hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
sequence | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
sequence | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
sequence | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
sequence | cell_types
sequence | cell_type_groups
sequence |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
e78b31d3b76556aba97a5a6960aee261b95c34eb | 132,738 | ipynb | Jupyter Notebook | AdvancedDataAnalysis/Session12 Geospatial/Notebook-4-Data-Borrowing.ipynb | robretoarenal/BTS2 | deeafc3892d5be5d2e65bea9ab1613761ab26499 | [
"MIT"
] | 1 | 2022-02-02T16:58:10.000Z | 2022-02-02T16:58:10.000Z | AdvancedDataAnalysis/Session12 Geospatial/Notebook-4-Data-Borrowing.ipynb | jlsjefferson/BTS2 | deeafc3892d5be5d2e65bea9ab1613761ab26499 | [
"MIT"
] | null | null | null | AdvancedDataAnalysis/Session12 Geospatial/Notebook-4-Data-Borrowing.ipynb | jlsjefferson/BTS2 | deeafc3892d5be5d2e65bea9ab1613761ab26499 | [
"MIT"
] | 1 | 2022-02-02T16:58:01.000Z | 2022-02-02T16:58:01.000Z | 99.058209 | 77,236 | 0.742734 | [
[
[
"# Concepts in Spatial Linear Modelling\n\n### Data Borrowing in Supervised Learning",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport libpysal.weights as lp\nimport geopandas as gpd\nimport pandas as pd\nimport shapely.geometry as shp\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n%matplotlib inline",
"_____no_output_____"
],
[
"listings = pd.read_csv('./data/berlin-listings.csv.gz')\nlistings['geometry'] = listings[['longitude','latitude']].apply(shp.Point, axis=1)\nlistings = gpd.GeoDataFrame(listings)\nlistings.crs = {'init':'epsg:4269'}\nlistings = listings.to_crs(epsg=3857)",
"/usr/local/lib/python3.7/site-packages/pyproj/crs/crs.py:68: FutureWarning: '+init=<authority>:<code>' syntax is deprecated. '<authority>:<code>' is the preferred initialization method. When making the change, be mindful of axis order changes: https://pyproj4.github.io/pyproj/stable/gotchas.html#axis-order-changes-in-proj-6\n return _prepare_from_string(\" \".join(pjargs))\n"
],
[
"listings[['scrape_id', 'name',\n 'summary', 'space', 'description', 'experiences_offered',\n 'neighborhood_overview', 'notes', 'transit', 'access', 'interaction',\n 'house_rules', 'thumbnail_url', 'medium_url', 'picture_url',\n 'xl_picture_url', 'host_id', 'host_url', 'host_name', 'host_since',\n 'host_location', 'host_about']].head()",
"_____no_output_____"
],
[
"listings.sort_values('price').plot('price', cmap='plasma')",
"_____no_output_____"
]
],
[
[
"# Kernel Regressions",
"_____no_output_____"
],
[
"Kernel regressions are one exceptionally common way to allow observations to \"borrow strength\" from nearby observations. \n\nHowever, when working with spatial data, there are *two simultaneous senses of what is near:* \n- things that similar in attribute (classical kernel regression)\n- things that are similar in spatial position (spatial kernel regression)\n\nBelow, we'll walk through how to use scikit to fit these two types of kernel regressions, show how it's not super simple to mix the two approaches together, and refer to an approach that does this correctly in another package. ",
"_____no_output_____"
],
[
"First, though, let's try to predict the log of an Airbnb's nightly price based on a few factors:\n- accommodates: the number of people the airbnb can accommodate\n- review_scores_rating: the aggregate rating of the listing\n- bedrooms: the number of bedrooms the airbnb has\n- bathrooms: the number of bathrooms the airbnb has\n- beds: the number of beds the airbnb offers",
"_____no_output_____"
]
],
[
[
"model_data = listings[['accommodates', 'review_scores_rating', \n 'bedrooms', 'bathrooms', 'beds', \n 'price', 'geometry']].dropna()",
"_____no_output_____"
],
[
"Xnames = ['accommodates', 'review_scores_rating', \n 'bedrooms', 'bathrooms', 'beds' ]\nX = model_data[Xnames].values\nX = X.astype(float)\ny = np.log(model_data[['price']].values)",
"_____no_output_____"
]
],
[
[
"Further, since each listing has a location, I'll extract the set of spatial coordinates coordinates for each listing.",
"_____no_output_____"
]
],
[
[
"coordinates = np.vstack(model_data.geometry.apply(lambda p: np.hstack(p.xy)).values)",
"_____no_output_____"
]
],
[
[
"scikit neighbor regressions are contained in the `sklearn.neighbors` module, and there are two main types:\n- `KNeighborsRegressor`, which uses a k-nearest neighborhood of observations around each focal site\n- `RadiusNeighborsRegressor`, which considers all observations within a fixed radius around each focal site.\n\nFurther, these methods can use inverse distance weighting to rank the relative importance of sites around each focal; in this way, near things are given more weight than far things, even when there's a lot of near things. ",
"_____no_output_____"
]
],
[
[
"import sklearn.neighbors as skn\nimport sklearn.metrics as skm",
"_____no_output_____"
],
[
"shuffle = np.random.permutation(len(y))\ntrain,test = shuffle[:14000],shuffle[14000:]",
"_____no_output_____"
]
],
[
[
"So, let's fit three models:\n- `spatial`: using inverse distance weighting on the nearest 500 neighbors geograpical space\n- `attribute`: using inverse distance weighting on the nearest 500 neighbors in attribute space\n- `both`: using inverse distance weighting in both geographical and attribute space. ",
"_____no_output_____"
]
],
[
[
"KNNR = skn.KNeighborsRegressor(weights='distance', n_neighbors=500)\nspatial = KNNR.fit(coordinates[train,:],\n y[train,:])\nKNNR = skn.KNeighborsRegressor(weights='distance', n_neighbors=500)\nattribute = KNNR.fit(X[train,:],\n y[train,])\nKNNR = skn.KNeighborsRegressor(weights='distance', n_neighbors=500)\nboth = KNNR.fit(np.hstack((coordinates,X))[train,:],\n y[train,:])",
"_____no_output_____"
]
],
[
[
"To score them, I'm going to grab their out of sample prediction accuracy and get their % explained variance:",
"_____no_output_____"
]
],
[
[
"sp_ypred = spatial.predict(coordinates[test,:])\natt_ypred = attribute.predict(X[test,:])\nboth_ypred = both.predict(np.hstack((X,coordinates))[test,:])",
"_____no_output_____"
],
[
"(skm.explained_variance_score(y[test,], sp_ypred),\n skm.explained_variance_score(y[test,], att_ypred),\n skm.explained_variance_score(y[test,], both_ypred))",
"_____no_output_____"
]
],
[
[
"If you don't know $X$, using $Wy$ would be better than nothing, but it works nowhere near as well... less than half of the variance that is explained by nearness in feature/attribute space is explained by nearness in geographical space. \n\nMaking things even worse, simply glomming on the geographical information to the feature set makes the model perform horribly. \n\n*There must be another way!*\n\nOne method that can exploit the fact that local data may be more informative in predicting $y$ at site $i$ than distant data is Geographically Weighted Regression, a type of Generalized Additive Spatial Model. Kind of like a Kernel Regression, GWR conducts a bunch of regressions at each training site only considering data near that site. This means it works like the kernel regressions above, but uses *both* the coordinates *and* the data in $X$ to predict $y$ at each site. It optimizes its sense of \"local\" depending on some information criteria or fit score.\n\nYou can find this in the `gwr` package, and significant development is ongoing on this at `https://github.com/pysal/gwr`.",
"_____no_output_____"
],
[
"# Data Borrowing",
"_____no_output_____"
],
[
"Another common case where these weights are used are in \"feature engineering.\" Using the weights matrix, you can construct neighbourhood averages of the data matrix and use these as synthetic features in your model. These often have a strong relationship to the outcome as well, since spatial data is often smooth and attributes of nearby sites often have a spillover impact on each other. ",
"_____no_output_____"
],
[
"First, we'll construct a Kernel weight from the data that we have, make it an adaptive Kernel bandwidth, and make sure that our kernel weights don't have any self-neighbors. Since we've got the data at each site anyway, we probably shouldn't use that data *again* when we construct our neighborhood-smoothed syntetic features. ",
"_____no_output_____"
]
],
[
[
"from libpysal.weights.util import fill_diagonal\n\nkW = lp.Kernel.from_dataframe(model_data, fixed=False, function='gaussian', k=100)\nkW = fill_diagonal(kW, 0)\nWX = lp.lag_spatial(kW, X)",
"_____no_output_____"
],
[
"WX",
"_____no_output_____"
],
[
"kW.to_adjlist()[kW.to_adjlist()[\"focal\"]== 1]",
"_____no_output_____"
]
],
[
[
"I like `statsmodels` regression summary tables, so I'll pop it up here. ",
"_____no_output_____"
],
[
"Below are the results for the model with only the covariates used above:\n- accommodates: the number of people the airbnb can accommodate\n- review_scores_rating: the aggregate rating of the listing\n- bedrooms: the number of bedrooms the airbnb has\n- bathrooms: the number of bathrooms the airbnb has\n- beds: the number of beds the airbnb offers\n\nWe've not used any of our synthetic features in `WX`. ",
"_____no_output_____"
]
],
[
[
"import statsmodels.api as sm\nXtable = pd.DataFrame(X, columns=Xnames)\nonlyX = sm.OLS(y,sm.add_constant(Xtable)).fit()\nonlyX.summary()",
"_____no_output_____"
]
],
[
[
"Then, we could fit a model using the neighbourhood average synthetic features as well:",
"_____no_output_____"
]
],
[
[
"WXtable = pd.DataFrame(WX, columns=['lag_{}'.format(name) for name in Xnames])\nWXtable.head()",
"_____no_output_____"
],
[
"XWXtable = pd.concat((Xtable,WXtable),axis=1)\nXWXtable.head()",
"_____no_output_____"
],
[
"withWX = sm.OLS(y,sm.add_constant(XWXtable)).fit()\nwithWX.summary()",
"_____no_output_____"
]
],
[
[
"This gains a nice bump in the model fit with no significant hit to model complexity. ",
"_____no_output_____"
],
[
"## Going Further",
"_____no_output_____"
],
[
"We could also use a spatial autoregressive model to further improve fit. This ceases to be estimatable in `statsmodels` and instead requires `pysal.spreg`, the spatial regression submodule of PySAL. Generalized method of moments estimators are available in `pysal.spreg.GM_Lag`, and maximum likelihood methods in `pysal.spreg.ML_Lag`. \n\nThese methods are often harder to fit, though, so like `gwr`, they may be less performant on big data. But, you can usually achieve a gain in fit for no significant increase in the number of terms by using these models. ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
e78b3495f55cdec1c5f9afaf779dfc3d92a06fed | 11,385 | ipynb | Jupyter Notebook | Alignment.ipynb | TomAugspurger/pandorable-pandas | 853725ba272dd519ed181445eb6330f3f7a23dc1 | [
"CC-BY-4.0"
] | 2 | 2020-11-13T17:26:59.000Z | 2020-11-16T17:53:43.000Z | Alignment.ipynb | TomAugspurger/pandorable-pandas | 853725ba272dd519ed181445eb6330f3f7a23dc1 | [
"CC-BY-4.0"
] | 1 | 2020-11-13T21:28:32.000Z | 2020-11-14T20:16:33.000Z | Alignment.ipynb | TomAugspurger/pandorable-pandas | 853725ba272dd519ed181445eb6330f3f7a23dc1 | [
"CC-BY-4.0"
] | 3 | 2020-11-13T06:03:53.000Z | 2020-11-13T19:16:01.000Z | 25.875 | 259 | 0.565569 | [
[
[
"# Alignment\n\nThis notebook covers [*alignment*](https://pandas.pydata.org/docs/user_guide/dsintro.html#dsintro-alignment), a feature of pandas that's crucial to using it well. It relies on the pandas' handling of *labels*.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"## Goal: Compute Real GDP\n\nLet's learn through an example: Gross Domestic Product (the total output of a country) is measured in dollars. This means we we can't just compare the GDP from 1950 to the GDP from 2000, since the value of a dollar changed over that time (inflation).\n\nIn the US, the Bureau of Economic Analysis already provides an estimate of real GDP, but we'll calculate something similar using the formula:\n\n$$\nreal\\_GDP = \\frac{nominal\\_GDP}{price\\_index}\n$$\n\nI've downloaded a couple time series from [FRED](https://fred.stlouisfed.org), one for GDP and one for the Consumer Price Index.\n\n* U.S. Bureau of Labor Statistics, Consumer Price Index for All Urban Consumers: All Items in U.S. City Average [CPIAUCSL], retrieved from FRED, Federal Reserve Bank of St. Louis; https://fred.stlouisfed.org/series/CPIAUCSL, October 31, 2020.\n* U.S. Bureau of Economic Analysis, Gross Domestic Product [GDP], retrieved from FRED, Federal Reserve Bank of St. Louis; https://fred.stlouisfed.org/series/GDP, October 31, 2020.\n\n\nWe're going to do things the wrong way first.",
"_____no_output_____"
]
],
[
[
"gdp_bad = pd.read_csv(\"data/GDP.csv.gz\", parse_dates=[\"DATE\"])\ncpi_bad = pd.read_csv(\"data/CPIAUCSL.csv.gz\", parse_dates=[\"DATE\"])",
"_____no_output_____"
]
],
[
[
"Our formula says `real_gdp = gdp / cpi`, so, let's try it!",
"_____no_output_____"
]
],
[
[
"%xmode plain\n\ngdp_bad / cpi_bad",
"_____no_output_____"
]
],
[
[
"Whoops, what happened? We should probably look at our data:",
"_____no_output_____"
]
],
[
[
"gdp_bad",
"_____no_output_____"
],
[
"gdp_bad.dtypes",
"_____no_output_____"
],
[
"gdp_bad['DATE'][0]",
"_____no_output_____"
]
],
[
[
"So, we've tried to divide a datetime by a datetime, and pandas has correctly raised a type error. That raises another issue though. These two timeseries have different frequencies.",
"_____no_output_____"
]
],
[
[
"cpi_bad.head()",
"_____no_output_____"
]
],
[
[
"CPI is measured monthly, while GDP is quarterly. What we'd really need to do is *join* the two timeseries on the `DATE` variable, and then do the operation. We could do that, but let's do things the pandorable way first.\n\nA DataFrame is a 2-D data structure composed of three components:\n\n1. The *values*, the actual data\n2. The *row labels*, stored in a `pandas.Index` class, accessible with `.index`\n3. The *column labels*, stored in a `pandas.Index` class, accessible with `.columns`\n\n\n\n\nWe'll use the *index* to store our *labels* (the dates). Then the only thing in the values is our observations (the GDP or CPI).",
"_____no_output_____"
]
],
[
[
"# Notice that we select the GDP column to convert the\n# 1-column DataFrame to a 1D Series\ngdp = pd.read_csv('data/GDP.csv.gz', index_col='DATE',\n parse_dates=['DATE'])[\"GDP\"]\ngdp.head()",
"_____no_output_____"
]
],
[
[
"Notice that we selected the single column `\"GDP\"` using `[]`. This returns a `pandas.Series` object, a 1-D array *with row labels*.\n\n",
"_____no_output_____"
]
],
[
[
"type(gdp)",
"_____no_output_____"
],
[
"gdp.index",
"_____no_output_____"
]
],
[
[
"The actual values are a NumPy array of floats.",
"_____no_output_____"
]
],
[
[
"gdp.to_numpy()[:10]",
"_____no_output_____"
]
],
[
[
"Let's read in CPI as well.",
"_____no_output_____"
]
],
[
[
"cpi = pd.read_csv('data/CPIAUCSL.csv.gz', index_col='DATE',\n parse_dates=['DATE'])[\"CPIAUCSL\"]\ncpi.head()",
"_____no_output_____"
]
],
[
[
"And let's try the formula again.",
"_____no_output_____"
]
],
[
[
"rgdp = gdp / cpi\nrgdp",
"_____no_output_____"
]
],
[
[
"**What happened?**\n\nWe've gotten our answer, but is there anything in the output that's surprising? What are these `NaN`s?\n\nIn pandas, any time you do an operation involving multiple pandas objects (dataframes, series), pandas will *align* the inputs. Alignment is a two-step process:\n\n1. Take the union of the labels\n2. Reindex the all inputs to the union of the labels, introducing NAs where there's new values.\n\nOnly after that does the operation (division in this case) happen.\n\nLooking at the raw data, we see that CPI is measured monthly, while GDP is just measured quarterly. So pandas has aligned the two (to monthly frequency, since that's the union), inserting missing values where there weren't any previously.",
"_____no_output_____"
]
],
[
[
"# manual alignment, just for demonstration:\n\nall_dates = gdp.index.union(cpi.index)\nall_dates",
"_____no_output_____"
],
[
"gdp2 = gdp.reindex(all_dates)\ngdp2",
"_____no_output_____"
],
[
"cpi2 = cpi.reindex(all_dates)\ncpi2",
"_____no_output_____"
],
[
"rgdp2 = gdp2 / cpi2\nrgdp2",
"_____no_output_____"
]
],
[
[
"So when we wrote\n\n```python\nrgdp = gdp / cpi\n```\n\npandas performs\n\n```python\nall_dates = gdp.index.union(cpi.index)\nrgdp = gdp.reindex(all_dates) / cpi.reindex(all_dates)\n```\n\nThis behavior is somewhat peculiar to pandas. But once you're used to it it's hard to go back. pandas handling the labels / alignment elimiates a class of errors that come from datasets not being aligned.",
"_____no_output_____"
],
[
"## Missing Data\n\nJust a quick aside on handling missing data: pandas provides tools for detecting and dealing with missing data. We'll use these throughout the tutorial.",
"_____no_output_____"
]
],
[
[
"rgdp.isna()",
"_____no_output_____"
],
[
"rgdp.dropna()",
"_____no_output_____"
],
[
"rgdp.fillna(method='ffill') # or fill with a scalar.",
"_____no_output_____"
]
],
[
[
"## Exercise:\n\nNormalize real GDP to year **2000** dollars.\n\nRight now, the unit on the `CPI` variable is \"Index 1982-1984=100\". This means that \"index value\" for the Consumer Price *Index* show year is the average of 1982 - 1984.",
"_____no_output_____"
]
],
[
[
"# use `.loc[start:end]` or `.loc[\"<year>\"]` to slice a subset of *rows*\ncpi.loc['1982':'1984'].mean() # close enough to 100",
"_____no_output_____"
]
],
[
[
"To *renormalize* an index like CPI, divide the series by the average of a different timespan (say the year 2000) and multiply by 100.",
"_____no_output_____"
]
],
[
[
"# Get the mean CPI for the year 2000\ncpi_2000_average = cpi.loc[...]...\n\n# *renormalize* the entire `cpi` series to \"Index 2000\" units.\ncpi_2000 = 100 * (... / ...)\n\n# Compute real GDP again, this time in \"year 2000 dollars\".\nrgdp_2000 = ...\nrgdp_2000",
"_____no_output_____"
],
[
"%load solutions/alignment-cpi2000.py",
"_____no_output_____"
]
],
[
[
"## Summary\n\nIn pandas, you generally want to have *meaningful row labels*. They should uniquely identify each observation.\nHaving a unique identifier is just good data hygenie. And since they're in the index they stay out of the way in operations.\n\n## Next Steps\n\nNow we'll discuss [Tidy Data](Tidy.ipynb).",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
e78b4cef89890535967496e606dbc0854a8f3f50 | 6,178 | ipynb | Jupyter Notebook | nbs/18b_callback.preds.ipynb | EmbraceLife/fastai | 85258502eff144708d657aa4b4d2ab4c2a2b3a0b | [
"Apache-2.0"
] | 1 | 2022-03-13T00:09:58.000Z | 2022-03-13T00:09:58.000Z | nbs/18b_callback.preds.ipynb | EmbraceLife/fastai | 85258502eff144708d657aa4b4d2ab4c2a2b3a0b | [
"Apache-2.0"
] | null | null | null | nbs/18b_callback.preds.ipynb | EmbraceLife/fastai | 85258502eff144708d657aa4b4d2ab4c2a2b3a0b | [
"Apache-2.0"
] | null | null | null | 28.33945 | 201 | 0.592748 | [
[
[
"#|hide\n#|skip\n! [ -e /content ] && pip install -Uqq fastai # upgrade fastai on colab",
"_____no_output_____"
],
[
"#|default_exp callback.preds",
"_____no_output_____"
],
[
"#|export\nfrom __future__ import annotations\nfrom fastai.basics import *",
"_____no_output_____"
],
[
"#|hide\nfrom nbdev.showdoc import *\nfrom fastai.test_utils import *",
"_____no_output_____"
]
],
[
[
"# Predictions callbacks\n\n> Various callbacks to customize get_preds behaviors",
"_____no_output_____"
],
[
"## MCDropoutCallback\n\n> Turns on dropout during inference, allowing you to call Learner.get_preds multiple times to approximate your model uncertainty using [Monte Carlo Dropout](https://arxiv.org/pdf/1506.02142.pdf).",
"_____no_output_____"
]
],
[
[
"#|export\nclass MCDropoutCallback(Callback):\n def before_validate(self):\n for m in [m for m in flatten_model(self.model) if 'dropout' in m.__class__.__name__.lower()]:\n m.train()\n \n def after_validate(self):\n for m in [m for m in flatten_model(self.model) if 'dropout' in m.__class__.__name__.lower()]:\n m.eval()",
"_____no_output_____"
],
[
"learn = synth_learner()\n\n# Call get_preds 10 times, then stack the predictions, yielding a tensor with shape [# of samples, batch_size, ...]\ndist_preds = []\nfor i in range(10):\n preds, targs = learn.get_preds(cbs=[MCDropoutCallback()])\n dist_preds += [preds]\n\ntorch.stack(dist_preds).shape",
"_____no_output_____"
]
],
[
[
"## Export -",
"_____no_output_____"
]
],
[
[
"#|hide\nfrom nbdev.export import notebook2script\nnotebook2script()",
"Converted 00_torch_core.ipynb.\nConverted 01_layers.ipynb.\nConverted 02_data.load.ipynb.\nConverted 03_data.core.ipynb.\nConverted 04_data.external.ipynb.\nConverted 05_data.transforms.ipynb.\nConverted 06_data.block.ipynb.\nConverted 07_vision.core.ipynb.\nConverted 08_vision.data.ipynb.\nConverted 09_vision.augment.ipynb.\nConverted 09b_vision.utils.ipynb.\nConverted 09c_vision.widgets.ipynb.\nConverted 10_tutorial.pets.ipynb.\nConverted 11_vision.models.xresnet.ipynb.\nConverted 12_optimizer.ipynb.\nConverted 13_callback.core.ipynb.\nConverted 13a_learner.ipynb.\nConverted 13b_metrics.ipynb.\nConverted 14_callback.schedule.ipynb.\nConverted 14a_callback.data.ipynb.\nConverted 15_callback.hook.ipynb.\nConverted 15a_vision.models.unet.ipynb.\nConverted 16_callback.progress.ipynb.\nConverted 17_callback.tracker.ipynb.\nConverted 18_callback.fp16.ipynb.\nConverted 18a_callback.training.ipynb.\nConverted 19_callback.mixup.ipynb.\nConverted 20_interpret.ipynb.\nConverted 20a_distributed.ipynb.\nConverted 21_vision.learner.ipynb.\nConverted 22_tutorial.imagenette.ipynb.\nConverted 23_tutorial.vision.ipynb.\nConverted 24_tutorial.siamese.ipynb.\nConverted 24_vision.gan.ipynb.\nConverted 30_text.core.ipynb.\nConverted 31_text.data.ipynb.\nConverted 32_text.models.awdlstm.ipynb.\nConverted 33_text.models.core.ipynb.\nConverted 34_callback.rnn.ipynb.\nConverted 35_tutorial.wikitext.ipynb.\nConverted 36_text.models.qrnn.ipynb.\nConverted 37_text.learner.ipynb.\nConverted 38_tutorial.text.ipynb.\nConverted 39_tutorial.transformers.ipynb.\nConverted 40_tabular.core.ipynb.\nConverted 41_tabular.data.ipynb.\nConverted 42_tabular.model.ipynb.\nConverted 43_tabular.learner.ipynb.\nConverted 44_tutorial.tabular.ipynb.\nConverted 45_collab.ipynb.\nConverted 46_tutorial.collab.ipynb.\nConverted 50_tutorial.datablock.ipynb.\nConverted 60_medical.imaging.ipynb.\nConverted 61_tutorial.medical_imaging.ipynb.\nConverted 65_medical.text.ipynb.\nConverted 70_callback.wandb.ipynb.\nConverted 71_callback.tensorboard.ipynb.\nConverted 72_callback.neptune.ipynb.\nConverted 73_callback.captum.ipynb.\nConverted 74_callback.cutmix.ipynb.\nConverted 97_test_utils.ipynb.\nConverted 99_pytorch_doc.ipynb.\nConverted index.ipynb.\nConverted tutorial.ipynb.\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
e78b4e48d17191066b8d4301ce354907c4cd9c21 | 14,264 | ipynb | Jupyter Notebook | course_2/course_material/Part_7_Deep_Learning/S54_L390/5. TensorFlow_MNIST_Activation_functions_Part_2_Solution.ipynb | Alexander-Meldrum/learning-data-science | a87cf6be80c67a8d1b57a96c042bdf423ba0a142 | [
"MIT"
] | null | null | null | course_2/course_material/Part_7_Deep_Learning/S54_L390/5. TensorFlow_MNIST_Activation_functions_Part_2_Solution.ipynb | Alexander-Meldrum/learning-data-science | a87cf6be80c67a8d1b57a96c042bdf423ba0a142 | [
"MIT"
] | null | null | null | course_2/course_material/Part_7_Deep_Learning/S54_L390/5. TensorFlow_MNIST_Activation_functions_Part_2_Solution.ipynb | Alexander-Meldrum/learning-data-science | a87cf6be80c67a8d1b57a96c042bdf423ba0a142 | [
"MIT"
] | null | null | null | 49.186207 | 911 | 0.656688 | [
[
[
"# Exercises\n\n### 5. Fiddle with the activation functions. Try applying a ReLu to the first hidden layer and tanh to the second one. The tanh activation is given by the method: tf.nn.tanh()\n\n**Solution** \n\nAnalogically to the previous lecture, we can change the activation functions. This time though, we will use different activators for the different layers.\n\nThe result should not be significantly different. However, with different width and depth, that may change.\n\n*Additional exercise: Try to find a better combination of activation functions*",
"_____no_output_____"
],
[
"## Deep Neural Network for MNIST Classification\n\nWe'll apply all the knowledge from the lectures in this section to write a deep neural network. The problem we've chosen is referred to as the \"Hello World\" for machine learning because for most students it is their first example. The dataset is called MNIST and refers to handwritten digit recognition. You can find more about it on Yann LeCun's website (Director of AI Research, Facebook). He is one of the pioneers of what we've been talking about and of more complex approaches that are widely used today, such as covolutional networks. The dataset provides 28x28 images of handwritten digits (1 per image) and the goal is to write an algorithm that detects which digit is written. Since there are only 10 digits, this is a classification problem with 10 classes. In order to exemplify what we've talked about in this section, we will build a network with 2 hidden layers between inputs and outputs.",
"_____no_output_____"
],
[
"## Import the relevant packages",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport tensorflow as tf\nfrom tensorflow.examples.tutorials.mnist import input_data\n\n# TensorFLow includes a data provider for MNIST that we'll use.\n# This function automatically downloads the MNIST dataset to the chosen directory. \n# The dataset is already split into training, validation, and test subsets. \n# Furthermore, it preprocess it into a particularly simple and useful format.\n# Every 28x28 image is flattened into a vector of length 28x28=784, where every value\n# corresponds to the intensity of the color of the corresponding pixel.\n# The samples are grayscale (but standardized from 0 to 1), so a value close to 0 is almost white and a value close to\n# 1 is almost purely black. This representation (flattening the image row by row into\n# a vector) is slightly naive but as you'll see it works surprisingly well.\n# Since this is a classification problem, our targets are categorical.\n# Recall from the lecture on that topic that one way to deal with that is to use one-hot encoding.\n# With it, the target for each individual sample is a vector of length 10\n# which has nine 0s and a single 1 at the position which corresponds to the correct answer.\n# For instance, if the true answer is \"1\", the target will be [0,0,0,1,0,0,0,0,0,0] (counting from 0).\n# Have in mind that the very first time you execute this command it might take a little while to run\n# because it has to download the whole dataset. Following commands only extract it so they're faster.\nmnist = input_data.read_data_sets(\"MNIST_data/\", one_hot=True)",
"_____no_output_____"
]
],
[
[
"## Outline the model\n\nThe whole code is in one cell, so you can simply rerun this cell (instead of the whole notebook) and train a new model.\nThe tf.reset_default_graph() function takes care of clearing the old parameters. From there on, a completely new training starts.",
"_____no_output_____"
]
],
[
[
"input_size = 784\noutput_size = 10\n# Use same hidden layer size for both hidden layers. Not a necessity.\nhidden_layer_size = 50\n\n# Reset any variables left in memory from previous runs.\ntf.reset_default_graph()\n\n# As in the previous example - declare placeholders where the data will be fed into.\ninputs = tf.placeholder(tf.float32, [None, input_size])\ntargets = tf.placeholder(tf.float32, [None, output_size])\n\n# Weights and biases for the first linear combination between the inputs and the first hidden layer.\n# Use get_variable in order to make use of the default TensorFlow initializer which is Xavier.\nweights_1 = tf.get_variable(\"weights_1\", [input_size, hidden_layer_size])\nbiases_1 = tf.get_variable(\"biases_1\", [hidden_layer_size])\n\n# Operation between the inputs and the first hidden layer.\n# We've chosen ReLu as our activation function. You can try playing with different non-linearities.\noutputs_1 = tf.nn.relu(tf.matmul(inputs, weights_1) + biases_1)\n\n# Weights and biases for the second linear combination.\n# This is between the first and second hidden layers.\nweights_2 = tf.get_variable(\"weights_2\", [hidden_layer_size, hidden_layer_size])\nbiases_2 = tf.get_variable(\"biases_2\", [hidden_layer_size])\n\n# Operation between the first and the second hidden layers. Again, we use ReLu.\noutputs_2 = tf.nn.tanh(tf.matmul(outputs_1, weights_2) + biases_2)\n\n# Weights and biases for the final linear combination.\n# That's between the second hidden layer and the output layer.\nweights_3 = tf.get_variable(\"weights_3\", [hidden_layer_size, output_size])\nbiases_3 = tf.get_variable(\"biases_3\", [output_size])\n\n# Operation between the second hidden layer and the final output.\n# Notice we have not used an activation function because we'll use the trick to include it directly in \n# the loss function. This works for softmax and sigmoid with cross entropy.\noutputs = tf.matmul(outputs_2, weights_3) + biases_3\n\n# Calculate the loss function for every output/target pair.\n# The function used is the same as applying softmax to the last layer and then calculating cross entropy\n# with the function we've seen in the lectures. This function, however, combines them in a clever way, \n# which makes it both faster and more numerically stable (when dealing with very small numbers).\n# Logits here means: unscaled probabilities (so, the outputs, before they are scaled by the softmax)\n# Naturally, the labels are the targets.\nloss = tf.nn.softmax_cross_entropy_with_logits(logits=outputs, labels=targets)\n\n# Get the average loss\nmean_loss = tf.reduce_mean(loss)\n\n# Define the optimization step. Using adaptive optimizers such as Adam in TensorFlow\n# is as simple as that.\noptimize = tf.train.AdamOptimizer(learning_rate=0.001).minimize(mean_loss)\n\n# Get a 0 or 1 for every input in the batch indicating whether it output the correct answer out of the 10.\nout_equals_target = tf.equal(tf.argmax(outputs, 1), tf.argmax(targets, 1))\n\n# Get the average accuracy of the outputs.\naccuracy = tf.reduce_mean(tf.cast(out_equals_target, tf.float32))\n\n# Declare the session variable.\nsess = tf.InteractiveSession()\n\n# Initialize the variables. Default initializer is Xavier.\ninitializer = tf.global_variables_initializer()\nsess.run(initializer)\n\n# Batching\nbatch_size = 100\n\n# Calculate the number of batches per epoch for the training set.\nbatches_number = mnist.train._num_examples // batch_size\n\n# Basic early stopping. Set a miximum number of epochs.\nmax_epochs = 15\n\n# Keep track of the validation loss of the previous epoch.\n# If the validation loss becomes increasing, we want to trigger early stopping.\n# We initially set it at some arbitrarily high number to make sure we don't trigger it\n# at the first epoch\nprev_validation_loss = 9999999.\n\nimport time\nstart_time = time.time()\n\n# Create a loop for the epochs. Epoch_counter is a variable which automatically starts from 0.\nfor epoch_counter in range(max_epochs):\n \n # Keep track of the sum of batch losses in the epoch.\n curr_epoch_loss = 0.\n \n # Iterate over the batches in this epoch.\n for batch_counter in range(batches_number):\n \n # Input batch and target batch are assigned values from the train dataset, given a batch size\n input_batch, target_batch = mnist.train.next_batch(batch_size)\n \n # Run the optimization step and get the mean loss for this batch.\n # Feed it with the inputs and the targets we just got from the train dataset\n _, batch_loss = sess.run([optimize, mean_loss], \n feed_dict={inputs: input_batch, targets: target_batch})\n \n # Increment the sum of batch losses.\n curr_epoch_loss += batch_loss\n \n # So far curr_epoch_loss contained the sum of all batches inside the epoch\n # We want to find the average batch losses over the whole epoch\n # The average batch loss is a good proxy for the current epoch loss\n curr_epoch_loss /= batches_number\n \n # At the end of each epoch, get the validation loss and accuracy\n # Get the input batch and the target batch from the validation dataset\n input_batch, target_batch = mnist.validation.next_batch(mnist.validation._num_examples)\n \n # Run without the optimization step (simply forward propagate)\n validation_loss, validation_accuracy = sess.run([mean_loss, accuracy], \n feed_dict={inputs: input_batch, targets: target_batch})\n \n # Print statistics for the current epoch\n # Epoch counter + 1, because epoch_counter automatically starts from 0, instead of 1\n # We format the losses with 3 digits after the dot\n # We format the accuracy in percentages for easier interpretation\n print('Epoch '+str(epoch_counter+1)+\n '. Mean loss: '+'{0:.3f}'.format(curr_epoch_loss)+\n '. Validation loss: '+'{0:.3f}'.format(validation_loss)+\n '. Validation accuracy: '+'{0:.2f}'.format(validation_accuracy * 100.)+'%')\n \n # Trigger early stopping if validation loss begins increasing.\n if validation_loss > prev_validation_loss:\n break\n \n # Store this epoch's validation loss to be used as previous validation loss in the next iteration.\n prev_validation_loss = validation_loss\n\n# Not essential, but it is nice to know when the algorithm stopped working in the output section, rather than check the kernel\nprint('End of training.')\n\n#Add the time it took the algorithm to train\nprint(\"Training time: %s seconds\" % (time.time() - start_time))",
"_____no_output_____"
]
],
[
[
"## Test the model\n\nAs we discussed in the lectures, after training on the training and validation sets, we test the final prediction power of our model by running it on the test dataset that the algorithm has not seen before.\n\nIt is very important to realize that fiddling with the hyperparameters overfits the validation dataset. The test is the absolute final instance. You should not test before you are completely done with adjusting your model.",
"_____no_output_____"
]
],
[
[
"input_batch, target_batch = mnist.test.next_batch(mnist.test._num_examples)\ntest_accuracy = sess.run([accuracy], \n feed_dict={inputs: input_batch, targets: target_batch})\n\n# Test accuracy is a list with 1 value, so we want to extract the value from it, using x[0]\n# Uncomment the print to see how it looks before the manipulation\n# print (test_accuracy)\ntest_accuracy_percent = test_accuracy[0] * 100.\n\n# Print the test accuracy formatted in percentages\nprint('Test accuracy: '+'{0:.2f}'.format(test_accuracy_percent)+'%')",
"_____no_output_____"
]
],
[
[
"Using the initial model and hyperparameters given in this notebook, the final test accuracy should be roughly between 97% and 98%. Each time the code is rerunned, we get a different accuracy as the batches are shuffled, the weights are initialized in a different way, etc.\n\nFinally, we have intentionally reached a suboptimal solution, so you can have space to build on it.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e78b578468f92251b717228292a3f2c038cf2cfe | 50,192 | ipynb | Jupyter Notebook | theta.ipynb | cyneuro/theta | f0d0e96461cc102347eb2f5fed4c59e4df7b3929 | [
"MIT"
] | null | null | null | theta.ipynb | cyneuro/theta | f0d0e96461cc102347eb2f5fed4c59e4df7b3929 | [
"MIT"
] | null | null | null | theta.ipynb | cyneuro/theta | f0d0e96461cc102347eb2f5fed4c59e4df7b3929 | [
"MIT"
] | 1 | 2020-12-21T12:03:38.000Z | 2020-12-21T12:03:38.000Z | 46.259908 | 1,683 | 0.600355 | [
[
[
"# Modeling Theta: Muti-population recurrent network (with BMTK BioNet) \n\nHere we will create a heterogenous yet relatively small network consisting of hundreds of cells recurrently connected. All cells will belong to one of four \"cell-types\". Two of these cell types will be biophysically detailed cells, i.e. containing a full morphology and somatic and dendritic channels and receptors. The other two will be point-neuron models, which lack a full morphology or channels but still act to provide inhibitory and excitory dynamics.\n\nAs input to drive the simulation, we will also create an external network of \"virtual cells\" that synapse directly onto our internal cells and provide spike trains stimulus\n\n**Note** - scripts and files for running this tutorial can be found in the directory [theta](https://github.com/cyneuro/theta)\n\nrequirements:\n* bmtk\n* NEURON 7.4+",
"_____no_output_____"
],
[
"## 1. Building the network\n\n#### cells\n\nThis network will loosely resemble the rodent hippocampal CA3 region. Along the center of the column will be a population of 50 biophysically detailed neurons: 40 excitatory Scnn1a cells and 10 inhibitory PV cells.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nfrom bmtk.builder.networks import NetworkBuilder\nfrom bmtk.builder.auxi.node_params import positions_list\n\ndef pos_CA3e():\n # Create the possible x,y,z coordinates for CA3e cells\n x_start, x_end, x_stride = 0.5,15,2.3\n y_start, y_end, y_stride = 0.5,3,1\n z_start, z_end, z_stride = 1,4,1\n x_grid = np.arange(x_start,x_end,x_stride)\n y_grid = np.arange(y_start,y_end,y_stride)\n z_grid = np.arange(z_start,z_end,z_stride)\n xx, yy, zz = np.meshgrid(x_grid, y_grid, z_grid)\n pos_list = np.vstack([xx.ravel(), yy.ravel(), zz.ravel()]).T\n \n return positions_list(pos_list)\n\ndef pos_CA3o():\n # Create the possible x,y,z coordinates for CA3 OLM cells\n x_start, x_end, x_stride = 1,15,4\n y_start, y_end, y_stride = 0.75,3,1.5\n z_start, z_end, z_stride = 2,2.5,2\n x_grid = np.arange(x_start,x_end,x_stride)\n y_grid = np.arange(y_start,y_end,y_stride)\n z_grid = np.arange(z_start,z_end,z_stride)\n xx, yy, zz = np.meshgrid(x_grid, y_grid, z_grid)\n pos_list = np.vstack([xx.ravel(), yy.ravel(), zz.ravel()]).T\n \n return positions_list(pos_list)\n\nCA3eTotal = 63 # number of CA3 principal cells\nCA3oTotal = 8 # number OLM inhibitory cells\n \n\nnet = NetworkBuilder('hippocampus')\n\nnet.add_nodes(N=CA3eTotal, pop_name='CA3e', pop_group='CA3',\n positions=pos_CA3e(),\n model_type='biophysical',\n model_template='hoc:CA3PyramidalCell',\n morphology='blank.swc')\n\nnet.add_nodes(N=CA3oTotal, pop_name='CA3o', pop_group='CA3',\n positions=pos_CA3o(),\n model_type='biophysical',\n model_template='hoc:IzhiCell_OLM',\n morphology='blank.swc')\n",
"_____no_output_____"
]
],
[
[
"To set the position and rotation of each cell, we use the built in function positions_columinar and xiter_random, which returns a list of values given the parameters. A user could set the values themselves using a list (or function that returns a list) of size N. The parameters like location, ei (potential), params_file, etc. are cell-type parameters, and will be used for all N cells of that type.\n\nThe excitory cells are also given a tuning_angle parameter. An instrinsic \"tuning angle\" is a property found in some cells in the visual cortex. In this model, we will use this property to determine number of strenght of connections between subsets of cells by using custom functions. But in general most models will not have or use a tuning angle, but they may require some other parameter. In general, users can assign whatever custom parameters they want to cells and cell-types and use them as properties for creating connections and running simulations.\n\nNext we continue to create our point (integrate-and-fire) neurons. Notice they don't have properities like y/z rotation or morphology, as they wouldn't apply to point neurons.",
"_____no_output_____"
]
],
[
[
"\"\"\"\nnet.add_nodes(N=200, pop_name='LIF_exc',\n positions=positions_columinar(N=200, center=[0, 50.0, 0], min_radius=30.0, max_radius=60.0, height=100.0),\n tuning_angle=np.linspace(start=0.0, stop=360.0, num=200, endpoint=False),\n location='VisL4',\n ei='e',\n model_type='point_process',\n model_template='nrn:IntFire1',\n dynamics_params='IntFire1_exc_1.json')\n\nnet.add_nodes(N=100, pop_name='LIF_inh',\n positions=positions_columinar(N=100, center=[0, 50.0, 0], min_radius=30.0, max_radius=60.0, height=100.0),\n location='VisL4',\n ei='i',\n model_type='point_process',\n model_template='nrn:IntFire1',\n dynamics_params='IntFire1_inh_1.json')\n\"\"\"\nimport glob\nimport json\nimport os\n\ndef syn_params_dicts(syn_dir='sim_theta/components/synaptic_models'):\n \"\"\"\n returns: A dictionary of dictionaries containing all\n properties in the synapse json files\n \"\"\"\n files = glob.glob(os.path.join(syn_dir,'*.json'))\n data = {}\n for fh in files:\n print(fh)\n with open(fh) as f:\n data[os.path.basename(fh)] = json.load(f) #data[\"filename.json\"] = {\"prop1\":\"val1\",...}\n return data\n\nsyn = syn_params_dicts()",
"sim_theta/components/synaptic_models\\AMPA_ExcToExc.json\nsim_theta/components/synaptic_models\\AMPA_ExcToInh.json\nsim_theta/components/synaptic_models\\CA3e2CA3o.exc.json\nsim_theta/components/synaptic_models\\CA3e2CA3o.exc.sfn19exp2d.json\nsim_theta/components/synaptic_models\\CA3o2CA3e.inh.json\nsim_theta/components/synaptic_models\\GABA_InhToExc.json\nsim_theta/components/synaptic_models\\GABA_InhToInh.json\nsim_theta/components/synaptic_models\\instanteneousExc.json\nsim_theta/components/synaptic_models\\instanteneousInh.json\nsim_theta/components/synaptic_models\\NetCon2EC.exc.json\n"
],
[
"from bmtk.simulator.bionet.pyfunction_cache import add_synapse_model\nfrom neuron import h\nimport random\n\ndef Pyr2Pyr(syn_params, sec_x, sec_id):\n \"\"\"Create a pyr2pyr synapse\n :param syn_params: parameters of a synapse\n :param sec_x: normalized distance along the section\n :param sec_id: target section\n :return: NEURON synapse object\n \"\"\"\n\n lsyn = h.pyr2pyr(sec_x, sec=sec_id)\n\n if syn_params.get('AlphaTmax_ampa'):\n lsyn.AlphaTmax_ampa = float(syn_params['AlphaTmax_ampa']) # par.x(21)\n if syn_params.get('Beta_ampa'):\n lsyn.Beta_ampa = float(syn_params['Beta_ampa']) # par.x(22)\n if syn_params.get('Cdur_ampa'):\n lsyn.Cdur_ampa = float(syn_params['Cdur_ampa']) # par.x(23)\n if syn_params.get('gbar_ampa'):\n lsyn.gbar_ampa = float(syn_params['gbar_ampa']) # par.x(24)\n if syn_params.get('Erev_ampa'):\n lsyn.Erev_ampa = float(syn_params['Erev_ampa']) # par.x(16)\n\n if syn_params.get('AlphaTmax_nmda'):\n lsyn.AlphaTmax_nmda = float(syn_params['AlphaTmax_nmda']) # par.x(25)\n if syn_params.get('Beta_nmda'):\n lsyn.Beta_nmda = float(syn_params['Beta_nmda']) # par.x(26)\n if syn_params.get('Cdur_nmda'):\n lsyn.Cdur_nmda = float(syn_params['Cdur_nmda']) # par.x(27)\n if syn_params.get('gbar_nmda'):\n lsyn.gbar_nmda = float(syn_params['gbar_nmda']) # par.x(28)\n if syn_params.get('Erev_nmda'):\n lsyn.Erev_nmda = float(syn_params['Erev_nmda']) # par.x(16)\n \n if syn_params.get('initW'):\n lsyn.initW = float(syn_params['initW']) * random.uniform(0.5,1.0) # par.x(0) * rC.uniform(0.5,1.0)//rand.normal(0.5,1.5) //`rand.repick() \n\n if syn_params.get('Wmax'):\n lsyn.Wmax = float(syn_params['Wmax']) * lsyn.initW # par.x(1) * lsyn.initW\n if syn_params.get('Wmin'):\n lsyn.Wmin = float(syn_params['Wmin']) * lsyn.initW # par.x(2) * lsyn.initW\n #delay = float(syn_params['initW']) # par.x(3) + delayDistance\n #lcon = new NetCon(&v(0.5), lsyn, 0, delay, 1)\n\n if syn_params.get('lambda1'):\n lsyn.lambda1 = float(syn_params['lambda1']) # par.x(6)\n if syn_params.get('lambda2'):\n lsyn.lambda2 = float(syn_params['lambda2']) # par.x(7)\n if syn_params.get('threshold1'):\n lsyn.threshold1 = float(syn_params['threshold1']) # par.x(8)\n if syn_params.get('threshold2'):\n lsyn.threshold2 = float(syn_params['threshold2']) # par.x(9)\n if syn_params.get('tauD1'):\n lsyn.tauD1 = float(syn_params['tauD1']) # par.x(10)\n if syn_params.get('d1'):\n lsyn.d1 = float(syn_params['d1']) # par.x(11)\n if syn_params.get('tauD2'):\n lsyn.tauD2 = float(syn_params['tauD2']) # par.x(12)\n if syn_params.get('d2'):\n lsyn.d2 = float(syn_params['d2']) # par.x(13)\n if syn_params.get('tauF'):\n lsyn.tauF = float(syn_params['tauF']) # par.x(14)\n if syn_params.get('f'):\n lsyn.f = float(syn_params['f']) # par.x(15)\n\n if syn_params.get('bACH'):\n lsyn.bACH = float(syn_params['bACH']) # par.x(17)\n if syn_params.get('aDA'):\n lsyn.aDA = float(syn_params['aDA']) # par.x(18)\n if syn_params.get('bDA'):\n lsyn.bDA = float(syn_params['bDA']) # par.x(19)\n if syn_params.get('wACH'):\n lsyn.wACH = float(syn_params['wACH']) # par.x(20)\n \n return lsyn\n\n\ndef pyr2pyr(syn_params, xs, secs):\n \"\"\"Create a list of pyr2pyr synapses\n :param syn_params: parameters of a synapse\n :param xs: list of normalized distances along the section\n :param secs: target sections\n :return: list of NEURON synpase objects\n \"\"\"\n syns = []\n for x, sec in zip(xs, secs):\n syn = Pyr2Pyr(syn_params, x, sec)\n syns.append(syn)\n return syns\n\ndef Inter2Pyr(syn_params, sec_x, sec_id):\n lsyn = h.inter2pyr(sec_x, sec=sec_id)\n\n #target.soma lsyn = new inter2pyr(0.9)\n if syn_params.get('AlphaTmax_gabaa'):\n lsyn.AlphaTmax_gabaa = float(syn_params['AlphaTmax_gabaa']) # par.x(21)\n if syn_params.get('Beta_gabaa'):\n lsyn.Beta_gabaa = float(syn_params['Beta_gabaa']) # par.x(22)\n if syn_params.get('Cdur_gabaa'):\n lsyn.Cdur_gabaa = float(syn_params['Cdur_gabaa']) # par.x(23)\n if syn_params.get('gbar_gabaa'):\n lsyn.gbar_gabaa = float(syn_params['gbar_gabaa']) # par.x(24)\n if syn_params.get('Erev_gabaa'):\n lsyn.Erev_gabaa = float(syn_params['Erev_gabaa']) # par.x(16)\n\n if syn_params.get('AlphaTmax_gabab'):\n lsyn.AlphaTmax_gabab = float(syn_params['AlphaTmax_gabab']) # par.x(25)\n if syn_params.get('Beta_gabab'):\n lsyn.Beta_gabab = float(syn_params['Beta_gabab']) # par.x(26)\n if syn_params.get('Cdur_gabab'):\n lsyn.Cdur_gabab = float(syn_params['Cdur_gabab']) # par.x(27)\n if syn_params.get('gbar_gabab'):\n lsyn.gbar_gabab = float(syn_params['gbar_gabab']) # par.x(28)\n if syn_params.get('Erev_gabab'):\n lsyn.Erev_gabab = float(syn_params['Erev_gabab']) # par.x(16)\n \n\n if syn_params.get('initW'):\n lsyn.initW = float(syn_params['initW']) * random.uniform(0.5,1.0) # par.x(0) * rC.uniform(0.5,1.0)//rand.normal(0.5,1.5) //`rand.repick() \n\n if syn_params.get('Wmax'):\n lsyn.Wmax = float(syn_params['Wmax']) * lsyn.initW # par.x(1) * lsyn.initW\n if syn_params.get('Wmin'):\n lsyn.Wmin = float(syn_params['Wmin']) * lsyn.initW # par.x(2) * lsyn.initW\n #delay = float(syn_params['initW']) # par.x(3) + delayDistance\n #lcon = new NetCon(&v(0.5), lsyn, 0, delay, 1)\n\n if syn_params.get('lambda1'):\n lsyn.lambda1 = float(syn_params['lambda1']) # par.x(6)\n if syn_params.get('lambda2'):\n lsyn.lambda2 = float(syn_params['lambda2']) # par.x(7)\n if syn_params.get('threshold1'):\n lsyn.threshold1 = float(syn_params['threshold1']) # par.x(8)\n if syn_params.get('threshold2'):\n lsyn.threshold2 = float(syn_params['threshold2']) # par.x(9)\n if syn_params.get('tauD1'):\n lsyn.tauD1 = float(syn_params['tauD1']) # par.x(10)\n if syn_params.get('d1'):\n lsyn.d1 = float(syn_params['d1']) # par.x(11)\n if syn_params.get('tauD2'):\n lsyn.tauD2 = float(syn_params['tauD2']) # par.x(12)\n if syn_params.get('d2'):\n lsyn.d2 = float(syn_params['d2']) # par.x(13)\n if syn_params.get('tauF'):\n lsyn.tauF = float(syn_params['tauF']) # par.x(14)\n if syn_params.get('f'):\n lsyn.f = float(syn_params['f']) # par.x(15)\n\n if syn_params.get('bACH'):\n lsyn.bACH = float(syn_params['bACH']) # par.x(17)\n if syn_params.get('aDA'):\n lsyn.aDA = float(syn_params['aDA']) # par.x(18)\n if syn_params.get('bDA'):\n lsyn.bDA = float(syn_params['bDA']) # par.x(19)\n if syn_params.get('wACH'):\n lsyn.wACH = float(syn_params['wACH']) # par.x(20)\n\n return lsyn\n\ndef inter2pyr(syn_params, xs, secs):\n \"\"\"Create a list of pyr2pyr synapses\n :param syn_params: parameters of a synapse\n :param xs: list of normalized distances along the section\n :param secs: target sections\n :return: list of NEURON synpase objects\n \"\"\"\n syns = []\n for x, sec in zip(xs, secs):\n syn = Inter2Pyr(syn_params, x, sec)\n syns.append(syn)\n return syns\n\n\ndef load_synapses():\n add_synapse_model(Pyr2Pyr, 'pyr2pyr', overwrite=False)\n add_synapse_model(Pyr2Pyr, overwrite=False)\n add_synapse_model(Inter2Pyr, 'inter2pyr', overwrite=False)\n add_synapse_model(Inter2Pyr, overwrite=False)\n return\n\nload_synapses()",
"_____no_output_____"
]
],
[
[
"#### connections\n\nNow we want to create connections between the cells. Depending on the model type, and whether or not the presynpatic \"source\" cell is excitory or inhibitory, we will have different synpatic model and parameters. Using the source and target filter parameters, we can create different connection types.\n\nTo determine excitory-to-excitory connection matrix we want to use distance and tuning_angle property. To do this we create a customized function \"dist_tuning_connector\"",
"_____no_output_____"
]
],
[
[
"import random\nimport math\n\n# list of all synapses created - used for recurrent connections\nsyn_list = []\n\n###########################################################\n# Build custom connection rules\n###########################################################\n#See bmtk.builder.auxi.edge_connectors\ndef hipp_dist_connector(source, target, con_pattern, ratio=1, gaussa=0, min_syn=1, max_syn=1):\n \"\"\"\n :returns: number of synapses per connection\n \"\"\"\n\n ratio = float(ratio)\n gaussa = float(gaussa)\n\n Lamellar = \"0\"\n Homogenous = \"1\"\n AxonalPlexus = \"2\"\n IntPyrFeedback = \"3\"\n\n x_ind,y_ind = 0,1\n dx = target['positions'][x_ind] - source['positions'][x_ind]\n dy = target['positions'][y_ind] - source['positions'][y_ind]\n distxy = math.sqrt(dx**2 + dy**2)\n prob = 1\n\n if con_pattern == Lamellar:\n prob = ratio/(math.exp(((abs(dx)-0)**2)/(2*(3**2))))\n if con_pattern == Homogenous:\n prob = ratio\n if con_pattern == IntPyrFeedback or con_pattern == AxonalPlexus:\n c = ratio\n a = gaussa\n prob = a /(math.exp(((abs(distxy)-0)**2)/(2*(c**2))))\n\n if random.random() < prob:\n #Since there will be recurrect connections we need to keep track externally to BMTK\n #BMTK will call build_edges twice if we use net.edges() before net.build()\n #Resulting in double the edge count\n syn_list.append({'source_gid':source['node_id'],'target_gid':target['node_id']})\n return random.randint(min_syn,max_syn)\n else:\n return 0\n\n###########################################################\n# Build recurrent connection rules\n###########################################################\ndef hipp_recurrent_connector(source,target,all_edges=[],min_syn=1, max_syn=1):\n \"\"\"\n General logic:\n 1. Given a *potential* source and target\n 2. Look through all edges currently made\n 3. If any of the current edges contains \n a. the current source as a previous target of \n b. the current target as a prevous source\n 4. Return number of synapses per this connection, 0 otherwise (no connection)\n \"\"\"\n for e in all_edges:\n #if source['node_id'] == e.target_gid and target['node_id'] == e.source_gid:\n if source['node_id'] == e['target_gid'] and target['node_id'] == e['source_gid']:\n return random.randint(min_syn,max_syn)\n\n return 0\n\ndef syn_dist_delay(source, target, base_delay, dist_delay=None):#, min_weight, max_weight):\n \"\"\"\n Original Code:\n distDelay = 0.1* (0.5*dist + rC.normal(0,1.5)*(1-exp(-dist^2/3)) ) \n \"\"\"\n base_delay = float(base_delay)\n if dist_delay:\n dist_delay = float(dist_delay)\n\n if dist_delay: #An override of sorts\n return base_delay + dist_delay\n\n x_ind,y_ind,z_ind = 0,1,2\n\n dx = target['positions'][x_ind] - source['positions'][x_ind]\n dy = target['positions'][y_ind] - source['positions'][y_ind]\n dz = target['positions'][z_ind] - source['positions'][z_ind]\n\n dist = math.sqrt(dx**2 + dy**2 + dz**2)\n distDelay = 0.1* (0.5*dist + np.random.normal(0,1.5,1)[0]*(1-math.exp(-dist**2/3)) ) \n return float(base_delay) + distDelay\n\ndef syn_dist_delay_section(source, target, base_delay, dist_delay=None, sec_id=0, sec_x=0.9):\n return syn_dist_delay(source, target, base_delay, dist_delay), sec_id, sec_x\n",
"_____no_output_____"
]
],
[
[
"This first two parameters of this function is \"source\" and \"target\" and are required for all custom connector functions. These are node objects which gives a representation of a single source and target cell, with properties that can be accessed like a python dictionary. When The Network Builder is creating the connection matrix, it will call this function for all possible source-target pairs. The user doesn't call this function directly.\n\nThe remaining parameters are optional. Using these parameters, plus the distance and angles between source and target cells, this function determines the number of connections between each given source and target cell. If there are none you can return either None or 0.\n\nTo create these connections we call add_edges method of the builder. We use the source and target parameter to filter out only excitory-to-excitory connections. We must also take into consideration the model type (biophysical or integrate-and-fire) of the target when setting parameters. We pass in the function throught the connection_rule parameter, and the function parameters (except source and target) through connection_params. (If our dist_tuning_connector function didn't have any parameters other than source and target, we could just not set connection_params).",
"_____no_output_____"
]
],
[
[
"dynamics_file = 'CA3o2CA3e.inh.json'\n\nconn = net.add_edges(source={'pop_name': 'CA3o'}, target={'pop_name': 'CA3e'},\n connection_rule=hipp_dist_connector,\n connection_params={'con_pattern':syn[dynamics_file]['con_pattern'],\n 'ratio':syn[dynamics_file]['ratio'],\n 'gaussa':syn[dynamics_file]['gaussa']},\n syn_weight=1,\n dynamics_params=dynamics_file,\n model_template=syn[dynamics_file]['level_of_detail'],\n distance_range=[0.0, 300.0],\n target_sections=['soma'])\nconn.add_properties(names=['delay', 'sec_id', 'sec_x'],\n rule=syn_dist_delay_section,\n rule_params={'base_delay':syn[dynamics_file]['delay'], 'sec_id':0, 'sec_x':0.9},\n dtypes=[np.float, np.int32, np.float])\n",
"_____no_output_____"
]
],
[
[
"Similarly we create the other types of connections. But since either the source, target, or both cells will not have the tuning_angle parameter, we don't want to use dist_tuning_connector. Instead we can use the built-in distance_connector function which just creates connections determined by distance.",
"_____no_output_____"
]
],
[
[
"dynamics_file = 'CA3e2CA3o.exc.json'\n\nexperiment = 'original'\nif experiment == \"SFN19-D\": #Weight of the synapses are set to 6 from max weight of 2\n dynamics_file = 'CA3e2CA3o.exc.sfn19exp2d.json'\n\nconn = net.add_edges(source={'pop_name': 'CA3e'}, target={'pop_name': 'CA3o'},\n connection_rule=hipp_recurrent_connector,\n connection_params={'all_edges':syn_list}, #net.edges()},\n syn_weight=1,\n dynamics_params=dynamics_file,\n model_template=syn[dynamics_file]['level_of_detail'],\n distance_range=[0.0, 300.0],\n target_sections=['soma'])\nconn.add_properties(names=['delay', 'sec_id', 'sec_x'],\n rule=syn_dist_delay_section,\n rule_params={'base_delay':syn[dynamics_file]['delay'], 'dist_delay':0.1, 'sec_id':0, 'sec_x':0.9}, #Connect.hoc:274 0.1 dist delay\n dtypes=[np.float, np.int32, np.float])\n",
"_____no_output_____"
]
],
[
[
"Finally we build the network (this may take a bit of time since it's essentially iterating over all 400x400 possible connection combinations), and save the nodes and edges.",
"_____no_output_____"
]
],
[
[
"net.build()\nnet.save_nodes(output_dir='sim_theta/network')\nnet.save_edges(output_dir='sim_theta/network')",
"_____no_output_____"
]
],
[
[
"### Building external network\n\nNext we want to create an external network consisting of virtual cells that form a feedforward network onto our V1, which will provide input during the simulation. We will call this LGN, since the LGN is the primary input the layer 4 cells of the V1 (if we wanted to we could also create multiple external networks and run simulations on any number of them). \n\nFirst we build our LGN nodes. Then we must import the V1 network nodes, and create connections between LGN --> V1.",
"_____no_output_____"
]
],
[
[
"from bmtk.builder.networks import NetworkBuilder\n\nexp0net = NetworkBuilder('exp0net')\n\nexp0net.add_nodes(N=CA3eTotal, model_type='virtual', pop_name='bgnoisevirtCA3', pop_group='bgnoisevirtCA3')\n",
"_____no_output_____"
]
],
[
[
"As before, we will use a customized function to determine the number of connections between each source and target pair, however this time our connection_rule is a bit different\n\nIn the previous example, our connection_rule function's first two arguments were the presynaptic and postsynaptic cells, which allowed us to choose how many synaptic connections between the pairs existed based on individual properties:\n```python\ndef connection_fnc(source, target, ...):\n source['param'] # presynaptic cell params\n target['param'] # postsynaptic cell params\n ...\n return nsyns # number of connections between pair\n```\n\nBut for our LGN --> V1 connection, we do things a bit differently. We want to make sure that for every source cell, there are a limited number of presynaptic targets. This is a not really possible with a function that iterates on a one-to-one basis. So instead we have a connector function who's first parameter is a list of all N source cell, and the second parameter is a single target cell. We return an array of integers, size N; which each index representing the number of synaptics between sources and the target. \n\nTo tell the builder to use this schema, we must set iterator='all_to_one' in the add_edges method. (By default this is set to 'one_to_one'. You can also use 'one_to_all' iterator which will pass in a single source and all possible targets).",
"_____no_output_____"
]
],
[
[
"def target_ind_equals_source_ind(source, targets, offset=0, min_syn=1,max_syn=1):\n # Creates a 1 to 1 mapping between source and destination nodes\n total_targets = len(targets)\n syns = np.zeros(total_targets)\n target_index = source['node_id']\n syns[target_index-offset] = 1\n return syns\n \n\nconn = exp0net.add_edges(target=net.nodes(pop_name='CA3e'),\n source={'pop_name':'bgnoisevirtCA3'},\n iterator='one_to_all',\n connection_rule=target_ind_equals_source_ind,\n connection_params={'offset':0},\n dynamics_params='NetCon2EC.exc.json',\n model_template='pyr2pyr',\n delay=0,\n syn_weight=1,\n )\nconn.add_properties(['sec_id','sec_x'],rule=(0, 0.9), dtypes=[np.int32,np.float])\n\n\nexp0net.build()\nexp0net.save_nodes(output_dir='sim_theta/network')\nexp0net.save_edges(output_dir='sim_theta/network')",
"_____no_output_____"
]
],
[
[
"## 2. Setting up BioNet\n\n#### file structure.\n\nBefore running a simulation, we will need to create the runtime environment, including parameter files, run-script and configuration files. You can copy the files from an existing simuatlion, execute the following command:\n\n```bash\n$ python -m bmtk.utils.sim_setup \\\n --report-vars v \\\n --report-nodes 10,80 \\\n --network sim_theta/network \\\n --dt 0.1 \\\n --tstop 3000.0 \\ \n --include-examples \\\n --compile-mechanisms \\ \n bionet sim_ch04\n```\n\n$ python -m bmtk.utils.sim_setup --report-vars v --report-nodes 0,80,100,300 --network sim_theta/network --dt 0.1 --tstop 3000.0 --include-examples --compile-mechanisms bionet sim_theta\n\nor run it directly in python",
"_____no_output_____"
]
],
[
[
"from bmtk.utils.sim_setup import build_env_bionet\n\nbuild_env_bionet(base_dir='sim_theta', \n network_dir='sim_theta/network',\n tstop=3000.0, dt=0.1,\n report_vars=['v'], # Record membrane potential (default soma)\n include_examples=True, # Copies components files\n compile_mechanisms=True # Will try to compile NEURON mechanisms\n )",
"ERROR:bmtk.utils.sim_setup: Was unable to compile mechanism in C:\\Users\\Tyler\\Desktop\\git_stage\\theta\\sim_theta\\components\\mechanisms\n"
]
],
[
[
"This will fill out the **sim_ch04** with all the files we need to get started to run the simulation. Of interest includes\n\n* **circuit_config.json** - A configuration file that contains the location of the network files we created above. Plus location of neuron and synpatic models, templates, morphologies and mechanisms required to build our instantiate individual cell models.\n\n\n* **simulation_config.json** - contains information about the simulation. Including initial conditions and run-time configuration (_run_ and _conditions_). In the _inputs_ section we define what external sources we will use to drive the network (in this case a current clamp). And in the _reports_ section we define the variables (soma membrane potential and calcium) that will be recorded during the simulation \n\n\n* **run_bionent.py** - A script for running our simulation. Usually this file doesn't need to be modified.\n\n\n* **components/biophysical_neuron_models/** - The parameter file for the cells we're modeling. Originally [downloaded from the Allen Cell Types Database](http://celltypes.brain-map.org/neuronal_model/download/482934212). These files were automatically copies over when we used the _include-examples_ directive. If using a differrent or extended set of cell models place them here\n\n\n* **components/biophysical_neuron_models/** - The morphology file for our cells. Originally [downloaded from the Allen Cell Types Database](http://celltypes.brain-map.org/neuronal_model/download/482934212) and copied over using the _include_examples_.\n\n\n* **components/point_neuron_models/** - The parameter file for our LIF_exc and LIF_inh cells.\n\n\n* **components/synaptic_models/** - Parameter files used to create different types of synapses.\n",
"_____no_output_____"
],
[
"#### lgn input\n\nWe need to provide our LGN external network cells with spike-trains so they can activate our recurrent network. Previously we showed how to do this by generating csv files. We can also use NWB files, which are a common format for saving electrophysiological data in neuroscience.\n\nWe can use any NWB file generated experimentally or computationally, but for this example we will use a preexsting one. First download the file:\n```bash\n $ wget https://github.com/AllenInstitute/bmtk/blob/develop/docs/examples/spikes_inputs/lgn_spikes.nwb?raw=true\n```\nor copy from [here](https://github.com/AllenInstitute/bmtk/tree/develop/docs/examples/spikes_inputs/lgn_spikes.nwb).\n\n\nThen we must edit the **simulation_config.json** file to tell the simulator to find the nwb file and which network to associate it with.\n\n```json\n{\n \"inputs\": {\n \"LGN_spikes\": {\n \"input_type\": \"spikes\",\n \"module\": \"nwb\",\n \"input_file\": \"$BASE_DIR/lgn_spikes.nwb\",\n \"node_set\": \"LGN\",\n \"trial\": \"trial_0\"\n }\n }\n}\n```\n",
"_____no_output_____"
],
[
"## 3. Running the simulation\n\n\nWe are close to running our simulation, however unlike in previous chapters we need a little more programming before we can begin. \n\nFor most of the connections we added the parameter weight_function='wmax'. This is a built-in function that tells the simulator when creating a connection between two cells, just use the 'weight_max' value assigned to that given edge-type. \n\nHowever, when creating excitatory-to-excitatory connections we used weight_function='gaussianLL'. This is because we want to use the tuning_angle parameter, when avaiable, to determine the synaptic strength between two connections. First we create the function which takes in target, source and connection properties (which are just the edge-type and properties set in the add_edges method). Then we must register the function with the BioNet simulator:",
"_____no_output_____"
]
],
[
[
"import math\nfrom bmtk.simulator.bionet.pyfunction_cache import add_weight_function\n\ndef gaussianLL(edge_props, source, target):\n src_tuning = source['tuning_angle']\n tar_tuning = target['tuning_angle']\n w0 = edge_props[\"syn_weight\"]\n sigma = edge_props[\"weight_sigma\"]\n\n delta_tuning = abs(abs(abs(180.0 - abs(float(tar_tuning) - float(src_tuning)) % 360.0) - 90.0) - 90.0)\n return w0 * math.exp(-(delta_tuning / sigma) ** 2)\n\nadd_weight_function(gaussianLL)",
"_____no_output_____"
]
],
[
[
"The weights will be adjusted before each simulation, and the function can be changed between different runs.. Simply opening the edge_types.csv file with a text editor and altering the weight_function column allows users to take an existing network and readjust weights on-the-fly.\n\nFinally we are ready to run the simulation. Note that because this is a 400 cell simulation, this may be computationally intensive for some older computers and may take anywhere between a few minutes to half-an-hour to complete.",
"_____no_output_____"
]
],
[
[
"from bmtk.simulator import bionet\n\n\nconf = bionet.Config.from_json('sim_theta/simulation_config.json')\nconf.build_env()\nnet = bionet.BioNetwork.from_config(conf)\nsim = bionet.BioSimulator.from_config(conf, network=net)\nsim.run()",
"2020-09-28 22:46:28,632 [INFO] Created log file\n"
]
],
[
[
"## 4. Analyzing results\n\nResults of the simulation, as specified in the config, are saved into the output directory. Using the analyzer functions, we can do things like plot the raster plot",
"_____no_output_____"
]
],
[
[
"from bmtk.analyzer.spike_trains import plot_raster, plot_rates_boxplot\n\nplot_raster(config_file='sim_theta/simulation_config.json', group_by='pop_name')",
"c:\\users\\tyler\\desktop\\git_stage\\bmtk\\bmtk\\simulator\\utils\\config.py:4: UserWarning: Please use bmtk.simulator.core.simulation_config instead.\n warnings.warn('Please use bmtk.simulator.core.simulation_config instead.')\n"
]
],
[
[
"and the rates of each node",
"_____no_output_____"
]
],
[
[
"plot_rates_boxplot(config_file='sim_ch04/simulation_config.json', group_by='pop_name')",
"_____no_output_____"
]
],
[
[
"In our config file we used the cell_vars and node_id_selections parameters to save the calcium influx and membrane potential of selected cells. We can also use the analyzer to display these traces:",
"_____no_output_____"
]
],
[
[
"from bmtk.analyzer.compartment import plot_traces\n\n_ = plot_traces(config_file='sim_ch04/simulation_config.json', group_by='pop_name', report_name='v_report')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e78b5df783ab2d4d228ea3def083294fcabc0cdd | 12,288 | ipynb | Jupyter Notebook | note/DQN.ipynb | Ceruleanacg/Learning-Notes | 1b2718dc85e622e35670fffbb525bb50d385f9a3 | [
"MIT"
] | 95 | 2018-06-01T03:57:39.000Z | 2021-12-31T04:51:21.000Z | note/DQN.ipynb | Ceruleanacg/Descent | 1b2718dc85e622e35670fffbb525bb50d385f9a3 | [
"MIT"
] | 1 | 2020-02-28T13:27:15.000Z | 2020-02-28T13:27:15.000Z | note/DQN.ipynb | Ceruleanacg/Descent | 1b2718dc85e622e35670fffbb525bb50d385f9a3 | [
"MIT"
] | 15 | 2018-06-24T07:33:29.000Z | 2020-10-03T04:12:27.000Z | 32 | 219 | 0.467855 | [
[
[
"# 问题设定",
"_____no_output_____"
],
[
"在小车倒立杆(CartPole)游戏中,我们希望通过强化学习训练一个智能体(agent),尽可能不断地左右移动小车,使得小车上的杆不倒,我们首先定义CartPole游戏:",
"_____no_output_____"
],
[
"CartPole游戏即是强化学习模型的enviorment,它与agent交互,实时更新state,内部定义了reward function,其中state有以下定义:",
"_____no_output_____"
],
[
"state每一个维度分别代表了:\n\n- 小车位置,它的取值范围是-2.4到2.4\n- 小车速度,它的取值范围是负无穷到正无穷\n- 杆的角度,它的取值范围是-41.8°到41.8°\n- 杆的角速,它的取值范围是负无穷到正无穷",
"_____no_output_____"
],
[
"action是一个2维向量,每一个维度分别代表向左和向右移动。",
"_____no_output_____"
],
[
"$$\naction \\in \\mathbb{R}^2\n$$",
"_____no_output_____"
],
[
"# DQN",
"_____no_output_____"
],
[
"我们将设计一个网络,作为状态-动作值函数(state-action value function),其输入是state,输出是对应各个action的value,并TD(Temporal Difference)进行迭代训练直至收敛。我们将定义两个这样的网络,分别记作$\\theta$和$\\theta^-$,分别代表估计网络与目标网络。",
"_____no_output_____"
],
[
"我们希望最小化:",
"_____no_output_____"
],
[
"$$\n\\left( y_j - Q \\left( \\phi_j, a_j; \\theta \\right) \\right)^2\n$$",
"_____no_output_____"
],
[
"其中,$a_j$具有以下形式:",
"_____no_output_____"
],
[
"$$\na_j = \\mathrm{argmax}_{a} Q \\left( \\phi(s_j), a; \\theta\\right)\n$$",
"_____no_output_____"
],
[
"其中,$y_j$具有以下形式:",
"_____no_output_____"
],
[
"$$\nf(x)=\n\\begin{cases}\nr_j & \\text{if episode ends at j + 1}\\\\\nr_j + \\gamma \\max_{a^{\\prime}} \\hat{Q} \\left( \\phi_{j+1}, a^{\\prime}; \\theta^{-} \\right)& \\text{otherwise}\n\\end{cases}$$\n\n",
"_____no_output_____"
],
[
"在最小化TD-Error时,我们将固定目标网络,只对估计网络做梯度反向传播,每次到达一定迭代次数后,将估计网络的权重复制到目标网络。在这个过程中,需要用到经验回放(Experience Replay)技术,即将每一次迭代观测到的$s_t, r_t, a_t, s_{t+1}$作为一个元组缓存,然后在这些缓存中随机抽取元组做批次梯度下降。",
"_____no_output_____"
],
[
"# 代码实现",
"_____no_output_____"
]
],
[
[
"# coding=utf-8\n\nimport tensorflow as tf\nimport numpy as np\nimport gym\nimport sys\n\nsys.path.append('..')\n\nfrom base.model import *\n\n%matplotlib inline",
"/Users/shuyu/anaconda3/envs/quant/lib/python3.5/importlib/_bootstrap.py:222: RuntimeWarning: compiletime version 3.6 of module 'tensorflow.python.framework.fast_tensor_util' does not match runtime version 3.5\n return f(*args, **kwds)\n"
],
[
"class Agent(BaseRLModel):\n\n def __init__(self, session, env, a_space, s_space, **options):\n super(Agent, self).__init__(session, env, a_space, s_space, **options)\n\n self._init_input()\n self._init_nn()\n self._init_op()\n self._init_saver()\n\n self.buffer = np.zeros((self.buffer_size, self.s_space + 1 + 1 + self.s_space))\n self.buffer_count = 0\n\n self.total_train_step = 0\n\n self.update_target_net_step = 200\n\n self.session.run(tf.global_variables_initializer())\n\n def _init_input(self, *args):\n with tf.variable_scope('input'):\n self.s_n = tf.placeholder(tf.float32, [None, self.s_space])\n self.s = tf.placeholder(tf.float32, [None, self.s_space])\n self.r = tf.placeholder(tf.float32, [None, ])\n self.a = tf.placeholder(tf.int32, [None, ])\n\n def _init_nn(self, *args):\n with tf.variable_scope('actor_net'):\n # w,b initializer\n w_initializer = tf.random_normal_initializer(mean=0.0, stddev=0.3)\n b_initializer = tf.constant_initializer(0.1)\n\n with tf.variable_scope('predict_q_net'):\n phi_state = tf.layers.dense(self.s,\n 32,\n tf.nn.relu,\n kernel_initializer=w_initializer,\n bias_initializer=b_initializer)\n\n self.q_predict = tf.layers.dense(phi_state,\n self.a_space,\n kernel_initializer=w_initializer,\n bias_initializer=b_initializer)\n\n with tf.variable_scope('target_q_net'):\n phi_state_next = tf.layers.dense(self.s_n,\n 32,\n tf.nn.relu,\n kernel_initializer=w_initializer,\n bias_initializer=b_initializer)\n\n self.q_target = tf.layers.dense(phi_state_next,\n self.a_space,\n kernel_initializer=w_initializer,\n bias_initializer=b_initializer)\n\n def _init_op(self):\n with tf.variable_scope('q_real'):\n # size of q_value_real is [BATCH_SIZE, 1]\n max_q_value = tf.reduce_max(self.q_target, axis=1)\n q_next = self.r + self.gamma * max_q_value\n self.q_next = tf.stop_gradient(q_next)\n\n with tf.variable_scope('q_predict'):\n # size of q_value_predict is [BATCH_SIZE, 1]\n action_indices = tf.stack([tf.range(tf.shape(self.a)[0], dtype=tf.int32), self.a], axis=1)\n self.q_eval = tf.gather_nd(self.q_predict, action_indices)\n\n with tf.variable_scope('loss'):\n self.loss_func = tf.reduce_mean(tf.squared_difference(self.q_next, self.q_eval, name='mse'))\n\n with tf.variable_scope('train'):\n self.train_op = tf.train.AdamOptimizer(self.learning_rate).minimize(self.loss_func)\n\n with tf.variable_scope('update_target_net'):\n t_params = tf.get_collection(tf.GraphKeys.GLOBAL_VARIABLES, scope='target_q_net')\n p_params = tf.get_collection(tf.GraphKeys.GLOBAL_VARIABLES, scope='predict_q_net')\n self.update_q_net = [tf.assign(t, e) for t, e in zip(t_params, p_params)]\n\n def predict(self, s):\n if np.random.uniform() < self.epsilon:\n a = np.argmax(self.session.run(self.q_predict, feed_dict={self.s: s[np.newaxis, :]}))\n else:\n a = np.random.randint(0, self.a_space)\n return a\n\n def snapshot(self, s, a, r, s_n):\n self.buffer[self.buffer_count % self.buffer_size, :] = np.hstack((s, [a, r], s_n))\n self.buffer_count += 1\n\n def train(self):\n if self.total_train_step % self.update_target_net_step == 0:\n self.session.run(self.update_q_net)\n\n batch = self.buffer[np.random.choice(self.buffer_size, size=self.batch_size), :]\n\n s = batch[:, :self.s_space]\n s_n = batch[:, -self.s_space:]\n a = batch[:, self.s_space].reshape((-1))\n r = batch[:, self.s_space + 1]\n\n _, cost = self.session.run([self.train_op, self.loss_func], {\n self.s: s, self.a: a, self.r: r, self.s_n: s_n\n })\n\n def run(self):\n if self.mode == 'train':\n for episode in range(self.train_episodes):\n s, r_episode = self.env.reset(), 0\n while True:\n # if episode > 400:\n # self.env.render()\n a = self.predict(s)\n s_n, r, done, _ = self.env.step(a)\n if done:\n r = -5\n r_episode += r\n self.snapshot(s, a, r_episode, s_n)\n s = s_n\n if done:\n break\n if self.buffer_count > self.buffer_size:\n self.train()\n if episode % 200 == 0:\n self.logger.warning('Episode: {} | Rewards: {}'.format(episode, r_episode))\n self.save()\n else:\n for episode in range(self.eval_episodes):\n s, r_episode = self.env.reset()\n while True:\n a = self.predict(s)\n s_n, r, done, _ = self.env.step(a)\n r_episode += r\n s = s_n\n if done:\n break",
"_____no_output_____"
],
[
"def main(_):\n # Make env.\n env = gym.make('CartPole-v0')\n env.seed(1)\n env = env.unwrapped\n # Init session.\n session = tf.Session()\n # Init agent.\n agent = Agent(session, env, env.action_space.n, env.observation_space.shape[0], **{\n KEY_MODEL_NAME: 'DQN',\n KEY_TRAIN_EPISODE: 3000\n })\n agent.run()\n",
"_____no_output_____"
],
[
"main(_)",
"\u001b[33mWARN: gym.spaces.Box autodetected dtype as <class 'numpy.float32'>. Please provide explicit dtype.\u001b[0m\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
e78b6bf6e9e17c21563c9c986530761fe12f7764 | 5,427 | ipynb | Jupyter Notebook | Day_4/Jupyter-Notebook.ipynb | akshatvg/NLP-Tasks | 609b54a76b7ba4cd080c21b5f3dc6b6ea0c6557a | [
"MIT"
] | 1 | 2020-08-31T15:39:06.000Z | 2020-08-31T15:39:06.000Z | Day_4/Jupyter-Notebook.ipynb | akshatvg/NLTK-Tasks | 609b54a76b7ba4cd080c21b5f3dc6b6ea0c6557a | [
"MIT"
] | null | null | null | Day_4/Jupyter-Notebook.ipynb | akshatvg/NLTK-Tasks | 609b54a76b7ba4cd080c21b5f3dc6b6ea0c6557a | [
"MIT"
] | null | null | null | 21.621514 | 206 | 0.513728 | [
[
[
"# Task 2: Simple Text Classification\ndef gender_features(word):\n return {'last_letter': word[-1]}",
"_____no_output_____"
],
[
"gender_features(\"Obama\")",
"_____no_output_____"
],
[
"from nltk.corpus import names",
"_____no_output_____"
],
[
"names.word()\nprint(len(names.words()))\nlabelled_names = ([(name,'male') for name in names.words('male.txt')] + [(name,'female') for name in names.words('female.txt')])",
"7944\n"
],
[
"import random\nrandom.shuffle(labelled_names)\nfeaturesets = [(gender_features(n),gender) for (n,gender) in labelled_names]",
"_____no_output_____"
],
[
"train_set, test_set=featuresets[:5000], featuresets[5000:]\nimport nltk\nclassifier = nltk.NaiveBayesClassifier.train(train_set)",
"_____no_output_____"
],
[
"classifier.classify(gender_features('Muskan'))",
"_____no_output_____"
],
[
"print(nltk.classify.accuracy(classifier,test_set))",
"0.7584918478260869\n"
],
[
"# Task 3: Count Vectorizer\nfrom sklearn.feature_extraction.text import CountVectorizer\nvect = CountVectorizer(binary=True)\ncorpus = [\"Tessaract is good optical character recognition engine \", \"optical character recognition is significant \"]\nvect.fit(corpus)",
"_____no_output_____"
],
[
"vocab = vect.vocabulary_",
"_____no_output_____"
],
[
"for key in sorted(vocab.keys()):\n print(\"{}:{}\".format(key, vocab[key]))",
"character:0\nengine:1\ngood:2\nis:3\noptical:4\nrecognition:5\nsignificant:6\ntessaract:7\n"
],
[
"print(vect.transform([\"This is a good optical illusion\"]).toarray())",
"[[0 0 1 1 1 0 0 0]]\n"
],
[
"# Finding Similarity Between Documents\nfrom sklearn.metrics.pairwise import cosine_similarity\nsimilarity = cosine_similarity(vect.transform([\"Google Cloud Vision is a character recognition engine\"]).toarray(), vect.transform([\"OCR is an optical character recognition engine\"]).toarray())\nprint(similarity)",
"[[0.89442719]]\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78b6d147eff3bee6433d6e3d8c9aa0b9ecc9867 | 3,617 | ipynb | Jupyter Notebook | notebooks/Chapter_13/00_Variance_Via_Covariance.ipynb | choldgraf/prob140 | 0750fc62fb114220035278ed2161e4b82ddca15f | [
"MIT"
] | null | null | null | notebooks/Chapter_13/00_Variance_Via_Covariance.ipynb | choldgraf/prob140 | 0750fc62fb114220035278ed2161e4b82ddca15f | [
"MIT"
] | null | null | null | notebooks/Chapter_13/00_Variance_Via_Covariance.ipynb | choldgraf/prob140 | 0750fc62fb114220035278ed2161e4b82ddca15f | [
"MIT"
] | null | null | null | 28.706349 | 412 | 0.545203 | [
[
[
"# HIDDEN\nfrom datascience import *\nfrom prob140 import *\nimport numpy as np\nimport matplotlib.pyplot as plt\nplt.style.use('fivethirtyeight')\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"# Variance Via Covariance #",
"_____no_output_____"
],
[
"In this chapter we return to random sampling and study the variability in the sum of a random sample. It is worth taking some time to understand how sample sums behave, because many interesting random variables like counts of successes can be written as sums. Binomial and hypergeometric random variables are such sums. Also, the mean of a random sample is a straightforward function of the sample sum. \n\nLet $X$ be a random variable. We will use some familiar shorthand:\n\n- $\\mu_X = E(X)$\n- $\\sigma_X = SD(X)$\n\nLet $D_X = X - \\mu_X$ denote the deviation of $X$ from its mean. Then\n\n$$\nVar(X) = \\sigma_X^2 = E(D_X^2)\n$$",
"_____no_output_____"
],
[
"### Variance of a Sum ###\nLet $X$ and $Y$ be two random variables on the same space, and let $S = X+Y$. Then $E(S) = \\mu_X + \\mu_Y$, and the deviation of $S$ is the sum of the deviations of $X$ and $Y$:\n\n$$\nD_S ~ = ~ S - \\mu_S ~ = ~ X + Y - (\\mu_X + \\mu_Y) ~ = ~ D_X + D_Y\n$$\n\nThis gives us some insight into the variance of the sum $S$.\n\n\\begin{align*}\nVar(S) &= E(D_S^2) \\\\\n&= E[(D_X + D_Y)^2] \\\\\n&= E(D_X^2) + E(D_Y^2) + 2E(D_XD_Y) \\\\\n&= Var(X) + Var(Y) + 2E(D_XD_Y)\n\\end{align*}\n\nThe first thing to note is that while the expectation of a sum is the sum of the expectations, the calculation above shows that the variance of a sum is in general **not** the sum of the variances. There's an extra term. \n\nTo calculate the variance of a sum, we have to understand that extra term. ",
"_____no_output_____"
],
[
"### Covariance ###\nThe *covariance of $X$ and $Y$*, denoted $Cov(X, Y)$, is the expected product of the deviations of $X$ and $Y$:\n\n$$\nCov(X, Y) ~ = ~ E(D_XD_Y) ~=~ E[(X - \\mu_X)(Y - \\mu_Y)]\n$$",
"_____no_output_____"
],
[
"In this chapter we will learn how to utilize covariance to find variances of sums. The fundamental calculation is the one we did above; here is the result again, using the language of covariance.\n\n$$\nVar(X+Y) ~ = ~ Var(X) + Var(Y) + 2Cov(X, Y)\n$$",
"_____no_output_____"
]
]
] | [
"code",
"markdown"
] | [
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
e78b7b56155cfdb02cc05fab30a0878482b40311 | 32,617 | ipynb | Jupyter Notebook | Day4.ipynb | JoachimMakowski/DataScienceMatrix2 | 7a817665dcd979f35acecbbfb3d6536d8f4a4f26 | [
"MIT"
] | null | null | null | Day4.ipynb | JoachimMakowski/DataScienceMatrix2 | 7a817665dcd979f35acecbbfb3d6536d8f4a4f26 | [
"MIT"
] | null | null | null | Day4.ipynb | JoachimMakowski/DataScienceMatrix2 | 7a817665dcd979f35acecbbfb3d6536d8f4a4f26 | [
"MIT"
] | null | null | null | 32,617 | 32,617 | 0.619094 | [
[
[
"!pip install --upgrade tables\n!pip install eli5\n!pip install xgboost",
"Collecting tables\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/ed/c3/8fd9e3bb21872f9d69eb93b3014c86479864cca94e625fd03713ccacec80/tables-3.6.1-cp36-cp36m-manylinux1_x86_64.whl (4.3MB)\n\u001b[K |████████████████████████████████| 4.3MB 4.8MB/s \n\u001b[?25hRequirement already satisfied, skipping upgrade: numpy>=1.9.3 in /usr/local/lib/python3.6/dist-packages (from tables) (1.17.5)\nRequirement already satisfied, skipping upgrade: numexpr>=2.6.2 in /usr/local/lib/python3.6/dist-packages (from tables) (2.7.1)\nInstalling collected packages: tables\n Found existing installation: tables 3.4.4\n Uninstalling tables-3.4.4:\n Successfully uninstalled tables-3.4.4\nSuccessfully installed tables-3.6.1\nCollecting eli5\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/97/2f/c85c7d8f8548e460829971785347e14e45fa5c6617da374711dec8cb38cc/eli5-0.10.1-py2.py3-none-any.whl (105kB)\n\u001b[K |████████████████████████████████| 112kB 4.9MB/s \n\u001b[?25hRequirement already satisfied: graphviz in /usr/local/lib/python3.6/dist-packages (from eli5) (0.10.1)\nRequirement already satisfied: jinja2 in /usr/local/lib/python3.6/dist-packages (from eli5) (2.11.1)\nRequirement already satisfied: tabulate>=0.7.7 in /usr/local/lib/python3.6/dist-packages (from eli5) (0.8.6)\nRequirement already satisfied: scipy in /usr/local/lib/python3.6/dist-packages (from eli5) (1.4.1)\nRequirement already satisfied: scikit-learn>=0.18 in /usr/local/lib/python3.6/dist-packages (from eli5) (0.22.1)\nRequirement already satisfied: numpy>=1.9.0 in /usr/local/lib/python3.6/dist-packages (from eli5) (1.17.5)\nRequirement already satisfied: attrs>16.0.0 in /usr/local/lib/python3.6/dist-packages (from eli5) (19.3.0)\nRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from eli5) (1.12.0)\nRequirement already satisfied: MarkupSafe>=0.23 in /usr/local/lib/python3.6/dist-packages (from jinja2->eli5) (1.1.1)\nRequirement already satisfied: joblib>=0.11 in /usr/local/lib/python3.6/dist-packages (from scikit-learn>=0.18->eli5) (0.14.1)\nInstalling collected packages: eli5\nSuccessfully installed eli5-0.10.1\nRequirement already satisfied: xgboost in /usr/local/lib/python3.6/dist-packages (0.90)\nRequirement already satisfied: scipy in /usr/local/lib/python3.6/dist-packages (from xgboost) (1.4.1)\nRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from xgboost) (1.17.5)\n"
],
[
"import pandas as pd\nimport numpy as np\n\nfrom sklearn.dummy import DummyRegressor\nfrom sklearn.tree import DecisionTreeRegressor\nfrom sklearn.ensemble import RandomForestRegressor\n\nimport xgboost as xgb\n\nfrom sklearn.metrics import mean_absolute_error\nfrom sklearn.model_selection import cross_val_score,KFold\n\nimport eli5\nfrom eli5.sklearn import PermutationImportance",
"/usr/local/lib/python3.6/dist-packages/sklearn/utils/deprecation.py:144: FutureWarning: The sklearn.metrics.scorer module is deprecated in version 0.22 and will be removed in version 0.24. The corresponding classes / functions should instead be imported from sklearn.metrics. Anything that cannot be imported from sklearn.metrics is now part of the private API.\n warnings.warn(message, FutureWarning)\n/usr/local/lib/python3.6/dist-packages/sklearn/utils/deprecation.py:144: FutureWarning: The sklearn.feature_selection.base module is deprecated in version 0.22 and will be removed in version 0.24. The corresponding classes / functions should instead be imported from sklearn.feature_selection. Anything that cannot be imported from sklearn.feature_selection is now part of the private API.\n warnings.warn(message, FutureWarning)\nUsing TensorFlow backend.\n"
],
[
"cd \"/content/drive/My Drive/Colab Notebooks/DataScienceMatrix/Matrix_two/DataScienceMatrix2\"",
"/content/drive/My Drive/Colab Notebooks/DataScienceMatrix/Matrix_two/DataScienceMatrix2\n"
],
[
"df = pd.read_hdf('Data/car.h5')\ndf.shape",
"_____no_output_____"
],
[
"SUFFIX_CAT = '__cat'\nfor feat in df.columns:\n if isinstance(df[feat][0],list): continue\n factorized_values = df[feat].factorize()[0]\n if SUFFIX_CAT in feat:\n df[feat] = factorized_values\n else:\n df[feat + SUFFIX_CAT] = factorized_values",
"_____no_output_____"
],
[
"cat_feats = [x for x in df.columns if SUFFIX_CAT in x]\ncat_feats = [x for x in cat_feats if 'price' not in x]\nlen(cat_feats)",
"_____no_output_____"
],
[
"def run_model(model,feats):\n X = df[feats].values\n Y = df['price_value'].values\n\n scores = cross_val_score(model,X,Y,cv = 3, scoring = 'neg_mean_absolute_error')\n return np.mean(scores),np.std(scores)",
"_____no_output_____"
],
[
"\nrun_model(DecisionTreeRegressor(max_depth=5),cat_feats)",
"_____no_output_____"
],
[
"run_model(RandomForestRegressor(max_depth=5,n_estimators=50,random_state=0),cat_feats)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"##XGBBOOST",
"_____no_output_____"
]
],
[
[
"xgb_params = {\n 'max_depth' : 5,\n 'n_estimators' : 50,\n 'learning_rate' : 0.1,\n 'seed' : 0\n}\n\nmodel = xgb.XGBRegressor(**xgb_params)\nrun_model(model,cat_feats)",
"[16:17:38] WARNING: /workspace/src/objective/regression_obj.cu:152: reg:linear is now deprecated in favor of reg:squarederror.\n[16:17:57] WARNING: /workspace/src/objective/regression_obj.cu:152: reg:linear is now deprecated in favor of reg:squarederror.\n[16:18:16] WARNING: /workspace/src/objective/regression_obj.cu:152: reg:linear is now deprecated in favor of reg:squarederror.\n"
],
[
"m = xgb.XGBRegressor(**xgb_params)\nm.fit(X,Y)\n\nimp = PermutationImportance(m,random_state = 0).fit(X,Y)\neli5.show_weights(imp,feature_names = cat_feats)",
"[16:19:44] WARNING: /workspace/src/objective/regression_obj.cu:152: reg:linear is now deprecated in favor of reg:squarederror.\n"
],
[
"feats = ['param_napęd__cat',\n'param_rok-produkcji__cat',\n'param_stan__cat',\n'param_skrzynia-biegów__cat',\n'param_faktura-vat__cat',\n'param_moc__cat',\n'param_marka-pojazdu__cat',\n'feature_kamera-cofania__cat',\n'param_typ__cat',\n'seller_name__cat',\n'param_pojemność-skokowa__cat',\n'feature_wspomaganie-kierownicy__cat',\n'param_model-pojazdu__cat',\n'param_wersja__cat',\n'param_kod-silnika__cat',\n'feature_system-start-stop__cat',\n'feature_asystent-pasa-ruchu__cat',\n'feature_czujniki-parkowania-przednie__cat',\n'feature_łopatki-zmiany-biegów__cat',\n'feature_regulowane-zawieszenie__cat']\nrun_model(xgb.XGBRegressor(**xgb_params),feats)",
"[16:30:41] WARNING: /workspace/src/objective/regression_obj.cu:152: reg:linear is now deprecated in favor of reg:squarederror.\n[16:30:45] WARNING: /workspace/src/objective/regression_obj.cu:152: reg:linear is now deprecated in favor of reg:squarederror.\n[16:30:49] WARNING: /workspace/src/objective/regression_obj.cu:152: reg:linear is now deprecated in favor of reg:squarederror.\n"
],
[
"df['param_napęd'].unique()\n\n",
"_____no_output_____"
],
[
"df['param_rok-produkcji'] = df['param_rok-produkcji'].map(lambda x: -1 if str(x) == 'None' else int(x))",
"_____no_output_____"
],
[
"feats = ['param_napęd__cat',\n'param_rok-produkcji',\n'param_stan__cat',\n'param_skrzynia-biegów__cat',\n'param_faktura-vat__cat',\n'param_moc__cat',\n'param_marka-pojazdu__cat',\n'feature_kamera-cofania__cat',\n'param_typ__cat',\n'seller_name__cat',\n'param_pojemność-skokowa__cat',\n'feature_wspomaganie-kierownicy__cat',\n'param_model-pojazdu__cat',\n'param_wersja__cat',\n'param_kod-silnika__cat',\n'feature_system-start-stop__cat',\n'feature_asystent-pasa-ruchu__cat',\n'feature_czujniki-parkowania-przednie__cat',\n'feature_łopatki-zmiany-biegów__cat',\n'feature_regulowane-zawieszenie__cat']\nrun_model(xgb.XGBRegressor(**xgb_params),feats)",
"[16:35:08] WARNING: /workspace/src/objective/regression_obj.cu:152: reg:linear is now deprecated in favor of reg:squarederror.\n[16:35:12] WARNING: /workspace/src/objective/regression_obj.cu:152: reg:linear is now deprecated in favor of reg:squarederror.\n[16:35:16] WARNING: /workspace/src/objective/regression_obj.cu:152: reg:linear is now deprecated in favor of reg:squarederror.\n"
],
[
"df['param_moc'] = df['param_moc'].map(lambda x: -1 if str(x) == 'None' else int(x.split(' ')[0]))",
"_____no_output_____"
],
[
"feats = ['param_napęd__cat',\n'param_rok-produkcji',\n'param_stan__cat',\n'param_skrzynia-biegów__cat',\n'param_faktura-vat__cat',\n'param_moc',\n'param_marka-pojazdu__cat',\n'feature_kamera-cofania__cat',\n'param_typ__cat',\n'seller_name__cat',\n'param_pojemność-skokowa__cat',\n'feature_wspomaganie-kierownicy__cat',\n'param_model-pojazdu__cat',\n'param_wersja__cat',\n'param_kod-silnika__cat',\n'feature_system-start-stop__cat',\n'feature_asystent-pasa-ruchu__cat',\n'feature_czujniki-parkowania-przednie__cat',\n'feature_łopatki-zmiany-biegów__cat',\n'feature_regulowane-zawieszenie__cat']\nrun_model(xgb.XGBRegressor(**xgb_params),feats)",
"[16:38:14] WARNING: /workspace/src/objective/regression_obj.cu:152: reg:linear is now deprecated in favor of reg:squarederror.\n[16:38:18] WARNING: /workspace/src/objective/regression_obj.cu:152: reg:linear is now deprecated in favor of reg:squarederror.\n[16:38:22] WARNING: /workspace/src/objective/regression_obj.cu:152: reg:linear is now deprecated in favor of reg:squarederror.\n"
],
[
"df['param_pojemność-skokowa'] = df['param_pojemność-skokowa'].map(lambda x: -1 if str(x) == 'None' else int(str(x).split('cm')[0].replace(' ','')))",
"_____no_output_____"
],
[
"feats = ['param_napęd__cat',\n'param_rok-produkcji',\n'param_stan__cat',\n'param_skrzynia-biegów__cat',\n'param_faktura-vat__cat',\n'param_moc',\n'param_marka-pojazdu__cat',\n'feature_kamera-cofania__cat',\n'param_typ__cat',\n'seller_name__cat',\n'param_pojemność-skokowa',\n'feature_wspomaganie-kierownicy__cat',\n'param_model-pojazdu__cat',\n'param_wersja__cat',\n'param_kod-silnika__cat',\n'feature_system-start-stop__cat',\n'feature_asystent-pasa-ruchu__cat',\n'feature_czujniki-parkowania-przednie__cat',\n'feature_łopatki-zmiany-biegów__cat',\n'feature_regulowane-zawieszenie__cat']\nrun_model(xgb.XGBRegressor(**xgb_params),feats)",
"[16:42:27] WARNING: /workspace/src/objective/regression_obj.cu:152: reg:linear is now deprecated in favor of reg:squarederror.\n[16:42:31] WARNING: /workspace/src/objective/regression_obj.cu:152: reg:linear is now deprecated in favor of reg:squarederror.\n[16:42:35] WARNING: /workspace/src/objective/regression_obj.cu:152: reg:linear is now deprecated in favor of reg:squarederror.\n"
],
[
"",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78b7c8e6a027385bd7da55e08edd92e3f354319 | 244,391 | ipynb | Jupyter Notebook | DS7_Unit_1_Sprint_Challenge_2_Data_Wrangling_and_Storytelling_(3).ipynb | johanaluna/DS-Unit-1-Sprint-2-Data-Wrangling-and-Storytelling | e3b1025254f8abebb3a16e101fa091922f6ca3d0 | [
"MIT"
] | null | null | null | DS7_Unit_1_Sprint_Challenge_2_Data_Wrangling_and_Storytelling_(3).ipynb | johanaluna/DS-Unit-1-Sprint-2-Data-Wrangling-and-Storytelling | e3b1025254f8abebb3a16e101fa091922f6ca3d0 | [
"MIT"
] | null | null | null | DS7_Unit_1_Sprint_Challenge_2_Data_Wrangling_and_Storytelling_(3).ipynb | johanaluna/DS-Unit-1-Sprint-2-Data-Wrangling-and-Storytelling | e3b1025254f8abebb3a16e101fa091922f6ca3d0 | [
"MIT"
] | null | null | null | 59.753301 | 50,622 | 0.572165 | [
[
[
"<a href=\"https://colab.research.google.com/github/johanaluna/DS-Unit-1-Sprint-2-Data-Wrangling-and-Storytelling/blob/master/DS7_Unit_1_Sprint_Challenge_2_Data_Wrangling_and_Storytelling_(3).ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Data Science Unit 1 Sprint Challenge 2\n\n## Data Wrangling and Storytelling\n\nTaming data from its raw form into informative insights and stories.",
"_____no_output_____"
],
[
"## Data Wrangling\n\nIn this Sprint Challenge you will first \"wrangle\" some data from [Gapminder](https://www.gapminder.org/about-gapminder/), a Swedish non-profit co-founded by Hans Rosling. \"Gapminder produces free teaching resources making the world understandable based on reliable statistics.\"\n- [Cell phones (total), by country and year](https://raw.githubusercontent.com/open-numbers/ddf--gapminder--systema_globalis/master/ddf--datapoints--cell_phones_total--by--geo--time.csv)\n- [Population (total), by country and year](https://raw.githubusercontent.com/open-numbers/ddf--gapminder--systema_globalis/master/ddf--datapoints--population_total--by--geo--time.csv)\n- [Geo country codes](https://github.com/open-numbers/ddf--gapminder--systema_globalis/blob/master/ddf--entities--geo--country.csv)\n\nThese two links have everything you need to successfully complete the first part of this sprint challenge.\n- [Pandas documentation: Working with Text Data](https://pandas.pydata.org/pandas-docs/stable/text.html) (one question)\n- [Pandas Cheat Sheet](https://github.com/pandas-dev/pandas/blob/master/doc/cheatsheet/Pandas_Cheat_Sheet.pdf) (everything else)",
"_____no_output_____"
],
[
"### Part 0. Load data\n\nYou don't need to add or change anything here. Just run this cell and it loads the data for you, into three dataframes.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\ncell_phones = pd.read_csv('https://raw.githubusercontent.com/open-numbers/ddf--gapminder--systema_globalis/master/ddf--datapoints--cell_phones_total--by--geo--time.csv')\n\npopulation = pd.read_csv('https://raw.githubusercontent.com/open-numbers/ddf--gapminder--systema_globalis/master/ddf--datapoints--population_total--by--geo--time.csv')\n\ngeo_country_codes = (pd.read_csv('https://raw.githubusercontent.com/open-numbers/ddf--gapminder--systema_globalis/master/ddf--entities--geo--country.csv')\n .rename(columns={'country': 'geo', 'name': 'country'}))",
"_____no_output_____"
]
],
[
[
"### Part 1. Join data",
"_____no_output_____"
],
[
"First, join the `cell_phones` and `population` dataframes (with an inner join on `geo` and `time`).\n\nThe resulting dataframe's shape should be: (8590, 4)",
"_____no_output_____"
]
],
[
[
"cell_phones.head()",
"_____no_output_____"
],
[
"population.head()",
"_____no_output_____"
],
[
"geo_country_codes.head()",
"_____no_output_____"
],
[
"#join the cell_phones and population dataframes (with an inner join on geo and time).\ndf=pd.merge(cell_phones,population, on=['geo','time'], how='inner')\nprint(df.shape)\ndf.head()",
"(8590, 4)\n"
]
],
[
[
"Then, select the `geo` and `country` columns from the `geo_country_codes` dataframe, and join with your population and cell phone data.\n\nThe resulting dataframe's shape should be: (8590, 5)",
"_____no_output_____"
]
],
[
[
"geo_country= geo_country_codes [['geo','country']]\ngeo_country.head()",
"_____no_output_____"
],
[
"df_merged = pd.merge(df, geo_country, on='geo')\nprint(df_merged.shape)\ndf_merged.head()",
"(8590, 5)\n"
]
],
[
[
"### Part 2. Make features",
"_____no_output_____"
],
[
"Calculate the number of cell phones per person, and add this column onto your dataframe.\n\n(You've calculated correctly if you get 1.220 cell phones per person in the United States in 2017.)",
"_____no_output_____"
]
],
[
[
"df_merged['cellphone_person']=df_merged['cell_phones_total']/df_merged['population_total']\ndf_merged.head()",
"_____no_output_____"
]
],
[
[
"Modify the `geo` column to make the geo codes uppercase instead of lowercase.",
"_____no_output_____"
]
],
[
[
"df_merged[(df_merged['country'] == 'United States') & (df_merged['time'] ==2017 )]",
"_____no_output_____"
]
],
[
[
"### Part 3. Process data",
"_____no_output_____"
],
[
"Use the describe function, to describe your dataframe's numeric columns, and then its non-numeric columns.\n\n(You'll see the time period ranges from 1960 to 2017, and there are 195 unique countries represented.)",
"_____no_output_____"
]
],
[
[
"import numpy as np\n# describe your dataframe's numeric columns\ndf_merged.describe()\n",
"_____no_output_____"
],
[
"df_merged.describe(exclude = [np.number])",
"_____no_output_____"
]
],
[
[
"In 2017, what were the top 5 countries with the most cell phones total?\n\nYour list of countries should have these totals:\n\n| country | cell phones total |\n|:-------:|:-----------------:|\n| ? | 1,474,097,000 |\n| ? | 1,168,902,277 |\n| ? | 458,923,202 |\n| ? | 395,881,000 |\n| ? | 236,488,548 |\n\n",
"_____no_output_____"
]
],
[
[
"# This optional code formats float numbers with comma separators\npd.options.display.float_format = '{:,}'.format",
"_____no_output_____"
],
[
"year2017 = df_merged[df_merged.time == 2017]\nyear2017.head()",
"_____no_output_____"
],
[
"#code to check the values\nyear2017.sort_values('cell_phones_total', ascending=False)",
"_____no_output_____"
],
[
"# Make top5 \ntop5_all=year2017.nlargest(5,'cell_phones_total')\ntop5=top5_all[['country','cell_phones_total']]\ntop5.head()",
"_____no_output_____"
]
],
[
[
"2017 was the first year that China had more cell phones than people.\n\nWhat was the first year that the USA had more cell phones than people?",
"_____no_output_____"
]
],
[
[
"order_celphones=df_merged.sort_values('cell_phones_total', ascending=False)\norder_celphones.head(10)",
"_____no_output_____"
],
[
"country_usa=df_merged[(df_merged['country'] == 'United States')]\ncountry_usa.head()\n# country_usa.country.unique()",
"_____no_output_____"
],
[
"condition_usa= country_usa[(country_usa['cell_phones_total'] > country_usa['population_total'])]\ncondition_usa.head()",
"_____no_output_____"
],
[
"#one way to do : What was the first year that the USA had more cell phones than people?\norder_usa=condition_usa.sort_values('time', ascending=True)\norder_usa.head(1)",
"_____no_output_____"
],
[
"#Second way to do: What was the first year that the USA had more cell phones than people?\ncondition_usa.nsmallest(1,'time')",
"_____no_output_____"
]
],
[
[
"### Part 4. Reshape data",
"_____no_output_____"
],
[
"\nCreate a pivot table:\n- Columns: Years 2007—2017\n- Rows: China, India, United States, Indonesia, Brazil (order doesn't matter)\n- Values: Cell Phones Total\n\nThe table's shape should be: (5, 11)",
"_____no_output_____"
]
],
[
[
"years=[2007,2008,2009,2010,2011,2012,2013,20014,2015,2016,2017]\ncountries=['China', 'India', 'United States', 'Indonesia', 'Brazil']\n#countries_pivot=df_merged.loc[df_merged['country'].isin(countries)]",
"_____no_output_____"
],
[
"years_merged=df_merged.loc[df_merged['time'].isin(years)& df_merged['country'].isin(countries)]\nyears_merged.head()",
"_____no_output_____"
],
[
"pivot_years=years_merged.pivot_table(index='country',columns='time',values='cell_phones_total',aggfunc='sum')\npivot_years.head()",
"_____no_output_____"
]
],
[
[
"Sort these 5 countries, by biggest increase in cell phones \n\nfrom 2007 to 2017.\n\nWhich country had 935,282,277 more cell phones in 2017 versus 2007?",
"_____no_output_____"
]
],
[
[
"flat_pivot= pd.DataFrame(pivot_years.to_records())\nflat_pivot.head()",
"_____no_output_____"
],
[
"flat_pivot['Percentage Change']=(flat_pivot['2017']-flat_pivot['2007'])/flat_pivot['2017']\nflat_pivot.head()",
"_____no_output_____"
],
[
"#ANSWER= Sort these 5 countries, by biggest increase in cell phones from 2007 to 2017.\nflat_pivot.sort_values('Percentage Change', ascending=False)",
"_____no_output_____"
],
[
"#Which country had 935,282,277 more cell phones in 2017 versus 2007?\nflat_pivot['Change']=(flat_pivot['2017']-flat_pivot['2007'])\nflat_pivot.head()",
"_____no_output_____"
],
[
"flat_pivot[flat_pivot['Change']==935282277]",
"_____no_output_____"
]
],
[
[
"## Data Storytelling\n\nIn this part of the sprint challenge you'll work with a dataset from **FiveThirtyEight's article, [Every Guest Jon Stewart Ever Had On ‘The Daily Show’](https://fivethirtyeight.com/features/every-guest-jon-stewart-ever-had-on-the-daily-show/)**!",
"_____no_output_____"
],
[
"### Part 0 — Run this starter code\n\nYou don't need to add or change anything here. Just run this cell and it loads the data for you, into a dataframe named `df`.\n\n(You can explore the data if you want, but it's not required to pass the Sprint Challenge.)",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\n\nurl = 'https://raw.githubusercontent.com/fivethirtyeight/data/master/daily-show-guests/daily_show_guests.csv'\ndf1 = pd.read_csv(url).rename(columns={'YEAR': 'Year', 'Raw_Guest_List': 'Guest'})\n\ndef get_occupation(group):\n if group in ['Acting', 'Comedy', 'Musician']:\n return 'Acting, Comedy & Music'\n elif group in ['Media', 'media']:\n return 'Media'\n elif group in ['Government', 'Politician', 'Political Aide']:\n return 'Government and Politics'\n else:\n return 'Other'\n \ndf1['Occupation'] = df1['Group'].apply(get_occupation)",
"_____no_output_____"
]
],
[
[
"### Part 1 — What's the breakdown of guests’ occupations per year?\n\nFor example, in 1999, what percentage of guests were actors, comedians, or musicians? What percentage were in the media? What percentage were in politics? What percentage were from another occupation?\n\nThen, what about in 2000? In 2001? And so on, up through 2015.\n\nSo, **for each year of _The Daily Show_, calculate the percentage of guests from each occupation:**\n- Acting, Comedy & Music\n- Government and Politics\n- Media\n- Other\n\n#### Hints:\nYou can make a crosstab. (See pandas documentation for examples, explanation, and parameters.)\n\nYou'll know you've calculated correctly when the percentage of \"Acting, Comedy & Music\" guests is 90.36% in 1999, and 45% in 2015.",
"_____no_output_____"
]
],
[
[
"print(df1.shape)\ndf1.head()",
"(2693, 6)\n"
],
[
"crosstab_profession=pd.crosstab(df1['Year'], df1['Occupation'], normalize='index').round(4)*100\ncrosstab_profession.head(20)",
"_____no_output_____"
]
],
[
[
"### Part 2 — Recreate this explanatory visualization:",
"_____no_output_____"
],
[
"**Hints:**\n- You can choose any Python visualization library you want. I've verified the plot can be reproduced with matplotlib, pandas plot, or seaborn. I assume other libraries like altair or plotly would work too.\n- If you choose to use seaborn, you may want to upgrade the version to 0.9.0.\n\n**Expectations:** Your plot should include:\n- 3 lines visualizing \"occupation of guests, by year.\" The shapes of the lines should look roughly identical to 538's example. Each line should be a different color. (But you don't need to use the _same_ colors as 538.)\n- Legend or labels for the lines. (But you don't need each label positioned next to its line or colored like 538.)\n- Title in the upper left: _\"Who Got To Be On 'The Daily Show'?\"_ with more visual emphasis than the subtitle. (Bolder and/or larger font.)\n- Subtitle underneath the title: _\"Occupation of guests, by year\"_\n\n**Optional Bonus Challenge:**\n- Give your plot polished aesthetics, with improved resemblance to the 538 example.\n- Any visual element not specifically mentioned in the expectations is an optional bonus.",
"_____no_output_____"
]
],
[
[
"from IPython.display import display, Image\npng = 'https://fivethirtyeight.com/wp-content/uploads/2015/08/hickey-datalab-dailyshow.png'\nexample = Image(png, width=500)\ndisplay(example)",
"_____no_output_____"
],
[
"import seaborn as sns\nsns.__version__",
"_____no_output_____"
],
[
"flat_df1= pd.DataFrame(crosstab_profession.to_records())\nflat_df1",
"_____no_output_____"
],
[
"flat_df1=flat_df1.drop(['Other'],axis=1)\nflat_df1.head()",
"_____no_output_____"
],
[
"from matplotlib.ticker import AutoMinorLocator, MultipleLocator, FuncFormatter\nsns.set(style=\"ticks\")\nsns.set(rc={'axes.facecolor':'#F0F0F0', 'figure.facecolor':'#F0F0F0'})\nplt.figure(figsize=(8,5.5))\nax=sns.lineplot(data=flat_df1, y='Media',x='Year', color='#6f2aa1', linewidth=2.5)\nax=sns.lineplot(data=flat_df1, y='Government and Politics',x='Year', color='red', linewidth=2.5)\nax=sns.lineplot(data=flat_df1, y='Acting, Comedy & Music',x='Year', color='#2bb1f0', linewidth=2.5)\n\nplt.suptitle('Who Got To Be On The Daily Show?', x=0.35, y=1.04,fontweight=\"bold\", )\nplt.title('Occupation of guests, by year', x=0.2, y=1.06)\nax.spines['right'].set_color('#F0F0F0')\nax.spines['left'].set_color('#F0F0F0')\nax.spines['bottom'].set_color('#bdbdbd')\nax.spines['top'].set_color('#F0F0F0')\n\nax.set_xlim(1999, 2015)\nax.set_ylim(0, 108)\n\nax.set_xlabel(\"\")\nax.set_ylabel(\"\")\nax.grid(linestyle=\"-\", linewidth=0.5, color='#bdbdbd', zorder=-10)\n\nax.yaxis.set_major_locator(MultipleLocator(25))\nax.yaxis.set_minor_locator(AutoMinorLocator(100))\n\n\nax.tick_params(which='major', width=0.25)\nax.tick_params(which='major', length=1.0)\nax.tick_params(which='minor', width=0.1, labelsize=10)\nax.tick_params(which='minor', length=5, labelsize=10, labelcolor='0.25')\n\nplt.text(x=2008, y= 50, s='Media' ,color='#6f2aa1', weight='bold')\nplt.text(x=2010, y= 5, s='Government and Politics' ,color='red', weight='bold')\nplt.text(x=2001, y= 75, s='Acting, Comedy & Music' ,color='#2bb1f0',weight='bold');\n",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78ba1229682cd8317dfd7b8f7db23af3706b520 | 77,238 | ipynb | Jupyter Notebook | ipynb/Sicherman Dice.ipynb | awesome-archive/pytudes | 650b64134f9f38787b2775abcf8e915f0e2d9da1 | [
"MIT"
] | null | null | null | ipynb/Sicherman Dice.ipynb | awesome-archive/pytudes | 650b64134f9f38787b2775abcf8e915f0e2d9da1 | [
"MIT"
] | null | null | null | ipynb/Sicherman Dice.ipynb | awesome-archive/pytudes | 650b64134f9f38787b2775abcf8e915f0e2d9da1 | [
"MIT"
] | null | null | null | 36.991379 | 11,884 | 0.584596 | [
[
[
"# Sicherman Dice\n\n*Note: This notebook takes the form of a conversation between two problem solvers. One speaks in* **bold**, *the other in* plain. *Also note, for those who are not native speakers of English: \"dice\" is the plural form; \"die\" is the singular.*\n\nHuh. <a href=\"http://wordplay.blogs.nytimes.com/2014/06/16/dice-3/\">This</a> is interesting. You know how in many games, such as craps or Monopoly, you roll two regular dice and add them up. Only the sum matters, not what either of the individual dice shows.\n\n**Right.**\n\nAnd some of those sums, like 8, can be made multiple ways, while 2 and 12 can only be made one way. \n\n**Yeah. 8 can be made 5 ways, so it has a 5/36 probability of occuring.**\n\nThe interesting thing is that people have been playing dice games for 7,000 years. But it wasn't until 1977 that <a href=\"http://userpages.monmouth.com/~colonel/\">Colonel George Sicherman</a> asked whether is is possible to have a pair of dice that are not regular dice—that is, they don't have (1, 2, 3, 4, 5, 6) on the six sides—but have the same distribution of sums as a regular pair—so the pair of dice would also have to have 5 ways of making 8, but it could be different ways; maybe 7+1 could be one way. Sicherman assumes that each side bears a positive integer.\n\n**And what did he find?**\n\nWouldn't it be more fun to figure it out for ourselves?\n\n**OK!**\n\nHow could we proceed?\n\n**When in doubt, [use brute force](http://quotes.lifehack.org/quote/ken-thompson/when-in-doubt-use-brute-force/): we can write a program to enumerate the possibilities:**\n\n\n- **Generate all dice that could possibly be part of a solution, such as (1, 2, 2, 4, 8, 9).**\n- **Consider all pairs of these dice, such as ((1, 3, 4, 4, 5, 8), (1, 2, 2, 3, 3, 4))**\n- **See if we find any pairs that are not the regular pair, but do have the same distribution of sums as the regular pair.**\n\n\nThat's great. I can code up your description almost verbatim. I'll also keep track of our TO DO list:",
"_____no_output_____"
]
],
[
[
"def sicherman():\n \"\"\"The set of pairs of 6-sided dice that have the same\n distribution of sums as a regular pair of dice.\"\"\"\n return {pair for pair in pairs(all_dice())\n if pair != regular_pair\n and sums(pair) == regular_sums}\n\n# TODO: pairs, all_dice, regular_pair, sums, regular_sums",
"_____no_output_____"
]
],
[
[
"**Looks good to me.**\n\nNow we can tick off the items in the TO DO list. The function `pairs` is first, and it is easy:",
"_____no_output_____"
]
],
[
[
"def pairs(collection): \n \"Return all pairs (A, B) from collection where A <= B.\"\n return [(A, B) for A in collection for B in collection if A <= B]\n\n# TODO: all_dice, regular_pair, sums, regular_sums",
"_____no_output_____"
]
],
[
[
"**That's good. We could have used the library function `itertools.combinations_with_replacement`, but let's just leave it as is. We should test to make sure it works:**",
"_____no_output_____"
]
],
[
[
"pairs(['A', 'B', 'C'])",
"_____no_output_____"
]
],
[
[
"# TO DO: `sums(pair)`\n\n\nNow for `sums`: we need some way to represent all the 36 possible sums from a pair of dice. We want a representation that will be the same for two different pairs if all 36 sums are the same, but where the order or composition of the sums doesn't matter. \n\n**So we want a set of the sums?**\n\nWell, it can't be a set, because we need to know that 8 can be made 5 ways, not just that 8 is a member of the set. The technical term for a collection where order doesn't matter but where you can have repeated elements is a **bag**, or sometimes called a [**multiset**](https://en.wikipedia.org/wiki/Multiset). For example, the regular pair of dice makes two 11s with 5+6 and 6+5, and another pair could make two 11s with 7+4 and 3+8. Can you think of a representation that will do that?\n\n**Well the easiest is just a sorted list or tuple—if we don't want order to matter, sorting takes care of that. Another choice would be a dictionary of {sum: count} entries, like {2: 1, 3: 2, ... 11: 2, 12: 1}. There is even a library class, `collections.Counter`, that does exactly that.**\n\nHow do we choose between the two representations?\n\n**I don't think it matters much. Since there are only 36 entries, I think the sorted list will be simpler, and probably more efficient. For 100-sided dice I'd probably go with the Counter.**\n\nOK, here's some code implementing `sums` as a sorted list, and definitions for regular die pair, and sums.\nBy the way, I could have used `range(1, 7)` to define a regular die, but `range` is zero-based, and regular dice are one-based, so I defined the function `ints` instead.",
"_____no_output_____"
]
],
[
[
"def sums(pair):\n \"All possible sums of a side from one die plus a side from the other.\"\n (A, B) = pair\n return Bag(a + b for a in A for b in B)\n\nBag = sorted # Implement a bag as a sorted list\n\ndef ints(start, end): \n \"A tuple of the integers from start to end, inclusive.\"\n return tuple(range(start, end + 1))\n\nregular_die = ints(1, 6)\nregular_pair = (regular_die, regular_die)\nregular_sums = sums(regular_pair)\n\n# TODO: all_dice",
"_____no_output_____"
]
],
[
[
"Let's check the `regular_sums`:",
"_____no_output_____"
]
],
[
[
"len(regular_sums)",
"_____no_output_____"
],
[
"print(regular_sums)",
"[2, 3, 3, 4, 4, 4, 5, 5, 5, 5, 6, 6, 6, 6, 6, 7, 7, 7, 7, 7, 7, 8, 8, 8, 8, 8, 9, 9, 9, 9, 10, 10, 10, 11, 11, 12]\n"
]
],
[
[
"**And we can see what that would look like to a `Counter`:**",
"_____no_output_____"
]
],
[
[
"from collections import Counter\n\nCounter(regular_sums)",
"_____no_output_____"
]
],
[
[
"**Looks good! Now only one more thing on our TODO list:**\n\n# TO DO: `all_dice()`\n\n\n`all_dice` should generate all possible dice, where by \"possible\" I mean the dice that could feasibly be part of a pair that is a solution to the Sicherman problem. Do we know how many dice that will be? Is it a large enough number that efficiency will be a problem?\n\n**Let's see. A die has six sides each. If each side can be a number from, say, 1 to 10, that's 10<sup>6</sup> or a million possible dice; a million is a small number for a computer.**\n\nTrue, a million is a relatively small number for `all_dice()`, but how many `pairs(all_dice())` will there be?\n\n**Ah. A million squared is a trillion. That's a large number even for a computer. Just counting to a trillion takes hours in Python; checking a trillion pairs will take days.**\n\nSo we need to get rid of most of the dice. What about permutations?\n\n**Good point. If I have the die (1, 2, 3, 4, 5, 6), then I don't need the 6! = 720 different permutations of this die— that is, dice like (2, 4, 6, 1, 3, 5).\nEach die should be a bag (I learned a new word!) of sides. So we've already eliminated 719/720 = 99.9% of the work.**\n\nOne other thing bothers me ... how do you know that the sides can range from 1 to 10? Are you sure that 11 can't be part of a solution? Or 12?\n\n**Every side on every die must be a positive integer, right?**\n\nRight. No zeroes, no negative numbers, no fractions.\n\n**Then I know for sure that 12 can't be on any die, because when you add 12 to whatever is on the other die, you would get at least 13, and 13 is not allowed in the regular distribution of sums.**\n\nGood. How about 11?\n\n**We can't have a sum that is bigger than 12. So if one die had an 11, the other would have to have all 1s. That wouldn't work, because then we'd have six 12s, but we only want one. So 10 is the biggest allowable number on a die.**\n\nWhat else can we say about the biggest number on a die?\n\n**There's one 12 in the sums. But there are several ways to make a 12: 6+6 or 7+5 or 8+4, and so on. So I can't say for sure what the biggest number on any one die will be. But I can say that whatever the biggest number on a die is, it will be involved in summing to 12, so there can be only one of them, because we only want to make one 12.**\n\nWhat about the smallest number on a die?\n\n**Well, there's only one 2 allowed in the sums. The only way to sum to 2 is 1+1: a 1 from each of the dice in the pair. If a die had no 1s, we wouldn't get a 2; if a die had more than one 1, we would get too many 2s. So every die has to have exactly one 1.**\n\nGood. So each die has exactly one 1, and exactly one of whatever the biggest number is, something in the range up to 10. Here's a picture of the six sides of any one die:\n\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\"> 1 </span> <\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2-10</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2-10</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2-10</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2-10</span> <\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2-10</span>\n\nThe bag of sides is always listed in non-decreasing order; the first side, 1, is less than the next, and the last side, whatever it is, is greater than the one before it.\n\n**Wait a minute: you have [2-10] < [2-10]. But 2 is not less than 2, and 10 is not less than 10. I think it should be [2-9] < [3-10]. So the picture should be like this:**\n\n\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\"> 1 </span> <\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2-9</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2-9</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2-9</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2-9</span> <\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">3-10</span>\n\nGood! We're making progress in cutting down the range. But it That this bothers me because it says the range for the biggest number is 3 to 10. But if one die has a 3 and the other a 10, that adds to 13. So I'm thinking that it is not possible to have a 10 after all—because if one die had a 10, then the other would have to have a 2 as the biggest number, and that can't be. Therefore the biggest number is in the range of 3 to 9. But then the others have to be \nless, so make them 2 to 8:\n\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\"> 1 </span> <\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2-8</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2-8</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2-8</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2-8</span> <\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">3-9</span>\n\n**I can turn this picture into code:**",
"_____no_output_____"
]
],
[
[
"def all_dice():\n \"A list of all feasible 6-sided dice for the Sicherman problem.\"\n return [(1, s2, s3, s4, s5, s6)\n for s2 in ints(2, 8) \n for s3 in ints(s2, 8)\n for s4 in ints(s3, 8)\n for s5 in ints(s4, 8) \n for s6 in ints(s5+1, 9)]",
"_____no_output_____"
]
],
[
[
"I think we're ready to run `sicherman()`. Any bets on what we'll find out?\n\n**I bet that Sicherman is remembered because he discovered a pair of dice that works. If he just proved the non-existence of a pair, I don't think that would be noteworthy.**\n\nMakes sense. Here goes:\n\n# The Answer",
"_____no_output_____"
]
],
[
[
"sicherman()",
"_____no_output_____"
]
],
[
[
"**Look at that!**\n\nIt turns out you can <a href=\"http://www.grand-illusions.com/acatalog/Sicherman_Dice.html\">buy</a> a pair of dice with just these numbers.\n\n<a href=\"http://www.grand-illusions.com/acatalog/Sicherman_Dice.html\"><img src=\"http://www.grand-illusions.com/acatalog/lge-sicherman_dice.jpg\"></a>\n\nHere's a table I borrowed from [Wikipedia](https://en.wikipedia.org/wiki/Sicherman_dice) that shows both pairs of dice have the same sums. \n\n<table class=\"wikitable\">\n<tr>\n<td align=\"centre\"></td>\n<td align=\"centre\">2</td>\n<td align=\"centre\">3</td>\n<td align=\"centre\">4</td>\n<td align=\"centre\">5</td>\n<td align=\"centre\">6</td>\n<td align=\"centre\">7</td>\n<td align=\"centre\">8</td>\n<td align=\"centre\">9</td>\n<td align=\"centre\">10</td>\n<td align=\"centre\">11</td>\n<td align=\"centre\">12</td>\n</tr>\n<tr>\n<td>Regular dice:\n<br>(1, 2, 3, 4, 5, 6)\n<br>(1, 2, 3, 4, 5, 6)</td>\n<td>1+1</td>\n<td>1+2<br />\n2+1</td>\n<td>1+3<br />\n2+2<br />\n3+1</td>\n<td>1+4<br />\n2+3<br />\n3+2<br />\n4+1</td>\n<td>1+5<br />\n2+4<br />\n3+3<br />\n4+2<br />\n5+1</td>\n<td>1+6<br />\n2+5<br />\n3+4<br />\n4+3<br />\n5+2<br />\n6+1</td>\n<td>2+6<br />\n3+5<br />\n4+4<br />\n5+3<br />\n6+2</td>\n<td>3+6<br />\n4+5<br />\n5+4<br />\n6+3</td>\n<td>4+6<br />\n5+5<br />\n6+4</td>\n<td>5+6<br />\n6+5</td>\n<td>6+6</td>\n</tr>\n<tr>\n<td>Sicherman dice:\n<br>(<b><span style=\"color:orange;\">1</span>, <span style=\"color:red;\">2</span>, <span style=\"color:blue;\"><i>2</i></span>, <span style=\"color:red;\">3</span>, <span style=\"color:blue;\"><i>3</i></span>, <span style=\"color:orange;\">4</span></b>)\n<br>(1, 3, 4, 5, 6, 8)</td>\n<td><b><span style=\"color:orange;\">1</span></b>+1</td>\n<td><b><span style=\"color:red;\">2</span></b>+1<br />\n<b><span style=\"color:blue;\"><i>2</i></span></b>+1</td>\n<td><b><span style=\"color:red;\">3</span></b>+1<br />\n<b><span style=\"color:blue;\"><i>3</i></span></b>+1<br />\n<b><span style=\"color:orange;\">1</span></b>+3</td>\n<td><b><span style=\"color:orange;\">1</span></b>+4<br />\n<b><span style=\"color:red;\">2</span></b>+3<br />\n<b><span style=\"color:blue;\"><i>2</i></span></b>+3<br />\n<b><span style=\"color:orange;\">4</span></b>+1</td>\n<td><b><span style=\"color:orange;\">1</span></b>+5<br />\n<b><span style=\"color:red;\">2</span></b>+4<br />\n<b><span style=\"color:blue;\"><i>2</i></span></b>+4<br />\n<b><span style=\"color:red;\">3</span></b>+3<br />\n<b><span style=\"color:blue;\"><i>3</i></span></b>+3</td>\n<td><b><span style=\"color:orange;\">1</span></b>+6<br />\n<b><span style=\"color:red;\">2</span></b>+5<br />\n<b><span style=\"color:blue;\"><i>2</i></span></b>+5<br />\n<b><span style=\"color:red;\">3</span></b>+4<br />\n<b><span style=\"color:blue;\"><i>3</i></span></b>+4<br />\n<b><span style=\"color:orange;\">4</span></b>+3</td>\n<td><b><span style=\"color:red;\">2</span></b>+6<br />\n<b><span style=\"color:blue;\"><i>2</i></span></b>+6<br />\n<b><span style=\"color:red;\">3</span></b>+5<br />\n<b><span style=\"color:blue;\"><i>3</i></span></b>+5<br />\n<b><span style=\"color:orange;\">4</span></b>+4</td>\n<td><b><span style=\"color:orange;\">1</span></b>+8<br />\n<b><span style=\"color:red;\">3</span></b>+6<br />\n<b><span style=\"color:blue;\"><i>3</i></span></b>+6<br />\n<b><span style=\"color:orange;\">4</span></b>+5</td>\n<td><b><span style=\"color:red;\">2</span></b>+8<br />\n<b><span style=\"color:blue;\"><i>2</i></span></b>+8<br />\n<b><span style=\"color:orange;\">4</span></b>+6<br /></td>\n<td><b><span style=\"color:red;\">3</span></b>+8<br />\n<b><span style=\"color:blue;\"><i>3</i></span></b>+8</td>\n<td><b><span style=\"color:orange;\">4</span></b>+8</td>\n</tr>\n</table>\n\nWe could stop here. Or we could try to solve it for *N*-sided dice.\n\n# Why stop now? Onward!",
"_____no_output_____"
],
[
"OK. I know 4-, 12-, and 20-sided dice are common, but we'll try to handle any *N* > 1. My guess is we won't go too far before our program becomes too slow. So, before we try *N*-sided dice, let's analyze six-sided dice a little better, to see if we can eliminate some of the pairs before we start. The picture says that (1, 2, 2, 2, 2, 3) could be a valid die. Could it?\n\n**No! If a die had four 2s, then we know that since the other die has one 1, we could make 2 + 1 = 3 four ways. But the `regular_sums` has only two 3s. So that means that a die can have no more than two 2s. New picture:**\n\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\"> 1 </span> <\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2-8</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2-8</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">3-8</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">3-8</span> <\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">3-9</span>\n\nNow we've got [3-8] < [3-9]; that's not right. If a die can only have one 1 and two 2s, then it must have at least one number that is a 3 or more, followed by the biggest number, which must be 4 or more, and we know a pair of biggest numbers must sum to 12, so the range of the biggest can't be [4-9], it must be [4-8]:\n\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\"> 1 </span> <\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2-7</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2-7</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">3-7</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">3-7</span> <\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">4-8</span>",
"_____no_output_____"
]
],
[
[
"def all_dice():\n \"A list of all feasible 6-sided dice for the Sicherman problem.\"\n return [(1, s2, s3, s4, s5, s6)\n for s2 in ints(2, 7) \n for s3 in ints(s2, 7)\n for s4 in ints(max(s3, 3), 7)\n for s5 in ints(s4, 7) \n for s6 in ints(s5+1, 8)]",
"_____no_output_____"
]
],
[
[
"I'll count how many dice and how many pairs there are now:",
"_____no_output_____"
]
],
[
[
"len(all_dice())",
"_____no_output_____"
],
[
"len(pairs(all_dice()))",
"_____no_output_____"
]
],
[
[
"**Nice—we got down from a trillion pairs to 26,000. I don't want to print `all_dice()`, but I can sample a few:**",
"_____no_output_____"
]
],
[
[
"import random\n\nrandom.sample(all_dice(), 10)",
"_____no_output_____"
]
],
[
[
"# `sicherman(N)`\n\nOK, I think we're ready to update `sicherman()` to `sicherman(N)`. \n\n**Sure, most of that will be easy, just parameterizing with `N`:**",
"_____no_output_____"
]
],
[
[
"def sicherman(N=6):\n \"\"\"The set of pairs of N-sided dice that have the same\n distribution of sums as a regular pair of N-sided dice.\"\"\"\n reg_sums = regular_sums(N)\n reg_pair = regular_pair(N)\n return {pair for pair in pairs(all_dice(N))\n if pair != reg_pair\n and sums(pair) == reg_sums}\n\ndef regular_die(N): return ints(1, N)\ndef regular_pair(N): return (regular_die(N), regular_die(N))\ndef regular_sums(N): return sums(regular_pair(N))\n\n# TODO: all_dice(N)",
"_____no_output_____"
]
],
[
[
"Good. I think it would be helpful for me to look at a table of `regular_sums`:",
"_____no_output_____"
]
],
[
[
"for N in ints(1, 7):\n print(\"N:\", N, dict(Counter(regular_sums(N))))",
"N: 1 {2: 1}\nN: 2 {2: 1, 3: 2, 4: 1}\nN: 3 {2: 1, 3: 2, 4: 3, 5: 2, 6: 1}\nN: 4 {2: 1, 3: 2, 4: 3, 5: 4, 6: 3, 7: 2, 8: 1}\nN: 5 {2: 1, 3: 2, 4: 3, 5: 4, 6: 5, 7: 4, 8: 3, 9: 2, 10: 1}\nN: 6 {2: 1, 3: 2, 4: 3, 5: 4, 6: 5, 7: 6, 8: 5, 9: 4, 10: 3, 11: 2, 12: 1}\nN: 7 {2: 1, 3: 2, 4: 3, 5: 4, 6: 5, 7: 6, 8: 7, 9: 6, 10: 5, 11: 4, 12: 3, 13: 2, 14: 1}\n"
]
],
[
[
"**That is helpful. I can see that any `regular_sums` must have one 2 and two 3s, and three 4s, and so on, not just for `N=6` but for any `N` (except for trivially small `N`). And that means that any regular die can have at most two 2s, three 3s, four 4s, and so on. So we have this picture:**\n\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\"> 1 </span> <\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2+</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2+</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">3+</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">3+</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">3+</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">4+</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">4+</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">4+</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">4+</span> ≤ ...\n\n**where [2+] means the lower bound is 2, but we haven't figured out yet what the upper bound is.**\n\nLet's figure out upper bounds starting from the biggest number. What can the biggest number be?\n\n**For a pair of *N*-sided die, the biggest sides from each one must add up to 2*N*. Let's take *N*=10 as an example. The biggest numbers on two 10-sided Sicherman dice must sum to 20. According to the picture above, the lower bound on the biggest number would be 4, but because there can only be one of the biggest number, the lower bound is 5. So to add up to 20, the range must be [5-15]:**\n\n\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\"> 1 </span> <\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2+</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2+</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">3+</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">3+</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">3+</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">4+</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">4+</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">4+</span> <\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">5-15</span> \n\nThere's probably some tricky argument for the upper bounds of the other sides, but I'm just going to say the upper bound is one less than the upper bound of the biggest number:\n\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\"> 1 </span> <\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2-14</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">2-14</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">3-14</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">3-14</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">3-14</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">4-14</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">4-14</span> ≤\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">4-14</span> <\n<span style=\"border-style: solid; width: 5em; height: 5em; padding: 3px\">5-15</span> \n\n\nLet's start by coding up `lower_bounds(N)`:",
"_____no_output_____"
]
],
[
[
"def lower_bounds(N):\n \"A list of lower bounds for respective sides of an N-sided die.\"\n lowers = [1]\n for _ in range(N-1):\n m = lowers[-1] # The last number in lowers so far\n lowers.append(m if (lowers.count(m) < m) else m + 1)\n lowers[-1] = lowers[-2] + 1\n return lowers",
"_____no_output_____"
],
[
"lower_bounds(6)",
"_____no_output_____"
],
[
"lower_bounds(10)",
"_____no_output_____"
]
],
[
[
"And `upper_bounds(N)`:",
"_____no_output_____"
]
],
[
[
"def upper_bounds(N):\n \"A list of upper bounds for respective sides of an N-sided die.\"\n U = 2 * N - lower_bounds(N)[-1]\n return [1] + (N - 2) * [U - 1] + [U]",
"_____no_output_____"
],
[
"upper_bounds(6)",
"_____no_output_____"
],
[
"upper_bounds(10)",
"_____no_output_____"
]
],
[
[
"Now, what do we have to do for `all_dice(N)`? When we knew we had six sides, we wrote six nested loops. We can't do that for *N*, so what do we do?\n\n**Here's an iterative approach: we keep track of a list of partially-formed dice, and on each iteration, we add a side to all the partially-formed dice in all possible ways, until the dice all have `N` sides. So for eaxmple, we'd start with:**\n\n dice = [(1,)]\n \n**and then on the next iteration (let's assume *N*=6, so the lower bound is 2 and the upper bound is 7), we'd get this:**\n\n dice = [(1, 2), (1, 3), (1, 4), (1, 5), (1, 6), (1, 7)]\n \n**on the next iteration, we find all the ways of adding a third side, and so on. Like this:**",
"_____no_output_____"
]
],
[
[
"def all_dice(N):\n \"Return a list of all possible N-sided dice for the Sicherman problem.\"\n lowers = lower_bounds(N)\n uppers = upper_bounds(N)\n def possibles(die, i):\n \"The possible numbers for the ith side of an N-sided die.\"\n return ints(max(lowers[i], die[-1] + int(i == N-1)),\n uppers[i])\n dice = [(1,)]\n for i in range(1, N):\n dice = [die + (side,)\n for die in dice\n for side in possibles(die, i)]\n return dice",
"_____no_output_____"
]
],
[
[
"**The tricky part was with the `max`: the actual lower bound at least `lowers[i]`, but it must be as big as the previous side, `die[-1]`. And just to make things complicated, the very last side has to be strictly bigger than the previous; `\" + int(i == N-1)\"` does that by adding 1 just in case we're on the last side, and 0 otherwise.**\n\n**Let's check it out:**",
"_____no_output_____"
]
],
[
[
"len(all_dice(6))",
"_____no_output_____"
]
],
[
[
"Reassuring that we get the same number we got with the old version of `all_dice()`.",
"_____no_output_____"
]
],
[
[
"random.sample(all_dice(6), 8)",
"_____no_output_____"
]
],
[
[
"# Running `sicherman(N)` for small `N`\n\nLet's try `sicherman` for some small values of `N`:",
"_____no_output_____"
]
],
[
[
"{N: sicherman(N)\n for N in ints(2, 6)}",
"_____no_output_____"
]
],
[
[
"Again, reassuring that we get the same result for `sicherman(6)`. And interesting that there is a result for `sicherman(4)` but not for the other *N*.\n\nLet's go onwards from *N*=6, but let's check the timing as we go:",
"_____no_output_____"
]
],
[
[
"%time sicherman(6)",
"CPU times: user 304 ms, sys: 2.73 ms, total: 307 ms\nWall time: 519 ms\n"
],
[
"%time sicherman(7)",
"CPU times: user 18.2 s, sys: 209 ms, total: 18.4 s\nWall time: 21.4 s\n"
]
],
[
[
"# Estimating run time of `sicherman(N)` for larger `N`\n\nOK, it takes 50 or 60 times longer to do 7, compared to 6. At this rate, *N*=8 will take 15 minutes, 9 will take 15 hours, and 10 will take a month.\n\n**Do we know it will continue to rise at the same rate? You're saying the run time is exponential in *N*? **\n\nI think so. The run time is proportional to the number of pairs. The number of pairs is proportional to the square of the number of dice. The number of dice is roughly exponential in *N*, because each time you increase *N* by 1, you have to try a number of new sides that is similar to the number for the previous side (but not quite the same). I should plot the number of pairs on a log scale and see if it looks like a straight line.\n\nI can count the number of pairs without explicitly generating the pairs. If there are *D* dice, then the number of pairs is what? Something like *D* × (*D* + 1) / 2? Or is it *D* × (*D* - 1) / 2?\n\n**Let's draw a picture. With *D* = 4, here are all the ways to pair one die with another to yield 10 distinct pairs:**\n \n 11 .. .. ..\n 21 22 .. ..\n 31 32 33 ..\n 41 42 43 44\n \n**To figure out the formula, add a row to the top:**\n\n .. .. .. ..\n 11 .. .. ..\n 21 22 .. ..\n 31 32 33 ..\n 41 42 43 44\n \n**Now we have a *D* × (*D* + 1) rectangle, and we can see that half (10) of them are pairs, and half (the other 10) are not pairs (because they would be repetitions). So the formula is *D* × (*D* + 1)/2, and checking for *D*=4, (4 × 5) / 2 = 10, so we're good.**\n \nOK, let's try it. First some boilerplate for plotting:",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport matplotlib\nimport numpy as np\nimport matplotlib.pyplot as plt\n\ndef logplot(X, Y, *options):\n \"Plot Y on a log scale vs X.\"\n fig, ax = plt.subplots()\n ax.set_yscale('log')\n ax.plot(X, Y, *options)",
"_____no_output_____"
]
],
[
[
"Now we can plot and display the number of pairs:",
"_____no_output_____"
]
],
[
[
"def plot_pairs(Ns):\n \"Given a list of N values, plot the number of pairs and return a dict of them.\"\n Ds = [len(all_dice(N)) for N in Ns]\n Npairs = [D * (D + 1) // 2 for D in Ds]\n logplot(Ns, Npairs, 'bo-')\n return {Ns[i]: Npairs[i] for i in range(len(Ns))}\n\nplot_pairs(ints(2, 12))",
"_____no_output_____"
]
],
[
[
"OK, we've learned two things. One, it *is* roughly a straight line, so the number of pairs is roughly exponential. Two, there are a *lot* of pairs. 10<sup>14</sup>, just for *N*=12. I don't want to even think about *N*=20.\n\n**So if we want to get much beyond *N*=8, we're either going to need a brand new approach, or we need to make far fewer pairs of dice.**\n\n\n\n# Making Fewer `pairs`\n\nMaybe we could tighten up the upper bounds, but I don't think that will help very much.\nHow about if we concentrate on making fewer pairs, without worrying about making fewer dice?\n\n**How could we do that? Isn't the number of pairs always (*D*<sup>2</sup> + *D*)/2 ?**\n\nRemember, we're looking for *feasible* pairs. So if there was some way of knowing ahead of time that two dice were incompatible as a pair, we wouldn't even need to consider the pair.\n\n**By incompatible, you mean they can't form a pair that is a solution.**\n\nRight. Consider this: in any valid pair, the sum of the biggest number on each die must be 2*N*. For example, with *N* = 6:\n\n ((1, 2, 2, 3, 3, 4), (1, 3, 4, 5, 6, 8)) sum of biggests = 4 + 8 = 12\n ((1, 2, 3, 4, 5, 6), (1, 2, 3, 4, 5, 6)) sum of biggests = 6 + 6 = 12\n \nSo if we have a die with biggest number 7, what dice should we consider pairing it with?\n\n**Only ones with biggest number 5.**\n\n**I get it: we sort all the die into bins labeled with their biggest number. Then we look at each bin, and for the \"7\" bin, we pair them up with the dice in the \"5\" bin. In general, the *B* bin can only pair with the 2*N* - *B* bin.**\n\nExactly. \n\n**Cool. I can see how that can cut the amount of work by a factor of 10 or so. But I was hoping for a factor of a million or so.**\n\nThere are other properties of a feasible pair.\n\n**Like what?**\n\nWell, what about the number of 2s in a pair?\n\n**Let's see. We know that any `regular_sums` has to have two 3s, and the only way to make a 3 is 2+1. And each die has only one 1, so that means that each pair of dice has to have a total of exactly two 2s.**\n\nDoes it have to be one 2 on each die?\n\n**No. It could be one each, or it could be two on one die and none on the other. So a die with *T* twos can only pair with dice that have 2 - *T* twos.**\n\nGreat. Can you think of another property?\n\n**Give me a hint.**\n\nLet's look at the sums of 6-sided Sicherman and regular pairs:",
"_____no_output_____"
]
],
[
[
"sum((1, 2, 2, 3, 3, 4) + (1, 3, 4, 5, 6, 8))",
"_____no_output_____"
],
[
"sum((1, 2, 3, 4, 5, 6) + (1, 2, 3, 4, 5, 6))",
"_____no_output_____"
]
],
[
[
"**They're the same. Is that [the question](http://hitchhikers.wikia.com/wiki/42) that 42 is the answer to? But does a Sicherman pair always have to have the same sum as a regular pair? I guess it doea, because the sum of `sums(pair)` is just all the sides added up *N* times each, so two pairs have the same sum of `sums(pair)` if and only if they have the same sum.**\n\nSo consider the die (1, 3, 3, 3, 4, 5). What do we know about the dice that it can possibly pair with?\n\n**OK, that die has a biggest side of 5, so it can only pair with dice that have a biggest side of 12 - 5 = 7. It has a sum of 19, so it can only pair with dice that have a sum of 42 - 19 = 23. And it has no 2s, so it can only pair with dice that have two 2s.**\n\nI wonder how many such dice there are, out of all 231 `all_dice(6)`?",
"_____no_output_____"
]
],
[
[
"{die for die in all_dice(6) if max(die) == 12 - 5 and sum(die) == 42 - 19 and die.count(2) == 2}",
"_____no_output_____"
]
],
[
[
"**There's only 1. So, (1, 3, 3, 3, 4, 5) only has to try to pair with one die, rather than 230. Nice improvement!**\n\nIn general, I wonder what the sum of the sides of a regular pair is?\n\n**Easy, that's `N * (N + 1)`. [Gauss](http://betterexplained.com/articles/techniques-for-adding-the-numbers-1-to-100/) knew that when he was in elementary school!**\n\n# More efficient `pairs(dice)`\n\n**OK, we can code this up easily enough**:",
"_____no_output_____"
]
],
[
[
"from collections import defaultdict\n\ndef tabulate(dice):\n \"\"\"Put all dice into bins in a hash table, keyed by bin_label(die).\n Each bin holds a list of dice with that key.\"\"\"\n # Example: {(21, 6, 1): [(1, 2, 3, 4, 5, 6), (1, 2, 3, 4, 4, 7), ...]\n table = defaultdict(list)\n for die in dice:\n table[bin_label(die)].append(die)\n return table\n\ndef pairs(dice):\n \"Return all pairs of dice that could possibly be a solution to the Sicherman problem.\"\n table = tabulate(dice)\n N = len(dice[0])\n for bin1 in table:\n bin2 = compatible_bin(bin1, N)\n if bin2 in table and bin1 <= bin2:\n for A in table[bin1]:\n for B in table[bin2]:\n yield (A, B)\n\ndef bin_label(die): return sum(die), max(die), die.count(2)\n\ndef compatible_bin(bin1, N): \n \"Return a bin label that is compatible with bin1.\"\n (S1, M1, T1) = bin1\n return (N * (N + 1) - S1, 2 * N - M1, 2 - T1)",
"_____no_output_____"
]
],
[
[
"**Let's make sure it works:**",
"_____no_output_____"
]
],
[
[
"{N: sicherman(N)\n for N in ints(2, 6)}",
"_____no_output_____"
]
],
[
[
"Good, those are the same answers as before. But how much faster is it?",
"_____no_output_____"
]
],
[
[
"%time sicherman(7)",
"CPU times: user 24.9 ms, sys: 1.23 ms, total: 26.1 ms\nWall time: 153 ms\n"
]
],
[
[
"Wow, that's 1000 times faster than before.",
"_____no_output_____"
],
[
"**I want to take a peek at what some of the bins look like:**",
"_____no_output_____"
]
],
[
[
"tabulate(all_dice(5))",
"_____no_output_____"
]
],
[
[
"**Pretty good: four of the bins have two dice, but the rest have only one die.**\n\nAnd let's see how many pairs we're producing now. We'll tabulate *N* (the number of sides); *D* (the number of *N*-sided dice), the number `pairs(dice)` using the new `pairs`, and the number using the old `pairs`:",
"_____no_output_____"
]
],
[
[
"print(' N: D #pairs(dice) D*(D-1)/2')\nfor N in ints(2, 11):\n dice = list(all_dice(N))\n D = len(dice)\n print('{:2}: {:9,d} {:12,d} {:17,d}'.format(N, D, len(list(pairs(dice))), D*(D-1)//2))",
" N: D #pairs(dice) D*(D-1)/2\n 2: 1 1 0\n 3: 1 1 0\n 4: 10 3 45\n 5: 31 9 465\n 6: 231 71 26,565\n 7: 1,596 670 1,272,810\n 8: 5,916 6,614 17,496,570\n 9: 40,590 76,215 823,753,755\n10: 274,274 920,518 37,612,976,401\n11: 1,837,836 11,506,826 1,688,819,662,530\n"
]
],
[
[
"OK, we're doing 100,000 times better for *N*=11. But it would still take a long time to test 11 million pairs. Let's just get the answers up to *N*=10:",
"_____no_output_____"
]
],
[
[
"%%time\n{N: sicherman(N)\n for N in ints(2, 10)}",
"CPU times: user 26.2 s, sys: 129 ms, total: 26.3 s\nWall time: 26.6 s\n"
]
],
[
[
"Not bad; we solved it for all *N* up to 10 in just a few seconds more than it took for just *N*=7 with the old version. Interestingly, there are solutions for all composite *N* but no prime *N*. So I have some questions:\n\n- Do all composire *N* have solutions?\n- Do no prime *N* have solutions?\n- Do some *N* have more than the 3 solutions that 8 has?\n- Could we handle larger *N* if we worked with [generating functions](https://en.wikipedia.org/wiki/Sicherman_dice)?\n\n\n**Good questions, but I think this is a good place to stop for now.**\n\nAgreed!",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e78bb17b4043ce49982042689d5b2e7855166160 | 23,130 | ipynb | Jupyter Notebook | src/patterns/dosdp_workshop/lch5_subclasses/.ipynb_checkpoints/defined-class-generator-checkpoint.ipynb | cthoyt/drosophila-anatomy-developmental-ontology | 1186e61c3aede5991112fdd4b496bad53a3ec21e | [
"CC-BY-4.0"
] | 13 | 2016-07-27T15:42:58.000Z | 2022-02-12T02:42:52.000Z | src/patterns/dosdp_workshop/lch5_subclasses/.ipynb_checkpoints/defined-class-generator-checkpoint.ipynb | cthoyt/drosophila-anatomy-developmental-ontology | 1186e61c3aede5991112fdd4b496bad53a3ec21e | [
"CC-BY-4.0"
] | 725 | 2015-06-15T14:16:14.000Z | 2022-03-31T11:42:52.000Z | src/patterns/dosdp_workshop/lch5_subclasses/.ipynb_checkpoints/defined-class-generator-checkpoint.ipynb | mmc46/drosophila-anatomy-developmental-ontology | 9494694456ca0e6b8bfb173740d5e5daa6e1bf3a | [
"CC-BY-4.0"
] | 7 | 2016-03-07T13:33:48.000Z | 2022-03-09T11:07:20.000Z | 32.036011 | 85 | 0.348854 | [
[
[
"import pandas as pd\nfillers = pd.read_table('segment_fillers.tsv')",
"_____no_output_____"
],
[
"fillers[:5]",
"_____no_output_____"
],
[
"#change start to match value in protege (without FBbt:000)\n#then advance protege value\nstart = 47273\ndefined_class = list()\n\nfor i in range(fillers.shape[0]):\n x = start + i\n ID = \"FBbt:000\"+str(x)\n defined_class.append(ID)",
"_____no_output_____"
],
[
"defined_class",
"_____no_output_____"
],
[
"fillers.defined_class = defined_class",
"_____no_output_____"
],
[
"fillers",
"_____no_output_____"
],
[
"fillers.to_csv('segment_fillers.tsv',sep='\\t',index=False)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78bccd0bf9ac5699104e84d6a8dc09cacb6f62e | 31,242 | ipynb | Jupyter Notebook | paper-figures/fig3a-apg-mle-data.ipynb | quantshah/qst-cgan | 703e7c486baccee7a830bf067ab6382d0953621e | [
"MIT"
] | 17 | 2020-08-20T19:20:05.000Z | 2022-02-24T06:52:24.000Z | paper-figures/fig3a-apg-mle-data.ipynb | quantshah/qst-cgan | 703e7c486baccee7a830bf067ab6382d0953621e | [
"MIT"
] | 3 | 2021-08-04T13:49:49.000Z | 2021-08-15T08:46:26.000Z | paper-figures/fig3a-apg-mle-data.ipynb | quantshah/qst-cgan | 703e7c486baccee7a830bf067ab6382d0953621e | [
"MIT"
] | 4 | 2021-02-23T06:58:56.000Z | 2022-03-10T09:10:13.000Z | 142.657534 | 14,062 | 0.891684 | [
[
[
"# APG-MLE performance\n\nReconstructing the `cat` state from measurements of the Husimi Q function. The cat state is defined as:\n\n$$|\\psi_{\\text{cat}} \\rangle = \\frac{1}{\\mathcal N} ( |\\alpha \\rangle + |-\\alpha \\rangle \\big ) $$\n\nwith $\\alpha=2$ and normalization $\\mathcal N$.\n\n## Husimi Q function measurements\n\nThe Husimi Q function can be obtained by calculating the expectation value of measuring the following operator:\n\n$$\\mathcal O_i = \\frac{1}{\\pi}|\\beta_i \\rangle \\langle \\beta_i|$$\n\nwhere $|\\beta_i \\rangle $ are coherent states written in the Fock basis.\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nfrom qutip import coherent, coherent_dm, expect, Qobj, fidelity, rand_dm\nfrom qutip.wigner import wigner, qfunc\n\nfrom scipy.io import savemat, loadmat\n\nimport matplotlib\nimport matplotlib.pyplot as plt\nfrom mpl_toolkits.axes_grid1.inset_locator import inset_axes\n\n%load_ext autoreload\ntf.keras.backend.set_floatx('float64') # Set float64 as the default",
"The autoreload extension is already loaded. To reload it, use:\n %reload_ext autoreload\n"
],
[
"hilbert_size = 32\nalpha = 2\n\npsi = coherent(hilbert_size, alpha) + coherent(hilbert_size, -alpha)\npsi = psi.unit() # The .unit() function normalizes the state to have unit trace\nrho = psi*psi.dag()\n\ngrid = 32\nxvec = np.linspace(-3, 3, grid)\nyvec = np.linspace(-3, 3, grid)\n\nq = qfunc(rho, xvec, yvec, g=2)\n\ncmap = \"Blues\"\nim = plt.pcolor(xvec, yvec, q, vmin=0, vmax=np.max(q), cmap=cmap, shading='auto')\nplt.colorbar(im)\nplt.xlabel(r\"Re($\\beta$)\")\nplt.ylabel(r\"Im($\\beta$)\")\nplt.title(\"Husimi Q function\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Construct the measurement operators and simulated data (without any noise)",
"_____no_output_____"
]
],
[
[
"X, Y = np.meshgrid(xvec, yvec)\nbetas = (X + 1j*Y).ravel()\nm_ops = [coherent_dm(hilbert_size, beta) for beta in betas]\ndata = expect(m_ops, rho)",
"_____no_output_____"
]
],
[
[
"# APG-MLE\n\nThe APG-MLE method implementation in MATLAB provided in https://github.com/qMLE/qMLE requires an input density matrix and a set of measurement operators. Here, we will export the same data to a matlab format and use the APG-MLE method for reconstruction of the density matrix of the state.",
"_____no_output_____"
]
],
[
[
"ops_numpy = np.array([op.data.toarray() for op in m_ops]) # convert the QuTiP Qobj to numpy arrays\nops = np.transpose(ops_numpy, [1, 2, 0])\n\nmdic = {\"measurements\": ops}\nsavemat(\"data/measurements.mat\", mdic)",
"_____no_output_____"
],
[
"mdic = {\"rho\": rho.full()}\nsavemat(\"data/rho.mat\", mdic)",
"_____no_output_____"
]
],
[
[
"# Reconstruct using the APG-MLE MATLAB code",
"_____no_output_____"
]
],
[
[
"fidelities = loadmat(\"data/fidelities-apg-mle.mat\")\nfidelities = fidelities['flist1'].ravel()",
"_____no_output_____"
],
[
"iterations = np.arange(len(fidelities))\nplt.plot(iterations, fidelities, color=\"black\", label=\"APG-MLE\")\nplt.legend()\nplt.xlabel(\"Iterations\")\nplt.ylabel(\"Fidelity\")\nplt.ylim(0, 1)\nplt.grid(which='minor', alpha=0.2)\nplt.grid(which='major', alpha=0.2)\nplt.xscale('log')\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
e78bda0117e40e1897bc35ebaeedccdb9ef81479 | 40,093 | ipynb | Jupyter Notebook | notebooks/metrics_tutorial.ipynb | RUrlus/ModelMetricUncertainty | f401a25dd196d6e4edf4901fcfee4b56ebd7c10b | [
"Apache-2.0"
] | null | null | null | notebooks/metrics_tutorial.ipynb | RUrlus/ModelMetricUncertainty | f401a25dd196d6e4edf4901fcfee4b56ebd7c10b | [
"Apache-2.0"
] | 11 | 2021-12-08T10:34:17.000Z | 2022-01-20T13:40:05.000Z | notebooks/metrics_tutorial.ipynb | RUrlus/ModelMetricUncertainty | f401a25dd196d6e4edf4901fcfee4b56ebd7c10b | [
"Apache-2.0"
] | null | null | null | 28.254405 | 266 | 0.404011 | [
[
[
"# MMU Confusion matrix & Metrics walkthrough\n\nThis notebook briefly demonstrates the various capabilities of the package on the computation of confusion matrix/matrices and binary classification metrics.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n\nimport mmu",
"_____no_output_____"
]
],
[
[
"### Data generation\n\nWe generate predictions and true labels where:\n* `scores`: classifier scores\n* `yhat`: estimated labels\n* `y`: true labels",
"_____no_output_____"
]
],
[
[
"scores, yhat, y = mmu.generate_data(n_samples=10000)",
"_____no_output_____"
]
],
[
[
"### Confusion matrix only\n\nWe can compute the confusion matrix for a single run using the estimated labels or based on the probability and a classification threshold.\n\nBased on the esstimated labels `yhat`",
"_____no_output_____"
]
],
[
[
"# based on yhat\nmmu.confusion_matrix(y, yhat)",
"_____no_output_____"
]
],
[
[
"based on classifier score with classification threshold",
"_____no_output_____"
]
],
[
[
"mmu.confusion_matrix(y, scores=scores, threshold=0.5)",
"_____no_output_____"
]
],
[
[
"## Precision-Recall\n\nmmu has a specialised function for the positive precision and recall",
"_____no_output_____"
]
],
[
[
"cm, prec_rec = mmu.precision_recall(y, scores=scores, threshold=0.5, return_df=True)",
"_____no_output_____"
]
],
[
[
"Next to the point precision and recall there is also a function to compute the precision recall curve.\n\n`precision_recall_curve` also available under alias `pr_curve` requires you to pass the discrimination/classification thresholds.\n\n## Auto thresholds\n\nmmu provides an utility function `auto_thresholds` that returns the all the thresholds that result in a different confusion matrix.\n\nFor large test sets this can be a bit much. `auto_thresholds` has an optional parameter `max_steps` that limits the number of thresholds.\nThis is done by weighted sampling where the scaled inverse proximity to the next score is used as a weight. In practice this means that the extremes of the scores are oversampled. `auto_thresholds` always ensures that the lowest and highest score are included.",
"_____no_output_____"
]
],
[
[
"thresholds = mmu.auto_thresholds(scores)",
"_____no_output_____"
]
],
[
[
"## Confusion matrix and metrics\n\nThe ``binary_metrics*`` functions compute ten classification metrics:\n * 0 - neg.precision aka Negative Predictive Value\n * 1 - pos.precision aka Positive Predictive Value\n * 2 - neg.recall aka True Negative Rate & Specificity\n * 3 - pos.recall aka True Positive Rate aka Sensitivity\n * 4 - neg.f1 score\n * 5 - pos.f1 score\n * 6 - False Positive Rate\n * 7 - False Negative Rate\n * 8 - Accuracy\n * 9 - MCC\n\nThese metrics were chosen as they are the most commonly used metrics and most other metrics can be compute from these. We don't provide individual functions at the moment as the overhead of computing all of them vs one or two is negligable.\n\nThis index can be retrieved using:",
"_____no_output_____"
]
],
[
[
"col_index = mmu.metrics.col_index\ncol_index",
"_____no_output_____"
]
],
[
[
"### For a single test set",
"_____no_output_____"
]
],
[
[
"cm, metrics = mmu.binary_metrics(y, yhat)",
"_____no_output_____"
],
[
"# the confusion matrix\ncm",
"_____no_output_____"
],
[
"metrics",
"_____no_output_____"
]
],
[
[
"We can also request dataframes back:",
"_____no_output_____"
]
],
[
[
"cm, metrics = mmu.binary_metrics(y, yhat, return_df=True)",
"_____no_output_____"
],
[
"metrics",
"_____no_output_____"
]
],
[
[
"### A single run using probabilities",
"_____no_output_____"
]
],
[
[
"cm, metrics = mmu.binary_metrics(y, scores=scores, threshold=0.5, return_df=True)",
"_____no_output_____"
],
[
"cm",
"_____no_output_____"
],
[
"metrics",
"_____no_output_____"
]
],
[
[
"### A single run using multiple thresholds\n\nCan be used when you want to compute a precision-recall curve for example",
"_____no_output_____"
]
],
[
[
"thresholds = mmu.auto_thresholds(scores)",
"_____no_output_____"
],
[
"cm, metrics = mmu.binary_metrics_thresholds(\n y=y,\n scores=scores,\n thresholds=thresholds,\n return_df=True\n)",
"_____no_output_____"
]
],
[
[
"The confusion matrix is now an 2D array where the rows contain the confusion matrix for a single threshold",
"_____no_output_____"
]
],
[
[
"cm",
"_____no_output_____"
]
],
[
[
"Similarly, `metrics` is now an 2D array where the rows contain the metrics for a single threshold",
"_____no_output_____"
]
],
[
[
"metrics",
"_____no_output_____"
]
],
[
[
"Generate multiple runs for the below functions",
"_____no_output_____"
]
],
[
[
"scores, yhat, y = mmu.generate_data(n_samples=10000, n_sets=100)",
"_____no_output_____"
]
],
[
[
"### Multiple runs using a single threshold\n\nYou have performed bootstrap or multiple train-test runs and want to evaluate the distribution of the metrics you can use `binary_metrics_runs`.\n\n`cm` and `metrics` are now two dimensional arrays where the rows are the confusion matrices/metrics for that a run",
"_____no_output_____"
]
],
[
[
"cm, metrics = mmu.binary_metrics_runs(\n y=y,\n scores=scores,\n threshold=0.5,\n)",
"_____no_output_____"
],
[
"cm[:5, :]",
"_____no_output_____"
]
],
[
[
"### Multiple runs using multiple thresholds\n\nYou have performed bootstrap or multiple train-test runs and, for example, want to evaluate the different precision recall curves",
"_____no_output_____"
]
],
[
[
"cm, metrics = mmu.binary_metrics_runs_thresholds(\n y=y,\n scores=scores,\n thresholds=thresholds,\n fill=1.0\n)",
"_____no_output_____"
]
],
[
[
"The confusion matrix and metrics are now cubes.\n\nFor the confusion matrix the:\n* row -- the thresholds\n* colomns -- the confusion matrix elements\n* slices -- the runs\n\nFor the metrics:\n\n* row -- thresholds\n* colomns -- the metrics\n* slices -- the runs\n\nThe stride is such that the biggest stride is over the thresholds for the confusion matrix and over the metrics for the metrics.\n\nThe argument being that you will want to model the confusion matrices over the runs\nand the metrics individually over the thresholds and runs",
"_____no_output_____"
]
],
[
[
"print('shape confusion matrix: ', cm.shape)\nprint('strides confusion matrix: ', cm.strides)",
"shape confusion matrix: (10000, 4, 100)\nstrides confusion matrix: (3200, 8, 32)\n"
],
[
"print('shape metrics: ', metrics.shape)\nprint('strides metrics: ', metrics.strides)",
"shape metrics: (10000, 10, 100)\nstrides metrics: (800, 8000000, 8)\n"
],
[
"pos_recalls = metrics[:, mmu.metrics.col_index['pos.rec'], :]\npos_precisions = metrics[:, mmu.metrics.col_index['pos.prec'], :]",
"_____no_output_____"
]
],
[
[
"### Binary metrics over confusion matrices\n\nThis can be used when you have a methodology where you model and generate confusion matrices",
"_____no_output_____"
]
],
[
[
"# We use confusion_matrices to create confusion matrices based on some output\ncm = mmu.confusion_matrices(\n y=y,\n scores=scores,\n threshold=0.5,\n)",
"_____no_output_____"
],
[
"metrics = mmu.binary_metrics_confusion_matrices(cm, 0.0)",
"_____no_output_____"
],
[
"mmu.metrics_to_dataframe(metrics)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
e78be5fc42a2aa657cfc5a538f9b5c78ad1aa915 | 99,555 | ipynb | Jupyter Notebook | stackoverflow-data-science-tools.ipynb | ClaudiuCreanga/kaggle | 49ed31cd12040a7996611d9aa5379acfb61c0192 | [
"MIT"
] | 5 | 2017-11-29T16:37:07.000Z | 2021-01-08T00:45:04.000Z | stackoverflow-data-science-tools.ipynb | ClaudiuCreanga/kaggle | 49ed31cd12040a7996611d9aa5379acfb61c0192 | [
"MIT"
] | null | null | null | stackoverflow-data-science-tools.ipynb | ClaudiuCreanga/kaggle | 49ed31cd12040a7996611d9aa5379acfb61c0192 | [
"MIT"
] | null | null | null | 366.011029 | 70,388 | 0.91687 | [
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport datetime\n\n%matplotlib inline",
"_____no_output_____"
],
[
"jobs_data = pd.read_csv(\n \"data/stackoverflow/data-science-jobs.csv\"\n)",
"_____no_output_____"
],
[
"jobs_data.head()",
"_____no_output_____"
],
[
"from collections import Counter\n\nskills = jobs_data[\"skills\"].dropna()\nskills_list = skills.str.split(\",\").as_matrix().flat\nsingle_skills_list = [item.strip(\" \") for sublist in skills_list for item in sublist]\n\noccurences = Counter(single_skills_list)\nsorted_occurences = occurences.most_common()",
"_____no_output_____"
],
[
"sorted_occurences_first_10 = sorted_occurences[:10]\nsorted_occurences_dict = dict((x, y) for x, y in sorted_occurences_first_10)\nexplode = (0.1, 0, 0, 0, 0, 0, 0, 0, 0, 0)\nfig = plt.figure(figsize=(10,10))\nplt.pie([float(v) for v in sorted_occurences_dict.values()], labels=[k for k in sorted_occurences_dict],\n autopct='%1.1f%%', explode=explode, shadow=True, startangle=140)\nplt.savefig('data/stackoverflow/skills.png')\n",
"_____no_output_____"
],
[
"description = jobs_data[\"description\"].dropna()\ndescription_degree = description.str.lower().str.contains('degree').value_counts()\ndescription_degree",
"_____no_output_____"
],
[
"\nfig = plt.figure(figsize=(10,10))\ndescription_degree.plot(kind=\"pie\", labels=[\"Degree not required\",\"Degree required\"], autopct='%1.1f%%')\nplt.savefig('data/stackoverflow/degree.png')\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78bf8dd17e9b8408163bd067859f85d6ecb7178 | 127,537 | ipynb | Jupyter Notebook | 01-notes/04-tidy.ipynb | chilperic/scipy-2017-tutorial-pandas | 6b52344ad58d6dfa4aaaad6327bb5054fb2a5b93 | [
"MIT"
] | 164 | 2017-06-27T19:20:26.000Z | 2022-01-09T03:31:02.000Z | 01-notes/04-tidy.ipynb | chilperic/scipy-2017-tutorial-pandas | 6b52344ad58d6dfa4aaaad6327bb5054fb2a5b93 | [
"MIT"
] | 2 | 2017-11-28T17:03:33.000Z | 2018-03-13T15:23:04.000Z | 01-notes/04-tidy.ipynb | chilperic/scipy-2017-tutorial-pandas | 6b52344ad58d6dfa4aaaad6327bb5054fb2a5b93 | [
"MIT"
] | 167 | 2017-06-29T21:16:19.000Z | 2021-10-01T22:53:29.000Z | 31.482844 | 93 | 0.320777 | [
[
[
"import pandas as pd",
"_____no_output_____"
]
],
[
[
" \n - https://github.com/tidyverse/tidyr/tree/master/vignettes\n - https://cran.r-project.org/web/packages/tidyr/vignettes/tidy-data.html\n - https://github.com/cmrivers/ebola\n",
"_____no_output_____"
]
],
[
[
"pd.read_csv('../data/preg.csv')",
"_____no_output_____"
],
[
"pd.read_csv('../data/preg2.csv')",
"_____no_output_____"
],
[
"pd.melt(pd.read_csv('../data/preg.csv'), 'name')",
"_____no_output_____"
]
],
[
[
"```\n Each variable forms a column.\n\n Each observation forms a row.\n\n Each type of observational unit forms a table.\n```",
"_____no_output_____"
],
[
"Some common data problems\n\n```\n\n Column headers are values, not variable names.\n\n Multiple variables are stored in one column.\n\n Variables are stored in both rows and columns.\n\n Multiple types of observational units are stored in the same table.\n\n A single observational unit is stored in multiple tables.\n\n```",
"_____no_output_____"
],
[
"## Column contain values, not variables",
"_____no_output_____"
]
],
[
[
"pew = pd.read_csv('../data/pew.csv')",
"_____no_output_____"
],
[
"pew.head()",
"_____no_output_____"
],
[
"pd.melt(pew, id_vars=['religion'])",
"_____no_output_____"
],
[
"pd.melt(pew, id_vars='religion', var_name='income', value_name='count')",
"_____no_output_____"
]
],
[
[
"### Keep multiple columns fixed",
"_____no_output_____"
]
],
[
[
"billboard = pd.read_csv('../data/billboard.csv')",
"_____no_output_____"
],
[
"billboard.head()",
"_____no_output_____"
],
[
"pd.melt(billboard,\n id_vars=['year', 'artist', 'track', 'time', 'date.entered'],\n value_name='rank', var_name='week')",
"_____no_output_____"
]
],
[
[
"## Multiple variables are stored in one column.",
"_____no_output_____"
]
],
[
[
"tb = pd.read_csv('../data/tb.csv')\ntb.head()",
"_____no_output_____"
],
[
"ebola = pd.read_csv('../data/ebola_country_timeseries.csv')",
"_____no_output_____"
],
[
"ebola.head()",
"_____no_output_____"
],
[
"# first let's melt the data down\nebola_long = ebola.melt(id_vars=['Date', 'Day'],\n value_name='count',\n var_name='cd_country')\nebola_long.head()",
"_____no_output_____"
],
[
"var_split = ebola_long['cd_country'].str.split('_')\nvar_split",
"_____no_output_____"
],
[
"type(var_split)",
"_____no_output_____"
],
[
"var_split[0]",
"_____no_output_____"
],
[
"var_split[0][0]",
"_____no_output_____"
],
[
"var_split[0][1]",
"_____no_output_____"
],
[
"# save each part to a separate variable\nstatus_values = var_split.str.get(0)\ncountry_values = var_split.str.get(1)",
"_____no_output_____"
],
[
"status_values.head()",
"_____no_output_____"
],
[
"country_values.head()",
"_____no_output_____"
],
[
"# assign the parts to new dataframe columns\nebola_long['status'] = status_values\nebola_long['country'] = country_values",
"_____no_output_____"
],
[
"ebola_long.head()",
"_____no_output_____"
]
],
[
[
"### above in a single step",
"_____no_output_____"
]
],
[
[
"variable_split = ebola_long['cd_country'].str.split('_', expand=True)\nvariable_split.head()",
"_____no_output_____"
],
[
"variable_split.columns = ['status1', 'country1']",
"_____no_output_____"
],
[
"ebola = pd.concat([ebola_long, variable_split], axis=1)\nebola.head()",
"_____no_output_____"
]
],
[
[
"## Variables in both rows and columns",
"_____no_output_____"
]
],
[
[
"weather = pd.read_csv('../data/weather.csv')",
"_____no_output_____"
],
[
"weather.head()",
"_____no_output_____"
],
[
"weather_melt = pd.melt(weather,\n id_vars=['id', 'year', 'month', 'element'],\n var_name='day',\n value_name='temp')",
"_____no_output_____"
],
[
"weather_melt.head()",
"_____no_output_____"
],
[
"weather_tidy = weather_melt.pivot_table(\n index=['id', 'year', 'month', 'day'],\n columns='element',\n values='temp')\nweather_tidy.head()",
"_____no_output_____"
],
[
"weather_flat = weather_tidy.reset_index()\nweather_flat.head()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78c02651a037e3b470d248452c36a724b9b9b9a | 4,800 | ipynb | Jupyter Notebook | graphs_trees/graph_bfs/bfs_challenge.ipynb | janhak/ica-answers | 82891eed97dc320d7cfc10ad4ac1c5c461956f0b | [
"Apache-2.0"
] | null | null | null | graphs_trees/graph_bfs/bfs_challenge.ipynb | janhak/ica-answers | 82891eed97dc320d7cfc10ad4ac1c5c461956f0b | [
"Apache-2.0"
] | null | null | null | graphs_trees/graph_bfs/bfs_challenge.ipynb | janhak/ica-answers | 82891eed97dc320d7cfc10ad4ac1c5c461956f0b | [
"Apache-2.0"
] | null | null | null | 22.325581 | 185 | 0.490625 | [
[
[
"This notebook was prepared by [Donne Martin](https://github.com/donnemartin). Source and license info is on [GitHub](https://github.com/donnemartin/interactive-coding-challenges).",
"_____no_output_____"
],
[
"# Challenge Notebook",
"_____no_output_____"
],
[
"## Problem: Implement breadth-first search on a graph.\n\n* [Constraints](#Constraints)\n* [Test Cases](#Test-Cases)\n* [Algorithm](#Algorithm)\n* [Code](#Code)\n* [Unit Test](#Unit-Test)\n* [Solution Notebook](#Solution-Notebook)",
"_____no_output_____"
],
[
"## Constraints\n\n* Is the graph directed?\n * Yes\n* Can we assume we already have Graph and Node classes?\n * Yes\n* Can we assume the inputs are valid?\n * Yes\n* Can we assume this fits memory?\n * Yes",
"_____no_output_____"
],
[
"## Test Cases\n\nInput:\n* `add_edge(source, destination, weight)`\n\n```\ngraph.add_edge(0, 1, 5)\ngraph.add_edge(0, 4, 3)\ngraph.add_edge(0, 5, 2)\ngraph.add_edge(1, 3, 5)\ngraph.add_edge(1, 4, 4)\ngraph.add_edge(2, 1, 6)\ngraph.add_edge(3, 2, 7)\ngraph.add_edge(3, 4, 8)\n```\n\nResult:\n* Order of nodes visited: [0, 1, 4, 5, 3, 2]",
"_____no_output_____"
],
[
"## Algorithm\n\nRefer to the [Solution Notebook](). If you are stuck and need a hint, the solution notebook's algorithm discussion might be a good place to start.",
"_____no_output_____"
],
[
"## Code",
"_____no_output_____"
]
],
[
[
"%run ../graph/graph.py",
"_____no_output_____"
],
[
"class GraphBfs(Graph):\n\n def bfs(self, root, visit_func):\n # TODO: Implement me\n pass",
"_____no_output_____"
]
],
[
[
"## Unit Test",
"_____no_output_____"
],
[
"**The following unit test is expected to fail until you solve the challenge.**",
"_____no_output_____"
]
],
[
[
"%run ../utils/results.py",
"_____no_output_____"
],
[
"# %load test_bfs.py\nfrom nose.tools import assert_equal\n\n\nclass TestBfs(object):\n\n def __init__(self):\n self.results = Results()\n\n def test_bfs(self):\n nodes = []\n graph = GraphBfs()\n for id in range(0, 6):\n nodes.append(graph.add_node(id))\n graph.add_edge(0, 1, 5)\n graph.add_edge(0, 4, 3)\n graph.add_edge(0, 5, 2)\n graph.add_edge(1, 3, 5)\n graph.add_edge(1, 4, 4)\n graph.add_edge(2, 1, 6)\n graph.add_edge(3, 2, 7)\n graph.add_edge(3, 4, 8)\n graph.bfs(nodes[0], self.results.add_result)\n assert_equal(str(self.results), \"[0, 1, 4, 5, 3, 2]\")\n\n print('Success: test_bfs')\n\n\ndef main():\n test = TestBfs()\n test.test_bfs()\n\n\nif __name__ == '__main__':\n main()",
"_____no_output_____"
]
],
[
[
"## Solution Notebook\n\nReview the [Solution Notebook]() for a discussion on algorithms and code solutions.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
e78c0f3562ffd12e1ad6d8032436d66660127862 | 9,327 | ipynb | Jupyter Notebook | machine/training/blackjack_training.ipynb | eleniums/blackjack | 2a9876f3584ccc83b9d12989e9e82832494f1de0 | [
"MIT"
] | null | null | null | machine/training/blackjack_training.ipynb | eleniums/blackjack | 2a9876f3584ccc83b9d12989e9e82832494f1de0 | [
"MIT"
] | null | null | null | machine/training/blackjack_training.ipynb | eleniums/blackjack | 2a9876f3584ccc83b9d12989e9e82832494f1de0 | [
"MIT"
] | null | null | null | 38.069388 | 459 | 0.5899 | [
[
[
"%%time \nimport pickle, gzip, urllib.request, json\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport os\nimport boto3\nimport re\nimport copy\nimport time\nimport io\nimport struct\nfrom time import gmtime, strftime\nfrom sagemaker import get_execution_role\n\nrole = get_execution_role()\n\nregion = boto3.Session().region_name\n\nbucket='sagemaker-blackjack' # Replace with your s3 bucket name\nprefix = 'sagemaker/blackjack' # Used as part of the path in the bucket where you store data\nbucket_path = 'https://s3-{}.amazonaws.com/{}'.format(region,bucket) # The URL to access the bucket",
"CPU times: user 68.2 ms, sys: 8.45 ms, total: 76.6 ms\nWall time: 144 ms\n"
],
[
"import sagemaker\n\nfrom sagemaker.amazon.amazon_estimator import get_image_uri\n\ncontainer = get_image_uri(boto3.Session().region_name, 'xgboost', '0.90-1')",
"_____no_output_____"
],
[
"train_data = 's3://{}/{}/{}'.format(bucket, prefix, 'train')\nprint(train_data)\n\ns3_output_location = 's3://{}/{}/{}'.format(bucket, prefix, 'xgboost_model_sdk')\nprint(s3_output_location)",
"s3://sagemaker-blackjack/sagemaker/blackjack/train\ns3://sagemaker-blackjack/sagemaker/blackjack/xgboost_model_sdk\n"
],
[
"xgb_model = sagemaker.estimator.Estimator(container,\n role, \n train_instance_count=1, # number of ML compute instances to use for training\n train_instance_type='ml.m4.xlarge', # type of ML computer instance for training\n train_volume_size = 1, # size of storage to attach to training instance\n output_path=s3_output_location,\n sagemaker_session=sagemaker.Session())",
"_____no_output_____"
],
[
"# https://docs.aws.amazon.com/sagemaker/latest/dg/xgboost_hyperparameters.html\nxgb_model.set_hyperparameters(max_depth = 5, # (Default: 6) Maximum depth of a tree. Increasing this value makes the model more complex and likely to be overfit. 0 indicates no limit.\n eta = .2, # (Default: 0.3) Step size shrinkage used in updates to prevent overfitting. After each boosting step, you can directly get the weights of new features. The eta parameter actually shrinks the feature weights to make the boosting process more conservative.\n gamma = 4, # (Default: 0) Minimum loss reduction required to make a further partition on a leaf node of the tree. The larger, the more conservative the algorithm is.\n min_child_weight = 6, # (Default: 1) Minimum sum of instance weight (hessian) needed in a child. If the tree partition step results in a leaf node with the sum of instance weight less than min_child_weight, the building process gives up further partitioning. In linear regression models, this simply corresponds to a minimum number of instances needed in each node. The larger the algorithm, the more conservative it is.\n silent = 0, # (Default: 0) 0 means print running messages, 1 means silent mode.\n objective = \"multi:softmax\", # (Default: reg:squarederror) Specifies the learning task and the corresponding learning objective. Examples: reg:logistic, multi:softmax, reg:squarederror. For a full list of valid inputs, refer to XGBoost Parameters (https://github.com/dmlc/xgboost/blob/master/doc/parameter.rst).\n num_class = 12, # The number of classes. Required if objective is set to multi:softmax or multi:softprob.\n num_round = 10) # The number of rounds to run the training. Required.",
"_____no_output_____"
],
[
"train_channel = sagemaker.session.s3_input(train_data, content_type='text/csv')\n\ndata_channels = {'train': train_channel} # can also add validation channel here",
"_____no_output_____"
],
[
"# Start model training\nxgb_model.fit(inputs=data_channels, logs=True)",
"2020-03-31 20:14:14 Starting - Starting the training job...\n2020-03-31 20:14:15 Starting - Launching requested ML instances......\n2020-03-31 20:15:15 Starting - Preparing the instances for training...\n2020-03-31 20:16:07 Downloading - Downloading input data...\n2020-03-31 20:16:25 Training - Downloading the training image...\n2020-03-31 20:16:55 Training - Training image download completed. Training in progress.\u001b[34mINFO:sagemaker-containers:Imported framework sagemaker_xgboost_container.training\u001b[0m\n\u001b[34mINFO:sagemaker-containers:Failed to parse hyperparameter objective value multi:softmax to Json.\u001b[0m\n\u001b[34mReturning the value itself\u001b[0m\n\u001b[34mINFO:sagemaker-containers:No GPUs detected (normal if no gpus installed)\u001b[0m\n\u001b[34mINFO:sagemaker_xgboost_container.training:Running XGBoost Sagemaker in algorithm mode\u001b[0m\n\u001b[34mINFO:root:Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34mINFO:root:Determined delimiter of CSV input is ','\u001b[0m\n\u001b[34m[20:16:57] 2501256x2 matrix with 5002512 entries loaded from /opt/ml/input/data/train?format=csv&label_column=0&delimiter=,\u001b[0m\n\u001b[34mINFO:root:Single node training.\u001b[0m\n\u001b[34mINFO:root:Train matrix has 2501256 rows\u001b[0m\n\u001b[34m[0]#011train-merror:0.60371\u001b[0m\n\u001b[34m[1]#011train-merror:0.596739\u001b[0m\n\u001b[34m[2]#011train-merror:0.590699\u001b[0m\n\u001b[34m[3]#011train-merror:0.585564\u001b[0m\n\u001b[34m[4]#011train-merror:0.58367\u001b[0m\n\u001b[34m[5]#011train-merror:0.581655\u001b[0m\n\u001b[34m[6]#011train-merror:0.579526\u001b[0m\n\u001b[34m[7]#011train-merror:0.579152\u001b[0m\n\n2020-03-31 20:18:12 Uploading - Uploading generated training model\u001b[34m[8]#011train-merror:0.577729\u001b[0m\n\u001b[34m[9]#011train-merror:0.574415\u001b[0m\n\n2020-03-31 20:18:19 Completed - Training job completed\nTraining seconds: 132\nBillable seconds: 132\n"
],
[
"# Deploy model for testing\nxgb_predictor = xgb_model.deploy(initial_instance_count=1,\n content_type='text/csv',\n instance_type='ml.t2.medium'\n )",
"-------------!"
