hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
e7948dbda068ae852c9b4ee58aa0b0da65570403 | 127,693 | ipynb | Jupyter Notebook | dev/01_core.ipynb | nareshr8/fastai_dev | c34bf4e0fe296cb4ff8410dea3895c0ad2f6fe93 | [
"Apache-2.0"
]
| null | null | null | dev/01_core.ipynb | nareshr8/fastai_dev | c34bf4e0fe296cb4ff8410dea3895c0ad2f6fe93 | [
"Apache-2.0"
]
| null | null | null | dev/01_core.ipynb | nareshr8/fastai_dev | c34bf4e0fe296cb4ff8410dea3895c0ad2f6fe93 | [
"Apache-2.0"
]
| null | null | null | 30.381394 | 3,276 | 0.557219 | [
[
[
"#default_exp core",
"_____no_output_____"
],
[
"#export\nfrom local.test import *\nfrom local.imports import *\nfrom local.notebook.showdoc import show_doc",
"_____no_output_____"
],
[
"#export\ntorch.cuda.set_device(int(os.environ.get('DEFAULT_GPU') or 0))",
"_____no_output_____"
]
],
[
[
"# Core\n\n> Basic functions used in the fastai library",
"_____no_output_____"
]
],
[
[
"# export\ndefaults = SimpleNamespace()",
"_____no_output_____"
]
],
[
[
"## Metaclasses",
"_____no_output_____"
]
],
[
[
"#export\nclass PrePostInitMeta(type):\n \"A metaclass that calls optional `__pre_init__` and `__post_init__` methods\"\n def __new__(cls, name, bases, dct):\n x = super().__new__(cls, name, bases, dct)\n def _pass(self, *args,**kwargs): pass\n for o in ('__init__', '__pre_init__', '__post_init__'):\n if not hasattr(x,o): setattr(x,o,_pass)\n old_init = x.__init__\n \n @functools.wraps(old_init)\n def _init(self,*args,**kwargs):\n self.__pre_init__()\n old_init(self, *args,**kwargs)\n self.__post_init__()\n setattr(x, '__init__', _init)\n return x",
"_____no_output_____"
],
[
"show_doc(PrePostInitMeta, title_level=3)",
"_____no_output_____"
],
[
"class _T(metaclass=PrePostInitMeta):\n def __pre_init__(self): self.a = 0; assert self.a==0\n def __init__(self): self.a += 1; assert self.a==1\n def __post_init__(self): self.a += 1; assert self.a==2\n\nt = _T()\nt.a",
"_____no_output_____"
],
[
"#export\nclass BaseObj(metaclass=PrePostInitMeta):\n \"Base class that provides `PrePostInitMeta` metaclass to subclasses\"\n pass",
"_____no_output_____"
],
[
"class _T(BaseObj):\n def __pre_init__(self): self.a = 0; assert self.a==0\n def __init__(self): self.a += 1; assert self.a==1\n def __post_init__(self): self.a += 1; assert self.a==2\n\nt = _T()\nt.a",
"_____no_output_____"
],
[
"#export\nclass NewChkMeta(PrePostInitMeta):\n \"Metaclass to avoid recreating object passed to constructor (plus all `PrePostInitMeta` functionality)\"\n def __new__(cls, name, bases, dct):\n x = super().__new__(cls, name, bases, dct)\n old_init,old_new = x.__init__,x.__new__\n\n @functools.wraps(old_init)\n def _new(cls, x=None, *args, **kwargs):\n if x is not None and isinstance(x,cls):\n x._newchk = 1\n return x\n res = old_new(cls)\n res._newchk = 0\n return res\n\n @functools.wraps(old_init)\n def _init(self,*args,**kwargs):\n if self._newchk: return\n old_init(self, *args, **kwargs)\n\n x.__init__,x.__new__ = _init,_new\n return x",
"_____no_output_____"
],
[
"class _T(metaclass=NewChkMeta):\n \"Testing\"\n def __init__(self, o=None): self.foo = getattr(o,'foo',0) + 1\n\nclass _T2():\n def __init__(self, o): self.foo = getattr(o,'foo',0) + 1\n\nt = _T(1)\ntest_eq(t.foo,1)\nt2 = _T(t)\ntest_eq(t2.foo,1)\ntest_is(t,t2)\n\nt = _T2(1)\ntest_eq(t.foo,1)\nt2 = _T2(t)\ntest_eq(t2.foo,2)\n\ntest_eq(_T.__doc__, \"Testing\")\ntest_eq(str(inspect.signature(_T)), '(o=None)')",
"_____no_output_____"
],
[
"#export\nclass BypassNewMeta(type):\n \"Metaclass: casts `x` to this class, initializing with `_new_meta` if available\"\n def __call__(cls, x, *args, **kwargs):\n if hasattr(cls, '_new_meta'): x = cls._new_meta(x, *args, **kwargs)\n if cls!=x.__class__: x.__class__ = cls\n return x",
"_____no_output_____"
],
[
"class T0: pass\nclass _T(T0, metaclass=BypassNewMeta): pass\n\nt = T0()\nt.a = 1\nt2 = _T(t)\ntest_eq(type(t2), _T)\ntest_eq(t2.a,1)",
"_____no_output_____"
]
],
[
[
"## Foundational functions",
"_____no_output_____"
],
[
"### Decorators",
"_____no_output_____"
]
],
[
[
"#export\ndef patch_to(cls, as_prop=False):\n \"Decorator: add `f` to `cls`\"\n def _inner(f):\n nf = copy(f)\n # `functools.update_wrapper` when passing patched function to `Pipeline`, so we do it manually\n for o in functools.WRAPPER_ASSIGNMENTS: setattr(nf, o, getattr(f,o))\n nf.__qualname__ = f\"{cls.__name__}.{f.__name__}\"\n setattr(cls, f.__name__, property(nf) if as_prop else nf)\n return f\n return _inner",
"_____no_output_____"
],
[
"class _T3(int): pass\n\n@patch_to(_T3)\ndef func1(x, a:bool): return x+2\n\nt = _T3(1)\ntest_eq(t.func1(1), 3)",
"_____no_output_____"
],
[
"#export\ndef patch(f):\n \"Decorator: add `f` to the first parameter's class (based on f's type annotations)\"\n cls = next(iter(f.__annotations__.values()))\n return patch_to(cls)(f)",
"_____no_output_____"
],
[
"@patch\ndef func(x:_T3, a:bool):\n \"test\"\n return x+2\n\nt = _T3(1)\ntest_eq(t.func(1), 3)\ntest_eq(t.func.__qualname__, '_T3.func')",
"_____no_output_____"
],
[
"#export\ndef patch_property(f):\n \"Decorator: add `f` as a property to the first parameter's class (based on f's type annotations)\"\n cls = next(iter(f.__annotations__.values()))\n return patch_to(cls, as_prop=True)(f)",
"_____no_output_____"
],
[
"@patch_property\ndef prop(x:_T3): return x+1\n\nt = _T3(1)\ntest_eq(t.prop, 2)",
"_____no_output_____"
],
[
"#export\ndef _mk_param(n,d=None): return inspect.Parameter(n, inspect.Parameter.KEYWORD_ONLY, default=d)",
"_____no_output_____"
],
[
"def test_sig(f, b): test_eq(str(inspect.signature(f)), b)",
"_____no_output_____"
],
[
"#export\ndef use_kwargs(names, keep=False):\n \"Decorator: replace `**kwargs` in signature with `names` params\"\n def _f(f):\n sig = inspect.signature(f)\n sigd = dict(sig.parameters)\n k = sigd.pop('kwargs')\n s2 = {n:_mk_param(n) for n in names if n not in sigd}\n sigd.update(s2)\n if keep: sigd['kwargs'] = k\n f.__signature__ = sig.replace(parameters=sigd.values())\n return f\n return _f",
"_____no_output_____"
],
[
"@use_kwargs(['y', 'z'])\ndef foo(a, b=1, **kwargs): pass\ntest_sig(foo, '(a, b=1, *, y=None, z=None)')\n\n@use_kwargs(['y', 'z'], keep=True)\ndef foo(a, *args, b=1, **kwargs): pass\ntest_sig(foo, '(a, *args, b=1, y=None, z=None, **kwargs)')",
"_____no_output_____"
],
[
"#export\ndef delegates(to=None, keep=False):\n \"Decorator: replace `**kwargs` in signature with params from `to`\"\n def _f(f):\n if to is None: to_f,from_f = f.__base__.__init__,f.__init__\n else: to_f,from_f = to,f\n sig = inspect.signature(from_f)\n sigd = dict(sig.parameters)\n k = sigd.pop('kwargs')\n s2 = {k:v for k,v in inspect.signature(to_f).parameters.items()\n if v.default != inspect.Parameter.empty and k not in sigd}\n sigd.update(s2)\n if keep: sigd['kwargs'] = k\n from_f.__signature__ = sig.replace(parameters=sigd.values())\n return f\n return _f",
"_____no_output_____"
],
[
"def basefoo(e, c=2): pass\n\n@delegates(basefoo)\ndef foo(a, b=1, **kwargs): pass\ntest_sig(foo, '(a, b=1, c=2)')\n\n@delegates(basefoo, keep=True)\ndef foo(a, b=1, **kwargs): pass\ntest_sig(foo, '(a, b=1, c=2, **kwargs)')",
"_____no_output_____"
],
[
"class BaseFoo:\n def __init__(self, e, c=2): pass\n\n@delegates()\nclass Foo(BaseFoo):\n def __init__(self, a, b=1, **kwargs): super().__init__(**kwargs)\n\ntest_sig(Foo, '(a, b=1, c=2)')",
"_____no_output_____"
],
[
"#export\ndef funcs_kwargs(cls):\n \"Replace methods in `self._methods` with those from `kwargs`\"\n old_init = cls.__init__\n def _init(self, *args, **kwargs):\n for k in cls._methods:\n arg = kwargs.pop(k,None)\n if arg is not None:\n if isinstance(arg,types.MethodType): arg = types.MethodType(arg.__func__, self)\n setattr(self, k, arg)\n old_init(self, *args, **kwargs)\n functools.update_wrapper(_init, old_init)\n cls.__init__ = use_kwargs(cls._methods)(_init)\n return cls",
"_____no_output_____"
],
[
"#export\ndef method(f):\n \"Mark `f` as a method\"\n # `1` is a dummy instance since Py3 doesn't allow `None` any more\n return types.MethodType(f, 1)",
"_____no_output_____"
],
[
"@funcs_kwargs\nclass T:\n _methods=['b']\n def __init__(self, f=1, **kwargs): assert not kwargs\n def a(self): return 1\n def b(self): return 2\n \nt = T()\ntest_eq(t.a(), 1)\ntest_eq(t.b(), 2)\nt = T(b = lambda:3)\ntest_eq(t.b(), 3)\ntest_sig(T, '(f=1, *, b=None)')\ntest_fail(lambda: T(a = lambda:3))\n\n@method\ndef _f(self,a=1): return a+1\nt = T(b = _f)\ntest_eq(t.b(2), 3)\n\nclass T2(T):\n def __init__(self,a):\n super().__init__(b = lambda:3)\n self.a=a\nt = T2(a=1)\ntest_eq(t.b(), 3)\ntest_sig(T2, '(a)')\n\ndef _g(a=1): return a+1\nclass T3(T): b = staticmethod(_g)\nt = T3()\ntest_eq(t.b(2), 3)",
"_____no_output_____"
]
],
[
[
"### Type checking",
"_____no_output_____"
],
[
"Runtime type checking is handy, so let's make it easy!",
"_____no_output_____"
]
],
[
[
"#export core\n#NB: Please don't move this to a different line or module, since it's used in testing `get_source_link`\ndef chk(f): return typechecked(always=True)(f)",
"_____no_output_____"
]
],
[
[
"Decorator for a function to check that type-annotated arguments receive arguments of the right type.",
"_____no_output_____"
]
],
[
[
"@chk\ndef test_chk(a:int=1): return a\n\ntest_eq(test_chk(2), 2)\ntest_eq(test_chk(), 1)\ntest_fail(lambda: test_chk('a'), contains='\"a\" must be int')",
"_____no_output_____"
]
],
[
[
"Decorated functions will pickle correctly.",
"_____no_output_____"
]
],
[
[
"t = pickle.loads(pickle.dumps(test_chk))\ntest_eq(t(2), 2)\ntest_eq(t(), 1)",
"_____no_output_____"
]
],
[
[
"### Context managers",
"_____no_output_____"
]
],
[
[
"@contextmanager\ndef working_directory(path):\n \"Change working directory to `path` and return to previous on exit.\"\n prev_cwd = Path.cwd()\n os.chdir(path)\n try: yield\n finally: os.chdir(prev_cwd)",
"_____no_output_____"
]
],
[
[
"### Monkey-patching",
"_____no_output_____"
]
],
[
[
"def is_listy(x): return isinstance(x,(list,tuple,Generator))",
"_____no_output_____"
],
[
"#export\ndef tensor(x, *rest, **kwargs):\n \"Like `torch.as_tensor`, but handle lists too, and can pass multiple vector elements directly.\"\n if len(rest): x = (x,)+rest\n # Pytorch bug in dataloader using num_workers>0\n if isinstance(x, (tuple,list)) and len(x)==0: return tensor(0)\n res = (torch.tensor(x, **kwargs) if isinstance(x, (tuple,list))\n else as_tensor(x, **kwargs) if hasattr(x, '__array__')\n else as_tensor(x, **kwargs) if is_listy(x)\n else as_tensor(x, **kwargs) if is_iter(x)\n else None)\n if res is None:\n res = as_tensor(array(x), **kwargs)\n if res.dtype is torch.float64: return res.float()\n if res.dtype is torch.int32:\n warn('Tensor is int32: upgrading to int64; for better performance use int64 input')\n return res.long()\n return res",
"_____no_output_____"
],
[
"test_eq(tensor(array([1,2,3])), torch.tensor([1,2,3]))\ntest_eq(tensor(1,2,3), torch.tensor([1,2,3]))\ntest_eq_type(tensor(1.0), torch.tensor(1.0))",
"_____no_output_____"
]
],
[
[
"#### `Tensor.ndim`",
"_____no_output_____"
],
[
"We add an `ndim` property to `Tensor` with same semantics as [numpy ndim](https://docs.scipy.org/doc/numpy/reference/generated/numpy.ndarray.ndim.html), which allows tensors to be used in matplotlib and other places that assume this property exists.",
"_____no_output_____"
]
],
[
[
"test_eq(torch.tensor([1,2]).ndim,1)\ntest_eq(torch.tensor(1).ndim,0)\ntest_eq(torch.tensor([[1]]).ndim,2)",
"_____no_output_____"
]
],
[
[
"### Documentation functions",
"_____no_output_____"
]
],
[
[
"#export core\ndef add_docs(cls, cls_doc=None, **docs):\n \"Copy values from `docs` to `cls` docstrings, and confirm all public methods are documented\"\n if cls_doc is not None: cls.__doc__ = cls_doc\n for k,v in docs.items():\n f = getattr(cls,k)\n if hasattr(f,'__func__'): f = f.__func__ # required for class methods\n f.__doc__ = v\n # List of public callables without docstring\n nodoc = [c for n,c in vars(cls).items() if isinstance(c,Callable)\n and not n.startswith('_') and c.__doc__ is None]\n assert not nodoc, f\"Missing docs: {nodoc}\"\n assert cls.__doc__ is not None, f\"Missing class docs: {cls}\"",
"_____no_output_____"
],
[
"#export core\ndef docs(cls):\n \"Decorator version of `add_docs`, using `_docs` dict\"\n add_docs(cls, **cls._docs)\n return cls",
"_____no_output_____"
],
[
"class _T:\n def f(self): pass\n @classmethod\n def g(cls): pass\nadd_docs(_T, \"a\", f=\"f\", g=\"g\")\n\ntest_eq(_T.__doc__, \"a\")\ntest_eq(_T.f.__doc__, \"f\")\ntest_eq(_T.g.__doc__, \"g\")",
"_____no_output_____"
],
[
"#export\ndef custom_dir(c, add:List):\n \"Implement custom `__dir__`, adding `add` to `cls`\"\n return dir(type(c)) + list(c.__dict__.keys()) + add",
"_____no_output_____"
],
[
"show_doc(is_iter)",
"_____no_output_____"
],
[
"assert is_iter([1])\nassert not is_iter(torch.tensor(1))\nassert is_iter(torch.tensor([1,2]))\nassert (o for o in range(3))",
"_____no_output_____"
]
],
[
[
"## GetAttr -",
"_____no_output_____"
]
],
[
[
"#export\nclass GetAttr(BaseObj):\n \"Inherit from this to have all attr accesses in `self._xtra` passed down to `self.default`\"\n @property\n def _xtra(self): return [o for o in dir(self.default) if not o.startswith('_')]\n def __getattr__(self,k):\n if k in self._xtra: return getattr(self.default, k)\n raise AttributeError(k)\n def __dir__(self): return custom_dir(self, self._xtra)",
"_____no_output_____"
],
[
"class _C(GetAttr): default,_xtra = 'Hi',['lower']\n\nt = _C()\ntest_eq(t.lower(), 'hi')\ntest_fail(lambda: t.upper())\nassert 'lower' in dir(t)",
"_____no_output_____"
],
[
"#export\ndef delegate_attr(self, k, to):\n \"Use in `__getattr__` to delegate to attr `to` without inheriting from `GetAttr`\"\n if k.startswith('_') or k==to: raise AttributeError(k)\n try: return getattr(getattr(self,to), k)\n except AttributeError: raise AttributeError(k) from None",
"_____no_output_____"
],
[
"class _C:\n f = 'Hi'\n def __getattr__(self, k): return delegate_attr(self, k, 'f')\n\nt = _C()\ntest_eq(t.lower(), 'hi')",
"_____no_output_____"
]
],
[
[
"## L -",
"_____no_output_____"
]
],
[
[
"# export\ndef coll_repr(c, max_n=10):\n \"String repr of up to `max_n` items of (possibly lazy) collection `c`\"\n return f'(#{len(c)}) [' + ','.join(itertools.islice(map(str,c), max_n)) + ('...'\n if len(c)>10 else '') + ']'",
"_____no_output_____"
],
[
"test_eq(coll_repr(range(1000), 5), '(#1000) [0,1,2,3,4...]')",
"_____no_output_____"
],
[
"# export\ndef mask2idxs(mask):\n \"Convert bool mask or index list to index `L`\"\n mask = list(mask)\n if len(mask)==0: return []\n if isinstance(mask[0],bool): return [i for i,m in enumerate(mask) if m]\n return [int(i) for i in mask]",
"_____no_output_____"
],
[
"test_eq(mask2idxs([False,True,False,True]), [1,3])\ntest_eq(mask2idxs(torch.tensor([1,2,3])), [1,2,3])",
"_____no_output_____"
],
[
"# export\ndef _listify(o):\n if o is None: return []\n if isinstance(o, list): return o\n if isinstance(o, (str,np.ndarray,Tensor)): return [o]\n if is_iter(o): return list(o)\n return [o]",
"_____no_output_____"
],
[
"#export\nclass CollBase(GetAttr, metaclass=NewChkMeta):\n \"Base class for composing a list of `items`\"\n _xtra = [o for o in dir([]) if not o.startswith('_')]\n\n def __init__(self, items): self.items = items\n def __len__(self): return len(self.items)\n def __getitem__(self, k): return self.items[k]\n def __setitem__(self, k, v): self.items[k] = v\n def __delitem__(self, i): del(self.items[i])\n def __repr__(self): return self.items.__repr__()\n def __iter__(self): return self.items.__iter__()\n def _new(self, items, *args, **kwargs): return self.__class__(items, *args, **kwargs)\n @property\n def default(self): return self.items",
"_____no_output_____"
],
[
"#export\nclass L(CollBase):\n \"Behaves like a list of `items` but can also index with list of indices or masks\"\n def __init__(self, items=None, *rest, use_list=False, match=None):\n if rest: items = (items,)+rest\n if items is None: items = []\n if (use_list is not None) or not isinstance(items,(Tensor,ndarray,pd.DataFrame,pd.Series)):\n items = list(items) if use_list else _listify(items)\n if match is not None:\n if len(items)==1: items = items*len(match)\n else: assert len(items)==len(match), 'Match length mismatch'\n super().__init__(items)\n \n def __getitem__(self, idx): return L(self._gets(idx), use_list=None) if is_iter(idx) else self._get(idx)\n def _get(self, i): return getattr(self.items,'iloc',self.items)[i]\n def _gets(self, i):\n i = mask2idxs(i)\n return (self.items.iloc[list(i)] if hasattr(self.items,'iloc')\n else self.items.__array__()[(i,)] if hasattr(self.items,'__array__')\n else [self.items[i_] for i_ in i])\n \n def __setitem__(self, idx, o):\n \"Set `idx` (can be list of indices, or mask, or int) items to `o` (which is broadcast if not iterable)\"\n idx = idx if isinstance(idx,L) else _listify(idx) \n if not is_iter(o): o = [o]*len(idx)\n for i,o_ in zip(idx,o): self.items[i] = o_\n \n def __repr__(self): return coll_repr(self)\n def __eq__(self,b): return all_equal(b,self)\n def __iter__(self): return (self[i] for i in range(len(self)))\n \n def __invert__(self): return self._new(not i for i in self)\n def __mul__ (a,b): return a._new(a.items*b)\n def __add__ (a,b): return a._new(a.items+_listify(b))\n def __radd__(a,b): return a._new(b)+a\n def __addi__(a,b):\n a.items += list(b)\n return a\n\n def sorted(self, key=None, reverse=False):\n \"New `L` sorted by `key`. If key is str then use `attrgetter`. If key is int then use `itemgetter`.\"\n if isinstance(key,str): k=lambda o:getattr(o,key,0)\n elif isinstance(key,int): k=itemgetter(key)\n else: k=key\n return self._new(sorted(self.items, key=k, reverse=reverse))\n \n @classmethod\n def range(cls, a, b=None, step=None):\n \"Same as builtin `range`, but returns an `L`. Can pass a collection for `a`, to use `len(a)`\"\n if is_coll(a): a = len(a)\n return cls(range(a,b,step) if step is not None else range(a,b) if b is not None else range(a))\n \n def itemgot(self, idx): return self.mapped(itemgetter(idx))\n def attrgot(self, k, default=None): return self.mapped(lambda o:getattr(o,k,default))\n def tensored(self): return self.mapped(tensor)\n def stack(self, dim=0): return torch.stack(list(self.tensored()), dim=dim)\n def cat (self, dim=0): return torch.cat (list(self.tensored()), dim=dim)\n def cycle(self): return itertools.cycle(self) if len(self) > 0 else itertools.cycle([None])\n def filtered(self, f, *args, **kwargs): return self._new(filter(partial(f,*args,**kwargs), self))\n def mapped(self, f, *args, **kwargs): return self._new(map(partial(f,*args,**kwargs), self))\n def mapped_dict(self, f, *args, **kwargs): return {k:f(k, *args,**kwargs) for k in self}\n def starmapped(self, f, *args, **kwargs): return self._new(itertools.starmap(partial(f,*args,**kwargs), self))\n def zipped(self, longest=False): return self._new((zip_longest if longest else zip)(*self))\n def zipwith(self, *rest, longest=False): return self._new([self, *rest]).zipped(longest=longest)\n def mapped_zip(self, f, longest=False): return self.zipped(longest=longest).starmapped(f)\n def mapped_zipwith(self, f, *rest, longest=False): return self.zipwith(*rest, longest=longest).starmapped(f)\n def shuffled(self):\n it = copy(self.items)\n random.shuffle(it)\n return self._new(it)",
"_____no_output_____"
],
[
"#export\nadd_docs(L,\n __getitem__=\"Retrieve `idx` (can be list of indices, or mask, or int) items\",\n filtered=\"Create new `L` filtered by predicate `f`, passing `args` and `kwargs` to `f`\",\n mapped=\"Create new `L` with `f` applied to all `items`, passing `args` and `kwargs` to `f`\",\n mapped_dict=\"Like `mapped`, but creates a dict from `items` to function results\",\n starmapped=\"Like `mapped`, but use `itertools.starmap`\",\n itemgot=\"Create new `L` with item `idx` of all `items`\",\n attrgot=\"Create new `L` with attr `k` of all `items`\",\n tensored=\"`mapped(tensor)`\",\n cycle=\"Same as `itertools.cycle`\",\n stack=\"Same as `torch.stack`\",\n cat=\"Same as `torch.cat`\",\n zipped=\"Create new `L` with `zip(*items)`\",\n zipwith=\"Create new `L` with `self` zipped with each of `*rest`\",\n mapped_zip=\"Combine `zipped` and `starmapped`\",\n mapped_zipwith=\"Combine `zipwith` and `starmapped`\",\n shuffled=\"Same as `random.shuffle`, but not inplace\")",
"_____no_output_____"
]
],
[
[
"You can create an `L` from an existing iterable (e.g. a list, range, etc) and access or modify it with an int list/tuple index, mask, int, or slice. All `list` methods can also be used with `L`.",
"_____no_output_____"
]
],
[
[
"t = L(range(12))\ntest_eq(t, list(range(12)))\ntest_ne(t, list(range(11)))\nt.reverse()\ntest_eq(t[0], 11)\nt[3] = \"h\"\ntest_eq(t[3], \"h\")\nt[3,5] = (\"j\",\"k\")\ntest_eq(t[3,5], [\"j\",\"k\"])\ntest_eq(t, L(t))\nt",
"_____no_output_____"
]
],
[
[
"There are optimized indexers for arrays, tensors, and DataFrames.",
"_____no_output_____"
]
],
[
[
"arr = np.arange(9).reshape(3,3)\nt = L(arr, use_list=None)\ntest_eq(t[1,2], arr[[1,2]])\n\narr = torch.arange(9).view(3,3)\nt = L(arr, use_list=None)\ntest_eq(t[1,2], arr[[1,2]])\n\ndf = pd.DataFrame({'a':[1,2,3]})\nt = L(df, use_list=None)\ntest_eq(t[1,2], L(pd.DataFrame({'a':[2,3]}), use_list=None))",
"_____no_output_____"
]
],
[
[
"You can also modify an `L` with `append`, `+`, and `*`.",
"_____no_output_____"
]
],
[
[
"t = L()\ntest_eq(t, [])\nt.append(1)\ntest_eq(t, [1])\nt += [3,2]\ntest_eq(t, [1,3,2])\nt = t + [4]\ntest_eq(t, [1,3,2,4])\nt = 5 + t\ntest_eq(t, [5,1,3,2,4])\ntest_eq(L(1,2,3), [1,2,3])\ntest_eq(L(1,2,3), L(1,2,3))\nt = L(1)*5\nt = t.mapped(operator.neg)\ntest_eq(t,[-1]*5)\ntest_eq(~L([True,False,False]), L([False,True,True]))\nt = L(range(4))\ntest_eq(zip(t, L(1).cycle()), zip(range(4),(1,1,1,1)))\nt = L.range(100)\nt2 = t.shuffled()\ntest_ne(t,t2)\ntest_eq(L.range(100), t)\ntest_eq(set(t),set(t2))",
"_____no_output_____"
],
[
"def _f(x,a=0): return x+a\nt = L(1)*5\ntest_eq(t.mapped(_f), t)\ntest_eq(t.mapped(_f,1), [2]*5)\ntest_eq(t.mapped(_f,a=2), [3]*5)",
"_____no_output_____"
]
],
[
[
"An `L` can be constructed from anything iterable, although tensors and arrays will not be iterated over on construction, unless you pass `use_list` to the constructor.",
"_____no_output_____"
]
],
[
[
"test_eq(L([1,2,3]),[1,2,3])\ntest_eq(L(L([1,2,3])),[1,2,3])\ntest_ne(L([1,2,3]),[1,2,])\ntest_eq(L('abc'),['abc'])\ntest_eq(L(range(0,3)),[0,1,2])\ntest_eq(L(o for o in range(0,3)),[0,1,2])\ntest_eq(L(tensor(0)),[tensor(0)])\ntest_eq(L([tensor(0),tensor(1)]),[tensor(0),tensor(1)])\ntest_eq(L(tensor([0.,1.1]))[0],tensor([0.,1.1]))\ntest_eq(L(tensor([0.,1.1]), use_list=True), [0.,1.1]) # `use_list=True` to unwrap arrays/tensors",
"_____no_output_____"
]
],
[
[
"If `match` is not `None` then the created list is same len as `match`, either by:\n\n- If `len(items)==1` then `items` is replicated,\n- Otherwise an error is raised if `match` and `items` are not already the same size.",
"_____no_output_____"
]
],
[
[
"test_eq(L(1,match=[1,2,3]),[1,1,1])\ntest_eq(L([1,2],match=[2,3]),[1,2])\ntest_fail(lambda: L([1,2],match=[1,2,3]))",
"_____no_output_____"
]
],
[
[
"If you create an `L` from an existing `L` then you'll get back the original object (since `L` uses the `NewChkMeta` metaclass).",
"_____no_output_____"
]
],
[
[
"test_is(L(t), t)",
"_____no_output_____"
]
],
[
[
"### Methods",
"_____no_output_____"
]
],
[
[
"show_doc(L.__getitem__)",
"_____no_output_____"
],
[
"t = L(range(12))\ntest_eq(t[1,2], [1,2]) # implicit tuple\ntest_eq(t[[1,2]], [1,2]) # list\ntest_eq(t[:3], [0,1,2]) # slice\ntest_eq(t[[False]*11 + [True]], [11]) # mask\ntest_eq(t[tensor(3)], 3)",
"_____no_output_____"
],
[
"show_doc(L.__setitem__)",
"_____no_output_____"
],
[
"t[4,6] = 0\ntest_eq(t[4,6], [0,0])\nt[4,6] = [1,2]\ntest_eq(t[4,6], [1,2])",
"_____no_output_____"
],
[
"show_doc(L.filtered)",
"_____no_output_____"
],
[
"test_eq(t.filtered(lambda o:o<5), [0,1,2,3,1,2])",
"_____no_output_____"
],
[
"show_doc(L.mapped)",
"_____no_output_____"
],
[
"test_eq(L(range(4)).mapped(operator.neg), [0,-1,-2,-3])",
"_____no_output_____"
],
[
"show_doc(L.mapped_dict)",
"_____no_output_____"
],
[
"test_eq(L(range(1,5)).mapped_dict(operator.neg), {1:-1, 2:-2, 3:-3, 4:-4})",
"_____no_output_____"
],
[
"show_doc(L.zipped)",
"_____no_output_____"
],
[
"t = L([[1,2,3],'abc'])\ntest_eq(t.zipped(), [(1, 'a'),(2, 'b'),(3, 'c')])",
"_____no_output_____"
],
[
"t = L([[1,2],'abc'])\ntest_eq(t.zipped(longest=True ), [(1, 'a'),(2, 'b'),(None, 'c')])\ntest_eq(t.zipped(longest=False), [(1, 'a'),(2, 'b')])",
"_____no_output_____"
],
[
"show_doc(L.mapped_zip)",
"_____no_output_____"
],
[
"t = L([1,2,3],[2,3,4])\ntest_eq(t.mapped_zip(operator.mul), [2,6,12])",
"_____no_output_____"
],
[
"show_doc(L.zipwith)",
"_____no_output_____"
],
[
"b = [[0],[1],[2,2]]\nt = L([1,2,3]).zipwith(b)\ntest_eq(t, [(1,[0]), (2,[1]), (3,[2,2])])",
"_____no_output_____"
],
[
"show_doc(L.mapped_zipwith)",
"_____no_output_____"
],
[
"test_eq(L(1,2,3).mapped_zipwith(operator.mul, [2,3,4]), [2,6,12])",
"_____no_output_____"
],
[
"show_doc(L.itemgot)",
"_____no_output_____"
],
[
"test_eq(t.itemgot(1), b)",
"_____no_output_____"
],
[
"show_doc(L.attrgot)",
"_____no_output_____"
],
[
"a = [SimpleNamespace(a=3,b=4),SimpleNamespace(a=1,b=2)]\ntest_eq(L(a).attrgot('b'), [4,2])",
"_____no_output_____"
],
[
"show_doc(L.sorted)",
"_____no_output_____"
],
[
"test_eq(L(a).sorted('a').attrgot('b'), [2,4])",
"_____no_output_____"
],
[
"show_doc(L.range)",
"_____no_output_____"
],
[
"test_eq_type(L.range([1,1,1]), L(range(3)))\ntest_eq_type(L.range(5,2,2), L(range(5,2,2)))",
"_____no_output_____"
],
[
"show_doc(L.tensored)",
"_____no_output_____"
]
],
[
[
"There are shortcuts for `torch.stack` and `torch.cat` if your `L` contains tensors or something convertible. You can manually convert with `tensored`.",
"_____no_output_____"
]
],
[
[
"t = L(([1,2],[3,4]))\ntest_eq(t.tensored(), [tensor(1,2),tensor(3,4)])",
"_____no_output_____"
],
[
"show_doc(L.stack)",
"_____no_output_____"
],
[
"test_eq(t.stack(), tensor([[1,2],[3,4]]))",
"_____no_output_____"
],
[
"show_doc(L.cat)",
"_____no_output_____"
],
[
"test_eq(t.cat(), tensor([1,2,3,4]))",
"_____no_output_____"
]
],
[
[
"## Utility functions",
"_____no_output_____"
],
[
"### Basics",
"_____no_output_____"
]
],
[
[
"# export\ndef ifnone(a, b):\n \"`b` if `a` is None else `a`\"\n return b if a is None else a",
"_____no_output_____"
]
],
[
[
"Since `b if a is None else a` is such a common pattern, we wrap it in a function. However, be careful, because python will evaluate *both* `a` and `b` when calling `ifnone` (which it doesn't do if using the `if` version directly).",
"_____no_output_____"
]
],
[
[
"test_eq(ifnone(None,1), 1)\ntest_eq(ifnone(2 ,1), 2)",
"_____no_output_____"
],
[
"#export\ndef get_class(nm, *fld_names, sup=None, doc=None, funcs=None, **flds):\n \"Dynamically create a class, optionally inheriting from `sup`, containing `fld_names`\"\n attrs = {}\n for f in fld_names: attrs[f] = None\n for f in L(funcs): attrs[f.__name__] = f\n for k,v in flds.items(): attrs[k] = v\n sup = ifnone(sup, ())\n if not isinstance(sup, tuple): sup=(sup,)\n \n def _init(self, *args, **kwargs):\n for i,v in enumerate(args): setattr(self, list(attrs.keys())[i], v)\n for k,v in kwargs.items(): setattr(self,k,v)\n \n def _repr(self):\n return '\\n'.join(f'{o}: {getattr(self,o)}' for o in set(dir(self))\n if not o.startswith('_') and not isinstance(getattr(self,o), types.MethodType))\n \n if not sup: attrs['__repr__'] = _repr\n attrs['__init__'] = _init\n res = type(nm, sup, attrs)\n if doc is not None: res.__doc__ = doc\n return res",
"_____no_output_____"
],
[
"_t = get_class('_t', 'a', b=2)\nt = _t()\ntest_eq(t.a, None)\ntest_eq(t.b, 2)\nt = _t(1, b=3)\ntest_eq(t.a, 1)\ntest_eq(t.b, 3)\nt = _t(1, 3)\ntest_eq(t.a, 1)\ntest_eq(t.b, 3)",
"_____no_output_____"
]
],
[
[
"Most often you'll want to call `mk_class`, since it adds the class to your module. See `mk_class` for more details and examples of use (which also apply to `get_class`).",
"_____no_output_____"
]
],
[
[
"#export\ndef mk_class(nm, *fld_names, sup=None, doc=None, funcs=None, mod=None, **flds):\n \"Create a class using `get_class` and add to the caller's module\"\n if mod is None: mod = inspect.currentframe().f_back.f_locals\n res = get_class(nm, *fld_names, sup=sup, doc=doc, funcs=funcs, **flds)\n mod[nm] = res",
"_____no_output_____"
]
],
[
[
"Any `kwargs` will be added as class attributes, and `sup` is an optional (tuple of) base classes.",
"_____no_output_____"
]
],
[
[
"mk_class('_t', a=1, sup=GetAttr)\nt = _t()\ntest_eq(t.a, 1)\nassert(isinstance(t,GetAttr))",
"_____no_output_____"
]
],
[
[
"A `__init__` is provided that sets attrs for any `kwargs`, and for any `args` (matching by position to fields), along with a `__repr__` which prints all attrs. The docstring is set to `doc`. You can pass `funcs` which will be added as attrs with the function names.",
"_____no_output_____"
]
],
[
[
"def foo(self): return 1\nmk_class('_t', 'a', sup=GetAttr, doc='test doc', funcs=foo)\n\nt = _t(3, b=2)\ntest_eq(t.a, 3)\ntest_eq(t.b, 2)\ntest_eq(t.foo(), 1)\ntest_eq(t.__doc__, 'test doc')\nt",
"_____no_output_____"
],
[
"#export\ndef wrap_class(nm, *fld_names, sup=None, doc=None, funcs=None, **flds):\n \"Decorator: makes function a method of a new class `nm` passing parameters to `mk_class`\"\n def _inner(f):\n mk_class(nm, *fld_names, sup=sup, doc=doc, funcs=L(funcs)+f, mod=f.__globals__, **flds)\n return f\n return _inner",
"_____no_output_____"
],
[
"@wrap_class('_t', a=2)\ndef bar(self,x): return x+1\n\nt = _t()\ntest_eq(t.a, 2)\ntest_eq(t.bar(3), 4)",
"_____no_output_____"
],
[
"show_doc(noop)",
"_____no_output_____"
],
[
"noop()\ntest_eq(noop(1),1)",
"_____no_output_____"
],
[
"show_doc(noops)",
"_____no_output_____"
],
[
"mk_class('_t', foo=noops)\ntest_eq(_t().foo(1),1)",
"_____no_output_____"
],
[
"#export\ndef set_seed(s):\n \"Set random seed for `random`, `torch`, and `numpy` (where available)\"\n try: torch.manual_seed(s)\n except NameError: pass\n try: np.random.seed(s%(2**32-1))\n except NameError: pass\n random.seed(s)",
"_____no_output_____"
],
[
"set_seed(2*33)\na1 = np.random.random()\na2 = torch.rand(())\na3 = random.random()\nset_seed(2*33)\nb1 = np.random.random()\nb2 = torch.rand(())\nb3 = random.random()\ntest_eq(a1,b1)\ntest_eq(a2,b2)\ntest_eq(a3,b3)",
"_____no_output_____"
],
[
"#export\ndef store_attr(self, nms):\n \"Store params named in comma-separated `nms` from calling context into attrs in `self`\"\n mod = inspect.currentframe().f_back.f_locals\n for n in re.split(', *', nms): setattr(self,n,mod[n])",
"_____no_output_____"
],
[
"class T:\n def __init__(self, a,b,c): store_attr(self, 'a,b, c')\n\nt = T(1,c=2,b=3)\nassert t.a==1 and t.b==3 and t.c==2",
"_____no_output_____"
]
],
[
[
"### Subclassing `Tensor`",
"_____no_output_____"
]
],
[
[
"#export\nclass TensorBase(Tensor, metaclass=BypassNewMeta):\n def _new_meta(self, *args, **kwargs): return tensor(self)",
"_____no_output_____"
],
[
"#export\ndef _patch_tb():\n def get_f(fn):\n def _f(self, *args, **kwargs):\n cls = self.__class__\n res = getattr(super(TensorBase, self), fn)(*args, **kwargs)\n return cls(res) if isinstance(res,Tensor) else res\n return _f\n \n t = tensor([1])\n skips = '__class__ __deepcopy__ __delattr__ __dir__ __doc__ __getattribute__ __hash__ __init__ \\\n __init_subclass__ __new__ __reduce__ __module__ __setstate__'.split()\n\n for fn in dir(t):\n if fn in skips: continue\n f = getattr(t, fn)\n if isinstance(f, (types.MethodWrapperType, types.BuiltinFunctionType, types.BuiltinMethodType, types.MethodType, types.FunctionType)):\n setattr(TensorBase, fn, get_f(fn))\n\n_patch_tb()",
"_____no_output_____"
],
[
"t = TensorBase(range(5))\ntest_eq_type(t[0], TensorBase(0))\ntest_eq_type(t[:2], TensorBase([0,1]))\ntest_eq_type(t+1, TensorBase(range(1,6)))",
"_____no_output_____"
],
[
"class _T(TensorBase): pass\n\nt = _T(range(5))\ntest_eq_type(t[0], _T(0))\ntest_eq_type(t[:2], _T([0,1]))\ntest_eq_type(t+1, _T(range(1,6)))",
"_____no_output_____"
],
[
"#export\ndef retain_type(new, old=None, typ=None):\n \"Cast `new` to type of `old` if it's a superclass\"\n # e.g. old is TensorImage, new is Tensor - if not subclass then do nothing\n assert old is not None or typ is not None\n if typ is None:\n if not isinstance(old, type(new)): return new\n typ = old if isinstance(old,type) else type(old)\n # Do nothing the new type is already an instance of requested type (i.e. same type)\n return typ(new) if typ!=NoneType and not isinstance(new, typ) else new",
"_____no_output_____"
],
[
"class _T(tuple): pass\na = _T((1,2))\nb = tuple((1,2))\ntest_eq_type(retain_type(b, typ=_T), a)",
"_____no_output_____"
],
[
"#export\ndef retain_types(new, old=None, typs=None):\n \"Cast each item of `new` to type of matching item in `old` if it's a superclass\"\n assert old is not None or typs is not None\n return tuple(L(new,L(old),L(typs)).mapped_zip(retain_type, longest=True))",
"_____no_output_____"
],
[
"class T(tuple): pass\n\nt1,t2 = retain_types((tensor(1),(tensor(1),)), (TensorBase(2),T((2,))))\ntest_eq_type(t1, TensorBase(1))\ntest_eq_type(t2, T((tensor(1),)))",
"_____no_output_____"
]
],
[
[
"### Collection functions",
"_____no_output_____"
]
],
[
[
"#export\ndef tuplify(o, use_list=False, match=None):\n \"Make `o` a tuple\"\n return tuple(L(o, use_list=use_list, match=match))",
"_____no_output_____"
],
[
"test_eq(tuplify(None),())\ntest_eq(tuplify([1,2,3]),(1,2,3))\ntest_eq(tuplify(1,match=[1,2,3]),(1,1,1))",
"_____no_output_____"
],
[
"#export\ndef replicate(item,match):\n \"Create tuple of `item` copied `len(match)` times\"\n return (item,)*len(match)",
"_____no_output_____"
],
[
"t = [1,1]\ntest_eq(replicate([1,2], t),([1,2],[1,2]))\ntest_eq(replicate(1, t),(1,1))",
"_____no_output_____"
],
[
"#export\ndef uniqueify(x, sort=False, bidir=False, start=None):\n \"Return the unique elements in `x`, optionally `sort`-ed, optionally return the reverse correspondance.\"\n res = list(OrderedDict.fromkeys(x).keys())\n if start is not None: res = L(start)+res\n if sort: res.sort()\n if bidir: return res, {v:k for k,v in enumerate(res)}\n return res",
"_____no_output_____"
],
[
"# test\ntest_eq(set(uniqueify([1,1,0,5,0,3])),{0,1,3,5})\ntest_eq(uniqueify([1,1,0,5,0,3], sort=True),[0,1,3,5])\nv,o = uniqueify([1,1,0,5,0,3], bidir=True)\ntest_eq(v,[1,0,5,3])\ntest_eq(o,{1:0, 0: 1, 5: 2, 3: 3})\nv,o = uniqueify([1,1,0,5,0,3], sort=True, bidir=True)\ntest_eq(v,[0,1,3,5])\ntest_eq(o,{0:0, 1: 1, 3: 2, 5: 3})",
"_____no_output_____"
],
[
"# export\ndef setify(o): return o if isinstance(o,set) else set(L(o))",
"_____no_output_____"
],
[
"# test\ntest_eq(setify(None),set())\ntest_eq(setify('abc'),{'abc'})\ntest_eq(setify([1,2,2]),{1,2})\ntest_eq(setify(range(0,3)),{0,1,2})\ntest_eq(setify({1,2}),{1,2})",
"_____no_output_____"
],
[
"#export\ndef is_listy(x):\n \"`isinstance(x, (tuple,list,L))`\"\n return isinstance(x, (tuple,list,L,slice,Generator))",
"_____no_output_____"
],
[
"assert is_listy([1])\nassert is_listy(L([1]))\nassert is_listy(slice(2))\nassert not is_listy(torch.tensor([1]))",
"_____no_output_____"
],
[
"#export\ndef range_of(x):\n \"All indices of collection `x` (i.e. `list(range(len(x)))`)\"\n return list(range(len(x)))",
"_____no_output_____"
],
[
"test_eq(range_of([1,1,1,1]), [0,1,2,3])",
"_____no_output_____"
],
[
"#export\ndef groupby(x, key):\n \"Like `itertools.groupby` but doesn't need to be sorted, and isn't lazy\"\n res = {}\n for o in x: res.setdefault(key(o), []).append(o)\n return res",
"_____no_output_____"
],
[
"test_eq(groupby('aa ab bb'.split(), itemgetter(0)), {'a':['aa','ab'], 'b':['bb']})",
"_____no_output_____"
],
[
"#export\ndef merge(*ds):\n \"Merge all dictionaries in `ds`\"\n return {k:v for d in ds for k,v in d.items()}",
"_____no_output_____"
],
[
"test_eq(merge(), {})\ntest_eq(merge(dict(a=1,b=2)), dict(a=1,b=2))\ntest_eq(merge(dict(a=1,b=2), dict(b=3,c=4)), dict(a=1, b=3, c=4))",
"_____no_output_____"
],
[
"#export\ndef shufflish(x, pct=0.04):\n \"Randomly relocate items of `x` up to `pct` of `len(x)` from their starting location\"\n n = len(x)\n return L(x[i] for i in sorted(range_of(x), key=lambda o: o+n*(1+random.random()*pct)))",
"_____no_output_____"
],
[
"l = list(range(100))\nl2 = array(shufflish(l))\ntest_close(l2[:50 ].mean(), 25, eps=5)\ntest_close(l2[-50:].mean(), 75, eps=5)\ntest_ne(l,l2)",
"_____no_output_____"
],
[
"#export\nclass IterLen:\n \"Base class to add iteration to anything supporting `len` and `__getitem__`\"\n def __iter__(self): return (self[i] for i in range_of(self))",
"_____no_output_____"
],
[
"#export\n@docs\nclass ReindexCollection(GetAttr, IterLen):\n \"Reindexes collection `coll` with indices `idxs` and optional LRU cache of size `cache`\"\n def __init__(self, coll, idxs=None, cache=None):\n self.default,self.coll,self.idxs,self.cache = coll,coll,ifnone(idxs,L.range(coll)),cache\n def _get(self, i): return self.coll[i]\n self._get = types.MethodType(_get,self)\n if cache is not None: self._get = functools.lru_cache(maxsize=cache)(self._get)\n\n def __getitem__(self, i): return self._get(self.idxs[i])\n def __len__(self): return len(self.coll)\n def reindex(self, idxs): self.idxs = idxs\n def shuffle(self): random.shuffle(self.idxs)\n def cache_clear(self): self._get.cache_clear()\n \n _docs = dict(reindex=\"Replace `self.idxs` with idxs\",\n shuffle=\"Randomly shuffle indices\",\n cache_clear=\"Clear LRU cache\")",
"_____no_output_____"
],
[
"sz = 50\nt = ReindexCollection(L.range(sz), cache=2)\ntest_eq(list(t), range(sz))\ntest_eq(t[sz-1], sz-1)\ntest_eq(t._get.cache_info().hits, 1)\nt.shuffle()\ntest_eq(t._get.cache_info().hits, 1)\ntest_ne(list(t), range(sz))\ntest_eq(set(t), set(range(sz)))\nt.cache_clear()\ntest_eq(t._get.cache_info().hits, 0)\ntest_eq(t.count(0), 1)",
"_____no_output_____"
],
[
"#export\ndef _oper(op,a,b=None): return (lambda o:op(o,a)) if b is None else op(a,b)\n\ndef _mk_op(nm, mod=None):\n \"Create an operator using `oper` and add to the caller's module\"\n if mod is None: mod = inspect.currentframe().f_back.f_locals\n op = getattr(operator,nm)\n def _inner(a,b=None): return _oper(op, a,b)\n _inner.__name__ = _inner.__qualname__ = nm\n _inner.__doc__ = f'Same as `operator.{nm}`, or returns partial if 1 arg'\n mod[nm] = _inner",
"_____no_output_____"
],
[
"#export\n_all_ = ['lt', 'gt', 'le', 'ge', 'eq', 'ne', 'add', 'sub', 'mul', 'truediv']",
"_____no_output_____"
],
[
"#export\nfor op in 'lt gt le ge eq ne add sub mul truediv'.split(): _mk_op(op)",
"_____no_output_____"
]
],
[
[
"The following functions are provided matching the behavior of the equivalent versions in `operator`:\n\n - *lt gt le ge eq ne add sub mul truediv*",
"_____no_output_____"
]
],
[
[
"lt(3,5),gt(3,5)",
"_____no_output_____"
]
],
[
[
"However, they also have additional functionality: if you only pass one param, they return a partial function that passes that param as the second positional parameter.",
"_____no_output_____"
]
],
[
[
"lt(5)(3),gt(5)(3)",
"_____no_output_____"
],
[
"#export\nclass _InfMeta(type):\n @property\n def count(self): return itertools.count()\n @property\n def zeros(self): return itertools.cycle([0])\n @property\n def ones(self): return itertools.cycle([1])\n @property\n def nones(self): return itertools.cycle([None])",
"_____no_output_____"
],
[
"#export\nclass Inf(metaclass=_InfMeta):\n \"Infinite lists\"\n pass",
"_____no_output_____"
]
],
[
[
"`Inf` defines the following properties:\n \n- `count: itertools.count()`\n- `zeros: itertools.cycle([0])`\n- `ones : itertools.cycle([1])`\n- `nones: itertools.cycle([None])`",
"_____no_output_____"
]
],
[
[
"test_eq([o for i,o in zip(range(5), Inf.count)],\n [0, 1, 2, 3, 4])\n\ntest_eq([o for i,o in zip(range(5), Inf.zeros)],\n [0, 0, 0, 0, 0])",
"_____no_output_____"
],
[
"#export\ndef true(*args, **kwargs):\n \"Predicate: always `True`\"\n return True",
"_____no_output_____"
],
[
"#export\ndef stop(e=StopIteration):\n \"Raises exception `e` (by default `StopException`) even if in an expression\"\n raise e",
"_____no_output_____"
],
[
"#export\ndef gen(func, seq, cond=true):\n \"Like `(func(o) for o in seq if cond(func(o)))` but handles `StopIteration`\"\n return itertools.takewhile(cond, map(func,seq))",
"_____no_output_____"
],
[
"test_eq(gen(noop, Inf.count, lt(5)),\n range(5))\ntest_eq(gen(operator.neg, Inf.count, gt(-5)),\n [0,-1,-2,-3,-4])\ntest_eq(gen(lambda o:o if o<5 else stop(), Inf.count),\n range(5))",
"_____no_output_____"
],
[
"#export\ndef chunked(it, cs, drop_last=False):\n if not isinstance(it, Iterator): it = iter(it)\n while True:\n res = list(itertools.islice(it, cs))\n if res and (len(res)==cs or not drop_last): yield res\n if len(res)<cs: return",
"_____no_output_____"
],
[
"t = L.range(10)\ntest_eq(chunked(t,3), [[0,1,2], [3,4,5], [6,7,8], [9]])\ntest_eq(chunked(t,3,True), [[0,1,2], [3,4,5], [6,7,8], ])\n\nt = map(lambda o:stop() if o==6 else o, Inf.count)\ntest_eq(chunked(t,3), [[0, 1, 2], [3, 4, 5]])\nt = map(lambda o:stop() if o==7 else o, Inf.count)\ntest_eq(chunked(t,3), [[0, 1, 2], [3, 4, 5], [6]])\n\nt = tensor(range(10))\ntest_eq(chunked(t,3), [[0,1,2], [3,4,5], [6,7,8], [9]])\ntest_eq(chunked(t,3,True), [[0,1,2], [3,4,5], [6,7,8], ])",
"_____no_output_____"
],
[
"#export\ndef concat(*ls):\n \"Concatenate tensors, arrays, lists, or tuples\"\n if not len(ls): return []\n it = ls[0]\n if isinstance(it,torch.Tensor): res = torch.cat(ls) \n elif isinstance(it,ndarray): res = np.concatenate(ls)\n else:\n res = [o for x in ls for o in L(x)]\n if isinstance(it,(tuple,list)): res = type(it)(res)\n else: res = L(res)\n return retain_type(res, it)",
"_____no_output_____"
],
[
"a,b,c = [1],[1,2],[1,1,2]\ntest_eq(concat(a,b), c)\ntest_eq_type(concat(tuple (a),tuple (b)), tuple (c))\ntest_eq_type(concat(array (a),array (b)), array (c))\ntest_eq_type(concat(tensor(a),tensor(b)), tensor(c))\ntest_eq_type(concat(TensorBase(a),TensorBase(b)), TensorBase(c))\ntest_eq_type(concat([1,1],1), [1,1,1])\ntest_eq_type(concat(1,1,1), L(1,1,1))\ntest_eq_type(concat(L(1,2),1), L(1,2,1))",
"_____no_output_____"
]
],
[
[
"### Chunks -",
"_____no_output_____"
]
],
[
[
"#export\nclass Chunks:\n \"Slice and int indexing into a list of lists\"\n def __init__(self, chunks, lens=None):\n self.chunks = chunks\n self.lens = L(map(len,self.chunks) if lens is None else lens)\n self.cumlens = np.cumsum(0+self.lens)\n self.totlen = self.cumlens[-1]\n\n def __getitem__(self,i):\n if isinstance(i,slice): return self.getslice(i)\n di,idx = self.doc_idx(i)\n return self.chunks[di][idx]\n\n def getslice(self, i):\n st_d,st_i = self.doc_idx(ifnone(i.start,0))\n en_d,en_i = self.doc_idx(ifnone(i.stop,self.totlen+1))\n res = [self.chunks[st_d][st_i:(en_i if st_d==en_d else sys.maxsize)]]\n for b in range(st_d+1,en_d): res.append(self.chunks[b])\n if st_d!=en_d and en_d<len(self.chunks): res.append(self.chunks[en_d][:en_i])\n return concat(*res)\n \n def doc_idx(self, i):\n if i<0: i=self.totlen+i # count from end\n docidx = np.searchsorted(self.cumlens, i+1)-1\n cl = self.cumlens[docidx]\n return docidx,i-cl",
"_____no_output_____"
],
[
"docs = L(list(string.ascii_lowercase[a:b]) for a,b in ((0,3),(3,7),(7,8),(8,16),(16,24),(24,26)))\n\nb = Chunks(docs)\ntest_eq([b[ o] for o in range(0,5)], ['a','b','c','d','e'])\ntest_eq([b[-o] for o in range(1,6)], ['z','y','x','w','v'])\ntest_eq(b[6:13], 'g,h,i,j,k,l,m'.split(','))\ntest_eq(b[20:77], 'u,v,w,x,y,z'.split(','))\ntest_eq(b[:5], 'a,b,c,d,e'.split(','))\ntest_eq(b[:2], 'a,b'.split(','))",
"_____no_output_____"
],
[
"t = torch.arange(26)\ndocs = L(t[a:b] for a,b in ((0,3),(3,7),(7,8),(8,16),(16,24),(24,26)))\nb = Chunks(docs)\ntest_eq([b[ o] for o in range(0,5)], range(0,5))\ntest_eq([b[-o] for o in range(1,6)], [25,24,23,22,21])\ntest_eq(b[6:13], torch.arange(6,13))\ntest_eq(b[20:77], torch.arange(20,26))\ntest_eq(b[:5], torch.arange(5))\ntest_eq(b[:2], torch.arange(2))",
"_____no_output_____"
],
[
"docs = L(TensorBase(t[a:b]) for a,b in ((0,3),(3,7),(7,8),(8,16),(16,24),(24,26)))\nb = Chunks(docs)\ntest_eq_type(b[:2], TensorBase(range(2)))\ntest_eq_type(b[:5], TensorBase(range(5)))\ntest_eq_type(b[9:13], TensorBase(range(9,13)))",
"_____no_output_____"
],
[
"type(b[9:13])",
"_____no_output_____"
]
],
[
[
"### Functions on functions",
"_____no_output_____"
]
],
[
[
"#export\ndef trace(f):\n \"Add `set_trace` to an existing function `f`\"\n def _inner(*args,**kwargs):\n set_trace()\n return f(*args,**kwargs)\n return _inner",
"_____no_output_____"
],
[
"# export\ndef compose(*funcs, order=None):\n \"Create a function that composes all functions in `funcs`, passing along remaining `*args` and `**kwargs` to all\"\n funcs = L(funcs)\n if order is not None: funcs = funcs.sorted(order)\n def _inner(x, *args, **kwargs):\n for f in L(funcs): x = f(x, *args, **kwargs)\n return x\n return _inner",
"_____no_output_____"
],
[
"f1 = lambda o,p=0: (o*2)+p\nf2 = lambda o,p=1: (o+1)/p\ntest_eq(f2(f1(3)), compose(f1,f2)(3))\ntest_eq(f2(f1(3,p=3),p=3), compose(f1,f2)(3,p=3))\ntest_eq(f2(f1(3, 3), 3), compose(f1,f2)(3, 3))\n\nf1.order = 1\ntest_eq(f1(f2(3)), compose(f1,f2, order=\"order\")(3))",
"_____no_output_____"
],
[
"#export\ndef maps(*args, retain=noop):\n \"Like `map`, except funcs are composed first\"\n f = compose(*args[:-1])\n def _f(b): return retain(f(b), b)\n return map(_f, args[-1])",
"_____no_output_____"
],
[
"test_eq(maps([1]), [1])\ntest_eq(maps(operator.neg, [1,2]), [-1,-2])\ntest_eq(maps(operator.neg, operator.neg, [1,2]), [1,2])\n\ntest_eq_type(list(maps(operator.neg, [TensorBase(1), 2], retain=retain_type)), \n [TensorBase(-1), -2])",
"_____no_output_____"
],
[
"#export\ndef mapper(f):\n \"Create a function that maps `f` over an input collection\"\n return lambda o: [f(o_) for o_ in o]",
"_____no_output_____"
],
[
"func = mapper(lambda o:o*2)\ntest_eq(func(range(3)),[0,2,4])",
"_____no_output_____"
],
[
"#export\ndef partialler(f, *args, order=None, **kwargs):\n \"Like `functools.partial` but also copies over docstring\"\n fnew = partial(f,*args,**kwargs)\n fnew.__doc__ = f.__doc__\n if order is not None: fnew.order=order\n elif hasattr(f,'order'): fnew.order=f.order\n return fnew",
"_____no_output_____"
],
[
"def _f(x,a=1):\n \"test func\"\n return x+a\n_f.order=1\n\nf = partialler(_f, a=2)\ntest_eq(f.order, 1)\nf = partialler(_f, a=2, order=3)\ntest_eq(f.__doc__, \"test func\")\ntest_eq(f.order, 3)\ntest_eq(f(3), _f(3,2))",
"_____no_output_____"
],
[
"#export\ndef instantiate(t):\n \"Instantiate `t` if it's a type, otherwise do nothing\"\n return t() if isinstance(t, type) else t",
"_____no_output_____"
],
[
"test_eq_type(instantiate(int), 0)\ntest_eq_type(instantiate(1), 1)",
"_____no_output_____"
],
[
"#export\nmk_class('_Arg', 'i')\n_0,_1,_2,_3,_4 = _Arg(0),_Arg(1),_Arg(2),_Arg(3),_Arg(4)",
"_____no_output_____"
],
[
"#export\nclass bind:\n \"Same as `partial`, except you can use `_0` `_1` etc param placeholders\"\n def __init__(self, fn, *pargs, **pkwargs):\n store_attr(self, 'fn,pargs,pkwargs')\n self.maxi = max((x.i for x in pargs if isinstance(x, _Arg)), default=-1)\n\n def __call__(self, *args, **kwargs):\n fargs = L(args[x.i] if isinstance(x, _Arg) else x for x in self.pargs) + args[self.maxi+1:]\n return self.fn(*fargs, **{**self.pkwargs, **kwargs})",
"_____no_output_____"
],
[
"def myfn(a,b,c,d=1,e=2): return(a,b,c,d,e)\ntest_eq(bind(myfn, _1, 17, _0, e=3)(19,14), (14,17,19,1,3))\ntest_eq(bind(myfn, 17, _0, e=3)(19,14), (17,19,14,1,3))\ntest_eq(bind(myfn, 17, e=3)(19,14), (17,19,14,1,3))\ntest_eq(bind(myfn)(17,19,14), (17,19,14,1,2))",
"_____no_output_____"
]
],
[
[
"### File and network functions",
"_____no_output_____"
]
],
[
[
"#export\n#NB: Please don't move this to a different line or module, since it's used in testing `get_source_link`\n@patch\ndef ls(self:Path, file_type=None, file_exts=None):\n \"Contents of path as a list\"\n extns=L(file_exts)\n if file_type: extns += L(k for k,v in mimetypes.types_map.items() if v.startswith(file_type))\n return L(self.iterdir()).filtered(lambda x: len(extns)==0 or x.suffix in extns)",
"_____no_output_____"
]
],
[
[
"We add an `ls()` method to `pathlib.Path` which is simply defined as `list(Path.iterdir())`, mainly for convenience in REPL environments such as notebooks.",
"_____no_output_____"
]
],
[
[
"path = Path()\nt = path.ls()\nassert len(t)>0\nt[0]",
"_____no_output_____"
]
],
[
[
"You can also pass an optional `file_type` MIME prefix and/or a list of file extensions.",
"_____no_output_____"
]
],
[
[
"txt_files=path.ls(file_type='text')\nassert len(txt_files) > 0 and txt_files[0].suffix=='.py'\nipy_files=path.ls(file_exts=['.ipynb'])\nassert len(ipy_files) > 0 and ipy_files[0].suffix=='.ipynb'\ntxt_files[0],ipy_files[0]",
"_____no_output_____"
],
[
"#hide\npkl = pickle.dumps(path)\np2 =pickle.loads(pkl)\ntest_eq(path.ls()[0], p2.ls()[0])",
"_____no_output_____"
],
[
"def bunzip(fn):\n \"bunzip `fn`, raising exception if output already exists\"\n fn = Path(fn)\n assert fn.exists(), f\"{fn} doesn't exist\"\n out_fn = fn.with_suffix('')\n assert not out_fn.exists(), f\"{out_fn} already exists\"\n with bz2.BZ2File(fn, 'rb') as src, out_fn.open('wb') as dst:\n for d in iter(lambda: src.read(1024*1024), b''): dst.write(d)",
"_____no_output_____"
],
[
"f = Path('files/test.txt')\nif f.exists(): f.unlink()\nbunzip('files/test.txt.bz2')\nt = f.open().readlines()\ntest_eq(len(t),1)\ntest_eq(t[0], 'test\\n')\nf.unlink()",
"_____no_output_____"
]
],
[
[
"### Tensor functions",
"_____no_output_____"
]
],
[
[
"#export\ndef apply(func, x, *args, **kwargs):\n \"Apply `func` recursively to `x`, passing on args\"\n if is_listy(x): return type(x)(apply(func, o, *args, **kwargs) for o in x)\n if isinstance(x,dict): return {k: apply(func, v, *args, **kwargs) for k,v in x.items()}\n return retain_type(func(x, *args, **kwargs), x)",
"_____no_output_____"
],
[
"#export\ndef to_detach(b, cpu=True):\n \"Recursively detach lists of tensors in `b `; put them on the CPU if `cpu=True`.\"\n def _inner(x, cpu=True):\n if not isinstance(x,Tensor): return x\n x = x.detach()\n return x.cpu() if cpu else x\n return apply(_inner, b, cpu=cpu)",
"_____no_output_____"
],
[
"#export\ndef to_half(b):\n \"Recursively map lists of tensors in `b ` to FP16.\"\n return apply(lambda x: x.half() if torch.is_floating_point(x) else x, b)",
"_____no_output_____"
],
[
"#export\ndef to_float(b):\n \"Recursively map lists of int tensors in `b ` to float.\"\n return apply(lambda x: x.float() if torch.is_floating_point(x) else x, b)",
"_____no_output_____"
],
[
"#export\n# None: True if available; True: error if not availabe; False: use CPU\ndefaults.use_cuda = None",
"_____no_output_____"
],
[
"#export\ndef default_device(use_cuda=-1):\n \"Return or set default device; `use_cuda`: None - CUDA if available; True - error if not availabe; False - CPU\"\n if use_cuda != -1: defaults.use_cuda=use_cuda\n use = defaults.use_cuda or (torch.cuda.is_available() and defaults.use_cuda is None)\n assert torch.cuda.is_available() or not use\n return torch.device(torch.cuda.current_device()) if use else torch.device('cpu')",
"_____no_output_____"
],
[
"#cuda\n_td = torch.device(torch.cuda.current_device())\ntest_eq(default_device(None), _td)\ntest_eq(default_device(True), _td)\ntest_eq(default_device(False), torch.device('cpu'))\ndefault_device(None);",
"_____no_output_____"
],
[
"#export\ndef to_device(b, device=None):\n \"Recursively put `b` on `device`.\"\n if device is None: device=default_device()\n def _inner(o): return o.to(device, non_blocking=True) if isinstance(o,Tensor) else o\n return apply(_inner, b)",
"_____no_output_____"
],
[
"t = to_device((3,(tensor(3),tensor(2))))\nt1,(t2,t3) = t\ntest_eq_type(t,(3,(tensor(3).cuda(),tensor(2).cuda())))\ntest_eq(t2.type(), \"torch.cuda.LongTensor\")\ntest_eq(t3.type(), \"torch.cuda.LongTensor\")",
"_____no_output_____"
],
[
"#export\ndef to_cpu(b):\n \"Recursively map lists of tensors in `b ` to the cpu.\"\n return to_device(b,'cpu')",
"_____no_output_____"
],
[
"t3 = to_cpu(t3)\ntest_eq(t3.type(), \"torch.LongTensor\")\ntest_eq(t3, 2)",
"_____no_output_____"
],
[
"def to_np(x):\n \"Convert a tensor to a numpy array.\"\n return x.data.cpu().numpy()",
"_____no_output_____"
],
[
"t3 = to_np(t3)\ntest_eq(type(t3), np.ndarray)\ntest_eq(t3, 2)",
"_____no_output_____"
],
[
"#export\ndef item_find(x, idx=0):\n \"Recursively takes the `idx`-th element of `x`\"\n if is_listy(x): return item_find(x[idx])\n if isinstance(x,dict): \n key = list(x.keys())[idx] if isinstance(idx, int) else idx\n return item_find(x[key])\n return x",
"_____no_output_____"
],
[
"#export\ndef find_device(b):\n \"Recursively search the device of `b`.\"\n return item_find(b).device",
"_____no_output_____"
],
[
"dev = default_device()\ntest_eq(find_device(t2), dev)\ntest_eq(find_device([t2,t2]), dev)\ntest_eq(find_device({'a':t2,'b':t2}), dev)\ntest_eq(find_device({'a':[[t2],[t2]],'b':t2}), dev)",
"_____no_output_____"
],
[
"#export\ndef find_bs(b):\n \"Recursively search the batch size of `b`.\"\n return item_find(b).shape[0]",
"_____no_output_____"
],
[
"x = torch.randn(4,5)\ntest_eq(find_bs(x), 4)\ntest_eq(find_bs([x, x]), 4)\ntest_eq(find_bs({'a':x,'b':x}), 4)\ntest_eq(find_bs({'a':[[x],[x]],'b':x}), 4)",
"_____no_output_____"
],
[
"def np_func(f):\n \"Convert a function taking and returning numpy arrays to one taking and returning tensors\"\n def _inner(*args, **kwargs):\n nargs = [to_np(arg) if isinstance(arg,Tensor) else arg for arg in args]\n return tensor(f(*nargs, **kwargs))\n functools.update_wrapper(_inner, f)\n return _inner",
"_____no_output_____"
]
],
[
[
"This decorator is particularly useful for using numpy functions as fastai metrics, for instance:",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import f1_score\n\n@np_func\ndef f1(inp,targ): return f1_score(targ, inp)\n\na1,a2 = array([0,1,1]),array([1,0,1])\nt = f1(tensor(a1),tensor(a2))\ntest_eq(f1_score(a1,a2), t)\nassert isinstance(t,Tensor)",
"_____no_output_____"
],
[
"class Module(nn.Module, metaclass=PrePostInitMeta):\n \"Same as `nn.Module`, but no need for subclasses to call `super().__init__`\"\n def __pre_init__(self): super().__init__()\n def __init__(self): pass",
"_____no_output_____"
],
[
"show_doc(Module, title_level=3)",
"_____no_output_____"
],
[
"class _T(Module):\n def __init__(self): self.f = nn.Linear(1,1)\n def forward(self,x): return self.f(x)\n\nt = _T()\nt(tensor([1.]))",
"_____no_output_____"
]
],
[
[
"### Sorting objects from before/after",
"_____no_output_____"
],
[
"Transforms and callbacks will have run_after/run_before attributes, this function will sort them to respect those requirements (if it's possible). Also, sometimes we want a tranform/callback to be run at the end, but still be able to use run_after/run_before behaviors. For those, the function checks for a toward_end attribute (that needs to be True).",
"_____no_output_____"
]
],
[
[
"#export\ndef _is_instance(f, gs):\n tst = [g if type(g) in [type, 'function'] else g.__class__ for g in gs]\n for g in tst:\n if isinstance(f, g) or f==g: return True\n return False\n\ndef _is_first(f, gs):\n for o in L(getattr(f, 'run_after', None)): \n if _is_instance(o, gs): return False\n for g in gs:\n if _is_instance(f, L(getattr(g, 'run_before', None))): return False\n return True\n\ndef sort_by_run(fs):\n end = L(getattr(f, 'toward_end', False) for f in fs)\n inp,res = L(fs)[~end] + L(fs)[end], []\n while len(inp) > 0:\n for i,o in enumerate(inp):\n if _is_first(o, inp): \n res.append(inp.pop(i))\n break\n else: raise Exception(\"Impossible to sort\")\n return res",
"_____no_output_____"
],
[
"class Tst(): pass \nclass Tst1():\n run_before=[Tst]\nclass Tst2():\n run_before=Tst\n run_after=Tst1\n \ntsts = [Tst(), Tst1(), Tst2()]\ntest_eq(sort_by_run(tsts), [tsts[1], tsts[2], tsts[0]])\n\nTst2.run_before,Tst2.run_after = Tst1,Tst\ntest_fail(lambda: sort_by_run([Tst(), Tst1(), Tst2()]))\n\ndef tst1(x): return x\ntst1.run_before = Tst\ntest_eq(sort_by_run([tsts[0], tst1]), [tst1, tsts[0]])\n \nclass Tst1():\n toward_end=True\nclass Tst2():\n toward_end=True\n run_before=Tst1\ntsts = [Tst(), Tst1(), Tst2()]\ntest_eq(sort_by_run(tsts), [tsts[0], tsts[2], tsts[1]])",
"_____no_output_____"
]
],
[
[
"### Other helpers",
"_____no_output_____"
]
],
[
[
"#export\ndef round_multiple(x, mult, round_down=False):\n \"Round `x` to nearest multiple of `mult`\"\n def _f(x_): return (int if round_down else round)(x_/mult)*mult\n res = L(x).mapped(_f)\n return res if is_listy(x) else res[0]",
"_____no_output_____"
],
[
"test_eq(round_multiple(63,32), 64)\ntest_eq(round_multiple(50,32), 64)\ntest_eq(round_multiple(40,32), 32)\ntest_eq(round_multiple( 0,32), 0)\ntest_eq(round_multiple(63,32, round_down=True), 32)\ntest_eq(round_multiple((63,40),32), (64,32))",
"_____no_output_____"
],
[
"#export\ndef num_cpus():\n \"Get number of cpus\"\n try: return len(os.sched_getaffinity(0))\n except AttributeError: return os.cpu_count()\n \ndefaults.cpus = min(16, num_cpus())",
"_____no_output_____"
],
[
"#export\ndef add_props(f, n=2):\n \"Create properties passing each of `range(n)` to f\"\n return (property(partial(f,i)) for i in range(n))",
"_____no_output_____"
],
[
"class _T(): a,b = add_props(lambda i,x:i*2)\n\nt = _T()\ntest_eq(t.a,0)\ntest_eq(t.b,2)",
"_____no_output_____"
]
],
[
[
"### Image helpers",
"_____no_output_____"
],
[
"This is a quick way to generate, for instance, *train* and *valid* versions of a property. See `DataBunch` definition for an example of this.",
"_____no_output_____"
]
],
[
[
"#export\ndef make_cross_image(bw=True):\n \"Create a tensor containing a cross image, either `bw` (True) or color\"\n if bw:\n im = torch.zeros(5,5)\n im[2,:] = 1.\n im[:,2] = 1.\n else:\n im = torch.zeros(3,5,5)\n im[0,2,:] = 1.\n im[1,:,2] = 1.\n return im",
"_____no_output_____"
],
[
"plt.imshow(make_cross_image(), cmap=\"Greys\");",
"_____no_output_____"
],
[
"plt.imshow(make_cross_image(False).permute(1,2,0));",
"_____no_output_____"
],
[
"#export\ndef show_title(o, ax=None, ctx=None, label=None, **kwargs):\n \"Set title of `ax` to `o`, or print `o` if `ax` is `None`\"\n ax = ifnone(ax,ctx)\n if ax is None: print(o)\n elif hasattr(ax, 'set_title'): ax.set_title(o)\n elif isinstance(ax, pd.Series):\n while label in ax: label += '_'\n ax = ax.append(pd.Series({label: o}))\n return ax",
"_____no_output_____"
],
[
"test_stdout(lambda: show_title(\"title\"), \"title\")\n# ensure that col names are unique when showing to a pandas series\nassert show_title(\"title\", ctx=pd.Series(dict(a=1)), label='a').equals(pd.Series(dict(a=1,a_='title')))",
"_____no_output_____"
],
[
"#export\ndef show_image(im, ax=None, figsize=None, title=None, ctx=None, **kwargs):\n \"Show a PIL or PyTorch image on `ax`.\"\n ax = ifnone(ax,ctx)\n if ax is None: _,ax = plt.subplots(figsize=figsize)\n # Handle pytorch axis order\n if isinstance(im,Tensor):\n im = to_cpu(im)\n if im.shape[0]<5: im=im.permute(1,2,0)\n elif not isinstance(im,np.ndarray): im=array(im)\n # Handle 1-channel images\n if im.shape[-1]==1: im=im[...,0]\n ax.imshow(im, **kwargs)\n if title is not None: ax.set_title(title)\n ax.axis('off')\n return ax",
"_____no_output_____"
]
],
[
[
"`show_image` can show b&w images...",
"_____no_output_____"
]
],
[
[
"im = make_cross_image()\nax = show_image(im, cmap=\"Greys\", figsize=(2,2))",
"_____no_output_____"
]
],
[
[
"...and color images with standard `c*h*w` dim order...",
"_____no_output_____"
]
],
[
[
"im2 = make_cross_image(False)\nax = show_image(im2, figsize=(2,2))",
"_____no_output_____"
]
],
[
[
"...and color images with `h*w*c` dim order...",
"_____no_output_____"
]
],
[
[
"im3 = im2.permute(1,2,0)\nax = show_image(im3, figsize=(2,2))",
"_____no_output_____"
],
[
"ax = show_image(im, cmap=\"Greys\", figsize=(2,2))\nshow_title(\"Cross\", ax)",
"_____no_output_____"
],
[
"#export\ndef show_titled_image(o, **kwargs):\n \"Call `show_image` destructuring `o` to `(img,title)`\"\n show_image(o[0], title=str(o[1]), **kwargs)",
"_____no_output_____"
],
[
"#export\ndef show_image_batch(b, show=show_titled_image, items=9, cols=3, figsize=None, **kwargs):\n \"Display batch `b` in a grid of size `items` with `cols` width\"\n rows = (items+cols-1) // cols\n if figsize is None: figsize = (cols*3, rows*3)\n fig,axs = plt.subplots(rows, cols, figsize=figsize)\n for *o,ax in zip(*to_cpu(b), axs.flatten()): show(o, ax=ax, **kwargs)",
"_____no_output_____"
],
[
"show_image_batch(([im,im2,im3],['bw','chw','hwc']), items=3)",
"_____no_output_____"
]
],
[
[
"# Export -",
"_____no_output_____"
]
],
[
[
"#hide\nfrom local.notebook.export import notebook2script\nnotebook2script(all_fs=True)",
"Converted 00_test.ipynb.\nConverted 01_core.ipynb.\nConverted 01a_dataloader.ipynb.\nConverted 01a_script.ipynb.\nConverted 02_transforms.ipynb.\nConverted 03_pipeline.ipynb.\nConverted 04_data_external.ipynb.\nConverted 05_data_core.ipynb.\nConverted 06_data_source.ipynb.\nConverted 07_vision_core.ipynb.\nConverted 08_pets_tutorial.ipynb.\nConverted 09_vision_augment.ipynb.\nConverted 11_layers.ipynb.\nConverted 12_optimizer.ipynb.\nConverted 13_learner.ipynb.\nConverted 14_callback_schedule.ipynb.\nConverted 15_callback_hook.ipynb.\nConverted 16_callback_progress.ipynb.\nConverted 17_callback_tracker.ipynb.\nConverted 18_callback_fp16.ipynb.\nConverted 19_callback_mixup.ipynb.\nConverted 20_metrics.ipynb.\nConverted 21_tutorial_imagenette.ipynb.\nConverted 30_text_core.ipynb.\nConverted 31_text_data.ipynb.\nConverted 32_text_models_awdlstm.ipynb.\nConverted 33_test_models_core.ipynb.\nConverted 34_callback_rnn.ipynb.\nConverted 35_tutorial_wikitext.ipynb.\nConverted 36_text_models_qrnn.ipynb.\nConverted 40_tabular_core.ipynb.\nConverted 41_tabular_model.ipynb.\nConverted 50_data_block.ipynb.\nConverted 60_vision_models_xresnet.ipynb.\nConverted 90_notebook_core.ipynb.\nConverted 91_notebook_export.ipynb.\nConverted 92_notebook_showdoc.ipynb.\nConverted 93_notebook_export2html.ipynb.\nConverted 94_index.ipynb.\nConverted 95_synth_learner.ipynb.\nConverted notebook2jekyll.ipynb.\n"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
]
|
e794928616ea32d31c674d7390870f05a5937b17 | 1,605 | ipynb | Jupyter Notebook | Chapter01/Activity03/Activity03.ipynb | stuffstuffstuf1/The-Python-Workshop | b529995980a7a8f8f09e9d2f8dd20d6e4d6acb80 | [
"MIT"
]
| 238 | 2019-12-13T15:44:34.000Z | 2022-03-21T05:38:21.000Z | Chapter01/Activity03/Activity03.ipynb | stuffstuffstuf1/The-Python-Workshop | b529995980a7a8f8f09e9d2f8dd20d6e4d6acb80 | [
"MIT"
]
| 8 | 2020-05-04T03:33:29.000Z | 2022-03-12T00:47:26.000Z | Chapter01/Activity03/Activity03.ipynb | stuffstuffstuf1/The-Python-Workshop | b529995980a7a8f8f09e9d2f8dd20d6e4d6acb80 | [
"MIT"
]
| 345 | 2019-10-08T09:15:11.000Z | 2022-03-31T18:28:03.000Z | 18.448276 | 83 | 0.497819 | [
[
[
"# Choose a question to ask\nprint('How would you rate your day on a scale of 1 to 10?')",
"How would you rate your day on a scale of 1 to 10?\n"
],
[
"# Set a variable equal to input()\nday_rating = input()",
"9\n"
],
[
"# Select an appropriate output.\nprint('You feel like a ' + day_rating + ' today. Thanks for letting me know')",
"You feel like a 9 today. Thanks for letting me know\n"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code"
]
]
|
e794b473480366c465a5470981236bda88f559f0 | 7,469 | ipynb | Jupyter Notebook | 15_Optimization/015_two_dimensional_optimization.ipynb | kangwonlee/2109eca-nmisp-template | 2e078870757fa06222df62d0ff8f4f4f288af51a | [
"BSD-3-Clause"
]
| null | null | null | 15_Optimization/015_two_dimensional_optimization.ipynb | kangwonlee/2109eca-nmisp-template | 2e078870757fa06222df62d0ff8f4f4f288af51a | [
"BSD-3-Clause"
]
| null | null | null | 15_Optimization/015_two_dimensional_optimization.ipynb | kangwonlee/2109eca-nmisp-template | 2e078870757fa06222df62d0ff8f4f4f288af51a | [
"BSD-3-Clause"
]
| null | null | null | 23.340625 | 139 | 0.472219 | [
[
[
"import numpy as np\nimport scipy.optimize as so\n\n",
"_____no_output_____"
],
[
"# ref : https://matplotlib.org/stable/gallery/\nimport matplotlib.pyplot as plt\nfrom mpl_toolkits.mplot3d import Axes3D\nfrom matplotlib import cm\n\n",
"_____no_output_____"
]
],
[
[
"## 2차원 최적화<br>Two dimensional optimizations\n\n다음과 같은 비용 함수를 생각해 보자.<br>Let's think about a cost function as follows.\n\n$$\nC(x_0, x_1) = \\frac{x_0^2}{2^2} + \\frac{x_1^2}{1^2}\n$$\n\n파이썬으로는 다음과 같이 구현할 수 있을 것이다.<br>We may implement in python as follows.\n\n",
"_____no_output_____"
]
],
[
[
"def c(x:np.ndarray, a:float=2, b:float=1) -> float:\n x0 = x[0]\n x1 = x[1]\n \n return (x0 * x0) / (a * a) + (x1 * x1) / (b * b)\n\n",
"_____no_output_____"
]
],
[
[
"시각화 해 보자.<br>Let's visualize.\n\n",
"_____no_output_____"
]
],
[
[
"def plot_cost():\n # ref : https://matplotlib.org/stable/gallery/\n\n fig = plt.figure(figsize=(15, 6))\n ax1 = plt.subplot(1, 2, 1)\n ax2 = plt.subplot(1, 2, 2, projection=\"3d\")\n\n x = np.linspace(-4, 4)\n y = np.linspace(-2, 2)\n X, Y = np.meshgrid(x, y)\n\n Z = c((X, Y))\n\n cset = ax1.contour(X, Y, Z, cmap=cm.coolwarm)\n\n surf = ax2.plot_surface(X, Y, Z, antialiased=True, cmap=cm.viridis, alpha=0.5)\n fig.colorbar(surf)\n\n return ax1, ax2\n\n",
"_____no_output_____"
],
[
"plot_cost()\nplt.show()\n\n",
"_____no_output_____"
]
],
[
[
"중간 과정의 그래프를 그려 주는 비용 함수를 선언<br>Declare another cost function that will plot intermediate results\n\n",
"_____no_output_____"
]
],
[
[
"def get_cost_with_plot(a=2, b=1, b_triangle=True):\n\n x0_history = []\n x1_history = []\n c_history = []\n\n def cost_with_plot(x, a=a, b=b):\n '''\n 이런 함수를 클로져 라고 부름. 다른 함수의 내부 함수이면서 해당 함수의 반환값.\n This is a closuer; an internal function being a return value\n '''\n ax1, ax2 = plot_cost()\n\n result = c(x)\n\n x0_history.append(x[0])\n x1_history.append(x[1])\n c_history.append(result)\n\n ax1.plot(x0_history, x1_history, '.')\n ax2.plot(x0_history, x1_history, c_history, '.')\n\n if b_triangle and (3 <= len(x0_history)):\n ax1.plot(\n x0_history[-3:]+[x0_history[-3]],\n x1_history[-3:]+[x1_history[-3]],\n '-'\n )\n ax2.plot(\n x0_history[-3:]+[x0_history[-3]],\n x1_history[-3:]+[x1_history[-3]],\n c_history[-3:]+[c_history[-3]],\n '-'\n )\n\n plt.show()\n\n return result\n\n return cost_with_plot\n\n",
"_____no_output_____"
],
[
"cost_with_plot = get_cost_with_plot()\n\n",
"_____no_output_____"
]
],
[
[
"### Nelder-Mead 법\nref : [[0]](https://en.wikipedia.org/wiki/Nelder-Mead_method)<br>\nNelder-Mead 법은 비용함수의 독립변수가 $n$ 차원인 경우, $n+1$ 개의 점으로 이루어진 **simplex**를 이용한다.<br>\nIf the independend variables of the cost function is $n$-dimensional, the Nelder-Mead method uses a **simplex** of $n+1$ vertices.\n\n",
"_____no_output_____"
]
],
[
[
"fmin_result = so.fmin(cost_with_plot, [3.0, 1.0])\n\n",
"_____no_output_____"
],
[
"fmin_result\n\n",
"_____no_output_____"
]
],
[
[
"### Newton-CG 법\n비용함수를 각각 $x_0$, $x_1$에 대해 편미분 해 보자.<br>Let's get the partial derivatives of the cost function over $x_0$ and $x_1$.\n$$\nC(x_0, x_1) = \\frac{x_0^2}{2^2} + \\frac{x_1^2}{1^2} \\\\\n\\frac{\\partial C}{\\partial x_0} = 2 \\cdot \\frac{x_0}{2^2} \\\\\n\\frac{\\partial C}{\\partial x_1} = 2 \\cdot \\frac{x_1}{1^2}\n$$\n파이썬으로는 다음과 같이 구현할 수 있을 것이다.<br>One may implement in python as follows.\n\n",
"_____no_output_____"
]
],
[
[
"def jacobian(x, a=2, b=1):\n x0 = x[0]\n x1 = x[1]\n return (2 * x0 / (a*a), 2 * x1 / (b*b),)\n\n",
"_____no_output_____"
]
],
[
[
"최적화에도 기울기를 사용할 수 있다.<br>We can also use the slopes in the optimization.\n\n",
"_____no_output_____"
]
],
[
[
"cost_with_plot = get_cost_with_plot(b_triangle=False)\n\n",
"_____no_output_____"
],
[
"fmin_newton = so.minimize(cost_with_plot, [3.0, 1.0], jac=jacobian, method=\"newton-cg\")\n\n",
"_____no_output_____"
],
[
"fmin_newton\n\n",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
]
|
e794c08a51cbb6efb3b046708fcc7bb081dff800 | 22,218 | ipynb | Jupyter Notebook | examples/ex2_0.ipynb | vzkqwvku/kglab | d339caf60511e54d2c62488003778584368b0cd1 | [
"MIT"
]
| 388 | 2020-11-06T23:35:04.000Z | 2022-03-30T06:59:56.000Z | examples/ex2_0.ipynb | drahnreb/kglab | 00b3fcb4094cad1c4d99732356ff9e11db825299 | [
"MIT"
]
| 76 | 2020-11-23T19:59:19.000Z | 2022-03-30T10:44:28.000Z | examples/ex2_0.ipynb | drahnreb/kglab | 00b3fcb4094cad1c4d99732356ff9e11db825299 | [
"MIT"
]
| 45 | 2020-11-23T19:20:10.000Z | 2022-03-27T10:44:37.000Z | 32.818316 | 252 | 0.450041 | [
[
[
"# for use in tutorial and development; do not include this `sys.path` change in production:\nimport sys ; sys.path.insert(0, \"../\")",
"_____no_output_____"
]
],
[
[
"# Build a medium size KG from a CSV dataset",
"_____no_output_____"
],
[
"First let's initialize the KG object as we did previously:",
"_____no_output_____"
]
],
[
[
"import kglab\n\nnamespaces = {\n \"wtm\": \"http://purl.org/heals/food/\",\n \"ind\": \"http://purl.org/heals/ingredient/\",\n \"skos\": \"http://www.w3.org/2004/02/skos/core#\",\n }\n\nkg = kglab.KnowledgeGraph(\n name = \"A recipe KG example based on Food.com\",\n base_uri = \"https://www.food.com/recipe/\",\n namespaces = namespaces,\n )",
"_____no_output_____"
]
],
[
[
"Here's a way to describe the namespaces that are available to use:",
"_____no_output_____"
]
],
[
[
"kg.describe_ns()",
"_____no_output_____"
]
],
[
[
"Next, we'll define a dictionary that maps (somewhat magically) from strings (i.e., \"labels\") to ingredients defined in the <http://purl.org/heals/ingredient/> vocabulary:",
"_____no_output_____"
]
],
[
[
"common_ingredient = {\n \"water\": kg.get_ns(\"ind\").Water,\n \"salt\": kg.get_ns(\"ind\").Salt,\n \"pepper\": kg.get_ns(\"ind\").BlackPepper,\n \"black pepper\": kg.get_ns(\"ind\").BlackPepper,\n \"dried basil\": kg.get_ns(\"ind\").Basil,\n\n \"butter\": kg.get_ns(\"ind\").Butter,\n \"milk\": kg.get_ns(\"ind\").CowMilk,\n \"egg\": kg.get_ns(\"ind\").ChickenEgg,\n \"eggs\": kg.get_ns(\"ind\").ChickenEgg,\n \"bacon\": kg.get_ns(\"ind\").Bacon,\n\n \"sugar\": kg.get_ns(\"ind\").WhiteSugar,\n \"brown sugar\": kg.get_ns(\"ind\").BrownSugar,\n \"honey\": kg.get_ns(\"ind\").Honey,\n \"vanilla\": kg.get_ns(\"ind\").VanillaExtract,\n \"vanilla extract\": kg.get_ns(\"ind\").VanillaExtract,\n\n \"flour\": kg.get_ns(\"ind\").AllPurposeFlour,\n \"all-purpose flour\": kg.get_ns(\"ind\").AllPurposeFlour,\n \"whole wheat flour\": kg.get_ns(\"ind\").WholeWheatFlour,\n\n \"olive oil\": kg.get_ns(\"ind\").OliveOil,\n \"vinegar\": kg.get_ns(\"ind\").AppleCiderVinegar,\n\n \"garlic\": kg.get_ns(\"ind\").Garlic,\n \"garlic clove\": kg.get_ns(\"ind\").Garlic,\n \"garlic cloves\": kg.get_ns(\"ind\").Garlic,\n\n \"onion\": kg.get_ns(\"ind\").Onion,\n \"onions\": kg.get_ns(\"ind\").Onion,\n \"cabbage\": kg.get_ns(\"ind\").Cabbage,\n \"carrot\": kg.get_ns(\"ind\").Carrot,\n \"carrots\": kg.get_ns(\"ind\").Carrot,\n \"celery\": kg.get_ns(\"ind\").Celery,\n \"potato\": kg.get_ns(\"ind\").Potato,\n \"potatoes\": kg.get_ns(\"ind\").Potato,\n \"tomato\": kg.get_ns(\"ind\").Tomato,\n \"tomatoes\": kg.get_ns(\"ind\").Tomato,\n \n \"baking powder\": kg.get_ns(\"ind\").BakingPowder,\n \"baking soda\": kg.get_ns(\"ind\").BakingSoda,\n}",
"_____no_output_____"
]
],
[
[
"This is where use of NLP work to produce *annotations* begins to overlap with KG pratices.",
"_____no_output_____"
],
[
"Now let's load our dataset of recipes – the `dat/recipes.csv` file in CSV format – into a `pandas` dataframe:",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\ndf = pd.read_csv(\"../dat/recipes.csv\")\ndf.head()",
"_____no_output_____"
]
],
[
[
"Then iterate over the rows in the dataframe, representing a recipe in the KG for each row:",
"_____no_output_____"
]
],
[
[
"import rdflib\n\nfor index, row in df.iterrows():\n recipe_id = row[\"id\"]\n node = rdflib.URIRef(\"https://www.food.com/recipe/{}\".format(recipe_id))\n kg.add(node, kg.get_ns(\"rdf\").type, kg.get_ns(\"wtm\").Recipe)\n\n recipe_name = row[\"name\"]\n kg.add(node, kg.get_ns(\"skos\").definition, rdflib.Literal(recipe_name))\n \n cook_time = row[\"minutes\"]\n cook_time_literal = \"PT{}M\".format(int(cook_time))\n code_time_node = rdflib.Literal(cook_time_literal, datatype=kg.get_ns(\"xsd\").duration)\n kg.add(node, kg.get_ns(\"wtm\").hasCookTime, code_time_node)\n \n ind_list = eval(row[\"ingredients\"])\n\n for ind in ind_list:\n ingredient = ind.strip()\n ingredient_obj = common_ingredient[ingredient]\n kg.add(node, kg.get_ns(\"wtm\").hasIngredient, ingredient_obj)",
"_____no_output_____"
]
],
[
[
"Notice how the `xsd:duration` literal is now getting used to represent cooking times.\n\nWe've structured this example such that each of the recipes in the CSV file has a known representation for all of its ingredients.\nThere are nearly 250K recipes in the full dataset from <https://food.com/> so the `common_ingredient` dictionary would need to be extended quite a lot to handle all of those possible ingredients.",
"_____no_output_____"
],
[
"At this stage, our graph has grown by a couple orders of magnitude, so its visualization should be more interesting now.\nLet's take a look:",
"_____no_output_____"
]
],
[
[
"VIS_STYLE = {\n \"wtm\": {\n \"color\": \"orange\",\n \"size\": 20,\n },\n \"ind\":{\n \"color\": \"blue\",\n \"size\": 35,\n },\n}\n\nsubgraph = kglab.SubgraphTensor(kg)\npyvis_graph = subgraph.build_pyvis_graph(notebook=True, style=VIS_STYLE)\n\npyvis_graph.force_atlas_2based()\npyvis_graph.show(\"tmp.fig01.html\")",
"_____no_output_____"
]
],
[
[
"Given the defaults for this kind of visualization, there's likely a dense center mass of orange (recipes) at the center, with a close cluster of common ingredients (dark blue), surrounded by less common ingredients and cooking times (light blue).",
"_____no_output_____"
],
[
"## Performance analysis of serialization methods",
"_____no_output_____"
],
[
"Let's serialize this recipe KG constructed from the CSV dataset to a local TTL file, while measuring the time and disk space required:",
"_____no_output_____"
]
],
[
[
"import time\n\nwrite_times = []\n\nt0 = time.time()\nkg.save_rdf(\"tmp.ttl\")\nwrite_times.append(round((time.time() - t0) * 1000.0, 2))",
"_____no_output_____"
]
],
[
[
"Let's also serialize the KG into the other formats that we've been using, to compare relative sizes for a medium size KG:",
"_____no_output_____"
]
],
[
[
"t0 = time.time()\nkg.save_rdf(\"tmp.xml\", format=\"xml\")\nwrite_times.append(round((time.time() - t0) * 1000.0, 2))\n\nt0 = time.time()\nkg.save_jsonld(\"tmp.jsonld\")\nwrite_times.append(round((time.time() - t0) * 1000.0, 2))\n\nt0 = time.time()\nkg.save_parquet(\"tmp.parquet\")\nwrite_times.append(round((time.time() - t0) * 1000.0, 2))",
"_____no_output_____"
],
[
"import pandas as pd\nimport os\n\nfile_paths = [\"tmp.ttl\", \"tmp.xml\", \"tmp.jsonld\", \"tmp.parquet\"]\nfile_sizes = [os.path.getsize(file_path) for file_path in file_paths]\n\ndf = pd.DataFrame({\"file_path\": file_paths, \"file_size\": file_sizes, \"write_time\": write_times})\ndf[\"ms_per_byte\"] = df[\"write_time\"] / df[\"file_size\"]\ndf",
"_____no_output_____"
]
],
[
[
"Notice the relative sizes and times?\n[Parquet](https://parquet.apache.org/) provides for compression in a way that works well with RDF.\nThe same KG stored as a Parquet file is ~10% the size of the same KG stored as JSON-LD.\nAlso the XML version is quite large.\n\nLooking at the write times, Parquet is relatively fast (after its first invocation) and its reads are faster.\nThe eponymous Turtle format is human-readable although relatively slow.\nXML is fast to write, but much larger on disk and difficult to read.\nJSON-LD is interesting in that any JSON library can read and use these files, without needing semantic technologies, *per se*; however, it's also large on disk.",
"_____no_output_____"
],
[
"---\n\n## Exercises",
"_____no_output_____"
],
[
"**Exercise 1:**\n\nSelect another ingredient in the <http://purl.org/heals/ingredient/> vocabulary that is not in the `common_ingredient` dictionary, for which you can find at least one simple recipe within <https://food.com/> searches.\nThen add this recipe into the KG.",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
]
|
e794c875937b433e13cb61007eb1a87705cde650 | 5,360 | ipynb | Jupyter Notebook | Segmenting and Clustering Neighborhoods in Toronto Part 2.ipynb | grepppo/Coursera_Capstone | 65faef39ae68afc9012fdc2a55bd2ee7785ee92c | [
"Apache-2.0"
]
| null | null | null | Segmenting and Clustering Neighborhoods in Toronto Part 2.ipynb | grepppo/Coursera_Capstone | 65faef39ae68afc9012fdc2a55bd2ee7785ee92c | [
"Apache-2.0"
]
| null | null | null | Segmenting and Clustering Neighborhoods in Toronto Part 2.ipynb | grepppo/Coursera_Capstone | 65faef39ae68afc9012fdc2a55bd2ee7785ee92c | [
"Apache-2.0"
]
| null | null | null | 30.11236 | 134 | 0.485821 | [
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"#Scraping the Values from the Wikipedia change\nurl = 'https://en.wikipedia.org/wiki/List_of_postal_codes_of_Canada:_M'\ndfs = pd.read_html(url)\npc = dfs[0]",
"_____no_output_____"
],
[
"#Removing Not Assigned Boroughs\npost_codes = pc.loc[(pc.Borough != 'Not assigned')]\n\n#Updating Column Names\npost_codes.columns = ['PostalCode', 'Borough', 'Neighbourhood']\n\n#Defaulting Nos assigned Neighbourhood to Borough Values\npost_codes.Neighbourhood = np.where(post_codes.Neighbourhood == 'Not assigned', post_codes.Borough, post_codes.Neighbourhood)\n\n#Skipping the step to merger post code records, as this case is no longer present in the source data \npost_codes.shape",
"_____no_output_____"
],
[
"#Loading from local file, had issues with the Geocode API\nlonglat = pd.read_csv('./Geospatial_Coordinates.csv')\nlonglat.columns = ['PostalCode', 'Latitude', 'Logitude']",
"_____no_output_____"
],
[
"longlat.head",
"_____no_output_____"
],
[
"#Merging the post code and positional data\npost_code_long_lat = post_codes.merge(longlat, on='PostalCode', how='inner')\n\npost_code_long_lat.head",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e794d20342626c5429861b4495db15e4904bd297 | 71,725 | ipynb | Jupyter Notebook | training_testing_GRID_SEARCH-backup.ipynb | gkovacs/crypocurrency-trading | 74edbea2a3c3585b7df976bbe5bb0bab911cd030 | [
"MIT"
]
| 5 | 2018-05-02T19:41:55.000Z | 2019-06-24T12:04:12.000Z | training_testing_GRID_SEARCH-backup.ipynb | gkovacs/crypocurrency-trading | 74edbea2a3c3585b7df976bbe5bb0bab911cd030 | [
"MIT"
]
| null | null | null | training_testing_GRID_SEARCH-backup.ipynb | gkovacs/crypocurrency-trading | 74edbea2a3c3585b7df976bbe5bb0bab911cd030 | [
"MIT"
]
| 1 | 2018-11-15T16:25:22.000Z | 2018-11-15T16:25:22.000Z | 58.123987 | 8,284 | 0.6226 | [
[
[
"#!/usr/bin/env python3\nimport pandas as pd\nimport lz4.frame\nimport gzip\nimport io\nimport pyarrow.parquet as pq\nimport pyarrow as pa\nimport numpy as np\nfrom glob import glob\nfrom plumbum.cmd import rm\nfrom keras.layers.core import Dense, Activation, Dropout\nfrom keras.layers.recurrent import LSTM\nfrom keras.layers import TimeDistributed\nfrom keras.models import Sequential\nfrom keras import regularizers\nfrom sklearn.preprocessing import MinMaxScaler\nfrom sklearn.metrics import mean_squared_error\nimport matplotlib.pyplot as plt",
"/home/ubuntu/anaconda3/envs/tensorflow_p36/lib/python3.6/site-packages/matplotlib/__init__.py:962: UserWarning: Duplicate key in file \"/home/ubuntu/.config/matplotlib/matplotlibrc\", line #2\n (fname, cnt))\n/home/ubuntu/anaconda3/envs/tensorflow_p36/lib/python3.6/site-packages/matplotlib/__init__.py:962: UserWarning: Duplicate key in file \"/home/ubuntu/.config/matplotlib/matplotlibrc\", line #3\n (fname, cnt))\n/home/ubuntu/anaconda3/envs/tensorflow_p36/lib/python3.6/site-packages/h5py/__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\nUsing TensorFlow backend.\n"
],
[
"def plotline(data):\n plt.figure()\n plt.plot(data)\n plt.legend()\n plt.show()\n\ndef event_count(time_series, data_name):\n time_series = time_series[['Fill Price (USD)']].values\n upevents = 0\n downevents = 0\n sameprice = 0\n prev_obv = time_series[0]\n for obv in time_series[1:]:\n if obv > prev_obv:\n upevents += 1\n elif obv < prev_obv:\n downevents += 1\n elif obv == prev_obv:\n sameprice += 1\n prev_obv = obv\n print('=== Event counts on %s ===' % data_name)\n print('upevents')\n print(upevents)\n print('downevents')\n print(downevents)\n print('sameprice')\n print(sameprice)\n print()\n\ndef mse(time_series, data_name):\n time_series = time_series[['Fill Price (USD)']].values\n total_squared_error = 0\n total_absolute_error = 0\n prev_obv = time_series[0]\n for obv in time_series[1:]:\n total_squared_error += (obv - prev_obv)**2\n total_absolute_error += abs(obv - prev_obv)\n prev_obv = obv\n num_predictions = len(time_series) - 1\n mean_squared_error = total_squared_error / num_predictions\n mean_absolute_error = total_absolute_error / num_predictions\n root_mean_squared_error = np.sqrt(mean_squared_error)\n print('=== baseline on %s ===' % data_name)\n print('total squared error')\n print(total_squared_error)\n print('total absolute error')\n print(total_absolute_error)\n print('mean squared error')\n print(mean_squared_error)\n print('mean absolute error')\n print(mean_absolute_error) \n print('root mean squared error')\n print(root_mean_squared_error) \n print()",
"_____no_output_____"
],
[
"def show_summary_statistics():\n #event_count(small_set, 'small')\n train_set = df.iloc[0:num_samples_training]\n dev_set = df.iloc[num_samples_training:num_samples_training+num_samples_dev]\n test_set = df.iloc[num_samples_training+num_samples_dev:]\n event_count(train_set, 'train')\n event_count(dev_set, 'dev')\n event_count(test_set, 'test')\n mse(train_set, 'train')\n mse(dev_set, 'dev')\n mse(test_set, 'test')\n#show_summary_statistics()",
"_____no_output_____"
],
[
"def preprocess(data):\n values = np.array(data)\n values = values.reshape(-1,1)\n values = values.astype('float32') \n return values",
"_____no_output_____"
],
[
"def plot_losses(model_history, title):\n plt.figure()\n plt.plot(model_history.history['loss'], label='Train')\n plt.plot(model_history.history['val_loss'], label='Dev')\n plt.xlabel('Epochs'); plt.ylabel('Loss (mse)')\n plt.title(title)\n plt.legend(); plt.show()",
"_____no_output_____"
],
[
"def inverse_transform_pricescaler(data, Y_prevrawprice, fitted_scaler):\n return fitted_scaler.inverse_transform(preprocess(data))\n\ndef inverse_transform_percentdiff(data, Y_prevrawprice, fitted_scaler=None):\n orig_prices = Y_prevrawprice\n change = orig_prices * data\n return orig_prices + change\n #return fitted_scaler.inverse_transform(preprocess(data))\n\n#print(Y_test_prevrawprice)\n#print(inverse_transform_percentdiff(Y_test, Y_test_prevrawprice))\n\ninverse_transform = inverse_transform_percentdiff",
"_____no_output_____"
],
[
"def plot_predictions(model, X_test, Y_test, Y_prevrawprice, title, inverse=False, scaler=None):\n y_hat = model.predict(X_test)\n\n if inverse:\n y_hat = inverse_transform(y_hat, Y_prevrawprice, scaler)\n Y_test = inverse_transform(Y_test, Y_prevrawprice, scaler)\n\n plt.plot(y_hat, label='Predicted')\n plt.plot(Y_test, label='True')\n plt.xlabel('Time'); \n\n if inverse:\n plt.ylabel('Price')\n else:\n plt.ylabel('RESCALED Price')\n\n plt.title(title)\n plt.legend(); plt.show()",
"_____no_output_____"
],
[
"def calculate_MSE_RMSE(model, scaler, X_test, Y_test, Y_prevrawprice, model_name):\n y_hat = model.predict(X_test)\n y_hat_inverse = inverse_transform(y_hat, Y_prevrawprice, scaler)\n Y_test_inverse = inverse_transform(Y_test, Y_prevrawprice, scaler)\n mse = mean_squared_error(Y_test_inverse, y_hat_inverse)\n rmse = np.sqrt(mean_squared_error(Y_test_inverse, y_hat_inverse))\n print('%s:' % model_name)\n print('Test MSE: %.3f' % mse)\n print('Test RMSE: %.3f' % rmse)\n print()",
"_____no_output_____"
],
[
"def train_evaluate(model, model_name, \n X_train, Y_train, Y_train_prevrawprice, X_dev, Y_dev, Y_dev_prevrawprice, X_test, Y_test, Y_test_prevrawprice,\n lag=10, batch_size=100, epochs=10, verbose=1):\n\n # Train model\n history = model.fit(X_train, Y_train, batch_size=batch_size, epochs=epochs,\n validation_split=0.05, verbose=verbose, shuffle=False)\n #train_evaluate_showresults(history, model, model_name, \n # X_train, Y_train, X_dev, Y_dev, X_test, Y_test,\n # lag, batch_size, epochs, verbose)\n return history",
"_____no_output_____"
],
[
"def train_evaluate_showresults(history, model, model_name, \n X_train, Y_train, Y_train_prevrawprice, X_dev, Y_dev, Y_dev_prevrawprice, X_test, Y_test, Y_test_prevrawprice,\n lag=10, batch_size=100, epochs=10, verbose=1):\n # Plot losses, predictions, and calculate MSE and RMSE\n plot_losses(history, 'Loss\\n(%s)' % model_name)\n plot_predictions(model, X_dev, Y_dev, Y_dev_prevrawprice, 'Test Predictions\\n(%s)' % model_name)\n plot_predictions(model, X_dev, Y_dev, Y_dev_prevrawprice, 'Test Predictions\\n(%s)' % model_name, inverse=True, scaler=price_scaler)\n calculate_MSE_RMSE(model, price_scaler, X_dev, Y_dev, Y_dev_prevrawprice, '%s' % model_name)",
"_____no_output_____"
],
[
"def evaluate_test(model, model_name, \n X_train, Y_train, Y_train_prevrawprice, X_dev, Y_dev, Y_dev_prevrawprice, X_test, Y_test, Y_test_prevrawprice,\n lag=10, batch_size=100, epochs=10, verbose=1):\n # Plot losses, predictions, and calculate MSE and RMSE\n #plot_losses(history, 'Loss\\n(%s)' % model_name)\n plot_predictions(model, X_test, Y_test, Y_test_prevrawprice, 'Test Predictions\\n(%s)' % model_name)\n plot_predictions(model, X_test, Y_test, Y_test_prevrawprice, 'Test Predictions\\n(%s)' % model_name, inverse=True, scaler=price_scaler)\n calculate_MSE_RMSE(model, price_scaler, X_test, Y_test, Y_test_prevrawprice, '%s' % model_name)",
"_____no_output_____"
],
[
"def initialize_model(X_train, loss, optimizer, num_LSTMs, num_units, dropout):\n \n LSTM_input_shape = [X_train.shape[1], X_train.shape[2]]\n print('input shape is')\n print(LSTM_input_shape)\n\n # DEFINE MODEL\n model = Sequential()\n\n if num_LSTMs == 2:\n model.add(LSTM(num_units[0], input_shape=LSTM_input_shape, return_sequences=True))\n model.add(Dropout(dropout))\n\n model.add(LSTM(num_units[1], return_sequences=True))\n \n if num_LSTMs == 3:\n model.add(LSTM(num_units[0], input_shape=LSTM_input_shape, return_sequences=True))\n model.add(Dropout(dropout))\n\n model.add(LSTM(num_units[1], return_sequences=True))\n model.add(Dropout(dropout))\n \n model.add(LSTM(num_units[2], return_sequences=True))\n\n model.add(TimeDistributed(Dense(1)))\n model.add(Activation('linear'))\n\n \n model.compile(loss=loss, optimizer=optimizer)\n \n return model",
"_____no_output_____"
],
[
"import os.path\n\ndef load_data():\n if not os.path.isfile('cboe/parquet_preprocessed_subset_only_BTCUSD_merged.parquet'):\n files = sorted(glob('cboe/parquet_preprocessed_BTCUSD_merged/*.parquet'))\n all_dataframes = []\n for file in files:\n print(file)\n df = pq.read_table(file).to_pandas()\n all_dataframes.append(df)\n result = pd.concat(all_dataframes)\n pq.write_table(pa.Table.from_pandas(result), 'cboe/parquet_preprocessed_subset_only_BTCUSD_merged.parquet', compression='snappy')\n df = pq.read_table('cboe/parquet_preprocessed_subset_only_BTCUSD_merged.parquet').to_pandas();\n print(df.dtypes)\n print(df.shape)\n return df",
"_____no_output_____"
],
[
"def split_X(df):\n n_all = df.shape[0]\n n_train = round(n_all * 0.9)\n n_dev = round(n_all * 0.05)\n n_test = round(n_all * 0.05)\n print('n_all: ', n_all)\n print('n_train:', n_train)\n print('n_dev: ', n_dev)\n print('n_test: ', n_test)\n\n X_train = df.iloc[:n_train, 1:16].values.astype('float32')\n X_dev = df.iloc[n_train:n_train+n_dev, 1:16].values.astype('float32')\n X_test = df.iloc[n_train+n_dev:, 1:16].values.astype('float32')\n print(X_train.shape)\n print(X_dev.shape)\n print(X_test.shape)\n\n return X_train, X_dev, X_test",
"_____no_output_____"
],
[
"def split_Y(df):\n n_all = df.shape[0]\n n_train = round(n_all * 0.9)\n n_dev = round(n_all * 0.05)\n n_test = round(n_all * 0.05)\n Y_train = df.iloc[:n_train, -1:].values.astype('float32')\n Y_dev = df.iloc[n_train:n_train+n_dev, -1:].values.astype('float32')\n Y_test = df.iloc[n_train+n_dev:, -1:].values.astype('float32')\n print(Y_train.shape)\n print(Y_dev.shape)\n print(Y_test.shape)\n \n return Y_train, Y_dev, Y_test",
"_____no_output_____"
],
[
"def df_to_parquet(df, outfile):\n pq.write_table(pa.Table.from_pandas(df), outfile, compression='snappy')",
"_____no_output_____"
],
[
"def evaluate_model(model, history, X_train, X_dev, X_test, Y_train, Y_dev, Y_test):\n train_loss = history.history['loss'][-1]\n dev_loss = history.history['val_loss'][-1]\n test_loss = model.evaluate(X_test, Y_test, verbose=0)\n \n y_hat_train = model.predict(X_train)\n y_hat_dev = model.predict(X_dev)\n y_hat_test = model.predict(X_test)\n \n train_prop_correct = np.sum(np.sign(y_hat_train) == np.sign(Y_train)) / (Y_train_final.shape[0] * Y_train_final.shape[1])\n dev_prop_correct = np.sum(np.sign(y_hat_dev) == np.sign(Y_dev)) / (Y_dev_final.shape[0] * Y_dev_final.shape[1])\n test_prop_correct = np.sum(np.sign(y_hat_test) == np.sign(Y_test)) / (Y_test_final.shape[0] * Y_test_final.shape[1])\n \n evaluation = [train_loss, dev_loss, test_loss, train_prop_correct, dev_prop_correct, test_prop_correct]\n return evaluation",
"_____no_output_____"
],
[
"def create_sequenced_data(data, window, step, y=True):\n sequenced = []\n for minute in range(0, len(data) - window, step):\n chunk = data[minute:minute+window]\n sequenced.append(chunk)\n sequenced = np.array(sequenced)\n return sequenced",
"_____no_output_____"
],
[
"batch_size = 8192 #16384 #32768 #4096\nepochs = 100\nverbose = 2\nloss = 'mean_squared_error'\noptimizer = 'adagrad' #'adam'\n#num_LSTM = 2\n#n_units = [256, 256]\nnum_LSTM = 3\nn_units = [256, 256, 256]\ndropout = 0.1\n\nmodel = initialize_model(X_train_final, loss, optimizer, num_LSTM, n_units, dropout)\n\nhistory = model.fit(X_train_final, Y_train_final, batch_size=batch_size, epochs=epochs,\n validation_data=(X_dev_final, Y_dev_final), verbose=verbose, shuffle=False) ",
"input shape is\n[30, 15]\nTrain on 35264 samples, validate on 1958 samples\nEpoch 1/200\n - 4s - loss: 10.9643 - val_loss: 0.0129\nEpoch 2/200\n - 2s - loss: 0.0088 - val_loss: 0.0063\nEpoch 3/200\n - 2s - loss: 0.0072 - val_loss: 0.0055\nEpoch 4/200\n - 2s - loss: 0.0063 - val_loss: 0.0049\nEpoch 5/200\n - 2s - loss: 0.0056 - val_loss: 0.0044\nEpoch 6/200\n - 2s - loss: 0.0051 - val_loss: 0.0040\nEpoch 7/200\n - 2s - loss: 0.0046 - val_loss: 0.0036\nEpoch 8/200\n - 2s - loss: 0.0042 - val_loss: 0.0033\nEpoch 9/200\n - 2s - loss: 0.0039 - val_loss: 0.0031\nEpoch 10/200\n - 2s - loss: 0.0036 - val_loss: 0.0029\nEpoch 11/200\n - 2s - loss: 0.0034 - val_loss: 0.0027\nEpoch 12/200\n - 2s - loss: 0.0032 - val_loss: 0.0025\nEpoch 13/200\n - 2s - loss: 0.0030 - val_loss: 0.0023\nEpoch 14/200\n - 2s - loss: 0.0028 - val_loss: 0.0022\nEpoch 15/200\n - 2s - loss: 0.0026 - val_loss: 0.0020\nEpoch 16/200\n - 2s - loss: 0.0025 - val_loss: 0.0019\nEpoch 17/200\n - 2s - loss: 0.0024 - val_loss: 0.0018\nEpoch 18/200\n - 2s - loss: 0.0023 - val_loss: 0.0017\nEpoch 19/200\n - 2s - loss: 0.0021 - val_loss: 0.0016\nEpoch 20/200\n - 2s - loss: 0.0020 - val_loss: 0.0016\nEpoch 21/200\n - 2s - loss: 0.0020 - val_loss: 0.0015\nEpoch 22/200\n - 2s - loss: 0.0019 - val_loss: 0.0014\nEpoch 23/200\n - 2s - loss: 0.0018 - val_loss: 0.0013\nEpoch 24/200\n - 2s - loss: 0.0017 - val_loss: 0.0013\nEpoch 25/200\n - 2s - loss: 0.0017 - val_loss: 0.0012\nEpoch 26/200\n - 2s - loss: 0.0016 - val_loss: 0.0012\nEpoch 27/200\n - 2s - loss: 0.0015 - val_loss: 0.0011\nEpoch 28/200\n - 2s - loss: 0.0015 - val_loss: 0.0011\nEpoch 29/200\n - 2s - loss: 0.0014 - val_loss: 0.0010\nEpoch 30/200\n - 2s - loss: 0.0014 - val_loss: 9.9952e-04\nEpoch 31/200\n - 2s - loss: 0.0013 - val_loss: 9.6192e-04\nEpoch 32/200\n - 2s - loss: 0.0013 - val_loss: 9.2679e-04\nEpoch 33/200\n - 2s - loss: 0.0013 - val_loss: 8.9353e-04\nEpoch 34/200\n - 2s - loss: 0.0012 - val_loss: 8.6235e-04\nEpoch 35/200\n - 2s - loss: 0.0012 - val_loss: 8.3256e-04\nEpoch 36/200\n - 2s - loss: 0.0012 - val_loss: 8.0466e-04\nEpoch 37/200\n - 2s - loss: 0.0011 - val_loss: 7.7834e-04\nEpoch 38/200\n - 2s - loss: 0.0011 - val_loss: 7.5309e-04\nEpoch 39/200\n - 2s - loss: 0.0011 - val_loss: 7.2906e-04\nEpoch 40/200\n - 2s - loss: 0.0010 - val_loss: 7.0632e-04\nEpoch 41/200\n - 2s - loss: 0.0010 - val_loss: 6.8486e-04\nEpoch 42/200\n - 2s - loss: 9.8930e-04 - val_loss: 6.6416e-04\nEpoch 43/200\n - 2s - loss: 9.6677e-04 - val_loss: 6.4445e-04\nEpoch 44/200\n - 2s - loss: 9.4354e-04 - val_loss: 6.2587e-04\nEpoch 45/200\n - 2s - loss: 9.2251e-04 - val_loss: 6.0793e-04\nEpoch 46/200\n - 2s - loss: 9.0079e-04 - val_loss: 5.9102e-04\nEpoch 47/200\n - 2s - loss: 8.8291e-04 - val_loss: 5.7466e-04\nEpoch 48/200\n - 2s - loss: 8.6111e-04 - val_loss: 5.5911e-04\nEpoch 49/200\n - 2s - loss: 8.4414e-04 - val_loss: 5.4423e-04\nEpoch 50/200\n - 2s - loss: 8.2643e-04 - val_loss: 5.2992e-04\nEpoch 51/200\n - 2s - loss: 8.0776e-04 - val_loss: 5.1618e-04\nEpoch 52/200\n - 2s - loss: 7.9379e-04 - val_loss: 5.0307e-04\nEpoch 53/200\n - 2s - loss: 7.7774e-04 - val_loss: 4.9054e-04\nEpoch 54/200\n - 2s - loss: 7.6358e-04 - val_loss: 4.7851e-04\nEpoch 55/200\n - 2s - loss: 7.4859e-04 - val_loss: 4.6688e-04\nEpoch 56/200\n - 2s - loss: 7.3544e-04 - val_loss: 4.5585e-04\nEpoch 57/200\n - 2s - loss: 7.2261e-04 - val_loss: 4.4501e-04\nEpoch 58/200\n - 2s - loss: 7.1029e-04 - val_loss: 4.3474e-04\nEpoch 59/200\n - 2s - loss: 6.9662e-04 - val_loss: 4.2481e-04\nEpoch 60/200\n - 2s - loss: 6.8573e-04 - val_loss: 4.1528e-04\nEpoch 61/200\n - 2s - loss: 6.7265e-04 - val_loss: 4.0622e-04\nEpoch 62/200\n - 2s - loss: 6.6380e-04 - val_loss: 3.9728e-04\nEpoch 63/200\n - 2s - loss: 6.5129e-04 - val_loss: 3.8873e-04\nEpoch 64/200\n - 2s - loss: 6.4071e-04 - val_loss: 3.8057e-04\nEpoch 65/200\n - 2s - loss: 6.3138e-04 - val_loss: 3.7264e-04\nEpoch 66/200\n - 2s - loss: 6.2177e-04 - val_loss: 3.6503e-04\nEpoch 67/200\n - 2s - loss: 6.1285e-04 - val_loss: 3.5762e-04\nEpoch 68/200\n - 2s - loss: 6.0125e-04 - val_loss: 3.5065e-04\nEpoch 69/200\n - 2s - loss: 5.9412e-04 - val_loss: 3.4368e-04\nEpoch 70/200\n - 2s - loss: 5.8569e-04 - val_loss: 3.3686e-04\nEpoch 71/200\n - 2s - loss: 5.7761e-04 - val_loss: 3.3043e-04\nEpoch 72/200\n - 2s - loss: 5.6914e-04 - val_loss: 3.2425e-04\nEpoch 73/200\n - 2s - loss: 5.6240e-04 - val_loss: 3.1823e-04\nEpoch 74/200\n - 2s - loss: 5.5471e-04 - val_loss: 3.1247e-04\nEpoch 75/200\n - 2s - loss: 5.4737e-04 - val_loss: 3.0677e-04\nEpoch 76/200\n - 2s - loss: 5.3940e-04 - val_loss: 3.0133e-04\nEpoch 77/200\n - 2s - loss: 5.3261e-04 - val_loss: 2.9621e-04\nEpoch 78/200\n - 2s - loss: 5.2577e-04 - val_loss: 2.9096e-04\nEpoch 79/200\n - 2s - loss: 5.2029e-04 - val_loss: 2.8608e-04\nEpoch 80/200\n - 2s - loss: 5.1361e-04 - val_loss: 2.8129e-04\nEpoch 81/200\n - 2s - loss: 5.0846e-04 - val_loss: 2.7661e-04\nEpoch 82/200\n - 2s - loss: 5.0160e-04 - val_loss: 2.7211e-04\nEpoch 83/200\n - 2s - loss: 4.9636e-04 - val_loss: 2.6769e-04\nEpoch 84/200\n - 2s - loss: 4.9059e-04 - val_loss: 2.6358e-04\nEpoch 85/200\n - 2s - loss: 4.8425e-04 - val_loss: 2.5929e-04\nEpoch 86/200\n - 2s - loss: 4.8051e-04 - val_loss: 2.5532e-04\nEpoch 87/200\n - 2s - loss: 4.7417e-04 - val_loss: 2.5135e-04\nEpoch 88/200\n - 2s - loss: 4.6836e-04 - val_loss: 2.4772e-04\nEpoch 89/200\n - 2s - loss: 4.6455e-04 - val_loss: 2.4393e-04\nEpoch 90/200\n - 2s - loss: 4.5965e-04 - val_loss: 2.4044e-04\nEpoch 91/200\n - 2s - loss: 4.5454e-04 - val_loss: 2.3703e-04\nEpoch 92/200\n - 2s - loss: 4.4997e-04 - val_loss: 2.3355e-04\nEpoch 93/200\n - 2s - loss: 4.4598e-04 - val_loss: 2.3041e-04\nEpoch 94/200\n - 2s - loss: 4.4064e-04 - val_loss: 2.2722e-04\nEpoch 95/200\n - 2s - loss: 4.3740e-04 - val_loss: 2.2413e-04\nEpoch 96/200\n - 2s - loss: 4.3303e-04 - val_loss: 2.2119e-04\nEpoch 97/200\n - 2s - loss: 4.2972e-04 - val_loss: 2.1834e-04\nEpoch 98/200\n - 2s - loss: 4.2530e-04 - val_loss: 2.1537e-04\nEpoch 99/200\n - 2s - loss: 4.2100e-04 - val_loss: 2.1267e-04\nEpoch 100/200\n - 2s - loss: 4.1785e-04 - val_loss: 2.0980e-04\nEpoch 101/200\n - 2s - loss: 4.1316e-04 - val_loss: 2.0728e-04\nEpoch 102/200\n - 2s - loss: 4.0985e-04 - val_loss: 2.0470e-04\nEpoch 103/200\n - 2s - loss: 4.0717e-04 - val_loss: 2.0218e-04\nEpoch 104/200\n - 2s - loss: 4.0238e-04 - val_loss: 1.9977e-04\nEpoch 105/200\n - 2s - loss: 3.9979e-04 - val_loss: 1.9730e-04\nEpoch 106/200\n - 2s - loss: 3.9662e-04 - val_loss: 1.9500e-04\nEpoch 107/200\n - 2s - loss: 3.9347e-04 - val_loss: 1.9277e-04\nEpoch 108/200\n - 2s - loss: 3.9002e-04 - val_loss: 1.9063e-04\nEpoch 109/200\n - 2s - loss: 3.8662e-04 - val_loss: 1.8858e-04\nEpoch 110/200\n - 2s - loss: 3.8514e-04 - val_loss: 1.8631e-04\nEpoch 111/200\n - 2s - loss: 3.8223e-04 - val_loss: 1.8428e-04\nEpoch 112/200\n - 2s - loss: 3.7782e-04 - val_loss: 1.8223e-04\nEpoch 113/200\n - 2s - loss: 3.7592e-04 - val_loss: 1.8041e-04\nEpoch 114/200\n - 2s - loss: 3.7269e-04 - val_loss: 1.7853e-04\nEpoch 115/200\n - 2s - loss: 3.7074e-04 - val_loss: 1.7648e-04\nEpoch 116/200\n - 2s - loss: 3.6691e-04 - val_loss: 1.7489e-04\nEpoch 117/200\n - 2s - loss: 3.6295e-04 - val_loss: 1.7311e-04\nEpoch 118/200\n - 2s - loss: 3.6174e-04 - val_loss: 1.7127e-04\nEpoch 119/200\n - 2s - loss: 3.5955e-04 - val_loss: 1.6955e-04\nEpoch 120/200\n - 2s - loss: 3.5755e-04 - val_loss: 1.6803e-04\nEpoch 121/200\n - 2s - loss: 3.5408e-04 - val_loss: 1.6626e-04\nEpoch 122/200\n - 2s - loss: 3.5216e-04 - val_loss: 1.6458e-04\nEpoch 123/200\n - 2s - loss: 3.5050e-04 - val_loss: 1.6309e-04\nEpoch 124/200\n - 2s - loss: 3.4688e-04 - val_loss: 1.6165e-04\nEpoch 125/200\n - 2s - loss: 3.4492e-04 - val_loss: 1.6023e-04\nEpoch 126/200\n - 2s - loss: 3.4258e-04 - val_loss: 1.5873e-04\nEpoch 127/200\n - 2s - loss: 3.4069e-04 - val_loss: 1.5728e-04\nEpoch 128/200\n - 2s - loss: 3.3946e-04 - val_loss: 1.5587e-04\nEpoch 129/200\n - 2s - loss: 3.3669e-04 - val_loss: 1.5457e-04\nEpoch 130/200\n - 2s - loss: 3.3425e-04 - val_loss: 1.5326e-04\nEpoch 131/200\n - 2s - loss: 3.3309e-04 - val_loss: 1.5191e-04\nEpoch 132/200\n - 2s - loss: 3.3096e-04 - val_loss: 1.5083e-04\nEpoch 133/200\n - 2s - loss: 3.2909e-04 - val_loss: 1.4949e-04\nEpoch 134/200\n - 2s - loss: 3.2612e-04 - val_loss: 1.4816e-04\nEpoch 135/200\n - 2s - loss: 3.2492e-04 - val_loss: 1.4705e-04\nEpoch 136/200\n - 2s - loss: 3.2387e-04 - val_loss: 1.4586e-04\nEpoch 137/200\n - 2s - loss: 3.2065e-04 - val_loss: 1.4473e-04\nEpoch 138/200\n"
],
[
"a = evaluate_model(model, history, X_train_final, X_dev_final, X_test_final, Y_train_final, Y_dev_final, Y_test_final)",
"_____no_output_____"
],
[
"# Concatenate dataframes\nfiles = sorted(glob('cboe/parquet_preprocessed_BTCUSD_merged/*.parquet'))[451:]\nall_dataframes = []\nfor file in files:\n df = pq.read_table(file).to_pandas()\n all_dataframes.append(df)\ndf = pd.concat(all_dataframes)\n\n#\nX_train, X_dev, X_test = split_X(df)\nY_train, Y_dev, Y_test = split_Y(df)\n\nwindow_size = 30\nstep = 30\n\nX_train = create_sequenced_data(X_train, window=window_size, step=step, y=False)\nX_dev = create_sequenced_data(X_dev, window=window_size, step=step, y=False)\nX_test = create_sequenced_data(X_test, window=window_size, step=step, y=False)\n\nY_train = create_sequenced_data(Y_train, window=window_size, step=step, y=True)\nY_dev = create_sequenced_data(Y_dev, window=window_size, step=step, y=True)\nY_test = create_sequenced_data(Y_test, window=window_size, step=step, y=True)\n\nprint('Train, dev, test shapes:')\nprint(X_train_final.shape)\nprint(X_dev_final.shape)\nprint(X_test_final.shape)\nprint(Y_train_final.shape)\nprint(Y_dev_final.shape)\nprint(Y_test_final.shape)",
"n_all: 587753\nn_train: 528978\nn_dev: 29388\nn_test: 29388\n(528978, 15)\n(29388, 15)\n(29387, 15)\n(528978, 1)\n(29388, 1)\n(29387, 1)\nTrain, dev, test shapes:\n(17632, 30, 15)\n(979, 30, 15)\n(979, 30, 15)\n(17632, 30, 1)\n(979, 30, 1)\n(979, 30, 1)\n"
],
[
"# Initialize output dataframe\noutfile = 'cboe/grid_search.parquet'\ncolumns = ['num_epochs', 'loss', 'optimizer', 'batch_size', 'num_LSTMs', 'num_units',\n 'train_loss', 'dev_loss', 'test_loss', 'train_prop_correct', 'dev_prop_correct', 'test_prop_correct']\ndf_output = pd.DataFrame(columns=columns)\npq.write_table(pa.Table.from_pandas(df_output), outfile, compression='snappy')",
"_____no_output_____"
],
[
"batch_size = 8192\nnum_epochs = 100\nverbose = 1\nloss = 'mean_squared_error'\noptimizers = ['adagrad', 'adam', 'rmsprop']\nnum_LSTMs = [2,3]\nnum_units_2 = [[128, 256], [256, 256]]\nnum_units_3 = [[128, 256, 256], [256, 256, 256], [256, 512, 512]]\ndropout = 0.1\n\ncount = 0\nfor optimizer in optimizers:\n for num_LSTM in num_LSTMs:\n if num_LSTM == 2:\n num_units = num_units_2\n elif num_LSTM == 3:\n num_units = num_units_3\n for n_units in num_units:\n # Load output dataframe\n df_output = pq.read_table(outfile).to_pandas()\n\n # Initialize model\n model = initialize_model(X_train, loss, optimizer, num_LSTM, n_units, dropout)\n\n # Train model\n if verbose:\n verbose=1\n print(count, '/', 15)\n else:\n verbose=0\n history = model.fit(X_train, Y_train, batch_size=batch_size, epochs=num_epochs,\n validation_data=(X_dev, Y_dev), verbose=0, shuffle=False) \n\n # Evaluate model\n evaluate = evaluate_model(model, history, X_train, X_dev, X_test, Y_train, Y_dev, Y_test)\n\n # Write to dataframe and save\n row = [num_epochs, loss, optimizer, batch_size, num_LSTM, str(n_units)]\n row.extend(evaluate)\n df_output.loc[len(df_output)] = row\n df_to_parquet(df_output, outfile)\n\n count += 1",
"input shape is\n[30, 15]\n0 / 15\ninput shape is\n[30, 15]\n1 / 15\ninput shape is\n[30, 15]\n2 / 15\ninput shape is\n[30, 15]\n3 / 15\ninput shape is\n[30, 15]\n4 / 15\n"
],
[
"y_hat_train = model.predict(X_train)\ny_hat_dev = model.predict(X_dev)\ny_hat_test = model.predict(X_test)\n\ntrain_prop_correct = np.sum(np.sign(y_hat_train) == np.sign(Y_train)) / (Y_train.shape[0] * Y_train.shape[1])\ndev_prop_correct = np.sum(np.sign(y_hat_dev) == np.sign(Y_dev)) / (Y_dev.shape[0] * Y_dev.shape[1])\ntest_prop_correct = np.sum(np.sign(y_hat_test) == np.sign(Y_test)) / (Y_test.shape[0] * Y_test.shape[1])",
"_____no_output_____"
],
[
"y_hat_test = model.predict(X_test)",
"_____no_output_____"
],
[
"a = np.sign(y_hat_test) == np.sign(Y_test)",
"_____no_output_____"
],
[
"np.sign(Y_test).shape",
"_____no_output_____"
],
[
"last = []\nfor i in range(len(a)):\n last.append(a[i][-1])",
"_____no_output_____"
],
[
"np.sum(last) / 979",
"_____no_output_____"
],
[
"np.sum(a) / (979*30)",
"_____no_output_____"
],
[
"np.sum(np.sign(Y_test)==1)",
"_____no_output_____"
],
[
"13615 / (979*30)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e794d5f52202f41c240d3b9f19e3d83f5bd48282 | 21,469 | ipynb | Jupyter Notebook | Chapter04/Issue_with_image_translation.ipynb | PacktPublishing/Neural-Networks-with-Keras-Cookbook | 406b95f981a8ce66ac22605f4a5f0eaa35ab3aee | [
"MIT"
]
| 53 | 2019-04-23T20:58:08.000Z | 2022-02-22T11:11:08.000Z | Chapter04/Issue_with_image_translation.ipynb | PacktPublishing/Neural-Networks-with-Keras-Cookbook | 406b95f981a8ce66ac22605f4a5f0eaa35ab3aee | [
"MIT"
]
| 3 | 2020-01-29T15:41:35.000Z | 2020-08-17T07:04:20.000Z | Chapter04/Issue_with_image_translation.ipynb | PacktPublishing/Neural-Networks-with-Keras-Cookbook | 406b95f981a8ce66ac22605f4a5f0eaa35ab3aee | [
"MIT"
]
| 44 | 2019-03-23T07:19:36.000Z | 2022-02-22T11:11:12.000Z | 66.467492 | 5,690 | 0.725185 | [
[
[
"https://colab.research.google.com/drive/1OmAdxU_Lw7r-tMXiTOeSI7NWgB3AF9QF",
"_____no_output_____"
],
[
"# Issue with image translation",
"_____no_output_____"
]
],
[
[
"from keras.datasets import mnist\nimport numpy\nfrom keras.datasets import mnist\nfrom keras.models import Sequential\nfrom keras.layers import Dense\nfrom keras.layers import Dropout\nfrom keras.utils import np_utils",
"Using TensorFlow backend.\n"
],
[
"import matplotlib.pyplot as plt\n%matplotlib inline\n(X_train, y_train), (X_test, y_test) = mnist.load_data()",
"Downloading data from https://s3.amazonaws.com/img-datasets/mnist.npz\n11493376/11490434 [==============================] - 1s 0us/step\n"
],
[
"X_train1 = X_train[y_train==1]",
"_____no_output_____"
],
[
"num_pixels = X_train.shape[1] * X_train.shape[2]\nX_train = X_train.reshape(X_train.shape[0],num_pixels).astype('float32')\nX_test = X_test.reshape(X_test.shape[0],num_pixels).astype('float32')\nX_train = X_train / 255\nX_test = X_test / 255",
"_____no_output_____"
],
[
"y_train = np_utils.to_categorical(y_train)\ny_test = np_utils.to_categorical(y_test)\nnum_classes = y_train.shape[1]",
"_____no_output_____"
],
[
"model = Sequential()\nmodel.add(Dense(1000, input_dim=num_pixels, activation='relu'))\nmodel.add(Dense(num_classes, activation='softmax'))\nmodel.compile(loss='categorical_crossentropy', optimizer='adam',metrics=['accuracy'])\nhistory = model.fit(X_train, y_train, validation_data=(X_test, y_test),epochs=5, batch_size=1024, verbose=1)",
"WARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:66: The name tf.get_default_graph is deprecated. Please use tf.compat.v1.get_default_graph instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:541: The name tf.placeholder is deprecated. Please use tf.compat.v1.placeholder instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:4432: The name tf.random_uniform is deprecated. Please use tf.random.uniform instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/optimizers.py:793: The name tf.train.Optimizer is deprecated. Please use tf.compat.v1.train.Optimizer instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:3576: The name tf.log is deprecated. Please use tf.math.log instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/ops/math_grad.py:1250: add_dispatch_support.<locals>.wrapper (from tensorflow.python.ops.array_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.where in 2.0, which has the same broadcast rule as np.where\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:1033: The name tf.assign_add is deprecated. Please use tf.compat.v1.assign_add instead.\n\nTrain on 60000 samples, validate on 10000 samples\nEpoch 1/5\n60000/60000 [==============================] - 5s 78us/step - loss: 0.4730 - acc: 0.8733 - val_loss: 0.2302 - val_acc: 0.9334\nEpoch 2/5\n60000/60000 [==============================] - 1s 10us/step - loss: 0.1954 - acc: 0.9451 - val_loss: 0.1620 - val_acc: 0.9531\nEpoch 3/5\n60000/60000 [==============================] - 1s 10us/step - loss: 0.1387 - acc: 0.9614 - val_loss: 0.1261 - val_acc: 0.9635\nEpoch 4/5\n60000/60000 [==============================] - 1s 10us/step - loss: 0.1056 - acc: 0.9706 - val_loss: 0.1057 - val_acc: 0.9685\nEpoch 5/5\n60000/60000 [==============================] - 1s 10us/step - loss: 0.0838 - acc: 0.9770 - val_loss: 0.0971 - val_acc: 0.9711\n"
],
[
"import numpy as np\npic=np.zeros((28,28))\npic2=np.copy(pic)\nfor i in range(X_train1.shape[0]):\n pic2=X_train1[i,:,:]\n pic=pic+pic2\npic=(pic/X_train1.shape[0])\nplt.imshow(pic)",
"_____no_output_____"
],
[
"for i in range(pic.shape[0]):\n if i<20:\n pic[:,i]=pic[:,i+1]\n plt.imshow(pic)",
"_____no_output_____"
],
[
"model.predict(pic.reshape(1,784)/255)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e794ee2eff8e62cb81118e87880bc4e792d74736 | 91,280 | ipynb | Jupyter Notebook | Trajectory_Prediction.ipynb | Gianfranco-98/Trajectory-Prediction-PyTorch | 4cc700a23110b9354bdad5758e0c62c7c5c0f451 | [
"MIT"
]
| 2 | 2022-01-13T13:34:55.000Z | 2022-02-03T09:47:43.000Z | Trajectory_Prediction.ipynb | Gianfranco-98/Trajectory-Prediction-PyTorch | 4cc700a23110b9354bdad5758e0c62c7c5c0f451 | [
"MIT"
]
| null | null | null | Trajectory_Prediction.ipynb | Gianfranco-98/Trajectory-Prediction-PyTorch | 4cc700a23110b9354bdad5758e0c62c7c5c0f451 | [
"MIT"
]
| null | null | null | 41.191336 | 435 | 0.502914 | [
[
[
"## Deep Learining project\n\n\n* Gianfranco Di Marco - 1962292\n* Giacomo Colizzi Coin - 1794538\n\n\n\\\n**- Trajectory Prediction -**\n\nIs the problem of predicting the short-term (1-3 seconds) and long-term (3-5 seconds) spatial coordinates of various road-agents such as cars, buses, pedestrians, rickshaws, and animals, etc. These road-agents have different dynamic behaviors that may correspond to aggressive or conservative driving styles.\n\n**- nuScenes Dataset -**\n\nAvailable at. https://www.nuscenes.org/nuscenes. The nuScenes\ndataset is a large-scale autonomous driving dataset. The dataset has 3D bounding boxes for 1000 scenes collected in Boston and Singapore. Each scene is 20 seconds long and annotated at 2Hz. This results in a total of 28130 samples for training, 6019 samples for validation and 6008 samples for testing. The dataset has the full autonomous vehicle data suite: 32-beam LiDAR, 6 cameras and radars with complete 360° coverage\n\n\n> Holger Caesar and Varun Bankiti and Alex H. Lang and Sourabh Vora and Venice Erin Liong and Qiang Xu and Anush Krishnan and Yu Pan and Giancarlo Baldan and Oscar Beijbom: \"*nuScenes: A multimodal dataset for autonomous driving*\", arXiv preprint arXiv:1903.11027, 2019.\n\nThe most important part of this dataset for our project is the Map Expansion Pack, which simplify the trajectory prediction problem",
"_____no_output_____"
],
[
"## Requirements",
"_____no_output_____"
],
[
"**Environment**",
"_____no_output_____"
]
],
[
[
"# Necessary since Google Colab supports only Python 3.7\n# -> some libraries can be different from local and Colab\ntry:\n import google.colab\n from google.colab import drive\n ENVIRONMENT = 'colab'\n %pip install tf-estimator-nightly==2.8.0.dev2021122109\n %pip install folium==0.2.1\nexcept:\n ENVIRONMENT = 'local'",
"_____no_output_____"
]
],
[
[
"**Libraries**",
"_____no_output_____"
]
],
[
[
"%pip install nuscenes-devkit\n%pip install pytorch-lightning",
"_____no_output_____"
],
[
"# Learning\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nfrom torchvision.models import resnet50\nfrom torchvision.transforms import Normalize\nfrom torchmetrics import functional\nimport pytorch_lightning as pl\nfrom pytorch_lightning.callbacks import ModelCheckpoint\n\n# Math\nimport numpy as np\n\n# Dataset\nfrom nuscenes.nuscenes import NuScenes\nfrom nuscenes.prediction import PredictHelper\nfrom nuscenes.prediction.input_representation.static_layers import StaticLayerRasterizer\nfrom nuscenes.prediction.input_representation.agents import AgentBoxesWithFadedHistory\nfrom nuscenes.prediction.input_representation.interface import InputRepresentation\nfrom nuscenes.prediction.input_representation.combinators import Rasterizer\nfrom nuscenes.eval.prediction.config import PredictionConfig, load_prediction_config\nfrom nuscenes.eval.prediction.splits import get_prediction_challenge_split\nfrom nuscenes.eval.prediction import metrics, data_classes\n\n# File system\nimport os\nimport shutil\nimport pickle\nimport zipfile\nimport tarfile\nimport urllib.request\n\n# Generic\nimport time\nfrom tqdm import tqdm\nfrom typing import List, Dict, Tuple, Any\nfrom collections import defaultdict\nfrom abc import abstractmethod\nimport multiprocessing as mp\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"## Configuration",
"_____no_output_____"
],
[
"**Generic Parameters**",
"_____no_output_____"
]
],
[
[
"# Environment-dependent parameters\nif ENVIRONMENT == 'colab':\n ROOT = '/content/drive/MyDrive/DL/Trajectory-Prediction-PyTorch/'\n MAX_NUM_WORKERS = 0\n MAX_BATCH_SIZE = 8\n PROGRESS_BAR_REFRESH_RATE = 20\nelif ENVIRONMENT == 'local':\n ROOT = os.getcwd()\n # TODO: solve problem with VRAM with PL\n if os.name == 'nt':\n MAX_NUM_WORKERS = 0\n MAX_BATCH_SIZE = 16\n else:\n MAX_NUM_WORKERS = 4\n MAX_BATCH_SIZE = 8\n PROGRESS_BAR_REFRESH_RATE = 10\nelse:\n raise ValueError(\"Wrong 'environment' value\")\n\n# Train parameters\nBATCH_SIZE = MAX_BATCH_SIZE\nNUM_WORKERS = MAX_NUM_WORKERS\nLEARNING_RATE = 1e-4\nMOMENTUM = 0.9\nTRAIN_EPOCHES = 20 \nPLOT_PERIOD = 1 # 1 = plot at each epoch\nCHECKPOINT_DIR = os.path.join(ROOT, 'checkpoints')\nBEST_CHECKPOINT_DIR = os.path.join(CHECKPOINT_DIR, 'best')\nCHECKPOINT_MONITOR = \"val_loss\"\nTOP_K_SAVE = 10\n\n# Test parameters\nDEBUG_MODE = False\n\n# Hardcoded parameters\nHELPER_NEEDED = False",
"_____no_output_____"
]
],
[
[
"**Network Parameters**",
"_____no_output_____"
]
],
[
[
"# TODO: add other baselines\nPREDICTION_MODEL = 'CoverNet'\nif PREDICTION_MODEL == 'CoverNet':\n # - Architecture parameters\n BACKBONE_WEIGHTS = 'ImageNet'\n BACKBONE_MODEL = 'ResNet18'\n K_SIZE = 20000\n # - Trajectory parameters\n AGENT_HISTORY = 1\n SHORT_TERM_HORIZON = 3\n LONG_TERM_HORIZON = 6\n TRAJ_HORIZON = SHORT_TERM_HORIZON\n TRAJ_LINK = 'https://www.nuscenes.org/public/nuscenes-prediction-challenge-trajectory-sets.zip'\n TRAJ_DIR = os.path.join(ROOT, 'trajectory_sets')\n EPSILON = 2 ",
"_____no_output_____"
]
],
[
[
"**Dataset Parameters**",
"_____no_output_____"
]
],
[
[
"# Organization parameters\nPREPARE_DATASET = False\nPREPROCESSED = True\n\n# File system parameters\nPL_SEED = 42\nDATAROOT = os.path.join(ROOT, 'data', 'sets', 'nuscenes')\nPREPROCESSED_FOLDER = 'preprocessed'\nGT_SUFFIX = '-gt'\nFILENAME_EXT = '.pt'\nDATASET_VERSION = 'v1.0-trainval'\nAGGREGATORS = [{'name': \"RowMean\"}]\n\n# Other parameters\nMAX_PREDICTED_MODES = 25\nSAMPLES_PER_SECOND = 2\nNORMALIZATION = 'imagenet'",
"_____no_output_____"
]
],
[
[
"## Dataset",
"_____no_output_____"
],
[
"**Initialization**\n\nN.B: The download links in function *urllib.request.urlretrieve()* should be replaced periodically because it expires. Steps to download correctly are (on Firefox):\n\n\n1. Dowload Map Expansion pack (or Trainval metadata) from the website\n2. Stop the download\n3. Right-click on the file -> copy download link\n4. Paste the copied link into the first argument of the urlretrieve function. The second argument is the final name of the file",
"_____no_output_____"
]
],
[
[
"# Drive initialization\nif ENVIRONMENT == 'colab':\n drive.mount('/content/drive')",
"_____no_output_____"
],
[
"if PREPARE_DATASET:\n\n # Creating dataset dir\n os.makedirs(DATAROOT, exist_ok=True)\n os.chdir(DATAROOT)\n\n # Downloading Map Expansion Pack\n os.mkdir('maps')\n os.chdir('maps')\n print(\"Downloading and extracting Map Expansion pack ...\")\n urllib.request.urlretrieve('https://s3.amazonaws.com/data.nuscenes.org/public/v1.0/nuScenes-map-expansion-v1.3.zip?AWSAccessKeyId=AKIA6RIK4RRMFUKM7AM2&Signature=AvzxB6d7CxtpCUYIUChItvDSA3Q%3D&Expires=1651141974', 'nuScenes-map-expansion-v1.3.zip')\n with zipfile.ZipFile('nuScenes-map-expansion-v1.3.zip', 'r') as zip_ref:\n zip_ref.extractall(os.getcwd())\n os.remove('nuScenes-map-expansion-v1.3.zip')\n\n # Downloading Trainval Metadata\n os.chdir('..')\n print(\"Downloading and extracting TrainVal metadata ...\")\n urllib.request.urlretrieve('https://s3.amazonaws.com/data.nuscenes.org/public/v1.0/v1.0-trainval_meta.tgz?AWSAccessKeyId=AKIA6RIK4RRMFUKM7AM2&Signature=ZDr9UgOoV3UpYCI5RCY%2BNKiZVZ4%3D&Expires=1651142002', 'v1.0-trainval_meta.tgz')\n tar_ref = tarfile.open('v1.0-trainval_meta.tgz', 'r:gz')\n tar_ref.extractall(os.getcwd())\n tar_ref.close()\n os.remove('v1.0-trainval_meta.tgz')\n os.chdir(DATAROOT)",
"_____no_output_____"
]
],
[
[
"**Dataset definition**",
"_____no_output_____"
]
],
[
[
"class TrajPredDataset(torch.utils.data.Dataset):\n \"\"\" Trajectory Prediction Dataset\n\n Base Class for Trajectory Prediction Datasets\n \"\"\"\n def __init__(self, dataset, name, data_type, preprocessed, split,\n dataroot, preprocessed_folder, filename_ext,\n gt_suffix, traj_horizon, max_traj_horizon, num_workers):\n \"\"\" Dataset Initialization\n\n Parameters\n ----------\n dataset: the instantiated dataset\n name: name of the dataset\n data_type: data type of the dataset elements\n preprocessed: True if data has already been preprocessed\n split: the dataset split ('train', 'train_val', 'val')\n dataroot: the root directory of the dataset\n preprocessed_folder: the folder containing preprocessed data\n filename_ext: the extension of the generated filenames\n gt_suffix: the suffix added after each GT filename (before ext)\n traj_horizon: horizon (in seconds) for the future trajectory\n max_traj_horizon: maximum trajectory horizon possible (in seconds)\n num_workers: num of processes that collect data\n \"\"\"\n super(TrajPredDataset, self).__init__()\n self.dataset = dataset\n self.name = name\n self.data_type = data_type\n self.preprocessed = preprocessed\n self.split = split\n self.dataroot = dataroot\n self.preprocessed_folder = preprocessed_folder\n self.filename_ext = filename_ext\n self.gt_suffix = gt_suffix\n self.traj_horizon = traj_horizon\n self.max_traj_horizon = max_traj_horizon\n self.num_workers = num_workers\n self.helper = None\n self.tokens = None\n self.static_layer_rasterizer = None\n self.agent_rasterizer = None\n self.input_representation = None\n\n def __len__(self):\n \"\"\" Return the size of the dataset \"\"\"\n raise NotImplementedError\n\n def __getitem__(self, idx):\n \"\"\" Return an element of the dataset \"\"\"\n raise NotImplementedError\n\n @abstractmethod\n def generate_data(self):\n \"\"\" Data generation\n\n If self.preprocessed, directly collect data.\n Otherwise, generate data without preprocess it.\n \"\"\"\n raise NotImplementedError\n\n @abstractmethod\n def get_raster(self, token):\n \"\"\" Convert a token split into a raster\n\n Parameters\n ----------\n token: token containing instance token and sample token\n\n Return\n ------\n raster: the raster image\n \"\"\"\n raise NotImplementedError\n\n\nclass nuScenesDataset(TrajPredDataset):\n \"\"\" nuScenes Dataset for Trajectory Prediction challenge \"\"\"\n def __init__(self, helper, data_type='raster', preprocessed=False, split='train',\n dataroot=DATAROOT, preprocessed_folder=PREPROCESSED_FOLDER,\n filename_ext=FILENAME_EXT, gt_suffix=GT_SUFFIX,\n traj_horizon=TRAJ_HORIZON, max_traj_horizon=LONG_TERM_HORIZON,\n samples_per_second=SAMPLES_PER_SECOND,\n agent_history=AGENT_HISTORY, normalization=NORMALIZATION, \n num_workers=NUM_WORKERS):\n \"\"\" nuScenes Dataset Initialization\n\n Parameters\n ----------\n helper: the helper of the instantiated nuScenes dataset (None if not needed)\n data_type: data type of the dataset elements\n preprocessed: True if data has already been preprocessed\n split: the dataset split ('train', 'train_val', 'val')\n dataroot: the root directory of the dataset\n preprocessed_folder: the folder containing preprocessed data\n filename_ext: the extension of the generated filenames\n gt_suffix: the suffix added after each GT filename (before ext)\n traj_horizon: horizon (in seconds) for the future trajectory\n max_traj_horizon: maximum trajectory horizon possible (in seconds)\n samples_per_second: sampling frequency (in Hertz)\n agent_history: the seconds of considered agent history\n normalization: which kind of normalization to apply to input\n num_workers: num of processes that collect data\n \"\"\"\n # General initialization\n super(nuScenesDataset, self).__init__(\n None, 'nuScenes', data_type, preprocessed, split, dataroot, preprocessed_folder, \n filename_ext, gt_suffix, traj_horizon, max_traj_horizon, num_workers)\n self.helper = helper\n self.tokens = get_prediction_challenge_split(\n split, dataroot=dataroot)\n self.samples_per_second = samples_per_second\n if data_type == 'raster':\n if helper is not None:\n self.static_layer_rasterizer = StaticLayerRasterizer(self.helper)\n self.agent_rasterizer = AgentBoxesWithFadedHistory(\n self.helper, seconds_of_history=agent_history)\n self.input_representation = InputRepresentation(\n self.static_layer_rasterizer, self.agent_rasterizer, Rasterizer())\n else:\n self.static_layer_rasterizer = None\n self.agent_rasterizer = None\n self.input_representation = None\n else: # NOTE: possible also other type of input data\n pass\n if not self.preprocessed:\n print(\"Preprocessing data ...\")\n self.generate_data()\n\n # Normalization function\n if normalization == 'imagenet':\n self.normalization = Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])\n else:\n raise ValueError(\"Available only 'imagenet' normalization\")\n \n def __len__(self) -> int:\n \"\"\" Return the size of the dataset \"\"\"\n return len(self.tokens)\n\n def __getitem__(self, idx) -> Tuple[torch.Tensor, torch.Tensor, torch.Tensor, int]:\n \"\"\" Return an element of the dataset \"\"\"\n # Select subfolder\n if idx < 0:\n idx = len(self) + idx\n subfolder = f'batch_{idx//128}'\n # Load files\n complete_tensor = torch.load(\n os.path.join(self.dataroot, self.preprocessed_folder, self.split,\n subfolder, self.tokens[idx] + self.filename_ext))\n gt_trajectory = torch.load(\n os.path.join(self.dataroot, self.preprocessed_folder, self.split, subfolder,\n self.tokens[idx] + self.gt_suffix + self.filename_ext))\n # Adjust tensors\n # NOTE: maybe it's better to handle this section in data generation \n while gt_trajectory.shape[0] < self.samples_per_second * self.max_traj_horizon:\n gt_trajectory = torch.concat((gt_trajectory, gt_trajectory[-1].unsqueeze(0)))\n gt_trajectory = gt_trajectory[:(self.samples_per_second * self.traj_horizon)]\n agent_state_vector, raster_img = self.tensor_io_conversion(\n \"read\", None, None, complete_tensor)\n raster_img = self.normalization(raster_img)\n nan_mask = agent_state_vector != agent_state_vector\n if nan_mask.any():\n agent_state_vector[nan_mask] = 0\n return agent_state_vector, raster_img, gt_trajectory, idx\n\n def generate_data(self):\n \"\"\" Data generation\n\n If self.preprocessed, directly collect data.\n Otherwise, generate data without preprocess it.\n \"\"\"\n # Generate directories if don't exist\n preprocessed_dir = os.path.join(self.dataroot, self.preprocessed_folder)\n split_dir = os.path.join(preprocessed_dir, self.split)\n if self.preprocessed_folder not in os.listdir(self.dataroot):\n os.mkdir(preprocessed_dir)\n if self.split not in os.listdir(preprocessed_dir):\n os.mkdir(split_dir)\n # Variable useful to restore interrupted preprocessing\n preprocessed_batches = os.listdir(split_dir)\n already_preproc = \\\n len([f for f in preprocessed_batches\n if os.path.isfile(os.path.join(split_dir, f))])\n\n # Create subfolders\n if len(preprocessed_batches) == 0:\n n_subfolders = len(self.tokens) // 128 + int(len(self.tokens) % 128 != 0)\n for i in range(n_subfolders):\n subfolder = 'batch_' + str(i)\n os.mkdir(os.path.join(split_dir, subfolder))\n\n # Generate data\n if self.data_type == 'raster':\n for i, t in enumerate(tqdm(self.tokens)):\n subfolder = f'batch_{i//128}'\n if i >= int(already_preproc/2):\n self.generate_raster_data(t, split_dir, subfolder)\n else:\n pass\n\n def generate_raster_data(self, token, batches_dir, subfolder):\n \"\"\" Generate a raster map and agent state vector from token split \n\n The generated input data consists in a tensor like this:\n [raster map | agent state vector]\n The generated ground truth data is the future agent trajectory tensor\n\n Parameters\n ----------\n token: token containing instance token and sample token\n batches_dir: the directory in which the batches will be generated\n subfolder: the data is divided into subfolders in order to avoid Drive timeouts;\n this parameter tells which is the actual subfolder towhere place data\n \"\"\"\n # Generate and concatenate input tensors\n instance_token, sample_token = token.split(\"_\")\n raster_img = self.input_representation.make_input_representation(\n instance_token, sample_token)\n raster_tensor = torch.Tensor(raster_img).permute(2, 0, 1) / 255.\n agent_state_vector = torch.Tensor(\n [[self.helper.get_velocity_for_agent(instance_token, sample_token),\n self.helper.get_acceleration_for_agent(instance_token, sample_token),\n self.helper.get_heading_change_rate_for_agent(instance_token, sample_token)]])\n raster_agent_tensor, _ = \\\n self.tensor_io_conversion('write', raster_tensor, agent_state_vector)\n\n # Generate ground truth\n gt_trajectory = torch.Tensor(\n self.helper.get_future_for_agent(instance_token, sample_token,\n seconds=self.max_traj_horizon, in_agent_frame=True))\n\n # Save to disk\n torch.save(raster_agent_tensor, os.path.join(\n batches_dir, subfolder, token + self.filename_ext))\n torch.save(gt_trajectory, os.path.join(\n batches_dir, subfolder, token + self.gt_suffix + self.filename_ext))\n \n @staticmethod\n def tensor_io_conversion(mode, big_t=None, small_t=None, complete_t=None) -> Tuple[torch.Tensor, torch.Tensor]:\n \"\"\" Utility IO function to concatenate tensors of different shape\n\n Normally used to concatenate (or separate) raster map and agent state vector in order to speed up IO\n\n Parameters\n ----------\n mode: 'write' (concatenate) or 'read' (separate)\n big_t: the bigger tensor (None if we are going to separate tensors)\n small_t: the smaller tensor (None if we are going to separate tensors)\n complete_t: the concatenated tensor (None if we are going to concatenate tensors)\n\n Return\n ------\n out1: big tensor (mode == 'read') or complete tensor (mode == 'write')\n out2: small tensor (mode == 'read') or empty tensor (mode == 'write') \n \"\"\"\n out1, out2 = None, None\n if mode == 'write': # concatenate\n if big_t is None or small_t is None:\n raise ValueError(\"Wrong argument: 'big_t' and 'small_t' cannot be None\")\n small_t = small_t.permute(1, 0).unsqueeze(2)\n small_t = small_t.expand(-1, -1, big_t.shape[-1])\n out1 = torch.cat((big_t, small_t), dim=1)\n out2 = torch.empty(small_t.shape)\n elif mode == 'read': # separate\n if complete_t is None:\n raise ValueError(\"Wrong argument: 'complete_t' cannot be None\")\n out1 = complete_t[..., -1, -1].unsqueeze(0)\n out2 = complete_t[..., :-1, :]\n else:\n raise ValueError(\n \"Wrong argument 'mode'; available 'read' or 'write'\")\n return out1, out2\n\nclass nuScenesDataModule(pl.LightningDataModule):\n \"\"\" PyTorch Lightning Data Module for the nuScenes dataset \"\"\"\n def __init__(self, nuscenes_train, nuscenes_val, batch_size=BATCH_SIZE, num_workers=NUM_WORKERS):\n \"\"\" Data Module initialization\n\n Parameters\n ----------\n nuscenes_train: instance of the nuScenesDataset class (split='train')\n nuscenes_val: instance of the nuScenesDataset class (split='val')\n batch_size: number of samples to extract from the dataset at each step\n num_workers: number of cores implied in data collection\n \"\"\"\n super(nuScenesDataModule, self).__init__()\n self.batch_size = batch_size\n self.num_workers = num_workers\n self.nuscenes_train = nuscenes_train\n self.nuscenes_val = nuscenes_val\n\n def setup(self, stage=None):\n \"\"\" Setup the data module \"\"\"\n if stage == \"fit\" or stage is None:\n self.nusc_train = self.nuscenes_train\n self.nusc_val = self.nuscenes_val\n\n if stage == \"test\" or stage is None:\n self.nusc_test = self.nuscenes_val\n\n def train_dataloader(self):\n \"\"\" Dataloader for the training part \"\"\"\n return torch.utils.data.DataLoader(self.nusc_train, self.batch_size, shuffle=True,\n num_workers=self.num_workers, drop_last=True)\n\n def val_dataloader(self):\n \"\"\" Dataloader for the validation part \"\"\"\n return torch.utils.data.DataLoader(self.nusc_val, self.batch_size, shuffle=False, \n num_workers=self.num_workers, drop_last=True)\n\n def test_dataloader(self):\n \"\"\" Dataloader for the testing part \"\"\"\n return torch.utils.data.DataLoader(self.nusc_test, self.batch_size, shuffle=False,\n num_workers=self.num_workers, drop_last=True)",
"_____no_output_____"
]
],
[
[
"## Models",
"_____no_output_____"
],
[
"**Covernet**",
"_____no_output_____"
]
],
[
[
"class CoverNet(pl.LightningModule):\n \"\"\" CoverNet model for Trajectory Prediction \"\"\"\n def __init__(self, K_size, epsilon, traj_link, traj_dir, device, \n lr=LEARNING_RATE, momentum=MOMENTUM,\n traj_samples=SAMPLES_PER_SECOND*TRAJ_HORIZON):\n \"\"\" CoverNet initialization\n\n Parameters\n ----------\n K_size: number of modes (trajectories) (needed ?)\n epsilon: value (in meters) relative to the space coverage\n traj_link: link from which to download the trajectories\n device: target device of the model (e.g. 'cuda:0')\n lr: learning rate of the optimizer\n momentum: momentum of the optimizer\n traj_samples: number of samples to consider in the trajectory\n \"\"\"\n super().__init__()\n self.K_size = K_size\n self.convModel = resnet50(pretrained=True)\n self.activation = {}\n def get_activation(name):\n def hook(model, input, output):\n self.activation[name] = output\n return hook\n self.convModel.layer4.register_forward_hook(get_activation('layer4'))\n self.trajectories = prepare_trajectories(epsilon, traj_link, traj_dir)\n self.fc1 = nn.Linear(2051, 4096)\n self.fc2 = nn.Linear(4096, self.trajectories.size()[0])\n self.traj_samples = traj_samples\n self.tgt_device = device\n self.momentum = momentum\n self.lr = lr\n\n def forward(self, x) -> torch.Tensor:\n \"\"\" Network inference \"\"\"\n img, state = x\n self.convModel(img)\n resnet_output = torch.flatten(self.convModel.avgpool(self.activation['layer4']),start_dim=1)\n x = torch.cat([resnet_output, state], 1)\n x = self.fc1(x)\n x = self.fc2(x)\n return x\n\n def training_step(self, batch, batch_idx):\n \"\"\" Training step of the model\n\n Parameters\n ----------\n batch: batch of data\n batch_idx: index of the actual batch (from 0 to len(dataset))\n \"\"\"\n # Collect data\n x_state, x_img, gt, _ = batch\n x_state = torch.flatten(x_state, 0, 1)\n reduced_traj = self.trajectories[:, :self.traj_samples]\n # Prepare positive samples\n with torch.no_grad():\n y = get_positives(reduced_traj, gt.to('cpu'))\n y = y.to(self.tgt_device)\n # Inference\n y_hat = self((x_img, x_state))\n loss = F.cross_entropy(y_hat, y)\n # Log\n self.log('train_loss', loss.item(), on_step=True)\n \n return loss\n\n def validation_step(self, batch, batch_idx):\n \"\"\" Validation step of the model\n\n Parameters\n ----------\n batch: batch of data\n batch_idx: index of the actual batch (from 0 to len(dataset))\n \"\"\"\n with torch.no_grad():\n # Collect data\n x_state, x_img, gt, _ = batch\n x_state = torch.flatten(x_state, 0, 1)\n reduced_traj = self.trajectories[:, :self.traj_samples]\n # Prepare positive samples\n y = get_positives(reduced_traj, gt.to('cpu'))\n y = y.to(self.tgt_device)\n # Inference\n y_hat = self((x_img, x_state))\n loss = F.cross_entropy(y_hat, y)\n # Log\n self.log('val_loss', loss.item(), on_epoch=True)\n \n return loss\n\n def configure_optimizers(self):\n \"\"\" Set the optimizer for the model \"\"\"\n # TODO: find best optimizer and parameters\n #return torch.optim.Adam(self.parameters(), lr=self.lr)\n return torch.optim.SGD(self.parameters(), lr=self.lr, momentum=self.momentum)\n\n# TODO: check if generated trajectory are expressed in the same frame of the agent\ndef get_positives(trajectories, ground_truth) -> torch.Tensor:\n \"\"\" Get positive samples wrt the actual GT\n\n Parameters\n ----------\n trajectories: the pre-generated set of trajectories\n ground_truth: the future trajectory for the agent\n\n Return\n ------\n positive_traj: as defined in the original CoverNet paper, \n 'positive samples determined by the element in the trajectory set\n closest to the actual ground truth in minimum average \n of point-wise Euclidean distances'\n \"\"\"\n euclidean_dist = torch.stack([torch.pow(torch.sub(trajectories, gt), 2) \n for gt in ground_truth]).sum(dim=3).sqrt() \n mean_euclidean_dist = euclidean_dist.mean(dim=2)\n positive_traj = mean_euclidean_dist.argmin(dim=1)\n return positive_traj\n\ndef prepare_trajectories(epsilon, download_link, directory) -> torch.Tensor:\n \"\"\" Function to download and extract trajectory sets for CoverNet \n\n Parameters\n ----------\n epsilon: value (in meters) relative to the space coverage\n download_link: link from which to download trajectory sets\n directory: directory where to download trajectory sets\n\n Return\n ------\n trajectories: tensor of the trajectory set for the specified epsilon\n \"\"\"\n # 1. Download and extract trajectories\n filename_zip = 'nuscenes-prediction-challenge-trajectory-sets.zip'\n filename = filename_zip[:-4]\n filename_dir = os.path.join(directory, filename)\n filename_zipdir = os.path.join(directory, filename_zip)\n if (not os.path.isdir(filename_dir) \n or any(e not in os.listdir(filename_dir)\n for e in ['epsilon_2.pkl', 'epsilon_4.pkl', 'epsilon_8.pkl'])):\n print(\"Downloading trajectories ...\")\n os.makedirs(directory, exist_ok=True)\n urllib.request.urlretrieve(download_link, filename_zipdir)\n with zipfile.ZipFile(filename_zipdir, 'r') as archive:\n archive.extractall(directory)\n os.remove(filename_zipdir)\n\n # 2. Generate trajectories\n traj_set_path = os.path.join(filename_dir, 'epsilon_' + str(epsilon) + '.pkl')\n trajectories = pickle.load(open(traj_set_path, 'rb'))\n return torch.Tensor(trajectories)",
"_____no_output_____"
]
],
[
[
"## Utilities",
"_____no_output_____"
],
[
"**Metrics**",
"_____no_output_____"
]
],
[
[
"def compute_metrics(predictions: List[data_classes.Prediction], ground_truths: List[np.ndarray], \n helper, aggregators=AGGREGATORS) -> Dict[str, Any]:#Dict[str, Dict[str, List[float]]]:\n \"\"\" Utility eval function to compute dataset metrics\n\n Parameters\n ----------\n predictions: list of predictions made by the model (in Prediction class format)\n ground_truths: the real trajectories of the agent (SHAPE -> [len(dataset), n_samples, state_dim])\n helper: nuScenes dataset helper\n aggregators: functions to aggregate metrics (e.g. mean)\n\n Return\n ------\n metric_output: dictionary of the computed metrics:\n - minADE_5: The average of pointwise L2 distances between the predicted trajectory \n and ground truth over the 5 most likely predictions.\n - minADE_10: The average of pointwise L2 distances between the predicted trajectory \n and ground truth over the 10 most likely predictions.\n - missRateTop_2_5: Proportion of misses relative to the 5 most likely trajectories\n over all agents\n - missRateTop_2_10: Proportion of misses relative to the 10 most likely trajectories\n over all agents\n - minFDE_1: The final displacement error (FDE) is the L2 distance \n between the final points of the prediction and ground truth, computed\n on the most likely trajectory\n - offRoadRate: the fraction of trajectories that are not entirely contained\n in the drivable area of the map.\n \"\"\"\n # 1. Define metrics\n print(\"\\t - Metrics definition ...\")\n aggregators = \\\n [metrics.deserialize_aggregator(agg) for agg in aggregators]\n min_ade = metrics.MinADEK([5, 10], aggregators)\n miss_rate = metrics.MissRateTopK([5, 10], aggregators)\n min_fde = metrics.MinFDEK([1], aggregators)\n if helper is not None:\n # FIXME: instantiating offRoadRate class makes RAM explode\n #offRoadRate = metrics.OffRoadRate(self.helper, self.aggregators)\n pass\n else:\n offRoadRate = None\n\n # 2. Compute metrics\n metric_list = []\n print(\"\\t - Effective metrics computation ...\")\n for p, pred in enumerate(tqdm(predictions)):\n # TODO: check for argument shapes\n minADE_5 = min_ade(ground_truths[p], pred)[0][0]\n minADE_10 = min_ade(ground_truths[p], pred)[0][1]\n missRateTop_2_5 = miss_rate(ground_truths[p], pred)[0][0]\n missRateTop_2_10 = miss_rate(ground_truths[p], pred)[0][1]\n minFDE_1 = min_fde(ground_truths[p], pred)\n #offRoadRate = offRoadRate(ground_truth[i], prediction)\n metric = {'minADE_5': minADE_5, 'missRateTop_2_5': missRateTop_2_5,\n 'minADE_10': minADE_10, 'missRateTop_2_10': missRateTop_2_10,\n 'minFDE_1': minFDE_1}#, 'offRoadRate': offRoadRate}\n metric_list.append(metric)\n\n # 3. Aggregate\n print(\"\\t - Metrics aggregation ...\")\n aggregations: Dict[str, Dict[str, List[float]]] = defaultdict(dict)\n metric_names = list(metric_list[0].keys())\n metrics_dict = {name: np.array([metric_list[i][name] for i in range(len(metric_list))]) \n for name in metric_names}\n for metric in metric_names:\n for agg in aggregators:\n aggregations[metric][agg.name] = agg(metrics_dict[metric])\n\n return aggregations ",
"_____no_output_____"
]
],
[
[
"**Plotting**",
"_____no_output_____"
]
],
[
[
"def plot_train_data(train_iterations, val_iterations, epoches, train_losses, val_losses):\n \"\"\" Plot a graph with the training trend\n\n Parameters\n ----------\n train_iterations: number of iterations for each epoch [train]\n val_iterations: number of iterations for each epoch [val]\n epoches: actual epoch number (starting from 1)\n train_losses: array of loss values [train]\n val_losses: array of loss values [val]\n \"\"\"\n # Data preparation\n train_iterations_list = list(range(epoches*(train_iterations)))\n val_iterations_list = list(range(epoches*(val_iterations)))\n epoches_list = list(range(epoches))\n\n # Adjust validation array dimension\n val_error = len(val_losses) - len(val_iterations_list)\n if val_error > 0:\n val_losses = val_losses[:-val_error]\n\n # Per-iteration plot\n fig = plt.figure()\n plt.title('Per-iteration Loss [train]')\n plt.xlabel('Iterations')\n plt.ylabel('Value')\n l1, = plt.plot(train_iterations_list, train_losses, c='blue')\n plt.legend(handles=[l1], labels=['Train loss'], loc='best')\n plt.show()\n fig = plt.figure()\n plt.title('Per-iteration Loss [val]')\n plt.xlabel('Iterations')\n plt.ylabel('Value')\n l2, = plt.plot(val_iterations_list, val_losses, c='red')\n plt.legend(handles=[l2], labels=['Validation loss'], loc='best')\n plt.show()\n\n # Per-epoch plot\n fig = plt.figure()\n plt.title('Per-epoch Loss')\n plt.xlabel('Epoches')\n plt.ylabel('Value')\n train_avg_losses = [np.array(train_losses[i:i+train_iterations]).mean() \n for i in range(0, len(train_losses), train_iterations)]\n val_avg_losses = [np.array(val_losses[i:i+val_iterations]).mean() \n for i in range(0, len(val_losses), val_iterations)]\n l1, = plt.plot(epoches_list, train_avg_losses, c='blue')\n l2, = plt.plot(epoches_list, val_avg_losses, c='red')\n plt.legend(handles=[l1, l2], labels=['Train loss', 'Validation loss'], loc='best')\n plt.show()\n\ndef plot_agent_future(raster, future, agent_pos=(0,0), reference_frame='local', color='green'):\n \"\"\" Plot agent's future trajectory\n\n Parameters\n ----------\n raster: raster map tensor (image)\n future: future trajectory of the agent (predicted or GT) [x,y]\n agent_pos: position of the agent (needed in case of local coords)\n reference_frame: frame to which future coordinates refer\n color: color of the plotted trajectory\n \"\"\"\n # Show raster map\n plt.imshow(raster.permute(1, 2, 0))\n\n # Show trajectory\n x, y = [], []\n for i in range(len(future)):\n point = (agent_pos[0], agent_pos[1]) if i == 0 else future[i].numpy()\n if reference_frame == 'local' and i > 0:\n point = (point[0] + agent_pos[0], -point[1] + agent_pos[1])\n x.append(point[0])\n y.append(point[1])\n \n plt.plot(x, y, color=color, markersize=10, linewidth=5)\n plt.show()",
"_____no_output_____"
]
],
[
[
"## Main",
"_____no_output_____"
],
[
"**Initialization**",
"_____no_output_____"
]
],
[
[
"# ---------- Dataset initialization ---------- #\n# Initialize nuScenes helper\nprint(\"nuScenes Helper initialization ...\")\nstart_time = time.time()\npl.seed_everything(PL_SEED)\nif ENVIRONMENT == 'local':\n if PREPARE_DATASET:\n nusc = NuScenes(version=DATASET_VERSION, dataroot=DATAROOT, verbose=True)\n with open(os.path.join(ROOT, 'nuscenes_checkpoint'+FILENAME_EXT), 'wb') as f:\n pickle.dump(nusc, f, protocol=pickle.HIGHEST_PROTOCOL)\n elif not 'nusc' in locals():\n if HELPER_NEEDED:\n with open(os.path.join(ROOT, 'nuscenes_checkpoint'+FILENAME_EXT), 'rb') as f:\n nusc = pickle.load(f)\nelif ENVIRONMENT == 'colab':\n if PREPARE_DATASET or HELPER_NEEDED:\n nusc = NuScenes(version=DATASET_VERSION, dataroot=DATAROOT, verbose=True)\nhelper = PredictHelper(nusc) if HELPER_NEEDED else None\nprint(\"nuScenes Helper initialization done in %f s\\n\" % (time.time() - start_time))\n\n# Initialize dataset and data module\nprint(\"\\nDataset and Data Module initialization ...\")\nstart_time = time.time()\ntrain_dataset = nuScenesDataset(helper, preprocessed=PREPROCESSED, split='train')\nval_dataset = nuScenesDataset(helper, preprocessed=PREPROCESSED, split='val')\ntrainval_dm = nuScenesDataModule(train_dataset, val_dataset, num_workers=NUM_WORKERS)\ntrainval_dm.setup(stage='fit')\nprint(\"Dataset and Data Module initialization done in %f s\\n\" % (time.time() - start_time))\n\n# ---------- Network initialization ---------- #\nprint(\"\\nCoverNet model initialization ...\")\nstart_time = time.time()\ndevice = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\nmodel = CoverNet(K_SIZE, EPSILON, TRAJ_LINK, TRAJ_DIR, device)\nprint(\"CoverNet model intialization done in %f s\\n\" % (time.time() - start_time))\n\n# ---------- Training initialization ---------- #\nprint(\"\\nTrainer initialization ...\")\nstart_time = time.time()\nGPUS = min(1, torch.cuda.device_count())\ncheckpoint_callback = ModelCheckpoint(dirpath=CHECKPOINT_DIR,\n save_top_k=TOP_K_SAVE,\n monitor=CHECKPOINT_MONITOR)\ntrainer = pl.Trainer(callbacks=[checkpoint_callback],\n progress_bar_refresh_rate=PROGRESS_BAR_REFRESH_RATE, \n gpus=GPUS, max_epochs=TRAIN_EPOCHES)\nprint(\"Trainer intialization done in %f s\\n\" % (time.time() - start_time))",
"_____no_output_____"
]
],
[
[
"**Training loop**",
"_____no_output_____"
]
],
[
[
"trainer.fit(model, trainval_dm)",
"_____no_output_____"
]
],
[
[
"**Testing**",
"_____no_output_____"
]
],
[
[
"# Dataloader initialization\nprint(\"Loading test dataloader ...\")\ntrainval_dm.setup(stage='test')\ntest_dataloader = trainval_dm.test_dataloader()\ntest_generator = iter(test_dataloader)\n\n# Trained model initialization\n# TODO: istantiate kwargs for network in a better way\nprint(\"\\nCoverNet trained model initialization ...\")\ncheckpoint_name = 'epoch=19-step=80460.ckpt'\nnet_args = {'K_size': K_SIZE, 'epsilon': EPSILON, 'traj_link': TRAJ_LINK, 'traj_dir': TRAJ_DIR, 'device': device}\nmodel = CoverNet.load_from_checkpoint(checkpoint_path=os.path.join(BEST_CHECKPOINT_DIR, checkpoint_name), \n map_location=None, hparams_file=None, strict=True, \n K_size=K_SIZE, epsilon=EPSILON, traj_link=TRAJ_LINK, traj_dir=TRAJ_DIR, device=device).to(device)\nmodel.eval()\n\n# ---------- CoverNet Metrics computation ---------- #\n# TODO: generalize metrics computation\npredictions = []\nground_truths = []\nstart = time.time()\nreduced_traj = model.trajectories[:, :model.traj_samples].numpy()\nprint(\"\\nCoverNet metrics computation ...\")\nprint(\"1 - Producing predictions ...\")\nfor i, token in enumerate(tqdm(val_dataset.tokens)):\n with torch.no_grad():\n x_state, x_img, gt, _ = val_dataset[i]\n x_state = x_state.to(device)\n x_img = x_img.to(device)\n x_state = torch.unsqueeze(torch.flatten(x_state, 0, 1), 0)\n x_img = torch.unsqueeze(x_img, 0)\n pred_logits = model((x_img, x_state))\n pred_probs = F.softmax(pred_logits, dim=1)[0]\n top_indices = pred_probs.argsort()[-MAX_PREDICTED_MODES:]\n cutted_probs = pred_probs[top_indices].cpu().numpy()\n cutted_traj = reduced_traj[top_indices.cpu()]\n i_t, s_t = token.split(\"_\")\n ground_truths.append(gt.numpy())\n predictions.append(data_classes.Prediction(i_t, s_t, cutted_traj, cutted_probs))\nprint(\"2 - Computing metrics ...\")\nconvernet_metrics = compute_metrics(predictions, ground_truths, helper)\nprint(\"Metric computation done in %f s\" % (time.time() - start))\n",
"_____no_output_____"
],
[
"convernet_metrics",
"_____no_output_____"
]
],
[
[
"## Code Debugging",
"_____no_output_____"
],
[
"**Training loop** (manual - debug only)",
"_____no_output_____"
]
],
[
[
"if DEBUG_MODE:\n\n # Dataset preparation\n train_dataloader = torch.utils.data.DataLoader(train_dataset, BATCH_SIZE, shuffle=True, num_workers=NUM_WORKERS, drop_last=True)\n val_dataloader = torch.utils.data.DataLoader(val_dataset, BATCH_SIZE, shuffle=False, num_workers=NUM_WORKERS, drop_last=True)\n\n # Training preparation\n optimizer = torch.optim.SGD(model.parameters(), lr=LEARNING_RATE, momentum=MOMENTUM)\n model = model.to(device)\n\n # Plotting preparation\n train_loss_arr = []\n val_loss_arr = []\n train_iterations = len(train_dataset) // BATCH_SIZE\n val_iterations = len(val_dataset) // BATCH_SIZE\n\n # Training loop\n for i in range(TRAIN_EPOCHES):\n print(\"-------- Epoch %d --------\" % i)\n model.train()\n\n # Training\n for j, data in enumerate(train_dataloader):\n \n # Data preparation\n x_state, x_img, gt, idx = data\n x_state = x_state.to(device)\n x_img = x_img.to(device)\n x_state = torch.flatten(x_state, 0, 1)\n with torch.no_grad():\n reduced_traj = model.trajectories[:, :SAMPLES_PER_SECOND*TRAJ_HORIZON]\n y = get_positives(reduced_traj, gt)\n\n # Inference\n optimizer.zero_grad()\n traj_logits = model((x_img, x_state))\n y = y.to(device)\n loss = F.cross_entropy(traj_logits, y)\n loss.backward()\n optimizer.step()\n\n # Logging\n loss_val = loss.item()\n train_loss_arr.append(loss_val)\n print(\"[%d] %d - train loss = %f\" % (i, j, loss_val))\n\n # Validation\n model.train(mode=False)\n for j, data in enumerate(val_dataloader):\n\n # Data preparation\n x_state, x_img, gt, idx = data\n x_state = x_state.to(device)\n x_img = x_img.to(device)\n x_state = torch.flatten(x_state, 0, 1)\n reduced_traj = model.trajectories[:, :SAMPLES_PER_SECOND*TRAJ_HORIZON]\n y = get_positives(reduced_traj, gt)\n\n # Inference\n traj_logits = model((x_img, x_state))\n y = y.to(device)\n loss = F.cross_entropy(traj_logits, y)\n\n # Logging\n loss_val = loss.item()\n val_loss_arr.append(loss_val)\n print(\"[%d] %d - val loss = %f\" % (i, j, loss_val))\n\n # Plotting\n if (i+1) % PLOT_PERIOD == 0:\n plot_train_data(train_iterations, val_iterations, i+1, train_loss_arr, val_loss_arr)\n a = input(\"Press Enter to continue...\")\n plt.close('all')\n ",
"_____no_output_____"
]
],
[
[
"**Dataset debugging**",
"_____no_output_____"
]
],
[
[
"# Initialize nuScenes\nHELPER_NEEDED = True\nif ENVIRONMENT == 'local':\n if PREPARE_DATASET:\n nusc = NuScenes(version=DATASET_VERSION, dataroot=DATAROOT, verbose=True)\n with open(os.path.join(ROOT, 'nuscenes_checkpoint'+FILENAME_EXT), 'wb') as f:\n pickle.dump(nusc, f, protocol=pickle.HIGHEST_PROTOCOL)\n elif not 'nusc' in locals():\n if HELPER_NEEDED:\n with open(os.path.join(ROOT, 'nuscenes_checkpoint'+FILENAME_EXT), 'rb') as f:\n nusc = pickle.load(f)\nelif ENVIRONMENT == 'colab':\n if PREPARE_DATASET or HELPER_NEEDED:\n nusc = NuScenes(version=DATASET_VERSION, dataroot=DATAROOT, verbose=True)",
"_____no_output_____"
],
[
"helper = PredictHelper(nusc)\ndataset = nuScenesDataset(helper, preprocessed=PREPROCESSED)\ntrain_dataloader = torch.utils.data.DataLoader(dataset, BATCH_SIZE, True, num_workers=NUM_WORKERS)\ntrain_generator = iter(train_dataloader)",
"_____no_output_____"
],
[
"# Useful to check ideal number of workers and batch size\nx = time.time()\ntry:\n state, img, gt, idxs = next(train_generator)\nexcept StopIteration:\n train_generator = iter(train_dataloader)\n state, img, gt, idxs = next(train_generator)\nprint(time.time() - x)",
"_____no_output_____"
],
[
"state, img, gt, idx = dataset[np.random.randint(len(dataset))]\nplt.imshow(img.permute(1, 2, 0))\nplt.show()\nprint(\"State input size:\", state.shape)\nprint(\"Ground truth size:\", gt.shape)",
"_____no_output_____"
],
[
"instance_token, sample_token = dataset.tokens[idx].split(\"_\")\nlong_gt = torch.Tensor(\n dataset.helper.get_future_for_agent(instance_token, sample_token,\n seconds=100, in_agent_frame=True))\n# TODO: check how to get agent position in the map \nplot_agent_future(img, long_gt, agent_pos=(250,400), reference_frame='local')",
"_____no_output_____"
]
],
[
[
"**Network debugging**",
"_____no_output_____"
]
],
[
[
"test_states, test_imgs, test_gts, _ = next(train_generator)\ntest_states = torch.flatten(test_states, 0, 1)\n\nprint(test_imgs.size())\nprint(test_states.size())",
"_____no_output_____"
],
[
"# Prediction\nmodel = CoverNet(K_SIZE, EPSILON, TRAJ_LINK, TRAJ_DIR, device='cuda:0')\ntraj_logits = model((test_imgs, test_states))\n\n# Output 5 and 10 most likely trajectories for this batch\ntop_5_trajectories = model.trajectories[traj_logits.argsort(descending=True)[:5]]\ntop_10_trajectories = model.trajectories[traj_logits.argsort(descending=True)[:10]]",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
e794fba6af7c16a53b32b0c39640f4222beb852b | 6,406 | ipynb | Jupyter Notebook | notebooks/Create cifar_datasets.ipynb | sinhaharsh/pytorch-CycleGAN-and-pix2pix | 7a38c79f4344c954dd28d041c82c121c92465d3d | [
"BSD-3-Clause"
]
| 1 | 2021-03-29T03:10:32.000Z | 2021-03-29T03:10:32.000Z | notebooks/Create cifar_datasets.ipynb | sinhaharsh/pytorch-CycleGAN-and-pix2pix | 7a38c79f4344c954dd28d041c82c121c92465d3d | [
"BSD-3-Clause"
]
| null | null | null | notebooks/Create cifar_datasets.ipynb | sinhaharsh/pytorch-CycleGAN-and-pix2pix | 7a38c79f4344c954dd28d041c82c121c92465d3d | [
"BSD-3-Clause"
]
| null | null | null | 29.657407 | 101 | 0.506712 | [
[
[
"import os\nimport shutil",
"_____no_output_____"
],
[
"labels = os.listdir('../datasets/cifar10png/train/')",
"_____no_output_____"
],
[
"labels",
"_____no_output_____"
],
[
"labels.index('automobile')",
"_____no_output_____"
],
[
"partition = 'train'\ndataset = 'cifar10'\nDIR = os.path.join('../datasets/', dataset+'png', partition)\nOUT_DIR = os.path.join('../datasets/', dataset, partition+'A')\nfor folder in os.listdir(DIR):\n input_path = os.path.join(DIR, folder)\n for image in os.listdir(input_path):\n if image.endswith('.png'):\n img_path = os.path.join(input_path, image)\n filename, ext = image.split('.')\n out_path = os.path.join(OUT_DIR, filename+'_'+str(labels.index(folder))+'.'+ext)\n shutil.copy(img_path, out_path)",
"_____no_output_____"
],
[
"partition = 'test'\ndataset = 'cifar10'\nDIR = os.path.join('../datasets/', dataset+'png', partition)\nOUT_DIR = os.path.join('../datasets/', dataset, partition+'A')\nfor folder in os.listdir(DIR):\n input_path = os.path.join(DIR, folder)\n for image in os.listdir(input_path):\n if image.endswith('.png'):\n img_path = os.path.join(input_path, image)\n filename, ext = image.split('.')\n out_path = os.path.join(OUT_DIR, filename+'_'+str(labels.index(folder))+'.'+ext)\n shutil.copy(img_path, out_path)",
"_____no_output_____"
],
[
"partition = 'train'\ndataset = 'cifar100'\nDIR = os.path.join('../datasets/', dataset+'png', partition)\nOUT_DIR = os.path.join('../datasets/', dataset, partition+'A')\nlabels = os.listdir(DIR)\n\nfor folder in os.listdir(DIR):\n input_path = os.path.join(DIR, folder)\n for image in os.listdir(input_path):\n if image.endswith('.png'):\n img_path = os.path.join(input_path, image)\n filename, ext = image.split('.')\n out_path = os.path.join(OUT_DIR, filename+'_'+str(labels.index(folder))+'.'+ext)\n shutil.copy(img_path, out_path)",
"_____no_output_____"
],
[
"!ls ../datasets/cifar100png/train",
"apple\t castle\t fox\t\tmushroom road\t tank\r\naquarium_fish caterpillar girl\toak_tree rocket\t telephone\r\nbaby\t cattle\t hamster\torange\t rose\t television\r\nbear\t chair\t house\torchid\t sea\t tiger\r\nbeaver\t chimpanzee kangaroo\totter\t seal\t tractor\r\nbed\t clock\t keyboard\tpalm_tree shark\t train\r\nbee\t cloud\t lamp\tpear\t shrew\t trout\r\nbeetle\t cockroach lawn_mower\tpickup_truck skunk\t tulip\r\nbicycle couch\t leopard\tpine_tree skyscraper turtle\r\nbottle\t crab\t lion\tplain\t snail\t wardrobe\r\nbowl\t crocodile lizard\tplate\t snake\t whale\r\nboy\t cup\t lobster\tpoppy\t spider\t willow_tree\r\nbridge\t dinosaur man\t\tporcupine squirrel\t wolf\r\nbus\t dolphin\t maple_tree\tpossum\t streetcar woman\r\nbutterfly elephant motorcycle\trabbit\t sunflower worm\r\ncamel\t flatfish mountain\traccoon sweet_pepper\r\ncan\t forest\t mouse\tray\t table\r\n"
],
[
"partition = 'test'\ndataset = 'cifar100'\nDIR = os.path.join('../datasets/', dataset+'png', partition)\nOUT_DIR = os.path.join('../datasets/', dataset, partition+'A')\nfor folder in os.listdir(DIR):\n input_path = os.path.join(DIR, folder)\n for image in os.listdir(input_path):\n if image.endswith('.png'):\n img_path = os.path.join(input_path, image)\n filename, ext = image.split('.')\n out_path = os.path.join(OUT_DIR, filename+'_'+str(labels.index(folder))+'.'+ext)\n shutil.copy(img_path, out_path)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e794fe9e2af6036fb518ac7473276b7495fe5636 | 2,471 | ipynb | Jupyter Notebook | jupyter/OpenVisus-Template.ipynb | ComputingElevatedLab/sciviscourse | e7a71be9aee3cd4fc591d9d868445a0840994785 | [
"MIT"
]
| null | null | null | jupyter/OpenVisus-Template.ipynb | ComputingElevatedLab/sciviscourse | e7a71be9aee3cd4fc591d9d868445a0840994785 | [
"MIT"
]
| null | null | null | jupyter/OpenVisus-Template.ipynb | ComputingElevatedLab/sciviscourse | e7a71be9aee3cd4fc591d9d868445a0840994785 | [
"MIT"
]
| null | null | null | 24.465347 | 131 | 0.532578 | [
[
[
"## OpenVisus Enabled Jupyter Notebook",
"_____no_output_____"
],
[
"### OpenViSUS: imports and utilities",
"_____no_output_____"
]
],
[
[
"%matplotlib notebook\n\nimport os,sys\n\nimport matplotlib.pyplot as plt\nimport numpy as np\nfrom ipywidgets import *\n\nimport OpenVisus as ov\n\n# Enable I/O component of OpenVisus\nov.DbModule.attach()",
"Requirement already satisfied: OpenVisus in /Users/steve/opt/anaconda3/lib/python3.7/site-packages (1.3.70)\nRequirement already satisfied: numpy in /Users/steve/opt/anaconda3/lib/python3.7/site-packages (from OpenVisus) (1.17.2)\nPythonEngine is working fine\n"
],
[
"# function to plot the image data with matplotlib\n# optional parameters: colormap, existing plot to reuse (for more interactivity)\ndef showData(data, cmap=None, plot=None):\n if len(data.shape)==3 and data.shape[0]==1: data=data[0,:,:]\n if len(data.shape)==3 and data.shape[1]==1: data=data[:,0,:] \n if len(data.shape)==3 and data.shape[2]==1: data=data[:,:,0]\n if(plot==None or cmap!=None):\n fig=plt.figure(figsize = (7,5))\n plot = plt.imshow(data, origin='lower', cmap=cmap)\n plt.show()\n return plot\n else:\n plot.set_data(data)\n plt.show()\n return plot\n ",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code"
]
]
|
e79513fe9267a586fc857d1a687125dee3235351 | 19,855 | ipynb | Jupyter Notebook | T2.ProcHilosPy/T2-MultiprocessingPython.ipynb | patoba/ComputacionConcurrente | 07a9adf80fe30e6dfb9fdd82015d0436d4210b50 | [
"MIT"
]
| 1 | 2020-12-31T17:53:28.000Z | 2020-12-31T17:53:28.000Z | T2.ProcHilosPy/T2-MultiprocessingPython.ipynb | patoba/ComputacionConcurrente | 07a9adf80fe30e6dfb9fdd82015d0436d4210b50 | [
"MIT"
]
| null | null | null | T2.ProcHilosPy/T2-MultiprocessingPython.ipynb | patoba/ComputacionConcurrente | 07a9adf80fe30e6dfb9fdd82015d0436d4210b50 | [
"MIT"
]
| 1 | 2021-09-10T04:41:44.000Z | 2021-09-10T04:41:44.000Z | 25.164766 | 385 | 0.496953 | [
[
[
"# Computo concurrente\n\n## Multiprocessing",
"_____no_output_____"
],
[
"El modulo 'multiprocessing' de Python permite la manipulacion y sincronizacion de procesos, tambien ofrece concurrencia local como remota.\n\nEjemplo de motivacion...",
"_____no_output_____"
]
],
[
[
"import time\ndef calc_cuad(numeros):\n print('Calcula el cuadrado:')\n for n in numeros:\n time.sleep(0.2)\n print('cuadrado:', n*n)\ndef calc_cubo(numeros):\n print('Calcula el cubo:')\n for n in numeros:\n time.sleep(0.2)\n print('cubo:', n*n*n)\n \nnums = range(10)\n\nt = time.time()\ncalc_cuad(nums)\ncalc_cubo(nums)\n\nprint('Finaliza la ejecucion')\nprint('Tiempo de ejecucion', time.time()-t)",
"Calcula el cuadrado:\ncuadrado: 0\ncuadrado: 1\ncuadrado: 4\ncuadrado: 9\ncuadrado: 16\ncuadrado: 25\ncuadrado: 36\ncuadrado: 49\ncuadrado: 64\ncuadrado: 81\nCalcula el cubo:\ncubo: 0\ncubo: 1\ncubo: 8\ncubo: 27\ncubo: 64\ncubo: 125\ncubo: 216\ncubo: 343\ncubo: 512\ncubo: 729\nFinaliza la ejecucion\nTiempo de ejecucion 4.024327278137207\n"
]
],
[
[
"Una manera sencilla de generar procesos en Python es por medio de la creacion del objeto `Process` y llamarlo por medio del metodo `start()`.",
"_____no_output_____"
]
],
[
[
"import multiprocessing as mp\n\ndef tarea(nombre):\n print('Hola', nombre)\n for n in range(10000):\n n**(1/(n+1))\n \nif __name__ == '__main__': # Esta condicion se interpreta como una verificacion de si este proceso es el principal\n p = mp.Process(target=tarea, args=('Saul', )) ## Ejecuta la funcion tarea con el los argumentos de args\n p.start() ## Ejecuta p el objeto multiprocess \n p.join()",
"Hola Saul\n"
],
[
"import multiprocessing as mp\nimport time\n\n\ndef calc_cuad(numeros):\n print('Calcula el cuadrado:')\n for n in numeros:\n time.sleep(0.2)\n print('cuadrado:', n*n)\ndef calc_cubo(numeros):\n print('Calcula el cubo:')\n for n in numeros:\n time.sleep(0.2)\n print('cubo:', n*n*n)\n \nnums = range(10)\n\n\nt1 = time.time()\n\np1 = mp.Process(target=calc_cuad, args=(nums,))\np2 = mp.Process(target=calc_cubo, args=(nums,))\n\np1.start()\np2.start()\np1.join()\np2.join()\n\n\nprint('Tiempo de ejecucion', time.time()-t1)\nprint('Finaliza la ejecucion')",
"Calcula el cuadrado:\nCalcula el cubo:\ncuadrado: cubo:0 \n0\ncuadrado: cubo:1 \n1\ncuadrado:cubo: 48\n\ncuadrado:cubo: 927\n\ncuadrado: cubo:16 \n64\ncuadrado: cubo:25\n 125\ncuadrado: 36\ncubo: 216\ncuadrado: 49\ncubo: 343\ncuadrado: 64\ncubo: 512\ncuadrado: 81\ncubo: 729\nTiempo de ejecucion 2.1406450271606445\nFinaliza la ejecucion\n"
]
],
[
[
"## Tarea \n\nInvestiga en la documentacion del modulo `Multiprocessing` cual es su funcionamiento y todos los metodos o funciones que estan implementados en el.",
"_____no_output_____"
],
[
"## Identificadores PID, PPID",
"_____no_output_____"
]
],
[
[
"import multiprocessing as mp\nimport os\n\nprint('Nombre del proceso:', __name__)\nprint('Proceso padre:', os.getppid())\nprint('Proceso actual:', os.getpid())",
"Nombre del proceso: __main__\nProceso padre: 7448\nProceso actual: 8016\n"
],
[
"import multiprocessing as mp\nimport os\n\ndef info(titulo):\n print(titulo)\n print('Nombre del proceso:', __name__)\n print('Proceso padre:', os.getppid())\n print('Proceso actual:', os.getpid())\n\ndef f(nombre):\n info('Funcion f')\n print('Hola', nombre)\n print('------------')\n \ninfo('Inicio')\n\np = mp.Process(target = f, args=('Valeriano',))\np.start()\np.join()",
"Inicio\nNombre del proceso: __main__\nProceso padre: 7448\nProceso actual: 8016\nFuncion f\nNombre del proceso: __main__\nProceso padre: 8016\nProceso actual: 9690\nHola Valeriano\n------------\n"
]
],
[
[
"## Ejercicio\n\nCrea 3 procesos hijos, donde:\n- El primero multiplique 3 numeros (a,b,c)\n- El segundo sume (a,b,c)\n- El tercero haga (a+b)/c\n- Todos devolveran el valor calculado, el nombre de cada proceso hijo y el id del proceso padre.",
"_____no_output_____"
]
],
[
[
"import multiprocessing as mp\nimport os\n\ndef info(titulo):\n print(titulo)\n print('Nombre del proceso:', __name__)\n print('Proceso actual:', os.getpid())\n print('Proceso padre:', os.getppid())\n \n\ndef primero(a,b,c):\n info('a*b*c =')\n print(a*b*c)\n \ndef segundo(a,b,c):\n info('a+b+c =')\n print(a+b+c)\n \ndef tercero(a,b,c):\n info('(a+b)/c =')\n print((a+b)/c)\n \ninfo('Inicio')\n\n\na = 3\nb = 5\nc = 1\n\np1 = mp.Process(target = primero, args=(a,b,c))\np2 = mp.Process(target = segundo, args=(a,b,c))\np3 = mp.Process(target = tercero, args=(a,b,c))\n\np1.start()\np1.join()\np2.start()\np2.join()\np3.start()\np3.join()",
"Inicio\nNombre del proceso: __main__\nProceso actual: 8016\nProceso padre: 7448\na*b*c =\nNombre del proceso: __main__\nProceso actual: 10829\nProceso padre: 8016\n15\na+b+c =\nNombre del proceso: __main__\nProceso actual: 10862\nProceso padre: 8016\n9\n(a+b)/c =\nNombre del proceso: __main__\nProceso actual: 10895\nProceso padre: 8016\n8.0\n"
],
[
"import time\n\nnums_res = []\n\ndef calc_cuad(numeros):\n global nums_res\n for n in numeros:\n print('Cuadrado:', n*n)\n nums_res.append(n*n)\n \n \nnums = range(10)\nt = time.time()\np1 = mp.Process(target=calc_cuad, args = (nums,))\n\n\np1.start()\np1.join()\n\nprint('Tiempo de ejecucion: ', time.time()-t)\nprint('Resultado del proceso:', nums_res)\nprint('Finaliza ejecucion')",
"Cuadrado: 0\nCuadrado: 1\nCuadrado: 4\nCuadrado: 9\nCuadrado: 16\nCuadrado: 25\nCuadrado: 36\nCuadrado: 49\nCuadrado: 64\nCuadrado: 81\nTiempo de ejecucion: 0.04141974449157715\nResultado del proceso: []\nFinaliza ejecucion\n"
]
],
[
[
"# 2020-10-27\n\n## Nombres y terminación de procesos",
"_____no_output_____"
]
],
[
[
"import multiprocessing\nmultiprocessing.cpu_count()",
"_____no_output_____"
]
],
[
[
"Con el método `cpu_count()",
"_____no_output_____"
]
],
[
[
"import time\ndef TareaHijo():\n print(\"Proceso HIJO con PID: {}\".format(multiprocessing.current_process().pid))\n time.sleep(3)\n print(\"Fin del proceso hijo\")\ndef main():\n print(\"Proceso Padre PID: {}\".format(multiprocessing.current_process().pid))\n myProcess = multiprocessing.Process(target=TareaHijo) # Define el objeto myProcess como el objeto que llam[a al proceso]\n myProcess.start()\n myProcess.join()\n# Se acostumbra usar la variable __name__\n# para hacer la ejecución desde el progragrama\n# principal, puede omitirse en los notebooks \nif __name__ == '__main__':\n main()",
"Proceso Padre PID: 6703\nProceso HIJO con PID: 7714\nFin del proceso hijo\n"
]
],
[
[
"Es posible asignar un nombre a un proceso hijo que ha sido creado, por medio del medio del argumento `name` se asigna el nombre del proceso hijo.",
"_____no_output_____"
]
],
[
[
"def myProcess():\n print(\"Proceso con nombre: {}\".format(multiprocessing.current_process().name)) ## Metodo current process para obtener el nombre del proceso\n \ndef main():\n childProcess = multiprocessing.Process(target=myProcess, name='Proceso-LCD-cc')\n childProcess.start()\n childProcess.join()\n \nmain()",
"Proceso con nombre: Proceso-LCD-cc\n"
],
[
"from multiprocessing import Process, current_process\nimport time\n\n\ndef f1():\n pname = current_process().name\n print('Starting process %s...' % pname)\n time.sleep(2)\n print('Exiting process %s...' % pname)\n \n \n\ndef f2():\n pname = current_process().name\n print('Starting process %s...' % pname)\n time.sleep(4)\n print('Exiting process %s...' % pname)\n \n \nif __name__ == '__main__':\n p1 = Process(name='Worker 1', target=f1)\n p2 = Process(name='Worker 2', target=f2)\n p3 = Process(target=f1)\n p1.start()\n p2.start()\n p3.start()\n \n p1.join()\n p2.join()\n p3.join()",
"Starting process Worker 1...\nStarting process Worker 2...\nStarting process Process-23...\nExiting process Worker 1...\nExiting process Process-23...\nExiting process Worker 2...\n"
],
[
"def TareaProceso():\n proceso_actual = multiprocessing.current_process()\n print(\"Procesos Hijo PID: {}\".format(proceso_actual.pid))\n time.sleep(20)\n proceso_actual = multiprocessing.current_process()\n print(\"Procesos Padre PID: {}\".format(proceso_actual.pid))\n \nmiProceso = multiprocessing.Process(target=TareaProceso)\nmiProceso.start()\n\n\n\nprint(\"Proceso Padre ha terminado, termina el proceso main\")\nprint(\"Terminando el proceso Hijo...\")\ntime.sleep(1)\nmiProceso.terminate()\n#miProceso.join()\nprint(\"Proceso Hijo ha terminado exitosamente\")\n",
"Procesos Hijo PID: 8953\nProceso Padre ha terminado, termina el proceso main\nTerminando el proceso Hijo...\nProceso Hijo ha terminado exitosamente\n"
],
[
"multiprocessing.cpu_count()",
"_____no_output_____"
]
],
[
[
"### Ejercicio:\n\n1. Vamos a crear 3 procesos los cuales tendrán nombre y código definido como funP1, funP2, funP3. Cada hijo escribirá su nombre, su PID y el PID del padre, además de hacer un cálculo sobre tres valores a, b y c.\n2. El proceso 1 calcula a*b + c, el proceso 2 calcula a*b*c y el proceso 3 calcula (a*b)/c\n3. Crea un mecanismo para terminar alguno de los procesos de manera aleatoria.",
"_____no_output_____"
]
],
[
[
"import multiprocessing as mp\nimport os\nimport random\n\ndef info(titulo):\n pname = current_process().name\n print('Nombre del proceso: %s...' % pname)\n print(titulo)\n print('Proceso actual:', os.getpid())\n print('Proceso padre:', os.getppid())\n \n\ndef funP1(a,b,c):\n info('a*b + c =')\n print(a*b + c)\n \ndef funP2(a,b,c):\n info('a*b*c =')\n print(a*b*c)\n \ndef funP3(a,b,c):\n info('(a+b)/c =')\n print((a+b)/c)\n\n\na = 3\nb = 5\nc = 1\n\np1 = mp.Process(name = 'funP1',target = funP1, args=(a,b,c))\np2 = mp.Process(name = 'funP2',target = funP2, args=(a,b,c))\np3 = mp.Process(name = 'funP3',target = funP3, args=(a,b,c))\n\n\np1.start()\np2.start()\np3.start()\n\ni = random.randint(1,3)\n\nif i==1:\n p1.terminate()\nelif i == 2:\n p2.terminate()\nelif i == 3:\n p3.terminate()\n\n\np1.join()\np2.join()\np3.join()",
"Nombre del proceso: funP2...\na*b*c =\nProceso actual: 15004\nProceso padre: 6703\n15\nNombre del proceso: funP1...\na*b + c =\nProceso actual: 15003\nProceso padre: 6703\n16\n"
]
],
[
[
"No obstante , a veces se requiere crear procesos que corran en silencio (*background*) y no bloquear el proceso principal al finalizarlos. Esta especificación es comunmente utilizada cuando el proceso principal no tiene la certeza de interrumpir un proceso después de esperar cierto tiempo o finalizar sin que haya terminado el proceso hijo sin afectaciones al resultado final\n\nEstos procesos se llaman **Procesos demonio** (*daemon processes*). Por medio del atributo `daemon` del método `Process` se crea un proceso de este tipo.\n\nEl valor por defecto del atributo `daemon` es False, por tanto se establece a `True`para crear el proceso demonio.",
"_____no_output_____"
]
],
[
[
"from multiprocessing import Process, current_process\nimport time\n\ndef f1():\n p = current_process()\n print('Starting process %s, ID %s....' %(p.name, p.pid))\n time.sleep(8)\n print('Starting process %s, ID, %s....' %(p.name, p.pid))\n \ndef f2():\n p = current_process()\n print('Starting process %s, ID %s....' %(p.name, p.pid))\n time.sleep(2)\n print('Starting process %s, ID %s....' %(p.name, p.pid))\n \n \nif __name__ == '__main__':\n p1 = Process(name='Worker 1', target=f1)\n p1.daemon = True\n p2 = Process(name='Worker 2', target=f2)\n \n p1.start()\n time.sleep(1)\n p2.start()\n \n # p1.join()\n # p2.join()\n # p3.join()",
"Starting process Worker 1, ID 15700....\nStarting process Worker 2, ID 15705....\nStarting process Worker 2, ID 15705....\nStarting process Worker 1, ID, 15700....\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
e7952170348c0f54bbe1344d025b1b66381374cf | 401,029 | ipynb | Jupyter Notebook | sentiment-network/sentiment-classification-project.ipynb | eugli/udacity-deep-learning | e426a2662e0981583c1e4fec95c7509a66c0ae75 | [
"MIT"
]
| null | null | null | sentiment-network/sentiment-classification-project.ipynb | eugli/udacity-deep-learning | e426a2662e0981583c1e4fec95c7509a66c0ae75 | [
"MIT"
]
| null | null | null | sentiment-network/sentiment-classification-project.ipynb | eugli/udacity-deep-learning | e426a2662e0981583c1e4fec95c7509a66c0ae75 | [
"MIT"
]
| null | null | null | 57.478716 | 37,692 | 0.656601 | [
[
[
"# Sentiment Classification & How To \"Frame Problems\" for a Neural Network\n\nby Andrew Trask\n\n- **Twitter**: @iamtrask\n- **Blog**: http://iamtrask.github.io",
"_____no_output_____"
],
[
"### What You Should Already Know\n\n- neural networks, forward and back-propagation\n- stochastic gradient descent\n- mean squared error\n- and train/test splits\n\n### Where to Get Help if You Need it\n- Re-watch previous Udacity Lectures\n- Leverage the recommended Course Reading Material - [Grokking Deep Learning](https://www.manning.com/books/grokking-deep-learning) (Check inside your classroom for a discount code)\n- Shoot me a tweet @iamtrask\n\n\n### Tutorial Outline:\n\n- Intro: The Importance of \"Framing a Problem\" (this lesson)\n\n- [Curate a Dataset](#lesson_1)\n- [Developing a \"Predictive Theory\"](#lesson_2)\n- [**PROJECT 1**: Quick Theory Validation](#project_1)\n\n\n- [Transforming Text to Numbers](#lesson_3)\n- [**PROJECT 2**: Creating the Input/Output Data](#project_2)\n\n\n- Putting it all together in a Neural Network (video only - nothing in notebook)\n- [**PROJECT 3**: Building our Neural Network](#project_3)\n\n\n- [Understanding Neural Noise](#lesson_4)\n- [**PROJECT 4**: Making Learning Faster by Reducing Noise](#project_4)\n\n\n- [Analyzing Inefficiencies in our Network](#lesson_5)\n- [**PROJECT 5**: Making our Network Train and Run Faster](#project_5)\n\n\n- [Further Noise Reduction](#lesson_6)\n- [**PROJECT 6**: Reducing Noise by Strategically Reducing the Vocabulary](#project_6)\n\n\n- [Analysis: What's going on in the weights?](#lesson_7)",
"_____no_output_____"
],
[
"# Lesson: Curate a Dataset<a id='lesson_1'></a>\nThe cells from here until Project 1 include code Andrew shows in the videos leading up to mini project 1. We've included them so you can run the code along with the videos without having to type in everything.",
"_____no_output_____"
]
],
[
[
"def pretty_print_review_and_label(i):\n print(labels[i] + \"\\t:\\t\" + reviews[i][:80] + \"...\")\n\ng = open('reviews.txt','r') # What we know!\nreviews = list(map(lambda x:x[:-1],g.readlines()))\ng.close()\n\ng = open('labels.txt','r') # What we WANT to know!\nlabels = list(map(lambda x:x[:-1].upper(),g.readlines()))\ng.close()",
"_____no_output_____"
]
],
[
[
"**Note:** The data in `reviews.txt` we're using has already been preprocessed a bit and contains only lower case characters. If we were working from raw data, where we didn't know it was all lower case, we would want to add a step here to convert it. That's so we treat different variations of the same word, like `The`, `the`, and `THE`, all the same way.",
"_____no_output_____"
]
],
[
[
"len(reviews)",
"_____no_output_____"
],
[
"reviews[0]",
"_____no_output_____"
],
[
"labels[0]",
"_____no_output_____"
]
],
[
[
"# Lesson: Develop a Predictive Theory<a id='lesson_2'></a>",
"_____no_output_____"
]
],
[
[
"print(\"labels.txt \\t : \\t reviews.txt\\n\")\npretty_print_review_and_label(2137)\npretty_print_review_and_label(12816)\npretty_print_review_and_label(6267)\npretty_print_review_and_label(21934)\npretty_print_review_and_label(5297)\npretty_print_review_and_label(4998)",
"labels.txt \t : \t reviews.txt\n\nNEGATIVE\t:\tthis movie is terrible but it has some good effects . ...\nPOSITIVE\t:\tadrian pasdar is excellent is this film . he makes a fascinating woman . ...\nNEGATIVE\t:\tcomment this movie is impossible . is terrible very improbable bad interpretat...\nPOSITIVE\t:\texcellent episode movie ala pulp fiction . days suicides . it doesnt get more...\nNEGATIVE\t:\tif you haven t seen this it s terrible . it is pure trash . i saw this about ...\nPOSITIVE\t:\tthis schiffer guy is a real genius the movie is of excellent quality and both e...\n"
]
],
[
[
"# Project 1: Quick Theory Validation<a id='project_1'></a>\n\nThere are multiple ways to implement these projects, but in order to get your code closer to what Andrew shows in his solutions, we've provided some hints and starter code throughout this notebook.\n\nYou'll find the [Counter](https://docs.python.org/2/library/collections.html#collections.Counter) class to be useful in this exercise, as well as the [numpy](https://docs.scipy.org/doc/numpy/reference/) library.",
"_____no_output_____"
]
],
[
[
"from collections import Counter\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"We'll create three `Counter` objects, one for words from postive reviews, one for words from negative reviews, and one for all the words.",
"_____no_output_____"
]
],
[
[
"# Create three Counter objects to store positive, negative and total counts\npositive_counts = Counter()\nnegative_counts = Counter()\ntotal_counts = Counter()",
"_____no_output_____"
]
],
[
[
"**TODO:** Examine all the reviews. For each word in a positive review, increase the count for that word in both your positive counter and the total words counter; likewise, for each word in a negative review, increase the count for that word in both your negative counter and the total words counter.\n\n**Note:** Throughout these projects, you should use `split(' ')` to divide a piece of text (such as a review) into individual words. If you use `split()` instead, you'll get slightly different results than what the videos and solutions show.",
"_____no_output_____"
]
],
[
[
"for i in range(len(reviews)):\n if(labels[i] == 'POSITIVE'):\n for word in reviews[i].split(\" \"):\n positive_counts[word] += 1\n total_counts[word] += 1\n else:\n for word in reviews[i].split(\" \"):\n negative_counts[word] += 1\n total_counts[word] += 1",
"_____no_output_____"
]
],
[
[
"Run the following two cells to list the words used in positive reviews and negative reviews, respectively, ordered from most to least commonly used. ",
"_____no_output_____"
]
],
[
[
"# Examine the counts of the most common words in positive reviews\npositive_counts.most_common()",
"_____no_output_____"
],
[
"# Examine the counts of the most common words in negative reviews\nnegative_counts.most_common()",
"_____no_output_____"
]
],
[
[
"As you can see, common words like \"the\" appear very often in both positive and negative reviews. Instead of finding the most common words in positive or negative reviews, what you really want are the words found in positive reviews more often than in negative reviews, and vice versa. To accomplish this, you'll need to calculate the **ratios** of word usage between positive and negative reviews.\n\n**TODO:** Check all the words you've seen and calculate the ratio of postive to negative uses and store that ratio in `pos_neg_ratios`. \n>Hint: the positive-to-negative ratio for a given word can be calculated with `positive_counts[word] / float(negative_counts[word]+1)`. Notice the `+1` in the denominator – that ensures we don't divide by zero for words that are only seen in positive reviews.",
"_____no_output_____"
]
],
[
[
"# Create Counter object to store positive/negative ratios\npos_neg_ratios = Counter()\n\n# TODO: Calculate the ratios of positive and negative uses of the most common words\n# Consider words to be \"common\" if they've been used at least 100 times\n# for word, count in total_counts.most_common():\n# if count > 100:\n# neg_ratio = positive_counts[word] / float(negative_counts[word] + 1)\n# pos_neg_ratios[word] = neg_ratio\n \nfor term,count in total_counts.most_common():\n if(count > 100):\n pos_neg_ratio = positive_counts[term] / float(negative_counts[term]+1)\n pos_neg_ratios[term] = pos_neg_ratio",
"_____no_output_____"
]
],
[
[
"Examine the ratios you've calculated for a few words:",
"_____no_output_____"
]
],
[
[
"print(\"Pos-to-neg ratio for 'the' = {}\".format(pos_neg_ratios[\"the\"]))\nprint(\"Pos-to-neg ratio for 'amazing' = {}\".format(pos_neg_ratios[\"amazing\"]))\nprint(\"Pos-to-neg ratio for 'terrible' = {}\".format(pos_neg_ratios[\"terrible\"]))",
"Pos-to-neg ratio for 'the' = 1.0607993145235326\nPos-to-neg ratio for 'amazing' = 4.022813688212928\nPos-to-neg ratio for 'terrible' = 0.17744252873563218\n"
]
],
[
[
"Looking closely at the values you just calculated, we see the following:\n\n* Words that you would expect to see more often in positive reviews – like \"amazing\" – have a ratio greater than 1. The more skewed a word is toward postive, the farther from 1 its positive-to-negative ratio will be.\n* Words that you would expect to see more often in negative reviews – like \"terrible\" – have positive values that are less than 1. The more skewed a word is toward negative, the closer to zero its positive-to-negative ratio will be.\n* Neutral words, which don't really convey any sentiment because you would expect to see them in all sorts of reviews – like \"the\" – have values very close to 1. A perfectly neutral word – one that was used in exactly the same number of positive reviews as negative reviews – would be almost exactly 1. The `+1` we suggested you add to the denominator slightly biases words toward negative, but it won't matter because it will be a tiny bias and later we'll be ignoring words that are too close to neutral anyway.\n\nOk, the ratios tell us which words are used more often in postive or negative reviews, but the specific values we've calculated are a bit difficult to work with. A very positive word like \"amazing\" has a value above 4, whereas a very negative word like \"terrible\" has a value around 0.18. Those values aren't easy to compare for a couple of reasons:\n\n* Right now, 1 is considered neutral, but the absolute value of the postive-to-negative rations of very postive words is larger than the absolute value of the ratios for the very negative words. So there is no way to directly compare two numbers and see if one word conveys the same magnitude of positive sentiment as another word conveys negative sentiment. So we should center all the values around netural so the absolute value from neutral of the postive-to-negative ratio for a word would indicate how much sentiment (positive or negative) that word conveys.\n* When comparing absolute values it's easier to do that around zero than one. \n\nTo fix these issues, we'll convert all of our ratios to new values using logarithms.\n\n**TODO:** Go through all the ratios you calculated and convert them to logarithms. (i.e. use `np.log(ratio)`)\n\nIn the end, extremely positive and extremely negative words will have positive-to-negative ratios with similar magnitudes but opposite signs.",
"_____no_output_____"
]
],
[
[
"# TODO: Convert ratios to logs\nfor word, ratio in pos_neg_ratios.most_common():\n pos_neg_ratios[word] = np.log(ratio)",
"_____no_output_____"
]
],
[
[
"Examine the new ratios you've calculated for the same words from before:",
"_____no_output_____"
]
],
[
[
"print(\"Pos-to-neg ratio for 'the' = {}\".format(pos_neg_ratios[\"the\"]))\nprint(\"Pos-to-neg ratio for 'amazing' = {}\".format(pos_neg_ratios[\"amazing\"]))\nprint(\"Pos-to-neg ratio for 'terrible' = {}\".format(pos_neg_ratios[\"terrible\"]))",
"Pos-to-neg ratio for 'the' = 0.05902269426102881\nPos-to-neg ratio for 'amazing' = 1.3919815802404802\nPos-to-neg ratio for 'terrible' = -1.7291085042663878\n"
]
],
[
[
"If everything worked, now you should see neutral words with values close to zero. In this case, \"the\" is near zero but slightly positive, so it was probably used in more positive reviews than negative reviews. But look at \"amazing\"'s ratio - it's above `1`, showing it is clearly a word with positive sentiment. And \"terrible\" has a similar score, but in the opposite direction, so it's below `-1`. It's now clear that both of these words are associated with specific, opposing sentiments.\n\nNow run the following cells to see more ratios. \n\nThe first cell displays all the words, ordered by how associated they are with postive reviews. (Your notebook will most likely truncate the output so you won't actually see *all* the words in the list.)\n\nThe second cell displays the 30 words most associated with negative reviews by reversing the order of the first list and then looking at the first 30 words. (If you want the second cell to display all the words, ordered by how associated they are with negative reviews, you could just write `reversed(pos_neg_ratios.most_common())`.)\n\nYou should continue to see values similar to the earlier ones we checked – neutral words will be close to `0`, words will get more positive as their ratios approach and go above `1`, and words will get more negative as their ratios approach and go below `-1`. That's why we decided to use the logs instead of the raw ratios.",
"_____no_output_____"
]
],
[
[
"# words most frequently seen in a review with a \"POSITIVE\" label\npos_neg_ratios.most_common()",
"_____no_output_____"
],
[
"# words most frequently seen in a review with a \"NEGATIVE\" label\nlist(reversed(pos_neg_ratios.most_common()))[0:30]\n\n# Note: Above is the code Andrew uses in his solution video, \n# so we've included it here to avoid confusion.\n# If you explore the documentation for the Counter class, \n# you will see you could also find the 30 least common\n# words like this: pos_neg_ratios.most_common()[:-31:-1]",
"_____no_output_____"
]
],
[
[
"# End of Project 1. \n## Watch the next video to see Andrew's solution, then continue on to the next lesson.\n\n# Transforming Text into Numbers<a id='lesson_3'></a>\nThe cells here include code Andrew shows in the next video. We've included it so you can run the code along with the video without having to type in everything.",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image\n\nreview = \"This was a horrible, terrible movie.\"\n\nImage(filename='sentiment_network.png')",
"_____no_output_____"
],
[
"review = \"The movie was excellent\"\n\nImage(filename='sentiment_network_pos.png')",
"_____no_output_____"
]
],
[
[
"# Project 2: Creating the Input/Output Data<a id='project_2'></a>\n\n**TODO:** Create a [set](https://docs.python.org/3/tutorial/datastructures.html#sets) named `vocab` that contains every word in the vocabulary.",
"_____no_output_____"
]
],
[
[
"# TODO: Create set named \"vocab\" containing all of the words from all of the reviews\nvocab = list(total_counts)",
"_____no_output_____"
]
],
[
[
"Run the following cell to check your vocabulary size. If everything worked correctly, it should print **74074**",
"_____no_output_____"
]
],
[
[
"vocab_size = len(vocab)\nprint(vocab_size)",
"74074\n"
]
],
[
[
"Take a look at the following image. It represents the layers of the neural network you'll be building throughout this notebook. `layer_0` is the input layer, `layer_1` is a hidden layer, and `layer_2` is the output layer.",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image\nImage(filename='sentiment_network_2.png')",
"_____no_output_____"
]
],
[
[
"**TODO:** Create a numpy array called `layer_0` and initialize it to all zeros. You will find the [zeros](https://docs.scipy.org/doc/numpy/reference/generated/numpy.zeros.html) function particularly helpful here. Be sure you create `layer_0` as a 2-dimensional matrix with 1 row and `vocab_size` columns. ",
"_____no_output_____"
]
],
[
[
"# TODO: Create layer_0 matrix with dimensions 1 by vocab_size, initially filled with zeros\nlayer_0 = np.zeros((1, vocab_size))",
"_____no_output_____"
]
],
[
[
"Run the following cell. It should display `(1, 74074)`",
"_____no_output_____"
]
],
[
[
"layer_0.shape",
"_____no_output_____"
],
[
"from IPython.display import Image\nImage(filename='sentiment_network.png')",
"_____no_output_____"
]
],
[
[
"`layer_0` contains one entry for every word in the vocabulary, as shown in the above image. We need to make sure we know the index of each word, so run the following cell to create a lookup table that stores the index of every word.",
"_____no_output_____"
]
],
[
[
"# Create a dictionary of words in the vocabulary mapped to index positions\n# (to be used in layer_0)\nword2index = {}\nfor i, word in enumerate(vocab):\n word2index[word] = i\n \n# display the map of words to indices\nword2index",
"_____no_output_____"
]
],
[
[
"**TODO:** Complete the implementation of `update_input_layer`. It should count \n how many times each word is used in the given review, and then store\n those counts at the appropriate indices inside `layer_0`.",
"_____no_output_____"
]
],
[
[
"def update_input_layer(review):\n \"\"\" Modify the global layer_0 to represent the vector form of review.\n The element at a given index of layer_0 should represent\n how many times the given word occurs in the review.\n Args:\n review(string) - the string of the review\n Returns:\n None\n \"\"\"\n global layer_0\n # clear out previous state by resetting the layer to be all 0s\n layer_0 *= 0\n \n # TODO: count how many times each word is used in the given review and store the results in layer_0 \n# print(total_counts.most_common())\n for word in review.split(\" \"):\n layer_0[0][word2index[word]] += 1",
"_____no_output_____"
]
],
[
[
"Run the following cell to test updating the input layer with the first review. The indices assigned may not be the same as in the solution, but hopefully you'll see some non-zero values in `layer_0`. ",
"_____no_output_____"
]
],
[
[
"update_input_layer(reviews[0])\nlayer_0",
"_____no_output_____"
]
],
[
[
"**TODO:** Complete the implementation of `get_target_for_labels`. It should return `0` or `1`, \n depending on whether the given label is `NEGATIVE` or `POSITIVE`, respectively.",
"_____no_output_____"
]
],
[
[
"def get_target_for_label(label):\n \"\"\"Convert a label to `0` or `1`.\n Args:\n label(string) - Either \"POSITIVE\" or \"NEGATIVE\".\n Returns:\n `0` or `1`.\n \"\"\"\n if label == 'POSITIVE':\n return 1\n \n return 0\n # TODO: Your code here",
"_____no_output_____"
]
],
[
[
"Run the following two cells. They should print out`'POSITIVE'` and `1`, respectively.",
"_____no_output_____"
]
],
[
[
"labels[0]",
"_____no_output_____"
],
[
"get_target_for_label(labels[0])",
"_____no_output_____"
]
],
[
[
"Run the following two cells. They should print out `'NEGATIVE'` and `0`, respectively.",
"_____no_output_____"
]
],
[
[
"labels[1]",
"_____no_output_____"
],
[
"get_target_for_label(labels[1])",
"_____no_output_____"
]
],
[
[
"# End of Project 2. \n## Watch the next video to see Andrew's solution, then continue on to the next lesson.",
"_____no_output_____"
],
[
"# Project 3: Building a Neural Network<a id='project_3'></a>",
"_____no_output_____"
],
[
"**TODO:** We've included the framework of a class called `SentimentNetork`. Implement all of the items marked `TODO` in the code. These include doing the following:\n- Create a basic neural network much like the networks you've seen in earlier lessons and in Project 1, with an input layer, a hidden layer, and an output layer. \n- Do **not** add a non-linearity in the hidden layer. That is, do not use an activation function when calculating the hidden layer outputs.\n- Re-use the code from earlier in this notebook to create the training data (see `TODO`s in the code)\n- Implement the `pre_process_data` function to create the vocabulary for our training data generating functions\n- Ensure `train` trains over the entire corpus",
"_____no_output_____"
],
[
"### Where to Get Help if You Need it\n- Re-watch earlier Udacity lectures\n- Chapters 3-5 - [Grokking Deep Learning](https://www.manning.com/books/grokking-deep-learning) - (Check inside your classroom for a discount code)",
"_____no_output_____"
]
],
[
[
