\n result = self.dense1(input)\n result = self.dense2(result)\n result = self.dense2(result) # reuse variables from dense2 layer\n return result\n\nmodel = MNISTModel()",
"_____no_output_____"
]
],
[
[
"It's not required to set an input shape for the `tf.keras.Model` class since\nthe parameters are set the first time input is passed to the layer.\n\n`tf.keras.layers` classes create and contain their own model variables that\nare tied to the lifetime of their layer objects. To share layer variables, share\ntheir objects.",
"_____no_output_____"
],
[
"## Eager training",
"_____no_output_____"
],
[
"### Computing gradients\n\n[Automatic differentiation](https://en.wikipedia.org/wiki/Automatic_differentiation)\nis useful for implementing machine learning algorithms such as\n[backpropagation](https://en.wikipedia.org/wiki/Backpropagation) for training\nneural networks. During eager execution, use `tf.GradientTape` to trace\noperations for computing gradients later.\n\n`tf.GradientTape` is an opt-in feature to provide maximal performance when\nnot tracing. Since different operations can occur during each call, all\nforward-pass operations get recorded to a \"tape\". To compute the gradient, play\nthe tape backwards and then discard. A particular `tf.GradientTape` can only\ncompute one gradient; subsequent calls throw a runtime error.",
"_____no_output_____"
]
],
[
[
"w = tf.Variable([[1.0]])\nwith tf.GradientTape() as tape:\n loss = w * w\n\ngrad = tape.gradient(loss, w)\nprint(grad) # => tf.Tensor([[ 2.]], shape=(1, 1), dtype=float32)",
"_____no_output_____"
]
],
[
[
"### Train a model\n\nThe following example creates a multi-layer model that classifies the standard\nMNIST handwritten digits. It demonstrates the optimizer and layer APIs to build\ntrainable graphs in an eager execution environment.",
"_____no_output_____"
]
],
[
[
"# Fetch and format the mnist data\n(mnist_images, mnist_labels), _ = tf.keras.datasets.mnist.load_data()\n\ndataset = tf.data.Dataset.from_tensor_slices(\n (tf.cast(mnist_images[...,tf.newaxis]/255, tf.float32),\n tf.cast(mnist_labels,tf.int64)))\ndataset = dataset.shuffle(1000).batch(32)",
"_____no_output_____"
],
[
"# Build the model\nmnist_model = tf.keras.Sequential([\n tf.keras.layers.Conv2D(16,[3,3], activation='relu'),\n tf.keras.layers.Conv2D(16,[3,3], activation='relu'),\n tf.keras.layers.GlobalAveragePooling2D(),\n tf.keras.layers.Dense(10)\n])",
"_____no_output_____"
]
],
[
[
"Even without training, call the model and inspect the output in eager execution:",
"_____no_output_____"
]
],
[
[
"for images,labels in dataset.take(1):\n print(\"Logits: \", mnist_model(images[0:1]).numpy())",
"_____no_output_____"
]
],
[
[
"While keras models have a builtin training loop (using the `fit` method), sometimes you need more customization. Here's an example, of a training loop implemented with eager:",
"_____no_output_____"
]
],
[
[
"optimizer = tf.train.AdamOptimizer()\n\nloss_history = []",
"_____no_output_____"
],
[
"for (batch, (images, labels)) in enumerate(dataset.take(400)):\n if batch % 10 == 0:\n print('.', end='')\n with tf.GradientTape() as tape:\n logits = mnist_model(images, training=True)\n loss_value = tf.losses.sparse_softmax_cross_entropy(labels, logits)\n\n loss_history.append(loss_value.numpy())\n grads = tape.gradient(loss_value, mnist_model.trainable_variables)\n optimizer.apply_gradients(zip(grads, mnist_model.trainable_variables),\n global_step=tf.train.get_or_create_global_step())",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n\nplt.plot(loss_history)\nplt.xlabel('Batch #')\nplt.ylabel('Loss [entropy]')",
"_____no_output_____"
]
],
[
[
"### Variables and optimizers\n\n`tf.Variable` objects store mutable `tf.Tensor` values accessed during\ntraining to make automatic differentiation easier. The parameters of a model can\nbe encapsulated in classes as variables.\n\nBetter encapsulate model parameters by using `tf.Variable` with\n`tf.GradientTape`. For example, the automatic differentiation example above\ncan be rewritten:",
"_____no_output_____"
]
],
[
[
"class Model(tf.keras.Model):\n def __init__(self):\n super(Model, self).__init__()\n self.W = tf.Variable(5., name='weight')\n self.B = tf.Variable(10., name='bias')\n def call(self, inputs):\n return inputs * self.W + self.B\n\n# A toy dataset of points around 3 * x + 2\nNUM_EXAMPLES = 2000\ntraining_inputs = tf.random_normal([NUM_EXAMPLES])\nnoise = tf.random_normal([NUM_EXAMPLES])\ntraining_outputs = training_inputs * 3 + 2 + noise\n\n# The loss function to be optimized\ndef loss(model, inputs, targets):\n error = model(inputs) - targets\n return tf.reduce_mean(tf.square(error))\n\ndef grad(model, inputs, targets):\n with tf.GradientTape() as tape:\n loss_value = loss(model, inputs, targets)\n return tape.gradient(loss_value, [model.W, model.B])\n\n# Define:\n# 1. A model.\n# 2. Derivatives of a loss function with respect to model parameters.\n# 3. A strategy for updating the variables based on the derivatives.\nmodel = Model()\noptimizer = tf.train.GradientDescentOptimizer(learning_rate=0.01)\n\nprint(\"Initial loss: {:.3f}\".format(loss(model, training_inputs, training_outputs)))\n\n# Training loop\nfor i in range(300):\n grads = grad(model, training_inputs, training_outputs)\n optimizer.apply_gradients(zip(grads, [model.W, model.B]),\n global_step=tf.train.get_or_create_global_step())\n if i % 20 == 0:\n print(\"Loss at step {:03d}: {:.3f}\".format(i, loss(model, training_inputs, training_outputs)))\n\nprint(\"Final loss: {:.3f}\".format(loss(model, training_inputs, training_outputs)))\nprint(\"W = {}, B = {}\".format(model.W.numpy(), model.B.numpy()))",
"_____no_output_____"
]
],
[
[
"## Use objects for state during eager execution\n\nWith graph execution, program state (such as the variables) is stored in global\ncollections and their lifetime is managed by the `tf.Session` object. In\ncontrast, during eager execution the lifetime of state objects is determined by\nthe lifetime of their corresponding Python object.\n\n### Variables are objects\n\nDuring eager execution, variables persist until the last reference to the object\nis removed, and is then deleted.",
"_____no_output_____"
]
],
[
[
"if tf.test.is_gpu_available():\n with tf.device(\"gpu:0\"):\n v = tf.Variable(tf.random_normal([1000, 1000]))\n v = None # v no longer takes up GPU memory",
"_____no_output_____"
]
],
[
[
"### Object-based saving\n\n`tf.train.Checkpoint` can save and restore `tf.Variable`s to and from\ncheckpoints:",
"_____no_output_____"
]
],
[
[
"x = tf.Variable(10.)\ncheckpoint = tf.train.Checkpoint(x=x)",
"_____no_output_____"
],
[
"x.assign(2.) # Assign a new value to the variables and save.\ncheckpoint_path = './ckpt/'\ncheckpoint.save('./ckpt/')",
"_____no_output_____"
],
[
"x.assign(11.) # Change the variable after saving.\n\n# Restore values from the checkpoint\ncheckpoint.restore(tf.train.latest_checkpoint(checkpoint_path))\n\nprint(x) # => 2.0",
"_____no_output_____"
]
],
[
[
"To save and load models, `tf.train.Checkpoint` stores the internal state of objects,\nwithout requiring hidden variables. To record the state of a `model`,\nan `optimizer`, and a global step, pass them to a `tf.train.Checkpoint`:",
"_____no_output_____"
]
],
[
[
"import os\nimport tempfile\n\nmodel = tf.keras.Sequential([\n tf.keras.layers.Conv2D(16,[3,3], activation='relu'),\n tf.keras.layers.GlobalAveragePooling2D(),\n tf.keras.layers.Dense(10)\n])\noptimizer = tf.train.AdamOptimizer(learning_rate=0.001)\ncheckpoint_dir = tempfile.mkdtemp()\ncheckpoint_prefix = os.path.join(checkpoint_dir, \"ckpt\")\nroot = tf.train.Checkpoint(optimizer=optimizer,\n model=model,\n optimizer_step=tf.train.get_or_create_global_step())\n\nroot.save(checkpoint_prefix)\nroot.restore(tf.train.latest_checkpoint(checkpoint_dir))",
"_____no_output_____"
]
],
[
[
"### Object-oriented metrics\n\n`tf.metrics` are stored as objects. Update a metric by passing the new data to\nthe callable, and retrieve the result using the `tf.metrics.result` method,\nfor example:",
"_____no_output_____"
]
],
[
[
"m = tf.keras.metrics.Mean(\"loss\")\nm(0)\nm(5)\nm.result() # => 2.5\nm([8, 9])\nm.result() # => 5.5",
"_____no_output_____"
]
],
[
[
"#### Summaries and TensorBoard\n\n[TensorBoard](https://tensorflow.org/tensorboard) is a visualization tool for\nunderstanding, debugging and optimizing the model training process. It uses\nsummary events that are written while executing the program.\n\nTensorFlow 1 summaries only work in eager mode, but can be run with the `compat.v2` module:",
"_____no_output_____"
]
],
[
[
"from tensorflow.compat.v2 import summary\n\nglobal_step = tf.train.get_or_create_global_step()\n\nlogdir = \"./tb/\"\nwriter = summary.create_file_writer(logdir)\nwriter.set_as_default()\n\nfor _ in range(10):\n global_step.assign_add(1)\n # your model code goes here\n summary.scalar('global_step', global_step, step=global_step)",
"_____no_output_____"
],
[
"!ls tb/",
"_____no_output_____"
]
],
[
[
"## Advanced automatic differentiation topics\n\n### Dynamic models\n\n`tf.GradientTape` can also be used in dynamic models. This example for a\n[backtracking line search](https://wikipedia.org/wiki/Backtracking_line_search)\nalgorithm looks like normal NumPy code, except there are gradients and is\ndifferentiable, despite the complex control flow:",
"_____no_output_____"
]
],
[
[
"def line_search_step(fn, init_x, rate=1.0):\n with tf.GradientTape() as tape:\n # Variables are automatically recorded, but manually watch a tensor\n tape.watch(init_x)\n value = fn(init_x)\n grad = tape.gradient(value, init_x)\n grad_norm = tf.reduce_sum(grad * grad)\n init_value = value\n while value > init_value - rate * grad_norm:\n x = init_x - rate * grad\n value = fn(x)\n rate /= 2.0\n return x, value",
"_____no_output_____"
]
],
[
[
"### Custom gradients\n\nCustom gradients are an easy way to override gradients in eager and graph\nexecution. Within the forward function, define the gradient with respect to the\ninputs, outputs, or intermediate results. For example, here's an easy way to clip\nthe norm of the gradients in the backward pass:",
"_____no_output_____"
]
],
[
[
"@tf.custom_gradient\ndef clip_gradient_by_norm(x, norm):\n y = tf.identity(x)\n def grad_fn(dresult):\n return [tf.clip_by_norm(dresult, norm), None]\n return y, grad_fn",
"_____no_output_____"
]
],
[
[
"Custom gradients are commonly used to provide a numerically stable gradient for a\nsequence of operations:",
"_____no_output_____"
]
],
[
[
"def log1pexp(x):\n return tf.log(1 + tf.exp(x))\n\nclass Grad(object):\n def __init__(self, f):\n self.f = f\n\n def __call__(self, x):\n x = tf.convert_to_tensor(x)\n with tf.GradientTape() as tape:\n tape.watch(x)\n r = self.f(x)\n g = tape.gradient(r, x)\n return g",
"_____no_output_____"
],
[
"grad_log1pexp = Grad(log1pexp)",
"_____no_output_____"
],
[
"# The gradient computation works fine at x = 0.\ngrad_log1pexp(0.).numpy()",
"_____no_output_____"
],
[
"# However, x = 100 fails because of numerical instability.\ngrad_log1pexp(100.).numpy()",
"_____no_output_____"
]
],
[
[
"Here, the `log1pexp` function can be analytically simplified with a custom\ngradient. The implementation below reuses the value for `tf.exp(x)` that is\ncomputed during the forward pass—making it more efficient by eliminating\nredundant calculations:",
"_____no_output_____"
]
],
[
[
"@tf.custom_gradient\ndef log1pexp(x):\n e = tf.exp(x)\n def grad(dy):\n return dy * (1 - 1 / (1 + e))\n return tf.log(1 + e), grad\n\ngrad_log1pexp = Grad(log1pexp)",
"_____no_output_____"
],
[
"# As before, the gradient computation works fine at x = 0.\ngrad_log1pexp(0.).numpy()",
"_____no_output_____"
],
[
"# And the gradient computation also works at x = 100.\ngrad_log1pexp(100.).numpy()",
"_____no_output_____"
]
],
[
[
"## Performance\n\nComputation is automatically offloaded to GPUs during eager execution. If you\nwant control over where a computation runs you can enclose it in a\n`tf.device('/gpu:0')` block (or the CPU equivalent):",
"_____no_output_____"
]
],
[
[
"import time\n\ndef measure(x, steps):\n # TensorFlow initializes a GPU the first time it's used, exclude from timing.\n tf.matmul(x, x)\n start = time.time()\n for i in range(steps):\n x = tf.matmul(x, x)\n # tf.matmul can return before completing the matrix multiplication\n # (e.g., can return after enqueing the operation on a CUDA stream).\n # The x.numpy() call below will ensure that all enqueued operations\n # have completed (and will also copy the result to host memory,\n # so we're including a little more than just the matmul operation\n # time).\n _ = x.numpy()\n end = time.time()\n return end - start\n\nshape = (1000, 1000)\nsteps = 200\nprint(\"Time to multiply a {} matrix by itself {} times:\".format(shape, steps))\n\n# Run on CPU:\nwith tf.device(\"/cpu:0\"):\n print(\"CPU: {} secs\".format(measure(tf.random_normal(shape), steps)))\n\n# Run on GPU, if available:\nif tf.test.is_gpu_available():\n with tf.device(\"/gpu:0\"):\n print(\"GPU: {} secs\".format(measure(tf.random_normal(shape), steps)))\nelse:\n print(\"GPU: not found\")",
"_____no_output_____"
]
],
[
[
"A `tf.Tensor` object can be copied to a different device to execute its\noperations:",