],
[
"# Test the deployed model\nresult = xgb_predictor.predict('803,1108')\nprint(result)\n\n# this data point accurately predicts a split!\nresult = xgb_predictor.predict('803,1111')\nprint(result)\n\nresult = xgb_predictor.predict('203,1007')\nprint(result)",
"b'6.0'\nb'11.0'\nb'1.0'\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78c295f67fb7c6ba9383cc57ecdaea201affd70 | 12,600 | ipynb | Jupyter Notebook | src/ScottPlot4/ScottPlot.Sandbox/Notebook/ScottPlotQuickstart.ipynb | p-rakash/ScottPlot | d3b9c13b67d0344cf68e6e1cb7893fc0f1785e7f | [
"MIT"
] | null | null | null | src/ScottPlot4/ScottPlot.Sandbox/Notebook/ScottPlotQuickstart.ipynb | p-rakash/ScottPlot | d3b9c13b67d0344cf68e6e1cb7893fc0f1785e7f | [
"MIT"
] | null | null | null | src/ScottPlot4/ScottPlot.Sandbox/Notebook/ScottPlotQuickstart.ipynb | p-rakash/ScottPlot | d3b9c13b67d0344cf68e6e1cb7893fc0f1785e7f | [
"MIT"
] | null | null | null | 112.5 | 10,359 | 0.875476 | [
[
[
"# ScottPlot Notebook Quickstart\n\n_How to use ScottPlot to plot data in a Jupyter / .NET Interactive notebook_",
"_____no_output_____"
]
],
[
[
"// Install the ScottPlot NuGet package\n#r \"nuget:ScottPlot\"",
"_____no_output_____"
],
[
"// Plot some data\ndouble[] dataX = new double[] { 1, 2, 3, 4, 5 };\ndouble[] dataY = new double[] { 1, 4, 9, 16, 25 };\nvar plt = new ScottPlot.Plot(400, 300);\nplt.AddScatter(dataX, dataY);\n\n// Display the result as a HTML image\ndisplay(HTML(plt.GetImageHTML()));",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
]
] |
e78c2eff7996ca7fb715903587adf3640a59034c | 6,097 | ipynb | Jupyter Notebook | .ipynb_checkpoints/PandasBasics-checkpoint.ipynb | kamlesh-p/myNotebooks | a65224402927432b4e4060f3f9ce8e3aa0407b38 | [
"Apache-2.0"
] | null | null | null | .ipynb_checkpoints/PandasBasics-checkpoint.ipynb | kamlesh-p/myNotebooks | a65224402927432b4e4060f3f9ce8e3aa0407b38 | [
"Apache-2.0"
] | null | null | null | .ipynb_checkpoints/PandasBasics-checkpoint.ipynb | kamlesh-p/myNotebooks | a65224402927432b4e4060f3f9ce8e3aa0407b38 | [
"Apache-2.0"
] | null | null | null | 23.909804 | 165 | 0.394128 | [
[
[
"import numpy as np\nimport pandas as pd",
"_____no_output_____"
],
[
"s = pd.Series([1, 3, 5, np.nan, 6, 8])\n\ns",
"_____no_output_____"
],
[
"dates = pd.date_range('20190101', periods=6)\ndates",
"_____no_output_____"
],
[
"df = pd.DataFrame(np.random.randn(6, 4), index=dates, columns=list('ABCD'))\ndf\n",
"_____no_output_____"
],
[
"df2 = pd.DataFrame({'A': 1., 'B': pd.Timestamp('20130102'), 'C': pd.Series(1, index=list(range(4)), dtype='float32'), 'D': np.array([3] * 4, dtype='int32'),\n'E': pd.Categorical([\"test\", \"train\", \"test\", \"train\"]),'F': 'foo'})",
"_____no_output_____"
],
[
"df2",
"_____no_output_____"
],
[
"df2.dtypes",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78c2f03c25d0d39d4e85d401517356208f81c7e | 10,373 | ipynb | Jupyter Notebook | community-content/pytorch_image_classification_single_gpu_with_vertex_sdk_and_torchserve/vertex_training_with_custom_container.ipynb | nayaknishant/vertex-ai-samples | 3ce120b953f1cdc2ec2c5a3f4509cfeab106b7d0 | [
"Apache-2.0"
] | 213 | 2021-06-10T20:05:20.000Z | 2022-03-31T16:09:29.000Z | community-content/pytorch_image_classification_single_gpu_with_vertex_sdk_and_torchserve/vertex_training_with_custom_container.ipynb | nayaknishant/vertex-ai-samples | 3ce120b953f1cdc2ec2c5a3f4509cfeab106b7d0 | [
"Apache-2.0"
] | 343 | 2021-07-25T22:55:25.000Z | 2022-03-31T23:58:47.000Z | community-content/pytorch_image_classification_single_gpu_with_vertex_sdk_and_torchserve/vertex_training_with_custom_container.ipynb | nayaknishant/vertex-ai-samples | 3ce120b953f1cdc2ec2c5a3f4509cfeab106b7d0 | [
"Apache-2.0"
] | 143 | 2021-07-21T17:27:47.000Z | 2022-03-29T01:20:43.000Z | 24.011574 | 231 | 0.472669 | [
[
[
"# Copyright 2021 Google LLC\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# PyTorch Image Classification Single GPU using Vertex Training with Custom Container",
"_____no_output_____"
],
[
"<table align=\"left\">\n <td>\n <a href=\"https://github.com/GoogleCloudPlatform/vertex-ai-samples/blob/master/community-content/pytorch_image_classification_single_gpu_with_vertex_sdk_and_torchserve/vertex_training_with_custom_container.ipynb\">\n <img src=\"https://cloud.google.com/ml-engine/images/github-logo-32px.png\" alt=\"GitHub logo\">\n View on GitHub\n </a>\n </td>\n</table>",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
]
],
[
[
"PROJECT_ID = \"YOUR PROJECT ID\"\nBUCKET_NAME = \"gs://YOUR BUCKET NAME\"\nREGION = \"YOUR REGION\"\nSERVICE_ACCOUNT = \"YOUR SERVICE ACCOUNT\"",
"_____no_output_____"
],
[
"content_name = \"pt-img-cls-gpu-cust-cont-torchserve\"",
"_____no_output_____"
]
],
[
[
"## Local Training",
"_____no_output_____"
]
],
[
[
"! ls trainer",
"_____no_output_____"
],
[
"! cat trainer/requirements.txt",
"_____no_output_____"
],
[
"! pip install -r trainer/requirements.txt",
"_____no_output_____"
],
[
"! cat trainer/task.py",
"_____no_output_____"
],
[
"%run trainer/task.py --epochs 5 --local-mode",
"_____no_output_____"
],
[
"! ls ./tmp",
"_____no_output_____"
],
[
"! rm -rf ./tmp",
"_____no_output_____"
]
],
[
[
"## Vertex Training using Vertex SDK and Custom Container",
"_____no_output_____"
],
[
"### Build Custom Container",
"_____no_output_____"
]
],
[
[
"hostname = \"gcr.io\"\nimage_name_train = content_name\ntag = \"latest\"\n\ncustom_container_image_uri_train = f\"{hostname}/{PROJECT_ID}/{image_name_train}:{tag}\"",
"_____no_output_____"
],
[
"! cd trainer && docker build -t $custom_container_image_uri_train -f Dockerfile .",
"_____no_output_____"
],
[
"! docker run --rm $custom_container_image_uri_train --epochs 5 --local-mode",
"_____no_output_____"
],
[
"! docker push $custom_container_image_uri_train",
"_____no_output_____"
],
[
"! gcloud container images list --repository $hostname/$PROJECT_ID",
"_____no_output_____"
]
],
[
[
"### Initialize Vertex SDK",
"_____no_output_____"
]
],
[
[
"! pip install -r requirements.txt",
"_____no_output_____"
],
[
"from google.cloud import aiplatform\n\naiplatform.init(\n project=PROJECT_ID,\n staging_bucket=BUCKET_NAME,\n location=REGION,\n)",
"_____no_output_____"
]
],
[
[
"### Create a Vertex Tensorboard Instance",
"_____no_output_____"
]
],
[
[
"tensorboard = aiplatform.Tensorboard.create(\n display_name=content_name,\n)",
"_____no_output_____"
]
],
[
[
"#### Option: Use a Previously Created Vertex Tensorboard Instance\n\n```\ntensorboard_name = \"Your Tensorboard Resource Name or Tensorboard ID\"\ntensorboard = aiplatform.Tensorboard(tensorboard_name=tensorboard_name)\n```",
"_____no_output_____"
],
[
"### Run a Vertex SDK CustomContainerTrainingJob",
"_____no_output_____"
]
],
[
[
"display_name = content_name\ngcs_output_uri_prefix = f\"{BUCKET_NAME}/{display_name}\"\n\nmachine_type = \"n1-standard-4\"\naccelerator_count = 1\naccelerator_type = \"NVIDIA_TESLA_K80\"\n\ncontainer_args = [\n \"--batch-size\",\n \"256\",\n \"--epochs\",\n \"100\",\n]",
"_____no_output_____"
],
[
"custom_container_training_job = aiplatform.CustomContainerTrainingJob(\n display_name=display_name,\n container_uri=custom_container_image_uri_train,\n)",
"_____no_output_____"
],
[
"custom_container_training_job.run(\n args=container_args,\n base_output_dir=gcs_output_uri_prefix,\n machine_type=machine_type,\n accelerator_type=accelerator_type,\n accelerator_count=accelerator_count,\n tensorboard=tensorboard.resource_name,\n service_account=SERVICE_ACCOUNT,\n)",
"_____no_output_____"
],
[
"print(f\"Custom Training Job Name: {custom_container_training_job.resource_name}\")\nprint(f\"GCS Output URI Prefix: {gcs_output_uri_prefix}\")",
"_____no_output_____"
]
],
[
[
"### Training Artifact",
"_____no_output_____"
]
],
[
[
"! gsutil ls $gcs_output_uri_prefix",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
e78c374cbb53e42f9f779e5006623bea89e37a4d | 4,031 | ipynb | Jupyter Notebook | notebooks/uploader/postimages.ipynb | TheYoke/PngBin | d00291980cbcb7639599906fa2edc3e1dc5516da | [
"MIT"
] | 16 | 2022-01-21T08:36:28.000Z | 2022-03-06T14:06:59.000Z | notebooks/uploader/postimages.ipynb | TheYoke/PngBin | d00291980cbcb7639599906fa2edc3e1dc5516da | [
"MIT"
] | null | null | null | notebooks/uploader/postimages.ipynb | TheYoke/PngBin | d00291980cbcb7639599906fa2edc3e1dc5516da | [
"MIT"
] | 4 | 2022-01-24T07:36:35.000Z | 2022-03-05T09:11:05.000Z | 22.027322 | 125 | 0.521211 | [
[
[
"# PostImages Uploader\n\nThis notebook provide an easy way to upload your images to [postimages.org](https://postimages.org).",
"_____no_output_____"
],
[
"#### How to use:\n- Modify the **configurations** if needed.\n- At menu bar, select Run > Run All Cells.\n- Scroll to the end of this notebook for outputs.",
"_____no_output_____"
],
[
"## Configurations",
"_____no_output_____"
],
[
"---\nPath to a directory which contains your images you want to upload.\n> Only files in top level directory will be uploaded. \n> Limit: 24 MiB per file or image dimension of 2508 squared pixels.",
"_____no_output_____"
]
],
[
[
"INPUT_DIR = '../outputs/images'",
"_____no_output_____"
]
],
[
[
"---\nPath to a file which will contain a list of uploaded image urls used for updating metadata database file.\n> Append if exists.",
"_____no_output_____"
]
],
[
[
"URLS_PATH = '../outputs/urls.txt'",
"_____no_output_____"
]
],
[
[
"---",
"_____no_output_____"
],
[
"### Import",
"_____no_output_____"
]
],
[
[
"import os\nimport sys\n\nfrom modules.PostImages import PostImages",
"_____no_output_____"
]
],
[
[
"### Basic Configuration Validation",
"_____no_output_____"
]
],
[
[
"assert os.path.isdir(INPUT_DIR), 'INPUT_DIR must exist and be a directory.'\nassert any(x.is_file() for x in os.scandir(INPUT_DIR)), 'INPUT_DIR top level directory must have at least one file.'\nassert not os.path.exists(URLS_PATH) or os.path.isfile(URLS_PATH), 'URLS_PATH must be a file if it exists.'",
"_____no_output_____"
]
],
[
[
"### Upload",
"_____no_output_____"
]
],
[
[
"uploader = PostImages()\n\nextras = []\ntry:\n with open(URLS_PATH, 'a') as f_urls:\n for entry in os.scandir(INPUT_DIR):\n if entry.is_file():\n image_url, extra = uploader.upload_file(entry.path)\n extras.append(extra)\n\n print(entry.name, image_url, '', sep='\\n', file=f_urls)\n print(entry.name, image_url, extra)\n\n print('DONE!')\nexcept KeyboardInterrupt:\n print('\\nStopped!')",
"_____no_output_____"
]
],
[
[
"---\n---\n### Delete Uploaded Images [EXTRA]\n> Change below cell to code mode to run the cell.",
"_____no_output_____"
]
],
[
[
"for extra in extras:\n result = uploader.delete(extra['removal_link'])\n print(result)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"raw"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"raw"
]
] |
e78c4b34ea50237b78d0ea1893965be6491a5b8e | 2,122 | ipynb | Jupyter Notebook | Chapter_5/.ipynb_checkpoints/Section_5.3.2.5-checkpoint.ipynb | godfanmiao/ML-Kaggle-Github-2022 | 19c9fd0fe5db432f43f5844e170f952eaaaeaefd | [
"BSD-3-Clause"
] | 8 | 2021-10-15T12:27:01.000Z | 2022-02-21T13:50:04.000Z | Chapter_5/.ipynb_checkpoints/Section_5.3.2.5-checkpoint.ipynb | godfanmiao/ML-Kaggle-Github-2022 | 19c9fd0fe5db432f43f5844e170f952eaaaeaefd | [
"BSD-3-Clause"
] | null | null | null | Chapter_5/.ipynb_checkpoints/Section_5.3.2.5-checkpoint.ipynb | godfanmiao/ML-Kaggle-Github-2022 | 19c9fd0fe5db432f43f5844e170f952eaaaeaefd | [
"BSD-3-Clause"
] | 1 | 2022-02-04T07:25:34.000Z | 2022-02-04T07:25:34.000Z | 21.22 | 101 | 0.543355 | [
[
[
"import pandas as pd\n\n\n#使用pandas,读取数据文件。\ndata = pd.read_csv('../datasets/white_wine/white_wine_quality.csv', sep=';')\n\n#从数据中拆分出特征与目标值。\nX = data[data.columns[:-1]]\ny = data[data.columns[-1]]",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\n\n\n#拆分训练集和测试集。\nX_train, X_test, y_train, y_test = train_test_split(X, y, train_size=0.8, random_state=2022)",
"_____no_output_____"
],
[
"from sklearn.tree import DecisionTreeRegressor\n\n\n#初始化决策树回归器模型。\ndtr = DecisionTreeRegressor()\n\n#使用训练数据,训练决策树回归器模型。\ndtr.fit(X_train, y_train)\n\n#使用训练好的回归模型,依据测试数据的特征,进行数值回归。\ny_predict = dtr.predict(X_test)",
"_____no_output_____"
],
[
"from sklearn.metrics import mean_squared_error\n\n\n#评估回归器的误差。\nprint ('Scikit-learn的决策树回归器在white_wine测试集上的均方误差为:%.4f。' %mean_squared_error(y_test, y_predict))",
"Scikit-learn的决策树回归器在white_wine_quality测试集上的均方误差为:0.6286。\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
e78c4c681e9b94dd56a76fa8b5d5f1d23e367566 | 14,899 | ipynb | Jupyter Notebook | Books and Courses/PyTorch/1 - tensor creation, reshaping, squeezing, flattening.ipynb | FairlyTales/Machine_Learning_Courses | 8fdbe477c05670ec1bb4832df193a1c9f1c76a7e | [
"MIT"
] | null | null | null | Books and Courses/PyTorch/1 - tensor creation, reshaping, squeezing, flattening.ipynb | FairlyTales/Machine_Learning_Courses | 8fdbe477c05670ec1bb4832df193a1c9f1c76a7e | [
"MIT"
] | null | null | null | Books and Courses/PyTorch/1 - tensor creation, reshaping, squeezing, flattening.ipynb | FairlyTales/Machine_Learning_Courses | 8fdbe477c05670ec1bb4832df193a1c9f1c76a7e | [
"MIT"
] | null | null | null | 26.323322 | 344 | 0.477146 | [
[
[
"import torch\nimport torchvision\nfrom torchvision import transforms, datasets\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"## 3 аттрибута тензора в pytorch:\n\ndtype - тип данных хранимых в тензоре (float32, etc.)\n\ndevice - на чём обрабатывается данный тензор (тензоры которые взаимодействую должны быть на одном проце/видяхе)\n\nlayout - как данные расположены в памяти (не трогай это, дефолтная настройка норм)\n\n# Создание тензоров (прямо как в NumPy):",
"_____no_output_____"
]
],
[
[
"# единичный тензор\nprint(torch.eye(3))",
"tensor([[1., 0., 0.],\n [0., 1., 0.],\n [0., 0., 1.]])\n"
],
[
"# нулевой\ntorch.zeros(2,2)",
"_____no_output_____"
],
[
"# единичный\ntorch.ones(2,2)",
"_____no_output_____"
]
],
[
[
"# Создание тензоров из данных:\n\nFactory function has more tweaking parameters than class constructor, so we will use them more often. They also infere the dtype (they get data in i.e. int32, they save it as int32).\n\nBeware that torch.Tensor and torch.tensor CREATE a copy of input data in memory, when torch.as_tensor and torch.from_numpy just mirror the existing data (so the last is more memory efficient, but if the original data is changed, tensor changed too. Data MUST be in NumPy array).\n\nFor casual use - torch.tensor()\n\nFor speed boost - torch.as_tensor()",
"_____no_output_____"
]
],
[
[
"data = np.array([1,2,3])\n\nt1 = torch.Tensor(data) # class constructor\nt2 = torch.tensor(data) # factory function\nt3 = torch.as_tensor(data) # factory function\nt4 = torch.from_numpy(data) # factory function",
"_____no_output_____"
]
],
[
[
"# Reshaping the the tensors, squeezing\n\n.reshape() or .view()(одно и то же, просто разные названия) - прямое изменение размеров тензора\n\n.squeeze() - удаление всех осей с длинной 1.\n\n.unsqueeze() - добавляет ось с длиной 1, это позволяет изменять ранг тензора.\n\n",
"_____no_output_____"
]
],
[
[
"t = torch.tensor([\n [1,1,1,1],\n [2,2,2,2],\n [3,3,3,3]\n], dtype=torch.float32)\nt.shape",
"_____no_output_____"
],
[
"# number of elements check\nprint(torch.tensor(t.shape).prod())\nprint(t.numel())",
"tensor(12)\n12\n"
],
[
"print(t.reshape(2, 1, 2, 3)) # reshape to rank-4 tensor (from rank-2)\nprint(t.reshape(2, 6))",
"tensor([[[[1., 1., 1.],\n [1., 2., 2.]]],\n\n\n [[[2., 2., 3.],\n [3., 3., 3.]]]])\ntensor([[1., 1., 1., 1., 2., 2.],\n [2., 2., 3., 3., 3., 3.]])\n"
],
[
"print(t.reshape(1,12)) # tensor rank 2\nprint(t.reshape(1,12).squeeze()) # tensor rank 1\n\n# маленький трюк - если не знаем сколько компонентов будет в тензоре который мы хотим\n# \"вытянуть\" сохранив второй ранг мы пишем \"1, -1\" во втором атрибуте .reshape\nprint(t.reshape(1,-1))\n\n# если хотим перевести его в тензор первого ранга пишем \"-1\"\nprint(t.reshape(-1))",
"tensor([[1., 1., 1., 1., 2., 2., 2., 2., 3., 3., 3., 3.]])\ntensor([1., 1., 1., 1., 2., 2., 2., 2., 3., 3., 3., 3.])\ntensor([[1., 1., 1., 1., 2., 2., 2., 2., 3., 3., 3., 3.]])\ntensor([1., 1., 1., 1., 2., 2., 2., 2., 3., 3., 3., 3.])\n"
]
],
[
[
"# Flattening, Tensor size for neural network\n\n.flatten() - позволяет зарешейпить тензор любого ранга в тензор первого ранга\n\nОднако для нейросети абсолютно \"плоские\" данные нам не нужны, т.к. она должна различать batch (какие данные к какой введённой переменой относятся).\n\nНейронка работает с тензорами ранга 4, вот что каждая из осей представляет(на примере тензора ниже):\n\nBatch - contains 3 images\n\nImage - contains 1 color channel (cause grayscale)\n\nColor channel - contains 4 arrays(rows/hight)\n\n4 arrays(rows/hight) - contains 4 pixel values (columns/width)",
"_____no_output_____"
]
],
[
[
"# create 3 \"images\" 4x4 pixels, grayscale\n\nt1 = torch.tensor([\n [1,1,1,1],\n [1,1,1,1],\n [1,1,1,1],\n [1,1,1,1]\n])\nt2 = torch.tensor([\n [2,2,2,2],\n [2,2,2,2],\n [2,2,2,2],\n [2,2,2,2]\n])\nt3 = torch.tensor([\n [3,3,3,3],\n [3,3,3,3],\n [3,3,3,3],\n [3,3,3,3]\n])\n\n# stack them to make a tensor with rank 3 (new axis represents batch), now we have a 3\n# \"grayscale\" pictures in the resulting tensor\nt = torch.stack((t1, t2, t3))\nt.shape",
"_____no_output_____"
],
[
"# and a new axis to this tensor to create a \"batch\" axis for discening the \"pictures\"\nt = t.reshape(3, 1, 4, 4)\nt",
"_____no_output_____"
]
],
[
[
"## how to use flatten\n\nWe can't flatten the whole tensor, cause it will become a simple vector and we won't be able to know where are the \"start\" and the \"end\" of each \"picture\". So we flatten this tensor in such a way that wa preserve the \"batch\" axis (flattening the color channel with hight and width axes), which tells us which \"picture\" is which.",
"_____no_output_____"
]
],
[
[
"# bad idea - got vector with 48 pixel, no way to know from which picture each pixel comes from\na = t.flatten()\nprint('bad - ', a.shape)\n\n# good idea - got 3 images with 16 pixels\nt = t.flatten(start_dim=1) # start flattening from axis=1 (it`s an index)\nprint('good - ', t.shape)",
"bad - torch.Size([48])\ngood - torch.Size([3, 16])\n"
]
],
[
[
"# Broadcasting for element-wise operations\n\nBroadcasting is transforming a lower rank tensor to match the shape of the tensor with which we want it to perform an element-wise operation.\n\nAll element-wise operations works with tensors due to the broadcasting.",
"_____no_output_____"
]
],
[
[
"print(t.eq(1)) # equal 1\nprint(t.abs()) # взятие модуля\nprint(t + 3)\nprint(t.neg())",
"tensor([[ True, True, True, True, True, True, True, True, True, True,\n True, True, True, True, True, True],\n [False, False, False, False, False, False, False, False, False, False,\n False, False, False, False, False, False],\n [False, False, False, False, False, False, False, False, False, False,\n False, False, False, False, False, False]])\ntensor([[1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1],\n [2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2],\n [3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3]])\ntensor([[4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4],\n [5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5],\n [6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6]])\ntensor([[-1, -1, -1, -1, -1, -1, -1, -1, -1, -1, -1, -1, -1, -1, -1, -1],\n [-2, -2, -2, -2, -2, -2, -2, -2, -2, -2, -2, -2, -2, -2, -2, -2],\n [-3, -3, -3, -3, -3, -3, -3, -3, -3, -3, -3, -3, -3, -3, -3, -3]])\n"
]
],
[
[
"# Reduction operation - .ArgMax()\n\nReduction operation on tensor is an operation that reduced the number of elements contained within the tensor.\n\nWe can use .mean(),\n\n.argmax() returns the index of the max value inside the tensor",
"_____no_output_____"
]
],
[
[
"t4 = torch.tensor([\n [0,1,0],\n [2,0,2],\n [0,3,0]\n], dtype=torch.float32)\nprint(t4.shape)\n\nprint(t4.sum()) # sum all elements = reduce this tensor to one axis\nprint(t4.sum(dim=0))\nprint(t4.sum(dim=0).shape) # sum along axis 1 (sum columns) = reduce this tensor to one axis",
"torch.Size([3, 3])\ntensor(8.)\ntensor([2., 4., 2.])\ntorch.Size([3])\n"
],
[
"print('value = ', t4.max()) # MAX value \nprint('flatten = ', t4.flatten()) # flatten tensor (count indices from '0')\nprint('index = ', t4.argmax()) # index of the MAX value (yep, '3' has index=7)\n",
"value = tensor(3.)\nflatten = tensor([0., 1., 0., 2., 0., 2., 0., 3., 0.])\nindex = tensor(7)\n"
],
[
"print(t4.max(dim=0), '\\n') # max values on axis=0\nprint(t4.max(dim=1)) # first tensor - max values, second tensor - their indices",
"torch.return_types.max(\nvalues=tensor([2., 3., 2.]),\nindices=tensor([1, 2, 1])) \n\ntorch.return_types.max(\nvalues=tensor([1., 2., 3.]),\nindices=tensor([1, 2, 1]))\n"
],
[
"# if we wanna get the results not as tensors, but as ints/floats/np.arrays:\n\nprint(t4.mean()) # got tensor\nprint(t4.mean().item(), '\\n') # got float32\n\nprint(t4.mean(dim=0)) # got tensor\nprint(t4.mean(dim=0).tolist(), '\\n') # got np.array of float32",
"tensor(0.8889)\n0.8888888955116272 \n\ntensor([0.6667, 1.3333, 0.6667])\n[0.6666666865348816, 1.3333333730697632, 0.6666666865348816] \n\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
e78c54b62e12cdf5f6fb6558cf0154dce1621d97 | 25,604 | ipynb | Jupyter Notebook | 05-machine-learning-nao-tabular/00-tabular-auto-correlacionado/rnns.ipynb | abefukasawa/datascience_course | 919c715598fa9529703ba340039ad82b23221a0e | [
"MIT"
] | 331 | 2019-01-26T21:11:45.000Z | 2022-03-02T11:35:16.000Z | 05-machine-learning-nao-tabular/00-tabular-auto-correlacionado/rnns.ipynb | abefukasawa/datascience_course | 919c715598fa9529703ba340039ad82b23221a0e | [
"MIT"
] | 2 | 2019-11-02T22:32:13.000Z | 2020-04-13T10:31:11.000Z | 05-machine-learning-nao-tabular/00-tabular-auto-correlacionado/rnns.ipynb | sn3fru/datascience_course | ee0a505134383034e09020d9b1de18904d9b2665 | [
"MIT"
] | 88 | 2019-01-25T16:53:47.000Z | 2022-03-03T00:05:08.000Z | 41.097913 | 1,020 | 0.662592 | [
[
[
"# Aim and motivation\nThe primary reason I have chosen to create this kernel is to practice and use RNNs for various tasks and applications. First of which is time series data. RNNs have truly changed the way sequential data is forecasted. My goal here is to create the ultimate reference for RNNs here on kaggle.",
"_____no_output_____"
],
[
"## Things to remember\n* Please upvote(like button) and share this kernel if you like it. This would increase its visibility and more people will be able to learn about the awesomeness of RNNs.\n* I will use keras for this kernel. If you are not familiar with keras or neural networks, refer to this kernel/tutorial of mine: https://www.kaggle.com/thebrownviking20/intro-to-keras-with-breast-cancer-data-ann\n* Your doubts and curiousity about time series can be taken care of here: https://www.kaggle.com/thebrownviking20/everything-you-can-do-with-a-time-series\n* Don't let the explanations intimidate you. It's simpler than you think.\n* Eventually, I will add more applications of LSTMs. So stay tuned for more!\n* The code is inspired from Kirill Eremenko's Deep Learning Course: https://www.udemy.com/deeplearning/",
"_____no_output_____"
],
[
"## Recurrent Neural Networks\nIn a recurrent neural network we store the output activations from one or more of the layers of the network. Often these are hidden later activations. Then, the next time we feed an input example to the network, we include the previously-stored outputs as additional inputs. You can think of the additional inputs as being concatenated to the end of the “normal” inputs to the previous layer. For example, if a hidden layer had 10 regular input nodes and 128 hidden nodes in the layer, then it would actually have 138 total inputs (assuming you are feeding the layer’s outputs into itself à la Elman) rather than into another layer). Of course, the very first time you try to compute the output of the network you’ll need to fill in those extra 128 inputs with 0s or something.\n\nSource: [Quora](https://www.quora.com/What-is-a-simple-explanation-of-a-recurrent-neural-network)\n<img src=\"https://cdn-images-1.medium.com/max/1600/1*NKhwsOYNUT5xU7Pyf6Znhg.png\">\n\nSource: [Medium](https://medium.com/ai-journal/lstm-gru-recurrent-neural-networks-81fe2bcdf1f9)\n\nLet me give you the best explanation of Recurrent Neural Networks that I found on internet: https://www.youtube.com/watch?v=UNmqTiOnRfg&t=3s",
"_____no_output_____"
],
[
"Now, even though RNNs are quite powerful, they suffer from **Vanishing gradient problem ** which hinders them from using long term information, like they are good for storing memory 3-4 instances of past iterations but larger number of instances don't provide good results so we don't just use regular RNNs. Instead, we use a better variation of RNNs: **Long Short Term Networks(LSTM).**\n\n### What is Vanishing Gradient problem?\nVanishing gradient problem is a difficulty found in training artificial neural networks with gradient-based learning methods and backpropagation. In such methods, each of the neural network's weights receives an update proportional to the partial derivative of the error function with respect to the current weight in each iteration of training. The problem is that in some cases, the gradient will be vanishingly small, effectively preventing the weight from changing its value. In the worst case, this may completely stop the neural network from further training. As one example of the problem cause, traditional activation functions such as the hyperbolic tangent function have gradients in the range (0, 1), and backpropagation computes gradients by the chain rule. This has the effect of multiplying n of these small numbers to compute gradients of the \"front\" layers in an n-layer network, meaning that the gradient (error signal) decreases exponentially with n while the front layers train very slowly.\n\nSource: [Wikipedia](https://en.wikipedia.org/wiki/Vanishing_gradient_problem)\n\n<img src=\"https://cdn-images-1.medium.com/max/1460/1*FWy4STsp8k0M5Yd8LifG_Q.png\">\n\nSource: [Medium](https://medium.com/@anishsingh20/the-vanishing-gradient-problem-48ae7f501257)",
"_____no_output_____"
],
[
"## Long Short Term Memory(LSTM)\nLong short-term memory (LSTM) units (or blocks) are a building unit for layers of a recurrent neural network (RNN). A RNN composed of LSTM units is often called an LSTM network. A common LSTM unit is composed of a cell, an input gate, an output gate and a forget gate. The cell is responsible for \"remembering\" values over arbitrary time intervals; hence the word \"memory\" in LSTM. Each of the three gates can be thought of as a \"conventional\" artificial neuron, as in a multi-layer (or feedforward) neural network: that is, they compute an activation (using an activation function) of a weighted sum. Intuitively, they can be thought as regulators of the flow of values that goes through the connections of the LSTM; hence the denotation \"gate\". There are connections between these gates and the cell.\n\nThe expression long short-term refers to the fact that LSTM is a model for the short-term memory which can last for a long period of time. An LSTM is well-suited to classify, process and predict time series given time lags of unknown size and duration between important events. LSTMs were developed to deal with the exploding and vanishing gradient problem when training traditional RNNs.\n\nSource: [Wikipedia](https://en.wikipedia.org/wiki/Long_short-term_memory)\n\n<img src=\"https://cdn-images-1.medium.com/max/1600/0*LyfY3Mow9eCYlj7o.\">\n\nSource: [Medium](https://codeburst.io/generating-text-using-an-lstm-network-no-libraries-2dff88a3968)\n\nThe best LSTM explanation on internet: https://medium.com/deep-math-machine-learning-ai/chapter-10-1-deepnlp-lstm-long-short-term-memory-networks-with-math-21477f8e4235\n\nRefer above link for deeper insights.",
"_____no_output_____"
],
[
"## Components of LSTMs\nSo the LSTM cell contains the following components\n* Forget Gate “f” ( a neural network with sigmoid)\n* Candidate layer “C\"(a NN with Tanh)\n* Input Gate “I” ( a NN with sigmoid )\n* Output Gate “O”( a NN with sigmoid)\n* Hidden state “H” ( a vector )\n* Memory state “C” ( a vector)\n\n* Inputs to the LSTM cell at any step are X<sub>t</sub> (current input) , H<sub>t-1</sub> (previous hidden state ) and C<sub>t-1</sub> (previous memory state). \n* Outputs from the LSTM cell are H<sub>t</sub> (current hidden state ) and C<sub>t</sub> (current memory state)",
"_____no_output_____"
],
[
"## Working of gates in LSTMs\nFirst, LSTM cell takes the previous memory state C<sub>t-1</sub> and does element wise multiplication with forget gate (f) to decide if present memory state C<sub>t</sub>. If forget gate value is 0 then previous memory state is completely forgotten else f forget gate value is 1 then previous memory state is completely passed to the cell ( Remember f gate gives values between 0 and 1 ).\n\n**C<sub>t</sub> = C<sub>t-1</sub> * f<sub>t</sub>**\n\nCalculating the new memory state: \n\n**C<sub>t</sub> = C<sub>t</sub> + (I<sub>t</sub> * C\\`<sub>t</sub>)**\n\nNow, we calculate the output:\n\n**H<sub>t</sub> = tanh(C<sub>t</sub>)**",
"_____no_output_____"
],
[
"### And now we get to the code...\nI will use LSTMs for predicting the price of stocks of IBM for the year 2017",
"_____no_output_____"
]
],
[
[
"# Importing the libraries\nimport numpy as np\nimport matplotlib.pyplot as plt\nplt.style.use('fivethirtyeight')\nimport pandas as pd\nfrom sklearn.preprocessing import MinMaxScaler\nfrom keras.models import Sequential\nfrom keras.layers import Dense, LSTM, Dropout, GRU, Bidirectional\nfrom keras.optimizers import SGD\nimport math\nfrom sklearn.metrics import mean_squared_error",
"_____no_output_____"
],
[
"# Some functions to help out with\ndef plot_predictions(test,predicted):\n plt.plot(test, color='red',label='Real IBM Stock Price')\n plt.plot(predicted, color='blue',label='Predicted IBM Stock Price')\n plt.title('IBM Stock Price Prediction')\n plt.xlabel('Time')\n plt.ylabel('IBM Stock Price')\n plt.legend()\n plt.show()\n\ndef return_rmse(test,predicted):\n rmse = math.sqrt(mean_squared_error(test, predicted))\n print(\"The root mean squared error is {}.\".format(rmse))",
"_____no_output_____"
],
[
"# First, we get the data\ndataset = pd.read_csv('./data/stocks/IBM_2006-01-01_to_2018-01-01.csv', index_col='Date', parse_dates=['Date'])\ndataset.head()",
"_____no_output_____"
],
[
"# Checking for missing values\ntraining_set = dataset[:'2016'].iloc[:,1:2].values\ntest_set = dataset['2017':].iloc[:,1:2].values",
"_____no_output_____"
],
[
"# We have chosen 'High' attribute for prices. Let's see what it looks like\ndataset[\"High\"][:'2016'].plot(figsize=(16,4),legend=True)\ndataset[\"High\"]['2017':].plot(figsize=(16,4),legend=True)\nplt.legend(['Training set (Before 2017)','Test set (2017 and beyond)'])\nplt.title('IBM stock price')\nplt.show()",
"_____no_output_____"
],
[
"# Scaling the training set\nsc = MinMaxScaler(feature_range=(0,1))\ntraining_set_scaled = sc.fit_transform(training_set)",
"_____no_output_____"
],
[
"# Since LSTMs store long term memory state, we create a data structure with 60 timesteps and 1 output\n# So for each element of training set, we have 60 previous training set elements \nX_train = []\ny_train = []\nfor i in range(60,2769):\n X_train.append(training_set_scaled[i-60:i,0])\n y_train.append(training_set_scaled[i,0])\nX_train, y_train = np.array(X_train), np.array(y_train)",
"_____no_output_____"
],
[
"# Reshaping X_train for efficient modelling\nX_train = np.reshape(X_train, (X_train.shape[0],X_train.shape[1],1))",
"_____no_output_____"
],
[
"# The LSTM architecture\nregressor = Sequential()\n# First LSTM layer with Dropout regularisation\nregressor.add(LSTM(units=50, return_sequences=True, input_shape=(X_train.shape[1],1)))\nregressor.add(Dropout(0.2))\n# Second LSTM layer\nregressor.add(LSTM(units=50, return_sequences=True))\nregressor.add(Dropout(0.2))\n# Third LSTM layer\nregressor.add(LSTM(units=50, return_sequences=True))\nregressor.add(Dropout(0.2))\n# Fourth LSTM layer\nregressor.add(LSTM(units=50))\nregressor.add(Dropout(0.2))\n# The output layer\nregressor.add(Dense(units=1))\n\n# Compiling the RNN\nregressor.compile(optimizer='rmsprop',loss='mean_squared_error')\n# Fitting to the training set\nregressor.fit(X_train,y_train,epochs=50,batch_size=32)",
"_____no_output_____"
],
[
"# Now to get the test set ready in a similar way as the training set.\n# The following has been done so forst 60 entires of test set have 60 previous values which is impossible to get unless we take the whole \n# 'High' attribute data for processing\ndataset_total = pd.concat((dataset[\"High\"][:'2016'],dataset[\"High\"]['2017':]),axis=0)\ninputs = dataset_total[len(dataset_total)-len(test_set) - 60:].values\ninputs = inputs.reshape(-1,1)\ninputs = sc.transform(inputs)",
"_____no_output_____"
],
[
"# Preparing X_test and predicting the prices\nX_test = []\nfor i in range(60,311):\n X_test.append(inputs[i-60:i,0])\nX_test = np.array(X_test)\nX_test = np.reshape(X_test, (X_test.shape[0],X_test.shape[1],1))\npredicted_stock_price = regressor.predict(X_test)\npredicted_stock_price = sc.inverse_transform(predicted_stock_price)",
"_____no_output_____"
],
[
"# Visualizing the results for LSTM\nplot_predictions(test_set,predicted_stock_price)",
"_____no_output_____"
],
[
"# Evaluating our model\nreturn_rmse(test_set,predicted_stock_price)",
"_____no_output_____"
]
],
[
[
"Truth be told. That's one awesome score. \n\nLSTM is not the only kind of unit that has taken the world of Deep Learning by a storm. We have **Gated Recurrent Units(GRU)**. It's not known, which is better: GRU or LSTM becuase they have comparable performances. GRUs are easier to train than LSTMs.\n\n## Gated Recurrent Units\nIn simple words, the GRU unit does not have to use a memory unit to control the flow of information like the LSTM unit. It can directly makes use of the all hidden states without any control. GRUs have fewer parameters and thus may train a bit faster or need less data to generalize. But, with large data, the LSTMs with higher expressiveness may lead to better results.\n\nThey are almost similar to LSTMs except that they have two gates: reset gate and update gate. Reset gate determines how to combine new input to previous memory and update gate determines how much of the previous state to keep. Update gate in GRU is what input gate and forget gate were in LSTM. We don't have the second non linearity in GRU before calculating the outpu, .neither they have the output gate.\n\nSource: [Quora](https://www.quora.com/Whats-the-difference-between-LSTM-and-GRU-Why-are-GRU-efficient-to-train)\n\n<img src=\"https://cdnpythonmachinelearning.azureedge.net/wp-content/uploads/2017/11/GRU.png?x31195\">",
"_____no_output_____"
]
],
[
[
"# The GRU architecture\nregressorGRU = Sequential()\n# First GRU layer with Dropout regularisation\nregressorGRU.add(GRU(units=50, return_sequences=True, input_shape=(X_train.shape[1],1), activation='tanh'))\nregressorGRU.add(Dropout(0.2))\n# Second GRU layer\nregressorGRU.add(GRU(units=50, return_sequences=True, input_shape=(X_train.shape[1],1), activation='tanh'))\nregressorGRU.add(Dropout(0.2))\n# Third GRU layer\nregressorGRU.add(GRU(units=50, return_sequences=True, input_shape=(X_train.shape[1],1), activation='tanh'))\nregressorGRU.add(Dropout(0.2))\n# Fourth GRU layer\nregressorGRU.add(GRU(units=50, activation='tanh'))\nregressorGRU.add(Dropout(0.2))\n# The output layer\nregressorGRU.add(Dense(units=1))\n# Compiling the RNN\nregressorGRU.compile(optimizer=SGD(lr=0.01, decay=1e-7, momentum=0.9, nesterov=False),loss='mean_squared_error')\n# Fitting to the training set\nregressorGRU.fit(X_train,y_train,epochs=50,batch_size=150)",
"_____no_output_____"
]
],
[
[
"The current version version uses a dense GRU network with 100 units as opposed to the GRU network with 50 units in previous version",
"_____no_output_____"
]
],
[
[
"# Preparing X_test and predicting the prices\nX_test = []\nfor i in range(60,311):\n X_test.append(inputs[i-60:i,0])\nX_test = np.array(X_test)\nX_test = np.reshape(X_test, (X_test.shape[0],X_test.shape[1],1))\nGRU_predicted_stock_price = regressorGRU.predict(X_test)\nGRU_predicted_stock_price = sc.inverse_transform(GRU_predicted_stock_price)",
"_____no_output_____"
],
[
"# Visualizing the results for GRU\nplot_predictions(test_set,GRU_predicted_stock_price)",
"_____no_output_____"
],
[
"# Evaluating GRU\nreturn_rmse(test_set,GRU_predicted_stock_price)",
"_____no_output_____"
]
],
[
[
"## Sequence Generation\nHere, I will generate a sequence using just initial 60 values instead of using last 60 values for every new prediction. **Due to doubts in various comments about predictions making use of test set values, I have decided to include sequence generation.** The above models make use of test set so it is using last 60 true values for predicting the new value(I will call it a benchmark). This is why the error is so low. Strong models can bring similar results like above models for sequences too but they require more than just data which has previous values. In case of stocks, we need to know the sentiments of the market, the movement of other stocks and a lot more. So, don't expect a remotely accurate plot. The error will be great and the best I can do is generate the trend similar to the test set.\n\nI will use GRU model for predictions. You can try this using LSTMs also. I have modified GRU model above to get the best sequence possible. I have run the model four times and two times I got error of around 8 to 9. The worst case had an error of around 11. Let's see what this iterations.\n\nThe GRU model in the previous versions is fine too. Just a little tweaking was required to get good sequences. **The main goal of this kernel is to show how to build RNN models. How you predict data and what kind of data you predict is up to you. I can't give you some 100 lines of code where you put the destination of training and test set and get world-class results. That's something you have to do yourself.**",
"_____no_output_____"
]
],
[
[
"# Preparing sequence data\ninitial_sequence = X_train[2708,:]\nsequence = []\nfor i in range(251):\n new_prediction = regressorGRU.predict(initial_sequence.reshape(initial_sequence.shape[1],initial_sequence.shape[0],1))\n initial_sequence = initial_sequence[1:]\n initial_sequence = np.append(initial_sequence,new_prediction,axis=0)\n sequence.append(new_prediction)\nsequence = sc.inverse_transform(np.array(sequence).reshape(251,1))",
"_____no_output_____"
],
[
"# Visualizing the sequence\nplot_predictions(test_set,sequence)",
"_____no_output_____"
],
[
"# Evaluating the sequence\nreturn_rmse(test_set,sequence)",
"_____no_output_____"
]
],
[
[
"So, GRU works better than LSTM in this case. Bidirectional LSTM is also a good way so make the model stronger. But this may vary for different data sets. **Applying both LSTM and GRU together gave even better results.** ",
"_____no_output_____"
],
[
"#### I was going to cover text generation using LSTM but already an excellent kernel by [Shivam Bansal](https://www.kaggle.com/shivamb) on the mentioned topic exists. Link for that kernel here: https://www.kaggle.com/shivamb/beginners-guide-to-text-generation-using-lstms",
"_____no_output_____"
],
[
"#### This is certainly not the end. Stay tuned for more stuff!",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
e78c5526c0a98511fb9088177f867ae17a86f3cd | 31,709 | ipynb | Jupyter Notebook | Exercise/exercise_1.ipynb | TangJiahui/AC215-Advanced_Practical_Data_Science | 904e595630ddb9b2348889ce0d2cc777bb90f071 | [
"MIT"
] | null | null | null | Exercise/exercise_1.ipynb | TangJiahui/AC215-Advanced_Practical_Data_Science | 904e595630ddb9b2348889ce0d2cc777bb90f071 | [
"MIT"
] | null | null | null | Exercise/exercise_1.ipynb | TangJiahui/AC215-Advanced_Practical_Data_Science | 904e595630ddb9b2348889ce0d2cc777bb90f071 | [
"MIT"
] | null | null | null | 36.489068 | 369 | 0.415087 | [
[
[
"<h1 style=\"padding-top: 25px;padding-bottom: 25px;text-align: left; padding-left: 10px; background-color: #DDDDDD; \n color: black;\"> <img style=\"float: left; padding-right: 10px;\" src=\"https://raw.githubusercontent.com/Harvard-IACS/2018-CS109A/master/content/styles/iacs.png\" height=\"50px\"> <a href='https://harvard-iacs.github.io/2021-AC215/' target='_blank'><strong><font color=\"#A41034\">AC215: Advanced Practical Data Science, MLOps</font></strong></a></h1>\n\n# **<font color=\"#A41034\">Exercise 1 - Dask</font>**\n\n**Harvard University**<br/>\n**Fall 2021**<br/>\n**Instructor:**\nPavlos Protopapas<br/>\n\n**Students:**\nJiahui Tang, Max Li<br/>\n\n<hr style=\"height:2pt\">",
"_____no_output_____"
],
[
"## **<font color=\"#A41034\">Setup Notebook</font>**",
"_____no_output_____"
],
[
"**Copy & setup Colab**",
"_____no_output_____"
],
[
"1) Select \"File\" menu and pick \"Save a copy in Drive\"",
"_____no_output_____"
],
[
"**Installs**",
"_____no_output_____"
]
],
[
[
"!pip install dask dask[dataframe] dask-image",
"Requirement already satisfied: dask in /usr/local/lib/python3.7/dist-packages (2.12.0)\nRequirement already satisfied: dask-image in /usr/local/lib/python3.7/dist-packages (0.6.0)\nRequirement already satisfied: scipy>=0.19.1 in /usr/local/lib/python3.7/dist-packages (from dask-image) (1.4.1)\nRequirement already satisfied: pims>=0.4.1 in /usr/local/lib/python3.7/dist-packages (from dask-image) (0.5)\nRequirement already satisfied: numpy>=1.11.3 in /usr/local/lib/python3.7/dist-packages (from dask-image) (1.19.5)\nRequirement already satisfied: toolz>=0.7.3 in /usr/local/lib/python3.7/dist-packages (from dask) (0.11.1)\nRequirement already satisfied: slicerator>=0.9.8 in /usr/local/lib/python3.7/dist-packages (from pims>=0.4.1->dask-image) (1.0.0)\nRequirement already satisfied: six>=1.8 in /usr/local/lib/python3.7/dist-packages (from pims>=0.4.1->dask-image) (1.15.0)\nRequirement already satisfied: fsspec>=0.6.0 in /usr/local/lib/python3.7/dist-packages (from dask) (2021.8.1)\nRequirement already satisfied: pandas>=0.23.0 in /usr/local/lib/python3.7/dist-packages (from dask) (1.1.5)\nRequirement already satisfied: partd>=0.3.10 in /usr/local/lib/python3.7/dist-packages (from dask) (1.2.0)\nRequirement already satisfied: python-dateutil>=2.7.3 in /usr/local/lib/python3.7/dist-packages (from pandas>=0.23.0->dask) (2.8.2)\nRequirement already satisfied: pytz>=2017.2 in /usr/local/lib/python3.7/dist-packages (from pandas>=0.23.0->dask) (2018.9)\nRequirement already satisfied: locket in /usr/local/lib/python3.7/dist-packages (from partd>=0.3.10->dask) (0.2.1)\n"
]
],
[
[
"**Imports**",
"_____no_output_____"
]
],
[
[
"import os\nimport requests\nimport zipfile\nimport tarfile\nimport shutil\nimport math\nimport json\nimport time\nimport sys\nimport numpy as np\nimport pandas as pd\n\n# Dask\nimport dask\nimport dask.dataframe as dd\nimport dask.array as da\nfrom dask.diagnostics import ProgressBar",
"_____no_output_____"
]
],
[
[
"**Utils**",
"_____no_output_____"
],
[
"Here are some util functions that we will be using for this exercise",
"_____no_output_____"
]
],
[
[
"def download_file(packet_url, base_path=\"\", extract=False, headers=None):\n if base_path != \"\":\n if not os.path.exists(base_path):\n os.mkdir(base_path)\n packet_file = os.path.basename(packet_url)\n with requests.get(packet_url, stream=True, headers=headers) as r:\n r.raise_for_status()\n with open(os.path.join(base_path,packet_file), 'wb') as f:\n for chunk in r.iter_content(chunk_size=8192):\n f.write(chunk)\n \n if extract:\n if packet_file.endswith(\".zip\"):\n with zipfile.ZipFile(os.path.join(base_path,packet_file)) as zfile:\n zfile.extractall(base_path)\n else:\n packet_name = packet_file.split('.')[0]\n with tarfile.open(os.path.join(base_path,packet_file)) as tfile:\n tfile.extractall(base_path)",
"_____no_output_____"
]
],
[
[
"## **<font color=\"#A41034\">Dataset</font>**",
"_____no_output_____"
],
[
"### **Load Data**",
"_____no_output_____"
]
],
[
[
"start_time = time.time()\ndownload_file(\"https://github.com/dlops-io/datasets/releases/download/v1.0/Parking_Violations_Issued_-_Fiscal_Year_2017.csv.zip\", base_path=\"datasets\", extract=True)\nexecution_time = (time.time() - start_time)/60.0\nprint(\"Download execution time (mins)\",execution_time)",
"Download execution time (mins) 0.8145878473917644\n"
],
[
"parking_violation_csv = os.path.join(\"datasets\",\"Parking_Violations_Issued_-_Fiscal_Year_2017.csv\")",
"_____no_output_____"
]
],
[
[
"## Q1: Compute Pi with a Slowly Converging Series\n\nLeibniz published one of the oldest known series in 1676. While this is easy to understand and derive, it converges very slowly.\nhttps://en.wikipedia.org/wiki/Leibniz_formula_for_%CF%80 <br/>\n$$\\frac{\\pi}{4} = 1 - \\frac{1}{3} + \\frac{1}{5} - \\frac{1}{7} ...$$\n\nWhile this is a genuinely cruel way to compute the value of $\\pi$, it’s a fun opportunity to use brute force on a problem instead of thinking.\nCompute using at least four billion terms in this sequence. Compare your time taken with numpy and dask. On my mac, with numpy this took 44 seconds and with dask it took 5.7 seconds. \n\n*Hint:* Use dask array",
"_____no_output_____"
],
[
"**Checking 1e9 * 4 terms with numpy**\n\nIf 1e9 * 4 fails, try 1e9 * 2 or increase memory ",
"_____no_output_____"
]
],
[
[
"# Your code here\nstart_time = time.time()\nk = int(1e9*2)\npositive_sum = np.sum(1/np.arange(1, k ,4))\nnegative_sum = np.sum(-1/np.arange(3, k, 4))\npi_computed = (positive_sum + negative_sum) * 4\nexecution_time = time.time() - start_time",
"_____no_output_____"
],
[
"# Error \nerror = np.abs(pi_computed-np.pi)\n\n# Report Results\nprint(f'Pi real value = {np.pi:14.12f}')\nprint(f'Pi computed value = {pi_computed:14.12f}')\nprint(f'Error = {error:6.3e}')\nprint(\"Numpy execution time (sec)\",execution_time)",
"Pi real value = 3.141592653590\nPi computed value = 3.141592652590\nError = 9.998e-10\nNumpy execution time (sec) 8.094965696334839\n"
]
],
[
[
"**Checking 1e9 * 4 terms with Dask**",
"_____no_output_____"
]
],
[
[
"# Your code here\nstart_time = time.time()\nk = int(1e9*2)\npositive_sum_da = da.sum(1/da.arange(1, k, 4)).compute()\nnegative_sum_da = da.sum(-1/da.arange(3, k, 4)).compute()\nstep3_pi = (positive_sum_da + negative_sum_da) * 4\nexecution_time = time.time() - start_time",
"_____no_output_____"
],
[
"error = np.abs(step3_pi - np.pi)\n\n# Report Results\nprint(f'Pi real value = {np.pi:14.12f}')\nprint(f'Pi computed value = {step3_pi:14.12f}')\nprint(f'Error = {error:6.3e}')\nprint(\"Dask Array execution time (sec)\",execution_time)",
"Pi real value = 3.141592653590\nPi computed value = 3.141592652590\nError = 1.000e-09\nDask Array execution time (sec) 4.978763103485107\n"
]
],
[
[
"## Filter Parking Tickets Dataset\n\nAccording to the parking tickets data set documentation, the column called ‘Plate Type’ consists mainly of two different types, ‘PAS’ and ‘COM’; presumably for passenger and commercial vehicles, respectively. Maybe the rest are the famous parking tickets from the UN diplomats, who take advantage of diplomatic immunity not to pay their fines.\n\nCreate a filtered Dask DataFrame with only the commercial plates.\nPersist it, so it is available in memory for future computations. Count the number of summonses in 2017 (i.e., Issue Year in 2016, 2017) issued to commercial plate types. Compute them as a percentage of the total data set. \n\n*Hint*: This is easy; it is only about 5-7 lines of code.",
"_____no_output_____"
]
],
[
[
"dict_1 = {'Summons Number': 'int64', 'Plate ID': 'object', 'Registration State': 'object', 'Plate Type': 'object',\n 'Issue Date': 'object', 'Violation Code': 'int64', 'Vehicle Body Type': 'object', 'Vehicle Make': 'object',\n 'Issuing Agency': 'object', 'Street Code1': 'int64', 'Street Code2': 'int64', 'Street Code3': 'int64',\n 'Vehicle Expiration Date': 'int64', 'Violation Location': 'float64', 'Violation Precinct': 'int64', 'Issuer Precinct': 'int64',\n 'Issuer Code': 'int64', 'Issuer Command': 'object', 'Issuer Squad': 'object', 'Violation Time': 'object',\n 'Time First Observed': 'object', 'Violation County': 'object', 'Violation In Front Of Or Opposite': 'object', 'House Number': 'object',\n 'Street Name': 'object', 'Intersecting Street': 'object', 'Date First Observed': 'int64', 'Law Section': 'int64',\n 'Sub Division': 'object', 'Violation Legal Code': 'object', 'Days Parking In Effect ': 'object', 'From Hours In Effect': 'object',\n 'To Hours In Effect': 'object', 'Vehicle Color': 'object', 'Unregistered Vehicle?': 'float64', 'Vehicle Year': 'int64',\n 'Meter Number': 'object', 'Feet From Curb': 'int64', 'Violation Post Code': 'object', 'Violation Description': 'object',\n 'No Standing or Stopping Violation': 'float64', 'Hydrant Violation': 'float64', 'Double Parking Violation': 'float64'}\n\n# This is to avoid the DtypeWarning \ndf = dd.read_csv(parking_violation_csv, dtype=dict_1)\ndf.head()",
"_____no_output_____"
],
[
"# info Dask Dataframe\nprint('<index where df has been split>', df.divisions)\nprint('<# partitions>', df.npartitions)",
"<index where df has been split> (None, None, None, None, None, None, None, None, None, None, None, None, None, None, None, None, None, None, None, None, None, None, None, None, None, None, None, None, None, None, None, None, None, None)\n<# partitions> 33\n"
],
[
"# filter entries in dask dataframe with COM\n# and persist it\naction_filtered = df[df['Plate Type'] == 'COM'].persist()",
"_____no_output_____"
],
[
"# df with number of summonses in 2017 (i.e., Issue Year in 2016, 2017)\nsummonses_value_df = action_filtered[action_filtered['Issue Date'].str.contains('2016|2017')]\n\n#after reorganizing dataframe in one partition, check number of summonses\naction_filtered_reduced = summonses_value_df.repartition(npartitions=1)\ncommercial_2017_count = action_filtered_reduced.map_partitions(len).compute()",
"_____no_output_____"
],
[
"# Compute them as a percentage of the total data set\ndf_size = df.index.size \ncommercial_2017_percent = ((commercial_2017_count/df_size)*100)",
"_____no_output_____"
],
[
"num_commercial_2017 = int(commercial_2017_count)\npct_commercial = int(commercial_2017_percent)",
"_____no_output_____"
],
[
"# Percentage relative to all the parking tickets in 2017\nprint(f'Number of NYC summonses with commercial plates in 2017 was {num_commercial_2017}')\nprint(f'Percentage {pct_commercial:5.2f}%')",
"Number of NYC summonses with commercial plates in 2017 was 1838970\nPercentage 17.00%\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78c645c20de5f98fe80d96cb3f345e3d052188d | 120,227 | ipynb | Jupyter Notebook | benchmark-results/6class_results/FeatureSet2.ipynb | VedantKalbag/metal-vocal-vataset | 6814afb61aebc73b28f1b519629a43ae51c6836a | [
"MIT"
] | null | null | null | benchmark-results/6class_results/FeatureSet2.ipynb | VedantKalbag/metal-vocal-vataset | 6814afb61aebc73b28f1b519629a43ae51c6836a | [
"MIT"
] | null | null | null | benchmark-results/6class_results/FeatureSet2.ipynb | VedantKalbag/metal-vocal-vataset | 6814afb61aebc73b28f1b519629a43ae51c6836a | [
"MIT"
] | null | null | null | 318.060847 | 70,882 | 0.93039 | [
[
[
"import os\nimport pandas as pd\nimport numpy as np\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.metrics import classification_report\nfrom sklearn.metrics import confusion_matrix,accuracy_score, ConfusionMatrixDisplay, balanced_accuracy_score,f1_score\n\nfrom sklearn.svm import SVC\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.preprocessing import StandardScaler\n\nfrom sys import platform\n\nlabels={0:'clean',1:'high fry',2:'layered',3:'lowfry',4:'midfry',5:'no_vocals'}\n# labels={0:'clean',1:'scream',2:'no vocal'}",
"_____no_output_____"
],
[
"n_class = '6class'\n\nX_train=np.load(f'./resources/working_data/{n_class}_x_train-vggish.npy',allow_pickle=True)\nX_test=np.load(f'./resources/working_data/{n_class}_x_test-vggish.npy',allow_pickle=True)\nX_valid=np.load(f'./resources/working_data/{n_class}_x_valid-vggish.npy',allow_pickle=True)\n\ny_train_hot=np.load(f'./resources/working_data/{n_class}_y_train-vggish.npy',allow_pickle=True)\ny_test_hot=np.load(f'./resources/working_data/{n_class}_y_test-vggish.npy',allow_pickle=True)\ny_valid_hot=np.load(f'./resources/working_data/{n_class}_y_valid-vggish.npy',allow_pickle=True)\n\ny_train = np.argmax(y_train_hot,axis=1)\ny_test = np.argmax(y_test_hot,axis=1)\ny_valid = np.argmax(y_valid_hot,axis=1)\n#df = pd.DataFrame(d,columns=feature_cols)\n\nscaler=StandardScaler()\nX_train=scaler.fit_transform(X_train)\nX_test=scaler.transform(X_test)\nX_valid=scaler.transform(X_valid)",
"_____no_output_____"
],
[
"def classwise_accuracy(cm):\n a=np.zeros(cm.shape)\n for i,x in enumerate(cm):\n a[i]=x/sum(x)\n return a",
"_____no_output_____"
],
[
"def eval_metrics(y_test,y_pred):\n cm = confusion_matrix(y_test,y_pred)\n acc = accuracy_score(y_test,y_pred)\n f1 = f1_score(y_test,y_pred,average='macro')\n macro_acc = balanced_accuracy_score(y_test,y_pred)\n classwise_acc = classwise_accuracy(cm)\n return(cm,acc,f1,macro_acc,classwise_acc)",
"_____no_output_____"
]
],
[
[
"# kNN",
"_____no_output_____"
]
],
[
[
"k=4\nKNN_model = KNeighborsClassifier(n_neighbors=k)\nKNN_model.fit(X_train, y_train)\nKNN_prediction = KNN_model.predict(X_test)\n\ncm,acc,f1,macro_acc,classwise_acc = eval_metrics(y_test,KNN_prediction)\n\nprint(f\"Overall Accuracy Score: {acc}\")\nprint(f\"Macro Accuracy: {macro_acc}\")\nprint(f\"Class-wise accuracy: \\n{classwise_acc}\")\nl=labels.values()\nConfusionMatrixDisplay(confusion_matrix=cm,display_labels=l).plot()",
"Overall Accuracy Score: 0.4154929577464789\nMacro Accuracy: 0.42229983219064593\nClass-wise accuracy: \n[[0.8079096 0.02824859 0.05084746 0.05084746 0.01694915 0.04519774]\n [0.16071429 0.46428571 0.03571429 0.07142857 0.14285714 0.125 ]\n [0.35483871 0.29032258 0.08064516 0.10080645 0.13306452 0.04032258]\n [0.07462687 0.28358209 0.1641791 0.14925373 0.32835821 0. ]\n [0.10897436 0.35897436 0.08333333 0.12820513 0.28846154 0.03205128]\n [0.06756757 0.12837838 0.00675676 0.02702703 0.02702703 0.74324324]]\n"
]
],
[
[
"# SVM",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"SVM_model = SVC(gamma='scale',C=1.0533, kernel='poly', degree=2,coef0=2.1,random_state=42)\nSVM_model.fit(X_train, y_train)\n\nSVM_prediction = SVM_model.predict(X_test)\n\ncm,acc,f1,macro_acc,classwise_acc = eval_metrics(y_test,SVM_prediction)\n\nprint(f\"Overall Accuracy Score: {acc}\")\nprint(f\"Macro Accuracy: {macro_acc}\")\nprint(f\"Class-wise accuracy: \\n{classwise_acc}\")\nprint(f\"F1 score: {f1}\")\nl=labels.values()\n# ConfusionMatrixDisplay(confusion_matrix=cm,display_labels=l).plot()\nl=['Sing','High Fry Scream','Layered Screams', 'Low Fry Screams','Mid Fry Screams', 'No Vocal']\ncm_classwise=np.array([[0.84180791, 0.02824859, 0.03954802, 0.01694915, 0.02824859, 0.04519774],\n [0.08928571, 0.46428571, 0.08928571, 0.03571429, 0.17857143, 0.14285714],\n [0.31451613, 0.29032258, 0.08870968, 0.09677419, 0.15322581, 0.05645161],\n [0.02985075, 0.2238806 , 0.08955224, 0.1641791 , 0.49253731, 0. ],\n [0.12820513, 0.30128205, 0.08333333, 0.07051282, 0.37179487, 0.04487179],\n [0.05405405, 0.07432432, 0.00675676, 0.02027027, 0.02027027, 0.82432432]])\n# fig, ax = plt.subplots()\ndisp = ConfusionMatrixDisplay(confusion_matrix=cm,display_labels=l)\ndisp.plot(xticks_rotation=45)\nplt.rcParams.update({'font.size': 18})\nfig = plt.gcf()\nfig.set_size_inches(10, 8)\nplt.tight_layout()\ndisp.ax_.set_xticklabels(l,ha='right')\n# plt.savefig('/Users/vedant/Desktop/Programming/ScreamDetection/charts/svm-vggish-6class.pdf')",
"Overall Accuracy Score: 0.45539906103286387\nMacro Accuracy: 0.4591836003177328\nClass-wise accuracy: \n[[0.84180791 0.02824859 0.03954802 0.01694915 0.02824859 0.04519774]\n [0.08928571 0.46428571 0.08928571 0.03571429 0.17857143 0.14285714]\n [0.31451613 0.29032258 0.08870968 0.09677419 0.15322581 0.05645161]\n [0.02985075 0.2238806 0.08955224 0.1641791 0.49253731 0. ]\n [0.12820513 0.30128205 0.08333333 0.07051282 0.37179487 0.04487179]\n [0.05405405 0.07432432 0.00675676 0.02027027 0.02027027 0.82432432]]\nF1 score: 0.4013489207140873\n"
],
[
"cm/cm.sum(axis=1)",
"_____no_output_____"
]
],
[
[
"# RF",
"_____no_output_____"
]
],
[
[
"RF_model = RandomForestClassifier(n_estimators=90,criterion='gini',max_depth=None,\\\n min_samples_split=2,min_samples_leaf=1,max_features='auto',max_leaf_nodes=None,class_weight='balanced',random_state=42)\nRF_model.fit(X_train, y_train)\nRF_prediction = RF_model.predict(X_test)\n\ncm,acc,f1,macro_acc,classwise_acc = eval_metrics(y_test,RF_prediction)\n\nprint(f\"Overall Accuracy Score: {acc}\")\nprint(f\"Macro Accuracy: {macro_acc}\")\nprint(f\"F1 score: {f1}\")\nprint(f\"Class-wise accuracy: \\n{classwise_acc}\")\nl=labels.values()\nConfusionMatrixDisplay(confusion_matrix=cm,display_labels=l).plot()",
"Overall Accuracy Score: 0.8191881918819188\nMacro Accuracy: 0.8203617260744455\nF1 score: 0.8200458870746504\nClass-wise accuracy: \n[[0.83564815 0.13657407 0.02777778]\n [0.14315789 0.77052632 0.08631579]\n [0.05580357 0.08928571 0.85491071]]\n"
],
[
"cm/cm.sum(axis=1)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
e78c98d4c9ef8e61b3f3564da17477001b442cf7 | 782,929 | ipynb | Jupyter Notebook | notebooks/01_Beginners_guide/08_Intro_to_xarray.ipynb | miguelalejo/2021-Better-Working-World-Data-Challenge | 581d35de4488307d8343331f389578dcbf611b01 | [
"MIT"
] | 116 | 2021-03-23T23:43:13.000Z | 2021-12-13T22:49:41.000Z | notebooks/01_Beginners_guide/08_Intro_to_xarray.ipynb | miguelalejo/2021-Better-Working-World-Data-Challenge | 581d35de4488307d8343331f389578dcbf611b01 | [
"MIT"
] | 12 | 2021-03-26T07:01:46.000Z | 2021-06-15T22:20:50.000Z | notebooks/01_Beginners_guide/08_Intro_to_xarray.ipynb | miguelalejo/2021-Better-Working-World-Data-Challenge | 581d35de4488307d8343331f389578dcbf611b01 | [
"MIT"
] | 119 | 2021-03-22T05:52:56.000Z | 2022-03-04T04:58:46.000Z | 280.418696 | 376,520 | 0.896563 | [
[
[
"# Introduction to Xarray <img align=\"right\" src=\"../Supplementary_data/dea_logo.jpg\">\n\n* **Acknowledgement**: This notebook was originally created by [Digital Eath Australia (DEA)](https://www.ga.gov.au/about/projects/geographic/digital-earth-australia) and has been modified for use in the EY Data Science Program\n* **Prerequisites**: Users of this notebook should have a basic understanding of:\n * How to run a [Jupyter notebook](01_Jupyter_notebooks.ipynb)\n * How to work with [Numpy](07_Intro_to_numpy.ipynb)",
"_____no_output_____"
],
[
"## Background\n`Xarray` is a python library which simplifies working with labelled multi-dimension arrays. \n`Xarray` introduces labels in the forms of dimensions, coordinates and attributes on top of raw `numpy` arrays, allowing for more intitutive and concise development. \nMore information about `xarray` data structures and functions can be found [here](http://xarray.pydata.org/en/stable/).\n\nOnce you've completed this notebook, you may be interested in advancing your `xarray` skills further, this [external notebook](https://rabernat.github.io/research_computing/xarray.html) introduces more uses of `xarray` and may help you advance your skills further. ",
"_____no_output_____"
],
[
"## Description\nThis notebook is designed to introduce users to `xarray` using Python code in Jupyter Notebooks via JupyterLab.\n\nTopics covered include:\n\n* How to use `xarray` functions in a Jupyter Notebook cell\n* How to access `xarray` dimensions and metadata\n* Using indexing to explore multi-dimensional `xarray` data\n* Appliction of built-in `xarray` functions such as sum, std and mean\n\n***\n",
"_____no_output_____"
],
[
"## Getting started\nTo run this notebook, run all the cells in the notebook starting with the \"Load packages\" cell. For help with running notebook cells, refer back to the [Jupyter Notebooks notebook](01_Jupyter_notebooks.ipynb). ",
"_____no_output_____"
],
[
"### Load packages",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport xarray as xr",
"_____no_output_____"
]
],
[
[
"### Introduction to xarray\nDEA uses `xarray` as its core data model. \nTo better understand what it is, let's first do a simple experiment using a combination of plain `numpy` arrays and Python dictionaries.\n\nSuppose we have a satellite image with three bands: `Red`, `NIR` and `SWIR`. \nThese bands are represented as 2-dimensional `numpy` arrays and the latitude and longitude coordinates for each dimension are represented using 1-dimensional arrays. \nFinally, we also have some metadata that comes with this image. \nThe code below creates fake satellite data and structures the data as a `dictionary`.",
"_____no_output_____"
]
],
[
[
"# Create fake satellite data\nred = np.random.rand(250, 250)\nnir = np.random.rand(250, 250)\nswir = np.random.rand(250, 250)\n\n# Create some lats and lons\nlats = np.linspace(-23.5, -26.0, num=red.shape[0], endpoint=False)\nlons = np.linspace(110.0, 112.5, num=red.shape[1], endpoint=False)\n\n# Create metadata\ntitle = \"Image of the desert\"\ndate = \"2019-11-10\"\n\n# Stack into a dictionary\nimage = {\n \"red\": red,\n \"nir\": nir,\n \"swir\": swir,\n \"latitude\": lats,\n \"longitude\": lons,\n \"title\": title,\n \"date\": date,\n}",
"_____no_output_____"
]
],
[
[
"All our data is conveniently packed in a dictionary. Now we can use this dictionary to work with the data it contains:",
"_____no_output_____"
]
],
[
[
"# Date of satellite image\nprint(image[\"date\"])\n\n# Mean of red values\nimage[\"red\"].mean()",
"2019-11-10\n"
]
],
[
[
"Still, to select data we have to use `numpy` indexes. \nWouldn't it be convenient to be able to select data from the images using the coordinates of the pixels instead of their relative positions? \nThis is exactly what `xarray` solves! Let's see how it works:\n\nTo explore `xarray` we have a file containing some surface reflectance data extracted from the DEA platform. \nThe object that we get `ds` is a `xarray` `Dataset`, which in some ways is very similar to the dictionary that we created before, but with lots of convenient functionality available.",
"_____no_output_____"
]
],
[
[
"ds = xr.open_dataset(\"../Supplementary_data/08_Intro_to_xarray/example_netcdf.nc\")\nds",
"_____no_output_____"
]
],
[
[
"### Xarray dataset structure\nA `Dataset` can be seen as a dictionary structure packing up the data, dimensions and attributes. Variables in a `Dataset` object are called `DataArrays` and they share dimensions with the higher level `Dataset`. \nThe figure below provides an illustrative example:\n\n\n<img src=\"../Supplementary_data/08_Intro_to_xarray/dataset-diagram.png\" alt=\"drawing\" width=\"600\" align=\"left\"/>",
"_____no_output_____"
],
[
"To access a variable we can access as if it were a Python dictionary, or using the `.` notation, which is more convenient.",
"_____no_output_____"
]
],
[
[
"ds[\"green\"]\n\n# Or alternatively:\nds.green",
"_____no_output_____"
]
],
[
[
"Dimensions are also stored as numeric arrays that we can easily access:",
"_____no_output_____"
]
],
[
[
"ds[\"time\"]\n\n# Or alternatively:\nds.time",
"_____no_output_____"
]
],
[
[
"Metadata is referred to as attributes and is internally stored under `.attrs`, but the same convenient `.` notation applies to them.",
"_____no_output_____"
]
],
[
[
"ds.attrs[\"crs\"]\n\n# Or alternatively:\nds.crs",
"_____no_output_____"
]
],
[
[
"`DataArrays` store their data internally as multidimensional `numpy` arrays. \nBut these arrays contain dimensions or labels that make it easier to handle the data. \nTo access the underlaying numpy array of a `DataArray` we can use the `.values` notation.",
"_____no_output_____"
]
],
[
[
"arr = ds.green.values\n\ntype(arr), arr.shape",
"_____no_output_____"
]
],
[
[
"### Indexing\n\n`Xarray` offers two different ways of selecting data. This includes the `isel()` approach, where data can be selected based on its index (like `numpy`).\n",
"_____no_output_____"
]
],
[
[
"print(ds.time.values)\n\nss = ds.green.isel(time=0)\nss",
"['2018-01-03T08:31:05.000000000' '2018-01-08T08:34:01.000000000'\n '2018-01-13T08:30:41.000000000' '2018-01-18T08:30:42.000000000'\n '2018-01-23T08:33:58.000000000' '2018-01-28T08:30:20.000000000'\n '2018-02-07T08:30:53.000000000' '2018-02-12T08:31:43.000000000'\n '2018-02-17T08:23:09.000000000' '2018-02-17T08:35:40.000000000'\n '2018-02-22T08:34:52.000000000' '2018-02-27T08:31:36.000000000']\n"
]
],
[
[
"Or the `sel()` approach, used for selecting data based on its dimension of label value.",
"_____no_output_____"
]
],
[
[
"ss = ds.green.sel(time=\"2018-01-08\")\nss",
"_____no_output_____"
]
],
[
[
"Slicing data is also used to select a subset of data.",
"_____no_output_____"
]
],
[
[
"ss.x.values[100]",
"_____no_output_____"
],
[
"ss = ds.green.sel(time=\"2018-01-08\", x=slice(2378390, 2380390))\nss",
"_____no_output_____"
]
],
[
[
"Xarray exposes lots of functions to easily transform and analyse `Datasets` and `DataArrays`. \nFor example, to calculate the spatial mean, standard deviation or sum of the green band:",
"_____no_output_____"
]
],
[
[
"print(\"Mean of green band:\", ds.green.mean().values)\nprint(\"Standard deviation of green band:\", ds.green.std().values)\nprint(\"Sum of green band:\", ds.green.sum().values)",
"Mean of green band: 4141.488778766468\nStandard deviation of green band: 3775.5536474649584\nSum of green band: 14426445446\n"
]
],
[
[
"### Plotting data with Matplotlib\nPlotting is also conveniently integrated in the library.",
"_____no_output_____"
]
],
[
[
"ds[\"green\"].isel(time=0).plot()",
"_____no_output_____"
]
],
[
[
"...but we still can do things manually using `numpy` and `matplotlib` if you choose:",
"_____no_output_____"
]
],
[
[
"rgb = np.dstack((ds.red.isel(time=0).values,\n ds.green.isel(time=0).values,\n ds.blue.isel(time=0).values))\nrgb = np.clip(rgb, 0, 2000) / 2000\n\nplt.imshow(rgb);",
"_____no_output_____"
]
],
[
[
"But compare the above to elegantly chaining operations within `xarray`:",
"_____no_output_____"
]
],
[
[
"ds[[\"red\", \"green\", \"blue\"]].isel(time=0).to_array().plot.imshow(robust=True, figsize=(6, 6));",
"_____no_output_____"
]
],
[
[
"## Recommended next steps\n\nFor more advanced information about working with Jupyter Notebooks or JupyterLab, you can explore [JupyterLab documentation page](https://jupyterlab.readthedocs.io/en/stable/user/notebook.html).\n\nTo continue working through the notebooks in this beginner's guide, the following notebooks are designed to be worked through in the following order:\n\n1. [Jupyter Notebooks](01_Jupyter_notebooks.ipynb)\n2. [Digital Earth Australia](02_DEA.ipynb)\n3. [Products and Measurements](03_Products_and_measurements.ipynb)\n4. [Loading data](04_Loading_data.ipynb)\n5. [Plotting](05_Plotting.ipynb)\n6. [Performing a basic analysis](06_Basic_analysis.ipynb)\n7. [Introduction to Numpy](07_Intro_to_numpy.ipynb)\n8. **Introduction to Xarray (this notebook)**\n9. [Parallel processing with Dask](09_Parallel_processing_with_Dask.ipynb)",
"_____no_output_____"
],
[
"***\n## Additional information\n\n**License:** The code in this notebook is licensed under the [Apache License, Version 2.0](https://www.apache.org/licenses/LICENSE-2.0). \nDigital Earth Australia data is licensed under the [Creative Commons by Attribution 4.0](https://creativecommons.org/licenses/by/4.0/) license.\n\n**Contact:** If you need assistance, please review the FAQ section and support options on the [EY Data Science platform](https://datascience.ey.com/).",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
e78ca0d1b16554d00d6e8a46e402d4fbbbd56a0e | 1,698 | ipynb | Jupyter Notebook | presentaties/elm-repl.ipynb | eelcodijkstra/fpbook | 6fe0b538d8f30d3518894abc1346df5932ae52e5 | [
"MIT"
] | null | null | null | presentaties/elm-repl.ipynb | eelcodijkstra/fpbook | 6fe0b538d8f30d3518894abc1346df5932ae52e5 | [
"MIT"
] | null | null | null | presentaties/elm-repl.ipynb | eelcodijkstra/fpbook | 6fe0b538d8f30d3518894abc1346df5932ae52e5 | [
"MIT"
] | null | null | null | 18.258065 | 71 | 0.495878 | [
[
[
"%%bash\nelm repl\n\nimport List exposing (..)\n\nfoldr (::) [] [1,2,3]",
"_____no_output_____"
],
[
"!touch src/just-for-elm",
"_____no_output_____"
],
[
"!ls src",
"_____no_output_____"
],
[
"%%bash\nelm repl\n\nimport String exposing (..)\nimport List exposing (..)\n\nList.foldr (\\x lst -> (fromInt x) ++ \" \" ++ lst ) \"\" [1,2,3]",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
e78cc05f35ce593eebe412d4c00b690c56705d63 | 3,456 | ipynb | Jupyter Notebook | archive/0-pre-tutorial-exercises.ipynb | ChrisKeefe/Network-Analysis-Made-Simple | 98644f0d03aa3c1ece4aa2d4147835fa10a0fcf8 | [
"MIT"
] | 853 | 2015-04-08T01:58:34.000Z | 2022-03-28T15:39:30.000Z | archive/0-pre-tutorial-exercises.ipynb | a1ip/Network-Analysis-Made-Simple | 7404c35cab8cdc9c119961ba33baef0398a20adc | [
"MIT"
] | 177 | 2015-08-08T05:33:06.000Z | 2022-03-21T15:43:07.000Z | archive/0-pre-tutorial-exercises.ipynb | a1ip/Network-Analysis-Made-Simple | 7404c35cab8cdc9c119961ba33baef0398a20adc | [
"MIT"
] | 390 | 2015-03-28T02:22:34.000Z | 2022-03-24T18:47:43.000Z | 24.167832 | 111 | 0.532118 | [
[
[
"# Pre-Tutorial Exercises\n\nIf you've arrived early for the tutorial, please feel free to attempt the following exercises to warm-up.",
"_____no_output_____"
]
],
[
[
"# 1. Basic Python data structures\n# I have a list of dictionaries as such:\n\nnames = [{'name': 'Eric',\n 'surname': 'Ma'},\n {'name': 'Jeffrey',\n 'surname': 'Elmer'},\n {'name': 'Mike',\n 'surname': 'Lee'},\n {'name': 'Jennifer',\n 'surname': 'Elmer'}]\n\n# Write a function that takes in a list of dictionaries and a query surname, \n# and searches it for all individuals with a given surname.\n\ndef find_persons_with_surname(persons, query_surname):\n # Assert that the persons parameter is a list. \n # This is a good defensive programming practice.\n assert isinstance(persons, list) \n \n results = []\n for ______ in ______:\n if ___________ == __________:\n results.append(________)\n \n return results",
"_____no_output_____"
],
[
"# Test your result below.\nresults = find_persons_with_surname(names, 'Lee')\nassert len(results) == 1\n\nresults = find_persons_with_surname(names, 'Elmer')\nassert len(results) == 2",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
]
] |
e78cc07012749ab99fe95685df82be353392f453 | 5,018 | ipynb | Jupyter Notebook | StarterCode/Belly_Button_Biodiversity/.ipynb_checkpoints/belly_button-checkpoint.ipynb | rinatiwari/RT_HW_Plot.ly---Belly-Button-Biodiversity | c2189860e3601790efac5ed6bd1606fa8f32e79a | [
"ADSL"
] | null | null | null | StarterCode/Belly_Button_Biodiversity/.ipynb_checkpoints/belly_button-checkpoint.ipynb | rinatiwari/RT_HW_Plot.ly---Belly-Button-Biodiversity | c2189860e3601790efac5ed6bd1606fa8f32e79a | [
"ADSL"
] | null | null | null | StarterCode/Belly_Button_Biodiversity/.ipynb_checkpoints/belly_button-checkpoint.ipynb | rinatiwari/RT_HW_Plot.ly---Belly-Button-Biodiversity | c2189860e3601790efac5ed6bd1606fa8f32e79a | [
"ADSL"
] | 4 | 2020-09-26T00:48:03.000Z | 2021-10-29T21:38:56.000Z | 29.174419 | 87 | 0.430849 | [
[
[
"import os\nimport pandas as pd\nimport numpy as np\n",
"_____no_output_____"
],
[
"import sqlalchemy\nfrom sqlalchemy.ext.automap import automap_base\nfrom sqlalchemy.orm import Session\nfrom sqlalchemy import create_engine\n\nfrom flask import Flask, jsonify, render_template\nfrom flask_sqlalchemy import SQLAlchemy\n\napp = Flask(__name__)",
"_____no_output_____"
],
[
"#import and read json file\nimport json\n\njson_file = \"samples.json\"\nbelly_button_json_df = pd.read_json(json_file)\nbelly_button_json_df.head()",
"_____no_output_____"
],
[
"#app.config[\"SQLALCHEMY_DATABASE_URI\"] = \"sqlite:///db/bellybutton.sqlite\"\ndb = SQLAlchemy(app)",
"_____no_output_____"
],
[
"# reflect an existing database into a new model\nBase = automap_base()\n# reflect the tables\nBase.prepare(db.engine, reflect=True)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
e78cc63987205ad23ebb3bed764a943863be92aa | 110,156 | ipynb | Jupyter Notebook | graphs.ipynb | rafacheang/social_dilemmas_regulation | a684e20b62114365d1c5d8fb9c339eb43155aa13 | [
"MIT"
] | null | null | null | graphs.ipynb | rafacheang/social_dilemmas_regulation | a684e20b62114365d1c5d8fb9c339eb43155aa13 | [
"MIT"
] | null | null | null | graphs.ipynb | rafacheang/social_dilemmas_regulation | a684e20b62114365d1c5d8fb9c339eb43155aa13 | [
"MIT"
] | null | null | null | 493.973094 | 55,068 | 0.943743 | [
[
[
"import numpy as np\nfrom numpy import genfromtxt\n\nimport matplotlib.pyplot as plt\n#import seaborn as sns",
"_____no_output_____"
],
[
"episodes_rewards = genfromtxt('logs/episodes_rewards.csv', delimiter=', ')\nno_regulator = genfromtxt('logs/no_regulator.csv', delimiter=', ')",
"_____no_output_____"
],
[
"episodes_rewards",
"_____no_output_____"
],
[
"no_regulator",
"_____no_output_____"
],
[
"no_regulator = no_regulator[:3469]",
"_____no_output_____"
],
[
"def moving_average(arr, n=5) :\n ret = np.cumsum(arr, dtype=float)\n ret[n:] = ret[n:] - ret[:-n]\n return ret[n - 1:] / n",
"_____no_output_____"
],
[
"x1 = episodes_rewards[:,0]\ny1 = episodes_rewards[:,1]\n\nx2 = no_regulator[:,0]\ny2 = no_regulator[:,1]\n\nfig, ax = plt.subplots(figsize=(12,4))\nax.plot(x1, y1)\nax.plot(x2, y2)\n\nax.set(xlabel='episode', ylabel='total consumption',\n title='Rewards')\nax.grid()\n\nfig.savefig(\"test.png\")\nplt.show()",
"_____no_output_____"
],
[
"mov_avg_rewards = moving_average(episodes_rewards[:,1])\nmov_avg_no_regulator = moving_average(no_regulator[:,1])",
"_____no_output_____"
],
[
"x1 = episodes_rewards[:3465,0]\ny1 = mov_avg_rewards\n\nx2 = no_regulator[:3465,0]\ny2 = mov_avg_no_regulator\n\nfig, ax = plt.subplots(figsize=(12,4))\nax.plot(x1, y1)\nax.plot(x2, y2)\n\nax.set(xlabel='episode', ylabel='total consumption',\n title='Total consumption per episode')\nax.grid()\n\nfig.savefig(\"rewards_plot.png\")\nplt.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78ced4abff89b763261ed8a3283e7cc04a16f69 | 13,955 | ipynb | Jupyter Notebook | Arithmetique-Le_BN_pour_calculer.ipynb | ECaMorlaix-2SI-1718/CR | 10a2acac7e476ef22e74745dc5f1792d6c1a9631 | [
"MIT"
] | null | null | null | Arithmetique-Le_BN_pour_calculer.ipynb | ECaMorlaix-2SI-1718/CR | 10a2acac7e476ef22e74745dc5f1792d6c1a9631 | [
"MIT"
] | null | null | null | Arithmetique-Le_BN_pour_calculer.ipynb | ECaMorlaix-2SI-1718/CR | 10a2acac7e476ef22e74745dc5f1792d6c1a9631 | [
"MIT"
] | 2 | 2018-01-25T13:13:33.000Z | 2018-02-01T13:09:28.000Z | 19.85064 | 447 | 0.534289 | [
[
[
"# Le Bloc Note pour calculer",
"_____no_output_____"
],
[
"Python est un langage interprété, jupyter peut donc lui faire exécuter progressivement des calculs mathématiques entre des nombres : les opérations étant saisies dans des cellules de type code, le résultat s'affichera directement en dessous.",
"_____no_output_____"
],
[
"Ainsi, cellule après cellule, notre notebook jupyter devient un document pour rendre compte de calculs successifs que l'on peut modifier et refaire à tout moment et qui peuvent être enrichis de commentaires en langage naturel.",
"_____no_output_____"
],
[
"*** \n> Ce document est un notebook jupyter, pour bien vous familiariser avec cet environnement regardez cette rapide [Introduction](Introduction-Le_BN_pour_explorer.ipynb). \n\n***",
"_____no_output_____"
],
[
"## Les opérateurs arithmétiques\n\n\n\n\n",
"_____no_output_____"
],
[
"Les additions, soustractions et multiplications sont simples et se réalisent via les opérateurs +,-,*.",
"_____no_output_____"
]
],
[
[
"4+5-3*2",
"_____no_output_____"
]
],
[
[
"Le produit est prioritaire.",
"_____no_output_____"
]
],
[
[
"4+(5-3)*2",
"_____no_output_____"
]
],
[
[
"Pour ce qui est des divisions, il existe trois opérateurs :\n- l’opérateur de division “/”, qui donne toujours un résultat avec [virgule flottante](https://fr.wikipedia.org/wiki/Virgule_flottante) en Python 3 ;\n- l’opérateur de division entière “//” ;\n- l’opérateur modulo “%” donnant le reste de la division euclidienne.",
"_____no_output_____"
]
],
[
[
"8/2",
"_____no_output_____"
],
[
"9//2",
"_____no_output_____"
],
[
"9%2",
"_____no_output_____"
]
],
[
[
"Pour élever à la puissance on utilise l'opérateur “**”",
"_____no_output_____"
]
],
[
[
"2**3",
"_____no_output_____"
]
],
[
[
"Attention, des opérations mêlant des nombres entiers et flottant donneront des résultats flottants.",
"_____no_output_____"
]
],
[
[
"13.0//3",
"_____no_output_____"
],
[
"13.0%3",
"_____no_output_____"
]
],
[
[
"On peut utiliser l'écriture scientifique pour saisir des nombres flottants :",
"_____no_output_____"
]
],
[
[
"2e-3",
"_____no_output_____"
]
],
[
[
"Pour convertir un flottant en entier et inversement on utilise respectivement les fonctions int() et float()",
"_____no_output_____"
]
],
[
[
"int(3.9)",
"_____no_output_____"
],
[
"float(3)",
"_____no_output_____"
]
],
[
[
"Pour obtenir la valeur absolue d'un nombre :",
"_____no_output_____"
]
],
[
[
"abs(-3.3)",
"_____no_output_____"
]
],
[
[
"Pour arrondir un nombre flottant par exemple à deux chiffres après la virgule :",
"_____no_output_____"
]
],
[
[
"round(3.1415926535897932384626433832795,2)",
"_____no_output_____"
]
],
[
[
"## Autres fonctions mathématiques",
"_____no_output_____"
],
[
"Pour faire appel à des fonctions mathématiques plus évoluées, il faut importer une bibliothèque tel que :",
"_____no_output_____"
]
],
[
[
"from numpy import *",
"_____no_output_____"
]
],
[
[
"L' **`*`** veut dire que nous pouvons maintenant utiliser toutes les fonctions de cette bibliothèque, telle que :",
"_____no_output_____"
]
],
[
[
"sqrt(4)",
"_____no_output_____"
],
[
"sin(pi)",
"_____no_output_____"
]
],
[
[
"> Peut-être que le résultat de cette dernière cellule vous étonne ? \nTout comme celui que produisent les cellules suivantes :",
"_____no_output_____"
]
],
[
[
"0.1+0.7",
"_____no_output_____"
],
[
"4e0+2e-1+1e-3",
"_____no_output_____"
]
],
[
[
"> Cet écart est du à la représentation des nombres [flottants](https://fr.wikipedia.org/wiki/Virgule_flottante) dans la mémoire de l'ordinateur, ce ne sont pas des valeurs exactes mais approchées. \nIl faudra donc s'en souvenir lorsqu'il s'agira d'interpréter un résultat issu d'un calcul avec des flottants, tout dépend du niveau de précision attendu...",
"_____no_output_____"
]
],
[
[
"round(0.1+0.7,3)",
"_____no_output_____"
],
[
"round(sin(pi),3)",
"_____no_output_____"
]
],
[
[
"### Pour générer un nombre aléatoire :",
"_____no_output_____"
]
],
[
[
"from numpy.random import *",
"_____no_output_____"
],
[
"rand()",
"_____no_output_____"
]
],
[
[
"Par exemple pour simuler un Dé à 6 faces",
"_____no_output_____"
]
],
[
[
"int(rint(rand()*5+1))",
"_____no_output_____"
]
],
[
[
"## Représentation Graphique d'une fonction Mathématiques",
"_____no_output_____"
],
[
"Pour tracer des courbes, si vous exécutez la fonction magique %pylab inline, les bibliothèques Numpy et Matplotlib sont importées et il sera possible de dessiner des graphiques de façon intégrés au notebook.",
"_____no_output_____"
]
],
[
[
"%pylab inline",
"_____no_output_____"
]
],
[
[
"L'exemple de code suivant sera alors exécutable.",
"_____no_output_____"
]
],
[
[
"# Fait appel à numpy (linspace et pi)\nx = linspace(0, 3*pi, 500)\n\n# Fait appel à matplotlib (plot et title)\nplot(x, sin(x))\ntitle('Graphique sin(x)')",
"_____no_output_____"
]
],
[
[
"## Ressources :\n* [Aide mémoire numpy de David RENAULT](http://www.labri.fr/perso/renault/working/teaching/algonum/sheet.php)\n* https://matplotlib.org/tutorials/index.html\n",
"_____no_output_____"
],
[
"## A vous de jouer :\n\nSaisir votre opération dans la cellule suivante, jupyter affichera le résultat calculé par Python...",
"_____no_output_____"
],
[
"*** \n\n> **Félicitations ! ** Vous êtes parvenu au bout des activités de ce bloc note. \n> Vous êtes maintenant capable d'utiliser la **calculatrice graphique Python** de l'environnement interactif jupyter notebook.\n\n> Pour explorer plus avant d'autres fonctionnalités de jupyter notebook repassez par le [Sommaire](index.ipynb).\n\n***",
"_____no_output_____"
],
[
"<a rel=\"license\" href=\"http://creativecommons.org/licenses/by-sa/4.0/\"><img alt=\"Licence Creative Commons\" style=\"border-width:0\" src=\"https://i.creativecommons.org/l/by-sa/4.0/88x31.png\" /></a><br />Ce document est mis à disposition selon les termes de la <a rel=\"license\" href=\"http://creativecommons.org/licenses/by-sa/4.0/\">Licence Creative Commons Attribution - Partage dans les Mêmes Conditions 4.0 International</a>.\n\nPour toute question, suggestion ou commentaire : <a href=\"mailto:[email protected]\">[email protected]</a>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
e78ceec4180e48bed1f53874f3a48109f64f1c19 | 15,490 | ipynb | Jupyter Notebook | docs/_downloads/68e97c325bcdbd63f73a37dc6b8c656d/buildmodel_tutorial.ipynb | YonghyunRyu/PyTorch-tutorials-kr-exercise | bb527494f304f76a4d2cf5a689f00039336fc0c1 | [
"BSD-3-Clause"
] | 221 | 2018-04-06T01:42:58.000Z | 2021-11-28T10:12:45.000Z | docs/_downloads/68e97c325bcdbd63f73a37dc6b8c656d/buildmodel_tutorial.ipynb | YonghyunRyu/PyTorch-tutorials-kr-exercise | bb527494f304f76a4d2cf5a689f00039336fc0c1 | [
"BSD-3-Clause"
] | 280 | 2018-05-25T08:53:21.000Z | 2021-12-02T05:37:25.000Z | docs/_downloads/68e97c325bcdbd63f73a37dc6b8c656d/buildmodel_tutorial.ipynb | YonghyunRyu/PyTorch-tutorials-kr-exercise | bb527494f304f76a4d2cf5a689f00039336fc0c1 | [
"BSD-3-Clause"
] | 181 | 2018-05-25T02:00:28.000Z | 2021-11-19T11:56:39.000Z | 56.739927 | 1,937 | 0.60213 | [
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"\n`파이토치(PyTorch) 기본 익히기 <intro.html>`_ ||\n`빠른 시작 <quickstart_tutorial.html>`_ ||\n`텐서(Tensor) <tensorqs_tutorial.html>`_ ||\n`Dataset과 Dataloader <data_tutorial.html>`_ ||\n`변형(Transform) <transforms_tutorial.html>`_ ||\n**신경망 모델 구성하기** ||\n`Autograd <autogradqs_tutorial.html>`_ ||\n`최적화(Optimization) <optimization_tutorial.html>`_ ||\n`모델 저장하고 불러오기 <saveloadrun_tutorial.html>`_\n\n신경망 모델 구성하기\n==========================================================================\n\n신경망은 데이터에 대한 연산을 수행하는 계층(layer)/모듈(module)로 구성되어 있습니다.\n`torch.nn <https://pytorch.org/docs/stable/nn.html>`_ 네임스페이스는 신경망을 구성하는데 필요한 모든 구성 요소를 제공합니다.\nPyTorch의 모든 모듈은 `nn.Module <https://pytorch.org/docs/stable/generated/torch.nn.Module.html>`_ 의 하위 클래스(subclass)\n입니다. 신경망은 다른 모듈(계층; layer)로 구성된 모듈입니다. 이러한 중첩된 구조는 복잡한 아키텍처를 쉽게 구축하고 관리할 수 있습니다.\n\n이어지는 장에서는 FashionMNIST 데이터셋의 이미지들을 분류하는 신경망을 구성해보겠습니다.\n",
"_____no_output_____"
]
],
[
[
"import os\nimport torch\nfrom torch import nn\nfrom torch.utils.data import DataLoader\nfrom torchvision import datasets, transforms",
"_____no_output_____"
]
],
[
[
"학습을 위한 장치 얻기\n------------------------------------------------------------------------------------------\n\n가능한 경우 GPU와 같은 하드웨어 가속기에서 모델을 학습하려고 합니다.\n`torch.cuda <https://pytorch.org/docs/stable/notes/cuda.html>`_ 를 사용할 수 있는지\n확인하고 그렇지 않으면 CPU를 계속 사용합니다.\n\n",
"_____no_output_____"
]
],
[
[
"device = 'cuda' if torch.cuda.is_available() else 'cpu'\nprint('Using {} device'.format(device))",
"_____no_output_____"
]
],
[
[
"클래스 정의하기\n------------------------------------------------------------------------------------------\n\n신경망 모델을 ``nn.Module`` 의 하위클래스로 정의하고, ``__init__`` 에서 신경망 계층들을 초기화합니다.\n``nn.Module`` 을 상속받은 모든 클래스는 ``forward`` 메소드에 입력 데이터에 대한 연산들을 구현합니다.\n\n",
"_____no_output_____"
]
],
[
[
"class NeuralNetwork(nn.Module):\n def __init__(self):\n super(NeuralNetwork, self).__init__()\n self.flatten = nn.Flatten()\n self.linear_relu_stack = nn.Sequential(\n nn.Linear(28*28, 512),\n nn.ReLU(),\n nn.Linear(512, 512),\n nn.ReLU(),\n nn.Linear(512, 10),\n nn.ReLU()\n )\n\n def forward(self, x):\n x = self.flatten(x)\n logits = self.linear_relu_stack(x)\n return logits",
"_____no_output_____"
]
],
[
[
"``NeuralNetwork`` 의 인스턴스(instance)를 생성하고 이를 ``device`` 로 이동한 뒤,\n구조(structure)를 출력합니다.\n\n",
"_____no_output_____"
]
],
[
[
"model = NeuralNetwork().to(device)\nprint(model)",
"_____no_output_____"
]
],
[
[
"모델을 사용하기 위해 입력 데이터를 전달합니다. 이는 일부\n`백그라운드 연산들 <https://github.com/pytorch/pytorch/blob/270111b7b611d174967ed204776985cefca9c144/torch/nn/modules/module.py#L866>`_ 과 함께\n모델의 ``forward`` 를 실행합니다. ``model.forward()`` 를 직접 호출하지 마세요!\n\n모델에 입력을 호출하면 각 분류(class)에 대한 원시(raw) 예측값이 있는 10-차원 텐서가 반환됩니다.\n원시 예측값을 ``nn.Softmax`` 모듈의 인스턴스에 통과시켜 예측 확률을 얻습니다.\n\n",
"_____no_output_____"
]
],
[
[
"X = torch.rand(1, 28, 28, device=device)\nlogits = model(X)\npred_probab = nn.Softmax(dim=1)(logits)\ny_pred = pred_probab.argmax(1)\nprint(f\"Predicted class: {y_pred}\")",
"_____no_output_____"
]
],
[
[
"------------------------------------------------------------------------------------------\n\n\n",
"_____no_output_____"
],
[
"모델 계층(Layer)\n------------------------------------------------------------------------------------------\n\nFashionMNIST 모델의 계층들을 살펴보겠습니다. 이를 설명하기 위해, 28x28 크기의 이미지 3개로 구성된\n미니배치를 가져와, 신경망을 통과할 때 어떤 일이 발생하는지 알아보겠습니다.\n\n",
"_____no_output_____"
]
],
[
[
"input_image = torch.rand(3,28,28)\nprint(input_image.size())",
"_____no_output_____"
]
],
[
[
"nn.Flatten\n^^^^^^^^^^^^^^^^^^^^^^\n`nn.Flatten <https://pytorch.org/docs/stable/generated/torch.nn.Flatten.html>`_ 계층을 초기화하여\n각 28x28의 2D 이미지를 784 픽셀 값을 갖는 연속된 배열로 변환합니다. (dim=0의 미니배치 차원은 유지됩니다.)\n\n",
"_____no_output_____"
]
],
[
[
"flatten = nn.Flatten()\nflat_image = flatten(input_image)\nprint(flat_image.size())",
"_____no_output_____"
]
],
[
[
"nn.Linear\n^^^^^^^^^^^^^^^^^^^^^^\n`선형 계층 <https://pytorch.org/docs/stable/generated/torch.nn.Linear.html>`_ 은 저장된 가중치(weight)와\n편향(bias)을 사용하여 입력에 선형 변환(linear transformation)을 적용하는 모듈입니다.\n\n\n",
"_____no_output_____"
]
],
[
[
"layer1 = nn.Linear(in_features=28*28, out_features=20)\nhidden1 = layer1(flat_image)\nprint(hidden1.size())",
"_____no_output_____"
]
],
[
[
"nn.ReLU\n^^^^^^^^^^^^^^^^^^^^^^\n비선형 활성화(activation)는 모델의 입력과 출력 사이에 복잡한 관계(mapping)를 만듭니다.\n비선형 활성화는 선형 변환 후에 적용되어 *비선형성(nonlinearity)* 을 도입하고, 신경망이 다양한 현상을 학습할 수 있도록 돕습니다.\n\n이 모델에서는 `nn.ReLU <https://pytorch.org/docs/stable/generated/torch.nn.ReLU.html>`_ 를 선형 계층들 사이에 사용하지만,\n모델을 만들 때는 비선형성을 가진 다른 활성화를 도입할 수도 있습니다.\n\n",
"_____no_output_____"
]
],
[
[
"print(f\"Before ReLU: {hidden1}\\n\\n\")\nhidden1 = nn.ReLU()(hidden1)\nprint(f\"After ReLU: {hidden1}\")",
"_____no_output_____"
]
],
[
[
"nn.Sequential\n^^^^^^^^^^^^^^^^^^^^^^\n`nn.Sequential <https://pytorch.org/docs/stable/generated/torch.nn.Sequential.html>`_ 은 순서를 갖는\n모듈의 컨테이너입니다. 데이터는 정의된 것과 같은 순서로 모든 모듈들을 통해 전달됩니다. 순차 컨테이너(sequential container)를 사용하여\n아래의 ``seq_modules`` 와 같은 신경망을 빠르게 만들 수 있습니다.\n\n",
"_____no_output_____"
]
],
[
[
"seq_modules = nn.Sequential(\n flatten,\n layer1,\n nn.ReLU(),\n nn.Linear(20, 10)\n)\ninput_image = torch.rand(3,28,28)\nlogits = seq_modules(input_image)",
"_____no_output_____"
]
],
[
[
"nn.Softmax\n^^^^^^^^^^^^^^^^^^^^^^\n신경망의 마지막 선형 계층은 `nn.Softmax <https://pytorch.org/docs/stable/generated/torch.nn.Softmax.html>`_ 모듈에 전달될\n([-\\infty, \\infty] 범위의 원시 값(raw value)인) `logits` 를 반환합니다. logits는 모델의 각 분류(class)에 대한 예측 확률을 나타내도록\n[0, 1] 범위로 비례하여 조정(scale)됩니다. ``dim`` 매개변수는 값의 합이 1이 되는 차원을 나타냅니다.\n\n",
"_____no_output_____"
]
],
[
[
"softmax = nn.Softmax(dim=1)\npred_probab = softmax(logits)",
"_____no_output_____"
]
],
[
[
"모델 매개변수\n------------------------------------------------------------------------------------------\n\n신경망 내부의 많은 계층들은 *매개변수화(parameterize)* 됩니다. 즉, 학습 중에 최적화되는 가중치와 편향과 연관지어집니다.\n``nn.Module`` 을 상속하면 모델 객체 내부의 모든 필드들이 자동으로 추적(track)되며, 모델의 ``parameters()`` 및\n``named_parameters()`` 메소드로 모든 매개변수에 접근할 수 있게 됩니다.\n\n이 예제에서는 각 매개변수들을 순회하며(iterate), 매개변수의 크기와 값을 출력합니다.\n\n\n",
"_____no_output_____"
]
],
[
[
"print(\"Model structure: \", model, \"\\n\\n\")\n\nfor name, param in model.named_parameters():\n print(f\"Layer: {name} | Size: {param.size()} | Values : {param[:2]} \\n\")",
"_____no_output_____"
]
],
[
[
"------------------------------------------------------------------------------------------\n\n\n",
"_____no_output_____"
],
[
"더 읽어보기\n------------------------------------------------------------------------------------------\n- `torch.nn API <https://pytorch.org/docs/stable/nn.html>`_\n\n",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
e78d0d2f236e7a60d84ce2f9b5e65c3a072a5806 | 601,957 | ipynb | Jupyter Notebook | DEEP LEARNING/image classification/fastai/fastai satellite multilabel classif.ipynb | Diyago/ML-DL-scripts | 40718a9d4318d6d6531bcea5998c0a18afcd9cb3 | [
"Apache-2.0"
] | 142 | 2018-09-02T08:59:45.000Z | 2022-03-30T17:08:24.000Z | DEEP LEARNING/image classification/fastai/fastai satellite multilabel classif.ipynb | jerinka/ML-DL-scripts | eeb5c3c7c5841eb4cdb272690e14d6718f3685b2 | [
"Apache-2.0"
] | 4 | 2019-09-08T07:27:11.000Z | 2021-10-19T05:50:24.000Z | DEEP LEARNING/image classification/fastai/fastai satellite multilabel classif.ipynb | jerinka/ML-DL-scripts | eeb5c3c7c5841eb4cdb272690e14d6718f3685b2 | [
"Apache-2.0"
] | 75 | 2018-10-04T17:08:40.000Z | 2022-03-08T18:50:52.000Z | 701.581585 | 260,798 | 0.938992 | [
[
[
"## Multi-label classification",
"_____no_output_____"
]
],
[
[
"%reload_ext autoreload\n%autoreload 2\n%matplotlib inline",
"_____no_output_____"
],
[
"from fastai.conv_learner import *",
"_____no_output_____"
],
[
"PATH = 'data/planet/'",
"_____no_output_____"
],
[
"# Data preparation steps if you are using Crestle:\n\nos.makedirs('data/planet/models', exist_ok=True)\nos.makedirs('/cache/planet/tmp', exist_ok=True)\n\n!ln -s /datasets/kaggle/planet-understanding-the-amazon-from-space/train-jpg {PATH}\n!ln -s /datasets/kaggle/planet-understanding-the-amazon-from-space/test-jpg {PATH}\n!ln -s /datasets/kaggle/planet-understanding-the-amazon-from-space/train_v2.csv {PATH}\n!ln -s /cache/planet/tmp {PATH}",
"_____no_output_____"
],
[
"ls {PATH}",
"\u001b[0m\u001b[01;34mmodels\u001b[0m/ \u001b[01;34mtest-jpg\u001b[0m/ \u001b[01;34mtmp\u001b[0m/ \u001b[01;34mtrain-jpg\u001b[0m/ \u001b[01;32mtrain_v2.csv\u001b[0m*\r\n"
]
],
[
[
"## Multi-label versus single-label classification",
"_____no_output_____"
]
],
[
[
"from fastai.plots import *",
"_____no_output_____"
],
[
"def get_1st(path): return glob(f'{path}/*.*')[0]",
"_____no_output_____"
],
[
"dc_path = \"data/dogscats/valid/\"\nlist_paths = [get_1st(f\"{dc_path}cats\"), get_1st(f\"{dc_path}dogs\")]\nplots_from_files(list_paths, titles=[\"cat\", \"dog\"], maintitle=\"Single-label classification\")",
"_____no_output_____"
]
],
[
[
"In single-label classification each sample belongs to one class. In the previous example, each image is either a *dog* or a *cat*.",
"_____no_output_____"
]
],
[
[
"list_paths = [f\"{PATH}train-jpg/train_0.jpg\", f\"{PATH}train-jpg/train_1.jpg\"]\ntitles=[\"haze primary\", \"agriculture clear primary water\"]\nplots_from_files(list_paths, titles=titles, maintitle=\"Multi-label classification\")",
"_____no_output_____"
]
],
[
[
"In multi-label classification each sample can belong to one or more clases. In the previous example, the first images belongs to two clases: *haze* and *primary*. The second image belongs to four clases: *agriculture*, *clear*, *primary* and *water*.",
"_____no_output_____"
],
[
"## Multi-label models for Planet dataset",
"_____no_output_____"
]
],
[
[
"from planet import f2\n\nmetrics=[f2]\nf_model = resnet34",
"_____no_output_____"
],
[
"label_csv = f'{PATH}train_v2.csv'\nn = len(list(open(label_csv)))-1\nval_idxs = get_cv_idxs(n)",
"_____no_output_____"
]
],
[
[
"We use a different set of data augmentations for this dataset - we also allow vertical flips, since we don't expect vertical orientation of satellite images to change our classifications.",
"_____no_output_____"
]
],
[
[
"def get_data(sz):\n tfms = tfms_from_model(f_model, sz, aug_tfms=transforms_top_down, max_zoom=1.05)\n return ImageClassifierData.from_csv(PATH, 'train-jpg', label_csv, tfms=tfms,\n suffix='.jpg', val_idxs=val_idxs, test_name='test-jpg')",
"_____no_output_____"
],
[
"data = get_data(256)",
"_____no_output_____"
],
[
"x,y = next(iter(data.val_dl))",
"_____no_output_____"
],
[
"y",
"_____no_output_____"
],
[
"list(zip(data.classes, y[0]))",
"_____no_output_____"
],
[
"plt.imshow(data.val_ds.denorm(to_np(x))[0]*1.4);",
"_____no_output_____"
],
[
"sz=64",
"_____no_output_____"
],
[
"data = get_data(sz)",
"_____no_output_____"
],
[
"data = data.resize(int(sz*1.3), 'tmp')",
"_____no_output_____"
],
[
"learn = ConvLearner.pretrained(f_model, data, metrics=metrics)",
"_____no_output_____"
],
[
"lrf=learn.lr_find()\nlearn.sched.plot()",
"_____no_output_____"
],
[
"lr = 0.2",
"_____no_output_____"
],
[
"learn.fit(lr, 3, cycle_len=1, cycle_mult=2)",
"_____no_output_____"
],
[
"lrs = np.array([lr/9,lr/3,lr])",
"_____no_output_____"
],
[
"learn.unfreeze()\nlearn.fit(lrs, 3, cycle_len=1, cycle_mult=2)",
"_____no_output_____"
],
[
"learn.save(f'{sz}')",
"_____no_output_____"
],
[
"learn.sched.plot_loss()",
"_____no_output_____"
],
[
"sz=128",
"_____no_output_____"
],
[
"learn.set_data(get_data(sz))\nlearn.freeze()\nlearn.fit(lr, 3, cycle_len=1, cycle_mult=2)",
"_____no_output_____"
],
[
"learn.unfreeze()\nlearn.fit(lrs, 3, cycle_len=1, cycle_mult=2)\nlearn.save(f'{sz}')",
"_____no_output_____"
],
[
"sz=256",
"_____no_output_____"
],
[
"learn.set_data(get_data(sz))\nlearn.freeze()\nlearn.fit(lr, 3, cycle_len=1, cycle_mult=2)",
"_____no_output_____"
],
[
"learn.unfreeze()\nlearn.fit(lrs, 3, cycle_len=1, cycle_mult=2)\nlearn.save(f'{sz}')",
"_____no_output_____"
],
[
"multi_preds, y = learn.TTA()\npreds = np.mean(multi_preds, 0)",
"_____no_output_____"
],
[
"f2(preds,y)",
"_____no_output_____"
]
],
[
[
"### End",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
e78d2b5b15483e4363240879a6aa9f62a9a30403 | 74,649 | ipynb | Jupyter Notebook | notebooks/3_1_classif_results.ipynb | Avditvs/sentiment-analysis-test | fd34bbb4487ff84cd4a970f4d77d99539c6c04ad | [
"MIT"
] | null | null | null | notebooks/3_1_classif_results.ipynb | Avditvs/sentiment-analysis-test | fd34bbb4487ff84cd4a970f4d77d99539c6c04ad | [
"MIT"
] | null | null | null | notebooks/3_1_classif_results.ipynb | Avditvs/sentiment-analysis-test | fd34bbb4487ff84cd4a970f4d77d99539c6c04ad | [
"MIT"
] | 1 | 2022-01-07T13:52:33.000Z | 2022-01-07T13:52:33.000Z | 60.789088 | 16,356 | 0.688917 | [
[
[
"# Results Classification",
"_____no_output_____"
]
],
[
[
"import os\nimport sys\n\nsys.path.append('../')\n\nimport torch\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"DATA_DIR = \"../data\"\ndata_train = pd.read_csv(os.path.join(DATA_DIR, \"train_cleaned.csv\"), na_filter=False)\ndata_val = pd.read_csv(os.path.join(DATA_DIR, \"val_cleaned.csv\"), na_filter=False)",
"_____no_output_____"
],
[
"from transformers import AutoTokenizer, AutoModelForSequenceClassification\nfrom transformers import TrainingArguments\nfrom transformers import Trainer\nfrom datasets import load_metric\n\nfrom utils.preprocessing import make_labels, tokenize\n\nfrom utils.classes import SentimentDataset",
"_____no_output_____"
],
[
"MODEL = \"xlm-roberta-base\"\nMODEL_PRETRAINED = \"../models/xlm_roberta_classif/checkpoint-1758\"\nmodel = AutoModelForSequenceClassification.from_pretrained(MODEL_PRETRAINED, num_labels=3)\ntokenizer = AutoTokenizer.from_pretrained(MODEL)",
"_____no_output_____"
],
[
"X_train = tokenize(tokenizer, data_train.content)\nX_val = tokenize(tokenizer, data_val.content)\ny_train = data_train.sentiment\ny_val = data_val.sentiment",
"_____no_output_____"
],
[
"y_train_labels = make_labels(y_train)\ny_val_labels = make_labels(y_val)",
"_____no_output_____"
],
[
"data_train = data_train.assign(label=pd.Series(y_train_labels).values)\ndata_val = data_val.assign(label=pd.Series(y_val_labels).values)",
"_____no_output_____"
],
[
"train_dataset_torch = SentimentDataset(X_train, y_train_labels)\nval_dataset_torch = SentimentDataset(X_val, y_val_labels)",
"_____no_output_____"
],
[
"metric = load_metric(\"accuracy\")",
"_____no_output_____"
],
[
"def compute_metrics(eval_pred):\n logits, labels = eval_pred\n predictions = np.argmax(logits, axis=-1)\n return metric.compute(predictions=predictions, references=labels)",
"_____no_output_____"
],
[
"training_args = TrainingArguments(\n \"bert_base_uncased_classif\",\n per_device_train_batch_size=1,\n per_device_eval_batch_size=8,\n gradient_accumulation_steps=32,\n fp16 = True,\n fp16_opt_level = 'O1',\n evaluation_strategy = 'epoch',\n save_strategy=\"epoch\",\n num_train_epochs=4\n \n)",
"_____no_output_____"
],
[
"trainer = Trainer(\n model=model,\n args=training_args,\n train_dataset=train_dataset_torch,\n eval_dataset=val_dataset_torch,\n compute_metrics=compute_metrics\n)",
"Using amp fp16 backend\n"
],
[
"val_predictions = trainer.predict(val_dataset_torch)",
"***** Running Prediction *****\n Num examples = 6250\n Batch size = 8\n"
],
[
"val_predictions_labels = np.argmax(val_predictions.predictions, axis = 1)",
"_____no_output_____"
],
[
"metric.compute(predictions=val_predictions_labels, references=val_predictions.label_ids)",
"_____no_output_____"
],
[
"from sklearn.metrics import confusion_matrix, ConfusionMatrixDisplay",
"_____no_output_____"
],
[
"conf_matrix = confusion_matrix(val_predictions.label_ids, val_predictions_labels)\ndisp = ConfusionMatrixDisplay(confusion_matrix=conf_matrix, display_labels = [\"negative\", \"neutral\", \"positive\"],)\ndisp.plot( cmap=\"cividis\")",
"_____no_output_____"
],
[
"data_val = data_val.assign(predictions=pd.Series(val_predictions_labels).values)",
"_____no_output_____"
],
[
"data_val",
"_____no_output_____"
],
[
"results_by_language = []\nfor language in set(data_val.language):\n data = data_val[data_val.language == language]\n results_by_language.append((language, metric.compute(predictions=data.predictions, references=data.label)[\"accuracy\"], len(data)))\nresults_by_language = pd.DataFrame(results_by_language, columns=[\"language\", \"accuracy\", \"size\"]).sort_values(\"size\")[::-1]",
"_____no_output_____"
],
[
"results_by_language",
"_____no_output_____"
]
],
[
[
"## Results on the main languages",
"_____no_output_____"
]
],
[
[
"main_languages = [\"en\", \"id\", \"ru\", \"ar\", \"fr\", \"es\", \"pt\", \"ko\", \"zh-cn\", \"ja\", \"de\", \"it\", \"th\", \"tr\"]",
"_____no_output_____"
],
[
" metric.compute(predictions=data_val[data_val.language.isin(main_languages)].predictions, references=data_val[data_val.language.isin(main_languages)].label)",
"_____no_output_____"
],
[
"conf_matrix = confusion_matrix(data_val[data_val.language.isin(main_languages)].label, data_val[data_val.language.isin(main_languages)].predictions)\ndisp = ConfusionMatrixDisplay(confusion_matrix=conf_matrix, display_labels = [\"negative\", \"neutral\", \"positive\"],)\ndisp.plot( cmap=\"cividis\")",
"_____no_output_____"
]
],
[
[
"## Anlayse results",
"_____no_output_____"
],
[
"Let's see what element have been misclassified",
"_____no_output_____"
],
[
"### Positive classified as negative",
"_____no_output_____"
]
],
[
[
"for sentence in data_val[data_val.language==\"en\"][data_val.label==2][data_val.predictions==0].content:\n print(sentence)",
"Did you notice that zero has a karambit knife and also that they changed the number 1 in the scoreboard and timer\n Smackgobbed? New word for me...gonna start using all the time now. \nAnime Saturday about to start. New episode of bleach. Yea \nSo tired. Finally getting some sleep. Nighty \nIf it was up to me I would give more berry's it is tottly not cool having to buy your eggs with berries I mean for real not cool update it a lift will you.\njust came from the most romantic wedding ever. the groom almost cried, which made everyone else almost cry \n핸드폰 전체 세팅에서 말고 앱 내에서 개인이나 단톡방 알림끄기하면 소리는 안나는데 배너는 계속떠요. 아예 메세지가 안온거처럼 안뜨고 들어갈경우에만 받은 메세지가 보였으면 좋겠네요 Once i turn off the notification of a person/ group, as long as I don't check messages, i wish i do not see any notification(banner) and also number of accumulated messages on the list.\n8 cm dilated and her water just broke. Getting closer \nStill thinking of Moscato, sigh... \nTomorrow Demi in Madrid I didn't win the concert to meet her LOL!\nis going to look at guitars tomorrow... woo for having money and working 2 public holidays in the next week \n typical metro, can't even get that right. \nchrome falling off or missing on a steel stand and poor stainless steel quality made utensils , returned for refund\n I am in bed. \nu will liv the app I'm not going to spoil it but your going to have to get it to s33\n will you vome to spain? \nFor No Reason Pou Can Die.\nin France tmrw \n [br] Ah! Yes. The competition is ending soon. You have just as much chance as everyone else. There's no tomfoolery in our comps \nI love Birkenstock Arizona sandals and have been wearing them for nearly 20 years. I recently bought a pair of them in HABANA LEATHER and the sandals delivered do not match the color of the picture. The shade of brown is quite different.\n Yes very kind of him to do that for such a pain in the arse like me! \nI'm still here, sorry I haven't signed on in a while, just been really busy. \n No. Wally-World is the root of all evil. Costco is not to be feared. \nIt was in the 30's last night but I dom't think it hurt anything. The elephant ears are still standing \n Not me. I'm as pure as the snow. But I drifted. \nI clocked out early! \n lol i cant w8 till yoiur film out on friday is it any good?\n oh what happened?? \nDH has just run away from his pc saying that twitter is too addictive lmao - then the sound for new message went and he came running back \n aww dont make me blush!\nis going to the SKINS party on the 2nd of May wicked!!!\ndownloaded it on a whim before going out the door, the only thing I don't like about it is the constant ads for games I've already downloaded and twitter requests before or after every game.\nAhhhhh.... Back to work. *BreBre.net*\n No invit'. You have to follow the smell of hickory and pork fat on hot coals. Next time maybe?\neeeee my first follower can't concentrate on the essay.\nPre-pandemic top vaccine companies, and where they are in COVID-19 race: $AZN (out in UK) $GSK (partner w/Sanofi, delayed) $JNJ (coming soon) $MRK (dropped out) $PFE (succeeded via small biotech partner), $SNY (partner w/GSK; delayed) (partner w/Pfizer to manufacture BNT162b2)\n Try to sport once and awhile instead of sitting behind your computer \n no not the bbc link? but have been looking at the changes, thanx hope ur well \n IMO trad segmenting in communities is not very useful \n stupid phone! It's about zune covers and designs New ones are out for spring \n hahas sucks to be you \n must have been one heck of a cloud because it's falling here \n"
]
],
[
[
" ",
"_____no_output_____"
],
[
" From what we can see, these wrongly classified sentences are not obviously positive. Example : \"hahas sucks to be you\"",
"_____no_output_____"
],
[
"### Negative classified as positive",
"_____no_output_____"
]
],
[
[
"for sentence in data_val[data_val.language==\"en\"][data_val.label==0][data_val.predictions==2].content:\n print(sentence)",
"off to college bleurgh\nI know, I know, it's exactly like mine craft. But, IT KEEPS FREEZING!!!!!!!!!! You might think it is just nothing, but trust me, it freezes all the time. I hardly get the time to play it. I'd give it 0 stars if I could.\nRest in peace, Ping. Best hamster ever. 2007-2009 \nIt's a fucking holiday. I promise this will be my last day with this bank\n Mine are grown -- does reading to your dogs count? :-D\noff for a shower. i hope i drown... im so depressed right now thanks a lot sharks\n_ I think I�ll end up going alone But I will see it at some point...\nAt Gatwick. Watch on BST, body 8 hours behind on PDT \n Thanks. 7:30 here in CA but not on Versus. \nCan't believe she's up this early on a Sunday \n omg you poooooor thing!!!! don't worry you can use my itunes or something.....\n yeah got it, but was on my way with hubby to garage etc. \nit's beautiful out, and i have a 30 page outline to do \nCouldn't be happier with this application. Lots of good tools easy to use once you get used to the interface. Not something that your young kids would like, because the interface is somewhat complicated.\nSnow mounted up to a couple of inches overnight, very pretty but I hope I don't lose my peaches, tree was in bloom \nloves sleep... wishing I was back there \nI took in 52 hardbound books by James Patterson and Lee Child that were in very good to excellent condition and was paid 27 cents per book. Yes, 27 cents per book. .\nhad quite a productive day doing art work. not much left to do now! have to go to a r.s revision session tomorrow school in holidays!!\nJust had a lovely avacado, bacon and chicken baguette - now time for more work \n I'm afraid the Trekkies have all the glory now. It's like we're back in the early '90s. \nPlay Forces of War. Its better and it cane out first. World War= (TURD) Forces of War= OYUS FORCES OF WAR NIGG@@@@@\nIts beautiful\n Great idea with the iTunes promo codes - they don't work in the UK iTunes store though \nTopic : Illusions of Greatness Went from we thought we were Elite, to average, to mediocrity and in 2020 just plain pathetic disgrace! RT _Coverage: Episode Alert March 17th, Wednesday, its Penn State Nittany Lions Talk! Tune in to hear about 20', 21', & 22' #WeAreWednesday #PennStateFootball #PSU #WeAre #Big10 #CFB #NittanyLions #HappyValley _PSU On all listening streams!\nI have installed this game 3 times now and still have not been able to play. it just keeps going to force close on kindle fire. 1 star for teasing me with looks it would be a good game.\nSo sleepy this morning! \nWish they'd stop laying people off. My 9-5 is becoming a 6-10. \nI tried it in several foods. It just wasn't for me. I gave it to my daughter who is a health nut. I suggest someone to make it known that it is a strong taste that if you don't like it you will not use it.\n Congrats!! i totally forgot to submit photos \n$2.82 for a bagel with plain cream cheese & rude waitress - Thank you New York Bagel! You just gave me the perfect excuse to save my wallet and my waistline!!!............. or maybe I'll go to Basha's for $1.20 for just as good a bagel & down home smile & friendly service\nJust good..\nMy following, followers and updates were all multiples on 10 just now. Now I'm uneasy as 2 are uneven \nis ready for bed, tons of work later \nI can't smell anything coming from this at all. I was very disappointed. The spray one is great; very refreshing and calming for bedtime.\nWeekend like this I should be at Killington \nFridge shopping 2day.....YUCK! \n_harvey RIP Budgie "Sleep now little one, in a feathery heap. No longer going cheep."\nplss add scrobling\nSeeing 15 huge workitems come by in my rss reader. Thought they were commit messages. Turns out they're just new trac tickets \nNot a bad game except for the fact that the steering is all jacked up. its hard to control the car with the arrows and the tilt type is backwards. I try to turn left amenity goes right.\nRing Relief indeed. Perhaps the ring of the cash register is what they meant? I've used the product faithfully for over 60 days (they recommend giving it 45 days) and other than improving my aim of getting drops into my ear canal I've received no other benefit from this product.Don't waste your time and money on it.\n All i'll say about Wolverine is that I had severe brain pain on walking out of that movie.. it started pretty awesome \n: How is chap stiques knee? \nbout to go to bed, without her baebe ... but she had a good time with her #1 tonight!!!\nAlways had good fried chicken from Frank's in the past but not tonight. The chicken breasts were the size of the buffalo wings.\n\nMake sure you check your order before you leave.\nWow. ...s'posed to go to the Dudie's Burger Festival thing today...but it looks like thunderstorm weather. Maybe it will clear up, eh?\n word? Me too. Condolences \nUR SUGAR 7.5ml Acrylic Poly Extension Quick Building Gel Polish Clear Pink UV Gel Colis reçu au bout d’un mois.\nTotal freak in 3 a.m\n Aww Ate Lois ? Bawiin mo na lang sa 18th birthday. :> :> I bet it would be really fun. :>\n The old ones don't give me any problems, it's the new fast-growing hybrid ones that are out of control. \nI should send Apple an angry email along the lines of "Hey d-bags, 'we fixed your computer' generally means you actually did something.". \n i thinkk you guys should take a picture. and @ reply it to me in a twitpic because i love you both of u to death. miss u \n_ i spent a whole summer with them two years ago before they got big. I see them every now and then. Havent heard them play in awhile \n"
]
],
[
[
"Same conclusion here, some texts are not obviously negative.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e78d433e0208c1a1028d94383daae06aecdc364c | 3,023 | ipynb | Jupyter Notebook | Neelesh_Video-Basic_opencv.ipynb | Shreyansh-Gupta/Open-contributions | e72a9ce2b0aa6a48081921bf8138b91ad259c422 | [
"MIT"
] | 61 | 2020-09-10T05:16:19.000Z | 2021-11-07T00:22:46.000Z | Neelesh_Video-Basic_opencv.ipynb | Shreyansh-Gupta/Open-contributions | e72a9ce2b0aa6a48081921bf8138b91ad259c422 | [
"MIT"
] | 72 | 2020-09-12T09:34:19.000Z | 2021-08-01T17:48:46.000Z | Neelesh_Video-Basic_opencv.ipynb | Shreyansh-Gupta/Open-contributions | e72a9ce2b0aa6a48081921bf8138b91ad259c422 | [
"MIT"
] | 571 | 2020-09-10T01:52:56.000Z | 2022-03-26T17:26:23.000Z | 27.990741 | 115 | 0.540853 | [
[
[
"**Guide**\n\n* Create a draw_circle function for the callback function\n* Use two events cv2.EVENT_LBUTTONDOWN and cv2.EVENT_LBUTTONUP\n* Use a boolean variable to keep track if the mouse has been clicked up and down based on the events above\n* Use a tuple to keep track of the x and y where the mouse was clicked.\n* You should be able to then draw a circle on the frame based on the x,y coordinates from the Event \n\nCheck out the skeleton guide below:",
"_____no_output_____"
]
],
[
[
"# Create a function based on a CV2 Event (Left button click)\n\n# mouse callback function\ndef draw_circle(event,x,y,flags,param):\n\n global center,clicked\n\n # get mouse click on down and track center\n if event == cv2.EVENT_LBUTTONDOWN:\n center = (x, y)\n clicked = False\n \n # Use boolean variable to track if the mouse has been released\n if event == cv2.EVENT_LBUTTONUP:\n clicked = True\n\n \n# Haven't drawn anything yet!\ncenter = (0,0)\nclicked = False\n\n\n# Capture Video\ncap = cv2.VideoCapture(0) \n\n# Create a named window for connections\ncv2.namedWindow('Test')\n\n# Bind draw_rectangle function to mouse cliks\ncv2.setMouseCallback('Test', draw_circle) \n\n\nwhile True:\n # Capture frame-by-frame\n ret, frame = cap.read()\n\n # Use if statement to see if clicked is true\n if clicked==True:\n # Draw circle on frame\n cv2.circle(frame, center=center, radius=50, color=(255,0,0), thickness=5)\n \n # Display the resulting frame\n cv2.imshow('Test', frame)\n\n # This command let's us quit with the \"q\" button on a keyboard.\n # Simply pressing X on the window won't work!\n if cv2.waitKey(1) & 0xFF == ord('q'):\n break\n\n# When everything is done, release the capture\ncap.release()\ncv2.destroyAllWindows()",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
e78d600c8a2a7f033e76b06371a55522ab8a3f24 | 8,558 | ipynb | Jupyter Notebook | 6 Sentinel-1 SLC Parallel Processing.ipynb | jamesemwheeler/OSTParallel | 166fdbcbab7c71b69aa01e81f4138c8f3599967e | [
"MIT"
] | null | null | null | 6 Sentinel-1 SLC Parallel Processing.ipynb | jamesemwheeler/OSTParallel | 166fdbcbab7c71b69aa01e81f4138c8f3599967e | [
"MIT"
] | null | null | null | 6 Sentinel-1 SLC Parallel Processing.ipynb | jamesemwheeler/OSTParallel | 166fdbcbab7c71b69aa01e81f4138c8f3599967e | [
"MIT"
] | null | null | null | 25.244838 | 444 | 0.571979 | [
[
[
"# **Process S1 SLC data using parallel processing**\n",
"_____no_output_____"
],
[
"First import all necessary libraries",
"_____no_output_____"
]
],
[
[
"import ost \nimport ost.helpers as h\nfrom ost.helpers import onda, asf_wget, vector\nfrom ost import Sentinel1_SLCBatch\nimport os\nfrom os.path import join\nfrom pathlib import Path\nfrom pprint import pprint",
"_____no_output_____"
]
],
[
[
"Ingest shapefile data and set start and end dates",
"_____no_output_____"
]
],
[
[
"\n# create a processing directory\nproject_dir = '/home/ost/Data/jwheeler/Sydney_Fires'\n\n# apply function with buffer in meters\nfrom ost.helpers import vector\ninput_shp = \"/home/ost/Data/jwheeler/Shapefiles/Sydney_fires.shp\"\naoi = vector.shp_to_wkt(input_shp)\n#----------------------------\n# Time of interest\n#----------------------------\n# we set only the start date to today - 30 days\nstart = '2019-11-30'\nend = '2019-12-12'\n",
"_____no_output_____"
]
],
[
[
"Initiate class with above attributes",
"_____no_output_____"
]
],
[
[
"# create s1Project class instance\ns1_batch = Sentinel1_SLCBatch(\n project_dir=project_dir, \n aoi=aoi, \n start=start, \n end=end, \n product_type='SLC',\n ard_type='OST Plus')",
"_____no_output_____"
]
],
[
[
"Search for images on scihub and plot footprints",
"_____no_output_____"
]
],
[
[
"#---------------------------------------------------\n# for plotting purposes we use this iPython magic\n%matplotlib inline\n%pylab inline\npylab.rcParams['figure.figsize'] = (19, 19)\n#---------------------------------------------------\n\n# search command\ns1_batch.search()\n# we plot the full Inventory on a map\ns1_batch.plot_inventory(transparency=.1)",
"_____no_output_____"
]
],
[
[
"Refine image search",
"_____no_output_____"
]
],
[
[
"s1_batch.refine()",
"_____no_output_____"
]
],
[
[
"Select appropriate key and plot filtered images",
"_____no_output_____"
]
],
[
[
"pylab.rcParams['figure.figsize'] = (13, 13)\n\n\nkey = 'DESCENDING_VVVH'\ns1_batch.refined_inventory_dict[key]\ns1_batch.plot_inventory(s1_batch.refined_inventory_dict[key], 0.3)",
"_____no_output_____"
]
],
[
[
"Download using a selected S-1 mirror - ideally ASF (2 using request or 5 using wget) or onda (4) if accounts are set up correctly for fast, parallel downloading",
"_____no_output_____"
]
],
[
[
"s1_batch.download(s1_batch.refined_inventory_dict[key],concurrent=8)",
"_____no_output_____"
]
],
[
[
"Create inventory of bursts in downloaded images, plot them and print information",
"_____no_output_____"
]
],
[
[
"s1_batch.create_burst_inventory(key=key, refine=True)\npylab.rcParams['figure.figsize'] = (13, 13)\ns1_batch.plot_inventory(s1_batch.burst_inventory, transparency=0.1)\nprint('Our burst inventory holds {} bursts to process.'.format(len(s1_batch.burst_inventory)))\nprint('------------------------------------------')\nprint(s1_batch.burst_inventory.head())",
"_____no_output_____"
]
],
[
[
"Uncomment the below command to view the current ard parameters",
"_____no_output_____"
]
],
[
[
"#pprint(s1_batch.ard_parameters)",
"_____no_output_____"
]
],
[
[
"Run the s1SLCbatch class function bursts to ard to generate parameter text files for each step from burst to ard, ard to timeseries, timeseries to timescan and mosaic.\n\n**NB Use a base name for the exec file without a extension AND make sure to choose the number of cores that each process will use for parallel processing. ncores in this function x multiproc in the multiprocess function should not exceed the number of cores on your machine**",
"_____no_output_____"
]
],
[
[
"s1_batch.bursts_to_ard(timeseries=True, timescan=True, mosaic=True, overwrite=False, exec_file='/home/ost/Data/jwheeler/Sydney_Fires/test', ncores=2)\n#print(s1_batch.temp_dir)",
"_____no_output_____"
]
],
[
[
"Run the s1SLCbatch class function multiprocessing to run, sequentially, the parameters in the previously generated text files for each step from burst to ard, ard to timeseries, timeseries to timescan and mosaic.\n\n**NB Use the same base name for the exec file without a extension as before AND make sure to choose the number of cores that each process will use for parallel processing as well as the number of concurrent processes. ncores in the previous function x multiproc in this function should not exceed the number of cores on your machine. You should also include the ncores again as the text file generation is reiterated during this process**",
"_____no_output_____"
]
],
[
[
"s1_batch.multiprocess(timeseries=True, timescan=True, mosaic=True, overwrite=False, exec_file='/home/ost/Data/jwheeler/Sydney_Fires/test', ncores=2,multiproc=4)\n#burst_to_ard_batch(s1_batch.burst_inventory,s1_batch.download_dir,s1_batch.processing_dir,s1_batch.temp_dir,s1_batch.proc_file,exec_file='/home/ost/Data/jwheeler/Sydney_Fires/test.txt')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e78d6322e711f25ccbf66e9c28579adb011dce63 | 1,110 | ipynb | Jupyter Notebook | Anjani/Leetcode/Array/Kth Missing Positive Number.ipynb | Anjani100/competitive-coding | 229e4475487412c702e99a45d8ec4f46e6aea241 | [
"MIT"
] | null | null | null | Anjani/Leetcode/Array/Kth Missing Positive Number.ipynb | Anjani100/competitive-coding | 229e4475487412c702e99a45d8ec4f46e6aea241 | [
"MIT"
] | null | null | null | Anjani/Leetcode/Array/Kth Missing Positive Number.ipynb | Anjani100/competitive-coding | 229e4475487412c702e99a45d8ec4f46e6aea241 | [
"MIT"
] | 2 | 2020-10-07T13:48:02.000Z | 2022-03-31T16:10:36.000Z | 18.196721 | 42 | 0.440541 | [
[
[
"def findKthPositive(arr, k):\n i = 1\n count = 0\n while True:\n if i not in arr:\n count += 1\n if count == k:\n break\n i += 1\n return i\n\nprint(findKthPositive([1,2,3,4], 2))",
"6\n"
]
]
] | [
"code"
] | [
[
"code"
]
] |
e78d643c2c00df75d9ff9bbeedf96397063939db | 12,452 | ipynb | Jupyter Notebook | Lab 6 Perceptron/Perceptron.ipynb | xup5/Computational-Neuroscience-Class | bc0593b6895fc8d1f92097ab9a612722b754e422 | [
"MIT"
] | null | null | null | Lab 6 Perceptron/Perceptron.ipynb | xup5/Computational-Neuroscience-Class | bc0593b6895fc8d1f92097ab9a612722b754e422 | [
"MIT"
] | null | null | null | Lab 6 Perceptron/Perceptron.ipynb | xup5/Computational-Neuroscience-Class | bc0593b6895fc8d1f92097ab9a612722b754e422 | [
"MIT"
] | 3 | 2022-01-17T16:50:04.000Z | 2022-01-23T07:21:23.000Z | 32.342857 | 340 | 0.488195 | [
[
[
"<a href=\"https://colab.research.google.com/github/xup5/Computational-Neuroscience-Class/blob/main/Lab%206%20Perceptron/Perceptron.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Perceptron\n\nThis is a simple example illustrating the classic perceptron algorithm. A linear decision function parametrized by the weight vector \"w\" and bias paramter \"b\" is learned by making small adjustements to these parameters every time the predicted label \"f_i\" mismatches the true label \"y_i\" of an input data point \"x_i\".\n\nThe predicted label corresponds to the following function:\n\n f_i = sign( < w, x_i> + b),\n\nwhere \"< . , . >\" denotes the dot product operation.\nThe update rule is given as follows:\n\nif \"y_i != f_i\" (predicted label f_i different from true label y_i) then\n\n w <-- w + y_i*x_i; and \n b <-- b + y_i;\n\nelse continue with next data sample The above process is repeated over the set of samples several times.\n\nIf data points are linearly separable the above rule is guaranteed to converge in a finite number of iterations. Proof see: http://www.cs.columbia.edu/~mcollins/courses/6998-2012/notes/perc.converge.pdf\n\n\n2016 Luis G Sanchez Giraldo and Odelia Schwartz. Transcribed and modified to Python by Xu Pan, 2022.",
"_____no_output_____"
]
],
[
[
"# Construct a simple data set based on MNIST images\n# This is a data set of handwritten digits 0 to 9\n\n# Download MNIST dataset from keras\n\nfrom keras.datasets import mnist\nimport numpy as np\n\n(train_X, train_y), (test_X, test_y) = mnist.load_data()\nprint(test_X.shape)\nprint(test_y.shape)",
"_____no_output_____"
],
[
"# y are the digit labels\nprint(train_y[0:10])",
"_____no_output_____"
],
[
"# Sort dataset by label.\n\nsorted_test_X =[[], [], [], [], [], [], [], [], [], []]\nfor i, y in enumerate(test_y):\n sorted_test_X[y].append(test_X[i,:,:])",
"_____no_output_____"
],
[
"# Create a simple two-class problem using images of digits 0 and 5 from\n# the MNIST test data set \npos_class = 0 # 3\nneg_class = 5",
"_____no_output_____"
],
[
"# get samples from positive and negative classes \npos_data = np.array(sorted_test_X[pos_class])\nneg_data = np.array(sorted_test_X[neg_class])\nprint(pos_data.shape)\nprint(neg_data.shape)",
"_____no_output_____"
],
[
"# Look at some digits from the classes\n# Look at different samples from each class (here plotted just the first; try others)\n\nimport matplotlib.pyplot as plt\n\nplt.imshow(pos_data[0,:,:], cmap='binary')\nplt.show()\nplt.imshow(neg_data[0,:,:], cmap='binary')\nplt.show()",
"_____no_output_____"
],
[
"# Gather the samples from the two classes into one matrix X\n# Note that there the samples from each class are appended \n# to make up 1872 samples altogether\n\nX = np.concatenate((pos_data, neg_data), axis=0)/255\nX = np.reshape(X, (X.shape[0],-1))\nprint(X.shape)",
"_____no_output_____"
],
[
"# Label the two classes with 1 and -1 respectively\nY = np.concatenate((np.ones(pos_data.shape[0]), -np.ones(neg_data.shape[0])), axis=0);\nprint(Y.shape)\nprint(Y[0:10])\nprint(Y[981:991])",
"_____no_output_____"
],
[
"# Choose random samples from data. To do so:\n# permute data samples to run the learning algorithm \n# and take just n_samples from the permuted data (here 60 samples)\n\nn_samples = 60\n\np_idx = np.random.permutation(X.shape[0])\nX = X[p_idx[0:n_samples], :]\nY = Y[p_idx[0:n_samples]]",
"_____no_output_____"
],
[
"# Project the data onto the means of the two classes\n# First look at the mean of the two class\n\nplt.imshow(np.reshape(np.mean(X[Y == 1, :], axis=0),(28,28)), cmap='binary')\nplt.show()\n\n# mean of second class\nplt.imshow(np.reshape(np.mean(X[Y == -1, :], axis=0),(28,28)), cmap='binary')\nplt.show()\n",
"_____no_output_____"
],
[
"# Now project the data\n\nV = np.stack((np.mean(X[Y == 1, :], axis=0), np.mean(X[Y == -1, :], axis=0)), axis=1)\nV[:, 0] = V[:, 0]/np.linalg.norm(V[:, 0])\nV[:, 1] = V[:, 1]/np.linalg.norm(V[:, 1])\n\nZ = np.matmul(X,V)\nprint(Z.shape)\nplt.figure(figsize=(5,5))\nplt.scatter(Z[Y == 1,0], Z[Y == 1,1], c='g')\nplt.scatter(Z[Y == -1,0], Z[Y == -1,1], c='r')",
"_____no_output_____"
],
[
"# This is a helper function that plots the partition.\n\ndef display2DPartition(Z, Y, w, b):\n plt.figure(figsize=(5,5))\n ax = plt.axes()\n plt.scatter(Z[Y == 1,0], Z[Y == 1,1], c='g', zorder=1)\n plt.scatter(Z[Y == -1,0], Z[Y == -1,1], c='r', zorder=2)\n ax.set_facecolor('pink')\n x = ax.get_xlim()\n y = ax.get_ylim()\n y_line = -(np.array(x)*w[0]+b)/w[1]\n plt.fill_between(x,y_line, color='palegreen',zorder=0)\n for iSmp in range(Z.shape[0]):\n z_i = Z[iSmp, :]\n # compute prediction with current w and b\n f_i = np.sign(np.dot(w, z_i) + b)\n if f_i != Y[iSmp]:\n plt.scatter(z_i[0], z_i[1], marker='o', s=100, facecolors='none',linewidths=2, edgecolors='black', zorder=3)\n ax.set_xlim(x)\n ax.set_ylim(y)",
"_____no_output_____"
],
[
"# Simple Learning algorithm for Perceptron\n# here we denote two classes: the positive class by label \"1\" and the negative\n# class by label \"-1.\" \n# Any point in the plane colored as green will be classified as positive\n# class and any point falling within the red region as negative class.\n# Training samples are denoted by the green crosses (positive) and red dots\n# (negative). A missclassified training point, that is \"f_i != y_i\" is \n# marked with a circle \n\nfrom IPython.display import clear_output\nimport time\n\n# first initialize the parameters\nlr = 1 # Learning rate parameter (1 in the classic perceptron algorithm)\nw = np.random.randn(Z.shape[1]) # Initial guess for the hyperplane parameters\nprint(w)\nprint(b)\nb = 0 # bias is initially zero\nmax_epoch = 100 # Number of epoch (complete loops trough all data)\nepoch = 1 # epoch counter\n\n# display the starting decision hyhyperplane (partition of the space)\n# (compare this to the decision hyperplane that is learned!)\ndisplay2DPartition(Z, Y, w, b)\nplt.show()\n",
"_____no_output_____"
],
[
"# now run the perceptron training\nwhile epoch <= max_epoch:\n # loop trough all data points one time (an epoch)\n for iSmp in range(n_samples):\n z_i = Z[iSmp, :]\n # compute prediction with current w and b\n f_i = np.sign(np.dot(w, z_i) + b)\n # update w and b if missclassified\n if f_i != Y[iSmp]:\n w = w + lr*Y[iSmp]*z_i\n b = b + lr*Y[iSmp]\n # diplay current decision hyperplane (partition of the space)\n clear_output(wait=True)\n display2DPartition(Z, Y, w, b)\n plt.show()\n time.sleep(0.1)\n epoch = epoch + 1;",
"_____no_output_____"
],
[
"# display the learned decision hyperplane (partition of the space)\nprint(w)\nprint(b)\ndisplay2DPartition(Z, Y, w, b)\nplt.show()",
"_____no_output_____"
]
],
[
[
"###Exercise: \nAfter trying the code for the given classes,\ntry running the code again, but this time changing the digits of the\npositive or negative class. \nYou can do this by changing the following two lines above:\n\npos_class = 0\n\nneg_class = 5\n\nWhat classes are easier to learn?",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
e78d817988b17093aaa80a6505e4e6b8991d4e20 | 24,692 | ipynb | Jupyter Notebook | scrapings/americanas/americanas_scraper_diario.ipynb | Felipeecp/desafio-3-trilhas-webscraping | f3b8241a71243ab98db67d8a7437dcf527e2bffa | [
"MIT"
] | null | null | null | scrapings/americanas/americanas_scraper_diario.ipynb | Felipeecp/desafio-3-trilhas-webscraping | f3b8241a71243ab98db67d8a7437dcf527e2bffa | [
"MIT"
] | null | null | null | scrapings/americanas/americanas_scraper_diario.ipynb | Felipeecp/desafio-3-trilhas-webscraping | f3b8241a71243ab98db67d8a7437dcf527e2bffa | [
"MIT"
] | 4 | 2021-10-07T01:18:30.000Z | 2022-01-23T13:31:05.000Z | 39.131537 | 216 | 0.438928 | [
[
[
"import pandas as pd\nfrom bs4 import BeautifulSoup\nfrom selenium import webdriver\nfrom webdriver_manager.chrome import ChromeDriverManager\nfrom datetime import date\nimport time",
"_____no_output_____"
],
[
"def scraper(pesquisa):\n \n url = f'https://www.americanas.com.br/busca/{pesquisa}?limit=75&offset=0'\n driver = webdriver.Chrome(ChromeDriverManager().install())\n driver.get(url)\n time.sleep(5)\n soup = BeautifulSoup(driver.page_source, 'html.parser')\n itens = soup.body.find('div',{'id':'root'}).find('div',{'class':'grid__StyledGrid-sc-1man2hx-0 iFeuoP'}).findAll('div',{'class':'col__StyledCol-sc-1snw5v3-0 epVkvq src__ColGridItem-sc-122lblh-0 bvfSKS'})\n\n # 'https://www.americanas.com.br' + itens[0].a.get(\"href\")\n\n cards = []\n for iten in itens:\n card = {}\n card['Descrição'] = iten.h3.text\n card['Preço'] = iten.find('span',{'class':'src__Text-sc-154pg0p-0 src__Price-sc-1k0ejj6-7 dvVMTs'}).text\n # card['Preço Promocional'] = iten.find('span',{'class':'src__Text-sc-154pg0p-0 src__PromotionalPrice-sc-1k0ejj6-8 gxxqGt'}).text\n # card['Url'] = 'https://www.americanas.com.br' + itens[0].a.get(\"href\")\n card['Data'] = date.today()\n cards.append(card)\n \n dataset = pd.DataFrame(cards)\n dataset.to_csv(f'output/diario/dataset_{pesquisa}_{date.today().day}.csv'.replace(' ','_'), index = False, encoding = 'utf-8-sig', sep=\";\")",
"_____no_output_____"
],
[
"scraper('smartphone')",
"\n\n====== WebDriver manager ======\nCurrent google-chrome version is 95.0.4638\nGet LATEST driver version for 95.0.4638\nDriver [C:\\Users\\Felip\\.wdm\\drivers\\chromedriver\\win32\\95.0.4638.54\\chromedriver.exe] found in cache\n"