"import time\nimport sys\nimport numpy as np\n\n# Encapsulate our neural network in a class\nclass SentimentNetwork:\n def __init__(self, reviews, labels, hidden_nodes = 10, learning_rate = 0.001):\n \"\"\"Create a SentimenNetwork with the given settings\n Args:\n reviews(list) - List of reviews used for training\n labels(list) - List of POSITIVE/NEGATIVE labels associated with the given reviews\n hidden_nodes(int) - Number of nodes to create in the hidden layer\n learning_rate(float) - Learning rate to use while training\n \n \"\"\"\n # Assign a seed to our random number generator to ensure we get\n # reproducable results during development \n np.random.seed(1)\n\n # process the reviews and their associated labels so that everything\n # is ready for training\n self.pre_process_data(reviews, labels)\n \n # Build the network to have the number of hidden nodes and the learning rate that\n # were passed into this initializer. Make the same number of input nodes as\n # there are vocabulary words and create a single output node.\n self.init_network(len(self.review_vocab), hidden_nodes, 1, learning_rate)\n\n def pre_process_data(self, reviews, labels):\n # populate review_vocab with all of the words in the given reviews\n review_vocab = set()\n for review in reviews:\n for word in review.split(\" \"):\n review_vocab.add(word)\n\n # Convert the vocabulary set to a list so we can access words via indices\n self.review_vocab = list(review_vocab)\n \n # populate label_vocab with all of the words in the given labels.\n label_vocab = set()\n for label in labels:\n label_vocab.add(label)\n \n # Convert the label vocabulary set to a list so we can access labels via indices\n self.label_vocab = list(label_vocab)\n \n # Store the sizes of the review and label vocabularies.\n self.review_vocab_size = len(self.review_vocab)\n self.label_vocab_size = len(self.label_vocab)\n \n # Create a dictionary of words in the vocabulary mapped to index positions\n self.word2index = {}\n for i, word in enumerate(self.review_vocab):\n self.word2index[word] = i\n \n # Create a dictionary of labels mapped to index positions\n self.label2index = {}\n for i, label in enumerate(self.label_vocab):\n self.label2index[label] = i\n \n def init_network(self, input_nodes, hidden_nodes, output_nodes, learning_rate):\n # Store the number of nodes in input, hidden, and output layers.\n self.input_nodes = input_nodes\n self.hidden_nodes = hidden_nodes\n self.output_nodes = output_nodes\n\n # Store the learning rate\n self.learning_rate = learning_rate\n\n # Initialize weights\n \n self.weights_0_1 = np.zeros((self.input_nodes,self.hidden_nodes))\n \n # These are the weights between the hidden layer and the output layer.\n self.weights_1_2 = np.random.normal(0.0, self.output_nodes**-0.5, \n (self.hidden_nodes, self.output_nodes))\n \n # The input layer, a two-dimensional matrix with shape 1 x input_nodes\n self.layer_0 = np.zeros((1,input_nodes))\n \n \n def update_input_layer(self,review):\n\n # clear out previous state, reset the layer to be all 0s\n self.layer_0 *= 0\n \n for word in review.split(\" \"):\n # NOTE: This if-check was not in the version of this method created in Project 2,\n # and it appears in Andrew's Project 3 solution without explanation. \n # It simply ensures the word is actually a key in word2index before\n # accessing it, which is important because accessing an invalid key\n # with raise an exception in Python. This allows us to ignore unknown\n # words encountered in new reviews.\n if(word in self.word2index.keys()):\n self.layer_0[0][self.word2index[word]] += 1\n \n def get_target_for_label(self,label):\n if(label == 'POSITIVE'):\n return 1\n else:\n return 0\n \n def sigmoid(self,x):\n return 1 / (1 + np.exp(-x))\n \n def sigmoid_output_2_derivative(self,output):\n return output * (1 - output)\n\n def train(self, training_reviews, training_labels):\n \n # make sure out we have a matching number of reviews and labels\n assert(len(training_reviews) == len(training_labels))\n \n # Keep track of correct predictions to display accuracy during training \n correct_so_far = 0\n \n # Remember when we started for printing time statistics\n start = time.time()\n\n # loop through all the given reviews and run a forward and backward pass,\n # updating weights for every item\n for i in range(len(training_reviews)):\n review = training_reviews[i]\n label = training_labels[i]\n \n #### Implement the forward pass here ####\n ### Forward pass ###\n\n # Input Layer\n self.update_input_layer(review)\n\n # Hidden layer\n layer_1 = self.layer_0.dot(self.weights_0_1)\n\n # Output layer\n layer_2 = self.sigmoid(layer_1.dot(self.weights_1_2))\n \n layer_2_error = layer_2 - self.get_target_for_label(label) # Output layer error is the difference between desired target and actual output.\n layer_2_delta = layer_2_error * self.sigmoid_output_2_derivative(layer_2)\n\n # Backpropagated error\n layer_1_error = layer_2_delta.dot(self.weights_1_2.T) # errors propagated to the hidden layer\n layer_1_delta = layer_1_error # hidden layer gradients - no nonlinearity so it's the same as the error\n\n # Update the weights\n self.weights_1_2 -= layer_1.T.dot(layer_2_delta) * self.learning_rate # update hidden-to-output weights with gradient descent step\n self.weights_0_1 -= self.layer_0.T.dot(layer_1_delta) * self.learning_rate \n # TODO: Implement the back propagation pass here. \n # That means calculate the error for the forward pass's prediction\n # and update the weights in the network according to their\n # contributions toward the error, as calculated via the\n # gradient descent and back propagation algorithms you \n # learned in class.\n \n # TODO: Keep track of correct predictions. To determine if the prediction was\n # correct, check that the absolute value of the output error \n # is less than 0.5. If so, add one to the correct_so_far count.\n if(layer_2 >= 0.5 and label == 'POSITIVE'):\n correct_so_far += 1\n elif(layer_2 < 0.5 and label == 'NEGATIVE'):\n correct_so_far += 1\n \n # For debug purposes, print out our prediction accuracy and speed \n # throughout the training process. \n\n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(training_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct_so_far) + \" #Trained:\" + str(i+1) \\\n + \" Training Accuracy:\" + str(correct_so_far * 100 / float(i+1))[:4] + \"%\")\n if(i % 2500 == 0):\n print(\"\")\n \n def test(self, testing_reviews, testing_labels):\n \"\"\"\n Attempts to predict the labels for the given testing_reviews,\n and uses the test_labels to calculate the accuracy of those predictions.\n \"\"\"\n \n # keep track of how many correct predictions we make\n correct = 0\n\n # we'll time how many predictions per second we make\n start = time.time()\n\n # Loop through each of the given reviews and call run to predict\n # its label. \n for i in range(len(testing_reviews)):\n pred = self.run(testing_reviews[i])\n# print(layer_2.shape)\n if(pred == testing_labels[i]):\n correct += 1\n \n # For debug purposes, print out our prediction accuracy and speed \n # throughout the prediction process. \n\n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(testing_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct) + \" #Tested:\" + str(i+1) \\\n + \" Testing Accuracy:\" + str(correct * 100 / float(i+1))[:4] + \"%\")\n \n def run(self, review):\n \"\"\"\n Returns a POSITIVE or NEGATIVE prediction for the given review.\n \"\"\"\n # Run a forward pass through the network, like in the \"train\" function.\n \n # Input Layer\n self.update_input_layer(review.lower())\n\n # Hidden layer\n layer_1 = self.layer_0.dot(self.weights_0_1)\n\n # Output layer\n layer_2 = self.sigmoid(layer_1.dot(self.weights_1_2))\n \n # Return POSITIVE for values above greater-than-or-equal-to 0.5 in the output layer;\n # return NEGATIVE for other values\n if(layer_2 >= 0.5):\n return \"POSITIVE\"\n else:\n return \"NEGATIVE\"",
"_____no_output_____"
]
],
[
[
"Run the following cell to create a `SentimentNetwork` that will train on all but the last 1000 reviews (we're saving those for testing). Here we use a learning rate of `0.1`.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000], learning_rate=0.1)",
"_____no_output_____"
]
],
[
[
"Run the following cell to test the network's performance against the last 1000 reviews (the ones we held out from our training set). \n\n**We have not trained the model yet, so the results should be about 50% as it will just be guessing and there are only two possible values to choose from.**",
"_____no_output_____"
]
],
[
[
"mlp.test(reviews[-1000:],labels[-1000:])",
"Progress:48.8% Speed(reviews/sec):800.1 #Correct:245 #Tested:489 Testing Accuracy:50.1%Progress:99.9% Speed(reviews/sec):777.5 #Correct:500 #Tested:1000 Testing Accuracy:50.0%"
]
],
[
[
"Run the following cell to actually train the network. During training, it will display the model's accuracy repeatedly as it trains so you can see how well it's doing.",
"_____no_output_____"
]
],
[
[
"mlp.train(reviews[:-1000],labels[:-1000])",
"Progress:0.0% Speed(reviews/sec):0.0 #Correct:1 #Trained:1 Training Accuracy:100.%\nProgress:10.4% Speed(reviews/sec):246.6 #Correct:1251 #Trained:2501 Training Accuracy:50.0%\nProgress:11.4% Speed(reviews/sec):247.1 #Correct:1369 #Trained:2737 Training Accuracy:50.0%"
]
],
[
[
"That most likely didn't train very well. Part of the reason may be because the learning rate is too high. Run the following cell to recreate the network with a smaller learning rate, `0.01`, and then train the new network.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000], learning_rate=0.01)\nmlp.train(reviews[:-1000],labels[:-1000])",
"Progress:0.0% Speed(reviews/sec):0.0 #Correct:1 #Trained:1 Training Accuracy:100.%\nProgress:9.72% Speed(reviews/sec):239.4 #Correct:1165 #Trained:2334 Training Accuracy:49.9%"
]
],
[
[
"That probably wasn't much different. Run the following cell to recreate the network one more time with an even smaller learning rate, `0.001`, and then train the new network.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000], learning_rate=0.001)\nmlp.train(reviews[:-1000],labels[:-1000])",
"Progress:0.0% Speed(reviews/sec):0.0 #Correct:1 #Trained:1 Training Accuracy:100.%\nProgress:10.4% Speed(reviews/sec):249.0 #Correct:1267 #Trained:2501 Training Accuracy:50.6%\nProgress:20.8% Speed(reviews/sec):248.8 #Correct:2655 #Trained:5001 Training Accuracy:53.0%\nProgress:31.2% Speed(reviews/sec):249.5 #Correct:4087 #Trained:7501 Training Accuracy:54.4%\nProgress:41.6% Speed(reviews/sec):250.9 #Correct:5535 #Trained:10001 Training Accuracy:55.3%\nProgress:49.3% Speed(reviews/sec):248.1 #Correct:6674 #Trained:11835 Training Accuracy:56.3%"
]
],
[
[
"With a learning rate of `0.001`, the network should finall have started to improve during training. It's still not very good, but it shows that this solution has potential. We will improve it in the next lesson.",
"_____no_output_____"
],
[
"# End of Project 3. \n## Watch the next video to see Andrew's solution, then continue on to the next lesson.",
"_____no_output_____"
],
[
"# Understanding Neural Noise<a id='lesson_4'></a>\n\nThe following cells include includes the code Andrew shows in the next video. We've included it here so you can run the cells along with the video without having to type in everything.",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image\nImage(filename='sentiment_network.png')",
"_____no_output_____"
],
[
"def update_input_layer(review):\n \n global layer_0\n \n # clear out previous state, reset the layer to be all 0s\n layer_0 *= 0\n for word in review.split(\" \"):\n layer_0[0][word2index[word]] += 1\n\nupdate_input_layer(reviews[0])",
"_____no_output_____"
],
[
"layer_0",
"_____no_output_____"
],
[
"review_counter = Counter()",
"_____no_output_____"
],
[
"for word in reviews[0].split(\" \"):\n review_counter[word] += 1",
"_____no_output_____"
],
[
"review_counter.most_common()",
"_____no_output_____"
]
],
[
[
"# Project 4: Reducing Noise in Our Input Data<a id='project_4'></a>\n\n**TODO:** Attempt to reduce the noise in the input data like Andrew did in the previous video. Specifically, do the following:\n* Copy the `SentimentNetwork` class you created earlier into the following cell.\n* Modify `update_input_layer` so it does not count how many times each word is used, but rather just stores whether or not a word was used. ",
"_____no_output_____"
]
],
[
[
"# TODO: -Copy the SentimentNetwork class from Projet 3 lesson\n# -Modify it to reduce noise, like in the video \nimport time\nimport sys\nimport numpy as np\n\n# Encapsulate our neural network in a class\nclass SentimentNetwork:\n def __init__(self, reviews, labels, hidden_nodes = 10, learning_rate = 0.001):\n \"\"\"Create a SentimenNetwork with the given settings\n Args:\n reviews(list) - List of reviews used for training\n labels(list) - List of POSITIVE/NEGATIVE labels associated with the given reviews\n hidden_nodes(int) - Number of nodes to create in the hidden layer\n learning_rate(float) - Learning rate to use while training\n \n \"\"\"\n # Assign a seed to our random number generator to ensure we get\n # reproducable results during development \n np.random.seed(1)\n\n # process the reviews and their associated labels so that everything\n # is ready for training\n self.pre_process_data(reviews, labels)\n \n # Build the network to have the number of hidden nodes and the learning rate that\n # were passed into this initializer. Make the same number of input nodes as\n # there are vocabulary words and create a single output node.\n self.init_network(len(self.review_vocab), hidden_nodes, 1, learning_rate)\n\n def pre_process_data(self, reviews, labels):\n # populate review_vocab with all of the words in the given reviews\n review_vocab = set()\n for review in reviews:\n for word in review.split(\" \"):\n review_vocab.add(word)\n\n # Convert the vocabulary set to a list so we can access words via indices\n self.review_vocab = list(review_vocab)\n \n # populate label_vocab with all of the words in the given labels.\n label_vocab = set()\n for label in labels:\n label_vocab.add(label)\n \n # Convert the label vocabulary set to a list so we can access labels via indices\n self.label_vocab = list(label_vocab)\n \n # Store the sizes of the review and label vocabularies.\n self.review_vocab_size = len(self.review_vocab)\n self.label_vocab_size = len(self.label_vocab)\n \n # Create a dictionary of words in the vocabulary mapped to index positions\n self.word2index = {}\n for i, word in enumerate(self.review_vocab):\n self.word2index[word] = i\n \n # Create a dictionary of labels mapped to index positions\n self.label2index = {}\n for i, label in enumerate(self.label_vocab):\n self.label2index[label] = i\n \n def init_network(self, input_nodes, hidden_nodes, output_nodes, learning_rate):\n # Store the number of nodes in input, hidden, and output layers.\n self.input_nodes = input_nodes\n self.hidden_nodes = hidden_nodes\n self.output_nodes = output_nodes\n\n # Store the learning rate\n self.learning_rate = learning_rate\n\n # Initialize weights\n \n self.weights_0_1 = np.zeros((self.input_nodes,self.hidden_nodes))\n \n # These are the weights between the hidden layer and the output layer.\n self.weights_1_2 = np.random.normal(0.0, self.output_nodes**-0.5, \n (self.hidden_nodes, self.output_nodes))\n \n # The input layer, a two-dimensional matrix with shape 1 x input_nodes\n self.layer_0 = np.zeros((1,input_nodes))\n \n \n def update_input_layer(self,review):\n\n # clear out previous state, reset the layer to be all 0s\n self.layer_0 *= 0\n \n for word in review.split(\" \"):\n # NOTE: This if-check was not in the version of this method created in Project 2,\n # and it appears in Andrew's Project 3 solution without explanation. \n # It simply ensures the word is actually a key in word2index before\n # accessing it, which is important because accessing an invalid key\n # with raise an exception in Python. This allows us to ignore unknown\n # words encountered in new reviews.\n if(word in self.word2index.keys()):\n self.layer_0[0][self.word2index[word]] = 1\n \n def get_target_for_label(self,label):\n if(label == 'POSITIVE'):\n return 1\n else:\n return 0\n \n def sigmoid(self,x):\n return 1 / (1 + np.exp(-x))\n \n def sigmoid_output_2_derivative(self,output):\n return output * (1 - output)\n\n def train(self, training_reviews, training_labels):\n \n # make sure out we have a matching number of reviews and labels\n assert(len(training_reviews) == len(training_labels))\n \n # Keep track of correct predictions to display accuracy during training \n correct_so_far = 0\n \n # Remember when we started for printing time statistics\n start = time.time()\n\n # loop through all the given reviews and run a forward and backward pass,\n # updating weights for every item\n for i in range(len(training_reviews)):\n review = training_reviews[i]\n label = training_labels[i]\n \n #### Implement the forward pass here ####\n ### Forward pass ###\n\n # Input Layer\n self.update_input_layer(review)\n\n # Hidden layer\n layer_1 = self.layer_0.dot(self.weights_0_1)\n\n # Output layer\n layer_2 = self.sigmoid(layer_1.dot(self.weights_1_2))\n \n layer_2_error = layer_2 - self.get_target_for_label(label) # Output layer error is the difference between desired target and actual output.\n layer_2_delta = layer_2_error * self.sigmoid_output_2_derivative(layer_2)\n\n # Backpropagated error\n layer_1_error = layer_2_delta.dot(self.weights_1_2.T) # errors propagated to the hidden layer\n layer_1_delta = layer_1_error # hidden layer gradients - no nonlinearity so it's the same as the error\n\n # Update the weights\n self.weights_1_2 -= layer_1.T.dot(layer_2_delta) * self.learning_rate # update hidden-to-output weights with gradient descent step\n self.weights_0_1 -= self.layer_0.T.dot(layer_1_delta) * self.learning_rate \n # TODO: Implement the back propagation pass here. \n # That means calculate the error for the forward pass's prediction\n # and update the weights in the network according to their\n # contributions toward the error, as calculated via the\n # gradient descent and back propagation algorithms you \n # learned in class.\n \n # TODO: Keep track of correct predictions. To determine if the prediction was\n # correct, check that the absolute value of the output error \n # is less than 0.5. If so, add one to the correct_so_far count.\n if(layer_2 >= 0.5 and label == 'POSITIVE'):\n correct_so_far += 1\n elif(layer_2 < 0.5 and label == 'NEGATIVE'):\n correct_so_far += 1\n \n # For debug purposes, print out our prediction accuracy and speed \n # throughout the training process. \n\n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(training_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct_so_far) + \" #Trained:\" + str(i+1) \\\n + \" Training Accuracy:\" + str(correct_so_far * 100 / float(i+1))[:4] + \"%\")\n if(i % 2500 == 0):\n print(\"\")\n \n def test(self, testing_reviews, testing_labels):\n \"\"\"\n Attempts to predict the labels for the given testing_reviews,\n and uses the test_labels to calculate the accuracy of those predictions.\n \"\"\"\n \n # keep track of how many correct predictions we make\n correct = 0\n\n # we'll time how many predictions per second we make\n start = time.time()\n\n # Loop through each of the given reviews and call run to predict\n # its label. \n for i in range(len(testing_reviews)):\n pred = self.run(testing_reviews[i])\n# print(layer_2.shape)\n if(pred == testing_labels[i]):\n correct += 1\n \n # For debug purposes, print out our prediction accuracy and speed \n # throughout the prediction process. \n\n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(testing_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct) + \" #Tested:\" + str(i+1) \\\n + \" Testing Accuracy:\" + str(correct * 100 / float(i+1))[:4] + \"%\")\n \n def run(self, review):\n \"\"\"\n Returns a POSITIVE or NEGATIVE prediction for the given review.\n \"\"\"\n # Run a forward pass through the network, like in the \"train\" function.\n \n # Input Layer\n self.update_input_layer(review.lower())\n\n # Hidden layer\n layer_1 = self.layer_0.dot(self.weights_0_1)\n\n # Output layer\n layer_2 = self.sigmoid(layer_1.dot(self.weights_1_2))\n \n # Return POSITIVE for values above greater-than-or-equal-to 0.5 in the output layer;\n # return NEGATIVE for other values\n if(layer_2 >= 0.5):\n return \"POSITIVE\"\n else:\n return \"NEGATIVE\"",
"_____no_output_____"
]
],
[
[
"Run the following cell to recreate the network and train it. Notice we've gone back to the higher learning rate of `0.1`.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000], learning_rate=0.1)\nmlp.train(reviews[:-1000],labels[:-1000])",
"Progress:0.0% Speed(reviews/sec):0.0 #Correct:1 #Trained:1 Training Accuracy:100.%\nProgress:10.4% Speed(reviews/sec):83.64 #Correct:1838 #Trained:2501 Training Accuracy:73.4%\nProgress:20.8% Speed(reviews/sec):83.27 #Correct:3820 #Trained:5001 Training Accuracy:76.3%\nProgress:31.2% Speed(reviews/sec):83.09 #Correct:5911 #Trained:7501 Training Accuracy:78.8%\nProgress:41.6% Speed(reviews/sec):82.75 #Correct:8044 #Trained:10001 Training Accuracy:80.4%\nProgress:52.0% Speed(reviews/sec):82.98 #Correct:10188 #Trained:12501 Training Accuracy:81.4%\nProgress:62.5% Speed(reviews/sec):83.07 #Correct:12317 #Trained:15001 Training Accuracy:82.1%\nProgress:72.9% Speed(reviews/sec):83.04 #Correct:14438 #Trained:17501 Training Accuracy:82.4%\nProgress:83.3% Speed(reviews/sec):83.06 #Correct:16609 #Trained:20001 Training Accuracy:83.0%\nProgress:93.7% Speed(reviews/sec):82.96 #Correct:18788 #Trained:22501 Training Accuracy:83.4%\nProgress:99.9% Speed(reviews/sec):82.92 #Correct:20105 #Trained:24000 Training Accuracy:83.7%"
]
],
[
[
"That should have trained much better than the earlier attempts. It's still not wonderful, but it should have improved dramatically. Run the following cell to test your model with 1000 predictions.",
"_____no_output_____"
]
],
[
[
"mlp.test(reviews[-1000:],labels[-1000:])",
"Progress:99.9% Speed(reviews/sec):942.5 #Correct:849 #Tested:1000 Testing Accuracy:84.9%"
]
],
[
[
"# End of Project 4. \n## Andrew's solution was actually in the previous video, so rewatch that video if you had any problems with that project. Then continue on to the next lesson.\n# Analyzing Inefficiencies in our Network<a id='lesson_5'></a>\nThe following cells include the code Andrew shows in the next video. We've included it here so you can run the cells along with the video without having to type in everything.",
"_____no_output_____"
]
],
[
[
"Image(filename='sentiment_network_sparse.png')",
"_____no_output_____"
],
[
"layer_0 = np.zeros(10)",
"_____no_output_____"
],
[
"layer_0",
"_____no_output_____"
],
[
"layer_0[4] = 1\nlayer_0[9] = 1",
"_____no_output_____"
],
[
"layer_0",
"_____no_output_____"
],
[
"weights_0_1 = np.random.randn(10,5)",
"_____no_output_____"
],
[
"layer_0.dot(weights_0_1)",
"_____no_output_____"
],
[
"indices = [4,9]",
"_____no_output_____"
],
[
"layer_1 = np.zeros(5)",
"_____no_output_____"
],
[
"for index in indices:\n layer_1 += (1 * weights_0_1[index])",
"_____no_output_____"
],
[
"layer_1",
"_____no_output_____"
],
[
"Image(filename='sentiment_network_sparse_2.png')",
"_____no_output_____"
],
[
"layer_1 = np.zeros(5)",
"_____no_output_____"
],
[
"for index in indices:\n layer_1 += (weights_0_1[index])",
"_____no_output_____"
],
[
"layer_1",
"_____no_output_____"
]
],
[
[
"# Project 5: Making our Network More Efficient<a id='project_5'></a>\n**TODO:** Make the `SentimentNetwork` class more efficient by eliminating unnecessary multiplications and additions that occur during forward and backward propagation. To do that, you can do the following:\n* Copy the `SentimentNetwork` class from the previous project into the following cell.\n* Remove the `update_input_layer` function - you will not need it in this version.\n* Modify `init_network`:\n>* You no longer need a separate input layer, so remove any mention of `self.layer_0`\n>* You will be dealing with the old hidden layer more directly, so create `self.layer_1`, a two-dimensional matrix with shape 1 x hidden_nodes, with all values initialized to zero\n* Modify `train`:\n>* Change the name of the input parameter `training_reviews` to `training_reviews_raw`. This will help with the next step.\n>* At the beginning of the function, you'll want to preprocess your reviews to convert them to a list of indices (from `word2index`) that are actually used in the review. This is equivalent to what you saw in the video when Andrew set specific indices to 1. Your code should create a local `list` variable named `training_reviews` that should contain a `list` for each review in `training_reviews_raw`. Those lists should contain the indices for words found in the review.\n>* Remove call to `update_input_layer`\n>* Use `self`'s `layer_1` instead of a local `layer_1` object.\n>* In the forward pass, replace the code that updates `layer_1` with new logic that only adds the weights for the indices used in the review.\n>* When updating `weights_0_1`, only update the individual weights that were used in the forward pass.\n* Modify `run`:\n>* Remove call to `update_input_layer` \n>* Use `self`'s `layer_1` instead of a local `layer_1` object.\n>* Much like you did in `train`, you will need to pre-process the `review` so you can work with word indices, then update `layer_1` by adding weights for the indices used in the review.",
"_____no_output_____"
]
],
[
[
"import time\nimport sys\nimport numpy as np\n\n# Encapsulate our neural network in a class\nclass SentimentNetwork:\n def __init__(self, reviews,labels,hidden_nodes = 10, learning_rate = 0.1):\n \"\"\"Create a SentimenNetwork with the given settings\n Args:\n reviews(list) - List of reviews used for training\n labels(list) - List of POSITIVE/NEGATIVE labels associated with the given reviews\n hidden_nodes(int) - Number of nodes to create in the hidden layer\n learning_rate(float) - Learning rate to use while training\n \n \"\"\"\n # Assign a seed to our random number generator to ensure we get\n # reproducable results during development \n np.random.seed(1)\n\n # process the reviews and their associated labels so that everything\n # is ready for training\n self.pre_process_data(reviews, labels)\n \n # Build the network to have the number of hidden nodes and the learning rate that\n # were passed into this initializer. Make the same number of input nodes as\n # there are vocabulary words and create a single output node.\n self.init_network(len(self.review_vocab),hidden_nodes, 1, learning_rate)\n\n def pre_process_data(self, reviews, labels):\n \n # populate review_vocab with all of the words in the given reviews\n review_vocab = set()\n for review in reviews:\n for word in review.split(\" \"):\n review_vocab.add(word)\n\n # Convert the vocabulary set to a list so we can access words via indices\n self.review_vocab = list(review_vocab)\n \n # populate label_vocab with all of the words in the given labels.\n label_vocab = set()\n for label in labels:\n label_vocab.add(label)\n \n # Convert the label vocabulary set to a list so we can access labels via indices\n self.label_vocab = list(label_vocab)\n \n # Store the sizes of the review and label vocabularies.\n self.review_vocab_size = len(self.review_vocab)\n self.label_vocab_size = len(self.label_vocab)\n \n # Create a dictionary of words in the vocabulary mapped to index positions\n self.word2index = {}\n for i, word in enumerate(self.review_vocab):\n self.word2index[word] = i\n \n # Create a dictionary of labels mapped to index positions\n self.label2index = {}\n for i, label in enumerate(self.label_vocab):\n self.label2index[label] = i\n\n def init_network(self, input_nodes, hidden_nodes, output_nodes, learning_rate):\n # Set number of nodes in input, hidden and output layers.\n self.input_nodes = input_nodes\n self.hidden_nodes = hidden_nodes\n self.output_nodes = output_nodes\n\n # Store the learning rate\n self.learning_rate = learning_rate\n\n # Initialize weights\n\n # These are the weights between the input layer and the hidden layer.\n self.weights_0_1 = np.zeros((self.input_nodes,self.hidden_nodes))\n\n # These are the weights between the hidden layer and the output layer.\n self.weights_1_2 = np.random.normal(0.0, self.output_nodes**-0.5, \n (self.hidden_nodes, self.output_nodes))\n \n ## New for Project 5: Removed self.layer_0; added self.layer_1\n # The input layer, a two-dimensional matrix with shape 1 x hidden_nodes\n self.layer_1 = np.zeros((1,hidden_nodes))\n \n ## New for Project 5: Removed update_input_layer function\n \n def get_target_for_label(self,label):\n if(label == 'POSITIVE'):\n return 1\n else:\n return 0\n \n def sigmoid(self,x):\n return 1 / (1 + np.exp(-x))\n \n def sigmoid_output_2_derivative(self,output):\n return output * (1 - output)\n\n def train(self, training_reviews_raw, training_labels):\n\n ## New for Project 5: pre-process training reviews so we can deal \n # directly with the indices of non-zero inputs\n training_reviews = list()\n for review in training_reviews_raw:\n indices = set()\n for word in review.split(\" \"):\n if(word in self.word2index.keys()):\n indices.add(self.word2index[word])\n training_reviews.append(list(indices))\n\n # make sure out we have a matching number of reviews and labels\n assert(len(training_reviews) == len(training_labels))\n \n # Keep track of correct predictions to display accuracy during training \n correct_so_far = 0\n\n # Remember when we started for printing time statistics\n start = time.time()\n \n # loop through all the given reviews and run a forward and backward pass,\n # updating weights for every item\n for i in range(len(training_reviews)):\n \n # Get the next review and its correct label\n review = training_reviews[i]\n label = training_labels[i]\n \n #### Implement the forward pass here ####\n ### Forward pass ###\n\n ## New for Project 5: Removed call to 'update_input_layer' function\n # because 'layer_0' is no longer used\n\n # Hidden layer\n ## New for Project 5: Add in only the weights for non-zero items\n self.layer_1 *= 0\n for index in review:\n self.layer_1 += self.weights_0_1[index]\n\n # Output layer\n ## New for Project 5: changed to use 'self.layer_1' instead of 'local layer_1'\n layer_2 = self.sigmoid(self.layer_1.dot(self.weights_1_2)) \n \n #### Implement the backward pass here ####\n ### Backward pass ###\n\n # Output error\n layer_2_error = layer_2 - self.get_target_for_label(label) # Output layer error is the difference between desired target and actual output.\n layer_2_delta = layer_2_error * self.sigmoid_output_2_derivative(layer_2)\n\n # Backpropagated error\n layer_1_error = layer_2_delta.dot(self.weights_1_2.T) # errors propagated to the hidden layer\n layer_1_delta = layer_1_error # hidden layer gradients - no nonlinearity so it's the same as the error\n\n # Update the weights\n ## New for Project 5: changed to use 'self.layer_1' instead of local 'layer_1'\n self.weights_1_2 -= self.layer_1.T.dot(layer_2_delta) * self.learning_rate # update hidden-to-output weights with gradient descent step\n \n ## New for Project 5: Only update the weights that were used in the forward pass\n for index in review:\n self.weights_0_1[index] -= layer_1_delta[0] * self.learning_rate # update input-to-hidden weights with gradient descent step\n\n # Keep track of correct predictions.\n if(layer_2 >= 0.5 and label == 'POSITIVE'):\n correct_so_far += 1\n elif(layer_2 < 0.5 and label == 'NEGATIVE'):\n correct_so_far += 1\n \n # For debug purposes, print out our prediction accuracy and speed \n # throughout the training process. \n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(training_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct_so_far) + \" #Trained:\" + str(i+1) \\\n + \" Training Accuracy:\" + str(correct_so_far * 100 / float(i+1))[:4] + \"%\")\n if(i % 2500 == 0):\n print(\"\")\n \n def test(self, testing_reviews, testing_labels):\n correct = 0\n\n start = time.time()\n for i in range(len(testing_reviews)):\n pred = self.run(testing_reviews[i])\n if(pred == testing_labels[i]):\n correct += 1\n\n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(testing_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct) + \" #Tested:\" + str(i+1) \\\n + \" Testing Accuracy:\" + str(correct * 100 / float(i+1))[:4] + \"%\")\n \n def run(self, review):\n \"\"\"\n Returns a POSITIVE or NEGATIVE prediction for the given review.\n \"\"\"\n # Run a forward pass through the network, like in the \"train\" function.\n \n ## New for Project 5: Removed call to update_input_layer function\n # because layer_0 is no longer used\n\n # Hidden layer\n ## New for Project 5: Identify the indices used in the review and then add\n # just those weights to layer_1 \n self.layer_1 *= 0\n unique_indices = set()\n for word in review.lower().split(\" \"):\n if word in self.word2index.keys():\n unique_indices.add(self.word2index[word])\n for index in unique_indices:\n self.layer_1 += self.weights_0_1[index]\n \n # Output layer\n ## New for Project 5: changed to use self.layer_1 instead of local layer_1\n layer_2 = self.sigmoid(self.layer_1.dot(self.weights_1_2))\n \n # Return POSITIVE for values above greater-than-or-equal-to 0.5 in the output layer;\n # return NEGATIVE for other values\n if(layer_2[0] >= 0.5):\n return \"POSITIVE\"\n else:\n return \"NEGATIVE\"",
"_____no_output_____"
]
],
[
[
"Run the following cell to recreate the network and train it once again.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000], learning_rate=0.1)\nmlp.train(reviews[:-1000],labels[:-1000])",
"Progress:0.0% Speed(reviews/sec):0.0 #Correct:1 #Trained:1 Training Accuracy:100.%\nProgress:10.4% Speed(reviews/sec):1756. #Correct:1823 #Trained:2501 Training Accuracy:72.8%\nProgress:20.8% Speed(reviews/sec):1710. #Correct:3810 #Trained:5001 Training Accuracy:76.1%\nProgress:31.2% Speed(reviews/sec):1707. #Correct:5884 #Trained:7501 Training Accuracy:78.4%\nProgress:41.6% Speed(reviews/sec):1720. #Correct:8023 #Trained:10001 Training Accuracy:80.2%\nProgress:52.0% Speed(reviews/sec):1715. #Correct:10147 #Trained:12501 Training Accuracy:81.1%\nProgress:62.5% Speed(reviews/sec):1716. #Correct:12277 #Trained:15001 Training Accuracy:81.8%\nProgress:72.9% Speed(reviews/sec):1712. #Correct:14388 #Trained:17501 Training Accuracy:82.2%\nProgress:83.3% Speed(reviews/sec):1709. #Correct:16559 #Trained:20001 Training Accuracy:82.7%\nProgress:93.7% Speed(reviews/sec):1707. #Correct:18745 #Trained:22501 Training Accuracy:83.3%\nProgress:99.9% Speed(reviews/sec):1707. #Correct:20078 #Trained:24000 Training Accuracy:83.6%"
]
],
[
[
"That should have trained much better than the earlier attempts. Run the following cell to test your model with 1000 predictions.",
"_____no_output_____"
]
],
[
[
"mlp.test(reviews[-1000:],labels[-1000:])",
"Progress:99.9% Speed(reviews/sec):1970. #Correct:853 #Tested:1000 Testing Accuracy:85.3%"
]
],
[
[
"# End of Project 5. \n## Watch the next video to see Andrew's solution, then continue on to the next lesson.\n# Further Noise Reduction<a id='lesson_6'></a>",
"_____no_output_____"
]
],
[
[
"Image(filename='sentiment_network_sparse_2.png')",
"_____no_output_____"
],
[
"# words most frequently seen in a review with a \"POSITIVE\" label\npos_neg_ratios.most_common()",
"_____no_output_____"
],
[
"# words most frequently seen in a review with a \"NEGATIVE\" label\nlist(reversed(pos_neg_ratios.most_common()))[0:30]",
"_____no_output_____"
],
[
"from bokeh.models import ColumnDataSource, LabelSet\nfrom bokeh.plotting import figure, show, output_file\nfrom bokeh.io import output_notebook\noutput_notebook()",
"_____no_output_____"
],
[
"hist, edges = np.histogram(list(map(lambda x:x[1],pos_neg_ratios.most_common())), density=True, bins=100, normed=True)\n\np = figure(tools=\"pan,wheel_zoom,reset,save\",\n toolbar_location=\"above\",\n title=\"Word Positive/Negative Affinity Distribution\")\np.quad(top=hist, bottom=0, left=edges[:-1], right=edges[1:], line_color=\"#555555\")\nshow(p)",
"_____no_output_____"
],
[
"frequency_frequency = Counter()\n\nfor word, cnt in total_counts.most_common():\n frequency_frequency[cnt] += 1",
"_____no_output_____"
],
[
"hist, edges = np.histogram(list(map(lambda x:x[1],frequency_frequency.most_common())), density=True, bins=100, normed=True)\n\np = figure(tools=\"pan,wheel_zoom,reset,save\",\n toolbar_location=\"above\",\n title=\"The frequency distribution of the words in our corpus\")\np.quad(top=hist, bottom=0, left=edges[:-1], right=edges[1:], line_color=\"#555555\")\nshow(p)",
"_____no_output_____"
]
],
[
[
"# Project 6: Reducing Noise by Strategically Reducing the Vocabulary<a id='project_6'></a>\n\n**TODO:** Improve `SentimentNetwork`'s performance by reducing more noise in the vocabulary. Specifically, do the following:\n* Copy the `SentimentNetwork` class from the previous project into the following cell.\n* Modify `pre_process_data`:\n>* Add two additional parameters: `min_count` and `polarity_cutoff`\n>* Calculate the positive-to-negative ratios of words used in the reviews. (You can use code you've written elsewhere in the notebook, but we are moving it into the class like we did with other helper code earlier.)\n>* Andrew's solution only calculates a postive-to-negative ratio for words that occur at least 50 times. This keeps the network from attributing too much sentiment to rarer words. You can choose to add this to your solution if you would like. \n>* Change so words are only added to the vocabulary if they occur in the vocabulary more than `min_count` times.\n>* Change so words are only added to the vocabulary if the absolute value of their postive-to-negative ratio is at least `polarity_cutoff`\n* Modify `__init__`:\n>* Add the same two parameters (`min_count` and `polarity_cutoff`) and use them when you call `pre_process_data`",
"_____no_output_____"
]
],
[
[
"import time\nimport sys\nimport numpy as np\nfrom collections import Counter\n\n# Encapsulate our neural network in a class\nclass SentimentNetwork:\n def __init__(self, reviews,labels,hidden_nodes = 10, learning_rate = 0.1, min_count = 10, polarity_cutoff = 0.1):\n \"\"\"Create a SentimenNetwork with the given settings\n Args:\n reviews(list) - List of reviews used for training\n labels(list) - List of POSITIVE/NEGATIVE labels associated with the given reviews\n hidden_nodes(int) - Number of nodes to create in the hidden layer\n learning_rate(float) - Learning rate to use while training\n \n \"\"\"\n # Assign a seed to our random number generator to ensure we get\n # reproducable results during development \n np.random.seed(1)\n\n # process the reviews and their associated labels so that everything\n # is ready for training\n self.pre_process_data(reviews, labels, min_count, polarity_cutoff)\n \n # Build the network to have the number of hidden nodes and the learning rate that\n # were passed into this initializer. Make the same number of input nodes as\n # there are vocabulary words and create a single output node.\n self.init_network(len(self.review_vocab),hidden_nodes, 1, learning_rate)\n\n def pre_process_data(self, reviews, labels, min_count, polarity_cutoff):\n positive_counts = Counter()\n negative_counts = Counter()\n total_counts = Counter()\n\n for i in range(len(reviews)):\n if(labels[i] == 'POSITIVE'):\n for word in reviews[i].split(\" \"):\n positive_counts[word] += 1\n total_counts[word] += 1\n else:\n for word in reviews[i].split(\" \"):\n negative_counts[word] += 1\n total_counts[word] += 1\n \n pos_neg_ratios = Counter()\n \n for term,count in total_counts.most_common():\n if(count > 50):\n pos_neg_ratio = positive_counts[term] / float(negative_counts[term]+1)\n try:\n pos_neg_ratios[term] = np.log(pos_neg_ratio)\n except:\n pass\n \n review_vocab = set()\n for review in reviews:\n for word in review.split(\" \"):\n ## New for Project 6: only add words that occur at least min_count times\n # and for words with pos/neg ratios, only add words\n # that meet the polarity_cutoff\n if(total_counts[word] > min_count):\n if(word in pos_neg_ratios.keys() and np.abs([pos_neg_ratios[word]]) >= polarity_cutoff):\n# if((pos_neg_ratios[word] >= polarity_cutoff) or (pos_neg_ratios[word] <= -polarity_cutoff)):\n review_vocab.add(word)\n else:\n review_vocab.add(word)\n\n # Convert the vocabulary set to a list so we can access words via indices\n self.review_vocab = list(review_vocab)\n \n # populate label_vocab with all of the words in the given labels.\n label_vocab = set()\n for label in labels:\n label_vocab.add(label)\n \n # Convert the label vocabulary set to a list so we can access labels via indices\n self.label_vocab = list(label_vocab)\n \n # Store the sizes of the review and label vocabularies.\n self.review_vocab_size = len(self.review_vocab)\n self.label_vocab_size = len(self.label_vocab)\n \n # Create a dictionary of words in the vocabulary mapped to index positions\n self.word2index = {}\n for i, word in enumerate(self.review_vocab):\n self.word2index[word] = i\n \n # Create a dictionary of labels mapped to index positions\n self.label2index = {}\n for i, label in enumerate(self.label_vocab):\n self.label2index[label] = i\n\n def init_network(self, input_nodes, hidden_nodes, output_nodes, learning_rate):\n # Set number of nodes in input, hidden and output layers.\n self.input_nodes = input_nodes\n self.hidden_nodes = hidden_nodes\n self.output_nodes = output_nodes\n\n # Store the learning rate\n self.learning_rate = learning_rate\n\n # Initialize weights\n\n # These are the weights between the input layer and the hidden layer.\n self.weights_0_1 = np.zeros((self.input_nodes,self.hidden_nodes))\n\n # These are the weights between the hidden layer and the output layer.\n self.weights_1_2 = np.random.normal(0.0, self.output_nodes**-0.5, \n (self.hidden_nodes, self.output_nodes))\n \n ## New for Project 5: Removed self.layer_0; added self.layer_1\n # The input layer, a two-dimensional matrix with shape 1 x hidden_nodes\n self.layer_1 = np.zeros((1,hidden_nodes))\n \n ## New for Project 5: Removed update_input_layer function\n \n def get_target_for_label(self,label):\n if(label == 'POSITIVE'):\n return 1\n else:\n return 0\n \n def sigmoid(self,x):\n return 1 / (1 + np.exp(-x))\n \n def sigmoid_output_2_derivative(self,output):\n return output * (1 - output)\n\n def train(self, training_reviews_raw, training_labels):\n\n ## New for Project 5: pre-process training reviews so we can deal \n # directly with the indices of non-zero inputs\n training_reviews = list()\n for review in training_reviews_raw:\n indices = set()\n for word in review.split(\" \"):\n if(word in self.word2index.keys()):\n indices.add(self.word2index[word])\n training_reviews.append(list(indices))\n\n # make sure out we have a matching number of reviews and labels\n assert(len(training_reviews) == len(training_labels))\n \n # Keep track of correct predictions to display accuracy during training \n correct_so_far = 0\n\n # Remember when we started for printing time statistics\n start = time.time()\n \n # loop through all the given reviews and run a forward and backward pass,\n # updating weights for every item\n for i in range(len(training_reviews)):\n \n # Get the next review and its correct label\n review = training_reviews[i]\n label = training_labels[i]\n \n #### Implement the forward pass here ####\n ### Forward pass ###\n\n ## New for Project 5: Removed call to 'update_input_layer' function\n # because 'layer_0' is no longer used\n\n # Hidden layer\n ## New for Project 5: Add in only the weights for non-zero items\n self.layer_1 *= 0\n for index in review:\n self.layer_1 += self.weights_0_1[index]\n\n # Output layer\n ## New for Project 5: changed to use 'self.layer_1' instead of 'local layer_1'\n layer_2 = self.sigmoid(self.layer_1.dot(self.weights_1_2)) \n \n #### Implement the backward pass here ####\n ### Backward pass ###\n\n # Output error\n layer_2_error = layer_2 - self.get_target_for_label(label) # Output layer error is the difference between desired target and actual output.\n layer_2_delta = layer_2_error * self.sigmoid_output_2_derivative(layer_2)\n\n # Backpropagated error\n layer_1_error = layer_2_delta.dot(self.weights_1_2.T) # errors propagated to the hidden layer\n layer_1_delta = layer_1_error # hidden layer gradients - no nonlinearity so it's the same as the error\n\n # Update the weights\n ## New for Project 5: changed to use 'self.layer_1' instead of local 'layer_1'\n self.weights_1_2 -= self.layer_1.T.dot(layer_2_delta) * self.learning_rate # update hidden-to-output weights with gradient descent step\n \n ## New for Project 5: Only update the weights that were used in the forward pass\n for index in review:\n self.weights_0_1[index] -= layer_1_delta[0] * self.learning_rate # update input-to-hidden weights with gradient descent step\n\n # Keep track of correct predictions.\n if(layer_2 >= 0.5 and label == 'POSITIVE'):\n correct_so_far += 1\n elif(layer_2 < 0.5 and label == 'NEGATIVE'):\n correct_so_far += 1\n \n # For debug purposes, print out our prediction accuracy and speed \n # throughout the training process. \n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(training_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct_so_far) + \" #Trained:\" + str(i+1) \\\n + \" Training Accuracy:\" + str(correct_so_far * 100 / float(i+1))[:4] + \"%\")\n if(i % 2500 == 0):\n print(\"\")\n \n def test(self, testing_reviews, testing_labels):\n correct = 0\n\n start = time.time()\n for i in range(len(testing_reviews)):\n pred = self.run(testing_reviews[i])\n if(pred == testing_labels[i]):\n correct += 1\n\n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(testing_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct) + \" #Tested:\" + str(i+1) \\\n + \" Testing Accuracy:\" + str(correct * 100 / float(i+1))[:4] + \"%\")\n \n def run(self, review):\n self.layer_1 *= 0\n unique_indices = set()\n for word in review.lower().split(\" \"):\n if word in self.word2index.keys():\n unique_indices.add(self.word2index[word])\n for index in unique_indices:\n self.layer_1 += self.weights_0_1[index]\n \n # Output layer\n ## New for Project 5: changed to use self.layer_1 instead of local layer_1\n layer_2 = self.sigmoid(self.layer_1.dot(self.weights_1_2))\n \n # Return POSITIVE for values above greater-than-or-equal-to 0.5 in the output layer;\n # return NEGATIVE for other values\n if(layer_2[0] >= 0.5):\n return \"POSITIVE\"\n else:\n return \"NEGATIVE\"",
"_____no_output_____"
]
],
[
[
"Run the following cell to train your network with a small polarity cutoff.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000],min_count=20,polarity_cutoff=0.05,learning_rate=0.01)\nmlp.train(reviews[:-1000],labels[:-1000])",
"/opt/conda/lib/python3.6/site-packages/ipykernel_launcher.py:51: RuntimeWarning: divide by zero encountered in log\n"
]
],
[
[
"And run the following cell to test it's performance. It should be ",
"_____no_output_____"
]
],
[
[
"mlp.test(reviews[-1000:],labels[-1000:])",
"_____no_output_____"
]
],
[
[
"Run the following cell to train your network with a much larger polarity cutoff.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000],min_count=20,polarity_cutoff=0.8,learning_rate=0.01)\nmlp.train(reviews[:-1000],labels[:-1000])",
"_____no_output_____"
]
],
[
[
"And run the following cell to test it's performance.",
"_____no_output_____"
]
],
[
[
"mlp.test(reviews[-1000:],labels[-1000:])",
"_____no_output_____"
]
],
[
[
"# End of Project 6. \n## Watch the next video to see Andrew's solution, then continue on to the next lesson.",
"_____no_output_____"
],
[
"# Analysis: What's Going on in the Weights?<a id='lesson_7'></a>",
"_____no_output_____"
]
],
[
[
"mlp_full = SentimentNetwork(reviews[:-1000],labels[:-1000],min_count=0,polarity_cutoff=0,learning_rate=0.01)",
"_____no_output_____"
],
[
"mlp_full.train(reviews[:-1000],labels[:-1000])",
"_____no_output_____"
],
[
"Image(filename='sentiment_network_sparse.png')",
"_____no_output_____"
],
[
"def get_most_similar_words(focus = \"horrible\"):\n most_similar = Counter()\n\n for word in mlp_full.word2index.keys():\n most_similar[word] = np.dot(mlp_full.weights_0_1[mlp_full.word2index[word]],mlp_full.weights_0_1[mlp_full.word2index[focus]])\n \n return most_similar.most_common()",
"_____no_output_____"
],
[
"get_most_similar_words(\"excellent\")",
"_____no_output_____"
],
[
"get_most_similar_words(\"terrible\")",
"_____no_output_____"
],
[
"import matplotlib.colors as colors\n\nwords_to_visualize = list()\nfor word, ratio in pos_neg_ratios.most_common(500):\n if(word in mlp_full.word2index.keys()):\n words_to_visualize.append(word)\n \nfor word, ratio in list(reversed(pos_neg_ratios.most_common()))[0:500]:\n if(word in mlp_full.word2index.keys()):\n words_to_visualize.append(word)",
"_____no_output_____"
],
[
"pos = 0\nneg = 0\n\ncolors_list = list()\nvectors_list = list()\nfor word in words_to_visualize:\n if word in pos_neg_ratios.keys():\n vectors_list.append(mlp_full.weights_0_1[mlp_full.word2index[word]])\n if(pos_neg_ratios[word] > 0):\n pos+=1\n colors_list.append(\"#00ff00\")\n else:\n neg+=1\n colors_list.append(\"#000000\")",
"_____no_output_____"
],
[
"from sklearn.manifold import TSNE\ntsne = TSNE(n_components=2, random_state=0)\nwords_top_ted_tsne = tsne.fit_transform(vectors_list)",
"_____no_output_____"
],
[
"p = figure(tools=\"pan,wheel_zoom,reset,save\",\n toolbar_location=\"above\",\n title=\"vector T-SNE for most polarized words\")\n\nsource = ColumnDataSource(data=dict(x1=words_top_ted_tsne[:,0],\n x2=words_top_ted_tsne[:,1],\n names=words_to_visualize,\n color=colors_list))\n\np.scatter(x=\"x1\", y=\"x2\", size=8, source=source, fill_color=\"color\")\n\nword_labels = LabelSet(x=\"x1\", y=\"x2\", text=\"names\", y_offset=6,\n text_font_size=\"8pt\", text_color=\"#555555\",\n source=source, text_align='center')\np.add_layout(word_labels)\n\nshow(p)\n\n# green indicates positive words, black indicates negative words",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e7952523f41380e0826a45d132a73faf1d9c445a | 843,590 | ipynb | Jupyter Notebook | examples/02_advanced_usage.ipynb | gavehan/openTSNE | 96f177169519f5fae7db840bcb28dc54dbe49676 | [
"BSD-3-Clause"
]
| 622 | 2018-12-18T15:04:06.000Z | 2022-03-27T09:37:42.000Z | examples/02_advanced_usage.ipynb | gavehan/openTSNE | 96f177169519f5fae7db840bcb28dc54dbe49676 | [
"BSD-3-Clause"
]
| 115 | 2018-12-17T14:59:19.000Z | 2022-03-17T21:40:33.000Z | examples/02_advanced_usage.ipynb | gavehan/openTSNE | 96f177169519f5fae7db840bcb28dc54dbe49676 | [
"BSD-3-Clause"
]
| 101 | 2019-01-11T02:35:11.000Z | 2022-03-30T11:22:39.000Z | 1,761.148225 | 201,148 | 0.961469 | [
[
[
"# Advanced usage\n\nThis notebook replicates what was done in the *simple_usage* notebooks, but this time with the advanced API. The advanced API is required if we want to use non-standard affinity methods that better preserve global structure.\n\nIf you are comfortable with the advanced API, please refer to the *preserving_global_structure* notebook for a guide how obtain better embeddings and preserve more global structure.",
"_____no_output_____"
]
],
[
[
"from openTSNE import TSNEEmbedding\nfrom openTSNE import affinity\nfrom openTSNE import initialization\n\nfrom examples import utils\n\nimport numpy as np\nfrom sklearn.model_selection import train_test_split\n\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"## Load data",
"_____no_output_____"
]
],
[
[
"import gzip\nimport pickle\n\nwith gzip.open(\"data/macosko_2015.pkl.gz\", \"rb\") as f:\n data = pickle.load(f)\n\nx = data[\"pca_50\"]\ny = data[\"CellType1\"].astype(str)",
"_____no_output_____"
],
[
"print(\"Data set contains %d samples with %d features\" % x.shape)",
"Data set contains 44808 samples with 50 features\n"
]
],
[
[
"## Create train/test split",
"_____no_output_____"
]
],
[
[
"x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=.33, random_state=42)",
"_____no_output_____"
],
[
"print(\"%d training samples\" % x_train.shape[0])\nprint(\"%d test samples\" % x_test.shape[0])",
"30021 training samples\n14787 test samples\n"
]
],
[
[
"## Create a t-SNE embedding\n\nLike in the *simple_usage* notebook, we will run the standard t-SNE optimization.\n\nThis example shows the standard t-SNE optimization. Much can be done in order to better preserve global structure and improve embedding quality. Please refer to the *preserving_global_structure* notebook for some examples.",
"_____no_output_____"
],
[
"**1. Compute the affinities between data points**",
"_____no_output_____"
]
],
[
[
"%%time\naffinities_train = affinity.PerplexityBasedNN(\n x_train,\n perplexity=30,\n metric=\"euclidean\",\n n_jobs=8,\n random_state=42,\n verbose=True,\n)",
"===> Finding 90 nearest neighbors using Annoy approximate search using euclidean distance...\n --> Time elapsed: 3.78 seconds\n===> Calculating affinity matrix...\n --> Time elapsed: 0.43 seconds\nCPU times: user 19.3 s, sys: 794 ms, total: 20.1 s\nWall time: 4.22 s\n"
]
],
[
[
"**2. Generate initial coordinates for our embedding**",
"_____no_output_____"
]
],
[
[
"%time init_train = initialization.pca(x_train, random_state=42)",
"CPU times: user 448 ms, sys: 88.3 ms, total: 536 ms\nWall time: 86.9 ms\n"
]
],
[
[
"**3. Construct the `TSNEEmbedding` object**",
"_____no_output_____"
]
],
[
[
"embedding_train = TSNEEmbedding(\n init_train,\n affinities_train,\n negative_gradient_method=\"fft\",\n n_jobs=8,\n verbose=True,\n)",
"_____no_output_____"
]
],
[
[
"**4. Optimize embedding**",
"_____no_output_____"
],
[
"1. Early exaggeration phase",
"_____no_output_____"
]
],
[
[
"%time embedding_train_1 = embedding_train.optimize(n_iter=250, exaggeration=12, momentum=0.5)",
"===> Running optimization with exaggeration=12.00, lr=2501.75 for 250 iterations...\nIteration 50, KL divergence 5.8046, 50 iterations in 1.8747 sec\nIteration 100, KL divergence 5.2268, 50 iterations in 2.0279 sec\nIteration 150, KL divergence 5.1357, 50 iterations in 1.9912 sec\nIteration 200, KL divergence 5.0977, 50 iterations in 1.9626 sec\nIteration 250, KL divergence 5.0772, 50 iterations in 1.9759 sec\n --> Time elapsed: 9.83 seconds\nCPU times: user 1min 11s, sys: 2.04 s, total: 1min 13s\nWall time: 9.89 s\n"
],
[
"utils.plot(embedding_train_1, y_train, colors=utils.MACOSKO_COLORS)",
"_____no_output_____"
]
],
[
[
"2. Regular optimization",
"_____no_output_____"
]
],
[
[
"%time embedding_train_2 = embedding_train_1.optimize(n_iter=500, momentum=0.8)",
"===> Running optimization with exaggeration=1.00, lr=2501.75 for 500 iterations...\nIteration 50, KL divergence 3.5741, 50 iterations in 1.9240 sec\nIteration 100, KL divergence 3.1653, 50 iterations in 1.9942 sec\nIteration 150, KL divergence 2.9612, 50 iterations in 2.3730 sec\nIteration 200, KL divergence 2.8342, 50 iterations in 3.4895 sec\nIteration 250, KL divergence 2.7496, 50 iterations in 4.7873 sec\nIteration 300, KL divergence 2.6901, 50 iterations in 5.2739 sec\nIteration 350, KL divergence 2.6471, 50 iterations in 6.9968 sec\nIteration 400, KL divergence 2.6138, 50 iterations in 7.8137 sec\nIteration 450, KL divergence 2.5893, 50 iterations in 9.5210 sec\nIteration 500, KL divergence 2.5699, 50 iterations in 10.6958 sec\n --> Time elapsed: 54.87 seconds\nCPU times: user 6min 2s, sys: 20.3 s, total: 6min 23s\nWall time: 55.1 s\n"
],
[
"utils.plot(embedding_train_2, y_train, colors=utils.MACOSKO_COLORS)",
"_____no_output_____"
]
],
[
[
"## Transform",
"_____no_output_____"
]
],
[
[
"%%time\nembedding_test = embedding_train_2.prepare_partial(\n x_test,\n initialization=\"median\",\n k=25,\n perplexity=5,\n)",
"===> Finding 15 nearest neighbors in existing embedding using Annoy approximate search...\n --> Time elapsed: 1.11 seconds\n===> Calculating affinity matrix...\n --> Time elapsed: 0.03 seconds\nCPU times: user 3 s, sys: 192 ms, total: 3.19 s\nWall time: 1.15 s\n"
],
[
"utils.plot(embedding_test, y_test, colors=utils.MACOSKO_COLORS)",
"_____no_output_____"
],
[
"%time embedding_test_1 = embedding_test.optimize(n_iter=250, learning_rate=0.1, momentum=0.8)",
"===> Running optimization with exaggeration=1.00, lr=0.10 for 250 iterations...\nIteration 50, KL divergence 226760.6820, 50 iterations in 0.3498 sec\nIteration 100, KL divergence 221529.7066, 50 iterations in 0.4099 sec\nIteration 150, KL divergence 215464.6854, 50 iterations in 0.4285 sec\nIteration 200, KL divergence 211201.7247, 50 iterations in 0.4060 sec\nIteration 250, KL divergence 209022.1241, 50 iterations in 0.4211 sec\n --> Time elapsed: 2.02 seconds\nCPU times: user 10.7 s, sys: 889 ms, total: 11.6 s\nWall time: 2.74 s\n"
],
[
"utils.plot(embedding_test_1, y_test, colors=utils.MACOSKO_COLORS)",
"_____no_output_____"
]
],
[
[
"## Together\n\nWe superimpose the transformed points onto the original embedding with larger opacity.",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots(figsize=(8, 8))\nutils.plot(embedding_train_2, y_train, colors=utils.MACOSKO_COLORS, alpha=0.25, ax=ax)\nutils.plot(embedding_test_1, y_test, colors=utils.MACOSKO_COLORS, alpha=0.75, ax=ax)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
]
|
e79553d96e0de340321c29fb6efa2fc051496612 | 50,004 | ipynb | Jupyter Notebook | week05_transformer_pos_tagging/week05_bilstm_for_pos_tagging.ipynb | JustM57/natural-language-processing | 45cf112c741343736e34529c7fd985de921deef2 | [
"MIT"
]
| 8 | 2021-12-06T14:48:00.000Z | 2022-03-31T19:05:46.000Z | week05_transformer_pos_tagging/week05_bilstm_for_pos_tagging.ipynb | JustM57/natural-language-processing | 45cf112c741343736e34529c7fd985de921deef2 | [
"MIT"
]
| 1 | 2022-03-05T15:27:37.000Z | 2022-03-05T15:27:37.000Z | week05_transformer_pos_tagging/week05_bilstm_for_pos_tagging.ipynb | JustM57/natural-language-processing | 45cf112c741343736e34529c7fd985de921deef2 | [
"MIT"
]
| 4 | 2021-11-24T19:43:56.000Z | 2022-03-31T22:17:47.000Z | 32.597132 | 441 | 0.555736 | [
[
[
"## Practice: BiLSTM for PoS Tagging\n*This notebook is based on [open-source implementation](https://github.com/bentrevett/pytorch-pos-tagging) of PoS Tagging in PyTorch.*\n\n### Introduction\n\nIn this series we'll be building a machine learning model that produces an output for every element in an input sequence, using PyTorch and TorchText. Specifically, we will be inputting a sequence of text and the model will output a part-of-speech (PoS) tag for each token in the input text. This can also be used for named entity recognition (NER), where the output for each token will be what type of entity, if any, the token is.\n\nIn this notebook, we'll be implementing a multi-layer bi-directional LSTM (BiLSTM) to predict PoS tags using the Universal Dependencies English Web Treebank (UDPOS) dataset.\n\n### Preparing Data\n\nFirst, let's import the necessary Python modules.",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn as nn\nimport torch.optim as optim\n\nfrom torchtext.legacy import data\nfrom torchtext.legacy import datasets\n\nimport spacy\nimport numpy as np\n\nimport time\nimport random",
"_____no_output_____"
]
],
[
[
"Next, we'll set the random seeds for reproducability.",
"_____no_output_____"
]
],
[
[
"SEED = 1234\n\nrandom.seed(SEED)\nnp.random.seed(SEED)\ntorch.manual_seed(SEED)\ntorch.backends.cudnn.deterministic = True",
"_____no_output_____"
]
],
[
[
"One of the key parts of TorchText is the `Field`. The `Field` handles how your dataset is processed.\n\nOur `TEXT` field handles how the text that we need to tag is dealt with. All we do here is set `lower = True` which lowercases all of the text.\n\nNext we'll define the `Fields` for the tags. This dataset actually has two different sets of tags, [universal dependency (UD) tags](https://universaldependencies.org/u/pos/) and [Penn Treebank (PTB) tags](https://www.sketchengine.eu/penn-treebank-tagset/). We'll only train our model on the UD tags, but will load the PTB tags to show how they could be used instead.\n\n`UD_TAGS` handles how the UD tags should be handled. Our `TEXT` vocabulary - which we'll build later - will have *unknown* tokens in it, i.e. tokens that are not within our vocabulary. However, we won't have unknown tags as we are dealing with a finite set of possible tags. TorchText `Fields` initialize a default unknown token, `<unk>`, which we remove by setting `unk_token = None`.\n\n`PTB_TAGS` does the same as `UD_TAGS`, but handles the PTB tags instead.",
"_____no_output_____"
]
],
[
[
"TEXT = data.Field(lower = True)\nUD_TAGS = data.Field(unk_token = None)\nPTB_TAGS = data.Field(unk_token = None)",
"_____no_output_____"
]
],
[
[
"We then define `fields`, which handles passing our fields to the dataset.\n\nNote that order matters, if you only wanted to load the PTB tags your field would be:\n\n```\nfields = ((\"text\", TEXT), (None, None), (\"ptbtags\", PTB_TAGS))\n```\n\nWhere `None` tells TorchText to not load those tags.",
"_____no_output_____"
]
],
[
[
"fields = ((\"text\", TEXT), (\"udtags\", UD_TAGS), (\"ptbtags\", PTB_TAGS))",
"_____no_output_____"
]
],
[
[
"Next, we load the UDPOS dataset using our defined fields.",
"_____no_output_____"
]
],
[
[
"train_data, valid_data, test_data = datasets.UDPOS.splits(fields)",
"_____no_output_____"
]
],
[
[
"We can check how many examples are in each section of the dataset by checking their length.",
"_____no_output_____"
]
],
[
[
"print(f\"Number of training examples: {len(train_data)}\")\nprint(f\"Number of validation examples: {len(valid_data)}\")\nprint(f\"Number of testing examples: {len(test_data)}\")",
"_____no_output_____"
]
],
[
[
"Let's print out an example:",
"_____no_output_____"
]
],
[
[
"print(vars(train_data.examples[0]))",
"_____no_output_____"
]
],
[
[
"We can also view the text and tags separately:",
"_____no_output_____"
]
],
[
[
"print(vars(train_data.examples[0])['text'])",
"_____no_output_____"
],
[
"print(vars(train_data.examples[0])['udtags'])",
"_____no_output_____"
],
[
"print(vars(train_data.examples[0])['ptbtags'])",
"_____no_output_____"
]
],
[
[
"Next, we'll build the vocabulary - a mapping of tokens to integers. \n\nWe want some unknown tokens within our dataset in order to replicate how this model would be used in real life, so we set the `min_freq` to 2 which means only tokens that appear twice in the training set will be added to the vocabulary and the rest will be replaced by `<unk>` tokens.\n\nWe also load the [GloVe](https://nlp.stanford.edu/projects/glove/) pre-trained token embeddings. Specifically, the 100-dimensional embeddings that have been trained on 6 billion tokens. Using pre-trained embeddings usually leads to improved performance - although admittedly the dataset used in this tutorial is too small to take advantage of the pre-trained embeddings. \n\n`unk_init` is used to initialize the token embeddings which are not in the pre-trained embedding vocabulary. By default this sets those embeddings to zeros, however it is better to not have them all initialized to the same value, so we initialize them from a Normal/Gaussian distribution.\n\nThese pre-trained vectors are now loaded into our vocabulary and we will initialize our model with these values later.",
"_____no_output_____"
]
],
[
[
"MIN_FREQ = 2\n\nTEXT.build_vocab(train_data, \n min_freq = MIN_FREQ,\n vectors = \"glove.6B.100d\",\n unk_init = torch.Tensor.normal_)\n\n\nUD_TAGS.build_vocab(train_data)\nPTB_TAGS.build_vocab(train_data)",
"_____no_output_____"
]
],
[
[
"We can check how many tokens and tags are in our vocabulary by getting their length:",
"_____no_output_____"
]
],
[
[
"print(f\"Unique tokens in TEXT vocabulary: {len(TEXT.vocab)}\")\nprint(f\"Unique tokens in UD_TAG vocabulary: {len(UD_TAGS.vocab)}\")\nprint(f\"Unique tokens in PTB_TAG vocabulary: {len(PTB_TAGS.vocab)}\")",
"_____no_output_____"
]
],
[
[
"Exploring the vocabulary, we can check the most common tokens within our texts:",
"_____no_output_____"
]
],
[
[
"print(TEXT.vocab.freqs.most_common(20))",
"_____no_output_____"
]
],
[
[
"We can see the vocabularies for both of our tags:",
"_____no_output_____"
]
],
[
[
"print(UD_TAGS.vocab.itos)",
"_____no_output_____"
],
[
"print(PTB_TAGS.vocab.itos)",
"_____no_output_____"
]
],
[
[
"We can also see how many of each tag are in our vocabulary:",
"_____no_output_____"
]
],
[
[
"print(UD_TAGS.vocab.freqs.most_common())",
"_____no_output_____"
],
[
"print(PTB_TAGS.vocab.freqs.most_common())",
"_____no_output_____"
]
],
[
[
"We can also view how common each of the tags are within the training set:",
"_____no_output_____"
]
],
[
[
"def tag_percentage(tag_counts):\n \n total_count = sum([count for tag, count in tag_counts])\n \n tag_counts_percentages = [(tag, count, count/total_count) for tag, count in tag_counts]\n \n return tag_counts_percentages",
"_____no_output_____"
],
[
"print(\"Tag\\t\\tCount\\t\\tPercentage\\n\")\n\nfor tag, count, percent in tag_percentage(UD_TAGS.vocab.freqs.most_common()):\n print(f\"{tag}\\t\\t{count}\\t\\t{percent*100:4.1f}%\")",
"_____no_output_____"
],
[
"print(\"Tag\\t\\tCount\\t\\tPercentage\\n\")\n\nfor tag, count, percent in tag_percentage(PTB_TAGS.vocab.freqs.most_common()):\n print(f\"{tag}\\t\\t{count}\\t\\t{percent*100:4.1f}%\")",
"_____no_output_____"
]
],
[
[
"The final part of data preparation is handling the iterator. \n\nThis will be iterated over to return batches of data to process. Here, we set the batch size and the `device` - which is used to place the batches of tensors on our GPU, if we have one. ",
"_____no_output_____"
]
],
[
[
"BATCH_SIZE = 128\n\ndevice = torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')\n\ntrain_iterator, valid_iterator, test_iterator = data.BucketIterator.splits(\n (train_data, valid_data, test_data), \n batch_size = BATCH_SIZE,\n device = device)",
"_____no_output_____"
]
],
[
[
"## Building the Model\n\nNext up, we define our model - a multi-layer bi-directional LSTM. The image below shows a simplified version of the model with only one LSTM layer and omitting the LSTM's cell state for clarity.\n\n\n\nThe model takes in a sequence of tokens, $X = \\{x_1, x_2,...,x_T\\}$, passes them through an embedding layer, $e$, to get the token embeddings, $e(X) = \\{e(x_1), e(x_2), ..., e(x_T)\\}$.\n\nThese embeddings are processed - one per time-step - by the forward and backward LSTMs. The forward LSTM processes the sequence from left-to-right, whilst the backward LSTM processes the sequence right-to-left, i.e. the first input to the forward LSTM is $x_1$ and the first input to the backward LSTM is $x_T$. \n\nThe LSTMs also take in the the hidden, $h$, and cell, $c$, states from the previous time-step\n\n$$h^{\\rightarrow}_t = \\text{LSTM}^{\\rightarrow}(e(x^{\\rightarrow}_t), h^{\\rightarrow}_{t-1}, c^{\\rightarrow}_{t-1})$$\n$$h^{\\leftarrow}_t=\\text{LSTM}^{\\leftarrow}(e(x^{\\leftarrow}_t), h^{\\leftarrow}_{t-1}, c^{\\leftarrow}_{t-1})$$\n\nAfter the whole sequence has been processed, the hidden and cell states are then passed to the next layer of the LSTM.\n\nThe initial hidden and cell states, $h_0$ and $c_0$, for each direction and layer are initialized to a tensor full of zeros.\n\nWe then concatenate both the forward and backward hidden states from the final layer of the LSTM, $H = \\{h_1, h_2, ... h_T\\}$, where $h_1 = [h^{\\rightarrow}_1;h^{\\leftarrow}_T]$, $h_2 = [h^{\\rightarrow}_2;h^{\\leftarrow}_{T-1}]$, etc. and pass them through a linear layer, $f$, which is used to make the prediction of which tag applies to this token, $\\hat{y}_t = f(h_t)$.\n\nWhen training the model, we will compare our predicted tags, $\\hat{Y}$ against the actual tags, $Y$, to calculate a loss, the gradients w.r.t. that loss, and then update our parameters.\n\nWe implement the model detailed above in the `BiLSTMPOSTagger` class.\n\n`nn.Embedding` is an embedding layer and the input dimension should be the size of the input (text) vocabulary. We tell it what the index of the padding token is so it does not update the padding token's embedding entry.\n\n`nn.LSTM` is the LSTM. We apply dropout as regularization between the layers, if we are using more than one.\n\n`nn.Linear` defines the linear layer to make predictions using the LSTM outputs. We double the size of the input if we are using a bi-directional LSTM. The output dimensions should be the size of the tag vocabulary.\n\nWe also define a dropout layer with `nn.Dropout`, which we use in the `forward` method to apply dropout to the embeddings and the outputs of the final layer of the LSTM.",
"_____no_output_____"
]
],
[
[
"class BiLSTMPOSTagger(nn.Module):\n def __init__(self, \n input_dim, \n embedding_dim, \n hidden_dim, \n output_dim, \n n_layers, \n bidirectional, \n dropout, \n pad_idx):\n \n super().__init__()\n \n self.embedding = nn.Embedding(input_dim, embedding_dim, padding_idx = pad_idx)\n \n self.lstm = nn.LSTM(embedding_dim, \n hidden_dim, \n num_layers = n_layers, \n bidirectional = bidirectional,\n dropout = dropout if n_layers > 1 else 0)\n \n self.fc = nn.Linear(hidden_dim * 2 if bidirectional else hidden_dim, output_dim)\n \n self.dropout = nn.Dropout(dropout)\n \n def forward(self, text):\n\n #text = [sent len, batch size]\n \n #pass text through embedding layer\n embedded = self.dropout(self.embedding(text))\n \n #embedded = [sent len, batch size, emb dim]\n \n #pass embeddings into LSTM\n outputs, (hidden, cell) = self.lstm(embedded)\n \n #outputs holds the backward and forward hidden states in the final layer\n #hidden and cell are the backward and forward hidden and cell states at the final time-step\n \n #output = [sent len, batch size, hid dim * n directions]\n #hidden/cell = [n layers * n directions, batch size, hid dim]\n \n #we use our outputs to make a prediction of what the tag should be\n predictions = self.fc(self.dropout(outputs))\n \n #predictions = [sent len, batch size, output dim]\n \n return predictions",
"_____no_output_____"
]
],
[
[
"## Training the Model\n\nNext, we instantiate the model. We need to ensure the embedding dimensions matches that of the GloVe embeddings we loaded earlier.\n\nThe rest of the hyperparmeters have been chosen as sensible defaults, though there may be a combination that performs better on this model and dataset.\n\nThe input and output dimensions are taken directly from the lengths of the respective vocabularies. The padding index is obtained using the vocabulary and the `Field` of the text.",
"_____no_output_____"
]
],
[
[
"INPUT_DIM = len(TEXT.vocab)\nEMBEDDING_DIM = 100\nHIDDEN_DIM = 128\nOUTPUT_DIM = len(UD_TAGS.vocab)\nN_LAYERS = 2\nBIDIRECTIONAL = True\nDROPOUT = 0.25\nPAD_IDX = TEXT.vocab.stoi[TEXT.pad_token]\n\nmodel = BiLSTMPOSTagger(INPUT_DIM, \n EMBEDDING_DIM, \n HIDDEN_DIM, \n OUTPUT_DIM, \n N_LAYERS, \n BIDIRECTIONAL, \n DROPOUT, \n PAD_IDX)",
"_____no_output_____"
]
],
[
[
"We initialize the weights from a simple Normal distribution. Again, there may be a better initialization scheme for this model and dataset.",
"_____no_output_____"
]
],
[
[
"def init_weights(m):\n for name, param in m.named_parameters():\n nn.init.normal_(param.data, mean = 0, std = 0.1)\n \nmodel.apply(init_weights)",
"_____no_output_____"
]
],
[
[
"Next, a small function to tell us how many parameters are in our model. Useful for comparing different models.",
"_____no_output_____"
]
],
[
[
"def count_parameters(model):\n return sum(p.numel() for p in model.parameters() if p.requires_grad)\n\nprint(f'The model has {count_parameters(model):,} trainable parameters')",
"_____no_output_____"
]
],
[
[
"We'll now initialize our model's embedding layer with the pre-trained embedding values we loaded earlier.\n\nThis is done by getting them from the vocab's `.vectors` attribute and then performing a `.copy` to overwrite the embedding layer's current weights.",
"_____no_output_____"
]
],
[
[
"pretrained_embeddings = TEXT.vocab.vectors\n\nprint(pretrained_embeddings.shape)",
"_____no_output_____"
],
[
"model.embedding.weight.data.copy_(pretrained_embeddings)",
"_____no_output_____"
]
],
[
[
"It's common to initialize the embedding of the pad token to all zeros. This, along with setting the `padding_idx` in the model's embedding layer, means that the embedding should always output a tensor full of zeros when a pad token is input.",
"_____no_output_____"
]
],
[
[
"model.embedding.weight.data[PAD_IDX] = torch.zeros(EMBEDDING_DIM)\n\nprint(model.embedding.weight.data)",
"_____no_output_____"
]
],
[
[
"We then define our optimizer, used to update our parameters w.r.t. their gradients. We use Adam with the default learning rate.",
"_____no_output_____"
]
],
[
[
"optimizer = optim.Adam(model.parameters())",
"_____no_output_____"
]
],
[
[
"Next, we define our loss function, cross-entropy loss.\n\nEven though we have no `<unk>` tokens within our tag vocab, we still have `<pad>` tokens. This is because all sentences within a batch need to be the same size. However, we don't want to calculate the loss when the target is a `<pad>` token as we aren't training our model to recognize padding tokens.\n\nWe handle this by setting the `ignore_index` in our loss function to the index of the padding token in our tag vocabulary.",
"_____no_output_____"
]
],
[
[
"TAG_PAD_IDX = UD_TAGS.vocab.stoi[UD_TAGS.pad_token]\n\ncriterion = nn.CrossEntropyLoss(ignore_index = TAG_PAD_IDX)",
"_____no_output_____"
]
],
[
[
"We then place our model and loss function on our GPU, if we have one.",
"_____no_output_____"
]
],
[
[
"model = model.to(device)\ncriterion = criterion.to(device)",
"_____no_output_____"
]
],
[
[
"We will be using the loss value between our predicted and actual tags to train the network, but ideally we'd like a more interpretable way to see how well our model is doing - accuracy.\n\nThe issue is that we don't want to calculate accuracy over the `<pad>` tokens as we aren't interested in predicting them.\n\nThe function below only calculates accuracy over non-padded tokens. `non_pad_elements` is a tensor containing the indices of the non-pad tokens within an input batch. We then compare the predictions of those elements with the labels to get a count of how many predictions were correct. We then divide this by the number of non-pad elements to get our accuracy value over the batch.",
"_____no_output_____"
]
],
[
[
"def categorical_accuracy(preds, y, tag_pad_idx):\n \"\"\"\n Returns accuracy per batch, i.e. if you get 8/10 right, this returns 0.8, NOT 8\n \"\"\"\n max_preds = preds.argmax(dim = 1, keepdim = True) # get the index of the max probability\n non_pad_elements = (y != tag_pad_idx).nonzero()\n correct = max_preds[non_pad_elements].squeeze(1).eq(y[non_pad_elements])\n return correct.sum() / torch.FloatTensor([y[non_pad_elements].shape[0]]).to(device)",
"_____no_output_____"
]
],
[
[
"Next is the function that handles training our model.\n\nWe first set the model to `train` mode to turn on dropout/batch-norm/etc. (if used). Then we iterate over our iterator, which returns a batch of examples. \n\nFor each batch: \n- we zero the gradients over the parameters from the last gradient calculation\n- insert the batch of text into the model to get predictions\n- as PyTorch loss functions cannot handle 3-dimensional predictions we reshape our predictions\n- calculate the loss and accuracy between the predicted tags and actual tags\n- call `backward` to calculate the gradients of the parameters w.r.t. the loss\n- take an optimizer `step` to update the parameters\n- add to the running total of loss and accuracy",
"_____no_output_____"
]
],
[
[
"def train(model, iterator, optimizer, criterion, tag_pad_idx):\n \n epoch_loss = 0\n epoch_acc = 0\n \n model.train()\n \n for batch in iterator:\n \n text = batch.text\n tags = batch.udtags\n \n optimizer.zero_grad()\n \n #text = [sent len, batch size]\n \n predictions = model(text)\n \n #predictions = [sent len, batch size, output dim]\n #tags = [sent len, batch size]\n \n predictions = predictions.view(-1, predictions.shape[-1])\n tags = tags.view(-1)\n \n #predictions = [sent len * batch size, output dim]\n #tags = [sent len * batch size]\n \n loss = criterion(predictions, tags)\n \n acc = categorical_accuracy(predictions, tags, tag_pad_idx)\n \n loss.backward()\n \n optimizer.step()\n \n epoch_loss += loss.item()\n epoch_acc += acc.item()\n \n return epoch_loss / len(iterator), epoch_acc / len(iterator)",
"_____no_output_____"
]
],
[
[
"The `evaluate` function is similar to the `train` function, except with changes made so we don't update the model's parameters.\n\n`model.eval()` is used to put the model in evaluation mode, so dropout/batch-norm/etc. are turned off. \n\nThe iteration loop is also wrapped in `torch.no_grad` to ensure we don't calculate any gradients. We also don't need to call `optimizer.zero_grad()` and `optimizer.step()`.",
"_____no_output_____"
]
],
[
[
"def evaluate(model, iterator, criterion, tag_pad_idx):\n \n epoch_loss = 0\n epoch_acc = 0\n \n model.eval()\n \n with torch.no_grad():\n \n for batch in iterator:\n\n text = batch.text\n tags = batch.udtags\n \n predictions = model(text)\n \n predictions = predictions.view(-1, predictions.shape[-1])\n tags = tags.view(-1)\n \n loss = criterion(predictions, tags)\n \n acc = categorical_accuracy(predictions, tags, tag_pad_idx)\n\n epoch_loss += loss.item()\n epoch_acc += acc.item()\n \n return epoch_loss / len(iterator), epoch_acc / len(iterator)",
"_____no_output_____"
]
],
[
[
"Next, we have a small function that tells us how long an epoch takes.",
"_____no_output_____"
]
],
[
[
"def epoch_time(start_time, end_time):\n elapsed_time = end_time - start_time\n elapsed_mins = int(elapsed_time / 60)\n elapsed_secs = int(elapsed_time - (elapsed_mins * 60))\n return elapsed_mins, elapsed_secs",
"_____no_output_____"
]
],
[
[
"Finally, we train our model!\n\nAfter each epoch we check if our model has achieved the best validation loss so far. If it has then we save the parameters of this model and we will use these \"best\" parameters to calculate performance over our test set.",
"_____no_output_____"
]
],
[
[
"N_EPOCHS = 15\n\nbest_valid_loss = float('inf')\n\nfor epoch in range(N_EPOCHS):\n\n start_time = time.time()\n \n train_loss, train_acc = train(model, train_iterator, optimizer, criterion, TAG_PAD_IDX)\n valid_loss, valid_acc = evaluate(model, valid_iterator, criterion, TAG_PAD_IDX)\n \n end_time = time.time()\n\n epoch_mins, epoch_secs = epoch_time(start_time, end_time)\n \n if valid_loss < best_valid_loss:\n best_valid_loss = valid_loss\n torch.save(model.state_dict(), 'tut1-model.pt')\n \n print(f'Epoch: {epoch+1:02} | Epoch Time: {epoch_mins}m {epoch_secs}s')\n print(f'\\tTrain Loss: {train_loss:.3f} | Train Acc: {train_acc*100:.2f}%')\n print(f'\\t Val. Loss: {valid_loss:.3f} | Val. Acc: {valid_acc*100:.2f}%')",
"_____no_output_____"
]
],
[
[
"We then load our \"best\" parameters and evaluate performance on the test set.",
"_____no_output_____"
]
],
[
[
"model.load_state_dict(torch.load('tut1-model.pt'))\n\ntest_loss, test_acc = evaluate(model, test_iterator, criterion, TAG_PAD_IDX)\n\nprint(f'Test Loss: {test_loss:.3f} | Test Acc: {test_acc*100:.2f}%')",
"_____no_output_____"
]
],
[
[
"## Inference\n\n88% accuracy looks pretty good, but let's see our model tag some actual sentences.\n\nWe define a `tag_sentence` function that will:\n- put the model into evaluation mode\n- tokenize the sentence with spaCy if it is not a list\n- lowercase the tokens if the `Field` did\n- numericalize the tokens using the vocabulary\n- find out which tokens are not in the vocabulary, i.e. are `<unk>` tokens\n- convert the numericalized tokens into a tensor and add a batch dimension\n- feed the tensor into the model\n- get the predictions over the sentence\n- convert the predictions into readable tags\n\nAs well as returning the tokens and tags, it also returns which tokens were `<unk>` tokens.",
"_____no_output_____"
]
],
[
[
"def tag_sentence(model, device, sentence, text_field, tag_field):\n \n model.eval()\n \n if isinstance(sentence, str):\n nlp = spacy.load('en')\n tokens = [token.text for token in nlp(sentence)]\n else:\n tokens = [token for token in sentence]\n\n if text_field.lower:\n tokens = [t.lower() for t in tokens]\n \n numericalized_tokens = [text_field.vocab.stoi[t] for t in tokens]\n\n unk_idx = text_field.vocab.stoi[text_field.unk_token]\n \n unks = [t for t, n in zip(tokens, numericalized_tokens) if n == unk_idx]\n \n token_tensor = torch.LongTensor(numericalized_tokens)\n \n token_tensor = token_tensor.unsqueeze(-1).to(device)\n \n predictions = model(token_tensor)\n \n top_predictions = predictions.argmax(-1)\n \n predicted_tags = [tag_field.vocab.itos[t.item()] for t in top_predictions]\n \n return tokens, predicted_tags, unks",
"_____no_output_____"
]
],
[
[
"We'll get an already tokenized example from the training set and test our model's performance.",
"_____no_output_____"
]
],
[
[
"example_index = 1\n\nsentence = vars(train_data.examples[example_index])['text']\nactual_tags = vars(train_data.examples[example_index])['udtags']\n\nprint(sentence)",
"_____no_output_____"
]
],
[
[
"We can then use our `tag_sentence` function to get the tags. Notice how the tokens referring to subject of the sentence, the \"respected cleric\", are both `<unk>` tokens!",
"_____no_output_____"
]
],
[
[
"tokens, pred_tags, unks = tag_sentence(model, \n device, \n sentence, \n TEXT, \n UD_TAGS)\n\nprint(unks)",
"_____no_output_____"
]
],
[
[
"We can then check how well it did. Surprisingly, it got every token correct, including the two that were unknown tokens!",
"_____no_output_____"
]
],
[
[
"print(\"Pred. Tag\\tActual Tag\\tCorrect?\\tToken\\n\")\n\nfor token, pred_tag, actual_tag in zip(tokens, pred_tags, actual_tags):\n correct = '✔' if pred_tag == actual_tag else '✘'\n print(f\"{pred_tag}\\t\\t{actual_tag}\\t\\t{correct}\\t\\t{token}\")",
"_____no_output_____"
]
],
[
[
"Let's now make up our own sentence and see how well the model does.\n\nOur example sentence below has every token within the model's vocabulary.",
"_____no_output_____"
]
],
[
[
"sentence = 'The Queen will deliver a speech about the conflict in North Korea at 1pm tomorrow.'\n\ntokens, tags, unks = tag_sentence(model, \n device, \n sentence, \n TEXT, \n UD_TAGS)\n\nprint(unks)",
"_____no_output_____"
]
],
[
[
"Looking at the sentence it seems like it gave sensible tags to every token!",
"_____no_output_____"
]
],
[
[
"print(\"Pred. Tag\\tToken\\n\")\n\nfor token, tag in zip(tokens, tags):\n print(f\"{tag}\\t\\t{token}\")",
"_____no_output_____"
]
],
[
[
"We've now seen how to implement PoS tagging with PyTorch and TorchText! \n\nThe BiLSTM isn't a state-of-the-art model, in terms of performance, but is a strong baseline for PoS tasks and is a good tool to have in your arsenal.",
"_____no_output_____"
],
[
"### Going deeper\nWhat if we could combine word-level and char-level approaches? \n\n\n\nActually, we can. Let's use LSTM or GRU to generate embedding for every word on char-level.\n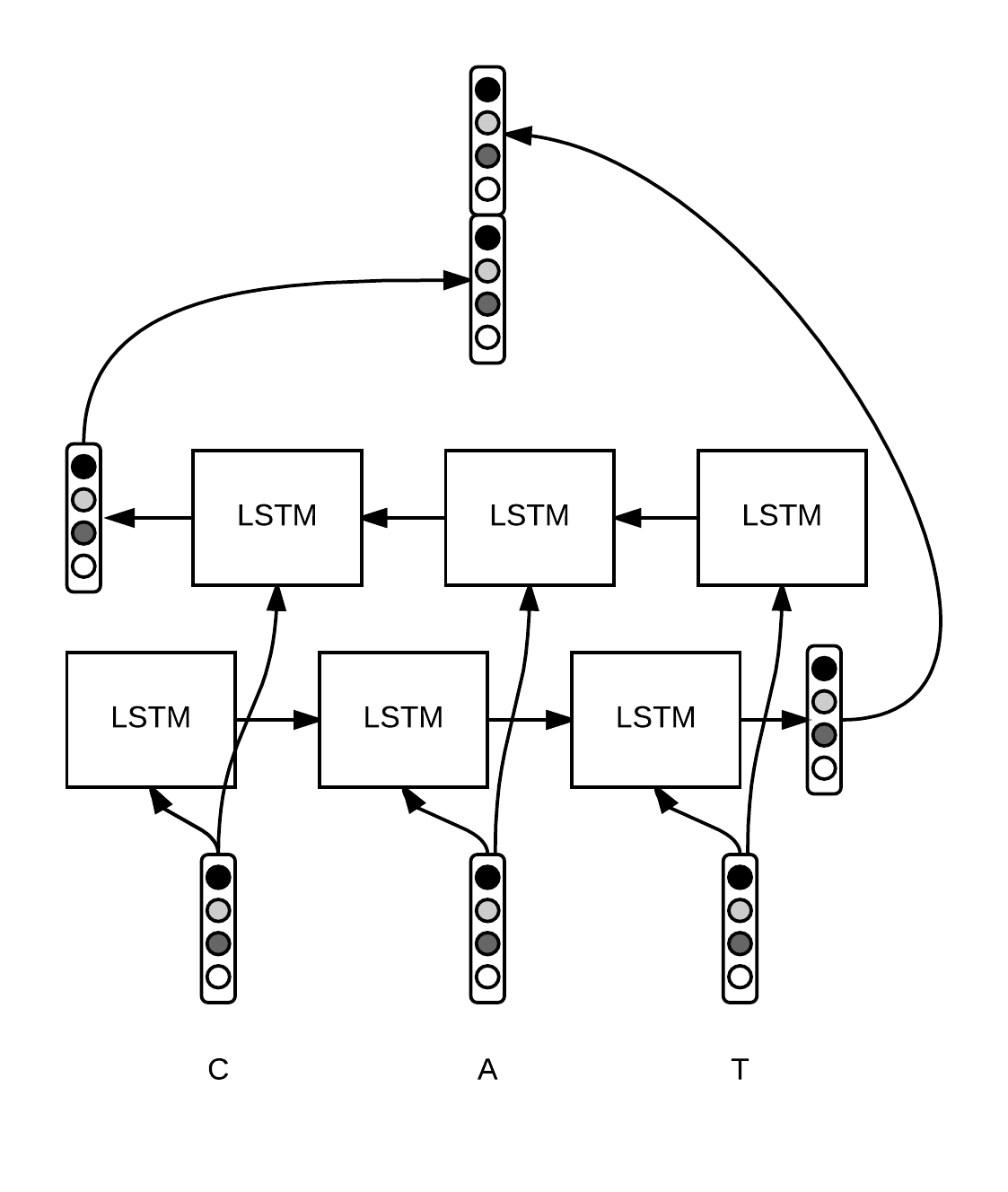\n*Image source: https://guillaumegenthial.github.io/sequence-tagging-with-tensorflow.html*\n\n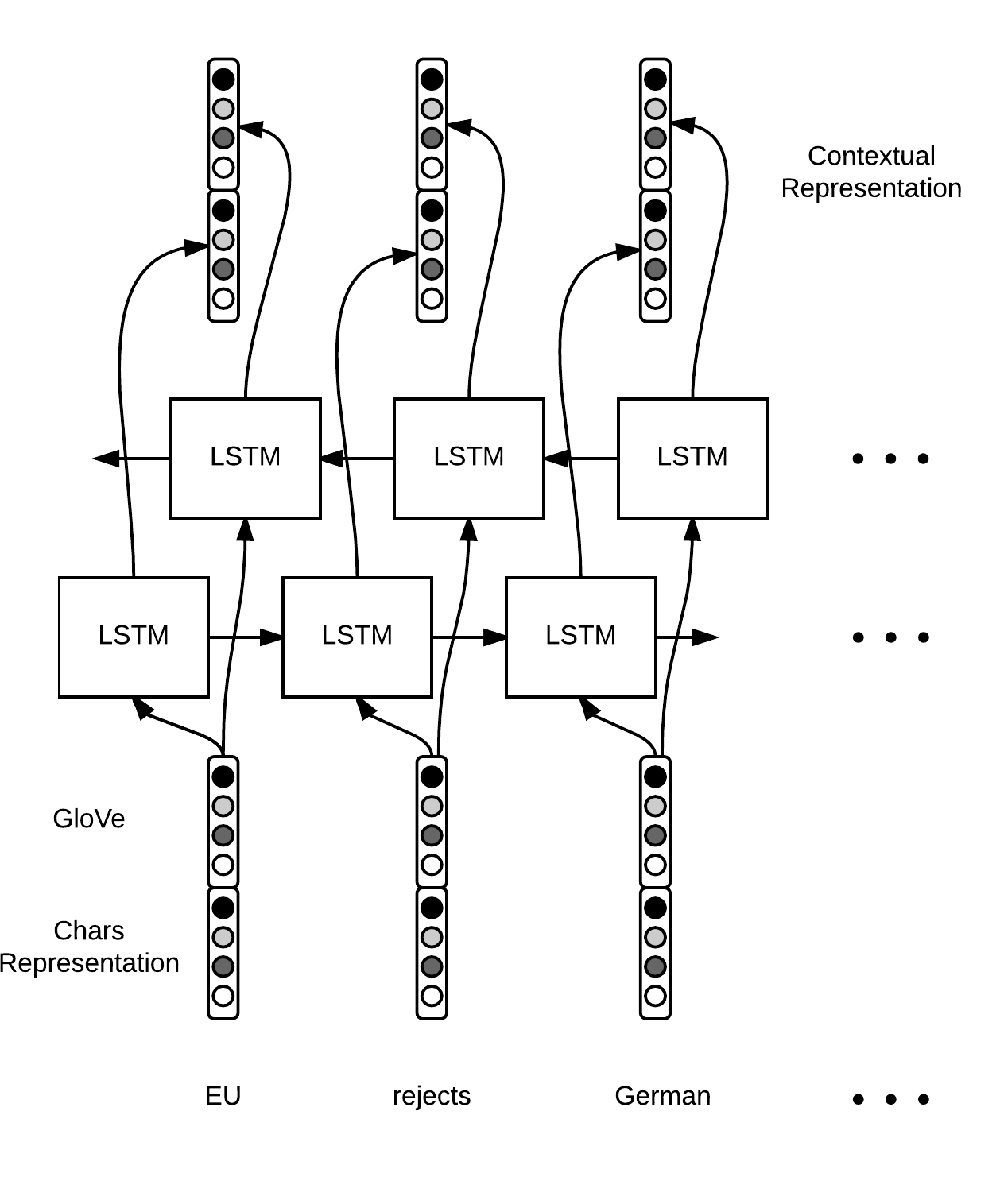\n*Image source: https://guillaumegenthial.github.io/sequence-tagging-with-tensorflow.html*",
"_____no_output_____"
],
[
"To do that we need to make few adjustments to the code above",
"_____no_output_____"
]
],
[
[
"# Now lets try both word and character embeddings\nWORD = data.Field(lower = True)\nUD_TAG = data.Field(unk_token = None)\nPTB_TAG = data.Field(unk_token = None)\n\n# We'll use NestedField to tokenize each word into list of chars\nCHAR_NESTING = data.Field(tokenize=list, init_token=\"<bos>\", eos_token=\"<eos>\")\nCHAR = data.NestedField(CHAR_NESTING)#, init_token=\"<bos>\", eos_token=\"<eos>\")\n\nfields = [(('word', 'char'), (WORD, CHAR)), ('udtag', UD_TAG), ('ptbtag', PTB_TAG)]\ntrain_data, valid_data, test_data = datasets.UDPOS.splits(fields)\n# train, val, test = datasets.UDPOS.splits(fields=fields)\n\nprint(train_data.fields)\nprint(len(train_data))\nprint(vars(train_data[0]))",
"_____no_output_____"
],
[
"WORD.build_vocab(\n train_data,\n min_freq = MIN_FREQ,\n vectors=\"glove.6B.100d\",\n unk_init = torch.Tensor.normal_\n)\n\n\nCHAR.build_vocab(train_data)\nUD_TAG.build_vocab(train_data)\nPTB_TAG.build_vocab(train_data)",
"_____no_output_____"
],
[
"print(f\"Unique tokens in WORD vocabulary: {len(WORD.vocab)}\")\nprint(f\"Unique tokens in CHAR vocabulary: {len(CHAR.vocab)}\")\nprint(f\"Unique tokens in UD_TAG vocabulary: {len(UD_TAG.vocab)}\")\nprint(f\"Unique tokens in PTB_TAG vocabulary: {len(PTB_TAG.vocab)}\")",
"_____no_output_____"
],
[
"BATCH_SIZE = 64\n\ndevice = torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')\n\ntrain_iterator, valid_iterator, test_iterator = data.BucketIterator.splits(\n (train_data, valid_data, test_data), \n batch_size = BATCH_SIZE,\n device = device)",
"_____no_output_____"
],
[
"batch = next(iter(train_iterator))",
"_____no_output_____"
],
[
"text = batch.word\nchars = batch.char\ntags = batch.udtag\n",
"_____no_output_____"
],
[
"class BiLSTMPOSTaggerWithChars(nn.Module):\n def __init__(self, \n word_input_dim, \n word_embedding_dim,\n char_input_dim,\n char_embedding_dim,\n char_hidden_dim,\n hidden_dim,\n output_dim, \n n_layers, \n bidirectional, \n dropout, \n pad_idx):\n \n super().__init__()\n \n self.char_embedding = # YOUR CODE HERE\n self.char_gru = # YOUR CODE HERE\n \n self.word_embedding = nn.Embedding(word_input_dim, word_embedding_dim, padding_idx = pad_idx)\n self.lstm = nn.LSTM(word_embedding_dim + # YOUR CODE HERE, \n hidden_dim, \n num_layers = n_layers, \n bidirectional = bidirectional,\n dropout = dropout if n_layers > 1 else 0)\n \n self.fc = nn.Linear(hidden_dim * 2 if bidirectional else hidden_dim, output_dim)\n \n self.dropout = nn.Dropout(dropout)\n \n def forward(self, text, chars):\n\n #text = [sent len, batch size]\n \n #pass text through embedding layer\n embedded = self.dropout(self.word_embedding(text))\n #embedded = [sent len, batch size, emb dim]\n \n chars_embedded = # YOUR CODE HERE\n hid_from_chars = # YOUR CODE HERE\n \n embedded_with_chars = torch.cat([embedded, hid_from_chars], dim=2)\n \n \n #pass embeddings into LSTM\n outputs, (hidden, cell) = self.lstm(embedded_with_chars)\n# outputs, (hidden, cell) = self.lstm(hid)\n\n \n #outputs holds the backward and forward hidden states in the final layer\n #hidden and cell are the backward and forward hidden and cell states at the final time-step\n \n #output = [sent len, batch size, hid dim * n directions]\n #hidden/cell = [n layers * n directions, batch size, hid dim]\n \n #we use our outputs to make a prediction of what the tag should be\n predictions = self.fc(self.dropout(outputs))\n \n #predictions = [sent len, batch size, output dim]\n \n return predictions",
"_____no_output_____"
],
[
"INPUT_DIM = len(WORD.vocab)\nEMBEDDING_DIM = 100\nHIDDEN_DIM = 160\nCHAR_INPUT_DIM = 112\nCHAR_EMBEDDING_DIM = 30\nCHAR_HIDDEN_DIM = 30\nOUTPUT_DIM = len(UD_TAGS.vocab)\nN_LAYERS = 2\nBIDIRECTIONAL = True\nDROPOUT = 0.25\nPAD_IDX = TEXT.vocab.stoi[TEXT.pad_token]\n\nmodel = BiLSTMPOSTaggerWithChars(\n INPUT_DIM, \n EMBEDDING_DIM,\n CHAR_INPUT_DIM,\n CHAR_EMBEDDING_DIM,\n CHAR_HIDDEN_DIM,\n HIDDEN_DIM, \n OUTPUT_DIM, \n N_LAYERS, \n BIDIRECTIONAL, \n DROPOUT, \n PAD_IDX\n)",
"_____no_output_____"
]
],
[
[
"**Congratulations, you've got LSTM which relies on GRU output on each step.**\n\nNow we need only to train it. Same actions, very small adjustments.",
"_____no_output_____"
]
],
[
[
"def init_weights(m):\n for name, param in m.named_parameters():\n nn.init.normal_(param.data, mean = 0, std = 0.1)\n \nmodel.apply(init_weights)",
"_____no_output_____"
],
[
"def count_parameters(model):\n return sum(p.numel() for p in model.parameters() if p.requires_grad)\n\nprint(f'The model has {count_parameters(model):,} trainable parameters')",
"_____no_output_____"
],
[
"pretrained_embeddings = TEXT.vocab.vectors\n\nprint(pretrained_embeddings.shape)",
"_____no_output_____"
],
[
"model.word_embedding.weight.data.copy_(pretrained_embeddings)\nmodel.word_embedding.weight.data[PAD_IDX] = torch.zeros(EMBEDDING_DIM)\n\nprint(model.word_embedding.weight.data)",
"_____no_output_____"
],
[
"optimizer = optim.Adam(model.parameters())\n\nTAG_PAD_IDX = UD_TAGS.vocab.stoi[UD_TAGS.pad_token]\n\ncriterion = nn.CrossEntropyLoss(ignore_index = TAG_PAD_IDX)\n\nmodel = model.to(device)\ncriterion = criterion.to(device)",
"_____no_output_____"
],
[
"def train(model, iterator, optimizer, criterion, tag_pad_idx):\n \n epoch_loss = 0\n epoch_acc = 0\n \n model.train()\n \n for batch in iterator:\n \n text = batch.word\n chars = batch.char\n tags = batch.udtag\n \n optimizer.zero_grad()\n \n #text = [sent len, batch size]\n \n predictions = model(text, chars)\n \n #predictions = [sent len, batch size, output dim]\n #tags = [sent len, batch size]\n \n predictions = predictions.view(-1, predictions.shape[-1])\n tags = tags.view(-1)\n \n #predictions = [sent len * batch size, output dim]\n #tags = [sent len * batch size]\n \n loss = criterion(predictions, tags)\n \n acc = categorical_accuracy(predictions, tags, tag_pad_idx)\n \n loss.backward()\n \n optimizer.step()\n \n epoch_loss += loss.item()\n epoch_acc += acc.item()\n \n return epoch_loss / len(iterator), epoch_acc / len(iterator)\n\n\ndef evaluate(model, iterator, criterion, tag_pad_idx):\n \n epoch_loss = 0\n epoch_acc = 0\n \n model.eval()\n \n with torch.no_grad():\n \n for batch in iterator:\n\n text = batch.word\n chars = batch.char\n tags = batch.udtag\n \n predictions = model(text, chars)\n \n predictions = predictions.view(-1, predictions.shape[-1])\n tags = tags.view(-1)\n \n loss = criterion(predictions, tags)\n \n acc = categorical_accuracy(predictions, tags, tag_pad_idx)\n\n epoch_loss += loss.item()\n epoch_acc += acc.item()\n \n return epoch_loss / len(iterator), epoch_acc / len(iterator)\n",
"_____no_output_____"
],
[
"N_EPOCHS = 15\n\nbest_valid_loss = float('inf')\n\nfor epoch in range(N_EPOCHS):\n\n start_time = time.time()\n \n train_loss, train_acc = train(model, train_iterator, optimizer, criterion, TAG_PAD_IDX)\n valid_loss, valid_acc = evaluate(model, valid_iterator, criterion, TAG_PAD_IDX)\n \n end_time = time.time()\n\n epoch_mins, epoch_secs = epoch_time(start_time, end_time)\n \n if valid_loss < best_valid_loss:\n best_valid_loss = valid_loss\n torch.save(model.state_dict(), 'tut2-model.pt')\n \n print(f'Epoch: {epoch+1:02} | Epoch Time: {epoch_mins}m {epoch_secs}s')\n print(f'\\tTrain Loss: {train_loss:.3f} | Train Acc: {train_acc*100:.2f}%')\n print(f'\\t Val. Loss: {valid_loss:.3f} | Val. Acc: {valid_acc*100:.2f}%')",
"_____no_output_____"
],
[
"# Let's take a look at the model from the last epoch\ntest_loss, test_acc = evaluate(model, test_iterator, criterion, TAG_PAD_IDX)\n\nprint(f'Test Loss: {test_loss:.3f} | Test Acc: {test_acc*100:.2f}%')",
"_____no_output_____"
],
[
"# And at the best checkpoint (based on validation score)\nmodel.load_state_dict(torch.load('tut2-model.pt'))\n\ntest_loss, test_acc = evaluate(model, test_iterator, criterion, TAG_PAD_IDX)\n\nprint(f'Test Loss: {test_loss:.3f} | Test Acc: {test_acc*100:.2f}%')",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e795541a70a777f5bbe44e5d7efadd9cdf73ce1b | 26,908 | ipynb | Jupyter Notebook | src/main.ipynb | kevinlinxc/ArtRater | 9d72419752b71288443d530735db46e64852039d | [
"MIT"
]
| 1 | 2021-01-12T08:31:17.000Z | 2021-01-12T08:31:17.000Z | src/main.ipynb | kevinlinxc/ArtRater | 9d72419752b71288443d530735db46e64852039d | [
"MIT"
]
| null | null | null | src/main.ipynb | kevinlinxc/ArtRater | 9d72419752b71288443d530735db46e64852039d | [
"MIT"
]
| null | null | null | 50.01487 | 9,340 | 0.681656 | [
[
[
"## 1. Imports",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nfrom PIL.Image import DecompressionBombError\nimport matplotlib.pyplot as plt\nimport json\nimport numpy as np\nimport cv2\nimport praw,requests\nimport psaw\nimport datetime as dt\nimport os\nimport sys\nfrom tensorflow.keras import models\nfrom tensorflow.keras import layers\nfrom tensorflow.keras import optimizers\nimport random",
"_____no_output_____"
]
],
[
[
"## 2. Functions",
"_____no_output_____"
]
],
[
[
"#Checks if two images are identical, returns true if they are, returns true if a file is blank\ndef compare2images(original,duplicate):\n if original is None or duplicate is None:\n return True #delete emtpy pictures\n if original.shape == duplicate.shape:\n #print(\"The images have same size and channels\")\n difference = cv2.subtract(original, duplicate)\n b, g, r = cv2.split(difference)\n if cv2.countNonZero(b) == 0 and cv2.countNonZero(g) == 0 and cv2.countNonZero(r) == 0:\n return True\n else:\n return False\n else:\n return False\n\n#A simple progress bar for transparency on the 20000 image processing tasks\ndef progress(purpose,currentcount, maxcount):\n sys.stdout.write('\\r')\n sys.stdout.write(\"{}: {:.1f}%\".format(purpose,(100/(maxcount-1)*currentcount)))\n sys.stdout.flush()\n\n#custom image data generator following this example https://www.pyimagesearch.com/2018/12/24/how-to-use-keras-fit-and-fit_generator-a-hands-on-tutorial/\ndef custom_file_image_generator(inputPath,bs,mode=\"train\",aug=None, max = 1, frompath=\"picsnew\"):\n f = open(inputPath, \"r\")\n while True:\n images = []\n labels = []\n while len(images)<bs:\n line = f.readline()\n if line == \"\":\n f.seek(0)\n line = f.readline()\n # if we are evaluating we should now break from our\n # loop to ensure we don't continue to fill up the\n # batch from samples at the beginning of the file\n if mode == \"eval\":\n break\n label = int(line.split(\".\")[0].split(\"_\")[0])\n stripped = line.strip('\\n')\n image = plt.imread(f\"{frompath}{stripped}\")\n #Removes alpha channel\n image = np.float32(image)[:,:,:3]\n #Neceesary resizing to avoid PIL pixel cap\n while image.shape[0] * image.shape[1]>89478485:\n image = cv2.resize(image, (0,0), fx=0.5, fy=0.5)\n cv2.cvtColor(image,cv2.COLOR_RGB2BGR)\n images.append(image)\n labels.append(label/max)\n labels = np.asarray(labels).T\n yield(np.asarray(images),labels)",
"_____no_output_____"
]
],
[
[
"## 3. Set up Reddit api for downloading images",
"_____no_output_____"
]
],
[
[
"#Set up API keys from .gitignored file\nwith open('config.json') as config_file:\n config = json.load(config_file)['keys']\n\n# Sign into Reddit using API Key\nreddit = praw.Reddit(user_agent=\"Downloading images from r/art for a machine learning project\",\n client_id=config['client_id'],\n client_secret=config['client_secret'],\n username=config['username'],\n password=config['password'])",
"_____no_output_____"
]
],
[
[
"## 4. Downloading pictures from Reddit r/art using PSAW and PRAW",
"_____no_output_____"
]
],
[
[
"#187mb for 200 pics, approx 18.7gb for 20000\n#Time periods to choose to download from\nJan12018 = int(dt.datetime(2018,1,1).timestamp())\nJan12019 = int(dt.datetime(2019,1,1).timestamp())\nJan12020 = int(dt.datetime(2020,1,1).timestamp())\nJan12021 = int(dt.datetime(2021,1,1).timestamp())\n\n#Pass a PRAW instance into PSAW so that scores are available\napi = psaw.PushshiftAPI(reddit)\n#Number of posts to try and download\nn = 30000\n#Path to download to\ndlpath = \"pics2/\"\n\nprint(\"Looking for posts using Pushshift...\")\n#this step takes a while\nposts = list(api.search_submissions(after = Jan12019, before=Jan12020, subreddit='art', limit = n*10))\nnumpostsfound = len(posts)\nprint(f\"Number of posts found: {numpostsfound}\")\ncounter = 0\n\nfor post in posts:\n if post.score>1:\n progress(\"Downloading\",counter,numpostsfound)\n counter +=1\n url = (post.url)\n #Save score for ML training, and post id for unique file names\n file_name = str(post.score) + \"_\" + str(post.id) + \".jpg\"\n try:\n #use requests to get image\n r = requests.get(url)\n fullfilename = dlpath + file_name\n #save image\n with open(fullfilename,\"wb\") as f:\n f.write(r.content)\n except (\n requests.ConnectionError,\n requests.exceptions.ReadTimeout,\n requests.exceptions.Timeout,\n requests.exceptions.ConnectTimeout,\n ) as e:\n print(e)\n \nfiles = [f for f in os.listdir(dlpath) if os.path.isfile(os.path.join(dlpath, f))]\n#Number of files downloaded not always the same as requested due to connection errors\nprint(f'\\nNumber of files downloaded: {len(files)}')",
"_____no_output_____"
]
],
[
[
"## 5. Processing code that removes pictures that are deleted/corrupt\nMight need to run multiple times if low on ram",
"_____no_output_____"
]
],
[
[
"#Path to delete bad pictures from\npath = \"pics2/\"\n\nfiles = [f for f in os.listdir(path) if os.path.isfile(os.path.join(path, f))]\ncull = []\ncounter = 0\nlength = len(files)\nprint(f\"Original Number of files: {length}\")\n#Template of a bad picture\ndeletedtemplate = cv2.imread(\"exampledeleted.jpg\")\ndeletedtemplate2 = cv2.imread(\"exampledeleted2.jpg\")\nfor file in files:\n progress(\"Deleting bad files\",counter,length)\n counter+=1\n fullfilename = path + file\n candidate = cv2.imread(fullfilename)\n #if it's the same picture as the template or the picture is None\n if compare2images(deletedtemplate,candidate) or compare2images(deletedtemplate2,candidate):\n #delete\n os.remove(fullfilename)\nfiles2 = [f for f in os.listdir(path) if os.path.isfile(os.path.join(path, f))]\nprint(f\"\\nFinal Number of files: {len(files2)}\")",
"Original Number of files: 11377\nDeleting bad files: 100.0%\nFinal Number of files: 11364\n"
]
],
[
[
"## 6. Preprocessing code that corrects grayscale images to RGB and rescales pictures to have maximum width or height of 1000\nIf I ran nn training with large images, it would take too long, and if I ran on google colab,\nI wouldn't have the drive space for all the pictrues",
"_____no_output_____"
]
],
[
[
"#Path being read from\npath = 'pics2/'\n#Path writing to\npath2 = 'picsfix/'\nfiles = [f for f in os.listdir(path) if os.path.isfile(os.path.join(path, f))]\nlength = len(files)\ncounter = 0\nfailures = []\nfor file in files:\n try:\n progress(\"Resizing and fixing pictures\",counter,length)\n #OpenCV doesn't open jpegs\n img = plt.imread(f'{path}{file}')\n if len(img.shape) <3:\n # print(file)\n # print(img.shape)\n img= cv2.cvtColor(img,cv2.COLOR_GRAY2RGB)\n #Resize to 1000 max\n largestdim = max(img.shape)\n targetlargestdim = 1000\n scaling= targetlargestdim / largestdim\n #print(scaling)\n if(scaling<1): #If image is already smaller, don't bother upscaling\n smaller = cv2.resize(img, (0,0), fx=scaling, fy=scaling)\n else:\n smaller = img\n filename = path2+file\n plt.imsave(filename,smaller)\n counter += 1\n except DecompressionBombError as e:\n print(file)\n print(\"Decomp error\")\nprint(\"\\ndone\")",
"_____no_output_____"
]
],
[
[
"## 7. Plot histogram of scores to see how bad the bias towards lower scores is",
"_____no_output_____"
]
],
[
[
"path = \"picsfix2\"\nfiles = [f for f in os.listdir(path) if os.path.isfile(os.path.join(path, f))]\nnumpics = len(files)\nlabelsall = []\nfor file in files:\n labelsall.append(int(file.split(\".\")[0].split(\"_\")[0]))\n#print(labelsall)\nplt.hist(labelsall)\nplt.yscale(\"log\")\nplt.ylabel(\"Frequency\")\nplt.xlabel(\"Score\")\nplt.title(\"Post score distribution, log scale\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 7. Split files into training and testing sets and write the names of files to txt files\nI'm following [this guide](https://www.pyimagesearch.com/2018/12/24/how-to-use-keras-fit-and-fit_generator-a-hands-on-tutorial/)\nand using a file reader to reset the index to 0 seemed like the easiest solution to mimic what the author set up.",
"_____no_output_____"
]
],
[
[
"#Path of pictures to split and write txts for\npath = \"picsfix2/\"\ntrainingpath = \"training.txt\"\ntestingpath = \"testing.txt\"\nfiles = [f for f in os.listdir(path) if os.path.isfile(os.path.join(path, f))]\n#randomize to avoid passing pictures to the neural net in alphabetical order\nrandom.shuffle(files)\n#print(files)\ntrainindex = int(np.round(0.8 * len(files)))\ntraining = files[0:trainindex]\ntesting = files[trainindex:]\nwith open(trainingpath, 'w') as f:\n for item in training:\n f.write(\"%s\\n\" % item)\nwith open(testingpath, 'w') as f:\n for item in testing:\n f.write(\"%s\\n\" % item)",
"_____no_output_____"
]
],
[
[
"## 8. Actual neural net training using Convolutional Neural Net\nI didn't have enough ram to train locally, so I ended porting to Google Colab and training there.\nThe trainin has not been succesful so far, and I haven't taken the time to diagnose why yet.",
"_____no_output_____"
]
],
[
[
"#Path of preprocessed pictures\npath = \"picsfix2/\"\n#Paths of training and testing txts that have file names\ntrainPath = 'training.txt'\ntestpath = 'testing.txt'\n#Get all file names\nfiles = [f for f in os.listdir(path) if os.path.isfile(os.path.join(path, f))]\nnumpics = len(files)\nlabelsall = []\nfor file in files:\n labelsall.append(int(file.split(\".\")[0].split(\"_\")[0]))\nhighestScore = max(labelsall)\nprint(f'Highest score: {highestScore}')\n\n\n#Store all image arrays and image names in a list\ninput_shape=(None, None,3)\n\nNUM_EPOCHS = 12\nBS = 1\nNUM_TRAIN_IMAGES = int(np.round(0.8 * len(files)))\nNUM_TEST_IMAGES = len(files)-NUM_TRAIN_IMAGES\ntraingen = custom_file_image_generator(trainPath,BS, \"train\" , None,highestScore, path)\ntestgen = custom_file_image_generator(testpath,BS, \"train\", None,highestScore, path)\ntf.keras.backend.clear_session()\nconv_model = models.Sequential()\n#normalize pictures to [0 1]\nconv_model.add(layers.experimental.preprocessing.Rescaling(1./255))\nconv_model.add(layers.Conv2D(32, (3, 3), activation='relu',\n input_shape=input_shape))\nconv_model.add(layers.GlobalMaxPooling2D())\n# conv_model.add(layers.Conv2D(64, (3, 3), activation='relu'))\n# conv_model.add(layers.MaxPooling2D(pool_size=(2, 2)))\nconv_model.add(layers.Flatten())\n#conv_model.add(layers.Dropout(0.2))\nconv_model.add(layers.Dense(512, activation='relu'))\nconv_model.add(layers.Dense(1, activation='linear'))\nLEARNING_RATE = 1e-4\nconv_model.compile(loss=tf.keras.losses.MeanSquaredError(),\n optimizer=optimizers.RMSprop(lr=LEARNING_RATE),\n metrics=['acc'])\nhistory_conv = conv_model.fit(traingen,\n steps_per_epoch= NUM_TRAIN_IMAGES // BS,\n validation_data=testgen,\n validation_steps = NUM_TEST_IMAGES // BS,\n epochs=NUM_EPOCHS)\nmodelfilename = 'art2.h5'\nconv_model.save(modelfilename)\n# plt.plot(history_conv.history['loss'])\n# plt.plot(history_conv.history['val_loss'])\n# plt.title('model loss')\n# plt.ylabel('loss')\n# plt.xlabel('epoch')\n# plt.legend(['train loss', 'val loss'], loc='upper right')\n# plt.show()\n#\n#\n# plt.plot(history_conv.history['acc'])\n# plt.plot(history_conv.history['val_acc'])\n# plt.title('model accuracy')\n# plt.ylabel('accuracy (%)')\n# plt.xlabel('epoch')\n# plt.legend(['train accuracy', 'val accuracy'], loc='lower right')\n# plt.show()\n",
"Highest score: 54157\nEpoch 1/12\n 1443/14918 [=>............................] - ETA: 1:37:21 - loss: 0.0014 - acc: 0.0049"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
e7955839b6cd4ce6e2b57a801841ec221cff0397 | 150,911 | ipynb | Jupyter Notebook | notebook/experiment/gan_style_model.ipynb | skywalker0803r/c620 | 84e944f4ef09b9722672d0627bd90e63a5e32cac | [
"MIT"
]
| null | null | null | notebook/experiment/gan_style_model.ipynb | skywalker0803r/c620 | 84e944f4ef09b9722672d0627bd90e63a5e32cac | [
"MIT"
]
| null | null | null | notebook/experiment/gan_style_model.ipynb | skywalker0803r/c620 | 84e944f4ef09b9722672d0627bd90e63a5e32cac | [
"MIT"
]
| 1 | 2020-12-09T11:53:49.000Z | 2020-12-09T11:53:49.000Z | 98.505875 | 29,124 | 0.488897 | [
[
[
"<a href=\"https://colab.research.google.com/github/skywalker0803r/c620/blob/main/notebook/experiment/gan_style_model.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# load data",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport joblib\nfrom urllib.request import urlopen\nimport numpy as np\n\ndf_url = 'https://c620.s3-ap-northeast-1.amazonaws.com/c620_train.csv'\nc_url = 'https://c620.s3-ap-northeast-1.amazonaws.com/c620_col_names.pkl'\n\ndf = pd.read_csv(df_url,index_col=0)\nc = joblib.load(urlopen(c_url))\n\ncase_col = c['case']\nfeed_col = c['x41']\nop_col = c['density']+c['yRefluxRate']+c['yHeatDuty']+c['yControl']\nsp_col = c['vent_gas_sf'] + c['distillate_sf'] + c['sidedraw_sf'] + c['bottoms_sf']\nwt_col = c['vent_gas_x']+c['distillate_x']+c['sidedraw_x']+c['bottoms_x']\nall_col = case_col + feed_col + op_col + sp_col + wt_col\n\nprint(len(case_col))\nprint(len(feed_col))\nprint(len(op_col))\nprint(len(sp_col))\nprint(len(wt_col))\n\ndf[all_col].head(1)",
"3\n41\n10\n164\n164\n"
],
[
"case_col_idx = [all_col.index(i) for i in case_col]\nfeed_col_idx = [all_col.index(i) for i in feed_col]\nop_col_idx = [all_col.index(i) for i in op_col]\nsp_col_idx = [all_col.index(i) for i in sp_col]\nwt_col_idx = [all_col.index(i) for i in wt_col]",
"_____no_output_____"
]
],
[
[
"# preprocess data",
"_____no_output_____"
]
],
[
[
"from sklearn.utils import shuffle\nfrom torch.utils.data import TensorDataset,DataLoader\nfrom sklearn.preprocessing import MinMaxScaler\nimport torch\n\n# split data\ndf = shuffle(df)\np1 = int(len(df)*0.8)\np2 = int(len(df)*0.9)\n\n# to FloatTensor\ntrain = torch.FloatTensor(df[all_col].values[:p1])\nvaild = torch.FloatTensor(df[all_col].values[p1:p2])\ntest = torch.FloatTensor(df[all_col].values[p2:])\n\n# create DataLoader\ntrainset = TensorDataset(train[:,case_col_idx],train[:,feed_col_idx],train[:,op_col_idx],train[:,sp_col_idx],train[:,wt_col_idx])\ntrain_iter = DataLoader(trainset,batch_size=64)\n\nvaildset = TensorDataset(vaild[:,case_col_idx],vaild[:,feed_col_idx],vaild[:,op_col_idx],vaild[:,sp_col_idx],vaild[:,wt_col_idx])\nvaild_iter = DataLoader(vaildset,batch_size=64)\n\ntestset = TensorDataset(test[:,case_col_idx],test[:,feed_col_idx],test[:,op_col_idx],test[:,sp_col_idx],test[:,wt_col_idx])\ntest_iter = DataLoader(testset,batch_size=64)",
"_____no_output_____"
]
],
[
[
"# def model,loss,optimizer",
"_____no_output_____"
]
],
[
[
"import torch\nfrom torch import nn\nimport torch.nn.functional as F\n\ndef mlp(sizes, activation, output_activation=nn.Identity):\n layers = []\n for j in range(len(sizes)-1):\n act = activation if j < len(sizes)-2 else output_activation\n layers += [nn.Linear(sizes[j], sizes[j+1]), act()]\n return nn.Sequential(*layers)\n\nclass Model(nn.Module):\n def __init__(self):\n super(Model, self).__init__()\n self.op_model = mlp(\n [len(case_col)+len(feed_col),128,len(op_col)],\n nn.ReLU\n )\n self.sp_model = mlp(\n [len(case_col)+len(feed_col)+len(op_col),128,len(wt_col)],\n nn.ReLU,\n nn.Sigmoid\n )\n\n def forward(self,case,feed):\n op = self.op_model(torch.cat((case,feed),dim=-1)).clone()\n sp = self.sp_model(torch.cat((case,feed,op),dim=-1)).clone()\n for idx in range(41):\n sp[:,[idx,idx+41,idx+41*2,idx+41*3]] = self.normalize(sp[:,[idx,idx+41,idx+41*2,idx+41*3]])\n s1,s2,s3,s4 = sp[:,:41],sp[:,41:41*2],sp[:,41*2:41*3],sp[:,41*3:41*4]\n w1,w2,w3,w4 = self.sp2wt(feed,s1),self.sp2wt(feed,s2),self.sp2wt(feed,s3),self.sp2wt(feed,s4)\n wt = torch.cat((w1,w2,w3,w4),dim=-1)\n return op,sp,wt\n\n @staticmethod\n def normalize(x):\n return x / x.sum(dim=1).reshape(-1,1)\n \n @staticmethod\n def sp2wt(x,s):\n a = 100*x*s\n b = torch.diag([email protected]).reshape(-1,1)\n b = torch.clamp(b,1e-8,float('inf'))\n return a/b\n\n# model optimizer loss_fn\nmodel = Model()\noptimizer = torch.optim.Adam(model.parameters())\nloss_fn = nn.SmoothL1Loss()\n\n# forward test\nfor case,feed,op,sp,wt in train_iter:\n op_hat,sp_hat,wt_hat = model(case,feed)\n print(op_hat.shape)\n print(sp_hat.shape)\n print(wt_hat.shape)\n break",
"torch.Size([64, 10])\ntorch.Size([64, 164])\ntorch.Size([64, 164])\n"
]
],
[
[
"# tensorboard",
"_____no_output_____"
]
],
[
[
"from torch.utils.tensorboard import SummaryWriter\nwriter = SummaryWriter()\ncase,feed,op,sp,wt = next(iter(train_iter))\nwriter.add_graph(model,[case,feed])\nwriter.close()",
"_____no_output_____"
],
[
"%load_ext tensorboard\n%tensorboard --logdir runs",
"The tensorboard extension is already loaded. To reload it, use:\n %reload_ext tensorboard\n"
]
],
[
[
"# train model",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nfrom tqdm import tqdm_notebook as tqdm\nfrom copy import deepcopy\n\n# train step\ndef train_step(model):\n model.train()\n total_loss = 0\n for t,(case,feed,op,sp,wt) in enumerate(train_iter):\n op_hat,sp_hat,wt_hat = model(case,feed)\n op_loss = loss_fn(op_hat,op)\n sp_loss = loss_fn(sp_hat,sp)\n wt_loss = loss_fn(wt_hat,wt)\n Sidedraw_Benzene_loss = loss_fn(\n wt_hat[:,wt_col.index('Tatoray Stripper C620 Operation_Sidedraw Production Rate and Composition_Benzene_wt%')],\n case[:,case_col.index('Tatoray Stripper C620 Operation_Specifications_Spec 3 : Benzene in Sidedraw_wt%')])\n loss = op_loss + sp_loss + wt_loss + Sidedraw_Benzene_loss\n # update model\n loss.backward()\n optimizer.step()\n optimizer.zero_grad()\n total_loss += loss.item()\n return total_loss/(t+1)\n\n# valid step\ndef valid_step(model):\n model.eval()\n total_loss = 0\n for t,(case,feed,op,sp,wt) in enumerate(vaild_iter):\n op_hat,sp_hat,wt_hat = model(case,feed)\n op_loss = loss_fn(op_hat,op)\n sp_loss = loss_fn(sp_hat,sp)\n wt_loss = loss_fn(wt_hat,wt)\n Sidedraw_Benzene_loss = loss_fn(\n wt_hat[:,wt_col.index('Tatoray Stripper C620 Operation_Sidedraw Production Rate and Composition_Benzene_wt%')],\n case[:,case_col.index('Tatoray Stripper C620 Operation_Specifications_Spec 3 : Benzene in Sidedraw_wt%')])\n loss = op_loss + sp_loss + wt_loss + Sidedraw_Benzene_loss\n total_loss += loss.item()\n return total_loss/(t+1)\n\ndef train(model,max_epochs): \n history = {'train_loss':[],'valid_loss':[]}\n current_loss = np.inf\n best_model = None\n for i in tqdm(range(max_epochs)):\n history['train_loss'].append(train_step(model))\n history['valid_loss'].append(valid_step(model))\n if i % 10 == 0:\n print(\"epoch:{} train_loss:{:.4f} valid_loss:{:.4f}\".format(i,history['train_loss'][-1],history['valid_loss'][-1]))\n if history['valid_loss'][-1] <= current_loss:\n best_model = deepcopy(model.eval())\n current_loss = history['valid_loss'][-1]\n model = deepcopy(best_model.eval())\n plt.plot(history['train_loss'],label='train_loss')\n plt.plot(history['valid_loss'],label='valid_loss')\n plt.legend()\n plt.show()\n return model",
"_____no_output_____"
],
[
"model = train(model,max_epochs=250)",
"_____no_output_____"
],
[
"op_pred,sp_pred,wt_pred = model(test[:,case_col_idx],test[:,feed_col_idx])\n\nwt_pred = pd.DataFrame(wt_pred.detach().cpu().numpy(),columns=wt_col)\nop_pred = pd.DataFrame(op_pred.detach().cpu().numpy(),columns=op_col)\n\nwt_real = pd.DataFrame(test[:,wt_col_idx].detach().cpu().numpy(),columns=wt_col)\nop_real = pd.DataFrame(test[:,op_col_idx].detach().cpu().numpy(),columns=op_col)",
"_____no_output_____"
],
[
"from sklearn.metrics import r2_score,mean_squared_error\nimport numpy as np\nimport warnings \nwarnings.filterwarnings('ignore')\n\ndef mape(y_true, y_pred, e = 2e-2):\n y_true, y_pred = np.array(y_true), np.array(y_pred)\n mask = y_true > e\n y_true, y_pred = y_true[mask], y_pred[mask]\n return np.mean(np.abs((y_true - y_pred) / y_true)) * 100\n\ndef show_metrics(y_real,y_pred,e=2e-2):\n res = pd.DataFrame(index=y_pred.columns,columns=['R2','MSE','MAPE'])\n for i in y_pred.columns:\n res.loc[i,'R2'] = np.clip(r2_score(y_real[i],y_pred[i]),0,1)\n res.loc[i,'MSE'] = mean_squared_error(y_real[i],y_pred[i])\n res.loc[i,'MAPE'] = mape(y_real[i],y_pred[i],e)\n res.loc['AVG'] = res.mean(axis=0)\n return res",
"_____no_output_____"
],
[
"show_metrics(op_real,op_pred)",
"_____no_output_____"
],
[
"show_metrics(wt_real,wt_pred)",
"_____no_output_____"
],
[
"case = pd.DataFrame(test[:,case_col_idx].detach().cpu().numpy(),columns=case_col)\ncase.iloc[:,[2]]",
"_____no_output_____"
],
[
"wt_pred.iloc[:,[89]]",
"_____no_output_____"
],
[
"wt_real.head()",
"_____no_output_____"
],
[
"wt_pred.head()",
"_____no_output_____"
],
[
"op_real.head()",
"_____no_output_____"
],
[
"op_pred.head()",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e7956141b00bd1ea39d0de49cbf06230df8bba3a | 2,080 | ipynb | Jupyter Notebook | 1/1-3.ipynb | petrluner/utpython_lab | 46b39390ca75c58414cd372aa17085ef0f7ce7d3 | [
"MIT"
]
| null | null | null | 1/1-3.ipynb | petrluner/utpython_lab | 46b39390ca75c58414cd372aa17085ef0f7ce7d3 | [
"MIT"
]
| null | null | null | 1/1-3.ipynb | petrluner/utpython_lab | 46b39390ca75c58414cd372aa17085ef0f7ce7d3 | [
"MIT"
]
| null | null | null | 18.086957 | 69 | 0.457212 | [
[
[
"## 練習\n\n1. 数値 `x` の絶対値を求める関数 `absolute(x)` を定義してください。\n (Pythonには `abs` という組み込み関数が用意されていますが。)\n2. `x` が正ならば 1、負ならば -1、ゼロならば 0 を返す `sign(x)` という関数を定義してください。\n\n定義ができたら、その次のセルを実行して、`True` のみが表示されることを確認してください。",
"_____no_output_____"
]
],
[
[
"def absolute(x):\n ...",
"_____no_output_____"
],
[
"def sign(x):\n ...",
"_____no_output_____"
],
[
"print(absolute(5) == 5)\nprint(absolute(-5) == 5)\nprint(absolute(0) == 0)\nprint(sign(5) == 1)\nprint(sign(-5) == -1)\nprint(sign(0) == 0)",
"_____no_output_____"
]
],
[
[
"## 練習の解答",
"_____no_output_____"
]
],
[
[
"def absolute(x):\n if x<0:\n return -x\n else:\n return x",
"_____no_output_____"
],
[
"def sign(x):\n if x<0:\n return -1\n if x>0:\n return 1\n return 0",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
e79577053d4436d7941c3bc5d9450c3f0b867d0f | 17,370 | ipynb | Jupyter Notebook | cudf/notebooks_numba_cuDF_integration.ipynb | rocketmlhq/rapids-notebooks | 5934516f87e906e065d9dcc06d7ed0ce564ed5e6 | [
"Apache-2.0"
]
| null | null | null | cudf/notebooks_numba_cuDF_integration.ipynb | rocketmlhq/rapids-notebooks | 5934516f87e906e065d9dcc06d7ed0ce564ed5e6 | [
"Apache-2.0"
]
| null | null | null | cudf/notebooks_numba_cuDF_integration.ipynb | rocketmlhq/rapids-notebooks | 5934516f87e906e065d9dcc06d7ed0ce564ed5e6 | [
"Apache-2.0"
]
| 1 | 2019-10-06T19:08:12.000Z | 2019-10-06T19:08:12.000Z | 37.76087 | 947 | 0.556246 | [
[
[
"## Objective\n\nIn my previous tutorial, I showed how to use `apply_rows` and `apply_chunks` methods in cuDF to implement customized data transformations. Under the hood, they are all using [Numba library](https://numba.pydata.org/) to compile the normal python code into GPU kernels. Numba is an excellent python library that accelerates the numerical computations. Most importantly, Numba has direct CUDA programming support. For detailed information, please check out this [Numba CUDA document](https://numba.pydata.org/numba-doc/dev/cuda/index.html). As we know, the underlying data structure of cuDF is a GPU version of Apache Arrow. We can directly pass the GPU array around without the copying operation. Once we have the nice Numba library and standard GPU array, the sky is the limit. In this tutorial, I will show how to use Numba CUDA to accelerate cuDF data transformation and how to step by step accelerate it using CUDA programming tricks. \n\nThe following experiments are performed at DGX V100 node.",
"_____no_output_____"
],
[
"## A simple example\nAs usual, I am going to start with a simple example of doubling the numbers in an array:",
"_____no_output_____"
]
],
[
[
"import cudf\nimport numpy as np\nfrom numba import cuda\n \narray_len = 1000\nnumber_of_threads = 128\nnumber_of_blocks = (array_len + (number_of_threads - 1)) // number_of_threads\ndf = cudf.DataFrame()\ndf['in'] = np.arange(array_len, dtype=np.float64)\n \n \[email protected]\ndef double_kernel(result, array_len):\n \"\"\"\n double each element of the array\n \"\"\"\n i = cuda.grid(1)\n if i < array_len:\n result[i] = result[i] * 2.0\n \n \nbefore = df['in'].sum()\ngpu_array = df['in'].to_gpu_array()\nprint(type(gpu_array))\ndouble_kernel[(number_of_blocks,), (number_of_threads,)](gpu_array, array_len)\nafter = df['in'].sum()\nassert(np.isclose(before * 2.0, after))",
"<class 'numba.cuda.cudadrv.devicearray.DeviceNDArray'>\n"
]
],
[
[
"From the output of this code, it shows the underlying GPU array is of type `numba.cuda.cudadrv.devicearray.DeviceNDArray`. We can directly pass it to the kernel function that is compiled by the `cuda.jit`. Because we passed in the reference, the effect of number transformation will automatically show up in the original cuDF Dataframe. Note we have to manually enter the block size and grid size, which gives us the maximum of GPU programming control. The `cuda.grid` is a convenient method to compute the absolute position for the threads. It is equivalent to the normal `block_id * block_dim + thread_id` formula.",
"_____no_output_____"
],
[
"## Practical example\n\n### Baseline\n\nWe will work on the moving average problem as the last time. Because we have the full control of the grid and block size allocation, the vanilla moving average implementation code is much simpler compared to the `apply_chunks` implementation. ",
"_____no_output_____"
]
],
[
[
"%reset -s -f",
"_____no_output_____"
],
[
"import cudf\nimport numpy as np\nimport pandas as pd\nfrom numba import cuda\nimport numba\nimport time\n \narray_len = int(5e8)\naverage_window = 3000\nnumber_of_threads = 128\nnumber_of_blocks = (array_len + (number_of_threads - 1)) // number_of_threads\ndf = cudf.DataFrame()\ndf['in'] = np.arange(array_len, dtype=np.float64)\ndf['out'] = np.arange(array_len, dtype=np.float64)\n \n \[email protected]\ndef kernel1(in_arr, out_arr, average_length, arr_len):\n s = numba.cuda.local.array(1, numba.float64)\n s[0] = 0.0\n i = cuda.grid(1)\n if i < arr_len:\n if i < average_length-1:\n out_arr[i] = np.inf\n else:\n for j in range(0, average_length):\n s[0] += in_arr[i-j]\n out_arr[i] = s[0] / np.float64(average_length)\n \n \ngpu_in = df['in'].to_gpu_array()\ngpu_out = df['out'].to_gpu_array()\nstart = time.time()\nkernel1[(number_of_blocks,), (number_of_threads,)](gpu_in, gpu_out,\n average_window, array_len)\ncuda.synchronize()\nend = time.time()\nprint('Numba with comipile time', end-start)\n \nstart = time.time()\nkernel1[(number_of_blocks,), (number_of_threads,)](gpu_in, gpu_out,\n average_window, array_len)\ncuda.synchronize()\nend = time.time()\nprint('Numba without comipile time', end-start)\n \npdf = pd.DataFrame()\npdf['in'] = np.arange(array_len, dtype=np.float64)\nstart = time.time()\npdf['out'] = pdf.rolling(average_window).mean()\nend = time.time()\nprint('pandas time', end-start)\n \nassert(np.isclose(pdf.out.values[average_window:].mean(),\n df.out.to_array()[average_window:].mean()))",
"Numba with comipile time 2.067620038986206\nNumba without comipile time 1.9229750633239746\npandas time 5.2932703495025635\n"
]
],
[
[
"Note, in order to compare the computation time accurately, I launch the kernel twice. The first time kernel launching will include the kernel compilation time. In this example, it takes 1.9s for the kernel to run without compilation. ",
"_____no_output_____"
],
[
"### Use shared memory\n\nIn the baseline code, each thread is reading the numbers from the global memory. When doing the moving average, the same number is read multiple times by different threads. GPU global memory IO, in this case, is the speed bottleneck. To mitigate it, we load the data into shared memory for each of the computation blocks. Then the threads are doing summation from the numbers in the cache. To do the moving average for the elements at the beginning of the array, we make sure to load the `average_window` more data in the shared_memory. ",
"_____no_output_____"
]
],
[
[
"%reset -s -f",
"_____no_output_____"
],
[
"import cudf\nimport numpy as np\nimport pandas as pd\nfrom numba import cuda\nimport numba\nimport time\n \narray_len = int(5e8)\naverage_window = 3000\nnumber_of_threads = 128\nnumber_of_blocks = (array_len + (number_of_threads - 1)) // number_of_threads\nshared_buffer_size = number_of_threads + average_window - 1\ndf = cudf.DataFrame()\ndf['in'] = np.arange(array_len, dtype=np.float64)\ndf['out'] = np.arange(array_len, dtype=np.float64)\n \n \[email protected]\ndef kernel1(in_arr, out_arr, average_length, arr_len):\n block_size = cuda.blockDim.x\n shared = cuda.shared.array(shape=(shared_buffer_size),\n dtype=numba.float64)\n i = cuda.grid(1)\n tx = cuda.threadIdx.x\n # Block id in a 1D grid\n bid = cuda.blockIdx.x\n starting_id = bid * block_size\n \n shared[tx + average_length - 1] = in_arr[i]\n cuda.syncthreads()\n for j in range(0, average_length - 1, block_size):\n if (tx + j) < average_length - 1:\n shared[tx + j] = in_arr[starting_id -\n average_length + 1 +\n tx + j]\n cuda.syncthreads()\n \n s = numba.cuda.local.array(1, numba.float64)\n s[0] = 0.0\n if i < arr_len:\n if i < average_length-1:\n out_arr[i] = np.inf\n else:\n for j in range(0, average_length):\n s[0] += shared[tx + average_length - 1 - j]\n out_arr[i] = s[0] / np.float64(average_length)\n \n \ngpu_in = df['in'].to_gpu_array()\ngpu_out = df['out'].to_gpu_array()\nstart = time.time()\nkernel1[(number_of_blocks,), (number_of_threads,)](gpu_in, gpu_out,\n average_window, array_len)\ncuda.synchronize()\nend = time.time()\n \nprint('Numba with comipile time', end-start)\n \nstart = time.time()\nkernel1[(number_of_blocks,), (number_of_threads,)](gpu_in, gpu_out,\n average_window, array_len)\ncuda.synchronize()\nend = time.time()\nprint('Numba without comipile time', end-start)\n \npdf = pd.DataFrame()\npdf['in'] = np.arange(array_len, dtype=np.float64)\nstart = time.time()\npdf['out'] = pdf.rolling(average_window).mean()\nend = time.time()\nprint('pandas time', end-start)\n \nassert(np.isclose(pdf.out.values[average_window:].mean(),\n df.out.to_array()[average_window:].mean()))",
"Numba with comipile time 1.3115026950836182\nNumba without comipile time 1.085998773574829\npandas time 5.594487428665161\n"
]
],
[
[
"Running this, the computation time is reduced to 1.09s without kernel compilation time. ",
"_____no_output_____"
],
[
"### Reduced redundant summations\n\nEach thread in the above code is doing one moving average in a for-loop. It is easy to see that there are a lot of redundant summation operations done by different threads. To reduce the redundancy, the following code is changed to let each thread to compute a consecutive number of moving averages. The later moving average step is able to reuse the sum of the previous steps. This eliminated `thread_tile` number of for-loops. ",
"_____no_output_____"
]
],
[
[
"%reset -s -f",
"_____no_output_____"
],
[
"import cudf\nimport numpy as np\nimport pandas as pd\nfrom numba import cuda\nimport numba\nimport time\n \narray_len = int(5e8)\naverage_window = 3000\nnumber_of_threads = 64\nthread_tile = 48\nnumber_of_blocks = (array_len + (number_of_threads * thread_tile - 1)) // (number_of_threads * thread_tile)\nshared_buffer_size = number_of_threads * thread_tile + average_window - 1\ndf = cudf.DataFrame()\ndf['in'] = np.arange(array_len, dtype=np.float64)\ndf['out'] = np.arange(array_len, dtype=np.float64)\n \n \[email protected]\ndef kernel1(in_arr, out_arr, average_length, arr_len):\n block_size = cuda.blockDim.x\n shared = cuda.shared.array(shape=(shared_buffer_size),\n dtype=numba.float64)\n tx = cuda.threadIdx.x\n # Block id in a 1D grid\n bid = cuda.blockIdx.x\n starting_id = bid * block_size * thread_tile\n \n for j in range(thread_tile):\n shared[tx + j * block_size + average_length - 1] = in_arr[starting_id\n + tx +\n j * block_size]\n cuda.syncthreads()\n for j in range(0, average_length - 1, block_size):\n if (tx + j) < average_length - 1:\n shared[tx + j] = in_arr[starting_id -\n average_length + 1 +\n tx + j]\n cuda.syncthreads()\n \n s = numba.cuda.local.array(1, numba.float64)\n first = False\n s[0] = 0.0\n for k in range(thread_tile):\n i = starting_id + tx * thread_tile + k\n if i < arr_len:\n if i < average_length-1:\n out_arr[i] = np.inf\n else:\n if not first:\n for j in range(0, average_length):\n s[0] += shared[tx * thread_tile + k + average_length - 1 - j]\n s[0] = s[0] / np.float64(average_length)\n out_arr[i] = s[0]\n first = True\n else:\n s[0] = s[0] + (shared[tx * thread_tile + k + average_length - 1]\n - shared[tx * thread_tile + k + average_length - 1 - average_length]) / np.float64(average_length)\n \n out_arr[i] = s[0]\n \n \ngpu_in = df['in'].to_gpu_array()\ngpu_out = df['out'].to_gpu_array()\nstart = time.time()\nkernel1[(number_of_blocks,), (number_of_threads,)](gpu_in, gpu_out,\n average_window, array_len)\ncuda.synchronize()\nend = time.time()\nprint('Numba with comipile time', end-start)\n \nstart = time.time()\nkernel1[(number_of_blocks,), (number_of_threads,)](gpu_in, gpu_out,\n average_window, array_len)\ncuda.synchronize()\nend = time.time()\nprint('Numba without comipile time', end-start)\n \npdf = pd.DataFrame()\npdf['in'] = np.arange(array_len, dtype=np.float64)\nstart = time.time()\npdf['out'] = pdf.rolling(average_window).mean()\nend = time.time()\nprint('pandas time', end-start)\n \nassert(np.isclose(pdf.out.values[average_window:].mean(),\n df.out.to_array()[average_window:].mean()))",
"Numba with comipile time 0.6331000328063965\nNumba without comipile time 0.30219364166259766\npandas time 6.03054666519165\n"
]
],
[
[
"After this change, the computation time is reduced to 0.3s without kernel compilation time, we achieved a total of 6x speedup compared with the baseline.",
"_____no_output_____"
],
[
"## Conclusion\n\nIn this tutorial, we take advantage of CUDA programming model in the Numba library to do moving average computation. We show by using a few CUDA programming tricks, we can achieve **6x** speed up in moving average computations for long arrays.\n\ncuDF is a powerful tool for data scientists to use. It provides the high-level API that covers most of the use cases. However, it also exposes its low-level components. Those components including gpu_array and Numba integration make the cuDF library to be very flexible to process data in a customized way. ",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
]
|
e7957dd29e4b23d70807db603f5bd876275c4d5e | 270,513 | ipynb | Jupyter Notebook | train-slimming.ipynb | jsg921019/pruning | b596e3ff2edc2e3bbd8db8be628e421be1f26054 | [
"MIT"
]
| null | null | null | train-slimming.ipynb | jsg921019/pruning | b596e3ff2edc2e3bbd8db8be628e421be1f26054 | [
"MIT"
]
| null | null | null | train-slimming.ipynb | jsg921019/pruning | b596e3ff2edc2e3bbd8db8be628e421be1f26054 | [
"MIT"
]
| null | null | null | 112.807756 | 200,316 | 0.85614 | [
[
[
"import glob\nimport os\nfrom typing import Any, Dict, List, Tuple, Union\n\nimport torch\nimport yaml\nfrom torch.utils.data import DataLoader, random_split\nfrom torchvision.datasets import ImageFolder, VisionDataset\n\nfrom torchvision import transforms\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom tqdm import tqdm\n\nfrom model import CustomVGG\nimport torchvision",
"_____no_output_____"
],
[
"device = 'cuda'\nlr = 0.0001\nnum_epoch = 100\nl1_weight = 0.01\n\ninput_size = 128\nbatch_size = 128\nn_worker = 8",
"_____no_output_____"
],
[
"import torchvision.transforms.functional as TF\nimport random\n\nclass MyRotationTransform:\n \"\"\"Rotate by one of the given angles.\"\"\"\n\n def __init__(self, angles):\n self.angles = angles\n\n def __call__(self, x):\n angle = random.choice(self.angles)\n return TF.rotate(x, angle)\n\nrotation_transform = MyRotationTransform(angles=[0, 180, 90, 270])",
"_____no_output_____"
],
[
"normalize = transforms.Normalize(mean= [0.485, 0.456, 0.406],\n std= [0.229, 0.224, 0.225])\n\ntrain_dataset = ImageFolder(\n \"/opt/ml/data/train\", transforms.Compose([\n transforms.Resize((input_size, input_size)),\n #transforms.RandomCrop(input_size),\n transforms.RandomHorizontalFlip(),\n #transforms.ColorJitter(brightness=0.5, contrast=0.2, saturation=0.5, hue=0.1),\n MyRotationTransform(angles=[0, 180, 90, 270]),\n transforms.ToTensor(),\n normalize,\n ]))\n\nval_dataset = ImageFolder(\"/opt/ml/data/val\", transforms.Compose([\n transforms.Resize((input_size, input_size)),\n #transforms.Resize(int(input_size/0.875)),\n #transforms.CenterCrop(input_size),\n transforms.ToTensor(),\n normalize,\n ]))",
"_____no_output_____"
],
[
"from torch.utils.data.sampler import WeightedRandomSampler\n\nsample_freq = [1169, 4826, 1020, 2655, 4879, 1092] #[0] * len(train_dataset.classes) #df_ff.gender.value_counts().sort_index().to_numpy()\nsample_weight = np.concatenate([[1/f]*f for f in sample_freq])\nsample_weight = torch.from_numpy(sample_weight)\nsampler = WeightedRandomSampler(sample_weight.type('torch.DoubleTensor'), len(sample_weight)//2)",
"_____no_output_____"
],
[
"idx = np.random.randint(0, len(train_dataset))\n\nfig, axes = plt.subplots(1, 5, figsize=(15, 3))\nfor i in range(5):\n img, label = train_dataset[idx]\n img = img.permute(1,2,0) * np.array([0.229, 0.224, 0.225]) + np.array([0.485, 0.456, 0.406])\n axes[i].imshow(img)\n axes[i].set_title(train_dataset.classes[label])\n axes[i].axis('off')\nplt.show()",
"Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\nClipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\nClipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\nClipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\nClipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\n"
],
[
"train_loader = torch.utils.data.DataLoader(\n train_dataset,\n batch_size=batch_size, drop_last=True, sampler = sampler,\n num_workers=n_worker)\n\nval_loader = torch.utils.data.DataLoader(\n val_dataset,\n batch_size=batch_size, shuffle=False,\n num_workers=n_worker)\n\ndataloaders = { 'train' : train_loader, 'valid' : val_loader}",
"_____no_output_____"
],
[
"model = CustomVGG()\nmodel.to(device)\nmodel.load_state_dict(torch.load('save/vgg9_112.pt'))",
"_____no_output_____"
],
[
"model = torchvision.models.vgg19_bn(pretrained=True)\nwith torch.no_grad():\n for m in model.features:\n if isinstance(m, torch.nn.Conv2d):\n m.bias = None\n # elif isinstance(m, torch.nn.BatchNorm2d):\n # m.weight.fill_(0.5)\nmodel.avgpool = torch.nn.AvgPool2d(4)\nmodel.classifier = torch.nn.Linear(512, 6)\nmodel.to(device)",
"_____no_output_____"
],
[
"import torch.optim as optim\nfrom torch.nn import CrossEntropyLoss\n\ncriterion = CrossEntropyLoss()\noptimizer = optim.Adam(model.parameters(), lr=lr)\nlr_scheduler = optim.lr_scheduler.ReduceLROnPlateau(optimizer, 'max', verbose=True, patience=5) # optim.lr_scheduler.CosineAnnealingLR(optimizer, T_max=60, eta_min=0.000001)#",
"_____no_output_____"
],
[
"from trainer_reg import Trainer\n\ntrainer = Trainer('/opt/ml/code/save', seed=42)",
"_____no_output_____"
],
[
"trainer.train(model, dataloaders, criterion, optimizer, lr_scheduler, num_epoch, 10, l1_weight, 'test')",
"Epoch 1/100\n----------\n"
],
[
"bn = []\n\nfor m in model.modules():\n if isinstance(m, torch.nn.BatchNorm2d):\n #print(m.weight)\n #break\n bn += m.weight.tolist()",
"_____no_output_____"
],
[
"model_base = CustomVGG()\nmodel_base.load_state_dict(torch.load('save/vgg9_112_reg.pt'))\nmodel_base = model_base.eval()",
"_____no_output_____"
],
[
"bn_base = []\n\nfor m in model_base.modules():\n if isinstance(m, torch.nn.BatchNorm2d):\n #print(m.weight)\n #break\n bn_base += m.weight.tolist()",
"_____no_output_____"
],
[
"plt.plot(sorted(bn), label='regularized')\nplt.plot(sorted(bn_base), label='base')\nplt.legend()\nplt.show()",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e795817627ee1b090f542e84f8ba1f6e0a022c8f | 89,436 | ipynb | Jupyter Notebook | Data Science and Machine Learning/Machine-Learning-In-Python-THOROUGH/RECAP_DS/06_BIG_DATA/L05.ipynb | okara83/Becoming-a-Data-Scientist | f09a15f7f239b96b77a2f080c403b2f3e95c9650 | [
"MIT"
]
| null | null | null | Data Science and Machine Learning/Machine-Learning-In-Python-THOROUGH/RECAP_DS/06_BIG_DATA/L05.ipynb | okara83/Becoming-a-Data-Scientist | f09a15f7f239b96b77a2f080c403b2f3e95c9650 | [
"MIT"
]
| null | null | null | Data Science and Machine Learning/Machine-Learning-In-Python-THOROUGH/RECAP_DS/06_BIG_DATA/L05.ipynb | okara83/Becoming-a-Data-Scientist | f09a15f7f239b96b77a2f080c403b2f3e95c9650 | [
"MIT"
]
| 2 | 2022-02-09T15:41:33.000Z | 2022-02-11T07:47:40.000Z | 44.495522 | 831 | 0.583736 | [
[
[
"# DS107 Big Data : Lesson Five Companion Notebook",
"_____no_output_____"
],
[
"### Table of Contents <a class=\"anchor\" id=\"DS107L5_toc\"></a>\n\n* [Table of Contents](#DS107L5_toc)\n * [Page 1 - Introduction](#DS107L5_page_1)\n * [Page 2 - Spark](#DS107L5_page_2)\n * [Page 3 - Running Spark in Hadoop](#DS107L5_page_3)\n * [Page 4 - Spark Data Storage](#DS107L5_page_4)\n * [Page 5 - Introduction to Scala](#DS107L5_page_5)\n * [Page 6 - Using Spark 2.0](#DS107L5_page_6)\n * [Page 7 - Using Spark SQL](#DS107L5_page_7)\n * [Page 8 - Spark Shell](#DS107L5_page_8)\n * [Page 9 - Decision Trees in Spark MLLib](#DS107L5_page_9)\n * [Page 10 - Decision Trees and Accuracy](#DS107L5_page_10)\n * [Page 11 - Hyperparameter Tuning](#DS107L5_page_11)\n * [Page 12 - Best Fit Model](#DS107L5_page_12)\n * [Page 13 - Key Terms](#DS107L5_page_13)\n * [Page 14 - Lesson 5 Practice Hands-On](#DS107L5_page_14)\n * [Page 15 - Lesson 5 Practice Hands-On Solution](#DS107L5_page_15)\n * [Page 16 - Lesson 5 Practice Hands-On Solution - Alternative Assignment](#DS107L5_page_16)\n ",
"_____no_output_____"
],
[
"<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n\n# Page 1 - Overview of this Module<a class=\"anchor\" id=\"DS107L5_page_1\"></a>\n\n[Back to Top](#DS107L5_toc)\n\n<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">",
"_____no_output_____"
]
],
[
[
"from IPython.display import VimeoVideo\n# Tutorial Video Name: Spark 2.0 and Zeppelin\nVimeoVideo('388865681', width=720, height=480)",
"_____no_output_____"
]
],
[
[
"The transcript for the above overview video **[is located here](https://repo.exeterlms.com/documents/V2/DataScience/Video-Transcripts/DSO107L05overview.zip)**.\n\n# Introduction\n\nProbably the most useful and versatile big data program you could utilize is *Spark*. It has wide-reaching functionality, including a SQL and a machine learning module, and can be used in many different languages. In this lesson, you will learn about: \n\n* Different components of Spark\n* Ways to run Spark on your Hadoop cluster\n* Apache Zeppelin, a notebook interface for Hadoop\n* Three ways to store data in Spark\n* Scala basics\n* Using Spark 2.0\n* Using Spark SQL\n* Launching the Spark Shell\n* Decision Trees in Spark using Scala\n\nThis lesson will culminate in a hands-on in which you perform your own decision tree machine learning model in Spark and Scala.\n\n<div class=\"panel panel-success\">\n <div class=\"panel-heading\">\n <h3 class=\"panel-title\">Additional Info!</h3>\n </div>\n <div class=\"panel-body\">\n <p>You may want to watch this <a href=\"https://vimeo.com/458401059\"> recorded live workshop on the concepts in this lesson. </a> </p>\n </div>\n</div>",
"_____no_output_____"
],
[
"<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n\n# Page 2 - Spark<a class=\"anchor\" id=\"DS107L5_page_2\"></a>\n\n[Back to Top](#DS107L5_toc)\n\n<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n",
"_____no_output_____"
],
[
"# Spark\n\n*Spark* is a data processing program built on top of MapReduce in the Hadoop ecosystem. Among many, many other things, it allows you to perform machine learning, data mining, and data streaming. It is powerful, fast, and scalable. How fast, you ask? 100 times faster than MapReduce, which is why MapReduce has pretty much become obsolete as an actual tool, though it remains an incredibly important and foundational big data concept. It uses the same kind of framework as TEZ to work backwards and find the fastest solution to your queries. Spark is actually written in *Scala*, but you can code in Spark using Python, Scala, or Java, with Python and Scala being the most popular languages. When using Spark, there are a lot of similarities between Python and Scala.\n\n---\n\n## Spark Components \n\nSpark has come a long way in the last several years, and now has a lot of different components that you can utilize within Spark to get varying data science tasks completed. These components include:\n\n* **Spark Core:** This is the base program for Spark. It is also known as *Spark 1.0*. \n* **Spark Streaming:** Allows you to feed in real-time data and provide real-time output. \n* **Spark SQL:** Write SQL queries and use SQLite functions in Spark. This is part of *Spark 2.0.* \n* **MLLib:** A library of machine learning and data mining tools you can use in Spark. This is also part of *Spark 2.0*.\n* **GraphX:** Allows you to create social network graphs and determine the degrees of separation in your data.\n\nOf these Spark components, you will be introduced to all but GraphX, which is quite specialized.\n\n---\n\n## Basic Programming Steps in Spark\n\nAlthough you will be using Spark to do all kinds of big data work, there are a few general steps that you will most likely always take when working in Spark:\n\n* Run transformations on the input data set\n* Run actions on the transformed data that can be stored or used\n* Work further with the results in a distributed fashion and see where to go next.\n\n---\n\n## Spark SQL\n\nSpark SQL allows you to transform your Spark DataFrames and DataSets into SQL tables, so that you can run SQL queries in Spark. With Spark SQL, you have the ability to read and write to a variety of file types, including JSON, Hive, and Parquet, and you can communicate with database connectors and with Tableau. \n\n---",
"_____no_output_____"
],
[
"<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n\n# Page 3 - Running Spark in Hadoop<a class=\"anchor\" id=\"DS107L5_page_3\"></a>\n\n[Back to Top](#DS107L5_toc)\n\n<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n",
"_____no_output_____"
],
[
"# Running Spark in Hadoop\n\nThere are three ways you can interact with Spark on your Hadoop cluster: \n\n1. Write files in a text editor and then run them through the command prompt. \n2. Use the Spark Shell.\n3. Interact with Spark in *Zeppelin*, which is a notebook system similar to Jupyter Notebook that you can access on Hadoop.\n\nYou will make use of Apache Zeppelin over the next few pages, then you'll switch to using the Spark Shell.\n\n---\n\n## Apache Zeppelin\n\nLuckily for you, Zeppelin comes pre-installed on your Hortonworks instance of Hadoop, and Zeppelin even comes with interaction to Spark 1.0 and 2.0, so that you can hit the ground running! You will access your Zeppelin notebook by typing **[http://127.0.0.1:9995](http://127.0.0.1:9995)** into your browser. \n\nHere's what it will look like when you get there:\n\n\n\n<div class=\"panel panel-success\">\n <div class=\"panel-heading\">\n <h3 class=\"panel-title\">Additional Info!</h3>\n </div>\n <div class=\"panel-body\">\n <p>You will learn the basics here, but if you want more tutorials, Zeppelin just happens to have them for you here - in particular look at the ones labeled Lab... and the ones labeled Zeppelin Tutorial if you want to go into more detail!</p>\n </div>\n</div>\n\nYou can get started by clicking on `Create new note`. When you do, it will ask you to give it a name:\n\n\n\nOnce you do, you'll get to this section here:\n\n\n\nYou will be able to type code into the cell, and just like Jupyter Notebook, either pressing the arrow button or typing `shift + enter` will run the cell.\n\n---\n\n### Ensure it Interprets Spark and Markdown\n\nClick the little black gear icon in the upper right hand corner of your Zeppelin notebook, which will bring up a menu looking something like this:\n\n\n\nYou will want to ensure that `spark` is first on that list, followed by `md`, for Markdown. Changing these settings makes sure that the Notebook processes the right type of languages or programs.\n\n---\n\n### Tell Zeppelin the Language\n\nThe `%` sign tells Zeppelin what language you want to use for each cell. Try out something in Markdown, just to get a feel for it. Type in:\n\n```\n%md\n# Testing out Markdown\nThis is text\n```\n\nAnd hit `shift + enter`. It should start running for you, and when it's done, the information you have should be output in Markdown, which can make web text much prettier. You can use Markdown for all your notes, just like you do in Jupyter.\n\n\n\n---",
"_____no_output_____"
],
[
"<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n\n# Page 4 - Spark Data Storage<a class=\"anchor\" id=\"DS107L5_page_4\"></a>\n\n[Back to Top](#DS107L5_toc)\n\n<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n",
"_____no_output_____"
],
[
"# Spark Data Storage\n\nThere are three main ways that data can be stored in Spark:\n\n* Resilient Distributed Datasets (RDDs)\n* DataSets\n* DataFrames\n\nRDDs are an artifact of Spark 1.0, and they will covered in more detail later on. This lesson will cover both DataSets and DataFrames, which are part of Spark 2.0's architecture. *RDDs* are a type of data storage that distribute your data across your Hadoop cluster, but they are somewhat slow. Mostly, you will only utilize them in Spark 1.0. \n\n*DataSets* are similar to RDDs, but they speed up the process, by utilizing more efficient memory representation in Spark. \n\nA *DataFrame* is a subclass of a DataSet, and they are specifically meant for relational data. A DataFrame keeps ahold of rows and columns in Spark, which allows you to do more with your data.\n\n<div class=\"panel panel-info\">\n <div class=\"panel-heading\">\n <h3 class=\"panel-title\">Tip!</h3>\n </div>\n <div class=\"panel-body\">\n <p>If you ever need to really optimize something, and pick up the speed of your work, use DataSets instead of DataFrames, since they don't have to store a structure.</p>\n </div>\n</div>\n\n---",
"_____no_output_____"
],
[
"<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n\n# Page 5 - Introduction to Scala<a class=\"anchor\" id=\"DS107L5_page_5\"></a>\n\n[Back to Top](#DS107L5_toc)\n\n<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n",
"_____no_output_____"
],
[
"# Introduction to Scala\n\nAlthough you can and will utilize Spark through Python in later lessons, which together is called *PySpark*, this lesson will make use of Scala. That means you can check another language off your bucket list! There are a few advantages to using Scala when it comes to Spark, since Spark is natively written in Scala. Those advantages include:\n\n* Your program is more likely to run as intended, because your code does not have to get translated between languages or environments.\n* Access to the most recent updates. It takes time to translate the most recent changes into other languages, so you might be a step or two behind in technology without using Scala.\n* You will understand Spark better, because it is natively written in Scala.\n* You save time and are more efficient because you will never need to switch between languages when using Spark.\n\n<div class=\"panel panel-success\">\n <div class=\"panel-heading\">\n <h3 class=\"panel-title\">Additional Info!</h3>\n </div>\n <div class=\"panel-body\">\n <p>Here is a list of <a href=\"https://alvinalexander.com/scala/scala-data-types-bits-ranges-int-short-long-float-double\"> all the different data types in Scala! </a></p>\n </div>\n</div>\n\n---\n\n## Values and Variables in Scala\n\nOften you will see in Scala code the designations of `val` and `var`. `val` stands for a value, and it is something that cannot be changed once it has been assigned. `var` stands for variable, and you can change it. When creating objects, you will need to designate them as either `val` or `var`, with `val` being by far the most common.\n\n---\n\n## How to Comment in Scala\n\nYou can comment out code in scala for a single line using `//` at the beginning. It would look something like this:\n\n```scala\n//This is a comment! Scala won't read it as code.\n```\n\n---",
"_____no_output_____"
],
[
"<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n\n# Page 6 - Using Spark 2.0<a class=\"anchor\" id=\"DS107L5_page_6\"></a>\n\n[Back to Top](#DS107L5_toc)\n\n<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n",
"_____no_output_____"
],
[
"# Using Spark 2.0\n\nNow that you've been introduced to Zeppelin, and thus have the environment you'll use to interact with Spark, you will really start to get into Spark 2.0 and Scala! You will do the work on this page in Zeppelin.\n\n---\n\n## Reading in Data\n\nThe first thing you will do is read in your data. For learning Spark 2.0 and Spark SQL, you will be looking at **[data with movie ratings](https://repo.exeterlms.com/documents/V2/DataScience/Big-Data/u.data.zip)**. There is also a file that has the **[movie names](https://repo.exeterlms.com/documents/V2/DataScience/Big-Data/u.item.zip)** contained in it. Make sure you upload these files to the `files` view, which will allow you to follow along with the lesson.\n\nYou'll start by creating a line that identifies the structure that your data will have:\n\n```scala\nfinal case class Table(movieID: Int, rating: Int)\n```\n\n`Table` is just the name you will give to your data; it could be anything you like. Within the parentheses, you have key-value pairs that represents the header and data type of each column. In this case, the data only has two rows, so that keeps it nice and simple! Further, both are integers (`Int`), making this even easier.\n\nThen, you will need to transform your file into an RDD, by mapping. In Scala, you will always start this with `val`. Next, you'll give your RDD a name; in this case, `MappedTable`. Then you will call `sc` for `Spark Context`, which just tells Zeppelin you are using Spark, and use the `textFile.map` command to map a flat text file into a usable RDD. You will put the file pathway in for the data. Since you uploaded it earlier to your `maria_dev` section of your `Files View` in Ambari, the pathway below should work for you.\n\nNext, call the `map` function. Here, you are basically separating your data into columns. `x =>` basically tells it that you are going to define the way to split things next, and in the curly brackets `{}`, you will find that map of how to split, based on `val fields`. This uses the `.split` function to break at the delimiter specified in the parentheses - in this case, tab: `\\t`. You could also split by other things, including, but not limited to, commas. Then, you will call the `Table` structure you created earlier, and map each field to the appropriate data structure. Each field will be numbered in parentheses (i.e.`fields(1)`), and should be followed by `.to` the data type. Since these were both `Int`, they are going to `Int`. The data type you specified above for `Table` must match what you call here.\n\n```scala\nval MappedTable = sc.textFile(\"hdfs:///user/maria_dev/u.data\").map(x => {val fields = x.split(\"\\t\"); Table(fields(1).toInt, fields(2).toInt)})\n```\n\nWhen you run those two lines together up above, you will most likely get a warning looking something like this:\n\n```text\nwarning: there were 1 unchecked warning(s); re-run with -unchecked for details\ndefined class Rating\nlines: org.apache.spark.rdd.RDD[Rating] = MapPartitionsRDD[85] at map at <console>:43\n```\n\nThat is perfectly fine; just a warning and something you can ignore.\n\n---\n\n## Making Data into a Spark 2.0 DataFrame\n\nYou now have your data in an RDD, which is serviceable, but slow and clunky compared to Spark 2.0's concept of a DataFrame. So, from this RDD, you will transfer to a DataFrame. Start by importing `sqlContext.implicits._`, which is what tells Zeppelin that you are now working with Spark 2.0.\n\nThen, you will use the function `.toDF()` to turn the `MappedTable` from above into a DataFrame. You will need to start the line with `val` and give it a name; in this case `DFtable` is being used.\n\nLastly, you can call the `printSchema()` function on your newly created DataFrame, which will provide you with the structure of the data.\n\n```scala\nimport sqlContext.implicits._\nval DFtable = MappedTable.toDF()\n\nDFtable.printSchema()\n```\n\nHere is the end result: \n\n```text\n |-- movieID: integer (nullable = false)\n |-- rating: integer (nullable = false)\n```\n\nAs you can see, they are both integers and they do not allow null values.\n\n---\n\n## Find the Most Popular Movies\n\nNow, you can leverage the power of Spark 2.0 and Scala to find the highest rated movies! You'll make use of the functions `groupBy()`, `count()` and `orderBy()` to produce some meaningful results: \n\n```scala\nval MostPopularMovies = DFtable.groupBy(\"movieID\").count().orderBy(desc(\"count\")).cache()\nMostPopularMovies.show()\n```\n\nThe code above first creates a table named `MostPopularMovies`. Then it uses the `DFtable` you created in the previously code line to group the data by `movieID` and get a count of how many times each `movieID` was referenced. Then, it orders the data by that `count` that was just created. Finally, this newly created table is cached (`.cache()`), so that it will stay in memory and will make referencing it faster.\n\n<div class=\"panel panel-info\">\n <div class=\"panel-heading\">\n <h3 class=\"panel-title\">To Cache or Not to Cache?</h3>\n </div>\n <div class=\"panel-body\">\n <p>That 'tis the question! Typically, if you are going to use data more than once and it is relatively small, you will save more memory and data usage on your cluster than if you didn't cache.</p>\n </div>\n</div>\n\nWith all that done, it is easy to use the `.show()` command, which is much like printing something, to display your results:\n\n```text\ntopMovieIDS: org.apache.spark.sql.DataFrame = [movieID: int, count: bigint]\n+-------+-----+\n|movieID|count|\n+-------+-----+\n| 50| 583|\n| 258| 509|\n| 100| 508|\n| 181| 507|\n| 294| 485|\n| 286| 481|\n| 288| 478|\n| 1| 452|\n| 300| 431|\n| 121| 429|\n| 174| 420|\n| 127| 413|\n| 56| 394|\n| 7| 392|\n| 98| 390|\n| 237| 384|\n| 117| 378|\n| 172| 367|\n| 222| 365|\n| 313| 350|\n+-------+-----+\nonly showing top 20 rows\n```\n\nAnd there you have it! Of course, without a key for what the `movieID`s are, this is not incredibly useful, but you should still be proud of your first execution in Spark 2.0!\n\n---",
"_____no_output_____"
],
[
"<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n\n# Page 7 - Using Spark SQL<a class=\"anchor\" id=\"DS107L5_page_7\"></a>\n\n[Back to Top](#DS107L5_toc)\n\n<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n",
"_____no_output_____"
],
[
"# Using Spark SQL\n\nInstead of using SQL through Pig or Hive, you could also use SQL through Spark! In a lovely, in-line interface like Zeppelin! Keep reading to get in on the fun.\n\n---\n\n## Create a Temporary Table\n\nFirst, you need to create a temporary table from the data frame you had created on the last page, using the function `registerTempTable`. You'll give this new table a name called `MovieRatings`.\n\n```scala\nDFtable.registerTempTable(\"MovieRatings\")\n```\n\n---\n\n## Run SQL Queries\n\nThen you're all set to run SQL queries! \n\n---\n\n### Show the First Ten Movies\n\nThese can be as simple as just showing the first ten movie ratings:\n\n```sql\n%sql\n\nselect * from MovieRatings limit 10\n```\n\nIn order to use Spark SQL, you'll need to include the `%sql` in your notebook, so that Zeppelin knows what type of code it is! And here is the result:\n\n\n\n<div class=\"panel panel-info\">\n <div class=\"panel-heading\">\n <h3 class=\"panel-title\">Tip!</h3>\n </div>\n <div class=\"panel-body\">\n <p>In Spark SQL, the only thing that differs is that you DON'T end your lines with a semi-colon! It won't run if you do.</p>\n </div>\n</div>\n\n---\n\n### Show the Frequency of Ratings\n\nNow, try and step up the complexity a notch. How about using the `count` function to get the frequency of each rating type? Ratings range from 1-5. \n\n```sql\n%sql\n\nselect rating, count(*) as count from MovieRatings group by rating\n```\n\nAnd here is that result:\n\n\n\n---\n\n## Visualize your Data\n\nIf your mind isn't blown already, get ready to hang on to something! Because Zeppelin in conjunction with Spark 2.0 SQL can automatically visualize your results! You just need to press a button! \n\n\n\nThe buttons from left to right show the following:\n\n* Table\n* Bar graph (like above)\n* Pie chart\n* Area graph\n* Line plot\n* Scatter plot\n\nYou can play with them here, but with frequencies and a category, bar chart is definitely the appropriate visual for this data.\n\n---\n\n## Find the Most Popular Movie\n\nWhat if you wanted to join some data, so you can actually find out the name of the most popular movie? Well, you can bring in data from the **[u.item]()** file as well, which contains movie IDs and titles, and merge it with your current data!\n\nThe first step is to read in that file:\n\n```scala\nfinal case class Movie(movieID: Int, title: String)\n\nval lines = sc.textFile(\"hdfs:///user/maria_dev/u.item\").map(x => {val fields = x.split('|'); Movie(fields(0).toInt, fields(1))})\n\nimport sqlContext.implicits._\nval moviesDF = lines.toDF()\n\nmoviesDF.show()\n```\n\nYou should get something like this:\n\n```text\nines: org.apache.spark.rdd.RDD[Movie] = MapPartitionsRDD[7] at map at <console>:34\nimport sqlContext.implicits._\nmoviesDF: org.apache.spark.sql.DataFrame = [movieID: int, title: string]\n+-------+--------------------+\n|movieID| title|\n+-------+--------------------+\n| 1| Toy Story (1995)|\n| 2| GoldenEye (1995)|\n| 3| Four Rooms (1995)|\n| 4| Get Shorty (1995)|\n| 5| Copycat (1995)|\n| 6|Shanghai Triad (Y...|\n| 7|Twelve Monkeys (1...|\n| 8| Babe (1995)|\n| 9|Dead Man Walking ...|\n| 10| Richard III (1995)|\n| 11|Seven (Se7en) (1995)|\n| 12|Usual Suspects, T...|\n| 13|Mighty Aphrodite ...|\n| 14| Postino, Il (1994)|\n| 15|Mr. Holland's Opu...|\n| 16|French Twist (Gaz...|\n| 17|From Dusk Till Da...|\n| 18|White Balloon, Th...|\n| 19|Antonia's Line (1...|\n| 20|Angels and Insect...|\n+-------+--------------------+\nonly showing top 20 rows\n```\n\nThen the next step is to create a temporary table:\n\n```scala\nmoviesDF.registerTempTable(\"MovieTitles\")\n```\n\nAnd then lastly, you can do your SQL query:\n\n```sql\n%sql \n\nselect t.title, count(*) count from MovieRatings r join MovieTitles t on r.movieID = t.movieID group by t.title order by count desc limit 20\n```\n\nYou will give the table `MovieRatings` an alias of `r` and the `MovieTitles` table an alias of `t` to make this a little easier to join. Then you'll count the ratings, group by the title, and order by the count. Results should look like this:\n\n\n\nNote that you can't see all 20 results, but that there is a scroll bar on the right that can help.\n\n---",
"_____no_output_____"
],
[
"<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n\n# Page 8 - Spark Shell<a class=\"anchor\" id=\"DS107L5_page_8\"></a>\n\n[Back to Top](#DS107L5_toc)\n\n<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n",
"_____no_output_____"
],
[
"# Spark Shell\n\nNext, you will move into using the Spark MLLib, and for best results, this must be done in the Spark Shell and in Scala. The next several pages contain directions for how run decision trees in Spark MLLib, using Scala. Get your swagger on, because this officially makes you cool! Or geeky. Do you find the line is so fine?\n\n---\n\n## Specify the Spark Version\n\nYour Hadoop Cluster comes with both versions of Spark, 1.0 and 2.0, so you will need to specify which version you want to use. If you don't, it will run 1.0 by default, which won't be any help to you, since Spark MLLib is contained within Spark 2.0. \n\n```bash\nexport SPARK_MAJOR_VERSION=2\n```\n\nNothing should happen when you run this code, so if you get a clean line, you are good to go.\n\n<div class=\"panel panel-danger\">\n <div class=\"panel-heading\">\n <h3 class=\"panel-title\">Caution!</h3>\n </div>\n <div class=\"panel-body\">\n <p>You will need to run the export line every time you run the shell - it is not a permanent setting that sticks around.</p>\n </div>\n</div>\n\n---\n\n## Start Spark Shell Locally\n\nThen you can open up the Spark Shell, using ths command. This has you opening Spark on your local machine, and not through your Hadoop cluster, because you don't actually have a real cluster with multiple nodes here.\n\n```bash\nspark-shell --master local[*]\n```\n\nThis may take a little while - so don't be alarmed if you have enough time to grab a cup o' tea!\n\n---\n\n### Start Spark through YARN\n\nIf you were using this in a real big data situation, in which you had multiple nodes, you would use a command like this, which has you open Spark through YARN:\n\n```bash\nspark-shell --master yarn --deploy-mode client\n```\n\nNow that you are in the Spark Shell, there are all sorts of things you can do to interact with Spark. You will know you are in and ready to roll when you see the `scala>` prompt.\n\n---\n\n## Exiting Spark Shell\n\nTo exit the Spark Shell, use `Ctrl + C`. \n\n---",
"_____no_output_____"
],
[
"<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n\n# Page 9 - Decision Trees in Spark MLLib<a class=\"anchor\" id=\"DS107L5_page_9\"></a>\n\n[Back to Top](#DS107L5_toc)\n\n<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n",
"_____no_output_____"
],
[
"# Decision Trees in Spark MLLib\n\nNow that you're into Spark Shell, you will need to do a fair amount of data wrangling and prep work before you can actually launch into your decision tree model. You'll need to read in your data, change the outcome variable data type, split your data up into testing and training data sets, and create a feature vector which contains all of your predictor variables.\n\n---\n\n## Read in Data\n\nFirst, you will need to read in your data. Spark 2.0 allows you to easily read-in CSVs, with options to bring with it the schema, or structure, of the data, and the headers. Brilliant! You'll be using **[a dataset](https://repo.exeterlms.com/documents/V2/DataScience/Big-Data/glass1.zip)** that will allow you to predict what kind of glass you have based on the component elements in it. \n\nThe `Type` variable is the type of glass, and the options are:\n\n* 1: Building Windows Float Processed\n* 2: Building Windows Non-Float Processed\n* 3: Vehicle Windows Float Processed\n* 4: Vehicle Windows Non-Float Processed\n* 5: Containers\n* 6: Tableware\n* 7: Headlamps\n\nFor those of you who care, float processed glass is made by floating molten glass along on a bed of molten metal. Hot stuff! \n\nMake sure you add this dataset to your `Files` view in Ambari in the `user/maria_dev` folder, so that you can use the code below to read in your data.\n\n```scala\nval data = spark.read.\noption(\"inferSchema\", true).\noption(\"header\", true).\ncsv(\"hdfs:///user/maria_dev/glass1.csv\")\n```\n\nThe code above tells Spark to pull in the structure of your data with the `inferSchema` option, and that your data has headers (`header` option). You'll also use the argument `csv` because your data is a CSV file.\n\nIf this works for you, Scala should spit out some basic information about your structure:\n\n```text\ndata: org.apache.spark.sql.DataFrame = [RI: double, Na: double ... 8 more fields]\n```\n\n---\n\n## Convert Outcome Variable to Double\n\nWhen you actually get to running your decision trees, all of the variables must be doubles. So, you'll need to convert any that aren't. You can do that by *casting* the variable to `double`. \n\n```scala\nval data1 = data.\nwithColumn(\"Type\", $\"Type\".cast(\"double\"))\n```\n\n---\n\n## Train Test Split\n\nThen, you need to split your data into training and testing data. In this case, you are keeping 90% of the data for training, and 10% for testing. Of the 90% of the training data, you will actually later be reserving 10% for additional testing, so if the high percentage of training data surprised you, it's actually really an 80-20 split rather than 90-10; it just doesn't look like it here.\n\n```scala\nval Array(trainData, testData) = data1.randomSplit(Array(0.9, 0.1))\ntrainData.cache()\ntestData.cache()\n```\n\nIf that has worked, Scala will echo back the fields for the training and the testing data:\n\n```text\nres0: trainData.type = [RI: double, Na: double ... 8 more fields]\nres1: testData.type = [RI: double, Na: double ... 8 more fields]\n```\n\n---\n\n## Create a Feature Vector\n\nNext, you will prep your data for machine learning in Spark MLLib. When you feed in data, it does not allow more than one column, so you will need to arrange all your data into only one column, which has a value of vector. Luckily, there is a function for this: `VectorAssembler`. \n\nIn the second line of the code below, you will state that you want to utilize all columns except for `Type`, which is what you are going to predict - the type of glass. The `_!=` is what specifies the exception. Then, in line 3, you will make use of the `VectorAssembler()` function to put the rest of the columns altogether in one vector. You'll then actually run this on your `trainData`, and then `show()` it, so you know it worked. \n\n```scala\nimport org.apache.spark.ml.feature.VectorAssembler\nval inputCols = trainData.columns.filter(_ != \"Type\")\nval assembler = new VectorAssembler().\nsetInputCols(inputCols).\nsetOutputCol(\"featureVector\")\nval assembledTrainData = assembler.transform(trainData)\nassembledTrainData.select(\"featureVector\").show(truncate = false)\n```\n\nYou should get output looking like this back, showing your vector:\n\n```text\n19/11/15 04:09:05 WARN Executor: 1 block locks were not released by TID = 4:\n[rdd_9_0]\n+--------------------------------------------------+\n|featureVector |\n+--------------------------------------------------+\n|[1.51115,17.38,0.0,0.34,75.41,0.0,6.65,0.0,0.0] |\n|[1.51131,13.69,3.2,1.81,72.81,1.76,5.43,1.19,0.0] |\n|[1.51215,12.99,3.47,1.12,72.98,0.62,8.35,0.0,0.31]|\n|[1.51299,14.4,1.74,1.54,74.55,0.0,7.59,0.0,0.0] |\n|[1.51316,13.02,0.0,3.04,70.48,6.21,6.96,0.0,0.0] |\n|[1.51321,13.0,0.0,3.02,70.7,6.21,6.93,0.0,0.0] |\n|[1.51409,14.25,3.09,2.08,72.28,1.1,7.08,0.0,0.0] |\n|[1.51508,15.15,0.0,2.25,73.5,0.0,8.34,0.63,0.0] |\n|[1.51514,14.01,2.68,3.5,69.89,1.68,5.87,2.2,0.0] |\n|[1.51514,14.85,0.0,2.42,73.72,0.0,8.39,0.56,0.0] |\n|[1.51531,14.38,0.0,2.66,73.1,0.04,9.08,0.64,0.0] |\n|[1.51545,14.14,0.0,2.68,73.39,0.08,9.07,0.61,0.05]|\n|[1.51556,13.87,0.0,2.54,73.23,0.14,9.41,0.81,0.01]|\n|[1.51567,13.29,3.45,1.21,72.74,0.56,8.57,0.0,0.0] |\n|[1.51569,13.24,3.49,1.47,73.25,0.38,8.03,0.0,0.0] |\n|[1.51571,12.72,3.46,1.56,73.2,0.67,8.09,0.0,0.24] |\n|[1.51574,14.86,3.67,1.74,71.87,0.16,7.36,0.0,0.12]|\n|[1.5159,12.82,3.52,1.9,72.86,0.69,7.97,0.0,0.0] |\n|[1.5159,13.24,3.34,1.47,73.1,0.39,8.22,0.0,0.0] |\n|[1.51593,13.09,3.59,1.52,73.1,0.67,7.83,0.0,0.0] |\n+--------------------------------------------------+\nonly showing top 20 rows\n```\n\n---",
"_____no_output_____"
],
[
"<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n\n# Page 10 - Decision Trees and Accuracy<a class=\"anchor\" id=\"DS107L5_page_10\"></a>\n\n[Back to Top](#DS107L5_toc)\n\n<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">",
"_____no_output_____"
],
[
"# Decision Trees and Accuracy\n\nNow that you've done all the prep work, it's time to actually run your decision tree and see how accurate it is! A decision tree is a type of machine learning model in which the computer finds the best way to differentiate between outcomes.\n\n---\n\n## Decision Tree Classifier\n\nIt's time to actually run the decision tree classifier! You'll save it in a `val` named `model`, and if you print those lines out (`println`), you can see the different branches of the decision tree.\n\n\n```scala\nimport org.apache.spark.ml.classification.DecisionTreeClassifier\nimport scala.util.Random\nval classifier = new DecisionTreeClassifier().\nsetSeed(Random.nextLong()).\nsetLabelCol(\"Type\").\nsetFeaturesCol(\"featureVector\").\nsetPredictionCol(\"prediction\")\nval model = classifier.fit(assembledTrainData)\nprintln(model.toDebugString)\n```\n\nHere are the branching results, meaning all the steps that the algorithm took to separate out the different types of glass:\n\n```text\nDecisionTreeClassificationModel (uid=dtc_5e57af65a40f) of depth 5 with 37 nodes\n If (feature 7 <= 0.27)\n If (feature 3 <= 1.38)\n If (feature 2 <= 3.25)\n If (feature 0 <= 1.5202)\n If (feature 1 <= 13.78)\n Predict: 2.0\n Else (feature 1 > 13.78)\n Predict: 6.0\n Else (feature 0 > 1.5202)\n Predict: 2.0\n Else (feature 2 > 3.25)\n If (feature 0 <= 1.5167)\n If (feature 0 <= 1.51567)\n Predict: 1.0\n Else (feature 0 > 1.51567)\n Predict: 3.0\n Else (feature 0 > 1.5167)\n If (feature 2 <= 3.61)\n Predict: 1.0\n Else (feature 2 > 3.61)\n Predict: 1.0\n Else (feature 3 > 1.38)\n If (feature 2 <= 1.88)\n If (feature 1 <= 13.44)\n If (feature 0 <= 1.52172)\n Predict: 5.0\n Else (feature 0 > 1.52172)\n Predict: 2.0\n Else (feature 1 > 13.44)\n If (feature 0 <= 1.519)\n Predict: 6.0\n Else (feature 0 > 1.519)\n Predict: 2.0\n Else (feature 2 > 1.88)\n If (feature 5 <= 0.0)\n Predict: 6.0\n Else (feature 5 > 0.0)\n If (feature 6 <= 8.31)\n Predict: 2.0\n Else (feature 6 > 8.31)\n Predict: 2.0\n Else (feature 7 > 0.27)\n If (feature 1 <= 14.01)\n If (feature 4 <= 71.76)\n If (feature 0 <= 1.51567)\n Predict: 5.0\n Else (feature 0 > 1.51567)\n If (feature 0 <= 1.5202)\n Predict: 1.0\n Else (feature 0 > 1.5202)\n Predict: 2.0\n Else (feature 4 > 71.76)\n Predict: 7.0\n Else (feature 1 > 14.01)\n Predict: 7.0\n```\n\nYou'll notice that the features are just numbered, which makes this a little difficult to interpret, but since you have neither fed it a codebook nor could use one in a decision tree, since they all have to be doubles, this makes sense.\n\n---\n\n## Assess the Importance of Features\n\nNext, you can use the `model` you created to assess the importance of the features, or variables, in your decision tree. You'll use the function `featureImportances` to do so, and then can sort them in reverse order and print, so you get the most important feature first on your list!\n\n```scala\nmodel.featureImportances.toArray.zip(inputCols).\nsorted.reverse.foreach(println)\n```\n\nHere are the results:\n\n```text\n(0.2694959533428122,Mg)\n(0.22949708599977559,Ba)\n(0.17868395220045094,Al)\n(0.1575592774341163,RI)\n(0.08972321273293513,Na)\n(0.03538920228307928,K)\n(0.022147581913725102,Ca)\n(0.0175037340931054,Si)\n(0.0,Fe)\n```\n\nThe higher the number, the better, which is why they have been printed in reverse order. The weight is printed first for the feature, and then the feature name. So, you can see up above that the most important feature is whether Magnesium (Mg) is present in the glass, followed by whether Barium (Ba) is present in the glass, etc. The least important feature is Iron (Fe). \n\n---\n\n## See the Accuracy of the Training Data\n\nNow you can start investigating how well your model is doing. The first thing to do is to examine the training data, and see if the actual glass `Type` matches the `prediction` that the decision tree made:\n\n```scala\nval predictions = model.transform(assembledTrainData)\npredictions.select(\"Type\", \"prediction\", \"probability\").\nshow(truncate = false)\n```\n\nHere are the results from the first 20 rows:\n\n```text\n19/11/15 06:53:07 WARN Executor: 1 block locks were not released by TID = 24:\n[rdd_24_0]\n+----+----------+-------------------------------------------------------------------------------+\n|Type|prediction|probability |\n+----+----------+-------------------------------------------------------------------------------+\n|6.0 |6.0 |[0.0,0.0,0.0,0.0,0.0,0.0,1.0,0.0] |\n|1.0 |1.0 |[0.0,1.0,0.0,0.0,0.0,0.0,0.0,0.0] |\n|6.0 |6.0 |[0.0,0.0,0.0,0.0,0.0,0.0,1.0,0.0] |\n|5.0 |5.0 |[0.0,0.0,0.0,0.0,0.0,1.0,0.0,0.0] |\n|5.0 |5.0 |[0.0,0.0,0.0,0.0,0.0,1.0,0.0,0.0] |\n|2.0 |2.0 |[0.0,0.08571428571428572,0.9142857142857143,0.0,0.0,0.0,0.0,0.0] |\n|7.0 |7.0 |[0.0,0.0,0.0,0.0,0.0,0.0,0.0,1.0] |\n|5.0 |5.0 |[0.0,0.0,0.0,0.0,0.0,1.0,0.0,0.0] |\n|7.0 |7.0 |[0.0,0.0,0.0,0.0,0.0,0.0,0.0,1.0] |\n|7.0 |7.0 |[0.0,0.0,0.0,0.0,0.0,0.0,0.0,1.0] |\n|7.0 |7.0 |[0.0,0.0,0.0,0.0,0.0,0.0,0.0,1.0] |\n|1.0 |1.0 |[0.0,1.0,0.0,0.0,0.0,0.0,0.0,0.0] |\n|2.0 |2.0 |[0.0,0.08571428571428572,0.9142857142857143,0.0,0.0,0.0,0.0,0.0] |\n|1.0 |2.0 |[0.0,0.08571428571428572,0.9142857142857143,0.0,0.0,0.0,0.0,0.0] |\n|2.0 |2.0 |[0.0,0.08571428571428572,0.9142857142857143,0.0,0.0,0.0,0.0,0.0] |\n|2.0 |2.0 |[0.0,0.14285714285714285,0.5714285714285714,0.2857142857142857,0.0,0.0,0.0,0.0]|\n|2.0 |2.0 |[0.0,0.08571428571428572,0.9142857142857143,0.0,0.0,0.0,0.0,0.0] |\n|2.0 |2.0 |[0.0,0.08571428571428572,0.9142857142857143,0.0,0.0,0.0,0.0,0.0] |\n|2.0 |2.0 |[0.0,0.08571428571428572,0.9142857142857143,0.0,0.0,0.0,0.0,0.0] |\n|2.0 |2.0 |[0.0,0.08571428571428572,0.9142857142857143,0.0,0.0,0.0,0.0,0.0] |\n+----+----------+-------------------------------------------------------------------------------+\nonly showing top 20 rows\n```\n\nThe `Type` column is the actual data, and the `prediction` column is what the decision tree predicted based on the model. The `probability` column shows the likelihood that each `Type` is correct. So you can read the first row as: \n\n```text\nIndex - ignore\n0% chance that the glass type is 1\n0% chance that the glass type is 2\n0% chance that the glass type is 3\n0% chance that the glass type is 4\n0% chance that the glass type is 5\n100% chance that the glass type is 6\n0% chance that the glass type is 7\n```\n\nYou'll notice that there are actually eight numbers, not seven, even though there are only seven glass types. This is because the first probability value is just the zero index, and it will always show a probability of zero.\n\nLooking at just the first 20 rows, it looks like you have created a decently accurate decision tree, but you'll also want to take a look at the accuracy values as well. \n\n---\n\n## Print Total Model Accuracy Values\n\nJust looking at the first twenty rows is a good eyeball check, but doesn't give you the total accuracy for the model. Good thing the function `MulticlassClassificationEvaluator` has your back! \n\n```scala\nimport org.apache.spark.ml.evaluation.MulticlassClassificationEvaluator\nval evaluator = new MulticlassClassificationEvaluator().\nsetLabelCol(\"Type\").\nsetPredictionCol(\"prediction\")\nevaluator.setMetricName(\"accuracy\").evaluate(predictions)\n```\n\nHere is the result:\n\n```text\nres25: Double = 0.8201058201058201\n```\n\nThis shows that the model accurately predicts the glass type 82% of the time. That's not bad...but it could be much higher! \n\n<div class=\"panel panel-info\">\n <div class=\"panel-heading\">\n <h3 class=\"panel-title\">Tip!</h3>\n </div>\n <div class=\"panel-body\">\n <p>Your accuracy value may come out slightly different than what is here, because by default, the decisions in a decision tree are random, and your computer may have done it slightly differently than the instructor's!</p>\n </div>\n</div>\n\n---\n\n## Examine the Classification Matrix\n\nAnother way you can examine accuracy, which will give you a little more detail about what is going well and what isn't, is to look at a confusion matrix. This will compare the predictions to the actual data, so that you can see where the decision tree got it right, and if it didn't, as what type of glass it was misclassified.\n\n```scala\nval confusionMatrix = predictions.\ngroupBy(\"Type\").\npivot(\"prediction\", (1 to 7)).\ncount().\nna.fill(0.0).\norderBy(\"Type\")\nconfusionMatrix.show()\n```\n\nThe output shows the predicted versus actual types. Along the diagonal (54, 56, etc.) you will find the number that was correctly classified. So in reading this matrix, you find that for type 1, 54 pieces of glass were correctly classified as 1s and 8 were incorrectly classified as type 2. You want to see lots of zeros for things not on the diagonal, so at a glance, this it looking pretty decent.\n\n```text\n+----+---+---+---+---+---+---+---+\n|Type| 1| 2| 3| 4| 5| 6| 7|\n+----+---+---+---+---+---+---+---+\n| 1.0| 54| 8| 0| 0| 0| 0| 0|\n| 2.0| 11| 56| 2| 0| 0| 0| 0|\n| 3.0| 6| 4| 6| 0| 0| 0| 0|\n| 5.0| 0| 1| 0| 0| 11| 0| 0|\n| 6.0| 0| 0| 0| 0| 0| 7| 0|\n| 7.0| 1| 1| 0| 0| 0| 0| 21|\n+----+---+---+---+---+---+---+---+\n```\n\n---\n\n## Is Your Accuracy Better than Random?\n\nIt is pretty difficult to benchmark accuracy. Is 82% good? Bad? Ugly? One way to determine at first blush whether your accuracy is any good at all is to find out what the accuracy would be like if you were random guessing. Here's the code to attempt that:\n\n```scala\nimport org.apache.spark.sql.DataFrame\ndef classProbabilities(data: DataFrame): Array[Double] = {\nval total = data.count()\ndata.groupBy(\"Type\").count().\norderBy(\"Type\").\nselect(\"count\").as[Double].\nmap(_ / total).\ncollect()\n}\n\nval trainPriorProbabilities = classProbabilities(trainData)\nval testPriorProbabilities = classProbabilities(testData)\ntrainPriorProbabilities.zip(testPriorProbabilities).map {\ncase (trainProb, cvProb) => trainProb * cvProb\n}.sum\n```\n\nAnd here is the result:\n\n```text\nres33: Double = 0.2452910052910053\n```\n\nLooks like random guessing will give you an accuracy of 25%, so 82% accuracy with your decision tree is looking spectacular now, isn't it?! \n\n---",
"_____no_output_____"
],
[
"<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n\n# Page 11 - Hyperparameter Tuning<a class=\"anchor\" id=\"DS107L5_page_11\"></a>\n\n[Back to Top](#DS107L5_toc)\n\n<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">",
"_____no_output_____"
],
[
"# Hyperparameter Tuning\n\nThe decision tree was fun, and it was decently accurate. But why stop there? Don't you want to be the best you possibly can? Well, you can probably improve things by playing with the *hyperparameters*. Hyperparameters are the components of the way your model has been created, and by changing them, you can get a better model fit. A decision tree has the following hyperparameters:\n\n* **Maximum depth:** Limits the number of decisions you can make in a decision tree. Sometimes having too many can lead to overfitting of data.\n* **Maximum bins:** Limits the number of decision rules the decision tree can have. A lot will probably make your decision tree more accurate, but could take up too much processing power.\n* **Impurity measure:** *Purity* is how good you are at classifying accurately. If you have two categories, and each group only contains the appropriate data for that category, then you have complete purity. If you have some mix-up in there, then you have *impurity*. You want to have low impurity.\n* **Minimum information gain:** If you include a decision rule that does not make the data more pure, then what good is it? Including a hyperparameter of minimum information gain allows you to only keep levels that will actually add to the accuracy of your data, and it can help ensure you don't overfit the model.\n\n\n---\n\n## Create a Pipeline for Hyperparameter Tuning\n\nNow that you know what the hyperparameters are for decision trees, you can start playing with them! The code below will create a *pipeline* for your decision tree data, and then the next set of code after that will be for trying all different varieties of these hyperparameters, to see which fits the best. A pipeline is when you chain two operations together, so that you don't need to keep running them individually over and over again. Using something like this can take some time and processing power on the computer's end, but will save you the trouble of having to tune everything manually, one at a time, which would take forever!\n\n```scala\nimport org.apache.spark.ml.Pipeline\nval inputCols = trainData.columns.filter(_ != \"Type\")\nval assembler = new VectorAssembler().\nsetInputCols(inputCols).\nsetOutputCol(\"featureVector\")\nval classifier = new DecisionTreeClassifier().\nsetSeed(Random.nextLong()).\nsetLabelCol(\"Type\").\nsetFeaturesCol(\"featureVector\").\nsetPredictionCol(\"prediction\")\nval pipeline = new Pipeline().setStages(Array(assembler, classifier))\n```\n\n---\n\n## Set the Hyperparameter Boundaries and How to Determine Which is Best\n\nNext, you will set the hyperparameter boundaries for impurity, maximum depth, maximum bins, and minimum information gain. For `impurity`, you will try two different versions of impurity: `gini` and `entropy`. For `maxDepth`, you will try the values of 1 and 20. For `maxBins`, you will try all the values of 40 and 300, and for `minInfoGain`, you will try all the values of 0 and .05. All in all, you will be testing 16 models, since you have two values for each hyperparameter. Then, you'll get the accuracy for each of them!\n\n```scala\nimport org.apache.spark.ml.tuning.ParamGridBuilder\nval paramGrid = new ParamGridBuilder().\naddGrid(classifier.impurity, Seq(\"gini\", \"entropy\")).\naddGrid(classifier.maxDepth, Seq(1, 20)).\naddGrid(classifier.maxBins, Seq(40, 300)).\naddGrid(classifier.minInfoGain, Seq(0.0, 0.05)).\nbuild()\nval multiclassEval = new MulticlassClassificationEvaluator().\nsetLabelCol(\"Type\").\nsetPredictionCol(\"prediction\").\nsetMetricName(\"accuracy\")\n```\n\nThe output you will receive back just basically acknowledges your model creation. \n\n---\n\n## Train Test Split for Hyperparameter Tuning\n\nNow that you know what all you want to run, and have it saved as `multiclassEval`, you can run train test split again, so that you have a little bit of data in reserve to test the hyperparameter tuning. This ensures that you don't accidentally overfit your hyperparameters as well.\n\n```scala\nimport org.apache.spark.ml.tuning.TrainValidationSplit\nval validator = new TrainValidationSplit().\nsetSeed(Random.nextLong()).\nsetEstimator(pipeline).\nsetEvaluator(multiclassEval).\nsetEstimatorParamMaps(paramGrid).\nsetTrainRatio(0.9)\nval validatorModel = validator.fit(trainData)\n```\n\n<div class=\"panel panel-info\">\n <div class=\"panel-heading\">\n <h3 class=\"panel-title\">Tip!</h3>\n </div>\n <div class=\"panel-body\">\n <p>Running this may take a while, depending on your computing power, and you may get many warnings about \"1 block locks were not released by TID =\". Just keep waiting, and know that you'll still get results, even though all those scary warnings show. Remember that you're checking 16 different models for accuracy!</p>\n </div>\n</div>\n\nThe `validatorModel` contains the best fit model, but you'll have to wait to see what it is. The suspense builds...\n\n---",
"_____no_output_____"
],
[
"<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n\n# Page 12 - Best Fit Model<a class=\"anchor\" id=\"DS107L5_page_12\"></a>\n\n[Back to Top](#DS107L5_toc)\n\n<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">",
"_____no_output_____"
],
[
"# Best Fit Model\n\nThe result of your hyperparameter tuning is the best-fit model for your data. This page will show you what that model is and the accuracy of said model.\n\n---\n\n## Determine the Best Fit Model\n\nNow you can actually extract the best fit model and find out what it is:\n\n```scala\nimport org.apache.spark.ml.PipelineModel\nval bestModel = validatorModel.bestModel\nbestModel.asInstanceOf[PipelineModel].stages.last.extractParamMap\n```\n\nAnd here is the output:\n\n```text\nres34: org.apache.spark.ml.param.ParamMap =\n{\n dtc_0e1e4da37cb7-cacheNodeIds: false,\n dtc_0e1e4da37cb7-checkpointInterval: 10,\n dtc_0e1e4da37cb7-featuresCol: featureVector,\n dtc_0e1e4da37cb7-impurity: gini,\n dtc_0e1e4da37cb7-labelCol: Type,\n dtc_0e1e4da37cb7-maxBins: 300,\n dtc_0e1e4da37cb7-maxDepth: 20,\n dtc_0e1e4da37cb7-maxMemoryInMB: 256,\n dtc_0e1e4da37cb7-minInfoGain: 0.0,\n dtc_0e1e4da37cb7-minInstancesPerNode: 1,\n dtc_0e1e4da37cb7-predictionCol: prediction,\n dtc_0e1e4da37cb7-probabilityCol: probability,\n dtc_0e1e4da37cb7-rawPredictionCol: rawPrediction,\n dtc_0e1e4da37cb7-seed: 8849016365946518463\n}\n```\n\nLooks like you want to use the `gini` method of calculating impurity, want a `maxBins` of 300, a `maxDepth` of 20, and want to use zero `minInfoGain`.\n\n---\n\n## Determine the Accuracy of the Models\n\nBut you want to know what the accuracy is for your best fit model? Too crazy; best go back to bed. Or maybe just run the code below:\n\n```scala\nval validatorModel = validator.fit(trainData)\nval paramsAndMetrics = validatorModel.validationMetrics.\nzip(validatorModel.getEstimatorParamMaps).sortBy(-_._1)\nparamsAndMetrics.foreach { case (metric, params) =>\nprintln(metric)\nprintln(params)\nprintln()\n}\n```\n\n<div class=\"panel panel-info\">\n <div class=\"panel-heading\">\n <h3 class=\"panel-title\">Tip!</h3>\n </div>\n <div class=\"panel-body\">\n <p>This may also take a minute, and you may see some of the same warnings up above. But as the Brits say, keep calm and carry on!</p>\n </div>\n</div>\n\nThe output of the code above gives you the accuracy of the models in descending order, followed by the hyperparameters for that model: \n\n```text\n0.8571428571428571\n{\n dtc_0e1e4da37cb7-impurity: gini,\n dtc_0e1e4da37cb7-maxBins: 300,\n dtc_0e1e4da37cb7-maxDepth: 20,\n dtc_0e1e4da37cb7-minInfoGain: 0.0\n}\n\n0.8571428571428571\n{\n dtc_0e1e4da37cb7-impurity: gini,\n dtc_0e1e4da37cb7-maxBins: 300,\n dtc_0e1e4da37cb7-maxDepth: 20,\n dtc_0e1e4da37cb7-minInfoGain: 0.05\n}\n\n0.7857142857142857\n{\n dtc_0e1e4da37cb7-impurity: entropy,\n dtc_0e1e4da37cb7-maxBins: 40,\n dtc_0e1e4da37cb7-maxDepth: 20,\n dtc_0e1e4da37cb7-minInfoGain: 0.0\n}\n\n0.7857142857142857\n{\n dtc_0e1e4da37cb7-impurity: entropy,\n dtc_0e1e4da37cb7-maxBins: 40,\n dtc_0e1e4da37cb7-maxDepth: 20,\n dtc_0e1e4da37cb7-minInfoGain: 0.05\n}\n\n0.7142857142857143\n{\n dtc_0e1e4da37cb7-impurity: gini,\n dtc_0e1e4da37cb7-maxBins: 40,\n dtc_0e1e4da37cb7-maxDepth: 1,\n dtc_0e1e4da37cb7-minInfoGain: 0.0\n}\n\n0.7142857142857143\n{\n dtc_0e1e4da37cb7-impurity: gini,\n dtc_0e1e4da37cb7-maxBins: 40,\n dtc_0e1e4da37cb7-maxDepth: 1,\n dtc_0e1e4da37cb7-minInfoGain: 0.05\n}\n\n0.7142857142857143\n{\n dtc_0e1e4da37cb7-impurity: gini,\n dtc_0e1e4da37cb7-maxBins: 300,\n dtc_0e1e4da37cb7-maxDepth: 1,\n dtc_0e1e4da37cb7-minInfoGain: 0.0\n}\n\n0.7142857142857143\n{\n dtc_0e1e4da37cb7-impurity: gini,\n dtc_0e1e4da37cb7-maxBins: 300,\n dtc_0e1e4da37cb7-maxDepth: 1,\n dtc_0e1e4da37cb7-minInfoGain: 0.05\n}\n\n0.7142857142857143\n{\n dtc_0e1e4da37cb7-impurity: gini,\n dtc_0e1e4da37cb7-maxBins: 40,\n dtc_0e1e4da37cb7-maxDepth: 20,\n dtc_0e1e4da37cb7-minInfoGain: 0.05\n}\n\n0.7142857142857143\n{\n dtc_0e1e4da37cb7-impurity: entropy,\n dtc_0e1e4da37cb7-maxBins: 300,\n dtc_0e1e4da37cb7-maxDepth: 20,\n dtc_0e1e4da37cb7-minInfoGain: 0.0\n}\n\n0.7142857142857143\n{\n dtc_0e1e4da37cb7-impurity: entropy,\n dtc_0e1e4da37cb7-maxBins: 300,\n dtc_0e1e4da37cb7-maxDepth: 20,\n dtc_0e1e4da37cb7-minInfoGain: 0.05\n}\n\n0.6428571428571429\n{\n dtc_0e1e4da37cb7-impurity: gini,\n dtc_0e1e4da37cb7-maxBins: 40,\n dtc_0e1e4da37cb7-maxDepth: 20,\n dtc_0e1e4da37cb7-minInfoGain: 0.0\n}\n\n0.35714285714285715\n{\n dtc_0e1e4da37cb7-impurity: entropy,\n dtc_0e1e4da37cb7-maxBins: 40,\n dtc_0e1e4da37cb7-maxDepth: 1,\n dtc_0e1e4da37cb7-minInfoGain: 0.0\n}\n\n0.35714285714285715\n{\n dtc_0e1e4da37cb7-impurity: entropy,\n dtc_0e1e4da37cb7-maxBins: 40,\n dtc_0e1e4da37cb7-maxDepth: 1,\n dtc_0e1e4da37cb7-minInfoGain: 0.05\n}\n\n0.35714285714285715\n{\n dtc_0e1e4da37cb7-impurity: entropy,\n dtc_0e1e4da37cb7-maxBins: 300,\n dtc_0e1e4da37cb7-maxDepth: 1,\n dtc_0e1e4da37cb7-minInfoGain: 0.0\n}\n\n0.35714285714285715\n{\n dtc_0e1e4da37cb7-impurity: entropy,\n dtc_0e1e4da37cb7-maxBins: 300,\n dtc_0e1e4da37cb7-maxDepth: 1,\n dtc_0e1e4da37cb7-minInfoGain: 0.05\n}\n```\n\nAs noted the first time, the best model is gini/300/20/0, and it yields an accuracy of 87%, which is an improvement upon the original accuracy of 82%. You gained an extra 5% accuracy utilizing your hyperparameters! High five!\n\n---\n\n## Use Testing Data to Evaluate the Model\n\nNow that you've split the data, and trained with it, it's time to test that bad boy out! You know that you're 87% accurate when training, but does that hold up in testing? The first line below is for evaluating the 10% for hyperparameter testing, and the second line below is for evaluating the 10% for testing as a whole.\n\n```scala\nvalidatorModel.validationMetrics.max\nmulticlassEval.evaluate(bestModel.transform(testData))\n```\n\nHere are the hyperparameter tuning results:\n\n```text\nres52: Double = 0.8571428571428571\n```\n\nAnd here are the overall testing results:\n\n```text\nres53: Double = 0.76\n```\n\nSo it looks like tuning those hyperparameters was a good call! A 9% increase in accuracy is good to have! \n\n---",
"_____no_output_____"
],
[
"<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n\n# Page 13 - Key Terms<a class=\"anchor\" id=\"DS107L5_page_13\"></a>\n\n[Back to Top](#DS107L5_toc)\n\n<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">",
"_____no_output_____"
],
[
"# Key Terms\n\nBelow is a list and short description of the important keywords learned in this lesson. Please read through and go back and review any concepts you do not fully understand. Great Work!\n\n<table class=\"table table-striped\">\n <tr>\n <th>Keyword</th>\n <th>Description</th>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>Spark</td>\n <td>Data processing program built on top of MapReduce.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>Spark Core</td>\n <td>Spark base; also known as Spark 1.0.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>Spark Streaming</td>\n <td>Program to feed in real-time data and receive real-time output.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>Spark SQL</td>\n <td>Use SQL within Spark for a speed boost.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>MLLib</td>\n <td>Machine learning library for Spark.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>GraphX</td>\n <td>Social networking graphs in Spark.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>Apache Zeppelin</td>\n <td>Notebook interface for your Hadoop cluster.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>Resilient Distributed Datasets (RDDs)</td>\n <td>Data stored across your Hadoop cluster.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>DataSets</td>\n <td>Spark 2.0 data storage that allows for efficiency.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>DataFrames</td>\n <td>Spark 2.0 data storage that maintains data structure. For relational data.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>Scala</td>\n <td>Programming language that Spark was built in.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>Hyperparameter</td>\n <td>Components to the way you create your machine learning model.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>Maximum Depth</td>\n <td>The number of decisions you allow your decision tree to make.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>Maximum Bins</td>\n <td>The number of decision rules you allow your decision tree to have.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>Impurity Measure</td>\n <td>When you sort your outcomes with some mix-up.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>Purity</td>\n <td>Having separate groups that only contain the specified category.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>Minimum Information Gain</td>\n <td>Don't include a decision rule that won't make the data more pure.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>Pipeline</td>\n <td>Automatically chaining operations together.</td>\n </tr>\n</table>\n\n---\n\n## Key Scala Code\n\n<table class=\"table table-striped\">\n <tr>\n <th>Keyword</th>\n <th>Description</th>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>val</td>\n <td>A value that cannot be changed once it has been assigned.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>var</td>\n <td>A variable that can be changed after it has been assigned.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>sc</td>\n <td>Spark Context; environment in which you can use Spark.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>.map()</td>\n <td>Provides structure for text files in Spark.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>.toDF()</td>\n <td>Changes data from an RDD to a DataFrame.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>printSchema()</td>\n <td>Prints the structure of your DataFrame.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>.cache()</td>\n <td>Keeps your data in memory so you can access it faster.</td>\n </tr>\n <tr>\n <td style=\"font-weight: bold;\" nowrap>registerTempTable()</td>\n <td>Creates a temporary table that you can use with Spark SQL.</td>\n </tr>\n</table>",