"_____no_output_____"
]
],
[
[
"if tf.test.is_gpu_available():\n x = tf.random_normal([10, 10])\n\n x_gpu0 = x.gpu()\n x_cpu = x.cpu()\n\n _ = tf.matmul(x_cpu, x_cpu) # Runs on CPU\n _ = tf.matmul(x_gpu0, x_gpu0) # Runs on GPU:0",
"_____no_output_____"
]
],
[
[
"### Benchmarks\n\nFor compute-heavy models, such as\n[ResNet50](https://github.com/tensorflow/tensorflow/tree/master/tensorflow/contrib/eager/python/examples/resnet50)\ntraining on a GPU, eager execution performance is comparable to graph execution.\nBut this gap grows larger for models with less computation and there is work to\nbe done for optimizing hot code paths for models with lots of small operations.",
"_____no_output_____"
],
[
"## Work with graphs\n\nWhile eager execution makes development and debugging more interactive,\nTensorFlow graph execution has advantages for distributed training, performance\noptimizations, and production deployment. However, writing graph code can feel\ndifferent than writing regular Python code and more difficult to debug.\n\nFor building and training graph-constructed models, the Python program first\nbuilds a graph representing the computation, then invokes `Session.run` to send\nthe graph for execution on the C++-based runtime. This provides:\n\n* Automatic differentiation using static autodiff.\n* Simple deployment to a platform independent server.\n* Graph-based optimizations (common subexpression elimination, constant-folding, etc.).\n* Compilation and kernel fusion.\n* Automatic distribution and replication (placing nodes on the distributed system).\n\nDeploying code written for eager execution is more difficult: either generate a\ngraph from the model, or run the Python runtime and code directly on the server.",
"_____no_output_____"
],
[
"### Write compatible code\n\nThe same code written for eager execution will also build a graph during graph\nexecution. Do this by simply running the same code in a new Python session where\neager execution is not enabled.\n\nMost TensorFlow operations work during eager execution, but there are some things\nto keep in mind:\n\n* Use `tf.data` for input processing instead of queues. It's faster and easier.\n* Use object-oriented layer APIs—like `tf.keras.layers` and\n `tf.keras.Model`—since they have explicit storage for variables.\n* Most model code works the same during eager and graph execution, but there are\n exceptions. (For example, dynamic models using Python control flow to change the\n computation based on inputs.)\n* Once eager execution is enabled with `tf.enable_eager_execution`, it\n cannot be turned off. Start a new Python session to return to graph execution.\n\nIt's best to write code for both eager execution *and* graph execution. This\ngives you eager's interactive experimentation and debuggability with the\ndistributed performance benefits of graph execution.\n\nWrite, debug, and iterate in eager execution, then import the model graph for\nproduction deployment. Use `tf.train.Checkpoint` to save and restore model\nvariables, this allows movement between eager and graph execution environments.\nSee the examples in:\n[tensorflow/contrib/eager/python/examples](https://github.com/tensorflow/tensorflow/tree/master/tensorflow/contrib/eager/python/examples).\n",
"_____no_output_____"
],
[
"### Use eager execution in a graph environment\n\nSelectively enable eager execution in a TensorFlow graph environment using\n`tfe.py_func`. This is used when `` has *not*\nbeen called.",
"_____no_output_____"
]
],
[
[
"def my_py_func(x):\n x = tf.matmul(x, x) # You can use tf ops\n print(x) # but it's eager!\n return x\n\nwith tf.Session() as sess:\n x = tf.placeholder(dtype=tf.float32)\n # Call eager function in graph!\n pf = tf.py_func(my_py_func, [x], tf.float32)\n\n sess.run(pf, feed_dict={x: [[2.0]]}) # [[4.0]]",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
e7943560144b19443772737f3d351e19112c9161 | 37,898 | ipynb | Jupyter Notebook | interactive/InteractiveVariantAnnotation.ipynb | bashir2/variant-annotation | bec91ba9b6ea8eadea0a438f61bf4e19e7ae06f8 | [
"Apache-2.0"
] | 11 | 2018-05-02T07:35:16.000Z | 2021-06-17T10:30:35.000Z | interactive/InteractiveVariantAnnotation.ipynb | bashir2/variant-annotation | bec91ba9b6ea8eadea0a438f61bf4e19e7ae06f8 | [
"Apache-2.0"
] | 5 | 2018-04-25T03:38:28.000Z | 2020-10-20T14:34:21.000Z | interactive/InteractiveVariantAnnotation.ipynb | bashir2/variant-annotation | bec91ba9b6ea8eadea0a438f61bf4e19e7ae06f8 | [
"Apache-2.0"
] | 7 | 2018-04-05T23:58:03.000Z | 2021-06-04T16:29:13.000Z | 207.092896 | 17,059 | 0.593039 | [
[
[
"# Interactive Variant Annotation\n\nThe following query retrieves variants from [DeepVariant-called Platinum Genomes](http://googlegenomics.readthedocs.io/en/latest/use_cases/discover_public_data/platinum_genomes_deepvariant.html) and interactively JOINs them with [ClinVar](http://googlegenomics.readthedocs.io/en/latest/use_cases/discover_public_data/clinvar_annotations.html). \n\nTo run this on your own table of variants, change the table name and call_set_name in the `sample_variants` sub query below.\n\nFor an ongoing investigation, you may wish to repeat this query each time a new version of ClinVar is released and [loaded into BigQuery](https://github.com/verilylifesciences/variant-annotation/tree/master/curation/tables/README.md) by changing the table name in the `rare_pathenogenic_variants` sub query.\n\nSee also similar examples for GRCh37 in https://github.com/googlegenomics/bigquery-examples/tree/master/platinumGenomes ",
"_____no_output_____"
]
],
[
[
"%%bq query\n#standardSQL\n --\n -- Return variants for sample NA12878 that are:\n -- annotated as 'pathogenic' or 'other' in ClinVar\n -- with observed population frequency less than 5%\n --\n WITH sample_variants AS (\n SELECT\n -- Remove the 'chr' prefix from the reference name.\n REGEXP_EXTRACT(reference_name, r'chr(.+)') AS chr,\n start,\n reference_bases,\n alt,\n call.call_set_name\n FROM\n `genomics-public-data.platinum_genomes_deepvariant.single_sample_genome_calls` v,\n v.call call,\n v.alternate_bases alt WITH OFFSET alt_offset\n WHERE\n call_set_name = 'NA12878_ERR194147'\n -- Require that at least one genotype matches this alternate.\n AND EXISTS (SELECT gt FROM UNNEST(call.genotype) gt WHERE gt = alt_offset+1)\n ),\n --\n --\n rare_pathenogenic_variants AS (\n SELECT\n -- ClinVar does not use the 'chr' prefix for reference names.\n reference_name AS chr,\n start,\n reference_bases,\n alt,\n CLNHGVS,\n CLNALLE,\n CLNSRC,\n CLNORIGIN,\n CLNSRCID,\n CLNSIG,\n CLNDSDB,\n CLNDSDBID,\n CLNDBN,\n CLNREVSTAT,\n CLNACC\n FROM\n `bigquery-public-data.human_variant_annotation.ncbi_clinvar_hg38_20170705` v,\n v.alternate_bases alt\n WHERE\n -- Variant Clinical Significance, 0 - Uncertain significance, 1 - not provided,\n -- 2 - Benign, 3 - Likely benign, 4 - Likely pathogenic, 5 - Pathogenic,\n -- 6 - drug response, 7 - histocompatibility, 255 - other\n EXISTS (SELECT sig FROM UNNEST(CLNSIG) sig WHERE REGEXP_CONTAINS(sig, '(4|5|255)'))\n -- TRUE if >5% minor allele frequency in 1+ populations\n AND G5 IS NULL\n)\n --\n --\nSELECT\n *\nFROM\n sample_variants\nJOIN\n rare_pathenogenic_variants USING(chr,\n start,\n reference_bases,\n alt)\nORDER BY\n chr,\n start,\n reference_bases,\n alt",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
e794475740c484122949e2a60bee5bf13bb488e3 | 2,872 | ipynb | Jupyter Notebook | docs/html/notebooks/h5_compound_dataset_as_dataframe.ipynb | sainjacobs/pydsm | 1f2175ae0e6d2a545a22b641617fd225a6a09bd1 | [
"MIT"
] | 3 | 2020-05-11T22:55:34.000Z | 2021-05-19T19:49:11.000Z | docs/html/notebooks/h5_compound_dataset_as_dataframe.ipynb | sainjacobs/pydsm | 1f2175ae0e6d2a545a22b641617fd225a6a09bd1 | [
"MIT"
] | 6 | 2020-02-12T23:18:38.000Z | 2022-02-18T20:31:53.000Z | docs/html/notebooks/h5_compound_dataset_as_dataframe.ipynb | sainjacobs/pydsm | 1f2175ae0e6d2a545a22b641617fd225a6a09bd1 | [
"MIT"
] | 1 | 2021-07-07T18:55:44.000Z | 2021-07-07T18:55:44.000Z | 20.225352 | 110 | 0.539694 | [
[
[
"## Dealing with compound data set\nUsing dtypes one can detect the names for the dtype and then copy into an array and convert to np.str\nThen pandas DataFrame can parse those properly as a table",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport h5py\nh5 = h5py.File('../../tests/historical_v82.h5')",
"_____no_output_____"
],
[
"x=h5.get('/hydro/geometry/reservoir_node_connect')",
"_____no_output_____"
]
],
[
[
"See below on how to use dtype on returned array to see the names",
"_____no_output_____"
]
],
[
[
"x[0].dtype.names",
"_____no_output_____"
]
],
[
[
"Now the names can be used to get the value for that dtype",
"_____no_output_____"
]
],
[
[
"x[0]['res_name']",
"_____no_output_____"
]
],
[
[
"Using generative expressions to get the values as arrays of arrays with everything converted to strings",
"_____no_output_____"
]
],
[
[
"pd.DataFrame([[v[name].astype(np.str) for name in v.dtype.names] for v in x])\n ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e79449bbb931193179f1721f1c85e9effe8c264e | 45,445 | ipynb | Jupyter Notebook | src/4AnalysingText/analyzing_text.ipynb | Densuke-fitness/MDandAAnalysisFlow | 520af7747bd0d07008eb83f56acad586f15e7fa7 | [
"MIT"
] | null | null | null | src/4AnalysingText/analyzing_text.ipynb | Densuke-fitness/MDandAAnalysisFlow | 520af7747bd0d07008eb83f56acad586f15e7fa7 | [
"MIT"
] | null | null | null | src/4AnalysingText/analyzing_text.ipynb | Densuke-fitness/MDandAAnalysisFlow | 520af7747bd0d07008eb83f56acad586f15e7fa7 | [
"MIT"
] | null | null | null | 62.083333 | 8,820 | 0.770228 | [
[
[
"# 4.3.4 抽出した文章群から日本語極性辞書にマッチする単語を特定しトーンを算出\n抽出した文章群から乾・鈴木(2008)で公開された日本語評価極性辞書を用いて、マッチする単語を特定しトーンを算出する。ここでは、osetiと呼ばれる日本語評価極性辞書を用いて極性の判定を行うPythonのライブラリを用いた。",
"_____no_output_____"
]
],
[
[
"import glob\n\ndef call_sample_dir_name(initial_name):\n if initial_name == \"a\":\n return \"AfterSample\"\n elif initial_name == \"t\":\n return \"TransitionPeriodSample\"\n else:\n return \"BeforeSample\"\n\n\ndef call_csv_files(sample_dir_name=\"AfterSample\", data_frame_spec=None, industry_spec=None):\n \n if data_frame_spec is None:\n \n if industry_spec is None:\n csv_files = glob.glob('/home/jovyan/3FetchingMDandA' + f\"/**/{sample_dir_name}/*.csv\", recursive=True)\n else:\n csv_files = glob.glob(f'/home/jovyan/3FetchingMDandA' + f\"/**/{industry_spec}/{sample_dir_name}/*.csv\", recursive=True)\n else:\n if industry_spec is None:\n csv_files = glob.glob(f'/home/jovyan/3FetchingMDandA/{data_frame_spec}' + f\"/**/{sample_dir_name}/*.csv\", recursive=True)\n else:\n csv_files = glob.glob(f\"/home/jovyan/3FetchingMDandA/{data_frame_spec}/{industry_spec}/{sample_dir_name}/*.csv\", recursive=True)\n \n return csv_files",
"_____no_output_____"
],
[
"import glob\nimport pandas as pd\nimport os \nimport oseti\n\n\n# analyzer = oseti.Analyzer()\ndef make_atb_li(atb_file, analyzer):\n\n atb_df = pd.read_csv(atb_file, index_col=0)\n if len(atb_df) < 1:\n return 0\n texts_joined = \"\".join(list(atb_df[\"Text\"].values))\n #parse error対策\n texts_joined = texts_joined.replace(\"\\n\", \"\")\n \n scores = analyzer.count_polarity(texts_joined)\n sum_plus = 0\n sum_minus = 0\n for score in scores:\n sum_plus += score[\"positive\"]\n sum_minus += score[\"negative\"]\n\n ret_val = (sum_plus - sum_minus)/(sum_plus + sum_minus)\n return ret_val",
"_____no_output_____"
],
[
"#今回は全企業を抽出して分析\ndata_frame_spec=None\nindustry_spec=None\n\n\n#before\ndir_name_b = call_sample_dir_name(\"b\")\nbefore_csv_files = call_csv_files(dir_name_b, data_frame_spec, industry_spec)\n#transition\ndir_name_t = call_sample_dir_name(\"t\")\ntransition_period_csv_files = call_csv_files(dir_name_t, data_frame_spec, industry_spec)\n#after\ndir_name_a = call_sample_dir_name(\"a\")\nafter_csv_files = call_csv_files(dir_name_a, data_frame_spec, industry_spec)\nprint(\"--------ここまで終わりました1------\")",
"--------ここまで終わりました1------\n"
],
[
"analyzer = oseti.Analyzer()",
"_____no_output_____"
],
[
"#cache化して速度向上\nbefore_li = []\nf = before_li.append\n\nfor b_file in before_csv_files :\n tmp = make_atb_li(b_file, analyzer) \n f(tmp)\nprint(\"-------ここまで終わりました2-------\")",
"-------ここまで終わりました2-------\n"
],
[
"transition_period_li = []\nf = transition_period_li.append\n\nfor t_file in transition_period_csv_files :\n tmp = make_atb_li(t_file, analyzer) \n f(tmp)\nprint(\"-------ここまで終わりました3-------\")",
"-------ここまで終わりました3-------\n"
],
[
"after_li = []\nf = after_li.append\n\nfor a_file in after_csv_files :\n tmp = make_atb_li(a_file, analyzer) \n f(tmp)\nprint(\"-------ここまで終わりました4-------\")\n",
"-------ここまで終わりました4-------\n"
],
[
"#各年度のサンプルサイズ\nprint(len(after_li), len(transition_period_li) ,len(before_li))",
"2277 2277 2277\n"
],
[
"#beforeのヒストグラム\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nfig = plt.figure()\nax = fig.add_subplot(1,1,1)\nplt.ylabel(\"Number of companies\")\nplt.xlabel(\"TONE\")\nax.hist(before_li, bins=50)",
"_____no_output_____"
],
[
"#transitionのヒストグラム\nfig = plt.figure()\nax = fig.add_subplot(1,1,1)\nplt.ylabel(\"Number of companies\")\nplt.xlabel(\"TONE\")\nax.hist(transition_period_li, bins=50)",
"_____no_output_____"
],
[