],
[
"dataset = pd.read_csv(f'output/diario/dataset_smartphone_{date.today().day}.csv', sep=';')\ndataset.dropna(inplace=True)\ndataset.reset_index(drop=True, inplace=True)\ndataset",
"_____no_output_____"
],
[
"scraper('notebook')",
"\n\n====== WebDriver manager ======\nCurrent google-chrome version is 95.0.4638\nGet LATEST driver version for 95.0.4638\nDriver [C:\\Users\\Felip\\.wdm\\drivers\\chromedriver\\win32\\95.0.4638.54\\chromedriver.exe] found in cache\n"
],
[
"dataset = pd.read_csv(f'output/diario/dataset_notebook_{date.today().day}.csv', sep=';')\ndataset.dropna(inplace=True)\ndataset.reset_index(drop=True, inplace=True)\ndataset",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78d9a4cf3800b0d7e29af06b26603a9b4f0b296 | 51,868 | ipynb | Jupyter Notebook | _doc/notebooks/2016/pydata/im_biopython.ipynb | sdpython/jupytalk | 34abdf128de24becb21a9f08f243c3a74dadbfd9 | [
"MIT"
] | null | null | null | _doc/notebooks/2016/pydata/im_biopython.ipynb | sdpython/jupytalk | 34abdf128de24becb21a9f08f243c3a74dadbfd9 | [
"MIT"
] | 16 | 2016-11-13T19:52:35.000Z | 2021-12-29T10:59:41.000Z | _doc/notebooks/2016/pydata/im_biopython.ipynb | sdpython/jupytalk | 34abdf128de24becb21a9f08f243c3a74dadbfd9 | [
"MIT"
] | 4 | 2016-09-10T10:44:50.000Z | 2021-09-22T16:28:56.000Z | 124.983133 | 2,924 | 0.641031 | [
[
[
"# biopython\n\nThe [Biopython](http://biopython.org/) Project is an international association of developers of freely available [Python](http://www.python.org) tools for computational molecular biology.",
"_____no_output_____"
],
[
"[documentation](http://biopython.org/wiki/Documentation) [source](https://github.com/biopython/biopython) [installation](http://biopython.org/wiki/Download) [tutorial](http://biopython.org/DIST/docs/tutorial/Tutorial.html) ",
"_____no_output_____"
]
],
[
[
"from jyquickhelper import add_notebook_menu\nadd_notebook_menu()",
"_____no_output_____"
]
],
[
[
"## example",
"_____no_output_____"
]
],
[
[
"from pyquickhelper.filehelper import download\ndownload(\"https://raw.githubusercontent.com/biopython/biopython/master/Tests/GenBank/NC_005816.gb\",\n outfile=\"NC_005816.gb\")",
"_____no_output_____"
],
[
"from reportlab.lib import colors\nfrom reportlab.lib.units import cm\nfrom Bio.Graphics import GenomeDiagram\nfrom Bio import SeqIO\nrecord = SeqIO.read(\"NC_005816.gb\", \"genbank\")\n\ngd_diagram = GenomeDiagram.Diagram(\"Yersinia pestis biovar Microtus plasmid pPCP1\")\ngd_track_for_features = gd_diagram.new_track(1, name=\"Annotated Features\")\ngd_feature_set = gd_track_for_features.new_set()\n\nfor feature in record.features:\n if feature.type != \"gene\":\n #Exclude this feature\n continue\n if len(gd_feature_set) % 2 == 0:\n color = colors.blue\n else:\n color = colors.lightblue\n gd_feature_set.add_feature(feature, color=color, label=True)\n\ngd_diagram.draw(format=\"linear\", orientation=\"landscape\", pagesize='A4',\n fragments=4, start=0, end=len(record))\ngd_diagram.write(\"plasmid_linear.svg\", \"svg\")",
"_____no_output_____"
],
[
"from IPython.display import SVG\nSVG(\"plasmid_linear.svg\")",
"_____no_output_____"
],
[
"gd_diagram.draw(format=\"circular\", circular=True, pagesize=(20*cm,20*cm),\n start=0, end=len(record), circle_core=0.7)\ngd_diagram.write(\"plasmid_circular.svg\", \"svg\")\nSVG(\"plasmid_circular.svg\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
e78da1d21d1ba24bec350a0977b5145acee35542 | 15,586 | ipynb | Jupyter Notebook | covid19_who_situation_reports_importer/notebook.ipynb | aarohijohal/covid19-who-situation-reports-importer | e124de725d4aaaa9ebaef178c5efd39bbee47e59 | [
"Apache-2.0"
] | 2 | 2020-03-31T07:19:58.000Z | 2020-03-31T10:10:03.000Z | covid19_who_situation_reports_importer/notebook.ipynb | aarohijohal/covid19-who-situation-reports-import-parsr | e124de725d4aaaa9ebaef178c5efd39bbee47e59 | [
"Apache-2.0"
] | null | null | null | covid19_who_situation_reports_importer/notebook.ipynb | aarohijohal/covid19-who-situation-reports-import-parsr | e124de725d4aaaa9ebaef178c5efd39bbee47e59 | [
"Apache-2.0"
] | null | null | null | 32.202479 | 122 | 0.325805 | [
[
[
"# Fetching WHO's situation reports on COVID-19 as DataFrames",
"_____no_output_____"
],
[
"## Get the data",
"_____no_output_____"
]
],
[
[
"pdf_save_location = '../data/pdf'\ncsv_save_location = '../data/csv'\n\nfrom who_covid_scraper import WHOCovidScraper\nscraper = WHOCovidScraper('https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports')\n\nscraper.df",
"_____no_output_____"
]
],
[
[
"## Download report for a given date",
"_____no_output_____"
]
],
[
[
"download = scraper.download_for_date(datearg='23rd of Feb', folder=pdf_save_location)",
"report for the date 2020/02/23 already exists at ../data/pdf/20200223-sitrep-34-covid-19.pdf. didn't re-download\n"
]
],
[
[
"## Send report for extraction",
"_____no_output_____"
]
],
[
[
"job = scraper.send_document_to_parsr(download['file'])\njob",
"> Polling server for the job f214dea6618020da1a446307879c1f...\n>> Job done!\n"
]
],
[
[
"## Assemble the stats from the report",
"_____no_output_____"
]
],
[
[
"scraper.assemble_data(job['server_response'])",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e78db385eee2308bad99741508f87bc69d74d56f | 128,597 | ipynb | Jupyter Notebook | boards/Pynq-Z2/logictools/notebooks/pattern_generator_and_trace_analyzer.ipynb | jackrosenthal/PYNQ | 788bf18529bc7a0564af4033ef3e246c03fc5b10 | [
"BSD-3-Clause"
] | 1,537 | 2016-09-26T22:51:50.000Z | 2022-03-31T13:33:54.000Z | boards/Pynq-Z2/logictools/notebooks/pattern_generator_and_trace_analyzer.ipynb | MakarenaLabs/PYNQ | 6f3113278e62b23315cf4e000df8f57fb53c4f6d | [
"BSD-3-Clause"
] | 414 | 2016-10-03T21:12:10.000Z | 2022-03-21T14:55:02.000Z | boards/Pynq-Z2/logictools/notebooks/pattern_generator_and_trace_analyzer.ipynb | MakarenaLabs/PYNQ | 6f3113278e62b23315cf4e000df8f57fb53c4f6d | [
"BSD-3-Clause"
] | 826 | 2016-09-23T22:29:43.000Z | 2022-03-29T11:02:09.000Z | 152.546856 | 59,660 | 0.909905 | [
[
[
"# Pattern Generator and Trace Analyzer\n\nThis notebook will show how to use the Pattern Generator to generate patterns on I/O pins. The pattern that will be generated is 3-bit up count performed 4 times. ",
"_____no_output_____"
],
[
"### Step 1: Download the `logictools` overlay",
"_____no_output_____"
]
],
[
[
"from pynq.overlays.logictools import LogicToolsOverlay\n\nlogictools_olay = LogicToolsOverlay('logictools.bit')",
"_____no_output_____"
]
],
[
[
"### Step 2: Create WaveJSON waveform\nThe pattern to be generated is specified in the waveJSON format \n\nThe pattern is applied to the Arduino interface, pins **D0**, **D1** and **D2** are set to generate a 3-bit count. \nTo check the generated pattern we loop them back to pins **D19**, **D18** and **D17** respectively and use the the trace analyzer to view the loopback signals\n\nThe Waveform class is used to display the specified waveform.",
"_____no_output_____"
]
],
[
[
"from pynq.lib.logictools import Waveform\n\nup_counter = {'signal': [\n ['stimulus',\n {'name': 'bit0', 'pin': 'D0', 'wave': 'lh' * 8},\n {'name': 'bit1', 'pin': 'D1', 'wave': 'l.h.' * 4},\n {'name': 'bit2', 'pin': 'D2', 'wave': 'l...h...' * 2}], \n \n ['analysis',\n {'name': 'bit2_loopback', 'pin': 'D17'},\n {'name': 'bit1_loopback', 'pin': 'D18'},\n {'name': 'bit0_loopback', 'pin': 'D19'}]], \n\n 'foot': {'tock': 1},\n 'head': {'text': 'up_counter'}}\n\nwaveform = Waveform(up_counter)\nwaveform.display()",
"_____no_output_____"
]
],
[
[
"**Note:** Since there are no captured samples at this moment, the analysis group will be empty.",
"_____no_output_____"
],
[
"### Step 3: Instantiate the pattern generator and trace analyzer objects\nUsers can choose whether to use the trace analyzer by calling the `trace()` method. \nThe analyzer can be set to trace a specific number of samples using, `num_analyzer_samples` argument.",
"_____no_output_____"
]
],
[
[
"pattern_generator = logictools_olay.pattern_generator\npattern_generator.trace(num_analyzer_samples=16)",
"_____no_output_____"
]
],
[
[
"### Step 4: Setup the pattern generator\nThe pattern generator will work at the default frequency of 10MHz. This can be modified using a `frequency` argument in the `setup()` method. ",
"_____no_output_____"
]
],
[
[
"pattern_generator.setup(up_counter,\n stimulus_group_name='stimulus',\n analysis_group_name='analysis')",
"_____no_output_____"
]
],
[
[
"__Set the loopback connections using jumper wires on the Arduino Interface__\n\n\n* __Output pins D0, D1 and D2 are connected to pins D19, D18 and D17 respectively__ \n* __Loopback/Input pins D19, D18 and D17 are observed using the trace analyzer as shown below__\n* __After setup, the pattern generator should be ready to run__\n\n**Note:** Make sure all other pins are disconnected.",
"_____no_output_____"
],
[
"### Step 5: Run and display waveform\n\nThe ` run()` method will execute all the samples, `show_waveform()` method is used to display the waveforms. \nAlternatively, we can also use `step()` method to single step the pattern.",
"_____no_output_____"
]
],
[
[
"pattern_generator.run()\npattern_generator.show_waveform()",
"_____no_output_____"
]
],
[
[
"### Step 6: Stop the pattern generator\nCalling `stop()` will clear the logic values on output pins; however, the waveform will be recorded locally in the pattern generator instance.",
"_____no_output_____"
]
],
[
[
"pattern_generator.stop()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e78dbc6181e14ced9be0c60d30a30faf92eb20d0 | 16,441 | ipynb | Jupyter Notebook | algorithms/KNearestNeighbor.ipynb | viga111/machine-learning | 9b2fe3f2399709745f16f4b8c480a811961773aa | [
"MIT"
] | 1 | 2021-09-23T11:56:42.000Z | 2021-09-23T11:56:42.000Z | algorithms/KNearestNeighbor.ipynb | viga111/machine-learning | 9b2fe3f2399709745f16f4b8c480a811961773aa | [
"MIT"
] | null | null | null | algorithms/KNearestNeighbor.ipynb | viga111/machine-learning | 9b2fe3f2399709745f16f4b8c480a811961773aa | [
"MIT"
] | null | null | null | 28.84386 | 370 | 0.484216 | [
[
[
"**<font color=black size=5>K近邻</font>**",
"_____no_output_____"
],
[
"**1** 介绍K近邻算法的基本原理\n\n**2** 介绍K近邻算法的KDTree实现方法\n\n**3** 通过简单案例和鸢尾花分类案例,对比分析本文算法和sklearn的结果。",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import load_iris\nfrom sklearn.neighbors import KNeighborsClassifier\nimport numpy as np\nimport heapq",
"_____no_output_____"
]
],
[
[
"**<font color=black size=4>1 K近邻算法</font>** \n\nK近邻算法多用于分类任务,本文仅就其分类问题进行讨论。**基本思想**:给定一个已知标签的数据集,对一个输入的未知标签的样本,计算其与数据集中各样本之间的距离,找出与之距离最小的$k$个样本;统计这$k$个样本的类别,以多数服从少数的原则,最终确定未知样本的分类标签。算法的三个基本要素如下:\n\n+ **$k$值:** $k$值越大,模型越简单;反之,越复杂。试想:若$k$取数据集的样本总数$n$,那么无论输入的未知样本是什么,最终得到的其分类标签都是数据集中占比最大的那一类的标签,这样的模型是非常简单的。\n\n+ **距离度量方法**:不同的距离度量确定的最邻近点通常是不同的。一般选择$Minkowski$距离,其定义为:$$ L_p(x_i, x_j)=(\\sum_{l=1}^{m}|x_i^{(l)}- x_j^{(l)}|^p)^{\\frac{1}{p}} $$\n其中:$p>=1$;$m$为特征的维数;$l$为第几维。<br>\n当$p=2$时,为欧氏距离:$ L_2(x_i, x_j)=(\\sum_{l=1}^{m}|x_i^{(l)}- x_j^{(l)}|^2)^{\\frac{1}{2}} $<br>\n当$p=1$时,为曼哈顿距离:$ L_1(x_i, x_j)=\\sum_{l=1}^{m}|x_i^{(l)}- x_j^{(l)}| $<br>\n当$p=\\infty$时,为各个维度下距离的最大值:$ L_\\infty(x_i, x_j)=max|x_i^{(l)}- x_j^{(l)}| $\n\n+ **分类决策规则**:一般是采用多数表决的规则,即由输入的未知样本的k个邻近样本中的多数类决定未知样本的类。",
"_____no_output_____"
],
[
"**<font color=black size=4>2 KDTree</font>** \n\nK近邻算法的思想很简单,但存在的问题是:当样本数量较大时,为预测一个未知样本的类别标签,需要遍历计算其与所有\n\n样本之间的距离(线性搜索),这无疑会造成较大的时间开销。通过构造KDTree,减少计算距离的次数,提高算法效率。\n\n\n**<font color=black size=3.5>2.1 KDTree构造</font>** \n\n+ **两个基本操作:**<br>\n1) 计算划分维度: $ l=j \\ \\% \\ m $。$\\%$为取模运算;$j$的起始值为0,每完成一次划分$j=j+1$;$m$为特征的维数<br>\n2) 计算划分点: 按$l$维的值对$X$从小到大排序,取中间点作为划分点。排序后中间点索引的计算方法为:$len(X)\\ // \\ 2$<br>\n+ **示例:** 给定数据集$X = [[2, 3], [5, 4], [9, 6], [4, 7], [8, 1], [7, 2]]$,构造一个kd树。<br>\n1) 第1次划分: <br>\n$ l_1=0 \\ \\% \\ 2 = 0$,也就是划分维度为第0维;<br>\n按第0维元素值对$X$排序的结果为$X = [[2, 3], [4, 7], [5, 4], [7, 2], [8, 1], [9, 6]]$;<br>\n划分点的索引为$len(X)\\ // \\ 2 = 6 \\ // \\ 2 = 3$,对应$[7, 2]$,存储到根节点;\n将$[7, 2]$左侧的点存储到左子树,右侧的点存储到右子树。<br>\n<img src=\"../src/kdtree1.png\" width=280, heigth=200>\n2) 第2次划分: <br>\n$j = j + 1$,即$ l_2=1 \\ \\% \\ 2 = 1$;<br>\n对第1次划分得到的左右子树,分别进行同样的划分操作,得到新的左右子树。<br>\n<img src=\"../src/kdtree2.png\" width=280, heigth=200>\n3) 第3次划分: <br>\n$j = j + 1$,即$ l_3=2 \\ \\% \\ 2 = 0$;<br>\n划分后发现所有叶节点中只有一个点,因而无法继续划分,划分停止,得到最终的KDTree。\n<img src=\"../src/kdtree3.png\" width=280, heigth=200><br>\n<font color=red>注:可以发现上述过程,划分维度是交替进行的,因此可以称为交替维度划分法。除此之外,还可以\n采用最大方差法,即:每一次划分计算各个维度上的方差,取方差最大的维度作为该次的划分维度。</font>",
"_____no_output_____"
],
[
"**<font color=black size=3.5>2.2 KDTree搜索</font>** <br>\n+ 上述的二维数据集构造的KDTree可以对应用如下二维平面的划分来表达,每一次划分即对应在该次的划分维度上根据划分点将平面切割。如下图所示,第$1$次划分在$l=0$维度的$x^{(1)}=7$处将平面切割为左右两个区域。\n<img src=\"../src/kdtree4.png\" width=280, heigth=200><br>\n+ **搜索过程**<br>\n以寻找点(2.5, 4.5)的最邻近点为例:<br>\n1) 按上述创建的KDTree从根节点开始判断,直至达到叶节点,可以得到(2.5, 4.5)的第一个候选最邻近点为(4, 7),对应的欧式距离$d_{min}$为2.915;<br>\n2) 回溯。(4, 7)的根节点为(5, 4),计算(2.5, 4.5)与(5, 4)的距离为2.550<2.915,因此(2.5, 4.5)的最邻近点更新为(5, 4),$d_{min}$更新为2.550;由于不清楚(5, 4)的左子树中有没有更近的点,因此需要做一步判断:计算(5, 4)的划分维度$l$下,(2.5, 4.5)与(5, 4)在$l$上的距离$d_l$,若$d_l<d_{min}$,则需要进入(5, 4)的右子树重复1)、2)的搜索过程,反之则不需要;在本例中,(5, 4)的划分维度$l=1$,则$d_l=|4.5-4|=0.5<2.550$,因此进入左子树进行搜索,计算(2.5, 4.5)与(2, 3)的距离为1.581,因此更新最邻近点为(2, 3),$d_{min}=1.581$。<br>\n<img src=\"../src/kdtree5.png\" width=280, heigth=200><br>\n3)继续向上回溯。(5, 4)的根节点为(7, 2),计算(2.5, 4.5)与(7, 2)的距离为5.148>1.581,因此不更新最邻近点与$d_{min}$。同样地,判断$d_l$和$d_{min}$的大小关系,发现$d_l= 4.5 > d_{min}=1.581$,因此不需要再进入\n(7, 2)的右子树进行搜索。本例中(7, 2)为整体树的根节点,因此到此搜索也就停止,最终得到的最邻近点为(2, 3),最邻近距离为1.581。<br>",
"_____no_output_____"
]
],
[
[
"class Node(object):\n \"\"\"kd树中的一个节点\"\"\"\n\n def __init__(self, seg_axis, seg_point, left=None, right=None):\n self.seg_axis = seg_axis # int 划分维度\n self.seg_point = seg_point # list 划分点\n self.left = left # class 左子树\n self.right = right # class 右子树",
"_____no_output_____"
],
[
"class KDTree(object):\n \"\"\"kd树类\"\"\"\n\n def __init__(self, X):\n self.X = X\n\n def create(X, depth=0):\n \"\"\"递归创建kd树\n Parameters\n ----------\n X: list 样本\n depth: int 树的深度 默认为0\n \"\"\"\n if not X:\n return\n\n # 交替轴法确定划分维度\n k = len(X[0])\n ax = depth % k\n sort_X = sorted(X, key=lambda x: x[ax])\n mid = len(X) // 2\n\n return Node(seg_axis=ax,\n seg_point=sort_X[mid],\n left=create(sort_X[:mid], depth + 1),\n right=create(sort_X[mid + 1:], depth + 1))\n\n self.root = create(X, 0)\n\n def query(self, x, k=1, p=2):\n \"\"\"kd树查询目标点的k个最邻近点\n Parameters\n ----------\n x: list 目标点\n k: int 最邻近点个数 默认为1\n p: int 距离度量方法 默认为2 代表欧式距离\n\n Return\n ------\n dist: list k个最邻近点距离(由大到小排列)\n ind: list 对应dist的k个最邻近点在X中的索引\n \"\"\"\n self.knn = [(-np.inf, None)] * k\n\n def search_knn(node):\n \"\"\"query方法的辅助方法。用于访问kd树的结点,并计算结点与目标的距离,\n 确定k个最邻近的点。\n Parameters\n ----------\n node: class kd树的结点\n \"\"\"\n if node is not None:\n ax = node.seg_axis\n ax_diff = abs(x[ax]-node.seg_point[ax]) # 目标点和结点在ax维度下的距离\n\n if x[ax] < node.seg_point[ax]:\n search_knn(node.left)\n else:\n search_knn(node.right)\n\n # 计算目标点与结点的距离\n d = np.linalg.norm(np.array(x)-np.array(node.seg_point), p)\n\n # 利用堆结构存储距离和划分点信息\n heapq.heappushpop(self.knn, (-d, node.seg_point))\n\n # 回溯过程中,是否需要进入另一子区域的判断\n if ax_diff < -self.knn[0][0]:\n if x[ax] < node.seg_point[ax]:\n search_knn(node.right)\n else:\n search_knn(node.left)\n\n search_knn(self.root)\n self.knn.sort(reverse=True) # 对k个邻近点进行排序(也可不加这行代码)\n \n dist = []\n ind = []\n for item in self.knn:\n dist.append(abs(item[0]))\n ind.append(self.X.index(item[1]))\n\n return dist, ind",
"_____no_output_____"
],
[
"class MyKNeighborsClassifier(KDTree):\n \"\"\"K近邻分类器\"\"\"\n\n def __init__(self, X, y, k=1, p=2):\n super(MyKNeighborsClassifier, self).__init__(X) # 继承父类的构造方法\n self.y = y\n self.k = k\n self.p = p\n\n def kneighbors(self, x, k=1):\n \"\"\"获取目标点的k个最邻近点\"\"\"\n neigh_dist, neigh_ind = self.query(x, k)\n\n return neigh_dist, neigh_ind\n\n def predict(self, x):\n \"\"\"预测目标点的分类标签\"\"\"\n _, neigh_ind = self.query(x, self.k)\n\n # 多数服从少数的投票方法\n vote = {}\n for i in neigh_ind:\n vote[self.y[i]] = vote.get(self.y[i], 0) + 1\n out = sorted(vote.items(), key=lambda s: s[1], reverse=True)\n\n return out[0][0]",
"_____no_output_____"
]
],
[
[
"**<font color=black size=4>3 案例</font>** \n\n**<font color=black size=3.5>3.1 简单案例</font>** ",
"_____no_output_____"
]
],
[
[
"# 数据准备\nX = [[2, 3], [5, 4], [9, 6], [4, 7], [8, 1], [7, 2]]\ny = [0, 0, 1, 1, 2, 2]",
"_____no_output_____"
],
[
"# 本文算法\nclf = MyKNeighborsClassifier(X, y, k=3)\nneigh_dist, neigh_ind = clf.kneighbors([2.5, 4.5], k=3)\nprint('本文算法')\nprint('-------')\nprint('最邻近点的距离:', neigh_dist)\nprint('最邻近点的索引:', neigh_ind)\nprint('预测类别:', clf.predict([2.5, 4.5]))",
"本文算法\n-------\n最邻近点的距离: [1.5811388300841898, 2.5495097567963922, 2.9154759474226504]\n最邻近点的索引: [0, 1, 3]\n预测类别: 0\n"
],
[
"# sklearn\nneigh = KNeighborsClassifier(n_neighbors=3, algorithm='kd_tree')\nneigh.fit(X, y)\ndist, ind = neigh.kneighbors(np.array([[2.5, 4.5]]), n_neighbors=3)\nprint('sklearn')\nprint('-------')\nprint('最邻近点的距离:', dist)\nprint('最邻近点的索引:', ind)\nprint('预测类别:', neigh.predict([[2.5, 4.5]]))",
"sklearn\n-------\n最邻近点的距离: [[1.58113883 2.54950976 2.91547595]]\n最邻近点的索引: [[0 1 3]]\n预测类别: [0]\n"
]
],
[
[
"**<font color=black size=3.5>3.2 鸢尾花分类</font>** <br>",
"_____no_output_____"
]
],
[
[
"# 数据准备\niris = load_iris()\nX2 = iris.data\ny2 = iris.target",
"_____no_output_____"
],
[
"# 本文算法\nX2_copy = X2.copy().tolist() # 转换成本文算法要求的数据类型\ny2_copy = y2.copy().tolist()\n\nclf2 = MyKNeighborsClassifier(X2_copy, y2_copy, k=3)\nneigh_dist2, neigh_ind2 = clf2.kneighbors([2, 3, 2, 3], 3)\nprint('本文算法')\nprint('-------')\nprint('最邻近点的距离:', neigh_dist2)\nprint('最邻近点的索引:', neigh_ind2)\nprint('预测类别:', clf2.predict([2, 3, 2, 3]))",
"本文算法\n-------\n最邻近点的距离: [3.7376463182061515, 3.753664875824692, 3.7589892258425004]\n最邻近点的索引: [8, 38, 42]\n预测类别: 0\n"
],
[
"# sklearn\nneigh2 = KNeighborsClassifier(n_neighbors=3, algorithm='kd_tree')\nneigh2.fit(X2, y2)\ndist2, ind2 = neigh2.kneighbors(np.array([[2, 3, 2, 3]]), n_neighbors=3)\nprint('sklearn')\nprint('-------')\nprint('最邻近点的距离:', dist2)\nprint('最邻近点的索引:', ind2)\nprint('预测类别:', neigh2.predict([[2, 3, 2, 3]]))",
"sklearn\n-------\n最邻近点的距离: [[3.73764632 3.75366488 3.75898923]]\n最邻近点的索引: [[ 8 38 42]]\n预测类别: [0]\n"
]
],
[
[
"可以看出:本文算法与sklearn算法得到的结果一致。",
"_____no_output_____"
],
[
"<font color=red>*注:本文重在通过代码理解算法的工作原理,仅供学习使用。更多关于算法相关的资料可以在参考里找到。</font>",
"_____no_output_____"
],
[
"**<font color=black size=4>参考</font>**\n\n1 李航. (2012) 统计学习方法. 清华大学出版社, 北京.\n\n2 [sklearn文档](https://scikit-learn.org/stable/modules/generated/sklearn.neighbors.KNeighborsClassifier.html)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
e78de06c19d2871d782bd61eeef236248e049b38 | 94,196 | ipynb | Jupyter Notebook | CodingChallenge_S2021.ipynb | pkramadhati/ISMTE111 | 64727e394e8ea0474e4d0fa58f8d5e3ed64df8f7 | [
"MIT"
] | null | null | null | CodingChallenge_S2021.ipynb | pkramadhati/ISMTE111 | 64727e394e8ea0474e4d0fa58f8d5e3ed64df8f7 | [
"MIT"
] | null | null | null | CodingChallenge_S2021.ipynb | pkramadhati/ISMTE111 | 64727e394e8ea0474e4d0fa58f8d5e3ed64df8f7 | [
"MIT"
] | null | null | null | 38.940058 | 13,400 | 0.451049 | [
[
[
"# ISMT E-111 Coding Challenge Spring 2021\n### Make sure to include any relevant code when providing all answers :)\n### Save this as a `.ipynb` file with all results visible and send it to Vasya when you're done. \n\n### Challenge is out of 150 points \n * **100 points for the main challenge**\n * **50 bonus points for extra credit**",
"_____no_output_____"
],
[
"# PRABHA KRAMADHATI ",
"_____no_output_____"
],
[
"### 1. Load your Data and Take a Look\n\n **a.** **(`15 pts`)** Load the training data (found in the same github repo as this notebook) into this notebook using Pandas. \n\n **b.** **(`5 pts`)** Show the first 10 lines of the data.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nimport pandas as pd",
"_____no_output_____"
],
[
"df = pd.read_csv('codingdata.csv')",
"_____no_output_____"
],
[
"df.shape",
"_____no_output_____"
],
[
"df.head(10)",
"_____no_output_____"
]
],
[
[
"### 2. Define and describe your target variable\nThe `email status` column contains three values with the following definitions. \n`0=ignored` `1=read` and `2=converted`\n* Ignored means that the customer did not interact with the email\n* Read means that a customer opened the email\n* Converted means that the customer clicked on the link for the product page within the email. \n\nThe company considers status=2 as a conversion, (statuses 0 and 1 are non-conversions).",
"_____no_output_____"
],
[
" **a.** **(`10 pts`)** Create a new column called `conversion` that has a value of `1` when the email led to a conversion and is 0 otherwise.",
"_____no_output_____"
]
],
[
[
"df['conversion'] = df['Email_Status'].apply(lambda x: 1 if x == 2 else 0)",
"_____no_output_____"
],
[
"df.tail(10)",
"_____no_output_____"
]
],
[
[
" **b.** **(`10 pts`)** How many conversions are in this dataset?",
"_____no_output_____"
]
],
[
[
"print('total conversions: %i out of %i' % (df.conversion.sum(), df.shape[0]))",
"total conversions: 2373 out of 68353\n"
]
],
[
[
" **c.** **(`10 pts`)** What percent of all emails resulted in a conversion?",
"_____no_output_____"
]
],
[
[
"print('conversion rate: %0.2f%%' % (df.conversion.sum() / df.shape[0] * 100.0))",
"conversion rate: 3.47%\n"
]
],
[
[
"### 3 Exploration",
"_____no_output_____"
],
[
" **a.** The `Email_Campaign_Type` column captures the campaign under which the email was sent. A campaign is a marketing strategy.\n\n **i.** **(`15 pts`)** Which campaign led to the most conversions?",
"_____no_output_____"
]
],
[
[
"most_conversions_df=pd.DataFrame(\n df.groupby(\n by='Email_Campaign_Type'\n )['conversion'].sum()\n)",
"_____no_output_____"
],
[
"most_conversions_df",
"_____no_output_____"
],
[
"#CAMPAIGN 3 has the most number of converstions ",
"_____no_output_____"
]
],
[
[
" **ii.** **(`15 pts`)** Which campaign has the highest conversion rate?",
"_____no_output_____"
]
],
[
[
"pd.DataFrame(\n df.groupby(\n by='Email_Campaign_Type'\n )['conversion'].count()\n)",
"_____no_output_____"
],
[
"conversion_rate = df.groupby(\n by='Email_Campaign_Type'\n)['conversion'].sum() / df.groupby(\n by='Email_Campaign_Type'\n)['conversion'].count() * 100.0",
"_____no_output_____"
],
[
"pd.DataFrame(conversion_rate)",
"_____no_output_____"
],
[
"#CAMPAIGN 1 has the highest conversion rate ",
"_____no_output_____"
],
[
"df.isnull().sum()",
"_____no_output_____"
],
[
"df.dropna(subset=['Total_Past_Communications'])",
"_____no_output_____"
],
[
"df['con_version'] = df['conversion'].apply(lambda x: 'Converted(LinkCliked)' if x == 1 else 'Ignored/Read')",
"_____no_output_____"
]
],
[
[
" **b**.\n`Total_Past_Communications` is a count of the number of times the customer has been contacted prior to the current email. \n\n **i.** **(`11 pts`)** Create a box plot showing the distribution of `Total_Past_Communications` by `conversion`. \n\n **ii** **(`5 pts`)** How do conversions relate to the number of times a customer had been emailed? You can provide your answer as a comment in its own cell (remember a comment is preceded by a hash symbol `#here is my comment`)\n\n **iii.** **(`3 pts`)** Set the title to something that briefly summarizes your key insight from the preceding question (3bii). Set the x-label to be `Email Status` and the y-label to be `Previous Emails`\n\n **iv** **(`1 pts`)** Prevent the default text `'Boxplot grouped by conversion'` from displaying in the title of the boxplot, and do not show extreme values / outliers. Then show your plot!",
"_____no_output_____"
]
],
[
[
"ax = df[['con_version', 'Total_Past_Communications']].boxplot(\n by='con_version',\n showfliers=False,\n figsize=(7,5)\n)\n\nax.set_xlabel('Email Status')\nax.set_ylabel('Previous Emails')\nax.set_title('First couple of contacts unlikely to lead to conversion')\n\nplt.suptitle(\"\")\nplt.show()",
"_____no_output_____"
],
[
"#Almost no converstions were made with the first 7-8 emails.\n#75% of the converstion were made after the 30th email\n#Median number of emails at which customers converted is 10 more than the median of non-converted emails.\n",
"_____no_output_____"
]
],
[
[
"***\nYou're done! You can relax and submit this assignment! <br>If you're feeling ambitious, you can try your hand at the _extra credit_ portion below. ",
"_____no_output_____"
],
[
"***\n# Extra Credit: Build, Interpret and Assess a Machine Learning Model to Predict Conversion Rate\n\n**1. Data Cleaning & Prep** \nWe'll be encoding a categorical variable, joining it to all of our numerical variables and generating a train-test split to train and test our model.<br>",
"_____no_output_____"
],
[
" **a.** **(`5 pts`)** Which columns are categorical columns and which are numerical? Create a list of categorical column names and a list of numerical column names. Ignore the following columns: `Email_ID`, `Email_Status`, `conversion`. You should have ten columns to categorize. Save each list to a variable (e.g., `my_var = ['a', 'b', 'c']`)",
"_____no_output_____"
]
],
[
[
"df.dtypes",
"_____no_output_____"
],
[
"categorical_var=['Email_Type','Email_Source_Type','Customer_Location','Email_Campaign_Type','Time_Email_sent_Category']",
"_____no_output_____"
],
[
"numerical_var=['Subject_Hotness_Score','Total_Past_Communications','Word_Count','Total_Links','Total_Images']",
"_____no_output_____"
]
],
[
[
" **b.** **(`2.5 pts`)** A few of the columns have missing values. Let's just drop them. To do that you can run `df.dropna(inplace=True)` where `df` is the name of your dataframe. That's it, this one's easy, no tricks.",
"_____no_output_____"
]
],
[
[
"df.dropna(inplace=True)",
"_____no_output_____"
]
],
[
[
" **c.** **(`10 pts`)** Add Dummy Variables! (Hint: Follow the code in chapter 4)<br>\n **i.** Using `pd.get_dummies(<your_column_of_data>, drop_first=True)`, create a new dataframe that contains the dummy-variable version of `Email_Campaign_Type`. **Note/Warning:** This is a bit different than we did in class since we use the `drop_first` argument. `drop_first` means: Don't encode Campaign 1, just tell me how good Campaigns 2 & 3 are in relation to Campaign 1. This is necessary for the model to produce sensible results<br>\n **ii.** Rename the columns of this dataframe to be `'Campaign_2','Campaign_3'`<br>\n **iii.** Concatenate this dataframes to the email data dataframe.<br>\n **iv.** Create a variable called `all_features` that is a list of all the variables we want to use in our model. This will be all numerical variables and the dummy variables `'Campaign_2','Campaign_3'`.<br>\n",
"_____no_output_____"
]
],
[
[
"df['Email_Campaign_Type'].unique()",
"_____no_output_____"
],
[
"Email_Campaign_Type_encoded_df = pd.get_dummies(df['Email_Campaign_Type'], drop_first=True)\nEmail_Campaign_Type_encoded_df.columns = ['Campaign_%s' % x for x in Email_Campaign_Type_encoded_df.columns]",
"_____no_output_____"
],
[
"Email_Campaign_Type_encoded_df.head()",
"_____no_output_____"
],
[
"df = pd.concat([df, Email_Campaign_Type_encoded_df], axis=1)",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"all_features=['Subject_Hotness_Score','Total_Past_Communications','Word_Count','Total_Links','Total_Images','Campaign_2','Campaign_3']\nresponse='conversion'",
"_____no_output_____"
],
[
"sample_df = df[all_features + [response]]",
"_____no_output_____"
],
[
"sample_df.head()",
"_____no_output_____"
]
],
[
[
" **d.** **(`7.5 pts`)** Split the dataframe into 80% training data and 20% testing data using the `train_test_split` function (see section 3 of the notebook from Chapter 8 for an example of how to do this.) When you pass the data to be split, be sure to add an intercept! To do this simply pass `sm.add_constant(df[all_features])` instead of just `df[all_features]`. Finally, add the following argument to train_test_split: `random_state=42`",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"import statsmodels.api as sm",
"_____no_output_____"
],
[
"x_train, x_test, y_train, y_test = train_test_split(sm.add_constant(df[all_features]), df[response], test_size=0.2, random_state=42)",
"_____no_output_____"
]
],
[
[
"**2. Build, interpret and score a logistic regression model!** <br>\n **a.** **(`5 pts`)** Build a Logistic Regression using statsmodels (statsmodels.api) as in Chapter 3. Only include the *train* data you created in question 1d.<br>",
"_____no_output_____"
]
],
[
[
"x_train.head()",
"_____no_output_____"
],
[
"logit = sm.Logit(\n y_train,\n x_train\n)",
"_____no_output_____"
],
[
"fit = logit.fit()",
"Optimization terminated successfully.\n Current function value: 0.130311\n Iterations 8\n"
],
[
"fit.summary()",
"_____no_output_____"
]
],
[
[
" **b.** **(`10 pts`)** Take a look at the model summary and interpret the results. <br>Remember that `Campaign_2` & `Campaign_3` are encoded in relation to `Campaign_1`, when interpretting them remember to mention that their effect is relative to `Campaign_1` (think in terms of negative vs positive coefficients). <br> \nYou can provide your answer as a comment (`# Anything following a hash mark is a comment`) or in any other manner you like.<br>",
"_____no_output_____"
]
],
[
[
"#Word count and total images have no corelation to conversion.\n#Total links and Total past communications have some positive corelation with conversion.\n#The coefficent of the subject hotness score indicates a positive co-relation with conversion but the p value indicates that this co-relation may not be significant.\n# Compared to campaign_1, campaign_2 is lot less likely than campaign_3 to lead to a converstion\n",
"_____no_output_____"
]
],
[
[
" **c.** **(`5 pts`)** Use your model to get predictions using the test data. Remember that your fit model is the result of the `.fit()` call (e.g., `my_fit = my_model.fit()`). The resulting object has a `.predict` method (e.g., `my_fit.predict()`) that will generate predictions using your model on the specified data. <br>Get predictions by applying the model you just fit to the _test_ data. Save the predictions to a variable.<br>",
"_____no_output_____"
]
],
[
[
"x_pred=fit.predict(x_test)",
"_____no_output_____"
]
],
[
[
" **d.** **(`5 pts`)** The ROC AUC score informs you as to how good your model is at telling which emails are more likely to result in conversions than others. You can use the Scikit-Learn function `sklearn.metrics.roc_auc_score` to compute this. This function has two inputs: predictions and the actual value from the data. You already have both of these variables. *You may want to take a look at Chapter 8's Notebook for more details on this metric*",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import roc_auc_score\nroc_auc_score(y_test, x_pred)",
"_____no_output_____"
]
],
[
[
"# PRABHA KRAMADHATI",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e78de1a1dd8fb6dfbf7722854688353f15f21831 | 68,430 | ipynb | Jupyter Notebook | tests/BBConvLSTM_emotion_predict.ipynb | geek-yang/NEmo | 4f310535c4865f3816155b99b4a2bbb891672cc9 | [
"Apache-2.0"
] | 1 | 2020-05-25T19:06:15.000Z | 2020-05-25T19:06:15.000Z | tests/BBConvLSTM_emotion_predict.ipynb | geek-yang/IIIDL | 4f310535c4865f3816155b99b4a2bbb891672cc9 | [
"Apache-2.0"
] | null | null | null | tests/BBConvLSTM_emotion_predict.ipynb | geek-yang/IIIDL | 4f310535c4865f3816155b99b4a2bbb891672cc9 | [
"Apache-2.0"
] | null | null | null | 80.791027 | 34,248 | 0.690238 | [
[
[
"# Copyright Netherlands eScience Center and Centrum Wiskunde & Informatica <br>\n** Function : Emotion recognition and forecast with ConvLSTM** <br>\n** Author : Yang Liu & Tianyi Zhang** <br>\n** First Built : 2020.05.17 ** <br>\n** Last Update : 2020.05.22 ** <br>\n** Library : Pytorth, Numpy, os, DLACs, matplotlib **<br>\nDescription : This notebook serves to test the prediction skill of deep neural networks in emotion recognition and forecast. The convolutional Long Short Time Memory neural network is used to deal with this spatial-temporal sequence problem. We use Pytorch as the deep learning framework. <br>\n<br>\n** Many to one prediction.** <br>\n\nReturn Values : Time series and figures <br>\n\n**This project is a joint venture between NLeSC and CWI** <br>\n\nThe method comes from the study by Shi et. al. (2015) Convolutional LSTM Network: A Machine Learning Approach for Precipitation Nowcasting. <br>",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport sys\nimport numbers\n\n# for data loading\nimport os\n# for pre-processing and machine learning\nimport numpy as np\nimport csv\n#import sklearn\n#import scipy\nimport torch\nimport torch.nn.functional\n\nsys.path.append(\"C:\\\\Users\\\\nosta\\\\ConvLSTM_emotion\\\\Scripts\\\\DLACs\")\n#sys.path.append(\"../\")\nimport dlacs\nimport dlacs.BBConvLSTM\nimport dlacs.ConvLSTM\nimport dlacs.preprocess\nimport dlacs.function\nimport dlacs.saveNetCDF\nimport dlacs.metric\n\n# for visualization\nimport dlacs.visual\nimport matplotlib\nimport matplotlib.pyplot as plt\nfrom matplotlib.pyplot import cm",
"_____no_output_____"
]
],
[
[
"The testing device is Dell Inspirion 5680 with Intel Core i7-8700 x64 CPU and Nvidia GTX 1060 6GB GPU.<br>\nHere is a benchmark about cpu v.s. gtx 1060 <br>\nhttps://www.analyticsindiamag.com/deep-learning-tensorflow-benchmark-intel-i5-4210u-vs-geforce-nvidia-1060-6gb/",
"_____no_output_____"
]
],
[
[
"################################################################################# \n######### datapath ########\n#################################################################################\n# please specify data path\ndatapath = 'C:\\\\Users\\\\nosta\\\\ConvLSTM_emotion\\\\Data_CASE'\noutput_path = 'C:\\\\Users\\\\nosta\\\\ConvLSTM_emotion\\\\results'\nmodel_path = 'C:\\\\Users\\\\nosta\\\\ConvLSTM_emotion\\\\models'",
"_____no_output_____"
],
[
"if __name__==\"__main__\":\n print ('*********************** extract variables *************************')\n data = np.load(os.path.join(datapath, \"data_10s.npz\"))\n #data = np.load(os.path.join(datapath, \"data_2s.npz\"))\n #data = np.load(os.path.join(datapath, \"data_0.5s.npz\"))\n #################################################################################\n ######### data gallery #########\n #################################################################################\n sample = data[\"Samples\"][:] # (batch_size, sample_size, channels)\n label_c = data[\"Labels_c\"][:] # (batch_size, sample_size, 2)\n label = data[\"Labels\"][:] # (batch_size, 2)\n subject = data[\"Subject_id\"][:] # (batch_size, 2)\n video_label = data[\"Video_labels\"][:] # (batch_size,1)\n \n # leave-one-out training and testing\n num_s = 2\n sample_train = sample[np.where(subject!=num_s)[0],:,0:5]\n sample_test = sample[np.where(subject==num_s)[0],:,0:5]\n \n label_c_train = label_c[np.where(subject!=num_s)[0],:,:] / 10 # normalize\n label_c_test = label_c[np.where(subject==num_s)[0],:,:] / 10 # normalize",
"*********************** extract variables *************************\n"
],
[
" #################################################################################\n ######### pre-processing #########\n #################################################################################\n # choose the target dimension for reshaping of the signals\n batch_train_size, sample_size, channels = sample_train.shape\n batch_test_size, _, _ = sample_test.shape\n _, _, label_channels = label_c_train.shape\n x_dim = 5\n y_dim = 5\n series_len = sample_size // (y_dim * x_dim)\n # reshape the input and labels\n sample_train_xy = np.reshape(sample_train,[batch_train_size, series_len, y_dim, x_dim, channels])\n sample_test_xy = np.reshape(sample_test,[batch_test_size, series_len, y_dim, x_dim, channels])\n label_c_train_xy = np.reshape(label_c_train,[batch_train_size, series_len, y_dim, x_dim, label_channels])\n label_c_test_xy = np.reshape(label_c_test,[batch_test_size, series_len, y_dim, x_dim, label_channels])\n #################################################################################\n ######### normalization #########\n #################################################################################\n print('================ extract individual variables =================')\n sample_1 = sample_train_xy[:,:,:,:,0]\n sample_2 = sample_train_xy[:,:,:,:,1]\n sample_3 = sample_train_xy[:,:,:,:,2]\n sample_4 = sample_train_xy[:,:,:,:,3]\n sample_5 = sample_train_xy[:,:,:,:,4]\n \n label_c_valance = label_c_train_xy[:,:,:,:,0]\n label_c_arousal = label_c_train_xy[:,:,:,:,1]\n \n label_c_test_valance = label_c_test_xy[:,:,:,:,0]\n label_c_test_arousal = label_c_test_xy[:,:,:,:,1]\n \n # using indicator for training\n # video_label_3D = np.repeat(video_label[:,np.newaxis,:],series_len,1)\n # video_label_4D = np.repeat(video_label_3D[:,:,np.newaxis,:],y_dim,2)\n # video_label_xy = np.repeat(video_label_4D[:,:,:,np.newaxis,:],x_dim,3)\n # video_label_xy.astype(float)",
"================ extract individual variables =================\n"
],
[
" # first check of data shape\n print(sample.shape)\n print(label_c.shape)\n print(label.shape)\n print(subject.shape)\n print(video_label.shape)\n # check of reshape\n print(label_c_train_xy.shape)",
"(3690, 1000, 8)\n(3690, 1000, 2)\n(3690, 2)\n(3690, 1)\n(3690, 1)\n(3567, 40, 5, 5, 2)\n"
]
],
[
[
"# Procedure for LSTM <br>\n** We use Pytorth to implement LSTM neural network with time series of climate data. ** <br>",
"_____no_output_____"
]
],
[
[
" print ('******************* create basic dimensions for tensor and network *********************')\n # specifications of neural network\n input_channels = 5\n hidden_channels = [4, 3, 2] # number of channels & hidden layers, the channels of last layer is the channels of output, too\n #hidden_channels = [3, 3, 3, 3, 2]\n #hidden_channels = [2]\n kernel_size = 3\n # here we input a sequence and predict the next step only\n learning_rate = 0.01\n num_epochs = 20\n # probability of dropout\n p = 0.5 # 0.5 for Bernoulli (binary) distribution\n print (torch.__version__)\n # check if CUDA is available\n use_cuda = torch.cuda.is_available()\n print(\"Is CUDA available? {}\".format(use_cuda))\n # CUDA settings torch.__version__ must > 0.4\n # !!! This is important for the model!!! The first option is gpu\n device = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\n print ('******************* cross validation and testing data *********************')\n # mini-batch\n mini_batch_size = 64\n # iterations\n iterations = batch_train_size // mini_batch_size\n if batch_train_size % mini_batch_size != 0:\n extra_loop = \"True\"\n iterations += 1",
"******************* create basic dimensions for tensor and network *********************\n1.1.0\nIs CUDA available? True\n******************* cross validation and testing data *********************\n"
],
[
" %%time\n print ('******************* load exsited LSTM model *********************')\n # load model parameters\n model = dlacs.BBConvLSTM.BBConvLSTM(input_channels, hidden_channels, kernel_size).to(device)\n model.load_state_dict(torch.load(os.path.join(model_path, 'BBconvlstm_emotion_hl_3_kernel_3_lr_0.01_epoch_20_mini_64_validSIC.pkl'),\n map_location=device))\n #model = torch.load(os.path.join(output_path, 'Barents','convlstm_emotion_hl_1_kernel_3_lr_0.01_epoch_500_validSIC.pkl'))\n print(model)\n # check the sequence length (dimension in need for post-processing)\n _, sequence_len, height, width = sample_1.shape",
"******************* load exsited LSTM model *********************\nBBConvLSTM(\n (cell0): BBConvLSTMCell(\n (Wxi): Conv2d(5, 4, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (Whi): Conv2d(4, 4, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (Wxf): Conv2d(5, 4, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (Whf): Conv2d(4, 4, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (Wxc): Conv2d(5, 4, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (Whc): Conv2d(4, 4, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (Wxo): Conv2d(5, 4, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (Who): Conv2d(4, 4, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (dropout): Dropout2d(p=0.5)\n )\n (cell1): BBConvLSTMCell(\n (Wxi): Conv2d(4, 3, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (Whi): Conv2d(3, 3, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (Wxf): Conv2d(4, 3, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (Whf): Conv2d(3, 3, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (Wxc): Conv2d(4, 3, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (Whc): Conv2d(3, 3, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (Wxo): Conv2d(4, 3, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (Who): Conv2d(3, 3, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (dropout): Dropout2d(p=0.5)\n )\n (cell2): BBConvLSTMCell(\n (Wxi): Conv2d(3, 2, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (Whi): Conv2d(2, 2, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (Wxf): Conv2d(3, 2, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (Whf): Conv2d(2, 2, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (Wxc): Conv2d(3, 2, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (Whc): Conv2d(2, 2, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (Wxo): Conv2d(3, 2, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))\n (Who): Conv2d(2, 2, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)\n (dropout): Dropout2d(p=0.5)\n )\n)\nWall time: 2.13 s\n"
],
[
" for name, param in model.named_parameters():\n if param.requires_grad:\n print (name)\n print (param.data)\n print (param.size())\n print (\"=========================\")",
"cell0.Wxi.weight\ntensor([[[[-0.0462, 0.0080, -0.0835],\n [ 0.1415, 0.1050, -0.0149],\n [-0.0819, -0.1153, -0.1139]],\n\n [[-0.1280, 0.0669, 0.0994],\n [ 0.0641, -0.0091, -0.1008],\n [ 0.0709, 0.0190, -0.1305]],\n\n [[ 0.1098, 0.0286, -0.0751],\n [ 0.1438, 0.0837, 0.1393],\n [-0.0610, -0.0658, 0.1107]],\n\n [[ 0.1442, 0.1400, -0.1337],\n [ 0.0109, -0.0077, 0.0062],\n [-0.1268, 0.0479, -0.0059]],\n\n [[-0.0267, 0.0680, -0.1253],\n [ 0.0656, 0.1415, 0.1333],\n [-0.1431, -0.0730, 0.0952]]],\n\n\n [[[ 0.0676, 0.0405, -0.1113],\n [-0.1024, 0.0473, -0.0706],\n [ 0.0761, -0.1360, -0.0666]],\n\n [[-0.0577, -0.1429, -0.0058],\n [ 0.0075, -0.1271, -0.1442],\n [-0.0007, -0.0369, -0.1170]],\n\n [[ 0.0250, -0.1097, 0.1011],\n [ 0.0515, -0.0578, -0.0964],\n [ 0.0106, -0.0379, -0.0005]],\n\n [[-0.1165, 0.0069, -0.0311],\n [ 0.0714, 0.0320, -0.0287],\n [ 0.0258, -0.0779, -0.0689]],\n\n [[ 0.0335, -0.0163, -0.0335],\n [-0.0620, -0.1270, 0.0463],\n [ 0.1068, -0.1040, 0.0804]]]], device='cuda:0')\ntorch.Size([2, 5, 3, 3])\n=========================\ncell0.Wxi.bias\ntensor([ 0.0082, -0.1239], device='cuda:0')\ntorch.Size([2])\n=========================\ncell0.Whi.weight\ntensor([[[[ 0.2224, 0.1471, 0.0304],\n [ 0.0607, 0.0100, -0.2149],\n [-0.0449, 0.1890, -0.1897]],\n\n [[ 0.1189, -0.1652, -0.2044],\n [ 0.0362, -0.2103, 0.0074],\n [-0.1596, -0.0616, -0.0757]]],\n\n\n [[[ 0.1746, -0.2193, -0.2345],\n [ 0.1731, -0.1000, -0.1208],\n [ 0.0935, 0.1443, 0.0904]],\n\n [[ 0.0321, 0.2075, 0.1338],\n [-0.1763, -0.1791, 0.0465],\n [ 0.0636, 0.0350, -0.1402]]]], device='cuda:0')\ntorch.Size([2, 2, 3, 3])\n=========================\ncell0.Wxf.weight\ntensor([[[[ 0.0846, -0.0249, 0.1231],\n [ 0.0277, 0.1230, -0.1356],\n [ 0.0794, 0.1106, 0.0791]],\n\n [[-0.0382, -0.0726, -0.0146],\n [ 0.0425, 0.0961, 0.0817],\n [ 0.0215, -0.0311, 0.1280]],\n\n [[ 0.0467, 0.0017, 0.0971],\n [-0.0766, 0.0427, -0.0276],\n [-0.0191, 0.1107, 0.0313]],\n\n [[-0.1322, 0.0281, 0.1349],\n [ 0.0764, 0.0447, 0.0841],\n [-0.0465, -0.0052, 0.0620]],\n\n [[-0.0353, -0.1239, 0.1257],\n [ 0.1212, -0.0448, -0.0784],\n [ 0.0123, 0.1102, 0.1447]]],\n\n\n [[[ 0.0189, 0.0620, 0.0537],\n [ 0.0617, -0.1400, -0.0074],\n [-0.1221, 0.1279, -0.1453]],\n\n [[ 0.0398, -0.0964, 0.1285],\n [ 0.0178, -0.0264, -0.1364],\n [-0.1378, 0.0805, -0.0332]],\n\n [[-0.0947, 0.0275, 0.0552],\n [-0.0204, -0.0919, -0.0397],\n [-0.1211, 0.1146, -0.0017]],\n\n [[-0.0099, -0.1444, 0.1223],\n [ 0.0151, -0.1067, 0.1366],\n [-0.1220, -0.1405, 0.0344]],\n\n [[ 0.1216, -0.1008, 0.0369],\n [ 0.0342, 0.0308, -0.0613],\n [-0.0354, 0.1004, 0.0143]]]], device='cuda:0')\ntorch.Size([2, 5, 3, 3])\n=========================\ncell0.Wxf.bias\ntensor([0.0604, 0.0464], device='cuda:0')\ntorch.Size([2])\n=========================\ncell0.Whf.weight\ntensor([[[[-0.0601, -0.2224, 0.0307],\n [-0.1135, -0.0086, 0.2317],\n [-0.2251, 0.1502, -0.1222]],\n\n [[-0.0416, -0.1507, 0.1001],\n [-0.0661, -0.0733, -0.1883],\n [ 0.1059, 0.1007, -0.1971]]],\n\n\n [[[-0.0342, -0.0231, -0.0395],\n [-0.2218, -0.1310, -0.0613],\n [ 0.1150, 0.1609, -0.2300]],\n\n [[ 0.0488, -0.0485, 0.1643],\n [-0.2117, -0.1217, -0.1539],\n [ 0.1114, 0.0064, -0.0573]]]], device='cuda:0')\ntorch.Size([2, 2, 3, 3])\n=========================\ncell0.Wxc.weight\ntensor([[[[ 0.1292, -0.0217, 0.0176],\n [ 0.0170, -0.0857, 0.1131],\n [-0.0478, -0.0048, 0.0327]],\n\n [[ 0.0330, 0.0473, -0.1361],\n [-0.0706, 0.0540, -0.1171],\n [ 0.1402, -0.1387, -0.0684]],\n\n [[ 0.1434, -0.0495, -0.0745],\n [-0.0975, -0.0504, -0.1333],\n [-0.1237, -0.0203, 0.0489]],\n\n [[-0.1398, -0.1467, -0.0359],\n [ 0.0635, 0.0701, 0.1306],\n [ 0.0870, -0.1229, 0.0004]],\n\n [[ 0.0287, 0.0910, 0.1108],\n [-0.1367, -0.0110, 0.1406],\n [-0.1184, -0.0726, 0.0806]]],\n\n\n [[[ 0.1378, -0.0895, -0.0164],\n [ 0.1307, -0.0862, -0.0147],\n [-0.0948, 0.0875, 0.0478]],\n\n [[ 0.0188, 0.0013, -0.0509],\n [ 0.0965, 0.1028, 0.1364],\n [ 0.0340, -0.1144, -0.1375]],\n\n [[ 0.1468, -0.1245, 0.0749],\n [ 0.1027, 0.1067, -0.1119],\n [ 0.1276, 0.0189, 0.1259]],\n\n [[-0.0707, -0.0634, 0.0373],\n [ 0.0888, 0.0652, 0.0766],\n [ 0.0656, 0.0357, 0.0840]],\n\n [[-0.0590, 0.0525, 0.1357],\n [ 0.0312, -0.0457, -0.0139],\n [-0.1429, 0.0383, 0.1230]]]], device='cuda:0')\ntorch.Size([2, 5, 3, 3])\n=========================\ncell0.Wxc.bias\ntensor([0.0967, 0.0781], device='cuda:0')\ntorch.Size([2])\n=========================\ncell0.Whc.weight\ntensor([[[[ 0.1495, 0.1515, -0.1610],\n [-0.1455, -0.0252, 0.1255],\n [-0.0410, -0.1620, -0.1051]],\n\n [[-0.1956, -0.0981, 0.1482],\n [ 0.0134, 0.2175, 0.0450],\n [ 0.1258, 0.0933, -0.1179]]],\n\n\n [[[ 0.1660, -0.0034, 0.0398],\n [ 0.2135, -0.2146, 0.0008],\n [ 0.1689, 0.2210, 0.1980]],\n\n [[ 0.0264, 0.0478, -0.1663],\n [-0.1807, -0.1513, 0.1744],\n [ 0.1550, -0.0313, -0.1623]]]], device='cuda:0')\ntorch.Size([2, 2, 3, 3])\n=========================\ncell0.Wxo.weight\ntensor([[[[-0.1236, -0.1040, -0.1368],\n [-0.1006, 0.0352, -0.0746],\n [-0.0211, -0.1006, 0.0798]],\n\n [[ 0.0863, -0.0068, 0.0152],\n [ 0.0512, -0.1200, -0.1088],\n [-0.0394, -0.0340, 0.1409]],\n\n [[ 0.1152, 0.1070, 0.0159],\n [-0.0422, 0.0856, 0.0131],\n [ 0.0440, 0.0865, -0.0477]],\n\n [[-0.0195, -0.0859, -0.1122],\n [ 0.0520, -0.0151, 0.0851],\n [ 0.0461, 0.1119, 0.0687]],\n\n [[ 0.1160, 0.0200, 0.1299],\n [ 0.0797, 0.0916, 0.0573],\n [-0.0064, 0.0644, 0.0101]]],\n\n\n [[[-0.0856, 0.1446, -0.1237],\n [-0.1321, -0.0099, -0.0715],\n [ 0.0247, 0.0314, -0.1413]],\n\n [[-0.1151, -0.0905, -0.1125],\n [-0.0275, 0.0119, 0.0818],\n [ 0.0837, 0.0225, 0.0822]],\n\n [[ 0.0192, -0.0298, 0.1030],\n [ 0.0796, -0.0172, -0.1171],\n [ 0.0346, -0.0463, 0.0649]],\n\n [[-0.0478, -0.1436, 0.1092],\n [-0.0079, 0.1125, 0.1472],\n [ 0.0189, -0.1366, -0.0857]],\n\n [[ 0.1188, -0.0563, 0.0460],\n [ 0.0185, 0.0902, 0.0849],\n [ 0.0789, 0.0668, -0.1056]]]], device='cuda:0')\ntorch.Size([2, 5, 3, 3])\n=========================\ncell0.Wxo.bias\ntensor([-0.0465, 0.0640], device='cuda:0')\ntorch.Size([2])\n=========================\ncell0.Who.weight\ntensor([[[[ 0.0942, 0.1899, -0.0379],\n [-0.1165, -0.0863, -0.0424],\n [ 0.2276, -0.0903, -0.0356]],\n\n [[ 0.0406, -0.1165, -0.0082],\n [-0.2335, 0.1486, -0.2275],\n [ 0.1587, -0.0119, -0.0848]]],\n\n\n [[[-0.0354, -0.0480, 0.0453],\n [-0.0244, 0.0434, 0.0499],\n [-0.1977, -0.1257, 0.1449]],\n\n [[-0.2031, 0.1514, -0.0061],\n [-0.2024, -0.0998, -0.0148],\n [-0.0308, 0.0064, -0.1589]]]], device='cuda:0')\ntorch.Size([2, 2, 3, 3])\n=========================\n"
],
[
" print('##############################################################')\n print('############# preview model parameters matrix ###############')\n print('##############################################################')\n print('Number of parameter matrices: ', len(list(model.parameters())))\n for i in range(len(list(model.parameters()))):\n print(list(model.parameters())[i].size())",
"##############################################################\n############# preview model parameters matrix ###############\n##############################################################\nNumber of parameter matrices: 12\ntorch.Size([2, 5, 3, 3])\ntorch.Size([2])\ntorch.Size([2, 2, 3, 3])\ntorch.Size([2, 5, 3, 3])\ntorch.Size([2])\ntorch.Size([2, 2, 3, 3])\ntorch.Size([2, 5, 3, 3])\ntorch.Size([2])\ntorch.Size([2, 2, 3, 3])\ntorch.Size([2, 5, 3, 3])\ntorch.Size([2])\ntorch.Size([2, 2, 3, 3])\n"
],
[
" print ('******************* evaluation matrix *********************')\n # The prediction will be evaluated through RMSE against climatology\n \n # error score for temporal-spatial fields, without keeping spatial pattern\n def RMSE(x,y):\n \"\"\"\n Calculate the RMSE. x is input series and y is reference series.\n It calculates RMSE over the domain, not over time. The spatial structure\n will not be kept.\n Parameter\n ----------------------\n x: input time series with the shape [time, lat, lon]\n \"\"\"\n x_series = x.reshape(x.shape[0],-1)\n y_series = y.reshape(y.shape[0],-1)\n rmse = np.sqrt(np.mean((x_series - y_series)**2,1))\n rmse_std = np.sqrt(np.std((x_series - y_series)**2,1))\n \n return rmse, rmse_std\n \n # error score for temporal-spatial fields, keeping spatial pattern\n def MAE(x,y):\n \"\"\"\n Calculate the MAE. x is input series and y is reference series.\n It calculate MAE over time and keeps the spatial structure.\n \"\"\"\n mae = np.mean(np.abs(x-y),0)\n \n return mae\n \n def MSE(x, y):\n \"\"\"\n Calculate the MSE. x is input series and y is reference series.\n \"\"\"\n mse = np.mean((x-y)**2)\n \n return mse",
"******************* evaluation matrix *********************\n"
],
[
" %%time\n #################################################################################\n ######## prediction ########\n #################################################################################\n print('##############################################################')\n print('################### start prediction loop ###################')\n print('##############################################################')\n # forecast array\n pred_valance = np.zeros((batch_test_size, series_len, y_dim, x_dim),dtype=float)\n pred_arousal = np.zeros((batch_test_size, series_len, y_dim, x_dim),dtype=float)\n # calculate loss for each sample\n hist_valance = np.zeros(batch_test_size)\n hist_arousal = np.zeros(batch_test_size)\n for n in range(batch_test_size):\n # Clear stored gradient\n model.zero_grad()\n for timestep in range(sequence_len):\n x_input = np.stack((sample_1[n,timestep,:,:],\n sample_2[n,timestep,:,:],\n sample_3[n,timestep,:,:],\n sample_4[n,timestep,:,:],\n sample_5[n,timestep,:,:]))\n x_var_pred = torch.autograd.Variable(torch.Tensor(x_input).view(-1,input_channels,height,width),\n requires_grad=False).to(device)\n # make prediction\n last_pred, _ = model(x_var_pred, timestep)\n # GPU data should be transferred to CPU\n pred_valance[n,timestep,:,:] = last_pred[0,0,:,:].cpu().data.numpy()\n pred_arousal[n,timestep,:,:] = last_pred[0,1,:,:].cpu().data.numpy()\n # compute the error for each sample\n hist_valance[n] = MSE(label_c_test_valance[n,:,:,:], pred_valance[n,:,:,:])\n hist_arousal[n] = MSE(label_c_test_arousal[n,:,:,:], pred_arousal[n,:,:,:]) \n \n # save prediction as npz file\n np.savez_compressed(os.path.join(output_path,'BBConvLSTM_emotion_pred.npz'),\n valance=pred_valance, arousal=pred_arousal)\n # plot the error\n print (\"******************* Loss with time **********************\")\n fig00 = plt.figure()\n plt.plot(hist_valance, 'r', label=\"Training loss - valance\")\n plt.plot(hist_arousal, 'b', label=\"Training loss - arousal\")\n plt.xlabel('Sample')\n plt.ylabel('MSE Error')\n plt.legend()\n fig00.savefig(os.path.join(output_path,'BBConvLSTM_pred_mse_error.png'),dpi=150) ",
"##############################################################\n################### start prediction loop ###################\n##############################################################\n******************* Loss with time **********************\nWall time: 51.8 s\n"
],
[
" #####################################################################################\n ######## visualization of prediction and implement metrics ########\n #####################################################################################\n # compute mse\n mse_valance = MSE(label_c_test_valance, pred_valance)\n print(mse_valance)\n mse_arousal = MSE(label_c_test_arousal, pred_arousal)\n print(mse_arousal)\n # save output as csv file\n with open(os.path.join(output_path, \"MSE_BBConvLSTM_emotion.csv\"), \"wt+\") as fp:\n writer = csv.writer(fp, delimiter=\",\")\n writer.writerow([\"emotion prediction\"]) # write header\n writer.writerow([\"label valance\"])\n writer.writerow([mse_valance])\n writer.writerow([\"label arousal\"])\n writer.writerow([mse_arousal])",
"0.04842006886017442\n0.05531430360448294\n"
],
[
" %%time\n #################################################################################\n ######## prediction of single subject ########\n #################################################################################\n ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78deaba69a61a1bf26c84a5bf661dcc590cc248 | 171,740 | ipynb | Jupyter Notebook | MOOCS/GAN_Specialization/Course 3 - Apply GANs/Week 1/C3W1_GANs4Augmentation.ipynb | itismesam/Courses-1 | 7669c4460be02b8bbaea2ae79182af2667e9e6b2 | [
"MIT"
] | 40 | 2020-09-30T13:45:50.000Z | 2022-03-10T10:22:19.000Z | MOOCS/GAN_Specialization/Course 3 - Apply GANs/Week 1/C3W1_GANs4Augmentation.ipynb | itismesam/Courses-1 | 7669c4460be02b8bbaea2ae79182af2667e9e6b2 | [
"MIT"
] | null | null | null | MOOCS/GAN_Specialization/Course 3 - Apply GANs/Week 1/C3W1_GANs4Augmentation.ipynb | itismesam/Courses-1 | 7669c4460be02b8bbaea2ae79182af2667e9e6b2 | [
"MIT"
] | 24 | 2020-10-06T07:05:38.000Z | 2022-03-10T10:23:29.000Z | 171.74 | 112,028 | 0.868097 | [
[
[
"# Data Augmentation",
"_____no_output_____"
],
[
"### Goals\nIn this notebook you're going to build a generator that can be used to help create data to train a classifier. There are many cases where this might be useful. If you are interested in any of these topics, you are welcome to explore the linked papers and articles! \n\n- With smaller datasets, GANs can provide useful data augmentation that substantially [improve classifier performance](https://arxiv.org/abs/1711.04340). \n- You have one type of data already labeled and would like to make predictions on [another related dataset for which you have no labels](https://www.nature.com/articles/s41598-019-52737-x). (You'll learn about the techniques for this use case in future notebooks!)\n- You want to protect the privacy of the people who provided their information so you can provide access to a [generator instead of real data](https://www.ahajournals.org/doi/full/10.1161/CIRCOUTCOMES.118.005122). \n- You have [input data with many missing values](https://arxiv.org/abs/1806.02920), where the input dimensions are correlated and you would like to train a model on complete inputs. \n- You would like to be able to identify a real-world abnormal feature in an image [for the purpose of diagnosis](https://link.springer.com/chapter/10.1007/978-3-030-00946-5_11), but have limited access to real examples of the condition. \n\nIn this assignment, you're going to be acting as a bug enthusiast — more on that later. \n\n### Learning Objectives\n1. Understand some use cases for data augmentation and why GANs suit this task.\n2. Implement a classifier that takes a mixed dataset of reals/fakes and analyze its accuracy.",
"_____no_output_____"
],
[
"## Getting Started\n\n### Data Augmentation\nBefore you implement GAN-based data augmentation, you should know a bit about data augmentation in general, specifically for image datasets. It is [very common practice](https://arxiv.org/abs/1712.04621) to augment image-based datasets in ways that are appropriate for a given dataset. This may include having your dataloader randomly flipping images across their vertical axis, randomly cropping your image to a particular size, randomly adding a bit of noise or color to an image in ways that are true-to-life. \n\nIn general, data augmentation helps to stop your model from overfitting to the data, and allows you to make small datasets many times larger. However, a sufficiently powerful classifier often still overfits to the original examples which is why GANs are particularly useful here. They can generate new images instead of simply modifying existing ones.\n\n### CIFAR\nThe [CIFAR-10 and CIFAR-100](https://www.cs.toronto.edu/~kriz/learning-features-2009-TR.pdf) datasets are extremely widely used within machine learning -- they contain many thousands of “tiny” 32x32 color images of different classes representing relatively common real-world objects like airplanes and dogs, with 10 classes in CIFAR-10 and 100 classes in CIFAR-100. In CIFAR-100, there are 20 “superclasses” which each contain five classes. For example, the “fish” superclass contains “aquarium fish, flatfish, ray, shark, trout”. For the purposes of this assignment, you’ll be looking at a small subset of these images to simulate a small data regime, with only 40 images of each class for training.\n\n\n\n### Initializations\nYou will begin by importing some useful libraries and packages and defining a visualization function that has been provided. You will also be re-using your conditional generator and functions code from earlier assignments. This will let you control what class of images to augment for your classifier.",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn.functional as F\nimport matplotlib.pyplot as plt\nfrom torch import nn\nfrom tqdm.auto import tqdm\nfrom torchvision import transforms\nfrom torchvision.utils import make_grid\nfrom torch.utils.data import DataLoader\ntorch.manual_seed(0) # Set for our testing purposes, please do not change!\n\ndef show_tensor_images(image_tensor, num_images=25, size=(3, 32, 32), nrow=5, show=True):\n '''\n Function for visualizing images: Given a tensor of images, number of images, and\n size per image, plots and prints the images in an uniform grid.\n '''\n image_tensor = (image_tensor + 1) / 2\n image_unflat = image_tensor.detach().cpu()\n image_grid = make_grid(image_unflat[:num_images], nrow=nrow)\n plt.imshow(image_grid.permute(1, 2, 0).squeeze())\n if show:\n plt.show()",
"_____no_output_____"
]
],
[
[
"#### Generator",
"_____no_output_____"
]
],
[
[
"class Generator(nn.Module):\n '''\n Generator Class\n Values:\n input_dim: the dimension of the input vector, a scalar\n im_chan: the number of channels of the output image, a scalar\n (CIFAR100 is in color (red, green, blue), so 3 is your default)\n hidden_dim: the inner dimension, a scalar\n '''\n def __init__(self, input_dim=10, im_chan=3, hidden_dim=64):\n super(Generator, self).__init__()\n self.input_dim = input_dim\n # Build the neural network\n self.gen = nn.Sequential(\n self.make_gen_block(input_dim, hidden_dim * 4, kernel_size=4),\n self.make_gen_block(hidden_dim * 4, hidden_dim * 2, kernel_size=4, stride=1),\n self.make_gen_block(hidden_dim * 2, hidden_dim, kernel_size=4),\n self.make_gen_block(hidden_dim, im_chan, kernel_size=2, final_layer=True),\n )\n\n def make_gen_block(self, input_channels, output_channels, kernel_size=3, stride=2, final_layer=False):\n '''\n Function to return a sequence of operations corresponding to a generator block of DCGAN;\n a transposed convolution, a batchnorm (except in the final layer), and an activation.\n Parameters:\n input_channels: how many channels the input feature representation has\n output_channels: how many channels the output feature representation should have\n kernel_size: the size of each convolutional filter, equivalent to (kernel_size, kernel_size)\n stride: the stride of the convolution\n final_layer: a boolean, true if it is the final layer and false otherwise \n (affects activation and batchnorm)\n '''\n if not final_layer:\n return nn.Sequential(\n nn.ConvTranspose2d(input_channels, output_channels, kernel_size, stride),\n nn.BatchNorm2d(output_channels),\n nn.ReLU(inplace=True),\n )\n else:\n return nn.Sequential(\n nn.ConvTranspose2d(input_channels, output_channels, kernel_size, stride),\n nn.Tanh(),\n )\n\n def forward(self, noise):\n '''\n Function for completing a forward pass of the generator: Given a noise tensor, \n returns generated images.\n Parameters:\n noise: a noise tensor with dimensions (n_samples, input_dim)\n '''\n x = noise.view(len(noise), self.input_dim, 1, 1)\n return self.gen(x)\n\n\ndef get_noise(n_samples, input_dim, device='cpu'):\n '''\n Function for creating noise vectors: Given the dimensions (n_samples, input_dim)\n creates a tensor of that shape filled with random numbers from the normal distribution.\n Parameters:\n n_samples: the number of samples to generate, a scalar\n input_dim: the dimension of the input vector, a scalar\n device: the device type\n '''\n return torch.randn(n_samples, input_dim, device=device)\n\ndef combine_vectors(x, y):\n '''\n Function for combining two vectors with shapes (n_samples, ?) and (n_samples, ?)\n Parameters:\n x: (n_samples, ?) the first vector. \n In this assignment, this will be the noise vector of shape (n_samples, z_dim), \n but you shouldn't need to know the second dimension's size.\n y: (n_samples, ?) the second vector.\n Once again, in this assignment this will be the one-hot class vector \n with the shape (n_samples, n_classes), but you shouldn't assume this in your code.\n '''\n return torch.cat([x, y], 1)\n\ndef get_one_hot_labels(labels, n_classes):\n '''\n Function for combining two vectors with shapes (n_samples, ?) and (n_samples, ?)\n Parameters:\n labels: (n_samples, 1) \n n_classes: a single integer corresponding to the total number of classes in the dataset\n '''\n return F.one_hot(labels, n_classes)",
"_____no_output_____"
]
],
[
[
"## Training\nNow you can begin training your models.\nFirst, you will define some new parameters:\n\n* cifar100_shape: the number of pixels in each CIFAR image, which has dimensions 32 x 32 and three channel (for red, green, and blue) so 3 x 32 x 32\n* n_classes: the number of classes in CIFAR100 (e.g. airplane, automobile, bird, cat, deer, dog, frog, horse, ship, truck)",
"_____no_output_____"
]
],
[
[
"cifar100_shape = (3, 32, 32)\nn_classes = 100",
"_____no_output_____"
]
],
[
[
"And you also include the same parameters from previous assignments:\n\n * criterion: the loss function\n * n_epochs: the number of times you iterate through the entire dataset when training\n * z_dim: the dimension of the noise vector\n * display_step: how often to display/visualize the images\n * batch_size: the number of images per forward/backward pass\n * lr: the learning rate\n * device: the device type",
"_____no_output_____"
]
],
[
[
"# Note: n_epochs need to be much larger! \n# I just changed so it would be a small file when I upload to Github\nn_epochs = 1\nz_dim = 64\ndisplay_step = 500\nbatch_size = 64\nlr = 0.0002\ndevice = 'cuda'",
"_____no_output_____"
]
],
[
[
"Then, you want to set your generator's input dimension. Recall that for conditional GANs, the generator's input is the noise vector concatenated with the class vector.",
"_____no_output_____"
]
],
[
[
"generator_input_dim = z_dim + n_classes",
"_____no_output_____"
]
],
[
[
"#### Classifier\n\nFor the classifier, you will use the same code that you wrote in an earlier assignment (the same as previous code for the discriminator as well since the discriminator is a real/fake classifier).",
"_____no_output_____"
]
],
[
[
"class Classifier(nn.Module):\n '''\n Classifier Class\n Values:\n im_chan: the number of channels of the output image, a scalar\n n_classes: the total number of classes in the dataset, an integer scalar\n hidden_dim: the inner dimension, a scalar\n '''\n def __init__(self, im_chan, n_classes, hidden_dim=32):\n super(Classifier, self).__init__()\n self.disc = nn.Sequential(\n self.make_classifier_block(im_chan, hidden_dim),\n self.make_classifier_block(hidden_dim, hidden_dim * 2),\n self.make_classifier_block(hidden_dim * 2, hidden_dim * 4),\n self.make_classifier_block(hidden_dim * 4, n_classes, final_layer=True),\n )\n\n def make_classifier_block(self, input_channels, output_channels, kernel_size=3, stride=2, final_layer=False):\n '''\n Function to return a sequence of operations corresponding to a classifier block; \n a convolution, a batchnorm (except in the final layer), and an activation (except in the final\n Parameters:\n input_channels: how many channels the input feature representation has\n output_channels: how many channels the output feature representation should have\n kernel_size: the size of each convolutional filter, equivalent to (kernel_size, kernel_size)\n stride: the stride of the convolution\n final_layer: a boolean, true if it is the final layer and false otherwise \n (affects activation and batchnorm)\n '''\n if not final_layer:\n return nn.Sequential(\n nn.Conv2d(input_channels, output_channels, kernel_size, stride),\n nn.BatchNorm2d(output_channels),\n nn.LeakyReLU(0.2, inplace=True),\n )\n else:\n return nn.Sequential(\n nn.Conv2d(input_channels, output_channels, kernel_size, stride),\n )\n\n def forward(self, image):\n '''\n Function for completing a forward pass of the classifier: Given an image tensor, \n returns an n_classes-dimension tensor representing fake/real.\n Parameters:\n image: a flattened image tensor with im_chan channels\n '''\n class_pred = self.disc(image)\n return class_pred.view(len(class_pred), -1)",
"_____no_output_____"
]
],
[
[
"#### Pre-training (Optional)\n\nYou are provided the code to pre-train the models (GAN and classifier) given to you in this assignment. However, this is intended only for your personal curiosity -- for the assignment to run as intended, you should not use any checkpoints besides the ones given to you.",
"_____no_output_____"
]
],
[
[
"# This code is here for you to train your own generator or classifier \n# outside the assignment on the full dataset if you'd like -- for the purposes \n# of this assignment, please use the provided checkpoints\nclass Discriminator(nn.Module):\n '''\n Discriminator Class\n Values:\n im_chan: the number of channels of the output image, a scalar\n (MNIST is black-and-white, so 1 channel is your default)\n hidden_dim: the inner dimension, a scalar\n '''\n def __init__(self, im_chan=3, hidden_dim=64):\n super(Discriminator, self).__init__()\n self.disc = nn.Sequential(\n self.make_disc_block(im_chan, hidden_dim, stride=1),\n self.make_disc_block(hidden_dim, hidden_dim * 2),\n self.make_disc_block(hidden_dim * 2, hidden_dim * 4),\n self.make_disc_block(hidden_dim * 4, 1, final_layer=True),\n )\n\n def make_disc_block(self, input_channels, output_channels, kernel_size=4, stride=2, final_layer=False):\n '''\n Function to return a sequence of operations corresponding to a discriminator block of the DCGAN; \n a convolution, a batchnorm (except in the final layer), and an activation (except in the final layer).\n Parameters:\n input_channels: how many channels the input feature representation has\n output_channels: how many channels the output feature representation should have\n kernel_size: the size of each convolutional filter, equivalent to (kernel_size, kernel_size)\n stride: the stride of the convolution\n final_layer: a boolean, true if it is the final layer and false otherwise \n (affects activation and batchnorm)\n '''\n if not final_layer:\n return nn.Sequential(\n nn.Conv2d(input_channels, output_channels, kernel_size, stride),\n nn.BatchNorm2d(output_channels),\n nn.LeakyReLU(0.2, inplace=True),\n )\n else:\n return nn.Sequential(\n nn.Conv2d(input_channels, output_channels, kernel_size, stride),\n )\n\n def forward(self, image):\n '''\n Function for completing a forward pass of the discriminator: Given an image tensor, \n returns a 1-dimension tensor representing fake/real.\n Parameters:\n image: a flattened image tensor with dimension (im_chan)\n '''\n disc_pred = self.disc(image)\n return disc_pred.view(len(disc_pred), -1)\n\ndef train_generator():\n gen = Generator(generator_input_dim).to(device)\n gen_opt = torch.optim.Adam(gen.parameters(), lr=lr)\n discriminator_input_dim = cifar100_shape[0] + n_classes\n disc = Discriminator(discriminator_input_dim).to(device)\n disc_opt = torch.optim.Adam(disc.parameters(), lr=lr)\n\n def weights_init(m):\n if isinstance(m, nn.Conv2d) or isinstance(m, nn.ConvTranspose2d):\n torch.nn.init.normal_(m.weight, 0.0, 0.02)\n if isinstance(m, nn.BatchNorm2d):\n torch.nn.init.normal_(m.weight, 0.0, 0.02)\n torch.nn.init.constant_(m.bias, 0)\n gen = gen.apply(weights_init)\n disc = disc.apply(weights_init)\n\n criterion = nn.BCEWithLogitsLoss()\n cur_step = 0\n mean_generator_loss = 0\n mean_discriminator_loss = 0\n for epoch in range(n_epochs):\n # Dataloader returns the batches and the labels\n for real, labels in dataloader:\n cur_batch_size = len(real)\n # Flatten the batch of real images from the dataset\n real = real.to(device)\n\n # Convert the labels from the dataloader into one-hot versions of those labels\n one_hot_labels = get_one_hot_labels(labels.to(device), n_classes).float()\n\n image_one_hot_labels = one_hot_labels[:, :, None, None]\n image_one_hot_labels = image_one_hot_labels.repeat(1, 1, cifar100_shape[1], cifar100_shape[2])\n\n ### Update discriminator ###\n # Zero out the discriminator gradients\n disc_opt.zero_grad()\n # Get noise corresponding to the current batch_size \n fake_noise = get_noise(cur_batch_size, z_dim, device=device)\n \n # Combine the vectors of the noise and the one-hot labels for the generator\n noise_and_labels = combine_vectors(fake_noise, one_hot_labels)\n fake = gen(noise_and_labels)\n # Combine the vectors of the images and the one-hot labels for the discriminator\n fake_image_and_labels = combine_vectors(fake.detach(), image_one_hot_labels)\n real_image_and_labels = combine_vectors(real, image_one_hot_labels)\n disc_fake_pred = disc(fake_image_and_labels)\n disc_real_pred = disc(real_image_and_labels)\n\n disc_fake_loss = criterion(disc_fake_pred, torch.zeros_like(disc_fake_pred))\n disc_real_loss = criterion(disc_real_pred, torch.ones_like(disc_real_pred))\n disc_loss = (disc_fake_loss + disc_real_loss) / 2\n disc_loss.backward(retain_graph=True)\n disc_opt.step() \n\n # Keep track of the average discriminator loss\n mean_discriminator_loss += disc_loss.item() / display_step\n\n ### Update generator ###\n # Zero out the generator gradients\n gen_opt.zero_grad()\n\n # Pass the discriminator the combination of the fake images and the one-hot labels\n fake_image_and_labels = combine_vectors(fake, image_one_hot_labels)\n\n disc_fake_pred = disc(fake_image_and_labels)\n gen_loss = criterion(disc_fake_pred, torch.ones_like(disc_fake_pred))\n gen_loss.backward()\n gen_opt.step()\n\n # Keep track of the average generator loss\n mean_generator_loss += gen_loss.item() / display_step\n\n if cur_step % display_step == 0 and cur_step > 0:\n print(f\"Step {cur_step}: Generator loss: {mean_generator_loss}, discriminator loss: {mean_discriminator_loss}\")\n show_tensor_images(fake)\n show_tensor_images(real)\n mean_generator_loss = 0\n mean_discriminator_loss = 0\n cur_step += 1\n\ndef train_classifier():\n criterion = nn.CrossEntropyLoss()\n n_epochs = 10\n\n validation_dataloader = DataLoader(\n CIFAR100(\".\", train=False, download=True, transform=transform),\n batch_size=batch_size)\n\n display_step = 10\n batch_size = 512\n lr = 0.0002\n device = 'cuda'\n classifier = Classifier(cifar100_shape[0], n_classes).to(device)\n classifier_opt = torch.optim.Adam(classifier.parameters(), lr=lr)\n cur_step = 0\n for epoch in range(n_epochs):\n for real, labels in tqdm(dataloader):\n cur_batch_size = len(real)\n real = real.to(device)\n labels = labels.to(device)\n\n ### Update classifier ###\n # Get noise corresponding to the current batch_size\n classifier_opt.zero_grad()\n labels_hat = classifier(real.detach())\n classifier_loss = criterion(labels_hat, labels)\n classifier_loss.backward()\n classifier_opt.step()\n\n if cur_step % display_step == 0:\n classifier_val_loss = 0\n classifier_correct = 0\n num_validation = 0\n for val_example, val_label in validation_dataloader:\n cur_batch_size = len(val_example)\n num_validation += cur_batch_size\n val_example = val_example.to(device)\n val_label = val_label.to(device)\n labels_hat = classifier(val_example)\n classifier_val_loss += criterion(labels_hat, val_label) * cur_batch_size\n classifier_correct += (labels_hat.argmax(1) == val_label).float().sum()\n\n print(f\"Step {cur_step}: \"\n f\"Classifier loss: {classifier_val_loss.item() / num_validation}, \"\n f\"classifier accuracy: {classifier_correct.item() / num_validation}\")\n cur_step += 1\n",
"_____no_output_____"
]
],
[
[
"## Tuning the Classifier\nAfter two courses, you've probably had some fun debugging your GANs and have started to consider yourself a bug master. For this assignment, your mastery will be put to the test on some interesting bugs... well, bugs as in insects.\n\nAs a bug master, you want a classifier capable of classifying different species of bugs: bees, beetles, butterflies, caterpillar, and more. Luckily, you found a great dataset with a lot of animal species and objects, and you trained your classifier on that.\n\nBut the bug classes don't do as well as you would like. Now your plan is to train a GAN on the same data so it can generate new bugs to make your classifier better at distinguishing between all of your favorite bugs!\n\nYou will fine-tune your model by augmenting the original real data with fake data and during that process, observe how to increase the accuracy of your classifier with these fake, GAN-generated bugs. After this, you will prove your worth as a bug master.",
"_____no_output_____"
],
[
"#### Sampling Ratio\n\nSuppose that you've decided that although you have this pre-trained general generator and this general classifier, capable of identifying 100 classes with some accuracy (~17%), what you'd really like is a model that can classify the five different kinds of bugs in the dataset. You'll fine-tune your model by augmenting your data with the generated images. Keep in mind that both the generator and the classifier were trained on the same images: the 40 images per class you painstakingly found so your generator may not be great. This is the caveat with data augmentation, ultimately you are still bound by the real data that you have but you want to try and create more. To make your models even better, you would need to take some more bug photos, label them, and add them to your training set and/or use higher quality photos.\n\nTo start, you'll first need to write some code to sample a combination of real and generated images. Given a probability, `p_real`, you'll need to generate a combined tensor where roughly `p_real` of the returned images are sampled from the real images. Note that you should not interpolate the images here: you should choose each image from the real or fake set with a given probability. For example, if your real images are a tensor of `[[1, 2, 3, 4, 5]]` and your fake images are a tensor of `[[-1, -2, -3, -4, -5]]`, and `p_real = 0.2`, two potential return values are `[[1, -2, 3, -4, -5]]` or `[[-1, 2, -3, -4, -5]]`\n\nIn addition, we will expect the images to remain in the same order to maintain their alignment with their labels (this applies to the fake images too!). \n\n<details>\n<summary>\n<font size=\"3\" color=\"green\">\n<b>Optional hints for <code><font size=\"4\">combine_sample</font></code></b>\n</font>\n</summary>\n\n1. This code probably shouldn't be much longer than 3 lines\n2. You can index using a set of booleans which have the same length as your tensor\n3. You want to generate an unbiased sample, which you can do (for example) with `torch.rand(length_reals) > p`.\n4. There are many approaches here that will give a correct answer here. You may find [`torch.rand`](https://pytorch.org/docs/stable/generated/torch.rand.html) or [`torch.bernoulli`](https://pytorch.org/docs/master/generated/torch.bernoulli.html) useful. \n5. You don't want to edit an argument in place, so you may find [`cur_tensor.clone()`](https://pytorch.org/docs/stable/tensors.html) useful too, which makes a copy of `cur_tensor`. \n\n</details>",
"_____no_output_____"
]
],
[
[
"# UNQ_C1 (UNIQUE CELL IDENTIFIER, DO NOT EDIT)\n# GRADED FUNCTION: combine_sample\ndef combine_sample(real, fake, p_real):\n '''\n Function to take a set of real and fake images of the same length (x)\n and produce a combined tensor with length (x) and sampled at the target probability\n Parameters:\n real: a tensor of real images, length (x)\n fake: a tensor of fake images, length (x)\n p_real: the probability the images are sampled from the real set\n '''\n #### START CODE HERE ####\n assert real.device == fake.device\n assert p_real>=0 and p_real<=1\n assert real.shape == fake.shape\n device = real.device\n chosen = torch.rand(real.shape[0], device=device) < p_real\n target_images = torch.zeros((real.shape), device=device)\n target_images[chosen] = real[chosen]\n target_images[~chosen] = fake[~chosen]\n #### END CODE HERE ####\n return target_images",
"_____no_output_____"
],
[
"n_test_samples = 9999\ntest_combination = combine_sample(\n torch.ones(n_test_samples, 1), \n torch.zeros(n_test_samples, 1), \n 0.3\n)\n# Check that the shape is right\nassert tuple(test_combination.shape) == (n_test_samples, 1)\n# Check that the ratio is right\nassert torch.abs(test_combination.mean() - 0.3) < 0.05\n# Make sure that no mixing happened\nassert test_combination.median() < 1e-5\n\ntest_combination = combine_sample(\n torch.ones(n_test_samples, 10, 10), \n torch.zeros(n_test_samples, 10, 10), \n 0.8\n)\n# Check that the shape is right\nassert tuple(test_combination.shape) == (n_test_samples, 10, 10)\n# Make sure that no mixing happened\nassert torch.abs((test_combination.sum([1, 2]).median()) - 100) < 1e-5\n\ntest_reals = torch.arange(n_test_samples)[:, None].float()\ntest_fakes = torch.zeros(n_test_samples, 1)\ntest_saved = (test_reals.clone(), test_fakes.clone())\ntest_combination = combine_sample(test_reals, test_fakes, 0.3)\n# Make sure that the sample isn't biased\nassert torch.abs((test_combination.mean() - 1500)) < 100\n# Make sure no inputs were changed\nassert torch.abs(test_saved[0] - test_reals).sum() < 1e-3\nassert torch.abs(test_saved[1] - test_fakes).sum() < 1e-3\n\ntest_fakes = torch.arange(n_test_samples)[:, None].float()\ntest_combination = combine_sample(test_reals, test_fakes, 0.3)\n# Make sure that the order is maintained\nassert torch.abs(test_combination - test_reals).sum() < 1e-4\nif torch.cuda.is_available():\n # Check that the solution matches the input device\n assert str(combine_sample(\n torch.ones(n_test_samples, 10, 10).cuda(), \n torch.zeros(n_test_samples, 10, 10).cuda(),\n 0.8\n ).device).startswith(\"cuda\")\nprint(\"Success!\")",
"Success!\n"
]
],
[
[
"Now you have a challenge: find a `p_real` and a generator image such that your classifier gets an average of a 51% accuracy or higher on the insects, when evaluated with the `eval_augmentation` function. **You'll need to fill in `find_optimal` to find these parameters to solve this part!** Note that if your answer takes a very long time to run, you may need to hard-code the solution it finds. \n\nWhen you're training a generator, you will often have to look at different checkpoints and choose one that does the best (either empirically or using some evaluation method). Here, you are given four generator checkpoints: `gen_1.pt`, `gen_2.pt`, `gen_3.pt`, `gen_4.pt`. You'll also have some scratch area to write whatever code you'd like to solve this problem, but you must return a `p_real` and an image name of your selected generator checkpoint. You can hard-code/brute-force these numbers if you would like, but you are encouraged to try to solve this problem in a more general way. In practice, you would also want a test set (since it is possible to overfit on a validation set), but for simplicity you can just focus on the validation set.",
"_____no_output_____"
]
],
[
[
"# UNQ_C2 (UNIQUE CELL IDENTIFIER, DO NOT EDIT)\n# GRADED FUNCTION: find_optimal\ndef find_optimal():\n # In the following section, you can write the code to choose your optimal answer\n # You can even use the eval_augmentation function in your code if you'd like!\n gen_names = [\n \"gen_1.pt\",\n \"gen_2.pt\",\n \"gen_3.pt\",\n \"gen_4.pt\"\n ]\n p_real = [0.1*x for x in range(1,10)]\n best_so_far = 0\n #### START CODE HERE #### \n for p in p_real:\n for gen in gen_names:\n avg_acc = eval_augmentation(p, gen, n_test=3)\n if avg_acc > best_so_far:\n best_so_far = avg_acc\n best_p_real = p\n best_gen_name = gen\n \n #### END CODE HERE ####\n return best_p_real, best_gen_name\n\ndef augmented_train(p_real, gen_name):\n gen = Generator(generator_input_dim).to(device)\n gen.load_state_dict(torch.load(gen_name))\n\n classifier = Classifier(cifar100_shape[0], n_classes).to(device)\n classifier.load_state_dict(torch.load(\"class.pt\"))\n criterion = nn.CrossEntropyLoss()\n batch_size = 256\n\n train_set = torch.load(\"insect_train.pt\")\n val_set = torch.load(\"insect_val.pt\")\n dataloader = DataLoader(\n torch.utils.data.TensorDataset(train_set[\"images\"], train_set[\"labels\"]),\n batch_size=batch_size,\n shuffle=True\n )\n validation_dataloader = DataLoader(\n torch.utils.data.TensorDataset(val_set[\"images\"], val_set[\"labels\"]),\n batch_size=batch_size\n )\n\n display_step = 1\n lr = 0.0002\n n_epochs = 20\n classifier_opt = torch.optim.Adam(classifier.parameters(), lr=lr)\n cur_step = 0\n best_score = 0\n for epoch in range(n_epochs):\n for real, labels in dataloader:\n real = real.to(device)\n # Flatten the image\n labels = labels.to(device)\n one_hot_labels = get_one_hot_labels(labels.to(device), n_classes).float()\n\n ### Update classifier ###\n # Get noise corresponding to the current batch_size\n classifier_opt.zero_grad()\n cur_batch_size = len(labels)\n fake_noise = get_noise(cur_batch_size, z_dim, device=device)\n noise_and_labels = combine_vectors(fake_noise, one_hot_labels)\n fake = gen(noise_and_labels)\n\n target_images = combine_sample(real.clone(), fake.clone(), p_real)\n labels_hat = classifier(target_images.detach())\n classifier_loss = criterion(labels_hat, labels)\n classifier_loss.backward()\n classifier_opt.step()\n\n # Calculate the accuracy on the validation set\n if cur_step % display_step == 0 and cur_step > 0:\n classifier_val_loss = 0\n classifier_correct = 0\n num_validation = 0\n with torch.no_grad():\n for val_example, val_label in validation_dataloader:\n cur_batch_size = len(val_example)\n num_validation += cur_batch_size\n val_example = val_example.to(device)\n val_label = val_label.to(device)\n labels_hat = classifier(val_example)\n classifier_val_loss += criterion(labels_hat, val_label) * cur_batch_size\n classifier_correct += (labels_hat.argmax(1) == val_label).float().sum()\n accuracy = classifier_correct.item() / num_validation\n if accuracy > best_score:\n best_score = accuracy\n cur_step += 1\n return best_score\n\ndef eval_augmentation(p_real, gen_name, n_test=20):\n total = 0\n for i in range(n_test):\n total += augmented_train(p_real, gen_name)\n return total / n_test\n\nbest_p_real, best_gen_name = find_optimal()\nperformance = eval_augmentation(best_p_real, best_gen_name)\nprint(f\"Your model had an accuracy of {performance:0.1%}\")\nassert performance > 0.51\nprint(\"Success!\")",
"Your model had an accuracy of 52.0%\nSuccess!\n"
]
],
[
[
"You'll likely find that the worst performance is when the generator is performing alone: this corresponds to the case where you might be trying to hide the underlying examples from the classifier. Perhaps you don't want other people to know about your specific bugs!",
"_____no_output_____"
]
],
[
[
"accuracies = []\np_real_all = torch.linspace(0, 1, 21)\nfor p_real_vis in tqdm(p_real_all):\n accuracies += [eval_augmentation(p_real_vis, best_gen_name, n_test=4)]\nplt.plot(p_real_all.tolist(), accuracies)\nplt.ylabel(\"Accuracy\")\n_ = plt.xlabel(\"Percent Real Images\")",
"_____no_output_____"
]
],
[
[
"Here's a visualization of what the generator is actually generating, with real examples of each class above the corresponding generated image. ",
"_____no_output_____"
]
],
[
[
"examples = [4, 41, 80, 122, 160]\ntrain_images = torch.load(\"insect_train.pt\")[\"images\"][examples]\ntrain_labels = torch.load(\"insect_train.pt\")[\"labels\"][examples]\n\none_hot_labels = get_one_hot_labels(train_labels.to(device), n_classes).float()\nfake_noise = get_noise(len(train_images), z_dim, device=device)\nnoise_and_labels = combine_vectors(fake_noise, one_hot_labels)\ngen = Generator(generator_input_dim).to(device)\ngen.load_state_dict(torch.load(best_gen_name))\n\nfake = gen(noise_and_labels)\nshow_tensor_images(torch.cat([train_images.cpu(), fake.cpu()]))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e78df73d983e28476a9fe79350df83f772d7f85a | 39,028 | ipynb | Jupyter Notebook | 03_TensorBoard/lab-02-3-linear_regression_tensorflow.org.ipynb | jetsonworld/TensorFlow_On_JetsonNano | 216b21f63cb0a33e1a2a4d39be8d250c5ce1b504 | [
"MIT"
] | 4 | 2021-01-30T14:35:42.000Z | 2021-07-19T06:55:34.000Z | 03_TensorBoard/lab-02-3-linear_regression_tensorflow.org.ipynb | jetsonworld/TensorFlow_On_JetsonNano | 216b21f63cb0a33e1a2a4d39be8d250c5ce1b504 | [
"MIT"
] | 1 | 2020-06-12T15:05:14.000Z | 2020-06-23T03:40:46.000Z | 03_TensorBoard/lab-02-3-linear_regression_tensorflow.org.ipynb | jetsonworld/TensorFlow_On_JetsonNano | 216b21f63cb0a33e1a2a4d39be8d250c5ce1b504 | [
"MIT"
] | 4 | 2021-01-30T14:35:46.000Z | 2021-08-13T02:57:39.000Z | 133.201365 | 29,923 | 0.556831 | [
[
[
"# Lab-02-3 linear regression tensorflow.org",
"_____no_output_____"
]
],
[
[
"# From https://www.tensorflow.org/get_started/get_started\nimport tensorflow as tf",
"/home/ubuntu/.local/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:526: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint8 = np.dtype([(\"qint8\", np.int8, 1)])\n/home/ubuntu/.local/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:527: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint8 = np.dtype([(\"quint8\", np.uint8, 1)])\n/home/ubuntu/.local/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:528: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint16 = np.dtype([(\"qint16\", np.int16, 1)])\n/home/ubuntu/.local/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:529: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint16 = np.dtype([(\"quint16\", np.uint16, 1)])\n/home/ubuntu/.local/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:530: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint32 = np.dtype([(\"qint32\", np.int32, 1)])\n/home/ubuntu/.local/lib/python3.6/site-packages/tensorflow/python/framework/dtypes.py:535: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n np_resource = np.dtype([(\"resource\", np.ubyte, 1)])\n"
]
],
[
[
"## Variable",
"_____no_output_____"
]
],
[
[
"# Model parameters\nW = tf.Variable([.3], tf.float32)\nb = tf.Variable([-.3], tf.float32)\n\n# Model input and output\nx = tf.placeholder(tf.float32)\ny = tf.placeholder(tf.float32)",
"WARNING:tensorflow:From /home/ubuntu/.local/lib/python3.6/site-packages/tensorflow/python/framework/op_def_library.py:263: colocate_with (from tensorflow.python.framework.ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nColocations handled automatically by placer.\n"
]
],
[
[
"## Our Model",
"_____no_output_____"
]
],
[
[
"linear_model = x * W + b\n\n# cost/loss function\nloss = tf.reduce_sum(tf.square(linear_model - y)) # sum of the squares",
"_____no_output_____"
]
],
[
[
"## Minimize",
"_____no_output_____"
]
],
[
[
"# optimizer\noptimizer = tf.train.GradientDescentOptimizer(0.01)\ntrain = optimizer.minimize(loss)",
"WARNING:tensorflow:From /home/ubuntu/.local/lib/python3.6/site-packages/tensorflow/python/ops/math_ops.py:3066: to_int32 (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.cast instead.\n"
]
],
[
[
"## X and Y data",
"_____no_output_____"
]
],
[
[
"# training data\nx_train = [1, 2, 3, 4]\ny_train = [0, -1, -2, -3]",
"_____no_output_____"
]
],
[
[
"## Fit the line",
"_____no_output_____"
]
],
[
[
"# training loop\ninit = tf.global_variables_initializer()\nsess = tf.Session()\nsess.run(init) # reset values to wrong\nfor i in range(1000):\n sess.run(train, {x: x_train, y: y_train})",
"_____no_output_____"
]
],
[
[
"## evaluate training accuracy",
"_____no_output_____"
]
],
[
[
"# evaluate training accuracy\ncurr_W, curr_b, curr_loss = sess.run([W, b, loss], {x: x_train, y: y_train})\nprint(\"W: %s b: %s loss: %s\" % (curr_W, curr_b, curr_loss))",
"W: [-0.9999969] b: [0.9999908] loss: 5.6999738e-11\n"
],
[
"# Functions to show the Graphs\nimport numpy as np\nfrom IPython.display import clear_output, Image, display, HTML\n\ndef strip_consts(graph_def, max_const_size=32):\n \"\"\"Strip large constant values from graph_def.\"\"\"\n strip_def = tf.GraphDef()\n for n0 in graph_def.node:\n n = strip_def.node.add() \n n.MergeFrom(n0)\n if n.op == 'Const':\n tensor = n.attr['value'].tensor\n size = len(tensor.tensor_content)\n if size > max_const_size:\n tensor.tensor_content = b\"<stripped %d bytes>\"%size\n return strip_def\n\ndef show_graph(graph_def, max_const_size=32):\n \"\"\"Visualize TensorFlow graph.\"\"\"\n if hasattr(graph_def, 'as_graph_def'):\n graph_def = graph_def.as_graph_def()\n strip_def = strip_consts(graph_def, max_const_size=max_const_size)\n code = \"\"\"\n <script>\n function load() {{\n document.getElementById(\"{id}\").pbtxt = {data};\n }}\n </script>\n <link rel=\"import\" href=\"https://tensorboard.appspot.com/tf-graph-basic.build.html\" onload=load()>\n <div style=\"height:600px\">\n <tf-graph-basic id=\"{id}\"></tf-graph-basic>\n </div>\n \"\"\".format(data=repr(str(strip_def)), id='graph'+str(np.random.rand()))\n\n iframe = \"\"\"\n <iframe seamless style=\"width:1200px;height:620px;border:0\" srcdoc=\"{}\"></iframe>\n \"\"\".format(code.replace('\"', '"'))\n display(HTML(iframe))",
"_____no_output_____"
],
[
"show_graph(tf.get_default_graph())",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
e78dfcbbaa5c84e51990936b0a1fbe22e1b1a06a | 1,030,067 | ipynb | Jupyter Notebook | head_view_xlnet.ipynb | chingheng113/bertviz | 948a3c0e2c27053b14a1a78bfabe59f0dc1e98d9 | [
"Apache-2.0"
] | 1 | 2019-11-26T07:01:35.000Z | 2019-11-26T07:01:35.000Z | head_view_xlnet.ipynb | minoriwww/bertviz | 331a6cd1108a020610a46e448b07a89e53c6bee4 | [
"Apache-2.0"
] | null | null | null | head_view_xlnet.ipynb | minoriwww/bertviz | 331a6cd1108a020610a46e448b07a89e53c6bee4 | [
"Apache-2.0"
] | null | null | null | 2,072.569416 | 1,011,405 | 0.842194 | [
[
[
"from bertviz.pytorch_transformers_attn import XLNetTokenizer, XLNetModel\nfrom bertviz.head_view import show",
"_____no_output_____"
],
[
"%%javascript\nrequire.config({\n paths: {\n d3: '//cdnjs.cloudflare.com/ajax/libs/d3/3.4.8/d3.min',\n jquery: '//ajax.googleapis.com/ajax/libs/jquery/2.0.0/jquery.min',\n }\n});",
"_____no_output_____"
],
[
"model_type = 'xlnet'\nmodel_version = 'xlnet-large-cased'\nmodel = XLNetModel.from_pretrained(model_version)\ntokenizer = XLNetTokenizer.from_pretrained(model_version)\ntext = \"At the store she bought apples, oranges\"\nshow(model, model_type, tokenizer, text)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
e78dfd0d846fb8aebb9e3fffe8f9f9e0405a36d5 | 352,670 | ipynb | Jupyter Notebook | examples/notebooks/0.0-example-usage.ipynb | jonluntzel/pulse2percept | dbe17230be0354c4cfc57a72b649c611fe496464 | [
"BSD-3-Clause"
] | null | null | null | examples/notebooks/0.0-example-usage.ipynb | jonluntzel/pulse2percept | dbe17230be0354c4cfc57a72b649c611fe496464 | [
"BSD-3-Clause"
] | null | null | null | examples/notebooks/0.0-example-usage.ipynb | jonluntzel/pulse2percept | dbe17230be0354c4cfc57a72b649c611fe496464 | [
"BSD-3-Clause"
] | null | null | null | 918.411458 | 327,956 | 0.942666 | [
[
[
"# pulse2percept: Example Usage\n\nThis notebook illustrates a simple used-case of the software.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nimport pulse2percept as p2p",
"/home/mbeyeler/anaconda3/lib/python3.5/site-packages/skvideo/__init__.py:356: UserWarning: avconv/avprobe not found in path: \n warnings.warn(\"avconv/avprobe not found in path: \" + str(path), UserWarning)\n2018-08-15 14:30:31,105 [pulse2percept] [INFO] Welcome to pulse2percept\n"
]
],
[
[
"## 1. Setting up the Simulation\n\n### 1.1 Setting up the Implant\n\nAll retinal implants live in the `implants` module.\n\nYou can either create your own `p2p.implants.ElectrodeArray`, or choose from one of the pre-defined types:\n\n- `p2p.implants.ArgusI`: The A16 array has 16 electrodes arranged in a 4x4 grid.\n Electrodes are 800um apart and either 260um or 520um in diameter.\n- `p2p.implants.ArgusII`: The A60 array has 60 electrodes arranged in a 6x10 grid.\n Electrodes are 525um apart and 200um in diameter.\n\nIf you are running this code in a Jupyter Notebook, you can see what arguments these objects take by starting to type the object's name, then hitting <tt>Shift+TAB</tt>:\n\n argus = p2p.implants.ArgusII(Shift+TAB\n \nIf you want to see more of the documentation, hit <tt>TAB</tt> again while still holding down <tt>Shift</tt> (so it's <tt>Shift+TAB+TAB</tt>).",
"_____no_output_____"
]
],
[
[
"# Load an Argus II array\nargus = p2p.implants.ArgusI(x_center=-800, y_center=-400, h=0, rot=np.deg2rad(-35), eye='RE')",
"_____no_output_____"
]
],
[
[
"### 1.2 Starting the Simulation Framework\n\nAll simulations revolve around the `p2p.Simulation` object, which starts a new simulation framework.\nOnce an implant `argus` has been passed, it cannot be changed.\n\nYou can also specify on which backend to run the simulation via `engine`:\n\n- `'serial'`: Run a single-thread computation.\n- `'joblib'`: Use JobLib for paralellization. Specify the number of jobs via `n_jobs`.\n- `'dask'`: Use Dask for parallelization. Specify the number of jobs via `n_jobs`",
"_____no_output_____"
]
],
[
[
"# Start the simulation framework\nsim = p2p.Simulation(argus, engine='joblib', n_jobs=1)",
"_____no_output_____"
]
],
[
[
"### 1.3 Specifying the Retinal Model",
"_____no_output_____"
],
[
"The `p2p.Simulation` object allows you access every layer of the retinal model via setter functions:\n\n- `set_optic_fiber_layer`: Specify parameters of the optic fiber layer (OFL), where the ganglion cell axons live.\n- `set_ganglion_cell_layer`: Specify parameters of the ganglion cell layer (GCL), where the ganglion cell bodies live.\n- `set_bipolar_cell_layer`: Coming soon.",
"_____no_output_____"
],
[
"In `set_optic_fiber_layer`, you can specify parameters such as the spatial sampling rate (`s_sample` in microns), or the dimensions of the retinal grid to simulate:",
"_____no_output_____"
]
],
[
[
"# Set parameters of the optic fiber layer (OFL)\n# In previous versions of the model, this used to be called the `Retina`\n# object, which created a spatial grid and generated the axtron streak map.\n\n# Set the spatial sampling step (microns) of the retinal grid\ns_sample = 100\nsim.set_optic_fiber_layer(sampling=s_sample, x_range=(-3500, 1500),\n n_axons=501, n_rho=801, rho_range=(4, 45),\n sensitivity_rule='decay', decay_const=5.0,\n contribution_rule='max')",
"2018-08-15 14:30:31,260 [pulse2percept.retina] [INFO] Loading file \"./retina_RE_s100_a501_r801_5000x5000.npz\".\n"
]
],
[
[
"In `set_ganglion_cell_layer`, you can choose from one of the available ganglion cell models:\n- `'latest'`: The latest ganglion cell model for epiretinal and subretinal devices (experimental).\n- `'Nanduri2012'`: A model of temporal sensitivity as described in Nanduri et al. (IOVS, 2012).\n- `'Horsager2009'`: A model of temporal sensitivity as described in Horsager et al. (IOVS, 2009).\n\nYou can also create your own custom model (see [0.3-add-your-own-model.ipynb](0.3-add-your-own-model.ipynb)).",
"_____no_output_____"
]
],
[
[
"# Set parameters of the ganglion cell layer (GCL)\n# In previous versions of the model, this used to be called `TemporalModel`.\n\n# Set the temporal sampling step (seconds)\nt_sample = 0.01 / 1000\nsim.set_ganglion_cell_layer('Nanduri2012', tsample=t_sample)",
"_____no_output_____"
]
],
[
[
"### 1.4 Specifying the input stimulus",
"_____no_output_____"
],
[
"All stimulation protocols live in `p2p.stimuli`. You can choose from the following:\n\n- `MonophasicPulse`: A single monophasic pulse of either cathodic (negative) or anodic (positive) current.\n- `BiphasicPulse`: A single biphasic pulse going either cathodic-first or anodic-first.\n- `PulseTrain`: A pulse train made from individual biphasic pulses with a specific amplitude, frequency, and duration.\n- `image2pulsetrain`: Converts an image to a series of pulse trains (pixel-by-pixel).\n- `video2pulsetrain`: Converts a video to a series of pulse trains (pixel-by-pixel).",
"_____no_output_____"
]
],
[
[
"# Send a pulse train to two specific electrodes, set all others to zero\nstim = {\n 'C1': p2p.stimuli.PulseTrain(t_sample, freq=50, amp=20, dur=0.5), \n 'B3': p2p.stimuli.PulseTrain(t_sample, freq=50, amp=20, dur=0.5)\n}",
"_____no_output_____"
]
],
[
[
"We can visualize the specified electrode array and its location on the retina with respect to the optic disc:",
"_____no_output_____"
]
],
[
[
"p2p.viz.plot_fundus(argus, stim, upside_down=False);",
"_____no_output_____"
]
],
[
[
"# 2. Running the Simulation",
"_____no_output_____"
],
[
"Simulations are run by passing an input stimulus `stim` to `p2p.pulse2percept`. Output is a `p2p.utils.TimeSeries` objects that contains the brightness changes over time for each pixel.",
"_____no_output_____"
]
],
[
[
"# Run a simulation\n# - tol: ignore pixels whose efficient current is smaller than 10% of the max\n# - layers: simulate ganglion cell layer (GCL) and optic fiber layer (OFL),\n# but ignore inner nuclear layer (INL) for now\npercept = sim.pulse2percept(stim, tol=0.1, layers=['GCL', 'OFL'])",
"2018-08-15 14:30:32,377 [pulse2percept.api] [INFO] Starting pulse2percept...\n2018-08-15 14:30:33,885 [pulse2percept.api] [INFO] tol=10.0%, 2089/2601 px selected\n2018-08-15 14:30:58,988 [pulse2percept.api] [INFO] Done.\n"
]
],
[
[
"# 3. Analyzing Output",
"_____no_output_____"
],
[
"You can look at the brightness time course of every pixel, or you can simply plot the brightest frame in the whole brightness \"movie\":",
"_____no_output_____"
]
],
[
[
"frame = p2p.get_brightest_frame(percept)\nplt.figure(figsize=(8, 5))\nplt.imshow(frame.data, cmap='gray')\nplt.colorbar()",
"_____no_output_____"
]
],
[
[
"Finally, you can also dump the percept to file, by creating MP4 or MOV videos (requires Scikit-Video).",
"_____no_output_____"
]
],
[
[
"# This requires ffmpeg or libav-tools\np2p.files.save_video(percept, 'percept.mp4', fps=30)",
"2018-08-15 14:31:00,422 [pulse2percept.files] [INFO] Saved video to file 'percept.mp4'.\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e78dfe6bbac32910bf444b14c4372cf120a1dd3b | 4,382 | ipynb | Jupyter Notebook | Re/REGEX_Exercises.ipynb | reddyprasade/Natural-Language-Processing | d02ef63832f595635645ea3468bf5dc8cafc0e76 | [
"Apache-2.0"
] | null | null | null | Re/REGEX_Exercises.ipynb | reddyprasade/Natural-Language-Processing | d02ef63832f595635645ea3468bf5dc8cafc0e76 | [
"Apache-2.0"
] | null | null | null | Re/REGEX_Exercises.ipynb | reddyprasade/Natural-Language-Processing | d02ef63832f595635645ea3468bf5dc8cafc0e76 | [
"Apache-2.0"
] | null | null | null | 27.217391 | 402 | 0.588544 | [
[
[
"# Regular Expressions Exercises\n\nLets download the complete works of sherlock holmes and do some detective work using REGEX.\n\nFor many of these exercises it is helpful to compile a regular expression p and then use p.findall() followed by some additional simple python processing to answer the questions\n\nstart by importing re",
"_____no_output_____"
]
],
[
[
"import re",
"_____no_output_____"
]
],
[
[
"now lets download [sherlock holmes](https://sherlock-holm.es/stories/plain-text/cnus.txt)",
"_____no_output_____"
],
[
"If we open that file we will get access to the complete works of Sherlock Holmes",
"_____no_output_____"
]
],
[
[
"text = ''\nwith open('Data/cnus.txt','r') as f:\n text = \" \".join([l.strip() for l in f.readlines()])",
"_____no_output_____"
],
[
"text[2611:3000]",
"_____no_output_____"
]
],
[
[
"## Question 1\n\nOne of Sherlock Holmes' famous catch phrases is the use of the word 'undoubtedly'\n\n* How many times is the word 'undoubtedly' used?",
"_____no_output_____"
],
[
"## Question 2\n\nCharacters are announced very deliberatly in the language of the setting in Victorian England. We can use this later to find characters in the book. But for now let's practice on a character we know\n\nHow often is Sherlock Holmes refered to by 'Mr. Sherlock Holmes' vs 'Sherlock Holmes' vs. 'Mr. Holmes' vs 'Sherlock'",
"_____no_output_____"
],
[
"## Question 3\n\n* Find all the doctors in the collection\n \n* make a list of all the characters that appear in the collection (hint: Mrs. Mr. Miss Dr. etc)",
"_____no_output_____"
],
[
"## Question 4\n\n* Search out all the years and dates that appear in the story",
"_____no_output_____"
],
[
"## Question 5\n\nSherlock holmes is frequently smoking his pipe. But like many verbs in english, there are many ways that the word smoking can be conjugated depending on the context.\n\n* capture all sentences that take about smoking (smoke, smokes, smoking, smoked)\n* capture the two words that appear after the smoking word\n* capture the two words that appear before the smoking word",
"_____no_output_____"
],
[
" \n## Question 6\n\nOften we will recieve a block of unstructured text and want to use REGEX to provide some structure. In this case, we may want to split the book by chapter.\n\n* use the re.split() function to split the books by chapter.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
e78e023824742672332c5bc9b97f8099c569339a | 2,941 | ipynb | Jupyter Notebook | (Not important)TicTacToe Board render.ipynb | tejasrajanna/Discord_TicTacToe_Bot | 5280a3a9a39b3609947589c6fb72ff3f23f6ef7f | [
"MIT"
] | null | null | null | (Not important)TicTacToe Board render.ipynb | tejasrajanna/Discord_TicTacToe_Bot | 5280a3a9a39b3609947589c6fb72ff3f23f6ef7f | [
"MIT"
] | 2 | 2022-01-13T03:47:35.000Z | 2022-03-12T00:57:52.000Z | (Not important)TicTacToe Board render.ipynb | tejasrajanna/Discord_TicTacToe_Bot | 5280a3a9a39b3609947589c6fb72ff3f23f6ef7f | [
"MIT"
] | 2 | 2021-04-29T18:06:29.000Z | 2021-05-14T16:22:03.000Z | 21.158273 | 71 | 0.466848 | [
[
[
"from PIL import Image\n \n# Opening the primary image (used in background)\nimg0 = Image.open(r\"Green grid.png\")\n# Opening the secondary images (overlay image)\nimg1 = Image.open(r\"cross white.png\")\nimg2 = Image.open(r\"circle white.png\") \nimar=[img1,img2]",
"_____no_output_____"
],
[
"\n \n# Pasting img2 image on top of img0 \n# starting at coordinates (0, 0)\nw=131\nh=129\n#[0,0]\nimg0.paste(img2, (0,0), mask = img2)\n\n#[0,1]\nimg0.paste(img2, (w,0), mask = img2)\n\n#[0,2]\nimg0.paste(img2, (2*w,0), mask = img2)\n\n#[1,0]\nimg0.paste(img2, (0,h), mask = img2)\n\n#[1,1]\nimg0.paste(img2, (w,h), mask = img2)\n\n#[1,2]\nimg0.paste(img2, (2*w,h), mask = img2)\n\n#[2,0]\nimg0.paste(img2, (0,2*h), mask = img2)\n\n#[2,1]\nimg0.paste(img2, (w,2*h), mask = img2)\n\n#[2,2]\nimg0.paste(img2, (2*w,2*h), mask = img2)\n \n# Displaying the image\nimg0.show()",
"_____no_output_____"
],
[
"img0 = Image.open(r\"Green grid.png\")\nimg1 = Image.open(r\"cross white.png\")\nimg2 = Image.open(r\"circle white.png\") \nimar=[img1,img2]\nw=131;h=129\nPos=[[(0,0),(w,0),(2*w,0)],\n [(0,h),(w,h),(2*w,h)],\n [(0,2*h),(w,2*h),(2*w,2*h)]]\n\ndef render(row, col, piece):\n img0.paste(imar[piece], Pos[row][col], mask = imar[piece])\n img0.show()",
"_____no_output_____"
],
[
"img0.show()",
"_____no_output_____"
],
[
"render(0,2,0)",
"_____no_output_____"
],
[
"render(2,1,1)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78e21cdcb13a0c3394a631791ec4a8516d489a0 | 7,804 | ipynb | Jupyter Notebook | 02-bqplot/examples/Index.ipynb | dushyantkhosla/dataviz | 05a004a390d180d87be2d09873c3f7283c2a2e27 | [
"MIT"
] | null | null | null | 02-bqplot/examples/Index.ipynb | dushyantkhosla/dataviz | 05a004a390d180d87be2d09873c3f7283c2a2e27 | [
"MIT"
] | 2 | 2021-03-25T22:11:43.000Z | 2022-03-02T22:43:47.000Z | 02-bqplot/examples/Index.ipynb | dushyantkhosla/viz4ds | 05a004a390d180d87be2d09873c3f7283c2a2e27 | [
"MIT"
] | null | null | null | 29.673004 | 317 | 0.621989 | [
[
[
"# bqplot",
"_____no_output_____"
],
[
"`bqplot` is a [Grammar of Graphics](https://www.cs.uic.edu/~wilkinson/TheGrammarOfGraphics/GOG.html) based interactive plotting framework for the Jupyter notebook. The library offers a simple bridge between `Python` and `d3.js` allowing users to quickly and easily build complex GUI's with layered interactions.",
"_____no_output_____"
],
[
"## Basic Plotting",
"_____no_output_____"
],
[
"To begin start by investigating the introductory notebooks:",
"_____no_output_____"
],
[
"1. [Introduction](Introduction.ipynb) - If you're new to `bqplot`, get started with our Introduction notebook\n2. [Basic Plotting](Basic Plotting/Basic Plotting.ipynb) - which demonstrates some basic `bqplot` plotting commands and how to use them\n3. [Pyplot](Basic Plotting/Pyplot.ipynb) - which introduces the simpler `pyplot` API",
"_____no_output_____"
],
[
"## Marks",
"_____no_output_____"
],
[
"Move to exploring the different `Marks` that you can use to represent your data. You have two options for rendering marks:\n* Object Model, which is a verbose API but gives you full flexibility and customizability\n* Pyplot, which is a simpler API (similar to matplotlib's pyplot) and sets meaningul defaults for the user\n\n\n1. Bars: Bar mark ([Object Model](Marks/Object Model/Bars.ipynb), [Pyplot](Marks/Pyplot/Bars.ipynb))\n* Bins: Backend histogram mark ([Object Model](Marks/Object Model/Bins.ipynb), [Pyplot](Marks/Pyplot/Bins.ipynb))\n* Boxplot: Boxplot mark ([Object Model](Marks/Object Model/Boxplot.ipynb), [Pyplot](Marks/Pyplot/Boxplot.ipynb))\n* Candles: OHLC mark ([Object Model](Marks/Object Model/Candles.ipynb), [Pyplot](Marks/Pyplot/Candles.ipynb))\n* FlexLine: Flexible lines mark ([Object Model](Marks/Object Model/Flexline.ipynb), Pyplot)\n* Graph: Network mark ([Object Model](Marks/Object Model/Graph.ipynb), Pyplot)\n* GridHeatMap: Grid heatmap mark ([Object Model](Marks/Object Model/GridHeatMap.ipynb), [Pyplot](Marks/Pyplot/GridHeatMap.ipynb))\n* HeatMap: Heatmap mark ([Object Model](Marks/Object Model/HeatMap.ipynb), [Pyplot](Marks/Pyplot/HeatMap.ipynb))\n* Hist: Histogram mark ([Object Model](Marks/Object Model/Hist.ipynb), [Pyplot](Marks/Pyplot/Hist.ipynb))\n* Image: Image mark ([Object Model](Marks/Object Model/Image.ipynb), [Pyplot](Marks/Pyplot/Image.ipynb))\n* Label: Label mark ([Object Model](Marks/Object Model/Label.ipynb), [Pyplot](Marks/Pyplot/Label.ipynb))\n* Lines: Lines mark ([Object Model](Marks/Object Model/Lines.ipynb), [Pyplot](Marks/Pyplot/Lines.ipynb))\n* Map: Geographical map mark ([Object Model](Marks/Object Model/Map.ipynb), [Pyplot](Marks/Pyplot/Map.ipynb))\n* Market Map: Tile map mark ([Object Model](Marks/Object Model/Market Map.ipynb), Pyplot)\n* Pie: Pie mark ([Object Model](Marks/Object Model/Pie.ipynb), [Pyplot](Marks/Pyplot/Pie.ipynb))\n* Scatter: Scatter mark ([Object Model](Marks/Object Model/Scatter.ipynb), [Pyplot](Marks/Pyplot/Scatter.ipynb))",
"_____no_output_____"
],
[
"## Interactions",
"_____no_output_____"
],
[
"Learn how to use `bqplot` interactions to convert your plots into interactive applications:",
"_____no_output_____"
],
[
"13. [Mark Interactions](Interactions/Mark Interactions.ipynb) - which describes the mark specific interactions and how to use them\n14. [Interaction Layer](Interactions/Interaction Layer.ipynb) - which describes the use of the interaction layers, including selectors and how they can be used for facilitating better interaction",
"_____no_output_____"
],
[
"## Advanced Plotting",
"_____no_output_____"
],
[
"Once you've mastered the basics of `bqplot`, you can use these notebooks to learn about some of it's more advanced features.",
"_____no_output_____"
],
[
"15. [Plotting Dates](Advanced Plotting/Plotting Dates.ipynb)\n16. [Advanced Plotting](Advanced Plotting/Advanced Plotting.ipynb)\n17. [Animations](Advanced Plotting/Animations.ipynb)\n18. [Axis Properties](Advanced Plotting/Axis Properties.ipynb)",
"_____no_output_____"
],
[
"## Applications",
"_____no_output_____"
],
[
"Finally, we have a collection of notebooks that demonstrate how to use `bqplot` and `ipywidgets` to create advanced interactive applications.",
"_____no_output_____"
],
[
"19. [Wealth of Nations](Applications/Wealth of Nations.ipynb) - a recreation of [Hans Rosling's famous TED Talk](http://www.ted.com/talks/hans_rosling_shows_the_best_stats_you_ve_ever_seen)",
"_____no_output_____"
],
[
"## Help",
"_____no_output_____"
],
[
"For more help, \n- Reach out to us via the `ipywidgets` gitter chat \n[](https://gitter.im/ipython/ipywidgets?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge&utm_content=badge)\n\n\n\n- Or take a look at a talk given on Interactive visualizations in Jupyter at PyData",
"_____no_output_____"
]
],
[
[
"from IPython.display import YouTubeVideo\nYouTubeVideo('eVET9IYgbao')",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
e78e3d658b35489c2fb54210d50448e1556c6cce | 19,602 | ipynb | Jupyter Notebook | site/en/guide/basic_training_loops.ipynb | jcjveraa/docs-2 | b5c78b0e9ab52094321153991d33534383d99610 | [
"Apache-2.0"
] | 2 | 2021-02-09T19:29:52.000Z | 2021-04-02T15:53:37.000Z | site/en/guide/basic_training_loops.ipynb | jcjveraa/docs-2 | b5c78b0e9ab52094321153991d33534383d99610 | [
"Apache-2.0"
] | null | null | null | site/en/guide/basic_training_loops.ipynb | jcjveraa/docs-2 | b5c78b0e9ab52094321153991d33534383d99610 | [
"Apache-2.0"
] | null | null | null | 35.770073 | 376 | 0.538823 | [
[
[
"##### Copyright 2020 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Basic training loops",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/guide/basic_training_loops\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/guide/basic_training_loops.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs/blob/master/site/en/guide/basic_training_loops.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/docs/site/en/guide/basic_training_loops.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"In the previous guides, you have learned about [tensors](./tensor.ipynb), [variables](./variable.ipynb), [gradient tape](autodiff.ipynb), and [modules](./intro_to_modules.ipynb). In this guide, you will fit these all together to train models.\n\nTensorFlow also includes the [tf.Keras API](keras/overview.ipynb), a high-level neural network API that provides useful abstractions to reduce boilerplate. However, in this guide, you will use basic classes.",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf",
"_____no_output_____"
]
],
[
[
"## Solving machine learning problems\n\nSolving a machine learning problem usually consists of the following steps:\n\n - Obtain training data.\n - Define the model.\n - Define a loss function.\n - Run through the training data, calculating loss from the ideal value\n - Calculate gradients for that loss and use an *optimizer* to adjust the variables to fit the data.\n - Evaluate your results.\n\nFor illustration purposes, in this guide you'll develop a simple linear model, $f(x) = x * W + b$, which has two variables: $W$ (weights) and $b$ (bias).\n\nThis is the most basic of machine learning problems: Given $x$ and $y$, try to find the slope and offset of a line via [simple linear regression](https://en.wikipedia.org/wiki/Linear_regression#Simple_and_multiple_linear_regression).",
"_____no_output_____"
],
[
"## Data\n\nSupervised learning uses *inputs* (usually denoted as *x*) and *outputs* (denoted *y*, often called *labels*). The goal is to learn from paired inputs and outputs so that you can predict the value of an output from an input.\n\nEach input of your data, in TensorFlow, is almost always represented by a tensor, and is often a vector. In supervised training, the output (or value you'd like to predict) is also a tensor.\n\nHere is some data synthesized by adding Gaussian (Normal) noise to points along a line.",
"_____no_output_____"
]
],
[
[
"# The actual line\nTRUE_W = 3.0\nTRUE_B = 2.0\n\nNUM_EXAMPLES = 1000\n\n# A vector of random x values\nx = tf.random.normal(shape=[NUM_EXAMPLES])\n\n# Generate some noise\nnoise = tf.random.normal(shape=[NUM_EXAMPLES])\n\n# Calculate y\ny = x * TRUE_W + TRUE_B + noise",
"_____no_output_____"
],
[
"# Plot all the data\nimport matplotlib.pyplot as plt\n\nplt.scatter(x, y, c=\"b\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"Tensors are usually gathered together in *batches*, or groups of inputs and outputs stacked together. Batching can confer some training benefits and works well with accelerators and vectorized computation. Given how small this dataset is, you can treat the entire dataset as a single batch.",
"_____no_output_____"
],
[
"## Define the model\n\nUse `tf.Variable` to represent all weights in a model. A `tf.Variable` stores a value and provides this in tensor form as needed. See the [variable guide](./variable.ipynb) for more details.\n\nUse `tf.Module` to encapsulate the variables and the computation. You could use any Python object, but this way it can be easily saved.\n\nHere, you define both *w* and *b* as variables.",
"_____no_output_____"
]
],
[
[
"class MyModel(tf.Module):\n def __init__(self, **kwargs):\n super().__init__(**kwargs)\n # Initialize the weights to `5.0` and the bias to `0.0`\n # In practice, these should be randomly initialized\n self.w = tf.Variable(5.0)\n self.b = tf.Variable(0.0)\n\n def __call__(self, x):\n return self.w * x + self.b\n\nmodel = MyModel()\n\n# List the variables tf.modules's built-in variable aggregation.\nprint(\"Variables:\", model.variables)\n\n# Verify the model works\nassert model(3.0).numpy() == 15.0",
"_____no_output_____"
]
],
[
[
"The initial variables are set here in a fixed way, but Keras comes with any of a number of [initalizers](https://www.tensorflow.org/api_docs/python/tf/keras/initializers) you could use, with or without the rest of Keras.",
"_____no_output_____"
],
[
"### Define a loss function\n\nA loss function measures how well the output of a model for a given input matches the target output. The goal is to minimize this difference during training. Define the standard L2 loss, also known as the \"mean squared\" error:",
"_____no_output_____"
]
],
[
[
"# This computes a single loss value for an entire batch\ndef loss(target_y, predicted_y):\n return tf.reduce_mean(tf.square(target_y - predicted_y))",
"_____no_output_____"
]
],
[
[
"Before training the model, you can visualize the loss value by plotting the model's predictions in red and the training data in blue:",
"_____no_output_____"
]
],
[
[
"plt.scatter(x, y, c=\"b\")\nplt.scatter(x, model(x), c=\"r\")\nplt.show()\n\nprint(\"Current loss: %1.6f\" % loss(model(x), y).numpy())",
"_____no_output_____"
]
],
[
[
"### Define a training loop\n\nThe training loop consists of repeatedly doing three tasks in order:\n\n* Sending a batch of inputs through the model to generate outputs\n* Calculating the loss by comparing the outputs to the output (or label)\n* Using gradient tape to find the gradients\n* Optimizing the variables with those gradients\n\nFor this example, you can train the model using [gradient descent](https://en.wikipedia.org/wiki/Gradient_descent).\n\nThere are many variants of the gradient descent scheme that are captured in `tf.keras.optimizers`. But in the spirit of building from first principles, here you will implement the basic math yourself with the help of `tf.GradientTape` for automatic differentiation and `tf.assign_sub` for decrementing a value (which combines `tf.assign` and `tf.sub`):",
"_____no_output_____"
]
],
[
[
"# Given a callable model, inputs, outputs, and a learning rate...\ndef train(model, x, y, learning_rate):\n\n with tf.GradientTape() as t:\n # Trainable variables are automatically tracked by GradientTape\n current_loss = loss(y, model(x))\n\n # Use GradientTape to calculate the gradients with respect to W and b\n dw, db = t.gradient(current_loss, [model.w, model.b])\n\n # Subtract the gradient scaled by the learning rate\n model.w.assign_sub(learning_rate * dw)\n model.b.assign_sub(learning_rate * db)",
"_____no_output_____"
]
],
[
[
"For a look at training, you can send the same batch of *x* an *y* through the training loop, and see how `W` and `b` evolve.",
"_____no_output_____"
]
],
[
[
"model = MyModel()\n\n# Collect the history of W-values and b-values to plot later\nWs, bs = [], []\nepochs = range(10)\n\n# Define a training loop\ndef training_loop(model, x, y):\n\n for epoch in epochs:\n # Update the model with the single giant batch\n train(model, x, y, learning_rate=0.1)\n\n # Track this before I update\n Ws.append(model.w.numpy())\n bs.append(model.b.numpy())\n current_loss = loss(y, model(x))\n\n print(\"Epoch %2d: W=%1.2f b=%1.2f, loss=%2.5f\" %\n (epoch, Ws[-1], bs[-1], current_loss))\n",
"_____no_output_____"
],
[
"print(\"Starting: W=%1.2f b=%1.2f, loss=%2.5f\" %\n (model.w, model.b, loss(y, model(x))))\n\n# Do the training\ntraining_loop(model, x, y)\n\n# Plot it\nplt.plot(epochs, Ws, \"r\",\n epochs, bs, \"b\")\n\nplt.plot([TRUE_W] * len(epochs), \"r--\",\n [TRUE_B] * len(epochs), \"b--\")\n\nplt.legend([\"W\", \"b\", \"True W\", \"True b\"])\nplt.show()\n",
"_____no_output_____"
],
[
"# Visualize how the trained model performs\nplt.scatter(x, y, c=\"b\")\nplt.scatter(x, model(x), c=\"r\")\nplt.show()\n\nprint(\"Current loss: %1.6f\" % loss(model(x), y).numpy())",
"_____no_output_____"
]
],
[
[
"## The same solution, but with Keras\n\nIt's useful to contrast the code above with the equivalent in Keras.\n\nDefining the model looks exactly the same if you subclass `tf.keras.Model`. Remember that Keras models inherit ultimately from module.",
"_____no_output_____"
]
],
[
[
"class MyModelKeras(tf.keras.Model):\n def __init__(self, **kwargs):\n super().__init__(**kwargs)\n # Initialize the weights to `5.0` and the bias to `0.0`\n # In practice, these should be randomly initialized\n self.w = tf.Variable(5.0)\n self.b = tf.Variable(0.0)\n\n def __call__(self, x, **kwargs):\n return self.w * x + self.b\n\nkeras_model = MyModelKeras()\n\n# Reuse the training loop with a Keras model\ntraining_loop(keras_model, x, y)\n\n# You can also save a checkpoint using Keras's built-in support\nkeras_model.save_weights(\"my_checkpoint\")",
"_____no_output_____"
]
],
[
[
"Rather than write new training loops each time you create a model, you can use the built-in features of Keras as a shortcut. This can be useful when you do not want to write or debug Python training loops.\n\nIf you do, you will need to use `model.compile()` to set the parameters, and `model.fit()` to train. It can be less code to use Keras implementations of L2 loss and gradient descent, again as a shortcut. Keras losses and optimizers can be used outside of these convenience functions, too, and the previous example could have used them.",
"_____no_output_____"
]
],
[
[
"keras_model = MyModelKeras()\n\n# compile sets the training paramaeters\nkeras_model.compile(\n # By default, fit() uses tf.function(). You can\n # turn that off for debugging, but it is on now.\n run_eagerly=False,\n\n # Using a built-in optimizer, configuring as an object\n optimizer=tf.keras.optimizers.SGD(learning_rate=0.1),\n\n # Keras comes with built-in MSE error\n # However, you could use the loss function\n # defined above\n loss=tf.keras.losses.mean_squared_error,\n)",
"_____no_output_____"
]
],
[
[
"Keras `fit` expects batched data or a complete dataset as a NumPy array. NumPy arrays are chopped into batches and default to a batch size of 32.\n\nIn this case, to match the behavior of the hand-written loop, you should pass `x` in as a single batch of size 1000.",
"_____no_output_____"
]
],
[
[
"print(x.shape[0])\nkeras_model.fit(x, y, epochs=10, batch_size=1000)",
"_____no_output_____"
]
],
[
[
"Note that Keras prints out the loss after training, not before, so the first loss appears lower, but otherwise this shows essentially the same training performance.",
"_____no_output_____"
],
[
"## Next steps\n\nIn this guide, you have seen how to use the core classes of tensors, variables, modules, and gradient tape to build and train a model, and further how those ideas map to Keras.\n\nThis is, however, an extremely simple problem. For a more practical introduction, see [Custom training walkthrough](../tutorials/customization/custom_training_walkthrough.ipynb).\n\nFor more on using built-in Keras training loops, see [this guide](keras/train_and_evaluate.ipynb). For more on training loops and Keras, see [this guide](keras/writing_a_training_loop_from_scratch.ipynb). For writing custom distributed training loops, see [this guide](./guide/distributed_training.ipynb#using_tfdistributestrategy_with_basic_training_loops_loops).",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
e78e4439aa37aa8574af3aee238a8becfe51100f | 27,035 | ipynb | Jupyter Notebook | Module3 (1).ipynb | Manassss/pythonassignments | 864e8f86d8faeaeb0d368c47f54929fea634614c | [
"MIT"
] | null | null | null | Module3 (1).ipynb | Manassss/pythonassignments | 864e8f86d8faeaeb0d368c47f54929fea634614c | [
"MIT"
] | null | null | null | Module3 (1).ipynb | Manassss/pythonassignments | 864e8f86d8faeaeb0d368c47f54929fea634614c | [
"MIT"
] | null | null | null | 74.068493 | 6,356 | 0.709525 | [
[
[
"65# Python Basics2",
"_____no_output_____"
],
[
"These assignments aim to get you acquainted with Python, which is an important requirement for all the research done at Solarillion Foundation. Apart from teaching you Python, these assignments also aim to make you a better programmer and cultivate better coding practices. \n\nVisit these links for more details: <br>\nPEP8 Practices: https://www.python.org/dev/peps/pep-0008/ <br>\nCheck PEP8: http://pep8online.com <br>\nPython Reference: https://www.py4e.com/lessons <br>\n\nDo use Google efficiently, and refer to StackOverflow for clarifying any programming doubts. If you're still stuck, feel free to ask a TA to help you.\n\nEach task in the assignment comprises of at least two cells. There are function definitions wherein you will name the function(s), and write code to solve the problem at hand. You will call the function(s) in the last cell of each task, and check your output.\n\nWe encourage you to play around and learn as much as possible, and be as creative as you can get. More than anything, have fun doing these assignments. Enjoy!",
"_____no_output_____"
],
[
"# Important\n* **Only the imports and functions must be present when you upload this notebook to GitHub for verification.** \n* **Do not upload it until you want to get it verified. Do not change function names or add extra cells or code, or remove anything.**\n* **For your rough work and four showing your code to TAs, use a different notebook with the name Module2Playground.ipynb and copy only the final functions to this notebook for verification.**",
"_____no_output_____"
],
[
"# Module 3\nScope: Algorithmic Thinking, Programming",
"_____no_output_____"
],
[
"## Imports - Always Execute First!\nImport any modules and turn on any magic here:",
"_____no_output_____"
]
],
[
[
"from IPython import get_ipython\nipy = get_ipython()\nif ipy is not None:\n ipy.run_line_magic(\"load_ext\", \"pycodestyle_magic\")\n ipy.run_line_magic(\"pycodestyle_on\", \"\")",
"_____no_output_____"
]
],
[
[
"## Burger Mania",
"_____no_output_____"
]
],
[
[
"\"\"\"\nImagine that you are a restaurant's cashier and are trying to keep records for analysing profits.\n\nYour restaurant sells 7 different items:\n 1. Burgers - $4.25\n 2. Nuggets - $2.50\n 3. French Fries - $2.00\n 4. Small Drink - $1.25\n 5. Medium Drink - $1.50\n 6. Large Drink - $1.75\n 7. Salad - $3.75\n\nCreate a program to randomly generate the orders of each customer as a string of numbers\n(corresponding to the item) and calculate the cost of the order. For example, if the generated\nstring is 5712335, the program should understand that the customer has ordered 1 burger, 1 \nportion of nuggets, 2 portions of fries, 2 medium drinks and 1 salad. It should then compute the\ncost ($17.50). The final cost is calculated after considering discounts for combo offers and\nadding 18% GST.\n\nThe combo offers are:\nA) 1 Burger + 1 Portion of Fries + 1 Drink -> 20% discount\nB) 1 Burger + 1 Portion of Nuggets + 1 Salad + 1 Drink -> 35% discount\n\nThe final cost of the 5712335 order is $13.4225. The profit gained each day has to be recorded for\n30 days and plotted for analysis.\n\nNote:\n - There will be at least 20 customers and not more than 50 customers per day. Each customer\n orders at least 3 items and not more than 7 items.\n - If there is a possibility of availing multiple combo offers in an order, the program\n should select the offer with maximum discount.\n\"\"\"",
"_____no_output_____"
],
[
"def generate_order():\n \"\"\"\n Function 1: generate_order()\n Return: A randomly generated order string\n \"\"\"\n import random\n s=random.randint(3,7)\n num_list=[]\n for i in range(s):\n num_list.append(random.randint(0,7))\n# print(num_list)\n list_to_str=\"\".join(map(str,num_list))\n# list_to_str=\" \".join(map(str,num_list))\n return list_to_str\n \n \n ",
"_____no_output_____"
],
[
" generate_order()",
"_____no_output_____"
],
[
"def compute_cost(order):\n \"\"\"\n Function 2: compute_cost(order)\n Parameters: order (String)\n Return: Final cost of order\n \"\"\"\n\n int_list=[]\n for ch in order:\n int_list.append(int(ch))\n# print(int_list)\n l=[4.25,2.50,2.00,1.25,1.50,1.75,3.75] \n total=0\n for i in int_list:\n for j in range(len(l)):\n if i==j+1:\n total=total+l[j]\n# print(total)\n discount=0\n\n if all(x in int_list for x in [1,3,5]) or all(x in int_list for x in [1,3,4]) or all(x in int_list for x in [1,3,6]):\n discount=total*0.8\n if all(x in int_list for x in [1,2,7,4]) or all(x in int_list for x in [1,2,7,5]) or all(x in int_list for x in [1,2,7,6]):\n discount=total*0.65\n if all(x in int_list for x in [1,2,7,3,4] or all(x in int_list for x in [1,2,7,3,5]) or all(x in int_list for x in [1,2,7,3,6])):\n discount=total*0.65\n else:\n discount=total\n# print(discount)\n\n sum=discount*1.18\n return sum\n \n ",
"_____no_output_____"
],
[
"order=generate_order()\ncompute_cost(order)",
"_____no_output_____"
],
[