"_____no_output_____"
],
[
"<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n\n# Page 14 - Lesson 5 Practice Hands-On<a class=\"anchor\" id=\"DS107L5_page_14\"></a>\n\n[Back to Top](#DS107L5_toc)\n\n<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">",
"_____no_output_____"
],
[
"This Hands-On will **not** be graded, but you are encouraged to complete it. The best way to become a great data scientist is to practice. Once you have submitted your project, you will be able to access the solution on the next page. Note that the solution will be slightly different from yours, but should look similar.\n\n<div class=\"panel panel-danger\">\n <div class=\"panel-heading\">\n <h3 class=\"panel-title\">Caution!</h3>\n </div>\n <div class=\"panel-body\">\n <p>Do not submit your project until you have completed all requirements, as you will not be able to resubmit.</p>\n </div>\n</div>\n\n---\n\n## Description\n\nUsing **[this data on boardgame ratings](https://repo.exeterlms.com/documents/V2/DataScience/Big-Data/boardgames3.zip)**, perform a decision tree to predict the average rating of boardgames (`average_rating`). You will need to upload this data file to your HDFS.\n\nPlease copy your Scala code into a text file, and include at the bottom the answer to the following questions:\n\n* What was the best model after hyperparameter tuning? \n* What is the overall accuracy?\n\n---\n\n## Alternative Assignment if You Can't Run Hadoop and/or Ambari\n\nIf your computer refuses to run Hadoop and/or Ambari, **[here](https://repo.exeterlms.com/documents/V2/DataScience/Big-Data/L5exam.zip)** is an alternative exam to test your understanding of the material. Please attach it instead.\n\n<div class=\"panel panel-danger\">\n <div class=\"panel-heading\">\n <h3 class=\"panel-title\">Caution!</h3>\n </div>\n <div class=\"panel-body\">\n <p>Be sure to zip and submit your entire directory when finished!</p>\n </div>\n</div>\n\n\n",
"_____no_output_____"
],
[
"<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n\n# Page 15 - Lesson 5 Practice Hands-On Solution<a class=\"anchor\" id=\"DS107L5_page_15\"></a>\n\n[Back to Top](#DS107L5_toc)\n\n<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">",
"_____no_output_____"
],
[
"# Lesson 5 Practice Hands-On Solution\n\n---\n\n## Best Model After Hyperparameter Tuning\n\nThe best model after hyperparamter tuning was the one that used entropy, a max of 300 bins, a depth of 20, and .05 of minimum information gain.\n\n---\n\n## Overall Accuracy\n\nThe overall accuracy was 62% with the best hyperparamter model.\n\n---\n\n## Code\n\nBelow you will find all the code to provide the answers above.\n\n---\n\n### Done in the Command Prompt\n\n```bash\nexport SPARK_MAJOR_VERSION=2\n\nspark-shell --master local[*]\n```\n\n---\n\n### Done in Spark Shell\n\n```scala\nval data = spark.read.\noption(\"inferSchema\", true).\noption(\"header\", true).\ncsv(\"hdfs:///user/maria_dev/boardgames3.csv\")\n\ndata.printSchema()\n\nval Array(trainData, testData) = data.randomSplit(Array(0.9, 0.1))\ntrainData.cache()\ntestData.cache()\n\nimport org.apache.spark.ml.feature.VectorAssembler\nval inputCols = trainData.columns.filter(_ != \"average_rating\")\nval assembler = new VectorAssembler().\nsetInputCols(inputCols).\nsetOutputCol(\"featureVector\")\nval assembledTrainData = assembler.transform(trainData)\nassembledTrainData.select(\"featureVector\").show(truncate = false)\n\nimport org.apache.spark.ml.classification.DecisionTreeClassifier\nimport scala.util.Random\nval classifier = new DecisionTreeClassifier().\nsetSeed(Random.nextLong()).\nsetLabelCol(\"average_rating\").\nsetFeaturesCol(\"featureVector\").\nsetPredictionCol(\"prediction\")\nval model = classifier.fit(assembledTrainData)\nprintln(model.toDebugString)\n\nmodel.featureImportances.toArray.zip(inputCols).\nsorted.reverse.foreach(println)\n\n//The best feature for prediction is users_rated.\n\nval predictions = model.transform(assembledTrainData)\npredictions.select(\"average_rating\", \"prediction\", \"probability\").\nshow(truncate = false)\n\n//Eyeball analysis shows that right now it is not predicting very well.\n\nimport org.apache.spark.ml.evaluation.MulticlassClassificationEvaluator\nval evaluator = new MulticlassClassificationEvaluator().\nsetLabelCol(\"average_rating\").\nsetPredictionCol(\"prediction\")\nevaluator.setMetricName(\"accuracy\").evaluate(predictions)\n\n//Right now the model has 57% accuracy.\n\nval confusionMatrix = predictions.\ngroupBy(\"average_rating\").\npivot(\"prediction\", (0 to 10)).\ncount().\nna.fill(0.0).\norderBy(\"average_rating\")\nconfusionMatrix.show()\n\n//Looks like most ratings, both higher and lower, are getting misclassified as 5s, 6s, and 7s. There were no accurate predictions of 1-4 or 8-10.\n\nimport org.apache.spark.sql.DataFrame\ndef classProbabilities(data: DataFrame): Array[Double] = {\nval total = data.count()\ndata.groupBy(\"average_rating\").count().\norderBy(\"average_rating\").\nselect(\"count\").as[Double].\nmap(_ / total).\ncollect()\n}\n\nval trainPriorProbabilities = classProbabilities(trainData)\nval testPriorProbabilities = classProbabilities(testData)\ntrainPriorProbabilities.zip(testPriorProbabilities).map {\ncase (trainProb, cvProb) => trainProb * cvProb\n}.sum\n\n//Random guessing accuracy is 18%, so the current model is better than just guessing. Yay!\n\nimport org.apache.spark.ml.Pipeline\nval inputCols = trainData.columns.filter(_ != \"average_rating\")\nval assembler = new VectorAssembler().\nsetInputCols(inputCols).\nsetOutputCol(\"featureVector\")\nval classifier = new DecisionTreeClassifier().\nsetSeed(Random.nextLong()).\nsetLabelCol(\"average_rating\").\nsetFeaturesCol(\"featureVector\").\nsetPredictionCol(\"prediction\")\nval pipeline = new Pipeline().setStages(Array(assembler, classifier))\n\nimport org.apache.spark.ml.tuning.ParamGridBuilder\nval paramGrid = new ParamGridBuilder().\naddGrid(classifier.impurity, Seq(\"gini\", \"entropy\")).\naddGrid(classifier.maxDepth, Seq(1, 20)).\naddGrid(classifier.maxBins, Seq(40, 300)).\naddGrid(classifier.minInfoGain, Seq(0.0, 0.05)).\nbuild()\nval multiclassEval = new MulticlassClassificationEvaluator().\nsetLabelCol(\"average_rating\").\nsetPredictionCol(\"prediction\").\nsetMetricName(\"accuracy\")\n\nimport org.apache.spark.ml.tuning.TrainValidationSplit\nval validator = new TrainValidationSplit().\nsetSeed(Random.nextLong()).\nsetEstimator(pipeline).\nsetEvaluator(multiclassEval).\nsetEstimatorParamMaps(paramGrid).\nsetTrainRatio(0.9)\nval validatorModel = validator.fit(trainData)\n\nimport org.apache.spark.ml.PipelineModel\nval bestModel = validatorModel.bestModel\nbestModel.asInstanceOf[PipelineModel].stages.last.extractParamMap\n\n//The best model uses entropy, a max of 300 bins, a depth of 20, and .05 of minimum information gain.\n\nval validatorModel = validator.fit(trainData)\nval paramsAndMetrics = validatorModel.validationMetrics.\nzip(validatorModel.getEstimatorParamMaps).sortBy(-_._1)\nparamsAndMetrics.foreach { case (metric, params) =>\nprintln(metric)\nprintln(params)\nprintln()\n}\n\nvalidatorModel.validationMetrics.max\nmulticlassEval.evaluate(bestModel.transform(testData))\n```",
"_____no_output_____"
],
[
"<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">\n\n# Page 16 - Lesson 5 Practice Hands-On Solution - Alternative Assignment<a class=\"anchor\" id=\"DS107L5_page_16\"></a>\n\n[Back to Top](#DS107L5_toc)\n\n<hr style=\"height:10px;border-width:0;color:gray;background-color:gray\">",
"_____no_output_____"
],
[
"# Lesson 5 Practice Hands-On Solution - Alternative Assignment\n\nThis exam serves as the assessment for those students who cannot utilize the Hadoop system and/or Ambari GUI. Correct answers are show in bold.\n\n1.\tWhich of the Spark components most interests you and why?\n\n **Spark ML seems so powerful! It also seems the most like \"regular\" programming that isn't done with big data, which makes it a little easier.**\n\n2.\tTrue or False? \"Zeppelin is very similar in structure and function to Jupyter Notebook.\"\n **a.\tTrue**\n b.\tFalse\n\n3.\tHow do the three types of Spark data storage differ from each other?\n\n **RDDs are the original way to store data, but they are slow. DataSets are more efficient. DataFrames are meant specially for relational data and hold row and column data.**\n\n4.\tHow do you denote comments in Scala?\n a.\t# \n b.\tChange it to markdown\n **c.\t//**\n d.\t/#\n\n5.\tWhat are the four hyperparameters for decision trees? Give both names and descriptions.\n\n **Maximum Depth: The number of decisions you can make in a tree.**\n\n **Maximum Bins: The number of decision rules you can use in a tree.**\n\n **Impurity Measure: How much mix-up you allow between categories.**\n\n **MInimum Information Gain: Keep only things that add to the accuracy of your data.**",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
e79592db92a45d90d69613d9b86840e28894a12f | 13,165 | ipynb | Jupyter Notebook | similarity-functions/SimilarityFunctions.ipynb | ansegura7/Algorithms | 4788c183ff42964dacc2bb51d7715f120d79c447 | [
"MIT"
]
| 112 | 2020-01-08T17:10:32.000Z | 2022-02-20T07:34:25.000Z | similarity-functions/SimilarityFunctions.ipynb | suanhwee1234/Algorithms | 7ac304fac42a8dec50580c78e623b0f5c021373b | [
"MIT"
]
| null | null | null | similarity-functions/SimilarityFunctions.ipynb | suanhwee1234/Algorithms | 7ac304fac42a8dec50580c78e623b0f5c021373b | [
"MIT"
]
| 19 | 2019-07-15T20:14:17.000Z | 2021-09-28T03:06:38.000Z | 20.160796 | 275 | 0.478162 | [
[
[
"# 6. Similarity Functions",
"_____no_output_____"
],
[
"- **Created by Andrés Segura Tinoco**\n- **Created on May 20, 2019**\n- **Updated on Mar 19, 2021**",
"_____no_output_____"
],
[
"In statistics and related fields, a **similarity measure** or similarity function is a real-valued function that quantifies the similarity between two objects. In short, a similarity function quantifies how much alike two data objects are <a href=\"#link_one\">[1]</a>.",
"_____no_output_____"
],
[
"## 6.1. Common similarity functions",
"_____no_output_____"
]
],
[
[
"# Load the Python libraries\nfrom math import *\nfrom decimal import Decimal\nfrom scipy import stats as ss\nimport sklearn.metrics.pairwise as sm\nimport math",
"_____no_output_____"
]
],
[
[
"\\begin{align}\n similarity(X, Y) = d(X, Y) = \\sqrt{\\sum_{i=1}^n (X_i - Y_i)^2} \\tag{1}\n\\end{align}",
"_____no_output_____"
]
],
[
[
"# (1) Euclidean distance function\ndef euclidean_distance(x, y):\n return sqrt(sum(pow(a-b,2) for a, b in zip(x, y)))",
"_____no_output_____"
]
],
[
[
"\\begin{align}\n similarity(X, Y) = d(X, Y) = \\sum_{i=1}^n |X_i - Y_i| \\tag{2}\n\\end{align}",
"_____no_output_____"
]
],
[
[
"# (2) manhattan distance function\ndef manhattan_distance(x, y):\n return sum(abs(a-b) for a,b in zip(x,y))",
"_____no_output_____"
]
],
[
[
"\\begin{align}\n similarity(X, Y) = d(X, Y) = (\\sum_{i=1}^n |X_i - Y_i|^p)^\\frac{1}{p} \\tag{3}\n\\end{align}",
"_____no_output_____"
]
],
[
[
"# (3) Minkowski distance function\ndef _nth_root(value, n_root):\n root_value = 1/float(n_root)\n return round(Decimal(value) ** Decimal(root_value),3)\n\ndef minkowski_distance(x, y, p = 3):\n return float(_nth_root(sum(pow(abs(a-b), p) for a,b in zip(x, y)), p))",
"_____no_output_____"
]
],
[
[
"\\begin{align}\n similarity(X, Y) = cos(\\theta) = \\frac{\\vec{X}.\\vec{Y}}{\\|\\vec{X}\\|.\\|\\vec{Y}\\|} = \\frac{\\sum_{i=1}^n X_i.Y_i}{\\sqrt{\\sum_{i=1}^n X_i^2}.\\sqrt{\\sum_{i=1}^n Y_i^2}} \\tag{4}\n\\end{align}",
"_____no_output_____"
]
],
[
[
"# (4) Cosine similarity function\ndef _square_rooted(x):\n return round(sqrt(sum([a*a for a in x])),3)\n\ndef cosine_similarity(x, y):\n numerator = sum(a*b for a,b in zip(x,y))\n denominator = _square_rooted(x) * _square_rooted(y)\n return round(numerator/float(denominator),3)",
"_____no_output_____"
]
],
[
[
"\\begin{align}\n similarity(X, Y) = \\frac{cov(X, Y)}{\\sigma_X . \\sigma_Y} = \\frac{\\sum_{i=1}^n (X_i - \\bar{X}).(Y_i - \\bar{Y})}{\\sqrt{\\sum_{i=1}^n (X_i - \\bar{X})^2 . (Y_i - \\bar{Y})^2}} \\tag{5}\n\\end{align}",
"_____no_output_____"
]
],
[
[
"# (5) Pearson similarity function\ndef _avg(x):\n assert len(x) > 0\n return float(sum(x)) / len(x)\n\ndef pearson_similarity(x, y):\n assert len(x) == len(y)\n n = len(x)\n assert n > 0\n avg_x = _avg(x)\n avg_y = _avg(y)\n diffprod = 0\n xdiff2 = 0\n ydiff2 = 0\n for idx in range(n):\n xdiff = x[idx] - avg_x\n ydiff = y[idx] - avg_y\n diffprod += xdiff * ydiff\n xdiff2 += xdiff * xdiff\n ydiff2 += ydiff * ydiff\n\n return diffprod / math.sqrt(xdiff2 * ydiff2)",
"_____no_output_____"
]
],
[
[
"\\begin{align}\n similarity(X, Y) = J(X, Y) = \\frac{|X \\cap Y|}{|X \\cup Y|} = \\frac{|X \\cap Y|}{|X| + |Y| - |X \\cap Y|} \\tag{6}\n\\end{align}",
"_____no_output_____"
]
],
[
[
"# (6) Jaccard similarity function\ndef jaccard_similarity(x, y):\n intersection_cardinality = len(set.intersection(*[set(x), set(y)]))\n union_cardinality = len(set.union(*[set(x), set(y)]))\n return intersection_cardinality / float(union_cardinality)",
"_____no_output_____"
]
],
[
[
"## 6.2. Manual examples",
"_____no_output_____"
]
],
[
[
"# Vectors\nx = [-4.593481, -5.478033, 1.127111, 1.252885, -2.286953] # Messi\ny = [-4.080334, -3.406618, 4.334073, -0.485612, -2.817897] # CR\nz = [-4.048185, -5.546171, 0.505673, 0.616553, -1.730906] # Neymar",
"_____no_output_____"
]
],
[
[
"### Euclidean distance",
"_____no_output_____"
]
],
[
[
"euclidean_distance(x, y)",
"_____no_output_____"
],
[
"euclidean_distance(x, z)",
"_____no_output_____"
]
],
[
[
"### Manhattan distance",
"_____no_output_____"
]
],
[
[
"manhattan_distance(x, y)",
"_____no_output_____"
],
[
"manhattan_distance(x, z)",
"_____no_output_____"
]
],
[
[
"### Minkowski distance",
"_____no_output_____"
]
],
[
[
"minkowski_distance(x, y)",
"_____no_output_____"
],
[
"minkowski_distance(x, z)",
"_____no_output_____"
]
],
[
[
"### Cosine similarity",
"_____no_output_____"
]
],
[
[
"cosine_similarity(x, y)",
"_____no_output_____"
],
[
"cosine_similarity(x, z)",
"_____no_output_____"
]
],
[
[
"### Pearson similarity",
"_____no_output_____"
]
],
[
[
"pearson_similarity(x, y)",
"_____no_output_____"
],
[
"pearson_similarity(x, z)",
"_____no_output_____"
]
],
[
[
"### Jaccard similarity",
"_____no_output_____"
]
],
[
[
"a = [0, 1, 2, 3, 4, 5]\nb = [-1, 1, 2, 0, 3, 5]",
"_____no_output_____"
],
[
"jaccard_similarity(a, b)",
"_____no_output_____"
]
],
[
[
"## 6.3. Sklearn examples",
"_____no_output_____"
]
],
[
[
"corr = sm.euclidean_distances([x], [y])\nfloat(corr[0])",
"_____no_output_____"
],
[
"corr = sm.manhattan_distances([x], [y])\nfloat(corr[0])",
"_____no_output_____"
],
[
"corr = sm.cosine_similarity([x], [y])\nfloat(corr[0])",
"_____no_output_____"
],
[
"corr, p_value = ss.pearsonr(x, y)\ncorr",
"_____no_output_____"
]
],
[
[
"## Reference",
"_____no_output_____"
],
[
"<a name='link_one' href='https://en.wikipedia.org/wiki/Similarity_measure' target='_blank' >[1]</a> Wikipedia - Similarity measure. ",
"_____no_output_____"
],
[
"---\n<a href=\"https://ansegura7.github.io/Algorithms/\">« Home</a>",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
]
|
e795acd35f62d232654574f549c5fbcdccc504fc | 8,587 | ipynb | Jupyter Notebook | .ipynb_checkpoints/YoutTube Scaper-checkpoint.ipynb | Vic-the-Legend/yt-trends | b3aed53a8493e10736487852200e22fca0c980f1 | [
"BSD-2-Clause"
]
| 1 | 2020-02-14T02:02:40.000Z | 2020-02-14T02:02:40.000Z | .ipynb_checkpoints/YoutTube Scaper-checkpoint.ipynb | Vic-the-Legend/yt-trends | b3aed53a8493e10736487852200e22fca0c980f1 | [
"BSD-2-Clause"
]
| null | null | null | .ipynb_checkpoints/YoutTube Scaper-checkpoint.ipynb | Vic-the-Legend/yt-trends | b3aed53a8493e10736487852200e22fca0c980f1 | [
"BSD-2-Clause"
]
| null | null | null | 45.433862 | 217 | 0.553162 | [
[
[
"import requests, sys, time, os, argparse\n\n# List of simple to collect features\nsnippet_features = [\"title\",\n \"publishedAt\",\n \"channelId\",\n \"channelTitle\",\n \"categoryId\"]\n\n# Any characters to exclude, generally these are things that become problematic in CSV files\nunsafe_characters = ['\\n', '\"']\n\n# Used to identify columns, currently hardcoded order\nheader = [\"video_id\"] + snippet_features + [\"trending_date\", \"tags\", \"view_count\", \"likes\", \"dislikes\",\n \"comment_count\", \"thumbnail_link\", \"comments_disabled\",\n \"ratings_disabled\", \"description\"]\n\n\ndef setup(api_path, code_path):\n with open(api_path, 'r') as file:\n api_key = file.readline()\n\n\n with open(code_path) as file:\n country_codes = [x.rstrip() for x in file]\n\n return api_key, country_codes\n\n\ndef prepare_feature(feature):\n # Removes any character from the unsafe characters list and surrounds the whole item in quotes\n for ch in unsafe_characters:\n feature = str(feature).replace(ch, \"\")\n return f'\"{feature}\"'\n\n\ndef api_request(page_token, country_code):\n # Builds the URL and requests the JSON from it\n request_url = f\"https://www.googleapis.com/youtube/v3/videos?part=id,statistics,snippet{page_token}chart=mostPopular®ionCode={country_code}&maxResults=50&key={api_key}\"\n request = requests.get(request_url)\n if request.status_code == 429:\n print(\"Temp-Banned due to excess requests, please wait and continue later\")\n sys.exit()\n return request.json()\n\n\ndef get_tags(tags_list):\n # Takes a list of tags, prepares each tag and joins them into a string by the pipe character\n return prepare_feature(\"|\".join(tags_list))\n\n\ndef get_videos(items):\n lines = []\n for video in items:\n comments_disabled = False\n ratings_disabled = False\n\n # We can assume something is wrong with the video if it has no statistics, often this means it has been deleted\n # so we can just skip it\n if \"statistics\" not in video:\n continue\n\n # A full explanation of all of these features can be found on the GitHub page for this project\n video_id = prepare_feature(video['id'])\n\n # Snippet and statistics are sub-dicts of video, containing the most useful info\n snippet = video['snippet']\n statistics = video['statistics']\n\n # This list contains all of the features in snippet that are 1 deep and require no special processing\n features = [prepare_feature(snippet.get(feature, \"\")) for feature in snippet_features]\n\n # The following are special case features which require unique processing, or are not within the snippet dict\n description = snippet.get(\"description\", \"\")\n thumbnail_link = snippet.get(\"thumbnails\", dict()).get(\"default\", dict()).get(\"url\", \"\")\n trending_date = time.strftime(\"%y.%d.%m\")\n tags = get_tags(snippet.get(\"tags\", [\"[none]\"]))\n view_count = statistics.get(\"viewCount\", 0)\n\n # This may be unclear, essentially the way the API works is that if a video has comments or ratings disabled\n # then it has no feature for it, thus if they don't exist in the statistics dict we know they are disabled\n if 'likeCount' in statistics and 'dislikeCount' in statistics:\n likes = statistics['likeCount']\n dislikes = statistics['dislikeCount']\n else:\n ratings_disabled = True\n likes = 0\n dislikes = 0\n\n if 'commentCount' in statistics:\n comment_count = statistics['commentCount']\n else:\n comments_disabled = True\n comment_count = 0\n\n # Compiles all of the various bits of info into one consistently formatted line\n line = [video_id] + features + [prepare_feature(x) for x in [trending_date, tags, view_count, likes, dislikes,\n comment_count, thumbnail_link, comments_disabled,\n ratings_disabled, description]]\n lines.append(\",\".join(line))\n return lines\n\n\ndef get_pages(country_code, next_page_token=\"&\"):\n country_data = []\n\n # Because the API uses page tokens (which are literally just the same function of numbers everywhere) it is much\n # more inconvenient to iterate over pages, but that is what is done here.\n while next_page_token is not None:\n # A page of data i.e. a list of videos and all needed data\n video_data_page = api_request(next_page_token, country_code)\n\n # Get the next page token and build a string which can be injected into the request with it, unless it's None,\n # then let the whole thing be None so that the loop ends after this cycle\n next_page_token = video_data_page.get(\"nextPageToken\", None)\n next_page_token = f\"&pageToken={next_page_token}&\" if next_page_token is not None else next_page_token\n\n # Get all of the items as a list and let get_videos return the needed features\n items = video_data_page.get('items', [])\n country_data += get_videos(items)\n\n return country_data\n\n\ndef write_to_file(country_code, country_data):\n\n print(f\"Writing {country_code} data to file...\")\n\n if not os.path.exists(output_dir):\n os.makedirs(output_dir)\n\n with open(f\"{output_dir}/{time.strftime('%y.%m.%d')}_{country_code}_videos.csv\", \"w+\", encoding='utf-8') as file:\n for row in country_data:\n file.write(f\"{row}\\n\")\n\n\ndef get_data():\n for country_code in country_codes:\n country_data = [\",\".join(header)] + get_pages(country_code)\n write_to_file(country_code, country_data)\n\n\nif __name__ == \"__main__\":\n\n parser = argparse.ArgumentParser()\n parser.add_argument('--key_path', help='Path to the file containing the api key, by default will use api_key.txt in the same directory', default='api_key.txt')\n parser.add_argument('--country_code_path', help='Path to the file containing the list of country codes to scrape, by default will use country_codes.txt in the same directory', default='country_codes.txt')\n parser.add_argument('--output_dir', help='Path to save the outputted files in', default='output/')\n\n args = parser.parse_args()\n\n output_dir = args.output_dir\n api_key, country_codes = setup(args.key_path, args.country_code_path)\n\n get_data()",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code"
]
]
|
e795bf266f013ff7fe4adc0a52e9c30b7c926d0d | 6,394 | ipynb | Jupyter Notebook | doc/pre-executed.ipynb | gehuazhen/nbsphinx | e31c0b0af52dfa3deddcc5f578277ab3f918c97d | [
"MIT"
]
| null | null | null | doc/pre-executed.ipynb | gehuazhen/nbsphinx | e31c0b0af52dfa3deddcc5f578277ab3f918c97d | [
"MIT"
]
| null | null | null | doc/pre-executed.ipynb | gehuazhen/nbsphinx | e31c0b0af52dfa3deddcc5f578277ab3f918c97d | [
"MIT"
]
| null | null | null | 28.672646 | 466 | 0.582421 | [
[
[
"This notebook is part of the `nbsphinx` documentation: https://nbsphinx.readthedocs.io/.",
"_____no_output_____"
],
[
"# Pre-Executing Notebooks\n\nAutomatically executing notebooks during the Sphinx build process is an important feature of `nbsphinx`.\nHowever, there are a few use cases where pre-executing a notebook and storing the outputs might be preferable.\nStoring any output will, by default, stop ``nbsphinx`` from executing the notebook.",
"_____no_output_____"
],
[
"## Long-Running Cells\n\nIf you are doing some very time-consuming computations, it might not be feasible to re-execute the notebook every time you build your Sphinx documentation.\n\nSo just do it once -- when you happen to have the time -- and then just keep the output.",
"_____no_output_____"
]
],
[
[
"import time",
"_____no_output_____"
],
[
"%time time.sleep(60 * 60)\n6 * 7",
"CPU times: user 160 ms, sys: 56 ms, total: 216 ms\nWall time: 1h 1s\n"
]
],
[
[
"If you *do* want to execute your notebooks, but some cells run for a long time, you can change the timeout, see [Cell Execution Timeout](timeout.ipynb).",
"_____no_output_____"
],
[
"## Rare Libraries\n\nYou might have created results with a library that's hard to install and therefore you have only managed to install it on one very old computer in the basement, so you probably cannot run this whenever you build your Sphinx docs.",
"_____no_output_____"
]
],
[
[
"from a_very_rare_library import calculate_the_answer",
"_____no_output_____"
],
[
"calculate_the_answer()",
"_____no_output_____"
]
],
[
[
"## Exceptions\n\nIf an exception is raised during the Sphinx build process, it is stopped (the build process, not the exception!).\nIf you want to show to your audience how an exception looks like, you have two choices:\n\n1. Allow errors -- either generally or on a per-notebook or per-cell basis -- see [Ignoring Errors](allow-errors.ipynb) ([per cell](allow-errors-per-cell.ipynb)).\n\n1. Execute the notebook beforehand and save the results, like it's done in this example notebook:",
"_____no_output_____"
]
],
[
[
"1 / 0",
"_____no_output_____"
]
],
[
[
"## Client-specific Outputs\n\nWhen `nbsphinx` executes notebooks,\nit uses the `nbconvert` module to do so.\nCertain Jupyter clients might produce output\nthat differs from what `nbconvert` would produce.\nTo preserve those original outputs,\nthe notebook has to be executed and saved\nbefore running Sphinx.\n\nFor example,\nthe JupyterLab help system shows the help text as cell outputs,\nwhile executing with `nbconvert` doesn't produce any output.",
"_____no_output_____"
]
],
[
[
"sorted?",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
e795c6cb121470b8c6e08e6e44e24fcf5f4b4a5a | 1,094 | ipynb | Jupyter Notebook | Modules/Bonus Module - Arduino-ROS Interface/2. Understanding ROS node APIs in Arduino.ipynb | mlsdpk/ROS_Basic_Course | 885c71214634ca907addf881fc0a99b53a60672c | [
"MIT"
]
| 1 | 2018-02-27T19:24:37.000Z | 2018-02-27T19:24:37.000Z | Modules/Bonus Module - Arduino-ROS Interface/2. Understanding ROS node APIs in Arduino.ipynb | mlsdpk/ROS_Basic_Course | 885c71214634ca907addf881fc0a99b53a60672c | [
"MIT"
]
| null | null | null | Modules/Bonus Module - Arduino-ROS Interface/2. Understanding ROS node APIs in Arduino.ipynb | mlsdpk/ROS_Basic_Course | 885c71214634ca907addf881fc0a99b53a60672c | [
"MIT"
]
| 2 | 2018-05-04T10:27:42.000Z | 2020-11-04T20:17:39.000Z | 17.934426 | 100 | 0.491773 | [
[
[
"# Understanding ROS node APIs in Arduino",
"_____no_output_____"
],
[
"Following is a basic structure ROS Arduino node. We can see the function of each line of code:",
"_____no_output_____"
]
],
[
[
"#include <ros.h>\n\nros::NodeHandle nh;\n \nvoid setup()\n{\n nh.initNode();\n}\n\nvoid loop()\n{\n nh.spinOnce();\n}",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
]
]
|
e795e016dd2419f3630f17cd44757e9c7db85ab1 | 2,950 | ipynb | Jupyter Notebook | datasetsforeverything/mergeTwoFolderContents.ipynb | akshat2048/MSOE_ML | 78d8d5753d70bb72b36df44926c95a808ad12f53 | [
"MIT"
]
| null | null | null | datasetsforeverything/mergeTwoFolderContents.ipynb | akshat2048/MSOE_ML | 78d8d5753d70bb72b36df44926c95a808ad12f53 | [
"MIT"
]
| 2 | 2022-01-19T19:11:32.000Z | 2022-01-19T19:13:14.000Z | datasetsforeverything/mergeTwoFolderContents.ipynb | akshat2048/MSOE_ML | 78d8d5753d70bb72b36df44926c95a808ad12f53 | [
"MIT"
]
| null | null | null | 40.410959 | 1,032 | 0.637288 | [
[
[
"import shutil\nimport os",
"_____no_output_____"
],
[
"FOLDER_1 = \"C:/Users/akash/Desktop/Akash/MSOEML/CheXpert/NIH/Abnormal/AP\"\nFOLDER_2 = \"C:/Users/akash/Desktop/Akash/MSOEML/NIH_data/AP/abnormal\"",
"_____no_output_____"
],
[
"# move files from folder 1 to folder 2\nfor file in os.listdir(FOLDER_1):\n _ = shutil.move(os.path.join(FOLDER_1, file), FOLDER_2)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code"
]
]
|
e795e20ca0bfdcf4787094b7cabd8a1ac26134c1 | 53,374 | ipynb | Jupyter Notebook | tackling_overfitting.ipynb | Chiebukar/Machine-Learning | 804b6bc675d5638aeb22b75c8efc047e0f98cbb9 | [
"MIT"
]
| 1 | 2021-06-27T05:58:43.000Z | 2021-06-27T05:58:43.000Z | tackling_overfitting.ipynb | Chiebukar/Machine-Learning | 804b6bc675d5638aeb22b75c8efc047e0f98cbb9 | [
"MIT"
]
| null | null | null | tackling_overfitting.ipynb | Chiebukar/Machine-Learning | 804b6bc675d5638aeb22b75c8efc047e0f98cbb9 | [
"MIT"
]
| null | null | null | 125.882075 | 17,922 | 0.85699 | [
[
[
"import numpy as np \r\nfrom sklearn.model_selection import train_test_split\r\nfrom tensorflow import keras\r\nfrom keras.models import Sequential\r\nfrom keras.layers import Dense, Embedding, GlobalAveragePooling1D, Dropout\r\nimport matplotlib.pyplot as plt\r\n%matplotlib inline",
"_____no_output_____"
],
[
"from keras.datasets import imdb\r\n(train_data, train_label), (test_data, test_label) = imdb.load_data(num_words= 10000)",
"_____no_output_____"
],
[
"train_data.shape, train_label.shape, test_data.shape, test_label.shape",
"_____no_output_____"
],
[
"list_len = [len(i) for i in train_data]\r\nmax(list_len)",
"_____no_output_____"
],
[
"from keras.preprocessing.sequence import pad_sequences\r\ntrain_data = pad_sequences(train_data,\r\n value = 0,\r\n padding ='post',\r\n maxlen = 2494)",
"_____no_output_____"
],
[
"test_data = pad_sequences(test_data,\r\n value = 0,\r\n padding = 'post',\r\n maxlen = 2494)",
"_____no_output_____"
],
[
"x_train, x_val, y_train, y_val = train_test_split(train_data, train_label, test_size= 0.2, random_state = 4, stratify = train_label )",
"_____no_output_____"
],
[
"def build_model(units= 64):\r\n model = Sequential()\r\n model.add(Embedding(10000, 16))\r\n model.add(GlobalAveragePooling1D())\r\n model.add(Dense(units, activation = 'relu'))\r\n model.add(Dense(units, activation = 'relu'))\r\n model.add(Dense(1, activation = 'sigmoid'))\r\n return model",
"_____no_output_____"
],
[
"model = build_model()\r\nmodel.compile(loss = 'binary_crossentropy',\r\n optimizer = 'rmsprop',\r\n metrics = ['accuracy'])\r\nmodel.summary()\r\nhistory = model.fit(x_train, y_train, validation_data = (x_val, y_val), epochs = 50, batch_size = 512)\r\nmodel.evaluate(test_data, test_label)",
"_____no_output_____"
],
[
"initial_val_loss = history.history['val_loss']\r\nepochs =range(1, len(initial_val_loss)+1)",
"_____no_output_____"
],
[
"plt.plot(epochs, initial_val_loss, 'bo')\r\nplt.title('validation loss')\r\nplt.xlabel('Epochs')\r\nplt.ylabel('Loss')\r\n\r\nplt.show",
"_____no_output_____"
],
[
"model = build_model(units = 4)\r\nmodel.compile(loss = 'binary_crossentropy',\r\n optimizer = 'rmsprop',\r\n metrics = ['accuracy'])\r\nmodel.summary()\r\nhistory = model.fit(x_train, y_train, validation_data = (x_val, y_val), epochs = 21, batch_size = 512)\r\nmodel.evaluate(test_data, test_label)",
"_____no_output_____"
],
[
"val_loss = history.history['val_loss']\r\nepochs =range(1, len(val_loss)+1)",
"_____no_output_____"
],
[
"plt.plot(epochs, val_loss, 'b')\r\nplt.title('validation loss')\r\nplt.xlabel('Epochs')\r\nplt.ylabel('Loss')\r\nplt.legend()\r\n\r\nplt.show",
"No handles with labels found to put in legend.\n"
],
[
"from keras.regularizers import l2",
"_____no_output_____"
],
[
"model = Sequential()\r\nmodel.add(Embedding(10000, 16))\r\nmodel.add(GlobalAveragePooling1D())\r\nmodel.add(Dense(64, kernel_regularizer = l2(0.001), activation = 'relu'))\r\nmodel.add(Dense(64, kernel_regularizer = l2(0.001), activation = 'relu'))\r\nmodel.add(Dense(1, activation = 'sigmoid'))\r\n\r\nmodel.compile(loss = 'binary_crossentropy',\r\n optimizer = 'rmsprop',\r\n metrics = ['accuracy'])\r\n\r\nhistory = model.fit(x_train, y_train, validation_data = (x_val, y_val), epochs = 50, batch_size = 512)\r\nmodel.evaluate(test_data, test_label)",
"_____no_output_____"
],
[
"val_loss = history.history['val_loss']\r\nepochs =range(1, len(val_loss)+1)\r\n\r\nplt.plot(epochs, val_loss, 'b')\r\nplt.plot(epochs, initial_val_loss, 'b-')\r\nplt.title('validation loss')\r\nplt.xlabel('Epochs')\r\nplt.ylabel('Loss')\r\n\r\n\r\nplt.show",
"_____no_output_____"
],
[
" model = Sequential()\r\nmodel.add(Embedding(10000, 16))\r\nmodel.add(GlobalAveragePooling1D())\r\nmodel.add(Dense(units, activation = 'relu'))\r\nmodel.add(Dropou(0.5))\r\nmodel.add(Dense(units, activation = 'relu'))\r\nmodel.add(Dropou(0.5))\r\nmodel.add(Dense(1, activation = 'sigmoid'))",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e795e6f61ae952546522ca863d67d2285046e068 | 6,474 | ipynb | Jupyter Notebook | Data Visualization with python/Data Visualization with python 3 (list).ipynb | al-sha/Handling-data-with-python | 4b2de88b6b4e634ca6afd1af23896ddc27b67d8f | [
"MIT"
]
| null | null | null | Data Visualization with python/Data Visualization with python 3 (list).ipynb | al-sha/Handling-data-with-python | 4b2de88b6b4e634ca6afd1af23896ddc27b67d8f | [
"MIT"
]
| null | null | null | Data Visualization with python/Data Visualization with python 3 (list).ipynb | al-sha/Handling-data-with-python | 4b2de88b6b4e634ca6afd1af23896ddc27b67d8f | [
"MIT"
]
| null | null | null | 109.728814 | 5,452 | 0.896664 | [
[
[
"#Visualization data in a list.",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nplt.plot([1,2,3,4], [1,4,9,16], 'ro')\nplt.show()",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code"
]
]
|
e795fc58facb9f80d841d986c9e1fac526f1ea9e | 193,150 | ipynb | Jupyter Notebook | src/train_setnetcond_on_mono_disc.ipynb | mcomunita/gfx_classifier | 94e5f09084837886990e05821da9ff4a55ed180a | [
"BSD-3-Clause"
]
| 6 | 2021-04-07T17:28:12.000Z | 2021-09-11T07:55:42.000Z | src/train_setnetcond_on_mono_disc.ipynb | mcomunita/gfx-classifier | 94e5f09084837886990e05821da9ff4a55ed180a | [
"BSD-3-Clause"
]
| 1 | 2021-04-22T08:23:02.000Z | 2021-04-22T08:23:02.000Z | src/train_setnetcond_on_mono_disc.ipynb | mcomunita/gfx-classifier | 94e5f09084837886990e05821da9ff4a55ed180a | [
"BSD-3-Clause"
]
| null | null | null | 78.26175 | 1,086 | 0.624432 | [
[
[
"import torch\nimport torch.optim as optim\nimport torchvision.transforms as transforms\nimport torch.nn as nn\nimport torch.nn.functional as F\n\nimport numpy as np\nimport time\nimport matplotlib\nimport matplotlib.pyplot as plt\n\nimport dataset.dataset as dataset\nimport datasplit.datasplit as datasplit\nimport model.models as models\nimport trainer.trainer as trainer\nimport utils.utils as utils\n\ntorch.cuda.device_count()\n\ncuda0 = torch.device('cuda:0')\ncuda1 = torch.device('cuda:1')\ncuda2 = torch.device('cuda:2')\ncuda3 = torch.device('cuda:3')\n\ndevice = torch.device(cuda0 if torch.cuda.is_available() else \"cpu\")",
"_____no_output_____"
]
],
[
[
"# INIT",
"_____no_output_____"
]
],
[
[
"# transforms\ntransform = transforms.Compose([\n transforms.ToTensor(),\n ])\n\n# dataset\nroot = '/Users/Marco/Documents/DATASETS/GUITAR-FX-DIST/Mono_Discrete/Features'\nexcl_folders = ['MT2']\nspectra_folder= 'mel_22050_1024_512'\nproc_settings_csv = 'proc_settings.csv'\nmax_num_settings=3\n\ndataset = dataset.FxDataset(root=root,\n excl_folders=excl_folders, \n spectra_folder=spectra_folder, \n processed_settings_csv=proc_settings_csv,\n max_num_settings=max_num_settings,\n transform=transform)\ndataset.init_dataset()\n# dataset.generate_mel()\n\n# split\nsplit = datasplit.DataSplit(dataset, shuffle=True)\n\n# loaders\ntrain_loader, val_loader, test_loader = split.get_split(batch_size=100)\n\nprint('dataset size: ', len(dataset))\nprint('train set size: ', len(split.train_sampler))\nprint('val set size: ', len(split.val_sampler))\nprint('test set size: ', len(split.test_sampler))\ndataset.fx_to_label",
"dataset size: 123552\ntrain set size: 88956\nval set size: 9885\ntest set size: 24711\n"
]
],
[
[
"# TRAIN SetNetCond",
"_____no_output_____"
]
],
[
[
"# model\nsetnetcond = models.SettingsNetCond(n_settings= dataset.max_num_settings,\n mel_shape=dataset.mel_shape, \n num_embeddings=dataset.num_fx, \n embedding_dim=50)\n# optimizer\noptimizer = optim.Adam(setnetcond.parameters(), lr=0.001)\n# loss function\nloss_func = nn.MSELoss(reduction='mean')\n\nprint(setnetcond)",
"SettingsNetCond(\n (emb): Embedding(13, 50)\n (fc0): Linear(in_features=50, out_features=11136, bias=True)\n (conv1): Conv2d(2, 6, kernel_size=(5, 5), stride=(1, 1))\n (batchNorm1): BatchNorm2d(6, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (conv2): Conv2d(6, 12, kernel_size=(5, 5), stride=(1, 1))\n (batchNorm2): BatchNorm2d(12, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (fc1): Linear(in_features=6264, out_features=120, bias=True)\n (batchNorm3): BatchNorm1d(120, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (fc2): Linear(in_features=120, out_features=60, bias=True)\n (batchNorm4): BatchNorm1d(60, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)\n (out): Linear(in_features=60, out_features=3, bias=True)\n)\n"
],
[
"# SAVE\nmodels_folder = '../../models_and_results/models'\nmodel_name = '20210409_setnetcond_mono_disc_best'\nresults_folder = '../../models_and_results/results'\nresults_subfolder = '20210409_setnetcond_mono_disc'",
"_____no_output_____"
],
[
"# TRAIN and TEST SettingsNetCond OVER MULTIPLE EPOCHS\ntrain_set_size = len(split.train_sampler)\nval_set_size = len(split.val_sampler)\ntest_set_size = len(split.test_sampler)\n\nall_train_losses, all_val_losses, all_test_losses = [],[],[]\nall_train_correct, all_val_correct, all_test_correct = [],[],[]\nall_train_results, all_val_results, all_test_results = [],[],[]\n\nbest_val_correct = 0\nearly_stop_counter = 0\n\nstart = time.time()\n\nfor epoch in range(100):\n train_loss, train_correct, train_results = trainer.train_settings_cond_net(\n model=setnetcond,\n optimizer=optimizer, \n train_loader=train_loader, \n train_sampler=split.train_sampler, \n epoch=epoch,\n loss_function=loss_func, \n device=device\n )\n \n val_loss, val_correct, val_results = trainer.val_settings_cond_net(\n model=setnetcond, \n val_loader=val_loader, \n val_sampler=split.val_sampler,\n loss_function=loss_func, \n device='cpu'\n )\n \n test_loss, test_correct, test_results = trainer.test_settings_cond_net(\n model=setnetcond, \n test_loader=test_loader, \n test_sampler=split.test_sampler,\n loss_function=loss_func, \n device='cpu'\n )\n # save model\n if val_correct > best_val_correct:\n best_val_correct = val_correct\n torch.save(setnetcond, '%s/%s' % (models_folder, model_name))\n early_stop_counter = 0\n print('\\n=== saved best model ===\\n')\n else:\n early_stop_counter += 1\n \n # append results\n all_train_losses.append(train_loss)\n all_val_losses.append(val_loss)\n all_test_losses.append(test_loss)\n \n all_train_correct.append(train_correct)\n all_val_correct.append(val_correct)\n all_test_correct.append(test_correct)\n \n all_train_results.append(train_results)\n all_val_results.append(val_results)\n all_test_results.append(test_results)\n\n if early_stop_counter == 15:\n print('\\n--- early stop ---\\n')\n break\n\nstop = time.time()\nprint(f\"Training time: {stop - start}s\")",
"/Users/Marco/Documents/OneDrive - Queen Mary, University of London/PHD/REPOS/venv_repos/lib/python3.8/site-packages/torch/nn/functional.py:1698: UserWarning: nn.functional.tanh is deprecated. Use torch.tanh instead.\n warnings.warn(\"nn.functional.tanh is deprecated. Use torch.tanh instead.\")\nTrain Epoch: 0\t[5000/88956 (6%)]\tTotal Loss: 6.1375\tAvg Loss: 0.0012\nTrain Epoch: 0\t[10000/88956 (11%)]\tTotal Loss: 9.8844\tAvg Loss: 0.0010\nTrain Epoch: 0\t[15000/88956 (17%)]\tTotal Loss: 13.4050\tAvg Loss: 0.0009\nTrain Epoch: 0\t[20000/88956 (22%)]\tTotal Loss: 16.8900\tAvg Loss: 0.0008\nTrain Epoch: 0\t[25000/88956 (28%)]\tTotal Loss: 20.2709\tAvg Loss: 0.0008\nTrain Epoch: 0\t[30000/88956 (34%)]\tTotal Loss: 23.6125\tAvg Loss: 0.0008\nTrain Epoch: 0\t[35000/88956 (39%)]\tTotal Loss: 26.7927\tAvg Loss: 0.0008\nTrain Epoch: 0\t[40000/88956 (45%)]\tTotal Loss: 29.9134\tAvg Loss: 0.0007\nTrain Epoch: 0\t[45000/88956 (51%)]\tTotal Loss: 32.8748\tAvg Loss: 0.0007\nTrain Epoch: 0\t[50000/88956 (56%)]\tTotal Loss: 35.6804\tAvg Loss: 0.0007\nTrain Epoch: 0\t[55000/88956 (62%)]\tTotal Loss: 38.3389\tAvg Loss: 0.0007\nTrain Epoch: 0\t[60000/88956 (67%)]\tTotal Loss: 40.8897\tAvg Loss: 0.0007\nTrain Epoch: 0\t[65000/88956 (73%)]\tTotal Loss: 43.2377\tAvg Loss: 0.0007\nTrain Epoch: 0\t[70000/88956 (79%)]\tTotal Loss: 45.5952\tAvg Loss: 0.0007\nTrain Epoch: 0\t[75000/88956 (84%)]\tTotal Loss: 47.7889\tAvg Loss: 0.0006\nTrain Epoch: 0\t[80000/88956 (90%)]\tTotal Loss: 49.9903\tAvg Loss: 0.0006\nTrain Epoch: 0\t[85000/88956 (96%)]\tTotal Loss: 52.0634\tAvg Loss: 0.0006\n====> Epoch: 0\tTotal Loss: 53.6313\t Avg Loss: 0.0006\tCorrect: 5097/88956\tPercentage Correct: 5.73\n====> Val Loss: 4.4819\t Avg Loss: 0.0005\tCorrect: 1015/9885\tPercentage Correct: 10.27\n====> Test Loss: 11.0885\t Avg Loss: 0.0004\tCorrect: 2741/24711\tPercentage Correct: 11.09\n\n=== saved best model ===\n\nTrain Epoch: 1\t[5000/88956 (6%)]\tTotal Loss: 1.9776\tAvg Loss: 0.0004\nTrain Epoch: 1\t[10000/88956 (11%)]\tTotal Loss: 3.8780\tAvg Loss: 0.0004\nTrain Epoch: 1\t[15000/88956 (17%)]\tTotal Loss: 5.8699\tAvg Loss: 0.0004\nTrain Epoch: 1\t[20000/88956 (22%)]\tTotal Loss: 7.7793\tAvg Loss: 0.0004\nTrain Epoch: 1\t[25000/88956 (28%)]\tTotal Loss: 9.6628\tAvg Loss: 0.0004\nTrain Epoch: 1\t[30000/88956 (34%)]\tTotal Loss: 11.4394\tAvg Loss: 0.0004\nTrain Epoch: 1\t[35000/88956 (39%)]\tTotal Loss: 13.1000\tAvg Loss: 0.0004\nTrain Epoch: 1\t[40000/88956 (45%)]\tTotal Loss: 14.8210\tAvg Loss: 0.0004\nTrain Epoch: 1\t[45000/88956 (51%)]\tTotal Loss: 16.4479\tAvg Loss: 0.0004\nTrain Epoch: 1\t[50000/88956 (56%)]\tTotal Loss: 18.0647\tAvg Loss: 0.0004\nTrain Epoch: 1\t[55000/88956 (62%)]\tTotal Loss: 19.6713\tAvg Loss: 0.0004\nTrain Epoch: 1\t[60000/88956 (67%)]\tTotal Loss: 21.2287\tAvg Loss: 0.0004\nTrain Epoch: 1\t[65000/88956 (73%)]\tTotal Loss: 22.8143\tAvg Loss: 0.0004\nTrain Epoch: 1\t[70000/88956 (79%)]\tTotal Loss: 24.3103\tAvg Loss: 0.0003\nTrain Epoch: 1\t[75000/88956 (84%)]\tTotal Loss: 25.8164\tAvg Loss: 0.0003\nTrain Epoch: 1\t[80000/88956 (90%)]\tTotal Loss: 27.2200\tAvg Loss: 0.0003\nTrain Epoch: 1\t[85000/88956 (96%)]\tTotal Loss: 28.6676\tAvg Loss: 0.0003\n====> Epoch: 1\tTotal Loss: 29.8095\t Avg Loss: 0.0003\tCorrect: 14041/88956\tPercentage Correct: 15.78\n====> Val Loss: 2.7384\t Avg Loss: 0.0003\tCorrect: 1847/9885\tPercentage Correct: 18.68\n====> Test Loss: 6.7626\t Avg Loss: 0.0003\tCorrect: 4788/24711\tPercentage Correct: 19.38\n\n=== saved best model ===\n\nTrain Epoch: 2\t[5000/88956 (6%)]\tTotal Loss: 1.3968\tAvg Loss: 0.0003\nTrain Epoch: 2\t[10000/88956 (11%)]\tTotal Loss: 2.7492\tAvg Loss: 0.0003\nTrain Epoch: 2\t[15000/88956 (17%)]\tTotal Loss: 4.1301\tAvg Loss: 0.0003\nTrain Epoch: 2\t[20000/88956 (22%)]\tTotal Loss: 5.4359\tAvg Loss: 0.0003\nTrain Epoch: 2\t[25000/88956 (28%)]\tTotal Loss: 6.8046\tAvg Loss: 0.0003\nTrain Epoch: 2\t[30000/88956 (34%)]\tTotal Loss: 8.1012\tAvg Loss: 0.0003\nTrain Epoch: 2\t[35000/88956 (39%)]\tTotal Loss: 9.4020\tAvg Loss: 0.0003\nTrain Epoch: 2\t[40000/88956 (45%)]\tTotal Loss: 10.6716\tAvg Loss: 0.0003\nTrain Epoch: 2\t[45000/88956 (51%)]\tTotal Loss: 12.0141\tAvg Loss: 0.0003\nTrain Epoch: 2\t[50000/88956 (56%)]\tTotal Loss: 13.2262\tAvg Loss: 0.0003\nTrain Epoch: 2\t[55000/88956 (62%)]\tTotal Loss: 14.4014\tAvg Loss: 0.0003\nTrain Epoch: 2\t[60000/88956 (67%)]\tTotal Loss: 15.6034\tAvg Loss: 0.0003\nTrain Epoch: 2\t[65000/88956 (73%)]\tTotal Loss: 16.7968\tAvg Loss: 0.0003\nTrain Epoch: 2\t[70000/88956 (79%)]\tTotal Loss: 17.9803\tAvg Loss: 0.0003\nTrain Epoch: 2\t[75000/88956 (84%)]\tTotal Loss: 19.1101\tAvg Loss: 0.0003\nTrain Epoch: 2\t[80000/88956 (90%)]\tTotal Loss: 20.2609\tAvg Loss: 0.0003\nTrain Epoch: 2\t[85000/88956 (96%)]\tTotal Loss: 21.3568\tAvg Loss: 0.0003\n====> Epoch: 2\tTotal Loss: 22.2575\t Avg Loss: 0.0003\tCorrect: 19408/88956\tPercentage Correct: 21.82\n====> Val Loss: 2.1658\t Avg Loss: 0.0002\tCorrect: 2455/9885\tPercentage Correct: 24.84\n====> Test Loss: 5.3747\t Avg Loss: 0.0002\tCorrect: 6091/24711\tPercentage Correct: 24.65\n\n=== saved best model ===\n\nTrain Epoch: 3\t[5000/88956 (6%)]\tTotal Loss: 1.0867\tAvg Loss: 0.0002\nTrain Epoch: 3\t[10000/88956 (11%)]\tTotal Loss: 2.1829\tAvg Loss: 0.0002\nTrain Epoch: 3\t[15000/88956 (17%)]\tTotal Loss: 3.2782\tAvg Loss: 0.0002\nTrain Epoch: 3\t[20000/88956 (22%)]\tTotal Loss: 4.3122\tAvg Loss: 0.0002\nTrain Epoch: 3\t[25000/88956 (28%)]\tTotal Loss: 5.3561\tAvg Loss: 0.0002\nTrain Epoch: 3\t[30000/88956 (34%)]\tTotal Loss: 6.3691\tAvg Loss: 0.0002\nTrain Epoch: 3\t[35000/88956 (39%)]\tTotal Loss: 7.3799\tAvg Loss: 0.0002\nTrain Epoch: 3\t[40000/88956 (45%)]\tTotal Loss: 8.4150\tAvg Loss: 0.0002\nTrain Epoch: 3\t[45000/88956 (51%)]\tTotal Loss: 9.4368\tAvg Loss: 0.0002\nTrain Epoch: 3\t[50000/88956 (56%)]\tTotal Loss: 10.4496\tAvg Loss: 0.0002\nTrain Epoch: 3\t[55000/88956 (62%)]\tTotal Loss: 11.4615\tAvg Loss: 0.0002\nTrain Epoch: 3\t[60000/88956 (67%)]\tTotal Loss: 12.4327\tAvg Loss: 0.0002\nTrain Epoch: 3\t[65000/88956 (73%)]\tTotal Loss: 13.3920\tAvg Loss: 0.0002\nTrain Epoch: 3\t[70000/88956 (79%)]\tTotal Loss: 14.3419\tAvg Loss: 0.0002\nTrain Epoch: 3\t[75000/88956 (84%)]\tTotal Loss: 15.3015\tAvg Loss: 0.0002\nTrain Epoch: 3\t[80000/88956 (90%)]\tTotal Loss: 16.2466\tAvg Loss: 0.0002\nTrain Epoch: 3\t[85000/88956 (96%)]\tTotal Loss: 17.2058\tAvg Loss: 0.0002\n====> Epoch: 3\tTotal Loss: 17.9554\t Avg Loss: 0.0002\tCorrect: 24034/88956\tPercentage Correct: 27.02\n====> Val Loss: 1.8258\t Avg Loss: 0.0002\tCorrect: 2659/9885\tPercentage Correct: 26.90\n====> Test Loss: 4.5769\t Avg Loss: 0.0002\tCorrect: 6656/24711\tPercentage Correct: 26.94\n\n=== saved best model ===\n\nTrain Epoch: 4\t[5000/88956 (6%)]\tTotal Loss: 0.8722\tAvg Loss: 0.0002\nTrain Epoch: 4\t[10000/88956 (11%)]\tTotal Loss: 1.7825\tAvg Loss: 0.0002\nTrain Epoch: 4\t[15000/88956 (17%)]\tTotal Loss: 2.6655\tAvg Loss: 0.0002\nTrain Epoch: 4\t[20000/88956 (22%)]\tTotal Loss: 3.5432\tAvg Loss: 0.0002\nTrain Epoch: 4\t[25000/88956 (28%)]\tTotal Loss: 4.4424\tAvg Loss: 0.0002\nTrain Epoch: 4\t[30000/88956 (34%)]\tTotal Loss: 5.2878\tAvg Loss: 0.0002\nTrain Epoch: 4\t[35000/88956 (39%)]\tTotal Loss: 6.1413\tAvg Loss: 0.0002\nTrain Epoch: 4\t[40000/88956 (45%)]\tTotal Loss: 6.9908\tAvg Loss: 0.0002\nTrain Epoch: 4\t[45000/88956 (51%)]\tTotal Loss: 7.9296\tAvg Loss: 0.0002\nTrain Epoch: 4\t[50000/88956 (56%)]\tTotal Loss: 8.8092\tAvg Loss: 0.0002\nTrain Epoch: 4\t[55000/88956 (62%)]\tTotal Loss: 9.6564\tAvg Loss: 0.0002\nTrain Epoch: 4\t[60000/88956 (67%)]\tTotal Loss: 10.4852\tAvg Loss: 0.0002\nTrain Epoch: 4\t[65000/88956 (73%)]\tTotal Loss: 11.3305\tAvg Loss: 0.0002\nTrain Epoch: 4\t[70000/88956 (79%)]\tTotal Loss: 12.1401\tAvg Loss: 0.0002\nTrain Epoch: 4\t[75000/88956 (84%)]\tTotal Loss: 12.9724\tAvg Loss: 0.0002\nTrain Epoch: 4\t[80000/88956 (90%)]\tTotal Loss: 13.8192\tAvg Loss: 0.0002\nTrain Epoch: 4\t[85000/88956 (96%)]\tTotal Loss: 14.6315\tAvg Loss: 0.0002\n====> Epoch: 4\tTotal Loss: 15.2525\t Avg Loss: 0.0002\tCorrect: 27883/88956\tPercentage Correct: 31.34\n====> Val Loss: 1.5898\t Avg Loss: 0.0002\tCorrect: 3156/9885\tPercentage Correct: 31.93\n====> Test Loss: 3.9769\t Avg Loss: 0.0002\tCorrect: 7842/24711\tPercentage Correct: 31.73\n\n=== saved best model ===\n\nTrain Epoch: 5\t[5000/88956 (6%)]\tTotal Loss: 0.7676\tAvg Loss: 0.0002\nTrain Epoch: 5\t[10000/88956 (11%)]\tTotal Loss: 1.5563\tAvg Loss: 0.0002\nTrain Epoch: 5\t[15000/88956 (17%)]\tTotal Loss: 2.3460\tAvg Loss: 0.0002\nTrain Epoch: 5\t[20000/88956 (22%)]\tTotal Loss: 3.1102\tAvg Loss: 0.0002\nTrain Epoch: 5\t[25000/88956 (28%)]\tTotal Loss: 3.8788\tAvg Loss: 0.0002\nTrain Epoch: 5\t[30000/88956 (34%)]\tTotal Loss: 4.6131\tAvg Loss: 0.0002\nTrain Epoch: 5\t[35000/88956 (39%)]\tTotal Loss: 5.3539\tAvg Loss: 0.0002\nTrain Epoch: 5\t[40000/88956 (45%)]\tTotal Loss: 6.0962\tAvg Loss: 0.0002\nTrain Epoch: 5\t[45000/88956 (51%)]\tTotal Loss: 6.8327\tAvg Loss: 0.0002\nTrain Epoch: 5\t[50000/88956 (56%)]\tTotal Loss: 7.5881\tAvg Loss: 0.0002\nTrain Epoch: 5\t[55000/88956 (62%)]\tTotal Loss: 8.3215\tAvg Loss: 0.0002\nTrain Epoch: 5\t[60000/88956 (67%)]\tTotal Loss: 9.0697\tAvg Loss: 0.0002\nTrain Epoch: 5\t[65000/88956 (73%)]\tTotal Loss: 9.7713\tAvg Loss: 0.0002\nTrain Epoch: 5\t[70000/88956 (79%)]\tTotal Loss: 10.4986\tAvg Loss: 0.0001\nTrain Epoch: 5\t[75000/88956 (84%)]\tTotal Loss: 11.2276\tAvg Loss: 0.0001\nTrain Epoch: 5\t[80000/88956 (90%)]\tTotal Loss: 11.9699\tAvg Loss: 0.0001\nTrain Epoch: 5\t[85000/88956 (96%)]\tTotal Loss: 12.7535\tAvg Loss: 0.0002\n====> Epoch: 5\tTotal Loss: 13.3569\t Avg Loss: 0.0002\tCorrect: 31237/88956\tPercentage Correct: 35.12\n====> Val Loss: 1.4198\t Avg Loss: 0.0001\tCorrect: 3554/9885\tPercentage Correct: 35.95\n====> Test Loss: 3.5379\t Avg Loss: 0.0001\tCorrect: 8611/24711\tPercentage Correct: 34.85\n\n=== saved best model ===\n\nTrain Epoch: 6\t[5000/88956 (6%)]\tTotal Loss: 0.7317\tAvg Loss: 0.0001\nTrain Epoch: 6\t[10000/88956 (11%)]\tTotal Loss: 1.3953\tAvg Loss: 0.0001\nTrain Epoch: 6\t[15000/88956 (17%)]\tTotal Loss: 2.0883\tAvg Loss: 0.0001\nTrain Epoch: 6\t[20000/88956 (22%)]\tTotal Loss: 2.7592\tAvg Loss: 0.0001\nTrain Epoch: 6\t[25000/88956 (28%)]\tTotal Loss: 3.5022\tAvg Loss: 0.0001\nTrain Epoch: 6\t[30000/88956 (34%)]\tTotal Loss: 4.2041\tAvg Loss: 0.0001\nTrain Epoch: 6\t[35000/88956 (39%)]\tTotal Loss: 4.8542\tAvg Loss: 0.0001\nTrain Epoch: 6\t[40000/88956 (45%)]\tTotal Loss: 5.5028\tAvg Loss: 0.0001\nTrain Epoch: 6\t[45000/88956 (51%)]\tTotal Loss: 6.1751\tAvg Loss: 0.0001\nTrain Epoch: 6\t[50000/88956 (56%)]\tTotal Loss: 6.8542\tAvg Loss: 0.0001\nTrain Epoch: 6\t[55000/88956 (62%)]\tTotal Loss: 7.5185\tAvg Loss: 0.0001\nTrain Epoch: 6\t[60000/88956 (67%)]\tTotal Loss: 8.1715\tAvg Loss: 0.0001\nTrain Epoch: 6\t[65000/88956 (73%)]\tTotal Loss: 8.8069\tAvg Loss: 0.0001\nTrain Epoch: 6\t[70000/88956 (79%)]\tTotal Loss: 9.4547\tAvg Loss: 0.0001\nTrain Epoch: 6\t[75000/88956 (84%)]\tTotal Loss: 10.1022\tAvg Loss: 0.0001\nTrain Epoch: 6\t[80000/88956 (90%)]\tTotal Loss: 10.7471\tAvg Loss: 0.0001\nTrain Epoch: 6\t[85000/88956 (96%)]\tTotal Loss: 11.3885\tAvg Loss: 0.0001\n====> Epoch: 6\tTotal Loss: 11.9062\t Avg Loss: 0.0001\tCorrect: 34403/88956\tPercentage Correct: 38.67\n====> Val Loss: 1.2699\t Avg Loss: 0.0001\tCorrect: 3841/9885\tPercentage Correct: 38.86\n====> Test Loss: 3.1077\t Avg Loss: 0.0001\tCorrect: 9647/24711\tPercentage Correct: 39.04\n\n=== saved best model ===\n\nTrain Epoch: 7\t[5000/88956 (6%)]\tTotal Loss: 0.6402\tAvg Loss: 0.0001\nTrain Epoch: 7\t[10000/88956 (11%)]\tTotal Loss: 1.2666\tAvg Loss: 0.0001\nTrain Epoch: 7\t[15000/88956 (17%)]\tTotal Loss: 1.8738\tAvg Loss: 0.0001\nTrain Epoch: 7\t[20000/88956 (22%)]\tTotal Loss: 2.5410\tAvg Loss: 0.0001\nTrain Epoch: 7\t[25000/88956 (28%)]\tTotal Loss: 3.1713\tAvg Loss: 0.0001\nTrain Epoch: 7\t[30000/88956 (34%)]\tTotal Loss: 3.8084\tAvg Loss: 0.0001\nTrain Epoch: 7\t[35000/88956 (39%)]\tTotal Loss: 4.4107\tAvg Loss: 0.0001\nTrain Epoch: 7\t[40000/88956 (45%)]\tTotal Loss: 5.0511\tAvg Loss: 0.0001\nTrain Epoch: 7\t[45000/88956 (51%)]\tTotal Loss: 5.6628\tAvg Loss: 0.0001\nTrain Epoch: 7\t[50000/88956 (56%)]\tTotal Loss: 6.2486\tAvg Loss: 0.0001\nTrain Epoch: 7\t[55000/88956 (62%)]\tTotal Loss: 6.8582\tAvg Loss: 0.0001\nTrain Epoch: 7\t[60000/88956 (67%)]\tTotal Loss: 7.4545\tAvg Loss: 0.0001\nTrain Epoch: 7\t[65000/88956 (73%)]\tTotal Loss: 8.0619\tAvg Loss: 0.0001\nTrain Epoch: 7\t[70000/88956 (79%)]\tTotal Loss: 8.6483\tAvg Loss: 0.0001\nTrain Epoch: 7\t[75000/88956 (84%)]\tTotal Loss: 9.2695\tAvg Loss: 0.0001\nTrain Epoch: 7\t[80000/88956 (90%)]\tTotal Loss: 9.8805\tAvg Loss: 0.0001\nTrain Epoch: 7\t[85000/88956 (96%)]\tTotal Loss: 10.5084\tAvg Loss: 0.0001\n====> Epoch: 7\tTotal Loss: 10.9845\t Avg Loss: 0.0001\tCorrect: 36474/88956\tPercentage Correct: 41.00\n====> Val Loss: 1.3034\t Avg Loss: 0.0001\tCorrect: 3976/9885\tPercentage Correct: 40.22\n====> Test Loss: 3.2811\t Avg Loss: 0.0001\tCorrect: 9985/24711\tPercentage Correct: 40.41\n\n=== saved best model ===\n\nTrain Epoch: 8\t[5000/88956 (6%)]\tTotal Loss: 0.5984\tAvg Loss: 0.0001\nTrain Epoch: 8\t[10000/88956 (11%)]\tTotal Loss: 1.1801\tAvg Loss: 0.0001\nTrain Epoch: 8\t[15000/88956 (17%)]\tTotal Loss: 1.7651\tAvg Loss: 0.0001\nTrain Epoch: 8\t[20000/88956 (22%)]\tTotal Loss: 2.3255\tAvg Loss: 0.0001\nTrain Epoch: 8\t[25000/88956 (28%)]\tTotal Loss: 2.8950\tAvg Loss: 0.0001\nTrain Epoch: 8\t[30000/88956 (34%)]\tTotal Loss: 3.5203\tAvg Loss: 0.0001\nTrain Epoch: 8\t[35000/88956 (39%)]\tTotal Loss: 4.0717\tAvg Loss: 0.0001\nTrain Epoch: 8\t[40000/88956 (45%)]\tTotal Loss: 4.6241\tAvg Loss: 0.0001\nTrain Epoch: 8\t[45000/88956 (51%)]\tTotal Loss: 5.2163\tAvg Loss: 0.0001\nTrain Epoch: 8\t[50000/88956 (56%)]\tTotal Loss: 5.7571\tAvg Loss: 0.0001\nTrain Epoch: 8\t[55000/88956 (62%)]\tTotal Loss: 6.3202\tAvg Loss: 0.0001\nTrain Epoch: 8\t[60000/88956 (67%)]\tTotal Loss: 6.8837\tAvg Loss: 0.0001\nTrain Epoch: 8\t[65000/88956 (73%)]\tTotal Loss: 7.4566\tAvg Loss: 0.0001\nTrain Epoch: 8\t[70000/88956 (79%)]\tTotal Loss: 8.0217\tAvg Loss: 0.0001\nTrain Epoch: 8\t[75000/88956 (84%)]\tTotal Loss: 8.5845\tAvg Loss: 0.0001\nTrain Epoch: 8\t[80000/88956 (90%)]\tTotal Loss: 9.1474\tAvg Loss: 0.0001\nTrain Epoch: 8\t[85000/88956 (96%)]\tTotal Loss: 9.7318\tAvg Loss: 0.0001\n====> Epoch: 8\tTotal Loss: 10.1764\t Avg Loss: 0.0001\tCorrect: 38730/88956\tPercentage Correct: 43.54\n====> Val Loss: 1.1481\t Avg Loss: 0.0001\tCorrect: 4338/9885\tPercentage Correct: 43.88\n====> Test Loss: 2.8417\t Avg Loss: 0.0001\tCorrect: 10821/24711\tPercentage Correct: 43.79\n\n=== saved best model ===\n\nTrain Epoch: 9\t[5000/88956 (6%)]\tTotal Loss: 0.5408\tAvg Loss: 0.0001\nTrain Epoch: 9\t[10000/88956 (11%)]\tTotal Loss: 1.0671\tAvg Loss: 0.0001\nTrain Epoch: 9\t[15000/88956 (17%)]\tTotal Loss: 1.6250\tAvg Loss: 0.0001\nTrain Epoch: 9\t[20000/88956 (22%)]\tTotal Loss: 2.1611\tAvg Loss: 0.0001\nTrain Epoch: 9\t[25000/88956 (28%)]\tTotal Loss: 2.7056\tAvg Loss: 0.0001\nTrain Epoch: 9\t[30000/88956 (34%)]\tTotal Loss: 3.2230\tAvg Loss: 0.0001\nTrain Epoch: 9\t[35000/88956 (39%)]\tTotal Loss: 3.7777\tAvg Loss: 0.0001\nTrain Epoch: 9\t[40000/88956 (45%)]\tTotal Loss: 4.3258\tAvg Loss: 0.0001\nTrain Epoch: 9\t[45000/88956 (51%)]\tTotal Loss: 4.8621\tAvg Loss: 0.0001\nTrain Epoch: 9\t[50000/88956 (56%)]\tTotal Loss: 5.3797\tAvg Loss: 0.0001\nTrain Epoch: 9\t[55000/88956 (62%)]\tTotal Loss: 5.9372\tAvg Loss: 0.0001\nTrain Epoch: 9\t[60000/88956 (67%)]\tTotal Loss: 6.4886\tAvg Loss: 0.0001\nTrain Epoch: 9\t[65000/88956 (73%)]\tTotal Loss: 7.0183\tAvg Loss: 0.0001\nTrain Epoch: 9\t[70000/88956 (79%)]\tTotal Loss: 7.5451\tAvg Loss: 0.0001\nTrain Epoch: 9\t[75000/88956 (84%)]\tTotal Loss: 8.0993\tAvg Loss: 0.0001\nTrain Epoch: 9\t[80000/88956 (90%)]\tTotal Loss: 8.6668\tAvg Loss: 0.0001\nTrain Epoch: 9\t[85000/88956 (96%)]\tTotal Loss: 9.2081\tAvg Loss: 0.0001\n====> Epoch: 9\tTotal Loss: 9.6262\t Avg Loss: 0.0001\tCorrect: 40324/88956\tPercentage Correct: 45.33\n====> Val Loss: 1.1321\t Avg Loss: 0.0001\tCorrect: 4395/9885\tPercentage Correct: 44.46\n====> Test Loss: 2.7960\t Avg Loss: 0.0001\tCorrect: 11090/24711\tPercentage Correct: 44.88\n\n=== saved best model ===\n\nTrain Epoch: 10\t[5000/88956 (6%)]\tTotal Loss: 0.5213\tAvg Loss: 0.0001\nTrain Epoch: 10\t[10000/88956 (11%)]\tTotal Loss: 1.0344\tAvg Loss: 0.0001\nTrain Epoch: 10\t[15000/88956 (17%)]\tTotal Loss: 1.5557\tAvg Loss: 0.0001\nTrain Epoch: 10\t[20000/88956 (22%)]\tTotal Loss: 2.0602\tAvg Loss: 0.0001\nTrain Epoch: 10\t[25000/88956 (28%)]\tTotal Loss: 2.5849\tAvg Loss: 0.0001\nTrain Epoch: 10\t[30000/88956 (34%)]\tTotal Loss: 3.1137\tAvg Loss: 0.0001\nTrain Epoch: 10\t[35000/88956 (39%)]\tTotal Loss: 3.6090\tAvg Loss: 0.0001\nTrain Epoch: 10\t[40000/88956 (45%)]\tTotal Loss: 4.1093\tAvg Loss: 0.0001\nTrain Epoch: 10\t[45000/88956 (51%)]\tTotal Loss: 4.6069\tAvg Loss: 0.0001\nTrain Epoch: 10\t[50000/88956 (56%)]\tTotal Loss: 5.1041\tAvg Loss: 0.0001\nTrain Epoch: 10\t[55000/88956 (62%)]\tTotal Loss: 5.6027\tAvg Loss: 0.0001\nTrain Epoch: 10\t[60000/88956 (67%)]\tTotal Loss: 6.1281\tAvg Loss: 0.0001\nTrain Epoch: 10\t[65000/88956 (73%)]\tTotal Loss: 6.6708\tAvg Loss: 0.0001\nTrain Epoch: 10\t[70000/88956 (79%)]\tTotal Loss: 7.2366\tAvg Loss: 0.0001\nTrain Epoch: 10\t[75000/88956 (84%)]\tTotal Loss: 7.7518\tAvg Loss: 0.0001\nTrain Epoch: 10\t[80000/88956 (90%)]\tTotal Loss: 8.2607\tAvg Loss: 0.0001\nTrain Epoch: 10\t[85000/88956 (96%)]\tTotal Loss: 8.7811\tAvg Loss: 0.0001\n====> Epoch: 10\tTotal Loss: 9.1886\t Avg Loss: 0.0001\tCorrect: 41810/88956\tPercentage Correct: 47.00\n====> Val Loss: 1.0966\t Avg Loss: 0.0001\tCorrect: 4539/9885\tPercentage Correct: 45.92\n====> Test Loss: 2.6882\t Avg Loss: 0.0001\tCorrect: 11491/24711\tPercentage Correct: 46.50\n\n=== saved best model ===\n\nTrain Epoch: 11\t[5000/88956 (6%)]\tTotal Loss: 0.4782\tAvg Loss: 0.0001\nTrain Epoch: 11\t[10000/88956 (11%)]\tTotal Loss: 0.9729\tAvg Loss: 0.0001\nTrain Epoch: 11\t[15000/88956 (17%)]\tTotal Loss: 1.4493\tAvg Loss: 0.0001\nTrain Epoch: 11\t[20000/88956 (22%)]\tTotal Loss: 1.9835\tAvg Loss: 0.0001\nTrain Epoch: 11\t[25000/88956 (28%)]\tTotal Loss: 2.4994\tAvg Loss: 0.0001\nTrain Epoch: 11\t[30000/88956 (34%)]\tTotal Loss: 2.9829\tAvg Loss: 0.0001\nTrain Epoch: 11\t[35000/88956 (39%)]\tTotal Loss: 3.4429\tAvg Loss: 0.0001\nTrain Epoch: 11\t[40000/88956 (45%)]\tTotal Loss: 3.9539\tAvg Loss: 0.0001\nTrain Epoch: 11\t[45000/88956 (51%)]\tTotal Loss: 4.4668\tAvg Loss: 0.0001\nTrain Epoch: 11\t[50000/88956 (56%)]\tTotal Loss: 4.9771\tAvg Loss: 0.0001\nTrain Epoch: 11\t[55000/88956 (62%)]\tTotal Loss: 5.4401\tAvg Loss: 0.0001\nTrain Epoch: 11\t[60000/88956 (67%)]\tTotal Loss: 5.9218\tAvg Loss: 0.0001\nTrain Epoch: 11\t[65000/88956 (73%)]\tTotal Loss: 6.3999\tAvg Loss: 0.0001\nTrain Epoch: 11\t[70000/88956 (79%)]\tTotal Loss: 6.8985\tAvg Loss: 0.0001\nTrain Epoch: 11\t[75000/88956 (84%)]\tTotal Loss: 7.3888\tAvg Loss: 0.0001\nTrain Epoch: 11\t[80000/88956 (90%)]\tTotal Loss: 7.8632\tAvg Loss: 0.0001\nTrain Epoch: 11\t[85000/88956 (96%)]\tTotal Loss: 8.3524\tAvg Loss: 0.0001\n====> Epoch: 11\tTotal Loss: 8.7395\t Avg Loss: 0.0001\tCorrect: 43616/88956\tPercentage Correct: 49.03\n====> Val Loss: 1.0668\t Avg Loss: 0.0001\tCorrect: 4642/9885\tPercentage Correct: 46.96\n====> Test Loss: 2.6093\t Avg Loss: 0.0001\tCorrect: 11571/24711\tPercentage Correct: 46.83\n\n=== saved best model ===\n\nTrain Epoch: 12\t[5000/88956 (6%)]\tTotal Loss: 0.4672\tAvg Loss: 0.0001\nTrain Epoch: 12\t[10000/88956 (11%)]\tTotal Loss: 0.9101\tAvg Loss: 0.0001\nTrain Epoch: 12\t[15000/88956 (17%)]\tTotal Loss: 1.3653\tAvg Loss: 0.0001\nTrain Epoch: 12\t[20000/88956 (22%)]\tTotal Loss: 1.8379\tAvg Loss: 0.0001\nTrain Epoch: 12\t[25000/88956 (28%)]\tTotal Loss: 2.2886\tAvg Loss: 0.0001\nTrain Epoch: 12\t[30000/88956 (34%)]\tTotal Loss: 2.7221\tAvg Loss: 0.0001\nTrain Epoch: 12\t[35000/88956 (39%)]\tTotal Loss: 3.1756\tAvg Loss: 0.0001\nTrain Epoch: 12\t[40000/88956 (45%)]\tTotal Loss: 3.6333\tAvg Loss: 0.0001\nTrain Epoch: 12\t[45000/88956 (51%)]\tTotal Loss: 4.1147\tAvg Loss: 0.0001\nTrain Epoch: 12\t[50000/88956 (56%)]\tTotal Loss: 4.5791\tAvg Loss: 0.0001\nTrain Epoch: 12\t[55000/88956 (62%)]\tTotal Loss: 5.0502\tAvg Loss: 0.0001\nTrain Epoch: 12\t[60000/88956 (67%)]\tTotal Loss: 5.4980\tAvg Loss: 0.0001\nTrain Epoch: 12\t[65000/88956 (73%)]\tTotal Loss: 5.9607\tAvg Loss: 0.0001\nTrain Epoch: 12\t[70000/88956 (79%)]\tTotal Loss: 6.4468\tAvg Loss: 0.0001\nTrain Epoch: 12\t[75000/88956 (84%)]\tTotal Loss: 6.9110\tAvg Loss: 0.0001\nTrain Epoch: 12\t[80000/88956 (90%)]\tTotal Loss: 7.3582\tAvg Loss: 0.0001\nTrain Epoch: 12\t[85000/88956 (96%)]\tTotal Loss: 7.8495\tAvg Loss: 0.0001\n====> Epoch: 12\tTotal Loss: 8.2338\t Avg Loss: 0.0001\tCorrect: 45205/88956\tPercentage Correct: 50.82\n====> Val Loss: 1.0261\t Avg Loss: 0.0001\tCorrect: 4811/9885\tPercentage Correct: 48.67\n====> Test Loss: 2.4850\t Avg Loss: 0.0001\tCorrect: 12177/24711\tPercentage Correct: 49.28\n\n=== saved best model ===\n\nTrain Epoch: 13\t[5000/88956 (6%)]\tTotal Loss: 0.4528\tAvg Loss: 0.0001\nTrain Epoch: 13\t[10000/88956 (11%)]\tTotal Loss: 0.8972\tAvg Loss: 0.0001\nTrain Epoch: 13\t[15000/88956 (17%)]\tTotal Loss: 1.3625\tAvg Loss: 0.0001\nTrain Epoch: 13\t[20000/88956 (22%)]\tTotal Loss: 1.8081\tAvg Loss: 0.0001\nTrain Epoch: 13\t[25000/88956 (28%)]\tTotal Loss: 2.2551\tAvg Loss: 0.0001\nTrain Epoch: 13\t[30000/88956 (34%)]\tTotal Loss: 2.6895\tAvg Loss: 0.0001\nTrain Epoch: 13\t[35000/88956 (39%)]\tTotal Loss: 3.1535\tAvg Loss: 0.0001\nTrain Epoch: 13\t[40000/88956 (45%)]\tTotal Loss: 3.5944\tAvg Loss: 0.0001\nTrain Epoch: 13\t[45000/88956 (51%)]\tTotal Loss: 4.0354\tAvg Loss: 0.0001\nTrain Epoch: 13\t[50000/88956 (56%)]\tTotal Loss: 4.4670\tAvg Loss: 0.0001\nTrain Epoch: 13\t[55000/88956 (62%)]\tTotal Loss: 4.9296\tAvg Loss: 0.0001\nTrain Epoch: 13\t[60000/88956 (67%)]\tTotal Loss: 5.4050\tAvg Loss: 0.0001\nTrain Epoch: 13\t[65000/88956 (73%)]\tTotal Loss: 5.8736\tAvg Loss: 0.0001\nTrain Epoch: 13\t[70000/88956 (79%)]\tTotal Loss: 6.3439\tAvg Loss: 0.0001\nTrain Epoch: 13\t[75000/88956 (84%)]\tTotal Loss: 6.7933\tAvg Loss: 0.0001\nTrain Epoch: 13\t[80000/88956 (90%)]\tTotal Loss: 7.2431\tAvg Loss: 0.0001\nTrain Epoch: 13\t[85000/88956 (96%)]\tTotal Loss: 7.7342\tAvg Loss: 0.0001\n====> Epoch: 13\tTotal Loss: 8.0745\t Avg Loss: 0.0001\tCorrect: 45855/88956\tPercentage Correct: 51.55\n====> Val Loss: 0.9728\t Avg Loss: 0.0001\tCorrect: 5030/9885\tPercentage Correct: 50.89\n====> Test Loss: 2.3779\t Avg Loss: 0.0001\tCorrect: 12726/24711\tPercentage Correct: 51.50\n\n=== saved best model ===\n\nTrain Epoch: 14\t[5000/88956 (6%)]\tTotal Loss: 0.4295\tAvg Loss: 0.0001\nTrain Epoch: 14\t[10000/88956 (11%)]\tTotal Loss: 0.8894\tAvg Loss: 0.0001\nTrain Epoch: 14\t[15000/88956 (17%)]\tTotal Loss: 1.3302\tAvg Loss: 0.0001\nTrain Epoch: 14\t[20000/88956 (22%)]\tTotal Loss: 1.7415\tAvg Loss: 0.0001\nTrain Epoch: 14\t[25000/88956 (28%)]\tTotal Loss: 2.1652\tAvg Loss: 0.0001\nTrain Epoch: 14\t[30000/88956 (34%)]\tTotal Loss: 2.5884\tAvg Loss: 0.0001\nTrain Epoch: 14\t[35000/88956 (39%)]\tTotal Loss: 3.0227\tAvg Loss: 0.0001\nTrain Epoch: 14\t[40000/88956 (45%)]\tTotal Loss: 3.4478\tAvg Loss: 0.0001\nTrain Epoch: 14\t[45000/88956 (51%)]\tTotal Loss: 3.8529\tAvg Loss: 0.0001\nTrain Epoch: 14\t[50000/88956 (56%)]\tTotal Loss: 4.3061\tAvg Loss: 0.0001\nTrain Epoch: 14\t[55000/88956 (62%)]\tTotal Loss: 4.7464\tAvg Loss: 0.0001\nTrain Epoch: 14\t[60000/88956 (67%)]\tTotal Loss: 5.1684\tAvg Loss: 0.0001\nTrain Epoch: 14\t[65000/88956 (73%)]\tTotal Loss: 5.6223\tAvg Loss: 0.0001\nTrain Epoch: 14\t[70000/88956 (79%)]\tTotal Loss: 6.0678\tAvg Loss: 0.0001\nTrain Epoch: 14\t[75000/88956 (84%)]\tTotal Loss: 6.5001\tAvg Loss: 0.0001\nTrain Epoch: 14\t[80000/88956 (90%)]\tTotal Loss: 6.9355\tAvg Loss: 0.0001\nTrain Epoch: 14\t[85000/88956 (96%)]\tTotal Loss: 7.3532\tAvg Loss: 0.0001\n====> Epoch: 14\tTotal Loss: 7.6884\t Avg Loss: 0.0001\tCorrect: 47337/88956\tPercentage Correct: 53.21\n====> Val Loss: 0.9701\t Avg Loss: 0.0001\tCorrect: 4877/9885\tPercentage Correct: 49.34\n====> Test Loss: 2.4333\t Avg Loss: 0.0001\tCorrect: 12402/24711\tPercentage Correct: 50.19\nTrain Epoch: 15\t[5000/88956 (6%)]\tTotal Loss: 0.4267\tAvg Loss: 0.0001\nTrain Epoch: 15\t[10000/88956 (11%)]\tTotal Loss: 0.8457\tAvg Loss: 0.0001\nTrain Epoch: 15\t[15000/88956 (17%)]\tTotal Loss: 1.2614\tAvg Loss: 0.0001\nTrain Epoch: 15\t[20000/88956 (22%)]\tTotal Loss: 1.6857\tAvg Loss: 0.0001\nTrain Epoch: 15\t[25000/88956 (28%)]\tTotal Loss: 2.1049\tAvg Loss: 0.0001\nTrain Epoch: 15\t[30000/88956 (34%)]\tTotal Loss: 2.5143\tAvg Loss: 0.0001\nTrain Epoch: 15\t[35000/88956 (39%)]\tTotal Loss: 2.9451\tAvg Loss: 0.0001\nTrain Epoch: 15\t[40000/88956 (45%)]\tTotal Loss: 3.3672\tAvg Loss: 0.0001\nTrain Epoch: 15\t[45000/88956 (51%)]\tTotal Loss: 3.8128\tAvg Loss: 0.0001\nTrain Epoch: 15\t[50000/88956 (56%)]\tTotal Loss: 4.2301\tAvg Loss: 0.0001\nTrain Epoch: 15\t[55000/88956 (62%)]\tTotal Loss: 4.6467\tAvg Loss: 0.0001\nTrain Epoch: 15\t[60000/88956 (67%)]\tTotal Loss: 5.0583\tAvg Loss: 0.0001\nTrain Epoch: 15\t[65000/88956 (73%)]\tTotal Loss: 5.4747\tAvg Loss: 0.0001\nTrain Epoch: 15\t[70000/88956 (79%)]\tTotal Loss: 5.9109\tAvg Loss: 0.0001\nTrain Epoch: 15\t[75000/88956 (84%)]\tTotal Loss: 6.3437\tAvg Loss: 0.0001\nTrain Epoch: 15\t[80000/88956 (90%)]\tTotal Loss: 6.7620\tAvg Loss: 0.0001\nTrain Epoch: 15\t[85000/88956 (96%)]\tTotal Loss: 7.1799\tAvg Loss: 0.0001\n====> Epoch: 15\tTotal Loss: 7.5157\t Avg Loss: 0.0001\tCorrect: 48141/88956\tPercentage Correct: 54.12\n====> Val Loss: 0.9582\t Avg Loss: 0.0001\tCorrect: 5073/9885\tPercentage Correct: 51.32\n====> Test Loss: 2.3570\t Avg Loss: 0.0001\tCorrect: 12922/24711\tPercentage Correct: 52.29\n\n=== saved best model ===\n\nTrain Epoch: 16\t[5000/88956 (6%)]\tTotal Loss: 0.3983\tAvg Loss: 0.0001\nTrain Epoch: 16\t[10000/88956 (11%)]\tTotal Loss: 0.7862\tAvg Loss: 0.0001\nTrain Epoch: 16\t[15000/88956 (17%)]\tTotal Loss: 1.1910\tAvg Loss: 0.0001\nTrain Epoch: 16\t[20000/88956 (22%)]\tTotal Loss: 1.5944\tAvg Loss: 0.0001\nTrain Epoch: 16\t[25000/88956 (28%)]\tTotal Loss: 1.9827\tAvg Loss: 0.0001\nTrain Epoch: 16\t[30000/88956 (34%)]\tTotal Loss: 2.3778\tAvg Loss: 0.0001\nTrain Epoch: 16\t[35000/88956 (39%)]\tTotal Loss: 2.7851\tAvg Loss: 0.0001\nTrain Epoch: 16\t[40000/88956 (45%)]\tTotal Loss: 3.1733\tAvg Loss: 0.0001\nTrain Epoch: 16\t[45000/88956 (51%)]\tTotal Loss: 3.5664\tAvg Loss: 0.0001\nTrain Epoch: 16\t[50000/88956 (56%)]\tTotal Loss: 3.9614\tAvg Loss: 0.0001\nTrain Epoch: 16\t[55000/88956 (62%)]\tTotal Loss: 4.3554\tAvg Loss: 0.0001\nTrain Epoch: 16\t[60000/88956 (67%)]\tTotal Loss: 4.7612\tAvg Loss: 0.0001\nTrain Epoch: 16\t[65000/88956 (73%)]\tTotal Loss: 5.1656\tAvg Loss: 0.0001\nTrain Epoch: 16\t[70000/88956 (79%)]\tTotal Loss: 5.5505\tAvg Loss: 0.0001\nTrain Epoch: 16\t[75000/88956 (84%)]\tTotal Loss: 5.9476\tAvg Loss: 0.0001\nTrain Epoch: 16\t[80000/88956 (90%)]\tTotal Loss: 6.3594\tAvg Loss: 0.0001\nTrain Epoch: 16\t[85000/88956 (96%)]\tTotal Loss: 6.7885\tAvg Loss: 0.0001\n====> Epoch: 16\tTotal Loss: 7.1481\t Avg Loss: 0.0001\tCorrect: 49573/88956\tPercentage Correct: 55.73\n====> Val Loss: 0.9247\t Avg Loss: 0.0001\tCorrect: 4951/9885\tPercentage Correct: 50.09\n====> Test Loss: 2.3046\t Avg Loss: 0.0001\tCorrect: 12527/24711\tPercentage Correct: 50.69\nTrain Epoch: 17\t[5000/88956 (6%)]\tTotal Loss: 0.3963\tAvg Loss: 0.0001\nTrain Epoch: 17\t[10000/88956 (11%)]\tTotal Loss: 0.8107\tAvg Loss: 0.0001\nTrain Epoch: 17\t[15000/88956 (17%)]\tTotal Loss: 1.2190\tAvg Loss: 0.0001\nTrain Epoch: 17\t[20000/88956 (22%)]\tTotal Loss: 1.6042\tAvg Loss: 0.0001\nTrain Epoch: 17\t[25000/88956 (28%)]\tTotal Loss: 1.9929\tAvg Loss: 0.0001\nTrain Epoch: 17\t[30000/88956 (34%)]\tTotal Loss: 2.4081\tAvg Loss: 0.0001\nTrain Epoch: 17\t[35000/88956 (39%)]\tTotal Loss: 2.7734\tAvg Loss: 0.0001\nTrain Epoch: 17\t[40000/88956 (45%)]\tTotal Loss: 3.1698\tAvg Loss: 0.0001\nTrain Epoch: 17\t[45000/88956 (51%)]\tTotal Loss: 3.5681\tAvg Loss: 0.0001\nTrain Epoch: 17\t[50000/88956 (56%)]\tTotal Loss: 3.9387\tAvg Loss: 0.0001\nTrain Epoch: 17\t[55000/88956 (62%)]\tTotal Loss: 4.3207\tAvg Loss: 0.0001\nTrain Epoch: 17\t[60000/88956 (67%)]\tTotal Loss: 4.6952\tAvg Loss: 0.0001\nTrain Epoch: 17\t[65000/88956 (73%)]\tTotal Loss: 5.0685\tAvg Loss: 0.0001\nTrain Epoch: 17\t[70000/88956 (79%)]\tTotal Loss: 5.4773\tAvg Loss: 0.0001\nTrain Epoch: 17\t[75000/88956 (84%)]\tTotal Loss: 5.8766\tAvg Loss: 0.0001\nTrain Epoch: 17\t[80000/88956 (90%)]\tTotal Loss: 6.2653\tAvg Loss: 0.0001\nTrain Epoch: 17\t[85000/88956 (96%)]\tTotal Loss: 6.6541\tAvg Loss: 0.0001\n====> Epoch: 17\tTotal Loss: 6.9743\t Avg Loss: 0.0001\tCorrect: 50256/88956\tPercentage Correct: 56.50\n====> Val Loss: 0.8467\t Avg Loss: 0.0001\tCorrect: 5466/9885\tPercentage Correct: 55.30\n====> Test Loss: 2.0827\t Avg Loss: 0.0001\tCorrect: 13786/24711\tPercentage Correct: 55.79\n\n=== saved best model ===\n\nTrain Epoch: 18\t[5000/88956 (6%)]\tTotal Loss: 0.3815\tAvg Loss: 0.0001\nTrain Epoch: 18\t[10000/88956 (11%)]\tTotal Loss: 0.7472\tAvg Loss: 0.0001\nTrain Epoch: 18\t[15000/88956 (17%)]\tTotal Loss: 1.1246\tAvg Loss: 0.0001\nTrain Epoch: 18\t[20000/88956 (22%)]\tTotal Loss: 1.4924\tAvg Loss: 0.0001\nTrain Epoch: 18\t[25000/88956 (28%)]\tTotal Loss: 1.8886\tAvg Loss: 0.0001\nTrain Epoch: 18\t[30000/88956 (34%)]\tTotal Loss: 2.2631\tAvg Loss: 0.0001\nTrain Epoch: 18\t[35000/88956 (39%)]\tTotal Loss: 2.6374\tAvg Loss: 0.0001\nTrain Epoch: 18\t[40000/88956 (45%)]\tTotal Loss: 2.9955\tAvg Loss: 0.0001\nTrain Epoch: 18\t[45000/88956 (51%)]\tTotal Loss: 3.3736\tAvg Loss: 0.0001\nTrain Epoch: 18\t[50000/88956 (56%)]\tTotal Loss: 3.7698\tAvg Loss: 0.0001\nTrain Epoch: 18\t[55000/88956 (62%)]\tTotal Loss: 4.1585\tAvg Loss: 0.0001\nTrain Epoch: 18\t[60000/88956 (67%)]\tTotal Loss: 4.5411\tAvg Loss: 0.0001\nTrain Epoch: 18\t[65000/88956 (73%)]\tTotal Loss: 4.9313\tAvg Loss: 0.0001\nTrain Epoch: 18\t[70000/88956 (79%)]\tTotal Loss: 5.3037\tAvg Loss: 0.0001\nTrain Epoch: 18\t[75000/88956 (84%)]\tTotal Loss: 5.6688\tAvg Loss: 0.0001\nTrain Epoch: 18\t[80000/88956 (90%)]\tTotal Loss: 6.0630\tAvg Loss: 0.0001\nTrain Epoch: 18\t[85000/88956 (96%)]\tTotal Loss: 6.4342\tAvg Loss: 0.0001\n====> Epoch: 18\tTotal Loss: 6.7185\t Avg Loss: 0.0001\tCorrect: 51390/88956\tPercentage Correct: 57.77\n====> Val Loss: 0.8802\t Avg Loss: 0.0001\tCorrect: 5351/9885\tPercentage Correct: 54.13\n====> Test Loss: 2.1771\t Avg Loss: 0.0001\tCorrect: 13316/24711\tPercentage Correct: 53.89\nTrain Epoch: 19\t[5000/88956 (6%)]\tTotal Loss: 0.3710\tAvg Loss: 0.0001\nTrain Epoch: 19\t[10000/88956 (11%)]\tTotal Loss: 0.7405\tAvg Loss: 0.0001\nTrain Epoch: 19\t[15000/88956 (17%)]\tTotal Loss: 1.1105\tAvg Loss: 0.0001\nTrain Epoch: 19\t[20000/88956 (22%)]\tTotal Loss: 1.4663\tAvg Loss: 0.0001\nTrain Epoch: 19\t[25000/88956 (28%)]\tTotal Loss: 1.8271\tAvg Loss: 0.0001\nTrain Epoch: 19\t[30000/88956 (34%)]\tTotal Loss: 2.1883\tAvg Loss: 0.0001\nTrain Epoch: 19\t[35000/88956 (39%)]\tTotal Loss: 2.5518\tAvg Loss: 0.0001\nTrain Epoch: 19\t[40000/88956 (45%)]\tTotal Loss: 2.9218\tAvg Loss: 0.0001\nTrain Epoch: 19\t[45000/88956 (51%)]\tTotal Loss: 3.2880\tAvg Loss: 0.0001\nTrain Epoch: 19\t[50000/88956 (56%)]\tTotal Loss: 3.6487\tAvg Loss: 0.0001\nTrain Epoch: 19\t[55000/88956 (62%)]\tTotal Loss: 4.0259\tAvg Loss: 0.0001\nTrain Epoch: 19\t[60000/88956 (67%)]\tTotal Loss: 4.3879\tAvg Loss: 0.0001\nTrain Epoch: 19\t[65000/88956 (73%)]\tTotal Loss: 4.7490\tAvg Loss: 0.0001\nTrain Epoch: 19\t[70000/88956 (79%)]\tTotal Loss: 5.1095\tAvg Loss: 0.0001\nTrain Epoch: 19\t[75000/88956 (84%)]\tTotal Loss: 5.4809\tAvg Loss: 0.0001\nTrain Epoch: 19\t[80000/88956 (90%)]\tTotal Loss: 5.8428\tAvg Loss: 0.0001\nTrain Epoch: 19\t[85000/88956 (96%)]\tTotal Loss: 6.2150\tAvg Loss: 0.0001\n====> Epoch: 19\tTotal Loss: 6.5102\t Avg Loss: 0.0001\tCorrect: 52561/88956\tPercentage Correct: 59.09\n====> Val Loss: 0.8437\t Avg Loss: 0.0001\tCorrect: 5433/9885\tPercentage Correct: 54.96\n====> Test Loss: 2.1135\t Avg Loss: 0.0001\tCorrect: 13667/24711\tPercentage Correct: 55.31\nTrain Epoch: 20\t[5000/88956 (6%)]\tTotal Loss: 0.3545\tAvg Loss: 0.0001\nTrain Epoch: 20\t[10000/88956 (11%)]\tTotal Loss: 0.7078\tAvg Loss: 0.0001\nTrain Epoch: 20\t[15000/88956 (17%)]\tTotal Loss: 1.0710\tAvg Loss: 0.0001\nTrain Epoch: 20\t[20000/88956 (22%)]\tTotal Loss: 1.4022\tAvg Loss: 0.0001\nTrain Epoch: 20\t[25000/88956 (28%)]\tTotal Loss: 1.7508\tAvg Loss: 0.0001\nTrain Epoch: 20\t[30000/88956 (34%)]\tTotal Loss: 2.0913\tAvg Loss: 0.0001\nTrain Epoch: 20\t[35000/88956 (39%)]\tTotal Loss: 2.4301\tAvg Loss: 0.0001\nTrain Epoch: 20\t[40000/88956 (45%)]\tTotal Loss: 2.7660\tAvg Loss: 0.0001\nTrain Epoch: 20\t[45000/88956 (51%)]\tTotal Loss: 3.1256\tAvg Loss: 0.0001\nTrain Epoch: 20\t[50000/88956 (56%)]\tTotal Loss: 3.5102\tAvg Loss: 0.0001\nTrain Epoch: 20\t[55000/88956 (62%)]\tTotal Loss: 3.8704\tAvg Loss: 0.0001\nTrain Epoch: 20\t[60000/88956 (67%)]\tTotal Loss: 4.2402\tAvg Loss: 0.0001\nTrain Epoch: 20\t[65000/88956 (73%)]\tTotal Loss: 4.5959\tAvg Loss: 0.0001\nTrain Epoch: 20\t[70000/88956 (79%)]\tTotal Loss: 4.9956\tAvg Loss: 0.0001\nTrain Epoch: 20\t[75000/88956 (84%)]\tTotal Loss: 5.3660\tAvg Loss: 0.0001\nTrain Epoch: 20\t[80000/88956 (90%)]\tTotal Loss: 5.7465\tAvg Loss: 0.0001\nTrain Epoch: 20\t[85000/88956 (96%)]\tTotal Loss: 6.1265\tAvg Loss: 0.0001\n====> Epoch: 20\tTotal Loss: 6.4268\t Avg Loss: 0.0001\tCorrect: 53142/88956\tPercentage Correct: 59.74\n====> Val Loss: 0.8534\t Avg Loss: 0.0001\tCorrect: 5327/9885\tPercentage Correct: 53.89\n====> Test Loss: 2.1481\t Avg Loss: 0.0001\tCorrect: 13328/24711\tPercentage Correct: 53.94\nTrain Epoch: 21\t[5000/88956 (6%)]\tTotal Loss: 0.3417\tAvg Loss: 0.0001\nTrain Epoch: 21\t[10000/88956 (11%)]\tTotal Loss: 0.6552\tAvg Loss: 0.0001\nTrain Epoch: 21\t[15000/88956 (17%)]\tTotal Loss: 0.9893\tAvg Loss: 0.0001\nTrain Epoch: 21\t[20000/88956 (22%)]\tTotal Loss: 1.3172\tAvg Loss: 0.0001\nTrain Epoch: 21\t[25000/88956 (28%)]\tTotal Loss: 1.6725\tAvg Loss: 0.0001\nTrain Epoch: 21\t[30000/88956 (34%)]\tTotal Loss: 1.9993\tAvg Loss: 0.0001\nTrain Epoch: 21\t[35000/88956 (39%)]\tTotal Loss: 2.3530\tAvg Loss: 0.0001\nTrain Epoch: 21\t[40000/88956 (45%)]\tTotal Loss: 2.7163\tAvg Loss: 0.0001\nTrain Epoch: 21\t[45000/88956 (51%)]\tTotal Loss: 3.0764\tAvg Loss: 0.0001\nTrain Epoch: 21\t[50000/88956 (56%)]\tTotal Loss: 3.4260\tAvg Loss: 0.0001\nTrain Epoch: 21\t[55000/88956 (62%)]\tTotal Loss: 3.7616\tAvg Loss: 0.0001\nTrain Epoch: 21\t[60000/88956 (67%)]\tTotal Loss: 4.0975\tAvg Loss: 0.0001\nTrain Epoch: 21\t[65000/88956 (73%)]\tTotal Loss: 4.4253\tAvg Loss: 0.0001\nTrain Epoch: 21\t[70000/88956 (79%)]\tTotal Loss: 4.7615\tAvg Loss: 0.0001\nTrain Epoch: 21\t[75000/88956 (84%)]\tTotal Loss: 5.1332\tAvg Loss: 0.0001\nTrain Epoch: 21\t[80000/88956 (90%)]\tTotal Loss: 5.4836\tAvg Loss: 0.0001\nTrain Epoch: 21\t[85000/88956 (96%)]\tTotal Loss: 5.8399\tAvg Loss: 0.0001\n====> Epoch: 21\tTotal Loss: 6.1098\t Avg Loss: 0.0001\tCorrect: 54516/88956\tPercentage Correct: 61.28\n====> Val Loss: 0.8075\t Avg Loss: 0.0001\tCorrect: 5681/9885\tPercentage Correct: 57.47\n====> Test Loss: 1.9594\t Avg Loss: 0.0001\tCorrect: 14275/24711\tPercentage Correct: 57.77\n\n=== saved best model ===\n\nTrain Epoch: 22\t[5000/88956 (6%)]\tTotal Loss: 0.3325\tAvg Loss: 0.0001\nTrain Epoch: 22\t[10000/88956 (11%)]\tTotal Loss: 0.6515\tAvg Loss: 0.0001\nTrain Epoch: 22\t[15000/88956 (17%)]\tTotal Loss: 0.9724\tAvg Loss: 0.0001\nTrain Epoch: 22\t[20000/88956 (22%)]\tTotal Loss: 1.3025\tAvg Loss: 0.0001\nTrain Epoch: 22\t[25000/88956 (28%)]\tTotal Loss: 1.6293\tAvg Loss: 0.0001\nTrain Epoch: 22\t[30000/88956 (34%)]\tTotal Loss: 1.9471\tAvg Loss: 0.0001\nTrain Epoch: 22\t[35000/88956 (39%)]\tTotal Loss: 2.2914\tAvg Loss: 0.0001\nTrain Epoch: 22\t[40000/88956 (45%)]\tTotal Loss: 2.6252\tAvg Loss: 0.0001\nTrain Epoch: 22\t[45000/88956 (51%)]\tTotal Loss: 2.9539\tAvg Loss: 0.0001\nTrain Epoch: 22\t[50000/88956 (56%)]\tTotal Loss: 3.3032\tAvg Loss: 0.0001\nTrain Epoch: 22\t[55000/88956 (62%)]\tTotal Loss: 3.6547\tAvg Loss: 0.0001\nTrain Epoch: 22\t[60000/88956 (67%)]\tTotal Loss: 4.0030\tAvg Loss: 0.0001\nTrain Epoch: 22\t[65000/88956 (73%)]\tTotal Loss: 4.3340\tAvg Loss: 0.0001\nTrain Epoch: 22\t[70000/88956 (79%)]\tTotal Loss: 4.6677\tAvg Loss: 0.0001\nTrain Epoch: 22\t[75000/88956 (84%)]\tTotal Loss: 5.0141\tAvg Loss: 0.0001\nTrain Epoch: 22\t[80000/88956 (90%)]\tTotal Loss: 5.3584\tAvg Loss: 0.0001\nTrain Epoch: 22\t[85000/88956 (96%)]\tTotal Loss: 5.7014\tAvg Loss: 0.0001\n====> Epoch: 22\tTotal Loss: 5.9602\t Avg Loss: 0.0001\tCorrect: 55138/88956\tPercentage Correct: 61.98\n====> Val Loss: 0.7774\t Avg Loss: 0.0001\tCorrect: 5782/9885\tPercentage Correct: 58.49\n====> Test Loss: 1.9316\t Avg Loss: 0.0001\tCorrect: 14365/24711\tPercentage Correct: 58.13\n\n=== saved best model ===\n\nTrain Epoch: 23\t[5000/88956 (6%)]\tTotal Loss: 0.3410\tAvg Loss: 0.0001\nTrain Epoch: 23\t[10000/88956 (11%)]\tTotal Loss: 0.6558\tAvg Loss: 0.0001\nTrain Epoch: 23\t[15000/88956 (17%)]\tTotal Loss: 0.9771\tAvg Loss: 0.0001\nTrain Epoch: 23\t[20000/88956 (22%)]\tTotal Loss: 1.3058\tAvg Loss: 0.0001\nTrain Epoch: 23\t[25000/88956 (28%)]\tTotal Loss: 1.6294\tAvg Loss: 0.0001\nTrain Epoch: 23\t[30000/88956 (34%)]\tTotal Loss: 1.9792\tAvg Loss: 0.0001\nTrain Epoch: 23\t[35000/88956 (39%)]\tTotal Loss: 2.3028\tAvg Loss: 0.0001\nTrain Epoch: 23\t[40000/88956 (45%)]\tTotal Loss: 2.6156\tAvg Loss: 0.0001\nTrain Epoch: 23\t[45000/88956 (51%)]\tTotal Loss: 2.9409\tAvg Loss: 0.0001\nTrain Epoch: 23\t[50000/88956 (56%)]\tTotal Loss: 3.2530\tAvg Loss: 0.0001\nTrain Epoch: 23\t[55000/88956 (62%)]\tTotal Loss: 3.5714\tAvg Loss: 0.0001\nTrain Epoch: 23\t[60000/88956 (67%)]\tTotal Loss: 3.9028\tAvg Loss: 0.0001\nTrain Epoch: 23\t[65000/88956 (73%)]\tTotal Loss: 4.2155\tAvg Loss: 0.0001\nTrain Epoch: 23\t[70000/88956 (79%)]\tTotal Loss: 4.5547\tAvg Loss: 0.0001\nTrain Epoch: 23\t[75000/88956 (84%)]\tTotal Loss: 4.8651\tAvg Loss: 0.0001\nTrain Epoch: 23\t[80000/88956 (90%)]\tTotal Loss: 5.1978\tAvg Loss: 0.0001\nTrain Epoch: 23\t[85000/88956 (96%)]\tTotal Loss: 5.5196\tAvg Loss: 0.0001\n====> Epoch: 23\tTotal Loss: 5.7805\t Avg Loss: 0.0001\tCorrect: 56110/88956\tPercentage Correct: 63.08\n====> Val Loss: 0.7871\t Avg Loss: 0.0001\tCorrect: 5614/9885\tPercentage Correct: 56.79\n====> Test Loss: 1.9534\t Avg Loss: 0.0001\tCorrect: 14112/24711\tPercentage Correct: 57.11\nTrain Epoch: 24\t[5000/88956 (6%)]\tTotal Loss: 0.3119\tAvg Loss: 0.0001\nTrain Epoch: 24\t[10000/88956 (11%)]\tTotal Loss: 0.6254\tAvg Loss: 0.0001\nTrain Epoch: 24\t[15000/88956 (17%)]\tTotal Loss: 0.9469\tAvg Loss: 0.0001\nTrain Epoch: 24\t[20000/88956 (22%)]\tTotal Loss: 1.2570\tAvg Loss: 0.0001\nTrain Epoch: 24\t[25000/88956 (28%)]\tTotal Loss: 1.5647\tAvg Loss: 0.0001\nTrain Epoch: 24\t[30000/88956 (34%)]\tTotal Loss: 1.8798\tAvg Loss: 0.0001\nTrain Epoch: 24\t[35000/88956 (39%)]\tTotal Loss: 2.2102\tAvg Loss: 0.0001\nTrain Epoch: 24\t[40000/88956 (45%)]\tTotal Loss: 2.5297\tAvg Loss: 0.0001\nTrain Epoch: 24\t[45000/88956 (51%)]\tTotal Loss: 2.8648\tAvg Loss: 0.0001\nTrain Epoch: 24\t[50000/88956 (56%)]\tTotal Loss: 3.1795\tAvg Loss: 0.0001\nTrain Epoch: 24\t[55000/88956 (62%)]\tTotal Loss: 3.5076\tAvg Loss: 0.0001\nTrain Epoch: 24\t[60000/88956 (67%)]\tTotal Loss: 3.8329\tAvg Loss: 0.0001\nTrain Epoch: 24\t[65000/88956 (73%)]\tTotal Loss: 4.1703\tAvg Loss: 0.0001\nTrain Epoch: 24\t[70000/88956 (79%)]\tTotal Loss: 4.5024\tAvg Loss: 0.0001\nTrain Epoch: 24\t[75000/88956 (84%)]\tTotal Loss: 4.8421\tAvg Loss: 0.0001\nTrain Epoch: 24\t[80000/88956 (90%)]\tTotal Loss: 5.1586\tAvg Loss: 0.0001\nTrain Epoch: 24\t[85000/88956 (96%)]\tTotal Loss: 5.4659\tAvg Loss: 0.0001\n====> Epoch: 24\tTotal Loss: 5.7214\t Avg Loss: 0.0001\tCorrect: 56410/88956\tPercentage Correct: 63.41\n====> Val Loss: 0.7978\t Avg Loss: 0.0001\tCorrect: 5597/9885\tPercentage Correct: 56.62\n====> Test Loss: 2.0053\t Avg Loss: 0.0001\tCorrect: 14030/24711\tPercentage Correct: 56.78\nTrain Epoch: 25\t[5000/88956 (6%)]\tTotal Loss: 0.3075\tAvg Loss: 0.0001\nTrain Epoch: 25\t[10000/88956 (11%)]\tTotal Loss: 0.6178\tAvg Loss: 0.0001\nTrain Epoch: 25\t[15000/88956 (17%)]\tTotal Loss: 0.9193\tAvg Loss: 0.0001\nTrain Epoch: 25\t[20000/88956 (22%)]\tTotal Loss: 1.2238\tAvg Loss: 0.0001\nTrain Epoch: 25\t[25000/88956 (28%)]\tTotal Loss: 1.5298\tAvg Loss: 0.0001\nTrain Epoch: 25\t[30000/88956 (34%)]\tTotal Loss: 1.8138\tAvg Loss: 0.0001\nTrain Epoch: 25\t[35000/88956 (39%)]\tTotal Loss: 2.1382\tAvg Loss: 0.0001\nTrain Epoch: 25\t[40000/88956 (45%)]\tTotal Loss: 2.4682\tAvg Loss: 0.0001\nTrain Epoch: 25\t[45000/88956 (51%)]\tTotal Loss: 2.7873\tAvg Loss: 0.0001\nTrain Epoch: 25\t[50000/88956 (56%)]\tTotal Loss: 3.1122\tAvg Loss: 0.0001\nTrain Epoch: 25\t[55000/88956 (62%)]\tTotal Loss: 3.4236\tAvg Loss: 0.0001\nTrain Epoch: 25\t[60000/88956 (67%)]\tTotal Loss: 3.7401\tAvg Loss: 0.0001\nTrain Epoch: 25\t[65000/88956 (73%)]\tTotal Loss: 4.0755\tAvg Loss: 0.0001\nTrain Epoch: 25\t[70000/88956 (79%)]\tTotal Loss: 4.3982\tAvg Loss: 0.0001\nTrain Epoch: 25\t[75000/88956 (84%)]\tTotal Loss: 4.7106\tAvg Loss: 0.0001\nTrain Epoch: 25\t[80000/88956 (90%)]\tTotal Loss: 5.0386\tAvg Loss: 0.0001\nTrain Epoch: 25\t[85000/88956 (96%)]\tTotal Loss: 5.3567\tAvg Loss: 0.0001\n====> Epoch: 25\tTotal Loss: 5.5908\t Avg Loss: 0.0001\tCorrect: 57128/88956\tPercentage Correct: 64.22\n====> Val Loss: 0.7365\t Avg Loss: 0.0001\tCorrect: 5902/9885\tPercentage Correct: 59.71\n====> Test Loss: 1.8116\t Avg Loss: 0.0001\tCorrect: 14804/24711\tPercentage Correct: 59.91\n\n=== saved best model ===\n\nTrain Epoch: 26\t[5000/88956 (6%)]\tTotal Loss: 0.3021\tAvg Loss: 0.0001\nTrain Epoch: 26\t[10000/88956 (11%)]\tTotal Loss: 0.5855\tAvg Loss: 0.0001\nTrain Epoch: 26\t[15000/88956 (17%)]\tTotal Loss: 0.8829\tAvg Loss: 0.0001\nTrain Epoch: 26\t[20000/88956 (22%)]\tTotal Loss: 1.1727\tAvg Loss: 0.0001\nTrain Epoch: 26\t[25000/88956 (28%)]\tTotal Loss: 1.4682\tAvg Loss: 0.0001\nTrain Epoch: 26\t[30000/88956 (34%)]\tTotal Loss: 1.7610\tAvg Loss: 0.0001\nTrain Epoch: 26\t[35000/88956 (39%)]\tTotal Loss: 2.0505\tAvg Loss: 0.0001\nTrain Epoch: 26\t[40000/88956 (45%)]\tTotal Loss: 2.3525\tAvg Loss: 0.0001\nTrain Epoch: 26\t[45000/88956 (51%)]\tTotal Loss: 2.6643\tAvg Loss: 0.0001\nTrain Epoch: 26\t[50000/88956 (56%)]\tTotal Loss: 2.9743\tAvg Loss: 0.0001\nTrain Epoch: 26\t[55000/88956 (62%)]\tTotal Loss: 3.2939\tAvg Loss: 0.0001\nTrain Epoch: 26\t[60000/88956 (67%)]\tTotal Loss: 3.5952\tAvg Loss: 0.0001\nTrain Epoch: 26\t[65000/88956 (73%)]\tTotal Loss: 3.8924\tAvg Loss: 0.0001\nTrain Epoch: 26\t[70000/88956 (79%)]\tTotal Loss: 4.1868\tAvg Loss: 0.0001\nTrain Epoch: 26\t[75000/88956 (84%)]\tTotal Loss: 4.4879\tAvg Loss: 0.0001\nTrain Epoch: 26\t[80000/88956 (90%)]\tTotal Loss: 4.7898\tAvg Loss: 0.0001\nTrain Epoch: 26\t[85000/88956 (96%)]\tTotal Loss: 5.0808\tAvg Loss: 0.0001\n====> Epoch: 26\tTotal Loss: 5.3263\t Avg Loss: 0.0001\tCorrect: 58271/88956\tPercentage Correct: 65.51\n====> Val Loss: 0.7613\t Avg Loss: 0.0001\tCorrect: 5872/9885\tPercentage Correct: 59.40\n====> Test Loss: 1.8751\t Avg Loss: 0.0001\tCorrect: 14783/24711\tPercentage Correct: 59.82\nTrain Epoch: 27\t[5000/88956 (6%)]\tTotal Loss: 0.3083\tAvg Loss: 0.0001\nTrain Epoch: 27\t[10000/88956 (11%)]\tTotal Loss: 0.6042\tAvg Loss: 0.0001\nTrain Epoch: 27\t[15000/88956 (17%)]\tTotal Loss: 0.8920\tAvg Loss: 0.0001\nTrain Epoch: 27\t[20000/88956 (22%)]\tTotal Loss: 1.1892\tAvg Loss: 0.0001\nTrain Epoch: 27\t[25000/88956 (28%)]\tTotal Loss: 1.4853\tAvg Loss: 0.0001\nTrain Epoch: 27\t[30000/88956 (34%)]\tTotal Loss: 1.7653\tAvg Loss: 0.0001\nTrain Epoch: 27\t[35000/88956 (39%)]\tTotal Loss: 2.0422\tAvg Loss: 0.0001\nTrain Epoch: 27\t[40000/88956 (45%)]\tTotal Loss: 2.3441\tAvg Loss: 0.0001\nTrain Epoch: 27\t[45000/88956 (51%)]\tTotal Loss: 2.6455\tAvg Loss: 0.0001\nTrain Epoch: 27\t[50000/88956 (56%)]\tTotal Loss: 2.9536\tAvg Loss: 0.0001\nTrain Epoch: 27\t[55000/88956 (62%)]\tTotal Loss: 3.3474\tAvg Loss: 0.0001\nTrain Epoch: 27\t[60000/88956 (67%)]\tTotal Loss: 3.6827\tAvg Loss: 0.0001\nTrain Epoch: 27\t[65000/88956 (73%)]\tTotal Loss: 4.0153\tAvg Loss: 0.0001\nTrain Epoch: 27\t[70000/88956 (79%)]\tTotal Loss: 4.3201\tAvg Loss: 0.0001\nTrain Epoch: 27\t[75000/88956 (84%)]\tTotal Loss: 4.6208\tAvg Loss: 0.0001\nTrain Epoch: 27\t[80000/88956 (90%)]\tTotal Loss: 4.9349\tAvg Loss: 0.0001\nTrain Epoch: 27\t[85000/88956 (96%)]\tTotal Loss: 5.2372\tAvg Loss: 0.0001\n====> Epoch: 27\tTotal Loss: 5.4803\t Avg Loss: 0.0001\tCorrect: 57541/88956\tPercentage Correct: 64.68\n====> Val Loss: 0.6967\t Avg Loss: 0.0001\tCorrect: 6087/9885\tPercentage Correct: 61.58\n====> Test Loss: 1.7500\t Avg Loss: 0.0001\tCorrect: 15239/24711\tPercentage Correct: 61.67\n\n=== saved best model ===\n\nTrain Epoch: 28\t[5000/88956 (6%)]\tTotal Loss: 0.2944\tAvg Loss: 0.0001\nTrain Epoch: 28\t[10000/88956 (11%)]\tTotal Loss: 0.5649\tAvg Loss: 0.0001\nTrain Epoch: 28\t[15000/88956 (17%)]\tTotal Loss: 0.8530\tAvg Loss: 0.0001\nTrain Epoch: 28\t[20000/88956 (22%)]\tTotal Loss: 1.1277\tAvg Loss: 0.0001\nTrain Epoch: 28\t[25000/88956 (28%)]\tTotal Loss: 1.3948\tAvg Loss: 0.0001\nTrain Epoch: 28\t[30000/88956 (34%)]\tTotal Loss: 1.6858\tAvg Loss: 0.0001\nTrain Epoch: 28\t[35000/88956 (39%)]\tTotal Loss: 1.9791\tAvg Loss: 0.0001\nTrain Epoch: 28\t[40000/88956 (45%)]\tTotal Loss: 2.2861\tAvg Loss: 0.0001\nTrain Epoch: 28\t[45000/88956 (51%)]\tTotal Loss: 2.5761\tAvg Loss: 0.0001\nTrain Epoch: 28\t[50000/88956 (56%)]\tTotal Loss: 2.8774\tAvg Loss: 0.0001\nTrain Epoch: 28\t[55000/88956 (62%)]\tTotal Loss: 3.1554\tAvg Loss: 0.0001\nTrain Epoch: 28\t[60000/88956 (67%)]\tTotal Loss: 3.4332\tAvg Loss: 0.0001\nTrain Epoch: 28\t[65000/88956 (73%)]\tTotal Loss: 3.7392\tAvg Loss: 0.0001\nTrain Epoch: 28\t[70000/88956 (79%)]\tTotal Loss: 4.0581\tAvg Loss: 0.0001\nTrain Epoch: 28\t[75000/88956 (84%)]\tTotal Loss: 4.3580\tAvg Loss: 0.0001\nTrain Epoch: 28\t[80000/88956 (90%)]\tTotal Loss: 4.6507\tAvg Loss: 0.0001\nTrain Epoch: 28\t[85000/88956 (96%)]\tTotal Loss: 4.9437\tAvg Loss: 0.0001\n====> Epoch: 28\tTotal Loss: 5.1858\t Avg Loss: 0.0001\tCorrect: 58765/88956\tPercentage Correct: 66.06\n====> Val Loss: 0.7277\t Avg Loss: 0.0001\tCorrect: 6016/9885\tPercentage Correct: 60.86\n====> Test Loss: 1.7436\t Avg Loss: 0.0001\tCorrect: 15126/24711\tPercentage Correct: 61.21\nTrain Epoch: 29\t[5000/88956 (6%)]\tTotal Loss: 0.2878\tAvg Loss: 0.0001\nTrain Epoch: 29\t[10000/88956 (11%)]\tTotal Loss: 0.5686\tAvg Loss: 0.0001\nTrain Epoch: 29\t[15000/88956 (17%)]\tTotal Loss: 0.8364\tAvg Loss: 0.0001\nTrain Epoch: 29\t[20000/88956 (22%)]\tTotal Loss: 1.1345\tAvg Loss: 0.0001\nTrain Epoch: 29\t[25000/88956 (28%)]\tTotal Loss: 1.4444\tAvg Loss: 0.0001\nTrain Epoch: 29\t[30000/88956 (34%)]\tTotal Loss: 1.7165\tAvg Loss: 0.0001\nTrain Epoch: 29\t[35000/88956 (39%)]\tTotal Loss: 1.9990\tAvg Loss: 0.0001\nTrain Epoch: 29\t[40000/88956 (45%)]\tTotal Loss: 2.2759\tAvg Loss: 0.0001\nTrain Epoch: 29\t[45000/88956 (51%)]\tTotal Loss: 2.5559\tAvg Loss: 0.0001\nTrain Epoch: 29\t[50000/88956 (56%)]\tTotal Loss: 2.8402\tAvg Loss: 0.0001\nTrain Epoch: 29\t[55000/88956 (62%)]\tTotal Loss: 3.1267\tAvg Loss: 0.0001\nTrain Epoch: 29\t[60000/88956 (67%)]\tTotal Loss: 3.4120\tAvg Loss: 0.0001\nTrain Epoch: 29\t[65000/88956 (73%)]\tTotal Loss: 3.6835\tAvg Loss: 0.0001\nTrain Epoch: 29\t[70000/88956 (79%)]\tTotal Loss: 3.9660\tAvg Loss: 0.0001\nTrain Epoch: 29\t[75000/88956 (84%)]\tTotal Loss: 4.2491\tAvg Loss: 0.0001\nTrain Epoch: 29\t[80000/88956 (90%)]\tTotal Loss: 4.5245\tAvg Loss: 0.0001\nTrain Epoch: 29\t[85000/88956 (96%)]\tTotal Loss: 4.8162\tAvg Loss: 0.0001\n====> Epoch: 29\tTotal Loss: 5.0273\t Avg Loss: 0.0001\tCorrect: 59780/88956\tPercentage Correct: 67.20\n====> Val Loss: 0.7901\t Avg Loss: 0.0001\tCorrect: 5874/9885\tPercentage Correct: 59.42\n====> Test Loss: 1.9130\t Avg Loss: 0.0001\tCorrect: 14636/24711\tPercentage Correct: 59.23\nTrain Epoch: 30\t[5000/88956 (6%)]\tTotal Loss: 0.2768\tAvg Loss: 0.0001\nTrain Epoch: 30\t[10000/88956 (11%)]\tTotal Loss: 0.5494\tAvg Loss: 0.0001\nTrain Epoch: 30\t[15000/88956 (17%)]\tTotal Loss: 0.8213\tAvg Loss: 0.0001\nTrain Epoch: 30\t[20000/88956 (22%)]\tTotal Loss: 1.1055\tAvg Loss: 0.0001\nTrain Epoch: 30\t[25000/88956 (28%)]\tTotal Loss: 1.3879\tAvg Loss: 0.0001\nTrain Epoch: 30\t[30000/88956 (34%)]\tTotal Loss: 1.6689\tAvg Loss: 0.0001\nTrain Epoch: 30\t[35000/88956 (39%)]\tTotal Loss: 1.9593\tAvg Loss: 0.0001\nTrain Epoch: 30\t[40000/88956 (45%)]\tTotal Loss: 2.2352\tAvg Loss: 0.0001\nTrain Epoch: 30\t[45000/88956 (51%)]\tTotal Loss: 2.5072\tAvg Loss: 0.0001\nTrain Epoch: 30\t[50000/88956 (56%)]\tTotal Loss: 2.7891\tAvg Loss: 0.0001\nTrain Epoch: 30\t[55000/88956 (62%)]\tTotal Loss: 3.0781\tAvg Loss: 0.0001\nTrain Epoch: 30\t[60000/88956 (67%)]\tTotal Loss: 3.3509\tAvg Loss: 0.0001\nTrain Epoch: 30\t[65000/88956 (73%)]\tTotal Loss: 3.6519\tAvg Loss: 0.0001\nTrain Epoch: 30\t[70000/88956 (79%)]\tTotal Loss: 3.9386\tAvg Loss: 0.0001\nTrain Epoch: 30\t[75000/88956 (84%)]\tTotal Loss: 4.2081\tAvg Loss: 0.0001\nTrain Epoch: 30\t[80000/88956 (90%)]\tTotal Loss: 4.4986\tAvg Loss: 0.0001\nTrain Epoch: 30\t[85000/88956 (96%)]\tTotal Loss: 4.7903\tAvg Loss: 0.0001\n====> Epoch: 30\tTotal Loss: 5.0219\t Avg Loss: 0.0001\tCorrect: 59672/88956\tPercentage Correct: 67.08\n====> Val Loss: 0.6963\t Avg Loss: 0.0001\tCorrect: 6195/9885\tPercentage Correct: 62.67\n====> Test Loss: 1.6857\t Avg Loss: 0.0001\tCorrect: 15542/24711\tPercentage Correct: 62.90\n\n=== saved best model ===\n\nTrain Epoch: 31\t[5000/88956 (6%)]\tTotal Loss: 0.2597\tAvg Loss: 0.0001\nTrain Epoch: 31\t[10000/88956 (11%)]\tTotal Loss: 0.5186\tAvg Loss: 0.0001\nTrain Epoch: 31\t[15000/88956 (17%)]\tTotal Loss: 0.7914\tAvg Loss: 0.0001\nTrain Epoch: 31\t[20000/88956 (22%)]\tTotal Loss: 1.0620\tAvg Loss: 0.0001\nTrain Epoch: 31\t[25000/88956 (28%)]\tTotal Loss: 1.3261\tAvg Loss: 0.0001\nTrain Epoch: 31\t[30000/88956 (34%)]\tTotal Loss: 1.6061\tAvg Loss: 0.0001\nTrain Epoch: 31\t[35000/88956 (39%)]\tTotal Loss: 1.8880\tAvg Loss: 0.0001\nTrain Epoch: 31\t[40000/88956 (45%)]\tTotal Loss: 2.1926\tAvg Loss: 0.0001\nTrain Epoch: 31\t[45000/88956 (51%)]\tTotal Loss: 2.4544\tAvg Loss: 0.0001\nTrain Epoch: 31\t[50000/88956 (56%)]\tTotal Loss: 2.7433\tAvg Loss: 0.0001\nTrain Epoch: 31\t[55000/88956 (62%)]\tTotal Loss: 3.0156\tAvg Loss: 0.0001\nTrain Epoch: 31\t[60000/88956 (67%)]\tTotal Loss: 3.2994\tAvg Loss: 0.0001\nTrain Epoch: 31\t[65000/88956 (73%)]\tTotal Loss: 3.5678\tAvg Loss: 0.0001\nTrain Epoch: 31\t[70000/88956 (79%)]\tTotal Loss: 3.8561\tAvg Loss: 0.0001\nTrain Epoch: 31\t[75000/88956 (84%)]\tTotal Loss: 4.1230\tAvg Loss: 0.0001\nTrain Epoch: 31\t[80000/88956 (90%)]\tTotal Loss: 4.3989\tAvg Loss: 0.0001\nTrain Epoch: 31\t[85000/88956 (96%)]\tTotal Loss: 4.6775\tAvg Loss: 0.0001\n====> Epoch: 31\tTotal Loss: 4.9020\t Avg Loss: 0.0001\tCorrect: 60356/88956\tPercentage Correct: 67.85\n====> Val Loss: 0.6804\t Avg Loss: 0.0001\tCorrect: 6212/9885\tPercentage Correct: 62.84\n====> Test Loss: 1.6827\t Avg Loss: 0.0001\tCorrect: 15504/24711\tPercentage Correct: 62.74\n\n=== saved best model ===\n\nTrain Epoch: 32\t[5000/88956 (6%)]\tTotal Loss: 0.2772\tAvg Loss: 0.0001\nTrain Epoch: 32\t[10000/88956 (11%)]\tTotal Loss: 0.5173\tAvg Loss: 0.0001\nTrain Epoch: 32\t[15000/88956 (17%)]\tTotal Loss: 0.7991\tAvg Loss: 0.0001\nTrain Epoch: 32\t[20000/88956 (22%)]\tTotal Loss: 1.0786\tAvg Loss: 0.0001\nTrain Epoch: 32\t[25000/88956 (28%)]\tTotal Loss: 1.3394\tAvg Loss: 0.0001\nTrain Epoch: 32\t[30000/88956 (34%)]\tTotal Loss: 1.6063\tAvg Loss: 0.0001\nTrain Epoch: 32\t[35000/88956 (39%)]\tTotal Loss: 1.8738\tAvg Loss: 0.0001\nTrain Epoch: 32\t[40000/88956 (45%)]\tTotal Loss: 2.1441\tAvg Loss: 0.0001\nTrain Epoch: 32\t[45000/88956 (51%)]\tTotal Loss: 2.4215\tAvg Loss: 0.0001\nTrain Epoch: 32\t[50000/88956 (56%)]\tTotal Loss: 2.6920\tAvg Loss: 0.0001\nTrain Epoch: 32\t[55000/88956 (62%)]\tTotal Loss: 3.0641\tAvg Loss: 0.0001\nTrain Epoch: 32\t[60000/88956 (67%)]\tTotal Loss: 3.3769\tAvg Loss: 0.0001\nTrain Epoch: 32\t[65000/88956 (73%)]\tTotal Loss: 3.6471\tAvg Loss: 0.0001\nTrain Epoch: 32\t[70000/88956 (79%)]\tTotal Loss: 3.9082\tAvg Loss: 0.0001\nTrain Epoch: 32\t[75000/88956 (84%)]\tTotal Loss: 4.1866\tAvg Loss: 0.0001\nTrain Epoch: 32\t[80000/88956 (90%)]\tTotal Loss: 4.4743\tAvg Loss: 0.0001\nTrain Epoch: 32\t[85000/88956 (96%)]\tTotal Loss: 4.7424\tAvg Loss: 0.0001\n====> Epoch: 32\tTotal Loss: 4.9524\t Avg Loss: 0.0001\tCorrect: 59861/88956\tPercentage Correct: 67.29\n====> Val Loss: 0.6906\t Avg Loss: 0.0001\tCorrect: 6177/9885\tPercentage Correct: 62.49\n====> Test Loss: 1.6714\t Avg Loss: 0.0001\tCorrect: 15612/24711\tPercentage Correct: 63.18\nTrain Epoch: 33\t[5000/88956 (6%)]\tTotal Loss: 0.2645\tAvg Loss: 0.0001\nTrain Epoch: 33\t[10000/88956 (11%)]\tTotal Loss: 0.5288\tAvg Loss: 0.0001\nTrain Epoch: 33\t[15000/88956 (17%)]\tTotal Loss: 0.7883\tAvg Loss: 0.0001\nTrain Epoch: 33\t[20000/88956 (22%)]\tTotal Loss: 1.0529\tAvg Loss: 0.0001\nTrain Epoch: 33\t[25000/88956 (28%)]\tTotal Loss: 1.3005\tAvg Loss: 0.0001\nTrain Epoch: 33\t[30000/88956 (34%)]\tTotal Loss: 1.5470\tAvg Loss: 0.0001\nTrain Epoch: 33\t[35000/88956 (39%)]\tTotal Loss: 1.8205\tAvg Loss: 0.0001\nTrain Epoch: 33\t[40000/88956 (45%)]\tTotal Loss: 2.1005\tAvg Loss: 0.0001\nTrain Epoch: 33\t[45000/88956 (51%)]\tTotal Loss: 2.3774\tAvg Loss: 0.0001\nTrain Epoch: 33\t[50000/88956 (56%)]\tTotal Loss: 2.6509\tAvg Loss: 0.0001\nTrain Epoch: 33\t[55000/88956 (62%)]\tTotal Loss: 2.9235\tAvg Loss: 0.0001\nTrain Epoch: 33\t[60000/88956 (67%)]\tTotal Loss: 3.2000\tAvg Loss: 0.0001\nTrain Epoch: 33\t[65000/88956 (73%)]\tTotal Loss: 3.4738\tAvg Loss: 0.0001\nTrain Epoch: 33\t[70000/88956 (79%)]\tTotal Loss: 3.7448\tAvg Loss: 0.0001\nTrain Epoch: 33\t[75000/88956 (84%)]\tTotal Loss: 4.0023\tAvg Loss: 0.0001\nTrain Epoch: 33\t[80000/88956 (90%)]\tTotal Loss: 4.2624\tAvg Loss: 0.0001\nTrain Epoch: 33\t[85000/88956 (96%)]\tTotal Loss: 4.5277\tAvg Loss: 0.0001\n====> Epoch: 33\tTotal Loss: 4.7325\t Avg Loss: 0.0001\tCorrect: 60996/88956\tPercentage Correct: 68.57\n====> Val Loss: 0.6645\t Avg Loss: 0.0001\tCorrect: 6164/9885\tPercentage Correct: 62.36\n====> Test Loss: 1.6093\t Avg Loss: 0.0001\tCorrect: 15535/24711\tPercentage Correct: 62.87\nTrain Epoch: 34\t[5000/88956 (6%)]\tTotal Loss: 0.2501\tAvg Loss: 0.0001\nTrain Epoch: 34\t[10000/88956 (11%)]\tTotal Loss: 0.5145\tAvg Loss: 0.0001\nTrain Epoch: 34\t[15000/88956 (17%)]\tTotal Loss: 0.7569\tAvg Loss: 0.0001\nTrain Epoch: 34\t[20000/88956 (22%)]\tTotal Loss: 0.9966\tAvg Loss: 0.0000\nTrain Epoch: 34\t[25000/88956 (28%)]\tTotal Loss: 1.2505\tAvg Loss: 0.0001\nTrain Epoch: 34\t[30000/88956 (34%)]\tTotal Loss: 1.4956\tAvg Loss: 0.0000\nTrain Epoch: 34\t[35000/88956 (39%)]\tTotal Loss: 1.7564\tAvg Loss: 0.0001\nTrain Epoch: 34\t[40000/88956 (45%)]\tTotal Loss: 2.0303\tAvg Loss: 0.0001\nTrain Epoch: 34\t[45000/88956 (51%)]\tTotal Loss: 2.2865\tAvg Loss: 0.0001\nTrain Epoch: 34\t[50000/88956 (56%)]\tTotal Loss: 2.5623\tAvg Loss: 0.0001\nTrain Epoch: 34\t[55000/88956 (62%)]\tTotal Loss: 2.8231\tAvg Loss: 0.0001\nTrain Epoch: 34\t[60000/88956 (67%)]\tTotal Loss: 3.0774\tAvg Loss: 0.0001\nTrain Epoch: 34\t[65000/88956 (73%)]\tTotal Loss: 3.3181\tAvg Loss: 0.0001\nTrain Epoch: 34\t[70000/88956 (79%)]\tTotal Loss: 3.5727\tAvg Loss: 0.0001\nTrain Epoch: 34\t[75000/88956 (84%)]\tTotal Loss: 3.8401\tAvg Loss: 0.0001\nTrain Epoch: 34\t[80000/88956 (90%)]\tTotal Loss: 4.1015\tAvg Loss: 0.0001\nTrain Epoch: 34\t[85000/88956 (96%)]\tTotal Loss: 4.3543\tAvg Loss: 0.0001\n====> Epoch: 34\tTotal Loss: 4.5646\t Avg Loss: 0.0001\tCorrect: 61818/88956\tPercentage Correct: 69.49\n====> Val Loss: 0.7065\t Avg Loss: 0.0001\tCorrect: 6221/9885\tPercentage Correct: 62.93\n====> Test Loss: 1.7242\t Avg Loss: 0.0001\tCorrect: 15654/24711\tPercentage Correct: 63.35\n\n=== saved best model ===\n\nTrain Epoch: 35\t[5000/88956 (6%)]\tTotal Loss: 0.2607\tAvg Loss: 0.0001\nTrain Epoch: 35\t[10000/88956 (11%)]\tTotal Loss: 0.5104\tAvg Loss: 0.0001\nTrain Epoch: 35\t[15000/88956 (17%)]\tTotal Loss: 0.7587\tAvg Loss: 0.0001\nTrain Epoch: 35\t[20000/88956 (22%)]\tTotal Loss: 1.0187\tAvg Loss: 0.0001\nTrain Epoch: 35\t[25000/88956 (28%)]\tTotal Loss: 1.2670\tAvg Loss: 0.0001\nTrain Epoch: 35\t[30000/88956 (34%)]\tTotal Loss: 1.5270\tAvg Loss: 0.0001\nTrain Epoch: 35\t[35000/88956 (39%)]\tTotal Loss: 1.7809\tAvg Loss: 0.0001\nTrain Epoch: 35\t[40000/88956 (45%)]\tTotal Loss: 2.0329\tAvg Loss: 0.0001\nTrain Epoch: 35\t[45000/88956 (51%)]\tTotal Loss: 2.2978\tAvg Loss: 0.0001\nTrain Epoch: 35\t[50000/88956 (56%)]\tTotal Loss: 2.5598\tAvg Loss: 0.0001\nTrain Epoch: 35\t[55000/88956 (62%)]\tTotal Loss: 2.8226\tAvg Loss: 0.0001\nTrain Epoch: 35\t[60000/88956 (67%)]\tTotal Loss: 3.0993\tAvg Loss: 0.0001\nTrain Epoch: 35\t[65000/88956 (73%)]\tTotal Loss: 3.3533\tAvg Loss: 0.0001\nTrain Epoch: 35\t[70000/88956 (79%)]\tTotal Loss: 3.6044\tAvg Loss: 0.0001\nTrain Epoch: 35\t[75000/88956 (84%)]\tTotal Loss: 3.8522\tAvg Loss: 0.0001\nTrain Epoch: 35\t[80000/88956 (90%)]\tTotal Loss: 4.1110\tAvg Loss: 0.0001\nTrain Epoch: 35\t[85000/88956 (96%)]\tTotal Loss: 4.4206\tAvg Loss: 0.0001\n====> Epoch: 35\tTotal Loss: 4.6429\t Avg Loss: 0.0001\tCorrect: 61651/88956\tPercentage Correct: 69.31\n====> Val Loss: 0.7070\t Avg Loss: 0.0001\tCorrect: 6054/9885\tPercentage Correct: 61.24\n====> Test Loss: 1.7595\t Avg Loss: 0.0001\tCorrect: 15077/24711\tPercentage Correct: 61.01\nTrain Epoch: 36\t[5000/88956 (6%)]\tTotal Loss: 0.2465\tAvg Loss: 0.0000\nTrain Epoch: 36\t[10000/88956 (11%)]\tTotal Loss: 0.4869\tAvg Loss: 0.0000\nTrain Epoch: 36\t[15000/88956 (17%)]\tTotal Loss: 0.7437\tAvg Loss: 0.0000\nTrain Epoch: 36\t[20000/88956 (22%)]\tTotal Loss: 0.9927\tAvg Loss: 0.0000\nTrain Epoch: 36\t[25000/88956 (28%)]\tTotal Loss: 1.2478\tAvg Loss: 0.0000\nTrain Epoch: 36\t[30000/88956 (34%)]\tTotal Loss: 1.4768\tAvg Loss: 0.0000\nTrain Epoch: 36\t[35000/88956 (39%)]\tTotal Loss: 1.7228\tAvg Loss: 0.0000\nTrain Epoch: 36\t[40000/88956 (45%)]\tTotal Loss: 1.9902\tAvg Loss: 0.0000\nTrain Epoch: 36\t[45000/88956 (51%)]\tTotal Loss: 2.2449\tAvg Loss: 0.0000\nTrain Epoch: 36\t[50000/88956 (56%)]\tTotal Loss: 2.4886\tAvg Loss: 0.0000\nTrain Epoch: 36\t[55000/88956 (62%)]\tTotal Loss: 2.7476\tAvg Loss: 0.0000\nTrain Epoch: 36\t[60000/88956 (67%)]\tTotal Loss: 2.9981\tAvg Loss: 0.0000\nTrain Epoch: 36\t[65000/88956 (73%)]\tTotal Loss: 3.2530\tAvg Loss: 0.0001\nTrain Epoch: 36\t[70000/88956 (79%)]\tTotal Loss: 3.5209\tAvg Loss: 0.0001\nTrain Epoch: 36\t[75000/88956 (84%)]\tTotal Loss: 3.7773\tAvg Loss: 0.0001\nTrain Epoch: 36\t[80000/88956 (90%)]\tTotal Loss: 4.0338\tAvg Loss: 0.0001\nTrain Epoch: 36\t[85000/88956 (96%)]\tTotal Loss: 4.2764\tAvg Loss: 0.0001\n====> Epoch: 36\tTotal Loss: 4.4848\t Avg Loss: 0.0001\tCorrect: 62260/88956\tPercentage Correct: 69.99\n====> Val Loss: 0.6664\t Avg Loss: 0.0001\tCorrect: 6325/9885\tPercentage Correct: 63.99\n====> Test Loss: 1.6711\t Avg Loss: 0.0001\tCorrect: 15829/24711\tPercentage Correct: 64.06\n\n=== saved best model ===\n\nTrain Epoch: 37\t[5000/88956 (6%)]\tTotal Loss: 0.2458\tAvg Loss: 0.0000\nTrain Epoch: 37\t[10000/88956 (11%)]\tTotal Loss: 0.4980\tAvg Loss: 0.0000\nTrain Epoch: 37\t[15000/88956 (17%)]\tTotal Loss: 0.7194\tAvg Loss: 0.0000\nTrain Epoch: 37\t[20000/88956 (22%)]\tTotal Loss: 0.9638\tAvg Loss: 0.0000\nTrain Epoch: 37\t[25000/88956 (28%)]\tTotal Loss: 1.1971\tAvg Loss: 0.0000\nTrain Epoch: 37\t[30000/88956 (34%)]\tTotal Loss: 1.4413\tAvg Loss: 0.0000\nTrain Epoch: 37\t[35000/88956 (39%)]\tTotal Loss: 1.6799\tAvg Loss: 0.0000\nTrain Epoch: 37\t[40000/88956 (45%)]\tTotal Loss: 1.9211\tAvg Loss: 0.0000\nTrain Epoch: 37\t[45000/88956 (51%)]\tTotal Loss: 2.1835\tAvg Loss: 0.0000\nTrain Epoch: 37\t[50000/88956 (56%)]\tTotal Loss: 2.4332\tAvg Loss: 0.0000\nTrain Epoch: 37\t[55000/88956 (62%)]\tTotal Loss: 2.6999\tAvg Loss: 0.0000\nTrain Epoch: 37\t[60000/88956 (67%)]\tTotal Loss: 2.9804\tAvg Loss: 0.0000\nTrain Epoch: 37\t[65000/88956 (73%)]\tTotal Loss: 3.2409\tAvg Loss: 0.0000\nTrain Epoch: 37\t[70000/88956 (79%)]\tTotal Loss: 3.4839\tAvg Loss: 0.0000\nTrain Epoch: 37\t[75000/88956 (84%)]\tTotal Loss: 3.7267\tAvg Loss: 0.0000\nTrain Epoch: 37\t[80000/88956 (90%)]\tTotal Loss: 3.9699\tAvg Loss: 0.0000\nTrain Epoch: 37\t[85000/88956 (96%)]\tTotal Loss: 4.2283\tAvg Loss: 0.0000\n====> Epoch: 37\tTotal Loss: 4.4379\t Avg Loss: 0.0000\tCorrect: 62365/88956\tPercentage Correct: 70.11\n====> Val Loss: 0.6322\t Avg Loss: 0.0001\tCorrect: 6380/9885\tPercentage Correct: 64.54\n====> Test Loss: 1.5677\t Avg Loss: 0.0001\tCorrect: 15949/24711\tPercentage Correct: 64.54\n\n=== saved best model ===\n\nTrain Epoch: 38\t[5000/88956 (6%)]\tTotal Loss: 0.2254\tAvg Loss: 0.0000\nTrain Epoch: 38\t[10000/88956 (11%)]\tTotal Loss: 0.4597\tAvg Loss: 0.0000\nTrain Epoch: 38\t[15000/88956 (17%)]\tTotal Loss: 0.7045\tAvg Loss: 0.0000\nTrain Epoch: 38\t[20000/88956 (22%)]\tTotal Loss: 0.9450\tAvg Loss: 0.0000\nTrain Epoch: 38\t[25000/88956 (28%)]\tTotal Loss: 1.1926\tAvg Loss: 0.0000\nTrain Epoch: 38\t[30000/88956 (34%)]\tTotal Loss: 1.4425\tAvg Loss: 0.0000\nTrain Epoch: 38\t[35000/88956 (39%)]\tTotal Loss: 1.6929\tAvg Loss: 0.0000\nTrain Epoch: 38\t[40000/88956 (45%)]\tTotal Loss: 1.9403\tAvg Loss: 0.0000\nTrain Epoch: 38\t[45000/88956 (51%)]\tTotal Loss: 2.1877\tAvg Loss: 0.0000\nTrain Epoch: 38\t[50000/88956 (56%)]\tTotal Loss: 2.4372\tAvg Loss: 0.0000\nTrain Epoch: 38\t[55000/88956 (62%)]\tTotal Loss: 2.6868\tAvg Loss: 0.0000\nTrain Epoch: 38\t[60000/88956 (67%)]\tTotal Loss: 2.9373\tAvg Loss: 0.0000\nTrain Epoch: 38\t[65000/88956 (73%)]\tTotal Loss: 3.1765\tAvg Loss: 0.0000\nTrain Epoch: 38\t[70000/88956 (79%)]\tTotal Loss: 3.4184\tAvg Loss: 0.0000\nTrain Epoch: 38\t[75000/88956 (84%)]\tTotal Loss: 3.6689\tAvg Loss: 0.0000\nTrain Epoch: 38\t[80000/88956 (90%)]\tTotal Loss: 3.9216\tAvg Loss: 0.0000\nTrain Epoch: 38\t[85000/88956 (96%)]\tTotal Loss: 4.1663\tAvg Loss: 0.0000\n====> Epoch: 38\tTotal Loss: 4.3583\t Avg Loss: 0.0000\tCorrect: 62843/88956\tPercentage Correct: 70.65\n====> Val Loss: 0.6734\t Avg Loss: 0.0001\tCorrect: 6248/9885\tPercentage Correct: 63.21\n====> Test Loss: 1.6548\t Avg Loss: 0.0001\tCorrect: 15755/24711\tPercentage Correct: 63.76\nTrain Epoch: 39\t[5000/88956 (6%)]\tTotal Loss: 0.2411\tAvg Loss: 0.0000\nTrain Epoch: 39\t[10000/88956 (11%)]\tTotal Loss: 0.4730\tAvg Loss: 0.0000\nTrain Epoch: 39\t[15000/88956 (17%)]\tTotal Loss: 0.6981\tAvg Loss: 0.0000\nTrain Epoch: 39\t[20000/88956 (22%)]\tTotal Loss: 0.9238\tAvg Loss: 0.0000\nTrain Epoch: 39\t[25000/88956 (28%)]\tTotal Loss: 1.1602\tAvg Loss: 0.0000\nTrain Epoch: 39\t[30000/88956 (34%)]\tTotal Loss: 1.4723\tAvg Loss: 0.0000\nTrain Epoch: 39\t[35000/88956 (39%)]\tTotal Loss: 1.7496\tAvg Loss: 0.0000\nTrain Epoch: 39\t[40000/88956 (45%)]\tTotal Loss: 2.0096\tAvg Loss: 0.0001\nTrain Epoch: 39\t[45000/88956 (51%)]\tTotal Loss: 2.2569\tAvg Loss: 0.0001\nTrain Epoch: 39\t[50000/88956 (56%)]\tTotal Loss: 2.4949\tAvg Loss: 0.0000\nTrain Epoch: 39\t[55000/88956 (62%)]\tTotal Loss: 2.7197\tAvg Loss: 0.0000\nTrain Epoch: 39\t[60000/88956 (67%)]\tTotal Loss: 2.9630\tAvg Loss: 0.0000\nTrain Epoch: 39\t[65000/88956 (73%)]\tTotal Loss: 3.1959\tAvg Loss: 0.0000\nTrain Epoch: 39\t[70000/88956 (79%)]\tTotal Loss: 3.4379\tAvg Loss: 0.0000\nTrain Epoch: 39\t[75000/88956 (84%)]\tTotal Loss: 3.6793\tAvg Loss: 0.0000\nTrain Epoch: 39\t[80000/88956 (90%)]\tTotal Loss: 3.9233\tAvg Loss: 0.0000\nTrain Epoch: 39\t[85000/88956 (96%)]\tTotal Loss: 4.1751\tAvg Loss: 0.0000\n====> Epoch: 39\tTotal Loss: 4.3669\t Avg Loss: 0.0000\tCorrect: 62824/88956\tPercentage Correct: 70.62\n====> Val Loss: 0.6597\t Avg Loss: 0.0001\tCorrect: 6301/9885\tPercentage Correct: 63.74\n====> Test Loss: 1.5935\t Avg Loss: 0.0001\tCorrect: 15850/24711\tPercentage Correct: 64.14\nTrain Epoch: 40\t[5000/88956 (6%)]\tTotal Loss: 0.2302\tAvg Loss: 0.0000\nTrain Epoch: 40\t[10000/88956 (11%)]\tTotal Loss: 0.4587\tAvg Loss: 0.0000\nTrain Epoch: 40\t[15000/88956 (17%)]\tTotal Loss: 0.6836\tAvg Loss: 0.0000\nTrain Epoch: 40\t[20000/88956 (22%)]\tTotal Loss: 0.9244\tAvg Loss: 0.0000\nTrain Epoch: 40\t[25000/88956 (28%)]\tTotal Loss: 1.1631\tAvg Loss: 0.0000\nTrain Epoch: 40\t[30000/88956 (34%)]\tTotal Loss: 1.3889\tAvg Loss: 0.0000\nTrain Epoch: 40\t[35000/88956 (39%)]\tTotal Loss: 1.6191\tAvg Loss: 0.0000\nTrain Epoch: 40\t[40000/88956 (45%)]\tTotal Loss: 1.8594\tAvg Loss: 0.0000\nTrain Epoch: 40\t[45000/88956 (51%)]\tTotal Loss: 2.0846\tAvg Loss: 0.0000\nTrain Epoch: 40\t[50000/88956 (56%)]\tTotal Loss: 2.3329\tAvg Loss: 0.0000\nTrain Epoch: 40\t[55000/88956 (62%)]\tTotal Loss: 2.5796\tAvg Loss: 0.0000\nTrain Epoch: 40\t[60000/88956 (67%)]\tTotal Loss: 2.8138\tAvg Loss: 0.0000\nTrain Epoch: 40\t[65000/88956 (73%)]\tTotal Loss: 3.0363\tAvg Loss: 0.0000\nTrain Epoch: 40\t[70000/88956 (79%)]\tTotal Loss: 3.2768\tAvg Loss: 0.0000\nTrain Epoch: 40\t[75000/88956 (84%)]\tTotal Loss: 3.5059\tAvg Loss: 0.0000\nTrain Epoch: 40\t[80000/88956 (90%)]\tTotal Loss: 3.7270\tAvg Loss: 0.0000\nTrain Epoch: 40\t[85000/88956 (96%)]\tTotal Loss: 3.9666\tAvg Loss: 0.0000\n====> Epoch: 40\tTotal Loss: 4.1468\t Avg Loss: 0.0000\tCorrect: 63985/88956\tPercentage Correct: 71.93\n====> Val Loss: 0.6079\t Avg Loss: 0.0001\tCorrect: 6543/9885\tPercentage Correct: 66.19\n====> Test Loss: 1.4701\t Avg Loss: 0.0001\tCorrect: 16365/24711\tPercentage Correct: 66.23\n\n=== saved best model ===\n\nTrain Epoch: 41\t[5000/88956 (6%)]\tTotal Loss: 0.2189\tAvg Loss: 0.0000\nTrain Epoch: 41\t[10000/88956 (11%)]\tTotal Loss: 0.4267\tAvg Loss: 0.0000\nTrain Epoch: 41\t[15000/88956 (17%)]\tTotal Loss: 0.6431\tAvg Loss: 0.0000\nTrain Epoch: 41\t[20000/88956 (22%)]\tTotal Loss: 0.8664\tAvg Loss: 0.0000\nTrain Epoch: 41\t[25000/88956 (28%)]\tTotal Loss: 1.0943\tAvg Loss: 0.0000\nTrain Epoch: 41\t[30000/88956 (34%)]\tTotal Loss: 1.3299\tAvg Loss: 0.0000\nTrain Epoch: 41\t[35000/88956 (39%)]\tTotal Loss: 1.5626\tAvg Loss: 0.0000\nTrain Epoch: 41\t[40000/88956 (45%)]\tTotal Loss: 1.7802\tAvg Loss: 0.0000\nTrain Epoch: 41\t[45000/88956 (51%)]\tTotal Loss: 2.0378\tAvg Loss: 0.0000\nTrain Epoch: 41\t[50000/88956 (56%)]\tTotal Loss: 2.2790\tAvg Loss: 0.0000\nTrain Epoch: 41\t[55000/88956 (62%)]\tTotal Loss: 2.5157\tAvg Loss: 0.0000\nTrain Epoch: 41\t[60000/88956 (67%)]\tTotal Loss: 2.7317\tAvg Loss: 0.0000\nTrain Epoch: 41\t[65000/88956 (73%)]\tTotal Loss: 2.9459\tAvg Loss: 0.0000\nTrain Epoch: 41\t[70000/88956 (79%)]\tTotal Loss: 3.1912\tAvg Loss: 0.0000\nTrain Epoch: 41\t[75000/88956 (84%)]\tTotal Loss: 3.4379\tAvg Loss: 0.0000\nTrain Epoch: 41\t[80000/88956 (90%)]\tTotal Loss: 3.6719\tAvg Loss: 0.0000\nTrain Epoch: 41\t[85000/88956 (96%)]\tTotal Loss: 3.9084\tAvg Loss: 0.0000\n====> Epoch: 41\tTotal Loss: 4.0977\t Avg Loss: 0.0000\tCorrect: 64213/88956\tPercentage Correct: 72.19\n====> Val Loss: 0.6462\t Avg Loss: 0.0001\tCorrect: 6273/9885\tPercentage Correct: 63.46\n====> Test Loss: 1.5748\t Avg Loss: 0.0001\tCorrect: 15810/24711\tPercentage Correct: 63.98\nTrain Epoch: 42\t[5000/88956 (6%)]\tTotal Loss: 0.2317\tAvg Loss: 0.0000\nTrain Epoch: 42\t[10000/88956 (11%)]\tTotal Loss: 0.4689\tAvg Loss: 0.0000\nTrain Epoch: 42\t[15000/88956 (17%)]\tTotal Loss: 0.6852\tAvg Loss: 0.0000\nTrain Epoch: 42\t[20000/88956 (22%)]\tTotal Loss: 0.8994\tAvg Loss: 0.0000\nTrain Epoch: 42\t[25000/88956 (28%)]\tTotal Loss: 1.1316\tAvg Loss: 0.0000\nTrain Epoch: 42\t[30000/88956 (34%)]\tTotal Loss: 1.3537\tAvg Loss: 0.0000\nTrain Epoch: 42\t[35000/88956 (39%)]\tTotal Loss: 1.5847\tAvg Loss: 0.0000\nTrain Epoch: 42\t[40000/88956 (45%)]\tTotal Loss: 1.8063\tAvg Loss: 0.0000\nTrain Epoch: 42\t[45000/88956 (51%)]\tTotal Loss: 2.0403\tAvg Loss: 0.0000\nTrain Epoch: 42\t[50000/88956 (56%)]\tTotal Loss: 2.2704\tAvg Loss: 0.0000\nTrain Epoch: 42\t[55000/88956 (62%)]\tTotal Loss: 2.5164\tAvg Loss: 0.0000\nTrain Epoch: 42\t[60000/88956 (67%)]\tTotal Loss: 2.7415\tAvg Loss: 0.0000\nTrain Epoch: 42\t[65000/88956 (73%)]\tTotal Loss: 2.9628\tAvg Loss: 0.0000\nTrain Epoch: 42\t[70000/88956 (79%)]\tTotal Loss: 3.2077\tAvg Loss: 0.0000\nTrain Epoch: 42\t[75000/88956 (84%)]\tTotal Loss: 3.4480\tAvg Loss: 0.0000\nTrain Epoch: 42\t[80000/88956 (90%)]\tTotal Loss: 3.6803\tAvg Loss: 0.0000\nTrain Epoch: 42\t[85000/88956 (96%)]\tTotal Loss: 3.9081\tAvg Loss: 0.0000\n====> Epoch: 42\tTotal Loss: 4.0915\t Avg Loss: 0.0000\tCorrect: 64348/88956\tPercentage Correct: 72.34\n====> Val Loss: 0.6336\t Avg Loss: 0.0001\tCorrect: 6537/9885\tPercentage Correct: 66.13\n====> Test Loss: 1.5483\t Avg Loss: 0.0001\tCorrect: 16292/24711\tPercentage Correct: 65.93\nTrain Epoch: 43\t[5000/88956 (6%)]\tTotal Loss: 0.2278\tAvg Loss: 0.0000\nTrain Epoch: 43\t[10000/88956 (11%)]\tTotal Loss: 0.4582\tAvg Loss: 0.0000\nTrain Epoch: 43\t[15000/88956 (17%)]\tTotal Loss: 0.6779\tAvg Loss: 0.0000\nTrain Epoch: 43\t[20000/88956 (22%)]\tTotal Loss: 0.8915\tAvg Loss: 0.0000\nTrain Epoch: 43\t[25000/88956 (28%)]\tTotal Loss: 1.1040\tAvg Loss: 0.0000\nTrain Epoch: 43\t[30000/88956 (34%)]\tTotal Loss: 1.3233\tAvg Loss: 0.0000\nTrain Epoch: 43\t[35000/88956 (39%)]\tTotal Loss: 1.5352\tAvg Loss: 0.0000\nTrain Epoch: 43\t[40000/88956 (45%)]\tTotal Loss: 1.7643\tAvg Loss: 0.0000\nTrain Epoch: 43\t[45000/88956 (51%)]\tTotal Loss: 1.9874\tAvg Loss: 0.0000\nTrain Epoch: 43\t[50000/88956 (56%)]\tTotal Loss: 2.2219\tAvg Loss: 0.0000\nTrain Epoch: 43\t[55000/88956 (62%)]\tTotal Loss: 2.4760\tAvg Loss: 0.0000\nTrain Epoch: 43\t[60000/88956 (67%)]\tTotal Loss: 2.7210\tAvg Loss: 0.0000\nTrain Epoch: 43\t[65000/88956 (73%)]\tTotal Loss: 2.9479\tAvg Loss: 0.0000\nTrain Epoch: 43\t[70000/88956 (79%)]\tTotal Loss: 3.1778\tAvg Loss: 0.0000\nTrain Epoch: 43\t[75000/88956 (84%)]\tTotal Loss: 3.4166\tAvg Loss: 0.0000\nTrain Epoch: 43\t[80000/88956 (90%)]\tTotal Loss: 3.6569\tAvg Loss: 0.0000\nTrain Epoch: 43\t[85000/88956 (96%)]\tTotal Loss: 3.8856\tAvg Loss: 0.0000\n====> Epoch: 43\tTotal Loss: 4.0613\t Avg Loss: 0.0000\tCorrect: 64286/88956\tPercentage Correct: 72.27\n====> Val Loss: 0.6107\t Avg Loss: 0.0001\tCorrect: 6447/9885\tPercentage Correct: 65.22\n====> Test Loss: 1.4657\t Avg Loss: 0.0001\tCorrect: 16316/24711\tPercentage Correct: 66.03\nTrain Epoch: 44\t[5000/88956 (6%)]\tTotal Loss: 0.2244\tAvg Loss: 0.0000\nTrain Epoch: 44\t[10000/88956 (11%)]\tTotal Loss: 0.4462\tAvg Loss: 0.0000\nTrain Epoch: 44\t[15000/88956 (17%)]\tTotal Loss: 0.6585\tAvg Loss: 0.0000\nTrain Epoch: 44\t[20000/88956 (22%)]\tTotal Loss: 0.8857\tAvg Loss: 0.0000\nTrain Epoch: 44\t[25000/88956 (28%)]\tTotal Loss: 1.0989\tAvg Loss: 0.0000\nTrain Epoch: 44\t[30000/88956 (34%)]\tTotal Loss: 1.3109\tAvg Loss: 0.0000\nTrain Epoch: 44\t[35000/88956 (39%)]\tTotal Loss: 1.5231\tAvg Loss: 0.0000\nTrain Epoch: 44\t[40000/88956 (45%)]\tTotal Loss: 1.7573\tAvg Loss: 0.0000\nTrain Epoch: 44\t[45000/88956 (51%)]\tTotal Loss: 1.9841\tAvg Loss: 0.0000\nTrain Epoch: 44\t[50000/88956 (56%)]\tTotal Loss: 2.2031\tAvg Loss: 0.0000\nTrain Epoch: 44\t[55000/88956 (62%)]\tTotal Loss: 2.4173\tAvg Loss: 0.0000\nTrain Epoch: 44\t[60000/88956 (67%)]\tTotal Loss: 2.6512\tAvg Loss: 0.0000\nTrain Epoch: 44\t[65000/88956 (73%)]\tTotal Loss: 2.8748\tAvg Loss: 0.0000\nTrain Epoch: 44\t[70000/88956 (79%)]\tTotal Loss: 3.0924\tAvg Loss: 0.0000\nTrain Epoch: 44\t[75000/88956 (84%)]\tTotal Loss: 3.3178\tAvg Loss: 0.0000\nTrain Epoch: 44\t[80000/88956 (90%)]\tTotal Loss: 3.5487\tAvg Loss: 0.0000\nTrain Epoch: 44\t[85000/88956 (96%)]\tTotal Loss: 3.7845\tAvg Loss: 0.0000\n====> Epoch: 44\tTotal Loss: 3.9721\t Avg Loss: 0.0000\tCorrect: 64834/88956\tPercentage Correct: 72.88\n====> Val Loss: 0.6023\t Avg Loss: 0.0001\tCorrect: 6534/9885\tPercentage Correct: 66.10\n====> Test Loss: 1.4468\t Avg Loss: 0.0001\tCorrect: 16347/24711\tPercentage Correct: 66.15\nTrain Epoch: 45\t[5000/88956 (6%)]\tTotal Loss: 0.2164\tAvg Loss: 0.0000\nTrain Epoch: 45\t[10000/88956 (11%)]\tTotal Loss: 0.4357\tAvg Loss: 0.0000\nTrain Epoch: 45\t[15000/88956 (17%)]\tTotal Loss: 0.6536\tAvg Loss: 0.0000\nTrain Epoch: 45\t[20000/88956 (22%)]\tTotal Loss: 0.8644\tAvg Loss: 0.0000\nTrain Epoch: 45\t[25000/88956 (28%)]\tTotal Loss: 1.0881\tAvg Loss: 0.0000\nTrain Epoch: 45\t[30000/88956 (34%)]\tTotal Loss: 1.3337\tAvg Loss: 0.0000\nTrain Epoch: 45\t[35000/88956 (39%)]\tTotal Loss: 1.5538\tAvg Loss: 0.0000\nTrain Epoch: 45\t[40000/88956 (45%)]\tTotal Loss: 1.7643\tAvg Loss: 0.0000\nTrain Epoch: 45\t[45000/88956 (51%)]\tTotal Loss: 1.9828\tAvg Loss: 0.0000\nTrain Epoch: 45\t[50000/88956 (56%)]\tTotal Loss: 2.2185\tAvg Loss: 0.0000\nTrain Epoch: 45\t[55000/88956 (62%)]\tTotal Loss: 2.4409\tAvg Loss: 0.0000\nTrain Epoch: 45\t[60000/88956 (67%)]\tTotal Loss: 2.6634\tAvg Loss: 0.0000\nTrain Epoch: 45\t[65000/88956 (73%)]\tTotal Loss: 2.8906\tAvg Loss: 0.0000\nTrain Epoch: 45\t[70000/88956 (79%)]\tTotal Loss: 3.1230\tAvg Loss: 0.0000\nTrain Epoch: 45\t[75000/88956 (84%)]\tTotal Loss: 3.3335\tAvg Loss: 0.0000\nTrain Epoch: 45\t[80000/88956 (90%)]\tTotal Loss: 3.5465\tAvg Loss: 0.0000\nTrain Epoch: 45\t[85000/88956 (96%)]\tTotal Loss: 3.7635\tAvg Loss: 0.0000\n====> Epoch: 45\tTotal Loss: 3.9342\t Avg Loss: 0.0000\tCorrect: 64978/88956\tPercentage Correct: 73.05\n====> Val Loss: 0.5804\t Avg Loss: 0.0001\tCorrect: 6592/9885\tPercentage Correct: 66.69\n====> Test Loss: 1.4051\t Avg Loss: 0.0001\tCorrect: 16597/24711\tPercentage Correct: 67.16\n\n=== saved best model ===\n\nTrain Epoch: 46\t[5000/88956 (6%)]\tTotal Loss: 0.2118\tAvg Loss: 0.0000\nTrain Epoch: 46\t[10000/88956 (11%)]\tTotal Loss: 0.4254\tAvg Loss: 0.0000\nTrain Epoch: 46\t[15000/88956 (17%)]\tTotal Loss: 0.6372\tAvg Loss: 0.0000\nTrain Epoch: 46\t[20000/88956 (22%)]\tTotal Loss: 0.8358\tAvg Loss: 0.0000\nTrain Epoch: 46\t[25000/88956 (28%)]\tTotal Loss: 1.0705\tAvg Loss: 0.0000\nTrain Epoch: 46\t[30000/88956 (34%)]\tTotal Loss: 1.2681\tAvg Loss: 0.0000\nTrain Epoch: 46\t[35000/88956 (39%)]\tTotal Loss: 1.4672\tAvg Loss: 0.0000\nTrain Epoch: 46\t[40000/88956 (45%)]\tTotal Loss: 1.6709\tAvg Loss: 0.0000\nTrain Epoch: 46\t[45000/88956 (51%)]\tTotal Loss: 1.8897\tAvg Loss: 0.0000\nTrain Epoch: 46\t[50000/88956 (56%)]\tTotal Loss: 2.1086\tAvg Loss: 0.0000\nTrain Epoch: 46\t[55000/88956 (62%)]\tTotal Loss: 2.3359\tAvg Loss: 0.0000\nTrain Epoch: 46\t[60000/88956 (67%)]\tTotal Loss: 2.5561\tAvg Loss: 0.0000\nTrain Epoch: 46\t[65000/88956 (73%)]\tTotal Loss: 2.7785\tAvg Loss: 0.0000\nTrain Epoch: 46\t[70000/88956 (79%)]\tTotal Loss: 3.0050\tAvg Loss: 0.0000\nTrain Epoch: 46\t[75000/88956 (84%)]\tTotal Loss: 3.2257\tAvg Loss: 0.0000\nTrain Epoch: 46\t[80000/88956 (90%)]\tTotal Loss: 3.4466\tAvg Loss: 0.0000\nTrain Epoch: 46\t[85000/88956 (96%)]\tTotal Loss: 3.6624\tAvg Loss: 0.0000\n====> Epoch: 46\tTotal Loss: 3.8376\t Avg Loss: 0.0000\tCorrect: 65288/88956\tPercentage Correct: 73.39\n====> Val Loss: 0.6389\t Avg Loss: 0.0001\tCorrect: 6460/9885\tPercentage Correct: 65.35\n====> Test Loss: 1.5784\t Avg Loss: 0.0001\tCorrect: 16101/24711\tPercentage Correct: 65.16\nTrain Epoch: 47\t[5000/88956 (6%)]\tTotal Loss: 0.2392\tAvg Loss: 0.0000\nTrain Epoch: 47\t[10000/88956 (11%)]\tTotal Loss: 0.4474\tAvg Loss: 0.0000\nTrain Epoch: 47\t[15000/88956 (17%)]\tTotal Loss: 0.6627\tAvg Loss: 0.0000\nTrain Epoch: 47\t[20000/88956 (22%)]\tTotal Loss: 0.8881\tAvg Loss: 0.0000\nTrain Epoch: 47\t[25000/88956 (28%)]\tTotal Loss: 1.0779\tAvg Loss: 0.0000\nTrain Epoch: 47\t[30000/88956 (34%)]\tTotal Loss: 1.2888\tAvg Loss: 0.0000\nTrain Epoch: 47\t[35000/88956 (39%)]\tTotal Loss: 1.4925\tAvg Loss: 0.0000\nTrain Epoch: 47\t[40000/88956 (45%)]\tTotal Loss: 1.6880\tAvg Loss: 0.0000\nTrain Epoch: 47\t[45000/88956 (51%)]\tTotal Loss: 1.8931\tAvg Loss: 0.0000\nTrain Epoch: 47\t[50000/88956 (56%)]\tTotal Loss: 2.1088\tAvg Loss: 0.0000\nTrain Epoch: 47\t[55000/88956 (62%)]\tTotal Loss: 2.3279\tAvg Loss: 0.0000\nTrain Epoch: 47\t[60000/88956 (67%)]\tTotal Loss: 2.5436\tAvg Loss: 0.0000\nTrain Epoch: 47\t[65000/88956 (73%)]\tTotal Loss: 2.7568\tAvg Loss: 0.0000\nTrain Epoch: 47\t[70000/88956 (79%)]\tTotal Loss: 2.9793\tAvg Loss: 0.0000\nTrain Epoch: 47\t[75000/88956 (84%)]\tTotal Loss: 3.2070\tAvg Loss: 0.0000\nTrain Epoch: 47\t[80000/88956 (90%)]\tTotal Loss: 3.4361\tAvg Loss: 0.0000\nTrain Epoch: 47\t[85000/88956 (96%)]\tTotal Loss: 3.6642\tAvg Loss: 0.0000\n====> Epoch: 47\tTotal Loss: 3.8295\t Avg Loss: 0.0000\tCorrect: 65103/88956\tPercentage Correct: 73.19\n====> Val Loss: 0.6225\t Avg Loss: 0.0001\tCorrect: 6513/9885\tPercentage Correct: 65.89\n====> Test Loss: 1.4947\t Avg Loss: 0.0001\tCorrect: 16344/24711\tPercentage Correct: 66.14\nTrain Epoch: 48\t[5000/88956 (6%)]\tTotal Loss: 0.2064\tAvg Loss: 0.0000\nTrain Epoch: 48\t[10000/88956 (11%)]\tTotal Loss: 0.4103\tAvg Loss: 0.0000\nTrain Epoch: 48\t[15000/88956 (17%)]\tTotal Loss: 0.6141\tAvg Loss: 0.0000\nTrain Epoch: 48\t[20000/88956 (22%)]\tTotal Loss: 0.8301\tAvg Loss: 0.0000\nTrain Epoch: 48\t[25000/88956 (28%)]\tTotal Loss: 1.0400\tAvg Loss: 0.0000\nTrain Epoch: 48\t[30000/88956 (34%)]\tTotal Loss: 1.2428\tAvg Loss: 0.0000\nTrain Epoch: 48\t[35000/88956 (39%)]\tTotal Loss: 1.4770\tAvg Loss: 0.0000\nTrain Epoch: 48\t[40000/88956 (45%)]\tTotal Loss: 1.6843\tAvg Loss: 0.0000\nTrain Epoch: 48\t[45000/88956 (51%)]\tTotal Loss: 1.8703\tAvg Loss: 0.0000\nTrain Epoch: 48\t[50000/88956 (56%)]\tTotal Loss: 2.0753\tAvg Loss: 0.0000\nTrain Epoch: 48\t[55000/88956 (62%)]\tTotal Loss: 2.2935\tAvg Loss: 0.0000\nTrain Epoch: 48\t[60000/88956 (67%)]\tTotal Loss: 2.5099\tAvg Loss: 0.0000\nTrain Epoch: 48\t[65000/88956 (73%)]\tTotal Loss: 2.7381\tAvg Loss: 0.0000\nTrain Epoch: 48\t[70000/88956 (79%)]\tTotal Loss: 2.9560\tAvg Loss: 0.0000\nTrain Epoch: 48\t[75000/88956 (84%)]\tTotal Loss: 3.1613\tAvg Loss: 0.0000\nTrain Epoch: 48\t[80000/88956 (90%)]\tTotal Loss: 3.3660\tAvg Loss: 0.0000\nTrain Epoch: 48\t[85000/88956 (96%)]\tTotal Loss: 3.6086\tAvg Loss: 0.0000\n====> Epoch: 48\tTotal Loss: 3.7756\t Avg Loss: 0.0000\tCorrect: 65512/88956\tPercentage Correct: 73.65\n====> Val Loss: 0.5973\t Avg Loss: 0.0001\tCorrect: 6550/9885\tPercentage Correct: 66.26\n====> Test Loss: 1.4591\t Avg Loss: 0.0001\tCorrect: 16445/24711\tPercentage Correct: 66.55\nTrain Epoch: 49\t[5000/88956 (6%)]\tTotal Loss: 0.2013\tAvg Loss: 0.0000\nTrain Epoch: 49\t[10000/88956 (11%)]\tTotal Loss: 0.4017\tAvg Loss: 0.0000\nTrain Epoch: 49\t[15000/88956 (17%)]\tTotal Loss: 0.6200\tAvg Loss: 0.0000\nTrain Epoch: 49\t[20000/88956 (22%)]\tTotal Loss: 0.8291\tAvg Loss: 0.0000\nTrain Epoch: 49\t[25000/88956 (28%)]\tTotal Loss: 1.0224\tAvg Loss: 0.0000\nTrain Epoch: 49\t[30000/88956 (34%)]\tTotal Loss: 1.2123\tAvg Loss: 0.0000\nTrain Epoch: 49\t[35000/88956 (39%)]\tTotal Loss: 1.4260\tAvg Loss: 0.0000\nTrain Epoch: 49\t[40000/88956 (45%)]\tTotal Loss: 1.6338\tAvg Loss: 0.0000\nTrain Epoch: 49\t[45000/88956 (51%)]\tTotal Loss: 1.8434\tAvg Loss: 0.0000\nTrain Epoch: 49\t[50000/88956 (56%)]\tTotal Loss: 2.0697\tAvg Loss: 0.0000\nTrain Epoch: 49\t[55000/88956 (62%)]\tTotal Loss: 2.2769\tAvg Loss: 0.0000\nTrain Epoch: 49\t[60000/88956 (67%)]\tTotal Loss: 2.4917\tAvg Loss: 0.0000\nTrain Epoch: 49\t[65000/88956 (73%)]\tTotal Loss: 2.7015\tAvg Loss: 0.0000\nTrain Epoch: 49\t[70000/88956 (79%)]\tTotal Loss: 2.8975\tAvg Loss: 0.0000\nTrain Epoch: 49\t[75000/88956 (84%)]\tTotal Loss: 3.0933\tAvg Loss: 0.0000\nTrain Epoch: 49\t[80000/88956 (90%)]\tTotal Loss: 3.3067\tAvg Loss: 0.0000\nTrain Epoch: 49\t[85000/88956 (96%)]\tTotal Loss: 3.5233\tAvg Loss: 0.0000\n====> Epoch: 49\tTotal Loss: 3.6998\t Avg Loss: 0.0000\tCorrect: 66066/88956\tPercentage Correct: 74.27\n====> Val Loss: 0.5792\t Avg Loss: 0.0001\tCorrect: 6669/9885\tPercentage Correct: 67.47\n====> Test Loss: 1.4253\t Avg Loss: 0.0001\tCorrect: 16716/24711\tPercentage Correct: 67.65\n\n=== saved best model ===\n\nTrain Epoch: 50\t[5000/88956 (6%)]\tTotal Loss: 0.2106\tAvg Loss: 0.0000\nTrain Epoch: 50\t[10000/88956 (11%)]\tTotal Loss: 0.4014\tAvg Loss: 0.0000\nTrain Epoch: 50\t[15000/88956 (17%)]\tTotal Loss: 0.6024\tAvg Loss: 0.0000\nTrain Epoch: 50\t[20000/88956 (22%)]\tTotal Loss: 0.7838\tAvg Loss: 0.0000\nTrain Epoch: 50\t[25000/88956 (28%)]\tTotal Loss: 0.9706\tAvg Loss: 0.0000\nTrain Epoch: 50\t[30000/88956 (34%)]\tTotal Loss: 1.1766\tAvg Loss: 0.0000\nTrain Epoch: 50\t[35000/88956 (39%)]\tTotal Loss: 1.3785\tAvg Loss: 0.0000\nTrain Epoch: 50\t[40000/88956 (45%)]\tTotal Loss: 1.5801\tAvg Loss: 0.0000\nTrain Epoch: 50\t[45000/88956 (51%)]\tTotal Loss: 1.8011\tAvg Loss: 0.0000\nTrain Epoch: 50\t[50000/88956 (56%)]\tTotal Loss: 2.0003\tAvg Loss: 0.0000\nTrain Epoch: 50\t[55000/88956 (62%)]\tTotal Loss: 2.2144\tAvg Loss: 0.0000\nTrain Epoch: 50\t[60000/88956 (67%)]\tTotal Loss: 2.4274\tAvg Loss: 0.0000\nTrain Epoch: 50\t[65000/88956 (73%)]\tTotal Loss: 2.6406\tAvg Loss: 0.0000\nTrain Epoch: 50\t[70000/88956 (79%)]\tTotal Loss: 2.8536\tAvg Loss: 0.0000\nTrain Epoch: 50\t[75000/88956 (84%)]\tTotal Loss: 3.0650\tAvg Loss: 0.0000\nTrain Epoch: 50\t[80000/88956 (90%)]\tTotal Loss: 3.2711\tAvg Loss: 0.0000\nTrain Epoch: 50\t[85000/88956 (96%)]\tTotal Loss: 3.4768\tAvg Loss: 0.0000\n====> Epoch: 50\tTotal Loss: 3.6304\t Avg Loss: 0.0000\tCorrect: 66440/88956\tPercentage Correct: 74.69\n====> Val Loss: 0.5813\t Avg Loss: 0.0001\tCorrect: 6567/9885\tPercentage Correct: 66.43\n====> Test Loss: 1.4502\t Avg Loss: 0.0001\tCorrect: 16382/24711\tPercentage Correct: 66.29\nTrain Epoch: 51\t[5000/88956 (6%)]\tTotal Loss: 0.2027\tAvg Loss: 0.0000\nTrain Epoch: 51\t[10000/88956 (11%)]\tTotal Loss: 0.3963\tAvg Loss: 0.0000\nTrain Epoch: 51\t[15000/88956 (17%)]\tTotal Loss: 0.5839\tAvg Loss: 0.0000\nTrain Epoch: 51\t[20000/88956 (22%)]\tTotal Loss: 0.7643\tAvg Loss: 0.0000\nTrain Epoch: 51\t[25000/88956 (28%)]\tTotal Loss: 0.9588\tAvg Loss: 0.0000\nTrain Epoch: 51\t[30000/88956 (34%)]\tTotal Loss: 1.1440\tAvg Loss: 0.0000\nTrain Epoch: 51\t[35000/88956 (39%)]\tTotal Loss: 1.3550\tAvg Loss: 0.0000\nTrain Epoch: 51\t[40000/88956 (45%)]\tTotal Loss: 1.5403\tAvg Loss: 0.0000\nTrain Epoch: 51\t[45000/88956 (51%)]\tTotal Loss: 1.7457\tAvg Loss: 0.0000\nTrain Epoch: 51\t[50000/88956 (56%)]\tTotal Loss: 1.9489\tAvg Loss: 0.0000\nTrain Epoch: 51\t[55000/88956 (62%)]\tTotal Loss: 2.1560\tAvg Loss: 0.0000\nTrain Epoch: 51\t[60000/88956 (67%)]\tTotal Loss: 2.3545\tAvg Loss: 0.0000\nTrain Epoch: 51\t[65000/88956 (73%)]\tTotal Loss: 2.5642\tAvg Loss: 0.0000\nTrain Epoch: 51\t[70000/88956 (79%)]\tTotal Loss: 2.7809\tAvg Loss: 0.0000\nTrain Epoch: 51\t[75000/88956 (84%)]\tTotal Loss: 2.9880\tAvg Loss: 0.0000\nTrain Epoch: 51\t[80000/88956 (90%)]\tTotal Loss: 3.2054\tAvg Loss: 0.0000\nTrain Epoch: 51\t[85000/88956 (96%)]\tTotal Loss: 3.4181\tAvg Loss: 0.0000\n====> Epoch: 51\tTotal Loss: 3.5815\t Avg Loss: 0.0000\tCorrect: 66574/88956\tPercentage Correct: 74.84\n====> Val Loss: 0.6213\t Avg Loss: 0.0001\tCorrect: 6553/9885\tPercentage Correct: 66.29\n====> Test Loss: 1.4904\t Avg Loss: 0.0001\tCorrect: 16478/24711\tPercentage Correct: 66.68\nTrain Epoch: 52\t[5000/88956 (6%)]\tTotal Loss: 0.1987\tAvg Loss: 0.0000\nTrain Epoch: 52\t[10000/88956 (11%)]\tTotal Loss: 0.3982\tAvg Loss: 0.0000\nTrain Epoch: 52\t[15000/88956 (17%)]\tTotal Loss: 0.5808\tAvg Loss: 0.0000\nTrain Epoch: 52\t[20000/88956 (22%)]\tTotal Loss: 0.7875\tAvg Loss: 0.0000\nTrain Epoch: 52\t[25000/88956 (28%)]\tTotal Loss: 0.9820\tAvg Loss: 0.0000\nTrain Epoch: 52\t[30000/88956 (34%)]\tTotal Loss: 1.1651\tAvg Loss: 0.0000\nTrain Epoch: 52\t[35000/88956 (39%)]\tTotal Loss: 1.3550\tAvg Loss: 0.0000\nTrain Epoch: 52\t[40000/88956 (45%)]\tTotal Loss: 1.5526\tAvg Loss: 0.0000\nTrain Epoch: 52\t[45000/88956 (51%)]\tTotal Loss: 1.7469\tAvg Loss: 0.0000\nTrain Epoch: 52\t[50000/88956 (56%)]\tTotal Loss: 1.9491\tAvg Loss: 0.0000\nTrain Epoch: 52\t[55000/88956 (62%)]\tTotal Loss: 2.1412\tAvg Loss: 0.0000\nTrain Epoch: 52\t[60000/88956 (67%)]\tTotal Loss: 2.3432\tAvg Loss: 0.0000\nTrain Epoch: 52\t[65000/88956 (73%)]\tTotal Loss: 2.5388\tAvg Loss: 0.0000\nTrain Epoch: 52\t[70000/88956 (79%)]\tTotal Loss: 2.7443\tAvg Loss: 0.0000\nTrain Epoch: 52\t[75000/88956 (84%)]\tTotal Loss: 2.9598\tAvg Loss: 0.0000\nTrain Epoch: 52\t[80000/88956 (90%)]\tTotal Loss: 3.1692\tAvg Loss: 0.0000\nTrain Epoch: 52\t[85000/88956 (96%)]\tTotal Loss: 3.3717\tAvg Loss: 0.0000\n====> Epoch: 52\tTotal Loss: 3.5253\t Avg Loss: 0.0000\tCorrect: 66885/88956\tPercentage Correct: 75.19\n====> Val Loss: 0.5978\t Avg Loss: 0.0001\tCorrect: 6557/9885\tPercentage Correct: 66.33\n====> Test Loss: 1.5083\t Avg Loss: 0.0001\tCorrect: 16417/24711\tPercentage Correct: 66.44\nTrain Epoch: 53\t[5000/88956 (6%)]\tTotal Loss: 0.1977\tAvg Loss: 0.0000\nTrain Epoch: 53\t[10000/88956 (11%)]\tTotal Loss: 0.3857\tAvg Loss: 0.0000\nTrain Epoch: 53\t[15000/88956 (17%)]\tTotal Loss: 0.5643\tAvg Loss: 0.0000\nTrain Epoch: 53\t[20000/88956 (22%)]\tTotal Loss: 0.7639\tAvg Loss: 0.0000\nTrain Epoch: 53\t[25000/88956 (28%)]\tTotal Loss: 0.9586\tAvg Loss: 0.0000\nTrain Epoch: 53\t[30000/88956 (34%)]\tTotal Loss: 1.1615\tAvg Loss: 0.0000\nTrain Epoch: 53\t[35000/88956 (39%)]\tTotal Loss: 1.3498\tAvg Loss: 0.0000\nTrain Epoch: 53\t[40000/88956 (45%)]\tTotal Loss: 1.5387\tAvg Loss: 0.0000\nTrain Epoch: 53\t[45000/88956 (51%)]\tTotal Loss: 1.7327\tAvg Loss: 0.0000\nTrain Epoch: 53\t[50000/88956 (56%)]\tTotal Loss: 1.9324\tAvg Loss: 0.0000\nTrain Epoch: 53\t[55000/88956 (62%)]\tTotal Loss: 2.1208\tAvg Loss: 0.0000\nTrain Epoch: 53\t[60000/88956 (67%)]\tTotal Loss: 2.3229\tAvg Loss: 0.0000\nTrain Epoch: 53\t[65000/88956 (73%)]\tTotal Loss: 2.5301\tAvg Loss: 0.0000\nTrain Epoch: 53\t[70000/88956 (79%)]\tTotal Loss: 2.7187\tAvg Loss: 0.0000\nTrain Epoch: 53\t[75000/88956 (84%)]\tTotal Loss: 2.9318\tAvg Loss: 0.0000\nTrain Epoch: 53\t[80000/88956 (90%)]\tTotal Loss: 3.1214\tAvg Loss: 0.0000\nTrain Epoch: 53\t[85000/88956 (96%)]\tTotal Loss: 3.3221\tAvg Loss: 0.0000\n====> Epoch: 53\tTotal Loss: 3.4734\t Avg Loss: 0.0000\tCorrect: 66952/88956\tPercentage Correct: 75.26\n====> Val Loss: 0.5899\t Avg Loss: 0.0001\tCorrect: 6655/9885\tPercentage Correct: 67.32\n====> Test Loss: 1.4157\t Avg Loss: 0.0001\tCorrect: 16800/24711\tPercentage Correct: 67.99\nTrain Epoch: 54\t[5000/88956 (6%)]\tTotal Loss: 0.1779\tAvg Loss: 0.0000\nTrain Epoch: 54\t[10000/88956 (11%)]\tTotal Loss: 0.3384\tAvg Loss: 0.0000\nTrain Epoch: 54\t[15000/88956 (17%)]\tTotal Loss: 0.5198\tAvg Loss: 0.0000\nTrain Epoch: 54\t[20000/88956 (22%)]\tTotal Loss: 0.7082\tAvg Loss: 0.0000\nTrain Epoch: 54\t[25000/88956 (28%)]\tTotal Loss: 0.8910\tAvg Loss: 0.0000\nTrain Epoch: 54\t[30000/88956 (34%)]\tTotal Loss: 1.0820\tAvg Loss: 0.0000\nTrain Epoch: 54\t[35000/88956 (39%)]\tTotal Loss: 1.2802\tAvg Loss: 0.0000\nTrain Epoch: 54\t[40000/88956 (45%)]\tTotal Loss: 1.4900\tAvg Loss: 0.0000\nTrain Epoch: 54\t[45000/88956 (51%)]\tTotal Loss: 1.6914\tAvg Loss: 0.0000\nTrain Epoch: 54\t[50000/88956 (56%)]\tTotal Loss: 1.8842\tAvg Loss: 0.0000\nTrain Epoch: 54\t[55000/88956 (62%)]\tTotal Loss: 2.0646\tAvg Loss: 0.0000\nTrain Epoch: 54\t[60000/88956 (67%)]\tTotal Loss: 2.2644\tAvg Loss: 0.0000\nTrain Epoch: 54\t[65000/88956 (73%)]\tTotal Loss: 2.4769\tAvg Loss: 0.0000\nTrain Epoch: 54\t[70000/88956 (79%)]\tTotal Loss: 2.6777\tAvg Loss: 0.0000\nTrain Epoch: 54\t[75000/88956 (84%)]\tTotal Loss: 2.8752\tAvg Loss: 0.0000\nTrain Epoch: 54\t[80000/88956 (90%)]\tTotal Loss: 3.0547\tAvg Loss: 0.0000\nTrain Epoch: 54\t[85000/88956 (96%)]\tTotal Loss: 3.2568\tAvg Loss: 0.0000\n====> Epoch: 54\tTotal Loss: 3.4059\t Avg Loss: 0.0000\tCorrect: 67510/88956\tPercentage Correct: 75.89\n====> Val Loss: 0.5884\t Avg Loss: 0.0001\tCorrect: 6601/9885\tPercentage Correct: 66.78\n====> Test Loss: 1.4337\t Avg Loss: 0.0001\tCorrect: 16671/24711\tPercentage Correct: 67.46\nTrain Epoch: 55\t[5000/88956 (6%)]\tTotal Loss: 0.1808\tAvg Loss: 0.0000\nTrain Epoch: 55\t[10000/88956 (11%)]\tTotal Loss: 0.3480\tAvg Loss: 0.0000\nTrain Epoch: 55\t[15000/88956 (17%)]\tTotal Loss: 0.5334\tAvg Loss: 0.0000\nTrain Epoch: 55\t[20000/88956 (22%)]\tTotal Loss: 0.7129\tAvg Loss: 0.0000\nTrain Epoch: 55\t[25000/88956 (28%)]\tTotal Loss: 0.8994\tAvg Loss: 0.0000\nTrain Epoch: 55\t[30000/88956 (34%)]\tTotal Loss: 1.0759\tAvg Loss: 0.0000\nTrain Epoch: 55\t[35000/88956 (39%)]\tTotal Loss: 1.2787\tAvg Loss: 0.0000\nTrain Epoch: 55\t[40000/88956 (45%)]\tTotal Loss: 1.4658\tAvg Loss: 0.0000\nTrain Epoch: 55\t[45000/88956 (51%)]\tTotal Loss: 1.6503\tAvg Loss: 0.0000\nTrain Epoch: 55\t[50000/88956 (56%)]\tTotal Loss: 1.8430\tAvg Loss: 0.0000\nTrain Epoch: 55\t[55000/88956 (62%)]\tTotal Loss: 2.0538\tAvg Loss: 0.0000\nTrain Epoch: 55\t[60000/88956 (67%)]\tTotal Loss: 2.2511\tAvg Loss: 0.0000\nTrain Epoch: 55\t[65000/88956 (73%)]\tTotal Loss: 2.4378\tAvg Loss: 0.0000\nTrain Epoch: 55\t[70000/88956 (79%)]\tTotal Loss: 2.6212\tAvg Loss: 0.0000\nTrain Epoch: 55\t[75000/88956 (84%)]\tTotal Loss: 2.8119\tAvg Loss: 0.0000\nTrain Epoch: 55\t[80000/88956 (90%)]\tTotal Loss: 3.0103\tAvg Loss: 0.0000\nTrain Epoch: 55\t[85000/88956 (96%)]\tTotal Loss: 3.2069\tAvg Loss: 0.0000\n====> Epoch: 55\tTotal Loss: 3.3485\t Avg Loss: 0.0000\tCorrect: 67822/88956\tPercentage Correct: 76.24\n====> Val Loss: 0.5585\t Avg Loss: 0.0001\tCorrect: 6736/9885\tPercentage Correct: 68.14\n====> Test Loss: 1.3850\t Avg Loss: 0.0001\tCorrect: 16770/24711\tPercentage Correct: 67.86\n\n=== saved best model ===\n\nTrain Epoch: 56\t[5000/88956 (6%)]\tTotal Loss: 0.1687\tAvg Loss: 0.0000\nTrain Epoch: 56\t[10000/88956 (11%)]\tTotal Loss: 0.3473\tAvg Loss: 0.0000\nTrain Epoch: 56\t[15000/88956 (17%)]\tTotal Loss: 0.5250\tAvg Loss: 0.0000\nTrain Epoch: 56\t[20000/88956 (22%)]\tTotal Loss: 0.7040\tAvg Loss: 0.0000\nTrain Epoch: 56\t[25000/88956 (28%)]\tTotal Loss: 0.8850\tAvg Loss: 0.0000\nTrain Epoch: 56\t[30000/88956 (34%)]\tTotal Loss: 1.0673\tAvg Loss: 0.0000\nTrain Epoch: 56\t[35000/88956 (39%)]\tTotal Loss: 1.2582\tAvg Loss: 0.0000\nTrain Epoch: 56\t[40000/88956 (45%)]\tTotal Loss: 1.4585\tAvg Loss: 0.0000\nTrain Epoch: 56\t[45000/88956 (51%)]\tTotal Loss: 1.6527\tAvg Loss: 0.0000\nTrain Epoch: 56\t[50000/88956 (56%)]\tTotal Loss: 1.8575\tAvg Loss: 0.0000\nTrain Epoch: 56\t[55000/88956 (62%)]\tTotal Loss: 2.0381\tAvg Loss: 0.0000\nTrain Epoch: 56\t[60000/88956 (67%)]\tTotal Loss: 2.2223\tAvg Loss: 0.0000\nTrain Epoch: 56\t[65000/88956 (73%)]\tTotal Loss: 2.4196\tAvg Loss: 0.0000\nTrain Epoch: 56\t[70000/88956 (79%)]\tTotal Loss: 2.6135\tAvg Loss: 0.0000\nTrain Epoch: 56\t[75000/88956 (84%)]\tTotal Loss: 2.8033\tAvg Loss: 0.0000\nTrain Epoch: 56\t[80000/88956 (90%)]\tTotal Loss: 2.9941\tAvg Loss: 0.0000\nTrain Epoch: 56\t[85000/88956 (96%)]\tTotal Loss: 3.1751\tAvg Loss: 0.0000\n====> Epoch: 56\tTotal Loss: 3.3204\t Avg Loss: 0.0000\tCorrect: 67717/88956\tPercentage Correct: 76.12\n====> Val Loss: 0.5702\t Avg Loss: 0.0001\tCorrect: 6667/9885\tPercentage Correct: 67.45\n====> Test Loss: 1.3819\t Avg Loss: 0.0001\tCorrect: 16784/24711\tPercentage Correct: 67.92\nTrain Epoch: 57\t[5000/88956 (6%)]\tTotal Loss: 0.2900\tAvg Loss: 0.0001\nTrain Epoch: 57\t[10000/88956 (11%)]\tTotal Loss: 0.5517\tAvg Loss: 0.0001\nTrain Epoch: 57\t[15000/88956 (17%)]\tTotal Loss: 0.7557\tAvg Loss: 0.0001\nTrain Epoch: 57\t[20000/88956 (22%)]\tTotal Loss: 0.9567\tAvg Loss: 0.0000\nTrain Epoch: 57\t[25000/88956 (28%)]\tTotal Loss: 1.1494\tAvg Loss: 0.0000\nTrain Epoch: 57\t[30000/88956 (34%)]\tTotal Loss: 1.3319\tAvg Loss: 0.0000\nTrain Epoch: 57\t[35000/88956 (39%)]\tTotal Loss: 1.5173\tAvg Loss: 0.0000\nTrain Epoch: 57\t[40000/88956 (45%)]\tTotal Loss: 1.6972\tAvg Loss: 0.0000\nTrain Epoch: 57\t[45000/88956 (51%)]\tTotal Loss: 1.8737\tAvg Loss: 0.0000\nTrain Epoch: 57\t[50000/88956 (56%)]\tTotal Loss: 2.0562\tAvg Loss: 0.0000\nTrain Epoch: 57\t[55000/88956 (62%)]\tTotal Loss: 2.2299\tAvg Loss: 0.0000\nTrain Epoch: 57\t[60000/88956 (67%)]\tTotal Loss: 2.4137\tAvg Loss: 0.0000\nTrain Epoch: 57\t[65000/88956 (73%)]\tTotal Loss: 2.6073\tAvg Loss: 0.0000\nTrain Epoch: 57\t[70000/88956 (79%)]\tTotal Loss: 2.8050\tAvg Loss: 0.0000\nTrain Epoch: 57\t[75000/88956 (84%)]\tTotal Loss: 2.9954\tAvg Loss: 0.0000\nTrain Epoch: 57\t[80000/88956 (90%)]\tTotal Loss: 3.1814\tAvg Loss: 0.0000\nTrain Epoch: 57\t[85000/88956 (96%)]\tTotal Loss: 3.3723\tAvg Loss: 0.0000\n====> Epoch: 57\tTotal Loss: 3.5196\t Avg Loss: 0.0000\tCorrect: 66622/88956\tPercentage Correct: 74.89\n====> Val Loss: 0.5659\t Avg Loss: 0.0001\tCorrect: 6774/9885\tPercentage Correct: 68.53\n====> Test Loss: 1.4158\t Avg Loss: 0.0001\tCorrect: 16955/24711\tPercentage Correct: 68.61\n\n=== saved best model ===\n\nTrain Epoch: 58\t[5000/88956 (6%)]\tTotal Loss: 0.1694\tAvg Loss: 0.0000\nTrain Epoch: 58\t[10000/88956 (11%)]\tTotal Loss: 0.3414\tAvg Loss: 0.0000\nTrain Epoch: 58\t[15000/88956 (17%)]\tTotal Loss: 0.5076\tAvg Loss: 0.0000\nTrain Epoch: 58\t[20000/88956 (22%)]\tTotal Loss: 0.6751\tAvg Loss: 0.0000\nTrain Epoch: 58\t[25000/88956 (28%)]\tTotal Loss: 0.8473\tAvg Loss: 0.0000\nTrain Epoch: 58\t[30000/88956 (34%)]\tTotal Loss: 1.0221\tAvg Loss: 0.0000\nTrain Epoch: 58\t[35000/88956 (39%)]\tTotal Loss: 1.2104\tAvg Loss: 0.0000\nTrain Epoch: 58\t[40000/88956 (45%)]\tTotal Loss: 1.3863\tAvg Loss: 0.0000\nTrain Epoch: 58\t[45000/88956 (51%)]\tTotal Loss: 1.5408\tAvg Loss: 0.0000\nTrain Epoch: 58\t[50000/88956 (56%)]\tTotal Loss: 1.7171\tAvg Loss: 0.0000\nTrain Epoch: 58\t[55000/88956 (62%)]\tTotal Loss: 1.8822\tAvg Loss: 0.0000\nTrain Epoch: 58\t[60000/88956 (67%)]\tTotal Loss: 2.0664\tAvg Loss: 0.0000\nTrain Epoch: 58\t[65000/88956 (73%)]\tTotal Loss: 2.2387\tAvg Loss: 0.0000\nTrain Epoch: 58\t[70000/88956 (79%)]\tTotal Loss: 2.4157\tAvg Loss: 0.0000\nTrain Epoch: 58\t[75000/88956 (84%)]\tTotal Loss: 2.6065\tAvg Loss: 0.0000\nTrain Epoch: 58\t[80000/88956 (90%)]\tTotal Loss: 2.7982\tAvg Loss: 0.0000\nTrain Epoch: 58\t[85000/88956 (96%)]\tTotal Loss: 2.9986\tAvg Loss: 0.0000\n====> Epoch: 58\tTotal Loss: 3.1505\t Avg Loss: 0.0000\tCorrect: 68877/88956\tPercentage Correct: 77.43\n====> Val Loss: 0.5391\t Avg Loss: 0.0001\tCorrect: 6749/9885\tPercentage Correct: 68.28\n====> Test Loss: 1.3066\t Avg Loss: 0.0001\tCorrect: 16982/24711\tPercentage Correct: 68.72\nTrain Epoch: 59\t[5000/88956 (6%)]\tTotal Loss: 0.1674\tAvg Loss: 0.0000\nTrain Epoch: 59\t[10000/88956 (11%)]\tTotal Loss: 0.3265\tAvg Loss: 0.0000\nTrain Epoch: 59\t[15000/88956 (17%)]\tTotal Loss: 0.5070\tAvg Loss: 0.0000\nTrain Epoch: 59\t[20000/88956 (22%)]\tTotal Loss: 0.6870\tAvg Loss: 0.0000\nTrain Epoch: 59\t[25000/88956 (28%)]\tTotal Loss: 0.8631\tAvg Loss: 0.0000\nTrain Epoch: 59\t[30000/88956 (34%)]\tTotal Loss: 1.0436\tAvg Loss: 0.0000\nTrain Epoch: 59\t[35000/88956 (39%)]\tTotal Loss: 1.2112\tAvg Loss: 0.0000\nTrain Epoch: 59\t[40000/88956 (45%)]\tTotal Loss: 1.3726\tAvg Loss: 0.0000\nTrain Epoch: 59\t[45000/88956 (51%)]\tTotal Loss: 1.5444\tAvg Loss: 0.0000\nTrain Epoch: 59\t[50000/88956 (56%)]\tTotal Loss: 1.7263\tAvg Loss: 0.0000\nTrain Epoch: 59\t[55000/88956 (62%)]\tTotal Loss: 1.9116\tAvg Loss: 0.0000\nTrain Epoch: 59\t[60000/88956 (67%)]\tTotal Loss: 2.0937\tAvg Loss: 0.0000\nTrain Epoch: 59\t[65000/88956 (73%)]\tTotal Loss: 2.2852\tAvg Loss: 0.0000\nTrain Epoch: 59\t[70000/88956 (79%)]\tTotal Loss: 2.4733\tAvg Loss: 0.0000\nTrain Epoch: 59\t[75000/88956 (84%)]\tTotal Loss: 2.6523\tAvg Loss: 0.0000\nTrain Epoch: 59\t[80000/88956 (90%)]\tTotal Loss: 2.8331\tAvg Loss: 0.0000\nTrain Epoch: 59\t[85000/88956 (96%)]\tTotal Loss: 3.0173\tAvg Loss: 0.0000\n====> Epoch: 59\tTotal Loss: 3.1645\t Avg Loss: 0.0000\tCorrect: 68478/88956\tPercentage Correct: 76.98\n====> Val Loss: 0.5300\t Avg Loss: 0.0001\tCorrect: 6683/9885\tPercentage Correct: 67.61\n====> Test Loss: 1.2828\t Avg Loss: 0.0001\tCorrect: 16992/24711\tPercentage Correct: 68.76\nTrain Epoch: 60\t[5000/88956 (6%)]\tTotal Loss: 0.1655\tAvg Loss: 0.0000\nTrain Epoch: 60\t[10000/88956 (11%)]\tTotal Loss: 0.3302\tAvg Loss: 0.0000\nTrain Epoch: 60\t[15000/88956 (17%)]\tTotal Loss: 0.4880\tAvg Loss: 0.0000\nTrain Epoch: 60\t[20000/88956 (22%)]\tTotal Loss: 0.6556\tAvg Loss: 0.0000\nTrain Epoch: 60\t[25000/88956 (28%)]\tTotal Loss: 0.8460\tAvg Loss: 0.0000\nTrain Epoch: 60\t[30000/88956 (34%)]\tTotal Loss: 1.0238\tAvg Loss: 0.0000\nTrain Epoch: 60\t[35000/88956 (39%)]\tTotal Loss: 1.1883\tAvg Loss: 0.0000\nTrain Epoch: 60\t[40000/88956 (45%)]\tTotal Loss: 1.3506\tAvg Loss: 0.0000\nTrain Epoch: 60\t[45000/88956 (51%)]\tTotal Loss: 1.5408\tAvg Loss: 0.0000\nTrain Epoch: 60\t[50000/88956 (56%)]\tTotal Loss: 1.7240\tAvg Loss: 0.0000\nTrain Epoch: 60\t[55000/88956 (62%)]\tTotal Loss: 1.9030\tAvg Loss: 0.0000\nTrain Epoch: 60\t[60000/88956 (67%)]\tTotal Loss: 2.0765\tAvg Loss: 0.0000\nTrain Epoch: 60\t[65000/88956 (73%)]\tTotal Loss: 2.2510\tAvg Loss: 0.0000\nTrain Epoch: 60\t[70000/88956 (79%)]\tTotal Loss: 2.4651\tAvg Loss: 0.0000\nTrain Epoch: 60\t[75000/88956 (84%)]\tTotal Loss: 2.6453\tAvg Loss: 0.0000\nTrain Epoch: 60\t[80000/88956 (90%)]\tTotal Loss: 2.8148\tAvg Loss: 0.0000\nTrain Epoch: 60\t[85000/88956 (96%)]\tTotal Loss: 2.9865\tAvg Loss: 0.0000\n====> Epoch: 60\tTotal Loss: 3.1230\t Avg Loss: 0.0000\tCorrect: 68959/88956\tPercentage Correct: 77.52\n====> Val Loss: 0.5127\t Avg Loss: 0.0001\tCorrect: 6787/9885\tPercentage Correct: 68.66\n====> Test Loss: 1.2490\t Avg Loss: 0.0001\tCorrect: 17097/24711\tPercentage Correct: 69.19\n\n=== saved best model ===\n\nTrain Epoch: 61\t[5000/88956 (6%)]\tTotal Loss: 0.1565\tAvg Loss: 0.0000\nTrain Epoch: 61\t[10000/88956 (11%)]\tTotal Loss: 0.3220\tAvg Loss: 0.0000\nTrain Epoch: 61\t[15000/88956 (17%)]\tTotal Loss: 0.4960\tAvg Loss: 0.0000\nTrain Epoch: 61\t[20000/88956 (22%)]\tTotal Loss: 0.6820\tAvg Loss: 0.0000\nTrain Epoch: 61\t[25000/88956 (28%)]\tTotal Loss: 0.8583\tAvg Loss: 0.0000\nTrain Epoch: 61\t[30000/88956 (34%)]\tTotal Loss: 1.0310\tAvg Loss: 0.0000\nTrain Epoch: 61\t[35000/88956 (39%)]\tTotal Loss: 1.2117\tAvg Loss: 0.0000\nTrain Epoch: 61\t[40000/88956 (45%)]\tTotal Loss: 1.3826\tAvg Loss: 0.0000\nTrain Epoch: 61\t[45000/88956 (51%)]\tTotal Loss: 1.5633\tAvg Loss: 0.0000\nTrain Epoch: 61\t[50000/88956 (56%)]\tTotal Loss: 1.7380\tAvg Loss: 0.0000\nTrain Epoch: 61\t[55000/88956 (62%)]\tTotal Loss: 1.9104\tAvg Loss: 0.0000\nTrain Epoch: 61\t[60000/88956 (67%)]\tTotal Loss: 2.0906\tAvg Loss: 0.0000\nTrain Epoch: 61\t[65000/88956 (73%)]\tTotal Loss: 2.2767\tAvg Loss: 0.0000\nTrain Epoch: 61\t[70000/88956 (79%)]\tTotal Loss: 2.4674\tAvg Loss: 0.0000\nTrain Epoch: 61\t[75000/88956 (84%)]\tTotal Loss: 2.6301\tAvg Loss: 0.0000\nTrain Epoch: 61\t[80000/88956 (90%)]\tTotal Loss: 2.8051\tAvg Loss: 0.0000\nTrain Epoch: 61\t[85000/88956 (96%)]\tTotal Loss: 2.9909\tAvg Loss: 0.0000\n====> Epoch: 61\tTotal Loss: 3.1356\t Avg Loss: 0.0000\tCorrect: 68819/88956\tPercentage Correct: 77.36\n====> Val Loss: 0.5462\t Avg Loss: 0.0001\tCorrect: 6776/9885\tPercentage Correct: 68.55\n====> Test Loss: 1.3123\t Avg Loss: 0.0001\tCorrect: 17040/24711\tPercentage Correct: 68.96\nTrain Epoch: 62\t[5000/88956 (6%)]\tTotal Loss: 0.1738\tAvg Loss: 0.0000\nTrain Epoch: 62\t[10000/88956 (11%)]\tTotal Loss: 0.3447\tAvg Loss: 0.0000\nTrain Epoch: 62\t[15000/88956 (17%)]\tTotal Loss: 0.5115\tAvg Loss: 0.0000\nTrain Epoch: 62\t[20000/88956 (22%)]\tTotal Loss: 0.6727\tAvg Loss: 0.0000\nTrain Epoch: 62\t[25000/88956 (28%)]\tTotal Loss: 0.8508\tAvg Loss: 0.0000\nTrain Epoch: 62\t[30000/88956 (34%)]\tTotal Loss: 1.0132\tAvg Loss: 0.0000\nTrain Epoch: 62\t[35000/88956 (39%)]\tTotal Loss: 1.1725\tAvg Loss: 0.0000\nTrain Epoch: 62\t[40000/88956 (45%)]\tTotal Loss: 1.3479\tAvg Loss: 0.0000\nTrain Epoch: 62\t[45000/88956 (51%)]\tTotal Loss: 1.5228\tAvg Loss: 0.0000\nTrain Epoch: 62\t[50000/88956 (56%)]\tTotal Loss: 1.6956\tAvg Loss: 0.0000\nTrain Epoch: 62\t[55000/88956 (62%)]\tTotal Loss: 1.8784\tAvg Loss: 0.0000\nTrain Epoch: 62\t[60000/88956 (67%)]\tTotal Loss: 2.0496\tAvg Loss: 0.0000\nTrain Epoch: 62\t[65000/88956 (73%)]\tTotal Loss: 2.2256\tAvg Loss: 0.0000\nTrain Epoch: 62\t[70000/88956 (79%)]\tTotal Loss: 2.3938\tAvg Loss: 0.0000\nTrain Epoch: 62\t[75000/88956 (84%)]\tTotal Loss: 2.5995\tAvg Loss: 0.0000\nTrain Epoch: 62\t[80000/88956 (90%)]\tTotal Loss: 2.7920\tAvg Loss: 0.0000\nTrain Epoch: 62\t[85000/88956 (96%)]\tTotal Loss: 2.9569\tAvg Loss: 0.0000\n====> Epoch: 62\tTotal Loss: 3.0819\t Avg Loss: 0.0000\tCorrect: 68959/88956\tPercentage Correct: 77.52\n====> Val Loss: 0.5074\t Avg Loss: 0.0001\tCorrect: 6855/9885\tPercentage Correct: 69.35\n====> Test Loss: 1.2607\t Avg Loss: 0.0001\tCorrect: 17168/24711\tPercentage Correct: 69.48\n\n=== saved best model ===\n\nTrain Epoch: 63\t[5000/88956 (6%)]\tTotal Loss: 0.1542\tAvg Loss: 0.0000\nTrain Epoch: 63\t[10000/88956 (11%)]\tTotal Loss: 0.3174\tAvg Loss: 0.0000\nTrain Epoch: 63\t[15000/88956 (17%)]\tTotal Loss: 0.4923\tAvg Loss: 0.0000\nTrain Epoch: 63\t[20000/88956 (22%)]\tTotal Loss: 0.6703\tAvg Loss: 0.0000\nTrain Epoch: 63\t[25000/88956 (28%)]\tTotal Loss: 0.8290\tAvg Loss: 0.0000\nTrain Epoch: 63\t[30000/88956 (34%)]\tTotal Loss: 1.0046\tAvg Loss: 0.0000\nTrain Epoch: 63\t[35000/88956 (39%)]\tTotal Loss: 1.1819\tAvg Loss: 0.0000\nTrain Epoch: 63\t[40000/88956 (45%)]\tTotal Loss: 1.3634\tAvg Loss: 0.0000\nTrain Epoch: 63\t[45000/88956 (51%)]\tTotal Loss: 1.5451\tAvg Loss: 0.0000\nTrain Epoch: 63\t[50000/88956 (56%)]\tTotal Loss: 1.7077\tAvg Loss: 0.0000\nTrain Epoch: 63\t[55000/88956 (62%)]\tTotal Loss: 1.8793\tAvg Loss: 0.0000\nTrain Epoch: 63\t[60000/88956 (67%)]\tTotal Loss: 2.0524\tAvg Loss: 0.0000\nTrain Epoch: 63\t[65000/88956 (73%)]\tTotal Loss: 2.2233\tAvg Loss: 0.0000\nTrain Epoch: 63\t[70000/88956 (79%)]\tTotal Loss: 2.4013\tAvg Loss: 0.0000\nTrain Epoch: 63\t[75000/88956 (84%)]\tTotal Loss: 2.5846\tAvg Loss: 0.0000\nTrain Epoch: 63\t[80000/88956 (90%)]\tTotal Loss: 2.7610\tAvg Loss: 0.0000\nTrain Epoch: 63\t[85000/88956 (96%)]\tTotal Loss: 2.9332\tAvg Loss: 0.0000\n====> Epoch: 63\tTotal Loss: 3.0685\t Avg Loss: 0.0000\tCorrect: 69114/88956\tPercentage Correct: 77.69\n====> Val Loss: 0.5051\t Avg Loss: 0.0001\tCorrect: 6874/9885\tPercentage Correct: 69.54\n====> Test Loss: 1.2483\t Avg Loss: 0.0001\tCorrect: 17415/24711\tPercentage Correct: 70.47\n\n=== saved best model ===\n\nTrain Epoch: 64\t[5000/88956 (6%)]\tTotal Loss: 0.1581\tAvg Loss: 0.0000\nTrain Epoch: 64\t[10000/88956 (11%)]\tTotal Loss: 0.3324\tAvg Loss: 0.0000\nTrain Epoch: 64\t[15000/88956 (17%)]\tTotal Loss: 0.5014\tAvg Loss: 0.0000\nTrain Epoch: 64\t[20000/88956 (22%)]\tTotal Loss: 0.6819\tAvg Loss: 0.0000\nTrain Epoch: 64\t[25000/88956 (28%)]\tTotal Loss: 0.8432\tAvg Loss: 0.0000\nTrain Epoch: 64\t[30000/88956 (34%)]\tTotal Loss: 1.0005\tAvg Loss: 0.0000\nTrain Epoch: 64\t[35000/88956 (39%)]\tTotal Loss: 1.1768\tAvg Loss: 0.0000\nTrain Epoch: 64\t[40000/88956 (45%)]\tTotal Loss: 1.3461\tAvg Loss: 0.0000\nTrain Epoch: 64\t[45000/88956 (51%)]\tTotal Loss: 1.5073\tAvg Loss: 0.0000\nTrain Epoch: 64\t[50000/88956 (56%)]\tTotal Loss: 1.7214\tAvg Loss: 0.0000\nTrain Epoch: 64\t[55000/88956 (62%)]\tTotal Loss: 1.9545\tAvg Loss: 0.0000\nTrain Epoch: 64\t[60000/88956 (67%)]\tTotal Loss: 2.1445\tAvg Loss: 0.0000\nTrain Epoch: 64\t[65000/88956 (73%)]\tTotal Loss: 2.3254\tAvg Loss: 0.0000\nTrain Epoch: 64\t[70000/88956 (79%)]\tTotal Loss: 2.4978\tAvg Loss: 0.0000\nTrain Epoch: 64\t[75000/88956 (84%)]\tTotal Loss: 2.6675\tAvg Loss: 0.0000\nTrain Epoch: 64\t[80000/88956 (90%)]\tTotal Loss: 2.8280\tAvg Loss: 0.0000\nTrain Epoch: 64\t[85000/88956 (96%)]\tTotal Loss: 3.0079\tAvg Loss: 0.0000\n====> Epoch: 64\tTotal Loss: 3.1785\t Avg Loss: 0.0000\tCorrect: 68625/88956\tPercentage Correct: 77.14\n====> Val Loss: 0.5679\t Avg Loss: 0.0001\tCorrect: 6608/9885\tPercentage Correct: 66.85\n====> Test Loss: 1.4302\t Avg Loss: 0.0001\tCorrect: 16524/24711\tPercentage Correct: 66.87\nTrain Epoch: 65\t[5000/88956 (6%)]\tTotal Loss: 0.1896\tAvg Loss: 0.0000\nTrain Epoch: 65\t[10000/88956 (11%)]\tTotal Loss: 0.3628\tAvg Loss: 0.0000\nTrain Epoch: 65\t[15000/88956 (17%)]\tTotal Loss: 0.5334\tAvg Loss: 0.0000\nTrain Epoch: 65\t[20000/88956 (22%)]\tTotal Loss: 0.6903\tAvg Loss: 0.0000\nTrain Epoch: 65\t[25000/88956 (28%)]\tTotal Loss: 0.8407\tAvg Loss: 0.0000\nTrain Epoch: 65\t[30000/88956 (34%)]\tTotal Loss: 0.9989\tAvg Loss: 0.0000\nTrain Epoch: 65\t[35000/88956 (39%)]\tTotal Loss: 1.1546\tAvg Loss: 0.0000\nTrain Epoch: 65\t[40000/88956 (45%)]\tTotal Loss: 1.3349\tAvg Loss: 0.0000\nTrain Epoch: 65\t[45000/88956 (51%)]\tTotal Loss: 1.5081\tAvg Loss: 0.0000\nTrain Epoch: 65\t[50000/88956 (56%)]\tTotal Loss: 1.6685\tAvg Loss: 0.0000\nTrain Epoch: 65\t[55000/88956 (62%)]\tTotal Loss: 1.8240\tAvg Loss: 0.0000\nTrain Epoch: 65\t[60000/88956 (67%)]\tTotal Loss: 1.9809\tAvg Loss: 0.0000\nTrain Epoch: 65\t[65000/88956 (73%)]\tTotal Loss: 2.1393\tAvg Loss: 0.0000\nTrain Epoch: 65\t[70000/88956 (79%)]\tTotal Loss: 2.3137\tAvg Loss: 0.0000\nTrain Epoch: 65\t[75000/88956 (84%)]\tTotal Loss: 2.4668\tAvg Loss: 0.0000\nTrain Epoch: 65\t[80000/88956 (90%)]\tTotal Loss: 2.6337\tAvg Loss: 0.0000\nTrain Epoch: 65\t[85000/88956 (96%)]\tTotal Loss: 2.7962\tAvg Loss: 0.0000\n====> Epoch: 65\tTotal Loss: 2.9261\t Avg Loss: 0.0000\tCorrect: 69660/88956\tPercentage Correct: 78.31\n====> Val Loss: 0.5135\t Avg Loss: 0.0001\tCorrect: 7012/9885\tPercentage Correct: 70.94\n====> Test Loss: 1.2487\t Avg Loss: 0.0001\tCorrect: 17451/24711\tPercentage Correct: 70.62\n\n=== saved best model ===\n\nTrain Epoch: 66\t[5000/88956 (6%)]\tTotal Loss: 0.1494\tAvg Loss: 0.0000\nTrain Epoch: 66\t[10000/88956 (11%)]\tTotal Loss: 0.2985\tAvg Loss: 0.0000\nTrain Epoch: 66\t[15000/88956 (17%)]\tTotal Loss: 0.4603\tAvg Loss: 0.0000\nTrain Epoch: 66\t[20000/88956 (22%)]\tTotal Loss: 0.6202\tAvg Loss: 0.0000\nTrain Epoch: 66\t[25000/88956 (28%)]\tTotal Loss: 0.7856\tAvg Loss: 0.0000\nTrain Epoch: 66\t[30000/88956 (34%)]\tTotal Loss: 0.9448\tAvg Loss: 0.0000\nTrain Epoch: 66\t[35000/88956 (39%)]\tTotal Loss: 1.0939\tAvg Loss: 0.0000\nTrain Epoch: 66\t[40000/88956 (45%)]\tTotal Loss: 1.2653\tAvg Loss: 0.0000\nTrain Epoch: 66\t[45000/88956 (51%)]\tTotal Loss: 1.4241\tAvg Loss: 0.0000\nTrain Epoch: 66\t[50000/88956 (56%)]\tTotal Loss: 1.5858\tAvg Loss: 0.0000\nTrain Epoch: 66\t[55000/88956 (62%)]\tTotal Loss: 1.7310\tAvg Loss: 0.0000\nTrain Epoch: 66\t[60000/88956 (67%)]\tTotal Loss: 1.9044\tAvg Loss: 0.0000\nTrain Epoch: 66\t[65000/88956 (73%)]\tTotal Loss: 2.0660\tAvg Loss: 0.0000\nTrain Epoch: 66\t[70000/88956 (79%)]\tTotal Loss: 2.2380\tAvg Loss: 0.0000\nTrain Epoch: 66\t[75000/88956 (84%)]\tTotal Loss: 2.4037\tAvg Loss: 0.0000\nTrain Epoch: 66\t[80000/88956 (90%)]\tTotal Loss: 2.5829\tAvg Loss: 0.0000\nTrain Epoch: 66\t[85000/88956 (96%)]\tTotal Loss: 2.7482\tAvg Loss: 0.0000\n====> Epoch: 66\tTotal Loss: 2.8721\t Avg Loss: 0.0000\tCorrect: 70254/88956\tPercentage Correct: 78.98\n====> Val Loss: 0.5092\t Avg Loss: 0.0001\tCorrect: 6840/9885\tPercentage Correct: 69.20\n====> Test Loss: 1.2521\t Avg Loss: 0.0001\tCorrect: 17164/24711\tPercentage Correct: 69.46\nTrain Epoch: 67\t[5000/88956 (6%)]\tTotal Loss: 0.1489\tAvg Loss: 0.0000\nTrain Epoch: 67\t[10000/88956 (11%)]\tTotal Loss: 0.3174\tAvg Loss: 0.0000\nTrain Epoch: 67\t[15000/88956 (17%)]\tTotal Loss: 0.4604\tAvg Loss: 0.0000\nTrain Epoch: 67\t[20000/88956 (22%)]\tTotal Loss: 0.6139\tAvg Loss: 0.0000\nTrain Epoch: 67\t[25000/88956 (28%)]\tTotal Loss: 0.7618\tAvg Loss: 0.0000\nTrain Epoch: 67\t[30000/88956 (34%)]\tTotal Loss: 0.9261\tAvg Loss: 0.0000\nTrain Epoch: 67\t[35000/88956 (39%)]\tTotal Loss: 1.0891\tAvg Loss: 0.0000\nTrain Epoch: 67\t[40000/88956 (45%)]\tTotal Loss: 1.2401\tAvg Loss: 0.0000\nTrain Epoch: 67\t[45000/88956 (51%)]\tTotal Loss: 1.3865\tAvg Loss: 0.0000\nTrain Epoch: 67\t[50000/88956 (56%)]\tTotal Loss: 1.5461\tAvg Loss: 0.0000\nTrain Epoch: 67\t[55000/88956 (62%)]\tTotal Loss: 1.7063\tAvg Loss: 0.0000\nTrain Epoch: 67\t[60000/88956 (67%)]\tTotal Loss: 1.8541\tAvg Loss: 0.0000\nTrain Epoch: 67\t[65000/88956 (73%)]\tTotal Loss: 2.0070\tAvg Loss: 0.0000\nTrain Epoch: 67\t[70000/88956 (79%)]\tTotal Loss: 2.1724\tAvg Loss: 0.0000\nTrain Epoch: 67\t[75000/88956 (84%)]\tTotal Loss: 2.3488\tAvg Loss: 0.0000\nTrain Epoch: 67\t[80000/88956 (90%)]\tTotal Loss: 2.5475\tAvg Loss: 0.0000\nTrain Epoch: 67\t[85000/88956 (96%)]\tTotal Loss: 2.7097\tAvg Loss: 0.0000\n====> Epoch: 67\tTotal Loss: 2.8310\t Avg Loss: 0.0000\tCorrect: 70280/88956\tPercentage Correct: 79.01\n====> Val Loss: 0.4968\t Avg Loss: 0.0001\tCorrect: 6954/9885\tPercentage Correct: 70.35\n====> Test Loss: 1.1989\t Avg Loss: 0.0000\tCorrect: 17560/24711\tPercentage Correct: 71.06\nTrain Epoch: 68\t[5000/88956 (6%)]\tTotal Loss: 0.1571\tAvg Loss: 0.0000\nTrain Epoch: 68\t[10000/88956 (11%)]\tTotal Loss: 0.3183\tAvg Loss: 0.0000\nTrain Epoch: 68\t[15000/88956 (17%)]\tTotal Loss: 0.4735\tAvg Loss: 0.0000\nTrain Epoch: 68\t[20000/88956 (22%)]\tTotal Loss: 0.6143\tAvg Loss: 0.0000\nTrain Epoch: 68\t[25000/88956 (28%)]\tTotal Loss: 0.7670\tAvg Loss: 0.0000\nTrain Epoch: 68\t[30000/88956 (34%)]\tTotal Loss: 0.9273\tAvg Loss: 0.0000\nTrain Epoch: 68\t[35000/88956 (39%)]\tTotal Loss: 1.1000\tAvg Loss: 0.0000\nTrain Epoch: 68\t[40000/88956 (45%)]\tTotal Loss: 1.2689\tAvg Loss: 0.0000\nTrain Epoch: 68\t[45000/88956 (51%)]\tTotal Loss: 1.4366\tAvg Loss: 0.0000\nTrain Epoch: 68\t[50000/88956 (56%)]\tTotal Loss: 1.5998\tAvg Loss: 0.0000\nTrain Epoch: 68\t[55000/88956 (62%)]\tTotal Loss: 1.7552\tAvg Loss: 0.0000\nTrain Epoch: 68\t[60000/88956 (67%)]\tTotal Loss: 1.9178\tAvg Loss: 0.0000\nTrain Epoch: 68\t[65000/88956 (73%)]\tTotal Loss: 2.0767\tAvg Loss: 0.0000\nTrain Epoch: 68\t[70000/88956 (79%)]\tTotal Loss: 2.2436\tAvg Loss: 0.0000\nTrain Epoch: 68\t[75000/88956 (84%)]\tTotal Loss: 2.4010\tAvg Loss: 0.0000\nTrain Epoch: 68\t[80000/88956 (90%)]\tTotal Loss: 2.5770\tAvg Loss: 0.0000\nTrain Epoch: 68\t[85000/88956 (96%)]\tTotal Loss: 2.7448\tAvg Loss: 0.0000\n====> Epoch: 68\tTotal Loss: 2.8712\t Avg Loss: 0.0000\tCorrect: 69975/88956\tPercentage Correct: 78.66\n====> Val Loss: 0.4959\t Avg Loss: 0.0001\tCorrect: 6774/9885\tPercentage Correct: 68.53\n====> Test Loss: 1.2209\t Avg Loss: 0.0000\tCorrect: 16966/24711\tPercentage Correct: 68.66\nTrain Epoch: 69\t[5000/88956 (6%)]\tTotal Loss: 0.1451\tAvg Loss: 0.0000\nTrain Epoch: 69\t[10000/88956 (11%)]\tTotal Loss: 0.2886\tAvg Loss: 0.0000\nTrain Epoch: 69\t[15000/88956 (17%)]\tTotal Loss: 0.4325\tAvg Loss: 0.0000\nTrain Epoch: 69\t[20000/88956 (22%)]\tTotal Loss: 0.5790\tAvg Loss: 0.0000\nTrain Epoch: 69\t[25000/88956 (28%)]\tTotal Loss: 0.7395\tAvg Loss: 0.0000\nTrain Epoch: 69\t[30000/88956 (34%)]\tTotal Loss: 0.8900\tAvg Loss: 0.0000\nTrain Epoch: 69\t[35000/88956 (39%)]\tTotal Loss: 1.0492\tAvg Loss: 0.0000\nTrain Epoch: 69\t[40000/88956 (45%)]\tTotal Loss: 1.2035\tAvg Loss: 0.0000\nTrain Epoch: 69\t[45000/88956 (51%)]\tTotal Loss: 1.3736\tAvg Loss: 0.0000\nTrain Epoch: 69\t[50000/88956 (56%)]\tTotal Loss: 1.5250\tAvg Loss: 0.0000\nTrain Epoch: 69\t[55000/88956 (62%)]\tTotal Loss: 1.6839\tAvg Loss: 0.0000\nTrain Epoch: 69\t[60000/88956 (67%)]\tTotal Loss: 1.8387\tAvg Loss: 0.0000\nTrain Epoch: 69\t[65000/88956 (73%)]\tTotal Loss: 1.9975\tAvg Loss: 0.0000\nTrain Epoch: 69\t[70000/88956 (79%)]\tTotal Loss: 2.1545\tAvg Loss: 0.0000\nTrain Epoch: 69\t[75000/88956 (84%)]\tTotal Loss: 2.3164\tAvg Loss: 0.0000\nTrain Epoch: 69\t[80000/88956 (90%)]\tTotal Loss: 2.4862\tAvg Loss: 0.0000\nTrain Epoch: 69\t[85000/88956 (96%)]\tTotal Loss: 2.6547\tAvg Loss: 0.0000\n====> Epoch: 69\tTotal Loss: 2.7785\t Avg Loss: 0.0000\tCorrect: 70540/88956\tPercentage Correct: 79.30\n====> Val Loss: 0.4996\t Avg Loss: 0.0001\tCorrect: 6807/9885\tPercentage Correct: 68.86\n====> Test Loss: 1.2632\t Avg Loss: 0.0001\tCorrect: 17123/24711\tPercentage Correct: 69.29\nTrain Epoch: 70\t[5000/88956 (6%)]\tTotal Loss: 0.1503\tAvg Loss: 0.0000\nTrain Epoch: 70\t[10000/88956 (11%)]\tTotal Loss: 0.3019\tAvg Loss: 0.0000\nTrain Epoch: 70\t[15000/88956 (17%)]\tTotal Loss: 0.4502\tAvg Loss: 0.0000\nTrain Epoch: 70\t[20000/88956 (22%)]\tTotal Loss: 0.6094\tAvg Loss: 0.0000\nTrain Epoch: 70\t[25000/88956 (28%)]\tTotal Loss: 0.7676\tAvg Loss: 0.0000\nTrain Epoch: 70\t[30000/88956 (34%)]\tTotal Loss: 0.9183\tAvg Loss: 0.0000\nTrain Epoch: 70\t[35000/88956 (39%)]\tTotal Loss: 1.0677\tAvg Loss: 0.0000\nTrain Epoch: 70\t[40000/88956 (45%)]\tTotal Loss: 1.2212\tAvg Loss: 0.0000\nTrain Epoch: 70\t[45000/88956 (51%)]\tTotal Loss: 1.3808\tAvg Loss: 0.0000\nTrain Epoch: 70\t[50000/88956 (56%)]\tTotal Loss: 1.5309\tAvg Loss: 0.0000\nTrain Epoch: 70\t[55000/88956 (62%)]\tTotal Loss: 1.6967\tAvg Loss: 0.0000\nTrain Epoch: 70\t[60000/88956 (67%)]\tTotal Loss: 1.8599\tAvg Loss: 0.0000\nTrain Epoch: 70\t[65000/88956 (73%)]\tTotal Loss: 2.0219\tAvg Loss: 0.0000\nTrain Epoch: 70\t[70000/88956 (79%)]\tTotal Loss: 2.1793\tAvg Loss: 0.0000\nTrain Epoch: 70\t[75000/88956 (84%)]\tTotal Loss: 2.3447\tAvg Loss: 0.0000\nTrain Epoch: 70\t[80000/88956 (90%)]\tTotal Loss: 2.4984\tAvg Loss: 0.0000\nTrain Epoch: 70\t[85000/88956 (96%)]\tTotal Loss: 2.6471\tAvg Loss: 0.0000\n====> Epoch: 70\tTotal Loss: 2.7677\t Avg Loss: 0.0000\tCorrect: 70706/88956\tPercentage Correct: 79.48\n====> Val Loss: 0.5056\t Avg Loss: 0.0001\tCorrect: 6960/9885\tPercentage Correct: 70.41\n====> Test Loss: 1.2190\t Avg Loss: 0.0000\tCorrect: 17402/24711\tPercentage Correct: 70.42\nTrain Epoch: 71\t[5000/88956 (6%)]\tTotal Loss: 0.1558\tAvg Loss: 0.0000\nTrain Epoch: 71\t[10000/88956 (11%)]\tTotal Loss: 0.2872\tAvg Loss: 0.0000\nTrain Epoch: 71\t[15000/88956 (17%)]\tTotal Loss: 0.4230\tAvg Loss: 0.0000\nTrain Epoch: 71\t[20000/88956 (22%)]\tTotal Loss: 0.5767\tAvg Loss: 0.0000\nTrain Epoch: 71\t[25000/88956 (28%)]\tTotal Loss: 0.7226\tAvg Loss: 0.0000\nTrain Epoch: 71\t[30000/88956 (34%)]\tTotal Loss: 0.8707\tAvg Loss: 0.0000\nTrain Epoch: 71\t[35000/88956 (39%)]\tTotal Loss: 1.0258\tAvg Loss: 0.0000\nTrain Epoch: 71\t[40000/88956 (45%)]\tTotal Loss: 1.1745\tAvg Loss: 0.0000\nTrain Epoch: 71\t[45000/88956 (51%)]\tTotal Loss: 1.3412\tAvg Loss: 0.0000\nTrain Epoch: 71\t[50000/88956 (56%)]\tTotal Loss: 1.4868\tAvg Loss: 0.0000\nTrain Epoch: 71\t[55000/88956 (62%)]\tTotal Loss: 1.6384\tAvg Loss: 0.0000\nTrain Epoch: 71\t[60000/88956 (67%)]\tTotal Loss: 1.7943\tAvg Loss: 0.0000\nTrain Epoch: 71\t[65000/88956 (73%)]\tTotal Loss: 1.9481\tAvg Loss: 0.0000\nTrain Epoch: 71\t[70000/88956 (79%)]\tTotal Loss: 2.1025\tAvg Loss: 0.0000\nTrain Epoch: 71\t[75000/88956 (84%)]\tTotal Loss: 2.2641\tAvg Loss: 0.0000\nTrain Epoch: 71\t[80000/88956 (90%)]\tTotal Loss: 2.5076\tAvg Loss: 0.0000\nTrain Epoch: 71\t[85000/88956 (96%)]\tTotal Loss: 2.6999\tAvg Loss: 0.0000\n====> Epoch: 71\tTotal Loss: 2.8258\t Avg Loss: 0.0000\tCorrect: 70401/88956\tPercentage Correct: 79.14\n====> Val Loss: 0.4836\t Avg Loss: 0.0000\tCorrect: 6915/9885\tPercentage Correct: 69.95\n====> Test Loss: 1.1758\t Avg Loss: 0.0000\tCorrect: 17346/24711\tPercentage Correct: 70.20\nTrain Epoch: 72\t[5000/88956 (6%)]\tTotal Loss: 0.1367\tAvg Loss: 0.0000\nTrain Epoch: 72\t[10000/88956 (11%)]\tTotal Loss: 0.2849\tAvg Loss: 0.0000\nTrain Epoch: 72\t[15000/88956 (17%)]\tTotal Loss: 0.4330\tAvg Loss: 0.0000\nTrain Epoch: 72\t[20000/88956 (22%)]\tTotal Loss: 0.5689\tAvg Loss: 0.0000\nTrain Epoch: 72\t[25000/88956 (28%)]\tTotal Loss: 0.7190\tAvg Loss: 0.0000\nTrain Epoch: 72\t[30000/88956 (34%)]\tTotal Loss: 0.8677\tAvg Loss: 0.0000\nTrain Epoch: 72\t[35000/88956 (39%)]\tTotal Loss: 1.0200\tAvg Loss: 0.0000\nTrain Epoch: 72\t[40000/88956 (45%)]\tTotal Loss: 1.1790\tAvg Loss: 0.0000\nTrain Epoch: 72\t[45000/88956 (51%)]\tTotal Loss: 1.3342\tAvg Loss: 0.0000\nTrain Epoch: 72\t[50000/88956 (56%)]\tTotal Loss: 1.4968\tAvg Loss: 0.0000\nTrain Epoch: 72\t[55000/88956 (62%)]\tTotal Loss: 1.6448\tAvg Loss: 0.0000\nTrain Epoch: 72\t[60000/88956 (67%)]\tTotal Loss: 1.8064\tAvg Loss: 0.0000\nTrain Epoch: 72\t[65000/88956 (73%)]\tTotal Loss: 1.9527\tAvg Loss: 0.0000\nTrain Epoch: 72\t[70000/88956 (79%)]\tTotal Loss: 2.1114\tAvg Loss: 0.0000\nTrain Epoch: 72\t[75000/88956 (84%)]\tTotal Loss: 2.2611\tAvg Loss: 0.0000\nTrain Epoch: 72\t[80000/88956 (90%)]\tTotal Loss: 2.4133\tAvg Loss: 0.0000\nTrain Epoch: 72\t[85000/88956 (96%)]\tTotal Loss: 2.5643\tAvg Loss: 0.0000\n====> Epoch: 72\tTotal Loss: 2.6877\t Avg Loss: 0.0000\tCorrect: 71250/88956\tPercentage Correct: 80.10\n====> Val Loss: 0.4874\t Avg Loss: 0.0000\tCorrect: 6864/9885\tPercentage Correct: 69.44\n====> Test Loss: 1.2117\t Avg Loss: 0.0000\tCorrect: 17371/24711\tPercentage Correct: 70.30\nTrain Epoch: 73\t[5000/88956 (6%)]\tTotal Loss: 0.1489\tAvg Loss: 0.0000\nTrain Epoch: 73\t[10000/88956 (11%)]\tTotal Loss: 0.2895\tAvg Loss: 0.0000\nTrain Epoch: 73\t[15000/88956 (17%)]\tTotal Loss: 0.4306\tAvg Loss: 0.0000\nTrain Epoch: 73\t[20000/88956 (22%)]\tTotal Loss: 0.5652\tAvg Loss: 0.0000\nTrain Epoch: 73\t[25000/88956 (28%)]\tTotal Loss: 0.7045\tAvg Loss: 0.0000\nTrain Epoch: 73\t[30000/88956 (34%)]\tTotal Loss: 0.8452\tAvg Loss: 0.0000\nTrain Epoch: 73\t[35000/88956 (39%)]\tTotal Loss: 0.9945\tAvg Loss: 0.0000\nTrain Epoch: 73\t[40000/88956 (45%)]\tTotal Loss: 1.1387\tAvg Loss: 0.0000\nTrain Epoch: 73\t[45000/88956 (51%)]\tTotal Loss: 1.2717\tAvg Loss: 0.0000\nTrain Epoch: 73\t[50000/88956 (56%)]\tTotal Loss: 1.4117\tAvg Loss: 0.0000\nTrain Epoch: 73\t[55000/88956 (62%)]\tTotal Loss: 1.5738\tAvg Loss: 0.0000\nTrain Epoch: 73\t[60000/88956 (67%)]\tTotal Loss: 1.7158\tAvg Loss: 0.0000\nTrain Epoch: 73\t[65000/88956 (73%)]\tTotal Loss: 1.8773\tAvg Loss: 0.0000\nTrain Epoch: 73\t[70000/88956 (79%)]\tTotal Loss: 2.0374\tAvg Loss: 0.0000\nTrain Epoch: 73\t[75000/88956 (84%)]\tTotal Loss: 2.1933\tAvg Loss: 0.0000\nTrain Epoch: 73\t[80000/88956 (90%)]\tTotal Loss: 2.3412\tAvg Loss: 0.0000\nTrain Epoch: 73\t[85000/88956 (96%)]\tTotal Loss: 2.4877\tAvg Loss: 0.0000\n====> Epoch: 73\tTotal Loss: 2.6095\t Avg Loss: 0.0000\tCorrect: 71608/88956\tPercentage Correct: 80.50\n====> Val Loss: 0.4817\t Avg Loss: 0.0000\tCorrect: 6942/9885\tPercentage Correct: 70.23\n====> Test Loss: 1.1657\t Avg Loss: 0.0000\tCorrect: 17553/24711\tPercentage Correct: 71.03\nTrain Epoch: 74\t[5000/88956 (6%)]\tTotal Loss: 0.1427\tAvg Loss: 0.0000\nTrain Epoch: 74\t[10000/88956 (11%)]\tTotal Loss: 0.2820\tAvg Loss: 0.0000\nTrain Epoch: 74\t[15000/88956 (17%)]\tTotal Loss: 0.4282\tAvg Loss: 0.0000\nTrain Epoch: 74\t[20000/88956 (22%)]\tTotal Loss: 0.5714\tAvg Loss: 0.0000\nTrain Epoch: 74\t[25000/88956 (28%)]\tTotal Loss: 0.7150\tAvg Loss: 0.0000\nTrain Epoch: 74\t[30000/88956 (34%)]\tTotal Loss: 0.8786\tAvg Loss: 0.0000\nTrain Epoch: 74\t[35000/88956 (39%)]\tTotal Loss: 1.0259\tAvg Loss: 0.0000\nTrain Epoch: 74\t[40000/88956 (45%)]\tTotal Loss: 1.1727\tAvg Loss: 0.0000\nTrain Epoch: 74\t[45000/88956 (51%)]\tTotal Loss: 1.3188\tAvg Loss: 0.0000\nTrain Epoch: 74\t[50000/88956 (56%)]\tTotal Loss: 1.4707\tAvg Loss: 0.0000\nTrain Epoch: 74\t[55000/88956 (62%)]\tTotal Loss: 1.6363\tAvg Loss: 0.0000\nTrain Epoch: 74\t[60000/88956 (67%)]\tTotal Loss: 1.7839\tAvg Loss: 0.0000\nTrain Epoch: 74\t[65000/88956 (73%)]\tTotal Loss: 1.9390\tAvg Loss: 0.0000\nTrain Epoch: 74\t[70000/88956 (79%)]\tTotal Loss: 2.0872\tAvg Loss: 0.0000\nTrain Epoch: 74\t[75000/88956 (84%)]\tTotal Loss: 2.2464\tAvg Loss: 0.0000\nTrain Epoch: 74\t[80000/88956 (90%)]\tTotal Loss: 2.3921\tAvg Loss: 0.0000\nTrain Epoch: 74\t[85000/88956 (96%)]\tTotal Loss: 2.5496\tAvg Loss: 0.0000\n====> Epoch: 74\tTotal Loss: 2.6696\t Avg Loss: 0.0000\tCorrect: 71435/88956\tPercentage Correct: 80.30\n====> Val Loss: 0.4557\t Avg Loss: 0.0000\tCorrect: 7040/9885\tPercentage Correct: 71.22\n====> Test Loss: 1.1191\t Avg Loss: 0.0000\tCorrect: 17688/24711\tPercentage Correct: 71.58\n\n=== saved best model ===\n\nTrain Epoch: 75\t[5000/88956 (6%)]\tTotal Loss: 0.1480\tAvg Loss: 0.0000\nTrain Epoch: 75\t[10000/88956 (11%)]\tTotal Loss: 0.2874\tAvg Loss: 0.0000\nTrain Epoch: 75\t[15000/88956 (17%)]\tTotal Loss: 0.4273\tAvg Loss: 0.0000\nTrain Epoch: 75\t[20000/88956 (22%)]\tTotal Loss: 0.5627\tAvg Loss: 0.0000\nTrain Epoch: 75\t[25000/88956 (28%)]\tTotal Loss: 0.6979\tAvg Loss: 0.0000\nTrain Epoch: 75\t[30000/88956 (34%)]\tTotal Loss: 0.8487\tAvg Loss: 0.0000\nTrain Epoch: 75\t[35000/88956 (39%)]\tTotal Loss: 0.9958\tAvg Loss: 0.0000\nTrain Epoch: 75\t[40000/88956 (45%)]\tTotal Loss: 1.1372\tAvg Loss: 0.0000\nTrain Epoch: 75\t[45000/88956 (51%)]\tTotal Loss: 1.2900\tAvg Loss: 0.0000\nTrain Epoch: 75\t[50000/88956 (56%)]\tTotal Loss: 1.4363\tAvg Loss: 0.0000\nTrain Epoch: 75\t[55000/88956 (62%)]\tTotal Loss: 1.5890\tAvg Loss: 0.0000\nTrain Epoch: 75\t[60000/88956 (67%)]\tTotal Loss: 1.7661\tAvg Loss: 0.0000\nTrain Epoch: 75\t[65000/88956 (73%)]\tTotal Loss: 1.9141\tAvg Loss: 0.0000\nTrain Epoch: 75\t[70000/88956 (79%)]\tTotal Loss: 2.0611\tAvg Loss: 0.0000\nTrain Epoch: 75\t[75000/88956 (84%)]\tTotal Loss: 2.2064\tAvg Loss: 0.0000\nTrain Epoch: 75\t[80000/88956 (90%)]\tTotal Loss: 2.3600\tAvg Loss: 0.0000\nTrain Epoch: 75\t[85000/88956 (96%)]\tTotal Loss: 2.5074\tAvg Loss: 0.0000\n====> Epoch: 75\tTotal Loss: 2.6383\t Avg Loss: 0.0000\tCorrect: 71480/88956\tPercentage Correct: 80.35\n====> Val Loss: 0.4682\t Avg Loss: 0.0000\tCorrect: 7027/9885\tPercentage Correct: 71.09\n====> Test Loss: 1.1422\t Avg Loss: 0.0000\tCorrect: 17652/24711\tPercentage Correct: 71.43\nTrain Epoch: 76\t[5000/88956 (6%)]\tTotal Loss: 0.1355\tAvg Loss: 0.0000\nTrain Epoch: 76\t[10000/88956 (11%)]\tTotal Loss: 0.2671\tAvg Loss: 0.0000\nTrain Epoch: 76\t[15000/88956 (17%)]\tTotal Loss: 0.3997\tAvg Loss: 0.0000\nTrain Epoch: 76\t[20000/88956 (22%)]\tTotal Loss: 0.5302\tAvg Loss: 0.0000\nTrain Epoch: 76\t[25000/88956 (28%)]\tTotal Loss: 0.6598\tAvg Loss: 0.0000\nTrain Epoch: 76\t[30000/88956 (34%)]\tTotal Loss: 0.8010\tAvg Loss: 0.0000\nTrain Epoch: 76\t[35000/88956 (39%)]\tTotal Loss: 0.9426\tAvg Loss: 0.0000\nTrain Epoch: 76\t[40000/88956 (45%)]\tTotal Loss: 1.0988\tAvg Loss: 0.0000\nTrain Epoch: 76\t[45000/88956 (51%)]\tTotal Loss: 1.2419\tAvg Loss: 0.0000\nTrain Epoch: 76\t[50000/88956 (56%)]\tTotal Loss: 1.3887\tAvg Loss: 0.0000\nTrain Epoch: 76\t[55000/88956 (62%)]\tTotal Loss: 1.5306\tAvg Loss: 0.0000\nTrain Epoch: 76\t[60000/88956 (67%)]\tTotal Loss: 1.6766\tAvg Loss: 0.0000\nTrain Epoch: 76\t[65000/88956 (73%)]\tTotal Loss: 1.8242\tAvg Loss: 0.0000\nTrain Epoch: 76\t[70000/88956 (79%)]\tTotal Loss: 1.9871\tAvg Loss: 0.0000\nTrain Epoch: 76\t[75000/88956 (84%)]\tTotal Loss: 2.1240\tAvg Loss: 0.0000\nTrain Epoch: 76\t[80000/88956 (90%)]\tTotal Loss: 2.2721\tAvg Loss: 0.0000\nTrain Epoch: 76\t[85000/88956 (96%)]\tTotal Loss: 2.4165\tAvg Loss: 0.0000\n====> Epoch: 76\tTotal Loss: 2.5365\t Avg Loss: 0.0000\tCorrect: 71947/88956\tPercentage Correct: 80.88\n====> Val Loss: 0.4544\t Avg Loss: 0.0000\tCorrect: 7086/9885\tPercentage Correct: 71.68\n====> Test Loss: 1.1302\t Avg Loss: 0.0000\tCorrect: 17706/24711\tPercentage Correct: 71.65\n\n=== saved best model ===\n\nTrain Epoch: 77\t[5000/88956 (6%)]\tTotal Loss: 0.1331\tAvg Loss: 0.0000\nTrain Epoch: 77\t[10000/88956 (11%)]\tTotal Loss: 0.2787\tAvg Loss: 0.0000\nTrain Epoch: 77\t[15000/88956 (17%)]\tTotal Loss: 0.4147\tAvg Loss: 0.0000\nTrain Epoch: 77\t[20000/88956 (22%)]\tTotal Loss: 0.5517\tAvg Loss: 0.0000\nTrain Epoch: 77\t[25000/88956 (28%)]\tTotal Loss: 0.6907\tAvg Loss: 0.0000\nTrain Epoch: 77\t[30000/88956 (34%)]\tTotal Loss: 0.8375\tAvg Loss: 0.0000\nTrain Epoch: 77\t[35000/88956 (39%)]\tTotal Loss: 1.0011\tAvg Loss: 0.0000\nTrain Epoch: 77\t[40000/88956 (45%)]\tTotal Loss: 1.1391\tAvg Loss: 0.0000\nTrain Epoch: 77\t[45000/88956 (51%)]\tTotal Loss: 1.2750\tAvg Loss: 0.0000\nTrain Epoch: 77\t[50000/88956 (56%)]\tTotal Loss: 1.4093\tAvg Loss: 0.0000\nTrain Epoch: 77\t[55000/88956 (62%)]\tTotal Loss: 1.5552\tAvg Loss: 0.0000\nTrain Epoch: 77\t[60000/88956 (67%)]\tTotal Loss: 1.6967\tAvg Loss: 0.0000\nTrain Epoch: 77\t[65000/88956 (73%)]\tTotal Loss: 1.8269\tAvg Loss: 0.0000\nTrain Epoch: 77\t[70000/88956 (79%)]\tTotal Loss: 1.9648\tAvg Loss: 0.0000\nTrain Epoch: 77\t[75000/88956 (84%)]\tTotal Loss: 2.1124\tAvg Loss: 0.0000\nTrain Epoch: 77\t[80000/88956 (90%)]\tTotal Loss: 2.2653\tAvg Loss: 0.0000\nTrain Epoch: 77\t[85000/88956 (96%)]\tTotal Loss: 2.4129\tAvg Loss: 0.0000\n====> Epoch: 77\tTotal Loss: 2.5255\t Avg Loss: 0.0000\tCorrect: 71960/88956\tPercentage Correct: 80.89\n====> Val Loss: 0.4471\t Avg Loss: 0.0000\tCorrect: 7085/9885\tPercentage Correct: 71.67\n====> Test Loss: 1.0974\t Avg Loss: 0.0000\tCorrect: 17804/24711\tPercentage Correct: 72.05\nTrain Epoch: 78\t[5000/88956 (6%)]\tTotal Loss: 0.1220\tAvg Loss: 0.0000\nTrain Epoch: 78\t[10000/88956 (11%)]\tTotal Loss: 0.2484\tAvg Loss: 0.0000\nTrain Epoch: 78\t[15000/88956 (17%)]\tTotal Loss: 0.3678\tAvg Loss: 0.0000\nTrain Epoch: 78\t[20000/88956 (22%)]\tTotal Loss: 0.5514\tAvg Loss: 0.0000\nTrain Epoch: 78\t[25000/88956 (28%)]\tTotal Loss: 0.8216\tAvg Loss: 0.0000\nTrain Epoch: 78\t[30000/88956 (34%)]\tTotal Loss: 0.9903\tAvg Loss: 0.0000\nTrain Epoch: 78\t[35000/88956 (39%)]\tTotal Loss: 1.1463\tAvg Loss: 0.0000\nTrain Epoch: 78\t[40000/88956 (45%)]\tTotal Loss: 1.2862\tAvg Loss: 0.0000\nTrain Epoch: 78\t[45000/88956 (51%)]\tTotal Loss: 1.4339\tAvg Loss: 0.0000\nTrain Epoch: 78\t[50000/88956 (56%)]\tTotal Loss: 1.5642\tAvg Loss: 0.0000\nTrain Epoch: 78\t[55000/88956 (62%)]\tTotal Loss: 1.7031\tAvg Loss: 0.0000\nTrain Epoch: 78\t[60000/88956 (67%)]\tTotal Loss: 1.8508\tAvg Loss: 0.0000\nTrain Epoch: 78\t[65000/88956 (73%)]\tTotal Loss: 2.0047\tAvg Loss: 0.0000\nTrain Epoch: 78\t[70000/88956 (79%)]\tTotal Loss: 2.1519\tAvg Loss: 0.0000\nTrain Epoch: 78\t[75000/88956 (84%)]\tTotal Loss: 2.3002\tAvg Loss: 0.0000\nTrain Epoch: 78\t[80000/88956 (90%)]\tTotal Loss: 2.4347\tAvg Loss: 0.0000\nTrain Epoch: 78\t[85000/88956 (96%)]\tTotal Loss: 2.5690\tAvg Loss: 0.0000\n====> Epoch: 78\tTotal Loss: 2.6750\t Avg Loss: 0.0000\tCorrect: 71282/88956\tPercentage Correct: 80.13\n====> Val Loss: 0.4763\t Avg Loss: 0.0000\tCorrect: 7087/9885\tPercentage Correct: 71.69\n====> Test Loss: 1.1612\t Avg Loss: 0.0000\tCorrect: 17832/24711\tPercentage Correct: 72.16\n\n=== saved best model ===\n\nTrain Epoch: 79\t[5000/88956 (6%)]\tTotal Loss: 0.1308\tAvg Loss: 0.0000\nTrain Epoch: 79\t[10000/88956 (11%)]\tTotal Loss: 0.2621\tAvg Loss: 0.0000\nTrain Epoch: 79\t[15000/88956 (17%)]\tTotal Loss: 0.3957\tAvg Loss: 0.0000\nTrain Epoch: 79\t[20000/88956 (22%)]\tTotal Loss: 0.5321\tAvg Loss: 0.0000\nTrain Epoch: 79\t[25000/88956 (28%)]\tTotal Loss: 0.6760\tAvg Loss: 0.0000\nTrain Epoch: 79\t[30000/88956 (34%)]\tTotal Loss: 0.8151\tAvg Loss: 0.0000\nTrain Epoch: 79\t[35000/88956 (39%)]\tTotal Loss: 0.9379\tAvg Loss: 0.0000\nTrain Epoch: 79\t[40000/88956 (45%)]\tTotal Loss: 1.0894\tAvg Loss: 0.0000\nTrain Epoch: 79\t[45000/88956 (51%)]\tTotal Loss: 1.3160\tAvg Loss: 0.0000\nTrain Epoch: 79\t[50000/88956 (56%)]\tTotal Loss: 1.4700\tAvg Loss: 0.0000\nTrain Epoch: 79\t[55000/88956 (62%)]\tTotal Loss: 1.6087\tAvg Loss: 0.0000\nTrain Epoch: 79\t[60000/88956 (67%)]\tTotal Loss: 1.7558\tAvg Loss: 0.0000\nTrain Epoch: 79\t[65000/88956 (73%)]\tTotal Loss: 1.8931\tAvg Loss: 0.0000\nTrain Epoch: 79\t[70000/88956 (79%)]\tTotal Loss: 2.0367\tAvg Loss: 0.0000\nTrain Epoch: 79\t[75000/88956 (84%)]\tTotal Loss: 2.1725\tAvg Loss: 0.0000\nTrain Epoch: 79\t[80000/88956 (90%)]\tTotal Loss: 2.3232\tAvg Loss: 0.0000\nTrain Epoch: 79\t[85000/88956 (96%)]\tTotal Loss: 2.4621\tAvg Loss: 0.0000\n====> Epoch: 79\tTotal Loss: 2.5868\t Avg Loss: 0.0000\tCorrect: 71630/88956\tPercentage Correct: 80.52\n====> Val Loss: 0.4619\t Avg Loss: 0.0000\tCorrect: 7041/9885\tPercentage Correct: 71.23\n====> Test Loss: 1.1051\t Avg Loss: 0.0000\tCorrect: 17736/24711\tPercentage Correct: 71.77\nTrain Epoch: 80\t[5000/88956 (6%)]\tTotal Loss: 0.1243\tAvg Loss: 0.0000\nTrain Epoch: 80\t[10000/88956 (11%)]\tTotal Loss: 0.2481\tAvg Loss: 0.0000\nTrain Epoch: 80\t[15000/88956 (17%)]\tTotal Loss: 0.3707\tAvg Loss: 0.0000\nTrain Epoch: 80\t[20000/88956 (22%)]\tTotal Loss: 0.4897\tAvg Loss: 0.0000\nTrain Epoch: 80\t[25000/88956 (28%)]\tTotal Loss: 0.6080\tAvg Loss: 0.0000\nTrain Epoch: 80\t[30000/88956 (34%)]\tTotal Loss: 0.7322\tAvg Loss: 0.0000\nTrain Epoch: 80\t[35000/88956 (39%)]\tTotal Loss: 0.8823\tAvg Loss: 0.0000\nTrain Epoch: 80\t[40000/88956 (45%)]\tTotal Loss: 1.0247\tAvg Loss: 0.0000\nTrain Epoch: 80\t[45000/88956 (51%)]\tTotal Loss: 1.1713\tAvg Loss: 0.0000\nTrain Epoch: 80\t[50000/88956 (56%)]\tTotal Loss: 1.2977\tAvg Loss: 0.0000\nTrain Epoch: 80\t[55000/88956 (62%)]\tTotal Loss: 1.4360\tAvg Loss: 0.0000\nTrain Epoch: 80\t[60000/88956 (67%)]\tTotal Loss: 1.5715\tAvg Loss: 0.0000\nTrain Epoch: 80\t[65000/88956 (73%)]\tTotal Loss: 1.7072\tAvg Loss: 0.0000\nTrain Epoch: 80\t[70000/88956 (79%)]\tTotal Loss: 1.8380\tAvg Loss: 0.0000\nTrain Epoch: 80\t[75000/88956 (84%)]\tTotal Loss: 1.9777\tAvg Loss: 0.0000\nTrain Epoch: 80\t[80000/88956 (90%)]\tTotal Loss: 2.1176\tAvg Loss: 0.0000\nTrain Epoch: 80\t[85000/88956 (96%)]\tTotal Loss: 2.2530\tAvg Loss: 0.0000\n====> Epoch: 80\tTotal Loss: 2.3549\t Avg Loss: 0.0000\tCorrect: 72838/88956\tPercentage Correct: 81.88\n====> Val Loss: 0.4734\t Avg Loss: 0.0000\tCorrect: 7063/9885\tPercentage Correct: 71.45\n====> Test Loss: 1.1425\t Avg Loss: 0.0000\tCorrect: 17812/24711\tPercentage Correct: 72.08\nTrain Epoch: 81\t[5000/88956 (6%)]\tTotal Loss: 0.1346\tAvg Loss: 0.0000\nTrain Epoch: 81\t[10000/88956 (11%)]\tTotal Loss: 0.2466\tAvg Loss: 0.0000\nTrain Epoch: 81\t[15000/88956 (17%)]\tTotal Loss: 0.3775\tAvg Loss: 0.0000\nTrain Epoch: 81\t[20000/88956 (22%)]\tTotal Loss: 0.5076\tAvg Loss: 0.0000\nTrain Epoch: 81\t[25000/88956 (28%)]\tTotal Loss: 0.6417\tAvg Loss: 0.0000\nTrain Epoch: 81\t[30000/88956 (34%)]\tTotal Loss: 0.7688\tAvg Loss: 0.0000\nTrain Epoch: 81\t[35000/88956 (39%)]\tTotal Loss: 0.8989\tAvg Loss: 0.0000\nTrain Epoch: 81\t[40000/88956 (45%)]\tTotal Loss: 1.0376\tAvg Loss: 0.0000\nTrain Epoch: 81\t[45000/88956 (51%)]\tTotal Loss: 1.1633\tAvg Loss: 0.0000\nTrain Epoch: 81\t[50000/88956 (56%)]\tTotal Loss: 1.2902\tAvg Loss: 0.0000\nTrain Epoch: 81\t[55000/88956 (62%)]\tTotal Loss: 1.4112\tAvg Loss: 0.0000\nTrain Epoch: 81\t[60000/88956 (67%)]\tTotal Loss: 1.5474\tAvg Loss: 0.0000\nTrain Epoch: 81\t[65000/88956 (73%)]\tTotal Loss: 1.7041\tAvg Loss: 0.0000\nTrain Epoch: 81\t[70000/88956 (79%)]\tTotal Loss: 1.8348\tAvg Loss: 0.0000\nTrain Epoch: 81\t[75000/88956 (84%)]\tTotal Loss: 1.9742\tAvg Loss: 0.0000\nTrain Epoch: 81\t[80000/88956 (90%)]\tTotal Loss: 2.1097\tAvg Loss: 0.0000\nTrain Epoch: 81\t[85000/88956 (96%)]\tTotal Loss: 2.2470\tAvg Loss: 0.0000\n====> Epoch: 81\tTotal Loss: 2.3513\t Avg Loss: 0.0000\tCorrect: 73049/88956\tPercentage Correct: 82.12\n====> Val Loss: 0.4332\t Avg Loss: 0.0000\tCorrect: 7219/9885\tPercentage Correct: 73.03\n====> Test Loss: 1.0628\t Avg Loss: 0.0000\tCorrect: 18015/24711\tPercentage Correct: 72.90\n\n=== saved best model ===\n\nTrain Epoch: 82\t[5000/88956 (6%)]\tTotal Loss: 0.1269\tAvg Loss: 0.0000\nTrain Epoch: 82\t[10000/88956 (11%)]\tTotal Loss: 0.2489\tAvg Loss: 0.0000\nTrain Epoch: 82\t[15000/88956 (17%)]\tTotal Loss: 0.3834\tAvg Loss: 0.0000\nTrain Epoch: 82\t[20000/88956 (22%)]\tTotal Loss: 0.4981\tAvg Loss: 0.0000\nTrain Epoch: 82\t[25000/88956 (28%)]\tTotal Loss: 0.6355\tAvg Loss: 0.0000\nTrain Epoch: 82\t[30000/88956 (34%)]\tTotal Loss: 0.7646\tAvg Loss: 0.0000\nTrain Epoch: 82\t[35000/88956 (39%)]\tTotal Loss: 0.8960\tAvg Loss: 0.0000\nTrain Epoch: 82\t[40000/88956 (45%)]\tTotal Loss: 1.0348\tAvg Loss: 0.0000\nTrain Epoch: 82\t[45000/88956 (51%)]\tTotal Loss: 1.1624\tAvg Loss: 0.0000\nTrain Epoch: 82\t[50000/88956 (56%)]\tTotal Loss: 1.2979\tAvg Loss: 0.0000\nTrain Epoch: 82\t[55000/88956 (62%)]\tTotal Loss: 1.4294\tAvg Loss: 0.0000\nTrain Epoch: 82\t[60000/88956 (67%)]\tTotal Loss: 1.5739\tAvg Loss: 0.0000\nTrain Epoch: 82\t[65000/88956 (73%)]\tTotal Loss: 1.7077\tAvg Loss: 0.0000\nTrain Epoch: 82\t[70000/88956 (79%)]\tTotal Loss: 1.8551\tAvg Loss: 0.0000\nTrain Epoch: 82\t[75000/88956 (84%)]\tTotal Loss: 1.9849\tAvg Loss: 0.0000\nTrain Epoch: 82\t[80000/88956 (90%)]\tTotal Loss: 2.1141\tAvg Loss: 0.0000\nTrain Epoch: 82\t[85000/88956 (96%)]\tTotal Loss: 2.2625\tAvg Loss: 0.0000\n====> Epoch: 82\tTotal Loss: 2.3654\t Avg Loss: 0.0000\tCorrect: 72853/88956\tPercentage Correct: 81.90\n====> Val Loss: 0.5101\t Avg Loss: 0.0001\tCorrect: 6963/9885\tPercentage Correct: 70.44\n====> Test Loss: 1.2397\t Avg Loss: 0.0001\tCorrect: 17523/24711\tPercentage Correct: 70.91\nTrain Epoch: 83\t[5000/88956 (6%)]\tTotal Loss: 0.1310\tAvg Loss: 0.0000\nTrain Epoch: 83\t[10000/88956 (11%)]\tTotal Loss: 0.2447\tAvg Loss: 0.0000\nTrain Epoch: 83\t[15000/88956 (17%)]\tTotal Loss: 0.3517\tAvg Loss: 0.0000\nTrain Epoch: 83\t[20000/88956 (22%)]\tTotal Loss: 0.4836\tAvg Loss: 0.0000\nTrain Epoch: 83\t[25000/88956 (28%)]\tTotal Loss: 0.6169\tAvg Loss: 0.0000\nTrain Epoch: 83\t[30000/88956 (34%)]\tTotal Loss: 0.7496\tAvg Loss: 0.0000\nTrain Epoch: 83\t[35000/88956 (39%)]\tTotal Loss: 0.8775\tAvg Loss: 0.0000\nTrain Epoch: 83\t[40000/88956 (45%)]\tTotal Loss: 1.0101\tAvg Loss: 0.0000\nTrain Epoch: 83\t[45000/88956 (51%)]\tTotal Loss: 1.1342\tAvg Loss: 0.0000\nTrain Epoch: 83\t[50000/88956 (56%)]\tTotal Loss: 1.2656\tAvg Loss: 0.0000\nTrain Epoch: 83\t[55000/88956 (62%)]\tTotal Loss: 1.4159\tAvg Loss: 0.0000\nTrain Epoch: 83\t[60000/88956 (67%)]\tTotal Loss: 1.5595\tAvg Loss: 0.0000\nTrain Epoch: 83\t[65000/88956 (73%)]\tTotal Loss: 1.6987\tAvg Loss: 0.0000\nTrain Epoch: 83\t[70000/88956 (79%)]\tTotal Loss: 1.8431\tAvg Loss: 0.0000\nTrain Epoch: 83\t[75000/88956 (84%)]\tTotal Loss: 1.9824\tAvg Loss: 0.0000\nTrain Epoch: 83\t[80000/88956 (90%)]\tTotal Loss: 2.1180\tAvg Loss: 0.0000\nTrain Epoch: 83\t[85000/88956 (96%)]\tTotal Loss: 2.2638\tAvg Loss: 0.0000\n====> Epoch: 83\tTotal Loss: 2.3667\t Avg Loss: 0.0000\tCorrect: 72833/88956\tPercentage Correct: 81.88\n====> Val Loss: 0.4665\t Avg Loss: 0.0000\tCorrect: 7051/9885\tPercentage Correct: 71.33\n====> Test Loss: 1.1316\t Avg Loss: 0.0000\tCorrect: 17765/24711\tPercentage Correct: 71.89\nTrain Epoch: 84\t[5000/88956 (6%)]\tTotal Loss: 0.1298\tAvg Loss: 0.0000\nTrain Epoch: 84\t[10000/88956 (11%)]\tTotal Loss: 0.2519\tAvg Loss: 0.0000\nTrain Epoch: 84\t[15000/88956 (17%)]\tTotal Loss: 0.3678\tAvg Loss: 0.0000\nTrain Epoch: 84\t[20000/88956 (22%)]\tTotal Loss: 0.4948\tAvg Loss: 0.0000\nTrain Epoch: 84\t[25000/88956 (28%)]\tTotal Loss: 0.6189\tAvg Loss: 0.0000\nTrain Epoch: 84\t[30000/88956 (34%)]\tTotal Loss: 0.7470\tAvg Loss: 0.0000\nTrain Epoch: 84\t[35000/88956 (39%)]\tTotal Loss: 0.8888\tAvg Loss: 0.0000\nTrain Epoch: 84\t[40000/88956 (45%)]\tTotal Loss: 1.0303\tAvg Loss: 0.0000\nTrain Epoch: 84\t[45000/88956 (51%)]\tTotal Loss: 1.1616\tAvg Loss: 0.0000\nTrain Epoch: 84\t[50000/88956 (56%)]\tTotal Loss: 1.2981\tAvg Loss: 0.0000\nTrain Epoch: 84\t[55000/88956 (62%)]\tTotal Loss: 1.4244\tAvg Loss: 0.0000\nTrain Epoch: 84\t[60000/88956 (67%)]\tTotal Loss: 1.5533\tAvg Loss: 0.0000\nTrain Epoch: 84\t[65000/88956 (73%)]\tTotal Loss: 1.6857\tAvg Loss: 0.0000\nTrain Epoch: 84\t[70000/88956 (79%)]\tTotal Loss: 1.8078\tAvg Loss: 0.0000\nTrain Epoch: 84\t[75000/88956 (84%)]\tTotal Loss: 1.9445\tAvg Loss: 0.0000\nTrain Epoch: 84\t[80000/88956 (90%)]\tTotal Loss: 2.0779\tAvg Loss: 0.0000\nTrain Epoch: 84\t[85000/88956 (96%)]\tTotal Loss: 2.2049\tAvg Loss: 0.0000\n====> Epoch: 84\tTotal Loss: 2.3089\t Avg Loss: 0.0000\tCorrect: 73114/88956\tPercentage Correct: 82.19\n====> Val Loss: 0.4646\t Avg Loss: 0.0000\tCorrect: 7084/9885\tPercentage Correct: 71.66\n====> Test Loss: 1.1139\t Avg Loss: 0.0000\tCorrect: 17885/24711\tPercentage Correct: 72.38\nTrain Epoch: 85\t[5000/88956 (6%)]\tTotal Loss: 0.1248\tAvg Loss: 0.0000\nTrain Epoch: 85\t[10000/88956 (11%)]\tTotal Loss: 0.2445\tAvg Loss: 0.0000\nTrain Epoch: 85\t[15000/88956 (17%)]\tTotal Loss: 0.3661\tAvg Loss: 0.0000\nTrain Epoch: 85\t[20000/88956 (22%)]\tTotal Loss: 0.4879\tAvg Loss: 0.0000\nTrain Epoch: 85\t[25000/88956 (28%)]\tTotal Loss: 0.6122\tAvg Loss: 0.0000\nTrain Epoch: 85\t[30000/88956 (34%)]\tTotal Loss: 0.7398\tAvg Loss: 0.0000\nTrain Epoch: 85\t[35000/88956 (39%)]\tTotal Loss: 0.8732\tAvg Loss: 0.0000\nTrain Epoch: 85\t[40000/88956 (45%)]\tTotal Loss: 1.0016\tAvg Loss: 0.0000\nTrain Epoch: 85\t[45000/88956 (51%)]\tTotal Loss: 1.1356\tAvg Loss: 0.0000\nTrain Epoch: 85\t[50000/88956 (56%)]\tTotal Loss: 1.2586\tAvg Loss: 0.0000\nTrain Epoch: 85\t[55000/88956 (62%)]\tTotal Loss: 1.3939\tAvg Loss: 0.0000\nTrain Epoch: 85\t[60000/88956 (67%)]\tTotal Loss: 1.5236\tAvg Loss: 0.0000\nTrain Epoch: 85\t[65000/88956 (73%)]\tTotal Loss: 1.6642\tAvg Loss: 0.0000\nTrain Epoch: 85\t[70000/88956 (79%)]\tTotal Loss: 1.7939\tAvg Loss: 0.0000\nTrain Epoch: 85\t[75000/88956 (84%)]\tTotal Loss: 1.9217\tAvg Loss: 0.0000\nTrain Epoch: 85\t[80000/88956 (90%)]\tTotal Loss: 2.0545\tAvg Loss: 0.0000\nTrain Epoch: 85\t[85000/88956 (96%)]\tTotal Loss: 2.2056\tAvg Loss: 0.0000\n====> Epoch: 85\tTotal Loss: 2.3261\t Avg Loss: 0.0000\tCorrect: 73216/88956\tPercentage Correct: 82.31\n====> Val Loss: 0.5435\t Avg Loss: 0.0001\tCorrect: 6703/9885\tPercentage Correct: 67.81\n====> Test Loss: 1.3366\t Avg Loss: 0.0001\tCorrect: 16833/24711\tPercentage Correct: 68.12\nTrain Epoch: 86\t[5000/88956 (6%)]\tTotal Loss: 0.1705\tAvg Loss: 0.0000\nTrain Epoch: 86\t[10000/88956 (11%)]\tTotal Loss: 0.3549\tAvg Loss: 0.0000\nTrain Epoch: 86\t[15000/88956 (17%)]\tTotal Loss: 0.4954\tAvg Loss: 0.0000\nTrain Epoch: 86\t[20000/88956 (22%)]\tTotal Loss: 0.6266\tAvg Loss: 0.0000\nTrain Epoch: 86\t[25000/88956 (28%)]\tTotal Loss: 0.7535\tAvg Loss: 0.0000\nTrain Epoch: 86\t[30000/88956 (34%)]\tTotal Loss: 0.8915\tAvg Loss: 0.0000\nTrain Epoch: 86\t[35000/88956 (39%)]\tTotal Loss: 1.0188\tAvg Loss: 0.0000\nTrain Epoch: 86\t[40000/88956 (45%)]\tTotal Loss: 1.1415\tAvg Loss: 0.0000\nTrain Epoch: 86\t[45000/88956 (51%)]\tTotal Loss: 1.2681\tAvg Loss: 0.0000\nTrain Epoch: 86\t[50000/88956 (56%)]\tTotal Loss: 1.3896\tAvg Loss: 0.0000\nTrain Epoch: 86\t[55000/88956 (62%)]\tTotal Loss: 1.5131\tAvg Loss: 0.0000\nTrain Epoch: 86\t[60000/88956 (67%)]\tTotal Loss: 1.6401\tAvg Loss: 0.0000\nTrain Epoch: 86\t[65000/88956 (73%)]\tTotal Loss: 1.7713\tAvg Loss: 0.0000\nTrain Epoch: 86\t[70000/88956 (79%)]\tTotal Loss: 1.8957\tAvg Loss: 0.0000\nTrain Epoch: 86\t[75000/88956 (84%)]\tTotal Loss: 2.0247\tAvg Loss: 0.0000\nTrain Epoch: 86\t[80000/88956 (90%)]\tTotal Loss: 2.1610\tAvg Loss: 0.0000\nTrain Epoch: 86\t[85000/88956 (96%)]\tTotal Loss: 2.2843\tAvg Loss: 0.0000\n====> Epoch: 86\tTotal Loss: 2.3906\t Avg Loss: 0.0000\tCorrect: 72534/88956\tPercentage Correct: 81.54\n====> Val Loss: 0.4558\t Avg Loss: 0.0000\tCorrect: 7048/9885\tPercentage Correct: 71.30\n====> Test Loss: 1.0953\t Avg Loss: 0.0000\tCorrect: 17807/24711\tPercentage Correct: 72.06\nTrain Epoch: 87\t[5000/88956 (6%)]\tTotal Loss: 0.1366\tAvg Loss: 0.0000\nTrain Epoch: 87\t[10000/88956 (11%)]\tTotal Loss: 0.2497\tAvg Loss: 0.0000\nTrain Epoch: 87\t[15000/88956 (17%)]\tTotal Loss: 0.3655\tAvg Loss: 0.0000\nTrain Epoch: 87\t[20000/88956 (22%)]\tTotal Loss: 0.4791\tAvg Loss: 0.0000\nTrain Epoch: 87\t[25000/88956 (28%)]\tTotal Loss: 0.6029\tAvg Loss: 0.0000\nTrain Epoch: 87\t[30000/88956 (34%)]\tTotal Loss: 0.7220\tAvg Loss: 0.0000\nTrain Epoch: 87\t[35000/88956 (39%)]\tTotal Loss: 0.8366\tAvg Loss: 0.0000\nTrain Epoch: 87\t[40000/88956 (45%)]\tTotal Loss: 0.9523\tAvg Loss: 0.0000\nTrain Epoch: 87\t[45000/88956 (51%)]\tTotal Loss: 1.0706\tAvg Loss: 0.0000\nTrain Epoch: 87\t[50000/88956 (56%)]\tTotal Loss: 1.2045\tAvg Loss: 0.0000\nTrain Epoch: 87\t[55000/88956 (62%)]\tTotal Loss: 1.3314\tAvg Loss: 0.0000\nTrain Epoch: 87\t[60000/88956 (67%)]\tTotal Loss: 1.4504\tAvg Loss: 0.0000\nTrain Epoch: 87\t[65000/88956 (73%)]\tTotal Loss: 1.5673\tAvg Loss: 0.0000\nTrain Epoch: 87\t[70000/88956 (79%)]\tTotal Loss: 1.6869\tAvg Loss: 0.0000\nTrain Epoch: 87\t[75000/88956 (84%)]\tTotal Loss: 1.8142\tAvg Loss: 0.0000\nTrain Epoch: 87\t[80000/88956 (90%)]\tTotal Loss: 1.9484\tAvg Loss: 0.0000\nTrain Epoch: 87\t[85000/88956 (96%)]\tTotal Loss: 2.0764\tAvg Loss: 0.0000\n====> Epoch: 87\tTotal Loss: 2.1788\t Avg Loss: 0.0000\tCorrect: 74028/88956\tPercentage Correct: 83.22\n====> Val Loss: 0.4331\t Avg Loss: 0.0000\tCorrect: 7080/9885\tPercentage Correct: 71.62\n====> Test Loss: 1.0872\t Avg Loss: 0.0000\tCorrect: 17753/24711\tPercentage Correct: 71.84\nTrain Epoch: 88\t[5000/88956 (6%)]\tTotal Loss: 0.1170\tAvg Loss: 0.0000\nTrain Epoch: 88\t[10000/88956 (11%)]\tTotal Loss: 0.2377\tAvg Loss: 0.0000\nTrain Epoch: 88\t[15000/88956 (17%)]\tTotal Loss: 0.3623\tAvg Loss: 0.0000\nTrain Epoch: 88\t[20000/88956 (22%)]\tTotal Loss: 0.4873\tAvg Loss: 0.0000\nTrain Epoch: 88\t[25000/88956 (28%)]\tTotal Loss: 0.6091\tAvg Loss: 0.0000\nTrain Epoch: 88\t[30000/88956 (34%)]\tTotal Loss: 0.7322\tAvg Loss: 0.0000\nTrain Epoch: 88\t[35000/88956 (39%)]\tTotal Loss: 0.8518\tAvg Loss: 0.0000\nTrain Epoch: 88\t[40000/88956 (45%)]\tTotal Loss: 0.9682\tAvg Loss: 0.0000\nTrain Epoch: 88\t[45000/88956 (51%)]\tTotal Loss: 1.1029\tAvg Loss: 0.0000\nTrain Epoch: 88\t[50000/88956 (56%)]\tTotal Loss: 1.2412\tAvg Loss: 0.0000\nTrain Epoch: 88\t[55000/88956 (62%)]\tTotal Loss: 1.3570\tAvg Loss: 0.0000\nTrain Epoch: 88\t[60000/88956 (67%)]\tTotal Loss: 1.4684\tAvg Loss: 0.0000\nTrain Epoch: 88\t[65000/88956 (73%)]\tTotal Loss: 1.5979\tAvg Loss: 0.0000\nTrain Epoch: 88\t[70000/88956 (79%)]\tTotal Loss: 1.7211\tAvg Loss: 0.0000\nTrain Epoch: 88\t[75000/88956 (84%)]\tTotal Loss: 1.8404\tAvg Loss: 0.0000\nTrain Epoch: 88\t[80000/88956 (90%)]\tTotal Loss: 1.9699\tAvg Loss: 0.0000\nTrain Epoch: 88\t[85000/88956 (96%)]\tTotal Loss: 2.0928\tAvg Loss: 0.0000\n====> Epoch: 88\tTotal Loss: 2.1949\t Avg Loss: 0.0000\tCorrect: 73751/88956\tPercentage Correct: 82.91\n====> Val Loss: 0.4483\t Avg Loss: 0.0000\tCorrect: 7128/9885\tPercentage Correct: 72.11\n====> Test Loss: 1.0827\t Avg Loss: 0.0000\tCorrect: 17856/24711\tPercentage Correct: 72.26\nTrain Epoch: 89\t[5000/88956 (6%)]\tTotal Loss: 0.1094\tAvg Loss: 0.0000\nTrain Epoch: 89\t[10000/88956 (11%)]\tTotal Loss: 0.2199\tAvg Loss: 0.0000\nTrain Epoch: 89\t[15000/88956 (17%)]\tTotal Loss: 0.3329\tAvg Loss: 0.0000\nTrain Epoch: 89\t[20000/88956 (22%)]\tTotal Loss: 0.4542\tAvg Loss: 0.0000\nTrain Epoch: 89\t[25000/88956 (28%)]\tTotal Loss: 0.5681\tAvg Loss: 0.0000\nTrain Epoch: 89\t[30000/88956 (34%)]\tTotal Loss: 0.6899\tAvg Loss: 0.0000\nTrain Epoch: 89\t[35000/88956 (39%)]\tTotal Loss: 0.8145\tAvg Loss: 0.0000\nTrain Epoch: 89\t[40000/88956 (45%)]\tTotal Loss: 0.9387\tAvg Loss: 0.0000\nTrain Epoch: 89\t[45000/88956 (51%)]\tTotal Loss: 1.0820\tAvg Loss: 0.0000\nTrain Epoch: 89\t[50000/88956 (56%)]\tTotal Loss: 1.2041\tAvg Loss: 0.0000\nTrain Epoch: 89\t[55000/88956 (62%)]\tTotal Loss: 1.3254\tAvg Loss: 0.0000\nTrain Epoch: 89\t[60000/88956 (67%)]\tTotal Loss: 1.4525\tAvg Loss: 0.0000\nTrain Epoch: 89\t[65000/88956 (73%)]\tTotal Loss: 1.6065\tAvg Loss: 0.0000\nTrain Epoch: 89\t[70000/88956 (79%)]\tTotal Loss: 1.7313\tAvg Loss: 0.0000\nTrain Epoch: 89\t[75000/88956 (84%)]\tTotal Loss: 1.8684\tAvg Loss: 0.0000\nTrain Epoch: 89\t[80000/88956 (90%)]\tTotal Loss: 1.9998\tAvg Loss: 0.0000\nTrain Epoch: 89\t[85000/88956 (96%)]\tTotal Loss: 2.1214\tAvg Loss: 0.0000\n====> Epoch: 89\tTotal Loss: 2.2094\t Avg Loss: 0.0000\tCorrect: 73819/88956\tPercentage Correct: 82.98\n====> Val Loss: 0.4076\t Avg Loss: 0.0000\tCorrect: 7260/9885\tPercentage Correct: 73.44\n====> Test Loss: 1.0189\t Avg Loss: 0.0000\tCorrect: 18203/24711\tPercentage Correct: 73.66\n\n=== saved best model ===\n\nTrain Epoch: 90\t[5000/88956 (6%)]\tTotal Loss: 0.1069\tAvg Loss: 0.0000\nTrain Epoch: 90\t[10000/88956 (11%)]\tTotal Loss: 0.2153\tAvg Loss: 0.0000\nTrain Epoch: 90\t[15000/88956 (17%)]\tTotal Loss: 0.3289\tAvg Loss: 0.0000\nTrain Epoch: 90\t[20000/88956 (22%)]\tTotal Loss: 0.4538\tAvg Loss: 0.0000\nTrain Epoch: 90\t[25000/88956 (28%)]\tTotal Loss: 0.5776\tAvg Loss: 0.0000\nTrain Epoch: 90\t[30000/88956 (34%)]\tTotal Loss: 0.7103\tAvg Loss: 0.0000\nTrain Epoch: 90\t[35000/88956 (39%)]\tTotal Loss: 0.8307\tAvg Loss: 0.0000\nTrain Epoch: 90\t[40000/88956 (45%)]\tTotal Loss: 0.9444\tAvg Loss: 0.0000\nTrain Epoch: 90\t[45000/88956 (51%)]\tTotal Loss: 1.0782\tAvg Loss: 0.0000\nTrain Epoch: 90\t[50000/88956 (56%)]\tTotal Loss: 1.1935\tAvg Loss: 0.0000\nTrain Epoch: 90\t[55000/88956 (62%)]\tTotal Loss: 1.3224\tAvg Loss: 0.0000\nTrain Epoch: 90\t[60000/88956 (67%)]\tTotal Loss: 1.4508\tAvg Loss: 0.0000\nTrain Epoch: 90\t[65000/88956 (73%)]\tTotal Loss: 1.5858\tAvg Loss: 0.0000\nTrain Epoch: 90\t[70000/88956 (79%)]\tTotal Loss: 1.7189\tAvg Loss: 0.0000\nTrain Epoch: 90\t[75000/88956 (84%)]\tTotal Loss: 1.8429\tAvg Loss: 0.0000\nTrain Epoch: 90\t[80000/88956 (90%)]\tTotal Loss: 1.9714\tAvg Loss: 0.0000\nTrain Epoch: 90\t[85000/88956 (96%)]\tTotal Loss: 2.0990\tAvg Loss: 0.0000\n====> Epoch: 90\tTotal Loss: 2.2022\t Avg Loss: 0.0000\tCorrect: 73783/88956\tPercentage Correct: 82.94\n====> Val Loss: 0.4338\t Avg Loss: 0.0000\tCorrect: 7166/9885\tPercentage Correct: 72.49\n====> Test Loss: 1.0610\t Avg Loss: 0.0000\tCorrect: 18024/24711\tPercentage Correct: 72.94\nTrain Epoch: 91\t[5000/88956 (6%)]\tTotal Loss: 0.1172\tAvg Loss: 0.0000\nTrain Epoch: 91\t[10000/88956 (11%)]\tTotal Loss: 0.2156\tAvg Loss: 0.0000\nTrain Epoch: 91\t[15000/88956 (17%)]\tTotal Loss: 0.3326\tAvg Loss: 0.0000\nTrain Epoch: 91\t[20000/88956 (22%)]\tTotal Loss: 0.4595\tAvg Loss: 0.0000\nTrain Epoch: 91\t[25000/88956 (28%)]\tTotal Loss: 0.5738\tAvg Loss: 0.0000\nTrain Epoch: 91\t[30000/88956 (34%)]\tTotal Loss: 0.7053\tAvg Loss: 0.0000\nTrain Epoch: 91\t[35000/88956 (39%)]\tTotal Loss: 0.8197\tAvg Loss: 0.0000\nTrain Epoch: 91\t[40000/88956 (45%)]\tTotal Loss: 0.9346\tAvg Loss: 0.0000\nTrain Epoch: 91\t[45000/88956 (51%)]\tTotal Loss: 1.0522\tAvg Loss: 0.0000\nTrain Epoch: 91\t[50000/88956 (56%)]\tTotal Loss: 1.1897\tAvg Loss: 0.0000\nTrain Epoch: 91\t[55000/88956 (62%)]\tTotal Loss: 1.3045\tAvg Loss: 0.0000\nTrain Epoch: 91\t[60000/88956 (67%)]\tTotal Loss: 1.4294\tAvg Loss: 0.0000\nTrain Epoch: 91\t[65000/88956 (73%)]\tTotal Loss: 1.5657\tAvg Loss: 0.0000\nTrain Epoch: 91\t[70000/88956 (79%)]\tTotal Loss: 1.6901\tAvg Loss: 0.0000\nTrain Epoch: 91\t[75000/88956 (84%)]\tTotal Loss: 1.8151\tAvg Loss: 0.0000\nTrain Epoch: 91\t[80000/88956 (90%)]\tTotal Loss: 1.9567\tAvg Loss: 0.0000\nTrain Epoch: 91\t[85000/88956 (96%)]\tTotal Loss: 2.0858\tAvg Loss: 0.0000\n====> Epoch: 91\tTotal Loss: 2.1864\t Avg Loss: 0.0000\tCorrect: 73919/88956\tPercentage Correct: 83.10\n====> Val Loss: 0.4247\t Avg Loss: 0.0000\tCorrect: 7145/9885\tPercentage Correct: 72.28\n====> Test Loss: 1.0853\t Avg Loss: 0.0000\tCorrect: 17766/24711\tPercentage Correct: 71.90\nTrain Epoch: 92\t[5000/88956 (6%)]\tTotal Loss: 0.1078\tAvg Loss: 0.0000\nTrain Epoch: 92\t[10000/88956 (11%)]\tTotal Loss: 0.2207\tAvg Loss: 0.0000\nTrain Epoch: 92\t[15000/88956 (17%)]\tTotal Loss: 0.3398\tAvg Loss: 0.0000\nTrain Epoch: 92\t[20000/88956 (22%)]\tTotal Loss: 0.4589\tAvg Loss: 0.0000\nTrain Epoch: 92\t[25000/88956 (28%)]\tTotal Loss: 0.5687\tAvg Loss: 0.0000\nTrain Epoch: 92\t[30000/88956 (34%)]\tTotal Loss: 0.6843\tAvg Loss: 0.0000\nTrain Epoch: 92\t[35000/88956 (39%)]\tTotal Loss: 0.8074\tAvg Loss: 0.0000\nTrain Epoch: 92\t[40000/88956 (45%)]\tTotal Loss: 0.9327\tAvg Loss: 0.0000\nTrain Epoch: 92\t[45000/88956 (51%)]\tTotal Loss: 1.0545\tAvg Loss: 0.0000\nTrain Epoch: 92\t[50000/88956 (56%)]\tTotal Loss: 1.1696\tAvg Loss: 0.0000\nTrain Epoch: 92\t[55000/88956 (62%)]\tTotal Loss: 1.2994\tAvg Loss: 0.0000\nTrain Epoch: 92\t[60000/88956 (67%)]\tTotal Loss: 1.4204\tAvg Loss: 0.0000\nTrain Epoch: 92\t[65000/88956 (73%)]\tTotal Loss: 1.5326\tAvg Loss: 0.0000\nTrain Epoch: 92\t[70000/88956 (79%)]\tTotal Loss: 1.6661\tAvg Loss: 0.0000\nTrain Epoch: 92\t[75000/88956 (84%)]\tTotal Loss: 1.7987\tAvg Loss: 0.0000\nTrain Epoch: 92\t[80000/88956 (90%)]\tTotal Loss: 1.9242\tAvg Loss: 0.0000\nTrain Epoch: 92\t[85000/88956 (96%)]\tTotal Loss: 2.0682\tAvg Loss: 0.0000\n====> Epoch: 92\tTotal Loss: 2.1708\t Avg Loss: 0.0000\tCorrect: 74172/88956\tPercentage Correct: 83.38\n====> Val Loss: 0.4604\t Avg Loss: 0.0000\tCorrect: 6988/9885\tPercentage Correct: 70.69\n====> Test Loss: 1.1180\t Avg Loss: 0.0000\tCorrect: 17552/24711\tPercentage Correct: 71.03\nTrain Epoch: 93\t[5000/88956 (6%)]\tTotal Loss: 0.1112\tAvg Loss: 0.0000\nTrain Epoch: 93\t[10000/88956 (11%)]\tTotal Loss: 0.2165\tAvg Loss: 0.0000\nTrain Epoch: 93\t[15000/88956 (17%)]\tTotal Loss: 0.3381\tAvg Loss: 0.0000\nTrain Epoch: 93\t[20000/88956 (22%)]\tTotal Loss: 0.4520\tAvg Loss: 0.0000\nTrain Epoch: 93\t[25000/88956 (28%)]\tTotal Loss: 0.5745\tAvg Loss: 0.0000\nTrain Epoch: 93\t[30000/88956 (34%)]\tTotal Loss: 0.6911\tAvg Loss: 0.0000\nTrain Epoch: 93\t[35000/88956 (39%)]\tTotal Loss: 0.8050\tAvg Loss: 0.0000\nTrain Epoch: 93\t[40000/88956 (45%)]\tTotal Loss: 0.9167\tAvg Loss: 0.0000\nTrain Epoch: 93\t[45000/88956 (51%)]\tTotal Loss: 1.0434\tAvg Loss: 0.0000\nTrain Epoch: 93\t[50000/88956 (56%)]\tTotal Loss: 1.1757\tAvg Loss: 0.0000\nTrain Epoch: 93\t[55000/88956 (62%)]\tTotal Loss: 1.2989\tAvg Loss: 0.0000\nTrain Epoch: 93\t[60000/88956 (67%)]\tTotal Loss: 1.4166\tAvg Loss: 0.0000\nTrain Epoch: 93\t[65000/88956 (73%)]\tTotal Loss: 1.5440\tAvg Loss: 0.0000\nTrain Epoch: 93\t[70000/88956 (79%)]\tTotal Loss: 1.6795\tAvg Loss: 0.0000\nTrain Epoch: 93\t[75000/88956 (84%)]\tTotal Loss: 1.8003\tAvg Loss: 0.0000\nTrain Epoch: 93\t[80000/88956 (90%)]\tTotal Loss: 1.9228\tAvg Loss: 0.0000\nTrain Epoch: 93\t[85000/88956 (96%)]\tTotal Loss: 2.0556\tAvg Loss: 0.0000\n====> Epoch: 93\tTotal Loss: 2.1525\t Avg Loss: 0.0000\tCorrect: 74006/88956\tPercentage Correct: 83.19\n====> Val Loss: 0.4049\t Avg Loss: 0.0000\tCorrect: 7190/9885\tPercentage Correct: 72.74\n====> Test Loss: 0.9922\t Avg Loss: 0.0000\tCorrect: 18131/24711\tPercentage Correct: 73.37\nTrain Epoch: 94\t[5000/88956 (6%)]\tTotal Loss: 0.0997\tAvg Loss: 0.0000\nTrain Epoch: 94\t[10000/88956 (11%)]\tTotal Loss: 0.2111\tAvg Loss: 0.0000\nTrain Epoch: 94\t[15000/88956 (17%)]\tTotal Loss: 0.3178\tAvg Loss: 0.0000\nTrain Epoch: 94\t[20000/88956 (22%)]\tTotal Loss: 0.4265\tAvg Loss: 0.0000\nTrain Epoch: 94\t[25000/88956 (28%)]\tTotal Loss: 0.5384\tAvg Loss: 0.0000\nTrain Epoch: 94\t[30000/88956 (34%)]\tTotal Loss: 0.6824\tAvg Loss: 0.0000\nTrain Epoch: 94\t[35000/88956 (39%)]\tTotal Loss: 0.8035\tAvg Loss: 0.0000\nTrain Epoch: 94\t[40000/88956 (45%)]\tTotal Loss: 0.9065\tAvg Loss: 0.0000\nTrain Epoch: 94\t[45000/88956 (51%)]\tTotal Loss: 1.0160\tAvg Loss: 0.0000\nTrain Epoch: 94\t[50000/88956 (56%)]\tTotal Loss: 1.1295\tAvg Loss: 0.0000\nTrain Epoch: 94\t[55000/88956 (62%)]\tTotal Loss: 1.2380\tAvg Loss: 0.0000\nTrain Epoch: 94\t[60000/88956 (67%)]\tTotal Loss: 1.3534\tAvg Loss: 0.0000\nTrain Epoch: 94\t[65000/88956 (73%)]\tTotal Loss: 1.4775\tAvg Loss: 0.0000\nTrain Epoch: 94\t[70000/88956 (79%)]\tTotal Loss: 1.6008\tAvg Loss: 0.0000\nTrain Epoch: 94\t[75000/88956 (84%)]\tTotal Loss: 1.7225\tAvg Loss: 0.0000\nTrain Epoch: 94\t[80000/88956 (90%)]\tTotal Loss: 1.8481\tAvg Loss: 0.0000\nTrain Epoch: 94\t[85000/88956 (96%)]\tTotal Loss: 1.9699\tAvg Loss: 0.0000\n====> Epoch: 94\tTotal Loss: 2.0579\t Avg Loss: 0.0000\tCorrect: 74715/88956\tPercentage Correct: 83.99\n====> Val Loss: 0.4250\t Avg Loss: 0.0000\tCorrect: 7174/9885\tPercentage Correct: 72.57\n====> Test Loss: 1.0493\t Avg Loss: 0.0000\tCorrect: 18069/24711\tPercentage Correct: 73.12\nTrain Epoch: 95\t[5000/88956 (6%)]\tTotal Loss: 0.1013\tAvg Loss: 0.0000\nTrain Epoch: 95\t[10000/88956 (11%)]\tTotal Loss: 0.2188\tAvg Loss: 0.0000\nTrain Epoch: 95\t[15000/88956 (17%)]\tTotal Loss: 0.3273\tAvg Loss: 0.0000\nTrain Epoch: 95\t[20000/88956 (22%)]\tTotal Loss: 0.4464\tAvg Loss: 0.0000\nTrain Epoch: 95\t[25000/88956 (28%)]\tTotal Loss: 0.5619\tAvg Loss: 0.0000\nTrain Epoch: 95\t[30000/88956 (34%)]\tTotal Loss: 0.6801\tAvg Loss: 0.0000\nTrain Epoch: 95\t[35000/88956 (39%)]\tTotal Loss: 0.7868\tAvg Loss: 0.0000\nTrain Epoch: 95\t[40000/88956 (45%)]\tTotal Loss: 0.9061\tAvg Loss: 0.0000\nTrain Epoch: 95\t[45000/88956 (51%)]\tTotal Loss: 1.0174\tAvg Loss: 0.0000\nTrain Epoch: 95\t[50000/88956 (56%)]\tTotal Loss: 1.1248\tAvg Loss: 0.0000\nTrain Epoch: 95\t[55000/88956 (62%)]\tTotal Loss: 1.2362\tAvg Loss: 0.0000\nTrain Epoch: 95\t[60000/88956 (67%)]\tTotal Loss: 1.3470\tAvg Loss: 0.0000\nTrain Epoch: 95\t[65000/88956 (73%)]\tTotal Loss: 1.4793\tAvg Loss: 0.0000\nTrain Epoch: 95\t[70000/88956 (79%)]\tTotal Loss: 1.5944\tAvg Loss: 0.0000\nTrain Epoch: 95\t[75000/88956 (84%)]\tTotal Loss: 1.7137\tAvg Loss: 0.0000\nTrain Epoch: 95\t[80000/88956 (90%)]\tTotal Loss: 1.8704\tAvg Loss: 0.0000\nTrain Epoch: 95\t[85000/88956 (96%)]\tTotal Loss: 1.9973\tAvg Loss: 0.0000\n====> Epoch: 95\tTotal Loss: 2.0850\t Avg Loss: 0.0000\tCorrect: 74593/88956\tPercentage Correct: 83.85\n====> Val Loss: 0.4209\t Avg Loss: 0.0000\tCorrect: 7229/9885\tPercentage Correct: 73.13\n====> Test Loss: 1.0305\t Avg Loss: 0.0000\tCorrect: 18159/24711\tPercentage Correct: 73.49\nTrain Epoch: 96\t[5000/88956 (6%)]\tTotal Loss: 0.1080\tAvg Loss: 0.0000\nTrain Epoch: 96\t[10000/88956 (11%)]\tTotal Loss: 0.2162\tAvg Loss: 0.0000\nTrain Epoch: 96\t[15000/88956 (17%)]\tTotal Loss: 0.3185\tAvg Loss: 0.0000\nTrain Epoch: 96\t[20000/88956 (22%)]\tTotal Loss: 0.4186\tAvg Loss: 0.0000\nTrain Epoch: 96\t[25000/88956 (28%)]\tTotal Loss: 0.5281\tAvg Loss: 0.0000\nTrain Epoch: 96\t[30000/88956 (34%)]\tTotal Loss: 0.6405\tAvg Loss: 0.0000\nTrain Epoch: 96\t[35000/88956 (39%)]\tTotal Loss: 0.7536\tAvg Loss: 0.0000\nTrain Epoch: 96\t[40000/88956 (45%)]\tTotal Loss: 0.8661\tAvg Loss: 0.0000\nTrain Epoch: 96\t[45000/88956 (51%)]\tTotal Loss: 0.9849\tAvg Loss: 0.0000\nTrain Epoch: 96\t[50000/88956 (56%)]\tTotal Loss: 1.1136\tAvg Loss: 0.0000\nTrain Epoch: 96\t[55000/88956 (62%)]\tTotal Loss: 1.2249\tAvg Loss: 0.0000\nTrain Epoch: 96\t[60000/88956 (67%)]\tTotal Loss: 1.3314\tAvg Loss: 0.0000\nTrain Epoch: 96\t[65000/88956 (73%)]\tTotal Loss: 1.4448\tAvg Loss: 0.0000\nTrain Epoch: 96\t[70000/88956 (79%)]\tTotal Loss: 1.5674\tAvg Loss: 0.0000\nTrain Epoch: 96\t[75000/88956 (84%)]\tTotal Loss: 1.6836\tAvg Loss: 0.0000\nTrain Epoch: 96\t[80000/88956 (90%)]\tTotal Loss: 1.8001\tAvg Loss: 0.0000\nTrain Epoch: 96\t[85000/88956 (96%)]\tTotal Loss: 1.9152\tAvg Loss: 0.0000\n====> Epoch: 96\tTotal Loss: 2.0132\t Avg Loss: 0.0000\tCorrect: 74966/88956\tPercentage Correct: 84.27\n====> Val Loss: 0.4193\t Avg Loss: 0.0000\tCorrect: 7244/9885\tPercentage Correct: 73.28\n====> Test Loss: 1.0105\t Avg Loss: 0.0000\tCorrect: 18207/24711\tPercentage Correct: 73.68\nTrain Epoch: 97\t[5000/88956 (6%)]\tTotal Loss: 0.1031\tAvg Loss: 0.0000\nTrain Epoch: 97\t[10000/88956 (11%)]\tTotal Loss: 0.2214\tAvg Loss: 0.0000\nTrain Epoch: 97\t[15000/88956 (17%)]\tTotal Loss: 0.3156\tAvg Loss: 0.0000\nTrain Epoch: 97\t[20000/88956 (22%)]\tTotal Loss: 0.4252\tAvg Loss: 0.0000\nTrain Epoch: 97\t[25000/88956 (28%)]\tTotal Loss: 0.5335\tAvg Loss: 0.0000\nTrain Epoch: 97\t[30000/88956 (34%)]\tTotal Loss: 0.6569\tAvg Loss: 0.0000\nTrain Epoch: 97\t[35000/88956 (39%)]\tTotal Loss: 0.7648\tAvg Loss: 0.0000\nTrain Epoch: 97\t[40000/88956 (45%)]\tTotal Loss: 0.8841\tAvg Loss: 0.0000\nTrain Epoch: 97\t[45000/88956 (51%)]\tTotal Loss: 0.9976\tAvg Loss: 0.0000\nTrain Epoch: 97\t[50000/88956 (56%)]\tTotal Loss: 1.1076\tAvg Loss: 0.0000\nTrain Epoch: 97\t[55000/88956 (62%)]\tTotal Loss: 1.2394\tAvg Loss: 0.0000\nTrain Epoch: 97\t[60000/88956 (67%)]\tTotal Loss: 1.3766\tAvg Loss: 0.0000\nTrain Epoch: 97\t[65000/88956 (73%)]\tTotal Loss: 1.5080\tAvg Loss: 0.0000\nTrain Epoch: 97\t[70000/88956 (79%)]\tTotal Loss: 1.6160\tAvg Loss: 0.0000\nTrain Epoch: 97\t[75000/88956 (84%)]\tTotal Loss: 1.7325\tAvg Loss: 0.0000\nTrain Epoch: 97\t[80000/88956 (90%)]\tTotal Loss: 1.8582\tAvg Loss: 0.0000\nTrain Epoch: 97\t[85000/88956 (96%)]\tTotal Loss: 1.9753\tAvg Loss: 0.0000\n====> Epoch: 97\tTotal Loss: 2.0696\t Avg Loss: 0.0000\tCorrect: 74579/88956\tPercentage Correct: 83.84\n====> Val Loss: 0.4400\t Avg Loss: 0.0000\tCorrect: 7157/9885\tPercentage Correct: 72.40\n====> Test Loss: 1.0631\t Avg Loss: 0.0000\tCorrect: 18144/24711\tPercentage Correct: 73.42\nTrain Epoch: 98\t[5000/88956 (6%)]\tTotal Loss: 0.1077\tAvg Loss: 0.0000\nTrain Epoch: 98\t[10000/88956 (11%)]\tTotal Loss: 0.2236\tAvg Loss: 0.0000\nTrain Epoch: 98\t[15000/88956 (17%)]\tTotal Loss: 0.3248\tAvg Loss: 0.0000\nTrain Epoch: 98\t[20000/88956 (22%)]\tTotal Loss: 0.4296\tAvg Loss: 0.0000\nTrain Epoch: 98\t[25000/88956 (28%)]\tTotal Loss: 0.5244\tAvg Loss: 0.0000\nTrain Epoch: 98\t[30000/88956 (34%)]\tTotal Loss: 0.6319\tAvg Loss: 0.0000\nTrain Epoch: 98\t[35000/88956 (39%)]\tTotal Loss: 0.7539\tAvg Loss: 0.0000\nTrain Epoch: 98\t[40000/88956 (45%)]\tTotal Loss: 0.8657\tAvg Loss: 0.0000\nTrain Epoch: 98\t[45000/88956 (51%)]\tTotal Loss: 0.9776\tAvg Loss: 0.0000\nTrain Epoch: 98\t[50000/88956 (56%)]\tTotal Loss: 1.0843\tAvg Loss: 0.0000\nTrain Epoch: 98\t[55000/88956 (62%)]\tTotal Loss: 1.1985\tAvg Loss: 0.0000\nTrain Epoch: 98\t[60000/88956 (67%)]\tTotal Loss: 1.3121\tAvg Loss: 0.0000\nTrain Epoch: 98\t[65000/88956 (73%)]\tTotal Loss: 1.4266\tAvg Loss: 0.0000\nTrain Epoch: 98\t[70000/88956 (79%)]\tTotal Loss: 1.5438\tAvg Loss: 0.0000\nTrain Epoch: 98\t[75000/88956 (84%)]\tTotal Loss: 1.6618\tAvg Loss: 0.0000\nTrain Epoch: 98\t[80000/88956 (90%)]\tTotal Loss: 1.7689\tAvg Loss: 0.0000\nTrain Epoch: 98\t[85000/88956 (96%)]\tTotal Loss: 1.8807\tAvg Loss: 0.0000\n====> Epoch: 98\tTotal Loss: 1.9690\t Avg Loss: 0.0000\tCorrect: 75057/88956\tPercentage Correct: 84.38\n====> Val Loss: 0.4483\t Avg Loss: 0.0000\tCorrect: 6996/9885\tPercentage Correct: 70.77\n====> Test Loss: 1.1005\t Avg Loss: 0.0000\tCorrect: 17673/24711\tPercentage Correct: 71.52\nTrain Epoch: 99\t[5000/88956 (6%)]\tTotal Loss: 0.1067\tAvg Loss: 0.0000\nTrain Epoch: 99\t[10000/88956 (11%)]\tTotal Loss: 0.2160\tAvg Loss: 0.0000\nTrain Epoch: 99\t[15000/88956 (17%)]\tTotal Loss: 0.3199\tAvg Loss: 0.0000\nTrain Epoch: 99\t[20000/88956 (22%)]\tTotal Loss: 0.4298\tAvg Loss: 0.0000\nTrain Epoch: 99\t[25000/88956 (28%)]\tTotal Loss: 0.5524\tAvg Loss: 0.0000\nTrain Epoch: 99\t[30000/88956 (34%)]\tTotal Loss: 0.6710\tAvg Loss: 0.0000\nTrain Epoch: 99\t[35000/88956 (39%)]\tTotal Loss: 0.7758\tAvg Loss: 0.0000\nTrain Epoch: 99\t[40000/88956 (45%)]\tTotal Loss: 0.8952\tAvg Loss: 0.0000\nTrain Epoch: 99\t[45000/88956 (51%)]\tTotal Loss: 1.0147\tAvg Loss: 0.0000\nTrain Epoch: 99\t[50000/88956 (56%)]\tTotal Loss: 1.1347\tAvg Loss: 0.0000\nTrain Epoch: 99\t[55000/88956 (62%)]\tTotal Loss: 1.2514\tAvg Loss: 0.0000\nTrain Epoch: 99\t[60000/88956 (67%)]\tTotal Loss: 1.3701\tAvg Loss: 0.0000\nTrain Epoch: 99\t[65000/88956 (73%)]\tTotal Loss: 1.5041\tAvg Loss: 0.0000\nTrain Epoch: 99\t[70000/88956 (79%)]\tTotal Loss: 1.6226\tAvg Loss: 0.0000\nTrain Epoch: 99\t[75000/88956 (84%)]\tTotal Loss: 1.7349\tAvg Loss: 0.0000\nTrain Epoch: 99\t[80000/88956 (90%)]\tTotal Loss: 1.8937\tAvg Loss: 0.0000\nTrain Epoch: 99\t[85000/88956 (96%)]\tTotal Loss: 2.0273\tAvg Loss: 0.0000\n====> Epoch: 99\tTotal Loss: 2.1260\t Avg Loss: 0.0000\tCorrect: 74504/88956\tPercentage Correct: 83.75\n====> Val Loss: 0.3969\t Avg Loss: 0.0000\tCorrect: 7277/9885\tPercentage Correct: 73.62\n====> Test Loss: 0.9654\t Avg Loss: 0.0000\tCorrect: 18340/24711\tPercentage Correct: 74.22\n\n=== saved best model ===\n\nTraining time: 69013.07139778137s\n"