"#afterのヒストグラム\nfig = plt.figure()\nax = fig.add_subplot(1,1,1)\nplt.ylabel(\"Number of companies\")\nplt.xlabel(\"TONE\")\nax.hist(after_li, bins=50)",
"_____no_output_____"
],
[
"# 正規性の検定\n #before\nimport scipy.stats as stats\nprint(stats.shapiro(before_li)) \nprint(stats.kstest(before_li, \"norm\"))",
"ShapiroResult(statistic=0.9717572927474976, pvalue=8.834468272238198e-21)\nKstestResult(statistic=0.6367143659091989, pvalue=0.0)\n"
],
[
"# 正規性の検定\n #transition\nprint(stats.shapiro(transition_period_li)) \nprint(stats.kstest(transition_period_li, \"norm\"))",
"ShapiroResult(statistic=0.9743578433990479, pvalue=8.687457274754784e-20)\nKstestResult(statistic=0.6323944243273957, pvalue=0.0)\n"
],
[
"# 正規性の検定\n #after\nprint(stats.shapiro(after_li)) \nprint(stats.kstest(after_li, \"norm\"))",
"ShapiroResult(statistic=0.9883420467376709, pvalue=1.1569712202175175e-12)\nKstestResult(statistic=0.5844571248123066, pvalue=0.0)\n"
],
[
" import numpy as np\n # 等分散性の検定\n #ex) A=before_li, B=transition_period_li\ndef exec_f_test(A, B):\n A_var = np.var(A, ddof=1) # Aの不偏分散\n B_var = np.var(B, ddof=1) # Bの不偏分散\n A_df = len(A) - 1 # Aの自由度\n B_df = len(B) - 1 # Bの自由度\n f = A_var / B_var # F比の値\n one_sided_pval1 = stats.f.cdf(f, A_df, B_df) # 片側検定のp値 1\n one_sided_pval2 = stats.f.sf(f, A_df, B_df) # 片側検定のp値 2\n two_sided_pval = min(one_sided_pval1, one_sided_pval2) * 2 # 両側検定のp値\n print('F: ', round(f, 3))\n print('p-value: ', round(two_sided_pval, 3))",
"_____no_output_____"
],
[
"A=before_li\nB=transition_period_li\nexec_f_test(A, B)",
"F: 0.931\np-value: 0.087\n"
],
[
"A=transition_period_li\nB=after_li\nexec_f_test(A, B)",
"F: 0.815\np-value: 0.0\n"
],
[
"A=before_li\nB=after_li\nexec_f_test(A, B)",
"F: 0.758\np-value: 0.0\n"
],
[
"import numpy\nprint(\"before_li: \", numpy.average(before_li))\nprint(\"transition_period_lii: \", numpy.average(transition_period_li))\nprint(\"after_li: \", numpy.average(after_li))",
"before_li: 0.652211639700913\ntransition_period_lii: 0.6424370778702994\nafter_li: 0.5339956014025147\n"
],
[
"import numpy\nprint(\"before_li: \", numpy.average(before_li))\nprint(\"transition_period_lii: \", numpy.average(transition_period_li))\nprint(\"after_li: \", numpy.average(after_li))",
"before_li: 0.652211639700913\ntransition_period_lii: 0.6424370778702994\nafter_li: 0.5339956014025147\n"
],
[
"#ウェルチのt検定\nstats.ttest_ind(before_li, transition_period_li, equal_var=False)",
"_____no_output_____"
],
[
"#スチューデントのt検定\nstats.ttest_ind(transition_period_li, after_li, axis=0, equal_var=True, nan_policy='propagate')",
"_____no_output_____"
],
[
"#スチューデントのt検定\nstats.ttest_ind(before_li, after_li, axis=0, equal_var=True, nan_policy='propagate')",
"_____no_output_____"
],
[
"#マンホイットニーのu検定\nstats.mannwhitneyu( before_li, transition_period_li, alternative='two-sided')",
"_____no_output_____"
],
[
"stats.mannwhitneyu(transition_period_li, after_li, alternative='two-sided')",
"_____no_output_____"
],
[
"stats.mannwhitneyu(before_li, after_li, alternative='two-sided')",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e79453002bf7b2594627fa80eb18f516d4e472a5 | 23,664 | ipynb | Jupyter Notebook | markdown_generator/publications.ipynb | mczielinski/mczielinski.github.io | e8638cb543ca8a926fb7411467c70811ecbec630 | [
"MIT"
] | null | null | null | markdown_generator/publications.ipynb | mczielinski/mczielinski.github.io | e8638cb543ca8a926fb7411467c70811ecbec630 | [
"MIT"
] | null | null | null | markdown_generator/publications.ipynb | mczielinski/mczielinski.github.io | e8638cb543ca8a926fb7411467c70811ecbec630 | [
"MIT"
] | null | null | null | 48.591376 | 1,505 | 0.541582 | [
[
[
"# Publications markdown generator for academicpages\n\nTakes a TSV of publications with metadata and converts them for use with [academicpages.github.io](academicpages.github.io). This is an interactive Jupyter notebook ([see more info here](http://jupyter-notebook-beginner-guide.readthedocs.io/en/latest/what_is_jupyter.html)). The core python code is also in `publications.py`. Run either from the `markdown_generator` folder after replacing `publications.tsv` with one containing your data.\n\nTODO: Make this work with BibTex and other databases of citations, rather than Stuart's non-standard TSV format and citation style.\n",
"_____no_output_____"
],
[
"## Data format\n\nThe TSV needs to have the following columns: pub_date, title, venue, excerpt, citation, site_url, and paper_url, with a header at the top. \n\n- `excerpt` and `paper_url` can be blank, but the others must have values. \n- `pub_date` must be formatted as YYYY-MM-DD.\n- `url_slug` will be the descriptive part of the .md file and the permalink URL for the page about the paper. The .md file will be `YYYY-MM-DD-[url_slug].md` and the permalink will be `https://[yourdomain]/publications/YYYY-MM-DD-[url_slug]`\n\nThis is how the raw file looks (it doesn't look pretty, use a spreadsheet or other program to edit and create).",
"_____no_output_____"
]
],
[
[
"!cat publications.tsv",
"pub_date\ttitle\tvenue\texcerpt\tcitation\turl_slug\tpaper_url\r\n2012\tThe effect of surface wave propagation on neural responses to vibration in primate glabrous skin.\tPloS one\t\tManfredi LR, Baker AT, Elias DO, Dammann III JF, Zielinski MC, Polashock VS, Bensmaia SJ. The effect of surface wave propagation on neural responses to vibration in primate glabrous skin. PloS one. 2012;7(2):e31203. \tNeuro-1\t\r\n2012\tQuantification of islet size and architecture.\tIslets\t\tKilimnik G, Jo J, Periwal V, Zielinski MC, Hara M. Quantification of islet size and architecture. Islets. 2012;4(2):167–172.\tDiabetes-1\t\r\n2013\tQuantitative analysis of pancreatic polypeptide cell distribution in the human pancreas.\tPloS one\t\tWang X, Zielinski MC, Misawa R, Wen P, Wang T-Y, Wang C-Z, Witkowski P, Hara M. Quantitative analysis of pancreatic polypeptide cell distribution in the human pancreas. PloS one. 2013;8(1):e55501. \tDiabetes-2\t\r\n2013\tRegional differences in islet distribution in the human pancreas--preferential beta-cell loss in the head region in patients with type 2 diabetes.\tPloS one\t\tWang X, Misawa R, Zielinski MC, Cowen P, Jo J, Periwal V, Ricordi C, Khan A, Szust J, Shen J. Regional differences in islet distribution in the human pancreas--preferential beta-cell loss in the head region in patients with type 2 diabetes. PLoS One. 2013;8(6). \tDiabetes-3\t\r\n2013\tDistinct function of the head region of human pancreas in the pathogenesis of diabetes.\tIslets\t\tSavari O, Zielinski MC, Wang X, Misawa R, Millis JM, Witkowski P, Hara M. Distinct function of the head region of human pancreas in the pathogenesis of diabetes. Islets. 2013;5(5):226–228.\tDiabetes-4\t\r\n2014\tNatural scenes in tactile texture.\tJournal of neurophysiology\t\tManfredi LR, Saal HP, Brown KJ, Zielinski MC, Dammann JF, Polashock VS, Bensmaia SJ. Natural scenes in tactile texture. Journal of neurophysiology. 2014;111(9):1792–1802. \tNeuro-2\t\r\n2014\tImproved coating of pancreatic islets with regulatory T cells to create local immunosuppression by using the biotin-polyethylene glycol-succinimidyl valeric acid ester molecule.\tTransplantation proceedings\t\tGołąb K, Kizilel S, Bal T, Hara M, Zielinski M, Grose R, Savari O, Wang X-J, Wang L-J, Tibudan M. Improved Coating of Pancreatic Islets With Regulatory T cells to Create Local Immunosuppression by Using the Biotin-polyethylene Glycol-succinimidyl Valeric Acid Ester Molecule. Transplantation proceedings. 2014;46(6):1967–1971.\tDiabetes-4\t\r\n2015\tEvidence of non-pancreatic beta cell-dependent roles of Tcf7l2 in the regulation of glucose metabolism in mice.\tHuman molecular genetics\t\tBailey KA, Savic D, Zielinski M, Park S-Y, Wang L, Witkowski P, Brady M, Hara M, Bell GI, Nobrega MA. Evidence of non-pancreatic beta cell-dependent roles of Tcf7l2 in the regulation of glucose metabolism in mice. Human molecular genetics. 2015;24(6):1646–1654. \tDiabetes-5\t\r\n2016\tStereological analyses of the whole human pancreas.\tScientific reports\t\tA Poudel, JL Fowler, MC Zielinski, G Kilimnik, M Hara. Stereological analyses of the whole human pancreas. Scientific Reports. 2016;6:34049. \tDiabetes-6\t\r\n2016\tInterplay between Hippocampal Sharp-Wave-Ripple Events and Vicarious Trial and Error Behaviors in Decision Making.\tNeuron\t\tAE Papale, MC Zielinski, LM Frank, SP Jadhav, AD Redish. Interplay between hippocampal sharp wave ripple events and vicarious trial and error behaviors in decision making. Neuron. 2016;92(5):975-982.\tNeuro-3\t\r\n2017\tPreservation of Reduced Numbers of Insulin-Positive Cells in Sulfonylurea-Unresponsive KCNJ11-Related Diabetes.\tThe Journal of clinical endocrinology and metabolism\t\tSA Greeley, MC Zielinski, A Poudel, H Ye, S Berry, JB Taxy, D Carmody, DF Steiner, LH Philipson, JR Wood, M Hara. Preservation of Reduced Numbers of Insulin-Positive Cells in Sulfonylurea-Unresponsive KCNJ11-Related Diabetes. Journal of Clinical Endocrinology and Metabolism. 2017;102(1):1-5.\tDiabetes-7\t\r\n2017\tThe role of replay and theta sequences in mediating hippocampal-prefrontal interactions for memory and cognition.\tHippocampus\t\tMC Zielinski, W Tang, SP Jadhav. The role of replay and theta sequences in mediating hippocampal-prefrontal interactions for memory and cognition Hippocampus. 2017;10.1002/hipo.22821\tNeuro-4\t\r\n"
]
],
[
[
"## Import pandas\n\nWe are using the very handy pandas library for dataframes.",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
]
],
[
[
"## Import TSV\n\nPandas makes this easy with the read_csv function. We are using a TSV, so we specify the separator as a tab, or `\\t`.\n\nI found it important to put this data in a tab-separated values format, because there are a lot of commas in this kind of data and comma-separated values can get messed up. However, you can modify the import statement, as pandas also has read_excel(), read_json(), and others.",
"_____no_output_____"
]
],
[
[
"publications = pd.read_csv(\"publications.tsv\", sep=\"\\t\", header=0)\npublications\n",
"_____no_output_____"
]
],
[
[
"## Escape special characters\n\nYAML is very picky about how it takes a valid string, so we are replacing single and double quotes (and ampersands) with their HTML encoded equivilents. This makes them look not so readable in raw format, but they are parsed and rendered nicely.",
"_____no_output_____"
]
],
[
[
"html_escape_table = {\n \"&\": \"&\",\n '\"': \""\",\n \"'\": \"'\"\n }\n\ndef html_escape(text):\n \"\"\"Produce entities within text.\"\"\"\n return \"\".join(html_escape_table.get(c,c) for c in text)",
"_____no_output_____"
]
],
[
[
"## Creating the markdown files\n\nThis is where the heavy lifting is done. This loops through all the rows in the TSV dataframe, then starts to concatentate a big string (```md```) that contains the markdown for each type. It does the YAML metadata first, then does the description for the individual page.",
"_____no_output_____"
]
],
[
[
"import os\nfor row, item in publications.iterrows():\n \n md_filename = str(item.pub_date) + \"-\" + item.url_slug + \".md\"\n html_filename = str(item.pub_date) + \"-\" + item.url_slug\n year = item.pub_date[:4]\n \n ## YAML variables\n \n md = \"---\\ntitle: \\\"\" + item.title + '\"\\n'\n \n md += \"\"\"collection: publications\"\"\"\n \n md += \"\"\"\\npermalink: /publication/\"\"\" + html_filename\n \n if len(str(item.excerpt)) > 5:\n md += \"\\nexcerpt: '\" + html_escape(item.excerpt) + \"'\"\n \n md += \"\\ndate: \" + str(item.pub_date) \n \n md += \"\\nvenue: '\" + html_escape(item.venue) + \"'\"\n \n if len(str(item.paper_url)) > 5:\n md += \"\\npaperurl: '\" + item.paper_url + \"'\"\n \n md += \"\\ncitation: '\" + html_escape(item.citation) + \"'\"\n \n md += \"\\n---\"\n \n ## Markdown description for individual page\n \n if len(str(item.excerpt)) > 5:\n md += \"\\n\" + html_escape(item.excerpt) + \"\\n\"\n \n if len(str(item.paper_url)) > 5:\n md += \"\\n[Download paper here](\" + item.paper_url + \")\\n\" \n \n md += \"\\nRecommended citation: \" + item.citation\n \n md_filename = os.path.basename(md_filename)\n \n with open(\"../_publications/\" + md_filename, 'w') as f:\n f.write(md)",
"_____no_output_____"
]
],
[
[
"These files are in the publications directory, one directory below where we're working from.",
"_____no_output_____"
]
],
[
[
"!ls ../_publications/",
"2009-10-01-paper-title-number-1.md 2015-10-01-paper-title-number-3.md\r\n2010-10-01-paper-title-number-2.md\r\n"
],
[
"!cat ../_publications/2009-10-01-paper-title-number-1.md",
"---\r\ntitle: \"Paper Title Number 1\"\r\ncollection: publications\r\npermalink: /publication/2009-10-01-paper-title-number-1\r\nexcerpt: 'This paper is about the number 1. The number 2 is left for future work.'\r\ndate: 2009-10-01\r\nvenue: 'Journal 1'\r\npaperurl: 'http://academicpages.github.io/files/paper1.pdf'\r\ncitation: 'Your Name, You. (2009). "Paper Title Number 1." <i>Journal 1</i>. 1(1).'\r\n---\r\nThis paper is about the number 1. The number 2 is left for future work.\r\n\r\n[Download paper here](http://academicpages.github.io/files/paper1.pdf)\r\n\r\nRecommended citation: Your Name, You. (2009). \"Paper Title Number 1.\" <i>Journal 1</i>. 1(1)."