"def simulate_restaurant():\n \"\"\"\n Function 3: simulate_restaurant()\n Purpose: Simulate the restaurant's operation using the previously declared functions,\n based on the constraints mentioned in the question\n Output: Plot of profit over 30 days\n \"\"\"\n import random\n import matplotlib.pyplot as plt\n profit=[]\n t=list(range(1,31))\n# print(t)\n daily=0\n total=0\n for i in range(0,30):\n n=random.randint(20,51)\n daily=0\n \n \n for j in range(0,n):\n x=generate_order()\n daily+=compute_cost(x)\n \n \n profit.append(daily)\n \n \n \n print(profit)\n plt.scatter(profit,t)\n \n \n \n\n",
"_____no_output_____"
],
[
"\nsimulate_restaurant()",
"[296.4749999999999, 571.8722499999999, 574.955, 422.1449999999999, 551.6499999999999, 609.1749999999998, 449.97825000000006, 583.02325, 640.8137500000001, 233.97925, 377.01, 551.119, 565.0872499999999, 615.37, 273.76, 438.901, 605.93, 446.03999999999985, 386.745, 613.01, 319.78, 396.47999999999985, 682.9250000000002, 459.6099999999999, 289.09999999999997, 597.0800000000002, 335.7099999999999, 441.90999999999997, 369.34000000000003, 467.86999999999995]\n"
]
],
[
[
"You're done with the Basics of Python! Give yourself a pat on the back.\n\nNow, choose an area you want to work on - Machine Learning, Internet of Things or Microgrids - and get started with the assignments. You could also choose to do assignments from multiple areas, it's entirely up to you. Hope you have fun!",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
e78e4a879fdb12d081bc8bdd6cf5dbccd59db252 | 22,893 | ipynb | Jupyter Notebook | how-to-use-azureml/explain-model/azure-integration/scoring-time/train-explain-model-locally-and-deploy.ipynb | angeltorresoa/MachineLearningNotebooks | a0e947c31bbe2f542f4a5c6b63c9631dcd16853f | [
"MIT"
] | 1 | 2021-11-25T05:33:34.000Z | 2021-11-25T05:33:34.000Z | how-to-use-azureml/explain-model/azure-integration/scoring-time/train-explain-model-locally-and-deploy.ipynb | angeltorresoa/MachineLearningNotebooks | a0e947c31bbe2f542f4a5c6b63c9631dcd16853f | [
"MIT"
] | null | null | null | how-to-use-azureml/explain-model/azure-integration/scoring-time/train-explain-model-locally-and-deploy.ipynb | angeltorresoa/MachineLearningNotebooks | a0e947c31bbe2f542f4a5c6b63c9631dcd16853f | [
"MIT"
] | 3 | 2021-06-22T11:38:12.000Z | 2022-02-28T03:50:14.000Z | 43.605714 | 1,061 | 0.568078 | [
[
[
"Copyright (c) Microsoft Corporation. All rights reserved.\n\nLicensed under the MIT License.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"# Train and explain models locally and deploy model and scoring explainer\n\n\n_**This notebook illustrates how to use the Azure Machine Learning Interpretability SDK to deploy a locally-trained model and its corresponding scoring explainer to Azure Container Instances (ACI) as a web service.**_\n\n\n\n\n\nProblem: IBM employee attrition classification with scikit-learn (train and explain a model locally and use Azure Container Instances (ACI) for deploying your model and its corresponding scoring explainer as a web service.)\n\n---\n\n## Table of Contents\n\n1. [Introduction](#Introduction)\n1. [Setup](#Setup)\n1. [Run model explainer locally at training time](#Explain)\n 1. Apply feature transformations\n 1. Train a binary classification model\n 1. Explain the model on raw features\n 1. Generate global explanations\n 1. Generate local explanations\n1. [Visualize explanations](#Visualize)\n1. [Deploy model and scoring explainer](#Deploy)\n1. [Next steps](#Next)",
"_____no_output_____"
],
[
"## Introduction\n\n\nThis notebook showcases how to train and explain a classification model locally, and deploy the trained model and its corresponding explainer to Azure Container Instances (ACI).\nIt demonstrates the API calls that you need to make to submit a run for training and explaining a model to AMLCompute, download the compute explanations remotely, and visualizing the global and local explanations via a visualization dashboard that provides an interactive way of discovering patterns in model predictions and downloaded explanations. It also demonstrates how to use Azure Machine Learning MLOps capabilities to deploy your model and its corresponding explainer.\n\nWe will showcase one of the tabular data explainers: TabularExplainer (SHAP) and follow these steps:\n1.\tDevelop a machine learning script in Python which involves the training script and the explanation script.\n2.\tRun the script locally.\n3.\tUse the interpretability toolkit’s visualization dashboard to visualize predictions and their explanation. If the metrics and explanations don't indicate a desired outcome, loop back to step 1 and iterate on your scripts.\n5.\tAfter a satisfactory run is found, create a scoring explainer and register the persisted model and its corresponding explainer in the model registry.\n6.\tDevelop a scoring script.\n7.\tCreate an image and register it in the image registry.\n8.\tDeploy the image as a web service in Azure.\n\n\n",
"_____no_output_____"
],
[
"## Setup\nMake sure you go through the [configuration notebook](../../../../configuration.ipynb) first if you haven't.",
"_____no_output_____"
]
],
[
[
"# Check core SDK version number\nimport azureml.core\n\nprint(\"SDK version:\", azureml.core.VERSION)",
"_____no_output_____"
]
],
[
[
"## Initialize a Workspace\n\nInitialize a workspace object from persisted configuration",
"_____no_output_____"
]
],
[
[
"from azureml.core import Workspace\n\nws = Workspace.from_config()\nprint(ws.name, ws.resource_group, ws.location, ws.subscription_id, sep='\\n')",
"_____no_output_____"
]
],
[
[
"## Explain\nCreate An Experiment: **Experiment** is a logical container in an Azure ML Workspace. It hosts run records which can include run metrics and output artifacts from your experiments.",
"_____no_output_____"
]
],
[
[
"from azureml.core import Experiment\nexperiment_name = 'explain_model_at_scoring_time'\nexperiment = Experiment(workspace=ws, name=experiment_name)\nrun = experiment.start_logging()",
"_____no_output_____"
],
[
"# Get IBM attrition data\nimport os\nimport pandas as pd\n\noutdirname = 'dataset.6.21.19'\ntry:\n from urllib import urlretrieve\nexcept ImportError:\n from urllib.request import urlretrieve\nimport zipfile\nzipfilename = outdirname + '.zip'\nurlretrieve('https://publictestdatasets.blob.core.windows.net/data/' + zipfilename, zipfilename)\nwith zipfile.ZipFile(zipfilename, 'r') as unzip:\n unzip.extractall('.')\nattritionData = pd.read_csv('./WA_Fn-UseC_-HR-Employee-Attrition.csv')",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\nimport joblib\nfrom sklearn.compose import ColumnTransformer\nfrom sklearn.preprocessing import StandardScaler, OneHotEncoder\nfrom sklearn.impute import SimpleImputer\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.ensemble import RandomForestClassifier\n\nfrom interpret.ext.blackbox import TabularExplainer\n\nos.makedirs('./outputs', exist_ok=True)\n\n# Dropping Employee count as all values are 1 and hence attrition is independent of this feature\nattritionData = attritionData.drop(['EmployeeCount'], axis=1)\n# Dropping Employee Number since it is merely an identifier\nattritionData = attritionData.drop(['EmployeeNumber'], axis=1)\nattritionData = attritionData.drop(['Over18'], axis=1)\n# Since all values are 80\nattritionData = attritionData.drop(['StandardHours'], axis=1)\n\n# Converting target variables from string to numerical values\ntarget_map = {'Yes': 1, 'No': 0}\nattritionData[\"Attrition_numerical\"] = attritionData[\"Attrition\"].apply(lambda x: target_map[x])\ntarget = attritionData[\"Attrition_numerical\"]\n\nattritionXData = attritionData.drop(['Attrition_numerical', 'Attrition'], axis=1)\n\n# Creating dummy columns for each categorical feature\ncategorical = []\nfor col, value in attritionXData.iteritems():\n if value.dtype == 'object':\n categorical.append(col)\n\n# Store the numerical columns in a list numerical\nnumerical = attritionXData.columns.difference(categorical)\n\n# We create the preprocessing pipelines for both numeric and categorical data.\nnumeric_transformer = Pipeline(steps=[\n ('imputer', SimpleImputer(strategy='median')),\n ('scaler', StandardScaler())])\n\ncategorical_transformer = Pipeline(steps=[\n ('imputer', SimpleImputer(strategy='constant', fill_value='missing')),\n ('onehot', OneHotEncoder(handle_unknown='ignore'))])\n\ntransformations = ColumnTransformer(\n transformers=[\n ('num', numeric_transformer, numerical),\n ('cat', categorical_transformer, categorical)])\n\n# Append classifier to preprocessing pipeline.\n# Now we have a full prediction pipeline.\nclf = Pipeline(steps=[('preprocessor', transformations),\n ('classifier', RandomForestClassifier())])\n\n# Split data into train and test\nfrom sklearn.model_selection import train_test_split\nx_train, x_test, y_train, y_test = train_test_split(attritionXData,\n target,\n test_size=0.2,\n random_state=0,\n stratify=target)\n\n# Preprocess the data and fit the classification model\nclf.fit(x_train, y_train)\nmodel = clf.steps[-1][1]\n\nmodel_file_name = 'log_reg.pkl'\n\n# Save model in the outputs folder so it automatically get uploaded\nwith open(model_file_name, 'wb') as file:\n joblib.dump(value=clf, filename=os.path.join('./outputs/',\n model_file_name))",
"_____no_output_____"
],
[
"# Explain predictions on your local machine\ntabular_explainer = TabularExplainer(model, \n initialization_examples=x_train, \n features=attritionXData.columns, \n classes=[\"Not leaving\", \"leaving\"], \n transformations=transformations)\n\n# Explain overall model predictions (global explanation)\n# Passing in test dataset for evaluation examples - note it must be a representative sample of the original data\n# x_train can be passed as well, but with more examples explanations it will\n# take longer although they may be more accurate\nglobal_explanation = tabular_explainer.explain_global(x_test)",
"_____no_output_____"
],
[
"from azureml.interpret.scoring.scoring_explainer import TreeScoringExplainer, save\n# ScoringExplainer\nscoring_explainer = TreeScoringExplainer(tabular_explainer)\n# Pickle scoring explainer locally\nsave(scoring_explainer, exist_ok=True)\n\n# Register original model\nrun.upload_file('original_model.pkl', os.path.join('./outputs/', model_file_name))\noriginal_model = run.register_model(model_name='local_deploy_model', \n model_path='original_model.pkl')\n\n# Register scoring explainer\nrun.upload_file('IBM_attrition_explainer.pkl', 'scoring_explainer.pkl')\nscoring_explainer_model = run.register_model(model_name='IBM_attrition_explainer', model_path='IBM_attrition_explainer.pkl')",
"_____no_output_____"
]
],
[
[
"## Visualize\nVisualize the explanations",
"_____no_output_____"
]
],
[
[
"from interpret_community.widget import ExplanationDashboard",
"_____no_output_____"
],
[
"ExplanationDashboard(global_explanation, clf, datasetX=x_test)",
"_____no_output_____"
]
],
[
[
"## Deploy \n\nDeploy Model and ScoringExplainer.\n\nPlease note that you must indicate azureml-defaults with verion >= 1.0.45 as a pip dependency, because it contains the functionality needed to host the model as a web service.",
"_____no_output_____"
]
],
[
[
"from azureml.core.conda_dependencies import CondaDependencies \n\n# azureml-defaults is required to host the model as a web service.\nazureml_pip_packages = [\n 'azureml-defaults', 'azureml-core', 'azureml-telemetry',\n 'azureml-interpret'\n]\n \n\n# Note: this is to pin the scikit-learn and pandas versions to be same as notebook.\n# In production scenario user would choose their dependencies\nimport pkg_resources\navailable_packages = pkg_resources.working_set\nsklearn_ver = None\npandas_ver = None\nfor dist in available_packages:\n if dist.key == 'scikit-learn':\n sklearn_ver = dist.version\n elif dist.key == 'pandas':\n pandas_ver = dist.version\nsklearn_dep = 'scikit-learn'\npandas_dep = 'pandas'\nif sklearn_ver:\n sklearn_dep = 'scikit-learn=={}'.format(sklearn_ver)\nif pandas_ver:\n pandas_dep = 'pandas=={}'.format(pandas_ver)\n# Specify CondaDependencies obj\n# The CondaDependencies specifies the conda and pip packages that are installed in the environment\n# the submitted job is run in. Note the remote environment(s) needs to be similar to the local\n# environment, otherwise if a model is trained or deployed in a different environment this can\n# cause errors. Please take extra care when specifying your dependencies in a production environment.\nmyenv = CondaDependencies.create(pip_packages=['pyyaml', sklearn_dep, pandas_dep] + azureml_pip_packages,\n pin_sdk_version=False)\n\nwith open(\"myenv.yml\",\"w\") as f:\n f.write(myenv.serialize_to_string())\n\nwith open(\"myenv.yml\",\"r\") as f:\n print(f.read())",
"_____no_output_____"
],
[
"from azureml.core.model import Model\n# Retrieve scoring explainer for deployment\nscoring_explainer_model = Model(ws, 'IBM_attrition_explainer')",
"_____no_output_____"
],
[
"from azureml.core.webservice import Webservice\nfrom azureml.core.model import InferenceConfig\nfrom azureml.core.webservice import AciWebservice\nfrom azureml.core.model import Model\nfrom azureml.core.environment import Environment\n\n\naciconfig = AciWebservice.deploy_configuration(cpu_cores=1, \n memory_gb=1, \n tags={\"data\": \"IBM_Attrition\", \n \"method\" : \"local_explanation\"}, \n description='Get local explanations for IBM Employee Attrition data')\n\nmyenv = Environment.from_conda_specification(name=\"myenv\", file_path=\"myenv.yml\")\ninference_config = InferenceConfig(entry_script=\"score_local_explain.py\", environment=myenv)\n\n# Use configs and models generated above\nservice = Model.deploy(ws, 'model-scoring-deploy-local', [scoring_explainer_model, original_model], inference_config, aciconfig)\ntry:\n service.wait_for_deployment(show_output=True)\nexcept WebserviceException as e:\n print(e.message)\n print(service.get_logs())\n raise",
"_____no_output_____"
],
[
"import requests\nimport json\n\n\n# Create data to test service with\nsample_data = '{\"Age\":{\"899\":49},\"BusinessTravel\":{\"899\":\"Travel_Rarely\"},\"DailyRate\":{\"899\":1098},\"Department\":{\"899\":\"Research & Development\"},\"DistanceFromHome\":{\"899\":4},\"Education\":{\"899\":2},\"EducationField\":{\"899\":\"Medical\"},\"EnvironmentSatisfaction\":{\"899\":1},\"Gender\":{\"899\":\"Male\"},\"HourlyRate\":{\"899\":85},\"JobInvolvement\":{\"899\":2},\"JobLevel\":{\"899\":5},\"JobRole\":{\"899\":\"Manager\"},\"JobSatisfaction\":{\"899\":3},\"MaritalStatus\":{\"899\":\"Married\"},\"MonthlyIncome\":{\"899\":18711},\"MonthlyRate\":{\"899\":12124},\"NumCompaniesWorked\":{\"899\":2},\"OverTime\":{\"899\":\"No\"},\"PercentSalaryHike\":{\"899\":13},\"PerformanceRating\":{\"899\":3},\"RelationshipSatisfaction\":{\"899\":3},\"StockOptionLevel\":{\"899\":1},\"TotalWorkingYears\":{\"899\":23},\"TrainingTimesLastYear\":{\"899\":2},\"WorkLifeBalance\":{\"899\":4},\"YearsAtCompany\":{\"899\":1},\"YearsInCurrentRole\":{\"899\":0},\"YearsSinceLastPromotion\":{\"899\":0},\"YearsWithCurrManager\":{\"899\":0}}'\n\n\n\nheaders = {'Content-Type':'application/json'}\n\n# Send request to service\nprint(\"POST to url\", service.scoring_uri)\nresp = requests.post(service.scoring_uri, sample_data, headers=headers)\n\n# Can covert back to Python objects from json string if desired\nprint(\"prediction:\", resp.text)\nresult = json.loads(resp.text)",
"_____no_output_____"
],
[
"# Plot the feature importance for the prediction\nimport numpy as np\nimport matplotlib.pyplot as plt; plt.rcdefaults()\n\nlabels = json.loads(sample_data)\nlabels = labels.keys()\nobjects = labels\ny_pos = np.arange(len(objects))\nperformance = result[\"local_importance_values\"][0][0]\n\nplt.bar(y_pos, performance, align='center', alpha=0.5)\nplt.xticks(y_pos, objects)\nlocs, labels = plt.xticks()\nplt.setp(labels, rotation=90)\nplt.ylabel('Feature impact - leaving vs not leaving')\nplt.title('Local feature importance for prediction')\n\nplt.show()",
"_____no_output_____"
],
[
"service.delete()",
"_____no_output_____"
]
],
[
[
"## Next\nLearn about other use cases of the explain package on a:\n1. [Training time: regression problem](https://github.com/interpretml/interpret-community/blob/master/notebooks/explain-regression-local.ipynb) \n1. [Training time: binary classification problem](https://github.com/interpretml/interpret-community/blob/master/notebooks/explain-binary-classification-local.ipynb)\n1. [Training time: multiclass classification problem](https://github.com/interpretml/interpret-community/blob/master/notebooks/explain-multiclass-classification-local.ipynb)\n1. Explain models with engineered features:\n 1. [Simple feature transformations](https://github.com/interpretml/interpret-community/blob/master/notebooks/simple-feature-transformations-explain-local.ipynb)\n 1. [Advanced feature transformations](https://github.com/interpretml/interpret-community/blob/master/notebooks/advanced-feature-transformations-explain-local.ipynb)\n1. [Save model explanations via Azure Machine Learning Run History](../run-history/save-retrieve-explanations-run-history.ipynb)\n1. [Run explainers remotely on Azure Machine Learning Compute (AMLCompute)](../remote-explanation/explain-model-on-amlcompute.ipynb)\n1. [Inferencing time: deploy a remotely-trained model and explainer](./train-explain-model-on-amlcompute-and-deploy.ipynb)\n1. [Inferencing time: deploy a locally-trained keras model and explainer](./train-explain-model-keras-locally-and-deploy.ipynb)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
e78e4c56bb10a2bbe7c31c29fe51d75ff51ff277 | 1,316 | ipynb | Jupyter Notebook | my_first_python_notebook.ipynb | PetersonZou/astr-119-session-5 | 90308dfc52d381d33cc3e0347b64e9095f1d233e | [
"MIT"
] | null | null | null | my_first_python_notebook.ipynb | PetersonZou/astr-119-session-5 | 90308dfc52d381d33cc3e0347b64e9095f1d233e | [
"MIT"
] | null | null | null | my_first_python_notebook.ipynb | PetersonZou/astr-119-session-5 | 90308dfc52d381d33cc3e0347b64e9095f1d233e | [
"MIT"
] | null | null | null | 16.871795 | 58 | 0.465046 | [
[
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"#THIS IS MARKDOWN\n\nIt's great",
"_____no_output_____"
]
],
[
[
"n=10 #define an integer\nx=np.arange(n,dtype=float) #define an array",
"_____no_output_____"
],
[
"print(x)",
"[0. 1. 2. 3. 4. 5. 6. 7. 8. 9.]\n"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
e78e5022d86c026b179f43d992e07e12d4ab4ed3 | 130,970 | ipynb | Jupyter Notebook | 0-introduction/cbsp_hackathon.ipynb | jxu8/cbsp-hackathon | 010c5869dcbc88e4fd5245cdda884f32ddd65b9b | [
"MIT"
] | null | null | null | 0-introduction/cbsp_hackathon.ipynb | jxu8/cbsp-hackathon | 010c5869dcbc88e4fd5245cdda884f32ddd65b9b | [
"MIT"
] | null | null | null | 0-introduction/cbsp_hackathon.ipynb | jxu8/cbsp-hackathon | 010c5869dcbc88e4fd5245cdda884f32ddd65b9b | [
"MIT"
] | null | null | null | 89.338336 | 32,236 | 0.840322 | [
[
[
"# Computation Biology Summer Program Hackathon",
"_____no_output_____"
],
[
"This [Jupyter notebook](https://jupyter.org/) gives examples on how to use the various [REST](https://en.wikipedia.org/wiki/Representational_state_transfer) web services from the [Knowledge Systems Group](https://www.mskcc.org/research-areas/labs/nikolaus-schultz). In this hackathon we will pull data from those APIs to make visualizations.",
"_____no_output_____"
],
[
"## How to run the notebook",
"_____no_output_____"
],
[
"This notebook can be executed on your own machine after installing Jupyter. Please install the Python 3 version of anaconda: https://www.anaconda.com/download/. After having that set up you can install Jupyter with:\n\n```bash\nconda install jupyter\n\n```\n\nFor these examples we also require the [Swagger API](https://swagger.io/specification/) client `bravado`.\n\n```bash\nconda install -c conda-forge bravado\n```\n\nAnd the popular data analysis libraries pandas, matplotlib and seaborn:\n\n```\nconda install pandas matplotlib seaborn\n```\n\nThen clone this repo:\n\n```\ngit clone https://github.com/mskcc/cbsp-hackathon\n```\n\nAnd run Jupyter in this folder\n```\ncd cbsp-hackathon/0-introduction\njupyter\n```\nThat should open Jupyter in a new browser window and you should be able to open this notebook using the web interface. You can then follow along with the next steps.",
"_____no_output_____"
],
[
"## How to use the notebook",
"_____no_output_____"
],
[
"The notebook consists of cells which can be executed by clicking on one and pressing shift+f. In the toolbar at the top there is a dropdown which indicates what type of cell you have selected e.g. `Code` or [Markdown](https://en.wikipedia.org/wiki/Markdown). The former will be executed as raw Python code the latter is a markup language and will be run through a Markdown parser. Both generate HTML that will be printed directly to the notebook page.\n\nThere a few keyboard shortcuts that are good to know. That is: `b` creates a new cell below the one you've selected and `a` above the one you selected. Editing a cell can be done with a single click for a code cell and a double click for a Markdown cell. A complete list of all keyboard shortcuts can be found by pressing the keyboard icon in the toolbar at the top.\n\n Give it a shot by editing one of the cells and pressing shift+f.",
"_____no_output_____"
],
[
"## Using the REST APIs",
"_____no_output_____"
],
[
"All [REST](https://en.wikipedia.org/wiki/Representational_state_transfer) web services from the [Knowledge Systems Group](https://www.mskcc.org/research-areas/labs/nikolaus-schultz) we will be using in this tutorial have their REST APIs defined following the [Open API / Swagger specification](https://swagger.io/specification/). This allows us to use `bravado` to connect to them directly, and explore the API interactively.\n\nFor example this is how to connect to the [cBioPortal](https://www.cbioportal.org) API:",
"_____no_output_____"
]
],
[
[
"from bravado.client import SwaggerClient\n\ncbioportal = SwaggerClient.from_url('https://www.cbioportal.org/api/api-docs',\n config={\"validate_requests\":False,\"validate_responses\":False})\nprint(cbioportal)",
"SwaggerClient(https://www.cbioportal.org/api)\n"
]
],
[
[
"You can now explore the API by using code completion, press `Tab` after typing `cbioportal.`:",
"_____no_output_____"
]
],
[
[
"cbioportal.B_Studies",
"_____no_output_____"
]
],
[
[
"This will give a dropdown with all the different APIs, similar to how you can see them here on the cBioPortal website: https://www.cbioportal.org/api/swagger-ui.html#/.\n\nYou can also get the parameters to a specific endpoint by pressing shift+tab twice after typing the name of the specific endpoint e.g.:",
"_____no_output_____"
]
],
[
[
"cbioportal.A_Cancer_Types.getCancerTypeUsingGET(",
"_____no_output_____"
]
],
[
[
"That shows one of the parameters is `cancerTypeId` of type `string`, the example `acc` is mentioned:",
"_____no_output_____"
]
],
[
[
"acc = cbioportal.A_Cancer_Types.getCancerTypeUsingGET(cancerTypeId='acc').result()\nprint(acc)",
"TypeOfCancer(cancerTypeId='acc', clinicalTrialKeywords='adrenocortical carcinoma', dedicatedColor='Purple', name='Adrenocortical Carcinoma', parent='adrenal_gland', shortName='ACC')\n"
]
],
[
[
"You can see that the JSON output returned by the cBioPortal API gets automatically converted into an object called `TypeOfCancer`. This object can be explored interactively as well by pressing tab after typing `acc.`:",
"_____no_output_____"
]
],
[
[
"acc.dedicatedColor",
"_____no_output_____"
]
],
[
[
"### cBioPortal API",
"_____no_output_____"
],
[
"[cBioPortal](https://www.cbioportal.org) stores cancer genomics data from a large number of published studies. Let's figure out:\n\n- how many studies are there?\n- how many cancer types do they span?\n- how many samples in total?\n- which study has the largest number of samples?",
"_____no_output_____"
]
],
[
[
"studies = cbioportal.B_Studies.getAllStudiesUsingGET().result()\ncancer_types = cbioportal.A_Cancer_Types.getAllCancerTypesUsingGET().result()\n\nprint(\"In total there are {} studies in cBioPortal, spanning {} different types of cancer.\".format(\n len(studies),\n len(cancer_types)\n))",
"In total there are 256 studies in cBioPortal, spanning 855 different types of cancer.\n"
]
],
[
[
"To get the total number of samples in each study we have to look a bit more at the response of the studies endpoint:",
"_____no_output_____"
]
],
[
[
"dir(studies[0])",
"_____no_output_____"
]
],
[
[
"We can sum the `allSampleCount` values of each study in cBioPortal:",
"_____no_output_____"
]
],
[
[
"print(\"The total number of samples in all studies is: {}\".format(sum([x.allSampleCount for x in studies])))",
"The total number of samples in all studies is: 77901\n"
]
],
[
[
"Let's see which study has the largest number of samples:",
"_____no_output_____"
]
],
[
[
"sorted_studies = sorted(studies, key=lambda x: x.allSampleCount)\nsorted_studies[-1]",
"_____no_output_____"
]
],
[
[
"Make it as little easier to read using pretty print:",
"_____no_output_____"
]
],
[
[
"from pprint import pprint\npprint(vars(sorted_studies[-1])['_Model__dict'])",
"{'allSampleCount': 10945,\n 'cancerType': None,\n 'cancerTypeId': 'mixed',\n 'citation': 'Zehir et al. Nat Med 2017',\n 'cnaSampleCount': None,\n 'completeSampleCount': None,\n 'description': 'Targeted sequencing of 10,000 clinical cases using the '\n 'MSK-IMPACT assay',\n 'groups': 'PUBLIC',\n 'importDate': '2019-05-07 00:00:00',\n 'methylationHm27SampleCount': None,\n 'miRnaSampleCount': None,\n 'mrnaMicroarraySampleCount': None,\n 'mrnaRnaSeqSampleCount': None,\n 'mrnaRnaSeqV2SampleCount': None,\n 'name': 'MSK-IMPACT Clinical Sequencing Cohort (MSKCC, Nat Med 2017)',\n 'pmid': '28481359',\n 'publicStudy': True,\n 'rppaSampleCount': None,\n 'sequencedSampleCount': None,\n 'shortName': 'MSK-IMPACT',\n 'status': 0,\n 'studyId': 'msk_impact_2017'}\n"
]
],
[
[
"Now that we've answered the inital questions we can dig a little deeper into this specific study:\n\n- How many patients are in this study?\n- What gene is most commonly mutated across the different samples?\n- Does this study span one or more types of cancer?\n\nThe description of the study with id `msk_impact_2017` study mentions there are 10,000 patients sequenced. Can we find this data in the cBioPortal?",
"_____no_output_____"
]
],
[
[
"patients = cbioportal.C_Patients.getAllPatientsInStudyUsingGET(studyId='msk_impact_2017').result()\nprint(\"The msk_impact_2017 study spans {} patients\".format(len(patients)))",
"The msk_impact_2017 study spans 10336 patients\n"
]
],
[
[
"Now let's try to figure out what gene is most commonly mutated. For this we can check the endpoints in the group `K_Mutations`. When looking at these endpoints it seems that a study can have multiple molecular profiles. This is because samples might have been sequenced using different assays (e.g. targeting a subset of genes or all genes). An example for the `acc_tcga` study is given for a molecular profile (`acc_tcga_mutations`) and a collection of samples (`msk_impact_2017_all`). We can use the same approach for the `msk_impact_2017` study. This will take a few seconds. You can use the command `%%time` to time a cell):",
"_____no_output_____"
]
],
[
[
"%%time\n\nmutations = cbioportal.K_Mutations.getMutationsInMolecularProfileBySampleListIdUsingGET(\n molecularProfileId='msk_impact_2017_mutations',\n sampleListId='msk_impact_2017_all'\n).result()",
"CPU times: user 10.9 s, sys: 407 ms, total: 11.3 s\nWall time: 14.8 s\n"
]
],
[
[
"We can explore what the mutation data structure looks like:",
"_____no_output_____"
]
],
[
[
"pprint(vars(mutations[0])['_Model__dict'])",
"{'aminoAcidChange': None,\n 'center': 'NA',\n 'driverFilter': '',\n 'driverFilterAnnotation': '',\n 'driverTiersFilter': '',\n 'driverTiersFilterAnnotation': '',\n 'endPosition': 36252995,\n 'entrezGeneId': 861,\n 'fisValue': 1.4013e-45,\n 'functionalImpactScore': '[Not Available]',\n 'gene': None,\n 'keyword': 'RUNX1 truncating',\n 'linkMsa': '[Not Available]',\n 'linkPdb': '[Not Available]',\n 'linkXvar': '[Not Available]',\n 'molecularProfileId': 'msk_impact_2017_mutations',\n 'mutationStatus': 'NA',\n 'mutationType': 'Frame_Shift_Ins',\n 'ncbiBuild': 'GRCh37',\n 'normalAltCount': None,\n 'normalRefCount': None,\n 'patientId': 'P-0008845',\n 'proteinChange': 'D96Gfs*11',\n 'proteinPosEnd': 96,\n 'proteinPosStart': 96,\n 'referenceAllele': 'NA',\n 'refseqMrnaId': 'NM_001001890.2',\n 'sampleId': 'P-0008845-T01-IM5',\n 'startPosition': 36252994,\n 'studyId': 'msk_impact_2017',\n 'tumorAltCount': 98,\n 'tumorRefCount': 400,\n 'uniquePatientKey': 'UC0wMDA4ODQ1Om1za19pbXBhY3RfMjAxNw',\n 'uniqueSampleKey': 'UC0wMDA4ODQ1LVQwMS1JTTU6bXNrX2ltcGFjdF8yMDE3',\n 'validationStatus': 'NA',\n 'variantAllele': 'CC',\n 'variantType': 'INS'}\n"
]
],
[
[
"It seems that the `gene` field is not filled in. To keep the response size of the API small, the API uses a parameter called `projection` that indicates whether or not to return all fields of an object or only a portion of the fields. By default it will use the `SUMMARY` projection. But because in this case we want to `gene` information, we'll use the `DETAILED` projection instead, so let's update the previous statement:",
"_____no_output_____"
]
],
[
[
"%%time \n\nmutations = cbioportal.K_Mutations.getMutationsInMolecularProfileBySampleListIdUsingGET(\n molecularProfileId='msk_impact_2017_mutations',\n sampleListId='msk_impact_2017_all',\n projection='DETAILED'\n).result()",
"CPU times: user 14.4 s, sys: 549 ms, total: 14.9 s\nWall time: 20.8 s\n"
]
],
[
[
"You can see the response time is slightly slower. Let's check if the gene field is filled in now:",
"_____no_output_____"
]
],
[
[
"pprint(vars(mutations[0])['_Model__dict'])",
"{'aminoAcidChange': None,\n 'center': 'NA',\n 'driverFilter': '',\n 'driverFilterAnnotation': '',\n 'driverTiersFilter': '',\n 'driverTiersFilterAnnotation': '',\n 'endPosition': 36252995,\n 'entrezGeneId': 861,\n 'fisValue': 1.4013e-45,\n 'functionalImpactScore': '[Not Available]',\n 'gene': Gene(chromosome='21', cytoband='21q22.12', entrezGeneId=861, hugoGeneSymbol='RUNX1', length=261534, type='protein-coding'),\n 'keyword': 'RUNX1 truncating',\n 'linkMsa': '[Not Available]',\n 'linkPdb': '[Not Available]',\n 'linkXvar': '[Not Available]',\n 'molecularProfileId': 'msk_impact_2017_mutations',\n 'mutationStatus': 'NA',\n 'mutationType': 'Frame_Shift_Ins',\n 'ncbiBuild': 'GRCh37',\n 'normalAltCount': None,\n 'normalRefCount': None,\n 'patientId': 'P-0008845',\n 'proteinChange': 'D96Gfs*11',\n 'proteinPosEnd': 96,\n 'proteinPosStart': 96,\n 'referenceAllele': 'NA',\n 'refseqMrnaId': 'NM_001001890.2',\n 'sampleId': 'P-0008845-T01-IM5',\n 'startPosition': 36252994,\n 'studyId': 'msk_impact_2017',\n 'tumorAltCount': 98,\n 'tumorRefCount': 400,\n 'uniquePatientKey': 'UC0wMDA4ODQ1Om1za19pbXBhY3RfMjAxNw',\n 'uniqueSampleKey': 'UC0wMDA4ODQ1LVQwMS1JTTU6bXNrX2ltcGFjdF8yMDE3',\n 'validationStatus': 'NA',\n 'variantAllele': 'CC',\n 'variantType': 'INS'}\n"
]
],
[
[
"Now that we have the gene field we can check what gene is most commonly mutated: ",
"_____no_output_____"
]
],
[
[
"from collections import Counter\nmutation_counts = Counter([m.gene.hugoGeneSymbol for m in mutations])\nmutation_counts.most_common(5)",
"_____no_output_____"
]
],
[
[
"We can verify that these results are correct by looking at the study view of the MSK-IMPACT study on the cBioPortal website: https://www.cbioportal.org/study/summary?id=msk_impact_2017. Note that the website uses the REST API we've been using in this hackathon, so we would expect those numbers to be the same, but good to do a sanity check. We see that the number of patients is indeed 10,336. But the number of samples with a mutation in TP53 is 4,561 instead of 4,985. Can you spot why they differ?\n\nNext question:\n\n- How many samples have a TP53 mutation?\n\nFor this exercise it might be useful to use a [pandas dataframe](https://pandas.pydata.org/) to be able to do grouping operations. You can convert the mutations result to a dataframe like this:",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\nmdf = pd.DataFrame.from_dict([\n # python magic that combines two dictionaries:\n dict(\n m.__dict__['_Model__dict'],\n **m.__dict__['_Model__dict']['gene'].__dict__['_Model__dict']) \n # create one item in the list for each mutation\n for m in mutations\n])",
"_____no_output_____"
]
],
[
[
"The DataFrame is a data type originally from `Matlab` and `R` that makes it easier to work with columnar data. Pandas brings that data type to Python. There are also several performance optimizations by it using the data types from [numpy](https://www.numpy.org/).\n\nNow that you have the data in a Dataframe you can group the mutations by the gene name and count the number of unique samples in TP53:",
"_____no_output_____"
]
],
[
[
"sample_count_per_gene = mdf.groupby('hugoGeneSymbol')['uniqueSampleKey'].nunique()\n\nprint(\"There are {} samples with a mutation in TP53\".format(\n sample_count_per_gene['TP53']\n))",
"There are 4561 samples with a mutation in TP53\n"
]
],
[
[
"It would be nice to visualize this result in context of the other genes by plotting the top 10 most mutated genes. For this you can use the matplotlib interface that integrates with pandas.\n\nFirst inline plotting in the notebook:",
"_____no_output_____"
]
],
[
[
"%matplotlib inline",
"_____no_output_____"
],
[
"sample_count_per_gene.sort_values(ascending=False).head(10).plot(kind='bar')",
"_____no_output_____"
]
],
[
[
"Make it look a little nicer by importing seaborn:",
"_____no_output_____"
]
],
[
[
"import seaborn as sns\nsns.set_style(\"white\")\nsns.set_context('notebook')",
"_____no_output_____"
],
[
"sample_count_per_gene.sort_values(ascending=False).head(10).plot(kind='bar')\nsns.despine(trim=False)",
"_____no_output_____"
]
],
[
[
"You can further change the plot a bit by using the arguments to the plot function or using the matplotlib interface directly:",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n\nsample_count_per_gene.sort_values(ascending=False).head(10).plot(\n kind='bar',\n ylim=[0,5000],\n color='green'\n)\nsns.despine(trim=False)\nplt.xlabel('')\nplt.xticks(rotation=300)\nplt.ylabel('Number of samples',labelpad=20)\nplt.title('Number of mutations in genes in MSK-IMPACT (2017)',pad=25)",
"_____no_output_____"
]
],
[
[
"A further extension of this plot could be to color the bar chart by the type of mutation in that sample (`mdf.mutationType`) and to include copy number alterations (see `L. Discrete Copy Number Alterations` endpoints).",
"_____no_output_____"
],
[
"### Genome Nexus API",
"_____no_output_____"
],
[
"[Genome Nexus](https://www.genomenexus.org) is a web service that aggregates all cancer related information about a particular mutation. Similarly to cBioPortal it provides a REST API following the [Swagger / OpenAPI specification](https://swagger.io/specification/).",
"_____no_output_____"
]
],
[
[
"from bravado.client import SwaggerClient\n\ngn = SwaggerClient.from_url('https://www.genomenexus.org/v2/api-docs',\n config={\"validate_requests\":False,\n \"validate_responses\":False,\n \"validate_swagger_spec\":False})\nprint(gn)",
"SwaggerClient(https://www.genomenexus.org/)\n"
]
],
[
[
"To look up annotations for a single variant, one can use the following endpoint:",
"_____no_output_____"
]
],
[
[
"variant = gn.annotation_controller.fetchVariantAnnotationByGenomicLocationGET(\n genomicLocation='7,140453136,140453136,A,T',\n # adds extra annotation resources, not included in default response:\n fields='hotspots mutation_assessor annotation_summary'.split()\n).result()",
"_____no_output_____"
]
],
[
[
"You can see a lot of information is provided for that particular variant if you type tab after `variant.`:",
"_____no_output_____"
]
],
[
[
"variant.",
"_____no_output_____"
]
],
[
[
"For this example we will focus on the hotspot annotation and ignore the others. [Cancer hotspots](https://www.cancerhotspots.org/) is a popular web resource which indicates whether particular variants have been found to be recurrently mutated in large scale cancer genomics data.\n\nThe example variant above is a hotspot:",
"_____no_output_____"
]
],
[
[
"variant.hotspots",
"_____no_output_____"
]
],
[
[
"Let's see how many hotspot mutations there are in the Cholangiocarcinoma (TCGA, PanCancer Atlas) study with study id `chol_tcga_pan_can_atlas_2018` from the cBioPortal:",
"_____no_output_____"
]
],
[
[
"%%time\n\ncbioportal = SwaggerClient.from_url('https://www.cbioportal.org/api/api-docs',\n config={\"validate_requests\":False,\"validate_responses\":False})\n\nmutations = cbioportal.K_Mutations.getMutationsInMolecularProfileBySampleListIdUsingGET(\n molecularProfileId='chol_tcga_pan_can_atlas_2018_mutations',\n sampleListId='chol_tcga_pan_can_atlas_2018_all',\n projection='DETAILED'\n).result()",
"CPU times: user 766 ms, sys: 20.1 ms, total: 786 ms\nWall time: 1.03 s\n"
]
],
[
[
"Convert the results to a dataframe again:",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\nmdf = pd.DataFrame.from_dict([\n # python magic that combines two dictionaries:\n dict(\n m.__dict__['_Model__dict'],\n **m.__dict__['_Model__dict']['gene'].__dict__['_Model__dict']) \n # create one item in the list for each mutation\n for m in mutations\n])",
"_____no_output_____"
]
],
[
[
"Then get only the unique mutations, to avoid calling the web service with the same variants:",
"_____no_output_____"
]
],
[
[
"variants = mdf['chromosome startPosition endPosition referenceAllele variantAllele'.split()]\\\n .drop_duplicates()\\\n .dropna(how='any',axis=0)\\\n .reset_index()",
"_____no_output_____"
]
],
[
[
"Convert them to input that genome nexus will understand:",
"_____no_output_____"
]
],
[
[
"variants = variants.rename(columns={'startPosition':'start','endPosition':'end'})\\\n .to_dict(orient='records')\n# remove the index field\nfor v in variants:\n del v['index']",
"_____no_output_____"
],
[
"print(\"There are {} mutations left to annotate\".format(len(variants)))",
"There are 1991 mutations left to annotate\n"
]
],
[
[
"Annotate them with genome nexus:",
"_____no_output_____"
]
],
[
[
"%%time \n\nvariants_annotated = gn.annotation_controller.fetchVariantAnnotationByGenomicLocationPOST(\n genomicLocations=variants,\n fields='hotspots annotation_summary'.split()\n).result()",
"CPU times: user 3.22 s, sys: 522 ms, total: 3.75 s\nWall time: 6.61 s\n"
]
],
[
[
"Index the variants to make it easier to query them:",
"_____no_output_____"
]
],
[
[
"gn_dict = {\n \"{},{},{},{},{}\".format(\n v.annotation_summary.genomicLocation.chromosome,\n v.annotation_summary.genomicLocation.start,\n v.annotation_summary.genomicLocation.end,\n v.annotation_summary.genomicLocation.referenceAllele,\n v.annotation_summary.genomicLocation.variantAllele)\n :\n v for v in variants_annotated\n}",
"_____no_output_____"
]
],
[
[
"Add a new column to indicate whether something is a hotspot",
"_____no_output_____"
]
],
[
[
"def is_hotspot(x):\n \"\"\"TODO: Current structure for hotspots in Genome Nexus is a little funky.\n Need to check whether all lists in the annotation field are empty.\"\"\"\n if x:\n return sum([len(a) for a in x.hotspots.annotation]) > 0\n else:\n return False\n\ndef create_dict_query_key(x):\n return \"{},{},{},{},{}\".format(\n x.chromosome, x.startPosition, x.endPosition, x.referenceAllele, x.variantAllele\n )",
"_____no_output_____"
],
[
"mdf['is_hotspot'] = mdf.apply(lambda x: is_hotspot(gn_dict.get(create_dict_query_key(x), None)), axis=1)",
"_____no_output_____"
]
],
[
[
"Then plot the results:",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport seaborn as sns\nsns.set_style(\"white\")\nsns.set_context('notebook')\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"mdf.groupby('hugoGeneSymbol').is_hotspot.sum().sort_values(ascending=False).head(10).plot(kind='bar')\n\nsns.despine(trim=False)\nplt.xlabel('')\nplt.xticks(rotation=300)\nplt.ylabel('Number of non-unique hotspots',labelpad=20)\nplt.title('Hotspots in Cholangiocarcinoma (TCGA, PanCancer Atlas)',pad=25)",
"_____no_output_____"
]
],
[
[
"### OncoKB API",
"_____no_output_____"
],
[
"[OncoKB](https://oncokb.org) is is a precision oncology knowledge base and contains information about the effects and treatment implications of specific cancer gene alterations. Similarly to cBioPortal and Genome Nexus it provides a REST API following the [Swagger / OpenAPI specification](https://swagger.io/specification/).",
"_____no_output_____"
]
],
[
[
"oncokb = SwaggerClient.from_url('https://www.oncokb.org/api/v1/v2/api-docs',\n config={\"validate_requests\":False,\n \"validate_responses\":False,\n \"validate_swagger_spec\":False})\nprint(oncokb)",
"SwaggerClient(https://www.oncokb.org:443/api/v1)\n"
]
],
[
[
"To look up annotations for a variant, one can use the following endpoint:",
"_____no_output_____"
]
],
[
[
"variant = oncokb.Annotations.annotateMutationsByGenomicChangeGetUsingGET(\n genomicLocation='7,140453136,140453136,A,T',\n).result()",
"_____no_output_____"
],
[
"drugs = oncokb.Drugs.drugsGetUsingGET().result()",
"_____no_output_____"
]
],
[
[
"You can see a lot of information is provided for that particular variant if you type tab after `variant.`:",
"_____no_output_____"
]
],
[
[
"drugs.count",
"_____no_output_____"
],
[
"variant.hotspot",
"_____no_output_____"
]
],
[
[
"For instance we can see the summary information about it:",
"_____no_output_____"
]
],
[
[
"variant.variantSummary",
"_____no_output_____"
]
],
[
[
"If you look up this variant on the OncoKB website: https://www.oncokb.org/gene/BRAF/V600E. You can see that there are various combinations of drugs and their level of evidence listed. This is a classification system for indicating how much we know about whether or not a patient might respond to a particular treatment. Please see https://www.oncokb.org/levels for more information about the levels of evidence for therapeutic biomarkers.\n\nWe can use the same `variants` we pulled from cBioPortal in the previous section to figure out the highest level of each variant.",
"_____no_output_____"
]
],
[
[
"%%time \n\nvariants_annotated = oncokb.Annotations.annotateMutationsByGenomicChangePostUsingPOST(\n body=[\n {\"genomicLocation\":\"{chromosome},{start},{end},{referenceAllele},{variantAllele}\".format(**v)} \n for v in variants\n ],\n).result()",
"CPU times: user 363 ms, sys: 16.4 ms, total: 379 ms\nWall time: 9.89 s\n"
]
],
[
[
"Count the highes level for each variant",
"_____no_output_____"
]
],
[
[
"from collections import Counter\n\ncounts_per_level = Counter([va.highestSensitiveLevel for va in variants_annotated if va.highestSensitiveLevel])",
"_____no_output_____"
]
],
[
[
"Then plot them",
"_____no_output_____"
]
],
[
[
"pd.DataFrame(counts_per_level,index=[0]).plot(kind='bar', colors=['#4D8834','#2E2E2C','#753579'])\nplt.xticks([])\nplt.ylabel('Number of variants')\nplt.title('Actionable variants in chol_tcga_pan_can_atlas_2018')\nsns.despine()",
"_____no_output_____"
]
],
[
[
"The current plot could be more useful. See the idea listed here for one example of how to improve it: https://github.com/mskcc/cbsp-hackathon/tree/master/1-ideas/annotate-oncokb-barchart.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e78e5baebc60b3d6501d60e3c267639379cb6ea3 | 10,378 | ipynb | Jupyter Notebook | Fetching_data_google_places_API.ipynb | vivek1240/Fetching-the-raw-data-from-google-places-API | d47a642cd4f449c81f6a0130520dfb4029c2b608 | [
"MIT"
] | null | null | null | Fetching_data_google_places_API.ipynb | vivek1240/Fetching-the-raw-data-from-google-places-API | d47a642cd4f449c81f6a0130520dfb4029c2b608 | [
"MIT"
] | null | null | null | Fetching_data_google_places_API.ipynb | vivek1240/Fetching-the-raw-data-from-google-places-API | d47a642cd4f449c81f6a0130520dfb4029c2b608 | [
"MIT"
] | null | null | null | 28.988827 | 247 | 0.386683 | [
[
[
"# Fetching the raw data from google places API by passing the coordinates of the cluster centroid and delivery radius as 5km ",
"_____no_output_____"
]
],
[
[
"import pandas as pd \nimport googlemaps\nimport pprint\nimport json\nimport time\nimport xlsxwriter\nimport functools\nimport operator\nfrom collections import Counter \nfrom itertools import chain ",
"_____no_output_____"
],
[
"#IMPORTING DATA\ndf = pd.read_excel('lat_long_google_api.xlsx')\nrslt_df= df.copy()",
"_____no_output_____"
],
[
"# Define our API Key\nAPI_KEY = 'Enter your API key'",
"_____no_output_____"
],
[
"# Define our Client \ngmaps = googlemaps.Client(key = API_KEY)",
"_____no_output_____"
]
],
[
[
"## Below code is the Call for the Places API and the result will be stored in a dictionary, we will take the key = store_id, value = result fetched for the corresponding store from the places API",
"_____no_output_____"
],
[
"### Dict \"d\" will contain the raw data corresponding to the coordinate(lat,long)",
"_____no_output_____"
]
],
[
[
"d= dict() #ALL THE RAW DATA WOULD BE STORED IN DICTIONARY CORRESPONDING TO THE LATITUDE OF THE STORE\nd[rslt_df['store_id'][0]]=dict() #Taking store latitude as the \nfor i in range(len(rslt_df)):\n lat= rslt_df['store_latitude'][i]\n lon= rslt_df['store_longitude'][i] \n d[rslt_df['store_id'][i]] = gmaps.places_nearby(location='{},{}'.format(lat,lon), radius = 5000, open_now =False ) #not giving type parameter so it will give all types in the result\n time.sleep(5) \n print(d[rslt_df['store_id'][i]]) ",
"_____no_output_____"
]
],
[
[
"# Fetched data with the index as cluster number",
"_____no_output_____"
]
],
[
[
" def no_of_lodges_or_eqv(d,key):\n \n \n rawdata=[] \n for i in range(len(d[key]['results'])):\n rawdata.append(d[key]['results'][i]['types'])\n \n rawdata = functools.reduce(operator.iconcat, rawdata, [])\n rawdata = CountFrequency(rawdata) \n return rawdata",
"_____no_output_____"
],
[
" def CountFrequency(my_list): \n \n # Creating an empty dictionary \n freq = {} \n for item in my_list: \n if (item in freq): \n freq[item] += 1\n else: \n freq[item] = 1\n \n return freq \n\n",
"_____no_output_____"
],
[
"#############\ndf_podcast=pd.DataFrame(columns=['atm', 'bakery', 'bank', 'establishment', 'finance', 'food', 'health', 'hospital', 'locality', 'point_of_interest', 'political', 'real_estate_agency', 'spa', 'store', 'sublocality', 'sublocality_level_1'])\n###########",
"_____no_output_____"
],
[
"df_podcast.head()",
"_____no_output_____"
]
],
[
[
"This is the final transformed data fetched from the google places API",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
e78e5d76b90e803df50c7e52ff0e366e744488ac | 105,139 | ipynb | Jupyter Notebook | MobileNetSSD_OpenCV.ipynb | hurutoriya/yolov2_api | 9e463a0b5ddbbcbc0c3853f533f87e2985931066 | [
"MIT"
] | null | null | null | MobileNetSSD_OpenCV.ipynb | hurutoriya/yolov2_api | 9e463a0b5ddbbcbc0c3853f533f87e2985931066 | [
"MIT"
] | null | null | null | MobileNetSSD_OpenCV.ipynb | hurutoriya/yolov2_api | 9e463a0b5ddbbcbc0c3853f533f87e2985931066 | [
"MIT"
] | null | null | null | 661.251572 | 99,670 | 0.932765 | [
[
[
"## MobileNetSSD with OpenCV\n\n- you can get trained model and prototxt : https://www.pyimagesearch.com/2017/09/11/object-detection-with-deep-learning-and-opencv/",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n \n# import the necessary packages\nimport numpy as np\nimport sys\nfrom logging import getLogger, DEBUG, StreamHandler\nimport matplotlib.pyplot as plt\nimport cv2\n\ndef deep_learning_object_detection(image, prototxt, model):\n logger = getLogger(__name__)\n logger.setLevel(DEBUG)\n handler = StreamHandler(sys.stderr)\n handler.setLevel(DEBUG)\n logger.addHandler(handler)\n\n # construct the argument parse and parse the arguments\n CONFIDENCE = 0.2\n\n # initialize the list of class labels MobileNet SSD was trained to\n # detect, then generate a set of bounding box colors for each class\n CLASSES = [\"background\", \"aeroplane\", \"bicycle\", \"bird\", \"boat\",\"bottle\", \"bus\", \"car\", \"cat\", \"chair\", \"cow\", \"diningtable\",\n \"dog\", \"horse\", \"motorbike\", \"person\", \"pottedplant\", \"sheep\", \"sofa\", \"train\", \"tvmonitor\"]\n COLORS = np.random.uniform(0, 255, size=(len(CLASSES), 3))\n \n # load our serialized model from disk\n logger.info(\"Loading model...\")\n\n net = cv2.dnn.readNetFromCaffe(prototxt, model)\n \n # load the input image and construct an input blob for the image\n # by resizing to a fixed 300x300 pixels and then normalizing it\n # (note: normalization is done via the authors of the MobileNet SSD\n # implementation)\n image = cv2.imread(image)\n (h, w) = image.shape[:2]\n blob = cv2.dnn.blobFromImage(cv2.resize(image, (300, 300)), 0.007843, (300, 300), 127.5)\n \n # pass the blob through the network and obtain the detections and\n # predictions\n logger.info(\"computing object detections...\")\n\n net.setInput(blob)\n detections = net.forward()\n\n # loop over the detections\n for i in np.arange(0, detections.shape[2]):\n # extract the confidence (i.e., probability) associated with the\n # prediction\n confidence = detections[0, 0, i, 2]\n\n # filter out weak detections by ensuring the `confidence` is\n # greater than the minimum confidence\n if confidence > CONFIDENCE:\n # extract the index of the class label from the `detections`,\n # then compute the (x, y)-coordinates of the bounding box for\n # the object\n idx = int(detections[0, 0, i, 1])\n box = detections[0, 0, i, 3:7] * np.array([w, h, w, h])\n (startX, startY, endX, endY) = box.astype(\"int\")\n \n # display the prediction\n label = \"{}: {:.2f}%\".format(CLASSES[idx], confidence * 100)\n logger.info(label)\n\n cv2.rectangle(image, (startX, startY), (endX, endY), COLORS[idx], 2)\n y = startY - 15 if startY - 15 > 15 else startY + 15\n cv2.putText(image, label, (startX, y), cv2.FONT_HERSHEY_SIMPLEX, 0.5, COLORS[idx], 2)\n\n # show the output image\n plt.imshow(image)",
"_____no_output_____"
],
[
"image = \"images/example_01.jpg\"\nprototxt=\"MobileNetSSD_deploy.prototxt.txt\"\nmodel=\"MobileNetSSD_deploy.caffemodel\"\n\ndeep_learning_object_detection(image, prototxt, model)",
"Loading model...\ncomputing object detections...\ncar: 99.96%\ncar: 95.68%\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
]
] |
e78e69e36b8e5016b1e4164a77a19ff3006e4d37 | 79,269 | ipynb | Jupyter Notebook | scratch/002_test_perlin.ipynb | ANaka/genpen | 08f811dde40596a774ab4343af45a0ac0896840e | [
"MIT"
] | null | null | null | scratch/002_test_perlin.ipynb | ANaka/genpen | 08f811dde40596a774ab4343af45a0ac0896840e | [
"MIT"
] | null | null | null | scratch/002_test_perlin.ipynb | ANaka/genpen | 08f811dde40596a774ab4343af45a0ac0896840e | [
"MIT"
] | null | null | null | 28.47306 | 126 | 0.524505 | [
[
[
"import itertools\nimport numpy as np\nimport os\nimport seaborn as sns\nfrom tqdm import tqdm\nfrom dataclasses import asdict, dataclass, field\nimport vsketch\nimport shapely.geometry as sg\nfrom shapely.geometry import box, MultiLineString, Point, MultiPoint, Polygon, MultiPolygon, LineString\nimport shapely.affinity as sa\nimport shapely.ops as so\nimport matplotlib.pyplot as plt\nimport pandas as pd\n\nimport vpype_cli\nfrom typing import List, Generic\nfrom genpen import genpen as gp, utils as utils\nfrom scipy import stats as ss\nimport geopandas\nfrom shapely.errors import TopologicalError\nimport functools\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"def occlude(top, bottom, distance=1e-6):\n try:\n return bottom.difference(top)\n except TopologicalError:\n return bottom.buffer(distance).difference(top.buffer(distance))",
"_____no_output_____"
],
[
"class ParticleCluster(object):\n \n def __init__(\n self,\n pos,\n perlin_grid,\n ):\n self.pos = Point(pos)\n self.pg = perlin_grid\n self.particles = []\n \n def gen_start_pts_gaussian(\n self,\n n_particles=10,\n xloc=0.,\n xscale=1.,\n yloc=0.,\n yscale=1.,\n ):\n xs = self.pos.x + ss.norm(loc=xloc, scale=xscale).rvs(n_particles)\n ys = self.pos.y + ss.norm(loc=yloc, scale=yscale).rvs(n_particles)\n self.start_pts = [Point((x,y)) for x,y in zip(xs, ys)]\n \n def init_particles(self, start_bounds=None):\n for pt in self.start_pts:\n p = gp.Particle(pos=pt, grid=self.pg)\n if start_bounds == None:\n self.particles.append(p)\n elif start_area.contains(p.pos):\n self.particles.append(p)\n \n @functools.singledispatchmethod \n def step(self, n_steps):\n for p,n in zip(self.particles, n_steps):\n for i in range(n):\n p.step()\n \n @step.register\n def _(self, n_steps: int):\n n_steps = [n_steps] * len(self.particles)\n for p,n in zip(self.particles, n_steps):\n for i in range(n):\n p.step()\n \n @property\n def lines(self):\n return MultiLineString([p.line for p in self.particles])\n ",
"_____no_output_____"
],
[
"@dataclass\nclass ScaleTransPrms(gp.DataClassBase):\n \n n_iters: int = 100\n d_buffer: float = -0.25\n d_translate_factor: float = 0.9\n d_translate: float = None\n angles: float = 0.\n d_translates: list = field(default=None, init=False)\n def __post_init__(self):\n self.d_buffers = np.array([self.d_buffer] * self.n_iters)\n \n if self.d_translates == None:\n if self.d_translate != None:\n self.d_translates = np.array([self.d_translate] * self.n_iters)\n else:\n self.d_translates = self.d_buffers * self.d_translate_factor\n \n @property\n def prms(self):\n varnames = ['d_buffers', 'd_translates', 'angles']\n return {var: getattr(self, var) for var in varnames}",
"_____no_output_____"
],
[
"def individual_difference(multipolygon0, multipolygon1, dist=1e-6):\n diffs = []\n for p0, p1 in itertools.product(multipolygon0, multipolygon1):\n if p0.overlaps(p1):\n try:\n diff = p0.difference(p1)\n except TopologicalError:\n diff = p0.buffer(dist).difference(p1.buffer(dist))\n diffs.append(diff)\n return diffs",
"_____no_output_____"
]
],
[
[
"## try 1",
"_____no_output_____"
]
],
[
[
"page_x_inches: float = 11. # inches\npage_y_inches: float = 8.5 # inches\nborder:float = 0.\n\nperlin_grid_params = {\n 'xstep':3,\n 'ystep':3,\n 'lod':10,\n 'falloff':None,\n 'noise_scale':0.0073,\n 'noiseSeed':6\n}\n\nparticle_init_grid_params = {\n 'xstep':16,\n 'ystep':16,\n}\n\nbuffer_style = 2",
"_____no_output_____"
],
[
"px = utils.DistanceConverter(page_x_inches, 'inches').mm\npy = utils.DistanceConverter(page_y_inches, 'inches').mm\npage_format = f'{px}mmx{py}mm'\ndrawbox = sg.box(border, border, px-border, py-border)\n\nxmin, ymin, xmax, ymax = drawbox.bounds\nbrad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])\npg = gp.PerlinGrid(drawbox, **perlin_grid_params)",
"_____no_output_____"
],
[
"start_area = sa.scale(drawbox.centroid.buffer(brad*0.45), xfact=1.3)",
"_____no_output_____"
],
[
"start_area = drawbox.buffer(-20)",
"_____no_output_____"
],
[
"xcs, ycs = gp.overlay_grid(start_area, xstep=19, ystep=15)\nstart_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]\nstart_pts = [p for p in start_pts if start_area.contains(p)]",
"_____no_output_____"
],
[
"gpolys = []\nfor p in start_pts:\n\n pc = ParticleCluster(pos=p, perlin_grid=pg)\n pc.gen_start_pts_gaussian(n_particles=20, xscale=4, yscale=4)\n pc.init_particles()\n n_steps = np.random.randint(low=10, high=50, size=len(pc.particles))\n pc.step(n_steps)\n\n buffer_distances = np.random.uniform(low=0.3, high=1.3) + np.random.uniform(low=0., high=0.7, size=len(pc.lines))\n polys = []\n for line, bd in zip(pc.lines, buffer_distances):\n poly = line.buffer(bd,\n cap_style=buffer_style,\n join_style=buffer_style)\n polys.append(poly)\n\n P = gp.Poly(so.unary_union(polys))\n\n stp = ScaleTransPrms(\n d_buffer=np.random.uniform(low=-0.45, high=-0.25), \n angles=-90,\n\n )\n stp.d_buffers += np.random.uniform(-0.03, 0.03, size=stp.d_buffers.shape)\n P.fill_scale_trans(**stp.prms)\n gpolys.append(P)",
"_____no_output_____"
],
[
"zorder = np.random.permutation(len(gpolys))\n\nfor z, gpoly in zip(zorder, gpolys):\n gpoly.z = z\n\nfor gp0, gp1 in itertools.combinations(gpolys, r=2):\n try:\n overlaps = gp0.p.overlaps(gp1.p)\n except :\n gp0.p = gp0.p.buffer(1e-6)\n gp1.p = gp1.p.buffer(1e-6)\n overlaps = gp0.p.overlaps(gp1.p)\n if overlaps:\n if gp0.z > gp1.z:\n gp1.p = occlude(top=gp0.p, bottom=gp1.p)\n elif gp0.z < gp1.z:\n gp0.p = occlude(top=gp1.p, bottom=gp0.p)",
"_____no_output_____"
],
[
"ifills = []\nfor p in gpolys:\n try:\n ifills.append(p.intersection_fill)\n except:\n pass\n\nsplits = utils.random_split(ifills, n_layers=5)\nlayers = [utils.merge_LineStrings(split) for split in splits]",
"_____no_output_____"
],
[
"sk = vsketch.Vsketch()\nsk.size(page_format)\nsk.scale('1mm')\nsk.penWidth('0.25mm')\nfor i, layer in enumerate(layers):\n sk.stroke(i+1)\n sk.geometry(layer)\nsk.display(color_mode='layer')",
"_____no_output_____"
],
[
"savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0083_perlin_flow_erode_frays_occlude.svg'\n\nsk.save(savepath)\n\nvpype_commands = 'linesimplify --tolerance 0.01mm linemerge --tolerance 0.1mm reloop linesort'\nvpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'\n\nos.system(vpype_str)",
"_____no_output_____"
]
],
[
[
"## try 2",
"_____no_output_____"
]
],
[
[
"page_x_inches: float = 11. # inches\npage_y_inches: float = 8.5 # inches\nborder:float = 0.\n\nperlin_grid_params = {\n 'xstep':1,\n 'ystep':1,\n 'lod':10,\n 'falloff':None,\n 'noise_scale':0.0073,\n 'noiseSeed':8\n}\nbuffer_style = 2",
"_____no_output_____"
],
[
"px = utils.DistanceConverter(page_x_inches, 'inches').mm\npy = utils.DistanceConverter(page_y_inches, 'inches').mm\npage_format = f'{px}mmx{py}mm'\ndrawbox = sg.box(border, border, px-border, py-border)\n\nxmin, ymin, xmax, ymax = drawbox.bounds\nbrad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])\npg = gp.PerlinGrid(drawbox, **perlin_grid_params)",
"_____no_output_____"
],
[
"start_area = drawbox",
"_____no_output_____"
],
[
"xcs, ycs = gp.overlay_grid(start_area, xstep=15, ystep=15)\nstart_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]\nstart_pts = [p for p in start_pts if start_area.contains(p)]",
"_____no_output_____"
],
[
"gpolys = []\nfor p in start_pts:\n\n pc = ParticleCluster(pos=p, perlin_grid=pg)\n pc.gen_start_pts_gaussian(n_particles=30, xscale=7, yscale=7)\n pc.init_particles(start_bounds=drawbox)\n n_steps = np.random.randint(low=10, high=90, size=len(pc.particles))\n pc.step(n_steps)\n\n buffer_distances = np.random.uniform(low=0.3, high=1.3) + np.random.uniform(low=0., high=0.7, size=len(pc.lines))\n polys = []\n for line, bd in zip(pc.lines, buffer_distances):\n poly = line.buffer(bd,\n cap_style=buffer_style,\n join_style=buffer_style)\n polys.append(poly)\n\n P = gp.Poly(so.unary_union(polys))\n\n stp = ScaleTransPrms(\n d_buffer=np.random.uniform(low=-0.4, high=-0.2),\n d_translate_factor=0.7,\n angles=-240,\n\n )\n stp.d_buffers += np.random.uniform(-0.03, 0.03, size=stp.d_buffers.shape)\n P.fill_scale_trans(**stp.prms)\n gpolys.append(P)",
"_____no_output_____"
],
[
"zorder = np.random.permutation(len(gpolys))\n\nfor z, gpoly in zip(zorder, gpolys):\n gpoly.z = z\n\nfor gp0, gp1 in itertools.combinations(gpolys, r=2):\n try:\n overlaps = gp0.p.overlaps(gp1.p)\n except:\n gp0.p = gp0.p.buffer(1e-6)\n gp1.p = gp1.p.buffer(1e-6)\n overlaps = gp0.p.overlaps(gp1.p)\n if overlaps:\n if gp0.z > gp1.z:\n gp1.p = occlude(top=gp0.p, bottom=gp1.p)\n elif gp0.z < gp1.z:\n gp0.p = occlude(top=gp1.p, bottom=gp0.p)",
"_____no_output_____"
],
[
"ifills = []\nfor p in gpolys:\n ifills.append(p.intersection_fill)",
"_____no_output_____"
],
[
"splits = utils.random_split(ifills, n_layers=4)\nlayers = [utils.merge_LineStrings(split) for split in splits]",
"_____no_output_____"
],
[
"sk = vsketch.Vsketch()\nsk.size(page_format)\nsk.scale('1mm')\nsk.penWidth('0.25mm')\nfor i, layer in enumerate(layers):\n sk.stroke(i+1)\n sk.geometry(layer)\nsk.display(color_mode='layer')",
"_____no_output_____"
],
[
"savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0084_perlin_flow_erode_frays_occlude.svg'\n\nsk.save(savepath)\n\nvpype_commands = 'linesimplify --tolerance 0.05mm linemerge --tolerance 0.1mm reloop linesort'\nvpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'\n\nos.system(vpype_str)",
"_____no_output_____"
]
],
[
[
"## try 3",
"_____no_output_____"
]
],
[
[
"page_x_inches: float = 6 # inches\npage_y_inches: float = 6 # inches\nborder:float = 20.\n\nperlin_grid_params = {\n 'xstep':1,\n 'ystep':1,\n 'lod':10,\n 'falloff':None,\n 'noise_scale':0.0063,\n 'noiseSeed':8\n}\nbuffer_style = 2",
"_____no_output_____"
],
[
"px = utils.DistanceConverter(page_x_inches, 'inches').mm\npy = utils.DistanceConverter(page_y_inches, 'inches').mm\npage_format = f'{px}mmx{py}mm'\ndrawbox = sg.box(border, border, px-border, py-border)\n\nxmin, ymin, xmax, ymax = drawbox.bounds\nbrad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])\npg = gp.PerlinGrid(drawbox, **perlin_grid_params)\nstart_area = drawbox\nxcs, ycs = gp.overlay_grid(start_area, xstep=45, ystep=45)\nstart_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]\nstart_pts = [p for p in start_pts if start_area.contains(p)]",
"_____no_output_____"
],
[
"gpolys = []\nfor p in start_pts:\n\n pc = ParticleCluster(pos=p, perlin_grid=pg)\n pc.gen_start_pts_gaussian(n_particles=30, xscale=6, yscale=6)\n pc.init_particles(start_bounds=drawbox)\n n_steps = np.random.randint(low=80, high=190, size=len(pc.particles))\n pc.step(n_steps)\n\n buffer_distances = np.random.uniform(low=0.3, high=1.3) + np.random.uniform(low=0., high=0.7, size=len(pc.lines))\n polys = []\n for line, bd in zip(pc.lines, buffer_distances):\n poly = line.buffer(bd,\n cap_style=buffer_style,\n join_style=buffer_style)\n polys.append(poly)\n\n P = gp.Poly(so.unary_union(polys))\n \n stp = ScaleTransPrms(\n d_buffer=np.random.uniform(low=-0.4, high=-0.2),\n d_translate_factor=0.7,\n angles=-240,\n\n )\n stp.d_buffers += np.random.uniform(-0.03, 0.03, size=stp.d_buffers.shape)\n P.fill0 = gp.merge_LineStrings([p.boundary for p in gp.scale_trans(P.p, **stp.prms)])\n \n stp = ScaleTransPrms(\n d_buffer=np.random.uniform(low=-0.4, high=-0.2),\n d_translate_factor=0.7,\n angles=120,\n\n )\n stp.d_buffers += np.random.uniform(-0.03, 0.03, size=stp.d_buffers.shape)\n P.fill1 = gp.merge_LineStrings([p.boundary for p in gp.scale_trans(P.p, **stp.prms)])\n gpolys.append(P)",
"_____no_output_____"
],
[
"zorder = np.random.permutation(len(gpolys))\n\nfor z, gpoly in zip(zorder, gpolys):\n gpoly.z = z\n\nfor gp0, gp1 in itertools.combinations(gpolys, r=2):\n try:\n overlaps = gp0.p.overlaps(gp1.p)\n except:\n gp0.p = gp0.p.buffer(1e-6)\n gp1.p = gp1.p.buffer(1e-6)\n overlaps = gp0.p.overlaps(gp1.p)\n if overlaps:\n if gp0.z > gp1.z:\n gp1.p = occlude(top=gp0.p, bottom=gp1.p)\n elif gp0.z < gp1.z:\n gp0.p = occlude(top=gp1.p, bottom=gp0.p)",
"_____no_output_____"
],
[
"ifills0 = []\nifills1 = []\nfor p in gpolys:\n ifills0.append(p.fill0.intersection(p.p.buffer(1e-6)))\n ifills1.append(p.fill1.intersection(p.p.buffer(1e-6)))",
"_____no_output_____"
],
[
"layers = []\nlayers.append(gp.merge_LineStrings(ifills0))\nlayers.append(gp.merge_LineStrings(ifills1))",
"_____no_output_____"
],
[
"sk = vsketch.Vsketch()\nsk.size(page_format)\nsk.scale('1mm')\nsk.penWidth('0.25mm')\nfor i, layer in enumerate(layers):\n sk.stroke(i+1)\n sk.geometry(layer)\nsk.display(color_mode='layer')",
"_____no_output_____"
],
[
"savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0085_perlin_flow_erode_frays_color_mix.svg'\n\nsk.save(savepath)\n\nvpype_commands = 'linesimplify --tolerance 0.2mm linemerge --tolerance 0.2mm reloop linesort'\nvpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'\n\nos.system(vpype_str)",
"_____no_output_____"
]
],
[
[
"## try 4 for fabiano black black",
"_____no_output_____"
]
],
[
[
"\nborder:float = 25.\n\nperlin_grid_params = {\n 'xstep':1,\n 'ystep':1,\n 'lod':10,\n 'falloff':None,\n 'noise_scale':0.0063,\n 'noiseSeed':8\n}\nbuffer_style = 2",
"_____no_output_____"
],
[
"px = 200\npy = 200\npage_format = f'{px}mmx{py}mm'\ndrawbox = sg.box(border, border, px-border, py-border)\n\nxmin, ymin, xmax, ymax = drawbox.bounds\nbrad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])\n\nstart_area = drawbox.centroid.buffer(brad/2)\npg = gp.PerlinGrid(start_area, **perlin_grid_params)\nxcs, ycs = gp.overlay_grid(start_area, xstep=11, ystep=11)\nstart_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]\nstart_pts = [p for p in start_pts if start_area.contains(p)]",
"_____no_output_____"
],
[
"gpolys = []\nfor p in start_pts:\n\n pc = ParticleCluster(pos=p, perlin_grid=pg)\n pc.gen_start_pts_gaussian(n_particles=35, xscale=4, yscale=4)\n pc.init_particles(start_bounds=drawbox)\n n_steps = np.random.randint(low=10, high=60, size=len(pc.particles))\n pc.step(n_steps)\n\n buffer_distances = np.random.uniform(low=0.3, high=1.7) + np.random.uniform(low=0., high=0.7, size=len(pc.lines))\n polys = []\n for line, bd in zip(pc.lines, buffer_distances):\n poly = line.buffer(bd,\n cap_style=buffer_style,\n join_style=buffer_style)\n polys.append(poly)\n\n P = gp.Poly(so.unary_union(polys))\n \n stp = ScaleTransPrms(\n d_buffer=np.random.uniform(low=-0.45, high=-0.25),\n d_translate_factor=0.7,\n angles=-240,\n\n )\n stp.d_buffers += np.random.uniform(-0.06, 0.06, size=stp.d_buffers.shape)\n P.fill0 = gp.merge_LineStrings([p.boundary for p in gp.scale_trans(P.p, **stp.prms)])\n \n \n gpolys.append(P)",
"_____no_output_____"
],
[
"zorder = np.random.permutation(len(gpolys))\n\nfor z, gpoly in zip(zorder, gpolys):\n gpoly.z = z\n\nfor gp0, gp1 in itertools.combinations(gpolys, r=2):\n try:\n overlaps = gp0.p.overlaps(gp1.p)\n except:\n gp0.p = gp0.p.buffer(1e-6)\n gp1.p = gp1.p.buffer(1e-6)\n overlaps = gp0.p.overlaps(gp1.p)\n if overlaps:\n if gp0.z > gp1.z:\n gp1.p = occlude(top=gp0.p, bottom=gp1.p)\n elif gp0.z < gp1.z:\n gp0.p = occlude(top=gp1.p, bottom=gp0.p)",
"_____no_output_____"
],
[
"ifills0 = []\nfor p in gpolys:\n ifills0.append(p.fill0.intersection(p.p.buffer(1e-6)))\n\nifills0 = [l for l in ifills0 if l.length > 1e-1]\n\nlayers = []\nlayers.append(gp.merge_LineStrings(ifills0))",
"_____no_output_____"
],
[
"sk = vsketch.Vsketch()\nsk.size(page_format)\nsk.scale('1mm')\nsk.penWidth('0.25mm')\nfor i, layer in enumerate(layers):\n sk.stroke(i+1)\n sk.geometry(layer)\nsk.display(color_mode='layer')",
"_____no_output_____"
],
[
"savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0086_perlin_flow_erode_frays_color_mix.svg'\n\nsk.save(savepath)\n\nvpype_commands = 'linesimplify --tolerance 0.2mm linemerge --tolerance 0.2mm reloop linesort'\nvpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'\n\nos.system(vpype_str)",
"_____no_output_____"
]
],
[
[
"## try 5 two color",
"_____no_output_____"
]
],
[
[
"page_x_inches: float = 8.5 # inches\npage_y_inches: float = 11 # inches\nborder:float = 0.\n\nperlin_grid_params = {\n 'xstep':1,\n 'ystep':1,\n 'lod':10,\n 'falloff':None,\n 'noise_scale':0.0063,\n 'noiseSeed':8\n}\nbuffer_style = 3",
"_____no_output_____"
],
[
"px = utils.DistanceConverter(page_x_inches, 'inches').mm\npy = utils.DistanceConverter(page_y_inches, 'inches').mm\npage_format = f'{px}mmx{py}mm'\ndrawbox = sg.box(border, border, px-border, py-border)\n\nxmin, ymin, xmax, ymax = drawbox.bounds\nbrad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])\n\nstart_area = drawbox.centroid.buffer(brad*0.47)\npg = gp.PerlinGrid(start_area, **perlin_grid_params)\nxcs, ycs = gp.overlay_grid(start_area, xstep=15, ystep=15)\nstart_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]\nstart_pts = [p for p in start_pts if start_area.contains(p)]",
"_____no_output_____"
],
[
"gpolys = []\nfor p in start_pts:\n\n pc = ParticleCluster(pos=p, perlin_grid=pg)\n pc.gen_start_pts_gaussian(n_particles=30, xscale=7, yscale=7)\n pc.init_particles(start_bounds=drawbox)\n n_steps = np.random.randint(low=10, high=100, size=len(pc.particles))\n pc.step(n_steps)\n\n buffer_distances = np.random.uniform(low=0.3, high=2.7) + np.random.uniform(low=0., high=0.7, size=len(pc.lines))\n polys = []\n for line, bd in zip(pc.lines, buffer_distances):\n poly = line.buffer(bd,\n cap_style=buffer_style,\n join_style=buffer_style)\n polys.append(poly)\n\n P = gp.Poly(so.unary_union(polys))\n \n stp = ScaleTransPrms(\n d_buffer=np.random.uniform(low=-0.5, high=-0.25),\n d_translate_factor=0.7,\n angles=np.radians(-60),\n\n )\n stp.d_buffers += np.random.uniform(-0.06, 0.06, size=stp.d_buffers.shape)\n P.fill0 = gp.merge_LineStrings([p.boundary for p in gp.scale_trans(P.p, **stp.prms)])\n \n stp = ScaleTransPrms(\n d_buffer=np.random.uniform(low=-0.5, high=-0.25),\n d_translate_factor=0.7,\n angles=np.radians(-120),\n\n )\n stp.d_buffers += np.random.uniform(-0.06, 0.06, size=stp.d_buffers.shape)\n P.fill1 = gp.merge_LineStrings([p.boundary for p in gp.scale_trans(P.p, **stp.prms)])\n \n gpolys.append(P)",
"_____no_output_____"
],
[
"zorder = np.random.permutation(len(gpolys))\n\nfor z, gpoly in zip(zorder, gpolys):\n gpoly.z = z\n\nfor gp0, gp1 in itertools.combinations(gpolys, r=2):\n try:\n overlaps = gp0.p.overlaps(gp1.p)\n except:\n gp0.p = gp0.p.buffer(1e-6)\n gp1.p = gp1.p.buffer(1e-6)\n overlaps = gp0.p.overlaps(gp1.p)\n if overlaps:\n if gp0.z > gp1.z:\n gp1.p = occlude(top=gp0.p, bottom=gp1.p)\n elif gp0.z < gp1.z:\n gp0.p = occlude(top=gp1.p, bottom=gp0.p)",
"_____no_output_____"
],
[
"ifills0 = []\nifills1 = []\nfor p in gpolys:\n ifills0.append(p.fill0.intersection(p.p.buffer(1e-6)))\n ifills1.append(p.fill1.intersection(p.p.buffer(1e-6)))\n \nifills0 = [l for l in ifills0 if l.length > 1e-1]\nifills1 = [l for l in ifills1 if l.length > 1e-1]\n\nlayers = []\nlayers.append(gp.merge_LineStrings(ifills0))\nlayers.append(gp.merge_LineStrings(ifills1))",
"_____no_output_____"
],
[
"sk = vsketch.Vsketch()\nsk.size(page_format)\nsk.scale('1mm')\nsk.penWidth('0.25mm')\nfor i, layer in enumerate(layers):\n sk.stroke(i+1)\n sk.geometry(layer)\nsk.display(color_mode='layer')",
"_____no_output_____"
],
[
"savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0087_perlin_flow_erode_frays_color_mix.svg'\n\nsk.save(savepath)\n\nvpype_commands = 'linesimplify --tolerance 0.2mm linemerge --tolerance 0.2mm reloop linesort'\nvpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'\n\nos.system(vpype_str)",
"_____no_output_____"
]
],
[
[
"## try 6 three color",
"_____no_output_____"
]
],
[
[
"page_x_inches: float = 8.5 # inches\npage_y_inches: float = 11 # inches\nborder:float = 0.\n\nperlin_grid_params = {\n 'xstep':1,\n 'ystep':1,\n 'lod':10,\n 'falloff':None,\n 'noise_scale':0.0063,\n 'noiseSeed':8\n}\nbuffer_style = 3",
"_____no_output_____"
],
[
"px = utils.DistanceConverter(page_x_inches, 'inches').mm\npy = utils.DistanceConverter(page_y_inches, 'inches').mm\npage_format = f'{px}mmx{py}mm'\ndrawbox = sg.box(border, border, px-border, py-border)\n\nxmin, ymin, xmax, ymax = drawbox.bounds\nbrad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])\n\nstart_area = drawbox.centroid.buffer(brad*0.47)\npg = gp.PerlinGrid(start_area, **perlin_grid_params)\nxcs, ycs = gp.overlay_grid(start_area, xstep=15, ystep=15)\nstart_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]\nstart_pts = [p for p in start_pts if start_area.contains(p)]",
"_____no_output_____"
],
[
"n_fills = 3\nfill_angles = [-30, -90, -150]\nn_iter_choices = np.array(list(itertools.product(*[[2, 100]] * n_fills)))",
"_____no_output_____"
],
[
"gpolys = []\nfor p in start_pts:\n\n pc = ParticleCluster(pos=p, perlin_grid=pg)\n pc.gen_start_pts_gaussian(n_particles=30, xscale=7, yscale=7)\n pc.init_particles(start_bounds=drawbox)\n n_steps = np.random.randint(low=10, high=100, size=len(pc.particles))\n pc.step(n_steps)\n\n buffer_distances = np.random.uniform(low=0.3, high=2.7) + np.random.uniform(low=0., high=0.7, size=len(pc.lines))\n polys = []\n for line, bd in zip(pc.lines, buffer_distances):\n poly = line.buffer(bd,\n cap_style=buffer_style,\n join_style=buffer_style)\n polys.append(poly)\n\n P = gp.Poly(so.unary_union(polys))\n P.fills = []\n n_iter_choice = n_iter_choices[np.random.choice(n_iter_choices.shape[0])]\n for i in range(n_fills):\n stp = ScaleTransPrms(\n n_iters=n_iter_choice[i],\n d_buffer=np.random.uniform(low=-0.6, high=-0.25),\n d_translate_factor=0.7,\n angles=np.radians(fill_angles[i]),)\n stp.d_buffers += np.random.uniform(-0.1, 0.1, size=stp.d_buffers.shape)\n fill = gp.merge_LineStrings([p.boundary for p in gp.scale_trans(P.p, **stp.prms)])\n P.fills.append(fill)\n \n gpolys.append(P)",
"_____no_output_____"
],
[
"zorder = np.random.permutation(len(gpolys))\n\nfor z, gpoly in zip(zorder, gpolys):\n gpoly.z = z\n\nfor gp0, gp1 in itertools.combinations(gpolys, r=2):\n try:\n overlaps = gp0.p.overlaps(gp1.p)\n except:\n gp0.p = gp0.p.buffer(1e-6)\n gp1.p = gp1.p.buffer(1e-6)\n overlaps = gp0.p.overlaps(gp1.p)\n if overlaps:\n if gp0.z > gp1.z:\n gp1.p = occlude(top=gp0.p, bottom=gp1.p)\n elif gp0.z < gp1.z:\n gp0.p = occlude(top=gp1.p, bottom=gp0.p)",
"_____no_output_____"
],
[
"fill_sets = []\nfor i in range(n_fills):\n fill_sets.append([])\n \nfor p in gpolys:\n for i in range(n_fills):\n fill_sets[i].append(p.fills[i].intersection(p.p.buffer(1e-6)))\n\nlayers = []\nfor fill_set in fill_sets:\n filter_fills = [l for l in fill_set if l.length > 0.2]\n layers.append(gp.merge_LineStrings(filter_fills))",
"_____no_output_____"
],
[
"sk = vsketch.Vsketch()\nsk.size(page_format)\nsk.scale('1mm')\nsk.penWidth('0.25mm')\nfor i, layer in enumerate(layers):\n sk.stroke(i+1)\n sk.geometry(layer)\nsk.display(color_mode='layer')",
"_____no_output_____"
],
[
"savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0088_perlin_flow_erode_frays_color_mix.svg'\n\nsk.save(savepath)\n\nvpype_commands = 'linesimplify --tolerance 0.2mm linemerge --tolerance 0.2mm reloop linesort'\nvpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'\n\nos.system(vpype_str)",
"_____no_output_____"
]
],
[
[
"# try 7\n",
"_____no_output_____"
]
],
[
[
"page_x_inches: float = 8.5 # inches\npage_y_inches: float = 11 # inches\nborder:float = 0.\n\nperlin_grid_params = {\n 'xstep':1,\n 'ystep':1,\n 'lod':10,\n 'falloff':None,\n 'noise_scale':0.0063,\n 'noiseSeed':8\n}\nbuffer_style = 3",
"_____no_output_____"
],
[
"px = utils.DistanceConverter(page_x_inches, 'inches').mm\npy = utils.DistanceConverter(page_y_inches, 'inches').mm\npage_format = f'{px}mmx{py}mm'\ndrawbox = sg.box(border, border, px-border, py-border)\n\nxmin, ymin, xmax, ymax = drawbox.bounds\nbrad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])\n\n# start_area = drawbox.centroid.buffer(brad*0.47)\nstart_area = drawbox\npg = gp.PerlinGrid(start_area, **perlin_grid_params)\nxcs, ycs = gp.overlay_grid(start_area, xstep=35, ystep=35)\nstart_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]\nstart_pts = [p for p in start_pts if start_area.contains(p)]",
"_____no_output_____"
],
[
"n_fills = 3\nfill_angles = [-30, -90, -150]\nn_iter_choices = np.array(list(itertools.product(*[[0, 100]] * n_fills)))\nn_iter_choices = n_iter_choices[1:-1,:]",
"_____no_output_____"
],
[
"gpolys = []\nfor p in start_pts:\n\n pc = ParticleCluster(pos=p, perlin_grid=pg)\n pc.gen_start_pts_gaussian(n_particles=60, xscale=6, yscale=6)\n pc.init_particles(start_bounds=drawbox)\n n_steps = np.random.randint(low=4, high=7, size=len(pc.particles))\n pc.step(n_steps)\n\n buffer_distances = np.random.uniform(low=0.3, high=0.7) + np.random.uniform(low=0., high=3.7, size=len(pc.lines))\n polys = []\n for line, bd in zip(pc.lines, buffer_distances):\n poly = line.buffer(bd,\n cap_style=buffer_style,\n join_style=buffer_style)\n polys.append(poly)\n\n P = gp.Poly(so.unary_union(polys))\n P.fills = []\n n_iter_choice = n_iter_choices[np.random.choice(n_iter_choices.shape[0])]\n for i in range(n_fills):\n stp = ScaleTransPrms(\n n_iters=n_iter_choice[i],\n d_buffer=np.random.uniform(low=-0.6, high=-0.25),\n d_translate_factor=0.7,\n angles=np.radians(fill_angles[i]),)\n stp.d_buffers += np.random.uniform(-0.1, 0.1, size=stp.d_buffers.shape)\n fill = gp.merge_LineStrings([p.boundary for p in gp.scale_trans(P.p, **stp.prms)])\n P.fills.append(fill)\n \n gpolys.append(P)",
"_____no_output_____"
],
[
"zorder = np.random.permutation(len(gpolys))\n\nfor z, gpoly in zip(zorder, gpolys):\n gpoly.z = z\n\nfor gp0, gp1 in itertools.combinations(gpolys, r=2):\n try:\n overlaps = gp0.p.overlaps(gp1.p)\n except:\n gp0.p = gp0.p.buffer(1e-6)\n gp1.p = gp1.p.buffer(1e-6)\n overlaps = gp0.p.overlaps(gp1.p)\n if overlaps:\n if gp0.z > gp1.z:\n gp1.p = occlude(top=gp0.p, bottom=gp1.p)\n elif gp0.z < gp1.z:\n gp0.p = occlude(top=gp1.p, bottom=gp0.p)",
"_____no_output_____"
],
[
"fill_sets = []\nfor i in range(n_fills):\n fill_sets.append([])\n \nfor p in gpolys:\n for i in range(n_fills):\n fill_sets[i].append(p.fills[i].intersection(p.p.buffer(1e-6)))\n\nlayers = []\nfor fill_set in fill_sets:\n filter_fills = [l for l in fill_set if l.length > 0.2]\n layers.append(gp.merge_LineStrings(filter_fills))",
"_____no_output_____"
],
[
"sk = vsketch.Vsketch()\nsk.size(page_format)\nsk.scale('1mm')\nsk.penWidth('0.25mm')\nfor i, layer in enumerate(layers):\n sk.stroke(i+1)\n sk.geometry(layer)\nsk.display(color_mode='layer')",
"_____no_output_____"
],
[
"savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0090_perlin_flow_erode_frays_color_mix.svg'\n\nsk.save(savepath)\n\nvpype_commands = 'linesimplify --tolerance 0.2mm linemerge --tolerance 0.2mm reloop linesort'\nvpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'\n\nos.system(vpype_str)",
"_____no_output_____"
]
],
[
[
"## try 8",
"_____no_output_____"
]
],
[
[
"page_x_inches: float = 8.5 # inches\npage_y_inches: float = 11 # inches\nborder:float = 0.\n\nperlin_grid_params = {\n 'xstep':3,\n 'ystep':3,\n 'lod':10,\n 'falloff':None,\n 'noise_scale':0.0013,\n 'noiseSeed':8\n}\nbuffer_style = 3",
"_____no_output_____"
],
[
"px = utils.DistanceConverter(page_x_inches, 'inches').mm\npy = utils.DistanceConverter(page_y_inches, 'inches').mm\npage_format = f'{px}mmx{py}mm'\ndrawbox = sg.box(border, border, px-border, py-border)\n\nxmin, ymin, xmax, ymax = drawbox.bounds\nbrad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])\n\n# start_area = drawbox.centroid.buffer(brad*0.47)\nstart_area = drawbox.buffer(-5, cap_style=3, join_style=3)\npg = gp.PerlinGrid(start_area, **perlin_grid_params)\nxcs, ycs = gp.overlay_grid(start_area, xstep=15, ystep=15)\nstart_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]\nstart_pts = [p for p in start_pts if start_area.contains(p)]",
"_____no_output_____"
],
[
"f,ax = plt.subplots(figsize=(6,6))\nax.quiver(np.cos(pg.a), np.sin(pg.a), scale=50)",
"_____no_output_____"
],
[
"n_fills = 3\nfill_angles = [-30, -40, -50]\nn_iter_choices = np.array(list(itertools.product(*[[0, 100]] * n_fills)))\nn_iter_choices = n_iter_choices[1:-1,:]",
"_____no_output_____"
],
[
"gpolys = []\nfor p in start_pts:\n\n pc = ParticleCluster(pos=p, perlin_grid=pg)\n pc.gen_start_pts_gaussian(n_particles=15, xscale=3, yscale=3)\n pc.init_particles(start_bounds=drawbox)\n n_steps = np.random.randint(low=1, high=15, size=len(pc.particles))\n pc.step(n_steps)\n\n buffer_distances = np.random.uniform(low=0.1, high=1.7) + np.random.uniform(low=0., high=0.3, size=len(pc.lines))\n polys = []\n for line, bd in zip(pc.lines, buffer_distances):\n poly = line.buffer(bd,\n cap_style=buffer_style,\n join_style=buffer_style)\n polys.append(poly)\n\n P = gp.Poly(so.unary_union(polys))\n P.fills = []\n n_iter_choice = n_iter_choices[np.random.choice(n_iter_choices.shape[0])]\n for i in range(n_fills):\n stp = ScaleTransPrms(\n n_iters=n_iter_choice[i],\n d_buffer=np.random.uniform(low=-0.6, high=-0.2),\n d_translate_factor=0.8,\n angles=np.radians(fill_angles[i]),)\n stp.d_buffers += np.random.uniform(-0.1, 0.1, size=stp.d_buffers.shape)\n fill = gp.merge_LineStrings([p.boundary for p in gp.scale_trans(P.p, **stp.prms)])\n P.fills.append(fill)\n \n gpolys.append(P)",
"_____no_output_____"
],
[
"zorder = np.random.permutation(len(gpolys))\n\nfor z, gpoly in zip(zorder, gpolys):\n gpoly.z = z\n\nfor gp0, gp1 in itertools.combinations(gpolys, r=2):\n try:\n overlaps = gp0.p.overlaps(gp1.p)\n except:\n gp0.p = gp0.p.buffer(1e-6)\n gp1.p = gp1.p.buffer(1e-6)\n overlaps = gp0.p.overlaps(gp1.p)\n if overlaps:\n if gp0.z > gp1.z:\n gp1.p = occlude(top=gp0.p, bottom=gp1.p)\n elif gp0.z < gp1.z:\n gp0.p = occlude(top=gp1.p, bottom=gp0.p)",
"_____no_output_____"
],
[
"fill_sets = []\nfor i in range(n_fills):\n fill_sets.append([])\n \nfor p in gpolys:\n for i in range(n_fills):\n fill_sets[i].append(p.fills[i].intersection(p.p.buffer(1e-6)))\n\nlayers = []\nfor fill_set in fill_sets:\n filter_fills = [l for l in fill_set if l.length > 0.2]\n layers.append(gp.merge_LineStrings(filter_fills))",
"_____no_output_____"
],
[
"sk = vsketch.Vsketch()\nsk.size(page_format)\nsk.scale('1mm')\nsk.penWidth('0.25mm')\nfor i, layer in enumerate(layers):\n sk.stroke(i+1)\n sk.geometry(layer)\nsk.display(color_mode='layer')",
"_____no_output_____"
],
[
"savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0091_perlin_flow_erode_frays_color_mix.svg'\n\nsk.save(savepath)\n\nvpype_commands = 'reloop linesimplify --tolerance 0.2mm linemerge --tolerance 0.2mm linesort'\nvpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'\n\nos.system(vpype_str)",
"_____no_output_____"
]
],
[
[
"# try 9\n",
"_____no_output_____"
]
],
[
[
"page_x_inches: float = 6 # inches\npage_y_inches: float = 6 # inches\nborder:float = 0.\n\nperlin_grid_params = {\n 'xstep':1,\n 'ystep':1,\n 'lod':10,\n 'falloff':None,\n 'noise_scale':0.0063,\n 'noiseSeed':8\n}\nbuffer_style = 3",
"_____no_output_____"
],
[
"px = utils.DistanceConverter(page_x_inches, 'inches').mm\npy = utils.DistanceConverter(page_y_inches, 'inches').mm\npage_format = f'{px}mmx{py}mm'\ndrawbox = sg.box(border, border, px-border, py-border)\n\nxmin, ymin, xmax, ymax = drawbox.bounds\nbrad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])\n\n# start_area = drawbox.centroid.buffer(brad*0.47)\nstart_area = drawbox.buffer(-10)\npg = gp.PerlinGrid(start_area, **perlin_grid_params)\nxcs, ycs = gp.overlay_grid(start_area, xstep=25, ystep=25)\nstart_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]\nstart_pts = [p for p in start_pts if start_area.contains(p)]",
"_____no_output_____"
],
[
"n_fills = 3\nfill_angles = [-30, -90, -150]\nn_iter_choices = np.array(list(itertools.product(*[[0, 100]] * n_fills)))\nn_iter_choices = n_iter_choices[1:-1,:]",
"_____no_output_____"
],
[
"gpolys = []\nfor p in start_pts:\n\n pc = ParticleCluster(pos=p, perlin_grid=pg)\n pc.gen_start_pts_gaussian(n_particles=30, xscale=5, yscale=5)\n pc.init_particles(start_bounds=start_area)\n n_steps = np.random.randint(low=4, high=17, size=len(pc.particles))\n pc.step(n_steps)\n\n buffer_distances = np.random.uniform(low=0.3, high=0.7) + np.random.uniform(low=0., high=3.7, size=len(pc.lines))\n polys = []\n for line, bd in zip(pc.lines, buffer_distances):\n poly = line.buffer(bd,\n cap_style=buffer_style,\n join_style=buffer_style)\n polys.append(poly)\n\n P = gp.Poly(so.unary_union(polys))\n P.fills = []\n n_iter_choice = n_iter_choices[np.random.choice(n_iter_choices.shape[0])]\n for i in range(n_fills):\n stp = ScaleTransPrms(\n n_iters=n_iter_choice[i],\n d_buffer=np.random.uniform(low=-0.6, high=-0.25),\n d_translate_factor=0.7,\n angles=np.radians(fill_angles[i]),)\n stp.d_buffers += np.random.uniform(-0.1, 0.1, size=stp.d_buffers.shape)\n fill = gp.merge_LineStrings([p.boundary for p in gp.scale_trans(P.p, **stp.prms)])\n P.fills.append(fill)\n \n gpolys.append(P)",
"_____no_output_____"
],
[
"zorder = np.random.permutation(len(gpolys))\n\nfor z, gpoly in zip(zorder, gpolys):\n gpoly.z = z\n\nfor gp0, gp1 in itertools.combinations(gpolys, r=2):\n try:\n overlaps = gp0.p.overlaps(gp1.p)\n except:\n gp0.p = gp0.p.buffer(1e-6)\n gp1.p = gp1.p.buffer(1e-6)\n overlaps = gp0.p.overlaps(gp1.p)\n if overlaps:\n if gp0.z > gp1.z:\n gp1.p = occlude(top=gp0.p, bottom=gp1.p)\n elif gp0.z < gp1.z:\n gp0.p = occlude(top=gp1.p, bottom=gp0.p)",
"_____no_output_____"
],
[
"fill_sets = []\nfor i in range(n_fills):\n fill_sets.append([])\n \nfor p in gpolys:\n for i in range(n_fills):\n fill_sets[i].append(p.fills[i].intersection(p.p.buffer(1e-6)))\n\nlayers = []\nfor fill_set in fill_sets:\n filter_fills = [l for l in fill_set if l.length > 0.2]\n layers.append(gp.merge_LineStrings(filter_fills))",
"_____no_output_____"
],
[
"sk = vsketch.Vsketch()\nsk.size(page_format)\nsk.scale('1mm')\nsk.penWidth('0.25mm')\nfor i, layer in enumerate(layers):\n sk.stroke(i+1)\n sk.geometry(layer)\nsk.display(color_mode='layer')",
"_____no_output_____"
],
[
"savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0092_perlin_flow_erode_frays_color_mix.svg'\n\nsk.save(savepath)\n\nvpype_commands = 'reloop linesimplify --tolerance 0.2mm linemerge --tolerance 0.2mm linesort'\nvpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'\n\nos.system(vpype_str)",
"_____no_output_____"
]
],
[
[
"# try 10",
"_____no_output_____"
]
],
[
[
"page_x_inches: float = 8.5 # inches\npage_y_inches: float = 11 # inches\nborder:float = 0.\n\nperlin_grid_params = {\n 'xstep':3,\n 'ystep':3,\n 'lod':10,\n 'falloff':None,\n 'noise_scale':0.0013,\n 'noiseSeed':8\n}\nbuffer_style = 3",
"_____no_output_____"
],
[
"px = utils.DistanceConverter(page_x_inches, 'inches').mm\npy = utils.DistanceConverter(page_y_inches, 'inches').mm\npage_format = f'{px}mmx{py}mm'\ndrawbox = sg.box(border, border, px-border, py-border)\n\nxmin, ymin, xmax, ymax = drawbox.bounds\nbrad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])\n\n# start_area = drawbox.centroid.buffer(brad*0.47)\nstart_area = drawbox.buffer(-5, cap_style=3, join_style=3)\npg = gp.PerlinGrid(start_area, **perlin_grid_params)\nxcs, ycs = gp.overlay_grid(start_area, xstep=25, ystep=25)\nstart_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]\nstart_pts = [p for p in start_pts if start_area.contains(p)]",
"_____no_output_____"
],
[
"f,ax = plt.subplots(figsize=(6,6))\nax.quiver(np.cos(pg.a), np.sin(pg.a), scale=50)",
"_____no_output_____"
],
[
"n_fills = 2\nfill_angles = [-30, -40, ]\nn_iter_choices = np.array(list(itertools.product(*[[0, 100]] * n_fills)))\nn_iter_choices = n_iter_choices[[-1], :]",
"_____no_output_____"
],
[
"gpolys = []\nfor p in start_pts:\n\n pc = ParticleCluster(pos=p, perlin_grid=pg)\n pc.gen_start_pts_gaussian(n_particles=35, xscale=5, yscale=5)\n pc.init_particles(start_bounds=drawbox)\n n_steps = np.random.randint(low=1, high=15, size=len(pc.particles))\n pc.step(n_steps)\n\n buffer_distances = np.random.uniform(low=0.1, high=1.7) + np.random.uniform(low=0., high=0.3, size=len(pc.lines))\n polys = []\n for line, bd in zip(pc.lines, buffer_distances):\n poly = line.buffer(bd,\n cap_style=buffer_style,\n join_style=buffer_style)\n polys.append(poly)\n\n P = gp.Poly(so.unary_union(polys))\n P.fills = []\n n_iter_choice = n_iter_choices[np.random.choice(n_iter_choices.shape[0])]\n for i in range(n_fills):\n stp = ScaleTransPrms(\n n_iters=n_iter_choice[i],\n d_buffer=np.random.uniform(low=-0.6, high=-0.2),\n d_translate_factor=0.8,\n angles=np.radians(fill_angles[i]),)\n stp.d_buffers += np.random.uniform(-0.1, 0.1, size=stp.d_buffers.shape)\n fill = gp.merge_LineStrings([p.boundary for p in gp.scale_trans(P.p, **stp.prms)])\n P.fills.append(fill)\n \n gpolys.append(P)",
"_____no_output_____"
],
[
"zorder = np.random.permutation(len(gpolys))\n\nfor z, gpoly in zip(zorder, gpolys):\n gpoly.z = z\n\nfor gp0, gp1 in itertools.combinations(gpolys, r=2):\n try:\n overlaps = gp0.p.overlaps(gp1.p)\n except:\n gp0.p = gp0.p.buffer(1e-6)\n gp1.p = gp1.p.buffer(1e-6)\n overlaps = gp0.p.overlaps(gp1.p)\n if overlaps:\n if gp0.z > gp1.z:\n gp1.p = occlude(top=gp0.p, bottom=gp1.p)\n elif gp0.z < gp1.z:\n gp0.p = occlude(top=gp1.p, bottom=gp0.p)",
"_____no_output_____"
],
[
"fill_sets = []\nfor i in range(n_fills):\n fill_sets.append([])\n \nfor p in gpolys:\n for i in range(n_fills):\n fill_sets[i].append(p.fills[i].intersection(p.p.buffer(1e-6)))\n\nlayers = []\nfor fill_set in fill_sets:\n filter_fills = [l for l in fill_set if l.length > 0.2]\n layers.append(gp.merge_LineStrings(filter_fills))",
"_____no_output_____"
],
[
"sk = vsketch.Vsketch()\nsk.size(page_format)\nsk.scale('1mm')\nsk.penWidth('0.25mm')\nfor i, layer in enumerate(layers):\n sk.stroke(i+1)\n sk.geometry(layer)\nsk.display(color_mode='layer')",
"_____no_output_____"
],
[
"savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0093_perlin_flow_erode_frays.svg'\n\nsk.save(savepath)\n\nvpype_commands = 'reloop linesimplify --tolerance 0.2mm linemerge --tolerance 0.2mm linesort'\nvpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'\n\nos.system(vpype_str)",
"_____no_output_____"
]
],
[
[
"# try 11",
"_____no_output_____"
]
],
[
[
"page_x_inches: float = 11 # inches\npage_y_inches: float = 8.5 # inches\nborder:float = 0.\n\nperlin_grid_params = {\n 'xstep':3,\n 'ystep':3,\n 'lod':10,\n 'falloff':None,\n 'noise_scale':0.0053,\n 'noiseSeed':3\n}\nbuffer_style = 3",
"_____no_output_____"
],
[
"px = utils.DistanceConverter(page_x_inches, 'inches').mm\npy = utils.DistanceConverter(page_y_inches, 'inches').mm\npage_format = f'{px}mmx{py}mm'\ndrawbox = sg.box(border, border, px-border, py-border)\n\nxmin, ymin, xmax, ymax = drawbox.bounds\nbrad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])\n\nstart_area = drawbox.buffer(-10, cap_style=3, join_style=3)\npg = gp.PerlinGrid(start_area, **perlin_grid_params)\nxcs, ycs = gp.overlay_grid(start_area, xstep=25, ystep=35)\nstart_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]\nstart_pts = [p for p in start_pts if start_area.contains(p)]",
"_____no_output_____"
],
[
"f,ax = plt.subplots(figsize=(6,6))\nax.quiver(np.cos(pg.a), np.sin(pg.a), scale=50)",
"_____no_output_____"
],
[
"gpolys = []\nfor p in start_pts:\n\n pc = ParticleCluster(pos=p, perlin_grid=pg)\n pc.gen_start_pts_gaussian(n_particles=40, xscale=5, yscale=5)\n pc.init_particles(start_bounds=start_area)\n n_steps = np.random.randint(low=3, high=20, size=len(pc.particles))\n pc.step(n_steps)\n\n buffer_distances = np.random.uniform(low=0.3, high=3.3) + np.random.uniform(low=0., high=1.7, size=len(pc.lines))\n polys = []\n for line, bd in zip(pc.lines, buffer_distances):\n poly = line.buffer(bd,\n cap_style=buffer_style,\n join_style=buffer_style)\n polys.append(poly)\n\n P = gp.Poly(so.unary_union(polys))\n\n stp = ScaleTransPrms(\n d_buffer=np.random.uniform(low=-0.75, high=-0.3), \n angles=-90,\n\n )\n stp.d_buffers += np.random.uniform(-0.03, 0.03, size=stp.d_buffers.shape)\n P.fill_scale_trans(**stp.prms)\n gpolys.append(P)",
"_____no_output_____"
],
[
"zorder = np.random.permutation(len(gpolys))\n\nfor z, gpoly in zip(zorder, gpolys):\n gpoly.z = z\n\nfor gp0, gp1 in itertools.combinations(gpolys, r=2):\n try:\n overlaps = gp0.p.overlaps(gp1.p)\n except :\n gp0.p = gp0.p.buffer(1e-6)\n gp1.p = gp1.p.buffer(1e-6)\n overlaps = gp0.p.overlaps(gp1.p)\n if overlaps:\n if gp0.z > gp1.z:\n gp1.p = occlude(top=gp0.p, bottom=gp1.p)\n elif gp0.z < gp1.z:\n gp0.p = occlude(top=gp1.p, bottom=gp0.p)",
"_____no_output_____"
],
[
"ifills = []\nfor p in gpolys:\n try:\n ifills.append(p.intersection_fill)\n except:\n pass\n\nsplits = utils.random_split(ifills, n_layers=5)\nlayers = [gp.merge_LineStrings(split) for split in splits]",
"_____no_output_____"
],
[
"sk = vsketch.Vsketch()\nsk.size(page_format)\nsk.scale('1mm')\nsk.penWidth('0.25mm')\nfor i, layer in enumerate(layers):\n sk.stroke(i+1)\n sk.geometry(layer)\nsk.display(color_mode='layer')",
"_____no_output_____"
],
[
"savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0100_perlin_flow_erode_frays_occlude.svg'\n\nsk.save(savepath)\n\nvpype_commands = 'reloop linesimplify --tolerance 0.01mm linemerge --tolerance 0.1mm reloop linesort'\nvpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'\n\nos.system(vpype_str)",
"_____no_output_____"
]
],
[
[
"# try 12",
"_____no_output_____"
]
],
[
[
"page_x_inches: float = 11 # inches\npage_y_inches: float = 8.5 # inches\nborder:float = 0.\n\nperlin_grid_params = {\n 'xstep':3,\n 'ystep':3,\n 'lod':10,\n 'falloff':None,\n 'noise_scale':0.0083,\n 'noiseSeed':3\n}\nbuffer_style = 3",
"_____no_output_____"
],
[
"px = utils.DistanceConverter(page_x_inches, 'inches').mm\npy = utils.DistanceConverter(page_y_inches, 'inches').mm\npage_format = f'{px}mmx{py}mm'\ndrawbox = sg.box(border, border, px-border, py-border)\n\nxmin, ymin, xmax, ymax = drawbox.bounds\nbrad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])\n\nstart_area = drawbox.buffer(-10, cap_style=3, join_style=3)\npg = gp.PerlinGrid(start_area, **perlin_grid_params)\nxcs, ycs = gp.overlay_grid(start_area, xstep=15, ystep=55)\nstart_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]\nstart_pts = [p for p in start_pts if start_area.contains(p)]",
"_____no_output_____"
],
[
"f,ax = plt.subplots(figsize=(6,6))\nax.quiver(np.cos(pg.a), np.sin(pg.a), scale=50)",
"_____no_output_____"
],
[
"gpolys = []\nfor p in start_pts:\n\n pc = ParticleCluster(pos=p, perlin_grid=pg)\n pc.gen_start_pts_gaussian(n_particles=60, xscale=7, yscale=12)\n pc.init_particles(start_bounds=start_area)\n n_steps = np.random.randint(low=1, high=15, size=len(pc.particles))\n pc.step(n_steps)\n \n buffer_distances = np.random.uniform(low=0.3, high=6.3) + np.random.uniform(low=0., high=0.5, size=len(pc.lines))\n polys = []\n for line, bd in zip(pc.lines, buffer_distances):\n bd = np.interp(line.centroid.x, [10, 270], [0.1, 3])\n poly = line.buffer(bd,\n cap_style=buffer_style,\n join_style=buffer_style)\n polys.append(poly)\n\n P = gp.Poly(so.unary_union(polys))\n\n stp = ScaleTransPrms(\n d_buffer=np.random.uniform(low=-0.65, high=-0.3), \n angles=-90,\n\n )\n stp.d_buffers += np.random.uniform(-0.03, 0.03, size=stp.d_buffers.shape)\n P.fill_scale_trans(**stp.prms)\n gpolys.append(P)",
"_____no_output_____"
],
[
"zorder = np.random.permutation(len(gpolys))\n\nfor z, gpoly in zip(zorder, gpolys):\n gpoly.z = z\n\nfor gp0, gp1 in itertools.combinations(gpolys, r=2):\n try:\n overlaps = gp0.p.overlaps(gp1.p)\n except :\n gp0.p = gp0.p.buffer(1e-6)\n gp1.p = gp1.p.buffer(1e-6)\n overlaps = gp0.p.overlaps(gp1.p)\n if overlaps:\n if gp0.z > gp1.z:\n gp1.p = occlude(top=gp0.p, bottom=gp1.p)\n elif gp0.z < gp1.z:\n gp0.p = occlude(top=gp1.p, bottom=gp0.p)",
"_____no_output_____"
],
[
"ifills = []\nfor p in gpolys:\n try:\n ifills.append(p.intersection_fill)\n except:\n pass\n\nsplits = utils.random_split(ifills, n_layers=5)\nlayers = [gp.merge_LineStrings(split) for split in splits]",
"_____no_output_____"
],
[
"sk = vsketch.Vsketch()\nsk.size(page_format)\nsk.scale('1mm')\nsk.penWidth('0.25mm')\nfor i, layer in enumerate(layers):\n sk.stroke(i+1)\n sk.geometry(layer)\nsk.display(color_mode='none')",
"_____no_output_____"
],
[
"savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0101_perlin_flow_erode_frays_occlude.svg'\n\nsk.save(savepath)\n\nvpype_commands = 'reloop linesimplify --tolerance 0.01mm linemerge --tolerance 0.1mm reloop linesort'\nvpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write --page-format {page_format} {savepath}'\n\nos.system(vpype_str)",
"_____no_output_____"
]
],
[
[
"# try 13",
"_____no_output_____"
]
],
[
[
"page_x_inches: float = 11 # inches\npage_y_inches: float = 8.5 # inches\nborder:float = 0.\n\nperlin_grid_params = {\n 'xstep':1,\n 'ystep':1,\n 'lod':10,\n 'falloff':None,\n 'noise_scale':0.0193,\n 'noiseSeed':3\n}\nbuffer_style = 3",
"_____no_output_____"
],
[
"px = utils.DistanceConverter(page_x_inches, 'inches').mm\npy = utils.DistanceConverter(page_y_inches, 'inches').mm\npage_format = f'{px}mmx{py}mm'\ndrawbox = sg.box(border, border, px-border, py-border)\n\nxmin, ymin, xmax, ymax = drawbox.bounds\nbrad = np.min([gp.get_width(drawbox), gp.get_height(drawbox)])\n\nstart_area = drawbox.buffer(-10, cap_style=3, join_style=3)\npg = gp.PerlinGrid(start_area, **perlin_grid_params)\nxcs, ycs = gp.overlay_grid(start_area, xstep=20, ystep=75)\nstart_pts = [Point(x,y) for x,y in itertools.product(xcs, ycs)]\nstart_pts = [p for p in start_pts if start_area.contains(p)]",
"_____no_output_____"
],
[
"# f,ax = plt.subplots(figsize=(6,6))\n# ax.quiver(np.cos(pg.a), np.sin(pg.a), scale=50)",
"_____no_output_____"
],
[
"gpolys = []\nfor p in start_pts:\n\n pc = ParticleCluster(pos=p, perlin_grid=pg)\n pc.gen_start_pts_gaussian(n_particles=60, xscale=8, yscale=13)\n pc.init_particles(start_bounds=start_area)\n n_steps = int(np.interp(pc.pos.centroid.x, [1, 270], [1, 30]))\n# n_steps = np.random.randint(low=1, high=15, size=len(pc.particles))\n pc.step(n_steps)\n \n buffer_distances = np.random.uniform(low=0.3, high=6.3) + np.random.uniform(low=0., high=0.5, size=len(pc.lines))\n polys = []\n for line, bd in zip(pc.lines, buffer_distances):\n bd = np.interp(line.centroid.x, [10, 270], [0.1, 3.5])\n poly = line.buffer(bd,\n cap_style=buffer_style,\n join_style=buffer_style)\n polys.append(poly)\n\n P = gp.Poly(so.unary_union(polys))\n\n stp = ScaleTransPrms(\n d_buffer=np.random.uniform(low=-0.4, high=-0.2), \n angles=-60,\n\n )\n stp.d_buffers += np.random.uniform(-0.06, 0.06, size=stp.d_buffers.shape)\n P.fill_scale_trans(**stp.prms)\n gpolys.append(P)",
"_____no_output_____"
],
[
"zorder = np.random.permutation(len(gpolys))\n\nfor z, gpoly in zip(zorder, gpolys):\n gpoly.z = z\n\nfor gp0, gp1 in itertools.combinations(gpolys, r=2):\n try:\n overlaps = gp0.p.overlaps(gp1.p)\n except :\n gp0.p = gp0.p.buffer(1e-6)\n gp1.p = gp1.p.buffer(1e-6)\n overlaps = gp0.p.overlaps(gp1.p)\n if overlaps:\n if gp0.z > gp1.z:\n gp1.p = occlude(top=gp0.p, bottom=gp1.p)\n elif gp0.z < gp1.z:\n gp0.p = occlude(top=gp1.p, bottom=gp0.p)",
"_____no_output_____"
],
[
"ifills = []\nfor p in gpolys:\n try:\n ifills.append(p.intersection_fill)\n except:\n pass\n\nsplits = utils.random_split(ifills, n_layers=5)\nlayers = [gp.merge_LineStrings(split) for split in splits]",
"_____no_output_____"
],
[
"sk = vsketch.Vsketch()\nsk.size(page_format)\nsk.scale('1mm')\nsk.penWidth('0.25mm')\nfor i, layer in enumerate(layers):\n sk.stroke(i+1)\n sk.geometry(layer)\nsk.display(color_mode='none')",
"_____no_output_____"
],
[
"savepath = '/mnt/c/code/side/plotter_images/oned_outputs/0102_perlin_flow_erode_frays_occlude.svg'\n\nsk.save(savepath)",
"_____no_output_____"
],
[
"vpype_commands = 'reloop linesimplify --tolerance 0.01mm linemerge --tolerance 0.1mm reloop linesort'\nvpype_str = f'vpype read -q 0.05mm {savepath} {vpype_commands} write {savepath}'\n\nos.system(vpype_str)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78e6e8ce277486161cb3b19fd1f2125e409d580 | 21,111 | ipynb | Jupyter Notebook | Assignment7Groundwater.ipynb | LaurelOak/hydro-teaching-resources | 062e493ad0468bdc46728b09929930f9bca8de9b | [
"BSD-3-Clause"
] | 6 | 2018-04-07T19:28:17.000Z | 2021-12-20T11:40:42.000Z | Assignment7Groundwater.ipynb | LaurelOak/hydro-teaching-resources | 062e493ad0468bdc46728b09929930f9bca8de9b | [
"BSD-3-Clause"
] | null | null | null | Assignment7Groundwater.ipynb | LaurelOak/hydro-teaching-resources | 062e493ad0468bdc46728b09929930f9bca8de9b | [
"BSD-3-Clause"
] | 4 | 2018-01-20T00:28:44.000Z | 2021-01-01T02:52:54.000Z | 75.128114 | 1,198 | 0.683767 | [
[
[
"# Assignment 7: Groundwater and the Woburn Toxics Trial\n*Due 4/25/17 <br>\n5 pts <br>*\n\nPlease **submit your assignment as an html export**, and for written responses, please type them in a cell that is of type `Markdown.` The final part of the exercise involves drawing a flow net by hand (actually, you could tackle this part of the assignment first). For that, you may turn in a hard copy of your answer.",
"_____no_output_____"
]
],
[
[
"# Import numerical tools\nimport numpy as np\n\n#Import pandas for reading in and managing data\nimport pandas as pd\n\n# Import pyplot for plotting\nimport matplotlib.pyplot as plt\n\n#Import seaborn (useful for plotting)\nimport seaborn as sns\n\n# Magic function to make matplotlib inline; other style specs must come AFTER\n%matplotlib inline\n\n%config InlineBackend.figure_formats = {'svg',}",
"_____no_output_____"
]
],
[
[
"## Background\nThis investigation of groundwater flow and drawdown in wells is based on the lawsuit described in the book and movie A Civil Action (a true story). For the background behind this story, read [the Wikipedia page](http://en.wikipedia.org/wiki/A_Civil_Action). Then look at the map and animation of the study site [here](http://researchnews.osu.edu/archive/woburnpics.htm). Wells G and H shown on the map are the municipal wells from which Woburn’s water supply was withdrawn. \n\nIn this assignment you will be investigating some of the very problems that the hydrologists hired to the case worked on. As usual, you may work with another person, but each person must turn in separate assignments and specify whom they worked with. Below I provide additional information specific to this assignment. \n\n\n## Introduction\nOne of the important questions raised during the trial centered on the drawdown of the water table when wells G and H (see map linked above) were operating and the effects of the Riley tannery well (also included in the model). If, when wells G and/or H operated, the water table was drawn down on the east side of the Aberjona River, then contaminants from west side of the river valley (Beatrice 15 acres and Olympia Trucking) could flow under the river to wells G and H. If the water table was not drawn down on the east side of the river when wells G, H, and the Riley Tannery well operated, then it is unlikely that contaminants on the west side of the river valley could flow to wells G and H. Thus, the interaction of these three wells with the Aberjona River and the groundwater flow system was critical to determining which of the contaminated properties contributed TCE and PCE to wells G and H.\n\n## Evaluating sources of impact to wells G and H\nProving the connection between the contaminants migrating from the defendants’ properties to wells G and H during the first phase of the trial was a requirement to proceed to the next phase. To make arguments regarding the potential sources of TCE to wells G and H, the plaintiffs' expert had to define the subsurface geology, the surface and subsurface, and show that TCE was used at both the Riley/Beatrice and W.R. Grace properties. Counsel for the plaintiffs was able to identify Grace employees involved with on-site TCE disposal activities. However, the plaintiffs' counsel was not able to directly link disposal of wastes on the Riley 15-acre property with activities at the tannery itself. The [ATSDR report](http://www.atsdr.cdc.gov/HAC/PHA/wellsgh/wgh_p1.html), which was prepared three years after the trial, is a good illustration of how difficult it is to conclusively assess risk from a specific contaminant source at the Woburn site.\n\n## Understanding groundwater flow to wells\nThe U.S. Geological Survey first performed a seven-hour pumping test to characterize aquifer properties, and then, in December 1985-January 1986, a 30-day pumping test to evaluate the effects of pumping on the distribution of hydraulic head. This was the first time wells G and H operated since their closure in May 1979. It was also the first time synoptic sets of water levels had ever been measured in the network of monitoring wells surrounding wells G and H. During the 30-day pumping test, wells G and H operated at their average rates of 700 gpm (well G) and 400 gpm (well H). However, pumping records kept by the City of Woburn showed that between 1964 and 1979 the wells rarely operated together and that the two municipal wells were frequently not in use for months at a time. As a result of changes in the pumping rates of wells G and H and their periodic (discontinuous) use, the water table in the buried valley aquifer was dynamic, rising and falling as wells G and H were turned on and off and pumped heavily or lightly. These dynamic changes in water levels result in the groundwater flow system being transient in character – as opposed to being steady state in character.\n\nUnder steady-state conditions, water levels at any location in the flow system do not change with time, which results in hydraulic gradients and flow velocities at any location not changing with time, and therein no net change in the amount of groundwater in storage. Under transient conditions, water levels at any location in the flow system change with time, which causes hydraulic gradients, flow velocities, and the amount of groundwater in storage (water levels) to change with time. In this exercise we assume that the wells G and H have been pumping together for a long enough time that the water table is stationary and flow is steady state. This is an assumption that we recognize is not true over periods of time greater than a few months. This simplistic assumption lets us learn about drawdown created by a single pumping well, the formation of a cone of depression, superposition of drawdowns from one or more pumping wells, and the creation of groundwater divides between pumping wells, all of which are important to understanding the responses of the groundwater flow system to the pumping history of wells G and H. \n\nThe operation of the Riley well was estimated by de Lima and Olimpio ([USGS, 1989](https://pubs.usgs.gov/wri/1989/4059/report.pdf)) to operate at an average pumping rate of 200 gallons per minute, continuously. This was based on the wastewater discharge emanating from the tannery; actual pumping rates were not available. It is believed that pumping volumes fluctuated depending on the number of shifts working, the volume of leather orders, actual type of leather being processed, etc. \n\n## Part I of assignment: Solving for aquifer properties\n\n1.\tThe hydrologists who worked on the Woburn case conducted a pumping test in well H to determine the aquifer’s properties (storativity and hydraulic conductivity). Although I couldn’t access their data, I generated synthetic data that will give you the same results, in the cell below. During the pumping test, well H was pumped at a rate of 475 gallons per minute. Drawdown was measured in an observation well 75 feet away from the pumping well. The thickness of the aquifer was measured to be 140 ft. Using the Cooper-Jacob equation and the regression method we discussed in class, determine the aquifer’s hydraulic conductivity (in feet per day) and storativity. In your answer, show the plot of your regression (don't forget to label the axes, or to take the log of time), and mathematically show how you arrived at your final answer. Make sure to keep track of your units! *[1 pt.]*\n\nHelpful commands for linear regression: `np.polyfit()`, `np.polyval()`.\nSee an example of their usage in the Feb 2 tutorial, found under *Files/Materials* on bCouses.",
"_____no_output_____"
]
],
[
[
"# The data\ntime_minutes = np.arange(170,570,10) #Times (minutes) at which drawdown was observed after the start of pumping\ns = np.array([0.110256896, 0.122567503, 0.180480293, 0.214489433, 0.356304352, 0.554603882, 0.49240574, 0.524209467, 0.562727644, 0.754849464, 0.718713002, 0.752910019, 0.73903658, 0.89009263, 0.967712464, 0.910807162, 0.986238396, 1.042178187, 1.081114186, 1.080825045, 1.196565491, 1.264971986, 1.430805272, 1.377858223, 1.376787182, 1.340970634, 1.466832052, 1.528405086, 1.44136224, 1.610936499, 1.503519725, 1.55398487, 1.628028833, 1.675649643, 1.672772239, 1.730410501, 1.730935188, 1.756850444, 1.731143013, 1.818924173])#Drawdown in observation well, ft",
"_____no_output_____"
]
],
[
[
"## Part II: Estimating hydraulic heads with the Thiem Equation\nWith knowledge of the hydraulic conductivity, you can now solve the Thiem equation for the equilibrium potentiometric surface under different combinations of well pumping rates. For this part of the assignment, you will be working with a module that solves the Thiem equation over a two-dimensional spatial domain.\n\nThe module requires the user to input several parameters, as described in the red text. When you first run it, use the hydraulic conductivity that you just solved for. Assume that the Riley well (QR) pumps at an average rate of 200 gallons per minute (gpm), Well G (QG) pumps at an average rate of 700 gpm, and Well H (QH) pumps at an average rate of 400 gpm. When you run the module, you will see two plots. The first plot is a filled contour plot that depicts the drawdown relative to non-pumping conditions. The value shown is the composite amount of drawdown related to pumping from each well (drawdowns are additive). Note how the values increase in the cells immediately around the wells and decrease toward the margins of the graph. \n\nThe second plot is a cross section that shows the drawdown associated with the north-south transect that passes through wells G and H under steady state conditions.\n\n**2.** Take a look at the model below. What is a key assumption that is being made in the implementation of the Theim equation, beyond the assumptions we talked about in class? Hint: compare the formulas under the `Calculate Drawdown for Each Well` section to Eq. 5.49 in the Fetter handout. *[1/3 pt.]*\n",
"_____no_output_____"
]
],
[
[
"def PlotWoburnDD(K,b,QG,QH,QR, returnval=0):\n \"\"\"\n This routine uses the Thiem equation for unCONFINED aquifers to generate a plot of drawdown\n contours around the Aberjona River in Woburn, Massachusetts. The Riley well is the source of\n the contamination. Wells G and H are wells for the town's municipal water supply. Users can\n evaluate how the potentiometric surface changes with changes in hydraulic conductivity (K),\n saturated thickness (b), and well pumping rates (QG for well G, QH for well H, and QR for the\n Riley well). Note that K is in units of ft/day, b is in units of ft, and QG, QH, and QR are\n in units of gallons per minute. returnval is an optional argument. If you set it equal to 1,\n the function will return the value of drawdown at well G. If unspecified, the function returns\n just the plots.\n \"\"\"\n #CONVERT GALLONS PER MINUTE TO CUBIC FEET PER DAY\n QG = QG*192.5\n QH = QH*192.5\n QR = QR*192.5\n \n #OTHER CONSTANTS\n delx = 100 #Cell size in feet\n r0 = 2700 #You figure this out! (It is in feet)\n \n #SET UP THE EXPERIMENTAL GRID\n nrows = 25 #number of rows\n ncols = 27 #number of columns\n rowvect = np.arange(nrows) #A vector of row coordinates\n colvect = np.arange(ncols) #A vector of column coordinates\n colcoords, rowcoords = np.meshgrid(colvect,rowvect) #Creates two matrices. In colcoords, each \n #entry is the column index of the cell (i.e., point in space). In rowcoords, each entry is the \n #row index of the cell (i.e., point in space).\n \n #SPECIFY WELL LOCATIONS AND RIVER COORDINATES WITHIN THE EXPERIMENTAL GRID\n loc_R = np.array([20,8]) #Index locations for the Riley well (row, column)\n loc_H = np.array([10,13]) #Index locations for H well\n loc_G = np.array([17,13]) #Index locations for G well\n river_row = np.array([0, 1, 2, 3, 4, 5, 6, 6.4, 7, 8, 9, 9.9, 10, 11, 11.5, 12, 13, 14, 15, 16, 16.1, 17, 17.5, 18, 18.4, 19, 19.6, 20, 20.3, 21, 21.7, 22, 22.4, 23, 23.1, 23.9, 24, 24.8, 25])\n river_col = np.array([8.6, 9, 9.1, 9.3, 9.5, 9.6, 9.9, 10, 10.3, 11, 11.7, 12, 12.1, 12.7, 12.9, 12.8, 12.6, 12, 11.6, 11.1, 11, 10.8, 10.7, 10.8, 11, 11.4, 12, 12.6, 13, 14, 15, 15.5, 16, 16.9, 17, 18, 18.1, 19, 19.2])\n \n #CALCULATE DISTANCES BETWEEN EACH WELL AND EVERY OTHER CELL\n d_R = np.sqrt((rowcoords-loc_R[0])**2+(colcoords-loc_R[1])**2)*delx #Solve for distance (feet) using the Pythagorean theorem\n d_H = np.sqrt((rowcoords-loc_H[0])**2+(colcoords-loc_H[1])**2)*delx #Solve for distance (feet) using the Pythagorean theorem\n d_G = np.sqrt((rowcoords-loc_G[0])**2+(colcoords-loc_G[1])**2)*delx #Solve for distance (feet) using the Pythagorean theorem\n\n #SET DISTANCES TO WELL IN CELLS WITH WELL TO 25 FT (TO AVOID SINGULARITY AT 0)\n d_R[20,8] = 25 #feet\n d_H[10,13] = 25 #feet\n d_G[17,13] = 25 #feet\n \n #CALCULATE DRAWDOWN FROM EACH WELL\n s_R = b-np.sqrt(b**2-QR*np.log(r0/d_R)/(np.pi*K)) #Drawdown from the Riley well\n s_H = b-np.sqrt(b**2-QH*np.log(r0/d_H)/(np.pi*K)) #Drawdown from well H\n s_G = b-np.sqrt(b**2-QG*np.log(r0/d_G)/(np.pi*K)) #Drawadown from well G\n s_total = s_R+s_H+s_G #Combined drawdown from all of the wells, feet\n \n #NOW GENERATE PLOTS\n plt.figure(figsize=(10,10))\n s_plot = plt.contourf(np.transpose((colcoords+0.5)*delx), np.transpose((rowcoords+0.5)*delx), np.transpose(s_total), cmap=plt.cm.plasma)\n plt.gca().invert_yaxis() #This puts the origin of the plot on the upper left\n cb = plt.colorbar(s_plot, orientation='horizontal')\n cb.set_label('Drawdown, ft')\n riv_plot = plt.plot(river_col*delx, river_row*delx, 'b-',linewidth=3.0)\n wH = plt.plot((loc_H[1]+0.5)*delx, (loc_H[0]+0.5)*delx, 'ko')\n ax = plt.gca()\n ax.annotate('H', xy = ((loc_H[1]+0.5)*delx, (loc_H[0]+0.5)*delx))\n wG = plt.plot((loc_G[1]+0.5)*delx, (loc_G[0]+0.5)*delx, 'ko')\n wR = plt.plot((loc_R[1]+0.5)*delx, (loc_R[0]+0.5)*delx, 'ko')\n ax.annotate('G', xy = ((loc_G[1]+0.5)*delx, (loc_G[0]+0.5)*delx))\n ax.annotate('Riley', xy = ((loc_R[1]+0.5)*delx, (loc_R[0]+0.5)*delx))\n #plt.axis('equal') #Uncomment this to make the x- and y- axis display on the same scale.\n plt.title('Map of Aberjona River and Drawdown Contours')\n plt.show()\n \n plt.figure()\n plt.fill_between(rowvect*delx, max(np.ceil(s_total[:,13])), s_total[:,13])\n plt.plot([loc_H[0]*delx, loc_H[0]*delx], [min(s_total[:,13]), max(s_total[:,13])], 'r')\n plt.plot([loc_G[0]*delx, loc_G[0]*delx], [min(s_total[:,13]), max(s_total[:,13])], 'r')\n plt.gca().invert_yaxis()\n plt.xlabel('y distance, feet')\n plt.ylabel('Drawdown, feet')\n plt.title('Cross-section across column with wells G and H')\n \n if returnval==1:\n return s_total[17,13]",
"_____no_output_____"
]
],
[
[
"**3.** Now run the model! (You might want to check back to the Jan 31 class tutorial for a refresher on how to run modules/functions.) Using the pumping rates and aquifer thickness specified above, and the hydraulic conductivity that you solved for (in feet per day), how do pumping from the Riley well and Well H affect the shape of the drawdown contours? Why does the contour shape become uniformly round as you move away from the wells? *[1/3 pt.]*",
"_____no_output_____"
],
[
"**4.** Set the pumping rates for wells G and H to 0. How does this change the shape of drawdowns on the map and the contours? How would this pumping condition affect the direction and rate of movement of a contaminant plume? *[1/3 pt.]*",
"_____no_output_____"
],
[
"**5.** Now reset the pumping rate for well G to 700 gpm and keep the Riley well value at 200 gpm. <br><br>\na. How does this pumping configuration change the shape of drawdowns on the cross section and contour map? *[0.25 pts.]* <br><br>\nb. What is the net change in drawdown in well G when the value used in question 3 (0 GPM) is compared to that used for question 4 (700 gpm)? Note if you want the figures from both runs of the module to show up together, you need to insert the line `plt.figure()` between your first call to `PlotWoburnDD` and your second call. *[0.5 pts.]* <br><br>\nc. Could the new pumping conditions (well G = 700 gpm) pull TCE-contaminated water from the Riley property to well G? Explain why or why not. *[0.25 pts.]*",
"_____no_output_____"
],
[
"**6.** How sensitive are your results to hydraulic conductivity? To aquifer thickness? In other words, how does the potentiometric surface change when you use higher or lower values of these parameters? Write your answer in paragraph format. *Hint: If you generate several plots in a row, you might want to separate them with `print` statements so that you will know which is which.* *[0.5 pts.]*",
"_____no_output_____"
],
[
"**7.** Although this Thiem module can be an effective tool for examining hypothetical pumping rates and groundwater conditions, there are some severe limitations to using this simplistic treatment in a trial. What are these limitations? Hint: review the assumptions of the Thiem equation and assess the geology/geography of the area, carefully considering the role of the river and the selection of the model’s domain. *[0.25 pts.]*",
"_____no_output_____"
],
[
"**8.** Numerical groundwater flow models commonly are used today to address many of the limitations assumptions of Thiem Equation as applied to the flow system at Woburn (see [MODFLOW information](http://water.usgs.gov/ogw/modflow/MODFLOW.html)). Why would a numerical groundwater flow model, such as the USGS model, MODFLOW, which was in development during the trial, have been advantageous to the experts? *[0.25 pts]*",
"_____no_output_____"
],
[
"## Part III: Flow Net Challenge\n\n**9.** Let's say you are not allowed to access any data about well pumping rates. However, you are able to make measurements in piezometers around the well of interest, which generates a very nice contour plot that looks like this:\n\n\n\nAssume that *K* = 100 ft/day, and the saturated thickness of the aquifer away from the cone of depression is 80 ft. Drawing a flow net by hand, estimate the **volumetric** pumping rate of the well. (It may be difficult to do this precisely, so you also might want to describe what your intent is.) The person whose estimate is closest gets extra credit points! *[1 pt, + 1 pt E.C.]*",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
e78e7ed6d31bc8d221a1d17a3fbb029b7e4190c4 | 208,699 | ipynb | Jupyter Notebook | toybox/old/bvbq_test3.ipynb | DFNaiff/BVBQ | 48f0eb624483f67b748d791efc0c06ddfb6e0646 | [
"MIT"
] | null | null | null | toybox/old/bvbq_test3.ipynb | DFNaiff/BVBQ | 48f0eb624483f67b748d791efc0c06ddfb6e0646 | [
"MIT"
] | null | null | null | toybox/old/bvbq_test3.ipynb | DFNaiff/BVBQ | 48f0eb624483f67b748d791efc0c06ddfb6e0646 | [
"MIT"
] | null | null | null | 124.447823 | 87,728 | 0.819855 | [
[
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"import sys\nsys.path.append('..')\n\nimport collections\nimport functools\nimport abc\nimport random\n\nimport numpy as np\nimport jax\nimport jax.numpy as jnp\nimport jax.experimental.optimizers\nimport matplotlib.pyplot as plt\nfrom mpl_toolkits.mplot3d import Axes3D\nimport sobol_seq\n\nimport bvbq\n# import bvbq.gp\n# import bvbq.bvi\n# import bvbq.distributions",
"_____no_output_____"
],
[
"ndim = 2\ndef exponential_quartic(x):\n x1,x2 = x[...,0],x[...,1]\n res = -0.1*x1**4 -0.5*(2*x2-x1**2)**2\n# return -0.5*((x1+1)**2 + (x2 - 1)**2)\n return res",
"_____no_output_____"
],
[
"def test_function(params):\n x = params['x']\n return jnp.sum(x**2)\n\nlearning_rate = 1e-1\noptwrapper = bvbq.optwrapper.JaxOptWrapper(\"adam\",learning_rate)\noptwrapper.init({'x':jnp.ones(2)},jax.value_and_grad(test_function))\nfor i in range(50):\n value,_ = optwrapper.step()\n# print(value)\nprint(optwrapper.params)",
"{'x': DeviceArray([-0.00481431, -0.00481431], dtype=float32)}\n"
],
[
"fig = plt.figure()\nax = fig.add_subplot(111,projection='3d')\nx = np.linspace(-1,1)\ny = np.linspace(-1,1)\nX,Y = np.meshgrid(x,y)\nZ = exponential_quartic(np.stack([X,Y],axis=-1))\nax.plot_surface(X,Y,Z,alpha=0.7)\n\nxys = (2*(sobol_seq.i4_sobol_generate(2,20)-0.5)) #[-1,1] cube\nzs = exponential_quartic(xys)\nax.scatter(xys[:,0],xys[:,1],zs,color='red')\nax.set_xlabel('x');\nax.set_ylabel('y');\n\ngp = bvbq.gp.SimpleGP(2,kind='smatern32',noise=0)\ngp.set_data(xys,zs,empirical_params=True)\ngp.optimize_params()\nZreg = gp.predict(np.stack([X,Y],axis=-1))[0]\nax.plot_surface(X,Y,Zreg,alpha=0.7,color='green')",
"If tensor has more than 2 dimensions, only diagonal of covariance is returned\n"
],
[
"x = np.random.rand(5,2)\n\ndistrib = bvbq.distributions.DiagonalNormalDistribution(-jnp.ones(2),0.1*jnp.ones(2))\nprint(distrib.sample(100).mean(axis=0))\nprint(distrib.logprob(x))\nprint(distrib.sample(101).mean(axis=0))",
"[-0.99044454 -1.0060383 ]\n[-301.80716 -158.27931 -294.80444 -237.84756 -310.4032 ]\n[-0.9890031 -1.015915 ]\n"
],
[
"distrib.sample(1000)",
"_____no_output_____"
],
[
"mixdistrib = distrib.make_mixture()\nmixdistrib.add_component(jnp.ones(2),0.1*jnp.ones(2),0.1)\nmixdistrib.add_component(2*jnp.ones(2),0.1*jnp.ones(2),0.1)\nprint(mixdistrib.logprob(x))\nprint(mixdistrib.sample(100).mean())\nprint(mixdistrib.sample(101).mean())",
"[-2.702716 -7.656293 -3.2022219 -6.9514666 -3.0097933]\n-0.37293068\n-0.6136273\n"
],
[
"x = mixdistrib.sample(1000)\nplt.scatter(x[:,0],x[:,1])",
"_____no_output_____"
],
[
"@jax.jit\ndef logbound(logx,logdelta):\n clipx = jnp.clip(logx,logdelta,None)\n boundx = clipx + jnp.log(jnp.exp(logx-clipx) + \\\n jnp.exp(logdelta-clipx))\n return boundx",
"_____no_output_____"
],
[
"bvbq.bvi",
"_____no_output_____"
],
[
"def logprob_t(x,nu):\n normalizer = jax.scipy.special.gammaln((nu+1)/2) \\\n -jax.scipy.special.gammaln(nu/2) \\\n -0.5*jnp.log(jnp.pi*nu)\n main_term = -(nu+1)/2*jnp.log(1+x**2/nu)\n return main_term + normalizer\n\ndef objective_function_relbo_2(params,mixdistrib):\n mean,rawvar = params['mean'],params['rawvar']\n var = jax.nn.softplus(rawvar)\n newcomp = bvbq.distributions.DiagonalNormalDistribution(mean,var)\n logprob = functools.partial(logprob_t,nu=1)\n res = -bvbq.bvi.relbo(logprob,mixdistrib,newcomp,1000,reg=1e0,logdelta=-5)\n return res\n\ndef objective_function_gradboost_2(weight,distrib,mixdistrib):\n logprob = functools.partial(logprob_t,nu=1000)\n res = -bvbq.bvi.boosted_elbo(logprob,mixdistrib,distrib,weight,1000)\n return res\n\nparams = {'mean':jnp.zeros(1),'rawvar':0.1*jnp.zeros(1)}\ndistrib = bvbq.distributions.DiagonalNormalDistribution(5*jnp.ones(1),jnp.ones(1))\nmixdistrib = distrib.make_mixture()\nfunc_and_grad_relbo = jax.value_and_grad(objective_function_relbo_2,argnums=0)\nfunc_and_grad_relbo(params,mixdistrib)\nprint(func_and_grad_relbo(params,mixdistrib))\nweight = 0.5\nfunc_and_grad_gradboost = jax.value_and_grad(objective_function_gradboost_2)\nprint(func_and_grad_gradboost(0.5,distrib,mixdistrib))",
"(DeviceArray(-4.66147, dtype=float32), {'mean': DeviceArray([0.05045595], dtype=float32), 'rawvar': DeviceArray([-0.12207514], dtype=float32)})\n(DeviceArray(12.308316, dtype=float32), DeviceArray(0.00424576, dtype=float32))\n"
],
[
"alpha_boost = 1e0\nmaxiter_boost = 100\nalpha_relbo = 3*1e-1\nmaxiter_relbo = 100\nmaxiter = 5\noptwrapper = bvbq.optwrapper.JaxOptWrapper(\"adam\",alpha_relbo)\nfor k in range(maxiter):\n params = {'mean':jax.random.normal(mixdistrib.split_key(),(1,)),\n 'rawvar':3*jax.random.normal(mixdistrib.split_key(),(1,))}\n optwrapper.init(params,func_and_grad_relbo)\n for i in range(maxiter_relbo):\n value,grads = optwrapper.step(mixdistrib)\n params = optwrapper.params\n# print(value,params['mean'],jax.nn.softplus(params['rawvar']))\n mean,rawvar = params['mean'],params['rawvar']\n var = jax.nn.softplus(rawvar)\n distrib = bvbq.distributions.DiagonalNormalDistribution(mean,var)\n weight = 0.01\n print('--')\n print(mean,var)\n for i in range(maxiter_boost):\n res,gradweight = func_and_grad_gradboost(weight,distrib,mixdistrib)\n weight -= alpha_boost*(i/10+1)**(-0.55)*gradweight\n weight = jnp.clip(weight,1e-4,1-1e-4)\n if abs(gradweight) < 1e-2:\n break\n# print(weight,res,gradweight)\n mixdistrib.add_component(mean,var,weight)\n print(k,mean,var,weight)\n print('--')",
"--\n[3.8478475] [8.775109]\n0 [3.8478475] [8.775109] 1e-04\n--\n--\n[2.8661525] [15.713153]\n1 [2.8661525] [15.713153] 1e-04\n--\n--\n[2.5462508] [18.28128]\n"
],
[
"x = np.linspace(-10,10,201)\ny = logprob_t(x,1000)\ny2 = -0.5*x**2 - 0.5*np.log(2*np.pi)\nyvi = mixdistrib.logprob(x.reshape(-1,1)).flatten()\nplt.plot(x,np.exp(y),'b')\nplt.plot(x,np.exp(y2),'r')\nplt.plot(x,np.exp(yvi),'g')",
"_____no_output_____"
],
[
"mixdistrib.weights.sum()",
"_____no_output_____"
],
[
"mixdistrib.add_component(np.zeros(1),np.ones(1),0.5)",
"_____no_output_____"
],
[
"def exponential_quartic(x):\n x1,x2 = x[...,0],x[...,1]\n res = -0.1*x1**4 -0.5*(2*x2-x1**2)**2\n# return -0.5*((x1+1)**2 + (x2 - 1)**2)\n return res\n\ndef objective_function_relbo_2(params,mixdistrib):\n mean,rawvar = params['mean'],params['rawvar']\n var = jax.nn.softplus(rawvar)\n newcomp = bvbq.distributions.DiagonalNormalDistribution(mean,var)\n logprob = functools.partial(logprob_t,nu=1)\n res = -bvbq.bvi.relbo(logprob,mixdistrib,newcomp,1000,reg=1e0,logdelta=-5)\n return res\n\ndef objective_function_gradboost_2(weight,distrib,mixdistrib):\n# logprob = functools.partial(logprob_t,nu=1000)\n logprob = exponential_quartic\n res = -bvbq.bvi.boosted_elbo(logprob,mixdistrib,distrib,weight,1000)\n return res\n\nparams = {'mean':jnp.zeros(2),'rawvar':0.1*jnp.zeros(2)}\ndistrib = bvbq.distributions.DiagonalNormalDistribution(jnp.zeros(2),jnp.ones(2))\nmixdistrib = distrib.make_mixture()\nfunc_and_grad_relbo = jax.value_and_grad(objective_function_relbo_2,argnums=0)\nfunc_and_grad_relbo(params,mixdistrib)\nprint(func_and_grad_relbo(params,mixdistrib))\nweight = 0.5\nfunc_and_grad_gradboost = jax.value_and_grad(objective_function_gradboost_2)\nprint(func_and_grad_gradboost(0.5,distrib,mixdistrib))",
"(DeviceArray(-3.3161716, dtype=float32), {'mean': DeviceArray([-0.0060608 , -0.00559549], dtype=float32), 'rawvar': DeviceArray([-0.45201987, -0.45770144], dtype=float32)})\n(DeviceArray(0.835276, dtype=float32), DeviceArray(-0.02615952, dtype=float32))\n"
],
[
"alpha_boost = 1e0\nmaxiter_boost = 100\nalpha_relbo = 1e-3\nmaxiter_relbo = 100\nmaxiter = 10\noptwrapper = bvbq.optwrapper.JaxOptWrapper(\"adam\",alpha_relbo)\nfor k in range(maxiter):\n params = {'mean':jax.random.normal(mixdistrib.split_key(),(2,)),\n 'rawvar':3*jax.random.normal(mixdistrib.split_key(),(2,))}\n optwrapper.init(params,func_and_grad_relbo)\n for i in range(maxiter_relbo):\n value,grads = optwrapper.step(mixdistrib)\n params = optwrapper.params\n# print(value,params['mean'],jax.nn.softplus(params['rawvar']))\n mean,rawvar = params['mean'],params['rawvar']\n var = jax.nn.softplus(rawvar)\n# mean,var = jnp.zeros(1),jnp.ones(1)\n distrib = bvbq.distributions.DiagonalNormalDistribution(mean,var)\n weight = 1e-12\n print('--')\n print(mean,var)\n for i in range(maxiter_boost):\n res,gradweight = func_and_grad_gradboost(weight,distrib,mixdistrib)\n weight -= alpha_boost*(i/10+1)**(-0.55)*gradweight\n weight = jnp.clip(weight,1e-12,1-1e12)\n if abs(gradweight) < 1e-2:\n break\n print(weight,res,gradweight)\n mixdistrib.add_component(mean,var,weight)\n print(k,mean,var,weight)\n print('--')",
"--\n[ 1.0887313 -0.76624817] [0.00164976 0.3564104 ]\n-1000000000000.0 1.0079429 0.77146375\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\n0 [ 1.0887313 -0.76624817] [0.00164976 0.3564104 ] nan\n--\n--\n[nan nan] [nan nan]\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\n1 [nan nan] [nan nan] nan\n--\n--\n[nan nan] [nan nan]\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\n2 [nan nan] [nan nan] nan\n--\n--\n[nan nan] [nan nan]\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\n3 [nan nan] [nan nan] nan\n--\n--\n[nan nan] [nan nan]\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\n4 [nan nan] [nan nan] nan\n--\n--\n[nan nan] [nan nan]\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\n5 [nan nan] [nan nan] nan\n--\n--\n[nan nan] [nan nan]\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\nnan nan nan\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78e90a88a28b292f25bb918e6f86e650b54224d | 608,079 | ipynb | Jupyter Notebook | Probability/5. Simpson's Paradox.ipynb | febinsathar/goodreads | 9f1c72721849de8db53316204305b0b436033c33 | [