],
[
"# BEST RESULTS\nprint('Accuracy: ', 100 * max(all_train_correct) / train_set_size)\nprint('Epoch: ', np.argmax(all_train_correct))\nprint()\nprint('Accuracy: ', 100 * max(all_val_correct) / val_set_size)\nprint('Epoch: ', np.argmax(all_val_correct))\nprint()\nprint('Accuracy: ', 100 * max(all_test_correct) / test_set_size)\nprint('Epoch: ', np.argmax(all_test_correct))\nprint()",
"Accuracy: 84.37542155672467\nEpoch: 98\n\nAccuracy: 73.61659079413252\nEpoch: 99\n\nAccuracy: 74.21795961312776\nEpoch: 99\n\n"
],
[
"# SAVE RESULTS - all losses, all correct, best results\nall_train_losses_npy = np.array(all_train_losses)\nall_train_correct_npy = np.array(all_train_correct)\nbest_train_results_npy = np.array(all_train_results[95])\n\nall_val_losses_npy = np.array(all_val_losses)\nall_val_correct_npy = np.array(all_val_correct)\nbest_val_results_npy = np.array(all_val_results[95])\n\nall_test_losses_npy = np.array(all_test_losses)\nall_test_correct_npy = np.array(all_test_correct)\nbest_test_results_npy = np.array(all_test_results[95])\n\nfx_labels_npy = np.array(list(dataset.fx_to_label.keys()))\n\nnp.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'all_train_losses')), arr=all_train_losses_npy)\nnp.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'all_train_correct')), arr=all_train_correct_npy)\nnp.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'best_train_results')), arr=best_train_results_npy)\n\nnp.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'all_val_losses')), arr=all_val_losses_npy)\nnp.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'all_val_correct')), arr=all_val_correct_npy)\nnp.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'best_val_results')), arr=best_val_results_npy)\n\nnp.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'all_test_losses')), arr=all_test_losses_npy)\nnp.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'all_test_correct')), arr=all_test_correct_npy)\nnp.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'best_test_results')), arr=best_test_results_npy)\n\nnp.save(file=('%s/%s/%s' % (results_folder, results_subfolder, 'fx_labels')), arr=fx_labels_npy)",
"<ipython-input-7-490bdd120e03>:4: VisibleDeprecationWarning: Creating an ndarray from ragged nested sequences (which is a list-or-tuple of lists-or-tuples-or ndarrays with different lengths or shapes) is deprecated. If you meant to do this, you must specify 'dtype=object' when creating the ndarray.\n best_train_results_npy = np.array(all_train_results[95])\n<ipython-input-7-490bdd120e03>:8: VisibleDeprecationWarning: Creating an ndarray from ragged nested sequences (which is a list-or-tuple of lists-or-tuples-or ndarrays with different lengths or shapes) is deprecated. If you meant to do this, you must specify 'dtype=object' when creating the ndarray.\n best_val_results_npy = np.array(all_val_results[95])\n<ipython-input-7-490bdd120e03>:12: VisibleDeprecationWarning: Creating an ndarray from ragged nested sequences (which is a list-or-tuple of lists-or-tuples-or ndarrays with different lengths or shapes) is deprecated. If you meant to do this, you must specify 'dtype=object' when creating the ndarray.\n best_test_results_npy = np.array(all_test_results[95])\n"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
]
|
e79612519e07dbfc59cb23addfa2874e2d3d194e | 38,077 | ipynb | Jupyter Notebook | examples/quickstart/multiclass.ipynb | nmasahiro/zr-obp | dde815dfe75fc6cc3c9ee6479d97db1e5567de6d | [
"Apache-2.0"
]
| null | null | null | examples/quickstart/multiclass.ipynb | nmasahiro/zr-obp | dde815dfe75fc6cc3c9ee6479d97db1e5567de6d | [
"Apache-2.0"
]
| null | null | null | examples/quickstart/multiclass.ipynb | nmasahiro/zr-obp | dde815dfe75fc6cc3c9ee6479d97db1e5567de6d | [
"Apache-2.0"
]
| null | null | null | 92.870732 | 25,048 | 0.832891 | [
[
[
"# Quickstart Example with Multi-class Classificatoin Data\n---\nThis notebook provides an example of conducting OPE of an evaluate policy using multi-class classification dataset as logged bandit feedback data.\n\nOur example with multi-class classification data contains the follwoing four major steps:\n- (1) Bandit Reduction\n- (2) Off-Policy Learning\n- (3) Off-Policy Evaluation\n- (4) Evaluation of OPE Estimators\n\nPlease see [../examples/multiclass](../examples/multiclass) for a more sophisticated example of the evaluation of OPE with multi-class classification datasets.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom sklearn.datasets import load_digits\nfrom sklearn.ensemble import RandomForestClassifier as RandomForest\nfrom sklearn.linear_model import LogisticRegression\n# import open bandit pipeline (obp)\nimport obp\nfrom obp.dataset import MultiClassToBanditReduction\nfrom obp.ope import (\n OffPolicyEvaluation, \n RegressionModel,\n InverseProbabilityWeighting,\n DirectMethod,\n DoublyRobust\n)",
"_____no_output_____"
],
[
"# obp version\nprint(obp.__version__)",
"0.3.3\n"
]
],
[
[
"## (1) Bandit Reduction\nWe prepare easy-to-use interface for bandit reduction of multi-class classificatoin dataset: `MultiClassToBanditReduction` class in the dataset module.\n\nIt takes feature vectors (`X`), class labels (`y`), classifier to construct behavior policy (`base_classifier_b`), paramter of behavior policy (`alpha_b`) as inputs and generates a bandit dataset that can be used to evaluate the performance of decision making policies (obtained by `off-policy learning`) and OPE estimators.",
"_____no_output_____"
]
],
[
[
"# load raw digits data\n# `return_X_y` splits feature vectors and labels, instead of returning a Bunch object\nX, y = load_digits(return_X_y=True)\n\n# convert the raw classification data into a logged bandit dataset\n# we construct a behavior policy using Logistic Regression and parameter alpha_b\n# given a pair of a feature vector and a label (x, c), create a pair of a context vector and reward (x, r)\n# where r = 1 if the output of the behavior policy is equal to c and r = 0 otherwise\n# please refer to https://zr-obp.readthedocs.io/en/latest/_autosummary/obp.dataset.multiclass.html for the details\ndataset = MultiClassToBanditReduction(\n X=X,\n y=y,\n base_classifier_b=LogisticRegression(max_iter=1000, random_state=12345),\n alpha_b=0.8,\n dataset_name=\"digits\",\n)\n# split the original data into training and evaluation sets\ndataset.split_train_eval(eval_size=0.7, random_state=12345)\n# obtain logged bandit feedback generated by behavior policy\nbandit_feedback = dataset.obtain_batch_bandit_feedback(random_state=12345)\n\n# `bandit_feedback` is a dictionary storing logged bandit feedback\nbandit_feedback",
"_____no_output_____"
]
],
[
[
"## (2) Off-Policy Learning\nAfter generating logged bandit feedback, we now obtain an evaluation policy using the training set. <br>",
"_____no_output_____"
]
],
[
[
"# obtain action choice probabilities by an evaluation policy\n# we construct an evaluation policy using Random Forest and parameter alpha_e\naction_dist = dataset.obtain_action_dist_by_eval_policy(\n base_classifier_e=RandomForest(random_state=12345),\n alpha_e=0.9,\n)",
"_____no_output_____"
]
],
[
[
"## (3) Off-Policy Evaluation (OPE)\nOPE attempts to estimate the performance of evaluation policies using their action choice probabilities.\n\nHere, we use the **InverseProbabilityWeighting (IPW)**, **DirectMethod (DM)**, and **Doubly Robust (DR)** estimators and visualize the OPE results.",
"_____no_output_____"
]
],
[
[
"# estimate the mean reward function by using ML model (Logistic Regression here)\n# the estimated rewards are used by model-dependent estimators such as DM and DR\nregression_model = RegressionModel(\n n_actions=dataset.n_actions,\n base_model=LogisticRegression(random_state=12345, max_iter=1000),\n)\n# please refer to https://arxiv.org/abs/2002.08536 about the details of the cross-fitting procedure.\nestimated_rewards_by_reg_model = regression_model.fit_predict(\n context=bandit_feedback[\"context\"],\n action=bandit_feedback[\"action\"],\n reward=bandit_feedback[\"reward\"],\n n_folds=3, # use 3-fold cross-fitting\n random_state=12345,\n)",
"_____no_output_____"
],
[
"# estimate the policy value of the evaluation policy based on their action choice probabilities\n# it is possible to set multiple OPE estimators to the `ope_estimators` argument\nope = OffPolicyEvaluation(\n bandit_feedback=bandit_feedback,\n ope_estimators=[InverseProbabilityWeighting(), DirectMethod(), DoublyRobust()]\n)",
"_____no_output_____"
],
[
"# estimate the policy value of IPWLearner with Logistic Regression\nestimated_policy_value, estimated_interval = ope.summarize_off_policy_estimates(\n action_dist=action_dist,\n estimated_rewards_by_reg_model=estimated_rewards_by_reg_model\n)\nprint(estimated_interval, '\\n')\n# visualize estimated policy values of the evaluation policy with Logistic Regression by the three OPE estimators\n# and their 95% confidence intervals (estimated by nonparametric bootstrap method)\nope.visualize_off_policy_estimates(\n action_dist=action_dist,\n estimated_rewards_by_reg_model=estimated_rewards_by_reg_model,\n n_bootstrap_samples=10000, # number of resampling performed in the bootstrap procedure\n random_state=12345,\n)",
" mean 95.0% CI (lower) 95.0% CI (upper)\nipw 0.890339 0.826724 0.975248\ndm 0.787085 0.779634 0.793370\ndr 0.882536 0.808637 0.937305 \n\n"
]
],
[
[
"## (4) Evaluation of OPE estimators\nOur final step is **the evaluation of OPE**, which evaluates and compares the estimation accuracy of OPE estimators.\n\nWith the multi-class classification data, we can calculate the ground-truth policy value of the evaluation policy. \nTherefore, we can compare the policy values estimated by OPE estimators with the ground-turth to evaluate OPE estimators.",
"_____no_output_____"
]
],
[
[
"# calculate the ground-truth performance of the evaluation policy\nground_truth = dataset.calc_ground_truth_policy_value(action_dist=action_dist)\n\nprint(f'ground-truth policy value (classification accuracy): {ground_truth}')",
"ground-truth policy value: 0.8770906200317964\n"
],
[
"# evaluate the estimation performances of OPE estimators \n# by comparing the estimated policy value of the evaluation policy and its ground-truth.\n# `evaluate_performance_of_estimators` returns a dictionary containing estimation performances of given estimators \nrelative_ee = ope.summarize_estimators_comparison(\n ground_truth_policy_value=ground_truth,\n action_dist=action_dist,\n estimated_rewards_by_reg_model=estimated_rewards_by_reg_model,\n metric=\"relative-ee\", # \"relative-ee\" (relative estimation error) or \"se\" (squared error)\n)\n\n# estimation performances of the three estimators (lower means accurate)\nrelative_ee",
"_____no_output_____"
]
],
[
[
"Please see [../examples/multiclass](../examples/multiclass) for a more sophisticated example of the evaluation of OPE with multi-class classification data.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
]
|
e7961e5f0cee7c73a85b391ade5e7b51131c8727 | 132,376 | ipynb | Jupyter Notebook | FailurePrediction/VariableRotationalSpeed/GraphicalComparisons/FilterComparison.ipynb | judithspd/predictive-maintenance | 47a8c9936f9c0d332a0b520de1b8c33053e50a55 | [
"MIT"
]
| 7 | 2021-05-10T09:41:40.000Z | 2022-03-22T01:20:04.000Z | FailurePrediction/VariableRotationalSpeed/GraphicalComparisons/FilterComparison.ipynb | judithspd/predictive-maintenance | 47a8c9936f9c0d332a0b520de1b8c33053e50a55 | [
"MIT"
]
| null | null | null | FailurePrediction/VariableRotationalSpeed/GraphicalComparisons/FilterComparison.ipynb | judithspd/predictive-maintenance | 47a8c9936f9c0d332a0b520de1b8c33053e50a55 | [
"MIT"
]
| 3 | 2021-06-08T04:25:30.000Z | 2021-11-21T17:17:49.000Z | 945.542857 | 128,776 | 0.955377 | [
[
[
"## Filter comparison",
"_____no_output_____"
]
],
[
[
"import scipy\nfrom scipy import signal\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport scipy.io as sio\nimport matplotlib.pyplot as plt\nimport statistics as stats\nimport pandas as pd\nfrom scipy.fft import fft, fftfreq, fftshift\nfrom scipy import signal\nfrom scipy.signal import savgol_filter\nfrom scipy.signal.signaltools import wiener\n\ndef highfilter(input_signal):\n #filtro: 'hp' high pass, 'low': low pass\n \n b, a = signal.butter(3, 0.05, 'hp') \n y = signal.filtfilt(b, a, input_signal)\n \n return y\n\ndef lowfilter(input_signal):\n #filtro: 'hp' high pass, 'low': low pass\n \n b, a = signal.butter(3, 0.05, 'low') \n y = signal.filtfilt(b, a, input_signal)\n \n return y",
"_____no_output_____"
],
[
"HA1 = sio.loadmat('H-A-1.mat')\nChannel1 = HA1['Channel_1']\ncanal1 = Channel1.T[0]\nt = np.linspace(0, 9, len(canal1))",
"_____no_output_____"
],
[
"fig, ((ax1, ax2), (ax3, ax4)) = plt.subplots(2, 2)\nfig.set_size_inches(18,9.5)\nfig.suptitle('Comparison of different filters', fontsize = 15)\nax1.plot(t, canal1, alpha = 0.5)\nax1.plot(t, highfilter(canal1), 'tab:blue')\nax1.set_xlabel('Time')\nax1.set_ylabel('Amplitude')\nax1.set_title('Highpass-filter')\nax2.plot(t, canal1, 'tab:orange', alpha = 0.5)\nax2.plot(t, lowfilter(canal1), 'tab:orange')\nax2.set_xlabel('Time')\nax2.set_ylabel('Amplitude')\nax2.set_title('Lowpass-filter')\nax3.plot(t, canal1, 'tab:green', alpha = 0.5)\nax3.plot(t, savgol_filter(canal1, 5, 2), 'tab:green')\nax3.set_xlabel('Time')\nax3.set_ylabel('Amplitude')\nax3.set_title('Savitzky-Golay filter')\nax4.plot(t, canal1, 'tab:red', alpha = 0.5)\nfiltered_img = wiener(canal1, 99)\nax4.plot(t, filtered_img, 'tab:red')\nax4.set_xlabel('Time')\nax4.set_ylabel('Amplitude')\nax4.set_title('Wiener filter')",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code"
]
]
|
e79644d7e245275cf2b80e5742e9a49b50105406 | 125,238 | ipynb | Jupyter Notebook | sequence models,time series,predictions/sunspot_dataset.ipynb | kartikay-99k/Notebooks-resources | 960da1e0ef8a541894bea01c80c97fed986c9208 | [
"MIT"
]
| null | null | null | sequence models,time series,predictions/sunspot_dataset.ipynb | kartikay-99k/Notebooks-resources | 960da1e0ef8a541894bea01c80c97fed986c9208 | [
"MIT"
]
| null | null | null | sequence models,time series,predictions/sunspot_dataset.ipynb | kartikay-99k/Notebooks-resources | 960da1e0ef8a541894bea01c80c97fed986c9208 | [
"MIT"
]
| 1 | 2021-02-10T15:27:43.000Z | 2021-02-10T15:27:43.000Z | 125,238 | 125,238 | 0.940585 | [
[
[
"!pip install tensorflow==2.0.0b1",
"_____no_output_____"
],
[
"import tensorflow as tf\nprint(tf.__version__)",
"2.0.0-beta1\n"
],
[
"import numpy as np\nimport matplotlib.pyplot as plt\ndef plot_series(time, series, format=\"-\", start=0, end=None):\n plt.plot(time[start:end], series[start:end], format)\n plt.xlabel(\"Time\")\n plt.ylabel(\"Value\")\n plt.grid(True)",
"_____no_output_____"
],
[
"!wget --no-check-certificate \\\n https://storage.googleapis.com/laurencemoroney-blog.appspot.com/Sunspots.csv \\\n -O /tmp/sunspots.csv",
"--2019-07-01 15:07:56-- https://storage.googleapis.com/laurencemoroney-blog.appspot.com/Sunspots.csv\nResolving storage.googleapis.com (storage.googleapis.com)... 74.125.195.128, 2607:f8b0:400e:c08::80\nConnecting to storage.googleapis.com (storage.googleapis.com)|74.125.195.128|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 70827 (69K) [application/octet-stream]\nSaving to: ‘/tmp/sunspots.csv’\n\n\r/tmp/sunspots.csv 0%[ ] 0 --.-KB/s \r/tmp/sunspots.csv 100%[===================>] 69.17K --.-KB/s in 0.001s \n\n2019-07-01 15:07:57 (95.3 MB/s) - ‘/tmp/sunspots.csv’ saved [70827/70827]\n\n"
],
[
"import csv\ntime_step = []\nsunspots = []\n\nwith open('/tmp/sunspots.csv') as csvfile:\n reader = csv.reader(csvfile, delimiter=',')\n next(reader)\n for row in reader:\n sunspots.append(float(row[2]))\n time_step.append(int(row[0]))\n\nseries = np.array(sunspots)\ntime = np.array(time_step)\nplt.figure(figsize=(10, 6))\nplot_series(time, series)",
"_____no_output_____"
],
[
"split_time = 3000\ntime_train = time[:split_time]\nx_train = series[:split_time]\ntime_valid = time[split_time:]\nx_valid = series[split_time:]\n\nwindow_size = 60\nbatch_size = 32\nshuffle_buffer_size = 1000\n\n",
"_____no_output_____"
],
[
"def windowed_dataset(series, window_size, batch_size, shuffle_buffer):\n dataset = tf.data.Dataset.from_tensor_slices(series)\n dataset = dataset.window(window_size + 1, shift=1, drop_remainder=True)\n dataset = dataset.flat_map(lambda window: window.batch(window_size + 1))\n dataset = dataset.shuffle(shuffle_buffer).map(lambda window: (window[:-1], window[-1]))\n dataset = dataset.batch(batch_size).prefetch(1)\n return dataset",
"_____no_output_____"
],
[
"dataset = windowed_dataset(x_train, window_size, batch_size, shuffle_buffer_size)\n\nmodel = tf.keras.models.Sequential([\n tf.keras.layers.Dense(20, input_shape=[window_size], activation=\"relu\"), \n tf.keras.layers.Dense(10, activation=\"relu\"),\n tf.keras.layers.Dense(1)\n])\n\nmodel.compile(loss=\"mse\", optimizer=tf.keras.optimizers.SGD(lr=1e-7, momentum=0.9))\nmodel.fit(dataset,epochs=100,verbose=0)\n\n\n",
"_____no_output_____"
],
[
"forecast=[]\nfor time in range(len(series) - window_size):\n forecast.append(model.predict(series[time:time + window_size][np.newaxis]))\n\nforecast = forecast[split_time-window_size:]\nresults = np.array(forecast)[:, 0, 0]\n\n\nplt.figure(figsize=(10, 6))\n\nplot_series(time_valid, x_valid)\nplot_series(time_valid, results)",
"_____no_output_____"
],
[
"tf.keras.metrics.mean_absolute_error(x_valid, results).numpy()",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e7964561309f5a60be59b1798339ffc3096687a7 | 8,777 | ipynb | Jupyter Notebook | Programming_Assingment13.ipynb | 14vpankaj/iNeuron_Programming_Assignments | 88e5c735dc896073324d335194875b7e74f1c272 | [
"CNRI-Python"
]
| null | null | null | Programming_Assingment13.ipynb | 14vpankaj/iNeuron_Programming_Assignments | 88e5c735dc896073324d335194875b7e74f1c272 | [
"CNRI-Python"
]
| null | null | null | Programming_Assingment13.ipynb | 14vpankaj/iNeuron_Programming_Assignments | 88e5c735dc896073324d335194875b7e74f1c272 | [
"CNRI-Python"
]
| null | null | null | 24.448468 | 208 | 0.50564 | [
[
[
"### Question 1:\n\nWrite a program that calculates and prints the value according to the given formula:\n\nQ = Square root of [(2 * C * D)/H]\n\nFollowing are the fixed values of C and H:\n\nC is 50. H is 30.\n\nD is the variable whose values should be input to your program in a comma-separated sequence.\n\nExample\n\nLet us assume the following comma separated input sequence is given to the program:\n\n100,150,180\n\nThe output of the program should be:\n\n18,22,24\n",
"_____no_output_____"
]
],
[
[
"import math\n\nC = 50\nH = 30\nnumbers = input('Please enter the comma-separated values of D: ').split(',')\noutput = []\n\nfor D in numbers:\n Q = math.sqrt((2*C*int(D))/H)\n output.append(i)\n print(round(Q), end=' ')\n ",
"Please enter the comma-separated values of D: 23,50,86\n9 13 17 "
]
],
[
[
"### Question 2:\n\nWrite a program which takes 2 digits, X,Y as input and generates a 2-dimensional array.\nThe element value in the i-th row and j-th column of the array should be i*j.\n\nNote: i=0,1.., X-1; j=0,1,¡¬Y-1.\n\nExample\n\nSuppose the following inputs are given to the program:\n\n3,5\n\nThen, the output of the program should be:\n\n[[0, 0, 0, 0, 0], [0, 1, 2, 3, 4], [0, 2, 4, 6, 8]]\n",
"_____no_output_____"
]
],
[
[
"def Matrix(x,y):\n M = []\n for i in range(x):\n row = []\n for j in range(y):\n row.append(i*j)\n M.append(row)\n return M\n\nX = int(input('Enter the value of X: '))\nY = int(input('Enter the value of Y: '))\n\nMatrix(X,Y)",
"Enter the value of X: 3\nEnter the value of Y: 5\n"
]
],
[
[
"### Question 3:\n\nWrite a program that accepts a comma separated sequence of words as input and prints the words in a comma-separated sequence after sorting them alphabetically.\n\nSuppose the following input is supplied to the program:\n\nwithout,hello,bag,world\n\nThen, the output should be:\n\nbag,hello,without,world\n",
"_____no_output_____"
]
],
[
[
"string = input('Please enter a comma separated sequence of words: ').split(',')\nstring.sort()\nprint(','.join(string))",
"Please enter a comma separated sequence of words: without,hello,bag,world\nbag,hello,without,world\n"
]
],
[
[
"### Question 4:\n\nWrite a program that accepts a sequence of whitespace separated words as input and prints the words after removing all duplicate words and sorting them alphanumerically.\n\nSuppose the following input is supplied to the program:\n\nhello world and practice makes perfect and hello world again\n\nThen, the output should be:\n\nagain and hello makes perfect practice world\n",
"_____no_output_____"
]
],
[
[
"string = input('Enter the sequence of white separated words: ').split(' ')\nprint(' '.join(sorted(set(string))))",
"Enter the sequence of white separated words: hello world and practice makes perfect and hello world again\nagain and hello makes perfect practice world\n"
]
],
[
[
"### Question 5:\n\nWrite a program that accepts a sentence and calculate the number of letters and digits.\n\nSuppose the following input is supplied to the program:\n\nhello world! 123\n\nThen, the output should be:\n\nLETTERS 10\n\nDIGITS 3\n",
"_____no_output_____"
]
],
[
[
"string = input('Enter a sentence: ')\nletter = 0\ndigit = 0\n\nfor i in string:\n if i.isalpha():\n letter += 1\n elif i.isdigit():\n digit += 1\n else:\n pass\n\nprint('LETTERS', letter)\nprint('DIGITS', digit)\n",
"Enter a sentence: hello world! 123\nLETTERS 10\nDIGITS 3\n"
]
],
[
[
"### Question 6:\n\nA website requires the users to input username and password to register. Write a program to check the validity of password input by users.\n\nFollowing are the criteria for checking the password:\n\n1. At least 1 letter between [a-z]\n2. At least 1 number between [0-9]\n1. At least 1 letter between [A-Z]\n3. At least 1 character from [$#@]\n4. Minimum length of transaction password: 6\n5. Maximum length of transaction password: 12\n\nYour program should accept a sequence of comma separated passwords and will check them according to the above criteria. Passwords that match the criteria are to be printed, each separated by a comma.\n\nExample\n\nIf the following passwords are given as input to the program:\n\nABd1234@1,a F1#,2w3E*,2We3345\n\nThen, the output of the program should be:\n\nABd1234@1\n",
"_____no_output_____"
]
],
[
[
"import re\n\npassword= input('Enter your password: ').split(',')\n\nvalid = []\n\nfor i in password:\n if len(i) < 6 or len(i) > 12:\n break\n elif not re.search('([a-z])+', i):\n break\n elif not re.search(\"([A-Z])+\", i):\n break\n elif not re.search(\"([0-9])+\", i):\n break\n elif not re.search(\"([!@$%^&])+\", i):\n break\n else:\n valid.append(i)\n print(' '.join(valid))\n break\n \nelse:\n print('Invalid Password')",
"Enter your password: ABd1234@1,a F1#,2w3E*,2We3345\nABd1234@1\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
e7964cb61d7e76dab8bbd6d102e7f057f2da0617 | 296,640 | ipynb | Jupyter Notebook | mini-project 2/CSPRJ2_9816603_abrehforoush.ipynb | Alireza-Abrehforoush/Mathematical-Foundations-of-Data-Science | cdfeb12a58ac1e21064841470c6fc8b9c47344cf | [
"MIT"
]
| null | null | null | mini-project 2/CSPRJ2_9816603_abrehforoush.ipynb | Alireza-Abrehforoush/Mathematical-Foundations-of-Data-Science | cdfeb12a58ac1e21064841470c6fc8b9c47344cf | [
"MIT"
]
| null | null | null | mini-project 2/CSPRJ2_9816603_abrehforoush.ipynb | Alireza-Abrehforoush/Mathematical-Foundations-of-Data-Science | cdfeb12a58ac1e21064841470c6fc8b9c47344cf | [
"MIT"
]
| null | null | null | 354.832536 | 89,924 | 0.930674 | [
[
[
"<p style='direction:rtl; text-align: right'>ابتدا باید کتابخانه های زیر را وارد کنیم:\n <ul style='direction:rtl; text-align: right'>\n <li>numpy: برای کار با ماتریس ها</li>\n <li>matplotlib: برای رسم نمودار</li>\n <li>PCA: برای کاهش بعد</li>\n <li>OpenCV: برای کار با عکس</li>\n <li>special_ortho_group: برای تولید پایه اورتونرمال </li>\n </ul>\n</p>\n\n</p>\n<p style='direction:rtl; text-align: right'>تذکر: اگر کتابخانه cv2 اجرا نشد باید آن را نصب کنید. در command prompt دستور زیر را اجرا کنید.\n</p>\n<p style='direction:rtl; text-align: right'> pip install opencv-python\n</p>",
"_____no_output_____"
]
],
[
[
"!pip install opencv-python",
"Requirement already satisfied: opencv-python in c:\\programdata\\anaconda3\\lib\\site-packages (4.5.5.64)\nRequirement already satisfied: numpy>=1.17.3; python_version >= \"3.8\" in c:\\programdata\\anaconda3\\lib\\site-packages (from opencv-python) (1.19.2)\n"
],
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom sklearn.decomposition import PCA\nimport cv2\nfrom scipy.stats import special_ortho_group as sog",
"_____no_output_____"
]
],
[
[
"<h1 style='direction:rtl; text-align: right'>\nپروژه ۲: استفاده از کاهش بعد\n</h1>",
"_____no_output_____"
],
[
"<h2 style='direction:rtl; text-align: right'>\nقسمت ۱.۱: تولید دیتا با استفاده از پایه اورتونرمال\n</h2>\n<p style='direction:rtl; text-align: right'> \n عملیات زیر را انجام دهید:\n <ul style='direction:rtl; text-align: right'>\n <li>ابتدا با استفاده از تابع np.zeros آلفا وکتور هایی با ابعاد dim و N بسازید.</li>\n <li>سعی کنید متغیر آلفا وکتور را طوری پر کنید که به ازای هر اندیس از بعد صفر آن، آرایه ای از توزیع نرمال با میانگین ۰ و انحراف معیار i+1 قرار گیرد.</li>\n <li> بردار پایه V را با استفاده از تابع special_ortho_group.rvs(dim) بسازید.</li>\n <li> مشخص کنید که در ده مولفه اول چند درصد دیتا برای هر کدام از ماتریس ها حفظ شده اند. </li>\n <li> حال بردار زیر را تولید کنید و در alpha_v قرار دهید. </li>\n $$\\alpha_1 V_1 + \\alpha_2 V_2 + ... + \\alpha_d V_d $$\n </ul>",
"_____no_output_____"
]
],
[
[
"dim = 20\nN = 1000\n\nalpha_vectors = np.zeros((N, dim))\n\nfor i in range(N):\n alpha_vectors[i] = np.random.normal(0, i + 1, dim)\n\nV = sog.rvs(dim)\nalpha_v = np.matmul(alpha_vectors, V)\nprint(alpha_v)",
"[[ 1.85796782e-01 5.95693503e-01 -1.06141413e+00 ... 1.25360933e+00\n -1.49196549e+00 -1.71645212e+00]\n [-5.52355256e-01 -2.32208128e-01 -1.45257747e+00 ... -5.75353815e-01\n 5.32574186e-01 -1.13204072e+00]\n [ 1.62991497e+00 3.05093383e-01 1.85496227e+00 ... -2.02565163e+00\n 3.66097985e+00 3.27154903e+00]\n ...\n [ 1.37340414e+03 1.19011377e+02 -1.16850578e+02 ... 1.26333703e+03\n -6.42699005e+02 -1.42814042e+03]\n [ 6.25504714e+01 -1.17225432e+03 2.19318481e+03 ... -6.21643322e+02\n 6.56490839e+02 -1.19807228e+03]\n [-3.01445100e+02 5.06836655e+02 -1.09693110e+03 ... 7.73835725e+02\n -1.90212911e+03 6.61624113e+02]]\n"
]
],
[
[
"<h2 style='direction:rtl; text-align: right'>\nقسمت ۱.۲:استفاده از PCA برای کاهش بعد\n</h2>\n<p style='direction:rtl; text-align: right'> \n عملیات زیر را انجام دهید:\n <ul style='direction:rtl; text-align: right'>\n <li>ابتدا یک شیی از PCA بسازید.</li>\n <li>با استفاده از تابع fit موجود در شیی PCA عملیات pca را روی دیتا alpha_v انجام دهید.</li>\n <li> با استفاده از تابع components_ موجود در شیی pca بردار های تکین را مشاهده کنید.</li>\n <li> با استفاده از تابع explained_variance_ موجود در شیی pca مقدار های تکین را مشاهده کنید.</li>\n </ul>",
"_____no_output_____"
]
],
[
[
"pca = PCA()\npca.fit(alpha_v)\nprint(pca.components_)\nprint(pca.explained_variance_)\n\n\n",
"[[ 3.34017742e-02 -1.18904961e-01 -4.25147294e-01 1.26947115e-01\n 3.13449083e-01 2.54046525e-01 2.71726916e-01 1.80475336e-01\n -1.14252489e-01 -5.92359744e-02 -7.92421226e-02 -3.44459207e-01\n 2.23677760e-01 4.90444010e-01 -1.12590124e-01 1.54180027e-01\n -5.84606532e-03 1.51139626e-01 -6.85230914e-02 1.50337823e-01]\n [-2.79972536e-01 -3.81875071e-04 -3.04331575e-01 3.97839316e-03\n 6.49960142e-02 2.92714369e-02 -3.76792291e-01 -3.10187582e-02\n -2.83348780e-01 -1.72045979e-01 4.50357740e-01 1.19864404e-01\n -2.44852022e-01 2.43681415e-02 -4.27746871e-01 -2.69710242e-01\n 1.16043399e-01 1.08055343e-01 -9.89675561e-02 5.19226613e-02]\n [ 5.66013430e-02 1.62255138e-01 1.89629385e-01 -7.90057936e-02\n -4.49016462e-01 -2.72328881e-03 -2.47581955e-01 -2.08425381e-01\n 2.48299359e-01 -7.02058223e-02 -1.62835979e-01 -2.72148420e-01\n -2.34592829e-01 4.40363626e-01 -1.12763612e-01 4.19896907e-03\n -7.33795257e-03 3.30866712e-01 -2.58390114e-02 2.87861015e-01]\n [ 1.16596502e-01 2.34476477e-01 2.22083735e-01 2.05160752e-01\n 2.69413528e-01 7.75101413e-02 1.37592325e-01 1.27251116e-01\n 5.81002707e-02 2.35254818e-01 3.90541178e-02 9.31818119e-02\n 7.40481424e-02 -1.54927152e-01 -1.80188308e-01 -4.24604153e-01\n -2.67317884e-01 3.11901416e-01 2.96512308e-01 3.98078448e-01]\n [ 3.59855417e-02 -5.17626653e-02 1.40630251e-01 -8.94043250e-03\n -4.58134241e-02 2.05479443e-01 5.48871842e-06 -3.20580899e-01\n -2.65748599e-01 1.47602854e-01 2.26601677e-01 -1.99249596e-01\n 1.99395675e-02 1.42541763e-02 1.17379666e-01 -3.99412283e-02\n -3.69565553e-01 -4.88639589e-01 -3.12771623e-01 3.95564094e-01]\n [-3.20531938e-01 -1.68911883e-01 1.35162555e-01 -3.63467493e-01\n 1.90352682e-01 2.25246225e-01 -1.76695982e-01 2.20292901e-01\n 1.91407295e-01 1.20663205e-01 -1.01932209e-02 -3.25768669e-02\n 7.86288460e-02 -5.91545549e-02 3.12916229e-01 -1.73479473e-01\n -1.32131024e-01 3.50213609e-01 -4.66191163e-01 -6.42863412e-02]\n [ 2.32335584e-02 -3.54696901e-01 1.06423776e-01 9.98315966e-02\n 1.20139914e-01 -1.55926954e-01 3.65203573e-01 -1.28949230e-01\n -1.42193192e-01 2.87579897e-01 -1.20196670e-01 1.39835624e-01\n -5.52001633e-01 -6.27135404e-02 -1.02557983e-01 1.59260237e-01\n 1.90991522e-01 2.18048498e-01 -2.75346628e-01 1.49424910e-01]\n [ 2.66607071e-01 -1.31786634e-01 9.42340353e-02 -1.10547064e-01\n 4.24367897e-01 -4.56475974e-01 -2.47516296e-01 -3.38312843e-01\n -6.68681288e-02 6.82036170e-02 5.14200233e-02 1.64392102e-01\n 1.34767759e-01 4.03926834e-01 5.40829871e-02 -4.29992410e-02\n -2.25672584e-01 8.94344569e-02 2.53358839e-02 -2.04500309e-01]\n [-4.08446169e-02 9.59503796e-03 -6.59448189e-02 -6.29938856e-02\n 1.67424070e-01 9.39861666e-02 -1.08784937e-01 1.44636550e-01\n 3.08735874e-01 -2.76628434e-01 2.11080379e-02 4.30848437e-01\n -1.86817356e-01 -2.15032615e-03 -8.65690559e-02 5.27572346e-01\n -3.96089734e-01 -7.74597731e-02 3.25106951e-02 2.78767583e-01]\n [ 3.45583546e-01 3.81938476e-01 -4.41654295e-02 -1.11420514e-01\n -1.00706496e-01 3.77177770e-02 8.90994314e-02 4.27785135e-01\n -9.20911845e-02 2.76733882e-01 3.83200526e-01 5.89571753e-02\n -3.39218846e-01 2.62329787e-01 2.02288902e-01 4.24411959e-02\n -6.28788470e-02 -2.77013631e-02 -7.94458466e-02 -2.11670649e-01]\n [-4.01362930e-01 2.14021551e-01 1.80599136e-02 1.40353418e-01\n -3.62893585e-01 -1.69450347e-01 2.38455836e-01 -3.81773246e-02\n -3.66398069e-01 5.67647017e-02 -2.15380061e-02 3.51446668e-01\n 3.13098232e-01 1.57750842e-01 3.37929823e-02 1.43191165e-01\n -2.52062742e-01 2.55866994e-01 -1.32335316e-01 -4.27475643e-02]\n [ 4.69884055e-01 -2.47952452e-01 7.82992081e-02 2.17049833e-01\n -1.87178286e-01 2.90337555e-01 -6.10153074e-02 1.04029127e-01\n -1.50266499e-01 -3.74698139e-01 -1.56568656e-01 4.13465140e-01\n 7.89671357e-02 9.58532374e-02 1.07083606e-01 -2.88326384e-01\n 6.86962104e-02 4.73320970e-02 -2.34077780e-01 1.97531559e-02]\n [-2.41003628e-01 5.71705090e-02 2.47957958e-01 -2.55132140e-01\n 1.02457970e-01 6.49639303e-02 1.03406771e-01 9.02330524e-02\n 1.04937013e-01 1.46428165e-01 -4.13585987e-02 3.47270001e-01\n 1.32369584e-01 4.17429785e-01 -9.97202666e-02 -1.26198961e-01\n 4.65691046e-01 -3.69967967e-01 5.90622794e-02 2.21106089e-01]\n [ 6.21256199e-02 4.13005918e-01 -2.41887713e-01 -3.43042911e-01\n 1.67873370e-01 -1.75186686e-01 -7.36081558e-02 5.00482981e-02\n -3.56101437e-01 -1.35878533e-01 -5.59437282e-01 2.09597019e-03\n -1.55355825e-01 -1.33788858e-01 9.32025284e-02 -1.18009025e-01\n 2.91333398e-04 -7.31636155e-02 -1.14657702e-01 2.02955129e-01]\n [ 1.13340224e-01 -2.90703634e-01 -3.30730196e-01 7.88363473e-03\n -2.81952095e-01 -5.09164234e-01 -1.34108566e-01 3.63603497e-01\n 1.91448426e-01 2.50411291e-01 1.40108620e-02 8.74435726e-03\n 2.04821198e-01 -5.62295383e-02 -5.22170894e-02 -1.10040652e-01\n -3.90952853e-02 -1.15627321e-01 -1.44339731e-01 3.30076886e-01]\n [ 9.08519506e-02 2.49577158e-01 7.14979119e-02 2.52513184e-01\n 1.76386132e-01 -1.17496871e-01 -2.12082684e-01 -6.40664908e-02\n -4.39238249e-02 -6.92512954e-02 2.67867830e-01 -1.78795097e-02\n 2.03895436e-01 -1.36030773e-01 3.69568817e-01 3.08116672e-01\n 4.62306457e-01 2.00492029e-01 -1.47473729e-01 3.47153741e-01]\n [-1.93508996e-02 -1.89717339e-01 3.98482905e-01 8.09161586e-02\n 1.25367916e-02 6.57541972e-02 -4.38008703e-01 4.06516596e-01\n -4.45337861e-01 1.77397772e-01 -1.91001312e-01 -1.44560579e-01\n 3.16491616e-02 8.49143890e-03 -1.80651355e-01 2.91253728e-01\n -1.81369450e-02 -2.55535792e-02 1.71126756e-01 -2.70168328e-02]\n [ 1.29105615e-01 -2.93623107e-01 -7.66601312e-03 -5.90494645e-01\n -1.61729863e-01 2.41921287e-02 1.91687616e-01 -2.96486105e-02\n -2.67072290e-01 -1.53130362e-01 2.62630756e-01 2.05393792e-03\n 7.47165945e-02 -8.15624057e-02 1.51842084e-01 7.16019864e-02\n 3.95853045e-02 2.36257499e-01 4.12561592e-01 2.28570232e-01]\n [-1.03071231e-01 -1.07042472e-01 -4.02887507e-01 1.46197268e-01\n -7.13175209e-02 3.05054740e-01 -2.90292365e-01 -2.12756804e-01\n -2.14447218e-02 4.42104580e-01 -1.69544225e-01 2.26855307e-01\n -1.26069659e-01 7.62996882e-02 3.83517564e-01 -1.64042413e-02\n 1.74308774e-02 2.95504760e-02 3.43425715e-01 4.72580985e-02]\n [-3.38168767e-01 -1.50358236e-01 1.21204491e-01 2.73435240e-01\n 6.98776545e-02 -2.71569663e-01 7.62004119e-02 2.23359419e-01\n -5.04083794e-02 -3.58344131e-01 3.48821749e-02 -1.41264091e-01\n -3.13295579e-01 2.02242187e-01 4.60755751e-01 -2.31049642e-01\n -1.00620747e-01 -1.18946628e-01 2.21317324e-01 9.55729060e-02]]\n[459338.51256464 426099.91399403 410847.21917205 392410.42768038\n 380751.72253453 369528.46403096 362403.79314679 351653.36336259\n 344206.96335909 327495.31605779 315513.97763167 309901.14676994\n 300603.60437195 295271.1739031 287504.79954667 271718.6479452\n 264138.64297627 254663.79547324 236554.66406725 223683.16717356]\n"
]
],
[
[
"<h2 style='direction:rtl; text-align: right'>\nقسمت ۱.۳: کاهش بعد به ۳ بعد\n</h2>\n <ul style='direction:rtl; text-align: right'>\n <li>ابتدا یک شیی از PCA با ورودی n_components=3 بسازید.</li>\n <li>با استفاده از تابع fit موجود در شیی PCA عملیات pca را روی دیتا alpha_v انجام دهید.</li>\n <li> تابع explained_variance_ratio_ موجود در شیی pca درصد حفظ دیتا به ازای هر کدام از بعد ها را می دهد.</li>\n <li>با کاهش بعد به ۳، چند درصد از اطلاعات حفظ می شود؟</li>\n </ul>",
"_____no_output_____"
]
],
[
[
"pca = PCA(n_components = 3)\npca.fit(alpha_v)\nprint(str(100 * np.sum(pca.explained_variance_ratio_)) + \" percent of data is preserved in 3 dimensions!\")\n",
"19.544901432955598 percent of data is preserved in 3 dimensions!\n"
]
],
[
[
"<p style='direction:rtl; text-align: right'> برای حفظ ۹۰ درصد از اطلاعات به چند بعد نیاز داریم؟ </p>",
"_____no_output_____"
]
],
[
[
"min_dim = 0\nfor i in range(1, dim):\n pca = PCA(n_components = i)\n pca.fit(alpha_v)\n if (np.sum(pca.explained_variance_ratio_) >= 0.9):\n min_dim = i\n break\nprint(\"Almost \" + str(100 * np.sum(pca.explained_variance_ratio_)) + \" percent of data is preserved in at least \" + str(min_dim) + \" dimensions!\")",
"Almost 93.01006062812166 percent of data is preserved in at least 18 dimensions!\n"
]
],
[
[
"<h2 style='direction:rtl; text-align: right'>\nقسمت ۲.۱: خواندن فایل تصویر\n</h2>\n<p style='direction:rtl; text-align: right'>ابتدا فایل تصویری رنگی باکیفیتی را از گوگل دانلود کنید.</p>\n<p style='direction:rtl; text-align: right'>با استفاده از تابع imread موجود در کتابخانه <a href=\"https://www.geeksforgeeks.org/python-opencv-cv2-imread-method/\">OpenCV</a> عکس مربوطه را فراخوانی کنید:</p>",
"_____no_output_____"
]
],
[
[
"image1 = cv2.imread(\"mona.jpg\")",
"_____no_output_____"
]
],
[
[
"<p style='direction:rtl; text-align: right'>عکس خوانده شده را به فرمت <a href=\"https://www.w3schools.com/colors/colors_rgb.asp\">RGB</a> در می آوریم:</p>",
"_____no_output_____"
]
],
[
[
"image = cv2.cvtColor(image1, cv2.COLOR_BGR2RGB)",
"_____no_output_____"
]
],
[
[
"<p style='direction:rtl; text-align: right'>\n همانطور که می بینید عکس خوانده شده به ازای هر پیکسل ۳ عدد دارد: بنابراین برای هر عکس رنگی x*y یک آرایه x*y*3 خواهیم داشت.</p>",
"_____no_output_____"
]
],
[
[
"dim = image.shape\nprint('Image shape =', dim)",
"Image shape = (720, 483, 3)\n"
]
],
[
[
"<h2 style='direction:rtl; text-align: right'>\nقسمت ۲.۲: نمایش تصویر\n</h2>\n<p style='direction:rtl; text-align: right'>با استفاده از تابع imshow موجود در <a href=\"https://www.geeksforgeeks.org/matplotlib-pyplot-imshow-in-python/\">matplotlib</a> تصویر خوانده شده را نمایش دهید:</p>",
"_____no_output_____"
]
],
[
[
"plt.imshow(image)\nplt.show()",
"_____no_output_____"
]
],
[
[
"<h2 style='direction:rtl; text-align: right'>\nقسمت ۲.۳: آماده سازی تصویر برای کاهش بعد\n</h2>\n<p style='direction:rtl; text-align: right'>سه ماتریس رنگ را در ماتریس های R,G,B ذخیره کنید:</p>",
"_____no_output_____"
]
],
[
[
"R = image[:, :, 0]\nG = image[:, :, 1]\nB = image[:, :, 2]\nprint(R.shape)\nprint(G.shape)\nprint(B.shape)",
"(720, 483)\n(720, 483)\n(720, 483)\n"
]
],
[
[
"<h2 style='direction:rtl; text-align: right'>\nقسمت ۲.۴:استفاده از PCA برای کاهش بعد\n</h2>\n\n<p style='direction:rtl; text-align: right'> \nبا استفاده از کلاس PCA در کتابخانه sklearn کاهش بعد را انجام میدهیم.\n عملیات زیر را انجام دهید:\n <a href=\"https://scikit-learn.org/stable/modules/generated/sklearn.decomposition.PCA.html\">راهنمایی</a>\n <ul style='direction:rtl; text-align: right'>\n <li>برای هر یک از ماتریس های R,G,B یک شی PCA ایجاد کنید. تعداد مولفه ها را ۱۰ قرار دهید.</li>\n <li>با استفاده از تابع fit موجود در pca الگوریتم را روی ماتریس ها فیت کنید.</li>\n <li> با استفاده از دستور _explained_variance_ratio میتوانید ببینید هرکدام از مولفه ها چند درصد دیتای ماتریس را دارند. </li>\n <li> مشخص کنید که در ده مولفه اول چند درصد دیتا برای هر کدام از ماتریس ها حفظ شده اند. </li>\n <li> با استفاده از دستور bar مقادیر _explained_variance_ratio را رسم کنید </li> \n </ul>",
"_____no_output_____"
]
],
[
[
"k = 10\nrpca = PCA(n_components = k)\ngpca = PCA(n_components = k)\nbpca = PCA(n_components = k)\n\nrpca.fit(R)\ngpca.fit(G)\nbpca.fit(B)\n\nprint(\"First \" + str(k) + \" components of Red Matrix have \" + str(100 * np.sum(rpca.explained_variance_ratio_)) + \" percent of data.\")\nprint(\"First \" + str(k) + \" components of Green Matrix have \" + str(100 * np.sum(gpca.explained_variance_ratio_)) + \" percent of data.\")\nprint(\"First \" + str(k) + \" components of Blue Matrix have \" + str(100 * np.sum(bpca.explained_variance_ratio_)) + \" percent of data.\")",
"First 10 components of Red Matrix have 91.5284949220633 percent of data.\nFirst 10 components of Green Matrix have 92.72653593218749 percent of data.\nFirst 10 components of Blue Matrix have 79.66273622738387 percent of data.\n"
],
[
"plt.bar([i for i in range(k)], rpca.explained_variance_ratio_, color ='red', width = 0.4)\nplt.xlabel(\"Red Components\")\nplt.ylabel(\"Variance %\")\nplt.show()",
"_____no_output_____"
],
[
"\nplt.bar([i for i in range(k)], gpca.explained_variance_ratio_, color ='green', width = 0.4)\nplt.xlabel(\"Green Components\")\nplt.ylabel(\"Variance %\")\nplt.show()",
"_____no_output_____"
],
[
"plt.bar([i for i in range(k)], bpca.explained_variance_ratio_, color ='blue', width = 0.4)\nplt.xlabel(\"Blue Components\")\nplt.ylabel(\"Variance %\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"<p style='direction:rtl; text-align: right'>عملیات زیر را انجام دهید:\n <ul style='direction:rtl; text-align: right'>\n <li>با استفاده از تابع transform موجود در pca دیتا با بعد کمتر را تولید کنید</li>\n <li> با استفاده از تابع inverse_transform دیتا را به بعد اولیه برگردانید </li>\n </ul>\n</p>",
"_____no_output_____"
]
],
[
[
"Transform_R = rpca.transform(R)\nTransform_B = gpca.transform(G)\nTransform_G = bpca.transform(B)\nReduced_R = rpca.inverse_transform(Transform_R)\nReduced_G = gpca.inverse_transform(Transform_G)\nReduced_B = bpca.inverse_transform(Transform_B)\n\nprint('Transform Matrix Shape = ', Transform_R.shape)\nprint('Inverse Transform Matrix Shape = ', Reduced_R.shape)",
"Transform Matrix Shape = (720, 10)\nInverse Transform Matrix Shape = (720, 483)\n"
]
],
[
[
"<p style='direction:rtl; text-align: right'>با استفاده از دستور concatenate سه ماتریس ً Reduced_R,Reduced_G,Reduced_B را کنار هم قرار دهید تا یک آرایه x*y*3 ایجاد شود. x , y همان ابعاد تصویر اولیه (image) هستند </p>\n<p style='direction:rtl; text-align: right'>با استفاده از دستور astype ماتریس بدست آمده را به عدد صحیح تبدیل کنید.</p>\n\n<p style='direction:rtl; text-align: right'>عکس بدست آمده را با imshow نمایش دهید.</p>",
"_____no_output_____"
]
],
[
[
"Reduced_R = Reduced_R.reshape((dim[0], dim[1], 1))\nReduced_G = Reduced_G.reshape((dim[0], dim[1], 1))\nReduced_B = Reduced_B.reshape((dim[0], dim[1], 1))\n\nreduced_image = np.dstack((Reduced_R, Reduced_G, Reduced_B))\nfinal_image = reduced_image.astype(int)\nprint('final_image shape = ', final_image.shape)\nplt.imshow(final_image)\nplt.show()",
"Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\n"
]
],
[
[
"<h2 style='direction:rtl; text-align: right'>\nقسمت ۲.۵:استفاده از PCA برای کاهش بعد و حفظ ۹۹ درصد داده ها\n</h2>\n\n<p style='direction:rtl; text-align: right'> \nکل قسمت ۲.۴ را مجددا اجرا کنید. این بار تعداد مولفه ها را عددی قرار دهید که در هر سه ماتریس R,G,B حداقل ۹۹ درصد داده ها حفظ شود.\n ",
"_____no_output_____"
]
],
[
[
"k = 188\nrpca = PCA(n_components = k)\ngpca = PCA(n_components = k)\nbpca = PCA(n_components = k)\n\nrpca.fit(R)\ngpca.fit(G)\nbpca.fit(B)",
"_____no_output_____"
],
[
"Transform_R = rpca.transform(R)\nTransform_B = gpca.transform(G)\nTransform_G = bpca.transform(B)\nReduced_R = rpca.inverse_transform(Transform_R)\nReduced_G = gpca.inverse_transform(Transform_G)\nReduced_B = bpca.inverse_transform(Transform_B)\n\nprint('Transform Matrix Shape = ', Transform_R.shape)\nprint('Inverse Transform Matrix Shape = ', Reduced_R.shape)",
"Transform Matrix Shape = (720, 188)\nInverse Transform Matrix Shape = (720, 483)\n"
],
[
"Reduced_R = Reduced_R.reshape((dim[0], dim[1], 1))\nReduced_G = Reduced_G.reshape((dim[0], dim[1], 1))\nReduced_B = Reduced_B.reshape((dim[0], dim[1], 1))\n\nreduced_image = np.dstack((Reduced_R, Reduced_G, Reduced_B))\nfinal_image = reduced_image.astype(int)\nprint('final_image shape = ', final_image.shape)\nplt.imshow(final_image)\nplt.show()\nprint(np.sum(rpca.explained_variance_ratio_))",
"Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
]
|
e7964d2e10a0f85d50bf44ea44d08dee23811289 | 115,658 | ipynb | Jupyter Notebook | example.ipynb | UeFan/Heatmap-with-flexible-size-cell | 63a39a224df7766e159141752d057edca9f928c9 | [
"MIT"
]
| 1 | 2020-08-14T18:13:54.000Z | 2020-08-14T18:13:54.000Z | example.ipynb | UeFan/Heatmap-for-visualizing-clustering-result | 63a39a224df7766e159141752d057edca9f928c9 | [
"MIT"
]
| null | null | null | example.ipynb | UeFan/Heatmap-for-visualizing-clustering-result | 63a39a224df7766e159141752d057edca9f928c9 | [
"MIT"
]
| null | null | null | 347.321321 | 87,696 | 0.896972 | [
[
[
"import numpy as np\nimport pandas as pd\nimport seaborn as sns\nfrom new_heatmap import multi_group_heatmap\nimport matplotlib.pyplot as plt\n\nimport scipy.spatial.distance as distance\nimport scipy.cluster.hierarchy as sch",
"_____no_output_____"
],
[
"\n\na0 = np.arange(0,1, 1/600).reshape(30,20)\na1 = np.arange(0,1, 1/1200).reshape(30,40)\n\n\n\ns0 = np.arange(0,1, 1/600).reshape(20,30).T\ns1 = np.arange(0,1, 1/1200).reshape(40,30).T\n\nylabels = (np.arange(30)*10).astype(str)\nx_label0 = (np.arange(20)).astype(int).astype(str)\nx_label1 = (np.arange(40)).astype(int).astype(str)\n\n\nval = np.random.rand(30,2)",
"_____no_output_____"
],
[
"col_pairwise_dists_0 = distance.squareform(distance.pdist(a0.T))\ncol_pairwise_dists_1 = distance.squareform(distance.pdist(a1.T))\ncol_clusters_0 = sch.linkage(col_pairwise_dists_0,method='complete')\ncol_clusters_1 = sch.linkage(col_pairwise_dists_1,method='complete')\ncol_denD_0 = sch.dendrogram(col_clusters_0,color_threshold=np.inf)\ncol_denD_1 = sch.dendrogram(col_clusters_1,color_threshold=np.inf)\n\nx_label0 = np.array(x_label0)[[col_denD_0['leaves']]]\nx_label1 = np.array(x_label1)[[col_denD_1['leaves']]]\na0 = np.array(a0.T[col_denD_0['leaves'],:]).T\na1 = np.array(a1.T[col_denD_1['leaves'],:]).T\n\ns0 = np.array(s0.T[col_denD_0['leaves'],:]).T\ns1 = np.array(s1.T[col_denD_1['leaves'],:]).T",
"/usr/local/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:3: ClusterWarning: scipy.cluster: The symmetric non-negative hollow observation matrix looks suspiciously like an uncondensed distance matrix\n This is separate from the ipykernel package so we can avoid doing imports until\n/usr/local/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:4: ClusterWarning: scipy.cluster: The symmetric non-negative hollow observation matrix looks suspiciously like an uncondensed distance matrix\n after removing the cwd from sys.path.\n/usr/local/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:8: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.\n \n/usr/local/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:9: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.\n if __name__ == '__main__':\n"
],
[
"a0",
"_____no_output_____"
],
[
"%reload_ext autoreload\n%load_ext autoreload\n%autoreload 2\n\n\nfig0 = plt.figure(figsize=(15, 10))\nmulti_group_heatmap(\n index_group = val,\n index_group_x_ticks = ['index1', 'index2'],\n group0 = a0,\n group1 = a1,\n col_pairwise_dists_0 = col_pairwise_dists_0,\n col_pairwise_dists_1 = col_pairwise_dists_1,\n group0_x_ticks = x_label0,\n group1_x_ticks = x_label1,\n size0 = s0,\n size1 = s1,\n size_scale=60, # Change the overall cube size \n x_axis_label0 = '\\nGroup0',\n x_axis_label1 =' ' + \\\n ' ' + \\\n ' X_labels' + '\\nGroup1',\n y_ticks = ylabels, \n color_range = [0,1],\n size_range = [0,1],\n chart = np.zeros((30,2)),\n chart_x_ticks = ['col_1','col_2'],\n palette=(\"RdYlGn\", 256),\n color_bar = True,\n size_bar = True,\n space_in_size_bar = 1,\n high_ligh_y_ticks = (0,5)\n)",
"The autoreload extension is already loaded. To reload it, use:\n %reload_ext autoreload\n"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code"
]
]
|
e796576f9c4fcfe1bbbf557d84feff2faa44f8e6 | 11,467 | ipynb | Jupyter Notebook | Missions_to_Mars/.ipynb_checkpoints/mission_to_mars-checkpoint.ipynb | goldenMJ/web-scraping-challenge | 9808976c1ec7481851c37cfa3ade04237c05d645 | [
"ADSL"
]
| null | null | null | Missions_to_Mars/.ipynb_checkpoints/mission_to_mars-checkpoint.ipynb | goldenMJ/web-scraping-challenge | 9808976c1ec7481851c37cfa3ade04237c05d645 | [
"ADSL"
]
| null | null | null | Missions_to_Mars/.ipynb_checkpoints/mission_to_mars-checkpoint.ipynb | goldenMJ/web-scraping-challenge | 9808976c1ec7481851c37cfa3ade04237c05d645 | [
"ADSL"
]
| null | null | null | 32.951149 | 2,015 | 0.581233 | [
[
[
"from splinter import Browser\nfrom bs4 import BeautifulSoup\nimport time\nimport requests\nimport pymongo\nfrom selenium import webdriver\n",
"_____no_output_____"
]
],
[
[
"# **Create Database in MongoDB**\n",
"_____no_output_____"
],
[
"# **Connect to Mongo DB Mars DB**",
"_____no_output_____"
]
],
[
[
"conn = 'mongodb://localhost:27017'\nclient = pymongo.MongoClient(conn)",
"_____no_output_____"
],
[
"# Define database and collection\ndb = client.mars\ncollection = db.items",
"_____no_output_____"
]
],
[
[
"**Get executable_path**",
"_____no_output_____"
]
],
[
[
"!which chromedriver",
"/usr/local/bin/chromedriver\r\n"
]
],
[
[
"# **Step 1 - Scraping**",
"_____no_output_____"
],
[
"**NASA Mars News**\n\nScrape the NASA Mars News Site and collect the latest News Title and Paragraph Text. Assign the text to variables that you can reference later.",
"_____no_output_____"
]
],
[
[
"def latest_nasa_news():\n \n executable_path = {'executable_path': '/usr/local/bin/chromedriver'}\n browser = Browser('chrome', **executable_path, headless=False)\n url = \"https://mars.nasa.gov/news/?page=0&per_page=40&order=publish_date+desc%2Ccreated_at+desc&search=&category=19%2C165%2C184%2C204&blank_scope=Latest\"\n browser.visit(url)\n #need timer to ensure page has load before scraping?\n time.sleep(5)\n html = browser.html\n soup = BeautifulSoup(html, 'html.parser')\n news_date = soup.find('div', class_='list_date').text\n news_title = soup.find('div', class_='content_title').text\n news_p = soup.find('div', class_='article_teaser_body').text\n print(news_date)\n print(news_title)\n print(news_p)\n \n#how to print multiple variables?\n\nlatest_nasa_news()\n",
"November 27, 2019\nNASA's Briefcase-Size MarCO Satellite Picks Up Honors\nThe twin spacecraft, the first of their kind to fly into deep space, earn a Laureate from Aviation Week & Space Technology.\n"
]
],
[
[
"**JPL Mars Space Images - Featured Image**\n\nLatest Mars image",
"_____no_output_____"
]
],
[
[
"def latest_mars_image():\n \n executable_path = {'executable_path': '/usr/local/bin/chromedriver'}\n browser = Browser('chrome', **executable_path, headless=False)\n url_mars_image = \"https://www.jpl.nasa.gov/spaceimages/?search=&category=Mars\"\n browser.visit(url_mars_image)\n #need timer to ensure page has load before scraping?\n time.sleep(5)\n html = browser.html\n soup = BeautifulSoup(html, 'html.parser')\n image = soup.find('img', class_='thumb')\n #image output <img alt=\"Indus Vallis\" class=\"thumb\" src=\"/spaceimages/images/wallpaper/PIA23573-640x350.jpg\" title=\"Indus Vallis\"/>\n #how to save image url and path to diplay in webpage?\n\n#need to call image?\nlatest_mars_image()",
"_____no_output_____"
]
],
[
[
"**Twitter Latest Mars Weather**",
"_____no_output_____"
]
],
[
[
"def latest_mars_weather():\n \n executable_path = {'executable_path': '/usr/local/bin/chromedriver'}\n browser = Browser('chrome', **executable_path, headless=False)\n url_mars_weather = \"https://twitter.com/marswxreport?lang=en\"\n browser.visit(url_mars_weather)\n #need timer to ensure page has load before scraping?\n time.sleep(5)\n soup = BeautifulSoup(browser.html, 'html.parser')\n latest_weather = soup.find('p', class_='TweetTextSize').text\n print('Current Weather on Mars')\n print(latest_weather)\n\n#how to print multiple variables?\n\nlatest_mars_weather()",
"Current Weather on Mars\nInSight sol 366 (2019-12-07) low -98.9ºC (-146.1ºF) high -20.4ºC (-4.8ºF)\nwinds from the SSE at 5.7 m/s (12.6 mph) gusting to 20.4 m/s (45.5 mph)\npressure at 6.60 hPapic.twitter.com/BYqMmSLmWr\n"
],
[
"import requests\nimport lxml.html as lh\nimport pandas as pd",
"_____no_output_____"
],
[
"def mars_facts():\n executable_path = {'executable_path': '/usr/local/bin/chromedriver'}\n browser = Browser('chrome', **executable_path, headless=False)\n url_mars_facts = \"http://space-facts.com/mars/\"\n browser.visit(url_mars_facts)\n #need timer to ensure page has load before scraping?\n time.sleep(5)\n soup = BeautifulSoup(html, 'html.parser')\n mars_facts_table = soup.find(\"table\", {\"class\": \"tablepress tablepress-id-p-mars\"})\n df_mars_facts = pd.read_html(str(mars_facts_table))\n print(df_mars_facts)\n\nmars_facts()\n\n\n \n \n \n ",
"_____no_output_____"
],
[
" latest_weather = soup.find('td', class_='column-2')\n for weather in latest_weather:\n print('----------------------------------')\n print(weather)",
"----------------------------------\n6,792 km\n----------------------------------\n<br/>\n"
]
],
[
[
"**Mars Hemispheres**\n\nVisit the USGS Astrogeology site here to obtain high resolution images for each of Mar's hemispheres.\n\n\nYou will need to click each of the links to the hemispheres in order to find the image url to the full resolution image.\n\n\nSave both the image url string for the full resolution hemisphere image, and the Hemisphere title containing the hemisphere name. Use a Python dictionary to store the data using the keys img_url and title.\n\n\nAppend the dictionary with the image url string and the hemisphere title to a list. This list will contain one dictionary for each hemisphere.",
"_____no_output_____"
]
],
[
[
"def mars_image():\n executable_path = {'executable_path': '/usr/local/bin/chromedriver'}\n browser = Browser('chrome', **executable_path, headless=False)\n url = \"https://astrogeology.usgs.gov/search/results?q=hemisphere+enhanced&k1=target&v1=Mars\"\n browser.visit(url)\n #need a pause to ensure page has load before scraping?\n soup = BeautifulSoup(browser.html, 'html.parser')\n div = soup.find('div', class_='results').findAll('div', class_='description')\n print(div)\nmars_image()",
"[<div class=\"description\"><a class=\"itemLink product-item\" href=\"/search/map/Mars/Viking/cerberus_enhanced\"><h3>Cerberus Hemisphere Enhanced</h3></a><span class=\"subtitle\" style=\"float:left\">image/tiff 21 MB</span><span class=\"pubDate\" style=\"float:right\"></span><br/><p>Mosaic of the Cerberus hemisphere of Mars projected into point perspective, a view similar to that which one would see from a spacecraft. This mosaic is composed of 104 Viking Orbiter images acquired…</p></div>, <div class=\"description\"><a class=\"itemLink product-item\" href=\"/search/map/Mars/Viking/schiaparelli_enhanced\"><h3>Schiaparelli Hemisphere Enhanced</h3></a><span class=\"subtitle\" style=\"float:left\">image/tiff 35 MB</span><span class=\"pubDate\" style=\"float:right\"></span><br/><p>Mosaic of the Schiaparelli hemisphere of Mars projected into point perspective, a view similar to that which one would see from a spacecraft. The images were acquired in 1980 during early northern…</p></div>, <div class=\"description\"><a class=\"itemLink product-item\" href=\"/search/map/Mars/Viking/syrtis_major_enhanced\"><h3>Syrtis Major Hemisphere Enhanced</h3></a><span class=\"subtitle\" style=\"float:left\">image/tiff 25 MB</span><span class=\"pubDate\" style=\"float:right\"></span><br/><p>Mosaic of the Syrtis Major hemisphere of Mars projected into point perspective, a view similar to that which one would see from a spacecraft. This mosaic is composed of about 100 red and violet…</p></div>, <div class=\"description\"><a class=\"itemLink product-item\" href=\"/search/map/Mars/Viking/valles_marineris_enhanced\"><h3>Valles Marineris Hemisphere Enhanced</h3></a><span class=\"subtitle\" style=\"float:left\">image/tiff 27 MB</span><span class=\"pubDate\" style=\"float:right\"></span><br/><p>Mosaic of the Valles Marineris hemisphere of Mars projected into point perspective, a view similar to that which one would see from a spacecraft. The distance is 2500 kilometers from the surface of…</p></div>]\n"
]
],
[
[
"**Step 2 - MongoDB and Flask Application**",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
e796749539bb68c9596269ddb201ff990aeb4c11 | 60,451 | ipynb | Jupyter Notebook | notebooks/demo_ellipsoidal_nfw.ipynb | aphearin/ellipsoidal_nfw | d5de00f3e1042896cb7ad02e68fa485f443727b7 | [
"BSD-3-Clause"
]
| null | null | null | notebooks/demo_ellipsoidal_nfw.ipynb | aphearin/ellipsoidal_nfw | d5de00f3e1042896cb7ad02e68fa485f443727b7 | [
"BSD-3-Clause"
]
| null | null | null | notebooks/demo_ellipsoidal_nfw.ipynb | aphearin/ellipsoidal_nfw | d5de00f3e1042896cb7ad02e68fa485f443727b7 | [
"BSD-3-Clause"
]
| null | null | null | 422.734266 | 57,172 | 0.944534 | [
[
[
"%matplotlib inline",
"_____no_output_____"
],
[
"import numpy as np\nfrom matplotlib import pyplot as plt",
"_____no_output_____"
]
],
[
[
"### Generate a realization of 5000 points within a single halo of conc=5",
"_____no_output_____"
]
],
[
[
"from ellipsoidal_nfw import random_nfw_ellipsoid\n\nnpts = 5_000\nconc = np.zeros(npts)+5.\nx, y, z = random_nfw_ellipsoid(conc, b=2, c=3)\n\nfig, (ax0, ax1, ax2) = plt.subplots(1, 3, figsize=(12, 4))\nfig.tight_layout(pad=3.0)\n\nfor ax in ax0, ax1, ax2:\n xlim = ax.set_xlim(-4, 4)\n ylim = ax.set_ylim(-4, 4)\n\n__=ax0.scatter(x, y, s=1)\n__=ax1.scatter(x, z, s=1)\n__=ax2.scatter(y, z, s=1)\n\nxlabel = ax0.set_xlabel(r'$x$')\nylabel = ax0.set_ylabel(r'$y$')\nxlabel = ax1.set_xlabel(r'$x$')\nylabel = ax1.set_ylabel(r'$z$')\nxlabel = ax2.set_xlabel(r'$y$')\nylabel = ax2.set_ylabel(r'$z$')\n\nfig.savefig('ellipsoidal_nfw.png', bbox_extra_artists=[xlabel, ylabel], bbox_inches='tight', dpi=200)",
"_____no_output_____"
]
],
[
[
"### Generate a realization of a collection of 10 halos, each with 5000 points, each with different concentrations",
"_____no_output_____"
]
],
[
[
"npts_per_halo = 5_000\nn_halos = 10\nconc = np.linspace(5, 25, n_halos)\nconc_halopop = np.repeat(conc, npts_per_halo)\nx, y, z = random_nfw_ellipsoid(conc_halopop, b=2, c=3)\nx = x.reshape((n_halos, npts_per_halo))\ny = y.reshape((n_halos, npts_per_halo))\nz = z.reshape((n_halos, npts_per_halo))\n",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
e7967a029f8d70e5a43ccd7d9a75d698488f57ec | 62,383 | ipynb | Jupyter Notebook | #Task_3/task_3.ipynb | ViKrAm-Bais/sparks_foundation_grip | f19049c0d105c4e3de1c93e25b3d267ea0134143 | [
"MIT"
]
| null | null | null | #Task_3/task_3.ipynb | ViKrAm-Bais/sparks_foundation_grip | f19049c0d105c4e3de1c93e25b3d267ea0134143 | [
"MIT"
]
| null | null | null | #Task_3/task_3.ipynb | ViKrAm-Bais/sparks_foundation_grip | f19049c0d105c4e3de1c93e25b3d267ea0134143 | [
"MIT"
]
| null | null | null | 142.102506 | 29,446 | 0.874116 | [
[
[
"#This is Task 2 of GRIP internship\nTo Explore Supervised Machine Learning",
"_____no_output_____"
]
],
[
[
"#Importing all the libraries required for the code\nimport pandas as pd\nimport numpy as np \nimport matplotlib.pyplot as plt \n%matplotlib inline",
"_____no_output_____"
],
[
"# Loading Data from the given URL\nurl = \"http://bit.ly/w-data\"\ndata = pd.read_csv(url)\nprint(\"shape of dataset: {}\".format(data.shape))\nprint(data.head(5))",
"shape of dataset: (25, 2)\n Hours Scores\n0 2.5 21\n1 5.1 47\n2 3.2 27\n3 8.5 75\n4 3.5 30\n"
],
[
"# Plotting the distribution of score using matplotlib\nplt.figure(figsize=(10, 6), dpi=100)\nplt.title(\"Distribution of Score\")\nplt.xlabel(\"Hours\")\nplt.ylabel(\"Scores\")\nplt.scatter(data.Hours,data.Scores,color=\"b\",marker=\"*\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Preparing the data for training",
"_____no_output_____"
],
[
"Dividing the data into attributes (Inputs) and labels (Outputs)",
"_____no_output_____"
]
],
[
[
"# Dividing the data into attributes(Inputs) and label(Outputs)\nx = data.iloc[:, :-1].values \ny = data.iloc[:, 1].values ",
"_____no_output_____"
]
],
[
[
"splitting the data set into tranining and testing data",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split \nx_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2, random_state=0) \nprint(\"training dataset shape: {}\".format(x_train.shape))\nprint(\"testing dataset shape: {}\".format(x_test.shape))",
"training dataset shape: (20, 1)\ntesting dataset shape: (5, 1)\n"
]
],
[
[
"#Training the model",
"_____no_output_____"
]
],
[
[
"# Importing library for linear regression\nfrom sklearn.linear_model import LinearRegression \nmodel = LinearRegression() \nmodel.fit(x_train, y_train)",
"_____no_output_____"
]
],
[
[
"Plotting the linear regression line with training data",
"_____no_output_____"
]
],
[
[
"# Defining the equation of line\nprint(\"coefficient: {}, intercept: {}\".format(model.coef_, model.intercept_))\nline = model.coef_*x + model.intercept_\n# plotting line with data\nplt.figure(figsize=(10, 6), dpi=100)\n#plotting training data\nplt.scatter(x_train, y_train, color=\"c\",marker=\"*\")\n#plotting testing data\nplt.scatter(x_test, y_test, color=\"m\",marker=\"+\")\nplt.plot(x, line)\nplt.title(\"Distribution of Score and Regression line\")\nplt.xlabel(\"Hours\")\nplt.ylabel(\"Scores\")\nplt.show()",
"coefficient: [9.91065648], intercept: 2.018160041434662\n"
]
],
[
[
"# Results",
"_____no_output_____"
]
],
[
[
"# getting predictions for test data\ny_predicted = model.predict(x_test)",
"_____no_output_____"
],
[
"# Comparing Actual vs Predicted\ndf = pd.DataFrame({'Actual': y_test, 'Predicted': y_predicted}) \nprint(df)",
" Actual Predicted\n0 20 16.884145\n1 27 33.732261\n2 69 75.357018\n3 30 26.794801\n4 62 60.491033\n"
]
],
[
[
"#predicted score if a student study for 9.25 hrs in a day",
"_____no_output_____"
]
],
[
[
"s_hours = 9.25\ns_score = model.predict([[shours]])\nprint(\"predicted score if a student study for {} hrs in a day is {}\".format(s_hours, s_score[0]))",
"predicted score if a student study for 9.25 hrs in a day is 93.69173248737539\n"
]
],
[
[
"# Calculating error for the model",
"_____no_output_____"
]
],
[
[
"from sklearn import metrics \nprint('Mean Absolute Error: {}'.format(metrics.mean_absolute_error(y_test, y_predicted)))\naccuracy = 100 * model.score(x_test, y_test)\nprint(\"Accuracy(%): \", accuracy)",
"Mean Absolute Error: 4.183859899002982\nAccuracy(%): 94.54906892105353\n"
],
[
"",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
e7967b8639289ed8f93dd492bce8867565f6d1d9 | 20,365 | ipynb | Jupyter Notebook | part-4.ipynb | willingc/intro-to-python | 9822315d28c603b17d7475c09925146d704a571c | [
"CC-BY-4.0"
]
| null | null | null | part-4.ipynb | willingc/intro-to-python | 9822315d28c603b17d7475c09925146d704a571c | [
"CC-BY-4.0"
]
| null | null | null | part-4.ipynb | willingc/intro-to-python | 9822315d28c603b17d7475c09925146d704a571c | [
"CC-BY-4.0"
]
| null | null | null | 24.043684 | 497 | 0.489123 | [
[
[
"empty"
]
]
]
| [
"empty"
]
| [
[
"empty"
]
]
|
e796a857250f527529fd444e931eb38151a3d7d1 | 489,178 | ipynb | Jupyter Notebook | Project_2/Spotify/Spotify_Solutions.ipynb | ds-modules/BUDS-SU21-Dev | 04c8dd0973efec728b22916767a1a7396b3baece | [
"BSD-3-Clause"
]
| 3 | 2021-06-14T19:27:45.000Z | 2021-06-21T03:19:08.000Z | Project_2/Spotify/Spotify_Solutions.ipynb | ds-modules/BUDS-SU21-Dev | 04c8dd0973efec728b22916767a1a7396b3baece | [
"BSD-3-Clause"
]
| null | null | null | Project_2/Spotify/Spotify_Solutions.ipynb | ds-modules/BUDS-SU21-Dev | 04c8dd0973efec728b22916767a1a7396b3baece | [
"BSD-3-Clause"
]
| 1 | 2021-07-06T00:01:44.000Z | 2021-07-06T00:01:44.000Z | 229.230553 | 121,992 | 0.833508 | [
[
[
"#Run this cell to install the necessary dependencies\nimport pandas as pd\nimport numpy as np\nfrom datascience import *\n\n%matplotlib inline\nimport matplotlib.pyplot as plt\nplt.style.use('fivethirtyeight')",
"_____no_output_____"
]
],
[
[
"# Project 2: Spotify",
"_____no_output_____"
],
[
"## Table of Contents\n<a href='#section 0'>Background Knowledge: Topic</a>\n\n1. <a href='#section 1'> The Data Science Life Cycle</a>\n\n a. <a href='#subsection 1a'>Formulating a question or problem</a> \n\n b. <a href='#subsection 1b'>Acquiring and cleaning data</a>\n\n c. <a href='#subsection 1c'>Conducting exploratory data analysis</a>\n\n d. <a href='#subsection 1d'>Using prediction and inference to draw conclusions</a>\n<br><br>",
"_____no_output_____"
],
[
"### Background Knowledge <a id='section 0'></a>\n",
"_____no_output_____"
],
[
"If you listen to music, chances are you use Spotify, Apple Music, or another similar streaming service. This new era of the music industry curates playlists, recommends new artists, and is based on the number of streams more than the number of albums sold. The way these streaming services do this is (you guessed it) data!\n\nSpotify, like many other companies, hire many full-time data scientists to analyze all the incoming user data and use it to make predictions and recommendations for users. If you're interested, feel free to check out [Spotify's Engineering Page](https://engineering.atspotify.com/) for more information!",
"_____no_output_____"
],
[
"<img src=\"images/spotify.png\" width = 700/>\n\n<center><a href=https://hrblog.spotify.com/2018/02/08/amping-up-diversity-inclusion-at-spotify/>Image Reference</a></center> ",
"_____no_output_____"
],
[
"# The Data Science Life Cycle <a id='section 1'></a>",
"_____no_output_____"
],
[
"## Formulating a Question or Problem <a id='subsection 1a'></a>\nIt is important to ask questions that will be informative and can be answered using the data. There are many different questions we could ask about music data. For example, there are many artists who want to find out how to get their music on Spotify's Discover Weekly playlist in order to gain exposure. Similarly, users love to see their *Spotify Wrapped* listening reports at the end of each year.",
"_____no_output_____"
],
[
"<div class=\"alert alert-warning\">\n<b>Question:</b> Recall the questions you developed with your group on Tuesday. Write down that question below, and try to add on to it with the context from the articles from Wednesday. Think about what data you would need to answer your question. You can review the articles on the bCourses page under Module 4.3.\n </div>",
"_____no_output_____"
],
[
"**Original Question(s):** *here*\n\n\n**Updated Question(s):** *here*\n\n\n**Data you would need:** *here*\n",
"_____no_output_____"
],
[
"## Acquiring and Cleaning Data <a id='subsection 1b'></a>\n\nWe'll be looking at song data from Spotify. You can find the raw data [here](https://github.com/rfordatascience/tidytuesday/tree/master/data/2020/2020-01-21). We've cleaned up the datasets a bit, and we will be investigating the popularity and the qualities of songs from this dataset.\n\nThe following table, `spotify`, contains a list of tracks identified by their unique song ID along with attributes about that track.\n\nHere are the descriptions of the columns for your reference. (We will not be using all of these fields):\n\n|Variable Name | Description |\n|--------------|------------|\n|`track_id` | \tSong unique ID |\n|`track_name` | Song Name |\n|`track_artist\t`| Song Artist |\n|`track_popularity` | Song Popularity (0-100) where higher is better |\n|`track_album_id`| Album unique ID |\n|`track_album_name` | Song album name |\n|`track_album_release_date`| Date when album released |\n|`playlist_name`| Name of playlist |\n|`playlist_id`| Playlist ID |\n|`playlist_genre`| Playlist genre |\n|`playlist_subgenre\t`| Playlist subgenre |\n|`danceability`| Danceability describes how suitable a track is for dancing based on a combination of musical elements including tempo, rhythm stability, beat strength, and overall regularity. A value of 0.0 is least danceable and 1.0 is most danceable. |\n|`energy`| Energy is a measure from 0.0 to 1.0 and represents a perceptual measure of intensity and activity. Typically, energetic tracks feel fast, loud, and noisy. For example, death metal has high energy, while a Bach prelude scores low on the scale. Perceptual features contributing to this attribute include dynamic range, perceived loudness, timbre, onset rate, and general entropy. |\n|`key`| The estimated overall key of the track. Integers map to pitches using standard Pitch Class notation . E.g. 0 = C, 1 = C♯/D♭, 2 = D, and so on. If no key was detected, the value is -1. |\n|`loudness`| The overall loudness of a track in decibels (dB). Loudness values are averaged across the entire track and are useful for comparing relative loudness of tracks. Loudness is the quality of a sound that is the primary psychological correlate of physical strength (amplitude). Values typical range between -60 and 0 db. |\n|`mode`| Mode indicates the modality (major or minor) of a track, the type of scale from which its melodic content is derived. Major is represented by 1 and minor is 0. |\n|`speechiness`| Speechiness detects the presence of spoken words in a track. The more exclusively speech-like the recording (e.g. talk show, audio book, poetry), the closer to 1.0 the attribute value. Values above 0.66 describe tracks that are probably made entirely of spoken words. Values between 0.33 and 0.66 describe tracks that may contain both music and speech, either in sections or layered, including such cases as rap music. Values below 0.33 most likely represent music and other non-speech-like tracks. |\n|`acousticness`| A confidence measure from 0.0 to 1.0 of whether the track is acoustic. 1.0 represents high confidence the track is acoustic. |\n|`instrumentalness`| Predicts whether a track contains no vocals. “Ooh” and “aah” sounds are treated as instrumental in this context. Rap or spoken word tracks are clearly “vocal”. The closer the instrumentalness value is to 1.0, the greater likelihood the track contains no vocal content. Values above 0.5 are intended to represent instrumental tracks, but confidence is higher as the value approaches 1.0. |\n|`liveness`| Detects the presence of an audience in the recording. Higher liveness values represent an increased probability that the track was performed live. A value above 0.8 provides strong likelihood that the track is live. |\n|`valence`| A measure from 0.0 to 1.0 describing the musical positiveness conveyed by a track. Tracks with high valence sound more positive (e.g. happy, cheerful, euphoric), while tracks with low valence sound more negative (e.g. sad, depressed, angry). |\n|`tempo`| The overall estimated tempo of a track in beats per minute (BPM). In musical terminology, tempo is the speed or pace of a given piece and derives directly from the average beat duration. |\n|`duration_ms`| Duration of song in milliseconds |\n|`creation_year`| Year when album was released |\n\n\n",
"_____no_output_____"
]
],
[
[
"spotify = Table.read_table('data/spotify.csv')\nspotify.show(10)",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-info\">\n<b>Question:</b> It's important to evalute our data source. What do you know about the source? What motivations do they have for collecting this data? What data is missing?\n </div>",
"_____no_output_____"
],
[
"*Insert answer here*",
"_____no_output_____"
],
[
"<div class=\"alert alert-info\">\n<b>Question:</b> Do you see any missing (nan) values? Why might they be there?\n </div>",
"_____no_output_____"
],
[
"*Insert answer here*",
"_____no_output_____"
],
[
"<div class=\"alert alert-info\">\n<b>Question:</b> We want to learn more about the dataset. First, how many total rows are in this table? What does each row represent?\n \n </div>",
"_____no_output_____"
]
],
[
[
"total_rows = spotify.num_rows\ntotal_rows",
"_____no_output_____"
]
],
[
[
"*Insert answer here*",
"_____no_output_____"
],
[
"## Conducting Exploratory Data Analysis <a id='subsection 1c'></a>",
"_____no_output_____"
],
[
"Visualizations help us to understand what the dataset is telling us. We will be using bar charts, scatter plots, and line plots to try to answer questions like the following:\n> What audio features make a song popular and which artists have these songs? How have features changed over time?",
"_____no_output_____"
],
[
"### Part 1: We'll start by looking at the length of songs using the `duration_ms` column.",
"_____no_output_____"
],
[
"Right now, the `duration` array contains the length of each song in milliseconds. However, that's not a common measurement when describing the length of a song - often, we use minutes and seconds. Using array arithmetic, we can find the length of each song in seconds and in minutes. There are 1000 milliseconds in a second, and 60 seconds in a minute. First, we will convert milliseconds to seconds.\n",
"_____no_output_____"
]
],
[
[
"#Access the duration column as an array.\nduration = spotify.column(\"duration_ms\")\nduration",
"_____no_output_____"
],
[
"#Divide the milliseconds by 1000\nduration_seconds = duration / 1000\nduration_seconds",
"_____no_output_____"
],
[
"#Now convert duration_seconds to minutes.\nduration_minutes = duration_seconds / 60\nduration_minutes ",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-info\">\n<b>Question:</b> How would we find the average duration (in minutes) of the songs in this dataset?\n </div>",
"_____no_output_____"
]
],
[
[
"avg_song_length_mins = np.mean(duration_minutes)\navg_song_length_mins",
"_____no_output_____"
]
],
[
[
"Now, we can add in the duration for each song (in minutes) by adding a column to our `spotify` table called `duration_min`. Run the following cell to do so.",
"_____no_output_____"
]
],
[
[
"#This cell will add the duration in minutes column we just created to our dataset.\nspotify = spotify.with_columns('duration_min', duration_minutes)\nspotify",
"_____no_output_____"
]
],
[
[
"### Artist Comparison",
"_____no_output_____"
],
[
"Let's see if we can find any meaningful difference in the average length of song for different artists.",
"_____no_output_____"
],
[
"<div class=\"alert alert-success\">\n <b>Note: </b>Now that we have the average duration for each song, you can compare average song length between two artists. Below is an example!\n </div>",
"_____no_output_____"
]
],
[
[
"sam_smith = spotify.where(\"track_artist\", are.equal_to(\"Sam Smith\"))\nsam_smith",
"_____no_output_____"
],
[
"sam_smith_mean = sam_smith.column(\"duration_min\").mean()\nsam_smith_mean",
"_____no_output_____"
],
[
"#In this cell, choose an artist you want to look at.\nartist_name = spotify.where(\"track_artist\", \"Kanye West\").column(\"duration_min\").mean()\nartist_name",
"_____no_output_____"
],
[
"#In this cell, choose another artist you want to compare it to.\nartist_name_2 = spotify.where(\"track_artist\", \"Justin Bieber\").column(\"duration_min\").mean()\nartist_name_2",
"_____no_output_____"
]
],
[
[
"This exercise was just one example of how you can play around with data and answer questions.",
"_____no_output_____"
],
[
"### Top Genres and Artists\nIn this section, we are interested in the categorical information in our dataset, such as the playlist each song comes from or the genre. There are almost 33,000 songs in our dataset, so let's do some investigating. What are the most popular genres? We can figure this out by grouping by the playlist genre.",
"_____no_output_____"
],
[
"<div class=\"alert alert-info\">\n<b>Question:</b> How can we group our data by unique genres?\n </div>",
"_____no_output_____"
]
],
[
[
"genre_counts = spotify.group('playlist_genre')\ngenre_counts",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-info\">\n<b>Question:</b> In our dataset, it looks like the most popular genre is EDM. Make a barchart below to show how the other genres compare.\n </div>",
"_____no_output_____"
]
],
[
[
"genre_counts.barh('playlist_genre', 'count')",
"_____no_output_____"
]
],
[
[
"Notice that it was difficult to analyze the above bar chart because the data wasn't sorted first. Let's sort our data and make a new bar chart so that it is much easier to make comparisons.",
"_____no_output_____"
]
],
[
[
"genre_counts_sorted = genre_counts.sort('count', descending = True)\ngenre_counts_sorted",
"_____no_output_____"
],
[
"genre_counts_sorted.barh('playlist_genre', 'count')",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-info\">\n<b>Question:</b> Was this what you expected? Which genre did you think would be the most popular?\n </div>",
"_____no_output_____"
],
[
"*Insert answer here.*",
"_____no_output_____"
],
[
"<div class=\"alert alert-info\">\n<b>Question:</b> Let's take a look at all the artists in the dataset. We can take a look at the top 25 artists based on the number of songs they have in our dataset. We'll follow a similar method as we did when grouping by genre above. First, we will group our data by artist and sort by count.\n </div>",
"_____no_output_____"
]
],
[
[
"#Here, we will group and sort in the same line.\n\nartists_grouped = spotify.group('track_artist').sort('count', descending=True)\nartists_grouped",
"_____no_output_____"
],
[
"top_artists = artists_grouped.take(np.arange(0, 25))\ntop_artists",
"_____no_output_____"
],
[
"top_artists.barh('track_artist', 'count')",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-info\">\n<b>Question:</b> What do you notice about the top 25 artists in our dataset?\n </div>",
"_____no_output_____"
],
[
"*insert answer here*",
"_____no_output_____"
],
[
"### Playlist Popularity",
"_____no_output_____"
],
[
"In our dataset, each song is listed as belonging to a particular playlist, and each song is given a \"popularity score\", called the `track_popularity`. Using the `track_popularity`, we can calculate an *aggregate popularity* for each playlist, which is just the sum of all the popularity scores for the songs on the playlist.\n\nIn order to create this aggregate popularity score, we need to group our data by playlist, and sum all of the popularity scores. First, we will create a subset of our `spotify` table using the `select` method. This lets us create a table with only the relevant columns we want. In this case, we only care about the name of the playlist and the popularity of each track. Keep in mind that each row still represents one track, even though we no longer have the track title in our table.",
"_____no_output_____"
]
],
[
[
"spotify_subset = spotify.select(['playlist_name', 'track_popularity'])\nspotify_subset",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-success\">\n<b>Note:</b> By grouping, we can get the number of songs from each playlist.\n </div>",
"_____no_output_____"
]
],
[
[
"playlists = spotify_subset.group('playlist_name')\nplaylists",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-info\">\n <b>Question:</b> We can use the <code>group</code> method again, this time passing in a second argument <code>collect</code>, which says that we want to take the sum rather than the count when grouping. This results in a table with the total aggregate popularity of each playlist.\n </div>",
"_____no_output_____"
]
],
[
[
"#Run this cell.\ntotal_playlist_popularity = spotify_subset.group('playlist_name', collect = sum)\ntotal_playlist_popularity",
"_____no_output_____"
]
],
[
[
"Similar to when we found duration in minutes, we can once again use the `column` method to access just the `track_popularity sum` column, and add it to our playlists table using the `with_column` method.",
"_____no_output_____"
]
],
[
[
"agg_popularity = total_playlist_popularity.column('track_popularity sum')\nplaylists = playlists.with_column('aggregate_popularity', agg_popularity)\nplaylists",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-info\">\n<b>Question:</b> Do you think that the most popular playlist would be the one with the highest aggregate_popularity score, or the one with the highest number of songs? We can sort our playlists table and compare the outputs.",
"_____no_output_____"
]
],
[
[
"playlists.sort('count', descending=True)",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-info\">\n<b>Question:</b> Now sort by aggregate popularity.\n </div>",
"_____no_output_____"
]
],
[
[
"playlists.sort('aggregate_popularity', descending=True)",
"_____no_output_____"
]
],
[
[
"Comparing these two outputs shows us that the \"most popular playlist\" depends on how we judge popularity. If we have a playlist that has only a few songs, but each of those songs are really popular, should that playlist be higher on the popularity rankings? By way of calculation, playlists with more songs will have a higher aggregate popularity, since more popularity values are being added together. We want a metric that will let us judge the actual quality and popularity of a playlist, not just how many songs it has.\n\nIn order to take into account the number of songs on each playlist, we can calculate the \"average popularity\" of each song on the playlist, or the proportion of aggregate popularity that each song takes up. We can do this by dividing `aggregate_popularity` by `count`. Remember, since the columns are just arrays, we can use array arithmetic to calculate these values.",
"_____no_output_____"
]
],
[
[
"#Run this cell to get the average.\navg_popularity = playlists.column('aggregate_popularity') / playlists.column('count')",
"_____no_output_____"
],
[
"#Now add it to the playlists table.\nplaylists = playlists.with_column('average_popularity', avg_popularity)\nplaylists",
"_____no_output_____"
]
],
[
[
"Let's see if our \"most popular playlist\" changes when we judge popularity by the average popularity of the songs on a playlist.",
"_____no_output_____"
]
],
[
[
"playlists.sort('average_popularity', descending=True)",
"_____no_output_____"
]
],
[
[
"Looking at the table above, we notice that 8/10 of the top 10 most popular playlists by the `average_popularity` metric are playlists with less than 100 songs. Just because a playlist has a lot of songs, or a high aggregate popularity, doesn't mean that the average popularity of a song on that playlist is high. Our new metric of `average_popularity` lets us rank playlists where the size of a playlist has no effect on it's overall score. We can visualize the top 25 playlists by average popularity in a bar chart.",
"_____no_output_____"
]
],
[
[
"top_25_playlists = playlists.sort('average_popularity', descending=True).take(np.arange(25))\ntop_25_playlists.barh('playlist_name', 'average_popularity')",
"_____no_output_____"
]
],
[
[
"Creating a new metric like `average_popularity` helps us more accurately and fairly measure the popularity of a playlist. \n\nWe saw before when looking at the top 25 artists that they were all male. Now looking at the top playlists, we see that the current landscape of popular playlists and music may have an effect on the artists that are popular. For example, the RapCaviar is the second most popular playlist, and generally there tends to be fewer female rap artists than male. This shows that the current landscape of popular music can affect the types of artists topping the charts.",
"_____no_output_____"
],
[
"## Using prediction and inference to draw conclusions <a id='subsection 1a'></a>",
"_____no_output_____"
],
[
"Now that we have some experience making these visualizations, let's go back to the visualizations others are working on to analyze Spotify data using more complex techniques.\n\n[Streaming Dashboard](https://public.tableau.com/profile/vaibhavi.gaekwad#!/vizhome/Spotify_15858686831320/Dashboard1)\n\n[Audio Analysis Visualizer](https://developer.spotify.com/community/showcase/spotify-audio-analysis/)",
"_____no_output_____"
],
[
"Music and culture are very intertwined so it's interesting to look at when songs are released and what is popular during that time. In this last exercise, you will be looking at the popularity of artists and tracks based on the dates you choose.\n\nLet's look back at the first five rows of our `spotify` table once more.",
"_____no_output_____"
]
],
[
[
"spotify.show(5)",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-info\">\n <b>Question:</b> Fill in the following cell the data according to the <code>creation_year</code> you choose.\n </div>",
"_____no_output_____"
]
],
[
[
"#Fill in the year as an integer.\nby_year = spotify.where(\"creation_year\", are.equal_to(2018))\nby_year",
"_____no_output_____"
]
],
[
[
"Based on the dataset you have now, use previous techniques to find the most popular song during that year. First group by what you want to look at, for example, artist/playlist/track.",
"_____no_output_____"
]
],
[
[
"your_grouped = by_year.group(\"playlist_name\")\npop_track = your_grouped.sort(\"count\", descending = True)\npop_track",
"_____no_output_____"
],
[
"pop_track.take(np.arange(25)).barh(\"playlist_name\", \"count\")",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-info\">\n<b>Question:</b> Finally, use this cell if you want to look at the popularity of a track released on a specific date. It's very similar to the process above.\n </div>",
"_____no_output_____"
]
],
[
[
"by_date = spotify.where(\"track_album_release_date\", are.equal_to(\"2019-06-14\"))\nyour_grouped = by_date.group(\"track_artist\")\npop_track = your_grouped.sort(\"count\", descending = True)\npop_track.take(np.arange(10)).barh(\"track_artist\", \"count\")",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-info\">\n<b>Question:</b> Tell us something interesting about this data.\n </div>",
"_____no_output_____"
],
[
"*Insert answer here.*",
"_____no_output_____"
],
[
"Notebook Authors: Alleanna Clark",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
]
|
e796b76bd81d8017597339f0644ac8a5ac925df9 | 148,761 | ipynb | Jupyter Notebook | nbs/22_01_02__LTH_Data_Diet_2_Pass_Initial.ipynb | mansheej/open_lth | f835e97f67f46fb189825b643aa686bbc1064b64 | [
"MIT"
]
| null | null | null | nbs/22_01_02__LTH_Data_Diet_2_Pass_Initial.ipynb | mansheej/open_lth | f835e97f67f46fb189825b643aa686bbc1064b64 | [
"MIT"
]
| null | null | null | nbs/22_01_02__LTH_Data_Diet_2_Pass_Initial.ipynb | mansheej/open_lth | f835e97f67f46fb189825b643aa686bbc1064b64 | [
"MIT"
]
| null | null | null | 1,070.223022 | 144,694 | 0.952192 | [
[
[
"# [01/02/22] LTH on a Data Diet -- 2 Pass Initial Results",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nfrom pathlib import Path\nimport seaborn as sns\nplt.style.use('default')\nsns.set_theme(\n style='ticks',\n font_scale=1.2,\n rc={\n 'axes.linewidth': '0.8',\n 'axes.grid': True,\n 'figure.constrained_layout.use': True,\n 'grid.linewidth': '0.8',\n 'legend.edgecolor': '1.0',\n 'legend.fontsize': 'small',\n 'legend.title_fontsize': 'small',\n 'xtick.major.width': '0.8',\n 'ytick.major.width': '0.8'\n },\n)",
"_____no_output_____"
]
],
[
[
"## Figure 0016",
"_____no_output_____"
]
],
[
[
"exp_meta_paths = [\n Path(f'/home/mansheej/open_lth_data/lottery_b279562b990bac9b852b17b287fca1ef/'),\n Path(f'/home/mansheej/open_lth_data/lottery_78a119e24960764e0de0964887d2597f/'),\n Path(f'/home/mansheej/open_lth_data/lottery_2cb77ad7e940a06d07b04a4b63fd718d/'),\n Path(f'/home/mansheej/open_lth_data/lottery_511428cbe43275064244db39edd0f60f/'),\n]\nexp_paths = [[emp / f'replicate_{i}' for i in range(1, 5)] for emp in exp_meta_paths]\n\nplt.figure(figsize=(8.4, 4.8))\nls = []\n\nfor i, eps in enumerate(exp_paths):\n num_levels = 15\n acc_run_level = []\n for p in eps:\n acc_level = []\n for l in range(num_levels + 1):\n df = pd.read_csv(p / f'level_{l}/main/logger', header=None)\n acc_level.append(df[2].iloc[-2])\n acc_run_level.append(acc_level)\n acc_run_level = np.array(acc_run_level)\n x = np.arange(16)\n ys = acc_run_level\n y_mean, y_std = ys.mean(0), ys.std(0)\n c = f'C{i}'\n l = plt.plot(x, y_mean, c=c, alpha=0.9, linewidth=2)\n ls.append(l[0])\n plt.fill_between(x, y_mean + y_std, y_mean - y_std, color=c, alpha=0.2)\n\nplt.legend(\n ls, \n [ \n 'Pre-train 1 Pass -> All 50000 Examples',\n 'Pre-train 2 Passes -> All 50000 Examples',\n 'Pre-train 2 Passes -> 32000 Smallest Scores at Epoch 3',\n 'Pre-train 2 Passes -> 12800 Smallest Scores at Epoch 3',\n ],\n)\nplt.xlim(0, 15)\nplt.ylim(0.815, 0.925)\nplt.xticks(np.arange(0, 16, 2), [f'{f*100:.1f}' for f in 0.8**np.arange(0, 16, 2)])\nplt.xlabel('% Weights Remaining')\nplt.ylabel('Test Accuracy')\nplt.title('CIFAR10 ResNet20: Pre-train LR = 0.4')\nsns.despine()\nplt.savefig('/home/mansheej/open_lth/figs/0016.svg')\nplt.show()",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
e796bc2787a229267cfacd5dc2c4f95effed077a | 44,822 | ipynb | Jupyter Notebook | tutorials/NBandLR.ipynb | oeclint/BIDMach | 999f6b0a2317c8869151b253e66a0cc5b0fb9841 | [
"BSD-3-Clause-No-Nuclear-License-2014",
"BSD-3-Clause"
]
| 826 | 2015-01-01T04:08:08.000Z | 2022-03-07T04:43:28.000Z | tutorials/NBandLR.ipynb | oeclint/BIDMach | 999f6b0a2317c8869151b253e66a0cc5b0fb9841 | [
"BSD-3-Clause-No-Nuclear-License-2014",
"BSD-3-Clause"
]
| 128 | 2015-01-06T02:14:42.000Z | 2020-11-25T01:03:14.000Z | tutorials/NBandLR.ipynb | oeclint/BIDMach | 999f6b0a2317c8869151b253e66a0cc5b0fb9841 | [
"BSD-3-Clause-No-Nuclear-License-2014",
"BSD-3-Clause"
]
| 190 | 2015-01-08T07:14:56.000Z | 2022-01-22T08:14:37.000Z | 47.131441 | 1,050 | 0.503369 | [
[
[
"# Naive Bayes and Logistic Regression",
"_____no_output_____"
],
[
"In this tutorial, we'll explore training and evaluation of Naive Bayes and Logitistic Regression Classifiers.\n\nTo start, we import the standard BIDMach class definitions. ",
"_____no_output_____"
]
],
[
[
"import $exec.^.lib.bidmach_notebook_init",
"1 CUDA device found, CUDA version 8.0\n"
]
],
[
[
"Now we load some training and test data, and some category labels. The data come from a news collection from Reuters, and is a \"classic\" test set for classification. Each article belongs to one or more of 103 categories. The articles are represented as Bag-of-Words (BoW) column vectors. For a data matrix A, element A(i,j) holds the count of word i in document j. \n\nThe category matrices have 103 rows, and a category matrix C has a one in position C(i,j) if document j is tagged with category i, or zero otherwise. \n\nTo reduce the computing time and memory footprint, the training data have been sampled. The full collection has about 700k documents. Our training set has 60k. \n\nSince the document matrices contain counts of words, we use a min function to limit the count to \"1\", i.e. because we need binary features for naive Bayes. ",
"_____no_output_____"
]
],
[
[
"val dict = \"../data/rcv1/\"\nval traindata = loadSMat(dict+\"docs.smat.lz4\")\nval traincats = loadFMat(dict+\"cats.fmat.lz4\")\nval testdata = loadSMat(dict+\"testdocs.smat.lz4\")\nval testcats = loadFMat(dict+\"testcats.fmat.lz4\")\nmin(traindata, 1, traindata) // the first \"traindata\" argument is the input, the other is output\nmin(testdata, 1, testdata)",
"_____no_output_____"
]
],
[
[
"Get the word and document counts from the data. This turns out to be equivalent to a matrix multiply. For a data matrix A and category matrix C, we want all (cat, word) pairs (i,j) such that C(i,k) and A(j,k) are both 1 - this means that document k contains word j, and is also tagged with category i. Summing over all documents gives us\n\n$${\\rm wordcatCounts(i,j)} = \\sum_{k=1}^N C(i,k) A(j,k) = C * A^T$$\n\n\nBecause we are doing independent binary classifiers for each class, we need to construct the counts for words not in the class (negwcounts).\n\nFinally, we add a smoothing count 0.5 to counts that could be zero.",
"_____no_output_____"
]
],
[
[
"val truecounts = traincats *^ traindata\nval wcounts = truecounts + 0.5\nval negwcounts = sum(truecounts) - truecounts + 0.5\nval dcounts = sum(traincats,2)",
"_____no_output_____"
]
],
[
[
"Now compute the probabilities \n* pwordcat = probability that a word is in a cat, given the cat.\n* pwordncat = probability of a word, given the complement of the cat.\n* pcat = probability that doc is in a given cat. \n* spcat = sum of pcat probabilities (> 1 because docs can be in multiple cats)",
"_____no_output_____"
]
],
[
[
"val pwordcat = wcounts / sum(wcounts,2) // Normalize the rows to sum to 1.\nval pwordncat = negwcounts / sum(negwcounts,2) // Each row represents word probabilities conditioned on one cat. \nval pcat = dcounts / traindata.ncols\nval spcat = sum(pcat)",
"_____no_output_____"
]
],
[
[
"Now take the logs of those probabilities. Here we're using the formula presented <a href=\"https://bcourses.berkeley.edu/courses/1267848/files/51512989/download?wrap=1in\">here</a> to match Naive Bayes to Logistic Regression for independent data.\n\nFor each word, we compute the log of the ratio of the complementary word probability over the in-class word probability. \n\nFor each category, we compute the log of the ratio of the complementary category probability over the current category probability.\n\nlpwordcat(j,i) represents $\\log\\left(\\frac{{\\rm Pr}(X_i|\\neg c_j)}{{\\rm Pr}(X_i|c_j)}\\right)$\n\nwhile lpcat(j) represents $\\log\\left(\\frac{{\\rm Pr}(\\neg c)}{{\\rm Pr}(c)}\\right)$",
"_____no_output_____"
]
],
[
[
"val lpwordcat = ln(pwordncat/pwordcat) // ln is log to the base e (natural log)\nval lpcat = ln((spcat-pcat)/pcat)",
"_____no_output_____"
]
],
[
[
"Here's where we apply Naive Bayes. The formula we're using is borrowed from <a href=\"https://bcourses.berkeley.edu/courses/1267848/files/51512989/download?wrap=1in\">here</a>.\n\n$${\\rm Pr}(c|X_1,\\ldots,X_k) = \\frac{1}{1 + \\frac{{\\rm Pr}(\\neg c)}{{\\rm Pr}(c)}\\prod_{i-1}^k\\frac{{\\rm Pr}(X_i|\\neg c)}{{\\rm Pr}(X_i|c)}}$$\n\nand we can rewrite\n\n$$\\frac{{\\rm Pr}(\\neg c)}{{\\rm Pr}(c)}\\prod_{i-1}^k\\frac{{\\rm Pr}(X_i|\\neg c)}{{\\rm Pr}(X_i|c)}$$\n\nas\n\n$$\\exp\\left(\\log\\left(\\frac{{\\rm Pr}(\\neg c)}{{\\rm Pr}(c)}\\right) + \\sum_{i=1}^k\\log\\left(\\frac{{\\rm Pr}(X_i|\\neg c)}{{\\rm Pr}(X_i|c)}\\right)\\right) = \\exp({\\rm lpcat(j)} + {\\rm lpwordcat(j,?)} * X)$$\n\nfor class number j and an input column $X$. This follows because an input column $X$ is a sparse vector with ones in the positions of the input features. The product ${\\rm lpwordcat(i,?)} * X$ picks out the features occuring in the input document and adds the corresponding logs from lpwordcat. \n\nFinally, we take the exponential above and fold it into the formula $P(c_j|X_1,\\ldots,X_k) = 1/(1+\\exp(\\cdots))$. This gives us a matrix of predictions. preds(i,j) = prediction of membership in category i for test document j. ",
"_____no_output_____"
]
],
[
[
"val logodds = lpwordcat * testdata + lpcat\nval preds = 1 / (1 + exp(logodds))",
"_____no_output_____"
]
],
[
[
"To measure the accuracy of the predictions above, we can compute the probability that the classifier outputs the right label. We used this formula in class for the expected accuracy for logistic regression. The \"dot arrow\" operator takes dot product along rows:",
"_____no_output_____"
]
],
[
[
"val acc = ((preds ∙→ testcats) + ((1-preds) ∙→ (1-testcats)))/preds.ncols\nacc.t",
"_____no_output_____"
]
],
[
[
"Raw accuracy is not a good measure in most cases. When there are few positives (instances in the class vs. its complement), accuracy simply drives down false-positive rate at the expense of false-negative rate. In the worst case, the learner may always predict \"no\" and still achieve high accuracy. \n\nROC curves and ROC Area Under the Curve (AUC) are much better. Here we compute the ROC curves from the predictions above. We need:\n* scores - the predicted quality from the formula above.\n* good - 1 for positive instances, 0 for negative instances.\n* bad - complement of good. \n* npoints (100) - specifies the number of X-axis points for the ROC plot. \n\nitest specifies which of the categories to plot for. We chose itest=6 because that category has one of the highest positive rates, and gives the most stable accuracy plots. ",
"_____no_output_____"
]
],
[
[
"val itest = 6\nval scores = preds(itest,?)\nval good = testcats(itest,?)\nval bad = 1-testcats(itest,?)\nval rr =roc(scores,good,bad,100)",
"_____no_output_____"
]
],
[
[
"> TODO 1: In the cell below, write an expression to derive the ROC Area under the curve (AUC) given the curve rr. rr gives the ROC curve y-coordinates at 100 evenly-spaced X-values from 0 to 1.0. ",
"_____no_output_____"
]
],
[
[
"// auc = ",
"_____no_output_____"
]
],
[
[
"> TODO 2: In the cell below, write the value of AUC returned by the expression above.",
"_____no_output_____"
],
[
"## Logistic Regression",
"_____no_output_____"
],
[
"Now lets train a logistic classifier on the same data. BIDMach has an umbrella classifier called GLM for Generalized Linear Model. GLM includes linear regression, logistic regression (with log accuracy or direct accuracy optimization), and SVM. \n\nThe learner function accepts these arguments:\n* traindata: the training data in the same format as for Naive Bayes\n* traincats: the training category labels\n* testdata: the test input data\n* predcats: a container for the predictions generated by the model\n* modeltype (GLM.logistic here): an integer that specifies the type of model (0=linear, 1=logistic log accuracy, 2=logistic accuracy, 3=SVM). \n\nWe'll construct the learner and then look at its options:",
"_____no_output_____"
]
],
[
[
"val predcats = zeros(testcats.nrows, testcats.ncols)\nval (mm,mopts) = GLM.learner(traindata, traincats, GLM.maxp)\nmopts.what",
"Option Name Type Value\n=========== ==== =====\naddConstFeat boolean false\naopts Opts null\nautoReset boolean true\nbatchSize int 10000\ncheckPointFile String null\ncheckPointInterval float 0.0\nclipByValue float -1.0\ncumScore int 0\ndebug int 0\ndebugCPUmem boolean false\ndebugMem boolean false\ndim int 256\ndoAllReduce boolean false\ndoubleScore boolean false\ndoVariance boolean false\nepsilon float 1.0E-5\nevalStep int 11\nfeatThreshold Mat null\nfeatType int 1\ngsq_decay float -1.0\nhashBound1 int 1000000\nhashBound2 int 1000000\nhashFeatures int 0\ninitsumsq float 1.0E-5\niweight FMat null\nl2reg FMat null\nlangevin float 0.0\nlim float 0.0\nlinks IMat 2,2,2,2,2,2,2,2,2,2,...\nlogDataSink DataSink null\nlogfile String log.txt\nlogFuncs Function2[] null\nlr_policy Function3 null\nlrate FMat 1\nmask FMat null\nmatrixOfScores boolean false\nmax_grad_norm float -1.0\nmixinInterval int 1\nnaturalLambda float 0.0\nnesterov_vel_decay FMat null\nnNatural int 1\nnpasses int 2\nnzPerColumn int 0\npauseAt long -1\npexp FMat 0\npstep float 0.01\nputBack int -1\nr1nmats int 1\nreg1weight FMat 1.0000e-07\nresFile String null\nrmask FMat null\nsample float 1.0\nsizeMargin float 3.0\nstartBlock int 8000\ntargets FMat null\ntargmap FMat null\ntexp FMat 0.50000\ntrace int 0\nupdateAll boolean false\nuseCache boolean true\nuseDouble boolean false\nuseGPU boolean true\nuseGPUcache boolean true\nvel_decay FMat null\nvexp FMat 0.50000\nwaitsteps int 3\n"
]
],
[
[
"The most important options are:\n* lrate: the learning rate\n* batchSize: the minibatch size\n* npasses: the number of passes over the dataset\n\nWe'll use the following parameters for this training run. ",
"_____no_output_____"
]
],
[
[
"mopts.lrate=1.0\nmopts.batchSize=1000\nmopts.npasses=2\nmm.train",
"corpus perplexity=81528.088805\n"
],
[
"val (nn, nopts) = GLM.predictor(mm.model, testdata)\n\nnn.predict",
"_____no_output_____"
],
[
"val predcats = FMat(nn.preds(0))\nval lacc = (predcats ∙→ testcats + (1-predcats) ∙→ (1-testcats))/preds.ncols\nlacc.t\nmean(lacc)",
"_____no_output_____"
]
],
[
[
"Since we have the accuracy scores for both Naive Bayes and Logistic regression, we can plot both of them on the same axes. Naive Bayes is red, Logistic regression is blue. The x-axis is the category number from 0 to 102. The y-axis is the absolute accuracy of the predictor for that category. ",
"_____no_output_____"
]
],
[
[
"val axaxis = row(0 until 103)\nplot(axaxis, acc, axaxis, lacc)",
"_____no_output_____"
]
],
[
[
"> TODO 3: With the full training set (700k training documents), Logistic Regression is noticeably more accurate than Naive Bayes in every category. What do you observe in the plot above? Why do you think this is?",
"_____no_output_____"
],
[
"Next we'll compute the ROC plot and ROC area (AUC) for Logistic regression for category itest.",
"_____no_output_____"
]
],
[
[
"val lscores = predcats(itest,?)\nval lrr =roc(lscores,good,bad,100)\nval auc = mean(lrr) // Fill in using the formula you used before",
"_____no_output_____"
]
],
[
[
"We computed the ROC curve for Naive Bayes earlier, so now we can plot them on the same axes. Naive Bayes is once again in red, Logistic regression in blue. ",
"_____no_output_____"
]
],
[
[
"val rocxaxis = row(0 until 101)\nplot(rocxaxis, rr, rocxaxis, lrr)",
"_____no_output_____"
]
],
[
[
">TODO 4: In the cell below, compute and plot lift curves from the ROC curves for Naive Bayes and Logistic regression. The lift curves should show the ratio of ROC y-values over a unit slope diagonal line (Y=X). The X-values should be the same as for the ROC plots, except that X=0 will be omitted since the lift will be undefined. ",
"_____no_output_____"
],
[
"> TODO 5: Experiment with different values for learning rate and batchSize to get the best performance for absolute accuracy and ROC area on category 6. Write your optimal values below:",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
]
|
e796c09d319ac962ed1da81fb4c9b6552b66e643 | 58,640 | ipynb | Jupyter Notebook | Census_income.ipynb | umairnsr87/deploying-ml-model-with-django | b2e4c9f0e7e4618f5bf2cd8e5edaefe35e1e2235 | [
"Apache-2.0"
]
| 1 | 2020-09-21T08:32:14.000Z | 2020-09-21T08:32:14.000Z | Census_income.ipynb | umairnsr87/deploying-ml-model-with-django | b2e4c9f0e7e4618f5bf2cd8e5edaefe35e1e2235 | [
"Apache-2.0"
]
| null | null | null | Census_income.ipynb | umairnsr87/deploying-ml-model-with-django | b2e4c9f0e7e4618f5bf2cd8e5edaefe35e1e2235 | [
"Apache-2.0"
]
| null | null | null | 33.223796 | 323 | 0.354349 | [
[
[
"import pandas as pd\nimport numpy as np\nfrom sklearn import tree\nimport numpy as np\nfrom sklearn.metrics import roc_auc_score\n\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"I'll be demonstrating just the classification problems , you can build regression following a very similar process",
"_____no_output_____"
]
],
[
[
"# data prep from previous module\nci_train=pd.read_csv('census_income.csv')\n\n# if you have a test data, you can combine as shown in the earlier modules",
"_____no_output_____"
],
[
"ci_train.head()",
"_____no_output_____"
],
[
"pd.crosstab(ci_train['education'],ci_train['education.num'])",
"_____no_output_____"
],
[
"ci_train.drop(['education'],axis=1,inplace=True)",
"_____no_output_____"
],
[
"ci_train['Y'].value_counts().index",
"_____no_output_____"
],
[
"ci_train['Y']=(ci_train['Y']==' >50K').astype(int)",
"_____no_output_____"
],
[
"cat_cols=ci_train.select_dtypes(['object']).columns",
"_____no_output_____"
],
[
"cat_cols",
"_____no_output_____"
],
[
"ci_train.shape",
"_____no_output_____"
],
[
"for col in cat_cols:\n freqs=ci_train[col].value_counts()\n k=freqs.index[freqs>500][:-1]\n for cat in k:\n name=col+'_'+cat\n ci_train[name]=(ci_train[col]==cat).astype(int)\n del ci_train[col]\n print(col)",
"workclass\nmarital.status\noccupation\nrelationship\nrace\nsex\nnative.country\n"
],
[
"ci_train.shape",
"_____no_output_____"
],
[
"ci_train.isnull().sum()",
"_____no_output_____"
],
[
"x_train=ci_train.drop(['Y'],1)\ny_train=ci_train['Y']",
"_____no_output_____"
]
],
[
[
"## Hyper Parameters For Decision Trees\n\n* criterion : there are two options available , \"entropy\" and \"gini\". These are the homogeneity measures that we discussed. By default \"gini\" is used \n\n* The maximum depth of the tree. If None, then nodes are expanded until all leaves are pure or until all leaves contain less than min_samples_split samples. Ignored if ``max_leaf_nodes`` is not None. We'll finding optimal value for max_leaf_nodes [which is basically size of the tree] through cross validation.\n\n* min_sample_split : The minimum number of samples required to split an internal node. defaults to too, good idea is to keep it slightly higher in order to reduce overfitting of the data. recommended values is between 5 to 10\n\n* min_sample_leaf : The minimum number of samples required to be at a leaf node. This defaults to 1. If this number is higher and a split results in a leaf node having lesser number of samples than specified then that split is cancelled.\n\n* max_leaf_node : this parameter controlls size of the tree, we'll be finding optimal value of this through cross validation\n\n* class_weight : this default to None in which case each class is given equal weightage. If the goal of the problem is good classification instead of accuracy then you should set this to \"balanced\", in which case class weights are assigned inversely proportional to class frequencies in the input data.\n\n* random_state : this is used to reproduce random result\n",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import RandomizedSearchCV",
"_____no_output_____"
],
[
"params={ 'class_weight':[None,'balanced'], \n 'criterion':['entropy','gini'],\n 'max_depth':[None,5,10,15,20,30,50,70],\n 'min_samples_leaf':[1,2,5,10,15,20], \n 'min_samples_split':[2,5,10,15,20]\n }",
"_____no_output_____"
],
[
"2*2*8*6*5",
"_____no_output_____"
],
[
"from sklearn.tree import DecisionTreeClassifier",
"_____no_output_____"
],
[
"clf=DecisionTreeClassifier()",
"_____no_output_____"
],
[
"random_search=RandomizedSearchCV(clf,cv=10,\n param_distributions=params,\n scoring='roc_auc',\n n_iter=10\n )",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"x_train[\"capital.gain\"]=x_train[\"capital.gain\"].fillna(0)\nx_train[\"capital.loss\"]=x_train[\"capital.loss\"].fillna(0)\nx_train[\"hours.per.week\"]=x_train[\"hours.per.week\"].fillna(40)\nx_train.isna().sum()",
"_____no_output_____"
],
[
"random_search.fit(x_train,y_train)",
"_____no_output_____"
]
],
[
[
"Printing the tree model is a little tricky in python. We'll have to output our tree to a .dot file using graphviz package. From there using graphviz.Source function we can print our tree for display. Here is how :",
"_____no_output_____"
]
],
[
[
"random_search.best_estimator_",
"_____no_output_____"
],
[
"def report(results, n_top=3):\n for i in range(1, n_top + 1):\n candidates = np.flatnonzero(results['rank_test_score'] == i)\n for candidate in candidates:\n print(\"Model with rank: {0}\".format(i))\n print(\"Mean validation score: {0:.3f} (std: {1:.5f})\".format(\n results['mean_test_score'][candidate],\n results['std_test_score'][candidate]))\n print(\"Parameters: {0}\".format(results['params'][candidate]))\n print(\"\")",
"_____no_output_____"
],
[
"report(random_search.cv_results_,5)",
"Model with rank: 1\nMean validation score: 0.888 (std: 0.00728)\nParameters: {'min_samples_split': 5, 'min_samples_leaf': 5, 'max_depth': 10, 'criterion': 'entropy', 'class_weight': 'balanced'}\n\nModel with rank: 2\nMean validation score: 0.885 (std: 0.00802)\nParameters: {'min_samples_split': 15, 'min_samples_leaf': 20, 'max_depth': 20, 'criterion': 'gini', 'class_weight': 'balanced'}\n\nModel with rank: 3\nMean validation score: 0.884 (std: 0.00820)\nParameters: {'min_samples_split': 5, 'min_samples_leaf': 20, 'max_depth': None, 'criterion': 'entropy', 'class_weight': None}\n\nModel with rank: 4\nMean validation score: 0.884 (std: 0.00818)\nParameters: {'min_samples_split': 10, 'min_samples_leaf': 20, 'max_depth': 50, 'criterion': 'entropy', 'class_weight': None}\n\nModel with rank: 5\nMean validation score: 0.879 (std: 0.00580)\nParameters: {'min_samples_split': 10, 'min_samples_leaf': 15, 'max_depth': 70, 'criterion': 'gini', 'class_weight': None}\n\n"
],
[
"dtree=random_search.best_estimator_",
"_____no_output_____"
],
[
"dtree.fit(x_train,y_train)",
"_____no_output_____"
],
[
"import pickle\nfilename = 'census_income'\noutfile = open(filename,'wb')\npickle.dump(dtree,outfile)\noutfile.close()",
"_____no_output_____"
],
[
"filename = 'census_income'\ninfile = open(filename,'rb')\nnew_census_model = pickle.load(infile)\ninfile.close()",
"_____no_output_____"
],
[
"predict=new_census_model.predict(x_train)\nfrom sklearn.metrics import accuracy_score\naccuracy_score(predict,y_train)",
"_____no_output_____"
],
[
"dotfile = open(\"mytree.dot\", 'w')\n\ntree.export_graphviz(dtree,out_file=dotfile,\n feature_names=x_train.columns,\n class_names=[\"0\",\"1\"],\n proportion=True)\ndotfile.close()",
"_____no_output_____"
]
],
[
[
"Open mytree.dot file in a simple text editor and copy and paste the code here to visualise your tree : http://webgraphviz.com",
"_____no_output_____"
],
[
"## Additional Hyper paprameters for RandomForests\n\n* n_estimators : number of trees in the forest . defaults to 10. good starting point will be 100. Its one of the hyper parameters. We'll see how to search through mutidimensional hyper parameter space in order to find optimal combination through randomised grid search\n\n* max_features : Number of features being considered for rule selection at each split. Look at the documentation for defaults\n\n* bootstrap : boolean values, Whether bootstrap samples are used when building trees.\n",
"_____no_output_____"
]
],
[
[
"\nfrom sklearn.ensemble import RandomForestClassifier\n",
"_____no_output_____"
],
[
"clf = RandomForestClassifier()",
"_____no_output_____"
],
[
"# this here is the base classifier we are going to try\n# we will be supplying different parameter ranges to our randomSearchCV which in turn\n# will pass it on to this classifier\n\n# Utility function to report best scores. This simply accepts grid scores from \n# our randomSearchCV/GridSearchCV and picks and gives top few combination according to \n# their scores\n\n# RandomSearchCV/GridSearchCV accept parameters values as dictionaries.\n# In example given below we have constructed dictionary for \n#different parameter values that we want to\n# try for randomForest model\n\nparam_dist = {\"n_estimators\":[100,200,300,500,700,1000],\n \"max_features\": [5,10,20,25,30,35],\n \"bootstrap\": [True, False],\n 'class_weight':[None,'balanced'], \n 'criterion':['entropy','gini'],\n 'max_depth':[None,5,10,15,20,30,50,70],\n 'min_samples_leaf':[1,2,5,10,15,20], \n 'min_samples_split':[2,5,10,15,20]\n }\n\n",
"_____no_output_____"
],
[
"x_train.shape",
"_____no_output_____"
],
[
"960*6*6*2",
"_____no_output_____"
],
[
"# run randomized search\nn_iter_search = 10\n# n_iter parameter of RandomizedSeacrhCV controls, how many \n# parameter combination will be tried; out of all possible given values\n\nrandom_search = RandomizedSearchCV(clf, param_distributions=param_dist,\n n_iter=n_iter_search,scoring='roc_auc',cv=5)\nrandom_search.fit(x_train, y_train)",
"_____no_output_____"
],
[
"random_search.best_estimator_",
"_____no_output_____"
]
],
[
[
"RandomForestClassifier(bootstrap=True, class_weight=None, criterion='gini',\n max_depth=50, max_features=10, max_leaf_nodes=None,\n min_impurity_split=1e-07, min_samples_leaf=10,\n min_samples_split=20, min_weight_fraction_leaf=0.0,\n n_estimators=300, n_jobs=1, oob_score=False, random_state=None,\n verbose=0, warm_start=False)\n \n**Note: This is a result from one of the runs, you can very well get different results from a different run. Your results need not match with this.**",
"_____no_output_____"
]
],
[
[
"report(random_search.cv_results_,5)",
"_____no_output_____"
],
[
"# select the best values from results above, they will vary slightly with each run\nrf=RandomForestClassifier(bootstrap=True, class_weight=None, criterion='gini', max_depth=50, \n max_features=10, max_leaf_nodes=None, min_impurity_split=1e-07, \n min_samples_leaf=10, min_samples_split=20, min_weight_fraction_leaf=0.0, \n n_estimators=300, n_jobs=1, oob_score=False, random_state=None, verbose=0, warm_start=False)",
"_____no_output_____"
],
[
"rf.fit(x_train,y_train)",
"_____no_output_____"
]
],
[
[
"## Feature Importance",
"_____no_output_____"
]
],
[
[
"feat_imp_df=pd.DataFrame({'features':x_train.columns,'importance':rf.feature_importances_})\n\nfeat_imp_df.sort_values('importance',ascending=False)",
"_____no_output_____"
]
],
[
[
"## Partial Dependence Plot\n\n",
"_____no_output_____"
]
],
[
[
"var_name='education.num'\n\npreds=rf.predict_proba(x_train)[:,1]\n# part_dep_data",
"_____no_output_____"
],
[
"var_data=pd.DataFrame({'var':x_train[var_name],'response':preds})",
"_____no_output_____"
],
[
"import seaborn as sns\n\nsns.lmplot(x='var',y='response',data=var_data,fit_reg=False)",
"_____no_output_____"
],
[
"import statsmodels.api as sm\nsmooth_data=sm.nonparametric.lowess(var_data['response'],var_data['var'])\n\n# smooth_data",
"_____no_output_____"
],
[
"df=pd.DataFrame({'response':smooth_data[:,1],var_name:smooth_data[:,0]})\n\nsns.lmplot(x=var_name,y='response',data=df,fit_reg=False)",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
]
|
e796c53e85d8a319d86b784018ca051aa38eda51 | 62,327 | ipynb | Jupyter Notebook | Youtube_Comments-checkpoint.ipynb | Vineeta12345/spam-detection | da842fca2388d3042ab9ab47ff8a5e8db373a4db | [
"Unlicense"
]
| 3 | 2020-05-06T11:55:55.000Z | 2022-02-27T06:54:58.000Z | Youtube_Comments-checkpoint.ipynb | Vineeta12345/spam-detection | da842fca2388d3042ab9ab47ff8a5e8db373a4db | [
"Unlicense"
]
| null | null | null | Youtube_Comments-checkpoint.ipynb | Vineeta12345/spam-detection | da842fca2388d3042ab9ab47ff8a5e8db373a4db | [
"Unlicense"
]
| null | null | null | 26.590017 | 369 | 0.386173 | [
[
[
"#EDA Packages\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"# ML Packages For Vectorization of Text For Feature Extraction\nfrom sklearn.feature_extraction.text import CountVectorizer\nfrom sklearn.feature_extraction.text import TfidfVectorizer",
"_____no_output_____"
],
[
"# Visualization Packages\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"_____no_output_____"
],
[
"# Dataset from https://archive.ics.uci.edu/ml/datasets/YouTube+Spam+Collection#\ndf1 = pd.read_csv(\"Youtube01-Psy.csv\")",
"_____no_output_____"
],
[
"df1.head()",
"_____no_output_____"
],
[
"# Load all our dataset to merge them\ndf2 = pd.read_csv(\"Youtube02-KatyPerry.csv\")\ndf3 = pd.read_csv(\"Youtube03-LMFAO.csv\")\ndf4 = pd.read_csv(\"Youtube04-Eminem.csv\")\ndf5 = pd.read_csv(\"Youtube05-Shakira.csv\")",
"_____no_output_____"
],
[
"frames = [df1,df2,df3,df4,df5]",
"_____no_output_____"
],
[
"# Merging or Concatenating our DF\ndf_merged = pd.concat(frames)",
"_____no_output_____"
],
[
"# Total Size\ndf_merged.shape",
"_____no_output_____"
],
[
"# Merging with Keys\nkeys = [\"Psy\",\"KatyPerry\",\"LMFAO\",\"Eminem\",\"Shakira\"]",
"_____no_output_____"
],
[
"df_with_keys = pd.concat(frames,keys=keys)",
"_____no_output_____"
],
[
"df_with_keys",
"_____no_output_____"
],
[
"# Checking for Only Comments on Shakira\ndf_with_keys.loc['Shakira']",
"_____no_output_____"
],
[
"# Save and Write Merged Data to csv\ndf_with_keys.to_csv(\"YoutubeSpamMergeddata.csv\")",
"_____no_output_____"
],
[
"df = df_with_keys",
"_____no_output_____"
],
[
"df.size",
"_____no_output_____"
]
],
[
[
"## Data Cleaning",
"_____no_output_____"
]
],
[
[
"# Checking for Consistent Column Name\ndf.columns",
"_____no_output_____"
],
[
"# Checking for Datatypes\ndf.dtypes",
"_____no_output_____"
],
[
"# Check for missing nan\ndf.isnull().isnull().sum()",
"_____no_output_____"
],
[
"# Checking for Date\ndf[\"DATE\"]",
"_____no_output_____"
],
[
"df.AUTHOR\n# Convert the Author Name to First Name and Last Name\n#df[[\"FIRSTNAME\",\"LASTNAME\"]] = df['AUTHOR'].str.split(expand=True)",
"_____no_output_____"
]
],
[
[
"## Working With Text Content",
"_____no_output_____"
]
],
[
[
"df_data = df[[\"CONTENT\",\"CLASS\"]]",
"_____no_output_____"
],
[
"df_data.columns",
"_____no_output_____"
],
[
"df_x = df_data['CONTENT']\ndf_y = df_data['CLASS']",
"_____no_output_____"
]
],
[
[
"## Feature Extraction From Text\n## CountVectorizer\n## TfidfVectorizer",
"_____no_output_____"
]
],
[
[
"cv = CountVectorizer()\nex = cv.fit_transform([\"Great song but check this out\",\"What is this song?\"])",
"_____no_output_____"
],
[
"ex.toarray()",
"_____no_output_____"
],
[
"cv.get_feature_names()",
"_____no_output_____"
],
[
"# Extract Feature With CountVectorizer\ncorpus = df_x\ncv = CountVectorizer()\nX = cv.fit_transform(corpus) # Fit the Data",
"_____no_output_____"
],
[
"X.toarray()",
"_____no_output_____"
],
[
"# get the feature names\ncv.get_feature_names()",
"_____no_output_____"
]
],
[
[
"## Model Building",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(X, df_y, test_size=0.33, random_state=42)",
"_____no_output_____"
],
[
"X_train",
"_____no_output_____"
],
[
"# Naive Bayes Classifier\nfrom sklearn.naive_bayes import MultinomialNB\nclf = MultinomialNB()\nclf.fit(X_train,y_train)\nclf.score(X_test,y_test)",
"_____no_output_____"
],
[
"# Accuracy of our Model\nprint(\"Accuracy of Model\",clf.score(X_test,y_test)*100,\"%\")",
"Accuracy of Model 91.95046439628483 %\n"
],
[
"## Predicting with our model\nclf.predict(X_test)",
"_____no_output_____"
],
[
"# Sample Prediciton\ncomment = [\"Check this out\"]\nvect = cv.transform(comment).toarray()",
"_____no_output_____"
],
[
"clf.predict(vect)",
"_____no_output_____"
],
[
"class_dict = {'ham':0,'spam':1}",
"_____no_output_____"
],
[
"class_dict.values()",
"_____no_output_____"
],
[
"if clf.predict(vect) == 1:\n print(\"Spam\")\nelse:\n print(\"Ham\")",
"Spam\n"
],
[
"# Sample Prediciton 2\ncomment1 = [\"Great song Friend\"]\nvect = cv.transform(comment1).toarray()\nclf.predict(vect)",
"_____no_output_____"
]
],
[
[
"## Save The Model",
"_____no_output_____"
]
],
[
[
"import pickle",
"_____no_output_____"
],
[
"naivebayesML = open(\"YtbSpam_model.pkl\",\"wb\")",
"_____no_output_____"
],
[
"pickle.dump(clf,naivebayesML)",
"_____no_output_____"
],
[
"naivebayesML.close()",
"_____no_output_____"
],
[
"# Load the model",
"_____no_output_____"
],
[
"ytb_model = open(\"YtbSpam_model.pkl\",\"rb\")",
"_____no_output_____"
],
[
"new_model = pickle.load(ytb_model)",
"_____no_output_____"
],
[
"new_model",
"_____no_output_____"
],
[
"# Sample Prediciton 3\ncomment2 = [\"Hey Music Fans I really appreciate all of you,but see this song too\"]\nvect = cv.transform(comment2).toarray()\nnew_model.predict(vect)",
"_____no_output_____"
],
[
"if new_model.predict(vect) == 1:\n print(\"Spam\")\nelse:\n print(\"Ham\")",
"Spam\n"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e796d15704c285179883cecb0a1f53bc00e40765 | 148,046 | ipynb | Jupyter Notebook | P1.ipynb | RedaMokarrab/Nano_Degree_Self_Driving | 73aed4963c08b6062ccd7a1c5a76c4e366a4ec22 | [
"MIT"
]
| 1 | 2022-01-06T19:37:47.000Z | 2022-01-06T19:37:47.000Z | P1.ipynb | RedaMokarrab/Nano_Degree_Self_Driving | 73aed4963c08b6062ccd7a1c5a76c4e366a4ec22 | [
"MIT"
]
| null | null | null | P1.ipynb | RedaMokarrab/Nano_Degree_Self_Driving | 73aed4963c08b6062ccd7a1c5a76c4e366a4ec22 | [
"MIT"
]
| null | null | null | 167.472851 | 114,988 | 0.880794 | [
[
[
"# Self-Driving Car Engineer Nanodegree\n\n\n## Project: **Finding Lane Lines on the Road** \n***\nIn this project, you will use the tools you learned about in the lesson to identify lane lines on the road. You can develop your pipeline on a series of individual images, and later apply the result to a video stream (really just a series of images). Check out the video clip \"raw-lines-example.mp4\" (also contained in this repository) to see what the output should look like after using the helper functions below. \n\nOnce you have a result that looks roughly like \"raw-lines-example.mp4\", you'll need to get creative and try to average and/or extrapolate the line segments you've detected to map out the full extent of the lane lines. You can see an example of the result you're going for in the video \"P1_example.mp4\". Ultimately, you would like to draw just one line for the left side of the lane, and one for the right.\n\nIn addition to implementing code, there is a brief writeup to complete. The writeup should be completed in a separate file, which can be either a markdown file or a pdf document. There is a [write up template](https://github.com/udacity/CarND-LaneLines-P1/blob/master/writeup_template.md) that can be used to guide the writing process. Completing both the code in the Ipython notebook and the writeup template will cover all of the [rubric points](https://review.udacity.com/#!/rubrics/322/view) for this project.\n\n---\nLet's have a look at our first image called 'test_images/solidWhiteRight.jpg'. Run the 2 cells below (hit Shift-Enter or the \"play\" button above) to display the image.\n\n**Note: If, at any point, you encounter frozen display windows or other confounding issues, you can always start again with a clean slate by going to the \"Kernel\" menu above and selecting \"Restart & Clear Output\".**\n\n---",
"_____no_output_____"
],
[
"**The tools you have are color selection, region of interest selection, grayscaling, Gaussian smoothing, Canny Edge Detection and Hough Tranform line detection. You are also free to explore and try other techniques that were not presented in the lesson. Your goal is piece together a pipeline to detect the line segments in the image, then average/extrapolate them and draw them onto the image for display (as below). Once you have a working pipeline, try it out on the video stream below.**\n\n---\n\n<figure>\n <img src=\"examples/line-segments-example.jpg\" width=\"380\" alt=\"Combined Image\" />\n <figcaption>\n <p></p> \n <p style=\"text-align: center;\"> Your output should look something like this (above) after detecting line segments using the helper functions below </p> \n </figcaption>\n</figure>\n <p></p> \n<figure>\n <img src=\"examples/laneLines_thirdPass.jpg\" width=\"380\" alt=\"Combined Image\" />\n <figcaption>\n <p></p> \n <p style=\"text-align: center;\"> Your goal is to connect/average/extrapolate line segments to get output like this</p> \n </figcaption>\n</figure>",
"_____no_output_____"
],
[
"**Run the cell below to import some packages. If you get an `import error` for a package you've already installed, try changing your kernel (select the Kernel menu above --> Change Kernel). Still have problems? Try relaunching Jupyter Notebook from the terminal prompt. Also, consult the forums for more troubleshooting tips.** ",
"_____no_output_____"
],
[
"## Import Packages",
"_____no_output_____"
]
],
[
[
"#importing some useful packages\nimport matplotlib.pyplot as plt\nimport matplotlib.image as mpimg\nimport numpy as np\nimport cv2\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"## Read in an Image",
"_____no_output_____"
]
],
[
[
"#reading in an image\nimage = mpimg.imread('test_images/solidWhiteRight.jpg')\n\n#printing out some stats and plotting\nprint('This image is:', type(image), 'with dimensions:', image.shape)\nplt.imshow(image) # if you wanted to show a single color channel image called 'gray', for example, call as plt.imshow(gray, cmap='gray')",
"This image is: <class 'numpy.ndarray'> with dimensions: (540, 960, 3)\n"
]
],
[
[
"## Ideas for Lane Detection Pipeline",
"_____no_output_____"
],
[
"**Some OpenCV functions (beyond those introduced in the lesson) that might be useful for this project are:**\n\n`cv2.inRange()` for color selection \n`cv2.fillPoly()` for regions selection \n`cv2.line()` to draw lines on an image given endpoints \n`cv2.addWeighted()` to coadd / overlay two images \n`cv2.cvtColor()` to grayscale or change color \n`cv2.imwrite()` to output images to file \n`cv2.bitwise_and()` to apply a mask to an image\n\n**Check out the OpenCV documentation to learn about these and discover even more awesome functionality!**",
"_____no_output_____"
],
[
"## Helper Functions",
"_____no_output_____"
],
[
"Below are some helper functions to help get you started. They should look familiar from the lesson!",
"_____no_output_____"
]
],
[
[
"import math\n\ndef grayscale(img):\n \"\"\"Applies the Grayscale transform\n This will return an image with only one color channel\n but NOTE: to see the returned image as grayscale\n (assuming your grayscaled image is called 'gray')\n you should call plt.imshow(gray, cmap='gray')\"\"\"\n return cv2.cvtColor(img, cv2.COLOR_RGB2GRAY)\n # Or use BGR2GRAY if you read an image with cv2.imread()\n # return cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)\n \ndef canny(img, low_threshold, high_threshold):\n \"\"\"Applies the Canny transform\"\"\"\n return cv2.Canny(img, low_threshold, high_threshold)\n\ndef gaussian_blur(img, kernel_size):\n \"\"\"Applies a Gaussian Noise kernel\"\"\"\n return cv2.GaussianBlur(img, (kernel_size, kernel_size), 0)\n\ndef region_of_interest(img, vertices):\n \"\"\"\n Applies an image mask.\n \n Only keeps the region of the image defined by the polygon\n formed from `vertices`. The rest of the image is set to black.\n `vertices` should be a numpy array of integer points.\n \"\"\"\n #defining a blank mask to start with\n mask = np.zeros_like(img) \n \n #defining a 3 channel or 1 channel color to fill the mask with depending on the input image\n if len(img.shape) > 2:\n channel_count = img.shape[2] # i.e. 3 or 4 depending on your image\n ignore_mask_color = (255,) * channel_count\n else:\n ignore_mask_color = 255\n \n #filling pixels inside the polygon defined by \"vertices\" with the fill color \n cv2.fillPoly(mask, vertices, ignore_mask_color)\n \n #returning the image only where mask pixels are nonzero\n masked_image = cv2.bitwise_and(img, mask)\n return masked_image\n\ndef draw_lines(img, lines, color=[255, 0, 0], thickness=8):\n\n #create variables to hold the x, y and slopes for all the lines, \n left_x=[]\n left_y=[]\n left_slopes=[]\n right_x=[]\n right_y=[]\n right_slopes=[]\n right_limits=[5,0.5]\n left_limits=[-5,-0.5]\n \n #variables to lane points\n left_lane=[0,0,0,0]#x1,y1,x2,y2\n right_lane=[0,0,0,0]\n #get image size to use the left and right corners for starting the line\n imshape = img.shape\n y_size = img.shape[0]\n\n #loop over all the lines to extract the ones in interest ( filter out the ones that are not expected)\n for line in lines:\n for x1,y1,x2,y2 in line:\n [slope, intercept] = np.polyfit((x1,x2), (y1,y2), 1)\n #filter only on the slopes that make sense to a lane\n if((slope<left_limits[1]) and (slope>left_limits[0])): # if line slope < 0 then it belongs to left lane (y=mx+b)\n left_x+=[x1,x2]\n left_y+=[y1,y2]\n left_slopes+=[slope]\n \n elif((slope<right_limits[0]) and (slope>right_limits[1])): # if line slope > 0 then it belongs to right lane (y=mx+b) where m is - \n right_x+=[x1,x2]\n right_y+=[y1,y2]\n right_slopes+=[slope]\n\n \n #average each line points to get the line equation which best describes the lane\n left_x_mean= np.mean(left_x)\n left_y_mean= np.mean(left_y)\n left_slope_mean=np.mean(left_slopes)\n left_intercept_mean =left_y_mean - (left_slope_mean * left_x_mean)\n \n right_x_mean= np.mean(right_x)\n right_y_mean= np.mean(right_y)\n right_slope_mean=np.mean(right_slopes)\n right_intercept_mean =right_y_mean - (right_slope_mean * right_x_mean)\n\n\n #get start and end of each line to draw the left lane and right lane using the equations above\n #only process incase size is > 0 (to fix challenge error )\n if((np.size(left_y))>0) :\n left_lane[0]=int((np.min(left_y)-left_intercept_mean)/left_slope_mean)# x=(y-b)/m\n left_lane[2]=int((y_size-left_intercept_mean)/left_slope_mean)#\n left_lane[1]=int(np.min(left_y))\n left_lane[3]=y_size\n #got errors seems that function only takes int \n cv2.line(img, (left_lane[0],left_lane[1] ), (left_lane[2],left_lane[3]), color, thickness)\n if((np.size(right_y))>0):\n right_lane[0]=int((np.min(right_y)-right_intercept_mean)/right_slope_mean)# x=(y-b)/m\n right_lane[2]=int((y_size-right_intercept_mean)/right_slope_mean)#\n right_lane[1]=int(np.min(right_y))\n right_lane[3]=y_size\n #got errors seems that function only takes int \n cv2.line(img, (right_lane[0],right_lane[1]), (right_lane[2],right_lane[3]), color, thickness)\n \n\n \ndef hough_lines(img, rho, theta, threshold, min_line_len, max_line_gap):\n \"\"\"\n `img` should be the output of a Canny transform.\n \n Returns an image with hough lines drawn.\n \"\"\"\n lines = cv2.HoughLinesP(img, rho, theta, threshold, np.array([]), minLineLength=min_line_len, maxLineGap=max_line_gap)\n line_img = np.zeros((img.shape[0], img.shape[1], 3), dtype=np.uint8)\n draw_lines(line_img, lines)\n return line_img\n\n# Python 3 has support for cool math symbols.\n\ndef weighted_img(img, initial_img, α=0.8, β=1., γ=0.):\n \"\"\"\n `img` is the output of the hough_lines(), An image with lines drawn on it.\n Should be a blank image (all black) with lines drawn on it.\n \n `initial_img` should be the image before any processing.\n \n The result image is computed as follows:\n \n initial_img * α + img * β + γ\n NOTE: initial_img and img must be the same shape!\n \"\"\"\n return cv2.addWeighted(initial_img, α, img, β, γ)\n\ndef convert_hls(image):\n return cv2.cvtColor(image, cv2.COLOR_RGB2HLS)\n\ndef select_color(image ):\n \n # find colors that are in provided range and highlight it in red\n \n white_lo= np.array([100,100,200])\n white_hi= np.array([255,255,255])\n \n yellow_lo_RGB= np.array([225,180,0])\n yellow_hi_RGB= np.array([255,255,170]) \n \n yellow_lo_HLS= np.array([20,120,80])\n yellow_hi_HLS= np.array([45,200,255]) \n\n rgb_image = np.copy(image)\n hls_image = convert_hls(image)\n \n #plt.figure()\n #plt.imshow(rgb_image)\n \n #plt.figure()\n #plt.imshow(hls_image)\n \n mask_1=cv2.inRange(rgb_image,white_lo,white_hi) #filter on rgb white\n mask_2=cv2.inRange(hls_image,yellow_lo_RGB,yellow_hi_RGB) #filter on rgb yellow \n mask_3=cv2.inRange(hls_image,yellow_lo_HLS,yellow_hi_HLS) #filter on hls yellow \n \n mask = mask_1+mask_2+mask_3\n \n #plt.figure()\n #plt.imshow(mask)\n \n result = cv2.bitwise_and(image,image, mask= mask)\n #plt.figure()\n #plt.imshow(result)\n \n \n return result\n",
"_____no_output_____"
]
],
[
[
"## Test Images\n\nBuild your pipeline to work on the images in the directory \"test_images\" \n**You should make sure your pipeline works well on these images before you try the videos.**",
"_____no_output_____"
]
],
[
[
"import os\nos.listdir(\"test_images/\")",
"_____no_output_____"
]
],
[
[
"## Build a Lane Finding Pipeline\n\n",
"_____no_output_____"
],
[
"Build the pipeline and run your solution on all test_images. Make copies into the `test_images_output` directory, and you can use the images in your writeup report.\n\nTry tuning the various parameters, especially the low and high Canny thresholds as well as the Hough lines parameters.",
"_____no_output_____"
]
],
[
[
"# TODO: Build your pipeline that will draw lane lines on the test_images\n# then save them to the test_images_output directory.\n#output all processed images for documentation\nimages=os.listdir(\"test_images/\")\n\nfor filename in images: \n #first read image and change to gray scale \n image = mpimg.imread(\"test_images/\"+filename)\n \n #color select for lane to improve performance but didn't work \n #highlight white and yellow colors for better late detection\n highlighted_color=select_color(image)\n\n \n #change to gray scale\n image_gray = cv2.cvtColor(highlighted_color,cv2.COLOR_RGB2GRAY)\n cv2.imwrite(\"test_images_output/\"+filename[:-4]+\"_gray.jpg\",image_gray)\n\n #Apply guassian filter \n image_blur_gray= gaussian_blur(image_gray, 5)\n cv2.imwrite(\"test_images_output/\"+filename[:-4]+\"_blur.jpg\",image_blur_gray)\n\n # Using canny edges ( used threshoulds achieved from the exercise )\n image_edge = canny(image_blur_gray, 100, 200)\n cv2.imwrite(\"test_images_output/\"+filename[:-4]+\"_edge.jpg\",image_edge)\n\n # Add filter region (added vehicle hood area)\n imshape = image.shape\n vertices = np.array([[(200,imshape[0]-50),(420, 330), (580, 330), (imshape[1]-200,imshape[0]-50)]], dtype=np.int32)\n image_masked_edges = region_of_interest(image_edge,vertices)\n cv2.imwrite(\"test_images_output/\"+filename[:-4]+\"_masked_edge.jpg\",image_masked_edges)\n\n\n # Define the Hough transform parameters\n # Make a blank the same size as our image to draw on\n rho = 1 # distance resolution in pixels of the Hough grid\n theta = np.pi/180 # angular resolution in radians of the Hough grid\n threshold = 40 # minimum number of votes (intersections in Hough grid cell)\n min_line_length = 20 #minimum number of pixels making up a line\n max_line_gap = 100 # maximum gap in pixels between connectable line \n\n image_hough_lines = hough_lines(image_masked_edges, rho, theta, threshold, min_line_length, max_line_gap)\n image_hough_lines = cv2.cvtColor(image_hough_lines, cv2.COLOR_RGB2BGR)\n cv2.imwrite(\"test_images_output/\"+filename[:-4]+\"_Hough.jpg\",image_hough_lines)\n \n #create image with overlay lines\n overlayed_image = weighted_img(image_hough_lines,image)\n cv2.imwrite(\"test_images_output/\"+filename[:-4]+\"_Final_overlay.jpg\",overlayed_image)\n ",
"C:\\Users\\Redaaaaaa\\Anaconda3\\lib\\site-packages\\numpy\\core\\fromnumeric.py:3373: RuntimeWarning: Mean of empty slice.\n out=out, **kwargs)\nC:\\Users\\Redaaaaaa\\Anaconda3\\lib\\site-packages\\numpy\\core\\_methods.py:170: RuntimeWarning: invalid value encountered in double_scalars\n ret = ret.dtype.type(ret / rcount)\n"
]
],
[
[
"## Test on Videos\n\nYou know what's cooler than drawing lanes over images? Drawing lanes over video!\n\nWe can test our solution on two provided videos:\n\n`solidWhiteRight.mp4`\n\n`solidYellowLeft.mp4`\n\n**Note: if you get an import error when you run the next cell, try changing your kernel (select the Kernel menu above --> Change Kernel). Still have problems? Try relaunching Jupyter Notebook from the terminal prompt. Also, consult the forums for more troubleshooting tips.**\n\n**If you get an error that looks like this:**\n```\nNeedDownloadError: Need ffmpeg exe. \nYou can download it by calling: \nimageio.plugins.ffmpeg.download()\n```\n**Follow the instructions in the error message and check out [this forum post](https://discussions.udacity.com/t/project-error-of-test-on-videos/274082) for more troubleshooting tips across operating systems.**",
"_____no_output_____"
]
],
[
[
"# Import everything needed to edit/save/watch video clips\nfrom moviepy.editor import VideoFileClip\nfrom IPython.display import HTML",
"_____no_output_____"
],
[
"def process_image(image):\n # NOTE: The output you return should be a color image (3 channel) for processing video below\n # TODO: put your pipeline here,\n #color select for lane to improve performance but didn't work \n #highlight white and yellow colors for better late detection\n highlighted_color=select_color(image)\n\n #Change image to Gray\n image_gray = cv2.cvtColor(highlighted_color,cv2.COLOR_RGB2GRAY)\n\n #Apply guassian filter \n image_blur_gray= gaussian_blur(image_gray, 5)\n\n # Using canny edges ( used threshoulds achieved from the exercise )\n image_edge = canny(image_blur_gray, 100, 200)\n\n # Add filter region \n imshape = image.shape\n vertices = np.array([[(200,imshape[0]-50),(420, 330), (580, 330), (imshape[1]-200,imshape[0]-50)]], dtype=np.int32)\n image_masked_edges = region_of_interest(image_edge,vertices)\n\n\n # Define the Hough transform parameters\n # Make a blank the same size as our image to draw on\n rho = 1 # distance resolution in pixels of the Hough grid\n theta = np.pi/180 # angular resolution in radians of the Hough grid\n threshold = 40 # minimum number of votes (intersections in Hough grid cell)\n min_line_length = 20 #minimum number of pixels making up a line\n max_line_gap = 100 # maximum gap in pixels between connectable line \n\n image_hough_lines = hough_lines(image_masked_edges, rho, theta, threshold, min_line_length, max_line_gap)\n \n #create image with overlay lines\n overlayed_image = weighted_img(image_hough_lines,image)\n \n # you should return the final output (image where lines are drawn on lanes)\n\n return overlayed_image\n\n",
"_____no_output_____"
]
],
[
[
"Let's try the one with the solid white lane on the right first ...",
"_____no_output_____"
]
],
[
[
"white_output = 'test_videos_output/solidWhiteRight.mp4'\n## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video\n## To do so add .subclip(start_second,end_second) to the end of the line below\n## Where start_second and end_second are integer values representing the start and end of the subclip\n## You may also uncomment the following line for a subclip of the first 5 seconds\n##clip1 = VideoFileClip(\"test_videos/solidWhiteRight.mp4\").subclip(0,5)\nclip1 = VideoFileClip(\"test_videos/solidWhiteRight.mp4\")\nwhite_clip = clip1.fl_image(process_image) #NOTE: this function expects color images!!\n%time white_clip.write_videofile(white_output, audio=False)",
"\rt: 0%| | 0/221 [00:00<?, ?it/s, now=None]"
]
],
[
[
"Play the video inline, or if you prefer find the video in your filesystem (should be in the same directory) and play it in your video player of choice.",
"_____no_output_____"
]
],
[
[
"HTML(\"\"\"\n<video width=\"960\" height=\"540\" controls>\n <source src=\"{0}\">\n</video>\n\"\"\".format(white_output))",
"_____no_output_____"
]
],
[
[
"## Improve the draw_lines() function\n\n**At this point, if you were successful with making the pipeline and tuning parameters, you probably have the Hough line segments drawn onto the road, but what about identifying the full extent of the lane and marking it clearly as in the example video (P1_example.mp4)? Think about defining a line to run the full length of the visible lane based on the line segments you identified with the Hough Transform. As mentioned previously, try to average and/or extrapolate the line segments you've detected to map out the full extent of the lane lines. You can see an example of the result you're going for in the video \"P1_example.mp4\".**\n\n**Go back and modify your draw_lines function accordingly and try re-running your pipeline. The new output should draw a single, solid line over the left lane line and a single, solid line over the right lane line. The lines should start from the bottom of the image and extend out to the top of the region of interest.**",
"_____no_output_____"
],
[
"Now for the one with the solid yellow lane on the left. This one's more tricky!",
"_____no_output_____"
]
],
[
[
"yellow_output = 'test_videos_output/solidYellowLeft.mp4'\n## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video\n## To do so add .subclip(start_second,end_second) to the end of the line below\n## Where start_second and end_second are integer values representing the start and end of the subclip\n## You may also uncomment the following line for a subclip of the first 5 seconds\n##clip2 = VideoFileClip('test_videos/solidYellowLeft.mp4').subclip(0,5)\nclip2 = VideoFileClip('test_videos/solidYellowLeft.mp4')\nyellow_clip = clip2.fl_image(process_image)\n%time yellow_clip.write_videofile(yellow_output, audio=False)",
"t: 0%| | 2/681 [00:00<01:00, 11.14it/s, now=None]"
],
[
"HTML(\"\"\"\n<video width=\"960\" height=\"540\" controls>\n <source src=\"{0}\">\n</video>\n\"\"\".format(yellow_output))",
"_____no_output_____"
]
],
[
[
"## Writeup and Submission\n\nIf you're satisfied with your video outputs, it's time to make the report writeup in a pdf or markdown file. Once you have this Ipython notebook ready along with the writeup, it's time to submit for review! Here is a [link](https://github.com/udacity/CarND-LaneLines-P1/blob/master/writeup_template.md) to the writeup template file.\n",
"_____no_output_____"
],
[
"## Optional Challenge\n\nTry your lane finding pipeline on the video below. Does it still work? Can you figure out a way to make it more robust? If you're up for the challenge, modify your pipeline so it works with this video and submit it along with the rest of your project!",
"_____no_output_____"
]
],
[
[
"challenge_output = 'test_videos_output/challenge.mp4'\n## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video\n## To do so add .subclip(start_second,end_second) to the end of the line below\n## Where start_second and end_second are integer values representing the start and end of the subclip\n## You may also uncomment the following line for a subclip of the first 5 seconds\n##clip3 = VideoFileClip('test_videos/challenge.mp4').subclip(0,5)\nclip3 = VideoFileClip('test_videos/challenge.mp4')\nchallenge_clip = clip3.fl_image(process_image)\n%time challenge_clip.write_videofile(challenge_output, audio=False)",
"\rt: 0%| | 0/251 [00:00<?, ?it/s, now=None]"
],
[
"HTML(\"\"\"\n<video width=\"960\" height=\"540\" controls>\n <source src=\"{0}\">\n</video>\n\"\"\".format(challenge_output))",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
]
|
e796eca194df42938708dfa71150d91eb1dc022b | 158,039 | ipynb | Jupyter Notebook | Boris_Krant_DS_Unit_1_Sprint_Challenge_1.ipynb | bkrant/DS-Unit-1-Sprint-1-Dealing-With-Data | cce374b92bacc4785fd5c877a9c82616287a8bb3 | [
"MIT"
]
| null | null | null | Boris_Krant_DS_Unit_1_Sprint_Challenge_1.ipynb | bkrant/DS-Unit-1-Sprint-1-Dealing-With-Data | cce374b92bacc4785fd5c877a9c82616287a8bb3 | [
"MIT"
]
| null | null | null | Boris_Krant_DS_Unit_1_Sprint_Challenge_1.ipynb | bkrant/DS-Unit-1-Sprint-1-Dealing-With-Data | cce374b92bacc4785fd5c877a9c82616287a8bb3 | [
"MIT"
]
| null | null | null | 402.13486 | 110,982 | 0.916913 | [
[
[
"<a href=\"https://colab.research.google.com/github/bkrant/DS-Unit-1-Sprint-1-Dealing-With-Data/blob/master/Boris_Krant_DS_Unit_1_Sprint_Challenge_1.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Data Science Unit 1 Sprint Challenge 1\n\n## Loading, cleaning, visualizing, and analyzing data\n\nIn this sprint challenge you will look at a dataset of the survival of patients who underwent surgery for breast cancer.\n\nhttp://archive.ics.uci.edu/ml/datasets/Haberman%27s+Survival\n\nData Set Information:\nThe dataset contains cases from a study that was conducted between 1958 and 1970 at the University of Chicago's Billings Hospital on the survival of patients who had undergone surgery for breast cancer.\n\nAttribute Information:\n1. Age of patient at time of operation (numerical)\n2. Patient's year of operation (year - 1900, numerical)\n3. Number of positive axillary nodes detected (numerical)\n4. Survival status (class attribute)\n-- 1 = the patient survived 5 years or longer\n-- 2 = the patient died within 5 year\n\nSprint challenges are evaluated based on satisfactory completion of each part. It is suggested you work through it in order, getting each aspect reasonably working, before trying to deeply explore, iterate, or refine any given step. Once you get to the end, if you want to go back and improve things, go for it!",
"_____no_output_____"
],
[
"## Part 1 - Load and validate the data\n\n- Load the data as a `pandas` data frame.\n- Validate that it has the appropriate number of observations (you can check the raw file, and also read the dataset description from UCI).\n- Validate that you have no missing values.\n- Add informative names to the features.\n- The survival variable is encoded as 1 for surviving >5 years and 2 for not - change this to be 0 for not surviving and 1 for surviving >5 years (0/1 is a more traditional encoding of binary variables)\n\nAt the end, print the first five rows of the dataset to demonstrate the above.",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"breast = pd.read_csv('http://archive.ics.uci.edu/ml/machine-learning-databases/haberman/haberman.data',names=['age','year_operation','nodes','survived'])\nprint(breast.shape)\nprint(breast.isna().sum())",
"(306, 4)\nage 0\nyear_operation 0\nnodes 0\nsurvived 0\ndtype: int64\n"
],
[
"labels = {'survived': {2:0}}\nbreast.replace(labels, inplace=True)\nbreast.survived.value_counts()",
"_____no_output_____"
],
[
"print(breast.head())",
" age year_operation nodes survived\n0 30 64 1 1\n1 30 62 3 1\n2 30 65 0 1\n3 31 59 2 1\n4 31 65 4 1\n"
]
],
[
[
"## Part 2 - Examine the distribution and relationships of the features\n\nExplore the data - create at least *2* tables (can be summary statistics or crosstabulations) and *2* plots illustrating the nature of the data.\n\nThis is open-ended, so to remind - first *complete* this task as a baseline, then go on to the remaining sections, and *then* as time allows revisit and explore further.\n\nHint - you may need to bin some variables depending on your chosen tables/plots.",
"_____no_output_____"
]
],
[
[
"print(breast.corr())\nprint(breast.describe())",
" age year_operation nodes survived\nage 1.000000 0.089529 -0.063176 -0.067950\nyear_operation 0.089529 1.000000 -0.003764 0.004768\nnodes -0.063176 -0.003764 1.000000 -0.286768\nsurvived -0.067950 0.004768 -0.286768 1.000000\n age year_operation nodes survived\ncount 306.000000 306.000000 306.000000 306.000000\nmean 52.457516 62.852941 4.026144 0.735294\nstd 10.803452 3.249405 7.189654 0.441899\nmin 30.000000 58.000000 0.000000 0.000000\n25% 44.000000 60.000000 0.000000 0.000000\n50% 52.000000 63.000000 1.000000 1.000000\n75% 60.750000 65.750000 4.000000 1.000000\nmax 83.000000 69.000000 52.000000 1.000000\n"
],
[
"sns.heatmap(breast.corr());",
"_____no_output_____"
],
[
"import seaborn as sns\nimport matplotlib.pyplot as plt\nimport numpy as np\nsns.pairplot(breast);",
"_____no_output_____"
],
[
"g = sns.FacetGrid(breast, row=\"survived\", margin_titles=True)\nbins = np.linspace(0, breast.age.max())\ng.map(plt.hist, \"age\", color=\"steelblue\", bins=bins);",
"_____no_output_____"
],
[
"breast.boxplot();",
"_____no_output_____"
]
],
[
[
"## Part 3 - Analysis and Interpretation\n\nNow that you've looked at the data, answer the following questions:\n\n- What is at least one feature that looks to have a positive relationship with survival?\n- What is at least one feature that looks to have a negative relationship with survival?\n- How are those two features related with each other, and what might that mean?\n\nAnswer with text, but feel free to intersperse example code/results or refer to it from earlier.",
"_____no_output_____"
],
[
"According to the correlation matrix 'nodes' has a positive correlation of 0.287 with survival. 'years_operation' has a weak negative correlation with survival of -0.004768; probably statistically insignificant. 'nodes' and 'years_operation' have a correlation of -0.003764. This means there's no linear relationship between them - which is not the same as being independent because there might be a non-linear relationship. Have to look at scatter plot before drawing conclusions.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
]
|
e796eeb132907c19832dee9a4623273119811ad7 | 21,921 | ipynb | Jupyter Notebook | reinforcement_learning/rl_hvac_coach_energyplus/rl_hvac_coach_energyplus.ipynb | P15241328/amazon-sagemaker-examples | 00cba545be0822474f070321a62d22865187e09b | [
"Apache-2.0"
]
| 5 | 2019-01-19T23:53:35.000Z | 2022-01-29T14:04:31.000Z | reinforcement_learning/rl_hvac_coach_energyplus/rl_hvac_coach_energyplus.ipynb | P15241328/amazon-sagemaker-examples | 00cba545be0822474f070321a62d22865187e09b | [
"Apache-2.0"
]
| 1 | 2021-03-25T18:31:29.000Z | 2021-03-25T18:31:29.000Z | reinforcement_learning/rl_hvac_coach_energyplus/rl_hvac_coach_energyplus.ipynb | P15241328/amazon-sagemaker-examples | 00cba545be0822474f070321a62d22865187e09b | [
"Apache-2.0"
]
| 7 | 2020-03-04T22:23:51.000Z | 2021-07-13T14:05:46.000Z | 38.525483 | 556 | 0.614297 | [
[
[
"# HVAC with Amazon SageMaker RL\n\n---\n## Introduction\n\n\nHVAC stands for Heating, Ventilation and Air Conditioning and is responsible for keeping us warm and comfortable indoors. HVAC takes up a whopping 50% of the energy in a building and accounts for 40% of energy use in the US [1, 2]. Several control system optimizations have been proposed to reduce energy usage while ensuring thermal comfort.\n\nModern buildings collect data about the weather, occupancy and equipment use. All of this can be used to optimize HVAC energy usage. Reinforcement Learning (RL) is a good fit because it can learn how to interact with the environment and identify strategies to limit wasted energy. Several recent research efforts have shown that RL can reduce HVAC energy consumption by 15-20% [3, 4].\n\nAs training an RL algorithm in a real HVAC system can take time to converge as well as potentially lead to hazardous settings as the agent explores its state space, we turn to a simulator to train the agent. [EnergyPlus](https://energyplus.net/) is an open source, state of the art HVAC simulator from the US Department of Energy. We use a simple example with this simulator to showcase how we can train an RL model easily with Amazon SageMaker RL.\n\n<br>\n\n<img width=\"85%\" src=\"images/datacenter_env.png\" />\n\n<br>\n\n1. Objective: Control the data center HVAC system to reduce energy consumption while ensuring the room temperature stays within specified limits.\n2. Environment: We have a small single room datacenter that the HVAC system is cooling to ensure the compute equipment works properly. We will train our RL agent to control this HVAC system for one day subject to weather conditions in San Francisco. The agent takes actions every 5 minutes for a 24 hour period. Hence, the episode is a fixed 120 steps. \n3. State: The outdoor temperature, outdoor humidity and indoor room temperature.\n4. Action: The agent can set the heating and cooling setpoints. The cooling setpoint tells the HVAC system that it should start cooling the room if the room temperature goes above this setpoint. Likewise, the HVAC systems starts heating if the room temperature goes below the heating setpoint.\n5. Reward: The rewards has two components which are added together with coefficients: \n 1. It is proportional to the energy consumed by the HVAC system.\n 2. It gets a large penalty when the room temperature exceeds pre-specified lower or upper limits (as defined in `data_center_env.py`).\n\nReferences\n\n1. [sciencedirect.com](https://www.sciencedirect.com/science/article/pii/S0378778807001016)\n2. [environment.gov.au](https://www.environment.gov.au/system/files/energy/files/hvac-factsheet-energy-breakdown.pdf)\n3. Wei, Tianshu, Yanzhi Wang, and Qi Zhu. \"Deep reinforcement learning for building hvac control.\" In Proceedings of the 54th Annual Design Automation Conference 2017, p. 22. ACM, 2017.\n4. Zhang, Zhiang, and Khee Poh Lam. \"Practical implementation and evaluation of deep reinforcement learning control for a radiant heating system.\" In Proceedings of the 5th Conference on Systems for Built Environments, pp. 148-157. ACM, 2018.",
"_____no_output_____"
],
[
"## Pre-requisites \n\n### Imports\n\nTo get started, we'll import the Python libraries we need, set up the environment with a few prerequisites for permissions and configurations.",
"_____no_output_____"
]
],
[
[
"import sagemaker\nimport boto3\nimport sys\nimport os\nimport glob\nimport re\nimport subprocess\nimport numpy as np\nfrom IPython.display import HTML\nimport time\nfrom time import gmtime, strftime\nsys.path.append(\"common\")\nfrom misc import get_execution_role, wait_for_s3_object\nfrom docker_utils import build_and_push_docker_image\nfrom sagemaker.rl import RLEstimator, RLToolkit, RLFramework",
"_____no_output_____"
]
],
[
[
"### Setup S3 bucket\n\nCreate a reference to the default S3 bucket that will be used for model outputs.",
"_____no_output_____"
]
],
[
[
"sage_session = sagemaker.session.Session()\ns3_bucket = sage_session.default_bucket() \ns3_output_path = 's3://{}/'.format(s3_bucket)\nprint(\"S3 bucket path: {}\".format(s3_output_path))",
"_____no_output_____"
]
],
[
[
"### Define Variables \n\nWe define a job below that's used to identify our jobs.",
"_____no_output_____"
]
],
[
[
"# create unique job name \njob_name_prefix = 'rl-hvac'",
"_____no_output_____"
]
],
[
[
"### Configure settings\n\nYou can run your RL training jobs locally on the SageMaker notebook instance or on SageMaker training. In both of these scenarios, you can run in either 'local' (where you run the commands) or 'SageMaker' mode (on SageMaker training instances). 'local' mode uses the SageMaker Python SDK to run your code in Docker containers locally. It can speed up iterative testing and debugging while using the same familiar Python SDK interface. Just set `local_mode = True`. And when you're ready move to 'SageMaker' mode to scale things up.",
"_____no_output_____"
]
],
[
[
"# run local (on this machine)?\n# or on sagemaker training instances?\nlocal_mode = False\n\nif local_mode:\n instance_type = 'local'\nelse:\n # choose a larger instance to avoid running out of memory\n instance_type = \"ml.m4.4xlarge\"",
"_____no_output_____"
]
],
[
[
"### Create an IAM role\n\nEither get the execution role when running from a SageMaker notebook instance `role = sagemaker.get_execution_role()` or, when running from local notebook instance, use utils method `role = get_execution_role()` to create an execution role.",
"_____no_output_____"
]
],
[
[
"try:\n role = sagemaker.get_execution_role()\nexcept:\n role = get_execution_role()\n\nprint(\"Using IAM role arn: {}\".format(role))",
"_____no_output_____"
]
],
[
[
"### Install docker for `local` mode\n\nIn order to work in `local` mode, you need to have docker installed. When running from your local machine, please make sure that you have docker or docker-compose (for local CPU machines) and nvidia-docker (for local GPU machines) installed. Alternatively, when running from a SageMaker notebook instance, you can simply run the following script to install dependencies.\n\nNote, you can only run a single local notebook at one time.",
"_____no_output_____"
]
],
[
[
"# Only run from SageMaker notebook instance\nif local_mode:\n !/bin/bash ./common/setup.sh",
"_____no_output_____"
]
],
[
[
"## Build docker container\n\nSince we're working with a custom environment with custom dependencies, we create our own container for training. We:\n\n1. Fetch the base MXNet and Coach container image,\n2. Install EnergyPlus and its dependencies on top,\n3. Upload the new container image to AWS ECR.",
"_____no_output_____"
]
],
[
[
"cpu_or_gpu = 'gpu' if instance_type.startswith('ml.p') else 'cpu'\nrepository_short_name = \"sagemaker-hvac-coach-%s\" % cpu_or_gpu\ndocker_build_args = {\n 'CPU_OR_GPU': cpu_or_gpu, \n 'AWS_REGION': boto3.Session().region_name,\n}\ncustom_image_name = build_and_push_docker_image(repository_short_name, build_args=docker_build_args)\nprint(\"Using ECR image %s\" % custom_image_name)",
"_____no_output_____"
]
],
[
[
"## Setup the environment\n\nThe environment is defined in a Python file called `data_center_env.py` and for SageMaker training jobs, the file will be uploaded inside the `/src` directory.\n\nThe environment implements the init(), step() and reset() functions that describe how the environment behaves. This is consistent with Open AI Gym interfaces for defining an environment.\n\n1. `init()` - initialize the environment in a pre-defined state\n2. `step()` - take an action on the environment\n3. `reset()` - restart the environment on a new episode",
"_____no_output_____"
],
[
"## Configure the presets for RL algorithm \n\nThe presets that configure the RL training jobs are defined in the “preset-energy-plus-clipped-ppo.py” file which is also uploaded as part of the `/src` directory. Using the preset file, you can define agent parameters to select the specific agent algorithm. You can also set the environment parameters, define the schedule and visualization parameters, and define the graph manager. The schedule presets will define the number of heat up steps, periodic evaluation steps, training steps between evaluations, etc.\n\nAll of these can be overridden at run-time by specifying the `RLCOACH_PRESET` hyperparameter. Additionally, it can be used to define custom hyperparameters.",
"_____no_output_____"
]
],
[
[
"!pygmentize src/preset-energy-plus-clipped-ppo.py",
"_____no_output_____"
]
],
[
[
"## Write the Training Code \n\nThe training code is written in the file “train-coach.py” which is uploaded in the /src directory. \nFirst import the environment files and the preset files, and then define the main() function. ",
"_____no_output_____"
]
],
[
[
"!pygmentize src/train-coach.py",
"_____no_output_____"
]
],
[
[
"## Train the RL model using the Python SDK Script mode\n\nIf you are using local mode, the training will run on the notebook instance. When using SageMaker for training, you can select a GPU or CPU instance. The RLEstimator is used for training RL jobs. \n\n1. Specify the source directory where the environment, presets and training code is uploaded.\n2. Specify the entry point as the training code \n3. Specify the choice of RL toolkit and framework. This automatically resolves to the ECR path for the RL Container. \n4. Define the training parameters such as the instance count, job name, S3 path for output and job name. \n5. Specify the hyperparameters for the RL agent algorithm. The RLCOACH_PRESET can be used to specify the RL agent algorithm you want to use. \n6. [optional] Define the metrics definitions that you are interested in capturing in your logs. These can also be visualized in CloudWatch and SageMaker Notebooks. ",
"_____no_output_____"
]
],
[
[
"%%time\nestimator = RLEstimator(entry_point=\"train-coach.py\",\n source_dir='src',\n dependencies=[\"common/sagemaker_rl\"],\n image_uri=custom_image_name,\n role=role,\n instance_type=instance_type,\n instance_count=1,\n output_path=s3_output_path,\n base_job_name=job_name_prefix,\n hyperparameters = {\n 'save_model': 1\n }\n )\n\nestimator.fit(wait=local_mode)\njob_name = estimator.latest_training_job.job_name\nprint(\"Training job: %s\" % job_name)",
"_____no_output_____"
]
],
[
[
"## Store intermediate training output and model checkpoints \n\nThe output from the training job above is stored on S3. The intermediate folder contains gifs and metadata of the training.",
"_____no_output_____"
]
],
[
[
"s3_url = \"s3://{}/{}\".format(s3_bucket,job_name)\n\nif local_mode:\n output_tar_key = \"{}/output.tar.gz\".format(job_name)\nelse:\n output_tar_key = \"{}/output/output.tar.gz\".format(job_name)\n\nintermediate_folder_key = \"{}/output/intermediate/\".format(job_name)\noutput_url = \"s3://{}/{}\".format(s3_bucket, output_tar_key)\nintermediate_url = \"s3://{}/{}\".format(s3_bucket, intermediate_folder_key)\n\nprint(\"S3 job path: {}\".format(s3_url))\nprint(\"Output.tar.gz location: {}\".format(output_url))\nprint(\"Intermediate folder path: {}\".format(intermediate_url))\n \ntmp_dir = \"/tmp/{}\".format(job_name)\nos.system(\"mkdir {}\".format(tmp_dir))\nprint(\"Create local folder {}\".format(tmp_dir))",
"_____no_output_____"
]
],
[
[
"## Visualization",
"_____no_output_____"
],
[
"### Plot metrics for training job\nWe can pull the reward metric of the training and plot it to see the performance of the model over time.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport pandas as pd\n\ncsv_file_name = \"worker_0.simple_rl_graph.main_level.main_level.agent_0.csv\"\nkey = os.path.join(intermediate_folder_key, csv_file_name)\nwait_for_s3_object(s3_bucket, key, tmp_dir)\n\ncsv_file = \"{}/{}\".format(tmp_dir, csv_file_name)\ndf = pd.read_csv(csv_file)\ndf = df.dropna(subset=['Training Reward'])\nx_axis = 'Episode #'\ny_axis = 'Training Reward'\n\nplt = df.plot(x=x_axis,y=y_axis, figsize=(12,5), legend=True, style='b-')\nplt.set_ylabel(y_axis);\nplt.set_xlabel(x_axis);",
"_____no_output_____"
]
],
[
[
"## Evaluation of RL models\n\nWe use the last checkpointed model to run evaluation for the RL Agent. \n\n### Load checkpointed model\n\nCheckpointed data from the previously trained models will be passed on for evaluation / inference in the checkpoint channel. In local mode, we can simply use the local directory, whereas in the SageMaker mode, it needs to be moved to S3 first.",
"_____no_output_____"
]
],
[
[
"wait_for_s3_object(s3_bucket, output_tar_key, tmp_dir) \n\nif not os.path.isfile(\"{}/output.tar.gz\".format(tmp_dir)):\n raise FileNotFoundError(\"File output.tar.gz not found\")\nos.system(\"tar -xvzf {}/output.tar.gz -C {}\".format(tmp_dir, tmp_dir))\n\nif local_mode:\n checkpoint_dir = \"{}/data/checkpoint\".format(tmp_dir)\nelse:\n checkpoint_dir = \"{}/checkpoint\".format(tmp_dir)\n\nprint(\"Checkpoint directory {}\".format(checkpoint_dir))",
"_____no_output_____"
],
[
"if local_mode:\n checkpoint_path = 'file://{}'.format(checkpoint_dir)\n print(\"Local checkpoint file path: {}\".format(checkpoint_path))\nelse:\n checkpoint_path = \"s3://{}/{}/checkpoint/\".format(s3_bucket, job_name)\n if not os.listdir(checkpoint_dir):\n raise FileNotFoundError(\"Checkpoint files not found under the path\")\n os.system(\"aws s3 cp --recursive {} {}\".format(checkpoint_dir, checkpoint_path))\n print(\"S3 checkpoint file path: {}\".format(checkpoint_path))",
"_____no_output_____"
]
],
[
[
"### Run the evaluation step\n\nUse the checkpointed model to run the evaluation step. ",
"_____no_output_____"
]
],
[
[
"estimator_eval = RLEstimator(entry_point=\"evaluate-coach.py\",\n source_dir='src',\n dependencies=[\"common/sagemaker_rl\"],\n image_uri=custom_image_name,\n role=role,\n instance_type=instance_type,\n instance_count=1,\n output_path=s3_output_path,\n base_job_name=job_name_prefix+\"-evaluation\",\n hyperparameters = {\n \"RLCOACH_PRESET\": \"preset-energy-plus-clipped-ppo\",\n \"evaluate_steps\": 288*2, #2 episodes, i.e. 2 days\n }\n )\n\nestimator_eval.fit({'checkpoint': checkpoint_path})",
"_____no_output_____"
]
],
[
[
"# Model deployment",
"_____no_output_____"
],
[
"Since we specified MXNet when configuring the RLEstimator, the MXNet deployment container will be used for hosting.",
"_____no_output_____"
]
],
[
[
"from sagemaker.mxnet.model import MXNetModel\n\nmodel = MXNetModel(model_data=estimator.model_data,\n entry_point='src/deploy-mxnet-coach.py',\n framework_version='1.8.0',\n py_version=\"py37\",\n role=role)\n\npredictor = model.deploy(initial_instance_count=1,\n instance_type=instance_type)",
"_____no_output_____"
]
],
[
[
"We can test the endpoint with a samples observation, where the current room temperature is high. Since the environment vector was of the form `[outdoor_temperature, outdoor_humidity, indoor_humidity]` and we used observation normalization in our preset, we choose an observation of `[0, 0, 2]`. Since we're deploying a PPO model, our model returns both state value and actions.",
"_____no_output_____"
]
],
[
[
"action, action_mean, action_std = predictor.predict(np.array([0., 0., 2.,]))\naction_mean",
"_____no_output_____"
]
],
[
[
"We can see heating and cooling setpoints are returned from the model, and these can be used to control the HVAC system for efficient energy usage. More training iterations will help improve the model further.",
"_____no_output_____"
],
[
"### Clean up endpoint",
"_____no_output_____"
]
],
[
[
"predictor.delete_endpoint()",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
]
|
e796f8bb634c758d63748ee1645cac61bdb78a82 | 73,473 | ipynb | Jupyter Notebook | experiments/misspec-symmetry/plotter.ipynb | mfinzi/residual-pathway-priors | f1b1910bd9cb69f3d6121fdb9b68e82d55db9983 | [
"BSD-2-Clause"
]
| 9 | 2021-11-23T18:21:57.000Z | 2022-02-10T06:29:21.000Z | experiments/misspec-symmetry/plotter.ipynb | mfinzi/residual-pathway-priors | f1b1910bd9cb69f3d6121fdb9b68e82d55db9983 | [
"BSD-2-Clause"
]
| null | null | null | experiments/misspec-symmetry/plotter.ipynb | mfinzi/residual-pathway-priors | f1b1910bd9cb69f3d6121fdb9b68e82d55db9983 | [
"BSD-2-Clause"
]
| null | null | null | 290.407115 | 37,236 | 0.932397 | [
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport matplotlib as mpl\nimport sys\nimport os\nimport cmocean\nimport cmocean.cm as cmo",
"_____no_output_____"
],
[
"os.path.exists",
"_____no_output_____"
],
[
"full_df = pd.DataFrame()\ncols = ['Trial', \"tr_mse\", 'te_mse']",
"_____no_output_____"
],
[
"fname = \"./saved-outputs/inertia_log_mixedemlp_basic1e-05_equiv1e-05.pkl\"\nrpp_df = pd.read_pickle(fname)\nrpp_df.columns = cols\nrpp_df['type'] = 'RPP'\nfull_df = pd.concat((full_df, rpp_df))\n\n\nfname = \"./saved-outputs/inertia_log_mlp_basic100.0_equiv0.001.pkl\"\nmlp_df = pd.read_pickle(fname)\nmlp_df.columns = cols\nmlp_df['type'] = \"MLP\"\nfull_df = pd.concat((full_df, mlp_df))\n\n\nfname = \"./saved-outputs/inertia_log_emlp_basic100.0_equiv0.001.pkl\"\nemlp_df = pd.read_pickle(fname)\nemlp_df.columns = cols\nemlp_df['type'] = \"EMLP\"\nfull_df = pd.concat((full_df, emlp_df))",
"_____no_output_____"
],
[
"full_df['log_te_mse'] = np.log(full_df['te_mse'])",
"_____no_output_____"
],
[
"cpal = sns.color_palette(\"cmo.matter\", n_colors=3)\n\nfs = 30\nalpha = 0.75\nfig, ax = plt.subplots(1,1, dpi=150, figsize=(8, 3))\nvlns = sns.boxplot(x='type', y='log_te_mse', data=full_df, palette=\"Blues\", ax=ax)\n# for violin in vlns.collections[::2]:\n# violin.set_alpha(alpha)\nax.set_xlabel(\"\", fontsize=fs)\nax.set_ylabel(\"Log MSE\", fontsize=fs)\nax.tick_params(\"both\", labelsize=fs-2)\nax.set_title(\"Inertia; SL(3)\", fontsize=fs+2)\nsns.despine()\nplt.savefig(\"./misspec_inertia.pdf\", bbox_inches='tight')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Pendulum Data",
"_____no_output_____"
]
],
[
[
"full_df = pd.DataFrame()\ncols = ['Trial', \"tr_mse\", 'te_mse']\n\nfname = \"./saved-outputs/log_mixedemlp_basic1e-05_equiv1e-05.pkl\"\nrpp_df = pd.read_pickle(fname)\nrpp_df.columns = cols\nrpp_df['type'] = 'RPP'\nfull_df = pd.concat((full_df, rpp_df))\n\n\nfname = \"./saved-outputs/log_mlp_basic0.01_equiv0.0001.pkl\"\nmlp_df = pd.read_pickle(fname)\nmlp_df.columns = cols\nmlp_df['type'] = \"MLP\"\nfull_df = pd.concat((full_df, mlp_df))\n\n\nfname = \"./saved-outputs/log_emlp_basic0.01_equiv0.0001.pkl\"\nemlp_df = pd.read_pickle(fname)\nemlp_df.columns = cols\nemlp_df['type'] = \"EMLP\"\nfull_df = pd.concat((full_df, emlp_df))",
"_____no_output_____"
],
[
"full_df['log_te_mse'] = np.log(full_df['te_mse'])",
"_____no_output_____"
],
[
"cpal = sns.color_palette(\"cmo.matter\", n_colors=3)\n\nfs = 30\nalpha = 0.75\nfig, ax = plt.subplots(1,1, dpi=150, figsize=(8, 3))\nvlns = sns.boxplot(x='type', y='log_te_mse', data=full_df, palette=\"Blues\", ax=ax)\n# for violin in vlns.collections[::2]:\n# violin.set_alpha(alpha)\nax.set_xlabel(\"\", fontsize=fs)\nax.set_ylabel(\"Log MSE\", fontsize=fs)\nax.tick_params(\"both\", labelsize=fs-2)\nax.set_title(\"Pendulum; SO(3)\", fontsize=fs+2)\nsns.despine()\nplt.savefig(\"./misspec_pendulum.pdf\", bbox_inches='tight')\nplt.show()",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
]
|
e796ff2ef9cdb6cb3300922e62bfe8d7644b17f1 | 23,486 | ipynb | Jupyter Notebook | LSTM/Music_Generation_Train1.ipynb | AbhilashPal/MuseNet | ba19493ab50fe53356f3eae82f21251225b7b8b5 | [
"MIT"
]
| 49 | 2020-04-13T02:49:55.000Z | 2022-03-07T16:11:43.000Z | LSTM/Music_Generation_Train1.ipynb | AbhilashPal/MuseNet | ba19493ab50fe53356f3eae82f21251225b7b8b5 | [
"MIT"
]
| 12 | 2020-09-26T01:13:10.000Z | 2022-02-10T02:01:19.000Z | LSTM/Music_Generation_Train1.ipynb | AbhilashPal/MuseNet | ba19493ab50fe53356f3eae82f21251225b7b8b5 | [
"MIT"
]
| 9 | 2021-02-06T20:03:23.000Z | 2022-01-07T10:44:09.000Z | 33.890332 | 571 | 0.407775 | [
[
[
"# Music Generation Using Deep Learning",
"_____no_output_____"
],
[
"## Real World Problem\n\nThis case-study focuses on generating music automatically using Recurrent Neural Network(RNN).<br> \nWe do not necessarily have to be a music expert in order to generate music. Even a non expert can generate a decent quality music using RNN.<br>\nWe all like to listen interesting music and if there is some way to generate music automatically, particularly decent quality music then it's a big leap in the world of music industry.<br><br>\n<b>Task:</b> Our task here is to take some existing music data then train a model using this existing data. The model has to learn the patterns in music that we humans enjoy. Once it learns this, the model should be able to generate new music for us. It cannot simply copy-paste from the training data. It has to understand the patterns of music to generate new music. We here are not expecting our model to generate new music which is of professional quality, but we want it to generate a decent quality music which should be melodious and good to hear.<br><br>\nNow, what is music? In short music is nothing but a sequence of musical notes. Our input to the model is a sequence of musical events/notes. Our output will be new sequence of musical events/notes. In this case-study we have limited our self to single instrument music as this is our first cut model. In future, we will extend this to multiple instrument music. ",
"_____no_output_____"
],
[
"## Data Source:\n1. http://abc.sourceforge.net/NMD/\n2. http://trillian.mit.edu/~jc/music/book/oneills/1850/X/\n\n### From first data-source, we have downloaded first two files:\n* Jigs (340 tunes)\n* Hornpipes (65 tunes)",
"_____no_output_____"
]
],
[
[
"import os\nimport json\nimport numpy as np\nimport pandas as pd\nfrom keras.models import Sequential\nfrom keras.layers import LSTM, Dropout, TimeDistributed, Dense, Activation, Embedding",
"C:\\Users\\GauravP\\Anaconda3\\lib\\site-packages\\h5py\\__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\nUsing TensorFlow backend.\n"
],
[
"data_directory = \"../Data/\"\ndata_file = \"Data_Tunes.txt\"\ncharIndex_json = \"char_to_index.json\"\nmodel_weights_directory = '../Data/Model_Weights/'\nBATCH_SIZE = 16\nSEQ_LENGTH = 64",
"_____no_output_____"
],
[
"def read_batches(all_chars, unique_chars):\n length = all_chars.shape[0]\n batch_chars = int(length / BATCH_SIZE) #155222/16 = 9701\n \n for start in range(0, batch_chars - SEQ_LENGTH, 64): #(0, 9637, 64) #it denotes number of batches. It runs everytime when\n #new batch is created. We have a total of 151 batches.\n X = np.zeros((BATCH_SIZE, SEQ_LENGTH)) #(16, 64)\n Y = np.zeros((BATCH_SIZE, SEQ_LENGTH, unique_chars)) #(16, 64, 87)\n for batch_index in range(0, 16): #it denotes each row in a batch. \n for i in range(0, 64): #it denotes each column in a batch. Each column represents each character means \n #each time-step character in a sequence.\n X[batch_index, i] = all_chars[batch_index * batch_chars + start + i]\n Y[batch_index, i, all_chars[batch_index * batch_chars + start + i + 1]] = 1 #here we have added '1' because the\n #correct label will be the next character in the sequence. So, the next character will be denoted by\n #all_chars[batch_index * batch_chars + start + i + 1]\n yield X, Y",
"_____no_output_____"
],
[
"def built_model(batch_size, seq_length, unique_chars):\n model = Sequential()\n \n model.add(Embedding(input_dim = unique_chars, output_dim = 512, batch_input_shape = (batch_size, seq_length))) \n \n model.add(LSTM(256, return_sequences = True, stateful = True))\n model.add(Dropout(0.2))\n \n model.add(LSTM(128, return_sequences = True, stateful = True))\n model.add(Dropout(0.2))\n \n model.add(TimeDistributed(Dense(unique_chars)))\n\n model.add(Activation(\"softmax\"))\n \n return model",
"_____no_output_____"
],
[
"def training_model(data, epochs = 80):\n #mapping character to index\n char_to_index = {ch: i for (i, ch) in enumerate(sorted(list(set(data))))}\n print(\"Number of unique characters in our whole tunes database = {}\".format(len(char_to_index))) #87\n \n with open(os.path.join(data_directory, charIndex_json), mode = \"w\") as f:\n json.dump(char_to_index, f)\n \n index_to_char = {i: ch for (ch, i) in char_to_index.items()}\n unique_chars = len(char_to_index)\n \n model = built_model(BATCH_SIZE, SEQ_LENGTH, unique_chars)\n model.summary()\n model.compile(loss = \"categorical_crossentropy\", optimizer = \"adam\", metrics = [\"accuracy\"])\n \n all_characters = np.asarray([char_to_index[c] for c in data], dtype = np.int32)\n print(\"Total number of characters = \"+str(all_characters.shape[0])) #155222\n \n epoch_number, loss, accuracy = [], [], []\n \n for epoch in range(epochs):\n print(\"Epoch {}/{}\".format(epoch+1, epochs))\n final_epoch_loss, final_epoch_accuracy = 0, 0\n epoch_number.append(epoch+1)\n \n for i, (x, y) in enumerate(read_batches(all_characters, unique_chars)):\n final_epoch_loss, final_epoch_accuracy = model.train_on_batch(x, y) #check documentation of train_on_batch here: https://keras.io/models/sequential/\n print(\"Batch: {}, Loss: {}, Accuracy: {}\".format(i+1, final_epoch_loss, final_epoch_accuracy))\n #here, above we are reading the batches one-by-one and train our model on each batch one-by-one.\n loss.append(final_epoch_loss)\n accuracy.append(final_epoch_accuracy)\n \n #saving weights after every 10 epochs\n if (epoch + 1) % 10 == 0:\n if not os.path.exists(model_weights_directory):\n os.makedirs(model_weights_directory)\n model.save_weights(os.path.join(model_weights_directory, \"Weights_{}.h5\".format(epoch+1)))\n print('Saved Weights at epoch {} to file Weights_{}.h5'.format(epoch+1, epoch+1))\n \n #creating dataframe and record all the losses and accuracies at each epoch\n log_frame = pd.DataFrame(columns = [\"Epoch\", \"Loss\", \"Accuracy\"])\n log_frame[\"Epoch\"] = epoch_number\n log_frame[\"Loss\"] = loss\n log_frame[\"Accuracy\"] = accuracy\n log_frame.to_csv(\"../Data/log.csv\", index = False)",
"_____no_output_____"
],
[
"file = open(os.path.join(data_directory, data_file), mode = 'r')\ndata = file.read()\nfile.close()\nif __name__ == \"__main__\":\n training_model(data)",
"_____no_output_____"
],
[
"log = pd.read_csv(os.path.join(data_directory, \"log.csv\"))\nlog",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e79717fefb86939960da8fd542dd615d1c348451 | 11,512 | ipynb | Jupyter Notebook | src/datasets/ConvertMultipleClasses2BinaryClasses.ipynb | robertu94/mlsvm | aa9b8a1bdb4bcf487a827499e0cc039d590e2bb6 | [
"BSD-2-Clause"
]
| 25 | 2016-10-06T18:57:21.000Z | 2022-02-09T20:43:59.000Z | src/datasets/ConvertMultipleClasses2BinaryClasses.ipynb | robertu94/mlsvm | aa9b8a1bdb4bcf487a827499e0cc039d590e2bb6 | [
"BSD-2-Clause"
]
| 7 | 2017-08-22T18:45:19.000Z | 2019-09-20T20:09:35.000Z | src/datasets/ConvertMultipleClasses2BinaryClasses.ipynb | robertu94/mlsvm | aa9b8a1bdb4bcf487a827499e0cc039d590e2bb6 | [
"BSD-2-Clause"
]
| 14 | 2016-06-02T03:47:27.000Z | 2022-03-23T01:36:12.000Z | 24.650964 | 107 | 0.31263 | [
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"df = pd.read_csv('./test_men.csv', header=None, sep=',', names=['lbl']+[i for i in range(13)])",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.groupby('lbl').size()",
"_____no_output_____"
],
[
"df.loc[df.lbl==0, 'lbl'] = 1\ndf.loc[df.lbl==1, 'lbl'] = 1\ndf.loc[df.lbl==2, 'lbl'] = -1\ndf.loc[df.lbl==3, 'lbl'] = -1",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.groupby('lbl').size()",
"_____no_output_____"
]
],
[
[
"## Make sure the number of minority class (lbl==1) is smaller than number of majority class (lbl==-1)",
"_____no_output_____"
]
],
[
[
"sum(df.lbl)",
"_____no_output_____"
],
[
"len(df)",
"_____no_output_____"
],
[
"df.to_csv('test_men_binary.csv',header=None, sep=',', index=None)",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
]
|
e79729e106d1bb1e95c4f1a24ec10d29197ce9d9 | 258,736 | ipynb | Jupyter Notebook | resources/notebooks/OldScrapper/20Minutos-getContenido.ipynb | cfespinoza/comments-retriever | 6e46053d66297f59da2fa93a00a58158dec195ca | [
"Apache-2.0"
]
| null | null | null | resources/notebooks/OldScrapper/20Minutos-getContenido.ipynb | cfespinoza/comments-retriever | 6e46053d66297f59da2fa93a00a58158dec195ca | [
"Apache-2.0"
]
| 1 | 2021-12-13T20:23:17.000Z | 2021-12-13T20:23:17.000Z | resources/notebooks/OldScrapper/20Minutos-getContenido.ipynb | cfespinoza/comments-retriever | 6e46053d66297f59da2fa93a00a58158dec195ca | [
"Apache-2.0"
]
| null | null | null | 61.224799 | 1,594 | 0.58448 | [
[
[
"import sys\nfrom PyQt5 import QtCore, QtWidgets, QtWebEngineWidgets\nfrom lxml import html as htmlRenderer\nimport requests\nimport json\nfrom datetime import date\nimport datetime\nfrom random import *",
"_____no_output_____"
],
[