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
e794674495bb713835b79772adf2a8027a78b007 | 17,147 | ipynb | Jupyter Notebook | deep_learning/07-deep-learning-from-scratch.ipynb | drakearch/kaggle-courses | 206244caf38c98e9b6d2f37cbeb8b045460723c9 | [
"MIT"
] | 27 | 2021-08-15T01:06:40.000Z | 2022-03-18T02:26:29.000Z | deep_learning/07-deep-learning-from-scratch.ipynb | drakessn/Kaggle-Courses | db9fb7a73743a22f35d65ec435d9b5dfd89fc5a7 | [
"MIT"
] | 17 | 2019-12-29T23:33:28.000Z | 2020-05-04T00:07:59.000Z | deep_learning/07-deep-learning-from-scratch.ipynb | drakessn/Kaggle-Courses | db9fb7a73743a22f35d65ec435d9b5dfd89fc5a7 | [
"MIT"
] | 11 | 2021-08-16T16:07:53.000Z | 2022-03-27T02:55:40.000Z | 33.555773 | 369 | 0.552575 | [
[
[
"**[Deep Learning Course Home Page](https://www.kaggle.com/learn/deep-learning)**\n\n---\n",
"_____no_output_____"
],
[
"# Introduction\n\nYou've seen how to build a model from scratch to identify handwritten digits. You'll now build a model to identify different types of clothing. To make models that train quickly, we'll work with very small (low-resolution) images. \n\nAs an example, your model will take an images like this and identify it as a shoe:\n\n",
"_____no_output_____"
],
[
"# Data Preparation\nThis code is supplied, and you don't need to change it. Just run the cell below.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom sklearn.model_selection import train_test_split\nfrom tensorflow import keras\n\nimg_rows, img_cols = 28, 28\nnum_classes = 10\n\ndef prep_data(raw):\n y = raw[:, 0]\n out_y = keras.utils.to_categorical(y, num_classes)\n \n x = raw[:,1:]\n num_images = raw.shape[0]\n out_x = x.reshape(num_images, img_rows, img_cols, 1)\n out_x = out_x / 255\n return out_x, out_y\n\nfashion_file = \"../input/fashionmnist/fashion-mnist_train.csv\"\nfashion_data = np.loadtxt(fashion_file, skiprows=1, delimiter=',')\nx, y = prep_data(fashion_data)\n\n# Set up code checking\nfrom learntools.core import binder\nbinder.bind(globals())\nfrom learntools.deep_learning.exercise_7 import *\nprint(\"Setup Complete\")",
"Using TensorFlow version 2.1.0\nSetup Complete\n"
]
],
[
[
"# 1) Start the model\nCreate a `Sequential` model called `fashion_model`. Don't add layers yet.",
"_____no_output_____"
]
],
[
[
"from tensorflow import keras\nfrom tensorflow.keras.models import Sequential\nfrom tensorflow.keras.layers import Dense, Flatten, Conv2D\n\n# Your Code Here\nfashion_model = Sequential()\nq_1.check()",
"_____no_output_____"
],
[
"#q_1.solution()",
"_____no_output_____"
]
],
[
[
"# 2) Add the first layer\n\nAdd the first `Conv2D` layer to `fashion_model`. It should have 12 filters, a kernel_size of 3 and the `relu` activation function. The first layer always requires that you specify the `input_shape`. We have saved the number of rows and columns to the variables `img_rows` and `img_cols` respectively, so the input shape in this case is `(img_rows, img_cols, 1)`.",
"_____no_output_____"
]
],
[
[
"# Your code here\nfashion_model.add(Conv2D(12, kernel_size=(3, 3),\n activation='relu',\n input_shape=(img_rows, img_cols, 1)))\nq_2.check()",
"_____no_output_____"
],
[
"# q_2.hint()\n#q_2.solution()",
"_____no_output_____"
]
],
[
[
"# 3) Add the remaining layers\n\n1. Add 2 more convolutional (`Conv2D layers`) with 20 filters each, 'relu' activation, and a kernel size of 3. Follow that with a `Flatten` layer, and then a `Dense` layer with 100 neurons. \n2. Add your prediction layer to `fashion_model`. This is a `Dense` layer. We alrady have a variable called `num_classes`. Use this variable when specifying the number of nodes in this layer. The activation should be `softmax` (or you will have problems later).",
"_____no_output_____"
]
],
[
[
"# Your code here\nfashion_model.add(Conv2D(20, kernel_size=(3, 3), activation='relu'))\nfashion_model.add(Conv2D(20, kernel_size=(3, 3), activation='relu'))\nfashion_model.add(Flatten())\nfashion_model.add(Dense(100, activation='relu'))\nfashion_model.add(Dense(num_classes, activation='softmax'))\n\nq_3.check()",
"_____no_output_____"
],
[
"# q_3.solution()",
"_____no_output_____"
]
],
[
[
"# 4) Compile Your Model\nCompile fashion_model with the `compile` method. Specify the following arguments:\n1. `loss = \"categorical_crossentropy\"`\n2. `optimizer = 'adam'`\n3. `metrics = ['accuracy']`",
"_____no_output_____"
]
],
[
[
"# Your code to compile the model in this cell\nfashion_model.compile(loss=keras.losses.categorical_crossentropy,\n optimizer='adam',\n metrics=['accuracy'])\n\nq_4.check()",
"_____no_output_____"
],
[
"# q_4.solution()",
"_____no_output_____"
]
],
[
[
"# 5) Fit The Model\nRun the command `fashion_model.fit`. The arguments you will use are\n1. The data used to fit the model. First comes the data holding the images, and second is the data with the class labels to be predicted. Look at the first code cell (which was supplied to you) where we called `prep_data` to find the variable names for these.\n2. `batch_size = 100`\n3. `epochs = 4`\n4. `validation_split = 0.2`\n\nWhen you run this command, you can watch your model start improving. You will see validation accuracies after each epoch.",
"_____no_output_____"
]
],
[
[
"# Your code to fit the model here\nfashion_model.fit(x, y, batch_size = 100, epochs = 4, validation_split = 0.2)\nq_5.check()",
"Train on 48000 samples, validate on 12000 samples\nEpoch 1/4\n48000/48000 [==============================] - 7s 145us/sample - loss: 0.4670 - accuracy: 0.8340 - val_loss: 0.3726 - val_accuracy: 0.8659\nEpoch 2/4\n48000/48000 [==============================] - 3s 54us/sample - loss: 0.2970 - accuracy: 0.8928 - val_loss: 0.2943 - val_accuracy: 0.8960\nEpoch 3/4\n48000/48000 [==============================] - 3s 56us/sample - loss: 0.2399 - accuracy: 0.9115 - val_loss: 0.2696 - val_accuracy: 0.9046\nEpoch 4/4\n48000/48000 [==============================] - 3s 53us/sample - loss: 0.1945 - accuracy: 0.9283 - val_loss: 0.2652 - val_accuracy: 0.9088\n"
],
[
"#q_5.solution()",
"_____no_output_____"
]
],
[
[
"# 6) Create A New Model\n\nCreate a new model called `second_fashion_model` in the cell below. Make some changes so it is different than `fashion_model` that you've trained above. The change could be using a different number of layers, different number of convolutions in the layers, etc.\n\nDefine the model, compile it and fit it in the cell below. See how it's validation score compares to that of the original model.",
"_____no_output_____"
]
],
[
[
"# Your code below\nsecond_fashion_model = Sequential()\nsecond_fashion_model.add(Conv2D(16, kernel_size=3, activation='relu', input_shape=(img_rows, img_cols, 1)))\nsecond_fashion_model.add(Conv2D(24, kernel_size=2, activation='relu'))\nsecond_fashion_model.add(Conv2D(32, kernel_size=2, activation='relu'))\nsecond_fashion_model.add(Conv2D(24, kernel_size=3, activation='relu'))\nsecond_fashion_model.add(Flatten())\nsecond_fashion_model.add(Dense(100, activation='sigmoid'))\nsecond_fashion_model.add(Dense(num_classes, activation='softmax'))\n\nsecond_fashion_model.compile(loss=keras.losses.categorical_crossentropy,\n optimizer='adam',\n metrics=['accuracy'])\n\nsecond_fashion_model.fit(x, y, batch_size=100, epochs=6, validation_split=0.2)\n\n# q_6.check()\nsecond_fashion_model.summary()",
"Train on 48000 samples, validate on 12000 samples\nEpoch 1/6\n48000/48000 [==============================] - 4s 76us/sample - loss: 0.5270 - accuracy: 0.8152 - val_loss: 0.3613 - val_accuracy: 0.8753\nEpoch 2/6\n48000/48000 [==============================] - 3s 60us/sample - loss: 0.3193 - accuracy: 0.8860 - val_loss: 0.3118 - val_accuracy: 0.8883\nEpoch 3/6\n48000/48000 [==============================] - 3s 63us/sample - loss: 0.2618 - accuracy: 0.9053 - val_loss: 0.2774 - val_accuracy: 0.9009\nEpoch 4/6\n48000/48000 [==============================] - 3s 60us/sample - loss: 0.2162 - accuracy: 0.9230 - val_loss: 0.2579 - val_accuracy: 0.9093\nEpoch 5/6\n48000/48000 [==============================] - 3s 61us/sample - loss: 0.1797 - accuracy: 0.9364 - val_loss: 0.2494 - val_accuracy: 0.9129\nEpoch 6/6\n48000/48000 [==============================] - 3s 60us/sample - loss: 0.1452 - accuracy: 0.9501 - val_loss: 0.2467 - val_accuracy: 0.9146\nModel: \"sequential_1\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nconv2d_3 (Conv2D) (None, 26, 26, 16) 160 \n_________________________________________________________________\nconv2d_4 (Conv2D) (None, 25, 25, 24) 1560 \n_________________________________________________________________\nconv2d_5 (Conv2D) (None, 24, 24, 32) 3104 \n_________________________________________________________________\nconv2d_6 (Conv2D) (None, 22, 22, 24) 6936 \n_________________________________________________________________\nflatten_1 (Flatten) (None, 11616) 0 \n_________________________________________________________________\ndense_2 (Dense) (None, 100) 1161700 \n_________________________________________________________________\ndense_3 (Dense) (None, 10) 1010 \n=================================================================\nTotal params: 1,174,470\nTrainable params: 1,174,470\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"# q_6.solution()",
"_____no_output_____"
]
],
[
[
"# Keep Going\nYou are ready to learn about **[strides and dropout](https://www.kaggle.com/dansbecker/dropout-and-strides-for-larger-models)**, which become important as you start using bigger and more powerful models.\n",
"_____no_output_____"
],
[
"---\n**[Deep Learning Course Home Page](https://www.kaggle.com/learn/deep-learning)**\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
e794684a03204551ba7be031db737e02c15a06af | 174,550 | ipynb | Jupyter Notebook | source/hello_world_tutorial.ipynb | oskarflordal/tutorials | bd76d73f59e40e0a707ee0b50f73360cd00d504d | [
"Apache-2.0"
] | 15 | 2020-10-29T09:12:39.000Z | 2022-03-13T05:50:57.000Z | source/hello_world_tutorial.ipynb | oskarflordal/tutorials | bd76d73f59e40e0a707ee0b50f73360cd00d504d | [
"Apache-2.0"
] | 25 | 2020-10-28T23:53:32.000Z | 2022-03-09T06:40:26.000Z | source/hello_world_tutorial.ipynb | oskarflordal/tutorials | bd76d73f59e40e0a707ee0b50f73360cd00d504d | [
"Apache-2.0"
] | 17 | 2020-10-29T04:28:18.000Z | 2022-03-15T16:10:00.000Z | 585.738255 | 107,132 | 0.944325 | [
[
[
"[](https://colab.sandbox.google.com/github/kornia/tutorials/blob/master/source/hello_world_tutorial.ipynb)\n\n# Hello world: Planet Kornia\n\nWelcome to Planet Kornia: a set of tutorials to learn about **Computer Vision** in [PyTorch](https://pytorch.org).\n\nThis is the first tutorial that show how one can simply start loading images with [Torchvision](https://pytorch.org/vision), [Kornia](https://kornia.org) and [OpenCV](https://opencv.org).\n",
"_____no_output_____"
]
],
[
[
"%%capture\n!pip install kornia",
"_____no_output_____"
],
[
"import cv2\nfrom matplotlib import pyplot as plt\nimport numpy as np\n\nimport torch\nimport torchvision\nimport kornia as K",
"_____no_output_____"
]
],
[
[
"Download first an image form internet to start to work.\n",
"_____no_output_____"
]
],
[
[
"%%capture\n!wget https://github.com/kornia/data/raw/main/arturito.jpg",
"_____no_output_____"
]
],
[
[
"## Load an image with OpenCV\n\nWe can use OpenCV to load an image. By default, OpenCV loads images in BGR format and casts to a `numpy.ndarray` with the data layout `(H,W,C)`. \n\nHowever, because matplotlib saves an image in RGB format, in OpenCV you need to change the BGR to RGB so that an image is displayed properly.",
"_____no_output_____"
]
],
[
[
"img_bgr: np.array = cv2.imread('arturito.jpg') # HxWxC / np.uint8\nimg_rgb: np.array = cv2.cvtColor(img_bgr, cv2.COLOR_BGR2RGB)\n\nplt.imshow(img_rgb); plt.axis('off');",
"_____no_output_____"
]
],
[
[
"## Load an image with Torchvision\n\nThe images can be also loaded using `torchvision` which directly returns the images in a `torch.Tensor` in the shape `(C,H,W)`.\n\n",
"_____no_output_____"
]
],
[
[
"x_rgb: torch.tensor = torchvision.io.read_image('arturito.jpg') # CxHxW / torch.uint8\nx_rgb = x_rgb.unsqueeze(0) # BxCxHxW\nprint(x_rgb.shape)",
"torch.Size([1, 3, 144, 256])\n"
]
],
[
[
"## Load an image with Kornia\n\nWith Kornia we can do all the preceding.\n\nWe have a couple of utilities to cast the image to a `torch.Tensor` to make it compliant to the other Kornia components and arrange the data in `(B,C,H,W)`. \n\n The utility is [`kornia.image_to_tensor`](https://kornia.readthedocs.io/en/latest/utils.html#kornia.utils.image_to_tensor) which casts a `numpy.ndarray` to a `torch.Tensor` and permutes the channels to leave the image ready for being used with any other PyTorch or Kornia component. \nThe image is casted into a 4D `torch.Tensor` with zero-copy.\n\n",
"_____no_output_____"
]
],
[
[
"x_bgr: torch.tensor = K.image_to_tensor(img_bgr) # CxHxW / torch.uint8\nx_bgr = x_bgr.unsqueeze(0) # 1xCxHxW\nprint(f\"convert from '{img_bgr.shape}' to '{x_bgr.shape}'\")",
"convert from '(144, 256, 3)' to 'torch.Size([1, 3, 144, 256])'\n"
]
],
[
[
"We can convert from BGR to RGB with a [`kornia.color`](https://kornia.readthedocs.io/en/latest/color.html) component.\n",
"_____no_output_____"
]
],
[
[
"x_rgb: torch.tensor = K.color.bgr_to_rgb(x_bgr) # 1xCxHxW / torch.uint8",
"_____no_output_____"
]
],
[
[
"## Visualize an image with Matplotib",
"_____no_output_____"
],
[
"We will use [Matplotlib](https://matplotlib.org/) for the visualisation inside the notebook. Matplotlib requires a `numpy.ndarray` in the `(H,W,C)` format, and for doing so we will go back with [`kornia.tensor_to_image`](https://kornia.readthedocs.io/en/latest/utils.html#kornia.utils.image_to_tensor) which will convert the image to the correct format.\n\n",
"_____no_output_____"
]
],
[
[
"img_bgr: np.array = K.tensor_to_image(x_bgr)\nimg_rgb: np.array = K.tensor_to_image(x_rgb)",
"_____no_output_____"
]
],
[
[
"Create a subplot to visualize the original an a modified image\n\n",
"_____no_output_____"
]
],
[
[
"fig, axs = plt.subplots(1, 2, figsize=(32, 16))\naxs = axs.ravel()\n\naxs[0].axis('off')\naxs[0].imshow(img_rgb)\n\naxs[1].axis('off')\naxs[1].imshow(img_bgr)\n\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7946af704fa15279078a618686cfcf4b56af1fe | 122,527 | ipynb | Jupyter Notebook | src/data_cleaning.ipynb | ALotOfData/data-512-a5 | 450d6bf4ca657f2ba5485dc568fe2ea0264d4d53 | [
"MIT"
] | null | null | null | src/data_cleaning.ipynb | ALotOfData/data-512-a5 | 450d6bf4ca657f2ba5485dc568fe2ea0264d4d53 | [
"MIT"
] | null | null | null | src/data_cleaning.ipynb | ALotOfData/data-512-a5 | 450d6bf4ca657f2ba5485dc568fe2ea0264d4d53 | [
"MIT"
] | 3 | 2020-11-10T08:24:37.000Z | 2021-06-05T02:47:25.000Z | 47.089547 | 499 | 0.590001 | [
[
[
"# Data cleaning\n\n## Goal\n\nIn this notebook, we will be taking in raw.csv and cleaning/parsing its different columns. The notebook contains the transformations below in order:\n\n1. Read in the data\n2. Removing unused columns for this analysis\n3. Removing rows with certain null columns\n4. Cleaning of columns\n * ad_age\n * ad_impressions\n * ad_clicks\n * ad_creation_date\n * ad_end_date\n * ad_targeting_interests\n * ad_targeting_people_who_match\n5. Writing to file\n6. Summary of lost rows\n\nWe first import 3 packages which will be useful for our data cleaning:\n\n* pandas for handling csv as a table\n* numpy to handle mulitdimensional array operations\n* re to handle regular expression parsing",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport re\n\n# We read in the data\nads_df = pd.read_csv('../raw_data/raw.csv')\n\n# Output first 2 rows\nads_df.head(2)",