"MIT"
] | 8 | 2020-09-19T20:20:21.000Z | 2021-05-03T11:30:04.000Z | Probability/5. Simpson's Paradox.ipynb | febinsathar/goodreads | 9f1c72721849de8db53316204305b0b436033c33 | [
"MIT"
] | 1 | 2020-09-25T14:42:12.000Z | 2020-09-25T14:42:12.000Z | Probability/5. Simpson's Paradox.ipynb | febinsathar/goodreads | 9f1c72721849de8db53316204305b0b436033c33 | [
"MIT"
] | 1 | 2020-09-25T14:38:22.000Z | 2020-09-25T14:38:22.000Z | 1,194.654224 | 299,192 | 0.952962 | [
[
[
"<div style=\"width: 100%; overflow: hidden;\">\n <div style=\"width: 150px; float: left;\"> <img src=\"data/D4Sci_logo_ball.png\" alt=\"Data For Science, Inc\" align=\"left\" border=\"0\"> </div>\n <div style=\"float: left; margin-left: 10px;\"> \n <h1>Applied Probability Theory From Scratch</h1>\n <h1>Simpson's Paradox</h1>\n <p>Bruno Gonçalves<br/>\n <a href=\"http://www.data4sci.com/\">www.data4sci.com</a><br/>\n @bgoncalves, @data4sci</p></div>\n</div>",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\n\nimport matplotlib\nimport matplotlib.pyplot as plt\n\nimport sklearn\nfrom sklearn.linear_model import LinearRegression\n\nimport watermark\n\n%load_ext watermark\n%matplotlib inline",
"_____no_output_____"
],
[
"%watermark -i -n -v -m -g -iv",
"autopep8 1.5\nnumpy 1.18.1\njson 2.0.9\nwatermark 2.0.2\npandas 1.0.1\nmatplotlib 3.1.3\nsklearn 0.22.1\nFri Aug 21 2020 2020-08-21T11:50:16-04:00\n\nCPython 3.7.3\nIPython 6.2.1\n\ncompiler : Clang 4.0.1 (tags/RELEASE_401/final)\nsystem : Darwin\nrelease : 19.6.0\nmachine : x86_64\nprocessor : i386\nCPU cores : 8\ninterpreter: 64bit\nGit hash : 3029db88538474325720d5f3ece806f74c8330eb\n"
],
[
"plt.style.use('./d4sci.mplstyle')",
"_____no_output_____"
]
],
[
[
"## Load the iris dataset ",
"_____no_output_____"
]
],
[
[
"iris = pd.read_csv('data/iris.csv')",
"_____no_output_____"
],
[
"iris",
"_____no_output_____"
]
],
[
[
"Split the dataset across species for convenience",
"_____no_output_____"
]
],
[
[
"setosa = iris[['sepal_width', 'petal_width']][iris['species'] == 'setosa']\nversicolor = iris[['sepal_width', 'petal_width']][iris['species'] == 'versicolor']\nvirginica = iris[['sepal_width', 'petal_width']][iris['species'] == 'virginica']",
"_____no_output_____"
]
],
[
[
"## Perform the fits",
"_____no_output_____"
]
],
[
[
"lm_setosa = LinearRegression()\nlm_setosa.fit(setosa['sepal_width'].values.reshape(-1,1), setosa['petal_width'])\ny_setosa = lm_setosa.predict(setosa['sepal_width'].values.reshape(-1,1))\n\nlm_versicolor = LinearRegression()\nlm_versicolor.fit(versicolor['sepal_width'].values.reshape(-1,1), versicolor['petal_width'])\ny_versicolor = lm_versicolor.predict(versicolor['sepal_width'].values.reshape(-1,1))\n\nlm_virginica = LinearRegression()\nlm_virginica.fit(virginica['sepal_width'].values.reshape(-1,1), virginica['petal_width'])\ny_virginica = lm_virginica.predict(virginica['sepal_width'].values.reshape(-1,1))\n\nlm_full = LinearRegression()\nlm_full.fit(iris['sepal_width'].values.reshape(-1,1), iris['petal_width'])\ny_full = lm_full.predict(iris['sepal_width'].values.reshape(-1,1))",
"_____no_output_____"
]
],
[
[
"## Generate the plot",
"_____no_output_____"
]
],
[
[
"fig, axs = plt.subplots(ncols=2, sharey=True)\n\ncolors = plt.rcParams['axes.prop_cycle'].by_key()['color']\n\nsetosa.plot.scatter(x='sepal_width', y='petal_width', label='setosa', ax=axs[0], c=colors[0])\nversicolor.plot.scatter(x='sepal_width', y='petal_width', label='versicolor', ax=axs[0], c=colors[1])\nvirginica.plot.scatter(x='sepal_width', y='petal_width', label='virginica', ax=axs[0], c=colors[2])\nl4, = axs[0].plot(iris['sepal_width'].values.reshape(-1,1), y_full, '-', c=colors[3])\n\nsetosa.plot.scatter(x='sepal_width', y='petal_width', ax=axs[1], c=colors[0])\nversicolor.plot.scatter(x='sepal_width', y='petal_width', ax=axs[1], c=colors[1])\nvirginica.plot.scatter(x='sepal_width', y='petal_width', ax=axs[1], c=colors[2])\n\nl1, = axs[1].plot(setosa['sepal_width'].values.reshape(-1,1), y_setosa, '-', c=colors[0])\nl2, = axs[1].plot(versicolor['sepal_width'].values.reshape(-1,1), y_versicolor, '-', c=colors[1])\nl3, = axs[1].plot(virginica['sepal_width'].values.reshape(-1,1), y_virginica, '-', c=colors[2])\n\naxs[0].set_xlabel('Sepal Width')\naxs[1].set_xlabel('Sepal Width')\naxs[0].set_ylabel('Petal Width')\n\nfig.subplots_adjust(bottom=0.3, wspace=0.33)\n\naxs[0].legend(handles = [l1, l2, l3, l4] , labels=['Setosa', 'Versicolor', 'Virginica', 'Total'],\n loc='lower left', bbox_to_anchor=(0, -0.4), ncol=2, fancybox=True, shadow=False)",
"_____no_output_____"
]
],
[
[
"## Removing setosa",
"_____no_output_____"
]
],
[
[
"reduced = iris[iris['species'] != 'setosa'].copy()",
"_____no_output_____"
],
[
"lm_reduced = LinearRegression()\nlm_reduced.fit(reduced['sepal_width'].values.reshape(-1,1), reduced['petal_width'])\ny_reduced = lm_reduced.predict(reduced['sepal_width'].values.reshape(-1,1))",
"_____no_output_____"
],
[
"fig, axs = plt.subplots(ncols=1, sharey=True)\n\ncolors = plt.rcParams['axes.prop_cycle'].by_key()['color']\n\nversicolor.plot.scatter(x='sepal_width', y='petal_width', ax=axs, c=colors[1])\nvirginica.plot.scatter(x='sepal_width', y='petal_width', ax=axs, c=colors[2])\naxs.plot(reduced['sepal_width'].values.reshape(-1,1), y_reduced, '-', c=colors[3], label='reduced')\naxs.plot(versicolor['sepal_width'].values.reshape(-1,1), y_versicolor, '-', c=colors[1], label='versicolor')\naxs.plot(virginica['sepal_width'].values.reshape(-1,1), y_virginica, '-', c=colors[2], label='virginica')\n\naxs.set_xlabel('Sepal Width')\naxs.set_ylabel('Petal Width')\nplt.legend()",
"_____no_output_____"
]
],
[
[
"<div style=\"width: 100%; overflow: hidden;\">\n <img src=\"data/D4Sci_logo_full.png\" alt=\"Data For Science, Inc\" align=\"center\" border=\"0\" width=300px> \n</div>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
e78e9c298194960be708e2af10cc6dfdf2c4d89c | 13,077 | ipynb | Jupyter Notebook | methyl-valerate/figures/ignition-delays.ipynb | bryanwweber/ncm-2017 | 887aa7c26bd68abe5db9417c2cd5c92dafb389fc | [
"CC-BY-4.0"
] | null | null | null | methyl-valerate/figures/ignition-delays.ipynb | bryanwweber/ncm-2017 | 887aa7c26bd68abe5db9417c2cd5c92dafb389fc | [
"CC-BY-4.0"
] | null | null | null | methyl-valerate/figures/ignition-delays.ipynb | bryanwweber/ncm-2017 | 887aa7c26bd68abe5db9417c2cd5c92dafb389fc | [
"CC-BY-4.0"
] | null | null | null | 52.94332 | 451 | 0.607326 | [
[
[
"import matplotlib as mpl\nmpl.use(\"pgf\")\n# from palettable.tableau import Tableau_10\n# from cycler import cycler\nimport numpy as np\npgf_with_pdflatex = {\n \"pgf.texsystem\": \"pdflatex\",\n \"pgf.preamble\": [\n r\"\\usepackage[utf8x]{inputenc}\",\n r\"\\usepackage[T1]{fontenc}\",\n r\"\\usepackage{mathptmx}\",\n r\"\\usepackage{mathtools}\",\n# r\"\\usepackage{biblatex}\",\n ],\n \"text.usetex\": True,\n \"figure.figsize\": [6, 4],\n \"axes.labelsize\": 12,\n \"font.size\": 12,\n \"font.family\": \"serif\",\n \"legend.fontsize\": 10,\n \"xtick.labelsize\": 10,\n \"ytick.labelsize\": 10,\n \"lines.linewidth\": 1.5,\n \"lines.linestyle\": None,\n \"lines.markeredgewidth\": 0.15,\n# \"axes.prop_cycle\": cycler(color=Tableau_10.mpl_colors),\n \"xtick.major.size\": 5,\n \"xtick.major.width\": 1,\n \"xtick.minor.size\": 2.5,\n \"xtick.minor.width\": 1,\n \"ytick.major.size\": 5,\n \"ytick.major.width\": 1,\n \"ytick.minor.size\": 2.5,\n \"ytick.minor.width\": 1,\n}\nmpl.rcParams.update(pgf_with_pdflatex)\n\nimport gspread\nfrom oauth2client.service_account import ServiceAccountCredentials\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom matplotlib.legend_handler import HandlerLine2D\nfrom matplotlib.ticker import FormatStrFormatter, AutoMinorLocator\nfrom palettable.tableau import Tableau_10\nfrom cycler import cycler",
"_____no_output_____"
],
[
"markers = ['o', 's', 'd', '^', 'x', '+', 'v', '<', '>', '*']\n# assert len(markers) == len(Tableau_10.mpl_colors)\ndef mfunc(syms):\n while True:\n for s in syms:\n yield s",
"_____no_output_____"
],
[
"scope = ['https://spreadsheets.google.com/feeds']\ncredentials = ServiceAccountCredentials.from_json_keyfile_name('methyl-valerate-78135e2c2c7c.json',\n scope)\ngc = gspread.authorize(credentials)\nws = gc.open_by_key('1UFdGzEyx7PsKprPybi-lkbuWYS8BAS-cBm2IKSKY-UA').sheet1\nrecs = ws.get_all_records(empty2zero=True)\ndata = pd.DataFrame(recs)\n\nphi_200_15_bar = data[(data['Equivalence Ratio'] == 2.00) &\n (np.isclose(data['Compressed Pressure (bar)'], 15, rtol=1E-2))][::-1]\nphi_100_15_bar = data[(data['Equivalence Ratio'] == 1.00) &\n (np.isclose(data['Compressed Pressure (bar)'], 15, rtol=1E-2))][::-1]\nphi_100_30_bar = data[(data['Equivalence Ratio'] == 1.00) &\n (np.isclose(data['Compressed Pressure (bar)'], 30, rtol=1E-2))][::-1]\nphi_050_15_bar = data[(data['Equivalence Ratio'] == 0.50) &\n (np.isclose(data['Compressed Pressure (bar)'], 15, rtol=1E-2))][::-1]\nphi_050_30_bar = data[(data['Equivalence Ratio'] == 0.50) &\n (np.isclose(data['Compressed Pressure (bar)'], 30, rtol=1E-2))][::-1]\nphi_025_15_bar = data[(data['Equivalence Ratio'] == 0.25) &\n (np.isclose(data['Compressed Pressure (bar)'], 15, rtol=1E-2))][::-1]\nphi_025_30_bar = data[(data['Equivalence Ratio'] == 0.25) &\n (np.isclose(data['Compressed Pressure (bar)'], 30, rtol=1E-2))][::-1]",
"_____no_output_____"
],
[
"# phi_100_30_bar[['1000/Tc (1/K)', 'First Stage Delay (ms)']].loc[phi_100_30_bar['First Stage Delay (ms)'] != 0]",
"_____no_output_____"
],
[
"fig, (ax_15_bar, ax_30_bar) = plt.subplots(nrows=2)\nfig.tight_layout(h_pad=1.75)\nax_15_bar.set_yscale('log')\n\nplot_opts = {'markersize': 6, 'fmt': '', 'elinewidth': 1.5, 'capthick': 1.5}\n\nmark = mfunc(markers)\nax_15_bar.errorbar(phi_025_15_bar['1000/Tc (1/K)'], phi_025_15_bar['Ignition Delay (ms)'], yerr=np.array(phi_025_15_bar['Ignition Delay Error (ms)']), label=r'$\\phi=0.25$', marker=next(mark), **plot_opts)\nax_15_bar.errorbar(phi_025_15_bar['1000/Tc (1/K)'].iloc[[0, -1]], phi_025_15_bar['Ignition Delay (ms)'].iloc[[0, -1]], xerr=0.01*np.array(phi_025_15_bar['1000/Tc (1/K)'].iloc[[0, -1]]), label='_ignored', fmt='none', ecolor=Tableau_10.mpl_colors[0], elinewidth=plot_opts['elinewidth'], capthick=plot_opts['capthick'])\nax_15_bar.errorbar(phi_050_15_bar['1000/Tc (1/K)'], phi_050_15_bar['Ignition Delay (ms)'], yerr=np.array(phi_050_15_bar['Ignition Delay Error (ms)']), label=r'$\\phi=0.50$', marker=next(mark), **plot_opts)\nax_15_bar.errorbar(phi_050_15_bar['1000/Tc (1/K)'].iloc[[0, -1]], phi_050_15_bar['Ignition Delay (ms)'].iloc[[0, -1]], xerr=0.01*np.array(phi_050_15_bar['1000/Tc (1/K)'].iloc[[0, -1]]), label='_ignored', fmt='none', ecolor=Tableau_10.mpl_colors[1], elinewidth=plot_opts['elinewidth'], capthick=plot_opts['capthick'])\nax_15_bar.errorbar(phi_100_15_bar['1000/Tc (1/K)'], phi_100_15_bar['Ignition Delay (ms)'], yerr=np.array(phi_100_15_bar['Ignition Delay Error (ms)']), label=r'$\\phi=1.00$', marker=next(mark), **plot_opts)\nax_15_bar.errorbar(phi_100_15_bar['1000/Tc (1/K)'].iloc[[0, -1]], phi_100_15_bar['Ignition Delay (ms)'].iloc[[0, -1]], xerr=0.01*np.array(phi_100_15_bar['1000/Tc (1/K)'].iloc[[0, -1]]), label='_ignored', fmt='none', ecolor=Tableau_10.mpl_colors[2], elinewidth=plot_opts['elinewidth'], capthick=plot_opts['capthick'])\nax_15_bar.errorbar(phi_200_15_bar['1000/Tc (1/K)'], phi_200_15_bar['Ignition Delay (ms)'], yerr=np.array(phi_200_15_bar['Ignition Delay Error (ms)']), label=r'$\\phi=2.00$', marker=next(mark), **plot_opts)\nax_15_bar.errorbar(phi_200_15_bar['1000/Tc (1/K)'].iloc[[0, -1]], phi_200_15_bar['Ignition Delay (ms)'].iloc[[0, -1]], xerr=0.01*np.array(phi_200_15_bar['1000/Tc (1/K)'].iloc[[0, -1]]), label='_ignored', fmt='none', ecolor=Tableau_10.mpl_colors[3], elinewidth=plot_opts['elinewidth'], capthick=plot_opts['capthick'])\n(_, phi_200_tau1_caps, _) = ax_15_bar.errorbar(phi_200_15_bar['1000/Tc (1/K)'], phi_200_15_bar['First Stage Delay (ms)'], yerr=np.array(phi_200_15_bar['First Stage Error (ms)']), marker=markers[3], color=Tableau_10.mpl_colors[3], label='_ignored', markerfacecolor='none', markeredgewidth='1.0', **plot_opts)\n\nmark = mfunc(markers)\nax_30_bar.set_yscale('log')\nax_30_bar.errorbar(phi_025_30_bar['1000/Tc (1/K)'], phi_025_30_bar['Ignition Delay (ms)'], yerr=np.array(phi_025_30_bar['Ignition Delay Error (ms)']), label=r'$\\phi=0.25$', marker=next(mark), **plot_opts)\nax_30_bar.errorbar(phi_025_30_bar['1000/Tc (1/K)'].iloc[[0, -1]], phi_025_30_bar['Ignition Delay (ms)'].iloc[[0, -1]], xerr=0.01*np.array(phi_025_30_bar['1000/Tc (1/K)'].iloc[[0, -1]]), label='_ignored', fmt='none', ecolor=Tableau_10.mpl_colors[0], elinewidth=plot_opts['elinewidth'], capthick=plot_opts['capthick'])\nax_30_bar.errorbar(phi_050_30_bar['1000/Tc (1/K)'], phi_050_30_bar['Ignition Delay (ms)'], yerr=np.array(phi_050_30_bar['Ignition Delay Error (ms)']), label=r'$\\phi=0.50$', marker=next(mark), **plot_opts)\nax_30_bar.errorbar(phi_050_30_bar['1000/Tc (1/K)'].iloc[[0, -1]], phi_050_30_bar['Ignition Delay (ms)'].iloc[[0, -1]], xerr=0.01*np.array(phi_050_30_bar['1000/Tc (1/K)'].iloc[[0, -1]]), label='_ignored', fmt='none', ecolor=Tableau_10.mpl_colors[1], elinewidth=plot_opts['elinewidth'], capthick=plot_opts['capthick'])\nax_30_bar.errorbar(phi_100_30_bar['1000/Tc (1/K)'], phi_100_30_bar['Ignition Delay (ms)'], yerr=np.array(phi_100_30_bar['Ignition Delay Error (ms)']), label=r'$\\phi=1.00$', marker=next(mark), **plot_opts)\nax_30_bar.errorbar(phi_100_30_bar['1000/Tc (1/K)'].iloc[[0, -1]], phi_100_30_bar['Ignition Delay (ms)'].iloc[[0, -1]], xerr=0.01*np.array(phi_100_30_bar['1000/Tc (1/K)'].iloc[[0, -1]]), label='_ignored', fmt='none', ecolor=Tableau_10.mpl_colors[2], elinewidth=plot_opts['elinewidth'], capthick=plot_opts['capthick'])\n(_, phi_100_tau1_caps, _) = ax_30_bar.errorbar(phi_100_30_bar['1000/Tc (1/K)'].loc[phi_100_30_bar['First Stage Delay (ms)'] != 0], phi_100_30_bar['First Stage Delay (ms)'].loc[phi_100_30_bar['First Stage Delay (ms)'] != 0], yerr=np.array(phi_100_30_bar['First Stage Error (ms)'].loc[phi_100_30_bar['First Stage Delay (ms)'] != 0]), marker=markers[2], color=Tableau_10.mpl_colors[2], markerfacecolor='none', markeredgewidth='1.0', **plot_opts)\n\n# Set y limits\nax_15_bar.set_ylim(2, 200)\nax_30_bar.set_ylim(3, 50)\n\n# Set the formatting of the y tick labels\nax_15_bar.yaxis.set_major_formatter(FormatStrFormatter('%d'))\nax_30_bar.yaxis.set_major_formatter(FormatStrFormatter('%d'))\nax_15_bar.xaxis.set_minor_locator(AutoMinorLocator(4))\nax_30_bar.xaxis.set_minor_locator(AutoMinorLocator(4))\n\n# Set x limits\nax_15_bar.set_xlim(0.94, 1.55)\nax_30_bar.set_xlim(ax_15_bar.get_xlim())\n\n# Create the legend, removing the error bars\nhandles, labels = ax_15_bar.get_legend_handles_labels()\nhandles = [h[0] for h in handles]\nax_15_bar.legend(handles, labels, loc='lower left', bbox_to_anchor=(0., 1.2, 1., 0.102), numpoints=1, frameon=True, handletextpad=0.0, ncol=4, mode=\"expand\", borderaxespad=0.0)\n\n# Make the error bar caps thicker\nfor c in (phi_200_tau1_caps + phi_100_tau1_caps):\n c.set_markeredgewidth(1.5)\n\n# Set the axis labels\nfig.text(0.0, 0.5, 'Ignition Delay, ms', verticalalignment='center', rotation='vertical')\nfig.text(0.5, 0.0, '$1000/T_C$, 1/K', horizontalalignment='center')\n\n# Set the a) b) figure labels\nax_15_bar.text(0.75, 0.10, r'a) $P_C = 15\\ \\text{bar}$', transform=ax_15_bar.transAxes)\nax_30_bar.text(0.75, 0.10, r'b) $P_C = 30\\ \\text{bar}$', transform=ax_30_bar.transAxes)\n\n# Create the temperature axes on the top\ndef convert_inv_temp(temps):\n \"\"\"Convert a list of temperatures to inverse temperature\"\"\"\n return [1000.0/temp for temp in temps]\n\nax_15_temp = ax_15_bar.twiny()\nax_30_temp = ax_30_bar.twiny()\n\n# Set the major tick marks in the temperature scale and convert to inverse scale\nmajor_temps = np.arange(1100, 600, -100)\nmajor_ticks = convert_inv_temp(major_temps)\n\n# Set the interval for the minor ticks and compute the minor ticks\nminor_interval = 20\nminor_ticks = []\nfor maj in major_temps:\n minor_ticks.extend(convert_inv_temp([maj - i*minor_interval for i in range(5)]))\n \n# Set the ticks on the axis. Note that the limit setting must be present and must be after setting the ticks\n# so that the scale is correct\nax_15_temp.set_xticks(major_ticks)\nax_15_temp.set_xticks(minor_ticks, minor=True)\nax_15_temp.set_xticklabels(['{:d} K'.format(temp) for temp in major_temps])\nax_15_temp.set_xlim(ax_15_bar.get_xlim())\nax_30_temp.set_xticks(major_ticks)\nax_30_temp.set_xticks(minor_ticks, minor=True)\nax_30_temp.set_xticklabels(['{:d} K'.format(temp) for temp in major_temps])\nax_30_temp.set_xlim(ax_15_bar.get_xlim());",
"_____no_output_____"
],
[
"fig.savefig('ignition-delays.pgf', bbox_inches='tight')",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78ea460dbaf58a1e51dc77eaabb4835c07f43fc | 809 | ipynb | Jupyter Notebook | Untitled Folder/reddit.ipynb | kabir12345/DASM | 86f777c47a83f400c2f99e9f71dcc3a40b9d3143 | [
"Apache-2.0"
] | 1 | 2021-06-07T02:55:01.000Z | 2021-06-07T02:55:01.000Z | Untitled Folder/reddit.ipynb | kabir12345/DASM | 86f777c47a83f400c2f99e9f71dcc3a40b9d3143 | [
"Apache-2.0"
] | null | null | null | Untitled Folder/reddit.ipynb | kabir12345/DASM | 86f777c47a83f400c2f99e9f71dcc3a40b9d3143 | [
"Apache-2.0"
] | 1 | 2021-03-22T05:49:30.000Z | 2021-03-22T05:49:30.000Z | 18.386364 | 116 | 0.546354 | [
[
[
"import praw",
"_____no_output_____"
],
[
"reddit = praw.Reddit(client_id='my_client_id', client_secret='my_client_secret', user_agent='my_user_agent')\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
e78ea470ea07b1a451b9e8c0f4d7622f63ad6fa7 | 50,856 | ipynb | Jupyter Notebook | vanilla CNN - FullDataset.ipynb | subha231/cancer | 8109489a283480907736faaa3d4633acd0996e12 | [
"Apache-2.0"
] | 64 | 2018-03-12T10:21:40.000Z | 2022-02-23T08:39:25.000Z | vanilla CNN - FullDataset.ipynb | subha231/cancer | 8109489a283480907736faaa3d4633acd0996e12 | [
"Apache-2.0"
] | 5 | 2018-04-13T11:14:47.000Z | 2020-12-10T12:33:46.000Z | vanilla CNN - FullDataset.ipynb | subha231/cancer | 8109489a283480907736faaa3d4633acd0996e12 | [
"Apache-2.0"
] | 29 | 2018-04-14T05:13:11.000Z | 2021-03-09T05:59:48.000Z | 50.104433 | 270 | 0.550613 | [
[
[
"# With rgb images",
"_____no_output_____"
],
[
"### Load data",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nfrom glob import glob\nfrom tqdm import tqdm\nfrom sklearn.utils import shuffle\n\ndf = pd.read_csv('sample/Data_Entry_2017.csv')\n\ndiseases = ['Cardiomegaly','Emphysema','Effusion','Hernia','Nodule','Pneumothorax','Atelectasis','Pleural_Thickening','Mass','Edema','Consolidation','Infiltration','Fibrosis','Pneumonia']\n#Number diseases\nfor disease in diseases :\n df[disease] = df['Finding Labels'].apply(lambda x: 1 if disease in x else 0)\n\n# #test to perfect\n# df = df.drop(df[df['Emphysema']==0][:-127].index.values)\n \n#remove Y after age\ndf['Age']=df['Patient Age'].apply(lambda x: x[:-1]).astype(int)\ndf['Age Type']=df['Patient Age'].apply(lambda x: x[-1:])\ndf.loc[df['Age Type']=='M',['Age']] = df[df['Age Type']=='M']['Age'].apply(lambda x: round(x/12.)).astype(int)\ndf.loc[df['Age Type']=='D',['Age']] = df[df['Age Type']=='D']['Age'].apply(lambda x: round(x/365.)).astype(int)\n# remove outliers\ndf = df.drop(df['Age'].sort_values(ascending=False).head(16).index)\ndf['Age'] = df['Age']/df['Age'].max()\n\n#one hot data\n# df = df.drop(df.index[4242])\ndf = df.join(pd.get_dummies(df['Patient Gender']))\ndf = df.join(pd.get_dummies(df['View Position']))\n\n#random samples\ndf = shuffle(df)\n\n#get other data\ndata = df[['Age', 'F', 'M', 'AP', 'PA']]\ndata = np.array(data)\n\nlabels = df[diseases].as_matrix()\nfiles_list = ('sample/images/' + df['Image Index']).tolist()\n\n# #test to perfect\n# labelB = df['Emphysema'].tolist()\n\nlabelB = (df[diseases].sum(axis=1)>0).tolist()\nlabelB = np.array(labelB, dtype=int)",
"_____no_output_____"
],
[
"from keras.preprocessing import image \nfrom tqdm import tqdm\n\ndef path_to_tensor(img_path, shape):\n # loads RGB image as PIL.Image.Image type\n img = image.load_img(img_path, target_size=shape)\n # convert PIL.Image.Image type to 3D tensor with shape (224, 224, 3)\n x = image.img_to_array(img)/255\n # convert 3D tensor to 4D tensor with shape (1, 224, 224, 3) and return 4D tensor\n return np.expand_dims(x, axis=0)\n\ndef paths_to_tensor(img_paths, shape):\n list_of_tensors = [path_to_tensor(img_path, shape) for img_path in tqdm(img_paths)]\n return np.vstack(list_of_tensors)\n\ntrain_labels = labelB[:89600][:, np.newaxis]\nvalid_labels = labelB[89600:100800][:, np.newaxis]\ntest_labels = labelB[100800:][:, np.newaxis]\n\ntrain_data = data[:89600]\nvalid_data = data[89600:100800]\ntest_data = data[100800:]\n\nimg_shape = (64, 64)\ntrain_tensors = paths_to_tensor(files_list[:89600], shape = img_shape)\nvalid_tensors = paths_to_tensor(files_list[89600:100800], shape = img_shape)\ntest_tensors = paths_to_tensor(files_list[100800:], shape = img_shape)",
"Using TensorFlow backend.\n/home/aind2/anaconda3/lib/python3.6/importlib/_bootstrap.py:219: RuntimeWarning: compiletime version 3.5 of module 'tensorflow.python.framework.fast_tensor_util' does not match runtime version 3.6\n return f(*args, **kwds)\n100%|██████████| 89600/89600 [20:26<00:00, 73.03it/s]\n100%|██████████| 11200/11200 [02:33<00:00, 73.10it/s]\n100%|██████████| 11319/11319 [02:38<00:00, 71.35it/s]\n"
]
],
[
[
"### CNN model",
"_____no_output_____"
]
],
[
[
"import time\n\nfrom keras.layers import Conv2D, MaxPooling2D, GlobalAveragePooling2D, Dropout, Flatten, Dense\nfrom keras.models import Sequential\nfrom keras.layers.normalization import BatchNormalization\nfrom keras import regularizers, initializers, optimizers\n\nmodel = Sequential()\n\nmodel.add(Conv2D(filters=16, \n kernel_size=7,\n padding='same', \n activation='relu', \n input_shape=train_tensors.shape[1:]))\nmodel.add(MaxPooling2D(pool_size=2))\n\nmodel.add(Conv2D(filters=32, \n kernel_size=5,\n padding='same', \n activation='relu'))\nmodel.add(MaxPooling2D(pool_size=2))\n\nmodel.add(Conv2D(filters=64, \n kernel_size=5,\n padding='same', \n activation='relu'))\nmodel.add(MaxPooling2D(pool_size=2))\n\nmodel.add(Conv2D(filters=128, \n kernel_size=5,\n strides=2,\n padding='same', \n activation='relu'))\nmodel.add(MaxPooling2D(pool_size=2))\n\nmodel.add(Flatten())\nmodel.add(Dense(100, activation='relu'))\nmodel.add(Dense(1, activation='sigmoid'))\n\nmodel.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nconv2d_13 (Conv2D) (None, 64, 64, 16) 2368 \n_________________________________________________________________\nmax_pooling2d_13 (MaxPooling (None, 32, 32, 16) 0 \n_________________________________________________________________\nconv2d_14 (Conv2D) (None, 32, 32, 32) 12832 \n_________________________________________________________________\nmax_pooling2d_14 (MaxPooling (None, 16, 16, 32) 0 \n_________________________________________________________________\nconv2d_15 (Conv2D) (None, 16, 16, 64) 51264 \n_________________________________________________________________\nmax_pooling2d_15 (MaxPooling (None, 8, 8, 64) 0 \n_________________________________________________________________\nconv2d_16 (Conv2D) (None, 4, 4, 128) 204928 \n_________________________________________________________________\nmax_pooling2d_16 (MaxPooling (None, 2, 2, 128) 0 \n_________________________________________________________________\nflatten_3 (Flatten) (None, 512) 0 \n_________________________________________________________________\ndense_5 (Dense) (None, 100) 51300 \n_________________________________________________________________\ndense_6 (Dense) (None, 1) 101 \n=================================================================\nTotal params: 322,793\nTrainable params: 322,793\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"from keras import backend as K\n\ndef binary_accuracy(y_true, y_pred):\n return K.mean(K.equal(y_true, K.round(y_pred)))\n\ndef precision_threshold(threshold = 0.5):\n def precision(y_true, y_pred):\n threshold_value = threshold\n y_pred = K.cast(K.greater(K.clip(y_pred, 0, 1), threshold_value), K.floatx())\n true_positives = K.round(K.sum(K.clip(y_true * y_pred, 0, 1)))\n predicted_positives = K.sum(y_pred)\n precision_ratio = true_positives / (predicted_positives + K.epsilon())\n return precision_ratio\n return precision\n\ndef recall_threshold(threshold = 0.5):\n def recall(y_true, y_pred):\n threshold_value = threshold\n y_pred = K.cast(K.greater(K.clip(y_pred, 0, 1), threshold_value), K.floatx())\n true_positives = K.round(K.sum(K.clip(y_true * y_pred, 0, 1)))\n possible_positives = K.sum(K.clip(y_true, 0, 1))\n recall_ratio = true_positives / (possible_positives + K.epsilon())\n return recall_ratio\n return recall\n\ndef fbeta_score_threshold(beta = 1, threshold = 0.5):\n def fbeta_score(y_true, y_pred):\n threshold_value = threshold\n beta_value = beta\n p = precision_threshold(threshold_value)(y_true, y_pred)\n r = recall_threshold(threshold_value)(y_true, y_pred)\n bb = beta_value ** 2\n fbeta_score = (1 + bb) * (p * r) / (bb * p + r + K.epsilon())\n return fbeta_score\n return fbeta_score",
"_____no_output_____"
],
[
"model.compile(optimizer='sgd', loss='binary_crossentropy', \n metrics=[precision_threshold(threshold = 0.5), \n recall_threshold(threshold = 0.5), \n fbeta_score_threshold(beta=0.5, threshold = 0.5),\n 'accuracy'])",
"_____no_output_____"
],
[
"from keras.callbacks import ModelCheckpoint, CSVLogger, EarlyStopping\nimport numpy as np\n\nepochs = 20\nbatch_size = 32\n\nearlystop = EarlyStopping(monitor='val_loss', min_delta=0, patience=3, verbose=1, mode='auto')\nlog = CSVLogger('saved_models/log_bCNN_rgb.csv')\ncheckpointer = ModelCheckpoint(filepath='saved_models/bCNN.best.from_scratch.hdf5', \n verbose=1, save_best_only=True)\n\nstart = time.time()\n\nmodel.fit(train_tensors, train_labels, \n validation_data=(valid_tensors, valid_labels),\n epochs=epochs, batch_size=batch_size, callbacks=[checkpointer, log, earlystop], verbose=1)\n\n# Show total training time\nprint(\"training time: %.2f minutes\"%((time.time()-start)/60))",
"Train on 89600 samples, validate on 11200 samples\nEpoch 1/20\n89536/89600 [============================>.] - ETA: 0s - loss: 0.6561 - precision: 0.5974 - recall: 0.4632 - fbeta_score: 0.5386 - acc: 0.6178Epoch 00000: val_loss improved from inf to 0.65672, saving model to saved_models/bCNN.best.from_scratch.hdf5\n89600/89600 [==============================] - 52s - loss: 0.6561 - precision: 0.5974 - recall: 0.4632 - fbeta_score: 0.5387 - acc: 0.6178 - val_loss: 0.6567 - val_precision: 0.5574 - val_recall: 0.7224 - val_fbeta_score: 0.5805 - val_acc: 0.6148\nEpoch 2/20\n89504/89600 [============================>.] - ETA: 0s - loss: 0.6416 - precision: 0.6351 - recall: 0.5353 - fbeta_score: 0.6006 - acc: 0.6412Epoch 00001: val_loss improved from 0.65672 to 0.63831, saving model to saved_models/bCNN.best.from_scratch.hdf5\n89600/89600 [==============================] - 51s - loss: 0.6416 - precision: 0.6353 - recall: 0.5353 - fbeta_score: 0.6007 - acc: 0.6412 - val_loss: 0.6383 - val_precision: 0.6061 - val_recall: 0.6122 - val_fbeta_score: 0.6025 - val_acc: 0.6460\nEpoch 3/20\n89504/89600 [============================>.] - ETA: 0s - loss: 0.6354 - precision: 0.6417 - recall: 0.5549 - fbeta_score: 0.6115 - acc: 0.6487Epoch 00002: val_loss improved from 0.63831 to 0.63043, saving model to saved_models/bCNN.best.from_scratch.hdf5\n89600/89600 [==============================] - 51s - loss: 0.6354 - precision: 0.6417 - recall: 0.5549 - fbeta_score: 0.6115 - acc: 0.6487 - val_loss: 0.6304 - val_precision: 0.6347 - val_recall: 0.5533 - val_fbeta_score: 0.6106 - val_acc: 0.6556\nEpoch 4/20\n89504/89600 [============================>.] - ETA: 0s - loss: 0.6303 - precision: 0.6453 - recall: 0.5640 - fbeta_score: 0.6173 - acc: 0.6537Epoch 00003: val_loss improved from 0.63043 to 0.63033, saving model to saved_models/bCNN.best.from_scratch.hdf5\n89600/89600 [==============================] - 51s - loss: 0.6303 - precision: 0.6455 - recall: 0.5640 - fbeta_score: 0.6174 - acc: 0.6538 - val_loss: 0.6303 - val_precision: 0.6627 - val_recall: 0.4711 - val_fbeta_score: 0.6032 - val_acc: 0.6525\nEpoch 5/20\n89504/89600 [============================>.] - ETA: 0s - loss: 0.6267 - precision: 0.6506 - recall: 0.5799 - fbeta_score: 0.6259 - acc: 0.6603Epoch 00004: val_loss improved from 0.63033 to 0.62570, saving model to saved_models/bCNN.best.from_scratch.hdf5\n89600/89600 [==============================] - 52s - loss: 0.6267 - precision: 0.6506 - recall: 0.5800 - fbeta_score: 0.6260 - acc: 0.6603 - val_loss: 0.6257 - val_precision: 0.6350 - val_recall: 0.5960 - val_fbeta_score: 0.6213 - val_acc: 0.6642\nEpoch 6/20\n89504/89600 [============================>.] - ETA: 0s - loss: 0.6232 - precision: 0.6519 - recall: 0.5841 - fbeta_score: 0.6287 - acc: 0.6627Epoch 00005: val_loss improved from 0.62570 to 0.62252, saving model to saved_models/bCNN.best.from_scratch.hdf5\n89600/89600 [==============================] - 52s - loss: 0.6232 - precision: 0.6519 - recall: 0.5839 - fbeta_score: 0.6287 - acc: 0.6627 - val_loss: 0.6225 - val_precision: 0.6655 - val_recall: 0.5105 - val_fbeta_score: 0.6197 - val_acc: 0.6628\nEpoch 7/20\n89504/89600 [============================>.] - ETA: 0s - loss: 0.6204 - precision: 0.6558 - recall: 0.5892 - fbeta_score: 0.6332 - acc: 0.6668Epoch 00006: val_loss did not improve\n89600/89600 [==============================] - 51s - loss: 0.6204 - precision: 0.6558 - recall: 0.5893 - fbeta_score: 0.6333 - acc: 0.6669 - val_loss: 0.6237 - val_precision: 0.6176 - val_recall: 0.6639 - val_fbeta_score: 0.6217 - val_acc: 0.6624\nEpoch 8/20\n89504/89600 [============================>.] - ETA: 0s - loss: 0.6179 - precision: 0.6574 - recall: 0.5978 - fbeta_score: 0.6364 - acc: 0.6693Epoch 00007: val_loss did not improve\n89600/89600 [==============================] - 51s - loss: 0.6180 - precision: 0.6574 - recall: 0.5979 - fbeta_score: 0.6364 - acc: 0.6692 - val_loss: 0.6243 - val_precision: 0.6157 - val_recall: 0.6677 - val_fbeta_score: 0.6206 - val_acc: 0.6618\nEpoch 9/20\n89504/89600 [============================>.] - ETA: 0s - loss: 0.6151 - precision: 0.6565 - recall: 0.5990 - fbeta_score: 0.6364 - acc: 0.6702Epoch 00008: val_loss improved from 0.62252 to 0.61709, saving model to saved_models/bCNN.best.from_scratch.hdf5\n89600/89600 [==============================] - 51s - loss: 0.6152 - precision: 0.6564 - recall: 0.5988 - fbeta_score: 0.6362 - acc: 0.6701 - val_loss: 0.6171 - val_precision: 0.6748 - val_recall: 0.5154 - val_fbeta_score: 0.6274 - val_acc: 0.6702\nEpoch 10/20\n89504/89600 [============================>.] - ETA: 0s - loss: 0.6125 - precision: 0.6618 - recall: 0.6044 - fbeta_score: 0.6417 - acc: 0.6739Epoch 00009: val_loss improved from 0.61709 to 0.61645, saving model to saved_models/bCNN.best.from_scratch.hdf5\n89600/89600 [==============================] - 51s - loss: 0.6125 - precision: 0.6618 - recall: 0.6043 - fbeta_score: 0.6416 - acc: 0.6739 - val_loss: 0.6164 - val_precision: 0.6585 - val_recall: 0.5817 - val_fbeta_score: 0.6353 - val_acc: 0.6746\nEpoch 11/20\n89504/89600 [============================>.] - ETA: 0s - loss: 0.6102 - precision: 0.6636 - recall: 0.6092 - fbeta_score: 0.6443 - acc: 0.6762Epoch 00010: val_loss improved from 0.61645 to 0.61598, saving model to saved_models/bCNN.best.from_scratch.hdf5\n89600/89600 [==============================] - 52s - loss: 0.6102 - precision: 0.6636 - recall: 0.6092 - fbeta_score: 0.6443 - acc: 0.6762 - val_loss: 0.6160 - val_precision: 0.6263 - val_recall: 0.6632 - val_fbeta_score: 0.6286 - val_acc: 0.6686\nEpoch 12/20\n89504/89600 [============================>.] - ETA: 0s - loss: 0.6075 - precision: 0.6644 - recall: 0.6140 - fbeta_score: 0.6461 - acc: 0.6782Epoch 00011: val_loss improved from 0.61598 to 0.61276, saving model to saved_models/bCNN.best.from_scratch.hdf5\n89600/89600 [==============================] - 53s - loss: 0.6074 - precision: 0.6644 - recall: 0.6140 - fbeta_score: 0.6461 - acc: 0.6782 - val_loss: 0.6128 - val_precision: 0.6400 - val_recall: 0.6476 - val_fbeta_score: 0.6363 - val_acc: 0.6753\nEpoch 13/20\n89504/89600 [============================>.] - ETA: 0s - loss: 0.6052 - precision: 0.6678 - recall: 0.6175 - fbeta_score: 0.6498 - acc: 0.6812Epoch 00012: val_loss improved from 0.61276 to 0.60942, saving model to saved_models/bCNN.best.from_scratch.hdf5\n89600/89600 [==============================] - 53s - loss: 0.6052 - precision: 0.6678 - recall: 0.6174 - fbeta_score: 0.6498 - acc: 0.6812 - val_loss: 0.6094 - val_precision: 0.6545 - val_recall: 0.5914 - val_fbeta_score: 0.6348 - val_acc: 0.6748\nEpoch 14/20\n89504/89600 [============================>.] - ETA: 0s - loss: 0.6021 - precision: 0.6698 - recall: 0.6210 - fbeta_score: 0.6520 - acc: 0.6836Epoch 00013: val_loss did not improve\n89600/89600 [==============================] - 53s - loss: 0.6022 - precision: 0.6699 - recall: 0.6210 - fbeta_score: 0.6520 - acc: 0.6836 - val_loss: 0.6187 - val_precision: 0.6155 - val_recall: 0.6913 - val_fbeta_score: 0.6249 - val_acc: 0.6645\nEpoch 15/20\n89504/89600 [============================>.] - ETA: 0s - loss: 0.5997 - precision: 0.6727 - recall: 0.6242 - fbeta_score: 0.6547 - acc: 0.6855Epoch 00014: val_loss did not improve\n89600/89600 [==============================] - 53s - loss: 0.5999 - precision: 0.6724 - recall: 0.6239 - fbeta_score: 0.6544 - acc: 0.6852 - val_loss: 0.6147 - val_precision: 0.6373 - val_recall: 0.6321 - val_fbeta_score: 0.6307 - val_acc: 0.6702\nEpoch 16/20\n89504/89600 [============================>.] - ETA: 0s - loss: 0.5969 - precision: 0.6754 - recall: 0.6313 - fbeta_score: 0.6586 - acc: 0.6883Epoch 00015: val_loss did not improve\n89600/89600 [==============================] - 53s - loss: 0.5970 - precision: 0.6754 - recall: 0.6312 - fbeta_score: 0.6586 - acc: 0.6883 - val_loss: 0.6095 - val_precision: 0.6442 - val_recall: 0.6437 - val_fbeta_score: 0.6389 - val_acc: 0.6779\nEpoch 17/20\n89504/89600 [============================>.] - ETA: 0s - loss: 0.5937 - precision: 0.6794 - recall: 0.6334 - fbeta_score: 0.6622 - acc: 0.6916Epoch 00016: val_loss did not improve\n89600/89600 [==============================] - 53s - loss: 0.5937 - precision: 0.6793 - recall: 0.6334 - fbeta_score: 0.6622 - acc: 0.6916 - val_loss: 0.6133 - val_precision: 0.6844 - val_recall: 0.5338 - val_fbeta_score: 0.6403 - val_acc: 0.6790\nEpoch 00016: early stopping\ntraining time: 14.89 minutes\n"
]
],
[
[
"### Metric",
"_____no_output_____"
]
],
[
[
"model.load_weights('saved_models/bCNN.best.from_scratch.hdf5')\nprediction = model.predict(test_tensors)",
"_____no_output_____"
],
[
"threshold = 0.5\nbeta = 0.5\n\npre = K.eval(precision_threshold(threshold = threshold)(K.variable(value=test_labels),\n K.variable(value=prediction)))\nrec = K.eval(recall_threshold(threshold = threshold)(K.variable(value=test_labels),\n K.variable(value=prediction)))\nfsc = K.eval(fbeta_score_threshold(beta = beta, threshold = threshold)(K.variable(value=test_labels),\n K.variable(value=prediction)))\n\nprint (\"Precision: %f %%\\nRecall: %f %%\\nFscore: %f %%\"% (pre, rec, fsc))",
"Precision: 0.672176 %\nRecall: 0.594230 %\nFscore: 0.654993 %\n"
],
[
"K.eval(binary_accuracy(K.variable(value=test_labels),\n K.variable(value=prediction)))",
"_____no_output_____"
],
[
"prediction[:30]",
"_____no_output_____"
],
[
"threshold = 0.4\nbeta = 0.5\n\npre = K.eval(precision_threshold(threshold = threshold)(K.variable(value=test_labels),\n K.variable(value=prediction)))\nrec = K.eval(recall_threshold(threshold = threshold)(K.variable(value=test_labels),\n K.variable(value=prediction)))\nfsc = K.eval(fbeta_score_threshold(beta = beta, threshold = threshold)(K.variable(value=test_labels),\n K.variable(value=prediction)))\n\nprint (\"Precision: %f %%\\nRecall: %f %%\\nFscore: %f %%\"% (pre, rec, fsc))",
"Precision: 0.617976 %\nRecall: 0.735481 %\nFscore: 0.638374 %\n"
],
[
"threshold = 0.6\nbeta = 0.5\n\npre = K.eval(precision_threshold(threshold = threshold)(K.variable(value=test_labels),\n K.variable(value=prediction)))\nrec = K.eval(recall_threshold(threshold = threshold)(K.variable(value=test_labels),\n K.variable(value=prediction)))\nfsc = K.eval(fbeta_score_threshold(beta = beta, threshold = threshold)(K.variable(value=test_labels),\n K.variable(value=prediction)))\n\nprint (\"Precision: %f %%\\nRecall: %f %%\\nFscore: %f %%\"% (pre, rec, fsc))",
"Precision: 0.712731 %\nRecall: 0.404833 %\nFscore: 0.618630 %\n"
]
],
[
[
"# With gray images",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nfrom glob import glob\nfrom tqdm import tqdm\nfrom sklearn.utils import shuffle\n\ndf = pd.read_csv('sample/Data_Entry_2017.csv')\n\ndiseases = ['Cardiomegaly','Emphysema','Effusion','Hernia','Nodule','Pneumothorax','Atelectasis','Pleural_Thickening','Mass','Edema','Consolidation','Infiltration','Fibrosis','Pneumonia']\n#Number diseases\nfor disease in diseases :\n df[disease] = df['Finding Labels'].apply(lambda x: 1 if disease in x else 0)\n\n# #test to perfect\n# df = df.drop(df[df['Emphysema']==0][:-127].index.values)\n \n#remove Y after age\ndf['Age']=df['Patient Age'].apply(lambda x: x[:-1]).astype(int)\ndf['Age Type']=df['Patient Age'].apply(lambda x: x[-1:])\ndf.loc[df['Age Type']=='M',['Age']] = df[df['Age Type']=='M']['Age'].apply(lambda x: round(x/12.)).astype(int)\ndf.loc[df['Age Type']=='D',['Age']] = df[df['Age Type']=='D']['Age'].apply(lambda x: round(x/365.)).astype(int)\n# remove outliers\ndf = df.drop(df['Age'].sort_values(ascending=False).head(16).index)\ndf['Age'] = df['Age']/df['Age'].max()\n\n#one hot data\n# df = df.drop(df.index[4242])\ndf = df.join(pd.get_dummies(df['Patient Gender']))\ndf = df.join(pd.get_dummies(df['View Position']))\n\n#random samples\ndf = shuffle(df)\n\n#get other data\ndata = df[['Age', 'F', 'M', 'AP', 'PA']]\ndata = np.array(data)\n\nlabels = df[diseases].as_matrix()\nfiles_list = ('sample/images/' + df['Image Index']).tolist()\n\n# #test to perfect\n# labelB = df['Emphysema'].tolist()\n\nlabelB = (df[diseases].sum(axis=1)>0).tolist()\nlabelB = np.array(labelB, dtype=int)",
"_____no_output_____"
],
[
"from keras.preprocessing import image \nfrom tqdm import tqdm\n\ndef path_to_tensor(img_path, shape):\n # loads RGB image as PIL.Image.Image type\n img = image.load_img(img_path, grayscale=True, target_size=shape)\n # convert PIL.Image.Image type to 3D tensor with shape (224, 224, 1)\n x = image.img_to_array(img)/255\n # convert 3D tensor to 4D tensor with shape (1, 224, 224, 1) and return 4D tensor\n return np.expand_dims(x, axis=0)\n\ndef paths_to_tensor(img_paths, shape):\n list_of_tensors = [path_to_tensor(img_path, shape) for img_path in tqdm(img_paths)]\n return np.vstack(list_of_tensors)\n\ntrain_labels = labelB[:89600][:, np.newaxis]\nvalid_labels = labelB[89600:100800][:, np.newaxis]\ntest_labels = labelB[100800:][:, np.newaxis]\n\ntrain_data = data[:89600]\nvalid_data = data[89600:100800]\ntest_data = data[100800:]\n\nimg_shape = (64, 64)\ntrain_tensors = paths_to_tensor(files_list[:89600], shape = img_shape)\nvalid_tensors = paths_to_tensor(files_list[89600:100800], shape = img_shape)\ntest_tensors = paths_to_tensor(files_list[100800:], shape = img_shape)",
"Using TensorFlow backend.\n/home/aind2/anaconda3/lib/python3.6/importlib/_bootstrap.py:219: RuntimeWarning: compiletime version 3.5 of module 'tensorflow.python.framework.fast_tensor_util' does not match runtime version 3.6\n return f(*args, **kwds)\n100%|██████████| 89600/89600 [19:43<00:00, 75.69it/s]\n100%|██████████| 11200/11200 [02:32<00:00, 73.55it/s]\n100%|██████████| 11319/11319 [02:33<00:00, 73.74it/s]\n"
],
[
"import time\n\nfrom keras.layers import Conv2D, MaxPooling2D, GlobalAveragePooling2D, Dropout, Flatten, Dense\nfrom keras.models import Sequential\nfrom keras.layers.normalization import BatchNormalization\nfrom keras import regularizers, initializers, optimizers\n\nmodel = Sequential()\n\nmodel.add(Conv2D(filters=16, \n kernel_size=7,\n padding='same', \n activation='relu', \n input_shape=train_tensors.shape[1:]))\nmodel.add(MaxPooling2D(pool_size=2))\n\nmodel.add(Conv2D(filters=32, \n kernel_size=5,\n padding='same', \n activation='relu'))\nmodel.add(MaxPooling2D(pool_size=2))\n\nmodel.add(Conv2D(filters=64, \n kernel_size=5,\n padding='same', \n activation='relu'))\nmodel.add(MaxPooling2D(pool_size=2))\n\nmodel.add(Conv2D(filters=128, \n kernel_size=5,\n strides=2,\n padding='same', \n activation='relu'))\nmodel.add(MaxPooling2D(pool_size=2))\n\nmodel.add(Flatten())\nmodel.add(Dense(100, activation='relu'))\nmodel.add(Dense(1, activation='sigmoid'))\n\nmodel.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nconv2d_1 (Conv2D) (None, 64, 64, 16) 800 \n_________________________________________________________________\nmax_pooling2d_1 (MaxPooling2 (None, 32, 32, 16) 0 \n_________________________________________________________________\nconv2d_2 (Conv2D) (None, 32, 32, 32) 12832 \n_________________________________________________________________\nmax_pooling2d_2 (MaxPooling2 (None, 16, 16, 32) 0 \n_________________________________________________________________\nconv2d_3 (Conv2D) (None, 16, 16, 64) 51264 \n_________________________________________________________________\nmax_pooling2d_3 (MaxPooling2 (None, 8, 8, 64) 0 \n_________________________________________________________________\nconv2d_4 (Conv2D) (None, 4, 4, 128) 204928 \n_________________________________________________________________\nmax_pooling2d_4 (MaxPooling2 (None, 2, 2, 128) 0 \n_________________________________________________________________\nflatten_1 (Flatten) (None, 512) 0 \n_________________________________________________________________\ndense_1 (Dense) (None, 100) 51300 \n_________________________________________________________________\ndense_2 (Dense) (None, 1) 101 \n=================================================================\nTotal params: 321,225\nTrainable params: 321,225\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"from keras import backend as K\n\ndef binary_accuracy(y_true, y_pred):\n return K.mean(K.equal(y_true, K.round(y_pred)))\n\ndef precision_threshold(threshold = 0.5):\n def precision(y_true, y_pred):\n threshold_value = threshold\n y_pred = K.cast(K.greater(K.clip(y_pred, 0, 1), threshold_value), K.floatx())\n true_positives = K.round(K.sum(K.clip(y_true * y_pred, 0, 1)))\n predicted_positives = K.sum(y_pred)\n precision_ratio = true_positives / (predicted_positives + K.epsilon())\n return precision_ratio\n return precision\n\ndef recall_threshold(threshold = 0.5):\n def recall(y_true, y_pred):\n threshold_value = threshold\n y_pred = K.cast(K.greater(K.clip(y_pred, 0, 1), threshold_value), K.floatx())\n true_positives = K.round(K.sum(K.clip(y_true * y_pred, 0, 1)))\n possible_positives = K.sum(K.clip(y_true, 0, 1))\n recall_ratio = true_positives / (possible_positives + K.epsilon())\n return recall_ratio\n return recall\n\ndef fbeta_score_threshold(beta = 1, threshold = 0.5):\n def fbeta_score(y_true, y_pred):\n threshold_value = threshold\n beta_value = beta\n p = precision_threshold(threshold_value)(y_true, y_pred)\n r = recall_threshold(threshold_value)(y_true, y_pred)\n bb = beta_value ** 2\n fbeta_score = (1 + bb) * (p * r) / (bb * p + r + K.epsilon())\n return fbeta_score\n return fbeta_score",
"_____no_output_____"
],
[
"model.compile(optimizer='sgd', loss='binary_crossentropy', \n metrics=[precision_threshold(threshold = 0.5), \n recall_threshold(threshold = 0.5), \n fbeta_score_threshold(beta=0.5, threshold = 0.5),\n 'accuracy'])",
"_____no_output_____"
],
[
"from keras.callbacks import ModelCheckpoint, CSVLogger, EarlyStopping\nimport numpy as np\n\nepochs = 20\nbatch_size = 32\n\nearlystop = EarlyStopping(monitor='val_loss', min_delta=0, patience=3, verbose=1, mode='auto')\nlog = CSVLogger('saved_models/log_bCNN_gray.csv')\ncheckpointer = ModelCheckpoint(filepath='saved_models/bCNN_gray.best.from_scratch.hdf5', \n verbose=1, save_best_only=True)\n\nstart = time.time()\n\nmodel.fit(train_tensors, train_labels, \n validation_data=(valid_tensors, valid_labels),\n epochs=epochs, batch_size=batch_size, callbacks=[checkpointer, log, earlystop], verbose=1)\n\n# Show total training time\nprint(\"training time: %.2f minutes\"%((time.time()-start)/60))",
"Train on 89600 samples, validate on 11200 samples\nEpoch 1/20\n89472/89600 [============================>.] - ETA: 0s - loss: 0.6632 - precision: 0.5035 - recall: 0.3776 - fbeta_score: 0.4438 - acc: 0.6012Epoch 00000: val_loss improved from inf to 0.64742, saving model to saved_models/bCNN_gray.best.from_scratch.hdf5\n89600/89600 [==============================] - 51s - loss: 0.6632 - precision: 0.5035 - recall: 0.3778 - fbeta_score: 0.4439 - acc: 0.6012 - val_loss: 0.6474 - val_precision: 0.6516 - val_recall: 0.3998 - val_fbeta_score: 0.5670 - val_acc: 0.6283\nEpoch 2/20\n89568/89600 [============================>.] - ETA: 0s - loss: 0.6456 - precision: 0.6291 - recall: 0.5252 - fbeta_score: 0.5922 - acc: 0.6347Epoch 00001: val_loss improved from 0.64742 to 0.63779, saving model to saved_models/bCNN_gray.best.from_scratch.hdf5\n89600/89600 [==============================] - 48s - loss: 0.6456 - precision: 0.6291 - recall: 0.5252 - fbeta_score: 0.5922 - acc: 0.6346 - val_loss: 0.6378 - val_precision: 0.6263 - val_recall: 0.5591 - val_fbeta_score: 0.6054 - val_acc: 0.6468\nEpoch 3/20\n89568/89600 [============================>.] - ETA: 0s - loss: 0.6391 - precision: 0.6361 - recall: 0.5440 - fbeta_score: 0.6047 - acc: 0.6444Epoch 00002: val_loss did not improve\n89600/89600 [==============================] - 48s - loss: 0.6391 - precision: 0.6361 - recall: 0.5440 - fbeta_score: 0.6048 - acc: 0.6444 - val_loss: 0.6387 - val_precision: 0.6762 - val_recall: 0.4284 - val_fbeta_score: 0.5951 - val_acc: 0.6449\nEpoch 4/20\n89504/89600 [============================>.] - ETA: 0s - loss: 0.6341 - precision: 0.6433 - recall: 0.5593 - fbeta_score: 0.6146 - acc: 0.6520Epoch 00003: val_loss improved from 0.63779 to 0.62709, saving model to saved_models/bCNN_gray.best.from_scratch.hdf5\n89600/89600 [==============================] - 48s - loss: 0.6341 - precision: 0.6434 - recall: 0.5594 - fbeta_score: 0.6147 - acc: 0.6520 - val_loss: 0.6271 - val_precision: 0.6313 - val_recall: 0.6086 - val_fbeta_score: 0.6215 - val_acc: 0.6601\nEpoch 5/20\n89472/89600 [============================>.] - ETA: 0s - loss: 0.6300 - precision: 0.6482 - recall: 0.5680 - fbeta_score: 0.6209 - acc: 0.6565Epoch 00004: val_loss improved from 0.62709 to 0.62225, saving model to saved_models/bCNN_gray.best.from_scratch.hdf5\n89600/89600 [==============================] - 47s - loss: 0.6299 - precision: 0.6484 - recall: 0.5681 - fbeta_score: 0.6211 - acc: 0.6566 - val_loss: 0.6222 - val_precision: 0.6434 - val_recall: 0.5803 - val_fbeta_score: 0.6238 - val_acc: 0.6628\nEpoch 6/20\n89472/89600 [============================>.] - ETA: 0s - loss: 0.6266 - precision: 0.6516 - recall: 0.5729 - fbeta_score: 0.6244 - acc: 0.6599Epoch 00005: val_loss improved from 0.62225 to 0.61933, saving model to saved_models/bCNN_gray.best.from_scratch.hdf5\n89600/89600 [==============================] - 47s - loss: 0.6266 - precision: 0.6516 - recall: 0.5730 - fbeta_score: 0.6244 - acc: 0.6599 - val_loss: 0.6193 - val_precision: 0.6418 - val_recall: 0.5943 - val_fbeta_score: 0.6267 - val_acc: 0.6646\nEpoch 7/20\n89536/89600 [============================>.] - ETA: 0s - loss: 0.6234 - precision: 0.6541 - recall: 0.5796 - fbeta_score: 0.6289 - acc: 0.6633Epoch 00006: val_loss did not improve\n89600/89600 [==============================] - 47s - loss: 0.6234 - precision: 0.6539 - recall: 0.5796 - fbeta_score: 0.6287 - acc: 0.6633 - val_loss: 0.6254 - val_precision: 0.6851 - val_recall: 0.4468 - val_fbeta_score: 0.6094 - val_acc: 0.6534\nEpoch 8/20\n89568/89600 [============================>.] - ETA: 0s - loss: 0.6206 - precision: 0.6558 - recall: 0.5843 - fbeta_score: 0.6316 - acc: 0.6660Epoch 00007: val_loss did not improve\n89600/89600 [==============================] - 47s - loss: 0.6206 - precision: 0.6558 - recall: 0.5843 - fbeta_score: 0.6316 - acc: 0.6660 - val_loss: 0.6205 - val_precision: 0.6817 - val_recall: 0.4982 - val_fbeta_score: 0.6261 - val_acc: 0.6633\nEpoch 9/20\n89568/89600 [============================>.] - ETA: 0s - loss: 0.6182 - precision: 0.6580 - recall: 0.5909 - fbeta_score: 0.6348 - acc: 0.6679Epoch 00008: val_loss improved from 0.61933 to 0.61392, saving model to saved_models/bCNN_gray.best.from_scratch.hdf5\n89600/89600 [==============================] - 47s - loss: 0.6182 - precision: 0.6580 - recall: 0.5909 - fbeta_score: 0.6348 - acc: 0.6679 - val_loss: 0.6139 - val_precision: 0.6392 - val_recall: 0.6666 - val_fbeta_score: 0.6401 - val_acc: 0.6762\nEpoch 10/20\n89568/89600 [============================>.] - ETA: 0s - loss: 0.6158 - precision: 0.6609 - recall: 0.5961 - fbeta_score: 0.6388 - acc: 0.6712Epoch 00009: val_loss improved from 0.61392 to 0.61250, saving model to saved_models/bCNN_gray.best.from_scratch.hdf5\n89600/89600 [==============================] - 47s - loss: 0.6158 - precision: 0.6608 - recall: 0.5961 - fbeta_score: 0.6387 - acc: 0.6711 - val_loss: 0.6125 - val_precision: 0.6469 - val_recall: 0.6398 - val_fbeta_score: 0.6406 - val_acc: 0.6764\nEpoch 11/20\n89568/89600 [============================>.] - ETA: 0s - loss: 0.6134 - precision: 0.6610 - recall: 0.5980 - fbeta_score: 0.6392 - acc: 0.6727Epoch 00010: val_loss did not improve\n89600/89600 [==============================] - 47s - loss: 0.6134 - precision: 0.6610 - recall: 0.5980 - fbeta_score: 0.6392 - acc: 0.6727 - val_loss: 0.6131 - val_precision: 0.6434 - val_recall: 0.6591 - val_fbeta_score: 0.6417 - val_acc: 0.6771\nEpoch 12/20\n89568/89600 [============================>.] - ETA: 0s - loss: 0.6111 - precision: 0.6655 - recall: 0.6038 - fbeta_score: 0.6440 - acc: 0.6755Epoch 00011: val_loss improved from 0.61250 to 0.60702, saving model to saved_models/bCNN_gray.best.from_scratch.hdf5\n89600/89600 [==============================] - 47s - loss: 0.6111 - precision: 0.6655 - recall: 0.6038 - fbeta_score: 0.6440 - acc: 0.6755 - val_loss: 0.6070 - val_precision: 0.6684 - val_recall: 0.5950 - val_fbeta_score: 0.6462 - val_acc: 0.6793\nEpoch 13/20\n89536/89600 [============================>.] - ETA: 0s - loss: 0.6091 - precision: 0.6655 - recall: 0.6088 - fbeta_score: 0.6460 - acc: 0.6770Epoch 00012: val_loss improved from 0.60702 to 0.60598, saving model to saved_models/bCNN_gray.best.from_scratch.hdf5\n89600/89600 [==============================] - 47s - loss: 0.6091 - precision: 0.6655 - recall: 0.6087 - fbeta_score: 0.6459 - acc: 0.6769 - val_loss: 0.6060 - val_precision: 0.6706 - val_recall: 0.5808 - val_fbeta_score: 0.6443 - val_acc: 0.6776\nEpoch 14/20\n89504/89600 [============================>.] - ETA: 0s - loss: 0.6066 - precision: 0.6661 - recall: 0.6114 - fbeta_score: 0.6467 - acc: 0.6792Epoch 00013: val_loss did not improve\n89600/89600 [==============================] - 47s - loss: 0.6067 - precision: 0.6661 - recall: 0.6114 - fbeta_score: 0.6467 - acc: 0.6792 - val_loss: 0.6065 - val_precision: 0.6767 - val_recall: 0.5648 - val_fbeta_score: 0.6445 - val_acc: 0.6774\nEpoch 15/20\n89568/89600 [============================>.] - ETA: 0s - loss: 0.6043 - precision: 0.6677 - recall: 0.6155 - fbeta_score: 0.6490 - acc: 0.6804Epoch 00014: val_loss did not improve\n89600/89600 [==============================] - 47s - loss: 0.6043 - precision: 0.6677 - recall: 0.6155 - fbeta_score: 0.6490 - acc: 0.6803 - val_loss: 0.6122 - val_precision: 0.6375 - val_recall: 0.6991 - val_fbeta_score: 0.6449 - val_acc: 0.6806\nEpoch 16/20\n89472/89600 [============================>.] - ETA: 0s - loss: 0.6018 - precision: 0.6698 - recall: 0.6196 - fbeta_score: 0.6518 - acc: 0.6833Epoch 00015: val_loss improved from 0.60598 to 0.60483, saving model to saved_models/bCNN_gray.best.from_scratch.hdf5\n89600/89600 [==============================] - 47s - loss: 0.6017 - precision: 0.6698 - recall: 0.6196 - fbeta_score: 0.6518 - acc: 0.6833 - val_loss: 0.6048 - val_precision: 0.6760 - val_recall: 0.5804 - val_fbeta_score: 0.6481 - val_acc: 0.6796\nEpoch 17/20\n89568/89600 [============================>.] - ETA: 0s - loss: 0.5996 - precision: 0.6730 - recall: 0.6234 - fbeta_score: 0.6555 - acc: 0.6861Epoch 00016: val_loss did not improve\n89600/89600 [==============================] - 48s - loss: 0.5997 - precision: 0.6730 - recall: 0.6234 - fbeta_score: 0.6555 - acc: 0.6861 - val_loss: 0.6108 - val_precision: 0.6958 - val_recall: 0.5157 - val_fbeta_score: 0.6425 - val_acc: 0.6755\nEpoch 18/20\n89536/89600 [============================>.] - ETA: 0s - loss: 0.5973 - precision: 0.6737 - recall: 0.6247 - fbeta_score: 0.6562 - acc: 0.6867Epoch 00017: val_loss did not improve\n89600/89600 [==============================] - 50s - loss: 0.5972 - precision: 0.6737 - recall: 0.6248 - fbeta_score: 0.6563 - acc: 0.6868 - val_loss: 0.6143 - val_precision: 0.6934 - val_recall: 0.5248 - val_fbeta_score: 0.6437 - val_acc: 0.6756\nEpoch 19/20\n89568/89600 [============================>.] - ETA: 0s - loss: 0.5948 - precision: 0.6778 - recall: 0.6327 - fbeta_score: 0.6612 - acc: 0.6910Epoch 00018: val_loss did not improve\n89600/89600 [==============================] - 49s - loss: 0.5948 - precision: 0.6779 - recall: 0.6327 - fbeta_score: 0.6613 - acc: 0.6910 - val_loss: 0.6085 - val_precision: 0.6664 - val_recall: 0.6087 - val_fbeta_score: 0.6480 - val_acc: 0.6796\nEpoch 20/20\n89568/89600 [============================>.] - ETA: 0s - loss: 0.5929 - precision: 0.6769 - recall: 0.6336 - fbeta_score: 0.6603 - acc: 0.6906Epoch 00019: val_loss did not improve\n89600/89600 [==============================] - 49s - loss: 0.5929 - precision: 0.6769 - recall: 0.6336 - fbeta_score: 0.6603 - acc: 0.6906 - val_loss: 0.6067 - val_precision: 0.6810 - val_recall: 0.5719 - val_fbeta_score: 0.6497 - val_acc: 0.6806\nEpoch 00019: early stopping\ntraining time: 16.09 minutes\n"
],
[
"model.load_weights('saved_models/bCNN_gray.best.from_scratch.hdf5')\nprediction = model.predict(test_tensors)",
"_____no_output_____"
],
[
"threshold = 0.5\nbeta = 0.5\n\npre = K.eval(precision_threshold(threshold = threshold)(K.variable(value=test_labels),\n K.variable(value=prediction)))\nrec = K.eval(recall_threshold(threshold = threshold)(K.variable(value=test_labels),\n K.variable(value=prediction)))\nfsc = K.eval(fbeta_score_threshold(beta = beta, threshold = threshold)(K.variable(value=test_labels),\n K.variable(value=prediction)))\n\nprint (\"Precision: %f %%\\nRecall: %f %%\\nFscore: %f %%\"% (pre, rec, fsc))",
"Precision: 0.671851 %\nRecall: 0.572077 %\nFscore: 0.649206 %\n"
],
[
"K.eval(binary_accuracy(K.variable(value=test_labels),\n K.variable(value=prediction)))",
"_____no_output_____"
],
[
"threshold = 0.4\nbeta = 0.5\n\npre = K.eval(precision_threshold(threshold = threshold)(K.variable(value=test_labels),\n K.variable(value=prediction)))\nrec = K.eval(recall_threshold(threshold = threshold)(K.variable(value=test_labels),\n K.variable(value=prediction)))\nfsc = K.eval(fbeta_score_threshold(beta = beta, threshold = threshold)(K.variable(value=test_labels),\n K.variable(value=prediction)))\n\nprint (\"Precision: %f %%\\nRecall: %f %%\\nFscore: %f %%\"% (pre, rec, fsc))",
"Precision: 0.627903 %\nRecall: 0.710935 %\nFscore: 0.642921 %\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78eaa8e3d40ac1209d113c46132a92b1dcc83d5 | 432,013 | ipynb | Jupyter Notebook | WeatherPy/WeatherPy.ipynb | Kylee-Grant/python-api-challenge | 546e31fa0273ddde881b071e6d3dfb4209e8e2e4 | [
"ADSL"
] | null | null | null | WeatherPy/WeatherPy.ipynb | Kylee-Grant/python-api-challenge | 546e31fa0273ddde881b071e6d3dfb4209e8e2e4 | [
"ADSL"
] | null | null | null | WeatherPy/WeatherPy.ipynb | Kylee-Grant/python-api-challenge | 546e31fa0273ddde881b071e6d3dfb4209e8e2e4 | [
"ADSL"
] | null | null | null | 205.134378 | 38,328 | 0.888439 | [
[
[
"# WeatherPy\n----\n\n#### Note\n* Instructions have been included for each segment. You do not have to follow them exactly, but they are included to help you think through the steps.",
"_____no_output_____"
]
],
[
[
"# Dependencies and Setup\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport numpy as np\nimport requests\nimport time\nfrom scipy.stats import linregress\nfrom pprint import pprint\n\n# Import API key\nfrom api_keys import weather_api_key\n\n# Incorporated citipy to determine city based on latitude and longitude\nfrom citipy import citipy\n\n# Output File (CSV)\noutput_data_file = \"../output_data/cities.csv\"\n\n# Range of latitudes and longitudes\nlat_range = (-90, 90)\nlng_range = (-180, 180)",
"_____no_output_____"
]
],
[
[
"## Generate Cities List",
"_____no_output_____"
]
],
[
[
"# List for holding lat_lngs and cities\nlat_lngs = []\ncities = []\n\n# Create a set of random lat and lng combinations\nlats = np.random.uniform(lat_range[0], lat_range[1], size=1750) # increased the amount due to my logic below\nlngs = np.random.uniform(lng_range[0], lng_range[1], size=1750) # increased the amount due to my logic below\nlat_lngs = zip(lats, lngs)\n\n# Identify nearest city for each lat, lng combination\nfor lat_lng in lat_lngs:\n city = citipy.nearest_city(lat_lng[0], lat_lng[1]).city_name\n \n # If the city is unique, then add it to a our cities list\n if city not in cities:\n cities.append(city)\n\n# Print the city count to confirm sufficient count\nlen(cities)",
"_____no_output_____"
]
],
[
[
"### Perform API Calls\n* Perform a weather check on each city using a series of successive API calls.\n* Include a print log of each city as it'sbeing processed (with the city number and city name).\n",
"_____no_output_____"
]
],
[
[
"# Save config information.\nurl = \"http://api.openweathermap.org/data/2.5/weather?\"\nunits = \"imperial\"\n\n# Build partial query URL\nquery_url = f\"{url}appid={weather_api_key}&units={units}&q=\"\n\n# Set up lists to hold reponse information \ncity_list = []\ncity_lat = []\ncity_lng = []\ncity_temp = []\ncity_humidity = []\ncity_clouds = []\ncity_wind = []\ncity_country = []\ncity_date = []\ni = 1 \nsetof = 1 \n\n# Loop through the list of cities and perform a request for data on each\n\nprint(\"\"\"Beginning Data Retrieval\\n-----------------------------\"\"\")\nfor city in cities:\n if i < 51: \n response = requests.get(query_url + city).json() \n try: \n if response['name'].casefold() == city: # casefold helps us ignore case\n city_list.append(response['name'])\n city_lat.append(response['coord']['lat'])\n city_lng.append(response['coord']['lon'])\n city_temp.append(response['main']['temp_max'])\n city_humidity.append(response['main']['humidity'])\n city_clouds.append(response['clouds']['all'])\n city_wind.append(response['wind']['speed'])\n city_country.append(response['sys']['country'])\n city_date.append(response['dt'])\n print(f\"Processing Record {i} of Set {setof} | {city}\")\n else: \n print(\"City not found. Skipping...\") # filtering these out to be extra sure everything matches\n except KeyError:\n print(\"City not found. Skipping...\") \n i += 1\n else: \n time.sleep(60) #https://docs.python.org/3/library/time.html#time.sleep\n setof += 1 # add one to the set \n i = 1 # reset the ticker \n response = requests.get(query_url + city).json() \n try: \n if response['name'].casefold() == city: # casefold helps us ignore case\n city_list.append(response['name'])\n city_lat.append(response['coord']['lat'])\n city_lng.append(response['coord']['lon'])\n city_temp.append(response['main']['temp_max'])\n city_humidity.append(response['main']['humidity'])\n city_clouds.append(response['clouds']['all'])\n city_wind.append(response['wind']['speed'])\n city_country.append(response['sys']['country'])\n city_date.append(response['dt'])\n print(f\"Processing Record {i} of Set {setof} | {city}\")\n else: \n print(\"City not found. Skipping...\") # filtering these out to be extra sure everything matches\n except KeyError:\n print(\"City not found. Skipping...\") \n i += 1\nprint(\"\"\"-----------------------------\\nData Retrieval Complete\\n-----------------------------\"\"\")",
"Beginning Data Retrieval\n-----------------------------\nProcessing Record 1 of Set 1 | norman wells\nProcessing Record 2 of Set 1 | touros\nCity not found. Skipping...\nProcessing Record 4 of Set 1 | qaanaaq\nCity not found. Skipping...\nProcessing Record 6 of Set 1 | punta arenas\nProcessing Record 7 of Set 1 | ushuaia\nProcessing Record 8 of Set 1 | vaini\nProcessing Record 9 of Set 1 | busselton\nProcessing Record 10 of Set 1 | poum\nProcessing Record 11 of Set 1 | sheregesh\nProcessing Record 12 of Set 1 | mataura\nProcessing Record 13 of Set 1 | khatanga\nProcessing Record 14 of Set 1 | saint-augustin\nProcessing Record 15 of Set 1 | kavieng\nProcessing Record 16 of Set 1 | richards bay\nProcessing Record 17 of Set 1 | kahului\nCity not found. Skipping...\nProcessing Record 19 of Set 1 | pisco\nCity not found. Skipping...\nProcessing Record 21 of Set 1 | dalvik\nProcessing Record 22 of Set 1 | port hardy\nProcessing Record 23 of Set 1 | vostok\nProcessing Record 24 of Set 1 | airai\nCity not found. Skipping...\nCity not found. Skipping...\nProcessing Record 27 of Set 1 | rikitea\nCity not found. Skipping...\nProcessing Record 29 of Set 1 | kerman\nProcessing Record 30 of Set 1 | puerto ayora\nProcessing Record 31 of Set 1 | hasaki\nProcessing Record 32 of Set 1 | oussouye\nProcessing Record 33 of Set 1 | kapaa\nCity not found. Skipping...\nProcessing Record 35 of Set 1 | nome\nProcessing Record 36 of Set 1 | hofn\nCity not found. Skipping...\nCity not found. Skipping...\nProcessing Record 39 of Set 1 | bull savanna\nCity not found. Skipping...\nProcessing Record 41 of Set 1 | jamestown\nProcessing Record 42 of Set 1 | bang saphan\nCity not found. Skipping...\nCity not found. Skipping...\nProcessing Record 45 of Set 1 | san cristobal\nProcessing Record 46 of Set 1 | bluff\nCity not found. Skipping...\nProcessing Record 48 of Set 1 | albany\nProcessing Record 49 of Set 1 | bandarbeyla\nProcessing Record 50 of Set 1 | punta gorda\nCity not found. Skipping...\nProcessing Record 2 of Set 2 | lagoa\nProcessing Record 3 of Set 2 | port elizabeth\nProcessing Record 4 of Set 2 | hilo\nProcessing Record 5 of Set 2 | araouane\nProcessing Record 6 of Set 2 | alice springs\nProcessing Record 7 of Set 2 | vite\nProcessing Record 8 of Set 2 | nishihara\nProcessing Record 9 of Set 2 | tasiilaq\nProcessing Record 10 of Set 2 | inta\nCity not found. Skipping...\nCity not found. Skipping...\nProcessing Record 13 of Set 2 | grindavik\nProcessing Record 14 of Set 2 | hermanus\nProcessing Record 15 of Set 2 | garden city\nProcessing Record 16 of Set 2 | anju\nProcessing Record 17 of Set 2 | nadadores\nProcessing Record 18 of Set 2 | tuktoyaktuk\nProcessing Record 19 of Set 2 | motril\nCity not found. Skipping...\nProcessing Record 21 of Set 2 | new norfolk\nProcessing Record 22 of Set 2 | verkhoyansk\nProcessing Record 23 of Set 2 | bethel\nProcessing Record 24 of Set 2 | benguela\nProcessing Record 25 of Set 2 | ponta do sol\nProcessing Record 26 of Set 2 | hirara\nCity not found. Skipping...\nCity not found. Skipping...\nCity not found. Skipping...\nProcessing Record 30 of Set 2 | hvolsvollur\nProcessing Record 31 of Set 2 | akureyri\nProcessing Record 32 of Set 2 | verkhnyaya inta\nProcessing Record 33 of Set 2 | tabriz\nProcessing Record 34 of Set 2 | ribeira grande\nCity not found. Skipping...\nProcessing Record 36 of Set 2 | hobart\nProcessing Record 37 of Set 2 | castro\nProcessing Record 38 of Set 2 | rehoboth\nProcessing Record 39 of Set 2 | mount isa\nCity not found. Skipping...\nProcessing Record 41 of Set 2 | sisimiut\nProcessing Record 42 of Set 2 | matara\nProcessing Record 43 of Set 2 | ko samui\nProcessing Record 44 of Set 2 | dikson\nCity not found. Skipping...\nCity not found. Skipping...\nProcessing Record 47 of Set 2 | naze\nProcessing Record 48 of Set 2 | isperih\nProcessing Record 49 of Set 2 | nanortalik\nProcessing Record 50 of Set 2 | palmer\nProcessing Record 1 of Set 3 | bubaque\nProcessing Record 2 of Set 3 | cherskiy\nProcessing Record 3 of Set 3 | nouadhibou\nProcessing Record 4 of Set 3 | hithadhoo\nProcessing Record 5 of Set 3 | rhyl\nProcessing Record 6 of Set 3 | stornoway\nCity not found. Skipping...\nCity not found. Skipping...\nProcessing Record 9 of Set 3 | atuona\nProcessing Record 10 of Set 3 | samarai\nProcessing Record 11 of Set 3 | marchena\nProcessing Record 12 of Set 3 | mattru\nCity not found. Skipping...\nProcessing Record 14 of Set 3 | carnarvon\nCity not found. Skipping...\nProcessing Record 16 of Set 3 | namie\nProcessing Record 17 of Set 3 | mercedes\nCity not found. Skipping...\nProcessing Record 19 of Set 3 | saskylakh\nProcessing Record 20 of Set 3 | clyde river\nCity not found. Skipping...\nProcessing Record 22 of Set 3 | fortuna\nProcessing Record 23 of Set 3 | yumen\nProcessing Record 24 of Set 3 | mar del plata\nProcessing Record 25 of Set 3 | vao\nProcessing Record 26 of Set 3 | harnai\nProcessing Record 27 of Set 3 | pangnirtung\nProcessing Record 28 of Set 3 | avarua\nProcessing Record 29 of Set 3 | bereda\nProcessing Record 30 of Set 3 | longview\nProcessing Record 31 of Set 3 | genhe\nProcessing Record 32 of Set 3 | saint-philippe\nProcessing Record 33 of Set 3 | faanui\nProcessing Record 34 of Set 3 | meulaboh\nProcessing Record 35 of Set 3 | barra do corda\nProcessing Record 36 of Set 3 | sungai padi\nProcessing Record 37 of Set 3 | raglan\nProcessing Record 38 of Set 3 | yellowknife\nCity not found. Skipping...\nCity not found. Skipping...\nCity not found. Skipping...\nProcessing Record 42 of Set 3 | juneau\nProcessing Record 43 of Set 3 | carbonia\nProcessing Record 44 of Set 3 | saryg-sep\nProcessing Record 45 of Set 3 | sioux lookout\nProcessing Record 46 of Set 3 | gainesville\nProcessing Record 47 of Set 3 | cape town\nProcessing Record 48 of Set 3 | kaoma\nCity not found. Skipping...\nProcessing Record 50 of Set 3 | east london\nCity not found. Skipping...\nCity not found. Skipping...\nProcessing Record 3 of Set 4 | loa janan\nProcessing Record 4 of Set 4 | okha\nProcessing Record 5 of Set 4 | canchungo\nProcessing Record 6 of Set 4 | commerce\nProcessing Record 7 of Set 4 | bredasdorp\nCity not found. Skipping...\nCity not found. Skipping...\nCity not found. Skipping...\nCity not found. Skipping...\nCity not found. Skipping...\nProcessing Record 13 of Set 4 | arraial do cabo\nProcessing Record 14 of Set 4 | brae\nCity not found. Skipping...\nProcessing Record 16 of Set 4 | kruisfontein\nProcessing Record 17 of Set 4 | butaritari\nCity not found. Skipping...\nProcessing Record 19 of Set 4 | leningradskiy\nProcessing Record 20 of Set 4 | kismayo\nProcessing Record 21 of Set 4 | lebu\nProcessing Record 22 of Set 4 | saint-pierre\nCity not found. Skipping...\nProcessing Record 24 of Set 4 | lorengau\nProcessing Record 25 of Set 4 | katav-ivanovsk\nProcessing Record 26 of Set 4 | jalalabad\nProcessing Record 27 of Set 4 | pyu\nCity not found. Skipping...\nProcessing Record 29 of Set 4 | yaan\nProcessing Record 30 of Set 4 | tolaga bay\nProcessing Record 31 of Set 4 | beidao\nProcessing Record 32 of Set 4 | west wendover\nProcessing Record 33 of Set 4 | kavaratti\nProcessing Record 34 of Set 4 | geraldton\nProcessing Record 35 of Set 4 | guerrero negro\nProcessing Record 36 of Set 4 | port alfred\nProcessing Record 37 of Set 4 | tiksi\nProcessing Record 38 of Set 4 | eureka\nCity not found. Skipping...\nProcessing Record 40 of Set 4 | husavik\nProcessing Record 41 of Set 4 | bathsheba\nProcessing Record 42 of Set 4 | oskemen\nCity not found. Skipping...\nProcessing Record 44 of Set 4 | fernley\nProcessing Record 45 of Set 4 | longyearbyen\nProcessing Record 46 of Set 4 | senneterre\nCity not found. Skipping...\nCity not found. Skipping...\nProcessing Record 49 of Set 4 | torbay\nCity not found. Skipping...\nProcessing Record 1 of Set 5 | rawson\nCity not found. Skipping...\nCity not found. Skipping...\nProcessing Record 4 of Set 5 | aklavik\nProcessing Record 5 of Set 5 | chokurdakh\nProcessing Record 6 of Set 5 | buritizeiro\nProcessing Record 7 of Set 5 | normal\nProcessing Record 8 of Set 5 | manzhouli\nProcessing Record 9 of Set 5 | esperance\nProcessing Record 10 of Set 5 | balikpapan\nProcessing Record 11 of Set 5 | inuvik\nProcessing Record 12 of Set 5 | coquimbo\nCity not found. Skipping...\nProcessing Record 14 of Set 5 | gushikawa\nProcessing Record 15 of Set 5 | shimoda\nProcessing Record 16 of Set 5 | zhangjiakou\n"
]
],
[
[
"### Convert Raw Data to DataFrame\n* Export the city data into a .csv.\n* Display the DataFrame",
"_____no_output_____"
]
],
[
[
"# Create a data frame from cities info \nweather_dict = {\n \"City\": city_list,\n \"Lat\": city_lat,\n \"Lng\": city_lng,\n \"Max Temp\": city_temp,\n \"Humidity\": city_humidity,\n \"Cloudiness\": city_clouds,\n \"Wind Speed\": city_wind,\n \"Country\": city_country,\n \"Date\": city_date\n}\nweather_data = pd.DataFrame(weather_dict)\nweather_data",
"_____no_output_____"
],
[
"# Exporting to csv file \nweather_data.to_csv(output_data_file, index = False)\n",
"_____no_output_____"
],
[
"# Importing file, if needed \nweather_data = pd.read_csv(\"../output_data/cities.csv\")",
"_____no_output_____"
]
],
[
[
"## Inspect the data and remove the cities where the humidity > 100%.\n----\nSkip this step if there are no cities that have humidity > 100%. ",
"_____no_output_____"
]
],
[
[
"# No cities have humidity > 100%, although a few are exactly 100%\nhumid_weather_data = weather_data.loc[weather_data[\"Humidity\"] >= 100]\nhumid_weather_data",
"_____no_output_____"
]
],
[
[
"## Plotting the Data\n* Use proper labeling of the plots using plot titles (including date of analysis) and axes labels.\n* Save the plotted figures as .pngs.",
"_____no_output_____"
],
[
"## Latitude vs. Temperature Plot",
"_____no_output_____"
]
],
[
[
"# Setting x and y values \nx_values = weather_data[\"Lat\"]\ny_values = weather_data[\"Max Temp\"]\n\n# Plot the scatter plot \nplt.scatter(x_values, y_values, marker=\"o\", facecolors=\"teal\", edgecolors=\"black\")\nplt.title(\"City Latitude vs. Max Temperature\")\nplt.ylabel(\"Max Temperature (F)\", fontsize=12)\nplt.xlabel(\"Latitude\", fontsize=12)\nplt.grid(alpha=0.2)\nplt.show()\n\n# Save the image \n#fig_path = os.path.abspath() # Figures out the absolute path for you in case your working directory moves around.\n#fig.savefig(my_path + '/Sub Directory/graph.png')",
"_____no_output_____"
]
],
[
[
"### ONE-SENTENCE DESCRIPTION: \nThe graph above is displaying the max temperature (y-axis) for all cities in the dataset (the circles) organized by latitude (x-axis). This graph suggests that cities north of the equator (>0 Lat) might have a lower max temp. ",
"_____no_output_____"
],
[
"## Latitude vs. Humidity Plot",
"_____no_output_____"
]
],
[
[
"# Setting x and y values \nx_values = weather_data[\"Lat\"]\ny_values = weather_data[\"Humidity\"]\n\n# Plot the scatter plot \nplt.scatter(x_values, y_values, marker=\"o\", facecolors=\"teal\", edgecolors=\"black\")\nplt.title(\"City Latitude vs. Humidity\")\nplt.ylabel(\"Humidity (%)\", fontsize=12)\nplt.xlabel(\"Latitude\", fontsize=12)\nplt.grid(alpha=0.2)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### ONE-SENTENCE DESCRIPTION: \nThe graph above is displaying the recent humidity (%)(y-axis) for all cities in the dataset (the circles) organized by latitude (x-axis). This graph does not have an immediately discernible trend, other than cities tending to cluster at or greater than 60% humidity. ",
"_____no_output_____"
],
[
"## Latitude vs. Cloudiness Plot",
"_____no_output_____"
]
],
[
[
"# Setting x and y values \nx_values = weather_data[\"Lat\"]\ny_values = weather_data[\"Cloudiness\"]\n\n# Plot the scatter plot \nplt.scatter(x_values, y_values, marker=\"o\", facecolors=\"teal\", edgecolors=\"black\")\nplt.title(\"City Latitude vs. Cloudiness\")\nplt.ylabel(\"Cloudiness (%)\", fontsize=12)\nplt.xlabel(\"Latitude\", fontsize=12)\nplt.grid(alpha=0.2)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### ONE-SENTENCE DESCRIPTION:\nThe graph above is displaying the recent cloudiness (%)(y-axis) for all cities in the dataset (the circles) organized by latitude (x-axis). This graph also does not have an immediately discernible trend.",
"_____no_output_____"
],
[
"## Latitude vs. Wind Speed Plot",
"_____no_output_____"
]
],
[
[
"# Setting x and y values \nx_values = weather_data[\"Lat\"]\ny_values = weather_data[\"Wind Speed\"]\n\n# Plot the scatter plot \nplt.scatter(x_values, y_values, marker=\"o\", facecolors=\"teal\", edgecolors=\"black\")\nplt.title(\"City Latitude vs. Wind Speed\")\nplt.ylabel(\"Wind Speed (mph)\", fontsize=12)\nplt.xlabel(\"Latitude\", fontsize=12)\nplt.grid(alpha=0.2)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### ONE-SENTENCE DESCRIPTION:\nThe graph above is displaying the recent wind speed (mph)(y-axis) for all cities in the dataset (the circles) organized by latitude (x-axis). Perhaps one observable trend from this graph: it appears that there is greater range in the wind speeds of cities north of equator (>Lat 0). ",
"_____no_output_____"
],
[
"## Linear Regression",
"_____no_output_____"
],
[
"After each pair of plots, take the time to explain what the linear regression is modeling. For example, describe any relationships you notice and any other analysis you may have.\n\nYour final notebook must:\n\n* Randomly select **at least** 500 unique (non-repeat) cities based on latitude and longitude.\n* Perform a weather check on each of the cities using a series of successive API calls.\n* Include a print log of each city as it's being processed with the city number and city name.\n* Save a CSV of all retrieved data and a PNG image for each scatter plot.",
"_____no_output_____"
]
],
[
[
"# Making two separate dfs for north and south hemisphere \nnorth_hemi = weather_data.loc[weather_data[\"Lat\"] >= 0]\nsouth_hemi = weather_data.loc[weather_data[\"Lat\"] < 0]",
"_____no_output_____"
]
],
[
[
"#### Northern Hemisphere - Max Temp vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"# Setting x and y values \nx_values = north_hemi[\"Lat\"]\ny_values = north_hemi[\"Max Temp\"]\n\n# Linear Regression \n(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)\nregress_values = x_values * slope + intercept\nline_eq = \"y = \" + str(round(slope,2)) + \"x + \" + str(round(intercept,2))\nplt.plot(x_values, regress_values, \"r-\")\nplt.annotate(line_eq, (0,-10), fontsize=15, color=\"red\")\nprint(f\"The r-value is: {rvalue}\")\n\n# Plot the scatter plot \nplt.scatter(x_values, y_values, marker=\"o\", facecolors=\"aqua\", edgecolors=\"black\")\nplt.title(\"Northern Hemisphere: Max Temp vs. Latitude\")\nplt.ylabel(\"Max Temp (F)\", fontsize=12)\nplt.xlabel(\"Latitude\", fontsize=12)\nplt.grid(alpha=0.2)\nplt.show()",
"The r-value is: -0.8765697068804407\n"
]
],
[
[
"#### Southern Hemisphere - Max Temp vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"# Setting x and y values \nx_values = south_hemi[\"Lat\"]\ny_values = south_hemi[\"Max Temp\"]\n\n# Linear Regression \n(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)\nregress_values = x_values * slope + intercept\nline_eq = \"y = \" + str(round(slope,2)) + \"x + \" + str(round(intercept,2))\nplt.plot(x_values, regress_values, \"r-\")\nplt.annotate(line_eq, (-30,50), fontsize=15, color=\"red\")\nprint(f\"The r-value is: {rvalue}\")\n\n# Plot the scatter plot \nplt.scatter(x_values, y_values, marker=\"o\", facecolors=\"aqua\", edgecolors=\"black\")\nplt.title(\"Southern Hemisphere: Max Temp vs. Latitude\")\nplt.ylabel(\"Max Temp (F)\", fontsize=12)\nplt.xlabel(\"Latitude\", fontsize=12)\nplt.grid(alpha=0.2)\nplt.show()",
"The r-value is: 0.44590906959636567\n"
]
],
[
[
"### ANALYSIS OF NORTH/SOUTH-HEMI MAX TEMP:\nThe graphs above are displaying the max temperature (y-axis) of cities in the dataset arranged by latitude (x-axis) and divided into two groups--the southern hemisphere and the northern hemisphere. What we see from both of these graphs is that max temperatures are higher for cities closer to the equator. This relationship is weakly positively correlated for the southern hemisphere (moving towards the equator), and strongly negatively correlated for the northern hemisphere (moving away from the equator). It is plausible that the seasons may be playing a role, as the northern hemisphere is currently experiencing winter. ",
"_____no_output_____"
],
[
"#### Northern Hemisphere - Humidity (%) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"# Setting x and y values \nx_values = north_hemi[\"Lat\"]\ny_values = north_hemi[\"Humidity\"]\n\n# Linear Regression \n(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)\nregress_values = x_values * slope + intercept\nline_eq = \"y = \" + str(round(slope,2)) + \"x + \" + str(round(intercept,2))\nplt.plot(x_values, regress_values, \"r-\")\nplt.annotate(line_eq, (40,20), fontsize=15, color=\"red\")\nprint(f\"The r-value is: {rvalue}\")\n\n# Plot the scatter plot \nplt.scatter(x_values, y_values, marker=\"o\", facecolors=\"aqua\", edgecolors=\"black\")\nplt.title(\"Northern Hemisphere: Humidity vs. Latitude\")\nplt.ylabel(\"Humidity (%)\", fontsize=12)\nplt.xlabel(\"Latitude\", fontsize=12)\nplt.grid(alpha=0.2)\nplt.show()",
"The r-value is: 0.30945504958053144\n"
]
],
[
[
"#### Southern Hemisphere - Humidity (%) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"# Setting x and y values \nx_values = south_hemi[\"Lat\"]\ny_values = south_hemi[\"Humidity\"]\n\n# Linear Regression \n(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)\nregress_values = x_values * slope + intercept\nline_eq = \"y = \" + str(round(slope,2)) + \"x + \" + str(round(intercept,2))\nplt.plot(x_values, regress_values, \"r-\")\nplt.annotate(line_eq, (-30,20), fontsize=15, color=\"red\")\nprint(f\"The r-value is: {rvalue}\")\n\n# Plot the scatter plot \nplt.scatter(x_values, y_values, marker=\"o\", facecolors=\"aqua\", edgecolors=\"black\")\nplt.title(\"Southern Hemisphere: Humidity vs. Latitude\")\nplt.ylabel(\"Humidity (%)\", fontsize=12)\nplt.xlabel(\"Latitude\", fontsize=12)\nplt.grid(alpha=0.2)\nplt.show()",
"The r-value is: 0.22520675986668628\n"
]
],
[
[
"### ANALYSIS OF NORTH/SOUTH-HEMI HUMIDITY:\nThe graphs above are displaying the humidity (y-axis) of cities in the dataset arranged by latitude (x-axis) and divided into two groups--the southern hemisphere and the northern hemisphere. These graphs reflect a weak positive correlation between humidity and latitude. Rather than being centered around the equator as shown in the previous section, the humidity seems to increase the more north a city is located. As speculated on previously, the seasons may be playing a role, however it is difficult to produce a logical explanation as colder air tends to hold less humidity. ",
"_____no_output_____"
],
[
"#### Northern Hemisphere - Cloudiness (%) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"# Setting x and y values \nx_values = north_hemi[\"Lat\"]\ny_values = north_hemi[\"Cloudiness\"]\n\n# Linear Regression \n(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)\nregress_values = x_values * slope + intercept\nline_eq = \"y = \" + str(round(slope,2)) + \"x + \" + str(round(intercept,2))\nplt.plot(x_values, regress_values, \"r-\")\nplt.annotate(line_eq, (45,15), fontsize=15, color=\"red\")\nprint(f\"The r-value is: {rvalue}\")\n\n# Plot the scatter plot \nplt.scatter(x_values, y_values, marker=\"o\", facecolors=\"aqua\", edgecolors=\"black\")\nplt.title(\"Northern Hemisphere: Cloudiness vs. Latitude\")\nplt.ylabel(\"Cloudiness (%)\", fontsize=12)\nplt.xlabel(\"Latitude\", fontsize=12)\nplt.grid(alpha=0.2)\nplt.show()",
"The r-value is: 0.2989895417151042\n"
]
],
[
[
"#### Southern Hemisphere - Cloudiness (%) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"# Setting x and y values \nx_values = south_hemi[\"Lat\"]\ny_values = south_hemi[\"Cloudiness\"]\n\n# Linear Regression \n(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)\nregress_values = x_values * slope + intercept\nline_eq = \"y = \" + str(round(slope,2)) + \"x + \" + str(round(intercept,2))\nplt.plot(x_values, regress_values, \"r-\")\nplt.annotate(line_eq, (-50,30), fontsize=15, color=\"red\")\nprint(f\"The r-value is: {rvalue}\")\n\n# Plot the scatter plot \nplt.scatter(x_values, y_values, marker=\"o\", facecolors=\"aqua\", edgecolors=\"black\")\nplt.title(\"Southern Hemisphere: Cloudiness vs. Latitude\")\nplt.ylabel(\"Cloudiness (%)\", fontsize=12)\nplt.xlabel(\"Latitude\", fontsize=12)\nplt.grid(alpha=0.2)\nplt.show()",
"The r-value is: 0.23659107153505674\n"
]
],
[
[
"### ANALYSIS OF NORTH/SOUTH-HEMI CLOUDINESS:\nThe graphs above are displaying the cloudiness (y-axis) of cities in the dataset arranged by latitude (x-axis) and divided into two groups--the southern hemisphere and the northern hemisphere. These graphs also reflect a very weak positive correlation, if any, between cloudiness and latitude. Cloudiness seems to increase the more north a city is located.",
"_____no_output_____"
],
[
"#### Northern Hemisphere - Wind Speed (mph) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"# Setting x and y values \nx_values = north_hemi[\"Lat\"]\ny_values = north_hemi[\"Wind Speed\"]\n\n# Linear Regression \n(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)\nregress_values = x_values * slope + intercept\nline_eq = \"y = \" + str(round(slope,2)) + \"x + \" + str(round(intercept,2))\nplt.plot(x_values, regress_values, \"r-\")\nplt.annotate(line_eq, (10,40), fontsize=15, color=\"red\")\nprint(f\"The r-value is: {rvalue}\")\n\n# Plot the scatter plot \nplt.scatter(x_values, y_values, marker=\"o\", facecolors=\"aqua\", edgecolors=\"black\")\nplt.title(\"Northern Hemisphere: Wind Speed vs. Latitude\")\nplt.ylabel(\"Wind Speed (mph)\", fontsize=12)\nplt.xlabel(\"Latitude\", fontsize=12)\nplt.grid(alpha=0.2)\nplt.show()",
"The r-value is: 0.04083196500915729\n"
]
],
[
[
"#### Southern Hemisphere - Wind Speed (mph) vs. Latitude Linear Regression",
"_____no_output_____"
]
],
[
[
"# Setting x and y values \nx_values = south_hemi[\"Lat\"]\ny_values = south_hemi[\"Wind Speed\"]\n\n# Linear Regression \n(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)\nregress_values = x_values * slope + intercept\nline_eq = \"y = \" + str(round(slope,2)) + \"x + \" + str(round(intercept,2))\nplt.plot(x_values, regress_values, \"r-\")\nplt.annotate(line_eq, (-55,20), fontsize=15, color=\"red\")\nprint(f\"The r-value is: {rvalue}\")\n\n# Plot the scatter plot \nplt.scatter(x_values, y_values, marker=\"o\", facecolors=\"aqua\", edgecolors=\"black\")\nplt.title(\"Southern Hemisphere: Wind Speed vs. Latitude\")\nplt.ylabel(\"Wind Speed (mph)\", fontsize=12)\nplt.xlabel(\"Latitude\", fontsize=12)\nplt.grid(alpha=0.2)\nplt.show()",
"The r-value is: -0.1644284507948641\n"
]
],
[
[
"### ANALYSIS OF NORTH/SOUTH-HEMI WIND SPEED:\nThe graphs above are displaying the wind speed (y-axis) of cities in the dataset arranged by latitude (x-axis) and divided into two groups--the southern hemisphere and the northern hemisphere. These graphs do not reflect a notable relationship between wind speed and latitude; the r-values returned are too low. ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e78eae12167c68c576db79b92f9bd93909efafea | 31,359 | ipynb | Jupyter Notebook | site/en-snapshot/guide/intro_to_graphs.ipynb | gadagashwini/docs-l10n | 61d5f7ac3a9fa0964d077bcba6e3a6bb6c5efa41 | [
"Apache-2.0"
] | null | null | null | site/en-snapshot/guide/intro_to_graphs.ipynb | gadagashwini/docs-l10n | 61d5f7ac3a9fa0964d077bcba6e3a6bb6c5efa41 | [
"Apache-2.0"
] | null | null | null | site/en-snapshot/guide/intro_to_graphs.ipynb | gadagashwini/docs-l10n | 61d5f7ac3a9fa0964d077bcba6e3a6bb6c5efa41 | [
"Apache-2.0"
] | null | null | null | 37.199288 | 489 | 0.570554 | [
[
[
"##### Copyright 2020 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Introduction to graphs and tf.function",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/guide/intro_to_graphs\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/guide/intro_to_graphs.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs/blob/master/site/en/guide/intro_to_graphs.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/docs/site/en/guide/intro_to_graphs.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"## Overview\n\nThis guide goes beneath the surface of TensorFlow and Keras to demonstrate how TensorFlow works. If you instead want to immediately get started with Keras, check out the [collection of Keras guides](keras/).\n\nIn this guide, you'll learn how TensorFlow allows you to make simple changes to your code to get graphs, how graphs are stored and represented, and how you can use them to accelerate your models.\n\nNote: For those of you who are only familiar with TensorFlow 1.x, this guide demonstrates a very different view of graphs.\n\n**This is a big-picture overview that covers how `tf.function` allows you to switch from eager execution to graph execution.** For a more complete specification of `tf.function`, go to the [`tf.function` guide](function).\n",
"_____no_output_____"
],
[
"### What are graphs?\n\nIn the previous three guides, you ran TensorFlow **eagerly**. This means TensorFlow operations are executed by Python, operation by operation, and returning results back to Python.\n\nWhile eager execution has several unique advantages, graph execution enables portability outside Python and tends to offer better performance. **Graph execution** means that tensor computations are executed as a *TensorFlow graph*, sometimes referred to as a `tf.Graph` or simply a \"graph.\"\n\n**Graphs are data structures that contain a set of `tf.Operation` objects, which represent units of computation; and `tf.Tensor` objects, which represent the units of data that flow between operations.** They are defined in a `tf.Graph` context. Since these graphs are data structures, they can be saved, run, and restored all without the original Python code.\n\nThis is what a TensorFlow graph representing a two-layer neural network looks like when visualized in TensorBoard.\n",
"_____no_output_____"
],
[
"<img alt=\"A simple TensorFlow graph\" src=\"./images/intro_to_graphs/two-layer-network.png\">",
"_____no_output_____"
],
[
"### The benefits of graphs\n\nWith a graph, you have a great deal of flexibility. You can use your TensorFlow graph in environments that don't have a Python interpreter, like mobile applications, embedded devices, and backend servers. TensorFlow uses graphs as the format for [saved models](saved_model) when it exports them from Python.\n\nGraphs are also easily optimized, allowing the compiler to do transformations like:\n\n* Statically infer the value of tensors by folding constant nodes in your computation *(\"constant folding\")*.\n* Separate sub-parts of a computation that are independent and split them between threads or devices.\n* Simplify arithmetic operations by eliminating common subexpressions.\n",
"_____no_output_____"
],
[
"There is an entire optimization system, [Grappler](./graph_optimization.ipynb), to perform this and other speedups.\n\nIn short, graphs are extremely useful and let your TensorFlow run **fast**, run **in parallel**, and run efficiently **on multiple devices**.\n\nHowever, you still want to define your machine learning models (or other computations) in Python for convenience, and then automatically construct graphs when you need them.",
"_____no_output_____"
],
[
"### Non-strict execution\n\nGraph execution only executes the operations necessary to produce the observable effects, which includes:\n- the return value of the function,\n- documented well-known side-effects:\n * input/output operations, `tf.print`\n * debugging operations, such as the assert functions in `tf.debugging`,\n * mutations of `tf.Variable`.\n\nThis behavior is usually known as [Non-strict execution](https://en.wikipedia.org/wiki/Evaluation_strategy#Non-strict_evaluation), and differs from eager execution, which steps through all of the program operations, needed or not. \n\nIn particular, runtime error checking does not count as an observable effect. If an operation is skipped because it is unnecessary it cannot raise any runtime errors. \n\nIn the following example, the \"unnecessary\" operation `tf.math.bincount` is skipped during graph execution, so the runtime error `InvalidArgumentError` is not raised as it would be in eager execution. Do not rely on an error being raised while executing a graph.",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nimport timeit\nfrom datetime import datetime",
"_____no_output_____"
],
[
"@tf.function\ndef unused_return_graph(x):\n _ = tf.math.bincount(x)\n return x\n\ndef unused_return_eager(x):\n _ = tf.math.bincount(x)\n return x\n\n# `tf.math.bincount` in eager execution raises an error.\ntry:\n _ = unused_return_eager([-1])\n raise None\nexcept tf.errors.InvalidArgumentError as e:\n assert \"Input arr must be non-negative\" in str(e)\n\n# Only needed operations are run during graph exection. The error is not raised.\n_ = unused_return_graph([-1])",
"_____no_output_____"
]
],
[
[
"## Taking advantage of graphs\n\nYou create and run a graph in TensorFlow by using `tf.function`, either as a direct call or as a decorator. `tf.function` takes a regular function as input and returns a `Function`. **A `Function` is a Python callable that builds TensorFlow graphs from the Python function. You use a `Function` in the same way as its Python equivalent.**\n",
"_____no_output_____"
]
],
[
[
"# Define a Python function.\ndef a_regular_function(x, y, b):\n x = tf.matmul(x, y)\n x = x + b\n return x\n\n# `a_function_that_uses_a_graph` is a TensorFlow `Function`.\na_function_that_uses_a_graph = tf.function(a_regular_function)\n\n# Make some tensors.\nx1 = tf.constant([[1.0, 2.0]])\ny1 = tf.constant([[2.0], [3.0]])\nb1 = tf.constant(4.0)\n\norig_value = a_regular_function(x1, y1, b1).numpy()\n# Call a `Function` like a Python function.\ntf_function_value = a_function_that_uses_a_graph(x1, y1, b1).numpy()\nassert(orig_value == tf_function_value)",
"_____no_output_____"
]
],
[
[
"On the outside, a `Function` looks like a regular function you write using TensorFlow operations. [Underneath](https://github.com/tensorflow/tensorflow/blob/master/tensorflow/python/eager/def_function.py), however, it is *very different*. A `Function` **encapsulates [several `tf.Graph`s behind one API](#polymorphism_one_function_many_graphs).** That is how `Function` is able to give you the [benefits of graph execution](#the_benefits_of_graphs), like speed and deployability.",
"_____no_output_____"
],
[
"`tf.function` applies to a function *and all other functions it calls*:",
"_____no_output_____"
]
],
[
[
"def inner_function(x, y, b):\n x = tf.matmul(x, y)\n x = x + b\n return x\n\n# Use the decorator to make `outer_function` a `Function`.\[email protected]\ndef outer_function(x):\n y = tf.constant([[2.0], [3.0]])\n b = tf.constant(4.0)\n\n return inner_function(x, y, b)\n\n# Note that the callable will create a graph that\n# includes `inner_function` as well as `outer_function`.\nouter_function(tf.constant([[1.0, 2.0]])).numpy()",
"_____no_output_____"
]
],
[
[
"If you have used TensorFlow 1.x, you will notice that at no time did you need to define a `Placeholder` or `tf.Session`.",
"_____no_output_____"
],
[
"### Converting Python functions to graphs\n\nAny function you write with TensorFlow will contain a mixture of built-in TF operations and Python logic, such as `if-then` clauses, loops, `break`, `return`, `continue`, and more. While TensorFlow operations are easily captured by a `tf.Graph`, Python-specific logic needs to undergo an extra step in order to become part of the graph. `tf.function` uses a library called AutoGraph (`tf.autograph`) to convert Python code into graph-generating code.\n",
"_____no_output_____"
]
],
[
[
"def simple_relu(x):\n if tf.greater(x, 0):\n return x\n else:\n return 0\n\n# `tf_simple_relu` is a TensorFlow `Function` that wraps `simple_relu`.\ntf_simple_relu = tf.function(simple_relu)\n\nprint(\"First branch, with graph:\", tf_simple_relu(tf.constant(1)).numpy())\nprint(\"Second branch, with graph:\", tf_simple_relu(tf.constant(-1)).numpy())",
"_____no_output_____"
]
],
[
[
"Though it is unlikely that you will need to view graphs directly, you can inspect the outputs to check the exact results. These are not easy to read, so no need to look too carefully!",
"_____no_output_____"
]
],
[
[
"# This is the graph-generating output of AutoGraph.\nprint(tf.autograph.to_code(simple_relu))",
"_____no_output_____"
],
[
"# This is the graph itself.\nprint(tf_simple_relu.get_concrete_function(tf.constant(1)).graph.as_graph_def())",
"_____no_output_____"
]
],
[
[
"Most of the time, `tf.function` will work without special considerations. However, there are some caveats, and the [tf.function guide](./function.ipynb) can help here, as well as the [complete AutoGraph reference](https://github.com/tensorflow/tensorflow/blob/master/tensorflow/python/autograph/g3doc/reference/index.md)",
"_____no_output_____"
],
[
"### Polymorphism: one `Function`, many graphs\n\nA `tf.Graph` is specialized to a specific type of inputs (for example, tensors with a specific [`dtype`](https://www.tensorflow.org/api_docs/python/tf/dtypes/DType) or objects with the same [`id()`](https://docs.python.org/3/library/functions.html#id])).\n\nEach time you invoke a `Function` with new `dtypes` and shapes in its arguments, `Function` creates a new `tf.Graph` for the new arguments. The `dtypes` and shapes of a `tf.Graph`'s inputs are known as an **input signature** or just a **signature**.\n\nThe `Function` stores the `tf.Graph` corresponding to that signature in a `ConcreteFunction`. **A `ConcreteFunction` is a wrapper around a `tf.Graph`.**\n",
"_____no_output_____"
]
],
[
[
"@tf.function\ndef my_relu(x):\n return tf.maximum(0., x)\n\n# `my_relu` creates new graphs as it observes more signatures.\nprint(my_relu(tf.constant(5.5)))\nprint(my_relu([1, -1]))\nprint(my_relu(tf.constant([3., -3.])))",
"_____no_output_____"
]
],
[
[
"If the `Function` has already been called with that signature, `Function` does not create a new `tf.Graph`.",
"_____no_output_____"
]
],
[
[
"# These two calls do *not* create new graphs.\nprint(my_relu(tf.constant(-2.5))) # Signature matches `tf.constant(5.5)`.\nprint(my_relu(tf.constant([-1., 1.]))) # Signature matches `tf.constant([3., -3.])`.",
"_____no_output_____"
]
],
[
[
"Because it's backed by multiple graphs, a `Function` is **polymorphic**. That enables it to support more input types than a single `tf.Graph` could represent, as well as to optimize each `tf.Graph` for better performance.",
"_____no_output_____"
]
],
[
[
"# There are three `ConcreteFunction`s (one for each graph) in `my_relu`.\n# The `ConcreteFunction` also knows the return type and shape!\nprint(my_relu.pretty_printed_concrete_signatures())",
"_____no_output_____"
]
],
[
[
"## Using `tf.function`\n\nSo far, you've learned how to convert a Python function into a graph simply by using `tf.function` as a decorator or wrapper. But in practice, getting `tf.function` to work correctly can be tricky! In the following sections, you'll learn how you can make your code work as expected with `tf.function`.",
"_____no_output_____"
],
[
"### Graph execution vs. eager execution\n\nThe code in a `Function` can be executed both eagerly and as a graph. By default, `Function` executes its code as a graph:\n",
"_____no_output_____"
]
],
[
[
"@tf.function\ndef get_MSE(y_true, y_pred):\n sq_diff = tf.pow(y_true - y_pred, 2)\n return tf.reduce_mean(sq_diff)",
"_____no_output_____"
],
[
"y_true = tf.random.uniform([5], maxval=10, dtype=tf.int32)\ny_pred = tf.random.uniform([5], maxval=10, dtype=tf.int32)\nprint(y_true)\nprint(y_pred)",
"_____no_output_____"
],
[
"get_MSE(y_true, y_pred)",
"_____no_output_____"
]
],
[
[
"To verify that your `Function`'s graph is doing the same computation as its equivalent Python function, you can make it execute eagerly with `tf.config.run_functions_eagerly(True)`. This is a switch that **turns off `Function`'s ability to create and run graphs**, instead executing the code normally.",
"_____no_output_____"
]
],
[
[
"tf.config.run_functions_eagerly(True)",
"_____no_output_____"
],
[
"get_MSE(y_true, y_pred)",
"_____no_output_____"
],
[
"# Don't forget to set it back when you are done.\ntf.config.run_functions_eagerly(False)",
"_____no_output_____"
]
],
[
[
"However, `Function` can behave differently under graph and eager execution. The Python [`print`](https://docs.python.org/3/library/functions.html#print) function is one example of how these two modes differ. Let's check out what happens when you insert a `print` statement to your function and call it repeatedly.\n",
"_____no_output_____"
]
],
[
[
"@tf.function\ndef get_MSE(y_true, y_pred):\n print(\"Calculating MSE!\")\n sq_diff = tf.pow(y_true - y_pred, 2)\n return tf.reduce_mean(sq_diff)",
"_____no_output_____"
]
],
[
[
"Observe what is printed:",
"_____no_output_____"
]
],
[
[
"error = get_MSE(y_true, y_pred)\nerror = get_MSE(y_true, y_pred)\nerror = get_MSE(y_true, y_pred)",
"_____no_output_____"
]
],
[
[
"Is the output surprising? **`get_MSE` only printed once even though it was called *three* times.**\n\nTo explain, the `print` statement is executed when `Function` runs the original code in order to create the graph in a process known as [\"tracing\"](function.ipynb#tracing). **Tracing captures the TensorFlow operations into a graph, and `print` is not captured in the graph.** That graph is then executed for all three calls **without ever running the Python code again**.\n\nAs a sanity check, let's turn off graph execution to compare:",
"_____no_output_____"
]
],
[
[
"# Now, globally set everything to run eagerly to force eager execution.\ntf.config.run_functions_eagerly(True)",
"_____no_output_____"
],
[
"# Observe what is printed below.\nerror = get_MSE(y_true, y_pred)\nerror = get_MSE(y_true, y_pred)\nerror = get_MSE(y_true, y_pred)",
"_____no_output_____"
],
[
"tf.config.run_functions_eagerly(False)",
"_____no_output_____"
]
],
[
[
"`print` is a *Python side effect*, and there are [other differences](function#limitations) that you should be aware of when converting a function into a `Function`.",
"_____no_output_____"
],
[
"Note: If you would like to print values in both eager and graph execution, use `tf.print` instead.",
"_____no_output_____"
],
[
"###`tf.function` best practices\n\nIt may take some time to get used to the behavior of `Function`. To get started quickly, first-time users should play around with decorating toy functions with `@tf.function` to get experience with going from eager to graph execution.\n\n*Designing for `tf.function`* may be your best bet for writing graph-compatible TensorFlow programs. Here are some tips:\n- Toggle between eager and graph execution early and often with `tf.config.run_functions_eagerly` to pinpoint if/ when the two modes diverge.\n- Create `tf.Variable`s\noutside the Python function and modify them on the inside. The same goes for objects that use `tf.Variable`, like `keras.layers`, `keras.Model`s and `tf.optimizers`.\n- Avoid writing functions that [depend on outer Python variables](function#depending_on_python_global_and_free_variables), excluding `tf.Variable`s and Keras objects.\n- Prefer to write functions which take tensors and other TensorFlow types as input. You can pass in other object types but [be careful](function#depending_on_python_objects)!\n- Include as much computation as possible under a `tf.function` to maximize the performance gain. For example, decorate a whole training step or the entire training loop.\n",
"_____no_output_____"
],
[
"## Seeing the speed-up",
"_____no_output_____"
],
[
"`tf.function` usually improves the performance of your code, but the amount of speed-up depends on the kind of computation you run. Small computations can be dominated by the overhead of calling a graph. You can measure the difference in performance like so:",
"_____no_output_____"
]
],
[
[
"x = tf.random.uniform(shape=[10, 10], minval=-1, maxval=2, dtype=tf.dtypes.int32)\n\ndef power(x, y):\n result = tf.eye(10, dtype=tf.dtypes.int32)\n for _ in range(y):\n result = tf.matmul(x, result)\n return result",
"_____no_output_____"
],
[
"print(\"Eager execution:\", timeit.timeit(lambda: power(x, 100), number=1000))",
"_____no_output_____"
],
[
"power_as_graph = tf.function(power)\nprint(\"Graph execution:\", timeit.timeit(lambda: power_as_graph(x, 100), number=1000))",
"_____no_output_____"
]
],
[
[
"`tf.function` is commonly used to speed up training loops, and you can learn more about it in [Writing a training loop from scratch](keras/writing_a_training_loop_from_scratch#speeding-up_your_training_step_with_tffunction) with Keras.\n\nNote: You can also try [`tf.function(jit_compile=True)`](https://www.tensorflow.org/xla#explicit_compilation_with_tffunctionjit_compiletrue) for a more significant performance boost, especially if your code is heavy on TF control flow and uses many small tensors.",
"_____no_output_____"
],
[
"### Performance and trade-offs\n\nGraphs can speed up your code, but the process of creating them has some overhead. For some functions, the creation of the graph takes more time than the execution of the graph. **This investment is usually quickly paid back with the performance boost of subsequent executions, but it's important to be aware that the first few steps of any large model training can be slower due to tracing.**\n\nNo matter how large your model, you want to avoid tracing frequently. The `tf.function` guide discusses [how to set input specifications and use tensor arguments](function#controlling_retracing) to avoid retracing. If you find you are getting unusually poor performance, it's a good idea to check if you are retracing accidentally.",
"_____no_output_____"
],
[
"## When is a `Function` tracing?\n\nTo figure out when your `Function` is tracing, add a `print` statement to its code. As a rule of thumb, `Function` will execute the `print` statement every time it traces.",
"_____no_output_____"
]
],
[
[
"@tf.function\ndef a_function_with_python_side_effect(x):\n print(\"Tracing!\") # An eager-only side effect.\n return x * x + tf.constant(2)\n\n# This is traced the first time.\nprint(a_function_with_python_side_effect(tf.constant(2)))\n# The second time through, you won't see the side effect.\nprint(a_function_with_python_side_effect(tf.constant(3)))",
"_____no_output_____"
],
[
"# This retraces each time the Python argument changes,\n# as a Python argument could be an epoch count or other\n# hyperparameter.\nprint(a_function_with_python_side_effect(2))\nprint(a_function_with_python_side_effect(3))",
"_____no_output_____"
]
],
[
[
"New Python arguments always trigger the creation of a new graph, hence the extra tracing.\n",
"_____no_output_____"
],
[
"## Next steps\n\nYou can learn more about `tf.function` on the API reference page and by following the [Better performance with `tf.function`](function) guide.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
e78eb0f91799a388cf2a9adc1fa0a7a0d6ad9a4b | 20,506 | ipynb | Jupyter Notebook | posts/whaiout.ipynb | wcmckee/wcmckee.com | 44a7fe2d9ce64d115128fb29ac9a1457727f4315 | [
"MIT"
] | null | null | null | posts/whaiout.ipynb | wcmckee/wcmckee.com | 44a7fe2d9ce64d115128fb29ac9a1457727f4315 | [
"MIT"
] | null | null | null | posts/whaiout.ipynb | wcmckee/wcmckee.com | 44a7fe2d9ce64d115128fb29ac9a1457727f4315 | [
"MIT"
] | null | null | null | 25.956962 | 768 | 0.475519 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
e78ec914b5e1eb6fd6b5c7dcb128b6b9333ba5e0 | 3,678 | ipynb | Jupyter Notebook | 2020/paula/day3/advent_code_day3.ipynb | bbglab/adventofcode | 65b6d8331d10f229b59232882d60024b08d69294 | [
"MIT"
] | null | null | null | 2020/paula/day3/advent_code_day3.ipynb | bbglab/adventofcode | 65b6d8331d10f229b59232882d60024b08d69294 | [
"MIT"
] | null | null | null | 2020/paula/day3/advent_code_day3.ipynb | bbglab/adventofcode | 65b6d8331d10f229b59232882d60024b08d69294 | [
"MIT"
] | 3 | 2016-12-02T09:20:42.000Z | 2021-12-01T13:31:07.000Z | 18.298507 | 52 | 0.401849 | [
[
[
"# Day 3",
"_____no_output_____"
],
[
"Part 1",
"_____no_output_____"
]
],
[
[
"\"\"\"\nRight 3, down 1.\n\"\"\"\ni=1\ntrees=0\nwith open('input_day3.txt','r') as file:\n for line in file:\n if line.strip()[i%31-1] == '#':\n trees+=1\n i+=3\n ",
"_____no_output_____"
],
[
"trees",
"_____no_output_____"
]
],
[
[
"Part 2",
"_____no_output_____"
]
],
[
[
"\"\"\"\nRight 1, down 1.\n\"\"\"\ni=1\ntrees1=0\nwith open('input_day3.txt','r') as file:\n for line in file:\n if line.strip()[i%31-1] == '#':\n trees1+=1\n i+=1\n ",
"_____no_output_____"
],
[
"\"\"\"\nRight 5, down 1.\n\"\"\"\ni=1\ntrees2=0\nwith open('input_day3.txt','r') as file:\n for line in file:\n if line.strip()[i%31-1] == '#':\n trees2+=1\n i+=5\n ",
"_____no_output_____"
],
[
"\"\"\"\nRight 7, down 1.\n\"\"\"\ni=1\ntrees3=0\nwith open('input_day3.txt','r') as file:\n for line in file:\n if line.strip()[i%31-1] == '#':\n trees3+=1\n i+=7\n ",
"_____no_output_____"
],
[
"\"\"\"\nRight 1, down 2.\n\"\"\"\ni=1\ntrees4=0\nj=0\nwith open('input_day3.txt','r') as file:\n for line in file:\n if j%2 == 0:\n if line.strip()[i%31-1] == '#':\n trees4+=1\n i+=1\n j+=1",
"_____no_output_____"
],
[
"print(trees,trees1,trees2,trees3,trees4)",
"278 90 88 98 45\n"
],
[
"trees*trees1*trees2*trees3*trees4",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78ecc7682afc93edff8eaaa19ebc41a5ef1b143 | 6,670 | ipynb | Jupyter Notebook | Section 4/4.2/StockAnalysis-MovingAverage.ipynb | PacktPublishing/Getting-Started-with-Haskell-Data-Analysis-Video- | ceeb3af004397dbfbfb127a83172c12361d05308 | [
"MIT"
] | 4 | 2019-06-17T08:53:46.000Z | 2021-11-08T22:30:38.000Z | Chapter04/4.2/StockAnalysis-MovingAverage.ipynb | PacktPublishing/Getting-Started-with-Haskell-Data-Analysis | dd66dc857f972864494c5b7afa43ad84d524fd1a | [
"MIT"
] | null | null | null | Chapter04/4.2/StockAnalysis-MovingAverage.ipynb | PacktPublishing/Getting-Started-with-Haskell-Data-Analysis | dd66dc857f972864494c5b7afa43ad84d524fd1a | [
"MIT"
] | 1 | 2021-05-14T20:19:45.000Z | 2021-05-14T20:19:45.000Z | 19.97006 | 76 | 0.453373 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
e78ed16f6d103a984b1fe0cbb9b27a907545806d | 1,032 | ipynb | Jupyter Notebook | Day 2 - Operators.ipynb | naveen-kumar-123/30-Days-of-Code--with-python--HackerRank | af9e9dd9e9c980f95c66e7698b51ccf97a6789b8 | [
"MIT"
] | 1 | 2020-10-29T07:01:13.000Z | 2020-10-29T07:01:13.000Z | Day 2 - Operators.ipynb | naveen-kumar-123/30-Days-of-Code--with-python--HackerRank | af9e9dd9e9c980f95c66e7698b51ccf97a6789b8 | [
"MIT"
] | null | null | null | Day 2 - Operators.ipynb | naveen-kumar-123/30-Days-of-Code--with-python--HackerRank | af9e9dd9e9c980f95c66e7698b51ccf97a6789b8 | [
"MIT"
] | null | null | null | 20.235294 | 70 | 0.516473 | [
[
[
"#!/bin/python3\n\nimport math\nimport os\nimport random\nimport re\nimport sys\n\nmeal_cost = float(input())\ntip_perc = int(input())\ntax_perc = int(input())\n\ntip = ( meal_cost * (tip_perc /100))\ntax = (meal_cost * (tax_perc/100))\ntotal_bill = meal_cost + tip + tax\n\nprint(\"The total meal cost is %d dollars.\" %round(total_bill))",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code"
]
] |
e78ef5e572d96741e1dd8cce1b36cd75a79154f6 | 584,966 | ipynb | Jupyter Notebook | Mod2-3-ML-SL-SVM.ipynb | QianLab/ICPE-639 | 7d20369fcddcc9cd2fa89ebec061c68fbd796b3c | [
"MIT"
] | null | null | null | Mod2-3-ML-SL-SVM.ipynb | QianLab/ICPE-639 | 7d20369fcddcc9cd2fa89ebec061c68fbd796b3c | [
"MIT"
] | null | null | null | Mod2-3-ML-SL-SVM.ipynb | QianLab/ICPE-639 | 7d20369fcddcc9cd2fa89ebec061c68fbd796b3c | [
"MIT"
] | 3 | 2021-09-22T20:17:17.000Z | 2021-09-27T03:45:01.000Z | 584,966 | 584,966 | 0.94544 | [
[
[
"\n# <font color=#770000>ICPE 639 Introduction to Machine Learning </font>\n\n## ------ With Energy Applications\n\nSome of the examples and exercises of this course are based on several books as well as open-access materials on machine learning, including [Hands-on Machine Learning with Scikit-Learn, Keras and TensorFlow](https://www.oreilly.com/library/view/hands-on-machine-learning/9781492032632/)\n\n\n<p> © 2021: Xiaoning Qian </p>\n\n[Homepage](http://xqian37.github.io/)\n\n**<font color=blue>[Note]</font>** This is currently a work in progress, will be updated as the material is tested in the class room.\n\nAll material open source under a Creative Commons license and free for use in non-commercial applications.\n\nSource material used under the Creative Commons Attribution-NonCommercial 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc/3.0/ or send a letter to Creative Commons, PO Box 1866, Mountain View, CA 94042, USA.",
"_____no_output_____"
],
[
"# Support Vector Machine\n\nThis section will cover the content listed below: \n\n- [1 Support Vector Machine](#1-Support-Vector-Machine)\n- [2 Support Vector Regression](#2-Support-Vector-Regression)\n- [3 One-class SVM](#3-One-class-SVM)\n- [4 Hands-on Exercise](#4-Exercise)\n- [Reference](#Reference)",
"_____no_output_____"
]
],
[
[
"import warnings\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom IPython.display import Image\nfrom IPython.core.display import HTML\n\n%matplotlib inline\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
]
],
[
[
"## 1 Support Vector Machine\n\n### 1.1 Introduction\n\nWe will start the introduction of Support Vector Machine (SVM) for classification problems---Support Vector Classifier (SVC). Consider a simple binary classification problem. Assume we have a linearly separable data in 2-d feature space. We try to find a boundary that divides the data into two regions such that the misclassification can be minimized. Notice that different lines can be used as separators between samples. Depending on the line we choose, a new point marked by 'x' in the plot will be assigned to a different class label. \n\nThe problem is how *well* the derived boundaries *generalize* to the new testing points. ",
"_____no_output_____"
]
],
[
[
"from scipy import stats\nimport seaborn as sns\nfrom sklearn.datasets.samples_generator import make_blobs\n\nX, y = make_blobs(n_samples = 30, centers = 2, random_state = 0, cluster_std = 0.6)\n\nplt.figure(figsize=(9, 7))\nplt.scatter(X[:, 0], X[:, 1], c = y, s = 50, cmap = \"icefire\")\n\nxfit = np.arange(-0.5, 3.0, 0.1)\nfor m, b in [(0.0, 2.5), (0.5, 1.8), (-0.2, 3.0)]:\n plt.plot(xfit, m * xfit + b, '--k')\n\nplt.plot([0.0], [2.0], 'x', color = 'red', markersize = 10)\nplt.show()",
"_____no_output_____"
],
[
"make_blobs??",
"_____no_output_____"
]
],
[
[
"SVMs provide a way to achieve good generalizability with the intuition: rather than simply drawing a zero-width line between the classes, consider each line with a margin of certain width, meaning that we do not worry about the errors as long as the errors fall within the margin. In SVMs, the line that maximizes this margin is the one to be chosen as the optimal model. ",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(9, 7))\nplt.scatter(X[:, 0], X[:, 1], c = y, s = 50, cmap = \"icefire\")\n\n\nxfit = np.arange(-0.5, 3.0, 0.1)\nfor m, b, d in [(0.0, 2.5, 0.4), (0.5, 1.8, 0.2), (-0.2, 3.0, 0.1)]:\n yfit = m * xfit + b\n plt.plot(xfit, yfit, '-k')\n plt.fill_between(xfit, yfit - d, yfit + d, edgecolor='none',\n color='#AAAAAA', alpha=0.4)\n\nplt.plot([0.0], [2.0], 'x', color = 'red', markersize = 10)\nplt.show()",
"_____no_output_____"
]
],
[
[
"To fit a SVM model on this generated dataset: ",
"_____no_output_____"
]
],
[
[
"# for visualization\ndef plot_svc_decision_function(model, ax=None, plot_support=True):\n \"\"\"Plot the decision function for a 2D SVC\"\"\"\n if ax is None:\n ax = plt.gca()\n xlim = ax.get_xlim()\n ylim = ax.get_ylim()\n \n # create grid to evaluate model\n x = np.linspace(xlim[0], xlim[1], 30)\n y = np.linspace(ylim[0], ylim[1], 30)\n Y, X = np.meshgrid(y, x)\n xy = np.vstack([X.ravel(), Y.ravel()]).T\n P = model.decision_function(xy).reshape(X.shape)\n \n # plot decision boundary and margins\n ax.contour(X, Y, P, colors='k',\n levels=[-1, 0, 1], alpha=0.5,\n linestyles=['--', '-', '--'])\n \n # plot support vectors\n if plot_support:\n ax.scatter(model.support_vectors_[:, 0],\n model.support_vectors_[:, 1],\n s=300, linewidth=1, facecolors='none');\n ax.set_xlim(xlim)\n ax.set_ylim(ylim)",
"_____no_output_____"
],
[
"from sklearn.svm import SVC # \"Support vector classifier\"\n\nmodel = SVC(kernel='linear', C=1E10)\nmodel.fit(X, y)\n\nplt.figure(figsize = (9, 7))\nplt.scatter(X[:, 0], X[:, 1], c=y, s=50, cmap='icefire')\nplot_svc_decision_function(model)",
"_____no_output_____"
]
],
[
[
"This is the dividing line that maximizes the margin between two sets of points. There are some points touching the margin which are the pivotal elements of this fit known as the support vectors and can be returned by `support_vectors_`. A key to this classifier is that only the position of the support vectors matter. The points that are further away from the margin on the correct side do not change the fit. ",
"_____no_output_____"
]
],
[
[
"model.support_vectors_",
"_____no_output_____"
]
],
[
[
"This method can be extended to nonlinear boundaries with kernels which gives the Kernel SVM where we can map the data into higher-dimensional space defined by basis function and find a linear classifier for the nonlinear relationship. ",
"_____no_output_____"
],
[
"### 1.2 Math Formulation of SVC\n\nLet $w$ denote the model coefficient vector and $b$ intercept, which define the linear boundary. The original SVC formulation can be written as: \n\n$$\\max_{w,b} \\frac{1}{\\|w\\|},$$\n\n$$\\mbox{subject to: }\\quad y_i (w^T x_i - b) \\geq 1, \\quad \\forall i \\mbox{ in training data set.}$$\n\n\n\nIt can be rewritten as follows (allowing linearly nonseparable data):\n\n$$\\min_{w,b} \\|w\\|^2 + C \\sum_i \\epsilon_i, $$\n\n$$\\mbox{subject to: }\\quad y_i (w^T x_i - b) \\geq 1 - \\epsilon_i, \\quad \\forall i \\mbox{ in training data set.}$$\n\n",
"_____no_output_____"
],
[
"#### Primal-Dual (Optimization)\n\nThe above is a **convex programming** formulation, which can be equivalently solved in the dual form: \n\n$$\\max_{\\alpha} \\sum_i \\alpha_i - \\frac{1}{2}\\sum_{i,j} \\alpha_iy_i <x_i, x_j> y_j \\alpha_j, $$\n\n$$\\mbox{subject to: }\\quad \\sum_i \\alpha_i y_i = 0; \\quad \\alpha_i \\geq 0, \\quad \\forall i \\mbox{ in training data set.}$$\n\nOther **Karush–Kuhn–Tucker (KKT) conditions**: $w=\\sum_i \\alpha_i y_i x_i$ and $b$ derived by **support vectors**.\n\n**<font color=blue>[Note]</font>** Solving the dual form only requires the **inner product** term of the input features $<x_i, x_j>$, which can be replaced by any kernel to extend it nonlinear SVC. ",
"_____no_output_____"
],
[
"### 1.3 Example\n\nUse the labeled faces of various public figures in the Wild dataset as an example. Eight public figures are included. Each image is of size $62 \\times 47$. We can use the pixels directly as a feature, but it's more efficient to do some preprocessing before hand, e.g. extract some fundamental components. ",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import fetch_lfw_people\nfrom sklearn.svm import SVC\nfrom sklearn.decomposition import PCA\nfrom sklearn.pipeline import make_pipeline\nfrom sklearn.model_selection import train_test_split, GridSearchCV\nfrom sklearn.metrics import classification_report, confusion_matrix\n\nfaces = fetch_lfw_people(min_faces_per_person=60)\nprint(faces.target_names)\nprint(faces.images.shape)",
"Downloading LFW metadata: https://ndownloader.figshare.com/files/5976012\nDownloading LFW metadata: https://ndownloader.figshare.com/files/5976009\nDownloading LFW metadata: https://ndownloader.figshare.com/files/5976006\nDownloading LFW data (~200MB): https://ndownloader.figshare.com/files/5976015\n"
],
[
"fig, ax = plt.subplots(3, 5)\nfor i, axi in enumerate(ax.flat):\n axi.imshow(faces.images[i], cmap='bone')\n axi.set(xticks=[], yticks=[],\n xlabel=faces.target_names[faces.target[i]])",
"_____no_output_____"
],
[
"pca = PCA(n_components=150, whiten=True, random_state=42)\nsvc = SVC(kernel='rbf', class_weight='balanced')\nmodel = make_pipeline(pca, svc)\n\nXtrain, Xtest, ytrain, ytest = train_test_split(faces.data, faces.target,\n random_state=42)",
"_____no_output_____"
],
[
"param_grid = {'svc__C': [1, 5, 10],\n 'svc__gamma': [0.0001, 0.001, 0.005]}\ngrid = GridSearchCV(model, param_grid)\n\n%time grid.fit(Xtrain, ytrain)\nprint(grid.best_params_)",
"CPU times: user 40.7 s, sys: 20.8 s, total: 1min 1s\nWall time: 37.8 s\n{'svc__C': 10, 'svc__gamma': 0.001}\n"
],
[
"model = grid.best_estimator_\nyfit = model.predict(Xtest)",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(4, 6)\nfor i, axi in enumerate(ax.flat):\n axi.imshow(Xtest[i].reshape(62, 47), cmap='bone')\n axi.set(xticks=[], yticks=[])\n axi.set_ylabel(faces.target_names[yfit[i]].split()[-1],\n color='black' if yfit[i] == ytest[i] else 'red')\nfig.suptitle('Predicted Names; Incorrect Labels in Red', size=14);",
"_____no_output_____"
],
[
"print(classification_report(ytest, yfit,\n target_names=faces.target_names))",
" precision recall f1-score support\n\n Ariel Sharon 0.65 0.73 0.69 15\n Colin Powell 0.80 0.87 0.83 68\n Donald Rumsfeld 0.74 0.84 0.79 31\n George W Bush 0.92 0.83 0.88 126\nGerhard Schroeder 0.86 0.83 0.84 23\n Hugo Chavez 0.93 0.70 0.80 20\nJunichiro Koizumi 0.92 1.00 0.96 12\n Tony Blair 0.85 0.95 0.90 42\n\n accuracy 0.85 337\n macro avg 0.83 0.84 0.84 337\n weighted avg 0.86 0.85 0.85 337\n\n"
],
[
"mat = confusion_matrix(ytest, yfit)\nsns.heatmap(mat.T, square=True, annot=True, fmt='d', cbar=False,\n xticklabels=faces.target_names,\n yticklabels=faces.target_names)\nplt.xlabel('true label')\nplt.ylabel('predicted label');",
"_____no_output_____"
]
],
[
[
"## 2 Support Vector Regression\n\nThe Support Vector Regression uses the same principles as the SVM for classification. The differences are as follows:\n* The output is a real number, inifinite possibilities \n* A margin of tolerance is set in approximation to the SVM\n",
"_____no_output_____"
],
[
"\nHowever, if we only need to reduce the errors to a certain degree, meaning as long as the errors fall within an acceptable range.\n\nFor illustrution, consider the relationship between TV and Sales. The plot below shows the results of a trained SVR model on the Advertising dataset. The red line represents the line of simple linear regression fit. The gray dashed lines represent the margin of error $\\epsilon = 5$. ",
"_____no_output_____"
]
],
[
[
"from sklearn import linear_model\nadvertising = pd.read_csv('https://raw.githubusercontent.com/XiaomengYan/MachineLearning_dataset/main/Advertising.csv', usecols=[1,2,3,4])\n\n# Visualization\nX = advertising.TV\nX = X.values.reshape(-1, 1)\ny = advertising.Sales\n\n# simple linear regression\nregr = linear_model.LinearRegression()\nregr.fit(X,y)\n\nxfit = np.linspace(X.min(), X.max(), 1000).reshape(-1, 1)\nyfit = regr.predict(xfit)\nyfit_ub = yfit + 5\nyfit_lb = yfit - 5\n\nplt.figure(figsize = (10, 6))\nplt.suptitle('TV vs Sales')\nplt.scatter(X,y)\nplt.xlim(-10, 330)\nplt.xlabel(\"TV\")\nplt.ylabel(\"sales\")\nplt.plot(xfit, yfit, 'r',linewidth = 3)\nplt.plot(xfit, yfit_lb, 'g--', linewidth = 2)\nplt.plot(xfit, yfit_ub, 'g--', linewidth = 2)\nplt.vlines(x= xfit[50], ymin=yfit[50], ymax=yfit_ub[50], colors='gray', ls=':', lw=2)\ns = 'epsilon = 5'\nplt.text(xfit[50]-20, 0.5 * (yfit[50] + yfit_ub[50]), s)\nplt.vlines(xfit[50], ymin=yfit_lb[50], ymax=yfit[50], colors='gray', ls=':', lw=2)\nplt.text(xfit[50]-20, 0.5 * (yfit[50] + yfit_lb[50]), s)\n\ns = 'beta_i x_i'\nplt.text(300, yfit[-1], s, fontsize = 15)\ns = 'beta_i x_i + epsilon'\nplt.text(300, yfit_ub[-1], s, fontsize = 15)\ns = 'beta_i x_i - epsilon'\nplt.text(300, yfit_lb[-1], s, fontsize = 15)\n\nax = plt.axes()\nax.spines['top'].set_visible(False)\nax.spines['right'].set_visible(False)\nplt.show()",
"_____no_output_____"
]
],
[
[
"SVR gives us the flexibility to define how much error is acceptable in our model and will find an appropriate line (or hyperplane in higher dimensions) to fit the data. \n\nThe objective function of SVR is to minimize the $l_2$-norm of the coefficients,\n$$\\min \\frac{1}{2}||w||^2$$\nand use the error term as the constraints as following,\n$$|y_i - w^Tx_i|\\leq \\epsilon$$\n\n**<font color=blue>[Note]</font>** The math formulation of SVR is similar as SVC. Based on the same *primal-dual* trick, kernel-based SVR can be derived for nonlinear problems. \n",
"_____no_output_____"
],
[
"## 3 One-class SVM\n\nIn addition to **anomaly detection** based on the probabilistic methods, for exam these steming from hypothesis testing, SVM can also be extended for that. One of such formulations is **one-class SVM**: \n$$\\min_{R, c} R^2 + \\frac{1}{C}\\sum_i \\xi_i $$\nsubject to the following constraints: \n$$\\|x_i - c \\|^2 \\leq R^2 + \\xi_i, \\quad \\forall i; $$\nand \n$$\\xi_i \\geq 0, \\quad \\forall i.$$\n\nOr another formulation **$\\nu$-SVM**: \n$$\\min_{w,\\xi,\\rho} \\frac{1}{2}||w||^2 + \\frac{1}{\\nu n}\\sum_i \\xi_i -\\rho$$\nsubject to the following constraints: \n$$w^Tx_i \\geq \\rho - \\xi_i, \\quad \\forall i; $$\nand \n$$\\xi_i \\geq 0, \\quad \\forall i.$$\n\nhttp://rvlasveld.github.io/blog/2013/07/12/introduction-to-one-class-support-vector-machines/\n\n",
"_____no_output_____"
]
],
[
[
"from sklearn.svm import OneClassSVM\n#from sklearn.datasets import make_blobs\nfrom numpy import quantile, where, random\n#import matplotlib.pyplot as plt\n\nrandom.seed(13)\nx, _ = make_blobs(n_samples=200, centers=1, cluster_std=.3, center_box=(8, 8))\n\nplt.scatter(x[:,0], x[:,1])\nplt.show()\n\n",
"_____no_output_____"
],
[
"svm = OneClassSVM(kernel='rbf', gamma='auto', nu=0.05)\nprint(svm)\n\nsvm.fit(x)\npred = svm.predict(x)\nanom_index = where(pred==-1)\nvalues = x[anom_index]\n\nplt.scatter(x[:,0], x[:,1])\nplt.scatter(values[:,0], values[:,1], color='r')\nplt.show()\n\n",
"OneClassSVM(cache_size=200, coef0=0.0, degree=3, gamma='auto', kernel='rbf',\n max_iter=-1, nu=0.05, shrinking=True, tol=0.001, verbose=False)\n"
],
[
"svm = OneClassSVM(kernel='rbf', gamma='auto', nu=0.03)\nprint(svm)\n\npred = svm.fit_predict(x)\nscores = svm.score_samples(x)\n\nthresh = quantile(scores, 0.03)\nprint(thresh)\nindex = where(scores<=thresh)\nvalues = x[index]\n\nplt.scatter(x[:,0], x[:,1])\nplt.scatter(values[:,0], values[:,1], color='r')\nplt.show()",
"OneClassSVM(cache_size=200, coef0=0.0, degree=3, gamma='auto', kernel='rbf',\n max_iter=-1, nu=0.03, shrinking=True, tol=0.001, verbose=False)\n3.577526406228678\n"
]
],
[
[
"## Kitchen Power Usage Example\n\n[REDD](http://redd.csail.mit.edu/) dataset contains several weeks of power data for 6 different homes. Here we'll extract one house's kitchen power useage as a simple example for one-class SVM. For more implementations, please refer to [minhup's repo](https://github.com/minhup/Energy-Disaggregation) .",
"_____no_output_____"
]
],
[
[
"# Download the dataset\n!wget http://redd:[email protected]/data/low_freq.tar.bz2\n!tar -xf low_freq.tar.bz2",
"--2021-09-20 18:23:07-- http://redd:*password*@redd.csail.mit.edu/data/low_freq.tar.bz2\nResolving redd.csail.mit.edu (redd.csail.mit.edu)... 128.52.128.232\nConnecting to redd.csail.mit.edu (redd.csail.mit.edu)|128.52.128.232|:80... connected.\nHTTP request sent, awaiting response... 401 Unauthorized\nAuthentication selected: Basic realm=\"REDD Data Download\"\nReusing existing connection to redd.csail.mit.edu:80.\nHTTP request sent, awaiting response... 200 OK\nLength: 169367706 (162M) [application/x-bzip2]\nSaving to: ‘low_freq.tar.bz2’\n\nlow_freq.tar.bz2 100%[===================>] 161.52M 15.4MB/s in 14s \n\n2021-09-20 18:23:21 (11.8 MB/s) - ‘low_freq.tar.bz2’ saved [169367706/169367706]\n\n"
],
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n%matplotlib inline\nfrom IPython.display import display\nimport datetime\nimport time\nimport math\nimport warnings\nwarnings.filterwarnings(\"ignore\")\nimport glob\n\ndef read_label():\n label = {}\n for i in range(1, 7):\n hi = 'low_freq/house_{}/labels.dat'.format(i)\n label[i] = {}\n with open(hi) as f:\n for line in f:\n splitted_line = line.split(' ')\n label[i][int(splitted_line[0])] = splitted_line[1].strip() + '_' + splitted_line[0]\n return label\n\ndef read_merge_data(house):\n path = 'low_freq/house_{}/'.format(house)\n file = path + 'channel_1.dat'\n df = pd.read_table(file, sep = ' ', names = ['unix_time', labels[house][1]], \n dtype = {'unix_time': 'int64', labels[house][1]:'float64'}) \n \n num_apps = len(glob.glob(path + 'channel*'))\n for i in range(2, num_apps + 1):\n file = path + 'channel_{}.dat'.format(i)\n data = pd.read_table(file, sep = ' ', names = ['unix_time', labels[house][i]], \n dtype = {'unix_time': 'int64', labels[house][i]:'float64'})\n df = pd.merge(df, data, how = 'inner', on = 'unix_time')\n df['timestamp'] = df['unix_time'].astype(\"datetime64[s]\")\n df = df.set_index(df['timestamp'].values)\n df.drop(['unix_time','timestamp'], axis=1, inplace=True)\n return df\n\n# Extract labels and data from the dataset\nlabels = read_label()\ndf = {}\nfor i in range(1,2):\n print('House {}: '.format(i), labels[i], '\\n')\n df[i] = read_merge_data(i) ",
"House 1: {1: 'mains_1', 2: 'mains_2', 3: 'oven_3', 4: 'oven_4', 5: 'refrigerator_5', 6: 'dishwaser_6', 7: 'kitchen_outlets_7', 8: 'kitchen_outlets_8', 9: 'lighting_9', 10: 'washer_dryer_10', 11: 'microwave_11', 12: 'bathroom_gfi_12', 13: 'electric_heat_13', 14: 'stove_14', 15: 'kitchen_outlets_15', 16: 'kitchen_outlets_16', 17: 'lighting_17', 18: 'lighting_18', 19: 'washer_dryer_19', 20: 'washer_dryer_20'} \n\n"
],
[
"# Extract the time index\ndates = {}\nfor i in range(1, 2):\n dates[i] = [str(time)[:10] for time in df[i].index.values]\n dates[i] = sorted(list(set(dates[i])))\n print('House {0} data contain {1} days from {2} to {3}.'.format(i,len(dates[i]),dates[i][0], dates[i][-1]))\n print(dates[i], '\\n')",
"House 1 data contain 23 days from 2011-04-18 to 2011-05-24.\n['2011-04-18', '2011-04-19', '2011-04-20', '2011-04-21', '2011-04-22', '2011-04-23', '2011-04-24', '2011-04-25', '2011-04-26', '2011-04-27', '2011-04-28', '2011-04-30', '2011-05-01', '2011-05-02', '2011-05-03', '2011-05-06', '2011-05-07', '2011-05-11', '2011-05-12', '2011-05-13', '2011-05-22', '2011-05-23', '2011-05-24'] \n\n"
],
[
"# Plot the first 3 days power usage of house 1's kitchen\nhouse = 1\nn_days = 3\ndf1 = df[house].loc[:dates[house][n_days - 1]]\nplt.figure(figsize=(18,8))\nplt.title('kitchen_outlets_7', fontsize='15')\nplt.ylabel('Power Usage', fontsize='15')\nplt.xlabel('Time', fontsize='15')\nplt.plot(df1['kitchen_outlets_7'])",
"_____no_output_____"
],
[
"from sklearn.svm import OneClassSVM\nfrom numpy import quantile, where, random\n\nx = np.array(df1['kitchen_outlets_7']).reshape(-1, 1)\ntime = np.array(df1['kitchen_outlets_7'].index)\n\nsvm = OneClassSVM(kernel='rbf', gamma=0.001, nu=0.01)\nprint(svm)\n\nsvm.fit(x)\npred = svm.predict(x)\nanom_index = where(pred==-1)\nvalues = x[anom_index]",