"def render(source_url):\n \"\"\"Fully render HTML, JavaScript and all.\"\"\"\n\n import sys\n from PyQt5.QtWidgets import QApplication\n from PyQt5.QtCore import QUrl\n from PyQt5.QtWebEngineWidgets import QWebEngineView\n\n class Render(QWebEngineView):\n def __init__(self, url):\n self.html = None\n self.app = QApplication(sys.argv)\n QWebEngineView.__init__(self)\n self.loadFinished.connect(self._loadFinished)\n #self.setHtml(html)\n self.load(QUrl(url))\n self.app.exec_()\n\n def _loadFinished(self, result):\n # This is an async call, you need to wait for this\n # to be called before closing the app\n self.page().toHtml(self._callable)\n\n def _callable(self, data):\n self.html = data\n # Data has been stored, it's safe to quit the app\n self.app.quit()\n\n return Render(source_url).html",
"_____no_output_____"
],
[
"url=\"https://www.20minutos.es/deportes/noticia/casillas-critico-tuit-arbitro-champions-liverpool-3611469/0/\"\nrenderUrl = render(url)\nrenderedPage = htmlRenderer.fromstring(renderUrl)",
"_____no_output_____"
],
[
"auxLinks = renderedPage.xpath(\"//div[@class='gtm-article-text']//p\")",
"_____no_output_____"
],
[
"a = auxLinks[0]",
"_____no_output_____"
],
[
"a.text_content()",
"_____no_output_____"
],
[
"contentArr = []\nfor p in auxLinks:\n contentArr.append(p.text_content())\ncontentArr",
"_____no_output_____"
],
[
"\"\".join([ parrafo for parrafo in contentArr])",
"_____no_output_____"
],
[
"commentEl = auxLinks[0]\ncommentElVal = commentEl.get(\"id\").split(\"_\")\nidNoticia = commentElVal[len(commentElVal)-1]\nperfiloHiloId = \"_{}\".format(idNoticia)",
"_____no_output_____"
],
[
"# Get Comments Info\n\nurlInfoComments = \"https://elpais.com/ThreadeskupSimple\"\nrnd = random()\n\ninfoArg = {\n \"action\": \"info\",\n \"th\": idNoticia,\n \"rnd\": rnd\n}",
"_____no_output_____"
],
[
"responseInfoComments = requests.get(urlInfoComments, infoArg)",
"_____no_output_____"
],
[
"responseInfoComments",
"_____no_output_____"
],
[
"infoComments = json.loads(responseInfoComments.text)",
"_____no_output_____"
],
[
"infoComments",
"_____no_output_____"
],
[
"pages = infoComments[\"perfilesHilos\"][perfiloHiloId][\"numero_mensajes\"]//50\nif (infoComments[\"perfilesHilos\"][perfiloHiloId][\"numero_mensajes\"]%50 > 0) :\n pages = pages + 1\n\npages",
"_____no_output_____"
],
[
"# https://elpais.com/OuteskupSimple?s=&rnd=0.5448952094970048&th=2&msg=1564664936-bca025601586bc5a00ef0c26fdd878f6&p=2&nummsg=120&tt=1\n# s=\n# &rnd=0.5448952094970048\n# &th=2\n# &msg=1564664936-bca025601586bc5a00ef0c26fdd878f6\n# &p=2\n# &nummsg=120\n# &tt=1\nrnd = random()\ncommentsArgs = {\n \"s\": \"\",\n \"rnd\": rnd,\n \"th\": 1,\n \"msg\": idNoticia,\n \"nummsg\": infoComments[\"perfilesHilos\"][perfiloHiloId][\"numero_mensajes\"],\n \"tt\": 1\n}",
"_____no_output_____"
],
[
"urlGetComments = \"https://elpais.com/OuteskupSimple\"\nresponseComments = requests.get(urlGetComments, commentsArgs)",
"_____no_output_____"
],
[
"commentsResponse = json.loads(responseComments.text)",
"_____no_output_____"
],
[
"commentsResponse",
"_____no_output_____"
],
[
"\ndef extractComments(commentsObjectList, urlNoticia=\"\", specialCase = False):\n print(\" \\t -> parsing comments list with -{}- elements:\".format(len(commentsObjectList)))\n parsedComments = []\n listToParse = commentsObjectList[1] if specialCase else commentsObjectList\n for commentObj in listToParse:\n fechaObj = datetime.datetime.fromtimestamp(commentObj[\"tsMensaje\"])\n fechaStr = fechaObj.strftime(\"%d/%m/%Y-%H:%M:%S\")\n fechaArr = fechaStr.split(\"-\")\n parsedComment = {\n \"urlNoticia\": urlNoticia,\n \"fecha\": fechaArr[0],\n \"hora\": fechaArr[1],\n \"user\": commentObj['usuarioOrigen'],\n \"commentario\": commentObj['contenido']\n }\n parsedComments.append(parsedComment)\n return parsedComments",
"_____no_output_____"
],
[
"parsedComments = extractComments(commentsResponse[\"mensajes\"], url)",
" \t -> parsing comments list with -121- elements:\n"
],
[
"parsedComments",
"_____no_output_____"
],
[
"len(parsedComments)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e7972af015990f8c5b0c5edcb288716e784830db | 16,962 | ipynb | Jupyter Notebook | nesh/01_default_cluster_example.ipynb | ExaESM-WP4/Dask-jobqueue-configs | 2bbada956455b1701424fcadb4c2ca106c4208d7 | [
"MIT"
]
| 1 | 2020-08-24T11:52:31.000Z | 2020-08-24T11:52:31.000Z | nesh/01_default_cluster_example.ipynb | ExaESM-WP4/dask-jobqueue-configs | 2bbada956455b1701424fcadb4c2ca106c4208d7 | [
"MIT"
]
| 7 | 2020-01-24T18:00:18.000Z | 2020-02-14T16:47:10.000Z | nesh/01_default_cluster_example.ipynb | ExaESM-WP4/Dask-jobqueue-configs | 2bbada956455b1701424fcadb4c2ca106c4208d7 | [
"MIT"
]
| null | null | null | 34.40568 | 264 | 0.449711 | [
[
[
"# Dask jobqueue example for NEC Linux cluster\ncovers the following aspects, i.e. how to\n* load project and machine specific Dask jobqueue configurations\n* open, scale and close a default jobqueue cluster\n* do an example calculation on larger than memory data",
"_____no_output_____"
],
[
"## Load jobqueue configuration defaults",
"_____no_output_____"
]
],
[
[
"import os \nos.environ['DASK_CONFIG']='.' # use local directory to look up Dask configurations",
"_____no_output_____"
],
[
"import dask.config\ndask.config.get('jobqueue') # prints available jobqueue configurations",
"_____no_output_____"
]
],
[
[
"## Set up jobqueue cluster ...",
"_____no_output_____"
]
],
[
[
"import dask_jobqueue\ndefault_cluster = dask_jobqueue.PBSCluster(config_name='nesh-jobqueue-config')",
"_____no_output_____"
],
[
"print(default_cluster.job_script())",
"#!/bin/bash\n\n#PBS -N dask-worker\n#PBS -q clmedium\n#PBS -l elapstim_req=00:45:00,cpunum_job=4,memsz_job=24gb\n#PBS -o dask_jobqueue_logs/dask-worker.o%s\n#PBS -e dask_jobqueue_logs/dask-worker.e%s\nJOB_ID=${PBS_JOBID%%.*}\n\n/sfs/fs6/home-geomar/smomw260/miniconda3/envs/dask-minimal-20191218/bin/python -m distributed.cli.dask_worker tcp://192.168.31.10:32956 --nthreads 4 --memory-limit 24.00GB --name name --nanny --death-timeout 60 --local-directory /scratch --interface ib0\n\n"
]
],
[
[
"## ... and the client process",
"_____no_output_____"
]
],
[
[
"import dask.distributed as dask_distributed\ndefault_cluster_client = dask_distributed.Client(default_cluster)",
"_____no_output_____"
]
],
[
[
"## Start jobqueue workers",
"_____no_output_____"
]
],
[
[
"default_cluster.scale(jobs=2)",
"_____no_output_____"
],
[
"!qstat",
"RequestID ReqName UserName Queue Pri STT S Memory CPU Elapse R H M Jobs\n--------------- -------- -------- -------- ---- --- - -------- -------- -------- - - - ----\n182478.nesh-bat dask-wor smomw260 clmedium 0 RUN - 0.00B 0.00 8 Y Y Y 1 \n182479.nesh-bat dask-wor smomw260 clmedium 0 RUN - 0.00B 0.00 8 Y Y Y 1 \n"
],
[
"default_cluster_client",
"_____no_output_____"
]
],
[
[
"## Do calculation on larger than memory data",
"_____no_output_____"
]
],
[
[
"import dask.array as da",
"_____no_output_____"
],
[
"fake_data = da.random.uniform(0, 1, size=(365, 1e4, 1e4), chunks=(365,500,500)) # problem specific chunking\nfake_data",
"_____no_output_____"
],
[
"import time",
"_____no_output_____"
],
[
"start_time = time.time()\nfake_data.mean(axis=0).compute()\nelapsed = time.time() - start_time",
"_____no_output_____"
],
[
"print('elapse time ',elapsed,' in seconds')",
"elapse time 46.89112448692322 in seconds\n"
]
],
[
[
"## Close jobqueue cluster and client process",
"_____no_output_____"
]
],
[
[
"!qstat",
"RequestID ReqName UserName Queue Pri STT S Memory CPU Elapse R H M Jobs\n--------------- -------- -------- -------- ---- --- - -------- -------- -------- - - - ----\n182478.nesh-bat dask-wor smomw260 clmedium 0 RUN - 2.58G 157.12 94 Y Y Y 1 \n182479.nesh-bat dask-wor smomw260 clmedium 0 RUN - 804.87M 157.49 94 Y Y Y 1 \n"
],
[
"default_cluster.close()\ndefault_cluster_client.close()",
"_____no_output_____"
],
[
"!qstat",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
]
|
e79731d82e7b1f6f6be929cdcb6f540b75d70d87 | 190,502 | ipynb | Jupyter Notebook | qiskit/advanced/aqua/optimization/max_cut_and_tsp.ipynb | gvvynplaine/qiskit-iqx-tutorials | 40af3da7aa86ce190d04f147daf46fbc893a1966 | [
"Apache-2.0"
]
| 2 | 2020-01-08T22:16:04.000Z | 2020-02-03T12:10:25.000Z | qiskit/advanced/aqua/optimization/max_cut_and_tsp.ipynb | gvvynplaine/qiskit-iqx-tutorials | 40af3da7aa86ce190d04f147daf46fbc893a1966 | [
"Apache-2.0"
]
| null | null | null | qiskit/advanced/aqua/optimization/max_cut_and_tsp.ipynb | gvvynplaine/qiskit-iqx-tutorials | 40af3da7aa86ce190d04f147daf46fbc893a1966 | [
"Apache-2.0"
]
| 1 | 2019-06-16T04:38:05.000Z | 2019-06-16T04:38:05.000Z | 193.99389 | 21,748 | 0.895177 | [
[
[
"",
"_____no_output_____"
],
[
"# _*Qiskit Aqua: Experimenting with Max-Cut problem and Traveling Salesman problem with variational quantum eigensolver*_ \n\nThe latest version of this notebook is available on https://github.com/Qiskit/qiskit-tutorial.\n\n***\n### Contributors\nAntonio Mezzacapo<sup>[1]</sup>, Jay Gambetta<sup>[1]</sup>, Kristan Temme<sup>[1]</sup>, Ramis Movassagh<sup>[1]</sup>, Albert Frisch<sup>[1]</sup>, Takashi Imamichi<sup>[1]</sup>, Giacomo Nannicni<sup>[1]</sup>, Richard Chen<sup>[1]</sup>, Marco Pistoia<sup>[1]</sup>, Stephen Wood<sup>[1]</sup>\n### Affiliation\n- <sup>[1]</sup>IBMQ",
"_____no_output_____"
],
[
"## Introduction\n\nMany problems in quantitative fields such as finance and engineering are optimization problems. Optimization problems lie at the core of complex decision-making and definition of strategies. \n\nOptimization (or combinatorial optimization) means searching for an optimal solution in a finite or countably infinite set of potential solutions. Optimality is defined with respect to some criterion function, which is to be minimized or maximized. This is typically called cost function or objective function. \n\n**Typical optimization problems**\n\nMinimization: cost, distance, length of a traversal, weight, processing time, material, energy consumption, number of objects\n\nMaximization: profit, value, output, return, yield, utility, efficiency, capacity, number of objects \n\nWe consider here max-cut problems of practical interest in many fields, and show how they can be nmapped on quantum computers.\n\n\n### Weighted Max-Cut\n\nMax-Cut is an NP-complete problem, with applications in clustering, network science, and statistical physics. To grasp how practical applications are mapped into given Max-Cut instances, consider a system of many people that can interact and influence each other. Individuals can be represented by vertices of a graph, and their interactions seen as pairwise connections between vertices of the graph, or edges. With this representation in mind, it is easy to model typical marketing problems. For example, suppose that it is assumed that individuals will influence each other's buying decisions, and knowledge is given about how strong they will influence each other. The influence can be modeled by weights assigned on each edge of the graph. It is possible then to predict the outcome of a marketing strategy in which products are offered for free to some individuals, and then ask which is the optimal subset of individuals that should get the free products, in order to maximize revenues.\n\nThe formal definition of this problem is the following:\n\nConsider an $n$-node undirected graph *G = (V, E)* where *|V| = n* with edge weights $w_{ij}>0$, $w_{ij}=w_{ji}$, for $(i, j)\\in E$. A cut is defined as a partition of the original set V into two subsets. The cost function to be optimized is in this case the sum of weights of edges connecting points in the two different subsets, *crossing* the cut. By assigning $x_i=0$ or $x_i=1$ to each node $i$, one tries to maximize the global profit function (here and in the following summations run over indices 0,1,...n-1)\n\n$$\\tilde{C}(\\textbf{x}) = \\sum_{i,j} w_{ij} x_i (1-x_j).$$\n\nIn our simple marketing model, $w_{ij}$ represents the probability that the person $j$ will buy a product after $i$ gets a free one. Note that the weights $w_{ij}$ can in principle be greater than $1$, corresponding to the case where the individual $j$ will buy more than one product. Maximizing the total buying probability corresponds to maximizing the total future revenues. In the case where the profit probability will be greater than the cost of the initial free samples, the strategy is a convenient one. An extension to this model has the nodes themselves carry weights, which can be regarded, in our marketing model, as the likelihood that a person granted with a free sample of the product will buy it again in the future. With this additional information in our model, the objective function to maximize becomes \n\n$$C(\\textbf{x}) = \\sum_{i,j} w_{ij} x_i (1-x_j)+\\sum_i w_i x_i. $$\n \nIn order to find a solution to this problem on a quantum computer, one needs first to map it to an Ising Hamiltonian. This can be done with the assignment $x_i\\rightarrow (1-Z_i)/2$, where $Z_i$ is the Pauli Z operator that has eigenvalues $\\pm 1$. Doing this we find that \n\n$$C(\\textbf{Z}) = \\sum_{i,j} \\frac{w_{ij}}{4} (1-Z_i)(1+Z_j) + \\sum_i \\frac{w_i}{2} (1-Z_i) = -\\frac{1}{2}\\left( \\sum_{i<j} w_{ij} Z_i Z_j +\\sum_i w_i Z_i\\right)+\\mathrm{const},$$\n\nwhere const = $\\sum_{i<j}w_{ij}/2+\\sum_i w_i/2 $. In other terms, the weighted Max-Cut problem is equivalent to minimizing the Ising Hamiltonian \n\n$$ H = \\sum_i w_i Z_i + \\sum_{i<j} w_{ij} Z_iZ_j.$$\n\nAqua can generate the Ising Hamiltonian for the first profit function $\\tilde{C}$.\n\n\n### Approximate Universal Quantum Computing for Optimization Problems\n\nThere has been a considerable amount of interest in recent times about the use of quantum computers to find a solution to combinatorial problems. It is important to say that, given the classical nature of combinatorial problems, exponential speedup in using quantum computers compared to the best classical algorithms is not guaranteed. However, due to the nature and importance of the target problems, it is worth investigating heuristic approaches on a quantum computer that could indeed speed up some problem instances. Here we demonstrate an approach that is based on the Quantum Approximate Optimization Algorithm by Farhi, Goldstone, and Gutman (2014). We frame the algorithm in the context of *approximate quantum computing*, given its heuristic nature. \n\nThe Algorithm works as follows:\n1. Choose the $w_i$ and $w_{ij}$ in the target Ising problem. In principle, even higher powers of Z are allowed.\n2. Choose the depth of the quantum circuit $m$. Note that the depth can be modified adaptively.\n3. Choose a set of controls $\\theta$ and make a trial function $|\\psi(\\boldsymbol\\theta)\\rangle$, built using a quantum circuit made of C-Phase gates and single-qubit Y rotations, parameterized by the components of $\\boldsymbol\\theta$. \n4. Evaluate $C(\\boldsymbol\\theta) = \\langle\\psi(\\boldsymbol\\theta)~|H|~\\psi(\\boldsymbol\\theta)\\rangle = \\sum_i w_i \\langle\\psi(\\boldsymbol\\theta)~|Z_i|~\\psi(\\boldsymbol\\theta)\\rangle+ \\sum_{i<j} w_{ij} \\langle\\psi(\\boldsymbol\\theta)~|Z_iZ_j|~\\psi(\\boldsymbol\\theta)\\rangle$ by sampling the outcome of the circuit in the Z-basis and adding the expectation values of the individual Ising terms together. In general, different control points around $\\boldsymbol\\theta$ have to be estimated, depending on the classical optimizer chosen. \n5. Use a classical optimizer to choose a new set of controls.\n6. Continue until $C(\\boldsymbol\\theta)$ reaches a minimum, close enough to the solution $\\boldsymbol\\theta^*$.\n7. Use the last $\\boldsymbol\\theta$ to generate a final set of samples from the distribution $|\\langle z_i~|\\psi(\\boldsymbol\\theta)\\rangle|^2\\;\\forall i$ to obtain the answer.\n \nIt is our belief the difficulty of finding good heuristic algorithms will come down to the choice of an appropriate trial wavefunction. For example, one could consider a trial function whose entanglement best aligns with the target problem, or simply make the amount of entanglement a variable. In this tutorial, we will consider a simple trial function of the form\n\n$$|\\psi(\\theta)\\rangle = [U_\\mathrm{single}(\\boldsymbol\\theta) U_\\mathrm{entangler}]^m |+\\rangle$$\n\nwhere $U_\\mathrm{entangler}$ is a collection of C-Phase gates (fully entangling gates), and $U_\\mathrm{single}(\\theta) = \\prod_{i=1}^n Y(\\theta_{i})$, where $n$ is the number of qubits and $m$ is the depth of the quantum circuit. The motivation for this choice is that for these classical problems this choice allows us to search over the space of quantum states that have only real coefficients, still exploiting the entanglement to potentially converge faster to the solution.\n\nOne advantage of using this sampling method compared to adiabatic approaches is that the target Ising Hamiltonian does not have to be implemented directly on hardware, allowing this algorithm not to be limited to the connectivity of the device. Furthermore, higher-order terms in the cost function, such as $Z_iZ_jZ_k$, can also be sampled efficiently, whereas in adiabatic or annealing approaches they are generally impractical to deal with. \n\n\nReferences:\n- A. Lucas, Frontiers in Physics 2, 5 (2014)\n- E. Farhi, J. Goldstone, S. Gutmann e-print arXiv 1411.4028 (2014)\n- D. Wecker, M. B. Hastings, M. Troyer Phys. Rev. A 94, 022309 (2016)\n- E. Farhi, J. Goldstone, S. Gutmann, H. Neven e-print arXiv 1703.06199 (2017)",
"_____no_output_____"
]
],
[
[
"# useful additional packages \nimport matplotlib.pyplot as plt\nimport matplotlib.axes as axes\n%matplotlib inline\nimport numpy as np\nimport networkx as nx\n\nfrom qiskit import BasicAer\nfrom qiskit.tools.visualization import plot_histogram\nfrom qiskit.optimization.ising import max_cut, tsp\nfrom qiskit.aqua.algorithms import VQE, ExactEigensolver\nfrom qiskit.aqua.components.optimizers import SPSA\nfrom qiskit.aqua.components.variational_forms import RY\nfrom qiskit.aqua import QuantumInstance\nfrom qiskit.optimization.ising.common import sample_most_likely\n\n# setup aqua logging\nimport logging\nfrom qiskit.aqua import set_qiskit_aqua_logging\n# set_qiskit_aqua_logging(logging.DEBUG) # choose INFO, DEBUG to see the log",
"_____no_output_____"
]
],
[
[
"### [Optional] Setup token to run the experiment on a real device\nIf you would like to run the experiment on a real device, you need to setup your account first.\n\nNote: If you do not store your token yet, use `IBMQ.save_account('MY_API_TOKEN')` to store it first.",
"_____no_output_____"
]
],
[
[
"from qiskit import IBMQ\n# provider = IBMQ.load_account()",
"_____no_output_____"
]
],
[
[
"## Max-Cut problem",
"_____no_output_____"
]
],
[
[
"# Generating a graph of 4 nodes \n\nn=4 # Number of nodes in graph\nG=nx.Graph()\nG.add_nodes_from(np.arange(0,n,1))\nelist=[(0,1,1.0),(0,2,1.0),(0,3,1.0),(1,2,1.0),(2,3,1.0)]\n# tuple is (i,j,weight) where (i,j) is the edge\nG.add_weighted_edges_from(elist)\n\ncolors = ['r' for node in G.nodes()]\npos = nx.spring_layout(G)\ndefault_axes = plt.axes(frameon=True)\nnx.draw_networkx(G, node_color=colors, node_size=600, alpha=.8, ax=default_axes, pos=pos)",
"_____no_output_____"
],
[
"# Computing the weight matrix from the random graph\nw = np.zeros([n,n])\nfor i in range(n):\n for j in range(n):\n temp = G.get_edge_data(i,j,default=0)\n if temp != 0:\n w[i,j] = temp['weight'] \nprint(w)",
"[[0. 1. 1. 1.]\n [1. 0. 1. 0.]\n [1. 1. 0. 1.]\n [1. 0. 1. 0.]]\n"
]
],
[
[
"### Brute force approach\n\nTry all possible $2^n$ combinations. For $n = 4$, as in this example, one deals with only 16 combinations, but for n = 1000, one has 1.071509e+30 combinations, which is impractical to deal with by using a brute force approach. ",
"_____no_output_____"
]
],
[
[
"best_cost_brute = 0\nfor b in range(2**n):\n x = [int(t) for t in reversed(list(bin(b)[2:].zfill(n)))]\n cost = 0\n for i in range(n):\n for j in range(n):\n cost = cost + w[i,j]*x[i]*(1-x[j])\n if best_cost_brute < cost:\n best_cost_brute = cost\n xbest_brute = x \n print('case = ' + str(x)+ ' cost = ' + str(cost))\n\ncolors = ['r' if xbest_brute[i] == 0 else 'b' for i in range(n)]\nnx.draw_networkx(G, node_color=colors, node_size=600, alpha=.8, pos=pos)\nprint('\\nBest solution = ' + str(xbest_brute) + ' cost = ' + str(best_cost_brute)) ",
"case = [0, 0, 0, 0] cost = 0.0\ncase = [1, 0, 0, 0] cost = 3.0\ncase = [0, 1, 0, 0] cost = 2.0\ncase = [1, 1, 0, 0] cost = 3.0\ncase = [0, 0, 1, 0] cost = 3.0\ncase = [1, 0, 1, 0] cost = 4.0\ncase = [0, 1, 1, 0] cost = 3.0\ncase = [1, 1, 1, 0] cost = 2.0\ncase = [0, 0, 0, 1] cost = 2.0\ncase = [1, 0, 0, 1] cost = 3.0\ncase = [0, 1, 0, 1] cost = 4.0\ncase = [1, 1, 0, 1] cost = 3.0\ncase = [0, 0, 1, 1] cost = 3.0\ncase = [1, 0, 1, 1] cost = 2.0\ncase = [0, 1, 1, 1] cost = 3.0\ncase = [1, 1, 1, 1] cost = 0.0\n\nBest solution = [1, 0, 1, 0] cost = 4.0\n"
]
],
[
[
"### Mapping to the Ising problem",
"_____no_output_____"
]
],
[
[
"qubitOp, offset = max_cut.get_operator(w)",
"_____no_output_____"
]
],
[
[
"### [Optional] Using DOcplex for mapping to the Ising problem\nUsing ```docplex.get_qubitops``` is a different way to create an Ising Hamiltonian of Max-Cut. ```docplex.get_qubitops``` can create a corresponding Ising Hamiltonian from an optimization model of Max-Cut. An example of using ```docplex.get_qubitops``` is as below. ",
"_____no_output_____"
]
],
[
[
"from docplex.mp.model import Model\nfrom qiskit.optimization.ising import docplex\n\n# Create an instance of a model and variables.\nmdl = Model(name='max_cut')\nx = {i: mdl.binary_var(name='x_{0}'.format(i)) for i in range(n)}\n\n# Object function\nmax_cut_func = mdl.sum(w[i,j]* x[i] * ( 1 - x[j] ) for i in range(n) for j in range(n))\nmdl.maximize(max_cut_func)\n\n# No constraints for Max-Cut problems.",
"_____no_output_____"
],
[
"qubitOp_docplex, offset_docplex = docplex.get_operator(mdl)",
"_____no_output_____"
]
],
[
[
"### Checking that the full Hamiltonian gives the right cost ",
"_____no_output_____"
]
],
[
[
"#Making the Hamiltonian in its full form and getting the lowest eigenvalue and eigenvector\nee = ExactEigensolver(qubitOp, k=1)\nresult = ee.run()\n\nx = sample_most_likely(result['eigvecs'][0])\nprint('energy:', result['energy'])\nprint('max-cut objective:', result['energy'] + offset)\nprint('solution:', max_cut.get_graph_solution(x))\nprint('solution objective:', max_cut.max_cut_value(x, w))\n\ncolors = ['r' if max_cut.get_graph_solution(x)[i] == 0 else 'b' for i in range(n)]\nnx.draw_networkx(G, node_color=colors, node_size=600, alpha = .8, pos=pos)",
"energy: -1.5\nmax-cut objective: -4.0\nsolution: [0. 1. 0. 1.]\nsolution objective: 4.0\n"
]
],
[
[
"### Running it on quantum computer\nWe run the optimization routine using a feedback loop with a quantum computer that uses trial functions built with Y single-qubit rotations, $U_\\mathrm{single}(\\theta) = \\prod_{i=1}^n Y(\\theta_{i})$, and entangler steps $U_\\mathrm{entangler}$.",
"_____no_output_____"
]
],
[
[
"seed = 10598\n\nspsa = SPSA(max_trials=300)\nry = RY(qubitOp.num_qubits, depth=5, entanglement='linear')\nvqe = VQE(qubitOp, ry, spsa)\n\nbackend = BasicAer.get_backend('statevector_simulator')\nquantum_instance = QuantumInstance(backend, seed_simulator=seed, seed_transpiler=seed)\n\nresult = vqe.run(quantum_instance)\n\nx = sample_most_likely(result['eigvecs'][0])\nprint('energy:', result['energy'])\nprint('time:', result['eval_time'])\nprint('max-cut objective:', result['energy'] + offset)\nprint('solution:', max_cut.get_graph_solution(x))\nprint('solution objective:', max_cut.max_cut_value(x, w))\n\ncolors = ['r' if max_cut.get_graph_solution(x)[i] == 0 else 'b' for i in range(n)]\nnx.draw_networkx(G, node_color=colors, node_size=600, alpha = .8, pos=pos)",
"energy: -1.4999575960856462\ntime: 6.378453969955444\nmax-cut objective: -3.999957596085646\nsolution: [1. 0. 1. 0.]\nsolution objective: 4.0\n"
],
[
"# run quantum algorithm with shots\nseed = 10598\n\nspsa = SPSA(max_trials=300)\nry = RY(qubitOp.num_qubits, depth=5, entanglement='linear')\nvqe = VQE(qubitOp, ry, spsa)\n\nbackend = BasicAer.get_backend('qasm_simulator')\nquantum_instance = QuantumInstance(backend, shots=1024, seed_simulator=seed, seed_transpiler=seed)\n\nresult = vqe.run(quantum_instance)\n\nx = sample_most_likely(result['eigvecs'][0])\nprint('energy:', result['energy'])\nprint('time:', result['eval_time'])\nprint('max-cut objective:', result['energy'] + offset)\nprint('solution:', max_cut.get_graph_solution(x))\nprint('solution objective:', max_cut.max_cut_value(x, w))\nplot_histogram(result['eigvecs'][0])\n\ncolors = ['r' if max_cut.get_graph_solution(x)[i] == 0 else 'b' for i in range(n)]\nnx.draw_networkx(G, node_color=colors, node_size=600, alpha = .8, pos=pos)",
"energy: -1.5\ntime: 11.74726128578186\nmax-cut objective: -4.0\nsolution: [0 1 0 1]\nsolution objective: 4.0\n"
]
],
[
[
"### [Optional] Checking that the full Hamiltonian made by ```docplex.get_operator``` gives the right cost",
"_____no_output_____"
]
],
[
[
"#Making the Hamiltonian in its full form and getting the lowest eigenvalue and eigenvector\nee = ExactEigensolver(qubitOp_docplex, k=1)\nresult = ee.run()\n\nx = sample_most_likely(result['eigvecs'][0])\nprint('energy:', result['energy'])\nprint('max-cut objective:', result['energy'] + offset_docplex)\nprint('solution:', max_cut.get_graph_solution(x))\nprint('solution objective:', max_cut.max_cut_value(x, w))\n\ncolors = ['r' if max_cut.get_graph_solution(x)[i] == 0 else 'b' for i in range(n)]\nnx.draw_networkx(G, node_color=colors, node_size=600, alpha = .8, pos=pos)",
"energy: -1.5\nmax-cut objective: -4.0\nsolution: [0. 1. 0. 1.]\nsolution objective: 4.0\n"
]
],
[
[
"## Traveling Salesman Problem\n\nIn addition to being a notorious NP-complete problem that has drawn the attention of computer scientists and mathematicians for over two centuries, the Traveling Salesman Problem (TSP) has important bearings on finance and marketing, as its name suggests. Colloquially speaking, the traveling salesman is a person that goes from city to city to sell merchandise. The objective in this case is to find the shortest path that would enable the salesman to visit all the cities and return to its hometown, i.e. the city where he started traveling. By doing this, the salesman gets to maximize potential sales in the least amount of time. \n\nThe problem derives its importance from its \"hardness\" and ubiquitous equivalence to other relevant combinatorial optimization problems that arise in practice.\n \nThe mathematical formulation with some early analysis was proposed by W.R. Hamilton in the early 19th century. Mathematically the problem is, as in the case of Max-Cut, best abstracted in terms of graphs. The TSP on the nodes of a graph asks for the shortest *Hamiltonian cycle* that can be taken through each of the nodes. A Hamilton cycle is a closed path that uses every vertex of a graph once. The general solution is unknown and an algorithm that finds it efficiently (e.g., in polynomial time) is not expected to exist.\n\nFind the shortest Hamiltonian cycle in a graph $G=(V,E)$ with $n=|V|$ nodes and distances, $w_{ij}$ (distance from vertex $i$ to vertex $j$). A Hamiltonian cycle is described by $N^2$ variables $x_{i,p}$, where $i$ represents the node and $p$ represents its order in a prospective cycle. The decision variable takes the value 1 if the solution occurs at node $i$ at time order $p$. We require that every node can only appear once in the cycle, and for each time a node has to occur. This amounts to the two constraints (here and in the following, whenever not specified, the summands run over 0,1,...N-1)\n\n$$\\sum_{i} x_{i,p} = 1 ~~\\forall p$$\n$$\\sum_{p} x_{i,p} = 1 ~~\\forall i.$$\n\nFor nodes in our prospective ordering, if $x_{i,p}$ and $x_{j,p+1}$ are both 1, then there should be an energy penalty if $(i,j) \\notin E$ (not connected in the graph). The form of this penalty is \n\n$$\\sum_{i,j\\notin E}\\sum_{p} x_{i,p}x_{j,p+1}>0,$$ \n\nwhere it is assumed the boundary condition of the Hamiltonian cycles $(p=N)\\equiv (p=0)$. However, here it will be assumed a fully connected graph and not include this term. The distance that needs to be minimized is \n\n$$C(\\textbf{x})=\\sum_{i,j}w_{ij}\\sum_{p} x_{i,p}x_{j,p+1}.$$\n\nPutting this all together in a single objective function to be minimized, we get the following:\n\n$$C(\\textbf{x})=\\sum_{i,j}w_{ij}\\sum_{p} x_{i,p}x_{j,p+1}+ A\\sum_p\\left(1- \\sum_i x_{i,p}\\right)^2+A\\sum_i\\left(1- \\sum_p x_{i,p}\\right)^2,$$\n\nwhere $A$ is a free parameter. One needs to ensure that $A$ is large enough so that these constraints are respected. One way to do this is to choose $A$ such that $A > \\mathrm{max}(w_{ij})$.\n\nOnce again, it is easy to map the problem in this form to a quantum computer, and the solution will be found by minimizing a Ising Hamiltonian. ",
"_____no_output_____"
]
],
[
[
"# Generating a graph of 3 nodes\nn = 3\nnum_qubits = n ** 2\nins = tsp.random_tsp(n)\nG = nx.Graph()\nG.add_nodes_from(np.arange(0, n, 1))\ncolors = ['r' for node in G.nodes()]\npos = {k: v for k, v in enumerate(ins.coord)}\ndefault_axes = plt.axes(frameon=True)\nnx.draw_networkx(G, node_color=colors, node_size=600, alpha=.8, ax=default_axes, pos=pos)\nprint('distance\\n', ins.w)",
"distance\n [[ 0. 25. 19.]\n [25. 0. 27.]\n [19. 27. 0.]]\n"
]
],
[
[
"### Brute force approach",
"_____no_output_____"
]
],
[
[
"from itertools import permutations\n\ndef brute_force_tsp(w, N):\n a=list(permutations(range(1,N)))\n last_best_distance = 1e10\n for i in a:\n distance = 0\n pre_j = 0\n for j in i:\n distance = distance + w[j,pre_j]\n pre_j = j\n distance = distance + w[pre_j,0]\n order = (0,) + i\n if distance < last_best_distance:\n best_order = order\n last_best_distance = distance\n print('order = ' + str(order) + ' Distance = ' + str(distance))\n return last_best_distance, best_order\n \nbest_distance, best_order = brute_force_tsp(ins.w, ins.dim)\nprint('Best order from brute force = ' + str(best_order) + ' with total distance = ' + str(best_distance))\n\ndef draw_tsp_solution(G, order, colors, pos):\n G2 = G.copy()\n n = len(order)\n for i in range(n):\n j = (i + 1) % n\n G2.add_edge(order[i], order[j])\n default_axes = plt.axes(frameon=True)\n nx.draw_networkx(G2, node_color=colors, node_size=600, alpha=.8, ax=default_axes, pos=pos)\n\ndraw_tsp_solution(G, best_order, colors, pos)",
"order = (0, 1, 2) Distance = 71.0\nBest order from brute force = (0, 1, 2) with total distance = 71.0\n"
]
],
[
[
"### Mapping to the Ising problem",
"_____no_output_____"
]
],
[
[
"qubitOp, offset = tsp.get_operator(ins)",
"_____no_output_____"
]
],
[
[
"### [Optional] Using DOcplex for mapping to the Ising problem\nUsing ```docplex.get_qubitops``` is a different way to create an Ising Hamiltonian of TSP. ```docplex.get_qubitops``` can create a corresponding Ising Hamiltonian from an optimization model of TSP. An example of using ```docplex.get_qubitops``` is as below. ",
"_____no_output_____"
]
],
[
[
"# Create an instance of a model and variables\nmdl = Model(name='tsp')\nx = {(i,p): mdl.binary_var(name='x_{0}_{1}'.format(i,p)) for i in range(n) for p in range(n)}\n\n# Object function\ntsp_func = mdl.sum(ins.w[i,j] * x[(i,p)] * x[(j,(p+1)%n)] for i in range(n) for j in range(n) for p in range(n))\nmdl.minimize(tsp_func)\n\n# Constrains\nfor i in range(n):\n mdl.add_constraint(mdl.sum(x[(i,p)] for p in range(n)) == 1)\nfor p in range(n):\n mdl.add_constraint(mdl.sum(x[(i,p)] for i in range(n)) == 1)",
"_____no_output_____"
],
[
"qubitOp_docplex, offset_docplex = docplex.get_operator(mdl)",
"_____no_output_____"
]
],
[
[
"### Checking that the full Hamiltonian gives the right cost ",
"_____no_output_____"
]
],
[
[
"#Making the Hamiltonian in its full form and getting the lowest eigenvalue and eigenvector\nee = ExactEigensolver(qubitOp, k=1)\nresult = ee.run()\n\nprint('energy:', result['energy'])\nprint('tsp objective:', result['energy'] + offset)\nx = sample_most_likely(result['eigvecs'][0])\nprint('feasible:', tsp.tsp_feasible(x))\nz = tsp.get_tsp_solution(x)\nprint('solution:', z)\nprint('solution objective:', tsp.tsp_value(z, ins.w))\ndraw_tsp_solution(G, z, colors, pos)",
"energy: -600035.5\ntsp objective: 71.0\nfeasible: True\nsolution: [0, 1, 2]\nsolution objective: 71.0\n"
]
],
[
[
"### Running it on quantum computer\nWe run the optimization routine using a feedback loop with a quantum computer that uses trial functions built with Y single-qubit rotations, $U_\\mathrm{single}(\\theta) = \\prod_{i=1}^n Y(\\theta_{i})$, and entangler steps $U_\\mathrm{entangler}$.",
"_____no_output_____"
]
],
[
[
"seed = 10598\n\nspsa = SPSA(max_trials=300)\nry = RY(qubitOp.num_qubits, depth=5, entanglement='linear')\nvqe = VQE(qubitOp, ry, spsa)\n\nbackend = BasicAer.get_backend('statevector_simulator')\nquantum_instance = QuantumInstance(backend, seed_simulator=seed, seed_transpiler=seed)\n\nresult = vqe.run(quantum_instance)\n\nprint('energy:', result['energy'])\nprint('time:', result['eval_time'])\n#print('tsp objective:', result['energy'] + offset)\nx = sample_most_likely(result['eigvecs'][0])\nprint('feasible:', tsp.tsp_feasible(x))\nz = tsp.get_tsp_solution(x)\nprint('solution:', z)\nprint('solution objective:', tsp.tsp_value(z, ins.w))\ndraw_tsp_solution(G, z, colors, pos)",
"energy: -595934.1907769153\ntime: 17.446267127990723\nfeasible: True\nsolution: [1, 2, 0]\nsolution objective: 71.0\n"
],
[
"# run quantum algorithm with shots\n\nseed = 10598\n\nspsa = SPSA(max_trials=300)\nry = RY(qubitOp.num_qubits, depth=5, entanglement='linear')\nvqe = VQE(qubitOp, ry, spsa)\n\nbackend = BasicAer.get_backend('qasm_simulator')\nquantum_instance = QuantumInstance(backend, shots=1024, seed_simulator=seed, seed_transpiler=seed)\n\nresult = vqe.run(quantum_instance)\n\nprint('energy:', result['energy'])\nprint('time:', result['eval_time'])\n#print('tsp objective:', result['energy'] + offset)\nx = sample_most_likely(result['eigvecs'][0])\nprint('feasible:', tsp.tsp_feasible(x))\nz = tsp.get_tsp_solution(x)\nprint('solution:', z)\nprint('solution objective:', tsp.tsp_value(z, ins.w))\nplot_histogram(result['eigvecs'][0])\ndraw_tsp_solution(G, z, colors, pos)",
"_____no_output_____"
]
],
[
[
"### [Optional] Checking that the full Hamiltonian made by ```docplex.get_operator``` gives the right cost",
"_____no_output_____"
]
],
[
[
"ee = ExactEigensolver(qubitOp_docplex, k=1)\nresult = ee.run()\n\nprint('energy:', result['energy'])\nprint('tsp objective:', result['energy'] + offset_docplex)\n\nx = sample_most_likely(result['eigvecs'][0])\nprint('feasible:', tsp.tsp_feasible(x))\nz = tsp.get_tsp_solution(x)\nprint('solution:', z)\nprint('solution objective:', tsp.tsp_value(z, ins.w))\ndraw_tsp_solution(G, z, colors, pos)",
"_____no_output_____"
],
[
"import qiskit.tools.jupyter\n%qiskit_version_table\n%qiskit_copyright",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
e79733a46e2ad6d03305c395e482b88bbd3948df | 1,108 | ipynb | Jupyter Notebook | algo_practice/code.ipynb | julkar9/deep_learning_nano_degree | 6bb77313cb95e88dcf90eb16711a2b7fff9bb23d | [
"Apache-2.0"
]
| null | null | null | algo_practice/code.ipynb | julkar9/deep_learning_nano_degree | 6bb77313cb95e88dcf90eb16711a2b7fff9bb23d | [
"Apache-2.0"
]
| null | null | null | algo_practice/code.ipynb | julkar9/deep_learning_nano_degree | 6bb77313cb95e88dcf90eb16711a2b7fff9bb23d | [
"Apache-2.0"
]
| null | null | null | 21.307692 | 77 | 0.522563 | [
[
[
"from ds import DoubledLinkedList\n\nclass Stack:\n def __init__(self):\n self.__list = DoubledLinkedList()\n def push(self , val):\n self.__list.add(val)\n def pop(self):\n self.pop.remove_last()\n def is_empty(self):\n pass\n def peek(self):\n pass\n\n",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code"
]
]
|
e7975c956fa51369f75023cff131fd3f40d61d2b | 17,834 | ipynb | Jupyter Notebook | official_tutorial/lesson2b_autograd_tutorial_deep_learning_usage.ipynb | zhennongchen/pytorch-tutorial | 7221bad5f85c6a937e3ad8d195bb6d7daf7803b6 | [
"MIT"
]
| null | null | null | official_tutorial/lesson2b_autograd_tutorial_deep_learning_usage.ipynb | zhennongchen/pytorch-tutorial | 7221bad5f85c6a937e3ad8d195bb6d7daf7803b6 | [
"MIT"
]
| null | null | null | official_tutorial/lesson2b_autograd_tutorial_deep_learning_usage.ipynb | zhennongchen/pytorch-tutorial | 7221bad5f85c6a937e3ad8d195bb6d7daf7803b6 | [
"MIT"
]
| null | null | null | 31.123909 | 159 | 0.559998 | [
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"**Read Later:**\n\nModule Documentation\n\nhttps://pytorch.org/docs/stable/generated/torch.nn.Module.html",
"_____no_output_____"
],
[
"\nA Gentle Introduction to ``torch.autograd``\n---------------------------------\n\n``torch.autograd`` is PyTorch’s automatic differentiation engine that powers\nneural network training. In this section, you will get a conceptual\nunderstanding of how autograd helps a neural network train.\n\nBackground\n~~~~~~~~~~\nNeural networks (NNs) are a collection of nested functions that are\nexecuted on some input data. These functions are defined by *parameters*\n(consisting of weights and biases), which in PyTorch are stored in\ntensors.\n\nTraining a NN happens in two steps:\n\n**Forward Propagation**: In forward prop, the NN makes its best guess\nabout the correct output. It runs the input data through each of its\nfunctions to make this guess.\n\n**Backward Propagation**: In backprop, the NN adjusts its parameters\nproportionate to the error in its guess. It does this by traversing\nbackwards from the output, collecting the derivatives of the error with\nrespect to the parameters of the functions (*gradients*), and optimizing\nthe parameters using gradient descent. For a more detailed walkthrough\nof backprop, check out this `video from\n3Blue1Brown <https://www.youtube.com/watch?v=tIeHLnjs5U8>`__.\n\n\n\n\nUsage in PyTorch\n~~~~~~~~~~~\nLet's take a look at a single training step.\nFor this example, we load a pretrained resnet18 model from ``torchvision``.\nWe create a random data tensor to represent a single image with 3 channels, and height & width of 64,\nand its corresponding ``label`` initialized to some random values.\n\n",
"_____no_output_____"
]
],
[
[
"import torch, torchvision\nmodel = torchvision.models.resnet18(pretrained=True)\ndata = torch.rand(1, 3, 64, 64)\nlabels = torch.rand(1, 1000)\nprint(data.size(),labels.size())",
"torch.Size([1, 3, 64, 64]) torch.Size([1, 1000])\n"
]
],
[
[
"Next, we run the input data through the model through each of its layers to make a prediction.\nThis is the **forward pass**.\n\n\n",
"_____no_output_____"
]
],
[
[
"prediction = model(data) # forward pass\nprint(prediction.size())",
"torch.Size([1, 1000])\n"
]
],
[
[
"We use the model's prediction and the corresponding label to calculate the error (``loss``).\nThe next step is to backpropagate this error through the network.\nBackward propagation is kicked off when we call ``.backward()`` on the error tensor.\nAutograd then calculates and stores the gradients for each model parameter in the parameter's ``.grad`` attribute.\n\n\n",
"_____no_output_____"
]
],
[
[
"loss = (prediction - labels).sum()\nloss.backward() # backward pass",
"_____no_output_____"
]
],
[
[
"Next, we load an optimizer, in this case SGD with a learning rate of 0.01 and momentum of 0.9.\nWe register all the parameters of the model in the optimizer.\n\nmodel.parameters() can acesss all model's parameters\n\n",
"_____no_output_____"
]
],
[
[
"optim = torch.optim.SGD(model.parameters(), lr=1e-2, momentum=0.9)",
"_____no_output_____"
]
],
[
[
"Finally, we call ``.step()`` to initiate gradient descent. The optimizer adjusts each parameter by its gradient stored in ``.grad``.\n\n\n",
"_____no_output_____"
]
],
[
[
"optim.step() #gradient descent",
"_____no_output_____"
]
],
[
[
"At this point, you have everything you need to train your neural network.\nThe below sections detail the workings of autograd - feel free to skip them.\n\n\n",
"_____no_output_____"
],
[
"--------------\n\n\n",
"_____no_output_____"
],
[
"Differentiation in Autograd\n~~~~~~~~~~~~~~~~~~~~~~~~~~~\nLet's take a look at how ``autograd`` collects gradients. We create two tensors ``a`` and ``b`` with\n``requires_grad=True``. This signals to ``autograd`` that every operation on them should be tracked.\n\n\n",
"_____no_output_____"
]
],
[
[
"import torch\n\na = torch.tensor([2., 3.], requires_grad=True)\nb = torch.tensor([6., 4.], requires_grad=True)",
"_____no_output_____"
]
],
[
[
"We create another tensor ``Q`` from ``a`` and ``b``.\n\n\\begin{align}Q = 3a^3 - b^2\\end{align}\n\n",
"_____no_output_____"
]
],
[
[
"Q = 3*a**3 - b**2\nprint(Q)",
"tensor([-12., 65.], grad_fn=<SubBackward0>)\n"
]
],
[
[
"Let's assume ``a`` and ``b`` to be parameters of an NN, and ``Q``\nto be the error. In NN training, we want gradients of the error\nw.r.t. parameters, i.e.\n\n\\begin{align}\\frac{\\partial Q}{\\partial a} = 9a^2\\end{align}\n\n\\begin{align}\\frac{\\partial Q}{\\partial b} = -2b\\end{align}\n\n\nWhen we call ``.backward()`` on ``Q``, autograd calculates these gradients\nand stores them in the respective tensors' ``.grad`` attribute.\n\nWe need to explicitly pass a ``gradient`` argument in ``Q.backward()`` because it is a vector.\n``gradient`` is a tensor of the same shape as ``Q``, and it represents the\ngradient of Q w.r.t. itself, i.e.\n\n\\begin{align}\\frac{dQ}{dQ} = 1\\end{align}\n\nEquivalently, we can also aggregate Q into a scalar and call backward implicitly, like ``Q.sum().backward()``.\n\n\n",
"_____no_output_____"
]
],
[
[
"external_grad = torch.tensor([1,1])\nQ.backward(gradient=external_grad)",
"_____no_output_____"
]
],
[
[
"Gradients are now deposited in ``a.grad`` and ``b.grad``\n\n",
"_____no_output_____"
]
],
[
[
"# check if collected gradients are correct\nprint(a.grad)\nprint(9*a**2)\nprint(9*a**2 == a.grad)\nprint(-2*b == b.grad)",
"tensor([36., 81.])\ntensor([36., 81.], grad_fn=<MulBackward0>)\ntensor([True, True])\ntensor([True, True])\n"
]
],
[
[
"Optional Reading - Vector Calculus using ``autograd``\n^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^\n\nMathematically, if you have a vector valued function\n$\\vec{y}=f(\\vec{x})$, then the gradient of $\\vec{y}$ with\nrespect to $\\vec{x}$ is a Jacobian matrix $J$:\n\n\\begin{align}J\n =\n \\left(\\begin{array}{cc}\n \\frac{\\partial \\bf{y}}{\\partial x_{1}} &\n ... &\n \\frac{\\partial \\bf{y}}{\\partial x_{n}}\n \\end{array}\\right)\n =\n \\left(\\begin{array}{ccc}\n \\frac{\\partial y_{1}}{\\partial x_{1}} & \\cdots & \\frac{\\partial y_{1}}{\\partial x_{n}}\\\\\n \\vdots & \\ddots & \\vdots\\\\\n \\frac{\\partial y_{m}}{\\partial x_{1}} & \\cdots & \\frac{\\partial y_{m}}{\\partial x_{n}}\n \\end{array}\\right)\\end{align}\n\nGenerally speaking, ``torch.autograd`` is an engine for computing\nvector-Jacobian product. That is, given any vector $\\vec{v}$, compute the product\n$J^{T}\\cdot \\vec{v}$\n\nIf $\\vec{v}$ happens to be the gradient of a scalar function $l=g\\left(\\vec{y}\\right)$:\n\n\\begin{align}\\vec{v}\n =\n \\left(\\begin{array}{ccc}\\frac{\\partial l}{\\partial y_{1}} & \\cdots & \\frac{\\partial l}{\\partial y_{m}}\\end{array}\\right)^{T}\\end{align}\n\nthen by the chain rule, the vector-Jacobian product would be the\ngradient of $l$ with respect to $\\vec{x}$:\n\n\\begin{align}J^{T}\\cdot \\vec{v}=\\left(\\begin{array}{ccc}\n \\frac{\\partial y_{1}}{\\partial x_{1}} & \\cdots & \\frac{\\partial y_{m}}{\\partial x_{1}}\\\\\n \\vdots & \\ddots & \\vdots\\\\\n \\frac{\\partial y_{1}}{\\partial x_{n}} & \\cdots & \\frac{\\partial y_{m}}{\\partial x_{n}}\n \\end{array}\\right)\\left(\\begin{array}{c}\n \\frac{\\partial l}{\\partial y_{1}}\\\\\n \\vdots\\\\\n \\frac{\\partial l}{\\partial y_{m}}\n \\end{array}\\right)=\\left(\\begin{array}{c}\n \\frac{\\partial l}{\\partial x_{1}}\\\\\n \\vdots\\\\\n \\frac{\\partial l}{\\partial x_{n}}\n \\end{array}\\right)\\end{align}\n\nThis characteristic of vector-Jacobian product is what we use in the above example;\n``external_grad`` represents $\\vec{v}$.\n\n\n",
"_____no_output_____"
],
[
"Computational Graph\n~~~~~~~~~~~~~~~~~~~\n\nConceptually, autograd keeps a record of data (tensors) & all executed\noperations (along with the resulting new tensors) in a directed acyclic\ngraph (DAG) consisting of\n`Function <https://pytorch.org/docs/stable/autograd.html#torch.autograd.Function>`__\nobjects. In this DAG, leaves are the input tensors, roots are the output\ntensors. By tracing this graph from roots to leaves, you can\nautomatically compute the gradients using the chain rule.\n\nIn a forward pass, autograd does two things simultaneously:\n\n- run the requested operation to compute a resulting tensor, and\n- maintain the operation’s *gradient function* in the DAG.\n\nThe backward pass kicks off when ``.backward()`` is called on the DAG\nroot. ``autograd`` then:\n\n- computes the gradients from each ``.grad_fn``,\n- accumulates them in the respective tensor’s ``.grad`` attribute, and\n- using the chain rule, propagates all the way to the leaf tensors.\n\nBelow is a visual representation of the DAG in our example. In the graph,\nthe arrows are in the direction of the forward pass. The nodes represent the backward functions\nof each operation in the forward pass. The leaf nodes in blue represent our leaf tensors ``a`` and ``b``.\n\n.. figure:: /_static/img/dag_autograd.png\n\n<div class=\"alert alert-info\"><h4>Note</h4><p>**DAGs are dynamic in PyTorch**\n An important thing to note is that the graph is recreated from scratch; after each\n ``.backward()`` call, autograd starts populating a new graph. This is\n exactly what allows you to use control flow statements in your model;\n you can change the shape, size and operations at every iteration if\n needed.</p></div>\n\nExclusion from the DAG\n^^^^^^^^^^^^^^^^^^^^^^\n\n``torch.autograd`` tracks operations on all tensors which have their\n``requires_grad`` flag set to ``True``. For tensors that don’t require\ngradients, setting this attribute to ``False`` excludes it from the\ngradient computation DAG.\n\nThe output tensor of an operation will require gradients even if only a\nsingle input tensor has ``requires_grad=True``.\n\n\n",
"_____no_output_____"
]
],
[
[
"x = torch.rand(5, 5)\ny = torch.rand(5, 5)\nz = torch.rand((5, 5), requires_grad=True)\n\na = x + y\nprint(f\"Does `a` require gradients? : {a.requires_grad}\")\nb = x + z\nprint(f\"Does `b` require gradients?: {b.requires_grad}\")",
"_____no_output_____"
]
],
[
[
"In a NN, parameters that don't compute gradients are usually called **frozen parameters**.\nIt is useful to \"freeze\" part of your model if you know in advance that you won't need the gradients of those parameters\n(this offers some performance benefits by reducing autograd computations).\n\nAnother common usecase where exclusion from the DAG is important is for\n`finetuning a pretrained network <https://pytorch.org/tutorials/beginner/finetuning_torchvision_models_tutorial.html>`__\n\nIn finetuning, we freeze most of the model and typically only modify the classifier layers to make predictions on new labels.\nLet's walk through a small example to demonstrate this. As before, we load a pretrained resnet18 model, and freeze all the parameters.\n\n",
"_____no_output_____"
]
],
[
[
"from torch import nn, optim\n\nmodel = torchvision.models.resnet18(pretrained=True)\n\n# Freeze all the parameters in the network\nfor param in model.parameters():\n param.requires_grad = False",
"_____no_output_____"
]
],
[
[
"Let's say we want to finetune the model on a new dataset with 10 labels.\nIn resnet, the classifier is the last linear layer ``model.fc``.\nWe can simply replace it with a new linear layer (unfrozen by default)\nthat acts as our classifier.\n\n",
"_____no_output_____"
]
],
[
[
"model.fc = nn.Linear(512, 10)",
"_____no_output_____"
]
],
[
[
"Now all parameters in the model, except the parameters of ``model.fc``, are frozen.\nThe only parameters that compute gradients are the weights and bias of ``model.fc``.\n\n",
"_____no_output_____"
]
],
[
[
"# Optimize only the classifier\noptimizer = optim.SGD(model.parameters(), lr=1e-2, momentum=0.9)",
"_____no_output_____"
]
],
[
[
"Notice although we register all the parameters in the optimizer,\nthe only parameters that are computing gradients (and hence updated in gradient descent)\nare the weights and bias of the classifier.\n\nThe same exclusionary functionality is available as a context manager in\n`torch.no_grad() <https://pytorch.org/docs/stable/generated/torch.no_grad.html>`__\n\n\n",
"_____no_output_____"
],
[
"--------------\n\n\n",
"_____no_output_____"
],
[
"Further readings:\n~~~~~~~~~~~~~~~~~~~\n\n- `In-place operations & Multithreaded Autograd <https://pytorch.org/docs/stable/notes/autograd.html>`__\n- `Example implementation of reverse-mode autodiff <https://colab.research.google.com/drive/1VpeE6UvEPRz9HmsHh1KS0XxXjYu533EC>`__\n\n",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
]
|
e79772eb740b0e5bfa046907d4796b9bd9fb2e99 | 166,916 | ipynb | Jupyter Notebook | notebook/ulmfit_wongnai.ipynb | titipata/thai2vec | 783eb63e5a6e214d3116f26cf2494b89d86f5be0 | [
"MIT"
]
| null | null | null | notebook/ulmfit_wongnai.ipynb | titipata/thai2vec | 783eb63e5a6e214d3116f26cf2494b89d86f5be0 | [
"MIT"
]
| null | null | null | notebook/ulmfit_wongnai.ipynb | titipata/thai2vec | 783eb63e5a6e214d3116f26cf2494b89d86f5be0 | [
"MIT"
]
| null | null | null | 104.19226 | 44,212 | 0.830022 | [
[
[
"# Thai2Vec Classification Using ULMFit\n\nThis notebook demonstrates how to use the [ULMFit model](https://arxiv.org/abs/1801.06146) implemented by`thai2vec` for text classification. We use [Wongnai Challenge: Review Rating Prediction](https://www.kaggle.com/c/wongnai-challenge-review-rating-prediction) as our benchmark as it is the only sizeable and publicly available text classification dataset at the time of writing (June 21, 2018). It has 39,999 reviews for training and validation, and 6,203 reviews for testing. Our workflow is as follows:\n\n* Perform 75/15 train-validation split\n* Minimal text cleaning and tokenization using `newmm` engine of `pyThaiNLP`\n* Get embeddings of Wongnai dataset from all the data available (train and test sets)\n* Load pretrained Thai Wikipedia embeddings; for those embeddings which exist only in Wongnai dataset, we use the average of Wikipedia embeddings instead\n* Train language model based on all the data available on Wongnai dataset\n* Replace the top and train the classifier based on the training set by gradual unfreezing\n\nWe achieved validation perplexity at 35.75113 and validation micro F1 score at 0.598 for five-label classification. Micro F1 scores for public and private leaderboards are 0.61451 and 0.60925 respectively (supposedly we could train further with the 15% validation set we did not use), which are state-of-the-art as of the time of writing (June 21, 2018). FastText benchmark has the performance of 0.50483 and 0.49366 for public and private leaderboards respectively.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"## Imports",
"_____no_output_____"
]
],
[
[
"%reload_ext autoreload\n%autoreload 2\n%matplotlib inline\n\nimport re\nimport html\nimport numpy as np\nimport dill as pickle\nfrom IPython.display import Image\nfrom IPython.core.display import HTML \nfrom collections import Counter\nfrom sklearn.model_selection import train_test_split\n\nfrom fastai.text import *\nfrom pythainlp.tokenize import word_tokenize\nfrom utils import *\n\n\nDATA_PATH='/home/ubuntu/Projects/new2vec/wongnai_data/'\nRAW_PATH = f'{DATA_PATH}raw/'\nMODEL_PATH = f'{DATA_PATH}models/'\n\nraw_files = !ls {RAW_PATH}",
"_____no_output_____"
]
],
[
[
"## Train/Validation Sets\n\nWe use data from [Wongnai Challenge: Review Rating Prediction](https://www.kaggle.com/c/wongnai-challenge-review-rating-prediction). The training data consists of 39,999 restaurant reviews from unknown number of reviewers labeled one to five stars, with the schema `(label,review)`. We use 75/15 train-validation split. The test set has 6,203 reviews from the same number of reviewers. No information accuracy is 46.9%.",
"_____no_output_____"
]
],
[
[
"raw_train = pd.read_csv(f'{RAW_PATH}w_review_train.csv',sep=';',header=None)\nraw_train = raw_train.iloc[:,[1,0]]\nraw_train.columns = ['label','review']\nraw_test = pd.read_csv(f'{RAW_PATH}test_file.csv',sep=';')\nsubmission = pd.read_csv(f'{RAW_PATH}sample_submission.csv',sep=',')",
"_____no_output_____"
],
[
"print(raw_train.shape)\nraw_train.head()",
"(40000, 2)\n"
],
[
"cnt = Counter(raw_train['label'])\ncnt",
"_____no_output_____"
],
[
"#baseline\ncnt.most_common(1)[0][1] / raw_train.shape[0]",
"_____no_output_____"
],
[
"raw_test.head()",
"_____no_output_____"
],
[
"submission.head()",
"_____no_output_____"
],
[
"#test df\nraw_test = pd.read_csv(f'{RAW_PATH}test_file.csv',sep=';')\nraw_test.head()\ndf_tst = pd.DataFrame({'label':raw_test['reviewID'],'review':raw_test['review']})\ndf_tst.to_csv(f'{DATA_PATH}test.csv', header=False, index=False)",
"_____no_output_____"
],
[
"#train/validation/train_language_model split\ndf_trn, df_val = train_test_split(raw_train, test_size = 0.15, random_state = 1412)\ndf_lm = pd.concat([df_trn,df_tst])\n\ndf_lm.to_csv(f'{DATA_PATH}train_lm.csv', header=False, index=False)\ndf_trn.to_csv(f'{DATA_PATH}train.csv', header=False, index=False)\ndf_val.to_csv(f'{DATA_PATH}valid.csv', header=False, index=False)",
"_____no_output_____"
],
[
"df_trn.shape,df_val.shape,df_lm.shape",
"_____no_output_____"
]
],
[
[
"## Language Modeling",
"_____no_output_____"
],
[
"### Text Processing",
"_____no_output_____"
],
[
"We first determine the vocab for the reviews, then train a language model based on our training set. We perform the following minimal text processing:\n* The token `xbos` is used to note start of a text since we will be chaining them together for the language model training. \n* `pyThaiNLP`'s `newmm` word tokenizer is used to tokenize the texts.\n* `tkrep` is used to replace repetitive characters such as `อร่อยมากกกกกก` becoming `อรอ่ยมาtkrep6ก`",
"_____no_output_____"
]
],
[
[
"max_vocab = 60000\nmin_freq = 2\n\ndf_lm = pd.read_csv(f'{DATA_PATH}train_lm.csv',header=None,chunksize=30000)\ndf_val = pd.read_csv(f'{DATA_PATH}valid.csv',header=None,chunksize=30000)\n\ntrn_lm,trn_tok,trn_labels,itos_cls,stoi_cls,freq_trn = numericalizer(df_trn)\nval_lm,val_tok,val_labels,itos_cls,stoi_cls,freq_val = numericalizer(df_val,itos_cls)",
"_____no_output_____"
],
[
"# np.save(f'{MODEL_PATH}trn_tok.npy', trn_tok)\n# np.save(f'{MODEL_PATH}val_tok.npy', val_tok)\n# np.save(f'{MODEL_PATH}trn_lm.npy', trn_lm)\n# np.save(f'{MODEL_PATH}val_lm.npy', val_lm)\n# pickle.dump(itos_cls, open(f'{MODEL_PATH}itos_cls.pkl', 'wb'))",
"_____no_output_____"
],
[
"tok_lm = np.load(f'{MODEL_PATH}tok_lm.npy')\ntok_val = np.load(f'{MODEL_PATH}tok_val.npy')",
"_____no_output_____"
],
[
"tok_lm[:1]",
"_____no_output_____"
],
[
"#numericalized tokenized texts\ntrn_lm[:1]",
"_____no_output_____"
],
[
"#index to token\nitos_cls[:10]",
"_____no_output_____"
]
],
[
[
"### Load Pretrained Language Model",
"_____no_output_____"
],
[
"Instead of starting from random weights, we import the language model pretrained on Wikipedia (see `pretrained_wiki.ipynb`). For words that appear only in the Wongnai dataset but not Wikipedia, we start with the average of all embeddings instead. Max vocab size is set at 60,000 and minimum frequency of 2 consistent with the pretrained model. We ended up with 19,998 embeddings for all reviews.",
"_____no_output_____"
]
],
[
[
"em_sz = 300\nvocab_size = len(itos_cls)\nwgts = torch.load(f'{MODEL_PATH}thwiki_model2.h5', map_location=lambda storage, loc: storage)\nitos_pre = pickle.load(open(f'{MODEL_PATH}itos_pre.pkl','rb'))\nstoi_pre = collections.defaultdict(lambda:-1, {v:k for k,v in enumerate(itos_pre)})",
"_____no_output_____"
],
[
"#pretrained weights\nwgts = merge_wgts(em_sz, wgts, itos_pre, itos_cls)",
"_____no_output_____"
]
],
[
[
"### Train Language Model",
"_____no_output_____"
]
],
[
[
"em_sz,nh,nl = 300,1150,3\nwd=1e-7\nbptt=70\nbs=60\nopt_fn = partial(optim.Adam, betas=(0.8, 0.99))\nweight_factor = 0.7\ndrops = np.array([0.25, 0.1, 0.2, 0.02, 0.15])*weight_factor",
"_____no_output_____"
],
[
"#data loader\ntrn_dl = LanguageModelLoader(np.concatenate(trn_lm), bs, bptt)\nval_dl = LanguageModelLoader(np.concatenate(val_lm), bs, bptt)\nmd = LanguageModelData(path=DATA_PATH, pad_idx=1, n_tok=vocab_size, \n trn_dl=trn_dl, val_dl=val_dl, bs=bs, bptt=bptt)",
"_____no_output_____"
],
[
"#model fitter\nlearner= md.get_model(opt_fn, em_sz, nh, nl, \n dropouti=drops[0], dropout=drops[1], wdrop=drops[2], dropoute=drops[3], dropouth=drops[4])\nlearner.metrics = [accuracy]",
"_____no_output_____"
],
[
"#load the saved models + new embeddings\nlearner.model.load_state_dict(wgts)",
"_____no_output_____"
],
[
"#find optimal learning rate\nlearner.lr_find2(start_lr = 1e-6, end_lr=0.1)\nlearner.sched.plot()",
"_____no_output_____"
],
[
"#optimal learning rate\nlr=3e-3\nlr",
"_____no_output_____"
],
[
"#train while frozen once to warm up\nlearner.freeze_to(-1)\nlearner.fit(lr, 1, wds=wd, use_clr=(20,5), cycle_len=1)",
"_____no_output_____"
],
[
"#use_clr (peak as ratio of lr, iterations to grow and descend e.g. 10 is 1/10 grow and 9/10 descend)\nlearner.unfreeze()\nlearner.fit(lr, 1, wds=wd, use_clr=(20,5), cycle_len=5)",
"_____no_output_____"
],
[
"learner.sched.plot_loss()",
"_____no_output_____"
],
[
"learner.save('wongnai_lm')\nlearner.save_encoder('wongnai_enc')",
"_____no_output_____"
]
],
[
[
"## Classification",
"_____no_output_____"
],
[
"With the language model trained on Wongnai dataset, we use its embeddings to initialize the review classifier. We train the classifier using discriminative learning rates, slanted triangular learning rates, gradual unfreezing and a few other tricks detailed in the [ULMFit paper](https://arxiv.org/abs/1801.06146). We have found that training only the last two layers of the model seems to be the right balance for this dataset.",
"_____no_output_____"
],
[
"### Load Tokenized Texts and Labels",
"_____no_output_____"
]
],
[
[
"#load csvs and tokenizer\nmax_vocab = 60000\nmin_freq = 2\ndf_trn = pd.read_csv(f'{DATA_PATH}train.csv',header=None,chunksize=30000)\ndf_val = pd.read_csv(f'{DATA_PATH}valid.csv',header=None,chunksize=30000)\ndf_tst = pd.read_csv(f'{DATA_PATH}test.csv',header=None,chunksize=30000)\n\ntrn_cls,trn_tok,trn_labels,itos_cls,stoi_cls,freq = numericalizer(df_trn)\nval_cls,val_tok,val_labels,itos_cls,stoi_cls,freq = numericalizer(df_val,itos_cls)\ntst_cls,tst_tok,tst_labels,itos_cls,stoi_cls,freq = numericalizer(df_tst,itos_cls)",
"_____no_output_____"
],
[
"#get labels\ntrn_labels = np.squeeze(trn_labels)\nval_labels = np.squeeze(val_labels)\ntst_labels = np.squeeze(tst_labels)\nmin_lbl = trn_labels.min()\ntrn_labels -= min_lbl\nval_labels -= min_lbl",
"_____no_output_____"
]
],
[
[
"### Create Data Loader",
"_____no_output_____"
]
],
[
[
"#dataset object\nbs = 60\ntrn_ds = TextDataset(trn_cls, trn_labels)\nval_ds = TextDataset(val_cls, val_labels)\ntst_ds = TextDataset(tst_cls, tst_labels)\n\n#sampler\ntrn_samp = SortishSampler(trn_cls, key=lambda x: len(trn_cls[x]), bs=bs//2)\nval_samp = SortSampler(val_cls, key=lambda x: len(val_cls[x]))\ntst_samp = SortSampler(tst_cls, key=lambda x: len(tst_cls[x]))\n\n#data loader\ntrn_dl = DataLoader(trn_ds, bs//2, transpose=True, num_workers=1, pad_idx=1, sampler=trn_samp)\nval_dl = DataLoader(val_ds, bs, transpose=True, num_workers=1, pad_idx=1, sampler=val_samp)\ntst_dl = DataLoader(tst_ds, bs, transpose=True, num_workers=1, pad_idx=1, sampler=tst_samp)\nmd = ModelData(DATA_PATH, trn_dl, val_dl, tst_dl)",
"_____no_output_____"
]
],
[
[
"### Train Classifier",
"_____no_output_____"
]
],
[
[
"#parameters\nweight_factor = 0.5\ndrops = np.array([0.25, 0.1, 0.2, 0.02, 0.15])*weight_factor\nbptt = 70\nem_sz = 300\nnh = 1150\nnl = 3\nvocab_size = len(itos_cls)\nnb_class=int(trn_labels.max())+1\nopt_fn = partial(optim.Adam, betas=(0.7, 0.99))\nbs = 60\nwd = 1e-7\n\n#classifier model\n# em_sz*3 for max, mean, just activations\nm = get_rnn_classifer(bptt, max_seq=1000, n_class=nb_class, \n n_tok=vocab_size, emb_sz=em_sz, n_hid=nh, n_layers=nl, pad_token=1,\n layers=[em_sz*3, 50, nb_class], drops=[drops[4], 0.1],\n dropouti=drops[0], wdrop=drops[1], dropoute=drops[2], dropouth=drops[3])",
"_____no_output_____"
],
[
"#get learner\nlearner = RNN_Learner(md, TextModel(to_gpu(m)), opt_fn=opt_fn)\nlearner.reg_fn = partial(seq2seq_reg, alpha=2, beta=1)\nlearner.clip=25.\nlearner.metrics = [accuracy]\n#load encoder trained earlier\nlearner.load_encoder('wongnai_enc')",
"_____no_output_____"
],
[
"#find learning rate\nlearner.lr_find2()\nlearner.sched.plot()",
"_____no_output_____"
],
[
"#set learning rate\nlr=1e-2\nlrm = 2.6\nlrs = np.array([lr/(lrm**4), lr/(lrm**3), lr/(lrm**2), lr/lrm, lr])",
"_____no_output_____"
],
[
"#train last layer\nlearner.freeze_to(-1)\nlearner.fit(lrs, 1, wds=wd, cycle_len=10, use_clr=(10,10))\nlearner.save('last_layer')",
"_____no_output_____"
],
[
"#train last two layers\nlearner.load('last_layer')\nlearner.freeze_to(-2)\nlearner.fit(lrs, 1, wds=wd, cycle_len=3, use_clr=(8,3))\nlearner.save('last_two_layers')",
"_____no_output_____"
]
],
[
[
"## Validation Performance",
"_____no_output_____"
]
],
[
[
"learner.load('last_two_layers')\n#get validation performance\nprobs,y= learner.predict_with_targs()\npreds = np.argmax(np.exp(probs),1)",
"_____no_output_____"
],
[
"Counter(preds)",
"_____no_output_____"
],
[
"Counter(y)",
"_____no_output_____"
],
[
"from sklearn.metrics import confusion_matrix\nfrom sklearn.metrics import fbeta_score\n\nmost_frequent = np.array([4]*len(preds))\nprint(f'Baseline Micro F1: {fbeta_score(y,most_frequent,1,average=\"micro\")}')\nprint(f'Micro F1: {fbeta_score(y,preds,1,average=\"micro\")}')\ncm = confusion_matrix(y,preds)\nplot_confusion_matrix(cm,classes=[1,2,3,4,5])",
"Baseline Micro F1: 0.176\nMicro F1: 0.5976666666666667\nConfusion matrix, without normalization\n[[ 14 36 13 1 1]\n [ 13 80 150 22 2]\n [ 5 36 945 814 28]\n [ 0 0 344 2210 230]\n [ 0 1 22 696 337]]\n"
]
],
[
[
"## Submission",
"_____no_output_____"
]
],
[
[
"probs,y= learner.predict_with_targs(is_test=True)",
"_____no_output_____"
],
[
"preds = np.argmax(np.exp(probs),1) + 1",
"_____no_output_____"
],
[
"Counter(preds)",
"_____no_output_____"
],
[
"submit_df = pd.DataFrame({'a':y,'b':preds})\nsubmit_df.columns = ['reviewID','rating']\nsubmit_df.head()",
"_____no_output_____"
],
[
"submit_df.to_csv(f'{DATA_PATH}valid10_2layers_newmm.csv',index=False)",
"_____no_output_____"
]
],
[
[
"## Benchmark with FastText",
"_____no_output_____"
],
[
"We used [fastText](https://github.com/facebookresearch/fastText)'s own [pretrained embeddings](https://github.com/facebookresearch/fastText/blob/master/pretrained-vectors.md) and a relatively \"default\" settings in order to benchmark our results. This gave us the micro-averaged F1 score of 0.50483 and 0.49366 for the public and private leaderboard respectively.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"### Data Preparation",
"_____no_output_____"
]
],
[
[
"df_trn = pd.read_csv(f'{DATA_PATH}train.csv',header=None)\ndf_val = pd.read_csv(f'{DATA_PATH}valid.csv',header=None)\ndf_tst = pd.read_csv(f'{DATA_PATH}test.csv', header=None)",
"_____no_output_____"
],
[
"train_set = []\nfor i in range(df_trn.shape[0]):\n label = df_trn.iloc[i,0]\n line = df_trn.iloc[i,1].replace('\\n', ' ')\n train_set.append(f'__label__{label} {line}')\ntrain_doc = '\\n'.join(train_set)\nwith open(f'{DATA_PATH}train.txt','w') as f:\n f.write(train_doc)",
"_____no_output_____"
],
[
"valid_set = []\nfor i in range(df_val.shape[0]):\n label = df_val.iloc[i,0]\n line = df_val.iloc[i,1].replace('\\n', ' ')\n valid_set.append(f'__label__{label} {line}')\nvalid_doc = '\\n'.join(valid_set)\nwith open(f'{DATA_PATH}valid.txt','w') as f:\n f.write(valid_doc)",
"_____no_output_____"
],
[
"test_set = []\nfor i in range(df_tst.shape[0]):\n label = df_tst.iloc[i,0]\n line = df_tst.iloc[i,1].replace('\\n', ' ')\n test_set.append(f'__label__{label} {line}')\ntest_doc = '\\n'.join(test_set)\nwith open(f'{DATA_PATH}test.txt','w') as f:\n f.write(test_doc)",
"_____no_output_____"
]
],
[
[
"### Train FastText",
"_____no_output_____"
]
],
[
[
"!/home/ubuntu/theFastText/fastText-0.1.0/fasttext supervised -input '{DATA_PATH}train.txt' -pretrainedVectors '{MODEL_PATH}wiki.th.vec' -epoch 10 -dim 300 -wordNgrams 2 -output '{MODEL_PATH}fasttext_model'",
"Read 1M words\nNumber of words: 629560\nNumber of labels: 5\nProgress: 100.0% words/sec/thread: 524361 lr: 0.000000 loss: 0.612446 eta: 0h0m \n"
],
[
"!/home/ubuntu/theFastText/fastText-0.1.0/fasttext test '{MODEL_PATH}fasttext_model.bin' '{DATA_PATH}valid.txt'",
"N\t6000\nP@1\t0.473\nR@1\t0.473\nNumber of examples: 6000\n"
],
[
"preds = !/home/ubuntu/theFastText/fastText-0.1.0/fasttext predict '{MODEL_PATH}fasttext_model.bin' '{DATA_PATH}test.txt'",
"_____no_output_____"
]
],
[
[
"### Submission",
"_____no_output_____"
]
],
[
[
"submit_df = pd.DataFrame({'a':[i+1 for i in range(len(preds))],'b':preds})\nsubmit_df.columns = ['reviewID','rating']\nsubmit_df['rating'] = submit_df['rating'].apply(lambda x: x.split('__')[2])\nsubmit_df.head()",
"_____no_output_____"
],
[
"submit_df.to_csv(f'{DATA_PATH}fasttext.csv',index=False)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
e79775b7368a31a8a15b8ea0a898993c48544f65 | 104,113 | ipynb | Jupyter Notebook | analysis_setting/centroid/merge_centroid.ipynb | Betristor/Adaptive-clustering-for-ESOM | 207925b639abcda2bc7b0bcfc5a82bb5c553ac4c | [
"MIT"
]
| null | null | null | analysis_setting/centroid/merge_centroid.ipynb | Betristor/Adaptive-clustering-for-ESOM | 207925b639abcda2bc7b0bcfc5a82bb5c553ac4c | [
"MIT"
]
| null | null | null | analysis_setting/centroid/merge_centroid.ipynb | Betristor/Adaptive-clustering-for-ESOM | 207925b639abcda2bc7b0bcfc5a82bb5c553ac4c | [
"MIT"
]
| null | null | null | 433.804167 | 76,524 | 0.93947 | [
[
[
"import glob\nimport pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"extend_df = pd.read_csv('/home/jupyter-zyh/Gnhe/analysis_profile/global_setting/Linear/Texas/profile-1/all.csv')\nextend_df = extend_df.query(\"trial == 'simulated'\")\nextend_df['centroid'] = False\nextend_df_c = extend_df.copy()\nextend_df['centroid'] = True",
"_____no_output_____"
],
[
"df = pd.read_csv('/home/jupyter-zyh/Gnhe/analysis_setting/centroid/Linear/all.csv')\ndf = df.append(extend_df).append(extend_df_c)\ndf.to_csv('/home/jupyter-zyh/Gnhe/analysis_setting/centroid/Linear/merged.csv')",
"_____no_output_____"
],
[
"sns.catplot(data=df,col='method',row='trial',row_order=['hourly','simulated','combined'],x='ncluster',y='mae_ex_tc',hue='centroid',kind='box')",
"_____no_output_____"
],
[
"query_string = \"method == 'single' and trial == 'combined'\"\np = sns.boxplot(data=df.query(query_string),x='ncluster',y='mae_ex_tc',hue='centroid',palette='coolwarm')\nplt.title('Mae under different centroid setting',pad=18,fontdict={'size':14})\nplt.text(s=query_string.replace(\"'\",\"\").replace(\"==\",\"=\").replace(\"and\",\"&\"),x=2.25,y=0.49,color='#c04851')\nplt.xlabel('Number of representative weeks',fontdict={'size':14})\nplt.xticks([0,1,2,3,4,5],['5','10','15','20','25','30'])\nplt.ylabel('Mean absolute error',fontdict={'size':14})\nplt.legend(title='centroid',loc='upper right')",
"_____no_output_____"
],
[
"p.get_figure().savefig(\n '/home/jupyter-zyh/Gnhe/analysis/images/setting/centroid/mae_under_different_centroid_setting.png',\n dpi=300)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e797848c8057a8a7bae098e671da66a42ccb538b | 102,477 | ipynb | Jupyter Notebook | codes/ANN Results/Generate_ANN_Results.ipynb | architdatar/NewsArticleClassification | 64427e38d952adda1f4693a27fbbbe38d16e04b8 | [
"MIT"
]
| null | null | null | codes/ANN Results/Generate_ANN_Results.ipynb | architdatar/NewsArticleClassification | 64427e38d952adda1f4693a27fbbbe38d16e04b8 | [
"MIT"
]
| null | null | null | codes/ANN Results/Generate_ANN_Results.ipynb | architdatar/NewsArticleClassification | 64427e38d952adda1f4693a27fbbbe38d16e04b8 | [
"MIT"
]
| null | null | null | 51.187313 | 169 | 0.587625 | [
[
[
"########################################################################################################################\n# Filename: Generate_ANN_Results.ipynb\n#\n# Purpose: Generate results from different ANN models trained on paragraph \n# classification task\n#\n# Author(s): Bobby (Robert) Lumpkin\n#\n# Library Dependencies: numpy, pandas, tensorflow, bpmll, os, json, ast, random, \n# tensorflow_addons, skljson, sklearn, sys, threshold_learning\n########################################################################################################################",
"_____no_output_____"
]
],
[
[
"# Generate and Save Results from Different ANN Methods",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport os\nimport json\nimport ast\nimport random\nimport tensorflow as tf\nimport tensorflow_addons as tfa\nfrom bpmll import bp_mll_loss\nimport sklearn_json as skljson\nfrom sklearn.model_selection import train_test_split\nfrom sklearn import metrics\nimport sys\nos.chdir('C:\\\\Users\\\\rober\\\\OneDrive\\\\Documents\\\\STAT 6500\\\\Project\\\\NewsArticleClassification\\\\codes\\\\ANN Results') ## Set working directory\n ## to be 'ANN Results'\nsys.path.append('../ThresholdFunctionLearning') ## Append path to the ThresholdFunctionLearning directory to the interpreters\n ## search path\nfrom threshold_learning import predict_test_labels_binary ## Import the 'predict_test_labels_binary()' function from the \nfrom threshold_learning import predict_labels_binary ## threshold_learning library",
"_____no_output_____"
]
],
[
[
"## Models on Reduced Dataset (each instance has atleast one label)",
"_____no_output_____"
]
],
[
[
"## Load the reduced tfidf dataset\nfile_object = open('../BP-MLL Text Categorization/tfidf_trainTest_data_reduced.json',)\ntfidf_data_reduced = json.load(file_object)\nX_train_hasLabel = np.array(tfidf_data_reduced['X_train_hasLabel'])\nX_test_hasLabel = np.array(tfidf_data_reduced['X_test_hasLabel'])\nY_train_hasLabel = np.array(tfidf_data_reduced['Y_train_hasLabel'])\nY_test_hasLabel = np.array(tfidf_data_reduced['Y_test_hasLabel'])",
"_____no_output_____"
]
],
[
[
"### Feed-Forward Cross-Entropy Network",
"_____no_output_____"
]
],
[
[
"## Start by defining and compiling the cross-entropy loss network (bpmll used later)\ntf.random.set_seed(123)\nnum_labels = 13\n\nmodel_ce_FF = tf.keras.models.Sequential([\n tf.keras.layers.Dense(32, activation = 'relu'),\n tf.keras.layers.Dropout(0.5),\n tf.keras.layers.Dense(num_labels, activation = 'sigmoid')\n])\n\noptim_func = tf.keras.optimizers.Adam(lr=0.0001)\n\nmetric = tfa.metrics.HammingLoss(mode = 'multilabel', threshold = 0.5)\n\nmodel_ce_FF.compile(optimizer = optim_func,\n loss = 'binary_crossentropy', metrics = metric\n )",
"_____no_output_____"
],
[
"tf.random.set_seed(123)\nhistory_ce_FF_lr001 = model_ce_FF.fit(X_train_hasLabel, Y_train_hasLabel, epochs = 100,\n validation_data = (X_test_hasLabel, Y_test_hasLabel), verbose=2)",
"Epoch 1/100\n6/6 - 1s - loss: 0.7028 - hamming_loss: 0.4847 - val_loss: 0.6898 - val_hamming_loss: 0.4650\nEpoch 2/100\n6/6 - 0s - loss: 0.6970 - hamming_loss: 0.4742 - val_loss: 0.6878 - val_hamming_loss: 0.4572\nEpoch 3/100\n6/6 - 0s - loss: 0.6969 - hamming_loss: 0.4764 - val_loss: 0.6858 - val_hamming_loss: 0.4484\nEpoch 4/100\n6/6 - 0s - loss: 0.6936 - hamming_loss: 0.4642 - val_loss: 0.6838 - val_hamming_loss: 0.4414\nEpoch 5/100\n6/6 - 0s - loss: 0.6903 - hamming_loss: 0.4572 - val_loss: 0.6818 - val_hamming_loss: 0.4318\nEpoch 6/100\n6/6 - 0s - loss: 0.6836 - hamming_loss: 0.4489 - val_loss: 0.6799 - val_hamming_loss: 0.4274\nEpoch 7/100\n6/6 - 0s - loss: 0.6821 - hamming_loss: 0.4414 - val_loss: 0.6780 - val_hamming_loss: 0.4248\nEpoch 8/100\n6/6 - 0s - loss: 0.6825 - hamming_loss: 0.4427 - val_loss: 0.6761 - val_hamming_loss: 0.4205\nEpoch 9/100\n6/6 - 0s - loss: 0.6757 - hamming_loss: 0.4135 - val_loss: 0.6743 - val_hamming_loss: 0.4152\nEpoch 10/100\n6/6 - 0s - loss: 0.6670 - hamming_loss: 0.4091 - val_loss: 0.6724 - val_hamming_loss: 0.4082\nEpoch 11/100\n6/6 - 0s - loss: 0.6709 - hamming_loss: 0.4087 - val_loss: 0.6705 - val_hamming_loss: 0.4012\nEpoch 12/100\n6/6 - 0s - loss: 0.6657 - hamming_loss: 0.4126 - val_loss: 0.6686 - val_hamming_loss: 0.3942\nEpoch 13/100\n6/6 - 0s - loss: 0.6648 - hamming_loss: 0.3964 - val_loss: 0.6668 - val_hamming_loss: 0.3837\nEpoch 14/100\n6/6 - 0s - loss: 0.6559 - hamming_loss: 0.3776 - val_loss: 0.6649 - val_hamming_loss: 0.3794\nEpoch 15/100\n6/6 - 0s - loss: 0.6587 - hamming_loss: 0.3811 - val_loss: 0.6630 - val_hamming_loss: 0.3706\nEpoch 16/100\n6/6 - 0s - loss: 0.6551 - hamming_loss: 0.3767 - val_loss: 0.6611 - val_hamming_loss: 0.3654\nEpoch 17/100\n6/6 - 0s - loss: 0.6499 - hamming_loss: 0.3741 - val_loss: 0.6592 - val_hamming_loss: 0.3566\nEpoch 18/100\n6/6 - 0s - loss: 0.6474 - hamming_loss: 0.3580 - val_loss: 0.6573 - val_hamming_loss: 0.3523\nEpoch 19/100\n6/6 - 0s - loss: 0.6433 - hamming_loss: 0.3510 - val_loss: 0.6554 - val_hamming_loss: 0.3453\nEpoch 20/100\n6/6 - 0s - loss: 0.6441 - hamming_loss: 0.3501 - val_loss: 0.6535 - val_hamming_loss: 0.3392\nEpoch 21/100\n6/6 - 0s - loss: 0.6385 - hamming_loss: 0.3448 - val_loss: 0.6516 - val_hamming_loss: 0.3313\nEpoch 22/100\n6/6 - 0s - loss: 0.6348 - hamming_loss: 0.3326 - val_loss: 0.6497 - val_hamming_loss: 0.3260\nEpoch 23/100\n6/6 - 0s - loss: 0.6361 - hamming_loss: 0.3339 - val_loss: 0.6478 - val_hamming_loss: 0.3156\nEpoch 24/100\n6/6 - 0s - loss: 0.6341 - hamming_loss: 0.3352 - val_loss: 0.6459 - val_hamming_loss: 0.3138\nEpoch 25/100\n6/6 - 0s - loss: 0.6297 - hamming_loss: 0.3269 - val_loss: 0.6440 - val_hamming_loss: 0.3086\nEpoch 26/100\n6/6 - 0s - loss: 0.6267 - hamming_loss: 0.3147 - val_loss: 0.6420 - val_hamming_loss: 0.3033\nEpoch 27/100\n6/6 - 0s - loss: 0.6171 - hamming_loss: 0.3090 - val_loss: 0.6400 - val_hamming_loss: 0.2972\nEpoch 28/100\n6/6 - 0s - loss: 0.6220 - hamming_loss: 0.3204 - val_loss: 0.6380 - val_hamming_loss: 0.2955\nEpoch 29/100\n6/6 - 0s - loss: 0.6199 - hamming_loss: 0.3164 - val_loss: 0.6359 - val_hamming_loss: 0.2928\nEpoch 30/100\n6/6 - 0s - loss: 0.6075 - hamming_loss: 0.2858 - val_loss: 0.6339 - val_hamming_loss: 0.2885\nEpoch 31/100\n6/6 - 0s - loss: 0.6019 - hamming_loss: 0.2976 - val_loss: 0.6317 - val_hamming_loss: 0.2841\nEpoch 32/100\n6/6 - 0s - loss: 0.6088 - hamming_loss: 0.3073 - val_loss: 0.6295 - val_hamming_loss: 0.2815\nEpoch 33/100\n6/6 - 0s - loss: 0.6038 - hamming_loss: 0.2911 - val_loss: 0.6274 - val_hamming_loss: 0.2797\nEpoch 34/100\n6/6 - 0s - loss: 0.5995 - hamming_loss: 0.2832 - val_loss: 0.6252 - val_hamming_loss: 0.2788\nEpoch 35/100\n6/6 - 0s - loss: 0.5993 - hamming_loss: 0.2885 - val_loss: 0.6231 - val_hamming_loss: 0.2771\nEpoch 36/100\n6/6 - 0s - loss: 0.5996 - hamming_loss: 0.2972 - val_loss: 0.6209 - val_hamming_loss: 0.2762\nEpoch 37/100\n6/6 - 0s - loss: 0.5926 - hamming_loss: 0.2775 - val_loss: 0.6187 - val_hamming_loss: 0.2719\nEpoch 38/100\n6/6 - 0s - loss: 0.5895 - hamming_loss: 0.2810 - val_loss: 0.6165 - val_hamming_loss: 0.2692\nEpoch 39/100\n6/6 - 0s - loss: 0.5860 - hamming_loss: 0.2710 - val_loss: 0.6142 - val_hamming_loss: 0.2692\nEpoch 40/100\n6/6 - 0s - loss: 0.5845 - hamming_loss: 0.2666 - val_loss: 0.6119 - val_hamming_loss: 0.2666\nEpoch 41/100\n6/6 - 0s - loss: 0.5836 - hamming_loss: 0.2684 - val_loss: 0.6097 - val_hamming_loss: 0.2657\nEpoch 42/100\n6/6 - 0s - loss: 0.5820 - hamming_loss: 0.2688 - val_loss: 0.6074 - val_hamming_loss: 0.2640\nEpoch 43/100\n6/6 - 0s - loss: 0.5766 - hamming_loss: 0.2478 - val_loss: 0.6051 - val_hamming_loss: 0.2605\nEpoch 44/100\n6/6 - 0s - loss: 0.5689 - hamming_loss: 0.2526 - val_loss: 0.6027 - val_hamming_loss: 0.2552\nEpoch 45/100\n6/6 - 0s - loss: 0.5691 - hamming_loss: 0.2570 - val_loss: 0.6004 - val_hamming_loss: 0.2535\nEpoch 46/100\n6/6 - 0s - loss: 0.5695 - hamming_loss: 0.2653 - val_loss: 0.5981 - val_hamming_loss: 0.2535\nEpoch 47/100\n6/6 - 0s - loss: 0.5633 - hamming_loss: 0.2395 - val_loss: 0.5958 - val_hamming_loss: 0.2517\nEpoch 48/100\n6/6 - 0s - loss: 0.5536 - hamming_loss: 0.2343 - val_loss: 0.5934 - val_hamming_loss: 0.2465\nEpoch 49/100\n6/6 - 0s - loss: 0.5501 - hamming_loss: 0.2268 - val_loss: 0.5909 - val_hamming_loss: 0.2421\nEpoch 50/100\n6/6 - 0s - loss: 0.5601 - hamming_loss: 0.2461 - val_loss: 0.5886 - val_hamming_loss: 0.2395\nEpoch 51/100\n6/6 - 0s - loss: 0.5460 - hamming_loss: 0.2391 - val_loss: 0.5862 - val_hamming_loss: 0.2395\nEpoch 52/100\n6/6 - 0s - loss: 0.5441 - hamming_loss: 0.2334 - val_loss: 0.5839 - val_hamming_loss: 0.2386\nEpoch 53/100\n6/6 - 0s - loss: 0.5418 - hamming_loss: 0.2356 - val_loss: 0.5815 - val_hamming_loss: 0.2360\nEpoch 54/100\n6/6 - 0s - loss: 0.5415 - hamming_loss: 0.2351 - val_loss: 0.5792 - val_hamming_loss: 0.2325\nEpoch 55/100\n6/6 - 0s - loss: 0.5389 - hamming_loss: 0.2330 - val_loss: 0.5768 - val_hamming_loss: 0.2308\nEpoch 56/100\n6/6 - 0s - loss: 0.5391 - hamming_loss: 0.2299 - val_loss: 0.5745 - val_hamming_loss: 0.2308\nEpoch 57/100\n6/6 - 0s - loss: 0.5311 - hamming_loss: 0.2264 - val_loss: 0.5721 - val_hamming_loss: 0.2290\nEpoch 58/100\n6/6 - 0s - loss: 0.5304 - hamming_loss: 0.2247 - val_loss: 0.5697 - val_hamming_loss: 0.2290\nEpoch 59/100\n6/6 - 0s - loss: 0.5301 - hamming_loss: 0.2198 - val_loss: 0.5674 - val_hamming_loss: 0.2264\nEpoch 60/100\n6/6 - 0s - loss: 0.5215 - hamming_loss: 0.2124 - val_loss: 0.5651 - val_hamming_loss: 0.2255\nEpoch 61/100\n6/6 - 0s - loss: 0.5163 - hamming_loss: 0.2172 - val_loss: 0.5628 - val_hamming_loss: 0.2255\nEpoch 62/100\n6/6 - 0s - loss: 0.5165 - hamming_loss: 0.2159 - val_loss: 0.5605 - val_hamming_loss: 0.2247\nEpoch 63/100\n6/6 - 0s - loss: 0.5209 - hamming_loss: 0.2238 - val_loss: 0.5582 - val_hamming_loss: 0.2247\nEpoch 64/100\n6/6 - 0s - loss: 0.5172 - hamming_loss: 0.2163 - val_loss: 0.5559 - val_hamming_loss: 0.2255\nEpoch 65/100\n6/6 - 0s - loss: 0.5037 - hamming_loss: 0.2032 - val_loss: 0.5537 - val_hamming_loss: 0.2238\nEpoch 66/100\n6/6 - 0s - loss: 0.5044 - hamming_loss: 0.1989 - val_loss: 0.5514 - val_hamming_loss: 0.2212\nEpoch 67/100\n6/6 - 0s - loss: 0.5076 - hamming_loss: 0.2098 - val_loss: 0.5491 - val_hamming_loss: 0.2194\nEpoch 68/100\n6/6 - 0s - loss: 0.5066 - hamming_loss: 0.2032 - val_loss: 0.5470 - val_hamming_loss: 0.2203\nEpoch 69/100\n6/6 - 0s - loss: 0.5032 - hamming_loss: 0.1980 - val_loss: 0.5448 - val_hamming_loss: 0.2203\nEpoch 70/100\n6/6 - 0s - loss: 0.4897 - hamming_loss: 0.2032 - val_loss: 0.5426 - val_hamming_loss: 0.2185\nEpoch 71/100\n6/6 - 0s - loss: 0.4867 - hamming_loss: 0.2006 - val_loss: 0.5405 - val_hamming_loss: 0.2150\nEpoch 72/100\n6/6 - 0s - loss: 0.4865 - hamming_loss: 0.1980 - val_loss: 0.5383 - val_hamming_loss: 0.2142\nEpoch 73/100\n6/6 - 0s - loss: 0.4941 - hamming_loss: 0.2050 - val_loss: 0.5362 - val_hamming_loss: 0.2133\nEpoch 74/100\n6/6 - 0s - loss: 0.4873 - hamming_loss: 0.1901 - val_loss: 0.5341 - val_hamming_loss: 0.2124\nEpoch 75/100\n6/6 - 0s - loss: 0.4855 - hamming_loss: 0.2015 - val_loss: 0.5319 - val_hamming_loss: 0.2115\nEpoch 76/100\n6/6 - 0s - loss: 0.4857 - hamming_loss: 0.1993 - val_loss: 0.5298 - val_hamming_loss: 0.2115\nEpoch 77/100\n6/6 - 0s - loss: 0.4655 - hamming_loss: 0.1770 - val_loss: 0.5277 - val_hamming_loss: 0.2107\nEpoch 78/100\n6/6 - 0s - loss: 0.4769 - hamming_loss: 0.1906 - val_loss: 0.5256 - val_hamming_loss: 0.2089\nEpoch 79/100\n6/6 - 0s - loss: 0.4855 - hamming_loss: 0.2050 - val_loss: 0.5235 - val_hamming_loss: 0.2072\nEpoch 80/100\n6/6 - 0s - loss: 0.4750 - hamming_loss: 0.1971 - val_loss: 0.5215 - val_hamming_loss: 0.2045\nEpoch 81/100\n6/6 - 0s - loss: 0.4594 - hamming_loss: 0.1788 - val_loss: 0.5195 - val_hamming_loss: 0.2045\nEpoch 82/100\n6/6 - 0s - loss: 0.4640 - hamming_loss: 0.1823 - val_loss: 0.5175 - val_hamming_loss: 0.2002\nEpoch 83/100\n6/6 - 0s - loss: 0.4574 - hamming_loss: 0.1849 - val_loss: 0.5156 - val_hamming_loss: 0.1993\nEpoch 84/100\n6/6 - 0s - loss: 0.4538 - hamming_loss: 0.1696 - val_loss: 0.5136 - val_hamming_loss: 0.1967\nEpoch 85/100\n6/6 - 0s - loss: 0.4546 - hamming_loss: 0.1788 - val_loss: 0.5117 - val_hamming_loss: 0.1967\nEpoch 86/100\n6/6 - 0s - loss: 0.4556 - hamming_loss: 0.1805 - val_loss: 0.5098 - val_hamming_loss: 0.1941\nEpoch 87/100\n6/6 - 0s - loss: 0.4508 - hamming_loss: 0.1796 - val_loss: 0.5079 - val_hamming_loss: 0.1932\nEpoch 88/100\n6/6 - 0s - loss: 0.4524 - hamming_loss: 0.1753 - val_loss: 0.5060 - val_hamming_loss: 0.1923\nEpoch 89/100\n6/6 - 0s - loss: 0.4381 - hamming_loss: 0.1683 - val_loss: 0.5041 - val_hamming_loss: 0.1906\nEpoch 90/100\n6/6 - 0s - loss: 0.4421 - hamming_loss: 0.1753 - val_loss: 0.5023 - val_hamming_loss: 0.1906\nEpoch 91/100\n6/6 - 0s - loss: 0.4494 - hamming_loss: 0.1757 - val_loss: 0.5005 - val_hamming_loss: 0.1897\nEpoch 92/100\n6/6 - 0s - loss: 0.4400 - hamming_loss: 0.1674 - val_loss: 0.4987 - val_hamming_loss: 0.1888\nEpoch 93/100\n6/6 - 0s - loss: 0.4324 - hamming_loss: 0.1656 - val_loss: 0.4969 - val_hamming_loss: 0.1888\nEpoch 94/100\n6/6 - 0s - loss: 0.4400 - hamming_loss: 0.1753 - val_loss: 0.4951 - val_hamming_loss: 0.1862\nEpoch 95/100\n6/6 - 0s - loss: 0.4294 - hamming_loss: 0.1700 - val_loss: 0.4933 - val_hamming_loss: 0.1862\nEpoch 96/100\n6/6 - 0s - loss: 0.4338 - hamming_loss: 0.1696 - val_loss: 0.4916 - val_hamming_loss: 0.1827\nEpoch 97/100\n6/6 - 0s - loss: 0.4300 - hamming_loss: 0.1639 - val_loss: 0.4899 - val_hamming_loss: 0.1827\nEpoch 98/100\n6/6 - 0s - loss: 0.4241 - hamming_loss: 0.1569 - val_loss: 0.4882 - val_hamming_loss: 0.1783\nEpoch 99/100\n6/6 - 0s - loss: 0.4332 - hamming_loss: 0.1735 - val_loss: 0.4866 - val_hamming_loss: 0.1757\nEpoch 100/100\n6/6 - 0s - loss: 0.4285 - hamming_loss: 0.1696 - val_loss: 0.4850 - val_hamming_loss: 0.1757\n"
],
[
"## (CAUTION: DO NOT OVERWRITE EXISTING FILES) -- Convert training history to dataframe and write to a .json file \nhistory_ce_FF_lr001_df = pd.DataFrame(history_ce_FF_lr001.history)\n#with open(\"Reduced Data Eval Metrics/Cross Entropy Feed Forward/history_ce_FF_lr0001.json\", \"w\") as outfile: \n# history_ce_FF_lr001_df.to_json(outfile)",
"_____no_output_____"
],
[
"## Learn a threshold function and save the test error for use in future DF\nY_train_pred = model_ce_FF.predict(X_train_hasLabel)\nY_test_pred = model_ce_FF.predict(X_test_hasLabel)\nt_range = (0, 1)\n\ntest_labels_binary, threshold_function = predict_test_labels_binary(Y_train_pred, Y_train_hasLabel, Y_test_pred, t_range)\nce_FF_withThreshold = metrics.hamming_loss(Y_test_hasLabel, test_labels_binary)",
"_____no_output_____"
]
],
[
[
"### Feed-Forward BP-MLL Network",
"_____no_output_____"
]
],
[
[
"## Start by defining and compiling the bp-mll loss network \ntf.random.set_seed(123)\nmodel_bpmll_FF = tf.keras.models.Sequential([\n tf.keras.layers.Dense(32, activation = 'relu'),\n tf.keras.layers.Dropout(0.5),\n tf.keras.layers.Dense(num_labels, activation = 'sigmoid')\n])\n\noptim_func = tf.keras.optimizers.Adam(lr = 0.0001)\n\nmodel_bpmll_FF.compile(optimizer = optim_func,\n loss = bp_mll_loss, metrics = metric\n )",
"_____no_output_____"
],
[
"tf.random.set_seed(123)\nhistory_bpmll_FF_lr001 = model_bpmll_FF.fit(X_train_hasLabel, Y_train_hasLabel, epochs = 100,\n validation_data = (X_test_hasLabel, Y_test_hasLabel), verbose=2)",
"Epoch 1/100\n6/6 - 1s - loss: 0.9846 - hamming_loss: 0.3820 - val_loss: 0.9769 - val_hamming_loss: 0.4685\nEpoch 2/100\n6/6 - 0s - loss: 0.9782 - hamming_loss: 0.4808 - val_loss: 0.9751 - val_hamming_loss: 0.4624\nEpoch 3/100\n6/6 - 0s - loss: 0.9793 - hamming_loss: 0.4830 - val_loss: 0.9733 - val_hamming_loss: 0.4598\nEpoch 4/100\n6/6 - 0s - loss: 0.9722 - hamming_loss: 0.4672 - val_loss: 0.9715 - val_hamming_loss: 0.4537\nEpoch 5/100\n6/6 - 0s - loss: 0.9715 - hamming_loss: 0.4668 - val_loss: 0.9698 - val_hamming_loss: 0.4519\nEpoch 6/100\n6/6 - 0s - loss: 0.9645 - hamming_loss: 0.4563 - val_loss: 0.9680 - val_hamming_loss: 0.4449\nEpoch 7/100\n6/6 - 0s - loss: 0.9601 - hamming_loss: 0.4515 - val_loss: 0.9662 - val_hamming_loss: 0.4423\nEpoch 8/100\n6/6 - 0s - loss: 0.9639 - hamming_loss: 0.4567 - val_loss: 0.9644 - val_hamming_loss: 0.4406\nEpoch 9/100\n6/6 - 0s - loss: 0.9539 - hamming_loss: 0.4388 - val_loss: 0.9626 - val_hamming_loss: 0.4379\nEpoch 10/100\n6/6 - 0s - loss: 0.9491 - hamming_loss: 0.4226 - val_loss: 0.9607 - val_hamming_loss: 0.4318\nEpoch 11/100\n6/6 - 0s - loss: 0.9474 - hamming_loss: 0.4349 - val_loss: 0.9589 - val_hamming_loss: 0.4301\nEpoch 12/100\n6/6 - 0s - loss: 0.9459 - hamming_loss: 0.4375 - val_loss: 0.9570 - val_hamming_loss: 0.4240\nEpoch 13/100\n6/6 - 0s - loss: 0.9469 - hamming_loss: 0.4226 - val_loss: 0.9552 - val_hamming_loss: 0.4152\nEpoch 14/100\n6/6 - 0s - loss: 0.9365 - hamming_loss: 0.4043 - val_loss: 0.9534 - val_hamming_loss: 0.4108\nEpoch 15/100\n6/6 - 0s - loss: 0.9372 - hamming_loss: 0.4130 - val_loss: 0.9515 - val_hamming_loss: 0.4065\nEpoch 16/100\n6/6 - 0s - loss: 0.9343 - hamming_loss: 0.4165 - val_loss: 0.9496 - val_hamming_loss: 0.4056\nEpoch 17/100\n6/6 - 0s - loss: 0.9313 - hamming_loss: 0.4174 - val_loss: 0.9477 - val_hamming_loss: 0.4030\nEpoch 18/100\n6/6 - 0s - loss: 0.9284 - hamming_loss: 0.4017 - val_loss: 0.9459 - val_hamming_loss: 0.3977\nEpoch 19/100\n6/6 - 0s - loss: 0.9251 - hamming_loss: 0.3925 - val_loss: 0.9441 - val_hamming_loss: 0.3969\nEpoch 20/100\n6/6 - 0s - loss: 0.9247 - hamming_loss: 0.3916 - val_loss: 0.9422 - val_hamming_loss: 0.3934\nEpoch 21/100\n6/6 - 0s - loss: 0.9152 - hamming_loss: 0.3837 - val_loss: 0.9403 - val_hamming_loss: 0.3951\nEpoch 22/100\n6/6 - 0s - loss: 0.9112 - hamming_loss: 0.3986 - val_loss: 0.9384 - val_hamming_loss: 0.3916\nEpoch 23/100\n6/6 - 0s - loss: 0.9108 - hamming_loss: 0.3890 - val_loss: 0.9364 - val_hamming_loss: 0.3881\nEpoch 24/100\n6/6 - 0s - loss: 0.9103 - hamming_loss: 0.3789 - val_loss: 0.9345 - val_hamming_loss: 0.3837\nEpoch 25/100\n6/6 - 0s - loss: 0.9038 - hamming_loss: 0.3890 - val_loss: 0.9326 - val_hamming_loss: 0.3820\nEpoch 26/100\n6/6 - 0s - loss: 0.9020 - hamming_loss: 0.3785 - val_loss: 0.9307 - val_hamming_loss: 0.3767\nEpoch 27/100\n6/6 - 0s - loss: 0.8928 - hamming_loss: 0.3641 - val_loss: 0.9288 - val_hamming_loss: 0.3733\nEpoch 28/100\n6/6 - 0s - loss: 0.8969 - hamming_loss: 0.3680 - val_loss: 0.9269 - val_hamming_loss: 0.3741\nEpoch 29/100\n6/6 - 0s - loss: 0.8967 - hamming_loss: 0.3702 - val_loss: 0.9249 - val_hamming_loss: 0.3715\nEpoch 30/100\n6/6 - 0s - loss: 0.8813 - hamming_loss: 0.3497 - val_loss: 0.9229 - val_hamming_loss: 0.3698\nEpoch 31/100\n6/6 - 0s - loss: 0.8766 - hamming_loss: 0.3497 - val_loss: 0.9209 - val_hamming_loss: 0.3663\nEpoch 32/100\n6/6 - 0s - loss: 0.8827 - hamming_loss: 0.3684 - val_loss: 0.9189 - val_hamming_loss: 0.3645\nEpoch 33/100\n6/6 - 0s - loss: 0.8800 - hamming_loss: 0.3510 - val_loss: 0.9169 - val_hamming_loss: 0.3636\nEpoch 34/100\n6/6 - 0s - loss: 0.8750 - hamming_loss: 0.3409 - val_loss: 0.9149 - val_hamming_loss: 0.3593\nEpoch 35/100\n6/6 - 0s - loss: 0.8699 - hamming_loss: 0.3400 - val_loss: 0.9129 - val_hamming_loss: 0.3540\nEpoch 36/100\n6/6 - 0s - loss: 0.8780 - hamming_loss: 0.3571 - val_loss: 0.9109 - val_hamming_loss: 0.3497\nEpoch 37/100\n6/6 - 0s - loss: 0.8746 - hamming_loss: 0.3457 - val_loss: 0.9090 - val_hamming_loss: 0.3462\nEpoch 38/100\n6/6 - 0s - loss: 0.8647 - hamming_loss: 0.3440 - val_loss: 0.9071 - val_hamming_loss: 0.3444\nEpoch 39/100\n6/6 - 0s - loss: 0.8597 - hamming_loss: 0.3413 - val_loss: 0.9051 - val_hamming_loss: 0.3435\nEpoch 40/100\n6/6 - 0s - loss: 0.8596 - hamming_loss: 0.3392 - val_loss: 0.9030 - val_hamming_loss: 0.3392\nEpoch 41/100\n6/6 - 0s - loss: 0.8591 - hamming_loss: 0.3370 - val_loss: 0.9010 - val_hamming_loss: 0.3339\nEpoch 42/100\n6/6 - 0s - loss: 0.8571 - hamming_loss: 0.3479 - val_loss: 0.8989 - val_hamming_loss: 0.3304\nEpoch 43/100\n6/6 - 0s - loss: 0.8511 - hamming_loss: 0.3396 - val_loss: 0.8969 - val_hamming_loss: 0.3287\nEpoch 44/100\n6/6 - 0s - loss: 0.8443 - hamming_loss: 0.3177 - val_loss: 0.8948 - val_hamming_loss: 0.3278\nEpoch 45/100\n6/6 - 0s - loss: 0.8467 - hamming_loss: 0.3234 - val_loss: 0.8928 - val_hamming_loss: 0.3260\nEpoch 46/100\n6/6 - 0s - loss: 0.8455 - hamming_loss: 0.3326 - val_loss: 0.8908 - val_hamming_loss: 0.3226\nEpoch 47/100\n6/6 - 0s - loss: 0.8401 - hamming_loss: 0.3156 - val_loss: 0.8888 - val_hamming_loss: 0.3199\nEpoch 48/100\n6/6 - 0s - loss: 0.8294 - hamming_loss: 0.3134 - val_loss: 0.8868 - val_hamming_loss: 0.3191\nEpoch 49/100\n6/6 - 0s - loss: 0.8296 - hamming_loss: 0.3077 - val_loss: 0.8847 - val_hamming_loss: 0.3173\nEpoch 50/100\n6/6 - 0s - loss: 0.8348 - hamming_loss: 0.3278 - val_loss: 0.8827 - val_hamming_loss: 0.3129\nEpoch 51/100\n6/6 - 0s - loss: 0.8213 - hamming_loss: 0.2963 - val_loss: 0.8806 - val_hamming_loss: 0.3112\nEpoch 52/100\n6/6 - 0s - loss: 0.8191 - hamming_loss: 0.3116 - val_loss: 0.8786 - val_hamming_loss: 0.3059\nEpoch 53/100\n6/6 - 0s - loss: 0.8206 - hamming_loss: 0.3081 - val_loss: 0.8767 - val_hamming_loss: 0.3042\nEpoch 54/100\n6/6 - 0s - loss: 0.8194 - hamming_loss: 0.2990 - val_loss: 0.8747 - val_hamming_loss: 0.3024\nEpoch 55/100\n6/6 - 0s - loss: 0.8149 - hamming_loss: 0.3103 - val_loss: 0.8727 - val_hamming_loss: 0.2990\nEpoch 56/100\n6/6 - 0s - loss: 0.8172 - hamming_loss: 0.3024 - val_loss: 0.8708 - val_hamming_loss: 0.2937\nEpoch 57/100\n6/6 - 0s - loss: 0.8162 - hamming_loss: 0.2898 - val_loss: 0.8688 - val_hamming_loss: 0.2920\nEpoch 58/100\n6/6 - 0s - loss: 0.8073 - hamming_loss: 0.3068 - val_loss: 0.8669 - val_hamming_loss: 0.2920\nEpoch 59/100\n6/6 - 0s - loss: 0.8073 - hamming_loss: 0.3024 - val_loss: 0.8649 - val_hamming_loss: 0.2876\nEpoch 60/100\n6/6 - 0s - loss: 0.7974 - hamming_loss: 0.2841 - val_loss: 0.8630 - val_hamming_loss: 0.2867\nEpoch 61/100\n6/6 - 0s - loss: 0.7957 - hamming_loss: 0.2823 - val_loss: 0.8611 - val_hamming_loss: 0.2841\nEpoch 62/100\n6/6 - 0s - loss: 0.7957 - hamming_loss: 0.2981 - val_loss: 0.8592 - val_hamming_loss: 0.2815\nEpoch 63/100\n6/6 - 0s - loss: 0.8008 - hamming_loss: 0.2898 - val_loss: 0.8573 - val_hamming_loss: 0.2780\nEpoch 64/100\n6/6 - 0s - loss: 0.7946 - hamming_loss: 0.2845 - val_loss: 0.8555 - val_hamming_loss: 0.2762\nEpoch 65/100\n6/6 - 0s - loss: 0.7847 - hamming_loss: 0.2802 - val_loss: 0.8536 - val_hamming_loss: 0.2745\nEpoch 66/100\n6/6 - 0s - loss: 0.7864 - hamming_loss: 0.2815 - val_loss: 0.8517 - val_hamming_loss: 0.2745\nEpoch 67/100\n6/6 - 0s - loss: 0.7899 - hamming_loss: 0.2793 - val_loss: 0.8498 - val_hamming_loss: 0.2736\nEpoch 68/100\n6/6 - 0s - loss: 0.7885 - hamming_loss: 0.2819 - val_loss: 0.8481 - val_hamming_loss: 0.2710\nEpoch 69/100\n6/6 - 0s - loss: 0.7883 - hamming_loss: 0.2832 - val_loss: 0.8463 - val_hamming_loss: 0.2692\nEpoch 70/100\n6/6 - 0s - loss: 0.7744 - hamming_loss: 0.2740 - val_loss: 0.8445 - val_hamming_loss: 0.2684\nEpoch 71/100\n6/6 - 0s - loss: 0.7699 - hamming_loss: 0.2806 - val_loss: 0.8427 - val_hamming_loss: 0.2649\nEpoch 72/100\n6/6 - 0s - loss: 0.7728 - hamming_loss: 0.2653 - val_loss: 0.8409 - val_hamming_loss: 0.2640\nEpoch 73/100\n6/6 - 0s - loss: 0.7749 - hamming_loss: 0.2793 - val_loss: 0.8391 - val_hamming_loss: 0.2640\nEpoch 74/100\n6/6 - 0s - loss: 0.7695 - hamming_loss: 0.2596 - val_loss: 0.8373 - val_hamming_loss: 0.2631\nEpoch 75/100\n6/6 - 0s - loss: 0.7669 - hamming_loss: 0.2609 - val_loss: 0.8356 - val_hamming_loss: 0.2587\nEpoch 76/100\n6/6 - 0s - loss: 0.7674 - hamming_loss: 0.2592 - val_loss: 0.8338 - val_hamming_loss: 0.2579\nEpoch 77/100\n6/6 - 0s - loss: 0.7532 - hamming_loss: 0.2574 - val_loss: 0.8320 - val_hamming_loss: 0.2587\nEpoch 78/100\n6/6 - 0s - loss: 0.7614 - hamming_loss: 0.2675 - val_loss: 0.8303 - val_hamming_loss: 0.2552\nEpoch 79/100\n6/6 - 0s - loss: 0.7678 - hamming_loss: 0.2666 - val_loss: 0.8286 - val_hamming_loss: 0.2561\nEpoch 80/100\n6/6 - 0s - loss: 0.7572 - hamming_loss: 0.2631 - val_loss: 0.8269 - val_hamming_loss: 0.2552\nEpoch 81/100\n6/6 - 0s - loss: 0.7446 - hamming_loss: 0.2509 - val_loss: 0.8252 - val_hamming_loss: 0.2535\nEpoch 82/100\n6/6 - 0s - loss: 0.7484 - hamming_loss: 0.2461 - val_loss: 0.8235 - val_hamming_loss: 0.2509\nEpoch 83/100\n6/6 - 0s - loss: 0.7455 - hamming_loss: 0.2448 - val_loss: 0.8219 - val_hamming_loss: 0.2491\nEpoch 84/100\n6/6 - 0s - loss: 0.7400 - hamming_loss: 0.2557 - val_loss: 0.8202 - val_hamming_loss: 0.2474\nEpoch 85/100\n6/6 - 0s - loss: 0.7447 - hamming_loss: 0.2386 - val_loss: 0.8185 - val_hamming_loss: 0.2448\nEpoch 86/100\n6/6 - 0s - loss: 0.7366 - hamming_loss: 0.2399 - val_loss: 0.8169 - val_hamming_loss: 0.2430\nEpoch 87/100\n6/6 - 0s - loss: 0.7391 - hamming_loss: 0.2522 - val_loss: 0.8153 - val_hamming_loss: 0.2421\nEpoch 88/100\n6/6 - 0s - loss: 0.7427 - hamming_loss: 0.2465 - val_loss: 0.8137 - val_hamming_loss: 0.2413\nEpoch 89/100\n6/6 - 0s - loss: 0.7228 - hamming_loss: 0.2378 - val_loss: 0.8120 - val_hamming_loss: 0.2404\nEpoch 90/100\n6/6 - 0s - loss: 0.7271 - hamming_loss: 0.2382 - val_loss: 0.8104 - val_hamming_loss: 0.2369\nEpoch 91/100\n6/6 - 0s - loss: 0.7351 - hamming_loss: 0.2504 - val_loss: 0.8088 - val_hamming_loss: 0.2351\nEpoch 92/100\n6/6 - 0s - loss: 0.7295 - hamming_loss: 0.2308 - val_loss: 0.8072 - val_hamming_loss: 0.2325\nEpoch 93/100\n6/6 - 0s - loss: 0.7152 - hamming_loss: 0.2295 - val_loss: 0.8057 - val_hamming_loss: 0.2316\nEpoch 94/100\n6/6 - 0s - loss: 0.7256 - hamming_loss: 0.2338 - val_loss: 0.8041 - val_hamming_loss: 0.2299\nEpoch 95/100\n6/6 - 0s - loss: 0.7153 - hamming_loss: 0.2251 - val_loss: 0.8025 - val_hamming_loss: 0.2290\nEpoch 96/100\n6/6 - 0s - loss: 0.7214 - hamming_loss: 0.2351 - val_loss: 0.8010 - val_hamming_loss: 0.2264\nEpoch 97/100\n6/6 - 0s - loss: 0.7192 - hamming_loss: 0.2299 - val_loss: 0.7994 - val_hamming_loss: 0.2255\nEpoch 98/100\n6/6 - 0s - loss: 0.7101 - hamming_loss: 0.2225 - val_loss: 0.7979 - val_hamming_loss: 0.2255\nEpoch 99/100\n6/6 - 0s - loss: 0.7176 - hamming_loss: 0.2356 - val_loss: 0.7964 - val_hamming_loss: 0.2238\nEpoch 100/100\n6/6 - 0s - loss: 0.7169 - hamming_loss: 0.2281 - val_loss: 0.7950 - val_hamming_loss: 0.2238\n"
],
[
"## (CAUTION: DO NOT OVERWRITE EXISTING FILES) -- Convert training history to dataframe and write to a .json file \nhistory_bpmll_FF_lr001_df = pd.DataFrame(history_bpmll_FF_lr001.history)\n#with open(\"Reduced Data Eval Metrics/BPMLL Feed Forward/history_bpmll_FF_lr0001.json\", \"w\") as outfile: \n# history_bpmll_FF_lr001_df.to_json(outfile)",
"_____no_output_____"
],
[
"## Learn a threshold function and save the test error for use in future DF\nY_train_pred = model_bpmll_FF.predict(X_train_hasLabel)\nY_test_pred = model_bpmll_FF.predict(X_test_hasLabel)\nt_range = (0, 1)\n\ntest_labels_binary, threshold_function = predict_test_labels_binary(Y_train_pred, Y_train_hasLabel, Y_test_pred, t_range)\nbpmll_FF_withThreshold = metrics.hamming_loss(Y_test_hasLabel, test_labels_binary)",
"_____no_output_____"
]
],
[
[
"### BPMLL Bidirectional LSTM Recurrent Network",
"_____no_output_____"
]
],
[
[
"## Load the pre-processed data\nfile_object_reduced = open('../RNN Text Categorization/RNN_data_dict_reduced.json',)\nRNN_data_dict_reduced = json.load(file_object_reduced)\nRNN_data_dict_reduced = ast.literal_eval(RNN_data_dict_reduced)\ntrain_padded_hasLabel = np.array(RNN_data_dict_reduced['train_padded_hasLabel'])\ntest_padded_hasLabel = np.array(RNN_data_dict_reduced['test_padded_hasLabel'])\nY_train_hasLabel = np.array(RNN_data_dict_reduced['Y_train_hasLabel'])\nY_test_hasLabel = np.array(RNN_data_dict_reduced['Y_test_hasLabel'])",
"_____no_output_____"
],
[
"## Define the bidirectional LSTM RNN architecture\ntf.random.set_seed(123)\nnum_labels = 13\nmax_length = 100\nnum_unique_words = 2711\n\nmodel_bpmll_biLSTM = tf.keras.models.Sequential([\n tf.keras.layers.Embedding(num_unique_words, 32, input_length = max_length),\n tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(16, return_sequences = False, return_state = False)),\n #tf.keras.layers.Dropout(0.5),\n tf.keras.layers.Dense(num_labels, activation = 'sigmoid')\n])\n\noptim_func = tf.keras.optimizers.Adam(lr = 0.0001)\n\nmodel_bpmll_biLSTM.compile(loss = bp_mll_loss, optimizer = optim_func, metrics = metric)",
"_____no_output_____"
],
[
"tf.random.set_seed(123)\nhistory_bpmll_RNN_lr001 = model_bpmll_biLSTM.fit(train_padded_hasLabel, Y_train_hasLabel, epochs = 100, \n validation_data = (test_padded_hasLabel, Y_test_hasLabel), verbose=2)",
"Epoch 1/100\n6/6 - 4s - loss: 0.9990 - hamming_loss: 0.4350 - val_loss: 0.9986 - val_hamming_loss: 0.5411\nEpoch 2/100\n6/6 - 0s - loss: 0.9981 - hamming_loss: 0.5197 - val_loss: 0.9979 - val_hamming_loss: 0.5262\nEpoch 3/100\n6/6 - 0s - loss: 0.9973 - hamming_loss: 0.5017 - val_loss: 0.9971 - val_hamming_loss: 0.5026\nEpoch 4/100\n6/6 - 0s - loss: 0.9964 - hamming_loss: 0.4786 - val_loss: 0.9964 - val_hamming_loss: 0.4895\nEpoch 5/100\n6/6 - 0s - loss: 0.9956 - hamming_loss: 0.4576 - val_loss: 0.9956 - val_hamming_loss: 0.4764\nEpoch 6/100\n6/6 - 0s - loss: 0.9947 - hamming_loss: 0.4406 - val_loss: 0.9948 - val_hamming_loss: 0.4510\nEpoch 7/100\n6/6 - 0s - loss: 0.9938 - hamming_loss: 0.4156 - val_loss: 0.9940 - val_hamming_loss: 0.4266\nEpoch 8/100\n6/6 - 0s - loss: 0.9928 - hamming_loss: 0.3872 - val_loss: 0.9931 - val_hamming_loss: 0.4073\nEpoch 9/100\n6/6 - 0s - loss: 0.9919 - hamming_loss: 0.3663 - val_loss: 0.9922 - val_hamming_loss: 0.3881\nEpoch 10/100\n6/6 - 0s - loss: 0.9908 - hamming_loss: 0.3540 - val_loss: 0.9912 - val_hamming_loss: 0.3733\nEpoch 11/100\n6/6 - 0s - loss: 0.9897 - hamming_loss: 0.3444 - val_loss: 0.9902 - val_hamming_loss: 0.3689\nEpoch 12/100\n6/6 - 0s - loss: 0.9886 - hamming_loss: 0.3361 - val_loss: 0.9892 - val_hamming_loss: 0.3593\nEpoch 13/100\n6/6 - 0s - loss: 0.9874 - hamming_loss: 0.3365 - val_loss: 0.9880 - val_hamming_loss: 0.3505\nEpoch 14/100\n6/6 - 0s - loss: 0.9861 - hamming_loss: 0.3335 - val_loss: 0.9868 - val_hamming_loss: 0.3497\nEpoch 15/100\n6/6 - 0s - loss: 0.9847 - hamming_loss: 0.3313 - val_loss: 0.9855 - val_hamming_loss: 0.3540\nEpoch 16/100\n6/6 - 0s - loss: 0.9833 - hamming_loss: 0.3352 - val_loss: 0.9841 - val_hamming_loss: 0.3523\nEpoch 17/100\n6/6 - 0s - loss: 0.9817 - hamming_loss: 0.3365 - val_loss: 0.9826 - val_hamming_loss: 0.3523\nEpoch 18/100\n6/6 - 0s - loss: 0.9800 - hamming_loss: 0.3387 - val_loss: 0.9810 - val_hamming_loss: 0.3514\nEpoch 19/100\n6/6 - 0s - loss: 0.9782 - hamming_loss: 0.3405 - val_loss: 0.9792 - val_hamming_loss: 0.3514\nEpoch 20/100\n6/6 - 0s - loss: 0.9761 - hamming_loss: 0.3427 - val_loss: 0.9773 - val_hamming_loss: 0.3514\nEpoch 21/100\n6/6 - 0s - loss: 0.9739 - hamming_loss: 0.3453 - val_loss: 0.9751 - val_hamming_loss: 0.3566\nEpoch 22/100\n6/6 - 0s - loss: 0.9715 - hamming_loss: 0.3466 - val_loss: 0.9727 - val_hamming_loss: 0.3584\nEpoch 23/100\n6/6 - 0s - loss: 0.9688 - hamming_loss: 0.3497 - val_loss: 0.9699 - val_hamming_loss: 0.3575\nEpoch 24/100\n6/6 - 0s - loss: 0.9657 - hamming_loss: 0.3514 - val_loss: 0.9668 - val_hamming_loss: 0.3558\nEpoch 25/100\n6/6 - 0s - loss: 0.9622 - hamming_loss: 0.3518 - val_loss: 0.9633 - val_hamming_loss: 0.3584\nEpoch 26/100\n6/6 - 0s - loss: 0.9583 - hamming_loss: 0.3523 - val_loss: 0.9592 - val_hamming_loss: 0.3584\nEpoch 27/100\n6/6 - 0s - loss: 0.9538 - hamming_loss: 0.3518 - val_loss: 0.9546 - val_hamming_loss: 0.3566\nEpoch 28/100\n6/6 - 0s - loss: 0.9487 - hamming_loss: 0.3510 - val_loss: 0.9491 - val_hamming_loss: 0.3558\nEpoch 29/100\n6/6 - 0s - loss: 0.9426 - hamming_loss: 0.3488 - val_loss: 0.9426 - val_hamming_loss: 0.3514\nEpoch 30/100\n6/6 - 0s - loss: 0.9352 - hamming_loss: 0.3431 - val_loss: 0.9348 - val_hamming_loss: 0.3418\nEpoch 31/100\n6/6 - 0s - loss: 0.9265 - hamming_loss: 0.3361 - val_loss: 0.9254 - val_hamming_loss: 0.3313\nEpoch 32/100\n6/6 - 0s - loss: 0.9161 - hamming_loss: 0.3234 - val_loss: 0.9143 - val_hamming_loss: 0.3147\nEpoch 33/100\n6/6 - 0s - loss: 0.9044 - hamming_loss: 0.3142 - val_loss: 0.9017 - val_hamming_loss: 0.3068\nEpoch 34/100\n6/6 - 0s - loss: 0.8909 - hamming_loss: 0.3103 - val_loss: 0.8888 - val_hamming_loss: 0.3007\nEpoch 35/100\n6/6 - 0s - loss: 0.8778 - hamming_loss: 0.3055 - val_loss: 0.8759 - val_hamming_loss: 0.2981\nEpoch 36/100\n6/6 - 0s - loss: 0.8652 - hamming_loss: 0.2990 - val_loss: 0.8643 - val_hamming_loss: 0.2981\nEpoch 37/100\n6/6 - 0s - loss: 0.8539 - hamming_loss: 0.2972 - val_loss: 0.8542 - val_hamming_loss: 0.2955\nEpoch 38/100\n6/6 - 0s - loss: 0.8440 - hamming_loss: 0.2968 - val_loss: 0.8457 - val_hamming_loss: 0.2972\nEpoch 39/100\n6/6 - 0s - loss: 0.8360 - hamming_loss: 0.2968 - val_loss: 0.8383 - val_hamming_loss: 0.2990\nEpoch 40/100\n6/6 - 0s - loss: 0.8289 - hamming_loss: 0.2985 - val_loss: 0.8319 - val_hamming_loss: 0.3007\nEpoch 41/100\n6/6 - 0s - loss: 0.8227 - hamming_loss: 0.2985 - val_loss: 0.8264 - val_hamming_loss: 0.3007\nEpoch 42/100\n6/6 - 0s - loss: 0.8174 - hamming_loss: 0.2985 - val_loss: 0.8214 - val_hamming_loss: 0.3007\nEpoch 43/100\n6/6 - 0s - loss: 0.8125 - hamming_loss: 0.2985 - val_loss: 0.8170 - val_hamming_loss: 0.3007\nEpoch 44/100\n6/6 - 0s - loss: 0.8082 - hamming_loss: 0.2985 - val_loss: 0.8129 - val_hamming_loss: 0.3007\nEpoch 45/100\n6/6 - 0s - loss: 0.8042 - hamming_loss: 0.2985 - val_loss: 0.8091 - val_hamming_loss: 0.3007\nEpoch 46/100\n6/6 - 0s - loss: 0.8005 - hamming_loss: 0.2985 - val_loss: 0.8056 - val_hamming_loss: 0.3007\nEpoch 47/100\n6/6 - 0s - loss: 0.7970 - hamming_loss: 0.2985 - val_loss: 0.8024 - val_hamming_loss: 0.3007\nEpoch 48/100\n6/6 - 0s - loss: 0.7939 - hamming_loss: 0.2985 - val_loss: 0.7993 - val_hamming_loss: 0.3007\nEpoch 49/100\n6/6 - 0s - loss: 0.7909 - hamming_loss: 0.2985 - val_loss: 0.7964 - val_hamming_loss: 0.3007\nEpoch 50/100\n6/6 - 0s - loss: 0.7881 - hamming_loss: 0.2985 - val_loss: 0.7938 - val_hamming_loss: 0.3007\nEpoch 51/100\n6/6 - 0s - loss: 0.7855 - hamming_loss: 0.2985 - val_loss: 0.7913 - val_hamming_loss: 0.3007\nEpoch 52/100\n6/6 - 0s - loss: 0.7830 - hamming_loss: 0.2985 - val_loss: 0.7889 - val_hamming_loss: 0.3007\nEpoch 53/100\n6/6 - 0s - loss: 0.7807 - hamming_loss: 0.2985 - val_loss: 0.7866 - val_hamming_loss: 0.3007\nEpoch 54/100\n6/6 - 0s - loss: 0.7785 - hamming_loss: 0.2985 - val_loss: 0.7844 - val_hamming_loss: 0.3007\nEpoch 55/100\n6/6 - 0s - loss: 0.7764 - hamming_loss: 0.2985 - val_loss: 0.7824 - val_hamming_loss: 0.3007\nEpoch 56/100\n6/6 - 0s - loss: 0.7744 - hamming_loss: 0.2985 - val_loss: 0.7805 - val_hamming_loss: 0.3007\nEpoch 57/100\n6/6 - 0s - loss: 0.7725 - hamming_loss: 0.2985 - val_loss: 0.7787 - val_hamming_loss: 0.3007\nEpoch 58/100\n6/6 - 0s - loss: 0.7707 - hamming_loss: 0.2985 - val_loss: 0.7769 - val_hamming_loss: 0.3007\nEpoch 59/100\n6/6 - 0s - loss: 0.7690 - hamming_loss: 0.2985 - val_loss: 0.7753 - val_hamming_loss: 0.3007\nEpoch 60/100\n6/6 - 0s - loss: 0.7674 - hamming_loss: 0.2985 - val_loss: 0.7737 - val_hamming_loss: 0.3007\nEpoch 61/100\n6/6 - 0s - loss: 0.7658 - hamming_loss: 0.2985 - val_loss: 0.7721 - val_hamming_loss: 0.3007\nEpoch 62/100\n6/6 - 0s - loss: 0.7643 - hamming_loss: 0.2985 - val_loss: 0.7706 - val_hamming_loss: 0.3007\nEpoch 63/100\n6/6 - 0s - loss: 0.7628 - hamming_loss: 0.2985 - val_loss: 0.7692 - val_hamming_loss: 0.3007\nEpoch 64/100\n6/6 - 0s - loss: 0.7614 - hamming_loss: 0.2985 - val_loss: 0.7678 - val_hamming_loss: 0.3007\nEpoch 65/100\n6/6 - 0s - loss: 0.7601 - hamming_loss: 0.2985 - val_loss: 0.7665 - val_hamming_loss: 0.3007\nEpoch 66/100\n6/6 - 0s - loss: 0.7589 - hamming_loss: 0.2985 - val_loss: 0.7653 - val_hamming_loss: 0.3007\nEpoch 67/100\n6/6 - 0s - loss: 0.7576 - hamming_loss: 0.2985 - val_loss: 0.7641 - val_hamming_loss: 0.3007\nEpoch 68/100\n6/6 - 0s - loss: 0.7564 - hamming_loss: 0.2985 - val_loss: 0.7629 - val_hamming_loss: 0.3007\nEpoch 69/100\n6/6 - 0s - loss: 0.7553 - hamming_loss: 0.2985 - val_loss: 0.7618 - val_hamming_loss: 0.3007\nEpoch 70/100\n6/6 - 0s - loss: 0.7542 - hamming_loss: 0.2985 - val_loss: 0.7607 - val_hamming_loss: 0.3007\nEpoch 71/100\n6/6 - 0s - loss: 0.7532 - hamming_loss: 0.2985 - val_loss: 0.7596 - val_hamming_loss: 0.3007\nEpoch 72/100\n6/6 - 0s - loss: 0.7522 - hamming_loss: 0.2985 - val_loss: 0.7587 - val_hamming_loss: 0.3007\nEpoch 73/100\n6/6 - 0s - loss: 0.7511 - hamming_loss: 0.2985 - val_loss: 0.7577 - val_hamming_loss: 0.3007\nEpoch 74/100\n6/6 - 0s - loss: 0.7502 - hamming_loss: 0.2985 - val_loss: 0.7568 - val_hamming_loss: 0.3007\nEpoch 75/100\n6/6 - 0s - loss: 0.7493 - hamming_loss: 0.2985 - val_loss: 0.7559 - val_hamming_loss: 0.3007\nEpoch 76/100\n6/6 - 0s - loss: 0.7484 - hamming_loss: 0.2985 - val_loss: 0.7550 - val_hamming_loss: 0.3007\nEpoch 77/100\n6/6 - 0s - loss: 0.7475 - hamming_loss: 0.2985 - val_loss: 0.7541 - val_hamming_loss: 0.3007\nEpoch 78/100\n6/6 - 0s - loss: 0.7467 - hamming_loss: 0.2985 - val_loss: 0.7533 - val_hamming_loss: 0.3007\nEpoch 79/100\n6/6 - 0s - loss: 0.7459 - hamming_loss: 0.2985 - val_loss: 0.7525 - val_hamming_loss: 0.3007\nEpoch 80/100\n6/6 - 0s - loss: 0.7451 - hamming_loss: 0.2985 - val_loss: 0.7517 - val_hamming_loss: 0.3007\nEpoch 81/100\n6/6 - 0s - loss: 0.7443 - hamming_loss: 0.2985 - val_loss: 0.7509 - val_hamming_loss: 0.3007\nEpoch 82/100\n6/6 - 0s - loss: 0.7436 - hamming_loss: 0.2985 - val_loss: 0.7502 - val_hamming_loss: 0.3007\nEpoch 83/100\n6/6 - 0s - loss: 0.7429 - hamming_loss: 0.2985 - val_loss: 0.7495 - val_hamming_loss: 0.3007\nEpoch 84/100\n6/6 - 0s - loss: 0.7422 - hamming_loss: 0.2985 - val_loss: 0.7489 - val_hamming_loss: 0.3007\nEpoch 85/100\n6/6 - 0s - loss: 0.7415 - hamming_loss: 0.2985 - val_loss: 0.7482 - val_hamming_loss: 0.3007\nEpoch 86/100\n6/6 - 0s - loss: 0.7408 - hamming_loss: 0.2985 - val_loss: 0.7476 - val_hamming_loss: 0.3007\nEpoch 87/100\n6/6 - 0s - loss: 0.7402 - hamming_loss: 0.2985 - val_loss: 0.7470 - val_hamming_loss: 0.3007\nEpoch 88/100\n6/6 - 0s - loss: 0.7395 - hamming_loss: 0.2985 - val_loss: 0.7464 - val_hamming_loss: 0.3007\nEpoch 89/100\n6/6 - 0s - loss: 0.7389 - hamming_loss: 0.2985 - val_loss: 0.7458 - val_hamming_loss: 0.3007\nEpoch 90/100\n6/6 - 0s - loss: 0.7383 - hamming_loss: 0.2985 - val_loss: 0.7452 - val_hamming_loss: 0.3007\nEpoch 91/100\n6/6 - 0s - loss: 0.7377 - hamming_loss: 0.2985 - val_loss: 0.7446 - val_hamming_loss: 0.3007\nEpoch 92/100\n6/6 - 0s - loss: 0.7371 - hamming_loss: 0.2985 - val_loss: 0.7441 - val_hamming_loss: 0.3007\nEpoch 93/100\n6/6 - 0s - loss: 0.7366 - hamming_loss: 0.2985 - val_loss: 0.7435 - val_hamming_loss: 0.3007\nEpoch 94/100\n6/6 - 0s - loss: 0.7360 - hamming_loss: 0.2985 - val_loss: 0.7430 - val_hamming_loss: 0.3007\nEpoch 95/100\n6/6 - 0s - loss: 0.7355 - hamming_loss: 0.2985 - val_loss: 0.7425 - val_hamming_loss: 0.3007\nEpoch 96/100\n6/6 - 0s - loss: 0.7349 - hamming_loss: 0.2985 - val_loss: 0.7419 - val_hamming_loss: 0.3007\nEpoch 97/100\n6/6 - 0s - loss: 0.7344 - hamming_loss: 0.2985 - val_loss: 0.7415 - val_hamming_loss: 0.3007\nEpoch 98/100\n6/6 - 0s - loss: 0.7339 - hamming_loss: 0.2985 - val_loss: 0.7410 - val_hamming_loss: 0.3007\nEpoch 99/100\n6/6 - 0s - loss: 0.7334 - hamming_loss: 0.2985 - val_loss: 0.7405 - val_hamming_loss: 0.3007\nEpoch 100/100\n6/6 - 0s - loss: 0.7329 - hamming_loss: 0.2985 - val_loss: 0.7400 - val_hamming_loss: 0.3007\n"
],
[
"## (CAUTION: DO NOT OVERWRITE EXISTING FILES) -- Convert training history to dataframe and write to a .json file\nhistory_bpmll_RNN_lr001_df = pd.DataFrame(history_bpmll_RNN_lr001.history)\n#with open(\"Reduced Data Eval Metrics/BPMLL RNN/history_bpmll_RNN_lr0001.json\", \"w\") as outfile: \n# history_bpmll_RNN_lr001_df.to_json(outfile)",
"_____no_output_____"
],
[
"## Learn a threshold function and save the test error for use in future DF\nY_train_pred = model_bpmll_biLSTM.predict(train_padded_hasLabel)\nY_test_pred = model_bpmll_biLSTM.predict(test_padded_hasLabel)\nt_range = (0, 1)\n\ntest_labels_binary, threshold_function = predict_test_labels_binary(Y_train_pred, Y_train_hasLabel, Y_test_pred, t_range)\nbpmll_RNN_withThreshold = metrics.hamming_loss(Y_test_hasLabel, test_labels_binary)",
"_____no_output_____"
]
],
[
[
"### Cross-Entropy Bidirectional LSTM Recurrent Network",
"_____no_output_____"
]
],
[
[
"## Define the bidirectional LSTM RNN architecture\ntf.random.set_seed(123)\nnum_labels = 13\nmax_length = 100\nnum_unique_words = 2711\n\nmodel_ce_biLSTM = tf.keras.models.Sequential([\n tf.keras.layers.Embedding(num_unique_words, 32, input_length = max_length),\n tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(16, return_sequences = False, return_state = False)),\n #tf.keras.layers.Dropout(0.5),\n tf.keras.layers.Dense(num_labels, activation = 'sigmoid')\n])\n\noptim_func = tf.keras.optimizers.Adam(lr = 0.0001)\n\nmodel_ce_biLSTM.compile(loss = 'binary_crossentropy', optimizer = optim_func, metrics = metric)",
"_____no_output_____"
],
[
"tf.random.set_seed(123)\nhistory_ce_RNN_lr001 = model_ce_biLSTM.fit(train_padded_hasLabel, Y_train_hasLabel, epochs = 100, \n validation_data = (test_padded_hasLabel, Y_test_hasLabel), verbose=2)",
"Epoch 1/100\n6/6 - 5s - loss: 0.6407 - hamming_loss: 0.2946 - val_loss: 0.4737 - val_hamming_loss: 0.1783\nEpoch 2/100\n6/6 - 0s - loss: 0.4329 - hamming_loss: 0.1792 - val_loss: 0.4222 - val_hamming_loss: 0.1888\nEpoch 3/100\n6/6 - 0s - loss: 0.4174 - hamming_loss: 0.1849 - val_loss: 0.4183 - val_hamming_loss: 0.1783\nEpoch 4/100\n6/6 - 0s - loss: 0.4093 - hamming_loss: 0.1796 - val_loss: 0.4088 - val_hamming_loss: 0.1783\nEpoch 5/100\n6/6 - 0s - loss: 0.3969 - hamming_loss: 0.1788 - val_loss: 0.4083 - val_hamming_loss: 0.1897\nEpoch 6/100\n6/6 - 0s - loss: 0.3801 - hamming_loss: 0.1744 - val_loss: 0.4075 - val_hamming_loss: 0.1801\nEpoch 7/100\n6/6 - 0s - loss: 0.3577 - hamming_loss: 0.1591 - val_loss: 0.4027 - val_hamming_loss: 0.1731\nEpoch 8/100\n6/6 - 0s - loss: 0.3325 - hamming_loss: 0.1342 - val_loss: 0.4029 - val_hamming_loss: 0.1809\nEpoch 9/100\n6/6 - 0s - loss: 0.3072 - hamming_loss: 0.1289 - val_loss: 0.4015 - val_hamming_loss: 0.1818\nEpoch 10/100\n6/6 - 0s - loss: 0.2821 - hamming_loss: 0.1132 - val_loss: 0.3962 - val_hamming_loss: 0.1722\nEpoch 11/100\n6/6 - 0s - loss: 0.2591 - hamming_loss: 0.1049 - val_loss: 0.4009 - val_hamming_loss: 0.1722\nEpoch 12/100\n6/6 - 0s - loss: 0.2396 - hamming_loss: 0.0940 - val_loss: 0.4037 - val_hamming_loss: 0.1722\nEpoch 13/100\n6/6 - 0s - loss: 0.2230 - hamming_loss: 0.0822 - val_loss: 0.4074 - val_hamming_loss: 0.1740\nEpoch 14/100\n6/6 - 0s - loss: 0.2085 - hamming_loss: 0.0712 - val_loss: 0.4131 - val_hamming_loss: 0.1713\nEpoch 15/100\n6/6 - 0s - loss: 0.1947 - hamming_loss: 0.0664 - val_loss: 0.4272 - val_hamming_loss: 0.1792\nEpoch 16/100\n6/6 - 0s - loss: 0.1818 - hamming_loss: 0.0625 - val_loss: 0.4242 - val_hamming_loss: 0.1722\nEpoch 17/100\n6/6 - 0s - loss: 0.1712 - hamming_loss: 0.0586 - val_loss: 0.4322 - val_hamming_loss: 0.1713\nEpoch 18/100\n6/6 - 0s - loss: 0.1596 - hamming_loss: 0.0485 - val_loss: 0.4446 - val_hamming_loss: 0.1652\nEpoch 19/100\n6/6 - 0s - loss: 0.1497 - hamming_loss: 0.0389 - val_loss: 0.4518 - val_hamming_loss: 0.1643\nEpoch 20/100\n6/6 - 0s - loss: 0.1412 - hamming_loss: 0.0350 - val_loss: 0.4494 - val_hamming_loss: 0.1626\nEpoch 21/100\n6/6 - 0s - loss: 0.1328 - hamming_loss: 0.0310 - val_loss: 0.4564 - val_hamming_loss: 0.1661\nEpoch 22/100\n6/6 - 0s - loss: 0.1240 - hamming_loss: 0.0288 - val_loss: 0.4581 - val_hamming_loss: 0.1661\nEpoch 23/100\n6/6 - 0s - loss: 0.1158 - hamming_loss: 0.0293 - val_loss: 0.4670 - val_hamming_loss: 0.1670\nEpoch 24/100\n6/6 - 0s - loss: 0.1081 - hamming_loss: 0.0245 - val_loss: 0.4701 - val_hamming_loss: 0.1643\nEpoch 25/100\n6/6 - 0s - loss: 0.1014 - hamming_loss: 0.0201 - val_loss: 0.4867 - val_hamming_loss: 0.1678\nEpoch 26/100\n6/6 - 0s - loss: 0.0953 - hamming_loss: 0.0188 - val_loss: 0.4735 - val_hamming_loss: 0.1652\nEpoch 27/100\n6/6 - 0s - loss: 0.0905 - hamming_loss: 0.0201 - val_loss: 0.4778 - val_hamming_loss: 0.1635\nEpoch 28/100\n6/6 - 0s - loss: 0.0847 - hamming_loss: 0.0188 - val_loss: 0.4841 - val_hamming_loss: 0.1678\nEpoch 29/100\n6/6 - 0s - loss: 0.0785 - hamming_loss: 0.0175 - val_loss: 0.4895 - val_hamming_loss: 0.1617\nEpoch 30/100\n6/6 - 0s - loss: 0.0739 - hamming_loss: 0.0144 - val_loss: 0.5127 - val_hamming_loss: 0.1678\nEpoch 31/100\n6/6 - 0s - loss: 0.0701 - hamming_loss: 0.0122 - val_loss: 0.5200 - val_hamming_loss: 0.1696\nEpoch 32/100\n6/6 - 0s - loss: 0.0659 - hamming_loss: 0.0092 - val_loss: 0.5120 - val_hamming_loss: 0.1643\nEpoch 33/100\n6/6 - 0s - loss: 0.0616 - hamming_loss: 0.0092 - val_loss: 0.5221 - val_hamming_loss: 0.1678\nEpoch 34/100\n6/6 - 0s - loss: 0.0580 - hamming_loss: 0.0083 - val_loss: 0.5252 - val_hamming_loss: 0.1678\nEpoch 35/100\n6/6 - 0s - loss: 0.0544 - hamming_loss: 0.0070 - val_loss: 0.5308 - val_hamming_loss: 0.1635\nEpoch 36/100\n6/6 - 0s - loss: 0.0512 - hamming_loss: 0.0066 - val_loss: 0.5415 - val_hamming_loss: 0.1635\nEpoch 37/100\n6/6 - 0s - loss: 0.0483 - hamming_loss: 0.0052 - val_loss: 0.5476 - val_hamming_loss: 0.1635\nEpoch 38/100\n6/6 - 0s - loss: 0.0459 - hamming_loss: 0.0039 - val_loss: 0.5558 - val_hamming_loss: 0.1635\nEpoch 39/100\n6/6 - 0s - loss: 0.0435 - hamming_loss: 0.0052 - val_loss: 0.5556 - val_hamming_loss: 0.1652\nEpoch 40/100\n6/6 - 0s - loss: 0.0413 - hamming_loss: 0.0035 - val_loss: 0.5556 - val_hamming_loss: 0.1626\nEpoch 41/100\n6/6 - 0s - loss: 0.0393 - hamming_loss: 0.0031 - val_loss: 0.5702 - val_hamming_loss: 0.1626\nEpoch 42/100\n6/6 - 0s - loss: 0.0378 - hamming_loss: 0.0022 - val_loss: 0.5764 - val_hamming_loss: 0.1643\nEpoch 43/100\n6/6 - 0s - loss: 0.0363 - hamming_loss: 0.0022 - val_loss: 0.5775 - val_hamming_loss: 0.1661\nEpoch 44/100\n6/6 - 0s - loss: 0.0347 - hamming_loss: 0.0013 - val_loss: 0.5779 - val_hamming_loss: 0.1652\nEpoch 45/100\n6/6 - 0s - loss: 0.0332 - hamming_loss: 0.0017 - val_loss: 0.5786 - val_hamming_loss: 0.1635\nEpoch 46/100\n6/6 - 0s - loss: 0.0319 - hamming_loss: 0.0017 - val_loss: 0.5799 - val_hamming_loss: 0.1608\nEpoch 47/100\n6/6 - 0s - loss: 0.0306 - hamming_loss: 0.0013 - val_loss: 0.5865 - val_hamming_loss: 0.1591\nEpoch 48/100\n6/6 - 0s - loss: 0.0295 - hamming_loss: 0.0013 - val_loss: 0.5915 - val_hamming_loss: 0.1643\nEpoch 49/100\n6/6 - 0s - loss: 0.0285 - hamming_loss: 0.0013 - val_loss: 0.5916 - val_hamming_loss: 0.1652\nEpoch 50/100\n6/6 - 0s - loss: 0.0273 - hamming_loss: 8.7413e-04 - val_loss: 0.5947 - val_hamming_loss: 0.1617\nEpoch 51/100\n6/6 - 0s - loss: 0.0267 - hamming_loss: 0.0013 - val_loss: 0.5986 - val_hamming_loss: 0.1626\nEpoch 52/100\n6/6 - 0s - loss: 0.0255 - hamming_loss: 8.7413e-04 - val_loss: 0.6050 - val_hamming_loss: 0.1678\nEpoch 53/100\n6/6 - 0s - loss: 0.0246 - hamming_loss: 8.7413e-04 - val_loss: 0.6124 - val_hamming_loss: 0.1652\nEpoch 54/100\n6/6 - 0s - loss: 0.0235 - hamming_loss: 4.3706e-04 - val_loss: 0.6159 - val_hamming_loss: 0.1705\nEpoch 55/100\n6/6 - 0s - loss: 0.0226 - hamming_loss: 4.3706e-04 - val_loss: 0.6195 - val_hamming_loss: 0.1635\nEpoch 56/100\n6/6 - 0s - loss: 0.0219 - hamming_loss: 4.3706e-04 - val_loss: 0.6227 - val_hamming_loss: 0.1643\nEpoch 57/100\n6/6 - 0s - loss: 0.0211 - hamming_loss: 4.3706e-04 - val_loss: 0.6252 - val_hamming_loss: 0.1643\nEpoch 58/100\n6/6 - 0s - loss: 0.0205 - hamming_loss: 4.3706e-04 - val_loss: 0.6296 - val_hamming_loss: 0.1652\nEpoch 59/100\n6/6 - 0s - loss: 0.0198 - hamming_loss: 4.3706e-04 - val_loss: 0.6377 - val_hamming_loss: 0.1713\nEpoch 60/100\n6/6 - 0s - loss: 0.0192 - hamming_loss: 4.3706e-04 - val_loss: 0.6345 - val_hamming_loss: 0.1705\nEpoch 61/100\n6/6 - 0s - loss: 0.0186 - hamming_loss: 4.3706e-04 - val_loss: 0.6369 - val_hamming_loss: 0.1713\nEpoch 62/100\n6/6 - 0s - loss: 0.0181 - hamming_loss: 4.3706e-04 - val_loss: 0.6424 - val_hamming_loss: 0.1705\nEpoch 63/100\n6/6 - 0s - loss: 0.0176 - hamming_loss: 4.3706e-04 - val_loss: 0.6431 - val_hamming_loss: 0.1696\nEpoch 64/100\n6/6 - 0s - loss: 0.0171 - hamming_loss: 4.3706e-04 - val_loss: 0.6480 - val_hamming_loss: 0.1722\nEpoch 65/100\n6/6 - 0s - loss: 0.0167 - hamming_loss: 4.3706e-04 - val_loss: 0.6563 - val_hamming_loss: 0.1687\nEpoch 66/100\n6/6 - 0s - loss: 0.0162 - hamming_loss: 4.3706e-04 - val_loss: 0.6620 - val_hamming_loss: 0.1705\nEpoch 67/100\n6/6 - 0s - loss: 0.0158 - hamming_loss: 4.3706e-04 - val_loss: 0.6650 - val_hamming_loss: 0.1687\nEpoch 68/100\n6/6 - 0s - loss: 0.0153 - hamming_loss: 4.3706e-04 - val_loss: 0.6666 - val_hamming_loss: 0.1696\nEpoch 69/100\n6/6 - 0s - loss: 0.0149 - hamming_loss: 4.3706e-04 - val_loss: 0.6730 - val_hamming_loss: 0.1670\nEpoch 70/100\n6/6 - 0s - loss: 0.0145 - hamming_loss: 4.3706e-04 - val_loss: 0.6749 - val_hamming_loss: 0.1696\nEpoch 71/100\n6/6 - 0s - loss: 0.0143 - hamming_loss: 4.3706e-04 - val_loss: 0.6782 - val_hamming_loss: 0.1678\nEpoch 72/100\n6/6 - 0s - loss: 0.0138 - hamming_loss: 4.3706e-04 - val_loss: 0.6845 - val_hamming_loss: 0.1687\nEpoch 73/100\n6/6 - 0s - loss: 0.0135 - hamming_loss: 4.3706e-04 - val_loss: 0.6838 - val_hamming_loss: 0.1687\nEpoch 74/100\n6/6 - 0s - loss: 0.0131 - hamming_loss: 4.3706e-04 - val_loss: 0.6846 - val_hamming_loss: 0.1670\nEpoch 75/100\n6/6 - 0s - loss: 0.0128 - hamming_loss: 4.3706e-04 - val_loss: 0.6935 - val_hamming_loss: 0.1705\nEpoch 76/100\n6/6 - 0s - loss: 0.0125 - hamming_loss: 4.3706e-04 - val_loss: 0.6937 - val_hamming_loss: 0.1696\nEpoch 77/100\n6/6 - 0s - loss: 0.0122 - hamming_loss: 4.3706e-04 - val_loss: 0.6938 - val_hamming_loss: 0.1670\nEpoch 78/100\n6/6 - 0s - loss: 0.0119 - hamming_loss: 4.3706e-04 - val_loss: 0.7017 - val_hamming_loss: 0.1687\nEpoch 79/100\n6/6 - 0s - loss: 0.0116 - hamming_loss: 0.0000e+00 - val_loss: 0.7070 - val_hamming_loss: 0.1705\nEpoch 80/100\n6/6 - 0s - loss: 0.0114 - hamming_loss: 4.3706e-04 - val_loss: 0.7072 - val_hamming_loss: 0.1670\nEpoch 81/100\n6/6 - 0s - loss: 0.0111 - hamming_loss: 4.3706e-04 - val_loss: 0.7083 - val_hamming_loss: 0.1722\nEpoch 82/100\n6/6 - 0s - loss: 0.0109 - hamming_loss: 4.3706e-04 - val_loss: 0.7100 - val_hamming_loss: 0.1722\nEpoch 83/100\n6/6 - 0s - loss: 0.0106 - hamming_loss: 0.0000e+00 - val_loss: 0.7143 - val_hamming_loss: 0.1713\nEpoch 84/100\n6/6 - 0s - loss: 0.0104 - hamming_loss: 0.0000e+00 - val_loss: 0.7158 - val_hamming_loss: 0.1740\nEpoch 85/100\n6/6 - 0s - loss: 0.0102 - hamming_loss: 0.0000e+00 - val_loss: 0.7199 - val_hamming_loss: 0.1740\nEpoch 86/100\n6/6 - 0s - loss: 0.0100 - hamming_loss: 0.0000e+00 - val_loss: 0.7226 - val_hamming_loss: 0.1713\nEpoch 87/100\n6/6 - 0s - loss: 0.0097 - hamming_loss: 0.0000e+00 - val_loss: 0.7239 - val_hamming_loss: 0.1705\nEpoch 88/100\n6/6 - 0s - loss: 0.0095 - hamming_loss: 0.0000e+00 - val_loss: 0.7277 - val_hamming_loss: 0.1748\nEpoch 89/100\n6/6 - 0s - loss: 0.0094 - hamming_loss: 0.0000e+00 - val_loss: 0.7301 - val_hamming_loss: 0.1731\nEpoch 90/100\n6/6 - 0s - loss: 0.0092 - hamming_loss: 0.0000e+00 - val_loss: 0.7343 - val_hamming_loss: 0.1713\nEpoch 91/100\n6/6 - 0s - loss: 0.0090 - hamming_loss: 0.0000e+00 - val_loss: 0.7359 - val_hamming_loss: 0.1687\nEpoch 92/100\n6/6 - 0s - loss: 0.0088 - hamming_loss: 0.0000e+00 - val_loss: 0.7389 - val_hamming_loss: 0.1705\nEpoch 93/100\n6/6 - 0s - loss: 0.0087 - hamming_loss: 0.0000e+00 - val_loss: 0.7400 - val_hamming_loss: 0.1687\nEpoch 94/100\n6/6 - 0s - loss: 0.0085 - hamming_loss: 0.0000e+00 - val_loss: 0.7454 - val_hamming_loss: 0.1713\nEpoch 95/100\n6/6 - 0s - loss: 0.0084 - hamming_loss: 0.0000e+00 - val_loss: 0.7423 - val_hamming_loss: 0.1705\nEpoch 96/100\n6/6 - 0s - loss: 0.0083 - hamming_loss: 0.0000e+00 - val_loss: 0.7469 - val_hamming_loss: 0.1696\nEpoch 97/100\n6/6 - 0s - loss: 0.0081 - hamming_loss: 0.0000e+00 - val_loss: 0.7465 - val_hamming_loss: 0.1696\nEpoch 98/100\n6/6 - 0s - loss: 0.0080 - hamming_loss: 0.0000e+00 - val_loss: 0.7377 - val_hamming_loss: 0.1696\nEpoch 99/100\n6/6 - 0s - loss: 0.0079 - hamming_loss: 0.0000e+00 - val_loss: 0.7567 - val_hamming_loss: 0.1748\nEpoch 100/100\n6/6 - 0s - loss: 0.0077 - hamming_loss: 0.0000e+00 - val_loss: 0.7553 - val_hamming_loss: 0.1678\n"
],
[
"## (CAUTION: DO NOT OVERWRITE EXISTING FILES) -- Convert training history to dataframe and write to a .json file\nhistory_ce_RNN_lr001_df = pd.DataFrame(history_ce_RNN_lr001.history)\n#with open(\"Reduced Data Eval Metrics/Cross Entropy RNN/history_ce_RNN_lr0001.json\", \"w\") as outfile: \n# history_ce_RNN_lr001_df.to_json(outfile)",
"_____no_output_____"
],
[
"## Learn a threshold function and save the test error for use in future DF\nY_train_pred = model_ce_biLSTM.predict(train_padded_hasLabel)\nY_test_pred = model_ce_biLSTM.predict(test_padded_hasLabel)\nt_range = (0, 1)\n\ntest_labels_binary, threshold_function = predict_test_labels_binary(Y_train_pred, Y_train_hasLabel, Y_test_pred, t_range)\nce_RNN_withThreshold = metrics.hamming_loss(Y_test_hasLabel, test_labels_binary)",
"_____no_output_____"
],
[
"## (CAUTION: DO NOT OVERWRITE EXISTING FILES) -- Collect the test set hamming losses for the models \n## with learned threshold functions into a df and write to .json file\nval_hamming_loss_withThreshold_lr001_df = pd.DataFrame({'ce_FF_lr0001' : ce_FF_withThreshold,\n 'bpmll_FF_lr0001' : bpmll_FF_withThreshold,\n 'ce_RNN_lr0001' : ce_RNN_withThreshold,\n 'bpmll_RNN_lr0001' : bpmll_RNN_withThreshold},\n index = [0])\n\n#with open(\"Reduced Data Eval Metrics/val_hamming_loss_withThreshold_lr0001.json\", \"w\") as outfile: \n# val_hamming_loss_withThreshold_lr001_df.to_json(outfile)",
"_____no_output_____"
],
[
"val_hamming_loss_withThreshold_lr001_df",
"_____no_output_____"
]
],
[
[
"## Models on Full Dataset (some instances have no labels)",
"_____no_output_____"
]
],
[
[
"## Load the full tfidf dataset\nfile_object = open('../BP-MLL Text Categorization/tfidf_trainTest_data.json',)\ntfidf_data_full = json.load(file_object)\nX_train = np.array(tfidf_data_full['X_train'])\nX_test = np.array(tfidf_data_full['X_test'])\nY_train = np.array(tfidf_data_full['Y_train'])\nY_test = np.array(tfidf_data_full['Y_test'])",
"_____no_output_____"
]
],
[
[
"### Feed-Forward Cross-Entropy Network",
"_____no_output_____"
]
],
[
[
"## Use same architecture as the previous cross-entropy feed-forward network and train on full dataset\ntf.random.set_seed(123)\nnum_labels = 13\n\nmodel_ce_FF_full = tf.keras.models.Sequential([\n tf.keras.layers.Dense(32, activation = 'relu'),\n tf.keras.layers.Dropout(0.5),\n tf.keras.layers.Dense(num_labels, activation = 'sigmoid')\n])\n\noptim_func = tf.keras.optimizers.Adam(lr = 0.0001)\n\nmodel_ce_FF_full.compile(optimizer = optim_func,\n loss = 'binary_crossentropy', metrics = metric\n )",
"_____no_output_____"
],
[
"tf.random.set_seed(123)\nhistory_ce_FF_lr001_full = model_ce_FF_full.fit(X_train, Y_train, epochs = 100,\n validation_data = (X_test, Y_test), verbose=2)",
"Epoch 1/100\n7/7 - 1s - loss: 0.7023 - hamming_loss: 0.3895 - val_loss: 0.6944 - val_hamming_loss: 0.4808\nEpoch 2/100\n7/7 - 0s - loss: 0.7009 - hamming_loss: 0.4738 - val_loss: 0.6923 - val_hamming_loss: 0.4728\nEpoch 3/100\n7/7 - 0s - loss: 0.7003 - hamming_loss: 0.4762 - val_loss: 0.6902 - val_hamming_loss: 0.4663\nEpoch 4/100\n7/7 - 0s - loss: 0.6940 - hamming_loss: 0.4592 - val_loss: 0.6881 - val_hamming_loss: 0.4607\nEpoch 5/100\n7/7 - 0s - loss: 0.6914 - hamming_loss: 0.4592 - val_loss: 0.6860 - val_hamming_loss: 0.4511\nEpoch 6/100\n7/7 - 0s - loss: 0.6892 - hamming_loss: 0.4496 - val_loss: 0.6840 - val_hamming_loss: 0.4431\nEpoch 7/100\n7/7 - 0s - loss: 0.6862 - hamming_loss: 0.4437 - val_loss: 0.6820 - val_hamming_loss: 0.4343\nEpoch 8/100\n7/7 - 0s - loss: 0.6767 - hamming_loss: 0.4211 - val_loss: 0.6800 - val_hamming_loss: 0.4271\nEpoch 9/100\n7/7 - 0s - loss: 0.6723 - hamming_loss: 0.4132 - val_loss: 0.6781 - val_hamming_loss: 0.4183\nEpoch 10/100\n7/7 - 0s - loss: 0.6712 - hamming_loss: 0.4096 - val_loss: 0.6763 - val_hamming_loss: 0.4119\nEpoch 11/100\n7/7 - 0s - loss: 0.6707 - hamming_loss: 0.4029 - val_loss: 0.6745 - val_hamming_loss: 0.4062\nEpoch 12/100\n7/7 - 0s - loss: 0.6704 - hamming_loss: 0.4048 - val_loss: 0.6727 - val_hamming_loss: 0.4030\nEpoch 13/100\n7/7 - 0s - loss: 0.6698 - hamming_loss: 0.4076 - val_loss: 0.6709 - val_hamming_loss: 0.3958\nEpoch 14/100\n7/7 - 0s - loss: 0.6630 - hamming_loss: 0.3838 - val_loss: 0.6691 - val_hamming_loss: 0.3926\nEpoch 15/100\n7/7 - 0s - loss: 0.6594 - hamming_loss: 0.3767 - val_loss: 0.6673 - val_hamming_loss: 0.3886\nEpoch 16/100\n7/7 - 0s - loss: 0.6486 - hamming_loss: 0.3672 - val_loss: 0.6655 - val_hamming_loss: 0.3814\nEpoch 17/100\n7/7 - 0s - loss: 0.6517 - hamming_loss: 0.3620 - val_loss: 0.6637 - val_hamming_loss: 0.3742\nEpoch 18/100\n7/7 - 0s - loss: 0.6466 - hamming_loss: 0.3501 - val_loss: 0.6619 - val_hamming_loss: 0.3710\nEpoch 19/100\n7/7 - 0s - loss: 0.6440 - hamming_loss: 0.3505 - val_loss: 0.6601 - val_hamming_loss: 0.3678\nEpoch 20/100\n7/7 - 0s - loss: 0.6397 - hamming_loss: 0.3351 - val_loss: 0.6584 - val_hamming_loss: 0.3582\nEpoch 21/100\n7/7 - 0s - loss: 0.6379 - hamming_loss: 0.3442 - val_loss: 0.6566 - val_hamming_loss: 0.3542\nEpoch 22/100\n7/7 - 0s - loss: 0.6332 - hamming_loss: 0.3366 - val_loss: 0.6547 - val_hamming_loss: 0.3478\nEpoch 23/100\n7/7 - 0s - loss: 0.6328 - hamming_loss: 0.3295 - val_loss: 0.6529 - val_hamming_loss: 0.3446\nEpoch 24/100\n7/7 - 0s - loss: 0.6347 - hamming_loss: 0.3402 - val_loss: 0.6510 - val_hamming_loss: 0.3429\nEpoch 25/100\n7/7 - 0s - loss: 0.6305 - hamming_loss: 0.3335 - val_loss: 0.6492 - val_hamming_loss: 0.3429\nEpoch 26/100\n7/7 - 0s - loss: 0.6248 - hamming_loss: 0.3168 - val_loss: 0.6473 - val_hamming_loss: 0.3365\nEpoch 27/100\n7/7 - 0s - loss: 0.6230 - hamming_loss: 0.3081 - val_loss: 0.6454 - val_hamming_loss: 0.3309\nEpoch 28/100\n7/7 - 0s - loss: 0.6223 - hamming_loss: 0.3152 - val_loss: 0.6436 - val_hamming_loss: 0.3245\nEpoch 29/100\n7/7 - 0s - loss: 0.6151 - hamming_loss: 0.3109 - val_loss: 0.6417 - val_hamming_loss: 0.3229\nEpoch 30/100\n7/7 - 0s - loss: 0.6134 - hamming_loss: 0.3037 - val_loss: 0.6397 - val_hamming_loss: 0.3197\nEpoch 31/100\n7/7 - 0s - loss: 0.6110 - hamming_loss: 0.2922 - val_loss: 0.6378 - val_hamming_loss: 0.3157\nEpoch 32/100\n7/7 - 0s - loss: 0.6054 - hamming_loss: 0.2934 - val_loss: 0.6358 - val_hamming_loss: 0.3077\nEpoch 33/100\n7/7 - 0s - loss: 0.6090 - hamming_loss: 0.2835 - val_loss: 0.6339 - val_hamming_loss: 0.3053\nEpoch 34/100\n7/7 - 0s - loss: 0.5966 - hamming_loss: 0.2807 - val_loss: 0.6320 - val_hamming_loss: 0.3013\nEpoch 35/100\n7/7 - 0s - loss: 0.6028 - hamming_loss: 0.2807 - val_loss: 0.6301 - val_hamming_loss: 0.2989\nEpoch 36/100\n7/7 - 0s - loss: 0.6033 - hamming_loss: 0.2902 - val_loss: 0.6282 - val_hamming_loss: 0.2973\nEpoch 37/100\n7/7 - 0s - loss: 0.5909 - hamming_loss: 0.2760 - val_loss: 0.6262 - val_hamming_loss: 0.2965\nEpoch 38/100\n7/7 - 0s - loss: 0.5904 - hamming_loss: 0.2724 - val_loss: 0.6242 - val_hamming_loss: 0.2925\nEpoch 39/100\n7/7 - 0s - loss: 0.5874 - hamming_loss: 0.2661 - val_loss: 0.6223 - val_hamming_loss: 0.2909\nEpoch 40/100\n7/7 - 0s - loss: 0.5800 - hamming_loss: 0.2613 - val_loss: 0.6203 - val_hamming_loss: 0.2885\nEpoch 41/100\n7/7 - 0s - loss: 0.5808 - hamming_loss: 0.2661 - val_loss: 0.6182 - val_hamming_loss: 0.2861\nEpoch 42/100\n7/7 - 0s - loss: 0.5803 - hamming_loss: 0.2577 - val_loss: 0.6162 - val_hamming_loss: 0.2829\nEpoch 43/100\n7/7 - 0s - loss: 0.5782 - hamming_loss: 0.2581 - val_loss: 0.6143 - val_hamming_loss: 0.2804\nEpoch 44/100\n7/7 - 0s - loss: 0.5760 - hamming_loss: 0.2577 - val_loss: 0.6124 - val_hamming_loss: 0.2772\nEpoch 45/100\n7/7 - 0s - loss: 0.5734 - hamming_loss: 0.2466 - val_loss: 0.6105 - val_hamming_loss: 0.2756\nEpoch 46/100\n7/7 - 0s - loss: 0.5699 - hamming_loss: 0.2375 - val_loss: 0.6086 - val_hamming_loss: 0.2740\nEpoch 47/100\n7/7 - 0s - loss: 0.5646 - hamming_loss: 0.2446 - val_loss: 0.6066 - val_hamming_loss: 0.2732\nEpoch 48/100\n7/7 - 0s - loss: 0.5700 - hamming_loss: 0.2542 - val_loss: 0.6046 - val_hamming_loss: 0.2724\nEpoch 49/100\n7/7 - 0s - loss: 0.5602 - hamming_loss: 0.2431 - val_loss: 0.6026 - val_hamming_loss: 0.2700\nEpoch 50/100\n7/7 - 0s - loss: 0.5564 - hamming_loss: 0.2395 - val_loss: 0.6005 - val_hamming_loss: 0.2676\nEpoch 51/100\n7/7 - 0s - loss: 0.5556 - hamming_loss: 0.2486 - val_loss: 0.5984 - val_hamming_loss: 0.2668\nEpoch 52/100\n7/7 - 0s - loss: 0.5512 - hamming_loss: 0.2272 - val_loss: 0.5962 - val_hamming_loss: 0.2644\nEpoch 53/100\n7/7 - 0s - loss: 0.5473 - hamming_loss: 0.2335 - val_loss: 0.5941 - val_hamming_loss: 0.2628\nEpoch 54/100\n7/7 - 0s - loss: 0.5480 - hamming_loss: 0.2367 - val_loss: 0.5920 - val_hamming_loss: 0.2604\nEpoch 55/100\n7/7 - 0s - loss: 0.5358 - hamming_loss: 0.2209 - val_loss: 0.5898 - val_hamming_loss: 0.2588\nEpoch 56/100\n7/7 - 0s - loss: 0.5429 - hamming_loss: 0.2300 - val_loss: 0.5878 - val_hamming_loss: 0.2572\nEpoch 57/100\n7/7 - 0s - loss: 0.5358 - hamming_loss: 0.2228 - val_loss: 0.5857 - val_hamming_loss: 0.2564\nEpoch 58/100\n7/7 - 0s - loss: 0.5319 - hamming_loss: 0.2177 - val_loss: 0.5835 - val_hamming_loss: 0.2548\nEpoch 59/100\n7/7 - 0s - loss: 0.5348 - hamming_loss: 0.2236 - val_loss: 0.5814 - val_hamming_loss: 0.2532\nEpoch 60/100\n7/7 - 0s - loss: 0.5277 - hamming_loss: 0.2197 - val_loss: 0.5794 - val_hamming_loss: 0.2516\nEpoch 61/100\n7/7 - 0s - loss: 0.5287 - hamming_loss: 0.2165 - val_loss: 0.5774 - val_hamming_loss: 0.2468\nEpoch 62/100\n7/7 - 0s - loss: 0.5305 - hamming_loss: 0.2228 - val_loss: 0.5754 - val_hamming_loss: 0.2452\nEpoch 63/100\n7/7 - 0s - loss: 0.5137 - hamming_loss: 0.2105 - val_loss: 0.5734 - val_hamming_loss: 0.2444\nEpoch 64/100\n7/7 - 0s - loss: 0.5194 - hamming_loss: 0.2149 - val_loss: 0.5714 - val_hamming_loss: 0.2444\nEpoch 65/100\n7/7 - 0s - loss: 0.5295 - hamming_loss: 0.2320 - val_loss: 0.5695 - val_hamming_loss: 0.2396\nEpoch 66/100\n7/7 - 0s - loss: 0.5144 - hamming_loss: 0.2141 - val_loss: 0.5675 - val_hamming_loss: 0.2372\nEpoch 67/100\n7/7 - 0s - loss: 0.5127 - hamming_loss: 0.2006 - val_loss: 0.5656 - val_hamming_loss: 0.2340\nEpoch 68/100\n7/7 - 0s - loss: 0.5035 - hamming_loss: 0.2050 - val_loss: 0.5636 - val_hamming_loss: 0.2316\nEpoch 69/100\n7/7 - 0s - loss: 0.5062 - hamming_loss: 0.2070 - val_loss: 0.5617 - val_hamming_loss: 0.2292\nEpoch 70/100\n7/7 - 0s - loss: 0.5077 - hamming_loss: 0.2090 - val_loss: 0.5597 - val_hamming_loss: 0.2284\nEpoch 71/100\n7/7 - 0s - loss: 0.5069 - hamming_loss: 0.2034 - val_loss: 0.5577 - val_hamming_loss: 0.2276\nEpoch 72/100\n7/7 - 0s - loss: 0.5016 - hamming_loss: 0.1990 - val_loss: 0.5557 - val_hamming_loss: 0.2268\nEpoch 73/100\n7/7 - 0s - loss: 0.5043 - hamming_loss: 0.2018 - val_loss: 0.5537 - val_hamming_loss: 0.2276\nEpoch 74/100\n7/7 - 0s - loss: 0.4894 - hamming_loss: 0.1967 - val_loss: 0.5517 - val_hamming_loss: 0.2260\nEpoch 75/100\n7/7 - 0s - loss: 0.4957 - hamming_loss: 0.1975 - val_loss: 0.5498 - val_hamming_loss: 0.2252\nEpoch 76/100\n7/7 - 0s - loss: 0.4910 - hamming_loss: 0.1955 - val_loss: 0.5479 - val_hamming_loss: 0.2236\nEpoch 77/100\n7/7 - 0s - loss: 0.4825 - hamming_loss: 0.1852 - val_loss: 0.5461 - val_hamming_loss: 0.2220\nEpoch 78/100\n7/7 - 0s - loss: 0.4847 - hamming_loss: 0.1907 - val_loss: 0.5444 - val_hamming_loss: 0.2179\nEpoch 79/100\n7/7 - 0s - loss: 0.4894 - hamming_loss: 0.1899 - val_loss: 0.5425 - val_hamming_loss: 0.2171\nEpoch 80/100\n7/7 - 0s - loss: 0.4767 - hamming_loss: 0.1899 - val_loss: 0.5408 - val_hamming_loss: 0.2171\nEpoch 81/100\n7/7 - 0s - loss: 0.4819 - hamming_loss: 0.1872 - val_loss: 0.5390 - val_hamming_loss: 0.2163\nEpoch 82/100\n7/7 - 0s - loss: 0.4748 - hamming_loss: 0.1923 - val_loss: 0.5373 - val_hamming_loss: 0.2139\nEpoch 83/100\n7/7 - 0s - loss: 0.4732 - hamming_loss: 0.1860 - val_loss: 0.5355 - val_hamming_loss: 0.2131\nEpoch 84/100\n7/7 - 0s - loss: 0.4701 - hamming_loss: 0.1935 - val_loss: 0.5337 - val_hamming_loss: 0.2115\nEpoch 85/100\n7/7 - 0s - loss: 0.4691 - hamming_loss: 0.1875 - val_loss: 0.5319 - val_hamming_loss: 0.2099\nEpoch 86/100\n7/7 - 0s - loss: 0.4667 - hamming_loss: 0.1737 - val_loss: 0.5301 - val_hamming_loss: 0.2099\nEpoch 87/100\n7/7 - 0s - loss: 0.4622 - hamming_loss: 0.1745 - val_loss: 0.5283 - val_hamming_loss: 0.2059\nEpoch 88/100\n7/7 - 0s - loss: 0.4554 - hamming_loss: 0.1745 - val_loss: 0.5265 - val_hamming_loss: 0.2051\nEpoch 89/100\n7/7 - 0s - loss: 0.4682 - hamming_loss: 0.1832 - val_loss: 0.5248 - val_hamming_loss: 0.2043\nEpoch 90/100\n7/7 - 0s - loss: 0.4532 - hamming_loss: 0.1737 - val_loss: 0.5231 - val_hamming_loss: 0.2019\nEpoch 91/100\n7/7 - 0s - loss: 0.4563 - hamming_loss: 0.1725 - val_loss: 0.5213 - val_hamming_loss: 0.2019\nEpoch 92/100\n7/7 - 0s - loss: 0.4572 - hamming_loss: 0.1804 - val_loss: 0.5196 - val_hamming_loss: 0.2019\nEpoch 93/100\n7/7 - 0s - loss: 0.4501 - hamming_loss: 0.1697 - val_loss: 0.5180 - val_hamming_loss: 0.2011\nEpoch 94/100\n7/7 - 0s - loss: 0.4447 - hamming_loss: 0.1705 - val_loss: 0.5163 - val_hamming_loss: 0.2011\nEpoch 95/100\n7/7 - 0s - loss: 0.4420 - hamming_loss: 0.1721 - val_loss: 0.5146 - val_hamming_loss: 0.2003\nEpoch 96/100\n7/7 - 0s - loss: 0.4412 - hamming_loss: 0.1649 - val_loss: 0.5130 - val_hamming_loss: 0.2003\nEpoch 97/100\n7/7 - 0s - loss: 0.4385 - hamming_loss: 0.1701 - val_loss: 0.5114 - val_hamming_loss: 0.1979\nEpoch 98/100\n7/7 - 0s - loss: 0.4309 - hamming_loss: 0.1602 - val_loss: 0.5098 - val_hamming_loss: 0.1955\nEpoch 99/100\n7/7 - 0s - loss: 0.4288 - hamming_loss: 0.1614 - val_loss: 0.5082 - val_hamming_loss: 0.1947\nEpoch 100/100\n7/7 - 0s - loss: 0.4349 - hamming_loss: 0.1598 - val_loss: 0.5066 - val_hamming_loss: 0.1939\n"
],
[
"## (CAUTION: DO NOT OVERWRITE EXISTING FILES) -- Convert training history to dataframe and write to a .json file\nhistory_ce_FF_lr001_full_df = pd.DataFrame(history_ce_FF_lr001_full.history)\n#with open(\"Full Data Eval Metrics/Cross Entropy Feed Forward/history_ce_FF_lr0001_full.json\", \"w\") as outfile: \n# history_ce_FF_lr001_full_df.to_json(outfile)",
"_____no_output_____"
],
[
"## Learn a threshold function and save the test error for use in future DF\nY_train_pred = model_ce_FF_full.predict(X_train)\nY_test_pred = model_ce_FF_full.predict(X_test)\nt_range = (0, 1)\n\ntest_labels_binary, threshold_function = predict_test_labels_binary(Y_train_pred, Y_train, Y_test_pred, t_range)\nce_FF_full_withThreshold = metrics.hamming_loss(Y_test, test_labels_binary)",
"_____no_output_____"
]
],
[
[
"### LSTM Reccurrent Network",
"_____no_output_____"
]
],
[
[
"## Load the pre-processed data\nfile_object = open('../RNN Text Categorization/RNN_data_dict.json',)\nRNN_data_dict = json.load(file_object)\nRNN_data_dict = ast.literal_eval(RNN_data_dict)\ntrain_padded = np.array(RNN_data_dict['train_padded'])\ntest_padded = np.array(RNN_data_dict['test_padded'])\nY_train = np.array(RNN_data_dict['Y_train'])\nY_test = np.array(RNN_data_dict['Y_test'])",
"_____no_output_____"
],
[
"## Define the LSTM RNN architecture\ntf.random.set_seed(123)\nnum_labels = 13\n\nmodel_LSTM_full = tf.keras.models.Sequential([\n tf.keras.layers.Embedding(num_unique_words, 32, input_length = max_length),\n tf.keras.layers.LSTM(16, return_sequences = False, return_state = False),\n #tf.keras.layers.Dropout(0.5),\n tf.keras.layers.Dense(num_labels, activation = 'sigmoid')\n])\n\noptim_func = tf.keras.optimizers.Adam(lr = 0.0001)\n\nmodel_LSTM_full.compile(loss = 'binary_crossentropy', optimizer = optim_func, metrics = metric)",
"_____no_output_____"
],
[
"tf.random.set_seed(123)\nhistory_ce_RNN_lr001_full = model_LSTM_full.fit(train_padded, Y_train, epochs = 100, \n validation_data = (test_padded, Y_test), verbose=2)",
"Epoch 1/100\n7/7 - 3s - loss: 0.6953 - hamming_loss: 0.5159 - val_loss: 0.6949 - val_hamming_loss: 0.5921\nEpoch 2/100\n7/7 - 0s - loss: 0.6944 - hamming_loss: 0.5948 - val_loss: 0.6940 - val_hamming_loss: 0.5921\nEpoch 3/100\n7/7 - 0s - loss: 0.6935 - hamming_loss: 0.5948 - val_loss: 0.6931 - val_hamming_loss: 0.5921\nEpoch 4/100\n7/7 - 0s - loss: 0.6926 - hamming_loss: 0.5956 - val_loss: 0.6922 - val_hamming_loss: 0.5585\nEpoch 5/100\n7/7 - 0s - loss: 0.6917 - hamming_loss: 0.5757 - val_loss: 0.6913 - val_hamming_loss: 0.5585\nEpoch 6/100\n7/7 - 0s - loss: 0.6908 - hamming_loss: 0.5757 - val_loss: 0.6904 - val_hamming_loss: 0.4960\nEpoch 7/100\n7/7 - 0s - loss: 0.6899 - hamming_loss: 0.4734 - val_loss: 0.6894 - val_hamming_loss: 0.4431\nEpoch 8/100\n7/7 - 0s - loss: 0.6889 - hamming_loss: 0.4187 - val_loss: 0.6885 - val_hamming_loss: 0.3421\nEpoch 9/100\n7/7 - 0s - loss: 0.6879 - hamming_loss: 0.3466 - val_loss: 0.6875 - val_hamming_loss: 0.3421\nEpoch 10/100\n7/7 - 0s - loss: 0.6870 - hamming_loss: 0.3466 - val_loss: 0.6864 - val_hamming_loss: 0.3421\nEpoch 11/100\n7/7 - 0s - loss: 0.6859 - hamming_loss: 0.3148 - val_loss: 0.6854 - val_hamming_loss: 0.2813\nEpoch 12/100\n7/7 - 0s - loss: 0.6848 - hamming_loss: 0.2879 - val_loss: 0.6843 - val_hamming_loss: 0.2813\nEpoch 13/100\n7/7 - 0s - loss: 0.6837 - hamming_loss: 0.2879 - val_loss: 0.6831 - val_hamming_loss: 0.2813\nEpoch 14/100\n7/7 - 0s - loss: 0.6825 - hamming_loss: 0.2879 - val_loss: 0.6818 - val_hamming_loss: 0.2813\nEpoch 15/100\n7/7 - 0s - loss: 0.6813 - hamming_loss: 0.2879 - val_loss: 0.6806 - val_hamming_loss: 0.2813\nEpoch 16/100\n7/7 - 0s - loss: 0.6800 - hamming_loss: 0.2879 - val_loss: 0.6792 - val_hamming_loss: 0.2813\nEpoch 17/100\n7/7 - 0s - loss: 0.6786 - hamming_loss: 0.2871 - val_loss: 0.6777 - val_hamming_loss: 0.2043\nEpoch 18/100\n7/7 - 0s - loss: 0.6770 - hamming_loss: 0.2109 - val_loss: 0.6760 - val_hamming_loss: 0.2043\nEpoch 19/100\n7/7 - 0s - loss: 0.6753 - hamming_loss: 0.2109 - val_loss: 0.6742 - val_hamming_loss: 0.2043\nEpoch 20/100\n7/7 - 0s - loss: 0.6735 - hamming_loss: 0.2109 - val_loss: 0.6722 - val_hamming_loss: 0.2043\nEpoch 21/100\n7/7 - 0s - loss: 0.6715 - hamming_loss: 0.2109 - val_loss: 0.6700 - val_hamming_loss: 0.2043\nEpoch 22/100\n7/7 - 0s - loss: 0.6691 - hamming_loss: 0.1907 - val_loss: 0.6675 - val_hamming_loss: 0.1723\nEpoch 23/100\n7/7 - 0s - loss: 0.6665 - hamming_loss: 0.1792 - val_loss: 0.6646 - val_hamming_loss: 0.1723\nEpoch 24/100\n7/7 - 0s - loss: 0.6635 - hamming_loss: 0.1792 - val_loss: 0.6614 - val_hamming_loss: 0.1723\nEpoch 25/100\n7/7 - 0s - loss: 0.6602 - hamming_loss: 0.1792 - val_loss: 0.6577 - val_hamming_loss: 0.1723\nEpoch 26/100\n7/7 - 0s - loss: 0.6564 - hamming_loss: 0.1792 - val_loss: 0.6535 - val_hamming_loss: 0.1723\nEpoch 27/100\n7/7 - 0s - loss: 0.6520 - hamming_loss: 0.1792 - val_loss: 0.6486 - val_hamming_loss: 0.1723\nEpoch 28/100\n7/7 - 0s - loss: 0.6468 - hamming_loss: 0.1792 - val_loss: 0.6430 - val_hamming_loss: 0.1723\nEpoch 29/100\n7/7 - 0s - loss: 0.6409 - hamming_loss: 0.1792 - val_loss: 0.6364 - val_hamming_loss: 0.1723\nEpoch 30/100\n7/7 - 0s - loss: 0.6342 - hamming_loss: 0.1792 - val_loss: 0.6289 - val_hamming_loss: 0.1723\nEpoch 31/100\n7/7 - 0s - loss: 0.6265 - hamming_loss: 0.1792 - val_loss: 0.6206 - val_hamming_loss: 0.1723\nEpoch 32/100\n7/7 - 0s - loss: 0.6181 - hamming_loss: 0.1792 - val_loss: 0.6117 - val_hamming_loss: 0.1723\nEpoch 33/100\n7/7 - 0s - loss: 0.6092 - hamming_loss: 0.1792 - val_loss: 0.6020 - val_hamming_loss: 0.1723\nEpoch 34/100\n7/7 - 0s - loss: 0.5998 - hamming_loss: 0.1792 - val_loss: 0.5925 - val_hamming_loss: 0.1723\nEpoch 35/100\n7/7 - 0s - loss: 0.5907 - hamming_loss: 0.1792 - val_loss: 0.5835 - val_hamming_loss: 0.1723\nEpoch 36/100\n7/7 - 0s - loss: 0.5823 - hamming_loss: 0.1792 - val_loss: 0.5756 - val_hamming_loss: 0.1723\nEpoch 37/100\n7/7 - 0s - loss: 0.5752 - hamming_loss: 0.1792 - val_loss: 0.5689 - val_hamming_loss: 0.1723\nEpoch 38/100\n7/7 - 0s - loss: 0.5693 - hamming_loss: 0.1792 - val_loss: 0.5634 - val_hamming_loss: 0.1723\nEpoch 39/100\n7/7 - 0s - loss: 0.5642 - hamming_loss: 0.1792 - val_loss: 0.5587 - val_hamming_loss: 0.1723\nEpoch 40/100\n7/7 - 0s - loss: 0.5599 - hamming_loss: 0.1792 - val_loss: 0.5546 - val_hamming_loss: 0.1723\nEpoch 41/100\n7/7 - 0s - loss: 0.5560 - hamming_loss: 0.1792 - val_loss: 0.5508 - val_hamming_loss: 0.1723\nEpoch 42/100\n7/7 - 0s - loss: 0.5523 - hamming_loss: 0.1792 - val_loss: 0.5472 - val_hamming_loss: 0.1723\nEpoch 43/100\n7/7 - 0s - loss: 0.5489 - hamming_loss: 0.1792 - val_loss: 0.5439 - val_hamming_loss: 0.1723\nEpoch 44/100\n7/7 - 0s - loss: 0.5458 - hamming_loss: 0.1792 - val_loss: 0.5408 - val_hamming_loss: 0.1723\nEpoch 45/100\n7/7 - 0s - loss: 0.5429 - hamming_loss: 0.1792 - val_loss: 0.5380 - val_hamming_loss: 0.1723\nEpoch 46/100\n7/7 - 0s - loss: 0.5401 - hamming_loss: 0.1792 - val_loss: 0.5353 - val_hamming_loss: 0.1723\nEpoch 47/100\n7/7 - 0s - loss: 0.5375 - hamming_loss: 0.1792 - val_loss: 0.5327 - val_hamming_loss: 0.1723\nEpoch 48/100\n7/7 - 0s - loss: 0.5349 - hamming_loss: 0.1792 - val_loss: 0.5301 - val_hamming_loss: 0.1723\nEpoch 49/100\n7/7 - 0s - loss: 0.5323 - hamming_loss: 0.1792 - val_loss: 0.5276 - val_hamming_loss: 0.1723\nEpoch 50/100\n7/7 - 0s - loss: 0.5298 - hamming_loss: 0.1792 - val_loss: 0.5250 - val_hamming_loss: 0.1723\nEpoch 51/100\n7/7 - 0s - loss: 0.5273 - hamming_loss: 0.1792 - val_loss: 0.5226 - val_hamming_loss: 0.1723\nEpoch 52/100\n7/7 - 0s - loss: 0.5249 - hamming_loss: 0.1792 - val_loss: 0.5203 - val_hamming_loss: 0.1723\nEpoch 53/100\n7/7 - 0s - loss: 0.5227 - hamming_loss: 0.1792 - val_loss: 0.5181 - val_hamming_loss: 0.1723\nEpoch 54/100\n7/7 - 0s - loss: 0.5206 - hamming_loss: 0.1792 - val_loss: 0.5159 - val_hamming_loss: 0.1723\nEpoch 55/100\n7/7 - 0s - loss: 0.5184 - hamming_loss: 0.1792 - val_loss: 0.5138 - val_hamming_loss: 0.1723\nEpoch 56/100\n7/7 - 0s - loss: 0.5164 - hamming_loss: 0.1792 - val_loss: 0.5119 - val_hamming_loss: 0.1723\nEpoch 57/100\n7/7 - 0s - loss: 0.5145 - hamming_loss: 0.1792 - val_loss: 0.5097 - val_hamming_loss: 0.1723\nEpoch 58/100\n7/7 - 0s - loss: 0.5124 - hamming_loss: 0.1792 - val_loss: 0.5077 - val_hamming_loss: 0.1723\nEpoch 59/100\n7/7 - 0s - loss: 0.5104 - hamming_loss: 0.1792 - val_loss: 0.5057 - val_hamming_loss: 0.1723\nEpoch 60/100\n7/7 - 0s - loss: 0.5085 - hamming_loss: 0.1792 - val_loss: 0.5038 - val_hamming_loss: 0.1723\nEpoch 61/100\n7/7 - 0s - loss: 0.5067 - hamming_loss: 0.1792 - val_loss: 0.5020 - val_hamming_loss: 0.1723\nEpoch 62/100\n7/7 - 0s - loss: 0.5049 - hamming_loss: 0.1792 - val_loss: 0.5002 - val_hamming_loss: 0.1723\nEpoch 63/100\n7/7 - 0s - loss: 0.5032 - hamming_loss: 0.1792 - val_loss: 0.4984 - val_hamming_loss: 0.1723\nEpoch 64/100\n7/7 - 0s - loss: 0.5014 - hamming_loss: 0.1792 - val_loss: 0.4967 - val_hamming_loss: 0.1723\nEpoch 65/100\n7/7 - 0s - loss: 0.4998 - hamming_loss: 0.1792 - val_loss: 0.4951 - val_hamming_loss: 0.1723\nEpoch 66/100\n7/7 - 0s - loss: 0.4981 - hamming_loss: 0.1792 - val_loss: 0.4934 - val_hamming_loss: 0.1723\nEpoch 67/100\n7/7 - 0s - loss: 0.4965 - hamming_loss: 0.1792 - val_loss: 0.4918 - val_hamming_loss: 0.1723\nEpoch 68/100\n7/7 - 0s - loss: 0.4949 - hamming_loss: 0.1792 - val_loss: 0.4902 - val_hamming_loss: 0.1723\nEpoch 69/100\n7/7 - 0s - loss: 0.4933 - hamming_loss: 0.1792 - val_loss: 0.4887 - val_hamming_loss: 0.1723\nEpoch 70/100\n7/7 - 0s - loss: 0.4918 - hamming_loss: 0.1792 - val_loss: 0.4872 - val_hamming_loss: 0.1723\nEpoch 71/100\n7/7 - 0s - loss: 0.4903 - hamming_loss: 0.1792 - val_loss: 0.4857 - val_hamming_loss: 0.1723\nEpoch 72/100\n7/7 - 0s - loss: 0.4888 - hamming_loss: 0.1792 - val_loss: 0.4842 - val_hamming_loss: 0.1723\nEpoch 73/100\n7/7 - 0s - loss: 0.4873 - hamming_loss: 0.1792 - val_loss: 0.4827 - val_hamming_loss: 0.1723\nEpoch 74/100\n7/7 - 0s - loss: 0.4858 - hamming_loss: 0.1792 - val_loss: 0.4812 - val_hamming_loss: 0.1723\nEpoch 75/100\n7/7 - 0s - loss: 0.4844 - hamming_loss: 0.1792 - val_loss: 0.4799 - val_hamming_loss: 0.1723\nEpoch 76/100\n7/7 - 0s - loss: 0.4831 - hamming_loss: 0.1792 - val_loss: 0.4785 - val_hamming_loss: 0.1723\nEpoch 77/100\n7/7 - 0s - loss: 0.4818 - hamming_loss: 0.1792 - val_loss: 0.4773 - val_hamming_loss: 0.1723\nEpoch 78/100\n7/7 - 0s - loss: 0.4806 - hamming_loss: 0.1792 - val_loss: 0.4761 - val_hamming_loss: 0.1723\nEpoch 79/100\n7/7 - 0s - loss: 0.4794 - hamming_loss: 0.1792 - val_loss: 0.4749 - val_hamming_loss: 0.1723\nEpoch 80/100\n7/7 - 0s - loss: 0.4782 - hamming_loss: 0.1792 - val_loss: 0.4738 - val_hamming_loss: 0.1723\nEpoch 81/100\n7/7 - 0s - loss: 0.4770 - hamming_loss: 0.1792 - val_loss: 0.4726 - val_hamming_loss: 0.1723\nEpoch 82/100\n7/7 - 0s - loss: 0.4759 - hamming_loss: 0.1792 - val_loss: 0.4715 - val_hamming_loss: 0.1723\nEpoch 83/100\n7/7 - 0s - loss: 0.4748 - hamming_loss: 0.1792 - val_loss: 0.4705 - val_hamming_loss: 0.1723\nEpoch 84/100\n7/7 - 0s - loss: 0.4736 - hamming_loss: 0.1792 - val_loss: 0.4694 - val_hamming_loss: 0.1723\nEpoch 85/100\n7/7 - 0s - loss: 0.4725 - hamming_loss: 0.1792 - val_loss: 0.4684 - val_hamming_loss: 0.1723\nEpoch 86/100\n7/7 - 0s - loss: 0.4714 - hamming_loss: 0.1792 - val_loss: 0.4673 - val_hamming_loss: 0.1723\nEpoch 87/100\n7/7 - 0s - loss: 0.4704 - hamming_loss: 0.1792 - val_loss: 0.4663 - val_hamming_loss: 0.1723\nEpoch 88/100\n7/7 - 0s - loss: 0.4693 - hamming_loss: 0.1792 - val_loss: 0.4653 - val_hamming_loss: 0.1723\nEpoch 89/100\n7/7 - 0s - loss: 0.4682 - hamming_loss: 0.1792 - val_loss: 0.4643 - val_hamming_loss: 0.1723\nEpoch 90/100\n7/7 - 0s - loss: 0.4672 - hamming_loss: 0.1792 - val_loss: 0.4633 - val_hamming_loss: 0.1723\nEpoch 91/100\n7/7 - 0s - loss: 0.4662 - hamming_loss: 0.1792 - val_loss: 0.4624 - val_hamming_loss: 0.1723\nEpoch 92/100\n7/7 - 0s - loss: 0.4652 - hamming_loss: 0.1792 - val_loss: 0.4615 - val_hamming_loss: 0.1723\nEpoch 93/100\n7/7 - 0s - loss: 0.4643 - hamming_loss: 0.1792 - val_loss: 0.4607 - val_hamming_loss: 0.1723\nEpoch 94/100\n7/7 - 0s - loss: 0.4633 - hamming_loss: 0.1792 - val_loss: 0.4598 - val_hamming_loss: 0.1723\nEpoch 95/100\n7/7 - 0s - loss: 0.4624 - hamming_loss: 0.1792 - val_loss: 0.4590 - val_hamming_loss: 0.1723\nEpoch 96/100\n7/7 - 0s - loss: 0.4615 - hamming_loss: 0.1792 - val_loss: 0.4581 - val_hamming_loss: 0.1723\nEpoch 97/100\n7/7 - 0s - loss: 0.4606 - hamming_loss: 0.1792 - val_loss: 0.4573 - val_hamming_loss: 0.1723\nEpoch 98/100\n7/7 - 0s - loss: 0.4598 - hamming_loss: 0.1792 - val_loss: 0.4565 - val_hamming_loss: 0.1723\nEpoch 99/100\n7/7 - 0s - loss: 0.4589 - hamming_loss: 0.1792 - val_loss: 0.4557 - val_hamming_loss: 0.1723\nEpoch 100/100\n7/7 - 0s - loss: 0.4581 - hamming_loss: 0.1792 - val_loss: 0.4550 - val_hamming_loss: 0.1723\n"
],
[
"## (CAUTION: DO NOT OVERWRITE EXISTING FILES) -- Convert training history to dataframe and write to a .json file\nhistory_ce_RNN_lr001_full_df = pd.DataFrame(history_ce_RNN_lr001_full.history)\n#with open(\"Full Data Eval Metrics/Cross Entropy RNN/history_ce_RNN_lr0001_full.json\", \"w\") as outfile: \n# history_ce_RNN_lr001_full_df.to_json(outfile)",
"_____no_output_____"
],
[
"## Learn a threshold function and save the test error for use in future DF\nY_train_pred = model_LSTM_full.predict(train_padded)\nY_test_pred = model_LSTM_full.predict(test_padded)\nt_range = (0, 1)\n\ntest_labels_binary, threshold_function = predict_test_labels_binary(Y_train_pred, Y_train, Y_test_pred, t_range)\nce_RNN_full_withThreshold = metrics.hamming_loss(Y_test, test_labels_binary)",
"_____no_output_____"
],
[
"ce_RNN_full_withThreshold",
"_____no_output_____"
],
[
"## (CAUTION: DO NOT OVERWRITE EXISTING FILES) -- Collect the test set hamming losses for the models \n## with learned threshold functions into a df and write to .json file\nval_hamming_loss_withThreshold_lr001_df = pd.DataFrame({'ce_FF_full_lr0001' : ce_FF_full_withThreshold,\n 'ce_RNN_full_lr0001' : ce_RNN_full_withThreshold},\n index = [0])\n\n#with open(\"Full Data Eval Metrics/val_hamming_loss_withThreshold_lr0001.json\", \"w\") as outfile: \n# val_hamming_loss_withThreshold_lr001_df.to_json(outfile)",
"_____no_output_____"
],
[
"val_hamming_loss_withThreshold_lr001_df",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e797886be63593eafc800b1f9fb6432d19dc01c3 | 2,188 | ipynb | Jupyter Notebook | evaluate-text-with-azure-cognitive-language-services/content-moderator.ipynb | zkan/azure-ai-engineer-associate-workshop | d80d8da33e65b3cf0bf3f1a1df2236652b14e4df | [
"MIT"
]
| null | null | null | evaluate-text-with-azure-cognitive-language-services/content-moderator.ipynb | zkan/azure-ai-engineer-associate-workshop | d80d8da33e65b3cf0bf3f1a1df2236652b14e4df | [
"MIT"
]
| null | null | null | evaluate-text-with-azure-cognitive-language-services/content-moderator.ipynb | zkan/azure-ai-engineer-associate-workshop | d80d8da33e65b3cf0bf3f1a1df2236652b14e4df | [
"MIT"
]
| 1 | 2020-06-28T09:06:54.000Z | 2020-06-28T09:06:54.000Z | 19.890909 | 127 | 0.537477 | [
[
[
"# Azure Content Moderator API",
"_____no_output_____"
],
[
"Reference: https://docs.microsoft.com/en-us/azure/cognitive-services/Content-Moderator/api-reference",
"_____no_output_____"
]
],
[
[
"import requests",
"_____no_output_____"
],
[
"subscripiton_key = 'YOUR_SUBSCRIPTION_KEY'\nendpoint = 'YOUR_ENDPOINT_URL'\nrequest_url = f'{endpoint}/contentmoderator/moderate/v1.0/ProcessText/Screen'",
"_____no_output_____"
],
[
"headers = {\n 'Content-Type': 'text/plain',\n 'Ocp-Apim-Subscription-Key': subscripiton_key,\n}",
"_____no_output_____"
],
[
"params = {\n 'classify': True,\n}",
"_____no_output_____"
],
[
"body = 'Is this a crap email [email protected], phone: 6657789887, IP: 255.255.255.255, 1 Microsoft Way, Redmond, WA 98052'",
"_____no_output_____"
],
[
"response = requests.post(request_url, data=body, headers=headers, params=params)",
"_____no_output_____"
],
[
"response.json()",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e797a19401bbf6f647b7f284dccc8fbeb5ecb8b8 | 26,303 | ipynb | Jupyter Notebook | ETL_Project_Completed.ipynb | NormanLo4319/ETL-Project | de792d98d112e51a55fa29a4654fb386c3b67b79 | [
"MIT"
]
| 1 | 2020-07-19T07:09:59.000Z | 2020-07-19T07:09:59.000Z | ETL_Project_Completed.ipynb | NormanLo4319/Restaurant-Health-Score-vs.-Yelp-Customer-Based-Scores-ETL-Project | de792d98d112e51a55fa29a4654fb386c3b67b79 | [
"MIT"
]
| null | null | null | ETL_Project_Completed.ipynb | NormanLo4319/Restaurant-Health-Score-vs.-Yelp-Customer-Based-Scores-ETL-Project | de792d98d112e51a55fa29a4654fb386c3b67b79 | [
"MIT"
]
| null | null | null | 34.884615 | 304 | 0.542143 | [
[
[
"# Data Analytic Boot Camp - ETL Project\n\n## How is the restaurant's inspection score compare to the Yelp customer review rating?\n\n#### We always rely on the application on our digital device to look for high rating restaurant. However,does the high rating restaurants (rank by customers) provide a clearn and healthy food environment for their customer? This project is trying to answer this question by building an ETL flow.\n\n#### Resources:\n#### - Restaurant Inspection Scores, San Francisco Department of Public Health\n#### (After cleaning the data, n = 54314)\n\n#### - Customers Based Rating Scores, Yelp API\n#### (After cleaning the data, n = 4049)\n\n#### Note: Yelp partnered with the local city government to develop the Local Inspector Value_Entry Specification (LIVES) system. However, the system is partnered with other local web developers, which has no link to Yelp database.",
"_____no_output_____"
]
],
[
[
"# Dependencies\nimport pandas as pd\nimport os\nimport csv\nimport requests\nimport json\nimport numpy as np\nfrom config_1 import ykey\n\n# Database Connection Dependencies\nimport sqlalchemy\nfrom sqlalchemy.ext.automap import automap_base\nfrom sqlalchemy.orm import Session\nfrom sqlalchemy import create_engine, inspect\nimport sqlite3\n\n# Import Matplot Lib\nimport matplotlib\nfrom matplotlib import style\nstyle.use('seaborn')\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"# Loading the CSV file\n\ncsvpath = os.path.join(\".\", \"Resources\", \"Restaurant_Scores_-_LIVES_Standard.csv\")\ninspection_scores = pd.read_csv(csvpath)",
"_____no_output_____"
],
[
"# Rename the headers of the dataframe for merging in the database\ninspection_scores = inspection_scores.rename(index=str, columns={\"business_name\":\"name\", \"business_address\":\"address\"})\ninspection_scores[\"zip\"] = inspection_scores[\"business_postal_code\"].astype(str)\ninspection_scores['phone'] = inspection_scores['business_phone_number'].astype(str)",
"_____no_output_____"
],
[
"# Count the unique value of business id in the data\ninspection_scores['business_id'].value_counts()\ninspection_scores['business_id'].nunique()",
"_____no_output_____"
],
[
"# Make the business name and address lower case for merging in the database\ninspection_scores['name'] = inspection_scores['name'].str.lower()\ninspection_scores['address'] = inspection_scores['address'].str.lower()\n\n# Modify the zip code and business phone number for referencing the business in the database\ninspection_scores[\"zip\"] = inspection_scores[\"zip\"].str[:5]\ninspection_scores['phone'] = inspection_scores['phone'].map(lambda x: str(x)[:-2])",
"_____no_output_____"
],
[
"# Drop the unnecessary data in the dataframe\ninspection_df = inspection_scores.drop(['business_postal_code', \n 'business_latitude', 'business_longitude',\n 'business_location', 'business_phone_number',\n 'Neighborhoods', 'Police Districts', 'Supervisor Districts',\n 'Fire Prevention Districts', 'Zip Codes', 'Analysis Neighborhoods'], axis=1)",
"_____no_output_____"
],
[
"# Check the dataframe after cleaning the data\nprint(len(inspection_df))\ninspection_df.head()",
"_____no_output_____"
],
[
"# Clearning the zip codes in the data frame and create a list for API request\nzip_codes = inspection_scores[\"zip\"].unique()\n\nzip_codes = zip_codes[zip_codes != \"CA\"]\nzip_codes = zip_codes[zip_codes != \"Ca\"]\nzip_codes = zip_codes[zip_codes != \"0\"]\nzip_codes = zip_codes[zip_codes != \"941\"]\n\nzip_codes = zip_codes.tolist()\ndel zip_codes[7]\n\nzip_codes = [int(i) for i in zip_codes]\n\nprint(zip_codes)",
"_____no_output_____"
],
[
"# Save the cleaned data frame as a csf file for later use\nyelp_df.to_csv(\"Resources/Restaurant_Scores_-_LIVES_Standard_Cleaned.csv\", index=False, header=True)",
"_____no_output_____"
]
],
[
[
"## Yelp API Request\n#### We tried two ways to extract data from Yelp API request,\n#### 1. Search by city location, San Francisco\n#### 2. Search by zip codes\n#### This project choose to use method 1 because method 2 create bunch of duplicates that is difficult to clean in the later time.",
"_____no_output_____"
]
],
[
[
"# Testing Yelp API request for extracting the business related data\n\n# Yelp API key is stored in ykey\nheaders = {\"Authorization\": \"bearer %s\" % ykey}\nendpoint = \"https://api.yelp.com/v3/businesses/search\"\nname = []\nrating = []\nreview_count = []\naddress = []\ncity = []\nstate = []\nzip_ = []\nphone = []\n\n\n \n# Define the parameters\nparams = {\"term\": \"restaurants\", \"location\": \"San Francisco\", \"radius\": 5000,\n \"categories\": \"food\", \"limit\": 50, \"offset\":0}\nprint(params)\n\nfor j in range(0, 50):\n\n try:\n# Make a request to the Yelp API\n response = requests.get(url = endpoint, params = params, headers = headers)\n data_response = response.json()\n\n# Add the total counts of fast food stores to \"total\"\n# print(json.dumps(data_response, indent=4, sort_keys=True))\n \n\n print(data_response[\"businesses\"][j][\"name\"])\n name.append(data_response[\"businesses\"][j][\"name\"])\n print(data_response[\"businesses\"][j][\"rating\"])\n rating.append(data_response[\"businesses\"][j][\"rating\"])\n print(data_response[\"businesses\"][j][\"review_count\"])\n review_count.append(data_response[\"businesses\"][j][\"review_count\"])\n print(data_response[\"businesses\"][j][\"location\"][\"address1\"])\n address.append(data_response[\"businesses\"][j][\"location\"][\"address1\"])\n print(data_response[\"businesses\"][j][\"location\"][\"city\"])\n city.append(data_response[\"businesses\"][j][\"location\"][\"city\"])\n print(data_response[\"businesses\"][j][\"location\"][\"state\"])\n state.append(data_response[\"businesses\"][j][\"location\"][\"state\"])\n print(data_response[\"businesses\"][j][\"location\"][\"zip_code\"])\n zip_.append(data_response[\"businesses\"][j][\"location\"][\"zip_code\"])\n print(data_response[\"businesses\"][j][\"phone\"])\n phone.append(data_response[\"businesses\"][j][\"phone\"])\n \n \n except KeyError:\n print(\"no restaurant found!\")",
"_____no_output_____"
],
[
"# Print out the responses\nprint(data_response['businesses'][1]['name'])\nprint(data_response['businesses'][1]['rating'])\nprint(data_response['businesses'][1]['price'])\nprint(data_response['businesses'][1]['location']['address1'])\nprint(data_response['businesses'][1]['location']['state'])\nprint(data_response['businesses'][1]['location']['city'])\nprint(data_response['businesses'][1]['location']['zip_code'])",
"_____no_output_____"
],
[
"# Extract data from Yelp API by location = San Francisco\n\n# Yelp API key is stored in ykey\nheaders = {\"Authorization\": \"bearer %s\" % ykey}\nendpoint = \"https://api.yelp.com/v3/businesses/search\"\nname = []\nrating = []\nreview_count = []\naddress = []\ncity = []\nstate = []\nzip_ = []\nphone = []\n\n# Sending 100 requests and each request will return 50 restaurants\nfor i in range(0, 100):\n \n for j in range(50):\n\n try:\n# Define the parameters\n params = {\"term\": \"restaurants\", \"location\": \"San Francisco\", \"radius\": 40000, \n \"categories\": \"food\", \"limit\": 50, \"offset\":(i*5)}\n print(params)\n\n\n# Make a request to the Yelp API\n response = requests.get(url = endpoint, params = params, headers = headers)\n data_response = response.json()\n\n# Add the total counts of fast food stores to \"total\"\n print(data_response[\"businesses\"][j][\"name\"])\n name.append(data_response[\"businesses\"][j][\"name\"])\n print(data_response[\"businesses\"][j][\"rating\"])\n rating.append(data_response[\"businesses\"][j][\"rating\"])\n print(data_response[\"businesses\"][j][\"review_count\"])\n review_count.append(data_response[\"businesses\"][j][\"review_count\"])\n print(data_response[\"businesses\"][j][\"location\"][\"address1\"])\n address.append(data_response[\"businesses\"][j][\"location\"][\"address1\"])\n print(data_response[\"businesses\"][j][\"location\"][\"city\"])\n city.append(data_response[\"businesses\"][j][\"location\"][\"city\"])\n print(data_response[\"businesses\"][j][\"location\"][\"state\"])\n state.append(data_response[\"businesses\"][j][\"location\"][\"state\"])\n print(data_response[\"businesses\"][j][\"location\"][\"zip_code\"])\n zip_.append(data_response[\"businesses\"][j][\"location\"][\"zip_code\"])\n print(data_response[\"businesses\"][j][\"phone\"])\n phone.append(data_response[\"businesses\"][j][\"phone\"])\n \n \n \n except KeyError:\n print(\"no restaurant found!\")\n \n# print(json.dumps(data, indent=4, sort_keys=True))",
"_____no_output_____"
],
[
"# Extract data from Yelp API by location = zip code\n\n# Yelp API key is stored in ykey\nheaders = {\"Authorization\": \"bearer %s\" % ykey}\nendpoint = \"https://api.yelp.com/v3/businesses/search\"\nname = []\nrating = []\nreview_count = []\naddress = []\ncity = []\nstate = []\nzip_ = []\nphone = []\n\nfor k in zip_codes:\n \n for i in range(0, 20):\n \n for j in range(50):\n\n try:\n# Define the parameters\n params = {\"term\": \"restaurants\", \"location\": k, \"radius\": 5000, \n \"categories\": \"food\", \"limit\": 50, \"offset\":(i*5)}\n print(params)\n\n\n# Make a request to the Yelp API\n response = requests.get(url = endpoint, params = params, headers = headers)\n data_response = response.json()\n\n# Add the total counts of fast food stores to \"total\"\n print(data_response[\"businesses\"][j][\"name\"])\n name.append(data_response[\"businesses\"][j][\"name\"])\n print(data_response[\"businesses\"][j][\"rating\"])\n rating.append(data_response[\"businesses\"][j][\"rating\"])\n print(data_response[\"businesses\"][j][\"review_count\"])\n review_count.append(data_response[\"businesses\"][j][\"review_count\"])\n print(data_response[\"businesses\"][j][\"location\"][\"address1\"])\n address.append(data_response[\"businesses\"][j][\"location\"][\"address1\"])\n print(data_response[\"businesses\"][j][\"location\"][\"city\"])\n city.append(data_response[\"businesses\"][j][\"location\"][\"city\"])\n print(data_response[\"businesses\"][j][\"location\"][\"state\"])\n state.append(data_response[\"businesses\"][j][\"location\"][\"state\"])\n print(data_response[\"businesses\"][j][\"location\"][\"zip_code\"])\n zip_.append(data_response[\"businesses\"][j][\"location\"][\"zip_code\"])\n print(data_response[\"businesses\"][j][\"phone\"])\n phone.append(data_response[\"businesses\"][j][\"phone\"])\n \n \n \n except KeyError:\n print(\"no restaurant found!\")\n \n# print(json.dumps(data, indent=4, sort_keys=True))",
"_____no_output_____"
],
[
"# Assign keys to the json data\nkeys = {\"name\":name, \"rating\":rating, \"reviews\":review_count,\n \"address\":address, \"city\":city, \"state\":state, \"zip_code\":zip_, \"phone\":phone}\n\nprint(len(name))\n\n# Create a data frame for the json data\nyelp_df = pd.DataFrame(keys)\nyelp_df.head()",
"_____no_output_____"
],
[
"# Save the Yelp API data into a csv file for future work\nyelp_df.to_csv(\"Resources/yelp_api.csv\", index=False, header=True)",
"_____no_output_____"
],
[
"# Cleaning the data for merging in the database\n\nyelp_df['zip_code'].dtype\n\nyelp_df['name'] = yelp_df['name'].str.lower()\nyelp_df['address'] = yelp_df['address'].str.lower()\n\nyelp_df['zip_code'] = yelp_df['zip_code'].astype(str)\nyelp_df['phone'] = yelp_df['phone'].astype(str)\n\nyelp_df['zip'] = yelp_df['zip_code'].map(lambda x: str(x)[:-2])\nyelp_df['phone'] = yelp_df['phone'].map(lambda x: str(x)[:-2])\n\nyelp_df = yelp_df.drop(['zip_code'], axis=1)\n\nyelp_df.head()\n\n# print(len(yelp_df))",
"_____no_output_____"
]
],
[
[
"## Storing the data to SQLite database:\n\n#### There are two ways to store the data into SQLite database,\n#### 1. Using pandas method \"dataframe.to_sql()\"\n#### 2. Create metadata base and append data from data frames to the specific tables in the database\n#### This project use the second method to append data because to_sql() method does not allow database to create primary key for the data.\n\n## Storing the data to MySQL:\n\n#### The data is also stored in MySQL database and the sql commands are saved in a separated file",
"_____no_output_____"
]
],
[
[
"# Import SQL Alchemy\nfrom sqlalchemy import create_engine\n\n# Import and establish Base for which classes will be constructed \nfrom sqlalchemy.ext.declarative import declarative_base\nBase = declarative_base()\n\n# Import modules to declare columns and column data types\nfrom sqlalchemy import Column, Integer, String, Float",
"_____no_output_____"
],
[
"# Loading the csv file back to pandas dataframe\ninspect_csvpath = os.path.join(\".\", \"Resources\", \"Restaurant_Scores_-_LIVES_Standard.csv\")\ninspection_scores = pd.read_csv(inspect_csvpath)\n\nyelp_csvpath = os.path.join(\".\", \"Resources\", \"yelp_api.csv\")\nyelp_0 = pd.read_csv(yelp_csvpath)",
"_____no_output_____"
],
[
"# Create the inspection class\nclass inspection(Base):\n __tablename__ = 'inspection'\n id = Column(Integer, primary_key=True)\n business_id = Column(Integer)\n name = Column(String(255))\n address = Column(String(255))\n business_city = Column(String(255))\n business_state = Column(String(255))\n inspection_id = Column(String(255))\n inspection_date = Column(String(255))\n inspection_score = Column(Float)\n inspection_type = Column(String(500))\n violation_id = Column(String(255))\n violation_description = Column(String(800))\n risk_category = Column(String(255))\n zip = Column(String(255))\n phone = Column(String(255))",
"_____no_output_____"
],
[
"# Create a connection to a SQLite database\nengine = create_engine('sqlite:///ELT_Project.db')",
"_____no_output_____"
],
[
"Base.metadata.create_all(engine)",
"_____no_output_____"
],
[
"# To push the objects made and query the server we use a Session object\nfrom sqlalchemy.orm import Session\nsession = Session(bind=engine)",
"_____no_output_____"
],
[
"# Appending the dataframe into database\n\nfor i in range(len(inspection_df['name'])):\n inspect = inspection(business_id = inspection_scores['business_id'][i],\n address = inspection_scores['address'][i],\n business_city = inspection_scores['business_city'][i],\n business_state = inspection_scores['business_state'][i],\n inspection_id = inspection_scores['inspection_id'][i],\n inspection_date = inspection_scores['inspection_date'][i],\n inspection_score = inspection_scores['inspection_score'][i],\n inspection_type = inspection_scores['inspection_type'][i],\n violation_id = inspection_scores['violation_id'][i],\n violation_description = inspection_scores['violation_description'][i],\n risk_category = inspection_scores['risk_category'][i],\n zip = inspection_scores['zip'][i],\n phone = inspection_scores['phone'][i])\n session.add(inspect)\n session.commit()",
"_____no_output_____"
],
[
"# Create the inspection class\nclass yelp(Base):\n __tablename__ = 'yelp'\n id = Column(Integer, primary_key=True)\n name = Column(String(255))\n rating = Column(Float)\n reviews = Column(Integer)\n address = Column(String(255))\n city = Column(String(255))\n state = Column(String(255))\n phone = Column(String(255))\n zip = Column(String(255))",
"_____no_output_____"
],
[
"# Appending the dataframe into database\n\nfor j in range(len(yelp_df['name'])):\n y = yelp(name = yelp_df['name'][j],\n rating = yelp_df['rating'][j],\n reviews = yelp_df['reviews'][j],\n address = yelp_df['address'][j],\n city = yelp_df['city'][j],\n state = yelp_df['state'][j],\n phone = yelp_df['phone'][j],\n zip = yelp_df['zip'][j])\n \n print(y)\n session.add(y)\n session.commit()",
"_____no_output_____"
],
[
"# Checking the data in the database engine\nengine.execute(\"SELECT * FROM inspection\").fetchall()",
"_____no_output_____"
],
[
"engine.execute(\"SELECT * FROM yelp\").fetchall()",
"_____no_output_____"
],
[
"# Checking the table names\ninspector = inspect(engine)\ninspector.get_table_names()",
"_____no_output_____"
],
[
"# checking the header names in the inspection scores table\ncolumns = inspector.get_columns('inspection')\nfor i in columns:\n print(i['name'], i['type'])",
"_____no_output_____"
],
[
"# Checking the header names in the yelp rating table\ncolumns = inspector.get_columns('yelp')\nfor j in columns:\n print(j[\"name\"], j[\"type\"])",
"_____no_output_____"
]
],
[
[
"## Analysis on restaurant inspection scores and customer-based rating\n\n#### Using matplotly for ploting the joined data.\n#### After joining the data, only 122 business can be matched by the business name and it's zip code\n",
"_____no_output_____"
]
],
[
[
"joined_df = pd.merge(inspection_df, yelp_df, on=['name', 'zip'])\n# joined_df.head(100)\n# print(len(joined_df))\njoined_df['name'].nunique()",
"_____no_output_____"
],
[
"# Cleaning the data in the joined data frame\njoined_df = joined_df.dropna(subset=['inspection_score'])\njoined_df = joined_df.drop_duplicates(subset='business_id', keep='first')\njoined_df.head(10)",
"_____no_output_____"
],
[
"# Plotting the inspection scores from the joined data frame\nax_1 = joined_df.plot(x='name', y='inspection_score', style='o',\n title=\"Inspection Scores\")\nfig_1 = ax_1.get_figure()\nfig_1.savefig('./Images/Inspection_Scores.png')",
"_____no_output_____"
],
[
"# Plotting the yelp rating from the joined data frame\nax_2 = joined_df.plot(x='name', y='rating', style='^',\n title=\"Yelp Rating\")\nfig_2 = ax_2.get_figure()\nfig_2.savefig('./Images/Yelp_Scores.png')",
"_____no_output_____"
],
[
"# Plotting the inspection scores and yelp rating\nplt.scatter(joined_df['rating'], joined_df['inspection_score'])\nplt.title(\"Inspection Scores Vs. Yelp Rating\")\nplt.xlabel(\"Yelp Rating (Scale: 0 - 5)\")\nplt.ylabel(\"Inspection Score\")\nplt.savefig('./Images/inspection_scores_vs_yelp_rating.png')",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
]
|
e797a986e507c462425ff85fd4e7fb2fdf357869 | 1,030,511 | ipynb | Jupyter Notebook | notebooks/modulo_01/aula_02_primeiras_visualizacoes_de_dados.ipynb | daviramalho/Bootcamp-DS2-Alura | 6fdd9c1a20516989d1b0f789c24a1d8b3b3f71d7 | [
"MIT"
]
| 1 | 2021-05-22T16:28:53.000Z | 2021-05-22T16:28:53.000Z | notebooks/modulo_01/aula_02_primeiras_visualizacoes_de_dados.ipynb | daviramalho/Bootcamp-DS2-Alura | 6fdd9c1a20516989d1b0f789c24a1d8b3b3f71d7 | [
"MIT"
]
| null | null | null | notebooks/modulo_01/aula_02_primeiras_visualizacoes_de_dados.ipynb | daviramalho/Bootcamp-DS2-Alura | 6fdd9c1a20516989d1b0f789c24a1d8b3b3f71d7 | [
"MIT"
]
| 1 | 2021-05-28T20:46:54.000Z | 2021-05-28T20:46:54.000Z | 304.344654 | 284,244 | 0.896411 | [
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"dados = pd.read_csv(\"../../data/modulo_01/A002654189_28_143_208.csv\", encoding = \"ISO-8859-1\",\n skiprows = 3, sep = \";\", skipfooter = 12,\n thousands = \".\", decimal = \",\")\ndados",
"<ipython-input-2-371dc2bffa1d>:1: ParserWarning: Falling back to the 'python' engine because the 'c' engine does not support skipfooter; you can avoid this warning by specifying engine='python'.\n dados = pd.read_csv(\"../../data/modulo_01/A002654189_28_143_208.csv\", encoding = \"ISO-8859-1\",\n"
],
[
"dados.head()",
"_____no_output_____"
],
[
"dados.tail()",
"_____no_output_____"
],
[
"dados.mean()",
"_____no_output_____"
],
[
"dados.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 27 entries, 0 to 26\nColumns: 287 entries, Unidade da Federação to Total\ndtypes: float64(165), object(122)\nmemory usage: 60.7+ KB\n"
],
[
"pd.options.display.float_format = \"{:.2f}\".format",
"_____no_output_____"
],
[
"dados.mean()",
"_____no_output_____"
],
[
"dados[\"2008/Ago\"]",
"_____no_output_____"
],
[
"dados[\"2008/Ago\"].mean()",
"_____no_output_____"
],
[
"dados.plot(x = \"Unidade da Federação\", y = \"2008/Ago\")",
"_____no_output_____"
],
[
"dados.plot(x = \"Unidade da Federação\", y = \"2008/Ago\", kind = \"bar\", figsize = (9, 6))",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nimport matplotlib.ticker as ticker\n\n\nax = dados.plot(x = \"Unidade da Federação\", y = \"2008/Ago\", kind = \"bar\", figsize = (9, 6))\nax.yaxis.set_major_formatter(ticker.StrMethodFormatter(\"{x:,.2f}\"))\nplt.title(\"Valor por unidade da Federação\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Desafio 1 - Escolher um título mais descritivo que passe a mensagem adequada",
"_____no_output_____"
]
],
[
[
"UF = \"Unidade da Federação\"\nAno_Mes = \"2008/Ago\"\n\nax = dados.plot(x = UF, y = Ano_Mes, kind = \"bar\", figsize = (9, 6))\nax.yaxis.set_major_formatter(ticker.StrMethodFormatter(\"{x:,.2f}\"))\nplt.title(\"Despesas em procedimentos hospitalares do SUS \\n {} - Processados em {}\".format(UF, Ano_Mes))\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Desafio 01.2 - Faça a mesma análise para o mês mais recente que você possui.",
"_____no_output_____"
]
],
[
[
"UF = \"Unidade da Federação\"\nAno_Mes = \"2021/Mar\"\n\nax = dados.plot(x = UF, y = Ano_Mes, kind = \"bar\", figsize = (9, 6))\nax.yaxis.set_major_formatter(ticker.StrMethodFormatter(\"{x:,.2f}\"))\nplt.title(\"Despesas em procedimentos hospitalares do SUS \\n {} - Processados em {}\".format(UF, Ano_Mes))\n#plt.savefig(\"../../reports/figures/modulo_01/desafio01_2.jpg\", dpi=150, bbox_inches=\"tight\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"## AULA 02 - Primeiras Visualizações de Dados",
"_____no_output_____"
]
],
[
[
"dados[[\"2008/Ago\", \"2008/Set\"]].head()",
"_____no_output_____"
],
[
"dados.mean()",
"_____no_output_____"
],
[
"colunas_usaveis = dados.mean().index.tolist()\ncolunas_usaveis.insert(0, \"Unidade da Federação\")\ncolunas_usaveis",
"_____no_output_____"
],
[
"usaveis = dados[colunas_usaveis]\nusaveis.head()",
"_____no_output_____"
],
[
"usaveis = usaveis.set_index(\"Unidade da Federação\")\nusaveis.head()",
"_____no_output_____"
],
[
"usaveis[\"2019/Ago\"].head()",
"_____no_output_____"
],
[
"usaveis.loc[\"12 Acre\"]",
"_____no_output_____"
],
[
"usaveis.plot(figsize = (12, 6))",
"_____no_output_____"
],
[
"usaveis.T.head()",
"_____no_output_____"
],
[
"usaveis.T.plot(figsize = (12,6))\nplt.show()",
"_____no_output_____"
],
[
"usaveis.T.tail()",
"_____no_output_____"
],
[
"usaveis = usaveis.drop(\"Total\", axis = 1)\nusaveis.head()",
"_____no_output_____"
],
[
"usaveis.T.plot(figsize = (12,6))\nplt.show()",
"_____no_output_____"
]
],
[
[
"### DESAFIO 02.1 - Reposicionar a legenda fora do gráfico",
"_____no_output_____"
]
],
[
[
"estados = \"Todos os Estados\"\nano_selecionado = \"2007/Ago a 2021/Mar\"\n\nax2 = usaveis.T.plot(figsize = (12,6))\nax2.legend(loc = 6, bbox_to_anchor = (1, 0.5))\nax2.yaxis.set_major_formatter(ticker.StrMethodFormatter(\"R$ {x:,.2f}\"))\nplt.title(\"Despesas em procedimentos hospitalares do SUS por local de internação \\n {} - Processados em {}\".format(estados, ano_selecionado))\n#plt.savefig(\"../../reports/figures/modulo_01/desafio02_1.jpg\", dpi=150, bbox_inches=\"tight\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"### DESAFIO 02.2 - Plotar o Gráfico de linha com apenas 5 estados de sua preferência",
"_____no_output_____"
]
],
[
[
"estados = \"PA, MG, CE, RS e SP\"\nano_selecionado = \"2007/Ago a 2021/Mar\"\n\nusaveis_selecionados = usaveis.loc[[\"15 Pará\", \"31 Minas Gerais\", \"23 Ceará\", \"43 Rio Grande do Sul\", \"35 São Paulo\"]]\nax3 = usaveis_selecionados.T.plot(figsize = (12,6))\nax3.legend(loc = 6, bbox_to_anchor = (1, 0.5))\nax3.yaxis.set_major_formatter(ticker.StrMethodFormatter(\"R$ {x:,.2f}\"))\nplt.title(\"Despesas em procedimentos hospitalares do SUS por local de internação \\n {} - Processados em {}\".format(estados, ano_selecionado))\n#plt.savefig(\"../../reports/figures/modulo_01/desafio02_2.jpg\", dpi=150, bbox_inches=\"tight\")\nplt.show()",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
e797b697150f294a70c2c30e668a692651703f89 | 35,491 | ipynb | Jupyter Notebook | 1-Benson/Project 1/Project 1 - Top Station Frequency - Sakura.ipynb | iamryanmurray/metis | 97621bd7b9f7ed88cd27df877050926aa6425823 | [
"Apache-2.0"
]
| null | null | null | 1-Benson/Project 1/Project 1 - Top Station Frequency - Sakura.ipynb | iamryanmurray/metis | 97621bd7b9f7ed88cd27df877050926aa6425823 | [
"Apache-2.0"
]
| 6 | 2021-02-02T22:56:07.000Z | 2022-03-12T00:41:48.000Z | 1-Benson/Project 1/Project 1 - Top Station Frequency - Sakura.ipynb | iamryanmurray/metis | 97621bd7b9f7ed88cd27df877050926aa6425823 | [
"Apache-2.0"
]
| null | null | null | 146.657025 | 29,416 | 0.862923 | [
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfilename = 'top100_station.csv'\ndata = pd.read_csv(filename)",
"_____no_output_____"
],
[
"data = data.head(15)",
"_____no_output_____"
],
[
"data",
"_____no_output_____"
],
[
"fig,ax = plt.subplots(figsize=(7,8))\n\nax = sns.barplot(data['frequncy'], data['station'], palette='rocket')\n\nplt.xlim(2, None)\n\nax.set(ylabel = 'Stations',\n\nxlabel=('Frequency'),\ntitle=('Top Stations that are busy at least 5 out of 7 days a week'))",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code"
]
]
|
e797bf57ca7dad81cf702b34a6f8eb45d40433ba | 11,127 | ipynb | Jupyter Notebook | Past/DSS/Programming/Scraping/180220_selenium.ipynb | Moons08/TIL | e257854e1f7b9af5a6e349f38037f3010c07310f | [
"MIT"
]
| null | null | null | Past/DSS/Programming/Scraping/180220_selenium.ipynb | Moons08/TIL | e257854e1f7b9af5a6e349f38037f3010c07310f | [
"MIT"
]
| null | null | null | Past/DSS/Programming/Scraping/180220_selenium.ipynb | Moons08/TIL | e257854e1f7b9af5a6e349f38037f3010c07310f | [
"MIT"
]
| null | null | null | 19.976661 | 94 | 0.506695 | [
[
[
"from selenium import webdriver\nimport time",
"_____no_output_____"
]
],
[
[
"---",
"_____no_output_____"
]
],
[
[
"md = webdriver.Chrome() # 오픈",
"_____no_output_____"
],
[
"md.get('https://cloud.google.com/vision/') # 해당 주소로 이동",
"_____no_output_____"
],
[
"md.set_window_size(900,700) # size setting",
"_____no_output_____"
],
[
"md.execute_script(\"window.scrollTo(0, 1000);\") # 브라우저 스크롤 이동",
"_____no_output_____"
]
],
[
[
"#### 현재 윈도우 위치 저장",
"_____no_output_____"
]
],
[
[
"main = md.current_window_handle",
"_____no_output_____"
]
],
[
[
"#### 새로운 탭 오픈 (포커스는 변경x)",
"_____no_output_____"
]
],
[
[
"md.execute_script(\"window.open('https://www.google.com');\")",
"_____no_output_____"
],
[
"windows = md.window_handles # 윈도우 체크\nwindows",
"_____no_output_____"
]
],
[
[
"#### switch_to_window : focus 변경",
"_____no_output_____"
]
],
[
[
"md.switch_to_window(windows[1])\nmd.get('https://www.naver.com')",
"_____no_output_____"
],
[
"md.switch_to_window(main)",
"_____no_output_____"
],
[
"md.execute_script('location.reload()') #새로고침",
"_____no_output_____"
]
],
[
[
"#### control alert",
"_____no_output_____"
]
],
[
[
"md.execute_script('alert(\"selenium test\")')\nalert = md.switch_to_alert()\nprint(alert.text)\nalert.accept()",
"selenium test\n"
],
[
"md.execute_script('alert(\"selenium test\")')\nmd.switch_to_alert().accept()",
"_____no_output_____"
],
[
"md.execute_script(\"confirm('confirm?')\")\n# alert = md.switch_to_alert() \nprint(alert.text)\n# alert.accept()\nalert.dismiss()",
"confirm?\n"
]
],
[
[
"#### input key & button",
"_____no_output_____"
]
],
[
[
"md.switch_to_window(windows[1])\nmd.find_element_by_css_selector('#query').send_keys('test')",
"_____no_output_____"
],
[
"md.find_element_by_css_selector(\".ico_search_submit\").click()",
"_____no_output_____"
]
],
[
[
"#### close driver",
"_____no_output_____"
]
],
[
[
"md.close() # one for one",
"_____no_output_____"
],
[
"for i in md.window_handles:\n md.switch_to_window(i)\n md.close()",
"_____no_output_____"
]
],
[
[
"---",
"_____no_output_____"
],
[
"### file upload\n\nhttps://visual-recognition-demo.ng.bluemix.net\n\nhttps://cloud.google.com/vision/",
"_____no_output_____"
]
],
[
[
"cr = webdriver.Chrome()\ncr.get('https://cloud.google.com/vision/')",
"_____no_output_____"
]
],
[
[
"iframe의 경우 포커스 이동이 필요함",
"_____no_output_____"
]
],
[
[
"iframe = cr.find_element_by_css_selector('#vision_demo_section > iframe ')\ncr.switch_to_frame(iframe)",
"_____no_output_____"
],
[
"# Switch back default content\n# cr.switch_to_default_content() #아이프레임 밖으로 포커스 이동",
"_____no_output_____"
],
[
"path = !pwd #현재 디렉토리위치 리스트\nprint(type(path), path)",
"<class 'IPython.utils.text.SList'> ['/home/mk/documents/dev/TIL/DSS/Scraping']\n"
],
[
"file_path = path[0] + \"/screenshot_element.png\"\ncr.find_element_by_css_selector('#input').send_keys(file_path) # 파입 업로드",
"_____no_output_____"
],
[
"cr.find_element_by_css_selector('#safeSearchAnnotation').click()",
"_____no_output_____"
]
],
[
[
"#### safe search 항목 점수 출력",
"_____no_output_____"
]
],
[
[
"a = cr.find_elements_by_css_selector('#card div.row.style-scope.vs-safe') \nfor i in a:\n print(i.text)",
"Adult Very Unlikely\nSpoof Unlikely\nMedical Very Unlikely\nViolence Very Unlikely\nRacy Very Unlikely\n"
]
],
[
[
"---",
"_____no_output_____"
],
[
"#### 한번에 실행",
"_____no_output_____"
]
],
[
[
"driver = webdriver.Chrome()\ndriver.get('https://cloud.google.com/vision/')\n\niframe = driver.find_element_by_css_selector(\"#vision_demo_section iframe\")\ndriver.switch_to_frame(iframe)\nfile_path = path[0] + \"/screenshot_element.png\"\ndriver.find_element_by_css_selector(\"#input\").send_keys(file_path)\ntime.sleep(15) # 이미지를 업로드하고 데이터를 분석하는 시간\ndriver.find_element_by_css_selector(\"#safeSearchAnnotation\").click()\na = driver.find_elements_by_css_selector('#card div.row.style-scope.vs-safe')\n\nfor i in a:\n print(i.text)\ndriver.close()",
"Adult Very Unlikely\nSpoof Unlikely\nMedical Very Unlikely\nViolence Very Unlikely\nRacy Very Unlikely\n"
]
],
[
[
"#### element 체크하면서 실행",
"_____no_output_____"
]
],
[
[
"def check_element(driver, selector):\n try:\n driver.find_element_by_css_selector(selector)\n return True\n except:\n return False",
"_____no_output_____"
],
[
"driver = webdriver.Chrome()\ndriver.get('https://cloud.google.com/vision/')\n\niframe = driver.find_element_by_css_selector(\"#vision_demo_section iframe\")\ndriver.switch_to_frame(iframe)\nfile_path = path[0] + \"/screenshot_element.png\"\ndriver.find_element_by_css_selector(\"#input\").send_keys(file_path)\n\nselector = '#card div.row.style-scope.vs-safe'\nsec, limit_sec = 0, 10\nwhile True:\n sec += 1\n print(\"{}sec\".format(sec))\n time.sleep(1)\n \n # element 확인\n if check_element(driver, selector):\n driver.find_element_by_css_selector(\"#safeSearchAnnotation\").click()\n a = driver.find_elements_by_css_selector('#card div.row.style-scope.vs-safe')\n for i in a:\n print(i.text)\n driver.close()\n break;\n \n # limit_sec가 넘어가면 에러 처리\n if sec + 1 > limit_sec:\n print(\"error\")\n driver.close()\n break;",
"1sec\n2sec\n3sec\n4sec\nAdult Very Unlikely\nSpoof Unlikely\nMedical Very Unlikely\nViolence Very Unlikely\nRacy Very Unlikely\n"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
e797c4ba6de36a29624ce2d16a6615c4cb83c914 | 239,066 | ipynb | Jupyter Notebook | .ipynb_checkpoints/Python Implementation-checkpoint.ipynb | DatenBiene/Vector_Quantile_Regression | 749bd33ecb529607c7c6b13d7269525f499d219b | [
"BSD-3-Clause"
]
| 2 | 2020-11-13T08:52:55.000Z | 2021-06-28T18:27:44.000Z | .ipynb_checkpoints/Python Implementation-checkpoint.ipynb | DatenBiene/Vector_Quantile_Regression | 749bd33ecb529607c7c6b13d7269525f499d219b | [
"BSD-3-Clause"
]
| null | null | null | .ipynb_checkpoints/Python Implementation-checkpoint.ipynb | DatenBiene/Vector_Quantile_Regression | 749bd33ecb529607c7c6b13d7269525f499d219b | [
"BSD-3-Clause"
]
| 2 | 2021-02-07T07:55:56.000Z | 2021-06-03T17:06:52.000Z | 197.738627 | 107,272 | 0.897225 | [
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport cvxpy as cp\nimport pandas as pd\n\n\ndef distmat(x, y):\n return np.sum(x**2, 0)[:, None] + np.sum(y**2, 0)[None, :] - 2*x.transpose().dot(y)\n\n\ndef normalize(a):\n return a/np.sum(a)\n\n\ndef add_intercept(dfX):\n dfX['intercept'] = 1\n l_col = list(dfX.columns)\n l_col.remove('intercept')\n return dfX[['intercept']+l_col]\n\n\nclass VectorQuantileRegression:\n\n def __init__(self):\n\n self.X = None\n self.Y = None\n self.U = None\n self.d = None\n self.m = None\n self.n = None\n self.q = None\n self.step = None\n self.df = None\n\n def get_U(self, d, step):\n\n if d > 6:\n print(\"Only d<=6 is yet supported\")\n return None\n\n elif d == 1:\n u = np.arange(0, 1+step, step).T\n\n elif d == 2:\n x = np.arange(0, 1+step, step)\n x, y = np.meshgrid(x, x)\n u = np.array([x.flatten(), y.flatten()]).T\n\n elif d == 3:\n x = np.arange(0, 1+step, step)\n x, y, z = np.meshgrid(x, x, x)\n u = np.array([x.flatten(), y.flatten(), z.flatten()]).T\n\n elif d == 4:\n x = np.arange(0, 1+step, step)\n x, y, z, x1 = np.meshgrid(x, x, x, x)\n u = np.array([x.flatten(), y.flatten(),\n z.flatten(), x1.flatten()]).T\n\n elif d == 5:\n x = np.arange(0, 1+step, step)\n x, y, z, x1, y1 = np.meshgrid(x, x, x, x, x)\n u = np.array([x.flatten(), y.flatten(),\n z.flatten(), x1.flatten(), y1.flatten()]).T\n\n elif d == 6:\n x = np.arange(0, 1+step, step)\n x, y, z, x1, y1, z1 = np.meshgrid(x, x, x, x, x, x)\n u = np.array([x.flatten(), y.flatten(), z.flatten(), x1.flatten(),\n y1.flatten(), z1.flatten()]).T\n return u\n\n def fit(self, X, Y, step=0.05, verbose=False):\n Y = Y.to_numpy().T\n X = add_intercept(X).to_numpy()\n self.X = X\n self.Y = Y\n\n self.q = X.shape[1]\n\n d = Y.shape[0]\n self.d = d\n self.step = step\n\n u = self.get_U(d, step)\n U = u.T\n self.U = U\n\n n = Y.shape[1]\n m = U.shape[1]\n\n self.n = n\n self.m = m\n\n nu = normalize(np.random.rand(n, 1))\n mu = normalize(np.random.rand(m, 1))\n\n C = distmat(U, Y)\n P = cp.Variable((m, n))\n ind_m = np.ones((m, 1))\n constraints = [0 <= P,\n cp.matmul(P.T, ind_m) == nu,\n cp.matmul(P, X) == cp.matmul(cp.matmul(mu, nu.T), X)]\n\n objective = cp.Minimize(cp.sum(cp.multiply(P, C)))\n prob = cp.Problem(objective, constraints)\n result = prob.solve(verbose=verbose)\n psi = constraints[1].dual_value\n b = constraints[2].dual_value\n\n self.result = result\n self.b = b\n self.psi = psi\n\n def get_dfU(self, U, b, step):\n u = U.T\n d = u.shape[1]\n dfU = pd.DataFrame(u)\n dim = [i for i in range(d)]\n self.dim = dim\n dfU[[str(i)+\"_follower\" for i in list(dfU.columns)]] = dfU[dfU.columns]\n\n for k in range(d):\n\n dfU_temp = dfU.copy()\n dfU_temp[k] = dfU_temp[k].apply(\n lambda x: x+step if x < 1 else x-step)\n\n find_in = list(dfU[dim].apply(\n lambda x: list(np.around(x, 3)), axis=1))\n dfU[str(k)+\"_follower\"] = dfU_temp[dim].apply(\n lambda x: list(np.around(x, 3)), axis=1\n ).apply(\n lambda x: find_in.index(x)\n )\n\n dfU['b'] = pd.DataFrame(b).apply(np.array, axis=1)\n\n for i in range(d):\n dfU['beta_'+str(i)] = (dfU.loc[list(dfU[str(i)+\"_follower\"])][['b']].reset_index(drop=True) - dfU[['b']])/step\n\n beta = ['beta_'+str(i) for i in range(2)]\n dfU['beta'] = dfU[beta].apply(lambda x: np.vstack(x), axis=1)\n\n return dfU\n\n def predict(self, X=None, u_quantile=None, argument=\"U\"):\n\n '''\n argument in {\"U\", \"X\"}\n u_quantile liste with quantiles\n '''\n U = self.U\n b = self.b\n step = self.step\n X = add_intercept(X)\n\n if argument == \"X\":\n\n if self.df is None:\n df = self.get_dfU(U, b, step)\n self.df = df\n else:\n df = self.df\n\n ser = pd.Series([u_quantile]*m)\n\n pos = df[self.dim].apply(lambda x: list(np.around(x, 3)), axis=1)\n beta = df['beta'][pos == ser].iloc[0]\n\n xeval = X.apply(lambda x: np.array(x).reshape(-1,1), axis=1).to_frame()\n xeval.columns = ['X']\n df_res = xeval.copy()\n\n if self.q == 1:\n df_res['y_pred'] = df_res['X'].apply(lambda x: beta*x)\n else:\n df_res['y_pred'] = df_res['X'].apply(lambda x: np.matmul(beta,x))\n\n return df_res\n\n elif argument == \"U\":\n if xeval.shape != (self.q,):\n print(\"If argument = U then you can only give one observation.\")\n return\n\n if self.df is None:\n df = self.get_dfU(U, b, step)\n self.df = df\n else:\n df = self.df\n\n df['y_pred'] = df['beta'].apply(lambda x: np.matmul(x, X))\n\n return df[self.dim + ['y_pred']]\n\n else:\n print(\"argument not recognized\")\n return None\n\n# def plot_surface():\n# \"une fonction pour ploter les surfaces\"\n\n# def plot_lines():\n",
"_____no_output_____"
],
[
"%%javascript\nIPython.OutputArea.prototype._should_scroll = function(lines) {\n return false;\n}",
"_____no_output_____"
],
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport cvxpy as cp\nimport cv2\nimport random",
"_____no_output_____"
],
[
"from pylab import rcParams\nrcParams['figure.figsize'] = 15, 10",
"_____no_output_____"
],
[