"_____no_output_____"
]
],
[
[
"## Removing unused columns\n\nOur first step will be to remove columns we will not be using for this analysis.\n\n| Column name | Reason for removal|\n|-------------| ------------------|\n| Unnamed | Index column added by accident in the data production step. |\n| ad_id | We will be using the file_name column as identifier. |\n| ad_text | Although interesting its analysis is outside the scope of this project | \n| ad_landing_page | This will not be useful in answering our research questions. |\n| ad_targeting_location | We will not be studying the impact of location for the ads |\n| ad_targeting_custom_audience | This field doesn't contain information not already in the ad_targeting_interests column |\n| ad_targeting_language | This field is almost always US English and is not present on most of the dataset |\n| ad_targeting_placements | We will not be studying the impact of location on the page for the ads |",
"_____no_output_____"
]
],
[
[
"# Columns we will not be using\ncolumns_to_remove = ['Unnamed: 0', 'ad_id', 'ad_text', 'ad_landing_page', 'ad_targeting_location', 'ad_targeting_custom_audience', 'ad_targeting_excluded_connections', 'ad_targeting_language', 'ad_targeting_placements']\nads_df = ads_df.drop(columns=columns_to_remove)\n\nads_df.head(2)",
"_____no_output_____"
]
],
[
[
"## Removing rows with null columns\n\nWe will be removing rows with null for: ad_creation_date, ad_spend, ad_targeting_age, ad_impressions and ad_clicks. Our first step will be to create a dictionary which can keep track of the number of rows remaining after a given operation. We will be using this dictionary when summarizing the cleaning of the dataset and its repercussions on our analysis. We then create a function which remove the null values for a column name and adds the row count after removal to our summary dictionary.",
"_____no_output_____"
]
],
[
[
"# Dictionary to keep track of row removals\ncleaning_summary_format = {'before_cleaning_count': len(ads_df)}\n\n# Function to remove null rows for a given column\ndef remove_nulls(ads_df, column_name):\n # np.where returns a tuple, we want the first member (the indexes of rows)\n null_indexes = np.where(pd.isnull(ads_df[column_name]))[0]\n # We drop the list of indexes\n ads_df = ads_df.drop(null_indexes)\n # We add the entry to our summary\n cleaning_summary_format['null_' + column_name + '_count'] = len(ads_df)\n return ads_df\n\n# We remove nulls for given columns\nads_df = remove_nulls(ads_df, 'ad_creation_date')\nads_df = remove_nulls(ads_df, 'ad_spend')\nads_df = remove_nulls(ads_df, 'ad_targeting_age')\nads_df = remove_nulls(ads_df, 'ad_impressions')\nads_df = remove_nulls(ads_df, 'ad_clicks')",
"_____no_output_____"
],
[
"print('''Before cleaning our dataset had {before_cleaning_count} columns.\nAfter removing rows with null creation dates: {null_ad_creation_date_count} columns.\nAfter removing rows with null ad spending: {null_ad_spend_count} columns.\nAfter removing rows with null ad targeting age: {null_ad_targeting_age_count} columns.\nAfter removing rows with null ad impressions: {null_ad_impressions_count} columns.\nAfter removing rows with null ad clicks: {null_ad_clicks_count} columns.'''.format(**cleaning_summary_format))",
"Before cleaning our dataset had 3517 columns.\nAfter removing rows with null creation dates: 3497 columns.\nAfter removing rows with null ad spending: 3497 columns.\nAfter removing rows with null ad targeting age: 3497 columns.\nAfter removing rows with null ad impressions: 3497 columns.\nAfter removing rows with null ad clicks: 3497 columns.\n"
]
],
[
[
"### Cleaning ad_age\n\nFirst we look at the values for the field and whether we will be able to leverage them in our analysis.",
"_____no_output_____"
]
],
[
[
"ads_df.ad_targeting_age.value_counts().index",
"_____no_output_____"
]
],
[
[
"The initial parsing for this field was not perfect... Let's simplify this bucketing by removing gender information. To do so we crop the string at 8 characters.",
"_____no_output_____"
]
],
[
[
"# Crop the ad_targeting_age to 8 characters\nads_df.ad_targeting_age = ads_df.ad_targeting_age.apply(lambda s: s if len(s)<=8 else s[0:8])\n\n# Count rows for the different values\ncount_table = ads_df.ad_targeting_age.value_counts().to_frame()\n\n# Rename the column for clarity\ncount_table.columns = ['Ad count']\n\n# Output top 5\ncount_table.head(5)",
"_____no_output_____"
]
],
[
[
"As per this table, almost all ads targeted voting age facebook users (18+). Bucketing the ads by age groups will not result in significant/interesting analysis. We drop the column.",
"_____no_output_____"
]
],
[
[
"ads_df = ads_df.drop(columns=['ad_targeting_age'])",
"_____no_output_____"
]
],
[
[
"### Cleaning ad_impressions and ad_clicks\n\nBoth these columns are numerical and do not contain None and or NaN values. [The Oxford study](https://comprop.oii.ox.ac.uk/wp-content/uploads/sites/93/2018/12/IRA-Report.pdf), mention that ads without impressions or clicks where unlikely to have been shown to Facebook users.\n\n* We will first be parsing these fields that sometimes use . or , to separate thousands.\n* We will then remove 0 values.\n\nThe studies mention that this removed quite a few entries.",
"_____no_output_____"
]
],
[
[
"# Parsing of string to integer\ndef format_string_to_integer(string):\n # Removing dots and commas and semicolons\n s = string.replace(',', '').replace('.', '').replace(';', '')\n \n # Removing typos betwee o, O (lower, upper letter o) and 0 (zero digit)\n s = s.replace('o', '0').replace('O', '0')\n \n # Accidental whitespace\n s = s.replace(' ', '')\n \n # Coerce string to integer\n return int(s)\n\n# Remvoe rows with 0s and adds count after removal to summary\ndef remove_zeros(ads_df, column_name):\n ads_df = ads_df[ads_df[column_name] != 0]\n cleaning_summary_format['zeros_' + column_name + '_count'] = len(ads_df)\n return ads_df",
"_____no_output_____"
],
[
"# How many columns do we have before removing zeros\ncleaning_summary_format['before_zeros_count'] = len(ads_df)\n\n# Conversion to integers\nads_df['ad_clicks'] = ads_df['ad_clicks'].apply(format_string_to_integer)\nads_df['ad_impressions'] = ads_df['ad_impressions'].apply(format_string_to_integer)\n\n# Removing zeros values\nads_df = remove_zeros(ads_df, 'ad_impressions')\nads_df = remove_zeros(ads_df, 'ad_clicks')\n\n# Reporting\nprint('''Before removing 0 ad_impressions or ad_clicks our dataset had {before_zeros_count} columns.\nAfter removing rows with 0 ad impressions: {zeros_ad_impressions_count} columns.\nAfter removing rows with 0 ad clicks: {zeros_ad_clicks_count} columns.'''.format(**cleaning_summary_format))",
"Before removing 0 ad_impressions or ad_clicks our dataset had 3497 columns.\nAfter removing rows with 0 ad impressions: 2588 columns.\nAfter removing rows with 0 ad clicks: 2450 columns.\n"
]
],
[
[
"### Parsing creation date and end date\n\nCreation date and end date are written in a complex format: 04/13/16 07:48:33 AM PDT. Our analysis only requires the date. In this section, we will extract the first 8 characters mm/dd/yy and convert them to a datetime object. We take a look at the entries:",
"_____no_output_____"
]
],
[
[
"ads_df['ad_creation_date']",
"_____no_output_____"
]
],
[
[
"We find that sometimes the first few characters contain spaces. We write a regular expression for this and apply the removal of these white space as part of a function. We also need to complete the year to be 4 characters for later date parsing.",
"_____no_output_____"
]
],
[
[
"# We first compile our date extraction regex to improve performance\ndate_regex = re.compile(r'(?P<date>\\d\\s*\\d\\s*\\/\\s*\\d\\s*\\d\\s*\\/\\s*\\d\\s*\\d)')\n\n# Given a string beginning with mm/dd/yy we produce mm/dd/YYYY\n# Function returns 'parse_error' on failure to parse and null if the\n# input string was null\ndef extract_date_from_string(string):\n matches = None\n date = None\n \n # If the string is not null attempt to find matches\n if not pd.isnull(string):\n matches = date_regex.search(string)\n else:\n # null value for string in pandas\n date = np.nan \n\n # If the ?P<date> group was found\n if matches and matches.groupdict():\n group_dict = matches.groupdict()\n date = group_dict.get('date')\n if date:\n # Remove whitespace\n date = date.replace(' ', '')\n # We prefix '20' to the year to make 01/01/17 -> 01/01/2017\n date = date[:6] + '20' + date[6:]\n \n # We identify parsing errors with the 'parse_error'\n return date if date else 'parse_error'",
"_____no_output_____"
]
],
[
[
"We apply the function to every row and create a new column: 'ad_creation_date_parsed'",
"_____no_output_____"
]
],
[
[
"ads_df['ad_creation_date_parsed'] = ads_df.ad_creation_date.apply(extract_date_from_string)",
"_____no_output_____"
]
],
[
[
"We check how many dates could not be parsed:",
"_____no_output_____"
]
],
[
[
"(ads_df['ad_creation_date_parsed'] == 'parse_error').sum()",
"_____no_output_____"
]
],
[
[
"Since only one date could not be parsed we validate its value:",
"_____no_output_____"
]
],
[
[
"row = ads_df[ads_df['ad_creation_date_parsed'] == 'parse_error']\nrow",
"_____no_output_____"
]
],
[
[
"In this case the date should be 02/21/2017. An l was mistaken to a 1. We replace it manually.",
"_____no_output_____"
]
],
[
[
"ads_df.loc[row.index, 'ad_creation_date_parsed'] = '02/21/2017'\nads_df.loc[row.index]",
"_____no_output_____"
]
],
[
[
"Now that all dates have been parsed, we replace the original column with the parsed one and remove the temporary parsed column. Since no columns were lost in the process we will not be adding an entry to the summary.",
"_____no_output_____"
]
],
[
[
"# Replace original column with parsed\nads_df['ad_creation_date'] = ads_df['ad_creation_date_parsed']\n\n# Drop temporary parsed column\nads_df = ads_df.drop(columns=['ad_creation_date_parsed'])",
"_____no_output_____"
]
],
[
[
"We now execute the same steps for the end date.",
"_____no_output_____"
]
],
[
[
"ads_df['ad_end_date_parsed'] = ads_df.ad_end_date.apply(extract_date_from_string)",
"_____no_output_____"
]
],
[
[
"We check how many dates could not be parsed:",
"_____no_output_____"
]
],
[
[
"(ads_df['ad_end_date_parsed'] == 'parse_error').sum()",
"_____no_output_____"
],
[
"ads_df['ad_end_date'] = ads_df['ad_end_date_parsed']\nads_df = ads_df.drop(columns=['ad_end_date_parsed'])",
"_____no_output_____"
]
],
[
[
"Now that both ad_start_date and ad_end_date are properly parsed strings, we can apply a pandas function to transform them into datetime objects. This will make date handling easier during our analysis.",
"_____no_output_____"
]
],
[
[
"ads_df.ad_creation_date = ads_df.ad_creation_date.apply(lambda date_string : pd.to_datetime(date_string, format='%m/%d/%Y'))\nads_df.ad_end_date = ads_df.ad_end_date.apply(lambda date_string : pd.to_datetime(date_string, format='%m/%d/%Y'))\n\n# Output first 3 rows\nads_df.head(3)",
"_____no_output_____"
]
],
[
[
"### Parsing ad_spend\n\nSometimes the ad_spend field contains spaces, dots instead of commas to seperate thousands and the 'RUB' currency shorthand. We use a regular expression to extract the amount of the ad_spend field. We then convert the string to a float.",
"_____no_output_____"
]
],
[
[
"ads_df['ad_spend']",
"_____no_output_____"
],
[
"# Pre compile regex for performance\namount_regex = re.compile(r'(?P<amount>([0-9]{1,3}(\\.|,)?)+(\\.|,)?[0-9]{2})')\n\n# Function returns 'parse_error' on failure to parse and null if the\n# input string was null or the string 'None'\ndef extract_amount_from_string(string):\n matches = None\n amount = None\n \n # If the string is not null or 'None' search for matches\n if not pd.isnull(string) and string != 'None':\n matches = amount_regex.search(string)\n else:\n # null value for string in pandas\n amount = np.nan\n\n # If the amount was found\n if matches and matches.groupdict():\n group_dict = matches.groupdict()\n amount = group_dict.get('amount')\n if amount:\n # Remove whitespace\n amount = amount.replace(' ', '')\n \n # Remove dots and commas\n amount = amount.replace('.', '').replace(',', '')\n \n # Add a dot two digits form the end\n amount = amount[:-2] + '.' + amount[-2:]\n \n # Return a parse_error if amount parsing failed\n return amount if amount else 'parse_error'",
"_____no_output_____"
]
],
[
[
"We run the function over our dataset and output the number of parsing errors we've encountered.",
"_____no_output_____"
]
],
[
[
"ads_df['ad_spend_parsed'] = ads_df.ad_spend.apply(extract_amount_from_string)\n(ads_df['ad_spend_parsed'] == 'parse_error').sum()",
"_____no_output_____"
]
],
[
[
"We validate nan values and remove them from the dataset.",
"_____no_output_____"
]
],
[
[
"cleaning_summary_format['none_ad_spend_count'] = (~pd.isnull(ads_df['ad_spend_parsed'])).sum()\nprint('There are a total of ' + str(pd.isnull(ads_df['ad_spend_parsed']).sum()) + ' nan values.')",
"There are a total of 8 nan values.\n"
],
[
"ads_df[pd.isnull(ads_df['ad_spend_parsed'])]",
"_____no_output_____"
],
[
"# Remove nulls\nads_df = ads_df[~pd.isnull(ads_df['ad_spend_parsed'])]\n\n# Replace ad_spend with the parse column\nads_df['ad_spend'] = ads_df['ad_spend_parsed']\n\n# Drop the parsed column\nads_df = ads_df.drop(columns=['ad_spend_parsed'])",
"_____no_output_____"
]
],
[
[
"We transform the ad_spend field from string into a float.",
"_____no_output_____"
]
],
[
[
"ads_df['ad_spend'] = ads_df['ad_spend'].astype(float)",
"_____no_output_____"
]
],
[
[
"We validate that all values are positive and remove other values after validation.",
"_____no_output_____"
]
],
[
[
"print('There are ' + str((ads_df['ad_spend'] > 0).sum()) + ' positive values and a total of ' + str(len(ads_df)) + ' entries.')",
"There are 2440 positive values and a total of 2442 entries.\n"
],
[
"cleaning_summary_format['non_positive_ad_spend_count'] = (ads_df['ad_spend'] > 0).sum()\nads_df[ads_df['ad_spend'] <= 0]",
"_____no_output_____"
]
],
[
[
"We remove the two entries with values equal to zero and print out the summary.",
"_____no_output_____"
]
],
[
[
"ads_df = ads_df[ads_df['ad_spend'] > 0]\n\ncleaning_summary_format['before_ad_spend_count'] = cleaning_summary_format['zeros_ad_clicks_count']\n\n# Reporting\nprint('''Before formating ad_spend our dataset had {before_ad_spend_count} columns.\nAfter removing rows with 'None': {none_ad_spend_count} columns.\nAfter removing rows with 0 ad clicks: {non_positive_ad_spend_count} columns.'''.format(**cleaning_summary_format))",
"Before formating ad_spend our dataset had 2450 columns.\nAfter removing rows with 'None': 2442 columns.\nAfter removing rows with 0 ad clicks: 2440 columns.\n"
]
],
[
[