"OneClassSVM(cache_size=200, coef0=0.0, degree=3, gamma=0.001, kernel='rbf',\n max_iter=-1, nu=0.01, shrinking=True, tol=0.001, verbose=False)\n"
],
[
"# Plot the prediction\n\nplt.figure(figsize=(18,8))\nplt.title('kitchen_outlets_7', fontsize='15')\nplt.ylabel('Power Usage', fontsize='15')\nplt.xlabel('Time', fontsize='15')\nplt.plot(time, x)\nplt.scatter(np.array(time)[anom_index], values, color='r')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Hands-on Exercise",
"_____no_output_____"
],
[
"\nPlease try to implement the SVM for classification of MNIST hand written digits dataset. Remember that different hyperparameters can have affect the results. \n\n1. Prepare data: Load the MNIST dataset using `load_digits` from `sklearn.datasets`\n2. Prepare the tool: load `svm` from `sklearn`\n3. Split the data into training set and test set: use 70% for training and the remaining for testing, get help from `train_test_split` from `sklearn.model_selection`\n4. Select the evaluation metric to evaluate the classification result\n5. Try SVM with different settings and save the accuracy score in a dictionary with key being `kernel name_C`\n * $C = [0.001, 0.1, 0.5, 1, 10, 100]$ (Regularization parameter. The strength of the regularization is inversely proportional to C. Must be strictly positive. The penalty is a squared l2 penalty.)\n * kernel = \\{‘linear’, ‘poly’, ‘rbf’, ‘sigmoid’\\} (Specifies the kernel type to be used in the algorithm.)\n 6. Visualize the first 4 results in the test set using polynomial kernel with $C = 0.1$.",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.svm import SVC\n\n# loading data\nfrom sklearn.datasets import load_digits\n\ndata = load_digits()\nX, y = data.data, data.target\nX_train, X_test, y_train, y_test = train_test_split(\n X, y, test_size=0.3, random_state=17 #test_size controls the proportion of test data in the whole data\n)\nn_samples = len(data.images)\nn_train = X_train.shape[0]\nprint(n_samples, n_train)",
"1797 1257\n"
],
[
"# Use loop to do the cross-validation\nC_list = [0.001, 0.1, 0.5, 1, 10, 100]\nkernel_list = ['linear', 'poly', 'rbf', 'sigmoid']\n\naccuracy_score_dict = dict()\nfor cc in C_list:\n for kern in kernel_list:\n acc_i = []\n for i in np.arange(5):\n if i == 0:\n train_idx = np.arange(0, n_train) >= (i+1) * 251\n else:\n train_idx = (np.arange(0, n_train) < i * 251) + (np.arange(0, n_train) >= (i+1) * 251)\n train_idx = train_idx > 0\n \n val_idx = (np.arange(0, n_train) >= i * 251) * (np.arange(0, n_train) < (i+1) * 251)\n clf_svm = SVC(C = cc, kernel = kern)\n clf_svm.fit(X_train[train_idx, :], y_train[train_idx])\n svm_pred = clf_svm.predict(X_train[val_idx, :])\n acc_i.append(accuracy_score(y_train[val_idx], svm_pred))\n accuracy_score_dict[kern + '_' + str(cc)] = np.mean(acc_i)",
"_____no_output_____"
],
[
"accuracy_score_dict",
"_____no_output_____"
],
[
"clf_svm = SVC(C = 1, kernel = 'poly')\nclf_svm.fit(X_train, y_train)\nsvm_pred = clf_svm.predict(X_test)\n\n_, axes = plt.subplots(nrows=1, ncols=4, figsize=(10, 3))\nfor ax, image, prediction in zip(axes, X_test, svm_pred):\n ax.set_axis_off()\n image = image.reshape(8, 8)\n ax.imshow(image, cmap=plt.cm.gray_r, interpolation='nearest')\n ax.set_title(f'Prediction: {prediction}')",
"_____no_output_____"
],
[
"# Use GridSearchCV\nparams_grid = {\n 'C': [0.001, 0.1, 0.5, 1, 10, 100],\n 'kernel': ['linear', 'poly', 'rbf', 'sigmoid']\n}\n\ngrid = GridSearchCV(SVC(), params_grid, cv = 5, scoring = 'accuracy')\ngrid.fit(X_train, y_train)\nselsvc = grid.best_estimator_\nsvm_pred = selsvc.predict(X_test)\nprint(grid.best_estimator_)\n\n_, axes = plt.subplots(nrows=1, ncols=4, figsize=(10, 3))\nfor ax, image, prediction in zip(axes, X_test, svm_pred):\n ax.set_axis_off()\n image = image.reshape(8, 8)\n ax.imshow(image, cmap=plt.cm.gray_r, interpolation='nearest')\n ax.set_title(f'Prediction: {prediction}')",
"SVC(C=1, break_ties=False, cache_size=200, class_weight=None, coef0=0.0,\n decision_function_shape='ovr', degree=3, gamma='scale', kernel='poly',\n max_iter=-1, probability=False, random_state=None, shrinking=True,\n tol=0.001, verbose=False)\n"
]
],
[
[
"## Reference\n\n* [An Idiot's guide to Support vector machines - MIT](https://www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=&cad=rja&uact=8&ved=2ahUKEwiRtuO39ervAhXVK80KHTjLDGIQFjAMegQIERAD&url=http%3A%2F%2Fweb.mit.edu%2F6.034%2Fwwwbob%2Fsvm-notes-long-08.pdf&usg=AOvVaw3_uFIYSaBhhk_23fPFso52)\n* [Support Vector Machine — Simply Explained](https://towardsdatascience.com/support-vector-machine-simply-explained-fee28eba5496)\n* [Understaing support vector machine algorithm with example](https://www.analyticsvidhya.com/blog/2017/09/understaing-support-vector-machine-example-code/)",
"_____no_output_____"
],
[
"# Questions? ",
"_____no_output_____"
]
],
[
[
"Image(url= \"https://mirrors.creativecommons.org/presskit/buttons/88x31/png/by-nc-sa.png\", width=100)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
e78efee3a815989ec571a1a15783f2ee5895d05b | 9,001 | ipynb | Jupyter Notebook | 10a_viseme_tabular_identify_landmarks.ipynb | pete88b/expoco | 29c6673cba0647bb64242628a8507fc26fbdbdfd | [
"Apache-2.0"
] | null | null | null | 10a_viseme_tabular_identify_landmarks.ipynb | pete88b/expoco | 29c6673cba0647bb64242628a8507fc26fbdbdfd | [
"Apache-2.0"
] | null | null | null | 10a_viseme_tabular_identify_landmarks.ipynb | pete88b/expoco | 29c6673cba0647bb64242628a8507fc26fbdbdfd | [
"Apache-2.0"
] | null | null | null | 37.504167 | 478 | 0.583046 | [
[
[
"#default_exp viseme_tabular.identify_landmarks",
"_____no_output_____"
],
[
"#all_do_not_test",
"_____no_output_____"
]
],
[
[
"# Identify face mesh landmarks\n\n> Identify a subset of face mesh landmarks.",
"_____no_output_____"
],
[
"How can we identify all the landmarks around the mouth?\nWe could use [mesh_map.jpg](https://github.com/tensorflow/tfjs-models/blob/master/facemesh/mesh_map.jpg) and type out IDs of all the landmarks we're interested in but ... that'll take a while and will be hard to do without making any mistakes.\n\nHow about we,\n- specify just 4 landmarks\n - to specify the left, top, right and bottom of a bounding box\n- then find all other landmarks that are in this bounding box?",
"_____no_output_____"
]
],
[
[
"from expoco.core import *\nimport numpy as np\nimport cv2, time, math\nimport win32api, win32con\n\nimport mediapipe as mp\nmp_face_mesh = mp.solutions.face_mesh",
"_____no_output_____"
],
[
"from collections import namedtuple\nBoundingLandmarks = namedtuple('BoundingLandmarks', 'left, top, right, bottom')\nmouth_bounding_landmarks = BoundingLandmarks(57, 164, 287, 18)",
"_____no_output_____"
],
[
"class FacePointHelper:\n def __init__(self, image_height, image_width, bounding_landmarks):\n self.image_height, self.image_width = image_height, image_width\n self.bounding_landmarks = bounding_landmarks\n self.face_mesh = mp_face_mesh.FaceMesh(max_num_faces=1)\n \n def process(self, image):\n self.results = self.face_mesh.process(image) # cv2.cvtColor(image, cv2.COLOR_BGR2RGB) already done\n return self.results\n \n def get_bounding_box(self, pixel_coordinates=True):\n fn = self._landmark_to_pixel_coordinates if pixel_coordinates else self._landmark_to_x_y\n bls = BoundingLandmarks(*[fn(i) for i in self.bounding_landmarks])\n return [bls.left[0], bls.top[1]], [bls.right[0], bls.bottom[1]]\n \n def get_bound_landmarks(self):\n result = []\n [left, top], [right, bottom] = self.get_bounding_box(False)\n for i in range(468): # len(self.results.multi_face_landmarks[0])\n landmark = self._landmark_to_x_y(i)\n if left <= landmark[0] <= right and top <= landmark[1] <= bottom:\n result.append(i)\n return result\n \n def _landmark(self, i):\n return self.results.multi_face_landmarks[0].landmark[i] # [0] is OK as we're running with max_num_faces=1\n \n def _is_valid_normalized_value(self, value):\n return (value > 0 or math.isclose(0, value)) and (value < 1 or math.isclose(1, value))\n \n def _normalized_x_to_pixel(self, value):\n return math.floor(value * self.image_width)\n \n def _normalized_y_to_pixel(self, value):\n return math.floor(value * self.image_height)\n \n def _landmark_to_x_y(self, landmark):\n if isinstance(landmark, int):\n landmark = self._landmark(landmark)\n if not (self._is_valid_normalized_value(landmark.x) and self._is_valid_normalized_value(landmark.y)):\n print(f'WARNING: {landmark.x} or {landmark.y} is not a valid normalized value')\n return landmark.x, landmark.y\n \n def _landmark_to_pixel_coordinates(self, landmark):\n x, y = self._landmark_to_x_y(landmark)\n return self._normalized_x_to_pixel(x), self._normalized_y_to_pixel(y)",
"_____no_output_____"
],
[
"def annotate_image(face_point_helper, image):\n if not face_point_helper.results.multi_face_landmarks:\n return image\n image = cv2.rectangle(image, *face_point_helper.get_bounding_box(), (130, 0, 130))\n for i in face_point_helper.get_bound_landmarks():\n point = face_point_helper._landmark_to_pixel_coordinates(i)\n image = cv2.circle(image, point, radius=1, color=(100,0,0), thickness=-1)\n return image",
"_____no_output_____"
]
],
[
[
"run the following cell to see the bounding box and the landmarks it encloses.\n\npress `ESC` to print all landmarks enclosed by the bounding box and stop capture",
"_____no_output_____"
]
],
[
[
"try: video_capture.release()\nexcept: pass\nvideo_capture = cv2.VideoCapture(0) \nface_mesh = mp_face_mesh.FaceMesh(max_num_faces=1)\nfor vk in [win32con.VK_ESCAPE, ord('D')]: win32api.GetAsyncKeyState(vk)\nretval, image = video_capture.read()\nface_point_helper = FacePointHelper(*image.shape[:2], mouth_bounding_landmarks)\nimage_display_helper = ImageDisplayHelper(cv2.flip(image, 1), 'expoco: Dry Run')\nwhile True:\n retval, image = video_capture.read()\n image = cv2.flip(image, 1)\n results = face_point_helper.process(cv2.cvtColor(image, cv2.COLOR_BGR2RGB))\n image_display_helper.show(annotate_image(face_point_helper, image))\n if win32api.GetAsyncKeyState(win32con.VK_ESCAPE): \n print(face_point_helper.get_bound_landmarks())\n video_capture.release()\n break\n time.sleep(.05)",
"_____no_output_____"
]
],
[
[
"output for `mouth_bounding_landmarks` should be;\n```\n[0, 11, 12, 13, 14, 15, 16, 17, 18, 37, 38, 39, 40, 41, 42, 43, 57, 61, 62, 72, 73, 74, 76, 77, 78, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 95, 96, 106, 146, 164, 165, 167, 178, 179, 180, 181, 182, 183, 184, 185, 186, 191, 204, 267, 268, 269, 270, 271, 272, 273, 287, 291, 292, 302, 303, 304, 306, 307, 308, 310, 311, 312, 313, 314, 315, 316, 317, 318, 319, 320, 321, 322, 324, 325, 335, 375, 391, 393, 402, 403, 404, 405, 406, 407, 408, 409, 410, 415, 424]\n```",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e78f0d51c1187338144507797f1dc50277b1ea6f | 148,591 | ipynb | Jupyter Notebook | 2020_Workshop/Alex_Python/PrettyPlot.ipynb | imedan/AstroPAL_Coding_Workshop | ef4d018065d4c3da0bd734ec37d20800b6a74352 | [
"BSD-3-Clause"
] | null | null | null | 2020_Workshop/Alex_Python/PrettyPlot.ipynb | imedan/AstroPAL_Coding_Workshop | ef4d018065d4c3da0bd734ec37d20800b6a74352 | [
"BSD-3-Clause"
] | null | null | null | 2020_Workshop/Alex_Python/PrettyPlot.ipynb | imedan/AstroPAL_Coding_Workshop | ef4d018065d4c3da0bd734ec37d20800b6a74352 | [
"BSD-3-Clause"
] | null | null | null | 505.411565 | 96,196 | 0.934512 | [
[
[
"#import Python stuff\nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nddir='' #your data directory, like '/Users/ayep/research/data'; starting with \"/Users\" makes code easily portable to other computers :)\npdir='' #your plot directory, like '/Users/ayep/research/plot'\n\ndef opendat(dir,filename): #dir,'filename'. For opening a data file. Can then send through roundtable.\n f=open(dir+filename,'r')\n dat=f.readlines()\n f.close()\n labels=dat[0][0:-1].split()\n dat2=[[a.strip('\\n') for a in d.split('\\t')] for d in dat if d[0]!='#']\n dat3=[['nan' if a.strip()=='' else a for a in d] for d in dat2]\n return [dat3,labels]\ndef opendat2(dirr,filename,params): #Use as var,var,var...=opendat2(dir,'filename',['keys']).\n dat,label=opendat(dirr,filename) #Get keys by first leaving ['keys'] blank: opendat2(dirr,filename,[])\n print(label)\n varrs=[]\n for i in range(len(params)):\n j=label.index(params[i])\n try:\n var=np.array([float(d[j]) for d in dat]) #works for float.\n varrs.append(var)\n except ValueError:\n var=[d[j].strip() for d in dat] #works for strings.\n varrs.append(var)\n return varrs\n\ndef writedat(dirr,filename,label,pars): #.dat auto included. pars as [name,ra,dec] etc.\n datp=[[str(a[i]) for a in pars] for i in range(len(pars[0]))]\n f=open(dirr+filename+'.dat','w')\n print('\\t'.join(label),file=f)\n print(label)\n for d in datp:\n print('\\t'.join(d),file=f)\n f.close()\n print('It is written: '+filename+'.dat')\n \ndef erm(val,err): #list,list\n v=np.array(val)\n e=np.array(err)\n w=1.0/e**2.0\n avg=np.nansum(w*v)/np.nansum(w)\n avgerr=1.0/np.sqrt(np.nansum(w))\n return avg,avgerr",
"_____no_output_____"
]
],
[
[
"General plotting tip: You can get pretty named colors from https://python-graph-gallery.com/196-select-one-color-with-matplotlib/ and unnamed colors from https://htmlcolorcodes.com/.",
"_____no_output_____"
]
],
[
[
"#Load data\n\n#UPK 535\nra535,dec535,p535,pra535,pdec535,rv535a,G535,B535,R535,spt535,d535,binf535=opendat2(ddir,'UPK535_combined.dat',['#ra', 'dec', 'p','pra','pdec', 'rv', 'G', 'B', 'R', 'spt','d','binaryflag'])\np535err,pra535err,pdec535err,rv535aerr,G535err,B535err,R535err,spt535err,d535perr,d535merr=opendat2(ddir,'UPK535_combined.dat',['perr','praerr','pdecerr', 'rverr', 'Gerr', 'Berr', 'Rerr', 'spterr','dp','dm'])\n#clean rv for simulation: already threw out my rverr>10 but gotta check new Gaia rvs.\nrv535aa=[rv535a[i] if rv535aerr[i]<10. else np.float('nan') for i in range(len(ra535))]\nrv535aaerr=[rv535aerr[i] if rv535aerr[i]<10. else np.float('nan') for i in range(len(ra535))]\n#neutralize all binaries.\nbindev=5.\nrv535med=np.nanmedian(rv535aa)\nbinn535=[binf535[i] if binf535[i]!='nan' else 'SB1?' if abs(rv535aa[i]-rv535med)>bindev else 'nan' for i in range(len(ra535))]\n#calculate cluster rv from nonbinary rvs and replace all nans with cluster rv.\nrv535aaa=[rv535aa[i] if 'SB' not in binn535[i] else np.float('nan') for i in range(len(ra535))]\nrv535aaaerr=[rv535aaerr[i] if 'SB' not in binn535[i] else np.float('nan') for i in range(len(ra535))]\nrvcl535,rvcl535err=erm(rv535aaa,rv535aaaerr)[0],np.nanstd(rv535aaa)\nprint('UPK535:',rvcl535,'+/-',rvcl535err)\n#clean rv:\nrv535=[rvcl535 if np.isnan(r) else r for r in rv535aaa]\nrv535err=[rvcl535err if np.isnan(rv535aaa[i]) else rv535aaaerr[i] for i in range(len(ra535))]\n\n#Theia 120\nra120,dec120,p120,pra120,pdec120,rv120a,G120,B120,R120,spt120,d120,binf120=opendat2(ddir,'Theia120_combined.dat',['#ra', 'dec', 'p','pra','pdec', 'rv', 'G', 'B', 'R', 'spt','d','binaryflag'])\np120err,pra120err,pdec120err,rv120aerr,G120err,B120err,R120err,spt120err,d120perr,d120merr=opendat2(ddir,'Theia120_combined.dat',['perr','praerr','pdecerr', 'rverr', 'Gerr', 'Berr', 'Rerr', 'spterr','dp','dm'])\nBR120=np.array(B120)-np.array(R120)\nBR120err=np.sqrt(np.array(B120)**2.+np.array(R120)**2.)\n#clean rv for simulation: already threw out my rverr>10 but gotta check new Gaia rvs.\nrv120aa=[rv120a[i] if rv120aerr[i]<10. else np.float('nan') for i in range(len(ra120))]\nrv120aaerr=[rv120aerr[i] if rv120aerr[i]<10. else np.float('nan') for i in range(len(ra120))]\n#neutralize all binaries.\nbindev=5.\nrv120med=np.nanmedian(rv120aa)\nbinn120=[binf120[i] if binf120[i]!='nan' else 'SB1?' if abs(rv120aa[i]-rv120med)>bindev else 'nan' for i in range(len(ra120))]\n#calculate cluster rv from nonbinary rvs and replace all nans with cluster rv.\nrv120aaa=[rv120aa[i] if 'SB' not in binn120[i] else np.float('nan') for i in range(len(ra120))]\nrv120aaaerr=[rv120aaerr[i] if 'SB' not in binn120[i] else np.float('nan') for i in range(len(ra120))]\nrvcl120,rvcl120err=erm(rv120aaa,rv120aaaerr)[0],np.nanstd(rv120aaa)\nprint('Theia120:',rvcl120,'+/-',rvcl120err)\n#clean rv:\nrv120=[rvcl120 if np.isnan(r) else r for r in rv120aaa]\nrv120err=[rvcl120err if np.isnan(rv120aaa[i]) else rv120aaaerr[i] for i in range(len(ra120))]",
"['#ra', 'dec', 'p', 'perr', 'pra', 'praerr', 'pdec', 'pdecerr', 'rv', 'rverr', 'G', 'Gerr', 'B', 'Berr', 'R', 'Rerr', 'V', 'Verr', 'spt', 'spterr', 'binaryflag', 'd', 'dm', 'dp']\n['#ra', 'dec', 'p', 'perr', 'pra', 'praerr', 'pdec', 'pdecerr', 'rv', 'rverr', 'G', 'Gerr', 'B', 'Berr', 'R', 'Rerr', 'V', 'Verr', 'spt', 'spterr', 'binaryflag', 'd', 'dm', 'dp']\nUPK535: 10.149414529091349 +/- 1.5322115646924208\n['#ra', 'dec', 'p', 'perr', 'pra', 'praerr', 'pdec', 'pdecerr', 'rv', 'rverr', 'G', 'Gerr', 'B', 'Berr', 'R', 'Rerr', 'V', 'Verr', 'spt', 'spterr', 'binaryflag', 'd', 'dm', 'dp']\n['#ra', 'dec', 'p', 'perr', 'pra', 'praerr', 'pdec', 'pdecerr', 'rv', 'rverr', 'G', 'Gerr', 'B', 'Berr', 'R', 'Rerr', 'V', 'Verr', 'spt', 'spterr', 'binaryflag', 'd', 'dm', 'dp']\nTheia120: 19.206145691070176 +/- 1.5132951173143845\n"
],
[
"plt.rcParams.update({'font.size':22,'lines.linewidth':4, 'font.family':'serif','mathtext.fontset':'dejavuserif'})\n\n# Histogram\nf,((a11,a12),(a21,a22))=plt.subplots(2,2,figsize=(12,12),gridspec_kw = {'wspace':0.24,'hspace':0.28})\nf.add_subplot(111, frameon=False,xticks=[],yticks=[]) #for tight layout\n\nhpars1=[d535,[a for a in rv535aaa if np.isnan(a)==False],pra535,pdec535]\nhpars2=[d120,[a for a in rv120aaa if np.isnan(a)==False],pra120,pdec120]\nmins=[290,5,-15,1]\nmaxs=[380,22,-10,14]\nints=[10,1,0.5,1]\naxs=[a11,a21,a12,a22]\nlabs=['Distance (pc)','$v_r$ (km s$^{-1}$)','$\\mu_{\\\\alpha}$ (mas yr$^{-1}$)','$\\mu_{\\delta}$ (mas yr$^{-1}$)']\nfor i in range(4):\n par1=hpars1[i]\n par2=hpars2[i]\n minn=mins[i]\n maxx=maxs[i]\n intt=ints[i]\n ax=axs[i]\n beans=np.arange(minn,maxx,intt)\n h535,w535=np.histogram(par1,beans)\n h120,w120=np.histogram(par2,beans)\n #plt.figure(figsize=(10,10))\n print(len(h535),len(w535))\n ax.bar(w120[:-1],h120/len(par2),color='#008FFF',width=intt)\n ax.bar(w535[:-1],h535/len(par1),color='blue',width=intt)\n overlap=[np.min([h120[k]/len(par2),h535[k]/len(par1)]) for k in range(len(h120))]\n ax.bar(w120[:-1],overlap,color='#00068D',width=intt)\n ax.set_xlabel(labs[i])\nplt.ylabel('Normalized Frequency\\n\\n')\nplt.savefig(pdir+'CC_Histograms.png',bbox_inches='tight')",
"8 9\n16 17\n9 10\n12 13\n"
]
],
[
[
"## Color-Mapping!",
"_____no_output_____"
],
[
"This basically gives you a 3rd dimension. Go wild! Color maps can be chosen from here: https://matplotlib.org/3.1.1/gallery/color/colormap_reference.html",
"_____no_output_____"
],
[
"I personally like rainbow, warm, cool, and RdYlBu.",
"_____no_output_____"
],
[
"You can also use lists for sizes to give different sizes to each data point. :) That's yet another dimension you can use!",
"_____no_output_____"
],
[
"Be warned that the color bar can be a wily creature when you're adding it to plots with subplots. I often actually create a separate axis for it when that happens and calibrate its size by hand. D: With one plot though, it's easy. :)",
"_____no_output_____"
]
],
[
[
"#3-D position with ra, dec, AND distance:\n\nplt.rcParams.update({'font.size':22,'lines.linewidth':4, 'font.family':'serif','mathtext.fontset':'dejavuserif'})\nf=plt.figure(figsize=(10,10))\ncm=plt.scatter(ra535,dec535,c=d535,cmap='rainbow',s=[d if d<300 else 300 for d in 10000./(d535-300)])\ncbar = f.colorbar(cm,label='\\nDistance (pc)')\nplt.xlabel('RA ($^{o}$)')\nplt.ylabel('Dec ($^{o}$)')\n",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
e78f1c968c5df119ff1766b233961c11d306b986 | 7,717 | ipynb | Jupyter Notebook | notebooks/hw2_answers.ipynb | cstorm125/abtest | 1d47c7144d693e67d897e6939119b23f9764f67d | [
"Apache-2.0"
] | 12 | 2019-04-23T03:12:39.000Z | 2020-09-16T06:00:44.000Z | notebooks/hw2_answers.ipynb | peaky2000/abtestoo | 7c3cac8d18a333d4e398a7e75ffdcb1aaf0bebf4 | [
"Apache-2.0"
] | null | null | null | notebooks/hw2_answers.ipynb | peaky2000/abtestoo | 7c3cac8d18a333d4e398a7e75ffdcb1aaf0bebf4 | [
"Apache-2.0"
] | 27 | 2020-10-08T19:22:58.000Z | 2021-11-29T11:09:45.000Z | 58.022556 | 503 | 0.593365 | [
[
[
"# Homework #2 - A/B Testing in the Wild\n\n1. The Law of Large Numbers (LLN) says that sample mean will converge to expectation as sample size grows. Assuming that this is true, prove that sample variance will converge to variance as sample size grows. \n\n\\begin{align}\ns^2 &= \\frac{1}{n}\\sum_{i=1}^{n}(X_i - \\bar{X}^2) \\\\\n&= \\frac{1}{n}\\sum_{i=1}^{n}(X_i - \\mu)^2 \\text{; as }n\\rightarrow\\infty\\text{ }\\bar{X}\\rightarrow\\mu\\\\\n&=\\frac{1}{n}(\\sum_{i=1}^{n}{X_i}^2 - 2\\mu\\sum_{i=1}^{n}X_i + n\\mu^2) \\\\\n&=\\frac{\\sum_{i=1}^{n}{X_i}^2}{n} - \\frac{2\\mu\\sum_{i=1}^{n}X_i}{n} + \\mu^2 \\\\\n&= \\frac{\\sum_{i=1}^{n}{X_i}^2}{n} - 2\\mu\\bar{X} + \\mu^2\\text{; as }\\frac{\\sum_{i=1}^{n}X_i}{n} = \\bar{X}\\\\\n&= \\frac{\\sum_{i=1}^{n}{X_i}^2}{n} - 2\\mu^2 + \\mu^2 = \\frac{\\sum_{i=1}^{n}{X_i}^2}{n} - \\mu^2 \\text{; as }n\\rightarrow\\infty\\text{ }\\bar{X}\\rightarrow\\mu\\\\\n&= E[{X_i}^2] - E[X_i]^2 = Var(X_i) = \\sigma^2\n\\end{align}\n\n2. What is p-value? (Choose one or more)\n\n* [x] Assuming that the null hypothesis is true, what is the probability of observing the current or more extreme data.\n\n* [ ] Based on the observed data, what is the probability of the null hypothesis being true.\n\n* [ ] Based on the observed data, what is the probability of the null hypothesis being false.\n\n* [ ] Assuming that our hypothesis is true, what is the chance that we reject the null hypothesis.\n\n3. If we conduct a frequentist statistical test at 5% significance level repeatedly for 4,000 times, how many times can we expect to have statistically significant results even if group A and B are exactly the same? - 200 times as false positive rate is expected to be 5%\n\n4. Hamster Inc. once again wants to test the conversion rates between package colors of its sunflower seeds; this time it is Red Package vs Gold Package. The Red Package is the existing group with average conversion rate of 11%. If they think the minimum detectable effect is 1% and want to make a 80/20 control/test split, how many unique users should see each package color before we decide which one performs better? Assume that they are testing at significance level of 15%. Show your work.\n\n\\begin{align}\nZ_{\\alpha} &= \\frac{\\text{MDE}-\\mu}{\\sqrt{\\sigma^2 * (\\frac{1}{n} + \\frac{1}{mn})}} \\\\\n\\frac{(m+1)\\sigma^2}{mn} &= (\\frac{\\text{MDE}}{Z_{\\alpha}})^2 \\\\\nn &= \\frac{m+1}{m}(\\frac{Z_{\\alpha} \\sigma}{\\text{MDE}})^2 \\\\\nn &= \\frac{5}{4}(\\frac{Z_{\\alpha} \\sigma}{\\text{MDE}})^2; m=4 \\text{ due to 80/20 split} \\\\\nn &= \\frac{5}{4}(\\frac{1.03643 \\sigma}{\\text{MDE}})^2; Z_{0.15, one-tailed}=1.03643 \\text{ at 15% significance, find out which one is better} \\\\\nn &= \\frac{5}{4}(\\frac{1.03643}{\\text{MDE}})^2 * 0.0979; \\sigma^2 = p*(1-p) = 0.11*(1-0.11) = 0.0979 \\text{ assuming sample variance of control is equal to pooled variance} \\\\\nn &= \\frac{5}{4}(\\frac{1.03643}{0.01})^2 * 0.0979; MDE = 0.01 \\\\\nn &= 1314.5451165869708\n\\end{align}\n\nWe will need a test group of 1,315 unique visitors and 5,260 unique visitors for the control group.\n\n5. Let us say Hamster Inc. ran the experiment and got the following results. \n\n| campaign_id | clicks | conv_cnt | conv_per |\n|------------:|-------:|---------:|---------:|\n| Red | 59504 | 5901 | 0.099170 |\n| Gold | 58944 | 6012 | 0.101995 |\n\n5.1 At significance level of 7%, which variation should be chosen to run at 100% traffic? Show your work.\n\nNull hypothesis is that the conversion rate of Red package is higher than the conversion rate of Gold package. We run a one-tailed Z-test at 7% significance level and reject the null hypothesis. We should run the Gold package at 100% traffic.\n\n\\begin{align}\np_\\text{pooled} &= \\frac{5901+6012}{59504+58944} \\\\\n&= 0.10057578008915305 \\\\\n\\sigma^2_\\text{pooled} &= p_\\text{pooled} * (1-p_\\text{pooled}) \\\\\n&= 0.10057578008915305 * (1-0.10057578008915305) \\\\ \n&= 0.09046029254861138 \\\\\n\\bar{X_\\Delta} &= 0.101995 - 0.099170 = 0.002825\\\\\nZ_\\Delta &= \\frac{\\bar{X_\\Delta}-\\mu}{\\sqrt{\\frac{\\sigma^2_\\text{A}}{n_\\text{A}} + \\frac{\\sigma^2_\\text{B}}{n_\\text{B}}}} \\\\\n&= \\frac{0.002825-0}{\\sqrt{0.09046029254861138 * (\\frac{1}{59504} + \\frac{1}{58944})}} \\\\\n&= 1.6162869810704354 \\\\\np_\\Delta &= 0.053026 < 0.07\n\\end{align}\n\n5.2 What are the confidence intervals at 7% significance of conversion rates for Red and Gold? Show your work.\n\n\\begin{align}\nCI &= \\bar{x} \\pm Z_{0.07,one-tailed} * \\sqrt{\\frac{\\sigma^2}{n}} \\\\\nCI &= \\bar{x} \\pm 1.475791 * \\sqrt{\\frac{\\sigma^2}{n}} \\\\\nCI_{red} &= 0.099170 \\pm 1.475791 * \\sqrt{\\frac{(0.099170)*(1-0.099170)}{59504}} \\\\\n&=(0.09736172968006342, 0.10097827031993657) \\\\\nCI_{gold} &= 0.101995 \\pm 1.475791 * \\sqrt{\\frac{(0.101995)*(1-0.101995)}{58944}} \\\\\n&=(0.10015535563734503, 0.10383464436265498)\n\\end{align}\n\n6. Which of the following are true about frequentist A/B tests? (True/False)\n\n* [T] It does not tell us the magnitude of the difference between control and test groups. - we can only incorporate the concept of minimum detectable effect to say what is the minimum magnitude of the difference we want\n\n* [F] We can never know when to stop the experiments. - we know the required samples via minimum detectable effect\n\n* [T] We can never determine if the null hypothesis being true. - we always assume the null hypothesis is true when calculating p-value\n\n* [F] We can run one or as many experiments as we want using the same significance level. - this will lead to https://xkcd.com/882/\n\n* [T] If we have too many samples in each group, the validity of the test can be jeopardized. - thus we need to give minimum detectable effect\n\n* [T] If you have set up the experiment based on desired minimum detectable effect and significance level, statististical significance is the only factor in determining which group is the better one. - since we already incorporate the power we needed via those assumptions\n\n* [F] We can only test difference between two proportions. - there are other tests such as t-tests for continuous variables \n\n* [F] More samples in control and test groups are always better. - as it leads to statistical significance when there is none\n",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown"
]
] |
e78f28a04dc5be14e27c83d899d43e1b04365985 | 53,142 | ipynb | Jupyter Notebook | source/lesson04/script.ipynb | psteinb/deeplearning540.github.io | 1186266f187e7529423c0a1ecdaf2d9949074b55 | [
"MIT"
] | null | null | null | source/lesson04/script.ipynb | psteinb/deeplearning540.github.io | 1186266f187e7529423c0a1ecdaf2d9949074b55 | [
"MIT"
] | null | null | null | source/lesson04/script.ipynb | psteinb/deeplearning540.github.io | 1186266f187e7529423c0a1ecdaf2d9949074b55 | [
"MIT"
] | null | null | null | 83.034375 | 15,968 | 0.826126 | [
[
[
"# Lesson 04: Classification Performance ROCs\n\n- evaluating and comparing trained models is of extreme importance when deciding in favor/against\n + model architectures\n + hyperparameter sets\n \n- evaluating performance or quality of prediction is performed with a myriad of tests, figure-of-merits and even statistical hypothesis testing\n- in the following, the rather popular \"Receiver Operating Characteristic\" curve (spoken ROC curve)\n- the ROC was invented in WWII by radar engineers when seeking to detect enemy vessels and comparing different devices/techniques\n",
"_____no_output_____"
],
[
"## preface\n- two main ingredients to ROC:\n\n + TPR = True Positive Rate\n + FPR = False Positive Rate\n ",
"_____no_output_____"
],
[
"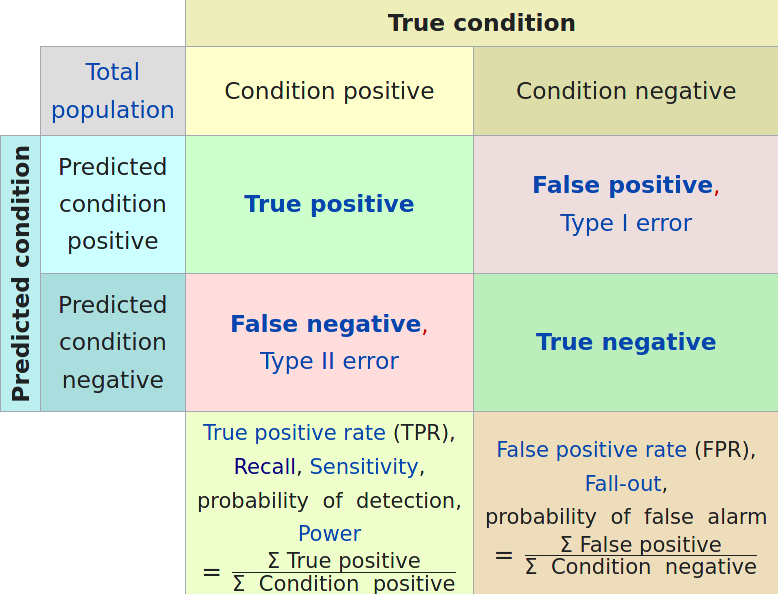\n\n- $TPR = \\frac{TP}{TP+FN}$ also known as `recall`, always within $[0,1]$\n- $FPR = \\frac{FP}{FP+TN}$ also known as `fall-out`, always within $[0,1]$",
"_____no_output_____"
],
[
"## Data\n\nFor the following, I will rely (again) on the Palmer penguin dataset obtained from [this repo](https://github.com/allisonhorst/palmerpenguins). To quote the repo:\n\n> Data were collected and made available by [Dr. Kristen Gorman](https://www.uaf.edu/cfos/people/faculty/detail/kristen-gorman.php)\n> and the [Palmer Station, Antarctica LTER](https://pal.lternet.edu/), a member of the [Long Term Ecological Research Network](https://lternet.edu/).\n",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"df = pd.read_csv(\"https://raw.githubusercontent.com/allisonhorst/palmerpenguins/master/inst/extdata/penguins.csv\")\n#let's remove the rows with NaN values\ndf = df[ df.bill_length_mm.notnull() ]\n#convert species column to \ndf[[\"species_\"]] = df[[\"species\"]].astype(\"category\")\n",
"_____no_output_____"
],
[
"print(df.shape)\nprint((df.species_.cat.codes < 1).shape)\n\n#create binary column\ndf[\"is_adelie\"] = (df.species_.cat.codes < 1).astype(np.int8)\n\nprint(df.head())\n",
"(342, 9)\n(342,)\n species island bill_length_mm bill_depth_mm flipper_length_mm \\\n0 Adelie Torgersen 39.1 18.7 181.0 \n1 Adelie Torgersen 39.5 17.4 186.0 \n2 Adelie Torgersen 40.3 18.0 195.0 \n4 Adelie Torgersen 36.7 19.3 193.0 \n5 Adelie Torgersen 39.3 20.6 190.0 \n\n body_mass_g sex year species_ is_adelie \n0 3750.0 male 2007 Adelie 1 \n1 3800.0 female 2007 Adelie 1 \n2 3250.0 female 2007 Adelie 1 \n4 3450.0 female 2007 Adelie 1 \n5 3650.0 male 2007 Adelie 1 \n"
],
[
"import matplotlib.pyplot as plt\nplt.style.use('dark_background')\nimport seaborn as sns\nprint(f'seaborn version: {sns.__version__}')",
"seaborn version: 0.10.1\n"
],
[
"from sklearn.neighbors import KNeighborsClassifier as knn\nfrom sklearn.model_selection import train_test_split\n\nkmeans = knn(n_neighbors=5)",
"_____no_output_____"
],
[
"#this time we train the knn algorithm, i.e. an unsupervised method is used in a supervised fashion\n#prepare the data\nX = np.stack((df.bill_length_mm, df.flipper_length_mm), axis=-1)\ny = df.is_adelie\n\nprint(X.shape)\nprint(y.shape)\n",
"(342, 2)\n(342,)\n"
],
[
"X_train, X_test, y_train, y_test = train_test_split(X, y,\n test_size = .15,\n random_state = 20210303)\n\nprint(X_train.shape)\nprint(y_train.shape)\n\n\nprint(X_test.shape)\nprint(y_test.shape)\n\n",
"(290, 2)\n(290,)\n(52, 2)\n(52,)\n"
],
[
"kmeans = kmeans.fit(X_train, y_train)\n",
"_____no_output_____"
],
[
"from sklearn.metrics import confusion_matrix, ConfusionMatrixDisplay\n\ny_test_hat = kmeans.predict(X_test)\n\ncm = confusion_matrix( y_test, y_test_hat )\n\nprint(cm)\n",
"[[26 1]\n [ 3 22]]\n"
],
[
"from sklearn.metrics import ConfusionMatrixDisplay\ndisp = ConfusionMatrixDisplay(confusion_matrix=cm)\ndisp.plot()\nprint(int(True))",
"1\n"
]
],
[
[
"# Starting to ROC\n\n- let's take 4 samples of different size from our test set (as if we would conduct 4 experiments)\n",
"_____no_output_____"
]
],
[
[
"n_experiments = 4\n\nX_test_exp = np.split(X_test[:32,...],n_experiments,axis=0)\ny_test_exp = np.split(y_test.values[:32,...],n_experiments,axis=0)\n\nprint(X_test_exp[0].shape)\nprint(y_test_exp[0].shape)\ny_test_exp",
"(8, 2)\n(8,)\n"
],
[
"y_test_hat = kmeans.predict(X_test)\ny_test_hat_exp = np.split(y_test_hat[:32,...],n_experiments,axis=0)\n\n",
"_____no_output_____"
],
[
"#let's compute tpr and fpr for each\nfrom sklearn.metrics import recall_score as tpr\n\ndef fpr(y_true, y_pred):\n \"\"\" compute the false positive rate using the confusion_matrix\"\"\"\n cm = confusion_matrix(y_true, y_pred)\n assert cm.shape == (2,2), f\"{y_true.shape, y_pred.shape} => {cm,cm.shape}\"\n cond_negative = cm[:,1].sum()\n value = cm[0,1] / cond_negative\n return value\n\ntpr_ = []\nfpr_ = []\n\nfor i in range(len(y_test_exp)):\n\n tpr_.append(tpr(y_test_exp[i], y_test_hat_exp[i]))\n fpr_.append(fpr(y_test_exp[i], y_test_hat_exp[i]))\n\nprint(tpr_)\nprint(fpr_)",
"[0.8333333333333334, 1.0, 1.0, 1.0]\n[0.0, 0.2, 0.0, 0.0]\n"
],
[
"f, ax = plt.subplots(1)\n\nax.plot(fpr_, tpr_, 'ro', markersize=10)\nax.set_xlabel('False Positive Rate')\nax.set_ylabel('True Positive Rate')\nax.set_xlim(0,1)\nax.set_ylim(0,1)\n",
"_____no_output_____"
]
],
[
[
"# But how to get from single entries to a full curve?\n\n- in our case, we can employ the positive class prediction probabilities\n- for KNN, this is given by the amount of N(true label)/N in the neighborhood around a query point",
"_____no_output_____"
]
],
[
[
"kmeans.predict_proba(X_test[:10])\n",
"_____no_output_____"
]
],
[
[
"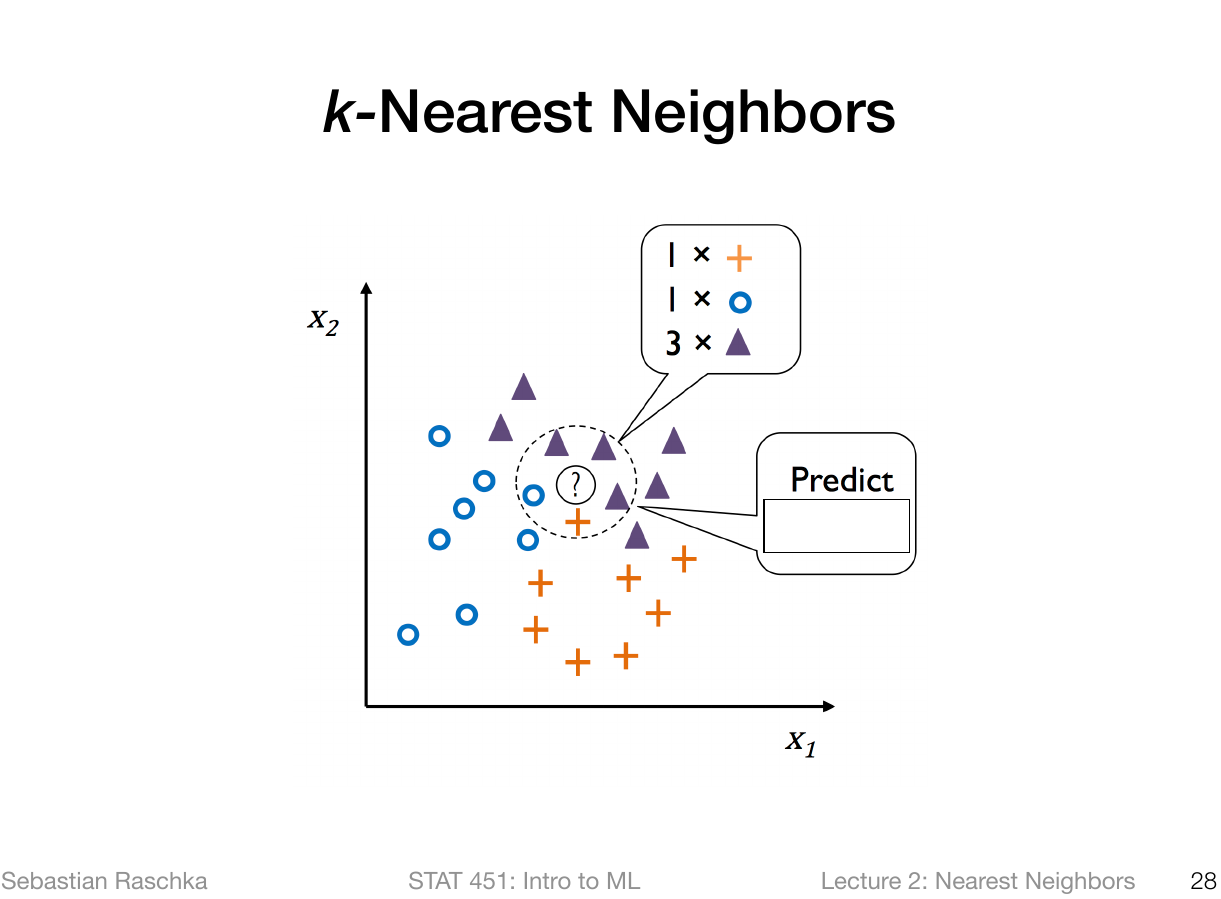\n- demonstrates how kNN classifyer is similar to `RandomForests`, `SVM`, ... :\n + spacial interpretation of the class prediction probability\n + the higher the probability for a sample, the more likely the sample belongs to `Adelie` in our case (i.e. the positive class in a binary classification setup)\n ",
"_____no_output_____"
],
[
"- relating this again to \n\n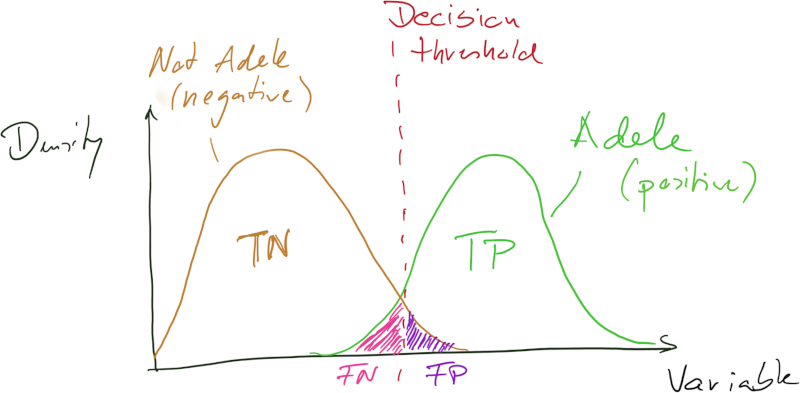\n\nthe decision threshold for a `5`-neighborhood with a binary classification task is `0.6`, i.e. 3 of 5 neighbors have the positive class (then our query point will get the positive class assigned)\n\n- knowing these positive class prediction probabilities, I can now draw an envelope that gives me the ROC from the test set as with these probabilites and the theoretical threshold, we can compute FPR and TPR\n\n",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import roc_curve\n\nprobs = kmeans.predict_proba(X_test)\npos_pred_probs = probs[:,-1]\n\nfpr, tpr, thr = roc_curve(y_test, pos_pred_probs)\n\nprint('false positive rate\\n',fpr)\nprint('true positive rate\\n',tpr)\nprint('thresholds\\n',thr)\n\n",
"false positive rate\n [0. 0. 0. 0.07407407 0.11111111 1. ]\ntrue positive rate\n [0. 0.8 0.88 0.88 0.92 1. ]\nthresholds\n [2. 1. 0.8 0.4 0.2 0. ]\n"
],
[
"from sklearn.metrics import RocCurveDisplay\n\nroc = RocCurveDisplay.from_estimator(kmeans, X_test, y_test)\n",
"_____no_output_____"
]
],
[
[
"- difference to plot with single entries? (size of test set -> only discrete values for single experiments, limited amount of samples)\n- summary of curve possible: AUC = area under curve\n- TPR and FPR used for ROC only -> similar plots possible with other variables, e.g. precision_recall_curve",
"_____no_output_____"
],
[
"# Take-Aways\n\n<p><a href=\"https://commons.wikimedia.org/wiki/File:Roc-draft-xkcd-style.svg#/media/File:Roc-draft-xkcd-style.svg\"><img src=\"https://upload.wikimedia.org/wikipedia/commons/thumb/3/36/Roc-draft-xkcd-style.svg/640px-Roc-draft-xkcd-style.svg.png\" alt=\"Roc-draft-xkcd-style.svg\"></a><br>By <a href=\"//commons.wikimedia.org/wiki/User:MartinThoma\" title=\"User:MartinThoma\">MartinThoma</a> - <span class=\"int-own-work\" lang=\"en\">Own work</span>, <a href=\"http://creativecommons.org/publicdomain/zero/1.0/deed.en\" title=\"Creative Commons Zero, Public Domain Dedication\">CC0</a>, <a href=\"https://commons.wikimedia.org/w/index.php?curid=70212136\">Link</a></p>\n\n- nearest neighbor clustering algorithms are able to offer a probabilistic score for each predicted datum based on the neighborhood chosen \n- the ROC is an envelope that describes how well a classifyer performs given a fixed testset\n- ROC expresses the balance between true-positives and false positives\n",
"_____no_output_____"
],
[
"# Further Reading\n\n- some parts of this material were inspired by [Sebastian Raschka](https://sebastianraschka.com)\n + [lecture 12.4, Receiver Operating Curve](https://youtu.be/GdSEkiArM3k)\n \n- a generally good resource \n + [Confusion_matrix](https://en.wikipedia.org/wiki/Confusion_matrix)\n \n- all of the above is nicely implemented and documented \n + [sklearn examples](https://scikit-learn.org/stable/auto_examples/model_selection/plot_roc.html#sphx-glr-auto-examples-model-selection-plot-roc-py)\n + [roc_curve API docs](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.roc_curve.html#sklearn.metrics.roc_curve)\n\n- [extensive discussion of ROC](https://stackabuse.com/understanding-roc-curves-with-python/)\n ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
e78f3bbae74f2e47bf69cb181146b4e5ae8559e7 | 8,074 | ipynb | Jupyter Notebook | debug_nvdbapilesv3/vegobjekter/sjekkmangler.ipynb | LtGlahn/diskusjon_diverse | 15e408e7acf78ffcf873f5fed79fd2996554ab51 | [
"MIT"
] | null | null | null | debug_nvdbapilesv3/vegobjekter/sjekkmangler.ipynb | LtGlahn/diskusjon_diverse | 15e408e7acf78ffcf873f5fed79fd2996554ab51 | [
"MIT"
] | null | null | null | debug_nvdbapilesv3/vegobjekter/sjekkmangler.ipynb | LtGlahn/diskusjon_diverse | 15e408e7acf78ffcf873f5fed79fd2996554ab51 | [
"MIT"
] | null | null | null | 24.246246 | 300 | 0.48811 | [
[
[
"# Mangelfulle spørringer mot NVDB api V3? \n\nher sammenligner vi resultatet av spørringen `/vegobjekter/904?kommune=5001&veg(system)referanse=K` mot NVDB api V2 og V3.",
"_____no_output_____"
]
],
[
[
"import json\nimport pandas as pd\nimport os",
"_____no_output_____"
],
[
"mappe = 'stavanger_904'\nfiler = os.listdir(mappe)",
"_____no_output_____"
],
[
"manglergeom = []\nfasit = [] \n\netterspurt_E = []\netterspurt_R = []\netterspurt_F = []\netterspurt_K = []\netterspurt_P = []\netterspurt_S = []\n\n\nfor eifil in filer: \n if 'json' in eifil: \n spurt_vegkat = eifil.split( '.')[0].split( '_')[-1]\n with open( os.path.join( mappe, eifil), encoding='utf-8') as f:\n data = json.load( f)\n for rad in data: \n rad['spurt_vegkat'] = spurt_vegkat\n \n if 'mengdeuttak_v2' in eifil:\n fasit.extend( data)\n elif 'mangler_geometriv3' in eifil: \n manglergeom.extend( data ) \n \n\nmanglergeom = pd.DataFrame( manglergeom )\nfasit = pd.DataFrame( fasit )",
"_____no_output_____"
]
],
[
[
"# Har vi duplikater i de ugyldige V3-objektene? ",
"_____no_output_____"
]
],
[
[
"duplikat = manglergeom[ manglergeom.duplicated(subset='id', keep=False) ]",
"_____no_output_____"
],
[
"print( len( duplikat), 'duplikater av', len( manglergeom), 'ugyldige og', len(fasit), 'totalt')",
"4 duplikater av 155 ugyldige og 5648 totalt\n"
]
],
[
[
"Relativt uinteressant, iallfall inntil videre",
"_____no_output_____"
],
[
"# Lær mer om ugyldige data",
"_____no_output_____"
],
[
"### Stemmer ønsket vegkategori med det vi fant? ",
"_____no_output_____"
]
],
[
[
"mangler = pd.merge( left=manglergeom, right=fasit, left_on='id', right_on='vegobjektid', how='inner')",
"_____no_output_____"
],
[
"mangler.columns",
"_____no_output_____"
],
[
"mangler[ mangler['spurt_vegkat_x'] == mangler['vegkat']]",
"_____no_output_____"
]
],
[
[
"### Er det en vegkategori som peker seg ut?",
"_____no_output_____"
]
],
[
[
"mangler.vegkat.value_counts()",
"_____no_output_____"
]
],
[
[
"Sjekker ut den skogsbilvegen og de to europaveg-objektene ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e78f43aac0221b6fda66be109862669946b0e247 | 944,200 | ipynb | Jupyter Notebook | wms_sample.ipynb | petercunning/notebook | 5b26f2dc96bcb36434542b397de6ca5fa3b61a0a | [
"MIT"
] | 32 | 2015-01-07T01:48:05.000Z | 2022-03-02T07:07:42.000Z | wms_sample.ipynb | petercunning/notebook | 5b26f2dc96bcb36434542b397de6ca5fa3b61a0a | [
"MIT"
] | 1 | 2015-04-13T21:00:18.000Z | 2015-04-13T21:00:18.000Z | wms_sample.ipynb | petercunning/notebook | 5b26f2dc96bcb36434542b397de6ca5fa3b61a0a | [
"MIT"
] | 30 | 2015-01-28T09:31:29.000Z | 2022-03-07T03:08:28.000Z | 1,136.22142 | 251,807 | 0.940153 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
e78f69c0e7d697475800db6c58a4f4ac5ae23b34 | 25,339 | ipynb | Jupyter Notebook | Model_Stephan_pH7.ipynb | HannahDi/EnzymeML_KineticModeling | bef23140353b984519d8e8e7f8306ecad2f1c52a | [
"BSD-2-Clause"
] | null | null | null | Model_Stephan_pH7.ipynb | HannahDi/EnzymeML_KineticModeling | bef23140353b984519d8e8e7f8306ecad2f1c52a | [
"BSD-2-Clause"
] | null | null | null | Model_Stephan_pH7.ipynb | HannahDi/EnzymeML_KineticModeling | bef23140353b984519d8e8e7f8306ecad2f1c52a | [
"BSD-2-Clause"
] | null | null | null | 83.078689 | 17,600 | 0.820238 | [
[
[
"# Estimate Paramters of system of ODEs for given time course data",
"_____no_output_____"
],
[
"imports",
"_____no_output_____"
]
],
[
[
"import pandas as pd # convert excel to dataframe\nimport numpy as np # convert dataframe to nparray for solver\nfrom scipy.integrate import odeint # solve ode\nfrom lmfit import minimize, Parameters, Parameter, report_fit # fitting\nimport matplotlib.pyplot as plt # plot data and results",
"_____no_output_____"
]
],
[
[
"### Get data from excel",
"_____no_output_____"
]
],
[
[
"data = './datasets/Stephan_pH7.xlsx'\ndf = pd.read_excel(data, sheet_name=1)",
"_____no_output_____"
]
],
[
[
"### Convert dataframe to np-array",
"_____no_output_____"
]
],
[
[
"# time:\ndata_time = df[df.columns[0]].to_numpy(np.float64) #shape: (60,)\n# substrate data (absorption):\ndata_s = np.transpose(df.iloc[:,1:].to_numpy(np.float64)) #shape: (3, 60)",
"_____no_output_____"
]
],
[
[
"## Fit data to system of odes",
"_____no_output_____"
],
[
"### define the ode functions",
"_____no_output_____"
]
],
[
[
"def f(w, t, paras):\n '''\n System of differential equations\n Arguments:\n w: vector of state variables: w = [v,s]\n t: time\n params: parameters\n '''\n v, s = w\n \n try:\n a = paras['a'].value\n vmax = paras['vmax'].value\n km = paras['km'].value\n\n except KeyError:\n a, vmax, km = paras\n \n \n # f(v',s'):\n f0 = a*(vmax-v) # v'\n f1 = -v*s/(km+s) # s'\n return [f0,f1]",
"_____no_output_____"
]
],
[
[
"### Solve ODE",
"_____no_output_____"
]
],
[
[
"def g(t, w0, paras):\n '''\n Solution to the ODE w'(t)=f(t,w,p) with initial condition w(0)= w0 (= [v0, s0])\n '''\n w = odeint(f, w0, t, args=(paras,))\n return w",
"_____no_output_____"
]
],
[
[
"### compute residual between actual data (s) and fitted data",
"_____no_output_____"
]
],
[
[
"def res_multi(params, t, data_s):\n ndata, nt = data_s.shape\n resid = 0.0*data_s[:]\n # residual per data set\n for i in range(ndata):\n w0 = params['v0'].value, params['s0'].value\n model = g(t, w0, params)\n # only have data for s not v\n s_model = model[:,1]\n s_model_b = s_model + params['b'].value\n resid[i,:]=data_s[i,:]-s_model_b\n return resid.flatten()",
"_____no_output_____"
]
],
[
[
"### Bringing everything together\nInitialize parameters",
"_____no_output_____"
]
],
[
[
"# initial conditions:\nv0 = 0\ns0 = np.mean(data_s,axis=0)[0]\n\n# Set parameters including bounds\nbias = 0.1\nparams = Parameters()\nparams.add('v0', value=v0, vary=False)\nparams.add('s0', value=s0-bias, min=0.1, max=s0)\nparams.add('a', value=1., min=0.0001, max=2.)\nparams.add('vmax', value=0.2, min=0.0001, max=1.)\nparams.add('km', value=0.05, min=0.0001, max=1.)\nparams.add('b', value=bias, min=0.01, max=0.5)\n\n# time\nt_measured = data_time",
"_____no_output_____"
]
],
[
[
"Fit model and visualize results",
"_____no_output_____"
]
],
[
[
"# fit model\nresult = minimize(res_multi , params, args=(t_measured, data_s), method='leastsq') # leastsq nelder\nreport_fit(result)\n# plot the data sets and fits\nw0 = params['v0'].value, params['s0'].value\ndata_fitted = g(t_measured, w0, result.params)\nplt.figure()\n\nfor i in range(data_s.shape[0]):\n plt.plot(t_measured, data_s[i, :], 'o')\nplt.plot(t_measured, data_fitted[:, 1]+params['b'].value, '-', linewidth=2, color='red', label='fitted data')\nplt.show()",
"[[Fit Statistics]]\n # fitting method = leastsq\n # function evals = 96\n # data points = 180\n # variables = 5\n chi-square = 0.04123983\n reduced chi-square = 2.3566e-04\n Akaike info crit = -1498.63538\n Bayesian info crit = -1482.67059\n[[Variables]]\n v0: 0 (fixed)\n s0: 0.86085398 +/- 0.00534285 (0.62%) (init = 0.8780667)\n a: 1.69791467 +/- 0.15515893 (9.14%) (init = 1)\n vmax: 0.28159258 +/- 0.01082300 (3.84%) (init = 0.2)\n km: 0.04761797 +/- 0.01094650 (22.99%) (init = 0.05)\n b: 0.09725106 +/- 0.00436845 (4.49%) (init = 0.1)\n[[Correlations]] (unreported correlations are < 0.100)\n C(vmax, km) = 0.947\n C(a, vmax) = -0.895\n C(a, km) = -0.743\n C(s0, b) = -0.736\n C(km, b) = -0.638\n C(vmax, b) = -0.504\n C(a, b) = 0.333\n C(s0, km) = 0.312\n C(s0, a) = 0.211\n C(s0, vmax) = 0.122\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e78f7893a279ca827705de4a8163a9bcf55faa15 | 73,592 | ipynb | Jupyter Notebook | courses/machine_learning/deepdive/03_model_performance/labs/d_hyperparameter_tuning.ipynb | aleistra/training-data-analyst | af6a59b56437c428f852d106de6e323717380ddb | [
"Apache-2.0"
] | null | null | null | courses/machine_learning/deepdive/03_model_performance/labs/d_hyperparameter_tuning.ipynb | aleistra/training-data-analyst | af6a59b56437c428f852d106de6e323717380ddb | [
"Apache-2.0"
] | null | null | null | courses/machine_learning/deepdive/03_model_performance/labs/d_hyperparameter_tuning.ipynb | aleistra/training-data-analyst | af6a59b56437c428f852d106de6e323717380ddb | [
"Apache-2.0"
] | null | null | null | 74.71269 | 553 | 0.720228 | [
[
[
"<h1> Hyper-parameter tuning </h1>\n\n**Learning Objectives**\n1. Understand various approaches to hyperparameter tuning\n2. Automate hyperparameter tuning using AI Platform HyperTune\n\n## Introduction\n\nIn the previous notebook we achieved an RMSE of **4.13**. Let's see if we can improve upon that by tuning our hyperparameters.\n\nHyperparameters are parameters that are set *prior* to training a model, as opposed to parameters which are learned *during* training. \n\nThese include learning rate and batch size, but also model design parameters such as type of activation function and number of hidden units.\n\nHere are the four most common ways to finding the ideal hyperparameters:\n1. Manual\n2. Grid Search\n3. Random Search\n4. Bayesian Optimzation\n\n**1. Manual**\n\nTraditionally, hyperparameter tuning is a manual trial and error process. A data scientist has some intution about suitable hyperparameters which they use as a starting point, then they observe the result and use that information to try a new set of hyperparameters to try to beat the existing performance. \n\nPros\n- Educational, builds up your intuition as a data scientist\n- Inexpensive because only one trial is conducted at a time\n\nCons\n- Requires alot of time and patience\n\n**2. Grid Search**\n\nOn the other extreme we can use grid search. Define a discrete set of values to try for each hyperparameter then try every possible combination. \n\nPros\n- Can run hundreds of trials in parallel using the cloud\n- Gauranteed to find the best solution within the search space\n\nCons\n- Expensive\n\n**3. Random Search**\n\nAlternatively define a range for each hyperparamter (e.g. 0-256) and sample uniformly at random from that range. \n\nPros\n- Can run hundreds of trials in parallel using the cloud\n- Requires less trials than Grid Search to find a good solution\n\nCons\n- Expensive (but less so than Grid Search)\n\n**4. Bayesian Optimization**\n\nUnlike Grid Search and Random Search, Bayesian Optimization takes into account information from past trials to select parameters for future trials. The details of how this is done is beyond the scope of this notebook, but if you're interested you can read how it works here [here](https://cloud.google.com/blog/products/gcp/hyperparameter-tuning-cloud-machine-learning-engine-using-bayesian-optimization). \n\nPros\n- Picks values intelligenty based on results from past trials\n- Less expensive because requires fewer trials to get a good result\n\nCons\n- Requires sequential trials for best results, takes longer\n\n**AI Platform HyperTune**\n\nAI Platform HyperTune, powered by [Google Vizier](https://ai.google/research/pubs/pub46180), uses Bayesian Optimization by default, but [also supports](https://cloud.google.com/ml-engine/docs/tensorflow/hyperparameter-tuning-overview#search_algorithms) Grid Search and Random Search. \n\n\nWhen tuning just a few hyperparameters (say less than 4), Grid Search and Random Search work well, but when tunining several hyperparameters and the search space is large Bayesian Optimization is best.",
"_____no_output_____"
]
],
[
[
"# Ensure that we have Tensorflow 1.13 installed.\n!pip3 freeze | grep tensorflow==1.13.1 || pip3 install tensorflow==1.13.1",
"tensorflow==1.13.1\n"
],
[
"PROJECT = \"qwiklabs-gcp-636667ae83e902b6\" # Replace with your PROJECT\nBUCKET = \"qwiklabs-gcp-636667ae83e902b6_al\" # Replace with your BUCKET\nREGION = \"us-east1\" # Choose an available region for AI Platform \nTFVERSION = \"1.13\" # TF version for AI Platform",
"_____no_output_____"
],
[
"import os \nos.environ[\"PROJECT\"] = PROJECT\nos.environ[\"BUCKET\"] = BUCKET\nos.environ[\"REGION\"] = REGION\nos.environ[\"TFVERSION\"] = TFVERSION ",
"_____no_output_____"
]
],
[
[
"## Move code into python package\n\nLet's package our updated code with feature engineering so it's AI Platform compatible.",
"_____no_output_____"
]
],
[
[
"%%bash\nmkdir taxifaremodel\ntouch taxifaremodel/__init__.py",
"mkdir: cannot create directory ‘taxifaremodel’: File exists\n"
]
],
[
[
"## Create model.py\n\nNote that any hyperparameters we want to tune need to be exposed as command line arguments. In particular note that the number of hidden units is now a parameter.",
"_____no_output_____"
]
],
[
[
"%%writefile taxifaremodel/model.py\nimport tensorflow as tf\nimport numpy as np\nimport shutil\nprint(tf.__version__)\n\n#1. Train and Evaluate Input Functions\nCSV_COLUMN_NAMES = [\"fare_amount\",\"dayofweek\",\"hourofday\",\"pickuplon\",\"pickuplat\",\"dropofflon\",\"dropofflat\"]\nCSV_DEFAULTS = [[0.0],[1],[0],[-74.0],[40.0],[-74.0],[40.7]]\n\ndef read_dataset(csv_path):\n def _parse_row(row):\n # Decode the CSV row into list of TF tensors\n fields = tf.decode_csv(records = row, record_defaults = CSV_DEFAULTS)\n\n # Pack the result into a dictionary\n features = dict(zip(CSV_COLUMN_NAMES, fields))\n \n # NEW: Add engineered features\n features = add_engineered_features(features)\n \n # Separate the label from the features\n label = features.pop(\"fare_amount\") # remove label from features and store\n\n return features, label\n \n # Create a dataset containing the text lines.\n dataset = tf.data.Dataset.list_files(file_pattern = csv_path) # (i.e. data_file_*.csv)\n dataset = dataset.flat_map(map_func = lambda filename:tf.data.TextLineDataset(filenames = filename).skip(count = 1))\n\n # Parse each CSV row into correct (features,label) format for Estimator API\n dataset = dataset.map(map_func = _parse_row)\n \n return dataset\n\ndef train_input_fn(csv_path, batch_size = 128):\n #1. Convert CSV into tf.data.Dataset with (features,label) format\n dataset = read_dataset(csv_path)\n \n #2. Shuffle, repeat, and batch the examples.\n dataset = dataset.shuffle(buffer_size = 1000).repeat(count = None).batch(batch_size = batch_size)\n \n return dataset\n\ndef eval_input_fn(csv_path, batch_size = 128):\n #1. Convert CSV into tf.data.Dataset with (features,label) format\n dataset = read_dataset(csv_path)\n\n #2.Batch the examples.\n dataset = dataset.batch(batch_size = batch_size)\n \n return dataset\n \nvocab = {}\nvocab['dayofweek'] = [i for i in range(7)]\nvocab['hourofday'] = [i for i in range(24)]\n\n# 1. One hot encode dayofweek and hourofday\nfc_dayofweek = tf.feature_column.indicator_column(tf.feature_column.categorical_column_with_vocabulary_list(key='dayofweek',vocabulary_list=vocab['dayofweek']))\nfc_hourofday = tf.feature_column.indicator_column(tf.feature_column.categorical_column_with_vocabulary_list(key='hourofday', vocabulary_list=vocab['hourofday']))\n\n# 2. Bucketize latitudes and longitude\nNBUCKETS = 16\nlatbuckets = np.linspace(start = 38.0, stop = 42.0, num = NBUCKETS).tolist()\nlonbuckets = np.linspace(start = -76.0, stop = -72.0, num = NBUCKETS).tolist()\nfc_bucketized_plat = tf.feature_column.bucketized_column(source_column = tf.feature_column.numeric_column('pickuplat'), boundaries = latbuckets)\nfc_bucketized_plon = tf.feature_column.bucketized_column(source_column = tf.feature_column.numeric_column('dropofflat'), boundaries = latbuckets)\nfc_bucketized_dlat = tf.feature_column.bucketized_column(source_column = tf.feature_column.numeric_column('pickuplon'), boundaries = lonbuckets)\nfc_bucketized_dlon = tf.feature_column.bucketized_column(source_column = tf.feature_column.numeric_column('dropofflon'), boundaries = lonbuckets)\n\nsignedbuckets = [-5,0,5]\n\nfc_northsouth = tf.feature_column.bucketized_column(source_column = tf.feature_column.numeric_column(key='latdiff'), boundaries = signedbuckets)\nfc_eastwest = tf.feature_column.bucketized_column(source_column = tf.feature_column.numeric_column(key='londiff'), boundaries = signedbuckets)\n\n# 3. Cross features to get combination of day and hour\nfc_crossed_day_hr = tf.feature_column.indicator_column(tf.feature_column.crossed_column(keys=['dayofweek', 'hourofday'], \n hash_bucket_size=len(vocab['dayofweek'] * len(vocab['hourofday']))))\n\nfc_crossed_ns_day_hr = tf.feature_column.indicator_column(tf.feature_column.crossed_column(keys=['fc_crossed_day_hr', 'fc_northsouth'], \n hash_bucket_size=len(vocab['dayofweek'] * len(vocab['hourofday']) * 2)))\n\nfc_crossed_ew_day_hr = tf.feature_column.indicator_column(tf.feature_column.crossed_column(keys=['fc_crossed_day_hr', 'fc_eastwest'], \n hash_bucket_size=len(vocab['dayofweek'] * len(vocab['hourofday']) * 2)))\n\ndef add_engineered_features(features):\n features[\"dayofweek\"] = features[\"dayofweek\"] - 1 # subtract one since our days of week are 1-7 instead of 0-6\n \n features[\"latdiff\"] = features[\"pickuplat\"] - features[\"dropofflat\"] # East/West\n features[\"londiff\"] = features[\"pickuplon\"] - features[\"dropofflon\"] # North/South\n features[\"euclidean_dist\"] = tf.sqrt(features[\"latdiff\"]**2 + features[\"londiff\"]**2)\n\n return features\n\nfeature_cols = [\n fc_dayofweek,\n fc_hourofday,\n fc_bucketized_plat,\n fc_bucketized_plon,\n fc_bucketized_dlat,\n fc_bucketized_dlon,\n fc_crossed_day_hr,\n fc_northsouth,\n fc_eastwest,\n tf.feature_column.numeric_column(key='latdiff'),\n tf.feature_column.numeric_column(key='londiff'),\n tf.feature_column.numeric_column(key='euclidean_dist')\n]\n\n#3. Serving Input Receiver Function\ndef serving_input_receiver_fn():\n receiver_tensors = {\n 'dayofweek' : tf.placeholder(dtype = tf.int32, shape = [None]), # shape is vector to allow batch of requests\n 'hourofday' : tf.placeholder(dtype = tf.int32, shape = [None]),\n 'pickuplon' : tf.placeholder(dtype = tf.float32, shape = [None]), \n 'pickuplat' : tf.placeholder(dtype = tf.float32, shape = [None]),\n 'dropofflat' : tf.placeholder(dtype = tf.float32, shape = [None]),\n 'dropofflon' : tf.placeholder(dtype = tf.float32, shape = [None]),\n }\n \n features = add_engineered_features(receiver_tensors) # 'features' is what is passed on to the model\n \n return tf.estimator.export.ServingInputReceiver(features = features, receiver_tensors = receiver_tensors)\n \n#4. Train and Evaluate\ndef train_and_evaluate(params):\n OUTDIR = params[\"output_dir\"]\n\n model = tf.estimator.DNNRegressor(\n hidden_units = params[\"hidden_units\"].split(\",\"), # NEW: paramaterize architecture\n feature_columns = feature_cols, \n model_dir = OUTDIR,\n config = tf.estimator.RunConfig(\n tf_random_seed = 1, # for reproducibility\n save_checkpoints_steps = max(100, params[\"train_steps\"] // 10) # checkpoint every N steps\n ) \n )\n\n # Add custom evaluation metric\n def my_rmse(labels, predictions):\n pred_values = tf.squeeze(input = predictions[\"predictions\"], axis = -1)\n return {\"rmse\": tf.metrics.root_mean_squared_error(labels = labels, predictions = pred_values)}\n \n model = tf.contrib.estimator.add_metrics(model, my_rmse) \n\n train_spec = tf.estimator.TrainSpec(\n input_fn = lambda: train_input_fn(params[\"train_data_path\"]),\n max_steps = params[\"train_steps\"])\n\n exporter = tf.estimator.BestExporter(name = \"exporter\", serving_input_receiver_fn = serving_input_receiver_fn) # export SavedModel once at the end of training\n # Note: alternatively use tf.estimator.BestExporter to export at every checkpoint that has lower loss than the previous checkpoint\n\n\n eval_spec = tf.estimator.EvalSpec(\n input_fn = lambda: eval_input_fn(params[\"eval_data_path\"]),\n steps = None,\n start_delay_secs = 1, # wait at least N seconds before first evaluation (default 120)\n throttle_secs = 1, # wait at least N seconds before each subsequent evaluation (default 600)\n exporters = exporter) # export SavedModel once at the end of training\n\n tf.logging.set_verbosity(v = tf.logging.INFO) # so loss is printed during training\n shutil.rmtree(path = OUTDIR, ignore_errors = True) # start fresh each time\n\n tf.estimator.train_and_evaluate(model, train_spec, eval_spec)",
"Overwriting taxifaremodel/model.py\n"
]
],
[
[
"## Create task.py",
"_____no_output_____"
],
[
"#### **Exercise 1**\n\nThe code cell below has two TODOs for you to complete. \n\nFirstly, in model.py above we set the number of hidden units in our model to be a hyperparameter. This means `hidden_units` must be exposed as a command line argument when we submit our training job to Cloud ML Engine. Modify the code below to add an flag for `hidden_units`. Be sure to include a description for the `help` field and specify the data `type` that the model should expect to receive. You can also include a `default` value. Look to the other parser arguments to make sure you have the formatting corret. \n\nSecond, when doing hyperparameter tuning we need to make sure the output directory is different for each run, otherwise successive runs will overwrite previous runs. In `task.py` below, add some code to append the trial_id to the output direcroty of the training job. \n\n**Hint**: You can use `json.loads(os.environ.get('TF_CONFIG', '{}')).get('task', {}).get('trial', '')` to extract the trial id of the training job. You will want to append this quanity to the output directory `args['output_dir']` to make sure the output directory is different for each run.",
"_____no_output_____"
]
],
[
[
"%%writefile taxifaremodel/task.py\nimport argparse\nimport json\nimport os\n\nfrom . import model\n\nif __name__ == \"__main__\":\n parser = argparse.ArgumentParser()\n parser.add_argument(\n \"--hidden_units\",\n help = \"Hidden layer sizes to use for DNN feature columns -- provide space-separated layers\",\n type = str,\n default = \"10,10\"\n )\n parser.add_argument(\n \"--train_data_path\",\n help = \"GCS or local path to training data\",\n required = True\n )\n parser.add_argument(\n \"--train_steps\",\n help = \"Steps to run the training job for (default: 1000)\",\n type = int,\n default = 1000\n )\n parser.add_argument(\n \"--eval_data_path\",\n help = \"GCS or local path to evaluation data\",\n required = True\n )\n parser.add_argument(\n \"--output_dir\",\n help = \"GCS location to write checkpoints and export models\",\n required = True\n )\n parser.add_argument(\n \"--job-dir\",\n help=\"This is not used by our model, but it is required by gcloud\",\n )\n args = parser.parse_args().__dict__\n \n # Append trial_id to path so trials don\"t overwrite each other\n args[\"output_dir\"] = os.path.join(\n args[\"output_dir\"],\n json.loads(\n os.environ.get(\"TF_CONFIG\", \"{}\")\n ).get(\"task\", {}).get(\"trial\", \"\")\n ) \n \n # Run the training job\n model.train_and_evaluate(args)",
"Overwriting taxifaremodel/task.py\n"
]
],
[
[
"## Create hypertuning configuration \n\nWe specify:\n1. How many trials to run (`maxTrials`) and how many of those trials can be run in parrallel (`maxParallelTrials`) \n2. Which algorithm to use (in this case `GRID_SEARCH`)\n3. Which metric to optimize (`hyperparameterMetricTag`)\n4. The search region in which to constrain the hyperparameter search\n\nFull specification options [here](https://cloud.google.com/ml-engine/reference/rest/v1/projects.jobs#HyperparameterSpec).\n\nHere we are just tuning one parameter, the number of hidden units, and we'll run all trials in parrallel. However more commonly you would tune multiple hyperparameters.",
"_____no_output_____"
],
[
"#### **Exercise 2**\n\nAdd some additional hidden units to the `hyperparam.yaml` file below to potetially explore during the hyperparameter job.",
"_____no_output_____"
]
],
[
[
"%%writefile hyperparam.yaml\ntrainingInput:\n scaleTier: BASIC\n hyperparameters:\n goal: MINIMIZE\n maxTrials: 5\n maxParallelTrials: 5\n hyperparameterMetricTag: rmse\n enableTrialEarlyStopping: True\n algorithm: GRID_SEARCH\n params:\n - parameterName: hidden_units\n type: CATEGORICAL\n categoricalValues:\n - 10,10\n - 64,32\n - 128,64,32\n - 32, 32\n - 128, 64, 64 ",
"Overwriting hyperparam.yaml\n"
]
],
[
[
"## Run the training job|\n\nSame as before with the addition of `--config=hyperpam.yaml` to reference the file we just created.\n\nThis will take about 20 minutes. Go to [cloud console](https://console.cloud.google.com/mlengine/jobs) and click on the job id. Once the job is completed, the choosen hyperparameters and resulting objective value (RMSE in this case) will be shown. Trials will sorted from best to worst. ",
"_____no_output_____"
],
[
"#### **Exercise 3**\n\nSubmit a hyperparameter tuning job to the cloud. Fill in the missing arguments below. This is similar to the exercise you completed in the `02_tensorlfow/g_distributed` notebook. Note that one difference here is that we now specify a `config` parameter giving the location of our .yaml file.",
"_____no_output_____"
]
],
[
[
"OUTDIR=\"gs://{}/taxifare/trained_hp_tune\".format(BUCKET)\n!gsutil -m rm -rf {OUTDIR} # start fresh each time\n!gcloud ai-platform jobs submit training taxifare_$(date -u +%y%m%d_%H%M%S) \\\n --package-path=taxifaremodel \\\n --module-name=taxifaremodel.task \\\n --config=hyperparam.yaml \\\n --job-dir=gs://{BUCKET}/taxifare \\\n --python-version=3.5 \\\n --runtime-version={TFVERSION} \\\n --region={REGION} \\\n -- \\\n --train_data_path=gs://{BUCKET}/taxifare/smallinput/taxi-train.csv \\\n --eval_data_path=gs://{BUCKET}/taxifare/smallinput/taxi-valid.csv \\\n --train_steps=5000 \\\n --output_dir={OUTDIR}",
"Removing gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/#1563484245754874...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/events.out.tfevents.1563484194.cmle-training-731225091109210751#1563484432093881...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/#1563484245617656...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/#1563484179915317...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484290/saved_model.pb#1563484296416930...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/eval/#1563484221723731...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484244/variables/#1563484250562059...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484244/#1563484250271431...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/#1563484422198624...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/checkpoint#1563484424012789...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484244/variables/variables.data-00000-of-00002#1563484250704999...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484290/variables/variables.data-00001-of-00002#1563484296817040...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/eval/events.out.tfevents.1563484221.cmle-training-731225091109210751#1563484431813894...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484290/variables/#1563484296549505...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484244/variables/variables.data-00001-of-00002#1563484250850781...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484244/saved_model.pb#1563484250419358...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484290/variables/variables.data-00000-of-00002#1563484296673502...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484290/#1563484296264391...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484244/variables/variables.index#1563484250974705...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484290/variables/variables.index#1563484296948921...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484337/#1563484342693000...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484337/saved_model.pb#1563484342830504...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484337/variables/#1563484342983018...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484337/variables/variables.data-00000-of-00002#1563484343137269...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484337/variables/variables.data-00001-of-00002#1563484343290159...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484337/variables/variables.index#1563484343427043...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484383/#1563484388730588...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484383/saved_model.pb#1563484388882431...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484383/variables/#1563484389007300...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484383/variables/variables.data-00000-of-00002#1563484389169653...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484383/variables/variables.data-00001-of-00002#1563484389324151...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/export/exporter/1563484383/variables/variables.index#1563484389473891...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/graph.pbtxt#1563484196248726...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/model.ckpt-3000.data-00000-of-00002#1563484328963452...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/model.ckpt-3000.data-00001-of-00002#1563484328757555...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/model.ckpt-3000.index#1563484329225429...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/model.ckpt-3000.meta#1563484330801327...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/model.ckpt-3500.data-00000-of-00002#1563484355145739...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/model.ckpt-3500.data-00001-of-00002#1563484354914714...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/model.ckpt-3500.index#1563484355346835...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/model.ckpt-3500.meta#1563484356828161...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/model.ckpt-4000.data-00000-of-00002#1563484374974893...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/model.ckpt-4000.data-00001-of-00002#1563484374746083...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/model.ckpt-4000.index#1563484375251856...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/model.ckpt-4000.meta#1563484376821830...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/model.ckpt-4500.data-00000-of-00002#1563484402466437...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/model.ckpt-4500.data-00001-of-00002#1563484402221124...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/model.ckpt-4500.index#1563484402697125...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/model.ckpt-4500.meta#1563484404434046...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/model.ckpt-5000.data-00000-of-00002#1563484423028688...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/model.ckpt-5000.data-00001-of-00002#1563484422803063...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/model.ckpt-5000.index#1563484423277025...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/1/model.ckpt-5000.meta#1563484425034903...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/checkpoint#1563484376190690...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/#1563484374709066...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/eval/events.out.tfevents.1563484205.cmle-training-12929692639216599982#1563484383348464...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/eval/#1563484205043343...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/export/#1563484244248422...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/events.out.tfevents.1563484180.cmle-training-12929692639216599982#1563484383566118...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/export/exporter/1563484243/#1563484248744457...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/export/exporter/#1563484244398170...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/export/exporter/1563484243/saved_model.pb#1563484248874194...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/export/exporter/1563484243/variables/#1563484249225632...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/export/exporter/1563484243/variables/variables.data-00000-of-00002#1563484249358059...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/export/exporter/1563484243/variables/variables.data-00001-of-00002#1563484249489227...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/export/exporter/1563484243/variables/variables.index#1563484249621916...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/export/exporter/1563484301/#1563484306419668...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/export/exporter/1563484301/saved_model.pb#1563484306552630...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/export/exporter/1563484301/variables/#1563484306733064...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/export/exporter/1563484301/variables/variables.data-00000-of-00002#1563484306867567...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/export/exporter/1563484301/variables/variables.data-00001-of-00002#1563484306999553...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/export/exporter/1563484343/saved_model.pb#1563484347695150...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/export/exporter/1563484301/variables/variables.index#1563484307113467...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/export/exporter/1563484343/variables/#1563484347801417...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/export/exporter/1563484343/#1563484347589201...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/export/exporter/1563484343/variables/variables.data-00000-of-00002#1563484347918820...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/graph.pbtxt#1563484181689138...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/export/exporter/1563484343/variables/variables.index#1563484348145772...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/export/exporter/1563484343/variables/variables.data-00001-of-00002#1563484348029085...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/model.ckpt-3000.data-00000-of-00002#1563484293360998...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/model.ckpt-3000.data-00001-of-00002#1563484292885587...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/model.ckpt-3000.index#1563484293553194...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/model.ckpt-3000.meta#1563484295070316...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/model.ckpt-3500.data-00000-of-00002#1563484317673600...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/model.ckpt-3500.data-00001-of-00002#1563484317473259...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/model.ckpt-3500.index#1563484317872837...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/model.ckpt-3500.meta#1563484319347611...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/model.ckpt-4000.data-00000-of-00002#1563484335095991...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/model.ckpt-4000.data-00001-of-00002#1563484334912284...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/model.ckpt-4000.index#1563484335312197...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/model.ckpt-4000.meta#1563484336753565...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/model.ckpt-4500.data-00000-of-00002#1563484358453052...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/model.ckpt-4500.data-00001-of-00002#1563484358268625...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/model.ckpt-4500.meta#1563484359941574...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/model.ckpt-4500.index#1563484358640561...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/model.ckpt-5000.meta#1563484377023540...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/model.ckpt-5000.index#1563484375638095...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/model.ckpt-5000.data-00000-of-00002#1563484375432522...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/2/model.ckpt-5000.data-00001-of-00002#1563484375235121...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/#1563484375644467...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/checkpoint#1563484381097479...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/eval/#1563484206049622...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/eval/events.out.tfevents.1563484206.cmle-training-10262658521264896257#1563484388064133...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/events.out.tfevents.1563484180.cmle-training-10262658521264896257#1563484388315477...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/#1563484224902536...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/#1563484225061414...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484224/#1563484229273832...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484224/saved_model.pb#1563484229407734...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484224/variables/#1563484229550094...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484224/variables/variables.data-00000-of-00002#1563484229673343...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484224/variables/variables.data-00001-of-00002#1563484229817562...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484224/variables/variables.index#1563484229958661...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484247/#1563484251529202...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484247/saved_model.pb#1563484251657075...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484247/variables/#1563484251824353...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484247/variables/variables.data-00000-of-00002#1563484251973306...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484247/variables/variables.data-00001-of-00002#1563484252116431...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484247/variables/variables.index#1563484252242940...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484304/#1563484309303863...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484304/saved_model.pb#1563484309421396...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484304/variables/#1563484309548087...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484304/variables/variables.data-00000-of-00002#1563484309705354...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484304/variables/variables.data-00001-of-00002#1563484309827817...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484304/variables/variables.index#1563484309951448...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484344/#1563484349303991...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484344/saved_model.pb#1563484349414154...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484344/variables/#1563484349531037...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484344/variables/variables.data-00000-of-00002#1563484349645873...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484344/variables/variables.data-00001-of-00002#1563484349762609...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/export/exporter/1563484344/variables/variables.index#1563484349872497...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/graph.pbtxt#1563484183000844...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/model.ckpt-3000.data-00000-of-00002#1563484296819223...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/model.ckpt-3000.data-00001-of-00002#1563484296619230...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/model.ckpt-3000.index#1563484297045905...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/model.ckpt-3000.meta#1563484298443644...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/model.ckpt-3500.data-00000-of-00002#1563484319923050...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/model.ckpt-3500.data-00001-of-00002#1563484319724037...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/model.ckpt-3500.meta#1563484321650722...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/model.ckpt-3500.index#1563484320116550...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/model.ckpt-4000.data-00000-of-00002#1563484337253547...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/model.ckpt-4000.data-00001-of-00002#1563484337047406...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/model.ckpt-4000.index#1563484337452247...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/model.ckpt-4000.meta#1563484338850438...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/model.ckpt-4500.data-00000-of-00002#1563484359302567...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/model.ckpt-4500.data-00001-of-00002#1563484359132401...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/model.ckpt-4500.index#1563484359484027...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/model.ckpt-4500.meta#1563484360914944...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/model.ckpt-5000.data-00000-of-00002#1563484380305813...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/model.ckpt-5000.data-00001-of-00002#1563484376120536...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/model.ckpt-5000.index#1563484380499366...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/3/model.ckpt-5000.meta#1563484382011616...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/#1563484416761122...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/checkpoint#1563484418329507...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/eval/#1563484241256373...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/eval/events.out.tfevents.1563484241.cmle-training-289151322071742182#1563484425230069...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/events.out.tfevents.1563484216.cmle-training-289151322071742182#1563484425447502...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/#1563484259707294...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/#1563484259837765...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484259/#1563484263893456...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484259/saved_model.pb#1563484264027996...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484259/variables/#1563484264142150...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484259/variables/variables.data-00000-of-00002#1563484264261719...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484259/variables/variables.index#1563484264497546...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484259/variables/variables.data-00001-of-00002#1563484264375700...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484281/#1563484286224229...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484281/saved_model.pb#1563484286340310...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484281/variables/#1563484286461425...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484281/variables/variables.data-00000-of-00002#1563484286580450...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484281/variables/variables.data-00001-of-00002#1563484286717986...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484281/variables/variables.index#1563484286845410...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484336/#1563484341015992...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484336/variables/#1563484341229010...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484336/variables/variables.data-00000-of-00002#1563484341339228...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484336/saved_model.pb#1563484341114302...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484336/variables/variables.data-00001-of-00002#1563484341462306...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484385/saved_model.pb#1563484390458842...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484336/variables/variables.index#1563484341574705...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484385/variables/#1563484390580196...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484385/#1563484390344098...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484385/variables/variables.data-00000-of-00002#1563484390708772...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484385/variables/variables.data-00001-of-00002#1563484390937401...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/export/exporter/1563484385/variables/variables.index#1563484391077987...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/graph.pbtxt#1563484218621545...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/model.ckpt-3000.data-00000-of-00002#1563484329147911...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/model.ckpt-3000.data-00001-of-00002#1563484328963816...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/model.ckpt-3000.index#1563484329346558...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/model.ckpt-3000.meta#1563484330624636...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/model.ckpt-3500.data-00000-of-00002#1563484352714406...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/model.ckpt-3500.data-00001-of-00002#1563484352540190...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/model.ckpt-3500.index#1563484352917577...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/model.ckpt-4000.data-00000-of-00002#1563484376649419...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/model.ckpt-4000.data-00001-of-00002#1563484376451643...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/model.ckpt-3500.meta#1563484354223608...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/model.ckpt-4000.index#1563484376821363...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/model.ckpt-4000.meta#1563484378128577...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/model.ckpt-4500.data-00000-of-00002#1563484400152487...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/model.ckpt-4500.data-00001-of-00002#1563484399861016...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/model.ckpt-4500.index#1563484400336409...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/model.ckpt-5000.data-00000-of-00002#1563484417563406...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/model.ckpt-4500.meta#1563484401765724...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/model.ckpt-5000.data-00001-of-00002#1563484417342324...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/model.ckpt-5000.index#1563484417784237...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/4/model.ckpt-5000.meta#1563484419112226...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/#1563484409924817...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/checkpoint#1563484411449108...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/eval/#1563484227258939...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/eval/events.out.tfevents.1563484227.cmle-training-3856349254539048287#1563484424346296...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/events.out.tfevents.1563484201.cmle-training-3856349254539048287#1563484424551849...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/#1563484247171469...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/#1563484247287596...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484246/#1563484251346528...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484246/saved_model.pb#1563484251476014...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484246/variables/variables.data-00000-of-00002#1563484251712054...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484246/variables/variables.data-00001-of-00002#1563484251817583...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484246/variables/#1563484251600701...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484270/#1563484274603991...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484246/variables/variables.index#1563484251923209...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484270/saved_model.pb#1563484274717488...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484270/variables/#1563484274827785...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484270/variables/variables.data-00000-of-00002#1563484274950999...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484270/variables/variables.data-00001-of-00002#1563484275062028...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484270/variables/variables.index#1563484275176893...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484330/#1563484335772288...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484330/saved_model.pb#1563484335884018...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484330/variables/#1563484336013128...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484330/variables/variables.data-00000-of-00002#1563484336123803...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484330/variables/variables.data-00001-of-00002#1563484336255692...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484330/variables/variables.index#1563484336370990...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484376/#1563484381085274...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484376/saved_model.pb#1563484381214917...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484376/variables/variables.data-00001-of-00002#1563484381562765...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484376/variables/variables.data-00000-of-00002#1563484381443357...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/graph.pbtxt#1563484203137946...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484376/variables/#1563484381332974...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/export/exporter/1563484376/variables/variables.index#1563484381681337...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/model.ckpt-3000.data-00000-of-00002#1563484322570048...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/model.ckpt-3000.data-00001-of-00002#1563484322377733...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/model.ckpt-3000.index#1563484322812373...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/model.ckpt-3000.meta#1563484324176992...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/model.ckpt-3500.data-00000-of-00002#1563484347151078...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/model.ckpt-3500.data-00001-of-00002#1563484346965836...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/model.ckpt-3500.index#1563484347363555...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/model.ckpt-3500.meta#1563484348801156...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/model.ckpt-4000.data-00000-of-00002#1563484368234852...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/model.ckpt-4000.data-00001-of-00002#1563484368045553...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/model.ckpt-4000.index#1563484368448018...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/model.ckpt-4000.meta#1563484369920700...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/model.ckpt-4500.data-00000-of-00002#1563484392172924...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/model.ckpt-4500.data-00001-of-00002#1563484391925249...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/model.ckpt-4500.meta#1563484393841966...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/model.ckpt-4500.index#1563484392359867...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/model.ckpt-5000.data-00000-of-00002#1563484410708980...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/model.ckpt-5000.data-00001-of-00002#1563484410498595...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/model.ckpt-5000.index#1563484410899374...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_hp_tune/5/model.ckpt-5000.meta#1563484412386035...\n/ [255/255 objects] 100% Done \nOperation completed over 255 objects. \nJob [taxifare_190719_131105] submitted successfully.\nYour job is still active. You may view the status of your job with the command\n\n $ gcloud ai-platform jobs describe taxifare_190719_131105\n\nor continue streaming the logs with the command\n\n $ gcloud ai-platform jobs stream-logs taxifare_190719_131105\njobId: taxifare_190719_131105\nstate: QUEUED\n"
]
],
[
[
"## Results\n\nThe best result is RMSE **4.02** with hidden units = 128,64,32. \n\nThis improvement is modest, but now that we have our hidden units tuned let's run on our larger dataset to see if it helps. \n\nNote the passing of hyperparameter values via command line",
"_____no_output_____"
]
],
[
[
"OUTDIR=\"gs://{}/taxifare/trained_large_tuned\".format(BUCKET)\n!gsutil -m rm -rf {OUTDIR} # start fresh each time\n!gcloud ai-platform jobs submit training taxifare_large_$(date -u +%y%m%d_%H%M%S) \\\n --package-path=taxifaremodel \\\n --module-name=taxifaremodel.task \\\n --job-dir=gs://{BUCKET}/taxifare \\\n --python-version=3.5 \\\n --runtime-version={TFVERSION} \\\n --region={REGION} \\\n --scale-tier=STANDARD_1 \\\n -- \\\n --train_data_path=gs://cloud-training-demos/taxifare/large/taxi-train*.csv \\\n --eval_data_path=gs://cloud-training-demos/taxifare/small/taxi-valid.csv \\\n --train_steps=200000 \\\n --output_dir={OUTDIR} \\\n --hidden_units=\"128,64,32\"",
"Removing gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/events.out.tfevents.1563484096.cmle-training-worker-285c128a20-0-kn9fp#1563485144189527...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/#1563485147333149...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/checkpoint#1563485150259049...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/#1563484319658283...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484528/variables/variables.data-00001-of-00002#1563484533435217...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/eval/events.out.tfevents.1563484217.cmle-training-master-285c128a20-0-2dgkd#1563485157570746...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484528/saved_model.pb#1563484532962349...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/eval/#1563484217291417...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/#1563484319800217...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484528/#1563484532833287...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484633/variables/#1563484638677121...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484528/variables/#1563484533113828...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/events.out.tfevents.1563484088.cmle-training-master-285c128a20-0-2dgkd#1563485044376692...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484633/saved_model.pb#1563484638544776...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484528/variables/variables.data-00000-of-00002#1563484533280453...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484528/variables/variables.index#1563484533564832...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484633/variables/variables.data-00000-of-00002#1563484638818566...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484633/variables/variables.data-00001-of-00002#1563484638949000...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484633/#1563484638417564...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484633/variables/variables.index#1563484639075867...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484736/#1563484741600907...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484736/saved_model.pb#1563484741745627...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484736/variables/#1563484741900597...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484736/variables/variables.data-00000-of-00002#1563484742036030...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484736/variables/variables.data-00001-of-00002#1563484742206744...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484736/variables/variables.index#1563484742343418...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484944/#1563484949505093...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484944/saved_model.pb#1563484949635794...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484944/variables/#1563484949772566...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484944/variables/variables.data-00000-of-00002#1563484949898220...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484944/variables/variables.data-00001-of-00002#1563484950094040...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563484944/variables/variables.index#1563484950271958...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563485051/#1563485056430632...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563485051/saved_model.pb#1563485056579291...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563485051/variables/#1563485056740246...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563485051/variables/variables.data-00000-of-00002#1563485056895302...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563485051/variables/variables.data-00001-of-00002#1563485057059242...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/export/exporter/1563485051/variables/variables.index#1563485057200913...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/graph.pbtxt#1563484090289612...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-120019.data-00000-of-00004#1563484728297956...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-120019.data-00001-of-00004#1563484728070330...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-120019.data-00002-of-00004#1563484727832446...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-120019.data-00003-of-00004#1563484727553393...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-120019.index#1563484728571392...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-120019.meta#1563484730474803...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-140022.data-00000-of-00004#1563484833349939...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-140022.data-00001-of-00004#1563484833133161...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-140022.data-00003-of-00004#1563484832548890...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-140022.data-00002-of-00004#1563484832896755...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-140022.index#1563484833598846...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-140022.meta#1563484835653301...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-160026.data-00000-of-00004#1563484936109525...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-160026.data-00001-of-00004#1563484935865504...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-160026.data-00002-of-00004#1563484935569969...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-160026.data-00003-of-00004#1563484935335460...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-160026.index#1563484936319481...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-160026.meta#1563484938500074...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-180032.data-00000-of-00004#1563485041773066...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-180032.data-00001-of-00004#1563485041524787...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-180032.data-00002-of-00004#1563485041274981...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-180032.data-00003-of-00004#1563485041035713...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-180032.index#1563485041983049...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-180032.meta#1563485044118660...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-200003.data-00000-of-00004#1563485149182394...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-200003.data-00001-of-00004#1563485148939219...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-200003.data-00002-of-00004#1563485148718153...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-200003.data-00003-of-00004#1563485148460511...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-200003.index#1563485149403992...\nRemoving gs://qwiklabs-gcp-636667ae83e902b6_al/taxifare/trained_large_tuned/model.ckpt-200003.meta#1563485151296972...\n/ [69/69 objects] 100% Done \nOperation completed over 69 objects. \nJob [taxifare_large_190719_132300] submitted successfully.\nYour job is still active. You may view the status of your job with the command\n\n $ gcloud ai-platform jobs describe taxifare_large_190719_132300\n\nor continue streaming the logs with the command\n\n $ gcloud ai-platform jobs stream-logs taxifare_large_190719_132300\njobId: taxifare_large_190719_132300\nstate: QUEUED\n"
]
],
[
[
"## Analysis\n\nOur RMSE improved to **3.85**\n\nI get **3.79** including direction of travel - crossing direction with DOW / time of day gives no extra benefit",
"_____no_output_____"
],
[
"## Challenge excercise\n\nTry to beat the current RMSE:\n\n- Try adding more engineered features or modifying existing ones\n- Try tuning additional hyperparameters ",
"_____no_output_____"
],
[
"Copyright 2019 Google Inc. Licensed under the Apache License, Version 2.0 (the \"License\"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0 Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an \"AS IS\" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
e78f7fc25c36d67c400a11f48adff138537c645d | 8,718 | ipynb | Jupyter Notebook | Sam/train_sam.ipynb | handertolium/lil-NLP-app | 10342a58034af2b343d3e993910fb98571b509d4 | [
"Apache-2.0"
] | null | null | null | Sam/train_sam.ipynb | handertolium/lil-NLP-app | 10342a58034af2b343d3e993910fb98571b509d4 | [
"Apache-2.0"
] | null | null | null | Sam/train_sam.ipynb | handertolium/lil-NLP-app | 10342a58034af2b343d3e993910fb98571b509d4 | [
"Apache-2.0"
] | null | null | null | 25.641176 | 83 | 0.487612 | [
[
[
"import nltk\nfrom nltk.stem.porter import PorterStemmer\n\nimport numpy as np\nimport torch\nimport torch.nn as nn\nimport random\nfrom torch.utils.data import Dataset, DataLoader\nimport json",
"_____no_output_____"
],
[
"with open('intents.json') as file:\n data = json.load(file)",
"_____no_output_____"
],
[
"stemmer = PorterStemmer()",
"_____no_output_____"
],
[
"all_words = []\nall_labels = []\ndocs_x = []\ndocs_y = []\n\nfor intent in data['intents']:\n for pattern in intent['patterns']:\n wrds = nltk.word_tokenize(pattern)\n all_words.extend(wrds)\n docs_x.append(wrds)\n docs_y.append(intent[\"tag\"])\n \n if intent['tag'] not in all_labels:\n all_labels.append(intent['tag'])",
"_____no_output_____"
],
[
"all_words = [stemmer.stem(w.lower()) for w in all_words if w != \"?\"]\nall_words = sorted(list(set(all_words)))\n",
"_____no_output_____"
],
[
"training = []\noutput = []\n\nout_empty = [0 for _ in range(len(all_labels))]\n\nfor x, doc in enumerate(docs_x):\n bag = []\n\n wrds = [stemmer.stem(w.lower()) for w in doc]\n\n for w in all_words:\n if w in wrds:\n bag.append(1)\n else:\n bag.append(0)\n\n output_row = out_empty[:]\n output_row[all_labels.index(docs_y[x])] = 1\n\n training.append(bag)\n output.append(output_row)",
"_____no_output_____"
],
[
"training = np.array(training, dtype=\"float32\")\noutput = np.array(output, dtype=\"float32\")\n",
"_____no_output_____"
],
[
"num_epochs = 1000\nbatch_size = 7\nlearning_rate = 0.001\ninput_size = len(training[0])\nhidden_size = 8\noutput_size = len(all_labels)\nprint(input_size, output_size)",
"41 7\n"
],
[
"class ChatDataset(Dataset):\n\n def __init__(self):\n self.n_samples = len(training)\n self.x_data = training\n self.y_data = output\n\n # support indexing such that dataset[i] can be used to get i-th sample\n def __getitem__(self, index):\n return self.x_data[index], self.y_data[index]\n\n # we can call len(dataset) to return the size\n def __len__(self):\n return self.n_samples",
"_____no_output_____"
],
[
"class NeuralNet(nn.Module):\n def __init__(self, input_size, hidden_size, num_classes):\n super(NeuralNet, self).__init__()\n self.l1 = nn.Linear(input_size, hidden_size) \n self.l2 = nn.Linear(hidden_size, hidden_size) \n self.l3 = nn.Linear(hidden_size, num_classes)\n self.relu = nn.ReLU()\n \n def forward(self, x):\n out = self.l1(x)\n out = self.relu(out)\n out = self.l2(out)\n out = self.relu(out)\n out = self.l3(out)\n # no activation and no softmax at the end\n return out",
"_____no_output_____"
],
[
"dataset = ChatDataset()\ntrain_loader = DataLoader(dataset=dataset,\n batch_size=batch_size,\n shuffle=True,\n num_workers=0)\n\ndevice = torch.device('cuda' if torch.cuda.is_available() else 'cpu')\n\nmodel = NeuralNet(input_size, hidden_size, output_size).to(device)",
"_____no_output_____"
],
[
"criterion = nn.CrossEntropyLoss()\noptimizer = torch.optim.Adam(model.parameters(), lr=learning_rate)\n\n# Train the model\nfor epoch in range(num_epochs):\n for (words, labels) in train_loader:\n words = words.to(device)\n labels = labels.to(dtype=torch.long).to(device)\n \n # Forward pass\n outputs = model(words)\n # if y would be one-hot, we must apply\n # labels = torch.max(labels, 1)[1]\n labels = torch.max(labels, 1)[1]\n loss = criterion(outputs, labels)\n \n # Backward and optimize\n optimizer.zero_grad()\n loss.backward()\n optimizer.step()\n \n if (epoch+1) % 100 == 0:\n print (f'Epoch [{epoch+1}/{num_epochs}], Loss: {loss.item():.4f}')",
"Epoch [100/1000], Loss: 0.1743\nEpoch [200/1000], Loss: 0.3836\nEpoch [300/1000], Loss: 0.0614\nEpoch [400/1000], Loss: 0.0068\nEpoch [500/1000], Loss: 0.0001\nEpoch [600/1000], Loss: 0.0042\nEpoch [700/1000], Loss: 0.0016\nEpoch [800/1000], Loss: 0.0009\nEpoch [900/1000], Loss: 0.0005\nEpoch [1000/1000], Loss: 0.0003\n"
],
[
"def bag_of_words(s, words):\n bag = [0 for _ in range(len(words))]\n\n s_words = nltk.word_tokenize(s)\n s_words = [stemmer.stem(word.lower()) for word in s_words]\n\n for se in s_words:\n for i, w in enumerate(words):\n if w == se:\n bag[i] = 1\n \n return torch.tensor(bag, dtype=torch.float, device=device)",
"_____no_output_____"
],
[
"tags = [x[\"tag\"] for x in data[\"intents\"]]\nsentence = \"how old are you\"\nwith torch.no_grad():\n output = model(bag_of_words(sentence, all_words))\n predicted = torch.argmax(output)\n \ntag = tags[predicted.item()]\nfor tg in data[\"intents\"]:\n if tg['tag'] == tag:\n responses = tg['responses']\n\nprint(random.choice(responses))",
"I don't know time runs so fast\n"
],
[
"data = {\n\"all_words\": all_words,\n\"tags\": tags\n}\n\nDATA_FILE = \"deploy_sam/data.pth\"\ntorch.save(data, DATA_FILE)",
"_____no_output_____"
],
[
"MODEL_FILE\ntorch.save(model, PATH)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78f89c8cc93524db449130fadd2dae476947ed2 | 60,419 | ipynb | Jupyter Notebook | Python/Introduction.ipynb | BuserLukas/Logic | cc0447554cfa75b213a10a2db37ce82c42afb91d | [
"MIT"
] | 13 | 2019-10-03T13:25:02.000Z | 2021-12-26T11:49:25.000Z | Python/Introduction.ipynb | BuserLukas/Logic | cc0447554cfa75b213a10a2db37ce82c42afb91d | [
"MIT"
] | 19 | 2015-01-14T15:36:24.000Z | 2019-04-21T02:13:23.000Z | Python/Introduction.ipynb | BuserLukas/Logic | cc0447554cfa75b213a10a2db37ce82c42afb91d | [
"MIT"
] | 18 | 2019-10-03T16:05:46.000Z | 2021-12-10T19:44:15.000Z | 26.499561 | 410 | 0.531356 | [
[
[
"from IPython.core.display import HTML\nwith open('style.css', 'r') as file:\n css = file.read()\nHTML(css)",
"_____no_output_____"
]
],
[
[
"# An Introduction to *Python*\n\nLet us begin by checking the version of our Python interpreter.",
"_____no_output_____"
]
],
[
[
"import sys\nsys.version",
"_____no_output_____"
]
],
[
[
"This notebook gives a short introduction to *Python*. We will start with the basics but as the **main goal** of this introduction is to show how *Python* supports *sets* we will quickly move to more advanced topics.\nIn order to show off the features of *Python* we will give some examples that are not fully explained at the point \nwhere we introduce them. However, rest assured that these features will be explained eventually.",
"_____no_output_____"
],
[
"## Evaluating expressions\n\nAs Python is an interactive language, expressions can be evaluated directly. In a *Jupyter* \nnotebook we just have to type <tt>Ctrl-Enter</tt> in the cell containing the expression. Instead\nof <tt>Ctrl-Enter</tt> we can also use <tt>Shift-Enter</tt>.",
"_____no_output_____"
]
],
[
[
"1 + 2",
"_____no_output_____"
]
],
[
[
"In *Python*, the precision of integers is not bounded. Hence, the following expression does **not** cause\nan <a href=\"https://en.wikipedia.org/wiki/Integer_overflow\">integer overflow</a>.",
"_____no_output_____"
]
],
[
[
"1 * 2 * 3 * 4 * 5 * 6 * 7 * 8 * 9 * 10 * 11 * 12 * 13 * 14 * 15 * 16 * 17 * 18 * 19 * 20 * 21 * 22 * 23 * 24 * 25",
"_____no_output_____"
]
],
[
[
"The next *cell* in this notebook shows how to compute the *factorial* of 1000, i.e. it shows how to compute \nthe product\n$$ 1000! = 1 \\cdot 2 \\cdot 3 \\cdot {\\dots} \\cdot 998 \\cdot 999 \\cdot 1000 $$\nIt uses some advanced features from *functional programming* that will be discussed later.",
"_____no_output_____"
]
],
[
[
"import functools \n\nfunctools.reduce(lambda x, y: (x*y), range(1, 1000+1))",
"_____no_output_____"
]
],
[
[
"The following command will stop the interpreter if executed. It is not useful inside a *Jupyter* notebook. \nHence, the next line should not be evaluated. \n\nHowever, if you evaluate the following line, nothing bad will happen as the interpreter is just restarted by *Jupyter*. ",
"_____no_output_____"
]
],
[
[
"exit()",
"_____no_output_____"
]
],
[
[
"In order to write something to the screen, we can use the function <tt>print</tt>. This function can print objects of any type. In the following example, this function prints a <tt>string</tt>. In *Python* any character sequence \nenclosed in single quotes is <tt>string</tt>.",
"_____no_output_____"
]
],
[
[
"print('Hello, World!')",
"_____no_output_____"
]
],
[
[
"The last expression in every notebook cell is automatically printed.",
"_____no_output_____"
]
],
[
[
"'Hello, World!'",
"_____no_output_____"
]
],
[
[
"If we want to supress the last expression from being printed, we can end the expression with a semicolon:",
"_____no_output_____"
]
],
[
[
"'Hello, World!';",
"_____no_output_____"
]
],
[
[
"Instead of using single quotes we can also use double quotes as seen in the next example. However, the convention is to use single quotes, unless the string itself contains a single quote.",
"_____no_output_____"
]
],
[
[
"\"Hello, World!\"",
"_____no_output_____"
]
],
[
[
"The function <tt>print</tt> accepts any number of arguments. For example, to print the string \"36 * 37 / 2 = \" followed by the value of the expression $36 \\cdot 37 / 2$ we can use the following <tt>print</tt> statement:",
"_____no_output_____"
]
],
[
[
"print('36 * 37 / 2 =', 36 * 37 // 2)",
"_____no_output_____"
]
],
[
[
"In the expression \"<tt>36 \\* 37 // 2</tt>\" we have used the operator \"<tt>//</tt>\" in order to enforce *integer division*. If we had used the operator \"<tt>/</tt>\" instead, *Python* would have used *floating point division* and therefore it would have printed the floating point number <tt>666.0</tt> instead of the integer <tt>666<tt>. ",
"_____no_output_____"
]
],
[
[
"print('36 * 37 / 2 =', 36 * 37 / 2)",
"_____no_output_____"
],
[
"5 // 3, 5 % 3",
"_____no_output_____"
],
[
"range(1, 9+1)",
"_____no_output_____"
],
[
"A = set(range(1, 9+1))\nA",
"_____no_output_____"
]
],
[
[
"The function `sum` can be used to some up all elements of a set (or a list).",
"_____no_output_____"
]
],
[
[
"sum(A)",
"_____no_output_____"
]
],
[
[
"The following script reads a natural number $n$ and computes the sum $\\sum\\limits_{i=1}^n i$.\n<ol>\n <li>The function <tt>input</tt> prompts the user to enter a string.\n </li>\n <li>This string is then converted into an integer using the function <tt>int</tt>.\n </li>\n <li>Next, the <em>set</em> <tt>S</tt> is created such that \n $$\\texttt{s} = \\{1, \\cdots, n\\}. $$ \n The set <tt>S</tt> is constructed using the function <tt>range</tt>. A function call \n of the form $\\texttt{range}(a, b + 1)$ returns a <em>generator</em> that produces the natural numbers starting\n from $a$ upto and including $b$. By using this generator as an argument to the function <tt>set</tt>, \n a set is created that contains all the natural number starting from $a$ upto and including $b$.\n The precise mechanics of <em>generators</em> will be explained later.\n </li>\n <li>The <tt>print</tt> statement uses the function <tt>sum</tt> to add up all the elements of the\n set <tt>s</tt>. The effect of the last argument <tt>sep=''</tt> is to prevent <tt>print</tt> from \n separating the previous arguments by spaces.\n </li>\n</ol>",
"_____no_output_____"
]
],
[
[
"n = input('Type a natural number and press return: ')\nn = int(n)\nS = set(range(1, n+1))\nprint('The sum 1 + 2 + ... + ', n, ' is equal to ', sum(S), '.', sep='')",
"_____no_output_____"
]
],
[
[
"If we input $36$, which happens to be equal to $6^2$ above, then we discover the following remarkable identity:\n$$ \\sum\\limits_{i=1}^{6^2} i = 666. $$",
"_____no_output_____"
],
[
"The following example shows how *functions* can be defined in *Python*. The function $\\texttt{sum}(n)$ is supposed \nto compute the sum of all the numbers in the set $\\{1, \\cdots, n\\}$. Therefore, we have\n$$\\texttt{sum}(n) = \\sum\\limits_{i=1}^n i. $$ \nThe function <tt>sum</tt> is defined *recursively*. The recursive implementation of the function <tt>sum</tt> can best by understood if we observe that it satisfies the following two equations:\n<ol>\n <li> $\\texttt{sum}(0) = 0$, </li>\n <li> $\\texttt{sum}(n) = \\texttt{sum}(n-1) + n \\quad$ provided that $n > 0$.</li>\n</ol>\nThe keyword <tt>def</tt> is used to signal that we define a function.",
"_____no_output_____"
]
],
[
[
"def sum(n):\n if n == 0:\n return 0\n return sum(n-1) + n",
"_____no_output_____"
],
[
"sum",
"_____no_output_____"
],
[
"sum(3)",
"_____no_output_____"
],
[
"def sum(n):\n print(f'calling sum({n})')\n if n == 0:\n print('sum(0) = 0')\n return 0\n snm1 = sum(n-1) \n print(f'sum({n}) = sum({n-1}) + {n} = {snm1} + {n} = {snm1 + n}')\n return snm1 + n",
"_____no_output_____"
],
[
"sum(3)",
"_____no_output_____"
]
],
[
[
"Let us discuss the implementation of the function <tt>sum</tt> line by line:\n<ol>\n <li> The keyword <tt>def</tt> starts the definition of the function. It is followed by the name of the function that is defined. The name is followed by the list of the parameters of the function. This list is enclosed in parentheses. If there had been more than one parameter, the parameters would have been separated by commas. Finally, there needs to be a colon at the end of the first line.\n </li>\n <li> The body of the function is indented. <b>Contrary</b> to most other programming languages, \n <u>Python is space sensitive</u>. \n The first statement of the body is a conditional statement, which starts with the keyword\n <tt>if</tt>. The keyword is followed by a test. In this case we test whether the variable $n$ \n is equal to the number $0$. Note that this test is followed by a colon.\n </li>\n <li> The next line contains a <tt>return</tt> statement. Note that this statement is again indented.\n All statements indented by the same amount that follow an <tt>if</tt>-statement are considered as\n the body of the <tt>if</tt>-statement. In this case the body contains only a single statement.\n </li>\n <li> The <tt>print</tt> statement in the penultimate line contains a so called \n <a href=\"https://realpython.com/python-f-strings/\">f-string</a>. Expressions enclosed in curly braces are evaluated, the result is converted to a string and inserted back into the f-string. This behaviour is known as \n <em style=\"color:blue;\">string interpolation</em>.\n </li>\n <li> The last line of the function definition contains the recursive invocation of the function <tt>sum</tt>.\n </li>\n</ol>\n\nUsing the function <tt>sum</tt>, we can compute the sum $\\sum\\limits_{i=1}^n i$ as follows:",
"_____no_output_____"
]
],
[
[
"n = int(input(\"Enter a natural number: \"))\ntotal = sum(n)\nif n > 2:\n print(\"0 + 1 + 2 + ... + \", n, \" = \", total, sep='')\nelse: \n print(total)",
"_____no_output_____"
]
],
[
[
"## Sets in *Python*",
"_____no_output_____"
],
[
"*Python* supports sets as a **native** datatype. This is one of the reasons that have lead me to choose *Python* as the programming language for this course. To get a first impression how sets are handled in *Python*, let us define two simple sets $A$ and $B$ and print them:",
"_____no_output_____"
]
],
[
[
"A = {1, 2, 3}\nB = {2, 3, 4}\nprint(f'A = {A}, B = {B}')",
"_____no_output_____"
]
],
[
[
"There is a caveat here, we cannot define the empty set using the expression <tt>{}</tt> since this expression \ncreates the empty <em style=\"color:blue;\">dictionary</em> instead. (We will discuss the data type of *dictionaries* later.) To define the empty set $\\emptyset$, we therefore have to use the following expression:",
"_____no_output_____"
]
],
[
[
"S = set()\nS",
"_____no_output_____"
]
],
[
[
"Note that the empty set is also printed as <tt>set()</tt> in *Python* and not as <tt>{}</tt>.",
"_____no_output_____"
],
[
"Next, let us compute the <em style=\"color:blue;\">union</em> $A \\cup B$. This is done using the operator \"<tt>|</tt>\". ",
"_____no_output_____"
]
],
[
[
"A | B",
"_____no_output_____"
]
],
[
[
"To compute the <em style=\"color:blue;\">intersection</em> $A \\cap B$, we use the operator \"<tt>&</tt>\":",
"_____no_output_____"
]
],
[
[
"A & B",
"_____no_output_____"
]
],
[
[
"The <em style=\"color:blue;\">set difference</em> $A \\backslash B$ is computed using the operator \"<tt>-</tt>\":",
"_____no_output_____"
]
],
[
[
"A - B, B - A",
"_____no_output_____"
]
],
[
[
"It is easy to test whether $A$ is a <em style=\"color:blue;\">subset</em> of $B$, i.e. whether $A \\subseteq B$ holds:",
"_____no_output_____"
]
],
[
[
"A <= B",
"_____no_output_____"
]
],
[
[
"Testing whether an object $x$ is an <em style=\"color:blue;\">element</em> of a set $M$, i.e. to test whether $x \\in M$ holds, is straightforward:",
"_____no_output_____"
]
],
[
[
"1 in A",
"_____no_output_____"
]
],
[
[
"On the other hand, the number $1$ is not an element of the set $B$, i.e. we have $1 \\not\\in B$:",
"_____no_output_____"
]
],
[
[
"1 not in B",
"_____no_output_____"
]
],
[
[
"It is important to know that sets are <b style=\"color:red; background-color:rgb(0,112,122)\">not ordered</b>. The reason is that sets are implemented as <em style=\"color:blue;\">hash tables</em>. We discuss hash tables in the lecture on algorithms in the second semester.",
"_____no_output_____"
]
],
[
[
"print({-1,2,-3,4})",
"_____no_output_____"
]
],
[
[
"However, if a set is displayed by a **Jupyter notebook** without a print statement, the elements are sorted.",
"_____no_output_____"
]
],
[
[
"{-1,2,-3,4}",
"_____no_output_____"
]
],
[
[
"## Defining Sets via Selection and Images",
"_____no_output_____"
],
[
"Remember how we can define subsets of a given set $M$ via the axiom of selection. If $p$ is a property such that for any object $x$ from the set $M$ the expression $p(x)$ is either <tt>True</tt> or <tt>False</tt>, the subset of all those elements of $M$ such that $p(x)$ is <tt>True</tt> can be defined as\n$$ \\{ x \\in M \\mid p(x) \\}. $$\nFor example, if $M$ is the set $\\{1, \\cdots, 100\\}$ and we want to compute the subset of this set that contains all numbers from $M$ that are divisible by $7$, then this set can be defined as\n$$ \\{ x \\in M \\mid x \\texttt{%} 7 = 0 \\}. $$\nIn *Python*, the definition of this set can be given as follows: ",
"_____no_output_____"
]
],
[
[
"M = set(range(1, 100+1))\n{ x for x in M if x % 7 == 0 }",
"_____no_output_____"
]
],
[
[
"In general, in *Python* the set\n$$ \\{ x \\in M \\mid p(x) \\} $$\nis computed by the expression\n$$ \\{\\; x\\; \\texttt{for}\\; x\\; \\texttt{in}\\; M\\; \\texttt{if}\\; p(x)\\; \\}. $$\nThis is called a *set comprehension*.\n\n*Image* sets can be computed in a similar way. If $f$ is a function defined for all elements of a set $M$, the image set \n$$ \\{ f(x) \\mid x \\in M \\} $$\ncan be computed in *Python* as follows:\n$$ \\{\\; f(x)\\; \\texttt{for}\\; x\\; \\texttt{in}\\; M\\; \\}. $$\nFor example, the following expression computes the set of all squares of numbers from the set $\\{1,\\cdots,10\\}$:",
"_____no_output_____"
]
],
[
[
"M = set(range(1,10+1))\n{ x*x for x in M }",
"_____no_output_____"
]
],
[
[
"The computation of image sets and selections can be *combined.* If $M$ is a set, $p$ is a property such that $p(x)$ is either <tt>True</tt> or <tt>False</tt> for elements of $M$, and $f$ is a function such that $f(x)$ is defined for all $x \\in M$ then we can compute set \n$$ \\{ f(x) \\mid x \\in M \\wedge p(x) \\} $$\nof all images $f(x)$ from those $x\\in M$ that satisfy the property $p(x)$ via the expression\n$$ \\{\\; f(x)\\; \\texttt{for}\\; x\\; \\texttt{in}\\; M\\; \\texttt{if}\\; p(x)\\; \\}. $$\nFor example, to compute the set of those squares of numbers from the set $\\{1,\\cdots,10\\}$ that are even we can write",
"_____no_output_____"
]
],
[
[
"{ x*x for x in M \n if x % 2 == 0 \n}",
"_____no_output_____"
]
],
[
[
"We can iterate over more than one set. For example, let us define the set of all products $p \\cdot q$ of numbers $p$ and $q$ from the set $\\{2, \\cdots, 10\\}$, i.e. we intend to define the set\n$$ \\bigl\\{ p \\cdot q \\bigm| p \\in \\{2,\\cdots,10\\} \\wedge q \\in \\{2,\\cdots,10\\} \\bigr\\}. $$\nIn *Python*, this set is defined as follows: ",
"_____no_output_____"
]
],
[
[
"{ p * q for p in range(2, 6+1) \n for q in range(2, 6+1) \n}",
"_____no_output_____"
]
],
[
[
"We can use this set to compute the set of *prime numbers*. After all, the set of prime numbers is the set of all those natural numbers bigger than $1$ that can not be written as a proper product, that is a number $x$ is *prime* if \n<ul>\n <li> $x$ is bigger than $1$ and </li>\n <li> there are no natural numbers $p$ and $q$ both bigger than $1$ such that \n $x = p \\cdot q$ holds.</li>\n</ul>\nMore formally, the set $\\mathbb{P}$ of prime numbers is defined as follows:\n$$ \\mathbb{P} = \\Bigl\\{ x \\in \\mathbb{N} \\;\\bigm|\\; x > 1 \\wedge \\neg \\exists p, q \\in \\mathbb{N}: \\bigl(x = p \\cdot q \\wedge p > 1 \\wedge q > 1\\bigr)\\Bigr\\}. $$\nHence the following code computes the set of all primes less \nthan 100: ",
"_____no_output_____"
]
],
[
[
"S = set(range(2, 100+1))\nprint(S - { p * q for p in S for q in S })",
"_____no_output_____"
]
],
[
[
"An alternative way to compute primes works by noting that a number $p$ is prime iff there is no number $t$ that is different from both $1$ and $p$ and, furthermore, divides the number $p$. The function `dividers` given below computes the set of all numbers dividing a given number $p$ evenly:",
"_____no_output_____"
]
],
[
[
"def dividers(p):\n '''\n Compute the set of numbers that divide the number p.\n This is just an auxiliary function.\n '''\n return { t for t in range(1, p+1) if p % t == 0 }\n\ndividers(20)",
"_____no_output_____"
],
[
"n = 100\nprimes = { p for p in range(2, n) if dividers(p) == {1, p} }\nprint(primes)",
"_____no_output_____"
]
],
[
[
"## Computing the Power Set",
"_____no_output_____"
],
[
"Unfortunately, there is no operator to compute the power set $2^M$ of a given set $M$. Since the power set is needed frequently, we have to implement a function <tt>power</tt> to compute this set ourselves. The easiest way to compute the power set $2^M$ of a set $M$ is to implement the following recursive equations:\n<ol>\n <li>The power set of the empty set contains only the empty set:\n $$2^{\\{\\}} = \\bigl\\{\\{\\}\\bigr\\}$$</li>\n <li>If a set $M$ can be written as $M = C \\cup \\{x\\}$, where the element $x$ does not occur in the set $C$, then \n the power set $2^M$ consists of two sets:</li>\n <ul>\n <li>Firstly, all subsets of $C$ are also subsets of $M$.</li>\n <li>Secondly, if A is a subset of $C$, then the set $A \\cup\\{x\\}$ is also a subset of $M$.</li>\n </ul>\n If we combine these parts we get the following equation:\n $$2^{C \\cup \\{x\\}} = 2^C \\cup \\bigl\\{ A \\cup \\{x\\} \\bigm| A \\in 2^C \\bigr\\}$$ \n</ol> \nBut there is another problem: In <em>Python</em> we can't create a set that has elements that are sets themselves! The expression <tt>{{1,2}, {2,3}}</tt> raises an exception when evaluated! Instead, we have to use <tt>frozenset</tt>s as shown below:",
"_____no_output_____"
]
],
[
[
"{{1,2}, {2,3}}",
"_____no_output_____"
],
[
"{frozenset({1,2}), frozenset({2,3})}",
"_____no_output_____"
]
],
[
[
"The reason we got the error when trying to evaluate the expression\n```\n {{1,2}, {2,3}}\n``` \nis that in *Python* sets are implemented via *hash tables* and therefore the elements of a set need \nto be *hashable*. (The notion of a *hash table* will be discussed in more detail in the lecture on *Algorithms*.) However, sets are *mutable* and *mutable* objects are not *hashable*. Fortunately, there is a\nworkaround: *Python* provides the data type of *frozen sets*. These sets behave like sets but are are lacking certain function and hence are **immutable**. So if we use *frozen sets* as elements of the power set, we can compute the power set of a given set. The function <tt>power</tt> given below shows how this works.",
"_____no_output_____"
]
],
[
[
"def power(M):\n \"This function computes the power set of the set M.\"\n if M == set():\n return { frozenset() }\n else:\n C = set(M) # C is a copy of M as we don't want to change the set M\n x = C.pop() # pop removes some element x from the set C\n P1 = power(C)\n P2 = { A | {x} for A in P1 }\n return P1 | P2",
"_____no_output_____"
],
[
"power(A)",
"_____no_output_____"
]
],
[
[
"Let us print this in a more readable way. To this end we implement a function <tt>prettify</tt> that turns a set of frozensets into a string that looks like a set of sets.",
"_____no_output_____"
]
],
[
[
"def prettify(M):\n \"\"\"Turn the set of frozen sets M into a string that looks like a set of sets.\n M is assumed to be the power set of some set.\n \"\"\"\n result = \"{{}, \" # The empty set is always an element of a power set.\n for A in M:\n if A == set(): # The empty set has already been taken care of.\n continue\n result += str(set(A)) + \", \" # A is converted from a frozen set to a set\n result = result[:-2] # remove the trailing substring \", \"\n result += \"}\"\n return result",
"_____no_output_____"
],
[
"prettify(power(A))",
"_____no_output_____"
]
],
[
[
"The function $S\\texttt{.add}(x)$ insert the element $x$ into the set $S$.",
"_____no_output_____"
]
],
[
[
"S = {1, 2, 3}\nS.add(4)\nS",
"_____no_output_____"
]
],
[
[
"In *Python*, variables are <em style=\"color:blue;\">references</em> and assignments \n<b style=\"color:red; background-color:rgb(0,112,122)\">do not copy values</b>. Instead, they just create new references to the old values! This is the reason that below both <tt>A</tt> and <tt>B</tt> change their value, even so it seems that we only change <tt>A</tt>. ",
"_____no_output_____"
]
],
[
[
"A = {1,2,3}\nB = A\nA.add(4)\nprint(f'A = {A}')\nprint(f'B = {B}')",
"_____no_output_____"
]
],
[
[
"In order to prevent this behaviour, we have to make a <em style=\"color:blue;\">copy</em> of the set <tt>A</tt>. This can be done by calling the function $\\texttt{set}(\\texttt{A})$.",
"_____no_output_____"
]
],
[
[
"A = {1,2,3}\nB = set(A)\nA.add(4)\nprint(f'A = {A}')\nprint(f'B = {B}')",
"_____no_output_____"
]
],
[
[
"## Pairs and Cartesian Products",
"_____no_output_____"
],
[
"In *Python*, pairs can be created by enclosing the components of the pair in parentheses. For example, to compute the pair $\\langle 1, 2 \\rangle$ we can write:",
"_____no_output_____"
]
],
[
[
"(1, 2)",
"_____no_output_____"
]
],
[
[
"It is not even necessary to enclose the components of a pair in parentheses. For example, to compute the pair $\\langle 1, 2 \\rangle$ we can also use the following expression:",
"_____no_output_____"
]
],
[
[
"1, 2",
"_____no_output_____"
]
],
[
[
"The Cartesian product $A \\times B$ of two sets $A$ and $B$ can now be computed via the following expression:\n$$ \\{\\; (x, y) \\;\\texttt{for}\\; x \\;\\texttt{in}\\; A\\; \\texttt{for}\\; y\\; \\texttt{in}\\; B\\; \\} $$ \nFor example, as we have defined $A$ as $\\{1,2,3\\}$ and $B$ as $\\{4,5\\}$, the Cartesian product of $A$ and $B$ is computed as follows:",
"_____no_output_____"
]
],
[
[
"A = {1, 2, 3}\nB = {4, 5}\n{ (x, y) for x in A \n for y in B \n}",
"_____no_output_____"
]
],
[
[
"## Tuples",
"_____no_output_____"
],
[
"*Tuples* are a generalization of pairs. For example, to compute the tuple $\\langle 1, 2, 3 \\rangle$ we can use the following expression: ",
"_____no_output_____"
]
],
[
[
"(1, 2, 3)",
"_____no_output_____"
]
],
[
[
"Tuples are not sets, the order of the elements matters:",
"_____no_output_____"
]
],
[
[
"(1, 2, 3) != (2, 3, 1)",
"_____no_output_____"
]
],
[
[
"Longer tuples can be build using the function <tt>range</tt> in combination with the function <tt>tuple</tt>:",
"_____no_output_____"
]
],
[
[
"tuple(range(1, 10+1))",
"_____no_output_____"
]
],
[
[
"Tuple can be *concatenated* using the operator <tt>+</tt>:",
"_____no_output_____"
]
],
[
[
"T1 = (1, 2, 3)\nT2 = (4, 5, 6)\nT3 = T1 + T2\nT3",
"_____no_output_____"
]
],
[
[
"The *length* of a tuple is computed using the function <tt>len</tt>:",
"_____no_output_____"
]
],
[
[
"len(T3)",
"_____no_output_____"
]
],
[
[
"The components of a tuple can be extracted using square brackets. <b>Note, that the first component of a tuple has the index $0$!</b> This is similar to the behaviour of *arrays* in the programming language <tt>C</tt>.",
"_____no_output_____"
]
],
[
[
"print(\"T3[0] =\", T3[0])\nprint(\"T3[1] =\", T3[1])\nprint(\"T3[2] =\", T3[2])",
"_____no_output_____"
]
],
[
[
"If we use negative indices, then we index from the back of the tuple, as shown in the following example:",
"_____no_output_____"
]
],
[
[
"print(f'T3[-1] = {T3[-1]}') # last element\nprint(f'T3[-2] = {T3[-2]}') # penultimate element\nprint(f'T3[-3] = {T3[-3]}') # antepenultimate element ",
"_____no_output_____"
],
[
"T3",
"_____no_output_____"
]
],
[
[
"The <em style=\"color:blue;\">slicing operator</em> extracts a subtuple from a given tuple. If $L$ is a tuple and $a$ and $b$ are natural numbers such that $a \\leq b$ and $a,b \\in \\{0, \\texttt{len}(L) \\}$, then the syntax of the slicing operator is as follows:\n$$ L[a:b] $$\nThe expression $L[a:b]$ extracts the subtuple that starts with the element $L[a]$ up to and <b>excluding</b> the element $L[b]$. The following shows an example:",
"_____no_output_____"
]
],
[
[
"L = tuple(range(1,10+1))\nprint(f'L = {L}, L[2:6] = {L[2:6]}')",
"_____no_output_____"
]
],
[
[
"Slicing works with negative indices, too: ",
"_____no_output_____"
]
],
[
[
"L[2:-2]",
"_____no_output_____"
]
],
[
[
"Slicing also works for strings.",
"_____no_output_____"
]
],
[
[
"s = 'abcdef'\ns[2:-2]",
"_____no_output_____"
]
],
[
[
"If we want to create a tuple of length $1$, we have to use the following syntax:",
"_____no_output_____"
]
],
[
[
"L = (1,)\nL",
"_____no_output_____"
]
],
[
[
"Note that above the comma is **not** optional as the expression $(1)$ would be interpreted as the number $1$.",