"X = pd.read_excel('Data/MVEngel.xls', 'X' ,header= None)\nY = pd.read_excel('Data/MVEngel.xls', 'Y' ,header= None).fillna(0)\n\nX.columns = ['income']\n\nY.columns = ['food',\n'clothing',\n'housing',\n'heating/lightening',\n'tools',\n'education',\n'public safety',\n'health',\n'services'\n]",
"_____no_output_____"
],
[
"X",
"_____no_output_____"
],
[
"Y = Y[['food',\n'clothing']]",
"_____no_output_____"
],
[
"Y.to_numpy().T.shape",
"_____no_output_____"
],
[
"clf = VectorQuantileRegression()",
"_____no_output_____"
],
[
"clf.fit(X,Y)",
"_____no_output_____"
],
[
"x_test = pd.DataFrame(np.arange(600,1400,100))",
"_____no_output_____"
],
[
"dr = clf.predict(X= x_test, u_quantile=[0.8,0.8], argument =\"X\")",
"_____no_output_____"
],
[
"dr['y_pred']#.iloc[0].shape",
"_____no_output_____"
],
[
"beta.shape",
"_____no_output_____"
],
[
"import pandas as pd\n\nX = pd.read_csv('Data/MVEngelX.csv', header= None)\nY = pd.read_csv('Data/MVEngelY.csv', header= None)\n\nX.columns = ['X']\nY.columns = ['Y1', 'Y2']\n\ndf = Y\ndf['Intercept'] =1\ndf['X'] = X\n#df['X2'] = X**2\n\ndf.head(5)",
"_____no_output_____"
],
[
"np.matmul(beta,df['X'].loc[0])",
"_____no_output_____"
],
[
"np.array([df['X'].loc[0]]).shape",
"_____no_output_____"
],
[
"df['X'].apply(lambda x: np.matmul(beta,np.array([x])))",
"_____no_output_____"
],
[
"X = df[['Intercept'\n , 'X'\n # , 'X2'\n ]].to_numpy()\n\nY = df[[\n 'Y1',\n 'Y2'\n ]].to_numpy()\n\nd = Y.shape[1]",
"_____no_output_____"
],
[
"Y.shape",
"_____no_output_____"
],
[
"def get_U(d, step):\n \n if d > 6 :\n print(\"Only d<=6 is yet supported\")\n return None\n \n elif d == 1 :\n u=np.arange(0, 1+step, step).T\n \n elif d ==2 :\n x = np.arange(0, 1+step, step)\n x,y = np.meshgrid(x,x)\n u=np.array([x.flatten(),y.flatten()]).T\n\n elif d ==3 :\n x = np.arange(0, 1+step, step)\n x,y,z = np.meshgrid(x,x,x)\n u=np.array([x.flatten(),y.flatten(), z.flatten()]).T\n\n elif d ==4 :\n x = np.arange(0, 1+step, step)\n x,y,z,x1 = np.meshgrid(x,x,x,x)\n u=np.array([x.flatten(),y.flatten(), z.flatten(),x1.flatten()]).T\n\n elif d == 5 :\n x = np.arange(0, 1+step, step)\n x,y,z,x1, y1 = np.meshgrid(x,x,x,x,x)\n u=np.array([x.flatten(),y.flatten(), z.flatten(), x1.flatten(), y1.flatten()]).T\n\n elif d == 6 :\n x = np.arange(0, 1+step, step)\n x,y,z,x1, y1, z1 = np.meshgrid(x,x,x,x,x,x)\n u=np.array([x.flatten(),y.flatten(), z.flatten(), x1.flatten(), y1.flatten(), z1.flatten()]).T\n \n return u",
"_____no_output_____"
],
[
"step = 0.05\nu = get_U(2, step)",
"_____no_output_____"
],
[
"U = u.T\nY = Y.T",
"_____no_output_____"
],
[
"U.shape , Y.shape, X.shape",
"_____no_output_____"
],
[
"n = Y.shape[1]\nm = U.shape[1]\n",
"_____no_output_____"
],
[
"normalize = lambda a: a/np.sum(a)\nnu = normalize(np.random.rand(n, 1))\nmu = normalize(np.random.rand(m, 1))",
"_____no_output_____"
],
[
"def distmat(x,y):\n return np.sum(x**2,0)[:,None] + np.sum(y**2,0)[None,:] - 2*x.transpose().dot(y)\nC = distmat(U,Y)",
"_____no_output_____"
],
[
"P = cp.Variable((m,n))\nind_n = np.ones((n,1))\nind_m = np.ones((m,1))",
"_____no_output_____"
],
[
"constraints = [0 <= P, cp.matmul(P.T,ind_m)==nu, cp.matmul(P,X)==cp.matmul(cp.matmul(mu,nu.T),X)]",
"_____no_output_____"
],
[
"%%time\nobjective = cp.Minimize(cp.sum(cp.multiply(P,C)))\nprob = cp.Problem(objective, constraints)\nresult = prob.solve()",
"Wall time: 1.31 s\n"
],
[
"print(\"Number of non-zero: %d (n + m-1 = %d)\" %(len(P.value[P.value>1e-5]), n + m-1))",
"Number of non-zero: 4535 (n + m-1 = 675)\n"
],
[
"psi = constraints[1].dual_value\nb = constraints[2].dual_value",
"_____no_output_____"
]
],
[
[
"# Precious function",
"_____no_output_____"
]
],
[
[
"U.shape",
"_____no_output_____"
],
[
"def get_dfU(U, b,step):\n d = U.T.shape[1]\n dfU = pd.DataFrame(u)\n dim = [i for i in range(d)]\n dfU[[str(i)+\"_follower\" for i in list(dfU.columns)]] = dfU[dfU.columns]\n\n for k in range(d):\n \n dfU_temp = dfU.copy()\n dfU_temp[k] = dfU_temp[k].apply(lambda x: x+step if x<1 else x-step )\n \n find_in = list(dfU[dim].apply(lambda x: list(np.around(x, 3)),axis=1))\n \n dfU[str(k)+\"_follower\"] = dfU_temp[dim].apply(lambda x: list(np.around(x, 3)), axis =1).apply(lambda x: find_in.index(x))\n \n dfU['b']=pd.DataFrame(b).apply(np.array,axis=1)\n \n for i in range(d):\n dfU['beta_'+str(i)]=(dfU.loc[list(dfU[str(i)+\"_follower\"])][['b']].reset_index(drop=True) - dfU[['b']])/step\n \n beta = ['beta_'+str(i) for i in range(2)]\n dfU['beta'] = dfU[beta].apply(lambda x : np.vstack(x), axis = 1)\n \n return dfU",
"_____no_output_____"
],
[
"xeval = np.array([1,\n 883.99,\n #883.99**2\n ])\nxeval.shape",
"_____no_output_____"
],
[
"xeval.shape ==(d,)",
"_____no_output_____"
],
[
"df = get_dfU(U, b,step)",
"_____no_output_____"
],
[
"y_hat = df['beta'].apply(lambda x : np.matmul(x, xeval))",
"_____no_output_____"
],
[
"y_1_hat = y_hat.apply(lambda x: x[0])\ny_2_hat = y_hat.apply(lambda x: x[1])\ny_1_hat = np.abs(y_1_hat)\ny_2_hat = np.abs(y_2_hat)",
"_____no_output_____"
],
[
"np.mean(y_1_hat + y_2_hat)",
"_____no_output_____"
],
[
"from mpl_toolkits.mplot3d import Axes3D\nimport matplotlib.pyplot as plt\nfrom matplotlib import cm\nfrom matplotlib.ticker import LinearLocator, FormatStrFormatter\nimport numpy as np\n\ng = int(np.sqrt(df.shape[0]))\n\nfig = plt.figure()\nax = fig.gca(projection='3d')\n\nx = np.reshape(df[0].ravel(), (g, g))\ny = np.reshape(df[1].ravel(), (g, g))\nz = np.reshape(y_1_hat.ravel(), (g, g))\n\nsurf = ax.plot_surface(-x, y, z, cmap=cm.coolwarm,\n linewidth=0, antialiased=False)\n\n\n \nplt.show()",
"_____no_output_____"
],
[
"from mpl_toolkits.mplot3d import Axes3D\nimport matplotlib.pyplot as plt\nfrom matplotlib import cm\nfrom matplotlib.ticker import LinearLocator, FormatStrFormatter\nimport numpy as np\n\ng = int(np.sqrt(df.shape[0]))\n\nfig = plt.figure()\nax = fig.gca(projection='3d')\n\nx = np.reshape(df[0].ravel(), (g, g))\ny = np.reshape(df[1].ravel(), (g, g))\nz = np.reshape(y_2_hat.ravel(), (g, g))\n\nsurf = ax.plot_surface(-x, y, z, cmap=cm.coolwarm,\n linewidth=0, antialiased=False)\n\n\nplt.show()",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e797c6fbee9886ab783b092a4eec72a74b1b66d4 | 266,181 | ipynb | Jupyter Notebook | Week7/july19_hierarchial_clustering.ipynb | Anantha-Rao12/NMR-quantumstates-GSOC21 | 33dbdb4b32ad4f25592e180c426c484984fad763 | [
"MIT"
]
| 1 | 2021-07-29T15:38:40.000Z | 2021-07-29T15:38:40.000Z | Week7/july19_hierarchial_clustering.ipynb | Anantha-Rao12/NMR-quantumstates-GSOC21 | 33dbdb4b32ad4f25592e180c426c484984fad763 | [
"MIT"
]
| null | null | null | Week7/july19_hierarchial_clustering.ipynb | Anantha-Rao12/NMR-quantumstates-GSOC21 | 33dbdb4b32ad4f25592e180c426c484984fad763 | [
"MIT"
]
| null | null | null | 266,181 | 266,181 | 0.915336 | [
[
[
"",
"_____no_output_____"
],
[
"import os, sys\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns ; sns.set()\n\nfrom google.colab import drive\ndrive.mount('/content/drive')\n\nsys.path.append(\"/content/drive/MyDrive/GSOC-NMR-project/Work/Notebooks\")\nfrom auxillary_functions import *\nfrom polynomial_featextract import poly_featextract",
"Mounted at /content/drive\n"
],
[
"# import raw data and params.txt file\ndatadir_path = \"/content/drive/MyDrive/GSOC-NMR-project/Work/Data/2021-06-21_classify_datagen_all_funcs\"\n\nrawdata = load_data(datadir_path)\nparams = load_params(datadir_path)\nker_integrals = load_wlist(datadir_path) # load wlist.txt file\n\n# Stencil type : {'0' : 'Gaussian', '1' : 'Power Law', '2' : 'RKKY'}",
"Finished loading rawdata into numpy array\nFinsihed loading parameters file\nfinished loading kernel-integrals file.\n"
],
[
"print(rawdata.shape)\noffset = 150\nshifted_data, center = get_window(rawdata,2/3,width=offset)\nprint(\"The Echo pulse occurs at timestep:\",center)\n\n# Rescaled data\nrscl_data = shifted_data / np.max(shifted_data,axis=1,keepdims=True)",
"(10500, 943)\nThe Echo pulse occurs at timestep: 628\n"
],
[
"y_classes = get_yclasses(params, ker_integrals)",
"_____no_output_____"
]
],
[
[
"## Building a RF model for $\\alpha _x$",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\nfrom sklearn.ensemble import RandomForestRegressor",
"_____no_output_____"
],
[
"rscl_df = pd.DataFrame(rscl_data, columns=[f\"Feat_{i}\" for i in range(300)])\nrscl_df.head()",
"_____no_output_____"
],
[
"model = RandomForestRegressor(n_estimators=40, max_features='sqrt', min_samples_split=5, n_jobs=-1)",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(rscl_df, y_classes['αx'], test_size=0.2, \n random_state=2, stratify = params['stencil_type'])",
"_____no_output_____"
],
[
"model.fit(X_train, y_train)",
"_____no_output_____"
],
[
"model.score(X_test ,y_test)",
"_____no_output_____"
],
[
"fi = pd.DataFrame(np.array([rscl_df.columns,model.feature_importances_]).T, \n columns=['timestamp','fi'])\n\nfi.sort_values('fi', ascending=False, inplace=True)",
"_____no_output_____"
],
[
"fi.head(10)",
"_____no_output_____"
],
[
"fi.plot('timestamp', 'fi', figsize=(10,6), legend=False);",
"_____no_output_____"
],
[
"def plot_fi(fi): return fi.plot('timestamp', 'fi', 'barh', legend=False)",
"_____no_output_____"
],
[
"plot_fi(fi[:60]);",
"_____no_output_____"
],
[
"to_keep = fi[fi.fi>0.005].timestamp; len(to_keep)",
"_____no_output_____"
],
[
"df_keep = rscl_df[to_keep].copy()\n\nX_train, X_test, y_train, y_test = train_test_split(df_keep, y_classes['αx'], test_size=0.2, \n random_state=2, stratify = params['stencil_type'])",
"_____no_output_____"
],
[
"model2 = RandomForestRegressor(n_estimators=40, max_features=\"sqrt\",\n n_jobs=-1, oob_score=True)\nmodel2.fit(X_train, y_train)",
"_____no_output_____"
],
[
"model2.score(X_test, y_test)",
"_____no_output_____"
],
[
"fi2 = pd.DataFrame(np.array([df_keep.columns, model2.feature_importances_]).T, \n columns=['timestamp','fi'])\n\nfi2.sort_values('fi', ascending=False, inplace=True)",
"_____no_output_____"
],
[
"fi2.plot('timestamp', 'fi', figsize=(10,6))",
"_____no_output_____"
],
[
"fig,ax = plt.subplots(figsize=(16,14))\nfi2.plot('timestamp', 'fi', 'barh', legend=False, ax=ax)",
"_____no_output_____"
]
],
[
[
"# Removing redundant features",
"_____no_output_____"
]
],
[
[
"import scipy \nfrom scipy.cluster import hierarchy as hc",
"_____no_output_____"
],
[
"corr = np.round(scipy.stats.spearmanr(df_keep).correlation, 4)\ncorr_condensed = hc.distance.squareform(1-corr)\nz = hc.linkage(corr_condensed, method='average')\n\n\nfig = plt.figure(figsize=(18,14))\ndendrogram = hc.dendrogram(z, labels=df_keep.columns, orientation='left', leaf_font_size=16)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Let's try removing some of these related features to see if the model can be simplified without impacting the accuracy.",
"_____no_output_____"
]
],
[
[
"def get_oob(df):\n m = RandomForestRegressor(n_estimators=30, min_samples_leaf=5, max_features=0.6, n_jobs=-1, oob_score=True)\n x, _ = split_vals(df, n_trn)\n m.fit(x, y_train)\n return m.oob_score_",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"!pip install pdpbox",
"Collecting pdpbox\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/d5/6b/8b214723ff134f6b3f69a77c5bbd9d80db86ce01d3a970a5dbbab45c57fc/PDPbox-0.2.1.tar.gz (34.0MB)\n\u001b[K |████████████████████████████████| 34.0MB 94kB/s \n\u001b[?25hRequirement already satisfied: pandas in /usr/local/lib/python3.7/dist-packages (from pdpbox) (1.1.5)\nRequirement already satisfied: numpy in /usr/local/lib/python3.7/dist-packages (from pdpbox) (1.19.5)\nRequirement already satisfied: scipy in /usr/local/lib/python3.7/dist-packages (from pdpbox) (1.4.1)\nRequirement already satisfied: joblib in /usr/local/lib/python3.7/dist-packages (from pdpbox) (1.0.1)\nRequirement already satisfied: psutil in /usr/local/lib/python3.7/dist-packages (from pdpbox) (5.4.8)\nCollecting matplotlib==3.1.1\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/19/7a/60bd79c5d79559150f8bba866dd7d434f0a170312e4d15e8aefa5faba294/matplotlib-3.1.1-cp37-cp37m-manylinux1_x86_64.whl (13.1MB)\n\u001b[K |████████████████████████████████| 13.1MB 238kB/s \n\u001b[?25hRequirement already satisfied: sklearn in /usr/local/lib/python3.7/dist-packages (from pdpbox) (0.0)\nRequirement already satisfied: pytz>=2017.2 in /usr/local/lib/python3.7/dist-packages (from pandas->pdpbox) (2018.9)\nRequirement already satisfied: python-dateutil>=2.7.3 in /usr/local/lib/python3.7/dist-packages (from pandas->pdpbox) (2.8.1)\nRequirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.7/dist-packages (from matplotlib==3.1.1->pdpbox) (0.10.0)\nRequirement already satisfied: kiwisolver>=1.0.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib==3.1.1->pdpbox) (1.3.1)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib==3.1.1->pdpbox) (2.4.7)\nRequirement already satisfied: scikit-learn in /usr/local/lib/python3.7/dist-packages (from sklearn->pdpbox) (0.22.2.post1)\nRequirement already satisfied: six>=1.5 in /usr/local/lib/python3.7/dist-packages (from python-dateutil>=2.7.3->pandas->pdpbox) (1.15.0)\nBuilding wheels for collected packages: pdpbox\n Building wheel for pdpbox (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for pdpbox: filename=PDPbox-0.2.1-cp37-none-any.whl size=35758227 sha256=a78ff86505cf6312b219f2a8ab84e13f965274ef73c438341deb9cbe198318bb\n Stored in directory: /root/.cache/pip/wheels/c8/13/11/ecac74c6192790c94f6f5d919f1a388f927b9febb690e45eef\nSuccessfully built pdpbox\n\u001b[31mERROR: albumentations 0.1.12 has requirement imgaug<0.2.7,>=0.2.5, but you'll have imgaug 0.2.9 which is incompatible.\u001b[0m\nInstalling collected packages: matplotlib, pdpbox\n Found existing installation: matplotlib 3.2.2\n Uninstalling matplotlib-3.2.2:\n Successfully uninstalled matplotlib-3.2.2\nSuccessfully installed matplotlib-3.1.1 pdpbox-0.2.1\n"
],
[
"from pdpbox import pdp\nfrom plotnine import *",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e797c9c8980635f6b573761982bfe98f05b4745b | 90,628 | ipynb | Jupyter Notebook | code/truncate_shipboard_adcp.ipynb | jtomfarrar/S-MODE_analysis | 9e2c8ed982febe4f26d6e6fd72910bd869f7c815 | [
"MIT"
]
| null | null | null | code/truncate_shipboard_adcp.ipynb | jtomfarrar/S-MODE_analysis | 9e2c8ed982febe4f26d6e6fd72910bd869f7c815 | [
"MIT"
]
| null | null | null | code/truncate_shipboard_adcp.ipynb | jtomfarrar/S-MODE_analysis | 9e2c8ed982febe4f26d6e6fd72910bd869f7c815 | [
"MIT"
]
| null | null | null | 69.875096 | 10,548 | 0.609094 | [
[
[
"## Examine sample file from shipboard real-time processed ADCP\n\nThis is a pre-cruise examination of the data file to see how to truncate it for sending to shore during the cruise.",
"_____no_output_____"
]
],
[
[
"import xarray as xr\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport matplotlib\nimport cftime\n",
"_____no_output_____"
],
[
"datapath = '../data/raw/shipboard_adcp_initial_look/'\nfile = datapath + 'wh300.nc' \n\nds = xr.open_dataset(file,drop_variables=['amp','pg','pflag','num_pings','tr_temp'])\n",
"_____no_output_____"
],
[
"ds",
"_____no_output_____"
],
[
"ds2=ds.sel(time=slice(\"2021-09-06\", \"2021-09-07\"))",
"_____no_output_____"
],
[
"ds2",
"_____no_output_____"
],
[
"ds2.to_netcdf(datapath + 'wh300_last_day.nc')",
"_____no_output_____"
],
[
"fig = plt.figure()\nplt.contourf(ds2.u)\n# plt.plot(ds2.uship)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e797ca4b469dba2a55facc7269fb92c77cec6994 | 32,683 | ipynb | Jupyter Notebook | empir19nrm02/Jupyter/IBudgetMETAS.ipynb | AndersThorseth/empir19nrm02 | 0af5f0d4e412980c2ba5eb291f1aa95805db00c0 | [
"CC0-1.0"
]
| null | null | null | empir19nrm02/Jupyter/IBudgetMETAS.ipynb | AndersThorseth/empir19nrm02 | 0af5f0d4e412980c2ba5eb291f1aa95805db00c0 | [
"CC0-1.0"
]
| null | null | null | empir19nrm02/Jupyter/IBudgetMETAS.ipynb | AndersThorseth/empir19nrm02 | 0af5f0d4e412980c2ba5eb291f1aa95805db00c0 | [
"CC0-1.0"
]
| null | null | null | 107.157377 | 14,643 | 0.880366 | [
[
[
"# METAS uncLib https://www.metas.ch/metas/en/home/fabe/hochfrequenz/unclib.html",
"_____no_output_____"
]
],
[
[
"from metas_unclib import *\nimport matplotlib.pyplot as plt\nfrom sigfig import round\n%matplotlib inline",
"_____no_output_____"
],
[
"use_mcprop(n=100000)\n#use_linprop()\n\ndef uncLib_PlotHist(mcValue, xLabel='Value / A.U.', yLabel='Probability', title='Histogram of value', bins=1001, coverage=0.95):\n hObject = mcValue.net_object \n hValues = [float(bi) for bi in hObject.values]\n y,x,_ = plt.hist(hValues, bins=bins, density=True)\n plt.xlabel(xLabel)\n plt.title(title)\n plt.ylabel(yLabel)\n \n # stat over all\n coverage_interval=[np.mean(hValues), np.percentile(hValues, ((1.0-coverage)/2.0) * 100), np.percentile(hValues, (coverage+((1.0-coverage)/2.0)) * 100)]\n plt.axvline( coverage_interval[0])\n plt.axvline( coverage_interval[1])\n plt.axvline( coverage_interval[2])\n outString = round(str(coverage_interval[0]), uncertainty=str((coverage_interval[2]-coverage_interval[1])/2))\n plt.text( coverage_interval[2], max(y)/2, outString)\n plt.show()\n return [[y,x], coverage_interval]",
"_____no_output_____"
]
],
[
[
"# Measurement Uncertainty Simplest Possible Example",
"_____no_output_____"
],
[
"## Define the parameter for the calibration factor",
"_____no_output_____"
]
],
[
[
"k_e = ufloat(0.01, 0.0000045)",
"_____no_output_____"
],
[
"k_e",
"_____no_output_____"
]
],
[
[
"## Define the parameter for the photometer reading",
"_____no_output_____"
]
],
[
[
"Y_e = ufloat(2673.3,1.)",
"_____no_output_____"
],
[
"Y_e",
"_____no_output_____"
]
],
[
[
"## Define the parameter for the distance measurement",
"_____no_output_____"
]
],
[
[
"d=ufloat(25.0000, 0.0025)",
"_____no_output_____"
],
[
"d",
"_____no_output_____"
]
],
[
[
"# The Model",
"_____no_output_____"
]
],
[
[
"I=k_e*Y_e*d**2",
"_____no_output_____"
],
[
"I",
"_____no_output_____"
],
[
"[h, result_vecotr] = uncLib_PlotHist(I, xLabel='Luminous intensity / cd')\nprint('Mean: {}, I0: {}, I1: {}'.format(result_vecotr[0], result_vecotr[1], result_vecotr[2]))\n",
"_____no_output_____"
],
[
"h=uncLib_PlotHist(k_e, xLabel='calibration factor / lx/LSB')",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
]
|
e797d3416d4b81ec470f0d100106fe1c5ba66624 | 83,166 | ipynb | Jupyter Notebook | world_series_prediction-master/ML/Logistic--2017.ipynb | kchhajed1/baseball_predictions | 5aea42659c72c04165758176d995d9f144ad49a6 | [
"MIT"
]
| 2 | 2018-12-12T00:38:22.000Z | 2019-01-25T02:01:06.000Z | world_series_prediction-master/ML/Logistic--2017.ipynb | kchhajed1/baseball_predictions | 5aea42659c72c04165758176d995d9f144ad49a6 | [
"MIT"
]
| 2 | 2018-10-27T18:01:30.000Z | 2018-10-27T18:02:15.000Z | ML/Logistic--2017.ipynb | knishina/World_Series_Prediction | a3d81bdd24fc26bafbdbe83318168d8b7f83ce2d | [
"MIT"
]
| null | null | null | 29.04855 | 405 | 0.292463 | [
[
[
"# Prediction of 2017 World Series winner given data 1905 dataset.",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"tot1905 = pd.read_csv(\"../clean_data/1905ML.csv\")\ntot1905 = tot1905.drop({\"Unnamed: 0\", \"H\", \"HR\", \"BB\", \"SB\", \"HA\", \"HRA\", \"BBA\", \"SOA\", \"E\"}, axis=1)\ntot1905",
"_____no_output_____"
],
[
"tot2017 = pd.read_csv(\"../clean_data/2017ML.csv\")\ntot2017 = tot2017.drop({\"Unnamed: 0\", \"WSWIN\"}, axis=1)\ntot2017",
"_____no_output_____"
],
[
"# Create a function to convert the bats and throws colums to numeric\ndef bats_throws(col):\n if col == \"Y\":\n return 1\n else:\n return 0\n\n# Use the `apply()` method to create numeric columns from the bats and throws columns\ntot1905['WSWin'] = tot1905['WSWin'].apply(bats_throws)\n\n# Print out the first rows of `master_df`\ntot1905",
"_____no_output_____"
],
[
"features = tot1905.drop({\"franchID\", \"WSWin\", \"yearID\"}, axis=1)\nfeatures",
"_____no_output_____"
],
[
"features2017 = tot2017.drop({\"franchID\"}, axis=1)\nfeatures2017",
"_____no_output_____"
],
[
"# Create `target` Series\ntarget = tot1905['WSWin']\ntarget",
"_____no_output_____"
],
[
"# Import cross_val_predict, KFold and LogisticRegression from 'sklearn'\nfrom sklearn.cross_validation import cross_val_predict, KFold\nfrom sklearn.linear_model import LogisticRegression\n\n# Create Logistic Regression model\nlr = LogisticRegression(class_weight='balanced')\n\n# Create an instance of the KFold class\nkf = KFold(features.shape[0], random_state=1)\n\n# Create predictions using cross validation\npredictions_lr = cross_val_predict(lr, features, target, cv=kf)",
"/Applications/anaconda3/envs/PythonData/lib/python3.6/site-packages/sklearn/cross_validation.py:41: DeprecationWarning: This module was deprecated in version 0.18 in favor of the model_selection module into which all the refactored classes and functions are moved. Also note that the interface of the new CV iterators are different from that of this module. This module will be removed in 0.20.\n \"This module will be removed in 0.20.\", DeprecationWarning)\n"
],
[
"import numpy as np\n\n# Convert predictions and target to NumPy arrays\nnp_predictions_lr = np.asarray(predictions_lr)\nnp_target = target.as_matrix()",
"_____no_output_____"
],
[
"# Determine True Positive count\ntp_filter_lr = (np_predictions_lr == 1) & (np_target == 1)\ntp_lr = len(np_predictions_lr[tp_filter_lr])\n\n# Determine False Negative count\nfn_filter_lr = (np_predictions_lr == 0) & (np_target == 1)\nfn_lr = len(np_predictions_lr[fn_filter_lr])\n\n# Determine False Positive count\nfp_filter_lr = (np_predictions_lr == 1) & (np_target == 0)\nfp_lr = len(np_predictions_lr[fp_filter_lr])\n\n# Determine True Negative count\ntn_filter_lr = (np_predictions_lr == 0) & (np_target == 0)\ntn_lr = len(np_predictions_lr[tn_filter_lr])\n\n# Determine True Positive rate\ntpr_lr = tp_lr / (tp_lr + fn_lr)\n\n# Determine False Negative rate\nfnr_lr = fn_lr / (fn_lr + tp_lr)\n\n# Determine False Positive rate\nfpr_lr = fp_lr / (fp_lr + tn_lr)\n\n# Print each count\nprint(tp_lr)\nprint(fn_lr)\nprint(fp_lr)\n\n# Print each rate\nprint(tpr_lr)\nprint(fnr_lr)\nprint(fpr_lr)",
"38\n8\n298\n0.8260869565217391\n0.17391304347826086\n0.24546952224052718\n"
],
[
"new_data = tot2017\nnew_features = features2017",
"_____no_output_____"
],
[
"# Fit the Random Forest model\nlr.fit(features, target)\n\n# Estimate probabilities of Hall of Fame induction\nprobabilities = lr.predict_proba(new_features)",
"_____no_output_____"
],
[
"# Convert predictions to a DataFrame\nWS_predictions = pd.DataFrame(probabilities[:,1])\n\n# Sort the DataFrame (descending)\nWS_predictions = WS_predictions.sort_values(0, ascending=False)\n\nWS_predictions['Probability'] = WS_predictions[0]\n\n# Print 50 highest probability HoF inductees from still eligible players\nfor i, row in WS_predictions.head(50).iterrows():\n prob = ' '.join(('WS Probability =', str(row['Probability'])))\n print('')\n print(prob)\n print(new_data.iloc[i,1:27])",
"\nWS Probability = 0.000567750444291\nR 818\nERA 3.3\nWP 0.63\nName: 7, dtype: object\n\nWS Probability = 0.000386572498575\nR 770\nERA 3.38\nWP 0.642\nName: 13, dtype: object\n\nWS Probability = 0.000337225795312\nR 858\nERA 3.72\nWP 0.562\nName: 18, dtype: object\n\nWS Probability = 0.000294728209055\nR 812\nERA 3.66\nWP 0.574\nName: 0, dtype: object\n\nWS Probability = 0.000239189716566\nR 785\nERA 3.7\nWP 0.574\nName: 3, dtype: object\n\nWS Probability = 0.000210695494912\nR 819\nERA 3.88\nWP 0.599\nName: 29, dtype: object\n\nWS Probability = 0.000209445957019\nR 896\nERA 4.12\nWP 0.623\nName: 10, dtype: object\n\nWS Probability = 0.000188917067286\nR 822\nERA 3.95\nWP 0.568\nName: 4, dtype: object\n\nWS Probability = 0.000123021808249\nR 761\nERA 4.01\nWP 0.512\nName: 25, dtype: object\n\nWS Probability = 0.000107954043486\nR 732\nERA 4\nWP 0.531\nName: 15, dtype: object\n\nWS Probability = 9.26971520401e-05\nR 694\nERA 3.97\nWP 0.494\nName: 26, dtype: object\n\nWS Probability = 7.2908164575e-05\nR 824\nERA 4.51\nWP 0.537\nName: 8, dtype: object\n\nWS Probability = 6.79856381284e-05\nR 710\nERA 4.2\nWP 0.494\nName: 12, dtype: object\n\nWS Probability = 6.05607902049e-05\nR 815\nERA 4.59\nWP 0.525\nName: 16, dtype: object\n\nWS Probability = 5.35534869266e-05\nR 750\nERA 4.46\nWP 0.481\nName: 23, dtype: object\n\nWS Probability = 5.2541114932e-05\nR 668\nERA 4.22\nWP 0.463\nName: 21, dtype: object\n\nWS Probability = 4.90864652238e-05\nR 799\nERA 4.66\nWP 0.481\nName: 27, dtype: object\n\nWS Probability = 4.25479454223e-05\nR 693\nERA 4.42\nWP 0.469\nName: 28, dtype: object\n\nWS Probability = 3.52144760617e-05\nR 739\nERA 4.67\nWP 0.463\nName: 19, dtype: object\n\nWS Probability = 3.34410987455e-05\nR 778\nERA 4.82\nWP 0.475\nName: 14, dtype: object\n\nWS Probability = 3.32039235179e-05\nR 690\nERA 4.55\nWP 0.407\nName: 20, dtype: object\n\nWS Probability = 3.23502443078e-05\nR 702\nERA 4.61\nWP 0.494\nName: 11, dtype: object\n\nWS Probability = 3.10768390315e-05\nR 732\nERA 4.72\nWP 0.444\nName: 1, dtype: object\n\nWS Probability = 2.76859423358e-05\nR 639\nERA 4.5\nWP 0.395\nName: 24, dtype: object\n\nWS Probability = 2.43796231571e-05\nR 706\nERA 4.78\nWP 0.414\nName: 5, dtype: object\n\nWS Probability = 2.15264503579e-05\nR 743\nERA 4.97\nWP 0.463\nName: 2, dtype: object\n\nWS Probability = 1.91870459094e-05\nR 735\nERA 5.01\nWP 0.432\nName: 17, dtype: object\n\nWS Probability = 1.73770791978e-05\nR 604\nERA 4.67\nWP 0.438\nName: 22, dtype: object\n\nWS Probability = 1.59958103859e-05\nR 753\nERA 5.17\nWP 0.42\nName: 6, dtype: object\n\nWS Probability = 1.04828404913e-05\nR 735\nERA 5.36\nWP 0.395\nName: 9, dtype: object\n"
]
],
[
[
"### Entry 7 is CLE.\n### Entry 13 (second place) is LAD. LAD played in the World Series but lost to HOU.\n### HOU won the World Series in 2017. The model failed. ",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
]
|
e797dc07b4ad8da8ca69025eb806e696d832bbed | 12,159 | ipynb | Jupyter Notebook | hypothesis_25.ipynb | IuriSly/DnA-POC-olist | 925a2392f84438a0d3906c9c8cd467cd461f5f8f | [
"MIT"
]
| null | null | null | hypothesis_25.ipynb | IuriSly/DnA-POC-olist | 925a2392f84438a0d3906c9c8cd467cd461f5f8f | [
"MIT"
]
| null | null | null | hypothesis_25.ipynb | IuriSly/DnA-POC-olist | 925a2392f84438a0d3906c9c8cd467cd461f5f8f | [
"MIT"
]
| null | null | null | 41.783505 | 175 | 0.541574 | [
[
[
"# Calculating the distance between the Customer's city and the Seller's city",
"_____no_output_____"
]
],
[
[
"from pyspark.sql import SparkSession, functions as F\nimport math",
"_____no_output_____"
],
[
"spark = SparkSession.builder.getOrCreate()",
"_____no_output_____"
],
[
"orders_items_df = spark.read \\\n .option('escape', '\\\"') \\\n .option('quote', '\\\"') \\\n .csv('./dataset/olist_order_items_dataset.csv', header=True, multiLine=True, inferSchema=True)\n\norders_df = spark.read \\\n .option('escape', '\\\"') \\\n .option('quote', '\\\"') \\\n .csv('./dataset/olist_orders_dataset.csv', header=True, multiLine=True, inferSchema=True)\n\ncustomers_df = spark.read \\\n .option('escape', '\\\"') \\\n .option('quote', '\\\"') \\\n .csv('./dataset/olist_customers_dataset.csv', header=True, multiLine=True, inferSchema=True)\n\nsellers_df = spark.read \\\n .option('escape', '\\\"') \\\n .option('quote', '\\\"') \\\n .csv('./dataset/olist_sellers_dataset.csv', header=True, multiLine=True, inferSchema=True)\n\n",
"_____no_output_____"
],
[
"geo_df = spark.read \\\n .option('escape', '\\\"') \\\n .option('quote', '\\\"') \\\n .csv('./dataset/olist_geolocation_dataset.csv', header=True, multiLine=True, inferSchema=True)",
"_____no_output_____"
]
],
[
[
"# Grouping data",
"_____no_output_____"
]
],
[
[
"data_df = orders_df.filter(F.col('order_status') == 'delivered').join(customers_df, 'customer_id')\n\ndata_df = orders_items_df.join(data_df, 'order_id') \\\n .join(sellers_df, 'seller_id') \\\n .select('customer_state', 'customer_city', 'customer_zip_code_prefix', 'seller_zip_code_prefix', 'freight_value')\n\ngeo_df = geo_df.groupBy('geolocation_zip_code_prefix').agg(F.min('geolocation_lat').alias('geolocation_lat'), F.min('geolocation_lng').alias('geolocation_lng'))\n\ndata_df = data_df.join(geo_df, data_df.customer_zip_code_prefix == geo_df.geolocation_zip_code_prefix) \\\n .select(F.col('geolocation_lat').alias('customer_lat'), F.col('geolocation_lng').alias('customer_lng'), 'seller_zip_code_prefix', 'freight_value') \\\n .join(geo_df, data_df.seller_zip_code_prefix == geo_df.geolocation_zip_code_prefix) \\\n .select('customer_lat', 'customer_lng', F.col('geolocation_lat').alias('seller_lat'), F.col('geolocation_lng').alias('seller_lng'),'freight_value')\ndata_df.count()\n",
"_____no_output_____"
],
[
"data_df.show()",
"+-------------------+-------------------+-------------------+-------------------+-------------+\n| customer_lat| customer_lng| seller_lat| seller_lng|freight_value|\n+-------------------+-------------------+-------------------+-------------------+-------------+\n| -23.50648246805157|-47.422068081741564|-23.545262137111173| -46.66134804356862| 14.43|\n| -23.82558722913311| -46.56982049999999| -23.51441473688614| -46.59097058895492| 9.34|\n|-21.213665497085813| -47.81670447259758| -23.51441473688614| -46.59097058895492| 11.74|\n|-21.445954952757404| -50.12641249999996| -23.51441473688614| -46.59097058895492| 3.07|\n|-21.445954952757404| -50.12641249999996| -23.51441473688614| -46.59097058895492| 3.06|\n|-23.635655999999997| -46.751535578894| -23.51441473688614| -46.59097058895492| 9.34|\n| -23.49878075214959|-46.632511331380975| -23.51441473688614| -46.59097058895492| 9.34|\n|-22.970853233039268|-43.671131559512865|-23.593123748530044| -46.64060056549716| 15.72|\n| -3.814121000836711| -38.59399257936673|-23.593123748530044| -46.64060056549716| 17.63|\n| -3.814121000836711| -38.59399257936673|-23.593123748530044| -46.64060056549716| 17.63|\n| -3.814121000836711| -38.59399257936673|-23.593123748530044| -46.64060056549716| 17.63|\n| -3.814121000836711| -38.59399257936673|-23.593123748530044| -46.64060056549716| 17.63|\n| -20.26779007607431| -56.7781600607055|-23.593123748530044| -46.64060056549716| 18.77|\n|-12.991703180012463| -38.45058108919243|-23.593123748530044| -46.64060056549716| 17.53|\n| -23.51441473688614| -46.59097058895492|-23.204420999999996|-46.590299999999985| 7.46|\n| -23.63496067770149| -46.75505959566482|-23.204420999999996|-46.590299999999985| 7.45|\n| -23.74259343710035| -46.60832671234592|-23.204420999999996|-46.590299999999985| 7.87|\n|-23.214238574995893| -49.40174227177037|-23.204420999999996|-46.590299999999985| 13.08|\n|-25.546951194558265| -49.29139473619366|-23.204420999999996|-46.590299999999985| 18.3|\n|-26.883628877564163| -49.08164479732078|-23.204420999999996|-46.590299999999985| 18.23|\n+-------------------+-------------------+-------------------+-------------------+-------------+\nonly showing top 20 rows\n\n"
]
],
[
[
"# Calculating distance",
"_____no_output_____"
]
],
[
[
"def d(c_lat, c_lng, s_lat, s_lng):\n radius = 6371 # km\n\n dlat = math.radians(s_lat-c_lat)\n dlon = math.radians(s_lng-c_lng)\n a = math.sin(dlat/2) * math.sin(dlat/2) + math.cos(math.radians(c_lat)) \\\n * math.cos(math.radians(s_lat)) * math.sin(dlon/2) * math.sin(dlon/2)\n c = 2 * math.atan2(math.sqrt(a), math.sqrt(1-a))\n d = radius * c;\n\n return d\n\ndistance = F.udf(d)\n\ndata_df = data_df.withColumn('distance', distance('customer_lat', 'customer_lng', 'seller_lat', 'seller_lng'))\n\ndata_df.show()",
"+-------------------+-------------------+-------------------+-------------------+-------------+------------------+\n| customer_lat| customer_lng| seller_lat| seller_lng|freight_value| distance|\n+-------------------+-------------------+-------------------+-------------------+-------------+------------------+\n| -23.50648246805157|-47.422068081741564|-23.545262137111173| -46.66134804356862| 14.43| 77.67691920579136|\n| -23.82558722913311| -46.56982049999999| -23.51441473688614| -46.59097058895492| 9.34|34.667779619845135|\n|-21.213665497085813| -47.81670447259758| -23.51441473688614| -46.59097058895492| 11.74| 285.1904555969043|\n|-21.445954952757404| -50.12641249999996| -23.51441473688614| -46.59097058895492| 3.07| 429.9117657164478|\n|-21.445954952757404| -50.12641249999996| -23.51441473688614| -46.59097058895492| 3.06| 429.9117657164478|\n|-23.635655999999997| -46.751535578894| -23.51441473688614| -46.59097058895492| 9.34|21.201989898325316|\n| -23.49878075214959|-46.632511331380975| -23.51441473688614| -46.59097058895492| 9.34| 4.57865588323346|\n|-22.970853233039268|-43.671131559512865|-23.593123748530044| -46.64060056549716| 15.72|311.08784955393077|\n| -3.814121000836711| -38.59399257936673|-23.593123748530044| -46.64060056549716| 17.63|2362.9895306493527|\n| -3.814121000836711| -38.59399257936673|-23.593123748530044| -46.64060056549716| 17.63|2362.9895306493527|\n| -3.814121000836711| -38.59399257936673|-23.593123748530044| -46.64060056549716| 17.63|2362.9895306493527|\n| -3.814121000836711| -38.59399257936673|-23.593123748530044| -46.64060056549716| 17.63|2362.9895306493527|\n| -20.26779007607431| -56.7781600607055|-23.593123748530044| -46.64060056549716| 18.77|1108.7398255326386|\n|-12.991703180012463| -38.45058108919243|-23.593123748530044| -46.64060056549716| 17.53|1460.9299074564092|\n| -23.51441473688614| -46.59097058895492|-23.204420999999996|-46.590299999999985| 7.46|34.469798805617216|\n| -23.63496067770149| -46.75505959566482|-23.204420999999996|-46.590299999999985| 7.45|50.739692068937146|\n| -23.74259343710035| -46.60832671234592|-23.204420999999996|-46.590299999999985| 7.87| 59.8702823591726|\n|-23.214238574995893| -49.40174227177037|-23.204420999999996|-46.590299999999985| 13.08|287.31589739648564|\n|-25.546951194558265| -49.29139473619366|-23.204420999999996|-46.590299999999985| 18.3|377.72000872499245|\n|-26.883628877564163| -49.08164479732078|-23.204420999999996|-46.590299999999985| 18.23| 479.920438191282|\n+-------------------+-------------------+-------------------+-------------------+-------------+------------------+\nonly showing top 20 rows\n\n"
],
[
"data_df = data_df.withColumn('distance', F.col('distance').cast('double'))",
"_____no_output_____"
],
[
"data_df.printSchema()",
"root\n |-- customer_lat: double (nullable = true)\n |-- customer_lng: double (nullable = true)\n |-- seller_lat: double (nullable = true)\n |-- seller_lng: double (nullable = true)\n |-- freight_value: double (nullable = true)\n |-- distance: double (nullable = true)\n\n"
],
[
"data_df.stat.corr('distance','freight_value')",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
]
|
e797ec16c9a36d99620b1a4d1a4106e52d2e2534 | 926,232 | ipynb | Jupyter Notebook | SCOPUS_data_analysis_Python3x_v1.ipynb | NeelShah18/scopus-analysis-for-indian-researcher | 94470992bc0155863541ffc593f9d1c7ce5df047 | [
"MIT"
]
| 6 | 2017-04-03T19:29:13.000Z | 2017-06-02T14:51:07.000Z | SCOPUS_data_analysis_Python3x_v1.ipynb | NeelShah18/scopus-analysis-for-indian-researcher | 94470992bc0155863541ffc593f9d1c7ce5df047 | [
"MIT"
]
| 1 | 2017-05-31T06:12:40.000Z | 2017-06-02T06:27:41.000Z | SCOPUS_data_analysis_Python3x_v1.ipynb | NeelShah18/scopus-analysis-for-indian-researcher | 94470992bc0155863541ffc593f9d1c7ce5df047 | [
"MIT"
]
| 6 | 2017-06-01T08:39:13.000Z | 2020-05-19T14:25:27.000Z | 876.283822 | 116,140 | 0.93819 | [
[
[
"# SCOPUS journal Data analysis of Indian Research",
"_____no_output_____"
],
[
"About: The main aim of this data analysis is to identify the ongoing research in Indian Universities and Indian Industry. It gives a basic answer about research source and trend with top authors and publication. It also shows the participation of Industry and Universities in research.",
"_____no_output_____"
],
[
"Created By : \n-------------\n**Neel Shah:** [Website](https://neelshah18.github.io/) | [Linkedin](https://www.linkedin.com/in/neel-shah-7b5495104/) | [GitHub](https://github.com/NeelShah18) | Email:**[email protected]**\n**Open to hire**\n\nEdited By:\n-----------\n1) Malaikannan Sankarasubbu - Know about him more: [Linkedin](https://www.linkedin.com/in/malaikannan/) | [GitHub](href=\"https://github.com/malaikannan)\n\nSpecial thanks to:\n--------------------\n1) Dr. Jacob Minz - Know about him more: [Linkedin](https://www.linkedin.com/in/jacob-minz-16762a3/) | [GitHub](https://github.com/jrminz)\n\n2) Anirban Santara - Know about him more: [Linkedin](https://www.linkedin.com/in/anirbansantara/) | [GitHub](https://github.com/Santara)\n\n-------------------------------------------------------------------------------------------------------------------\nTechnical Implementation - Open source license\n\nIt is implemented in a Jupyter notebook with a back of Anaconda and Python 3.6+ version. \n\nDataset and Jupyter notebook is available under MIT - open source license. If you want to use this code or data feel free to do it, But, Please cite me.\n\n* Link to the repository : [click here!](https://github.com/NeelShah18/scopus-analysis-for-indian-researcher)\n* Link to Code and Dataset(store in SQLite and CSV format) : [click here!](https://github.com/NeelShah18/scopus-analysis-for-indian-researcher)\n",
"_____no_output_____"
],
[
"### Top 20 research papers from 2001 to 2016 - Indian researchers\n\nIt is surprising that from TOP 20 almost 70% papers are related to AI, ML, CV and DL. But ",
"_____no_output_____"
]
],
[
[
"import sqlite3\nimport matplotlib.pyplot as plt\nimport operator\n\nsqlite_database = '/home/neel/scopus_data/scopus_data.sqlite'\nconn = sqlite3.connect(sqlite_database)\nc = conn.cursor()\nc.execute(\"SELECT `Title`,`cited_rank` FROM `AI_scopus` ORDER BY `cited_rank` DESC LIMIT 0, 20;\")\ndata = c.fetchall()\nconn.close()\ntop_paper = {}\n#print(data)\n\nfor x in data:\n text = (((str(x).replace(\"'\",\"\")).replace(\"(\",\"\")).replace(\")\",\"\"))\n lis = text.split(\",\")\n #print(lis[0])\n #print(lis[1].strip())\n top_paper[str(lis[0])]= int(lis[1])\n#print(top_paper)\nplt.barh(range(len(top_paper)),top_paper.values(),align='center')\nplt.yticks(range(len(top_paper)),list(top_paper.keys()))\nplt.xlabel('\\n Paper cited ')\nplt.title(\"Top 20 Indian researcher's paper in SCOPUS journal \\nfrom 2000 to 2016\\n\")\nplt.ylabel('---- Paper ---- \\n')\nfig_size = plt.rcParams[\"figure.figsize\"]\nfig_size[0] = 15\nfig_size[1] = 15\nplt.rcParams[\"figure.figsize\"] = fig_size\nplt.show()",
"_____no_output_____"
],
[
"import sqlite3\nimport matplotlib.pyplot as plt\nimport operator\n\ndata = []\nsqlite_database = '/home/neel/scopus_data/scopus_data.sqlite'\nconn = sqlite3.connect(sqlite_database)\nc = conn.cursor()\nc.execute(\"SELECT `Authors`,`cited_rank` FROM `AI_scopus` ORDER BY `cited_rank` DESC LIMIT 0, 20;\")\ndata = c.fetchall()\ntop_author = {}\ntext = str(data[0]).replace(\"'\",\"\")\nfor x in data:\n cite = (str(x)[-4:-1]).strip()\n authors = (str(x)[2:len(x)-7]).replace(\"'\",\"\")\n top_author[str(authors)] = int(cite)\n \n#print(top_author)\nconn.close()\n\n\nplt.barh(range(len(top_author)),top_author.values(),align='center')\nplt.yticks(range(len(top_author)),list(top_author.keys()))\nplt.xlabel('\\n Author cited ')\nplt.title('Top 20 Indian researcher in SCOPUS journal\\n from 2000 to 2016\\n')\nplt.ylabel('---- Authors ---- \\n')\nfig_size = plt.rcParams[\"figure.figsize\"]\nfig_size[0] = 15\nfig_size[1] = 15\nplt.rcParams[\"figure.figsize\"] = fig_size\nplt.show()\n",
"_____no_output_____"
],
[
"import sqlite3\nimport matplotlib.pyplot as plt\nimport operator\n\ndata = []\nsqlite_database = '/home/neel/scopus_data/scopus_data.sqlite'\nconn = sqlite3.connect(sqlite_database)\nc = conn.cursor()\nc.execute(\"SELECT `Year`,`IndexKeywords` FROM `AI_scopus` DESC LIMIT 0, 5000;\")\ndata = c.fetchall()\n#text = str(data[0])\n#print(text[2:6])\n#print(text[10:len(text)-2])\n#tr = []\n#tr = (text[10:len(text)-2]).split(\";\")\n#print(tr)\nconn.close()\ndata_dic = {}\n\nz = 0 \nword_lis = []\nwhile z < len(data):\n text = str(data[z])\n year = str(text[2:6])\n #print(year)\n lis_word = (text[10:len(text)-2].replace(\" \",\"\")).split(\";\")\n #print(lis_word)\n if year == '2016':\n for word in lis_word:\n try:\n data_dic[str(word)] = int(data_dic[str(word)]) + 1\n except:\n data_dic[str(word)] = 1\n z += 1\n\n#print(data_dic)\nlis_f = sorted(data_dic, key=data_dic.get, reverse=True)\ncount = 0\ndraw_word_dic = {}\n#print(lis_f)\nwhile count < 10:\n draw_word_dic[str(lis_f[count])] = data_dic[str(lis_f[count])]\n count += 1\n \n\nplt.barh(range(len(draw_word_dic)),draw_word_dic.values(),align='center')\nplt.yticks(range(len(draw_word_dic)),list(draw_word_dic.keys()))\nplt.xlabel('\\nNumber of Papers')\nplt.title('Trend of research in 2016 \"SCOPUS\" journal')\nplt.ylabel('---- Areas ---- \\n')\nplt.show()",
"_____no_output_____"
],
[
"import sqlite3\nimport matplotlib.pyplot as plt\nimport operator\n\ndata = []\nsqlite_database = '/home/neel/scopus_data/scopus_data.sqlite'\nconn = sqlite3.connect(sqlite_database)\nc = conn.cursor()\nc.execute(\"SELECT `Year`,`IndexKeywords` FROM `AI_scopus` DESC LIMIT 0, 5000;\")\ndata = c.fetchall()\n#text = str(data[0])\n#print(text[2:6])\n#print(text[10:len(text)-2])\n#tr = []\n#tr = (text[10:len(text)-2]).split(\";\")\n#print(tr)\nconn.close()\ndata_dic = {}\n\nz = 0 \nword_lis = []\nwhile z < len(data):\n text = str(data[z])\n year = str(text[2:6])\n #print(year)\n lis_word = (text[10:len(text)-2].replace(\" \",\"\")).split(\";\")\n #print(lis_word)\n if year == '2015':\n for word in lis_word:\n try:\n data_dic[str(word)] = int(data_dic[str(word)]) + 1\n except:\n data_dic[str(word)] = 1\n z += 1\n\n#print(data_dic)\nlis_f = sorted(data_dic, key=data_dic.get, reverse=True)\ncount = 0\ndraw_word_dic = {}\n#print(lis_f)\nwhile count < 10:\n draw_word_dic[str(lis_f[count])] = data_dic[str(lis_f[count])]\n count += 1\n \n\nplt.barh(range(len(draw_word_dic)),draw_word_dic.values(),align='center')\nplt.yticks(range(len(draw_word_dic)),list(draw_word_dic.keys()))\nplt.xlabel('\\nNumber of Papers')\nplt.title('Trend of research in 2015 \"SCOPUS\" journal')\nplt.ylabel('---- Areas ---- \\n')\nplt.show()",
"_____no_output_____"
],
[
"import sqlite3\nimport matplotlib.pyplot as plt\nimport operator\n\ndata = []\nsqlite_database = '/home/neel/scopus_data/scopus_data.sqlite'\nconn = sqlite3.connect(sqlite_database)\nc = conn.cursor()\nc.execute(\"SELECT `Year`,`IndexKeywords` FROM `AI_scopus` DESC LIMIT 0, 5000;\")\ndata = c.fetchall()\n#text = str(data[0])\n#print(text[2:6])\n#print(text[10:len(text)-2])\n#tr = []\n#tr = (text[10:len(text)-2]).split(\";\")\n#print(tr)\nconn.close()\ndata_dic = {}\n\nz = 0 \nword_lis = []\nwhile z < len(data):\n text = str(data[z])\n year = str(text[2:6])\n #print(year)\n lis_word = (text[10:len(text)-2].replace(\" \",\"\")).split(\";\")\n #print(lis_word)\n if year == '2014':\n for word in lis_word:\n try:\n data_dic[str(word)] = int(data_dic[str(word)]) + 1\n except:\n data_dic[str(word)] = 1\n z += 1\n\n#print(data_dic)\nlis_f = sorted(data_dic, key=data_dic.get, reverse=True)\ncount = 0\ndraw_word_dic = {}\n#print(lis_f)\nwhile count < 10:\n draw_word_dic[str(lis_f[count])] = data_dic[str(lis_f[count])]\n count += 1\n \n\nplt.barh(range(len(draw_word_dic)),draw_word_dic.values(),align='center')\nplt.yticks(range(len(draw_word_dic)),list(draw_word_dic.keys()))\nplt.xlabel('\\nNumber of Papers')\nplt.title('Trend of research in 2014 \"SCOPUS\" journal')\nplt.ylabel('---- Areas ---- \\n')\nplt.show()",
"_____no_output_____"
],
[
"import sqlite3\nimport matplotlib.pyplot as plt\nimport operator\n\ndata = []\nsqlite_database = '/home/neel/scopus_data/scopus_data.sqlite'\nconn = sqlite3.connect(sqlite_database)\nc = conn.cursor()\nc.execute(\"SELECT `Year`,`IndexKeywords` FROM `AI_scopus` DESC LIMIT 0, 5000;\")\ndata = c.fetchall()\n#text = str(data[0])\n#print(text[2:6])\n#print(text[10:len(text)-2])\n#tr = []\n#tr = (text[10:len(text)-2]).split(\";\")\n#print(tr)\nconn.close()\ndata_dic = {}\n\nz = 0 \nword_lis = []\nwhile z < len(data):\n text = str(data[z])\n year = str(text[2:6])\n #print(year)\n lis_word = (text[10:len(text)-2].replace(\" \",\"\")).split(\";\")\n #print(lis_word)\n if year == '2013':\n for word in lis_word:\n try:\n data_dic[str(word)] = int(data_dic[str(word)]) + 1\n except:\n data_dic[str(word)] = 1\n z += 1\n\n#print(data_dic)\nlis_f = sorted(data_dic, key=data_dic.get, reverse=True)\ncount = 0\ndraw_word_dic = {}\n#print(lis_f)\nwhile count < 10:\n draw_word_dic[str(lis_f[count])] = data_dic[str(lis_f[count])]\n count += 1\n \n\nplt.barh(range(len(draw_word_dic)),draw_word_dic.values(),align='center')\nplt.yticks(range(len(draw_word_dic)),list(draw_word_dic.keys()))\nplt.xlabel('\\nNumber of Papers')\nplt.title('Trend of research in 2013 \"SCOPUS\" journal')\nplt.ylabel('---- Areas ---- \\n')\nplt.show()",
"_____no_output_____"
],
[
"import sqlite3\nimport matplotlib.pyplot as plt\nimport operator\n\ndata = []\nsqlite_database = '/home/neel/scopus_data/scopus_data.sqlite'\nconn = sqlite3.connect(sqlite_database)\nc = conn.cursor()\nc.execute(\"SELECT `Year`,`IndexKeywords` FROM `AI_scopus` DESC LIMIT 0, 5000;\")\ndata = c.fetchall()\n#text = str(data[0])\n#print(text[2:6])\n#print(text[10:len(text)-2])\n#tr = []\n#tr = (text[10:len(text)-2]).split(\";\")\n#print(tr)\nconn.close()\ndata_dic = {}\n\nz = 0 \nword_lis = []\nwhile z < len(data):\n text = str(data[z])\n year = str(text[2:6])\n #print(year)\n lis_word = (text[10:len(text)-2].replace(\" \",\"\")).split(\";\")\n #print(lis_word)\n if year == '2012':\n for word in lis_word:\n try:\n data_dic[str(word)] = int(data_dic[str(word)]) + 1\n except:\n data_dic[str(word)] = 1\n z += 1\n\n#print(data_dic)\nlis_f = sorted(data_dic, key=data_dic.get, reverse=True)\ncount = 0\ndraw_word_dic = {}\n#print(lis_f)\nwhile count < 10:\n draw_word_dic[str(lis_f[count])] = data_dic[str(lis_f[count])]\n count += 1\n \n\nplt.barh(range(len(draw_word_dic)),draw_word_dic.values(),align='center')\nplt.yticks(range(len(draw_word_dic)),list(draw_word_dic.keys()))\nplt.xlabel('\\nNumber of Papers')\nplt.title('Trend of research in 2012 \"SCOPUS\" journal')\nplt.ylabel('---- Areas ---- \\n')\nplt.show()",
"_____no_output_____"
],
[
"import sqlite3\nimport matplotlib.pyplot as plt\nimport operator\n\ndata = []\nsqlite_database = '/home/neel/scopus_data/scopus_data.sqlite'\nconn = sqlite3.connect(sqlite_database)\nc = conn.cursor()\nc.execute(\"SELECT `Year`,`IndexKeywords` FROM `AI_scopus` DESC LIMIT 0, 5000;\")\ndata = c.fetchall()\n#text = str(data[0])\n#print(text[2:6])\n#print(text[10:len(text)-2])\n#tr = []\n#tr = (text[10:len(text)-2]).split(\";\")\n#print(tr)\nconn.close()\ndata_dic = {}\n\nz = 0 \nword_lis = []\nwhile z < len(data):\n text = str(data[z])\n year = str(text[2:6])\n #print(year)\n lis_word = (text[10:len(text)-2].replace(\" \",\"\")).split(\";\")\n #print(lis_word)\n if year == '2011':\n for word in lis_word:\n try:\n data_dic[str(word)] = int(data_dic[str(word)]) + 1\n except:\n data_dic[str(word)] = 1\n z += 1\n\n#print(data_dic)\nlis_f = sorted(data_dic, key=data_dic.get, reverse=True)\ncount = 0\ndraw_word_dic = {}\n#print(lis_f)\nwhile count < 10:\n draw_word_dic[str(lis_f[count])] = data_dic[str(lis_f[count])]\n count += 1\n \n\nplt.barh(range(len(draw_word_dic)),draw_word_dic.values(),align='center')\nplt.yticks(range(len(draw_word_dic)),list(draw_word_dic.keys()))\nplt.xlabel('\\nNumber of Papers')\nplt.title('Trend of research in 2011 \"SCOPUS\" journal')\nplt.ylabel('---- Areas ---- \\n')\nplt.show()",
"_____no_output_____"
],
[
"import sqlite3\nimport matplotlib.pyplot as plt\nimport operator\n\ndata = []\nsqlite_database = '/home/neel/scopus_data/scopus_data.sqlite'\nconn = sqlite3.connect(sqlite_database)\nc = conn.cursor()\nc.execute(\"SELECT `Year`,`IndexKeywords` FROM `AI_scopus` DESC LIMIT 0, 5000;\")\ndata = c.fetchall()\n#text = str(data[0])\n#print(text[2:6])\n#print(text[10:len(text)-2])\n#tr = []\n#tr = (text[10:len(text)-2]).split(\";\")\n#print(tr)\nconn.close()\ndata_dic = {}\n\nz = 0 \nword_lis = []\nwhile z < len(data):\n text = str(data[z])\n year = str(text[2:6])\n #print(year)\n lis_word = (text[10:len(text)-2].replace(\" \",\"\")).split(\";\")\n #print(lis_word)\n if year == '2010':\n for word in lis_word:\n try:\n data_dic[str(word)] = int(data_dic[str(word)]) + 1\n except:\n data_dic[str(word)] = 1\n z += 1\n\n#print(data_dic)\nlis_f = sorted(data_dic, key=data_dic.get, reverse=True)\ncount = 0\ndraw_word_dic = {}\n#print(lis_f)\nwhile count < 10:\n draw_word_dic[str(lis_f[count])] = data_dic[str(lis_f[count])]\n count += 1\n \n\nplt.barh(range(len(draw_word_dic)),draw_word_dic.values(),align='center')\nplt.yticks(range(len(draw_word_dic)),list(draw_word_dic.keys()))\nplt.xlabel('\\nNumber of Papers')\nplt.title('Trend of research in 2010 \"SCOPUS\" journal')\nplt.ylabel('---- Areas ---- \\n')\nplt.show()",
"_____no_output_____"
],
[
"import sqlite3\nimport matplotlib.pyplot as plt\n\n#fetching the name of different fields\nname = []\n#create the connection with database\nsqlite_database = '/home/neel/scopus_data/scopus_data.sqlite'\nconn = sqlite3.connect(sqlite_database)\nc = conn.cursor()\nc.execute(\"SELECT `name` FROM `university_data` ORDER BY `publish_paper` DESC LIMIT 0, 500;\")\n#store all name in as list\ninit_name = c.fetchall()\nfor each in init_name:\n text = (str(each)[2:len(each)-4]).replace(\"\\\\n\",\"\")\n name.append(text)\n#close the connection with database\nconn.close()\n\n#fetching the number of publication field wise\nsep = []\n#connection create with database\nsqlite_database = '/home/neel/scopus_data/scopus_data.sqlite'\nconn = sqlite3.connect(sqlite_database)\nc = conn.cursor()\nc.execute(\"SELECT `publish_paper` FROM `university_data` ORDER BY `publish_paper` DESC LIMIT 0, 500;\")\n#store the data in sep as list\nsep = c.fetchall()\n#connection close with databae\nconn.close()\n\n#create a list of realtive percentage for publish paper field wise\nper = []\nfor n in sep:\n text = str(n)[1:len(n)-3]\n n_to_per = int(text)\n val = (n_to_per*100)/1187\n val_2 = \"%.2f\"%val\n per.append(val_2)\n\n#---------------------------Graph code------------------------------\nlabel = []\nx = 0\nwhile x < len(per):\n label.append(str(name[x].upper())+\" : \"+str(per[x])+\"%\")\n x += 1\n\nlabels = label\nsizes = per\npatches, texts = plt.pie(sizes, startangle=90)\nplt.legend(patches, labels,bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)\n# Set aspect ratio to be equal so that pie is drawn as a circle.\nplt.axis('equal')\nplt.title('Research done Top 15 Universitites and other Universities\\n from 2001 to 2016\\n Source: SCOPUS journal ')\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
],
[
"import sqlite3\nfrom matplotlib import pyplot as plt\nfrom matplotlib import style\nimport matplotlib.pyplot as plt; plt.rcdefaults()\n\ndata = []\nsqlite_database = '/home/neel/scopus_data/scopus_data.sqlite'\nconn = sqlite3.connect(sqlite_database)\nc = conn.cursor()\nc.execute(\"SELECT `Year`,`IndexKeywords` FROM `AI_scopus` DESC LIMIT 0, 5000;\")\ndata = c.fetchall()\ntext = str(data[0])\n#print(text[2:6])\n#print(text[10:len(text)-2])\n#tr = []\ntr = ((text[10:len(text)-2]).replace(\" \",\"\")).split(\";\")\n#print(tr)\nconn.close()\ntred_word_dic = {}\n\ndata_ai = {}\ndata_nm = {}\ndata_ls = {}\ndata_algo = {}\ndata_cv = {}\n\nfield_lis = []\n\nfor line in data:\n text = str(line)\n year = text[2:6]\n field_lis = ((text[10:len(text)-2]).replace(\" \",\"\")).split(\";\")\n for field in field_lis:\n if field == 'Artificialintelligence':\n try:\n data_ai[year] = int(data_ai[year]) + 1\n except:\n data_ai[year] = 1\n if field == 'Neuralnetworks':\n try:\n data_nm[year] = int(data_nm[year]) + 1\n except:\n data_nm[year] = 1\n if field == 'Learningsystems':\n try:\n data_ls[year] = int(data_ls[year]) + 1\n except:\n data_ls[year] = 1\n if field == 'Algorithms':\n try:\n data_algo[year] = int(data_algo[year]) + 1\n except:\n data_algo[year] = 1\n if field == 'Computervision':\n try:\n data_cv[year] = int(data_cv[year]) + 1\n except:\n data_cv[year] = 1\n\nx_xix = []\ny_ai = []\ny_nm = []\ny_ls = []\ny_algo = []\ny_cv = []\n\nx = 2001\nzero = 0\nwhile x < 2017:\n try:\n #print(x)\n y_ai.append(data_ai[str(x)])\n #print(data_CV[x])\n except:\n y_ai.append(int(zero))\n pass\n try:\n #print(x)\n y_nm.append(data_nm[str(x)])\n #print(data_CV[x])\n except:\n y_nm.append(int(zero))\n pass\n try:\n #print(x)\n y_ls.append(data_ls[str(x)])\n #print(data_CV[x])\n except:\n y_ls.append(int(zero))\n pass\n try:\n #print(x)\n y_algo.append(data_algo[str(x)])\n #print(data_CV[x])\n except:\n y_algo.append(int(zero))\n pass\n try:\n #print(x)\n y_cv.append(data_cv[str(x)])\n #print(data_CV[x])\n except:\n y_cv.append(int(zero))\n pass\n x_xix.append(x)\n x += 1\n \nstyle.use('ggplot')\nplt.plot(x_xix,y_cv,label=\"Computer Vision\")\nplt.plot(x_xix,y_ai,label=\"Artificial Intelligence\")\nplt.plot(x_xix,y_algo,label=\"Algorithms\")\nplt.plot(x_xix,y_ls,label=\"Learning Systems\")\nplt.plot(x_xix,y_nm,label=\"Neural Networks\")\n\nplt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)\nplt.title('Trend of research in different realm of CS\\n from 2001 to 2016')\nplt.ylabel('Number of publish paper')\nplt.xlabel('\\nYears: 2001 - 2016')\n\nplt.show()",
"_____no_output_____"
],
[
"import sqlite3\nimport matplotlib.pyplot as plt\n\n#fetching the name of different fields\nname = []\n#create the connection with database\nsqlite_database = '/home/neel/scopus_data/scopus_data.sqlite'\nconn = sqlite3.connect(sqlite_database)\nc = conn.cursor()\nc.execute(\"SELECT `name` FROM `industry_data` ORDER BY `publish_paper` DESC LIMIT 0, 5000;\")\n#store all name in as list\ninit_name = c.fetchall()\nfor each in init_name:\n text = (str(each)[2:len(each)-4]).replace(\"\\\\n\",\"\")\n name.append(text)\n#close the connection with database\nconn.close()\n\n#fetching the number of publication field wise\nsep = []\n#connection create with database\nsqlite_database = '/home/neel/scopus_data/scopus_data.sqlite'\nconn = sqlite3.connect(sqlite_database)\nc = conn.cursor()\nc.execute(\"SELECT `publish_paper` FROM `industry_data` ORDER BY `publish_paper` DESC LIMIT 0, 5000;\")\n#store the data in sep as list\nsep = c.fetchall()\n#connection close with databae\nconn.close()\n\n#create a list of realtive percentage for publish paper field wise\nper = []\nfor n in sep:\n text = str(n)[1:len(n)-3]\n n_to_per = int(text)\n val = (n_to_per*100)/200\n val_2 = \"%.2f\"%val\n per.append(val_2)\n\n#---------------------------Graph code------------------------------\nlabel = []\nx = 0\nwhile x < len(per):\n label.append(str(name[x].upper())+\" : \"+str(per[x])+\"%\")\n x += 1\n\nlabels = label\nsizes = per\npatches, texts = plt.pie(sizes, startangle=90)\nplt.legend(patches, labels,bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)\n# Set aspect ratio to be equal so that pie is drawn as a circle.\nplt.axis('equal')\nplt.title('Research percentage of different Industries\\n from 2001 to 2016\\n Source: SCOPUS journal ')\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
],
[
"import sqlite3\nimport matplotlib.pyplot as plt\n\n#fetching the name of different fields\nname = []\n#create the connection with database\nsqlite_database = '/home/neel/scopus_data/scopus_data.sqlite'\nconn = sqlite3.connect(sqlite_database)\nc = conn.cursor()\nc.execute(\"SELECT `name` FROM `seprate` DESC LIMIT 0, 5000;\")\n#store all name in as list\ninit_name = c.fetchall()\nfor each in init_name:\n text = (str(each)[2:len(each)-4]).replace(\"\\\\n\",\"\")\n name.append(text)\n#close the connection with database\nconn.close()\n\n#fetching the number of publication field wise\nsep = []\n#connection create with database\nsqlite_database = '/home/neel/scopus_data/scopus_data.sqlite'\nconn = sqlite3.connect(sqlite_database)\nc = conn.cursor()\nc.execute(\"SELECT `number` FROM `seprate` DESC LIMIT 0, 5000;\")\n#store the data in sep as list\nsep = c.fetchall()\n#connection close with databae\nconn.close()\n\n#create a list of realtive percentage for publish paper field wise\nper = []\nfor n in sep:\n text = str(n)[1:len(n)-3]\n n_to_per = int(text)\n val = (n_to_per*100)/1387\n val_2 = \"%.2f\"%val\n per.append(val_2)\n\n#---------------------------Graph code------------------------------\nlabel = []\nx = 0\nwhile x < len(per):\n label.append(str(name[x].upper())+\" : \"+str(per[x])+\"%\")\n x += 1\n\nlabels = label\nsizes = per\npatches, texts = plt.pie(sizes, startangle=90)\nplt.legend(patches, labels,bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)\n# Set aspect ratio to be equal so that pie is drawn as a circle.\nplt.axis('equal')\nplt.title('Research done by Universities and Industries\\n from 2001 to 2016\\n Source: SCOPUS journal ')\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e798064a67dcc763f0bfad4d032613a1c2a7011b | 12,239 | ipynb | Jupyter Notebook | Fairies_NLP_Tutorial(ipython)/01-3.Stemmer_SearchEngine.ipynb | AdonisHan/Fairies_NLP_Tutorclass | 13d40e0742f383c58313cf5d8f6d2fc235c5bb1a | [
"Apache-2.0"
]
| null | null | null | Fairies_NLP_Tutorial(ipython)/01-3.Stemmer_SearchEngine.ipynb | AdonisHan/Fairies_NLP_Tutorclass | 13d40e0742f383c58313cf5d8f6d2fc235c5bb1a | [
"Apache-2.0"
]
| null | null | null | Fairies_NLP_Tutorial(ipython)/01-3.Stemmer_SearchEngine.ipynb | AdonisHan/Fairies_NLP_Tutorclass | 13d40e0742f383c58313cf5d8f6d2fc235c5bb1a | [
"Apache-2.0"
]
| null | null | null | 34.476056 | 1,540 | 0.590408 | [
[
[
"import nltk\nfrom nltk.util import ngrams\nfrom nltk.corpus import alpino\nfrom nltk.collocations import BigramCollocationFinder\nfrom nltk.corpus import webtext\nfrom nltk.corpus import stopwords\nfrom nltk.metrics import BigramAssocMeasures\nimport nltk\nfrom nltk.collocations import *",
"_____no_output_____"
],
[
"# Stemmer\nfrom nltk.stem import PorterStemmer\nfrom nltk.stem import LancasterStemmer\nfrom nltk.stem import RegexpStemmer\nstemmerregexp=RegexpStemmer('ing')\nstemmerlan=LancasterStemmer()\nstemmerporter = PorterStemmer()\nprint(stemmerporter.stem('eatting'))",
"eat\n"
],
[
"from nltk.stem import SnowballStemmer\n# 잘 안되는데 불어...\nfrenchstemmer=SnowballStemmer('french')\nprint(frenchstemmer.stem('mangent'))",
"mangent\n"
],
[
"# Lemmatizer\nfrom nltk.stem import WordNetLemmatizer\nlemmatizer_output=WordNetLemmatizer()\nprint(lemmatizer_output.lemmatize('working',pos='v'))\nprint(lemmatizer_output.lemmatize('works'))",
"work\nwork\n"
],
[
"# Sentence length\nimport nltk\ncorpus=u\"<s> hello how are you doing ? Hope you find the book interesting. </s>\".split()\nsentence=u\"<s>how are you doing</s>\".split()\nvocabulary=set(corpus)\nprint(len(vocabulary))",
"13\n"
],
[
"cfd = nltk.ConditionalFreqDist(nltk.bigrams(corpus))\nprint([cfd[a][b] for (a,b) in nltk.bigrams(sentence)])",
"[0, 1, 0]\n"
],
[
"print([cfd[a].N() for (a,b) in nltk.bigrams(sentence)])",
"[0, 1, 2]\n"
],
[
"print([cfd[a].freq(b) for (a,b) in nltk.bigrams(sentence)])",
"[0, 1.0, 0.0]\n"
],
[
"# 문맥 관점에서 자주 발생하고 중요하지 않은 단어 제거\ndef eliminatestopwords(self,list):\n return [ word for word in list if word not in self.stopwords]\n",
"_____no_output_____"
],
[
"# 텍스트를 불용어와 토큰으로 분할하는 작업을 수행\ndef tokenize(self, string):\n Str = self.clean(str)\n Words = str.split(\"\")\n return [self.stemmer.stem(word,0,len(word)-1) for word in words]\n ",
"_____no_output_____"
],
[
"# 키워드를 벡터 차원에 매핑\ndef obtainvectorkeywordindex(self, documentList):\n # Text 를 문자열로 매핑\n vocabstring = \"\".join(documentList)\n vocablist = self.parser.tokenise(vocabstring)\n \n # 검색의 중요성이 없는 일반적인 단어 제거\n vocablist = self.parser.eliminatestopwords(vocablist)\n uniqueVocablist = util.removeDuplicates(vocablist)\n \n vectorIndex = {}\n offset = 0\n # 이 토큰을 설명하는 데 사용되는 차원과의 매핑을 수행하는 키워드에 포지션을 연결.\n \n for word in uniqueVocablist:\n vectorIndex[word]=offset\n offset += 1\n return vectorIndex",
"_____no_output_____"
],
[
"# simple term count model\n# string to vector\ndef constructVector(self, wordString):\n # 0으로 벡터 초기화\n Vector_val = [0] * len(self.vectorKeywordIndex)\n tokList = self.parser.tokenize(tokString)\n tokList = self.parser.eliminatestopwords(tokList)\n for word in toklist:\n vector[self.vectorKeywordIndex[word]] += 1;\n return vector",
"_____no_output_____"
],
[
"# Cosine Similarity\n# cosine = (X * Y) / ||X|| x ||Y||\ndef cosine(vec1, vec2):\n return float(dot(vec1,vec2) / (normj(vec1) * norm(vec2)))\n ",
"_____no_output_____"
],
[
"# 키워드와 벡터 공간의 매핑을 수행 - 검색할 항목을 나타내는 임시 텍스트를 구성한 다음 코사인 측정을 통해 문서 벡터와 비교한다.\n\ndef searching(self, searchinglist):\n askVector = self.buildQueryVector(searchinglist)\n \n ratings = [utils.cosine(askVector, textVector) for textVector in self.documentVectors]\n ratings.sort(reverse=True)\n return ratings\n",
"_____no_output_____"
],
[
"\n# 소스 텍스트에서 언어를 탐지하는 데 사용\ndef _calculate_languages_ratios(text):\n # {'german':2, 'french':4, 'english':1}\n languages_ratios = {}\n \n tok = nltk.wordpunct_tokenize(text)\n wor = [word.lower() for word in tok]\n \n # 텍스트에서 고유 불용어의 발생을 계산\n for language in stopwords.fileids():\n stopwords_set = set(stopwords.words(language))\n words_set = set(words)\n common_elements = words_set.intersection(stopwords_set)\n languages_ratios[language] = len(common_elements)\n # 언어 \"점수\"\n return languages_ratios",
"_____no_output_____"
],
[
"def detect_language(text):\n ratios = _calculate_languages_ratios(text)\n most_rate_language = max(ratios, key = ratios.get)\n return most_rate_language\n\nif __name__=='__main__':\n text = '''\n WASHINGTON President Trump declared on Tuesday that he was withdrawing from the Iran nuclear deal, unraveling the signature foreign policy achievement of his predecessor Barack Obama, isolating the United States from its Western allies and sowing uncertainty before a risky nuclear negotiation with North Korea.The decision, while long anticipated and widely telegraphed, leaves the 2015 agreement reached by seven countries after more than two years of grueling negotiations in tatters. The United States will now reimpose the stringent sanctions it imposed on Iran before the deal and is considering new penalties.\n '''\n ",
"_____no_output_____"
],
[
"language = detect_language(text)\nprint(language)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e798131ce5e4c7a89515fcb7677c8f628683925d | 580,724 | ipynb | Jupyter Notebook | notebooks/evaluate.ipynb | kalufinnle/python_autocomplete | b51c7495a747c0756c0dc1458538243dad06d7b7 | [
"MIT"
]
| 1 | 2021-01-10T16:32:05.000Z | 2021-01-10T16:32:05.000Z | notebooks/evaluate.ipynb | hiteshkalwani/python_autocomplete | 70e528b5be22d276ca02affa38d6c71e472e2129 | [
"MIT"
]
| null | null | null | notebooks/evaluate.ipynb | hiteshkalwani/python_autocomplete | 70e528b5be22d276ca02affa38d6c71e472e2129 | [
"MIT"
]
| null | null | null | 707.337393 | 10,596 | 0.671569 | [
[
[
"[](https://github.com/lab-ml/python_autocomplete)\n[](https://colab.research.google.com/github/lab-ml/python_autocomplete/blob/master/notebooks/evaluate.ipynb)\n\n# Evaluate a model trained on predicting Python code\n\nThis notebook evaluates a model trained on Python code.\n\nHere's a link to [training notebook](https://github.com/lab-ml/python_autocomplete/blob/master/notebooks/train.ipynb)\n[](https://colab.research.google.com/github/lab-ml/python_autocomplete/blob/master/notebooks/train.ipynb)",
"_____no_output_____"
],
[
"### Install dependencies",
"_____no_output_____"
]
],
[
[
"%%capture\n!pip install labml labml_python_autocomplete",
"_____no_output_____"
]
],
[
[
"Imports",
"_____no_output_____"
]
],
[
[
"import string\n\nimport torch\nfrom torch import nn\n\nfrom labml import experiment, logger, lab\nfrom labml_helpers.module import Module\nfrom labml.logger import Text, Style\nfrom labml.utils.pytorch import get_modules\nfrom labml.utils.cache import cache\nfrom labml_helpers.datasets.text import TextDataset\n\nfrom python_autocomplete.train import Configs\nfrom python_autocomplete.evaluate import evaluate, anomalies, complete, Predictor",
"_____no_output_____"
]
],
[
[
"We load the model from a training run. For this demo I'm loading from a run I trained at home.\n\n[](https://web.lab-ml.com/run?uuid=39b03a1e454011ebbaff2b26e3148b3d)\n\n*If you want to try this on Colab you need to run this on the same space where you run the training, because models are saved locally.*",
"_____no_output_____"
]
],
[
[
"TRAINING_RUN_UUID = '39b03a1e454011ebbaff2b26e3148b3d'",
"_____no_output_____"
]
],
[
[
"We initialize `Configs` object defined in [`train.py`](https://github.com/lab-ml/python_autocomplete/blob/master/python_autocomplete/train.py).",
"_____no_output_____"
]
],
[
[
"conf = Configs()",
"_____no_output_____"
]
],
[
[
"Create a new experiment in evaluation mode. In evaluation mode a new training run is not created. ",
"_____no_output_____"
]
],
[
[
"experiment.evaluate()",
"_____no_output_____"
]
],
[
[
"Load custom configurations/hyper-parameters used in the training run.",
"_____no_output_____"
]
],
[
[
"custom_conf = experiment.load_configs(TRAINING_RUN_UUID)\ncustom_conf",
"_____no_output_____"
]
],
[
[
"Set the custom configurations",
"_____no_output_____"
]
],
[
[
"experiment.configs(conf, custom_conf)",
"_____no_output_____"
]
],
[
[
"Set models for saving and loading. This will load `conf.model` from the specified run.",
"_____no_output_____"
]
],
[
[
"experiment.add_pytorch_models({'model': conf.model})",
"_____no_output_____"
]
],
[
[
"Specify which run to load from",
"_____no_output_____"
]
],
[
[
"experiment.load(TRAINING_RUN_UUID)",
"_____no_output_____"
]
],
[
[
"Start the experiment",
"_____no_output_____"
]
],
[
[
"experiment.start()",
"_____no_output_____"
]
],
[
[
"Initialize the `Predictor` defined in [`evaluate.py`](https://github.com/lab-ml/python_autocomplete/blob/master/python_autocomplete/evaluate.py).\n\nWe load `stoi` and `itos` from cache, so that we don't have to read the dataset to generate them. `stoi` is the map for character to an integer index and `itos` is the map of integer to character map. These indexes are used in the model embeddings for each character.",
"_____no_output_____"
]
],
[
[
"p = Predictor(conf.model, cache('stoi', lambda: conf.text.stoi), cache('itos', lambda: conf.text.itos))",
"_____no_output_____"
]
],
[
[
"Set model to evaluation mode",
"_____no_output_____"
]
],
[
[
"_ = conf.model.eval()",
"_____no_output_____"
]
],
[
[
"A python prompt to test completion.",
"_____no_output_____"
]
],
[
[
"PROMPT = \"\"\"from torch import nn\n\nfrom labml_helpers.module import Module\nfrom labml_nn.lstm import LSTM\n\n\nclass LSTM(Module):\n def __init__(self, *,\n n_tokens: int,\n embedding_size: int,\n hidden_size int,\n n_layers int):\"\"\"",
"_____no_output_____"
]
],
[
[
"Get a token. `get_token` predicts character by character greedily (no beam search) until it find and end of token character (non alpha-numeric character).",
"_____no_output_____"
]
],
[
[
"%%time\nres = p.get_token(PROMPT)\nprint('\"' + res + '\"')",
"\"\n super\"\nCPU times: user 950 ms, sys: 34.7 ms, total: 984 ms\nWall time: 254 ms\n"
]
],
[
[
"Try another token",
"_____no_output_____"
]
],
[
[
"res = p.get_token(PROMPT + res)\nprint('\"' + res + '\"')",
"\"(LSTM\"\n"
]
],
[
[
"Load a sample python file to test our model",
"_____no_output_____"
]
],
[
[
"with open(str(lab.get_data_path() / 'sample.py'), 'r') as f:\n sample = f.read()\nprint(sample[-50:])",
"ckpoint()\n\n\nif __name__ == '__main__':\n main()\n\n"
]
],
[
[
"## Test the model on a sample python file\n\n`evaluate` function defined in\n[`evaluate.py`](https://github.com/lab-ml/python_autocomplete/blob/master/python_autocomplete/evaluate.py)\nwill predict token by token using the `Predictor`, and simulates an editor autocompletion.\n\nColors:\n* <span style=\"color:yellow\">yellow</span>: the token predicted is wrong and the user needs to type that character.\n* <span style=\"color:blue\">blue</span>: the token predicted is correct and the user selects it with a special key press, such as TAB or ENTER.\n* <span style=\"color:green\">green</span>: autocompleted characters based on the prediction",
"_____no_output_____"
]
],
[
[
"%%time\nevaluate(p, sample)",
"_____no_output_____"
]
],
[
[
"`accuracy` is the fraction of charactors predicted correctly. `key_strokes` is the number of key strokes required to write the code with help of the model and `length` is the number of characters in the code, that is the number of key strokes required without the model.\n\n*Note that this sample is a classic MNIST example, and the model must have overfitted to similar codes (exept for it's use of [LabML](https://github.com/lab-ml/labml) 😛).*",
"_____no_output_____"
],
[
"## Test anomalies in code\n\nWe run the model through the same sample code and visualize the probabilty of predicting each character.\n<span style=\"color:green\">green</span> means the probabilty of that character is high and \n<span style=\"color:red\">red</span> means the probability is low.",
"_____no_output_____"
]
],
[
[
"anomalies(p, sample)",
"_____no_output_____"
]
],
[
[
"Here we try to autocomplete 100 characters",
"_____no_output_____"
]
],
[
[
"sample = \"\"\"import torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torch.optim as optim\nimport torch.utils.data\nfrom torchvision import datasets, transforms\n\nfrom labml import lab\n\n\nclass Model(nn.Module):\n\"\"\"\n\ncomplete(p, sample, 100)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
e79816b2f80ab6d17fa15e5285e2f08946be982e | 354,581 | ipynb | Jupyter Notebook | Workshop/MLP_208.ipynb | ShepherdCode/ShepherdML | fd8d71c63f7bd788ea0052294d93e43246254a12 | [
"MIT"
]
| null | null | null | Workshop/MLP_208.ipynb | ShepherdCode/ShepherdML | fd8d71c63f7bd788ea0052294d93e43246254a12 | [
"MIT"
]
| 4 | 2020-03-24T18:05:09.000Z | 2020-12-22T17:42:54.000Z | Workshop/MLP_208.ipynb | ShepherdCode/ShepherdML | fd8d71c63f7bd788ea0052294d93e43246254a12 | [
"MIT"
]
| null | null | null | 132.652825 | 31,382 | 0.663688 | [
[
[
"# MLP 208\n* Operate on 16000 GenCode 34 seqs.\n* 5-way cross validation. Save best model per CV.\n* Report mean accuracy from final re-validation with best 5.\n* Use Adam with a learn rate decay schdule.",
"_____no_output_____"
]
],
[
[
"NC_FILENAME='ncRNA.gc34.processed.fasta'\nPC_FILENAME='pcRNA.gc34.processed.fasta'\nDATAPATH=\"\"\ntry:\n from google.colab import drive\n IN_COLAB = True\n PATH='/content/drive/'\n drive.mount(PATH)\n DATAPATH=PATH+'My Drive/data/' # must end in \"/\"\n NC_FILENAME = DATAPATH+NC_FILENAME\n PC_FILENAME = DATAPATH+PC_FILENAME\nexcept:\n IN_COLAB = False\n DATAPATH=\"\" \n\nEPOCHS=200\nSPLITS=5\nK=1\nVOCABULARY_SIZE=4**K+1 # e.g. K=3 => 64 DNA K-mers + 'NNN'\nEMBED_DIMEN=16\nFILENAME='MLP208'\nNEURONS=32",
"Drive already mounted at /content/drive/; to attempt to forcibly remount, call drive.mount(\"/content/drive/\", force_remount=True).\n"
],
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom sklearn.model_selection import ShuffleSplit\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.model_selection import RepeatedKFold\nfrom sklearn.model_selection import StratifiedKFold\nimport tensorflow as tf\nfrom tensorflow import keras\nfrom keras.wrappers.scikit_learn import KerasRegressor\nfrom keras.models import Sequential\nfrom keras.layers import Bidirectional\nfrom keras.layers import GRU\nfrom keras.layers import Dense\nfrom keras.layers import LayerNormalization\nimport time\ndt='float32'\ntf.keras.backend.set_floatx(dt)",
"_____no_output_____"
]
],
[
[
"## Build model",
"_____no_output_____"
]
],
[
[
"def compile_model(model):\n adam_default_learn_rate = 0.001\n schedule = tf.keras.optimizers.schedules.ExponentialDecay(\n initial_learning_rate = adam_default_learn_rate*10,\n #decay_steps=100000, decay_rate=0.96, staircase=True)\n decay_steps=10000, decay_rate=0.99, staircase=True)\n # learn rate = initial_learning_rate * decay_rate ^ (step / decay_steps)\n opt = tf.keras.optimizers.Adam(learning_rate=schedule)\n bc=tf.keras.losses.BinaryCrossentropy(from_logits=False)\n print(\"COMPILE...\")\n model.compile(loss=bc, optimizer=opt, metrics=[\"accuracy\"])\n print(\"...COMPILED\")\n return model\n\ndef build_model(maxlen):\n act=\"elu\"\n #embed_layer = keras.layers.Embedding(\n # VOCABULARY_SIZE,EMBED_DIMEN,input_length=maxlen);\n dense1_layer = keras.layers.Dense(NEURONS, activation=act,dtype=dt,\n input_dim=VOCABULARY_SIZE)\n dense2_layer = keras.layers.Dense(NEURONS, activation=act,dtype=dt)\n #dense3_layer = keras.layers.Dense(NEURONS, activation=act,dtype=dt)\n output_layer = keras.layers.Dense(1, activation=\"sigmoid\",dtype=dt)\n mlp = keras.models.Sequential()\n #mlp.add(embed_layer)\n mlp.add(dense1_layer)\n mlp.add(dense2_layer)\n #mlp.add(dense3_layer)\n mlp.add(output_layer)\n mlpc = compile_model(mlp)\n return mlpc",
"_____no_output_____"
]
],
[
[
"## Load and partition sequences",
"_____no_output_____"
]
],
[
[
"# Assume file was preprocessed to contain one line per seq.\n# Prefer Pandas dataframe but df does not support append.\n# For conversion to tensor, must avoid python lists.\ndef load_fasta(filename,label):\n DEFLINE='>'\n labels=[]\n seqs=[]\n lens=[]\n nums=[]\n num=0\n with open (filename,'r') as infile:\n for line in infile:\n if line[0]!=DEFLINE:\n seq=line.rstrip()\n num += 1 # first seqnum is 1\n seqlen=len(seq)\n nums.append(num)\n labels.append(label)\n seqs.append(seq)\n lens.append(seqlen)\n df1=pd.DataFrame(nums,columns=['seqnum'])\n df2=pd.DataFrame(labels,columns=['class'])\n df3=pd.DataFrame(seqs,columns=['sequence'])\n df4=pd.DataFrame(lens,columns=['seqlen'])\n df=pd.concat((df1,df2,df3,df4),axis=1)\n return df\n\ndef separate_X_and_y(data):\n y= data[['class']].copy()\n X= data.drop(columns=['class','seqnum','seqlen'])\n return (X,y)\n\n",
"_____no_output_____"
]
],
[
[
"## Make K-mers",
"_____no_output_____"
]
],
[
[
"def make_kmer_table(K):\n npad='N'*K\n shorter_kmers=['']\n for i in range(K):\n longer_kmers=[]\n for mer in shorter_kmers:\n longer_kmers.append(mer+'A')\n longer_kmers.append(mer+'C')\n longer_kmers.append(mer+'G')\n longer_kmers.append(mer+'T')\n shorter_kmers = longer_kmers\n all_kmers = shorter_kmers\n kmer_dict = {}\n kmer_dict[npad]=0\n value=1\n for mer in all_kmers:\n kmer_dict[mer]=value\n value += 1\n return kmer_dict\n\nKMER_TABLE=make_kmer_table(K)\n\ndef strings_to_vectors(data,uniform_len):\n all_seqs=[]\n for seq in data['sequence']:\n i=0\n seqlen=len(seq)\n kmers=[]\n while i < seqlen-K+1 -1: # stop at minus one for spaced seed\n #kmer=seq[i:i+2]+seq[i+3:i+5] # SPACED SEED 2/1/2 for K=4\n kmer=seq[i:i+K] \n i += 1\n value=KMER_TABLE[kmer]\n kmers.append(value)\n pad_val=0\n while i < uniform_len:\n kmers.append(pad_val)\n i += 1\n all_seqs.append(kmers)\n pd2d=pd.DataFrame(all_seqs)\n return pd2d # return 2D dataframe, uniform dimensions",
"_____no_output_____"
],
[
"def make_kmers(MAXLEN,train_set):\n (X_train_all,y_train_all)=separate_X_and_y(train_set)\n X_train_kmers=strings_to_vectors(X_train_all,MAXLEN)\n # From pandas dataframe to numpy to list to numpy\n num_seqs=len(X_train_kmers)\n tmp_seqs=[]\n for i in range(num_seqs):\n kmer_sequence=X_train_kmers.iloc[i]\n tmp_seqs.append(kmer_sequence)\n X_train_kmers=np.array(tmp_seqs)\n tmp_seqs=None\n labels=y_train_all.to_numpy()\n return (X_train_kmers,labels)",
"_____no_output_____"
],
[
"def make_frequencies(Xin):\n Xout=[]\n VOCABULARY_SIZE= 4**K + 1 # plus one for 'NNN'\n for seq in Xin:\n freqs =[0] * VOCABULARY_SIZE\n total = 0\n for kmerval in seq:\n freqs[kmerval] += 1\n total += 1\n for c in range(VOCABULARY_SIZE):\n freqs[c] = freqs[c]/total\n Xout.append(freqs)\n Xnum = np.asarray(Xout)\n return (Xnum)\ndef make_slice(data_set,min_len,max_len):\n slice = data_set.query('seqlen <= '+str(max_len)+' & seqlen>= '+str(min_len))\n return slice",
"_____no_output_____"
]
],
[
[
"## Cross validation",
"_____no_output_____"
]
],
[
[
"def do_cross_validation(X,y,given_model):\n cv_scores = []\n fold=0\n splitter = ShuffleSplit(n_splits=SPLITS, test_size=0.1, random_state=37863)\n for train_index,valid_index in splitter.split(X):\n fold += 1\n X_train=X[train_index] # use iloc[] for dataframe\n y_train=y[train_index]\n X_valid=X[valid_index]\n y_valid=y[valid_index] \n # Avoid continually improving the same model.\n model = compile_model(keras.models.clone_model(given_model))\n bestname=DATAPATH+FILENAME+\".cv.\"+str(fold)+\".best\"\n mycallbacks = [keras.callbacks.ModelCheckpoint(\n filepath=bestname, save_best_only=True, \n monitor='val_accuracy', mode='max')] \n print(\"FIT\")\n start_time=time.time()\n history=model.fit(X_train, y_train, # batch_size=10, default=32 works nicely\n epochs=EPOCHS, verbose=1, # verbose=1 for ascii art, verbose=0 for none\n callbacks=mycallbacks,\n validation_data=(X_valid,y_valid) )\n end_time=time.time()\n elapsed_time=(end_time-start_time) \n print(\"Fold %d, %d epochs, %d sec\"%(fold,EPOCHS,elapsed_time))\n pd.DataFrame(history.history).plot(figsize=(8,5))\n plt.grid(True)\n plt.gca().set_ylim(0,1)\n plt.show()\n best_model=keras.models.load_model(bestname)\n scores = best_model.evaluate(X_valid, y_valid, verbose=0)\n print(\"%s: %.2f%%\" % (best_model.metrics_names[1], scores[1]*100))\n cv_scores.append(scores[1] * 100) \n print()\n print(\"%d-way Cross Validation mean %.2f%% (+/- %.2f%%)\" % (fold, np.mean(cv_scores), np.std(cv_scores)))",
"_____no_output_____"
]
],
[
[
"## Train on RNA lengths 200-1Kb",
"_____no_output_____"
]
],
[
[
"MINLEN=200\nMAXLEN=1000\nprint(\"Load data from files.\")\nnc_seq=load_fasta(NC_FILENAME,0)\npc_seq=load_fasta(PC_FILENAME,1)\ntrain_set=pd.concat((nc_seq,pc_seq),axis=0)\nnc_seq=None\npc_seq=None\nprint(\"Ready: train_set\")\n#train_set\nprint (\"Compile the model\")\nmodel=build_model(MAXLEN)\nprint (\"Summarize the model\")\nprint(model.summary()) # Print this only once\nmodel.save(DATAPATH+FILENAME+'.model')\nprint (\"Data prep\")\nsubset=make_slice(train_set,MINLEN,MAXLEN)# One array to two: X and y\nprint (\"Data reshape\")\n(X_train,y_train)=make_kmers(MAXLEN,subset)\nX_train=make_frequencies(X_train)",
"Load data from files.\nReady: train_set\nCompile the model\nCOMPILE...\n...COMPILED\nSummarize the model\nModel: \"sequential_1\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_3 (Dense) (None, 32) 192 \n_________________________________________________________________\ndense_4 (Dense) (None, 32) 1056 \n_________________________________________________________________\ndense_5 (Dense) (None, 1) 33 \n=================================================================\nTotal params: 1,281\nTrainable params: 1,281\nNon-trainable params: 0\n_________________________________________________________________\nNone\nINFO:tensorflow:Assets written to: /content/drive/My Drive/data/MLP208.model/assets\nData prep\nData reshape\n"
],
[
"print (\"Cross valiation\")\ndo_cross_validation(X_train,y_train,model) \nprint (\"Done\")",
"Cross valiation\nCOMPILE...\n...COMPILED\nFIT\nEpoch 1/200\n434/453 [===========================>..] - ETA: 0s - loss: 0.5565 - accuracy: 0.7117INFO:tensorflow:Assets written to: /content/drive/My Drive/data/MLP208.cv.1.best/assets\n453/453 [==============================] - 2s 4ms/step - loss: 0.5565 - accuracy: 0.7119 - val_loss: 0.5254 - val_accuracy: 0.7387\nEpoch 2/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5465 - accuracy: 0.7206 - val_loss: 0.5226 - val_accuracy: 0.7381\nEpoch 3/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5481 - accuracy: 0.7172 - val_loss: 0.5295 - val_accuracy: 0.7281\nEpoch 4/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5428 - accuracy: 0.7223 - val_loss: 0.5512 - val_accuracy: 0.7169\nEpoch 5/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5464 - accuracy: 0.7195 - val_loss: 0.5237 - val_accuracy: 0.7343\nEpoch 6/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5435 - accuracy: 0.7214 - val_loss: 0.5268 - val_accuracy: 0.7331\nEpoch 7/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5430 - accuracy: 0.7186 - val_loss: 0.5251 - val_accuracy: 0.7325\nEpoch 8/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5416 - accuracy: 0.7219 - val_loss: 0.5298 - val_accuracy: 0.7356\nEpoch 9/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5424 - accuracy: 0.7201 - val_loss: 0.5321 - val_accuracy: 0.7331\nEpoch 10/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5413 - accuracy: 0.7236 - val_loss: 0.5238 - val_accuracy: 0.7343\nEpoch 11/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5416 - accuracy: 0.7208 - val_loss: 0.5841 - val_accuracy: 0.6946\nEpoch 12/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5414 - accuracy: 0.7223 - val_loss: 0.5320 - val_accuracy: 0.7318\nEpoch 13/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5405 - accuracy: 0.7221 - val_loss: 0.5438 - val_accuracy: 0.7287\nEpoch 14/200\n443/453 [============================>.] - ETA: 0s - loss: 0.5405 - accuracy: 0.7202INFO:tensorflow:Assets written to: /content/drive/My Drive/data/MLP208.cv.1.best/assets\n453/453 [==============================] - 2s 4ms/step - loss: 0.5395 - accuracy: 0.7213 - val_loss: 0.5251 - val_accuracy: 0.7449\nEpoch 15/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5398 - accuracy: 0.7244 - val_loss: 0.5225 - val_accuracy: 0.7399\nEpoch 16/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5396 - accuracy: 0.7252 - val_loss: 0.5271 - val_accuracy: 0.7343\nEpoch 17/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5400 - accuracy: 0.7228 - val_loss: 0.5282 - val_accuracy: 0.7318\nEpoch 18/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5398 - accuracy: 0.7261 - val_loss: 0.5270 - val_accuracy: 0.7343\nEpoch 19/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5379 - accuracy: 0.7241 - val_loss: 0.5292 - val_accuracy: 0.7381\nEpoch 20/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5388 - accuracy: 0.7221 - val_loss: 0.5321 - val_accuracy: 0.7399\nEpoch 21/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5384 - accuracy: 0.7215 - val_loss: 0.5220 - val_accuracy: 0.7343\nEpoch 22/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5366 - accuracy: 0.7250 - val_loss: 0.5253 - val_accuracy: 0.7368\nEpoch 23/200\n453/453 [==============================] - 2s 3ms/step - loss: 0.5370 - accuracy: 0.7243 - val_loss: 0.5450 - val_accuracy: 0.7244\nEpoch 24/200\n453/453 [==============================] - 2s 3ms/step - loss: 0.5388 - accuracy: 0.7234 - val_loss: 0.5277 - val_accuracy: 0.7294\nEpoch 25/200\n453/453 [==============================] - 2s 3ms/step - loss: 0.5376 - accuracy: 0.7248 - val_loss: 0.5237 - val_accuracy: 0.7368\nEpoch 26/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5376 - accuracy: 0.7246 - val_loss: 0.5284 - val_accuracy: 0.7356\nEpoch 27/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5354 - accuracy: 0.7266 - val_loss: 0.5237 - val_accuracy: 0.7362\nEpoch 28/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5353 - accuracy: 0.7272 - val_loss: 0.5356 - val_accuracy: 0.7232\nEpoch 29/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5362 - accuracy: 0.7265 - val_loss: 0.5442 - val_accuracy: 0.7300\nEpoch 30/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5356 - accuracy: 0.7266 - val_loss: 0.5226 - val_accuracy: 0.7381\nEpoch 31/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5347 - accuracy: 0.7287 - val_loss: 0.5247 - val_accuracy: 0.7325\nEpoch 32/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5346 - accuracy: 0.7264 - val_loss: 0.5275 - val_accuracy: 0.7287\nEpoch 33/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5361 - accuracy: 0.7257 - val_loss: 0.5220 - val_accuracy: 0.7368\nEpoch 34/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5361 - accuracy: 0.7237 - val_loss: 0.5256 - val_accuracy: 0.7337\nEpoch 35/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5341 - accuracy: 0.7263 - val_loss: 0.5214 - val_accuracy: 0.7356\nEpoch 36/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5337 - accuracy: 0.7272 - val_loss: 0.5313 - val_accuracy: 0.7362\nEpoch 37/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5349 - accuracy: 0.7266 - val_loss: 0.5269 - val_accuracy: 0.7356\nEpoch 38/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5353 - accuracy: 0.7262 - val_loss: 0.5215 - val_accuracy: 0.7405\nEpoch 39/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5340 - accuracy: 0.7290 - val_loss: 0.5251 - val_accuracy: 0.7343\nEpoch 40/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5343 - accuracy: 0.7285 - val_loss: 0.5336 - val_accuracy: 0.7244\nEpoch 41/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5341 - accuracy: 0.7252 - val_loss: 0.5225 - val_accuracy: 0.7418\nEpoch 42/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5342 - accuracy: 0.7264 - val_loss: 0.5208 - val_accuracy: 0.7399\nEpoch 43/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5343 - accuracy: 0.7277 - val_loss: 0.5233 - val_accuracy: 0.7318\nEpoch 44/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5344 - accuracy: 0.7301 - val_loss: 0.5245 - val_accuracy: 0.7281\nEpoch 45/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5336 - accuracy: 0.7259 - val_loss: 0.5228 - val_accuracy: 0.7368\nEpoch 46/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5329 - accuracy: 0.7272 - val_loss: 0.5242 - val_accuracy: 0.7387\nEpoch 47/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5344 - accuracy: 0.7277 - val_loss: 0.5268 - val_accuracy: 0.7300\nEpoch 48/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5352 - accuracy: 0.7241 - val_loss: 0.5232 - val_accuracy: 0.7362\nEpoch 49/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5322 - accuracy: 0.7286 - val_loss: 0.5295 - val_accuracy: 0.7387\nEpoch 50/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5321 - accuracy: 0.7268 - val_loss: 0.5236 - val_accuracy: 0.7374\nEpoch 51/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5324 - accuracy: 0.7289 - val_loss: 0.5198 - val_accuracy: 0.7405\nEpoch 52/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5335 - accuracy: 0.7308 - val_loss: 0.5266 - val_accuracy: 0.7343\nEpoch 53/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5327 - accuracy: 0.7291 - val_loss: 0.5269 - val_accuracy: 0.7374\nEpoch 54/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5325 - accuracy: 0.7287 - val_loss: 0.5197 - val_accuracy: 0.7405\nEpoch 55/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5326 - accuracy: 0.7279 - val_loss: 0.5322 - val_accuracy: 0.7312\nEpoch 56/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5320 - accuracy: 0.7279 - val_loss: 0.5230 - val_accuracy: 0.7337\nEpoch 57/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5317 - accuracy: 0.7284 - val_loss: 0.5225 - val_accuracy: 0.7399\nEpoch 58/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5326 - accuracy: 0.7272 - val_loss: 0.5191 - val_accuracy: 0.7436\nEpoch 59/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5315 - accuracy: 0.7305 - val_loss: 0.5352 - val_accuracy: 0.7219\nEpoch 60/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5311 - accuracy: 0.7269 - val_loss: 0.5264 - val_accuracy: 0.7318\nEpoch 61/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5330 - accuracy: 0.7292 - val_loss: 0.5261 - val_accuracy: 0.7374\nEpoch 62/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5309 - accuracy: 0.7297 - val_loss: 0.5230 - val_accuracy: 0.7405\nEpoch 63/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5326 - accuracy: 0.7288 - val_loss: 0.5209 - val_accuracy: 0.7300\nEpoch 64/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5323 - accuracy: 0.7282 - val_loss: 0.5244 - val_accuracy: 0.7393\nEpoch 65/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5319 - accuracy: 0.7277 - val_loss: 0.5180 - val_accuracy: 0.7424\nEpoch 66/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5324 - accuracy: 0.7289 - val_loss: 0.5242 - val_accuracy: 0.7436\nEpoch 67/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5303 - accuracy: 0.7287 - val_loss: 0.5194 - val_accuracy: 0.7436\nEpoch 68/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5309 - accuracy: 0.7281 - val_loss: 0.5273 - val_accuracy: 0.7393\nEpoch 69/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5302 - accuracy: 0.7292 - val_loss: 0.5239 - val_accuracy: 0.7362\nEpoch 70/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5314 - accuracy: 0.7285 - val_loss: 0.5217 - val_accuracy: 0.7362\nEpoch 71/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5303 - accuracy: 0.7287 - val_loss: 0.5316 - val_accuracy: 0.7331\nEpoch 72/200\n443/453 [============================>.] - ETA: 0s - loss: 0.5312 - accuracy: 0.7288INFO:tensorflow:Assets written to: /content/drive/My Drive/data/MLP208.cv.1.best/assets\n453/453 [==============================] - 2s 4ms/step - loss: 0.5319 - accuracy: 0.7280 - val_loss: 0.5288 - val_accuracy: 0.7474\nEpoch 73/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5315 - accuracy: 0.7288 - val_loss: 0.5203 - val_accuracy: 0.7368\nEpoch 74/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5321 - accuracy: 0.7275 - val_loss: 0.5163 - val_accuracy: 0.7424\nEpoch 75/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5315 - accuracy: 0.7312 - val_loss: 0.5170 - val_accuracy: 0.7418\nEpoch 76/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5307 - accuracy: 0.7282 - val_loss: 0.5210 - val_accuracy: 0.7393\nEpoch 77/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5318 - accuracy: 0.7306 - val_loss: 0.5167 - val_accuracy: 0.7455\nEpoch 78/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5300 - accuracy: 0.7297 - val_loss: 0.5194 - val_accuracy: 0.7405\nEpoch 79/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5304 - accuracy: 0.7284 - val_loss: 0.5242 - val_accuracy: 0.7312\nEpoch 80/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5308 - accuracy: 0.7305 - val_loss: 0.5286 - val_accuracy: 0.7430\nEpoch 81/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5302 - accuracy: 0.7283 - val_loss: 0.5379 - val_accuracy: 0.7381\nEpoch 82/200\n450/453 [============================>.] - ETA: 0s - loss: 0.5309 - accuracy: 0.7295INFO:tensorflow:Assets written to: /content/drive/My Drive/data/MLP208.cv.1.best/assets\n453/453 [==============================] - 2s 4ms/step - loss: 0.5307 - accuracy: 0.7297 - val_loss: 0.5193 - val_accuracy: 0.7480\nEpoch 83/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5314 - accuracy: 0.7310 - val_loss: 0.5181 - val_accuracy: 0.7381\nEpoch 84/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5298 - accuracy: 0.7317 - val_loss: 0.5227 - val_accuracy: 0.7362\nEpoch 85/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5308 - accuracy: 0.7299 - val_loss: 0.5195 - val_accuracy: 0.7412\nEpoch 86/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5300 - accuracy: 0.7290 - val_loss: 0.5204 - val_accuracy: 0.7381\nEpoch 87/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5304 - accuracy: 0.7305 - val_loss: 0.5194 - val_accuracy: 0.7381\nEpoch 88/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5308 - accuracy: 0.7280 - val_loss: 0.5288 - val_accuracy: 0.7381\nEpoch 89/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5313 - accuracy: 0.7294 - val_loss: 0.5231 - val_accuracy: 0.7368\nEpoch 90/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5295 - accuracy: 0.7305 - val_loss: 0.5222 - val_accuracy: 0.7418\nEpoch 91/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5295 - accuracy: 0.7315 - val_loss: 0.5225 - val_accuracy: 0.7374\nEpoch 92/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5303 - accuracy: 0.7299 - val_loss: 0.5229 - val_accuracy: 0.7461\nEpoch 93/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5300 - accuracy: 0.7282 - val_loss: 0.5259 - val_accuracy: 0.7362\nEpoch 94/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5293 - accuracy: 0.7305 - val_loss: 0.5285 - val_accuracy: 0.7312\nEpoch 95/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5297 - accuracy: 0.7279 - val_loss: 0.5261 - val_accuracy: 0.7393\nEpoch 96/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5299 - accuracy: 0.7280 - val_loss: 0.5222 - val_accuracy: 0.7356\nEpoch 97/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5291 - accuracy: 0.7304 - val_loss: 0.5297 - val_accuracy: 0.7331\nEpoch 98/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5294 - accuracy: 0.7304 - val_loss: 0.5197 - val_accuracy: 0.7405\nEpoch 99/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5292 - accuracy: 0.7322 - val_loss: 0.5245 - val_accuracy: 0.7356\nEpoch 100/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5299 - accuracy: 0.7301 - val_loss: 0.5172 - val_accuracy: 0.7430\nEpoch 101/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5293 - accuracy: 0.7329 - val_loss: 0.5287 - val_accuracy: 0.7430\nEpoch 102/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5285 - accuracy: 0.7312 - val_loss: 0.5193 - val_accuracy: 0.7424\nEpoch 103/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5288 - accuracy: 0.7302 - val_loss: 0.5210 - val_accuracy: 0.7356\nEpoch 104/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5283 - accuracy: 0.7289 - val_loss: 0.5239 - val_accuracy: 0.7474\nEpoch 105/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5299 - accuracy: 0.7307 - val_loss: 0.5208 - val_accuracy: 0.7368\nEpoch 106/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5290 - accuracy: 0.7305 - val_loss: 0.5203 - val_accuracy: 0.7387\nEpoch 107/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5295 - accuracy: 0.7293 - val_loss: 0.5204 - val_accuracy: 0.7405\nEpoch 108/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5300 - accuracy: 0.7317 - val_loss: 0.5203 - val_accuracy: 0.7393\nEpoch 109/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5291 - accuracy: 0.7299 - val_loss: 0.5243 - val_accuracy: 0.7412\nEpoch 110/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5300 - accuracy: 0.7303 - val_loss: 0.5205 - val_accuracy: 0.7430\nEpoch 111/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5288 - accuracy: 0.7317 - val_loss: 0.5244 - val_accuracy: 0.7393\nEpoch 112/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5290 - accuracy: 0.7308 - val_loss: 0.5260 - val_accuracy: 0.7318\nEpoch 113/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5286 - accuracy: 0.7339 - val_loss: 0.5211 - val_accuracy: 0.7449\nEpoch 114/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5287 - accuracy: 0.7301 - val_loss: 0.5176 - val_accuracy: 0.7455\nEpoch 115/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5294 - accuracy: 0.7288 - val_loss: 0.5232 - val_accuracy: 0.7405\nEpoch 116/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5284 - accuracy: 0.7295 - val_loss: 0.5236 - val_accuracy: 0.7424\nEpoch 117/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5281 - accuracy: 0.7328 - val_loss: 0.5193 - val_accuracy: 0.7325\nEpoch 118/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5272 - accuracy: 0.7326 - val_loss: 0.5270 - val_accuracy: 0.7337\nEpoch 119/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5280 - accuracy: 0.7299 - val_loss: 0.5274 - val_accuracy: 0.7412\nEpoch 120/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5287 - accuracy: 0.7332 - val_loss: 0.5311 - val_accuracy: 0.7393\nEpoch 121/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5283 - accuracy: 0.7310 - val_loss: 0.5220 - val_accuracy: 0.7374\nEpoch 122/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5292 - accuracy: 0.7324 - val_loss: 0.5242 - val_accuracy: 0.7368\nEpoch 123/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5279 - accuracy: 0.7305 - val_loss: 0.5187 - val_accuracy: 0.7424\nEpoch 124/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5280 - accuracy: 0.7321 - val_loss: 0.5261 - val_accuracy: 0.7306\nEpoch 125/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5270 - accuracy: 0.7305 - val_loss: 0.5174 - val_accuracy: 0.7461\nEpoch 126/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5286 - accuracy: 0.7331 - val_loss: 0.5234 - val_accuracy: 0.7449\nEpoch 127/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5265 - accuracy: 0.7355 - val_loss: 0.5193 - val_accuracy: 0.7405\nEpoch 128/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5273 - accuracy: 0.7350 - val_loss: 0.5218 - val_accuracy: 0.7405\nEpoch 129/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5283 - accuracy: 0.7308 - val_loss: 0.5433 - val_accuracy: 0.7312\nEpoch 130/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5288 - accuracy: 0.7313 - val_loss: 0.5174 - val_accuracy: 0.7412\nEpoch 131/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5289 - accuracy: 0.7314 - val_loss: 0.5232 - val_accuracy: 0.7393\nEpoch 132/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5273 - accuracy: 0.7324 - val_loss: 0.5158 - val_accuracy: 0.7467\nEpoch 133/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5275 - accuracy: 0.7346 - val_loss: 0.5188 - val_accuracy: 0.7405\nEpoch 134/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5266 - accuracy: 0.7339 - val_loss: 0.5195 - val_accuracy: 0.7436\nEpoch 135/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5272 - accuracy: 0.7323 - val_loss: 0.5177 - val_accuracy: 0.7374\nEpoch 136/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5276 - accuracy: 0.7323 - val_loss: 0.5217 - val_accuracy: 0.7412\nEpoch 137/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5286 - accuracy: 0.7306 - val_loss: 0.5183 - val_accuracy: 0.7412\nEpoch 138/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5265 - accuracy: 0.7315 - val_loss: 0.5187 - val_accuracy: 0.7393\nEpoch 139/200\n438/453 [============================>.] - ETA: 0s - loss: 0.5275 - accuracy: 0.7324INFO:tensorflow:Assets written to: /content/drive/My Drive/data/MLP208.cv.1.best/assets\n453/453 [==============================] - 2s 4ms/step - loss: 0.5272 - accuracy: 0.7321 - val_loss: 0.5138 - val_accuracy: 0.7492\nEpoch 140/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5284 - accuracy: 0.7327 - val_loss: 0.5228 - val_accuracy: 0.7467\nEpoch 141/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5265 - accuracy: 0.7329 - val_loss: 0.5224 - val_accuracy: 0.7325\nEpoch 142/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5281 - accuracy: 0.7317 - val_loss: 0.5220 - val_accuracy: 0.7412\nEpoch 143/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5275 - accuracy: 0.7333 - val_loss: 0.5167 - val_accuracy: 0.7480\nEpoch 144/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5279 - accuracy: 0.7323 - val_loss: 0.5188 - val_accuracy: 0.7405\nEpoch 145/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5274 - accuracy: 0.7326 - val_loss: 0.5234 - val_accuracy: 0.7412\nEpoch 146/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5272 - accuracy: 0.7306 - val_loss: 0.5252 - val_accuracy: 0.7467\nEpoch 147/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5262 - accuracy: 0.7345 - val_loss: 0.5200 - val_accuracy: 0.7399\nEpoch 148/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5268 - accuracy: 0.7339 - val_loss: 0.5196 - val_accuracy: 0.7424\nEpoch 149/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5267 - accuracy: 0.7349 - val_loss: 0.5220 - val_accuracy: 0.7331\nEpoch 150/200\n444/453 [============================>.] - ETA: 0s - loss: 0.5253 - accuracy: 0.7341INFO:tensorflow:Assets written to: /content/drive/My Drive/data/MLP208.cv.1.best/assets\n453/453 [==============================] - 2s 4ms/step - loss: 0.5259 - accuracy: 0.7337 - val_loss: 0.5214 - val_accuracy: 0.7511\nEpoch 151/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5257 - accuracy: 0.7321 - val_loss: 0.5313 - val_accuracy: 0.7449\nEpoch 152/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5278 - accuracy: 0.7329 - val_loss: 0.5240 - val_accuracy: 0.7362\nEpoch 153/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5266 - accuracy: 0.7361 - val_loss: 0.5230 - val_accuracy: 0.7387\nEpoch 154/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5275 - accuracy: 0.7319 - val_loss: 0.5147 - val_accuracy: 0.7498\nEpoch 155/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5257 - accuracy: 0.7340 - val_loss: 0.5214 - val_accuracy: 0.7393\nEpoch 156/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5258 - accuracy: 0.7348 - val_loss: 0.5204 - val_accuracy: 0.7449\nEpoch 157/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5263 - accuracy: 0.7340 - val_loss: 0.5259 - val_accuracy: 0.7467\nEpoch 158/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5260 - accuracy: 0.7329 - val_loss: 0.5189 - val_accuracy: 0.7498\nEpoch 159/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5265 - accuracy: 0.7278 - val_loss: 0.5209 - val_accuracy: 0.7349\nEpoch 160/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5265 - accuracy: 0.7310 - val_loss: 0.5232 - val_accuracy: 0.7474\nEpoch 161/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5255 - accuracy: 0.7316 - val_loss: 0.5238 - val_accuracy: 0.7480\nEpoch 162/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5263 - accuracy: 0.7333 - val_loss: 0.5157 - val_accuracy: 0.7455\nEpoch 163/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5258 - accuracy: 0.7328 - val_loss: 0.5251 - val_accuracy: 0.7467\nEpoch 164/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5270 - accuracy: 0.7328 - val_loss: 0.5197 - val_accuracy: 0.7461\nEpoch 165/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5261 - accuracy: 0.7346 - val_loss: 0.5152 - val_accuracy: 0.7430\nEpoch 166/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5258 - accuracy: 0.7321 - val_loss: 0.5200 - val_accuracy: 0.7393\nEpoch 167/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5256 - accuracy: 0.7340 - val_loss: 0.5217 - val_accuracy: 0.7393\nEpoch 168/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5251 - accuracy: 0.7335 - val_loss: 0.5180 - val_accuracy: 0.7480\nEpoch 169/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5251 - accuracy: 0.7333 - val_loss: 0.5238 - val_accuracy: 0.7300\nEpoch 170/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5259 - accuracy: 0.7344 - val_loss: 0.5263 - val_accuracy: 0.7443\nEpoch 171/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5255 - accuracy: 0.7333 - val_loss: 0.5197 - val_accuracy: 0.7443\nEpoch 172/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5254 - accuracy: 0.7353 - val_loss: 0.5224 - val_accuracy: 0.7387\nEpoch 173/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5264 - accuracy: 0.7335 - val_loss: 0.5176 - val_accuracy: 0.7480\nEpoch 174/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5245 - accuracy: 0.7341 - val_loss: 0.5182 - val_accuracy: 0.7412\nEpoch 175/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5249 - accuracy: 0.7333 - val_loss: 0.5235 - val_accuracy: 0.7412\nEpoch 176/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5260 - accuracy: 0.7310 - val_loss: 0.5191 - val_accuracy: 0.7318\nEpoch 177/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5240 - accuracy: 0.7335 - val_loss: 0.5172 - val_accuracy: 0.7436\nEpoch 178/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5248 - accuracy: 0.7324 - val_loss: 0.5165 - val_accuracy: 0.7381\nEpoch 179/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5254 - accuracy: 0.7322 - val_loss: 0.5194 - val_accuracy: 0.7455\nEpoch 180/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5261 - accuracy: 0.7324 - val_loss: 0.5193 - val_accuracy: 0.7430\nEpoch 181/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5235 - accuracy: 0.7333 - val_loss: 0.5160 - val_accuracy: 0.7505\nEpoch 182/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5254 - accuracy: 0.7346 - val_loss: 0.5183 - val_accuracy: 0.7455\nEpoch 183/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5260 - accuracy: 0.7331 - val_loss: 0.5207 - val_accuracy: 0.7480\nEpoch 184/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5247 - accuracy: 0.7370 - val_loss: 0.5218 - val_accuracy: 0.7362\nEpoch 185/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5248 - accuracy: 0.7360 - val_loss: 0.5213 - val_accuracy: 0.7362\nEpoch 186/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5247 - accuracy: 0.7320 - val_loss: 0.5241 - val_accuracy: 0.7455\nEpoch 187/200\n449/453 [============================>.] - ETA: 0s - loss: 0.5250 - accuracy: 0.7341INFO:tensorflow:Assets written to: /content/drive/My Drive/data/MLP208.cv.1.best/assets\n453/453 [==============================] - 2s 4ms/step - loss: 0.5248 - accuracy: 0.7341 - val_loss: 0.5198 - val_accuracy: 0.7536\nEpoch 188/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5240 - accuracy: 0.7363 - val_loss: 0.5249 - val_accuracy: 0.7467\nEpoch 189/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5244 - accuracy: 0.7335 - val_loss: 0.5200 - val_accuracy: 0.7443\nEpoch 190/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5254 - accuracy: 0.7329 - val_loss: 0.5160 - val_accuracy: 0.7424\nEpoch 191/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5241 - accuracy: 0.7346 - val_loss: 0.5207 - val_accuracy: 0.7374\nEpoch 192/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5245 - accuracy: 0.7349 - val_loss: 0.5208 - val_accuracy: 0.7405\nEpoch 193/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5260 - accuracy: 0.7339 - val_loss: 0.5180 - val_accuracy: 0.7443\nEpoch 194/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5241 - accuracy: 0.7357 - val_loss: 0.5158 - val_accuracy: 0.7480\nEpoch 195/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5248 - accuracy: 0.7337 - val_loss: 0.5143 - val_accuracy: 0.7505\nEpoch 196/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5246 - accuracy: 0.7363 - val_loss: 0.5164 - val_accuracy: 0.7443\nEpoch 197/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5242 - accuracy: 0.7337 - val_loss: 0.5203 - val_accuracy: 0.7455\nEpoch 198/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5240 - accuracy: 0.7322 - val_loss: 0.5227 - val_accuracy: 0.7443\nEpoch 199/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5241 - accuracy: 0.7344 - val_loss: 0.5161 - val_accuracy: 0.7430\nEpoch 200/200\n453/453 [==============================] - 1s 3ms/step - loss: 0.5247 - accuracy: 0.7326 - val_loss: 0.5179 - val_accuracy: 0.7511\nFold 1, 200 epochs, 270 sec\n"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
]
|
e7982b119d16377d504245f4dc39e2465b6c6993 | 18,405 | ipynb | Jupyter Notebook | _notebooks/2021-11-14-jax-random.ipynb | abap34/my-website | 7a2e76ceeae700a302f58eb5bf6f8d40f80bf08b | [
"Apache-2.0"
]
| null | null | null | _notebooks/2021-11-14-jax-random.ipynb | abap34/my-website | 7a2e76ceeae700a302f58eb5bf6f8d40f80bf08b | [
"Apache-2.0"
]
| 4 | 2021-03-12T07:02:04.000Z | 2022-02-26T09:49:12.000Z | _notebooks/2021-11-14-jax-random.ipynb | abap34/my-website | 7a2e76ceeae700a302f58eb5bf6f8d40f80bf08b | [
"Apache-2.0"
]
| 1 | 2020-12-19T23:02:28.000Z | 2020-12-19T23:02:28.000Z | 22.694205 | 226 | 0.510676 | [
[
[
"# JAXの乱数生成について調べてみたけどよくわからない\n> JAXにおける乱数生成について調べたけどよくわからない\n\n- toc: true \n- badges: true\n- comments: true\n- categories: [Python, JAX, DeepLearning]\n- image: images/jax-samune.png",
"_____no_output_____"
],
[
"JAX流行ってますね。JAXについての詳しい説明は、[たくさんの記事](https://www.google.com/search?q=jax%E3%81%A8%E3%81%AF)や[https://github.com/google/jax](https://github.com/google/jax) を参照していただくとして、JAXの乱数生成について勉強してみようと思います。",
"_____no_output_____"
],
[
"# Numpyにおける乱数の再現性の確保\nさて、JAXはNumpyをとても意識して作られたライブラリですが、乱数周りに関しては大きく異なる点があります。\nまずは, Numpyの例を見てみます。",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
],
[
"# numpy \nx = np.random.rand()\nprint('x:', x)",
"x: 0.26455561210462697\n"
],
[
"for i in range(10):\n x = np.random.rand()\n print('x:', x)",
"x: 0.7742336894342167\nx: 0.45615033221654855\nx: 0.5684339488686485\nx: 0.018789800436355142\nx: 0.6176354970758771\nx: 0.6120957227224214\nx: 0.6169339968747569\nx: 0.9437480785146242\nx: 0.6818202991034834\nx: 0.359507900573786\n"
]
],
[
[
"バラバラの結果が出てきました。これを固定するには、このようなコードを書きます。",
"_____no_output_____"
]
],
[
[
"for i in range(10):\n np.random.seed(0)\n x = np.random.rand()\n print('x:', x)",
"x: 0.5488135039273248\nx: 0.5488135039273248\nx: 0.5488135039273248\nx: 0.5488135039273248\nx: 0.5488135039273248\nx: 0.5488135039273248\nx: 0.5488135039273248\nx: 0.5488135039273248\nx: 0.5488135039273248\nx: 0.5488135039273248\n"
]
],
[
[
"ところでnumpyでは、`np.random.get_state()` で乱数生成器の状態が確認できます。",
"_____no_output_____"
]
],
[
[
"np.random.seed(0)\nstate = np.random.get_state()\nprint(state[0])\nprint('[', *state[1][:10], '...')\nprint(*state[1][-10:], ']')",
"MT19937\n[ 0 1 1812433255 1900727105 1208447044 2481403966 4042607538 337614300 3232553940 1018809052 ...\n2906783932 3668048733 2030009470 1910839172 1234925283 3575831445 123595418 2362440495 3048484911 1796872496 ]\n"
],
[
"np.random.seed(20040304)\nstate = np.random.get_state()\nprint(state[0])\nprint('[', *state[1][:10], '...')\nprint(*state[1][-10:], ']')",
"MT19937\n[ 20040304 3876245041 2868517820 934780921 2883411521 496831348 4198668490 1502140500 1427494545 3747657433 ...\n744972032 1872723303 3654422950 1926579586 2599193113 3757568530 3621035041 2338180567 2885432439 2647019928 ]\n"
]
],
[
[
"逆に言えば、Numpyの乱数生成はグローバルな一つの状態に依存しています。このことは次のような弊害を生みます。",
"_____no_output_____"
],
[
"# 並列実行と実行順序、再現性 \n簡単なゲームを作ってみます。 \n関数`a`, `b`が乱数を生成するので、大きい数を返した方が勝ちというゲームです。",
"_____no_output_____"
]
],
[
[
"a = lambda : np.random.rand()\nb = lambda : np.random.rand()\n\ndef battle():\n if a() > b():\n return 'A'\n else:\n return 'B'",
"_____no_output_____"
],
[
"for i in range(10):\n print('winner is', battle(), '!')",
"winner is B !\nwinner is A !\nwinner is B !\nwinner is A !\nwinner is A !\nwinner is A !\nwinner is B !\nwinner is B !\nwinner is B !\nwinner is A !\n"
]
],
[
[
"また実行すれば、結果は変化します。",
"_____no_output_____"
]
],
[
[
"for i in range(10):\n print('winner is', battle(), '!')",
"winner is B !\nwinner is A !\nwinner is A !\nwinner is B !\nwinner is B !\nwinner is B !\nwinner is A !\nwinner is A !\nwinner is A !\nwinner is B !\n"
]
],
[
[
"ではこの結果の再現性を持たせるにはどうすればいいでしょうか。簡単な例はこうなります。",
"_____no_output_____"
]
],
[
[
"res1 = []\nnp.random.seed(0)\nfor i in range(10):\n res1.append(battle())\n\n# もう一回\n\nres2 = []\nnp.random.seed(0)\nfor i in range(10):\n res2.append(battle())",
"_____no_output_____"
],
[
"print('1 | 2')\nprint('=====')\nfor i in range(10):\n print(res1[i], '|', res2[i])",
"1 | 2\n=====\nB | B\nA | A\nB | B\nB | B\nA | A\nA | A\nB | B\nB | B\nB | B\nB | B\n"
]
],
[
[
"というわけで同じ結果が得られました。しかし、この結果には落とし穴があります。\n関数`battle`の動作をもう少し詳しく確認してみましょう。\n`a`と`b`が呼び出されるタイミングを確認してみます。",
"_____no_output_____"
]
],
[
[
"def a():\n print('a is called!')\n return np.random.rand()\n\ndef b():\n print('b is called!')\n return np.random.rand()",
"_____no_output_____"
],
[
"for i in range(5):\n battle()\n print('======')",
"a is called!\nb is called!\n======\na is called!\nb is called!\n======\na is called!\nb is called!\n======\na is called!\nb is called!\n======\na is called!\nb is called!\n======\n"
]
],
[
[
"このように、aはbより常に先に呼び出されます。ここまでだと何の問題もないように見えますが、実際にはそうではありません。\nこのコードを高速に動作させたい、つまり並列化を行う時にはどうなるでしょうか。\n関数`a`, `b`に依存関係はありませんから、これらを並列に動作させても問題ないように感じます。\nですが、実際には `a`, `b`が返す関数は呼び出し順序に依存しています!従って、このままではせっかく`np.random.seed`をしても意味がなくなってしまいます。",
"_____no_output_____"
],
[
"# JAXの乱数生成\n\nでは、JAXにおける乱数生成を確認してみます。\n先ほどまでで述べたように、次のような条件を満たす乱数生成器を実装したいです。\n\n- 再現性があること\n- 並列化できること\n\nこれらを実現するために、JAXでは<b>key</b>という概念が用いられます。",
"_____no_output_____"
]
],
[
[
"key = jax.random.PRNGKey(0)\nkey",
"_____no_output_____"
]
],
[
[
"keyは単に二つの実数値からなるオブジェクトで、これを用いることによって、JAXでは乱数を生成します。",
"_____no_output_____"
]
],
[
[
"jax.random.normal(key)",
"_____no_output_____"
]
],
[
[
"そして、keyが同じであれば同じ値が生成されます。",
"_____no_output_____"
]
],
[
[
"print(key, jax.random.normal(key))\nprint(key, jax.random.normal(key))\nprint(key, jax.random.normal(key))\nprint(key, jax.random.normal(key))\nprint(key, jax.random.normal(key))\nprint(key, jax.random.normal(key))\nprint(key, jax.random.normal(key))\nprint(key, jax.random.normal(key))\nprint(key, jax.random.normal(key))",
"[0 0] -0.20584235\n[0 0] -0.20584235\n[0 0] -0.20584235\n[0 0] -0.20584235\n[0 0] -0.20584235\n[0 0] -0.20584235\n[0 0] -0.20584235\n[0 0] -0.20584235\n[0 0] -0.20584235\n"
]
],
[
[
"とはいえこれだけだとひとつの数字しか得ることができません。もっとたくさんの乱数が欲しくなった際には、`jax.random.split`を用います。",
"_____no_output_____"
]
],
[
[
"key1, key2 = jax.random.split(key)\nprint(key, '->', key1, key2)",
"[0 0] -> [4146024105 967050713] [2718843009 1272950319]\n"
]
],
[
[
"`jax.random.split`によって、ひとつのkeyから2つのkeyが作り出されます。\nこのkeyによって、また新しい乱数を生み出します。 ",
"_____no_output_____"
],
[
"ちなみに、この二つのkeyは等価ですが、慣例的に二つ目を新しい乱数生成につかい、一つ目はまた新しいkeyを使うために用いられるようです。(以下のコードを参照)",
"_____no_output_____"
]
],
[
[
"# 慣例的に二つ目をsub_keyとして新しい乱数生成に、一つ目をまた新しい乱数を作るために使用する(下のように書くことでsplit元の古いkeyも削除できる。keyが残ると誤って同じ乱数を作ってしまうので注意が必要。)\nkey, sub_key = jax.random.split(key)\nkey, subsub_key = jax.random.split(key)",
"_____no_output_____"
]
],
[
[
"また、同じkeyから分割されたkeyは、常に等しくなります。",
"_____no_output_____"
]
],
[
[
"def check_split(seed):\n key = jax.random.PRNGKey(seed)\n key, sub_key = jax.random.split(key)\n print(key, '->', key, sub_key)",
"_____no_output_____"
],
[
"check_split(0)\ncheck_split(0)\ncheck_split(0)\nprint('=============================================================================')\ncheck_split(2004)\ncheck_split(2004)\ncheck_split(2004)",
"[4146024105 967050713] -> [4146024105 967050713] [2718843009 1272950319]\n[4146024105 967050713] -> [4146024105 967050713] [2718843009 1272950319]\n[4146024105 967050713] -> [4146024105 967050713] [2718843009 1272950319]\n=============================================================================\n[2965909967 2346697052] -> [2965909967 2346697052] [2813626588 818499380]\n[2965909967 2346697052] -> [2965909967 2346697052] [2813626588 818499380]\n[2965909967 2346697052] -> [2965909967 2346697052] [2813626588 818499380]\n"
]
],
[
[
"また、一度に何個にもsplitできます。例えば1つのkeyから次のようにして10個のkeyを得ることができます。\n",
"_____no_output_____"
]
],
[
[
"# 何個にもsplitできる。\nkey = jax.random.PRNGKey(0)\nkey, *sub_keys = jax.random.split(key, num=10)",
"_____no_output_____"
],
[
"key",
"_____no_output_____"
],
[
"sub_keys",
"_____no_output_____"
]
],
[
[
"# sequential-equivalent",
"_____no_output_____"
],
[
"Numpyではsequential-equivalentが保障されています。(適切な訳語がわからない)\n\n簡単にいうと、まとめてN個の乱数を取得することと、ひとつひとつ乱数を取得して連結したものは等価である、ということが保障されています。(以下のコードを見るとわかりやすいです)",
"_____no_output_____"
]
],
[
[
"# ひとつずつ\nnp.random.seed(0)\nprint(np.array([np.random.rand() for i in range(10)]))\n\nprint('================================================')\n\n# まとめて \nnp.random.seed(0)\nprint(np.random.rand(10))",
"[0.5488135 0.71518937 0.60276338 0.54488318 0.4236548 0.64589411\n 0.43758721 0.891773 0.96366276 0.38344152]\n================================================\n[0.5488135 0.71518937 0.60276338 0.54488318 0.4236548 0.64589411\n 0.43758721 0.891773 0.96366276 0.38344152]\n"
]
],
[
[
"ところがJAXではその限りではありません。JAXで10個の配列を取得する方法としては、\n\n- keyを10個用意する\n- ひとつのkeyから10個作るということが考えられます。",
"_____no_output_____"
]
],
[
[
"# やり方 1: keyを10個用意\nkey = jax.random.PRNGKey(0)\nkey, *sub_keys = jax.random.split(key, 11)\nprint(np.array([jax.random.normal(sub_key) for sub_key in sub_keys]))",
"[-1.3700832 -1.6277806 1.2452871 -1.0201586 0.80342007 -1.5052081\n -1.2988805 0.3053512 -0.22334994 1.1694573 ]\n"
],
[
"# やり方 2: ひとつのkeyから10個作る\nkey = jax.random.PRNGKey(0)\nprint(np.array(jax.random.normal(key, shape=(10,))))",
"[-0.372111 0.2642311 -0.18252774 -0.7368198 -0.44030386 -0.15214427\n -0.6713536 -0.59086424 0.73168874 0.56730247]\n"
]
],
[
[
"しかし、見ての通り生成される乱数は異なっています。(JAXではsequential-equivalentは保障されません。)\nこの理由として、\n\n> As in NumPy, JAX's random module also allows sampling of vectors of numbers. However, JAX does not provide a sequential equivalence guarantee, because doing so would interfere with the vectorization on SIMD hardware\n\nが[ドキュメント](https://colab.research.google.com/github/google/jax/blob/main/docs/jax-101/05-random-numbers.ipynb#scrollTo=Fhu7ejhLB4R_)で挙げられています。\nどういうことだってばよ。\n\n[design_noteのdesignの項目](https://github.com/google/jax/blob/main/design_notes/prng.md#design)がよくわからない。この擬似コード的な何かはいったい何なんだ...",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
]
|
e7982f43818919f09c5d07e10b1b0a3edacf1369 | 10,190 | ipynb | Jupyter Notebook | doc/source/examples/circuit/Lumped Element Circuits.ipynb | nmaterise/scikit-rf | 779b173894a7b21c238039cee54e92436473cf5d | [
"BSD-3-Clause"
]
| 1 | 2021-12-15T09:34:13.000Z | 2021-12-15T09:34:13.000Z | doc/source/examples/circuit/Lumped Element Circuits.ipynb | nmaterise/scikit-rf | 779b173894a7b21c238039cee54e92436473cf5d | [
"BSD-3-Clause"
]
| null | null | null | doc/source/examples/circuit/Lumped Element Circuits.ipynb | nmaterise/scikit-rf | 779b173894a7b21c238039cee54e92436473cf5d | [
"BSD-3-Clause"
]
| null | null | null | 30.508982 | 591 | 0.579784 | [
[
[
"# Lumped Elements Circuits",
"_____no_output_____"
],
[
"In this notebook, we construct various network from basic lumped elements (resistor, capacitor, inductor), with the 'classic' and the `Circuit` approach. Generally the `Circuit` approach is more verbose than the 'classic' way for building a circuit. However, as the circuit complexity increases, in particular when components are connected in parallel, the `Circuit` approach is interesting as it increases the readability of the code. Moreover, `Circuit` object can be plotted using its `plot_graph()` method, which is usefull to rapidly control if the circuit is built as expected. ",
"_____no_output_____"
]
],
[
[
"import numpy as np # for np.allclose() to check that S-params are similar\nimport skrf as rf\nrf.stylely()",
"_____no_output_____"
]
],
[
[
"## LC Series Circuit",
"_____no_output_____"
],
[
"In this section we reproduce a simple equivalent model of a capacitor $C$, as illustrated by the figure below:\n\n<img src=\"designer_capacitor_simple.png\" width=\"700\">\n",
"_____no_output_____"
]
],
[
[
"# reference LC circuit made in Designer\nLC_designer = rf.Network('designer_capacitor_30_80MHz_simple.s2p')",
"_____no_output_____"
],
[
"# scikit-rf: manually connecting networks\nline = rf.media.DefinedGammaZ0(frequency=LC_designer.frequency, z0=50)\nLC_manual = line.inductor(24e-9) ** line.capacitor(70e-12)",
"_____no_output_____"
],
[
"# scikit-rf: using Circuit builder \nport1 = rf.Circuit.Port(frequency=LC_designer.frequency, name='port1', z0=50)\nport2 = rf.Circuit.Port(frequency=LC_designer.frequency, name='port2', z0=50)\ncap = rf.Circuit.SeriesImpedance(frequency=LC_designer.frequency, name='cap', z0=50,\n Z=1/(1j*LC_designer.frequency.w*70e-12))\nind = rf.Circuit.SeriesImpedance(frequency=LC_designer.frequency, name='ind', z0=50,\n Z=1j*LC_designer.frequency.w*24e-9)\n\n# NB: it is also possible to create 2-port lumped elements like:\n# line = rf.media.DefinedGammaZ0(frequency=LC_designer.frequency, z0=50)\n# cap = line.capacitor(70e-12, name='cap')\n# ind = line.inductor(24e-9, name='ind')\n\nconnections = [\n [(port1, 0), (cap, 0)],\n [(cap, 1), (ind, 0)],\n [(ind, 1), (port2, 0)]\n]\ncircuit = rf.Circuit(connections)\nLC_from_circuit = circuit.network",
"_____no_output_____"
],
[
"# testing the equivalence of the results\nprint(np.allclose(LC_designer.s, LC_manual.s))\nprint(np.allclose(LC_designer.s, LC_from_circuit.s))",
"_____no_output_____"
],
[
"circuit.plot_graph(network_labels=True, edge_labels=True, port_labels=True)",
"_____no_output_____"
]
],
[
[
"## A More Advanced Equivalent Model",
"_____no_output_____"
],
[
"In this section we reproduce an equivalent model of a capacitor $C$, as illustrated by the figure below:\n\n<img src=\"designer_capacitor_adv.png\" width=\"800\">",
"_____no_output_____"
]
],
[
[
"# Reference results from ANSYS Designer\nLCC_designer = rf.Network('designer_capacitor_30_80MHz_adv.s2p')",
"_____no_output_____"
],
[
"# scikit-rf: usual way, but this time this is more tedious to deal with connection and port number\nfreq = LCC_designer.frequency\nline = rf.media.DefinedGammaZ0(frequency=freq, z0=50)\nelements1 = line.resistor(1e-2) ** line.inductor(24e-9) ** line.capacitor(70e-12)\nelements2 = line.resistor(20e6)\nT_in = line.tee()\nT_out = line.tee()\nntw = rf.connect(T_in, 1, elements1, 0)\nntw = rf.connect(ntw, 2, elements2, 0)\nntw = rf.connect(ntw, 1, T_out, 1)\nntw = rf.innerconnect(ntw, 1, 2)\nLCC_manual = ntw ** line.shunt_capacitor(50e-12) ",
"_____no_output_____"
],
[
"# scikit-rf: using Circuit builder \nfreq = LCC_designer.frequency\nport1 = rf.Circuit.Port(frequency=freq, name='port1', z0=50)\nport2 = rf.Circuit.Port(frequency=freq, name='port2', z0=50)\nline = rf.media.DefinedGammaZ0(frequency=freq, z0=50)\ncap = line.capacitor(70e-12, name='cap')\nind = line.inductor(24e-9, name='ind')\nres_series = line.resistor(1e-2, name='res_series')\nres_parallel = line.resistor(20e6, name='res_parallel')\ncap_shunt = line.capacitor(50e-12, name='cap_shunt')\nground = rf.Circuit.Ground(frequency=freq, name='ground', z0=50)\n\nconnections = [\n [(port1, 0), (res_series, 0), (res_parallel, 0)],\n [(res_series, 1), (cap, 0)],\n [(cap, 1), (ind, 0)],\n [(ind, 1), (cap_shunt, 0), (res_parallel, 1), (port2, 0)],\n [(cap_shunt, 1), (ground, 0)],\n]\ncircuit = rf.Circuit(connections)\nLCC_from_circuit = circuit.network",
"_____no_output_____"
],
[
"# testing the equivalence of the results\nprint(np.allclose(LCC_designer.s, LCC_manual.s))\nprint(np.allclose(LCC_designer.s, LCC_from_circuit.s))",
"_____no_output_____"
],
[
"circuit.plot_graph(network_labels=True, edge_labels=True, port_labels=True)",
"_____no_output_____"
]
],
[
[
"## Pass band filter",
"_____no_output_____"
],
[
"Below we construct a pass-band filter, from an example given in [Microwaves101](https://www.microwaves101.com/encyclopedias/lumped-element-filter-calculator):",
"_____no_output_____"
],
[
"<img src=\"designer_bandpass_filter_450_550MHz.png\" width=\"800\">\n",
"_____no_output_____"
]
],
[
[
"# Reference result calculated from Designer\npassband_designer = rf.Network('designer_bandpass_filter_450_550MHz.s2p')",
"_____no_output_____"
],
[
"# scikit-rf: the filter by cascading all lumped-elements \nfreq = passband_designer.frequency\npassband_manual = line.shunt_capacitor(25.406e-12) ** line.shunt_inductor(4.154e-9) ** \\\n line.capacitor(2.419e-12) ** line.inductor(43.636e-9) ** \\\n line.shunt_capacitor(25.406e-12) ** line.shunt_inductor(4.154e-9)",
"_____no_output_____"
],
[
"# scikit-rf: the filter with the Circuit builder\nfreq = passband_designer.frequency\nline = rf.media.DefinedGammaZ0(frequency=freq)\nC1 = line.capacitor(25.406e-12, name='C1')\nC2 = line.capacitor(2.419e-12, name='C2')\nC3 = line.capacitor(25.406e-12, name='C3')\nL1 = line.inductor(4.154e-9, name='L1')\nL2 = line.inductor(43.636e-9, name='L2')\nL3 = line.inductor(4.154e-9, name='L3')\nport1 = rf.Circuit.Port(frequency=freq, name='port1', z0=50)\nport2 = rf.Circuit.Port(frequency=freq, name='port2', z0=50)\nground = rf.Circuit.Ground(frequency=freq, name='ground', z0=50)\n\nconnections = [\n [(port1, 0), (C1, 0), (L1, 0), (C2, 0)],\n [(C2, 1), (L2, 0)],\n [(L2, 1), (C3, 0), (L3, 0), (port2, 0)],\n [(C1, 1), (C3, 1), (L1, 1), (L3, 1), (ground, 0)],\n]\n\ncircuit = rf.Circuit(connections)\npassband_circuit = circuit.network\npassband_circuit.name = 'Pass-band circuit'",
"_____no_output_____"
],
[
"passband_circuit.plot_s_db(m=0, n=0, lw=2)\npassband_circuit.plot_s_db(m=1, n=0, lw=2)\npassband_designer.plot_s_db(m=0, n=0, lw=2, ls='-.')\npassband_designer.plot_s_db(m=1, n=0, lw=2, ls='-.')",
"_____no_output_____"
],
[
"circuit.plot_graph(network_labels=True, port_labels=True, edge_labels=True)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
]
|
e7982f60150fedf27f21c9270cf01aa928111638 | 3,113 | ipynb | Jupyter Notebook | ml_algorithms/06_Boosting/06_03/End/06_03.ipynb | joejoeyjoseph/playground | fa739d51635823b866fafd1e712760074cfc175c | [
"MIT"
]
| null | null | null | ml_algorithms/06_Boosting/06_03/End/06_03.ipynb | joejoeyjoseph/playground | fa739d51635823b866fafd1e712760074cfc175c | [
"MIT"
]
| null | null | null | ml_algorithms/06_Boosting/06_03/End/06_03.ipynb | joejoeyjoseph/playground | fa739d51635823b866fafd1e712760074cfc175c | [
"MIT"
]
| null | null | null | 31.765306 | 324 | 0.591712 | [
[
[
"## Boosting: Hyperparameters\n\nImport [`GradientBoostingClassifier`](https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.GradientBoostingClassifier.html) and [`GradientBoostingRegressor`](https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.GradientBoostingRegressor.html) from `sklearn` and explore the hyperparameters.",
"_____no_output_____"
],
[
"### Import Boosting Algorithm for Classification & Regression",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import GradientBoostingClassifier, GradientBoostingRegressor\n\nprint(GradientBoostingClassifier())\nprint(GradientBoostingRegressor())",
"GradientBoostingClassifier(criterion='friedman_mse', init=None,\n learning_rate=0.1, loss='deviance', max_depth=3,\n max_features=None, max_leaf_nodes=None,\n min_impurity_decrease=0.0, min_impurity_split=None,\n min_samples_leaf=1, min_samples_split=2,\n min_weight_fraction_leaf=0.0, n_estimators=100,\n n_iter_no_change=None, presort='auto', random_state=None,\n subsample=1.0, tol=0.0001, validation_fraction=0.1,\n verbose=0, warm_start=False)\nGradientBoostingRegressor(alpha=0.9, criterion='friedman_mse', init=None,\n learning_rate=0.1, loss='ls', max_depth=3, max_features=None,\n max_leaf_nodes=None, min_impurity_decrease=0.0,\n min_impurity_split=None, min_samples_leaf=1,\n min_samples_split=2, min_weight_fraction_leaf=0.0,\n n_estimators=100, n_iter_no_change=None, presort='auto',\n random_state=None, subsample=1.0, tol=0.0001,\n validation_fraction=0.1, verbose=0, warm_start=False)\n"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
]
]
|
e79833742ee9a911449eec68d2dda17838f28921 | 189,954 | ipynb | Jupyter Notebook | Models/AgentBasedModels.ipynb | ttrogers/frigo-chen-rogers | ddc8808f21a89259df83a161ee72faf2487623d4 | [
"MIT"
]
| null | null | null | Models/AgentBasedModels.ipynb | ttrogers/frigo-chen-rogers | ddc8808f21a89259df83a161ee72faf2487623d4 | [
"MIT"
]
| null | null | null | Models/AgentBasedModels.ipynb | ttrogers/frigo-chen-rogers | ddc8808f21a89259df83a161ee72faf2487623d4 | [
"MIT"
]
| null | null | null | 201.435843 | 43,332 | 0.888168 | [
[
[
"# Simulations: Dynamic learning with two learners, one oracle, and *heuristic evidence-weighting function*",
"_____no_output_____"
],
[
"This notebook provides code to simulate 1D boundary learning in two agents learning from each other and from an \"oracle\" that always tells the truth. Each agent receives labels from the other agent (based on her current belief) and from the oracle (based on the ground truth). Neither agent knows which source is telling the truth.\n\nEach agent begins with an initial belief about the location of the category boundary, so that the two agent beliefs together can be viewed as a point in a 2D plane. Across one \"batch\" of learning, it is assumed that each agent moves her boundary toward a weighted average of the difference between her current boundary and the boundaries of the other two sources (ie, the other learner and the oracle). So:\n\n$$\\Delta_b = w_c * d_c + (1 - w_c) * d_f$$\n\n...where $\\Delta_b$ is the change in the learner's boundary, $w_c$ is the (proportional) weight given to the closer source, $1-w_c$ is the (proportional) weight given to the more distal source, and $d_c$ and $d_f$ are the distances from the closer/more distal source to the learner's current boundary.\n\nThe amount moved on each batch is determined by a fixed rate parameter r:\n\n$$b_{t+1} = b_t + r * \\Delta_b$$\n\nAs shown in the empirical studies, the proportional weight given to each source is determined by a function that decays nonlinearly with the distance between the source's boundary and the learner's curent boundary:\n\n$$w_c = 1 - (d_c + o)/(d_c + o + s)$$\n\n...where $d_c$ is the distance from the learner's boundary to the closer source, $o$ is an offset term (distance considered 0 if less than $o$), and $s$ controls the slope of the decay. The defaults for $o$ and $s$ are the best-fitting parameters determined in Experiment 2.\n\nThe functions and code are as follows:\n\nget.pwt: given a distance between learner's boundary and source, and parameters for the trust decay, return weight given to source.\n\nupdate.bound: given the learner's boundary, a current boundary for each source, a rate constant, and parameters for the decay curve, return the learner's new boundary.\n\ndynamic.sim: given starting boundaries for each learner, the true boundary, a learning rate constant, number of batches, and parameters for the trust decay curves, reutrn a matrix indicating each learner's boundary at each epoch of learning.\n",
"_____no_output_____"
],
[
"## Heuristic evidence-weighting function",
"_____no_output_____"
],
[
"This function computes the weight given to a source as a function of its distance from the learner's current boundary, according to the heuristic evidence-weighting function determined in Experiment 2. This is a proportional weight $w_c$ relative to some fixed second source, which in turn receives a weight of $1 - w_c$. The empirical studies showed that $w_c ~= 1$ when the source boundary is within $o$ of the learner's boundary--therefore in simulations, we compute $w_c$ for the closest source boundary, then give the remaining $1-w_c$ weight to the second source.",
"_____no_output_____"
]
],
[
[
"get.pwt <- function(d, s=24, o=5, p=1){\n #Computes the proportion of weight given to a source\n #based on the distance between learner and source boundary\n #\n #d = vector of distances between learner and source boundary for n sources\n #s = slope of HEW curve\n #o = offsetof HEW curve---distances less than this value will return an output value of 1.0\n #p = power to which resulting proportion is raised. Experimental, use default of 1.0\n #out = returned vector of proportional weights for far source\n ###################\n \n d <- d - o #Shift distances by offset so d = o becomes d = 0\n tmp <- 1 - (d/(d+s)) #Proportional weight given to distal source\n tmp <- c(tmp)^p #Raise to power p, not currently used\n tmp[tmp > 1] <- 1.0 #If weight is larger than 1 replace with 1\n tmp\n}",
"_____no_output_____"
]
],
[
[
"Here is what the weighting function looks like with default parameters from experiment 2, where s = 24 and o = 5:",
"_____no_output_____"
]
],
[
[
"plot(0:150, get.pwt(0:150, s=24, o=5), type = \"l\", lwd = 3, pch = 16, ylab = \"Source weight\", xlab = \"Source distance\", ylim = c(0,1))\n\n",
"_____no_output_____"
]
],
[
[
"## Other possible weighting functions",
"_____no_output_____"
],
[
"Here we define some other weighting functions to investigate group learning dynamics under different hypotheses about weighting.",
"_____no_output_____"
],
[
"### Equal weight to both sources",
"_____no_output_____"
],
[
"This function can be used in place of get.pwt to simulate learning where both sources always get equal weight.",
"_____no_output_____"
]
],
[
[
"get.samewt <-function(d, s=NA, o=NA, p=NA){\n #Returns a vector of 0.5 for each element of d\n #essentially always giving the same .5 weight to each source\n #All parameters ignored except d, only included to work with other code\n #############\n \n out<-rep(.5, times=length(d))\n out\n}",
"_____no_output_____"
]
],
[
[
"### Rectified linear weighting",
"_____no_output_____"
],
[
"This function returns the weight of a source based on a linear decline of the source's distance from the learner's curent boundary, rectified at 0 and 1.",
"_____no_output_____"
]
],
[
[
"get.rlwt <- function(d, s=0.01, o=4.5,p=0){\n #Rectified linear weighting function\n #d = vector of distances for sources to be weighted\n #o = offset; distances less than this get weight 1\n #s = slope, rate at which weight diminishes with distance\n #p = proportion shrinkage from 1 and 0.\n #Returns vector of weights, one for each element in d\n #############\n \n out <- 1 - ((d - o) * s)\n out[d <= o] <- 1.0\n out[out < 0] <- 0.0\n out <- (out * (1-p)) + p/2\n out\n}",
"_____no_output_____"
]
],
[
[
"Here is what the rectified weighting function looks like. $o$ shifts it left/right, $s$ changes the slope.",
"_____no_output_____"
]
],
[
[
"plot(0:300, get.rlwt(0:300, s=.005, o=4.5, p=.0), type=\"l\", ylim = c(0,1),\n ylab=\"Source weight\", xlab=\"Source distance\")",
"_____no_output_____"
]
],
[
[
"## Sigmoid",
"_____no_output_____"
],
[
"This returns a source weight as the sigmoid of its distance from the learner's source. Like HEW and rectified linear, the function is bounded at ${0,1}$.",
"_____no_output_____"
]
],
[
[
"get.sigwt <- function(d, s=1, o=4.5,p=NA){\n #Sigmoidal weighting function\n #d = vector of distances for sources to be weighted\n #o = offset, shifts sigmoid left/right\n #s = slope of sigmoid\n #p = for compatibility, not used\n #Returns vector of weights, one for each element in d\n #############\n d <- c(max(d) - d) - max(d)/2\n out <- 1 / (1 + exp(-1 * (o + s*d)))\n out\n}",
"_____no_output_____"
]
],
[
[
"Here is the plot:",
"_____no_output_____"
]
],
[
[
"plot(0:300, get.sigwt(0:300, s=.05, o=-3), type=\"l\", ylim = c(0,1),\n ylab=\"Source weight\", xlab=\"Source distance\")",
"_____no_output_____"
]
],
[
[
"## Update the learner's current boundary according to evidence-weighting function.",
"_____no_output_____"
],
[
"Note that different results obtain depending on whether you use the curve to compute the close source weight first or the far source weight first. Both lead to stable states where learners disagree, but Experiment 2 shows that a source with a very close boundary receives all the learner's weight--so we typically compute the closest source weight first and attribute remaining weight to the second source.",
"_____no_output_____"
]
],
[
[
"update.bound <- function(i, s1, s2, r = 1, weightfirst =\"c\", closebig=T, f = get.pwt, fpars=c(24, 5, 1)){\n ##############\n #Updates learner's current boundary accourding to nonlinear weighting function\n #\n #i=learner's initial boundary\n #s1, s2 = source 1 and 2 boundaries\n #r = rate of boundary change\n #f = function to use for weighting, get.pwt by default\n #fpars = parameters for the weighting function: slope, offset, power\n #closebig: is the closer boundary toward the larger end of the stimulus range?\n #weightfirst = which source to weight first---default (c) is closest, use anything else for distal\n #returns the new boundary\n ######\n\n #Determine which is closer and which is distal source\n if(abs(i-s1) < abs(i-s2)){\n cs <- s1\n ds <- s2\n } \n else{\n cs <- s2\n ds <- s1\n }\n \n #Use function f to compute weights for two sources\n if(weightfirst==\"c\"){ #If close source is computed first\n cwt <- f(abs(cs-i), s=fpars[1], o=fpars[2], p=fpars[3]) #get weight for close source first\n if((cwt < 0.5) & closebig) cwt <- 1 - cwt #closer source gets larger weight if closebig is true\n dwt <- 1 - cwt #Weight for more distal source\n } \n else{ #If distal source is computed first\n dwt <- f(abs(ds-i), s=fpars[1], o=fpars[2], p=fpars[3]) #get weight for distal source first\n cwt <- 1 - dwt #Weight for closer source\n }\n \n delta_i <- (ds-i) * dwt + (cs-i) * cwt #Change in boundary\n i <- i + delta_i * r #update boundary\n i\n}",
"_____no_output_____"
]
],
[
[
"# Simulate two learners and one static source",
"_____no_output_____"
],
[
"This simulation involves two learners and one oracle, as reported in the main paper. The following function generates the sequence of belief-states occupied by each learner over the course of learning, given their starting states, the ground truth, one of the evidence-weighting functions defined above and its parameters, and a constant indicating how quickly beliefs are updated on each round.",
"_____no_output_____"
]
],
[
[
"dynamic.sim <- function(l1, l2, static, nsteps=100, r=1, f = get.pwt, fpars=c(25,5,1)){\n #Simulates two learners, learning from each other and from one static source\n #l1, l2, static = initial boundaries for learners 1 and 2 and static source\n #nsteps = number of learning steps to simulate\n #r = updating rate for each learner's boundary\n #fpars = parameters for nonlinear weighting function\n \n out <- matrix(0, nsteps, 2) #matrix to contain each learner's boundary at each step\n out <- rbind(c(l1,l2), out) #add initial boundaries to top of matrix\n\n #Loop to update each learner's bound based on other learner and static bound at previous step\n for(i1 in c(1:nsteps)){\n out[i1+1,1] <- update.bound(out[i1,1], out[i1,2], static, r=r, f=f, fpars=fpars)\n out[i1+1,2] <- update.bound(out[i1,2], out[i1,1], static, r=r, f=f, fpars=fpars)\n }\n out\n}",
"_____no_output_____"
]
],
[
[
"## Plot simulations for a grid of possible initial learning boundaries in the pair",
"_____no_output_____"
],
[
"The code below runs the 2-learer simulation several times, with the two learners each beginning with a belief about the category boundary lying somewhere between 0 and 300. For each pair of initial beliefs, it computes how the beliefs change over time and where they stop after 100 \"epochs\" of learning. Each such trajectory is plotted as a gray line, with the starting beliefs shown as a green point and the ending beliefs shown as a red point. \n\nParameters controlling the simulation are ste at the top of the block as follows:\n\n**gridpts** sets the number of starting beliefs simulated for both learners; these will be evenly-spaced in $[0-300]$.\n\n**gtruth** specifies the location of the ground truth provided by the oracle.\n\n**niter** indicates the number of learning iterations to run.\n\n**uprate** is a constant specifying the rate at which beliefs are updated. \n\n**upfunc** indicates which of the above-defined functions should be used to weight the sources\n\n**upfunc.pars** is a 3-element vector indicating the slope, offset, and power parameters for the weighting function in that order\n\nTo reproduce main paper Figure 4A use upfunc <- get.samewt; for panel B use upfunc <- get.pwt",
"_____no_output_____"
]
],
[
[
"gridpts <- 20 #Number of grid points in each dimension\ngtruth <- 150 #Location of ground-truth boundary\nniter <- 200 #Number of learning iterations\nuprate <- 0.1 #Proportional rate at which beliefs are updated on each iteration\nupfunc <- get.pwt #Function for computing source weights\nupfunc.pars <- c(24,5,1) #Parameters for weighting function\n\n#Create empty plot frame:\nplot(150, 150, type=\"n\", xlim = c(0,300), ylim = c(0,300), xlab = \"L1 belief\", ylab=\"L2 belief\")\nabline(0,1, lwd=2, col = \"blue\") #line where learners have same boundary\nabline(h=150, lwd=2, col = \"blue\") #ground truth at 150\nabline(v=150, lwd=2, col = \"blue\") #ground truth at 150\n\ngrspace <- floor(300/gridpts) #Compute grid spacing\n\nfor(i1 in c(0:gridpts)) for(i2 in c(0:gridpts)){ #loop over grid points for l1 and l2\n #for each iteration compute learners boundary trajectories\n tmp <- dynamic.sim(i1*grspace, i2*grspace, gtruth, nsteps = niter, r=uprate, f=upfunc, fpars = upfunc.pars)\n lines(tmp, col=gray(.5)) #add lines showing trajectory\n points(tmp[1,1], tmp[1,2], pch = 16, col = \"green\", cex=.5) #start point\n points(tmp[niter+1,1], tmp[niter+1,2], pch = 16, col = \"red\") #end point\n }\n",
"_____no_output_____"
]
],
[
[
"As you can see, when learner beliefs begin in the upper left or lower right quadrants, they converge on the truth. These are cases where each learner begins on a different side of the ground truth--so the oracle and the other source are always pulling in the same direction. The two learners do not perfectly agree until they meet at the ground truth. Something different happens in the upper right and lower left quadrants, where the two learners are on the same side of the ground truth. In these cases the two learners often end up agreeing with each other, and hence giving each other all their weight, before they meet at the ground truth. This then produces different groups of beliefs, depending on where the learners began. The same behavior is observed for all nonlinear weighting functions.",
"_____no_output_____"
],
[
"## Explaining the experimental data",
"_____no_output_____"
],
[
"Does this weighting curve allow us to explain the pattern of results in the 4 experiments? \n\nExperiments 1 and 2 are fairly straightforward: the parameters for the weighting function were derived from Experiment 2, where one source always appeared 15 units from the midline, and the other was sampled with uniform probability from $b_v \\in [0,150]$ units from the midline. Since experiment 1 also used a source 15 units from the midline, the same parameters for the weighting function should apply. Simply plugging in the experiment design parameters to the update.bound function gives us the predicted new boundary after learning, and hence the expected amount of shift, and the amount of weight given to the far source:",
"_____no_output_____"
]
],
[
[
"firstbound <- 150 #Expected initial boundary\nnewbound <- update.bound(150, 165, 50, fpars = c(25,4.5,1), weightfirst = \"d\") #new boundary after updating weight\nbshift <- newbound - firstbound #expected amount of shift\ndwt <- get.pwt(100) #expected weight given to far source\nprint(round(c(firstbound, newbound, bshift, dwt),2))",
"[1] 150.00 141.14 -8.86 0.20\n"
]
],
[
[
"So given an initial boundary at 150 and sources at 165 and 50, the weighting curve from Experiment 2 predicts a final boundary around 141, with a total shift of about 9 units toward the distal source. The observed shift wa 14 +/- 7.5 units toward the far source, a confidence interval that includes this prediction. The curve also predicts a weight of about 0.2 given to the far source, quite close to and within the confidence limits of the empirical estimate of 0.23 +/- 0.05.\n\nWhat about experiment 3? This was motivated by the observation that, according to the curve in Experiment 2, a learner should give all weight to a source that is v close to her initial belief. So that is a fairly trivial prediction---total weight given to each group's close source should be 1; that source is very near the learner's boundary; the boundary does not change, which is what we observed.\n\nExperiment 4 is the most challenging to think about. We want to know what the model predicts about boundary change when the learner has a boundary about 50 units from the midpoint and the two sources are symmetric around the midpoint but vary in their distance. Clearly when the two sources are both very close to the midpoint, the learner should shift her boundary toward the midpoint. And we know that, when one source grows near to the learner's boundary, it should get all the weight and the learner's boundary should not shift very much. But what happens as the sources grow more discrepant, with one pulling the learner toward one extreme and the other pulling the learner toward the other extreme?\n\nPredictions from the weighting model are complicated by the fact that we only estimated parameters for the case in which one source is about 15 units from the learner's initial boundary. We don't know how/whether the slope of the weighing function will change when both sources differ in their distance from the initial boundary. So, we don't know exactly how the boundary is expected to change. Here we consider whether the model makes predictions that apply across a wide range of parameterizations.\n\nThe code below considers models in which the slope parameter varies from 16 (quite steep) to 100 (fairly shallow). For each parameter, we consider what the model predicts about how the boundary should change for sources situated near the midpoint or increasingly distal to it. For each model parameter, we plot a line that indicates the expected movement of the boundary, with positive numbers indicating a shift toward the midpoint and the X-axis indicating source distance from the midpoint. The colors indicate the steepness of the weighting function, with the red end of the spectrum showing what happens when this slope is very steep and the violet end showing predictions for much shallower slopes",
"_____no_output_____"
]
],
[
[
"tmp <- rep(0, times = 100) #vector of zeros to contain predictions for one model\n#for(i1 in c(1:100)) tmp[i1] <- update.bound(100, 150-i1, 150+i1, fpars = c(24,5,1))\n\n#Create empy plot\nplot(0,0, type = \"n\", xaxt = \"n\", xlab = \"Source location\", pch = 16, \n ylab = \"Shift toward midpoint\", ylim = c(-10,100), xlim = c(1,100))\nmtext(side = 1, line = .5, at = c(100,50,1), adj = c(1,.5, 0), text = c(\"Midpoint\", \"Initial\", \"Pole\"))\n\n#Consider slope values ranging from 100 to 16\nfor(i2 in c(100:16)){\n #For each, compute prediction about how boundary changes as sources move\n #from midline to poles, and plot as a line\n for(i1 in c(1:100)) tmp[i1] <- update.bound(100, 150-i1, 150+i1, closebig=F, fpars = c(i2,5,1))\n lines(100:1, tmp-100, col = rainbow(100)[i2-15], lwd=10)\n }\n\nfor(i1 in c(1:100)) tmp[i1] <- update.bound(100, 150-i1, 150+i1, closebig=F, fpars = c(24,5,1))\nlines(100:1, tmp-100, col = 1, lwd=5)\n\n\nabline(h=50, lty=2) #Expected shift if they go to midpoint\nabline(h=0) #Points above this line indicate shift toward the midpoint\n\nabline(v = c(75, 50, 25,0), lty = 3)",
"_____no_output_____"
]
],
[
[
"The result shows that quantitative predictions about the amount of shift vary quite a bit with the parameters of the trust weighting function, especially as the sources grow further toward the poles. But all parameterizations yield the same U-shape: when the sources are closer to the midpoint than is the learner's initial boundary, the boundary is expected to shift toward the midpoint. For a \"bubble\" around the learner's initial boundary, little or no shift is expected, but as the sources grow even closer to the poles, all parameterizations predict that the boundary should again shift toward the midpoint. So the weighting model makes a qualitative prediction robust under several parameterizations: learner boundaries should shift toward the midpoint when the two sources are both near the midpoint, and *also* when they are far from the midpoint. In between there should be a bubble where learners shift their boundary less or not at all. \n\nThis is the prediction tested in Experiment 4, where we observed a boundary-shift toward the midpoint of about 20 units when the two sources were near the midpoint (strong agree condition) *and* when they were both far from the midpoint (strong disagree condition), with no shift observed when sources were at intermediate distances from the midpoint (moderate-agree and moderate-disagree conditions). In the main paper we note this pattern is qualitatively similar to the U-shaped curve shown by the HEW model under different parameterizations as shown above, but it is also quantitatively similar to predictions of a heuristic evidence-weighting function with a somewhat shallower slope than that estimated in Experiment 2. ",
"_____no_output_____"
],
[
"In case it is useful, here is some code plotting the shape of the weighting curves corresponding to the parameters that produce the above figure.",
"_____no_output_____"
]
],
[
[
"plot(0,0,type = \"n\", xlim = c(0,150), ylim = c(0,1), xlab = \"Distance of source\", ylab = \"Source weight\")\nfor(i1 in c(100:5)) lines(0:150, get.pwt(0:150, i1, 5, 1), col = rainbow(100)[i1-15], lwd = 5)\nlines(0:150, get.pwt(0:150, 24,5,1), col=1, lwd = 5)",
"_____no_output_____"
]
],
[
[
"# Effects of social connections amongst learning pairs",
"_____no_output_____"
],
[
"In the above simulation of dynamic learners we considered pairs of learners with many different initial beliefs, each learning from the other and from a static oracle. What happens in a group of learners, where the two sources for any single learner are selected according to some policy on each epoch? For instance, suppose you run a social media platform and you want to decide which opinions to \"share\" with each learner. On each epoch, for each learner, you must choose two opinions to share. Here are some of the ways you might pick:\n\n(1) two sources selected at random\n(2) the two closest sources\n(3) the closest and the fartherst source\n(4) the two farthest sources\n(5) the two closest sources *outside the bubble*\n\nThe following code considers these possibilities. The function get.srcs takes a set of source boundaries and a learner's current boundary, and returns two source boundaries according to one of the policies noted above. The subsequent code shows what happens to 10 learners when that policy is applied over successive learning batches.\n",
"_____no_output_____"
],
[
"### Function for selecting two sources for a learner under different policies",
"_____no_output_____"
],
[
"Given a learner's boundary and a set of source boundaries, return two sources for the learner according to some policy.",
"_____no_output_____"
]
],
[
[
"get.srcs <- function(l, s, p = \"r\", r=NA){\n #Function to get two source distances for a learner based on a policy\n #l = learner's current boundary\n #s = current boundary for all sources\n #p = policy for choosing 2 sources:\n #r = random\n #s = two most similar\n #m = mixed ie most similar and most distal\n #f = two farthest sources\n #n = not too similar: choose closest outside of bubble\n #r = radius of similarity to avoid for policy n\n #o = outputs, returns distances for the two selected sources\n ###################\n d <- s - l #vector of distances from learner's boundary\n s <- s[order(abs(d))] #Sort sources by magnitude of distance\n d <- d[order(abs(d))] #Sort distances by magnitude of distance\n s <- s[2:length(s)] #remove first source, which is the learner herself\n d <- d[2:length(d)] #remove first distance, which is the learner herself\n \n if(p==\"r\"){ #random policy\n s <- s[order(runif(length(s)))] #scramble order randomly\n o <- s[1:2] #take first two elements\n } else if(p==\"s\"){ #two most similar policy\n o <- s[1:2]\n } else if(p==\"m\"){ #closest and farthest\n o <- s[c(1, length(s))]\n } else if(p==\"f\"){ #farthest 2\n o <- s[c(length(s)-1, length(s))]\n } else if(p==\"n\"){\n if(is.na(r)) stop(\"Radius for policy n not specified\")\n if(sum(abs(d) > r) > 1){ #If there is at least 1 source outside radius\n s <- s[abs(d) > r] #Remove sources within exclusion radius\n o <- s[1:2] #Select closest two of those remaining\n } else o <- s[c(length(s)-1, length(s))] #otherwise take two farthest\n } else stop(\"Didn't recognize specified policy\")\n o\n}",
"_____no_output_____"
]
],
[
[
"Check to make sure the code works",
"_____no_output_____"
]
],
[
[
"get.srcs(5, 1:10, p=\"n\", r=2)",
"_____no_output_____"
]
],
[
[
"## Functions to simulate a population",
"_____no_output_____"
],
[
"The following code populates a matrix (out) in which columns indicate learners/sources and rows indicate learning epochs. For each epoch and learner, the code selects two sources according to some policy, as determined by the get.srcs function. The learner then updates her boundary according to the specified weighting function. Each learner updates her boundary once per epoch. This procedure iterates for 100 batches.\n\nTo see the results of different policies for selecting sources for a learner, use the following values for policy as an argument in the following function:\n\nr = random\ns = two most similar\nf = two farthest (most dissimilar)\nm = mixed (closest and farthest)\nn = not-too-similar: closest sources outside some exclusion radius\n**note**: for policy n, you need to specify the exclusion radius by also setting a value for exrad",
"_____no_output_____"
]
],
[
[
"sim.pop <- function(l=c(1:10)*14, o=150, nsteps=300, rate=.1, policy=\"r\", exrad=5){\n init<-c(l, o) #Initial boundaries for learners and oracles\n nl <- length(l) #number of learners\n no <- length(o) #number of oracles\n ns <- length(init) #total number of sources\n out <- matrix(0, nsteps+1, ns) #Initialize output matrix\n out[1,] <- init #Seed first row with starting boundaries\n for(i1 in c(1:nsteps)) { #loop over batches\n for(i2 in c(1:nl)) { #loop over learners\n sdists <- get.srcs(out[i1,i2], out[i1,], p=policy, r=exrad) #get sources for learner i2\n out[i1+1,i2] <- update.bound(out[i1,i2], sdists[1], sdists[2], r=rate) #update learner's boundary\n }\n out[i1+1,(nl+1):(nl+no)] <- out[i1,(nl+1):(nl+no)] #oracle boundaries are always the same\n }\n out\n}",
"_____no_output_____"
],
[
"out <- sim.pop(policy=\"r\")",
"_____no_output_____"
]
],
[
[
"The following code plots the change in learner boundaries over time generated by the preceding code.",
"_____no_output_____"
]
],
[
[
"plot.popsim <- function(d, nl=10){\n no <- dim(d)[2] - nl #Number of oracles\n nsteps <- dim(d)[1] -1\n\n #Plot initial boundaries and frame:\n plot(rep(0, times = nl), d[1,1:nl], pch=16, col = 3, xlim = c(0,nsteps), ylim = c(0,300), \n ylab = \"Boundary\", xlab = \"Time\")\n\n #Add lines showing how each learner's boundary changes over time:\n for(i1 in c(1:nl)) lines(c(0:nsteps), d[,i1])\n\n #Show final bondary as red dot:\n points(rep(nsteps, times = nl), d[nsteps+1,1:nl], pch = 16, col = 2, cex = 2)\n\n abline(h=d[1,c((nl+1):(nl+no))], lty = 2) #Dotted lines showing oracle boundaries\n}",
"_____no_output_____"
]
],
[
[
"Below code runs simulation with two oracles at 150 and random initial beliefs sampled from 10-200. Change the policy parameter to one of the above to see the result of different source-selection policies.",
"_____no_output_____"
]
],
[
[
"n <- 10 #Number of simulated agents\nno <- 2 #Number of oracles\ngt <- 150 #ground truth provided by oracles\nibspan <- 100 #Maximum span of initial learner belief distribution\nibshift <- 140 #Shift from 0 of initial learner belief distribution\np <- \"m\" #Policy for selecting sources, one of: \n #r = random s = two most similar, f = two farthest (most dissimilar)\n #m = mixed (closest and farthest), n = not-too-similar\n\n#Initial beliefs will be sampled uniformly from ibshift to (ibshift + ibspan)\n\nibounds <- runif(n)*ibspan + ibshift #Sample initial beliefs for n learners\n\n#Run simulation\nout <- sim.pop(l=ibounds , o=rep(gt, times = no), policy=p, rate = .1)\n\n#Plot results\nplot.popsim(out, nl=n)",
"_____no_output_____"
],
[
"#Compute mean absolute change by end of learning period\nchng <- out[2:301,] - out[1:300,]\nmean(abs(chng[300,1:10]))\n",
"_____no_output_____"
],
[
"sqrt(var(out[301,1:10]))",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
]
|
e79836abf5f35afba533a46ce549a7cc5fbf6e63 | 15,373 | ipynb | Jupyter Notebook | site/ja/hub/tutorials/tf2_arbitrary_image_stylization.ipynb | phoenix-fork-tensorflow/docs-l10n | 2287738c22e3e67177555e8a41a0904edfcf1544 | [
"Apache-2.0"
]
| 491 | 2020-01-27T19:05:32.000Z | 2022-03-31T08:50:44.000Z | site/ja/hub/tutorials/tf2_arbitrary_image_stylization.ipynb | phoenix-fork-tensorflow/docs-l10n | 2287738c22e3e67177555e8a41a0904edfcf1544 | [
"Apache-2.0"
]
| 511 | 2020-01-27T22:40:05.000Z | 2022-03-21T08:40:55.000Z | site/ja/hub/tutorials/tf2_arbitrary_image_stylization.ipynb | phoenix-fork-tensorflow/docs-l10n | 2287738c22e3e67177555e8a41a0904edfcf1544 | [
"Apache-2.0"
]
| 627 | 2020-01-27T21:49:52.000Z | 2022-03-28T18:11:50.000Z | 42.466851 | 353 | 0.585637 | [
[
[
"##### Copyright 2019 The TensorFlow Hub Authors.\n\nLicensed under the Apache License, Version 2.0 (the \"License\");",
"_____no_output_____"
]
],
[
[
"# Copyright 2019 The TensorFlow Hub Authors. All Rights Reserved.\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# http://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.\n# ==============================================================================",
"_____no_output_____"
]
],
[
[
"# 任意画風の高速画風変換\n",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td><a target=\"_blank\" href=\"https://www.tensorflow.org/hub/tutorials/tf2_arbitrary_image_stylization\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\">TensorFlow.org で表示</a></td>\n <td> <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs-l10n/blob/master/site/ja/hub/tutorials/tf2_arbitrary_image_stylization.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\">Google Colab で実行</a> </td>\n <td><a target=\"_blank\" href=\"https://github.com/tensorflow/docs-l10n/blob/master/site/ja/hub/tutorials/tf2_arbitrary_image_stylization.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\">GitHub でソースを表示</a></td>\n <td> <a href=\"https://storage.googleapis.com/tensorflow_docs/docs-l10n/site/ja/hub/tutorials/tf2_arbitrary_image_stylization.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\">ノートブックをダウンロード</a> </td>\n <td><a href=\"https://tfhub.dev/google/magenta/arbitrary-image-stylization-v1-256/2\"><img src=\"https://www.tensorflow.org/images/hub_logo_32px.png\">TF Hub モデルを見る</a></td>\n</table>",
"_____no_output_____"
],
[
"[magenta](https://github.com/tensorflow/magenta/tree/master/magenta/models/arbitrary_image_stylization) と次の発表のモデルコードに基づきます。\n\n[Exploring the structure of a real-time, arbitrary neural artistic stylization network](https://arxiv.org/abs/1705.06830). *Golnaz Ghiasi, Honglak Lee, Manjunath Kudlur, Vincent Dumoulin, Jonathon Shlens*, Proceedings of the British Machine Vision Conference (BMVC), 2017.\n",
"_____no_output_____"
],
[
"## セットアップ",
"_____no_output_____"
],
[
"はじめに、TF-2 とすべての関連する依存ファイルをインポートしましょう。",
"_____no_output_____"
]
],
[
[
"import functools\nimport os\n\nfrom matplotlib import gridspec\nimport matplotlib.pylab as plt\nimport numpy as np\nimport tensorflow as tf\nimport tensorflow_hub as hub\n\nprint(\"TF Version: \", tf.__version__)\nprint(\"TF-Hub version: \", hub.__version__)\nprint(\"Eager mode enabled: \", tf.executing_eagerly())\nprint(\"GPU available: \", tf.test.is_gpu_available())",
"_____no_output_____"
],
[
"# @title Define image loading and visualization functions { display-mode: \"form\" }\n\ndef crop_center(image):\n \"\"\"Returns a cropped square image.\"\"\"\n shape = image.shape\n new_shape = min(shape[1], shape[2])\n offset_y = max(shape[1] - shape[2], 0) // 2\n offset_x = max(shape[2] - shape[1], 0) // 2\n image = tf.image.crop_to_bounding_box(\n image, offset_y, offset_x, new_shape, new_shape)\n return image\n\[email protected]_cache(maxsize=None)\ndef load_image(image_url, image_size=(256, 256), preserve_aspect_ratio=True):\n \"\"\"Loads and preprocesses images.\"\"\"\n # Cache image file locally.\n image_path = tf.keras.utils.get_file(os.path.basename(image_url)[-128:], image_url)\n # Load and convert to float32 numpy array, add batch dimension, and normalize to range [0, 1].\n img = tf.io.decode_image(\n tf.io.read_file(image_path),\n channels=3, dtype=tf.float32)[tf.newaxis, ...]\n img = crop_center(img)\n img = tf.image.resize(img, image_size, preserve_aspect_ratio=True)\n return img\n\ndef show_n(images, titles=('',)):\n n = len(images)\n image_sizes = [image.shape[1] for image in images]\n w = (image_sizes[0] * 6) // 320\n plt.figure(figsize=(w * n, w))\n gs = gridspec.GridSpec(1, n, width_ratios=image_sizes)\n for i in range(n):\n plt.subplot(gs[i])\n plt.imshow(images[i][0], aspect='equal')\n plt.axis('off')\n plt.title(titles[i] if len(titles) > i else '')\n plt.show()\n",
"_____no_output_____"
]
],
[
[
"使用する画像を取得しましょう。",
"_____no_output_____"
]
],
[
[
"# @title Load example images { display-mode: \"form\" }\n\ncontent_image_url = 'https://upload.wikimedia.org/wikipedia/commons/thumb/f/fd/Golden_Gate_Bridge_from_Battery_Spencer.jpg/640px-Golden_Gate_Bridge_from_Battery_Spencer.jpg' # @param {type:\"string\"}\nstyle_image_url = 'https://upload.wikimedia.org/wikipedia/commons/0/0a/The_Great_Wave_off_Kanagawa.jpg' # @param {type:\"string\"}\noutput_image_size = 384 # @param {type:\"integer\"}\n\n# The content image size can be arbitrary.\ncontent_img_size = (output_image_size, output_image_size)\n# The style prediction model was trained with image size 256 and it's the \n# recommended image size for the style image (though, other sizes work as \n# well but will lead to different results).\nstyle_img_size = (256, 256) # Recommended to keep it at 256.\n\ncontent_image = load_image(content_image_url, content_img_size)\nstyle_image = load_image(style_image_url, style_img_size)\nstyle_image = tf.nn.avg_pool(style_image, ksize=[3,3], strides=[1,1], padding='SAME')\nshow_n([content_image, style_image], ['Content image', 'Style image'])",
"_____no_output_____"
]
],
[
[
"## TF-Hub モジュールをインポートする",
"_____no_output_____"
]
],
[
[
"# Load TF-Hub module.\n\nhub_handle = 'https://tfhub.dev/google/magenta/arbitrary-image-stylization-v1-256/2'\nhub_module = hub.load(hub_handle)",
"_____no_output_____"
]
],
[
[
"画風に使用する Hub モジュールのシグネチャは、次のとおりです。\n\n```\noutputs = hub_module(content_image, style_image) stylized_image = outputs[0]\n```\n\n上記の `content_image`、`style_image`、および `stylized_image` は、形状 `[batch_size, image_height, image_width, 3]` の 4-D テンソルです。\n\n現在の例では 1 つの画像のみを提供するためバッチの次元は 1 ですが、同じモジュールを使用して、同時に複数の画像を処理することができます。\n\n画像の入力と出力の値範囲は [0, 1] です。\n\nコンテンツとスタイル画像の形状が一致する必要はありません。出力画像の形状はコンテンツ画像の形状と同一です。",
"_____no_output_____"
],
[
"## 画風の実演",
"_____no_output_____"
]
],
[
[
"# Stylize content image with given style image.\n# This is pretty fast within a few milliseconds on a GPU.\n\noutputs = hub_module(tf.constant(content_image), tf.constant(style_image))\nstylized_image = outputs[0]",
"_____no_output_____"
],
[
"# Visualize input images and the generated stylized image.\n\nshow_n([content_image, style_image, stylized_image], titles=['Original content image', 'Style image', 'Stylized image'])",
"_____no_output_____"
]
],
[
[
"## 複数の画像で試してみる",
"_____no_output_____"
]
],
[
[
"# @title To Run: Load more images { display-mode: \"form\" }\n\ncontent_urls = dict(\n sea_turtle='https://upload.wikimedia.org/wikipedia/commons/d/d7/Green_Sea_Turtle_grazing_seagrass.jpg',\n tuebingen='https://upload.wikimedia.org/wikipedia/commons/0/00/Tuebingen_Neckarfront.jpg',\n grace_hopper='https://storage.googleapis.com/download.tensorflow.org/example_images/grace_hopper.jpg',\n )\nstyle_urls = dict(\n kanagawa_great_wave='https://upload.wikimedia.org/wikipedia/commons/0/0a/The_Great_Wave_off_Kanagawa.jpg',\n kandinsky_composition_7='https://upload.wikimedia.org/wikipedia/commons/b/b4/Vassily_Kandinsky%2C_1913_-_Composition_7.jpg',\n hubble_pillars_of_creation='https://upload.wikimedia.org/wikipedia/commons/6/68/Pillars_of_creation_2014_HST_WFC3-UVIS_full-res_denoised.jpg',\n van_gogh_starry_night='https://upload.wikimedia.org/wikipedia/commons/thumb/e/ea/Van_Gogh_-_Starry_Night_-_Google_Art_Project.jpg/1024px-Van_Gogh_-_Starry_Night_-_Google_Art_Project.jpg',\n turner_nantes='https://upload.wikimedia.org/wikipedia/commons/b/b7/JMW_Turner_-_Nantes_from_the_Ile_Feydeau.jpg',\n munch_scream='https://upload.wikimedia.org/wikipedia/commons/c/c5/Edvard_Munch%2C_1893%2C_The_Scream%2C_oil%2C_tempera_and_pastel_on_cardboard%2C_91_x_73_cm%2C_National_Gallery_of_Norway.jpg',\n picasso_demoiselles_avignon='https://upload.wikimedia.org/wikipedia/en/4/4c/Les_Demoiselles_d%27Avignon.jpg',\n picasso_violin='https://upload.wikimedia.org/wikipedia/en/3/3c/Pablo_Picasso%2C_1911-12%2C_Violon_%28Violin%29%2C_oil_on_canvas%2C_Kr%C3%B6ller-M%C3%BCller_Museum%2C_Otterlo%2C_Netherlands.jpg',\n picasso_bottle_of_rum='https://upload.wikimedia.org/wikipedia/en/7/7f/Pablo_Picasso%2C_1911%2C_Still_Life_with_a_Bottle_of_Rum%2C_oil_on_canvas%2C_61.3_x_50.5_cm%2C_Metropolitan_Museum_of_Art%2C_New_York.jpg',\n fire='https://upload.wikimedia.org/wikipedia/commons/3/36/Large_bonfire.jpg',\n derkovits_woman_head='https://upload.wikimedia.org/wikipedia/commons/0/0d/Derkovits_Gyula_Woman_head_1922.jpg',\n amadeo_style_life='https://upload.wikimedia.org/wikipedia/commons/8/8e/Untitled_%28Still_life%29_%281913%29_-_Amadeo_Souza-Cardoso_%281887-1918%29_%2817385824283%29.jpg',\n derkovtis_talig='https://upload.wikimedia.org/wikipedia/commons/3/37/Derkovits_Gyula_Talig%C3%A1s_1920.jpg',\n amadeo_cardoso='https://upload.wikimedia.org/wikipedia/commons/7/7d/Amadeo_de_Souza-Cardoso%2C_1915_-_Landscape_with_black_figure.jpg'\n)\n\ncontent_image_size = 384\nstyle_image_size = 256\ncontent_images = {k: load_image(v, (content_image_size, content_image_size)) for k, v in content_urls.items()}\nstyle_images = {k: load_image(v, (style_image_size, style_image_size)) for k, v in style_urls.items()}\nstyle_images = {k: tf.nn.avg_pool(style_image, ksize=[3,3], strides=[1,1], padding='SAME') for k, style_image in style_images.items()}\n",
"_____no_output_____"
],
[
"#@title Specify the main content image and the style you want to use. { display-mode: \"form\" }\n\ncontent_name = 'sea_turtle' # @param ['sea_turtle', 'tuebingen', 'grace_hopper']\nstyle_name = 'munch_scream' # @param ['kanagawa_great_wave', 'kandinsky_composition_7', 'hubble_pillars_of_creation', 'van_gogh_starry_night', 'turner_nantes', 'munch_scream', 'picasso_demoiselles_avignon', 'picasso_violin', 'picasso_bottle_of_rum', 'fire', 'derkovits_woman_head', 'amadeo_style_life', 'derkovtis_talig', 'amadeo_cardoso']\n\nstylized_image = hub_module(tf.constant(content_images[content_name]),\n tf.constant(style_images[style_name]))[0]\n\nshow_n([content_images[content_name], style_images[style_name], stylized_image],\n titles=['Original content image', 'Style image', 'Stylized image'])",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
e7983f5b9df9b7d60d5f625f6653fbe5763d1e72 | 813,515 | ipynb | Jupyter Notebook | Hackathon_ML Sample Problems/Amex Dataset/ML on Amex Dataset_EDA.ipynb | girishvankudre/hackathon_ml_sample | b60dde5f6ccfddc15f937fa19e14c15df8c61f28 | [
"MIT"
]
| null | null | null | Hackathon_ML Sample Problems/Amex Dataset/ML on Amex Dataset_EDA.ipynb | girishvankudre/hackathon_ml_sample | b60dde5f6ccfddc15f937fa19e14c15df8c61f28 | [
"MIT"
]
| null | null | null | Hackathon_ML Sample Problems/Amex Dataset/ML on Amex Dataset_EDA.ipynb | girishvankudre/hackathon_ml_sample | b60dde5f6ccfddc15f937fa19e14c15df8c61f28 | [
"MIT"
]
| 1 | 2020-03-11T08:25:32.000Z | 2020-03-11T08:25:32.000Z | 69.501495 | 56,367 | 0.668964 | [
[
[
"'''\nResampling strategies for imbalanced datasets\nhttps://www.kaggle.com/rafjaa/resampling-strategies-for-imbalanced-datasets\n\nhttps://www.kaggle.com/bharath901/amexpert-2019/data#\n'''\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\npd.set_option('display.max_columns', None)\npd.set_option('display.max_rows', None)\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.metrics import classification_report, precision_score, recall_score, f1_score, accuracy_score\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.model_selection import GridSearchCV\nfrom sklearn.metrics import confusion_matrix\nfrom sklearn.feature_selection import RFE\nimport csv",
"_____no_output_____"
],
[
"'''\nDefining a Class to import the Data into a pandas DataFrame for analysis\n- Method to storing the Data into a DataFrame\n- Method to extracting Information of the Data to understand datatype associated with each column\n- Method to describing the Data\n- Method to understanding Null values distribution\n- Method to understanding Unique values distribution\n'''\n\nclass import_data():\n \n '''\n Method to extract and store the data as pandas dataframe\n '''\n def __init__(self,path):\n self.raw_data = pd.read_csv(path)\n display (self.raw_data.head(10))\n \n\n '''\n Method to extract information about the data and display\n '''\n def get_info(self):\n display (self.raw_data.info())\n \n '''\n Method to describe the data\n '''\n def get_describe(self):\n display (self.raw_data.describe())\n \n '''\n Mehtod to understand Null values distribution\n '''\n def null_value(self):\n col_null = pd.DataFrame(self.raw_data.isnull().sum()).reset_index()\n col_null.columns = ['DataColumns','NullCount']\n col_null['NullCount_Pct'] = round((col_null['NullCount']/self.raw_data.shape[0])*100,2)\n display (col_null)\n \n '''\n Method to understand Unique values distribution\n '''\n def unique_value(self):\n col_uniq = pd.DataFrame(self.raw_data.nunique()).reset_index()\n col_uniq.columns = ['DataColumns','UniqCount']\n col_uniq_cnt = pd.DataFrame(self.raw_data.count(axis=0)).reset_index()\n col_uniq_cnt.columns = ['DataColumns','UniqCount']\n col_uniq['UniqCount_Pct'] = round((col_uniq['UniqCount']/col_uniq_cnt['UniqCount'])*100,2)\n display (col_uniq)\n '''\n Method to return the dataset as dataframe\n '''\n def return_data(self):\n base_loan_data = self.raw_data\n return (base_loan_data)\n \n\n'''\nEvaluation and Analysis starts here for train.csv\n'''\n#path = str(input('Enter the path to load the dataset:'))\npath = '/Users/pritigirishvankudre/Day7_BasicML/Amex/train.csv'\nprint ('='*100)\ndata = import_data(path)\n#data.get_info()\n#data.null_value()\n#data.unique_value()\n#data.get_describe()\ntrain_data = data.return_data()",
"====================================================================================================\n"
],
[
"'''\nEvaluation and Analysis starts here for Campaign Data\n'''\n#path = str(input('Enter the path to load the dataset:'))\npath = '/Users/pritigirishvankudre/Day7_BasicML/Amex/campaign_data.csv'\nprint ('='*100)\ndata = import_data(path)\n#data.get_info()\n#data.null_value()\n#data.unique_value()\n#data.get_describe()\ncampaign_data = data.return_data()",
"====================================================================================================\n"
],
[
"'''\nEvaluation and Analysis starts here for Coupon Data\n'''\n#path = str(input('Enter the path to load the dataset:'))\npath = '/Users/pritigirishvankudre/Day7_BasicML/Amex/coupon_item_mapping.csv'\nprint ('='*100)\ndata = import_data(path)\n#data.get_info()\n#data.null_value()\n#data.unique_value()\n#data.get_describe()\ncoupon_data = data.return_data()",
"====================================================================================================\n"
],
[
"'''\nEvaluation and Analysis starts here for Customer Demographic\n'''\n#path = str(input('Enter the path to load the dataset:'))\npath = '/Users/pritigirishvankudre/Day7_BasicML/Amex/customer_demographics.csv'\nprint ('='*100)\ndata = import_data(path)\n#data.get_info()\n#data.null_value()\n#data.unique_value()\n#data.get_describe()\ncust_demo_data = data.return_data()",
"====================================================================================================\n"
],
[
"'''\nEvaluation and Analysis starts here for Customer Transaction\n'''\n#path = str(input('Enter the path to load the dataset:'))\npath = '/Users/pritigirishvankudre/Day7_BasicML/Amex/customer_transaction_data.csv'\nprint ('='*100)\ndata = import_data(path)\n#data.get_info()\n#data.null_value()\n#data.unique_value()\n#data.get_describe()\ncust_tran_data = data.return_data()",
"====================================================================================================\n"
],
[
"'''\nEvaluation and Analysis starts here for Item Data\n'''\n#path = str(input('Enter the path to load the dataset:'))\npath = '/Users/pritigirishvankudre/Day7_BasicML/Amex/item_data.csv'\nprint ('='*100)\ndata = import_data(path)\n#data.get_info()\n#data.null_value()\n#data.unique_value()\n#data.get_describe()\nitem_data = data.return_data()",
"====================================================================================================\n"
],
[
"train_data['redemption_status'].value_counts()[0]/len(train_data)",
"_____no_output_____"
]
],
[
[
"Data seems imbalance hence need to be balanced basis resampling techniques.",
"_____no_output_____"
]
],
[
[
"def date_q(date):\n \"\"\"\n Convert Date to Quarter when separated with /\n \"\"\"\n qdate = date.strip().split('/')[1:]\n qdate1 = qdate[0]\n\n if qdate1 in ['01','02','03']:\n return (str('Q1' + '-' + qdate[1]))\n if qdate1 in ['04','05','06']:\n return (str('Q2' + '-' + qdate[1]))\n if qdate1 in ['07','08','09']:\n return (str('Q3' + '-' + qdate[1]))\n if qdate1 in ['10','11','12']:\n return (str('Q4' + '-' + qdate[1]))",
"_____no_output_____"
],
[
"def date_q1(date):\n \"\"\"\n Calculates Age in years from DOB and Disbursal Date\n \"\"\"\n qdate = date.strip().split('-')[0:2]\n qdate1 = qdate[1]\n qdate2 = str(qdate[0])\n if qdate1 in ['01','02','03']:\n return (str('Q1' + '-' + qdate2[2:]))\n if qdate1 in ['04','05','06']:\n return (str('Q2' + '-' + qdate2[2:]))\n if qdate1 in ['07','08','09']:\n return (str('Q3' + '-' + qdate2[2:]))\n if qdate1 in ['10','11','12']:\n return (str('Q4' + '-' + qdate2[2:]))",
"_____no_output_____"
]
],
[
[
"# EDA for merged file",
"_____no_output_____"
]
],
[
[
"campaign_data_DATE = campaign_data.copy()\ncampaign_data_DATE.head()\n\ncampaign_data_DATE['start_date_q'] = campaign_data_DATE['start_date'].map(lambda x: date_q(x))\ncampaign_data_DATE['end_date_q'] = campaign_data_DATE['end_date'].map(lambda x: date_q(x))\ncampaign_data_DATE.head()\n\ncampaign_data_DATE.drop(['start_date','end_date'],axis=1,inplace=True)\n\ncust_tran_data_4 = cust_tran_data.copy()\ncust_tran_data_4 = pd.merge(cust_tran_data_4,coupon_data,how='inner',on='item_id')\ncust_tran_data_4['tran_date_q'] = cust_tran_data_4['date'].map(lambda x: date_q1(x))\ncust_tran_data_4.drop('date',axis=1,inplace=True)\n\ncust_tran_data_4['tot_quantity'] = pd.DataFrame(cust_tran_data_4.groupby(['customer_id','item_id','coupon_id','tran_date_q'])['quantity'].transform('sum'))\ncust_tran_data_4['tot_coupon_disc'] = pd.DataFrame(cust_tran_data_4.groupby(['customer_id','item_id','coupon_id','tran_date_q'])['coupon_discount'].transform('sum'))\ncust_tran_data_4['tot_other_disc'] = pd.DataFrame(cust_tran_data_4.groupby(['customer_id','item_id','coupon_id','tran_date_q'])['other_discount'].transform('sum'))\ncust_tran_data_4['tot_sell_price'] = pd.DataFrame(cust_tran_data_4.groupby(['customer_id','item_id','coupon_id','tran_date_q'])['selling_price'].transform('sum'))\ncust_tran_data_4.drop(['quantity','coupon_discount','other_discount','selling_price'],axis=1,inplace=True)\ncust_tran_data_4.drop_duplicates(subset=['customer_id','item_id','coupon_id','tran_date_q'], keep='first', inplace=True)\ntrain_data_merge_DATE = pd.merge(train_data,cust_tran_data_4,how='inner',on=['customer_id','coupon_id'])\ntrain_data_merge_DATE = pd.merge(train_data_merge_DATE,cust_demo_data,how='left',on='customer_id')\ntrain_data_merge_DATE = pd.merge(train_data_merge_DATE,item_data,how='left',on='item_id')\ntrain_data_merge_DATE = pd.merge(train_data_merge_DATE,campaign_data_DATE,how='left',on='campaign_id')\n\ntrain_data_merge_EDA = train_data_merge_DATE.copy()\ntrain_data_merge_EDA['no_of_children'].fillna('Unspecified',inplace=True)\ntrain_data_merge_EDA['marital_status'].fillna('Unspecified',inplace=True)\ntrain_data_merge_EDA['rented'].fillna('Unspecified',inplace=True)\ntrain_data_merge_EDA['family_size'].fillna('Unspecified',inplace=True)\ntrain_data_merge_EDA['age_range'].fillna('Unspecified',inplace=True)\ntrain_data_merge_EDA['income_bracket'].fillna('Unspecified',inplace=True)",
"_____no_output_____"
],
[
"train_data_merge_EDA.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 103300 entries, 0 to 103299\nData columns (total 23 columns):\nid 103300 non-null int64\ncampaign_id 103300 non-null int64\ncoupon_id 103300 non-null int64\ncustomer_id 103300 non-null int64\nredemption_status 103300 non-null int64\nitem_id 103300 non-null int64\ntran_date_q 103300 non-null object\ntot_quantity 103300 non-null int64\ntot_coupon_disc 103300 non-null float64\ntot_other_disc 103300 non-null float64\ntot_sell_price 103300 non-null float64\nage_range 103300 non-null object\nmarital_status 103300 non-null object\nrented 103300 non-null object\nfamily_size 103300 non-null object\nno_of_children 103300 non-null object\nincome_bracket 103300 non-null object\nbrand 103300 non-null int64\nbrand_type 103300 non-null object\ncategory 103300 non-null object\ncampaign_type 103300 non-null object\nstart_date_q 103300 non-null object\nend_date_q 103300 non-null object\ndtypes: float64(3), int64(8), object(12)\nmemory usage: 18.9+ MB\n"
],
[
"train_data_merge_EDA.describe()",
"_____no_output_____"
],
[
"col_uniq = pd.DataFrame(train_data_merge_EDA.nunique()).reset_index()\ncol_uniq.columns = ['DataColumns','UniqCount']\ncol_uniq_cnt = pd.DataFrame(train_data_merge_EDA.count(axis=0)).reset_index()\ncol_uniq_cnt.columns = ['DataColumns','UniqCount']\ncol_uniq['UniqCount_Pct'] = round((col_uniq['UniqCount']/col_uniq_cnt['UniqCount'])*100,2)\ndisplay (col_uniq)",
"_____no_output_____"
]
],
[
[
"# Lets have a look for Customer Id's in terms of using coupons most and least number of times.",
"_____no_output_____"
]
],
[
[
"'''\nCustomer ids using coupons at least once with their demographic details\n'''\n\na = pd.DataFrame(train_data_merge_EDA[(train_data_merge_EDA['redemption_status']==1)])\nb = pd.DataFrame(a.groupby('customer_id')['redemption_status'].sum()).reset_index()\nb.columns = ['customer_id','redeem_count']\nb.sort_values(by='redeem_count',ascending=False,inplace=True)\nprint ('Top 5 Customers reediming coupons')\ndisplay (b.head())",
"Top 5 Customers reediming coupons\n"
],
[
"c = pd.DataFrame(train_data_merge_EDA[(train_data_merge_EDA['customer_id']==626)|(train_data_merge_EDA['customer_id']==1574)|(train_data_merge_EDA['customer_id']==1210)|(train_data_merge_EDA['customer_id']==235)|(train_data_merge_EDA['customer_id']==1534)][['customer_id','age_range','marital_status','rented','family_size','no_of_children','income_bracket']])\nc.drop_duplicates(subset=['customer_id'], keep='first', inplace=True)\ndisplay (c)",
"_____no_output_____"
]
],
[
[
"Hypothesis to test:\n\n1. Is age_range of 36 to 55 is mostly using coupons???\n\n2. Is Marital Status as Married are to redeem coupon???\n\n3. Is couple (family size of 2) are using coupons mostly???\n\n4. Is people not on rent are mostly using coupons???\n\n5. Is no_of_children irrelevant to redeem coupon???\n\n6. Is income bracket of 5 are using coupons mostly???",
"_____no_output_____"
]
],
[
[
"'''\n1. Is age_range of 36 to 55 is mostly using coupons???\n'''\nd = pd.DataFrame(train_data_merge_EDA.groupby(['age_range'])['redemption_status'].sum()).reset_index()\nd.columns = ['age_range','tot_redeem']\nd['percent'] = round(d['tot_redeem']/(d['tot_redeem'].sum())*100,2)\ndisplay (d)",
"_____no_output_____"
],
[
"%matplotlib notebook\ntrain_data_merge_EDA.groupby(['age_range','redemption_status']).size().unstack().plot(kind='barh',stacked=True, width=0.7)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Inference : Out of the age demographic data available, age range of 36 to 55 are mostly redeeming coupons.",
"_____no_output_____"
]
],
[
[
"'''\n2. Is Marital Status as Married are to redeem coupon???\n'''\n\ne = pd.DataFrame(train_data_merge_EDA.groupby(['marital_status'])['redemption_status'].sum()).reset_index()\ne.columns = ['marital_status','tot_redeem']\ne['percent'] = round(e['tot_redeem']/(e['tot_redeem'].sum())*100,2)\ndisplay (e)",
"_____no_output_____"
],
[
"%matplotlib notebook\ntrain_data_merge_EDA.groupby(['marital_status','redemption_status']).size().unstack().plot(kind='barh',stacked=True, width=0.7)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Inference : As most of the customers haven't mentioned their marital status it is difficult to support that Married customer redeem more. But basis available data, we could see Married people are mostly using coupons to redeem. So at this stage I could think of giving more weightage to a person which has discloses marital status as Married.",
"_____no_output_____"
]
],
[
[
"'''\n3. Is couple (family size of 2) are using coupons mostly???\n'''\n\nf = pd.DataFrame(train_data_merge_EDA.groupby(['family_size'])['redemption_status'].sum()).reset_index()\nf.columns = ['family_size','tot_redeem']\nf['percent'] = round(f['tot_redeem']/(f['tot_redeem'].sum())*100,2)\ndisplay (f)",
"_____no_output_____"
],
[
"%matplotlib notebook\ntrain_data_merge_EDA.groupby(['family_size','redemption_status']).size().unstack().plot(kind='barh',stacked=True, width=0.7)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Inference : Considering family size of 2 or more as a married couple (and transforming that ratio on to Marital Status data of Unspecified), then we could make an assumption over here that Married couple are mostly using the coupons to redeem.",
"_____no_output_____"
]
],
[
[
"'''\n4. Is people not on rent are mostly using coupons???\n'''\n\ng = pd.DataFrame(train_data_merge_EDA.groupby(['rented'])['redemption_status'].sum()).reset_index()\ng.columns = ['rented','tot_redeem']\ng['percent'] = round(g['tot_redeem']/(g['tot_redeem'].sum())*100,2)\ndisplay (g)",
"_____no_output_____"
],
[
"%matplotlib notebook\ntrain_data_merge_EDA.groupby(['rented','redemption_status']).size().unstack().plot(kind='barh',stacked=True, width=0.7)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Inference : Maximum people have provided status as not rented so we could assume here more weightage to such customers as they have shown greater tendency towards redemption of the coupons.",
"_____no_output_____"
]
],
[
[
"'''\n5. Is no_of_children irrelevant to redeem coupon???\n'''\n\nh = pd.DataFrame(train_data_merge_EDA.groupby(['no_of_children'])['redemption_status'].sum()).reset_index()\nh.columns = ['no_of_children','tot_redeem']\nh['percent'] = round(h['tot_redeem']/(h['tot_redeem'].sum())*100,2)\ndisplay (h)",
"_____no_output_____"
],
[
"%matplotlib notebook\ntrain_data_merge_EDA.groupby(['no_of_children','redemption_status']).size().unstack().plot(kind='barh',stacked=True, width=0.7)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Inference : Since most of the customers preferred not to disclose on number of children, at this point we can assume that this field has no significance with redemption of coupons.",
"_____no_output_____"
]
],
[
[
"'''\n6. Is income bracket of 5 are using coupons mostly???\n'''\n\nj = pd.DataFrame(train_data_merge_EDA.groupby(['income_bracket'])['redemption_status'].sum()).reset_index()\nj.columns = ['income_bracket','tot_redeem']\nj['percent'] = round(j['tot_redeem']/(j['tot_redeem'].sum())*100,2)\ndisplay (j)",
"_____no_output_____"
],
[
"%matplotlib notebook\ntrain_data_merge_EDA.groupby(['income_bracket','redemption_status']).size().unstack().plot(kind='barh',stacked=True, width=0.7)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Inference : Assuming 5 as mid income group, customers in this group clearly shows behaviour towards redemption of coupon.",
"_____no_output_____"
],
[
"# Let's explore from Coupon's perspective, in terms of most redeemed and attributes associated with coupons",
"_____no_output_____"
]
],
[
[
"'''\nCoupon ids getting redeemed very often and attributes associated with it\n'''\n\nk = pd.DataFrame(a.groupby('coupon_id')['redemption_status'].sum()).reset_index()\nk.columns = ['coupon_id','redeem_count']\nk.sort_values(by='redeem_count',ascending=False,inplace=True)\nprint ('Top 5 Coupon ids redeemed')\ndisplay (k.head())",
"Top 5 Coupon ids redeemed\n"
],
[
"l1 = pd.DataFrame(train_data_merge_EDA[(train_data_merge_EDA['coupon_id']==21)|(train_data_merge_EDA['coupon_id']==6)|(train_data_merge_EDA['coupon_id']==22)|(train_data_merge_EDA['coupon_id']==9)|(train_data_merge_EDA['coupon_id']==8)][['coupon_id','item_id','brand','brand_type','category']])\nl1.drop_duplicates(subset=['coupon_id','item_id'], keep='first', inplace=True)",
"_____no_output_____"
],
[
"l2 = pd.DataFrame(l1.groupby(['brand'])['brand'].count())\nl2.columns = ['tot_brand_cnt']\nl2['percent'] = round(l2['tot_brand_cnt']/(l2['tot_brand_cnt'].sum())*100,2)\nl2.sort_values(by='tot_brand_cnt',ascending=False,inplace=True)\ndisplay (l2.head(10))",
"_____no_output_____"
],
[
"l3 = pd.DataFrame(l1.groupby(['brand_type'])['brand_type'].count())\nl3.columns = ['tot_brand_type_cnt']\nl3['percent'] = round(l3['tot_brand_type_cnt']/(l3['tot_brand_type_cnt'].sum())*100,2)\nl3.sort_values(by='tot_brand_type_cnt',ascending=False,inplace=True)\ndisplay (l3)",
"_____no_output_____"
],
[
"l4 = pd.DataFrame(l1.groupby(['category'])['category'].count())\nl4.columns = ['tot_category_cnt']\nl4['percent'] = round(l4['tot_category_cnt']/(l4['tot_category_cnt'].sum())*100,2)\nl4.sort_values(by='tot_category_cnt',ascending=False,inplace=True)\ndisplay (l4)",
"_____no_output_____"
]
],
[
[
"Basis visualization for top 5 coupons redeemed, we could look for trend towards\n1. brand 56 is the top selling, let's verify if specific brand shows tendency towards coupon redemption???\n\n2. Verify brand type shows tendency towards coupon redemption???\n\n3. Verify category shows tendency towards coupon redemption???\n\n4. Verify campaign type shows tendency towards coupon redemption???\n",
"_____no_output_____"
]
],
[
[
"'''\n1. verify if specific brand shows tendency towards coupon redemption???\n'''\nm = pd.DataFrame(l1[(l1['brand']==56)|(l1['brand']==133)|(l1['brand']==1337)|(l1['brand']==544)|(l1['brand']==681)][['brand','brand_type','category']])\nm.drop_duplicates(subset=['brand','brand_type','category'], keep='first', inplace=True)\ndisplay(m)",
"_____no_output_____"
]
],
[
[
"Inference: For brand 56, there is a wide range of category available under the umbrella of Food products and displays greater tendency towards coupon redemption as well. Even rest other top 4 brands as well belong to general food product category under Grocery.",
"_____no_output_____"
]
],
[
[
"'''\n2. Verify brand type shows tendency towards coupon redemption???\n'''\n\nm1 = pd.DataFrame(train_data_merge_EDA.groupby(['brand_type'])['redemption_status'].sum()).reset_index()\nm1.columns = ['brand_type','tot_redeem']\nm1['percent'] = round(m1['tot_redeem']/(m1['tot_redeem'].sum())*100,2)\ndisplay (m1)",
"_____no_output_____"
],
[
"%matplotlib notebook\ntrain_data_merge_EDA.groupby(['brand_type','redemption_status']).size().unstack().plot(kind='barh',stacked=True, width=0.7)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Inference: Coupon redemption percentage seems high when associated with an Established brand type.",
"_____no_output_____"
]
],
[
[
"'''\n3. Verify category shows tendency towards coupon redemption???\n'''\n\nm2 = pd.DataFrame(train_data_merge_EDA.groupby(['category'])['redemption_status'].sum()).reset_index()\nm2.columns = ['category','tot_redeem']\nm2['percent'] = round(m2['tot_redeem']/(m2['tot_redeem'].sum())*100,2)\nm2.sort_values(by='tot_redeem',ascending=False,inplace=True)\ndisplay (m2)",
"_____no_output_____"
],
[
"%matplotlib notebook\ntrain_data_merge_EDA.groupby(['category','redemption_status']).size().unstack().plot(kind='barh',stacked=True, width=0.7)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Inference : Grocery, Packaged Meat, Pharmaceutical, Natural Prodcut, Dairy & Juice and Meat are the category seems more associated with coupon redemption.",