"### Parsing ad_targeting_interests & ad_targeting_people_who_match\n\nThe ad_targeting_interests column is split between its own column and a portion of the ad_targeting_people_who_match column's string. To make treatment of this column simpler, our first step will be to extract the 'interest' portion of ad_targeting_people_who_match. We will then parse the ad_targeting_interests column and combine the result.\n\nFirst we take a look at ad_targeting_people_who_match for entries with and without 'Interests'. We will investigate those without 'Interests' first.",
"_____no_output_____"
]
],
[
[
"count_null = 0\ncount_interests = 0\ncount_other = 0\nfor s in ads_df['ad_targeting_people_who_match']:\n if pd.isnull(s):\n count_null += 1\n elif 'Interests' in s:\n count_interests += 1\n else:\n count_other +=1\n print(s)",
"People who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like United Muslims of America, Friends of connections: Friends of people who are connected to United Muslims of America\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPolitics: Likely to engage with political content (liberal)\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nBehaviors: African American (US)\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Blacktivist, Friends of connections: Friends of people who are connected to Blacktivist\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nBehaviors: African American (US)\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPolitics: Likely to engage with political content (liberal)\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Blacktivist, Friends of connections: Friends of people who are connected to Blacktivist\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Williams&Kalvin, Friends of connections: Friends of people who are connected to Williams&Kalvin\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like United Muslims of America, Friends of connections: Friends of people who are connected to United Muslims of America\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nMulticultural Affinity: African American (US)\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Stop A.I., Friends of connections: Friends of people who are connected to Stop A.1.\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Blacktivist, Friends of connections: Friends of people who are connected to Blacktivist\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nMulticultural Affinity: African American (US)\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nBehaviors: African American (US)\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Born Liberal, Friends of connections: Friends of people who are connected to Born Liberal\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Williams&Kalvin, Friends of connections.Friends of people who are connected to Williams&Kalvin\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Stop A.I., Friends of connections: Friends of people who are connected to Stop A.1.\nPeople who like Stop A.I.. Friends of connections: Friends of people who are connected to Stop A.1.\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Williams&Kalvin, Friends of connections: Friends of people who are connected to Williams&Kalvin\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like United Muslims of America, Friends of connections: Friends of people who are connected to United Muslims of America\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Stop A.I., Friends of connections: Friends of people who are connected to Stop A.1.\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nBehaviors: African American (US)\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Stop A.I., Friends of connections: Friends of people who are connected to Stop A.1.\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPolitics: Likely to engage with political content (conservative)\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Williams&Kalvin, Friends of connections: Friends of people who are connected to Williams&Kalvin\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like United Muslims of America. Friends of connections: Friends of people who are connected to United Muslims of America\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nBehaviors: African American (US)\nPeople who like Williams&Kalvin, Friends of connections: Friends of people who are connected to Williams&Kalvin\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Heart of Texas, Friends of connections: Friends of people who are connected to Heart of Texas\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United. Friends of connections: Friends of people who are connected to LGBT United\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Williams&Kalvin, Friends of connections: Friends of people who are connected to Williams&Kalvin\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Born Liberal, Friends of connections: Friends of people who are connected to Born Liberal\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Williams&Kalvin, Friends of connections: Friends of people who are connected to Williams&Kalvin\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Blacktivist, Friends of connections: Friends of people who are connected to Blacktivist\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Stop A.I., Friends of connections: Friends of people who are connected to Stop A.1.\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Secured Borders; Friends of connections: Friends of people who are connected to Secured Borders\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nBehaviors: African American (US)\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nBehaviors: African American (US)\nPeople who like United Muslims of America.. Friends of connections: Friends of people who are connected to United Muslims of America\nEmployers: Jesus Christ is my King\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Secured Borders; Friends of connections: Friends of people who are connected to Secured Borders\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Memopolis. Friends of connections: Friends of people who are connected to Memopolis\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Stop A.I.; Friends of connections Friends of people who are connected to Stop A.1.\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United. Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nBehaviors: African American (US)\nPeople who like United Muslims of America, Friends of connections: Friends of people who are connected to United Muslims of America\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nBehaviors: African American (US)\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Stop A.I., Friends of connections: Friends of people who are connected to Stop A.1.\nBehaviors: African American (US)\nBehaviors: African American (US)\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Stop A.I., Friends of connections: Friends of people who are connected to Stop A.1.\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like BM, Friends of connections: Friends of people who are connected to BM\nPeople who like LGBT United. Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Williams&Kalvin, Friends of connections: Friends of people who are connected to Williams&Kalvin\nBehaviors: African American (US)\nBehaviors: African American (US)\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Williams&Kalvin, Friends of connections: Friends of people who are connected to Williams&Kalvin\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nBehaviors: African American (US)\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Blacktivist, Friends of connections: Friends of people who are connected to Blacktivist\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nMulticultural Affinity: African American (US)\nPeople who like Blacktivist, Friends of connections: Friends of people who are connected to Blacktivist\nPeople who like Born Liberal, Friends of connections: Friends of people who are connected to Born Liberal\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like United Muslims of America, Friends of connections: Friends of people who are connected to United Muslims of America\nMulticultural Affinity: African American (US)\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPolitics: US politics (conservative), US politics (very conservative) or Likely to engage with political content (conservative)\nBehaviors: African American (US)\nPeople who like United Muslims of America, Friends of connections: Friends of people who are connected to United Muslims of America\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like LGBT United. Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Williams&Kalvin, Friends of connections. Friends of people who are connected to Williams&Kalvin\nPeople who like Williams&Kalvin, Friends of connections: Friends of people who are connected to Williams&Kalvin\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Heart of Texas, Friends of connections: Friends of people who are connected to Heart of Texas\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Stop A.I., Friends of connections: Friends of people who are connected to Stop A.1.\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nMulticultural Affinity: African American (US)\nMulticultural Affinity: African American (US)\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Stop A.I., Friends of connections: Friends of people who are connected to Stop A. I.\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nMulticultural Affinity: African American (US)\nEmployers: Facebook\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Williams&Kalvin, Friends of connections: Friends of people who are connected to Williams&Kalvin\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like United Muslims of America, Friends of connections: Friends of people who are connected to United Muslims of America\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Don't Shoot, Friends of connections: Friends of people who are connected to Don't Shoot\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Black Matters, Friends of connections: Friends of people who are connected to Black Matters\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Williams&Kalvin, Friends of connections: Friends of people who are connected to Williams&Kalvin\nBehaviors: African American (US)\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like LGBT United, Friends of connections: Friends of people who are connected to LGBT United\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Being Patriotic, Friends of connections: Friends of people who are connected to Being Patriotic\nPeople who like Black Matters. Friends of connections: Friends of people who are connected to Black Matters\n"
],
[
"print('Null:' + str(count_null) +\n ' Interests: ' + str(count_interests) +\n ' Other: ' + str(count_other) +\n ' Total: ' + str(count_null + count_interests + count_other))",
"Null:822 Interests: 1243 Other: 375 Total: 2440\n"
]
],
[
[
"From this print out, we see that although some rows belonging to the \"Other\" category have the 'interests' field missing, we can grab a proxy by taking the \"like\" groups. We can grab the value of the \"like\" groups correctly by taking the string after 'Friends of people who are connected to'. We've created the function treat_string_with_friends below to treat these entries.\n\nTo treat the rows with 'Interests', we create the funciton treat_string_with_interest. After looking at a few of the raw_files, we see that ad_targeting_people_who_match sometimes contains other fields. To identify rows which had additonal fields, we looked for the number of ':' characters, we then identified patterns in those strings that didn't match the interests field. These patterns are used in the treat_string_with_interest function below.\n\nThe crop_to_interest function was created to dynamically use the method when interests are present or not.",
"_____no_output_____"
]
],
[
[
"# Utility function to crop everything after a given word\ndef crop_everything_after(string, contains):\n return string[:string.index(contains)] if contains in string else string\n\n# Returns a string containing everything after 'Friends of people who are connected to '\ndef treat_string_with_friends(string):\n friends_string = 'Friends of people who are connected to '\n start = string.index(friends_string)\n return string[start+len(friends_string):]\n\n# Returns a string containing everything in the Interests: marker, but nothing in the other markers (see the crop_after variable)\ndef treat_string_with_interest(string):\n # Crop everything before 'Interests'\n string = string[string.index('Interests'):]\n\n # Strings identified by visual inspections of entries\n crop_after = [\n 'And Must Also Match',\n 'School:',\n 'Behaviors:',\n 'expansion:',\n 'Job title:',\n 'Multicultural Affinity:',\n 'Politics:',\n 'Employers:',\n 'Field of study:'\n ]\n\n for to_crop in crop_after:\n string = crop_everything_after(string, to_crop)\n\n # Finally this substring had a typo\n if 'Stop Racism!:.' in string:\n string = string.replace('Stop Racism!:.', 'Stop Racism!!,')\n \n return string\n\n# If Interests is part of the string use the interest\n# method otherwise use the crop to like method.\ndef crop_to_interest(string):\n if not pd.isnull(string):\n \n if 'Interests' in string:\n string = treat_string_with_interest(string)\n elif 'Friends of people who are connected to ' in string:\n string = treat_string_with_friends(string)\n else:\n # pd.isnull or does not contain interests nor likes\n string = np.nan\n\n return string",
"_____no_output_____"
],
[
"ads_df['ad_targeting_people_who_match'] = ads_df['ad_targeting_people_who_match'].apply(crop_to_interest)",
"_____no_output_____"
]
],
[
[
"During this operation we lost a few rows that could not be parsed as it did not contain interests.",
"_____no_output_____"
]
],
[
[
"cleaning_summary_format['null_people_who_match_count'] = pd.isnull(ads_df['ad_targeting_people_who_match']).sum() - count_null\nprint(str(cleaning_summary_format['null_people_who_match_count']) + ' rows where lost.')",
"29 rows where lost.\n"
]
],
[
[
"The last cleaning step for this field is to remove the 'Interests' keyword which is sometimes followed by a colon. We use a regular expression to replace this string.",
"_____no_output_____"
]
],
[
[
"interests_regex = re.compile(r'Interests\\s*:?')\n\ndef remove_interests_marker(string):\n if not pd.isnull(string):\n string = interests_regex.sub('', string)\n return string",
"_____no_output_____"
],
[
"ads_df['ad_targeting_people_who_match'] = ads_df['ad_targeting_people_who_match'].apply(remove_interests_marker)",
"_____no_output_____"
],
[
"ads_df.head(3)",
"_____no_output_____"
]
],
[
[
"We now do the same exercise with ad_targeting_interests. We first identify non-null rows that may contain an extra field. We do so by looking for the ':' character and printing out these rows.",
"_____no_output_____"
]
],
[
[
"non_null_interests = ads_df[~pd.isnull(ads_df['ad_targeting_interests'])]['ad_targeting_interests']\n\nfor row_with_colon in non_null_interests[non_null_interests.str.contains(':')]:\n print(row_with_colon)",
"BlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nHumanitarianism, Human rights or Humanitarian aid Behaviors: African American (US)\nBlack Power Behaviors: Multicultural Affinity: African American (US)\nHuman rights or Malcolm X Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nHumanitarianism, Human rights or Humanitarian aid Behaviors: African American (US)\nMuslims Are Not Terrorists. Islamism or Muslim Brotherhood Connections: People who like United Muslims of America\nHistory Politics: US politics (conservative)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nHuffPost Black Voices Behaviors: African American (US)\nVeterans, United States Department of Veterans Affairs, Disabled American Veterans or Supporting Our Veterans Home Composition: Veterans in home\nTV talkshows or Black (Color) Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nThe Second Amendment, AR-15, 2nd Amendment or Guns & Ammo Politics: US politics (conservative)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nTax Behaviors: African American (US)\nHomeless shelter Politics: US politics (liberal) or US politics (moderate)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nHuman rights or Malcolm X Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nHuman rights, Police, Police officer or Order of Merit of the Police Forces Behaviors: Multicultural Affinity: Hispanic(US -All), Multicultural Affinity: Hispanic(US - English dominant) or Multicultural Affinity: African American (US)\nMuslims Are Not Terrorists, Islamism or Muslim Brotherhood Connections: People who like United Muslims of America\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nSports Behaviors: Facebook access (mobile): all mobile devices Job title: Combat medic, ???????????, Mercenary, Polisi militer, Engenharia militar or Soldado Generation: Millennials\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nFitness and wellness, Sports and outdoors or Family and relationships Behaviors: Facebook access (mobile): smartphones and tablets Generation: Millennials\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nThe Second Amendment, AR-15, Protect the Second Amendment, 2nd Amendment or Guns & Ammo Politics: US politics (conservative)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nBlackNews.com or HuffPost Black Voices Behaviors: African American (US)\nCop Block Behaviors: African American (US)\n"
]
],
[
[
"Most of these rows seem to contain an additional field \"Behaviors\" we will remove it from the rows as we can easily use the interest part to identify the targeted demographic.",
"_____no_output_____"
]
],
[
[
"# Removes additional section part of the ad_targeting_interests string\ndef treat_interest(string):\n if not pd.isnull(string):\n # Strings identified by visual inspections of entries\n crop_after = [\n 'And Must Also Match',\n 'School:',\n 'Behaviors:',\n 'expansion:',\n 'Job title:',\n 'Multicultural Affinity:',\n 'Politics:',\n 'Employers:',\n 'Field of study:',\n 'Connections:',\n 'Home Composition:'\n ]\n\n for to_crop in crop_after:\n string = crop_everything_after(string, to_crop)\n else:\n # pd.isnull value for strings\n string = np.nan\n \n return string\n\nads_df['ad_targeting_interests'] = ads_df['ad_targeting_interests'].apply(treat_interest)",
"_____no_output_____"
],
[
"non_null_interests = ads_df[~pd.isnull(ads_df['ad_targeting_interests'])]['ad_targeting_interests']\nprint('After treatment, there are ' + str(non_null_interests[non_null_interests.str.contains(':')].count()) +' rows with more than one field.')",
"After treatment, there are 0 rows with more than one field.\n"
]
],
[
[
"Now that both ad_targeting_interests and ad_targeting_people_who_match have been cleaned, we can now merge the two columns into one. First let's verify that there are no rows where both columns are non-null or both null.",
"_____no_output_____"
]
],
[
[
"def interests_both_null(row):\n return (pd.isnull(row.ad_targeting_interests) and pd.isnull(row.ad_targeting_people_who_match))\n\ndef interests_both_non_null(row):\n return (not pd.isnull(row.ad_targeting_interests) and not pd.isnull(row.ad_targeting_people_who_match))\n \n# How many rows have both columns as null \ncleaning_summary_format['null_all_interests_columns_count'] = ads_df.apply(interests_both_null, axis=1).sum()\n\n\n# How many rows have both columns populated\nnon_null_count = ads_df.apply(interests_both_non_null, axis=1).sum()\n\nprint('We have a total of ' + str(both_null) + ' rows with both columns null and a total of ' + str(non_null_count) + ' rows which have both values set.')",
"We have a total of 214 rows with both columns null and a total of 0 rows which have both values set.\n"
]
],
[
[
"We drop rows that do not contain interests information in both columns. We will merge the other rows by replacing the values of ad_targeting_interests with ad_targeting_people_who_match.",
"_____no_output_____"
]
],
[
[
"def merge_interests(row):\n return row.ad_targeting_interests if not pd.isnull(row.ad_targeting_interests) else row.ad_targeting_people_who_match\n\n# Merge interests\nads_df['ad_targeting_interests'] = ads_df.apply(merge_interests, axis=1)\n\n# Drop 'ad_targeting_people_who_match'\nads_df = ads_df.drop(columns=['ad_targeting_people_who_match'])\n\n# Drop null columns\nads_df = ads_df[(~pd.isnull(ads_df['ad_targeting_interests']))]",
"_____no_output_____"
]
],
[
[
"### Writing to file",
"_____no_output_____"
]
],
[
[
"ads_df.head(3)",
"_____no_output_____"
],
[
"ads_df.to_csv('../clean_data/clean_data.csv', index=None, header=True)",
"_____no_output_____"
]
],
[
[
"## Data cleaning summary\n\nWe have lost rows for various reasons during the cleaning:\n\n| Rows lost | Reason |\n|------|------|\n| 20 | Null ad_creation_date column|\n| 1047 | Missing creation_date|\n| 10 | Improper input of ad_spend|\n| 29 | Interests missing from people_who_match |\n|214 | Interests missing from both people_who_match and interests |\n\nFinally, we have decided to keep the following fields:\n\n| Field name | Type | Description |\n|------------------------|----------|-------------------------------------|\n| ad_targeting_interests | string | Interests used to target users |\n| ad_impressions | int | Number of users who saw the ads |\n| ad_clicks | int | Number of times the ads was clicked |\n| ad_spend | float | Money spent on the ad in RUB |\n| ad_creation_date | datetime | Creation date of the ad |\n| ad_end_date | datetime | Date at which the ad stopped |\n\n---\n\nYou can now proceed to the [demographic_labeling](demographic_labeling.ipynb) notebook.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
e79473638c579c9e5a5dec37cb6056dde015b44b | 42,277 | ipynb | Jupyter Notebook | Lyrics_generator.ipynb | NLP-Lyrics-Team/nlp-lyrics | dad129797e7d48a575b214d57c62c3032e70c05d | [
"MIT"
] | null | null | null | Lyrics_generator.ipynb | NLP-Lyrics-Team/nlp-lyrics | dad129797e7d48a575b214d57c62c3032e70c05d | [
"MIT"
] | null | null | null | Lyrics_generator.ipynb | NLP-Lyrics-Team/nlp-lyrics | dad129797e7d48a575b214d57c62c3032e70c05d | [
"MIT"
] | null | null | null | 33.740623 | 151 | 0.487334 | [
[
[
"# Initialize the framework",
"_____no_output_____"
],
[
"Import torch libraries and try to use the GPU device (if available)",
"_____no_output_____"
]
],
[
[
"import torch\nfrom torch import nn\n\nimport random\n\n# Try to use GPU device\ndevice = \"cuda\" if torch.cuda.is_available() else \"cpu\"\nprint(\"Using %s device\" %(device))",
"Using cpu device\n"
]
],
[
[
"Mount Google Drive to load\n* the lyrics dataset,\n* the word2vec pretrained embedding dictionaries,\n* the one hot encoding dictionary for the genres,\n* the lyrics generator neural network\n\n",
"_____no_output_____"
]
],
[
[
"from google.colab import drive\ndrive.mount(\"/content/drive\")",
"Mounted at /content/drive\n"
]
],
[
[
"Load the dictionaries to convert words to indices and viceversa",
"_____no_output_____"
]
],
[
[
"#import pickle\nimport json\n\nFILENAME_W2I = '/content/drive/MyDrive/DM project - NLP lyrics generation/Dictionaries/words2indices'\nFILENAME_I2W = '/content/drive/MyDrive/DM project - NLP lyrics generation/Dictionaries/indices2words'\n\n# Load a dictionary from a stored file converting keys to integers if needed\ndef load_dictionary(filename, convert_keys=False):\n #with open(filename + \".pkl\", \"rb\") as f:\n # return pickle.load(f) \n\n with open(filename + \".json\", \"r\") as f:\n d = json.load(f)\n\n if convert_keys:\n dd = {}\n for key, value in d.items():\n dd[int(key)] = value\n\n return dd\n \n return d\n\nwords2indices = load_dictionary(FILENAME_W2I)\nindices2words = load_dictionary(FILENAME_I2W, convert_keys=True) # JSON stores keys as strings while here we expect integers",
"_____no_output_____"
]
],
[
[
"Load the word vectors tensor (word2vec embedding)",
"_____no_output_____"
]
],
[
[
"FILENAME = '/content/drive/MyDrive/DM project - NLP lyrics generation/Dictionaries/word_vectors.pt'\n\nword_vectors = torch.load(FILENAME, map_location=device)",
"_____no_output_____"
]
],
[
[
"Load the one hot encoding dictionary for the genres",
"_____no_output_____"
]
],
[
[
"FILENAME = '/content/drive/MyDrive/DM project - NLP lyrics generation/Dictionaries/one_hot_encoding_genres'\n\none_hot_encoding_genres = load_dictionary(FILENAME)\nNUMBER_GENRES = len(one_hot_encoding_genres)",
"_____no_output_____"
]
],
[
[
"Define vocabulary functions",
"_____no_output_____"
]
],
[
[
"# Get word from index\ndef get_word_from_index(idx):\n # Use get to automatically return None if the index is not present in the dictionary\n return indices2words.get(idx)\n\n# Get index from word\ndef get_index_from_word(word):\n # Use get to automatically return None if the word is not present in the dictionary\n return words2indices.get(word)\n\n# Get word vector from word\ndef get_word_vector(word):\n idx = get_index_from_word(word)\n return word_vectors[idx] if idx != None else None",
"_____no_output_____"
]
],
[
[
"Define the generator neural network",
"_____no_output_____"
]
],
[
[
"class Generator(nn.Module):\n\n def __init__(\n self,\n word_vectors: torch.Tensor,\n lstm_hidden_size: int,\n dense_size: int,\n vocab_size: int\n ):\n super().__init__()\n \n # Embedding layer\n self.embedding = torch.nn.Embedding.from_pretrained(word_vectors)\n \n # Recurrent layer (LSTM)\n self.rnn = torch.nn.LSTM(input_size=word_vectors.size(1), hidden_size=lstm_hidden_size, num_layers=1, batch_first=True)\n\n # Dense layer\n self.lin1 = torch.nn.Linear(dense_size, vocab_size) # Legacy\n #self.dense = torch.nn.Linear(dense_size, vocab_size)\n #torch.nn.init.uniform_(self.dense.weight)\n\n # Dropout function\n self.dropout = nn.Dropout(p=0.1)\n\t\t\n\t\t# Loss function\n self.loss = torch.nn.CrossEntropyLoss()\n \n self.global_epoch = 0\n \n def forward(self, x, y=None, states=None):\n # Split input in lyrics and genre\n lyrics = x[0]\n genres = x[1]\n\n # Embedding words from indices\n out = self.embedding(lyrics)\n\n # Recurrent layer\n out, states = self.rnn(out, states)\n\n # Duplicate the genre vector associated to a sequence for each word in the sequence\n seq_length = lyrics.size()[1]\n\n if seq_length > 1:\n genres_duplicated = []\n for tensor in genres:\n duplicated = [list(tensor) for i in range(seq_length)]\n genres_duplicated.append(duplicated)\n\n genres = torch.tensor(genres_duplicated, device=device)\n else:\n # Just increment the genres vector dimension\n genres = genres.unsqueeze(0)\n\n\n # Concatenate the LSTM output with the encoding of genres\n out = torch.cat((out, genres), dim=-1)\n\n # Dense layer\n out = self.lin1(out) # Legacy\n #out = self.dense(out)\n\n # Use the last prediction\n logits = out[:, -1, :]\n \n # Scale logits in [0,1] to avoid negative logits\n logits = torch.softmax(logits, dim=-1)\n\n # Max likelihood can return repeated sequences over and over.\n # Sample from the multinomial probability distribution of 'logits' (after softmax). \n # Return the index of the sample (one for each row of the input matrix) \n # that corresponds to the index in the vocabulary as logits are calculated on the whole vocabulary\n sampled_indices = torch.multinomial(logits, num_samples=1)\n \n result = {'logits': logits, 'pred': sampled_indices, 'states': states}\n \n if y is not None:\n result['loss'] = self.loss(logits, y)\n result['accuracy'] = self.accuracy(sampled_indices, y.unsqueeze(-1))\n \n return result\n\n def accuracy(self, pred, target):\n return torch.sum(pred == target) / pred.size()[0]",
"_____no_output_____"
]
],
[
[
"Load the generator model",
"_____no_output_____"
]
],
[
[
"PATH = '/content/drive/MyDrive/DM project - NLP lyrics generation/Models/generator_model.pt'\n#PATH = '/content/drive/MyDrive/DM project - NLP lyrics generation/generator_model_GAN.pt'\n\ngen = Generator(\n word_vectors,\n lstm_hidden_size=256,\n dense_size=256+NUMBER_GENRES,\n vocab_size=len(word_vectors))\n\ncheckpoint = torch.load(PATH, map_location=device)\ngen.load_state_dict(checkpoint['model_state_dict'])\n\n# Try to move the model on the GPU \nif torch.cuda.is_available():\n gen.cuda()",
"_____no_output_____"
]
],
[
[
"# User input",
"_____no_output_____"
],
[
"Sort genres",
"_____no_output_____"
]
],
[
[
"genres = [key for key in one_hot_encoding_genres]\ngenres.sort()",
"_____no_output_____"
]
],
[
[
"Display input form",
"_____no_output_____"
]
],
[
[
"from ipywidgets import Layout, Box, Label, Dropdown, Text\n\nprint(\"Enter a word and a genre to generate a lyrics\\n\")\nform_item_layout = Layout(\n display='flex',\n flex_flow='row',\n justify_content='space-between'\n)\n\nword_widget = Text()\ngenres_widget = Dropdown(options=genres)\n\nform_items = [\n Box([Label(value='Word'), word_widget], layout=form_item_layout), \n Box([Label(value='Genre'), genres_widget], layout=form_item_layout),\n]\n\nform = Box(form_items, layout=Layout(\n display='flex',\n flex_flow='column',\n align_items='stretch',\n width='22%'\n))\nform",
"Enter a word and a genre to generate a lyrics\n\n"
]
],
[
[
"Get user input",
"_____no_output_____"
]
],
[
[
"word = word_widget.value\ngenre = genres_widget.value",
"_____no_output_____"
],
[
"##@title # Insert a word and a genre to generate a lyrics\n#word = \"\" #@param {type:\"string\", required: true}\n#genre = \"Country\" #@param [\"Country\", \"Electronic\", \"Folk\", \"Hip-Hop\", \"Indie\", \"Jazz\", \"Metal\", \"Pop\", \"Rock\", \"R&B\"]",
"_____no_output_____"
]
],
[
[
"Preprocess the user input",
"_____no_output_____"
]
],
[
[
"# Split entered words on whitespaces to support also sequences of words\ninput_words = word.strip().split()\n\nif not input_words:\n raise ValueError(\"No word entered\")\n\n# Check if every input word is present in the vocabulary (or in lowercase form)\nfor word in input_words:\n if word not in words2indices and word.lower() not in words2indices:\n raise ValueError(\"The entered word is not valid\")",
"_____no_output_____"
]
],
[
[
"Generate the lyrics",
"_____no_output_____"
]
],
[
[