"_____no_output_____"
],
[
"Similar to working with sets, it is often convenient to build long tuples using *comprehensions*.\nIn order to create all square numbers that are odd and $\\leq$ 100 we can do the following:",
"_____no_output_____"
]
],
[
[
"G = (x*x for x in range(10+1) if x % 2 == 1)\nG",
"_____no_output_____"
]
],
[
[
"Above, `G` is a *generator object*. To turn this into a tuple we use the predefined function `tuple`:",
"_____no_output_____"
]
],
[
[
"tuple(G)",
"_____no_output_____"
]
],
[
[
"## Lists",
"_____no_output_____"
],
[
"Next, we discuss the data type of lists. Lists are a lot like tuples, but in contrast to tuples, lists are \n<em style=\"color:blue;\">mutatable</em>, i.e. we can change lists. To construct a list, we use square backets:",
"_____no_output_____"
]
],
[
[
"L = [1, 2, 3]\nL",
"_____no_output_____"
]
],
[
[
"In a list, the order of the elements does matter:",
"_____no_output_____"
]
],
[
[
"[1, 2, 3] == [3, 2, 1]",
"_____no_output_____"
]
],
[
[
"The *index operator* $[\\cdot]$ is a postfix operator that is used to return the element that is stored at \na given index in a list.",
"_____no_output_____"
]
],
[
[
"L[0]",
"_____no_output_____"
]
],
[
[
"To change the first element of a list, we can use the *index operator* on the left hand side of an assignment:",
"_____no_output_____"
]
],
[
[
"L[0] = 7\nL",
"_____no_output_____"
]
],
[
[
"This last operation would not be possible if <tt>L</tt> had been a tuple instead of a list.\nLists support concatenation in the same way as tuples: ",
"_____no_output_____"
]
],
[
[
"[1,2,3] + [4,5,6]",
"_____no_output_____"
]
],
[
[
"The function `len` computes the length of a list:",
"_____no_output_____"
]
],
[
[
"len([4,5,6])",
"_____no_output_____"
]
],
[
[
"Lists support the functions <tt>max</tt> and <tt>min</tt>. The expression $\\texttt{max}(L)$ computes the maximum of all the elements of the list (or tuple) $L$, while $\\texttt{min}(L)$ computes the smallest element of $L$. These functions also operate on tuples or sets. They even work on list of lists or set of frozensets, but this is not really useful.",
"_____no_output_____"
]
],
[
[
"max([1,2,3])",
"_____no_output_____"
],
[
"min([1,2,3])",
"_____no_output_____"
],
[
"max({1,2,3})",
"_____no_output_____"
],
[
"max([[1,2], [2,3,4], [5]])",
"_____no_output_____"
],
[
"max({frozenset({1,2}), frozenset({2,3,4}), frozenset({5})})",
"_____no_output_____"
]
],
[
[
"## Boolean Values and Boolean Operators",
"_____no_output_____"
],
[
"In *Python*, the *truth values*, also known as *Boolean values*, are written as <tt>True</tt> and <tt>False</tt>. ",
"_____no_output_____"
]
],
[
[
"True",
"_____no_output_____"
],
[
"False",
"_____no_output_____"
]
],
[
[
"The following function is needed for pretty-printing. It assumes that the argument <tt>val</tt> is a truth value. This truth value is then turned into a string that has a size of exactly 5 characters.",
"_____no_output_____"
]
],
[
[
"def toStr(val):\n if val:\n return 'True '\n return 'False'",
"_____no_output_____"
]
],
[
[
"These values can be combined using the <em style=\"color:blue;\">Boolean operators</em> $\\wedge$, $\\vee$, and $\\neg$. In *Python*, these operators are denoted as `and`, `or`, and `not`. The following table shows how the operator `and` is defined:",
"_____no_output_____"
]
],
[
[
"B = (True, False)\nfor x in B:\n for y in B:\n print(toStr(x), 'and', toStr(y), '=', x and y)",
"_____no_output_____"
]
],
[
[
"The *disjunction* of two Boolean values is only *False* if both values are *False*:",
"_____no_output_____"
]
],
[
[
"for x in B:\n for y in B:\n print(toStr(x), 'or', toStr(y), '=', x or y)",
"_____no_output_____"
]
],
[
[
"Finally, the <em style=\"color:blue;\">negation</em> operator works as expected:",
"_____no_output_____"
]
],
[
[
"for x in B:\n print('not', toStr(x), '=', not x)",
"_____no_output_____"
]
],
[
[
"Boolean values are created by comparing numbers using the follwing comparison operators:\n<ol>\n <li>$a\\;\\texttt{==}\\;b$ is true iff $a$ is equal to $b$.</li>\n <li>$a\\;\\texttt{!=}\\;b$ is true iff $a$ is different from $b$.</li>\n <li>$a\\;\\texttt{<}\\;b$ is true iff $a$ is less than $b$.</li>\n <li>$a\\;\\texttt{<=}\\;b$ is true iff $a$ is less than or equal to $b$.</li>\n <li>$a\\;\\texttt{>=}\\;b$ is true iff $a$ is bigger than or equal to $b$.</li>\n <li>$a\\;\\texttt{>}\\;b$ is true iff $a$ is bigger than $b$.</li>\n</ol>",
"_____no_output_____"
]
],
[
[
"1 == 2",
"_____no_output_____"
],
[
"1 != 2",
"_____no_output_____"
],
[
"1 < 2",
"_____no_output_____"
],
[
"1 <= 2",
"_____no_output_____"
],
[
"1 > 2",
"_____no_output_____"
],
[
"1 >= 2",
"_____no_output_____"
]
],
[
[
"Comparison operators can be <em style=\"color:blue;\">chained</em> as shown in the following example:",
"_____no_output_____"
]
],
[
[
"1 < 2 > -1",
"_____no_output_____"
]
],
[
[
"*Python* supports the <em style=\"color:blue;\">universal quantifier</em> $\\forall$. If $L$ is a list of Boolean values, then we can check whether all elements of $L$ are <tt>True</tt> by writing\n$$ \\texttt{all}(L). $$\nFor example, to check whether all elements of a list $L$ are even we can write the following:",
"_____no_output_____"
]
],
[
[
"L = [2, 4, 6, 7]\nall(x % 2 == 0 for x in L)",
"_____no_output_____"
]
],
[
[
"*Python* also supports the <em style=\"color:blue;\">existential quantifier</em> $\\exists$. If $L$ is a list of Boolean values, the expression\n$$ \\texttt{any}(L) $$\nis true iff there exists an element $x \\in L$ such that $x$ is true.",
"_____no_output_____"
]
],
[
[
"any(x ** 2 > 2 ** x for x in range(1,4+1))",
"_____no_output_____"
]
],
[
[
"## Control Structures",
"_____no_output_____"
],
[
"First of all, *Python* supports *branching*. The following example is taken from the *Python* tutorial at [https://python.org](https://docs.python.org/3/tutorial/controlflow.html):",
"_____no_output_____"
]
],
[
[
"x = int(input(\"Please enter an integer: \"))\nif x < 0:\n print('The number is negative!')\nelif x == 0:\n print('The number is zero.')\nelif x == 1:\n print(\"It's a one.\")\nelse:\n print(\"It's more than one.\")",
"_____no_output_____"
]
],
[
[
"*Loops* can be used to iterate over sets, lists, tuples, or generators. The following example prints the numbers from 1 to 10.",
"_____no_output_____"
]
],
[
[
"for x in range(1, 10+1):\n print(x)",
"_____no_output_____"
]
],
[
[
"The same can be achieved with a `while` loop:",
"_____no_output_____"
]
],
[
[
"x = 1\nwhile x <= 10:\n print(x, end=' ')\n x += 1",
"_____no_output_____"
]
],
[
[
"The following program computes the prime numbers according to an\n[algorithm given by Eratosthenes](https://en.wikipedia.org/wiki/Sieve_of_Eratosthenes).\n 1. We set $n$ equal to 100 as we want to compute the set all prime numbers less or equal that 100.\n 2. `primes` is the list of numbers from 0 upto $n$, i.e. we have initially\n $$ \\texttt{primes} = [0,1,2,\\cdots,n] $$\n Therefore, we have\n $$ \\texttt{primes}[i] = i \\quad \\mbox{for all $i \\in \\{0,1,\\cdots,n\\}$.} $$\n The idea is to set $\\texttt{primes}[i]$ to zero iff $i$ is a proper product of two numbers.\n 3. To this end we iterate over all $i$ and $j$ from the set $\\{2,\\cdots,n\\}$\n and set the product $\\texttt{primes}[i\\cdot j]$ to zero. \n This is achieved by the two `for` loops in line 3 and 4 below.\n\n Note that we have to check that the product $i * j$ is not bigger than $n$ \n for otherwise we would get an *out of range error* when trying to assign $\\texttt{primes}[i*j]$. \n \n 4. After the iteration, all non-prime elements greater than one of the list primes have been set to zero.\n 5. Finally, we compute the set of primes by collecting those elements that have not been set to $0$.",
"_____no_output_____"
]
],
[
[
"n = 100\nprimes = list(range(0, n+1))\nprimes[1] = 0\nfor i in range(2, n+1):\n for j in range(2, n+1):\n if i * j <= n:\n primes[i * j] = 0\nprint(primes)\nprint({ i for i in range(2, n+1) if primes[i] != 0 })",
"_____no_output_____"
]
],
[
[
"The algorithm given above can be improved by using the following observations:\n 1. If a number $x$ can be written as a product $a \\cdot b$, then at least one of the numbers \n $a$ or $b$ has to be less than $\\sqrt{x}$. Therefore, the `for` loop in line 5 below \n iterates as long as $i \\leq \\sqrt{x}$. The function `ceil` is needed to cast the square \n root of $x$ to a natural number. In order to use the functions `sqrt` and `ceil` we have \n to import them from the module `math`. This is done in line 1 of the program shown below. \n 2. When we iterate over $j$ in the inner loop, it is sufficient if we start with $j = i$ \n since all products of the form $i \\cdot j$ where $j < i$ have already been eliminated at the time \n when the multiples of $i$ had been eliminated.\n 3. If $\\texttt{primes}[i] = 0$, then $i$ is not a prime and hence it has to be a product of two \n numbers $a$ and $b$ both of which are smaller than $i$. However, since all the multiples of \n $a$ and $b$ have already been eliminated, there is no point in eliminating the multiples of \n $i$ since these are also multiples of both $a$ and $b$ and hence they have already been eliminated. \n Therefore, if $\\texttt{primes}[i] = 0$ we can immediately jump to the next value of $i$. \n This is achieved by the *continue* statement in line 8 below. \n\nThe program shown below is easily capable of computing all prime numbers less than a million.",
"_____no_output_____"
]
],
[
[
"%%time\nfrom math import sqrt, ceil\n\nn = 1000000\nprimes = list(range(n+1))\nfor i in range(2, ceil(sqrt(n))):\n if primes[i] == 0:\n continue\n j = i\n while i * j <= n:\n primes[i * j] = 0\n j += 1\nP = { i for i in range(2, n+1) if primes[i] != 0 }\nprint(sorted(list(P)))",
"_____no_output_____"
]
],
[
[
"## Numerical Functions",
"_____no_output_____"
],
[
"*Python* provides all of the mathematical functions that you learned at school. A detailed listing of these functions can be found at \n[https://docs.python.org/3.6/library/math.html](https://docs.python.org/3.6/library/math.html). We just show the most important functions and constants. In order to make the module `math` available, we use \nthe following `import` statement: ",
"_____no_output_____"
]
],
[
[
"import math",
"_____no_output_____"
]
],
[
[
"The mathematical constant [Pi](https://en.wikipedia.org/wiki/Pi) which is most often written as $\\pi$ is available as `math.pi`.",
"_____no_output_____"
]
],
[
[
"math.pi",
"_____no_output_____"
]
],
[
[
"The <a href=\"https://en.wikipedia.org/wiki/Sine\">sine</a> function is called as follows:",
"_____no_output_____"
]
],
[
[
"math.sin(math.pi/6)",
"_____no_output_____"
]
],
[
[
"The <a href=\"https://en.wikipedia.org/wiki/Trigonometric_functions#cosine\">cosine</a> function is called as follows:",
"_____no_output_____"
]
],
[
[
"math.cos(0.0)",
"_____no_output_____"
]
],
[
[
"The tangent function is called as follows:",
"_____no_output_____"
]
],
[
[
"math.tan(math.pi/4)",
"_____no_output_____"
]
],
[
[
"The *arc sine*, *arc cosine*, and *arc tangent* are called by prefixing the character `'a'` to the name of the function as seen below:",
"_____no_output_____"
]
],
[
[
"math.asin(1.0)",
"_____no_output_____"
],
[
"math.acos(1.0)",
"_____no_output_____"
],
[
"math.atan(1.0)",
"_____no_output_____"
]
],
[
[
"<a href=\"https://en.wikipedia.org/wiki/E_(mathematical_constant)\">Euler's number $e$</a> can be computed as follows:",
"_____no_output_____"
]
],
[
[
"math.e",
"_____no_output_____"
]
],
[
[
"The exponential function $\\mathrm{exp}(x) := e^x$ is computed as follows:",
"_____no_output_____"
]
],
[
[
"math.exp(1)",
"_____no_output_____"
]
],
[
[
"The natural logarithm $\\ln(x)$, which is defined as the inverse of the function $\\exp(x)$, is called `log` (instead of `ln`):",
"_____no_output_____"
]
],
[
[
"math.log(math.e * math.e)",
"_____no_output_____"
]
],
[
[
"The square root $\\sqrt{x}$ of a number $x$ is computed using the function `sqrt`:",
"_____no_output_____"
]
],
[
[
"math.sqrt(2)",
"_____no_output_____"
]
],
[
[
"The <em style=\"color:blue;\">flooring function</em> $\\texttt{floor}(x)$ truncates a floating point number $x$ down to the biggest integer number less or equal to $x$:\n$$ \\texttt{floor}(x) = \\max(\\{ n \\in \\mathbb{Z} \\mid n \\leq x \\} $$",
"_____no_output_____"
]
],
[
[
"math.floor(1.999)",
"_____no_output_____"
]
],
[
[
"The <em style=\"color:blue;\">ceiling function</em> $\\texttt{ceil}(x)$ rounds a floating point number $x$ up to the next integer number bigger or equal to $x$:\n$$ \\texttt{ceil}(x) = \\min(\\{ n \\in \\mathbb{Z} \\mid x \\leq n \\} $$",
"_____no_output_____"
]
],
[
[
"math.ceil(1.001)",
"_____no_output_____"
],
[
"round(1.5), round(1.39), round(1.51)",
"_____no_output_____"
],
[
"abs(-1)",
"_____no_output_____"
]
],
[
[
"## The Help System",
"_____no_output_____"
],
[
"Typing a single question mark <tt>'?'</tt> starts the help system of *Jupyter*.",
"_____no_output_____"
]
],
[
[
"?",
"_____no_output_____"
]
],
[
[
"If the name of a module is followed by a question mark, a description of the module is printed.",
"_____no_output_____"
]
],
[
[
"math?",
"_____no_output_____"
]
],
[
[
"This also works for functions defined in a module.",
"_____no_output_____"
]
],
[
[
"math.sin?",
"_____no_output_____"
]
],
[
[
"The question mark operator only works inside a <em style=\"color:blue;\">Jupyter notebook</em>. If you are using an interpreted in the command line for executing *Python* commands, use the function `help` instead.",
"_____no_output_____"
]
],
[
[
"help(math)",
"_____no_output_____"
],
[
"help(math.sin)",
"_____no_output_____"
],
[
"help(dir)",
"_____no_output_____"
]
],
[
[
"We can use the function <tt>dir()</tt> to print the names of the variables that have been defined.",
"_____no_output_____"
]
],
[
[
"dir()",
"_____no_output_____"
],
[
"2 * 3",
"_____no_output_____"
],
[
"_",
"_____no_output_____"
]
],
[
[
"The <em style=\"color:blue;\">magic</em> command <tt>%quickref</tt> prints an overview of the so called \n<em style=\"color:blue;\">magic commands</em> that are available in <em style=\"color:blue;\">Jupyter notebooks</em>.",
"_____no_output_____"
]
],
[
[
"%quickref",
"_____no_output_____"
]
],
[
[
"We can use the command <tt>ls</tt> to list the files in the current directory.",
"_____no_output_____"
]
],
[
[
"ls -al",
"_____no_output_____"
]
],
[
[
"Prefixing a shell command with a `!` executes this command in a shell. Below, I have used the Windows command `dir`. On Linux, the corresponding command is called `ls`.",
"_____no_output_____"
]
],
[
[
"!ls",
"_____no_output_____"
],
[
"!dir",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
e78f8a8f939481fca497818686f4c5a8b966942d | 32,758 | ipynb | Jupyter Notebook | notebooks/Cat_ii.ipynb | nguyenanhtuan1008/Machine-Learning | 15eb109c0704f607af229d171aebe42ab84b9892 | [
"MIT"
] | null | null | null | notebooks/Cat_ii.ipynb | nguyenanhtuan1008/Machine-Learning | 15eb109c0704f607af229d171aebe42ab84b9892 | [
"MIT"
] | null | null | null | notebooks/Cat_ii.ipynb | nguyenanhtuan1008/Machine-Learning | 15eb109c0704f607af229d171aebe42ab84b9892 | [
"MIT"
] | null | null | null | 20.196054 | 209 | 0.248367 | [
[
[
"import pandas as pd\nfrom sklearn import preprocessing",
"_____no_output_____"
],
[
"df = pd.read_csv(\"../input/kalapa/train_kalapa.csv\")",
"C:\\Users\\NguyenAnhTuan\\Anaconda3\\lib\\site-packages\\IPython\\core\\interactiveshell.py:3058: DtypeWarning: Columns (51) have mixed types. Specify dtype option on import or set low_memory=False.\n interactivity=interactivity, compiler=compiler, result=result)\n"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.province.value_counts()",
"_____no_output_____"
],
[
"df.maCv.value_counts()",
"_____no_output_____"
],
[
"df.FIELD_3.value_counts()",
"_____no_output_____"
],
[
"df.FIELD_7.value_counts()",
"_____no_output_____"
],
[
"df.FIELD_32.value_counts()",
"_____no_output_____"
],
[
"df.FIELD_51.value_counts()",
"_____no_output_____"
],
[
"df.label.value_counts()",
"_____no_output_____"
],
[
"lbl = preprocessing.LabelEncoder()\nlbl.fit(df.bin_0.fillna(-1).values)",
"_____no_output_____"
],
[
"df.bin_0.fillna(-1).values\n# df.bin_0.fillna(-1).values.tolist()#show nan value",
"_____no_output_____"
],
[
"lbl.transform(df.bin_0.fillna(-1).values).tolist()",
"_____no_output_____"
],
[
"lbl = preprocessing.LabelBinarizer()\nlbl.fit(df.bin_0.fillna(-1).values)",
"_____no_output_____"
],
[
"lbl.transform(df.bin_0.fillna(-1).values)",
"_____no_output_____"
],
[
"lbl = preprocessing.OneHotEncoder(categories='auto')\nlbl.fit(df.bin_0.fillna(-1).values.reshape((-1, 1)))",
"_____no_output_____"
],
[
"lbl.transform(df.bin_0.fillna(-1).values.reshape((-1, 1)))",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78f907b0b4d28c95b318f50d5a63cdd49319205 | 271,547 | ipynb | Jupyter Notebook | Real_world_examples/Scalable_machine_learning/1_Extract_training_data.ipynb | anchor228/dea-notebooks | 72a7e1a38b3d6ed27f2c450b3eeba08212fd9727 | [
"Apache-2.0"
] | 1 | 2021-07-28T01:41:10.000Z | 2021-07-28T01:41:10.000Z | Real_world_examples/Scalable_machine_learning/1_Extract_training_data.ipynb | jgrovegeo/dea-notebooks | 87bd3f0a2e7d381f9bf235f5722a2ebb012d1aa5 | [
"Apache-2.0"
] | null | null | null | Real_world_examples/Scalable_machine_learning/1_Extract_training_data.ipynb | jgrovegeo/dea-notebooks | 87bd3f0a2e7d381f9bf235f5722a2ebb012d1aa5 | [
"Apache-2.0"
] | null | null | null | 23.421339 | 626 | 0.340958 | [
[
[
"# Extracting training data from the ODC <img align=\"right\" src=\"../../Supplementary_data/dea_logo.jpg\">\n\n* [**Sign up to the DEA Sandbox**](https://docs.dea.ga.gov.au/setup/sandbox.html) to run this notebook interactively from a browser\n* **Compatibility:** Notebook currently compatible with the `DEA Sandbox` environment\n* **Products used:** \n[ls8_nbart_geomedian_annual](https://explorer.sandbox.dea.ga.gov.au/products/ls8_nbart_geomedian_annual/extents),\n[ls8_nbart_tmad_annual](https://explorer.sandbox.dea.ga.gov.au/products/ls8_nbart_tmad_annual/extents),\n[fc_percentile_albers_annual](https://explorer.sandbox.dea.ga.gov.au/products/fc_percentile_albers_annual/extents)",
"_____no_output_____"
],
[
"## Background\n\n**Training data** is the most important part of any supervised machine learning workflow. The quality of the training data has a greater impact on the classification than the algorithm used. Large and accurate training data sets are preferable: increasing the training sample size results in increased classification accuracy ([Maxell et al 2018](https://www.tandfonline.com/doi/full/10.1080/01431161.2018.1433343)). A review of training data methods in the context of Earth Observation is available [here](https://www.mdpi.com/2072-4292/12/6/1034) \n\nWhen creating training labels, be sure to capture the **spectral variability** of the class, and to use imagery from the time period you want to classify (rather than relying on basemap composites). Another common problem with training data is **class imbalance**. This can occur when one of your classes is relatively rare and therefore the rare class will comprise a smaller proportion of the training set. When imbalanced data is used, it is common that the final classification will under-predict less abundant classes relative to their true proportion.\n\nThere are many platforms to use for gathering training labels, the best one to use depends on your application. GIS platforms are great for collection training data as they are highly flexible and mature platforms; [Geo-Wiki](https://www.geo-wiki.org/) and [Collect Earth Online](https://collect.earth/home) are two open-source websites that may also be useful depending on the reference data strategy employed. Alternatively, there are many pre-existing training datasets on the web that may be useful, e.g. [Radiant Earth](https://www.radiant.earth/) manages a growing number of reference datasets for use by anyone.\n",
"_____no_output_____"
],
[
"## Description\nThis notebook will extract training data (feature layers, in machine learning parlance) from the `open-data-cube` using labelled geometries within a geojson. The default example will use the crop/non-crop labels within the `'data/crop_training_WA.geojson'` file. This reference data was acquired and pre-processed from the USGS's Global Food Security Analysis Data portal [here](https://croplands.org/app/data/search?page=1&page_size=200) and [here](https://e4ftl01.cr.usgs.gov/MEASURES/GFSAD30VAL.001/2008.01.01/).\n\nTo do this, we rely on a custom `dea-notebooks` function called `collect_training_data`, contained within the [dea_tools.classification](../../Tools/dea_tools/classification.py) script. The principal goal of this notebook is to familarise users with this function so they can extract the appropriate data for their use-case. The default example also highlights extracting a set of useful feature layers for generating a cropland mask forWA.\n\n1. Preview the polygons in our training data by plotting them on a basemap\n2. Define a feature layer function to pass to `collect_training_data`\n3. Extract training data from the datacube using `collect_training_data`\n4. Export the training data to disk for use in subsequent scripts\n\n***",
"_____no_output_____"
],
[
"## Getting started\n\nTo run this analysis, run all the cells in the notebook, starting with the \"Load packages\" cell. ",
"_____no_output_____"
],
[
"### Load packages\n",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport os\nimport sys\nimport datacube\nimport numpy as np\nimport xarray as xr\nimport subprocess as sp\nimport geopandas as gpd\nfrom odc.io.cgroups import get_cpu_quota\nfrom datacube.utils.geometry import assign_crs\n\nsys.path.append('../../Scripts')\nfrom dea_plotting import map_shapefile\nfrom dea_bandindices import calculate_indices\nfrom dea_classificationtools import collect_training_data\n\nimport warnings\nwarnings.filterwarnings(\"ignore\")",
"/env/lib/python3.6/site-packages/geopandas/_compat.py:88: UserWarning: The Shapely GEOS version (3.7.2-CAPI-1.11.0 ) is incompatible with the GEOS version PyGEOS was compiled with (3.9.0-CAPI-1.16.2). Conversions between both will be slow.\n shapely_geos_version, geos_capi_version_string\n/env/lib/python3.6/site-packages/datacube/storage/masking.py:8: DeprecationWarning: datacube.storage.masking has moved to datacube.utils.masking\n category=DeprecationWarning)\n"
]
],
[
[
"## Analysis parameters\n\n* `path`: The path to the input vector file from which we will extract training data. A default geojson is provided.\n* `field`: This is the name of column in your shapefile attribute table that contains the class labels. **The class labels must be integers**\n",
"_____no_output_____"
]
],
[
[
"path = 'data/crop_training_WA.geojson' \nfield = 'class'",
"_____no_output_____"
]
],
[
[
"### Find the number of CPUs",
"_____no_output_____"
]
],
[
[
"ncpus = round(get_cpu_quota())\nprint('ncpus = ' + str(ncpus))",
"ncpus = 7\n"
]
],
[
[
"## Preview input data\n\nWe can load and preview our input data shapefile using `geopandas`. The shapefile should contain a column with class labels (e.g. 'class'). These labels will be used to train our model. \n\n> Remember, the class labels **must** be represented by `integers`.\n",
"_____no_output_____"
]
],
[
[
"# Load input data shapefile\ninput_data = gpd.read_file(path)\n\n# Plot first five rows\ninput_data.head()",
"_____no_output_____"
],
[
"# Plot training data in an interactive map\nmap_shapefile(input_data, attribute=field)",
"_____no_output_____"
]
],
[
[
"## Extracting training data\n\nThe function `collect_training_data` takes our geojson containing class labels and extracts training data (features) from the datacube over the locations specified by the input geometries. The function will also pre-process our training data by stacking the arrays into a useful format and removing any `NaN` or `inf` values. \n\nThe below variables can be set within the `collect_training_data` function:\n* `zonal_stats`: An optional string giving the names of zonal statistics to calculate across each polygon (if the geometries in the vector file are polygons and not points). Default is `None` (all pixel values are returned). Supported values are 'mean', 'median', 'max', and 'min'.\n* `return_coords`: If `True`, then the training data will contain two extra columns 'x_coord' and 'y_coord' corresponding to the x,y coordinate of each sample. This variable can be useful for handling spatial autocorrelation between samples later on in the ML workflow when we conduct k-fold cross validation.\n* `dc_query`: a datacube dictionary query object, This should not contain lat and long (x or y) variables as these are supplied by the 'gdf' geometries.\n\n> Note: `collect_training_data` also has a number of additional parameters for handling ODC I/O read failures, where polygons that return an excessive number of null values can be resubmitted to the multiprocessing queue. Check out the [docs](https://github.com/GeoscienceAustralia/dea-notebooks/blob/68d3526f73779f3316c5e28001c69f556c0d39ae/Tools/dea_tools/classification.py#L661) to learn more. \n\nIn addition to the parameters required for `collect_training_data`, we also need to set up a few parameters for the `dc_query` parameter, such as `measurements` (the bands to load from the satellite), the `resolution` (the cell size), and the `output_crs` (the output projection). ",
"_____no_output_____"
]
],
[
[
"# Set up our inputs to collect_training_data\ntime = ('2014')\nzonal_stats = None\nreturn_coords = True\n\n# Set up the inputs for the ODC query\nmeasurements = ['blue', 'green', 'red', 'nir', 'swir1', 'swir2']\nresolution = (-30, 30)\noutput_crs = 'epsg:3577'",
"_____no_output_____"
],
[
"# Generate a new datacube query object\nquery = {\n 'time': time,\n 'measurements': measurements,\n 'resolution': resolution,\n 'output_crs': output_crs,\n}",
"_____no_output_____"
]
],
[
[
"## Defining feature layers\n\nTo create the desired feature layers, we pass instructions to `collect training data` through the `feature_func` parameter. \n\n* `feature_func`: A function for generating feature layers that is applied to the data within the bounds of the input geometry. The 'feature_func' must accept a 'dc_query' object, and return a single xarray.Dataset or xarray.DataArray containing 2D coordinates (i.e x, y - no time dimension). e.g.\n\n def feature_function(query):\n dc = datacube.Datacube(app='feature_layers')\n ds = dc.load(**query)\n ds = ds.mean('time')\n return ds\n\nBelow, we will define a more complicated feature layer function than the brief example shown above. We will calculate some band indices on the Landsat 8 geomedian, append the ternary median aboslute deviation dataset from the same year: [ls8_nbart_tmad_annual](https://explorer.sandbox.dea.ga.gov.au/products/ls8_nbart_tmad_annual/extents), and append fractional cover percentiles for the photosynthetic vegetation band, also from the same year: [fc_percentile_albers_annual](https://explorer.sandbox.dea.ga.gov.au/products/fc_percentile_albers_annual/extents).",
"_____no_output_____"
]
],
[
[
"def feature_layers(query):\n #connect to the datacube\n dc = datacube.Datacube(app='custom_feature_layers')\n \n #load ls8 geomedian\n ds = dc.load(product='ls8_nbart_geomedian_annual',\n **query)\n \n # Calculate some band indices\n da = calculate_indices(ds,\n index=['NDVI', 'LAI', 'MNDWI'],\n drop=False,\n collection='ga_ls_2')\n \n # Add TMADs dataset\n tmad = dc.load(product='ls8_nbart_tmad_annual',\n measurements=['sdev','edev','bcdev'],\n like=ds.geobox, #will match geomedian extent\n time='2014' #same as geomedian\n )\n \n # Add Fractional cover percentiles\n fc = dc.load(product='fc_percentile_albers_annual',\n measurements=['PV_PC_10','PV_PC_50','PV_PC_90'], #only the PV band\n like=ds.geobox, #will match geomedian extent\n time='2014' #same as geomedian\n )\n\n # Merge results into single dataset \n result = xr.merge([da, tmad, fc],compat='override')\n \n return result",
"_____no_output_____"
]
],
[
[
"Now, we can pass this function to `collect_training_data`. This will take a few minutes to run all 430 samples on the default sandbox as it only has two cpus.",
"_____no_output_____"
]
],
[
[
"%%time\ncolumn_names, model_input = collect_training_data(\n gdf=input_data,\n dc_query=query,\n ncpus=ncpus,\n return_coords=return_coords,\n field=field,\n zonal_stats=zonal_stats,\n feature_func=feature_layers)",
"Collecting training data in parallel mode\n"
],
[
"print(column_names)\nprint('')\nprint(np.array_str(model_input, precision=2, suppress_small=True))",
"['class', 'blue', 'green', 'red', 'nir', 'swir1', 'swir2', 'NDVI', 'LAI', 'MNDWI', 'sdev', 'edev', 'bcdev', 'PV_PC_10', 'PV_PC_50', 'PV_PC_90', 'x_coord', 'y_coord']\n\n[[ 1. 809. 1249. ... 70. -1447515. -3510225.]\n [ 1. 1005. 1464. ... 68. -1393035. -3614685.]\n [ 1. 950. 1506. ... 70. -1430025. -3532245.]\n ...\n [ 0. 519. 744. ... 48. -698085. -1657005.]\n [ 0. 667. 1049. ... 22. -516825. -3463935.]\n [ 0. 232. 344. ... 71. -1468095. -3805095.]]\n"
]
],
[
[
"## Separate coordinate data\n\nBy setting `return_coords=True` in the `collect_training_data` function, our training data now has two extra columns called `x_coord` and `y_coord`. We need to separate these from our training dataset as they will not be used to train the machine learning model. Instead, these variables will be used to help conduct Spatial K-fold Cross validation (SKVC) in the notebook `3_Evaluate_optimize_fit_classifier`. For more information on why this is important, see this [article](https://www.tandfonline.com/doi/abs/10.1080/13658816.2017.1346255?journalCode=tgis20).",
"_____no_output_____"
]
],
[
[
"# Select the variables we want to use to train our model\ncoord_variables = ['x_coord', 'y_coord']\n\n# Extract relevant indices from the processed shapefile\nmodel_col_indices = [column_names.index(var_name) for var_name in coord_variables]\n\n# Export to coordinates to file\nnp.savetxt(\"results/training_data_coordinates.txt\", model_input[:, model_col_indices])\n",
"_____no_output_____"
]
],
[
[
"## Export training data\n\nOnce we've collected all the training data we require, we can write the data to disk. This will allow us to import the data in the next step(s) of the workflow.\n",
"_____no_output_____"
]
],
[
[
"# Set the name and location of the output file\noutput_file = \"results/test_training_data.txt\"",
"_____no_output_____"
],
[
"# Grab all columns except the x-y coords\nmodel_col_indices = [column_names.index(var_name) for var_name in column_names[0:-2]]\n\n# Export files to disk\nnp.savetxt(output_file, model_input[:, model_col_indices], header=\" \".join(column_names[0:-2]), fmt=\"%4f\")",
"_____no_output_____"
]
],
[
[
"## Recommended next steps\n\nTo continue working through the notebooks in this `Scalable Machine Learning on the ODC` workflow, go to the next notebook `2_Inspect_training_data.ipynb`.\n\n1. **Extracting training data from the ODC (this notebook)**\n2. [Inspecting training data](2_Inspect_training_data.ipynb)\n3. [Evaluate, optimize, and fit a classifier](3_Evaluate_optimize_fit_classifier.ipynb)\n4. [Classifying satellite data](4_Classify_satellite_data.ipynb)\n5. [Object-based filtering of pixel classifications](5_Object-based_filtering.ipynb)\n",
"_____no_output_____"
],
[
"***\n\n## Additional information\n\n**License:** The code in this notebook is licensed under the [Apache License, Version 2.0](https://www.apache.org/licenses/LICENSE-2.0). \nDigital Earth Australia data is licensed under the [Creative Commons by Attribution 4.0](https://creativecommons.org/licenses/by/4.0/) license.\n\n**Contact:** If you need assistance, please post a question on the [Open Data Cube Slack channel](http://slack.opendatacube.org/) or on the [GIS Stack Exchange](https://gis.stackexchange.com/questions/ask?tags=open-data-cube) using the `open-data-cube` tag (you can view previously asked questions [here](https://gis.stackexchange.com/questions/tagged/open-data-cube)).\nIf you would like to report an issue with this notebook, you can file one on [Github](https://github.com/GeoscienceAustralia/dea-notebooks).\n\n**Last modified:** March 2021\n\n**Compatible datacube version:** ",
"_____no_output_____"
]
],
[
[
"print(datacube.__version__)",
"1.8.4.dev52+g07bc51a5\n"
]
],
[
[
"## Tags\nBrowse all available tags on the DEA User Guide's [Tags Index](https://docs.dea.ga.gov.au/genindex.html)",
"_____no_output_____"
]
],
[
[
"**Tags** :index:`Landsat 8 geomedian`, :index:`Landsat 8 TMAD`, :index:`machine learning`, :index:`collect_training_data`, :index:`Fractional Cover`",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"raw"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"raw"
]
] |
e78f91c2c380b0691fa2dafdcc549522dedad285 | 4,296 | ipynb | Jupyter Notebook | Sort/0926/1122. Relative Sort Array.ipynb | YuHe0108/Leetcode | 90d904dde125dd35ee256a7f383961786f1ada5d | [
"Apache-2.0"
] | 1 | 2020-08-05T11:47:47.000Z | 2020-08-05T11:47:47.000Z | Sort/0926/1122. Relative Sort Array.ipynb | YuHe0108/LeetCode | b9e5de69b4e4d794aff89497624f558343e362ad | [
"Apache-2.0"
] | null | null | null | Sort/0926/1122. Relative Sort Array.ipynb | YuHe0108/LeetCode | b9e5de69b4e4d794aff89497624f558343e362ad | [
"Apache-2.0"
] | null | null | null | 27.363057 | 85 | 0.429469 | [
[
[
"说明:\n 给定两个数组arr1和arr2,arr2的元素是不同的,arr2中的所有元素也在arr1中。\n 对arr1的元素进行排序,以使arr1中项目的相对顺序与arr2中的相同。\n 不在arr2中出现的元素应按升序放置在arr1的末尾。\n\nExample 1:\n Input: arr1 = [2,3,1,3,2,4,6,7,9,2,19], arr2 = [2,1,4,3,9,6]\n Output: [2,2,2,1,4,3,3,9,6,7,19]\n\nConstraints:\n 1、arr1.length, arr2.length <= 1000\n 2、0 <= arr1[i], arr2[i] <= 1000\n 3、Each arr2[i] is distinct.\n 4、Each arr2[i] is in arr1.",
"_____no_output_____"
]
],
[
[
"class Solution:\n def relativeSortArray(self, arr1, arr2):\n res = sorted(arr1)\n idx_r = 0\n idx_2 = 0\n print(res)\n while idx_r < len(res) and idx_2 < len(arr2):\n if res[idx_r] == arr2[idx_2]:\n if idx_r < len(res) - 1 and res[idx_r + 1] != res[idx_r]:\n idx_2 += 1\n idx_r += 1\n else:\n while idx_r < len(res) - 1 and res[idx_r] == res[idx_r + 1]:\n idx_r += 1\n print(res)\n res[idx_r], res[idx_r+1] = res[idx_r+1], res[idx_r]\n return res",
"_____no_output_____"
],
[
"class Solution:\n def relativeSortArray(self, arr1, arr2):\n res = []\n arr1.sort()\n for n_2 in arr2:\n while n_2 in arr1:\n res.append(n_2)\n arr1.remove(n_2)\n res.extend(arr1)\n return res",
"_____no_output_____"
],
[
"class Solution:\n def relativeSortArray(self, arr1, arr2):\n frequency_dict = {}\n for i in range(len(arr1)):\n if arr1[i] in frequency_dict:\n frequency_dict[arr1[i]] += 1\n else:\n frequency_dict[arr1[i]] = 1 \n \n result = [] \n for i in range(len(arr2)):\n if arr2[i] in frequency_dict:\n for j in range(0,frequency_dict[arr2[i]]):\n result.append(arr2[i])\n frequency_dict.pop(arr2[i])\n \n if len(frequency_dict.keys()) == 0:\n return result\n else:\n excess = list(frequency_dict.keys())\n excess.sort()\n for i in range(len(excess)):\n for j in range(0,frequency_dict[excess[i]]):\n result.append(excess[i])\n return result",
"_____no_output_____"
],
[
"solution = Solution()\nsolution.relativeSortArray([2,3,1,3,2,4,6,7,9,2,19], [2,1,4,3,9,6])",
"{2: 3, 3: 2, 1: 1, 4: 1, 6: 1, 7: 1, 9: 1, 19: 1}\n"
]
]
] | [
"raw",
"code"
] | [
[
"raw"
],
[
"code",
"code",
"code",
"code"
]
] |
e78f96d03a94471f279f23eeab0aaab7def62941 | 952,940 | ipynb | Jupyter Notebook | Twitter_Sentiment.ipynb | prava1/The-Sparks-Foundation-Task | 7502ceb5e58929f8c2486a47d71a03ebfecf421c | [
"Apache-2.0"
] | null | null | null | Twitter_Sentiment.ipynb | prava1/The-Sparks-Foundation-Task | 7502ceb5e58929f8c2486a47d71a03ebfecf421c | [
"Apache-2.0"
] | null | null | null | Twitter_Sentiment.ipynb | prava1/The-Sparks-Foundation-Task | 7502ceb5e58929f8c2486a47d71a03ebfecf421c | [
"Apache-2.0"
] | null | null | null | 487.43734 | 308,342 | 0.915336 | [
[
[
"import numpy as np\nimport pandas as pd\n\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\nimport warnings",
"_____no_output_____"
],
[
"train = pd.read_csv('drive/My Drive/Projects/Twitter Sentiment/train_tweet.csv')\ntest = pd.read_csv('drive/My Drive/Projects/Twitter Sentiment/test_tweets.csv')\n\nprint(train.shape)\nprint(test.shape)",
"(31962, 3)\n(17197, 2)\n"
],
[
"train.head()",
"_____no_output_____"
],
[
"test.head()",
"_____no_output_____"
],
[
"train.isnull().any()\ntest.isnull().any()",
"_____no_output_____"
],
[
"# checking out the negative comments from the train set\n\ntrain[train['label'] == 0].head(10)",
"_____no_output_____"
],
[
"# checking out the postive comments from the train set \n\ntrain[train['label'] == 1].head(10)",
"_____no_output_____"
],
[
"train['label'].value_counts().plot.bar(color = 'pink', figsize = (6, 4))",
"_____no_output_____"
],
[
"# checking the distribution of tweets in the data\n\nlength_train = train['tweet'].str.len().plot.hist(color = 'pink', figsize = (6, 4))\nlength_test = test['tweet'].str.len().plot.hist(color = 'orange', figsize = (6, 4))",
"_____no_output_____"
],
[
"# adding a column to represent the length of the tweet\n\ntrain['len'] = train['tweet'].str.len()\ntest['len'] = test['tweet'].str.len()\n\ntrain.head(10)",
"_____no_output_____"
],
[
"\ntrain.groupby('label').describe()",
"_____no_output_____"
],
[
"train.groupby('len').mean()['label'].plot.hist(color = 'black', figsize = (6, 4),)\nplt.title('variation of length')\nplt.xlabel('Length')\nplt.show()",
"_____no_output_____"
],
[
"from sklearn.feature_extraction.text import CountVectorizer\n\n\ncv = CountVectorizer(stop_words = 'english')\nwords = cv.fit_transform(train.tweet)\n\nsum_words = words.sum(axis=0)\n\nwords_freq = [(word, sum_words[0, i]) for word, i in cv.vocabulary_.items()]\nwords_freq = sorted(words_freq, key = lambda x: x[1], reverse = True)\n\nfrequency = pd.DataFrame(words_freq, columns=['word', 'freq'])\n\nfrequency.head(30).plot(x='word', y='freq', kind='bar', figsize=(15, 7), color = 'blue')\nplt.title(\"Most Frequently Occuring Words - Top 30\")\n",
"_____no_output_____"
],
[
"from wordcloud import WordCloud\n\nwordcloud = WordCloud(background_color = 'white', width = 1000, height = 1000).generate_from_frequencies(dict(words_freq))\n\nplt.figure(figsize=(10,8))\nplt.imshow(wordcloud)\nplt.title(\"WordCloud - Vocabulary from Reviews\", fontsize = 22)",
"_____no_output_____"
],
[
"normal_words =' '.join([text for text in train['tweet'][train['label'] == 0]])\n\nwordcloud = WordCloud(width=800, height=500, random_state = 0, max_font_size = 110).generate(normal_words)\nplt.figure(figsize=(10, 7))\nplt.imshow(wordcloud, interpolation=\"bilinear\")\nplt.axis('off')\nplt.title('The Neutral Words')\nplt.show()\n",
"_____no_output_____"
],
[
"negative_words =' '.join([text for text in train['tweet'][train['label'] == 1]])\n\nwordcloud = WordCloud(background_color = 'cyan', width=800, height=500, random_state = 0, max_font_size = 110).generate(negative_words)\nplt.figure(figsize=(10, 7))\nplt.imshow(wordcloud, interpolation=\"bilinear\")\nplt.axis('off')\nplt.title('The Negative Words')\nplt.show()\n",
"_____no_output_____"
],
[
"# collecting the hashtags\n\ndef hashtag_extract(x):\n hashtags = []\n \n for i in x:\n ht = re.findall(r\"#(\\w+)\", i)\n hashtags.append(ht)\n\n return hashtags",
"_____no_output_____"
],
[
"# extracting hashtags from non racist/sexist tweets\nHT_regular = hashtag_extract(train['tweet'][train['label'] == 0])\n\n# extracting hashtags from racist/sexist tweets\nHT_negative = hashtag_extract(train['tweet'][train['label'] == 1])\n\n# unnesting list\nHT_regular = sum(HT_regular,[])\nHT_negative = sum(HT_negative,[])",
"_____no_output_____"
],
[
"a = nltk.FreqDist(HT_regular)\nd = pd.DataFrame({'Hashtag': list(a.keys()),\n 'Count': list(a.values())})\n\n# selecting top 20 most frequent hashtags \nd = d.nlargest(columns=\"Count\", n = 20) \nplt.figure(figsize=(16,5))\nax = sns.barplot(data=d, x= \"Hashtag\", y = \"Count\")\nax.set(ylabel = 'Count')\nplt.show()",
"/usr/local/lib/python3.6/dist-packages/seaborn/categorical.py:1428: FutureWarning: remove_na is deprecated and is a private function. Do not use.\n stat_data = remove_na(group_data)\n"
],
[
"a = nltk.FreqDist(HT_negative)\nd = pd.DataFrame({'Hashtag': list(a.keys()),\n 'Count': list(a.values())})\n\n# selecting top 20 most frequent hashtags \nd = d.nlargest(columns=\"Count\", n = 20) \nplt.figure(figsize=(16,5))\nax = sns.barplot(data=d, x= \"Hashtag\", y = \"Count\")\nax.set(ylabel = 'Count')\nplt.show()",
"/usr/local/lib/python3.6/dist-packages/seaborn/categorical.py:1428: FutureWarning: remove_na is deprecated and is a private function. Do not use.\n stat_data = remove_na(group_data)\n"
],
[
"# tokenizing the words present in the training set\ntokenized_tweet = train['tweet'].apply(lambda x: x.split()) \n\n# importing gensim\nimport gensim\n\n# creating a word to vector model\nmodel_w2v = gensim.models.Word2Vec(\n tokenized_tweet,\n size=200, # desired no. of features/independent variables \n window=5, # context window size\n min_count=2,\n sg = 1, # 1 for skip-gram model\n hs = 0,\n negative = 10, # for negative sampling\n workers= 2, # no.of cores\n seed = 34)\n\nmodel_w2v.train(tokenized_tweet, total_examples= len(train['tweet']), epochs=20)",
"_____no_output_____"
],
[
"model_w2v.wv.most_similar(positive = \"dinner\")",
"/usr/local/lib/python3.6/dist-packages/gensim/matutils.py:737: FutureWarning: Conversion of the second argument of issubdtype from `int` to `np.signedinteger` is deprecated. In future, it will be treated as `np.int64 == np.dtype(int).type`.\n if np.issubdtype(vec.dtype, np.int):\n"
],
[
"model_w2v.wv.most_similar(positive = \"cancer\")",
"/usr/local/lib/python3.6/dist-packages/gensim/matutils.py:737: FutureWarning: Conversion of the second argument of issubdtype from `int` to `np.signedinteger` is deprecated. In future, it will be treated as `np.int64 == np.dtype(int).type`.\n if np.issubdtype(vec.dtype, np.int):\n"
],
[
"model_w2v.wv.most_similar(positive = \"apple\")",
"/usr/local/lib/python3.6/dist-packages/gensim/matutils.py:737: FutureWarning: Conversion of the second argument of issubdtype from `int` to `np.signedinteger` is deprecated. In future, it will be treated as `np.int64 == np.dtype(int).type`.\n if np.issubdtype(vec.dtype, np.int):\n"
],
[
"model_w2v.wv.most_similar(negative = \"hate\")",
"/usr/local/lib/python3.6/dist-packages/gensim/matutils.py:737: FutureWarning: Conversion of the second argument of issubdtype from `int` to `np.signedinteger` is deprecated. In future, it will be treated as `np.int64 == np.dtype(int).type`.\n if np.issubdtype(vec.dtype, np.int):\n"
],
[
"from tqdm import tqdm\ntqdm.pandas(desc=\"progress-bar\")\nfrom gensim.models.doc2vec import LabeledSentence",
"_____no_output_____"
],
[
"def add_label(twt):\n output = []\n for i, s in zip(twt.index, twt):\n output.append(LabeledSentence(s, [\"tweet_\" + str(i)]))\n return output\n\n# label all the tweets\nlabeled_tweets = add_label(tokenized_tweet)\n\nlabeled_tweets[:6]",
"/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:4: DeprecationWarning: Call to deprecated `LabeledSentence` (Class will be removed in 4.0.0, use TaggedDocument instead).\n after removing the cwd from sys.path.\n"
],
[
"# removing unwanted patterns from the data\n\nimport re\nimport nltk\n\nnltk.download('stopwords')\nfrom nltk.corpus import stopwords\nfrom nltk.stem.porter import PorterStemmer\n",
"[nltk_data] Downloading package stopwords to /root/nltk_data...\n[nltk_data] Package stopwords is already up-to-date!\n"
],
[
"train_corpus = []\n\nfor i in range(0, 31962):\n review = re.sub('[^a-zA-Z]', ' ', train['tweet'][i])\n review = review.lower()\n review = review.split()\n \n ps = PorterStemmer()\n \n # stemming\n review = [ps.stem(word) for word in review if not word in set(stopwords.words('english'))]\n \n # joining them back with space\n review = ' '.join(review)\n train_corpus.append(review)",
"_____no_output_____"
],
[
"test_corpus = []\n\nfor i in range(0, 17197):\n review = re.sub('[^a-zA-Z]', ' ', test['tweet'][i])\n review = review.lower()\n review = review.split()\n \n ps = PorterStemmer()\n \n # stemming\n review = [ps.stem(word) for word in review if not word in set(stopwords.words('english'))]\n \n # joining them back with space\n review = ' '.join(review)\n test_corpus.append(review)",
"_____no_output_____"
],
[
"# creating bag of words\n\nfrom sklearn.feature_extraction.text import CountVectorizer\n\ncv = CountVectorizer(max_features = 2500)\nx = cv.fit_transform(train_corpus).toarray()\ny = train.iloc[:, 1]\n\nprint(x.shape)\nprint(y.shape)\n",
"(31962, 2500)\n(31962,)\n"
],
[
"# creating bag of words\n\nfrom sklearn.feature_extraction.text import CountVectorizer\n\ncv = CountVectorizer(max_features = 2500)\nx_test = cv.fit_transform(test_corpus).toarray()\n\nprint(x_test.shape)\n",
"(17197, 2500)\n"
],
[
"# splitting the training data into train and valid sets\n\nfrom sklearn.model_selection import train_test_split\n\nx_train, x_valid, y_train, y_valid = train_test_split(x, y, test_size = 0.25, random_state = 42)\n\nprint(x_train.shape)\nprint(x_valid.shape)\nprint(y_train.shape)\nprint(y_valid.shape)",
"(23971, 2500)\n(7991, 2500)\n(23971,)\n(7991,)\n"
],
[
"# standardization\n\nfrom sklearn.preprocessing import StandardScaler\n\nsc = StandardScaler()\n\nx_train = sc.fit_transform(x_train)\nx_valid = sc.transform(x_valid)\nx_test = sc.transform(x_test)\n",
"/usr/local/lib/python3.6/dist-packages/sklearn/utils/validation.py:595: DataConversionWarning: Data with input dtype int64 was converted to float64 by StandardScaler.\n warnings.warn(msg, DataConversionWarning)\n/usr/local/lib/python3.6/dist-packages/sklearn/utils/validation.py:595: DataConversionWarning: Data with input dtype int64 was converted to float64 by StandardScaler.\n warnings.warn(msg, DataConversionWarning)\n/usr/local/lib/python3.6/dist-packages/sklearn/utils/validation.py:595: DataConversionWarning: Data with input dtype int64 was converted to float64 by StandardScaler.\n warnings.warn(msg, DataConversionWarning)\n/usr/local/lib/python3.6/dist-packages/sklearn/utils/validation.py:595: DataConversionWarning: Data with input dtype int64 was converted to float64 by StandardScaler.\n warnings.warn(msg, DataConversionWarning)\n"
],
[
"from sklearn.ensemble import RandomForestClassifier\nfrom sklearn.metrics import confusion_matrix\nfrom sklearn.metrics import f1_score\n\nmodel = RandomForestClassifier()\nmodel.fit(x_train, y_train)\n\ny_pred = model.predict(x_valid)\n\nprint(\"Training Accuracy :\", model.score(x_train, y_train))\nprint(\"Validation Accuracy :\", model.score(x_valid, y_valid))\n\n# calculating the f1 score for the validation set\nprint(\"F1 score :\", f1_score(y_valid, y_pred))\n\n# confusion matrix\ncm = confusion_matrix(y_valid, y_pred)\nprint(cm)\n\n",
"/usr/local/lib/python3.6/dist-packages/sklearn/ensemble/forest.py:246: FutureWarning: The default value of n_estimators will change from 10 in version 0.20 to 100 in 0.22.\n \"10 in version 0.20 to 100 in 0.22.\", FutureWarning)\n"
],
[
"from sklearn.linear_model import LogisticRegression\n\nmodel = LogisticRegression()\nmodel.fit(x_train, y_train)\n\ny_pred = model.predict(x_valid)\n\nprint(\"Training Accuracy :\", model.score(x_train, y_train))\nprint(\"Validation Accuracy :\", model.score(x_valid, y_valid))\n\n# calculating the f1 score for the validation set\nprint(\"f1 score :\", f1_score(y_valid, y_pred))\n\n# confusion matrix\ncm = confusion_matrix(y_valid, y_pred)\nprint(cm)\n",
"/usr/local/lib/python3.6/dist-packages/sklearn/linear_model/logistic.py:433: FutureWarning: Default solver will be changed to 'lbfgs' in 0.22. Specify a solver to silence this warning.\n FutureWarning)\n"
],
[
"from sklearn.tree import DecisionTreeClassifier\n\nmodel = DecisionTreeClassifier()\nmodel.fit(x_train, y_train)\n\ny_pred = model.predict(x_valid)\n\nprint(\"Training Accuracy :\", model.score(x_train, y_train))\nprint(\"Validation Accuracy :\", model.score(x_valid, y_valid))\n\n# calculating the f1 score for the validation set\nprint(\"f1 score :\", f1_score(y_valid, y_pred))\n\n# confusion matrix\ncm = confusion_matrix(y_valid, y_pred)\nprint(cm)\n",
"Training Accuracy : 0.9991656585040257\nValidation Accuracy : 0.9326742585408585\nf1 score : 0.5393835616438356\n[[7138 294]\n [ 244 315]]\n"
],
[
"from sklearn.svm import SVC\n\nmodel = SVC()\nmodel.fit(x_train, y_train)\n\ny_pred = model.predict(x_valid)\n\nprint(\"Training Accuracy :\", model.score(x_train, y_train))\nprint(\"Validation Accuracy :\", model.score(x_valid, y_valid))\n\n# calculating the f1 score for the validation set\nprint(\"f1 score :\", f1_score(y_valid, y_pred))\n\n# confusion matrix\ncm = confusion_matrix(y_valid, y_pred)\nprint(cm)\n",
"_____no_output_____"
],
[
"from xgboost import XGBClassifier\n\nmodel = XGBClassifier()\nmodel.fit(x_train, y_train)\n\ny_pred = model.predict(x_valid)\n\nprint(\"Training Accuracy :\", model.score(x_train, y_train))\nprint(\"Validation Accuracy :\", model.score(x_valid, y_valid))\n\n# calculating the f1 score for the validation set\nprint(\"f1 score :\", f1_score(y_valid, y_pred))\n\n# confusion matrix\ncm = confusion_matrix(y_valid, y_pred)\nprint(cm)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78f99c123b8b1b54354012beb7f98f040d17970 | 62,106 | ipynb | Jupyter Notebook | tf_estimator_linearmodel.ipynb | Junhojuno/DeepLearning-Beginning | c9072c9504ed79d05273c88dae3ae4ac06367d9f | [
"MIT"
] | null | null | null | tf_estimator_linearmodel.ipynb | Junhojuno/DeepLearning-Beginning | c9072c9504ed79d05273c88dae3ae4ac06367d9f | [
"MIT"
] | 2 | 2018-09-23T13:40:43.000Z | 2018-10-08T05:04:13.000Z | tf_estimator_linearmodel.ipynb | Junhojuno/DeepLearning-Beginning | c9072c9504ed79d05273c88dae3ae4ac06367d9f | [
"MIT"
] | null | null | null | 55.451786 | 676 | 0.57368 | [
[
[
"<a href=\"https://colab.research.google.com/github/Junhojuno/DeepLearning-Beginning/blob/master/tf_estimator_linearmodel.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nimport tensorflow.feature_column as fc\n\nimport os\nimport sys\n\nimport matplotlib.pyplot as plt\nfrom IPython.display import clear_output",
"_____no_output_____"
],
[
"tf.enable_eager_execution()",
"_____no_output_____"
],
[
"!pip install -q requests\n!git clone --depth 1 https://github.com/tensorflow/models",
"Cloning into 'models'...\nremote: Enumerating objects: 3168, done.\u001b[K\nremote: Counting objects: 100% (3168/3168), done.\u001b[K\nremote: Compressing objects: 100% (2682/2682), done.\u001b[K\nremote: Total 3168 (delta 573), reused 2075 (delta 408), pack-reused 0\u001b[K\nReceiving objects: 100% (3168/3168), 370.56 MiB | 40.54 MiB/s, done.\nResolving deltas: 100% (573/573), done.\nChecking out files: 100% (2998/2998), done.\n"
],
[
"# add the root directory of the repo to your python path\nmodels_path = os.path.join(os.getcwd(), 'models')\nsys.path.append(models_path)",
"_____no_output_____"
],
[
"# download the dataset\nfrom official.wide_deep import census_dataset, census_main\ncensus_dataset.download(\"/tmp/census_data/\")",
"W0618 02:17:59.553497 140493422073728 deprecation_wrapper.py:119] From /content/models/official/wide_deep/census_dataset.py:78: The name tf.gfile.MakeDirs is deprecated. Please use tf.io.gfile.makedirs instead.\n\nW0618 02:17:59.555037 140493422073728 deprecation_wrapper.py:119] From /content/models/official/wide_deep/census_dataset.py:81: The name tf.gfile.Exists is deprecated. Please use tf.io.gfile.exists instead.\n\nW0618 02:18:00.228131 140493422073728 deprecation_wrapper.py:119] From /content/models/official/wide_deep/census_dataset.py:62: The name tf.gfile.Open is deprecated. Please use tf.io.gfile.GFile instead.\n\nW0618 02:18:00.459901 140493422073728 deprecation_wrapper.py:119] From /content/models/official/wide_deep/census_dataset.py:73: The name tf.gfile.Remove is deprecated. Please use tf.io.gfile.remove instead.\n\n"
],
[
"# for using in Command line\nif 'PYTHONPATH' in os.environ:\n os.environ['PYTHONPATH'] += os.pathsep + models_path\nelse:\n os.environ['PYTHONPATH'] = models_path",
"_____no_output_____"
],
[
"# Use --help to see what command line options are available:\n!python -m official.wide_deep.census_main --help",
"WARNING: Logging before flag parsing goes to stderr.\nW0618 02:20:33.613071 140282108643200 deprecation_wrapper.py:119] From /content/models/official/wide_deep/census_main.py:114: The name tf.logging.set_verbosity is deprecated. Please use tf.compat.v1.logging.set_verbosity instead.\n\nW0618 02:20:33.613363 140282108643200 deprecation_wrapper.py:119] From /content/models/official/wide_deep/census_main.py:114: The name tf.logging.INFO is deprecated. Please use tf.compat.v1.logging.INFO instead.\n\nTrain DNN on census income dataset.\nflags:\n\n/content/models/official/wide_deep/census_main.py:\n -bs,--batch_size:\n Batch size for training and evaluation. When using multiple gpus, this is\n the\n global batch size for all devices. For example, if the batch size is 32 and\n there are 4 GPUs, each GPU will get 8 examples on each step.\n (default: '40')\n (an integer)\n --[no]clean:\n If set, model_dir will be removed if it exists.\n (default: 'false')\n -dd,--data_dir:\n The location of the input data.\n (default: '/tmp/census_data')\n --[no]download_if_missing:\n Download data to data_dir if it is not already present.\n (default: 'true')\n -ebe,--epochs_between_evals:\n The number of training epochs to run between evaluations.\n (default: '2')\n (an integer)\n -ed,--export_dir:\n If set, a SavedModel serialization of the model will be exported to this\n directory at the end of training. See the README for more details and\n relevant\n links.\n -hk,--hooks:\n A list of (case insensitive) strings to specify the names of training hooks.\n Hook:\n loggingtensorhook\n profilerhook\n examplespersecondhook\n loggingmetrichook\n Example: `--hooks ProfilerHook,ExamplesPerSecondHook`\n See official.utils.logs.hooks_helper for details.\n (default: 'LoggingTensorHook')\n (a comma separated list)\n -md,--model_dir:\n The location of the model checkpoint files.\n (default: '/tmp/census_model')\n -mt,--model_type: <wide|deep|wide_deep>: Select model topology.\n (default: 'wide_deep')\n -te,--train_epochs:\n The number of epochs used to train.\n (default: '40')\n (an integer)\n\nTry --helpfull to get a list of all flags.\n"
],
[
"# run model\n!python -m official.wide_deep.census_main --model_type=wide --train_epochs=2",
"WARNING: Logging before flag parsing goes to stderr.\nW0618 02:21:41.690503 140374330308480 deprecation_wrapper.py:119] From /content/models/official/wide_deep/census_main.py:114: The name tf.logging.set_verbosity is deprecated. Please use tf.compat.v1.logging.set_verbosity instead.\n\nW0618 02:21:41.690805 140374330308480 deprecation_wrapper.py:119] From /content/models/official/wide_deep/census_main.py:114: The name tf.logging.INFO is deprecated. Please use tf.compat.v1.logging.INFO instead.\n\nW0618 02:21:41.693827 140374330308480 deprecation_wrapper.py:119] From /content/models/official/wide_deep/census_dataset.py:78: The name tf.gfile.MakeDirs is deprecated. Please use tf.io.gfile.makedirs instead.\n\nW0618 02:21:41.694007 140374330308480 deprecation_wrapper.py:119] From /content/models/official/wide_deep/census_dataset.py:81: The name tf.gfile.Exists is deprecated. Please use tf.io.gfile.exists instead.\n\nW0618 02:21:41.694878 140374330308480 deprecation_wrapper.py:119] From /content/models/official/wide_deep/census_main.py:49: The name tf.ConfigProto is deprecated. Please use tf.compat.v1.ConfigProto instead.\n\nI0618 02:21:41.695704 140374330308480 estimator.py:209] Using config: {'_model_dir': '/tmp/census_model', '_tf_random_seed': None, '_save_summary_steps': 100, '_save_checkpoints_steps': None, '_save_checkpoints_secs': 600, '_session_config': device_count {\n key: \"GPU\"\n value: 0\n}\n, '_keep_checkpoint_max': 5, '_keep_checkpoint_every_n_hours': 10000, '_log_step_count_steps': 100, '_train_distribute': None, '_device_fn': None, '_protocol': None, '_eval_distribute': None, '_experimental_distribute': None, '_experimental_max_worker_delay_secs': None, '_service': None, '_cluster_spec': <tensorflow.python.training.server_lib.ClusterSpec object at 0x7fab375045f8>, '_task_type': 'worker', '_task_id': 0, '_global_id_in_cluster': 0, '_master': '', '_evaluation_master': '', '_is_chief': True, '_num_ps_replicas': 0, '_num_worker_replicas': 1}\nW0618 02:21:41.696815 140374330308480 logger.py:391] 'cpuinfo' not imported. CPU info will not be logged.\n2019-06-18 02:21:41.745614: I tensorflow/core/platform/profile_utils/cpu_utils.cc:94] CPU Frequency: 2300000000 Hz\n2019-06-18 02:21:41.749830: I tensorflow/compiler/xla/service/service.cc:168] XLA service 0x265ad80 executing computations on platform Host. Devices:\n2019-06-18 02:21:41.749868: I tensorflow/compiler/xla/service/service.cc:175] StreamExecutor device (0): <undefined>, <undefined>\n2019-06-18 02:21:41.755555: I tensorflow/stream_executor/platform/default/dso_loader.cc:42] Successfully opened dynamic library libcuda.so.1\n2019-06-18 02:21:42.003599: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:1005] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n2019-06-18 02:21:42.004149: I tensorflow/compiler/xla/service/service.cc:168] XLA service 0x265b9c0 executing computations on platform CUDA. Devices:\n2019-06-18 02:21:42.004179: I tensorflow/compiler/xla/service/service.cc:175] StreamExecutor device (0): Tesla T4, Compute Capability 7.5\n2019-06-18 02:21:42.004397: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:1005] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n2019-06-18 02:21:42.004799: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1640] Found device 0 with properties: \nname: Tesla T4 major: 7 minor: 5 memoryClockRate(GHz): 1.59\npciBusID: 0000:00:04.0\n2019-06-18 02:21:42.021662: I tensorflow/stream_executor/platform/default/dso_loader.cc:42] Successfully opened dynamic library libcudart.so.10.0\n2019-06-18 02:21:42.191784: I tensorflow/stream_executor/platform/default/dso_loader.cc:42] Successfully opened dynamic library libcublas.so.10.0\n2019-06-18 02:21:42.268323: I tensorflow/stream_executor/platform/default/dso_loader.cc:42] Successfully opened dynamic library libcufft.so.10.0\n2019-06-18 02:21:42.295403: I tensorflow/stream_executor/platform/default/dso_loader.cc:42] Successfully opened dynamic library libcurand.so.10.0\n2019-06-18 02:21:42.494979: I tensorflow/stream_executor/platform/default/dso_loader.cc:42] Successfully opened dynamic library libcusolver.so.10.0\n2019-06-18 02:21:42.623572: I tensorflow/stream_executor/platform/default/dso_loader.cc:42] Successfully opened dynamic library libcusparse.so.10.0\n2019-06-18 02:21:42.969422: I tensorflow/stream_executor/platform/default/dso_loader.cc:42] Successfully opened dynamic library libcudnn.so.7\n2019-06-18 02:21:42.969626: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:1005] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n2019-06-18 02:21:42.970163: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:1005] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n2019-06-18 02:21:42.970542: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1763] Adding visible gpu devices: 0\n2019-06-18 02:21:42.972954: I tensorflow/stream_executor/platform/default/dso_loader.cc:42] Successfully opened dynamic library libcudart.so.10.0\n2019-06-18 02:21:42.974925: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1181] Device interconnect StreamExecutor with strength 1 edge matrix:\n2019-06-18 02:21:42.974959: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1187] 0 \n2019-06-18 02:21:42.974982: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1200] 0: N \n2019-06-18 02:21:42.982266: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:1005] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n2019-06-18 02:21:42.982774: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:1005] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n2019-06-18 02:21:42.983156: W tensorflow/core/common_runtime/gpu/gpu_bfc_allocator.cc:40] Overriding allow_growth setting because the TF_FORCE_GPU_ALLOW_GROWTH environment variable is set. Original config value was 0.\n2019-06-18 02:21:42.983206: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1326] Created TensorFlow device (/device:GPU:0 with 14202 MB memory) -> physical GPU (device: 0, name: Tesla T4, pci bus id: 0000:00:04.0, compute capability: 7.5)\nI0618 02:21:48.031453 140374330308480 logger.py:152] Benchmark run: {'model_name': 'wide_deep', 'dataset': {'name': 'Census Income'}, 'machine_config': {'gpu_info': {'count': 1, 'model': 'Tesla T4'}, 'memory_total': 13655322624, 'memory_available': 12568748032}, 'test_id': None, 'run_date': '2019-06-18T02:21:41.696408Z', 'tensorflow_version': {'version': '1.14.0-rc1', 'git_hash': 'v1.14.0-rc1-0-g648ea74ea0'}, 'tensorflow_environment_variables': [{'name': 'TF_FORCE_GPU_ALLOW_GROWTH', 'value': 'true'}], 'run_parameters': [{'name': 'batch_size', 'long_value': 40}, {'name': 'model_type', 'string_value': 'wide'}, {'name': 'train_epochs', 'long_value': 2}]}\nW0618 02:21:48.042725 140374330308480 deprecation.py:323] From /usr/local/lib/python3.6/dist-packages/tensorflow/python/training/training_util.py:236: Variable.initialized_value (from tensorflow.python.ops.variables) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse Variable.read_value. Variables in 2.X are initialized automatically both in eager and graph (inside tf.defun) contexts.\nW0618 02:21:48.071355 140374330308480 deprecation_wrapper.py:119] From /content/models/official/wide_deep/census_dataset.py:167: The name tf.logging.info is deprecated. Please use tf.compat.v1.logging.info instead.\n\nI0618 02:21:48.071504 140374330308480 census_dataset.py:167] Parsing /tmp/census_data/adult.data\nW0618 02:21:48.071606 140374330308480 deprecation_wrapper.py:119] From /content/models/official/wide_deep/census_dataset.py:168: The name tf.decode_csv is deprecated. Please use tf.io.decode_csv instead.\n\nI0618 02:21:48.113370 140374330308480 estimator.py:1145] Calling model_fn.\nW0618 02:21:48.467810 140374330308480 deprecation.py:323] From /usr/local/lib/python3.6/dist-packages/tensorflow/python/ops/sparse_ops.py:1719: add_dispatch_support.<locals>.wrapper (from tensorflow.python.ops.array_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.where in 2.0, which has the same broadcast rule as np.where\nW0618 02:21:48.932053 140374330308480 deprecation.py:323] From /usr/local/lib/python3.6/dist-packages/tensorflow_estimator/python/estimator/canned/linear.py:308: to_float (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse `tf.cast` instead.\nI0618 02:21:49.631353 140374330308480 estimator.py:1147] Done calling model_fn.\nI0618 02:21:49.631717 140374330308480 basic_session_run_hooks.py:541] Create CheckpointSaverHook.\nI0618 02:21:50.034778 140374330308480 monitored_session.py:240] Graph was finalized.\n2019-06-18 02:21:50.035308: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1181] Device interconnect StreamExecutor with strength 1 edge matrix:\n2019-06-18 02:21:50.035351: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1187] \n2019-06-18 02:21:50.127599: W tensorflow/compiler/jit/mark_for_compilation_pass.cc:1412] (One-time warning): Not using XLA:CPU for cluster because envvar TF_XLA_FLAGS=--tf_xla_cpu_global_jit was not set. If you want XLA:CPU, either set that envvar, or use experimental_jit_scope to enable XLA:CPU. To confirm that XLA is active, pass --vmodule=xla_compilation_cache=1 (as a proper command-line flag, not via TF_XLA_FLAGS) or set the envvar XLA_FLAGS=--xla_hlo_profile.\nI0618 02:21:50.179698 140374330308480 session_manager.py:500] Running local_init_op.\nI0618 02:21:50.201351 140374330308480 session_manager.py:502] Done running local_init_op.\nI0618 02:21:50.972360 140374330308480 basic_session_run_hooks.py:606] Saving checkpoints for 0 into /tmp/census_model/model.ckpt.\nI0618 02:21:51.833827 140374330308480 basic_session_run_hooks.py:262] average_loss = 0.6931472, loss = 27.725887\nI0618 02:21:51.834183 140374330308480 basic_session_run_hooks.py:262] loss = 27.725887, step = 1\nI0618 02:21:52.429172 140374330308480 basic_session_run_hooks.py:692] global_step/sec: 167.844\nI0618 02:21:52.429909 140374330308480 basic_session_run_hooks.py:260] average_loss = 0.2580979, loss = 10.3239155 (0.596 sec)\nI0618 02:21:52.430119 140374330308480 basic_session_run_hooks.py:260] loss = 10.3239155, step = 101 (0.596 sec)\nI0618 02:21:52.714958 140374330308480 basic_session_run_hooks.py:692] global_step/sec: 349.949\nI0618 02:21:52.715568 140374330308480 basic_session_run_hooks.py:260] average_loss = 0.3234958, loss = 12.939833 (0.286 sec)\nI0618 02:21:52.715775 140374330308480 basic_session_run_hooks.py:260] loss = 12.939833, step = 201 (0.286 sec)\nI0618 02:21:52.976084 140374330308480 basic_session_run_hooks.py:692] global_step/sec: 382.923\nI0618 02:21:52.976797 140374330308480 basic_session_run_hooks.py:260] average_loss = 0.25078636, loss = 10.031454 (0.261 sec)\nI0618 02:21:52.977014 140374330308480 basic_session_run_hooks.py:260] loss = 10.031454, step = 301 (0.261 sec)\nI0618 02:21:53.265064 140374330308480 basic_session_run_hooks.py:692] global_step/sec: 346.046\nI0618 02:21:53.265777 140374330308480 basic_session_run_hooks.py:260] average_loss = 0.30604935, loss = 12.241974 (0.289 sec)\nI0618 02:21:53.266076 140374330308480 basic_session_run_hooks.py:260] loss = 12.241974, step = 401 (0.289 sec)\nI0618 02:21:53.553257 140374330308480 basic_session_run_hooks.py:692] global_step/sec: 347.029\nI0618 02:21:53.554040 140374330308480 basic_session_run_hooks.py:260] average_loss = 0.24040127, loss = 9.616051 (0.288 sec)\nI0618 02:21:53.554281 140374330308480 basic_session_run_hooks.py:260] loss = 9.616051, step = 501 (0.288 sec)\nI0618 02:21:53.835034 140374330308480 basic_session_run_hooks.py:692] global_step/sec: 354.829\nI0618 02:21:53.835660 140374330308480 basic_session_run_hooks.py:260] average_loss = 0.26537886, loss = 10.615154 (0.282 sec)\nI0618 02:21:53.835870 140374330308480 basic_session_run_hooks.py:260] loss = 10.615154, step = 601 (0.282 sec)\nI0618 02:21:54.097162 140374330308480 basic_session_run_hooks.py:692] global_step/sec: 381.521\nI0618 02:21:54.097899 140374330308480 basic_session_run_hooks.py:260] average_loss = 0.34397393, loss = 13.758957 (0.262 sec)\nI0618 02:21:54.098104 140374330308480 basic_session_run_hooks.py:260] loss = 13.758957, step = 701 (0.262 sec)\nI0618 02:21:54.369472 140374330308480 basic_session_run_hooks.py:692] global_step/sec: 367.241\nI0618 02:21:54.370166 140374330308480 basic_session_run_hooks.py:260] average_loss = 0.40742293, loss = 16.296917 (0.272 sec)\nI0618 02:21:54.370376 140374330308480 basic_session_run_hooks.py:260] loss = 16.296917, step = 801 (0.272 sec)\nI0618 02:21:54.708876 140374330308480 basic_session_run_hooks.py:692] global_step/sec: 294.645\nI0618 02:21:54.710653 140374330308480 basic_session_run_hooks.py:260] average_loss = 0.30915707, loss = 12.366283 (0.340 sec)\nI0618 02:21:54.711043 140374330308480 basic_session_run_hooks.py:260] loss = 12.366283, step = 901 (0.341 sec)\nI0618 02:21:54.984972 140374330308480 basic_session_run_hooks.py:692] global_step/sec: 362.167\nI0618 02:21:54.985655 140374330308480 basic_session_run_hooks.py:260] average_loss = 0.39361295, loss = 15.744518 (0.275 sec)\nI0618 02:21:54.985872 140374330308480 basic_session_run_hooks.py:260] loss = 15.744518, step = 1001 (0.275 sec)\nI0618 02:21:55.254609 140374330308480 basic_session_run_hooks.py:692] global_step/sec: 370.86\nI0618 02:21:55.255295 140374330308480 basic_session_run_hooks.py:260] average_loss = 0.5208241, loss = 20.832962 (0.270 sec)\nI0618 02:21:55.255553 140374330308480 basic_session_run_hooks.py:260] loss = 20.832962, step = 1101 (0.270 sec)\nI0618 02:21:55.522802 140374330308480 basic_session_run_hooks.py:692] global_step/sec: 372.883\nI0618 02:21:55.523541 140374330308480 basic_session_run_hooks.py:260] average_loss = 0.28922597, loss = 11.569038 (0.268 sec)\nI0618 02:21:55.523917 140374330308480 basic_session_run_hooks.py:260] loss = 11.569038, step = 1201 (0.268 sec)\nI0618 02:21:55.802838 140374330308480 basic_session_run_hooks.py:692] global_step/sec: 357.082\nI0618 02:21:55.803584 140374330308480 basic_session_run_hooks.py:260] average_loss = 0.24933887, loss = 9.973555 (0.280 sec)\nI0618 02:21:55.803913 140374330308480 basic_session_run_hooks.py:260] loss = 9.973555, step = 1301 (0.280 sec)\nI0618 02:21:56.076504 140374330308480 basic_session_run_hooks.py:692] global_step/sec: 365.433\nI0618 02:21:56.077114 140374330308480 basic_session_run_hooks.py:260] average_loss = 0.38473326, loss = 15.38933 (0.274 sec)\nI0618 02:21:56.077326 140374330308480 basic_session_run_hooks.py:260] loss = 15.38933, step = 1401 (0.273 sec)\nI0618 02:21:56.357172 140374330308480 basic_session_run_hooks.py:692] global_step/sec: 356.265\nI0618 02:21:56.357934 140374330308480 basic_session_run_hooks.py:260] average_loss = 0.31031436, loss = 12.412575 (0.281 sec)\nI0618 02:21:56.358235 140374330308480 basic_session_run_hooks.py:260] loss = 12.412575, step = 1501 (0.281 sec)\nI0618 02:21:56.628048 140374330308480 basic_session_run_hooks.py:692] global_step/sec: 369.168\nI0618 02:21:56.628675 140374330308480 basic_session_run_hooks.py:260] average_loss = 0.32551187, loss = 13.020475 (0.271 sec)\nI0618 02:21:56.628884 140374330308480 basic_session_run_hooks.py:260] loss = 13.020475, step = 1601 (0.271 sec)\nI0618 02:21:56.722892 140374330308480 basic_session_run_hooks.py:606] Saving checkpoints for 1629 into /tmp/census_model/model.ckpt.\nI0618 02:21:56.881346 140374330308480 estimator.py:368] Loss for final step: 0.33970028.\nI0618 02:21:56.905838 140374330308480 census_dataset.py:167] Parsing /tmp/census_data/adult.test\nI0618 02:21:56.941754 140374330308480 estimator.py:1145] Calling model_fn.\nW0618 02:21:57.855187 140374330308480 deprecation.py:323] From /usr/local/lib/python3.6/dist-packages/tensorflow/python/ops/metrics_impl.py:2027: div (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nDeprecated in favor of operator or tf.math.divide.\nW0618 02:21:58.352527 140374330308480 metrics_impl.py:804] Trapezoidal rule is known to produce incorrect PR-AUCs; please switch to \"careful_interpolation\" instead.\nW0618 02:21:58.373770 140374330308480 metrics_impl.py:804] Trapezoidal rule is known to produce incorrect PR-AUCs; please switch to \"careful_interpolation\" instead.\nI0618 02:21:58.396237 140374330308480 estimator.py:1147] Done calling model_fn.\nI0618 02:21:58.416667 140374330308480 evaluation.py:255] Starting evaluation at 2019-06-18T02:21:58Z\nI0618 02:21:58.550940 140374330308480 monitored_session.py:240] Graph was finalized.\n2019-06-18 02:21:58.551406: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1181] Device interconnect StreamExecutor with strength 1 edge matrix:\n2019-06-18 02:21:58.551464: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1187] \nW0618 02:21:58.551583 140374330308480 deprecation.py:323] From /usr/local/lib/python3.6/dist-packages/tensorflow/python/training/saver.py:1276: checkpoint_exists (from tensorflow.python.training.checkpoint_management) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse standard file APIs to check for files with this prefix.\nI0618 02:21:58.552948 140374330308480 saver.py:1280] Restoring parameters from /tmp/census_model/model.ckpt-1629\nI0618 02:21:58.651469 140374330308480 session_manager.py:500] Running local_init_op.\nI0618 02:21:58.698924 140374330308480 session_manager.py:502] Done running local_init_op.\nI0618 02:22:00.488134 140374330308480 evaluation.py:275] Finished evaluation at 2019-06-18-02:22:00\nI0618 02:22:00.488391 140374330308480 estimator.py:2039] Saving dict for global step 1629: accuracy = 0.8360666, accuracy_baseline = 0.76377374, auc = 0.88413656, auc_precision_recall = 0.6953496, average_loss = 0.3516626, global_step = 1629, label/mean = 0.23622628, loss = 14.032889, precision = 0.70412767, prediction/mean = 0.22265631, recall = 0.5278211\nI0618 02:22:00.725699 140374330308480 estimator.py:2099] Saving 'checkpoint_path' summary for global step 1629: /tmp/census_model/model.ckpt-1629\nI0618 02:22:00.726319 140374330308480 wide_deep_run_loop.py:116] Results at epoch 2 / 2\nI0618 02:22:00.726489 140374330308480 wide_deep_run_loop.py:117] ------------------------------------------------------------\nI0618 02:22:00.726589 140374330308480 wide_deep_run_loop.py:120] accuracy: 0.8360666\nI0618 02:22:00.726665 140374330308480 wide_deep_run_loop.py:120] accuracy_baseline: 0.76377374\nI0618 02:22:00.726736 140374330308480 wide_deep_run_loop.py:120] auc: 0.88413656\nI0618 02:22:00.726807 140374330308480 wide_deep_run_loop.py:120] auc_precision_recall: 0.6953496\nI0618 02:22:00.726878 140374330308480 wide_deep_run_loop.py:120] average_loss: 0.3516626\nI0618 02:22:00.726952 140374330308480 wide_deep_run_loop.py:120] global_step: 1629\nI0618 02:22:00.727018 140374330308480 wide_deep_run_loop.py:120] label/mean: 0.23622628\nI0618 02:22:00.727084 140374330308480 wide_deep_run_loop.py:120] loss: 14.032889\nI0618 02:22:00.727148 140374330308480 wide_deep_run_loop.py:120] precision: 0.70412767\nI0618 02:22:00.727212 140374330308480 wide_deep_run_loop.py:120] prediction/mean: 0.22265631\nI0618 02:22:00.727276 140374330308480 wide_deep_run_loop.py:120] recall: 0.5278211\nI0618 02:22:00.727462 140374330308480 logger.py:147] Benchmark metric: {'name': 'accuracy', 'value': 0.8360666036605835, 'unit': None, 'global_step': 1629, 'timestamp': '2019-06-18T02:22:00.727405Z', 'extras': []}\nI0618 02:22:00.727608 140374330308480 logger.py:147] Benchmark metric: {'name': 'accuracy_baseline', 'value': 0.7637737393379211, 'unit': None, 'global_step': 1629, 'timestamp': '2019-06-18T02:22:00.727583Z', 'extras': []}\nI0618 02:22:00.727721 140374330308480 logger.py:147] Benchmark metric: {'name': 'auc', 'value': 0.8841365575790405, 'unit': None, 'global_step': 1629, 'timestamp': '2019-06-18T02:22:00.727697Z', 'extras': []}\nI0618 02:22:00.727825 140374330308480 logger.py:147] Benchmark metric: {'name': 'auc_precision_recall', 'value': 0.6953495740890503, 'unit': None, 'global_step': 1629, 'timestamp': '2019-06-18T02:22:00.727801Z', 'extras': []}\nI0618 02:22:00.727926 140374330308480 logger.py:147] Benchmark metric: {'name': 'average_loss', 'value': 0.35166260600090027, 'unit': None, 'global_step': 1629, 'timestamp': '2019-06-18T02:22:00.727903Z', 'extras': []}\nI0618 02:22:00.728036 140374330308480 logger.py:147] Benchmark metric: {'name': 'label/mean', 'value': 0.23622627556324005, 'unit': None, 'global_step': 1629, 'timestamp': '2019-06-18T02:22:00.728012Z', 'extras': []}\nI0618 02:22:00.728146 140374330308480 logger.py:147] Benchmark metric: {'name': 'loss', 'value': 14.032889366149902, 'unit': None, 'global_step': 1629, 'timestamp': '2019-06-18T02:22:00.728122Z', 'extras': []}\nI0618 02:22:00.728249 140374330308480 logger.py:147] Benchmark metric: {'name': 'precision', 'value': 0.7041276693344116, 'unit': None, 'global_step': 1629, 'timestamp': '2019-06-18T02:22:00.728227Z', 'extras': []}\nI0618 02:22:00.728351 140374330308480 logger.py:147] Benchmark metric: {'name': 'prediction/mean', 'value': 0.22265630960464478, 'unit': None, 'global_step': 1629, 'timestamp': '2019-06-18T02:22:00.728329Z', 'extras': []}\nI0618 02:22:00.728468 140374330308480 logger.py:147] Benchmark metric: {'name': 'recall', 'value': 0.5278211236000061, 'unit': None, 'global_step': 1629, 'timestamp': '2019-06-18T02:22:00.728445Z', 'extras': []}\n"
],
[
"# Read US census data\n!ls /tmp/census_data/",
"adult.data adult.test\n"
],
[
"train_file = '/tmp/census_data/adult.data'\ntest_file = '/tmp/census_data/adult.test'",
"_____no_output_____"
],
[
"import pandas as pd\n\ntrain_df = pd.read_csv(train_file, names=census_dataset._CSV_COLUMNS)\ntest_df = pd.read_csv(test_file, names=census_dataset._CSV_COLUMNS)\n\ntrain_df.tail()",
"_____no_output_____"
],
[
"# 위에서 eager execution enable했기 때문에 쉽게 데이터셋 보기 가능\n# tf.data.Dataset으로 slicing하는건 데이터가 작기때문에 가능\ndef easy_input_function(df, label_key, num_epochs, shuffle, batch_size):\n label = df[label_key]\n ds = tf.data.Dataset.from_tensor_slices((dict(df),label))\n\n if shuffle:\n ds = ds.shuffle(10000)\n\n ds = ds.batch(batch_size).repeat(num_epochs)\n\n return ds\n\nds = easy_input_function(train_df, label_key='income_bracket', num_epochs=5, shuffle=True, batch_size=10)\n\nfor feature_batch, label_batch in ds.take(1):\n print('Some feature keys:', list(feature_batch.keys())[:5])\n print()\n print('A batch of Ages :', feature_batch['age'])\n print()\n print('A batch of Labels:', label_batch )",
"Some feature keys: ['age', 'workclass', 'fnlwgt', 'education', 'education_num']\n\nA batch of Ages : tf.Tensor([22 33 23 36 45 18 41 53 45 27], shape=(10,), dtype=int32)\n\nA batch of Labels: tf.Tensor(\n[b'<=50K' b'>50K' b'<=50K' b'>50K' b'<=50K' b'<=50K' b'<=50K' b'>50K'\n b'<=50K' b'<=50K'], shape=(10,), dtype=string)\n"
],
[
"# 데이터가 커지면....tf.decode_csv와 tf.data.TextLineDataset를 사용한다. (이 둘을 사용한게 input_fn)\n# 위와 같은 결과가 나온다.\n# input_fn을 사용하여 batch와 epochs를 지정하여 계속 뽑을 수 있다.\nimport inspect\nprint(inspect.getsource(census_dataset.input_fn))\n\nds = census_dataset.input_fn(train_file, num_epochs=5, shuffle=True, batch_size=10)\n\nfor feature_batch, label_batch in ds.take(1):\n print('Feature keys:', list(feature_batch.keys())[:5])\n print()\n print('Age batch :', feature_batch['age'])\n print()\n print('Label batch :', label_batch )",
"def input_fn(data_file, num_epochs, shuffle, batch_size):\n \"\"\"Generate an input function for the Estimator.\"\"\"\n assert tf.gfile.Exists(data_file), (\n '%s not found. Please make sure you have run census_dataset.py and '\n 'set the --data_dir argument to the correct path.' % data_file)\n\n def parse_csv(value):\n tf.logging.info('Parsing {}'.format(data_file))\n columns = tf.decode_csv(value, record_defaults=_CSV_COLUMN_DEFAULTS)\n features = dict(zip(_CSV_COLUMNS, columns))\n labels = features.pop('income_bracket')\n classes = tf.equal(labels, '>50K') # binary classification\n return features, classes\n\n # Extract lines from input files using the Dataset API.\n dataset = tf.data.TextLineDataset(data_file)\n\n if shuffle:\n dataset = dataset.shuffle(buffer_size=_NUM_EXAMPLES['train'])\n\n dataset = dataset.map(parse_csv, num_parallel_calls=5)\n\n # We call repeat after shuffling, rather than before, to prevent separate\n # epochs from blending together.\n dataset = dataset.repeat(num_epochs)\n dataset = dataset.batch(batch_size)\n return dataset\n\nFeature keys: ['age', 'workclass', 'fnlwgt', 'education', 'education_num']\n\nAge batch : tf.Tensor([22 44 48 68 39 37 56 42 28 18], shape=(10,), dtype=int32)\n\nLabel batch : tf.Tensor([False True True True True True False True False False], shape=(10,), dtype=bool)\n"
],
[
"# Because Estimators expect an input_fn that takes no arguments, \n# we typically wrap configurable input function into an obejct with the expected signature. \n# For this notebook configure the train_inpf to iterate over the data twice:\nimport functools\n\ntrain_inpf = functools.partial(census_dataset.input_fn, train_file, num_epochs=2, shuffle=True, batch_size=64)\ntest_inpf = functools.partial(census_dataset.input_fn, test_file, num_epochs=1, shuffle=False, batch_size=64)",
"_____no_output_____"
],
[
"# model only using a column\nclassifier = tf.estimator.LinearClassifier(feature_columns=[fc.numeric_column('age')])\nclassifier.train(train_inpf)\nresult = classifier.evaluate(test_inpf)\n\nclear_output()\nprint(result)",
"{'accuracy': 0.7590443, 'accuracy_baseline': 0.76377374, 'auc': 0.67830986, 'auc_precision_recall': 0.31138018, 'average_loss': 0.52304345, 'label/mean': 0.23622628, 'loss': 33.394787, 'precision': 0.14678898, 'prediction/mean': 0.2417537, 'recall': 0.0041601663, 'global_step': 1018}\n"
],
[
"# Similarly, we can define a NumericColumn for each continuous feature column that we want to use in the model:\n# 이런식으로 numerical columns들만 뽑을 수도 있음.\nage = fc.numeric_column('age')\neducation_num = tf.feature_column.numeric_column('education_num')\ncapital_gain = tf.feature_column.numeric_column('capital_gain')\ncapital_loss = tf.feature_column.numeric_column('capital_loss')\nhours_per_week = tf.feature_column.numeric_column('hours_per_week')\n\nmy_numeric_columns = [age,education_num, capital_gain, capital_loss, hours_per_week]\n\nfc.input_layer(feature_batch, my_numeric_columns).numpy()",
"W0618 02:44:06.778625 140493422073728 deprecation.py:323] From /usr/local/lib/python3.6/dist-packages/tensorflow/python/feature_column/feature_column.py:205: NumericColumn._get_dense_tensor (from tensorflow.python.feature_column.feature_column_v2) is deprecated and will be removed in a future version.\nInstructions for updating:\nThe old _FeatureColumn APIs are being deprecated. Please use the new FeatureColumn APIs instead.\nW0618 02:44:06.781412 140493422073728 deprecation.py:323] From /usr/local/lib/python3.6/dist-packages/tensorflow/python/feature_column/feature_column.py:2115: NumericColumn._transform_feature (from tensorflow.python.feature_column.feature_column_v2) is deprecated and will be removed in a future version.\nInstructions for updating:\nThe old _FeatureColumn APIs are being deprecated. Please use the new FeatureColumn APIs instead.\nW0618 02:44:06.785332 140493422073728 deprecation.py:323] From /usr/local/lib/python3.6/dist-packages/tensorflow/python/feature_column/feature_column.py:206: NumericColumn._variable_shape (from tensorflow.python.feature_column.feature_column_v2) is deprecated and will be removed in a future version.\nInstructions for updating:\nThe old _FeatureColumn APIs are being deprecated. Please use the new FeatureColumn APIs instead.\n"
],
[
"classifier = tf.estimator.LinearClassifier(feature_columns=my_numeric_columns)\nclassifier.train(train_inpf)\n\nresult = classifier.evaluate(test_inpf)\n\nclear_output()\n\nfor key,value in sorted(result.items()):\n print('%s: %s' % (key, value))",
"accuracy: 0.78250724\naccuracy_baseline: 0.76377374\nauc: 0.71121407\nauc_precision_recall: 0.5095864\naverage_loss: 7.3911877\nglobal_step: 1018\nlabel/mean: 0.23622628\nloss: 471.9056\nprecision: 0.61526835\nprediction/mean: 0.26402935\nrecall: 0.21164846\n"
],
[
"# categorical columns들에 대해선...\n# 다음과 같이 뽑을 수 있음.\nrelationship = fc.categorical_column_with_vocabulary_list(\n 'relationship',\n ['Husband', 'Not-in-family', 'Wife', 'Own-child', 'Unmarried', 'Other-relative'])\n\nfc.input_layer(feature_batch, [age, fc.indicator_column(relationship)])",
"W0618 02:49:16.828300 140493422073728 deprecation.py:323] From /usr/local/lib/python3.6/dist-packages/tensorflow/python/feature_column/feature_column.py:205: IndicatorColumn._get_dense_tensor (from tensorflow.python.feature_column.feature_column_v2) is deprecated and will be removed in a future version.\nInstructions for updating:\nThe old _FeatureColumn APIs are being deprecated. Please use the new FeatureColumn APIs instead.\nW0618 02:49:16.831818 140493422073728 deprecation.py:323] From /usr/local/lib/python3.6/dist-packages/tensorflow/python/feature_column/feature_column.py:2115: IndicatorColumn._transform_feature (from tensorflow.python.feature_column.feature_column_v2) is deprecated and will be removed in a future version.\nInstructions for updating:\nThe old _FeatureColumn APIs are being deprecated. Please use the new FeatureColumn APIs instead.\nW0618 02:49:16.835200 140493422073728 deprecation.py:323] From /usr/local/lib/python3.6/dist-packages/tensorflow/python/feature_column/feature_column_v2.py:4236: VocabularyListCategoricalColumn._get_sparse_tensors (from tensorflow.python.feature_column.feature_column_v2) is deprecated and will be removed in a future version.\nInstructions for updating:\nThe old _FeatureColumn APIs are being deprecated. Please use the new FeatureColumn APIs instead.\nW0618 02:49:16.838194 140493422073728 deprecation.py:323] From /usr/local/lib/python3.6/dist-packages/tensorflow/python/feature_column/feature_column.py:2115: VocabularyListCategoricalColumn._transform_feature (from tensorflow.python.feature_column.feature_column_v2) is deprecated and will be removed in a future version.\nInstructions for updating:\nThe old _FeatureColumn APIs are being deprecated. Please use the new FeatureColumn APIs instead.\nW0618 02:49:16.846775 140493422073728 deprecation.py:323] From /usr/local/lib/python3.6/dist-packages/tensorflow/python/feature_column/feature_column_v2.py:4207: IndicatorColumn._variable_shape (from tensorflow.python.feature_column.feature_column_v2) is deprecated and will be removed in a future version.\nInstructions for updating:\nThe old _FeatureColumn APIs are being deprecated. Please use the new FeatureColumn APIs instead.\nW0618 02:49:16.847798 140493422073728 deprecation.py:323] From /usr/local/lib/python3.6/dist-packages/tensorflow/python/feature_column/feature_column_v2.py:4262: VocabularyListCategoricalColumn._num_buckets (from tensorflow.python.feature_column.feature_column_v2) is deprecated and will be removed in a future version.\nInstructions for updating:\nThe old _FeatureColumn APIs are being deprecated. Please use the new FeatureColumn APIs instead.\n"
],
[
"# 위는 category갯수를 아는 경우에 해당되고,\n# 모를 땐 다음을 활용한다. categorical_column_with_hash_bucket\noccupation = tf.feature_column.categorical_column_with_hash_bucket(\n 'occupation', hash_bucket_size=1000)\n\nfor item in feature_batch['occupation'].numpy():\n print(item.decode())",
"Priv-house-serv\nExec-managerial\nAdm-clerical\n?\nExec-managerial\nTransport-moving\nAdm-clerical\nCraft-repair\nCraft-repair\nSales\n"
],
[
"education = tf.feature_column.categorical_column_with_vocabulary_list(\n 'education', [\n 'Bachelors', 'HS-grad', '11th', 'Masters', '9th', 'Some-college',\n 'Assoc-acdm', 'Assoc-voc', '7th-8th', 'Doctorate', 'Prof-school',\n '5th-6th', '10th', '1st-4th', 'Preschool', '12th'])\n\nmarital_status = tf.feature_column.categorical_column_with_vocabulary_list(\n 'marital_status', [\n 'Married-civ-spouse', 'Divorced', 'Married-spouse-absent',\n 'Never-married', 'Separated', 'Married-AF-spouse', 'Widowed'])\n\nworkclass = tf.feature_column.categorical_column_with_vocabulary_list(\n 'workclass', [\n 'Self-emp-not-inc', 'Private', 'State-gov', 'Federal-gov',\n 'Local-gov', '?', 'Self-emp-inc', 'Without-pay', 'Never-worked'])\n\n\nmy_categorical_columns = [relationship, occupation, education, marital_status, workclass]",
"_____no_output_____"
],
[
"classifier = tf.estimator.LinearClassifier(feature_columns=my_numeric_columns+my_categorical_columns)\nclassifier.train(train_inpf)\nresult = classifier.evaluate(test_inpf)\n\nclear_output()\n\nfor key,value in sorted(result.items()):\n print('%s: %s' % (key, value))",
"accuracy: 0.82673055\naccuracy_baseline: 0.76377374\nauc: 0.87491494\nauc_precision_recall: 0.6558813\naverage_loss: 1.6820824\nglobal_step: 1018\nlabel/mean: 0.23622628\nloss: 107.39602\nprecision: 0.63922846\nprediction/mean: 0.25356495\nrecall: 0.6118045\n"
],
[
"# to be continue....",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78fa08678db40a436c9bf2796764a98b5ce5968 | 19,255 | ipynb | Jupyter Notebook | notebooks/connect_01_text_processing.ipynb | aws-samples/contact-lens-for-amazon-connect-data-gathering-mechanism | 0daddcc850ad62877a20868bd34f2f7d6903ac0d | [
"MIT-0"
] | 1 | 2021-11-01T21:18:00.000Z | 2021-11-01T21:18:00.000Z | notebooks/connect_01_text_processing.ipynb | aws-samples/contact-lens-for-amazon-connect-data-gathering-mechanism | 0daddcc850ad62877a20868bd34f2f7d6903ac0d | [
"MIT-0"
] | null | null | null | notebooks/connect_01_text_processing.ipynb | aws-samples/contact-lens-for-amazon-connect-data-gathering-mechanism | 0daddcc850ad62877a20868bd34f2f7d6903ac0d | [
"MIT-0"
] | 1 | 2021-10-03T12:50:03.000Z | 2021-10-03T12:50:03.000Z | 26.743056 | 140 | 0.554142 | [
[
[
"# Pre-processing the text for Object2Vec\n\nProcessing the text to fit Object2Vec algorithm.",
"_____no_output_____"
]
],
[
[
"import boto3\nimport pandas as pd\nimport re\nfrom sklearn import preprocessing\nimport numpy as np\nimport json\nimport os\nfrom sklearn.feature_extraction.text import CountVectorizer\nimport random\nrandom.seed(42)\nfrom random import sample\nfrom sklearn.utils import shuffle\nfrom nltk import word_tokenize",
"_____no_output_____"
]
],
[
[
"#### Functions",
"_____no_output_____"
]
],
[
[
"def get_filtered_objects(bucket_name, prefix):\n \"\"\"filter objects based on bucket and prefix\"\"\"\n s3 = boto3.client(\"s3\")\n files = s3.list_objects_v2(Bucket = bucket_name, Prefix =prefix)\n return files",
"_____no_output_____"
],
[
"def download_object(bucket_name, key, local_path):\n \"\"\"Download S3 object to local\"\"\"\n s3 = boto3.resource('s3')\n try:\n s3.Bucket(bucket_name).download_file(key,local_path)\n except botocore.exceptions.ClientError as e:\n if e.response['Error']['Code'] == \"404\":\n print(\"The object does not exist\")\n else:\n raise",
"_____no_output_____"
],
[
"def get_csv(files):\n \"\"\"Filter the files by selecting .csv extension\"\"\"\n paths = []\n for file in files:\n if file['Key'].endswith(\".csv\"):\n paths.append(file['Key'])\n return paths",
"_____no_output_____"
],
[
"def sentence_to_tokens(sentence, vocab_to_tokens):\n \"\"\"converts sentences to tokens\"\"\"\n words = word_tokenize(sentence)\n return [ vocab_to_tokens[w] for w in words if w in vocab_to_tokens]",
"_____no_output_____"
],
[
"def create_dir(directory):\n \"\"\"Create a directory\"\"\"\n if not os.path.exists(directory):\n os.makedirs(directory)",
"_____no_output_____"
],
[
"def remove_file(file_path):\n \"\"\"Remove locally the specified path\"\"\"\n if os.path.isfile(file_path):\n os.remove(file_path)\n else:\n print(\"Error, file not found.\")",
"_____no_output_____"
],
[
"def build_sentence_pairs(data):\n \"\"\"transform the dataframe into sentence pairs for Object2Vec algorithm.\"\"\"\n sentence_pairs = []\n for r in range(len(data)):\n row = data.iloc[r]\n sentence_pairs.append({'in0': row['encoded_content'], \\\n 'in1': row['labels'],\\\n 'label':1})\n return sentence_pairs",
"_____no_output_____"
],
[
"def build_negative_pairs(data, negative_labels_to_sample,sentence_pairs, n_neg_pairs_per_label=10):\n \"\"\"build negative pairs for training dataframe\"\"\"\n for r in negative_labels_to_sample:\n #news that have that label as tag\n selection = data.loc[data.labels.apply(lambda x: x is not None and r in x)]\n #news that do not have that label as tag.\n wrong_selection = data.loc[data.labels.apply(lambda x: x is not None and r not in x)]\n if len(wrong_selection)>0:\n for p in range(n_neg_pairs_per_label):\n negative_pair = {}\n negative_pair['in0'] = selection.sample(1)['encoded_content'].iloc[0]\n negative_pair['in1'] = wrong_selection.sample(1)['labels'].iloc[0]\n negative_pair['label'] = 0\n sentence_pairs.append(negative_pair)\n return sentence_pairs",
"_____no_output_____"
]
],
[
[
"##### Download the data locally",
"_____no_output_____"
]
],
[
[
"bucket_name = \"YOUR_BUCKET_HERE\"\nprefix = \"connect/\"",
"_____no_output_____"
],
[
"#save the files locally.\ncreate_dir(\"./data\")",
"_____no_output_____"
],
[
"files = get_filtered_objects(bucket_name, prefix)['Contents']\nfiles = get_csv(files)\nlocal_files=[]\nprint(files)\nfor file in files:\n full_prefix = \"/\".join(file.split(\"/\")[:-1])\n inner_folder = full_prefix.replace(prefix,'')\n local_path = \"./data/\" +file.split(\"/\")[-1]\n download_object(bucket_name, file, local_path)\n local_files.append(local_path)",
"_____no_output_____"
],
[
"local_files",
"_____no_output_____"
]
],
[
[
"##### Concatenate the .csv files",
"_____no_output_____"
]
],
[
[
"import pandas.errors\ncontent = []\nfor filename in local_files:\n try:\n df = pd.read_csv(filename, sep=\";\")\n print(df.columns)\n content.append(df)\n except pandas.errors.ParserError:\n print(\"File\", filename, \"cannot be parsed. Check its format\")\ndata = pd.concat(content)",
"_____no_output_____"
],
[
"customer_text = data.loc[data.ParticipantId=='CUSTOMER']",
"_____no_output_____"
],
[
"customer_text.shape",
"_____no_output_____"
]
],
[
[
"##### Create random labels\n\nChange this to use your own labels\nAlso: we are here replicating the texts to increase statistics",
"_____no_output_____"
]
],
[
[
"customer_text = pd.concat([customer_text]*300, ignore_index=True)",
"_____no_output_____"
],
[
"customer_text['labels']=np.random.randint(low=0, high=5, size=len(customer_text))",
"_____no_output_____"
],
[
"customer_text.labels.hist()",
"_____no_output_____"
]
],
[
[
"##### Get vocabulary from the corpus using sklearn for the heavy lifting\n\nThe vocabulary will be built only taking into account words that belong to news related to crimes.",
"_____no_output_____"
]
],
[
[
"counts = CountVectorizer(min_df=5, max_df=0.95, token_pattern=r'(?u)\\b[A-Za-z]{2,}\\b').fit(customer_text['Content'].values.tolist())",
"_____no_output_____"
],
[
"vocab = counts.get_feature_names()\nvocab_to_token_dict = dict(zip(vocab, range(len(vocab))))\ntoken_to_vocab_dict = dict(zip(range(len(vocab)), vocab))",
"_____no_output_____"
],
[
"len(vocab)",
"_____no_output_____"
],
[
"create_dir(\"./vocab\")\nvocab_filename = './vocab/vocab.json'\nwith open(vocab_filename, \"w\") as write_file:\n json.dump(vocab_to_token_dict, write_file)",
"_____no_output_____"
]
],
[
[
"##### Encode data body\n\nTransform the texts in the data to encodings from the vocabulary created.",
"_____no_output_____"
]
],
[
[
"import nltk\nnltk.download('punkt')",
"_____no_output_____"
],
[
"customer_text['encoded_content'] = customer_text['Content'].apply(lambda x: sentence_to_tokens(x, vocab_to_token_dict))",
"_____no_output_____"
],
[
"customer_text['labels']",
"_____no_output_____"
],
[
"customer_text['labels']=customer_text['labels'].apply(lambda x: [x])",
"_____no_output_____"
],
[
"customer_text[['labels','encoded_content']]",
"_____no_output_____"
],
[
"# remove entriews with no text",
"_____no_output_____"
],
[
"customer_text = customer_text.loc[customer_text['encoded_content'].apply(lambda x: len(x)>0)]",
"_____no_output_____"
],
[
"customer_text[['labels','encoded_content', 'Content']]",
"_____no_output_____"
]
],
[
[
"##### Build sentence pairs Object2Vec",
"_____no_output_____"
]
],
[
[
"#negative pairs for the algorithm: need to decide which lables we want to sample *against*. \nnegative_labels_to_sample = range(5)",
"_____no_output_____"
],
[
"sentence_pairs = build_sentence_pairs(customer_text)\n",
"_____no_output_____"
]
],
[
[
"##### Build negative sentence pairs for training Object2Vec\n\nNegative sampling for the Object2Vec algorithm - add negative and positive pairs (document,label)",
"_____no_output_____"
]
],
[
[
"sentence_pairs = build_negative_pairs(customer_text,negative_labels_to_sample,sentence_pairs)\n",
"_____no_output_____"
],
[
"print(\"Sample of input for Object2vec algorith: {}\".format(sentence_pairs[1]))",
"_____no_output_____"
],
[
"!pip install jsonlines",
"_____no_output_____"
]
],
[
[
"##### train/test/val split, save to file\n",
"_____no_output_____"
]
],
[
[
"# shuffle and split test/train/val\nrandom.seed(42)\nrandom.shuffle(sentence_pairs)\n\nn_train = int(0.7 * len(sentence_pairs))\n\n# split train and test\nsentence_pairs_train = sentence_pairs[:n_train]\nsentence_pairs_test = sentence_pairs[n_train:]\n\n# further split test set into validation set (val_vectors) and test set (test_vectors)\nn_test = len(sentence_pairs_test)\n\nsentence_pairs_val = sentence_pairs_test[:n_test//2]\nsentence_pairs_test = sentence_pairs_test[n_test//2:]\n",
"_____no_output_____"
],
[
"import jsonlines\nwith jsonlines.open('./data/train.jsonl', mode='w') as writer:\n writer.write_all(sentence_pairs_train)\n \nwith jsonlines.open('./data/test.jsonl', mode='w') as writer:\n writer.write_all(sentence_pairs_test)\n\nwith jsonlines.open('./data/val.jsonl', mode='w') as writer:\n writer.write_all(sentence_pairs_val)",
"_____no_output_____"
]
],
[
[
"##### 8. Upload to S3",
"_____no_output_____"
]
],
[
[
"import os\ns3_client = boto3.client('s3')\n\nout_prefix = \"connect/O2VInput\"\nfor n in ['train', 'test', 'val',]:\n s3_client.upload_file(\"./data/\"+n+'.jsonl', bucket_name, \\\n os.path.join(out_prefix, n, n+'.jsonl'),\\\n ExtraArgs = {'ServerSideEncryption':'AES256'}) #upload input files",
"_____no_output_____"
],
[
"print(vocab_filename)\nprint(out_prefix)\nprint( os.path.join(out_prefix, \"auxiliary/vocab.json\"))",
"_____no_output_____"
],
[
"s3_client.upload_file(vocab_filename,\n bucket_name, os.path.join(out_prefix, \"auxiliary/vocab.json\"),\n ExtraArgs = {'ServerSideEncryption':'AES256'}) #upload vocab file",
"_____no_output_____"
],
[
"import pickle\npickle.dump(vocab_to_token_dict, open('./vocab/vocab_to_token_dict.p', 'wb'))\npickle.dump(token_to_vocab_dict, open('./vocab/token_to_vocab_dict.p', 'wb'))\nfor f in ['vocab_to_token_dict.p','token_to_vocab_dict.p']:\n s3_client.upload_file(\"./vocab/\"+f, bucket_name, \\\n os.path.join(out_prefix, 'meta', f),ExtraArgs = {'ServerSideEncryption':'AES256'})",
"_____no_output_____"
],
[
"for f in local_files:\n remove_file(f)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
e78fa6b097441ff352724d8393bb403163491dc1 | 296,459 | ipynb | Jupyter Notebook | ASL Recognition with Deep Learning/notebook.ipynb | Shogun89/DataCamp-Python | 66f44152d557a1b212e6cd635194cf933bc58a5c | [
"MIT"
] | null | null | null | ASL Recognition with Deep Learning/notebook.ipynb | Shogun89/DataCamp-Python | 66f44152d557a1b212e6cd635194cf933bc58a5c | [
"MIT"
] | null | null | null | ASL Recognition with Deep Learning/notebook.ipynb | Shogun89/DataCamp-Python | 66f44152d557a1b212e6cd635194cf933bc58a5c | [
"MIT"
] | null | null | null | 296,459 | 296,459 | 0.953731 | [
[
[
"## 1. American Sign Language (ASL)\n<p>American Sign Language (ASL) is the primary language used by many deaf individuals in North America, and it is also used by hard-of-hearing and hearing individuals. The language is as rich as spoken languages and employs signs made with the hand, along with facial gestures and bodily postures.</p>\n<p><img src=\"https://s3.amazonaws.com/assets.datacamp.com/production/project_509/img/asl.png\" alt=\"american sign language\"></p>\n<p>A lot of recent progress has been made towards developing computer vision systems that translate sign language to spoken language. This technology often relies on complex neural network architectures that can detect subtle patterns in streaming video. However, as a first step, towards understanding how to build a translation system, we can reduce the size of the problem by translating individual letters, instead of sentences.</p>\n<p><strong>In this notebook</strong>, we will train a convolutional neural network to classify images of American Sign Language (ASL) letters. After loading, examining, and preprocessing the data, we will train the network and test its performance.</p>\n<p>In the code cell below, we load the training and test data. </p>\n<ul>\n<li><code>x_train</code> and <code>x_test</code> are arrays of image data with shape <code>(num_samples, 3, 50, 50)</code>, corresponding to the training and test datasets, respectively.</li>\n<li><code>y_train</code> and <code>y_test</code> are arrays of category labels with shape <code>(num_samples,)</code>, corresponding to the training and test datasets, respectively.</li>\n</ul>",
"_____no_output_____"
]
],
[
[
"# Import packages and set numpy random seed\nimport numpy as np\nnp.random.seed(5) \nimport tensorflow as tf\ntf.set_random_seed(2)\nfrom datasets import sign_language\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\n# Load pre-shuffled training and test datasets\n(x_train, y_train), (x_test, y_test) = sign_language.load_data()",
"_____no_output_____"
]
],
[
[
"## 2. Visualize the training data\n<p>Now we'll begin by creating a list of string-valued labels containing the letters that appear in the dataset. Then, we visualize the first several images in the training data, along with their corresponding labels.</p>",
"_____no_output_____"
]
],
[
[
"# Store labels of dataset\nlabels = ['A','B','C']\n\n# Print the first several training images, along with the labels\nfig = plt.figure(figsize=(20,5))\nfor i in range(36):\n ax = fig.add_subplot(3, 12, i + 1, xticks=[], yticks=[])\n ax.imshow(np.squeeze(x_train[i]))\n ax.set_title(\"{}\".format(labels[y_train[i]]))\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 3. Examine the dataset\n<p>Let's examine how many images of each letter can be found in the dataset.</p>\n<p>Remember that dataset has already been split into training and test sets for you, where <code>x_train</code> and <code>x_test</code> contain the images, and <code>y_train</code> and <code>y_test</code> contain their corresponding labels.</p>\n<p>Each entry in <code>y_train</code> and <code>y_test</code> is one of <code>0</code>, <code>1</code>, or <code>2</code>, corresponding to the letters <code>'A'</code>, <code>'B'</code>, and <code>'C'</code>, respectively.</p>\n<p>We will use the arrays <code>y_train</code> and <code>y_test</code> to verify that both the training and test sets each have roughly equal proportions of each letter.</p>",
"_____no_output_____"
]
],
[
[
"# Number of A's in the training dataset\nnum_A_train = sum(y_train==0)\n# Number of B's in the training dataset\nnum_B_train = sum(y_train==1)\n# Number of C's in the training dataset\nnum_C_train = sum(y_train==2)\n\n# Number of A's in the test dataset\nnum_A_test = sum(y_test==0)\n# Number of B's in the test dataset\nnum_B_test = sum(y_test==1)\n# Number of C's in the test dataset\nnum_C_test = sum(y_test==2)\n\n# Print statistics about the dataset\nprint(\"Training set:\")\nprint(\"\\tA: {}, B: {}, C: {}\".format(num_A_train, num_B_train, num_C_train))\nprint(\"Test set:\")\nprint(\"\\tA: {}, B: {}, C: {}\".format(num_A_test, num_B_test, num_C_test))",
"Training set:\n\tA: 540, B: 528, C: 532\nTest set:\n\tA: 118, B: 144, C: 138\n"
]
],
[
[
"## 4. One-hot encode the data\n<p>Currently, our labels for each of the letters are encoded as categorical integers, where <code>'A'</code>, <code>'B'</code> and <code>'C'</code> are encoded as <code>0</code>, <code>1</code>, and <code>2</code>, respectively. However, recall that Keras models do not accept labels in this format, and we must first one-hot encode the labels before supplying them to a Keras model.</p>\n<p>This conversion will turn the one-dimensional array of labels into a two-dimensional array.</p>\n<p><img src=\"https://s3.amazonaws.com/assets.datacamp.com/production/project_509/img/onehot.png\" alt=\"one-hot encoding\"></p>\n<p>Each row in the two-dimensional array of one-hot encoded labels corresponds to a different image. The row has a <code>1</code> in the column that corresponds to the correct label, and <code>0</code> elsewhere. </p>\n<p>For instance, </p>\n<ul>\n<li><code>0</code> is encoded as <code>[1, 0, 0]</code>, </li>\n<li><code>1</code> is encoded as <code>[0, 1, 0]</code>, and </li>\n<li><code>2</code> is encoded as <code>[0, 0, 1]</code>.</li>\n</ul>",
"_____no_output_____"
]
],
[
[
"from keras.utils import np_utils\n\n# One-hot encode the training labels\ny_train_OH = np_utils.to_categorical(y_train, 3)\n\n# One-hot encode the test labels\ny_test_OH = np_utils.to_categorical(y_test, 3)",
"_____no_output_____"
]
],
[
[
"## 5. Define the model\n<p>Now it's time to define a convolutional neural network to classify the data.</p>\n<p>This network accepts an image of an American Sign Language letter as input. The output layer returns the network's predicted probabilities that the image belongs in each category.</p>",
"_____no_output_____"
]
],
[
[
"from keras.layers import Conv2D, MaxPooling2D\nfrom keras.layers import Flatten, Dense\nfrom keras.models import Sequential\n\nmodel = Sequential()\n# First convolutional layer accepts image input\nmodel.add(Conv2D(filters=5, kernel_size=5, padding='same', activation='relu', \n input_shape=(50, 50, 3)))\n# Add a max pooling layer\nmodel.add(MaxPooling2D(pool_size=4))\n# Add a convolutional layer\nmodel.add(Conv2D(filters=15, kernel_size=5, padding='same', activation='relu', \n input_shape=(50, 50, 3)))\n# Add another max pooling layer\nmodel.add(MaxPooling2D(pool_size=4))\n# Flatten and feed to output layer\nmodel.add(Flatten())\nmodel.add(Dense(3, activation='softmax'))\n\n# Summarize the model\nmodel.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nconv2d_10 (Conv2D) (None, 50, 50, 5) 380 \n_________________________________________________________________\nmax_pooling2d_7 (MaxPooling2 (None, 12, 12, 5) 0 \n_________________________________________________________________\nconv2d_11 (Conv2D) (None, 12, 12, 15) 1890 \n_________________________________________________________________\nmax_pooling2d_8 (MaxPooling2 (None, 3, 3, 15) 0 \n_________________________________________________________________\nflatten_4 (Flatten) (None, 135) 0 \n_________________________________________________________________\ndense_4 (Dense) (None, 3) 408 \n=================================================================\nTotal params: 2,678\nTrainable params: 2,678\nNon-trainable params: 0\n_________________________________________________________________\n"
]
],
[
[
"## 6. Compile the model\n<p>After we have defined a neural network in Keras, the next step is to compile it! </p>",
"_____no_output_____"
]
],
[
[
"# Compile the model\nmodel.compile(optimizer='rmsprop', \n loss='categorical_crossentropy', \n metrics=['accuracy'])",
"_____no_output_____"
]
],
[
[
"## 7. Train the model\n<p>Once we have compiled the model, we're ready to fit it to the training data.</p>",
"_____no_output_____"
]
],
[
[
"# Train the model\nhist = model.fit(x_train, y_train_OH,\n validation_split=0.2,\n epochs=2,\n batch_size=32)",
"Train on 1280 samples, validate on 320 samples\nEpoch 1/2\n1280/1280 [==============================] - 4s 3ms/step - loss: 0.9623 - acc: 0.6102 - val_loss: 0.7729 - val_acc: 0.8688\nEpoch 2/2\n1280/1280 [==============================] - 3s 3ms/step - loss: 0.6252 - acc: 0.8656 - val_loss: 0.4826 - val_acc: 0.9406\n"
]
],
[
[
"## 8. Test the model\n<p>To evaluate the model, we'll use the test dataset. This will tell us how the network performs when classifying images it has never seen before!</p>\n<p>If the classification accuracy on the test dataset is similar to the training dataset, this is a good sign that the model did not overfit to the training data. </p>",
"_____no_output_____"
]
],
[
[
"# Obtain accuracy on test set\nscore = model.evaluate(x=x_test, \n y=y_test_OH,\n verbose=0)\nprint('Test accuracy:', score[1])",
"Test accuracy: 0.9475\n"
]
],
[
[
"## 9. Visualize mistakes\n<p>Hooray! Our network gets very high accuracy on the test set! </p>\n<p>The final step is to take a look at the images that were incorrectly classified by the model. Do any of the mislabeled images look relatively difficult to classify, even to the human eye? </p>\n<p>Sometimes, it's possible to review the images to discover special characteristics that are confusing to the model. However, it is also often the case that it's hard to interpret what the model had in mind!</p>",
"_____no_output_____"
]
],
[
[
"# Get predicted probabilities for test dataset\ny_probs = ...\n\n# Get predicted labels for test dataset\ny_preds = ...\n\n# Indices corresponding to test images which were mislabeled\nbad_test_idxs = ...\n\n# Print mislabeled examples\nfig = plt.figure(figsize=(25,4))\nfor i, idx in enumerate(bad_test_idxs):\n ax = fig.add_subplot(2, np.ceil(len(bad_test_idxs)/2), i + 1, xticks=[], yticks=[])\n ax.imshow(np.squeeze(x_test[idx]))\n ax.set_title(\"{} (pred: {})\".format(labels[y_test[idx]], labels[y_preds[idx]]))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e78fb3171008fd606a677cc3ffa75e73c66e1384 | 2,097 | ipynb | Jupyter Notebook | 7Diciembre.ipynb | samuelgh15/daa_2021_1 | 1cf0eaa5c70e6842c826c053def293fdd71a970f | [
"MIT"
] | null | null | null | 7Diciembre.ipynb | samuelgh15/daa_2021_1 | 1cf0eaa5c70e6842c826c053def293fdd71a970f | [
"MIT"
] | null | null | null | 7Diciembre.ipynb | samuelgh15/daa_2021_1 | 1cf0eaa5c70e6842c826c053def293fdd71a970f | [
"MIT"
] | null | null | null | 25.26506 | 230 | 0.419647 | [
[
[
"<a href=\"https://colab.research.google.com/github/samuelgh15/daa_2021_1/blob/master/7Diciembre.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"def fibonaci(n):\n print(\"Llamada\",n)\n if n== 1 or n ==0:\n return n\n else:\n return (fibonaci(n-1) + fibonaci (n-2))\n\nprint(fibonaci(6))",
"Llamada 6\nLlamada 5\nLlamada 4\nLlamada 3\nLlamada 2\nLlamada 1\nLlamada 0\nLlamada 1\nLlamada 2\nLlamada 1\nLlamada 0\nLlamada 3\nLlamada 2\nLlamada 1\nLlamada 0\nLlamada 1\nLlamada 4\nLlamada 3\nLlamada 2\nLlamada 1\nLlamada 0\nLlamada 1\nLlamada 2\nLlamada 1\nLlamada 0\n8\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
e78fbe2d3e20bdf5a2a6c99397da502452d45aec | 22,305 | ipynb | Jupyter Notebook | 1-Python-Basics.ipynb | aktgitrepo/python-for-datascience | d8bcc866a22e2093c9fd11e0ee6fc23f0add62ad | [
"Apache-2.0"
] | null | null | null | 1-Python-Basics.ipynb | aktgitrepo/python-for-datascience | d8bcc866a22e2093c9fd11e0ee6fc23f0add62ad | [
"Apache-2.0"
] | null | null | null | 1-Python-Basics.ipynb | aktgitrepo/python-for-datascience | d8bcc866a22e2093c9fd11e0ee6fc23f0add62ad | [
"Apache-2.0"
] | null | null | null | 17.549174 | 60 | 0.459224 | [
[
[
"# Printing basic types",
"_____no_output_____"
]
],
[
[
"print(\"Hello World!\")",
"_____no_output_____"
],
[
"print(\"Welcome to Foundations of Data Science!\")",
"_____no_output_____"
],
[
"print(2020)",
"_____no_output_____"
],
[
"print(1.314)",
"_____no_output_____"
],
[
"print(True)",
"_____no_output_____"
],
[
"print(False)",
"_____no_output_____"
],
[
"print(True or False)",
"_____no_output_____"
],
[
"print(True and False)",
"_____no_output_____"
]
],
[
[
"# Variables and inputs",
"_____no_output_____"
]
],
[
[
"var = \"value\"",
"_____no_output_____"
],
[
"print(var)",
"_____no_output_____"
],
[
"print(type(var))",
"_____no_output_____"
],
[
"print(id(var))",
"_____no_output_____"
],
[
"var_2 = \"_2\"",
"_____no_output_____"
],
[
"print(var_2)",
"_____no_output_____"
],
[
"print(id(var_2))",
"_____no_output_____"
],
[
"print(\"variable \" + var)",
"_____no_output_____"
],
[
"num = 2020",
"_____no_output_____"
],
[
"print(type(num))",
"_____no_output_____"
],
[
"print(type(num))",
"_____no_output_____"
],
[
"is_good = True",
"_____no_output_____"
],
[
"print(type(is_good))",
"_____no_output_____"
],
[
"my_name = input(\"What is your name \")",
"_____no_output_____"
],
[
"print(my_name, type(my_name))",
"_____no_output_____"
],
[
"hours_per_week = 24 * 7",
"_____no_output_____"
],
[
"print(\"hours_per_week\", hours_per_week)",
"_____no_output_____"
]
],
[
[
"# String Processing",
"_____no_output_____"
]
],
[
[
"topic = \"Foundations of Data Science\"",
"_____no_output_____"
],
[
"print(topic)",
"_____no_output_____"
],
[
"print(topic[0])",
"_____no_output_____"
],
[
"print(topic[1])",
"_____no_output_____"
],
[
"print(topic[10])",
"_____no_output_____"
],
[
"print(topic[-1])",
"_____no_output_____"
],
[
"print(topic[-2])",
"_____no_output_____"
],
[
"print(topic[0:10])",
"_____no_output_____"
],
[
"print(topic[12:16])",
"_____no_output_____"
],
[
"print(topic.lower())",
"_____no_output_____"
],
[
"print(topic)",
"_____no_output_____"
],
[
"topic = topic.lower()",
"_____no_output_____"
],
[
"print(topic.upper())",
"_____no_output_____"
],
[
"print(topic.islower())",
"_____no_output_____"
],
[
"topic = topic.upper()",
"_____no_output_____"
],
[
"print(topic.islower())",
"_____no_output_____"
],
[
"print(topic.isupper())",
"_____no_output_____"
],
[
"print(topic.find(\"DATA\"))",
"_____no_output_____"
],
[
"print(topic[15])",
"_____no_output_____"
],
[
"print(topic.find(\"AI\"))",
"_____no_output_____"
],
[
"print(topic.replace(\"SCIENCE\", \"ENGINEERING\"))",
"_____no_output_____"
],
[
"print(55/34)",
"_____no_output_____"
],
[
"golden_ratio = 55/34",
"_____no_output_____"
],
[
"print(type(golden_ratio))",
"_____no_output_____"
],
[
"print(golden_ratio.is_integer())",
"_____no_output_____"
],
[
"print(golden_ratio.as_integer_ratio())",
"_____no_output_____"
],
[
"print(3642617345667313/2251799813685248)",
"_____no_output_____"
],
[
"print(55 // 34, 55/34)",
"_____no_output_____"
],
[
"print(55 % 34)",
"_____no_output_____"
],
[
"print(2 ** 3)",
"_____no_output_____"
]
],
[
[
"# If, For, While Blocks",
"_____no_output_____"
]
],
[
[
"hours_per_week = 5",
"_____no_output_____"
],
[
"if hours_per_week > 10:\n print(my_name + \" you are doing well\")",
"_____no_output_____"
],
[
"if hours_per_week > 10:\n print(my_name + \" you are doing well\")\nprint(\"Outside If\")",
"_____no_output_____"
],
[
"if hours_per_week > 10:\n print(my_name + \" you are doing well\")\nelse:\n print(my_name + \" you need to study more\")",
"_____no_output_____"
],
[
"for i in range(5):\n print(i)",
"_____no_output_____"
],
[
"range?",
"_____no_output_____"
],
[
"for i in range(2, 5):\n print(i, i**2)",
"_____no_output_____"
],
[
"a = 1\nb = 1\nprint(a)\nprint(b)\nfor i in range(10):\n temp = a + b\n a = b\n b = temp\n print(temp)",
"_____no_output_____"
],
[
"a = 1\nb = 1\nprint(a)\nprint(b)\ni = 2\nwhile b < 1729:\n temp = a + b\n i += 1\n a = b\n b = temp\n print(temp)\nprint(\"Printed\", i, \"numbers\")",
"_____no_output_____"
]
],
[
[
"# Functions",
"_____no_output_____"
]
],
[
[
"def fibonacci(pos):\n a = 1\n b = 1\n for i in range(pos):\n temp = a + b\n a = b\n b = temp\n return temp",
"_____no_output_____"
],
[
"print(fibonacci(3))",
"_____no_output_____"
],
[
"print(fibonacci(8), fibonacci(7))",
"_____no_output_____"
],
[
"for i in range(2, 20):\n ratio = fibonacci(i) / fibonacci(i - 1)\n print(i, ratio)",
"_____no_output_____"
],
[
"def fibonacci_relative(pos, a, b):\n for i in range(pos):\n temp = a + b\n a = b\n b = temp\n return temp",
"_____no_output_____"
],
[
"print(fibonacci_relative(3, 34, 55))",
"_____no_output_____"
],
[
"def fibonacci_ol(pos, a = 1, b = 1):\n for i in range(pos):\n temp = a + b\n a = b\n b = temp\n return temp",
"_____no_output_____"
],
[
"print(fibonacci_ol(3, 34, 55))",
"_____no_output_____"
],
[
"print(fibonacci_relative(3))",
"_____no_output_____"
],
[
"def fibonacci_recursive(n, a = 1, b = 1):\n if n > 1:\n return fibonacci_recursive(n - 1, b, a + b)\n else:\n return a + b",
"_____no_output_____"
],
[
"print(fibonacci_recursive(3, 34, 55))",
"_____no_output_____"
],
[
"def fibonacci(pos):\n a = 1\n b = 1\n for i in range(pos):\n temp = a + b\n a = b\n b = temp\n return temp",
"_____no_output_____"
],
[
"print(fibonacci(3))",
"_____no_output_____"
],
[
"print(fibonacci(8), fibonacci(7))",
"_____no_output_____"
],
[
"for i in range(2, 20):\n ratio = fibonacci(i) / fibonacci(i - 1)\n print(i, ratio)",
"_____no_output_____"
],
[
"def fibonacci_relative(pos, a, b):\n for i in range(pos):\n temp = a + b\n a = b\n b = temp\n return temp",
"_____no_output_____"
],
[
"print(fibonacci_relative(3, 34, 55))",
"_____no_output_____"
],
[
"def fibonacci_ol(pos, a = 1, b = 1):\n for i in range(pos):\n temp = a + b\n a = b\n b = temp\n return temp",
"_____no_output_____"
],
[
"print(fibonacci_ol(3, 34, 55))",
"_____no_output_____"
],
[
"print(fibonacci_relative(3))",
"_____no_output_____"
],
[
"def fibonacci_recursive(n, a = 1, b = 1):\n if n > 1:\n return fibonacci_recursive(n - 1, b, a + b)\n else:\n return a + b",
"_____no_output_____"
],
[
"print(fibonacci_recursive(3, 34, 55))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78fcdab96e9e842e5f4e4720aeb9f05734d85cd | 987,894 | ipynb | Jupyter Notebook | Intro to Python Class Projects/Intro to Python ML Project D.ipynb | Ddottsai/Code-Storage | fe8753e3d93dfa69822ae06b64cc7d3b259a4434 | [
"MIT"
] | null | null | null | Intro to Python Class Projects/Intro to Python ML Project D.ipynb | Ddottsai/Code-Storage | fe8753e3d93dfa69822ae06b64cc7d3b259a4434 | [
"MIT"
] | null | null | null | Intro to Python Class Projects/Intro to Python ML Project D.ipynb | Ddottsai/Code-Storage | fe8753e3d93dfa69822ae06b64cc7d3b259a4434 | [
"MIT"
] | null | null | null | 499.188479 | 64,300 | 0.554506 | [
[
[
"# processing\nimport pandas as pd\nimport numpy as np\nimport sklearn as sk\nfrom sklearn import preprocessing\nfrom sklearn.preprocessing import LabelEncoder\nfrom scipy import stats\nfrom scipy.stats import mode\nfrom scipy.stats import skew\nfrom scipy.stats import kurtosis\nfrom numpy import linalg\nfrom sklearn.ensemble import AdaBoostRegressor\nimport math\nimport random\nfrom sklearn.decomposition import PCA\nfrom itertools import product\nfrom sklearn.preprocessing import MaxAbsScaler\nfrom sklearn.preprocessing import RobustScaler\n\n# graphs\nfrom scipy.stats import boxcox\n\nfrom matplotlib import pyplot as plt\nplt.rcParams['figure.figsize'] = (16, 9)\n\n# models\nfrom sklearn.cluster import KMeans\n\nfrom sklearn.neighbors import NearestNeighbors\nfrom sklearn.neighbors import KNeighborsClassifier\n\nfrom sklearn.linear_model import LinearRegression\nfrom sklearn.linear_model import Lasso\nfrom sklearn.linear_model import Ridge\nfrom sklearn.neighbors import KNeighborsRegressor\nfrom sklearn.svm import SVR\n\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.tree import DecisionTreeRegressor\nfrom sklearn.tree import export_graphviz\n\n\nfrom sklearn.linear_model import LogisticRegression\n\nimport pickle\n\n\n# accuracy\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.metrics import r2_score\nfrom sklearn.metrics import mean_squared_error\nfrom sklearn.model_selection import LeavePOut\nfrom sklearn.model_selection import ShuffleSplit\n\n#display\nfrom IPython.display import Markdown, display\nfrom __future__ import print_function\ndef printmd(string):\n display(Markdown(string))\nnp.set_printoptions(threshold=np.nan)\npd.set_option(\"mode.use_inf_as_na\", True)",
"_____no_output_____"
],
[
"dataFile = pd.read_csv(\"housing_filled.csv\")\ndata = pd.DataFrame(dataFile)\ndata.dropna(inplace=True)\nfinal_features = []",
"_____no_output_____"
]
],
[
[
"# <font size = 40 color = blue> KMeans </font>",
"_____no_output_____"
]
],
[
[
"X_cluster = pd.DataFrame(data['LotArea'])\nX_cluster['OverallCond'] = data['OverallCond']\nX_cluster['OverallQual'] = data['OverallQual']\n#X_svm['FullBath'] = data['FullBath']\nX_cluster['TotRmsAbvGrd'] = data['TotRmsAbvGrd']\n#X_cluster['SalePrice'] = data['SalePrice']\n#X_svm['GarageArea'] = data['GarageArea']\n\nk_range = np.arange(2,10)\nSSE = []\nfor i in k_range:\n kmeans_model = KMeans(i)\n clusters = kmeans_model.fit_predict(X_cluster)\n SSE.append(kmeans_model.inertia_)\n\nplt.scatter(k_range, SSE)",
"_____no_output_____"
],
[
"kmeans_model = KMeans(4)\nclusters = kmeans_model.fit_predict(X_cluster)\n\nplt.title('SalePrice Distribution for each cluster')\nplt.scatter(clusters,data['SalePrice'])\n\"\"\"plt.plot(clusters[0]['OverallCond'],clusters[0]['SalePrice'],'g',\n clusters[1]['OverallCond'],clusters[1]['SalePrice'],'b', \n clusters[2]['OverallCond'],clusters[2]['SalePrice'],'r',\n clusters[3]['OverallCond'],clusters[3]['SalePrice'],'b')\"\"\"\nplt.show()\n\nclust_0 = [i for i,d in enumerate(clusters) if d == 0]\nclust_1 = [i for i,d in enumerate(clusters) if d == 1]\nclust_2 = [i for i,d in enumerate(clusters) if d == 2]\nclust_3 = [i for i,d in enumerate(clusters) if d == 3]\n\nplt.title('SalePrice vs. OverallCond')\nplt.plot(data.iloc[clust_0]['OverallCond'],data.iloc[clust_0]['SalePrice'],'go',\n data.iloc[clust_1]['OverallCond'],data.iloc[clust_1]['SalePrice'],'bx', \n data.iloc[clust_2]['OverallCond'],data.iloc[clust_2]['SalePrice'],'ro',\n data.iloc[clust_3]['OverallCond'],data.iloc[clust_3]['SalePrice'],'m^')\nplt.show()\n\nplt.title('SalePrice vs. 1stFlrSF')\nplt.plot(data.iloc[clust_0]['1stFlrSF'],data.iloc[clust_0]['SalePrice'],'go',\n data.iloc[clust_1]['1stFlrSF'],data.iloc[clust_1]['SalePrice'],'bo', \n data.iloc[clust_2]['1stFlrSF'],data.iloc[clust_2]['SalePrice'],'ro',\n data.iloc[clust_3]['1stFlrSF'],data.iloc[clust_3]['SalePrice'],'m^')\nplt.rcParams['figure.figsize'] = (16, 16)\nplt.show()",
"_____no_output_____"
]
],
[
[
"We decided to cluster using several features that we previously saw had significant relationships with SalePrice. The clustering revealed several unique properties. In the first graph, which shows the distribution of SalePrice within each cluster, indicates that the variables we chose do indeed have a strong relationship with SalePrice; the distributions and means are notably different. The second graph compares SalePrice to one specific feature that was used in our KMeans, OverallCond. It appears that KMeans was able to separate out one cluster that had a large spread in OverallCond and a small spread in SalePrice (primarily in the low range of SalePrice). The third graph looks at two features that were both not included in our KMeans: 1stFlrSF and SalePrice. It appears that 1stFlrSF tends to have a positive relationship with SalePrice; however, that is not true for one cluster. For one cluster, the shape of the points has the shape of an inverse relationship.",
"_____no_output_____"
]
],
[
[
"<font color=blue size = 40>**Ensemble (Stacking) Model**</font>",
"_____no_output_____"
],
[
"init_prior = []\ndata['BldgType'] = LabelEncoder().fit_transform(data['BldgType'])\ninit_prior.append('OverallQual')\ninit_prior.append('BldgType')\ninit_prior.append('TotalBsmtSF')\ninit_prior.append('1stFlrSF')\ninit_prior.append('GrLivArea')\ninit_prior.append('GarageCars')\ninit_prior.append('GarageArea')\ninit_prior.append('2ndFlrSF')\ninit_prior.append('FullBath')\ninit_prior.append('TotRmsAbvGrd')\ninit_prior.append('LotArea')\ninit_prior.append('YearRemodAdd')\ninit_prior.append('YearBuilt')\ninit_prior.append('GarageYrBlt')\ninit_prior.append('FireplaceQu')\ninit_prior.append('MSSubClass')\ninit_prior.append('WoodDeckSF')\n\n# eliminate some dependent/redundant features\ncorr_within_prior = data[init_prior].corr()\nfor row_name,cols in corr_within_prior.iterrows():\n print(row_name)\n for (name,val) in cols.iteritems():\n if (row_name != name and ((val > 0.6) | (val < -0.6))):\n print(\" %s: %0.3f\" % (name,val))\n print()",
"_____no_output_____"
]
],
[
[
"Sets of somewhat correlated variables:\n\n['GarageCars', 'OverallQual', 'GarageArea']\n\n\n['TotalBsmtSF', '1stFlrSF']\n\n\n['GrLivArea', '2ndFlrSF', 'FullBath', 'TotRmsAbvGrd']\n\n\n['GarageYrBlt', 'YearRemodAdd', 'YearBuilt']\n",
"_____no_output_____"
],
[
"# <font size=40> **Feature Selection** </font>",
"_____no_output_____"
]
],
[
[
"new_prior = list(init_prior)\nnames = ['Garage','LowerSF','Living','Year']\nnew_prior.extend(names)\nfor dependents in [['GarageCars', 'OverallQual', 'GarageArea'],['TotalBsmtSF','1stFlrSF'],\n ['GrLivArea', '2ndFlrSF', 'FullBath', 'TotRmsAbvGrd'],['GarageYrBlt', 'YearRemodAdd', 'YearBuilt']]:\n pca = PCA(1).fit(data[dependents])\n factors = pca.components_\n data[names.pop()] = np.sum(np.multiply(factors, data[dependents]),axis=1)\n #plt.bar(np.arrange(len(dependents)),pca.explained_variance_ratio_)\n #plt.xticks(np.arrange(len(dependents)),dependents)\n #plt.title('Fraction of Variance Explained by Each Component')\n #plt.xlabel('Component')\n #plt.ylabel('Fraction of Total Variance')\n #plt.show()\n for i in dependents:\n new_prior.remove(i)\nprint(new_prior)",
"['BldgType', 'LotArea', 'FireplaceQu', 'MSSubClass', 'WoodDeckSF', 'Garage', 'LowerSF', 'Living', 'Year']\n"
],
[
"sale_corr = data.corr()['SalePrice']\nsale_corr.drop('SalePrice')\ndisplay(Markdown(\"**Correlation with SalePrice**\"))\ndisplay(sale_corr[new_prior])",
"_____no_output_____"
],
[
"new_prior_2 = list(new_prior)\nnew_prior_2.remove('BldgType')\nnew_prior_2.remove('LotArea')\nnew_prior_2.remove('FireplaceQu')\nnew_prior_2.remove('MSSubClass')\nnew_prior_2.remove('WoodDeckSF')\ndisplay(new_prior_2)",
"_____no_output_____"
]
],
[
[
"# <font size=40>**Preprocessing**</font>",
"_____no_output_____"
]
],
[
[
"plt.rcParams['figure.figsize'] = (16,4)\nfor feature_name in new_prior_2:\n plt.title(feature_name)\n plt.subplot(1, 3, 1)\n plt.hist(data[feature_name],density=True)\n plt.xlabel(feature_name)\n plt.ylabel('Frequency')\n \n stdev = np.std(data[feature_name])\n mean = np.mean(data[feature_name])\n col_distr = ((data[feature_name] - mean) / stdev)\n plt.subplot(1, 3, 2)\n plt.hist(col_distr,density=True)\n plt.xlabel('std. dev. distance of ' + feature_name)\n plt.ylabel('Frequency')\n \n plt.subplot(1, 3, 3)\n plt.boxplot(data[feature_name])\n plt.xlabel(feature_name)\n plt.ylabel('Value')\n\n plt.show()",
"_____no_output_____"
],
[
"scaler = RobustScaler()\ndata[\"Garage_scaled\"] = scaler.fit_transform(data['Garage'].values.reshape(-1,1))\ndata[\"LowerSF_scaled\"] = scaler.fit_transform(data['LowerSF'].values.reshape(-1,1))\ndata[\"Living_scaled\"] = scaler.fit_transform(data['Living'].values.reshape(-1,1))\ndata[\"Year_scaled\"] = scaler.fit_transform(data['Year'].values.reshape(-1,1))\nfor feature_name in new_prior_2:\n plt.title(feature_name + \"_scaled\")\n plt.subplot(1, 3, 1)\n plt.hist(data[feature_name + \"_scaled\"],density=True)\n plt.xlabel(feature_name + \"_scaled\")\n plt.ylabel('Frequency')\n plt.show()",
"_____no_output_____"
],
[
"final_features = ['Garage_scaled','LowerSF_scaled','Living_scaled','Year_scaled']",
"_____no_output_____"
]
],
[
[
"# <font size = 40> Clustering Algorithm for part of Stack </font>",
"_____no_output_____"
]
],
[
[