"_____no_output_____"
]
],
[
[
"'''\n4. Verify campaign type shows tendency towards coupon redemption???\n'''\n\nm3 = pd.DataFrame(train_data_merge_EDA.groupby(['campaign_type'])['redemption_status'].sum()).reset_index()\nm3.columns = ['campaign_type','tot_redeem']\nm3['percent'] = round(m3['tot_redeem']/(m3['tot_redeem'].sum())*100,2)\ndisplay (m3)",
"_____no_output_____"
],
[
"%matplotlib notebook\ntrain_data_merge_EDA.groupby(['campaign_type','redemption_status']).size().unstack().plot(kind='barh',stacked=True, width=0.7)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Inference : Campaign type of X seems to have more associated with coupon redemption.",
"_____no_output_____"
],
[
"# Let's explore coupon redemption trend basis campaign start date and transaction date",
"_____no_output_____"
]
],
[
[
"n = pd.DataFrame(train_data_merge_EDA.groupby(['start_date_q','end_date_q'])['redemption_status'].sum()).reset_index()\nn.columns = ['start_date_q','end_date_q','tot_redeem']\nn['percent'] = round(n['tot_redeem']/(n['tot_redeem'].sum())*100,2)\nn.sort_values(by='tot_redeem',ascending=False,inplace=True)\ndisplay (n)",
"_____no_output_____"
],
[
"%matplotlib notebook\ntrain_data_merge_EDA.groupby(['start_date_q','end_date_q','redemption_status']).size().unstack().plot(kind='barh',stacked=True, width=0.7)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Inference : Campaign started between Q2-13 to Q3-13 foolwed by Q1-13 to Q2-13, seems experience more association towards coupon redemption trend. So we could assume over here campaign spanned across early quarters of year seem to more association with coupon redemption.",
"_____no_output_____"
]
],
[
[
"n1 = pd.DataFrame(train_data_merge_EDA.groupby(['tran_date_q'])['redemption_status'].sum()).reset_index()\nn1.columns = ['tran_date_q','tot_redeem']\nn1['percent'] = round(n1['tot_redeem']/(n1['tot_redeem'].sum())*100,2)\nn1.sort_values(by='tot_redeem',ascending=False,inplace=True)\ndisplay (n1)",
"_____no_output_____"
]
],
[
[
"Inference : We could not clearly see association between transaction quarter and coupon redemption behaviour.",
"_____no_output_____"
],
[
"# Basis EDA, we could assume below fields to be more associated towards coupon redemption tendency\n- Age range of 36-55\n- Married couple (mostly with family size of just 2)\n- Income bracket of 5\n- Brand 56 covering wide variety of Category under a large umbrella of Food and Beverages\n- Brand Type of Established\n- Category sapnning mostly across Grocery, Packaged Meat, Pharmaceutical, Natural Prodcut, Dairy & Juice and Meat\n- Coupons associated with Campaign Type X seem to be redeemed more in terms of percentage\n- Campaign start and end date spanned across early quarters of year seem to more association with coupon redemption",
"_____no_output_____"
],
[
"['campaign_id', 'coupon_id', 'customer_id', 'item_id', 'tot_coupon_disc', 'tot_sell_price', 'age_range', 'family_size', 'income_bracket', 'brand']",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
]
|
e7984610983dc13956b961721fe93e75f70e3272 | 30,764 | ipynb | Jupyter Notebook | SVR/Python/svr.ipynb | arpit1920/Machine-Learning-all-Algorithms | cc4bc882741e1d8ef6a8af4a4b7e027079c22db1 | [
"MIT"
]
| 2 | 2020-08-09T22:46:01.000Z | 2021-07-14T14:15:37.000Z | SVR/Python/svr.ipynb | arpit1920/Machine-Learning-all-Algorithms | cc4bc882741e1d8ef6a8af4a4b7e027079c22db1 | [
"MIT"
]
| null | null | null | SVR/Python/svr.ipynb | arpit1920/Machine-Learning-all-Algorithms | cc4bc882741e1d8ef6a8af4a4b7e027079c22db1 | [
"MIT"
]
| 1 | 2021-11-29T11:03:38.000Z | 2021-11-29T11:03:38.000Z | 153.82 | 12,814 | 0.878397 | [
[
[
"# SVR\n\n# Importing the libraries\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd",
"_____no_output_____"
],
[
"# Importing the dataset\ndataset = pd.read_csv('Position_Salaries.csv')\nX = dataset.iloc[:, 1:2].values\nY = dataset.iloc[:, 2].values",
"_____no_output_____"
],
[
"# Feature Scaling\nfrom sklearn.preprocessing import StandardScaler\nsc_X = StandardScaler()\nsc_Y = StandardScaler()\nX = sc_X.fit_transform(X)\nY = sc_Y.fit_transform(Y.reshape(-1,1))",
"_____no_output_____"
],
[
"# Fitting SVR to the dataset\nfrom sklearn.svm import SVR\nregressor = SVR(kernel = 'rbf')\nregressor.fit(X, Y)",
"/usr/local/lib/python3.6/dist-packages/sklearn/utils/validation.py:760: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().\n y = column_or_1d(y, warn=True)\n"
],
[
"# Predicting a new result\nY_pred = regressor.predict(np.array([6.5]).reshape(1, 1))",
"_____no_output_____"
],
[
"# Visualising the SVR results\nplt.scatter(X, Y, color = 'red')\nplt.plot(X, regressor.predict(X), color = 'blue')\nplt.title('Truth or Bluff (SVR)')\nplt.xlabel('Position level')\nplt.ylabel('Salary')\nplt.show()",
"_____no_output_____"
],
[
"# Visualising the SVR results (for higher resolution and smoother curve)\nX_grid = np.arange(min(X), max(X), 0.01) # choice of 0.01 instead of 0.1 step because the data is feature scaled\nX_grid = X_grid.reshape((len(X_grid), 1))\nplt.scatter(X, Y, color = 'red')\nplt.plot(X_grid, regressor.predict(X_grid), color = 'blue')\nplt.title('Truth or Bluff (SVR)')\nplt.xlabel('Position level')\nplt.ylabel('Salary')\nplt.show()",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e79857cd85b405d4be814dad3c76f5fd6324b023 | 3,843 | ipynb | Jupyter Notebook | Informatics/Deep Learning/TensorFlow - deeplearning.ai/3. NLP/Course_3_Week_1_Lesson_1.ipynb | MarcosSalib/Cocktail_MOOC | 46279c2ec642554537c639702ed8e540ea49afdf | [
"MIT"
]
| null | null | null | Informatics/Deep Learning/TensorFlow - deeplearning.ai/3. NLP/Course_3_Week_1_Lesson_1.ipynb | MarcosSalib/Cocktail_MOOC | 46279c2ec642554537c639702ed8e540ea49afdf | [
"MIT"
]
| null | null | null | Informatics/Deep Learning/TensorFlow - deeplearning.ai/3. NLP/Course_3_Week_1_Lesson_1.ipynb | MarcosSalib/Cocktail_MOOC | 46279c2ec642554537c639702ed8e540ea49afdf | [
"MIT"
]
| null | null | null | 30.744 | 308 | 0.515483 | [
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"<a href=\"https://colab.research.google.com/github/lmoroney/dlaicourse/blob/master/TensorFlow%20In%20Practice/Course%203%20-%20NLP/Course%203%20-%20Week%201%20-%20Lesson%201.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"##### Copyright 2019 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
],
[
"from tensorflow.keras.preprocessing.text import Tokenizer\n\nsentences = [\n 'i love my dog',\n 'I, love my cat',\n 'You love my dog!'\n]\n\ntokenizer = Tokenizer(num_words = 100)\ntokenizer.fit_on_texts(sentences)\nword_index = tokenizer.word_index\nprint(word_index)",
"{'love': 1, 'my': 2, 'i': 3, 'dog': 4, 'cat': 5, 'you': 6}\n"
],
[
"",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
]
|
e7985bb4eaca45007470852016f6304858e1e4dc | 116,348 | ipynb | Jupyter Notebook | Extras/Exercicio de Graficos.ipynb | Cesarso/Python-para-DataScience | 6c0ff82240d439e9dc89cb68e3e2a87473435174 | [
"MIT"
]
| null | null | null | Extras/Exercicio de Graficos.ipynb | Cesarso/Python-para-DataScience | 6c0ff82240d439e9dc89cb68e3e2a87473435174 | [
"MIT"
]
| null | null | null | Extras/Exercicio de Graficos.ipynb | Cesarso/Python-para-DataScience | 6c0ff82240d439e9dc89cb68e3e2a87473435174 | [
"MIT"
]
| null | null | null | 668.666667 | 43,576 | 0.949548 | [
[
[
"%matplotlib inline\nimport pandas as pd\nimport matplotlib.pyplot as plt\nplt.rc('figure', figsize = (15, 7))\n\ndados = pd.read_csv('dados/aluguel_amostra.csv', sep = ';')",
"_____no_output_____"
],
[
"area = plt.figure()\ng1 = area.add_subplot(2, 2, 1)\ng2 = area.add_subplot(2, 2, 2)\ngrupo1 = dados.groupby('Tipo Agregado')['Valor']\nlabel = grupo1.count().index\nvalores = grupo1.count().values\ng1.pie(valores, labels = label, autopct='%1.1f%%')\ng1.set_title('Total de Imóveis por Tipo Agregado')\ngrupo2 = dados.groupby('Tipo')['Valor']\nlabel = grupo2.count().index\nvalores = grupo2.count().values\ng2.pie(valores, labels = label, autopct='%1.1f%%', explode = (.1, .1, .1, .1, .1))\ng2.set_title('Total de Imóveis por Tipo')",
"_____no_output_____"
],
[
"\narea = plt.figure()\ng1 = area.add_subplot(1, 2, 1)\ng2 = area.add_subplot(1, 2, 2)\ngrupo1 = dados.groupby('Tipo Agregado')['Valor']\nlabel = grupo1.mean().index\nvalores = grupo1.mean().values\ng1.pie(valores, labels = label, autopct='%1.1f%%')\ng1.set_title('Total de Imóveis por Tipo Agregado')\ngrupo2 = dados.groupby('Tipo')['Valor']\nlabel = grupo2.mean().index\nvalores = grupo2.mean().values\ng2.pie(valores, labels = label, autopct='%1.1f%%', explode = (.1, .1, .1, .1, .1))\ng2.set_title('Total de Imóveis por Tipo')",
"_____no_output_____"
],
[
"area = plt.figure()\ng1 = area.add_subplot(1, 2, 1)\ng2 = area.add_subplot(1, 2, 2)\ngrupo1 = dados.groupby('Tipo Agregado')['Valor']\nlabel = grupo1.count().index\nvalores = grupo1.count().values\ng1.pie(valores, labels = label, autopct='%1.1f%%')\ng1.set_title('Total de Imóveis por Tipo Agregado')\ngrupo2 = dados.groupby('Tipo')['Valor']\nlabel = grupo2.count().index\nvalores = grupo2.count().values\ng2.pie(valores, labels = label, autopct='%1.1f%%', explode = (.1, .1, .1, .1, .1))\ng2.set_title('Total de Imóveis por Tipo')",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code"
]
]
|
e798600003393369ea8df79bebe61963737512a1 | 171,169 | ipynb | Jupyter Notebook | train_t_shirts.ipynb | lumstery/maskrcnn | dd5008fcfdbaf46a61167214759b90dce0a3efd6 | [
"MIT"
]
| null | null | null | train_t_shirts.ipynb | lumstery/maskrcnn | dd5008fcfdbaf46a61167214759b90dce0a3efd6 | [
"MIT"
]
| null | null | null | train_t_shirts.ipynb | lumstery/maskrcnn | dd5008fcfdbaf46a61167214759b90dce0a3efd6 | [
"MIT"
]
| null | null | null | 255.09538 | 38,112 | 0.905842 | [
[
[
"# Mask R-CNN - Train on Custom Dataset\n\n\nThis notebook shows how to train Mask R-CNN on your own dataset. You'd still need a GPU, though, because the network backbone is a Resnet101, which would be too slow to train on a CPU. On a GPU, you can start to get okay-ish results in a few minutes, and good results in less than an hour.",
"_____no_output_____"
]
],
[
[
"import os\nimport sys\nimport random\nimport math\nimport re\nimport time\nimport numpy as np\nimport cv2\nimport matplotlib\nimport matplotlib.pyplot as plt\n\nfrom config import Config\nimport utils\nimport model as modellib\nimport visualize\nfrom model import log\n\n%matplotlib inline \n\n# Root directory of the project\nROOT_DIR = os.getcwd()\n# Directory to save logs and trained model\nMODEL_DIR = os.path.join(ROOT_DIR, \"logs\")\n\n# Local path to trained weights file\nCOCO_MODEL_PATH = os.path.join(ROOT_DIR, \"mask_rcnn_coco.h5\")\n# Download COCO trained weights from Releases if needed\nif not os.path.exists(COCO_MODEL_PATH):\n utils.download_trained_weights(COCO_MODEL_PATH)",
"c:\\users\\yaroslav_strontsitsk\\appdata\\local\\continuum\\anaconda3\\envs\\maskrcnn\\lib\\site-packages\\h5py\\__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\nUsing TensorFlow backend.\n"
]
],
[
[
"## Configurations",
"_____no_output_____"
]
],
[
[
"class ShapesConfig(Config):\n \"\"\"Configuration for training on the toy shapes dataset.\n Derives from the base Config class and overrides values specific\n to the toy shapes dataset.\n \"\"\"\n # Give the configuration a recognizable name\n NAME = \"clothes\"\n\n # Train on 1 GPU and 8 images per GPU. We can put multiple images on each\n # GPU because the images are small. Batch size is 8 (GPUs * images/GPU).\n GPU_COUNT = 1\n IMAGES_PER_GPU = 8\n\n # Number of classes (including background)\n NUM_CLASSES = 1 + 1 # background + 1 shapes\n\n # Use small images for faster training. Set the limits of the small side\n # the large side, and that determines the image shape.\n IMAGE_MIN_DIM = 128\n IMAGE_MAX_DIM = 128\n \n # Use smaller anchors because our image and objects are small\n RPN_ANCHOR_SCALES = (8, 16, 32, 64, 128) # anchor side in pixels\n\n # Reduce training ROIs per image because the images are small and have\n # few objects. Aim to allow ROI sampling to pick 33% positive ROIs.\n TRAIN_ROIS_PER_IMAGE = 64\n\n # Use a small epoch since the data is simple\n STEPS_PER_EPOCH = 100\n\n # use small validation steps since the epoch is small\n VALIDATION_STEPS = 6\n \nconfig = ShapesConfig()\nconfig.display()",
"\nConfigurations:\nBACKBONE_SHAPES [[32 32]\n [16 16]\n [ 8 8]\n [ 4 4]\n [ 2 2]]\nBACKBONE_STRIDES [4, 8, 16, 32, 64]\nBATCH_SIZE 8\nBBOX_STD_DEV [0.1 0.1 0.2 0.2]\nDETECTION_MAX_INSTANCES 100\nDETECTION_MIN_CONFIDENCE 0.7\nDETECTION_NMS_THRESHOLD 0.3\nGPU_COUNT 1\nIMAGES_PER_GPU 8\nIMAGE_MAX_DIM 128\nIMAGE_MIN_DIM 128\nIMAGE_PADDING True\nIMAGE_SHAPE [128 128 3]\nLEARNING_MOMENTUM 0.9\nLEARNING_RATE 0.001\nMASK_POOL_SIZE 14\nMASK_SHAPE [28, 28]\nMAX_GT_INSTANCES 100\nMEAN_PIXEL [123.7 116.8 103.9]\nMINI_MASK_SHAPE (56, 56)\nNAME clothes\nNUM_CLASSES 2\nPOOL_SIZE 7\nPOST_NMS_ROIS_INFERENCE 1000\nPOST_NMS_ROIS_TRAINING 2000\nROI_POSITIVE_RATIO 0.33\nRPN_ANCHOR_RATIOS [0.5, 1, 2]\nRPN_ANCHOR_SCALES (8, 16, 32, 64, 128)\nRPN_ANCHOR_STRIDE 1\nRPN_BBOX_STD_DEV [0.1 0.1 0.2 0.2]\nRPN_NMS_THRESHOLD 0.7\nRPN_TRAIN_ANCHORS_PER_IMAGE 256\nSTEPS_PER_EPOCH 100\nTRAIN_ROIS_PER_IMAGE 64\nUSE_MINI_MASK True\nUSE_RPN_ROIS True\nVALIDATION_STEPS 6\nWEIGHT_DECAY 0.0001\n\n\n"
]
],
[
[
"## Notebook Preferences",
"_____no_output_____"
]
],
[
[
"def get_ax(rows=1, cols=1, size=8):\n \"\"\"Return a Matplotlib Axes array to be used in\n all visualizations in the notebook. Provide a\n central point to control graph sizes.\n \n Change the default size attribute to control the size\n of rendered images\n \"\"\"\n _, ax = plt.subplots(rows, cols, figsize=(size*cols, size*rows))\n return ax",
"_____no_output_____"
]
],
[
[
"## Dataset\n\nLoad a dataset\n\nExtend the Dataset class and add a method to load the shapes dataset, `load_images()`, and override the following methods:\n\n* load_image()\n* load_mask()\n* image_reference()",
"_____no_output_____"
]
],
[
[
" class ShapesDataset(utils.Dataset):\n\n def load_images(self, count, prefix):\n \"\"\"Load the requested number of images.\n count: number of images to load.\n \"\"\"\n # Add classes\n self.add_class(\"clothes\", 1, \"t-shirt\")\n \n # Add images\n for i in range(count):\n self.add_image(\"clothes\", image_id=i, path=\"./t-shirt/\"+prefix+\"/\"+str(i+1)+\".jpg\",)\n\n def image_reference(self, image_id):\n info = self.image_info[image_id]\n if info[\"source\"] == \"clothes\":\n return info[\"clothes\"]\n else:\n super(self.__class__).image_reference(self, image_id)\n\n def load_mask(self, image_id):\n \"\"\"Load instance mask for shape of the given image ID.\"\"\"\n info = self.image_info[image_id]\n img = cv2.imread(info['path'], cv2.IMREAD_UNCHANGED)\n #print(\"img.shape=\"+str(image.shape))\n #resized_image = cv2.resize(img, (128, 128)) \n #print(\"resized_image.shape=\"+str(resized_image.shape))\n #img[np.where((image!=[255,255,255]).all(axis=2))] = [0,0,0]\n lower_black = np.array([235,235,235], dtype = \"uint8\")\n upper_black = np.array([255,255,255], dtype = \"uint8\")\n mask = cv2.inRange(img, lower_black, upper_black)\n mask = mask[..., np.newaxis]\n mask = mask.astype(dtype=bool)\n mask = np.logical_not(mask)\n #cv2.imshow('mask', mask)\n #cv2.waitKey()\n class_ids = np.array([1])\n return mask, class_ids.astype(np.int8)",
"_____no_output_____"
],
[
"# Training dataset\ndataset_train = ShapesDataset()\ndataset_train.load_images(20,\"train\")\ndataset_train.prepare()\n\n# Validation dataset\ndataset_val = ShapesDataset()\ndataset_val.load_images(5,\"validation\")\ndataset_val.prepare()",
"_____no_output_____"
],
[
"# Load and display random samples\nimage_ids = np.random.choice(dataset_train.image_ids, 3)\nfor image_id in image_ids:\n image = dataset_train.load_image(image_id)\n mask, class_ids = dataset_train.load_mask(image_id)\n visualize.display_top_masks(image, mask, class_ids, dataset_train.class_names,1)",
"_____no_output_____"
]
],
[
[
"## Ceate Model",
"_____no_output_____"
]
],
[
[
"# Create model in training mode\nmodel = modellib.MaskRCNN(mode=\"training\", config=config,\n model_dir=MODEL_DIR)",
"_____no_output_____"
],
[
"# Which weights to start with?\ninit_with = \"coco\" # imagenet, coco, or last\n\nif init_with == \"imagenet\":\n model.load_weights(model.get_imagenet_weights(), by_name=True)\nelif init_with == \"coco\":\n # Load weights trained on MS COCO, but skip layers that\n # are different due to the different number of classes\n # See README for instructions to download the COCO weights\n model.load_weights(COCO_MODEL_PATH, by_name=True,\n exclude=[\"mrcnn_class_logits\", \"mrcnn_bbox_fc\", \n \"mrcnn_bbox\", \"mrcnn_mask\"])\nelif init_with == \"last\":\n # Load the last model you trained and continue training\n model.load_weights(model.find_last()[1], by_name=True)\n ",
"_____no_output_____"
]
],
[
[
"## Training\n\nTrain in two stages:\n1. Only the heads. Here we're freezing all the backbone layers and training only the randomly initialized layers (i.e. the ones that we didn't use pre-trained weights from MS COCO). To train only the head layers, pass `layers='heads'` to the `train()` function.\n\n2. Fine-tune all layers. For this simple example it's not necessary, but we're including it to show the process. Simply pass `layers=\"all` to train all layers.",
"_____no_output_____"
]
],
[
[
"# Train the head branches\n# Passing layers=\"heads\" freezes all layers except the head\n# layers. You can also pass a regular expression to select\n# which layers to train by name pattern.\nmodel.train(dataset_train, dataset_val, \n learning_rate=config.LEARNING_RATE, \n epochs=1, \n layers='heads')",
"\nStarting at epoch 0. LR=0.001\n\nCheckpoint Path: D:\\MaskCNN\\Mask_RCNN\\logs\\clothes20180314T1303\\mask_rcnn_clothes_{epoch:04d}.h5\nSelecting layers to train\nfpn_c5p5 (Conv2D)\nfpn_c4p4 (Conv2D)\nfpn_c3p3 (Conv2D)\nfpn_c2p2 (Conv2D)\nfpn_p5 (Conv2D)\nfpn_p2 (Conv2D)\nfpn_p3 (Conv2D)\nfpn_p4 (Conv2D)\nIn model: rpn_model\n rpn_conv_shared (Conv2D)\n rpn_class_raw (Conv2D)\n rpn_bbox_pred (Conv2D)\nmrcnn_mask_conv1 (TimeDistributed)\nmrcnn_mask_bn1 (TimeDistributed)\nmrcnn_mask_conv2 (TimeDistributed)\nmrcnn_mask_bn2 (TimeDistributed)\nmrcnn_class_conv1 (TimeDistributed)\nmrcnn_class_bn1 (TimeDistributed)\nmrcnn_mask_conv3 (TimeDistributed)\nmrcnn_mask_bn3 (TimeDistributed)\nmrcnn_class_conv2 (TimeDistributed)\nmrcnn_class_bn2 (TimeDistributed)\nmrcnn_mask_conv4 (TimeDistributed)\nmrcnn_mask_bn4 (TimeDistributed)\nmrcnn_bbox_fc (TimeDistributed)\nmrcnn_mask_deconv (TimeDistributed)\nmrcnn_class_logits (TimeDistributed)\nmrcnn_mask (TimeDistributed)\nWARNING:tensorflow:From D:\\MaskCNN\\Mask_RCNN\\model.py:2073: calling reduce_mean (from tensorflow.python.ops.math_ops) with keep_dims is deprecated and will be removed in a future version.\nInstructions for updating:\nkeep_dims is deprecated, use keepdims instead\n"
],
[
"# Fine tune all layers\n# Passing layers=\"all\" trains all layers. You can also \n# pass a regular expression to select which layers to\n# train by name pattern.\n#model.train(dataset_train, dataset_val, \n# learning_rate=config.LEARNING_RATE / 10,\n# epochs=2, \n# layers=\"all\")",
"_____no_output_____"
],
[
"# Save weights\n# Typically not needed because callbacks save after every epoch\n# Uncomment to save manually\n# model_path = os.path.join(MODEL_DIR, \"mask_rcnn_shapes.h5\")\n# model.keras_model.save_weights(model_path)",
"_____no_output_____"
]
],
[
[
"## Detection",
"_____no_output_____"
]
],
[
[
"class InferenceConfig(ShapesConfig):\n GPU_COUNT = 1\n IMAGES_PER_GPU = 1\n\ninference_config = InferenceConfig()\n\n# Recreate the model in inference mode\nmodel = modellib.MaskRCNN(mode=\"inference\", \n config=inference_config,\n model_dir=MODEL_DIR)\n\n# Get path to saved weights\n# Either set a specific path or find last trained weights\n# model_path = os.path.join(ROOT_DIR, \".h5 file name here\")\nmodel_path = model.find_last()[1]\n\n# Load trained weights (fill in path to trained weights here)\nassert model_path != \"\", \"Provide path to trained weights\"\nprint(\"Loading weights from \", model_path)\nmodel.load_weights(model_path, by_name=True)",
"Loading weights from D:\\MaskCNN\\Mask_RCNN\\logs\\clothes20180314T1303\\mask_rcnn_clothes_0001.h5\n"
],
[
"# Test on a random image\nimage_id = random.choice(dataset_val.image_ids)\noriginal_image, image_meta, gt_class_id, gt_bbox, gt_mask =\\\n modellib.load_image_gt(dataset_val, inference_config, \n image_id, use_mini_mask=False)\n\nlog(\"original_image\", original_image)\nlog(\"image_meta\", image_meta)\nlog(\"gt_class_id\", gt_class_id)\nlog(\"gt_bbox\", gt_bbox)\nlog(\"gt_mask\", gt_mask)\nvisualize.display_instances(original_image, gt_bbox, gt_mask, gt_class_id, \n dataset_train.class_names, figsize=(8, 8))",
"original_image shape: (128, 128, 3) min: 5.00000 max: 255.00000\nimage_meta shape: (10,) min: 0.00000 max: 400.00000\ngt_class_id shape: (1,) min: 1.00000 max: 1.00000\ngt_bbox shape: (1, 4) min: 7.00000 max: 121.00000\ngt_mask shape: (128, 128, 1) min: 0.00000 max: 1.00000\n"
],
[
"results = model.detect([original_image], verbose=1)\n\nr = results[0]\nvisualize.display_instances(original_image, r['rois'], r['masks'], r['class_ids'], \n dataset_val.class_names, r['scores'], figsize=(8, 8))",
"Processing 1 images\nimage shape: (128, 128, 3) min: 5.00000 max: 255.00000\nmolded_images shape: (1, 128, 128, 3) min: -115.70000 max: 151.10000\nimage_metas shape: (1, 10) min: 0.00000 max: 128.00000\n"
]
],
[
[
"## Evaluation",
"_____no_output_____"
]
],
[
[
"# Compute VOC-Style mAP @ IoU=0.5\n# Running on 10 images. Increase for better accuracy.\nimage_ids = np.random.choice(dataset_val.image_ids, 10)\nAPs = []\nfor image_id in image_ids:\n # Load image and ground truth data\n image, image_meta, gt_class_id, gt_bbox, gt_mask =\\\n modellib.load_image_gt(dataset_val, inference_config,\n image_id, use_mini_mask=False)\n molded_images = np.expand_dims(modellib.mold_image(image, inference_config), 0)\n # Run object detection\n results = model.detect([image], verbose=0)\n r = results[0]\n # Compute AP\n AP, precisions, recalls, overlaps =\\\n utils.compute_ap(gt_bbox, gt_class_id, gt_mask,\n r[\"rois\"], r[\"class_ids\"], r[\"scores\"], r['masks'])\n APs.append(AP)\n \nprint(\"mAP: \", np.mean(APs))",
"mAP: 1.0\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
]
|
e79868ef875be242b6a4cc68d495da37a4afc618 | 74,668 | ipynb | Jupyter Notebook | 02_Filtering_&_Sorting/Euro12/Exercises_with_Solutions.ipynb | arscool3/pandas_exercises | 5ef128fd186c0be84ac5917e60642b83953ee56a | [
"BSD-3-Clause"
]
| null | null | null | 02_Filtering_&_Sorting/Euro12/Exercises_with_Solutions.ipynb | arscool3/pandas_exercises | 5ef128fd186c0be84ac5917e60642b83953ee56a | [
"BSD-3-Clause"
]
| null | null | null | 02_Filtering_&_Sorting/Euro12/Exercises_with_Solutions.ipynb | arscool3/pandas_exercises | 5ef128fd186c0be84ac5917e60642b83953ee56a | [
"BSD-3-Clause"
]
| null | null | null | 34.110553 | 185 | 0.258625 | [
[
[
"# Ex2 - Filtering and Sorting Data\nCheck out [Euro 12 Exercises Video Tutorial](https://youtu.be/iqk5d48Qisg) to watch a data scientist go through the exercises",
"_____no_output_____"
],
[
"This time we are going to pull data directly from the internet.\n\n### Step 1. Import the necessary libraries",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
]
],
[
[
"### Step 2. Import the dataset from this [address](https://raw.githubusercontent.com/guipsamora/pandas_exercises/master/02_Filtering_%26_Sorting/Euro12/Euro_2012_stats_TEAM.csv). ",
"_____no_output_____"
],
[
"### Step 3. Assign it to a variable called euro12.",
"_____no_output_____"
]
],
[
[
"euro12 = pd.read_csv('https://raw.githubusercontent.com/guipsamora/pandas_exercises/master/02_Filtering_%26_Sorting/Euro12/Euro_2012_stats_TEAM.csv', sep=',')\neuro12",
"_____no_output_____"
]
],
[
[
"### Step 4. Select only the Goal column.",
"_____no_output_____"
]
],
[
[
"euro12.Goals",
"_____no_output_____"
]
],
[
[
"### Step 5. How many team participated in the Euro2012?",
"_____no_output_____"
]
],
[
[
"euro12.shape[0]",
"_____no_output_____"
]
],
[
[
"### Step 6. What is the number of columns in the dataset?",
"_____no_output_____"
]
],
[
[
"euro12.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 16 entries, 0 to 15\nData columns (total 35 columns):\nTeam 16 non-null object\nGoals 16 non-null int64\nShots on target 16 non-null int64\nShots off target 16 non-null int64\nShooting Accuracy 16 non-null object\n% Goals-to-shots 16 non-null object\nTotal shots (inc. Blocked) 16 non-null int64\nHit Woodwork 16 non-null int64\nPenalty goals 16 non-null int64\nPenalties not scored 16 non-null int64\nHeaded goals 16 non-null int64\nPasses 16 non-null int64\nPasses completed 16 non-null int64\nPassing Accuracy 16 non-null object\nTouches 16 non-null int64\nCrosses 16 non-null int64\nDribbles 16 non-null int64\nCorners Taken 16 non-null int64\nTackles 16 non-null int64\nClearances 16 non-null int64\nInterceptions 16 non-null int64\nClearances off line 15 non-null float64\nClean Sheets 16 non-null int64\nBlocks 16 non-null int64\nGoals conceded 16 non-null int64\nSaves made 16 non-null int64\nSaves-to-shots ratio 16 non-null object\nFouls Won 16 non-null int64\nFouls Conceded 16 non-null int64\nOffsides 16 non-null int64\nYellow Cards 16 non-null int64\nRed Cards 16 non-null int64\nSubs on 16 non-null int64\nSubs off 16 non-null int64\nPlayers Used 16 non-null int64\ndtypes: float64(1), int64(29), object(5)\nmemory usage: 4.4+ KB\n"
]
],
[
[
"### Step 7. View only the columns Team, Yellow Cards and Red Cards and assign them to a dataframe called discipline",
"_____no_output_____"
]
],
[
[
"# filter only giving the column names\n\ndiscipline = euro12[['Team', 'Yellow Cards', 'Red Cards']]\ndiscipline",
"_____no_output_____"
]
],
[
[
"### Step 8. Sort the teams by Red Cards, then to Yellow Cards",
"_____no_output_____"
]
],
[
[
"discipline.sort_values(['Red Cards', 'Yellow Cards'], ascending = False)",
"_____no_output_____"
]
],
[
[
"### Step 9. Calculate the mean Yellow Cards given per Team",
"_____no_output_____"
]
],
[
[
"round(discipline['Yellow Cards'].mean())",
"_____no_output_____"
]
],
[
[
"### Step 10. Filter teams that scored more than 6 goals",
"_____no_output_____"
]
],
[
[
"euro12[euro12.Goals > 6]",
"_____no_output_____"
]
],
[
[
"### Step 11. Select the teams that start with G",
"_____no_output_____"
]
],
[
[
"euro12[euro12.Team.str.startswith('G')]",
"_____no_output_____"
]
],
[
[
"### Step 12. Select the first 7 columns",
"_____no_output_____"
]
],
[
[
"# use .iloc to slices via the position of the passed integers\n# : means all, 0:7 means from 0 to 7\n\neuro12.iloc[: , 0:7]",
"_____no_output_____"
]
],
[
[
"### Step 13. Select all columns except the last 3.",
"_____no_output_____"
]
],
[
[
"# use negative to exclude the last 3 columns\n\neuro12.iloc[: , :-3]",
"_____no_output_____"
]
],
[
[
"### Step 14. Present only the Shooting Accuracy from England, Italy and Russia",
"_____no_output_____"
]
],
[
[
"# .loc is another way to slice, using the labels of the columns and indexes\n\neuro12.loc[euro12.Team.isin(['England', 'Italy', 'Russia']), ['Team','Shooting Accuracy']]",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
e7986cf698bf3238bbd5c0951ebde6f08fe1c88c | 16,189 | ipynb | Jupyter Notebook | notebooks/LSTM4_pred_plots_12h.ipynb | harryli18/hybrid-rnn-models | 9baae52985cf21635b5c2e75b785ee6c2eac85d4 | [
"MIT"
]
| 1 | 2021-03-11T03:45:06.000Z | 2021-03-11T03:45:06.000Z | notebooks/LSTM4_pred_plots_12h.ipynb | harryli18/hybrid-rnn-models | 9baae52985cf21635b5c2e75b785ee6c2eac85d4 | [
"MIT"
]
| null | null | null | notebooks/LSTM4_pred_plots_12h.ipynb | harryli18/hybrid-rnn-models | 9baae52985cf21635b5c2e75b785ee6c2eac85d4 | [
"MIT"
]
| null | null | null | 33.727083 | 181 | 0.590957 | [
[
[
"import plaidml.keras\nplaidml.keras.install_backend()\nimport os\nos.environ[\"KERAS_BACKEND\"] = \"plaidml.keras.backend\"",
"_____no_output_____"
],
[
"# Importing useful libraries\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd\nfrom sklearn.preprocessing import MinMaxScaler\nfrom keras.models import Sequential\nfrom keras.layers import Dense, LSTM, Dropout, GRU, Bidirectional, Conv1D, Flatten, MaxPooling1D\nfrom keras.optimizers import SGD\nimport math\nfrom sklearn.metrics import mean_squared_error\nfrom sklearn.model_selection import train_test_split\nfrom keras import optimizers\n\nimport time ",
"_____no_output_____"
]
],
[
[
"### Data Processing",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv('../data/num_data.csv')",
"_____no_output_____"
],
[
"dataset = df",
"_____no_output_____"
],
[
"dataset.shape",
"_____no_output_____"
],
[
"def return_rmse(test,predicted):\n rmse = math.sqrt(mean_squared_error(test, predicted))\n return rmse",
"_____no_output_____"
],
[
"data_size = dataset.shape[0]\ntrain_size=int(data_size * 0.6)\ntest_size = 100\nvalid_size = data_size - train_size - test_size",
"_____no_output_____"
],
[
"training_set = dataset[:train_size].iloc[:,4:16].values\nvalid_set = dataset[train_size:train_size+valid_size].iloc[:,4:16].values\ntest_set = dataset[data_size-test_size:].iloc[:,4:16].values",
"_____no_output_____"
],
[
"y = dataset.iloc[:,4].values\ny = y.reshape(-1,1)\nn_feature = training_set.shape[1]\ny.shape",
"_____no_output_____"
],
[
"# Scaling the dataset\nsc = MinMaxScaler(feature_range=(0,1))\ntraining_set_scaled = sc.fit_transform(training_set)\nvalid_set_scaled = sc.fit_transform(valid_set)\ntest_set_scaled = sc.fit_transform(test_set)\n\nsc_y = MinMaxScaler(feature_range=(0,1))\ny_scaled = sc_y.fit_transform(y)",
"_____no_output_____"
],
[
"# split a multivariate sequence into samples\nposition_of_target = 4\ndef split_sequences(sequences, n_steps_in, n_steps_out):\n X_, y_ = list(), list()\n for i in range(len(sequences)):\n # find the end of this pattern\n end_ix = i + n_steps_in\n out_end_ix = end_ix + n_steps_out-1\n # check if we are beyond the dataset\n if out_end_ix > len(sequences):\n break\n # gather input and output parts of the pattern\n seq_x, seq_y = sequences[i:end_ix, :], sequences[end_ix-1:out_end_ix, position_of_target]\n X_.append(seq_x)\n y_.append(seq_y)\n return np.array(X_), np.array(y_)",
"_____no_output_____"
],
[
"n_steps_in = 12\nn_steps_out = 12\nX_train, y_train = split_sequences(training_set_scaled, n_steps_in, n_steps_out)\nX_valid, y_valid = split_sequences(valid_set_scaled, n_steps_in, n_steps_out)\nX_test, y_test = split_sequences(test_set_scaled, n_steps_in, n_steps_out)",
"_____no_output_____"
],
[
"LSTM_4 = Sequential()\n\nLSTM_4.add(LSTM(units=50, return_sequences=True, input_shape=(X_train.shape[1],n_feature), activation='tanh'))\nLSTM_4.add(LSTM(units=50, return_sequences=True, input_shape=(X_train.shape[1],n_feature), activation='tanh'))\nLSTM_4.add(LSTM(units=50, return_sequences=True, input_shape=(X_train.shape[1],n_feature), activation='tanh'))\nLSTM_4.add(LSTM(units=50, activation='tanh'))\nLSTM_4.add(Dense(units=n_steps_out))\n\n\n# Compiling the RNNs\nadam = optimizers.Adam(lr=0.01)\nLSTM_4.compile(optimizer=adam,loss='mean_squared_error')",
"INFO:plaidml:Opening device \"llvm_cpu.0\"\n"
],
[
"RnnModelDict = {'LSTM_4': LSTM_4}\n\nrmse_df = pd.DataFrame(columns=['Model', 'train_rmse', 'valid_rmse', 'train_time'])\n\n# RnnModelDict = {'LSTM_GRU': LSTM_GRU_reg}",
"_____no_output_____"
],
[
"for model in RnnModelDict:\n regressor = RnnModelDict[model]\n \n print('training start for', model) \n start = time.process_time()\n regressor.fit(X_train,y_train,epochs=50,batch_size=1024)\n train_time = round(time.process_time() - start, 2)\n \n print('results for training set')\n y_train_pred = regressor.predict(X_train)\n# plot_predictions(y_train,y_train_pred)\n train_rmse = return_rmse(y_train,y_train_pred)\n \n print('results for valid set')\n y_valid_pred = regressor.predict(X_valid)\n# plot_predictions(y_valid,y_valid_pred)\n valid_rmse = return_rmse(y_valid,y_valid_pred)\n \n \n# print('results for test set - 24 hours')\n# y_test_pred24 = regressor.predict(X_test_24)\n# plot_predictions(y_test_24,y_test_pred24)\n# test24_rmse = return_rmse(y_test_24,y_test_pred24)\n \n \n one_df = pd.DataFrame([[model, train_rmse, valid_rmse, train_time]], \n columns=['Model', 'train_rmse', 'valid_rmse', 'train_time'])\n rmse_df = pd.concat([rmse_df, one_df])\n\n# save the rmse results \n# rmse_df.to_csv('../rmse_24h_plus_time.csv')\n",
"training start for LSTM_4\nEpoch 1/50\n121856/252438 [=============>................] - ETA: 8:10 - loss: 0.0072"
],
[
"# history = regressor.fit(X_train, y_train, epochs=50, batch_size=1024, validation_data=(X_valid, y_valid),\n# verbose=2, shuffle=False)\n# # plot history\n\n# plt.figure(figsize=(30, 15))\n# plt.plot(history.history['loss'], label='Training')\n# plt.plot(history.history['val_loss'], label='Validation')\n# plt.xlabel('Epochs')\n# plt.ylabel('Loss')\n# plt.legend()\n# plt.show()",
"_____no_output_____"
],
[
"# Transform back and plot\ny_train_origin = y[:train_size-46]\ny_valid_origin = y[train_size:train_size+valid_size]\n\ny_train_pred = regressor.predict(X_train)\ny_train_pred_origin = sc_y.inverse_transform(y_train_pred)\n\ny_valid_pred = regressor.predict(X_valid)\ny_valid_pred_origin = sc_y.inverse_transform(y_valid_pred)\n\n_y_train_pred_origin = y_train_pred_origin[:, 0:1]\n_y_valid_pred_origin = y_valid_pred_origin[:, 0:1]\n\n",
"_____no_output_____"
],
[
"plt.figure(figsize=(20, 8));\nplt.plot(pd.to_datetime(valid_original.index), valid_original, \n alpha=0.5, color='red', label='Actual PM2.5 Concentration',)\nplt.plot(pd.to_datetime(valid_original.index), y_valid_pred_origin[:,0:1], \n alpha=0.5, color='blue', label='Predicted PM2.5 Concentation')\nplt.title('PM2.5 Concentration Prediction')\nplt.xlabel('Time')\nplt.ylabel('PM2.5 Concentration')\nplt.legend()\nplt.show()\n",
"_____no_output_____"
],
[
"sample = 500\nplt.figure(figsize=(20, 8));\nplt.plot(pd.to_datetime(valid_original.index[-500:]), valid_original[-500:], \n alpha=0.5, color='red', label='Actual PM2.5 Concentration',)\nplt.plot(pd.to_datetime(valid_original.index[-500:]), y_valid_pred_origin[:,11:12][-500:], \n alpha=0.5, color='blue', label='Predicted PM2.5 Concentation')\nplt.title('PM2.5 Concentration Prediction')\nplt.xlabel('Time')\nplt.ylabel('PM2.5 Concentration')\nplt.legend()\nplt.show()",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code"
]
| [
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e7986f8a6697222a0d696682d2dc04ff5daf3ed9 | 12,982 | ipynb | Jupyter Notebook | sentiment_analysis/training_classfiers.ipynb | dipjyotidas/NLP | c0f83596764dee292230e664a2a10fa1acd1fdc5 | [
"MIT"
]
| null | null | null | sentiment_analysis/training_classfiers.ipynb | dipjyotidas/NLP | c0f83596764dee292230e664a2a10fa1acd1fdc5 | [
"MIT"
]
| null | null | null | sentiment_analysis/training_classfiers.ipynb | dipjyotidas/NLP | c0f83596764dee292230e664a2a10fa1acd1fdc5 | [
"MIT"
]
| null | null | null | 34.80429 | 454 | 0.559544 | [
[
[
"#### Author : Dipjyoti Das (https://www.linkedin.com/in/dipjyotidas)",
"_____no_output_____"
],
[
"### This script is used to Train classifiers on the dataset and all the classifers are saved as pickle files.\n\n### The pickled classfiers are used in the sentiment_analysis.py file.",
"_____no_output_____"
],
[
"###### Import all the libraries",
"_____no_output_____"
]
],
[
[
"import nltk\nimport random\n#from nltk.corpus import movie_reviews\nfrom nltk.classify.scikitlearn import SklearnClassifier\nimport pickle\n\nfrom sklearn.naive_bayes import MultinomialNB, BernoulliNB\nfrom sklearn.linear_model import LogisticRegression, SGDClassifier\nfrom sklearn.svm import SVC, LinearSVC, NuSVC\n\nfrom nltk.classify import ClassifierI\nfrom statistics import mode\n\nfrom nltk.tokenize import word_tokenize\n\n\nclass VoteClassifier(ClassifierI):\n def __init__(self, *classifiers):\n self._classifiers = classifiers\n\n def classify(self, features):\n votes = []\n for c in self._classifiers:\n v = c.classify(features)\n votes.append(v)\n return mode(votes)\n\n def confidence(self, features):\n votes = []\n for c in self._classifiers:\n v = c.classify(features)\n votes.append(v)\n\n choice_votes = votes.count(mode(votes))\n conf = choice_votes / len(votes)\n return conf\n \n \n \nshort_pos = open(\"data/positive.txt\",\"r\").read()\nshort_neg = open(\"data/negative.txt\",\"r\").read()\n\n\n# using POS -parts of speech tag - allow only specific words\n#pos - tuple- word, parts of speech\n\nall_words = []\ndocuments = []\n\n# j is adject, r is adverb, and v is verb\n#allowed_word_types = [\"J\",\"R\",\"V\"]\nallowed_word_types = [\"J\"] # allowing only Adjectives\n\nfor p in short_pos.split('\\n'):\n documents.append((p, \"pos\") )\n words = word_tokenize(p)\n pos = nltk.pos_tag(words)\n for w in pos:\n if w[1][0] in allowed_word_types: # w - tuple, not getting Nouns, commas\n all_words.append(w[0].lower())\n\n \nfor p in short_neg.split('\\n'):\n documents.append((p, \"neg\"))\n words = word_tokenize(p)\n pos = nltk.pos_tag(words)\n for w in pos:\n if w[1][0] in allowed_word_types:\n all_words.append(w[0].lower())\n\n\n# pickle and store documents\n# pickled algos - folder created to store all the pickled objects :\n\n\nsave_documents = open(\"pickled_algos/documents.pickle\", \"wb\")\npickle.dump(documents, save_documents)\nsave_documents.close()\n\n\nall_words = nltk.FreqDist(all_words)\n\n\nword_features = list(all_words.keys())[:5000]\n\n# pickle and store word features\nsave_word_features = open(\"pickled_algos/word_features5k.pickle\",\"wb\")\npickle.dump(word_features, save_word_features)\nsave_word_features.close()\n\n\ndef find_features(document):\n words = word_tokenize(document)\n features = {}\n for w in word_features:\n features[w] = (w in words)\n\n return features\n\nfeaturesets = [(find_features(rev), category) for (rev, category) in documents]\n\n# Pickle and store - featuresets : occupies space of 300 MB, don't store it as pickle object\n\n#save_featuresets = open(\"pickled_algos/featuresets.pickle\", \"wb\")\n#pickle.dump(featuresets, save_featuresets)\n#save_featuresets.close()\n\nrandom.shuffle(featuresets)\nprint(len(featuresets))\n\n\n# Train and Test set:\n\n\ntesting_set = featuresets[10000:]\ntraining_set = featuresets[:10000]\n\n\n## List of Classifiers :\n\n## Naive Bayes classifier:\n\nclassifier = nltk.NaiveBayesClassifier.train(training_set)\nprint(\"Original Naive Bayes Algo accuracy percent:\", (nltk.classify.accuracy(classifier, testing_set))*100)\nclassifier.show_most_informative_features(15)\n\n## pickle and store - Naive Bayes classifier\nsave_classifier = open(\"pickled_algos/originalnaivebayes5k.pickle\",\"wb\")\npickle.dump(classifier, save_classifier)\nsave_classifier.close()\n\n\n## MNB classifier :\n\n\nMNB_classifier = SklearnClassifier(MultinomialNB())\nMNB_classifier.train(training_set)\nprint(\"MNB_classifier accuracy percent:\", (nltk.classify.accuracy(MNB_classifier, testing_set))*100)\n\n\n# pickle and store MNB classifier:\n\nsave_classifier = open(\"pickled_algos/MNB_classifier5k.pickle\",\"wb\")\npickle.dump(MNB_classifier, save_classifier)\nsave_classifier.close()\n\n## BernoulliNB classifier:\n\nBernoulliNB_classifier = SklearnClassifier(BernoulliNB())\nBernoulliNB_classifier.train(training_set)\nprint(\"BernoulliNB_classifier accuracy percent:\", (nltk.classify.accuracy(BernoulliNB_classifier, testing_set))*100)\n\n#pickle and store BernoulliNB classifier:\n\nsave_classifier = open(\"pickled_algos/BernoulliNB_classifier5k.pickle\",\"wb\")\npickle.dump(BernoulliNB_classifier, save_classifier)\nsave_classifier.close()\n\n\n## Logistic Regression classifier:\n\n\nLogisticRegression_classifier = SklearnClassifier(LogisticRegression())\nLogisticRegression_classifier.train(training_set)\nprint(\"LogisticRegression_classifier accuracy percent:\", (nltk.classify.accuracy(LogisticRegression_classifier, testing_set))*100)\n\n#pickle and store Logistic Regression classifier:\n\nsave_classifier = open(\"pickled_algos/LogisticRegression_classifier5k.pickle\",\"wb\")\npickle.dump(LogisticRegression_classifier, save_classifier)\nsave_classifier.close()\n\n\n## LinearSVC classifier\n\n\nLinearSVC_classifier = SklearnClassifier(LinearSVC())\nLinearSVC_classifier.train(training_set)\nprint(\"LinearSVC_classifier accuracy percent:\", (nltk.classify.accuracy(LinearSVC_classifier, testing_set))*100)\n\n\n# pickle and store LinearSVC classifier:\n\nsave_classifier = open(\"pickled_algos/LinearSVC_classifier5k.pickle\",\"wb\")\npickle.dump(LinearSVC_classifier, save_classifier)\nsave_classifier.close()\n\n## SGDC classifier:\n\nSGDC_classifier = SklearnClassifier(SGDClassifier())\nSGDC_classifier.train(training_set)\nprint(\"SGDClassifier accuracy percent:\",nltk.classify.accuracy(SGDC_classifier, testing_set)*100)\n\n# pickle and store SGDC classifier:\n\nsave_classifier = open(\"pickled_algos/SGDC_classifier5k.pickle\",\"wb\")\npickle.dump(SGDC_classifier, save_classifier)\nsave_classifier.close()\n\n\n## Can't pickle the Voted Classifier - class of its own\n",
"10664\nOriginal Naive Bayes Algo accuracy percent: 73.64457831325302\nMost Informative Features\n wonderful = True pos : neg = 21.8 : 1.0\n engrossing = True pos : neg = 19.7 : 1.0\n generic = True neg : pos = 16.9 : 1.0\n mediocre = True neg : pos = 16.9 : 1.0\n inventive = True pos : neg = 15.7 : 1.0\n routine = True neg : pos = 14.9 : 1.0\n flat = True neg : pos = 14.9 : 1.0\n refreshing = True pos : neg = 14.4 : 1.0\n boring = True neg : pos = 13.8 : 1.0\n warm = True pos : neg = 13.1 : 1.0\n intimate = True pos : neg = 11.7 : 1.0\n realistic = True pos : neg = 11.7 : 1.0\n stale = True neg : pos = 11.6 : 1.0\n mindless = True neg : pos = 11.6 : 1.0\n delicate = True pos : neg = 11.0 : 1.0\nMNB_classifier accuracy percent: 74.24698795180723\nBernoulliNB_classifier accuracy percent: 74.3975903614458\nLogisticRegression_classifier accuracy percent: 73.49397590361446\nLinearSVC_classifier accuracy percent: 72.13855421686746\n"
]
],
[
[
"We can run upto this cell one time. The sentiment analysis module uses the saved pickle objects and it also has the voting classfier and the sentiment function. The module saved is sentiment_analysis.py",
"_____no_output_____"
],
[
"#### After importing the sentiment analysis module :",
"_____no_output_____"
],
[
"### We can use this to check if any sentiment is positive or negative with the confidence level.",
"_____no_output_____"
],
[
"#### Examples :",
"_____no_output_____"
]
],
[
[
"import sentiment_analysis as s\n\n# referencing the sentiment function of the sentiment_analysis.py script\n\n# Example - Pass through our own positive review\nprint(s.sentiment(\"This movie was awesome! The story was great and performances were amazing, I really liked it!\"))\n\n# Example - Pass through a negative review\nprint(s.sentiment(\"This movie was junk. No story at all and acting sucked. Horrible movie, 1/10\"))\n\n",
"('pos', 1.0)\n('neg', 1.0)\n"
],
[
"## Both are at 100% confidence level",
"_____no_output_____"
]
],
[
[
"#### This module can be used to perform live sentiment analysis from Twitter!",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
]
|
e79889d315acc9cb825186923fa3e0e902669161 | 4,571 | ipynb | Jupyter Notebook | locale/examples/02-plot/pbr.ipynb | tkoyama010/pyvista-doc-translations | 23bb813387b7f8bfe17e86c2244d5dd2243990db | [
"MIT"
]
| 4 | 2020-08-07T08:19:19.000Z | 2020-12-04T09:51:11.000Z | locale/examples/02-plot/pbr.ipynb | tkoyama010/pyvista-doc-translations | 23bb813387b7f8bfe17e86c2244d5dd2243990db | [
"MIT"
]
| 19 | 2020-08-06T00:24:30.000Z | 2022-03-30T19:22:24.000Z | locale/examples/02-plot/pbr.ipynb | tkoyama010/pyvista-doc-translations | 23bb813387b7f8bfe17e86c2244d5dd2243990db | [
"MIT"
]
| 1 | 2021-03-09T07:50:40.000Z | 2021-03-09T07:50:40.000Z | 42.324074 | 756 | 0.564865 | [
[
[
"%matplotlib inline\nfrom pyvista import set_plot_theme\nset_plot_theme('document')",
"_____no_output_____"
]
],
[
[
"Physically Based Rendering {#pbr_example}\n==========================\n\nVTK 9 introduced Physically Based Rendering (PBR) and we have exposed\nthat functionality in PyVista. Read the [blog about\nPBR](https://blog.kitware.com/vtk-pbr/) for more details.\n\nPBR is only supported for `pyvista.PolyData`{.interpreted-text\nrole=\"class\"} and can be triggered via the `pbr` keyword argument of\n`add_mesh`. Also use the `metallic` and `roughness` arguments for\nfurther control.\n\nLet\\'s show off this functionality by rendering a high quality mesh of a\nstatue as though it were metallic.\n",
"_____no_output_____"
]
],
[
[
"import pyvista as pv\nfrom pyvista import examples\n\n# Load the statue mesh\nmesh = examples.download_nefertiti()\nmesh.rotate_x(-90.) # rotate to orient with the skybox\n\n# Download skybox\ncubemap = examples.download_sky_box_cube_map()",
"_____no_output_____"
]
],
[
[
"Let\\'s render the mesh with a base color of \\\"linen\\\" to give it a metal\nlooking finish.\n",
"_____no_output_____"
]
],
[
[
"p = pv.Plotter()\np.add_actor(cubemap.to_skybox())\np.set_environment_texture(cubemap) # For reflecting the environment off the mesh\np.add_mesh(mesh, color='linen',\n pbr=True, metallic=0.8, roughness=0.1,\n diffuse=1)\n\n# Define a nice camera perspective\ncpos = [(-313.40, 66.09, 1000.61),\n (0.0, 0.0, 0.0),\n (0.018, 0.99, -0.06)]\n\np.show(cpos=cpos)",
"_____no_output_____"
]
],
[
[
"Show the variation of the metallic and roughness parameters.\n\nPlot with metallic increasing from left to right and roughness\nincreasing from bottom to top.\n",
"_____no_output_____"
]
],
[
[
"colors = ['red', 'teal', 'black', 'orange', 'silver']\n\np = pv.Plotter()\np.set_environment_texture(cubemap)\n\nfor i in range(5):\n for j in range(6):\n sphere = pv.Sphere(radius=0.5, center=(0.0, 4 - i, j))\n p.add_mesh(sphere, color=colors[i],\n pbr=True, metallic=i/4, roughness=j/5)\n\np.view_vector((-1, 0, 0), (0, 1, 0))\np.show()",
"_____no_output_____"
]
],
[
[
"Combine custom lighting and physically based rendering.\n",
"_____no_output_____"
]
],
[
[
"# download louis model\nmesh = examples.download_louis_louvre()\nmesh.rotate_z(140)\n\n\nplotter = pv.Plotter(lighting=None)\nplotter.set_background('black')\nplotter.add_mesh(mesh, color='linen', pbr=True,\n metallic=0.5, roughness=0.5, diffuse=1)\n\n\n# setup lighting\nlight = pv.Light((-2, 2, 0), (0, 0, 0), 'white')\nplotter.add_light(light)\n\nlight = pv.Light((2, 0, 0), (0, 0, 0), (0.7, 0.0862, 0.0549))\nplotter.add_light(light)\n\nlight = pv.Light((0, 0, 10), (0, 0, 0), 'white')\nplotter.add_light(light)\n\n\n# plot with a good camera position\nplotter.camera_position = [(9.51, 13.92, 15.81),\n (-2.836, -0.93, 10.2),\n (-0.22, -0.18, 0.959)]\ncpos = plotter.show()",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
e79895cf9dc388ebcc50462e09c9dccf230811fa | 324,965 | ipynb | Jupyter Notebook | examples/overview.ipynb | werthm/becquerel | 975c522419eb4245efb0069b4aac23520703deb5 | [
"BSD-3-Clause-LBNL"
]
| 1 | 2022-02-24T23:27:07.000Z | 2022-02-24T23:27:07.000Z | examples/overview.ipynb | Am6er/becquerel | 975c522419eb4245efb0069b4aac23520703deb5 | [
"BSD-3-Clause-LBNL"
]
| null | null | null | examples/overview.ipynb | Am6er/becquerel | 975c522419eb4245efb0069b4aac23520703deb5 | [
"BSD-3-Clause-LBNL"
]
| null | null | null | 145.986074 | 75,328 | 0.865173 | [
[
[
"# Becquerel Overview\n\nThis notebook demonstrates some of the main features and functionalities of `becquerel`:\n\n1. [`bq.Spectrum`](#1.-bq.Spectrum)\n - [Constructor](#1.1-From-scratch)\n - [Energy Calibration Models](#1.2-Energy-Calibration-Models)\n - [File IO](#1.3-From-File)\n - [Backrgound Subtraction](#1.4-Background-Subtraction)\n - [Rebinning](#1.5-Rebinning)\n - [Scaling](#1.6-Scaling)\n - [Peak Finding + Auto Calibration](#1.7-Automatic-Calibration)\n1. [Nuclear-Data](#2.-Nuclear-Data)\n - [`bq.Element`](#2.1-bq.Element)\n - [`bq.Isotope`](#2.2-bq.Isotope)\n - [`bq.IsotopeQuantity`](#2.3-bq.IsotopeQuantity)\n - [`bq.materials`](#2.4-bq.materials)\n - [`bq.nndc`](#2.5-bq.nndc)\n - [`bq.xcom`](#2.6-bq.xcom)\n\nFor more details on particular features please see the other notebooks in this directory as noted. In addition, a few practical examples of using `becquerel` are given in the [misc notebook](./misc.ipynb)",
"_____no_output_____"
]
],
[
[
"%pylab inline\nimport pandas as pd\nimport becquerel as bq\nfrom pprint import pprint\nnp.random.seed(0)",
"Populating the interactive namespace from numpy and matplotlib\n"
]
],
[
[
"# 1. `bq.Spectrum`\n\nThe core class in `bq` is `Spectrum`. This class contains a variety of tools for handling **single spectrum** data.\n\nFurther details can be found in the [spectrum notebook](./spectrum.ipynb) and [spectrum plotting notebook](./plotting.ipynb).",
"_____no_output_____"
]
],
[
[
"bq.Spectrum?",
"_____no_output_____"
]
],
[
[
"## 1.1 From scratch",
"_____no_output_____"
]
],
[
[
"c, _ = np.histogram(np.random.poisson(50, 1000), bins=np.arange(101))\nspec = bq.Spectrum(counts=c, livetime=60.)\nspec",
"_____no_output_____"
],
[
"spec.plot(xmode='channels');",
"_____no_output_____"
],
[
"try:\n spec.plot(xmode='energy')\nexcept bq.PlottingError as e:\n print('ERROR:', e)\n plt.close('all')",
"ERROR: Spectrum is not calibrated, however x axis was requested as energy\n"
]
],
[
[
"## 1.2 Energy Calibration Models\n\nAll calibrations are instances of `Calibration`, which stores an arbitrary scalar function and its relevant parameters.\n\nFurther details can be found in the [energycal notebook](./energycal.ipynb).",
"_____no_output_____"
]
],
[
[
"chlist = (40, 80)\nkevlist = (661.7, 1460.83)\ncal = bq.Calibration.from_points(\"p[0] + p[1] * x\", chlist, kevlist, rng=(-1e3, 1e5))\nprint(cal.params)\nspec.apply_calibration(cal)\nprint(spec)\nspec.plot(xmode='keV');",
"[-137.43 19.97825]\nbecquerel.Spectrum\n start_time: None\n stop_time: None\n realtime: None\n livetime: 60.0\n is_calibrated: True\n num_bins: 100\n gross_counts: 1000+/-33\n gross_cps: 16.7+/-0.5\n filename: None\n"
],
[
"# New spec\nc, _ = np.histogram(np.random.poisson(50, 1000), bins=np.arange(101))\nspec2 = bq.Spectrum(counts=c, livetime=60.)\nspec2",
"_____no_output_____"
],
[
"spec2.calibrate_like(spec)\nspec2",
"_____no_output_____"
]
],
[
[
"## 1.3 From File\n\n`becquerel` currently provides parsers for:\n\n- `SPE`\n- `SPC`\n- `CNF`",
"_____no_output_____"
]
],
[
[
"spec = bq.Spectrum.from_file('../tests/samples/1110C NAA cave pottery.Spe')\nspec",
"SpeFile: Reading file ../tests/samples/1110C NAA cave pottery.Spe\n"
],
[
"spec.is_calibrated",
"_____no_output_____"
],
[
"spec.plot(yscale='log', linewidth=0.5, ymode='counts');",
"_____no_output_____"
],
[
"%%capture\nspec = bq.Spectrum.from_file('../tests/samples/01122014152731-GT01122014182338-GA37.4963000N-GO122.4633000W.cnf')",
"_____no_output_____"
],
[
"spec",
"_____no_output_____"
],
[
"%%capture\nspec = bq.Spectrum.from_file('../tests/samples/Alcatraz14.Spc')",
"_____no_output_____"
],
[
"spec",
"_____no_output_____"
]
],
[
[
"## 1.4 Background Subtraction",
"_____no_output_____"
]
],
[
[
"spec = bq.Spectrum.from_file('../tests/samples/1110C NAA cave pottery.Spe')\nprint(spec)\nbkg = bq.Spectrum.from_file('../tests/samples/1110C NAA cave background May 2017.spe')\nprint(bkg)",
"SpeFile: Reading file ../tests/samples/1110C NAA cave pottery.Spe\nbecquerel.Spectrum\n start_time: 2017-04-25 12:54:27\n stop_time: 2017-04-25 17:30:24\n realtime: 16557.0\n livetime: 16543.0\n is_calibrated: True\n num_bins: 16384\n gross_counts: (3.047+/-0.006)e+05\n gross_cps: 18.419+/-0.034\n filename: ../tests/samples/1110C NAA cave pottery.Spe\nSpeFile: Reading file ../tests/samples/1110C NAA cave background May 2017.spe\nbecquerel.Spectrum\n start_time: 2017-04-26 11:05:11\n stop_time: 2017-05-01 12:43:34\n realtime: 437903.0\n livetime: 437817.0\n is_calibrated: True\n num_bins: 16384\n gross_counts: (1.0529+/-0.0010)e+06\n gross_cps: 2.4049+/-0.0023\n filename: ../tests/samples/1110C NAA cave background May 2017.spe\n"
],
[
"bkgsub = spec - bkg\nprint('Total pottery countrate: {:6.3f}'.format(np.sum(spec.cps)))\nprint('Total background countrate: {:6.3f}'.format(np.sum(bkg.cps)))\nprint('Total subtracted countrate: {:6.3f}'.format(np.sum(bkgsub.cps)))",
"becquerel/core/spectrum.py:815: SpectrumWarning: Subtraction of counts-based specta, spectra have been converted to CPS\n warnings.warn(\n"
],
[
"fig, ax = plt.subplots(1, figsize=(12, 6))\nax = spec.plot(color='firebrick', linewidth=0.5, yscale='log', ax=ax, label='Measurement', ymode='cps')\nbkgsub.plot(ax=ax, color='dodgerblue', linewidth=0.5, label='Measurement - Background', ymode='cps')\nbkg.plot(ax=ax, color='olive', linewidth=0.5, label='Background', ymode='cps')\nax.set_ylim(bottom=1e-5)\nax.set_title('Background Subtraction')\nax.legend();",
"_____no_output_____"
],
[
"# Is there any Tl-208 in the background-subtracted spectrum?\nfig, ax = plt.subplots(1, figsize=(12, 6))\nax = spec.plot(color='firebrick', linewidth=1, yscale='linear', ax=ax, label='Measurement', ymode='cps')\nbkg.plot(ax=ax, color='olive', linewidth=1, label='Background', ymode='cps')\nbkgsub.plot(ax=ax, color='dodgerblue', linewidth=1, label='Measurement - Background', ymode='cps')\nax.set_ylim(bottom=1e-5)\nax.set_title('Background Subtraction')\nax.legend()\nplt.xlim(2600, 2630)\nplt.ylim(0, 0.0008);",
"_____no_output_____"
]
],
[
[
"## 1.5 Rebinning\n\n- deterministic (interpolation): `interpolation`\n- stochastic (convert to listmode): `listmode`\n\nFurther details can be found in the [rebinning notebook](./rebinning.ipynb).",
"_____no_output_____"
]
],
[
[
"spec = bq.Spectrum.from_file('../tests/samples/1110C NAA cave pottery.Spe')\nbkg = bq.Spectrum.from_file('../tests/samples/1110C NAA cave background May 2017.spe')\nbkg_rebin = bkg.rebin(np.linspace(0., 3000., 16000))",
"SpeFile: Reading file ../tests/samples/1110C NAA cave pottery.Spe\nSpeFile: Reading file ../tests/samples/1110C NAA cave background May 2017.spe\n"
],
[
"try:\n bkgsub = spec - bkg_rebin\nexcept bq.SpectrumError as e:\n print('ERROR:', e)",
"ERROR: Cannot add/subtract spectra of different lengths\n"
],
[
"spec_rebin = spec.rebin_like(bkg_rebin)",
"_____no_output_____"
],
[
"spec_rebin - bkg_rebin",
"becquerel/core/spectrum.py:815: SpectrumWarning: Subtraction of counts-based specta, spectra have been converted to CPS\n warnings.warn(\n"
]
],
[
[
"## 1.6 Scaling\n\nMultiplication or division will be applied to the data of the spectrum. The following decimates a spectrum by dividing by 10:",
"_____no_output_____"
]
],
[
[
"spec = bq.Spectrum.from_file('../tests/samples/1110C NAA cave background May 2017.spe')\nspec_div = spec / 10\nprint(spec_div)",
"SpeFile: Reading file ../tests/samples/1110C NAA cave background May 2017.spe\nbecquerel.Spectrum\n start_time: None\n stop_time: None\n realtime: None\n livetime: None\n is_calibrated: True\n num_bins: 16384\n gross_counts: (1.0529+/-0.0010)e+05\n gross_cps: None\n filename: None\n"
]
],
[
[
"One might however want to decimate a spectrum in a way consistent with Poisson statistics. For that there is the `downsample` method:",
"_____no_output_____"
]
],
[
[
"spec_downsample = spec.downsample(10, handle_livetime='reduce')\nprint(spec_downsample)\n\nax = spec_div.plot(label='div', ymode='counts', yscale='log')\nspec_downsample.plot(ax=ax, label='downsample', ymode='counts')\nax.legend()\nax.set_xlim(600, 620)\nax.set_ylim(1.3, 5e1);",
"becquerel.Spectrum\n start_time: None\n stop_time: None\n realtime: None\n livetime: 43781.7\n is_calibrated: True\n num_bins: 16384\n gross_counts: (1.0484+/-0.0033)e+05\n gross_cps: 2.395+/-0.008\n filename: None\n"
]
],
[
[
"## 1.7 Automatic Calibration\n\nThere are utilities in Becquerel for automatically finding peaks in a raw spectrum and matching them to a list of energies as a first pass at a full calibration.\n\nFurther details can be found in the [autocal notebook](./autocal.ipynb).\n\nLet's load an uncalibrated sodium iodide spectrum that has Cobalt-60 and background lines:",
"_____no_output_____"
]
],
[
[
"spec = bq.Spectrum.from_file('../tests/samples/digibase_5min_30_1.spe')\nfig, ax = plt.subplots(1, figsize=(12, 6))\nspec.plot(ax=ax, linewidth=0.5, xmode='channels', yscale='log')\nplt.xlim(0, len(spec));",
"SpeFile: Reading file ../tests/samples/digibase_5min_30_1.spe\n"
],
[
"# filter the spectrum\nkernel = bq.GaussianPeakFilter(400, 20, 3)\nfinder = bq.PeakFinder(spec, kernel)\nfinder.find_peaks(min_snr=10, xmin=50)\ncal = bq.AutoCalibrator(finder)\n\nplt.figure(figsize=(10, 5))\nplt.title('Signal-to-noise ratio of spectral lines after filter')\ncal.peakfinder.plot()\nplt.tight_layout()",
"_____no_output_____"
],
[
"# perform calibration\ncal.fit(\n [1173.2, 1332.5, 1460.8, 2614.5],\n gain_range=[5., 7.],\n de_max=100.,\n)\nspec.apply_calibration(cal.cal)\n\nfig, ax = plt.subplots(1, figsize=(12, 6))\nspec.plot(ax=ax, linewidth=0.5, xmode='energy', yscale='log')\nfor erg in cal.fit_energies:\n plt.plot([erg, erg], [1e-1, 1e4], 'r-', alpha=0.5)\n plt.text(erg, 1e4, '{:.1f} keV'.format(erg), rotation=90)\nplt.xlim(0, 3000);",
"found best gain: 6.371703 keV/channel\n"
]
],
[
[
"# 2. Nuclear Data",
"_____no_output_____"
],
[
"## 2.1 `bq.Element`",
"_____no_output_____"
]
],
[
[
"e1 = bq.Element('Cs')\ne2 = bq.Element(55)\ne3 = bq.Element('55')\nprint(e1, e2, e3)\nprint(e1 == e2 == e3)\nprint('{:%n(%s) Z=%z}'.format(e1))\npprint(e1.__dict__, width=10)",
"Cesium(Cs) Z=55 Cesium(Cs) Z=55 Cesium(Cs) Z=55\nTrue\nCesium(Cs) Z=55\n{'Z': 55,\n 'atomic_mass': 132.91,\n 'name': 'Cesium',\n 'symbol': 'Cs'}\n"
]
],
[
[
"## 2.2 `bq.Isotope`\n\nFurther examples of `Isotope` and `IsotopeQuantity` can be found in the [isotopes notebook](./isotopes.ipynb).",
"_____no_output_____"
]
],
[
[
"i1 = bq.Isotope('Cs-137')\ni2 = bq.Isotope('137CS')\ni3 = bq.Isotope('Cs', 137)\ni4 = bq.Isotope('Cesium-137')\ni5 = bq.Isotope('137CAESIUM')\nprint(i1, i2, i3, i4, i5)\nprint(i1 == i2 == i3 == i4 == i5)",
"Cs-137 Cs-137 Cs-137 Cs-137 Cs-137\nTrue\n"
]
],
[
[
"Isotope names and properties",
"_____no_output_____"
]
],
[
[
"iso = bq.Isotope('Tc-99m')\nprint(iso)\nprint('{:%n(%s)-%a%m Z=%z}'.format(iso))\npprint(iso.__dict__)\nprint('half-life: {:.2f} hr'.format(iso.half_life / 3600))",
"Tc-99m\nTechnetium(Tc)-99m Z=43\n{'A': 99,\n 'M': 1,\n 'N': 56,\n 'Z': 43,\n 'atomic_mass': 98,\n 'm': 'm',\n 'name': 'Technetium',\n 'symbol': 'Tc'}\nhalf-life: 6.01 hr\n"
]
],
[
[
"More isotope properties such as half-life, stability, and natural abundance are available:",
"_____no_output_____"
]
],
[
[
"for a in range(39, 42):\n iso = bq.Isotope('Potassium', a)\n print('')\n print('Isotope: {}'.format(iso))\n print(' Spin-parity: {}'.format(iso.j_pi))\n if iso.abundance is not None:\n print(' Abundance: {:.2f}%'.format(iso.abundance))\n print(' Stable? {}'.format(iso.is_stable))\n if not iso.is_stable:\n print(' Half-life: {:.3e} years'.format(iso.half_life / 365.25 / 24 / 3600))\n print(' Decay modes: {}'.format(iso.decay_modes))",
"\nIsotope: K-39\n Spin-parity: 3/2+\n Abundance: 93.26+/-0.00%\n Stable? True\n\nIsotope: K-40\n Spin-parity: 4-\n Abundance: 0.01+/-0.00%\n Stable? False\n Half-life: 1.248e+09 years\n Decay modes: (['EC', 'B-'], [10.72, 89.28])\n\nIsotope: K-41\n Spin-parity: 3/2+\n Abundance: 6.73+/-0.00%\n Stable? True\n"
]
],
[
[
"## 2.3 `bq.IsotopeQuantity`",
"_____no_output_____"
],
[
"Source activity on a given date\n\nHere's a check source activity on today's date:",
"_____no_output_____"
]
],
[
[
"ba133_chk = bq.IsotopeQuantity('ba133', date='2013-05-01', uci=10.02)\nba133_chk.uci_now()",
"_____no_output_____"
]
],
[
[
"Or for another date:",
"_____no_output_____"
]
],
[
[
"ba133_chk.uci_at('2018-02-16')",
"_____no_output_____"
]
],
[
[
"## 2.4 `bq.materials`\n\nAccess the [NIST X-ray mass attenuation coefficients database](https://www.nist.gov/pml/x-ray-mass-attenuation-coefficients) for [elements](https://physics.nist.gov/PhysRefData/XrayMassCoef/tab1.html) and [compounds](https://physics.nist.gov/PhysRefData/XrayMassCoef/tab2.html) and also data from the [PNNL Materials Compendium](https://compendium.cwmd.pnnl.gov).",
"_____no_output_____"
]
],
[
[
"mat_data = bq.materials.fetch_materials()\npprint(list(mat_data.keys()))",
"['15 mmol L-1 Ceric Ammonium Sulfate Solution',\n 'A-150 Tissue-Equivalent Plastic',\n 'Ac',\n 'Acetone',\n 'Acetylene',\n 'Actinium',\n 'Adipose Tissue (ICRU-44)',\n 'Ag',\n 'Air (dry, near sea level)',\n 'Air, Dry (near sea level)',\n 'Al',\n 'Alanine',\n 'Aluminum',\n 'Aluminum Oxide',\n 'Aluminum, alloy 2024-O',\n 'Aluminum, alloy 2090-T83',\n 'Aluminum, alloy 3003',\n 'Aluminum, alloy 4043-O',\n 'Aluminum, alloy 5086-O',\n 'Aluminum, alloy 6061-O',\n 'Aluminum, alloy 7075-O',\n 'Ammonia (liquid at T= -79 C)',\n 'Anthracene',\n 'Antimony',\n 'Ar',\n 'Argon',\n 'Arsenic',\n 'As',\n 'Asbestos (Chrysotile)',\n 'Asphalt',\n 'Asphalt pavement',\n 'Astatine',\n 'At',\n 'Au',\n 'B',\n 'B-100 Bone-Equivalent Plastic',\n 'Ba',\n 'Bakelite',\n 'Barium',\n 'Barium Fluoride',\n 'Barium sulfate',\n 'Be',\n 'Benzene',\n 'Beryllium',\n 'Beryllium Carbide',\n 'Beryllium Oxide',\n 'Bi',\n 'Bismuth',\n 'Bismuth Germanate (BGO)',\n 'Bismuth Iodide',\n 'Blood (ICRP)',\n 'Blood, Whole (ICRU-44)',\n 'Bone Equivalent Plastic, B-100',\n 'Bone Equivalent Plastic, B-110',\n 'Bone, Compact (ICRU)',\n 'Bone, Cortical (ICRP)',\n 'Bone, Cortical (ICRU-44)',\n 'Boral (65% Al-35% B4C)',\n 'Boral (Aluminum 10% boron alloy)',\n 'Boral (Aluminum 5% boron alloy)',\n 'Borax',\n 'Boric Acid',\n 'Boron',\n 'Boron Carbide',\n 'Boron Fluoride (B2F4)',\n 'Boron Fluoride (BF3)',\n 'Boron Oxide',\n 'Br',\n 'Brain (ICRP)',\n 'Brain, Grey/White Matter (ICRU-44)',\n 'Brass (typical composition)',\n 'Breast Tissue (ICRU-44)',\n 'Brick, Common Silica',\n 'Brick, Fire',\n 'Brick, Kaolin (white)',\n 'Bromine',\n 'Bronze (typical composition)',\n 'C',\n 'C-552 Air-Equivalent Plastic',\n 'C-552 Air-equivalent Plastic',\n 'CELOTEX (Lignocellulosic Fiberboard)',\n 'CLLB(Ce) - Cesium Lithium Lanthanum Bromide - 0.3 wt% Cerium doped',\n 'Ca',\n 'Cadmium',\n 'Cadmium Nitrate Tetrahydrate',\n 'Cadmium Telluride',\n 'Cadmium Tungstate (CWO)',\n 'Cadmium Zinc Telluride (CZT)',\n 'Calcium',\n 'Calcium Carbonate',\n 'Calcium Fluoride',\n 'Calcium Oxide',\n 'Calcium Sulfate',\n 'Calcium Tungstate',\n 'Carbon Dioxide',\n 'Carbon Tetrachloride',\n 'Carbon, Activated',\n 'Carbon, Amorphous',\n 'Carbon, Graphite',\n 'Carbon, Graphite (reactor grade)',\n 'Cat litter (clumping)',\n 'Cat litter (non-clumping)',\n 'Cd',\n 'Ce',\n 'Cellulose',\n 'Cellulose Acetate',\n 'Ceric Sulfate Dosimeter Solution',\n 'Cerium',\n 'Cerium Bromide',\n 'Cerium Fluoride',\n 'Cesium',\n 'Cesium Iodide',\n 'Cesium Iodide - 1 wt% Sodium doped',\n 'Cesium Iodide - 1 wt% Thalium doped',\n 'Cesium Lithium Yttrium Chloride (CLYC)',\n 'Cesium Lithium Yttrium Chloride (CLYC) with 95% Li6 Enrichment',\n 'Chlorine',\n 'Chromium',\n 'Cl',\n 'Clay',\n 'Co',\n 'Coal, Anthracite',\n 'Coal, Bituminous',\n 'Coal, Lignite',\n 'Cobalt',\n 'Concrete [Los Alamos (MCNP) Mix]',\n 'Concrete, Barite (TYPE BA)',\n 'Concrete, Barite (Type BA)',\n 'Concrete, Barytes-Limonite',\n 'Concrete, Boron Frits-baryte',\n 'Concrete, Colemanite-baryte',\n 'Concrete, Ferro-phosphorus',\n 'Concrete, Hanford Dry',\n 'Concrete, Hanford Wet',\n 'Concrete, Iron-Portland',\n 'Concrete, Iron-limonite',\n 'Concrete, Limonite and steel',\n 'Concrete, Luminite-Portland-colemanite-baryte',\n 'Concrete, Luminite-colemanite-baryte',\n 'Concrete, M-1',\n 'Concrete, MO',\n 'Concrete, Magnetite',\n 'Concrete, Magnetite and steel',\n \"Concrete, Magnuson's\",\n 'Concrete, Oak Ridge (ORNL)',\n 'Concrete, Ordinary',\n 'Concrete, Ordinary (NBS 03)',\n 'Concrete, Ordinary (NBS 04)',\n 'Concrete, Ordinary (NIST)',\n 'Concrete, Portland',\n 'Concrete, Regulatory Concrete (developed for U.S. NRC)',\n 'Concrete, Rocky Flats',\n 'Concrete, Serpentine',\n 'Copper',\n 'Cr',\n 'Cs',\n 'Cu',\n 'Diatomaceous Earth',\n 'Diesel Fuel',\n 'Dy',\n 'Dysprosium',\n 'Earth, Typical Western U.S.',\n 'Earth, U.S. Average',\n 'Er',\n 'Erbium',\n 'Ethane',\n 'Ethyl Acetate',\n 'Ethyl Alcohol',\n 'Ethylene',\n 'Ethylene Glycol',\n 'Eu',\n 'Europium',\n 'Explosive compound, AN',\n 'Explosive compound, EGDN',\n 'Explosive compound, HMX',\n 'Explosive compound, NC',\n 'Explosive compound, NG',\n 'Explosive compound, PETN',\n 'Explosive compound, RDX',\n 'Explosive compound, TNT',\n 'Eye Lens (ICRP)',\n 'Eye Lens (ICRU-44)',\n 'F',\n 'Fe',\n 'Felt',\n 'Ferric Oxide',\n 'Ferrous Sulfate Dosimeter Solution',\n 'Ferrous Sulfate Standard Fricke',\n 'Fertilizer (Muriate of Potash)',\n 'Fiberglass, Type C',\n 'Fiberglass, Type E',\n 'Fiberglass, Type R',\n 'Fluorine',\n 'Fr',\n 'Francium',\n 'Freon-12',\n 'Freon-12B2',\n 'Freon-13',\n 'Freon-13B1',\n 'Freon-13I1',\n 'GAGG(CE)',\n 'Ga',\n 'Gadolinium',\n 'Gadolinium Aluminum Galium Oxide - 0.5 atom% Cerium doped',\n 'Gadolinium Oxysulfide',\n 'Gadolinium Silicate (GSO)',\n 'Gafchromic Sensor',\n 'Gafchromic Sensor (GS)',\n 'Gallium',\n 'Gallium Arsenide',\n 'Gasoline',\n 'Gd',\n 'Ge',\n 'Germanium',\n 'Germanium, High Purity',\n 'Glass Scintillator, Li Doped (GS1, GS2, GS3)',\n 'Glass Scintillator, Li Doped (GS10, GS20, GS30)',\n 'Glass Scintillator, Li Doped (GSF1, GSF2, and GSF3)',\n 'Glass Scintillator, Li Doped (KG1, KG2, KG3)',\n 'Glass, Borosilicate (Pyrex Glass)',\n 'Glass, Borosilicate (Pyrex)',\n 'Glass, Foam',\n 'Glass, Lead',\n 'Glass, Plate',\n 'Glycerol',\n 'Gold',\n 'Gypsum (Plaster of Paris)',\n 'H',\n 'Hafnium',\n 'He',\n 'He-3 proportional gas',\n 'He-4 gas detector',\n 'Helium',\n 'Helium, Natural',\n 'Hf',\n 'Hg',\n 'Ho',\n 'Holmium',\n 'Hydrogen',\n 'I',\n 'In',\n 'Incoloy Alloy 800',\n 'Inconel Alloy 600',\n 'Inconel Alloy 625',\n 'Inconel Alloy 718',\n 'Indium',\n 'Iodine',\n 'Ir',\n 'Iridium',\n 'Iron',\n 'Iron Boride (Fe2B)',\n 'Iron Boride (FeB)',\n 'Iron, Armco Ingot',\n 'Iron, Cast (gray)',\n 'Iron, Wrought (Byers No.1)',\n 'K',\n 'Kaowool',\n 'Kapton Polyimide Film',\n 'Kennertium',\n 'Kernite',\n 'Kerosene',\n 'Kr',\n 'Krypton',\n 'Kynar',\n 'La',\n 'Lanthanum',\n 'Lanthanum Bromide - 0.5 wt% Cerium doped',\n 'Lanthanum Bromide - 10 wt% Cerium and 0.10 wt% Strontium doped',\n 'Lanthanum Bromide - 10 wt% Cerium doped',\n 'Lanthanum Bromide - 5 wt% Cerium doped',\n 'Lead',\n 'Lead Iodide',\n 'Lead Tungstate (PWO)',\n 'Li',\n 'Lithium',\n 'Lithium Amide',\n 'Lithium Fluoride',\n 'Lithium Fluride',\n 'Lithium Gadolinium Borate (LGB)',\n 'Lithium Hydride',\n 'Lithium Iodide (high density)',\n 'Lithium Iodide (low density)',\n 'Lithium Oxide',\n 'Lithium Tetraborate',\n 'Lu',\n 'Lucite',\n 'Lung Tissue (ICRU-44)',\n 'Lutetium',\n 'Lutetium Aluminum Garnet (LuAG)',\n 'Lutetium Iodide(Cerium)',\n 'Lutetium Orthoaluminate (LuAP)',\n 'Lutetium Oxyorthosilicate (LSO)',\n 'Lutetium Yttrium OxyorthoSilicate (LYSO)',\n 'Magnesium',\n 'Magnesium Oxide',\n 'Magnesium Tetraborate',\n 'Magnesium Tetroborate',\n 'Manganese',\n 'Masonite',\n 'Melamine',\n 'Melamine Formaldehyde',\n 'Mercuric Iodide',\n 'Mercury',\n 'Methane',\n 'Methanol',\n 'Methylene Chloride',\n 'Mg',\n 'Mn',\n 'Mo',\n 'Molybdenum',\n 'Monosodium Titanate, MST',\n 'Mortar',\n 'Muscle Equivalent-Liquid, with sucrose',\n 'Muscle Equivalent-Liquid, without sucrose',\n 'Muscle, Skeletal',\n 'Muscle, Skeletal (ICRU-44)',\n 'Muscle, Striated',\n 'N',\n 'NE-213 Equivalent',\n 'Na',\n 'Nb',\n 'Nd',\n 'Ne',\n 'Neodymium',\n 'Neon',\n 'Ni',\n 'Nickel',\n 'Niobium',\n 'Nitrogen',\n 'Nylon, Dupont ELVAmide 8062',\n 'Nylon, Type 11 (Rilsan)',\n 'Nylon, Type 6 and Type 6/6',\n 'Nylon, Type 6/10',\n 'O',\n 'Oil, Crude (Heavy, Cold Lake, Canada)',\n 'Oil, Crude (Heavy, Mexican)',\n 'Oil, Crude (Heavy, Qayarah, Iraq)',\n 'Oil, Crude (Light, Texas)',\n 'Oil, Fuel (Calif.)',\n 'Oil, Hydraulic',\n 'Oil, Lard',\n 'Os',\n 'Osmium',\n 'Ovary (ICRU-44)',\n 'Oxygen',\n 'P',\n 'P-10 gas',\n 'P-5 gas',\n 'P-terphenyl',\n 'Pa',\n 'Palladium',\n 'Paper, News print',\n 'Paper, glossy',\n 'Paper, printer',\n 'Pb',\n 'Pd',\n 'Phosphorus',\n 'Photographic Emulsion (Kodak Type AA)',\n 'Photographic Emulsion (Standard Nuclear)',\n 'Photographic Emulsion, Gel in',\n 'Photographic Emulsion, Kodak Type AA',\n 'Photographic Emulsion, Standard Nuclear',\n 'Plastic Scintillator, Vinyltoluene',\n 'Platinum',\n 'Plutonium Bromide',\n 'Plutonium Carbide',\n 'Plutonium Chloride',\n 'Plutonium Dioxide',\n 'Plutonium Fluoride (PuF3)',\n 'Plutonium Fluoride (PuF4)',\n 'Plutonium Fluoride (PuF6)',\n 'Plutonium Iodide',\n 'Plutonium Nitrate',\n 'Plutonium Nitride',\n 'Plutonium Oxide (Pu2O3)',\n 'Plutonium Oxide (PuO)',\n 'Plutonium, Aged WGPu (A: 4-7% Pu240)',\n 'Plutonium, Aged WGPu (B: 10-13% Pu240)',\n 'Plutonium, Aged WGPu (C: 16-19% Pu240)',\n 'Plutonium, DOE 3013 WGPu',\n 'Plutonium, Fuel Grade',\n 'Plutonium, Power Grade',\n 'Plutonium, Shefelbine WGPu',\n 'Pm',\n 'Po',\n 'Polonium',\n 'Polycarbonate',\n 'Polyethylene',\n 'Polyethylene Terephthalate (PET)',\n 'Polyethylene Terephthalate, (Mylar)',\n 'Polyethylene, Borated',\n 'Polyethylene, Non-borated',\n 'Polyisocyanurate (PIR)',\n 'Polymethyl Methacrylate',\n 'Polypropylene (PP)',\n 'Polystyrene',\n 'Polystyrene (PS)',\n 'Polytetrafluoroethylene (PTFE)',\n 'Polytetrafluoroethylene, (Teflon)',\n 'Polyurethane Foam (PUR)',\n 'Polyvinyl Acetate (PVA)',\n 'Polyvinyl Chloride',\n 'Polyvinyl Chloride (PVC)',\n 'Polyvinyl Toluene (PVT)',\n 'Polyvinylidene Chloride (PVDC)',\n 'Potassium',\n 'Potassium Aluminum Silicate',\n 'Potassium Iodide',\n 'Potassium Oxide',\n 'Pr',\n 'Praseodymium',\n 'Promethium',\n 'Propane (gas)',\n 'Propane (liquid)',\n 'Protactinium',\n 'Pt',\n 'Quartz',\n 'Quartz Glass',\n 'Ra',\n 'Radiochromic Dye Film, Nylon Base',\n 'Radiochromic Dye Film, Nylon Base (RDF: NB)',\n 'Radium',\n 'Radon',\n 'Rayon',\n 'Rb',\n 'Re',\n 'Rh',\n 'Rhenium',\n 'Rhodium',\n 'Rn',\n 'Rock (Average of 5 types)',\n 'Rock, Basalt',\n 'Rock, Granite',\n 'Rock, Limestone',\n 'Rock, Sandstone',\n 'Rock, Shale',\n 'Ru',\n 'Rubber, Butyl',\n 'Rubber, Natural',\n 'Rubber, Neoprene',\n 'Rubber, Silicon',\n 'Rubidium',\n 'Ruthenium',\n 'S',\n 'Salt Water (T=0 C)',\n 'Salt Water (T=20 C)',\n 'Samarium',\n 'Sand',\n 'Sb',\n 'Sc',\n 'Scandium',\n 'Se',\n 'Sea Water, Simple Artificial',\n 'Sea Water, Standard',\n 'Selenium',\n 'Sepiolite',\n 'Si',\n 'Silciate Yttrium - 0.5 atom% Cerium',\n 'Silicon',\n 'Silicon Carbide (hexagonal)',\n 'Silicon Dioxide (Alpha-quartz)',\n 'Silicon Dioxide (Silica)',\n 'Silver',\n 'Skin (ICRP)',\n 'Sm',\n 'Sn',\n 'Sodium',\n 'Sodium Bismuth Tungstate (NBWO)',\n 'Sodium Chloride',\n 'Sodium Iodide - 0.2 wt% Thalium Doped',\n 'Sodium Iodide with 0.8 wt% Lithium - 0.10 wt% Thalium doped',\n 'Sodium Nitrate',\n 'Sodium Oxide',\n 'Sr',\n 'Steel, Boron Stainless',\n 'Steel, HT9 Stainless',\n 'Steel, High Carbon (1095)',\n 'Steel, Low Carbon (1008)',\n 'Steel, Medium Carbon (1045)',\n 'Steel, Stainless 202',\n 'Steel, Stainless 302',\n 'Steel, Stainless 304',\n 'Steel, Stainless 304L',\n 'Steel, Stainless 316',\n 'Steel, Stainless 316L',\n 'Steel, Stainless 321',\n 'Steel, Stainless 347',\n 'Steel, Stainless 409',\n 'Steel, Stainless 440A',\n 'Steel, Stainless 440B',\n 'Steel, Stainless 440C',\n 'Sterotex',\n 'Stilbene (trans-stilbene isomer)',\n 'Strontium',\n 'Strontium Iodide - 2.5 atom% Europium doped',\n 'Sulfur',\n 'Sulfuric acid',\n 'Sulphur',\n 'TLYC',\n 'Ta',\n 'Tantalum',\n 'Tb',\n 'Tc',\n 'Te',\n 'Technetium',\n 'Tellurium',\n 'Terbium',\n 'Testis (ICRU-44)',\n 'Th',\n 'Thallium',\n 'Thorium',\n 'Thorium Dioxide',\n 'Thulium',\n 'Ti',\n 'Tin',\n 'Tissue Equivalent, MS20',\n 'Tissue Equivalent-Gas, methane based (TEG: MB)',\n 'Tissue Equivalent-Gas, propane based (TEG: PB)',\n 'Tissue, Adipose (ICRP)',\n 'Tissue, Breast',\n 'Tissue, Lung (ICRP)',\n 'Tissue, Ovary',\n 'Tissue, Soft (ICRP)',\n 'Tissue, Soft (ICRU Four-Component)',\n 'Tissue, Soft (ICRU four component)',\n 'Tissue, Soft (ICRU-44)',\n 'Tissue, Testes (ICRP)',\n 'Tissue, Testis (ICRU)',\n 'Tissue-Equivalent Gas, Methane Based',\n 'Tissue-Equivalent Gas, Propane Based',\n 'Titanium',\n 'Titanium Dioxide',\n 'Titanium Hydride',\n 'Titanium alloy, grade 5',\n 'Tl',\n 'Tm',\n 'Toluene',\n 'Tributyl Borate',\n 'Tributyl Phosphate (TBP)',\n 'Tungsten',\n 'U',\n 'Uranium',\n 'Uranium Carbide',\n 'Uranium Dicarbide',\n 'Uranium Dioxide',\n 'Uranium Hexafluoride',\n 'Uranium Hydride',\n 'Uranium Nitride',\n 'Uranium Oxide',\n 'Uranium Tetrafluoride',\n 'Uranium Trioxide',\n 'Uranium, Depleted, Typical',\n 'Uranium, Enriched, Typical Commercial',\n 'Uranium, HEU, Health Physics Society',\n 'Uranium, HEU, Russian Average',\n 'Uranium, HEU, US Average',\n 'Uranium, Low Enriched (LEU)',\n 'Uranium, Natural (NU)',\n 'Uranium-Plutonium, Mixed Oxide (MOX)',\n 'Uranyl Fluoride',\n 'Uranyl Nitrate',\n 'V',\n 'Vanadium',\n 'Vermiculite, Exfoliated',\n 'Viton Fluoroelastomer',\n 'W',\n 'Water, Heavy',\n 'Water, Liquid',\n 'Water, Vapor',\n 'Wax, M3',\n 'Wax, Mix D',\n 'Wax, Paraffin',\n 'Wood (Southern Pine)',\n 'Xe',\n 'Xenon',\n 'Y',\n 'Yb',\n 'Ytterbium',\n 'Yttrium',\n 'Yttrium Aluminum Oxide - 1 atom% Cerium',\n 'Yttrium Aluminum Perovslite - 0.5 atom% Cerium',\n 'Zeolite (Natrolite)',\n 'Zinc',\n 'Zinc Selenide',\n 'Zinc Sulfide',\n 'Zircaloy-2',\n 'Zircaloy-4',\n 'Zirconium',\n 'Zirconium Hydride (Zr5H8)',\n 'Zirconium Hydride (ZrH2)',\n 'Zn',\n 'ZnS(Ag):LiF 95wt% 6Li PHOSPHOR POWDER Neutron Detectors',\n 'ZnS:Ag PHOSPHOR POWDER EJ-600 for Neutron Detectors',\n 'Zr']\n"
],
[
"pprint(mat_data['Air, Dry (near sea level)'], indent=4)",
"{ 'density': 0.001205,\n 'formula': '-',\n 'source': 'NIST '\n '(http://physics.nist.gov/PhysRefData/XrayMassCoef/tab2.html)',\n 'weight_fractions': [ 'C 0.000124',\n 'N 0.755268',\n 'O 0.231781',\n 'Ar 0.012827']}\n"
]
],
[
[
"## 2.5 `bq.nndc`\n\nTools to query the [National Nuclear Data Center databases](https://www.nndc.bnl.gov/nudat2/) to obtain decay radiation, branching ratios, and many other types of nuclear data.\n\nFurther details and examples can be found in the [nndc notebook](./nndc.ipynb) and the [nndc_chart_of_nuclides notebook](./nndc_chart_of_nuclides.ipynb).\n\nHere are the gamma-ray lines above 5% branching ratio from Co-60:",
"_____no_output_____"
]
],
[
[
"rad = bq.nndc.fetch_decay_radiation(nuc='Co-60', type='Gamma', i_range=(5, None))\ncols = ['Z', 'Element', 'A', 'Decay Mode', 'Radiation', 'Radiation Energy (keV)',\n 'Radiation Intensity (%)', 'Energy Level (MeV)']\ndisplay(rad[cols])",
"_____no_output_____"
],
[
"# NNDC nuclear wallet cards are used by bq.Isotope but can be accessed directly like this:\ndata = bq.nndc.fetch_wallet_card(\n z_range=(19, 19),\n a_range=(37, 44),\n elevel_range=(0, 0), # ground states only\n)\ndisplay(data)",
"_____no_output_____"
]
],
[
[
"## 2.6 `bq.xcom`\n\nThe [NIST XCOM photon cross sections database](https://www.nist.gov/pml/xcom-photon-cross-sections-database) can be [queried](https://physics.nist.gov/PhysRefData/Xcom/html/xcom1.html) in `becquerel`.\n\nFurther details can be found in the [xcom notebook](./xcom.ipynb)\n\nFor example, here is how to access the cross section data for an element (Pb)",
"_____no_output_____"
]
],
[
[
"# query XCOM by element symbol\ndata = bq.xcom.fetch_xcom_data('Pb', e_range_kev=[10., 3000.])\n\nplt.figure()\nfor field in ['total_w_coh', 'total_wo_coh', 'coherent', 'incoherent',\n 'photoelec', 'pair_nuc', 'pair_elec']:\n plt.semilogy(data.energy, data[field], label=field)\nplt.xlim(0, 3000)\nplt.xlabel('Energy (keV)')\nplt.ylabel(r'Attenuation coefficient [cm$^2$/g]')\nplt.legend();",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
]
|
e7989bff1fec759134c627e96bf44438e1535420 | 40,564 | ipynb | Jupyter Notebook | 7_Change_Index and Column_Header/2_Set_Index.ipynb | sureshmecad/Pandas | 128091e7021158f39eb0ff97e0e63d76e778a52c | [
"CNRI-Python"
]
| null | null | null | 7_Change_Index and Column_Header/2_Set_Index.ipynb | sureshmecad/Pandas | 128091e7021158f39eb0ff97e0e63d76e778a52c | [
"CNRI-Python"
]
| null | null | null | 7_Change_Index and Column_Header/2_Set_Index.ipynb | sureshmecad/Pandas | 128091e7021158f39eb0ff97e0e63d76e778a52c | [
"CNRI-Python"
]
| null | null | null | 34.405428 | 150 | 0.344591 | [
[
[
"### 1. set_index\n\n### 2. reset_index",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"df = pd.read_csv(\"C:/Users/deepusuresh/Documents/Data Science/08. Data Sets/Pandas.csv\")\ndf",
"_____no_output_____"
],
[
"df.index",
"_____no_output_____"
],
[
"df.set_index('day') # df.set_index('day',inplace=True)\ndf",
"_____no_output_____"
],
[
"df.reset_index(inplace=True)\ndf",
"_____no_output_____"
],
[
"df.set_index('event',inplace=True)\ndf",
"_____no_output_____"
]
],
[
[
"-------------------------------------------------------------------------------------------------------------------------------\n-------------------------------------------------------------------------------------------------------------------------------",
"_____no_output_____"
]
],
[
[
"#url = \"C:/Users/deepusuresh/Documents/Data Science/01. Python/3. PANDAS/1. Data Frame\"\n\ndf = pd.read_csv('C:/Users/deepusuresh/Documents/Data Science/01. Python/3. PANDAS/1. Data Frame/IMDB-Movie-Data.csv')\ndf.head(3)",
"_____no_output_____"
],
[
"df = pd.read_csv(\"C:/Users/deepusuresh/Documents/Data Science/01. Python/3. PANDAS/1. Data Frame/IMDB-Movie-Data.csv\", index_col=0)\ndf.head(3)",
"_____no_output_____"
],
[
"df = pd.read_csv(\"C:/Users/deepusuresh/Documents/Data Science/01. Python/3. PANDAS/1. Data Frame/IMDB-Movie-Data.csv\", index_col=1)\ndf.head(3)",
"_____no_output_____"
],
[
"df = pd.read_csv(\"C:/Users/deepusuresh/Documents/Data Science/01. Python/3. PANDAS/1. Data Frame/IMDB-Movie-Data.csv\", index_col=2)\ndf.head(3)",
"_____no_output_____"
]
],
[
[
"### We're loading this dataset from a CSV and designating the movie titles to be our index.",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv(\"C:/Users/deepusuresh/Documents/Data Science/01. Python/3. PANDAS/1. Data Frame/IMDB-Movie-Data.csv\", index_col=\"Title\")\ndf.head(3)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
]
|
e7989c6eb4677f6de86ead4e23ea83bf3ef2b913 | 37,591 | ipynb | Jupyter Notebook | notebooks/visualizing-big-data-in-r-codealong-answers.ipynb | marsanul/visualizacionbigdata | 2d809a4665eac2469b644c8b672dddcfc5b8d6b2 | [
"MIT"
]
| 4 | 2020-07-28T17:27:02.000Z | 2021-02-03T22:55:07.000Z | notebooks/visualizing-big-data-in-r-codealong-answers.ipynb | datacamp/Visualizing-Big-Data-in-R-live-training4 | cfe78d71652529d4b11b28380986ce95df6e856b | [
"MIT"
]
| 2 | 2020-07-27T11:26:03.000Z | 2020-07-28T20:47:38.000Z | notebooks/visualizing-big-data-in-r-codealong-answers.ipynb | datacamp/Visualizing-Big-Data-in-R-live-training4 | cfe78d71652529d4b11b28380986ce95df6e856b | [
"MIT"
]
| 13 | 2020-07-28T15:12:57.000Z | 2020-10-24T20:09:19.000Z | 24.66601 | 296 | 0.553909 | [
[
[
"",
"_____no_output_____"
],
[
"# Visualizing Big Data in R (Answers)\n## by Richie Cotton",
"_____no_output_____"
],
[
"# Prelude",
"_____no_output_____"
],
[
"Install these additional R packages.",
"_____no_output_____"
]
],
[
[
"rlib <- \"~/lib\"\ndir.create(rlib)\n.libPaths(rlib)\nlibrary(remotes)\ninstall_version(\"hexbin\", \"1.28.1\", lib = rlib, upgrade = \"never\")\ninstall_version(\"fst\", \"0.9.2\", lib = rlib, upgrade = \"never\")\ninstall_version(\"ggcorrplot\", \"0.1.3\", lib = rlib, upgrade = \"never\")\ninstall_version(\"trelliscopejs\", \"0.2.5\", lib = rlib, upgrade = \"never\")\ninstall_version(\"plotly\", \"4.9.2.1\", lib = rlib, upgrade = \"never\")",
"_____no_output_____"
]
],
[
[
"# Chapter 1: Too many points - point size, transparency, transformation",
"_____no_output_____"
],
[
"Here, you'll look at the LA home prices dataset to explore ways of reducing overplotting.",
"_____no_output_____"
],
[
"## Learning objectives\n\n- Understands that one cause of overplotting in scatter plots is simply that there are too many points.\n- Can apply point size adjustments, transparency, and axis scale transformations to reduce overplotting problems.\n- Can draw and interpret a hex plot.",
"_____no_output_____"
],
[
"## Loading the packages\n\nYou need a way to import a CSV file, `tibble` and `ggplot2`.",
"_____no_output_____"
]
],
[
[
"# Load tibble, ggplot2, and a CSV reader\nlibrary(readr) # or library(data.table)\nlibrary(tibble)\nlibrary(ggplot2)",
"_____no_output_____"
]
],
[
[
"## Exploring the dataset\n\nThe dataset is here. Run this cell!",
"_____no_output_____"
]
],
[
[
"data_file <- \"https://raw.githubusercontent.com/datacamp/Visualizing-Big-Data-in-R-live-training/master/data/LAhomes.csv\"",
"_____no_output_____"
]
],
[
[
"Read in the dataset from `data_file`, assigning the result to `la_homes`. Explore it (using whichever functions you like).",
"_____no_output_____"
]
],
[
[
"# Read data_file from CSV\nla_homes <- read_csv(data_file)\n\n# Explore it\nglimpse(la_homes)",
"_____no_output_____"
]
],
[
[
"- `price` is the sale price of the home, in USD.\n- `sqft` is the area of the home in square feet (about 0.1 square meters).\n\nUsing `la_homes`, draw a scatter plot of `price` versus `sqft`.",
"_____no_output_____"
]
],
[
[
"# Using la_homes, plot price vs. sqft with point layer\nggplot(la_homes, aes(sqft, price)) +\n geom_point()",
"_____no_output_____"
]
],
[
[
"## Changing point size\n\nNotice that points in the plot are heavily overplotted in the bottom left corner.\n\nRedraw the basic scatter plot, changing the point size to `0.5`.",
"_____no_output_____"
]
],
[
[
"# Draw same scatter plot, with size 0.5\nggplot(la_homes, aes(sqft, price)) +\n geom_point(size = 0.5)",
"_____no_output_____"
]
],
[
[
"Redraw the basic scatter plot, changing the point shape to be \"pixel points\".",
"_____no_output_____"
]
],
[
[
"# Draw same scatter plot, with pixel shape\nggplot(la_homes, aes(sqft, price)) +\n geom_point(shape = \".\")",
"_____no_output_____"
]
],
[
[
"## Using transparency\n\nRedraw the basic scatter plot, changing the transparency level of points to `0.25`. Set a white background by using ggplot2's black and white theme.",
"_____no_output_____"
]
],
[
[
"# Draw same scatter plot, with transparency 0.25 and black & white theme\nggplot(la_homes, aes(sqft, price)) +\n geom_point(alpha = 0.25) +\n theme_bw()",
"_____no_output_____"
]
],
[
[
"## Transform the axes\n\nMost of the plots are stuck in the bottom-left corner. Transform the x and y axes to spread points more evenly throughout.\n\nRedraw the basic scatter plot, applying a log10 transformation to the x and y scales.",
"_____no_output_____"
]
],
[
[
"# Draw same scatter plot, with log-log scales\nggplot(la_homes, aes(sqft, price)) +\n geom_point() +\n scale_x_log10() +\n scale_y_log10()",
"_____no_output_____"
]
],
[
[
"Redraw the scatter plot using all three tricks at once.\n\n- Set the point size to `0.5`.\n- Set the point transparency to `0.25`.\n- Using log10 transformations for the x and y scales.\n- Use the black and white theme.",
"_____no_output_____"
]
],
[
[
"# Draw same scatter plot, with all 3 tricks\nggplot(la_homes, aes(sqft, price)) +\n geom_point(size = 0.5, alpha = 0.25) +\n scale_x_log10() +\n scale_y_log10() +\n theme_bw()",
"_____no_output_____"
]
],
[
[
"## Hex plots\n\nDraw a hex plot of `price` versus `sqft`.",
"_____no_output_____"
]
],
[
[
"# Using la_homes, plot price vs. sqft with hex layer\nggplot(la_homes, aes(sqft, price)) +\n geom_hex()",
"_____no_output_____"
]
],
[
[
"Redraw the hex plot, applying log10 transformations to the x and y scales.",
"_____no_output_____"
]
],
[
[
"# Draw same hex plot, with log-log scales\nggplot(la_homes, aes(sqft, price)) +\n geom_hex() +\n scale_x_log10() +\n scale_y_log10()",
"_____no_output_____"
]
],
[
[
"Which statement about the trend is true?\n\n- [ ] Price increases roughly linearly with area.\n- [ ] Price increases roughly linearly with log area.\n- [ ] Log price increases roughly linearly with area.\n- [x] Log price increases roughly linearly with log area.",
"_____no_output_____"
],
[
"Which statement about the overplotting is true?\n\n- [ ] The majority of the houses are found in the region of darkest blues on the hex plot.\n- [x] The majority of the houses are found in the region of lightest blues on the hex plot.\n- [ ] The hex plot tells us nothing about where the majority of the houses are found.",
"_____no_output_____"
],
[
"# Chapter 2: Aligned values - jittering",
"_____no_output_____"
],
[
"Here you'll take another look at overplotting in the LA homes dataset.",
"_____no_output_____"
],
[
"## Learning objectives\n\n- Understands that one cause of overplotting in scatter plots is low-precision, integer, or categorical variables taking exactly the same values.\n- Can apply jittering, transparency, and scale transformations to solve the problem.",
"_____no_output_____"
],
[
"## Loading the packages\n\nYou'll need `readr`, `dplyr` and `ggplot2`. Just run this code.",
"_____no_output_____"
]
],
[
[
"library(readr)\nlibrary(dplyr)\nlibrary(ggplot2)",
"_____no_output_____"
]
],
[
[
"## Importing and exploring the data\n\nThe dataset is here. Run this chunk!",
"_____no_output_____"
]
],
[
[
"data_file <- \"https://raw.githubusercontent.com/datacamp/Visualizing-Big-Data-in-R-live-training/master/data/LAhomes.csv\"",
"_____no_output_____"
]
],
[
[
"Import the LA homes dataset from `data_file`, assigning to `la_homes`.",
"_____no_output_____"
]
],
[
[
"# Import data_file\nla_homes <- read_csv(data_file)",
"_____no_output_____"
]
],
[
[
"- `bed` contains the number of bedrooms in the home.\n\nTake a look at the distinct values in `bed` using `distinct()`.",
"_____no_output_____"
]
],
[
[
"# Look at the distinct values of the bed column\nla_homes %>% \n distinct(bed)",
"_____no_output_____"
]
],
[
[
"Notice that the number of bedrooms is always an integer and sometimes zero.",
"_____no_output_____"
],
[
"## Scatter plots of price vs. bedrooms\n\nUsing `la_homes`, draw a scatter plot of `price` versus `bed`.",
"_____no_output_____"
]
],
[
[
"# Using la_homes, plot price vs. bed with a point layer\nggplot(la_homes, aes(bed, price)) +\n geom_point()",
"_____no_output_____"
]
],
[
[
"Draw the same plot again, this time jittering the points along the x-axis.\n\n- Use a maximum jitter distance of `0.4` in the x direction.\n- Don't jitter in the y direction.",
"_____no_output_____"
]
],
[
[
"# Draw the previous plot but jitter points with width 0.4\nggplot(la_homes, aes(bed, price)) +\n geom_jitter(width = 0.4)",
"_____no_output_____"
]
],
[
[
"Most of the points are near the bottom of the plot.\n\nDraw the same jittered plot again, this time using a log10 transformation on the y-scale.",
"_____no_output_____"
]
],
[
[
"# Draw the previous plot but use a log y-axis\nggplot(la_homes, aes(bed, price)) +\n geom_jitter(width = 0.4) +\n scale_y_log10()",
"_____no_output_____"
]
],
[
[
"## Scatter plots of bathrooms vs. bedrooms\n\n- `bath` contains the number of bathrooms in the home.\n\nTake a look at the distinct values in `bath` using `distinct()`.",
"_____no_output_____"
]
],
[
[
"# Look at the distinct values of the bath column\nla_homes %>% \n distinct(bath)",
"_____no_output_____"
]
],
[
[
"Notice that the dataset includes half and quarter bathrooms (whatever they are).\n\nDraw a scatter plot of `bath` versus `bed`.",
"_____no_output_____"
]
],
[
[
"# Using la_homes, plot bath vs. bed with a point layer\nggplot(la_homes, aes(bed, bath)) +\n geom_point()",
"_____no_output_____"
]
],
[
[
"Draw the same plot again, this time jittering the points.\n\n- Use a maximum jitter distance of `0.4` in the x direction.\n- Use a maximum jitter distance of `0.05` in the y direction.",
"_____no_output_____"
]
],
[
[
"# Using la_homes, plot price vs. bed with a jittered point layer\nggplot(la_homes, aes(bed, bath)) +\n geom_jitter(width = 0.4, height = 0.05)",
"_____no_output_____"
]
],
[
[
"## Filtering and transformation\n\nThere are three homes with 10 or more bedrooms. These constitute outliers, and for the purpose of drawing nicer plots, we're going to remove them.\n\nFilter `la_homes` for rows where `bed` is less than `10`, assigning to `la_homes10`. Count the number of rows you removed to check you've done it correctly.",
"_____no_output_____"
]
],
[
[
"# Filter for bed less than 10\nla_homes10 <- la_homes %>% \n filter(bed < 10)\n\n# Calculate the number of outliers you removed\nnrow(la_homes) - nrow(la_homes10)",
"_____no_output_____"
]
],
[
[
"Draw the same jittered scatter plot again, this time using the filtered dataset (`la_homes10`). As before, use a jitter width of `0.4` and a jitter height of `0.05`.",
"_____no_output_____"
]
],
[
[
"# Draw the previous plot, but with the la_homes10 dataset\nggplot(la_homes10, aes(bed, bath)) +\n geom_jitter(width = 0.4, height = 0.05)",
"_____no_output_____"
]
],
[
[
"Most of the points are towards the bottom left of the plot.\n\nDraw the same jittered scatter plot again, this time applying square-root transformations to the x and y scales.",
"_____no_output_____"
]
],
[
[
"# Draw the previous plot but with sqrt-sqrt scales\nggplot(la_homes10, aes(bed, bath)) +\n geom_jitter(width = 0.4, height = 0.05) +\n scale_x_sqrt() +\n scale_y_sqrt()",
"_____no_output_____"
]
],
[
[
"Refine the plot one more time, by making the points transparent.\n\nDraw the previous plot again, setting the transparency level to 0.25 (and using a black and white theme).",
"_____no_output_____"
]
],
[
[
"ggplot(la_homes10, aes(bed, bath)) +\n geom_jitter(width = 0.4, height = 0.05, alpha = 0.25) +\n scale_x_sqrt() +\n scale_y_sqrt() +\n theme_bw()",
"_____no_output_____"
]
],
[
[
"# Chapter 3: Too many variables - correlation heatmaps",
"_____no_output_____"
],
[
"Here you'll look at a dataset on Scotch whisky preferences.\n\n## Learning objectives\n\n- Can draw a correlation heatmap.\n- Can use hierarchical clustering to order cells in a correlation heatmap.\n- Can adjust the color scale in a correlation heatmap.\n- Can interpret a correlation heatmap.\n\n## Loading the packages\n\nYou'll need `fst`, `dplyr`, `ggplot2`, and `ggcorrplot`. Just run this code.",
"_____no_output_____"
]
],
[
[
"library(fst)\nlibrary(tibble)\nlibrary(ggplot2)\nlibrary(ggcorrplot)",
"_____no_output_____"
]
],
[
[
"## Get the dataset\n\nThe dataset is a modified version of `bayesm::Scotch`. \n\n- See https://www.rdocumentation.org/packages/bayesm/topics/Scotch for details.\n- Each observation is a survey response indicating the brands of Scotch consumed in the last year.",
"_____no_output_____"
],
[
"Run this to download the data file.",
"_____no_output_____"
]
],
[
[
"download.file(\n \"https://github.com/datacamp/Visualizing-Big-Data-in-R-live-training/raw/master/data/scotch.fst\",\n \"scotch.fst\"\n)",
"_____no_output_____"
]
],
[
[
"Import the dataset from `scotch.fst` and assign to `scotch`.",
"_____no_output_____"
]
],
[
[
"# Import from scotch.fst\nscotch <- read_fst(\"scotch.fst\")\n\n# Explore the dataset, however you wish\nglimpse(scotch)",
"_____no_output_____"
]
],
[
[
"## Draw a basic correlation heatmap\n\nCalculate the correlation matrix for `scotch`, assigning to `correl`.",
"_____no_output_____"
]
],
[
[
"# Calculate the correlation matrix\ncorrel <- cor(scotch)",
"_____no_output_____"
]
],
[
[
"Draw a correlation heatmap of it (no customization).",
"_____no_output_____"
]
],
[
[
"# Draw a correlation heatmap\nggcorrplot(correl)",
"_____no_output_____"
]
],
[
[
"## Drop redundant cells\n\nDraw the previous plot again, this time only showing the upper triangular portion of the correlation matrix.",
"_____no_output_____"
]
],
[
[
"# Draw a correlation heatmap of the upper triangular portion\nggcorrplot(correl, type = \"upper\")",
"_____no_output_____"
]
],
[
[
"## Use hierarchical clustering\n\nDraw the previous plot again, this time using hierarchical clustering to reorder cells.",
"_____no_output_____"
]
],
[
[
"# Draw a correlation heatmap of the upper triangular portion\nggcorrplot(correl, type = \"upper\", hc.order = TRUE)",
"_____no_output_____"
]
],
[
[
"# Override the color scale\n\nSet the diagonal values in the correlation matrix to `NA`, then calculate the range of the correlation matrix.",
"_____no_output_____"
]
],
[
[
"# Set the diagonals of correl to NA\ndiag(correl) <- NA\n\n# Calculate the range of correl (removing NAs)\nrange(correl, na.rm = TRUE)",
"_____no_output_____"
]
],
[
[
"We have both positive and negative correlations, so this is a slightly trickier situation than in the slides. We want a symmetric color scale centered on zero.\n\nDefine the limits of the color scale.\n\n- Calculate the `max`imum `abs`olute correlation (removing NAs). Assign to `max_abs_correl`.\n- Add some padding to `max_abs_correl` (any small number). Assign to `max_abs_correl_padded`.\n- Define the scale limits as the vector (`-max_abs_correl_padded`, `max_abs_correl_padded`).",
"_____no_output_____"
]
],
[
[
"# Calculate the largest absolute correlation (removing NAs)\nmax_abs_correl <- max(abs(correl), na.rm = TRUE)\n\n# Add some padding\nmax_abs_correl_padded <- max_abs_correl + 0.02\n\n# Define limits from -max_abs_correl_padded to max_abs_correl_padded\nscale_limits <- c(-max_abs_correl_padded, max_abs_correl_padded)",
"_____no_output_____"
]
],
[
[
"Draw the previous plot again, this time overriding the fill color scale.\n\n- Add `scale_fill_gradient2()`.\n- Pass the scale limits.\n- Set the `high` argument to `\"red\"`.\n- Set the `mid` argument to `\"white\"`.\n- Set the `low` argument to `\"blue\"`.",
"_____no_output_____"
]
],
[
[
"# Draw a correlation heatmap of the upper triangular portion\n# Override the fill scale to use a 2-way gradient\nggcorrplot(correl, type = \"upper\", hc.order = TRUE) +\n scale_fill_gradient2(\n limits = scale_limits,\n high = \"red\",\n mid = \"white\",\n low = \"blue\"\n )",
"_____no_output_____"
]
],
[
[
"## Interpreting correlation heatmaps\n\nDrinkers of Glenfiddich are most likely to also drink which other whisky?\n\n- [ ] Scoresby rare\n- [ ] J & B\n- [x] Glenlivet\n- [ ] Black & White\n- [ ] Chivas Regal",
"_____no_output_____"
],
[
"Drinkers of Knockando are most likely to also drink which other whisky?\n\n- [ ] Dewar's White Label\n- [ ] Johnny Walker Red Label\n- [ ] Johnny Walker Black Label\n- [x] Macallan\n- [ ] Chivas Regal",
"_____no_output_____"
],
[
"# Chapter 4: Too many facets - trelliscope plots\n\nHere, you'll explore the 30 stocks in the Dow Jones Industrial Average (DJIA).",
"_____no_output_____"
],
[
"## Learning objectives\n\n- Can convert a ggplot into a trelliscope plot.\n- Can use common arguments to control the appearance of a trelliscope plot.\n- Can use the interactive filter and sort tools to interpret a trelliscope plot.",
"_____no_output_____"
],
[
"## Load the packages\n\nYou'll need `fst`, `tibble`, `ggplot2`, and `trelliscopejs`. Just run this code.",
"_____no_output_____"
]
],
[
[
"library(fst)\nlibrary(tibble)\nlibrary(ggplot2)\nlibrary(trelliscopejs)",
"_____no_output_____"
]
],
[
[
"## Get the dataset",
"_____no_output_____"
],
[
"Run this to download the data file.",
"_____no_output_____"
]
],
[
[
"download.file(\n \"https://github.com/datacamp/Visualizing-Big-Data-in-R-live-training/raw/master/data/dow_stock_prices.fst\",\n \"dow_stock_prices.fst\"\n)",
"_____no_output_____"
]
],
[
[
"Import the DJIA data from `dow_stock_prices.fst`, assigning to `dow_stock_prices`. Explore it however you wish.",
"_____no_output_____"
]
],
[
[
"# Import the dataset from dow_stock_prices.fst\ndow_stock_prices <- read_fst(\"dow_stock_prices.fst\")\n\n# Explore the dataset, however you wish\nglimpse(dow_stock_prices)",
"_____no_output_____"
]
],
[
[
"- `symbol`: The stock ticker symbol (unique ID for company).\n- `company`: Human-readable company name.\n- `sector`: Business sector that the company participates in.\n- `date`: Date on which price and volume data was calculated for.\n- `volume`: Number of shares traded on `date`.\n- `adjusted`: Price of 1 share, after adjusting for dividends and splits.\n- `relative`: Price of 1 share, relative to the maximum of `adjusted` over the time period\n- `date_of_max_price`: For each stock, the date when the maximum share price was first achieved.\n- `date_of_min_price`: For each stock, the date when the maximum share price was first achieved.\n\nTake a look at the range of the dates in the dataset.",
"_____no_output_____"
]
],
[
[
"# Get the range of the dates in dow_stock_prices\nrange(dow_stock_prices$date)",
"_____no_output_____"
]
],
[
[
"## From ggplot2 to trelliscopejs\n\nUsing `dow_stock_prices`, draw a line plot of `relative` versus `date`, faceted by `symbol`.",
"_____no_output_____"
]
],
[
[
"# Using dow_stock_prices, plot relative vs. date\n# as a line plot\n# faceted by symbol\nggplot(dow_stock_prices, aes(date, relative)) +\n geom_line() +\n facet_wrap(vars(symbol))",
"_____no_output_____"
]
],
[
[
"Redraw the previous plot, this time as a trelliscope plot (no customization). \n\n- Set the `path` argument to `\"trelliscope/basic\"`.",
"_____no_output_____"
]
],
[
[
"# Same plot as before, using trelliscope\nggplot(dow_stock_prices, aes(date, relative)) +\n geom_line() +\n facet_trelliscope(\n vars(symbol),\n path = \"trelliscope/basic\"\n )",
"_____no_output_____"
]
],
[
[
"Run this next line to open the plot in a new browser tab.",
"_____no_output_____"
]
],
[
[
"# Browse for the plot URL\nbrowseURL(\"trelliscope/basic/index.html\")",
"_____no_output_____"
]
],
[
[
"# Improving the plot\n\nWe can improve on the previous plot by customizing it.\n\nRedraw the previous plot, with the following changes.\n\n- Set the `path` argument to `\"trelliscope/improved\"`.\n- Set the plot title to `Dow Jones Industrial Average`.\n- Set the plot description to `Share prices 2017-01-01 to 2020-01-01`.\n- Arrange the panels in `5` rows of `2` columns per page. \n- Increase the width of each panel to `1200` pixels.",
"_____no_output_____"
]
],
[
[
"# Draw the same plot again, customizing the display\n# Set path, name, desc, nrow, ncol, width\nggplot(dow_stock_prices, aes(date, relative)) +\n geom_line() +\n facet_trelliscope(\n vars(symbol),\n path = \"trelliscope/improved\",\n name = \"Dow Jones Industrial Average\",\n desc = \"Share prices 2017-01-01 to 2020-01-01\",\n nrow = 5, \n ncol = 2,\n width = 1200\n )",
"_____no_output_____"
]
],
[
[
"Open the plot in a new browser tab.",
"_____no_output_____"
]
],
[
[
"# Browse for the plot URL\nbrowseURL(\"trelliscope/improved/index.html\")",
"_____no_output_____"
]
],
[
[
"## Labels\n\nAdd the `company` to the labels shown on each panel.",
"_____no_output_____"
],
[
"## Filtering",
"_____no_output_____"
],
[
"Which `sector` contains the most companies?\n\n- [ ] Health Care\n- [x] Information Technology\n- [ ] Consumer Staples\n- [ ] Industrials\n- [ ] Financials",
"_____no_output_____"
],
[
"Which `Energy` sector company began 2020 with a lower share price than 2017?\n\n- [ ] CVX (Chevron)\n- [x] XOM (Exxon Mobil)",
"_____no_output_____"
],
[
"How many companies had a maximum price more than double the minimum price during the time period? That is, how many companies had a relative minimum less than `0.5`?\n\n- [ ] 4\n- [ ] 5\n- [x] 6\n- [ ] 7",
"_____no_output_____"
],
[
"# Sorting",
"_____no_output_____"
],
[
"Based on mean daily volume of trades, which company was the 3rd most traded during the time period?\n\n- [ ] AAPL (Apple)\n- [ ] MSFT (Microsoft)\n- [ ] CSCO (Cisco Systems)\n- [ ] PFE (Pfizer)\n- [x] INTC (Intel)",
"_____no_output_____"
],
[
"Which company's median realtive price during the time period was lowest?\n\n- [x] AAPL (Apple)\n- [ ] MSFT (Microsoft)\n- [ ] V (Verizon Communications)\n- [ ] MRK (Merck & Co.)\n- [ ] PG ()",
"_____no_output_____"
],
[
"## Free scales\n\nThe relative share prices were plotted to make it easier to compare performance between companies. If you want to plot the non-normalized `adjusted` prices, you need to give each panel its own y-axis.\n\nRedraw the previous plot, with these changes.\n\n- Set the `path` argument to `\"trelliscope/yscale\"`.\n- On the y-axis, plot `adjusted` rather than `relative`.\n- Give each panel a free y-axis scale (while keeping the x-axis scales the same).",
"_____no_output_____"
]
],
[
[
"# This time plot adjusted vs. date\n# Use a free y-scale\nggplot(dow_stock_prices, aes(date, adjusted)) +\n geom_line() +\n facet_trelliscope(\n vars(symbol),\n path = \"trelliscope/yscale\",\n name = \"Dow Jones Industrial Average\",\n desc = \"Share prices 2017-01-01 to 2020-01-01\",\n nrow = 5, \n ncol = 2,\n width = 1200,\n scales = c(\"same\", \"free\")\n )",
"_____no_output_____"
]
],
[
[
"Open the plot in a new browser tab.",
"_____no_output_____"
]
],
[
[
"# Browse for the plot URL\nbrowseURL(\"trelliscope/yscale/index.html\")",
"_____no_output_____"
]
],
[
[
"Which company, at it's maximum, had the highest price for 1 share?\n\n- [x] BA (Boeing)\n- [ ] GS (Goldman Sachs Group)\n- [ ] UNH (UnitedHealth Group)\n- [ ] AAPL (Apple)\n- [ ] MMM (3M)",
"_____no_output_____"
],
[
"## Interactive plotting with plotly\n\nBy using plotly to create each panel, each panel becomes interactive. Hover over the line to see the values of individual points.\n\nRedraw the previous plot, using plotly to create the panels.\n\n- Set the `path` argument to `\"trelliscope/plotly\"`.",
"_____no_output_____"
]
],
[
[
"# Redraw the last plot using plotly for panels\nggplot(dow_stock_prices, aes(date, adjusted)) +\n geom_line() +\n facet_trelliscope(\n vars(symbol),\n path = \"trelliscope/plotly\",\n name = \"Dow Jones Industrial Average\",\n desc = \"Share prices 2017-01-01 to 2020-01-01\",\n nrow = 5, \n ncol = 2,\n width = 1200,\n scales = c(\"same\", \"free\"),\n as_plotly = TRUE\n )",
"_____no_output_____"
]
],
[
[
"Open the plot in a new browser tab.",
"_____no_output_____"
]
],
[
[
"# Browse for the plot URL\nbrowseURL(\"trelliscope/plotly/index.html\")",
"_____no_output_____"
]
],
[
[
"`V` (Verizon) had a dip in its share price in December 2018. What was its adjusted share price on 2018-12-24?\n\n> 120.78",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
e7989d2e9fd019732c3e1d35467c883cd8837de8 | 275,500 | ipynb | Jupyter Notebook | Imaging/Test_distort.ipynb | CHEN-yongquan/Asteroid_CPO_seeker | b180d08228e51a11b87d0a579023f7f09e332a7c | [
"MIT"
]
| 2 | 2021-06-17T11:02:51.000Z | 2021-11-20T07:57:24.000Z | Imaging/Test_distort.ipynb | CHEN-yongquan/Asteroid_CPO_seeker | b180d08228e51a11b87d0a579023f7f09e332a7c | [
"MIT"
]
| null | null | null | Imaging/Test_distort.ipynb | CHEN-yongquan/Asteroid_CPO_seeker | b180d08228e51a11b87d0a579023f7f09e332a7c | [
"MIT"
]
| 1 | 2022-03-05T03:43:25.000Z | 2022-03-05T03:43:25.000Z | 75.520833 | 33,439 | 0.69457 | [
[
[
"import numpy as np\nimport os,sys\n\nsys.path.append('.')\nsys.path.append('../RL_lib/Utils')\n%load_ext autoreload\n%load_ext autoreload\n%autoreload 2\n%matplotlib nbagg\nimport os\nprint(os.getcwd())",
"The autoreload extension is already loaded. To reload it, use:\n %reload_ext autoreload\n/Users/briangaudet/Study/Subjects/MachineLearning/Projects/MetaLearning_SBIR-master/Imaging\n"
],
[
"%%html\n<style>\n.output_wrapper, .output {\n height:auto !important;\n max-height:1000px; /* your desired max-height here */\n}\n.output_scroll {\n box-shadow:none !important;\n webkit-box-shadow:none !important;\n}\n</style>",
"_____no_output_____"
]
],
[
[
"# Image a cube's vertices\n## grayscale: 0 is black, higher intensities are lighter\n## here intensities are ranges. so foreground should have smaller range, and therefore be darker\n## Optical axis +Z (default)",
"_____no_output_____"
],
[
"# Centered",
"_____no_output_____"
]
],
[
[
"from camera_model_distort import Camera_model\nimport attitude_utils as attu\nimport optics_utils as optu\nimport itertools\nfrom time import time\nap = attu.Euler_attitude()\n\nobject_locations = optu.make_cube(10.,1.0*np.asarray([0,0,200]))\nobject_locations = optu.make_grid()\nprint(object_locations.shape)\nz = np.expand_dims(400*np.ones(object_locations.shape[0]),axis=1)\nprint(z.shape)\nobject_locations=np.hstack((object_locations,z))\n#print(object_locations)\nagent_location = 1.0*np.asarray([0,0,100])\nobject_intensities = np.linalg.norm(object_locations-agent_location,axis=1)-50\nfov=np.pi/4\n\n\nyaw = 0.0\npitch = 0.0\nroll = 0.0\nagent_q = np.asarray([yaw,pitch,roll])\n\n\nC_cb = optu.rotate_optical_axis(0.0, 0.0, 0.0)\nr_cb = np.asarray([0,0,0])\n\nk=-0.9\np=0.1\nK0 = np.zeros(3)\nK1 = [2.0,0.5,0.0]\nK2 = [5.0,10.0,0.0]\nK3 = [20.,40.,0.0]\nK4 = [50,100,0]\n#K3 = [-1.0,-5.0,0.0]\nK = K3\nP0 = np.zeros(2)\nP1 = [0.1,0.1]\nP = P0\ncm = Camera_model(attitude_parameterization=ap, C_cb=C_cb, r_cb=r_cb, slant=20.0,\n fov=fov, debug=False, p1=P[0], p2=P[1], k1=K[0],k2=K[1],k3=K[2])\nt0 = time()\npix1 = cm.get_pixel_coords(agent_location, agent_q, object_locations, object_intensities)\nt1 = time()\nprint('ET: ',t1-t0)\n\ncm.render(agent_location, agent_q, object_locations, object_intensities)\n",
"Euler321 Attitude\n(738, 2)\n(738, 1)\nEuler321 Attitude\nOverriding focal length using FOV: 0.7853981633974483 13.361957121094465\nK: \n[[133.61957121 20. 50. ]\n [ 0. 133.61957121 50. ]\n [ 0. 0. 1. ]]\nC_cb: \n[[ 1. 0. -0.]\n [ 0. 1. 0.]\n [ 0. 0. 1.]]\nt: \nET: 0.004895210266113281\n"
]
],
[
[
"# Positive Roll, Image should move down in FOV",
"_____no_output_____"
]
],
[
[
"object_locations = optu.make_cube(10.,1.0*np.asarray([0,0,200]))\n\nagent_location = 1.0*np.asarray([0,0,100])\nobject_intensities = np.linalg.norm(object_locations-agent_location,axis=1)-50\nfov=np.pi/4\n\n\nyaw = 0.0\npitch = 0.0\nroll = np.pi/16\nagent_q = np.asarray([yaw,pitch,roll])\n\n\nC_cb = optu.rotate_optical_axis(0.0, 0.0, 0.0)\nr_cb = np.asarray([0,0,0])\n\n\ncm = Camera_model(attitude_parameterization=ap, C_cb=C_cb, r_cb=r_cb,\n fov=fov, debug=False)\nt0 = time()\npix1 = cm.get_pixel_coords(agent_location, agent_q, object_locations, object_intensities)\nt1 = time()\nprint('ET: ',t1-t0)\n\ncm.render(agent_location, agent_q, object_locations, object_intensities)\n",
"Euler321 Attitude\nOverriding focal length using FOV: 0.7853981633974483 13.361957121094465\nK: \n[[133.61957121 0. 50. ]\n [ 0. 133.61957121 50. ]\n [ 0. 0. 1. ]]\nC_cb: \n[[ 1. 0. -0.]\n [ 0. 1. 0.]\n [ 0. 0. 1.]]\nt: \npixel_locs.shape: (8, 2)\nET: 0.0016019344329833984\npixel_locs.shape: (8, 2)\n(8, 2) (8,) (8,)\n"
]
],
[
[
"# Negative Pitch, Image should move right",
"_____no_output_____"
]
],
[
[
"object_locations = optu.make_cube(10.,1.0*np.asarray([0,0,200]))\n\nagent_location = 1.0*np.asarray([0,0,100])\nobject_intensities = np.linalg.norm(object_locations-agent_location,axis=1)-50\nfov=np.pi/4\n\n\nyaw = 0.0\npitch = -np.pi/16\nroll = 0.0\nagent_q = np.asarray([yaw,pitch,roll])\n\n\nC_cb = optu.rotate_optical_axis(0.0, 0.0, 0.0)\nr_cb = np.asarray([0,0,0])\n\n\ncm = Camera_model(attitude_parameterization=ap, C_cb=C_cb, r_cb=r_cb,\n fov=fov, debug=False)\nt0 = time()\npix1 = cm.get_pixel_coords(agent_location, agent_q, object_locations, object_intensities)\nt1 = time()\nprint('ET: ',t1-t0)\n\ncm.render(agent_location, agent_q, object_locations, object_intensities)\n",
"Euler321 Attitude\nOverriding focal length using FOV: 0.7853981633974483 13.361957121094465\nK: \n[[133.61957121 0. 50. ]\n [ 0. 133.61957121 50. ]\n [ 0. 0. 1. ]]\nC_cb: \n[[ 1. 0. -0.]\n [ 0. 1. 0.]\n [ 0. 0. 1.]]\nt: \npixel_locs.shape: (8, 2)\nET: 0.0051801204681396484\npixel_locs.shape: (8, 2)\n(8, 2) (8,) (8,)\n"
]
],
[
[
"# Positive Yaw should rotate image",
"_____no_output_____"
]
],
[
[
"object_locations = optu.make_cube(10.,1.0*np.asarray([0,0,200]))\n\nagent_location = 1.0*np.asarray([0,0,100])\nobject_intensities = np.linalg.norm(object_locations-agent_location,axis=1)-50\nfov=np.pi/4\n\n\nyaw = np.pi/8\npitch = 0.0\nroll = 0.0\nagent_q = np.asarray([yaw,pitch,roll])\n\n\nC_cb = optu.rotate_optical_axis(0.0, 0.0, 0.0)\nr_cb = np.asarray([0,0,0])\n\n\ncm = Camera_model(attitude_parameterization=ap, C_cb=C_cb, r_cb=r_cb,\n fov=fov, debug=False)\nt0 = time()\npix1 = cm.get_pixel_coords(agent_location, agent_q, object_locations, object_intensities)\nt1 = time()\nprint('ET: ',t1-t0)\n\ncm.render(agent_location, agent_q, object_locations, object_intensities)\n",
"Euler321 Attitude\nOverriding focal length using FOV: 0.7853981633974483 13.361957121094465\nK: \n[[133.61957121 0. 50. ]\n [ 0. 133.61957121 50. ]\n [ 0. 0. 1. ]]\nC_cb: \n[[ 1. 0. -0.]\n [ 0. 1. 0.]\n [ 0. 0. 1.]]\nt: \npixel_locs.shape: (8, 2)\nET: 0.0015840530395507812\npixel_locs.shape: (8, 2)\n(8, 2) (8,) (8,)\n"
],
[
"print(np.linspace(-100,100,11))",
"[-100. -80. -60. -40. -20. 0. 20. 40. 60. 80. 100.]\n"
],
[
"print(np.linspace(-4,4,9))",
"[-4. -3. -2. -1. 0. 1. 2. 3. 4.]\n"
],
[
"x = np.linspace(-7,7,15)\np = np.linspace(-3,3,7)\nfor i in range(p.shape[0]):\n line = np.stack((x,p[i]*np.ones_like(x)))\n print(line)",
"[[-7. -6. -5. -4. -3. -2. -1. 0. 1. 2. 3. 4. 5. 6. 7.]\n [-3. -3. -3. -3. -3. -3. -3. -3. -3. -3. -3. -3. -3. -3. -3.]]\n[[-7. -6. -5. -4. -3. -2. -1. 0. 1. 2. 3. 4. 5. 6. 7.]\n [-2. -2. -2. -2. -2. -2. -2. -2. -2. -2. -2. -2. -2. -2. -2.]]\n[[-7. -6. -5. -4. -3. -2. -1. 0. 1. 2. 3. 4. 5. 6. 7.]\n [-1. -1. -1. -1. -1. -1. -1. -1. -1. -1. -1. -1. -1. -1. -1.]]\n[[-7. -6. -5. -4. -3. -2. -1. 0. 1. 2. 3. 4. 5. 6. 7.]\n [ 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]]\n[[-7. -6. -5. -4. -3. -2. -1. 0. 1. 2. 3. 4. 5. 6. 7.]\n [ 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]]\n[[-7. -6. -5. -4. -3. -2. -1. 0. 1. 2. 3. 4. 5. 6. 7.]\n [ 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.]]\n[[-7. -6. -5. -4. -3. -2. -1. 0. 1. 2. 3. 4. 5. 6. 7.]\n [ 3. 3. 3. 3. 3. 3. 3. 3. 3. 3. 3. 3. 3. 3. 3.]]\n"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
]
|
e798bd8e38c18b8ed02745c1e938b34e4ffa016f | 10,033 | ipynb | Jupyter Notebook | save/13-Structured-Query-Language/ClassNotes.ipynb | ecl95/LectureNotes | 45e01baa3ca2adee7188d0dc666a7b76429924e7 | [
"BSD-2-Clause"
]
| 103 | 2016-01-07T05:27:16.000Z | 2022-02-18T03:56:41.000Z | save/13-Structured-Query-Language/ClassNotes.ipynb | ecl95/LectureNotes | 45e01baa3ca2adee7188d0dc666a7b76429924e7 | [
"BSD-2-Clause"
]
| 4 | 2016-01-07T19:45:08.000Z | 2020-05-05T21:46:51.000Z | save/13-Structured-Query-Language/ClassNotes.ipynb | ecl95/LectureNotes | 45e01baa3ca2adee7188d0dc666a7b76429924e7 | [
"BSD-2-Clause"
]
| 103 | 2016-01-07T14:40:11.000Z | 2020-09-09T06:05:30.000Z | 21.858388 | 109 | 0.375062 | [
[
[
"# loading the special sql extension\n%load_ext sql",
"_____no_output_____"
],
[
"# connecting to a database which lives on the Amazon Cloud\n# need to substitute password with the one provided in the email!!!\n%sql postgresql://dssg_student:[email protected]/dssg2016",
"ERROR: Line magic function `%sql` not found.\n"
]
],
[
[
"Two interesting tables:\n seattlecrimeincidents first half of 2015\n census_data",
"_____no_output_____"
]
],
[
[
"# running a simple SQL command\n%sql select * from seattlecrimeincidents limit 10;",
"_____no_output_____"
],
[
"# Show specific columns\n%sql select \"Offense Type\",latitude,longitude from seattlecrimeincidents limit 10;",
"_____no_output_____"
],
[
"%%sql\n-- select rows\nselect \"Offense Type\", latitude, longitude, month from seattlecrimeincidents\n where \"Offense Type\" ='THEFT-BICYCLE' and month = 1",
"_____no_output_____"
],
[
"%%sql\nselect count(*) from seattlecrimeincidents;",
"_____no_output_____"
],
[
"%%sql\nselect count(*) from settlecrimeincidents",
"_____no_output_____"
],
[
"%%sql\nselect count(*) from (select \"Offense Type\", latitude, longitude, month from seattlecrimeincidents\n where \"Offense Type\" ='THEFT-BICYCLE' and month = 1) as small_table",
"_____no_output_____"
],
[
"# use max, min functions",
"_____no_output_____"
],
[
"%%sql \nselect min(latitude) as min_lat,max(latitude) as max_lat,\n min(longitude)as min_long,max(longitude) as max_long\n from seattlecrimeincidents;",
"1 rows affected.\n"
],
[
"%%sql\nselect year,count(*) from seattlecrimeincidents \n group by year\n order by year ASC;",
"17 rows affected.\n"
],
[
"%%sql\nselect distinct year from seattlecrimeincidents;",
"17 rows affected.\n"
]
]
]
| [
"code",
"markdown",
"code"
]
| [
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
e798bfb0aee3af609367ed55dcfc4556bb25867c | 1,374 | ipynb | Jupyter Notebook | notebooks/pyramid_pattern_1.ipynb | neso613/python_coding | 418a24ebb1ec30cdab251eb3f165920e71da5c29 | [
"Apache-2.0"
]
| null | null | null | notebooks/pyramid_pattern_1.ipynb | neso613/python_coding | 418a24ebb1ec30cdab251eb3f165920e71da5c29 | [
"Apache-2.0"
]
| null | null | null | notebooks/pyramid_pattern_1.ipynb | neso613/python_coding | 418a24ebb1ec30cdab251eb3f165920e71da5c29 | [
"Apache-2.0"
]
| null | null | null | 18.078947 | 42 | 0.442504 | [
[
[
" 1\n 212\n 32123\n 4321234\n543212345",
"_____no_output_____"
]
],
[
[
"num=5\nfor i in range(1,num+1):\n for j in range(1,num-i+1):\n print(end=\" \")\n for j in range(i,0,-1):\n print(j,end=\"\")\n for j in range(2,i+1):\n print(j,end=\"\")\n print() ",
" 1\n 212\n 32123\n 4321234\n543212345\n"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
]
]
|
e798c0a2a7218f5af892e1e7cc9e15c0a07d969f | 10,033 | ipynb | Jupyter Notebook | notebooks/Pre-Hydrogen-Demo.ipynb | jopasserat/PySonar | 877cad6ec1e180ff0f3831501bd3c30c5880731b | [
"Apache-2.0"
]
| 171 | 2017-07-29T21:51:07.000Z | 2018-04-07T10:04:12.000Z | notebooks/Pre-Hydrogen-Demo.ipynb | jopasserat/PySonar | 877cad6ec1e180ff0f3831501bd3c30c5880731b | [
"Apache-2.0"
]
| 36 | 2017-07-31T01:54:18.000Z | 2017-12-06T00:15:32.000Z | notebooks/Pre-Hydrogen-Demo.ipynb | jopasserat/PySonar | 877cad6ec1e180ff0f3831501bd3c30c5880731b | [
"Apache-2.0"
]
| 63 | 2017-08-06T18:52:35.000Z | 2018-03-29T12:56:41.000Z | 23.831354 | 346 | 0.545998 | [
[
[
"# Sonar - Decentralized Model Training Simulation (local)\n\nDISCLAIMER: This is a proof-of-concept implementation. It does not represent a remotely product ready implementation or follow proper conventions for security, convenience, or scalability. It is part of a broader proof-of-concept demonstrating the vision of the OpenMined project, its major moving parts, and how they might work together.\n",
"_____no_output_____"
],
[
"### Imports and Convenience Functions",
"_____no_output_____"
]
],
[
[
"import warnings\nimport numpy as np\nimport phe as paillier\nfrom sonar.contracts import ModelRepository,Model\nfrom syft.he.Paillier import KeyPair\nfrom syft.nn.linear import LinearClassifier\nimport numpy as np\nfrom sklearn.datasets import load_diabetes\n\ndef get_balance(account):\n return repo.web3.fromWei(repo.web3.eth.getBalance(account),'ether')\n\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
]
],
[
[
"### Setting up the Experiment",
"_____no_output_____"
]
],
[
[
"# for the purpose of the simulation, we're going to split our dataset up amongst\n# the relevant simulated users\n\ndiabetes = load_diabetes()\ny = diabetes.target\nX = diabetes.data\n\nvalidation = (X[0:42],y[0:42])\nanonymous_diabetes_users = (X[42:],y[42:])\n\n# we're also going to initialize the model trainer smart contract, which in the\n# real world would already be on the blockchain (managing other contracts) before\n# the simulation begins\n\n# ATTENTION: copy paste the correct address (NOT THE DEFAULT SEEN HERE) from truffle migrate output.\nrepo = ModelRepository('0xf30068fb49616db7d5afb89862d6b40d11389327', ipfs_host='localhost', web3_host='localhost') # blockchain hosted model repository",
"No account submitted... using default[2]\nConnected to OpenMined ModelRepository:0xf30068fb49616db7d5afb89862d6b40d11389327\n"
],
[
"\n\n# we're going to set aside 400 accounts for our 400 patients\n# Let's go ahead and pair each data point with each patient's \n# address so that we know we don't get them confused\npatient_addresses = repo.web3.eth.accounts[1:40]\nanonymous_diabetics = list(zip(patient_addresses,\n anonymous_diabetes_users[0],\n anonymous_diabetes_users[1]))\n\n# we're going to set aside 1 account for Cure Diabetes Inc\ncure_diabetes_inc = repo.web3.eth.accounts[0]",
"_____no_output_____"
]
],
[
[
"## Step 1: Cure Diabetes Inc Initializes a Model and Provides a Bounty",
"_____no_output_____"
]
],
[
[
"pubkey,prikey = KeyPair().generate(n_length=1024)\ndiabetes_classifier = LinearClassifier(desc=\"DiabetesClassifier\",n_inputs=10,n_labels=1)\ninitial_error = diabetes_classifier.evaluate(validation[0],validation[1])\ndiabetes_classifier.encrypt(pubkey)\n\ndiabetes_model = Model(owner=cure_diabetes_inc,\n syft_obj = diabetes_classifier,\n bounty = 1,\n initial_error = initial_error,\n target_error = 10000\n )",
"_____no_output_____"
],
[
"model_id = repo.submit_model(diabetes_model)",
"_____no_output_____"
],
[
"cure_diabetes_inc",
"_____no_output_____"
]
],
[
[
"## Step 2: An Anonymous Patient Downloads the Model and Improves It",
"_____no_output_____"
]
],
[
[
"model_id",
"_____no_output_____"
],
[
"model = repo[model_id]",
"_____no_output_____"
],
[
"diabetic_address,input_data,target_data = anonymous_diabetics[0]",
"_____no_output_____"
],
[
"repo[model_id].submit_gradient(diabetic_address,input_data,target_data)",
"_____no_output_____"
]
],
[
[
"## Step 3: Cure Diabetes Inc. Evaluates the Gradient ",
"_____no_output_____"
]
],
[
[
"repo[model_id]",
"_____no_output_____"
],
[
"old_balance = get_balance(diabetic_address)\nprint(old_balance)",
"100.007394085094304861\n"
],
[
"new_error = repo[model_id].evaluate_gradient(cure_diabetes_inc,repo[model_id][0],prikey,pubkey,validation[0],validation[1])",
"_____no_output_____"
],
[
"new_error",
"_____no_output_____"
],
[
"new_balance = get_balance(diabetic_address)\nincentive = new_balance - old_balance\nprint(incentive)",
"0.000840812917924814\n"
]
],
[
[
"## Step 4: Rinse and Repeat",
"_____no_output_____"
]
],
[
[
"model",
"_____no_output_____"
],
[
"for i,(addr, input, target) in enumerate(anonymous_diabetics):\n try:\n \n model = repo[model_id]\n \n # patient is doing this\n model.submit_gradient(addr,input,target)\n \n # Cure Diabetes Inc does this\n old_balance = get_balance(addr)\n new_error = model.evaluate_gradient(cure_diabetes_inc,model[i+1],prikey,pubkey,validation[0],validation[1],alpha=2)\n print(\"new error = \"+str(new_error))\n incentive = round(get_balance(addr) - old_balance,5)\n print(\"incentive = \"+str(incentive))\n except:\n \"Connection Reset\"",
"new error = 26580005\nincentive = 0.00162\nnew error = 26639344\nincentive = 0.00000\nnew error = 26536737\nincentive = 0.00163\nnew error = 26546235\nincentive = 0.00000\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
e798d9055eacca4735fd65ae18a99ec45a908d2e | 775,351 | ipynb | Jupyter Notebook | CS20_Tensorflow_for_Deep_learning_Research/Tensor2Tensor_Intro.ipynb | jungi21cc/DeepLearning | 677475aa321cca32ebc5bde84dddbbb06f53decf | [
"MIT"
]
| 3 | 2018-04-21T17:30:21.000Z | 2019-11-17T08:51:10.000Z | CS20_Tensorflow_for_Deep_learning_Research/Tensor2Tensor_Intro.ipynb | jungi21cc/DeepLearning | 677475aa321cca32ebc5bde84dddbbb06f53decf | [
"MIT"
]
| null | null | null | CS20_Tensorflow_for_Deep_learning_Research/Tensor2Tensor_Intro.ipynb | jungi21cc/DeepLearning | 677475aa321cca32ebc5bde84dddbbb06f53decf | [
"MIT"
]
| 3 | 2018-04-08T01:01:00.000Z | 2019-11-17T08:51:13.000Z | 481.285537 | 599,278 | 0.811089 | [
[
[
"# Welcome to the [Tensor2Tensor](https://github.com/tensorflow/tensor2tensor) Colab\n\nTensor2Tensor, or T2T for short, is a library of deep learning models and datasets designed to make deep learning more accessible and [accelerate ML research](https://research.googleblog.com/2017/06/accelerating-deep-learning-research.html). T2T is actively used and maintained by researchers and engineers within the [Google Brain team](https://research.google.com/teams/brain/) and a community of users. This colab shows you some datasets we have in T2T, how to download and use them, some models we have, how to download pre-trained models and use them, and how to create and train your own models.",
"_____no_output_____"
]
],
[
[
"#@title\n# Copyright 2018 Google LLC.\n\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n\n# https://www.apache.org/licenses/LICENSE-2.0\n\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
],
[
"# Install deps\n!pip install -q -U tensor2tensor\n!pip install -q tensorflow matplotlib",
"\u001b[31mxhtml2pdf 0.2.2 has requirement html5lib>=1.0, but you'll have html5lib 0.9999999 which is incompatible.\u001b[0m\n\u001b[31mweasyprint 0.42.3 has requirement html5lib>=0.999999999, but you'll have html5lib 0.9999999 which is incompatible.\u001b[0m\n\u001b[33mYou are using pip version 10.0.1, however version 18.0 is available.\nYou should consider upgrading via the 'pip install --upgrade pip' command.\u001b[0m\n\u001b[31mxhtml2pdf 0.2.2 has requirement html5lib>=1.0, but you'll have html5lib 0.9999999 which is incompatible.\u001b[0m\n\u001b[31mweasyprint 0.42.3 has requirement html5lib>=0.999999999, but you'll have html5lib 0.9999999 which is incompatible.\u001b[0m\n\u001b[33mYou are using pip version 10.0.1, however version 18.0 is available.\nYou should consider upgrading via the 'pip install --upgrade pip' command.\u001b[0m\n"
],
[
"# Imports we need.\nimport tensorflow as tf\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport os\nimport collections\n\nfrom tensor2tensor import models\nfrom tensor2tensor import problems\nfrom tensor2tensor.layers import common_layers\nfrom tensor2tensor.utils import trainer_lib\nfrom tensor2tensor.utils import t2t_model\nfrom tensor2tensor.utils import registry\nfrom tensor2tensor.utils import metrics\n\n# Enable TF Eager execution\ntfe = tf.contrib.eager\ntfe.enable_eager_execution()\n\n# Other setup\nModes = tf.estimator.ModeKeys\n\n# Setup some directories\ndata_dir = os.path.expanduser(\"~/t2t/data\")\ntmp_dir = os.path.expanduser(\"~/t2t/tmp\")\ntrain_dir = os.path.expanduser(\"~/t2t/train\")\ncheckpoint_dir = os.path.expanduser(\"~/t2t/checkpoints\")\ntf.gfile.MakeDirs(data_dir)\ntf.gfile.MakeDirs(tmp_dir)\ntf.gfile.MakeDirs(train_dir)\ntf.gfile.MakeDirs(checkpoint_dir)\ngs_data_dir = \"gs://tensor2tensor-data\"\ngs_ckpt_dir = \"gs://tensor2tensor-checkpoints/\"",
"/home/jk/anaconda3/lib/python3.6/site-packages/h5py/__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\n"
]
],
[
[
"# Download MNIST and inspect it",
"_____no_output_____"
]
],
[
[
"# A Problem is a dataset together with some fixed pre-processing.\n# It could be a translation dataset with a specific tokenization,\n# or an image dataset with a specific resolution.\n#\n# There are many problems available in Tensor2Tensor\nproblems.available()",
"_____no_output_____"
],
[
"# Fetch the MNIST problem\nmnist_problem = problems.problem(\"image_mnist\")\n# The generate_data method of a problem will download data and process it into\n# a standard format ready for training and evaluation.\nmnist_problem.generate_data(data_dir, tmp_dir)",
"INFO:tensorflow:Downloading http://yann.lecun.com/exdb/mnist/train-images-idx3-ubyte.gz to /home/jk/t2t/tmp/train-images-idx3-ubyte.gz\n100% completed\nINFO:tensorflow:Successfully downloaded train-images-idx3-ubyte.gz, 9912422 bytes.\nINFO:tensorflow:Downloading http://yann.lecun.com/exdb/mnist/train-labels-idx1-ubyte.gz to /home/jk/t2t/tmp/train-labels-idx1-ubyte.gz\n113% completed\nINFO:tensorflow:Successfully downloaded train-labels-idx1-ubyte.gz, 28881 bytes.\nINFO:tensorflow:Downloading http://yann.lecun.com/exdb/mnist/t10k-images-idx3-ubyte.gz to /home/jk/t2t/tmp/t10k-images-idx3-ubyte.gz\n100% completed\nINFO:tensorflow:Successfully downloaded t10k-images-idx3-ubyte.gz, 1648877 bytes.\nINFO:tensorflow:Downloading http://yann.lecun.com/exdb/mnist/t10k-labels-idx1-ubyte.gz to /home/jk/t2t/tmp/t10k-labels-idx1-ubyte.gz\n180% completed\nINFO:tensorflow:Successfully downloaded t10k-labels-idx1-ubyte.gz, 4542 bytes.\nINFO:tensorflow:Not downloading, file already found: /home/jk/t2t/tmp/train-images-idx3-ubyte.gz\nINFO:tensorflow:Not downloading, file already found: /home/jk/t2t/tmp/train-labels-idx1-ubyte.gz\nINFO:tensorflow:Not downloading, file already found: /home/jk/t2t/tmp/t10k-images-idx3-ubyte.gz\nINFO:tensorflow:Not downloading, file already found: /home/jk/t2t/tmp/t10k-labels-idx1-ubyte.gz\nINFO:tensorflow:Generating case 0.\nINFO:tensorflow:Generated 60000 Examples\nINFO:tensorflow:Generating case 0.\nINFO:tensorflow:Generated 10000 Examples\nINFO:tensorflow:Shuffling data...\nINFO:tensorflow:Data shuffled.\n"
],
[
"# Now let's see the training MNIST data as Tensors.\nmnist_example = tfe.Iterator(mnist_problem.dataset(Modes.TRAIN, data_dir)).next()\nimage = mnist_example[\"inputs\"]\nlabel = mnist_example[\"targets\"]\n\nplt.imshow(image.numpy()[:, :, 0].astype(np.float32), cmap=plt.get_cmap('gray'))\nprint(\"Label: %d\" % label.numpy())",
"INFO:tensorflow:Reading data files from /home/jk/t2t/data/image_mnist-train*\nINFO:tensorflow:partition: 0 num_data_files: 10\nLabel: 1\n"
]
],
[
[
"# Translate from English to German with a pre-trained model",
"_____no_output_____"
]
],
[
[
"# Fetch the problem\nende_problem = problems.problem(\"translate_ende_wmt32k\")\n\n# Copy the vocab file locally so we can encode inputs and decode model outputs\n# All vocabs are stored on GCS\nvocab_name = \"vocab.ende.32768\"\nvocab_file = os.path.join(gs_data_dir, vocab_name)\n!gsutil cp {vocab_file} {data_dir}\n\n# Get the encoders from the problem\nencoders = ende_problem.feature_encoders(data_dir)\n\n# Setup helper functions for encoding and decoding\ndef encode(input_str, output_str=None):\n \"\"\"Input str to features dict, ready for inference\"\"\"\n inputs = encoders[\"inputs\"].encode(input_str) + [1] # add EOS id\n batch_inputs = tf.reshape(inputs, [1, -1, 1]) # Make it 3D.\n return {\"inputs\": batch_inputs}\n\ndef decode(integers):\n \"\"\"List of ints to str\"\"\"\n integers = list(np.squeeze(integers))\n if 1 in integers:\n integers = integers[:integers.index(1)]\n return encoders[\"inputs\"].decode(np.squeeze(integers))",
"_____no_output_____"
],
[
"# # Generate and view the data\n# # This cell is commented out because WMT data generation can take hours\n\n# ende_problem.generate_data(data_dir, tmp_dir)\n# example = tfe.Iterator(ende_problem.dataset(Modes.TRAIN, data_dir)).next()\n# inputs = [int(x) for x in example[\"inputs\"].numpy()] # Cast to ints.\n# targets = [int(x) for x in example[\"targets\"].numpy()] # Cast to ints.\n\n\n\n# # Example inputs as int-tensor.\n# print(\"Inputs, encoded:\")\n# print(inputs)\n# print(\"Inputs, decoded:\")\n# # Example inputs as a sentence.\n# print(decode(inputs))\n# # Example targets as int-tensor.\n# print(\"Targets, encoded:\")\n# print(targets)\n# # Example targets as a sentence.\n# print(\"Targets, decoded:\")\n# print(decode(targets))",
"_____no_output_____"
],
[
"# There are many models available in Tensor2Tensor\nregistry.list_models()",
"_____no_output_____"
],
[
"# Create hparams and the model\nmodel_name = \"transformer\"\nhparams_set = \"transformer_base\"\n\nhparams = trainer_lib.create_hparams(hparams_set, data_dir=data_dir, problem_name=\"translate_ende_wmt32k\")\n\n# NOTE: Only create the model once when restoring from a checkpoint; it's a\n# Layer and so subsequent instantiations will have different variable scopes\n# that will not match the checkpoint.\ntranslate_model = registry.model(model_name)(hparams, Modes.EVAL)",
"INFO:tensorflow:Setting T2TModel mode to 'eval'\nINFO:tensorflow:Setting hparams.layer_prepostprocess_dropout to 0.0\nINFO:tensorflow:Setting hparams.symbol_dropout to 0.0\nINFO:tensorflow:Setting hparams.label_smoothing to 0.0\nINFO:tensorflow:Setting hparams.attention_dropout to 0.0\nINFO:tensorflow:Setting hparams.dropout to 0.0\nINFO:tensorflow:Setting hparams.relu_dropout to 0.0\n"
],
[
"# Copy the pretrained checkpoint locally\nckpt_name = \"transformer_ende_test\"\ngs_ckpt = os.path.join(gs_ckpt_dir, ckpt_name)\n!gsutil -q cp -R {gs_ckpt} {checkpoint_dir}\nckpt_path = tf.train.latest_checkpoint(os.path.join(checkpoint_dir, ckpt_name))\nckpt_path",
"_____no_output_____"
],
[
"# Restore and translate!\ndef translate(inputs):\n encoded_inputs = encode(inputs)\n with tfe.restore_variables_on_create(ckpt_path):\n model_output = translate_model.infer(encoded_inputs)[\"outputs\"]\n return decode(model_output)\n\ninputs = \"The animal didn't cross the street because it was too tired\"\noutputs = translate(inputs)\n\nprint(\"Inputs: %s\" % inputs)\nprint(\"Outputs: %s\" % outputs)",
"INFO:tensorflow:Greedy Decoding\nInputs: The animal didn't cross the street because it was too tired\nOutputs: Das Tier überquerte nicht die Straße, weil es zu müde war.\n"
]
],
[
[
"## Attention Viz Utils",
"_____no_output_____"
]
],
[
[
"from tensor2tensor.visualization import attention\nfrom tensor2tensor.data_generators import text_encoder\n\nSIZE = 35\n\ndef encode_eval(input_str, output_str):\n inputs = tf.reshape(encoders[\"inputs\"].encode(input_str) + [1], [1, -1, 1, 1]) # Make it 3D.\n outputs = tf.reshape(encoders[\"inputs\"].encode(output_str) + [1], [1, -1, 1, 1]) # Make it 3D.\n return {\"inputs\": inputs, \"targets\": outputs}\n\ndef get_att_mats():\n enc_atts = []\n dec_atts = []\n encdec_atts = []\n\n for i in range(hparams.num_hidden_layers):\n enc_att = translate_model.attention_weights[\n \"transformer/body/encoder/layer_%i/self_attention/multihead_attention/dot_product_attention\" % i][0]\n dec_att = translate_model.attention_weights[\n \"transformer/body/decoder/layer_%i/self_attention/multihead_attention/dot_product_attention\" % i][0]\n encdec_att = translate_model.attention_weights[\n \"transformer/body/decoder/layer_%i/encdec_attention/multihead_attention/dot_product_attention\" % i][0]\n enc_atts.append(resize(enc_att))\n dec_atts.append(resize(dec_att))\n encdec_atts.append(resize(encdec_att))\n return enc_atts, dec_atts, encdec_atts\n\ndef resize(np_mat):\n # Sum across heads\n np_mat = np_mat[:, :SIZE, :SIZE]\n row_sums = np.sum(np_mat, axis=0)\n # Normalize\n layer_mat = np_mat / row_sums[np.newaxis, :]\n lsh = layer_mat.shape\n # Add extra dim for viz code to work.\n layer_mat = np.reshape(layer_mat, (1, lsh[0], lsh[1], lsh[2]))\n return layer_mat\n\ndef to_tokens(ids):\n ids = np.squeeze(ids)\n subtokenizer = hparams.problem_hparams.vocabulary['targets']\n tokens = []\n for _id in ids:\n if _id == 0:\n tokens.append('<PAD>')\n elif _id == 1:\n tokens.append('<EOS>')\n elif _id == -1:\n tokens.append('<NULL>')\n else:\n tokens.append(subtokenizer._subtoken_id_to_subtoken_string(_id))\n return tokens",
"_____no_output_____"
],
[
"def call_html():\n import IPython\n display(IPython.core.display.HTML('''\n <script src=\"/static/components/requirejs/require.js\"></script>\n <script>\n requirejs.config({\n paths: {\n base: '/static/base',\n \"d3\": \"https://cdnjs.cloudflare.com/ajax/libs/d3/3.5.8/d3.min\",\n jquery: '//ajax.googleapis.com/ajax/libs/jquery/2.0.0/jquery.min',\n },\n });\n </script>\n '''))",
"_____no_output_____"
]
],
[
[
"## Display Attention",
"_____no_output_____"
]
],
[
[
"# Convert inputs and outputs to subwords\ninp_text = to_tokens(encoders[\"inputs\"].encode(inputs))\nout_text = to_tokens(encoders[\"inputs\"].encode(outputs))\n\n# Run eval to collect attention weights\nexample = encode_eval(inputs, outputs)\nwith tfe.restore_variables_on_create(tf.train.latest_checkpoint(checkpoint_dir)):\n translate_model.set_mode(Modes.EVAL)\n translate_model(example)\n# Get normalized attention weights for each layer\nenc_atts, dec_atts, encdec_atts = get_att_mats()\n\ncall_html()\nattention.show(inp_text, out_text, enc_atts, dec_atts, encdec_atts)",
"INFO:tensorflow:Transforming feature 'inputs' with symbol_modality_33708_512.bottom\nINFO:tensorflow:Transforming 'targets' with symbol_modality_33708_512.targets_bottom\nINFO:tensorflow:Building model body\nINFO:tensorflow:Transforming body output with symbol_modality_33708_512.top\n"
]
],
[
[
"# Train a custom model on MNIST",
"_____no_output_____"
]
],
[
[
"# Create your own model\n\nclass MySimpleModel(t2t_model.T2TModel):\n\n def body(self, features):\n inputs = features[\"inputs\"]\n filters = self.hparams.hidden_size\n h1 = tf.layers.conv2d(inputs, filters,\n kernel_size=(5, 5), strides=(2, 2))\n h2 = tf.layers.conv2d(tf.nn.relu(h1), filters,\n kernel_size=(5, 5), strides=(2, 2))\n return tf.layers.conv2d(tf.nn.relu(h2), filters,\n kernel_size=(3, 3))\n\nhparams = trainer_lib.create_hparams(\"basic_1\", data_dir=data_dir, problem_name=\"image_mnist\")\nhparams.hidden_size = 64\nmodel = MySimpleModel(hparams, Modes.TRAIN)",
"INFO:tensorflow:Setting T2TModel mode to 'train'\n"
],
[
"# Prepare for the training loop\n\n# In Eager mode, opt.minimize must be passed a loss function wrapped with\n# implicit_value_and_gradients\[email protected]_value_and_gradients\ndef loss_fn(features):\n _, losses = model(features)\n return losses[\"training\"]\n\n# Setup the training data\nBATCH_SIZE = 128\nmnist_train_dataset = mnist_problem.dataset(Modes.TRAIN, data_dir)\nmnist_train_dataset = mnist_train_dataset.repeat(None).batch(BATCH_SIZE)\n\noptimizer = tf.train.AdamOptimizer()",
"INFO:tensorflow:Reading data files from /content/t2t/data/image_mnist-train*\nINFO:tensorflow:partition: 0 num_data_files: 10\n"
],
[
"# Train\nNUM_STEPS = 500\n\nfor count, example in enumerate(tfe.Iterator(mnist_train_dataset)):\n example[\"targets\"] = tf.reshape(example[\"targets\"], [BATCH_SIZE, 1, 1, 1]) # Make it 4D.\n loss, gv = loss_fn(example)\n optimizer.apply_gradients(gv)\n\n if count % 50 == 0:\n print(\"Step: %d, Loss: %.3f\" % (count, loss.numpy()))\n if count >= NUM_STEPS:\n break",
"INFO:tensorflow:Transforming feature 'inputs' with image_modality.bottom\nINFO:tensorflow:Transforming 'targets' with class_label_modality_10_64.targets_bottom\nINFO:tensorflow:Transforming body output with class_label_modality_10_64.top\nStep: 0, Loss: 9.131\nStep: 50, Loss: 0.958\nStep: 100, Loss: 0.587\nStep: 150, Loss: 0.677\nStep: 200, Loss: 0.562\nStep: 250, Loss: 0.595\nStep: 300, Loss: 0.395\nStep: 350, Loss: 0.435\nStep: 400, Loss: 0.320\nStep: 450, Loss: 0.228\nStep: 500, Loss: 0.206\n"
],
[
"model.set_mode(Modes.EVAL)\nmnist_eval_dataset = mnist_problem.dataset(Modes.EVAL, data_dir)\n\n# Create eval metric accumulators for accuracy (ACC) and accuracy in\n# top 5 (ACC_TOP5)\nmetrics_accum, metrics_result = metrics.create_eager_metrics(\n [metrics.Metrics.ACC, metrics.Metrics.ACC_TOP5])\n\nfor count, example in enumerate(tfe.Iterator(mnist_eval_dataset)):\n if count >= 200:\n break\n\n # Make the inputs and targets 4D\n example[\"inputs\"] = tf.reshape(example[\"inputs\"], [1, 28, 28, 1])\n example[\"targets\"] = tf.reshape(example[\"targets\"], [1, 1, 1, 1])\n\n # Call the model\n predictions, _ = model(example)\n\n # Compute and accumulate metrics\n metrics_accum(predictions, example[\"targets\"])\n\n# Print out the averaged metric values on the eval data\nfor name, val in metrics_result().items():\n print(\"%s: %.2f\" % (name, val))",
"INFO:tensorflow:Reading data files from /content/t2t/data/image_mnist-dev*\nINFO:tensorflow:partition: 0 num_data_files: 1\naccuracy_top5: 0.99\naccuracy: 0.97\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
]
|
e7990c4790828716a02966c6f7fad92f8b95fc1f | 105,176 | ipynb | Jupyter Notebook | samples/5.analysis.ipynb | EsriJapan/arcgis-samples-python-api | 448eb4aa0bc281ab7b928777a09a8bafa7b7068b | [
"Apache-2.0"
]
| 4 | 2018-03-09T13:57:17.000Z | 2021-05-07T09:00:55.000Z | samples/5.analysis.ipynb | EsriJapan/arcgis-samples-python-api | 448eb4aa0bc281ab7b928777a09a8bafa7b7068b | [
"Apache-2.0"
]
| null | null | null | samples/5.analysis.ipynb | EsriJapan/arcgis-samples-python-api | 448eb4aa0bc281ab7b928777a09a8bafa7b7068b | [
"Apache-2.0"
]
| 3 | 2018-02-15T02:06:53.000Z | 2022-02-25T02:49:46.000Z | 74.172073 | 52,707 | 0.668299 | [
[
[
"# ArcGIS Online の解析機能を使用する\n### 使用するデータ\n* 栃木県のダム諸元表: https://www.geospatial.jp/ckan/dataset/09000-103\n",
"_____no_output_____"
],
[
"## データを確認する",
"_____no_output_____"
]
],
[
[
"# pandas を使用して csv ファイルの読み込み、中身を表示する\nimport pandas as pd\ndam_csv = pd.read_csv('https://www.geospatial.jp/ckan/dataset/d6a87e42-6e86-449e-9d76-1e40319bb99b/resource/b5d633c8-f2c8-4baa-88a9-fcf1872dcfcd/download/724522014tochiginodamsyogen04033.csv',encoding=\"SHIFT-JIS\")",
"_____no_output_____"
],
[
"dam_csv",
"_____no_output_____"
]
],
[
[
"## ArcGIS Online にログイン",
"_____no_output_____"
]
],
[
[
"# ArcGIS Online に開発者アカウントでサインインする\nfrom arcgis.gis import GIS\nimport getpass\n\ndeveloersUser = 'あなたのユーザー名'\ndeveloersPass = getpass.getpass('ユーザー['+ develoersUser + ']のパスワード=')\n\ngis = GIS(\"http://\"+ develoersUser +\".maps.arcgis.com/\",develoersUser,develoersPass)\nuser = gis.users.get(develoersUser)\nuser",
"ユーザー[ejpythondev]のパスワード=········\n"
]
],
[
[
"## ArcGIS Online にホスト フィーチャ サービスを公開する",
"_____no_output_____"
]
],
[
[
"# ArcGIS Online に CSV ファイルをアイテムとして追加する\ncsv_file = 'https://www.geospatial.jp/ckan/dataset/d6a87e42-6e86-449e-9d76-1e40319bb99b/resource/b5d633c8-f2c8-4baa-88a9-fcf1872dcfcd/download/724522014tochiginodamsyogen04033.csv'\ncsv_item = gis.content.add({}, csv_file)\ndisplay(csv_item)",
"_____no_output_____"
],
[
"# CSV にある緯度経度の情報を使用して、追加したアイテムからホスト フィーチャ サービス(ダムのポイント)を公開する\ncsv_lyr = csv_item.publish({'name':'dam','locationType':'coordinates', 'latitudeFieldName':'緯度', 'longitudeFieldName':'経度'})\ndisplay(csv_lyr)",
"_____no_output_____"
],
[
"# マップにホスト フィーチャ サービス追加して表示する\nmap = gis.map('栃木県')\nmap.add_layer(csv_lyr)\nmap",
"_____no_output_____"
]
],
[
[
"## ArcGIS Online の集水域解析を実行する",
"_____no_output_____"
]
],
[
[
"# ダムのポイントのホスト フィーチャ サービスを引数にして集水域の作成ツール(create_watersheds)を実行する\nfrom arcgis.features import analysis\nwatershedsResult = analysis.create_watersheds(csv_lyr, output_name='watersheds_result')\nwatershedsResult",
"_____no_output_____"
],
[
"# 解析結果の集水域ポリゴンをマップに追加して表示する\nmap.add_layer(watershedsResult)",
"_____no_output_____"
]
],
[
[
"## ArcGIS Online の下流解析を実行する",
"_____no_output_____"
]
],
[
[
"# 集水域の解析結果で出力された調整された入力ポイントを引数にして下流解析ツール(trace_downstream)を実行する\ninput_layer = watershedsResult.layers[0]\ndownstreamResult = analysis.trace_downstream(input_layer, output_name='downstream_result')\ndownstreamResult",
"_____no_output_____"
],
[
"# 解析結果の河川ラインをマップに追加して表示する\nmap.add_layer(downstreamResult)",
"_____no_output_____"
]
],
[
[
"## Web マップとして保存する",
"_____no_output_____"
]
],
[
[
"# Web マップのタイトルなどを定義する\nwebMap_properties = {'title':'栃木県のダム・河川',\n 'snippet':'Python API で作成した栃木県のダム・河川 Web マップ',\n 'tags':'栃木県, ダム, 河川',\n 'extent':downstreamResult.extent\n }\n# Web マップを保存する\nwebMap = map.save(item_properties=webMap_properties)\nwebMap",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
]
|
e799138547d0b5d9602b2e54a81f6b2c47f69ba5 | 19,704 | ipynb | Jupyter Notebook | examples/tests/test_snapshot.ipynb | bdice/openpathsampling | 8c7ab8cb1bd7f6ae388a49d441423e2332c8301b | [
"MIT"
]
| 64 | 2016-07-06T13:38:51.000Z | 2022-03-30T15:58:01.000Z | examples/tests/test_snapshot.ipynb | bdice/openpathsampling | 8c7ab8cb1bd7f6ae388a49d441423e2332c8301b | [
"MIT"
]
| 601 | 2016-06-13T10:22:01.000Z | 2022-03-25T00:10:40.000Z | examples/tests/test_snapshot.ipynb | bdice/openpathsampling | 8c7ab8cb1bd7f6ae388a49d441423e2332c8301b | [
"MIT"
]
| 45 | 2016-11-10T11:17:53.000Z | 2022-02-13T11:50:26.000Z | 28.391931 | 220 | 0.506953 | [
[
[
"## Some testing and analysis of the new `Snapshot` implementation",
"_____no_output_____"
]
],
[
[
"from __future__ import print_function\n\nimport numpy as np\nimport openpathsampling as paths\nimport openpathsampling.engines.features as features",
"_____no_output_____"
]
],
[
[
"Function to show the generated source code",
"_____no_output_____"
]
],
[
[
"from IPython.display import Markdown\n\ndef code_to_md(snapshot_class):\n md = '```py\\n'\n for f, s in snapshot_class.__features__.debug.items():\n if s is not None:\n md += s\n else:\n md += 'def ' + f + '(...):\\n # user defined\\n pass' \n md += '\\n\\n'\n md += '```'\n\n return md",
"_____no_output_____"
]
],
[
[
"### Check generated source code",
"_____no_output_____"
],
[
"Generate simple Snapshot without any features using factory",
"_____no_output_____"
]
],
[
[
"EmptySnap = paths.engines.snapshot.SnapshotFactory('no', [], 'Empty', use_lazy_reversed=False)",
"_____no_output_____"
]
],
[
[
"Generate Snapshot with overridden `.copy` method.",
"_____no_output_____"
]
],
[
[
"@features.base.attach_features([\n features.velocities,\n features.coordinates,\n features.box_vectors,\n features.topology\n])\nclass A(paths.BaseSnapshot):\n def copy(self):\n return 'copy'",
"_____no_output_____"
]
],
[
[
"Check that subclassing with overridden copy needs more overriding.",
"_____no_output_____"
]
],
[
[
"#! lazy\n# lazy because of some issue with Py3k comparing strings\ntry:\n @features.base.attach_features([\n ])\n class B(A):\n pass\nexcept RuntimeWarning as e:\n print(e)\nelse:\n raise RuntimeError('Should have raised a RUNTIME warning') ",
"Subclassing snapshots with overridden function \"copy\" is only possible if this function is overridden again, otherwise some features might not be copied. The general practise of overriding is not recommended.\n"
],
[
"a = A()\nassert(a.copy() == 'copy')",
"_____no_output_____"
],
[
"# NBVAL_IGNORE_OUTPUT\nMarkdown(code_to_md(A))",
"_____no_output_____"
],
[
"# NBVAL_IGNORE_OUTPUT\nMarkdown(code_to_md(EmptySnap))",
"_____no_output_____"
],
[
"SuperSnap = paths.engines.snapshot.SnapshotFactory(\n 'my', [\n paths.engines.features.coordinates,\n paths.engines.features.box_vectors,\n paths.engines.features.velocities\n ], 'No desc', use_lazy_reversed=False)",
"_____no_output_____"
],
[
"# NBVAL_IGNORE_OUTPUT\nMarkdown(code_to_md(SuperSnap))",
"_____no_output_____"
],
[
"MegaSnap = paths.engines.snapshot.SnapshotFactory(\n 'mega', [\n paths.engines.features.statics,\n paths.engines.features.kinetics,\n paths.engines.features.engine\n ], 'Long desc', use_lazy_reversed=False)",
"_____no_output_____"
],
[
"# NBVAL_IGNORE_OUTPUT\nMarkdown(code_to_md(MegaSnap))",
"_____no_output_____"
]
],
[
[
"Test subclassing",
"_____no_output_____"
]
],
[
[
"@features.base.attach_features([\n])\nclass HyperSnap(MegaSnap):\n pass",
"_____no_output_____"
]
],
[
[
"Test subclassing with redundant features (should work / be ignored)",
"_____no_output_____"
]
],
[
[
"@features.base.attach_features([\n paths.engines.features.statics,\n])\nclass HyperSnap(MegaSnap):\n pass",
"_____no_output_____"
]
],
[
[
"Test subclassing with conflicting features (should not work)",
"_____no_output_____"
]
],
[
[
"try:\n @features.base.attach_features([\n paths.engines.features.statics,\n paths.engines.features.coordinates\n ])\n class HyperSnap(MegaSnap):\n pass\nexcept RuntimeWarning as e:\n print(e)\nelse:\n raise RuntimeError('Should have raised a RUNTIME warning') ",
"Collision: Property \"xyz\" already exists.\n"
],
[
"# NBVAL_IGNORE_OUTPUT\nMarkdown(code_to_md(paths.engines.openmm.MDSnapshot))",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
e7992898ddd5a4315b0c9f8b968faee2ac45083b | 14,485 | ipynb | Jupyter Notebook | cv06/forest.ipynb | LukasForst/KO | e77d7ebe6cae4ab2494490adce6439cd1bed3b61 | [
"MIT"
]
| null | null | null | cv06/forest.ipynb | LukasForst/KO | e77d7ebe6cae4ab2494490adce6439cd1bed3b61 | [
"MIT"
]
| null | null | null | cv06/forest.ipynb | LukasForst/KO | e77d7ebe6cae4ab2494490adce6439cd1bed3b61 | [
"MIT"
]
| null | null | null | 32.995444 | 415 | 0.450811 | [
[
[
"# Find the Tents\n\n_Combinatorial Optimization course, FEE CTU in Prague. Created by [Industrial Informatics Department](http://industrialinformatics.fel.cvut.cz)._\n\nThe problem was taken from https://www.brainbashers.com/tents.asp ; there, you can try to solve some examples manually.\n\n## Task\n\nFind all of the hidden tents in the forest grid.\n\nYou know that:\n\n- Each tent is attached to one tree (so there are as many tents as there are trees).\n- A tent can only be found horizontally or vertically adjacent to a tree.\n- Tents are never adjacent to each other, neither vertically, horizontally, nor diagonally.\n- A tree might be next to two tents but is only connected to one.\n\nYou are also given two vectors indicating how many tents are in each respective row or column of the forest grid.\n\n\n## Input\n\nYou are given a positive integer $n \\geq 2$, representing the size of the forest grid (assume it is a square of size $(n \\times n$). You are also given vectors $\\mathbf r = (r_1, \\dots, r_n)$ and $\\mathbf c = (c_1, \\dots, c_n)$ representing the numbers of the tents in the rows and columns of the forest grid. Finally, you are given a list of coordinates of the trees $((x_1, y_1), \\dots, (x_k, y_k))$.",
"_____no_output_____"
]
],
[
[
"# 2x2 - Extra small (for debugging)\nn1 = 3\nr1 = (1, 1, 0)\nc1 = (1, 0, 1)\ntrees1 = [(1,1), (3,2)]\n",
"_____no_output_____"
],
[
"# 8x8 - Medium\nn2 = 8\nr2 = (3, 1, 1, 2, 0, 2, 0, 3)\nc2 = (2, 1, 2, 2 ,1, 1 ,2 ,1)\ntrees2 = [(2, 1), (5, 1), (6, 1),\n (1, 2),\n (3, 3),\n (3, 4), (6, 4),\n (4, 5), (6, 5),\n (8, 7),\n (2, 8), (4, 8)]",
"_____no_output_____"
],
[
"# Weekly special\nn3 = 20\nr3 = (7, 2, 3, 4, 3, 5, 4, 4, 4, 4, 3, 6, 3, 6, 2, 3, 6, 3, 3, 5)\nc3 = (6, 4, 3, 5, 4, 4, 4, 3, 5, 3, 4, 3, 4, 4, 6, 3 ,4, 3, 6, 2)\ntrees3 = [(3, 1), (4, 1), (8, 1), (13, 1), (15, 1),\n (1, 2), (9, 2), (18, 2), (19, 2),\n (5, 3), (12, 3), (15, 3),\n (2, 4), (4, 4), (9, 4), (17, 4),\n (6, 5), (10, 5), (13, 5), (17, 5), (20, 5),\n (1, 6), (7, 6), (10, 6), (12, 6), (16, 6),\n (20, 7),\n (1, 8), (4, 8), (5, 8), (11, 8), (13, 8), (14, 8), (19, 8),\n (4, 9), (6, 9), (9, 9), (15, 9), (17, 9),\n (8, 10), (17, 10), (19, 10),\n (12, 11),\n (5, 12), (7, 12), (14, 12), (16, 12),\n (1, 13), (2, 13), (6, 13), (19, 13),\n (11, 14), (14, 14), (20, 14),\n (3, 15), (5, 15), (6, 15), (8, 15), (13, 15), (20, 15),\n (2, 16), (3, 16), (10, 16),\n (8, 17), (11, 17), (14, 17), (15, 17),\n (2, 18), (6, 18), (9, 18), (12, 18), (13, 18), (18, 18),\n (2, 19), (7, 19), (15, 19), (17, 19), (20, 19),\n (5, 20), (10, 20)]",
"_____no_output_____"
]
],
[
[
"## Output\n\nYou should find the coordinates $(x_i, y_i), i \\in \\{1,\\dots,k\\}$, of the individual tents.\n\n## Model",
"_____no_output_____"
]
],
[
[
"from gurobipy import *\nfrom itertools import product as cartesian",
"_____no_output_____"
],
[
"def optimize(n, r, c, trees):\n m = Model()\n # n+2 -> extend the board such as we don't need check borders\n # this is really nice hack, disable all variables with uper bound 0 and\n # then allow them only in tree neighborhood -> we don't need second matrix\n X = m.addVars(n+2, n+2, vtype=GRB.BINARY, ub=0)\n \n # set sums per rows, iterate only through valid board (not extended)\n for i, val in enumerate(r, start=1):\n m.addConstr(sum(X[j, i] for j in range(1, n+1)) == val)\n # sums per columns\n for j, val in enumerate(c, start=1):\n m.addConstr(sum(X[j, i] for i in range(1, n+1)) == val)\n \n # no other tents near one tent\n tents_neighboorhood = {(-1, -1), (-1, 0), (-1, 1), (0, -1), (0, 1), (1, -1), (1, 0), (1, 1)}\n # sum max of neighberhood + 1\n M = 9\n for i in range(1, n+1):\n for j in range(1, n+1):\n # if there's a tent, there can't be another one around it\n m.addConstr(M * (1 - X[i, j]) >= sum(X[i+ii, j+jj] for ii, jj in tents_neighboorhood))\n \n # as we extended board to n+2, we need to know indicies that are ouside of the board\n outer_frame = set(cartesian(range(n+2), range(n+2))) - set(cartesian(range(1,n+1), range(1,n+1)))\n # tents can be only in these incidies\n allowed_tent_indicies = {(1,0), (-1,0), (0,1), (0,-1)}\n for i, j in trees:\n allowed_neighberhood = [(i+ii, j+jj) for (ii,jj) in allowed_tent_indicies]\n # allow to place tent only if there's tree\n for idx in allowed_neighberhood:\n # if index is inside the board, allow to place tent\n if idx not in outer_frame: \n # thanks Prokop\n X[idx].ub = 1\n \n # there must be at least one tent in the neighberhood\n m.addConstr(sum(X[idx] for idx in allowed_neighberhood) >= 1)\n \n m.optimize()\n \n return [coords for coords, var in X.items() if var.x > 0]",
"_____no_output_____"
]
],
[
[
" ## Visualization",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\n\ndef visualize(n, trees, tents, r, c):\n grid = [[\".\" for _ in range(n+2)] for _ in range(n+2)]\n \n for t_x, t_y in tents:\n grid[t_y][t_x] = \"X\"\n \n for t_x, t_y in trees:\n grid[t_y][t_x] = \"T\"\n\n print(\" \", end=\"\")\n for c_cur in c:\n print(c_cur, end=\" \")\n print()\n \n for y in range(1, n+1):\n print(r[y-1], end=\" \")\n for x in range(1, n+1):\n print(grid[y][x], end=\" \")\n \n print()",
"_____no_output_____"
],
[
"tents1 = optimize(n1, r1, c1, trees1)\nvisualize(n1, trees1, tents1, r1, c1)",
"Gurobi Optimizer version 9.0.1 build v9.0.1rc0 (mac64)\nOptimize a model with 17 rows, 25 columns and 107 nonzeros\nModel fingerprint: 0xd2371660\nVariable types: 0 continuous, 25 integer (25 binary)\nCoefficient statistics:\n Matrix range [1e+00, 9e+00]\n Objective range [0e+00, 0e+00]\n Bounds range [1e+00, 1e+00]\n RHS range [1e+00, 9e+00]\nPresolve removed 17 rows and 25 columns\nPresolve time: 0.00s\nPresolve: All rows and columns removed\n\nExplored 0 nodes (0 simplex iterations) in 0.01 seconds\nThread count was 1 (of 12 available processors)\n\nSolution count 1: 0 \n\nOptimal solution found (tolerance 1.00e-04)\nBest objective 0.000000000000e+00, best bound 0.000000000000e+00, gap 0.0000%\n 1 0 1 \n1 T . X \n1 X . T \n0 . . . \n"
],
[
"tents2 = optimize(n2, r2, c2, trees2)\nvisualize(n2, trees2, tents2, r2, c2)",
"Gurobi Optimizer version 9.0.1 build v9.0.1rc0 (mac64)\nOptimize a model with 92 rows, 100 columns and 752 nonzeros\nModel fingerprint: 0x134b16b8\nVariable types: 0 continuous, 100 integer (100 binary)\nCoefficient statistics:\n Matrix range [1e+00, 9e+00]\n Objective range [0e+00, 0e+00]\n Bounds range [1e+00, 1e+00]\n RHS range [1e+00, 9e+00]\nFound heuristic solution: objective 0.0000000\n\nExplored 0 nodes (0 simplex iterations) in 0.00 seconds\nThread count was 1 (of 12 available processors)\n\nSolution count 1: 0 \n\nOptimal solution found (tolerance 1.00e-04)\nBest objective 0.000000000000e+00, best bound 0.000000000000e+00, gap 0.0000%\n 2 1 2 2 1 1 2 1 \n3 X T X . T T X . \n1 T . . . X . . . \n1 . X T . . . . . \n2 . . T X . T X . \n0 . . . T . T . . \n2 . . . X . X . . \n0 . . . . . . . T \n3 X T X T . . . X \n"
],
[
"tents3 = optimize(n3, r3, c3, trees3)\nvisualize(n3, trees3, tents3, r3, c3)",
"Gurobi Optimizer version 9.0.1 build v9.0.1rc0 (mac64)\nOptimize a model with 520 rows, 484 columns and 4720 nonzeros\nModel fingerprint: 0xcc059f6b\nVariable types: 0 continuous, 484 integer (484 binary)\nCoefficient statistics:\n Matrix range [1e+00, 9e+00]\n Objective range [0e+00, 0e+00]\n Bounds range [1e+00, 1e+00]\n RHS range [1e+00, 9e+00]\nPresolve removed 198 rows and 283 columns\nPresolve time: 0.01s\nPresolved: 322 rows, 201 columns, 1586 nonzeros\nVariable types: 0 continuous, 201 integer (201 binary)\n\nRoot relaxation: objective 0.000000e+00, 600 iterations, 0.03 seconds\n\n Nodes | Current Node | Objective Bounds | Work\n Expl Unexpl | Obj Depth IntInf | Incumbent BestBd Gap | It/Node Time\n\n 0 0 0.00000 0 97 - 0.00000 - - 0s\nH 0 0 0.0000000 0.00000 0.00% - 0s\n 0 0 - 0 0.00000 0.00000 0.00% - 0s\n\nCutting planes:\n Gomory: 27\n Cover: 35\n Clique: 3\n MIR: 8\n StrongCG: 2\n Zero half: 16\n RLT: 19\n\nExplored 1 nodes (1257 simplex iterations) in 0.12 seconds\nThread count was 12 (of 12 available processors)\n\nSolution count 1: 0 \n\nOptimal solution found (tolerance 1.00e-04)\nBest objective 0.000000000000e+00, best bound 0.000000000000e+00, gap 0.0000%\n 6 4 3 5 4 4 4 3 5 3 4 3 4 4 6 3 4 3 6 2 \n7 . X T T X . X T X . . . T X T X . X . . \n2 T . . . . . . . T . . X . . . . . T T X \n3 X . . X T . . . . . . T . X T . . . . . \n4 . T . T . X . X T X . . . . . X T . . . \n3 . X . X . T . . . T . . T . . . T . X T \n5 T . . . . . T . X T X T X . X T X . . . \n4 X . . . X . X . . . . . . . . . . . X T \n4 T . X T T . . . X . T . T T X . X . T . \n4 X . . T . T . . T . X . X . T . T . X . \n4 . . . X . X . T X . . . . . X . T . T . \n3 . . . . . . . . . . X T . . . . X . X . \n6 . X . X T X T X . . . . X T X T . . . . \n3 T T . . . T . . . . X . . . . . . X T X \n6 X . X . . X . X . . T . X T X . . . . T \n2 . . T . T T . T . X . . T . . . . . X T \n3 X T T . X . . . . T . . . X . . . . . . \n6 . . X . . . X T X . T X . T T X . X . . \n3 X T . . X T . . T . . T T X . . . T . . \n3 . T . . . . T . . X . X . . T . T . X T \n5 . X . X T . X . . T . . . . X . X . . . \n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
]
|
e7995130964f7b50c702a8adb7ca8fc378fc450f | 127,073 | ipynb | Jupyter Notebook | Regression/Gradient Boosting Machine/GradientBoostingRegressor_Normalize.ipynb | devVipin01/ds-seed | 36b8559d49208003601d44176a44063be4e2fef6 | [
"Apache-2.0"
]
| 2 | 2021-07-28T15:26:40.000Z | 2021-07-29T04:14:35.000Z | Regression/Gradient Boosting Machine/GradientBoostingRegressor_Normalize.ipynb | devVipin01/ds-seed | 36b8559d49208003601d44176a44063be4e2fef6 | [
"Apache-2.0"
]
| 1 | 2021-07-30T06:00:30.000Z | 2021-07-30T06:00:30.000Z | Regression/Gradient Boosting Machine/GradientBoostingRegressor_Normalize.ipynb | devVipin01/ds-seed | 36b8559d49208003601d44176a44063be4e2fef6 | [
"Apache-2.0"
]
| null | null | null | 162.289911 | 74,979 | 0.86551 | [
[
[
"# GradientBoostingRegressor with Normalize\r\n\r\n### Required Packages",
"_____no_output_____"
]
],
[
[
"import warnings\r\nimport numpy as np \r\nimport pandas as pd \r\nimport matplotlib.pyplot as plt \r\nimport seaborn as se \r\nfrom sklearn.preprocessing import Normalizer\r\nfrom sklearn.model_selection import train_test_split\r\nfrom sklearn.ensemble import GradientBoostingRegressor \r\nfrom sklearn.metrics import r2_score, mean_absolute_error, mean_squared_error \r\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
]
],
[
[
"### Initialization\n\nFilepath of CSV file",
"_____no_output_____"
]
],
[
[
"#filepath\r\nfile_path = \"\"",
"_____no_output_____"
]
],
[
[
"List of features which are required for model training .",
"_____no_output_____"
]
],
[
[
"#x_values\r\nfeatures=[]",
"_____no_output_____"
]
],
[
[
"Target feature for prediction.",
"_____no_output_____"
]
],
[
[
"#y_value\r\ntarget = ''",
"_____no_output_____"
]
],
[
[
"### Data Fetching\n\nPandas is an open-source, BSD-licensed library providing high-performance, easy-to-use data manipulation and data analysis tools.\n\nWe will use panda's library to read the CSV file using its storage path.And we use the head function to display the initial row or entry.",
"_____no_output_____"
]
],
[
[
"df=pd.read_csv(file_path)\ndf.head()",
"_____no_output_____"
]
],
[
[
"### Feature Selections\n\nIt is the process of reducing the number of input variables when developing a predictive model. Used to reduce the number of input variables to both reduce the computational cost of modelling and, in some cases, to improve the performance of the model.\n\nWe will assign all the required input features to X and target/outcome to Y.",
"_____no_output_____"
]
],
[
[
"X = df[features]\nY = df[target]",
"_____no_output_____"
]
],
[
[
"### Data Preprocessing\n\nSince the majority of the machine learning models in the Sklearn library doesn't handle string category data and Null value, we have to explicitly remove or replace null values. The below snippet have functions, which removes the null value if any exists. And convert the string classes data in the datasets by encoding them to integer classes.\n",
"_____no_output_____"
]
],
[
[
"def NullClearner(df):\n if(isinstance(df, pd.Series) and (df.dtype in [\"float64\",\"int64\"])):\n df.fillna(df.mean(),inplace=True)\n return df\n elif(isinstance(df, pd.Series)):\n df.fillna(df.mode()[0],inplace=True)\n return df\n else:return df\ndef EncodeX(df):\n return pd.get_dummies(df)",
"_____no_output_____"
]
],
[
[
"Calling preprocessing functions on the feature and target set.",
"_____no_output_____"
]
],
[
[
"x=X.columns.to_list()\nfor i in x:\n X[i]=NullClearner(X[i])\nX=EncodeX(X)\nY=NullClearner(Y)\nX.head()",
"_____no_output_____"
]
],
[
[
"#### Correlation Map\n\nIn order to check the correlation between the features, we will plot a correlation matrix. It is effective in summarizing a large amount of data where the goal is to see patterns.",
"_____no_output_____"
]
],
[
[
"f,ax = plt.subplots(figsize=(18, 18))\nmatrix = np.triu(X.corr())\nse.heatmap(X.corr(), annot=True, linewidths=.5, fmt= '.1f',ax=ax, mask=matrix)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Data Splitting\n\nThe train-test split is a procedure for evaluating the performance of an algorithm. The procedure involves taking a dataset and dividing it into two subsets. The first subset is utilized to fit/train the model. The second subset is used for prediction. The main motive is to estimate the performance of the model on new data.",
"_____no_output_____"
]
],
[
[
"X_train, X_test, y_train, y_test = train_test_split(X, Y, test_size = 0.2, random_state = 123)#performing datasplitting",
"_____no_output_____"
]
],
[
[
"## Data Rescaling\nNormalizer normalizes samples (rows) individually to unit norm.\n\nEach sample with at least one non zero component is rescaled independently of other samples so that its norm (l1, l2 or inf) equals one.\n\nWe will fit an object of Normalizer to train data then transform the same data via fit_transform(X_train) method, following which we will transform test data via transform(X_test) method.",
"_____no_output_____"
]
],
[
[
"normalizer = Normalizer()\nX_train = normalizer.fit_transform(X_train)\nX_test = normalizer.transform(X_test)",
"_____no_output_____"
]
],
[
[
"### Model\n\nGradient Boosting builds an additive model in a forward stage-wise fashion; it allows for the optimization of arbitrary differentiable loss functions. In each stage a regression tree is fit on the negative gradient of the given loss function.\n\n#### Model Tuning Parameters\n\n 1. loss : {‘ls’, ‘lad’, ‘huber’, ‘quantile’}, default=’ls’\n> Loss function to be optimized. ‘ls’ refers to least squares regression. ‘lad’ (least absolute deviation) is a highly robust loss function solely based on order information of the input variables. ‘huber’ is a combination of the two. ‘quantile’ allows quantile regression (use `alpha` to specify the quantile).\n\n 2. learning_ratefloat, default=0.1\n> Learning rate shrinks the contribution of each tree by learning_rate. There is a trade-off between learning_rate and n_estimators.\n\n 3. n_estimators : int, default=100\n> The number of trees in the forest.\n\n 4. criterion : {‘friedman_mse’, ‘mse’, ‘mae’}, default=’friedman_mse’\n> The function to measure the quality of a split. Supported criteria are ‘friedman_mse’ for the mean squared error with improvement score by Friedman, ‘mse’ for mean squared error, and ‘mae’ for the mean absolute error. The default value of ‘friedman_mse’ is generally the best as it can provide a better approximation in some cases.\n\n 5. max_depth : int, default=3\n> The maximum depth of the individual regression estimators. The maximum depth limits the number of nodes in the tree. Tune this parameter for best performance; the best value depends on the interaction of the input variables.\n\n 6. max_features : {‘auto’, ‘sqrt’, ‘log2’}, int or float, default=None\n> The number of features to consider when looking for the best split: \n\n 7. random_state : int, RandomState instance or None, default=None\n> Controls both the randomness of the bootstrapping of the samples used when building trees (if <code>bootstrap=True</code>) and the sampling of the features to consider when looking for the best split at each node (if `max_features < n_features`).\n\n 8. verbose : int, default=0\n> Controls the verbosity when fitting and predicting.\n \n 9. n_iter_no_change : int, default=None\n> <code>n_iter_no_change</code> is used to decide if early stopping will be used to terminate training when validation score is not improving. By default it is set to None to disable early stopping. If set to a number, it will set aside <code>validation_fraction</code> size of the training data as validation and terminate training when validation score is not improving in all of the previous <code>n_iter_no_change</code> numbers of iterations. The split is stratified.\n \n 10. tol : float, default=1e-4\n> Tolerance for the early stopping. When the loss is not improving by at least tol for <code>n_iter_no_change</code> iterations (if set to a number), the training stops.",
"_____no_output_____"
]
],
[
[
"# Build Model here\nmodel = GradientBoostingRegressor(random_state = 123)\nmodel.fit(X_train, y_train)",
"_____no_output_____"
]
],
[
[
"#### Model Accuracy\n\nWe will use the trained model to make a prediction on the test set.Then use the predicted value for measuring the accuracy of our model.\n\n> **score**: The **score** function returns the coefficient of determination <code>R<sup>2</sup></code> of the prediction.",
"_____no_output_____"
]
],
[
[
"print(\"Accuracy score {:.2f} %\\n\".format(model.score(X_test,y_test)*100))",
"Accuracy score 93.80 %\n\n"
]
],
[
[
"> **r2_score**: The **r2_score** function computes the percentage variablility explained by our model, either the fraction or the count of correct predictions. \n\n> **mae**: The **mean abosolute error** function calculates the amount of total error(absolute average distance between the real data and the predicted data) by our model. \n\n> **mse**: The **mean squared error** function squares the error(penalizes the model for large errors) by our model. ",
"_____no_output_____"
]
],
[
[
"y_pred=model.predict(X_test)\nprint(\"R2 Score: {:.2f} %\".format(r2_score(y_test,y_pred)*100))\nprint(\"Mean Absolute Error {:.2f}\".format(mean_absolute_error(y_test,y_pred)))\nprint(\"Mean Squared Error {:.2f}\".format(mean_squared_error(y_test,y_pred)))",
"R2 Score: 93.80 %\nMean Absolute Error 3.24\nMean Squared Error 17.94\n"
]
],
[
[
"#### Feature Importances\nThe Feature importance refers to techniques that assign a score to features based on how useful they are for making the prediction.",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(8,6))\nn_features = len(X.columns)\nplt.barh(range(n_features), model.feature_importances_, align='center')\nplt.yticks(np.arange(n_features), X.columns)\nplt.xlabel(\"Feature importance\")\nplt.ylabel(\"Feature\")\nplt.ylim(-1, n_features)",
"_____no_output_____"
]
],
[
[
"#### Prediction Plot\n\nFirst, we make use of a plot to plot the actual observations, with x_train on the x-axis and y_train on the y-axis.\nFor the regression line, we will use x_train on the x-axis and then the predictions of the x_train observations on the y-axis.",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(14,10))\nplt.plot(range(20),y_test[0:20], color = \"blue\")\nplt.plot(range(20),model.predict(X_test[0:20]), color = \"red\")\nplt.legend([\"Actual\",\"prediction\"]) \nplt.title(\"Predicted vs True Value\")\nplt.xlabel(\"Record number\")\nplt.ylabel(target)\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### Creator: Ganapathi Thota , Github: [Profile](https://github.com/Shikiz)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
e79952c659666f205c8d2081e046551a9971db40 | 362,615 | ipynb | Jupyter Notebook | notebooks/Gene_Expression.ipynb | alliance-genome/ontobio | 0ec3aa6fea9d4492a9873a4b9b394c4866f741b6 | [
"BSD-3-Clause"
]
| 101 | 2017-04-19T20:54:49.000Z | 2022-03-14T02:32:11.000Z | notebooks/Gene_Expression.ipynb | valearna/ontobio | 460915df0beb0d4fbcd414cf4157769b08954857 | [
"BSD-3-Clause"
]
| 402 | 2017-04-24T19:53:12.000Z | 2022-03-31T20:27:59.000Z | notebooks/Gene_Expression.ipynb | valearna/ontobio | 460915df0beb0d4fbcd414cf4157769b08954857 | [
"BSD-3-Clause"
]
| 30 | 2017-04-20T17:59:12.000Z | 2022-02-25T22:26:08.000Z | 72.392693 | 4,111 | 0.582866 | [
[
[
"# Gene Expression Simple Demo\n\nThis shows how to query BgeeDb gene expression data ingested in Monarch",
"_____no_output_____"
]
],
[
[
"## Create an ontology factory in order to fetch Uberon\nfrom ontobio.ontol_factory import OntologyFactory\n\nofactory = OntologyFactory()\nont = ofactory.create(\"uberon\") ",
"_____no_output_____"
],
[
"## Create a sub-ontology that excludes all relations other than is-a and part-of\nsubont = ont.subontology(relations=['subClassOf', 'BFO:0000050'])",
"_____no_output_____"
],
[
"## Create an association factory to get mouse gene-expression associations (sourced from bgeedb)\nfrom ontobio.assoc_factory import AssociationSetFactory\nafactory = AssociationSetFactory()\naset = afactory.create(ontology=subont, subject_category='gene', object_category='anatomy', taxon='NCBITaxon:10090')\n",
"_____no_output_____"
],
[
"# show first 5\n[\"{} '{}'\".format(g, aset.label(g)) for g in aset.subjects[:5]]",
"_____no_output_____"
],
[
"# fetch uberon term\n[liver] = ont.search('liver')\nliver",
"_____no_output_____"
],
[
"liver_genes = aset.query([liver])\n[\"{} '{}'\".format(g, aset.label(g)) for g in liver_genes]",
"_____no_output_____"
],
[
"## NOTE: we currently lack rank scores, see https://github.com/monarch-initiative/monarch-app/issues/1271\n## For now let's do something naive\n\ndef specificity_score(g, t):\n \"\"\"\n Naive specificity score - penalize for every expression *not* in desired term, e.g. liver\n \"\"\"\n anns = aset.annotations(g)\n nonspecific = [a for a in anns if t!=a and t not in subont.ancestors(a) and a not in subont.ancestors(g)]\n return 1/(len(nonspecific)+1)\n\n## Tuples of (gene_id, gene_symbol, score)\ngscores = [(g,aset.label(g),specificity_score(g,liver)) for g in liver_genes]\ngscores\n ",
"_____no_output_____"
],
[
"sorted(gscores, key=lambda x: -x[2])",
"_____no_output_____"
],
[
"only_in_liver = [x for x in gscores if x[2] == 1.0]\nonly_in_liver",
"_____no_output_____"
],
[
"## get phenotype associations\nmp = ofactory.create(\"mp\")\n\npheno_aset = afactory.create(ontology=mp, subject_category='gene', object_category='phenotype', taxon='NCBITaxon:10090')\n",
"_____no_output_____"
],
[
"## Show phenotype anns for all genes in liver\n\nfor g in liver_genes:\n anns = pheno_aset.annotations(g)\n print(\"{} {} {}\".format(g,aset.label(g), [(a,mp.label(a)) for a in anns]))\n ",