"TEXT_LENGTH = 100 # Truncate the text when the goal text length has been generated (hard truncation)\nLINES = random.randrange(10, 50) # Truncate the text when the goal lines number has been generated (soft truncation)\n\nstates = None\ntext = \"\"\nprev_word = \"\"\nlines = 0\ngenerated_words = 0\n\nword2capitalize = [\"I\", \"I'm\", \"I'd\"]\npunctuation_subset = { '.', ',', ';', ':', '!', '?', ')', ']', '}', '$', '/', '…', '...', '..' }\n\n\n# Iterate input words\nfor i in range(len(input_words)):\n w = input_words[i]\n\n # Check if the word is not present in the vocabulary in the current form\n if w not in words2indices:\n # Use the lowercase version (as it must be present in one of the two forms)\n input_words[i] = w.lower()\n\n # Check if this is the first word\n if i == 0:\n # Capitalize the first letter of the word\n w = w[0].upper() + w[1:]\n text = w\n else:\n text += ' ' + w\n\n prev_word = w\n\n# Copy user input words to allow generating multiple lyrics with the same input\ninput_words_ = input_words.copy()\n\n# One hot encode the genre\ninput_genre = one_hot_encoding_genres[genre]\ninput_genre = torch.tensor(input_genre, device=device).unsqueeze(0)\n\n\ndef generate_next_word(input_words, states=None):\n # Convert words to indices\n indices = [get_index_from_word(w) for w in input_words]\n indices = torch.tensor(indices, device=device).unsqueeze(0)\n\n y = gen((indices, input_genre), states=states)\n\n next_word_index = y['pred'].item()\n #print(\"next_word_index:\", next_word_index)\n\n return get_word_from_index(next_word_index), y['states']\n\n\n#for i in range(TEXT_LENGTH):\nwhile lines < LINES:\n # Generate next word\n next_word, states = generate_next_word(input_words_, states)\n \n # Append at the end removing the head\n input_words_ = input_words_[1:]\n input_words_.append(next_word)\n\n #print(\"next word:\", next_word)\n\n # Check if next word must be capitalized in the output text\n for word in word2capitalize:\n if next_word == word.lower():\n # Replace the generated word with the capitalized version\n next_word = word\n break\n\n # Check if previous word is newline (i.e. the generated word belongs to a new line) or a dot\n if prev_word == '\\n' or prev_word == '.':\n # Capitalize the first letter of the generated word\n next_word = next_word[0].upper() + next_word[1:]\n \n # Check if previous word is newline or a parenthesis or next word is newline or punctuation\n if prev_word == '\\n' or prev_word == '(' or next_word == '\\n' or next_word in punctuation_subset:\n if next_word == '\\n':\n # Update generated lines\n lines += 1\n\n # Check if the number of lines has been achieved\n if lines == LINES:\n break\n\n # Add the generated word to the output text without prepending a space\n text += next_word\n\n else:\n # Add the generated word to the output text prepending a space\n text += ' ' + next_word\n\n prev_word = next_word\n generated_words += 1\n\n\nprint(\"Word:\", input_words)\nprint(\"Genre:\", genre)\nprint(\"\\nlines:\", LINES)\nprint(\"generated words:\", generated_words)\nprint(\"\\nLyrics:\")\nprint(text)",
"Word: ['saturday']\nGenre: Rock\n\nlines: 27\ngenerated words: 233\n\nLyrics:\nSaturday night\nIn the midnight room for the street\nShe walked dressed to themselves\nYou should have took her away, in a call as she cried for me\nI was given to: about your mom. Punisher's bad news\nYou living why a girl I told you before\nDon't you think that's the world?\nBlack space, smells blue?\nThere was her name and way: to the father french, out of the albums\nBring your blood underneath your gut bare\nIf you move your house\nDon't know me\nNo I don't really want I love you (just make you to catch me!)\nShe's in the zone\nI don't need you, half souls, forgive me, or nothing\n\nYou could ever leave it\nWhen all fired this ray right into town\nI don't even dream, fuck, step in my limousine\nI can swear your ways\nAnd leave me the questions I will know you are ready\nIn the wonderful dream\nI like my real strange skies would tell me to find that girl I laid to rest\nOn the telephone I just steal trouble\nMy whole loving life\nThough it's over, I can't go back\n\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7947546bd84765465798794225eaa0f42af55e1 | 1,438 | ipynb | Jupyter Notebook | 00_core.ipynb | Rahuketu86/Learnathon | 6a9d05c03be138edd238432543b7ce48840c9b1d | [
"Apache-2.0"
] | null | null | null | 00_core.ipynb | Rahuketu86/Learnathon | 6a9d05c03be138edd238432543b7ce48840c9b1d | [
"Apache-2.0"
] | 3 | 2021-05-20T12:34:27.000Z | 2022-02-26T06:24:23.000Z | 00_core.ipynb | Rahuketu86/learnathon | 6a9d05c03be138edd238432543b7ce48840c9b1d | [
"Apache-2.0"
] | null | null | null | 17.325301 | 52 | 0.497218 | [
[
[
"# default_exp core",
"_____no_output_____"
]
],
[
[
"# core\n\n> This is module which provide core utilities ",
"_____no_output_____"
]
],
[
[
"#hide\nfrom nbdev.showdoc import *",
"_____no_output_____"
],
[
"#export\ndef say_hello():\n return \"Hello From Learnathon Module\"",
"_____no_output_____"
],
[
"#export\ndef say_hello2():\n return \"This is a test for new function\"",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
e794825cd33e29b3e051c80857e0eb01889ad353 | 87,583 | ipynb | Jupyter Notebook | examples/090.sma-percent-band/optimize.ipynb | alialamiidrissi/pinkfish | c34920d970281b60ae4d46d6c52af13f6b3761f0 | [
"MIT"
] | 197 | 2015-10-03T13:23:05.000Z | 2022-03-27T03:26:34.000Z | examples/090.sma-percent-band/optimize.ipynb | alialamiidrissi/pinkfish | c34920d970281b60ae4d46d6c52af13f6b3761f0 | [
"MIT"
] | 30 | 2015-12-19T21:26:20.000Z | 2022-03-21T01:27:47.000Z | examples/090.sma-percent-band/optimize.ipynb | alialamiidrissi/pinkfish | c34920d970281b60ae4d46d6c52af13f6b3761f0 | [
"MIT"
] | 56 | 2015-12-19T02:02:53.000Z | 2022-03-26T19:36:51.000Z | 107.200734 | 32,224 | 0.794309 | [
[
[
"# SMA Percent Band\n\n 1. The SPY closes above its upper band, buy\n 2. If the SPY closes below its lower band, sell your long position.\n \n Optimize: sma, percent band.",
"_____no_output_____"
]
],
[
[
"import datetime\n\nimport matplotlib.pyplot as plt\nimport pandas as pd\nfrom talib.abstract import *\n\nimport pinkfish as pf\nimport strategy\n\n# Format price data\npd.options.display.float_format = '{:0.2f}'.format\n\n%matplotlib inline",
"_____no_output_____"
],
[
"# Set size of inline plots\n'''note: rcParams can't be in same cell as import matplotlib\n or %matplotlib inline\n \n %matplotlib notebook: will lead to interactive plots embedded within\n the notebook, you can zoom and resize the figure\n \n %matplotlib inline: only draw static images in the notebook\n'''\nplt.rcParams[\"figure.figsize\"] = (10, 7)",
"_____no_output_____"
]
],
[
[
"Some global data",
"_____no_output_____"
]
],
[
[
"symbol = '^GSPC'\n#symbol = 'SPY'\n#symbol = 'ES=F'\n#symbol = 'DIA'\n#symbol = 'QQQ'\n#symbol = 'IWM'\n#symbol = 'TLT'\n#symbol = 'GLD'\n#symbol = 'AAPL'\n#symbol = 'BBRY'\n#symbol = 'GDX'\ncapital = 10000\nstart = datetime.datetime(1900, 1, 1)\n#start = datetime.datetime(*pf.SP500_BEGIN)\nend = datetime.datetime.now()",
"_____no_output_____"
]
],
[
[
"Define Optimizations",
"_____no_output_____"
]
],
[
[
"# pick one\noptimize_sma = True\noptimize_band = False\n\n# define SMAs ranges\nif optimize_sma:\n Xs = range(50, 525, 25)\n Xs = [str(X) for X in Xs]\n\n# define band ranges\nelif optimize_band:\n Xs = range(0, 100, 5)\n Xs = [str(X) for X in Xs]",
"_____no_output_____"
],
[
"options = {\n 'use_adj' : True,\n 'use_cache' : True,\n 'sma' : 200,\n 'band' : 0.0\n}",
"_____no_output_____"
]
],
[
[
"Run Strategy",
"_____no_output_____"
]
],
[
[
"strategies = pd.Series(dtype=object)\nfor X in Xs:\n print(X, end=\" \")\n if optimize_sma:\n options['sma'] = int(X)\n elif optimize_band:\n options['band'] = int(X)/10\n \n strategies[X] = strategy.Strategy(symbol, capital, start, end, options) \n strategies[X].run()",
"50 75 100 125 150 175 200 225 250 275 300 325 350 375 400 425 450 475 500 "
]
],
[
[
"Summarize results",
"_____no_output_____"
]
],
[
[
"metrics = ('annual_return_rate',\n 'max_closed_out_drawdown',\n 'annualized_return_over_max_drawdown',\n 'drawdown_recovery_period',\n 'expected_shortfall',\n 'best_month',\n 'worst_month',\n 'sharpe_ratio',\n 'sortino_ratio',\n 'monthly_std',\n 'pct_time_in_market',\n 'total_num_trades',\n 'pct_profitable_trades',\n 'avg_points')\n\ndf = pf.optimizer_summary(strategies, metrics)\ndf",
"_____no_output_____"
]
],
[
[
"Bar graphs",
"_____no_output_____"
]
],
[
[
"pf.optimizer_plot_bar_graph(df, 'annual_return_rate')\npf.optimizer_plot_bar_graph(df, 'sharpe_ratio')\npf.optimizer_plot_bar_graph(df, 'max_closed_out_drawdown')",
"_____no_output_____"
]
],
[
[
"Run Benchmark",
"_____no_output_____"
]
],
[
[
"s = strategies[Xs[0]]\nbenchmark = pf.Benchmark(symbol, capital, s.start, s.end)\nbenchmark.run()",
"_____no_output_____"
]
],
[
[
"Equity curve",
"_____no_output_____"
]
],
[
[
"if optimize_sma : Y = '200'\nelif optimize_band: Y = '30'\n\npf.plot_equity_curve(strategies[Y].dbal, benchmark=benchmark.dbal)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e79487095c51bf1b201a78078c2505f6f4db6d86 | 10,831 | ipynb | Jupyter Notebook | examples/estimator/classifier/MLPClassifier/js/basics_imported.pct.ipynb | karoka/sklearn-porter | f57b6d042c9ae18c6bb8c027362f9e40bfd34d63 | [
"MIT"
] | 1,197 | 2016-08-30T14:49:34.000Z | 2022-03-30T05:38:52.000Z | examples/estimator/classifier/MLPClassifier/js/basics_imported.pct.ipynb | karoka/sklearn-porter | f57b6d042c9ae18c6bb8c027362f9e40bfd34d63 | [
"MIT"
] | 80 | 2016-11-18T17:37:19.000Z | 2022-03-25T12:41:40.000Z | examples/estimator/classifier/MLPClassifier/js/basics_imported.pct.ipynb | karoka/sklearn-porter | f57b6d042c9ae18c6bb8c027362f9e40bfd34d63 | [
"MIT"
] | 171 | 2016-08-25T20:05:27.000Z | 2022-03-28T07:39:54.000Z | 33.532508 | 158 | 0.419167 | [
[
[
"# sklearn-porter\n\nRepository: [https://github.com/nok/sklearn-porter](https://github.com/nok/sklearn-porter)\n\n## MLPClassifier\n\nDocumentation: [sklearn.neural_network.MLPClassifier](http://scikit-learn.org/stable/modules/generated/sklearn.neural_network.MLPClassifier.html)",
"_____no_output_____"
]
],
[
[
"import sys\nsys.path.append('../../../../..')",
"_____no_output_____"
]
],
[
[
"### Load data",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import load_iris\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.utils import shuffle\n\niris_data = load_iris()\nX = iris_data.data\ny = iris_data.target\n\nX = shuffle(X, random_state=0)\ny = shuffle(y, random_state=0)\n\nX_train, X_test, y_train, y_test = train_test_split(\n X, y, test_size=0.4, random_state=5)\n\nprint(X_train.shape, y_train.shape)\nprint(X_test.shape, y_test.shape)",
"((90, 4), (90,))\n((60, 4), (60,))\n"
]
],
[
[
"### Train classifier",
"_____no_output_____"
]
],
[
[
"from sklearn.neural_network import MLPClassifier\n\nclf = MLPClassifier(activation='relu', hidden_layer_sizes=50,\n max_iter=500, alpha=1e-4, solver='sgd',\n tol=1e-4, random_state=1, learning_rate_init=.1)\nclf.fit(X_train, y_train)",
"_____no_output_____"
]
],
[
[
"### Transpile classifier",
"_____no_output_____"
]
],
[
[
"from sklearn_porter import Porter\n\nporter = Porter(clf, language='js')\noutput = porter.export(export_data=True)\n\nprint(output)",
"if (typeof XMLHttpRequest === 'undefined') {\n var XMLHttpRequest = require(\"xmlhttprequest\").XMLHttpRequest;\n}\n\nvar MLPClassifier = function(jsonFile) {\n this.mdl = undefined;\n\n var promise = new Promise(function(resolve, reject) {\n var httpRequest = new XMLHttpRequest();\n httpRequest.onreadystatechange = function() {\n if (httpRequest.readyState === 4) {\n if (httpRequest.status === 200) {\n resolve(JSON.parse(httpRequest.responseText));\n } else {\n reject(new Error(httpRequest.status + ': ' + httpRequest.statusText));\n }\n }\n };\n httpRequest.open('GET', jsonFile, true);\n httpRequest.send();\n });\n\n // Return max index:\n var maxi = function(nums) {\n var index = 0;\n for (var i=0, l=nums.length; i < l; i++) {\n index = nums[i] > nums[index] ? i : index;\n }\n return index;\n };\n\n // Compute the activation function:\n var compute = function(activation, v) {\n switch (activation) {\n case 'LOGISTIC':\n for (var i = 0, l = v.length; i < l; i++) {\n v[i] = 1. / (1. + Math.exp(-v[i]));\n }\n break;\n case 'RELU':\n for (var i = 0, l = v.length; i < l; i++) {\n v[i] = Math.max(0, v[i]);\n }\n break;\n case 'TANH':\n for (var i = 0, l = v.length; i < l; i++) {\n v[i] = Math.tanh(v[i]);\n }\n break;\n case 'SOFTMAX':\n var max = Number.NEGATIVE_INFINITY;\n for (var i = 0, l = v.length; i < l; i++) {\n if (v[i] > max) {\n max = v[i];\n }\n }\n for (var i = 0, l = v.length; i < l; i++) {\n v[i] = Math.exp(v[i] - max);\n }\n var sum = 0.0;\n for (var i = 0, l = v.length; i < l; i++) {\n sum += v[i];\n }\n for (var i = 0, l = v.length; i < l; i++) {\n v[i] /= sum;\n }\n break;\n }\n return v;\n };\n\n this.predict = function(neurons) {\n return new Promise(function(resolve, reject) {\n promise.then(function(mdl) {\n\n // Initialization:\n if (typeof this.mdl === 'undefined') {\n mdl.hidden_activation = mdl.hidden_activation.toUpperCase();\n mdl.output_activation = mdl.output_activation.toUpperCase();\n mdl.network = new Array(mdl.layers.length + 1);\n for (var i = 0, l = mdl.layers.length; i < l; i++) {\n mdl.network[i + 1] = new Array(mdl.layers[i]).fill(0.);\n }\n this.mdl = mdl;\n }\n\n // Feed forward:\n this.mdl.network[0] = neurons;\n for (var i = 0; i < this.mdl.network.length - 1; i++) {\n for (var j = 0; j < this.mdl.network[i + 1].length; j++) {\n for (var l = 0; l < this.mdl.network[i].length; l++) {\n this.mdl.network[i + 1][j] += this.mdl.network[i][l] * this.mdl.weights[i][l][j];\n }\n this.mdl.network[i + 1][j] += this.mdl.bias[i][j];\n }\n if ((i + 1) < (this.mdl.network.length - 1)) {\n this.mdl.network[i + 1] = compute(this.mdl.hidden_activation, this.mdl.network[i + 1]);\n }\n }\n this.mdl.network[this.mdl.network.length - 1] = compute(this.mdl.output_activation, this.mdl.network[this.mdl.network.length - 1]);\n\n // Return result:\n if (this.mdl.network[this.mdl.network.length - 1].length == 1) {\n if (this.mdl.network[this.mdl.network.length - 1][0] > .5) {\n resolve(1);\n }\n resolve(0);\n } else {\n resolve(maxi(this.mdl.network[this.mdl.network.length - 1]));\n }\n }, function(error) {\n reject(error);\n });\n });\n };\n};\n\nif (typeof process !== 'undefined' && typeof process.argv !== 'undefined') {\n if (process.argv[2].trim().endsWith('.json')) {\n\n // Features:\n var features = process.argv.slice(3);\n\n // Parameters:\n var json = process.argv[2];\n\n // Estimator:\n var clf = new MLPClassifier(json);\n\n // Prediction:\n clf.predict(features).then(function(prediction) {\n console.log(prediction);\n }, function(error) {\n console.log(error);\n });\n\n }\n}\n"
]
],
[
[
"### Run classification in JavaScript",
"_____no_output_____"
]
],
[
[
"# Save classifier:\n# with open('MLPClassifier.js', 'w') as f:\n# f.write(output)\n\n# Check model data:\n# $ cat data.json\n\n# Run classification:\n# if hash node 2/dev/null; then\n# python -m SimpleHTTPServer 8877 & serve_pid=$!\n# node MLPClassifier.js http://127.0.0.1:8877/data.json 1 2 3 4\n# kill $serve_pid\n# fi",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.