" def cluster_from_stack_of_KMeans(data, K=8, num_weak_learners = 5,num_neighbors=1,\n distance_penalty= lambda x: x):\n \n assert K < len(data)\n assert num_neighbors <= len(data)\n \n C = np.empty((num_weak_learners,data.shape[0]))\n SSE = np.empty((num_weak_learners,))\n for iter_num in range(num_weak_learners):\n clustering = KMeans(K,init='random', n_init=1, n_jobs=-1)\n clustering.fit(data)\n #np.put(C,[iter_num,],clustering.labels_)\n C[iter_num] = clustering.labels_\n SSE[iter_num] = clustering.inertia_\n \n \n \n \n # calculate node connections\n \n C_scores = distance_penalty((1 - SSE)/ np.max(SSE)) \n \n connections = [[] for _ in range(len(data))]\n for i in range(len(connections)):\n connections[i] = np.argsort([np.sum([d for iter_num,d in enumerate(C_scores) if\n i != j and C[iter_num][i] == C[iter_num][j]])\n for j in range(len(data))])[-num_neighbors:]\n for _ in range(num_neighbors-1):\n if np.random.ranf() > 0.7:\n connections[i] = connections[i][:-1]\n \n # group together linked nodes to form (unequal) clusters\n \n cluster_sets = []\n cluster_dict = {} # set of (element index -> index in clusters)\n to_merge = {} # lowest cluster_set index in sets to be merged -> the rest of the sets that should be merged together\n #to_merge_not_keys = set()\n for i1,links in enumerate(connections):\n for i2 in links:\n if i2 not in cluster_dict:\n if i1 in cluster_dict:\n c = cluster_dict[i1]\n cluster_sets[c].add(i2)\n cluster_dict[i2]= c\n else:\n cluster_sets.append({i1,i2})\n cluster_dict.update({i1 : len(cluster_sets)-1, i2 : len(cluster_sets)-1})\n else:\n if i1 in cluster_dict:\n in_to_merge = 0\n temp_key = None\n for merge_key,merge_set in to_merge.items():\n if in_to_merge != 1 and (cluster_dict[i1] in merge_set or cluster_dict[i1] == merge_key):\n if in_to_merge == 0:\n in_to_merge = 1\n temp_key = merge_key\n else:\n merge_set.add(merge_key)\n to_merge[temp_key].add(merge_key)\n comb = merge_set.union(to_merge[temp_key])\n comb.add(cluster_dict[i2])\n comb.add(cluster_dict[i1])\n min_key = min(comb)\n comb.remove(min_key)\n to_merge.update({min_key:comb})\n del to_merge[merge_key]\n del to_merge[temp_key]\n in_to_merge=3\n break\n elif in_to_merge != 2 and (cluster_dict[i2] in merge_set or cluster_dict[i2] == merge_key):\n if in_to_merge == 0:\n in_to_merge = 2\n temp_key = merge_key\n else:\n merge_set.add(merge_key)\n to_merge[temp_key].add(merge_key)\n comb = merge_set.union(to_merge[temp_key])\n comb.add(cluster_dict[i2])\n comb.add(cluster_dict[i1])\n min_key = min(comb)\n comb.remove(min_key)\n to_merge.update({min_key:comb})\n del to_merge[merge_key]\n del to_merge[temp_key]\n in_to_merge=3\n break \n if in_to_merge==1:\n if cluster_dict[i2] > temp_key:\n to_merge[temp_key].add(cluster_dict[i2])\n else:\n to_merge[temp_key].add(temp_key)\n to_merge.update({cluster_dict[i2]:to_merge[temp_key]})\n del to_merge[temp_key]\n elif in_to_merge==2:\n if cluster_dict[i1] > temp_key:\n to_merge[temp_key].add(cluster_dict[i1])\n else:\n to_merge[temp_key].add(temp_key)\n to_merge.update({cluster_dict[i1]:to_merge[temp_key]})\n del to_merge[temp_key]\n elif in_to_merge==0:\n if cluster_dict[i1] < cluster_dict[i2]:\n to_merge.update({cluster_dict[i1]:{cluster_dict[i2]}})\n else:\n to_merge.update({cluster_dict[i2]:{cluster_dict[i1]}})\n #to_merge_not_keys.add(i2)\n else:\n c = cluster_dict[i2]\n cluster_dict[i1] = c\n cluster_sets[c].add(i1)\n \n combined_clusters = set()\n for i,c_set in enumerate(cluster_sets):\n if c_set is not None:\n if i not in to_merge:\n combined_clusters.add(frozenset(c_set))\n else:\n total_set = c_set\n for cluster_sets_index in to_merge[i]:\n total_set = total_set.union(cluster_sets[cluster_sets_index])\n cluster_sets[cluster_sets_index] = None\n combined_clusters.add(frozenset(total_set))\n return combined_clusters",
"_____no_output_____"
]
],
[
[
"# Finding Stable Parameters for clustering",
"_____no_output_____"
]
],
[
[
"k_ticks = [3,7,20]\nw_ticks = [5,20]\nn_ticks = [1,2,3]\n\ndef sample_clustering_params():\n cluster_sizes = {}\n for k in k_ticks:\n for w in w_ticks: \n for n in n_ticks:\n c_sizes = []\n for i in range(5):\n train, valid, _, _ = train_test_split(\n data[final_features],data['SalePrice'],test_size=0.2)\n clusters = cluster_from_stack_of_KMeans(train, K=k, num_weak_learners=w, num_neighbors=n)\n temp = []\n for c in clusters:\n temp.append(len(c))\n c_sizes.append(temp)\n print(' ...')\n cluster_sizes[(k,w,n)]= c_sizes\n print(\"----------\" + str(n))\n print(cluster_sizes)\n print(\"----\" + str(w))\n print(k)\n return cluster_sizes",
"_____no_output_____"
],
[
"cluster_sizes = sample_clustering_params()",
" ...\n ...\n ...\n ...\n ...\n----------1\n{(3, 5, 1): [[718, 418], [347, 789], [1, 34, 4, 10, 1, 225, 3, 724, 134], [1, 1, 1, 66, 4, 1, 1, 1, 215, 69, 757, 13, 6], [1136]]}\n ...\n ...\n ...\n ...\n ...\n----------2\n{(3, 5, 1): [[718, 418], [347, 789], [1, 34, 4, 10, 1, 225, 3, 724, 134], [1, 1, 1, 66, 4, 1, 1, 1, 215, 69, 757, 13, 6], [1136]], (3, 5, 2): [[1, 65, 1067, 2, 1], [31, 126, 1, 6, 102, 3, 122, 94, 6, 278, 2, 176, 4, 185], [39, 1, 46, 55, 668, 43, 5, 65, 112, 101, 1], [696, 1, 7, 39, 1, 3, 2, 92, 2, 108, 17, 166, 1, 1], [64, 358, 29, 179, 57, 7, 2, 8, 181, 10, 82, 157, 2]]}\n ...\n ...\n ...\n ...\n ...\n----------3\n{(3, 5, 1): [[718, 418], [347, 789], [1, 34, 4, 10, 1, 225, 3, 724, 134], [1, 1, 1, 66, 4, 1, 1, 1, 215, 69, 757, 13, 6], [1136]], (3, 5, 2): [[1, 65, 1067, 2, 1], [31, 126, 1, 6, 102, 3, 122, 94, 6, 278, 2, 176, 4, 185], [39, 1, 46, 55, 668, 43, 5, 65, 112, 101, 1], [696, 1, 7, 39, 1, 3, 2, 92, 2, 108, 17, 166, 1, 1], [64, 358, 29, 179, 57, 7, 2, 8, 181, 10, 82, 157, 2]], (3, 5, 3): [[72, 191, 4, 22, 2, 2, 79, 51, 4, 84, 38, 5, 7, 17, 4, 4, 403, 145, 2], [1, 88, 11, 5, 36, 448, 530, 17], [142, 3, 53, 4, 6, 200, 55, 16, 5, 46, 2, 46, 1, 2, 137, 1, 11, 3, 403], [181, 108, 117, 2, 254, 7, 2, 75, 6, 85, 231, 68], [87, 3, 1, 754, 4, 75, 2, 33, 6, 153, 6, 2, 10]]}\n----5\n ...\n ...\n ...\n ...\n ...\n----------1\n{(3, 5, 1): [[718, 418], [347, 789], [1, 34, 4, 10, 1, 225, 3, 724, 134], [1, 1, 1, 66, 4, 1, 1, 1, 215, 69, 757, 13, 6], [1136]], (3, 5, 2): [[1, 65, 1067, 2, 1], [31, 126, 1, 6, 102, 3, 122, 94, 6, 278, 2, 176, 4, 185], [39, 1, 46, 55, 668, 43, 5, 65, 112, 101, 1], [696, 1, 7, 39, 1, 3, 2, 92, 2, 108, 17, 166, 1, 1], [64, 358, 29, 179, 57, 7, 2, 8, 181, 10, 82, 157, 2]], (3, 5, 3): [[72, 191, 4, 22, 2, 2, 79, 51, 4, 84, 38, 5, 7, 17, 4, 4, 403, 145, 2], [1, 88, 11, 5, 36, 448, 530, 17], [142, 3, 53, 4, 6, 200, 55, 16, 5, 46, 2, 46, 1, 2, 137, 1, 11, 3, 403], [181, 108, 117, 2, 254, 7, 2, 75, 6, 85, 231, 68], [87, 3, 1, 754, 4, 75, 2, 33, 6, 153, 6, 2, 10]], (3, 20, 1): [[1, 1, 1, 1, 1, 4, 1, 1, 1125], [3, 153, 2, 1, 5, 5, 967], [2, 2, 2, 24, 2, 139, 897, 68], [1, 1135], [1, 1, 1, 1, 1132]]}\n ...\n ...\n ...\n ...\n ...\n----------2\n{(3, 5, 1): [[718, 418], [347, 789], [1, 34, 4, 10, 1, 225, 3, 724, 134], [1, 1, 1, 66, 4, 1, 1, 1, 215, 69, 757, 13, 6], [1136]], (3, 5, 2): [[1, 65, 1067, 2, 1], [31, 126, 1, 6, 102, 3, 122, 94, 6, 278, 2, 176, 4, 185], [39, 1, 46, 55, 668, 43, 5, 65, 112, 101, 1], [696, 1, 7, 39, 1, 3, 2, 92, 2, 108, 17, 166, 1, 1], [64, 358, 29, 179, 57, 7, 2, 8, 181, 10, 82, 157, 2]], (3, 5, 3): [[72, 191, 4, 22, 2, 2, 79, 51, 4, 84, 38, 5, 7, 17, 4, 4, 403, 145, 2], [1, 88, 11, 5, 36, 448, 530, 17], [142, 3, 53, 4, 6, 200, 55, 16, 5, 46, 2, 46, 1, 2, 137, 1, 11, 3, 403], [181, 108, 117, 2, 254, 7, 2, 75, 6, 85, 231, 68], [87, 3, 1, 754, 4, 75, 2, 33, 6, 153, 6, 2, 10]], (3, 20, 1): [[1, 1, 1, 1, 1, 4, 1, 1, 1125], [3, 153, 2, 1, 5, 5, 967], [2, 2, 2, 24, 2, 139, 897, 68], [1, 1135], [1, 1, 1, 1, 1132]], (3, 20, 2): [[10, 10, 252, 78, 381, 10, 3, 1, 2, 131, 4, 40, 11, 1, 5, 47, 7, 2, 15, 126], [23, 56, 5, 2, 15, 5, 90, 2, 6, 6, 1, 13, 7, 4, 4, 3, 3, 3, 6, 76, 1, 2, 28, 1, 103, 70, 1, 430, 158, 4, 1, 3, 2, 2], [2, 156, 4, 98, 4, 5, 2, 85, 13, 2, 619, 1, 3, 3, 113, 9, 3, 14], [3, 1, 140, 140, 5, 5, 1, 128, 11, 632, 5, 11, 40, 4, 2, 4, 2, 2], [6, 1128, 2]]}\n ...\n ...\n ...\n ...\n ...\n----------3\n{(3, 5, 1): [[718, 418], [347, 789], [1, 34, 4, 10, 1, 225, 3, 724, 134], [1, 1, 1, 66, 4, 1, 1, 1, 215, 69, 757, 13, 6], [1136]], (3, 5, 2): [[1, 65, 1067, 2, 1], [31, 126, 1, 6, 102, 3, 122, 94, 6, 278, 2, 176, 4, 185], [39, 1, 46, 55, 668, 43, 5, 65, 112, 101, 1], [696, 1, 7, 39, 1, 3, 2, 92, 2, 108, 17, 166, 1, 1], [64, 358, 29, 179, 57, 7, 2, 8, 181, 10, 82, 157, 2]], (3, 5, 3): [[72, 191, 4, 22, 2, 2, 79, 51, 4, 84, 38, 5, 7, 17, 4, 4, 403, 145, 2], [1, 88, 11, 5, 36, 448, 530, 17], [142, 3, 53, 4, 6, 200, 55, 16, 5, 46, 2, 46, 1, 2, 137, 1, 11, 3, 403], [181, 108, 117, 2, 254, 7, 2, 75, 6, 85, 231, 68], [87, 3, 1, 754, 4, 75, 2, 33, 6, 153, 6, 2, 10]], (3, 20, 1): [[1, 1, 1, 1, 1, 4, 1, 1, 1125], [3, 153, 2, 1, 5, 5, 967], [2, 2, 2, 24, 2, 139, 897, 68], [1, 1135], [1, 1, 1, 1, 1132]], (3, 20, 2): [[10, 10, 252, 78, 381, 10, 3, 1, 2, 131, 4, 40, 11, 1, 5, 47, 7, 2, 15, 126], [23, 56, 5, 2, 15, 5, 90, 2, 6, 6, 1, 13, 7, 4, 4, 3, 3, 3, 6, 76, 1, 2, 28, 1, 103, 70, 1, 430, 158, 4, 1, 3, 2, 2], [2, 156, 4, 98, 4, 5, 2, 85, 13, 2, 619, 1, 3, 3, 113, 9, 3, 14], [3, 1, 140, 140, 5, 5, 1, 128, 11, 632, 5, 11, 40, 4, 2, 4, 2, 2], [6, 1128, 2]], (3, 20, 3): [[35, 17, 165, 7, 3, 3, 56, 45, 207, 56, 212, 2, 12, 8, 304, 4], [40, 84, 70, 1, 12, 132, 196, 2, 1, 10, 6, 103, 4, 4, 33, 2, 19, 13, 1, 2, 1, 1, 1, 2, 392, 2, 2], [1, 4, 2, 5, 5, 9, 914, 35, 4, 4, 6, 9, 2, 1, 2, 1, 7, 3, 115, 7], [392, 181, 2, 3, 61, 5, 191, 4, 1, 2, 3, 3, 17, 2, 47, 4, 10, 3, 1, 2, 3, 2, 4, 4, 2, 9, 3, 2, 1, 7, 163, 2], [36, 46, 83, 3, 80, 2, 5, 92, 5, 2, 35, 1, 78, 3, 30, 5, 1, 469, 9, 112, 3, 7, 9, 20]]}\n----20\n3\n ...\n ...\n ...\n ...\n ...\n----------1\n{(3, 5, 1): [[718, 418], [347, 789], [1, 34, 4, 10, 1, 225, 3, 724, 134], [1, 1, 1, 66, 4, 1, 1, 1, 215, 69, 757, 13, 6], [1136]], (3, 5, 2): [[1, 65, 1067, 2, 1], [31, 126, 1, 6, 102, 3, 122, 94, 6, 278, 2, 176, 4, 185], [39, 1, 46, 55, 668, 43, 5, 65, 112, 101, 1], [696, 1, 7, 39, 1, 3, 2, 92, 2, 108, 17, 166, 1, 1], [64, 358, 29, 179, 57, 7, 2, 8, 181, 10, 82, 157, 2]], (3, 5, 3): [[72, 191, 4, 22, 2, 2, 79, 51, 4, 84, 38, 5, 7, 17, 4, 4, 403, 145, 2], [1, 88, 11, 5, 36, 448, 530, 17], [142, 3, 53, 4, 6, 200, 55, 16, 5, 46, 2, 46, 1, 2, 137, 1, 11, 3, 403], [181, 108, 117, 2, 254, 7, 2, 75, 6, 85, 231, 68], [87, 3, 1, 754, 4, 75, 2, 33, 6, 153, 6, 2, 10]], (3, 20, 1): [[1, 1, 1, 1, 1, 4, 1, 1, 1125], [3, 153, 2, 1, 5, 5, 967], [2, 2, 2, 24, 2, 139, 897, 68], [1, 1135], [1, 1, 1, 1, 1132]], (3, 20, 2): [[10, 10, 252, 78, 381, 10, 3, 1, 2, 131, 4, 40, 11, 1, 5, 47, 7, 2, 15, 126], [23, 56, 5, 2, 15, 5, 90, 2, 6, 6, 1, 13, 7, 4, 4, 3, 3, 3, 6, 76, 1, 2, 28, 1, 103, 70, 1, 430, 158, 4, 1, 3, 2, 2], [2, 156, 4, 98, 4, 5, 2, 85, 13, 2, 619, 1, 3, 3, 113, 9, 3, 14], [3, 1, 140, 140, 5, 5, 1, 128, 11, 632, 5, 11, 40, 4, 2, 4, 2, 2], [6, 1128, 2]], (3, 20, 3): [[35, 17, 165, 7, 3, 3, 56, 45, 207, 56, 212, 2, 12, 8, 304, 4], [40, 84, 70, 1, 12, 132, 196, 2, 1, 10, 6, 103, 4, 4, 33, 2, 19, 13, 1, 2, 1, 1, 1, 2, 392, 2, 2], [1, 4, 2, 5, 5, 9, 914, 35, 4, 4, 6, 9, 2, 1, 2, 1, 7, 3, 115, 7], [392, 181, 2, 3, 61, 5, 191, 4, 1, 2, 3, 3, 17, 2, 47, 4, 10, 3, 1, 2, 3, 2, 4, 4, 2, 9, 3, 2, 1, 7, 163, 2], [36, 46, 83, 3, 80, 2, 5, 92, 5, 2, 35, 1, 78, 3, 30, 5, 1, 469, 9, 112, 3, 7, 9, 20]], (7, 5, 1): [[1136], [1015, 121], [328, 808], [190, 946], [262, 874]]}\n ...\n ...\n ...\n ...\n ...\n----------2\n{(3, 5, 1): [[718, 418], [347, 789], [1, 34, 4, 10, 1, 225, 3, 724, 134], [1, 1, 1, 66, 4, 1, 1, 1, 215, 69, 757, 13, 6], [1136]], (3, 5, 2): [[1, 65, 1067, 2, 1], [31, 126, 1, 6, 102, 3, 122, 94, 6, 278, 2, 176, 4, 185], [39, 1, 46, 55, 668, 43, 5, 65, 112, 101, 1], [696, 1, 7, 39, 1, 3, 2, 92, 2, 108, 17, 166, 1, 1], [64, 358, 29, 179, 57, 7, 2, 8, 181, 10, 82, 157, 2]], (3, 5, 3): [[72, 191, 4, 22, 2, 2, 79, 51, 4, 84, 38, 5, 7, 17, 4, 4, 403, 145, 2], [1, 88, 11, 5, 36, 448, 530, 17], [142, 3, 53, 4, 6, 200, 55, 16, 5, 46, 2, 46, 1, 2, 137, 1, 11, 3, 403], [181, 108, 117, 2, 254, 7, 2, 75, 6, 85, 231, 68], [87, 3, 1, 754, 4, 75, 2, 33, 6, 153, 6, 2, 10]], (3, 20, 1): [[1, 1, 1, 1, 1, 4, 1, 1, 1125], [3, 153, 2, 1, 5, 5, 967], [2, 2, 2, 24, 2, 139, 897, 68], [1, 1135], [1, 1, 1, 1, 1132]], (3, 20, 2): [[10, 10, 252, 78, 381, 10, 3, 1, 2, 131, 4, 40, 11, 1, 5, 47, 7, 2, 15, 126], [23, 56, 5, 2, 15, 5, 90, 2, 6, 6, 1, 13, 7, 4, 4, 3, 3, 3, 6, 76, 1, 2, 28, 1, 103, 70, 1, 430, 158, 4, 1, 3, 2, 2], [2, 156, 4, 98, 4, 5, 2, 85, 13, 2, 619, 1, 3, 3, 113, 9, 3, 14], [3, 1, 140, 140, 5, 5, 1, 128, 11, 632, 5, 11, 40, 4, 2, 4, 2, 2], [6, 1128, 2]], (3, 20, 3): [[35, 17, 165, 7, 3, 3, 56, 45, 207, 56, 212, 2, 12, 8, 304, 4], [40, 84, 70, 1, 12, 132, 196, 2, 1, 10, 6, 103, 4, 4, 33, 2, 19, 13, 1, 2, 1, 1, 1, 2, 392, 2, 2], [1, 4, 2, 5, 5, 9, 914, 35, 4, 4, 6, 9, 2, 1, 2, 1, 7, 3, 115, 7], [392, 181, 2, 3, 61, 5, 191, 4, 1, 2, 3, 3, 17, 2, 47, 4, 10, 3, 1, 2, 3, 2, 4, 4, 2, 9, 3, 2, 1, 7, 163, 2], [36, 46, 83, 3, 80, 2, 5, 92, 5, 2, 35, 1, 78, 3, 30, 5, 1, 469, 9, 112, 3, 7, 9, 20]], (7, 5, 1): [[1136], [1015, 121], [328, 808], [190, 946], [262, 874]], (7, 5, 2): [[90, 2, 2, 277, 31, 9, 69, 34, 15, 2, 120, 39, 20, 213, 6, 3, 7, 15, 5, 5, 102, 2, 2, 66], [74, 373, 2, 67, 43, 92, 113, 367, 5], [17, 1, 18, 4, 2, 3, 2, 696, 16, 22, 73, 94, 23, 88, 8, 5, 4, 5, 2, 19, 2, 2, 30], [83, 4, 2, 99, 6, 73, 66, 6, 3, 5, 27, 2, 16, 519, 6, 2, 2, 31, 30, 8, 7, 3, 2, 7, 2, 7, 19, 48, 51], [8, 41, 3, 474, 31, 2, 52, 2, 2, 26, 10, 76, 3, 14, 8, 4, 2, 10, 182, 16, 2, 2, 3, 134, 5, 11, 13]]}\n"
],
[
"temp = cluster_sizes",
"_____no_output_____"
],
[
"other = sample_clustering_params()",
" ...\n ...\n ...\n ...\n ...\n----------1\n{(3, 5, 1): [[1136], [9, 1041, 81, 5], [1, 292, 842, 1], [1, 2, 382, 751], [6, 4, 2, 13, 1111]]}\n ...\n ...\n ...\n ...\n ...\n----------2\n{(3, 5, 1): [[1136], [9, 1041, 81, 5], [1, 292, 842, 1], [1, 2, 382, 751], [6, 4, 2, 13, 1111]], (3, 5, 2): [[2, 3, 9, 11, 54, 9, 520, 4, 43, 28, 1, 54, 36, 290, 1, 70, 1], [436, 302, 7, 1, 1, 142, 53, 95, 30, 7, 5, 57], [12, 12, 2, 4, 3, 23, 65, 2, 99, 84, 61, 2, 75, 207, 2, 346, 2, 2, 124, 2, 7], [166, 3, 112, 260, 25, 1, 2, 377, 50, 5, 53, 30, 8, 2, 42], [72, 619, 1, 120, 17, 4, 2, 5, 296]]}\n ...\n ...\n ...\n ...\n ...\n----------3\n{(3, 5, 1): [[1136], [9, 1041, 81, 5], [1, 292, 842, 1], [1, 2, 382, 751], [6, 4, 2, 13, 1111]], (3, 5, 2): [[2, 3, 9, 11, 54, 9, 520, 4, 43, 28, 1, 54, 36, 290, 1, 70, 1], [436, 302, 7, 1, 1, 142, 53, 95, 30, 7, 5, 57], [12, 12, 2, 4, 3, 23, 65, 2, 99, 84, 61, 2, 75, 207, 2, 346, 2, 2, 124, 2, 7], [166, 3, 112, 260, 25, 1, 2, 377, 50, 5, 53, 30, 8, 2, 42], [72, 619, 1, 120, 17, 4, 2, 5, 296]], (3, 5, 3): [[72, 5, 3, 1, 513, 1, 534, 1, 6], [246, 1, 362, 88, 432, 3, 4], [40, 2, 318, 1, 6, 73, 133, 36, 1, 15, 1, 1, 1, 148, 21, 3, 1, 3, 7, 230, 2, 1, 87, 4, 1], [671, 11, 278, 3, 9, 67, 93, 4], [4, 235, 9, 4, 111, 2, 10, 6, 3, 83, 4, 1, 1, 31, 321, 1, 4, 15, 1, 1, 166, 53, 6, 1, 3, 23, 37]]}\n ...\n ...\n ...\n ...\n ...\n----------6\n{(3, 5, 1): [[1136], [9, 1041, 81, 5], [1, 292, 842, 1], [1, 2, 382, 751], [6, 4, 2, 13, 1111]], (3, 5, 2): [[2, 3, 9, 11, 54, 9, 520, 4, 43, 28, 1, 54, 36, 290, 1, 70, 1], [436, 302, 7, 1, 1, 142, 53, 95, 30, 7, 5, 57], [12, 12, 2, 4, 3, 23, 65, 2, 99, 84, 61, 2, 75, 207, 2, 346, 2, 2, 124, 2, 7], [166, 3, 112, 260, 25, 1, 2, 377, 50, 5, 53, 30, 8, 2, 42], [72, 619, 1, 120, 17, 4, 2, 5, 296]], (3, 5, 3): [[72, 5, 3, 1, 513, 1, 534, 1, 6], [246, 1, 362, 88, 432, 3, 4], [40, 2, 318, 1, 6, 73, 133, 36, 1, 15, 1, 1, 1, 148, 21, 3, 1, 3, 7, 230, 2, 1, 87, 4, 1], [671, 11, 278, 3, 9, 67, 93, 4], [4, 235, 9, 4, 111, 2, 10, 6, 3, 83, 4, 1, 1, 31, 321, 1, 4, 15, 1, 1, 166, 53, 6, 1, 3, 23, 37]], (3, 5, 6): [[17, 11, 115, 7, 74, 62, 40, 1, 1, 101, 240, 1, 118, 348], [13, 130, 245, 12, 24, 92, 4, 269, 137, 64, 146], [6, 15, 41, 314, 20, 1, 11, 207, 65, 12, 10, 6, 183, 88, 144, 13], [60, 9, 5, 188, 191, 1, 14, 7, 89, 219, 1, 8, 210, 134], [6, 1, 542, 3, 130, 81, 18, 11, 344]]}\n----5\n ...\n ...\n ...\n ...\n ...\n----------1\n{(3, 5, 1): [[1136], [9, 1041, 81, 5], [1, 292, 842, 1], [1, 2, 382, 751], [6, 4, 2, 13, 1111]], (3, 5, 2): [[2, 3, 9, 11, 54, 9, 520, 4, 43, 28, 1, 54, 36, 290, 1, 70, 1], [436, 302, 7, 1, 1, 142, 53, 95, 30, 7, 5, 57], [12, 12, 2, 4, 3, 23, 65, 2, 99, 84, 61, 2, 75, 207, 2, 346, 2, 2, 124, 2, 7], [166, 3, 112, 260, 25, 1, 2, 377, 50, 5, 53, 30, 8, 2, 42], [72, 619, 1, 120, 17, 4, 2, 5, 296]], (3, 5, 3): [[72, 5, 3, 1, 513, 1, 534, 1, 6], [246, 1, 362, 88, 432, 3, 4], [40, 2, 318, 1, 6, 73, 133, 36, 1, 15, 1, 1, 1, 148, 21, 3, 1, 3, 7, 230, 2, 1, 87, 4, 1], [671, 11, 278, 3, 9, 67, 93, 4], [4, 235, 9, 4, 111, 2, 10, 6, 3, 83, 4, 1, 1, 31, 321, 1, 4, 15, 1, 1, 166, 53, 6, 1, 3, 23, 37]], (3, 5, 6): [[17, 11, 115, 7, 74, 62, 40, 1, 1, 101, 240, 1, 118, 348], [13, 130, 245, 12, 24, 92, 4, 269, 137, 64, 146], [6, 15, 41, 314, 20, 1, 11, 207, 65, 12, 10, 6, 183, 88, 144, 13], [60, 9, 5, 188, 191, 1, 14, 7, 89, 219, 1, 8, 210, 134], [6, 1, 542, 3, 130, 81, 18, 11, 344]], (3, 20, 1): [[1134, 1, 1], [691, 2, 4, 439], [1134, 1, 1], [2, 708, 11, 7, 2, 406], [11, 1, 75, 687, 3, 20, 3, 37, 1, 151, 1, 7, 1, 112, 7, 19]]}\n ...\n ...\n ...\n ...\n ...\n----------2\n{(3, 5, 1): [[1136], [9, 1041, 81, 5], [1, 292, 842, 1], [1, 2, 382, 751], [6, 4, 2, 13, 1111]], (3, 5, 2): [[2, 3, 9, 11, 54, 9, 520, 4, 43, 28, 1, 54, 36, 290, 1, 70, 1], [436, 302, 7, 1, 1, 142, 53, 95, 30, 7, 5, 57], [12, 12, 2, 4, 3, 23, 65, 2, 99, 84, 61, 2, 75, 207, 2, 346, 2, 2, 124, 2, 7], [166, 3, 112, 260, 25, 1, 2, 377, 50, 5, 53, 30, 8, 2, 42], [72, 619, 1, 120, 17, 4, 2, 5, 296]], (3, 5, 3): [[72, 5, 3, 1, 513, 1, 534, 1, 6], [246, 1, 362, 88, 432, 3, 4], [40, 2, 318, 1, 6, 73, 133, 36, 1, 15, 1, 1, 1, 148, 21, 3, 1, 3, 7, 230, 2, 1, 87, 4, 1], [671, 11, 278, 3, 9, 67, 93, 4], [4, 235, 9, 4, 111, 2, 10, 6, 3, 83, 4, 1, 1, 31, 321, 1, 4, 15, 1, 1, 166, 53, 6, 1, 3, 23, 37]], (3, 5, 6): [[17, 11, 115, 7, 74, 62, 40, 1, 1, 101, 240, 1, 118, 348], [13, 130, 245, 12, 24, 92, 4, 269, 137, 64, 146], [6, 15, 41, 314, 20, 1, 11, 207, 65, 12, 10, 6, 183, 88, 144, 13], [60, 9, 5, 188, 191, 1, 14, 7, 89, 219, 1, 8, 210, 134], [6, 1, 542, 3, 130, 81, 18, 11, 344]], (3, 20, 1): [[1134, 1, 1], [691, 2, 4, 439], [1134, 1, 1], [2, 708, 11, 7, 2, 406], [11, 1, 75, 687, 3, 20, 3, 37, 1, 151, 1, 7, 1, 112, 7, 19]], (3, 20, 2): [[83, 6, 3, 2, 3, 2, 118, 1, 268, 1, 133, 2, 94, 114, 50, 3, 8, 2, 2, 239, 2], [94, 5, 1, 12, 2, 4, 1, 693, 16, 1, 72, 46, 1, 1, 9, 121, 57], [3, 2, 8, 13, 4, 4, 2, 85, 5, 250, 7, 4, 106, 37, 63, 92, 20, 3, 2, 392, 18, 16], [93, 4, 106, 2, 2, 307, 3, 3, 49, 2, 22, 1, 4, 7, 50, 2, 1, 2, 24, 3, 49, 58, 116, 151, 2, 3, 68, 2], [75, 61, 7, 88, 1, 1, 6, 2, 15, 6, 8, 75, 2, 38, 7, 17, 2, 2, 121, 8, 3, 3, 171, 6, 83, 275, 53]]}\n ...\n ...\n ...\n ...\n ...\n----------3\n{(3, 5, 1): [[1136], [9, 1041, 81, 5], [1, 292, 842, 1], [1, 2, 382, 751], [6, 4, 2, 13, 1111]], (3, 5, 2): [[2, 3, 9, 11, 54, 9, 520, 4, 43, 28, 1, 54, 36, 290, 1, 70, 1], [436, 302, 7, 1, 1, 142, 53, 95, 30, 7, 5, 57], [12, 12, 2, 4, 3, 23, 65, 2, 99, 84, 61, 2, 75, 207, 2, 346, 2, 2, 124, 2, 7], [166, 3, 112, 260, 25, 1, 2, 377, 50, 5, 53, 30, 8, 2, 42], [72, 619, 1, 120, 17, 4, 2, 5, 296]], (3, 5, 3): [[72, 5, 3, 1, 513, 1, 534, 1, 6], [246, 1, 362, 88, 432, 3, 4], [40, 2, 318, 1, 6, 73, 133, 36, 1, 15, 1, 1, 1, 148, 21, 3, 1, 3, 7, 230, 2, 1, 87, 4, 1], [671, 11, 278, 3, 9, 67, 93, 4], [4, 235, 9, 4, 111, 2, 10, 6, 3, 83, 4, 1, 1, 31, 321, 1, 4, 15, 1, 1, 166, 53, 6, 1, 3, 23, 37]], (3, 5, 6): [[17, 11, 115, 7, 74, 62, 40, 1, 1, 101, 240, 1, 118, 348], [13, 130, 245, 12, 24, 92, 4, 269, 137, 64, 146], [6, 15, 41, 314, 20, 1, 11, 207, 65, 12, 10, 6, 183, 88, 144, 13], [60, 9, 5, 188, 191, 1, 14, 7, 89, 219, 1, 8, 210, 134], [6, 1, 542, 3, 130, 81, 18, 11, 344]], (3, 20, 1): [[1134, 1, 1], [691, 2, 4, 439], [1134, 1, 1], [2, 708, 11, 7, 2, 406], [11, 1, 75, 687, 3, 20, 3, 37, 1, 151, 1, 7, 1, 112, 7, 19]], (3, 20, 2): [[83, 6, 3, 2, 3, 2, 118, 1, 268, 1, 133, 2, 94, 114, 50, 3, 8, 2, 2, 239, 2], [94, 5, 1, 12, 2, 4, 1, 693, 16, 1, 72, 46, 1, 1, 9, 121, 57], [3, 2, 8, 13, 4, 4, 2, 85, 5, 250, 7, 4, 106, 37, 63, 92, 20, 3, 2, 392, 18, 16], [93, 4, 106, 2, 2, 307, 3, 3, 49, 2, 22, 1, 4, 7, 50, 2, 1, 2, 24, 3, 49, 58, 116, 151, 2, 3, 68, 2], [75, 61, 7, 88, 1, 1, 6, 2, 15, 6, 8, 75, 2, 38, 7, 17, 2, 2, 121, 8, 3, 3, 171, 6, 83, 275, 53]], (3, 20, 3): [[3, 4, 1, 1, 4, 96, 1, 1, 149, 3, 598, 3, 46, 2, 17, 102, 1, 3, 3, 8, 6, 73, 6, 1, 4], [652, 7, 15, 3, 7, 2, 3, 81, 60, 15, 6, 52, 3, 1, 50, 7, 1, 1, 1, 4, 13, 23, 10, 8, 111], [3, 105, 3, 6, 4, 4, 1, 27, 84, 5, 3, 44, 53, 1, 442, 169, 4, 4, 18, 5, 3, 6, 2, 95, 35, 4, 6], [5, 8, 18, 104, 7, 3, 4, 57, 5, 41, 1, 22, 3, 1, 21, 53, 11, 19, 9, 72, 8, 33, 306, 174, 6, 145], [127, 3, 2, 126, 4, 81, 4, 4, 26, 5, 16, 270, 28, 50, 1, 228, 161]]}\n ...\n ...\n ...\n ...\n ...\n----------6\n{(3, 5, 1): [[1136], [9, 1041, 81, 5], [1, 292, 842, 1], [1, 2, 382, 751], [6, 4, 2, 13, 1111]], (3, 5, 2): [[2, 3, 9, 11, 54, 9, 520, 4, 43, 28, 1, 54, 36, 290, 1, 70, 1], [436, 302, 7, 1, 1, 142, 53, 95, 30, 7, 5, 57], [12, 12, 2, 4, 3, 23, 65, 2, 99, 84, 61, 2, 75, 207, 2, 346, 2, 2, 124, 2, 7], [166, 3, 112, 260, 25, 1, 2, 377, 50, 5, 53, 30, 8, 2, 42], [72, 619, 1, 120, 17, 4, 2, 5, 296]], (3, 5, 3): [[72, 5, 3, 1, 513, 1, 534, 1, 6], [246, 1, 362, 88, 432, 3, 4], [40, 2, 318, 1, 6, 73, 133, 36, 1, 15, 1, 1, 1, 148, 21, 3, 1, 3, 7, 230, 2, 1, 87, 4, 1], [671, 11, 278, 3, 9, 67, 93, 4], [4, 235, 9, 4, 111, 2, 10, 6, 3, 83, 4, 1, 1, 31, 321, 1, 4, 15, 1, 1, 166, 53, 6, 1, 3, 23, 37]], (3, 5, 6): [[17, 11, 115, 7, 74, 62, 40, 1, 1, 101, 240, 1, 118, 348], [13, 130, 245, 12, 24, 92, 4, 269, 137, 64, 146], [6, 15, 41, 314, 20, 1, 11, 207, 65, 12, 10, 6, 183, 88, 144, 13], [60, 9, 5, 188, 191, 1, 14, 7, 89, 219, 1, 8, 210, 134], [6, 1, 542, 3, 130, 81, 18, 11, 344]], (3, 20, 1): [[1134, 1, 1], [691, 2, 4, 439], [1134, 1, 1], [2, 708, 11, 7, 2, 406], [11, 1, 75, 687, 3, 20, 3, 37, 1, 151, 1, 7, 1, 112, 7, 19]], (3, 20, 2): [[83, 6, 3, 2, 3, 2, 118, 1, 268, 1, 133, 2, 94, 114, 50, 3, 8, 2, 2, 239, 2], [94, 5, 1, 12, 2, 4, 1, 693, 16, 1, 72, 46, 1, 1, 9, 121, 57], [3, 2, 8, 13, 4, 4, 2, 85, 5, 250, 7, 4, 106, 37, 63, 92, 20, 3, 2, 392, 18, 16], [93, 4, 106, 2, 2, 307, 3, 3, 49, 2, 22, 1, 4, 7, 50, 2, 1, 2, 24, 3, 49, 58, 116, 151, 2, 3, 68, 2], [75, 61, 7, 88, 1, 1, 6, 2, 15, 6, 8, 75, 2, 38, 7, 17, 2, 2, 121, 8, 3, 3, 171, 6, 83, 275, 53]], (3, 20, 3): [[3, 4, 1, 1, 4, 96, 1, 1, 149, 3, 598, 3, 46, 2, 17, 102, 1, 3, 3, 8, 6, 73, 6, 1, 4], [652, 7, 15, 3, 7, 2, 3, 81, 60, 15, 6, 52, 3, 1, 50, 7, 1, 1, 1, 4, 13, 23, 10, 8, 111], [3, 105, 3, 6, 4, 4, 1, 27, 84, 5, 3, 44, 53, 1, 442, 169, 4, 4, 18, 5, 3, 6, 2, 95, 35, 4, 6], [5, 8, 18, 104, 7, 3, 4, 57, 5, 41, 1, 22, 3, 1, 21, 53, 11, 19, 9, 72, 8, 33, 306, 174, 6, 145], [127, 3, 2, 126, 4, 81, 4, 4, 26, 5, 16, 270, 28, 50, 1, 228, 161]], (3, 20, 6): [[326, 8, 6, 23, 39, 1, 10, 99, 1, 1, 401, 11, 7, 1, 17, 34, 151], [2, 8, 12, 2, 177, 33, 5, 50, 1, 132, 4, 220, 7, 4, 5, 125, 1, 348], [5, 315, 4, 9, 6, 2, 173, 7, 186, 68, 14, 7, 37, 25, 7, 194, 77], [8, 30, 4, 119, 34, 6, 27, 40, 14, 112, 9, 164, 316, 12, 241], [7, 1, 85, 8, 12, 3, 328, 176, 4, 11, 128, 102, 10, 10, 16, 84, 14, 124, 5, 7, 1]]}\n----20\n"
],
[
"# data from runnning sample_clustering_params \ncluster_sizes={(3, 5, 1): [[1136], [9, 1041, 81, 5], [1, 292, 842, 1], [1, 2, 382, 751], [6, 4, 2, 13, 1111]], (3, 5, 2): [[2, 3, 9, 11, 54, 9, 520, 4, 43, 28, 1, 54, 36, 290, 1, 70, 1], [436, 302, 7, 1, 1, 142, 53, 95, 30, 7, 5, 57], [12, 12, 2, 4, 3, 23, 65, 2, 99, 84, 61, 2, 75, 207, 2, 346, 2, 2, 124, 2, 7], [166, 3, 112, 260, 25, 1, 2, 377, 50, 5, 53, 30, 8, 2, 42], [72, 619, 1, 120, 17, 4, 2, 5, 296]], (3, 5, 3): [[72, 5, 3, 1, 513, 1, 534, 1, 6], [246, 1, 362, 88, 432, 3, 4], [40, 2, 318, 1, 6, 73, 133, 36, 1, 15, 1, 1, 1, 148, 21, 3, 1, 3, 7, 230, 2, 1, 87, 4, 1], [671, 11, 278, 3, 9, 67, 93, 4], [4, 235, 9, 4, 111, 2, 10, 6, 3, 83, 4, 1, 1, 31, 321, 1, 4, 15, 1, 1, 166, 53, 6, 1, 3, 23, 37]], (3, 5, 6): [[17, 11, 115, 7, 74, 62, 40, 1, 1, 101, 240, 1, 118, 348], [13, 130, 245, 12, 24, 92, 4, 269, 137, 64, 146], [6, 15, 41, 314, 20, 1, 11, 207, 65, 12, 10, 6, 183, 88, 144, 13], [60, 9, 5, 188, 191, 1, 14, 7, 89, 219, 1, 8, 210, 134], [6, 1, 542, 3, 130, 81, 18, 11, 344]], (3, 20, 1): [[1134, 1, 1], [691, 2, 4, 439], [1134, 1, 1], [2, 708, 11, 7, 2, 406], [11, 1, 75, 687, 3, 20, 3, 37, 1, 151, 1, 7, 1, 112, 7, 19]], (3, 20, 2): [[83, 6, 3, 2, 3, 2, 118, 1, 268, 1, 133, 2, 94, 114, 50, 3, 8, 2, 2, 239, 2], [94, 5, 1, 12, 2, 4, 1, 693, 16, 1, 72, 46, 1, 1, 9, 121, 57], [3, 2, 8, 13, 4, 4, 2, 85, 5, 250, 7, 4, 106, 37, 63, 92, 20, 3, 2, 392, 18, 16], [93, 4, 106, 2, 2, 307, 3, 3, 49, 2, 22, 1, 4, 7, 50, 2, 1, 2, 24, 3, 49, 58, 116, 151, 2, 3, 68, 2], [75, 61, 7, 88, 1, 1, 6, 2, 15, 6, 8, 75, 2, 38, 7, 17, 2, 2, 121, 8, 3, 3, 171, 6, 83, 275, 53]], (3, 20, 3): [[3, 4, 1, 1, 4, 96, 1, 1, 149, 3, 598, 3, 46, 2, 17, 102, 1, 3, 3, 8, 6, 73, 6, 1, 4], [652, 7, 15, 3, 7, 2, 3, 81, 60, 15, 6, 52, 3, 1, 50, 7, 1, 1, 1, 4, 13, 23, 10, 8, 111], [3, 105, 3, 6, 4, 4, 1, 27, 84, 5, 3, 44, 53, 1, 442, 169, 4, 4, 18, 5, 3, 6, 2, 95, 35, 4, 6], [5, 8, 18, 104, 7, 3, 4, 57, 5, 41, 1, 22, 3, 1, 21, 53, 11, 19, 9, 72, 8, 33, 306, 174, 6, 145], [127, 3, 2, 126, 4, 81, 4, 4, 26, 5, 16, 270, 28, 50, 1, 228, 161]], (3, 20, 6): [[326, 8, 6, 23, 39, 1, 10, 99, 1, 1, 401, 11, 7, 1, 17, 34, 151], [2, 8, 12, 2, 177, 33, 5, 50, 1, 132, 4, 220, 7, 4, 5, 125, 1, 348], [5, 315, 4, 9, 6, 2, 173, 7, 186, 68, 14, 7, 37, 25, 7, 194, 77], [8, 30, 4, 119, 34, 6, 27, 40, 14, 112, 9, 164, 316, 12, 241], [7, 1, 85, 8, 12, 3, 328, 176, 4, 11, 128, 102, 10, 10, 16, 84, 14, 124, 5, 7, 1]], (3, 40, 1): [[542, 2, 6, 3, 142, 4, 3, 19, 1, 13, 401], [9, 4, 2, 392, 2, 5, 6, 2, 714], [147, 18, 860, 2, 109], [904, 1, 1, 81, 102, 7, 2, 3, 2, 1, 17, 3, 2, 1, 1, 8], [13, 2, 1, 2, 1114, 2, 2]], (3, 40, 2): [[3, 3, 8, 5, 56, 29, 9, 2, 3, 458, 194, 4, 1, 3, 9, 1, 97, 3, 105, 7, 28, 32, 42, 2, 2, 3, 3, 3, 21], [228, 7, 5, 3, 54, 1, 533, 2, 4, 3, 3, 21, 170, 2, 2, 14, 1, 64, 3, 16], [55, 1, 2, 1, 1, 2, 54, 4, 207, 19, 8, 16, 2, 4, 36, 3, 145, 8, 251, 24, 141, 2, 1, 2, 3, 19, 11, 4, 2, 6, 6, 3, 2, 4, 41, 2, 7, 22, 2, 3, 6, 4], [4, 10, 66, 17, 6, 93, 227, 66, 125, 20, 41, 2, 2, 27, 4, 135, 2, 5, 3, 3, 4, 8, 46, 3, 177, 9, 7, 3, 21], [2, 16, 4, 3, 1, 4, 2, 11, 8, 3, 183, 70, 141, 11, 3, 67, 4, 2, 2, 3, 3, 570, 23]], (3, 40, 3): [[26, 13, 40, 26, 1, 3, 4, 5, 37, 2, 101, 32, 6, 93, 3, 2, 4, 1, 1, 4, 6, 106, 538, 1, 1, 6, 4, 4, 66], [1, 1, 4, 58, 5, 35, 165, 24, 331, 39, 42, 109, 3, 2, 2, 7, 1, 5, 1, 4, 111, 4, 4, 2, 16, 81, 6, 1, 72], [2, 4, 79, 3, 112, 120, 12, 20, 6, 1, 1, 1, 50, 28, 1, 5, 162, 1, 60, 162, 306], [3, 1, 3, 4, 5, 2, 2, 3, 113, 421, 25, 4, 114, 55, 122, 4, 6, 70, 166, 13], [166, 13, 31, 10, 36, 89, 2, 1, 1, 3, 6, 45, 1, 5, 262, 52, 91, 1, 1, 5, 223, 46, 8, 1, 4, 6, 3, 24]], (3, 40, 6): [[187, 228, 12, 57, 7, 1, 331, 16, 209, 88], [1, 136, 10, 95, 215, 11, 51, 38, 6, 107, 230, 55, 5, 1, 1, 156, 1, 17], [102, 20, 4, 5, 5, 365, 10, 13, 14, 15, 148, 168, 15, 6, 28, 39, 25, 5, 1, 3, 7, 1, 104, 33], [841, 4, 22, 85, 35, 8, 7, 6, 51, 1, 74, 1, 1], [7, 6, 112, 68, 1, 8, 1, 6, 12, 13, 7, 76, 1, 11, 2, 154, 1, 5, 338, 205, 8, 1, 1, 2, 50, 1, 1, 38]], (7, 5, 1): [[431, 705], [1136], [1010, 126], [5, 1131], [342, 794]], (7, 5, 2): [[33, 3, 16, 4, 2, 25, 1, 8, 3, 4, 211, 3, 2, 9, 20, 394, 5, 8, 165, 4, 12, 104, 14, 69, 2, 3, 9, 3], [9, 2, 2, 2, 64, 33, 6, 4, 2, 26, 43, 2, 2, 2, 3, 40, 39, 6, 9, 201, 3, 9, 2, 8, 26, 2, 2, 6, 49, 2, 9, 8, 2, 64, 18, 50, 269, 2, 2, 13, 2, 89, 2], [45, 28, 6, 2, 5, 4, 4, 4, 116, 237, 4, 210, 20, 2, 53, 135, 3, 2, 47, 4, 127, 1, 6, 1, 2, 12, 41, 2, 10, 1, 2], [3, 3, 85, 2, 221, 2, 2, 1, 155, 169, 60, 4, 5, 1, 67, 3, 2, 5, 4, 48, 11, 2, 26, 11, 2, 1, 62, 32, 6, 76, 65], [3, 50, 2, 238, 11, 2, 30, 2, 68, 57, 8, 31, 13, 7, 30, 15, 137, 70, 191, 4, 9, 7, 28, 31, 3, 23, 4, 8, 26, 5, 6, 13, 4]], (7, 5, 3): [[81, 18, 10, 49, 24, 113, 109, 3, 4, 27, 3, 3, 145, 6, 43, 4, 311, 116, 8, 2, 2, 3, 52], [11, 10, 8, 3, 6, 157, 166, 4, 63, 7, 7, 1, 99, 32, 20, 12, 12, 4, 16, 5, 7, 73, 148, 265], [6, 17, 24, 26, 14, 147, 23, 52, 151, 75, 91, 4, 5, 3, 24, 5, 4, 7, 8, 12, 4, 4, 140, 90, 3, 37, 3, 101, 9, 41, 6], [3, 13, 1, 3, 23, 2, 368, 41, 5, 116, 13, 4, 3, 67, 3, 8, 26, 3, 310, 111, 7, 4, 2], [2, 51, 3, 4, 106, 6, 3, 33, 14, 1, 18, 85, 16, 28, 11, 8, 12, 101, 57, 5, 15, 44, 129, 3, 63, 2, 5, 48, 3, 8, 128, 42, 82]], (7, 5, 6): [[61, 1, 11, 11, 6, 9, 346, 42, 14, 6, 157, 4, 154, 12, 9, 20, 85, 117, 9, 62], [10, 12, 7, 35, 227, 8, 7, 6, 5, 96, 6, 5, 14, 11, 222, 44, 4, 78, 71, 268], [45, 8, 46, 2, 5, 98, 65, 24, 4, 3, 50, 109, 41, 5, 246, 20, 6, 5, 124, 6, 7, 180, 10, 21, 6], [5, 7, 46, 125, 3, 104, 9, 164, 19, 68, 47, 60, 1, 179, 36, 30, 66, 56, 111], [28, 12, 15, 195, 181, 86, 43, 221, 350, 5]], (7, 20, 1): [[1123, 13], [242, 894], [1046, 90], [380, 756], [280, 854, 2]], (7, 20, 2): [[4, 6, 140, 87, 17, 82, 2, 2, 4, 17, 4, 139, 15, 4, 35, 2, 1, 41, 23, 4, 2, 4, 2, 3, 40, 12, 5, 3, 11, 2, 3, 4, 2, 3, 62, 4, 2, 3, 3, 6, 2, 4, 27, 2, 11, 11, 128, 43, 15, 3, 4, 2, 11, 3, 8, 4, 2, 4, 3, 21, 23], [11, 146, 131, 8, 2, 9, 16, 3, 2, 23, 25, 43, 3, 2, 2, 34, 7, 12, 14, 44, 13, 40, 5, 3, 2, 4, 12, 4, 2, 5, 7, 20, 4, 2, 2, 3, 3, 7, 6, 2, 4, 42, 6, 3, 3, 2, 3, 12, 3, 2, 5, 6, 11, 2, 3, 2, 2, 44, 11, 2, 4, 2, 72, 12, 9, 20, 34, 19, 18, 2, 53, 21, 16, 3], [5, 4, 18, 5, 3, 6, 4, 2, 3, 3, 91, 4, 5, 8, 18, 2, 1, 28, 2, 3, 193, 17, 15, 2, 7, 2, 2, 2, 103, 12, 9, 4, 2, 4, 38, 2, 23, 4, 3, 12, 2, 23, 3, 3, 4, 64, 44, 12, 4, 52, 3, 8, 2, 77, 2, 22, 3, 2, 4, 4, 27, 24, 23, 4, 7, 2, 23, 13, 4], [19, 53, 4, 4, 43, 72, 10, 28, 12, 4, 66, 23, 5, 2, 2, 3, 10, 2, 2, 2, 4, 47, 3, 23, 4, 3, 2, 20, 50, 6, 4, 3, 5, 2, 4, 7, 17, 2, 1, 2, 20, 5, 4, 2, 2, 11, 5, 5, 12, 2, 4, 20, 3, 9, 2, 72, 3, 14, 27, 3, 10, 3, 4, 2, 3, 2, 2, 308, 2], [2, 2, 2, 2, 23, 42, 2, 5, 2, 3, 3, 2, 48, 2, 4, 27, 5, 3, 65, 25, 16, 110, 4, 1, 2, 2, 47, 2, 5, 2, 2, 15, 19, 19, 101, 12, 4, 2, 2, 19, 47, 2, 4, 5, 4, 2, 73, 26, 4, 11, 19, 2, 11, 3, 2, 2, 3, 73, 2, 4, 3, 51, 3, 7, 3, 15, 21, 2, 11, 64, 2]], (7, 20, 3): [[167, 82, 27, 21, 6, 20, 3, 17, 4, 10, 22, 27, 4, 5, 39, 88, 24, 92, 3, 2, 3, 3, 29, 9, 16, 5, 72, 3, 27, 8, 19, 4, 3, 3, 6, 4, 64, 5, 4, 6, 3, 54, 10, 6, 51, 8, 38, 10], [6, 2, 1, 3, 70, 2, 104, 3, 3, 43, 5, 50, 4, 33, 10, 7, 2, 15, 9, 3, 36, 3, 5, 58, 11, 7, 4, 5, 4, 138, 13, 3, 14, 11, 47, 3, 1, 3, 29, 16, 8, 40, 127, 148, 2, 20, 5], [20, 2, 2, 5, 1, 4, 23, 138, 3, 9, 67, 32, 8, 7, 5, 3, 8, 6, 63, 7, 1, 2, 73, 58, 6, 8, 2, 36, 53, 4, 81, 23, 26, 1, 5, 5, 7, 26, 3, 11, 4, 3, 3, 5, 1, 47, 91, 58, 10, 7, 27, 4, 32], [3, 4, 3, 72, 73, 18, 12, 12, 22, 11, 60, 86, 3, 8, 9, 32, 25, 9, 8, 7, 16, 4, 6, 49, 2, 11, 66, 24, 13, 348, 2, 15, 5, 94, 4], [5, 1, 48, 2, 3, 6, 12, 3, 4, 5, 5, 89, 41, 10, 5, 24, 4, 4, 32, 2, 18, 30, 6, 2, 4, 50, 2, 59, 116, 5, 9, 6, 6, 2, 3, 10, 26, 18, 10, 8, 3, 48, 8, 3, 6, 14, 10, 4, 16, 3, 7, 46, 3, 30, 14, 10, 85, 11, 19, 3, 16, 58, 4, 20]], (7, 20, 6): [[18, 20, 7, 63, 9, 59, 251, 146, 4, 14, 7, 3, 24, 85, 6, 65, 6, 3, 5, 8, 13, 8, 19, 5, 6, 5, 41, 5, 27, 7, 145, 26, 9, 5, 6, 6], [6, 19, 123, 22, 42, 6, 45, 15, 3, 31, 18, 175, 14, 20, 11, 21, 216, 1, 7, 6, 109, 12, 6, 12, 5, 6, 5, 9, 3, 90, 4, 16, 58], [7, 12, 7, 15, 8, 5, 5, 7, 6, 7, 4, 20, 7, 8, 6, 72, 37, 8, 91, 30, 5, 58, 285, 3, 6, 6, 5, 21, 87, 3, 12, 113, 74, 17, 6, 6, 7, 5, 55], [16, 61, 8, 169, 8, 5, 4, 4, 13, 56, 4, 8, 38, 16, 3, 62, 21, 2, 80, 29, 93, 44, 3, 6, 3, 29, 9, 5, 8, 7, 38, 19, 6, 16, 19, 40, 110, 9, 19, 20, 19, 7], [91, 8, 7, 1, 24, 9, 86, 7, 12, 6, 174, 138, 34, 4, 4, 4, 20, 15, 57, 4, 26, 33, 10, 6, 7, 7, 4, 47, 57, 61, 173]], (7, 40, 1): [[7, 3, 952, 160, 5, 2, 7], [1136], [1136], [2, 3, 5, 39, 834, 2, 52, 2, 2, 5, 188, 2], [976, 158, 2]], (7, 40, 2): [[4, 12, 23, 45, 6, 62, 3, 3, 10, 2, 33, 11, 9, 15, 3, 2, 14, 2, 26, 8, 7, 37, 15, 2, 3, 2, 17, 3, 4, 4, 9, 2, 24, 3, 3, 45, 4, 7, 116, 1, 2, 2, 33, 7, 2, 29, 4, 4, 3, 33, 3, 5, 22, 3, 8, 41, 2, 4, 5, 13, 79, 3, 3, 2, 3, 22, 30, 3, 8, 11, 2, 4, 10, 40, 4, 3, 2, 2, 9, 4, 2, 27, 7, 3, 7, 3, 3, 14], [3, 2, 10, 6, 2, 15, 18, 5, 4, 2, 9, 29, 64, 6, 70, 22, 13, 45, 2, 3, 3, 3, 21, 80, 2, 116, 4, 29, 5, 6, 8, 9, 3, 43, 10, 1, 2, 2, 1, 48, 3, 2, 2, 2, 27, 2, 9, 19, 3, 2, 3, 5, 15, 2, 3, 2, 36, 16, 6, 13, 4, 3, 2, 19, 32, 3, 11, 5, 5, 10, 2, 15, 6, 45, 2, 2, 2, 4, 3, 6, 2, 4, 15, 7, 4, 4, 4, 3, 2, 6, 2, 2, 2], [6, 11, 44, 47, 5, 4, 2, 3, 2, 2, 54, 16, 3, 3, 4, 27, 2, 4, 157, 4, 28, 2, 12, 7, 2, 2, 66, 4, 2, 80, 16, 92, 3, 12, 8, 10, 11, 6, 5, 4, 2, 2, 17, 14, 14, 2, 2, 2, 3, 8, 7, 2, 10, 2, 6, 15, 5, 7, 5, 19, 70, 30, 7, 12, 2, 4, 41, 28, 9, 14, 3], [3, 3, 166, 8, 2, 3, 2, 5, 2, 7, 3, 15, 5, 2, 3, 6, 37, 4, 4, 1, 64, 2, 2, 6, 2, 7, 12, 5, 3, 19, 4, 2, 5, 29, 2, 2, 101, 2, 16, 4, 36, 19, 75, 9, 5, 2, 2, 4, 3, 7, 6, 2, 7, 20, 3, 2, 2, 2, 2, 21, 10, 3, 3, 15, 10, 31, 2, 3, 5, 5, 2, 2, 68, 3, 14, 22, 11, 4, 26, 2, 2, 5, 42, 2, 2, 2, 31, 9, 1], [15, 5, 2, 2, 5, 19, 10, 26, 2, 5, 2, 2, 23, 6, 18, 9, 23, 46, 2, 5, 21, 3, 2, 2, 8, 3, 7, 4, 3, 2, 4, 2, 7, 2, 3, 2, 22, 5, 49, 2, 55, 14, 6, 53, 24, 3, 3, 69, 3, 3, 12, 2, 10, 24, 15, 2, 4, 11, 30, 9, 10, 2, 21, 2, 14, 5, 5, 6, 21, 6, 14, 3, 28, 1, 17, 2, 3, 2, 8, 10, 25, 2, 3, 18, 62, 2, 3, 5, 67, 2, 6, 2, 11, 7, 3, 2, 2, 2]], (7, 40, 3): [[48, 5, 9, 20, 11, 9, 8, 17, 3, 4, 5, 5, 6, 2, 219, 68, 114, 4, 7, 17, 3, 7, 7, 19, 33, 6, 7, 32, 3, 6, 65, 3, 6, 5, 2, 83, 90, 9, 28, 3, 16, 1, 7, 8, 5, 18, 6, 3, 4, 4, 5, 56, 5], [72, 20, 4, 5, 7, 15, 3, 6, 4, 43, 2, 10, 9, 4, 3, 2, 53, 42, 89, 11, 26, 97, 6, 196, 47, 2, 3, 2, 2, 4, 76, 8, 3, 2, 35, 5, 2, 1, 5, 112, 3, 29, 4, 9, 2, 4, 47], [3, 30, 5, 43, 17, 3, 7, 6, 5, 8, 4, 20, 8, 240, 26, 2, 4, 21, 44, 8, 53, 2, 2, 1, 56, 15, 18, 7, 4, 5, 25, 21, 4, 5, 3, 14, 107, 3, 10, 7, 15, 5, 4, 30, 6, 58, 5, 9, 3, 3, 132], [13, 5, 7, 4, 45, 16, 31, 3, 11, 3, 3, 31, 3, 2, 2, 5, 8, 9, 3, 2, 6, 37, 6, 13, 14, 8, 28, 7, 18, 5, 2, 13, 42, 3, 8, 68, 26, 15, 6, 13, 9, 2, 8, 2, 3, 8, 6, 20, 3, 4, 4, 11, 28, 4, 42, 3, 101, 56, 28, 4, 3, 5, 2, 9, 26, 15, 5, 7, 5, 15, 52, 19, 5, 28, 3, 6, 19, 5, 7, 7, 3], [2, 2, 6, 3, 3, 17, 10, 4, 4, 2, 5, 4, 11, 2, 24, 157, 23, 12, 9, 86, 4, 21, 3, 4, 14, 21, 4, 4, 4, 4, 22, 14, 11, 31, 4, 4, 73, 3, 10, 6, 3, 5, 4, 33, 33, 17, 17, 3, 6, 157, 9, 7, 51, 3, 15, 6, 10, 10, 3, 19, 3, 4, 45, 24, 2]], (7, 40, 6): [[40, 5, 2, 183, 38, 47, 6, 136, 22, 8, 6, 7, 4, 13, 6, 7, 19, 5, 28, 5, 5, 4, 3, 219, 26, 5, 8, 6, 9, 46, 3, 89, 2, 40, 1, 52, 6, 10, 12, 3], [11, 6, 238, 7, 8, 72, 34, 12, 9, 100, 6, 4, 1, 15, 16, 4, 23, 18, 5, 6, 3, 9, 5, 24, 7, 13, 66, 21, 16, 5, 8, 12, 6, 23, 8, 4, 6, 108, 93, 42, 62], [2, 5, 5, 6, 30, 4, 13, 5, 8, 11, 6, 10, 4, 11, 3, 41, 7, 28, 7, 7, 191, 11, 6, 6, 13, 180, 46, 62, 39, 7, 255, 16, 38, 53], [16, 10, 22, 4, 10, 62, 3, 36, 6, 33, 14, 334, 4, 12, 105, 13, 9, 20, 2, 9, 11, 5, 10, 6, 50, 49, 70, 8, 52, 101, 28, 3, 11, 5, 3], [17, 58, 4, 30, 7, 6, 58, 34, 71, 7, 6, 8, 6, 177, 18, 14, 5, 12, 4, 6, 6, 3, 104, 18, 14, 8, 5, 6, 6, 34, 9, 5, 43, 17, 6, 11, 34, 152, 60, 19, 13, 15]], (10, 5, 1): [[2, 219, 915], [219, 917], [1135, 1], [1078, 58], [1072, 64]], (10, 5, 2): [[10, 50, 3, 13, 2, 8, 76, 50, 2, 116, 2, 74, 7, 1, 177, 85, 84, 11, 14, 13, 110, 4, 18, 19, 45, 2, 19, 59, 3, 13, 2, 16, 4, 3, 21], [217, 11, 1, 37, 192, 90, 11, 51, 2, 2, 2, 2, 5, 30, 103, 10, 11, 3, 2, 2, 5, 4, 15, 9, 44, 3, 12, 4, 4, 2, 57, 19, 37, 14, 76, 2, 7, 27, 6, 5], [143, 2, 17, 9, 3, 41, 117, 152, 5, 2, 18, 2, 123, 34, 20, 7, 3, 8, 4, 15, 67, 12, 25, 10, 3, 10, 5, 10, 3, 23, 5, 6, 17, 6, 3, 24, 44, 9, 77, 3, 31, 5, 3, 10], [33, 7, 5, 2, 49, 5, 70, 4, 12, 24, 2, 3, 2, 12, 9, 4, 80, 13, 1, 4, 51, 16, 127, 86, 4, 2, 12, 49, 2, 2, 107, 5, 4, 105, 2, 6, 18, 19, 3, 29, 1, 3, 3, 3, 11, 5, 21, 15, 2, 4, 2, 2, 66, 5, 3], [5, 6, 66, 120, 19, 9, 2, 33, 2, 32, 13, 17, 3, 5, 8, 12, 80, 34, 3, 34, 3, 215, 4, 1, 109, 3, 2, 6, 2, 19, 9, 4, 4, 6, 31, 11, 2, 30, 28, 61, 2, 7, 74]], (10, 5, 3): [[61, 186, 13, 93, 49, 18, 17, 98, 10, 115, 22, 6, 122, 7, 4, 4, 42, 48, 11, 29, 3, 21, 19, 5, 4, 3, 71, 18, 3, 3, 5, 26], [10, 147, 2, 49, 82, 52, 2, 5, 34, 3, 85, 10, 129, 32, 2, 2, 67, 7, 13, 69, 8, 6, 5, 9, 57, 5, 2, 13, 5, 47, 36, 48, 3, 15, 75], [41, 10, 17, 38, 108, 2, 130, 7, 12, 51, 3, 97, 19, 8, 4, 8, 3, 15, 3, 5, 25, 4, 4, 26, 7, 6, 2, 5, 60, 3, 126, 71, 28, 14, 6, 4, 164], [3, 101, 91, 38, 132, 4, 189, 3, 28, 6, 11, 183, 13, 36, 4, 5, 53, 3, 29, 7, 6, 52, 5, 9, 15, 9, 3, 2, 61, 5, 4, 26], [22, 4, 4, 3, 27, 2, 19, 20, 36, 4, 48, 7, 46, 14, 106, 102, 12, 10, 46, 4, 3, 6, 20, 3, 7, 98, 3, 98, 24, 15, 72, 2, 34, 11, 52, 18, 17, 62, 46, 7, 2]], (10, 5, 6): [[45, 204, 11, 8, 88, 153, 9, 14, 190, 4, 110, 26, 22, 69, 109, 74], [14, 122, 17, 4, 33, 82, 174, 3, 13, 201, 16, 346, 38, 3, 70], [10, 77, 18, 46, 35, 57, 113, 39, 170, 11, 148, 63, 13, 81, 5, 19, 36, 10, 9, 10, 17, 4, 54, 35, 45, 9, 2], [38, 6, 180, 7, 8, 6, 124, 57, 6, 219, 25, 43, 128, 7, 8, 9, 201, 60, 4], [157, 3, 168, 17, 11, 6, 25, 9, 44, 14, 19, 40, 22, 50, 4, 1, 52, 23, 7, 319, 145]], (10, 20, 1): [[2, 236, 898], [1136], [1136], [1, 1135], [216, 918, 2]], (10, 20, 2): [[2, 2, 3, 2, 38, 20, 2, 3, 5, 3, 18, 4, 8, 2, 2, 12, 2, 3, 77, 2, 19, 59, 7, 10, 10, 8, 10, 30, 6, 8, 3, 25, 14, 36, 46, 3, 2, 12, 8, 32, 3, 35, 3, 13, 11, 10, 7, 27, 15, 3, 22, 3, 2, 3, 4, 4, 6, 3, 7, 13, 16, 2, 2, 4, 38, 2, 11, 5, 2, 8, 51, 6, 9, 5, 2, 5, 6, 29, 8, 5, 8, 12, 2, 3, 51, 11, 75, 6], [13, 13, 2, 8, 4, 13, 7, 22, 78, 17, 2, 35, 2, 2, 8, 4, 2, 7, 10, 8, 70, 4, 9, 29, 7, 10, 2, 2, 6, 2, 3, 4, 2, 2, 2, 2, 4, 35, 10, 3, 16, 110, 8, 2, 8, 5, 4, 2, 2, 15, 2, 9, 7, 3, 4, 123, 63, 5, 82, 3, 7, 13, 10, 11, 27, 19, 48, 2, 8, 4, 4, 1, 29, 2, 2, 4, 7], [4, 50, 2, 4, 51, 16, 11, 10, 7, 4, 3, 12, 3, 18, 8, 2, 9, 24, 104, 60, 2, 3, 4, 3, 7, 24, 61, 3, 32, 3, 3, 7, 2, 14, 2, 4, 4, 4, 7, 2, 3, 2, 2, 3, 10, 3, 2, 2, 19, 17, 46, 9, 5, 21, 5, 5, 3, 6, 2, 19, 6, 7, 6, 2, 3, 2, 2, 4, 2, 2, 32, 6, 3, 1, 72, 25, 5, 6, 2, 28, 6, 2, 8, 1, 50, 2, 9, 17, 6, 10, 8, 24], [23, 27, 76, 18, 17, 14, 8, 30, 8, 10, 9, 53, 4, 23, 2, 20, 11, 3, 11, 85, 26, 2, 2, 9, 5, 3, 2, 2, 3, 4, 6, 37, 4, 5, 58, 10, 10, 3, 9, 3, 5, 2, 3, 4, 22, 2, 5, 10, 48, 2, 2, 15, 9, 5, 29, 3, 6, 8, 3, 4, 10, 72, 3, 2, 5, 4, 6, 10, 10, 44, 31, 75, 5, 8, 4, 3, 2], [32, 8, 3, 2, 2, 6, 8, 43, 34, 17, 15, 10, 2, 2, 3, 2, 11, 9, 4, 2, 4, 13, 51, 40, 3, 30, 4, 4, 28, 13, 2, 10, 8, 2, 6, 8, 6, 3, 32, 2, 3, 5, 29, 114, 3, 29, 7, 92, 61, 17, 18, 8, 11, 31, 2, 5, 5, 6, 2, 2, 5, 3, 5, 22, 6, 3, 2, 47, 15, 4, 7, 12, 58, 2, 4, 2, 5]], (10, 20, 3): [[2, 63, 7, 30, 5, 15, 38, 4, 3, 4, 5, 40, 2, 60, 36, 29, 4, 21, 31, 4, 19, 20, 2, 3, 12, 34, 22, 111, 7, 3, 2, 6, 31, 25, 70, 3, 6, 13, 4, 5, 134, 4, 39, 5, 7, 4, 39, 6, 3, 2, 13, 9, 10, 21, 39], [6, 24, 2, 3, 8, 44, 2, 52, 49, 2, 6, 10, 36, 15, 44, 8, 43, 8, 90, 5, 14, 4, 129, 13, 9, 2, 28, 68, 41, 65, 29, 3, 20, 2, 15, 3, 4, 72, 16, 7, 2, 68, 16, 4, 2, 3, 12, 5, 14, 5, 4], [14, 82, 3, 19, 13, 8, 12, 5, 10, 28, 2, 13, 10, 3, 4, 10, 2, 3, 9, 2, 2, 41, 18, 25, 3, 14, 17, 44, 13, 3, 5, 12, 10, 47, 3, 11, 4, 2, 21, 3, 3, 120, 13, 17, 6, 11, 11, 4, 42, 4, 9, 10, 15, 11, 8, 4, 29, 6, 3, 66, 4, 17, 5, 22, 4, 20, 15, 6, 9, 7, 36, 5, 18, 3, 5, 11, 3, 4], [3, 3, 33, 7, 2, 56, 3, 3, 3, 24, 3, 24, 8, 2, 90, 9, 29, 6, 4, 16, 3, 5, 60, 11, 29, 71, 3, 7, 4, 6, 43, 3, 18, 12, 6, 3, 6, 23, 3, 7, 6, 3, 6, 34, 24, 13, 3, 10, 3, 8, 11, 2, 3, 9, 17, 30, 4, 3, 29, 38, 4, 7, 21, 19, 3, 4, 11, 22, 7, 26, 4, 3, 37, 3, 4, 5, 49], [4, 3, 6, 28, 14, 3, 3, 46, 58, 17, 27, 3, 40, 48, 46, 5, 43, 15, 3, 5, 18, 29, 3, 4, 4, 5, 3, 59, 6, 2, 43, 3, 66, 6, 13, 6, 14, 3, 4, 44, 4, 2, 5, 60, 2, 15, 3, 3, 20, 12, 22, 6, 4, 71, 42, 7, 36, 4, 23, 43]], (10, 20, 6): [[19, 25, 69, 6, 7, 5, 119, 24, 25, 42, 133, 23, 13, 136, 6, 6, 5, 28, 173, 73, 102, 78, 6, 3, 10], [16, 18, 10, 93, 17, 5, 4, 7, 3, 12, 4, 23, 2, 21, 34, 51, 1, 221, 11, 19, 6, 109, 11, 16, 17, 10, 119, 28, 39, 188, 6, 15], [5, 10, 65, 4, 2, 151, 17, 5, 19, 5, 22, 12, 8, 79, 6, 1, 13, 4, 85, 184, 8, 11, 5, 4, 6, 21, 7, 148, 4, 8, 12, 72, 3, 24, 12, 5, 3, 17, 4, 3, 5, 9, 48], [9, 6, 33, 23, 6, 2, 7, 10, 8, 63, 277, 31, 6, 157, 6, 7, 6, 26, 10, 35, 6, 10, 35, 15, 7, 18, 86, 25, 6, 36, 8, 53, 29, 4, 37, 5, 16, 12], [51, 4, 15, 34, 8, 42, 11, 15, 134, 13, 90, 55, 107, 58, 31, 83, 89, 99, 4, 34, 40, 8, 13, 8, 23, 7, 7, 53]], (10, 40, 1): [[1135, 1], [322, 814], [882, 254], [254, 882], [252, 882, 2]], (10, 40, 2): [[6, 6, 3, 26, 4, 54, 6, 2, 2, 5, 14, 3, 2, 5, 35, 9, 47, 7, 2, 15, 6, 7, 2, 3, 9, 6, 4, 3, 3, 3, 3, 2, 5, 10, 31, 2, 22, 19, 27, 3, 18, 70, 19, 17, 6, 6, 3, 37, 6, 2, 5, 16, 3, 2, 31, 35, 44, 2, 11, 27, 20, 12, 2, 4, 80, 8, 14, 5, 25, 2, 2, 8, 2, 16, 10, 5, 7, 5, 11, 7, 5, 44, 4, 17, 2, 4, 25, 7], [14, 38, 19, 8, 38, 8, 2, 2, 3, 4, 4, 12, 13, 23, 44, 5, 2, 6, 18, 7, 3, 53, 6, 5, 6, 4, 12, 3, 2, 2, 7, 4, 5, 13, 12, 20, 10, 3, 14, 26, 30, 5, 38, 11, 81, 3, 23, 24, 9, 9, 2, 28, 5, 2, 2, 2, 11, 2, 9, 2, 8, 7, 24, 48, 5, 26, 39, 3, 17, 2, 5, 5, 2, 28, 4, 4, 8, 12, 2, 14, 3, 2, 8, 35, 4, 2, 4, 3, 1, 11, 5, 3, 12, 2], [2, 14, 3, 4, 9, 3, 16, 2, 2, 35, 5, 14, 14, 8, 2, 5, 2, 7, 2, 10, 2, 10, 19, 13, 5, 17, 2, 8, 15, 3, 48, 7, 8, 3, 4, 3, 34, 13, 96, 16, 3, 3, 5, 12, 2, 6, 19, 17, 26, 10, 11, 61, 7, 14, 69, 20, 5, 3, 10, 27, 2, 4, 11, 3, 8, 5, 5, 22, 5, 3, 4, 30, 16, 2, 4, 74, 12, 2, 43, 3, 2, 25, 2, 3, 5, 2, 6, 2, 2, 2, 2, 2, 8], [22, 53, 8, 3, 4, 23, 2, 10, 3, 65, 3, 6, 3, 3, 170, 2, 2, 14, 64, 11, 4, 20, 2, 3, 56, 4, 11, 2, 6, 24, 5, 29, 4, 19, 3, 4, 12, 8, 7, 6, 6, 4, 11, 2, 2, 8, 2, 4, 19, 11, 12, 50, 4, 18, 15, 25, 8, 46, 2, 28, 2, 16, 6, 11, 1, 2, 4, 3, 4, 5, 2, 43, 16, 44], [28, 2, 43, 14, 8, 77, 29, 16, 3, 4, 5, 3, 4, 9, 3, 2, 2, 4, 2, 12, 8, 14, 3, 7, 6, 12, 4, 4, 3, 23, 3, 3, 21, 32, 30, 2, 6, 2, 6, 2, 26, 21, 14, 43, 4, 6, 2, 35, 17, 5, 40, 32, 5, 5, 2, 4, 2, 8, 5, 4, 8, 8, 2, 8, 59, 15, 23, 7, 15, 36, 13, 3, 7, 2, 3, 12, 4, 46, 2, 12, 7, 3, 2, 5, 2, 3, 62, 7, 15, 4]], (10, 40, 3): [[31, 15, 13, 5, 9, 35, 9, 19, 40, 3, 52, 51, 8, 9, 11, 16, 121, 17, 3, 25, 11, 25, 6, 10, 5, 8, 55, 5, 21, 112, 3, 3, 8, 27, 4, 12, 4, 13, 3, 13, 25, 9, 4, 5, 6, 40, 26, 20, 3, 41, 3, 5, 2, 2, 9, 6, 8, 30, 7, 16, 3, 3, 4, 19], [11, 6, 2, 20, 7, 10, 23, 26, 14, 4, 16, 3, 2, 11, 33, 26, 23, 36, 14, 14, 2, 23, 28, 2, 4, 3, 70, 6, 3, 4, 24, 6, 2, 24, 27, 3, 4, 4, 5, 3, 2, 9, 2, 65, 10, 7, 2, 9, 5, 4, 10, 43, 3, 3, 4, 8, 3, 3, 4, 10, 13, 3, 16, 4, 6, 3, 66, 22, 20, 3, 16, 17, 7, 10, 13, 3, 4, 87, 5, 37, 15, 12], [21, 4, 22, 6, 3, 42, 26, 12, 39, 6, 6, 3, 10, 9, 8, 41, 3, 17, 24, 24, 2, 3, 3, 12, 7, 24, 7, 7, 11, 13, 1, 3, 162, 93, 21, 10, 19, 31, 12, 13, 40, 5, 3, 25, 3, 26, 17, 15, 6, 3, 20, 3, 9, 30, 3, 2, 8, 7, 22, 2, 27, 32, 48], [9, 46, 6, 3, 12, 57, 22, 29, 21, 3, 4, 4, 9, 4, 23, 2, 64, 6, 11, 5, 7, 9, 3, 16, 3, 10, 3, 42, 31, 4, 3, 4, 4, 28, 6, 15, 3, 13, 5, 32, 3, 17, 15, 60, 6, 34, 3, 23, 12, 12, 27, 6, 4, 4, 61, 28, 43, 12, 31, 9, 56, 2, 7, 2, 10, 10, 19, 17, 5, 2, 4, 11], [2, 61, 8, 2, 3, 6, 59, 37, 4, 5, 27, 11, 5, 7, 21, 22, 6, 3, 15, 3, 5, 2, 15, 4, 19, 26, 3, 9, 4, 25, 72, 22, 30, 41, 7, 7, 11, 3, 5, 8, 12, 30, 8, 8, 6, 5, 56, 5, 3, 7, 12, 22, 60, 4, 11, 21, 34, 44, 2, 11, 12, 62, 14, 13, 6, 30, 2, 3, 3, 5]], (10, 40, 6): [[36, 46, 130, 3, 6, 29, 6, 3, 10, 46, 6, 67, 6, 5, 4, 5, 76, 26, 4, 7, 4, 9, 2, 11, 14, 6, 7, 236, 4, 22, 140, 10, 4, 6, 57, 7, 14, 62], [53, 22, 11, 12, 17, 18, 96, 11, 2, 43, 3, 15, 8, 15, 4, 2, 263, 6, 6, 3, 9, 4, 26, 9, 72, 96, 66, 3, 2, 3, 19, 45, 74, 39, 52, 7], [7, 46, 55, 15, 9, 41, 12, 3, 3, 6, 13, 94, 127, 11, 4, 225, 101, 51, 9, 4, 13, 11, 13, 30, 88, 5, 24, 19, 90, 1, 6], [67, 4, 12, 14, 60, 18, 63, 19, 8, 2, 41, 108, 22, 4, 90, 70, 84, 54, 90, 42, 16, 7, 17, 58, 22, 26, 3, 110, 5], [5, 74, 13, 52, 45, 71, 62, 6, 8, 7, 31, 216, 6, 5, 12, 4, 72, 31, 20, 122, 46, 5, 117, 106]], (20, 5, 1): [[1136], [127, 1009], [118, 1018], [1031, 102, 3], [93, 1043]], (20, 5, 2): [[74, 19, 16, 6, 64, 2, 18, 5, 2, 24, 47, 20, 6, 9, 62, 2, 57, 22, 3, 44, 19, 46, 7, 3, 24, 6, 32, 39, 14, 76, 78, 50, 26, 47, 65, 20, 12, 12, 15, 16, 14, 2, 11], [2, 59, 12, 26, 3, 5, 4, 2, 12, 56, 2, 2, 28, 57, 5, 33, 2, 60, 13, 36, 3, 21, 22, 11, 17, 7, 26, 44, 2, 32, 2, 17, 49, 18, 4, 4, 34, 81, 8, 12, 49, 2, 120, 10, 18, 28, 8, 65, 3], [3, 12, 118, 5, 48, 84, 9, 91, 8, 45, 42, 8, 8, 13, 2, 12, 16, 38, 7, 3, 23, 29, 4, 34, 34, 83, 3, 49, 15, 18, 6, 5, 11, 27, 44, 2, 3, 58, 17, 4, 3, 28, 26, 24, 14], [2, 4, 41, 13, 3, 10, 38, 49, 6, 15, 48, 14, 53, 40, 8, 54, 29, 6, 5, 3, 2, 7, 3, 4, 1, 6, 35, 4, 13, 5, 2, 25, 10, 18, 90, 27, 24, 4, 2, 3, 20, 2, 50, 9, 2, 103, 22, 20, 24, 62, 16, 5, 6, 6, 10, 2, 9, 2, 10, 22, 5, 3], [11, 20, 22, 9, 4, 82, 29, 12, 14, 6, 38, 2, 3, 18, 6, 2, 5, 21, 41, 58, 3, 6, 8, 23, 47, 72, 3, 19, 40, 8, 28, 7, 2, 14, 135, 10, 184, 5, 6, 54, 16, 3, 40]], (20, 5, 3): [[17, 42, 6, 10, 30, 19, 32, 2, 20, 3, 25, 22, 60, 19, 16, 32, 5, 52, 98, 32, 28, 17, 9, 2, 13, 22, 39, 3, 53, 15, 92, 22, 19, 37, 4, 11, 97, 46, 65], [3, 6, 77, 21, 2, 53, 22, 5, 7, 63, 37, 7, 38, 33, 8, 13, 48, 73, 10, 35, 18, 69, 126, 23, 8, 40, 86, 116, 7, 82], [25, 96, 5, 21, 33, 44, 2, 17, 63, 4, 11, 22, 16, 26, 4, 64, 79, 4, 120, 55, 55, 13, 21, 33, 47, 3, 76, 16, 6, 5, 16, 10, 8, 32, 56, 28], [50, 20, 3, 102, 78, 38, 15, 2, 46, 3, 23, 7, 65, 69, 2, 86, 44, 36, 69, 32, 15, 9, 74, 12, 40, 22, 25, 36, 23, 6, 9, 22, 51, 2], [16, 35, 49, 8, 15, 12, 119, 36, 31, 30, 9, 2, 4, 7, 5, 42, 30, 2, 5, 2, 11, 52, 355, 150, 45, 11, 39, 14]], (20, 5, 6): [[143, 6, 9, 20, 17, 25, 268, 35, 102, 134, 98, 201, 25, 53], [251, 34, 61, 57, 443, 10, 88, 5, 67, 34, 25, 5, 56], [7, 10, 29, 6, 19, 39, 26, 13, 151, 106, 63, 100, 14, 12, 131, 13, 16, 11, 365, 5], [15, 6, 188, 262, 95, 432, 119, 19], [329, 274, 140, 63, 54, 15, 95, 84, 29, 2, 20, 31]], (20, 20, 1): [[187, 949], [955, 181], [1031, 105], [2, 239, 2, 891, 2], [905, 2, 204, 3, 3, 3, 2, 4, 2, 6, 2]], (20, 20, 2): [[74, 2, 2, 2, 2, 16, 3, 3, 17, 3, 2, 27, 10, 6, 5, 8, 7, 19, 4, 4, 14, 17, 14, 3, 3, 11, 9, 7, 27, 11, 17, 50, 4, 43, 13, 3, 9, 15, 13, 3, 19, 32, 13, 2, 15, 3, 31, 5, 4, 18, 8, 5, 12, 32, 6, 10, 2, 7, 5, 16, 42, 3, 4, 5, 4, 3, 3, 11, 10, 4, 11, 36, 4, 10, 32, 8, 3, 24, 17, 16, 6, 10, 19, 2, 6, 3, 34, 7, 5, 4, 2, 12, 11, 3, 10, 5], [9, 2, 5, 7, 17, 4, 6, 29, 49, 39, 15, 4, 2, 3, 2, 2, 32, 56, 7, 13, 4, 2, 18, 6, 3, 4, 11, 3, 25, 2, 4, 3, 2, 53, 20, 25, 10, 29, 4, 17, 24, 19, 2, 9, 16, 24, 16, 2, 55, 2, 10, 2, 13, 2, 6, 9, 9, 2, 4, 4, 5, 79, 33, 57, 54, 7, 29, 12, 7, 2, 31, 4, 10, 28], [3, 13, 11, 18, 10, 26, 25, 14, 22, 6, 33, 4, 10, 20, 17, 32, 31, 5, 6, 14, 2, 3, 3, 10, 2, 9, 26, 27, 17, 9, 6, 12, 3, 15, 5, 10, 35, 6, 30, 32, 4, 34, 2, 7, 4, 15, 2, 3, 27, 32, 19, 43, 4, 3, 2, 32, 3, 20, 18, 17, 8, 15, 3, 13, 13, 4, 8, 2, 11, 4, 25, 13, 2, 34, 8, 5, 8, 25, 5, 5, 5, 27, 6, 2, 12], [10, 3, 2, 17, 23, 45, 11, 16, 54, 16, 10, 2, 28, 15, 2, 25, 33, 2, 2, 2, 7, 39, 5, 4, 11, 9, 13, 17, 57, 7, 8, 32, 8, 47, 53, 30, 6, 13, 39, 2, 3, 9, 7, 3, 2, 46, 2, 8, 4, 9, 3, 24, 19, 3, 2, 16, 12, 3, 12, 5, 5, 5, 23, 13, 6, 21, 19, 9, 10, 19, 2, 2, 7, 8, 4, 35, 2, 13, 14, 2], [4, 26, 15, 13, 7, 1, 12, 3, 33, 3, 11, 38, 2, 49, 10, 57, 13, 2, 26, 6, 23, 38, 21, 3, 14, 25, 18, 6, 13, 5, 44, 4, 7, 2, 13, 11, 7, 21, 2, 4, 5, 66, 7, 53, 13, 16, 3, 2, 15, 5, 10, 6, 2, 8, 36, 5, 6, 28, 8, 17, 2, 17, 2, 35, 31, 10, 4, 82, 30]], (20, 20, 3): [[34, 64, 29, 2, 101, 11, 11, 3, 4, 10, 7, 11, 28, 66, 27, 115, 7, 16, 4, 5, 13, 3, 43, 8, 3, 21, 56, 4, 6, 53, 12, 16, 5, 23, 4, 9, 14, 10, 4, 16, 13, 39, 31, 13, 2, 86, 6, 7, 53, 2, 3, 3], [32, 3, 61, 4, 3, 17, 18, 24, 2, 27, 4, 32, 21, 31, 8, 17, 3, 15, 4, 14, 22, 4, 7, 11, 9, 109, 18, 106, 3, 13, 5, 27, 109, 22, 8, 21, 17, 36, 57, 20, 25, 10, 29, 59, 13, 9, 7, 20], [11, 86, 3, 8, 25, 20, 19, 46, 4, 6, 62, 22, 8, 1, 4, 31, 16, 9, 2, 23, 2, 8, 19, 46, 44, 42, 6, 3, 60, 3, 37, 11, 15, 4, 8, 69, 24, 28, 34, 50, 7, 5, 10, 11, 11, 49, 2, 4, 2, 16, 4, 2, 3, 36, 11, 6, 29, 6, 3], [48, 28, 75, 3, 12, 20, 37, 8, 5, 24, 21, 2, 56, 18, 110, 11, 54, 18, 10, 22, 68, 10, 46, 97, 48, 19, 37, 6, 41, 106, 8, 8, 60], [63, 12, 2, 15, 4, 3, 16, 30, 130, 45, 8, 9, 26, 5, 3, 44, 5, 2, 84, 2, 5, 4, 36, 4, 7, 18, 9, 4, 4, 37, 8, 31, 70, 11, 6, 73, 3, 6, 104, 3, 10, 47, 76, 6, 46]], (20, 20, 6): [[59, 14, 115, 244, 41, 57, 9, 265, 60, 21, 35, 147, 69], [31, 4, 97, 5, 237, 105, 8, 10, 8, 6, 7, 6, 6, 6, 4, 106, 110, 322, 4, 6, 7, 4, 12, 16, 9], [186, 38, 7, 21, 9, 110, 12, 26, 3, 8, 15, 157, 12, 115, 310, 8, 81, 9, 2, 3, 4], [5, 69, 12, 35, 9, 82, 37, 29, 26, 79, 16, 737], [261, 5, 128, 19, 67, 34, 3, 145, 98, 19, 49, 6, 302]], (20, 40, 1): [[1019, 117], [1136], [915, 217, 2, 2], [89, 1047], [55, 1081]], (20, 40, 2): [[5, 3, 6, 3, 3, 4, 2, 37, 8, 2, 16, 19, 6, 3, 2, 7, 3, 15, 12, 2, 3, 2, 15, 76, 12, 13, 18, 13, 9, 6, 8, 33, 2, 12, 3, 5, 4, 2, 2, 8, 5, 4, 38, 23, 4, 6, 3, 46, 3, 3, 13, 58, 13, 30, 3, 21, 4, 42, 7, 2, 11, 4, 12, 18, 2, 3, 4, 3, 34, 18, 11, 33, 17, 23, 2, 8, 2, 2, 3, 2, 23, 12, 9, 19, 21, 15, 7, 2, 3, 2, 33, 3, 5, 2, 17, 8, 6, 5], [2, 5, 44, 38, 24, 3, 10, 34, 2, 6, 3, 11, 5, 25, 4, 9, 9, 2, 38, 15, 4, 24, 25, 2, 6, 4, 15, 2, 4, 8, 8, 6, 6, 4, 4, 13, 4, 6, 11, 4, 34, 3, 2, 10, 9, 6, 10, 43, 2, 21, 6, 11, 24, 36, 4, 5, 2, 3, 8, 9, 2, 31, 26, 18, 2, 4, 15, 21, 2, 14, 23, 4, 2, 18, 51, 6, 2, 11, 14, 6, 9, 6, 3, 5, 24, 2, 21, 2, 5, 31, 2, 3, 8, 22, 10, 5, 7, 2], [16, 2, 3, 10, 3, 20, 10, 2, 36, 70, 2, 3, 5, 11, 10, 6, 5, 4, 23, 3, 6, 13, 3, 11, 5, 4, 29, 3, 2, 12, 6, 6, 4, 13, 11, 13, 2, 4, 3, 3, 16, 5, 26, 9, 37, 5, 3, 25, 12, 26, 7, 2, 4, 37, 29, 18, 6, 16, 4, 7, 2, 6, 6, 5, 6, 6, 3, 11, 2, 23, 8, 12, 2, 3, 23, 8, 27, 7, 8, 4, 5, 3, 22, 16, 5, 6, 3, 10, 7, 8, 11, 6, 32, 3, 5, 2, 20, 8, 8, 3, 12, 4, 12, 12, 14, 22, 14], [89, 4, 4, 14, 2, 6, 4, 3, 6, 7, 8, 2, 35, 3, 6, 8, 27, 12, 2, 15, 8, 26, 44, 14, 6, 27, 3, 4, 10, 3, 2, 2, 19, 18, 6, 42, 30, 6, 7, 3, 2, 5, 16, 12, 4, 2, 37, 6, 12, 63, 8, 37, 29, 32, 24, 3, 3, 5, 15, 14, 11, 5, 9, 2, 35, 20, 45, 10, 44, 9, 5, 12, 18, 4, 8, 10, 2, 8, 13], [2, 24, 5, 7, 40, 34, 9, 2, 24, 2, 7, 3, 3, 20, 8, 2, 2, 6, 11, 3, 5, 70, 3, 2, 17, 37, 7, 7, 13, 5, 12, 6, 22, 4, 13, 2, 15, 2, 23, 31, 6, 8, 5, 20, 15, 7, 7, 4, 26, 10, 3, 12, 26, 4, 5, 5, 2, 12, 8, 5, 4, 8, 3, 26, 3, 4, 25, 10, 5, 13, 12, 17, 17, 5, 2, 13, 2, 26, 6, 13, 31, 2, 4, 6, 11, 5, 39, 3, 4, 12, 5, 11, 33, 2, 2, 8, 10, 3, 9, 5, 6, 11]], (20, 40, 3): [[13, 39, 5, 7, 36, 3, 21, 50, 36, 58, 3, 80, 13, 10, 50, 20, 46, 52, 5, 58, 3, 48, 23, 9, 30, 5, 32, 7, 3, 13, 100, 3, 4, 24, 3, 16, 3, 4, 22, 28, 5, 62, 8, 32, 44], [45, 46, 86, 51, 62, 53, 23, 10, 15, 36, 7, 7, 23, 28, 7, 5, 63, 31, 34, 4, 14, 14, 13, 2, 77, 21, 2, 3, 4, 4, 71, 78, 3, 27, 8, 14, 55, 7, 72, 11], [20, 10, 60, 3, 46, 4, 43, 16, 22, 3, 25, 61, 10, 4, 119, 11, 32, 9, 60, 13, 2, 13, 18, 11, 13, 3, 83, 19, 45, 58, 5, 32, 3, 9, 61, 22, 40, 11, 5, 49, 3, 29, 31], [41, 9, 10, 51, 52, 45, 5, 6, 11, 8, 4, 3, 3, 15, 44, 21, 86, 34, 63, 28, 3, 83, 9, 58, 64, 5, 7, 9, 32, 17, 54, 24, 30, 25, 38, 3, 3, 28, 3, 13, 69, 11, 4, 5], [11, 11, 9, 12, 3, 11, 16, 16, 17, 27, 11, 14, 5, 19, 4, 4, 6, 9, 11, 14, 3, 11, 8, 19, 6, 6, 15, 23, 19, 15, 4, 4, 3, 45, 67, 7, 40, 24, 10, 2, 29, 2, 7, 6, 5, 56, 4, 43, 4, 5, 8, 4, 4, 17, 128, 4, 17, 77, 32, 64, 8, 13, 12, 4, 11, 11]], (20, 40, 6): [[11, 21, 7, 105, 63, 8, 11, 86, 4, 26, 12, 13, 7, 19, 40, 18, 6, 123, 4, 3, 40, 9, 4, 11, 31, 24, 6, 9, 63, 5, 6, 47, 75, 8, 11, 7, 26, 112, 6, 6, 43], [3, 15, 9, 16, 9, 23, 10, 410, 45, 5, 28, 6, 6, 12, 16, 25, 9, 9, 15, 121, 14, 29, 25, 33, 24, 18, 8, 10, 15, 13, 24, 25, 62, 44], [9, 2, 12, 3, 47, 227, 20, 29, 5, 32, 461, 219, 39, 24, 7], [368, 45, 10, 43, 124, 171, 44, 28, 27, 7, 220, 44, 5], [5, 78, 98, 49, 25, 42, 54, 81, 3, 19, 53, 43, 3, 34, 14, 7, 357, 171]]}",
"None\n"
],
[
"# results of runnning sample_clustering_params\ncluster_sizes= {(3, 5, 3): [[41, 179, 27, 61, 34, 209, 69, 2, 2, 23, 168, 55, 4, 2, 274], [6, 107, 23, 751, 130, 124], [489, 2, 355, 449, 187]], (3, 5, 6): [[16, 96, 410, 2, 1, 306, 59, 238, 7, 9], [240, 2, 543, 2, 56, 29, 2, 269], [13, 2, 2, 240, 2, 356, 8, 2, 118, 44, 7, 284, 2, 2, 8, 7, 9, 45, 2, 2]], (3, 5, 10): [[2, 301, 555, 12, 169, 2, 90, 12], [2, 136, 2, 331, 156, 2, 512, 2], [199, 267, 2, 2, 105, 1, 12, 379, 718, 1, 131, 1, 2, 2]], (3, 5, 40): [[211, 434, 128, 157, 210], [1, 2, 1, 68, 1, 238, 630, 325, 41, 2, 1, 1, 32, 250, 177], [1, 322, 33, 949, 269, 262]], (3, 5, 100): [[179, 221, 302, 435, 2], [2, 198, 2, 139, 2, 121, 129, 549], [371, 156, 341, 867, 1, 133, 75]], (3, 20, 3): [[10, 7, 9, 2, 3, 420, 180, 4, 236, 7, 266], [117, 835, 2, 2, 2, 180, 4], [2, 2, 102, 8, 436, 2, 2, 2, 106, 480, 4]], (3, 20, 6): [[186, 576, 2, 315, 61], [7, 22, 4, 131, 219, 9, 14, 382, 48, 19, 7, 15, 91, 7, 145, 2, 28], [8, 194, 6, 744, 213, 61, 2, 8, 155, 409, 31]], (3, 20, 10): [[553, 74, 301, 210, 2], [11, 149, 660, 2, 384, 82, 497, 12, 46, 108, 27], [11, 2, 316, 30, 8, 26, 68, 144, 105, 11, 110, 107, 4, 77, 2, 129]], (3, 20, 40): [[105, 40, 309, 934, 145], [42, 695, 36, 7, 136, 191, 35], [1, 917, 209, 51, 69, 443, 1, 1]], (3, 20, 100): [[64, 160, 20, 376, 103, 1000, 227], [2, 386, 102, 387, 2, 2, 260, 2], [44, 997, 129, 703, 48]], (3, 80, 3): [[12, 4, 7, 200, 249, 7, 601, 63], [9, 415, 77, 2, 21, 243, 107, 5, 4, 5, 257], [245, 20, 161, 64, 219, 2, 4, 4, 8, 7, 188, 234]], (3, 80, 6): [[542, 2, 2, 7, 2, 317, 2, 2, 2, 267], [11, 44, 14, 90, 41, 1, 49, 10, 1, 13, 1, 1, 13, 7, 12, 7, 1, 7, 1, 8, 27, 792, 2], [48, 8, 19, 40, 390, 144, 6, 4, 6, 198, 3, 163, 8, 84, 7, 9, 7, 9]], (3, 80, 10): [[38, 14, 6, 20, 106, 881, 370, 29, 68, 3, 108, 13, 26, 32, 76, 1, 5, 43, 182, 1], [76, 100, 2, 18, 76, 8, 139, 90, 2, 181, 93, 132, 46, 15, 124, 15, 34], [38, 10, 64, 148, 312, 2, 177, 33, 106, 2, 13, 24, 16, 16, 8, 654, 11, 5]], (3, 80, 40): [[415, 32, 62, 232, 103, 119, 26, 152], [40, 104, 190, 1, 139, 1051, 42, 43, 52, 15, 1, 202], [124, 342, 79, 43, 104, 50, 313, 3, 12, 73, 2]], (3, 80, 100): [[435, 109, 307, 287], [588, 20, 2, 271, 2, 22, 197, 41], [428, 460, 250]], (5, 5, 3): [[4, 102, 57, 89, 10, 61, 233, 11, 43, 62, 28, 4, 5, 9, 23, 96, 6, 309], [31, 23, 90, 2, 116, 2, 4, 253, 13, 5, 4, 2, 8, 31, 160, 5, 219, 41, 81, 4, 62], [91, 84, 160, 74, 29, 140, 15, 6, 10, 74, 204, 13, 5, 110, 135]], (5, 5, 6): [[13, 7, 7, 126, 36, 6, 28, 112, 245, 1, 6, 525, 24, 173, 117, 58, 6], [61, 229, 6, 2, 16, 18, 125, 2, 2, 14, 86, 80, 13, 14, 57, 10, 2, 134, 72, 2, 25, 68, 120], [8, 255, 7, 8, 112, 84, 113, 17, 13, 9, 7, 13, 95, 84, 296, 7, 5, 19]], (5, 5, 10): [[13, 128, 114, 37, 83, 10, 66, 18, 14, 320, 10, 265, 20, 11, 41], [10, 17, 10, 4, 7, 211, 54, 8, 43, 10, 12, 387, 96, 14, 50, 39, 19, 43, 2, 36, 83], [13, 299, 138, 362, 17, 2, 107, 2, 9, 7, 2, 7, 732, 384]], (5, 5, 40): [[69, 51, 873, 549, 45, 1, 8], [149, 40, 252, 445, 57, 155, 43], [539, 2, 7, 53, 168, 19, 330, 2, 24]], (5, 5, 100): [[50, 185, 202, 206, 134, 364], [54, 69, 207, 507, 5, 199, 72, 29], [917, 754, 194, 81, 85]], (5, 20, 3): [[277, 4, 4, 4, 50, 29, 7, 145, 5, 9, 6, 9, 9, 9, 4, 6, 4, 53, 30, 4, 8, 81, 9, 94, 115, 5, 6, 50, 34, 26, 36, 3, 30, 4], [7, 214, 22, 8, 246, 7, 4, 13, 43, 104, 6, 20, 17, 34, 5, 3, 12, 2, 20, 15, 90, 4, 24, 7, 4, 74, 36, 4, 13, 32, 17, 44, 17], [2, 14, 66, 17, 16, 30, 28, 222, 16, 3, 3, 4, 35, 8, 79, 34, 12, 4, 33, 10, 5, 267, 15, 6, 21, 5, 5, 101, 4, 5, 21, 70, 6]], (5, 20, 6): [[25, 2, 8, 112, 15, 38, 48, 7, 33, 8, 52, 274, 23, 104, 23, 2, 10, 30, 219, 85, 7, 32], [73, 120, 8, 5, 9, 12, 9, 2, 106, 210, 17, 138, 34, 7, 29, 153, 12, 160, 8, 43], [100, 94, 2, 18, 11, 6, 35, 28, 42, 10, 9, 10, 19, 5, 65, 9, 20, 369, 23, 253, 27]], (5, 20, 10): [[16, 12, 7, 129, 82, 11, 22, 110, 36, 4, 20, 29, 121, 17, 191, 4, 30, 7, 306], [13, 150, 10, 108, 38, 25, 106, 46, 223, 51, 13, 10, 16, 76, 5, 3, 22, 12, 80, 9, 73, 32, 24, 13], [1, 747, 78, 170, 8, 79, 1, 206, 115, 112, 36, 69, 12, 198]], (5, 20, 40): [[671, 8, 187, 36, 7, 29, 41, 38, 72, 11, 28, 19], [41, 266, 52, 147, 26, 190, 76, 24, 295, 27, 2], [8, 68, 50, 19, 39, 158, 66, 92, 193, 95, 357]], (5, 20, 100): [[263, 27, 266, 86, 498], [470, 182, 1085, 71, 40], [75, 237, 705, 4, 3, 117]], (5, 80, 3): [[12, 4, 3, 39, 51, 9, 6, 10, 78, 17, 4, 4, 4, 10, 12, 3, 9, 11, 131, 10, 31, 10, 9, 26, 9, 102, 31, 13, 4, 379, 4, 6, 11, 50, 5, 5, 11, 5, 4, 4, 4, 21, 7], [20, 18, 5, 12, 14, 5, 92, 11, 65, 1, 50, 10, 6, 5, 31, 11, 30, 4, 10, 9, 43, 4, 19, 5, 5, 21, 17, 22, 5, 247, 4, 15, 5, 12, 9, 9, 239, 18, 24, 6, 23, 2, 4, 11], [5, 4, 11, 30, 8, 9, 11, 4, 3, 21, 5, 11, 5, 68, 9, 31, 28, 46, 6, 4, 16, 21, 7, 108, 26, 3, 20, 43, 9, 25, 50, 107, 9, 6, 157, 9, 4, 52, 22, 76, 58, 30]], (5, 80, 6): [[9, 11, 4, 7, 183, 78, 22, 17, 7, 6, 88, 2, 10, 51, 139, 6, 28, 25, 9, 19, 24, 109, 6, 740, 42, 8, 31, 47, 2, 16, 8, 47, 56, 5], [5, 102, 140, 7, 7, 4, 53, 3, 6, 19, 261, 40, 25, 15, 10, 4, 199, 56, 49, 19, 23, 7, 36, 14, 6, 13, 7, 11, 12, 4, 8], [13, 11, 22, 9, 19, 38, 52, 5, 163, 6, 31, 38, 10, 26, 13, 45, 9, 15, 7, 20, 22, 7, 14, 58, 7, 18, 251, 5, 116, 31, 23, 8, 56]], (5, 80, 10): [[34, 23, 37, 2, 15, 66, 27, 65, 4, 8, 21, 118, 95, 7, 66, 12, 372, 153, 29], [30, 156, 5, 51, 85, 13, 38, 18, 110, 336, 15, 35, 38, 17, 5, 18, 10, 13, 26, 9, 5, 829, 5, 10], [14, 5, 10, 12, 10, 214, 79, 60, 38, 2, 88, 9, 147, 92, 14, 25, 760, 9, 9, 7, 66, 44, 3, 61, 44, 45, 3, 37]], (5, 80, 40): [[32, 119, 614, 103, 30, 38, 73, 43, 91], [169, 51, 89, 96, 110, 177, 196, 154, 101], [165, 2, 84, 284, 12, 17, 29, 209, 3, 337, 3]], (5, 80, 100): [[12, 122, 723, 36, 1026, 4, 90, 3], [27, 68, 218, 182, 620, 26], [104, 110, 131, 212, 42, 129, 11, 7, 21, 1, 486, 55, 1054, 106]], (7, 5, 3): [[169, 34, 4, 118, 4, 11, 17, 354, 2, 6, 103, 8, 167, 30, 2, 45, 59, 20], [36, 32, 4, 54, 10, 82, 58, 3, 54, 2, 8, 4, 5, 45, 118, 67, 84, 115, 25, 104, 116, 9, 123], [12, 3, 76, 26, 16, 383, 19, 17, 19, 29, 24, 106, 25, 35, 28, 64, 47, 4, 140, 4, 12, 31, 39]], (7, 5, 6): [[34, 8, 84, 9, 230, 13, 60, 65, 61, 249, 74, 13, 4, 34, 6, 17, 94, 93, 6], [7, 231, 900, 29, 56, 49, 6, 8, 66, 7, 9, 25, 6, 95], [213, 31, 130, 49, 48, 8, 2, 8, 43, 9, 62, 2, 5, 279, 162, 56, 6, 15, 6, 21]], (7, 5, 10): [[93, 45, 11, 27, 127, 59, 9, 73, 44, 12, 130, 12, 7, 900, 19, 137, 8], [64, 23, 4, 2, 24, 130, 30, 18, 4, 127, 35, 11, 172, 87, 9, 126, 113, 10, 54, 112], [27, 370, 3, 84, 101, 95, 58, 47, 228, 76, 45, 12]], (7, 5, 40): [[86, 184, 2, 76, 221, 203, 370], [375, 499, 18, 204, 44], [124, 44, 11, 22, 93, 381, 47, 111, 310]], (7, 5, 100): [[286, 37, 361, 1008, 53, 95, 48], [71, 296, 123, 311, 247, 93], [280, 126, 18, 227, 297, 124, 6, 64]], (7, 20, 3): [[6, 9, 40, 84, 7, 2, 128, 2, 6, 4, 134, 74, 27, 13, 40, 3, 4, 16, 12, 28, 6, 21, 21, 5, 88, 7, 4, 4, 10, 34, 20, 71, 2, 222, 15], [6, 29, 13, 63, 15, 87, 4, 16, 7, 231, 17, 45, 231, 17, 4, 6, 28, 17, 32, 58, 12, 2, 17, 5, 199], [6, 10, 96, 5, 10, 7, 2, 13, 19, 21, 31, 25, 19, 12, 43, 14, 43, 2, 10, 24, 10, 55, 2, 39, 37, 85, 40, 58, 30, 21, 16, 22, 26, 4, 31, 31, 6, 16, 6, 3, 225]], (7, 20, 6): [[30, 7, 101, 31, 57, 68, 37, 10, 18, 45, 30, 15, 7, 6, 22, 16, 9, 150, 44, 255, 7, 16, 4, 176], [87, 6, 8, 48, 16, 2, 10, 29, 142, 14, 36, 98, 35, 7, 7, 231, 251, 16, 14, 2, 47, 20, 8, 25], [6, 151, 32, 124, 14, 5, 5, 152, 113, 54, 57, 5, 105, 7, 16, 147, 10, 60, 78, 13, 2]], (7, 20, 10): [[64, 75, 210, 19, 66, 14, 54, 14, 74, 70, 71, 109, 157, 29, 10, 62, 53], [10, 20, 78, 30, 79, 39, 41, 17, 462, 14, 29, 10, 21, 14, 87, 160, 41], [28, 2, 78, 23, 14, 5, 14, 9, 311, 3, 51, 33, 213, 26, 43, 55, 24, 62, 160]], (7, 20, 40): [[62, 22, 932, 1, 95, 25, 7, 2, 214, 115, 48, 100, 284], [88, 46, 24, 10, 44, 207, 99, 56, 206, 145, 221], [25, 147, 182, 139, 9, 186, 943, 361, 102]], (7, 20, 100): [[359, 37, 221, 7, 21, 11, 1092], [1017, 34, 482, 147, 388, 92], [59, 44, 32, 9, 22, 81, 143, 40, 4, 635, 77]], (7, 80, 3): [[9, 54, 25, 10, 5, 60, 4, 5, 11, 6, 4, 30, 70, 7, 140, 33, 65, 15, 47, 5, 4, 24, 21, 21, 4, 5, 11, 84, 58, 20, 23, 13, 4, 28, 14, 46, 11, 65, 17, 16, 4, 4, 8, 4, 18, 46, 4], [28, 4, 58, 2, 7, 7, 13, 4, 4, 4, 17, 107, 10, 49, 21, 10, 4, 15, 21, 5, 180, 4, 66, 147, 30, 23, 7, 19, 3, 10, 45, 3, 29, 8, 10, 67, 5, 31, 55, 2, 19, 11, 3, 11], [26, 7, 59, 122, 18, 229, 16, 5, 30, 27, 24, 4, 37, 3, 13, 22, 4, 5, 17, 5, 33, 10, 41, 4, 10, 6, 4, 18, 32, 3, 12, 58, 11, 10, 15, 63, 35, 66, 4, 50, 6, 2, 12]], (7, 80, 6): [[68, 5, 11, 79, 54, 18, 8, 115, 43, 7, 26, 18, 4, 23, 35, 16, 8, 23, 4, 72, 6, 31, 43, 9, 104, 11, 111, 8, 51, 5, 7, 4, 23, 9, 111], [13, 24, 2, 13, 5, 7, 2, 103, 8, 42, 95, 8, 36, 3, 23, 11, 40, 213, 41, 8, 26, 42, 8, 24, 5, 26, 9, 51, 116, 6, 15, 5, 8, 4, 6, 49, 27, 13, 19, 18], [5, 26, 27, 2, 33, 3, 292, 2, 622, 169, 3, 5, 9, 30, 9, 229, 29, 8, 57, 56, 2, 106, 145, 54, 2]], (7, 80, 10): [[48, 13, 6, 58, 11, 11, 19, 59, 11, 10, 3, 94, 51, 236, 36, 86, 267, 11, 24, 50, 52], [99, 107, 12, 9, 82, 9, 24, 20, 8, 157, 56, 18, 17, 55, 47, 13, 11, 14, 31, 11, 17, 27, 42, 106, 15, 154], [18, 26, 100, 25, 50, 21, 9, 123, 19, 4, 20, 15, 17, 12, 34, 21, 17, 25, 29, 6, 473, 12, 78, 4]], (7, 80, 40): [[315, 9, 76, 29, 559, 15, 139], [206, 2, 107, 19, 86, 170, 13, 192, 5, 2, 62, 251, 15, 19], [34, 88, 85, 24, 397, 64, 957, 28, 180]], (7, 80, 100): [[101, 119, 765, 75, 80], [506, 4, 1054, 40, 15, 320, 245], [118, 9, 62, 5, 641, 166, 140]], (13, 5, 3): [[28, 25, 6, 6, 12, 56, 4, 39, 9, 24, 4, 4, 5, 22, 4, 77, 22, 3, 7, 65, 50, 12, 29, 30, 4, 10, 12, 132, 30, 10, 4, 115, 8, 55, 32, 48, 59, 6, 4, 4, 24, 44, 34], [80, 24, 34, 164, 92, 131, 69, 32, 5, 91, 138, 61, 34, 11, 6, 46, 26, 5, 8, 90, 9], [11, 7, 80, 32, 91, 9, 17, 12, 13, 109, 10, 8, 28, 10, 29, 41, 61, 2, 4, 42, 43, 24, 102, 18, 79, 11, 4, 71, 22, 72, 12, 45, 19, 14, 18]], (13, 5, 6): [[329, 27, 9, 54, 114, 38, 5, 104, 15, 84, 51, 83, 58, 40, 17, 6, 118], [18, 2, 53, 104, 19, 177, 234, 91, 6, 49, 52, 26, 49, 37, 9, 35, 70, 121], [8, 55, 45, 58, 7, 58, 9, 7, 20, 12, 146, 109, 320, 121, 65, 110]], (13, 5, 10): [[43, 31, 10, 51, 45, 19, 120, 317, 59, 38, 80, 107, 3, 74, 55, 5, 62, 6, 11, 19], [219, 39, 7, 19, 7, 45, 498, 20, 21, 84, 10, 23, 48, 93, 17], [8, 52, 72, 25, 128, 64, 96, 26, 91, 21, 8, 8, 16, 161, 70, 258, 33, 7, 9]], (13, 5, 40): [[29, 224, 80, 93, 190, 183, 14, 118, 46, 168], [40, 23, 330, 126, 327, 20, 54, 51, 127, 47], [230, 530, 136, 63, 96, 42, 10, 36]], (13, 5, 100): [[48, 85, 1005], [43, 1094], [125, 873, 140]], (13, 20, 3): [[4, 23, 7, 89, 14, 12, 19, 25, 3, 27, 4, 20, 8, 5, 5, 11, 44, 45, 27, 22, 3, 32, 44, 20, 26, 6, 7, 25, 32, 8, 4, 15, 20, 8, 19, 16, 22, 3, 12, 13, 4, 23, 32, 71, 3, 36, 3, 63, 23, 5, 7, 18, 10, 6, 19, 28, 3, 8, 83], [27, 60, 9, 83, 13, 4, 10, 19, 5, 6, 3, 5, 64, 7, 52, 4, 23, 6, 56, 16, 8, 9, 4, 21, 19, 43, 40, 58, 3, 6, 3, 32, 45, 4, 28, 109, 5, 42, 25, 4, 27, 33, 25, 15, 7, 4, 4, 4, 16, 14, 23, 34], [34, 5, 53, 30, 7, 46, 3, 32, 11, 4, 19, 10, 10, 22, 16, 5, 20, 13, 7, 15, 4, 3, 4, 44, 30, 8, 7, 10, 14, 11, 5, 117, 4, 21, 15, 23, 13, 9, 4, 24, 9, 24, 1, 15, 3, 3, 17, 3, 4, 22, 15, 8, 751, 13, 5, 62, 24, 6, 38, 10, 65, 21, 22, 3, 5, 21, 39]], (13, 20, 6): [[15, 31, 3, 17, 5, 78, 39, 9, 14, 14, 19, 29, 75, 90, 20, 4, 11, 52, 48, 100, 42, 12, 33, 198, 17, 74, 22, 75, 5, 12, 3], [13, 18, 37, 57, 19, 7, 21, 115, 63, 22, 9, 130, 4, 85, 142, 259, 144, 7], [17, 11, 207, 16, 7, 34, 9, 47, 82, 7, 12, 14, 13, 9, 76, 200, 5, 5, 21, 67, 13, 99, 11, 16, 27, 104, 14, 8, 13]], (13, 20, 10): [[11, 152, 849, 34, 9, 287, 96, 11, 5, 5, 28, 23, 259], [41, 3, 21, 4, 9, 18, 9, 108, 275, 579, 40, 10, 15, 15, 3], [285, 9, 24, 16, 287, 19, 6, 8, 231, 47, 5, 14, 783, 5, 16, 8, 18, 119, 16]], (13, 20, 40): [[18, 7, 136, 56, 79, 77, 517, 238, 16], [98, 887, 17, 50, 19, 75, 619], [43, 14, 44, 37, 20, 458, 118, 15, 63, 142, 194]], (13, 20, 100): [[27, 21, 1087, 4], [39, 18, 32, 1064, 702, 34, 17, 80, 48], [9, 10, 186, 850, 22, 64]], (13, 80, 3): [[3, 14, 27, 21, 10, 76, 119, 30, 63, 29, 24, 5, 4, 47, 37, 22, 60, 5, 25, 23, 24, 113, 3, 4, 25, 4, 33, 36, 8, 12, 11, 64, 43, 6, 68, 8, 45, 22], [44, 9, 18, 73, 41, 26, 52, 55, 3, 11, 8, 7, 21, 5, 47, 5, 17, 18, 61, 8, 11, 5, 43, 9, 14, 3, 4, 20, 13, 40, 5, 12, 6, 10, 49, 115, 39, 23, 3, 38, 26, 10, 22, 10, 12, 9, 12, 4, 7, 77, 3, 7], [4, 24, 4, 18, 33, 22, 11, 17, 33, 4, 12, 14, 30, 21, 47, 7, 21, 37, 4, 6, 4, 8, 77, 35, 11, 54, 59, 56, 13, 17, 30, 7, 28, 33, 43, 12, 30, 48, 11, 24, 8, 28, 20, 10, 11, 4, 28, 41, 68]], (13, 80, 6): [[121, 119, 13, 108, 4, 8, 11, 5, 29, 218, 303, 44, 15, 19, 12, 13, 4, 106], [14, 8, 164, 4, 9, 84, 11, 223, 3, 28, 92, 45, 68, 93, 25, 8, 41, 12, 13, 52, 69, 62, 2, 3, 8, 11, 10], [4, 11, 25, 6, 6, 13, 22, 15, 34, 10, 20, 225, 8, 164, 55, 5, 32, 5, 167, 9, 24, 13, 17, 16, 50, 3, 29, 97, 79]], (13, 80, 10): [[62, 52, 36, 98, 76, 171, 8, 9, 30, 14, 9, 89, 30, 25, 13, 12, 149, 7, 34, 40, 19, 8, 11, 21, 12, 9, 13, 11, 45, 52], [59, 32, 10, 434, 43, 57, 307, 12, 7, 43, 47, 23, 10, 12, 15, 8, 15, 19], [36, 16, 41, 4, 51, 18, 875, 54, 6, 181, 22, 102, 62, 18, 16, 11, 11, 9, 6, 9, 20, 19, 4, 267]], (13, 80, 40): [[8, 133, 86, 42, 124, 29, 60, 76, 333, 1081, 139, 75], [49, 22, 922, 79, 324, 48, 24, 228, 11, 30, 181, 51], [54, 35, 302, 24, 272, 384, 71]], (13, 80, 100): [[29, 5, 38, 7, 146, 12, 794, 14, 9, 8, 84], [5, 11, 15, 1108], [5, 54, 744, 1058, 15, 178, 26]], (20, 5, 3): [[23, 19, 24, 67, 106, 7, 15, 20, 48, 127, 4, 16, 38, 34, 28, 21, 12, 7, 4, 99, 12, 12, 15, 79, 237, 8, 46, 12, 6, 19], [90, 43, 48, 14, 107, 122, 10, 17, 5, 7, 5, 9, 5, 23, 8, 109, 66, 30, 15, 6, 37, 78, 19, 17, 7, 48, 16, 13, 21, 3, 10, 31, 13, 3, 56, 20, 41], [79, 6, 8, 19, 13, 3, 9, 8, 23, 21, 48, 29, 98, 4, 206, 17, 91, 18, 50, 52, 5, 5, 9, 8, 10, 3, 25, 14, 15, 14, 5, 7, 13, 39, 11, 20, 8, 49, 31, 17, 49, 18]], (20, 5, 6): [[45, 15, 7, 32, 298, 53, 27, 63, 7, 155, 160, 20, 107, 111, 36, 17], [31, 10, 2, 325, 165, 33, 6, 88, 128, 138, 22, 178, 22], [76, 7, 8, 9, 229, 104, 37, 17, 16, 446, 24, 54, 121]], (20, 5, 10): [[293, 14, 8, 27, 25, 6, 33, 10, 681, 30, 13, 7], [14, 791, 4, 37, 10, 23, 9, 334, 165, 31, 4, 39, 24, 401], [9, 71, 11, 3, 74, 18, 11, 83, 11, 108, 6, 462, 43, 54, 8, 50, 20, 13, 99]], (20, 5, 40): [[81, 20, 433, 331, 25, 29, 222], [875, 472, 472, 106, 64, 5], [32, 207, 65, 185, 234, 53, 208, 159]], (20, 5, 100): [[629, 315, 194], [30, 88, 524, 476, 22], [612, 96, 3, 272, 157]], (20, 20, 3): [[78, 8, 66, 83, 35, 14, 7, 17, 42, 20, 36, 6, 37, 78, 6, 7, 15, 30, 43, 60, 3, 4, 6, 26, 36, 10, 53, 13, 3, 12, 18, 4, 40, 52, 9, 14, 4, 21, 11, 24, 16, 12, 4, 14, 18, 50, 16], [5, 41, 5, 6, 6, 30, 4, 21, 6, 36, 5, 5, 8, 8, 32, 95, 17, 12, 18, 5, 25, 10, 5, 8, 5, 12, 42, 15, 66, 4, 151, 70, 16, 8, 4, 8, 63, 38, 25, 40, 6, 46, 40, 6, 17, 7, 5, 24, 7, 3, 22, 17, 8], [19, 4, 40, 3, 12, 15, 5, 28, 22, 9, 4, 34, 62, 34, 33, 12, 19, 3, 3, 17, 79, 78, 10, 30, 16, 4, 4, 37, 22, 17, 8, 30, 19, 17, 4, 6, 28, 16, 8, 12, 12, 17, 27, 12, 4, 4, 4, 11, 16, 76, 4, 17, 5, 7, 4, 9, 36, 5, 16, 35, 10, 43]], (20, 20, 6): [[7, 66, 130, 62, 5, 12, 144, 70, 16, 5, 6, 41, 16, 423, 53, 21, 42, 6, 5, 8, 7, 6, 8], [39, 7, 223, 9, 36, 6, 62, 801, 6, 7, 4, 29, 42, 15, 7, 21, 115, 12, 6, 357, 49, 55, 14], [28, 179, 5, 308, 28, 7, 5, 105, 40, 29, 58, 12, 14, 5, 13, 6, 52, 43, 20, 13, 782, 18, 34, 7, 102]], (20, 20, 10): [[23, 11, 10, 13, 18, 9, 3, 143, 103, 6, 19, 43, 39, 74, 47, 16, 10, 30, 300, 4, 199, 9, 3, 8, 12, 9], [15, 11, 11, 3, 200, 26, 16, 23, 8, 90, 304, 29, 7, 32, 368, 8], [481, 6, 5, 3, 9, 133, 15, 161, 38, 112, 164, 19]], (20, 20, 40): [[15, 117, 509, 115, 354, 31], [221, 91, 107, 78, 189, 70, 22, 327, 23, 11, 7], [125, 49, 432, 14, 271, 17, 96, 43, 58, 40]], (20, 20, 100): [[88, 426, 76, 531, 19], [22, 459, 206, 221, 108, 59, 11, 57], [355, 18, 1131, 8]], (20, 80, 3): [[27, 74, 8, 9, 3, 9, 26, 3, 4, 54, 12, 83, 47, 66, 60, 14, 9, 63, 4, 10, 7, 10, 7, 27, 15, 7, 4, 7, 42, 3, 24, 48, 12, 5, 44, 10, 25, 11, 65, 26, 4, 7, 3, 106, 61, 15], [35, 100, 44, 5, 5, 47, 66, 68, 13, 10, 18, 4, 13, 26, 12, 3, 9, 16, 49, 3, 84, 5, 83, 4, 4, 13, 58, 15, 111, 7, 13, 7, 3, 6, 3, 19, 65, 52, 5, 36, 36], [15, 7, 31, 15, 14, 16, 22, 48, 8, 9, 23, 7, 5, 6, 7, 14, 30, 4, 5, 36, 6, 27, 12, 30, 3, 112, 21, 13, 11, 18, 19, 50, 3, 9, 7, 7, 37, 51, 4, 5, 3, 13, 3, 8, 3, 27, 31, 67, 61, 4, 4, 25, 23, 3, 4, 6, 9, 4, 37, 42, 4, 29, 7, 15]], (20, 80, 6): [[77, 13, 71, 10, 114, 30, 10, 15, 28, 11, 41, 25, 39, 8, 29, 6, 62, 33, 64, 9, 3, 85, 11, 8, 7, 41, 88, 61, 4, 8, 57, 8, 37, 14, 7, 15, 12, 11], [8, 22, 24, 6, 91, 58, 85, 10, 34, 4, 7, 27, 4, 48, 35, 7, 12, 81, 9, 6, 6, 54, 179, 260, 8, 3, 71, 4], [65, 45, 14, 11, 8, 7, 33, 7, 19, 20, 14, 370, 3, 13, 8, 16, 791, 22, 30, 5, 39, 248, 40, 34, 12, 10, 9, 3, 5, 8]], (20, 80, 10): [[6, 3, 219, 53, 7, 584, 11, 54, 11, 11, 154, 9, 14, 13], [883, 150, 4, 7, 7, 16, 19, 3, 13, 115, 17, 420, 7, 17, 19, 9, 7, 3, 7, 11], [69, 694, 49, 16, 104, 18, 10, 121, 7, 8, 7, 38, 7]], (20, 80, 40): [[899, 747, 43, 52, 15], [12, 164, 4, 425, 60, 21, 181, 96, 54, 28, 101], [45, 216, 39, 12, 328, 75, 357, 70]], (20, 80, 100): [[71, 33, 1095, 128, 51, 482], [10, 10, 10, 63, 1047], [31, 969, 241, 834]]}",
"_____no_output_____"
],
[
"plt.rcParams['figure.figsize'] = (16,16)\nk_stats = [[] for i in range(4)] # rows are: k,mean,var,var of var\nweak_stats = [[] for i in range(4)]\nneighbor_stats = [[] for i in range(4)]\nfor params,c_sizes in cluster_sizes.items():\n stats = (np.mean([len(a) for a in c_sizes]),np.mean([np.var(a) for a in c_sizes]),\n np.var([np.var(a) for a in c_sizes]))\n k_stats[0].append(params[0])\n weak_stats[0].append(params[1])\n neighbor_stats[0].append(params[2])\n for i in [0,1,2]:\n k_stats[i+1].append(stats[i])\n weak_stats[i+1].append(stats[i])\n neighbor_stats[i+1].append(stats[i])\n\n_,subplt = plt.subplots(3, 3,gridspec_kw = {'wspace': 0.5,'hspace': 0.25}, sharey='col')\nfor row,d in enumerate([k_stats,weak_stats,neighbor_stats]):\n for col in range(3):\n subplt[row,col].scatter(d[0],d[col+1])\n\nfor i in [0,1,2]:\n subplt[0][i].set_xlabel('# clusters for each weak kNN (k)')\n subplt[1][i].set_xlabel('# weak kNN\\'s (w)')\n subplt[2][i].set_xlabel(\"# connections per node considered 'strong' (n)\")\nfor i in [0,1,2]:\n subplt[i][0].set_ylabel('mean of c_sizes')\n subplt[i][1].set_ylabel('var of c_size')\n subplt[i][2].set_ylabel('var of var of c_size')",
"_____no_output_____"
],
[
"#k_stats_2 = [d[1] for k in [3,5,7,13,20] for i,d in enumerate(k_stats) if k_stats[0]==k ]#,\nk_stats_2 = [[[k_stats[1][i] for i in range(len(k_stats[1])) if k_stats[0][i]==k] for k in k_ticks],\n [[k_stats[2][i] for i in range(len(k_stats[1])) if k_stats[0][i]==k] for k in k_ticks],\n [[k_stats[3][i] for i in range(len(k_stats[1])) if k_stats[0][i]==k] for k in k_ticks]]\nweak_stats_2 = [[[weak_stats[1][i] for i in range(len(weak_stats[1])) if weak_stats[0][i]==w] for w in w_ticks],\n [[weak_stats[2][i] for i in range(len(weak_stats[1])) if weak_stats[0][i]==w] for w in w_ticks],\n [[weak_stats[3][i] for i in range(len(weak_stats[1])) if weak_stats[0][i]==w] for w in w_ticks]]\nneighbor_stats_2 = [[[neighbor_stats[1][i] for i in range(len(neighbor_stats[1])) if neighbor_stats[0][i]==n] for n in n_ticks],\n [[neighbor_stats[2][i] for i in range(len(neighbor_stats[1])) if neighbor_stats[0][i]==n] for n in n_ticks],\n [[neighbor_stats[3][i] for i in range(len(neighbor_stats[1])) if neighbor_stats[0][i]==n] for n in n_ticks]]\n_,subplt = plt.subplots(3, 3,gridspec_kw = {'wspace': 0.5,'hspace': 0.25}, sharey='col')\nfor row,d in enumerate([k_stats_2,weak_stats_2,neighbor_stats_2]):\n for col in range(3):\n subplt[row,col].boxplot(d[col])\nfor i in [0,1,2]:\n subplt[0][i].set_xlabel('# clusters for each weak kNN (k)')\n subplt[0][i].set_xticklabels(k_ticks)\n subplt[1][i].set_xlabel('# weak kNN\\'s (w)')\n subplt[1][i].set_xticklabels(w_ticks)\n subplt[2][i].set_xlabel(\"# connections per node considered 'strong' (n)\")\n subplt[2][i].set_xticklabels(n_ticks)\nfor i in [0,1,2]:\n subplt[i][0].set_ylabel('mean of c_sizes')\n subplt[i][1].set_ylabel('var of c_size')\n subplt[i][2].set_ylabel('var of var of c_size')\n",
"_____no_output_____"
],
[
"def get_regression_performance(predict_func,args,iterations,bias_factor=1):\n \"\"\"\n predict_func: function that implements model. must return 3-tuple of rank 1 ndarrays, \n (_, test true values, test predicted values)\n args: tuple of arguments for predict_func\n iterations: number of validation iterations desired \n \"\"\"\n #cum_stats = np.zeros((3,testSize)) # rows: 0.sum(prediction); 1.sum(prediction^2); 2.sum(pred-real)\n #bias = ( sum(pred - real) )^2 = ( sum(pred) - sum(real) )^2\n #var = sum(pred^2) - sum(pred) ^2\n for i in range(iterations):\n (y_true,y_pred,_) = predict_func(*args)\n if i == 0:\n cum_stats = np.zeros((3,len(y_pred)))\n cum_stats[0] += y_pred\n cum_stats[1] += (y_pred - y_true)\n cum_stats[2] += y_pred**2\n mean_y_pred = np.sum(cum_stats[0]) / cum_stats.shape[1]\n bias = np.sum(cum_stats[1]) / (cum_stats.shape[1]-1)\n variance = (np.sum(cum_stats[2]) - mean_y_pred**2) / (cum_stats.shape[1]-1)\n return (mean_y_pred, predict_func(*args)[2],(bias*bias_factor)**2 + variance)",
"_____no_output_____"
],
[
"def get_regression_performance_shuffle_split(predict_func,data,goal,args,test_size=0.2,iterations=5,bias_factor=1):\n \"\"\"\n predict_func: function that implements model. must return 3-tuple of rank 1 ndarrays, \n (_, test true values, test predicted values)\n args: tuple of arguments for predict_func\n \"\"\"\n \n for train_index, test_index in ShuffleSplit(n_splits=5, test_size=.2).split(data):\n (y_true,y_pred,_) = predict_func(data[train_index],data[test_index],goal[train_index],goal[test_index],*args)\n if i == 0:\n cum_stats = np.zeros((3,len(y_pred)))\n cum_stats[0] += y_pred\n cum_stats[1] += (y_pred - y_true)\n cum_stats[2] += y_pred**2\n mean_y_pred = np.sum(cum_stats[0]) / cum_stats.shape[1]\n bias = np.sum(cum_stats[1]) / (cum_stats.shape[1]-1)\n variance = (np.sum(cum_stats[2]) - mean_y_pred**2) / (cum_stats.shape[1]-1)\n return (mean_y_pred, predict_func(*args)[2],(bias*bias_factor)**2 + variance)",
"_____no_output_____"
],
[
"def lasso_prediction(x,y,alpha=1.0,rand_state=(np.random.rand(1) * 10000).astype(int),silent=True):\n train_x, valid_x, train_y, valid_y = train_test_split(\n x, y, test_size=0.2, random_state=rand_state)\n \n model = Lasso(alpha=alpha)\n model.fit(train_x,train_y)\n y_pred = model.predict(valid_x)\n if not silent:\n plt.rcParams['figure.figsize'] = (5,2)\n plt.title(str(alpha))\n plt.violinplot(y_pred-valid_y, vert = False, showmeans=True, showextrema=True, showmedians=True)\n return(valid_y, y_pred, pickle.dumps(model))\n\ndef lasso_prediction_pre_split(train_x, valid_x, train_y, valid_y,alpha=1.0,silent=True): \n model = Lasso(alpha=alpha)\n model.fit(train_x,train_y)\n y_pred = model.predict(valid_x)\n if not silent:\n plt.rcParams['figure.figsize'] = (5,2)\n plt.title(str(alpha))\n plt.violinplot(y_pred-valid_y, vert = False, showmeans=True, showextrema=True, showmedians=True)\n return(valid_y, y_pred, pickle.dumps(model))\n\n\ndef ridge_prediction(x,y,alpha=1.0,rand_state=(np.random.rand(1) * 10000).astype(int),silent=True):\n train_x, valid_x, train_y, valid_y = train_test_split(\n x, y, test_size=0.2, random_state=rand_state)\n \n model = Ridge(alpha=alpha)\n model.fit(train_x,train_y)\n y_pred = model.predict(valid_x)\n if not silent:\n plt.rcParams['figure.figsize'] = (5,2)\n plt.title(str(alpha))\n plt.violinplot(y_pred-valid_y, vert = False, showmeans=True, showextrema=True, showmedians=True)\n return(valid_y, y_pred, pickle.dumps(model))\n \n \ndef ridge_prediction_pre_split(train_x, valid_x, train_y, valid_y,alpha=1.0,silent=True):\n\n model = Ridge(alpha=alpha)\n model.fit(train_x,train_y)\n y_pred = model.predict(valid_x)\n if not silent:\n plt.rcParams['figure.figsize'] = (5,2)\n plt.title(str(alpha))\n plt.violinplot(y_pred-valid_y, vert = False, showmeans=True, showextrema=True, showmedians=True)\n return(valid_y, y_pred, pickle.dumps(model))\n \n \n\n \ndef kNN_prediction(x,y,n_neighbors=5,rand_state=(np.random.rand(1) * 10000).astype(int),silent=True):\n train_x, valid_x, train_y, valid_y = train_test_split(\n x, y, test_size=0.2, random_state=rand_state)\n \n model = KNeighborsRegressor(n_neighbors=n_neighbors)\n model.fit(train_x,train_y)\n y_pred = model.predict(valid_x)\n if not silent:\n plt.rcParams['figure.figsize'] = (5,2)\n plt.title(str(alpha))\n plt.violinplot(y_pred-valid_y, vert = False, showmeans=True, showextrema=True, showmedians=True)\n return(valid_y, y_pred, pickle.dumps(model))\n \n\ndef kNN_prediction_pre_split(train_x, valid_x, train_y, valid_y,silent=True):\n \n model = KNeighborsRegressor(n_neighbors=n_neighbors)\n model.fit(train_x,train_y)\n y_pred = model.predict(valid_x)\n if not silent:\n plt.rcParams['figure.figsize'] = (5,2)\n plt.title(str(alpha))\n plt.violinplot(y_pred-valid_y, vert = False, showmeans=True, showextrema=True, showmedians=True)\n return(valid_y, y_pred, pickle.dumps(model))\n \n\n \ndef svr_prediction(x,y,kernel='rbf',rand_state=(np.random.rand(1) * 10000).astype(int), C=0.8,\n epsilon=0.1,gamma='auto',silent=True):\n train_x, valid_x, train_y, valid_y = train_test_split(\n x, y, test_size=0.2, random_state=rand_state)\n \n model = SVR(kernel=kernel,C=C,epsilon=epsilon,gamma=gamma)\n model.fit(train_x,train_y)\n y_pred = model.predict(valid_x)\n if not silent:\n plt.rcParams['figure.figsize'] = (5,2)\n plt.title(str(alpha))\n plt.violinplot(y_pred-valid_y, vert = False, showmeans=True, showextrema=True, showmedians=True)\n return(valid_y, y_pred, pickle.dumps(model))\n \n \n \n \ndef adatree_prediction(x,y,depth=4,n_estimators=50,,rand_state=(np.random.rand(1) * 10000).astype(int),silent=True):\n train_x, valid_x, train_y, valid_y = train_test_split(\n x, y, test_size=0.2, random_state=rand_state)\n \n model = AdaBoostRegressor(DecisionTreeRegressor(max_depth=depth),\n n_estimators=n_estimators,random_state=rand_state)\n model.fit(train_x,train_y)\n y_pred = model.predict(valid_x)\n if not silent:\n plt.rcParams['figure.figsize'] = (5,2)\n plt.title(str(alpha))\n plt.violinplot(y_pred-valid_y, vert = False, showmeans=True, showextrema=True, showmedians=True)\n return(valid_y, y_pred, pickle.dumps(model))\n \n",
"_____no_output_____"
],
[
"def get_regression(data, goal, bias_factor, lasso=True, ridge=True, \n kNN=True, svr=True, adatree=True):\n #rand_states = [1213,239485,7298345,143542,535]\n regression_options = [] # store tuples of (pickled model, error)\n if lasso:\n lst_alpha = [0.4,0.55,0.6,0.65,0.7,0.75,0.8,0.85,0.9,1,1.1]\n for alpha in lst_alpha:\n best = (None,1000000000000000000)\n args = (alpha,)\n _,pickled_model, model_score = get_regression_performance_shuffle_split(lasso_prediction_pre_split,data,\n goal,args,\n bias_factor=bias_factor)\n best = (best if (best[1] <= model_score) else (pickled_model,model_score))\n regression_options.append(best)\n\n if ridge:\n lst_alpha = [0.4,0.55,0.6,0.65,0.7,0.75,0.8,0.85,0.9,1,1.1]\n for alpha in lst_alpha:\n best = (None,1000000000000000000)\n args = (alpha,)\n _, pickled_model, model_score = get_regression_performance_shuffle_split(ridge_prediction_pre_split,args,\n bias_factor=bias_factor)\n best = (best if (best[1] <= model_score) else (pickled_model,model_score))\n regression_options.append(best)\n \n if kNN:\n if len(data) > 75: # otherwise kNN regression is probably not effective\n lst_n = np.arange(3,10)\n for n in lst_n:\n best = (None,1000000000000000000)\n args = (n,)\n _, pickled_model, model_score = get_regression_performance_shuffle_split(kNN_prediction_pre_split,data,\n goal,args,\n bias_factor=bias_factor)\n best = (best if (best[1] <= model_score) else (pickled_model,model_score))\n regression_options.append(best)\n \n \n if svr:\n lst_kernel=['rbf','linear']\n lst_C= [0.1,0.5,1,5,10,50,100]\n lst_epsilon = [0.01,0.03,0.05,0.07,0.1]\n lst_gamma = [0.001,0.005,0.01,0.05,0.1]\n combos = itertools.product(lst_kernel,lst_C,lst_epsilon)\n for comb in combos:\n args = (data,goal,*comb)\n _, pickled_model, model_score = get_regression_performance(svr_prediction,args,\n iterations=5,bias_factor=bias_factor)\n best = (best if (best[1] <= model_score) else (pickled_model,model_score))\n regression_options.append(best)\n \n if adatree:\n lst_depth=np.arange(2,6)\n lst_weak=[10,50,100]\n combos = itertools.product(lst_depth,lst_weak)\n for comb in combos:\n args = (data,goal,*comb)\n _, pickled_model, model_score = get_regression_performance(adatree_prediction,args,\n iterations=5,bias_factor=bias_factor)\n best = (best if (best[1] <= model_score) else (pickled_model,model_score))\n regression_options.append(best)\n \n return regression_options, regression_options[np.argmin(tup[1] for tup in regression_options)]",
"_____no_output_____"
],
[
"# estimate most similar cluster (using purely intuition)\ndef find_cluster(d,cluster_stats):\n return np.argmin([np.sum( (d - stat[0]) / (stat[1] * (d-stat[0])**(stat[2]*stat[3])) ) for stat in cluster_stats])",
"_____no_output_____"
],
[
"def regress_overall_stacked(features, goal, test_size, r_state=(np.random.rand(1) * 10000).astype(int),\n lasso=True, ridge=True, kNN=True, svm=True, adatree=True,silent=False):\n \n\n _, pickled_overall_weak = get_regression(data,goal,1,ridge=False)\n overall_weak = [pickle.loads(d) for d in pickled_overall_weak]\n \n train, valid, train_goal_true, valid_goal_true = train_test_split(\n features,goal,test_size=test_size,random_state=r_state)\n \n overall_weak_pred_train = []\n for weak in overall_weak:\n weak.fit_predict(train,train_goal)\n overall_weak_pred_train.append(weak.predict(train))\n pred_input = np.vstack(overall_weak_pred_train)\n overall_stack = LogisticRegression()\n overall_stack.fit(pred_input, train_goal)\n overall_pred = overall_stack.predict(valid)\n \n overall_SSE += mean_squared_error(valid_goal.values, overall_pred)\n overall_r2 += r2_score(valid_goal.values, overall_pred)\n \n if not silent:\n print(\"Ensemble model without clustering SSE: %f\" % overall_SSE)\n print(\"Ensemble model without clustering r2: %0.4f\" % overall_r2)\n print(\"\")\n \n ",
"_____no_output_____"
],
[
"def regress_by_cluster(features, goal, test_size, r_state=rand_state=(np.random.rand(1) * 10000).astype(int)\n lasso=True, ridge=True, kNN=True, svm=True, adatree=True,silent=False):\n \n\n _, pickled_overall_weak = get_regression(data,goal,1,ridge=False)\n overall_weak = [pickle.loads(d) for d in pickled_overall_weak]\n \n train, valid, train_goal_true, valid_goal_true = train_test_split(\n features,goal,test_size=test_size,random_state=r_state)\n \n overall_weak_pred_train = []\n for weak in overall_weak:\n overall_weak_pred_train.append(weak.fit_predict(train,train_goal))\n pred_input = np.vstack(overall_weak_pred_train)\n overall_stack = LogisticRegression()\n overall_stack.fit(pred_input, train_goal)\n overall_pred = overall_stack.predict(valid)\n \n overall_weak_pred = [weak.predict(valid) for weak in overall_weak]\n overall_weak_scores = [(type(weak), mean_squared_error(valid_goal.values, overall_weak_pred[i]),\n r2_score(valid_goal.values, overall_weak_pred[i])) for i,weak in enumerate(overall_weak)] \n overall_SSE += mean_squared_error(valid_goal.values, overall_pred)\n overall_r2 += r2_score(valid_goal.values, overall_pred)\n\n clusters = list(cluster_from_stack_of_KMeans(train, K=3, num_weak_learners = 20,num_neighbors=20))\n cluster_stats = np.empty((len(clusters),4,len(train.columns))) #dim2=mean,var,skew,kurtosis\n for i,cluster in enumerate(clusters):\n lst = list(cluster)\n cluster_stats[i,0,:] = np.mean(lst)\n cluster_stats[i,1,:] = np.std(lst)\n cluster_stats[i,2,:] = skew(lst)\n cluster_stats[i,3,:] = kurtosis(lst)\n\n #####\n if not silent:\n for i,d in enumerate(clusters):\n print(\"cluster size: \" + str(len(d)))\n display (pd.DataFrame(cluster_stats[i],columns=['mean','var','skew','kurtosis']))\n print('\\n')\n #####\n\n cluster_regressions = []\n\n for c in clusters:\n if (len(c) > 30):\n bias_factor=1\n #bias_factor = (1 if len(c) > 100 else (len(c)+100)/200)\n lst_c = list(c)\n #reg_options=get_regression(train.iloc[lst_c], train_goal.iloc[lst_c], bias_factor)\n #cluster_regressions.append(reg_options[np.argmin(tup[1] for tup in reg_options)])\n cluster_regressions.append(get_regression(train.iloc[lst_c], train_goal.iloc[lst_c], \n bias_factor,ridge=False))\n else:\n cluster_regressions.append(None)\n\n valid_clusters = [find_cluster(i[1],cluster_stats) for i in valid.iterrows()]\n\n y_pred = []\n for i,d in enumerate(valid.iterrows()):\n if not cluster_regressions[valid_clusters[i]] is None:\n cluster_prediction = pickle.loads((cluster_regressions[valid_clusters[i]])[0]).predict(d[1].values.reshape(1,-1))\n overall_prediction = overall_model.predict(d[1].values.reshape(1,-1))\n cluster_factor = 1 / (2 + 5 * math.exp(-len(clusters[valid_clusters[i]])/100))\n y_pred.append(cluster_prediction*cluster_factor + overall_prediction*(1-cluster_factor))\n else:\n y_pred.append(overall_model.predict(d[1].values.reshape(1,-1)))\n \n \n total_ensemble_SSE = mean_squared_error(valid_goal.values, y_pred)\n total_ensemble_r2 = r2_score(valid_goal.values, y_pred)\n \n if not silent:\n print(\"Overall weak, SSE, r2:\")\n print(overall_weak_scores)\n print(\"Ensemble model without clustering SSE: %f\" % overall_SSE)\n print(\"Ensemble model without clustering r2: %0.4f\" % overall_r2)\n print(\"Total Ensemble model SSE: %f\" % total_ensemble_SSE)\n print(\"Total Ensemble model r2: %0.4f\" % total_ensemble_r2)\n print(\"\")\n \n ",
"_____no_output_____"
],
[
"print(get_regression_performance(regress_overall_stacked,(data[final_features], data['SalePrice'], 0.2),iterations=5,bias_factor=1)):",
"_____no_output_____"
],
[
"print(get_regression_performance(regress_by_cluster,(data[final_features], data['SalePrice'], 0.2),iterations=5,bias_factor=1):",
"190325167167.65564\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e78fe0cc15d5026749edc6d330aa59ded94a3ac4 | 51,820 | ipynb | Jupyter Notebook | assignments/assignment1/Linear classifier.ipynb | DANTEpolaris/dlcourse_ai | 66604c9c7182c46e5b38d3650410c69ce85d931a | [
"MIT"
] | 3 | 2019-04-05T17:48:15.000Z | 2020-01-09T12:41:45.000Z | assignments/assignment1/Linear classifier.ipynb | DANTEpolaris/dlcourse_ai | 66604c9c7182c46e5b38d3650410c69ce85d931a | [
"MIT"
] | null | null | null | assignments/assignment1/Linear classifier.ipynb | DANTEpolaris/dlcourse_ai | 66604c9c7182c46e5b38d3650410c69ce85d931a | [
"MIT"
] | null | null | null | 57.196468 | 15,764 | 0.717734 | [
[
[
"# Задание 1.2 - Линейный классификатор (Linear classifier)\n\nВ этом задании мы реализуем другую модель машинного обучения - линейный классификатор. Линейный классификатор подбирает для каждого класса веса, на которые нужно умножить значение каждого признака и потом сложить вместе.\nТот класс, у которого эта сумма больше, и является предсказанием модели.\n\nВ этом задании вы:\n- потренируетесь считать градиенты различных многомерных функций\n- реализуете подсчет градиентов через линейную модель и функцию потерь softmax\n- реализуете процесс тренировки линейного классификатора\n- подберете параметры тренировки на практике\n\nНа всякий случай, еще раз ссылка на туториал по numpy: \nhttp://cs231n.github.io/python-numpy-tutorial/",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n\n%matplotlib inline\n\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"from dataset import load_svhn, random_split_train_val\nfrom gradient_check import check_gradient\nfrom metrics import multiclass_accuracy \nimport linear_classifer",
"_____no_output_____"
]
],
[
[
"# Как всегда, первым делом загружаем данные\n\nМы будем использовать все тот же SVHN.",
"_____no_output_____"
]
],
[
[
"def prepare_for_linear_classifier(train_X, test_X):\n train_flat = train_X.reshape(train_X.shape[0], -1).astype(np.float) / 255.0\n test_flat = test_X.reshape(test_X.shape[0], -1).astype(np.float) / 255.0\n \n # Subtract mean\n mean_image = np.mean(train_flat, axis = 0)\n train_flat -= mean_image\n test_flat -= mean_image\n \n # Add another channel with ones as a bias term\n train_flat_with_ones = np.hstack([train_flat, np.ones((train_X.shape[0], 1))])\n test_flat_with_ones = np.hstack([test_flat, np.ones((test_X.shape[0], 1))]) \n return train_flat_with_ones, test_flat_with_ones\n \ntrain_X, train_y, test_X, test_y = load_svhn(\"data\", max_train=10000, max_test=1000) \ntrain_X, test_X = prepare_for_linear_classifier(train_X, test_X)\n# Split train into train and val\ntrain_X, train_y, val_X, val_y = random_split_train_val(train_X, train_y, num_val = 1000)",
"_____no_output_____"
]
],
[
[
"# Играемся с градиентами!\n\nВ этом курсе мы будем писать много функций, которые вычисляют градиенты аналитическим методом.\n\nВсе функции, в которых мы будем вычислять градиенты будут написаны по одной и той же схеме. \nОни будут получать на вход точку, где нужно вычислить значение и градиент функции, а на выходе будут выдавать кортеж (tuple) из двух значений - собственно значения функции в этой точке (всегда одно число) и аналитического значения градиента в той же точке (той же размерности, что и вход).\n```\ndef f(x):\n \"\"\"\n Computes function and analytic gradient at x\n \n x: np array of float, input to the function\n \n Returns:\n value: float, value of the function \n grad: np array of float, same shape as x\n \"\"\"\n ...\n \n return value, grad\n```\n\nНеобходимым инструментом во время реализации кода, вычисляющего градиенты, является функция его проверки. Эта функция вычисляет градиент численным методом и сверяет результат с градиентом, вычисленным аналитическим методом.\n\nМы начнем с того, чтобы реализовать вычисление численного градиента (numeric gradient) в функции `check_gradient` в `gradient_check.py`. Эта функция будет принимать на вход функции формата, заданного выше, использовать значение `value` для вычисления численного градиента и сравнит его с аналитическим - они должны сходиться.\n\nНапишите часть функции, которая вычисляет градиент с помощью численной производной для каждой координаты. Для вычисления производной используйте так называемую two-point formula (https://en.wikipedia.org/wiki/Numerical_differentiation):\n\n\n\nВсе функции приведенные в следующей клетке должны проходить gradient check.",
"_____no_output_____"
]
],
[
[
"# TODO: Implement check_gradient function in gradient_check.py\n# All the functions below should pass the gradient check\n\ndef square(x):\n return float(x*x), 2*x\n\ncheck_gradient(square, np.array([3.0]))\n\ndef array_sum(x):\n assert x.shape == (2,), x.shape\n return np.sum(x), np.ones_like(x)\n\ncheck_gradient(array_sum, np.array([3.0, 2.0]))\n\ndef array_2d_sum(x):\n assert x.shape == (2,2)\n return np.sum(x), np.ones_like(x)\n\ncheck_gradient(array_2d_sum, np.array([[3.0, 2.0], [1.0, 0.0]]))",
"Gradient check passed!\nGradient check passed!\nGradient check passed!\n"
]
],
[
[
"## Начинаем писать свои функции, считающие аналитический градиент\n\nТеперь реализуем функцию softmax, которая получает на вход оценки для каждого класса и преобразует их в вероятности от 0 до 1:\n\n\n**Важно:** Практический аспект вычисления этой функции заключается в том, что в ней учавствует вычисление экспоненты от потенциально очень больших чисел - это может привести к очень большим значениям в числителе и знаменателе за пределами диапазона float.\n\nК счастью, у этой проблемы есть простое решение -- перед вычислением softmax вычесть из всех оценок максимальное значение среди всех оценок:\n```\npredictions -= np.max(predictions)\n```\n(подробнее здесь - http://cs231n.github.io/linear-classify/#softmax, секция `Practical issues: Numeric stability`)",
"_____no_output_____"
]
],
[
[
"# TODO Implement softmax and cross-entropy for single sample\nprobs = linear_classifer.softmax(np.array([-10, 0, 10]))\n\n# Make sure it works for big numbers too!\nprobs = linear_classifer.softmax(np.array([1000, 0, 0]))\nassert np.isclose(probs[0], 1.0)",
"_____no_output_____"
]
],
[
[
"Кроме этого, мы реализуем cross-entropy loss, которую мы будем использовать как функцию ошибки (error function).\nВ общем виде cross-entropy определена следующим образом:\n\n\nгде x - все классы, p(x) - истинная вероятность принадлежности сэмпла классу x, а q(x) - вероятность принадлежности классу x, предсказанная моделью. \nВ нашем случае сэмпл принадлежит только одному классу, индекс которого передается функции. Для него p(x) равна 1, а для остальных классов - 0. \n\nЭто позволяет реализовать функцию проще!",
"_____no_output_____"
]
],
[
[
"probs = linear_classifer.softmax(np.array([-5, 0, 5]))\ndisplay(probs)\nlinear_classifer.cross_entropy_loss(probs, 1)",
"_____no_output_____"
]
],
[
[
"После того как мы реализовали сами функции, мы можем реализовать градиент.\n\nОказывается, что вычисление градиента становится гораздо проще, если объединить эти функции в одну, которая сначала вычисляет вероятности через softmax, а потом использует их для вычисления функции ошибки через cross-entropy loss.\n\nЭта функция `softmax_with_cross_entropy` будет возвращает и значение ошибки, и градиент по входным параметрам. Мы проверим корректность реализации с помощью `check_gradient`.",
"_____no_output_____"
]
],
[
[
"# TODO Implement combined function or softmax and cross entropy and produces gradient\nloss, grad = linear_classifer.softmax_with_cross_entropy(np.array([1, 0, 0]), 1)\ncheck_gradient(lambda x: linear_classifer.softmax_with_cross_entropy(x, 1), np.array([1, 0, 0], np.float))",
"Gradient check passed!\n"
]
],
[
[
"В качестве метода тренировки мы будем использовать стохастический градиентный спуск (stochastic gradient descent или SGD), который работает с батчами сэмплов. \n\nПоэтому все наши фукнции будут получать не один пример, а батч, то есть входом будет не вектор из `num_classes` оценок, а матрица размерности `batch_size, num_classes`. Индекс примера в батче всегда будет первым измерением.\n\nСледующий шаг - переписать наши функции так, чтобы они поддерживали батчи.\n\nФинальное значение функции ошибки должно остаться числом, и оно равно среднему значению ошибки среди всех примеров в батче.",
"_____no_output_____"
],
[
"### Наконец, реализуем сам линейный классификатор!\n\nsoftmax и cross-entropy получают на вход оценки, которые выдает линейный классификатор.\n\nОн делает это очень просто: для каждого класса есть набор весов, на которые надо умножить пиксели картинки и сложить. Получившееся число и является оценкой класса, идущей на вход softmax.\n\nТаким образом, линейный классификатор можно представить как умножение вектора с пикселями на матрицу W размера `num_features, num_classes`. Такой подход легко расширяется на случай батча векторов с пикселями X размера `batch_size, num_features`:\n\n`predictions = X * W`, где `*` - матричное умножение.\n\nРеализуйте функцию подсчета линейного классификатора и градиентов по весам `linear_softmax` в файле `linear_classifer.py`",
"_____no_output_____"
]
],
[
[
"# TODO Extend combined function so it can receive a 2d array with batch of samples\nnp.random.seed(42)\n# Test batch_size = 1\nnum_classes = 4\nbatch_size = 1\npredictions = np.random.randint(-1, 3, size=(num_classes, batch_size)).astype(np.float)\ntarget_index = np.random.randint(0, num_classes, size=(batch_size, 1)).astype(np.int)\ncheck_gradient(lambda x: linear_classifer.softmax_with_cross_entropy(x, target_index), predictions)\n\n# Test batch_size = 3\nnum_classes = 4\nbatch_size = 3\npredictions = np.random.randint(-1, 3, size=(num_classes, batch_size)).astype(np.float)\ntarget_index = np.random.randint(0, num_classes, size=(batch_size, 1)).astype(np.int)\ncheck_gradient(lambda x: linear_classifer.softmax_with_cross_entropy(x, target_index), predictions)",
"Gradient check passed!\nGradient check passed!\n"
]
],
[
[
"### И теперь регуляризация\n\nМы будем использовать L2 regularization для весов как часть общей функции ошибки.\n\nНапомним, L2 regularization определяется как\n\nl2_reg_loss = regularization_strength * sum<sub>ij</sub> W[i, j]<sup>2</sup>\n\nРеализуйте функцию для его вычисления и вычисления соотвествующих градиентов.",
"_____no_output_____"
]
],
[
[
"# TODO Implement linear_softmax function that uses softmax with cross-entropy for linear classifier\nbatch_size = 2\nnum_classes = 2\nnum_features = 3\nnp.random.seed(42)\nW = np.random.randint(-1, 3, size=(num_features, num_classes)).astype(np.float)\nX = np.random.randint(-1, 3, size=(batch_size, num_features)).astype(np.float)\ntarget_index = np.ones(batch_size, dtype=np.int)\n\nloss, dW = linear_classifer.linear_softmax(X, W, target_index)\ncheck_gradient(lambda w: linear_classifer.linear_softmax(X, w, target_index), W)",
"Gradient check passed!\n"
],
[
"# TODO Implement l2_regularization function that implements loss for L2 regularization\nlinear_classifer.l2_regularization(W, 0.01)\ncheck_gradient(lambda w: linear_classifer.l2_regularization(w, 0.01), W)",
"Gradient check passed!\n"
]
],
[
[
"# Тренировка!",
"_____no_output_____"
],
[
"Градиенты в порядке, реализуем процесс тренировки!",
"_____no_output_____"
]
],
[
[
"# TODO: Implement LinearSoftmaxClassifier.fit function\nclassifier = linear_classifer.LinearSoftmaxClassifier()\nloss_history = classifier.fit(train_X, train_y, epochs=10, learning_rate=1e-3, batch_size=300, reg=1e1)",
"Epoch 0, loss: 2.609816\nEpoch 1, loss: 2.609771\nEpoch 2, loss: 2.609726\nEpoch 3, loss: 2.609682\nEpoch 4, loss: 2.609639\nEpoch 5, loss: 2.609597\nEpoch 6, loss: 2.609556\nEpoch 7, loss: 2.609516\nEpoch 8, loss: 2.609476\nEpoch 9, loss: 2.609438\n"
],
[
"# let's look at the loss history!\nplt.plot(loss_history)",
"_____no_output_____"
],
[
"# Let's check how it performs on validation set\npred = classifier.predict(val_X)\naccuracy = multiclass_accuracy(pred, val_y)\nprint(\"Accuracy: \", accuracy)\n\n# Now, let's train more and see if it performs better\nclassifier.fit(train_X, train_y, epochs=100, learning_rate=1e-3, batch_size=300, reg=1e1)\npred = classifier.predict(val_X)\naccuracy = multiclass_accuracy(pred, val_y)\nprint(\"Accuracy after training for 100 epochs: \", accuracy)",
"Accuracy: 0.09399999999999997\nEpoch 0, loss: 2.609400\nEpoch 1, loss: 2.609363\nEpoch 2, loss: 2.609327\nEpoch 3, loss: 2.609292\nEpoch 4, loss: 2.609257\nEpoch 5, loss: 2.609224\nEpoch 6, loss: 2.609191\nEpoch 7, loss: 2.609159\nEpoch 8, loss: 2.609128\nEpoch 9, loss: 2.609097\nEpoch 10, loss: 2.609068\nEpoch 11, loss: 2.609039\nEpoch 12, loss: 2.609011\nEpoch 13, loss: 2.608983\nEpoch 14, loss: 2.608957\nEpoch 15, loss: 2.608931\nEpoch 16, loss: 2.608906\nEpoch 17, loss: 2.608882\nEpoch 18, loss: 2.608859\nEpoch 19, loss: 2.608836\nEpoch 20, loss: 2.608814\nEpoch 21, loss: 2.608793\nEpoch 22, loss: 2.608773\nEpoch 23, loss: 2.608753\nEpoch 24, loss: 2.608735\nEpoch 25, loss: 2.608717\nEpoch 26, loss: 2.608699\nEpoch 27, loss: 2.608683\nEpoch 28, loss: 2.608667\nEpoch 29, loss: 2.608652\nEpoch 30, loss: 2.608637\nEpoch 31, loss: 2.608624\nEpoch 32, loss: 2.608611\nEpoch 33, loss: 2.608599\nEpoch 34, loss: 2.608587\nEpoch 35, loss: 2.608577\nEpoch 36, loss: 2.608567\nEpoch 37, loss: 2.608557\nEpoch 38, loss: 2.608549\nEpoch 39, loss: 2.608541\nEpoch 40, loss: 2.608534\nEpoch 41, loss: 2.608527\nEpoch 42, loss: 2.608521\nEpoch 43, loss: 2.608516\nEpoch 44, loss: 2.608512\nEpoch 45, loss: 2.608508\nEpoch 46, loss: 2.608505\nEpoch 47, loss: 2.608503\nEpoch 48, loss: 2.608502\nEpoch 49, loss: 2.608501\nEpoch 50, loss: 2.608501\nEpoch 51, loss: 2.608501\nEpoch 52, loss: 2.608502\nEpoch 53, loss: 2.608504\nEpoch 54, loss: 2.608507\nEpoch 55, loss: 2.608510\nEpoch 56, loss: 2.608514\nEpoch 57, loss: 2.608518\nEpoch 58, loss: 2.608524\nEpoch 59, loss: 2.608530\nEpoch 60, loss: 2.608536\nEpoch 61, loss: 2.608543\nEpoch 62, loss: 2.608551\nEpoch 63, loss: 2.608560\nEpoch 64, loss: 2.608569\nEpoch 65, loss: 2.608579\nEpoch 66, loss: 2.608590\nEpoch 67, loss: 2.608601\nEpoch 68, loss: 2.608613\nEpoch 69, loss: 2.608625\nEpoch 70, loss: 2.608639\nEpoch 71, loss: 2.608652\nEpoch 72, loss: 2.608667\nEpoch 73, loss: 2.608682\nEpoch 74, loss: 2.608698\nEpoch 75, loss: 2.608714\nEpoch 76, loss: 2.608731\nEpoch 77, loss: 2.608749\nEpoch 78, loss: 2.608767\nEpoch 79, loss: 2.608786\nEpoch 80, loss: 2.608806\nEpoch 81, loss: 2.608826\nEpoch 82, loss: 2.608847\nEpoch 83, loss: 2.608869\nEpoch 84, loss: 2.608891\nEpoch 85, loss: 2.608914\nEpoch 86, loss: 2.608937\nEpoch 87, loss: 2.608961\nEpoch 88, loss: 2.608986\nEpoch 89, loss: 2.609011\nEpoch 90, loss: 2.609037\nEpoch 91, loss: 2.609064\nEpoch 92, loss: 2.609091\nEpoch 93, loss: 2.609119\nEpoch 94, loss: 2.609147\nEpoch 95, loss: 2.609176\nEpoch 96, loss: 2.609206\nEpoch 97, loss: 2.609236\nEpoch 98, loss: 2.609267\nEpoch 99, loss: 2.609299\nAccuracy after training for 100 epochs: 0.125\n"
]
],
[
[
"### Как и раньше, используем кросс-валидацию для подбора гиперпараметтов.\n\nВ этот раз, чтобы тренировка занимала разумное время, мы будем использовать только одно разделение на тренировочные (training) и проверочные (validation) данные.\n\nТеперь нам нужно подобрать не один, а два гиперпараметра! Не ограничивайте себя изначальными значениями в коде. \nДобейтесь точности более чем **20%** на проверочных данных (validation data).",
"_____no_output_____"
]
],
[
[
"num_epochs = 200\nbatch_size = 300\n\nlearning_rates = [1e-1, 1e-2, 1e-3, 1e-4, 1e-5]\nreg_strengths = [1e-3, 1e-2, 1e-4, 1e-5, 1e-6, 1e-7]\nbest_val_accuracy = 0\nfor learning_rate in learning_rates:\n for reg_strength in reg_strengths:\n classifier.fit(train_X, train_y, batch_size, learning_rate, reg_strength, num_epochs)\n pred = classifier.predict(val_X)\n accuracy = multiclass_accuracy(pred, val_y)\n print('Accuracy %f reg_strength %f learning rate %f' % (accuracy, reg_strength, learning_rate))\n print('Accuracy %f reg_strength %f learning rate %f' % (accuracy, reg_strength, learning_rate), file=open(\"accuracy.txt\", \"a\"))\n best_val_accuracy = max(best_val_accuracy, accuracy)\n \nbest_classifier = None\n# TODO use validation set to find the best hyperparameters\n# hint: for best results, you might need to try more values for learning rate and regularization strength \n# than provided initially\n\nprint('best validation accuracy achieved: %f' % best_val_accuracy)",
"Epoch 0, loss: 2.299253\nEpoch 1, loss: 2.296205\nEpoch 2, loss: 2.293332\nEpoch 3, loss: 2.290550\nEpoch 4, loss: 2.287845\nEpoch 5, loss: 2.285214\nEpoch 6, loss: 2.282651\nEpoch 7, loss: 2.280155\nEpoch 8, loss: 2.277721\nEpoch 9, loss: 2.275346\nEpoch 10, loss: 2.273029\nEpoch 11, loss: 2.270765\nEpoch 12, loss: 2.268554\nEpoch 13, loss: 2.266393\nEpoch 14, loss: 2.264280\nEpoch 15, loss: 2.262214\nEpoch 16, loss: 2.260192\nEpoch 17, loss: 2.258214\nEpoch 18, loss: 2.256277\nEpoch 19, loss: 2.254381\nEpoch 20, loss: 2.252524\nEpoch 21, loss: 2.250705\nEpoch 22, loss: 2.248923\nEpoch 23, loss: 2.247177\nEpoch 24, loss: 2.245466\nEpoch 25, loss: 2.243788\nEpoch 26, loss: 2.242144\nEpoch 27, loss: 2.240532\nEpoch 28, loss: 2.238952\nEpoch 29, loss: 2.237402\nEpoch 30, loss: 2.235882\nEpoch 31, loss: 2.234391\nEpoch 32, loss: 2.232929\nEpoch 33, loss: 2.231495\nEpoch 34, loss: 2.230087\nEpoch 35, loss: 2.228706\nEpoch 36, loss: 2.227352\nEpoch 37, loss: 2.226022\nEpoch 38, loss: 2.224717\nEpoch 39, loss: 2.223437\nEpoch 40, loss: 2.222180\nEpoch 41, loss: 2.220946\nEpoch 42, loss: 2.219734\nEpoch 43, loss: 2.218545\nEpoch 44, loss: 2.217378\nEpoch 45, loss: 2.216231\nEpoch 46, loss: 2.215105\nEpoch 47, loss: 2.214000\nEpoch 48, loss: 2.212914\nEpoch 49, loss: 2.211847\nEpoch 50, loss: 2.210800\nEpoch 51, loss: 2.209770\nEpoch 52, loss: 2.208759\nEpoch 53, loss: 2.207766\nEpoch 54, loss: 2.206789\nEpoch 55, loss: 2.205830\nEpoch 56, loss: 2.204887\nEpoch 57, loss: 2.203960\nEpoch 58, loss: 2.203049\nEpoch 59, loss: 2.202154\nEpoch 60, loss: 2.201273\nEpoch 61, loss: 2.200408\nEpoch 62, loss: 2.199556\nEpoch 63, loss: 2.198719\nEpoch 64, loss: 2.197896\nEpoch 65, loss: 2.197086\nEpoch 66, loss: 2.196289\nEpoch 67, loss: 2.195505\nEpoch 68, loss: 2.194734\nEpoch 69, loss: 2.193975\nEpoch 70, loss: 2.193228\nEpoch 71, loss: 2.192493\nEpoch 72, loss: 2.191770\nEpoch 73, loss: 2.191058\nEpoch 74, loss: 2.190357\nEpoch 75, loss: 2.189666\nEpoch 76, loss: 2.188987\nEpoch 77, loss: 2.188317\nEpoch 78, loss: 2.187658\nEpoch 79, loss: 2.187008\nEpoch 80, loss: 2.186369\nEpoch 81, loss: 2.185738\nEpoch 82, loss: 2.185117\nEpoch 83, loss: 2.184505\nEpoch 84, loss: 2.183902\nEpoch 85, loss: 2.183308\nEpoch 86, loss: 2.182722\nEpoch 87, loss: 2.182144\nEpoch 88, loss: 2.181574\nEpoch 89, loss: 2.181013\nEpoch 90, loss: 2.180459\nEpoch 91, loss: 2.179913\nEpoch 92, loss: 2.179374\nEpoch 93, loss: 2.178842\nEpoch 94, loss: 2.178318\nEpoch 95, loss: 2.177801\nEpoch 96, loss: 2.177290\nEpoch 97, loss: 2.176786\nEpoch 98, loss: 2.176289\nEpoch 99, loss: 2.175799\nEpoch 100, loss: 2.175314\nEpoch 101, loss: 2.174836\nEpoch 102, loss: 2.174363\nEpoch 103, loss: 2.173897\nEpoch 104, loss: 2.173437\nEpoch 105, loss: 2.172982\nEpoch 106, loss: 2.172532\nEpoch 107, loss: 2.172089\nEpoch 108, loss: 2.171650\nEpoch 109, loss: 2.171217\nEpoch 110, loss: 2.170789\nEpoch 111, loss: 2.170366\nEpoch 112, loss: 2.169947\nEpoch 113, loss: 2.169534\nEpoch 114, loss: 2.169125\nEpoch 115, loss: 2.168722\nEpoch 116, loss: 2.168322\nEpoch 117, loss: 2.167927\nEpoch 118, loss: 2.167537\nEpoch 119, loss: 2.167151\nEpoch 120, loss: 2.166769\nEpoch 121, loss: 2.166391\nEpoch 122, loss: 2.166017\nEpoch 123, loss: 2.165647\nEpoch 124, loss: 2.165281\nEpoch 125, loss: 2.164919\nEpoch 126, loss: 2.164561\nEpoch 127, loss: 2.164206\nEpoch 128, loss: 2.163855\nEpoch 129, loss: 2.163508\nEpoch 130, loss: 2.163164\nEpoch 131, loss: 2.162824\nEpoch 132, loss: 2.162486\nEpoch 133, loss: 2.162153\nEpoch 134, loss: 2.161822\nEpoch 135, loss: 2.161495\nEpoch 136, loss: 2.161171\nEpoch 137, loss: 2.160850\nEpoch 138, loss: 2.160532\nEpoch 139, loss: 2.160217\nEpoch 140, loss: 2.159904\nEpoch 141, loss: 2.159595\nEpoch 142, loss: 2.159289\nEpoch 143, loss: 2.158985\nEpoch 144, loss: 2.158684\nEpoch 145, loss: 2.158386\nEpoch 146, loss: 2.158090\nEpoch 147, loss: 2.157797\n"
]
],
[
[
"# Какой же точности мы добились на тестовых данных?",
"_____no_output_____"
]
],
[
[
"test_pred = best_classifier.predict(test_X)\ntest_accuracy = multiclass_accuracy(test_pred, test_y)\nprint('Linear softmax classifier test set accuracy: %f' % (test_accuracy, ))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e78fe5bc8ca3a1eb96d4d39bec95975a19ce4a0c | 22,191 | ipynb | Jupyter Notebook | Examples/Overview.ipynb | IbHansen/Modelflow2 | 48c5a5c13746650c37d8af36250fd35cdd40b05b | [
"X11"
] | 1 | 2020-11-11T22:58:58.000Z | 2020-11-11T22:58:58.000Z | Examples/Overview.ipynb | IbHansen/Modelflow2 | 48c5a5c13746650c37d8af36250fd35cdd40b05b | [
"X11"
] | null | null | null | Examples/Overview.ipynb | IbHansen/Modelflow2 | 48c5a5c13746650c37d8af36250fd35cdd40b05b | [
"X11"
] | 1 | 2022-01-16T17:19:56.000Z | 2022-01-16T17:19:56.000Z | 31.079832 | 257 | 0.524537 | [
[
[
"# This notebook links to **ModelFlow** models and examples.\n\nPlease have patience until the notebook had been loaded and executed. ",
"_____no_output_____"
],
[
"# ModelFlow Abstract\nModelFlow is a Python toolkit which can handle a wide range of models from small to huge. This covers: on-boarding a model, analyze the logical structure, solve the model, analyze and compare the results. \n\nModelflow is located on Github [here](https://github.com/IbHansen/ModelFlow2). The repo includes some models implemented in ModelFlow: \n - FRB/US - Federal Reserve Board \n - Q-JEM - Bank of Japan\n - ADAM - Statistics Denmark\n - and other models. \n \nThe toolkit is fast, lean, and agile.\nModels and results can be analyzed and visualized using the full arsenal of Python data science tools. \nThis is achieved by leveraging on the Python ecosystem (especially the [Pandas](https://pandas.pydata.org/) and the [numpy](https://www.nature.com/articles/s41586-020-2649-2) libraries. \n**A model i this context is a system of non-linear equations** specified either as:\n\n - a general model: $\\textbf{0} = \\textbf{G}(\\textbf{y}_{t+u} \\cdots \\textbf{y}_t \\cdots \\textbf{y}_{t-r},\\textbf{x}_t \\cdots \\textbf{x}_{t-s})$ \n - or as a normalized model: $\\textbf{y}_t = \\textbf{F}(\\textbf{y}_{t+u} \\cdots \\textbf{y}_t \\cdots \\textbf{y}_{t-r},\\textbf{x}_t \\cdots \\textbf{x}_{t-s})$ \n \nMany stress test, liquidity, macro or other models conforms to this pattern - or can easy be made to conform.\n\nSimultaneous models can be solved by\n\n - Gauss-Seidle or Newton-Raphson algorithmes if no lead\n - Fair-Taylor or stacked Newton-Raphson if the model contains leads and lags\n - The Jacobian matrice used by Newton-Raphson solvers will be calculated either by symbolic or by numerical differentiation and then handled by sparse matrix libraries. \n \nNon-simultaneous models will be sequenced and calculated. \n \n**Models can be specified in a Business logic language**. The user can concentrate on the economic content. To specify models incorporating many banks or sectors a **Macro-business logic language** is part of the library. \n\nCreating a (Macro prudential) model often entails implementing models specified in different ways: Excel, Latex, Eviews, Dynare, Python or other languages. Python''s ecosystem makes it possible to transform such model specification into ModelFlow. \n\nThe core function of the library is a transpiler which first parse and analyze a model specified in the Business logic language then generates Python code which can solved the model. \n\nJupyter notebooks with the examples can be run on a Binder virtual machine [here](https://mybinder.org/v2/gh/Ibhansen/modelflow2/tobinder?filepath=Examples%2FOverview.ipynb). Which is probably where you are just now. ",
"_____no_output_____"
]
],
[
[
"from modelclass import model\nmodel.modelflow_auto()",
"_____no_output_____"
]
],
[
[
"# Gallery \nBelow you will find links Jupyter notebooks using ModelFlow to run different models. The purpose of the notebooks are primarily to illustrate how ModelFlow can be used to manage a fairly large range of models and to show some of the capabilities. ",
"_____no_output_____"
]
],
[
[
"model.display_toc()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e78ff142af7e8e8b9f886b1ab86c9a7eb5f685d1 | 22,566 | ipynb | Jupyter Notebook | notebooks/Usage_Filters.ipynb | binaryflesh/ATTACK-Python-Client | 1328878eaa9ea944505153aa025304e215572fff | [
"BSD-3-Clause"
] | 3 | 2020-08-18T20:15:34.000Z | 2020-12-21T14:33:38.000Z | notebooks/Usage_Filters.ipynb | Azrara/ATTACK-Python-Client | b851afcd7065186602e05782ad20a397beb2b31b | [
"BSD-3-Clause"
] | null | null | null | notebooks/Usage_Filters.ipynb | Azrara/ATTACK-Python-Client | b851afcd7065186602e05782ad20a397beb2b31b | [
"BSD-3-Clause"
] | 1 | 2021-04-15T17:18:28.000Z | 2021-04-15T17:18:28.000Z | 47.607595 | 3,398 | 0.675618 | [
[
[
"# **MITRE ATT&CK API FILTERS**: Python Client\n------------------",
"_____no_output_____"
],
[
"## Import ATTACK API Client",
"_____no_output_____"
]
],
[
[
"from attackcti import attack_client",
"_____no_output_____"
]
],
[
[
"## Import Extra Libraries",
"_____no_output_____"
]
],
[
[
"from pandas import *\nfrom pandas.io.json import json_normalize",
"_____no_output_____"
]
],
[
[
"## Initialize ATT&CK Client Variable",
"_____no_output_____"
]
],
[
[
"lift = attack_client()",
"_____no_output_____"
]
],
[
[
"## Get Technique by Name (TAXII)\nYou can use a custom method in the attack_client class to get a technique across all the matrices by its name. It is case sensitive.",
"_____no_output_____"
]
],
[
[
"technique_name = lift.get_technique_by_name('Rundll32')",
"_____no_output_____"
],
[
"technique_name",
"_____no_output_____"
]
],
[
[
"## Get Data Sources from All Techniques (TAXII)\n* You can also get all the data sources available in ATT&CK\n* Currently the only techniques with data sources are the ones in Enterprise ATT&CK.",
"_____no_output_____"
]
],
[
[
"data_sources = lift.get_data_sources()",
"_____no_output_____"
],
[
"len(data_sources)",
"_____no_output_____"
],
[
"data_sources",
"_____no_output_____"
]
],
[
[
"## Get Any STIX Object by ID (TAXII)\n* You can get any STIX object by its id across all the matrices. It is case sensitive.\n* You can use the following STIX Object Types:\n * attack-pattern > techniques\n * course-of-action > mitigations\n * intrusion-set > groups\n * malware\n * tool",
"_____no_output_____"
]
],
[
[
"object_by_id = lift.get_object_by_attack_id('attack-pattern', 'T1307')",
"_____no_output_____"
],
[
"object_by_id",
"_____no_output_____"
]
],
[
[
"## Get Any Group by Alias (TAXII)\nYou can get any Group by its Alias property across all the matrices. It is case sensitive.",
"_____no_output_____"
]
],
[
[
"group_name = lift.get_group_by_alias('Cozy Bear')",
"_____no_output_____"
],
[
"group_name",
"_____no_output_____"
]
],
[
[
"## Get Relationships by Any Object (TAXII)\n* You can get available relationships defined in ATT&CK of type **uses** and **mitigates** for specific objects across all the matrices.",
"_____no_output_____"
]
],
[
[
"groups = lift.get_groups()\none_group = groups[0]\nrelationships = lift.get_relationships_by_object(one_group)",
"_____no_output_____"
],
[
"relationships[0]",
"_____no_output_____"
]
],
[
[
"## Get All Techniques with Mitigations (TAXII)\nThe difference with this function and **get_all_techniques()** is that **get_techniques_mitigated_by_all_mitigations** returns techniques that have mitigations mapped to them.",
"_____no_output_____"
]
],
[
[
"techniques_mitigated = lift.get_techniques_mitigated_by_all_mitigations()",
"_____no_output_____"
],
[
"techniques_mitigated[0]",
"_____no_output_____"
]
],
[
[
"## Get Techniques Used by Software (TAXII)\nThis the function returns information about a specific software STIX object.",
"_____no_output_____"
]
],
[
[
"all_software = lift.get_software()\none_software = all_software[0]\nsoftware_techniques = lift.get_techniques_used_by_software(one_software)",
"_____no_output_____"
],
[
"software_techniques[0]",
"_____no_output_____"
]
],
[
[
"## Get Techniques Used by Group (TAXII)\nIf you do not provide the name of a specific **Group** (Case Sensitive), the function returns information about all the groups available across all the matrices.",
"_____no_output_____"
]
],
[
[
"groups = lift.get_groups()\none_group = groups[0]\ngroup_techniques = lift.get_techniques_used_by_group(one_group)",
"_____no_output_____"
],
[
"group_techniques[0]",
"_____no_output_____"
]
],
[
[
"## Get Software Used by Group (TAXII)\nYou can retrieve every software (malware or tool) mapped to a specific Group STIX object",
"_____no_output_____"
]
],
[
[
"groups = lift.get_groups()\none_group = groups[0]\ngroup_software = lift.get_software_used_by_group(one_group)",
"_____no_output_____"
],
[
"group_software[0]",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
e78ff712e73b28ee8c7b89bb378a69468e562d07 | 119,431 | ipynb | Jupyter Notebook | aqua/simulations_with_noise_and_measurement_error_mitigation.ipynb | lukasszz/qiskit-tutorials-community | 1cf11ad977aec8e0680e32c38110aa8cf3677e60 | [
"Apache-2.0"
] | 3 | 2020-09-12T01:50:58.000Z | 2021-05-09T20:52:19.000Z | aqua/simulations_with_noise_and_measurement_error_mitigation.ipynb | lukasszz/qiskit-tutorials-community | 1cf11ad977aec8e0680e32c38110aa8cf3677e60 | [
"Apache-2.0"
] | null | null | null | aqua/simulations_with_noise_and_measurement_error_mitigation.ipynb | lukasszz/qiskit-tutorials-community | 1cf11ad977aec8e0680e32c38110aa8cf3677e60 | [
"Apache-2.0"
] | 1 | 2019-09-06T08:54:17.000Z | 2019-09-06T08:54:17.000Z | 294.165025 | 35,348 | 0.919485 | [
[
[
"## _*Running simulations with noise and measurement error mitigation in Aqua*_\n\nThis notebook demonstrates using the [Qiskit Aer](https://qiskit.org/aer) `qasm_simulator` to run a simulation with noise, based on a noise model, in Aqua. This can be useful to investigate behavior under different noise conditions. Aer not only allows you to define your own custom noise model, but also allows a noise model to be easily created based on the properties of a real quantum device. The latter is what this notebook will demonstrate since the goal is to show how to do this in Aqua not how to build custom noise models.\n\nOn the other hand, [Qiskit Ignis](https://qiskit.org/ignis) provides a solution to mitigate the measurement error when running on a noise simulation or a real quantum device.\n\nFurther information on Qiskit Aer noise model can be found in the online Qiskit Aer documentation [here](https://qiskit.org/documentation/aer/device_noise_simulation.html) as well as in the [Qiskit Aer tutorials](https://github.com/Qiskit/qiskit-tutorials/tree/master/qiskit/aer).\n\nFurther information on measurement error mitigation in Qiskit Ignis can be found in the [Qiskit Ignis tutorial](https://github.com/Qiskit/qiskit-tutorials/blob/master/qiskit/ignis/measurement_error_mitigation.ipynb).\n\nNote: this tutorial requires Qiskit Aer and Qiskit Ignis if you intend to run it. This can be installed using pip if you do not have it installed using `pip install qiskit-aer qiskit-ignis`",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pylab\n\nfrom qiskit import Aer, IBMQ\nfrom qiskit.aqua import QuantumInstance, aqua_globals\nfrom qiskit.aqua.algorithms.adaptive import VQE\nfrom qiskit.aqua.algorithms.classical import ExactEigensolver\nfrom qiskit.aqua.components.optimizers import SPSA\nfrom qiskit.aqua.components.variational_forms import RY\nfrom qiskit.aqua.operators import WeightedPauliOperator\n",
"_____no_output_____"
]
],
[
[
"Noisy simulation will be demonstrated here with VQE, finding the minimum (ground state) energy of an Hamiltonian, but the technique applies to any quantum algorithm from Aqua.\n\nSo for VQE we need a qubit operator as input. Here we will take a set of paulis that were originally computed by qiskit-chemistry, for an H2 molecule, so we can quickly create an Operator.",
"_____no_output_____"
]
],
[
[
"pauli_dict = {\n 'paulis': [{\"coeff\": {\"imag\": 0.0, \"real\": -1.052373245772859}, \"label\": \"II\"},\n {\"coeff\": {\"imag\": 0.0, \"real\": 0.39793742484318045}, \"label\": \"ZI\"},\n {\"coeff\": {\"imag\": 0.0, \"real\": -0.39793742484318045}, \"label\": \"IZ\"},\n {\"coeff\": {\"imag\": 0.0, \"real\": -0.01128010425623538}, \"label\": \"ZZ\"},\n {\"coeff\": {\"imag\": 0.0, \"real\": 0.18093119978423156}, \"label\": \"XX\"}\n ]\n}\n\nqubit_op = WeightedPauliOperator.from_dict(pauli_dict)\nnum_qubits = qubit_op.num_qubits\nprint('Number of qubits: {}'.format(num_qubits))",
"Number of qubits: 2\n"
]
],
[
[
"As the above problem is still easily tractable classically we can use ExactEigensolver to compute a reference value so we can compare later the results. \n\n<span style=\"font-size:0.9em\">_(A copy of the operator is used below as what is passed to ExactEigensolver will be converted to matrix form and we want the operator we use later, on the Aer qasm simuator, to be in paulis form.)_</span>",
"_____no_output_____"
]
],
[
[
"ee = ExactEigensolver(qubit_op.copy())\nresult = ee.run()\nref = result['energy']\nprint('Reference value: {}'.format(ref))",
"Reference value: -1.8572750302023797\n"
]
],
[
[
"### Performance *without* noise\n\nFirst we will run on the simulator without adding noise to see the result. I have created the backend and QuantumInstance, which holds the backend as well as various other run time configuration, which are defaulted here, so it easy to compare when we get to the next section where noise is added. There is no attempt to mitigate noise or anything in this notebook so the latter setup and running of VQE is identical.",
"_____no_output_____"
]
],
[
[
"backend = Aer.get_backend('qasm_simulator')\nquantum_instance = QuantumInstance(backend=backend, seed_simulator=167, seed_transpiler=167) \n\ncounts = []\nvalues = []\ndef store_intermediate_result(eval_count, parameters, mean, std):\n counts.append(eval_count)\n values.append(mean)\n\naqua_globals.random_seed = 167\noptimizer = SPSA(max_trials=200)\nvar_form = RY(num_qubits)\nvqe = VQE(qubit_op, var_form, optimizer, callback=store_intermediate_result)\nvqe_result = vqe.run(quantum_instance)\nprint('VQE on Aer qasm simulator (no noise): {}'.format(vqe_result['energy']))\nprint('Delta from reference: {}'.format(vqe_result['energy']-ref))",
"VQE on Aer qasm simulator (no noise): -1.8598749159580135\nDelta from reference: -0.0025998857556337462\n"
]
],
[
[
"We captured the energy values above during the convergence so we can see what went on in the graph below.",
"_____no_output_____"
]
],
[
[
"pylab.rcParams['figure.figsize'] = (12, 4)\npylab.plot(counts, values)\npylab.xlabel('Eval count')\npylab.ylabel('Energy')\npylab.title('Convergence with no noise');",
"_____no_output_____"
]
],
[
[
"### Performance *with* noise\n\nNow we will add noise. Here we will create a noise model for Aer from an actual device. You can create custom noise models with Aer but that goes beyond the scope of this notebook. Links to further information on Aer noise model, for those that may be interested in doing this, were given in instruction above.\n\nFirst we need to get an actual device backend and from its `configuration` and `properties` we can setup a coupling map and a noise model to match the device. While we could leave the simulator with the default all to all map, this shows how to set the coupling map too. Note: We can also use this coupling map as the entanglement map for the variational form if we choose.\n\nNote: simulation with noise takes significantly longer than without noise.",
"_____no_output_____"
]
],
[
[
"from qiskit.providers.aer import noise\n\nprovider = IBMQ.load_account()\ndevice = provider.get_backend('ibmqx4')\ncoupling_map = device.configuration().coupling_map\nnoise_model = noise.device.basic_device_noise_model(device.properties())\nbasis_gates = noise_model.basis_gates\n\nprint(noise_model)\n\nbackend = Aer.get_backend('qasm_simulator')\nquantum_instance = QuantumInstance(backend=backend, seed_simulator=167, seed_transpiler=167,\n noise_model=noise_model,)\n\ncounts1 = []\nvalues1 = []\ndef store_intermediate_result1(eval_count, parameters, mean, std):\n counts1.append(eval_count)\n values1.append(mean)\n\naqua_globals.random_seed = 167\noptimizer = SPSA(max_trials=200)\nvar_form = RY(num_qubits)\nvqe = VQE(qubit_op, var_form, optimizer, callback=store_intermediate_result1)\nvqe_result1 = vqe.run(quantum_instance)\nprint('VQE on Aer qasm simulator (with noise): {}'.format(vqe_result1['energy']))\nprint('Delta from reference: {}'.format(vqe_result1['energy']-ref))",
"NoiseModel:\n Basis gates: ['cx', 'id', 'u2', 'u3']\n Instructions with noise: ['u2', 'cx', 'measure', 'u3']\n Qubits with noise: [0, 1, 2, 3, 4]\n Specific qubit errors: [('u2', [0]), ('u2', [1]), ('u2', [2]), ('u2', [3]), ('u2', [4]), ('u3', [0]), ('u3', [1]), ('u3', [2]), ('u3', [3]), ('u3', [4]), ('cx', [1, 0]), ('cx', [2, 0]), ('cx', [2, 1]), ('cx', [3, 2]), ('cx', [3, 4]), ('cx', [4, 2]), ('measure', [0]), ('measure', [1]), ('measure', [2]), ('measure', [3]), ('measure', [4])]\nVQE on Aer qasm simulator (with noise): -1.6782227276010517\nDelta from reference: 0.17905230260132798\n"
],
[
"pylab.rcParams['figure.figsize'] = (12, 4)\npylab.plot(counts1, values1)\npylab.xlabel('Eval count')\npylab.ylabel('Energy')\npylab.title('Convergence with noise');",
"_____no_output_____"
]
],
[
[
"### Declarative approach and noise model\n\nNote: if you are running an experiment using the declarative approach, with a dictionary/json, there are keywords in the `backend` section that let you define the noise model based on a device, as well as setup the coupling map too. The basis gate setup, that is shown above, will automatically be done. Here is an example of such a `backend` configuration:\n```\n 'backend': { \n\t 'provider': 'qiskit.Aer',\n 'name': 'qasm_simulator',\n\t 'coupling_map_from_device': 'qiskit.IBMQ:ibmqx4',\n 'noise_model': 'qiskit.IBMQ:ibmqx4',\n 'shots': 1024\n\t },\n```\n\nIf you call `run_algorithm` and override the `backend` section by explicity supplying a backend instance as a parameter to run_algorithm, please note that you can provide a QuantumInstance type there instead of BaseBackend. A QuantumInstance allows you to setup and define your own custom noise model and other run time configuration. \n\n<span style=\"font-size:0.9em\">(Note when a BaseBackend type is supplied to run_algorithm it is internally wrapped into a QuantumInstance, with default values supplied for noise, run time parameters etc., so you do not get the opportunity that way to set a noise model etc. But by explicitly providing a QuantumInstance you can setup these aspects to your choosing.)</span>",
"_____no_output_____"
],
[
"### Performance *with* noise and measurement error mitigation\n\nNow we will add method for measurement error mitigation, which increases the fidelity of measurement. Here we choose `CompleteMeasFitter` to mitigate the measurement error. The calibration matrix will be auto-refresh every 30 minute (default value).\n\nNote: simulation with noise takes significantly longer than without noise.",
"_____no_output_____"
]
],
[
[
"from qiskit.ignis.mitigation.measurement import CompleteMeasFitter\n\nquantum_instance = QuantumInstance(backend=backend, seed_simulator=167, seed_transpiler=167,\n noise_model=noise_model, \n measurement_error_mitigation_cls=CompleteMeasFitter, \n cals_matrix_refresh_period=30)\n\ncounts1 = []\nvalues1 = []\ndef store_intermediate_result1(eval_count, parameters, mean, std):\n counts1.append(eval_count)\n values1.append(mean)\n\naqua_globals.random_seed = 167\noptimizer = SPSA(max_trials=200)\nvar_form = RY(num_qubits)\nvqe = VQE(qubit_op, var_form, optimizer, callback=store_intermediate_result1)\nvqe_result1 = vqe.run(quantum_instance)\nprint('VQE on Aer qasm simulator (with noise and measurement error mitigation): {}'.format(vqe_result1['energy']))\nprint('Delta from reference: {}'.format(vqe_result1['energy']-ref))",
"VQE on Aer qasm simulator (with noise and measurement error mitigation): -1.8551030559231287\nDelta from reference: 0.0021719742792509766\n"
],
[
"pylab.rcParams['figure.figsize'] = (12, 4)\npylab.plot(counts1, values1)\npylab.xlabel('Eval count')\npylab.ylabel('Energy')\npylab.title('Convergence with noise, enabling measurement error mitigation');",
"_____no_output_____"
]
],
[
[
"### Declarative approach of measurement error mitigation\n\nNote: if you are running an experiment using the declarative approach, with a dictionary/json, there is a keyword `measurement_error_mitigation` in the `problem` section that let you turn ON/OFF the measurement error mitigation, simply set it to `True`.\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
e78ff7e4a165bef70bb3cb830aa9250a7ce4ee5a | 389,134 | ipynb | Jupyter Notebook | Experiments/20181204/Untitled.ipynb | j1c/multigraph_clustering | 681d2050aa5e5d4b9984396ee2fbf90cc2bb2bcc | [
"Apache-2.0"
] | null | null | null | Experiments/20181204/Untitled.ipynb | j1c/multigraph_clustering | 681d2050aa5e5d4b9984396ee2fbf90cc2bb2bcc | [
"Apache-2.0"
] | null | null | null | Experiments/20181204/Untitled.ipynb | j1c/multigraph_clustering | 681d2050aa5e5d4b9984396ee2fbf90cc2bb2bcc | [
"Apache-2.0"
] | 1 | 2019-02-13T05:12:15.000Z | 2019-02-13T05:12:15.000Z | 935.418269 | 217,368 | 0.956742 | [
[
[
"import scipy\n\nimport graspy\nfrom graspy.embed import OmnibusEmbed, ClassicalMDS\nfrom graspy.cluster import GaussianCluster\nfrom graspy.utils import get_multigraph_lcc\n\nimport os\n\nimport numpy as np\n\n%matplotlib inline",
"_____no_output_____"
],
[
"path = '../../data/HNU1/dwi/Talairach/'\nfiles = sorted([f for f in os.listdir(path) if 'npz' in f])",
"_____no_output_____"
],
[
"graphs = []\nlabels = []\nfor f in files:\n labels.append(f.split('_')[0])\n A = scipy.sparse.load_npz(path + f)\n graphs.append(A.toarray())",
"_____no_output_____"
]
],
[
[
"## Omni",
"_____no_output_____"
]
],
[
[
"ptr_before = [graspy.utils.pass_to_ranks(g) for g in graphs]",
"_____no_output_____"
],
[
"lccs = get_multigraph_lcc(graphs)",
"_____no_output_____"
],
[
"tensor = np.stack(lccs)",
"_____no_output_____"
],
[
"tensor.shape",
"_____no_output_____"
],
[
"ptr = [graspy.utils.pass_to_ranks(g) for g in lccs]",
"_____no_output_____"
],
[
"omni = OmnibusEmbed()\nZhat = omni.fit_transform(lccs[:100])",
"_____no_output_____"
],
[
"Zhat = Zhat.reshape(100, 670, -1)\n\ncmds = ClassicalMDS()\nXhat = cmds.fit_transform(Zhat)",
"_____no_output_____"
],
[
"graspy.plot.heatmap(cmds.dissimilarity_matrix_)",
"_____no_output_____"
],
[
"graspy.plot.pairplot(Xhat)",
"_____no_output_____"
],
[
"gclust = GaussianCluster(50)\ngclust.fit(Xhat, labels[:100])",
"_____no_output_____"
],
[
"omni = OmnibusEmbed(n_elbows=10)\nZhat_ptr = omni.fit_transform(ptr[:100])",
"_____no_output_____"
],
[
"Zhat_ptr = Zhat_ptr.reshape(100, 670, -1)\n\ncmds = ClassicalMDS()\nXhat_ptr = cmds.fit_transform(Zhat_ptr)",
"_____no_output_____"
],
[
"graspy.plot.heatmap(cmds.dissimilarity_matrix_)",
"_____no_output_____"
],
[
"graspy.plot.pairplot(Xhat_ptr, Y=labels[:100])",
"_____no_output_____"
],
[
"np.diag(graphs[5]).sum()",
"_____no_output_____"
],
[
"np.load('../../data/HNU1/dwi/desikan/sub-0025427_ses-10_dwi_desikan.npy')",
"_____no_output_____"
],
[
"import warnings",
"_____no_output_____"
],
[
"if True:\n warnings.warn('test', Warning)",
"/home/j1c/graphstats/venv/lib/python3.5/site-packages/ipykernel_launcher.py:2: Warning: test\n \n"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.