"MGI:95590 Ftl2-ps []\nMGI:1098641 Wasf2 [('MP:0002188', 'small heart'), ('MP:0020329', 'decreased capillary density'), ('MP:0011091', 'prenatal lethality, complete penetrance'), ('HP:0002170', None), ('HP:0000969', None), ('MP:0003984', 'embryonic growth retardation'), ('MP:0000260', 'abnormal angiogenesis'), ('MP:0011098', 'embryonic lethality during organogenesis, complete penetrance'), ('MP:0001722', 'pale yolk sac'), ('MP:0000295', 'trabecula carnea hypoplasia'), ('MP:0008803', 'abnormal placental labyrinth vasculature morphology'), ('GO:0001667PHENOTYPE', None), ('HP:0025016', None), ('MP:0000822', 'abnormal brain ventricle morphology'), ('GO:0006928PHENOTYPE', None), ('MP:0003974', 'abnormal endocardium morphology'), ('GO:0001525PHENOTYPE', None), ('MP:0004251', 'failure of heart looping')]\nMGI:3644452 Gm9083 []\nMGI:3651858 Gm11337 []\nMGI:3702318 Gm11989 []\nMGI:1916396 Gsdmd [('MP:0011073', 'abnormal macrophage apoptosis')]\nMGI:2443767 Aaas [('MP:0005379', 'endocrine/exocrine gland phenotype'), ('MP:0003631', 'nervous system phenotype'), ('MP:0005376', 'homeostasis/metabolism phenotype'), ('MP:0001417', 'decreased exploration in new environment'), ('MP:0005381', 'digestive/alimentary phenotype'), ('GO:0007612PHENOTYPE', None)]\nMGI:2137698 Ugt1a6a []\nMGI:1351659 Abcg5 [('HP:0001882', None), ('HP:0003010', None), ('HP:0003251', None), ('HP:0001638', None), ('HP:0004446', None), ('HP:0011875', None), ('MP:0002413', 'abnormal megakaryocyte progenitor cell morphology'), ('HP:0003540', None), ('MP:0006298', 'abnormal platelet activation'), ('HP:0011877', None), ('HP:0011273', None), ('HP:0001878', None), ('HP:0002155', None), ('HP:0008669', None), ('HP:0005513', None), ('HP:0001939', None), ('MP:0001265', 'decreased body size'), ('HP:0002240', None), ('HP:0003146', None)]\nMGI:3705426 Rpl21-ps15 []\nMGI:1914745 Tmem167b []\nMGI:1347249 Psg16 []\nMGI:3818630 Sco2 [('GO:0022904PHENOTYPE', None), ('HP:0001324', None), ('HP:0025321', None), ('MP:0010956', 'abnormal mitochondrial ATP synthesis coupled electron transport'), ('HP:0010836', None), ('GO:0003012PHENOTYPE', None), ('MP:0001392', 'abnormal locomotor behavior'), ('GO:0001701PHENOTYPE', None), ('MP:0011095', 'embryonic lethality between implantation and placentation, complete penetrance')]\nMGI:1919235 Acad10 []\nMGI:3780170 Gm2000 []\nMGI:1343095 Emc8 [('MP:0010024', 'increased total body fat amount')]\nMGI:5454530 Gm24753 []\nMGI:3649027 Gm7117 []\nMGI:5454475 Gm24698 []\nMGI:4415000 Gm16580 []\nMGI:3646089 Gm7803 []\nMGI:97987 Rnu3b3 []\nMGI:1353433 Timm8a1 []\nMGI:3649865 Rpl23a-ps14 []\nMGI:106686 Pon3 [('MP:0011086', 'postnatal lethality, incomplete penetrance'), ('MP:0004921', 'decreased placenta weight'), ('MP:0011109', 'lethality throughout fetal growth and development, incomplete penetrance'), ('MP:0003674', 'oxidative stress')]\nMGI:88216 Btk [('MP:0001844', 'autoimmune response'), ('HP:0004325', None), ('HP:0006270', None), ('GO:0007249PHENOTYPE', None), ('MP:0005093', 'decreased B cell proliferation'), ('MP:0008186', 'increased pro-B cell number'), ('MP:0008203', 'absent B-1a cells'), ('MP:0002401', 'abnormal lymphopoiesis'), ('HP:0011990', None), ('HP:0003496', None), ('HP:0002850', None), ('HP:0004313', None), ('MP:0002491', 'decreased IgD level'), ('MP:0002451', 'abnormal macrophage physiology'), ('HP:0010978', None), ('MP:0005153', 'abnormal B cell proliferation'), ('MP:0009339', 'decreased splenocyte number'), ('MP:0009788', 'increased susceptibility to bacterial infection induced morbidity/mortality'), ('HP:0010976', None), ('GO:0030889PHENOTYPE', None), ('MP:0005387', 'immune system phenotype'), ('HP:0001881', None), ('HP:0003212', None), ('MP:0008211', 'decreased mature B cell number')]\nMGI:3704359 Gm9803 []\nMGI:3780980 Gm2810 []\nMGI:2140962 Ugt2b34 []\nMGI:2444981 Phldb2 []\nMGI:3644778 Gm8738 []\nMGI:88054 Apoc2 [('MP:0003975', 'increased circulating VLDL triglyceride level'), ('MP:0000180', 'abnormal circulating cholesterol level'), ('HP:0003233', None)]\nMGI:3649201 Gm12396 []\nMGI:5451834 Gm22057 []\nMGI:1861354 Apbb1ip []\nMGI:5804868 Gm45753 []\nMGI:891967 Serpina1e []\nMGI:2138853 AI182371 []\nMGI:3041196 Fam198a []\nMGI:1913761 Chtop [('HP:0010442', None)]\nMGI:3783208 Gm15766 []\nMGI:2385276 Kctd15 [('MP:0011940', 'decreased food intake'), ('MP:0013294', 'prenatal lethality prior to heart atrial septation'), ('HP:0010683', None)]\nMGI:1927868 Pex14 [('MP:0011091', 'prenatal lethality, complete penetrance')]\nMGI:1888526 Xpo4 []\nMGI:5455017 Gm25240 []\nMGI:1913534 Gkn2 [('HP:0001640', None)]\nMGI:1930008 Ghrl [('MP:0005379', 'endocrine/exocrine gland phenotype'), ('MP:0005452', 'abnormal adipose tissue amount'), ('MP:0005381', 'digestive/alimentary phenotype'), ('MP:0005560', 'decreased circulating glucose level'), ('MP:0002169', 'no abnormal phenotype detected'), ('MP:0005292', 'improved glucose tolerance')]\nMGI:2385008 Ggact []\nMGI:1921821 Kcnk16 []\nMGI:96611 Itgb2 [('HP:0005404', None), ('MP:0001194', 'dermatitis'), ('HP:0003330', None), ('MP:0003156', 'abnormal leukocyte migration'), ('HP:0000938', None), ('MP:0001874', 'acanthosis'), ('HP:0000962', None), ('MP:0000702', 'enlarged lymph nodes'), ('GO:0045123PHENOTYPE', None), ('HP:0000509', None), ('HP:0200042', None), ('MP:0008623', 'increased circulating interleukin-3 level'), ('HP:0002732', None), ('HP:0011990', None), ('MP:0001222', 'epidermal hyperplasia'), ('HP:0003496', None), ('MP:0005094', 'abnormal T cell proliferation'), ('MP:0001246', 'mixed cellular infiltration to dermis'), ('MP:0002356', 'abnormal spleen red pulp morphology'), ('HP:0010987', None), ('MP:0008566', 'increased interferon-gamma secretion'), ('HP:0001744', None), ('MP:0005559', 'increased circulating glucose level'), ('HP:0001974', None), ('MP:0008126', 'increased dendritic cell number'), ('MP:0003434', 'decreased susceptibility to induced choroidal neovascularization'), ('MP:0000321', 'increased bone marrow cell number'), ('MP:0002606', 'increased basophil cell number'), ('MP:0000219', 'increased neutrophil cell number'), ('MP:0001191', 'abnormal skin condition'), ('HP:0012311', None), ('HP:0011840', None), ('MP:0002169', 'no abnormal phenotype detected'), ('HP:0000939', None), ('MP:0020137', 'decreased bone mineralization'), ('MP:0008567', 'decreased interferon-gamma secretion'), ('HP:0003765', None), ('MP:0002411', 'decreased susceptibility to bacterial infection'), ('HP:0001036', None), ('MP:0003813', 'abnormal hair follicle dermal papilla morphology'), ('HP:0001880', None), ('MP:0001186', 'pigmentation phenotype'), ('HP:0100828', None)]\nMGI:894286 P4ha2 []\nMGI:5455293 Gm25516 []\nMGI:1922466 Cep128 []\nMGI:3645079 Gm16470 []\nMGI:3645628 Gm8019 []\nMGI:3647773 Gm6498 []\nMGI:3819557 Snord83b []\nMGI:1914709 Nvl []\nMGI:4937849 Gm17022 []\nMGI:3583955 Rdh16f2 []\nMGI:2138968 Clp1 [('MP:0002083', 'premature death'), ('MP:0003631', 'nervous system phenotype'), ('HP:0010831', None), ('MP:0008412', 'increased cellular sensitivity to oxidative stress'), ('HP:0003307', None), ('HP:0003202', None), ('MP:0002175', 'decreased brain weight'), ('MP:0003203', 'increased neuron apoptosis'), ('HP:0003323', None), ('HP:0001322', None), ('HP:0001251', None), ('MP:0012055', 'abnormal phrenic nerve innervation pattern to diaphragm'), ('MP:0005498', 'hyporesponsive to tactile stimuli'), ('MP:0000819', 'abnormal olfactory bulb morphology'), ('HP:0002398', None), ('MP:0003718', 'maternal effect'), ('HP:0011017', None), ('MP:0011400', 'lethality, complete penetrance'), ('MP:0001053', 'abnormal neuromuscular synapse morphology'), ('MP:0011087', 'neonatal lethality, complete penetrance')]\nMGI:5453898 Gm24121 []\nMGI:1915442 Leprotl1 [('MP:0002953', 'thick ventricular wall'), ('HP:0010679', None)]\nMGI:3779824 Gm8941 []\nMGI:3650622 Gm12338 []\nMGI:2444508 Fitm2 [('MP:0002981', 'increased liver weight'), ('HP:0004324', None), ('MP:0005376', 'homeostasis/metabolism phenotype'), ('HP:0000842', None), ('MP:0005386', 'behavior/neurological phenotype'), ('HP:0011014', None), ('MP:0013294', 'prenatal lethality prior to heart atrial septation'), ('HP:0007703', None), ('MP:0009116', 'abnormal brown fat cell morphology'), ('MP:0009124', 'increased brown fat cell lipid droplet size'), ('HP:0000855', None), ('MP:0011100', 'preweaning lethality, complete penetrance'), ('MP:0005378', 'growth/size/body region phenotype')]\nMGI:5455452 Gm25675 []\nMGI:5454373 Gm24596 []\nMGI:1096324 Lst1 [('HP:0000035', None), ('HP:0011001', None), ('HP:0001640', None), ('MP:0009791', 'increased susceptibility to viral infection induced morbidity/mortality'), ('HP:0008734', None), ('MP:0010123', 'increased bone mineral content')]\nMGI:2159614 Mia2 [('HP:0040006', None), ('GO:0070328PHENOTYPE', None), ('HP:0003233', None), ('MP:0005389', 'reproductive system phenotype')]\nMGI:1918956 Slc46a3 [('MP:0011275', 'abnormal behavioral response to light')]\nMGI:3649696 Rps8-ps2 []\nMGI:3644876 Rps2-ps6 []\nMGI:5453824 Gm24047 []\nMGI:3649769 Gm12355 []\nMGI:2445040 Tyw3 []\nMGI:5452129 Gm22352 []\nMGI:1196423 Onecut1 [('HP:0001508', None), ('MP:0004201', 'fetal growth retardation'), ('HP:0100732', None), ('MP:0005559', 'increased circulating glucose level'), ('MP:0009181', 'decreased pancreatic delta cell number'), ('MP:0005379', 'endocrine/exocrine gland phenotype'), ('MP:0013221', 'pancreatic acinar-to-ductal metaplasia'), ('MP:0009164', 'exocrine pancreas atrophy'), ('HP:0006274', None), ('MP:0012242', 'abnormal hepatoblast differentiation'), ('GO:0048536PHENOTYPE', None), ('HP:0012090', None), ('HP:0005213', None), ('MP:0009254', 'disorganized pancreatic islets'), ('MP:0011932', 'abnormal endocrine pancreas development'), ('HP:0040216', None), ('MP:0009143', 'abnormal pancreatic duct morphology'), ('MP:0009178', 'absent pancreatic alpha cells'), ('GO:0031016PHENOTYPE', None), ('MP:0009145', 'abnormal pancreatic acinus morphology')]\nMGI:97620 Plg [('GO:0042246PHENOTYPE', None), ('MP:0002083', 'premature death'), ('HP:0002088', None), ('HP:0001395', None), ('MP:0003305', 'proctitis'), ('HP:0004325', None), ('HP:0002577', None), ('HP:0000105', None), ('HP:0002035', None), ('MP:0005602', 'decreased angiogenesis'), ('MP:0012331', 'increased circulating fibrinogen level'), ('MP:0010211', 'abnormal acute phase protein level'), ('MP:0000702', 'enlarged lymph nodes'), ('HP:0000509', None), ('MP:0011507', 'abnormal kidney thrombosis'), ('MP:0009507', 'abnormal mammary gland connective tissue morphology'), ('HP:0000502', None), ('MP:0006270', 'abnormal mammary gland growth during lactation'), ('HP:0011885', None), ('MP:0006137', 'venoocclusion'), ('HP:0002592', None), ('HP:0012647', None), ('MP:0009764', 'decreased sensitivity to induced morbidity/mortality'), ('MP:0001923', 'reduced female fertility'), ('HP:0002588', None), ('MP:0001139', 'abnormal vagina morphology'), ('HP:0001392', None), ('MP:0002249', 'abnormal larynx morphology'), ('MP:0010249', 'lactation failure'), ('MP:0001792', 'impaired wound healing'), ('MP:0005048', 'abnormal thrombosis'), ('MP:0009763', 'increased sensitivity to induced morbidity/mortality'), ('MP:0002282', 'abnormal trachea morphology'), ('MP:0001851', 'eye inflammation'), ('HP:0001824', None), ('MP:0005300', 'abnormal corneal stroma morphology'), ('MP:0000495', 'abnormal colon morphology'), ('MP:0008236', 'decreased susceptibility to neuronal excitotoxicity')]\nMGI:3643679 Gm8682 []\nMGI:95856 Gsta3 [('MP:0009766', 'increased sensitivity to xenobiotic induced morbidity/mortality'), ('MP:0008873', 'increased physiological sensitivity to xenobiotic')]\nMGI:5455590 Gm25813 []\nMGI:3651301 Gm14107 []\nMGI:105103 Rprl3 []\nMGI:5454298 Gm24521 []\nMGI:1915951 Ppp1r27 []\nMGI:106636 Atp5k []\nMGI:3704327 Gm10182 []\nMGI:3651595 Gm11295 []\nMGI:3652326 Gm13902 []\nMGI:3718464 Mir291b []\nMGI:1918982 Vps11 []\nMGI:5454076 Gm24299 []\nMGI:1350917 Rps3 []\nMGI:1913638 Cutc []\nMGI:1914195 Sdha [('HP:0003228', None)]\nMGI:1915254 Tmem9b []\nMGI:3779470 Ces1b []\nMGI:5454859 Gm25082 []\nMGI:1926264 Tspan6 []\nMGI:3651503 Gm13862 []\nMGI:95862 Gstm4 [('MP:0001415', 'increased exploration in new environment'), ('HP:0001743', None)]\nMGI:2444947 Mical2 []\nMGI:5504138 Gm27023 []\nMGI:5453406 Gm23629 []\nMGI:3651534 Gm12258 []\nMGI:3705806 Gm14536 []\nMGI:1921611 4931429L15Rik []\nMGI:3642824 Rpl9-ps7 []\nMGI:1278340 Rpl21 []\nMGI:4834232 Mir3060 []\nMGI:107508 Ereg [('MP:0001194', 'dermatitis'), ('HP:0001824', None), ('MP:0008873', 'increased physiological sensitivity to xenobiotic')]\nMGI:88343 Cd69 [('MP:0008075', 'decreased CD4-positive, alpha beta T cell number'), ('MP:0008719', 'impaired neutrophil recruitment'), ('HP:0003496', None), ('MP:0005387', 'immune system phenotype'), ('HP:0040238', None), ('MP:0008051', 'abnormal memory T cell physiology'), ('MP:0005463', 'abnormal CD4-positive, alpha-beta T cell physiology'), ('MP:0008561', 'decreased tumor necrosis factor secretion')]\nMGI:3650581 Gm12419 []\nMGI:3610314 Scimp [('HP:0001903', None), ('HP:0003124', None)]\nMGI:1098779 Cdk2ap2 []\nMGI:2144766 Slc25a47 []\nMGI:2138935 Fam102a []\nMGI:5434102 Ftl1-ps2 []\nMGI:1925560 1810024B03Rik []\nMGI:3704487 Amd-ps3 []\nMGI:101921 Ap2a1 []\nMGI:2685672 Gm826 []\nMGI:3648378 Sult2a5 []\nMGI:3648883 Rpl13a-ps1 []\nMGI:1354945 Plpp2 [('MP:0002169', 'no abnormal phenotype detected')]\nMGI:2146020 Mief1 []\nMGI:1921808 Gvin1 []\nMGI:2685015 Mtg1 []\nMGI:106499 Ppih []\nMGI:4421912 n-R5s67 []\nMGI:3801856 Gm15956 []\nMGI:1336892 Slc6a18 [('MP:0011417', 'abnormal renal transport'), ('HP:0000822', None)]\nMGI:1914411 Sclt1 []\nMGI:1915871 Mthfd2l []\nMGI:1919405 Cenpn []\nMGI:3612471 C330021F23Rik []\nMGI:5568573 Rubie [('MP:0011238', 'abnormal inner ear development'), ('HP:0040106', None), ('HP:0000752', None)]\nMGI:3648788 Gm5239 []\nMGI:1352495 Zfp385a [('GO:0007611PHENOTYPE', None), ('GO:0007626PHENOTYPE', None), ('MP:0002417', 'abnormal megakaryocyte morphology'), ('MP:0001265', 'decreased body size'), ('MP:0002551', 'abnormal blood coagulation'), ('HP:0002170', None), ('MP:0002169', 'no abnormal phenotype detected')]\nMGI:1914535 Cwc27 [('MP:0011110', 'preweaning lethality, incomplete penetrance'), ('HP:0003251', None), ('HP:0008222', None)]\nMGI:3644274 Gm5687 []\nMGI:99501 Fgb [('MP:0002059', 'abnormal seminal vesicle morphology')]\nMGI:2140940 Acacb [('HP:0000708', None), ('HP:0002591', None), ('MP:0002188', 'small heart'), ('HP:0003292', None), ('MP:0005376', 'homeostasis/metabolism phenotype'), ('MP:0005282', 'decreased fatty acid level'), ('HP:0000842', None), ('MP:0002575', 'increased circulating ketone body level'), ('HP:0012338', None), ('MP:0005439', 'decreased glycogen level'), ('HP:0011014', None), ('MP:0005289', 'increased oxygen consumption'), ('MP:0010379', 'decreased respiratory quotient'), ('MP:0002118', 'abnormal lipid homeostasis'), ('MP:0002169', 'no abnormal phenotype detected')]\nMGI:5455514 Gm25737 []\nMGI:1922948 Fam35a []\nMGI:2443301 Slc35c1 [('MP:0011086', 'postnatal lethality, incomplete penetrance'), ('MP:0020332', 'impaired leukocyte tethering or rolling'), ('MP:0011090', 'perinatal lethality, incomplete penetrance'), ('MP:0002345', 'abnormal lymph node primary follicle morphology'), ('HP:0004325', None), ('MP:0011084', 'lethality at weaning, incomplete penetrance'), ('MP:0001922', 'reduced male fertility'), ('MP:0003628', 'abnormal leukocyte adhesion'), ('MP:0000219', 'increased neutrophil cell number'), ('HP:0001939', None), ('HP:0012311', None), ('MP:0001183', 'overexpanded pulmonary alveoli'), ('MP:0000420', 'ruffled hair'), ('GO:0045746PHENOTYPE', None)]\nMGI:1927086 Ube4b [('MP:0000278', 'abnormal myocardial fiber morphology'), ('HP:0001698', None), ('MP:0001787', 'pericardial edema'), ('GO:0044257PHENOTYPE', None), ('MP:0000876', 'Purkinje cell degeneration'), ('MP:0003225', 'axonal dystrophy'), ('HP:0002624', None), ('MP:0000877', 'abnormal Purkinje cell morphology'), ('MP:0001405', 'impaired coordination'), ('HP:0001640', None), ('GO:0006511PHENOTYPE', None), ('HP:0011017', None), ('MP:0003222', 'increased cardiomyocyte apoptosis'), ('HP:0030746', None)]\nMGI:3819569 Snord98 []\nMGI:3651577 Gm13666 []\nMGI:98223 Saa3 []\nMGI:1270860 Plscr2 [('HP:0005815', None)]\nMGI:1921138 Ppp1r42 [('HP:0002948', None)]\nMGI:107180 Elf1 []\nMGI:3801877 Uckl1os []\nMGI:3646659 Fbxw24 []\nMGI:1923113 Clec4g [('HP:0012311', None), ('MP:0008078', 'increased CD8-positive, alpha-beta T cell number'), ('MP:0003131', 'increased erythrocyte cell number'), ('HP:0001873', None), ('HP:0100827', None), ('MP:0008074', 'increased CD4-positive, alpha beta T cell number')]\nMGI:1916003 Mybphl [('MP:0004647', 'decreased lumbar vertebrae number'), ('MP:0010101', 'increased sacral vertebrae number')]\nMGI:98625 Tcrg-C1 [('MP:0008757', 'abnormal T cell receptor gamma chain V-J recombination'), ('GO:0048873PHENOTYPE', None)]\nMGI:1918397 Oxsm []\nMGI:1923890 1700113H08Rik []\nMGI:1860137 Gp9 [('HP:0001873', None)]\nMGI:99207 Zfp60 []\nMGI:5453219 Gm23442 []\nMGI:2142174 4933405O20Rik []\nMGI:3809043 Rps8-ps4 []\nMGI:1914361 Naaa [('HP:0040216', None)]\nMGI:3643262 Gm9392 []\nMGI:1922264 4930503B20Rik []\nMGI:101899 Pla2g5 [('MP:0000343', 'altered response to myocardial infarction'), ('MP:0003038', 'decreased myocardial infarction size'), ('MP:0008874', 'decreased physiological sensitivity to xenobiotic')]\nMGI:3650064 Gm11517 []\nMGI:3705509 Gm14633 []\nMGI:1933161 Trim23 []\nMGI:3643578 Gm6793 []\nMGI:1923301 Ganc []\nMGI:3651124 Gm12248 []\nMGI:108414 Pafah1b3 [('MP:0005380', 'embryo phenotype'), ('GO:0007283PHENOTYPE', None)]\nMGI:1194499 Gsg1 []\nMGI:3649243 Gm13231 []\nMGI:1914616 Tmed11 []\nMGI:1918436 4933417D19Rik []\nMGI:3642408 Gm10250 []\nMGI:1926022 Hhipl2 []\nMGI:1196608 Slc4a1ap []\nMGI:1346051 Dut []\nMGI:1098533 Ints9 []\nMGI:3646055 Gm5801 []\nMGI:2140201 Slc5a9 []\nMGI:5456270 Gm26493 []\nMGI:5455464 Gm25687 []\nMGI:2444813 9030617O03Rik []\nMGI:3579898 Rhox8 []\nMGI:3704480 Gm10184 []\nMGI:5452188 Gm22411 []\nMGI:1914830 Lrrc18 []\nMGI:1201780 Atp6v1a []\nMGI:3642024 Gm10722 []\nMGI:2140910 AI480526 []\nMGI:1920603 Actrt2 []\nMGI:4421954 n-R5s106 []\nMGI:5455261 Gm25484 []\nMGI:1349763 Dpysl2 [('MP:0008143', 'abnormal dendrite morphology'), ('MP:0003631', 'nervous system phenotype')]\nMGI:1914391 Fbxo5 [('MP:0002663', 'absent blastocoele'), ('MP:0005031', 'abnormal trophoblast layer morphology'), ('MP:0001672', 'abnormal embryo development'), ('MP:0002718', 'abnormal inner cell mass morphology'), ('MP:0011094', 'embryonic lethality before implantation, complete penetrance')]\nMGI:1196368 Carhsp1 []\nMGI:5453712 Gm23935 []\nMGI:1924058 Rpl18a []\nMGI:1923539 Phf14 [('HP:0002093', None), ('MP:0000351', 'increased cell proliferation'), ('MP:0008277', 'abnormal sternum ossification'), ('MP:0010903', 'abnormal pulmonary alveolus wall morphology')]\nMGI:88518 Cryba1 [('HP:0100018', None), ('MP:0002840', 'abnormal lens fiber morphology'), ('HP:0008063', None), ('MP:0020378', 'abnormal cell cytoskeleton morphology'), ('MP:0012143', 'decreased a wave amplitude'), ('MP:0008260', 'abnormal autophagy'), ('HP:0012379', None), ('HP:0000568', None), ('MP:0012144', 'decreased b wave amplitude'), ('MP:0005201', 'abnormal retinal pigment epithelium morphology'), ('MP:0005058', 'abnormal lysosome morphology'), ('GO:0043010PHENOTYPE', None), ('HP:0000546', None), ('HP:0002171', None), ('HP:0010700', None), ('MP:0003172', 'abnormal lysosome physiology')]\nMGI:3644473 Gm7335 []\nMGI:1921406 Acot12 []\nMGI:99927 mt-Atp6 []\nMGI:3651700 Gm11752 []\nMGI:1922717 Stpg4 []\nMGI:3646150 Gm7964 []\nMGI:2146565 Nsun3 []\nMGI:104645 Hsd3b5 []\nMGI:103164 Mif-ps3 []\nMGI:109568 Wbp4 []\nMGI:3704271 Gm9769 []\nMGI:3641929 Rps19-ps9 []\nMGI:4937884 Gm17057 []\nMGI:108247 Tdg [('MP:0004181', 'abnormal carotid artery morphology'), ('MP:0006126', 'abnormal cardiac outflow tract development'), ('MP:0008877', 'abnormal DNA methylation'), ('MP:0003984', 'embryonic growth retardation'), ('MP:0001787', 'pericardial edema'), ('MP:0011098', 'embryonic lethality during organogenesis, complete penetrance'), ('MP:0003888', 'liver hemorrhage'), ('HP:0011851', None), ('MP:0009797', 'abnormal mismatch repair'), ('HP:0011029', None), ('MP:0006279', 'abnormal limb development'), ('MP:0003920', 'abnormal heart right ventricle morphology'), ('HP:0001892', None)]\nMGI:107741 Pvr [('MP:0005025', 'abnormal response to infection'), ('HP:0002720', None), ('MP:0008874', 'decreased physiological sensitivity to xenobiotic')]\nMGI:1923274 Gm11346 []\nMGI:2387217 Ift52 [('MP:0011098', 'embryonic lethality during organogenesis, complete penetrance')]\nMGI:4937289 Gm17655 []\nMGI:5455766 Gm25989 []\nMGI:3644711 Gm8163 []\nMGI:1919335 Osgepl1 []\nMGI:5454593 Gm24816 []\nMGI:1929259 Asic5 []\nMGI:3643448 Gm7931 []\nMGI:3650377 Gm12275 []\nMGI:3648582 Gm4895 []\nMGI:3652029 Gm11877 []\nMGI:5456022 Gm26245 []\nMGI:3650191 Gm14480 []\nMGI:3649722 Gm13007 []\nMGI:3642342 Gm10086 []\nMGI:3650060 Gm14269 []\nMGI:97604 Pklr [('MP:0002875', 'decreased erythrocyte cell number'), ('MP:0000245', 'abnormal erythropoiesis'), ('HP:0001877', None), ('MP:0011178', 'increased erythroblast number'), ('MP:0010035', 'increased erythrocyte clearance'), ('GO:0051707PHENOTYPE', None), ('MP:0002874', 'decreased hemoglobin content'), ('HP:0001878', None), ('MP:0002169', 'no abnormal phenotype detected'), ('MP:0000208', 'decreased hematocrit'), ('HP:0012132', None), ('HP:0001744', None), ('MP:0010751', 'decreased susceptibility to parasitic infection induced morbidity/mortality')]\nMGI:97810 Ptprc [('MP:0008351', 'decreased gamma-delta intraepithelial T cell number'), ('MP:0000693', 'spleen hyperplasia'), ('MP:0008168', 'decreased B-1a cell number'), ('MP:0003944', 'abnormal T cell subpopulation ratio'), ('MP:0002418', 'increased susceptibility to viral infection'), ('MP:0000702', 'enlarged lymph nodes'), ('MP:0002398', 'abnormal bone marrow cell morphology/development'), ('HP:0002014', None), ('MP:0005092', 'decreased double-positive T cell number'), ('HP:0002090', None), ('MP:0005597', 'decreased susceptibility to type I hypersensitivity reaction'), ('HP:0012177', None), ('MP:0010136', 'decreased DN4 thymocyte number'), ('MP:0010839', 'decreased CD8-positive, alpha-beta memory T cell number'), ('MP:0008894', 'abnormal intraepithelial T cell morphology'), ('HP:0010987', None), ('MP:0010133', 'increased DN3 thymocyte number'), ('MP:0008080', 'abnormal CD8-positive, alpha-beta T cell differentiation'), ('GO:0045577PHENOTYPE', None), ('HP:0040006', None), ('MP:0008044', 'increased NK cell number'), ('HP:0001744', None), ('HP:0010976', None), ('HP:0010975', None), ('MP:0009808', 'decreased oligodendrocyte number'), ('MP:0001541', 'abnormal osteoclast physiology'), ('MP:0000245', 'abnormal erythropoiesis'), ('HP:0001882', None), ('MP:0001800', 'abnormal humoral immune response'), ('MP:0000715', 'decreased thymocyte number'), ('HP:0000083', None), ('MP:0005350', 'increased susceptibility to autoimmune disorder'), ('MP:0005095', 'decreased T cell proliferation'), ('MP:0008895', 'abnormal intraepithelial T cell number'), ('HP:0011840', None), ('MP:0008048', 'abnormal memory T cell number'), ('HP:0012115', None), ('MP:0008076', 'abnormal CD4-positive T cell differentiation'), ('MP:0005397', 'hematopoietic system phenotype'), ('MP:0010836', 'decreased CD4-positive, alpha-beta memory T cell number'), ('MP:0011084', 'lethality at weaning, incomplete penetrance'), ('GO:0030183PHENOTYPE', None), ('MP:0001828', 'abnormal T cell activation'), ('MP:0008211', 'decreased mature B cell number'), ('HP:0002846', None), ('MP:0008207', 'decreased B-2 B cell number'), ('MP:0004977', 'increased B-1 B cell number'), ('HP:0011893', None), ('HP:0002716', None), ('HP:0005404', None), ('HP:0001978', None), ('HP:0011115', None)]\nMGI:3780193 Mup12 []\nMGI:2385955 Defb19 []\nMGI:3650009 Gm14052 []\nMGI:1096353 Khk [('MP:0005380', 'embryo phenotype')]\nMGI:5521097 Gm27254 []\nMGI:1201607 Blzf1 []\nMGI:5452551 Gm22774 []\nMGI:5477346 Gm26852 []\nMGI:5453718 Gm23941 []\nMGI:3642193 Rpl19-ps11 []\nMGI:1919104 Dpep3 []\nMGI:1201387 Nlk [('MP:0011091', 'prenatal lethality, complete penetrance'), ('MP:0002217', 'small lymph nodes'), ('MP:0008208', 'decreased pro-B cell number'), ('MP:0000715', 'decreased thymocyte number'), ('MP:0001602', 'impaired myelopoiesis'), ('MP:0000333', 'decreased bone marrow cell number'), ('MP:0001405', 'impaired coordination'), ('HP:0040218', None), ('MP:0002722', 'abnormal immune system organ morphology')]\nMGI:2387642 Cxcl17 [('HP:0001627', None), ('HP:0009887', None)]\nMGI:107161 Cst8 []\nMGI:5456092 Gm26315 []\nMGI:96945 Smcp []\nMGI:5454482 Gm24705 []\nMGI:5453509 Gm23732 []\nMGI:3819551 Snord69 []\nMGI:3801917 Hopxos []\nMGI:2138953 Fibcd1 []\nMGI:3651545 Rpsa-ps4 []\nMGI:3030536 Olfr702 []\nMGI:5455227 Gm25450 []\nMGI:1933156 Acox3 []\nMGI:5455431 Gm25654 []\nMGI:3708729 Gm9825 []\nMGI:1915112 2310033P09Rik []\nMGI:3651345 Gm11450 []\nMGI:3645317 Gm16412 []\nMGI:1913954 Rbm4b [('HP:0003074', None), ('GO:0035883PHENOTYPE', None), ('HP:0000833', None)]\nMGI:5453045 Gm23268 []\nMGI:1099439 Stk10 []\nMGI:2685142 Olfm4 [('HP:0002718', None), ('MP:0010377', 'abnormal gut flora balance'), ('MP:0008713', 'abnormal cytokine level')]\nMGI:1913756 Psmg3 []\nMGI:3041188 Fam69c []\nMGI:1925953 Zfp972 []\nMGI:1859822 Crtam [('MP:0008682', 'decreased interleukin-17 secretion'), ('MP:0005387', 'immune system phenotype'), ('MP:0008078', 'increased CD8-positive, alpha-beta T cell number'), ('MP:0005463', 'abnormal CD4-positive, alpha-beta T cell physiology'), ('HP:0002718', None), ('HP:0011117', None), ('MP:0008074', 'increased CD4-positive, alpha beta T cell number')]\nMGI:5452880 Gm23103 []\nMGI:1917951 Nipal1 []\nMGI:1338820 Bmp10 [('HP:0000003', None), ('HP:0012101', None), ('HP:0004971', None), ('MP:0010725', 'thin interventricular septum'), ('MP:0000291', 'enlarged pericardium'), ('HP:0000967', None), ('MP:0001730', 'embryonic growth arrest'), ('MP:0011100', 'preweaning lethality, complete penetrance'), ('MP:0004076', 'abnormal vitelline vascular remodeling'), ('HP:0000778', None)]\nMGI:5452925 Gm23148 []\nMGI:97473 Pah [('HP:0001508', None), ('MP:0005332', 'abnormal amino acid level'), ('MP:0001265', 'decreased body size'), ('MP:0001496', 'audiogenic seizures'), ('MP:0001525', 'impaired balance'), ('HP:0003112', None), ('HP:0000568', None), ('MP:0009358', 'environmentally induced seizures'), ('GO:0004505PHENOTYPE', None)]\nMGI:3647751 Gm4987 []\nMGI:1917895 Oip5 []\nMGI:4421901 n-R5s56 []\nMGI:1888978 Ntn4 [('HP:0010783', None), ('MP:0002792', 'abnormal retinal vasculature morphology'), ('HP:0000525', None), ('MP:0010144', 'abnormal tumor vascularization'), ('MP:0000351', 'increased cell proliferation'), ('MP:0001265', 'decreased body size'), ('HP:0002597', None), ('MP:0009450', 'abnormal axon fasciculation')]\nMGI:2152844 Slc2a9 [('HP:0000123', None), ('HP:0000803', None), ('HP:0001395', None), ('HP:0004325', None), ('HP:0003259', None), ('HP:0001959', None), ('MP:0011925', 'abnormal heart echocardiography feature'), ('MP:0009350', 'decreased urine pH'), ('HP:0003081', None), ('MP:0008965', 'increased basal metabolism'), ('HP:0000126', None), ('HP:0001397', None), ('MP:0010359', 'increased liver free fatty acids level'), ('MP:0005289', 'increased oxygen consumption'), ('HP:0002155', None), ('HP:0000855', None), ('MP:0010107', 'abnormal renal reabsorbtion'), ('MP:0005367', 'renal/urinary system phenotype'), ('MP:0008882', 'abnormal enterocyte physiology'), ('HP:0003149', None), ('HP:0000092', None), ('HP:0001939', None), ('MP:0010379', 'decreased respiratory quotient'), ('MP:0003868', 'abnormal feces composition'), ('MP:0002169', 'no abnormal phenotype detected'), ('HP:0002048', None), ('HP:0000787', None), ('HP:0012611', None), ('HP:0003124', None)]\nMGI:88599 Cyp2b13 [('MP:0010851', 'decreased effector memory CD8-positive, alpha-beta T cell number')]\nMGI:99613 Zap70 [('MP:0002432', 'abnormal CD4-positive, alpha beta T cell morphology'), ('MP:0004919', 'abnormal positive T cell selection'), ('MP:0002217', 'small lymph nodes'), ('GO:0046777PHENOTYPE', None), ('MP:0001829', 'increased activated T cell number'), ('GO:0035556PHENOTYPE', None), ('MP:0003944', 'abnormal T cell subpopulation ratio'), ('MP:0008826', 'abnormal splenic cell ratio'), ('HP:0002090', None), ('MP:0008075', 'decreased CD4-positive, alpha beta T cell number'), ('MP:0002408', 'abnormal double-positive T cell morphology'), ('MP:0003790', 'absent CD4-positive, alpha beta T cells'), ('MP:0008080', 'abnormal CD8-positive, alpha-beta T cell differentiation'), ('HP:0001370', None), ('MP:0004974', 'decreased regulatory T cell number'), ('MP:0008049', 'increased memory T cell number'), ('MP:0002145', 'abnormal T cell differentiation'), ('MP:0001825', 'arrested T cell differentiation'), ('MP:0008070', 'absent T cells'), ('HP:0005415', None), ('HP:0011840', None), ('MP:0004816', 'abnormal class switch recombination'), ('MP:0002169', 'no abnormal phenotype detected'), ('MP:0008828', 'abnormal lymph node cell ratio'), ('HP:0000939', None), ('MP:0008499', 'increased IgG1 level'), ('MP:0008827', 'abnormal thymus cell ratio'), ('GO:0004713PHENOTYPE', None), ('HP:0005403', None), ('MP:0001828', 'abnormal T cell activation'), ('MP:0004972', 'abnormal regulatory T cell number'), ('MP:0003725', 'increased autoantibody level'), ('HP:0001367', None), ('HP:0000778', None), ('MP:0001606', 'impaired hematopoiesis')]\nMGI:1339962 Ftcd []\nMGI:88603 Cyp2d11 []\nENSEMBL:ENSMUSG00000099077 Ppp2r3d []\nMGI:5456072 Gm26295 []\nMGI:3648632 Gm8814 []\nMGI:3648561 Gm9169 []\nMGI:1929217 Ap3s1-ps2 []\nMGI:2385211 Fam76a []\nMGI:2178742 Stab1 []\nMGI:1277143 Gtf2h3 []\nMGI:1920185 Ddx41 [('MP:0011100', 'preweaning lethality, complete penetrance'), ('MP:0013293', 'embryonic lethality prior to tooth bud stage')]\nMGI:1920648 Nipsnap3a []\nMGI:2442629 Mfsd7a []\nMGI:97429 Oas1g [('MP:0004651', 'increased thoracic vertebrae number'), ('HP:0005815', None)]\nMGI:97178 Map4 []\nMGI:3645529 Cyp21a2-ps []\nMGI:2679274 Adck5 []\nMGI:97828 Pygb []\nMGI:5452140 Gm22363 []\nMGI:2385046 Slc26a8 [('MP:0009243', 'hairpin sperm flagellum'), ('MP:0009831', 'abnormal sperm midpiece morphology'), ('MP:0009832', 'abnormal sperm mitochondrial sheath morphology'), ('MP:0004542', 'impaired acrosome reaction'), ('MP:0002675', 'asthenozoospermia')]\nMGI:3644440 Gm6175 []\nMGI:5530773 Mir6353 []\nMGI:2442858 Ddx58 [('MP:0011085', 'postnatal lethality, complete penetrance'), ('MP:0005387', 'immune system phenotype'), ('HP:0005403', None), ('MP:0011108', 'embryonic lethality during organogenesis, incomplete penetrance'), ('MP:0001829', 'increased activated T cell number'), ('HP:0001978', None), ('MP:0002462', 'abnormal granulocyte physiology'), ('HP:0002846', None), ('MP:0002418', 'increased susceptibility to viral infection'), ('HP:0005237', None), ('HP:0001911', None), ('MP:0008750', 'abnormal interferon level'), ('HP:0001744', None), ('MP:0008049', 'increased memory T cell number')]\nMGI:3643116 Gm8185 []\nMGI:894689 Ywhae [('MP:0004759', 'decreased mitotic index'), ('MP:0008022', 'dilated heart ventricle'), ('MP:0011390', 'abnormal fetal cardiomyocyte physiology'), ('MP:0000788', 'abnormal cerebral cortex morphology'), ('GO:0001764PHENOTYPE', None), ('MP:0008284', 'abnormal hippocampus pyramidal cell layer'), ('HP:0002269', None), ('HP:0000961', None), ('GO:0021766PHENOTYPE', None)]\nMGI:3652220 Hnf1aos2 []\nMGI:102504 mt-Co1 [('HP:0001508', None), ('MP:0000278', 'abnormal myocardial fiber morphology'), ('HP:0004325', None), ('MP:0001853', 'heart inflammation'), ('MP:0008775', 'abnormal heart ventricle pressure'), ('MP:0004215', 'abnormal myocardial fiber physiology'), ('HP:0002597', None), ('MP:0013405', 'increased circulating lactate level'), ('HP:0003198', None)]\nMGI:3780550 Mthfsl [('MP:0010053', 'decreased grip strength')]\nMGI:104768 Gast [('MP:0003892', 'abnormal gastric gland morphology'), ('MP:0004499', 'increased incidence of tumors by chemical induction'), ('HP:0000842', None), ('MP:0004140', 'abnormal gastric chief cell morphology'), ('HP:0011031', None), ('MP:0000495', 'abnormal colon morphology'), ('MP:0000501', 'abnormal digestive secretion'), ('MP:0003564', 'abnormal insulin secretion')]\nMGI:3652081 Rpsa-ps3 []\nMGI:3704287 Gm10175 []\nMGI:3783229 Gm15787 []\nMGI:3644566 Gm8731 []\nMGI:3650777 Gm14049 []\nMGI:3651698 Gm14239 []\nMGI:109565 Kmt2b [('MP:0003787', 'abnormal imprinting'), ('HP:0000969', None), ('MP:0008871', 'abnormal ovarian follicle number'), ('MP:0003059', 'decreased insulin secretion'), ('HP:0011969', None), ('MP:0009288', 'increased epididymal fat pad weight'), ('MP:0009648', 'abnormal superovulation'), ('HP:0001397', None), ('MP:0005181', 'decreased circulating estradiol level'), ('MP:0009289', 'decreased epididymal fat pad weight'), ('MP:0010359', 'increased liver free fatty acids level'), ('HP:0000855', None), ('MP:0012157', 'rostral body truncation'), ('MP:0011092', 'embryonic lethality, complete penetrance'), ('MP:0001923', 'reduced female fertility'), ('MP:0000929', 'open neural tube'), ('GO:0007613PHENOTYPE', None), ('HP:0000234', None), ('MP:0000269', 'abnormal heart looping'), ('MP:0002169', 'no abnormal phenotype detected'), ('MP:0003984', 'embryonic growth retardation'), ('HP:0000842', None), ('MP:0006042', 'increased apoptosis'), ('MP:0001688', 'abnormal somite development')]\nMGI:3037698 Gm1840 []\nMGI:3782787 Gm4604 []\nMGI:1921443 Ccdc9 []\nMGI:3779200 Gm10985 []\nMGI:103238 Cyp2c29 []\nMGI:5504161 Gm27046 []\nMGI:5455658 Gm25881 []\nMGI:1918017 Prpf3 [('MP:0005391', 'vision/eye phenotype'), ('MP:0011092', 'embryonic lethality, complete penetrance'), ('HP:0000546', None)]\nMGI:2152878 A1bg []\nMGI:4421983 n-R5s127 []\nMGI:4834247 Mir3074-1 []\nMGI:3643593 Gm8210 [('HP:0001508', None), ('MP:0011704', 'decreased fibroblast proliferation'), ('HP:0000823', None), ('MP:0011110', 'preweaning lethality, incomplete penetrance'), ('GO:0035264PHENOTYPE', None), ('MP:0000060', 'delayed bone ossification'), ('GO:0006412PHENOTYPE', None), ('GO:0008283PHENOTYPE', None), ('HP:0011018', None)]\nMGI:3612701 Spdye4b []\nMGI:1914006 Cfap97 []\nMGI:3644851 Gm7336 []\nMGI:1914722 Mtfr1 [('MP:0010956', 'abnormal mitochondrial ATP synthesis coupled electron transport')]\nMGI:1915317 Mrnip [('HP:0000752', None)]\nMGI:106651 Ly6e [('GO:0030325PHENOTYPE', None), ('GO:0055010PHENOTYPE', None), ('MP:0001740', 'failure of adrenal epinephrine secretion'), ('MP:0005663', 'abnormal circulating noradrenaline level'), ('MP:0008288', 'abnormal adrenal cortex morphology'), ('GO:0001701PHENOTYPE', None), ('GO:0035265PHENOTYPE', None)]\nMGI:3705695 Gm14516 []\nMGI:3650190 Gm14286 []\nMGI:3649910 Gm11619 []\nMGI:3651800 Gm14373 []\nMGI:2387201 Yrdc []\nMGI:4415005 Gm16585 []\nMGI:5452548 Gm22771 []\nMGI:1918464 Ppm1f [('HP:0000752', None)]\nMGI:1920905 Pex11g []\nMGI:95403 Stom []\nMGI:5453837 Gm24060 []\nMGI:2148248 1700102P08Rik []\nMGI:109266 Gzma [('MP:0005079', 'decreased cytotoxic T cell cytolysis'), ('MP:0005387', 'immune system phenotype'), ('MP:0003721', 'increased tumor growth/size')]\nMGI:1919519 Cda []\nMGI:2140998 Ube3c [('HP:0010679', None), ('MP:0005387', 'immune system phenotype'), ('HP:0005736', None), ('MP:0001258', 'decreased body length'), ('MP:0013513', 'decreased memory-marker CD4-negative NK T cell number'), ('MP:0003961', 'decreased lean body mass'), ('MP:0013522', 'decreased memory-marker CD4-positive NK T cell number'), ('MP:0013678', 'decreased Ly6C-positive NK T cell number'), ('MP:0011940', 'decreased food intake')]\nMGI:3649154 Rps12-ps22 []\nMGI:4422025 n-R5s161 []\nMGI:3708759 Gm10602 []\nMGI:3629652 Mir697 []\nMGI:1925106 Ankrd66 []\nMGI:3834078 Gm15832 []\nMGI:2443977 Gchfr []\nMGI:3651407 Gm13363 []\nMGI:88540 Csn1s1 [('MP:0004047', 'abnormal milk composition')]\nMGI:2385053 Rrp36 []\nMGI:3649935 Gm13320 []\nMGI:2444029 Ankrd52 []\nMGI:1913362 Marc1 []\nMGI:1922942 Nr2c2ap []\nMGI:2180203 Tmlhe []\nMGI:102475 mt-Ts1 []\nMGI:1931463 Sult5a1 []\nMGI:2142208 Faap24 []\nMGI:3651653 Gm11285 []\nMGI:4421907 n-R5s62 []\nMGI:3651744 Gm13141 []\nMGI:5452530 Gm22753 []\nMGI:2384812 Lpcat1 [('MP:0008450', 'retinal photoreceptor degeneration'), ('MP:0005551', 'abnormal eye electrophysiology'), ('HP:0000504', None), ('MP:0008444', 'retinal cone cell degeneration'), ('MP:0012029', 'abnormal electroretinogram waveform feature'), ('MP:0008451', 'retinal rod cell degeneration'), ('MP:0011087', 'neonatal lethality, complete penetrance'), ('MP:0001182', 'lung hemorrhage'), ('HP:0000961', None), ('MP:0011965', 'decreased total retina thickness'), ('HP:0000546', None)]\nMGI:1353627 Angptl3 [('MP:0002981', 'increased liver weight'), ('HP:0012153', None), ('MP:0005318', 'decreased triglyceride level'), ('MP:0009289', 'decreased epididymal fat pad weight'), ('MP:0000183', 'decreased circulating LDL cholesterol level'), ('MP:0005146', 'decreased circulating VLDL cholesterol level'), ('MP:0005375', 'adipose tissue phenotype'), ('HP:0003233', None)]\nMGI:5531399 Gm28017 []\nMGI:102563 Dct [('GO:0043473PHENOTYPE', None), ('GO:0048468PHENOTYPE', None), ('HP:0008034', None), ('MP:0005391', 'vision/eye phenotype'), ('HP:0009887', None)]\nMGI:1914652 Txndc8 []\nMGI:3646907 Gm6563 []\nMGI:3643270 Gm9386 []\nMGI:3652270 Gm13012 []\nMGI:96676 Kin []\nMGI:5530897 Mir7060 []\nMGI:3649301 Gm13577 []\nMGI:109254 Gzmf []\nMGI:1915271 Bphl [('MP:0011967', 'increased or absent threshold for auditory brainstem response'), ('HP:0012101', None)]\nMGI:5521072 Zfp-ps []\nMGI:5530891 Gm27509 []\nMGI:2181510 Dhrsx []\nMGI:2449939 Zgpat [('MP:0013279', 'increased fasted circulating glucose level')]\nMGI:99500 Ephx2 [('MP:0002792', 'abnormal retinal vasculature morphology'), ('HP:0001939', None)]\nMGI:5453401 Gm23624 []\nMGI:3782245 Gm4070 []\nMGI:3643790 Gm5451 []\nMGI:2387153 Npb []\nMGI:3652235 Rps19-ps7 []\nMGI:1913449 Commd4 []\nMGI:3782973 Gm15526 []\nMGI:3650507 Gm11868 []\nMGI:1202298 Nmt2 []\nMGI:3782325 Gm4149 []\nMGI:1914282 Snapc5 []\nMGI:97766 Prm2 [('HP:0000144', None), ('GO:0007286PHENOTYPE', None), ('HP:0000789', None), ('HP:0012206', None)]\nMGI:3651418 Gm11893 []\nMGI:1341284 Scnm1 [('GO:0008380PHENOTYPE', None)]\nMGI:2444491 Heatr3 []\nMGI:1347055 Vnn3 []\nMGI:1921587 Palm3 []\nMGI:5452105 Gm22328 []\nMGI:2445042 A230107N01Rik []\nMGI:1914246 Ceacam11 []\nMGI:3649742 Gm12226 []\nMGI:1891917 Ywhab []\nMGI:107283 Rab6b [('MP:0005387', 'immune system phenotype')]\nMGI:1315204 Slc40a1 [('HP:0001903', None), ('MP:0005562', 'decreased mean corpuscular hemoglobin'), ('MP:0013293', 'embryonic lethality prior to tooth bud stage'), ('MP:0009322', 'increased splenocyte apoptosis'), ('HP:0000938', None), ('MP:0004151', 'decreased circulating iron level'), ('HP:0000568', None), ('MP:0003998', 'decreased thermal nociceptive threshold'), ('MP:0011106', 'embryonic lethality between implantation and somite formation, incomplete penetrance'), ('MP:0005638', 'hemochromatosis'), ('MP:0005564', 'increased hemoglobin content'), ('HP:0001923', None), ('HP:0012465', None), ('MP:0005391', 'vision/eye phenotype'), ('MP:0011890', 'increased circulating ferritin level'), ('HP:0004840', None), ('MP:0009323', 'abnormal spleen development'), ('MP:0003131', 'increased erythrocyte cell number'), ('MP:0011897', 'decreased circulating unsaturated transferrin level'), ('MP:0020367', 'increased heart iron level'), ('HP:0040006', None), ('HP:0010504', None), ('MP:0005376', 'homeostasis/metabolism phenotype'), ('MP:0008737', 'abnormal spleen physiology'), ('MP:0010124', 'decreased bone mineral content'), ('HP:0001877', None), ('HP:0001392', None), ('MP:0013302', 'increased pancreas iron level'), ('HP:0012675', None), ('HP:0011031', None), ('HP:0011273', None), ('MP:0008809', 'increased spleen iron level'), ('HP:0002910', None), ('MP:0001698', 'decreased embryo size'), ('MP:0011097', 'embryonic lethality between somite formation and embryo turning, complete penetrance'), ('HP:0005518', None), ('MP:0005397', 'hematopoietic system phenotype'), ('MP:0003631', 'nervous system phenotype'), ('MP:0002591', 'decreased mean corpuscular volume'), ('GO:0055072PHENOTYPE', None), ('MP:0001265', 'decreased body size'), ('MP:0010375', 'increased kidney iron level'), ('MP:0000692', 'small spleen')]\nMGI:1913315 Hsd17b14 []\nMGI:1921272 Glyr1 []\nMGI:3032634 Ugt1a5 []\nMGI:3783155 Gm15713 []\nMGI:1922375 4930513O06Rik []\nMGI:1914386 Kbtbd4 []\nMGI:97518 Cdk18 []\nMGI:3646708 Gm16409 []\nMGI:1934682 Emc9 []\nMGI:3652253 Gm14414 []\nMGI:3702407 Gm14416 []\nMGI:96018 Hba-ps4 []\nMGI:2442653 Rbm48 []\nMGI:2682318 Zfp395 []\nMGI:2148523 Vmn1r29 []\nMGI:1917473 Nars []\nMGI:88095 Serpinc1 [('MP:0002083', 'premature death'), ('HP:0200023', None), ('MP:0001935', 'decreased litter size'), ('HP:0001397', None), ('HP:0012372', None), ('HP:0001627', None), ('MP:0011099', 'lethality throughout fetal growth and development, complete penetrance'), ('MP:0002658', 'abnormal liver regeneration'), ('MP:0002698', 'abnormal sclera morphology'), ('HP:0001744', None), ('MP:0002847', 'abnormal renal glomerular filtration rate'), ('MP:0005098', 'abnormal optic choroid morphology'), ('MP:0020407', 'abnormal placental thrombosis'), ('MP:0006137', 'venoocclusion'), ('MP:0011100', 'preweaning lethality, complete penetrance'), ('MP:0002551', 'abnormal blood coagulation'), ('MP:0005329', 'abnormal myocardium layer morphology'), ('HP:0012115', None)]\nMGI:2673990 Vaultrc5 []\nMGI:1915737 Tmem140 []\nMGI:1925680 Prr9 []\nMGI:1310000 Kcnj15 []\nMGI:3649994 Gm13586 []\nMGI:97370 Enpp1 [('MP:0004181', 'abnormal carotid artery morphology'), ('MP:0002083', 'premature death'), ('HP:0004325', None), ('MP:0006357', 'abnormal circulating mineral level'), ('HP:0000938', None), ('MP:0004231', 'abnormal calcium ion homeostasis'), ('HP:0002754', None), ('MP:0003653', 'decreased skin turgor'), ('HP:0100774', None), ('HP:0007862', None), ('HP:0005645', None), ('MP:0002066', 'abnormal motor capabilities/coordination/movement'), ('HP:0011729', None), ('MP:0001505', 'hunched posture'), ('MP:0020039', 'increased bone ossification'), ('HP:0007973', None), ('MP:0001565', 'abnormal circulating phosphate level'), ('MP:0005376', 'homeostasis/metabolism phenotype'), ('MP:0010124', 'decreased bone mineral content'), ('HP:0100671', None), ('MP:0011584', 'increased alkaline phosphatase activity'), ('MP:0010234', 'abnormal vibrissa follicle morphology'), ('MP:0000166', 'abnormal chondrocyte morphology'), ('MP:0009820', 'abnormal liver vasculature morphology'), ('HP:0002143', None), ('MP:0009763', 'increased sensitivity to induced morbidity/mortality'), ('HP:0001508', None), ('MP:0001533', 'abnormal skeleton physiology'), ('MP:0003196', 'calcified skin'), ('HP:0002650', None), ('MP:0005592', 'abnormal vascular smooth muscle morphology'), ('MP:0004173', 'abnormal intervertebral disk morphology'), ('HP:0004963', None), ('HP:0100769', None), ('HP:0000365', None), ('MP:0013941', 'abnormal enthesis morphology'), ('HP:0001367', None), ('HP:0002758', None), ('HP:0011138', None)]\nMGI:3779896 Ccnb2-ps []\nMGI:1927099 Rplp1 []\nMGI:1915930 Fitm1 []\nMGI:2181366 Capn8 []\nMGI:3643411 Gm5422 []\nMGI:3648259 Gm6768 []\nMGI:5455354 Gm25577 []\nMGI:1918049 Cep192 []\nMGI:5455351 Gm25574 []\nMGI:1918249 Naa40 [('MP:0005559', 'increased circulating glucose level'), ('HP:0004324', None), ('MP:0014171', 'increased fatty acid oxidation'), ('MP:0000183', 'decreased circulating LDL cholesterol level'), ('MP:0010360', 'decreased liver free fatty acids level'), ('MP:0002310', 'decreased susceptibility to hepatic steatosis'), ('MP:0004889', 'increased energy expenditure'), ('HP:0100738', None), ('MP:0010379', 'decreased respiratory quotient'), ('MP:0003976', 'decreased circulating VLDL triglyceride level'), ('MP:0005292', 'improved glucose tolerance')]\nMGI:5453825 Gm24048 []\nMGI:99182 Zscan21 [('MP:0009004', 'progressive hair loss'), ('HP:0001595', None), ('MP:0008925', 'increased cerebellar granule cell number'), ('MP:0003941', 'abnormal skin development'), ('MP:0000886', 'abnormal cerebellar granule layer morphology'), ('HP:0001006', None), ('MP:0002015', 'epithelioid cysts'), ('MP:0000855', 'accelerated formation of intralobular fissures')]\nMGI:97849 Rag2 [('HP:0001888', None), ('HP:0002720', None), ('MP:0009540', \"absent Hassall's corpuscle\"), ('MP:0008479', 'decreased spleen white pulp amount'), ('MP:0005092', 'decreased double-positive T cell number'), ('HP:0002664', None), ('HP:0002732', None), ('MP:0008213', 'absent immature B cells'), ('HP:0002090', None), ('MP:0002401', 'abnormal lymphopoiesis'), ('HP:0002719', None), ('MP:0000511', 'abnormal intestinal mucosa morphology'), ('MP:0008182', 'decreased marginal zone B cell number'), ('MP:0000333', 'decreased bone marrow cell number'), ('MP:0010133', 'increased DN3 thymocyte number'), ('HP:0004315', None), ('MP:0002722', 'abnormal immune system organ morphology'), ('HP:0010976', None), ('MP:0005671', 'abnormal response to transplant'), ('MP:0008356', 'abnormal gamma-delta T cell differentiation'), ('HP:0001019', None), ('MP:0005095', 'decreased T cell proliferation'), ('MP:0008174', 'decreased follicular B cell number'), ('MP:0002407', 'abnormal double-negative T cell morphology'), ('MP:0008097', 'increased plasma cell number'), ('MP:0002169', 'no abnormal phenotype detected'), ('HP:0004326', None), ('MP:0002403', 'abnormal pre-B cell morphology'), ('MP:0008209', 'decreased pre-B cell number'), ('HP:0004295', None), ('MP:0002435', 'abnormal effector T cell morphology'), ('HP:0002037', None), ('HP:0002846', None), ('HP:0002718', None), ('MP:0002831', \"absent Peyer's patches\"), ('MP:0001802', 'arrested B cell differentiation'), ('MP:0001265', 'decreased body size'), ('MP:0000696', \"abnormal Peyer's patch morphology\")]\nMGI:105305 Slc1a5 []\nMGI:1914846 Tespa1 [('MP:0013588', 'small thymus medulla'), ('MP:0008567', 'decreased interferon-gamma secretion'), ('MP:0008075', 'decreased CD4-positive, alpha beta T cell number'), ('MP:0008049', 'increased memory T cell number'), ('HP:0005403', None)]\nMGI:1096868 Cxcl5 [('MP:0000322', 'increased granulocyte number'), ('MP:0008597', 'decreased circulating interleukin-6 level'), ('MP:0010209', 'abnormal circulating chemokine level'), ('HP:0001875', None), ('MP:0008734', 'decreased susceptibility to endotoxin shock'), ('MP:0008706', 'decreased interleukin-6 secretion'), ('MP:0008563', 'decreased interferon-alpha secretion')]\nMGI:97072 Gbp4 []\nMGI:3646665 Gm5457 []\nMGI:2384581 Ces4a []\nMGI:3648043 Rybp-ps []\nMGI:3645843 Gm7765 []\nMGI:3028580 Cyp4a31 []\nMGI:2446144 March9 [('HP:0003330', None), ('HP:0004325', None)]\nMGI:2445210 Cyp4f39 []\nMGI:3612288 Cyp2c67 []\nMGI:3651005 Gm13436 []\nMGI:5455059 Gm25282 []\nMGI:2674130 Cnksr3 []\nMGI:3711310 9930111J21Rik2 []\nMGI:5452978 Gm23201 []\nMGI:5454374 Gm24597 []\nMGI:96211 Hp [('MP:0011086', 'postnatal lethality, incomplete penetrance'), ('MP:0000332', 'hemoglobinemia'), ('HP:0012211', None), ('MP:0002703', 'abnormal renal tubule morphology'), ('MP:0003917', 'increased kidney weight'), ('MP:0005325', 'abnormal renal glomerulus morphology'), ('MP:0004756', 'abnormal proximal convoluted tubule morphology')]\nMGI:1929278 Tmem59 []\nMGI:101788 Cisd3 []\nMGI:96247 Hsp90ab1 [('MP:0001716', 'abnormal placenta labyrinth morphology'), ('MP:0005031', 'abnormal trophoblast layer morphology')]\nMGI:2141165 Kpna7 []\nMGI:98812 Tpmt [('MP:0002429', 'abnormal blood cell morphology/development'), ('MP:0005389', 'reproductive system phenotype')]\nMGI:3651973 Gm14101 []\nMGI:109610 Enc1 [('MP:0008075', 'decreased CD4-positive, alpha beta T cell number'), ('HP:0008887', None), ('MP:0010168', 'increased CD4-positive, CD25-positive, alpha-beta regulatory T cell number'), ('HP:0008165', None), ('HP:0003233', None), ('HP:0003146', None)]\nMGI:3651784 Gm11878 []\nMGI:3704291 B230322F03Rik []\nMGI:1913378 Mrps36 []\nMGI:102501 mt-Cytb []\nMGI:3704311 Gm10284 []\nMGI:1919193 Tom1l1 []\nMGI:95499 Fcgr2b [('MP:0005596', 'increased susceptibility to type I hypersensitivity reaction'), ('GO:0007166PHENOTYPE', None), ('HP:0000123', None), ('HP:0001733', None), ('HP:0004325', None), ('GO:0009617PHENOTYPE', None), ('HP:0000969', None), ('MP:0005026', 'decreased susceptibility to parasitic infection'), ('HP:0012649', None), ('HP:0002090', None), ('MP:0005390', 'skeleton phenotype'), ('HP:0000093', None), ('MP:0005325', 'abnormal renal glomerulus morphology'), ('MP:0001505', 'hunched posture'), ('GO:0006955PHENOTYPE', None), ('MP:0008501', 'increased IgG2b level'), ('MP:0003724', 'increased susceptibility to induced arthritis'), ('MP:0004829', 'increased anti-chromatin antibody level'), ('HP:0002633', None), ('MP:0002148', 'abnormal hypersensitivity reaction'), ('MP:0008783', 'decreased B cell apoptosis'), ('GO:0006952PHENOTYPE', None)]\nMGI:3782952 Rpl15-ps3 []\nENSEMBL:ENSMUSG00000097394 AC109138.2 []\nMGI:3644020 Rpl7a-ps11 []\nMGI:1101055 Ifit3 []\nMGI:1916804 Klhdc2 [('HP:0007957', None), ('MP:0011953', 'prolonged PQ interval'), ('HP:0004324', None), ('MP:0000443', 'abnormal snout morphology'), ('MP:0001402', 'hypoactivity'), ('HP:0001337', None), ('HP:0001640', None), ('MP:0011100', 'preweaning lethality, complete penetrance')]\nMGI:1352481 Znhit2 []\nMGI:3643471 Gm7493 []\nMGI:3650525 Gm11737 []\nMGI:3819536 Snord45c []\nMGI:3651086 Gm13009 []\nMGI:5455417 Gm25640 []\nMGI:1336161 Klrc1 [('MP:0008561', 'decreased tumor necrosis factor secretion'), ('HP:0002090', None), ('HP:0012115', None)]\nMGI:1099443 Nnmt []\nMGI:3643814 Gm4841 []\nMGI:1914533 Slc25a19 [('HP:0012379', None), ('MP:0011098', 'embryonic lethality during organogenesis, complete penetrance'), ('MP:0012677', 'absent brain ventricles'), ('MP:0001698', 'decreased embryo size')]\nMGI:1913614 2310009A05Rik []\nMGI:3702070 Gm13697 []\nMGI:5434024 Gm21860 []\nMGI:3652217 Gm14130 []\nMGI:5452525 Gm22748 []\nMGI:3643433 Gm8062 []\nMGI:1933177 Pik3ap1 [('MP:0008497', 'decreased IgG2b level'), ('HP:0006270', None), ('HP:0010976', None), ('HP:0004313', None), ('HP:0002850', None), ('MP:0008174', 'decreased follicular B cell number')]\nMGI:1923245 5033417F24Rik []\nMGI:1338798 S100a11 []\nMGI:96963 Mep1a [('MP:0001935', 'decreased litter size'), ('HP:0100577', None), ('MP:0008641', 'increased circulating interleukin-1 beta level'), ('MP:0002703', 'abnormal renal tubule morphology'), ('MP:0009642', 'abnormal blood homeostasis'), ('MP:0008640', 'abnormal circulating interleukin-1 beta level'), ('MP:0008721', 'abnormal chemokine level'), ('MP:0008537', 'increased susceptibility to induced colitis')]\nMGI:1915756 Ctnnbip1 [('HP:0030769', None), ('GO:0001658PHENOTYPE', None), ('MP:0010983', 'abnormal ureteric bud invasion'), ('HP:0000175', None), ('HP:0000104', None), ('MP:0001648', 'abnormal apoptosis'), ('MP:0004936', 'impaired branching involved in ureteric bud morphogenesis'), ('HP:0005105', None)]\nMGI:3044162 Rsl1 []\nMGI:109336 Etv6 [('GO:0022008PHENOTYPE', None), ('MP:0002083', 'premature death'), ('MP:0000245', 'abnormal erythropoiesis'), ('MP:0002401', 'abnormal lymphopoiesis'), ('HP:0001508', None), ('MP:0002082', 'postnatal lethality'), ('MP:0010373', 'myeloid hyperplasia'), ('HP:0010976', None), ('MP:0001884', 'mammary gland alveolar hyperplasia'), ('MP:0011098', 'embryonic lethality during organogenesis, complete penetrance'), ('MP:0011110', 'preweaning lethality, incomplete penetrance'), ('HP:0001875', None), ('MP:0010299', 'increased mammary gland tumor incidence'), ('HP:0005506', None), ('HP:0001873', None), ('MP:0002414', 'abnormal myeloblast morphology/development'), ('MP:0006410', 'abnormal common myeloid progenitor cell morphology'), ('MP:0000229', 'abnormal megakaryocyte differentiation'), ('MP:0002123', 'abnormal definitive hematopoiesis'), ('HP:0002664', None)]\nMGI:3704317 Sap18b []\nMGI:3574098 Xlr4a []\nMGI:5454748 Gm24971 []\nMGI:2685663 Rbm44 [('MP:0001934', 'increased litter size')]\nMGI:2136976 Cecr5 []\nMGI:3643894 Gm7091 []\nMGI:1347023 Txnrd2 [('HP:0000969', None), ('HP:0001392', None), ('MP:0003140', 'dilated heart atrium'), ('GO:0007507PHENOTYPE', None), ('HP:0001635', None), ('MP:0011087', 'neonatal lethality, complete penetrance'), ('MP:0002123', 'abnormal definitive hematopoiesis'), ('MP:0002169', 'no abnormal phenotype detected'), ('GO:0030097PHENOTYPE', None)]\nMGI:3781073 Gm2895 []\nMGI:3648068 Gm6091 []\nMGI:3648656 Gm5507 []\nMGI:3651237 Gm13767 []\nMGI:1913741 Polr2l []\nMGI:108100 Dvl3 [('HP:0000126', None), ('MP:0011086', 'postnatal lethality, incomplete penetrance'), ('MP:0011666', 'double outlet right ventricle, ventricular defect committed to aorta'), ('HP:0100891', None), ('HP:0002098', None)]\nMGI:5453866 Gm24089 []\nMGI:3644269 C1s2 []\nMGI:3651485 Gm12583 []\nMGI:96269 Ranbp1 [('HP:0008669', None), ('HP:0001508', None), ('MP:0005389', 'reproductive system phenotype')]\nMGI:1276109 Cldn1 [('MP:0011100', 'preweaning lethality, complete penetrance'), ('HP:0000962', None), ('MP:0011087', 'neonatal lethality, complete penetrance'), ('HP:0100678', None), ('HP:0001944', None)]\nMGI:1261428 Chchd2 []\nMGI:3802001 Gm16124 []\nMGI:1925188 Fam53b [('HP:0005518', None), ('MP:0010087', 'increased circulating fructosamine level'), ('MP:0010068', 'decreased red blood cell distribution width')]\nMGI:1924139 Coq8b []\nMGI:1915881 Cryl1 []\nMGI:1916476 Snx24 []\nMGI:1918355 Mterf4 [('MP:0000278', 'abnormal myocardial fiber morphology'), ('MP:0011389', 'absent optic disc'), ('HP:0004325', None), ('MP:0001698', 'decreased embryo size'), ('HP:0001640', None), ('MP:0004215', 'abnormal myocardial fiber physiology'), ('GO:0007507PHENOTYPE', None), ('MP:0006036', 'abnormal mitochondrial physiology')]\nMGI:2159681 Timd2 [('HP:0100827', None)]\nMGI:1924270 Atl3 []\nMGI:1919602 Zfyve27 []\nMGI:3644570 Gm8724 []\nMGI:96534 Cd74 [('MP:0005616', 'decreased susceptibility to type IV hypersensitivity reaction'), ('MP:0005465', 'abnormal T-helper 1 physiology'), ('MP:0008567', 'decreased interferon-gamma secretion'), ('GO:0016064PHENOTYPE', None), ('MP:0005387', 'immune system phenotype'), ('MP:0004919', 'abnormal positive T cell selection'), ('MP:0005348', 'increased T cell proliferation'), ('MP:0005397', 'hematopoietic system phenotype'), ('MP:0001265', 'decreased body size'), ('GO:0006886PHENOTYPE', None), ('MP:0005094', 'abnormal T cell proliferation'), ('MP:0002452', 'abnormal professional antigen presenting cell physiology'), ('MP:0004800', 'decreased susceptibility to experimental autoimmune encephalomyelitis'), ('MP:0005042', 'abnormal level of surface class II molecules'), ('HP:0002846', None), ('GO:0019882PHENOTYPE', None), ('MP:0001835', 'abnormal antigen presentation'), ('HP:0010976', None), ('MP:0004799', 'increased susceptibility to experimental autoimmune encephalomyelitis')]\nMGI:3649557 Gm12366 []\nMGI:1913844 Uqcr11 []\nMGI:1338074 Ikbkg [('MP:0005343', 'increased circulating aspartate transaminase level'), ('MP:0009580', 'increased keratinocyte apoptosis'), ('HP:0000962', None), ('HP:0002605', None), ('HP:0200042', None), ('HP:0001397', None), ('MP:0011098', 'embryonic lethality during organogenesis, complete penetrance'), ('HP:0011115', None), ('MP:0005387', 'immune system phenotype'), ('HP:0001392', None), ('MP:0003638', 'abnormal response/metabolism to endogenous compounds'), ('MP:0008670', 'decreased interleukin-12b secretion'), ('HP:0012379', None), ('HP:0005415', None), ('MP:0011009', 'increased circulating glutamate dehydrogenase level'), ('MP:0008734', 'decreased susceptibility to endotoxin shock'), ('MP:0005560', 'decreased circulating glucose level'), ('MP:0002941', 'increased circulating alanine transaminase level'), ('MP:0009766', 'increased sensitivity to xenobiotic induced morbidity/mortality'), ('MP:0000607', 'abnormal hepatocyte morphology'), ('HP:0012115', None), ('MP:0002161', 'abnormal fertility/fecundity'), ('MP:0008174', 'decreased follicular B cell number'), ('MP:0011086', 'postnatal lethality, incomplete penetrance'), ('MP:0002169', 'no abnormal phenotype detected'), ('MP:0008873', 'increased physiological sensitivity to xenobiotic'), ('MP:0008735', 'increased susceptibility to endotoxin shock'), ('MP:0008553', 'increased circulating tumor necrosis factor level'), ('MP:0008874', 'decreased physiological sensitivity to xenobiotic'), ('HP:0005403', None), ('MP:0008211', 'decreased mature B cell number'), ('MP:0003887', 'increased hepatocyte apoptosis'), ('MP:0009385', 'abnormal dermal pigmentation'), ('MP:0001233', 'abnormal epidermis suprabasal layer morphology'), ('HP:0002037', None), ('HP:0002846', None), ('HP:0001337', None), ('HP:0011123', None), ('MP:0006042', 'increased apoptosis'), ('MP:0008561', 'decreased tumor necrosis factor secretion'), ('MP:0008215', 'decreased immature B cell number'), ('MP:0000692', 'small spleen')]\nMGI:88609 Cyp3a11 [('HP:0010683', None)]\nMGI:3819548 Snord66 []\nMGI:1921373 Foxp4 [('MP:0004187', 'cardia bifida'), ('MP:0006065', 'abnormal heart position or orientation'), ('MP:0002151', 'abnormal neural tube morphology'), ('HP:0001360', None), ('HP:0002414', None), ('MP:0003119', 'abnormal digestive system development'), ('GO:0007507PHENOTYPE', None), ('HP:0040006', None), ('HP:0011025', None)]\nMGI:3647101 Gm16368 []\nMGI:3650890 Gm13841 []\nMGI:2182335 Cables2 []\nMGI:2384767 BC023105 []\nMGI:1914490 Taok1 []\nMGI:3644593 Klhl33 []\nMGI:3707464 Gm15371 []\nMGI:3652108 Zeb2os []\nMGI:3650480 Rpl36-ps8 []\nMGI:5455269 Gm25492 []\nMGI:1924105 Slc17a5 [('HP:0001508', None), ('MP:0011085', 'postnatal lethality, complete penetrance'), ('HP:0012638', None), ('HP:0002069', None), ('HP:0012757', None), ('HP:0001093', None), ('HP:0001337', None), ('MP:0009358', 'environmentally induced seizures'), ('MP:0000746', 'weakness'), ('MP:0003871', 'abnormal myelin sheath morphology'), ('HP:0000365', None), ('MP:0008025', 'brain vacuoles'), ('HP:0001288', None), ('MP:0000358', 'abnormal cell morphology'), ('HP:0000833', None)]\nMGI:1914724 Snap29 [('MP:0003960', 'increased lean body mass'), ('MP:0020352', 'abnormal endoplasmic reticulum physiology'), ('HP:0011121', None), ('MP:0001239', 'abnormal epidermis stratum granulosum morphology'), ('MP:0009608', 'abnormal epidermal lamellar body morphology'), ('HP:0000962', None), ('MP:0009546', 'absent gastric milk in neonates'), ('HP:0100679', None), ('HP:0011122', None), ('MP:0010123', 'increased bone mineral content'), ('MP:0001240', 'abnormal epidermis stratum corneum morphology'), ('MP:0011087', 'neonatal lethality, complete penetrance'), ('MP:0009583', 'increased keratinocyte proliferation'), ('MP:0002656', 'abnormal keratinocyte differentiation')]\nMGI:106402 Tmem30a [('MP:0011100', 'preweaning lethality, complete penetrance')]\nMGI:2159680 Havcr1 [('MP:0001844', 'autoimmune response'), ('MP:0005387', 'immune system phenotype'), ('MP:0005090', 'increased double-negative T cell number'), ('MP:0005463', 'abnormal CD4-positive, alpha-beta T cell physiology'), ('MP:0008702', 'increased interleukin-5 secretion'), ('MP:0008660', 'increased interleukin-10 secretion'), ('MP:0002405', 'respiratory system inflammation'), ('MP:0002376', 'abnormal dendritic cell physiology'), ('HP:0001744', None)]\nMGI:5452582 Gm22805 []\nMGI:3647980 Gm5560 []\nMGI:1202880 Traf4 [('MP:0011101', 'prenatal lethality, incomplete penetrance'), ('HP:0004325', None), ('MP:0003120', 'abnormal tracheal cartilage morphology'), ('GO:0030323PHENOTYPE', None), ('MP:0003091', 'abnormal cell migration'), ('MP:0000141', 'abnormal vertebral body morphology'), ('HP:0002090', None), ('MP:0001935', 'decreased litter size'), ('MP:0008146', 'asymmetric sternocostal joints'), ('MP:0004703', 'abnormal vertebral column morphology'), ('MP:0012547', 'spina bifida cystica'), ('HP:0000902', None), ('MP:0001950', 'abnormal respiratory sounds'), ('HP:0010307', None), ('GO:0007585PHENOTYPE', None), ('MP:0003051', 'curly tail'), ('HP:0002777', None), ('MP:0002376', 'abnormal dendritic cell physiology'), ('MP:0008148', 'abnormal sternocostal joint morphology'), ('MP:0001258', 'decreased body length'), ('MP:0004552', 'fused tracheal cartilage rings'), ('MP:0004173', 'abnormal intervertebral disk morphology'), ('HP:0002751', None)]\nMGI:3842428 Pisd-ps1 []\nMGI:4421985 n-R5s129 []\nMGI:1915541 Mto1 [('HP:0001678', None), ('HP:0004325', None), ('MP:0001489', 'decreased startle reflex'), ('MP:0010632', 'cardiac muscle necrosis'), ('MP:0010394', 'decreased QRS amplitude'), ('MP:0003461', 'abnormal response to novel object'), ('MP:0001405', 'impaired coordination'), ('HP:0000752', None), ('MP:0002833', 'increased heart weight'), ('HP:0006682', None), ('MP:0011639', 'decreased mitochondrial DNA content')]\nMGI:4421989 n-R5s133 []\nMGI:1923875 Hoxd3os1 []\nMGI:3782916 Mup-ps4 []\nMGI:105061 Clcn2 [('MP:0001154', 'seminiferous tubule degeneration'), ('HP:0012863', None), ('MP:0004109', 'abnormal Sertoli cell development'), ('HP:0008669', None), ('MP:0005310', 'abnormal salivary gland physiology'), ('MP:0004022', 'abnormal cone electrophysiology'), ('MP:0008586', 'disorganized photoreceptor outer segment'), ('HP:0003134', None), ('HP:0012373', None), ('MP:0005201', 'abnormal retinal pigment epithelium morphology'), ('MP:0003871', 'abnormal myelin sheath morphology'), ('MP:0008025', 'brain vacuoles'), ('MP:0004021', 'abnormal rod electrophysiology'), ('MP:0008581', 'disorganized photoreceptor inner segment'), ('HP:0010791', None), ('MP:0003731', 'abnormal retinal outer nuclear layer morphology'), ('MP:0003690', 'abnormal glial cell physiology'), ('HP:0002143', None), ('HP:0006989', None), ('HP:0002363', None), ('MP:0002216', 'abnormal seminiferous tubule morphology'), ('MP:0005386', 'behavior/neurological phenotype'), ('MP:0003729', 'abnormal photoreceptor outer segment morphology'), ('HP:0002180', None), ('MP:0008917', 'abnormal oligodendrocyte physiology'), ('HP:0000546', None)]\nMGI:5452356 Gm22579 []\nMGI:5451854 Gm22077 []\nMGI:2145043 Cep170b []\nMGI:107784 Zfp74 []\nMGI:1346875 Map3k4 [('MP:0011090', 'perinatal lethality, incomplete penetrance'), ('HP:0003251', None), ('HP:0012861', None), ('HP:0000969', None), ('MP:0011099', 'lethality throughout fetal growth and development, complete penetrance'), ('MP:0002151', 'abnormal neural tube morphology'), ('HP:0000921', None), ('HP:0002414', None), ('MP:0003830', 'abnormal testis development'), ('MP:0001265', 'decreased body size'), ('MP:0002211', 'abnormal primary sex determination'), ('MP:0002995', 'primary sex reversal'), ('MP:0006418', 'abnormal testis cord formation'), ('GO:0010468PHENOTYPE', None)]\nMGI:3650297 Gm11967 []\nMGI:97876 Rbp1 [('MP:0005551', 'abnormal eye electrophysiology'), ('MP:0002216', 'abnormal seminiferous tubule morphology'), ('MP:0005444', 'abnormal retinol metabolism'), ('MP:0003011', 'delayed dark adaptation'), ('MP:0011750', 'abnormal seminiferous tubule epithelium morphology'), ('HP:0012373', None), ('MP:0011234', 'abnormal retinol level'), ('HP:0007973', None)]\nMGI:4422058 n-R5s193 []\nMGI:2443935 Snapc4 [('MP:0005296', 'abnormal humerus morphology')]\nMGI:1925300 4930570N18Rik []\nMGI:2182838 Serpina3f []\nMGI:1917115 A1cf [('MP:0000352', 'decreased cell proliferation'), ('MP:0011094', 'embryonic lethality before implantation, complete penetrance')]\nMGI:3705193 Gm15265 []\nMGI:3645107 Gm6506 []\nMGI:2180021 Cd207 [('MP:0005616', 'decreased susceptibility to type IV hypersensitivity reaction'), ('MP:0008496', 'decreased IgG2a level'), ('MP:0005387', 'immune system phenotype'), ('HP:0002850', None), ('MP:0008498', 'decreased IgG3 level'), ('HP:0002720', None), ('MP:0008127', 'decreased dendritic cell number'), ('MP:0000696', \"abnormal Peyer's patch morphology\"), ('MP:0008118', 'absent Langerhans cell')]\nMGI:1098791 Sepsecs []\nMGI:1861694 Cited4 []\nMGI:3819500 Snora31 []\nMGI:4937017 Gm17383 []\nMGI:3783088 Gm15644 []\nMGI:2181434 Phc3 []\nMGI:4421968 n-R5s118 []\nMGI:3646919 Fignl2 []\nMGI:2140623 Mob3c []\nMGI:1859559 Slc22a7 []\nMGI:3704247 Rpl35a-ps5 []\nMGI:88515 Cryaa [('GO:0007017PHENOTYPE', None), ('HP:0000517', None), ('MP:0003237', 'abnormal lens epithelium morphology'), ('HP:0008063', None), ('MP:0008796', 'increased lens fiber apoptosis'), ('HP:0000568', None), ('GO:0001666PHENOTYPE', None), ('GO:0006915PHENOTYPE', None), ('GO:0010629PHENOTYPE', None), ('MP:0003236', 'abnormal lens capsule morphology'), ('GO:0001934PHENOTYPE', None)]\nMGI:5531285 Gm27903 []\nMGI:5453796 Gm24019 []\nMGI:3646005 Gm5944 []\nMGI:1925841 Mir142hg [('MP:0005461', 'abnormal dendritic cell morphology'), ('MP:0008126', 'increased dendritic cell number'), ('MP:0008115', 'abnormal dendritic cell differentiation')]\nMGI:88057 Apoe [('MP:0003949', 'abnormal circulating lipid level'), ('MP:0004404', 'cochlear outer hair cell degeneration'), ('MP:0002270', 'abnormal pulmonary alveolus morphology'), ('MP:0005146', 'decreased circulating VLDL cholesterol level'), ('HP:0006482', None), ('MP:0009289', 'decreased epididymal fat pad weight'), ('MP:0001663', 'abnormal digestive system physiology'), ('MP:0001413', 'abnormal response to new environment'), ('HP:0012153', None), ('MP:0000788', 'abnormal cerebral cortex morphology'), ('HP:0002910', None), ('MP:0001392', 'abnormal locomotor behavior'), ('MP:0001745', 'increased circulating corticosterone level'), ('MP:0005341', 'decreased susceptibility to atherosclerosis'), ('HP:0002621', None), ('MP:0010026', 'decreased liver cholesterol level'), ('HP:0008216', None), ('MP:0005455', 'increased susceptibility to weight gain'), ('MP:0010451', 'kidney microaneurysm'), ('MP:0008288', 'abnormal adrenal cortex morphology'), ('MP:0005591', 'decreased vasodilation'), ('MP:0004398', 'cochlear inner hair cell degeneration'), ('MP:0001363', 'increased anxiety-related response'), ('MP:0001625', 'cardiac hypertrophy'), ('MP:0002891', 'increased insulin sensitivity'), ('HP:0002904', None), ('MP:0004148', 'increased compact bone thickness'), ('MP:0005167', 'abnormal blood-brain barrier function'), ('MP:0003871', 'abnormal myelin sheath morphology'), ('MP:0003674', 'oxidative stress'), ('MP:0005325', 'abnormal renal glomerulus morphology'), ('MP:0010895', 'increased lung compliance'), ('HP:0003233', None), ('MP:0002733', 'abnormal thermal nociception'), ('MP:0004879', 'decreased systemic vascular resistance'), ('MP:0008289', 'abnormal adrenal medulla morphology'), ('GO:0042157PHENOTYPE', None), ('HP:0002718', None), ('MP:0004952', 'increased spleen weight'), ('MP:0005412', 'vascular stenosis'), ('MP:0000181', 'abnormal circulating LDL cholesterol level'), ('MP:0001212', 'skin lesions'), ('MP:0001417', 'decreased exploration in new environment'), ('HP:0003330', None), ('HP:0003141', None), ('HP:0003146', None), ('MP:0005459', 'decreased percent body fat/body weight'), ('HP:0012184', None), ('HP:0012757', None), ('MP:0001547', 'abnormal lipid level'), ('HP:0011001', None), ('MP:0008182', 'decreased marginal zone B cell number'), ('HP:0002155', None), ('MP:0004630', 'spiral modiolar artery stenosis'), ('MP:0005376', 'homeostasis/metabolism phenotype'), ('MP:0008916', 'abnormal astrocyte physiology'), ('MP:0002376', 'abnormal dendritic cell physiology'), ('MP:0008174', 'decreased follicular B cell number'), ('MP:0005551', 'abnormal eye electrophysiology'), ('GO:0006979PHENOTYPE', None), ('HP:0002486', None), ('MP:0004769', 'abnormal synaptic vesicle morphology'), ('MP:0008706', 'decreased interleukin-6 secretion'), ('MP:0004629', 'abnormal spiral modiolar artery morphology'), ('HP:0002634', None), ('HP:0003464', None), ('HP:0004325', None), ('HP:0001006', None), ('MP:0009122', 'decreased white fat cell lipid droplet size'), ('HP:0030781', None), ('MP:0008515', 'thin retinal outer nuclear layer'), ('MP:0001489', 'decreased startle reflex'), ('MP:0011508', 'glomerular capillary thrombosis'), ('MP:0010868', 'increased bone trabecula number'), ('HP:0003073', None), ('MP:0005641', 'increased mean corpuscular hemoglobin concentration'), ('MP:0003976', 'decreased circulating VLDL triglyceride level'), ('MP:0011561', 'renal glomerulus lipidosis'), ('HP:0001114', None), ('MP:0000031', 'abnormal cochlea morphology'), ('HP:0000752', None), ('MP:0002164', 'abnormal gland physiology'), ('MP:0005560', 'decreased circulating glucose level'), ('HP:0003292', None), ('MP:0002421', 'abnormal cell-mediated immunity'), ('GO:0008203PHENOTYPE', None), ('HP:0010702', None), ('MP:0001942', 'abnormal lung volume'), ('MP:0005386', 'behavior/neurological phenotype'), ('MP:0000043', 'organ of Corti degeneration'), ('MP:0011309', 'abnormal kidney arterial blood vessel morphology'), ('MP:0006076', 'abnormal circulating homocysteine level'), ('HP:0002846', None), ('MP:0005317', 'increased triglyceride level'), ('MP:0011941', 'increased fluid intake')]\nMGI:2686991 Tmem120a []\nMGI:3026615 Mettl7a2 []\nMGI:3650055 Gm11972 []\nMGI:3781847 Gm3671 []\nMGI:3704266 Gm10243 []\nMGI:4422029 n-R5s165 []\nMGI:3652005 Gm12918 []\nMGI:3819484 Scarna2 []\nMGI:1923839 Unc5cl []\nMGI:4421886 n-R5s41 []\nMGI:2145733 Cphx1 []\nMGI:104913 Abi1 [('MP:0003229', 'abnormal vitelline vasculature morphology'), ('HP:0030769', None), ('MP:0001787', 'pericardial edema'), ('MP:0010545', 'abnormal heart layer morphology'), ('MP:0002836', 'abnormal chorion morphology'), ('MP:0001622', 'abnormal vasculogenesis'), ('MP:0003974', 'abnormal endocardium morphology'), ('MP:0000930', 'wavy neural tube')]\nMGI:2685587 Vmo1 []\nMGI:3619412 Mir451a [('HP:0001871', None), ('MP:0000693', 'spleen hyperplasia'), ('MP:0000245', 'abnormal erythropoiesis'), ('HP:0001903', None), ('MP:0008406', 'increased cellular sensitivity to hydrogen peroxide'), ('HP:0012517', None), ('HP:0011273', None), ('MP:0005097', 'polychromatophilia'), ('MP:0004952', 'increased spleen weight'), ('MP:0011241', 'abnormal fetal derived definitive erythrocyte cell number'), ('MP:0001586', 'abnormal erythrocyte cell number')]\nMGI:3819504 Snora36b []\nMGI:1313259 Slfn1 [('MP:0002169', 'no abnormal phenotype detected'), ('MP:0005387', 'immune system phenotype')]\nMGI:3645365 Gm9104 []\nMGI:3030535 Olfr701 []\nMGI:3650107 Gm14279 []\nMGI:3782703 Gm4518 []\nMGI:1924242 Fam219aos []\nMGI:3705781 Rpl39-ps []\nMGI:1099464 Ssr4 []\nMGI:2385708 Rin3 []\nMGI:95392 Eng [('MP:0000267', 'abnormal heart development'), ('MP:0002083', 'premature death'), ('MP:0001719', 'absent vitelline blood vessels'), ('MP:0012732', 'abnormal perineural vascular plexus morphology'), ('MP:0004086', 'absent heartbeat'), ('MP:0005602', 'decreased angiogenesis'), ('HP:0000252', None), ('MP:0004177', 'tail telangiectases'), ('GO:0003273PHENOTYPE', None), ('HP:0001698', None), ('MP:0003396', 'abnormal embryonic hematopoiesis'), ('HP:0011854', None), ('HP:0002098', None), ('HP:0001640', None), ('MP:0001689', 'incomplete somite formation'), ('HP:0001892', None), ('MP:0000265', 'atretic vasculature'), ('MP:0000259', 'abnormal vascular development'), ('MP:0000420', 'ruffled hair'), ('MP:0000269', 'abnormal heart looping'), ('HP:0002629', None), ('MP:0013241', 'embryo tissue necrosis'), ('MP:0002169', 'no abnormal phenotype detected'), ('MP:0013139', 'moribund'), ('MP:0004787', 'abnormal dorsal aorta morphology'), ('MP:0004883', 'abnormal vascular wound healing'), ('HP:0100026', None), ('HP:0011029', None), ('GO:0001525PHENOTYPE', None), ('MP:0002652', 'thin myocardium')]\nMGI:4421955 n-R5s107 []\nMGI:2136335 Klhl1 [('HP:0001288', None), ('MP:0000890', 'thin cerebellar molecular layer')]\nMGI:2139790 Clca4b []\nMGI:1916990 Dph5 []\nMGI:95561 Flt4 [('HP:0045006', None), ('MP:0002082', 'postnatal lethality'), ('MP:0005385', 'cardiovascular system phenotype'), ('MP:0001787', 'pericardial edema'), ('MP:0004076', 'abnormal vitelline vascular remodeling'), ('GO:0007585PHENOTYPE', None), ('MP:0003659', 'abnormal lymph circulation'), ('MP:0012732', 'abnormal perineural vascular plexus morphology'), ('HP:0002597', None), ('MP:0000013', 'abnormal adipose tissue distribution'), ('MP:0004784', 'abnormal anterior cardinal vein morphology'), ('GO:0003016PHENOTYPE', None), ('MP:0005389', 'reproductive system phenotype')]\nMGI:97771 Proc [('MP:0001656', 'focal hepatic necrosis'), ('HP:0001395', None), ('HP:0002170', None), ('GO:0007596PHENOTYPE', None), ('GO:0001889PHENOTYPE', None), ('MP:0011089', 'perinatal lethality, complete penetrance'), ('MP:0002551', 'abnormal blood coagulation'), ('MP:0011087', 'neonatal lethality, complete penetrance'), ('HP:0002033', None), ('MP:0005048', 'abnormal thrombosis'), ('MP:0002419', 'abnormal innate immunity'), ('HP:0000978', None)]\nMGI:4936934 Gm17300 []\nMGI:1925182 Sh2d4b []\nMGI:5453656 Gm23879 []\nMGI:1916780 2310002L09Rik []\nMGI:2146198 Ttc38 []\nMGI:1925246 Cutal [('HP:0010442', None)]\nMGI:3647437 Rps11-ps5 []\nMGI:5504031 Gm26916 []\nMGI:2385205 Ipo13 []\nMGI:1915646 Nat8 []\nMGI:3651234 Rpsa-ps12 []\nMGI:3645153 Gm7389 []\nMGI:3650208 Gm13167 []\nMGI:3605234 BC049715 []\nMGI:3704353 Gm10145 []\nMGI:2443267 Agfg2 []\nMGI:1341830 Eif2ak3 [('MP:0005215', 'abnormal pancreatic islet morphology'), ('MP:0009176', 'increased pancreatic alpha cell number'), ('MP:0009145', 'abnormal pancreatic acinus morphology'), ('MP:0000141', 'abnormal vertebral body morphology'), ('MP:0004986', 'abnormal osteoblast morphology'), ('MP:0011085', 'postnatal lethality, complete penetrance'), ('HP:0011842', None), ('MP:0001402', 'hypoactivity'), ('GO:0031016PHENOTYPE', None), ('MP:0001505', 'hunched posture'), ('MP:0005220', 'abnormal exocrine pancreas morphology'), ('MP:0003562', 'abnormal pancreatic beta cell physiology'), ('MP:0005376', 'homeostasis/metabolism phenotype'), ('GO:0004672PHENOTYPE', None), ('HP:0003074', None), ('HP:0009121', None), ('HP:0006568', None), ('HP:0002753', None), ('MP:0009174', 'absent pancreatic beta cells'), ('MP:0003564', 'abnormal insulin secretion'), ('HP:0000833', None), ('MP:0008873', 'increased physiological sensitivity to xenobiotic'), ('HP:0000939', None), ('MP:0020137', 'decreased bone mineralization'), ('MP:0009172', 'small pancreatic islets'), ('MP:0009164', 'exocrine pancreas atrophy'), ('MP:0003340', 'acute pancreas inflammation'), ('GO:0019722PHENOTYPE', None), ('HP:0003103', None)]\nMGI:4937850 Gm17023 []\nMGI:3643510 Gm8807 []\nMGI:1353496 Slc25a5 []\nMGI:3819521 Snord13 []\nMGI:5433902 Gm21738 []\nMGI:96244 Hspa1a []\nMGI:1919016 Raver1 [('MP:0001475', 'reduced long term depression'), ('MP:0005384', 'cellular phenotype')]\nMGI:3649470 Gm13292 []\nMGI:3805955 Gm6043 []\nMGI:3651428 Gm13522 []\nMGI:102503 mt-Co2 []\nMGI:3644689 Gm8623 []\nMGI:1858496 Deaf1 [('HP:0030769', None), ('MP:0003012', 'no phenotypic analysis'), ('MP:0000929', 'open neural tube'), ('MP:0003460', 'decreased fear-related response'), ('HP:0000892', None), ('HP:0000902', None)]\nMGI:2448403 Hist1h2bl []\nMGI:2684919 Gcnt4 [('GO:0048872PHENOTYPE', None), ('MP:0005478', 'decreased circulating thyroxine level'), ('GO:0048729PHENOTYPE', None), ('MP:0000219', 'increased neutrophil cell number')]\nMGI:3650675 Rpl9-ps3 []\nMGI:1923639 Ankrd36 []\nMGI:3647829 Gm10382 []\nMGI:1916946 2310057N15Rik []\nMGI:3603817 Appbp2os []\nMGI:3780066 Gm9658 []\nMGI:1931086 Tusc2 [('HP:0001974', None), ('HP:0004325', None), ('GO:0006909PHENOTYPE', None), ('HP:0012089', None), ('MP:0002019', 'abnormal tumor incidence'), ('MP:0011508', 'glomerular capillary thrombosis'), ('GO:2000377PHENOTYPE', None), ('MP:0013249', 'adipose tissue necrosis'), ('GO:0032618PHENOTYPE', None), ('MP:0004794', 'increased anti-nuclear antigen antibody level'), ('MP:0005325', 'abnormal renal glomerulus morphology'), ('HP:0002665', None), ('MP:0008676', 'decreased interleukin-15 secretion'), ('GO:0048469PHENOTYPE', None), ('HP:0002633', None)]\nMGI:1920949 Ppp2r1b [('HP:0003251', None), ('MP:0000219', 'increased neutrophil cell number')]\nMGI:5451833 Gm22056 []\nMGI:3650059 Snhg15 []\nMGI:3040693 Zmiz1 [('GO:0003007PHENOTYPE', None), ('MP:0002053', 'decreased incidence of induced tumors'), ('MP:0004787', 'abnormal dorsal aorta morphology'), ('HP:0005403', None), ('HP:0000980', None), ('MP:0002672', 'abnormal pharyngeal arch artery morphology'), ('MP:0011110', 'preweaning lethality, incomplete penetrance'), ('MP:0011098', 'embryonic lethality during organogenesis, complete penetrance'), ('MP:0012732', 'abnormal perineural vascular plexus morphology'), ('MP:0001722', 'pale yolk sac'), ('GO:0001570PHENOTYPE', None), ('GO:0048589PHENOTYPE', None), ('MP:0014224', 'increased ileal goblet cell number'), ('MP:0001698', 'decreased embryo size'), ('HP:0040218', None), ('MP:0000285', 'abnormal heart valve morphology'), ('GO:0001701PHENOTYPE', None), ('MP:0003105', 'abnormal heart atrium morphology'), ('HP:0010976', None), ('GO:0007296PHENOTYPE', None)]\nMGI:1917004 Fbxo7 [('MP:0002875', 'decreased erythrocyte cell number'), ('HP:0005518', None), ('HP:0002904', None), ('HP:0003251', None), ('MP:0008075', 'decreased CD4-positive, alpha beta T cell number'), ('MP:0002416', 'abnormal proerythroblast morphology'), ('HP:0001894', None), ('GO:0045620PHENOTYPE', None), ('MP:0010088', 'decreased circulating fructosamine level'), ('MP:0011179', 'decreased erythroblast number'), ('HP:0012132', None), ('MP:0010850', 'increased effector memory CD8-positive, alpha-beta T cell number'), ('MP:0005561', 'increased mean corpuscular hemoglobin')]\nMGI:2387894 Snord87 []\nMGI:1917685 Inf2 []\nMGI:5434641 Gm21286 []\nMGI:108093 Bid [('HP:0025100', None), ('MP:0003992', 'increased mortality induced by ionizing radiation'), ('MP:0005376', 'homeostasis/metabolism phenotype'), ('MP:0005370', 'liver/biliary system phenotype'), ('MP:0008734', 'decreased susceptibility to endotoxin shock'), ('MP:0002273', 'abnormal pulmonary alveolus epithelial cell morphology'), ('MP:0009764', 'decreased sensitivity to induced morbidity/mortality')]\nMGI:5455942 Gm26165 []\nMGI:3037684 Gm1826 []\nMGI:5452563 Gm22786 []\nMGI:2144284 Inca1 [('MP:0002357', 'abnormal spleen white pulp morphology'), ('HP:0005513', None), ('HP:0003261', None), ('MP:0011704', 'decreased fibroblast proliferation'), ('HP:0001978', None)]\nMGI:3801722 Gm15982 []\nMGI:3648580 Gm5863 []\nMGI:1926005 Fam122b []\nMGI:1916027 Tmem53 []\nMGI:2445363 Serpinb1c []\nMGI:3649405 Cd300ld2 []\nMGI:1345092 Pglyrp1 [('GO:0050728PHENOTYPE', None), ('HP:0011990', None), ('MP:0000488', 'abnormal intestinal epithelium morphology'), ('MP:0000490', 'abnormal crypts of Lieberkuhn morphology'), ('MP:0000512', 'intestinal ulcer'), ('MP:0005074', 'impaired granulocyte bactericidal activity'), ('MP:0003449', 'abnormal intestinal goblet cell morphology'), ('MP:0003724', 'increased susceptibility to induced arthritis'), ('MP:0000511', 'abnormal intestinal mucosa morphology'), ('MP:0010377', 'abnormal gut flora balance'), ('MP:0008705', 'increased interleukin-6 secretion'), ('HP:0001824', None), ('HP:0002718', None), ('MP:0008537', 'increased susceptibility to induced colitis'), ('MP:0009788', 'increased susceptibility to bacterial infection induced morbidity/mortality'), ('HP:0011115', None)]\nMGI:4439785 Gm16861 []\nMGI:103575 Skp1a []\nMGI:4421952 n-R5s104 []\nMGI:3649802 Rps11-ps3 []\nMGI:3650081 Rpl36-ps2 []\nMGI:3704493 Gm10053 []\nMGI:1201409 Pknox1 [('HP:0001903', None), ('HP:0000969', None), ('MP:0004196', 'abnormal prenatal growth/weight/body size'), ('MP:0005090', 'increased double-negative T cell number'), ('MP:0011096', 'embryonic lethality between implantation and somite formation, complete penetrance'), ('MP:0005391', 'vision/eye phenotype'), ('GO:0043010PHENOTYPE', None), ('MP:0006413', 'increased T cell apoptosis'), ('MP:0001693', 'failure of primitive streak formation'), ('HP:0010976', None), ('MP:0002429', 'abnormal blood cell morphology/development'), ('HP:0000708', None), ('HP:0012153', None), ('HP:0000980', None), ('MP:0009703', 'decreased birth body size'), ('MP:0001698', 'decreased embryo size'), ('MP:0008083', 'decreased single-positive T cell number'), ('MP:0011089', 'perinatal lethality, complete penetrance'), ('MP:0001672', 'abnormal embryo development'), ('GO:0030217PHENOTYPE', None), ('MP:0003984', 'embryonic growth retardation'), ('MP:0008211', 'decreased mature B cell number'), ('MP:0008209', 'decreased pre-B cell number'), ('GO:0001525PHENOTYPE', None), ('GO:0030097PHENOTYPE', None)]\nMGI:3652099 Gm11586 []\nMGI:1914267 Fam210b []\nMGI:3650486 Gm13121 []\nMGI:2143702 Enpp3 [('MP:0005596', 'increased susceptibility to type I hypersensitivity reaction'), ('HP:0002090', None), ('MP:0005387', 'immune system phenotype'), ('MP:0013956', 'decreased colon length'), ('HP:0100495', None), ('MP:0008699', 'increased interleukin-4 secretion'), ('MP:0002606', 'increased basophil cell number'), ('MP:0010180', 'increased susceptibility to weight loss'), ('MP:0002464', 'abnormal basophil physiology'), ('MP:0009763', 'increased sensitivity to induced morbidity/mortality')]\nMGI:1891389 Ccl27b []\nMGI:1918940 Esm1 []\nMGI:1929711 Anapc7 []\nMGI:2444345 Acnat2 []\nMGI:1921285 Nol9 []\nMGI:88597 Cyp2a5 []\nMGI:1923151 Dcp1a []\nMGI:1918095 4921508D12Rik []\nMGI:3606001 Apol9a []\nMGI:2445168 Elmod3 []\nMGI:107932 Ndufs6 [('MP:0002083', 'premature death'), ('MP:0003915', 'increased left ventricle weight'), ('GO:0006936PHENOTYPE', None), ('MP:0005281', 'increased fatty acid level'), ('MP:0001935', 'decreased litter size'), ('MP:0003819', 'increased left ventricle diastolic pressure'), ('MP:0003913', 'increased heart right ventricle weight'), ('MP:0003141', 'cardiac fibrosis'), ('MP:0004084', 'abnormal cardiac muscle relaxation'), ('MP:0010563', 'increased heart right ventricle size'), ('GO:0006631PHENOTYPE', None), ('MP:0008772', 'increased heart ventricle size'), ('GO:0072358PHENOTYPE', None), ('MP:0005598', 'decreased ventricle muscle contractility'), ('MP:0010579', 'increased heart left ventricle size'), ('HP:0008322', None), ('MP:0003822', 'decreased left ventricle systolic pressure'), ('HP:0001824', None), ('MP:0011631', 'decreased mitochondria size')]\nMGI:3646431 Rpl21-ps3 []\nMGI:1916998 Aldh16a1 [('HP:0003072', None), ('HP:0002902', None), ('HP:0007973', None)]\nMGI:5454744 Gm24967 []\nMGI:2141980 Scaf1 []\nMGI:1926230 Treh [('MP:0005367', 'renal/urinary system phenotype')]\nMGI:3649808 Gm11703 []\nMGI:96551 Il2rg [('MP:0000693', 'spleen hyperplasia'), ('MP:0000494', 'abnormal cecum morphology'), ('MP:0002217', 'small lymph nodes'), ('MP:0002416', 'abnormal proerythroblast morphology'), ('MP:0002378', 'abnormal gut-associated lymphoid tissue morphology'), ('MP:0000706', 'small thymus'), ('MP:0008479', 'decreased spleen white pulp amount'), ('MP:0008700', 'decreased interleukin-4 secretion'), ('MP:0005092', 'decreased double-positive T cell number'), ('MP:0001927', 'abnormal estrous cycle'), ('MP:0009011', 'prolonged diestrus'), ('MP:0005389', 'reproductive system phenotype'), ('MP:0013592', 'small thymus cortex'), ('HP:0011111', None), ('MP:0008075', 'decreased CD4-positive, alpha beta T cell number'), ('HP:0002850', None), ('MP:0005046', 'absent spleen white pulp'), ('HP:0004313', None), ('MP:0005425', 'increased macrophage cell number'), ('MP:0008688', 'decreased interleukin-2 secretion'), ('HP:0010987', None), ('MP:0001601', 'abnormal myelopoiesis'), ('MP:0008498', 'decreased IgG3 level'), ('HP:0010975', None), ('MP:0008348', 'absent gamma-delta T cells'), ('HP:0100494', None), ('MP:0000715', 'decreased thymocyte number'), ('MP:0008356', 'abnormal gamma-delta T cell differentiation'), ('MP:0000219', 'increased neutrophil cell number'), ('MP:0005095', 'decreased T cell proliferation'), ('MP:0012529', 'abnormal decidua basalis morphology'), ('MP:0004816', 'abnormal class switch recombination'), ('MP:0009020', 'prolonged metestrus'), ('MP:0002464', 'abnormal basophil physiology'), ('MP:0010776', 'abnormal placenta metrial gland morphology'), ('MP:0002421', 'abnormal cell-mediated immunity'), ('MP:0008122', 'decreased myeloid dendritic cell number'), ('MP:0008209', 'decreased pre-B cell number'), ('MP:0002831', \"absent Peyer's patches\"), ('MP:0008098', 'decreased plasma cell number')]\nMGI:3603401 A230005M16Rik []\nMGI:1931870 Zbtb22 []\nMGI:3647234 Rps15-ps2 []\nMGI:3644333 Gm8756 []\nMGI:2443701 B930095G15Rik []\nMGI:3645172 Gm9242 []\nMGI:1918141 Spdya []\nMGI:3801728 Gm16216 []\nMGI:2442153 Gak [('MP:0011085', 'postnatal lethality, complete penetrance'), ('MP:0000812', 'abnormal dentate gyrus morphology'), ('MP:0010900', 'abnormal pulmonary interalveolar septum morphology'), ('MP:0010902', 'abnormal pulmonary alveolar sac morphology'), ('MP:0004948', 'abnormal neuronal precursor proliferation'), ('GO:0072583PHENOTYPE', None), ('HP:0001392', None), ('MP:0012242', 'abnormal hepatoblast differentiation'), ('MP:0004782', 'abnormal surfactant physiology'), ('MP:0008458', 'abnormal cortical ventricular zone morphology'), ('MP:0002169', 'no abnormal phenotype detected'), ('MP:0011087', 'neonatal lethality, complete penetrance'), ('HP:0011124', None), ('MP:0004981', 'decreased neuronal precursor cell number')]\nMGI:3652124 Gm11793 []\nMGI:88384 F9 [('MP:0002083', 'premature death'), ('HP:0003010', None), ('MP:0005376', 'homeostasis/metabolism phenotype'), ('MP:0012305', 'umbilical cord hemorrhage'), ('MP:0002551', 'abnormal blood coagulation'), ('HP:0001892', None), ('MP:0005023', 'abnormal wound healing')]\nMGI:3809525 Gm9574 []\nMGI:4414990 Gm16570 []\nMGI:103572 Cebpe [('MP:0005012', 'decreased eosinophil cell number'), ('MP:0002083', 'premature death'), ('GO:0030225PHENOTYPE', None), ('HP:0011990', None), ('HP:0001978', None), ('MP:0008111', 'abnormal granulocyte differentiation'), ('HP:0011121', None), ('MP:0001602', 'impaired myelopoiesis'), ('HP:0011992', None), ('MP:0000321', 'increased bone marrow cell number'), ('HP:0012649', None), ('MP:0000702', 'enlarged lymph nodes'), ('HP:0002754', None), ('HP:0012384', None), ('HP:0000509', None), ('MP:0008097', 'increased plasma cell number'), ('MP:0006410', 'abnormal common myeloid progenitor cell morphology')]\nMGI:4421965 n-R5s115 []\nMGI:2687281 Fam105a []\nMGI:101782 Scnn1a [('MP:0001402', 'hypoactivity'), ('MP:0011417', 'abnormal renal transport'), ('HP:0100738', None), ('MP:0011100', 'preweaning lethality, complete penetrance'), ('MP:0011087', 'neonatal lethality, complete penetrance'), ('MP:0002169', 'no abnormal phenotype detected')]\nMGI:2140260 Pcsk9 [('HP:0003464', None), ('HP:0003146', None), ('HP:0012184', None)]\nMGI:700013 Sorbs3 []\nMGI:3648029 Gm7658 []\nMGI:96239 Hsf2 [('MP:0011086', 'postnatal lethality, incomplete penetrance'), ('MP:0001134', 'absent corpus luteum'), ('MP:0004901', 'decreased male germ cell number'), ('MP:0002216', 'abnormal seminiferous tubule morphology'), ('HP:0003228', None), ('MP:0002682', 'decreased mature ovarian follicle number'), ('HP:0001342', None), ('HP:0007082', None), ('HP:0008669', None), ('MP:0004929', 'decreased epididymis weight'), ('MP:0005431', 'decreased oocyte number'), ('MP:0001153', 'small seminiferous tubules')]\nMGI:104725 Atn1 [('GO:0008340PHENOTYPE', None), ('HP:0004325', None), ('MP:0003631', 'nervous system phenotype'), ('HP:0005736', None), ('GO:0008584PHENOTYPE', None), ('HP:0008887', None), ('HP:0001627', None), ('GO:0035264PHENOTYPE', None), ('MP:0002673', 'abnormal sperm number'), ('HP:0008734', None), ('GO:0007283PHENOTYPE', None), ('HP:0001288', None), ('MP:0002169', 'no abnormal phenotype detected'), ('GO:0009791PHENOTYPE', None)]\nMGI:3646182 Gm9385 []\nMGI:4421733 n-R5s1 []\nMGI:103009 Epb41l2 [('HP:0008669', None), ('MP:0011750', 'abnormal seminiferous tubule epithelium morphology'), ('MP:0004852', 'decreased testis weight'), ('HP:0008734', None), ('MP:0002169', 'no abnormal phenotype detected')]\nMGI:1315201 Grcc10 []\nMGI:3644263 Gm8648 []\nMGI:5453489 Gm23712 []\nMGI:3646180 Gm6361 []\nMGI:1916709 Ubl7 []\nMGI:109443 Gtpbp1 []\nMGI:1341171 Cnmd [('MP:0001533', 'abnormal skeleton physiology'), ('HP:0011001', None), ('MP:0005006', 'abnormal osteoblast physiology'), ('GO:0016525PHENOTYPE', None), ('MP:0002169', 'no abnormal phenotype detected'), ('HP:0010683', None)]\nMGI:894407 Tmem165 [('HP:0000525', None)]\nMGI:96513 Igkv15-103 []\nMGI:107750 Dync1i2 [('MP:0001392', 'abnormal locomotor behavior'), ('HP:0002902', None), ('HP:0003146', None)]\nMGI:5451819 Gm22042 []\nMGI:3781450 Gm3272 []\nMGI:4422013 n-R5s151 []\nMGI:5455409 Gm25632 []\nMGI:1919485 1700021J08Rik []\nMGI:1352754 Clic4 [('MP:0006055', 'abnormal vascular endothelial cell morphology'), ('HP:0200042', None), ('GO:0061299PHENOTYPE', None), ('GO:0035264PHENOTYPE', None), ('MP:0008734', 'decreased susceptibility to endotoxin shock'), ('GO:0007035PHENOTYPE', None), ('MP:0009703', 'decreased birth body size'), ('MP:0008554', 'decreased circulating tumor necrosis factor level'), ('HP:0002718', None), ('MP:0008706', 'decreased interleukin-6 secretion'), ('MP:0008770', 'decreased survivor rate'), ('MP:0004003', 'abnormal vascular endothelial cell physiology'), ('MP:0009764', 'decreased sensitivity to induced morbidity/mortality'), ('HP:0011115', None)]\nMGI:3644824 Gm5540 []\nMGI:2142980 Tmem266 []\nMGI:1336888 Prdx6b []\nMGI:3652245 Gm12989 []\nMGI:5455100 Gm25323 []\nMGI:1913788 Rps19bp1 [('HP:0001743', None)]\nMGI:2142572 Mtus1 [('MP:0002083', 'premature death'), ('HP:0001974', None), ('MP:0000688', 'lymphoid hyperplasia'), ('HP:0004324', None), ('MP:0005385', 'cardiovascular system phenotype'), ('MP:0004801', 'increased susceptibility to systemic lupus erythematosus'), ('HP:0000099', None), ('MP:0000351', 'increased cell proliferation'), ('HP:0001873', None), ('MP:0000358', 'abnormal cell morphology')]\nMGI:1915244 Mrps11 []\nMGI:2139258 Depdc7 []\nMGI:2149905 Ugt2a1 []\nMGI:1915686 Rpl34 []\nMGI:4421910 n-R5s65 []\nMGI:107682 Ppp1r14b []\nMGI:3781346 Rpl35a-ps7 []\nMGI:98105 Rps12 []\nMGI:5454285 Gm24508 []\nMGI:3645947 Mrip-ps []\nMGI:5477264 Gm26770 []\nMGI:3650759 Gm12715 []\nMGI:3802118 Mup-ps13 []\nMGI:1306775 Sucla2 [('MP:0011089', 'perinatal lethality, complete penetrance'), ('MP:0006036', 'abnormal mitochondrial physiology'), ('MP:0011638', 'abnormal mitochondrial chromosome morphology'), ('MP:0011639', 'decreased mitochondrial DNA content')]\nMGI:1923580 Sephs1 []\nMGI:1355310 Scly [('MP:0001463', 'abnormal spatial learning')]\nMGI:1347354 Homer2 [('HP:0012638', None), ('MP:0009754', 'enhanced behavioral response to cocaine'), ('MP:0005386', 'behavior/neurological phenotype'), ('MP:0009713', 'enhanced conditioned place preference behavior'), ('MP:0002062', 'abnormal associative learning')]\nMGI:3648534 Gm8973 []\nMGI:1923733 Lmf1 [('GO:0009306PHENOTYPE', None), ('MP:0002082', 'postnatal lethality'), ('HP:0000980', None), ('MP:0001402', 'hypoactivity'), ('HP:0002155', None), ('HP:0011029', None), ('HP:0003124', None), ('MP:0001182', 'lung hemorrhage'), ('HP:0000961', None)]\nMGI:4937867 Gm17040 []\nMGI:3644227 Gm8991 []\nMGI:5451977 Gm22200 []\nMGI:3645339 Gm5866 []\nMGI:1861232 Clec4e [('MP:0005616', 'decreased susceptibility to type IV hypersensitivity reaction'), ('MP:0008874', 'decreased physiological sensitivity to xenobiotic'), ('MP:0005025', 'abnormal response to infection'), ('GO:0002292PHENOTYPE', None), ('HP:0012312', None), ('MP:0004800', 'decreased susceptibility to experimental autoimmune encephalomyelitis'), ('MP:0008706', 'decreased interleukin-6 secretion'), ('MP:0008127', 'decreased dendritic cell number'), ('MP:0008615', 'decreased circulating interleukin-17 level')]\nMGI:2182607 Sdsl []\nMGI:88574 Cybb [('MP:0009873', 'abnormal aorta tunica media morphology'), ('MP:0002451', 'abnormal macrophage physiology'), ('MP:0001473', 'reduced long term potentiation'), ('HP:0011990', None), ('MP:0006317', 'decreased urine sodium level'), ('MP:0005385', 'cardiovascular system phenotype'), ('HP:0004325', None), ('MP:0010759', 'decreased right ventricle systolic pressure'), ('HP:0011037', None), ('MP:0003957', 'abnormal nitric oxide homeostasis'), ('HP:0002719', None), ('MP:0005381', 'digestive/alimentary phenotype'), ('MP:0002207', 'abnormal long term potentiation'), ('GO:0050665PHENOTYPE', None), ('MP:0000511', 'abnormal intestinal mucosa morphology'), ('MP:0005527', 'increased renal glomerular filtration rate'), ('MP:0003447', 'decreased tumor growth/size'), ('MP:0001405', 'impaired coordination'), ('MP:0005088', 'increased acute inflammation'), ('MP:0005526', 'decreased renal plasma flow rate'), ('HP:0012649', None)]\nMGI:1344415 Dmtf1 [('HP:0001508', None), ('MP:0013320', 'dilated seminal vesicles'), ('MP:0009546', 'absent gastric milk in neonates'), ('MP:0009703', 'decreased birth body size'), ('HP:0000021', None), ('MP:0002020', 'increased tumor incidence')]\nMGI:3705447 Gm15267 []\nMGI:3840152 Gm16335 []\nMGI:4936909 Gm17275 []\nMGI:3647495 Gm7399 []\nMGI:5434255 Gapdh-ps15 [('MP:0005397', 'hematopoietic system phenotype'), ('MP:0005385', 'cardiovascular system phenotype'), ('GO:0004365PHENOTYPE', None), ('MP:0002596', 'abnormal hematocrit'), ('MP:0011609', 'decreased glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity'), ('HP:0012379', None), ('MP:0002874', 'decreased hemoglobin content'), ('MP:0005367', 'renal/urinary system phenotype'), ('MP:0011100', 'preweaning lethality, complete penetrance'), ('MP:0008770', 'decreased survivor rate'), ('MP:0005378', 'growth/size/body region phenotype')]\nMGI:2139607 Usp53 [('MP:0004749', 'nonsyndromic hearing loss'), ('MP:0004398', 'cochlear inner hair cell degeneration'), ('MP:0001489', 'decreased startle reflex'), ('MP:0002857', 'cochlear ganglion degeneration'), ('GO:0001508PHENOTYPE', None), ('GO:0007605PHENOTYPE', None), ('MP:0005386', 'behavior/neurological phenotype'), ('GO:0010996PHENOTYPE', None), ('MP:0004399', 'abnormal cochlear outer hair cell morphology'), ('GO:0006915PHENOTYPE', None), ('HP:0000365', None), ('MP:0004528', 'fused outer hair cell stereocilia'), ('MP:0004411', 'decreased endocochlear potential')]\nMGI:3649489 Gm12247 []\nMGI:1354958 Hs6st1 [('MP:0011086', 'postnatal lethality, incomplete penetrance'), ('GO:0015012PHENOTYPE', None), ('MP:0005104', 'abnormal tarsal bone morphology'), ('MP:0003231', 'abnormal placenta vasculature'), ('MP:0005376', 'homeostasis/metabolism phenotype'), ('HP:0004325', None), ('HP:0012372', None), ('MP:0011099', 'lethality throughout fetal growth and development, complete penetrance'), ('MP:0011109', 'lethality throughout fetal growth and development, incomplete penetrance'), ('MP:0001698', 'decreased embryo size'), ('GO:0048286PHENOTYPE', None), ('MP:0001183', 'overexpanded pulmonary alveoli')]\nMGI:108057 Rpl6 []\nMGI:3643356 Rpsa-ps2 []\nMGI:2142985 Xylb [('MP:0005316', 'abnormal response to tactile stimuli'), ('MP:0003960', 'increased lean body mass'), ('HP:0011001', None), ('MP:0001486', 'abnormal startle reflex'), ('HP:0010683', None)]\nMGI:2685590 Rubcnl []\nMGI:2442443 Fsd1l []\nMGI:2441932 Prepl [('HP:0001252', None), ('MP:0001523', 'impaired righting response'), ('MP:0001258', 'decreased body length')]\nMGI:3705885 Rpl10a-ps1 []\nMGI:95525 Fgfr4 [('HP:0004325', None)]\nMGI:5454996 Gm25219 []\nMGI:97797 Ptgs1 [('MP:0004841', 'abnormal small intestine crypts of Lieberkuhn morphology'), ('MP:0012124', 'increased bronchoconstrictive response'), ('MP:0008800', 'increased small intestinal crypt cell apoptosis'), ('MP:0005389', 'reproductive system phenotype'), ('HP:0002664', None), ('MP:0005531', 'increased renal vascular resistance'), ('MP:0005527', 'increased renal glomerular filtration rate'), ('GO:0008217PHENOTYPE', None), ('HP:0001744', None), ('MP:0005186', 'increased circulating progesterone level'), ('MP:0009815', 'decreased prostaglandin level'), ('GO:0006693PHENOTYPE', None), ('MP:0003436', 'decreased susceptibility to induced arthritis'), ('MP:0009766', 'increased sensitivity to xenobiotic induced morbidity/mortality'), ('MP:0005378', 'growth/size/body region phenotype'), ('HP:0012533', None), ('MP:0011090', 'perinatal lethality, incomplete penetrance'), ('MP:0000233', 'abnormal blood flow velocity'), ('MP:0009818', 'abnormal thromboxane level'), ('MP:0009814', 'increased prostaglandin level'), ('MP:0011021', 'abnormal circadian regulation of heart rate'), ('MP:0003631', 'nervous system phenotype'), ('HP:0003110', None), ('HP:0011869', None), ('MP:0002736', 'abnormal nociception after inflammation'), ('MP:0003718', 'maternal effect'), ('MP:0002551', 'abnormal blood coagulation')]\nMGI:2442631 Mob1a []\nMGI:109246 Htr4 [('MP:0005449', 'abnormal food intake')]\nMGI:2139793 Cept1 []\nMGI:3650528 Gm13017 []\nMGI:2142885 Cog8 []\nMGI:5452840 Gm23063 []\nMGI:3783117 Gm15675 []\nMGI:5454950 Gm25173 []\nMGI:1860489 Ptbp2 [('HP:0001508', None), ('MP:0002199', 'abnormal brain commissure morphology'), ('MP:0008128', 'abnormal brain internal capsule morphology'), ('HP:0002180', None), ('MP:0008489', 'slow postnatal weight gain'), ('HP:0003811', None), ('MP:0008283', 'small hippocampus'), ('MP:0008458', 'abnormal cortical ventricular zone morphology'), ('MP:0001265', 'decreased body size'), ('MP:0008026', 'abnormal brain white matter morphology'), ('MP:0011087', 'neonatal lethality, complete penetrance'), ('MP:0004981', 'decreased neuronal precursor cell number')]\nMGI:1333759 Vmn1r51 []\nMGI:2138271 Gorab [('MP:0000379', 'decreased hair follicle number'), ('MP:0001243', 'abnormal dermal layer morphology'), ('MP:0009545', 'abnormal dermis papillary layer morphology'), ('HP:0002098', None), ('MP:0001786', 'skin edema')]\nMGI:3588216 Ube2u []\nMGI:4414979 Gm16559 []\nMGI:2443194 Harbi1 []\nMGI:1923822 Rbmxl2 []\nMGI:87886 Chrna2 [('MP:0002945', 'abnormal inhibitory postsynaptic currents'), ('MP:0002910', 'abnormal excitatory postsynaptic currents'), ('MP:0003631', 'nervous system phenotype')]\nMGI:5000466 Anpep [('MP:0003711', 'pathological neovascularization'), ('MP:0002451', 'abnormal macrophage physiology'), ('MP:0005387', 'immune system phenotype'), ('MP:0012075', 'impaired mammary gland growth during pregnancy')]\nMGI:1914588 Rffl []\nMGI:109501 Crat [('HP:0000816', None), ('MP:0005559', 'increased circulating glucose level'), ('MP:0002702', 'decreased circulating free fatty acid level'), ('MP:0004215', 'abnormal myocardial fiber physiology')]\nMGI:3645406 Gm8430 []\nMGI:3645521 Gm4735 []\nMGI:3583959 Fam188b []\nMGI:5477043 Gm26549 []\nMGI:1913839 Ube2v1 []\nMGI:5456093 Gm26316 []\nMGI:3650465 Gm13182 []\nMGI:3649532 Gm13675 []\nMGI:2147707 Kank1 []\nMGI:1202069 Cdk2ap1 [('GO:0001701PHENOTYPE', None), ('GO:0060325PHENOTYPE', None)]\nMGI:1925558 Gpr108 []\nMGI:5455394 Gm25617 []\nMGI:3642592 Gm10603 []\nMGI:3644647 Gm6071 []\nMGI:98224 Saa4 []\nMGI:1924753 Alg9 []\nMGI:3651632 Gm11222 []\nMGI:3826530 Gm16316 []\nMGI:1913417 Tma7 []\nMGI:2442862 Fcrl1 []\nMGI:1918619 Krtap16-3 []\nMGI:2676794 Mirlet7b []\nMGI:3704330 Gm10177 []\nMGI:2444308 Atg4d [('HP:0007703', None)]\nMGI:3651276 Gm11628 []\nMGI:96428 Ifi203 []\nMGI:1349215 Abcd1 [('MP:0008295', 'abnormal adrenal gland zona reticularis morphology'), ('MP:0000136', 'abnormal microglial cell morphology'), ('MP:0003313', 'abnormal locomotor activation'), ('HP:0002143', None), ('MP:0008294', 'abnormal adrenal gland zona fasciculata morphology'), ('MP:0001921', 'reduced fertility'), ('MP:0008288', 'abnormal adrenal cortex morphology'), ('HP:0002446', None), ('MP:0005284', 'increased saturated fatty acid level'), ('MP:0005281', 'increased fatty acid level'), ('MP:0002169', 'no abnormal phenotype detected')]\nMGI:1923558 Rab1b [('MP:0002169', 'no abnormal phenotype detected')]\nMGI:1341879 Zim1 []\nMGI:5453132 Gm23355 []\nMGI:2685446 Nuggc [('MP:0005387', 'immune system phenotype'), ('GO:0016446PHENOTYPE', None)]\nMGI:3650968 Gm12630 []\nMGI:1927667 Defb4 []\nMGI:96930 Mcc [('MP:0002169', 'no abnormal phenotype detected')]\nMGI:1289308 Tax1bp1 [('MP:0008687', 'increased interleukin-2 secretion'), ('MP:0008657', 'increased interleukin-1 beta secretion'), ('MP:0011098', 'embryonic lethality during organogenesis, complete penetrance'), ('MP:0002442', 'abnormal leukocyte physiology'), ('HP:0011017', None), ('HP:0001627', None), ('MP:0001853', 'heart inflammation'), ('MP:0008489', 'slow postnatal weight gain'), ('MP:0000285', 'abnormal heart valve morphology'), ('MP:0008560', 'increased tumor necrosis factor secretion'), ('MP:0002148', 'abnormal hypersensitivity reaction')]\nMGI:109240 Ube2l3 [('MP:0011089', 'perinatal lethality, complete penetrance'), ('MP:0003984', 'embryonic growth retardation'), ('MP:0011109', 'lethality throughout fetal growth and development, incomplete penetrance'), ('MP:0001711', 'abnormal placenta morphology')]\nMGI:2441809 A530099J19Rik []\nMGI:1933114 Bcl9l []\nMGI:1929237 Pus1 [('MP:0004819', 'decreased skeletal muscle mass'), ('HP:0011804', None)]\nMGI:1914498 Rpl39 []\nMGI:2144158 Nudcd3 []\nMGI:3651491 Gm13864 []\nMGI:3705508 Gm14648 []\nMGI:109544 Aim1 [('MP:0009142', 'decreased prepulse inhibition'), ('MP:0013293', 'embryonic lethality prior to tooth bud stage'), ('MP:0013292', 'embryonic lethality prior to organogenesis'), ('MP:0001402', 'hypoactivity'), ('MP:0003232', 'abnormal forebrain development'), ('HP:0045005', None), ('HP:0002143', None), ('MP:0006108', 'abnormal hindbrain development')]\nMGI:5313104 Gm20657 []\nMGI:5452900 Gm23123 []\nMGI:1353636 Abt1 []\nMGI:5477078 Gm26584 []\nMGI:4421975 n-R5s123 []\nMGI:3704189 Rpl31-ps14 []\nMGI:2387430 Trim60 []\nMGI:2138584 Gigyf2 [('GO:0050881PHENOTYPE', None), ('GO:0008344PHENOTYPE', None), ('HP:0004325', None), ('MP:0001258', 'decreased body length'), ('MP:0008946', 'abnormal neuron number'), ('GO:0044267PHENOTYPE', None), ('GO:0035264PHENOTYPE', None), ('HP:0002180', None), ('MP:0005449', 'abnormal food intake'), ('GO:0021522PHENOTYPE', None), ('MP:0001405', 'impaired coordination'), ('MP:0002066', 'abnormal motor capabilities/coordination/movement'), ('MP:0008493', 'alpha-synuclein inclusion body'), ('GO:0009791PHENOTYPE', None)]\nMGI:2388477 Igsf11 []\nMGI:104803 Siae [('MP:0008499', 'increased IgG1 level'), ('HP:0003496', None), ('HP:0006270', None), ('MP:0008501', 'increased IgG2b level'), ('HP:0005403', None), ('MP:0008502', 'increased IgG3 level'), ('MP:0008182', 'decreased marginal zone B cell number'), ('MP:0005154', 'increased B cell proliferation'), ('MP:0004771', 'increased anti-single stranded DNA antibody level'), ('MP:0008500', 'increased IgG2a level'), ('HP:0003212', None), ('MP:0008169', 'increased B-1b cell number'), ('MP:0004762', 'increased anti-double stranded DNA antibody level')]\nMGI:105932 Inhbc [('MP:0005370', 'liver/biliary system phenotype'), ('MP:0002169', 'no abnormal phenotype detected'), ('MP:0005389', 'reproductive system phenotype')]\nMGI:1925678 Pym1 []\nMGI:2142489 Ido2 [('MP:0005616', 'decreased susceptibility to type IV hypersensitivity reaction'), ('MP:0005376', 'homeostasis/metabolism phenotype'), ('MP:0005397', 'hematopoietic system phenotype'), ('MP:0004946', 'abnormal regulatory T cell physiology'), ('MP:0008706', 'decreased interleukin-6 secretion'), ('MP:0008561', 'decreased tumor necrosis factor secretion'), ('HP:0002664', None)]\nMGI:1914346 Mmachc [('MP:0005382', 'craniofacial phenotype'), ('MP:0002989', 'small kidney'), ('HP:0012372', None), ('HP:0000035', None), ('MP:0001697', 'abnormal embryo size'), ('MP:0004002', 'abnormal jejunum morphology'), ('MP:0000702', 'enlarged lymph nodes'), ('MP:0009931', 'abnormal skin appearance'), ('HP:0001789', None), ('HP:0010683', None)]\nMGI:1924356 Tmem181a []\nMGI:3705279 Gm14508 []\nMGI:1916510 Ing2 [('MP:0000693', 'spleen hyperplasia'), ('GO:2001234PHENOTYPE', None), ('MP:0004901', 'decreased male germ cell number'), ('MP:0001154', 'seminiferous tubule degeneration'), ('HP:0012205', None), ('MP:0009321', 'increased histiocytic sarcoma incidence'), ('MP:0009839', 'multiflagellated sperm'), ('MP:0009238', 'coiled sperm flagellum'), ('MP:0010769', 'abnormal survival'), ('HP:0012864', None), ('MP:0006042', 'increased apoptosis'), ('MP:0009239', 'short sperm flagellum'), ('MP:0013538', 'increased Harderian gland adenoma incidence')]\nMGI:3648357 Gm7393 []\nMGI:3779224 Gm11008 []\nMGI:1921519 1700063H04Rik []\nMGI:104717 Meis1 [('GO:0060216PHENOTYPE', None), ('HP:0008063', None), ('MP:0008253', 'absent megakaryocytes'), ('HP:0000980', None), ('MP:0020327', 'abnormal capillary branching pattern'), ('GO:0030097PHENOTYPE', None), ('HP:0001627', None), ('MP:0004809', 'increased hematopoietic stem cell number'), ('MP:0010763', 'abnormal hematopoietic stem cell physiology'), ('HP:0000568', None), ('HP:0025016', None), ('MP:0003227', 'abnormal vascular branching morphogenesis'), ('MP:0003714', 'absent platelets'), ('GO:0007626PHENOTYPE', None), ('MP:0002135', 'abnormal kidney morphology'), ('HP:0002597', None), ('MP:0002398', 'abnormal bone marrow cell morphology/development'), ('HP:0001629', None), ('GO:0048514PHENOTYPE', None)]\nMGI:1354698 Fbxw4 []\nMGI:1352464 Nr1h4 [('MP:0009305', 'decreased retroperitoneal fat pad weight'), ('MP:0003324', 'increased liver adenoma incidence'), ('MP:0009342', 'enlarged gallbladder'), ('MP:0011939', 'increased food intake'), ('HP:0001402', None), ('MP:0009289', 'decreased epididymal fat pad weight'), ('MP:0002944', 'increased lactate dehydrogenase level'), ('MP:0009642', 'abnormal blood homeostasis'), ('HP:0003073', None), ('HP:0040216', None), ('MP:0004774', 'abnormal bile salt level'), ('MP:0008988', 'abnormal liver perisinusoidal space morphology'), ('MP:0002981', 'increased liver weight'), ('MP:0005376', 'homeostasis/metabolism phenotype'), ('MP:0010124', 'decreased bone mineral content'), ('HP:0001392', None), ('MP:0009133', 'decreased white fat cell size'), ('MP:0008019', 'increased liver tumor incidence'), ('HP:0002910', None), ('HP:0005237', None), ('MP:0002941', 'increased circulating alanine transaminase level'), ('HP:0002240', None), ('MP:0005365', 'abnormal bile salt homeostasis'), ('HP:0004326', None), ('HP:0008887', None), ('MP:0008989', 'abnormal liver sinusoid morphology'), ('MP:0010182', 'decreased susceptibility to weight gain'), ('MP:0011941', 'increased fluid intake'), ('HP:0003124', None), ('MP:0010771', 'integument phenotype')]\nMGI:1919338 Ush1c [('MP:0001394', 'circling'), ('MP:0006358', 'absent pinna reflex'), ('HP:0003228', None), ('MP:0010053', 'decreased grip strength'), ('MP:0004522', 'abnormal orientation of cochlear hair cell stereociliary bundles'), ('MP:0004399', 'abnormal cochlear outer hair cell morphology'), ('MP:0008805', 'decreased circulating amylase level'), ('GO:0042491PHENOTYPE', None), ('MP:0004491', 'abnormal orientation of outer hair cell stereociliary bundles'), ('HP:0040216', None), ('MP:0010123', 'increased bone mineral content'), ('MP:0001410', 'head bobbing'), ('HP:0003233', None), ('MP:0001525', 'impaired balance'), ('MP:0004577', 'abnormal cochlear hair cell inter-stereocilial links morphology'), ('MP:0008141', 'decreased small intestinal microvillus size'), ('MP:0004431', 'abnormal hair cell mechanoelectric transduction'), ('MP:0003960', 'increased lean body mass'), ('HP:0008887', None), ('MP:0000043', 'organ of Corti degeneration'), ('HP:0011877', None), ('HP:0000733', None), ('MP:0000495', 'abnormal colon morphology'), ('HP:0000546', None)]\nMGI:3651359 Gm12508 []\nMGI:1916255 Ubald1 []\nMGI:2444853 Ttc26 [('MP:0011086', 'postnatal lethality, incomplete penetrance'), ('MP:0008892', 'abnormal sperm flagellum morphology'), ('MP:0005169', 'abnormal male meiosis'), ('MP:0002948', 'abnormal neuron specification'), ('MP:0003729', 'abnormal photoreceptor outer segment morphology'), ('MP:0002151', 'abnormal neural tube morphology'), ('MP:0002784', 'abnormal Sertoli cell morphology'), ('HP:0010442', None), ('MP:0004131', 'abnormal motile primary cilium morphology'), ('MP:0002084', 'abnormal developmental patterning'), ('MP:0004289', 'abnormal bony labyrinth'), ('HP:0000238', None)]\nMGI:1923434 Abca6 [('HP:0003075', None)]\nMGI:1913904 Tex12 [('HP:0000134', None), ('MP:0005168', 'abnormal female meiosis'), ('HP:0008222', None)]\nMGI:1916796 Mapk1ip1 []\nMGI:4421990 n-R5s134 []\nMGI:3650419 Snrpert []\nMGI:1913608 Adprm []\nMGI:2443738 BC022687 []\nMGI:3643406 Rps3a3 []\nMGI:1921076 Poldip3 []\nMGI:107995 Upf1 [('MP:0001672', 'abnormal embryo development')]\nMGI:3650839 Gm12168 []\nMGI:1914120 Serbp1 []\nMGI:2141314 Naa11 []\nMGI:1922238 4930480G23Rik []\nMGI:4439773 Igkv12-46 []\nMGI:3801742 Gm14925 []\nMGI:3576049 Ugt1a2 []\nMGI:3649338 Gm12219 []\nMGI:1915094 Rab32 [('MP:0011110', 'preweaning lethality, incomplete penetrance')]\nMGI:2683087 Zhx2 [('HP:0010876', None)]\nMGI:4421986 n-R5s130 []\nMGI:2142048 Rnf40 []\nMGI:3649231 Gm13199 []\nMGI:3802009 Gm16165 []\nMGI:5456101 Gm26324 []\nMGI:1924029 Cluap1 [('MP:0011998', 'decreased embryonic cilium length'), ('MP:0000929', 'open neural tube'), ('MP:0003400', 'kinked neural tube'), ('GO:0060972PHENOTYPE', None), ('GO:0021508PHENOTYPE', None), ('MP:0011099', 'lethality throughout fetal growth and development, complete penetrance'), ('MP:0000291', 'enlarged pericardium'), ('MP:0001700', 'abnormal embryo turning'), ('MP:0001265', 'decreased body size'), ('GO:0035082PHENOTYPE', None), ('GO:0007224PHENOTYPE', None), ('MP:0000358', 'abnormal cell morphology'), ('MP:0010117', 'abnormal lateral plate mesoderm morphology'), ('MP:0004132', 'absent embryonic cilia')]\nMGI:1891731 Stub1 [('MP:0009541', 'increased thymocyte apoptosis'), ('MP:0011090', 'perinatal lethality, incomplete penetrance'), ('MP:0004250', 'tau protein deposits'), ('HP:0004325', None), ('MP:0001407', 'short stride length'), ('MP:0001489', 'decreased startle reflex'), ('MP:0005165', 'increased susceptibility to injury'), ('MP:0012307', 'impaired spatial learning'), ('MP:0001921', 'reduced fertility'), ('MP:0005130', 'decreased follicle stimulating hormone level'), ('HP:0000778', None), ('MP:0001648', 'abnormal apoptosis'), ('HP:0011017', None), ('MP:0004852', 'decreased testis weight')]\nMGI:3782724 Gm4540 []\nMGI:2685586 Cyb5d1 [('HP:0003228', None), ('MP:0005561', 'increased mean corpuscular hemoglobin'), ('HP:0008887', None)]\nMGI:2444772 Zhx3 [('MP:0011110', 'preweaning lethality, incomplete penetrance'), ('MP:0010053', 'decreased grip strength'), ('HP:0010504', None), ('HP:0011001', None)]\nMGI:109561 Sp100 []\nMGI:5454663 Gm24886 []\nMGI:97371 Npr1 [('MP:0002083', 'premature death'), ('MP:0000278', 'abnormal myocardial fiber morphology'), ('HP:0030875', None), ('HP:0040171', None), ('MP:0001273', 'decreased metastatic potential'), ('MP:0006144', 'increased systemic arterial systolic blood pressure'), ('HP:0000822', None), ('MP:0006143', 'increased systemic arterial diastolic blood pressure'), ('MP:0002842', 'increased systemic arterial blood pressure'), ('MP:0005647', 'abnormal sex gland physiology'), ('MP:0000230', 'abnormal systemic arterial blood pressure'), ('MP:0001625', 'cardiac hypertrophy'), ('MP:0001613', 'abnormal vasodilation'), ('MP:0001272', 'increased metastatic potential'), ('HP:0001640', None), ('HP:0002647', None), ('MP:0004875', 'increased mean systemic arterial blood pressure'), ('HP:0001635', None), ('MP:0005329', 'abnormal myocardium layer morphology'), ('MP:0009764', 'decreased sensitivity to induced morbidity/mortality'), ('MP:0002843', 'decreased systemic arterial blood pressure')]\nMGI:3641755 Gm10804 []\nMGI:5455565 Gm25788 []\nMGI:2386964 St7l []\nMGI:4421933 n-R5s85 []\nMGI:1336212 Ncr1 [('MP:0005397', 'hematopoietic system phenotype'), ('HP:0012177', None), ('HP:0005415', None), ('GO:0051607PHENOTYPE', None), ('HP:0040218', None), ('MP:0009790', 'decreased susceptibility to viral infection induced morbidity/mortality'), ('MP:0005070', 'impaired natural killer cell mediated cytotoxicity')]\nMGI:1921898 S100pbp []\nMGI:3651313 Gm11824 []\nMGI:5531091 Mir6986 []\nMGI:1915207 March5 []\nMGI:5452882 Gm23105 []\nMGI:5313124 Gm20677 []\nMGI:1097706 Cetn3 []\nMGI:1929008 Copz2 []\nMGI:3650896 Rpl26-ps5 []\nMGI:1298386 Tpd52l1 []\nMGI:1914199 Trim59 []\nMGI:3648831 Rpl31-ps16 []\nMGI:102484 mt-Ti []\nMGI:3646682 Rpl13-ps3 []\nMGI:3650227 Gm13340 []\nMGI:1919027 Ing3 []\nMGI:894668 Serpinb9b []\nMGI:98815 Crisp2 []\nMGI:1915433 Bcas2 []\nMGI:2443764 Iqcb1 []\nMGI:2140175 Ldlrap1 [('HP:0002155', None), ('MP:0000181', 'abnormal circulating LDL cholesterol level'), ('GO:0042632PHENOTYPE', None), ('HP:0003124', None)]\nMGI:1926341 Sult1d1 []\nMGI:98287 Srsf5 []\nMGI:3652015 Gm11410 []\nMGI:3782979 Gm15531 []\nMGI:3641908 Gm10073 []\nMGI:1915500 Fam96a [('MP:0003047', 'abnormal thoracic vertebrae morphology'), ('MP:0010851', 'decreased effector memory CD8-positive, alpha-beta T cell number'), ('HP:0010683', None)]\nMGI:95420 Ces1c [('HP:0002045', None)]\nMGI:3643291 Gm5436 []\nMGI:95402 Epb42 [('HP:0001944', None), ('GO:0048536PHENOTYPE', None), ('MP:0002874', 'decreased hemoglobin content'), ('HP:0004444', None), ('MP:0008809', 'increased spleen iron level'), ('MP:0000208', 'decreased hematocrit'), ('HP:0001923', None)]\nMGI:3648523 Gm5453 []\nMGI:3650664 Gm13050 []\nMGI:5452063 Gm22286 []\nMGI:1929878 Smoc1 [('HP:0001440', None), ('HP:0000609', None), ('HP:0000708', None), ('MP:0000455', 'abnormal maxilla morphology'), ('MP:0011085', 'postnatal lethality, complete penetrance'), ('HP:0002990', None), ('HP:0001508', None), ('MP:0011110', 'preweaning lethality, incomplete penetrance'), ('MP:0000571', 'interdigital webbing'), ('HP:0005922', None), ('HP:0000175', None), ('MP:0009874', 'abnormal interdigital cell death'), ('HP:0000921', None), ('MP:0005201', 'abnormal retinal pigment epithelium morphology'), ('MP:0000556', 'abnormal hindlimb morphology'), ('HP:0012521', None), ('HP:0000546', None), ('HP:0011499', None), ('HP:0002982', None), ('MP:0011087', 'neonatal lethality, complete penetrance'), ('HP:0007973', None)]\nMGI:106362 Sco1 [('MP:0010244', 'decreased kidney copper level'), ('HP:0001392', None), ('HP:0040014', None), ('HP:0001824', None), ('MP:0001265', 'decreased body size'), ('MP:0003067', 'decreased liver copper level'), ('GO:0006878PHENOTYPE', None)]\nMGI:3643695 Gm8055 []\nMGI:2141142 Rpap2 []\nMGI:2447166 Cmtm7 []\nMGI:3650588 Gm12416 []\nMGI:90168 Dcaf11 [('MP:0004647', 'decreased lumbar vertebrae number'), ('HP:0005518', None)]\nMGI:3643810 Zscan18 []\nMGI:5456192 Gm26415 []\nMGI:1919057 Tars2 []\nMGI:1354184 Nox4 [('HP:0002088', None), ('MP:0003223', 'decreased cardiomyocyte apoptosis'), ('HP:0001712', None), ('MP:0006036', 'abnormal mitochondrial physiology'), ('MP:0003141', 'cardiac fibrosis'), ('MP:0004485', 'increased response of heart to induced stress'), ('MP:0003204', 'decreased neuron apoptosis'), ('MP:0010181', 'decreased susceptibility to weight loss'), ('MP:0001625', 'cardiac hypertrophy'), ('MP:0002833', 'increased heart weight'), ('MP:0005598', 'decreased ventricle muscle contractility'), ('MP:0004937', 'dilated heart'), ('MP:0003674', 'oxidative stress'), ('MP:0010724', 'thick interventricular septum'), ('MP:0009764', 'decreased sensitivity to induced morbidity/mortality')]\nMGI:98662 Tec []\nMGI:3648477 Gm14928 []\nMGI:3704451 Gm10076 []\nMGI:5530708 Gm27326 []\nMGI:2444070 Nlrc3 [('MP:0008560', 'increased tumor necrosis factor secretion'), ('MP:0008735', 'increased susceptibility to endotoxin shock'), ('MP:0008553', 'increased circulating tumor necrosis factor level')]\nMGI:3651796 Rps11-ps2 []\nMGI:3643597 Gm8213 []\nMGI:5455373 Gm25596 []\nMGI:3641693 Gm10335 []\nMGI:109505 Xlr3b []\nMGI:1923665 Fam187b []\nMGI:3650221 Gm13339 []\nMGI:1916949 2310079G19Rik []\nMGI:3646811 Gm8624 []\nMGI:5439751 Gm21994 []\nMGI:2148180 Snora69 []\nMGI:5477162 Gm26668 []\nMGI:97286 Ncl []\nMGI:109173 Dsc1 [('HP:0007957', None), ('HP:0004325', None), ('MP:0001194', 'dermatitis'), ('MP:0001195', 'flaky skin'), ('MP:0001236', 'abnormal epidermis stratum spinosum morphology'), ('HP:0001036', None), ('HP:0000962', None), ('HP:0100792', None), ('MP:0001511', 'disheveled coat'), ('MP:0002169', 'no abnormal phenotype detected'), ('MP:0002656', 'abnormal keratinocyte differentiation')]\nMGI:5452841 Gm23064 []\nMGI:5453907 Gm24130 []\nMGI:5453073 Gm23296 []\nMGI:1310002 Fmo1 []\nMGI:3651145 Gm12936 []\nMGI:1923786 Mmadhc []\nMGI:3705734 Gm14681 []\nMGI:2148251 Ddx19b []\nMGI:97279 Nat1 [('MP:0008875', 'abnormal xenobiotic pharmacokinetics'), ('MP:0001921', 'reduced fertility')]\nMGI:3039592 Fads6 []\nMGI:3781533 Gm3355 []\nMGI:4360056 Snora78 []\nMGI:3643039 Gm4852 []\nMGI:1921729 Snx11 []\nMGI:1918648 5430427O19Rik []\nMGI:1929745 Cdc42ep5 []\nMGI:3704272 Gm10443 []\nMGI:1914312 Slc25a53 []\nMGI:1920511 1700037C18Rik []\nMGI:3643536 Gm8508 []\nMGI:101813 Coq3 []\nMGI:2153045 Elmo2 []\nMGI:1918039 Kynu []\nMGI:3650926 Gm14388 []\nMGI:5453975 Gm24198 []\nMGI:3649569 Gm12781 []\nMGI:1201406 Slc10a2 [('HP:0002910', None), ('MP:0004773', 'abnormal bile composition')]\nMGI:2444989 Spg11 [('MP:0009940', 'abnormal hippocampus pyramidal cell morphology'), ('HP:0002062', None), ('MP:0000876', 'Purkinje cell degeneration'), ('HP:0012757', None), ('MP:0003224', 'neuron degeneration'), ('MP:0008260', 'abnormal autophagy'), ('MP:0005058', 'abnormal lysosome morphology'), ('MP:0001405', 'impaired coordination'), ('HP:0001824', None), ('GO:0007040PHENOTYPE', None)]\nMGI:1923511 0610040J01Rik []\nMGI:96213 Hpd [('HP:0003110', None)]\nMGI:3641865 Gm10357 []\nMGI:88192 Smarca4 [('MP:0006126', 'abnormal cardiac outflow tract development'), ('MP:0000740', 'impaired smooth muscle contractility'), ('HP:0030875', None), ('MP:0009476', 'enlarged cecum'), ('GO:0061626PHENOTYPE', None), ('HP:0006517', None), ('MP:0010656', 'thick myocardium'), ('HP:0000347', None), ('MP:0010819', 'primary atelectasis'), ('HP:0000958', None), ('MP:0005092', 'decreased double-positive T cell number'), ('HP:0002669', None), ('HP:0001629', None), ('MP:0002747', 'abnormal aortic valve morphology'), ('MP:0011684', 'coronary-cameral fistula to right ventricle'), ('MP:0000297', 'abnormal atrioventricular cushion morphology'), ('HP:0002623', None), ('MP:0011098', 'embryonic lethality during organogenesis, complete penetrance'), ('HP:0003826', None), ('MP:0002746', 'abnormal semilunar valve morphology'), ('MP:0011094', 'embryonic lethality before implantation, complete penetrance'), ('MP:0010602', 'abnormal pulmonary valve cusp morphology'), ('GO:0048562PHENOTYPE', None), ('HP:0000778', None), ('HP:0011804', None), ('HP:0001640', None), ('GO:0043966PHENOTYPE', None), ('GO:0060318PHENOTYPE', None), ('MP:0005294', 'abnormal heart ventricle morphology'), ('MP:0000727', 'absent CD8-positive, alpha-beta T cells'), ('HP:0001877', None), ('MP:0002796', 'impaired skin barrier function'), ('MP:0002145', 'abnormal T cell differentiation'), ('MP:0000715', 'decreased thymocyte number'), ('MP:0005384', 'cellular phenotype'), ('HP:0001647', None), ('MP:0010595', 'abnormal aortic valve cusp morphology'), ('MP:0008802', 'abnormal intestinal smooth muscle morphology'), ('MP:0008083', 'decreased single-positive T cell number'), ('MP:0004937', 'dilated heart'), ('GO:0001701PHENOTYPE', None), ('GO:0001832PHENOTYPE', None), ('HP:0001635', None), ('MP:0003857', 'abnormal hindlimb zeugopod morphology'), ('MP:0000556', 'abnormal hindlimb morphology'), ('HP:0000528', None), ('MP:0011086', 'postnatal lethality, incomplete penetrance'), ('MP:0010412', 'atrioventricular septal defect'), ('MP:0001196', 'shiny skin'), ('MP:0002731', 'megacolon'), ('GO:0003151PHENOTYPE', None), ('HP:0011121', None), ('MP:0011648', 'thick heart valve cusps'), ('MP:0001883', 'increased mammary adenocarcinoma incidence'), ('MP:0011109', 'lethality throughout fetal growth and development, incomplete penetrance'), ('HP:0011297', None), ('GO:0043066PHENOTYPE', None), ('MP:0008770', 'decreased survivor rate'), ('MP:0002020', 'increased tumor incidence')]\nMGI:3651089 Gm13171 []\nMGI:3802104 Gm16238 []\nMGI:2443361 6430550D23Rik []\nMGI:3707466 Gm15369 []\nMGI:102500 mt-Nd2 []\nMGI:5452861 Gm23084 []\nMGI:1918185 Speer4f1 []\nMGI:5012000 Gm19815 []\nMGI:3704312 Gm10282 []\nMGI:2652836 Gmeb2 []\nMGI:3796835 Gm10509 []\nMGI:3704203 Gm10171 []\nMGI:109351 Slc4a2 [('MP:0011086', 'postnatal lethality, incomplete penetrance'), ('MP:0001541', 'abnormal osteoclast physiology'), ('MP:0002083', 'premature death'), ('HP:0001508', None), ('HP:0003251', None), ('MP:0011090', 'perinatal lethality, incomplete penetrance'), ('MP:0005605', 'increased bone mass'), ('MP:0010878', 'increased trabecular bone volume'), ('MP:0000468', 'abnormal esophageal epithelium morphology'), ('MP:0010868', 'increased bone trabecula number'), ('MP:0013566', 'dilated gastric glands'), ('MP:0001438', 'aphagia'), ('MP:0002661', 'abnormal corpus epididymis morphology'), ('MP:0002662', 'abnormal cauda epididymis morphology'), ('MP:0000133', 'abnormal long bone metaphysis morphology'), ('MP:0009703', 'decreased birth body size'), ('HP:0000365', None), ('MP:0009347', 'increased trabecular bone thickness'), ('HP:0011002', None), ('MP:0002169', 'no abnormal phenotype detected')]\nMGI:3652225 Hnf1aos1 []\nMGI:1889842 Fam13a []\nMGI:2151045 Lsm10 []\nMGI:98430 Sult2a1 []\nMGI:4422060 n-R5s195 []\nMGI:1924963 Peli3 [('MP:0009790', 'decreased susceptibility to viral infection induced morbidity/mortality'), ('HP:0011017', None), ('MP:0002410', 'decreased susceptibility to viral infection'), ('MP:0011072', 'abnormal macrophage cytokine production')]\nMGI:2681861 Proser3 []\nMGI:3647156 Gm6682 []\nMGI:2685270 Pm20d2 []\nMGI:1913869 Atat1 [('MP:0008280', 'abnormal male germ cell apoptosis'), ('MP:0005397', 'hematopoietic system phenotype'), ('MP:0001922', 'reduced male fertility'), ('MP:0005386', 'behavior/neurological phenotype'), ('MP:0002675', 'asthenozoospermia'), ('HP:0012864', None), ('GO:0007283PHENOTYPE', None), ('MP:0004929', 'decreased epididymis weight'), ('MP:0009239', 'short sperm flagellum'), ('HP:0040006', None), ('MP:0002169', 'no abnormal phenotype detected'), ('MP:0004852', 'decreased testis weight')]\nMGI:1925127 6030458C11Rik []\nMGI:2444401 Snrnp200 [('MP:0011100', 'preweaning lethality, complete penetrance'), ('HP:0012165', None), ('MP:0013293', 'embryonic lethality prior to tooth bud stage'), ('MP:0011094', 'embryonic lethality before implantation, complete penetrance')]\nMGI:96161 Hmga1-rs1 []\nMGI:97172 Mt2 [('GO:0006882PHENOTYPE', None)]\nMGI:3649743 Gm12229 []\nMGI:1916560 Pacrg []\nMGI:2676856 Mir192 [('MP:0006315', 'abnormal urine protein level'), ('MP:0002135', 'abnormal kidney morphology'), ('MP:0008874', 'decreased physiological sensitivity to xenobiotic'), ('MP:0011338', 'abnormal mesangial matrix morphology')]\nMGI:3781339 Gm3160 []\nMGI:102480 mt-Tm []\nMGI:3629654 Mir692-2 []\nMGI:1923052 4930458D05Rik []\nMGI:3782854 Gm4673 []\nMGI:1916428 Snx5 [('MP:0005631', 'decreased lung weight'), ('HP:0001508', None), ('MP:0011090', 'perinatal lethality, incomplete penetrance'), ('MP:0011086', 'postnatal lethality, incomplete penetrance'), ('HP:0004325', None), ('MP:0002942', 'decreased circulating alanine transaminase level'), ('HP:0003468', None), ('MP:0000183', 'decreased circulating LDL cholesterol level'), ('HP:0009887', None), ('MP:0001265', 'decreased body size'), ('HP:0000961', None), ('HP:0003233', None), ('HP:0003146', None)]\nMGI:4421994 n-R5s138 []\nMGI:1915562 Gstm7 []\nMGI:1289301 Ubxn1 []\nMGI:1913664 Ndufa12 []\nMGI:1920708 Aldh3b3 []\nMGI:3647161 Mup-ps6 []\nMGI:3641838 Gm10247 []\nMGI:1196620 Hpn [('HP:0010679', None), ('HP:0012153', None), ('MP:0002855', 'abnormal cochlear ganglion morphology'), ('GO:0007605PHENOTYPE', None), ('MP:0003149', 'abnormal tectorial membrane morphology'), ('MP:0005471', 'decreased thyroxine level'), ('MP:0011967', 'increased or absent threshold for auditory brainstem response'), ('MP:0004434', 'abnormal cochlear outer hair cell physiology'), ('HP:0000365', None), ('HP:0000821', None), ('MP:0001158', 'abnormal prostate gland morphology')]\nMGI:2144724 Ttc7b []\nMGI:3705630 Gm15190 []\nMGI:3045379 4732419C18Rik []\nMGI:3781169 Gm2991 []\nMGI:1916856 Mtfmt [('MP:0011100', 'preweaning lethality, complete penetrance')]\nMGI:88373 Cebpb [('MP:0003355', 'decreased ovulation rate'), ('MP:0002083', 'premature death'), ('MP:0008472', 'abnormal spleen secondary B follicle morphology'), ('MP:0002357', 'abnormal spleen white pulp morphology'), ('HP:0003237', None), ('MP:0005666', 'abnormal adipose tissue physiology'), ('MP:0008395', 'abnormal osteoblast differentiation'), ('HP:0000962', None), ('MP:0004130', 'abnormal muscle cell glucose uptake'), ('MP:0010172', 'abnormal mammary gland epithelium physiology'), ('MP:0000702', 'enlarged lymph nodes'), ('MP:0001129', 'impaired ovarian folliculogenesis'), ('MP:0000628', 'abnormal mammary gland development'), ('MP:0002500', 'granulomatous inflammation'), ('MP:0009114', 'decreased pancreatic beta cell mass'), ('MP:0002451', 'abnormal macrophage physiology'), ('HP:0011842', None), ('MP:0001222', 'epidermal hyperplasia'), ('MP:0008944', 'decreased sensitivity to induced cell death'), ('MP:0002702', 'decreased circulating free fatty acid level'), ('MP:0010868', 'increased bone trabecula number'), ('MP:0013239', 'impaired skeletal muscle regeneration'), ('MP:0002344', 'abnormal lymph node B cell domain morphology'), ('GO:0060644PHENOTYPE', None), ('MP:0003058', 'increased insulin secretion'), ('MP:0002343', 'abnormal lymph node cortex morphology'), ('HP:0100249', None), ('MP:0002971', 'abnormal brown adipose tissue morphology'), ('MP:0005639', 'hemosiderosis'), ('MP:0005399', 'increased susceptibility to fungal infection'), ('MP:0004502', 'decreased incidence of tumors by chemical induction'), ('HP:0040216', None), ('MP:0005466', 'abnormal T-helper 2 physiology'), ('MP:0002742', 'enlarged submandibular lymph nodes'), ('MP:0004985', 'decreased osteoclast cell number'), ('MP:0011427', 'mesangial cell hyperplasia'), ('MP:0003383', 'abnormal gluconeogenesis'), ('MP:0000321', 'increased bone marrow cell number'), ('MP:0009419', 'skeletal muscle fibrosis'), ('MP:0008734', 'decreased susceptibility to endotoxin shock'), ('MP:0009307', 'decreased uterine fat pad weight'), ('MP:0005006', 'abnormal osteoblast physiology'), ('MP:0010876', 'decreased bone volume'), ('HP:0001967', None), ('MP:0008596', 'increased circulating interleukin-6 level'), ('MP:0005319', 'abnormal enzyme/coenzyme level'), ('MP:0002169', 'no abnormal phenotype detected'), ('MP:0008663', 'increased interleukin-12 secretion'), ('MP:0008873', 'increased physiological sensitivity to xenobiotic'), ('MP:0011086', 'postnatal lethality, incomplete penetrance'), ('MP:0002421', 'abnormal cell-mediated immunity'), ('MP:0004817', 'abnormal skeletal muscle mass'), ('MP:0002347', 'abnormal lymph node T cell domain morphology'), ('MP:0008033', 'impaired lipolysis'), ('MP:0020080', 'increased bone mineralization'), ('HP:0001547', None), ('MP:0011224', 'abnormal lymph node medullary cord morphology'), ('HP:0002718', None), ('MP:0002346', 'abnormal lymph node secondary follicle morphology'), ('MP:0005517', 'decreased liver regeneration'), ('MP:0009347', 'increased trabecular bone thickness'), ('HP:0001978', None), ('MP:0001882', 'abnormal lactation'), ('MP:0000352', 'decreased cell proliferation'), ('MP:0008618', 'decreased circulating interleukin-12 level')]\nMGI:5454350 Gm24573 []\nMGI:3036289 Cd200r4 []\nMGI:3652158 Gm13416 []\nMGI:1277962 Hus1 [('MP:0011204', 'abnormal visceral yolk sac blood island morphology'), ('MP:0003229', 'abnormal vitelline vasculature morphology'), ('GO:0006468PHENOTYPE', None), ('MP:0003984', 'embryonic growth retardation'), ('MP:0004030', 'induced chromosome breakage'), ('MP:0003400', 'kinked neural tube'), ('MP:0011098', 'embryonic lethality during organogenesis, complete penetrance'), ('MP:0003396', 'abnormal embryonic hematopoiesis'), ('MP:0001700', 'abnormal embryo turning'), ('MP:0002151', 'abnormal neural tube morphology'), ('MP:0004573', 'absent limb buds'), ('MP:0002084', 'abnormal developmental patterning'), ('MP:0001688', 'abnormal somite development'), ('MP:0003674', 'oxidative stress'), ('MP:0013504', 'increased embryonic tissue cell apoptosis'), ('MP:0001726', 'abnormal allantois morphology'), ('GO:0000077PHENOTYPE', None), ('MP:0009657', 'failure of chorioallantoic fusion')]\nMGI:3819487 Scarna6 []\nMGI:104899 Mob4 []\nMGI:5454701 Gm24924 []\nMGI:95797 Gpi1 [('MP:0011086', 'postnatal lethality, incomplete penetrance'), ('MP:0002981', 'increased liver weight'), ('GO:0006096PHENOTYPE', None), ('MP:0011091', 'prenatal lethality, complete penetrance'), ('MP:0002085', 'abnormal embryonic tissue morphology'), ('MP:0010831', 'lethality, incomplete penetrance'), ('MP:0003656', 'abnormal erythrocyte physiology'), ('MP:0003657', 'abnormal erythrocyte osmotic lysis'), ('HP:0012379', None), ('MP:0004952', 'increased spleen weight'), ('HP:0001923', None), ('MP:0002833', 'increased heart weight'), ('GO:0001701PHENOTYPE', None), ('MP:0011095', 'embryonic lethality between implantation and placentation, complete penetrance'), ('GO:0042593PHENOTYPE', None), ('GO:0001707PHENOTYPE', None)]\nMGI:3046938 Hrnr []\nMGI:3045249 A830005F24Rik [('MP:0002135', 'abnormal kidney morphology'), ('HP:0001640', None)]\nMGI:1196398 Champ1 []\nMGI:5454116 Gm24339 []\nMGI:1921772 Morc2a []\nMGI:88474 Cox5a []\nMGI:95628 Slc6a12 []\nMGI:4821183 Trim12c []\nMGI:3629885 Mir677 []\nMGI:3642236 Rps23-ps2 []\nMGI:1915968 Rnf166 []\nMGI:3033475 Vmn1r65 []\nMGI:3648836 Gm5822 []\nMGI:2148802 Sec16b [('HP:0003146', None)]\nMGI:2685483 Gsg1l []\nMGI:3651112 Gm11889 []\nMGI:95781 Gnb1 [('MP:0011090', 'perinatal lethality, incomplete penetrance'), ('MP:0004948', 'abnormal neuronal precursor proliferation'), ('MP:0006254', 'thin cerebral cortex'), ('MP:0009937', 'abnormal neuron differentiation'), ('HP:0001322', None), ('MP:0008458', 'abnormal cortical ventricular zone morphology'), ('HP:0002033', None)]\nMGI:88593 Cyp24a1 [('MP:0002135', 'abnormal kidney morphology'), ('MP:0002705', 'dilated renal tubules'), ('GO:0042359PHENOTYPE', None), ('MP:0011228', 'abnormal vitamin D level'), ('HP:0003072', None)]\nMGI:4415007 Gm16587 []\nMGI:3643792 Gm5449 []\nMGI:1349452 Polr3e []\nMGI:3641978 Gm10010 []\nMGI:3704348 Gm10052 []\nMGI:5455459 Gm25682 []\nMGI:1919140 Mad2l2 [('HP:0000135', None), ('HP:0004325', None), ('HP:0008669', None), ('MP:0000746', 'weakness'), ('MP:0005389', 'reproductive system phenotype'), ('MP:0004901', 'decreased male germ cell number'), ('MP:0003202', 'abnormal neuron apoptosis'), ('MP:0012167', 'abnormal epigenetic regulation of gene expression'), ('MP:0005384', 'cellular phenotype'), ('MP:0000339', 'decreased enterocyte cell number'), ('HP:0010791', None), ('HP:0001511', None), ('MP:0002777', 'absent ovarian follicles'), ('MP:0001155', 'arrest of spermatogenesis'), ('MP:0008392', 'decreased primordial germ cell number'), ('MP:0003631', 'nervous system phenotype'), ('MP:0002216', 'abnormal seminiferous tubule morphology'), ('MP:0004200', 'decreased fetal size'), ('MP:0008393', 'absent primordial germ cells'), ('MP:0008489', 'slow postnatal weight gain'), ('MP:0001265', 'decreased body size'), ('MP:0004852', 'decreased testis weight')]\nMGI:3709612 Gm14760 []\nMGI:3650350 Gm14277 []\nMGI:1338883 Gfpt2 [('HP:0006482', None), ('HP:0000517', None)]\nMGI:1202295 Entpd6 [('MP:0010124', 'decreased bone mineral content'), ('MP:0003443', 'increased circulating glycerol level'), ('HP:0003330', None), ('HP:0008887', None), ('MP:0010123', 'increased bone mineral content'), ('MP:0003442', 'decreased circulating glycerol level')]\nMGI:1924555 Wdr82 []\nMGI:5454235 Gm24458 []\nMGI:95666 Gbp2b [('MP:0002451', 'abnormal macrophage physiology'), ('HP:0002718', None), ('HP:0002014', None), ('HP:0001824', None), ('MP:0009788', 'increased susceptibility to bacterial infection induced morbidity/mortality')]\nMGI:1289225 Tmem41b []\nMGI:1891012 F12 [('MP:0003075', 'altered response to CNS ischemic injury'), ('GO:0005615PHENOTYPE', None), ('GO:0008233PHENOTYPE', None)]\nMGI:3650211 Gm12732 []\nMGI:3782988 Gm15540 []\nMGI:1919006 Cpn2 []\nMGI:1919216 Nkiras2 []\nMGI:894649 Ppfibp2 []\nMGI:3702953 Gm13594 []\nMGI:3583899 A330069E16Rik []\nMGI:1926421 Tcerg1 []\nMGI:108020 Ear2 []\nMGI:3650318 Gm14387 []\nMGI:3646830 Gm5135 []\nMGI:5456149 Gm26372 []\nMGI:3045359 C130073F10Rik []\nMGI:3802159 Gm15889 []\nMGI:1096345 Gckr [('HP:0040216', None), ('MP:0005439', 'decreased glycogen level'), ('MP:0005376', 'homeostasis/metabolism phenotype'), ('HP:0000833', None)]\nMGI:5455606 Gm25829 []\nMGI:87995 Aldob [('MP:0001415', 'increased exploration in new environment'), ('HP:0010679', None), ('HP:0001397', None), ('MP:0003921', 'abnormal heart left ventricle morphology'), ('HP:0001392', None), ('MP:0003961', 'decreased lean body mass'), ('MP:0011967', 'increased or absent threshold for auditory brainstem response'), ('MP:0000603', 'pale liver'), ('MP:0001265', 'decreased body size'), ('HP:0012115', None)]\nMGI:88285 Cbs [('MP:0002083', 'premature death'), ('HP:0004325', None), ('MP:0010098', 'abnormal retinal blood vessel pattern'), ('HP:0000568', None), ('HP:0000962', None), ('MP:0001613', 'abnormal vasodilation'), ('MP:0009018', 'short estrus'), ('MP:0001176', 'abnormal lung development'), ('MP:0008751', 'abnormal interleukin level'), ('MP:0014179', 'abnormal blood-retinal barrier function'), ('MP:0003070', 'increased vascular permeability'), ('MP:0001935', 'decreased litter size'), ('HP:0001397', None), ('MP:0002109', 'abnormal limb morphology'), ('MP:0009392', 'retinal gliosis'), ('MP:0003674', 'oxidative stress'), ('MP:0005639', 'hemosiderosis'), ('MP:0000652', 'enlarged sebaceous gland'), ('MP:0005186', 'increased circulating progesterone level'), ('MP:0008957', 'abnormal placenta junctional zone morphology'), ('GO:0001974PHENOTYPE', None), ('MP:0002182', 'abnormal astrocyte morphology'), ('HP:0001999', None), ('MP:0004126', 'thin hypodermis'), ('MP:0009020', 'prolonged metestrus'), ('HP:0012115', None), ('MP:0011086', 'postnatal lethality, incomplete penetrance'), ('GO:0001958PHENOTYPE', None), ('MP:0000377', 'abnormal hair follicle morphology'), ('MP:0005386', 'behavior/neurological phenotype'), ('MP:0002699', 'abnormal vitreous body morphology'), ('MP:0001286', 'abnormal eye development'), ('MP:0006076', 'abnormal circulating homocysteine level'), ('MP:0004921', 'decreased placenta weight'), ('MP:0010452', 'retina microaneurysm'), ('MP:0000528', 'delayed kidney development'), ('MP:0010771', 'integument phenotype'), ('MP:0001716', 'abnormal placenta labyrinth morphology'), ('MP:0002656', 'abnormal keratinocyte differentiation')]\nMGI:5454894 Gm25117 []\nMGI:1913509 Camk2n1 []\nMGI:2138939 Nat10 [('MP:0011100', 'preweaning lethality, complete penetrance'), ('MP:0008044', 'increased NK cell number')]\nMGI:1923616 Mtif3 []\nMGI:2135611 Immp2l [('GO:0007420PHENOTYPE', None), ('GO:0006974PHENOTYPE', None), ('MP:0001132', 'absent mature ovarian follicles'), ('MP:0001921', 'reduced fertility'), ('MP:0001125', 'abnormal oocyte morphology'), ('MP:0010954', 'abnormal cellular respiration'), ('GO:0007283PHENOTYPE', None), ('HP:0000798', None), ('GO:0008015PHENOTYPE', None), ('MP:0001129', 'impaired ovarian folliculogenesis')]\nMGI:1921063 Fam83e []\nMGI:3646407 Gm7363 []\nMGI:3643534 Angptl8 [('HP:0004325', None), ('MP:0005376', 'homeostasis/metabolism phenotype'), ('MP:0011578', 'increased lipoprotein lipase activity'), ('HP:0003119', None), ('HP:0008887', None), ('MP:0008489', 'slow postnatal weight gain'), ('MP:0003976', 'decreased circulating VLDL triglyceride level')]\nMGI:96777 Lgals1 [('HP:0011113', None), ('MP:0005386', 'behavior/neurological phenotype'), ('MP:0001973', 'increased thermal nociceptive threshold'), ('MP:0008566', 'increased interferon-gamma secretion'), ('HP:0040006', None), ('MP:0000972', 'abnormal mechanoreceptor morphology')]\nMGI:1922646 Spp2 []\nMGI:95286 Eed [('MP:0000371', 'diluted coat color'), ('MP:0002427', 'disproportionate dwarf'), ('GO:0016571PHENOTYPE', None)]\nMGI:2668347 C8a []\nMGI:1927186 Nt5c3 []\nMGI:3647055 Gm8580 []\nMGI:3781012 Gm2840 []\nMGI:3648232 Gm7855 []\nMGI:1931502 Snord82 []\nMGI:1919489 Parp14 [('HP:0002720', None), ('MP:0008782', 'increased B cell apoptosis'), ('MP:0008182', 'decreased marginal zone B cell number')]\nMGI:98818 Trap1a []\nMGI:5454156 Gm24379 []\nMGI:3651445 Gm13203 []\nMGI:3648396 Gm6472 []\nMGI:5663350 Gm43213 []\nMGI:5454727 Gm24950 []\nMGI:2442293 Usp28 [('MP:0009339', 'decreased splenocyte number'), ('MP:0005387', 'immune system phenotype')]\nMGI:1341217 Uba3 [('MP:0002718', 'abnormal inner cell mass morphology'), ('GO:0051726PHENOTYPE', None), ('MP:0002085', 'abnormal embryonic tissue morphology'), ('MP:0003988', 'disorganized embryonic tissue'), ('GO:0000278PHENOTYPE', None), ('MP:0010380', 'abnormal inner cell mass apoptosis'), ('MP:0003693', 'abnormal blastocyst hatching'), ('MP:0011095', 'embryonic lethality between implantation and placentation, complete penetrance')]\nMGI:3037679 Gm1821 []\nENSEMBL:ENSMUSG00000098615 Arvcf []\nMGI:1914015 4933411K16Rik []\nMGI:1352502 Limd1 [('MP:0001541', 'abnormal osteoclast physiology')]\nMGI:5521099 Gtpbp4-ps2 []\nMGI:3705843 Mup-ps17 []\nMGI:1261856 Ankle2 []\nMGI:1915602 Aspdh []\nMGI:1860517 Rdh7 []\nMGI:1917113 Ttc39b []\nMGI:1915393 Herpud2 []\nMGI:1917143 Coa7 []\nMGI:2151104 Akr1c20 []\nMGI:4938054 Gm17227 []\nMGI:3801875 Gm15883 []\nMGI:109581 S100a13 []\nMGI:5452348 Gm22571 []\nMGI:1347046 Rfwd2 [('MP:0000267', 'abnormal heart development'), ('HP:0004325', None), ('MP:0009219', 'increased prostate intraepithelial neoplasia incidence'), ('MP:0009321', 'increased histiocytic sarcoma incidence'), ('MP:0011109', 'lethality throughout fetal growth and development, incomplete penetrance'), ('HP:0010784', None), ('HP:0100616', None), ('MP:0003607', 'abnormal prostate gland physiology')]\nMGI:3646644 Gm6576 []\nMGI:1202713 Rhag [('MP:0009642', 'abnormal blood homeostasis'), ('MP:0003656', 'abnormal erythrocyte physiology'), ('MP:0008810', 'increased circulating iron level'), ('GO:0048821PHENOTYPE', None)]\nMGI:5452921 Gm23144 []\nMGI:107547 Zfp101 []\nMGI:3649792 Rpl27-ps2 []\nMGI:5452524 Gm22747 []\nMGI:3649506 Llph-ps1 []\nMGI:3783061 Gm15616 []\nMGI:4360046 Snora73a []\nMGI:3651860 Gm11336 []\nMGI:1915566 Apoo []\nMGI:1916109 Smim1 []\nMGI:3576090 Ugt1a8 []\nMGI:3704338 Gm10642 []\nMGI:1353620 Slamf6 [('MP:0008553', 'increased circulating tumor necrosis factor level'), ('HP:0011990', None)]\nMGI:1917111 2010003K11Rik []\nMGI:1196217 Tcaim [('HP:0003330', None)]\nMGI:5434131 Gm20775 []\nMGI:3646854 Gm8832 []\nMGI:3643856 Gm7582 []\nMGI:3646827 Rps6-ps2 []\nMGI:5456238 Gm26461 []\nMGI:3648525 Kansl2-ps []\nMGI:2147897 Cenpi []\nMGI:3652241 Gm11964 []\nMGI:1920912 Apol7c []\nMGI:5454316 Gm24539 []\nMGI:3644080 Gm14650 []\nMGI:5451813 Gm22036 []\nMGI:2449316 Syne2 [('MP:0008511', 'thin retinal inner nuclear layer'), ('GO:0005635PHENOTYPE', None), ('MP:0008582', 'short photoreceptor inner segment'), ('MP:0003732', 'abnormal retinal outer plexiform layer morphology'), ('MP:0008587', 'short photoreceptor outer segment'), ('GO:0034504PHENOTYPE', None), ('MP:0005391', 'vision/eye phenotype'), ('HP:0000962', None), ('MP:0004022', 'abnormal cone electrophysiology'), ('MP:0006068', 'abnormal horizontal cell morphology'), ('MP:0005369', 'muscle phenotype'), ('MP:0005547', 'abnormal Muller cell morphology'), ('GO:0006998PHENOTYPE', None), ('MP:0006072', 'abnormal retinal apoptosis'), ('MP:0002655', 'abnormal keratinocyte morphology'), ('MP:0001053', 'abnormal neuromuscular synapse morphology'), ('MP:0004021', 'abnormal rod electrophysiology')]\nMGI:1923805 Mmaa []\nMGI:1930124 Apom [('MP:0003983', 'decreased cholesterol level'), ('HP:0100878', None), ('HP:0003233', None)]\nMGI:1914492 Gemin6 []\nMGI:1342058 B4galnt2 [('HP:0010975', None)]\nMGI:1918367 Rbbp5 []\nMGI:4439831 Gm16907 []\nMGI:101775 Cd80 [('MP:0001800', 'abnormal humoral immune response'), ('MP:0008497', 'decreased IgG2b level'), ('MP:0004804', 'decreased susceptibility to autoimmune diabetes'), ('MP:0005376', 'homeostasis/metabolism phenotype'), ('HP:0004313', None), ('MP:0005042', 'abnormal level of surface class II molecules'), ('HP:0011839', None), ('MP:0008207', 'decreased B-2 B cell number'), ('MP:0008495', 'decreased IgG1 level'), ('MP:0004031', 'insulitis'), ('MP:0008498', 'decreased IgG3 level')]\nMGI:3804971 Gm5561 []\nMGI:892001 Slc22a6 [('MP:0006272', 'abnormal urine organic anion level'), ('MP:0008874', 'decreased physiological sensitivity to xenobiotic')]\nMGI:88583 Cyp11b1 [('MP:0011546', 'increased urine progesterone level'), ('MP:0011541', 'decreased urine aldosterone level'), ('MP:0011550', 'decreased urine corticosterone level'), ('MP:0008294', 'abnormal adrenal gland zona fasciculata morphology'), ('HP:0008221', None), ('MP:0009092', 'endometrium hyperplasia'), ('MP:0002833', 'increased heart weight'), ('HP:0100878', None), ('HP:0008222', None), ('HP:0000833', None)]\nMGI:2442326 Zbtb1 [('MP:0008075', 'decreased CD4-positive, alpha beta T cell number'), ('HP:0005403', None), ('MP:0000703', 'abnormal thymus morphology'), ('MP:0002145', 'abnormal T cell differentiation'), ('GO:0030183PHENOTYPE', None), ('MP:0010136', 'decreased DN4 thymocyte number'), ('HP:0002846', None), ('HP:0010978', None), ('MP:0010134', 'decreased DN3 thymocyte number'), ('GO:0033077PHENOTYPE', None), ('MP:0010132', 'decreased DN2 thymocyte number'), ('HP:0005404', None), ('MP:0005092', 'decreased double-positive T cell number'), ('HP:0010976', None)]\nMGI:1916296 Isca1 [('MP:0013278', 'decreased fasted circulating glucose level'), ('MP:0001417', 'decreased exploration in new environment'), ('MP:0013293', 'embryonic lethality prior to tooth bud stage'), ('MP:0011275', 'abnormal behavioral response to light'), ('MP:0002757', 'decreased vertical activity')]\nMGI:1353563 Snai3 []\nMGI:3644565 Gm8730 []\nMGI:2442610 A330035P11Rik []\nMGI:1919290 Cdhr5 []\nMGI:1346344 Nr0b2 [('MP:0002646', 'increased intestinal cholesterol absorption'), ('MP:0004884', 'abnormal testis physiology'), ('MP:0005365', 'abnormal bile salt homeostasis'), ('MP:0005376', 'homeostasis/metabolism phenotype'), ('MP:0004928', 'increased epididymis weight'), ('MP:0009355', 'increased liver triglyceride level'), ('HP:0003233', None), ('MP:0000183', 'decreased circulating LDL cholesterol level'), ('MP:0010180', 'increased susceptibility to weight loss'), ('MP:0004852', 'decreased testis weight'), ('MP:0004789', 'increased bile salt level'), ('MP:0002169', 'no abnormal phenotype detected'), ('HP:0003146', None)]\nMGI:1918632 Pex1 [('MP:0005282', 'decreased fatty acid level'), ('HP:0001397', None), ('HP:0001408', None), ('MP:0006084', 'abnormal circulating phospholipid level'), ('MP:0004022', 'abnormal cone electrophysiology'), ('MP:0005365', 'abnormal bile salt homeostasis'), ('MP:0004021', 'abnormal rod electrophysiology')]\nMGI:3714859 Cyp3a41b []\nMGI:96108 Hlf []\nMGI:1888505 Retnlb [('MP:0008561', 'decreased tumor necrosis factor secretion'), ('MP:0005026', 'decreased susceptibility to parasitic infection'), ('MP:0001663', 'abnormal digestive system physiology'), ('MP:0008499', 'increased IgG1 level'), ('MP:0008537', 'increased susceptibility to induced colitis')]\nMGI:1913489 Rsrc2 []\nMGI:3649326 Gm14448 []\nMGI:3649364 Atp5l2-ps []\nMGI:1913697 Mgst3 [('HP:0005518', None)]\nMGI:2685505 C2cd4d []\nMGI:1920999 Ttc7 [('MP:0010176', 'dacryocytosis'), ('MP:0002082', 'postnatal lethality'), ('MP:0009395', 'increased nucleated erythrocyte cell number'), ('HP:0004325', None), ('MP:0011242', 'increased fetal derived definitive erythrocyte cell number'), ('MP:0008476', 'increased spleen red pulp amount'), ('HP:0001006', None), ('MP:0008479', 'decreased spleen white pulp amount'), ('MP:0001586', 'abnormal erythrocyte cell number'), ('HP:0001923', None), ('MP:0002594', 'low mean erythrocyte cell number'), ('MP:0001246', 'mixed cellular infiltration to dermis'), ('MP:0011413', 'colorless urine'), ('MP:0008810', 'increased circulating iron level'), ('HP:0040162', None), ('MP:0002655', 'abnormal keratinocyte morphology'), ('MP:0000245', 'abnormal erythropoiesis'), ('HP:0001595', None), ('HP:0000980', None), ('HP:0001877', None), ('MP:0001243', 'abnormal dermal layer morphology'), ('HP:0005548', None), ('HP:0011273', None), ('MP:0005097', 'polychromatophilia'), ('MP:0004969', 'pale kidney'), ('HP:0004447', None), ('MP:0000472', 'abnormal stomach non-glandular epithelium morphology'), ('HP:0002240', None), ('MP:0000607', 'abnormal hepatocyte morphology'), ('MP:0002123', 'abnormal definitive hematopoiesis'), ('HP:0012115', None), ('HP:0001927', None), ('HP:0001036', None), ('MP:0002591', 'decreased mean corpuscular volume'), ('MP:0010771', 'integument phenotype'), ('GO:0030097PHENOTYPE', None)]\nMGI:3644887 Gm5812 []\nMGI:5454222 Gm24445 []\nMGI:3704357 Gm9855 []\nMGI:1351634 Abcc6 [('MP:0005239', 'abnormal Bruch membrane morphology'), ('HP:0003761', None), ('MP:0010234', 'abnormal vibrissa follicle morphology'), ('MP:0002838', 'decreased susceptibility to dystrophic cardiac calcinosis'), ('HP:0007862', None), ('MP:0002839', 'increased susceptibility to dystrophic cardiac calcinosis')]\nMGI:99175 Zfp28 []\nMGI:3649924 Gm13237 []\nMGI:2137092 Eefsec []\nMGI:3783082 Gm15638 []\nMGI:1919356 Jmjd8 [('HP:0005518', None)]\nMGI:3647036 Gm5931 []\nMGI:3651480 Gm11759 []\nMGI:1330304 Vps52 [('MP:0002085', 'abnormal embryonic tissue morphology'), ('MP:0001723', 'disorganized yolk sac vascular plexus'), ('MP:0011092', 'embryonic lethality, complete penetrance')]\nMGI:1916964 Tfpt []\nMGI:97350 Nkx2-5 [('MP:0006126', 'abnormal cardiac outflow tract development'), ('MP:0000298', 'absent atrioventricular cushions'), ('HP:0001903', None), ('HP:0012722', None), ('GO:0001947PHENOTYPE', None), ('MP:0005385', 'cardiovascular system phenotype'), ('HP:0000969', None), ('GO:0055005PHENOTYPE', None), ('MP:0001722', 'pale yolk sac'), ('MP:0002086', 'abnormal extraembryonic tissue morphology'), ('MP:0001625', 'cardiac hypertrophy'), ('GO:0060038PHENOTYPE', None), ('HP:0009555', None), ('GO:0060048PHENOTYPE', None), ('MP:0002747', 'abnormal aortic valve morphology'), ('HP:0001698', None), ('MP:0002085', 'abnormal embryonic tissue morphology'), ('HP:0003826', None), ('MP:0004114', 'abnormal atrioventricular node morphology'), ('MP:0001633', 'poor circulation'), ('HP:0001644', None), ('HP:0001750', None), ('GO:0007507PHENOTYPE', None), ('MP:0001689', 'incomplete somite formation'), ('MP:0011205', 'excessive folding of visceral yolk sac'), ('HP:0001719', None), ('HP:0001647', None), ('MP:0000282', 'abnormal interatrial septum morphology'), ('MP:0004117', 'abnormal atrioventricular bundle morphology'), ('MP:0000729', 'abnormal myogenesis'), ('MP:0005598', 'decreased ventricle muscle contractility'), ('MP:0003649', 'decreased heart right ventricle size'), ('HP:0001660', None), ('MP:0000278', 'abnormal myocardial fiber morphology'), ('MP:0004787', 'abnormal dorsal aorta morphology'), ('MP:0004124', 'abnormal Purkinje fiber morphology'), ('MP:0003872', 'absent heart right ventricle'), ('MP:0003984', 'embryonic growth retardation'), ('GO:0001570PHENOTYPE', None), ('MP:0006113', 'abnormal heart septum morphology'), ('MP:0003920', 'abnormal heart right ventricle morphology'), ('HP:0000961', None), ('HP:0001789', None), ('MP:0003140', 'dilated heart atrium')]\nMGI:95895 H2-Aa [('MP:0000702', 'enlarged lymph nodes'), ('MP:0008075', 'decreased CD4-positive, alpha beta T cell number'), ('MP:0008078', 'increased CD8-positive, alpha-beta T cell number')]\nMGI:5454177 Gm24400 []\nMGI:3780551 Gm2383 []\nMGI:2146156 Espl1 [('MP:0012570', 'increased mammary gland tumor incidence in breeding females'), ('MP:0002052', 'decreased tumor incidence'), ('MP:0000630', 'mammary gland hyperplasia'), ('MP:0004957', 'abnormal blastocyst morphology'), ('MP:0004024', 'aneuploidy'), ('MP:0008393', 'absent primordial germ cells'), ('HP:0008669', None), ('MP:0000333', 'decreased bone marrow cell number'), ('MP:0006271', 'abnormal involution of the mammary gland'), ('MP:0011411', 'abnormal gonadal ridge morphology'), ('MP:0004966', 'abnormal inner cell mass proliferation'), ('MP:0008866', 'chromosomal instability'), ('MP:0000607', 'abnormal hepatocyte morphology'), ('MP:0011092', 'embryonic lethality, complete penetrance'), ('MP:0008390', 'abnormal primordial germ cell proliferation')]\nMGI:2144585 Slc16a6 []\nMGI:1923388 Ccdc138 []\nMGI:1917059 1810046K07Rik []\nMGI:3710580 Gm9800 []\nMGI:1928138 Mrps23 []\nMGI:1298204 Ppt1 [('MP:0001513', 'limb grasping'), ('GO:0008344PHENOTYPE', None), ('HP:0004324', None), ('GO:0044257PHENOTYPE', None), ('HP:0001743', None), ('HP:0001392', None), ('HP:0012443', None), ('MP:0002175', 'decreased brain weight'), ('GO:0008306PHENOTYPE', None), ('MP:0000788', 'abnormal cerebral cortex morphology'), ('GO:0008474PHENOTYPE', None), ('GO:0044265PHENOTYPE', None), ('MP:0008713', 'abnormal cytokine level'), ('HP:0002446', None), ('GO:0005764PHENOTYPE', None), ('MP:0002135', 'abnormal kidney morphology'), ('HP:0010831', None), ('HP:0000718', None), ('MP:0003241', 'loss of cortex neurons')]\nMGI:1923764 Tmub1 [('MP:0008853', 'decreased abdominal adipose tissue amount'), ('HP:0012311', None), ('MP:0001501', 'abnormal sleep pattern')]\nMGI:3650962 Mup6 []\nMGI:5455456 Gm25679 []\nMGI:95679 Gpd1 [('GO:0006116PHENOTYPE', None), ('MP:0005378', 'growth/size/body region phenotype'), ('MP:0005389', 'reproductive system phenotype')]\nMGI:2451333 Khnyn []\nMGI:3783040 Gm15593 []\nMGI:1918920 Acy3 []\nMGI:5456164 Gm26387 []\nMGI:3652074 Gm11367 []\nMGI:4422070 n-R5s205 []\nMGI:1098258 Kif15 []\nMGI:1923028 Them4 [('MP:0005389', 'reproductive system phenotype')]\nMGI:3649491 Gm11852 []\nMGI:1914691 Isoc2b []\nMGI:96692 Krt18 [('GO:0097191PHENOTYPE', None), ('HP:0002240', None)]\nMGI:2676630 Nlrp12 [('MP:0005616', 'decreased susceptibility to type IV hypersensitivity reaction'), ('GO:0071345PHENOTYPE', None), ('HP:0040238', None), ('HP:0001875', None)]\nMGI:1918115 Lnpk [('HP:0003010', None), ('HP:0003251', None), ('HP:0005736', None), ('MP:0002109', 'abnormal limb morphology'), ('MP:0004355', 'short radius'), ('GO:0007596PHENOTYPE', None), ('MP:0003072', 'abnormal metatarsal bone morphology'), ('MP:0000060', 'delayed bone ossification'), ('MP:0002187', 'abnormal fibula morphology')]\nMGI:3643566 Gm5617 []\nMGI:2139354 Arfgef2 [('HP:0002269', None), ('HP:0004298', None), ('MP:0003631', 'nervous system phenotype'), ('MP:0011094', 'embryonic lethality before implantation, complete penetrance')]\nMGI:2443362 Lax1 [('MP:0002359', 'abnormal spleen germinal center morphology'), ('HP:0003212', None), ('MP:0001828', 'abnormal T cell activation'), ('MP:0008826', 'abnormal splenic cell ratio'), ('MP:0008499', 'increased IgG1 level')]\nMGI:2685466 1700024P16Rik []\nMGI:96177 Hoxa5 [('GO:0001501PHENOTYPE', None), ('MP:0003120', 'abnormal tracheal cartilage morphology'), ('MP:0010993', 'decreased surfactant secretion'), ('MP:0009018', 'short estrus'), ('MP:0001179', 'thick pulmonary interalveolar septum'), ('GO:0007389PHENOTYPE', None), ('MP:0009247', 'meteorism'), ('MP:0010943', 'abnormal bronchus epithelium morphology'), ('MP:0002267', 'abnormal bronchiole morphology'), ('MP:0013497', 'trachea occlusion'), ('MP:0001927', 'abnormal estrous cycle'), ('MP:0005122', 'increased circulating thyroid-stimulating hormone level'), ('HP:0000826', None), ('MP:0004615', 'cervical vertebral transformation'), ('MP:0011034', 'impaired branching involved in respiratory bronchiole morphogenesis'), ('MP:0004780', 'abnormal surfactant secretion'), ('GO:0060536PHENOTYPE', None), ('MP:0004551', 'decreased tracheal cartilage ring number'), ('GO:0030324PHENOTYPE', None), ('MP:0008146', 'asymmetric sternocostal joints'), ('GO:0060644PHENOTYPE', None), ('MP:0005388', 'respiratory system phenotype'), ('HP:0040006', None), ('HP:0000772', None), ('MP:0010900', 'abnormal pulmonary interalveolar septum morphology'), ('MP:0011088', 'neonatal lethality, incomplete penetrance'), ('MP:0000054', 'delayed ear emergence'), ('GO:0007585PHENOTYPE', None), ('HP:0000138', None), ('GO:0009952PHENOTYPE', None), ('HP:0002777', None), ('MP:0001183', 'overexpanded pulmonary alveoli'), ('HP:0005815', None), ('MP:0005629', 'abnormal lung weight'), ('MP:0009020', 'prolonged metestrus'), ('MP:0011086', 'postnatal lethality, incomplete penetrance'), ('HP:0001508', None), ('MP:0011090', 'perinatal lethality, incomplete penetrance'), ('GO:0030878PHENOTYPE', None), ('MP:0010911', 'abnormal pulmonary acinus morphology'), ('MP:0010082', 'sternebra fusion'), ('MP:0010810', 'increased type II pneumocyte number'), ('HP:0100750', None), ('MP:0011024', 'abnormal branching involved in lung morphogenesis'), ('GO:0048704PHENOTYPE', None), ('MP:0008245', 'abnormal alveolar macrophage morphology'), ('MP:0001290', 'delayed eyelid opening')]\nMGI:104992 Crybb1 []\nMGI:96114 Hmgb1-ps8 []\nMGI:1932389 Tlr9 [('MP:0005465', 'abnormal T-helper 1 physiology'), ('GO:0006955PHENOTYPE', None), ('MP:0008769', 'abnormal plasmacytoid dendritic cell physiology'), ('HP:0011113', None), ('MP:0008561', 'decreased tumor necrosis factor secretion'), ('MP:0005387', 'immune system phenotype'), ('MP:0008565', 'decreased interferon-beta secretion'), ('HP:0010978', None), ('MP:0002418', 'increased susceptibility to viral infection'), ('HP:0012648', None), ('MP:0002376', 'abnormal dendritic cell physiology')]\nMGI:97372 Npr2 [('HP:0011028', None), ('MP:0000097', 'short maxilla'), ('MP:0002427', 'disproportionate dwarf'), ('HP:0004325', None), ('MP:0005385', 'cardiovascular system phenotype'), ('MP:0001921', 'reduced fertility'), ('MP:0004386', 'enlarged interparietal bone'), ('HP:0003026', None), ('MP:0009677', 'abnormal spinal cord dorsal column morphology'), ('HP:0002983', None), ('HP:0003097', None), ('HP:0005105', None), ('HP:0000134', None), ('HP:0003270', None), ('MP:0006398', 'increased long bone epiphyseal plate size'), ('MP:0000165', 'abnormal long bone hypertrophic chondrocyte zone'), ('MP:0002637', 'small uterus'), ('MP:0010029', 'abnormal basicranium morphology'), ('MP:0004421', 'enlarged parietal bone'), ('MP:0005390', 'skeleton phenotype'), ('MP:0009088', 'thin uterine horn'), ('GO:0060348PHENOTYPE', None), ('HP:0002808', None), ('HP:0010504', None), ('HP:0040006', None), ('HP:0004422', None), ('HP:0009121', None), ('HP:0100671', None), ('HP:0000689', None), ('MP:0009861', 'abnormal pyloric sphincter morphology'), ('HP:0002766', None), ('HP:0000267', None), ('MP:0000592', 'short tail'), ('MP:0001392', 'abnormal locomotor behavior'), ('HP:0010824', None), ('HP:0003022', None), ('MP:0003662', 'abnormal long bone epiphyseal plate proliferative zone'), ('MP:0004470', 'small nasal bone'), ('HP:0002578', None), ('MP:0004595', 'abnormal mandibular condyloid process morphology'), ('MP:0000438', 'abnormal cranium morphology'), ('MP:0009009', 'absent estrous cycle'), ('HP:0001508', None), ('HP:0001291', None), ('HP:0005736', None), ('MP:0010155', 'abnormal intestine physiology'), ('MP:0004673', 'splayed ribs'), ('MP:0002113', 'abnormal skeleton development'), ('MP:0004607', 'abnormal cervical atlas morphology'), ('MP:0001265', 'decreased body size'), ('MP:0008272', 'abnormal endochondral bone ossification'), ('MP:0008770', 'decreased survivor rate'), ('HP:0011063', None), ('MP:0002657', 'chondrodystrophy')]\nMGI:95564 Fmr1 [('MP:0001363', 'increased anxiety-related response'), ('MP:0008871', 'abnormal ovarian follicle number'), ('HP:0000137', None), ('MP:0001458', 'abnormal object recognition memory'), ('MP:0009456', 'impaired cued conditioning behavior'), ('MP:0000877', 'abnormal Purkinje cell morphology'), ('MP:0002574', 'increased vertical activity'), ('MP:0002062', 'abnormal associative learning'), ('MP:0001364', 'decreased anxiety-related response'), ('MP:0009940', 'abnormal hippocampus pyramidal cell morphology'), ('MP:0005391', 'vision/eye phenotype'), ('MP:0003008', 'enhanced long term potentiation'), ('MP:0000947', 'convulsive seizures'), ('MP:0001463', 'abnormal spatial learning'), ('MP:0012144', 'decreased b wave amplitude'), ('MP:0001529', 'abnormal vocalization'), ('MP:0009363', 'abnormal secondary ovarian follicle morphology'), ('HP:0007973', None), ('MP:0005168', 'abnormal female meiosis'), ('HP:0004324', None), ('MP:0000812', 'abnormal dentate gyrus morphology'), ('MP:0012143', 'decreased a wave amplitude'), ('HP:0000138', None), ('MP:0002063', 'abnormal learning/memory/conditioning'), ('MP:0001415', 'increased exploration in new environment'), ('MP:0003702', 'abnormal chromosome morphology'), ('HP:0012443', None), ('MP:0004753', 'abnormal miniature excitatory postsynaptic currents'), ('MP:0009141', 'increased prepulse inhibition'), ('MP:0009937', 'abnormal neuron differentiation'), ('MP:0001360', 'abnormal social investigation'), ('MP:0002680', 'decreased corpora lutea number'), ('MP:0004851', 'increased testis weight'), ('GO:0007417PHENOTYPE', None), ('MP:0009712', 'impaired conditioned place preference behavior')]\nMGI:5454704 Gm24927 []\nMGI:3645626 Gm16433 []\nMGI:3645402 Gm6733 []\nMGI:5452049 Gm22272 []\nMGI:3645731 Gm5641 []\nMGI:3649615 Gm13443 []\nMGI:1926262 Irgm2 []\nMGI:5452146 Gm22369 []\nMGI:3773841 Mfsd4b3 []\nMGI:1202882 Rhd [('MP:0002591', 'decreased mean corpuscular volume'), ('MP:0002874', 'decreased hemoglobin content'), ('MP:0001191', 'abnormal skin condition'), ('MP:0005642', 'decreased mean corpuscular hemoglobin concentration')]\nMGI:2387602 Cd300e [('MP:0005560', 'decreased circulating glucose level'), ('MP:0002059', 'abnormal seminal vesicle morphology'), ('MP:0004931', 'enlarged epididymis')]\nMGI:2685565 Nup62cl []\nMGI:3783067 Gm15622 []\nMGI:3649359 Gm13886 []\nMGI:1919266 1600002H07Rik []\nMGI:1925579 2310010J17Rik []\nMGI:2663233 Muc6 []\nMGI:102944 Ube2b [('MP:0009230', 'abnormal sperm head morphology'), ('MP:0001510', 'abnormal coat appearance'), ('HP:0012153', None), ('MP:0005169', 'abnormal male meiosis'), ('MP:0011110', 'preweaning lethality, incomplete penetrance'), ('HP:0008669', None), ('HP:0001288', None), ('MP:0004852', 'decreased testis weight'), ('MP:0006379', 'abnormal spermatocyte morphology')]\nMGI:3648443 Gm7847 []\nMGI:3644644 Gm5883 []\nMGI:104664 Tcf15 [('GO:0042755PHENOTYPE', None), ('GO:0007517PHENOTYPE', None), ('GO:0043588PHENOTYPE', None), ('HP:0000973', None), ('GO:0001756PHENOTYPE', None), ('HP:0003468', None), ('HP:0004298', None), ('GO:0045198PHENOTYPE', None), ('MP:0004263', 'abnormal limb posture'), ('HP:0002098', None), ('MP:0002280', 'abnormal intercostal muscle morphology'), ('HP:0000921', None), ('GO:0009952PHENOTYPE', None), ('HP:0000892', None), ('MP:0004073', 'caudal body truncation'), ('GO:0003016PHENOTYPE', None), ('MP:0008277', 'abnormal sternum ossification'), ('MP:0000080', 'abnormal exoccipital bone morphology'), ('HP:0000902', None), ('HP:0000772', None)]\nMGI:3643873 Gm6594 []\nMGI:1921737 5430405H02Rik []\nMGI:95936 H2-Q7 []\nMGI:5454456 Gm24679 []\nMGI:1096341 E2f2 [('MP:0000313', 'abnormal cell death'), ('HP:0002090', None), ('MP:0002145', 'abnormal T cell differentiation'), ('HP:0000099', None), ('MP:0005350', 'increased susceptibility to autoimmune disorder'), ('MP:0004952', 'increased spleen weight'), ('MP:0003944', 'abnormal T cell subpopulation ratio'), ('MP:0011294', 'renal glomerulus hypertrophy'), ('HP:0011017', None), ('MP:0004762', 'increased anti-double stranded DNA antibody level')]\nMGI:3645972 Sult3a2 []\nMGI:3651135 Gm12909 []\nMGI:3646434 Gm4788 []\nMGI:1923455 Stard3nl []\nMGI:5454201 Gm24424 []\nMGI:1921080 Eif3k []\nMGI:2444899 5031439G07Rik []\nMGI:1919311 2010111I01Rik []\nMGI:1920179 Dip2c []\nMGI:1891713 Habp4 []\nMGI:3649539 Gm13880 []\nMGI:3036263 Zfp976 []\nMGI:3649578 Gm13623 []\nMGI:1919352 Dusp11 [('HP:0100689', None), ('MP:0001258', 'decreased body length')]\nMGI:98797 Tpi1 [('HP:0012379', None), ('HP:0001878', None), ('MP:0000208', 'decreased hematocrit'), ('MP:0011096', 'embryonic lethality between implantation and somite formation, complete penetrance')]\nMGI:1913513 1810014B01Rik []\nMGI:1861676 Nme6 [('MP:0003984', 'embryonic growth retardation'), ('MP:0013292', 'embryonic lethality prior to organogenesis'), ('MP:0001697', 'abnormal embryo size'), ('MP:0011100', 'preweaning lethality, complete penetrance')]\nMGI:2449929 Il1f9 []\nMGI:1913288 Sdhaf3 []\nMGI:3783232 Gm15790 []\nMGI:3648120 Gm5637 []\nMGI:1923666 Znrd1as []\nMGI:3769707 Cyp3a59 []\nMGI:3780787 Gm2619 []\nMGI:1860079 Olfr70 []\nMGI:1346527 Psmb8 [('MP:0001838', 'defective intracellular transport of class I molecules')]\nMGI:3649746 Gm12230 []\nMGI:5531106 Mir6963 []\nMGI:95629 Slc6a13 []\nMGI:2178743 Stab2 [('MP:0009642', 'abnormal blood homeostasis')]\nMGI:3644670 Gm6142 []\nMGI:3781329 Gm3150 []\nMGI:3648278 Gm7541 []\nMGI:5454607 Gm24830 []\nMGI:3649931 Gm11361 []\nMGI:2146052 Agxt2 [('MP:0005590', 'increased vasodilation'), ('HP:0003112', None)]\nMGI:1101758 H3f3a-ps2 []\nMGI:3651322 Gm12328 []\nMGI:3708691 Gm2564 []\nMGI:1333825 Dgat1 [('MP:0010026', 'decreased liver cholesterol level'), ('MP:0011232', 'abnormal vitamin A level'), ('MP:0001212', 'skin lesions'), ('MP:0001222', 'epidermal hyperplasia'), ('MP:0004047', 'abnormal milk composition'), ('HP:0004325', None), ('MP:0006270', 'abnormal mammary gland growth during lactation'), ('MP:0005459', 'decreased percent body fat/body weight'), ('MP:0008858', 'abnormal hair cycle anagen phase'), ('MP:0009504', 'abnormal mammary gland epithelium morphology'), ('MP:0005289', 'increased oxygen consumption'), ('HP:0000752', None), ('MP:0009356', 'decreased liver triglyceride level'), ('MP:0001191', 'abnormal skin condition'), ('MP:0000427', 'abnormal hair cycle'), ('MP:0000628', 'abnormal mammary gland development'), ('MP:0012075', 'impaired mammary gland growth during pregnancy')]\nMGI:1915720 Impad1 [('HP:0002795', None), ('HP:0005792', None), ('HP:0004324', None), ('GO:0001501PHENOTYPE', None), ('GO:0002063PHENOTYPE', None), ('GO:0001958PHENOTYPE', None), ('HP:0009121', None), ('MP:0002427', 'disproportionate dwarf'), ('HP:0000879', None), ('HP:0003826', None), ('HP:0001547', None), ('HP:0002098', None), ('MP:0005390', 'skeleton phenotype'), ('HP:0000267', None), ('HP:0008873', None), ('MP:0000071', 'axial skeleton hypoplasia'), ('MP:0006397', 'disorganized long bone epiphyseal plate'), ('MP:0011087', 'neonatal lethality, complete penetrance'), ('GO:0009791PHENOTYPE', None)]\nMGI:5455884 Gm26107 []\nMGI:3652203 Gm13604 []\nMGI:1918943 Colec11 []\nMGI:894297 Clta []\nMGI:1276535 Ncoa3 [('MP:0003355', 'decreased ovulation rate'), ('MP:0005379', 'endocrine/exocrine gland phenotype'), ('HP:0009124', None), ('MP:0012106', 'impaired exercise endurance'), ('MP:0002427', 'disproportionate dwarf'), ('MP:0009006', 'prolonged estrous cycle'), ('MP:0011110', 'preweaning lethality, incomplete penetrance'), ('GO:0035264PHENOTYPE', None), ('MP:0005290', 'decreased oxygen consumption'), ('MP:0005076', 'abnormal cell differentiation'), ('HP:0006482', None), ('MP:0002401', 'abnormal lymphopoiesis'), ('HP:0000823', None), ('MP:0006319', 'abnormal epididymal fat pad morphology'), ('MP:0009136', 'decreased brown fat cell size'), ('MP:0005181', 'decreased circulating estradiol level'), ('HP:0030759', None), ('MP:0005289', 'increased oxygen consumption'), ('MP:0002356', 'abnormal spleen red pulp morphology'), ('HP:0000855', None), ('HP:0040216', None), ('MP:0002696', 'decreased circulating glucagon level'), ('HP:0001974', None), ('HP:0004324', None), ('MP:0005376', 'homeostasis/metabolism phenotype'), ('HP:0003074', None), ('MP:0009133', 'decreased white fat cell size'), ('MP:0009341', 'decreased splenocyte apoptosis'), ('MP:0005154', 'increased B cell proliferation'), ('MP:0005560', 'decreased circulating glucose level'), ('MP:0008705', 'increased interleukin-6 secretion'), ('MP:0008483', 'increased spleen germinal center size'), ('MP:0008596', 'increased circulating interleukin-6 level'), ('MP:0003402', 'decreased liver weight'), ('HP:0011017', None), ('GO:0060068PHENOTYPE', None), ('MP:0002169', 'no abnormal phenotype detected'), ('HP:0010683', None), ('MP:0008074', 'increased CD4-positive, alpha beta T cell number'), ('HP:0001508', None), ('MP:0008735', 'increased susceptibility to endotoxin shock'), ('HP:0000369', None), ('MP:0008553', 'increased circulating tumor necrosis factor level'), ('HP:0002045', None), ('MP:0011049', 'impaired adaptive thermogenesis'), ('MP:0008641', 'increased circulating interleukin-1 beta level'), ('MP:0011630', 'increased mitochondria size'), ('HP:0008887', None), ('HP:0002846', None), ('GO:0048589PHENOTYPE', None), ('MP:0008658', 'decreased interleukin-1 beta secretion'), ('MP:0001265', 'decreased body size'), ('MP:0010378', 'increased respiratory quotient'), ('MP:0001780', 'decreased brown adipose tissue amount'), ('HP:0002665', None), ('MP:0000352', 'decreased cell proliferation'), ('MP:0000628', 'abnormal mammary gland development'), ('MP:0005292', 'improved glucose tolerance')]\nMGI:3772572 Xaf1 []\nMGI:1922471 Dpp3 []\nMGI:5452893 Gm23116 []\nMGI:1351627 Pdhx []\nMGI:1920081 Dhx30 [('MP:0004180', 'failure of initiation of embryo turning')]\nMGI:3651835 Gm13994 []\nMGI:108450 Adcy9 [('MP:0011110', 'preweaning lethality, incomplete penetrance'), ('HP:0003113', None)]\nMGI:3652298 Gm11349 []\nMGI:3648635 Gm8815 []\nMGI:3782393 Gm4217 []\nMGI:104982 Cebpg [('MP:0001183', 'overexpanded pulmonary alveoli'), ('MP:0005070', 'impaired natural killer cell mediated cytotoxicity'), ('MP:0011088', 'neonatal lethality, incomplete penetrance')]\nMGI:3650608 Gm11942 []\nMGI:2136934 Clec2h []\nMGI:1329005 Slfn3 []\nMGI:1915921 Pcyt2 [('MP:0011092', 'embryonic lethality, complete penetrance'), ('MP:0002118', 'abnormal lipid homeostasis'), ('MP:0010027', 'increased liver cholesterol level'), ('MP:0009355', 'increased liver triglyceride level')]\nMGI:1918937 Tmem25 []\nMGI:96031 Hc [('MP:0005376', 'homeostasis/metabolism phenotype'), ('MP:0001952', 'increased airway responsiveness'), ('MP:0005166', 'decreased susceptibility to injury'), ('MP:0011471', 'decreased urine creatinine level'), ('MP:0002833', 'increased heart weight'), ('MP:0011396', 'abnormal sleep behavior'), ('MP:0001257', 'increased body length')]\nMGI:3650694 Gm11979 []\nMGI:3614797 Ripply1 [('MP:0003048', 'abnormal cervical vertebrae morphology'), ('GO:0060349PHENOTYPE', None)]\nMGI:3652153 Gm12568 []\nMGI:97517 Cdk17 []\nMGI:3588218 F830016B08Rik []\nMGI:102483 mt-Tk [('HP:0001324', None), ('MP:0010955', 'abnormal respiratory electron transport chain'), ('MP:0001258', 'decreased body length')]\nMGI:1341190 Hmg20b []\nMGI:3651366 Gm13132 []\nMGI:108520 Fzd4 [('MP:0002083', 'premature death'), ('GO:0030947PHENOTYPE', None), ('MP:0004362', 'cochlear hair cell degeneration'), ('MP:0004404', 'cochlear outer hair cell degeneration'), ('MP:0004398', 'cochlear inner hair cell degeneration'), ('MP:0004368', 'abnormal stria vascularis vasculature morphology'), ('GO:0031987PHENOTYPE', None), ('HP:0000568', None), ('MP:0005602', 'decreased angiogenesis'), ('MP:0009619', 'abnormal optokinetic reflex'), ('HP:0001321', None), ('MP:0002792', 'abnormal retinal vasculature morphology'), ('MP:0000880', 'decreased Purkinje cell number'), ('HP:0007033', None), ('HP:0000573', None), ('MP:0004363', 'stria vascularis degeneration'), ('MP:0005185', 'decreased circulating progesterone level'), ('MP:0000469', 'abnormal esophageal squamous epithelium morphology'), ('MP:0001505', 'hunched posture'), ('HP:0001892', None), ('GO:0042701PHENOTYPE', None), ('HP:0012372', None), ('HP:0200057', None), ('GO:0001568PHENOTYPE', None), ('MP:0000259', 'abnormal vascular development'), ('HP:0008222', None), ('MP:0002169', 'no abnormal phenotype detected'), ('GO:0007605PHENOTYPE', None), ('MP:0000886', 'abnormal cerebellar granule layer morphology'), ('MP:0009402', 'decreased skeletal muscle fiber diameter'), ('MP:0003356', 'impaired luteinization'), ('HP:0000365', None), ('HP:0001789', None)]\nMGI:99260 Prkci [('MP:0012104', 'small amniotic cavity'), ('MP:0003949', 'abnormal circulating lipid level'), ('MP:0002083', 'premature death'), ('MP:0008140', 'podocyte foot process effacement'), ('HP:0003259', None), ('HP:0000568', None), ('MP:0011190', 'thick embryonic epiblast'), ('MP:0008060', 'abnormal podocyte slit diaphragm morphology'), ('MP:0004185', 'abnormal adipocyte glucose uptake'), ('MP:0004559', 'small allantois'), ('MP:0011402', 'renal cast'), ('MP:0011257', 'abnormal head fold morphology'), ('MP:0005669', 'increased circulating leptin level'), ('MP:0005325', 'abnormal renal glomerulus morphology'), ('MP:0011092', 'embryonic lethality, complete penetrance'), ('MP:0000358', 'abnormal cell morphology'), ('HP:0003233', None), ('MP:0012081', 'absent heart tube'), ('HP:0003138', None), ('MP:0000270', 'abnormal heart tube morphology'), ('MP:0011108', 'embryonic lethality during organogenesis, incomplete penetrance'), ('MP:0011483', 'renal glomerular synechia'), ('HP:0001967', None), ('MP:0001705', 'abnormal proximal-distal axis patterning'), ('MP:0001680', 'abnormal mesoderm development'), ('MP:0001318', 'pupil opacity'), ('MP:0001320', 'small pupils'), ('HP:0000238', None), ('HP:0000517', None), ('MP:0011869', 'detached podocyte'), ('MP:0005199', 'abnormal iris pigment epithelium'), ('HP:0000096', None), ('HP:0001513', None)]\nMGI:3643265 Gm9143 []\nMGI:96923 Mbl1 [('MP:0008497', 'decreased IgG2b level'), ('MP:0005387', 'immune system phenotype'), ('MP:0008502', 'increased IgG3 level'), ('HP:0010701', None), ('MP:0008554', 'decreased circulating tumor necrosis factor level'), ('MP:0009764', 'decreased sensitivity to induced morbidity/mortality')]\nMGI:1929216 Ap3s1-ps1 []\nMGI:1915650 0610043K17Rik []\nMGI:5530764 Mir8114 []\nMGI:3705130 Gm13166 []\nMGI:3704354 Gm10275 []\nMGI:1916846 Ap5s1 []\nMGI:3650439 Amd-ps4 []\nMGI:5477020 Gm26526 []\nMGI:95453 Smarcad1 [('HP:0030040', None), ('MP:0004174', 'abnormal spine curvature'), ('HP:0100891', None), ('MP:0011088', 'neonatal lethality, incomplete penetrance'), ('HP:0009121', None), ('HP:0000902', None), ('MP:0004200', 'decreased fetal size'), ('MP:0008146', 'asymmetric sternocostal joints'), ('MP:0001625', 'cardiac hypertrophy'), ('MP:0009703', 'decreased birth body size'), ('MP:0000462', 'abnormal digestive system morphology'), ('MP:0002161', 'abnormal fertility/fecundity'), ('MP:0002169', 'no abnormal phenotype detected'), ('MP:0001923', 'reduced female fertility')]\nMGI:1919004 Cyp2d40 []\nMGI:2442798 Dhrs9 []\nMGI:2443063 Mylk3 [('MP:0003915', 'increased left ventricle weight'), ('HP:0001712', None), ('MP:0009763', 'increased sensitivity to induced morbidity/mortality'), ('MP:0005385', 'cardiovascular system phenotype'), ('MP:0012735', 'abnormal response to exercise'), ('MP:0010632', 'cardiac muscle necrosis'), ('MP:0010724', 'thick interventricular septum'), ('MP:0002833', 'increased heart weight'), ('HP:0001635', None), ('MP:0005608', 'cardiac interstitial fibrosis'), ('MP:0010754', 'abnormal heart left ventricle pressure')]\nMGI:4834226 Mir3058 []\nMGI:3836982 Mir1943 []\nMGI:1306806 Cyp2c37 []\nMGI:105388 Cdkn2c [('MP:0000693', 'spleen hyperplasia'), ('MP:0002083', 'premature death'), ('HP:0000803', None), ('HP:0012505', None), ('HP:0001251', None), ('MP:0002364', 'abnormal thymus size'), ('MP:0000702', 'enlarged lymph nodes'), ('HP:0040171', None), ('MP:0006262', 'increased testis tumor incidence'), ('MP:0002048', 'increased lung adenoma incidence'), ('MP:0002041', 'increased pituitary adenoma incidence'), ('MP:0010299', 'increased mammary gland tumor incidence'), ('HP:0010516', None), ('HP:0000053', None), ('HP:0004324', None), ('MP:0013602', 'abnormal Leydig cell differentiation'), ('MP:0000889', 'abnormal cerebellar molecular layer'), ('HP:0200058', None), ('MP:0002348', 'abnormal lymph node medulla morphology'), ('MP:0000630', 'mammary gland hyperplasia'), ('MP:0001264', 'increased body size'), ('HP:0005404', None), ('MP:0000523', 'cortical renal glomerulopathies')]\nMGI:2448399 Hist1h2bk []\nMGI:3651950 Gm12435 []\nMGI:1351670 Hsd17b6 []\nMGI:3781871 Gm3695 []\nMGI:3649989 Rpl12-ps2 []\nMGI:3648820 Gm7129 []\nMGI:3651867 Gm13641 []\nMGI:1914948 Fam174a []\nMGI:3650794 Gm12271 []\nMGI:1914742 Zfand4 []\nMGI:2682935 Tmprss13 [('MP:0005376', 'homeostasis/metabolism phenotype'), ('MP:0001240', 'abnormal epidermis stratum corneum morphology'), ('MP:0001282', 'short vibrissae'), ('MP:0002169', 'no abnormal phenotype detected')]\nMGI:1309467 Gstm6 []\nMGI:3644053 Gm7676 []\nMGI:97976 Rnu2-10 []\nMGI:88328 Tnfsf8 [('GO:0045944PHENOTYPE', None), ('GO:0043374PHENOTYPE', None), ('MP:0005673', 'decreased susceptibility to graft versus host disease')]\nMGI:1913470 Zdhhc12 []\nMGI:3650076 Gm14400 []\nMGI:5453858 Gm24081 []\nMGI:1915690 Dusp23 []\nMGI:1098773 Tmem141 []\nMGI:3652187 Rps15a-ps4 []\nMGI:95486 Fbl [('MP:0011101', 'prenatal lethality, incomplete penetrance'), ('MP:0001730', 'embryonic growth arrest'), ('MP:0011094', 'embryonic lethality before implantation, complete penetrance')]\nMGI:5453488 Gm23711 []\nMGI:2387617 Obp2a [('HP:0009887', None)]\nMGI:3651166 Gm11843 []\nMGI:1313291 Vezf1 [('HP:0100763', None), ('MP:0011090', 'perinatal lethality, incomplete penetrance'), ('MP:0005380', 'embryo phenotype'), ('MP:0002672', 'abnormal pharyngeal arch artery morphology'), ('MP:0000260', 'abnormal angiogenesis'), ('GO:0001885PHENOTYPE', None), ('MP:0011109', 'lethality throughout fetal growth and development, incomplete penetrance'), ('MP:0001698', 'decreased embryo size'), ('HP:0011029', None), ('HP:0002597', None), ('MP:0000265', 'atretic vasculature'), ('GO:0001525PHENOTYPE', None)]\nMGI:3650537 Gm14165 []\nMGI:98304 St3gal1 []\nMGI:3648857 Gm6430 []\nMGI:3704220 Gm9762 []\nMGI:5456234 Gm26457 []\nMGI:106025 Gucy2g [('MP:0005367', 'renal/urinary system phenotype')]\nMGI:3780904 Gm2735 []\nMGI:105112 Hpx [('GO:0051246PHENOTYPE', None), ('MP:0012666', 'increased circulating haptoglobin level'), ('GO:0020027PHENOTYPE', None)]\nMGI:1888594 Icmt [('MP:0012235', 'abnormal liver bud morphology'), ('HP:0000980', None), ('GO:0008104PHENOTYPE', None), ('MP:0011098', 'embryonic lethality during organogenesis, complete penetrance'), ('MP:0012504', 'increased forebrain apoptosis'), ('MP:0000596', 'abnormal liver development'), ('MP:0001265', 'decreased body size'), ('GO:0001701PHENOTYPE', None), ('MP:0000352', 'decreased cell proliferation')]\nMGI:3780699 Gm2531 []\nMGI:1915071 Atp1b4 []\nMGI:1916078 Sync [('MP:0002841', 'impaired skeletal muscle contractility'), ('MP:0005369', 'muscle phenotype')]\nMGI:5453021 Gm23244 []\nMGI:1336893 Rnu12 []\nMGI:2158650 Idh3b []\nMGI:1929763 Cdc42ep1 []\nMGI:1919007 Selenoo []\nMGI:2181053 Dnajc28 [('MP:0011968', 'decreased threshold for auditory brainstem response'), ('MP:0002574', 'increased vertical activity'), ('MP:0002942', 'decreased circulating alanine transaminase level')]\nMGI:2442106 Lrtm1 []\nMGI:1916233 Rint1 [('HP:0001402', None), ('HP:0001028', None), ('MP:0002083', 'premature death'), ('MP:0013293', 'embryonic lethality prior to tooth bud stage'), ('MP:0003570', 'increased uterus leiomyoma incidence'), ('HP:0002904', None), ('MP:0002020', 'increased tumor incidence'), ('MP:0002702', 'decreased circulating free fatty acid level'), ('MP:0013488', 'increased keratoacanthoma incidence'), ('MP:0002014', 'increased papilloma incidence'), ('MP:0010299', 'increased mammary gland tumor incidence'), ('HP:0100615', None), ('MP:0011096', 'embryonic lethality between implantation and somite formation, complete penetrance'), ('HP:0006739', None)]\nMGI:2684939 Fam170a []\nMGI:3649320 Gm12912 []\nMGI:1915265 Trap1 [('MP:0000693', 'spleen hyperplasia'), ('HP:0004325', None), ('HP:0001392', None), ('MP:0010959', 'abnormal oxidative phosphorylation'), ('MP:0003674', 'oxidative stress'), ('HP:0012648', None), ('MP:0002052', 'decreased tumor incidence')]\nMGI:1858414 Ngef []\nMGI:109330 Prop1 [('HP:0000135', None), ('MP:0002083', 'premature death'), ('MP:0008338', 'decreased thyrotroph cell number'), ('MP:0002270', 'abnormal pulmonary alveolus morphology'), ('GO:0009953PHENOTYPE', None), ('MP:0005130', 'decreased follicle stimulating hormone level'), ('HP:0030341', None), ('MP:0008330', 'absent somatotrophs'), ('HP:0012503', None), ('HP:0002098', None), ('MP:0005132', 'decreased luteinizing hormone level'), ('GO:0021979PHENOTYPE', None), ('MP:0008321', 'small adenohypophysis'), ('MP:0011088', 'neonatal lethality, incomplete penetrance'), ('HP:0001254', None), ('MP:0002777', 'absent ovarian follicles'), ('HP:0008222', None), ('MP:0011086', 'postnatal lethality, incomplete penetrance'), ('GO:0048732PHENOTYPE', None), ('HP:0002795', None), ('MP:0003816', 'abnormal pituitary gland development'), ('HP:0100750', None), ('MP:0001265', 'decreased body size'), ('HP:0000961', None)]\nMGI:1913656 Sac3d1 [('HP:0100494', None), ('MP:0002398', 'abnormal bone marrow cell morphology/development'), ('GO:0050776PHENOTYPE', None), ('HP:0010516', None), ('MP:0005348', 'increased T cell proliferation')]\nMGI:108052 Bcl2l2 [('MP:0004996', 'abnormal CNS synapse formation'), ('MP:0002216', 'abnormal seminiferous tubule morphology'), ('MP:0008572', 'abnormal Purkinje cell dendrite morphology'), ('HP:0000027', None), ('MP:0004910', 'decreased seminal vesicle weight'), ('HP:0010791', None), ('MP:0020355', 'abnormal Sertoli cell barrier morphology'), ('HP:0012243', None), ('HP:0008322', None)]\nMGI:3642960 Cyp2c54 []\nMGI:3650756 Myadml2os []\nMGI:104593 Xcl1 [('MP:0001844', 'autoimmune response'), ('MP:0001870', 'salivary gland inflammation'), ('MP:0008880', 'lacrimal gland inflammation'), ('MP:0005387', 'immune system phenotype'), ('MP:0005079', 'decreased cytotoxic T cell cytolysis'), ('HP:0002715', None), ('HP:0012649', None), ('HP:0012115', None)]\nMGI:3704367 Gm10146 []\nMGI:3039594 A2ml1 []\nMGI:3643871 Gm6272 []\nMGI:95480 Fancc [('HP:0000134', None), ('MP:0002209', 'decreased germ cell number'), ('MP:0008813', 'decreased common myeloid progenitor cell number'), ('MP:0002216', 'abnormal seminiferous tubule morphology'), ('MP:0000239', 'absent common myeloid progenitor cells'), ('MP:0001935', 'decreased litter size'), ('MP:0004045', 'abnormal cell cycle checkpoint function'), ('MP:0004030', 'induced chromosome breakage'), ('MP:0001545', 'abnormal hematopoietic system physiology'), ('MP:0001154', 'seminiferous tubule degeneration'), ('HP:0000013', None), ('MP:0001921', 'reduced fertility'), ('MP:0008249', 'abnormal common lymphocyte progenitor cell morphology'), ('GO:0002262PHENOTYPE', None), ('HP:0000798', None), ('MP:0004029', 'spontaneous chromosome breakage'), ('HP:0008724', None), ('HP:0001873', None), ('GO:0007276PHENOTYPE', None), ('MP:0001155', 'arrest of spermatogenesis'), ('MP:0001923', 'reduced female fertility')]\nMGI:3646825 Rpl21-ps4 []\nMGI:5313103 Gm20656 []\nMGI:1924792 Nxpe4 []\nMGI:96761 Ldha-ps2 []\nMGI:3647791 Gm5257 []\nMGI:3779623 Gm6710 []\nMGI:1861755 Nsmf [('MP:0008335', 'decreased gonadotroph cell number'), ('MP:0001922', 'reduced male fertility'), ('HP:0000823', None), ('MP:0005561', 'increased mean corpuscular hemoglobin'), ('MP:0001923', 'reduced female fertility')]\nMGI:1354956 Tfr2 [('MP:0008955', 'increased cellular hemoglobin content'), ('HP:0004325', None), ('MP:0005397', 'hematopoietic system phenotype'), ('MP:0011890', 'increased circulating ferritin level'), ('MP:0008810', 'increased circulating iron level'), ('HP:0011031', None), ('MP:0008809', 'increased spleen iron level'), ('MP:0008808', 'decreased spleen iron level'), ('MP:0005638', 'hemochromatosis')]\nMGI:95928 H2-Q1 []\nMGI:2144518 Stac2 []\nMGI:3801854 Gm16005 []\nMGI:2183434 Wfdc12 []\nMGI:88082 Asgr2 [('MP:0009763', 'increased sensitivity to induced morbidity/mortality'), ('MP:0005376', 'homeostasis/metabolism phenotype')]\nMGI:1196295 Ptpn20 [('MP:0001513', 'limb grasping'), ('HP:0008887', None)]\nMGI:2180781 Slc8b1 [('MP:0001523', 'impaired righting response'), ('MP:0005560', 'decreased circulating glucose level')]\nMGI:1916083 Pdcl3 []\nMGI:1924165 Mnd1 [('HP:0008669', None)]\nMGI:1927498 Rgs18 [('MP:0002398', 'abnormal bone marrow cell morphology/development'), ('MP:0005386', 'behavior/neurological phenotype'), ('HP:0001873', None)]\nMGI:2143558 Chchd10 []\nMGI:4936885 Gm17251 []\nMGI:3648679 Rpl10-ps2 []\nMGI:5452422 Gm22645 []\nMGI:1914215 Ctu2 []\nMGI:4937303 Gm17669 []\nMGI:3781253 Gm3076 []\nMGI:1924750 Ppfia1 []\nMGI:2675256 Adm2 []\nMGI:2445361 Serpinb1b []\nMGI:3781646 Gm3470 []\nMGI:3782365 Gm4189 []\nMGI:3705106 Gm15318 []\nMGI:1913881 Mfsd14b []\nMGI:1925947 Pus7 []\nMGI:1924117 Rhbdd1 [('MP:0002896', 'abnormal bone mineralization')]\nMGI:5453405 Gm23628 []\nMGI:4938023 Gm17196 []\nMGI:102896 Sult1a1 [('MP:0009642', 'abnormal blood homeostasis')]\nMGI:2146071 Tubgcp6 []\nMGI:2443387 Zc3hav1l []\nMGI:3713585 Zfp993 []\nMGI:3783212 Gm15770 []\nMGI:1928486 Tdo2 [('MP:0005332', 'abnormal amino acid level'), ('MP:0004948', 'abnormal neuronal precursor proliferation'), ('MP:0013908', 'small lateral ventricles')]\nMGI:3648782 Rpl26-ps2 []\nMGI:3649297 Gm13140 []\nMGI:3704332 Gm9830 []\nMGI:1096381 Arntl [('MP:0002563', 'shortened circadian period'), ('MP:0002607', 'decreased basophil cell number'), ('MP:0002083', 'premature death'), ('MP:0005595', 'abnormal vascular smooth muscle physiology'), ('MP:0005489', 'vascular smooth muscle hyperplasia'), ('GO:0007623PHENOTYPE', None), ('HP:0100240', None), ('MP:0005664', 'decreased circulating noradrenaline level'), ('HP:0011014', None), ('MP:0003059', 'decreased insulin secretion'), ('HP:0001888', None), ('MP:0009168', 'decreased pancreatic islet number'), ('MP:0002560', 'arrhythmic circadian persistence'), ('MP:0008190', 'decreased transitional stage B cell number'), ('MP:0005370', 'liver/biliary system phenotype'), ('HP:0001288', None), ('MP:0002891', 'increased insulin sensitivity'), ('MP:0005591', 'decreased vasodilation'), ('HP:0001944', None), ('MP:0002907', 'abnormal parturition'), ('MP:0020332', 'impaired leukocyte tethering or rolling'), ('MP:0008908', 'increased total fat pad weight'), ('MP:0004953', 'decreased spleen weight'), ('HP:0012184', None), ('MP:0005185', 'decreased circulating progesterone level'), ('MP:0020408', 'altered susceptibility to induced thrombosis'), ('HP:0005645', None), ('MP:0005369', 'muscle phenotype'), ('HP:0010976', None), ('MP:0003976', 'decreased circulating VLDL triglyceride level'), ('MP:0005631', 'decreased lung weight'), ('HP:0003138', None), ('MP:0002840', 'abnormal lens fiber morphology'), ('MP:0003918', 'decreased kidney weight'), ('MP:0005376', 'homeostasis/metabolism phenotype'), ('MP:0004876', 'decreased mean systemic arterial blood pressure'), ('MP:0010686', 'abnormal hair follicle matrix region morphology'), ('HP:0001649', None), ('MP:0008858', 'abnormal hair cycle anagen phase'), ('MP:0009133', 'decreased white fat cell size'), ('MP:0004988', 'increased osteoblast cell number'), ('HP:0001662', None), ('MP:0003198', 'calcified tendon'), ('MP:0001728', 'failure of embryo implantation'), ('MP:0001392', 'abnormal locomotor behavior'), ('MP:0001391', 'abnormal tail movements'), ('HP:0012311', None), ('MP:0002941', 'increased circulating alanine transaminase level'), ('HP:0100827', None), ('MP:0001792', 'impaired wound healing'), ('HP:0008222', None), ('MP:0005048', 'abnormal thrombosis'), ('MP:0010213', 'abnormal circulating fibrinogen level'), ('HP:0000833', None), ('MP:0002834', 'decreased heart weight'), ('MP:0004905', 'decreased uterus weight'), ('MP:0005397', 'hematopoietic system phenotype'), ('MP:0004232', 'decreased muscle weight'), ('MP:0005551', 'abnormal eye electrophysiology'), ('MP:0005386', 'behavior/neurological phenotype'), ('MP:0005317', 'increased triglyceride level'), ('MP:0005300', 'abnormal corneal stroma morphology'), ('HP:0003124', None), ('MP:0000427', 'abnormal hair cycle'), ('HP:0001367', None), ('MP:0004852', 'decreased testis weight'), ('HP:0002533', None), ('HP:0000481', None), ('MP:0005504', 'abnormal ligament morphology')]\nMGI:3802146 Gm16023 []\nMGI:3649580 Rpl38-ps1 []\nMGI:1920257 Rnf169 [('HP:0000938', None), ('HP:0012311', None)]\nMGI:5521015 Obox4-ps2 []\nMGI:109269 Inhbe []\nMGI:3645637 Gm14794 []\nMGI:1332226 Soat2 [('MP:0003983', 'decreased cholesterol level'), ('HP:0012184', None), ('MP:0003982', 'increased cholesterol level'), ('MP:0002310', 'decreased susceptibility to hepatic steatosis'), ('MP:0009356', 'decreased liver triglyceride level'), ('MP:0004773', 'abnormal bile composition'), ('HP:0003146', None)]\nMGI:5477367 Gm26873 []\nMGI:4414958 Gm16538 []\nMGI:3648402 Gm15204 []\nMGI:2183691 Nav2 [('MP:0001489', 'decreased startle reflex'), ('GO:0021554PHENOTYPE', None), ('GO:0007605PHENOTYPE', None), ('MP:0001973', 'increased thermal nociceptive threshold'), ('HP:0001824', None), ('HP:0004408', None)]\nMGI:4937960 Gm17133 []\nMGI:107304 Rny3 []\nMGI:3652179 Gm13267 []\nMGI:2682302 Zbed4 [('MP:0011110', 'preweaning lethality, incomplete penetrance'), ('HP:0001627', None), ('HP:0000105', None), ('HP:0001640', None), ('HP:0001744', None), ('HP:0001743', None)]\nMGI:2676901 Mir26b []\nMGI:104767 Gpx4 [('HP:0004326', None), ('MP:0009230', 'abnormal sperm head morphology'), ('MP:0008267', 'abnormal hippocampus CA3 region morphology'), ('MP:0001674', 'abnormal germ layer development'), ('MP:0000859', 'abnormal somatosensory cortex morphology'), ('HP:0003251', None), ('MP:0001935', 'decreased litter size'), ('HP:0001254', None), ('MP:0005384', 'cellular phenotype'), ('MP:0009836', 'abnormal sperm principal piece morphology'), ('MP:0000947', 'convulsive seizures'), ('HP:0012206', None), ('MP:0002086', 'abnormal extraembryonic tissue morphology'), ('MP:0004543', 'abnormal sperm physiology'), ('MP:0001648', 'abnormal apoptosis'), ('HP:0002446', None), ('HP:0000735', None), ('MP:0002169', 'no abnormal phenotype detected'), ('MP:0005389', 'reproductive system phenotype'), ('MP:0009237', 'kinked sperm flagellum')]\nMGI:1913357 Wfdc21 []\nMGI:5454785 Gm25008 []\nMGI:97991 Rnu6-ps2 []\nMGI:1921692 Sgms2 [('MP:0009289', 'decreased epididymal fat pad weight'), ('MP:0010080', 'abnormal hepatocyte physiology'), ('MP:0001547', 'abnormal lipid level')]\nMGI:3642955 Gm5643 []\nMGI:4421973 n-R5s121 []\nMGI:105387 Cdkn2d [('HP:0000029', None), ('HP:0008232', None), ('GO:0007605PHENOTYPE', None), ('MP:0004398', 'cochlear inner hair cell degeneration')]\nMGI:5011755 Mroh6 []\nMGI:2183102 Sardh []\nMGI:1915781 1110020A21Rik []\nMGI:88331 Cd3d [('HP:0005415', None), ('MP:0001825', 'arrested T cell differentiation')]\nMGI:98511 Tfe3 [('MP:0002169', 'no abnormal phenotype detected')]\nMGI:3819494 Snora23 []\nMGI:2448407 Hist1h2bn []\nMGI:3708125 Gm15191 []\nMGI:3651881 Gm11246 []\nMGI:4439832 Carmn []\nMGI:107364 Il17a [('HP:0005479', None), ('MP:0000322', 'increased granulocyte number'), ('MP:0008348', 'absent gamma-delta T cells'), ('HP:0002090', None), ('MP:0008497', 'decreased IgG2b level'), ('HP:0011990', None), ('MP:0008075', 'decreased CD4-positive, alpha beta T cell number'), ('HP:0002850', None), ('HP:0004313', None), ('MP:0005562', 'decreased mean corpuscular hemoglobin'), ('HP:0001508', None), ('MP:0005362', 'abnormal Langerhans cell physiology'), ('MP:0001179', 'thick pulmonary interalveolar septum'), ('MP:0008495', 'decreased IgG1 level'), ('HP:0004315', None), ('MP:0002267', 'abnormal bronchiole morphology')]\nMGI:3650315 Gm11951 []\nMGI:1913344 Lsm7 []\nMGI:1891066 Aqp9 []\nMGI:3648907 Gm7666 []\nMGI:3704336 Rpl10-ps3 []\nMGI:105304 Il6ra [('MP:0005343', 'increased circulating aspartate transaminase level'), ('MP:0008553', 'increased circulating tumor necrosis factor level'), ('MP:0005376', 'homeostasis/metabolism phenotype'), ('MP:0003631', 'nervous system phenotype'), ('MP:0004185', 'abnormal adipocyte glucose uptake'), ('MP:0010214', 'abnormal circulating serum amyloid protein level'), ('MP:0005026', 'decreased susceptibility to parasitic infection'), ('MP:0010751', 'decreased susceptibility to parasitic infection induced morbidity/mortality'), ('MP:0003887', 'increased hepatocyte apoptosis'), ('MP:0004001', 'decreased hepatocyte proliferation'), ('MP:0010398', 'decreased liver glycogen level'), ('HP:0002457', None), ('MP:0005463', 'abnormal CD4-positive, alpha-beta T cell physiology'), ('MP:0005375', 'adipose tissue phenotype'), ('MP:0004502', 'decreased incidence of tumors by chemical induction'), ('MP:0008596', 'increased circulating interleukin-6 level'), ('MP:0005023', 'abnormal wound healing'), ('MP:0005378', 'growth/size/body region phenotype'), ('MP:0002169', 'no abnormal phenotype detected')]\nMGI:98270 Sds []\nMGI:3644479 Hnrnpa1l2-ps []\nMGI:2136459 Cdc42bpb []\nMGI:1915065 Sec14l2 [('HP:0003146', None)]\nMGI:105490 Gramd1a []\nMGI:1927343 Rps6kb2 [('MP:0002169', 'no abnormal phenotype detected')]\nMGI:1921402 Stra6l []\nMGI:4937268 Gm17634 []\nMGI:3782538 Gm4353 []\nMGI:97855 Rap2a []\nMGI:3647418 Cfhr3 []\nMGI:3819492 Snora20 []\nMGI:3782792 Gm4609 []\nMGI:5530911 Gm27529 []\nMGI:1916196 1500002C15Rik []\nMGI:894332 Ebf2 [('HP:0003330', None), ('MP:0002741', 'small olfactory bulb'), ('MP:0008789', 'abnormal olfactory epithelium morphology'), ('MP:0001940', 'testis hypoplasia'), ('HP:0000035', None), ('HP:0007033', None), ('MP:0000852', 'small cerebellum'), ('MP:0008152', 'decreased diameter of femur'), ('HP:0012243', None), ('HP:0011314', None), ('HP:0100671', None), ('MP:0012014', 'abnormal olfactory neuron innervation pattern'), ('MP:0002566', 'abnormal sexual interaction'), ('MP:0005236', 'abnormal olfactory nerve morphology'), ('MP:0002161', 'abnormal fertility/fecundity'), ('MP:0002651', 'abnormal sciatic nerve morphology'), ('MP:0011086', 'postnatal lethality, incomplete penetrance'), ('HP:0001508', None), ('HP:0000762', None), ('MP:0003631', 'nervous system phenotype'), ('MP:0002631', 'abnormal epididymis morphology'), ('HP:0012286', None)]\nMGI:3648626 Gm5417 []\nMGI:1920933 Atg16l2 []\nMGI:3646307 Gbp11 []\nMGI:3801721 Gm16157 []\nMGI:3643179 Gm5537 []\nMGI:98283 Srsf1 [('MP:0002083', 'premature death'), ('MP:0000280', 'thin ventricular wall'), ('MP:0008106', 'decreased amacrine cell number'), ('HP:0001093', None), ('MP:0010235', 'abnormal retina inner limiting membrane morphology'), ('GO:0001701PHENOTYPE', None), ('MP:0005241', 'abnormal retinal ganglion layer morphology'), ('HP:0000546', None)]\nMGI:3648597 Gm9095 []\nMGI:3645191 Gm8355 []\nMGI:3646640 Rplp1-ps1 []\nMGI:1919202 Riox1 [('MP:0004985', 'decreased osteoclast cell number'), ('MP:0004988', 'increased osteoblast cell number'), ('MP:0009673', 'increased birth weight'), ('HP:0012790', None), ('MP:0008272', 'abnormal endochondral bone ossification'), ('MP:0003408', 'increased width of hypertrophic chondrocyte zone')]\nMGI:1345963 Coro1b [('MP:0005387', 'immune system phenotype')]\nMGI:5455250 Gm25473 []\nMGI:3648695 Hmgb1-ps7 []\nMGI:1098808 Pex5 [('MP:0002083', 'premature death'), ('MP:0000478', 'delayed intestine development'), ('MP:0001154', 'seminiferous tubule degeneration'), ('MP:0005370', 'liver/biliary system phenotype'), ('HP:0011014', None), ('HP:0001251', None), ('MP:0011085', 'postnatal lethality, complete penetrance'), ('HP:0002098', None), ('MP:0010956', 'abnormal mitochondrial ATP synthesis coupled electron transport'), ('MP:0009642', 'abnormal blood homeostasis'), ('HP:0012087', None), ('MP:0010952', 'abnormal fatty acid beta-oxidation'), ('HP:0002808', None), ('HP:0012647', None), ('MP:0010955', 'abnormal respiratory electron transport chain'), ('HP:0001392', None), ('MP:0001889', 'delayed brain development'), ('MP:0008019', 'increased liver tumor incidence'), ('HP:0002240', None), ('HP:0001324', None), ('MP:0003984', 'embryonic growth retardation'), ('MP:0008489', 'slow postnatal weight gain'), ('MP:0000754', 'paresis'), ('MP:0000528', 'delayed kidney development'), ('MP:0008026', 'abnormal brain white matter morphology'), ('MP:0004852', 'decreased testis weight')]\nMGI:1919332 Cyp2c55 []\nMGI:3646410 Ifi206 []\nMGI:3826519 Gm16305 []\nMGI:5452357 Gm22580 []\nMGI:3646863 Gm8129 []\nMGI:2664996 Tpt1-ps2 []\nMGI:4422076 n-R5s211 []\nMGI:5453074 Gm23297 []\nMGI:5454980 Gm25203 []\nMGI:2136910 Hemgn [('MP:0011968', 'decreased threshold for auditory brainstem response')]\nMGI:1922484 Rnf19b [('MP:0008567', 'decreased interferon-gamma secretion'), ('MP:0001272', 'increased metastatic potential'), ('GO:0042267PHENOTYPE', None), ('GO:0072643PHENOTYPE', None)]\nMGI:3644144 Gm8034 []\nMGI:1345193 Nlrp5 [('MP:0003718', 'maternal effect'), ('GO:0043623PHENOTYPE', None), ('HP:0008222', None), ('GO:0043487PHENOTYPE', None), ('GO:0034613PHENOTYPE', None)]\nMGI:2443051 Egfros []\nMGI:1921160 Arhgap18 [('MP:0001257', 'increased body length'), ('HP:0003072', None)]\nMGI:3647121 Gm5611 []\nMGI:3646686 Gm5909 []\nMGI:5547772 Gm28036 []\nMGI:3649750 Hspd1-ps4 []\nMGI:2156391 Pramel7 []\nMGI:3781826 Gm3650 []\nMGI:1351596 Sh2d2a [('MP:0008567', 'decreased interferon-gamma secretion'), ('MP:0008699', 'increased interleukin-4 secretion'), ('GO:0008283PHENOTYPE', None)]\nMGI:3783235 Gm15793 []\nMGI:1351326 Nrk [('MP:0011086', 'postnatal lethality, incomplete penetrance'), ('MP:0012098', 'increased spongiotrophoblast size'), ('MP:0011088', 'neonatal lethality, incomplete penetrance'), ('GO:0008285PHENOTYPE', None), ('HP:0006267', None)]\nMGI:4415001 Gm16581 []\nMGI:1859609 Sfmbt1 []\nMGI:3649822 Gm12344 []\nMGI:2442750 Slc22a30 []\nMGI:1349410 Triobp [('MP:0011967', 'increased or absent threshold for auditory brainstem response'), ('MP:0008762', 'embryonic lethality'), ('HP:0000365', None), ('MP:0004523', 'decreased cochlear hair cell stereocilia number')]\nMGI:1347476 Foxa2 [('MP:0001394', 'circling'), ('MP:0009177', 'decreased pancreatic alpha cell number'), ('MP:0002082', 'postnatal lethality'), ('GO:0090009PHENOTYPE', None), ('MP:0010861', 'increased respiratory mucosa goblet cell number'), ('HP:0011014', None), ('MP:0010935', 'increased airway resistance'), ('MP:0008029', 'abnormal paraxial mesoderm morphology'), ('HP:0030781', None), ('MP:0009331', 'absent primitive node'), ('MP:0011939', 'increased food intake'), ('MP:0000313', 'abnormal cell death'), ('MP:0003935', 'abnormal craniofacial development'), ('MP:0011085', 'postnatal lethality, complete penetrance'), ('MP:0012082', 'delayed heart development'), ('MP:0002085', 'abnormal embryonic tissue morphology'), ('MP:0000926', 'absent floor plate'), ('MP:0011098', 'embryonic lethality during organogenesis, complete penetrance'), ('MP:0010903', 'abnormal pulmonary alveolus wall morphology'), ('MP:0006027', 'impaired lung alveolus development'), ('HP:0002155', None), ('MP:0005440', 'increased glycogen level'), ('MP:0013504', 'increased embryonic tissue cell apoptosis'), ('MP:0001505', 'hunched posture'), ('MP:0011092', 'embryonic lethality, complete penetrance'), ('HP:0001250', None), ('MP:0002696', 'decreased circulating glucagon level'), ('GO:0008344PHENOTYPE', None), ('MP:0005387', 'immune system phenotype'), ('MP:0011732', 'decreased somite size'), ('MP:0011088', 'neonatal lethality, incomplete penetrance'), ('MP:0010896', 'decreased lung compliance'), ('MP:0002275', 'abnormal type II pneumocyte morphology'), ('HP:0100547', None), ('MP:0001698', 'decreased embryo size'), ('MP:0005560', 'decreased circulating glucose level'), ('HP:0001939', None), ('GO:0032525PHENOTYPE', None), ('MP:0010856', 'dilated respiratory conducting tubes'), ('MP:0011733', 'fused somites'), ('MP:0002169', 'no abnormal phenotype detected'), ('MP:0001385', 'pup cannibalization'), ('MP:0001685', 'abnormal endoderm development'), ('MP:0003960', 'increased lean body mass'), ('GO:0071542PHENOTYPE', None), ('MP:0005217', 'abnormal pancreatic beta cell morphology'), ('MP:0003400', 'kinked neural tube'), ('MP:0003861', 'abnormal nervous system development'), ('MP:0002314', 'abnormal respiratory mechanics'), ('MP:0001691', 'abnormal somite shape'), ('MP:0009937', 'abnormal neuron differentiation'), ('MP:0001265', 'decreased body size'), ('MP:0001688', 'abnormal somite development'), ('MP:0011183', 'abnormal primitive endoderm morphology'), ('HP:0011063', None)]\nMGI:4421949 n-R5s101 []\nMGI:1861380 Sphk2 [('HP:0002846', None), ('MP:0005463', 'abnormal CD4-positive, alpha-beta T cell physiology')]\nMGI:1919258 Zfyve19 []\nMGI:5521100 Gtpbp4-ps1 []\n"
]
],
[
[
"## More advanced expression analyses\n\nFor more advanced analyses, see http://bgee.unil.ch",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
]
|
e79958003d9fcbbc38afd5eac27f09d4abcf7b7d | 15,700 | ipynb | Jupyter Notebook | examples/complaints example.ipynb | imperva/mal2vec | 6926034d4f6c816793cb95ccc0149840d2133507 | [
"MIT"
]
| 4 | 2020-02-12T16:36:37.000Z | 2020-06-08T13:05:42.000Z | examples/complaints example.ipynb | imperva/mal2vec | 6926034d4f6c816793cb95ccc0149840d2133507 | [
"MIT"
]
| null | null | null | examples/complaints example.ipynb | imperva/mal2vec | 6926034d4f6c816793cb95ccc0149840d2133507 | [
"MIT"
]
| null | null | null | 38.480392 | 489 | 0.574459 | [
[
[
"<img src=\"../logo.png\" align='center' width=80%>\n# Overview\nAs data scientists working in a cyber-security company, we wanted to show that Natural Language Processing (NLP) algorithms can be applied to security related events. For this task we used 2 algorithm developed by Google: **Word2vec** ([link](https://arxiv.org/abs/1301.3781)) and **Doc2vec** ([link](https://arxiv.org/abs/1405.4053)). These algorithms use the context of words to extract a vectorized representation (aka embedding) for each word/document in a given vocabulary. \nIf you want to learn about how **Word2vec** works, you can [start here](https://skymind.ai/wiki/word2vec).\n\nUsing these algorithms, we managed to model the behavior of common vulnerability scanners (and other client applications) based on their unique 'syntax' of malicious web requests. We named our implementation **Mal2vec**.\n\n### About this notebook\nThis notebook contains easy to use widgets to execute each step on your own data. We also include 3 datasets as examples of how to use this project.\n\n### Table of contents\n- [Load csv data file](#Load-CSV-data-file)\n- [Map columns](#Map-columns)\n- [Select additional grouping columns](#Select-additional-grouping-columns)\n- [Create sentences](#Create-sentences)\n- [Prepare dataset](#Prepare-dataset)\n- [Train classification model](#Train-classifictaion-model)\n- [Evaluate the model](#Evaluate-the-model)\n\n# Imports",
"_____no_output_____"
]
],
[
[
"import random\nfrom IPython.display import display, Markdown, clear_output, HTML\ndef hide_toggle():\n # @author: harshil\n # @Source: https://stackoverflow.com/a/28073228/6306692\n this_cell = \"\"\"$('div.cell.code_cell.rendered.selected')\"\"\"\n next_cell = this_cell + '.next()'\n\n toggle_text = 'Show/hide code' # text shown on toggle link\n target_cell = this_cell # target cell to control with toggle\n js_hide_current = '' # bit of JS to permanently hide code in current cell (only when toggling next cell)\n\n js_f_name = 'code_toggle_{}'.format(str(random.randint(1,2**64)))\n\n html = \"\"\"\n <script>\n function {f_name}() {{\n {cell_selector}.find('div.input').toggle();\n }}\n\n {js_hide_current}\n </script>\n\n <a href=\"javascript:{f_name}()\">{toggle_text}</a>\n \"\"\".format(\n f_name=js_f_name,\n cell_selector=target_cell,\n js_hide_current=js_hide_current, \n toggle_text=toggle_text\n )\n\n return HTML(html)\ndisplay(hide_toggle())\ndisplay(HTML('''<style>.text_cell {background: #E0E5EE;}\n.widget-inline-hbox .widget-label{width:120px;}</style>'''))\n\n%load_ext autoreload\n%autoreload 2\n\nimport os\nimport pandas as pd\nimport ipywidgets as widgets\n\nimport sys\nsys.path.append(\"..\")\nfrom classify import prepare_dataset, train_classifier\nfrom vizualize import draw_model, plot_model_results\nfrom sentensize import create_sentences, dump_sentences ",
"_____no_output_____"
]
],
[
[
"# Load CSV data file\n### Ready to use dataset - Customer Complaints\n- Open source dataset by U.S. gov ([link](https://catalog.data.gov/dataset/consumer-complaint-database))\n- **Events**: the first word in the column 'issue' \n- **Label**: the product\n- **Groupping by**: 'Zip code'",
"_____no_output_____"
]
],
[
[
"display(hide_toggle())\n\ndf = None\ndef load_csv(btn):\n global df\n clear_output()\n display(hide_toggle())\n display(widgets.VBox([filename_input, nrows_input]))\n display(HTML('<img src=\"../loading.gif\" alt=\"Drawing\" style=\"width: 50px;\"/>'))\n\n nrows = int(nrows_input.value)\n df = pd.read_csv(filename_input.value, nrows=nrows if nrows > 0 else None)\n\n clear_output()\n display(hide_toggle())\n display(widgets.VBox([filename_input, nrows_input, load_button]))\n print('Loaded {} rows'.format(df.shape[0]))\n display(df.sample(n=5))\n\nfilename_input = widgets.Text(description='CSV file:', value='data/complaints.gz')\nnrows_input = widgets.Text(description='Rows limit:', value='0')\n\nload_button = widgets.Button(description='Load CSV')\nload_button.on_click(load_csv)\n\nwidgets.VBox([filename_input, nrows_input, load_button])",
"_____no_output_____"
]
],
[
[
"# Map columns\nThe data should have at least 3 columns:\n- **Timestamp** (int) - if you don't have timestamps, it can also be a simple increasing index\n- **Event** (string) - rule name, event description, etc. Must be a single word containing only alpha-numeric characters\n- **Label** (string) - type of event. This will be later used to create the classification model",
"_____no_output_____"
]
],
[
[
"time_column_input, event_column_input, label_column_input = None, None, None\ndef show_dropdown(obj):\n global time_column_input, event_column_input, label_column_input\n time_column_input = widgets.Dropdown(options=df.columns, description='Time column:')\n event_column_input = widgets.Dropdown(options=df.columns, value='Issue', description='Event column:')\n label_column_input = widgets.Dropdown(options=df.columns, value='Product', description='Label column:')\n\n clear_output()\n display(hide_toggle())\n display(widgets.VBox([show_dropdown_button, time_column_input, event_column_input, label_column_input]))\n \nshow_dropdown_button = widgets.Button(description='Refresh')\nshow_dropdown_button.on_click(show_dropdown)\nshow_dropdown(None)",
"_____no_output_____"
]
],
[
[
"# Select additional grouping columns\nSelect those columns which represents unique sequences",
"_____no_output_____"
]
],
[
[
"checkboxes = None\ndef show_checkboxes(obj):\n global checkboxes\n checkboxes = {k:widgets.Checkbox(description=k) for k in df.columns if k not in [time_column_input.value, \n event_column_input.value, \n label_column_input.value\n ]}\n checkboxes['ZIP code'].value = True\n clear_output()\n display(hide_toggle())\n display(widgets.VBox([show_checkboxes_button] + [checkboxes[x] for x in checkboxes]))\n\nshow_checkboxes_button = widgets.Button(description='Refresh')\nshow_checkboxes_button.on_click(show_checkboxes)\nshow_checkboxes(None)",
"_____no_output_____"
]
],
[
[
"# Create sentences\nThis cell will group events into sentences (using the grouping columns selected). \nIt will then split sentences if to consecutive events are separated by more than the given timeout (default: 300 seconds)",
"_____no_output_____"
]
],
[
[
"display(hide_toggle())\n\ndataset_name = os.path.splitext(os.path.basename(filename_input.value))[0]\nsentences_df, sentences_filepath = None, None\ndef sentences(obj):\n global sentences_df, sentences_filepath\n clear_output()\n display(hide_toggle())\n display(HTML('<img src=\"../loading.gif\" alt=\"Drawing\" style=\"width: 50px;\"/>'))\n\n groupping_columns = [x for x in checkboxes if checkboxes[x].value]\n sentences_df = create_sentences(df, \n time_column_input.value, \n event_column_input.value, \n label_column_input.value, \n groupping_columns,\n timeout=300\n )\n sentences_filepath = dump_sentences(sentences_df, dataset_name)\n\n clear_output()\n display(hide_toggle())\n display(sentence_button)\n print('Created {} sentences. Showing 5 examples:'.format(sentences_df.shape[0]))\n display(sentences_df.sample(n=5))\n\nsentence_button = widgets.Button(description='Start')\n\ndisplay(sentence_button)\nsentence_button.on_click(sentences)",
"_____no_output_____"
]
],
[
[
"# Prepare dataset\n1) Train a doc2vec model to extract the embedding vector from each sentence. \n**Parameters**: \n*vector_size*: the size of embedding vector. Increasing this parameters might improve accuracy, but will take longer to train (int, default=30) \n*epochs*: how many epochs should be applied during training. Increasing this parameters might improve accuracy, but will take longer to train (int, default=50) \n*min_sentence_count*: don't classify labels with small amount of sentences (int, default=200) \n\n2) Prepare dataset\n- Infer the embedding vector for each sample in the data set\n- Perform [stratified sampling](https://en.wikipedia.org/wiki/Stratified_sampling) for each label\n- Split to train/test sets 80%-20%",
"_____no_output_____"
]
],
[
[
"display(hide_toggle())\n\nX_train, X_test, y_train, y_test, classes = None, None, None, None, None\ndef dataset(obj):\n global sentences_df, sentences_filepath, dataset_name, X_train, X_test, y_train, y_test, classes\n clear_output()\n display(hide_toggle())\n display(HTML('<img src=\"../loading.gif\" alt=\"Drawing\" style=\"width: 50px;\"/>'))\n\n X_train, X_test, y_train, y_test, classes = prepare_dataset(sentences_df, \n sentences_filepath, \n dataset_name,\n vector_size=30,\n epochs=50,\n min_sentence_count=200\n )\n\n dataset_button.description = 'Run Again'\n clear_output()\n display(hide_toggle())\n print('Dataset ready!')\n display(dataset_button)\n\ndataset_button = widgets.Button(description='Start')\n\ndisplay(dataset_button)\ndataset_button.on_click(dataset)",
"_____no_output_____"
]
],
[
[
"# Train classification model\nTrain a deep neural network to classify each sentence to its correct label for 500 epochs (automatically stop when training no longer improves results)\n\nFor the purpose of this demo, the network architecture and hyper-parameters are constant. Feel free the modify to code and improve the model",
"_____no_output_____"
]
],
[
[
"display(hide_toggle())\n\nhistory, report, df_cm = None, None, None\ndef train(obj):\n global dataset_name, X_train, X_test, y_train, y_test, classes, history, report, df_cm\n train_button.description = 'Train Again'\n\n clear_output()\n display(hide_toggle())\n display(train_button)\n\n history, report, df_cm = train_classifier(X_train, X_test, y_train, y_test, classes, dataset_name)\n \n\ntrain_button = widgets.Button(description='Start')\n\ndisplay(train_button)\ntrain_button.on_click(train)",
"_____no_output_____"
]
],
[
[
"# Evaluate the model\nPlot the results of the model:\n- **Loss** - how did the model progress during training (lower values mean better performance)\n- **Accuracy** - how did the model perform on the validation set (higher values are better)\n- **Confusion Matrix** - mapping each of the model's predictions (x-axis) to its true label (y-axis). Correct predictions are placed on the main diagonal (brighter is better)\n- **Detailed report** - for each label, show the following metrics: precision, recall, f1-score ([read more here](https://towardsdatascience.com/accuracy-precision-recall-or-f1-331fb37c5cb9)). The 'support' metric is the number of instances in that class",
"_____no_output_____"
]
],
[
[
"display(hide_toggle())\n\ndef evaluate(btn):\n global history, report, df_cm\n \n clear_output()\n evaluate_button.description = 'Refresh'\n display(hide_toggle())\n display(evaluate_button)\n plot_model_results(history, report, df_cm, classes)\n \nevaluate_button = widgets.Button(description='Evaluate Model')\ndisplay(evaluate_button)\nevaluate_button.on_click(evaluate)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.